markdown
stringlengths 0
1.02M
| code
stringlengths 0
832k
| output
stringlengths 0
1.02M
| license
stringlengths 3
36
| path
stringlengths 6
265
| repo_name
stringlengths 6
127
|
|---|---|---|---|---|---|
A Scientific Deep Dive Into SageMaker LDA1. [Introduction](Introduction)1. [Setup](Setup)1. [Data Exploration](DataExploration)1. [Training](Training)1. [Inference](Inference)1. [Epilogue](Epilogue) Introduction***Amazon SageMaker LDA is an unsupervised learning algorithm that attempts to describe a set of observations as a mixture of distinct categories. Latent Dirichlet Allocation (LDA) is most commonly used to discover a user-specified number of topics shared by documents within a text corpus. Here each observation is a document, the features are the presence (or occurrence count) of each word, and the categories are the topics. Since the method is unsupervised, the topics are not specified up front, and are not guaranteed to align with how a human may naturally categorize documents. The topics are learned as a probability distribution over the words that occur in each document. Each document, in turn, is described as a mixture of topics.This notebook is similar to **LDA-Introduction.ipynb** but its objective and scope are a different. We will be taking a deeper dive into the theory. The primary goals of this notebook are,* to understand the LDA model and the example dataset,* understand how the Amazon SageMaker LDA algorithm works,* interpret the meaning of the inference output.Former knowledge of LDA is not required. However, we will run through concepts rather quickly and at least a foundational knowledge of mathematics or machine learning is recommended. Suggested references are provided, as appropriate. | %matplotlib inline
import os, re, tarfile
import boto3
import matplotlib.pyplot as plt
import mxnet as mx
import numpy as np
np.set_printoptions(precision=3, suppress=True)
# some helpful utility functions are defined in the Python module
# "generate_example_data" located in the same directory as this
# notebook
from generate_example_data import (
generate_griffiths_data,
match_estimated_topics,
plot_lda,
plot_lda_topics,
)
# accessing the SageMaker Python SDK
import sagemaker
from sagemaker.amazon.common import RecordSerializer
from sagemaker.serializers import CSVSerializer
from sagemaker.deserializers import JSONDeserializer | _____no_output_____ | Apache-2.0 | scientific_details_of_algorithms/lda_topic_modeling/LDA-Science.ipynb | Amirosimani/amazon-sagemaker-examples |
Setup****This notebook was created and tested on an ml.m4.xlarge notebook instance.*We first need to specify some AWS credentials; specifically data locations and access roles. This is the only cell of this notebook that you will need to edit. In particular, we need the following data:* `bucket` - An S3 bucket accessible by this account. * Used to store input training data and model data output. * Should be withing the same region as this notebook instance, training, and hosting.* `prefix` - The location in the bucket where this notebook's input and and output data will be stored. (The default value is sufficient.)* `role` - The IAM Role ARN used to give training and hosting access to your data. * See documentation on how to create these. * The script below will try to determine an appropriate Role ARN. | from sagemaker import get_execution_role
role = get_execution_role()
bucket = sagemaker.Session().default_bucket()
prefix = "sagemaker/DEMO-lda-science"
print("Training input/output will be stored in {}/{}".format(bucket, prefix))
print("\nIAM Role: {}".format(role)) | _____no_output_____ | Apache-2.0 | scientific_details_of_algorithms/lda_topic_modeling/LDA-Science.ipynb | Amirosimani/amazon-sagemaker-examples |
The LDA ModelAs mentioned above, LDA is a model for discovering latent topics describing a collection of documents. In this section we will give a brief introduction to the model. Let,* $M$ = the number of *documents* in a corpus* $N$ = the average *length* of a document.* $V$ = the size of the *vocabulary* (the total number of unique words)We denote a *document* by a vector $w \in \mathbb{R}^V$ where $w_i$ equals the number of times the $i$th word in the vocabulary occurs within the document. This is called the "bag-of-words" format of representing a document.$$\underbrace{w}_{\text{document}} = \overbrace{\big[ w_1, w_2, \ldots, w_V \big] }^{\text{word counts}},\quadV = \text{vocabulary size}$$The *length* of a document is equal to the total number of words in the document: $N_w = \sum_{i=1}^V w_i$.An LDA model is defined by two parameters: a topic-word distribution matrix $\beta \in \mathbb{R}^{K \times V}$ and a Dirichlet topic prior $\alpha \in \mathbb{R}^K$. In particular, let,$$\beta = \left[ \beta_1, \ldots, \beta_K \right]$$be a collection of $K$ *topics* where each topic $\beta_k \in \mathbb{R}^V$ is represented as probability distribution over the vocabulary. One of the utilities of the LDA model is that a given word is allowed to appear in multiple topics with positive probability. The Dirichlet topic prior is a vector $\alpha \in \mathbb{R}^K$ such that $\alpha_k > 0$ for all $k$. Data Exploration--- An Example DatasetBefore explaining further let's get our hands dirty with an example dataset. The following synthetic data comes from [1] and comes with a very useful visual interpretation.> [1] Thomas Griffiths and Mark Steyvers. *Finding Scientific Topics.* Proceedings of the National Academy of Science, 101(suppl 1):5228-5235, 2004. | print("Generating example data...")
num_documents = 6000
known_alpha, known_beta, documents, topic_mixtures = generate_griffiths_data(
num_documents=num_documents, num_topics=10
)
num_topics, vocabulary_size = known_beta.shape
# separate the generated data into training and tests subsets
num_documents_training = int(0.9 * num_documents)
num_documents_test = num_documents - num_documents_training
documents_training = documents[:num_documents_training]
documents_test = documents[num_documents_training:]
topic_mixtures_training = topic_mixtures[:num_documents_training]
topic_mixtures_test = topic_mixtures[num_documents_training:]
print("documents_training.shape = {}".format(documents_training.shape))
print("documents_test.shape = {}".format(documents_test.shape)) | _____no_output_____ | Apache-2.0 | scientific_details_of_algorithms/lda_topic_modeling/LDA-Science.ipynb | Amirosimani/amazon-sagemaker-examples |
Let's start by taking a closer look at the documents. Note that the vocabulary size of these data is $V = 25$. The average length of each document in this data set is 150. (See `generate_griffiths_data.py`.) | print("First training document =\n{}".format(documents_training[0]))
print("\nVocabulary size = {}".format(vocabulary_size))
print("Length of first document = {}".format(documents_training[0].sum()))
average_document_length = documents.sum(axis=1).mean()
print("Observed average document length = {}".format(average_document_length)) | _____no_output_____ | Apache-2.0 | scientific_details_of_algorithms/lda_topic_modeling/LDA-Science.ipynb | Amirosimani/amazon-sagemaker-examples |
The example data set above also returns the LDA parameters,$$(\alpha, \beta)$$used to generate the documents. Let's examine the first topic and verify that it is a probability distribution on the vocabulary. | print("First topic =\n{}".format(known_beta[0]))
print(
"\nTopic-word probability matrix (beta) shape: (num_topics, vocabulary_size) = {}".format(
known_beta.shape
)
)
print("\nSum of elements of first topic = {}".format(known_beta[0].sum())) | _____no_output_____ | Apache-2.0 | scientific_details_of_algorithms/lda_topic_modeling/LDA-Science.ipynb | Amirosimani/amazon-sagemaker-examples |
Unlike some clustering algorithms, one of the versatilities of the LDA model is that a given word can belong to multiple topics. The probability of that word occurring in each topic may differ, as well. This is reflective of real-world data where, for example, the word *"rover"* appears in a *"dogs"* topic as well as in a *"space exploration"* topic.In our synthetic example dataset, the first word in the vocabulary belongs to both Topic 1 and Topic 6 with non-zero probability. | print("Topic #1:\n{}".format(known_beta[0]))
print("Topic #6:\n{}".format(known_beta[5])) | _____no_output_____ | Apache-2.0 | scientific_details_of_algorithms/lda_topic_modeling/LDA-Science.ipynb | Amirosimani/amazon-sagemaker-examples |
Human beings are visual creatures, so it might be helpful to come up with a visual representation of these documents.In the below plots, each pixel of a document represents a word. The greyscale intensity is a measure of how frequently that word occurs within the document. Below we plot the first few documents of the training set reshaped into 5x5 pixel grids. | %matplotlib inline
fig = plot_lda(documents_training, nrows=3, ncols=4, cmap="gray_r", with_colorbar=True)
fig.suptitle("$w$ - Document Word Counts")
fig.set_dpi(160) | _____no_output_____ | Apache-2.0 | scientific_details_of_algorithms/lda_topic_modeling/LDA-Science.ipynb | Amirosimani/amazon-sagemaker-examples |
When taking a close look at these documents we can see some patterns in the word distributions suggesting that, perhaps, each topic represents a "column" or "row" of words with non-zero probability and that each document is composed primarily of a handful of topics.Below we plots the *known* topic-word probability distributions, $\beta$. Similar to the documents we reshape each probability distribution to a $5 \times 5$ pixel image where the color represents the probability of that each word occurring in the topic. | %matplotlib inline
fig = plot_lda(known_beta, nrows=1, ncols=10)
fig.suptitle(r"Known $\beta$ - Topic-Word Probability Distributions")
fig.set_dpi(160)
fig.set_figheight(2) | _____no_output_____ | Apache-2.0 | scientific_details_of_algorithms/lda_topic_modeling/LDA-Science.ipynb | Amirosimani/amazon-sagemaker-examples |
These 10 topics were used to generate the document corpus. Next, we will learn about how this is done. Generating DocumentsLDA is a generative model, meaning that the LDA parameters $(\alpha, \beta)$ are used to construct documents word-by-word by drawing from the topic-word distributions. In fact, looking closely at the example documents above you can see that some documents sample more words from some topics than from others.LDA works as follows: given * $M$ documents $w^{(1)}, w^{(2)}, \ldots, w^{(M)}$,* an average document length of $N$,* and an LDA model $(\alpha, \beta)$.**For** each document, $w^{(m)}$:* sample a topic mixture: $\theta^{(m)} \sim \text{Dirichlet}(\alpha)$* **For** each word $n$ in the document: * Sample a topic $z_n^{(m)} \sim \text{Multinomial}\big( \theta^{(m)} \big)$ * Sample a word from this topic, $w_n^{(m)} \sim \text{Multinomial}\big( \beta_{z_n^{(m)}} \; \big)$ * Add to documentThe [plate notation](https://en.wikipedia.org/wiki/Plate_notation) for the LDA model, introduced in [2], encapsulates this process pictorially.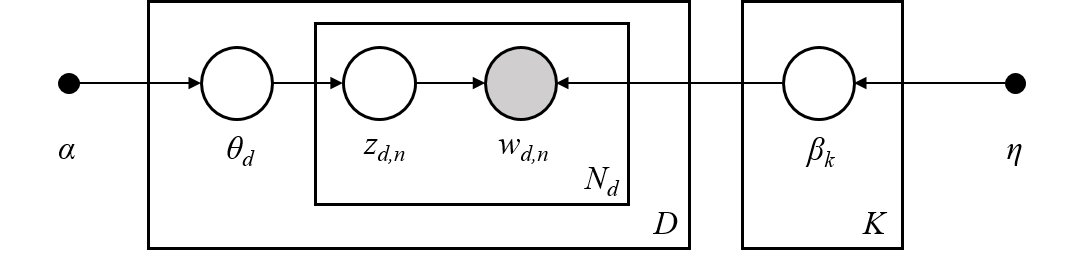> [2] David M Blei, Andrew Y Ng, and Michael I Jordan. Latent Dirichlet Allocation. Journal of Machine Learning Research, 3(Jan):993–1022, 2003. Topic MixturesFor the documents we generated above lets look at their corresponding topic mixtures, $\theta \in \mathbb{R}^K$. The topic mixtures represent the probablility that a given word of the document is sampled from a particular topic. For example, if the topic mixture of an input document $w$ is,$$\theta = \left[ 0.3, 0.2, 0, 0.5, 0, \ldots, 0 \right]$$then $w$ is 30% generated from the first topic, 20% from the second topic, and 50% from the fourth topic. In particular, the words contained in the document are sampled from the first topic-word probability distribution 30% of the time, from the second distribution 20% of the time, and the fourth disribution 50% of the time.The objective of inference, also known as scoring, is to determine the most likely topic mixture of a given input document. Colloquially, this means figuring out which topics appear within a given document and at what ratios. We will perform infernece later in the [Inference](Inference) section.Since we generated these example documents using the LDA model we know the topic mixture generating them. Let's examine these topic mixtures. | print("First training document =\n{}".format(documents_training[0]))
print("\nVocabulary size = {}".format(vocabulary_size))
print("Length of first document = {}".format(documents_training[0].sum()))
print("First training document topic mixture =\n{}".format(topic_mixtures_training[0]))
print("\nNumber of topics = {}".format(num_topics))
print("sum(theta) = {}".format(topic_mixtures_training[0].sum())) | _____no_output_____ | Apache-2.0 | scientific_details_of_algorithms/lda_topic_modeling/LDA-Science.ipynb | Amirosimani/amazon-sagemaker-examples |
We plot the first document along with its topic mixture. We also plot the topic-word probability distributions again for reference. | %matplotlib inline
fig, (ax1, ax2) = plt.subplots(2, 1)
ax1.matshow(documents[0].reshape(5, 5), cmap="gray_r")
ax1.set_title(r"$w$ - Document", fontsize=20)
ax1.set_xticks([])
ax1.set_yticks([])
cax2 = ax2.matshow(topic_mixtures[0].reshape(1, -1), cmap="Reds", vmin=0, vmax=1)
cbar = fig.colorbar(cax2, orientation="horizontal")
ax2.set_title(r"$\theta$ - Topic Mixture", fontsize=20)
ax2.set_xticks([])
ax2.set_yticks([])
fig.set_dpi(100)
%matplotlib inline
# pot
fig = plot_lda(known_beta, nrows=1, ncols=10)
fig.suptitle(r"Known $\beta$ - Topic-Word Probability Distributions")
fig.set_dpi(160)
fig.set_figheight(1.5) | _____no_output_____ | Apache-2.0 | scientific_details_of_algorithms/lda_topic_modeling/LDA-Science.ipynb | Amirosimani/amazon-sagemaker-examples |
Finally, let's plot several documents with their corresponding topic mixtures. We can see how topics with large weight in the document lead to more words in the document within the corresponding "row" or "column". | %matplotlib inline
fig = plot_lda_topics(documents_training, 3, 4, topic_mixtures=topic_mixtures)
fig.suptitle(r"$(w,\theta)$ - Documents with Known Topic Mixtures")
fig.set_dpi(160) | _____no_output_____ | Apache-2.0 | scientific_details_of_algorithms/lda_topic_modeling/LDA-Science.ipynb | Amirosimani/amazon-sagemaker-examples |
Training***In this section we will give some insight into how AWS SageMaker LDA fits an LDA model to a corpus, create an run a SageMaker LDA training job, and examine the output trained model. Topic Estimation using Tensor DecompositionsGiven a document corpus, Amazon SageMaker LDA uses a spectral tensor decomposition technique to determine the LDA model $(\alpha, \beta)$ which most likely describes the corpus. See [1] for a primary reference of the theory behind the algorithm. The spectral decomposition, itself, is computed using the CPDecomp algorithm described in [2].The overall idea is the following: given a corpus of documents $\mathcal{W} = \{w^{(1)}, \ldots, w^{(M)}\}, \; w^{(m)} \in \mathbb{R}^V,$ we construct a statistic tensor,$$T \in \bigotimes^3 \mathbb{R}^V$$such that the spectral decomposition of the tensor is approximately the LDA parameters $\alpha \in \mathbb{R}^K$ and $\beta \in \mathbb{R}^{K \times V}$ which maximize the likelihood of observing the corpus for a given number of topics, $K$,$$T \approx \sum_{k=1}^K \alpha_k \; (\beta_k \otimes \beta_k \otimes \beta_k)$$This statistic tensor encapsulates information from the corpus such as the document mean, cross correlation, and higher order statistics. For details, see [1].> [1] Animashree Anandkumar, Rong Ge, Daniel Hsu, Sham Kakade, and Matus Telgarsky. *"Tensor Decompositions for Learning Latent Variable Models"*, Journal of Machine Learning Research, 15:2773–2832, 2014.>> [2] Tamara Kolda and Brett Bader. *"Tensor Decompositions and Applications"*. SIAM Review, 51(3):455–500, 2009. Store Data on S3Before we run training we need to prepare the data.A SageMaker training job needs access to training data stored in an S3 bucket. Although training can accept data of various formats we convert the documents MXNet RecordIO Protobuf format before uploading to the S3 bucket defined at the beginning of this notebook. | # convert documents_training to Protobuf RecordIO format
recordio_protobuf_serializer = RecordSerializer()
fbuffer = recordio_protobuf_serializer.serialize(documents_training)
# upload to S3 in bucket/prefix/train
fname = "lda.data"
s3_object = os.path.join(prefix, "train", fname)
boto3.Session().resource("s3").Bucket(bucket).Object(s3_object).upload_fileobj(fbuffer)
s3_train_data = "s3://{}/{}".format(bucket, s3_object)
print("Uploaded data to S3: {}".format(s3_train_data)) | _____no_output_____ | Apache-2.0 | scientific_details_of_algorithms/lda_topic_modeling/LDA-Science.ipynb | Amirosimani/amazon-sagemaker-examples |
Next, we specify a Docker container containing the SageMaker LDA algorithm. For your convenience, a region-specific container is automatically chosen for you to minimize cross-region data communication | from sagemaker.image_uris import retrieve
region_name = boto3.Session().region_name
container = retrieve("lda", boto3.Session().region_name)
print("Using SageMaker LDA container: {} ({})".format(container, region_name)) | _____no_output_____ | Apache-2.0 | scientific_details_of_algorithms/lda_topic_modeling/LDA-Science.ipynb | Amirosimani/amazon-sagemaker-examples |
Training ParametersParticular to a SageMaker LDA training job are the following hyperparameters:* **`num_topics`** - The number of topics or categories in the LDA model. * Usually, this is not known a priori. * In this example, howevever, we know that the data is generated by five topics.* **`feature_dim`** - The size of the *"vocabulary"*, in LDA parlance. * In this example, this is equal 25.* **`mini_batch_size`** - The number of input training documents.* **`alpha0`** - *(optional)* a measurement of how "mixed" are the topic-mixtures. * When `alpha0` is small the data tends to be represented by one or few topics. * When `alpha0` is large the data tends to be an even combination of several or many topics. * The default value is `alpha0 = 1.0`.In addition to these LDA model hyperparameters, we provide additional parameters defining things like the EC2 instance type on which training will run, the S3 bucket containing the data, and the AWS access role. Note that,* Recommended instance type: `ml.c4`* Current limitations: * SageMaker LDA *training* can only run on a single instance. * SageMaker LDA does not take advantage of GPU hardware. * (The Amazon AI Algorithms team is working hard to provide these capabilities in a future release!) Using the above configuration create a SageMaker client and use the client to create a training job. | session = sagemaker.Session()
# specify general training job information
lda = sagemaker.estimator.Estimator(
container,
role,
output_path="s3://{}/{}/output".format(bucket, prefix),
instance_count=1,
instance_type="ml.c4.2xlarge",
sagemaker_session=session,
)
# set algorithm-specific hyperparameters
lda.set_hyperparameters(
num_topics=num_topics,
feature_dim=vocabulary_size,
mini_batch_size=num_documents_training,
alpha0=1.0,
)
# run the training job on input data stored in S3
lda.fit({"train": s3_train_data}) | _____no_output_____ | Apache-2.0 | scientific_details_of_algorithms/lda_topic_modeling/LDA-Science.ipynb | Amirosimani/amazon-sagemaker-examples |
If you see the message> `===== Job Complete =====`at the bottom of the output logs then that means training sucessfully completed and the output LDA model was stored in the specified output path. You can also view information about and the status of a training job using the AWS SageMaker console. Just click on the "Jobs" tab and select training job matching the training job name, below: | print("Training job name: {}".format(lda.latest_training_job.job_name)) | _____no_output_____ | Apache-2.0 | scientific_details_of_algorithms/lda_topic_modeling/LDA-Science.ipynb | Amirosimani/amazon-sagemaker-examples |
Inspecting the Trained ModelWe know the LDA parameters $(\alpha, \beta)$ used to generate the example data. How does the learned model compare the known one? In this section we will download the model data and measure how well SageMaker LDA did in learning the model.First, we download the model data. SageMaker will output the model in > `s3:////output//output/model.tar.gz`.SageMaker LDA stores the model as a two-tuple $(\alpha, \beta)$ where each LDA parameter is an MXNet NDArray. | # download and extract the model file from S3
job_name = lda.latest_training_job.job_name
model_fname = "model.tar.gz"
model_object = os.path.join(prefix, "output", job_name, "output", model_fname)
boto3.Session().resource("s3").Bucket(bucket).Object(model_object).download_file(fname)
with tarfile.open(fname) as tar:
tar.extractall()
print("Downloaded and extracted model tarball: {}".format(model_object))
# obtain the model file
model_list = [fname for fname in os.listdir(".") if fname.startswith("model_")]
model_fname = model_list[0]
print("Found model file: {}".format(model_fname))
# get the model from the model file and store in Numpy arrays
alpha, beta = mx.ndarray.load(model_fname)
learned_alpha_permuted = alpha.asnumpy()
learned_beta_permuted = beta.asnumpy()
print("\nLearned alpha.shape = {}".format(learned_alpha_permuted.shape))
print("Learned beta.shape = {}".format(learned_beta_permuted.shape)) | _____no_output_____ | Apache-2.0 | scientific_details_of_algorithms/lda_topic_modeling/LDA-Science.ipynb | Amirosimani/amazon-sagemaker-examples |
Presumably, SageMaker LDA has found the topics most likely used to generate the training corpus. However, even if this is case the topics would not be returned in any particular order. Therefore, we match the found topics to the known topics closest in L1-norm in order to find the topic permutation.Note that we will use the `permutation` later during inference to match known topic mixtures to found topic mixtures.Below plot the known topic-word probability distribution, $\beta \in \mathbb{R}^{K \times V}$ next to the distributions found by SageMaker LDA as well as the L1-norm errors between the two. | permutation, learned_beta = match_estimated_topics(known_beta, learned_beta_permuted)
learned_alpha = learned_alpha_permuted[permutation]
fig = plot_lda(np.vstack([known_beta, learned_beta]), 2, 10)
fig.set_dpi(160)
fig.suptitle("Known vs. Found Topic-Word Probability Distributions")
fig.set_figheight(3)
beta_error = np.linalg.norm(known_beta - learned_beta, 1)
alpha_error = np.linalg.norm(known_alpha - learned_alpha, 1)
print("L1-error (beta) = {}".format(beta_error))
print("L1-error (alpha) = {}".format(alpha_error)) | _____no_output_____ | Apache-2.0 | scientific_details_of_algorithms/lda_topic_modeling/LDA-Science.ipynb | Amirosimani/amazon-sagemaker-examples |
Not bad!In the eyeball-norm the topics match quite well. In fact, the topic-word distribution error is approximately 2%. Inference***A trained model does nothing on its own. We now want to use the model we computed to perform inference on data. For this example, that means predicting the topic mixture representing a given document.We create an inference endpoint using the SageMaker Python SDK `deploy()` function from the job we defined above. We specify the instance type where inference is computed as well as an initial number of instances to spin up. With this realtime endpoint at our fingertips we can finally perform inference on our training and test data.We can pass a variety of data formats to our inference endpoint. In this example we will demonstrate passing CSV-formatted data. Other available formats are JSON-formatted, JSON-sparse-formatter, and RecordIO Protobuf. We make use of the SageMaker Python SDK utilities `CSVSerializer` and `JSONDeserializer` when configuring the inference endpoint. | lda_inference = lda.deploy(
initial_instance_count=1,
instance_type="ml.m4.xlarge", # LDA inference may work better at scale on ml.c4 instances
serializer=CSVSerializer(),
deserializer=JSONDeserializer(),
) | _____no_output_____ | Apache-2.0 | scientific_details_of_algorithms/lda_topic_modeling/LDA-Science.ipynb | Amirosimani/amazon-sagemaker-examples |
Congratulations! You now have a functioning SageMaker LDA inference endpoint. You can confirm the endpoint configuration and status by navigating to the "Endpoints" tab in the AWS SageMaker console and selecting the endpoint matching the endpoint name, below: | print("Endpoint name: {}".format(lda_inference.endpoint_name)) | _____no_output_____ | Apache-2.0 | scientific_details_of_algorithms/lda_topic_modeling/LDA-Science.ipynb | Amirosimani/amazon-sagemaker-examples |
We pass some test documents to the inference endpoint. Note that the serializer and deserializer will atuomatically take care of the datatype conversion. | results = lda_inference.predict(documents_test[:12])
print(results) | _____no_output_____ | Apache-2.0 | scientific_details_of_algorithms/lda_topic_modeling/LDA-Science.ipynb | Amirosimani/amazon-sagemaker-examples |
It may be hard to see but the output format of SageMaker LDA inference endpoint is a Python dictionary with the following format.```{ 'predictions': [ {'topic_mixture': [ ... ] }, {'topic_mixture': [ ... ] }, {'topic_mixture': [ ... ] }, ... ]}```We extract the topic mixtures, themselves, corresponding to each of the input documents. | inferred_topic_mixtures_permuted = np.array(
[prediction["topic_mixture"] for prediction in results["predictions"]]
)
print("Inferred topic mixtures (permuted):\n\n{}".format(inferred_topic_mixtures_permuted)) | _____no_output_____ | Apache-2.0 | scientific_details_of_algorithms/lda_topic_modeling/LDA-Science.ipynb | Amirosimani/amazon-sagemaker-examples |
Inference AnalysisRecall that although SageMaker LDA successfully learned the underlying topics which generated the sample data the topics were in a different order. Before we compare to known topic mixtures $\theta \in \mathbb{R}^K$ we should also permute the inferred topic mixtures | inferred_topic_mixtures = inferred_topic_mixtures_permuted[:, permutation]
print("Inferred topic mixtures:\n\n{}".format(inferred_topic_mixtures)) | _____no_output_____ | Apache-2.0 | scientific_details_of_algorithms/lda_topic_modeling/LDA-Science.ipynb | Amirosimani/amazon-sagemaker-examples |
Let's plot these topic mixture probability distributions alongside the known ones. | %matplotlib inline
# create array of bar plots
width = 0.4
x = np.arange(10)
nrows, ncols = 3, 4
fig, ax = plt.subplots(nrows, ncols, sharey=True)
for i in range(nrows):
for j in range(ncols):
index = i * ncols + j
ax[i, j].bar(x, topic_mixtures_test[index], width, color="C0")
ax[i, j].bar(x + width, inferred_topic_mixtures[index], width, color="C1")
ax[i, j].set_xticks(range(num_topics))
ax[i, j].set_yticks(np.linspace(0, 1, 5))
ax[i, j].grid(which="major", axis="y")
ax[i, j].set_ylim([0, 1])
ax[i, j].set_xticklabels([])
if i == (nrows - 1):
ax[i, j].set_xticklabels(range(num_topics), fontsize=7)
if j == 0:
ax[i, j].set_yticklabels([0, "", 0.5, "", 1.0], fontsize=7)
fig.suptitle("Known vs. Inferred Topic Mixtures")
ax_super = fig.add_subplot(111, frameon=False)
ax_super.tick_params(labelcolor="none", top="off", bottom="off", left="off", right="off")
ax_super.grid(False)
ax_super.set_xlabel("Topic Index")
ax_super.set_ylabel("Topic Probability")
fig.set_dpi(160) | _____no_output_____ | Apache-2.0 | scientific_details_of_algorithms/lda_topic_modeling/LDA-Science.ipynb | Amirosimani/amazon-sagemaker-examples |
In the eyeball-norm these look quite comparable.Let's be more scientific about this. Below we compute and plot the distribution of L1-errors from **all** of the test documents. Note that we send a new payload of test documents to the inference endpoint and apply the appropriate permutation to the output. | %%time
# create a payload containing all of the test documents and run inference again
#
# TRY THIS:
# try switching between the test data set and a subset of the training
# data set. It is likely that LDA inference will perform better against
# the training set than the holdout test set.
#
payload_documents = documents_test # Example 1
known_topic_mixtures = topic_mixtures_test # Example 1
# payload_documents = documents_training[:600]; # Example 2
# known_topic_mixtures = topic_mixtures_training[:600] # Example 2
print("Invoking endpoint...\n")
results = lda_inference.predict(payload_documents)
inferred_topic_mixtures_permuted = np.array(
[prediction["topic_mixture"] for prediction in results["predictions"]]
)
inferred_topic_mixtures = inferred_topic_mixtures_permuted[:, permutation]
print("known_topics_mixtures.shape = {}".format(known_topic_mixtures.shape))
print("inferred_topics_mixtures_test.shape = {}\n".format(inferred_topic_mixtures.shape))
%matplotlib inline
l1_errors = np.linalg.norm((inferred_topic_mixtures - known_topic_mixtures), 1, axis=1)
# plot the error freqency
fig, ax_frequency = plt.subplots()
bins = np.linspace(0, 1, 40)
weights = np.ones_like(l1_errors) / len(l1_errors)
freq, bins, _ = ax_frequency.hist(l1_errors, bins=50, weights=weights, color="C0")
ax_frequency.set_xlabel("L1-Error")
ax_frequency.set_ylabel("Frequency", color="C0")
# plot the cumulative error
shift = (bins[1] - bins[0]) / 2
x = bins[1:] - shift
ax_cumulative = ax_frequency.twinx()
cumulative = np.cumsum(freq) / sum(freq)
ax_cumulative.plot(x, cumulative, marker="o", color="C1")
ax_cumulative.set_ylabel("Cumulative Frequency", color="C1")
# align grids and show
freq_ticks = np.linspace(0, 1.5 * freq.max(), 5)
freq_ticklabels = np.round(100 * freq_ticks) / 100
ax_frequency.set_yticks(freq_ticks)
ax_frequency.set_yticklabels(freq_ticklabels)
ax_cumulative.set_yticks(np.linspace(0, 1, 5))
ax_cumulative.grid(which="major", axis="y")
ax_cumulative.set_ylim((0, 1))
fig.suptitle("Topic Mixutre L1-Errors")
fig.set_dpi(110) | _____no_output_____ | Apache-2.0 | scientific_details_of_algorithms/lda_topic_modeling/LDA-Science.ipynb | Amirosimani/amazon-sagemaker-examples |
Machine learning algorithms are not perfect and the data above suggests this is true of SageMaker LDA. With more documents and some hyperparameter tuning we can obtain more accurate results against the known topic-mixtures.For now, let's just investigate the documents-topic mixture pairs that seem to do well as well as those that do not. Below we retreive a document and topic mixture corresponding to a small L1-error as well as one with a large L1-error. | N = 6
good_idx = l1_errors < 0.05
good_documents = payload_documents[good_idx][:N]
good_topic_mixtures = inferred_topic_mixtures[good_idx][:N]
poor_idx = l1_errors > 0.3
poor_documents = payload_documents[poor_idx][:N]
poor_topic_mixtures = inferred_topic_mixtures[poor_idx][:N]
%matplotlib inline
fig = plot_lda_topics(good_documents, 2, 3, topic_mixtures=good_topic_mixtures)
fig.suptitle("Documents With Accurate Inferred Topic-Mixtures")
fig.set_dpi(120)
%matplotlib inline
fig = plot_lda_topics(poor_documents, 2, 3, topic_mixtures=poor_topic_mixtures)
fig.suptitle("Documents With Inaccurate Inferred Topic-Mixtures")
fig.set_dpi(120) | _____no_output_____ | Apache-2.0 | scientific_details_of_algorithms/lda_topic_modeling/LDA-Science.ipynb | Amirosimani/amazon-sagemaker-examples |
In this example set the documents on which inference was not as accurate tend to have a denser topic-mixture. This makes sense when extrapolated to real-world datasets: it can be difficult to nail down which topics are represented in a document when the document uses words from a large subset of the vocabulary. Stop / Close the EndpointFinally, we should delete the endpoint before we close the notebook.To do so execute the cell below. Alternately, you can navigate to the "Endpoints" tab in the SageMaker console, select the endpoint with the name stored in the variable `endpoint_name`, and select "Delete" from the "Actions" dropdown menu. | sagemaker.Session().delete_endpoint(lda_inference.endpoint_name) | _____no_output_____ | Apache-2.0 | scientific_details_of_algorithms/lda_topic_modeling/LDA-Science.ipynb | Amirosimani/amazon-sagemaker-examples |
Automate loan approvals with Business rules in Apache Spark and Scala Automating at scale your business decisions in Apache Spark with IBM ODM 8.9.2This Scala notebook shows you how to execute locally business rules in DSX and Apache Spark. You'll learn how to call in Apache Spark a rule-based decision service. This decision service has been programmed with IBM Operational Decision Manager. This notebook puts in action a decision service named Miniloan that is part of the ODM tutorials. It determines with business rules whether a customer is eligible for a loan according to specific criteria. The criteria include the amount of the loan, the annual income of the borrower, and the duration of the loan.First we load an application data set that was captured as a CSV file. In scala we apply a map to this data set to automate a rule-based reasoning, in order to outcome a decision. The rule execution is performed locally in the Spark service. This notebook shows a complete Scala code that can execute any ruleset based on the public APIs.To get the most out of this notebook, you should have some familiarity with the Scala programming language. Contents This notebook contains the following main sections:1. [Load the loan validation request dataset.](loaddatatset)2. [Load the business rule execution and the simple loan application object model libraries.](loadjars)3. [Import Scala packages.](importpackages)4. [Implement a decision making function.](implementDecisionServiceMap)5. [Execute the business rules to approve or reject the loan applications.](executedecisions) 6. [View the automated decisions.](viewdecisions)7. [Summary and next steps.](summary) 1. Loading a loan application dataset fileA data set of simple loan applications is already available. You load it in the Notebook through its url. | // @hidden_cell
import scala.sys.process._
"wget https://raw.githubusercontent.com/ODMDev/decisions-on-spark/master/data/miniloan/miniloan-requests-10K.csv".!
val filename = "miniloan-requests-10K.csv" | _____no_output_____ | Apache-2.0 | notebooks/Automate loan approvals with business rules.ipynb | ODMDev/decisions-on-spark |
This following code loads the 10 000 simple loan application dataset written in CSV format. | val requestData = sc.textFile(filename)
val requestDataCount = requestData.count
println(s"$requestDataCount loan requests read in a CVS format")
println("The first 5 requests:")
requestData.take(20).foreach(println) | 10000 loan requests read in a CVS format
The first 5 requests:
John Doe, 550, 80000, 250000, 240, 0.05d
John Woo, 540, 100000, 250000, 240, 0.05d
Peter Woo, 540, 60000, 250000, 120, 0.05d
Peter Woo, 540, 60000, 250000, 120, 0.07d
John Doe, 550, 80000, 250000, 240, 0.05d
John Woo, 540, 100000, 250000, 240, 0.05d
Peter Woo, 540, 60000, 250000, 120, 0.05d
Peter Woo, 540, 60000, 250000, 120, 0.07d
John Doe, 550, 80000, 250000, 240, 0.05d
John Woo, 540, 100000, 250000, 240, 0.05d
Peter Woo, 540, 60000, 250000, 120, 0.05d
Peter Woo, 540, 60000, 250000, 120, 0.07d
John Doe, 550, 80000, 250000, 240, 0.05d
John Woo, 540, 100000, 250000, 240, 0.05d
Peter Woo, 540, 60000, 250000, 120, 0.05d
Peter Woo, 540, 60000, 250000, 120, 0.07d
John Doe, 550, 80000, 250000, 240, 0.05d
John Woo, 540, 100000, 250000, 240, 0.05d
Peter Woo, 540, 60000, 250000, 120, 0.05d
Peter Woo, 540, 60000, 250000, 120, 0.07d
| Apache-2.0 | notebooks/Automate loan approvals with business rules.ipynb | ODMDev/decisions-on-spark |
2. Add libraries for business rule execution and a loan application object modelThe XXX refers to your object storage or other place where you make available these jars.Add the following jars to execute the deployed decision service%AddJar https://XXX/j2ee_connector-1_5-fr.jar%AddJar https://XXX/jrules-engine.jar%AddJar https://XXX/jrules-res-execution.jarIn addition you need the Apache Jackson annotation lib%AddJar https://XXX/jackson-annotations-2.6.5.jarBusiness Rules apply on a Java executable Object Model packaged as a jar. We need these classes to create the decision requests, and to retreive the response from the rule engine.%AddJar https://XXX/miniloan-xom.jar | // @hidden_cell
// The urls below are accessible for an IBM internal usage only
%AddJar https://XXX/j2ee_connector-1_5-fr.jar
%AddJar https://XXX/jrules-engine.jar
%AddJar https://XXX/jrules-res-execution.jar
%AddJar https://XXX/jackson-annotations-2.6.5.jar -f
//Loan Application eXecutable Object Model
%AddJar https://XXX/miniloan-xom.jar -f
print("Your notebook is now ready to execute business rules to approve or reject loan applications") | _____no_output_____ | Apache-2.0 | notebooks/Automate loan approvals with business rules.ipynb | ODMDev/decisions-on-spark |
3. Import packagesImport ODM and Apache Spark packages. | import java.util.Map
import java.util.HashMap
import com.fasterxml.jackson.core.JsonGenerationException
import com.fasterxml.jackson.core.JsonProcessingException
import com.fasterxml.jackson.databind.JsonMappingException
import com.fasterxml.jackson.databind.ObjectMapper
import com.fasterxml.jackson.databind.SerializationFeature
import org.apache.spark.SparkConf
import org.apache.spark.api.java.JavaDoubleRDD
import org.apache.spark.api.java.JavaRDD
import org.apache.spark.api.java.JavaSparkContext
import org.apache.spark.api.java.function.Function
import org.apache.hadoop.fs.FileSystem
import org.apache.hadoop.fs.Path
import scala.collection.JavaConverters._
import ilog.rules.res.model._
import com.ibm.res.InMemoryJ2SEFactory
import com.ibm.res.InMemoryRepositoryDAO
import ilog.rules.res.session._
import miniloan.Borrower
import miniloan.Loan
import scala.io.Source
import java.net.URL
import java.io.InputStream | _____no_output_____ | Apache-2.0 | notebooks/Automate loan approvals with business rules.ipynb | ODMDev/decisions-on-spark |
4. Implement a Map function that executes a rule-based decision service | case class MiniLoanRequest(borrower: miniloan.Borrower,
loan: miniloan.Loan)
case class RESRunner(sessionFactory: com.ibm.res.InMemoryJ2SEFactory) {
def executeAsString(s: String): String = {
println("executeAsString")
val request = makeRequest(s)
val response = executeRequest(request)
response
}
private def makeRequest(s: String): MiniLoanRequest = {
val tokens = s.split(",")
// Borrower deserialization from CSV
val borrowerName = tokens(0)
val borrowerCreditScore = java.lang.Integer.parseInt(tokens(1).trim())
val borrowerYearlyIncome = java.lang.Integer.parseInt(tokens(2).trim())
val loanAmount = java.lang.Integer.parseInt(tokens(3).trim())
val loanDuration = java.lang.Integer.parseInt(tokens(4).trim())
val yearlyInterestRate = java.lang.Double.parseDouble(tokens(5).trim())
val borrower = new miniloan.Borrower(borrowerName, borrowerCreditScore, borrowerYearlyIncome)
// Loan request deserialization from CSV
val loan = new miniloan.Loan()
loan.setAmount(loanAmount)
loan.setDuration(loanDuration)
loan.setYearlyInterestRate(yearlyInterestRate)
val request = new MiniLoanRequest(borrower, loan)
request
}
def executeRequest(request: MiniLoanRequest): String = {
try {
val sessionRequest = sessionFactory.createRequest()
val rulesetPath = "/Miniloan/Miniloan"
sessionRequest.setRulesetPath(ilog.rules.res.model.IlrPath.parsePath(rulesetPath))
//sessionRequest.getTraceFilter.setInfoAllFilters(false)
val inputParameters = sessionRequest.getInputParameters
inputParameters.put("loan", request.loan)
inputParameters.put("borrower", request.borrower)
val session = sessionFactory.createStatelessSession()
val response = session.execute(sessionRequest)
var loan = response.getOutputParameters().get("loan").asInstanceOf[miniloan.Loan]
val mapper = new com.fasterxml.jackson.databind.ObjectMapper()
mapper.configure(com.fasterxml.jackson.databind.SerializationFeature.FAIL_ON_EMPTY_BEANS, false)
val results = new java.util.HashMap[String,Object]()
results.put("input", inputParameters)
results.put("output", response.getOutputParameters())
try {
//return mapper.writeValueAsString(results)
return mapper.writerWithDefaultPrettyPrinter().writeValueAsString(results);
} catch {
case e: Exception => return e.toString()
}
"Error"
} catch {
case exception: Exception => {
return exception.toString()
}
}
"Error"
}
}
val decisionService = new Function[String, String]() {
@transient private var ruleSessionFactory: InMemoryJ2SEFactory = null
private val rulesetURL = "https://odmlibserver.mybluemix.net/8901/decisionservices/miniloan-8901.dsar"
@transient private var rulesetStream: InputStream = null
def GetRuleSessionFactory(): InMemoryJ2SEFactory = {
if (ruleSessionFactory == null) {
ruleSessionFactory = new InMemoryJ2SEFactory()
// Create the Management Session
var repositoryFactory = ruleSessionFactory.createManagementSession().getRepositoryFactory()
var repository = repositoryFactory.createRepository()
// Deploy the Ruleapp with the Regular Management Session API.
var rapp = repositoryFactory.createRuleApp("Miniloan", IlrVersion.parseVersion("1.0"));
var rs = repositoryFactory.createRuleset("Miniloan",IlrVersion.parseVersion("1.1"));
rapp.addRuleset(rs);
//var fileStream = Source.fromResourceAsStream(RulesetFileName)
rulesetStream = new java.net.URL(rulesetURL).openStream()
rs.setRESRulesetArchive(IlrEngineType.DE,rulesetStream)
repository.addRuleApp(rapp)
}
ruleSessionFactory
}
def call(s: String): String = {
var runner = new RESRunner(GetRuleSessionFactory())
return runner.executeAsString(s)
}
def execute(s: String): String = {
try {
var runner = new RESRunner(GetRuleSessionFactory())
return runner.executeAsString(s)
} catch {
case exception: Exception => {
exception.printStackTrace(System.err)
}
}
"Execution error"
}
} | _____no_output_____ | Apache-2.0 | notebooks/Automate loan approvals with business rules.ipynb | ODMDev/decisions-on-spark |
5. Automate the decision making on the loan application datasetYou invoke a map on the decision function. While the map occurs rule engines are processing in parallel the loan applications to produce a data set of answers. | println("Start of Execution")
val answers = requestData.map(decisionService.execute)
printf("Number of rule based decisions: %s \n" , answers.count)
// Cleanup output file
//val fs = FileSystem.get(new URI(outputPath), sc.hadoopConfiguration);
//if (fs.exists(new Path(outputPath)))
// fs.delete(new Path(outputPath), true)
// Save RDD in a HDFS file
println("End of Execution ")
//answers.saveAsTextFile("swift://DecisionBatchExecution." + securedAccessName + "/miniloan-decisions-10.csv")
println("Decision automation job done") | Start of Execution
Number of rule based decisions: 10000
End of Execution
Decision automation job done
| Apache-2.0 | notebooks/Automate loan approvals with business rules.ipynb | ODMDev/decisions-on-spark |
6. View your automated decisionsEach decision is composed of output parameters and of a decision trace. The loan data contains the approval flag and the computed yearly repayment. The decision trace lists the business rules that have been executed in sequence to come to the conclusion. Each decision has been serialized in JSON. | //answers.toDF().show(false)
answers.take(1).foreach(println) | {
"output" : {
"ilog.rules.firedRulesCount" : 0,
"loan" : {
"amount" : 250000,
"duration" : 240,
"yearlyInterestRate" : 0.05,
"yearlyRepayment" : 19798,
"approved" : true,
"messages" : [ ]
}
},
"input" : {
"loan" : {
"amount" : 250000,
"duration" : 240,
"yearlyInterestRate" : 0.05,
"yearlyRepayment" : 19798,
"approved" : true,
"messages" : [ ]
},
"borrower" : {
"name" : "John Doe",
"creditScore" : 550,
"yearlyIncome" : 80000
}
}
}
| Apache-2.0 | notebooks/Automate loan approvals with business rules.ipynb | ODMDev/decisions-on-spark |
[this doc on github](https://github.com/dotnet/interactive/tree/master/samples/notebooks/fsharp/Samples) Machine Learning over House Prices with ML.NET Reference the packages | #r "nuget:Microsoft.ML,1.4.0"
#r "nuget:Microsoft.ML.AutoML,0.16.0"
#r "nuget:Microsoft.Data.Analysis,0.2.0"
#r "nuget: XPlot.Plotly.Interactive, 4.0.6"
open Microsoft.Data.Analysis
open XPlot.Plotly | _____no_output_____ | MIT | samples/notebooks/fsharp/Samples/HousingML.ipynb | BillWagner/interactive |
Adding better default formatting for data framesRegister a formatter for data frames and data frame rows. | module DateFrameFormatter =
// Locally open the F# HTML DSL.
open Html
let maxRows = 20
Formatter.Register<DataFrame>((fun (df: DataFrame) (writer: TextWriter) ->
let take = 20
table [] [
thead [] [
th [] [ str "Index" ]
for c in df.Columns do
th [] [ str c.Name]
]
tbody [] [
for i in 0 .. min maxRows (int df.Rows.Count - 1) do
tr [] [
td [] [ i ]
for o in df.Rows.[int64 i] do
td [] [ o ]
]
]
]
|> writer.Write
), mimeType = "text/html")
Formatter.Register<DataFrameRow>((fun (row: DataFrameRow) (writer: TextWriter) ->
table [] [
tbody [] [
tr [] [
for o in row do
td [] [ o ]
]
]
]
|> writer.Write
), mimeType = "text/html")
| _____no_output_____ | MIT | samples/notebooks/fsharp/Samples/HousingML.ipynb | BillWagner/interactive |
Download the data | open System.Net.Http
let housingPath = "housing.csv"
if not(File.Exists(housingPath)) then
let contents = HttpClient().GetStringAsync("https://raw.githubusercontent.com/ageron/handson-ml2/master/datasets/housing/housing.csv").Result
File.WriteAllText("housing.csv", contents) | _____no_output_____ | MIT | samples/notebooks/fsharp/Samples/HousingML.ipynb | BillWagner/interactive |
Add the data to the data frame | let housingData = DataFrame.LoadCsv(housingPath)
housingData
housingData.Description() | _____no_output_____ | MIT | samples/notebooks/fsharp/Samples/HousingML.ipynb | BillWagner/interactive |
Display the data | let graph =
Histogram(x = housingData.["median_house_value"],
nbinsx = 20)
graph |> Chart.Plot
let graph =
Scattergl(
x = housingData.["longitude"],
y = housingData.["latitude"],
mode = "markers",
marker =
Marker(
color = housingData.["median_house_value"],
colorscale = "Jet"))
let plot = Chart.Plot(graph)
plot.Width <- 600
plot.Height <- 600
display(plot) | _____no_output_____ | MIT | samples/notebooks/fsharp/Samples/HousingML.ipynb | BillWagner/interactive |
Prepare the training and validation sets | module Array =
let shuffle (arr: 'T[]) =
let rnd = Random()
let arr = Array.copy arr
for i in 0 .. arr.Length - 1 do
let r = i + rnd.Next(arr.Length - i)
let temp = arr.[r]
arr.[r] <- arr.[i]
arr.[i] <- temp
arr
let randomIndices = [| 0 .. int housingData.Rows.Count - 1 |] |> Array.shuffle
let testSize = int (float (housingData.Rows.Count) * 0.1)
let trainRows = randomIndices.[testSize..]
let testRows = randomIndices.[..testSize - 1]
let housing_train = housingData.[trainRows]
let housing_test = housingData.[testRows]
display(housing_train.Rows.Count)
display(housing_test.Rows.Count) | _____no_output_____ | MIT | samples/notebooks/fsharp/Samples/HousingML.ipynb | BillWagner/interactive |
Create the regression model and train it | #!time
open Microsoft.ML
open Microsoft.ML.Data
open Microsoft.ML.AutoML
let mlContext = MLContext()
let experiment = mlContext.Auto().CreateRegressionExperiment(maxExperimentTimeInSeconds = 15u)
let result = experiment.Execute(housing_train, labelColumnName = "median_house_value") | _____no_output_____ | MIT | samples/notebooks/fsharp/Samples/HousingML.ipynb | BillWagner/interactive |
Display the training results | let scatters =
result.RunDetails
|> Seq.filter (fun d -> not (isNull d.ValidationMetrics))
|> Seq.groupBy (fun r -> r.TrainerName)
|> Seq.map (fun (name, details) ->
Scattergl(
name = name,
x = (details |> Seq.map (fun r -> r.RuntimeInSeconds)),
y = (details |> Seq.map (fun r -> r.ValidationMetrics.MeanAbsoluteError)),
mode = "markers",
marker = Marker(size = 12)))
let chart = Chart.Plot(scatters)
chart.WithXTitle("Training Time")
chart.WithYTitle("Error")
display(chart)
Console.WriteLine("Best Trainer:{0}", result.BestRun.TrainerName); | _____no_output_____ | MIT | samples/notebooks/fsharp/Samples/HousingML.ipynb | BillWagner/interactive |
Validate and display the results | let testResults = result.BestRun.Model.Transform(housing_test)
let trueValues = testResults.GetColumn<float32>("median_house_value")
let predictedValues = testResults.GetColumn<float32>("Score")
let predictedVsTrue =
Scattergl(
x = trueValues,
y = predictedValues,
mode = "markers")
let maximumValue = Math.Max(Seq.max trueValues, Seq.max predictedValues)
let perfectLine =
Scattergl(
x = [| 0.0f; maximumValue |],
y = [| 0.0f; maximumValue |],
mode = "lines")
let chart = Chart.Plot([| predictedVsTrue; perfectLine |])
chart.WithXTitle("True Values")
chart.WithYTitle("Predicted Values")
chart.WithLegend(false)
chart.Width = 600
chart.Height = 600
display(chart) | _____no_output_____ | MIT | samples/notebooks/fsharp/Samples/HousingML.ipynb | BillWagner/interactive |
ML Course, Bogotá, Colombia (© Josh Bloom; June 2019) | %run ../talktools.py | _____no_output_____ | BSD-3-Clause | Lectures/1_ComputationalAndInferentialThinking/03_Featurization_and_NLP.ipynb | ijpulidos/ml_course |
Featurization and Dirty Data (and NLP) Source: [V. Singh](https://www.slideshare.net/hortonworks/data-science-workshop) Notes:Biased workflow :)* Labels → Answer* Feature extraction. Taking images/data and manipulate it, and do a feature matrix, each feature is a number (measurable). Domain-specific.* Run a ML model on the featurized data. Split in validation and test sets.* After predicting and validating results, the new results can become new training data. But you have to beware of skewing the results or the training data → introducing bias. One way to fight this is randomize sampling/results? (I didn't understand how to do this tbh). Source: Lightsidelabs Featurization ExamplesIn the real world, we are very rarely presented with a clean feature matrix. Raw data are missing, noisy, ugly and unfiltered. And sometimes we dont even have the data we need to make models and predictions. Indeed the conversion of raw data to data that's suitable for learning on is time consuming, difficult, and where a lot of the domain understanding is required.When we extract features from raw data (say PDF documents) we often are presented with a variety of data types: Categorical & Missing FeaturesOften times, we might be presented with raw data (say from an Excel spreadsheet) that looks like:| eye color | height | country of origin | gender || ------------| ---------| ---------------------| ------- || brown | 1.85 | Colombia | M || brown | 1.25 | USA | || blonde | 1.45 | Mexico | F || red | 2.01 | Mexico | F || | | Chile | F || Brown | 1.02 | Colombia | | What do you notice in this dataset? Since many ML learn algorithms require, as we'll see, a full matrix of numerical input features, there's often times a lot of preprocessing work that is needed before we can learn. | import numpy as np
import pandas as pd
df = pd.DataFrame({"eye color": ["brown", "brown", "blonde", "red", None, "Brown"],
"height": [1.85, 1.25, 1.45, 2.01, None, 1.02],
"country of origin": ["Colombia", "USA", "Mexico", "Mexico", "Chile", "Colombia"],
"gender": ["M", None, "F", "F","F", None]})
df | _____no_output_____ | BSD-3-Clause | Lectures/1_ComputationalAndInferentialThinking/03_Featurization_and_NLP.ipynb | ijpulidos/ml_course |
Let's first normalize the data so it's all lower case. This will handle the "Brown" and "brown" issue. | df_new = df.copy()
df_new["eye color"] = df_new["eye color"].str.lower()
df_new | _____no_output_____ | BSD-3-Clause | Lectures/1_ComputationalAndInferentialThinking/03_Featurization_and_NLP.ipynb | ijpulidos/ml_course |
Let's next handle the NaN in the height. What should we use here? | # mean of everyone?
np.nanmean(df_new["height"].values)
# mean of just females?
np.nanmean(df_new[df_new["gender"] == 'F']["height"])
df_new1 = df_new.copy()
df_new1.at[4, "height"] = np.nanmean(df_new[df_new["gender"] == 'F']["height"])
df_new1 | _____no_output_____ | BSD-3-Clause | Lectures/1_ComputationalAndInferentialThinking/03_Featurization_and_NLP.ipynb | ijpulidos/ml_course |
Let's next handle the eye color. What should we use? | df_new1["eye color"].mode()
df_new2 = df_new1.copy()
df_new2.at[4, "eye color"] = df_new1["eye color"].mode().values[0]
df_new2 | _____no_output_____ | BSD-3-Clause | Lectures/1_ComputationalAndInferentialThinking/03_Featurization_and_NLP.ipynb | ijpulidos/ml_course |
How should we handle the missing gender entries? | df_new3 = df_new2.fillna("N/A")
df_new3 | _____no_output_____ | BSD-3-Clause | Lectures/1_ComputationalAndInferentialThinking/03_Featurization_and_NLP.ipynb | ijpulidos/ml_course |
We're done, right? No. We fixed the dirty, missing data problem but we still dont have a numerical feature matrix.We could do a mapping such that "Colombia" -> 1, "USA" -> 2, ... etc. but then that would imply an ordering between what is fundamentally categories (without ordering). Instead we want to do `one-hot encoding`, where every unique value gets its own column. `pandas` as a method on DataFrames called `get_dummies` which does this for us. Notes:Many algorithms/methods cannot handle cathegorical data, that's why you have to do the `one-hot encoding` "trick". An example of one that can handle it is Random forest. | pd.get_dummies(df_new3, prefix=['eye color', 'country of origin', 'gender']) | _____no_output_____ | BSD-3-Clause | Lectures/1_ComputationalAndInferentialThinking/03_Featurization_and_NLP.ipynb | ijpulidos/ml_course |
Note: depending on the learning algorithm you use, you may want to do `drop_first=True` in `get_dummies`. Of course there are helpful tools that exist for us to deal with dirty, missing data. | %run transform
bt = BasicTransformer(return_df=True)
bt.fit_transform(df_new) | _____no_output_____ | BSD-3-Clause | Lectures/1_ComputationalAndInferentialThinking/03_Featurization_and_NLP.ipynb | ijpulidos/ml_course |
Time seriesThe [wafer dataset](http://www.timeseriesclassification.com/description.php?Dataset=Wafer) is a set of timeseries capturing sensor measurements (1000 training examples, 6164 test examples) of one silicon wafer during the manufacture of semiconductors. Each wafer has a classification of normal or abnormal. The abnormal wafers are representative of a range of problems commonly encountered during semiconductor manufacturing. | import requests
from io import StringIO
dat_file = requests.get("https://github.com/zygmuntz/time-series-classification/blob/master/data/wafer/Wafer.csv?raw=true")
data = StringIO(dat_file.text)
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
data.seek(0)
df = pd.read_csv(data, header=None)
df.head() | _____no_output_____ | BSD-3-Clause | Lectures/1_ComputationalAndInferentialThinking/03_Featurization_and_NLP.ipynb | ijpulidos/ml_course |
0 to 151 time measurements, the latest colum is the label we are trying to predict. | df[152].value_counts()
## save the data as numpy arrays
target = df.values[:,152].astype(int)
time_series = df.values[:,0:152]
normal_inds = np.argwhere(target == 1) ; np.random.shuffle(normal_inds)
abnormal_inds = np.argwhere(target == -1); np.random.shuffle(abnormal_inds)
num_to_plot = 3
fig, (ax1, ax2) = plt.subplots(nrows=1, ncols=2, sharey=True, figsize=(12,6))
for i in range(num_to_plot):
ax1.plot(time_series[normal_inds[i][0],:], label=f"#{normal_inds[i][0]}: {target[normal_inds[i][0]]}")
ax2.plot(time_series[abnormal_inds[i][0],:], label=f"#{abnormal_inds[i][0]}: {target[abnormal_inds[i][0]]}")
ax1.legend()
ax2.legend()
ax1.set_title("Normal") ; ax2.set_title("Abnormal")
ax1.set_xlabel("time") ; ax2.set_xlabel("time")
ax1.set_ylabel("Value") | _____no_output_____ | BSD-3-Clause | Lectures/1_ComputationalAndInferentialThinking/03_Featurization_and_NLP.ipynb | ijpulidos/ml_course |
What would be good features here? | f1 = np.mean(time_series, axis=1) # how about the mean?
f1.shape
import seaborn as sns, numpy as np
import warnings
warnings.filterwarnings("ignore")
ax = sns.distplot(f1)
# Plotting differences of means between normal and abnormal
ax = sns.distplot(f1[normal_inds], kde_kws={"label": "normal"})
sns.distplot(f1[abnormal_inds], ax=ax, kde_kws={"label": "abnormal"})
f2 = np.min(time_series, axis=1) # how about the mean?
f2.shape
# Differences in the minimum - Not much difference, just a small subset at the "end that can tell something interesting
ax = sns.distplot(f2[normal_inds], kde_kws={"label": "normal"})
sns.distplot(f2[abnormal_inds], ax=ax, kde_kws={"label": "abnormal"}) | _____no_output_____ | BSD-3-Clause | Lectures/1_ComputationalAndInferentialThinking/03_Featurization_and_NLP.ipynb | ijpulidos/ml_course |
Often there are entire python packages devoted to help us build features from certain types of datasets (timeseries, text, images, movies, etc.). In the case of timeseries, a popular package is `tsfresh` (*"It automatically calculates a large number of time series characteristics, the so called features. Further the package contains methods to evaluate the explaining power and importance of such characteristics for regression or classification tasks."*). See the [tsfresh docs](https://tsfresh.readthedocs.io/en/latest/) and the [list of features generated](https://tsfresh.readthedocs.io/en/latest/text/list_of_features.html). | # !pip install tsfresh
dfc = df.copy()
del dfc[152]
d = dfc.stack()
d = d.reset_index()
d = d.rename(columns={"level_0": "id", "level_1": "time", 0: "value"})
y = df[152]
from tsfresh import extract_features
max_num=300
from tsfresh import extract_relevant_features
features_filtered_direct = extract_relevant_features(d[d["id"] < max_num], y.iloc[0:max_num],
column_id='id', column_sort='time', n_jobs=4)
#extracted_features = extract_features(, column_id="id",
# column_sort="time", disable_progressbar=False, n_jobs=3)
feats = features_filtered_direct[features_filtered_direct.columns[0:4]].rename(lambda x: x[0:14], axis='columns')
feats["target"] = y.iloc[0:max_num]
sns.pairplot(feats, hue="target") | _____no_output_____ | BSD-3-Clause | Lectures/1_ComputationalAndInferentialThinking/03_Featurization_and_NLP.ipynb | ijpulidos/ml_course |
Text DataMany applications involve parsing and understanding something about natural language, ie. speech or text data. Categorization is a classic usage of Natural Language Processing (NLP): what bucket does this text belong to? Question: **What are some examples where learning on text has commerical or industrial applications?** A classic dataset in text processing is the [20,000+ newsgroup documents corpus](http://qwone.com/~jason/20Newsgroups/). These texts taken from old discussion threads in 20 different [newgroups](https://en.wikipedia.org/wiki/Usenet_newsgroup):comp.graphicscomp.os.ms-windows.misccomp.sys.ibm.pc.hardwarecomp.sys.mac.hardwarecomp.windows.x rec.autosrec.motorcyclesrec.sport.baseballrec.sport.hockey sci.cryptsci.electronicssci.medsci.spacemisc.forsale talk.politics.misctalk.politics.gunstalk.politics.mideast talk.religion.miscalt.atheismsoc.religion.christianOne of the tasks is to assign a document to the correct group, ie. classify which group this belongs to. `sklearn` has a download facility for this dataset: | from sklearn.datasets import fetch_20newsgroups
news_train = fetch_20newsgroups(subset='train', categories=['sci.space','rec.autos'], data_home='datatmp/')
news_train.target_names
print(news_train.data[1])
news_train.target_names[news_train.target[1]]
autos = np.argwhere(news_train.target == 1)
sci = np.argwhere(news_train.target == 0) | _____no_output_____ | BSD-3-Clause | Lectures/1_ComputationalAndInferentialThinking/03_Featurization_and_NLP.ipynb | ijpulidos/ml_course |
**How do you (as a human) classify text? What do you look for? How might we make these features?** | # total character count?
f1 = np.array([len(x) for x in news_train.data])
f1
ax = sns.distplot(f1[autos], kde_kws={"label": "autos"})
sns.distplot(f1[sci], ax=ax, kde_kws={"label": "sci"})
ax.set_xscale("log")
ax.set_xlabel("number of charaters")
# total character words?
f2 = np.array([len(x.split(" ")) for x in news_train.data])
f2
ax = sns.distplot(f2[autos], kde_kws={"label": "autos"})
sns.distplot(f2[sci], ax=ax, kde_kws={"label": "sci"})
ax.set_xscale("log")
ax.set_xlabel("number of words")
# number of questions asked or exclaimations?
f3 = np.array([x.count("?") + x.count("!") for x in news_train.data])
f3
ax = sns.distplot(f3[autos], kde_kws={"label": "autos"})
sns.distplot(f3[sci], ax=ax, kde_kws={"label": "sci"})
ax.set_xlabel("number of questions asked") | _____no_output_____ | BSD-3-Clause | Lectures/1_ComputationalAndInferentialThinking/03_Featurization_and_NLP.ipynb | ijpulidos/ml_course |
We've got three fairly uninformative features now. We should be able to do better. Unsurprisingly, what matters most in NLP is the content: the words used, the tone, the meaning from the ordering of those words. The basic components of NLP are: * Tokenization - intelligently splitting up words in sentences, paying attention to conjunctions, punctuation, etc. * Lemmization - reducing a word to its base form * Entity recognition - finding proper names, places, etc. in documents There a many Python packages that help with NLP, including `nltk`, `textblob`, `gensim`, etc. Here we'll use the fairly modern and battletested [`spaCy`](https://spacy.io/). Notes:* For tokenization you need to know the language you are dealing with.* Lemmization idea is to kind of normalize the whole text content, and maybe reduce words, such as adverbs to adjectives or plurals to singular, etc.* For lemmization not only needs the language but also the kind of content you are dealing with. | #!pip install spacy
#!python -m spacy download en
#!python -m spacy download es
import spacy
# Load English tokenizer, tagger, parser, NER and word vectors
nlp = spacy.load("en")
# the spanish model is
# nlp = spacy.load("es")
doc = nlp(u"Guido said that 'Python is one of the best languages for doing Data Science.' "
"Why he said that should be clear to anyone who knows Python.")
en_doc = doc | _____no_output_____ | BSD-3-Clause | Lectures/1_ComputationalAndInferentialThinking/03_Featurization_and_NLP.ipynb | ijpulidos/ml_course |
`doc` is now an `iterable ` with each word/item properly tokenized and tagged. This is done by applying rules specific to each language. Linguistic annotations are available as Token attributes. | for token in doc:
print(token.text, token.lemma_, token.pos_, token.tag_, token.dep_,
token.shape_, token.is_alpha, token.is_stop)
from spacy import displacy
displacy.serve(doc, style="dep")
displacy.render(doc, style = "ent", jupyter = True)
nlp = spacy.load("es")
# https://www.elespectador.com/noticias/ciencia/decenas-de-nuevas-supernovas-ayudaran-medir-la-expansion-del-universo-articulo-863683
doc = nlp(u'En los últimos años, los investigadores comenzaron a'
'informar un nuevo tipo de supernovas de cinco a diez veces'
'más brillantes que las supernovas de Tipo "IA". ')
for token in doc:
print(token.text, token.lemma_, token.pos_, token.tag_, token.dep_,
token.shape_, token.is_alpha, token.is_stop)
from spacy import displacy
displacy.serve(doc, style="dep")
[i for i in doc.sents] | _____no_output_____ | BSD-3-Clause | Lectures/1_ComputationalAndInferentialThinking/03_Featurization_and_NLP.ipynb | ijpulidos/ml_course |
One very powerful way to featurize text/documents is to count the frequency of words---this is called **bag of words**. Each individual token occurrence frequency is used to generate a feature. So the two sentences become:```json{"Guido": 1, "said": 2, "that": 2, "Python": 2, "is": 1, "one": 1, "of": 1, "best": 1, "languages": 1, "for": 1, "Data": 1, "Science": 1, "Why", 1, "he": 1, "should": 1, "be": 1, "anyone": 1, "who": 1 } ``` A corpus of documents can be represented as a matrix with one row per document and one column per token.Question: **What are some challenges you see with brute force BoW?** | from spacy.lang.en.stop_words import STOP_WORDS
STOP_WORDS | _____no_output_____ | BSD-3-Clause | Lectures/1_ComputationalAndInferentialThinking/03_Featurization_and_NLP.ipynb | ijpulidos/ml_course |
`sklearn` has a number of helper functions, include the [`CountVectorizer`](https://scikit-learn.org/stable/modules/generated/sklearn.feature_extraction.text.CountVectorizer.html):> Convert a collection of text documents to a matrix of token counts. This implementation produces a sparse representation of the counts using `scipy.sparse.csr_matrix`. | # the following is from https://www.dataquest.io/blog/tutorial-text-classification-in-python-using-spacy/
import pandas as pd
from sklearn.feature_extraction.text import CountVectorizer,TfidfVectorizer
from sklearn.base import TransformerMixin
from sklearn.pipeline import Pipeline
import string
from spacy.lang.en.stop_words import STOP_WORDS
from spacy.lang.en import English
# Create our list of punctuation marks
punctuations = string.punctuation
# Create our list of stopwords
nlp = spacy.load('en')
stop_words = spacy.lang.en.stop_words.STOP_WORDS
# Load English tokenizer, tagger, parser, NER and word vectors
parser = English()
# Creating our tokenizer function
def spacy_tokenizer(sentence):
# Creating our token object, which is used to create documents with linguistic annotations.
mytokens = parser(sentence)
# Lemmatizing each token and converting each token into lowercase
mytokens = [ word.lemma_.lower().strip() if word.lemma_ != "-PRON-" else word.lower_ for word in mytokens ]
# Removing stop words
mytokens = [ word for word in mytokens if word not in stop_words and word not in punctuations ]
# return preprocessed list of tokens
return mytokens
# Custom transformer using spaCy
class predictors(TransformerMixin):
def transform(self, X, **transform_params):
# Cleaning Text
return [clean_text(text) for text in X]
def fit(self, X, y=None, **fit_params):
return self
def get_params(self, deep=True):
return {}
# Basic function to clean the text
def clean_text(text):
# Removing spaces and converting text into lowercase
return text.strip().lower()
bow_vector = CountVectorizer(tokenizer = spacy_tokenizer, ngram_range=(1,1))
X = bow_vector.fit_transform([x.text for x in en_doc.sents])
X
bow_vector.get_feature_names() | _____no_output_____ | BSD-3-Clause | Lectures/1_ComputationalAndInferentialThinking/03_Featurization_and_NLP.ipynb | ijpulidos/ml_course |
Why did we get `datum` as one of our feature names? | X.toarray()
doc.text | _____no_output_____ | BSD-3-Clause | Lectures/1_ComputationalAndInferentialThinking/03_Featurization_and_NLP.ipynb | ijpulidos/ml_course |
Let's try a bigger corpus (the newsgroups): | news_train = fetch_20newsgroups(subset='train',
remove=('headers', 'footers', 'quotes'),
categories=['sci.space','rec.autos'], data_home='datatmp/')
%time X = bow_vector.fit_transform(news_train.data)
X
bow_vector.get_feature_names() | _____no_output_____ | BSD-3-Clause | Lectures/1_ComputationalAndInferentialThinking/03_Featurization_and_NLP.ipynb | ijpulidos/ml_course |
Most of those features will only appear once and we might not want to include them (as they add noise). In order to reweight the count features into floating point values suitable for usage by a classifier it is very common to use the *tf–idf* transform. From [`sklearn`](https://scikit-learn.org/stable/modules/generated/sklearn.feature_extraction.text.TfidfTransformer.htmlsklearn.feature_extraction.text.TfidfTransformer): > Tf means term-frequency while tf-idf means term-frequency times inverse document-frequency. This is a common term weighting scheme in information retrieval, that has also found good use in document classification.The goal of using tf-idf instead of the raw frequencies of occurrence of a token in a given document is to scale down the impact of tokens that occur very frequently in a given corpus and that are hence empirically less informative than features that occur in a small fraction of the training corpus.Let's keep only those terms that show up in at least 3% of the docs, but not those that show up in more than 90%. | tfidf_vector = TfidfVectorizer(tokenizer = spacy_tokenizer, min_df=0.03, max_df=0.9, max_features=1000)
%time X = tfidf_vector.fit_transform(news_train.data)
tfidf_vector.get_feature_names()
X
print(X[1,:])
y = news_train.target
np.savez("tfidf.npz", X=X.todense(), y=y) | _____no_output_____ | BSD-3-Clause | Lectures/1_ComputationalAndInferentialThinking/03_Featurization_and_NLP.ipynb | ijpulidos/ml_course |
One of the challenges with BoW and TF-IDF is that we lose context. "Me gusta esta clase, no" is the same as "No me gusta esta clase". One way to handle this is with N-grams -- not just frequencies of individual words but of groupings of n-words. Eg. "Me gusta", "gusta esta", "esta clase", "clase no", "no me" (bigrams). | bow_vector = CountVectorizer(tokenizer = spacy_tokenizer, ngram_range=(1,2))
X = bow_vector.fit_transform([x.text for x in en_doc.sents])
bow_vector.get_feature_names() | _____no_output_____ | BSD-3-Clause | Lectures/1_ComputationalAndInferentialThinking/03_Featurization_and_NLP.ipynb | ijpulidos/ml_course |
Calibration of non-isoplanatic low frequency dataThis uses an implementation of the SageCAL algorithm to calibrate a simulated SKA1LOW observation in which sources inside the primary beam have one set of calibration errors and sources outside have different errors.In this example, the peeler sources are held fixed in strength and location and only the gains solved. The other sources, inside the primary beam, are partitioned into weak (5Jy). The weak sources are processed collectively as an image. The bright sources are processed individually. | % matplotlib inline
import os
import sys
sys.path.append(os.path.join('..', '..'))
from data_models.parameters import arl_path
results_dir = arl_path('test_results')
import numpy
from astropy.coordinates import SkyCoord
from astropy import units as u
from astropy.wcs.utils import pixel_to_skycoord
from matplotlib import pyplot as plt
from data_models.memory_data_models import SkyModel
from data_models.polarisation import PolarisationFrame
from workflows.arlexecute.execution_support.arlexecute import arlexecute
from processing_components.skycomponent.operations import find_skycomponents
from processing_components.calibration.calibration import solve_gaintable
from processing_components.calibration.operations import apply_gaintable, create_gaintable_from_blockvisibility
from processing_components.visibility.base import create_blockvisibility, copy_visibility
from processing_components.image.deconvolution import restore_cube
from processing_components.skycomponent.operations import select_components_by_separation, insert_skycomponent, \
select_components_by_flux
from processing_components.image.operations import show_image, qa_image, copy_image, create_empty_image_like
from processing_components.simulation.testing_support import create_named_configuration, create_low_test_beam, \
simulate_gaintable, create_low_test_skycomponents_from_gleam
from processing_components.skycomponent.operations import apply_beam_to_skycomponent, find_skycomponent_matches
from processing_components.imaging.base import create_image_from_visibility, advise_wide_field, \
predict_skycomponent_visibility
from processing_components.imaging.imaging_functions import invert_function
from workflows.arlexecute.calibration.calskymodel_workflows import calskymodel_solve_workflow
from processing_components.image.operations import export_image_to_fits
import logging
def init_logging():
log = logging.getLogger()
logging.basicConfig(filename='%s/skymodel_cal.log' % results_dir,
filemode='a',
format='%(thread)s %(asctime)s,%(msecs)d %(name)s %(levelname)s %(message)s',
datefmt='%H:%M:%S',
level=logging.INFO)
log = logging.getLogger()
logging.info("Starting skymodel_cal")
| _____no_output_____ | Apache-2.0 | workflows/notebooks/calskymodel.ipynb | mfarrera/algorithm-reference-library |
Use Dask throughout | arlexecute.set_client(use_dask=True)
arlexecute.run(init_logging) | _____no_output_____ | Apache-2.0 | workflows/notebooks/calskymodel.ipynb | mfarrera/algorithm-reference-library |
We make the visibility. The parameter rmax determines the distance of the furthest antenna/stations used. All over parameters are determined from this number.We set the w coordinate to be zero for all visibilities so as not to have to do full w-term processing. This speeds up the imaging steps. | nfreqwin = 1
ntimes = 1
rmax = 750
frequency = numpy.linspace(0.8e8, 1.2e8, nfreqwin)
if nfreqwin > 1:
channel_bandwidth = numpy.array(nfreqwin * [frequency[1] - frequency[0]])
else:
channel_bandwidth = [0.4e8]
times = numpy.linspace(-numpy.pi / 3.0, numpy.pi / 3.0, ntimes)
phasecentre=SkyCoord(ra=+30.0 * u.deg, dec=-45.0 * u.deg, frame='icrs', equinox='J2000')
lowcore = create_named_configuration('LOWBD2', rmax=rmax)
block_vis = create_blockvisibility(lowcore, times, frequency=frequency,
channel_bandwidth=channel_bandwidth, weight=1.0, phasecentre=phasecentre,
polarisation_frame=PolarisationFrame("stokesI"), zerow=True)
wprojection_planes=1
advice=advise_wide_field(block_vis, guard_band_image=5.0, delA=0.02,
wprojection_planes=wprojection_planes)
vis_slices = advice['vis_slices']
npixel=advice['npixels2']
cellsize=advice['cellsize'] | _____no_output_____ | Apache-2.0 | workflows/notebooks/calskymodel.ipynb | mfarrera/algorithm-reference-library |
Generate the model from the GLEAM catalog, including application of the primary beam. | beam = create_image_from_visibility(block_vis, npixel=npixel, frequency=frequency,
nchan=nfreqwin, cellsize=cellsize, phasecentre=phasecentre)
original_gleam_components = create_low_test_skycomponents_from_gleam(flux_limit=1.0,
phasecentre=phasecentre, frequency=frequency,
polarisation_frame=PolarisationFrame('stokesI'),
radius=npixel * cellsize/2.0)
beam = create_low_test_beam(beam)
pb_gleam_components = apply_beam_to_skycomponent(original_gleam_components, beam,
flux_limit=0.5)
from matplotlib import pylab
pylab.rcParams['figure.figsize'] = (12.0, 12.0)
pylab.rcParams['image.cmap'] = 'rainbow'
show_image(beam, components=pb_gleam_components, cm='Greys', title='Primary beam plus GLEAM components')
print("Number of components %d" % len(pb_gleam_components)) | _____no_output_____ | Apache-2.0 | workflows/notebooks/calskymodel.ipynb | mfarrera/algorithm-reference-library |
Generate the template image | model = create_image_from_visibility(block_vis, npixel=npixel,
frequency=[numpy.average(frequency)],
nchan=1,
channel_bandwidth=[numpy.sum(channel_bandwidth)],
cellsize=cellsize, phasecentre=phasecentre) | _____no_output_____ | Apache-2.0 | workflows/notebooks/calskymodel.ipynb | mfarrera/algorithm-reference-library |
Create sources to be peeled | peel_distance = 0.16
peelers = select_components_by_separation(phasecentre, pb_gleam_components,
min=peel_distance)
gleam_components = select_components_by_separation(phasecentre, pb_gleam_components,
max=peel_distance)
print("There are %d sources inside the primary beam and %d sources outside"
% (len(gleam_components), len(peelers))) | _____no_output_____ | Apache-2.0 | workflows/notebooks/calskymodel.ipynb | mfarrera/algorithm-reference-library |
Create the model visibilities, applying a different gain table for peeled sources and other components | corrupted_vis = copy_visibility(block_vis, zero=True)
gt = create_gaintable_from_blockvisibility(block_vis, timeslice='auto')
components_errors = [(p, 1.0) for p in peelers]
components_errors.append((pb_gleam_components, 0.1))
for sc, phase_error in components_errors:
component_vis = copy_visibility(block_vis, zero=True)
gt = simulate_gaintable(gt, amplitude_error=0.0, phase_error=phase_error)
component_vis = predict_skycomponent_visibility(component_vis, sc)
component_vis = apply_gaintable(component_vis, gt)
corrupted_vis.data['vis'][...]+=component_vis.data['vis'][...]
dirty, sumwt = invert_function(corrupted_vis, model, context='2d')
qa=qa_image(dirty)
vmax=qa.data['medianabs']*20.0
vmin=-qa.data['medianabs']
print(qa)
export_image_to_fits(dirty, '%s/calskymodel_before_dirty.fits' % results_dir)
show_image(dirty, cm='Greys', components=peelers, vmax=vmax, vmin=vmin, title='Peelers')
show_image(dirty, cm='Greys', components=gleam_components, vmax=vmax, vmin=vmin, title='Targets')
plt.show() | _____no_output_____ | Apache-2.0 | workflows/notebooks/calskymodel.ipynb | mfarrera/algorithm-reference-library |
Find the components above the threshold | qa = qa_image(dirty)
vmax=qa.data['medianabs']*20.0
vmin=-qa.data['medianabs']*2.0
print(qa)
threshold = 10.0*qa.data['medianabs']
print("Selecting sources brighter than %f" % threshold)
initial_found_components= find_skycomponents(dirty, threshold=threshold)
show_image(dirty, components=initial_found_components, cm='Greys', vmax=vmax, vmin=vmin,
title='Dirty image plus found components')
plt.show()
peel_distance = 0.16
flux_threshold=5.0
peelers = select_components_by_separation(phasecentre, initial_found_components,
min=peel_distance)
inbeam_components = select_components_by_separation(phasecentre, initial_found_components,
max=peel_distance)
bright_components = select_components_by_flux(inbeam_components, fmin=flux_threshold)
faint_components = select_components_by_flux(inbeam_components, fmax=flux_threshold)
print("%d sources will be peeled (i.e. held fixed but gain solved)" % len(peelers))
print("%d bright sources will be processed as components (solved both as component and for gain)" % len(bright_components))
print("%d faint sources will be processed collectively as a fixed image and gain solved" % len(faint_components))
faint_model = create_empty_image_like(model)
faint_model = insert_skycomponent(faint_model, faint_components, insert_method='Lanczos')
show_image(faint_model, cm='Greys', title='Model for faint sources', vmax=0.3, vmin=-0.03)
plt.show()
calskymodel_graph = [arlexecute.execute(SkyModel, nout=1)(components=[p], fixed=True) for p in peelers] \
+ [arlexecute.execute(SkyModel, nout=1)(components=[b], fixed=False) for b in bright_components] \
+ [arlexecute.execute(SkyModel, nout=1)(images=[faint_model], fixed=True)] | _____no_output_____ | Apache-2.0 | workflows/notebooks/calskymodel.ipynb | mfarrera/algorithm-reference-library |
Run skymodel_cal using dask | corrupted_vis = arlexecute.scatter(corrupted_vis)
graph = calskymodel_solve_workflow(corrupted_vis, calskymodel_graph, niter=30, gain=0.25, tol=1e-8)
calskymodel, residual_vis = arlexecute.compute(graph, sync=True) | _____no_output_____ | Apache-2.0 | workflows/notebooks/calskymodel.ipynb | mfarrera/algorithm-reference-library |
Combine all components for display | skymodel_components = list()
for csm in calskymodel:
skymodel_components += csm[0].components | _____no_output_____ | Apache-2.0 | workflows/notebooks/calskymodel.ipynb | mfarrera/algorithm-reference-library |
Check that the peeled sources are not altered | recovered_peelers = find_skycomponent_matches(peelers, skymodel_components, 1e-5)
ok = True
for p in recovered_peelers:
ok = ok and numpy.abs(peelers[p[0]].flux[0,0] - skymodel_components[p[1]].flux[0,0]) < 1e-7
print("Peeler sources flux unchanged: %s" % ok)
ok = True
for p in recovered_peelers:
ok = ok and peelers[p[0]].direction.separation(skymodel_components[p[1]].direction).rad < 1e-15
print("Peeler sources directions unchanged: %s" % ok) | _____no_output_____ | Apache-2.0 | workflows/notebooks/calskymodel.ipynb | mfarrera/algorithm-reference-library |
Now we find the components in the residual image and add those to the existing model | residual, sumwt = invert_function(residual_vis, model, context='2d')
qa = qa_image(residual)
vmax=qa.data['medianabs']*30.0
vmin=-qa.data['medianabs']*3.0
print(qa)
threshold = 20.0*qa.data['medianabs']
print("Selecting sources brighter than %f" % threshold)
final_found_components = find_skycomponents(residual, threshold=threshold)
show_image(residual, components=final_found_components, cm='Greys',
title='Residual image after Sagecal with newly identified components', vmax=vmax, vmin=vmin)
plt.show()
final_components= skymodel_components + final_found_components | _____no_output_____ | Apache-2.0 | workflows/notebooks/calskymodel.ipynb | mfarrera/algorithm-reference-library |
Make a restored image | psf, _ = invert_function(residual_vis, model, dopsf=True, context='2d')
component_image = copy_image(faint_model)
component_image = insert_skycomponent(component_image, final_components)
restored = restore_cube(component_image, psf, residual)
export_image_to_fits(restored, '%s/calskymodel_restored.fits' % results_dir)
qa=qa_image(restored, context='Restored image after SageCal')
print(qa)
show_image(restored, components=final_components, cm='Greys',
title='Restored image after SageCal', vmax=vmax, vmin=vmin)
plt.show() | _____no_output_____ | Apache-2.0 | workflows/notebooks/calskymodel.ipynb | mfarrera/algorithm-reference-library |
Now match the recovered components to the originals | original_bright_components = peelers + bright_components
matches = find_skycomponent_matches(final_components, original_bright_components, 3*cellsize) | _____no_output_____ | Apache-2.0 | workflows/notebooks/calskymodel.ipynb | mfarrera/algorithm-reference-library |
Look at the range of separations found | separations = [match[2] for match in matches]
plt.clf()
plt.hist(separations/cellsize, bins=50)
plt.title('Separation between input and recovered source in pixels')
plt.xlabel('Separation in cells (cellsize = %g radians)' % cellsize)
plt.ylabel('Number')
plt.show() | _____no_output_____ | Apache-2.0 | workflows/notebooks/calskymodel.ipynb | mfarrera/algorithm-reference-library |
Now look at the matches between the original components and those recovered. | totalfluxin = numpy.sum([c.flux[0,0] for c in pb_gleam_components])
totalfluxout = numpy.sum([c.flux[0,0] for c in final_components]) + numpy.sum(faint_model.data)
print("Recovered %.3f (Jy) of original %.3f (Jy)" % (totalfluxout, totalfluxin))
found = [match[1] for match in matches]
notfound = list()
for c in range(len(original_bright_components)):
if c not in found:
notfound.append(c)
print("The following original components were not found", notfound) | _____no_output_____ | Apache-2.0 | workflows/notebooks/calskymodel.ipynb | mfarrera/algorithm-reference-library |
Look at the recovered flux and the location of the unmatched components. From the image display these seem to be blends of close components. | fluxin = [original_bright_components[match[1]].flux[0,0] for match in matches]
fluxout = [final_components[match[0]].flux[0,0] for match in matches]
missed_components = [original_bright_components[c] for c in notfound]
missed_flux = [match.flux[0,0] for match in missed_components]
plt.clf()
plt.plot(fluxin, fluxout, '.', color='blue')
plt.plot(missed_flux, len(missed_flux)*[0.0], '.', color='red')
plt.title('Recovered flux')
plt.xlabel('Component input flux')
plt.ylabel('Component recovered flux')
plt.show()
show_image(restored, components=missed_components, cm='Greys',
title='Restored original model with missing components', vmax=vmax, vmin=vmin)
plt.show()
arlexecute.close() | _____no_output_____ | Apache-2.0 | workflows/notebooks/calskymodel.ipynb | mfarrera/algorithm-reference-library |
Multiple linear regression**For Table 3 of the paper**Cell-based QUBICC R2B5 model | from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error
from tensorflow.keras import backend as K
from tensorflow.keras.regularizers import l1_l2
import tensorflow as tf
import tensorflow.nn as nn
import gc
import numpy as np
import pandas as pd
import importlib
import os
import sys
#Import sklearn before tensorflow (static Thread-local storage)
from sklearn.preprocessing import StandardScaler
from tensorflow.keras.optimizers import Nadam
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, BatchNormalization
path = '/pf/b/b309170'
path_data = path + '/my_work/icon-ml_data/cloud_cover_parameterization/grid_cell_based_QUBICC_R02B05/based_on_var_interpolated_data'
# Add path with my_classes to sys.path
sys.path.insert(0, path + '/workspace_icon-ml/cloud_cover_parameterization/')
import my_classes
importlib.reload(my_classes)
from my_classes import simple_sundqvist_scheme
from my_classes import write_infofile
from my_classes import load_data
import matplotlib.pyplot as plt
import time
NUM = 1
# Prevents crashes of the code
gpus = tf.config.list_physical_devices('GPU')
tf.config.set_visible_devices(gpus[0], 'GPU')
# Allow the growth of memory Tensorflow allocates (limits memory usage overall)
for gpu in gpus:
tf.config.experimental.set_memory_growth(gpu, True)
scaler = StandardScaler()
# Data is not yet normalized
input_data = np.load(path_data + '/cloud_cover_input_qubicc.npy', mmap_mode='r')
output_data = np.load(path_data + '/cloud_cover_output_qubicc.npy', mmap_mode='r')
(samples_total, no_of_features) = input_data.shape
assert no_of_features < samples_total # Making sure there's no mixup
# Scale the input data = (samples_total, no_of_features)
scaler.fit(input_data)
assert len(scaler.mean_) == no_of_features # Every feature has its own mean and std and we scale accordingly
input_data_scaled = scaler.transform(input_data) | _____no_output_____ | MIT | additional_content/baselines/multiple_linear_regression_cell_based-R2B5.ipynb | agrundner24/iconml_clc |
Training the multiple linear model on the entire data set | t0 = time.time()
# The optimal multiple linear regression model
lin_reg = LinearRegression()
lin_reg.fit(input_data_scaled, output_data)
print(time.time() - t0)
# Loss of this optimal multiple linear regression model
clc_predictions = lin_reg.predict(input_data_scaled)
lin_mse = mean_squared_error(output_data, clc_predictions)
print('The mean squared error of the linear model is %.2f.'%lin_mse) | The mean squared error of the linear model is 401.47.
| MIT | additional_content/baselines/multiple_linear_regression_cell_based-R2B5.ipynb | agrundner24/iconml_clc |
Zero Output Model | np.mean(output_data**2, dtype=np.float64) | _____no_output_____ | MIT | additional_content/baselines/multiple_linear_regression_cell_based-R2B5.ipynb | agrundner24/iconml_clc |
Constant Output Model | mean = np.mean(output_data, dtype=np.float64)
np.mean((output_data-mean)**2, dtype=np.float64) | _____no_output_____ | MIT | additional_content/baselines/multiple_linear_regression_cell_based-R2B5.ipynb | agrundner24/iconml_clc |
Randomly initialized neural network | # Create the model
model = Sequential()
# First hidden layer
model.add(Dense(units=64, activation='tanh', input_dim=no_of_features,
kernel_regularizer=l1_l2(l1=0.004749, l2=0.008732)))
# Second hidden layer
model.add(Dense(units=64, activation=nn.leaky_relu, kernel_regularizer=l1_l2(l1=0.004749, l2=0.008732)))
# model.add(Dropout(0.221)) # We drop 18% of the hidden nodes
model.add(BatchNormalization())
# Third hidden layer
model.add(Dense(units=64, activation='tanh', kernel_regularizer=l1_l2(l1=0.004749, l2=0.008732)))
# model.add(Dropout(0.221)) # We drop 18% of the hidden nodes
# Output layer
model.add(Dense(1, activation='linear', kernel_regularizer=l1_l2(l1=0.004749, l2=0.008732)))
model.compile(loss='mse', optimizer=Nadam())
# model_fold_3 is implemented in ICON-A
batch_size = 2**20
for i in range(1 + input_data_scaled.shape[0]//batch_size):
if i == 0:
clc_predictions = model.predict_on_batch(input_data_scaled[i*batch_size:(i+1)*batch_size])
else:
clc_predictions = np.concatenate((clc_predictions, model.predict_on_batch(input_data_scaled[i*batch_size:(i+1)*batch_size])), axis=0)
K.clear_session()
gc.collect()
lin_mse = mean_squared_error(output_data, clc_predictions[:, 0])
print('The mean squared error of the randomly initialized neural network is %.2f.'%lin_mse) | The mean squared error of the randomly initialized neural network is 913.91.
| MIT | additional_content/baselines/multiple_linear_regression_cell_based-R2B5.ipynb | agrundner24/iconml_clc |
Simplified Sundqvist functioninput_data is unscaled | qv = input_data[:, 0]
temp = input_data[:, 3]
pres = input_data[:, 4]
t0 = time.time()
# 0.001% of the data
ind = np.random.randint(0, samples_total, samples_total//10**5)
# Entries will be in [0, 1]
sundqvist = []
for i in ind:
sundqvist.append(simple_sundqvist_scheme(qv[i], temp[i], pres[i], ps=101325))
time.time() - t0
np.mean((output_data[ind] - 100*np.array(sundqvist))**2, dtype=np.float64) | _____no_output_____ | MIT | additional_content/baselines/multiple_linear_regression_cell_based-R2B5.ipynb | agrundner24/iconml_clc |
** Artistic Colorizer ** ◢ DeOldify - Colorize your own photos!**Credits:**Special thanks to:Matt Robinson and María Benavente for pioneering the DeOldify image colab notebook. Dana Kelley for doing things, breaking stuff & having an opinion on everything. ---◢ Verify Correct Runtime Settings** IMPORTANT **In the "Runtime" menu for the notebook window, select "Change runtime type." Ensure that the following are selected:* Runtime Type = Python 3* Hardware Accelerator = GPU ◢ Git clone and install DeOldify | !git clone https://github.com/jantic/DeOldify.git DeOldify
cd DeOldify | _____no_output_____ | MIT | ImageColorizerColab.ipynb | pabenz99/DeOldify |
◢ Setup | #NOTE: This must be the first call in order to work properly!
from deoldify import device
from deoldify.device_id import DeviceId
#choices: CPU, GPU0...GPU7
device.set(device=DeviceId.GPU0)
import torch
if not torch.cuda.is_available():
print('GPU not available.')
!pip install -r colab_requirements.txt
import fastai
from deoldify.visualize import *
!mkdir 'models'
!wget https://www.dropbox.com/s/zkehq1uwahhbc2o/ColorizeArtistic_gen.pth?dl=0 -O ./models/ColorizeArtistic_gen.pth
!wget https://media.githubusercontent.com/media/jantic/DeOldify/master/resource_images/watermark.png -O ./resource_images/watermark.png
colorizer = get_image_colorizer(artistic=True) | _____no_output_____ | MIT | ImageColorizerColab.ipynb | pabenz99/DeOldify |
◢ Instructions source_urlType in a url to a direct link of an image. Usually that means they'll end in .png, .jpg, etc. NOTE: If you want to use your own image, upload it first to a site like Imgur. render_factorThe default value of 35 has been carefully chosen and should work -ok- for most scenarios (but probably won't be the -best-). This determines resolution at which the color portion of the image is rendered. Lower resolution will render faster, and colors also tend to look more vibrant. Older and lower quality images in particular will generally benefit by lowering the render factor. Higher render factors are often better for higher quality images, but the colors may get slightly washed out. watermarkedSelected by default, this places a watermark icon of a palette at the bottom left corner of the image. This is intended to be a standard way to convey to others viewing the image that it is colorized by AI. We want to help promote this as a standard, especially as the technology continues to improve and the distinction between real and fake becomes harder to discern. This palette watermark practice was initiated and lead by the company MyHeritage in the MyHeritage In Color feature (which uses a newer version of DeOldify than what you're using here). How to Download a CopySimply right click on the displayed image and click "Save image as..."! Pro TipsYou can evaluate how well the image is rendered at each render_factor by using the code at the bottom (that cell under "See how well render_factor values perform on a frame here"). ◢ Colorize!! | source_url = '' #@param {type:"string"}
render_factor = 35 #@param {type: "slider", min: 7, max: 40}
watermarked = True #@param {type:"boolean"}
if source_url is not None and source_url !='':
image_path = colorizer.plot_transformed_image_from_url(url=source_url, render_factor=render_factor, compare=True, watermarked=watermarked)
show_image_in_notebook(image_path)
else:
print('Provide an image url and try again.') | _____no_output_____ | MIT | ImageColorizerColab.ipynb | pabenz99/DeOldify |
See how well render_factor values perform on the image here | for i in range(10,40,2):
colorizer.plot_transformed_image('test_images/image.png', render_factor=i, display_render_factor=True, figsize=(8,8)) | _____no_output_____ | MIT | ImageColorizerColab.ipynb | pabenz99/DeOldify |
M2.859 · Visualización de datos · Práctica, Parte 22021-1 · Máster universitario en Ciencia de datos (Data science)Estudios de Informática, Multimedia y Telecomunicación A9: Práctica Final (parte 2) - Wrangling dataEl [**wrangling data**](https://en.wikipedia.org/wiki/Data_wrangling) es el proceso de transformar y mapear datos de un formulario de datos "sin procesar" a otro formato con la intención de hacerlo más apropiado y valioso para una variedad de propósitos posteriores, como el análisis. El objetivo del wrangling data es garantizar la calidad y la utilidad de los datos. Los analistas de datos suelen pasar la mayor parte de su tiempo en el proceso de disputa de datos en comparación con el análisis real de los datos.El proceso de wrangling data puede incluir más manipulación, visualización de datos, agregación de datos, entrenamiento de un modelo estadístico, así como muchos otros usos potenciales. El wrangling data normalmente sigue un conjunto de pasos generales que comienzan con la extracción de los datos sin procesar de la fuente de datos, "removiendo" los datos sin procesar (por ejemplo, clasificación) o analizando los datos en estructuras de datos predefinidas y, finalmente, depositando el contenido resultante en un sumidero de datos para almacenamiento y uso futuro. Para ello vamos a necesitar las siguientes librerías: | from six import StringIO
from IPython.display import Image
from sklearn import datasets
from sklearn.decomposition import PCA
from sklearn.model_selection import train_test_split, cross_val_score
from sklearn.metrics import accuracy_score, confusion_matrix, classification_report
from sklearn.tree import DecisionTreeClassifier, export_graphviz
import pydotplus
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib inline
import pandas as pd
pd.set_option('display.max_columns', None) | _____no_output_____ | MIT | M2.859_20211_A9_gbonillas.ipynb | gpbonillas/stackoverflow_2021_wrangling_data |
1. Carga del conjunto de datos (1 punto)Se ha seleccionado un conjunto de datos desde el portal Stack Overflow Annual Developer Survey, que examina todos los aspectos de la experiencia de los programadores de la comunidad (Stack Overflow), desde la satisfacción profesional y la búsqueda de empleo hasta la educación y las opiniones sobre el software de código abierto; y los resultados se publican en la siguiente URL: https://insights.stackoverflow.com/survey.En este portal se encuentran publicados los resultados de los últimos 11 años. Para los fines de la práctica final de esta asignatura se usará el dataset del año 2021, cuyo link de descarga es: https://info.stackoverflowsolutions.com/rs/719-EMH-566/images/stack-overflow-developer-survey-2021.zip. | so2021_df = pd.read_csv('survey_results_public.csv', header=0)
so2021_df.sample(5) | _____no_output_____ | MIT | M2.859_20211_A9_gbonillas.ipynb | gpbonillas/stackoverflow_2021_wrangling_data |
Selección de variables: se realiza la selección de todas las variables del dataset que servirán para responder a todas las cuestiones planteadas en la primera parte de la práctica: | so2021_data = so2021_df[['MainBranch', 'Employment', 'Country', 'EdLevel', 'Age1stCode', 'YearsCode', 'YearsCodePro', 'DevType', 'CompTotal', 'LanguageHaveWorkedWith', 'DatabaseHaveWorkedWith', 'PlatformHaveWorkedWith', 'WebframeHaveWorkedWith', 'MiscTechHaveWorkedWith', 'ToolsTechHaveWorkedWith', 'NEWCollabToolsHaveWorkedWith', 'OpSys', 'Age', 'Gender', 'Trans', 'Ethnicity', 'MentalHealth', 'ConvertedCompYearly']]
so2021_data.head(5)
so2021_data.shape
so2021_data.info()
so2021_data.isnull().values.any() #valores perdidos en dataset
so2021_data.isnull().any() # valores perdidos por columnas en el dataset
data = so2021_data.dropna()
data.isnull().any() # valores perdidos por columnas en el dataset
data.info()
data.head()
data.to_csv('data.csv', index=False)
data_test = data.copy()
data_test.to_csv('data_test.csv', index=False) | _____no_output_____ | MIT | M2.859_20211_A9_gbonillas.ipynb | gpbonillas/stackoverflow_2021_wrangling_data |
Variable Ethnicity:. | from re import search
def choose_ethnia(cell_ethnia):
val_ethnia_exceptions = ["I don't know", "Or, in your own words:"]
if cell_ethnia == "I don't know;Or, in your own words:":
return val_ethnia_exceptions[0]
if search(";", cell_ethnia):
row_ethnia_values = cell_ethnia.split(';', 5)
first_val = row_ethnia_values[0]
if first_val not in val_ethnia_exceptions:
return first_val
if len(row_ethnia_values) > 1:
if row_ethnia_values[1] not in val_ethnia_exceptions:
return row_ethnia_values[1]
if len(row_ethnia_values) > 2:
if row_ethnia_values[2] not in val_ethnia_exceptions:
return row_ethnia_values[2]
else:
return cell_ethnia
data_test['Ethnicity'] = data_test['Ethnicity'].apply(choose_ethnia)
data_test.drop(index=data_test[data_test['Ethnicity'] == 'Or, in your own words:'].index, inplace=True)
data_test.drop(index=data_test[data_test['Ethnicity'] == 'Prefer not to say'].index, inplace=True)
data_test['Ethnicity'].drop_duplicates().sort_values()
data_test['Ethnicity'] = data_test['Ethnicity'].replace(['Black or of African descent'], 'Negro')
data_test['Ethnicity'] = data_test['Ethnicity'].replace(['East Asian'], 'Asiatico del este')
data_test['Ethnicity'] = data_test['Ethnicity'].replace(['Hispanic or Latino/a/x'], 'Latino')
data_test['Ethnicity'] = data_test['Ethnicity'].replace(["I don't know"], 'No Definido')
data_test['Ethnicity'] = data_test['Ethnicity'].replace(['Indigenous (such as Native American, Pacific Islander, or Indigenous Australian)'], 'Indigena')
data_test['Ethnicity'] = data_test['Ethnicity'].replace(['Middle Eastern'], 'Medio Oriente')
data_test['Ethnicity'] = data_test['Ethnicity'].replace(['South Asian'], 'Asiatico del Sur')
data_test['Ethnicity'] = data_test['Ethnicity'].replace(['Southeast Asian'], 'Asiatico del Sudeste')
data_test['Ethnicity'] = data_test['Ethnicity'].replace(['White or of European descent'], 'Blanco o Europeo')
data_test['Ethnicity'].drop_duplicates().sort_values()
data_test.to_csv('data_test.csv', index=False) | _____no_output_____ | MIT | M2.859_20211_A9_gbonillas.ipynb | gpbonillas/stackoverflow_2021_wrangling_data |
Variable Employment:. | data_test['Employment'].drop_duplicates().sort_values()
data_test['Employment'] = data_test['Employment'].replace(['Employed full-time'], 'Tiempo completo')
data_test['Employment'] = data_test['Employment'].replace(['Employed part-time'], 'Tiempo parcial')
data_test['Employment'] = data_test['Employment'].replace(['Independent contractor, freelancer, or self-employed'], 'Independiete')
data_test['Employment'].drop_duplicates().sort_values() | _____no_output_____ | MIT | M2.859_20211_A9_gbonillas.ipynb | gpbonillas/stackoverflow_2021_wrangling_data |
Variable EdLevel:. | data_test['EdLevel'].drop_duplicates().sort_values()
data_test['EdLevel'] = data_test['EdLevel'].replace(['Associate degree (A.A., A.S., etc.)'], 'Grado Asociado')
data_test['EdLevel'] = data_test['EdLevel'].replace(['Bachelor’s degree (B.A., B.S., B.Eng., etc.)'], 'Licenciatura')
data_test['EdLevel'] = data_test['EdLevel'].replace(['Master’s degree (M.A., M.S., M.Eng., MBA, etc.)'], 'Master')
data_test['EdLevel'] = data_test['EdLevel'].replace(['Other doctoral degree (Ph.D., Ed.D., etc.)'], 'Doctorado')
data_test['EdLevel'] = data_test['EdLevel'].replace(['Primary/elementary school'], 'Primaria')
data_test['EdLevel'] = data_test['EdLevel'].replace(['Professional degree (JD, MD, etc.)'], 'Grado Profesional')
data_test['EdLevel'] = data_test['EdLevel'].replace(['Secondary school (e.g. American high school, German Realschule or Gymnasium, etc.)'], 'Secundaria')
data_test['EdLevel'] = data_test['EdLevel'].replace(['Some college/university study without earning a degree'], 'Estudios sin grado')
data_test['EdLevel'] = data_test['EdLevel'].replace(['Something else'], 'Otro')
data_test['EdLevel'].drop_duplicates().sort_values()
data_test.to_csv('data_test.csv', index=False) | _____no_output_____ | MIT | M2.859_20211_A9_gbonillas.ipynb | gpbonillas/stackoverflow_2021_wrangling_data |
Variable DevType:. | data_test['DevType'].drop_duplicates().sort_values()
from re import search
def choose_devtype(cell_devtype):
val_devtype_exceptions = ["Other (please specify):"]
if cell_devtype == "Other (please specify):":
return val_devtype_exceptions[0]
if search(";", cell_devtype):
row_devtype_values = cell_devtype.split(';', 10)
first_val = row_devtype_values[0]
if first_val not in val_devtype_exceptions:
return first_val
if len(row_devtype_values) > 1:
if row_devtype_values[1] not in val_devtype_exceptions:
return row_devtype_values[1]
else:
return cell_devtype
data_test['DevType'] = data_test['DevType'].apply(choose_devtype)
data_test['DevType'].head()
data_test['DevType'].drop_duplicates().sort_values()
data_test['DevType'].value_counts()
data_test['DevType'] = data_test['DevType'].replace(['Developer, full-stack'], 'Desarrollador full-stack')
data_test['DevType'] = data_test['DevType'].replace(['Developer, front-end'], 'Desarrollador front-end')
data_test['DevType'] = data_test['DevType'].replace(['Developer, mobile'], 'Desarrollador móvil')
data_test['DevType'] = data_test['DevType'].replace(['Developer, back-end'], 'Desarrollador back-end')
data_test['DevType'] = data_test['DevType'].replace(['Developer, desktop or enterprise applications'], 'Desarrollador Escritorio')
data_test['DevType'] = data_test['DevType'].replace(['Engineer, data'], 'Ingeniero de datos')
data_test['DevType'] = data_test['DevType'].replace(['Data scientist or machine learning specialist'], 'Cientifico de datos')
data_test['DevType'] = data_test['DevType'].replace(['Other (please specify):'], 'Otro')
data_test['DevType'] = data_test['DevType'].replace(['Engineering manager'], 'Manager de Ingeniería')
data_test['DevType'] = data_test['DevType'].replace(['DevOps specialist'], 'Especialista en DevOps')
data_test['DevType'] = data_test['DevType'].replace(['Senior Executive (C-Suite, VP, etc.)'], 'Ejecutivo Senior')
data_test['DevType'] = data_test['DevType'].replace(['Academic researcher'], 'Investigador Académico')
data_test['DevType'] = data_test['DevType'].replace(['Developer, QA or test'], 'Desarrollador de QA o Test')
data_test['DevType'] = data_test['DevType'].replace(['Data or business analyst'], 'Analista de datos o negocio')
data_test['DevType'] = data_test['DevType'].replace(['Developer, embedded applications or devices'], 'Desarrollador de aplicaciones embebidas')
data_test['DevType'] = data_test['DevType'].replace(['System administrator'], 'Administrador de sistemas')
data_test['DevType'] = data_test['DevType'].replace(['Engineer, site reliability'], 'Ingeniero de confiabilidad del sitio')
data_test['DevType'] = data_test['DevType'].replace(['Product manager'], 'Gerente de producto')
data_test['DevType'] = data_test['DevType'].replace(['Database administrator'], 'Administrador de base de datos')
data_test['DevType'] = data_test['DevType'].replace(['Student'], 'Estudiante')
data_test['DevType'] = data_test['DevType'].replace(['Developer, game or graphics'], 'Desarrollador de juegos o gráfico')
data_test['DevType'] = data_test['DevType'].replace(['Scientist'], 'Científico')
data_test['DevType'] = data_test['DevType'].replace(['Designer'], 'Diseñador')
data_test['DevType'] = data_test['DevType'].replace(['Educator'], 'Educador')
data_test['DevType'] = data_test['DevType'].replace(['Marketing or sales professional'], 'Profesional en Marketing o ventas')
data_test['DevType'].drop_duplicates().sort_values()
data_test['DevType'].value_counts() | _____no_output_____ | MIT | M2.859_20211_A9_gbonillas.ipynb | gpbonillas/stackoverflow_2021_wrangling_data |
Variable MainBranch: | data_test['MainBranch'].drop_duplicates().sort_values()
data_test['MainBranch'] = data_test['MainBranch'].replace(['I am a developer by profession'], 'Desarrollador Profesional')
data_test['MainBranch'] = data_test['MainBranch'].replace(['I am not primarily a developer, but I write code sometimes as part of my work'], 'Desarrollador ocasional')
data_test['MainBranch'].drop_duplicates().sort_values()
data_test.to_csv('data_test.csv', index=False) | _____no_output_____ | MIT | M2.859_20211_A9_gbonillas.ipynb | gpbonillas/stackoverflow_2021_wrangling_data |
Variable Age1stCode: | data_test['Age1stCode'].drop_duplicates().sort_values()
data_test['Age1stCode'].value_counts()
data_test['Age1stCode'] = data_test['Age1stCode'].replace(['11 - 17 years'], '11-17')
data_test['Age1stCode'] = data_test['Age1stCode'].replace(['18 - 24 years'], '18-24')
data_test['Age1stCode'] = data_test['Age1stCode'].replace(['25 - 34 years'], '25-34')
data_test['Age1stCode'] = data_test['Age1stCode'].replace(['35 - 44 years'], '35-44')
data_test['Age1stCode'] = data_test['Age1stCode'].replace(['45 - 54 years'], '45-54')
data_test['Age1stCode'] = data_test['Age1stCode'].replace(['5 - 10 years'], '5-10')
data_test['Age1stCode'] = data_test['Age1stCode'].replace(['55 - 64 years'], '55-64')
data_test['Age1stCode'] = data_test['Age1stCode'].replace(['Older than 64 years'], '> 64')
data_test['Age1stCode'] = data_test['Age1stCode'].replace(['Younger than 5 years'], '< 5')
data_test['Age1stCode'].value_counts() | _____no_output_____ | MIT | M2.859_20211_A9_gbonillas.ipynb | gpbonillas/stackoverflow_2021_wrangling_data |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.