markdown
stringlengths 0
1.02M
| code
stringlengths 0
832k
| output
stringlengths 0
1.02M
| license
stringlengths 3
36
| path
stringlengths 6
265
| repo_name
stringlengths 6
127
|
|---|---|---|---|---|---|
ndarray による1次元配列の例 | a2 = np.array([[1, 2, 3],[4, 5, 6]], dtype='float32') # データ型 float32 の2次元配列を生成
print('データの型 (dtype):', a2.dtype)
print('要素の数 (size):', a2.size)
print('形状 (shape):', a2.shape)
print('次元の数 (ndim):', a2.ndim)
print('中身:', a2) | データの型 (dtype): float32
要素の数 (size): 6
形状 (shape): (2, 3)
次元の数 (ndim): 2
中身: [[1. 2. 3.]
[4. 5. 6.]]
| MIT | notebooks/Chapter03/math_numpy.ipynb | tagomaru/ai_security |
2. ベクトル(1次元配列) ベクトル a の生成(1次元配列の生成) | a = np.array([4, 1]) | _____no_output_____ | MIT | notebooks/Chapter03/math_numpy.ipynb | tagomaru/ai_security |
ベクトルのスカラー倍 | for k in (2, 0.5, -1):
print(k * a) | [8 2]
[2. 0.5]
[-4 -1]
| MIT | notebooks/Chapter03/math_numpy.ipynb | tagomaru/ai_security |
ベクトルの和と差 | b = np.array([1, 2]) # ベクトル b の生成
print('a + b =', a + b) # ベクトル a とベクトル b の和
print('a - b =', a - b) # ベクトル a とベクトル b の差 | a + b = [5 3]
a - b = [ 3 -1]
| MIT | notebooks/Chapter03/math_numpy.ipynb | tagomaru/ai_security |
3. 行列(2次元配列) 行列を2次元配列で生成 | A = np.array([[1, 2], [3 ,4], [5, 6]])
B = np.array([[5, 6], [7 ,8]])
print('A:\n', A)
print('A.shape:', A.shape )
print()
print('B:\n', B)
print('B.shape:', B.shape ) | A:
[[1 2]
[3 4]
[5 6]]
A.shape: (3, 2)
B:
[[5 6]
[7 8]]
B.shape: (2, 2)
| MIT | notebooks/Chapter03/math_numpy.ipynb | tagomaru/ai_security |
行列Aの i = 3, j = 2 にアクセス | print(A[2][1]) | 6
| MIT | notebooks/Chapter03/math_numpy.ipynb | tagomaru/ai_security |
A の転置行列 | print(A.T) | [[1 3 5]
[2 4 6]]
| MIT | notebooks/Chapter03/math_numpy.ipynb | tagomaru/ai_security |
行列のスカラー倍 | print(2 * A) | [[ 2 4]
[ 6 8]
[10 12]]
| MIT | notebooks/Chapter03/math_numpy.ipynb | tagomaru/ai_security |
行列の和と差 | print('A + A:\n', A + A) # 行列 A と行列 A の和
print()
print('A - A:\n', A - A) # 行列 A と行列 A の差 | A + A:
[[ 2 4]
[ 6 8]
[10 12]]
A - A:
[[0 0]
[0 0]
[0 0]]
| MIT | notebooks/Chapter03/math_numpy.ipynb | tagomaru/ai_security |
行列 A と行列 B の和 | print(A + B) | _____no_output_____ | MIT | notebooks/Chapter03/math_numpy.ipynb | tagomaru/ai_security |
行列の積 | print(np.dot(A, B)) | [[19 22]
[43 50]
[67 78]]
| MIT | notebooks/Chapter03/math_numpy.ipynb | tagomaru/ai_security |
積 BA | print(np.dot(B, A)) | _____no_output_____ | MIT | notebooks/Chapter03/math_numpy.ipynb | tagomaru/ai_security |
アダマール積 A $\circ$ A | print(A * A) | [[ 1 4]
[ 9 16]
[25 36]]
| MIT | notebooks/Chapter03/math_numpy.ipynb | tagomaru/ai_security |
行列 X と行ベクトル a の積 | X = np.array([[0, 1, 2, 3, 4],
[5, 6, 7, 8, 9]])
a = np.array([[1, 2, 3, 4, 5]])
print('X.shape:', X.shape)
print('a.shape:', a.shape)
print(np.dot(X, a)) | X.shape: (2, 5)
a.shape: (1, 5)
| MIT | notebooks/Chapter03/math_numpy.ipynb | tagomaru/ai_security |
行列 X と列ベクトル a の積 | X = np.array([[0, 1, 2, 3, 4],
[5, 6, 7, 8, 9]])
a = np.array([[1],
[2],
[3],
[4],
[5]])
print('X.shape:', X.shape)
print('a.shape:', a.shape)
Xa = np.dot(X, a)
print('Xa.shape:', Xa.shape)
print('Xa:\n', Xa) | X.shape: (2, 5)
a.shape: (5, 1)
Xa.shape: (2, 1)
Xa:
[[ 40]
[115]]
| MIT | notebooks/Chapter03/math_numpy.ipynb | tagomaru/ai_security |
NumPy による行列 X と1次元配列の積 | X = np.array([[0, 1, 2, 3, 4],
[5, 6, 7, 8, 9]])
a = np.array([1, 2, 3, 4, 5]) # 1次元配列で生成
print('X.shape:', X.shape)
print('a.shape:', a.shape)
Xa = np.dot(X, a)
print('Xa.shape:', Xa.shape)
print('Xa:\n', Xa)
import numpy as np
np.array([1, 0.1]) | _____no_output_____ | MIT | notebooks/Chapter03/math_numpy.ipynb | tagomaru/ai_security |
4. ndarray の 軸(axis)について Aの合計を計算 | np.sum(A) | _____no_output_____ | MIT | notebooks/Chapter03/math_numpy.ipynb | tagomaru/ai_security |
axis = 0 で A の合計を計算 | print(np.sum(A, axis=0).shape)
print(np.sum(A, axis=0)) | (2,)
[ 9 12]
| MIT | notebooks/Chapter03/math_numpy.ipynb | tagomaru/ai_security |
axis = 1 で A の合計を計算 | print(np.sum(A, axis=1).shape)
print(np.sum(A, axis=1)) | (3,)
[ 3 7 11]
| MIT | notebooks/Chapter03/math_numpy.ipynb | tagomaru/ai_security |
np.max 関数の利用例 | Y_hat = np.array([[3, 4], [6, 5], [7, 8]]) # 2次元配列を生成
print(np.max(Y_hat)) # axis 指定なし
print(np.max(Y_hat, axis=1)) # axix=1 を指定 | 8
[4 6 8]
| MIT | notebooks/Chapter03/math_numpy.ipynb | tagomaru/ai_security |
argmax 関数の利用例 | print(np.argmax(Y_hat)) # axis 指定なし
print(np.argmax(Y_hat, axis=1)) # axix=1 を指定 | 5
[1 0 1]
| MIT | notebooks/Chapter03/math_numpy.ipynb | tagomaru/ai_security |
5. 3次元以上の配列 行列 A を4つ持つ配列の生成 | A_arr = np.array([A, A, A, A])
print(A_arr.shape) | (4, 3, 2)
| MIT | notebooks/Chapter03/math_numpy.ipynb | tagomaru/ai_security |
A_arr の合計を計算 | np.sum(A_arr) | _____no_output_____ | MIT | notebooks/Chapter03/math_numpy.ipynb | tagomaru/ai_security |
axis = 0 を指定して A_arr の合計を計算 | print(np.sum(A_arr, axis=0).shape)
print(np.sum(A_arr, axis=0)) | (3, 2)
[[ 4 8]
[12 16]
[20 24]]
| MIT | notebooks/Chapter03/math_numpy.ipynb | tagomaru/ai_security |
axis = (1, 2) を指定して A_arr の合計を計算 | print(np.sum(A_arr, axis=(1, 2))) | [21 21 21 21]
| MIT | notebooks/Chapter03/math_numpy.ipynb | tagomaru/ai_security |
Simple Linear Regression using ANN.continuing from ["here"](https://colab.research.google.com/drive/1zTy_7Z5rfKHPKTTCWyou5EemqL8yBqih) | #importing libraries
import numpy as np
import torch
import torch.nn as nn
import matplotlib.pyplot as plt
from IPython import display
display.set_matplotlib_formats('svg')
print('modules imported')
def build_and_train(x, y, learning_rate, n_epochs):
## building
model = nn.Sequential(
nn.Linear(1,1),
nn.ReLU(),
nn.Linear(1,1)
)
## optimizer --> stochastic gradient descent
## loss--> Mean Squared Error
loss_fun = nn.MSELoss()
optimizer = torch.optim.SGD(model.parameters(), lr = learning_rate)
losses = torch.zeros(n_epochs)
for i in range(n_epochs):
y_hat = model(x)
##forward prop has been done
loss = loss_fun(y_hat, y)
losses[i] = loss
##loss has been computed
optimizer.zero_grad()
loss.backward()
optimizer.step()
##back prop has been done
## end for loop
predictions = model(x)
return predictions, losses
## we have to create data with a generic function such that the slope could be varied
def create_data(slope, N, scale=2):
x = torch.randn(N, 1)
y = slope*x + torch.randn(N,1)/scale
return x, y
x, y = create_data(0.75, 40)
yhat, losses = build_and_train(x, y, 0.5, 500)
fig, ax = plt.subplots(1,2, figsize=(10,4))
corr = np.corrcoef(y.T, yhat.detach().T)[0,1]
ax[0].plot(losses.detach(), 'o', markerfacecolor='w', linewidth=.15)
ax[0].set_xlabel('Epoch')
ax[0].set_title('Loss')
ax[1].plot(x, y, 'go', label='Actual Data')
ax[1].plot(x, yhat.detach(), 'rs', label='Predicted Data')
ax[1].set_xlabel('x')
ax[1].set_ylabel('y')
ax[1].set_title(f'Prediction-data-correlation {corr: .2f}')
| _____no_output_____ | MIT | ManipulatingRegressionSlopes.ipynb | ShashwatVv/naiveDL |
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import json
import tensorflow as tf
import csv
import random
import numpy as np
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
from tensorflow.keras.utils import to_categorical
from tensorflow.keras import regularizers
embedding_dim = 100
max_length = 16
trunc_type='post'
padding_type='post'
oov_tok = "<OOV>"
training_size= 160000#Your dataset size here. Experiment using smaller values (i.e. 16000), but don't forget to train on at least 160000 to see the best effects
test_portion=.1
corpus = []
# Note that I cleaned the Stanford dataset to remove LATIN1 encoding to make it easier for Python CSV reader
# You can do that yourself with:
# iconv -f LATIN1 -t UTF8 training.1600000.processed.noemoticon.csv -o training_cleaned.csv
# I then hosted it on my site to make it easier to use in this notebook
!wget --no-check-certificate \
https://storage.googleapis.com/laurencemoroney-blog.appspot.com/training_cleaned.csv \
-O /tmp/training_cleaned.csv
num_sentences = 0
with open("/tmp/training_cleaned.csv") as csvfile:
reader = csv.reader(csvfile, delimiter=',')
for row in reader:
# Your Code here. Create list items where the first item is the text, found in row[5], and the second is the label. Note that the label is a '0' or a '4' in the text. When it's the former, make
# your label to be 0, otherwise 1. Keep a count of the number of sentences in num_sentences
list_item=[]
list_item.append(row[5])
this_label=row[0]
if this_label == '0':
list_item.append(0)
else:
list_item.append(1)
# YOUR CODE HERE
num_sentences = num_sentences + 1
corpus.append(list_item)
print(num_sentences)
print(len(corpus))
print(corpus[1])
# Expected Output:
# 1600000
# 1600000
# ["is upset that he can't update his Facebook by texting it... and might cry as a result School today also. Blah!", 0]
sentences=[]
labels=[]
random.shuffle(corpus)
for x in range(training_size):
sentences.append(corpus[x][0])
labels.append(corpus[x][1])
tokenizer = Tokenizer(oov_token=oov_tok)
tokenizer.fit_on_texts(sentences)# YOUR CODE HERE
word_index = tokenizer.word_index
vocab_size=len(word_index)
sequences = tokenizer.texts_to_sequences(sentences)# YOUR CODE HERE
padded = pad_sequences(sequences,maxlen=max_length, padding=padding_type,truncating=trunc_type)# YOUR CODE HERE
split = int(test_portion * training_size)
print(split)
test_sequences = padded[0:split]
training_sequences = padded[split:training_size]
test_labels = labels[0:split]
training_labels = labels[split:training_size]
print(vocab_size)
print(word_index['i'])
# Expected Output
# 138858
# 1
!wget http://nlp.stanford.edu/data/glove.6B.zip
!unzip /content/glove.6B.zip
# Note this is the 100 dimension version of GloVe from Stanford
# I unzipped and hosted it on my site to make this notebook easier
#### NOTE - Below link is not working. So download and zip on your own
#!wget --no-check-certificate \
# https://storage.googleapis.com/laurencemoroney-blog.appspot.com/glove.6B.100d.txt \
# -O /tmp/glove.6B.100d.txt
embeddings_index = {};
with open('/content/glove.6B.100d.txt') as f:
for line in f:
values = line.split();
word = values[0];
coefs = np.asarray(values[1:], dtype='float32');
embeddings_index[word] = coefs;
embeddings_matrix = np.zeros((vocab_size+1, embedding_dim));
for word, i in word_index.items():
embedding_vector = embeddings_index.get(word);
if embedding_vector is not None:
embeddings_matrix[i] = embedding_vector;
print(len(embeddings_matrix))
# Expected Output
# 138859
training_padded = np.asarray(training_sequences)
training_labels_np = np.asarray(training_labels)
testing_padded = np.asarray(test_sequences)
testing_labels_np = np.asarray(test_labels)
print(training_labels)
model = tf.keras.Sequential([
tf.keras.layers.Embedding(vocab_size+1, embedding_dim, input_length=max_length, weights=[embeddings_matrix], trainable=False),
# YOUR CODE HERE - experiment with combining different types, such as convolutions and LSTMs
tf.keras.layers.Dropout(0.2),
tf.keras.layers.Conv1D(64, 5, activation='relu'),
tf.keras.layers.MaxPooling1D(pool_size=4),
#tf.keras.layers.LSTM(64),
tf.keras.layers.Bidirectional(tf.keras.layers.GRU(32)),
tf.keras.layers.Dense(1, activation='sigmoid')
])
model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])# YOUR CODE HERE
model.summary()
num_epochs = 50
history = model.fit(training_padded, training_labels_np, epochs=num_epochs, validation_data=(testing_padded, testing_labels_np), verbose=2)
print("Training Complete")
import matplotlib.image as mpimg
import matplotlib.pyplot as plt
#-----------------------------------------------------------
# Retrieve a list of list results on training and test data
# sets for each training epoch
#-----------------------------------------------------------
acc=history.history['accuracy']
val_acc=history.history['val_accuracy']
loss=history.history['loss']
val_loss=history.history['val_loss']
epochs=range(len(acc)) # Get number of epochs
#------------------------------------------------
# Plot training and validation accuracy per epoch
#------------------------------------------------
plt.plot(epochs, acc, 'r')
plt.plot(epochs, val_acc, 'b')
plt.title('Training and validation accuracy')
plt.xlabel("Epochs")
plt.ylabel("Accuracy")
plt.legend(["Accuracy", "Validation Accuracy"])
plt.figure()
#------------------------------------------------
# Plot training and validation loss per epoch
#------------------------------------------------
plt.plot(epochs, loss, 'r')
plt.plot(epochs, val_loss, 'b')
plt.title('Training and validation loss')
plt.xlabel("Epochs")
plt.ylabel("Loss")
plt.legend(["Loss", "Validation Loss"])
plt.figure()
# Expected Output
# A chart where the validation loss does not increase sharply!
| _____no_output_____ | Apache-2.0 | Natural Language Processing in TensorFlow/Week 3 Sequence models/NLP_Course_Week_3_Exercise_Question Exploring overfitting in NLP - Glove Embedding.ipynb | mohameddhameem/TensorflowCertification |
|
01.2 Scattering Compute Speed**NOT COMPLETED**In this notebook, the speed to extract scattering coefficients is computed. | import sys
import random
import os
sys.path.append('../src')
import warnings
warnings.filterwarnings("ignore")
import torch
from tqdm import tqdm
from kymatio.torch import Scattering2D
import time
import kymatio.scattering2d.backend as backend
###############################################################################
# Finally, we import the `Scattering2D` class that computes the scattering
# transform.
from kymatio import Scattering2D
| _____no_output_____ | RSA-MD | notebooks/01.2_scattering_compute_speed.ipynb | sgaut023/Chronic-Liver-Classification |
3. Scattering Speed Test | # From: https://github.com/kymatio/kymatio/blob/0.1.X/examples/2d/compute_speed.py
# Benchmark setup
# --------------------
J = 3
L = 8
times = 10
devices = ['cpu', 'gpu']
scattering = Scattering2D(J, shape=(M, N), L=L, backend='torch_skcuda')
data = np.concatenate(dataset['img'],axis=0)
data = torch.from_numpy(data)
x = data[0:batch_size]
%%time
#mlflow.set_experiment('compute_speed_scattering')
for device in devices:
#with mlflow.start_run():
fmt_str = '==> Testing Float32 with {} backend, on {}, forward'
print(fmt_str.format('torch', device.upper()))
if device == 'gpu':
scattering.cuda()
x = x.cuda()
else:
scattering.cpu()
x = x.cpu()
scattering.forward(x)
if device == 'gpu':
torch.cuda.synchronize()
t_start = time.time()
for _ in range(times):
scattering.forward(x)
if device == 'gpu':
torch.cuda.synchronize()
t_elapsed = time.time() - t_start
fmt_str = 'Elapsed time: {:2f} [s / {:d} evals], avg: {:.2f} (s/batch)'
print(fmt_str.format(t_elapsed, times, t_elapsed/times))
# mlflow.log_param('M',M)
# mlflow.log_param('N',N)
# mlflow.log_param('Backend', device.upper())
# mlflow.log_param('J', J)
# mlflow.log_param('L', L)
# mlflow.log_param('Batch Size', batch_size)
# mlflow.log_param('Times', times)
# mlflow.log_metric('Elapsed Time', t_elapsed)
# mlflow.log_metric('Average Time', times)
###############################################################################
# The resulting output should be something like
#
# .. code-block:: text
#
# ==> Testing Float32 with torch backend, on CPU, forward
# Elapsed time: 624.910853 [s / 10 evals], avg: 62.49 (s/batch)
# ==> Testing Float32 with torch backend, on GPU, forward
| ==> Testing Float32 with torch backend, on CPU, forward
Elapsed time: 523.081820 [s / 10 evals], avg: 52.31 (s/batch)
==> Testing Float32 with torch backend, on GPU, forward
Elapsed time: 16.777041 [s / 10 evals], avg: 1.68 (s/batch)
CPU times: user 53min 2s, sys: 4min 47s, total: 57min 50s
Wall time: 9min 54s
| RSA-MD | notebooks/01.2_scattering_compute_speed.ipynb | sgaut023/Chronic-Liver-Classification |
*This notebook contains material from [nbpages](https://jckantor.github.io/nbpages) by Jeffrey Kantor (jeff at nd.edu). The text is released under the[CC-BY-NC-ND-4.0 license](https://creativecommons.org/licenses/by-nc-nd/4.0/legalcode).The code is released under the [MIT license](https://opensource.org/licenses/MIT).* | # IMPORT DATA FILES USED BY THIS NOTEBOOK
import os, requests
file_links = [("data/Stock_Data.csv", "https://jckantor.github.io/nbpages/data/Stock_Data.csv")]
# This cell has been added by nbpages. Run this cell to download data files required for this notebook.
for filepath, fileurl in file_links:
stem, filename = os.path.split(filepath)
if stem:
if not os.path.exists(stem):
os.mkdir(stem)
if not os.path.isfile(filepath):
with open(filepath, 'wb') as f:
response = requests.get(fileurl)
f.write(response.content)
| _____no_output_____ | MIT | docs/02.04-Working-with-Data-and-Figures.ipynb | jckantor/nbcollection |
2.4 Working with Data and Figures 2.4.1 Importing dataThe following cell reads the data file `Stock_Data.csv` from the `data` subdirectory. The name of this file will appear in the data index. | import pandas as pd
df = pd.read_csv("data/Stock_Data.csv")
df.head() | _____no_output_____ | MIT | docs/02.04-Working-with-Data-and-Figures.ipynb | jckantor/nbcollection |
2.4.2 Creating and saving figuresThe following cell creates a figure `Stock_Data.png` in the `figures` subdirectory. The name of this file will appear in the figures index. | %matplotlib inline
import os
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style("darkgrid")
fig, ax = plt.subplots(2, 1, figsize=(8, 5))
(df/df.iloc[0]).drop('VIX', axis=1).plot(ax=ax[0])
df['VIX'].plot(ax=ax[1])
ax[0].set_title('Normalized Indices')
ax[1].set_title('Volatility VIX')
ax[1].set_xlabel('Days')
fig.tight_layout()
if not os.path.exists("figures"):
os.mkdir("figures")
plt.savefig("figures/Stock_Data.png") | _____no_output_____ | MIT | docs/02.04-Working-with-Data-and-Figures.ipynb | jckantor/nbcollection |
Homework 4These problem sets focus on list comprehensions, string operations and regular expressions. Problem set 1: List slices and list comprehensionsLet's start with some data. The following cell contains a string with comma-separated integers, assigned to a variable called `numbers_str`: | numbers_str = '496,258,332,550,506,699,7,985,171,581,436,804,736,528,65,855,68,279,721,120' | _____no_output_____ | MIT | homeworkdata/Homework_4_Paolo_Rivas_Legua.ipynb | paolorivas/homeworkfoundations |
In the following cell, complete the code with an expression that evaluates to a list of integers derived from the raw numbers in `numbers_str`, assigning the value of this expression to a variable `numbers`. If you do everything correctly, executing the cell should produce the output `985` (*not* `'985'`). | values = numbers_str.split(",")
numbers = [int(i) for i in values]
# numbers
max(numbers) | _____no_output_____ | MIT | homeworkdata/Homework_4_Paolo_Rivas_Legua.ipynb | paolorivas/homeworkfoundations |
Great! We'll be using the `numbers` list you created above in the next few problems.In the cell below, fill in the square brackets so that the expression evaluates to a list of the ten largest values in `numbers`. Expected output: [506, 528, 550, 581, 699, 721, 736, 804, 855, 985] (Hint: use a slice.) | #test
print(sorted(numbers))
sorted(numbers)[10:] | _____no_output_____ | MIT | homeworkdata/Homework_4_Paolo_Rivas_Legua.ipynb | paolorivas/homeworkfoundations |
In the cell below, write an expression that evaluates to a list of the integers from `numbers` that are evenly divisible by three, *sorted in numerical order*. Expected output: [120, 171, 258, 279, 528, 699, 804, 855] | [i for i in sorted(numbers) if i%3 == 0] | _____no_output_____ | MIT | homeworkdata/Homework_4_Paolo_Rivas_Legua.ipynb | paolorivas/homeworkfoundations |
Okay. You're doing great. Now, in the cell below, write an expression that evaluates to a list of the square roots of all the integers in `numbers` that are less than 100. In order to do this, you'll need to use the `sqrt` function from the `math` module, which I've already imported for you. Expected output: [2.6457513110645907, 8.06225774829855, 8.246211251235321] (These outputs might vary slightly depending on your platform.) | import math
from math import sqrt
[math.sqrt(i) for i in sorted(numbers) if i < 100] | _____no_output_____ | MIT | homeworkdata/Homework_4_Paolo_Rivas_Legua.ipynb | paolorivas/homeworkfoundations |
Problem set 2: Still more list comprehensionsStill looking good. Let's do a few more with some different data. In the cell below, I've defined a data structure and assigned it to a variable `planets`. It's a list of dictionaries, with each dictionary describing the characteristics of a planet in the solar system. Make sure to run the cell before you proceed. | planets = [
{'diameter': 0.382,
'mass': 0.06,
'moons': 0,
'name': 'Mercury',
'orbital_period': 0.24,
'rings': 'no',
'type': 'terrestrial'},
{'diameter': 0.949,
'mass': 0.82,
'moons': 0,
'name': 'Venus',
'orbital_period': 0.62,
'rings': 'no',
'type': 'terrestrial'},
{'diameter': 1.00,
'mass': 1.00,
'moons': 1,
'name': 'Earth',
'orbital_period': 1.00,
'rings': 'no',
'type': 'terrestrial'},
{'diameter': 0.532,
'mass': 0.11,
'moons': 2,
'name': 'Mars',
'orbital_period': 1.88,
'rings': 'no',
'type': 'terrestrial'},
{'diameter': 11.209,
'mass': 317.8,
'moons': 67,
'name': 'Jupiter',
'orbital_period': 11.86,
'rings': 'yes',
'type': 'gas giant'},
{'diameter': 9.449,
'mass': 95.2,
'moons': 62,
'name': 'Saturn',
'orbital_period': 29.46,
'rings': 'yes',
'type': 'gas giant'},
{'diameter': 4.007,
'mass': 14.6,
'moons': 27,
'name': 'Uranus',
'orbital_period': 84.01,
'rings': 'yes',
'type': 'ice giant'},
{'diameter': 3.883,
'mass': 17.2,
'moons': 14,
'name': 'Neptune',
'orbital_period': 164.8,
'rings': 'yes',
'type': 'ice giant'}] | _____no_output_____ | MIT | homeworkdata/Homework_4_Paolo_Rivas_Legua.ipynb | paolorivas/homeworkfoundations |
Now, in the cell below, write a list comprehension that evaluates to a list of names of the planets that have a radius greater than four earth radii. Expected output: ['Jupiter', 'Saturn', 'Uranus'] | earth_diameter = planets[2]['diameter']
#earth radius is = half diameter. In a multiplication equation the diameter value can be use as a parameter.
[i['name'] for i in planets if i['diameter'] >= earth_diameter*4] | _____no_output_____ | MIT | homeworkdata/Homework_4_Paolo_Rivas_Legua.ipynb | paolorivas/homeworkfoundations |
In the cell below, write a single expression that evaluates to the sum of the mass of all planets in the solar system. Expected output: `446.79` | mass_list = []
for planet in planets:
outcome = planet['mass']
mass_list.append(outcome)
total = sum(mass_list)
total | _____no_output_____ | MIT | homeworkdata/Homework_4_Paolo_Rivas_Legua.ipynb | paolorivas/homeworkfoundations |
Good work. Last one with the planets. Write an expression that evaluates to the names of the planets that have the word `giant` anywhere in the value for their `type` key. Expected output: ['Jupiter', 'Saturn', 'Uranus', 'Neptune'] | [i['name'] for i in planets if 'giant' in i['type']] | _____no_output_____ | MIT | homeworkdata/Homework_4_Paolo_Rivas_Legua.ipynb | paolorivas/homeworkfoundations |
*EXTREME BONUS ROUND*: Write an expression below that evaluates to a list of the names of the planets in ascending order by their number of moons. (The easiest way to do this involves using the [`key` parameter of the `sorted` function](https://docs.python.org/3.5/library/functions.htmlsorted), which we haven't yet discussed in class! That's why this is an EXTREME BONUS question.) Expected output: ['Mercury', 'Venus', 'Earth', 'Mars', 'Neptune', 'Uranus', 'Saturn', 'Jupiter'] | #Done in class | _____no_output_____ | MIT | homeworkdata/Homework_4_Paolo_Rivas_Legua.ipynb | paolorivas/homeworkfoundations |
Problem set 3: Regular expressions In the following section, we're going to do a bit of digital humanities. (I guess this could also be journalism if you were... writing an investigative piece about... early 20th century American poetry?) We'll be working with the following text, Robert Frost's *The Road Not Taken*. Make sure to run the following cell before you proceed. | import re
poem_lines = ['Two roads diverged in a yellow wood,',
'And sorry I could not travel both',
'And be one traveler, long I stood',
'And looked down one as far as I could',
'To where it bent in the undergrowth;',
'',
'Then took the other, as just as fair,',
'And having perhaps the better claim,',
'Because it was grassy and wanted wear;',
'Though as for that the passing there',
'Had worn them really about the same,',
'',
'And both that morning equally lay',
'In leaves no step had trodden black.',
'Oh, I kept the first for another day!',
'Yet knowing how way leads on to way,',
'I doubted if I should ever come back.',
'',
'I shall be telling this with a sigh',
'Somewhere ages and ages hence:',
'Two roads diverged in a wood, and I---',
'I took the one less travelled by,',
'And that has made all the difference.'] | _____no_output_____ | MIT | homeworkdata/Homework_4_Paolo_Rivas_Legua.ipynb | paolorivas/homeworkfoundations |
In the cell above, I defined a variable `poem_lines` which has a list of lines in the poem, and `import`ed the `re` library.In the cell below, write a list comprehension (using `re.search()`) that evaluates to a list of lines that contain two words next to each other (separated by a space) that have exactly four characters. (Hint: use the `\b` anchor. Don't overthink the "two words in a row" requirement.)Expected result:```['Then took the other, as just as fair,', 'Had worn them really about the same,', 'And both that morning equally lay', 'I doubted if I should ever come back.', 'I shall be telling this with a sigh']``` | [line for line in poem_lines if re.search(r"\b\w{4}\b\s\b\w{4}\b", line)] | _____no_output_____ | MIT | homeworkdata/Homework_4_Paolo_Rivas_Legua.ipynb | paolorivas/homeworkfoundations |
Good! Now, in the following cell, write a list comprehension that evaluates to a list of lines in the poem that end with a five-letter word, regardless of whether or not there is punctuation following the word at the end of the line. (Hint: Try using the `?` quantifier. Is there an existing character class, or a way to *write* a character class, that matches non-alphanumeric characters?) Expected output:```['And be one traveler, long I stood', 'And looked down one as far as I could', 'And having perhaps the better claim,', 'Though as for that the passing there', 'In leaves no step had trodden black.', 'Somewhere ages and ages hence:']``` | [line for line in poem_lines if re.search(r"(?:\s\w{5}\b$|\s\w{5}\b[.:;,]$)", line)] | _____no_output_____ | MIT | homeworkdata/Homework_4_Paolo_Rivas_Legua.ipynb | paolorivas/homeworkfoundations |
Okay, now a slightly trickier one. In the cell below, I've created a string `all_lines` which evaluates to the entire text of the poem in one string. Execute this cell. | all_lines = " ".join(poem_lines) | _____no_output_____ | MIT | homeworkdata/Homework_4_Paolo_Rivas_Legua.ipynb | paolorivas/homeworkfoundations |
Now, write an expression that evaluates to all of the words in the poem that follow the word 'I'. (The strings in the resulting list should *not* include the `I`.) Hint: Use `re.findall()` and grouping! Expected output: ['could', 'stood', 'could', 'kept', 'doubted', 'should', 'shall', 'took'] | [item[2:] for item in (re.findall(r"\bI\b\s\b[a-z]{1,}", all_lines))] | _____no_output_____ | MIT | homeworkdata/Homework_4_Paolo_Rivas_Legua.ipynb | paolorivas/homeworkfoundations |
Finally, something super tricky. Here's a list of strings that contains a restaurant menu. Your job is to wrangle this plain text, slightly-structured data into a list of dictionaries. | entrees = [
"Yam, Rosemary and Chicken Bowl with Hot Sauce $10.95",
"Lavender and Pepperoni Sandwich $8.49",
"Water Chestnuts and Peas Power Lunch (with mayonnaise) $12.95 - v",
"Artichoke, Mustard Green and Arugula with Sesame Oil over noodles $9.95 - v",
"Flank Steak with Lentils And Tabasco Pepper With Sweet Chilli Sauce $19.95",
"Rutabaga And Cucumber Wrap $8.49 - v"
] | _____no_output_____ | MIT | homeworkdata/Homework_4_Paolo_Rivas_Legua.ipynb | paolorivas/homeworkfoundations |
You'll need to pull out the name of the dish and the price of the dish. The `v` after the hyphen indicates that the dish is vegetarian---you'll need to include that information in your dictionary as well. I've included the basic framework; you just need to fill in the contents of the `for` loop.Expected output:```[{'name': 'Yam, Rosemary and Chicken Bowl with Hot Sauce ', 'price': 10.95, 'vegetarian': False}, {'name': 'Lavender and Pepperoni Sandwich ', 'price': 8.49, 'vegetarian': False}, {'name': 'Water Chestnuts and Peas Power Lunch (with mayonnaise) ', 'price': 12.95, 'vegetarian': True}, {'name': 'Artichoke, Mustard Green and Arugula with Sesame Oil over noodles ', 'price': 9.95, 'vegetarian': True}, {'name': 'Flank Steak with Lentils And Tabasco Pepper With Sweet Chilli Sauce ', 'price': 19.95, 'vegetarian': False}, {'name': 'Rutabaga And Cucumber Wrap ', 'price': 8.49, 'vegetarian': True}]``` | menu = []
for dish in entrees:
match = re.search(r"^(.*) \$(.*)", dish)
vegetarian = re.search(r"v$", match.group(2))
price = re.search(r"(?:\d\.\d\d|\d\d\.\d\d)", dish)
if vegetarian == None:
vegetarian = False
else:
vegetarian = True
if match:
dish = {
'name': match.group(1), 'price': price.group(), 'vegetarian': vegetarian
}
menu.append(dish)
menu | _____no_output_____ | MIT | homeworkdata/Homework_4_Paolo_Rivas_Legua.ipynb | paolorivas/homeworkfoundations |
Used https://github.com/GoogleCloudPlatform/cloudml-samples/blob/master/xgboost/notebooks/census_training/train.py as a starting point and adjusted to CatBoost | #Google Cloud Libraries
from google.cloud import storage
#System Libraries
import datetime
import subprocess
#Data Libraries
import pandas as pd
import numpy as np
#ML Libraries
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import train_test_split
import xgboost as xgb
from catboost import CatBoostClassifier, Pool, cv
from catboost import CatBoost, Pool
from catboost.utils import get_gpu_device_count
print('I see %i GPU devices' % get_gpu_device_count())
# Fill in your Cloud Storage bucket name
BUCKET_ID = "mchrestkha-demo-env-ml-examples"
census_data_filename = 'adult.data.csv'
# Public bucket holding the census data
bucket = storage.Client().bucket('cloud-samples-data')
# Path to the data inside the public bucket
data_dir = 'ai-platform/census/data/'
# Download the data
blob = bucket.blob(''.join([data_dir, census_data_filename]))
blob.download_to_filename(census_data_filename)
# these are the column labels from the census data files
COLUMNS = (
'age',
'workclass',
'fnlwgt',
'education',
'education-num',
'marital-status',
'occupation',
'relationship',
'race',
'sex',
'capital-gain',
'capital-loss',
'hours-per-week',
'native-country',
'income-level'
)
# categorical columns contain data that need to be turned into numerical values before being used by XGBoost
CATEGORICAL_COLUMNS = (
'workclass',
'education',
'marital-status',
'occupation',
'relationship',
'race',
'sex',
'native-country'
)
# Load the training census dataset
with open(census_data_filename, 'r') as train_data:
raw_training_data = pd.read_csv(train_data, header=None, names=COLUMNS)
# remove column we are trying to predict ('income-level') from features list
X = raw_training_data.drop('income-level', axis=1)
# create training labels list
#train_labels = (raw_training_data['income-level'] == ' >50K')
y = raw_training_data['income-level']
# Since the census data set has categorical features, we need to convert
# them to numerical values.
# convert data in categorical columns to numerical values
X_enc=X
encoders = {col:LabelEncoder() for col in CATEGORICAL_COLUMNS}
for col in CATEGORICAL_COLUMNS:
X_enc[col] = encoders[col].fit_transform(X[col])
y_enc=LabelEncoder().fit_transform(y)
X_train, X_validation, y_train, y_validation = train_test_split(X_enc, y_enc, train_size=0.75, random_state=42)
print(type(y))
print(type(y_enc))
%%time
#model = CatBoost({'iterations':50})
model=CatBoostClassifier(
od_type='Iter'
#iterations=5000,
#custom_loss=['Accuracy']
)
model.fit(
X_train,y_train,eval_set=(X_validation, y_validation),
verbose=50)
# # load data into DMatrix object
# dtrain = xgb.DMatrix(train_features, train_labels)
# # train model
# bst = xgb.train({}, dtrain, 20)
# Export the model to a file
fname = 'catboost_census_model.onnx'
model.save_model(fname, format='onnx')
# Upload the model to GCS
bucket = storage.Client().bucket(BUCKET_ID)
blob = bucket.blob('{}/{}'.format(
datetime.datetime.now().strftime('census/catboost_model_dir/catboost_census_%Y%m%d_%H%M%S'),
fname))
blob.upload_from_filename(fname)
!gsutil ls gs://$BUCKET_ID/census/* | gs://mchrestkha-demo-env-ml-examples/census/catboost_census_20200525_212707/:
gs://mchrestkha-demo-env-ml-examples/census/catboost_census_20200525_212707/<catboost.core.CatBoostClassifier object at 0x7fdb929aa6d0>
gs://mchrestkha-demo-env-ml-examples/census/catboost_census_20200525_212852/:
gs://mchrestkha-demo-env-ml-examples/census/catboost_census_20200525_212852/<catboost.core.CatBoostClassifier object at 0x7fdb929aa6d0>
gs://mchrestkha-demo-env-ml-examples/census/catboost_census_20200525_213004/:
gs://mchrestkha-demo-env-ml-examples/census/catboost_census_20200525_213004/<catboost.core.CatBoostClassifier object at 0x7fdb929aa6d0>
gs://mchrestkha-demo-env-ml-examples/census/xgboost_census_20200525_020526/:
gs://mchrestkha-demo-env-ml-examples/census/xgboost_census_20200525_020526/model.bst
gs://mchrestkha-demo-env-ml-examples/census/xgboost_census_20200525_021023/:
gs://mchrestkha-demo-env-ml-examples/census/xgboost_census_20200525_021023/model.bst
gs://mchrestkha-demo-env-ml-examples/census/xgboost_census_20200525_023122/:
gs://mchrestkha-demo-env-ml-examples/census/xgboost_census_20200525_023122/model.bst
gs://mchrestkha-demo-env-ml-examples/census/xgboost_job_dir/:
gs://mchrestkha-demo-env-ml-examples/census/xgboost_job_dir/packages/
| MIT | census/catboost/gcp_ai_platform/notebooks/catboost_census_notebook.ipynb | jared-burns/machine_learning_examples |
Final models with hyperparameters tuned for Logistics Regression and XGBoost with selected features. | #Import the libraries
import pandas as pd
import numpy as np
from tqdm import tqdm
from sklearn import linear_model, metrics, preprocessing, model_selection
from sklearn.preprocessing import StandardScaler
import xgboost as xgb
#Load the data
modeling_dataset = pd.read_csv('/content/drive/MyDrive/prediction/frac_cleaned_fod_data.csv', low_memory = False)
#All columns - except 'HasDetections', 'kfold', and 'MachineIdentifier'
train_features = [tf for tf in modeling_dataset.columns if tf not in ('HasDetections', 'kfold', 'MachineIdentifier')]
#The features selected based on the feature selection method earlier employed
train_features_after_selection = ['AVProductStatesIdentifier', 'Processor','AvSigVersion', 'Census_TotalPhysicalRAM', 'Census_InternalPrimaryDiagonalDisplaySizeInInches',
'Census_IsVirtualDevice', 'Census_PrimaryDiskTotalCapacity', 'Wdft_IsGamer', 'Census_IsAlwaysOnAlwaysConnectedCapable', 'EngineVersion',
'Census_ProcessorCoreCount', 'Census_OSEdition', 'Census_OSInstallTypeName', 'Census_OSSkuName', 'AppVersion', 'OsBuildLab', 'OsSuite',
'Firewall', 'IsProtected', 'Census_IsTouchEnabled', 'Census_ActivationChannel', 'LocaleEnglishNameIdentifier','Census_SystemVolumeTotalCapacity',
'Census_InternalPrimaryDisplayResolutionHorizontal','Census_HasOpticalDiskDrive', 'OsBuild', 'Census_InternalPrimaryDisplayResolutionVertical',
'CountryIdentifier', 'Census_MDC2FormFactor', 'GeoNameIdentifier', 'Census_PowerPlatformRoleName', 'Census_OSWUAutoUpdateOptionsName', 'SkuEdition',
'Census_OSVersion', 'Census_GenuineStateName', 'Census_OSBuildRevision', 'Platform', 'Census_ChassisTypeName', 'Census_FlightRing',
'Census_PrimaryDiskTypeName', 'Census_OSBranch', 'Census_IsSecureBootEnabled', 'OsPlatformSubRelease']
#Define the categorical features of the data
categorical_features = ['ProductName',
'EngineVersion',
'AppVersion',
'AvSigVersion',
'Platform',
'Processor',
'OsVer',
'OsPlatformSubRelease',
'OsBuildLab',
'SkuEdition',
'Census_MDC2FormFactor',
'Census_DeviceFamily',
'Census_PrimaryDiskTypeName',
'Census_ChassisTypeName',
'Census_PowerPlatformRoleName',
'Census_OSVersion',
'Census_OSArchitecture',
'Census_OSBranch',
'Census_OSEdition',
'Census_OSSkuName',
'Census_OSInstallTypeName',
'Census_OSWUAutoUpdateOptionsName',
'Census_GenuineStateName',
'Census_ActivationChannel',
'Census_FlightRing']
#XGBoost
"""
Best parameters set:
alpha: 1.0
colsample_bytree: 0.6
eta: 0.05
gamma: 0.1
lamda: 1.0
max_depth: 9
min_child_weight: 5
subsample: 0.7
"""
#XGBoost
def opt_run_xgboost(fold):
for col in train_features:
if col in categorical_features:
#Initialize the Label Encoder
lbl = preprocessing.LabelEncoder()
#Fit on the categorical features
lbl.fit(modeling_dataset[col])
#Transform
modeling_dataset.loc[:,col] = lbl.transform(modeling_dataset[col])
#Get training and validation data using folds
modeling_datasets_train = modeling_dataset[modeling_dataset.kfold != fold].reset_index(drop=True)
modeling_datasets_valid = modeling_dataset[modeling_dataset.kfold == fold].reset_index(drop=True)
#Get train data
X_train = modeling_datasets_train[train_features_after_selection].values
#Get validation data
X_valid = modeling_datasets_valid[train_features_after_selection].values
#Initialize XGboost model
xgb_model = xgb.XGBClassifier(
alpha= 1.0,
colsample_bytree= 0.6,
eta= 0.05,
gamma= 0.1,
lamda= 1.0,
max_depth= 9,
min_child_weight= 5,
subsample= 0.7,
n_jobs=-1)
#Fit the model on training data
xgb_model.fit(X_train, modeling_datasets_train.HasDetections.values)
#Predict on validation
valid_preds = xgb_model.predict_proba(X_valid)[:,1]
valid_preds_pc = xgb_model.predict(X_valid)
#Get the ROC AUC score
auc = metrics.roc_auc_score(modeling_datasets_valid.HasDetections.values, valid_preds)
#Get the precision score
pre = metrics.precision_score(modeling_datasets_valid.HasDetections.values, valid_preds_pc, average='binary')
#Get the Recall score
rc = metrics.recall_score(modeling_datasets_valid.HasDetections.values, valid_preds_pc, average='binary')
return auc, pre, rc
#LR
"""
'penalty': 'l2',
'C': 49.71967742639108,
'solver': 'lbfgs'
max_iter: 300
"""
#Function for Logistic Regression Classification
def opt_run_lr(fold):
#Get training and validation data using folds
cleaned_fold_datasets_train = modeling_dataset[modeling_dataset.kfold != fold].reset_index(drop=True)
cleaned_fold_datasets_valid = modeling_dataset[modeling_dataset.kfold == fold].reset_index(drop=True)
#Initialize OneHotEncoder from scikit-learn, and fit it on training and validation features
ohe = preprocessing.OneHotEncoder()
full_data = pd.concat(
[cleaned_fold_datasets_train[train_features_after_selection],cleaned_fold_datasets_valid[train_features_after_selection]],
axis = 0
)
ohe.fit(full_data[train_features_after_selection])
#transform the training and validation data
x_train = ohe.transform(cleaned_fold_datasets_train[train_features_after_selection])
x_valid = ohe.transform(cleaned_fold_datasets_valid[train_features_after_selection])
#Initialize the Logistic Regression Model
lr_model = linear_model.LogisticRegression(
penalty= 'l2',
C = 49.71967742639108,
solver= 'lbfgs',
max_iter= 300,
n_jobs=-1
)
#Fit model on training data
lr_model.fit(x_train, cleaned_fold_datasets_train.HasDetections.values)
#Predict on the validation data using the probability for the AUC
valid_preds = lr_model.predict_proba(x_valid)[:, 1]
#For precision and Recall
valid_preds_pc = lr_model.predict(x_valid)
#Get the ROC AUC score
auc = metrics.roc_auc_score(cleaned_fold_datasets_valid.HasDetections.values, valid_preds)
#Get the precision score
pre = metrics.precision_score(cleaned_fold_datasets_valid.HasDetections.values, valid_preds_pc, average='binary')
#Get the Recall score
rc = metrics.recall_score(cleaned_fold_datasets_valid.HasDetections.values, valid_preds_pc, average='binary')
return auc, pre, rc
#A list to hold the values of the XGB performance metrics
xg = []
for fold in tqdm(range(10)):
xg.append(opt_run_xgboost(fold))
#Run the Logistic regression model for all folds and hold their values
lr = []
for fold in tqdm(range(10)):
lr.append(opt_run_lr(fold))
xgb_auc = []
xgb_pre = []
xgb_rc = []
lr_auc = []
lr_pre = []
lr_rc = []
#Loop to get each of the performance metric for average computation
for i in lr:
lr_auc.append(i[0])
lr_pre.append(i[1])
lr_rc.append(i[2])
for j in xg:
xgb_auc.append(i[0])
xgb_pre.append(i[1])
xgb_rc.append(i[2])
#Dictionary to hold the basic model performance data
final_model_performance = {"logistic_regression": {"auc":"", "precision":"", "recall":""},
"xgb": {"auc":"","precision":"","recall":""}
}
#Calculate average of each of the lists of performance metrics and update the dictionary
final_model_performance['logistic_regression'].update({'auc':sum(lr_auc)/len(lr_auc)})
final_model_performance['xgb'].update({'auc':sum(xgb_auc)/len(xgb_auc)})
final_model_performance['logistic_regression'].update({'precision':sum(lr_pre)/len(lr_pre)})
final_model_performance['xgb'].update({'precision':sum(xgb_pre)/len(xgb_pre)})
final_model_performance['logistic_regression'].update({'recall':sum(lr_rc)/len(lr_rc)})
final_model_performance['xgb'].update({'recall':sum(xgb_rc)/len(xgb_rc)})
final_model_performance
#LR
"""
'penalty': 'l2',
'C': 49.71967742639108,
'solver': 'lbfgs'
max_iter: 100
"""
#Function for Logistic Regression Classification - max_iter = 100
def opt_run_lr100(fold):
#Get training and validation data using folds
cleaned_fold_datasets_train = modeling_dataset[modeling_dataset.kfold != fold].reset_index(drop=True)
cleaned_fold_datasets_valid = modeling_dataset[modeling_dataset.kfold == fold].reset_index(drop=True)
#Initialize OneHotEncoder from scikit-learn, and fit it on training and validation features
ohe = preprocessing.OneHotEncoder()
full_data = pd.concat(
[cleaned_fold_datasets_train[train_features_after_selection],cleaned_fold_datasets_valid[train_features_after_selection]],
axis = 0
)
ohe.fit(full_data[train_features_after_selection])
#transform the training and validation data
x_train = ohe.transform(cleaned_fold_datasets_train[train_features_after_selection])
x_valid = ohe.transform(cleaned_fold_datasets_valid[train_features_after_selection])
#Initialize the Logistic Regression Model
lr_model = linear_model.LogisticRegression(
penalty= 'l2',
C = 49.71967742639108,
solver= 'lbfgs',
max_iter= 100,
n_jobs=-1
)
#Fit model on training data
lr_model.fit(x_train, cleaned_fold_datasets_train.HasDetections.values)
#Predict on the validation data using the probability for the AUC
valid_preds = lr_model.predict_proba(x_valid)[:, 1]
#For precision and Recall
valid_preds_pc = lr_model.predict(x_valid)
#Get the ROC AUC score
auc = metrics.roc_auc_score(cleaned_fold_datasets_valid.HasDetections.values, valid_preds)
#Get the precision score
pre = metrics.precision_score(cleaned_fold_datasets_valid.HasDetections.values, valid_preds_pc, average='binary')
#Get the Recall score
rc = metrics.recall_score(cleaned_fold_datasets_valid.HasDetections.values, valid_preds_pc, average='binary')
return auc, pre, rc
#Run the Logistic regression model for all folds and hold their values
lr100 = []
for fold in tqdm(range(10)):
lr100.append(opt_run_lr100(fold))
lr100_auc = []
lr100_pre = []
lr100_rc = []
for k in lr100:
lr100_auc.append(k[0])
lr100_pre.append(k[1])
lr100_rc.append(k[2])
sum(lr100_auc)/len(lr100_auc)
sum(lr100_pre)/len(lr100_pre)
sum(lr100_rc)/len(lr100_rc)
"""
{'logistic_regression': {'auc': 0.660819451656712,
'precision': 0.6069858170181643,
'recall': 0.6646704904969867},
'xgb': {'auc': 0.6583717792973377,
'precision': 0.6042291042291044,
'recall': 0.6542422535211267}}
""" | _____no_output_____ | MIT | MS-malware-suspectibility-detection/6-final-model/FinalModel.ipynb | Semiu/malware-detector |
Dealing with errors after a run In this example, we run the model on a list of three glaciers:two of them will end with errors: one because it already failed atpreprocessing (i.e. prior to this run), and one during the run. We show how to analyze theses erros and solve (some) of them, as described in the OGGM documentation under [troubleshooting](https://docs.oggm.org/en/latest/faq.html?highlight=bordertroubleshooting). Run with `cfg.PARAMS['continue_on_error'] = True` | # Locals
import oggm.cfg as cfg
from oggm import utils, workflow, tasks
# Libs
import os
import xarray as xr
import pandas as pd
# Initialize OGGM and set up the default run parameters
cfg.initialize(logging_level='WARNING')
# Here we override some of the default parameters
# How many grid points around the glacier?
# We make it small because we want the model to error because
# of flowing out of the domain
cfg.PARAMS['border'] = 80
# This is useful since we have three glaciers
cfg.PARAMS['use_multiprocessing'] = True
# This is the important bit!
# We tell OGGM to continue despite of errors
cfg.PARAMS['continue_on_error'] = True
# Local working directory (where OGGM will write its output)
WORKING_DIR = utils.gettempdir('OGGM_Errors')
utils.mkdir(WORKING_DIR, reset=True)
cfg.PATHS['working_dir'] = WORKING_DIR
rgi_ids = ['RGI60-11.00897', 'RGI60-11.01450', 'RGI60-11.03295']
# Go - get the pre-processed glacier directories
gdirs = workflow.init_glacier_directories(rgi_ids, from_prepro_level=4)
# We can step directly to the experiment!
# Random climate representative for the recent climate (1985-2015)
# with a negative bias added to the random temperature series
workflow.execute_entity_task(tasks.run_random_climate, gdirs,
nyears=150, seed=0,
temperature_bias=-1) | _____no_output_____ | BSD-3-Clause | notebooks/deal_with_errors.ipynb | anoukvlug/tutorials |
Error diagnostics | # Write the compiled output
utils.compile_glacier_statistics(gdirs); # saved as glacier_statistics.csv in the WORKING_DIR folder
utils.compile_run_output(gdirs); # saved as run_output.nc in the WORKING_DIR folder
# Read it
with xr.open_dataset(os.path.join(WORKING_DIR, 'run_output.nc')) as ds:
ds = ds.load()
df_stats = pd.read_csv(os.path.join(WORKING_DIR, 'glacier_statistics.csv'), index_col=0)
# all possible statistics about the glaciers
df_stats | _____no_output_____ | BSD-3-Clause | notebooks/deal_with_errors.ipynb | anoukvlug/tutorials |
- in the column *error_task*, we can see whether an error occurred, and if yes during which task- *error_msg* describes the actual error message | df_stats[['error_task', 'error_msg']] | _____no_output_____ | BSD-3-Clause | notebooks/deal_with_errors.ipynb | anoukvlug/tutorials |
We can also check which glacier failed at which task by using [compile_task_log]('https://docs.oggm.org/en/latest/generated/oggm.utils.compile_task_log.htmloggm.utils.compile_task_log'). | # also saved as task_log.csv in the WORKING_DIR folder - "append=False" replaces the existing one
utils.compile_task_log(gdirs, task_names=['glacier_masks', 'compute_centerlines', 'flowline_model_run'], append=False) | _____no_output_____ | BSD-3-Clause | notebooks/deal_with_errors.ipynb | anoukvlug/tutorials |
Error solving RuntimeError: `Glacier exceeds domain boundaries, at year: 98.08333333333333` To remove this error just increase the domain boundary **before** running `init_glacier_directories` ! Attention, this means that more data has to be downloaded and the run takes more time. The available options for `cfg.PARAMS['border']` are **10, 40, 80 or 160** at the moment; the unit is number of grid points outside the glacier boundaries. More about that in the OGGM documentation under [preprocessed files](https://docs.oggm.org/en/latest/input-data.htmlpre-processed-directories). | # reset to recompute statistics
utils.mkdir(WORKING_DIR, reset=True)
# increase the amount of gridpoints outside the glacier
cfg.PARAMS['border'] = 160
gdirs = workflow.init_glacier_directories(rgi_ids, from_prepro_level=4)
workflow.execute_entity_task(tasks.run_random_climate, gdirs,
nyears=150, seed=0,
temperature_bias=-1);
# recompute the output
# we can also get the run output directly from the methods
df_stats = utils.compile_glacier_statistics(gdirs)
ds = utils.compile_run_output(gdirs)
# check again
df_stats[['error_task', 'error_msg']] | _____no_output_____ | BSD-3-Clause | notebooks/deal_with_errors.ipynb | anoukvlug/tutorials |
Now `RGI60-11.00897` runs without errors! Error: `Need a valid model_flowlines file.` This error message in the log is misleading: it does not really describe the source of the error, which happened earlier in the processing chain. Therefore we can look instead into the glacier_statistics via [compile_glacier_statistics](https://docs.oggm.org/en/latest/generated/oggm.utils.compile_glacier_statistics.html) or into the log output via [compile_task_log](https://docs.oggm.org/en/latest/generated/oggm.utils.compile_task_log.htmloggm.utils.compile_task_log): | print('error_task: {}, error_msg: {}'.format(df_stats.loc['RGI60-11.03295']['error_task'],
df_stats.loc['RGI60-11.03295']['error_msg'])) | _____no_output_____ | BSD-3-Clause | notebooks/deal_with_errors.ipynb | anoukvlug/tutorials |
Now we have a better understanding of the error: - OGGM can not work with this geometry of this glacier and could therefore not make a gridded mask of the glacier outlines. - there is no way to prevent this except you find a better way to pre-process the geometry of this glacier- these glaciers have to be ignored! Less than 0.5% of glacier area globally have errors during the geometry processing or failures in computing certain topographical properties by e.g. invalid DEM, see [Sect. 4.2 Invalid Glaciers of the OGGM paper (Maussion et al., 2019)](https://gmd.copernicus.org/articles/12/909/2019/section4) and [this tutorial](preprocessing_errors.ipynb) for more up-to-date numbers Ignoring those glaciers with errors that we can't solve In the run_output, you can for example just use `*.dropna` to remove these. For other applications (e.g. quantitative mass change evaluation), more will be needed (not available yet in the OGGM codebase): | ds.dropna(dim='rgi_id') # here we can e.g. find the volume evolution | _____no_output_____ | BSD-3-Clause | notebooks/deal_with_errors.ipynb | anoukvlug/tutorials |
Goals In the previous tutorial you studied the role of freezing models on a small dataset. Understand the role of freezing models in transfer learning on a fairly large dataset Why freeze/unfreeze base models in transfer learning Use comparison feature to appropriately set this parameter on custom dataset You will be using lego bricks dataset to train the classifiers What is freezing base network - To recap you have two parts in your network - One that already existed, the pretrained one, the base network - The new sub-network or a single layer you added -The hyper-parameter we can see here: Freeze base network - Freezing base network makes the base network untrainable - The base network now acts as a feature extractor and only the next half is trained - If you do not freeze the base network the entire network is trained Table of Contents [Install](0) [Freeze Base network in densenet121 and train a classifier](1) [Unfreeze base network in densenet121 and train another classifier](2) [Compare both the experiment](3) Install Monk Using pip (Recommended) - colab (gpu) - All bakcends: `pip install -U monk-colab` - kaggle (gpu) - All backends: `pip install -U monk-kaggle` - cuda 10.2 - All backends: `pip install -U monk-cuda102` - Gluon bakcned: `pip install -U monk-gluon-cuda102` - Pytorch backend: `pip install -U monk-pytorch-cuda102` - Keras backend: `pip install -U monk-keras-cuda102` - cuda 10.1 - All backend: `pip install -U monk-cuda101` - Gluon bakcned: `pip install -U monk-gluon-cuda101` - Pytorch backend: `pip install -U monk-pytorch-cuda101` - Keras backend: `pip install -U monk-keras-cuda101` - cuda 10.0 - All backend: `pip install -U monk-cuda100` - Gluon bakcned: `pip install -U monk-gluon-cuda100` - Pytorch backend: `pip install -U monk-pytorch-cuda100` - Keras backend: `pip install -U monk-keras-cuda100` - cuda 9.2 - All backend: `pip install -U monk-cuda92` - Gluon bakcned: `pip install -U monk-gluon-cuda92` - Pytorch backend: `pip install -U monk-pytorch-cuda92` - Keras backend: `pip install -U monk-keras-cuda92` - cuda 9.0 - All backend: `pip install -U monk-cuda90` - Gluon bakcned: `pip install -U monk-gluon-cuda90` - Pytorch backend: `pip install -U monk-pytorch-cuda90` - Keras backend: `pip install -U monk-keras-cuda90` - cpu - All backend: `pip install -U monk-cpu` - Gluon bakcned: `pip install -U monk-gluon-cpu` - Pytorch backend: `pip install -U monk-pytorch-cpu` - Keras backend: `pip install -U monk-keras-cpu` Install Monk Manually (Not recommended) Step 1: Clone the library - git clone https://github.com/Tessellate-Imaging/monk_v1.git Step 2: Install requirements - Linux - Cuda 9.0 - `cd monk_v1/installation/Linux && pip install -r requirements_cu90.txt` - Cuda 9.2 - `cd monk_v1/installation/Linux && pip install -r requirements_cu92.txt` - Cuda 10.0 - `cd monk_v1/installation/Linux && pip install -r requirements_cu100.txt` - Cuda 10.1 - `cd monk_v1/installation/Linux && pip install -r requirements_cu101.txt` - Cuda 10.2 - `cd monk_v1/installation/Linux && pip install -r requirements_cu102.txt` - CPU (Non gpu system) - `cd monk_v1/installation/Linux && pip install -r requirements_cpu.txt` - Windows - Cuda 9.0 (Experimental support) - `cd monk_v1/installation/Windows && pip install -r requirements_cu90.txt` - Cuda 9.2 (Experimental support) - `cd monk_v1/installation/Windows && pip install -r requirements_cu92.txt` - Cuda 10.0 (Experimental support) - `cd monk_v1/installation/Windows && pip install -r requirements_cu100.txt` - Cuda 10.1 (Experimental support) - `cd monk_v1/installation/Windows && pip install -r requirements_cu101.txt` - Cuda 10.2 (Experimental support) - `cd monk_v1/installation/Windows && pip install -r requirements_cu102.txt` - CPU (Non gpu system) - `cd monk_v1/installation/Windows && pip install -r requirements_cpu.txt` - Mac - CPU (Non gpu system) - `cd monk_v1/installation/Mac && pip install -r requirements_cpu.txt` - Misc - Colab (GPU) - `cd monk_v1/installation/Misc && pip install -r requirements_colab.txt` - Kaggle (GPU) - `cd monk_v1/installation/Misc && pip install -r requirements_kaggle.txt` Step 3: Add to system path (Required for every terminal or kernel run) - `import sys` - `sys.path.append("monk_v1/");` Dataset - LEGO Classification - https://www.kaggle.com/joosthazelzet/lego-brick-images/ | ! wget --load-cookies /tmp/cookies.txt "https://docs.google.com/uc?export=download&confirm=$(wget --save-cookies /tmp/cookies.txt --keep-session-cookies --no-check-certificate 'https://docs.google.com/uc?export=download&id=1MRC58-oCdR1agFTWreDFqevjEOIWDnYZ' -O- | sed -rn 's/.*confirm=([0-9A-Za-z_]+).*/\1\n/p')&id=1MRC58-oCdR1agFTWreDFqevjEOIWDnYZ" -O skin_cancer_mnist_dataset.zip && rm -rf /tmp/cookies.txt
! unzip -qq skin_cancer_mnist_dataset.zip | _____no_output_____ | Apache-2.0 | study_roadmaps/2_transfer_learning_roadmap/6_freeze_base_network/2.2) Understand the effect of freezing base model in transfer learning - 2 - pytorch.ipynb | take2rohit/monk_v1 |
Imports | #Using pytorch backend
# When installed using pip
from monk.pytorch_prototype import prototype
# When installed manually (Uncomment the following)
#import os
#import sys
#sys.path.append("monk_v1/");
#sys.path.append("monk_v1/monk/");
#from monk.pytorch_prototype import prototype | _____no_output_____ | Apache-2.0 | study_roadmaps/2_transfer_learning_roadmap/6_freeze_base_network/2.2) Understand the effect of freezing base model in transfer learning - 2 - pytorch.ipynb | take2rohit/monk_v1 |
Freeze Base network in densenet121 and train a classifier Creating and managing experiments - Provide project name - Provide experiment name - For a specific data create a single project - Inside each project multiple experiments can be created - Every experiment can be have diferent hyper-parameters attached to it | gtf = prototype(verbose=1);
gtf.Prototype("Project", "Freeze_Base_Network"); | Pytorch Version: 1.2.0
Experiment Details
Project: Project
Experiment: Freeze_Base_Network
Dir: /home/abhi/Desktop/Work/tess_tool/gui/v0.3/finetune_models/Organization/development/v5.0_blocks/study_roadmap/change_post_num_layers/5_transfer_learning_params/2_freezing_base_network/workspace/Project/Freeze_Base_Network/
| Apache-2.0 | study_roadmaps/2_transfer_learning_roadmap/6_freeze_base_network/2.2) Understand the effect of freezing base model in transfer learning - 2 - pytorch.ipynb | take2rohit/monk_v1 |
This creates files and directories as per the following structure workspace | |--------Project | | |-----Freeze_Base_Network | |-----experiment-state.json | |-----output | |------logs (All training logs and graphs saved here) | |------models (all trained models saved here) Set dataset and select the model Quick mode training - Using Default Function - dataset_path - model_name - freeze_base_network - num_epochs Sample Dataset folder structure parent_directory | | |------cats | |------img1.jpg |------img2.jpg |------.... (and so on) |------dogs | |------img1.jpg |------img2.jpg |------.... (and so on) Modifyable params - dataset_path: path to data - model_name: which pretrained model to use - freeze_base_network: Retrain already trained network or not - num_epochs: Number of epochs to train for | gtf.Default(dataset_path="skin_cancer_mnist_dataset/images",
path_to_csv="skin_cancer_mnist_dataset/train_labels.csv",
model_name="densenet121",
freeze_base_network=True, # Set this param as true
num_epochs=5);
#Read the summary generated once you run this cell. | Dataset Details
Train path: skin_cancer_mnist_dataset/images
Val path: None
CSV train path: skin_cancer_mnist_dataset/train_labels.csv
CSV val path: None
Dataset Params
Input Size: 224
Batch Size: 4
Data Shuffle: True
Processors: 4
Train-val split: 0.7
Delimiter: ,
Pre-Composed Train Transforms
[{'RandomHorizontalFlip': {'p': 0.8}}, {'Normalize': {'mean': [0.485, 0.456, 0.406], 'std': [0.229, 0.224, 0.225]}}]
Pre-Composed Val Transforms
[{'RandomHorizontalFlip': {'p': 0.8}}, {'Normalize': {'mean': [0.485, 0.456, 0.406], 'std': [0.229, 0.224, 0.225]}}]
Dataset Numbers
Num train images: 7010
Num val images: 3005
Num classes: 7
Model Params
Model name: densenet121
Use Gpu: True
Use pretrained: True
Freeze base network: True
Model Details
Loading pretrained model
Model Loaded on device
Model name: densenet121
Num layers in model: 242
Num trainable layers: 1
Optimizer
Name: sgd
Learning rate: 0.01
Params: {'lr': 0.01, 'momentum': 0, 'weight_decay': 0.0001, 'momentum_dampening_rate': 0, 'clipnorm': 0.0, 'clipvalue': 0.0}
Learning rate scheduler
Name: multisteplr
Params: {'milestones': [2, 3], 'gamma': 0.1, 'last_epoch': -1}
Loss
Name: softmaxcrossentropy
Params: {'weight': None, 'batch_axis': 0, 'axis_to_sum_over': -1, 'label_as_categories': True, 'label_smoothing': False}
Training params
Num Epochs: 5
Display params
Display progress: True
Display progress realtime: True
Save Training logs: True
Save Intermediate models: True
Intermediate model prefix: intermediate_model_
| Apache-2.0 | study_roadmaps/2_transfer_learning_roadmap/6_freeze_base_network/2.2) Understand the effect of freezing base model in transfer learning - 2 - pytorch.ipynb | take2rohit/monk_v1 |
From the summary above - Model Params Model name: densenet121 Use Gpu: True Use pretrained: True Freeze base network: True Another thing to notice from summary Model Details Loading pretrained model Model Loaded on device Model name: densenet121 Num of potentially trainable layers: 242 Num of actual trainable layers: 1 There are a total of 242 layers Since we have freezed base network only 1 is trainable, the final layer Train the classifier | #Start Training
gtf.Train();
#Read the training summary generated once you run the cell and training is completed | Training Start
Epoch 1/5
----------
| Apache-2.0 | study_roadmaps/2_transfer_learning_roadmap/6_freeze_base_network/2.2) Understand the effect of freezing base model in transfer learning - 2 - pytorch.ipynb | take2rohit/monk_v1 |
Best validation Accuracy achieved - 74.77 %(You may get a different result) Unfreeze Base network in densenet121 and train a classifier Creating and managing experiments - Provide project name - Provide experiment name - For a specific data create a single project - Inside each project multiple experiments can be created - Every experiment can be have diferent hyper-parameters attached to it | gtf = prototype(verbose=1);
gtf.Prototype("Project", "Unfreeze_Base_Network"); | Pytorch Version: 1.2.0
Experiment Details
Project: Project
Experiment: Unfreeze_Base_Network
Dir: /home/abhi/Desktop/Work/tess_tool/gui/v0.3/finetune_models/Organization/development/v5.0_blocks/study_roadmap/change_post_num_layers/5_transfer_learning_params/2_freezing_base_network/workspace/Project/Unfreeze_Base_Network/
| Apache-2.0 | study_roadmaps/2_transfer_learning_roadmap/6_freeze_base_network/2.2) Understand the effect of freezing base model in transfer learning - 2 - pytorch.ipynb | take2rohit/monk_v1 |
This creates files and directories as per the following structure workspace | |--------Project | | |-----Freeze_Base_Network (Previously created) | |-----experiment-state.json | |-----output | |------logs (All training logs and graphs saved here) | |------models (all trained models saved here) | | |-----Unfreeze_Base_Network (Created Now) | |-----experiment-state.json | |-----output | |------logs (All training logs and graphs saved here) | |------models (all trained models saved here) Set dataset and select the model Quick mode training - Using Default Function - dataset_path - model_name - freeze_base_network - num_epochs Sample Dataset folder structure parent_directory | | |------cats | |------img1.jpg |------img2.jpg |------.... (and so on) |------dogs | |------img1.jpg |------img2.jpg |------.... (and so on) Modifyable params - dataset_path: path to data - model_name: which pretrained model to use - freeze_base_network: Retrain already trained network or not - num_epochs: Number of epochs to train for | gtf.Default(dataset_path="skin_cancer_mnist_dataset/images",
path_to_csv="skin_cancer_mnist_dataset/train_labels.csv",
model_name="densenet121",
freeze_base_network=False, # Set this param as false
num_epochs=5);
#Read the summary generated once you run this cell. | Dataset Details
Train path: skin_cancer_mnist_dataset/images
Val path: None
CSV train path: skin_cancer_mnist_dataset/train_labels.csv
CSV val path: None
Dataset Params
Input Size: 224
Batch Size: 4
Data Shuffle: True
Processors: 4
Train-val split: 0.7
Delimiter: ,
Pre-Composed Train Transforms
[{'RandomHorizontalFlip': {'p': 0.8}}, {'Normalize': {'mean': [0.485, 0.456, 0.406], 'std': [0.229, 0.224, 0.225]}}]
Pre-Composed Val Transforms
[{'RandomHorizontalFlip': {'p': 0.8}}, {'Normalize': {'mean': [0.485, 0.456, 0.406], 'std': [0.229, 0.224, 0.225]}}]
Dataset Numbers
Num train images: 7010
Num val images: 3005
Num classes: 7
Model Params
Model name: densenet121
Use Gpu: True
Use pretrained: True
Freeze base network: False
Model Details
Loading pretrained model
Model Loaded on device
Model name: densenet121
Num layers in model: 242
Num trainable layers: 242
Optimizer
Name: sgd
Learning rate: 0.01
Params: {'lr': 0.01, 'momentum': 0, 'weight_decay': 0.0001, 'momentum_dampening_rate': 0, 'clipnorm': 0.0, 'clipvalue': 0.0}
Learning rate scheduler
Name: multisteplr
Params: {'milestones': [2, 3], 'gamma': 0.1, 'last_epoch': -1}
Loss
Name: softmaxcrossentropy
Params: {'weight': None, 'batch_axis': 0, 'axis_to_sum_over': -1, 'label_as_categories': True, 'label_smoothing': False}
Training params
Num Epochs: 5
Display params
Display progress: True
Display progress realtime: True
Save Training logs: True
Save Intermediate models: True
Intermediate model prefix: intermediate_model_
| Apache-2.0 | study_roadmaps/2_transfer_learning_roadmap/6_freeze_base_network/2.2) Understand the effect of freezing base model in transfer learning - 2 - pytorch.ipynb | take2rohit/monk_v1 |
From the summary above - Model Params Model name: densenet121 Use Gpu: True Use pretrained: True Freeze base network: False Another thing to notice from summary Model Details Loading pretrained model Model Loaded on device Model name: densenet121 Num of potentially trainable layers: 242 Num of actual trainable layers: 242 There are a total of 242 layers Since we have unfreezed base network all 242 layers are trainable including the final layer Train the classifier | #Start Training
gtf.Train();
#Read the training summary generated once you run the cell and training is completed | Training Start
Epoch 1/5
----------
| Apache-2.0 | study_roadmaps/2_transfer_learning_roadmap/6_freeze_base_network/2.2) Understand the effect of freezing base model in transfer learning - 2 - pytorch.ipynb | take2rohit/monk_v1 |
Best Val Accuracy achieved - 81.33 %(You may get a different result) Compare both the experiment | # Invoke the comparison class
from monk.compare_prototype import compare | _____no_output_____ | Apache-2.0 | study_roadmaps/2_transfer_learning_roadmap/6_freeze_base_network/2.2) Understand the effect of freezing base model in transfer learning - 2 - pytorch.ipynb | take2rohit/monk_v1 |
Creating and managing comparison experiments - Provide project name | # Create a project
gtf = compare(verbose=1);
gtf.Comparison("Compare-effect-of-freezing"); | Comparison: - Compare-effect-of-freezing
| Apache-2.0 | study_roadmaps/2_transfer_learning_roadmap/6_freeze_base_network/2.2) Understand the effect of freezing base model in transfer learning - 2 - pytorch.ipynb | take2rohit/monk_v1 |
This creates files and directories as per the following structure workspace | |--------comparison | | |-----Compare-effect-of-freezing | |------stats_best_val_acc.png |------stats_max_gpu_usage.png |------stats_training_time.png |------train_accuracy.png |------train_loss.png |------val_accuracy.png |------val_loss.png | |-----comparison.csv (Contains necessary details of all experiments) Add the experiments - First argument - Project name - Second argument - Experiment name | gtf.Add_Experiment("Project", "Freeze_Base_Network");
gtf.Add_Experiment("Project", "Unfreeze_Base_Network"); | Project - Project, Experiment - Freeze_Base_Network added
Project - Project, Experiment - Unfreeze_Base_Network added
| Apache-2.0 | study_roadmaps/2_transfer_learning_roadmap/6_freeze_base_network/2.2) Understand the effect of freezing base model in transfer learning - 2 - pytorch.ipynb | take2rohit/monk_v1 |
Run Analysis | gtf.Generate_Statistics(); | Generating statistics...
Generated
| Apache-2.0 | study_roadmaps/2_transfer_learning_roadmap/6_freeze_base_network/2.2) Understand the effect of freezing base model in transfer learning - 2 - pytorch.ipynb | take2rohit/monk_v1 |
Visualize and study comparison metrics Training Accuracy Curves | from IPython.display import Image
Image(filename="workspace/comparison/Compare-effect-of-freezing/train_accuracy.png") | _____no_output_____ | Apache-2.0 | study_roadmaps/2_transfer_learning_roadmap/6_freeze_base_network/2.2) Understand the effect of freezing base model in transfer learning - 2 - pytorch.ipynb | take2rohit/monk_v1 |
Training Loss Curves | from IPython.display import Image
Image(filename="workspace/comparison/Compare-effect-of-freezing/train_loss.png") | _____no_output_____ | Apache-2.0 | study_roadmaps/2_transfer_learning_roadmap/6_freeze_base_network/2.2) Understand the effect of freezing base model in transfer learning - 2 - pytorch.ipynb | take2rohit/monk_v1 |
Validation Accuracy Curves | from IPython.display import Image
Image(filename="workspace/comparison/Compare-effect-of-freezing/val_accuracy.png") | _____no_output_____ | Apache-2.0 | study_roadmaps/2_transfer_learning_roadmap/6_freeze_base_network/2.2) Understand the effect of freezing base model in transfer learning - 2 - pytorch.ipynb | take2rohit/monk_v1 |
Validation loss curves | from IPython.display import Image
Image(filename="workspace/comparison/Compare-effect-of-freezing/val_loss.png") | _____no_output_____ | Apache-2.0 | study_roadmaps/2_transfer_learning_roadmap/6_freeze_base_network/2.2) Understand the effect of freezing base model in transfer learning - 2 - pytorch.ipynb | take2rohit/monk_v1 |
Caesar CipherA Caesar cipher, also known as shift cipher is one of the simplest and most widely known encryption techniques. It is a type of substitution cipher in which each letter in the plaintext is replaced by a letter some fixed number of positions down the alphabet. For example, with a left shift of 3, D would be replaced by A, E would become B, and so on. 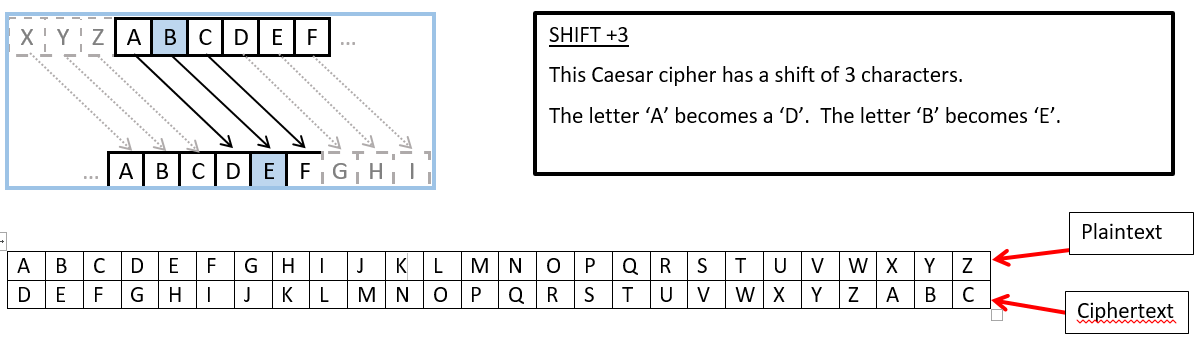 Insert message to encrypt and shift (0<= S <=26) below.By default, Caesar Cipher does a left shift of 3 | msg = "The quick brown fox jumps over the lazy dog 123 !@#"
shift = 3
def getmsg():
processedmsg = ''
for x in msg:
if x.isalpha():
num = ord(x)
num += shift
if x.isupper():
if num > ord('Z'):
num -= 26
elif num < ord('A'):
num += 26
elif x.islower():
if num > ord('z'):
num -= 26
elif num < ord('a'):
num += 26
processedmsg += chr(num)
else:
processedmsg += x
return processedmsg | _____no_output_____ | MIT | ipynb/Caesar Cipher.ipynb | davzoku/pyground |
The for loop above inspects each letter in the message.chr(), character function takes an integer ordinal and returns a character. ie. chr(65) outputs 'A' based on the ASCII tableord(), ordinal does the reverse. ie ord('A') gives 65.Based on the ASCII Table, 'Z' with a shift of 3 will give us ']', which is undesirable.Thus, we need the if-else statements to perform a "wraparound". If num has a value large than the ordinal value of 'Z', subtract 26.If num is less than 'a', add 26.**"else: processedmsg += x'** concatenates any spaces, numbers etc that are not encrypted or decrypted. | encrypted=getmsg()
print(encrypted)
| Wkh txlfn eurzq ira mxpsv ryhu wkh odcb grj 123 !@#
| MIT | ipynb/Caesar Cipher.ipynb | davzoku/pyground |
Note that only alphabets are encrypted.To decrypt, the algorithm is very similar. | shift=-shift
msg=encrypted
decrypted= getmsg()
print(decrypted) | The quick brown fox jumps over the lazy dog 123 !@#
| MIT | ipynb/Caesar Cipher.ipynb | davzoku/pyground |
使用TensorFlow的基本步骤以使用LinearRegression来预测房价为例。- 使用RMSE(均方根误差)评估模型预测的准确率- 通过调整超参数来提高模型的预测准确率 | from __future__ import print_function
import math
from IPython import display
from matplotlib import cm
from matplotlib import gridspec
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from sklearn import metrics
import tensorflow as tf
from tensorflow.python.data import Dataset
tf.logging.set_verbosity(tf.logging.ERROR)
pd.options.display.max_rows = 10
pd.options.display.float_format = '{:.1f}'.format
# 加载数据集
california_housing_df = pd.read_csv("https://download.mlcc.google.cn/mledu-datasets/california_housing_train.csv", sep=",")
# 将数据打乱
california_housing_df = california_housing_df.reindex(np.random.permutation(california_housing_df.index))
# 替换房价的单位为k
california_housing_df['median_house_value'] /=1000.0
print("california house dataframe: \n", california_housing_df) # 根据pd设置,只显示10条数据,以及保留小数点后一位 | california house dataframe:
longitude latitude housing_median_age total_rooms total_bedrooms \
840 -117.1 32.7 29.0 1429.0 293.0
15761 -122.4 37.8 52.0 3260.0 1535.0
2964 -117.8 34.1 23.0 7079.0 1381.0
5005 -118.1 33.8 36.0 1074.0 188.0
9816 -119.7 36.5 29.0 1702.0 301.0
... ... ... ... ... ...
1864 -117.3 34.7 28.0 1932.0 421.0
6257 -118.2 34.1 11.0 1281.0 418.0
4690 -118.1 34.1 52.0 1282.0 189.0
6409 -118.3 33.9 44.0 1103.0 265.0
11082 -121.0 38.7 5.0 5743.0 1074.0
population households median_income median_house_value
840 1091.0 317.0 3.5 118.0
15761 3260.0 1457.0 0.9 500.0
2964 3205.0 1327.0 3.1 212.3
5005 496.0 196.0 4.6 217.4
9816 914.0 280.0 2.8 79.2
... ... ... ... ...
1864 1156.0 404.0 1.9 55.6
6257 1584.0 330.0 2.9 153.1
4690 431.0 187.0 6.1 470.8
6409 760.0 247.0 1.7 99.6
11082 2651.0 962.0 4.1 172.5
[17000 rows x 9 columns]
| CC-BY-3.0 | code/first_step_with_tensorflow.ipynb | kevinleeex/notes-for-mlcc |
检查数据 | # 使用pd的describe方法来统计一些信息
california_housing_df.describe() | _____no_output_____ | CC-BY-3.0 | code/first_step_with_tensorflow.ipynb | kevinleeex/notes-for-mlcc |
构建模型我们将在这个例子中预测中位房价,将其作为学习的标签,使用房间总数作为输入特征。 第1步:定义特征并配置特征列为了把数据导入TensorFlow,我们需要指定每个特征包含的数据类型。我们主要使用以下两种类型:- 分类数据: 一种文字数据。- 数值数据:一种数字(整数或浮点数)数据或希望视为数字的数据。在TF中我们使用**特征列**的结构来表示特征的数据类型。特征列仅存储对特征数据的描述,不包含特征数据本身。 | # 定义输入特征
kl_feature = california_housing_df[['total_rooms']]
# 配置房间总数为数值特征列
feature_columns = [tf.feature_column.numeric_column('total_rooms')] | [_NumericColumn(key='total_rooms', shape=(1,), default_value=None, dtype=tf.float32, normalizer_fn=None)]
| CC-BY-3.0 | code/first_step_with_tensorflow.ipynb | kevinleeex/notes-for-mlcc |
第2步: 定义目标 | # 定义目标标签
targets = california_housing_df['median_house_value'] | _____no_output_____ | CC-BY-3.0 | code/first_step_with_tensorflow.ipynb | kevinleeex/notes-for-mlcc |
**梯度裁剪**是在应用梯度值之前设置其上限,梯度裁剪有助于确保数值稳定性,防止梯度爆炸。 第3步:配置线性回归器 | # 使用Linear Regressor配置线性回归模型,使用GradientDescentOptimizer优化器训练模型
kl_optimizer = tf.train.GradientDescentOptimizer(learning_rate=0.0000001)
# 使用clip_gradients_by_norm梯度裁剪我们的优化器,梯度裁剪可以确保我们的梯度大小在训练期间不会变得过大,梯度过大会导致梯度下降失败。
kl_optimizer = tf.contrib.estimator.clip_gradients_by_norm(kl_optimizer, 5.0)
# 使用我们的特征列和优化器配置线性回归模型
house_linear_regressor = tf.estimator.LinearRegressor(feature_columns=feature_columns, optimizer=kl_optimizer) | _____no_output_____ | CC-BY-3.0 | code/first_step_with_tensorflow.ipynb | kevinleeex/notes-for-mlcc |
第4步:定义输入函数要将数据导入LinearRegressor,我们需要定义一个输入函数,让它告诉TF如何对数据进行预处理,以及在模型训练期间如何批处理、随机处理和重复数据。首先我们将Pandas特征数据转换成NumPy数组字典,然后利用Dataset API构建Dataset对象,拆分数据为batch_size的批数据,以按照指定周期数(num_epochs)进行重复,**注意:**如果默认值num_epochs=None传递到repeat(),输入数据会无限期重复。shuffle: Bool, 是否打乱数据buffer_size: 指定shuffle从中随机抽样的数据集大小 | def kl_input_fn(features, targets, batch_size=1, shuffle=True, num_epochs=None):
"""使用单个特征训练房价预测模型
Args:
features: 特征DataFrame
targets: 目标DataFrame
batch_size: 批大小
shuffle: Bool. 是否打乱数据
Return:
下一个数据批次的元组(features, labels)
"""
# 把pandas数据转换成np.array构成的dict数据
features = {key: np.array(value) for key, value in dict(features).items()}
# 构建数据集,配置批和重复次数、
ds = Dataset.from_tensor_slices((features, targets)) # 数据大小 2GB 限制
ds = ds.batch(batch_size).repeat(num_epochs)
# 打乱数据
if shuffle:
ds = ds.shuffle(buffer_size=10000) # buffer_size指随机抽样的数据集大小
# 返回下一批次的数据
features, labels = ds.make_one_shot_iterator().get_next()
return features, labels | _____no_output_____ | CC-BY-3.0 | code/first_step_with_tensorflow.ipynb | kevinleeex/notes-for-mlcc |
**注意:** 更详细的输入函数和Dataset API参考:[TF Developer's Guide](https://www.tensorflow.org/programmers_guide/datasets) 第5步:训练模型在linear_regressor上调用train()来训练模型 | _ = house_linear_regressor.train(input_fn=lambda: kl_input_fn(kl_feature, targets), steps=100) | _____no_output_____ | CC-BY-3.0 | code/first_step_with_tensorflow.ipynb | kevinleeex/notes-for-mlcc |
第6步:评估模型**注意:**训练误差可以衡量训练的模型与训练数据的拟合情况,但**不能**衡量模型泛化到新数据的效果,我们需要拆分数据来评估模型的泛化能力。 | # 只做一次预测,所以把epoch设为1并关闭随机
prediction_input_fn = lambda: kl_input_fn(kl_feature, targets, num_epochs=1, shuffle=False)
# 调用predict进行预测
predictions = house_linear_regressor.predict(input_fn=prediction_input_fn)
# 把预测结果转换为numpy数组
predictions = np.array([item['predictions'][0] for item in predictions])
# 打印MSE和RMSE
mean_squared_error = metrics.mean_squared_error(predictions, targets)
root_mean_squared_error = math.sqrt(mean_squared_error)
print("均方误差 %0.3f" % mean_squared_error)
print("均方根误差: %0.3f" % root_mean_squared_error)
min_house_value = california_housing_df['median_house_value'].min()
max_house_value = california_housing_df['median_house_value'].max()
min_max_diff = max_house_value- min_house_value
print("最低中位房价: %0.3f" % min_house_value)
print("最高中位房价: %0.3f" % max_house_value)
print("中位房价最低最高差值: %0.3f" % min_max_diff)
print("均方根误差:%0.3f" % root_mean_squared_error) | 最低中位房价: 14.999
最高中位房价: 500.001
中位房价最低最高差值: 485.002
均方根误差:237.417
| CC-BY-3.0 | code/first_step_with_tensorflow.ipynb | kevinleeex/notes-for-mlcc |
由此结果可以看出模型的效果并不理想,我们可以使用一些基本的策略来降低误差。 | calibration_data = pd.DataFrame()
calibration_data["predictions"] = pd.Series(predictions)
calibration_data["targets"] = pd.Series(targets)
calibration_data.describe()
# 我们可以可视化数据和我们学到的线,
sample = california_housing_df.sample(n=300) # 得到均匀分布的sample数据df
# 得到房屋总数的最小最大值
x_0 = sample["total_rooms"].min()
x_1 = sample["total_rooms"].max()
# 获得训练后的最终权重和偏差
weight = house_linear_regressor.get_variable_value('linear/linear_model/total_rooms/weights')[0]
bias = house_linear_regressor.get_variable_value('linear/linear_model/bias_weights')
# 计算最低最高房间数(特征)对应的房价(标签)
y_0 = weight * x_0 + bias
y_1 = weight * x_1 +bias
# 画图
plt.plot([x_0,x_1], [y_0,y_1],c='r')
plt.ylabel('median_house_value')
plt.xlabel('total_rooms')
# 画出散点图
plt.scatter(sample["total_rooms"], sample["median_house_value"])
plt.show() | _____no_output_____ | CC-BY-3.0 | code/first_step_with_tensorflow.ipynb | kevinleeex/notes-for-mlcc |
模型调参以上代码封装调参 | def train_model(learning_rate, steps, batch_size, input_feature="total_rooms"):
"""Trains a linear regression model of one feature.
Args:
learning_rate: A `float`, the learning rate.
steps: A non-zero `int`, the total number of training steps. A training step
consists of a forward and backward pass using a single batch.
batch_size: A non-zero `int`, the batch size.
input_feature: A `string` specifying a column from `california_housing_df`
to use as input feature.
"""
periods = 10
steps_per_period = steps / periods
my_feature = input_feature
my_feature_data = california_housing_df[[my_feature]]
my_label = "median_house_value"
targets = california_housing_df[my_label]
# Create feature columns.
feature_columns = [tf.feature_column.numeric_column(my_feature)]
# Create input functions.
training_input_fn = lambda:kl_input_fn(my_feature_data, targets, batch_size=batch_size)
prediction_input_fn = lambda: kl_input_fn(my_feature_data, targets, num_epochs=1, shuffle=False)
# Create a linear regressor object.
my_optimizer = tf.train.GradientDescentOptimizer(learning_rate=learning_rate)
my_optimizer = tf.contrib.estimator.clip_gradients_by_norm(my_optimizer, 5.0)
linear_regressor = tf.estimator.LinearRegressor(
feature_columns=feature_columns,
optimizer=my_optimizer
)
# Set up to plot the state of our model's line each period.
plt.figure(figsize=(15, 6))
plt.subplot(1, 2, 1)
plt.title("Learned Line by Period")
plt.ylabel(my_label)
plt.xlabel(my_feature)
sample = california_housing_df.sample(n=300)
plt.scatter(sample[my_feature], sample[my_label])
colors = [cm.coolwarm(x) for x in np.linspace(-1, 1, periods)]
# Train the model, but do so inside a loop so that we can periodically assess
# loss metrics.
print("Training model...")
print("RMSE (on training data):")
root_mean_squared_errors = []
for period in range (0, periods):
# Train the model, starting from the prior state.
linear_regressor.train(
input_fn=training_input_fn,
steps=steps_per_period
)
# Take a break and compute predictions.
predictions = linear_regressor.predict(input_fn=prediction_input_fn)
predictions = np.array([item['predictions'][0] for item in predictions])
# Compute loss.
root_mean_squared_error = math.sqrt(
metrics.mean_squared_error(predictions, targets))
# Occasionally print the current loss.
print(" period %02d : %0.2f" % (period, root_mean_squared_error))
# Add the loss metrics from this period to our list.
root_mean_squared_errors.append(root_mean_squared_error)
# Finally, track the weights and biases over time.
# Apply some math to ensure that the data and line are plotted neatly.
y_extents = np.array([0, sample[my_label].max()])
weight = linear_regressor.get_variable_value('linear/linear_model/%s/weights' % input_feature)[0]
bias = linear_regressor.get_variable_value('linear/linear_model/bias_weights')
x_extents = (y_extents - bias) / weight
x_extents = np.maximum(np.minimum(x_extents,
sample[my_feature].max()),
sample[my_feature].min())
y_extents = weight * x_extents + bias
plt.plot(x_extents, y_extents, color=colors[period])
print("Model training finished.")
# Output a graph of loss metrics over periods.
plt.subplot(1, 2, 2)
plt.ylabel('RMSE')
plt.xlabel('Periods')
plt.title("Root Mean Squared Error vs. Periods")
plt.tight_layout()
plt.plot(root_mean_squared_errors)
# Output a table with calibration data.
calibration_data = pd.DataFrame()
calibration_data["predictions"] = pd.Series(predictions)
calibration_data["targets"] = pd.Series(targets)
display.display(calibration_data.describe())
print("Final RMSE (on training data): %0.2f" % root_mean_squared_error) | _____no_output_____ | CC-BY-3.0 | code/first_step_with_tensorflow.ipynb | kevinleeex/notes-for-mlcc |
**练习1: 使RMSE不超过180** | train_model(learning_rate=0.00002, steps=500, batch_size=5) | Training model...
RMSE (on training data):
period 00 : 225.63
period 01 : 214.42
period 02 : 204.04
period 03 : 194.97
period 04 : 186.60
period 05 : 180.80
period 06 : 175.66
period 07 : 171.74
period 08 : 168.96
period 09 : 167.23
Model training finished.
| CC-BY-3.0 | code/first_step_with_tensorflow.ipynb | kevinleeex/notes-for-mlcc |
模型调参的启发法> 不要死循规则- 训练误差应该稳步减小,刚开始是急剧减小,最终应随着训练收敛达到平稳状态。- 如果训练尚未收敛,尝试运行更长的时间。- 如果训练误差减小速度过慢,则提高学习速率也许有助于加快其减小速度。- 但有时如果学习速率过高,训练误差的减小速度反而会变慢。- 如果训练误差变化很大,尝试降低学习速率。- 较低的学习速率和较大的步数/较大的批量大小通常是不错的组合。- 批量大小过小也会导致不稳定情况。不妨先尝试 100 或 1000 等较大的值,然后逐渐减小值的大小,直到出现性能降低的情况。 **练习2:尝试其他特征**我们使用population特征替代。 | train_model(learning_rate=0.00005, steps=500, batch_size=5, input_feature="population") | Training model...
RMSE (on training data):
period 00 : 222.79
period 01 : 209.51
period 02 : 198.00
period 03 : 189.59
period 04 : 182.78
period 05 : 179.35
period 06 : 177.30
period 07 : 176.11
period 08 : 175.97
period 09 : 176.51
Model training finished.
| CC-BY-3.0 | code/first_step_with_tensorflow.ipynb | kevinleeex/notes-for-mlcc |
test note* jupyterはコンテナ起動すること* テストベッド一式起動済みであること | !pip install --upgrade pip
!pip install --force-reinstall ../lib/ait_sdk-0.1.7-py3-none-any.whl
from pathlib import Path
import pprint
from ait_sdk.test.hepler import Helper
import json
# settings cell
# mounted dir
root_dir = Path('/workdir/root/ait')
ait_name='eval_metamorphic_test_tf1.13'
ait_version='0.1'
ait_full_name=f'{ait_name}_{ait_version}'
ait_dir = root_dir / ait_full_name
td_name=f'{ait_name}_test'
# (dockerホスト側の)インベントリ登録用アセット格納ルートフォルダ
current_dir = %pwd
with open(f'{current_dir}/config.json', encoding='utf-8') as f:
json_ = json.load(f)
root_dir = json_['host_ait_root_dir']
is_container = json_['is_container']
invenotory_root_dir = f'{root_dir}\\ait\\{ait_full_name}\\local_qai\\inventory'
# entry point address
# コンテナ起動かどうかでポート番号が変わるため、切り替える
if is_container:
backend_entry_point = 'http://host.docker.internal:8888/qai-testbed/api/0.0.1'
ip_entry_point = 'http://host.docker.internal:8888/qai-ip/api/0.0.1'
else:
backend_entry_point = 'http://host.docker.internal:5000/qai-testbed/api/0.0.1'
ip_entry_point = 'http://host.docker.internal:6000/qai-ip/api/0.0.1'
# aitのデプロイフラグ
# 一度実施すれば、それ以降は実施しなくてOK
is_init_ait = True
# インベントリの登録フラグ
# 一度実施すれば、それ以降は実施しなくてOK
is_init_inventory = True
helper = Helper(backend_entry_point=backend_entry_point,
ip_entry_point=ip_entry_point,
ait_dir=ait_dir,
ait_full_name=ait_full_name)
# health check
helper.get_bk('/health-check')
helper.get_ip('/health-check')
# create ml-component
res = helper.post_ml_component(name=f'MLComponent_{ait_full_name}', description=f'Description of {ait_full_name}', problem_domain=f'ProbremDomain of {ait_full_name}')
helper.set_ml_component_id(res['MLComponentId'])
# deploy AIT
if is_init_ait:
helper.deploy_ait_non_build()
else:
print('skip deploy AIT')
res = helper.get_data_types()
model_data_type_id = [d for d in res['DataTypes'] if d['Name'] == 'model'][0]['Id']
dataset_data_type_id = [d for d in res['DataTypes'] if d['Name'] == 'dataset'][0]['Id']
res = helper.get_file_systems()
unix_file_system_id = [f for f in res['FileSystems'] if f['Name'] == 'UNIX_FILE_SYSTEM'][0]['Id']
windows_file_system_id = [f for f in res['FileSystems'] if f['Name'] == 'WINDOWS_FILE'][0]['Id']
# add inventories
if is_init_inventory:
inv1_name = helper.post_inventory('train_image', dataset_data_type_id, windows_file_system_id,
f'{invenotory_root_dir}\\mnist_dataset\\mnist_dataset.zip',
'MNIST_dataset are train image, train label, test image, test label', ['zip'])
inv2_name = helper.post_inventory('mnist_model', dataset_data_type_id, windows_file_system_id,
f'{invenotory_root_dir}\\mnist_model\\model_mnist.zip',
'MNIST_model', ['zip'])
else:
print('skip add inventories')
# get ait_json and inventory_jsons
res_json = helper.get_bk('/QualityMeasurements/RelationalOperators', is_print_json=False).json()
eq_id = int([r['Id'] for r in res_json['RelationalOperator'] if r['Expression'] == '=='][0])
nq_id = int([r['Id'] for r in res_json['RelationalOperator'] if r['Expression'] == '!='][0])
gt_id = int([r['Id'] for r in res_json['RelationalOperator'] if r['Expression'] == '>'][0])
ge_id = int([r['Id'] for r in res_json['RelationalOperator'] if r['Expression'] == '>='][0])
lt_id = int([r['Id'] for r in res_json['RelationalOperator'] if r['Expression'] == '<'][0])
le_id = int([r['Id'] for r in res_json['RelationalOperator'] if r['Expression'] == '<='][0])
res_json = helper.get_bk('/testRunners', is_print_json=False).json()
ait_json = [j for j in res_json['TestRunners'] if j['Name'] == ait_name][-1]
inv_1_json = helper.get_inventory(inv1_name)
inv_2_json = helper.get_inventory(inv2_name)
# add teast_descriptions
helper.post_td(td_name, ait_json['QualityDimensionId'],
quality_measurements=[
{"Id":ait_json['Report']['Measures'][0]['Id'], "Value":"0.25", "RelationalOperatorId":lt_id, "Enable":True}
],
target_inventories=[
{"Id":1, "InventoryId": inv_1_json['Id'], "TemplateInventoryId": ait_json['TargetInventories'][0]['Id']},
{"Id":2, "InventoryId": inv_2_json['Id'], "TemplateInventoryId": ait_json['TargetInventories'][1]['Id']}
],
test_runner={
"Id":ait_json['Id'],
"Params":[
{"TestRunnerParamTemplateId":ait_json['ParamTemplates'][0]['Id'], "Value":"10"},
{"TestRunnerParamTemplateId":ait_json['ParamTemplates'][1]['Id'], "Value":"500"},
{"TestRunnerParamTemplateId":ait_json['ParamTemplates'][2]['Id'], "Value":"train"}
]
})
# get test_description_jsons
td_1_json = helper.get_td(td_name)
# run test_descriptions
helper.post_run_and_wait(td_1_json['Id'])
res_json = helper.get_td_detail(td_1_json['Id'])
pprint.pprint(res_json)
# generate report
res = helper.post_report(td_1_json['Id']) | <Response [200]>
{'OutParams': {'ReportUrl': 'http://127.0.0.1:8888/qai-testbed/api/0.0.1/download/469'},
'Result': {'Code': 'D12000', 'Message': 'command invoke success.'}}
| Apache-2.0 | ait_repository/test/tests/eval_metamorphic_test_tf1.13_0.1.ipynb | ads-ad-itcenter/qunomon.forked |
Python APIProphet follows the `sklearn` model API. We create an instance of the `Prophet` class and then call its `fit` and `predict` methods. The input to Prophet is always a dataframe with two columns: `ds` and `y`. The `ds` (datestamp) column should be of a format expected by Pandas, ideally YYYY-MM-DD for a date or YYYY-MM-DD HH:MM:SS for a timestamp. The `y` column must be numeric, and represents the measurement we wish to forecast.As an example, let's look at a time series of the log daily page views for the Wikipedia page for [Peyton Manning](https://en.wikipedia.org/wiki/Peyton_Manning). We scraped this data using the [Wikipediatrend](https://cran.r-project.org/package=wikipediatrend) package in R. Peyton Manning provides a nice example because it illustrates some of Prophet's features, like multiple seasonality, changing growth rates, and the ability to model special days (such as Manning's playoff and superbowl appearances). The CSV is available [here](https://github.com/facebook/prophet/blob/master/examples/example_wp_log_peyton_manning.csv).First we'll import the data: | import pandas as pd
from fbprophet import Prophet
df = pd.read_csv('../examples/example_wp_log_peyton_manning.csv')
df.head() | _____no_output_____ | MIT | notebooks/quick_start.ipynb | timgates42/prophet |
We fit the model by instantiating a new `Prophet` object. Any settings to the forecasting procedure are passed into the constructor. Then you call its `fit` method and pass in the historical dataframe. Fitting should take 1-5 seconds. | m = Prophet()
m.fit(df) | INFO:fbprophet:Disabling daily seasonality. Run prophet with daily_seasonality=True to override this.
| MIT | notebooks/quick_start.ipynb | timgates42/prophet |
Predictions are then made on a dataframe with a column `ds` containing the dates for which a prediction is to be made. You can get a suitable dataframe that extends into the future a specified number of days using the helper method `Prophet.make_future_dataframe`. By default it will also include the dates from the history, so we will see the model fit as well. | future = m.make_future_dataframe(periods=365)
future.tail() | _____no_output_____ | MIT | notebooks/quick_start.ipynb | timgates42/prophet |
The `predict` method will assign each row in `future` a predicted value which it names `yhat`. If you pass in historical dates, it will provide an in-sample fit. The `forecast` object here is a new dataframe that includes a column `yhat` with the forecast, as well as columns for components and uncertainty intervals. | forecast = m.predict(future)
forecast[['ds', 'yhat', 'yhat_lower', 'yhat_upper']].tail() | _____no_output_____ | MIT | notebooks/quick_start.ipynb | timgates42/prophet |
You can plot the forecast by calling the `Prophet.plot` method and passing in your forecast dataframe. | fig1 = m.plot(forecast) | _____no_output_____ | MIT | notebooks/quick_start.ipynb | timgates42/prophet |
If you want to see the forecast components, you can use the `Prophet.plot_components` method. By default you'll see the trend, yearly seasonality, and weekly seasonality of the time series. If you include holidays, you'll see those here, too. | fig2 = m.plot_components(forecast) | _____no_output_____ | MIT | notebooks/quick_start.ipynb | timgates42/prophet |
An interactive figure of the forecast and components can be created with plotly. You will need to install plotly 4.0 or above separately, as it will not by default be installed with fbprophet. You will also need to install the `notebook` and `ipywidgets` packages. | from fbprophet.plot import plot_plotly, plot_components_plotly
plot_plotly(m, forecast)
plot_components_plotly(m, forecast) | _____no_output_____ | MIT | notebooks/quick_start.ipynb | timgates42/prophet |
More details about the options available for each method are available in the docstrings, for example, via `help(Prophet)` or `help(Prophet.fit)`. The [R reference manual](https://cran.r-project.org/web/packages/prophet/prophet.pdf) on CRAN provides a concise list of all of the available functions, each of which has a Python equivalent. R APIIn R, we use the normal model fitting API. We provide a `prophet` function that performs fitting and returns a model object. You can then call `predict` and `plot` on this model object. | %%R
library(prophet) | _____no_output_____ | MIT | notebooks/quick_start.ipynb | timgates42/prophet |
First we read in the data and create the outcome variable. As in the Python API, this is a dataframe with columns `ds` and `y`, containing the date and numeric value respectively. The ds column should be YYYY-MM-DD for a date, or YYYY-MM-DD HH:MM:SS for a timestamp. As above, we use here the log number of views to Peyton Manning's Wikipedia page, available [here](https://github.com/facebook/prophet/blob/master/examples/example_wp_log_peyton_manning.csv). | %%R
df <- read.csv('../examples/example_wp_log_peyton_manning.csv') | _____no_output_____ | MIT | notebooks/quick_start.ipynb | timgates42/prophet |
We call the `prophet` function to fit the model. The first argument is the historical dataframe. Additional arguments control how Prophet fits the data and are described in later pages of this documentation. | %%R
m <- prophet(df) | _____no_output_____ | MIT | notebooks/quick_start.ipynb | timgates42/prophet |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.