hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
sequence | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
sequence | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
sequence | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
sequence | cell_types
sequence | cell_type_groups
sequence |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
d091823ecdb07c4b1a5f429eb2e9c740182194f9 | 6,915 | ipynb | Jupyter Notebook | 1_Required_Python_Library_Installation.ipynb | chahatmax/World_VS_India_COVID-19 | b46091c7f2ec6f3302624f77385800473c3c4f7f | [
"MIT"
] | 1 | 2020-05-30T19:18:53.000Z | 2020-05-30T19:18:53.000Z | 1_Required_Python_Library_Installation.ipynb | chahatmax/World_VS_India_COVID-19 | b46091c7f2ec6f3302624f77385800473c3c4f7f | [
"MIT"
] | null | null | null | 1_Required_Python_Library_Installation.ipynb | chahatmax/World_VS_India_COVID-19 | b46091c7f2ec6f3302624f77385800473c3c4f7f | [
"MIT"
] | null | null | null | 30.733333 | 548 | 0.632972 | [
[
[
"!pip install plotly",
"_____no_output_____"
]
],
[
[
"<a href=\"https://plotly.com/python/\" target=\"_blank\">Plotly's</a> Python graphing library makes interactive, publication-quality graphs. Examples of how to make line plots, scatter plots, area charts, bar charts, error bars, box plots, histograms, heatmaps, subplots, multiple-axes, polar charts, and bubble charts.",
"_____no_output_____"
]
],
[
[
"!pip install plotly_express",
"_____no_output_____"
]
],
[
[
"<a href=\"https://pypi.org/project/plotly-express/0.1.9/\" target=\"_blank\">Plotly Express</a> is a terse, consistent, high-level wrapper around Plotly.py for rapid data exploration and figure generation.",
"_____no_output_____"
]
],
[
[
"!pip install calmap",
"_____no_output_____"
]
],
[
[
"<a href=\"https://pypi.org/project/calmap/\" target=\"_blank\">Calendar heatmaps (calmap)</a> Plot Pandas time series data sampled by day in a heatmap per calendar year, similar to GitHub’s contributions plot, using matplotlib.",
"_____no_output_____"
]
],
[
[
"!pip install squarify",
"_____no_output_____"
]
],
[
[
"Pure Python implementation of the <a href=\"https://pypi.org/project/squarify/0.1/\" target=\"_blank\">squarify</a> treemap layout algorithm. Based on algorithm from Bruls, Huizing, van Wijk, \"Squarified Treemaps\", but implements it differently.",
"_____no_output_____"
]
],
[
[
"!pip install pycountry_convert",
"_____no_output_____"
]
],
[
[
"Using country data derived from wikipedia, <a href=\"https://pypi.org/project/pycountry-convert/\" target=\"_blank\">pycountry-convert</a> provides conversion functions between ISO country names, country-codes, and continent names.",
"_____no_output_____"
]
],
[
[
"!pip install GoogleMaps",
"_____no_output_____"
]
],
[
[
"Use Python? Want to geocode something? Looking for directions? Maybe matrices of directions? This library brings the <a href=\"https://pypi.org/project/googlemaps/\" target=\"_blank\">Google Maps</a> Platform Web Services to your Python application.",
"_____no_output_____"
]
],
[
[
"!pip install xgboost",
"_____no_output_____"
]
],
[
[
"<a href=\"https://xgboost.readthedocs.io/en/latest/\" target=\"_blank\">XGBoost</a> is an optimized distributed gradient boosting library designed to be highly efficient, flexible and portable. It implements machine learning algorithms under the Gradient Boosting framework. XGBoost provides a parallel tree boosting (also known as GBDT, GBM) that solve many data science problems in a fast and accurate way. The same code runs on major distributed environment (Hadoop, SGE, MPI) and can solve problems beyond billions of examples.",
"_____no_output_____"
]
],
[
[
"!pip install lightgbm",
"_____no_output_____"
]
],
[
[
"<a href=\"https://lightgbm.readthedocs.io/en/latest/\" target=\"_blank\">LightGBM</a> is a gradient boosting framework that uses tree based learning algorithms. It is designed to be distributed and efficient with the following advantages:\n* Faster training speed and higher efficiency.\n* Lower memory usage.\n* Better accuracy.\n* Support of parallel and GPU learning.\n* Capable of handling large-scale data.",
"_____no_output_____"
]
],
[
[
"!pip install altair",
"_____no_output_____"
]
],
[
[
"<a href=\"https://pypi.org/project/altair/\" target=\"_blank\">Altair</a> is a declarative statistical visualization library for Python. With Altair, you can spend more time understanding your data and its meaning. Altair's API is simple, friendly and consistent and built on top of the powerful Vega-Lite JSON specification. This elegant simplicity produces beautiful and effective visualizations with a minimal amount of code. Altair is developed by Jake Vanderplas and Brian Granger in close collaboration with the UW Interactive Data Lab.",
"_____no_output_____"
]
],
[
[
"!pip install folium",
"_____no_output_____"
]
],
[
[
"<a href=\"https://pypi.org/project/folium/\" target=\"_blank\">folium</a> builds on the data wrangling strengths of the Python ecosystem and the mapping strengths of the Leaflet.js library. Manipulate your data in Python, then visualize it in a Leaflet map via folium.",
"_____no_output_____"
]
],
[
[
"!pip install fbprophet",
"_____no_output_____"
]
],
[
[
"<a href=\"https://pypi.org/project/fbprophet/\" target=\"_blank\">Prophet</a> is a procedure for forecasting time series data based on an additive model where non-linear trends are fit with yearly, weekly, and daily seasonality, plus holiday effects. It works best with time series that have strong seasonal effects and several seasons of historical data. Prophet is robust to missing data and shifts in the trend, and typically handles outliers well.",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0918d37dc57dc969fc767e9c97e5fe1506998da | 3,499 | ipynb | Jupyter Notebook | notebooks/Welcome.ipynb | TerrainBento/umami | ab6cf5aabd8ecfe22b3de8e762317ceb53236362 | [
"MIT"
] | 1 | 2019-11-05T17:49:05.000Z | 2019-11-05T17:49:05.000Z | notebooks/Welcome.ipynb | TerrainBento/umami | ab6cf5aabd8ecfe22b3de8e762317ceb53236362 | [
"MIT"
] | 9 | 2019-09-18T20:04:40.000Z | 2020-02-28T19:56:24.000Z | notebooks/Welcome.ipynb | TerrainBento/umami | ab6cf5aabd8ecfe22b3de8e762317ceb53236362 | [
"MIT"
] | 2 | 2019-09-11T17:08:32.000Z | 2019-10-29T11:57:29.000Z | 47.931507 | 610 | 0.682195 | [
[
[
"# Umami notebooks\n\nWelcome to the umami notebooks. This page provides links to notebooks that provide an introduction to umami and its use. We recommend that you look at them in the following order.\n\nFirst, look at two notebooks designed to introduce you to the core classes and methods of umami.\n\n * [Part 1: Introduction to umami and the `Metric` class](IntroductionToMetric.ipynb)\n * [Part 2: Introduction to the `Residual` class](IntroductionToResidual.ipynb)\n * [Part 3: Other IO options (using umami without Landlab or terrainbento)](OtherIO_options.ipynb)\n \nThen, look at two notebooks with example applications.\n \n * [Part 4: Example Application](ExampleApplication.ipynb)\n * [Part 5: Application using the Discretized Misfit calculation](DiscretizedMisfit.ipynb)\n \nIf you have comments or questions about the notebooks, the best place to get help is through [GitHub Issues](https://github.com/TerrainBento/umami/issues).\n\n## Some background\n\n### What is umami?\n\nUmami is a package for calculating objective functions or objective function components for Earth surface dynamics modeling. It was designed to work well with [terrainbento](https://github.com/TerrainBento/terrainbento) and other models built with the [Landlab Toolkit](https://github.com/landlab/landlab). However, it is not necessary to do modeling with either of these packages to use umami (this is described further in [Part 3 of the notebook series](OtherIO_options.ipynb).\n\nUmami offers two primary classes:\n* a[`Residual`](https://umami.readthedocs.io/en/latest/umami.residual.html#Residual),\nwhich represents the difference between model and data, and \n* a [`Metric`](https://umami.readthedocs.io/en/latest/umami.metric.html),\nwhich is a calculated value on either model or data. \n\nThe set of currently supported calculations can be found in the [`umami.calculations`](https://umami.readthedocs.io/en/latest/umami.calculations.html) submodule.\n\n### What does it do well?\n\nUmami was designed to provide an input-file based interface for calculating single-value landscape metrics for use in model analysis. This supports reproducible analysis and systematic variation in metric construction. When used with `terrainbento`, one input file can describe the model run, and one input file can describe the model assessment or model-data comparison. This streamlines model analysis applications. Umami also provides multiple output formats (YAML and Dakota), the latter of which is designed to interface with Sandia National Laboratory's [Dakota package](https://dakota.sandia.gov).",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown"
]
] |
d091960b66a94cf736584d2b57379f62a3cf142f | 24,906 | ipynb | Jupyter Notebook | materials/lecture_notebooks/lecture_1_old.ipynb | Rajk-Prog1/prog1_2021_fall | bf7983cfd146b91b5788ed9874f6b415c33cc4c4 | [
"MIT"
] | 1 | 2021-09-08T10:25:23.000Z | 2021-09-08T10:25:23.000Z | materials/lecture_notebooks/lecture_1_old.ipynb | Rajk-Prog1/prog1_2021_fall | bf7983cfd146b91b5788ed9874f6b415c33cc4c4 | [
"MIT"
] | null | null | null | materials/lecture_notebooks/lecture_1_old.ipynb | Rajk-Prog1/prog1_2021_fall | bf7983cfd146b91b5788ed9874f6b415c33cc4c4 | [
"MIT"
] | 2 | 2021-09-05T00:13:22.000Z | 2021-09-08T10:25:31.000Z | 29.614744 | 2,502 | 0.62491 | [
[
[
"### Mit kellene tudni?\n#### 1. Megfogalmazni egy programozási problémát <!-- .element: class=\"fragment\" -->\n",
"_____no_output_____"
],
[
"#### 1. Számításelmélet értelmét elmagyarázni <!-- .element: class=\"fragment\" -->\n",
"_____no_output_____"
],
[
"#### 1. Lebontani egy komplex problémát egyszerűbbekre <!-- .element: class=\"fragment\" -->\n",
"_____no_output_____"
],
[
"#### 1. Megérteni egy leírt programot <!-- .element: class=\"fragment\" -->\n",
"_____no_output_____"
],
[
"#### 1. Megírni egy programot <!-- .element: class=\"fragment\" -->\n",
"_____no_output_____"
],
[
"#### 1. Találni érdeklődést és folytatást <!-- .element: class=\"fragment\" -->\n",
"_____no_output_____"
],
[
"<center><img src=\"https://pics.me.me/doctors-googling-stuff-online-does-not-make-you-a-doctor-61282088.png\"></center> -->",
"_____no_output_____"
],
[
"### Deklaratív tudás\nA \"mit?\" kérdésre adott válasz egy probléma kapcsán",
"_____no_output_____"
],
[
"### Imperatív tudás\nA \"hogyan\" kérdésre adott válasz egy probléma kapcsán",
"_____no_output_____"
],
[
"### Deklaratív példák\n- tervrajz\n",
"_____no_output_____"
],
[
"- térképjelzés\n",
"_____no_output_____"
],
[
"- anatómiai ábra\n",
"_____no_output_____"
],
[
"### Imperatív példák\n- recept\n",
"_____no_output_____"
],
[
"- IKEA útmutató\n",
"_____no_output_____"
],
[
"- útvonalterv\n",
"_____no_output_____"
],
[
"### Mit adunk át a gépnek?\nEgy probléma megoldására szeretnénk megtanítani/megkérni:\n",
"_____no_output_____"
],
[
"tetszőleges $x$-re, számolja ki $ \\sqrt{x} $-et úgy, hogy csak osztani, szorozni, meg összeadni tud",
"_____no_output_____"
],
[
"### Tudás Gyökvonásról\n- deklaratív tudás:\n",
"_____no_output_____"
],
[
"- $ \\sqrt{x} = y \\Leftrightarrow y^2=x $\n",
"_____no_output_____"
],
[
"- $ x < y^2 \\Leftrightarrow \\sqrt{x} < y $\n",
"_____no_output_____"
],
[
"- $ ( (y_1 < x \\land y_2 < x) \\lor (y_1 > x \\land y_2 > x) ) \\Rightarrow $\n",
"_____no_output_____"
],
[
"- $ \\Rightarrow ( |x - y_1| < |x - y_2| \\Leftrightarrow |x^2 - y_1^2| < |x^2 - y_2^2| ) $\n",
"_____no_output_____"
],
[
"- imperatív tudás:\n",
"_____no_output_____"
],
[
"- tippelünk egy kezdeti $ G $ -t, és ameddig nem elég jó, $ G' = \\frac{G+\\frac{x}{G}}{2} $ -vel közelítünk, így tetszőlegesen közel kerülünk az eredményhez\n",
"_____no_output_____"
],
[
"(minden nemnegatív számok esetén)",
"_____no_output_____"
],
[
"### Ember\nokos, kreatív, tud következtetni, memóriája ködös,\n",
"_____no_output_____"
],
[
"szeretné minél kevesebbet nyomkodni a számológépet\n",
"_____no_output_____"
],
[
"- felhasznál csomó meglévő deklaratív tudást\n",
"_____no_output_____"
],
[
"- összeállítja egy komplex tippelési folyamattá\n",
"_____no_output_____"
],
[
"- tippek eredményéből továbbfejleszti a folyamatot\n",
"_____no_output_____"
],
[
"- felhasznál olyan komplex fogalmakat mint nagyságrend, vagy előjelváltás",
"_____no_output_____"
],
[
"### Gép\nbuta, nincs önálló gondolata, memóriája tökéletes\n",
"_____no_output_____"
],
[
"másodpercenként töbmilliárd műveletet el tud végezni,\n",
"_____no_output_____"
],
[
"- tippel egy nagyon rosszat, pl G = 1\n",
"_____no_output_____"
],
[
"- valaki megtanította neki hogy $G' = \\frac{G+\\frac{x}{G}}{2}$ közelít, szóval elkezdi\n",
"_____no_output_____"
],
[
"- Megoldandó probléma: $ \\sqrt{1366561} = ? $\n",
"_____no_output_____"
],
[
"### Specifikáció\nEgyfajta deklaratív leírása egy programnak\n",
"_____no_output_____"
],
[
"Miből, mit?\n",
"_____no_output_____"
],
[
"- állapottér - bemenő/kimenő adatok\n",
"_____no_output_____"
],
[
"- előfeltétel - amit tudunk a bemenetről\n",
"_____no_output_____"
],
[
"- utófeltétel - az eredmény kiszámítási szabálya (amit tudunk az eredményről)\n",
"_____no_output_____"
],
[
"legyen **teljes** és **részletes**",
"_____no_output_____"
],
[
"### Specifikáció I. példa\nValós számok halmazán megoldható másodfokú egyneletet oldjon meg a program, ami $ax^2+bx+c=0$ formában van megadva\n",
"_____no_output_____"
],
[
"- Á: $(a,b,c,x_1,x_2 \\in \\mathbb{R})$ <!-- .element: class=\"fragment\" data-fragment-index=\"1\" -->\n",
"_____no_output_____"
],
[
"- Ef: $(a \\neq 0 \\land b^2-4ac \\geq 0)$ <!-- .element: class=\"fragment\" data-fragment-index=\"2\" -->\n",
"_____no_output_____"
],
[
"- Uf: $ (x_1 = \\frac{-b + \\sqrt{b^2-4ac}}{2a} \\land x_2 = \\frac{-b - \\sqrt{b^2-4ac}}{2a})$ <!-- .element: class=\"fragment\" data-fragment-index=\"3\" -->",
"_____no_output_____"
],
[
"### Specifikáció példa 2\nValós számok halmazán megoldható másodfokú egyneletet oldjon meg a program, ami $ax^2+bx+c=0$ formában van megadva\n",
"_____no_output_____"
],
[
"- Á: $(a,b,c,x_1,x_2 \\in \\mathbb{R})$\n",
"_____no_output_____"
],
[
"- Ef: $(a \\neq 0 \\land b^2-4ac \\geq 0)$\n",
"_____no_output_____"
],
[
"- Uf: $ (\\forall i (ax_i^2+bx_i+c=0) \\land ( x_1 = x_2 \\iff b^2-4ac = 0 ))$",
"_____no_output_____"
],
[
"### Specifikáció teljessége\nAmit az előfeltétel megenged az állapottéren belül, azt az utófeltételnek kezelnie kell\n",
"_____no_output_____"
],
[
"Pl:\n",
"_____no_output_____"
],
[
"- Válassza ki két szám közül azt amelyiknek több prím osztója van\n",
"_____no_output_____"
],
[
"- több különböző vagy több összesen?\n",
"_____no_output_____"
],
[
"- mi van ha ugyanannyi?\n",
"_____no_output_____"
],
[
"- Válassza ki egy számsorból azt az 5 számot ami legjobban eltér az átlagtól\n",
"_____no_output_____"
],
[
"- mi van ha nincs 5 szám a számsorban?\n",
"_____no_output_____"
],
[
"- mi van ha minden szám ugyanaz?\n",
"_____no_output_____"
],
[
"- Sakktáblán szereplő állásról döntse el hogy matt-e.\n",
"_____no_output_____"
],
[
"- mi van ha mindkét fél sakkban van?\n",
"_____no_output_____"
],
[
"- lehet 11 királynő valakinél?",
"_____no_output_____"
],
[
"### Specifikáció részletessége\nA program főzzön rántottát:\n",
"_____no_output_____"
],
[
"- Á: (konyha)\n",
"_____no_output_____"
],
[
"- Ef: ( )\n",
"_____no_output_____"
],
[
"- Uf: (ha a konyha alkalmas ennek elkészítésére, a konyhában legyen egy finom rántotta)\n",
"_____no_output_____"
],
[
"vagy:\n",
"_____no_output_____"
],
[
"- Á: (serpenyő, főzőlap, 3 tojás, olaj, só, fakanál, tányér)\n",
"_____no_output_____"
],
[
"- Ef: (a serpenyő, tányér és fakanál tiszta, a tojás nem romlott, a főzőlap képes a serpenyőt 200 Celsius fokra felmelegíteni)\n",
"_____no_output_____"
],
[
"- Uf: (a tojások összekeverve kerültek a 200 fokos serpenyőbe az olaj után, több mint 4 de kevesebb mint 10 percet töltöttek ott, ebből 20 másodpercnél többet nem álltak kavarás nélkül, végül megsózva kikerültek a tányérra)",
"_____no_output_____"
],
[
"### Specifikáció II. példa\nKét természetes szám legnagyobb közös osztójának megtalálása\n",
"_____no_output_____"
],
[
"- Á: $(a,b,x \\in \\mathbb{N})$ <!-- .element: class=\"fragment\" data-fragment-index=\"1\" -->\n",
"_____no_output_____"
],
[
"- Ef: $(a \\neq 0 \\land b \\neq 0)$ <!-- .element: class=\"fragment\" data-fragment-index=\"2\" -->\n",
"_____no_output_____"
],
[
"- Uf: $ (x|a \\land x|b \\land \\nexists i(i|a \\land i|b \\land i > x))$ <!-- .element: class=\"fragment\" data-fragment-index=\"3\" -->",
"_____no_output_____"
],
[
"<a style='text-decoration:none;line-height:16px;display:flex;color:#5B5B62;padding:10px;justify-content:end;' href='https://deepnote.com?utm_source=created-in-deepnote-cell&projectId=978e47b7-a961-4dca-a945-499e8b781a34' target=\"_blank\">\n<img alt='Created in deepnote.com' style='display:inline;max-height:16px;margin:0px;margin-right:7.5px;' src='data:image/svg+xml;base64,PD94bWwgdmVyc2lvbj0iMS4wIiBlbmNvZGluZz0iVVRGLTgiPz4KPHN2ZyB3aWR0aD0iODBweCIgaGVpZ2h0PSI4MHB4IiB2aWV3Qm94PSIwIDAgODAgODAiIHZlcnNpb249IjEuMSIgeG1sbnM9Imh0dHA6Ly93d3cudzMub3JnLzIwMDAvc3ZnIiB4bWxuczp4bGluaz0iaHR0cDovL3d3dy53My5vcmcvMTk5OS94bGluayI+CiAgICA8IS0tIEdlbmVyYXRvcjogU2tldGNoIDU0LjEgKDc2NDkwKSAtIGh0dHBzOi8vc2tldGNoYXBwLmNvbSAtLT4KICAgIDx0aXRsZT5Hcm91cCAzPC90aXRsZT4KICAgIDxkZXNjPkNyZWF0ZWQgd2l0aCBTa2V0Y2guPC9kZXNjPgogICAgPGcgaWQ9IkxhbmRpbmciIHN0cm9rZT0ibm9uZSIgc3Ryb2tlLXdpZHRoPSIxIiBmaWxsPSJub25lIiBmaWxsLXJ1bGU9ImV2ZW5vZGQiPgogICAgICAgIDxnIGlkPSJBcnRib2FyZCIgdHJhbnNmb3JtPSJ0cmFuc2xhdGUoLTEyMzUuMDAwMDAwLCAtNzkuMDAwMDAwKSI+CiAgICAgICAgICAgIDxnIGlkPSJHcm91cC0zIiB0cmFuc2Zvcm09InRyYW5zbGF0ZSgxMjM1LjAwMDAwMCwgNzkuMDAwMDAwKSI+CiAgICAgICAgICAgICAgICA8cG9seWdvbiBpZD0iUGF0aC0yMCIgZmlsbD0iIzAyNjVCNCIgcG9pbnRzPSIyLjM3NjIzNzYyIDgwIDM4LjA0NzY2NjcgODAgNTcuODIxNzgyMiA3My44MDU3NTkyIDU3LjgyMTc4MjIgMzIuNzU5MjczOSAzOS4xNDAyMjc4IDMxLjY4MzE2ODMiPjwvcG9seWdvbj4KICAgICAgICAgICAgICAgIDxwYXRoIGQ9Ik0zNS4wMDc3MTgsODAgQzQyLjkwNjIwMDcsNzYuNDU0OTM1OCA0Ny41NjQ5MTY3LDcxLjU0MjI2NzEgNDguOTgzODY2LDY1LjI2MTk5MzkgQzUxLjExMjI4OTksNTUuODQxNTg0MiA0MS42NzcxNzk1LDQ5LjIxMjIyODQgMjUuNjIzOTg0Niw0OS4yMTIyMjg0IEMyNS40ODQ5Mjg5LDQ5LjEyNjg0NDggMjkuODI2MTI5Niw0My4yODM4MjQ4IDM4LjY0NzU4NjksMzEuNjgzMTY4MyBMNzIuODcxMjg3MSwzMi41NTQ0MjUgTDY1LjI4MDk3Myw2Ny42NzYzNDIxIEw1MS4xMTIyODk5LDc3LjM3NjE0NCBMMzUuMDA3NzE4LDgwIFoiIGlkPSJQYXRoLTIyIiBmaWxsPSIjMDAyODY4Ij48L3BhdGg+CiAgICAgICAgICAgICAgICA8cGF0aCBkPSJNMCwzNy43MzA0NDA1IEwyNy4xMTQ1MzcsMC4yNTcxMTE0MzYgQzYyLjM3MTUxMjMsLTEuOTkwNzE3MDEgODAsMTAuNTAwMzkyNyA4MCwzNy43MzA0NDA1IEM4MCw2NC45NjA0ODgyIDY0Ljc3NjUwMzgsNzkuMDUwMzQxNCAzNC4zMjk1MTEzLDgwIEM0Ny4wNTUzNDg5LDc3LjU2NzA4MDggNTMuNDE4MjY3Nyw3MC4zMTM2MTAzIDUzLjQxODI2NzcsNTguMjM5NTg4NSBDNTMuNDE4MjY3Nyw0MC4xMjg1NTU3IDM2LjMwMzk1NDQsMzcuNzMwNDQwNSAyNS4yMjc0MTcsMzcuNzMwNDQwNSBDMTcuODQzMDU4NiwzNy43MzA0NDA1IDkuNDMzOTE5NjYsMzcuNzMwNDQwNSAwLDM3LjczMDQ0MDUgWiIgaWQ9IlBhdGgtMTkiIGZpbGw9IiMzNzkzRUYiPjwvcGF0aD4KICAgICAgICAgICAgPC9nPgogICAgICAgIDwvZz4KICAgIDwvZz4KPC9zdmc+' > </img>\nCreated in <span style='font-weight:600;margin-left:4px;'>Deepnote</span></a>",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d0919648df8c4ebbe2f88630a8db07a6dd43dfc7 | 256,451 | ipynb | Jupyter Notebook | 09.Eleitorado_2020/09_csv_eleitores_2020.ipynb | alexfviana/Python_LabHacker | b3c75896be86d660e68ab502cffd96c43df44112 | [
"MIT"
] | null | null | null | 09.Eleitorado_2020/09_csv_eleitores_2020.ipynb | alexfviana/Python_LabHacker | b3c75896be86d660e68ab502cffd96c43df44112 | [
"MIT"
] | null | null | null | 09.Eleitorado_2020/09_csv_eleitores_2020.ipynb | alexfviana/Python_LabHacker | b3c75896be86d660e68ab502cffd96c43df44112 | [
"MIT"
] | 1 | 2021-03-27T17:20:45.000Z | 2021-03-27T17:20:45.000Z | 90.458907 | 119,160 | 0.708389 | [
[
[
"# Análise do eleitorado brasileiro\n\nFonte -> http://www.tse.jus.br/eleicoes/estatisticas/estatisticas-eleitorais",
"_____no_output_____"
]
],
[
[
"# importando as bibliotecas\nimport pandas as pd",
"_____no_output_____"
],
[
"# Carregando o arquivo csv\ndf = pd.read_csv('eleitorado_municipio_2020.csv', encoding='latin1', sep=';')\n\ndf.head().T",
"_____no_output_____"
],
[
"# Tamanho do arquivo\ndf.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 5568 entries, 0 to 5567\nData columns (total 43 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 NR_ANO_ELEICAO 5568 non-null int64 \n 1 CD_PAIS 5568 non-null int64 \n 2 NM_PAIS 5568 non-null object\n 3 SG_REGIAO 5568 non-null object\n 4 NM_REGIAO 5568 non-null object\n 5 SG_UF 5568 non-null object\n 6 NM_UF 5568 non-null object\n 7 CD_MUNICIPIO 5568 non-null int64 \n 8 NM_MUNICIPIO 5568 non-null object\n 9 QTD_ELEITORES 5568 non-null int64 \n 10 QTD_ELEITORES_FEMININO 5568 non-null int64 \n 11 QTD_ELEITORES_MASCULINO 5568 non-null int64 \n 12 QTD_ELEITORES_NAOINFORMADO 5568 non-null int64 \n 13 QTD_ELEITORES_MENOR16 5568 non-null int64 \n 14 QTD_ELEITORES_16 5568 non-null int64 \n 15 QTD_ELEITORES_17 5568 non-null int64 \n 16 QTD_ELEITORES_18 5568 non-null int64 \n 17 QTD_ELEITORES_19 5568 non-null int64 \n 18 QTD_ELEITORES_20 5568 non-null int64 \n 19 QTD_ELEITORES_21A24 5568 non-null int64 \n 20 QTD_ELEITORES_25A29 5568 non-null int64 \n 21 QTD_ELEITORES_30A34 5568 non-null int64 \n 22 QTD_ELEITORES_35A39 5568 non-null int64 \n 23 QTD_ELEITORES_40A44 5568 non-null int64 \n 24 QTD_ELEITORES_45A49 5568 non-null int64 \n 25 QTD_ELEITORES_50A54 5568 non-null int64 \n 26 QTD_ELEITORES_55A59 5568 non-null int64 \n 27 QTD_ELEITORES_60A64 5568 non-null int64 \n 28 QTD_ELEITORES_65A69 5568 non-null int64 \n 29 QTD_ELEITORES_70A74 5568 non-null int64 \n 30 QTD_ELEITORES_75A79 5568 non-null int64 \n 31 QTD_ELEITORES_80A84 5568 non-null int64 \n 32 QTD_ELEITORES_85A89 5568 non-null int64 \n 33 QTD_ELEITORES_90A94 5568 non-null int64 \n 34 QTD_ELEITORES_95A99 5568 non-null int64 \n 35 QTD_ELEITORES_MAIORIGUAL100 5568 non-null int64 \n 36 QTD_ELEITORES_IDADEINVALIDO 5568 non-null int64 \n 37 QTD_ELEITORES_IDADENAOSEAPLICA 5568 non-null int64 \n 38 QTD_ELEITORES_IDADENAOINFORMADA 5568 non-null int64 \n 39 QTD_ELEITORES_COMBIOMETRIA 5568 non-null int64 \n 40 QTD_ELEITORES_SEMBIOMETRIA 5568 non-null int64 \n 41 QTD_ELEITORES_DEFICIENTE 5568 non-null int64 \n 42 QUANTITATIVO_NOMESOCIAL 5568 non-null int64 \ndtypes: int64(37), object(6)\nmemory usage: 1.8+ MB\n"
],
[
"# As cinco cidades com o maior número de eleitores deficientes.\ndf.nlargest(5,'QTD_ELEITORES_DEFICIENTE')",
"_____no_output_____"
],
[
"# As cinco cidades com o menor número de eleitores.\ndf.nsmallest(5,'QTD_ELEITORES')",
"_____no_output_____"
],
[
"# Quantidade de eleitores por gênero\nprint('Eleitoras: ', df['QTD_ELEITORES_FEMININO'].sum())\nprint('Eleitores: ', df['QTD_ELEITORES_MASCULINO'].sum())\nprint('Não informado: ', df['QTD_ELEITORES_NAOINFORMADO'].sum())",
"Eleitoras: 77649569\nEleitores: 70228457\nNão informado: 40457\n"
],
[
"# Criar variáveis para calcular o percentual\ntot_eleitores = df['QTD_ELEITORES'].sum()\ntot_masc = df['QTD_ELEITORES_MASCULINO'].sum()\ntot_fem = df['QTD_ELEITORES_FEMININO'].sum()\ntot_ninf = df['QTD_ELEITORES_NAOINFORMADO'].sum()",
"_____no_output_____"
],
[
"# Mostrar valores percentuais\nprint('Eleitoras: ', ( ( tot_fem / tot_eleitores ) * 100) .round(2), '%' ) \nprint('Eleitores: ', ( ( tot_masc / tot_eleitores ) * 100) .round(2), '%' ) \nprint('Não informado: ', ( ( tot_ninf / tot_eleitores ) * 100) .round(2), '%' ) ",
"Eleitoras: 52.49 %\nEleitores: 47.48 %\nNão informado: 0.03 %\n"
],
[
"# Quantos municípios com mais homens do que mulheres\ndf[df['QTD_ELEITORES_MASCULINO'] > df['QTD_ELEITORES_FEMININO']].count()",
"_____no_output_____"
],
[
"# Vamos criar uma coluna nova para indicar a relação fem/masc\ndf['RELACAO_FM'] = df['QTD_ELEITORES_FEMININO'] / df['QTD_ELEITORES_MASCULINO']",
"_____no_output_____"
],
[
"# Quais os municípios com maior relação fem/masc?\ndf.nlargest(5,'RELACAO_FM').T",
"_____no_output_____"
],
[
"# Quais os municípios com menor relação fem/masc?\ndf.nsmallest(5,'RELACAO_FM')",
"_____no_output_____"
],
[
"# Vamos criar um DataFrame só com municípios de São Paulo\ndf_sp = df[ df['SG_UF'] == 'SP' ].copy()\ndf_sp.head()",
"_____no_output_____"
],
[
"# Quais os municípios com maior relação fem/masc de São Paulo?\ndf_sp.nlargest(5,'RELACAO_FM')",
"_____no_output_____"
],
[
"# Quais os municípios com menor relação fem/masc de São Paulo?\ndf_sp.nsmallest(5,'RELACAO_FM')",
"_____no_output_____"
],
[
"# Plotar um gráfico de distribuição fem/masc\ndf['RELACAO_FM'].plot.hist(bins=100)",
"_____no_output_____"
],
[
"# carregar biblioteca gráfica\nimport seaborn as sns\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"# Plotar um gráfico de distribuição fem/masc\nsns.distplot( df['RELACAO_FM'] , bins=100 , color='red', kde=False )\n\n# Embelezando o gráfico\nplt.title('Relação Eleitoras/Eleitores', fontsize=18)\nplt.xlabel('Eleitoras/Eleitores', fontsize=14)\nplt.ylabel('Frequência', fontsize=14)\nplt.axvline(1.0, color='black', linestyle='--')",
"_____no_output_____"
],
[
"# Plotar um gráfico de distribuição fem/masc, mas mostrando os pontos (municípios)\nsns.swarmplot( data=df , x='NM_REGIAO', y='RELACAO_FM' )\nplt.axhline(1.0, color='black', linestyle='--')",
"_____no_output_____"
],
[
"# Vamos plotar o gráfico de total de eleitores por faixa etária usando o gráfico de barras horizontal\n\n# primeiro vamos listar as colunas de nosso interesse\nlista = ['QTD_ELEITORES_16', 'QTD_ELEITORES_17', 'QTD_ELEITORES_18', 'QTD_ELEITORES_19', 'QTD_ELEITORES_20']\n\ntot_idade = df[lista].sum()",
"_____no_output_____"
],
[
"tot_idade",
"_____no_output_____"
],
[
"# Mostrando o gráfico de barras\n\ntot_idade.plot.barh()",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d09198fa24dc4b84337b7ca271e791bb5bc4687c | 9,938 | ipynb | Jupyter Notebook | dataquest/gun_deaths/.ipynb_checkpoints/Gun_Deaths-checkpoint.ipynb | arturbacu/data-science | 3c59ef788f38d5aa0f179024db1fe3190a684651 | [
"MIT"
] | null | null | null | dataquest/gun_deaths/.ipynb_checkpoints/Gun_Deaths-checkpoint.ipynb | arturbacu/data-science | 3c59ef788f38d5aa0f179024db1fe3190a684651 | [
"MIT"
] | null | null | null | dataquest/gun_deaths/.ipynb_checkpoints/Gun_Deaths-checkpoint.ipynb | arturbacu/data-science | 3c59ef788f38d5aa0f179024db1fe3190a684651 | [
"MIT"
] | null | null | null | 34.034247 | 1,567 | 0.522842 | [
[
[
"## Gun Deaths in the US, 2012-2014\n\nThis is an analysis of gun deaths in the US between the years 2012 and 2014. The data comes from FiveThirtyEight, specifcally [here](https://github.com/fivethirtyeight/guns-data).",
"_____no_output_____"
]
],
[
[
"import csv\n\nf = open(\"guns.csv\", \"r\")\ndata = list(csv.reader(f))\nprint(data[:5])",
"[['', 'year', 'month', 'intent', 'police', 'sex', 'age', 'race', 'hispanic', 'place', 'education'], ['1', '2012', '01', 'Suicide', '0', 'M', '34', 'Asian/Pacific Islander', '100', 'Home', '4'], ['2', '2012', '01', 'Suicide', '0', 'F', '21', 'White', '100', 'Street', '3'], ['3', '2012', '01', 'Suicide', '0', 'M', '60', 'White', '100', 'Other specified', '4'], ['4', '2012', '02', 'Suicide', '0', 'M', '64', 'White', '100', 'Home', '4']]\n"
],
[
"headers = data[0]\ndata = data[1:]\nprint(headers)\nprint(data[:5])",
"['', 'year', 'month', 'intent', 'police', 'sex', 'age', 'race', 'hispanic', 'place', 'education']\n[['1', '2012', '01', 'Suicide', '0', 'M', '34', 'Asian/Pacific Islander', '100', 'Home', '4'], ['2', '2012', '01', 'Suicide', '0', 'F', '21', 'White', '100', 'Street', '3'], ['3', '2012', '01', 'Suicide', '0', 'M', '60', 'White', '100', 'Other specified', '4'], ['4', '2012', '02', 'Suicide', '0', 'M', '64', 'White', '100', 'Home', '4'], ['5', '2012', '02', 'Suicide', '0', 'M', '31', 'White', '100', 'Other specified', '2']]\n"
],
[
"import datetime\n\ndates = [datetime.datetime(year=int(d[1]), month=int(d[2]), day=1) for d in data]\nprint(dates[:5])\n\ndate_counts = {}\nfor d in dates:\n if d in date_counts:\n date_counts[d] += 1\n else:\n date_counts[d] = 1\nprint(date_counts)",
"[datetime.datetime(2012, 1, 1, 0, 0), datetime.datetime(2012, 1, 1, 0, 0), datetime.datetime(2012, 1, 1, 0, 0), datetime.datetime(2012, 2, 1, 0, 0), datetime.datetime(2012, 2, 1, 0, 0)]\n{datetime.datetime(2012, 3, 1, 0, 0): 2743, datetime.datetime(2014, 8, 1, 0, 0): 2970, datetime.datetime(2014, 2, 1, 0, 0): 2361, datetime.datetime(2014, 7, 1, 0, 0): 2884, datetime.datetime(2014, 4, 1, 0, 0): 2862, datetime.datetime(2014, 6, 1, 0, 0): 2931, datetime.datetime(2012, 6, 1, 0, 0): 2826, datetime.datetime(2012, 11, 1, 0, 0): 2729, datetime.datetime(2014, 9, 1, 0, 0): 2914, datetime.datetime(2014, 3, 1, 0, 0): 2684, datetime.datetime(2014, 1, 1, 0, 0): 2651, datetime.datetime(2013, 10, 1, 0, 0): 2808, datetime.datetime(2014, 5, 1, 0, 0): 2864, datetime.datetime(2012, 1, 1, 0, 0): 2758, datetime.datetime(2012, 10, 1, 0, 0): 2733, datetime.datetime(2014, 10, 1, 0, 0): 2865, datetime.datetime(2013, 5, 1, 0, 0): 2806, datetime.datetime(2013, 3, 1, 0, 0): 2862, datetime.datetime(2014, 11, 1, 0, 0): 2756, datetime.datetime(2013, 1, 1, 0, 0): 2864, datetime.datetime(2012, 12, 1, 0, 0): 2791, datetime.datetime(2012, 4, 1, 0, 0): 2795, datetime.datetime(2012, 8, 1, 0, 0): 2954, datetime.datetime(2013, 7, 1, 0, 0): 3079, datetime.datetime(2012, 2, 1, 0, 0): 2357, datetime.datetime(2013, 6, 1, 0, 0): 2920, datetime.datetime(2012, 7, 1, 0, 0): 3026, datetime.datetime(2013, 11, 1, 0, 0): 2758, datetime.datetime(2012, 9, 1, 0, 0): 2852, datetime.datetime(2014, 12, 1, 0, 0): 2857, datetime.datetime(2012, 5, 1, 0, 0): 2999, datetime.datetime(2013, 2, 1, 0, 0): 2375, datetime.datetime(2013, 12, 1, 0, 0): 2765, datetime.datetime(2013, 9, 1, 0, 0): 2742, datetime.datetime(2013, 8, 1, 0, 0): 2859, datetime.datetime(2013, 4, 1, 0, 0): 2798}\n"
],
[
"sex_counts = {}\nfor d in data:\n if d[5] in sex_counts:\n sex_counts[d[5]] += 1\n else:\n sex_counts[d[5]] = 1\n\nrace_counts = {}\nfor d in data:\n if d[7] in race_counts:\n race_counts[d[7]] += 1\n else:\n race_counts[d[7]] = 1\n\nprint(sex_counts)\nprint(race_counts)",
"{'F': 14449, 'M': 86349}\n{'Native American/Native Alaskan': 917, 'Hispanic': 9022, 'White': 66237, 'Asian/Pacific Islander': 1326, 'Black': 23296}\n"
]
],
[
[
"## Patterns so far\n* Most gun deaths are for race = white and sex = M, but is there a further correlation here?\n* It may help to explore this further to find more patterns like age, education status, and if police were involved",
"_____no_output_____"
]
],
[
[
"f2 = open(\"census.csv\", \"r\")\ncensus = list(csv.reader(f2))\ncensus",
"_____no_output_____"
],
[
"mapping = {\"Asian/Pacific Islander\": 15834141, \n \"Black\": 40250635, \n \"Native American/Native Alaskan\": 3739506, \n \"Hispanic\": 44618105, \n \"White\": 197318956}\nrace_per_hundredk = {}\nfor r, v in race_counts.items():\n race_per_hundredk[r] = (v / mapping[r]) * 100000\nprint(race_per_hundredk)",
"{'Native American/Native Alaskan': 24.521955573811088, 'Hispanic': 20.220491210910907, 'White': 33.56849303419181, 'Asian/Pacific Islander': 8.374309664161762, 'Black': 57.8773477735196}\n"
],
[
"intent = [d[3] for d in data]\nraces = [d[7] for d in data]\nhomicide_race_counts = {}\nfor i, race in enumerate(races):\n if intent[i] == \"Suicide\":\n if race in homicide_race_counts:\n homicide_race_counts[race] += 1\n else:\n homicide_race_counts[race] = 1\nhomicide_race_per_hundredk = {}\nfor r, v in homicide_race_counts.items():\n homicide_race_per_hundredk[r] = (v / mapping[r]) * 100000\nprint(homicide_race_per_hundredk)",
"{'Native American/Native Alaskan': 14.841532544673013, 'Hispanic': 7.106980451097149, 'White': 28.06217969245692, 'Asian/Pacific Islander': 4.705023152187416, 'Black': 8.278130270491385}\n"
]
],
[
[
"# More findings\n* If Homicide is the intent, the black race is significantly more likely to be affected by gun death (over 10 times that of whites and 4 times that of Hispanic)\n* If Suicide is the intent, the white race is significantly more likely to be affected by gun death (2 times that of Native American and over 3 times that of Black or Hispanic)\n\n# Next steps\n* Investigate further if/where age comes into play as well as education",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
d0919a839265628cb87ae8b2d6826350419ba449 | 16,313 | ipynb | Jupyter Notebook | NoteBooks/Curso de WebScraping/Unificado/web-scraping-master/Clases/Módulo 3_ Scraping con Selenium/M3C5. Demoras inteligentes - Script.ipynb | Alejandro-sin/Learning_Notebooks | 161d6bed4c7b1d171b45f61c0cc6fa91e9894aad | [
"MIT"
] | 1 | 2021-02-26T13:12:22.000Z | 2021-02-26T13:12:22.000Z | NoteBooks/Curso de WebScraping/Unificado/web-scraping-master/Clases/Módulo 3_ Scraping con Selenium/M3C5. Demoras inteligentes - Script.ipynb | Alejandro-sin/Learning_Notebooks | 161d6bed4c7b1d171b45f61c0cc6fa91e9894aad | [
"MIT"
] | null | null | null | NoteBooks/Curso de WebScraping/Unificado/web-scraping-master/Clases/Módulo 3_ Scraping con Selenium/M3C5. Demoras inteligentes - Script.ipynb | Alejandro-sin/Learning_Notebooks | 161d6bed4c7b1d171b45f61c0cc6fa91e9894aad | [
"MIT"
] | null | null | null | 29.078431 | 310 | 0.560105 | [
[
[
"# Módulo 2: Scraping con Selenium\n## LATAM Airlines\n<a href=\"https://www.latam.com/es_ar/\"><img src=\"https://i.pinimg.com/originals/dd/52/74/dd5274702d1382d696caeb6e0f6980c5.png\" width=\"420\"></img></a>\n<br>\n\nVamos a scrapear el sitio de Latam para averiguar datos de vuelos en funcion el origen y destino, fecha y cabina. La información que esperamos obtener de cada vuelo es:\n- Precio(s) disponibles\n- Horas de salida y llegada (duración)\n- Información de las escalas\n\n¡Empecemos!",
"_____no_output_____"
]
],
[
[
"url = 'https://www.latam.com/es_ar/apps/personas/booking?fecha1_dia=20&fecha1_anomes=2019-12&auAvailability=1&ida_vuelta=ida&vuelos_origen=Buenos%20Aires&from_city1=BUE&vuelos_destino=Madrid&to_city1=MAD&flex=1&vuelos_fecha_salida_ddmmaaaa=20/12/2019&cabina=Y&nadults=1&nchildren=0&ninfants=0&cod_promo='",
"_____no_output_____"
],
[
"from selenium import webdriver",
"_____no_output_____"
],
[
"options = webdriver.ChromeOptions()\noptions.add_argument('--incognito')\ndriver = webdriver.Chrome(executable_path='../../chromedriver', options=options)\ndriver.get(url)",
"_____no_output_____"
],
[
"#Usaremos el Xpath para obtener la lista de vuelos\nvuelos = driver.find_elements_by_xpath('//li[@class=\"flight\"]')",
"_____no_output_____"
],
[
"vuelo = vuelos[0]",
"_____no_output_____"
]
],
[
[
"Obtenemos la información de la hora de salida, llegada y duración del vuelo",
"_____no_output_____"
]
],
[
[
"# Hora de salida\nvuelo.find_element_by_xpath('.//div[@class=\"departure\"]/time').get_attribute('datetime')",
"_____no_output_____"
],
[
"# Hora de llegada\nvuelo.find_element_by_xpath('.//div[@class=\"arrival\"]/time').get_attribute('datetime')",
"_____no_output_____"
],
[
"# Duración del vuelo\nvuelo.find_element_by_xpath('.//span[@class=\"duration\"]/time').get_attribute('datetime')",
"_____no_output_____"
],
[
"boton_escalas = vuelo.find_element_by_xpath('.//div[@class=\"flight-summary-stops-description\"]/button')\nboton_escalas",
"_____no_output_____"
],
[
"boton_escalas.click()",
"_____no_output_____"
],
[
"segmentos = vuelo.find_elements_by_xpath('//div[@class=\"segments-graph\"]/div[@class=\"segments-graph-segment\"]')\nsegmentos",
"_____no_output_____"
],
[
"escalas = len(segmentos) - 1 #0 escalas si es un vuelo directo",
"_____no_output_____"
],
[
"segmento = segmentos[0]",
"_____no_output_____"
],
[
"# Origen\nsegmento.find_element_by_xpath('.//div[@class=\"departure\"]/span[@class=\"ground-point-name\"]').text",
"_____no_output_____"
],
[
"# Hora de salida\nsegmento.find_element_by_xpath('.//div[@class=\"departure\"]/time').get_attribute('datetime')",
"_____no_output_____"
],
[
"# Destino\nsegmento.find_element_by_xpath('.//div[@class=\"arrival\"]/span[@class=\"ground-point-name\"]').text",
"_____no_output_____"
],
[
"# Hora de llegada\nsegmento.find_element_by_xpath('.//div[@class=\"arrival\"]/time').get_attribute('datetime')",
"_____no_output_____"
],
[
"# Duración del vuelo\nsegmento.find_element_by_xpath('.//span[@class=\"duration flight-schedule-duration\"]/time').get_attribute('datetime')",
"_____no_output_____"
],
[
"# Numero del vuelo\nsegmento.find_element_by_xpath('.//span[@class=\"equipment-airline-number\"]').text",
"_____no_output_____"
],
[
"# Modelo de avion\nsegmento.find_element_by_xpath('.//span[@class=\"equipment-airline-material\"]').text",
"_____no_output_____"
],
[
"# Duracion de la escala\nsegmento.find_element_by_xpath('.//div[@class=\"stop connection\"]//p[@class=\"stop-wait-time\"]//time').get_attribute('datetime')",
"_____no_output_____"
],
[
"vuelo.find_element_by_xpath('//div[@class=\"modal-dialog\"]//button[@class=\"close\"]').click()",
"_____no_output_____"
],
[
"vuelo.click()",
"_____no_output_____"
],
[
"tarifas = vuelo.find_elements_by_xpath('.//div[@class=\"fares-table-container\"]//tfoot//td[contains(@class, \"fare-\")]')",
"_____no_output_____"
],
[
"precios = []\nfor tarifa in tarifas:\n nombre = tarifa.find_element_by_xpath('.//label').get_attribute('for')\n moneda = tarifa.find_element_by_xpath('.//span[@class=\"price\"]/span[@class=\"currency-symbol\"]').text\n valor = tarifa.find_element_by_xpath('.//span[@class=\"price\"]/span[@class=\"value\"]').text \n dict_tarifa={nombre:{'moneda':moneda, 'valor':valor}}\n precios.append(dict_tarifa)\n print(dict_tarifa)",
"_____no_output_____"
],
[
"def obtener_tiempos(vuelo):\n # Hora de salida\n salida = vuelo.find_element_by_xpath('.//div[@class=\"departure\"]/time').get_attribute('datetime')\n # Hora de llegada\n llegada = vuelo.find_element_by_xpath('.//div[@class=\"arrival\"]/time').get_attribute('datetime')\n # Duracion\n duracion = vuelo.find_element_by_xpath('.//span[@class=\"duration\"]/time').get_attribute('datetime')\n return {'hora_salida': salida, 'hora_llegada': llegada, 'duracion': duracion}",
"_____no_output_____"
],
[
"def obtener_precios(vuelo):\n tarifas = vuelo.find_elements_by_xpath(\n './/div[@class=\"fares-table-container\"]//tfoot//td[contains(@class, \"fare-\")]')\n precios = []\n for tarifa in tarifas:\n nombre = tarifa.find_element_by_xpath('.//label').get_attribute('for')\n moneda = tarifa.find_element_by_xpath('.//span[@class=\"price\"]/span[@class=\"currency-symbol\"]').text\n valor = tarifa.find_element_by_xpath('.//span[@class=\"price\"]/span[@class=\"value\"]').text \n dict_tarifa={nombre:{'moneda':moneda, 'valor':valor}}\n precios.append(dict_tarifa)\n return precios",
"_____no_output_____"
],
[
"def obtener_datos_escalas(vuelo):\n segmentos = vuelo.find_elements_by_xpath('//div[@class=\"segments-graph\"]/div[@class=\"segments-graph-segment\"]')\n info_escalas = []\n for segmento in segmentos:\n # Origen\n origen = segmento.find_element_by_xpath(\n './/div[@class=\"departure\"]/span[@class=\"ground-point-name\"]').text\n # Hora de salida\n dep_time = segmento.find_element_by_xpath(\n './/div[@class=\"departure\"]/time').get_attribute('datetime')\n # Destino\n destino = segmento.find_element_by_xpath(\n './/div[@class=\"arrival\"]/span[@class=\"ground-point-name\"]').text\n # Hora de llegada\n arr_time = segmento.find_element_by_xpath(\n './/div[@class=\"arrival\"]/time').get_attribute('datetime')\n # Duración del vuelo\n duracion_vuelo = segmento.find_element_by_xpath(\n './/span[@class=\"duration flight-schedule-duration\"]/time').get_attribute('datetime')\n # Numero del vuelo\n numero_vuelo = segmento.find_element_by_xpath(\n './/span[@class=\"equipment-airline-number\"]').text\n # Modelo de avion\n modelo_avion = segmento.find_element_by_xpath(\n './/span[@class=\"equipment-airline-material\"]').text\n # Duracion de la escala\n if segmento != segmentos[-1]:\n duracion_escala = segmento.find_element_by_xpath(\n './/div[@class=\"stop connection\"]//p[@class=\"stop-wait-time\"]//time').get_attribute('datetime')\n else:\n duracion_escala = ''\n\n # Armo un diccionario para almacenar los datos\n data_dict={'origen': origen, \n 'dep_time': dep_time, \n 'destino': destino,\n 'arr_time': arr_time,\n 'duracion_vuelo': duracion_vuelo,\n 'numero_vuelo': numero_vuelo,\n 'modelo_avion': modelo_avion,\n 'duracion_escala': duracion_escala}\n info_escalas.append(data_dict)\n \n return info_escalas",
"_____no_output_____"
]
],
[
[
"## Clase 15\nYa tenemos el scraper casi listo. Unifiquemos las 3 funciones de la clase anterior en una sola",
"_____no_output_____"
]
],
[
[
"def obtener_info(driver):\n vuelos = driver.find_elements_by_xpath('//li[@class=\"flight\"]')\n print(f'Se encontraron {len(vuelos)} vuelos.')\n print('Iniciando scraping...')\n info = []\n for vuelo in vuelos:\n # Obtenemos los tiempos generales del vuelo\n tiempos = obtener_tiempos(vuelo)\n # Clickeamos el botón de escalas para ver los detalles\n vuelo.find_element_by_xpath('.//div[@class=\"flight-summary-stops-description\"]/button').click()\n escalas = obtener_datos_escalas(vuelo)\n # Cerramos el pop-up con los detalles\n vuelo.find_element_by_xpath('//div[@class=\"modal-dialog\"]//button[@class=\"close\"]').click()\n # Clickeamos el vuelo para ver los precios\n vuelo.click()\n precios = obtener_precios(vuelo)\n # Cerramos los precios del vuelo\n vuelo.click()\n info.append({'precios':precios, 'tiempos':tiempos , 'escalas': escalas})\n return info",
"_____no_output_____"
]
],
[
[
"Ahora podemos cargar la página con el driver y pasárselo a esta función",
"_____no_output_____"
]
],
[
[
"driver = webdriver.Chrome(executable_path='../../chromedriver', options=options)\ndriver.get(url)\nobtener_info(driver)",
"_____no_output_____"
]
],
[
[
"Se encontraron 0 vuelos porque la página no terminó de cargar",
"_____no_output_____"
],
[
"Lo más simple que podemos hacer es agregar una demora fija lo suficientemente grande para asegurarnos que la página terminó de cargar.",
"_____no_output_____"
]
],
[
[
"import time",
"_____no_output_____"
],
[
"options = webdriver.ChromeOptions()\noptions.add_argument('--incognito')\ndriver = webdriver.Chrome(executable_path='../../chromedriver', options=options)\ndriver.get(url)",
"_____no_output_____"
],
[
"time.sleep(10)",
"_____no_output_____"
],
[
"vuelos = driver.find_elements_by_xpath('//li[@class=\"flight\"]')\nvuelos",
"_____no_output_____"
],
[
"driver.close()",
"_____no_output_____"
]
],
[
[
"Esto funciona pero no es muy eficiente. Lo mejor sería esperar a que la página termine de cargar y luego recuperar los elementos.",
"_____no_output_____"
]
],
[
[
"from selenium.webdriver.support.ui import WebDriverWait\nfrom selenium.webdriver.support import expected_conditions as EC\nfrom selenium.webdriver.common.by import By\nfrom selenium.common.exceptions import TimeoutException",
"_____no_output_____"
],
[
"options = webdriver.ChromeOptions()\noptions.add_argument('--incognito')\ndriver = webdriver.Chrome(executable_path='../../chromedriver', options=options)\ndriver.get(url)\ndelay = 10\ntry:\n vuelo = WebDriverWait(driver, delay).until(EC.presence_of_element_located((By.XPATH, '//li[@class=\"flight\"]')))\n print(\"La página terminó de cargar\")\n info_vuelos = obtener_info(driver)\nexcept TimeoutException:\n print(\"La página tardó demasiado en cargar\")\ndriver.close()",
"_____no_output_____"
],
[
"info_vuelos",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d091ae9bc88d6e27db0efb4e6857ae4df3e58241 | 13,209 | ipynb | Jupyter Notebook | Reinforcement_learning/UCB_model.ipynb | likki0207/Machine-learning-projects | 74c62a60ae6f4dc2f0d7cc7236dbdebf743d9cce | [
"CC0-1.0"
] | null | null | null | Reinforcement_learning/UCB_model.ipynb | likki0207/Machine-learning-projects | 74c62a60ae6f4dc2f0d7cc7236dbdebf743d9cce | [
"CC0-1.0"
] | null | null | null | Reinforcement_learning/UCB_model.ipynb | likki0207/Machine-learning-projects | 74c62a60ae6f4dc2f0d7cc7236dbdebf743d9cce | [
"CC0-1.0"
] | null | null | null | 121.183486 | 10,752 | 0.889772 | [
[
[
"# Importing the required libraries\nimport math\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"# Importing the dataset\ndf=pd.read_csv('Ads_CTR_Optimisation.csv')",
"_____no_output_____"
],
[
"# Implementing Upper confidence bound model\nN=10000\nd=10\ntotal_ads_selected=[]\ntotal_selections=[0]*d\nreward_sum=[0]*d\ntotal_reward=0\nfor n in range(0,N):\n ad=0\n max_upper_bound=0\n for i in range(0,d):\n if (total_selections[i]>0):\n avg_reward=reward_sum[i]/total_selections[i]\n deltai=math.sqrt(3/2*math.log(n+1)/total_selections[i])\n upper_bound=avg_reward+deltai\n else:\n upper_bound=1e400\n if (upper_bound>max_upper_bound):\n max_upper_bound=upper_bound\n ad=i\n total_ads_selected.append(ad)\n total_selections[ad]=total_selections[ad]+1\n reward=df.values[n,ad]\n reward_sum[ad]=reward_sum[ad]+reward\n total_reward=total_reward+reward",
"_____no_output_____"
],
[
"# Visualising the results\n\nplt.hist(total_ads_selected)\nplt.xlabel('Advertisements')\nplt.ylabel('Total number of times each ad was clicked')\nplt.show()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code"
]
] |
d091bb5f2b540af164218b26167c504c793243e7 | 240,697 | ipynb | Jupyter Notebook | 1_APIs/3_api_workbook.ipynb | ucla-data-archive/scrape-interwebz | d6af70491ec09e71422ec69de447abd673408344 | [
"MIT"
] | 2 | 2021-02-09T18:07:29.000Z | 2021-02-16T22:13:24.000Z | 1_APIs/3_api_workbook.ipynb | ucla-data-archive/scrape-interwebz | d6af70491ec09e71422ec69de447abd673408344 | [
"MIT"
] | null | null | null | 1_APIs/3_api_workbook.ipynb | ucla-data-archive/scrape-interwebz | d6af70491ec09e71422ec69de447abd673408344 | [
"MIT"
] | null | null | null | 184.019113 | 183,280 | 0.908823 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d091c9ff42dab48a4374e15853a948694cac5de2 | 21,675 | ipynb | Jupyter Notebook | Data Science Professional/2 - Python For Data Science/5.2 Numpy2D.ipynb | 2series/DataScience-Courses | 5ee71305721a61dfc207d8d7de67a9355530535d | [
"MIT"
] | 1 | 2020-02-23T07:40:39.000Z | 2020-02-23T07:40:39.000Z | Data Science Professional/2 - Python For Data Science/5.2 Numpy2D.ipynb | 2series/DataScience-Courses | 5ee71305721a61dfc207d8d7de67a9355530535d | [
"MIT"
] | null | null | null | Data Science Professional/2 - Python For Data Science/5.2 Numpy2D.ipynb | 2series/DataScience-Courses | 5ee71305721a61dfc207d8d7de67a9355530535d | [
"MIT"
] | 3 | 2019-12-05T11:04:58.000Z | 2020-02-26T10:42:08.000Z | 21.567164 | 716 | 0.507636 | [
[
[
"## RIHAD VARIAWA, Data Scientist - Who has fun LEARNING, EXPLORING & GROWING\n<h1>2D <code>Numpy</code> in Python</h1>",
"_____no_output_____"
],
[
"<p><strong>Welcome!</strong> This notebook will teach you about using <code>Numpy</code> in the Python Programming Language. By the end of this lab, you'll know what <code>Numpy</code> is and the <code>Numpy</code> operations.</p>",
"_____no_output_____"
],
[
"<h2>Table of Contents</h2>\n<div class=\"alert alert-block alert-info\" style=\"margin-top: 20px\">\n <ul>\n <li><a href=\"create\">Create a 2D Numpy Array</a></li>\n <li><a href=\"access\">Accessing different elements of a Numpy Array</a></li>\n <li><a href=\"op\">Basic Operations</a></li>\n </ul>\n <p>\n Estimated time needed: <strong>20 min</strong>\n </p>\n</div>\n\n<hr>",
"_____no_output_____"
],
[
"<h2 id=\"create\">Create a 2D Numpy Array</h2>",
"_____no_output_____"
]
],
[
[
"# Import the libraries\n\nimport numpy as np \nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"Consider the list <code>a</code>, the list contains three nested lists **each of equal size**. ",
"_____no_output_____"
]
],
[
[
"# Create a list\n\na = [[11, 12, 13], [21, 22, 23], [31, 32, 33]]\na",
"_____no_output_____"
]
],
[
[
"We can cast the list to a Numpy Array as follow",
"_____no_output_____"
]
],
[
[
"# Convert list to Numpy Array\n# Every element is the same type\n\nA = np.array(a)\nA",
"_____no_output_____"
]
],
[
[
"We can use the attribute <code>ndim</code> to obtain the number of axes or dimensions referred to as the rank. ",
"_____no_output_____"
]
],
[
[
"# Show the numpy array dimensions\n\nA.ndim",
"_____no_output_____"
]
],
[
[
"Attribute <code>shape</code> returns a tuple corresponding to the size or number of each dimension.",
"_____no_output_____"
]
],
[
[
"# Show the numpy array shape\n\nA.shape",
"_____no_output_____"
]
],
[
[
"The total number of elements in the array is given by the attribute <code>size</code>.",
"_____no_output_____"
]
],
[
[
"# Show the numpy array size\n\nA.size",
"_____no_output_____"
]
],
[
[
"<hr>",
"_____no_output_____"
],
[
"<h2 id=\"access\">Accessing different elements of a Numpy Array</h2>",
"_____no_output_____"
],
[
"We can use rectangular brackets to access the different elements of the array. The correspondence between the rectangular brackets and the list and the rectangular representation is shown in the following figure for a 3x3 array: ",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%205/Images/NumTwoEg.png\" width=\"500\" />",
"_____no_output_____"
],
[
"We can access the 2nd-row 3rd column as shown in the following figure:",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%205/Images/NumTwoFT.png\" width=\"400\" />",
"_____no_output_____"
],
[
" We simply use the square brackets and the indices corresponding to the element we would like:",
"_____no_output_____"
]
],
[
[
"# Access the element on the second row and third column\n\nA[1, 2]",
"_____no_output_____"
]
],
[
[
" We can also use the following notation to obtain the elements: ",
"_____no_output_____"
]
],
[
[
"# Access the element on the second row and third column\n\nA[1][2]",
"_____no_output_____"
]
],
[
[
" Consider the elements shown in the following figure ",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%205/Images/NumTwoFF.png\" width=\"400\" />",
"_____no_output_____"
],
[
"We can access the element as follows ",
"_____no_output_____"
]
],
[
[
"# Access the element on the first row and first column\n\nA[0][0]",
"_____no_output_____"
]
],
[
[
"We can also use slicing in numpy arrays. Consider the following figure. We would like to obtain the first two columns in the first row",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%205/Images/NumTwoFSF.png\" width=\"400\" />",
"_____no_output_____"
],
[
" This can be done with the following syntax ",
"_____no_output_____"
]
],
[
[
"# Access the element on the first row and first and second columns\n\nA[0][0:2]",
"_____no_output_____"
]
],
[
[
"Similarly, we can obtain the first two rows of the 3rd column as follows:",
"_____no_output_____"
]
],
[
[
"# Access the element on the first and second rows and third column\n\nA[0:2, 2]",
"_____no_output_____"
]
],
[
[
"Corresponding to the following figure: ",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%205/Images/NumTwoTST.png\" width=\"400\" />",
"_____no_output_____"
],
[
"<hr>",
"_____no_output_____"
],
[
"<h2 id=\"op\">Basic Operations</h2>",
"_____no_output_____"
],
[
"We can also add arrays. The process is identical to matrix addition. Matrix addition of <code>X</code> and <code>Y</code> is shown in the following figure:",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%205/Images/NumTwoAdd.png\" width=\"500\" />",
"_____no_output_____"
],
[
"The numpy array is given by <code>X</code> and <code>Y</code>",
"_____no_output_____"
]
],
[
[
"# Create a numpy array X\n\nX = np.array([[1, 0], [0, 1]]) \nX",
"_____no_output_____"
],
[
"# Create a numpy array Y\n\nY = np.array([[2, 1], [1, 2]]) \nY",
"_____no_output_____"
]
],
[
[
" We can add the numpy arrays as follows.",
"_____no_output_____"
]
],
[
[
"# Add X and Y\n\nZ = X + Y\nZ",
"_____no_output_____"
]
],
[
[
"Multiplying a numpy array by a scaler is identical to multiplying a matrix by a scaler. If we multiply the matrix <code>Y</code> by the scaler 2, we simply multiply every element in the matrix by 2 as shown in the figure.",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%205/Images/NumTwoDb.png\" width=\"500\" />",
"_____no_output_____"
],
[
"We can perform the same operation in numpy as follows ",
"_____no_output_____"
]
],
[
[
"# Create a numpy array Y\n\nY = np.array([[2, 1], [1, 2]]) \nY",
"_____no_output_____"
],
[
"# Multiply Y with 2\n\nZ = 2 * Y\nZ",
"_____no_output_____"
]
],
[
[
"Multiplication of two arrays corresponds to an element-wise product or Hadamard product. Consider matrix <code>X</code> and <code>Y</code>. The Hadamard product corresponds to multiplying each of the elements in the same position, i.e. multiplying elements contained in the same color boxes together. The result is a new matrix that is the same size as matrix <code>Y</code> or <code>X</code>, as shown in the following figure.",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%205/Images/NumTwoMul.png\" width=\"500\" />",
"_____no_output_____"
],
[
"We can perform element-wise product of the array <code>X</code> and <code>Y</code> as follows:",
"_____no_output_____"
]
],
[
[
"# Create a numpy array Y\n\nY = np.array([[2, 1], [1, 2]]) \nY",
"_____no_output_____"
],
[
"# Create a numpy array X\n\nX = np.array([[1, 0], [0, 1]]) \nX",
"_____no_output_____"
],
[
"# Multiply X with Y\n\nZ = X * Y\nZ",
"_____no_output_____"
]
],
[
[
"We can also perform matrix multiplication with the numpy arrays <code>A</code> and <code>B</code> as follows:",
"_____no_output_____"
],
[
"First, we define matrix <code>A</code> and <code>B</code>:",
"_____no_output_____"
]
],
[
[
"# Create a matrix A\n\nA = np.array([[0, 1, 1], [1, 0, 1]])\nA",
"_____no_output_____"
],
[
"# Create a matrix B\n\nB = np.array([[1, 1], [1, 1], [-1, 1]])\nB",
"_____no_output_____"
]
],
[
[
"We use the numpy function <code>dot</code> to multiply the arrays together.",
"_____no_output_____"
]
],
[
[
"# Calculate the dot product\n\nZ = np.dot(A,B)\nZ",
"_____no_output_____"
],
[
"# Calculate the sine of Z\n\nnp.sin(Z)",
"_____no_output_____"
]
],
[
[
"We use the numpy attribute <code>T</code> to calculate the transposed matrix",
"_____no_output_____"
]
],
[
[
"# Create a matrix C\n\nC = np.array([[1,1],[2,2],[3,3]])\nC",
"_____no_output_____"
],
[
"# Get the transposed of C\n\nC.T",
"_____no_output_____"
]
],
[
[
"<hr>\n<h2>The last exercise!</h2>\n<p>Congratulations, you have completed your first lesson and hands-on lab in Python. However, there is one more thing you need to do. The Data Science community encourages sharing work. The best way to share and showcase your work is to share it on GitHub. By sharing your notebook on GitHub you are not only building your reputation with fellow data scientists, but you can also show it off when applying for a job. Even though this was your first piece of work, it is never too early to start building good habits. So, please read and follow <a href=\"https://cognitiveclass.ai/blog/data-scientists-stand-out-by-sharing-your-notebooks/\" target=\"_blank\">this article</a> to learn how to share your work.\n<hr>",
"_____no_output_____"
],
[
"<div class=\"alert alert-block alert-info\" style=\"margin-top: 20px\">\n<h2>Get IBM Watson Studio free of charge!</h2>\n <p><a href=\"https://cocl.us/bottemNotebooksPython101Coursera\"><img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Ad/BottomAd.png\" width=\"750\" align=\"center\"></a></p>\n</div>",
"_____no_output_____"
],
[
"<h3>About the Authors:</h3> \n<p><a href=\"https://www.linkedin.com/in/joseph-s-50398b136/\" target=\"_blank\">Joseph Santarcangelo</a> is a Data Scientist at IBM, and holds a PhD in Electrical Engineering. His research focused on using Machine Learning, Signal Processing, and Computer Vision to determine how videos impact human cognition. Joseph has been working for IBM since he completed his PhD.</p>",
"_____no_output_____"
],
[
"Other contributors: <a href=\"www.linkedin.com/in/jiahui-mavis-zhou-a4537814a\">Mavis Zhou</a>",
"_____no_output_____"
],
[
"<hr>",
"_____no_output_____"
],
[
"<p>Copyright © 2018 IBM Developer Skills Network. This notebook and its source code are released under the terms of the <a href=\"https://cognitiveclass.ai/mit-license/\">MIT License</a>.</p>",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d091d1b6d8fbe4b5a0d34581c663d5bfab01aba5 | 10,155 | ipynb | Jupyter Notebook | rf_master.ipynb | MidoriR/Antimicrobial_compound_classifier | 7b6d0e469ea0fb1b5b72f208d9a0ad393485b602 | [
"MIT"
] | null | null | null | rf_master.ipynb | MidoriR/Antimicrobial_compound_classifier | 7b6d0e469ea0fb1b5b72f208d9a0ad393485b602 | [
"MIT"
] | null | null | null | rf_master.ipynb | MidoriR/Antimicrobial_compound_classifier | 7b6d0e469ea0fb1b5b72f208d9a0ad393485b602 | [
"MIT"
] | null | null | null | 28.525281 | 726 | 0.579911 | [
[
[
"# Master Notebook: Bosques aleatorios",
"_____no_output_____"
],
[
"Como ya vímos en scikit-learn gran parte de codigo es reciclable. Particularmente, leyendo variables y preparando los datos es lo mismo, independientemente del clasificador que usamos.",
"_____no_output_____"
],
[
"## Leyendo datos",
"_____no_output_____"
],
[
"Para que no esta tan aburrido (también para mi) esta vez nos vamos a escribir una función para leer los datos que podemos reciclar para otros datos.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom collections import Counter\n\ndef lea_datos(archivo, i_clase=-1, encabezado=True, delim=\",\"):\n '''Una funcion para leer archivos con datos de clasificación. \n Argumentos:\n archivo - direccion del archivo\n i_clase - indice de columna que contiene las clases. \n default es -1 y significa la ultima fila.\n header - si hay un encabezado\n delim - que separa los datos\n Regresa:\n Un tuple de los datos, clases y cabezazo en caso que hay.'''\n \n todo = np.loadtxt(archivo, dtype=\"S\", delimiter=delim) # para csv\n if(encabezado):\n encabezado = todo[0,:]\n todo = todo[1:,:]\n else: \n encabezado = None\n \n clases = todo[:, i_clase]\n datos = np.delete(todo, i_clase, axis=1)\n print (\"Clases\")\n for k,v in Counter(clases).items(): print (k,\":\",v)\n return (datos, clases, encabezado) ",
"_____no_output_____"
]
],
[
[
"Ahora importando datos se hace muy simple en el futuro. Para datos de csv con cabezazo por ejemplo:",
"_____no_output_____"
]
],
[
[
"datos, clases, encabezado = lea_datos(\"datos_peña.csv\") # _ significa que no nos interesa este valor \nclases.shape",
"Clases\nb'1' : 9\nb'0' : 14\n"
]
],
[
[
"## Balanceando datos",
"_____no_output_____"
],
[
"Normalizacion de datos no es necesario para bosques aleatorios o arboles porque son invariantes al respeto de los magnitudes de variables. Lo unico que podría ser un problema es los clases son imbalanceados bajo nuestra percepción. Esto significa que a veces ni esperamos que nuestros datos van a ser balanceados y, en este caso, es permisible dejarlos sin balancear porque queremos que el clasificador incluye este probabilidad \"prior\" de la vida real. Si por ejemplo clasifico por ejemplo evaluaciones del vino, puede ser que lo encuentro normal que muchos vinos tienen una evaluacion promedia y solamente poco una muy buena. Si quiero que mi clasificador también tenga esta \"tendencía\" no tengo que balancear. \nEn otro caso, si diseño una prueba para una enfermeda y no quiero que mi clasificador hace asumpciones sobre el estado del paciente no basados en mis variables, mejor balanceo.\n\nScikit-learn no llega con una función para balancear datos entonces porque no lo hacemos nosotros?",
"_____no_output_____"
]
],
[
[
"def balance(datos, clases, estrategia=\"down\"):\n '''Balancea unos datos así que cada clase aparece en las mismas proporciones.\n Argumentos:\n datos - los datos. Filas son muestras y columnas variables.\n clases - las clases para cada muestra.\n estrategia - \"up\" para up-scaling y \"down\" para down-scaling'''\n \n import numpy as np\n from collections import Counter\n \n # Decidimos los nuevos numeros de muestras para cada clase\n conteos = Counter(clases)\n if estrategia==\"up\": muestras = max( conteos.values() )\n else: muestras = min( conteos.values() )\n \n datos_nuevo = np.array([]).reshape( 0, datos.shape[1] )\n clases_nuevo = []\n \n for c in conteos:\n c_i = np.where(clases==c)[0]\n new_i = np.random.choice(c_i, muestras, replace=(estrategia==\"up\") )\n datos_nuevo = np.append( datos_nuevo, datos[new_i,:], axis=0 )\n clases_nuevo = np.append( clases_nuevo, clases[new_i] )\n \n return (datos_nuevo, clases_nuevo)\n ",
"_____no_output_____"
]
],
[
[
"Y vamos a ver si funciona...",
"_____no_output_____"
]
],
[
[
"datos_b, clases_b = balance(datos, clases)\nprint Counter(clases_b)\nprint datos_b.shape",
"_____no_output_____"
]
],
[
[
"## Entrenando el bosque",
"_____no_output_____"
],
[
"Corriendo un bosque aleatorio para la clasificación es casi lo mismo como todos los otros clasificadores. Nada más importamos de una libreria diferente.",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import RandomForestClassifier\n\nclf = RandomForestClassifier(oob_score=True)\nclf.fit(datos_b, clases_b)\nprint \"Exactitud estimado:\", clf.oob_score_",
"_____no_output_____"
]
],
[
[
"Ya sabemos que los hiper-parametros mas importantes para los bosques son el numero de arboles dado por `n_estimators` y la profundidad del arbol dado por `max_depth`. Porque son monotonos los podemos variar por separado. ",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nfrom sklearn.cross_validation import StratifiedKFold\nfrom sklearn.grid_search import GridSearchCV\nimport matplotlib.pyplot as plt\n\ncv = StratifiedKFold(y=clases, n_folds=4)\narboles_vals = np.arange(5,200,5)\nbusqueda = GridSearchCV(clf, param_grid=dict(n_estimators=arboles_vals), cv=cv)\nbusqueda.fit(datos, clases)\n\nprint 'Mejor numero de arboles=',busqueda.best_params_,',exactitud =',busqueda.best_score_\n\nscores = [x[1] for x in busqueda.grid_scores_]\n\nplt.plot(arboles_vals, scores)\nplt.xlabel('C')\nplt.ylabel('exactitud cv')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Y para la profundidad:",
"_____no_output_____"
]
],
[
[
"prof_vals = np.arange(1,12)\nbusqueda = GridSearchCV(clf, param_grid=dict(max_depth=prof_vals), cv=cv)\nbusqueda.fit(datos, clases)\n\nprint 'Mejor profundidad=',busqueda.best_params_,',exactitud =',busqueda.best_score_\n\nscores = [x[1] for x in busqueda.grid_scores_]\nplt.plot(prof_vals, scores)\nplt.xlabel('profundidad maxima')\nplt.ylabel('exactitud cv')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Viendo unas buenas valores podemos escoger un bosque con la menor numero de arboles y profundidad para una buena exactitud. También esta vez vamos a sacar las importancias de variables de una vez.",
"_____no_output_____"
]
],
[
[
"clf = RandomForestClassifier(n_estimators=101, oob_score=True) \nclf.fit(datos, clases)\nprint \"Exactitud estimada:\", clf.oob_score_\n\nprint cabeza\nprint clf.feature_importances_",
"_____no_output_____"
]
],
[
[
"## Predecir nuevas variables",
"_____no_output_____"
]
],
[
[
"datos_test, clases_test, _ = lea_datos(\"titanic_test.csv\")\nclases_pred = clf.predict(datos_test)\nprint \"Predicho:\", clases_pred\nprint \"Verdad: \", clases_test",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d091d7fcacca00c23535aae59d43089f5b1fbcce | 26,961 | ipynb | Jupyter Notebook | challenge/data_challenge_1.ipynb | rasegiIML/IML.HUJI | 1f3a33f48642527d6a4bcc276fbc56e90ff166e3 | [
"MIT"
] | null | null | null | challenge/data_challenge_1.ipynb | rasegiIML/IML.HUJI | 1f3a33f48642527d6a4bcc276fbc56e90ff166e3 | [
"MIT"
] | null | null | null | challenge/data_challenge_1.ipynb | rasegiIML/IML.HUJI | 1f3a33f48642527d6a4bcc276fbc56e90ff166e3 | [
"MIT"
] | null | null | null | 52.148936 | 149 | 0.62761 | [
[
[
"Initialize estimator class",
"_____no_output_____"
]
],
[
[
"from __future__ import annotations\n\nfrom typing import NoReturn\n\nimport numpy as np\nimport pandas as pd\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.metrics import RocCurveDisplay, accuracy_score\n\nfrom IMLearn.base import BaseEstimator\n\nimport re\nfrom copy import copy\nfrom datetime import datetime\nfrom typing import NoReturn\n\nfrom sklearn.model_selection import GridSearchCV\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.preprocessing import FunctionTransformer\n\nfrom IMLearn import BaseEstimator\nfrom challenge.agoda_cancellation_estimator import AgodaCancellationEstimator\n\n\n\nclass AgodaCancellationEstimator(BaseEstimator):\n def __init__(self, threshold: float = None) -> AgodaCancellationEstimator:\n super().__init__()\n self.__fit_model: RandomForestClassifier = None\n self.thresh = threshold\n\n def get_params(self, deep=False):\n return {'threshold': self.thresh}\n\n def set_params(self, threshold) -> AgodaCancellationEstimator:\n self.thresh = threshold\n\n return self\n\n def _fit(self, X: np.ndarray, y: np.ndarray) -> NoReturn:\n self.__fit_model = RandomForestClassifier(random_state=0).fit(X, y)\n\n def _predict(self, X: pd.DataFrame) -> np.ndarray:\n probs = self.__fit_model.predict_proba(X)[:, 1]\n return probs > self.thresh if self.thresh is not None else probs\n\n def _loss(self, X: np.ndarray, y: np.ndarray) -> float:\n pass\n\n def plot_roc_curve(self, X: np.ndarray, y: np.ndarray):\n RocCurveDisplay.from_estimator(self.__fit_model, X, y)\n\n def score(self, X: pd.DataFrame, y: pd.Series):\n return accuracy_score(y, self._predict(X))",
"_____no_output_____"
]
],
[
[
"Helper Functions",
"_____no_output_____"
]
],
[
[
"def read_data_file(path: str) -> pd.DataFrame:\n return pd.read_csv(path).drop_duplicates() \\\n .astype({'checkout_date': 'datetime64',\n 'checkin_date': 'datetime64',\n 'hotel_live_date': 'datetime64',\n 'booking_datetime': 'datetime64'})\n\n\ndef get_days_between_dates(dates1: pd.Series, dates2: pd.Series):\n return (dates1 - dates2).apply(lambda period: period.days)\n\n\ndef create_col_prob_mapper(col: str, mapper: dict):\n mapper = copy(mapper)\n\n def map_col_to_prob(df):\n df[col] = df[col].apply(mapper.get)\n\n return df\n\n return map_col_to_prob\n\n\ndef add_categorical_prep_to_pipe(train_features: pd.DataFrame, pipeline: Pipeline, cat_vars: list, one_hot=False,\n calc_probs=True) -> Pipeline:\n assert one_hot ^ calc_probs, \\\n 'Error: can only do either one-hot encoding or probability calculations, not neither/both!'\n # one-hot encoding\n if one_hot:\n # TODO - use sklearn OneHotEncoder\n pipeline.steps.append(('one-hot encoding',\n FunctionTransformer(lambda df: pd.get_dummies(df, columns=cat_vars))))\n\n # category probability preprocessing - make each category have its success percentage\n if calc_probs:\n for cat_var in cat_vars:\n map_cat_to_prob: dict = train_features.groupby(cat_var, dropna=False).labels.mean().to_dict()\n\n pipeline.steps.append((f'map {cat_var} to prob',\n FunctionTransformer(create_col_prob_mapper(cat_var, map_cat_to_prob))))\n\n return pipeline\n\n\ndef get_week_of_year(dates):\n return dates.apply(lambda d: d.weekofyear)\n\n\ndef get_booked_on_weekend(dates):\n return dates.apply(lambda d: d.day_of_week >= 4)\n\n\ndef get_weekend_holiday(in_date, out_date):\n return list(map(lambda d: (d[1] - d[0]).days <= 3 and d[0].dayofweek >= 4, zip(in_date, out_date)))\n\n\ndef get_local_holiday(col1, col2):\n return list(map(lambda x: x[0] == x[1], zip(col1, col2)))\n\n\ndef get_days_until_policy(policy_code: str) -> list:\n policies = policy_code.split('_')\n return [int(policy.split('D')[0]) if 'D' in policy else 0 for policy in policies]\n\n\ndef get_policy_cost(policy, stay_cost, stay_length, time_until_checkin):\n \"\"\"\n returns tuple of the format (max lost, min lost, part min lost)\n \"\"\"\n if policy == 'UNKNOWN':\n return 0, 0, 0\n nums = tuple(map(int, re.split('[a-zA-Z]', policy)[:-1]))\n if 'D' not in policy: # no show is suppressed\n return 0, 0, 0\n if 'N' in policy:\n nights_cost = stay_cost / stay_length * nums[0]\n min_cost = nights_cost if time_until_checkin <= nums[1] else 0\n return nights_cost, min_cost, min_cost / stay_cost\n elif 'P' in policy:\n nights_cost = stay_cost * nums[0] / 100\n min_cost = nights_cost if time_until_checkin <= nums[1] else 0\n return nights_cost, min_cost, min_cost / stay_cost\n else:\n raise Exception(\"Invalid Input\")\n\n\ndef get_money_lost_per_policy(features: pd.Series) -> list:\n policies = features.cancellation_policy_code.split('_')\n stay_cost = features.original_selling_amount\n stay_length = features.stay_length\n time_until_checkin = features.booking_to_arrival_time\n policy_cost = [get_policy_cost(policy, stay_cost, stay_length, time_until_checkin) for policy in policies]\n\n return list(map(list, zip(*policy_cost)))\n\n\ndef add_cancellation_policy_features(features: pd.DataFrame) -> pd.DataFrame:\n cancellation_policy = features.cancellation_policy_code\n features['n_policies'] = cancellation_policy.apply(lambda policy: len(policy.split('_')))\n days_until_policy = cancellation_policy.apply(get_days_until_policy)\n\n features['min_policy_days'] = days_until_policy.apply(min)\n features['max_policy_days'] = days_until_policy.apply(max)\n\n x = features.apply(get_money_lost_per_policy, axis='columns')\n features['max_policy_cost'], features['min_policy_cost'], features['part_min_policy_cost'] = list(\n map(list, zip(*x)))\n\n features['min_policy_cost'] = features['min_policy_cost'].apply(min)\n features['part_min_policy_cost'] = features['part_min_policy_cost'].apply(min)\n features['max_policy_cost'] = features['max_policy_cost'].apply(max)\n\n return features\n\n\ndef add_time_based_cols(df: pd.DataFrame) -> pd.DataFrame:\n df['stay_length'] = get_days_between_dates(df.checkout_date, df.checkin_date)\n df['time_registered_pre_book'] = get_days_between_dates(df.checkin_date, df.hotel_live_date)\n df['booking_to_arrival_time'] = get_days_between_dates(df.checkin_date, df.booking_datetime)\n df['checkin_week_of_year'] = get_week_of_year(df.checkin_date)\n df['booking_week_of_year'] = get_week_of_year(df.booking_datetime)\n df['booked_on_weekend'] = get_booked_on_weekend(df.booking_datetime)\n df['is_weekend_holiday'] = get_weekend_holiday(df.checkin_date, df.checkout_date)\n df['is_local_holiday'] = get_local_holiday(df.origin_country_code, df.hotel_country_code)\n\n return df\n",
"_____no_output_____"
]
],
[
[
"Define pipeline",
"_____no_output_____"
]
],
[
[
"def create_pipeline_from_data(filename: str):\n NONE_OUTPUT_COLUMNS = ['checkin_date',\n 'checkout_date',\n 'booking_datetime',\n 'hotel_live_date',\n 'hotel_country_code',\n 'origin_country_code',\n 'cancellation_policy_code']\n CATEGORICAL_COLUMNS = ['hotel_star_rating',\n 'guest_nationality_country_name',\n 'charge_option',\n 'accommadation_type_name',\n 'language',\n 'is_first_booking',\n 'customer_nationality',\n 'original_payment_currency',\n 'is_user_logged_in',\n ]\n RELEVANT_COLUMNS = ['no_of_adults',\n 'no_of_children',\n 'no_of_extra_bed',\n 'no_of_room',\n 'original_selling_amount'] + NONE_OUTPUT_COLUMNS + CATEGORICAL_COLUMNS\n features = read_data_file(filename)\n\n features['labels'] = features[\"cancellation_datetime\"].isna()\n\n pipeline_steps = [('columns selector', FunctionTransformer(lambda df: df[RELEVANT_COLUMNS])),\n ('add time based columns', FunctionTransformer(add_time_based_cols)),\n ('add cancellation policy features', FunctionTransformer(add_cancellation_policy_features))\n ]\n\n pipeline = Pipeline(pipeline_steps)\n pipeline = add_categorical_prep_to_pipe(features, pipeline, CATEGORICAL_COLUMNS)\n\n pipeline.steps.append( ('drop irrelevant columns', FunctionTransformer(lambda df: df.drop(NONE_OUTPUT_COLUMNS, axis='columns'))))\n\n return features.drop('labels', axis='columns'), features.labels, pipeline\n",
"_____no_output_____"
]
],
[
[
"Train, predict and export",
"_____no_output_____"
]
],
[
[
"def evaluate_and_export(estimator: BaseEstimator, X: pd.DataFrame, filename: str):\n preds = (~estimator.predict(X)).astype(int)\n pd.DataFrame(preds, columns=[\"predicted_values\"]).to_csv(filename, index=False)\n\n\ndef create_estimator_from_data(path=\"../datasets/agoda_cancellation_train.csv\", threshold: float = 0.47,\n optimize_threshold=False, debug=False) -> Pipeline:\n np.random.seed(0)\n\n # Load data\n raw_df, cancellation_labels, pipeline = create_pipeline_from_data(path)\n\n train_X = raw_df\n train_y = cancellation_labels\n train_X = pipeline.transform(train_X)\n\n # Fit model over data\n estimator = AgodaCancellationEstimator(threshold).fit(train_X, train_y)\n pipeline.steps.append(('estimator', estimator))\n\n return pipeline\n\ndef export_test_data(pipeline: Pipeline, path=\"../datasets/test_set_week_1.csv\") -> NoReturn:\n data = read_data_file(path)\n\n # Store model predictions over test set\n id1, id2, id3 = 209855253, 205843964, 212107536\n evaluate_and_export(pipeline, data, f\"{id1}_{id2}_{id3}.csv\")\n\npipeline = create_estimator_from_data()\nexport_test_data(pipeline)\n",
"C:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:117: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:118: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:119: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:120: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:121: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:122: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:123: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:124: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:99: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:102: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:103: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:107: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:109: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:110: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:111: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:17: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:117: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:118: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:119: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:120: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:121: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:122: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:123: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:124: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:99: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:102: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:103: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:107: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:109: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:110: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:111: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\nC:\\conda\\envs\\iml.env\\lib\\site-packages\\ipykernel_launcher.py:17: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d091decf9525bd6bf16bf0c3ea01811d96522a27 | 49,695 | ipynb | Jupyter Notebook | notebooks/.ipynb_checkpoints/machine_learning_tutorial-checkpoint.ipynb | orchid00/nipype_arcana_workshop | 2ead3a07da24e429c65277513502c7c4da702b70 | [
"BSD-3-Clause"
] | 5 | 2019-09-19T09:56:06.000Z | 2020-12-22T10:05:02.000Z | notebooks/.ipynb_checkpoints/machine_learning_tutorial-checkpoint.ipynb | orchid00/nipype_arcana_workshop | 2ead3a07da24e429c65277513502c7c4da702b70 | [
"BSD-3-Clause"
] | null | null | null | notebooks/.ipynb_checkpoints/machine_learning_tutorial-checkpoint.ipynb | orchid00/nipype_arcana_workshop | 2ead3a07da24e429c65277513502c7c4da702b70 | [
"BSD-3-Clause"
] | 3 | 2019-10-15T00:05:49.000Z | 2021-12-22T18:23:36.000Z | 29.882742 | 514 | 0.565349 | [
[
[
"# Let's keep our notebook clean, so it's a little more readable!\nimport warnings\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
],
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"# Machine learning to predict age from rs-fmri\n\nThe goal is to extract data from several rs-fmri images, and use that data as features in a machine learning model. We will integrate what we've learned in the previous machine learning lecture to build an unbiased model and test it on a left out sample.\n\nWe're going to use a dataset that was prepared for this tutorial by [Elizabeth Dupre](https://elizabeth-dupre.com/#/), [Jake Vogel](https://github.com/illdopejake) and [Gael Varoquaux](http://gael-varoquaux.info/), by preprocessing [ds000228](https://openneuro.org/datasets/ds000228/versions/1.0.0) (from [Richardson et al. (2018)](https://dx.doi.org/10.1038%2Fs41467-018-03399-2)) through [fmriprep](https://github.com/poldracklab/fmriprep). They also created this tutorial and should be credited for it. ",
"_____no_output_____"
],
[
"## Load the data\n\n<img src=\"data/SampFeat.png\" alt=\"terms\" width=\"300\"/>\n",
"_____no_output_____"
]
],
[
[
"# change this to the location where you downloaded the data\nwdir = '/data/ml_tutorial/'\n",
"_____no_output_____"
],
[
"# Now fetch the data\n\nfrom glob import glob\nimport os\ndata = sorted(glob(os.path.join(wdir,'*.gz')))\nconfounds = sorted(glob(os.path.join(wdir,'*regressors.tsv')))",
"_____no_output_____"
]
],
[
[
"How many individual subjects do we have?",
"_____no_output_____"
]
],
[
[
"#len(data.func)\nlen(data)",
"_____no_output_____"
]
],
[
[
"## Extract features\n\n",
"_____no_output_____"
],
[
"In order to do our machine learning, we will need to extract feature from our rs-fmri images.\n\nSpecifically, we will extract signals from a brain parcellation and compute a correlation matrix, representing regional coactivation between regions.\n\nWe will practice on one subject first, then we'll extract data for all subjects",
"_____no_output_____"
],
[
"#### Retrieve the atlas for extracting features and an example subject",
"_____no_output_____"
],
[
"Since we're using rs-fmri data, it makes sense to use an atlas defined using rs-fmri data\n\nThis paper has many excellent insights about what kind of atlas to use for an rs-fmri machine learning task. See in particular Figure 5.\n\nhttps://www.sciencedirect.com/science/article/pii/S1053811919301594?via%3Dihub\n\nLet's use the MIST atlas, created here in Montreal using the BASC method. This atlas has multiple resolutions, for larger networks or finer-grained ROIs. Let's use a 64-ROI atlas to allow some detail, but to ultimately keep our connectivity matrices manageable\n\nHere is a link to the MIST paper: https://mniopenresearch.org/articles/1-3",
"_____no_output_____"
]
],
[
[
"from nilearn import datasets\n\nparcellations = datasets.fetch_atlas_basc_multiscale_2015(version='sym')\natlas_filename = parcellations.scale064\n\n\nprint('Atlas ROIs are located in nifti image (4D) at: %s' %\n atlas_filename)",
"_____no_output_____"
]
],
[
[
"Let's have a look at that atlas",
"_____no_output_____"
]
],
[
[
"from nilearn import plotting\n\nplotting.plot_roi(atlas_filename, draw_cross=False)",
"_____no_output_____"
]
],
[
[
"Great, let's load an example 4D fmri time-series for one subject",
"_____no_output_____"
]
],
[
[
"fmri_filenames = data[0]\nprint(fmri_filenames)",
"_____no_output_____"
]
],
[
[
"Let's have a look at the image! Because it is a 4D image, we can only look at one slice at a time. Or, better yet, let's look at an average image!",
"_____no_output_____"
]
],
[
[
"from nilearn import image \n\naveraged_Img = image.mean_img(image.mean_img(fmri_filenames))\nplotting.plot_stat_map(averaged_Img)",
"_____no_output_____"
]
],
[
[
"#### Extract signals on a parcellation defined by labels\nUsing the NiftiLabelsMasker\n\nSo we've loaded our atlas and 4D data for a single subject. Let's practice extracting features!\n",
"_____no_output_____"
]
],
[
[
"from nilearn.input_data import NiftiLabelsMasker\n\nmasker = NiftiLabelsMasker(labels_img=atlas_filename, standardize=True, \n memory='nilearn_cache', verbose=1)\n\n# Here we go from nifti files to the signal time series in a numpy\n# array. Note how we give confounds to be regressed out during signal\n# extraction\nconf = confounds[0]\ntime_series = masker.fit_transform(fmri_filenames, confounds=conf)",
"_____no_output_____"
]
],
[
[
"So what did we just create here?",
"_____no_output_____"
]
],
[
[
"type(time_series)",
"_____no_output_____"
],
[
"time_series.shape",
"_____no_output_____"
]
],
[
[
"What are these \"confounds\" and how are they used? ",
"_____no_output_____"
]
],
[
[
"import pandas\nconf_df = pandas.read_table(conf)\nconf_df.head()",
"_____no_output_____"
],
[
"conf_df.shape",
"_____no_output_____"
]
],
[
[
"#### Compute and display a correlation matrix\n",
"_____no_output_____"
]
],
[
[
"from nilearn.connectome import ConnectivityMeasure\n\ncorrelation_measure = ConnectivityMeasure(kind='correlation')\ncorrelation_matrix = correlation_measure.fit_transform([time_series])[0]\ncorrelation_matrix.shape",
"_____no_output_____"
]
],
[
[
"Plot the correlation matrix",
"_____no_output_____"
]
],
[
[
"import numpy as np\n# Mask the main diagonal for visualization:\nnp.fill_diagonal(correlation_matrix, 0)\n\n# The labels we have start with the background (0), hence we skip the\n# first label\nplotting.plot_matrix(correlation_matrix, figure=(10, 8), \n labels=range(time_series.shape[-1]),\n vmax=0.8, vmin=-0.8, reorder=False)\n\n# matrices are ordered for block-like representation",
"_____no_output_____"
]
],
[
[
"#### Extract features from the whole dataset\n\nHere, we are going to use a for loop to iterate through each image and use the same techniques we learned above to extract rs-fmri connectivity features from every subject.\n\n",
"_____no_output_____"
]
],
[
[
"# Here is a really simple for loop\n\nfor i in range(10):\n print('the number is', i)",
"_____no_output_____"
],
[
"container = []\nfor i in range(10):\n container.append(i)\n\ncontainer",
"_____no_output_____"
]
],
[
[
"Now lets construct a more complicated loop to do what we want",
"_____no_output_____"
],
[
"First we do some things we don't need to do in the loop. Let's reload our atlas, and re-initiate our masker and correlation_measure",
"_____no_output_____"
]
],
[
[
"from nilearn.input_data import NiftiLabelsMasker\nfrom nilearn.connectome import ConnectivityMeasure\nfrom nilearn import datasets\n\n# load atlas\nmultiscale = datasets.fetch_atlas_basc_multiscale_2015()\natlas_filename = multiscale.scale064\n\n# initialize masker (change verbosity)\nmasker = NiftiLabelsMasker(labels_img=atlas_filename, standardize=True, \n memory='nilearn_cache', verbose=0)\n\n# initialize correlation measure, set to vectorize\ncorrelation_measure = ConnectivityMeasure(kind='correlation', vectorize=True,\n discard_diagonal=True)",
"_____no_output_____"
]
],
[
[
"Okay -- now that we have that taken care of, let's run our big loop!",
"_____no_output_____"
],
[
"**NOTE**: On a laptop, this might a few minutes.",
"_____no_output_____"
]
],
[
[
"all_features = [] # here is where we will put the data (a container)\n\nfor i,sub in enumerate(data):\n # extract the timeseries from the ROIs in the atlas\n time_series = masker.fit_transform(sub, confounds=confounds[i])\n # create a region x region correlation matrix\n correlation_matrix = correlation_measure.fit_transform([time_series])[0]\n # add to our container\n all_features.append(correlation_matrix)\n # keep track of status\n print('finished %s of %s'%(i+1,len(data)))",
"_____no_output_____"
],
[
"# Let's save the data to disk\nimport numpy as np\n\nnp.savez_compressed('MAIN_BASC064_subsamp_features',a = all_features)",
"_____no_output_____"
]
],
[
[
"In case you do not want to run the full loop on your computer, you can load the output of the loop here!",
"_____no_output_____"
]
],
[
[
"feat_file = 'MAIN_BASC064_subsamp_features.npz'\nX_features = np.load(feat_file)['a']",
"_____no_output_____"
],
[
"X_features.shape",
"_____no_output_____"
]
],
[
[
"<img src=\"data/SampFeat.png\" alt=\"terms\" width=\"300\"/>",
"_____no_output_____"
],
[
"Okay so we've got our features.",
"_____no_output_____"
],
[
"We can visualize our feature matrix",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n\nplt.imshow(X_features, aspect='auto')\nplt.colorbar()\nplt.title('feature matrix')\nplt.xlabel('features')\nplt.ylabel('subjects')",
"_____no_output_____"
]
],
[
[
"## Get Y (our target) and assess its distribution",
"_____no_output_____"
]
],
[
[
"# Let's load the phenotype data\n\npheno_path = os.path.join(wdir, 'participants.tsv')",
"_____no_output_____"
],
[
"import pandas\n\npheno = pandas.read_csv(pheno_path, sep='\\t').sort_values('participant_id')\npheno.head()",
"_____no_output_____"
]
],
[
[
"Looks like there is a column labeling age. Let's capture it in a variable",
"_____no_output_____"
]
],
[
[
"y_age = pheno['Age']",
"_____no_output_____"
]
],
[
[
"Maybe we should have a look at the distribution of our target variable",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport seaborn as sns\nsns.distplot(y_age)",
"_____no_output_____"
]
],
[
[
"## Prepare data for machine learning\n\nHere, we will define a \"training sample\" where we can play around with our models. We will also set aside a \"validation\" sample that we will not touch until the end",
"_____no_output_____"
],
[
"We want to be sure that our training and test sample are matched! We can do that with a \"stratified split\". This dataset has a variable indicating AgeGroup. We can use that to make sure our training and testing sets are balanced!",
"_____no_output_____"
]
],
[
[
"age_class = pheno['AgeGroup']\nage_class.value_counts()",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\n\n# Split the sample to training/validation with a 60/40 ratio, and \n# stratify by age class, and also shuffle the data.\n\nX_train, X_val, y_train, y_val = train_test_split(\n X_features, # x\n y_age, # y\n test_size = 0.4, # 60%/40% split \n shuffle = True, # shuffle dataset\n # before splitting\n stratify = age_class, # keep\n # distribution\n # of ageclass\n # consistent\n # betw. train\n # & test sets.\n random_state = 123 # same shuffle each\n # time\n )\n\n# print the size of our training and test groups\nprint('training:', len(X_train),\n 'testing:', len(X_val))",
"_____no_output_____"
]
],
[
[
"Let's visualize the distributions to be sure they are matched",
"_____no_output_____"
]
],
[
[
"sns.distplot(y_train)\nsns.distplot(y_val)",
"_____no_output_____"
]
],
[
[
"## Run your first model!\n\nMachine learning can get pretty fancy pretty quickly. We'll start with a fairly standard regression model called a Support Vector Regressor (SVR). \n\nWhile this may seem unambitious, simple models can be very robust. And we probably don't have enough data to create more complex models (but we can try later).\n\nFor more information, see this excellent resource:\nhttps://hal.inria.fr/hal-01824205",
"_____no_output_____"
],
[
"Let's fit our first model!",
"_____no_output_____"
]
],
[
[
"from sklearn.svm import SVR\nl_svr = SVR(kernel='linear') # define the model\n\nl_svr.fit(X_train, y_train) # fit the model",
"_____no_output_____"
]
],
[
[
"Well... that was easy. Let's see how well the model learned the data!\n\n\n<img src=\"data/modval.png\" alt=\"terms\" width=\"800\"/>",
"_____no_output_____"
]
],
[
[
"\n# predict the training data based on the model\ny_pred = l_svr.predict(X_train) \n\n# caluclate the model accuracy\nacc = l_svr.score(X_train, y_train) \n",
"_____no_output_____"
]
],
[
[
"Let's view our results and plot them all at once!",
"_____no_output_____"
]
],
[
[
"# print results\nprint('accuracy (R2)', acc)\n\nsns.regplot(y_pred,y_train)\nplt.xlabel('Predicted Age')",
"_____no_output_____"
]
],
[
[
"HOLY COW! Machine learning is amazing!!! Almost a perfect fit!\n\n...which means there's something wrong. What's the problem here?",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\n\n# Split the sample to training/test with a 75/25 ratio, and \n# stratify by age class, and also shuffle the data.\n\nage_class2 = pheno.loc[y_train.index,'AgeGroup']\n\nX_train2, X_test, y_train2, y_test = train_test_split(\n X_train, # x\n y_train, # y\n test_size = 0.25, # 75%/25% split \n shuffle = True, # shuffle dataset\n # before splitting\n stratify = age_class2, # keep\n # distribution\n # of ageclass\n # consistent\n # betw. train\n # & test sets.\n random_state = 123 # same shuffle each\n # time\n )\n\n# print the size of our training and test groups\nprint('training:', len(X_train2),\n 'testing:', len(X_test))",
"_____no_output_____"
],
[
"from sklearn.metrics import mean_absolute_error\n\n# fit model just to training data\nl_svr.fit(X_train2,y_train2)\n\n# predict the *test* data based on the model trained on X_train2\ny_pred = l_svr.predict(X_test) \n\n# caluclate the model accuracy\nacc = l_svr.score(X_test, y_test) \nmae = mean_absolute_error(y_true=y_test,y_pred=y_pred)",
"_____no_output_____"
],
[
"# print results\nprint('accuracy (R2) = ', acc)\nprint('MAE = ',mae)\n\nsns.regplot(y_pred,y_test)\nplt.xlabel('Predicted Age')",
"_____no_output_____"
]
],
[
[
"Not perfect, but as predicting with unseen data goes, not too bad! Especially with a training sample of \"only\" 69 subjects. But we can do better! \n\nFor example, we can increase the size our training set while simultaneously reducing bias by instead using 10-fold cross-validation\n\n<img src=\"data/KCV.png\" alt=\"terms\" width=\"500\"/>",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import cross_val_predict, cross_val_score\n\n# predict\ny_pred = cross_val_predict(l_svr, X_train, y_train, cv=10)\n# scores\nacc = cross_val_score(l_svr, X_train, y_train, cv=10)\nmae = cross_val_score(l_svr, X_train, y_train, cv=10, scoring='neg_mean_absolute_error')",
"_____no_output_____"
]
],
[
[
"We can look at the accuracy of the predictions for each fold of the cross-validation",
"_____no_output_____"
]
],
[
[
"for i in range(10):\n print('Fold {} -- Acc = {}, MAE = {}'.format(i, acc[i],-mae[i]))",
"_____no_output_____"
]
],
[
[
"We can also look at the overall accuracy of the model",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import r2_score\n\noverall_acc = r2_score(y_train, y_pred)\noverall_mae = mean_absolute_error(y_train,y_pred)\nprint('R2:',overall_acc)\nprint('MAE:',overall_mae)\n\nsns.regplot(y_pred, y_train)\nplt.xlabel('Predicted Age')",
"_____no_output_____"
]
],
[
[
"Not too bad at all! But more importantly, this is a more accurate estimation of our model's predictive efficacy. Our sample size is larger and this is based on several rounds of prediction of unseen data. \n\nFor example, we can now see that the effect is being driven by the model's successful parsing of adults vs. children, but is not performing so well within the adult or children group. This was not evident during our previous iteration of the model",
"_____no_output_____"
],
[
"## Tweak your model\n\nIt's very important to learn when and where its appropriate to \"tweak\" your model.\n\nSince we have done all of the previous analysis in our training data, it's fine to try out different models. But we **absolutely cannot** \"test\" it on our left out data. If we do, we are in great danger of overfitting.\n\nIt is not uncommon to try other models, or tweak hyperparameters. In this case, due to our relatively small sample size, we are probably not powered sufficiently to do so, and we would once again risk overfitting. However, for the sake of demonstration, we will do some tweaking. \n\n<img src=\"data/KCV2.png\" alt=\"terms\" width=\"500\"/>\n",
"_____no_output_____"
],
[
"We will try a few different examples:\n* Normalizing our target data\n* Tweaking our hyperparameters\n* Trying a more complicated model\n* Feature selection",
"_____no_output_____"
],
[
"#### Normalize the target data",
"_____no_output_____"
]
],
[
[
"# Create a log transformer function and log transform Y (age)\n\nfrom sklearn.preprocessing import FunctionTransformer\n\nlog_transformer = FunctionTransformer(func = np.log, validate=True)\nlog_transformer.fit(y_train.values.reshape(-1,1))\ny_train_log = log_transformer.transform(y_train.values.reshape(-1,1))[:,0]\n\nsns.distplot(y_train_log)\nplt.title(\"Log-Transformed Age\")",
"_____no_output_____"
]
],
[
[
"Now let's go ahead and cross-validate our model once again with this new log-transformed target",
"_____no_output_____"
]
],
[
[
"# predict\ny_pred = cross_val_predict(l_svr, X_train, y_train_log, cv=10)\n\n# scores\nacc = r2_score(y_train_log, y_pred)\nmae = mean_absolute_error(y_train_log,y_pred)\n\nprint('R2:',acc)\nprint('MAE:',mae)\n\nsns.regplot(y_pred, y_train_log)\nplt.xlabel('Predicted Log Age')\nplt.ylabel('Log Age')",
"_____no_output_____"
]
],
[
[
"Seems like a definite improvement, right? I think we can agree on that. \n\nBut we can't forget about interpretability? The MAE is much less interpretable now.",
"_____no_output_____"
],
[
"#### Tweak the hyperparameters\n\nMany machine learning algorithms have hyperparameters that can be \"tuned\" to optimize model fitting. Careful parameter tuning can really improve a model, but haphazard tuning will often lead to overfitting.\n\nOur SVR model has multiple hyperparameters. Let's explore some approaches for tuning them",
"_____no_output_____"
]
],
[
[
"SVR?",
"_____no_output_____"
]
],
[
[
"One way is to plot a \"Validation Curve\" -- this will let us view changes in training and validation accuracy of a model as we shift its hyperparameters. We can do this easily with sklearn.",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import validation_curve\n\nC_range = 10. ** np.arange(-3, 8) # A range of different values for C\n\ntrain_scores, valid_scores = validation_curve(l_svr, X_train, y_train_log, \n param_name= \"C\",\n param_range = C_range,\n cv=10)\n",
"_____no_output_____"
],
[
"# A bit of pandas magic to prepare the data for a seaborn plot\n\ntScores = pandas.DataFrame(train_scores).stack().reset_index()\ntScores.columns = ['C','Fold','Score']\ntScores.loc[:,'Type'] = ['Train' for x in range(len(tScores))]\n\nvScores = pandas.DataFrame(valid_scores).stack().reset_index()\nvScores.columns = ['C','Fold','Score']\nvScores.loc[:,'Type'] = ['Validate' for x in range(len(vScores))]\n\nValCurves = pandas.concat([tScores,vScores]).reset_index(drop=True)\nValCurves.head()",
"_____no_output_____"
],
[
"# And plot!\n# g = sns.lineplot(x='C',y='Score',hue='Type',data=ValCurves)\n# g.set_xticks(range(10))\n# g.set_xticklabels(C_range, rotation=90)\n\ng = sns.factorplot(x='C',y='Score',hue='Type',data=ValCurves)\nplt.xticks(range(10))\ng.set_xticklabels(C_range, rotation=90)",
"_____no_output_____"
]
],
[
[
"It looks like accuracy is better for higher values of C, and plateaus somewhere between 0.1 and 1. The default setting is C=1, so it looks like we can't really improve by changing C.",
"_____no_output_____"
],
[
"But our SVR model actually has two hyperparameters, C and epsilon. Perhaps there is an optimal combination of settings for these two parameters.\n\nWe can explore that somewhat quickly with a grid search, which is once again easily achieved with sklearn. Because we are fitting the model multiple times witih cross-validation, this will take some time",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import GridSearchCV\n\nC_range = 10. ** np.arange(-3, 8)\nepsilon_range = 10. ** np.arange(-3, 8)\n\nparam_grid = dict(epsilon=epsilon_range, C=C_range)\n\ngrid = GridSearchCV(l_svr, param_grid=param_grid, cv=10)\n\ngrid.fit(X_train, y_train_log)",
"_____no_output_____"
]
],
[
[
"Now that the grid search has completed, let's find out what was the \"best\" parameter combination",
"_____no_output_____"
]
],
[
[
"print(grid.best_params_)",
"_____no_output_____"
]
],
[
[
"And if redo our cross-validation with this parameter set?",
"_____no_output_____"
]
],
[
[
"y_pred = cross_val_predict(SVR(kernel='linear',C=0.10,epsilon=0.10, gamma='auto'), \n X_train, y_train_log, cv=10)\n\n# scores\nacc = r2_score(y_train_log, y_pred)\nmae = mean_absolute_error(y_train_log,y_pred)\n\nprint('R2:',acc)\nprint('MAE:',mae)\n\nsns.regplot(y_pred, y_train_log)\nplt.xlabel('Predicted Log Age')\nplt.ylabel('Log Age')",
"_____no_output_____"
]
],
[
[
"Perhaps unsurprisingly, the model fit is actually exactly the same as what we had with our defaults. There's a reason they are defaults ;-)\n\nGrid search can be a powerful and useful tool. But can you think of a way that, if not properly utilized, it could lead to overfitting?",
"_____no_output_____"
],
[
"You can find a nice set of tutorials with links to very helpful content regarding how to tune hyperparameters and while be aware of over- and under-fitting here:\n\nhttps://scikit-learn.org/stable/modules/learning_curve.html",
"_____no_output_____"
],
[
"#### Trying a more complicated model",
"_____no_output_____"
],
[
"In principle, there is no real reason to do this. Perhaps one could make an argument for quadratic relationship with age, but we probably don't have enough subjects to learn a complicated non-linear model. But for the sake of demonstration, we can give it a shot.\n\nWe'll use a validation curve to see the result of our model if, instead of fitting a linear model, we instead try to the fit a 2nd, 3rd, ... 8th order polynomial.",
"_____no_output_____"
]
],
[
[
"validation_curve?",
"_____no_output_____"
],
[
"from sklearn.model_selection import validation_curve\n\ndegree_range = list(range(1,8)) # A range of different values for C\n\ntrain_scores, valid_scores = validation_curve(SVR(kernel='poly',\n gamma='scale'\n ), \n X=X_train, y=y_train_log, \n param_name= \"degree\",\n param_range = degree_range,\n cv=10)\n",
"_____no_output_____"
],
[
"# A bit of pandas magic to prepare the data for a seaborn plot\n\ntScores = pandas.DataFrame(train_scores).stack().reset_index()\ntScores.columns = ['Degree','Fold','Score']\ntScores.loc[:,'Type'] = ['Train' for x in range(len(tScores))]\n\nvScores = pandas.DataFrame(valid_scores).stack().reset_index()\nvScores.columns = ['Degree','Fold','Score']\nvScores.loc[:,'Type'] = ['Validate' for x in range(len(vScores))]\n\nValCurves = pandas.concat([tScores,vScores]).reset_index(drop=True)\nValCurves.head()",
"_____no_output_____"
],
[
"# And plot!\n\n# g = sns.lineplot(x='Degree',y='Score',hue='Type',data=ValCurves)\n# g.set_xticks(range(10))\n# g.set_xticklabels(degree_range, rotation=90)\n\ng = sns.factorplot(x='Degree',y='Score',hue='Type',data=ValCurves)\nplt.xticks(range(10))\ng.set_xticklabels(degree_range, rotation=90)",
"_____no_output_____"
]
],
[
[
"It appears that we cannot improve our model by increasing the complexity of the fit. If one looked only at the training data, one might surmise that a 2nd order fit could be a slightly better model. But that improvement does not generalize to the validation data.",
"_____no_output_____"
]
],
[
[
"# y_pred = cross_val_predict(SVR(kernel='rbf', gamma='scale'), X_train, y_train_log, cv=10)\n\n# # scores\n# acc = r2_score(y_train_log, y_pred)\n# mae = mean_absolute_error(y_train_log,y_pred)\n\n# print('R2:',acc)\n# print('MAE:',mae)\n\n# sns.regplot(y_pred, y_train_log)\n# plt.xlabel('Predicted Log Age')\n# plt.ylabel('Log Age')",
"_____no_output_____"
]
],
[
[
"#### Feature selection\n\nRight now, we have 2016 features. Are all of those really going to contribute to the model stably? \n\nIntuitively, models tend to perform better when there are fewer, more important features than when there are many, less imortant features. The tough part is figuring out which features are useful or important.\n\nHere will quickly try a basic feature seclection strategy\n\n<img src=\"data/FeatSel.png\" alt=\"terms\" width=\"400\"/>",
"_____no_output_____"
],
[
"The SelectPercentile() function will select the top X% of features based on univariate tests. This is a way of identifying theoretically more useful features. But remember, significance != prediction! \n\nWe are also in danger of overfitting here. For starters, if we want to test this with 10-fold cross-validation, we will need to do a separate feature selection within each fold! That means we'll need to do the cross-validation manually instead of using cross_val_predict(). ",
"_____no_output_____"
]
],
[
[
"from sklearn.feature_selection import SelectPercentile, f_regression\nfrom sklearn.model_selection import KFold\nfrom sklearn.pipeline import Pipeline\n\n# Build a tiny pipeline that does feature selection (top 20% of features), \n# and then prediction with our linear svr model.\nmodel = Pipeline([\n ('feature_selection',SelectPercentile(f_regression,percentile=20)),\n ('prediction', l_svr)\n ])\n\ny_pred = [] # a container to catch the predictions from each fold\ny_index = [] # just in case, the index for each prediciton\n\n# First we create 10 splits of the data\nskf = KFold(n_splits=10, shuffle=True, random_state=123)\n\n# For each split, assemble the train and test samples \nfor tr_ind, te_ind in skf.split(X_train):\n X_tr = X_train[tr_ind]\n y_tr = y_train_log[tr_ind]\n X_te = X_train[te_ind]\n y_index += list(te_ind) # store the index of samples to predict\n \n # and run our pipeline \n model.fit(X_tr, y_tr) # fit the data to the model using our mini pipeline\n predictions = model.predict(X_te).tolist() # get the predictions for this fold\n y_pred += predictions # add them to the list of predictions\n\n\n ",
"_____no_output_____"
]
],
[
[
"Alrighty, let's see if only using the top 20% of features improves the model at all...",
"_____no_output_____"
]
],
[
[
"acc = r2_score(y_train_log[y_index], y_pred)\nmae = mean_absolute_error(y_train_log[y_index],y_pred)\n\nprint('R2:',acc)\nprint('MAE:',mae)\n\nsns.regplot(np.array(y_pred), y_train_log[y_index])\nplt.xlabel('Predicted Log Age')\nplt.ylabel('Log Age')\n",
"_____no_output_____"
]
],
[
[
"Nope, in fact it got a bit worse. It seems we're getting \"value at the margins\" so to speak. This is a very good example of how significance != prediction, as demonstrated in this figure from Bzdok et al., 2018 *bioRxiv*\n\n",
"_____no_output_____"
],
[
"See here for an explanation of different feature selection options and how to implement them in sklearn: https://scikit-learn.org/stable/modules/feature_selection.html\n\nAnd here is a thoughtful tutorial covering feature selection for novel machine learners: https://www.datacamp.com/community/tutorials/feature-selection-python",
"_____no_output_____"
],
[
"So there you have it. We've tried many different strategies, but most of our \"tweaks\" haven't really lead to improvements in the model. This is not always the case, but it is not uncommon. Can you think of some reasons why?\n\nMoving on to our validation data, we probably should just stick to a basic model, though predicting log age might be a good idea!",
"_____no_output_____"
],
[
"## Can our model predict age in completely un-seen data?\nNow that we've fit a model we think has possibly learned how to decode age based on rs-fmri signal, let's put it to the test. We will train our model on all of the training data, and try to predict the age of the subjects we left out at the beginning of this section.",
"_____no_output_____"
],
[
"Because we performed a log transformation on our training data, we will need to transform our testing data using the *same information!* But that's easy because we stored our transformation in an object!\n",
"_____no_output_____"
]
],
[
[
"# Notice how we use the Scaler that was fit to X_train and apply to X_test,\n# rather than creating a new Scaler for X_test\ny_val_log = log_transformer.transform(y_val.values.reshape(-1,1))[:,0]",
"_____no_output_____"
]
],
[
[
"And now for the moment of truth! \n\nNo cross-validation needed here. We simply fit the model with the training data and use it to predict the testing data\n\nI'm so nervous. Let's just do it all in one cell",
"_____no_output_____"
]
],
[
[
"l_svr.fit(X_train, y_train_log) # fit to training data\ny_pred = l_svr.predict(X_val) # classify age class using testing data\nacc = l_svr.score(X_val, y_val_log) # get accuracy (r2)\nmae = mean_absolute_error(y_val_log, y_pred) # get mae\n\n# print results\nprint('accuracy (r2) =', acc)\nprint('mae = ',mae)\n\n# plot results\nsns.regplot(y_pred, y_val_log)\nplt.xlabel('Predicted Log Age')\nplt.ylabel('Log Age')",
"_____no_output_____"
]
],
[
[
"***Wow!!*** Congratulations. You just trained a machine learning model that used real rs-fmri data to predict the age of real humans.\n\nThe proper thing to do at this point would be to repeat the train-validation split multiple times. This will ensure the results are not specific to this validation set, and will give you some confidence intervals around your results.\n\nAs an assignment, you can give that a try below. Create 10 different splits of the entire dataset, fit the model and get your predictions. Then, plot the range of predictions.\n",
"_____no_output_____"
]
],
[
[
"# SPACE FOR YOUR ASSIGNMENT\n\n\n",
"_____no_output_____"
]
],
[
[
"So, it seems like something in this data does seem to be systematically related to age ... but what? ",
"_____no_output_____"
],
[
"#### Interpreting model feature importances\nInterpreting the feature importances of a machine learning model is a real can of worms. This is an area of active research. Unfortunately, it's hard to trust the feature importance of some models. \n\nYou can find a whole tutorial on this subject here:\nhttp://gael-varoquaux.info/interpreting_ml_tuto/index.html\n\nFor now, we'll just eschew better judgement and take a look at our feature importances. While we can't ascribe any biological relevance to the features, it can still be helpful to know what the model is using to make its predictions. This is a good way to, for example, establish whether your model is actually learning based on a confound! Could you think of some examples?",
"_____no_output_____"
],
[
"We can access the feature importances (weights) used my the model",
"_____no_output_____"
]
],
[
[
"l_svr.coef_",
"_____no_output_____"
]
],
[
[
"lets plot these weights to see their distribution better",
"_____no_output_____"
]
],
[
[
"plt.bar(range(l_svr.coef_.shape[-1]),l_svr.coef_[0])\nplt.title('feature importances')\nplt.xlabel('feature')\nplt.ylabel('weight')",
"_____no_output_____"
]
],
[
[
"Or perhaps it will be easier to visualize this information as a matrix similar to the one we started with\n\nWe can use the correlation measure from before to perform an inverse transform",
"_____no_output_____"
]
],
[
[
"correlation_measure.inverse_transform(l_svr.coef_).shape",
"_____no_output_____"
],
[
"from nilearn import plotting\n\nfeat_exp_matrix = correlation_measure.inverse_transform(l_svr.coef_)[0]\n\nplotting.plot_matrix(feat_exp_matrix, figure=(10, 8), \n labels=range(feat_exp_matrix.shape[0]),\n reorder=False,\n tri='lower')",
"_____no_output_____"
]
],
[
[
"Let's see if we can throw those features onto an actual brain.\n\nFirst, we'll need to gather the coordinates of each ROI of our atlas",
"_____no_output_____"
]
],
[
[
"coords = plotting.find_parcellation_cut_coords(atlas_filename)",
"_____no_output_____"
]
],
[
[
"And now we can use our feature matrix and the wonders of nilearn to create a connectome map where each node is an ROI, and each connection is weighted by the importance of the feature to the model",
"_____no_output_____"
]
],
[
[
"plotting.plot_connectome(feat_exp_matrix, coords, colorbar=True)",
"_____no_output_____"
]
],
[
[
"Whoa!! That's...a lot to process. Maybe let's threshold the edges so that only the most important connections are visualized",
"_____no_output_____"
]
],
[
[
"plotting.plot_connectome(feat_exp_matrix, coords, colorbar=True, edge_threshold=0.035)",
"_____no_output_____"
]
],
[
[
"That's definitely an improvement, but it's still a bit hard to see what's going on.\nNilearn has a new feature that let's use view this data interactively!",
"_____no_output_____"
]
],
[
[
"plotting.view_connectome(feat_exp_matrix, coords, threshold='98%')",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d091e56db2d0ac66ae9b33ed5faf3cb88066af93 | 471,244 | ipynb | Jupyter Notebook | Example/Example.ipynb | jvines/Ceres-plusplus | d7f248130beba373691e10ba6b63f7e3bd7ed70f | [
"MIT"
] | null | null | null | Example/Example.ipynb | jvines/Ceres-plusplus | d7f248130beba373691e10ba6b63f7e3bd7ed70f | [
"MIT"
] | null | null | null | Example/Example.ipynb | jvines/Ceres-plusplus | d7f248130beba373691e10ba6b63f7e3bd7ed70f | [
"MIT"
] | null | null | null | 2,908.91358 | 253,899 | 0.958699 | [
[
[
"import cerespp\nimport glob",
"_____no_output_____"
],
[
"files = glob.glob('HD10700*.fits')\nfiles",
"_____no_output_____"
],
[
"lines = ['CaHK', 'Ha', 'HeI', 'NaID1D2']\ncerespp.line_plot_from_file(files[0], lines, './', 'HD10700')",
"Correcting spectrum to rest frame. (This could take a while)\nMerging echelle spectra.\nPlotting line CaHK\nPlotting line Ha\nPlotting line HeI\nPlotting line NaID1D2\n"
],
[
"cerespp.get_activities(files, './HD10700_activities.dat')",
"100%|██████████| 2/2 [06:15<00:00, 187.67s/it]\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code"
]
] |
d091f49de736a5d47212000be7b853027aaa6c6b | 18,627 | ipynb | Jupyter Notebook | 10-Iterators.ipynb | jas076100/WhirlwindTourOfPython-master | 05f435e0391563c23fdf2222edc1312bd102dcc8 | [
"CC0-1.0"
] | null | null | null | 10-Iterators.ipynb | jas076100/WhirlwindTourOfPython-master | 05f435e0391563c23fdf2222edc1312bd102dcc8 | [
"CC0-1.0"
] | null | null | null | 10-Iterators.ipynb | jas076100/WhirlwindTourOfPython-master | 05f435e0391563c23fdf2222edc1312bd102dcc8 | [
"CC0-1.0"
] | 1 | 2021-05-25T14:13:10.000Z | 2021-05-25T14:13:10.000Z | 23.110422 | 372 | 0.527514 | [
[
[
"<!--NAVIGATION-->\n< [Errors and Exceptions](09-Errors-and-Exceptions.ipynb) | [Contents](Index.ipynb) | [List Comprehensions](11-List-Comprehensions.ipynb) >",
"_____no_output_____"
],
[
"# Iterators",
"_____no_output_____"
],
[
"Often an important piece of data analysis is repeating a similar calculation, over and over, in an automated fashion.\nFor example, you may have a table of a names that you'd like to split into first and last, or perhaps of dates that you'd like to convert to some standard format.\nOne of Python's answers to this is the *iterator* syntax.\nWe've seen this already with the ``range`` iterator:",
"_____no_output_____"
]
],
[
[
"for i in range(10):\n print(i, end=' ')",
"0 1 2 3 4 5 6 7 8 9 "
]
],
[
[
"Here we're going to dig a bit deeper.\nIt turns out that in Python 3, ``range`` is not a list, but is something called an *iterator*, and learning how it works is key to understanding a wide class of very useful Python functionality.",
"_____no_output_____"
],
[
"## Iterating over lists\nIterators are perhaps most easily understood in the concrete case of iterating through a list.\nConsider the following:",
"_____no_output_____"
]
],
[
[
"for value in [2, 4, 6, 8, 10]:\n # do some operation\n print(value + 1, end=' ')",
"3 5 7 9 11 "
]
],
[
[
"The familiar \"``for x in y``\" syntax allows us to repeat some operation for each value in the list.\nThe fact that the syntax of the code is so close to its English description (\"*for [each] value in [the] list*\") is just one of the syntactic choices that makes Python such an intuitive language to learn and use.\n\nBut the face-value behavior is not what's *really* happening.\nWhen you write something like \"``for val in L``\", the Python interpreter checks whether it has an *iterator* interface, which you can check yourself with the built-in ``iter`` function:",
"_____no_output_____"
]
],
[
[
"iter([2, 4, 6, 8, 10])",
"_____no_output_____"
]
],
[
[
"It is this iterator object that provides the functionality required by the ``for`` loop.\nThe ``iter`` object is a container that gives you access to the next object for as long as it's valid, which can be seen with the built-in function ``next``:",
"_____no_output_____"
]
],
[
[
"I = iter([2, 4, 6, 8, 10])",
"_____no_output_____"
],
[
"print(next(I))",
"2\n"
],
[
"print(next(I))",
"4\n"
],
[
"print(next(I))",
"6\n"
]
],
[
[
"What is the purpose of this level of indirection?\nWell, it turns out this is incredibly useful, because it allows Python to treat things as lists that are *not actually lists*.",
"_____no_output_____"
],
[
"## ``range()``: A List Is Not Always a List\nPerhaps the most common example of this indirect iteration is the ``range()`` function in Python 3 (named ``xrange()`` in Python 2), which returns not a list, but a special ``range()`` object:",
"_____no_output_____"
]
],
[
[
"range(10)",
"_____no_output_____"
]
],
[
[
"``range``, like a list, exposes an iterator:",
"_____no_output_____"
]
],
[
[
"iter(range(10))",
"_____no_output_____"
]
],
[
[
"So Python knows to treat it *as if* it's a list:",
"_____no_output_____"
]
],
[
[
"for i in range(10):\n print(i, end=' ')",
"0 1 2 3 4 5 6 7 8 9 "
]
],
[
[
"The benefit of the iterator indirection is that *the full list is never explicitly created!*\nWe can see this by doing a range calculation that would overwhelm our system memory if we actually instantiated it (note that in Python 2, ``range`` creates a list, so running the following will not lead to good things!):",
"_____no_output_____"
]
],
[
[
"N = 10 ** 12\nfor i in range(N):\n if i >= 10: break\n print(i, end=', ')",
"0, 1, 2, 3, 4, 5, 6, 7, 8, 9, "
]
],
[
[
"If ``range`` were to actually create that list of one trillion values, it would occupy tens of terabytes of machine memory: a waste, given the fact that we're ignoring all but the first 10 values!\n\nIn fact, there's no reason that iterators ever have to end at all!\nPython's ``itertools`` library contains a ``count`` function that acts as an infinite range:",
"_____no_output_____"
]
],
[
[
"from itertools import count\n\nfor i in count():\n if i >= 10:\n break\n print(i, end=', ')",
"0, 1, 2, 3, 4, 5, 6, 7, 8, 9, "
]
],
[
[
"Had we not thrown-in a loop break here, it would go on happily counting until the process is manually interrupted or killed (using, for example, ``ctrl-C``).",
"_____no_output_____"
],
[
"## Useful Iterators\nThis iterator syntax is used nearly universally in Python built-in types as well as the more data science-specific objects we'll explore in later sections.\nHere we'll cover some of the more useful iterators in the Python language:",
"_____no_output_____"
],
[
"### ``enumerate``\nOften you need to iterate not only the values in an array, but also keep track of the index.\nYou might be tempted to do things this way:",
"_____no_output_____"
]
],
[
[
"L = [2, 4, 6, 8, 10]\nfor i in range(len(L)):\n print(i, L[i])",
"0 2\n1 4\n2 6\n3 8\n4 10\n"
]
],
[
[
"Although this does work, Python provides a cleaner syntax using the ``enumerate`` iterator:",
"_____no_output_____"
]
],
[
[
"for i, val in enumerate(L):\n print(i, val)",
"0 2\n1 4\n2 6\n3 8\n4 10\n"
]
],
[
[
"This is the more \"Pythonic\" way to enumerate the indices and values in a list.",
"_____no_output_____"
],
[
"### ``zip``\nOther times, you may have multiple lists that you want to iterate over simultaneously.\nYou could certainly iterate over the index as in the non-Pythonic example we looked at previously, but it is better to use the ``zip`` iterator, which zips together iterables:",
"_____no_output_____"
]
],
[
[
"L = [2, 4, 6, 8, 10]\nR = [3, 6, 9, 12, 15]\nfor lval, rval in zip(L, R):\n print(lval, rval)",
"2 3\n4 6\n6 9\n8 12\n10 15\n"
]
],
[
[
"Any number of iterables can be zipped together, and if they are different lengths, the shortest will determine the length of the ``zip``.",
"_____no_output_____"
],
[
"### ``map`` and ``filter``\nThe ``map`` iterator takes a function and applies it to the values in an iterator:",
"_____no_output_____"
]
],
[
[
"# find the first 10 square numbers\nsquare = lambda x: x ** 2\nfor val in map(square, range(10)):\n print(val, end=' ')",
"0 1 4 9 16 25 36 49 64 81 "
]
],
[
[
"The ``filter`` iterator looks similar, except it only passes-through values for which the filter function evaluates to True:",
"_____no_output_____"
]
],
[
[
"# find values up to 10 for which x % 2 is zero\nis_even = lambda x: x % 2 == 0\nfor val in filter(is_even, range(10)):\n print(val, end=' ')",
"0 2 4 6 8 "
]
],
[
[
"The ``map`` and ``filter`` functions, along with the ``reduce`` function (which lives in Python's ``functools`` module) are fundamental components of the *functional programming* style, which, while not a dominant programming style in the Python world, has its outspoken proponents (see, for example, the [pytoolz](https://toolz.readthedocs.org/en/latest/) library).",
"_____no_output_____"
],
[
"### Iterators as function arguments\n\nWe saw in [``*args`` and ``**kwargs``: Flexible Arguments](#*args-and-**kwargs:-Flexible-Arguments). that ``*args`` and ``**kwargs`` can be used to pass sequences and dictionaries to functions.\nIt turns out that the ``*args`` syntax works not just with sequences, but with any iterator:",
"_____no_output_____"
]
],
[
[
"print(*range(10))",
"0 1 2 3 4 5 6 7 8 9\n"
]
],
[
[
"So, for example, we can get tricky and compress the ``map`` example from before into the following:",
"_____no_output_____"
]
],
[
[
"print(*map(lambda x: x ** 2, range(10)))",
"0 1 4 9 16 25 36 49 64 81\n"
]
],
[
[
"Using this trick lets us answer the age-old question that comes up in Python learners' forums: why is there no ``unzip()`` function which does the opposite of ``zip()``?\nIf you lock yourself in a dark closet and think about it for a while, you might realize that the opposite of ``zip()`` is... ``zip()``! The key is that ``zip()`` can zip-together any number of iterators or sequences. Observe:",
"_____no_output_____"
]
],
[
[
"L1 = (1, 2, 3, 4)\nL2 = ('a', 'b', 'c', 'd')",
"_____no_output_____"
],
[
"z = zip(L1, L2)\nprint(*z)",
"(1, 'a') (2, 'b') (3, 'c') (4, 'd')\n"
],
[
"z = zip(L1, L2)\nnew_L1, new_L2 = zip(*z)\nprint(new_L1, new_L2)",
"(1, 2, 3, 4) ('a', 'b', 'c', 'd')\n"
]
],
[
[
"Ponder this for a while. If you understand why it works, you'll have come a long way in understanding Python iterators!",
"_____no_output_____"
],
[
"## Specialized Iterators: ``itertools``\n\nWe briefly looked at the infinite ``range`` iterator, ``itertools.count``.\nThe ``itertools`` module contains a whole host of useful iterators; it's well worth your while to explore the module to see what's available.\nAs an example, consider the ``itertools.permutations`` function, which iterates over all permutations of a sequence:",
"_____no_output_____"
]
],
[
[
"from itertools import permutations\np = permutations(range(3))\nprint(*p)",
"(0, 1, 2) (0, 2, 1) (1, 0, 2) (1, 2, 0) (2, 0, 1) (2, 1, 0)\n"
]
],
[
[
"Similarly, the ``itertools.combinations`` function iterates over all unique combinations of ``N`` values within a list:",
"_____no_output_____"
]
],
[
[
"from itertools import combinations\nc = combinations(range(4), 2)\nprint(*c)",
"(0, 1) (0, 2) (0, 3) (1, 2) (1, 3) (2, 3)\n"
]
],
[
[
"Somewhat related is the ``product`` iterator, which iterates over all sets of pairs between two or more iterables:",
"_____no_output_____"
]
],
[
[
"from itertools import product\np = product('ab', range(3))\nprint(*p)",
"('a', 0) ('a', 1) ('a', 2) ('b', 0) ('b', 1) ('b', 2)\n"
]
],
[
[
"Many more useful iterators exist in ``itertools``: the full list can be found, along with some examples, in Python's [online documentation](https://docs.python.org/3.5/library/itertools.html).",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
"<!--NAVIGATION-->\n< [Errors and Exceptions](09-Errors-and-Exceptions.ipynb) | [Contents](Index.ipynb) | [List Comprehensions](11-List-Comprehensions.ipynb) >",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
d091f923d809575310567e0f3089d2a5e814b27a | 74,156 | ipynb | Jupyter Notebook | lessons/04_spreadout/04_04_Heat_Equation_2D_Implicit.ipynb | Fluidentity/numerical-mooc | 083bbe9dc923b0ada6db2ebfbe13392fb66c6fbc | [
"CC-BY-3.0"
] | null | null | null | lessons/04_spreadout/04_04_Heat_Equation_2D_Implicit.ipynb | Fluidentity/numerical-mooc | 083bbe9dc923b0ada6db2ebfbe13392fb66c6fbc | [
"CC-BY-3.0"
] | null | null | null | lessons/04_spreadout/04_04_Heat_Equation_2D_Implicit.ipynb | Fluidentity/numerical-mooc | 083bbe9dc923b0ada6db2ebfbe13392fb66c6fbc | [
"CC-BY-3.0"
] | null | null | null | 71.717602 | 32,856 | 0.716031 | [
[
[
"###### Content under Creative Commons Attribution license CC-BY 4.0, code under MIT license (c)2014 L.A. Barba, G.F. Forsyth, C.D. Cooper.",
"_____no_output_____"
],
[
"# Spreading out",
"_____no_output_____"
],
[
"We're back! This is the fourth notebook of _Spreading out: parabolic PDEs,_ Module 4 of the course [**\"Practical Numerical Methods with Python\"**](https://openedx.seas.gwu.edu/courses/course-v1:MAE+MAE6286+2017/about). \n\nIn the [previous notebook](https://nbviewer.jupyter.org/github/numerical-mooc/numerical-mooc/blob/master/lessons/04_spreadout/04_03_Heat_Equation_2D_Explicit.ipynb), we solved a 2D problem for the first time, using an explicit scheme. We know explicit schemes have stability constraints that might make them impractical in some cases, due to requiring a very small time step. Implicit schemes are unconditionally stable, offering the advantage of larger time steps; in [notebook 2](https://nbviewer.jupyter.org/github/numerical-mooc/numerical-mooc/blob/master/lessons/04_spreadout/04_02_Heat_Equation_1D_Implicit.ipynb), we look at the 1D implicit solution of diffusion. Already, that was quite a lot of work: setting up a matrix of coefficients and a right-hand-side vector, while taking care of the boundary conditions, and then solving the linear system. And now, we want to do implicit schemes in 2D—are you ready for this challenge?",
"_____no_output_____"
],
[
"## 2D Heat conduction",
"_____no_output_____"
],
[
"We already studied 2D heat conduction in the previous lesson, but now we want to work out how to build an implicit solution scheme. To refresh your memory, here is the heat equation again:\n\n$$\n\\begin{equation}\n\\frac{\\partial T}{\\partial t} = \\alpha \\left(\\frac{\\partial^2 T}{\\partial x^2} + \\frac{\\partial^2 T}{\\partial y^2} \\right)\n\\end{equation}\n$$\n\nOur previous solution used a Dirichlet boundary condition on the left and bottom boundaries, with $T(x=0)=T(y=0)=100$, and a Neumann boundary condition with zero flux on the top and right edges, with $q_x=q_y=0$.\n\n$$\n\\left( \\left.\\frac{\\partial T}{\\partial y}\\right|_{y=0.1} = q_y \\right) \\quad \\text{and} \\quad \\left( \\left.\\frac{\\partial T}{\\partial x}\\right|_{x=0.1} = q_x \\right)\n$$\n\nFigure 1 shows a sketch of the problem set up for our hypothetical computer chip with two hot edges and two insulated edges.",
"_____no_output_____"
],
[
"#### <img src=\"./figures/2dchip.svg\" width=\"400px\"> Figure 1: Simplified microchip problem setup.",
"_____no_output_____"
],
[
"### Implicit schemes in 2D",
"_____no_output_____"
],
[
"An implicit discretization will evaluate the spatial derivatives at the next time level, $t^{n+1}$, using the unknown values of the solution variable. For the 2D heat equation with central difference in space, that is written as:\n\n$$\n\\begin{equation}\n \\begin{split}\n & \\frac{T^{n+1}_{i,j} - T^n_{i,j}}{\\Delta t} = \\\\\n & \\quad \\alpha \\left( \\frac{T^{n+1}_{i+1, j} - 2T^{n+1}_{i,j} + T^{n+1}_{i-1,j}}{\\Delta x^2} + \\frac{T^{n+1}_{i, j+1} - 2T^{n+1}_{i,j} + T^{n+1}_{i,j-1}}{\\Delta y^2} \\right) \\\\\n \\end{split}\n\\end{equation}\n$$\n\nThis equation looks better when we put what we *don't know* on the left and what we *do know* on the right. Make sure to work this out yourself on a piece of paper.\n\n$$\n\\begin{equation}\n \\begin{split}\n & -\\frac{\\alpha \\Delta t}{\\Delta x^2} \\left( T^{n+1}_{i-1,j} + T^{n+1}_{i+1,j} \\right) + \\left( 1 + 2 \\frac{\\alpha \\Delta t}{\\Delta x^2} + 2 \\frac{\\alpha \\Delta t}{\\Delta y^2} \\right) T^{n+1}_{i,j} \\\\\n& \\quad \\quad \\quad -\\frac{\\alpha \\Delta t}{\\Delta y^2} \\left( T^{n+1}_{i,j-1} + T^{n+1}_{i,j+1} \\right) = T^n_{i,j} \\\\\n \\end{split}\n\\end{equation}\n$$\n\nTo make this discussion easier, let's assume that the mesh spacing is the same in both directions and $\\Delta x=\\Delta y = \\delta$:\n\n$$\n\\begin{equation}\n-T^{n+1}_{i-1,j} - T^{n+1}_{i+1,j} + \\left(\\frac{\\delta^2}{\\alpha \\Delta t} + 4 \\right) T^{n+1}_{i,j} - T^{n+1}_{i,j-1}-T^{n+1}_{i,j+1} = \\frac{\\delta^2}{\\alpha \\Delta t}T^n_{i,j}\n\\end{equation}\n$$\n\nJust like in the one-dimensional case, $T_{i,j}$ appears in the equation for $T_{i-1,j}$, $T_{i+1,j}$, $T_{i,j+1}$ and $T_{i,j-1}$, and we can form a linear system to advance in time. But, how do we construct the matrix in this case? What are the $(i+1,j)$, $(i-1,j)$, $(i,j+1)$, and $(i,j-1)$ positions in the matrix?\n\nWith explicit schemes we don't need to worry about these things. We can lay out the data just as it is in the physical problem. We had an array `T` that was a 2-dimensional matrix. To fetch the temperature in the next node in the $x$ direction $(T_{i+1,j})$ we just did `T[j,i+1]`, and likewise in the $y$ direction $(T_{i,j+1})$ was in `T[j+1,i]`. In implicit schemes, we need to think a bit harder about how the data is mapped to the physical problem.\n\nAlso, remember from the [notebook on 1D-implicit schemes](https://nbviewer.jupyter.org/github/numerical-mooc/numerical-mooc/blob/master/lessons/04_spreadout/04_02_Heat_Equation_1D_Implicit.ipynb) that the linear system had $N-2$ elements? We applied boundary conditions on nodes $i=0$ and $i=N-1$, and they were not modified by the linear system. In 2D, this becomes a bit more complicated.\n\nLet's use Figure 1, representing a set of grid nodes in two dimensions, to guide the discussion.",
"_____no_output_____"
],
[
"#### <img src=\"./figures/2D_discretization.png\"> Figure 2: Layout of matrix elements in 2D problem",
"_____no_output_____"
],
[
"Say we have the 2D domain of size $L_x\\times L_y$ discretized in $n_x$ and $n_y$ points. We can divide the nodes into boundary nodes (empty circles) and interior nodes (filled circles).\n\nThe boundary nodes, as the name says, are on the boundary. They are the nodes with indices $(i=0,j)$, $(i=n_x-1,j)$, $(i,j=0)$, and $(i,j=n_y-1)$, and boundary conditions are enforced there.\n\nThe interior nodes are not on the boundary, and the finite-difference equation acts on them. If we leave the boundary nodes aside for the moment, then the grid will have $(n_x-2)\\cdot(n_y-2)$ nodes that need to be updated on each time step. This is the number of unknowns in the linear system. The matrix of coefficients will have $\\left( (n_x-2)\\cdot(n_y-2) \\right)^2$ elements (most of them zero!).\n\nTo construct the matrix, we will iterate over the nodes in an x-major order: index $i$ will run faster. The order will be \n\n* $(i=1,j=1)$\n* $(i=2,j=1)$ ...\n* $(i=nx-2,j=1)$\n* $(i=1,j=2)$\n* $(i=2,j=2)$ ... \n* $(i=n_x-2,j=n_y-2)$. \n\nThat is the ordering represented by dotted line on Figure 1. Of course, if you prefer to organize the nodes differently, feel free to do so!\n\nBecause we chose this ordering, the equation for nodes $(i-1,j)$ and $(i+1,j)$ will be just before and after $(i,j)$, respectively. But what about $(i,j-1)$ and $(i,j+1)$? Even though in the physical problem they are very close, the equations are $n_x-2$ places apart! This can tie your head in knots pretty quickly. \n\n_The only way to truly understand it is to make your own diagrams and annotations on a piece of paper and reconstruct this argument!_",
"_____no_output_____"
],
[
"### Boundary conditions",
"_____no_output_____"
],
[
"Before we attempt to build the matrix, we need to think about boundary conditions. There is some bookkeeping to be done here, so bear with us for a moment.\n\nSay, for example, that the left and bottom boundaries have Dirichlet boundary conditions, and the top and right boundaries have Neumann boundary conditions.\n\nLet's look at each case:\n\n**Bottom boundary:**\n \nThe equation for $j=1$ (interior points adjacent to the bottom boundary) uses values from $j=0$, which are known. Let's put that on the right-hand side of the equation. We get this equation for all points across the $x$-axis that are adjacent to the bottom boundary:\n\n$$\n\\begin{equation}\n \\begin{split}\n -T^{n+1}_{i-1,1} - T^{n+1}_{i+1,1} + \\left( \\frac{\\delta^2}{\\alpha \\Delta t} + 4 \\right) T^{n+1}_{i,1} - T^{n+1}_{i,j+1} \\qquad & \\\\\n = \\frac{\\delta^2}{\\alpha \\Delta t} T^n_{i,1} + T^{n+1}_{i,0} & \\\\\n \\end{split}\n\\end{equation}\n$$\n\n**Left boundary:**\n\nLike for the bottom boundary, the equation for $i=1$ (interior points adjacent to the left boundary) uses known values from $i=0$, and we will put that on the right-hand side:\n\n$$\n\\begin{equation}\n \\begin{split}\n -T^{n+1}_{2,j} + \\left( \\frac{\\delta^2}{\\alpha \\Delta t} + 4 \\right) T^{n+1}_{1,j} - T^{n+1}_{1,j-1} - T^{n+1}_{1,j+1} \\qquad & \\\\\n = \\frac{\\delta^2}{\\alpha \\Delta t} T^n_{1,j} + T^{n+1}_{0,j} & \\\\\n \\end{split}\n\\end{equation}\n$$\n\n**Right boundary:**\n\nSay the boundary condition is $\\left. \\frac{\\partial T}{\\partial x} \\right|_{x=L_x} = q_x$. Its finite-difference approximation is\n\n$$\n\\begin{equation}\n \\frac{T^{n+1}_{n_x-1,j} - T^{n+1}_{n_x-2,j}}{\\delta} = q_x\n\\end{equation}\n$$\n\nWe can write $T^{n+1}_{n_x-1,j} = \\delta q_x + T^{n+1}_{n_x-2,j}$ to get the finite difference equation for $i=n_x-2$:\n\n$$\n\\begin{equation}\n \\begin{split}\n -T^{n+1}_{n_x-3,j} + \\left( \\frac{\\delta^2}{\\alpha \\Delta t} + 3 \\right) T^{n+1}_{n_x-2,j} - T^{n+1}_{n_x-2,j-1} - T^{n+1}_{n_x-2,j+1} \\qquad & \\\\\n = \\frac{\\delta^2}{\\alpha \\Delta t} T^n_{n_x-2,j} + \\delta q_x & \\\\\n \\end{split}\n\\end{equation}\n$$\n\nNot sure about this? Grab pen and paper! _Please_, check this yourself. It will help you understand!\n\n**Top boundary:**\n\nNeumann boundary conditions specify the derivative normal to the boundary: $\\left. \\frac{\\partial T}{\\partial y} \\right|_{y=L_y} = q_y$. No need to repeat what we did for the right boundary, right? The equation for $j=n_y-2$ is\n\n$$\n\\begin{equation}\n \\begin{split}\n -T^{n+1}_{i-1,n_y-2} - T^{n+1}_{i+1,n_y-2} + \\left( \\frac{\\delta^2}{\\alpha \\Delta t} + 3 \\right) T^{n+1}_{i,n_y-2} - T^{n+1}_{i,n_y-3} \\qquad & \\\\\n = \\frac{\\delta^2}{\\alpha \\Delta t} T^n_{i,n_y-2} + \\delta q_y & \\\\\n \\end{split}\n\\end{equation}\n$$\n\nSo far, we have then 5 possible cases: bottom, left, right, top, and interior points. Does this cover everything? What about corners?",
"_____no_output_____"
],
[
"**Bottom-left corner**\n\nAt $T_{1,1}$ there is a Dirichlet boundary condition at $i=0$ and $j=0$. This equation is:\n\n$$\n\\begin{equation}\n \\begin{split}\n -T^{n+1}_{2,1} + \\left( \\frac{\\delta^2}{\\alpha \\Delta t} + 4 \\right) T^{n+1}_{1,1} - T^{n+1}_{1,2} \\qquad & \\\\\n = \\frac{\\delta^2}{\\alpha \\Delta t} T^n_{1,1} + T^{n+1}_{0,1} + T^{n+1}_{1,0} & \\\\\n \\end{split}\n\\end{equation}\n$$\n\n**Top-left corner:**\n\nAt $T_{1,n_y-2}$ there is a Dirichlet boundary condition at $i=0$ and a Neumann boundary condition at $i=n_y-1$. This equation is:\n\n$$\n\\begin{equation}\n \\begin{split}\n -T^{n+1}_{2,n_y-2} + \\left( \\frac{\\delta^2}{\\alpha \\Delta t} + 3 \\right) T^{n+1}_{1,n_y-2} - T^{n+1}_{1,n_y-3} \\qquad & \\\\\n = \\frac{\\delta^2}{\\alpha \\Delta t} T^n_{1,n_y-2} + T^{n+1}_{0,n_y-2} + \\delta q_y & \\\\\n \\end{split}\n\\end{equation}\n$$\n\n**Top-right corner**\n\nAt $T_{n_x-2,n_y-2}$, there are Neumann boundary conditions at both $i=n_x-1$ and $j=n_y-1$. The finite difference equation is then\n\n$$\n\\begin{equation}\n \\begin{split}\n -T^{n+1}_{n_x-3,n_y-2} + \\left( \\frac{\\delta^2}{\\alpha \\Delta t} + 2 \\right) T^{n+1}_{n_x-2,n_y-2} - T^{n+1}_{n_x-2,n_y-3} \\qquad & \\\\\n = \\frac{\\delta^2}{\\alpha \\Delta t} T^n_{n_x-2,n_y-2} + \\delta(q_x + q_y) & \\\\\n \\end{split}\n\\end{equation}\n$$\n\n**Bottom-right corner**\n\nTo calculate $T_{n_x-2,1}$ we need to consider a Dirichlet boundary condition to the bottom and a Neumann boundary condition to the right. We will get a similar equation to the top-left corner!\n\n$$\n\\begin{equation}\n \\begin{split}\n -T^{n+1}_{n_x-3,1} + \\left( \\frac{\\delta^2}{\\alpha \\Delta t} + 3 \\right) T^{n+1}_{n_x-2,1} - T^{n+1}_{n_x-2,2} \\qquad & \\\\\n = \\frac{\\delta^2}{\\alpha \\Delta t} T^n_{n_x-2,1} + T^{n+1}_{n_x-2,0} + \\delta q_x & \\\\\n \\end{split}\n\\end{equation}\n$$\n\nOkay, now we are actually ready. We have checked every possible case!",
"_____no_output_____"
],
[
"### The linear system",
"_____no_output_____"
],
[
"Like in the previous lesson introducing implicit schemes, we will solve a linear system at every time step:\n\n$$\n[A][T^{n+1}_\\text{int}] = [b]+[b]_{b.c.}\n$$\n\nThe coefficient matrix now takes some more work to figure out and to build in code. There is no substitute for you working this out patiently on paper!\n\nThe structure of the matrix can be described as a series of diagonal blocks, and lots of zeros elsewhere. Look at Figure 3, representing the block structure of the coefficient matrix, and refer back to Figure 2, showing the discretization grid in physical space. The first row of interior points, adjacent to the bottom boundary, generates the matrix block labeled $A_1$. The top row of interior points, adjacent to the top boundary generates the matrix block labeled $A_3$. All other interior points in the grid generate similar blocks, labeled $A_2$ on Figure 3.",
"_____no_output_____"
],
[
"#### <img src=\"./figures/implicit-matrix-blocks.png\"> Figure 3: Sketch of coefficient-matrix blocks.",
"_____no_output_____"
],
[
"#### <img src=\"./figures/matrix-blocks-on-grid.png\"> Figure 4: Grid points corresponding to each matrix-block type.",
"_____no_output_____"
],
[
"The matrix block $A_1$ is\n\n<img src=\"./figures/A_1.svg\" width=\"640px\">\n\nThe block matrix $A_2$ is\n\n<img src=\"./figures/A_2.svg\" width=\"640px\">\n\nThe block matrix $A_3$ is\n\n<img src=\"./figures/A_3.svg\" width=\"640px\">",
"_____no_output_____"
],
[
"Vector $T^{n+1}_\\text{int}$ contains the temperature of the interior nodes in the next time step. It is:\n\n$$\n\\begin{equation}\nT^{n+1}_\\text{int} = \\left[\n\\begin{array}{c}\nT^{n+1}_{1,1}\\\\\nT^{n+1}_{2,1} \\\\\n\\vdots \\\\\nT^{n+1}_{n_x-2,1} \\\\\nT^{n+1}_{2,1} \\\\\n\\vdots \\\\\nT^{n+1}_{n_x-2,n_y-2}\n\\end{array}\n\\right]\n\\end{equation}\n$$\n\nRemember the x-major ordering we chose!",
"_____no_output_____"
],
[
"Finally, the right-hand side is\n\\begin{equation}\n[b]+[b]_{b.c.} = \n\\left[\\begin{array}{c}\n\\sigma^\\prime T^n_{1,1} + T^{n+1}_{0,1} + T^{n+1}_{1,0} \\\\\n\\sigma^\\prime T^n_{2,0} + T^{n+1}_{2,0} \\\\\n\\vdots \\\\\n\\sigma^\\prime T^n_{n_x-2,1} + T^{n+1}_{n_x-2,0} + \\delta q_x \\\\\n\\sigma^\\prime T^n_{1,2} + T^{n+1}_{0,2} \\\\\n\\vdots \\\\\n\\sigma^\\prime T^n_{n_x-2,n_y-2} + \\delta(q_x + q_y)\n\\end{array}\\right]\n\\end{equation}",
"_____no_output_____"
],
[
"where $\\sigma^\\prime = 1/\\sigma = \\delta^2/\\alpha \\Delta t$. The matrix looks very ugly, but it is important you understand it! Think about it. Can you answer:\n * Why a -1 factor appears $n_x-2$ columns after the diagonal? What about $n_x-2$ columns before the diagonal?\n * Why in row $n_x-2$ the position after the diagonal contains a 0?\n * Why in row $n_x-2$ the diagonal is $\\sigma^\\prime + 3$ rather than $\\sigma^\\prime + 4$?\n * Why in the last row the diagonal is $\\sigma^\\prime + 2$ rather than $\\sigma^\\prime + 4$?\n \nIf you can answer those questions, you are in good shape to continue!",
"_____no_output_____"
],
[
"Let's write a function that will generate the matrix and right-hand side for the heat conduction problem in the previous notebook. Remember, we had Dirichlet boundary conditions in the left and bottom, and zero-flux Neumann boundary condition on the top and right $(q_x=q_y=0)$. \n\nAlso, we'll import `scipy.linalg.solve` because we need to solve a linear system.",
"_____no_output_____"
]
],
[
[
"import numpy\nfrom scipy import linalg",
"_____no_output_____"
],
[
"def lhs_operator(M, N, sigma):\n \"\"\"\n Assembles and returns the implicit operator\n of the system for the 2D diffusion equation.\n We use a Dirichlet condition at the left and\n bottom boundaries and a Neumann condition\n (zero-gradient) at the right and top boundaries.\n \n Parameters\n ----------\n M : integer\n Number of interior points in the x direction.\n N : integer\n Number of interior points in the y direction.\n sigma : float\n Value of alpha * dt / dx**2.\n \n Returns\n -------\n A : numpy.ndarray\n The implicit operator as a 2D array of floats\n of size M*N by M*N.\n \"\"\"\n A = numpy.zeros((M * N, M * N))\n for j in range(N):\n for i in range(M):\n I = j * M + i # row index\n # Get index of south, west, east, and north points.\n south, west, east, north = I - M, I - 1, I + 1, I + M\n # Setup coefficients at corner points.\n if i == 0 and j == 0: # bottom-left corner\n A[I, I] = 1.0 / sigma + 4.0\n A[I, east] = -1.0\n A[I, north] = -1.0\n elif i == M - 1 and j == 0: # bottom-right corner\n A[I, I] = 1.0 / sigma + 3.0\n A[I, west] = -1.0\n A[I, north] = -1.0\n elif i == 0 and j == N - 1: # top-left corner\n A[I, I] = 1.0 / sigma + 3.0\n A[I, south] = -1.0\n A[I, east] = -1.0\n elif i == M - 1 and j == N - 1: # top-right corner\n A[I, I] = 1.0 / sigma + 2.0\n A[I, south] = -1.0\n A[I, west] = -1.0\n # Setup coefficients at side points (excluding corners).\n elif i == 0: # left side\n A[I, I] = 1.0 / sigma + 4.0\n A[I, south] = -1.0\n A[I, east] = -1.0\n A[I, north] = -1.0\n elif i == M - 1: # right side\n A[I, I] = 1.0 / sigma + 3.0\n A[I, south] = -1.0\n A[I, west] = -1.0\n A[I, north] = -1.0\n elif j == 0: # bottom side\n A[I, I] = 1.0 / sigma + 4.0\n A[I, west] = -1.0\n A[I, east] = -1.0\n A[I, north] = -1.0\n elif j == N - 1: # top side\n A[I, I] = 1.0 / sigma + 3.0\n A[I, south] = -1.0\n A[I, west] = -1.0\n A[I, east] = -1.0\n # Setup coefficients at interior points.\n else:\n A[I, I] = 1.0 / sigma + 4.0\n A[I, south] = -1.0\n A[I, west] = -1.0\n A[I, east] = -1.0\n A[I, north] = -1.0\n return A",
"_____no_output_____"
],
[
"def rhs_vector(T, M, N, sigma, Tb):\n \"\"\"\n Assembles and returns the right-hand side vector\n of the system for the 2D diffusion equation.\n We use a Dirichlet condition at the left and\n bottom boundaries and a Neumann condition\n (zero-gradient) at the right and top boundaries.\n \n Parameters\n ----------\n T : numpy.ndarray\n The temperature distribution as a 1D array of floats.\n M : integer\n Number of interior points in the x direction.\n N : integer\n Number of interior points in the y direction.\n sigma : float\n Value of alpha * dt / dx**2.\n Tb : float\n Boundary value for Dirichlet conditions.\n \n Returns\n -------\n b : numpy.ndarray\n The right-hand side vector as a 1D array of floats\n of size M*N.\n \"\"\"\n b = 1.0 / sigma * T\n # Add Dirichlet term at points located next\n # to the left and bottom boundaries.\n for j in range(N):\n for i in range(M):\n I = j * M + i\n if i == 0:\n b[I] += Tb\n if j == 0:\n b[I] += Tb\n return b",
"_____no_output_____"
]
],
[
[
"The solution of the linear system $(T^{n+1}_\\text{int})$ contains the temperatures of the interior points at the next time step in a 1D array. We will also create a function that will take the values of $T^{n+1}_\\text{int}$ and put them in a 2D array that resembles the physical domain.",
"_____no_output_____"
]
],
[
[
"def map_1d_to_2d(T_1d, nx, ny, Tb):\n \"\"\"\n Maps a 1D array of the temperature at the interior points\n to a 2D array that includes the boundary values.\n \n Parameters\n ----------\n T_1d : numpy.ndarray\n The temperature at the interior points as a 1D array of floats.\n nx : integer\n Number of points in the x direction of the domain.\n ny : integer\n Number of points in the y direction of the domain.\n Tb : float\n Boundary value for Dirichlet conditions.\n \n Returns\n -------\n T : numpy.ndarray\n The temperature distribution in the domain\n as a 2D array of size ny by nx.\n \"\"\"\n T = numpy.zeros((ny, nx))\n # Get the value at interior points.\n T[1:-1, 1:-1] = T_1d.reshape((ny - 2, nx - 2))\n # Use Dirichlet condition at left and bottom boundaries.\n T[:, 0] = Tb\n T[0, :] = Tb\n # Use Neumann condition at right and top boundaries.\n T[:, -1] = T[:, -2]\n T[-1, :] = T[-2, :]\n return T",
"_____no_output_____"
]
],
[
[
"And to advance in time, we will use",
"_____no_output_____"
]
],
[
[
"def btcs_implicit_2d(T0, nt, dt, dx, alpha, Tb):\n \"\"\"\n Computes and returns the distribution of the\n temperature after a given number of time steps.\n \n The 2D diffusion equation is integrated using\n Euler implicit in time and central differencing\n in space, with a Dirichlet condition at the left\n and bottom boundaries and a Neumann condition\n (zero-gradient) at the right and top boundaries.\n \n Parameters\n ----------\n T0 : numpy.ndarray\n The initial temperature distribution as a 2D array of floats.\n nt : integer\n Number of time steps to compute.\n dt : float\n Time-step size.\n dx : float\n Grid spacing in the x and y directions.\n alpha : float\n Thermal diffusivity of the plate.\n Tb : float\n Boundary value for Dirichlet conditions.\n \n Returns\n -------\n T : numpy.ndarray\n The temperature distribution as a 2D array of floats.\n \"\"\"\n # Get the number of points in each direction.\n ny, nx = T0.shape\n # Get the number of interior points in each direction.\n M, N = nx - 2, ny - 2\n # Compute the constant sigma.\n sigma = alpha * dt / dx**2\n # Create the implicit operator of the system.\n A = lhs_operator(M, N, sigma)\n # Integrate in time.\n T = T0[1:-1, 1:-1].flatten() # interior points as a 1D array\n I, J = int(M / 2), int(N / 2) # indices of the center\n for n in range(nt):\n # Compute the right-hand side of the system.\n b = rhs_vector(T, M, N, sigma, Tb)\n # Solve the system with scipy.linalg.solve.\n T = linalg.solve(A, b)\n # Check if the center of the domain has reached T = 70C.\n if T[J * M + I] >= 70.0:\n break\n print('[time step {}] Center at T={:.2f} at t={:.2f} s'\n .format(n + 1, T[J * M + I], (n + 1) * dt))\n # Returns the temperature in the domain as a 2D array.\n return map_1d_to_2d(T, nx, ny, Tb)",
"_____no_output_____"
]
],
[
[
"Remember, we want the function to tell us when the center of the plate reaches $70^\\circ C$.",
"_____no_output_____"
],
[
"##### Dig deeper",
"_____no_output_____"
],
[
"For demonstration purposes, these functions are very explicit. But you can see a trend here, right? \n\nSay we start with a matrix with `1/sigma+4` in the main diagonal, and `-1` on the 4 other corresponding diagonals. Now, we have to modify the matrix only where the boundary conditions are affecting. We saw the impact of the Dirichlet and Neumann boundary condition on each position of the matrix, we just need to know in which position to perform those changes. \n\nA function that maps `i` and `j` into `row_number` would be handy, right? How about `row_number = (j-1)*(nx-2)+(i-1)`? By feeding `i` and `j` to that equation, you know exactly where to operate on the matrix. For example, `i=nx-2, j=2`, which is in row `row_number = 2*nx-5`, is next to a Neumann boundary condition: we have to substract one out of the main diagonal (`A[2*nx-5,2*nx-5]-=1`), and put a zero in the next column (`A[2*nx-5,2*nx-4]=0`). This way, the function can become much simpler!\n\nCan you use this information to construct a more general function `lhs_operator`? Can you make it such that the type of boundary condition is an input to the function? ",
"_____no_output_____"
],
[
"## Heat diffusion in 2D",
"_____no_output_____"
],
[
"Let's recast the 2D heat conduction from the previous notebook, and solve it with an implicit scheme. ",
"_____no_output_____"
]
],
[
[
"# Set parameters.\nLx = 0.01 # length of the plate in the x direction\nLy = 0.01 # length of the plate in the y direction\nnx = 21 # number of points in the x direction\nny = 21 # number of points in the y direction\ndx = Lx / (nx - 1) # grid spacing in the x direction\ndy = Ly / (ny - 1) # grid spacing in the y direction\nalpha = 1e-4 # thermal diffusivity\n\n# Define the locations along a gridline.\nx = numpy.linspace(0.0, Lx, num=nx)\ny = numpy.linspace(0.0, Ly, num=ny)\n\n# Compute the initial temperature distribution.\nTb = 100.0 # temperature at the left and bottom boundaries\nT0 = 20.0 * numpy.ones((ny, nx))\nT0[:, 0] = Tb\nT0[0, :] = Tb",
"_____no_output_____"
]
],
[
[
"We are ready to go!",
"_____no_output_____"
]
],
[
[
"# Set the time-step size based on CFL limit.\nsigma = 0.25\ndt = sigma * min(dx, dy)**2 / alpha # time-step size\nnt = 300 # number of time steps to compute\n\n# Compute the temperature along the rod.\nT = btcs_implicit_2d(T0, nt, dt, dx, alpha, Tb)",
"[time step 257] Center at T=70.00 at t=0.16 s\n"
]
],
[
[
"And plot,",
"_____no_output_____"
]
],
[
[
"from matplotlib import pyplot\n%matplotlib inline",
"_____no_output_____"
],
[
"# Set the font family and size to use for Matplotlib figures.\npyplot.rcParams['font.family'] = 'serif'\npyplot.rcParams['font.size'] = 16",
"_____no_output_____"
],
[
"# Plot the filled contour of the temperature.\npyplot.figure(figsize=(8.0, 5.0))\npyplot.xlabel('x [m]')\npyplot.ylabel('y [m]')\nlevels = numpy.linspace(20.0, 100.0, num=51)\ncontf = pyplot.contourf(x, y, T, levels=levels)\ncbar = pyplot.colorbar(contf)\ncbar.set_label('Temperature [C]')\npyplot.axis('scaled', adjustable='box');",
"_____no_output_____"
]
],
[
[
"Try this out with different values of `sigma`! You'll see that it will always give a stable solution!\n\nDoes this result match the explicit scheme from the previous notebook? Do they take the same amount of time to reach $70^\\circ C$ in the center of the plate? Now that we can use higher values of `sigma`, we need fewer time steps for the center of the plate to reach $70^\\circ C$! Of course, we need to be careful that `dt` is small enough to resolve the physics correctly.",
"_____no_output_____"
],
[
"---\n###### The cell below loads the style of the notebook",
"_____no_output_____"
]
],
[
[
"from IPython.core.display import HTML\ncss_file = '../../styles/numericalmoocstyle.css'\nHTML(open(css_file, 'r').read())",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
d09212d5f7a2298486edbd560d600111992b279c | 4,014 | ipynb | Jupyter Notebook | notebook/re_sub_subn.ipynb | puyopop/python-snippets | 9d70aa3b2a867dd22f5a5e6178a5c0c5081add73 | [
"MIT"
] | 174 | 2018-05-30T21:14:50.000Z | 2022-03-25T07:59:37.000Z | notebook/re_sub_subn.ipynb | puyopop/python-snippets | 9d70aa3b2a867dd22f5a5e6178a5c0c5081add73 | [
"MIT"
] | 5 | 2019-08-10T03:22:02.000Z | 2021-07-12T20:31:17.000Z | notebook/re_sub_subn.ipynb | puyopop/python-snippets | 9d70aa3b2a867dd22f5a5e6178a5c0c5081add73 | [
"MIT"
] | 53 | 2018-04-27T05:26:35.000Z | 2022-03-25T07:59:37.000Z | 18.162896 | 100 | 0.449925 | [
[
[
"import re",
"_____no_output_____"
],
[
"s = '[email protected], [email protected], [email protected]'",
"_____no_output_____"
],
[
"result = re.sub(r'[a-z]+@[a-z]+\\.com', 'new-address', s)\nprint(result)",
"new-address, new-address, [email protected]\n"
],
[
"print(type(result))",
"<class 'str'>\n"
],
[
"result = re.sub(r'([a-z]+)@([a-z]+)\\.com', r'\\1@\\2.net', s)\nprint(result)",
"[email protected], [email protected], [email protected]\n"
],
[
"result = re.sub(r'(?P<local>[a-z]+)@(?P<SLD>[a-z]+)\\.com', r'\\g<local>@\\g<SLD>.net', s)\nprint(result)",
"[email protected], [email protected], [email protected]\n"
],
[
"result = re.sub(r'[a-z]+@[a-z]+\\.com', 'new-address', s, count=1)\nprint(result)",
"new-address, [email protected], [email protected]\n"
],
[
"result = re.subn(r'[a-z]+@[a-z]+\\.com', 'new-address', s)\nprint(result)",
"('new-address, new-address, [email protected]', 2)\n"
],
[
"print(result[0])",
"new-address, new-address, [email protected]\n"
],
[
"print(result[1])",
"2\n"
],
[
"result = re.subn(r'(?P<local>[a-z]+)@(?P<SLD>[a-z]+)\\.com', r'\\g<local>@\\g<SLD>.net', s)\nprint(result)",
"('[email protected], [email protected], [email protected]', 2)\n"
],
[
"result = re.subn(r'[a-z]+@[a-z]+\\.com', 'new-address', s, count=1)\nprint(result)",
"('new-address, [email protected], [email protected]', 1)\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d09217fd2b7eb0c3474495989e5cb6776f4df730 | 119,905 | ipynb | Jupyter Notebook | data-512-a1/StepByStepNotebook.ipynb | NalaniKai/data-512 | 7173817e7521624855498cb3902ed2e6c153a631 | [
"MIT"
] | null | null | null | data-512-a1/StepByStepNotebook.ipynb | NalaniKai/data-512 | 7173817e7521624855498cb3902ed2e6c153a631 | [
"MIT"
] | null | null | null | data-512-a1/StepByStepNotebook.ipynb | NalaniKai/data-512 | 7173817e7521624855498cb3902ed2e6c153a631 | [
"MIT"
] | null | null | null | 131.330778 | 90,144 | 0.840674 | [
[
[
"# DATA 512: A1 Data Curation Assignment\nBy: Megan Nalani Chun",
"_____no_output_____"
],
[
"## Step 1: Gathering the data <br>\nGather Wikipedia traffic from Jan 1, 2008 - August 30, 2020 <br>\n- Legacy Pagecounts API provides desktop and mobile traffic data from Dec. 2007 - July 2016 <br>\n- Pageviews API provides desktop, mobile web, and mobile app traffic data from July 2015 - last month. \n",
"_____no_output_____"
],
[
"First, import the json and requests libraries to call the Pagecounts and Pageviews APIs and save the output in json format.",
"_____no_output_____"
]
],
[
[
"import json\nimport requests",
"_____no_output_____"
]
],
[
[
"Second, set the location of the endpoints and header information. This information is needed to call the Pagecounts and Pageviews APIs.",
"_____no_output_____"
]
],
[
[
"endpoint_legacy = 'https://wikimedia.org/api/rest_v1/metrics/legacy/pagecounts/aggregate/{project}/{access-site}/{granularity}/{start}/{end}'\n\nendpoint_pageviews = 'https://wikimedia.org/api/rest_v1/metrics/pageviews/aggregate/{project}/{access}/{agent}/{granularity}/{start}/{end}'\n\nheaders = {\n 'User-Agent': 'https://github.com/NalaniKai/',\n 'From': '[email protected]'\n}",
"_____no_output_____"
]
],
[
[
"Third, define a function to call the APIs taking in the endpoint and parameters. This function returns the data in json format. ",
"_____no_output_____"
]
],
[
[
"def api_call(endpoint, parameters):\n call = requests.get(endpoint.format(**parameters), headers=headers)\n response = call.json()\n \n return response",
"_____no_output_____"
]
],
[
[
"Fourth, define a function to make the api_call() with the correct parameters and save the output to json file in format apiname_accesstype_startyearmonth_endyearmonth.json",
"_____no_output_____"
]
],
[
[
"def get_data(api, access_dict, params, endpoint, access_name):\n start = 0 #index of start date\n end = 1 #index of end data\n year_month = 6 #size of YYYYMM\n for access_type, start_end_dates in access_dict.items(): #get data for all access types in API\n params[access_name] = access_type\n data = api_call(endpoint, params)\n with open(api + \"_\" + access_type + \"_\" + start_end_dates[start][:year_month] + \"_\" + start_end_dates[end][:year_month] + \".json\", 'w') as f:\n json.dump(data, f) #save data",
"_____no_output_____"
]
],
[
[
"Fifth, define the parameters for the legacy page count API and call the get_data function. This will pull the data and save it.",
"_____no_output_____"
]
],
[
[
"api = \"pagecounts\"\naccess_type_legacy = {\"desktop-site\": [\"2008010100\", \"2016070100\"], #access type: start year_month, end year_month\n \"mobile-site\": [\"2014100100\", \"2016070100\"]} #used to save outputs with correct filenames\n#https://wikimedia.org/api/rest_v1/#!/Legacy_data/get_metrics_legacy_pagecounts_aggregate_project_access_site_granularity_start_end\nparams_legacy = {\"project\" : \"en.wikipedia.org\",\n \"granularity\" : \"monthly\",\n \"start\" : \"2008010100\",\n \"end\" : \"2016080100\" #will get data through July 2016\n }\nget_data(api, access_type_legacy, params_legacy, endpoint_legacy, \"access-site\")",
"_____no_output_____"
]
],
[
[
"Sixth, define the parameters for the page views API and call the get_data function. This will pull the data and save it.",
"_____no_output_____"
]
],
[
[
"api = \"pageviews\"\nstart_end_dates = [\"2015070100\", \"2020080100\"] #start year_month, end year_month\naccess_type_pageviews = {\"desktop\": start_end_dates, #access type: start year_month, end year_month\n \"mobile-app\": start_end_dates, \n \"mobile-web\": start_end_dates }\n#https://wikimedia.org/api/rest_v1/#!/Pageviews_data/get_metrics_pageviews_aggregate_project_access_agent_granularity_start_end\nparams_pageviews = {\"project\" : \"en.wikipedia.org\",\n \"access\" : \"mobile-web\",\n \"agent\" : \"user\", #remove crawler traffic\n \"granularity\" : \"monthly\",\n \"start\" : \"2008010100\",\n \"end\" : '2020090100' #will get data through August 2020\n }\nget_data(api, access_type_pageviews, params_pageviews, endpoint_pageviews, \"access\") ",
"_____no_output_____"
]
],
[
[
"## Step 2: Processing the data",
"_____no_output_____"
],
[
"First, create a function to read in all data files and extract the list of records from items.",
"_____no_output_____"
]
],
[
[
"def read_json(filename):\n with open(filename, 'r') as f:\n return json.load(f)[\"items\"]",
"_____no_output_____"
]
],
[
[
"Second, use the read_json function to get a list of records for each file.",
"_____no_output_____"
]
],
[
[
"pageviews_mobile_web = read_json(\"pageviews_mobile-web_201507_202008.json\")\npageviews_mobile_app = read_json(\"pageviews_mobile-app_201507_202008.json\")\npageviews_desktop = read_json(\"pageviews_desktop_201507_202008.json\")\npagecounts_mobile = read_json(\"pagecounts_mobile-site_201410_201607.json\")\npagecounts_desktop = read_json(\"pagecounts_desktop-site_200801_201607.json\")",
"_____no_output_____"
]
],
[
[
"Third, create a total mobile traffic count for each month using the mobile-app and mobile-web data from the pageviews API. The list of [timestamp, view_count] pairs data structure will enable easy transformation into a dataframe.",
"_____no_output_____"
]
],
[
[
"pageviews_mobile = [[r1[\"timestamp\"], r0[\"views\"] + r1[\"views\"]] for r0 in pageviews_mobile_web for r1 in pageviews_mobile_app if r0[\"timestamp\"] == r1[\"timestamp\"]]",
"_____no_output_____"
]
],
[
[
"Fourth, get the timestamps and values in the [timestamp, view_count] format for the desktop pageviews, desktop pagecounts, and mobile pagecounts. ",
"_____no_output_____"
]
],
[
[
"pageviews_desktop = [[record[\"timestamp\"], record[\"views\"]] for record in pageviews_desktop]\npagecounts_desktop = [[record[\"timestamp\"], record[\"count\"]] for record in pagecounts_desktop]\npagecounts_mobile = [[record[\"timestamp\"], record[\"count\"]] for record in pagecounts_mobile]",
"_____no_output_____"
]
],
[
[
"Fifth, import pandas library and transform data into dataframes.",
"_____no_output_____"
]
],
[
[
"import pandas as pd \npageview_desktop_views = pd.DataFrame(pageviews_desktop, columns=[\"timestamp\", \"pageview_desktop_views\"])\npageview_mobile_views = pd.DataFrame(pageviews_mobile, columns=[\"timestamp\", \"pageview_mobile_views\"])\npagecounts_desktop = pd.DataFrame(pagecounts_desktop, columns=[\"timestamp\", \"pagecount_desktop_views\"])\npagecounts_mobile = pd.DataFrame(pagecounts_mobile, columns=[\"timestamp\", \"pagecount_mobile_views\"])",
"_____no_output_____"
]
],
[
[
"Sixth, join page view dataframes and calculate total for all views.",
"_____no_output_____"
]
],
[
[
"df_pageviews = pd.merge(pageview_desktop_views, pageview_mobile_views, how=\"outer\", on=\"timestamp\")\ndf_pageviews[\"pageview_all_views\"] = df_pageviews[\"pageview_desktop_views\"] + df_pageviews[\"pageview_mobile_views\"]\ndf_pageviews.head()",
"_____no_output_____"
]
],
[
[
"Seventh, join page count dataframes. Then fill in NaN values with 0 to calculate total for all counts.",
"_____no_output_____"
]
],
[
[
"df_pagecounts = pd.merge(pagecounts_desktop, pagecounts_mobile, how=\"outer\", on=\"timestamp\")\ndf_pagecounts[\"pagecount_mobile_views\"] = df_pagecounts[\"pagecount_mobile_views\"].fillna(0)\ndf_pagecounts[\"pagecount_all_views\"] = df_pagecounts[\"pagecount_desktop_views\"] + df_pagecounts[\"pagecount_mobile_views\"]\ndf_pagecounts.head()",
"_____no_output_____"
]
],
[
[
"Eighth, join page count and page view dataframes into one table. Filling in missing values with 0.",
"_____no_output_____"
]
],
[
[
"df = pd.merge(df_pagecounts, df_pageviews, how=\"outer\", on=\"timestamp\")\ndf = df.fillna(0)\ndf.head()",
"_____no_output_____"
]
],
[
[
"Ninth, separate the timestamp into the year and month in YYYY and MM format for all the data. Remove the timestamp column.",
"_____no_output_____"
]
],
[
[
"df[\"year\"] = df[\"timestamp\"].apply(lambda date: date[:4])\ndf[\"month\"] = df[\"timestamp\"].apply(lambda date: date[4:6])\ndf.drop(\"timestamp\", axis=1, inplace=True)\ndf.head()",
"_____no_output_____"
]
],
[
[
"Tenth, save processed data to csv file without the index column. ",
"_____no_output_____"
]
],
[
[
"df.to_csv(\"en-wikipedia_traffic_200801-202008.csv\", index=False)",
"_____no_output_____"
]
],
[
[
"## Step 3: Analyze the data",
"_____no_output_____"
],
[
"First, fill 0 values with numpy.nan values so these values are not plotted on the chart.",
"_____no_output_____"
]
],
[
[
"import numpy as np\ndf.replace(0, np.nan, inplace=True)",
"_____no_output_____"
]
],
[
[
"Second, transform the year and month into a datetime.date type which will be used for the x-axis in the chart.",
"_____no_output_____"
]
],
[
[
"from datetime import date\ndate = df.apply(lambda r: date(int(r[\"year\"]), int(r[\"month\"]), 1), axis=1) ",
"_____no_output_____"
]
],
[
[
"Third, divide all page view counts by 1e6 so the chart is easier to read. Y-axis will be the values shown x 1,000,000.",
"_____no_output_____"
]
],
[
[
"pc_mobile = df[\"pagecount_mobile_views\"] / 1e6\npv_mobile = df[\"pageview_mobile_views\"] / 1e6\npc_desktop = df[\"pagecount_desktop_views\"] / 1e6\npv_desktop = df[\"pageview_desktop_views\"] / 1e6\npv_total = df[\"pageview_all_views\"] / 1e6\npc_total = df[\"pagecount_all_views\"] / 1e6",
"_____no_output_____"
]
],
[
[
"Fourth, plot the data in a time series for desktop (main site), mobile, and the total all up. The dashed lines are data from the pagecount API and the solid lines are the data from the pageview API without crawler traffic.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt \nfrom matplotlib.dates import YearLocator\n\n#create plot and assign time series to plot\nfig, ax1 = plt.subplots(figsize=(18,6))\nax1.plot(date, pc_desktop, label=\"main site\", color=\"green\", ls=\"--\")\nax1.plot(date, pv_desktop, label=\"_Hidden label\", color=\"green\", ls=\"-\")\nax1.plot(date, pc_mobile, label=\"mobile site\", color=\"blue\", ls=\"--\")\nax1.plot(date, pv_mobile, label=\"_Hidden label\", color=\"blue\", ls=\"-\")\nax1.plot(date, pc_total, label=\"total\", color=\"black\", ls=\"--\")\nax1.plot(date, pv_total, label=\"_Hidden label\", color=\"black\", ls=\"-\")\n\nax1.xaxis.set_major_locator(YearLocator()) #show every year on the x-axis\n\n#set caption\ncaption = \"May 2015: a new pageview definition took effect, which eliminated all crawler traffic. Solid lines mark new definition.\"\nfig.text(.5, .01, caption, ha='center', color=\"red\")\n\n#set labels for x-axis, y-axis, and title\nplt.xlabel(\"Date\")\nplt.ylabel(\"Page Views (x 1,000,000)\")\nplt.title(\"Page Views on English Wikipedia (x 1,000,000)\")\n\nplt.ylim(ymin=0) #start y-axis at 0\n\nplt.grid(True) #turn on background grid\nplt.legend(loc=\"upper left\")\n\n#save chart to png file\nfilename = \"Time Series.png\"\nplt.savefig(filename)\n\nplt.show() #display chart",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0921c8d40c4e6b9ef9eb1691dc18cb860f904da | 39,475 | ipynb | Jupyter Notebook | tr/Sekil_3_26.ipynb | bosmanoglu/Digital_Image_Processing_Book | 14d11fe34ba9b6cd0f2b7b67c87e4c7f8845d89f | [
"Apache-2.0"
] | null | null | null | tr/Sekil_3_26.ipynb | bosmanoglu/Digital_Image_Processing_Book | 14d11fe34ba9b6cd0f2b7b67c87e4c7f8845d89f | [
"Apache-2.0"
] | null | null | null | tr/Sekil_3_26.ipynb | bosmanoglu/Digital_Image_Processing_Book | 14d11fe34ba9b6cd0f2b7b67c87e4c7f8845d89f | [
"Apache-2.0"
] | null | null | null | 506.089744 | 37,976 | 0.934034 | [
[
[
"## İki saniye periyotlu kare dalga\n### Sekil 3.24\n",
"_____no_output_____"
]
],
[
[
"# Tanimlar\nimport matplotlib.pyplot as plt\nimport numpy as np\n\nt=np.linspace(0,10, 1000)\nfreqs=np.r_[0.5:6.5]\n\nfig, ax=plt.subplots(7,1, sharex=True, sharey=True)\nsumSines=np.zeros(t.shape);\ncnt=1;\nfor f in freqs:\n s= np.sin(2*np.pi*f*t)/(2*f)\n ax[cnt].plot(t,s)\n sumSines+=s\n cnt+=1\n \nax[0].plot(t,sumSines, 'r')\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
]
] |
d0922407ca01574d7ff3ef2b67ee43febd81daae | 4,526 | ipynb | Jupyter Notebook | train.ipynb | jsandersen/WU-RNN | 6ee7e419a9837498c79ee7859bca5a8737126c5d | [
"Apache-2.0"
] | 1 | 2021-03-04T13:09:38.000Z | 2021-03-04T13:09:38.000Z | train.ipynb | jsandersen/WU-RNN | 6ee7e419a9837498c79ee7859bca5a8737126c5d | [
"Apache-2.0"
] | null | null | null | train.ipynb | jsandersen/WU-RNN | 6ee7e419a9837498c79ee7859bca5a8737126c5d | [
"Apache-2.0"
] | 1 | 2021-03-04T13:09:33.000Z | 2021-03-04T13:09:33.000Z | 24.333333 | 97 | 0.557225 | [
[
[
"import tensorflow\nimport pickle\nfrom tensorflow.keras.utils import to_categorical\nimport numpy as np\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.utils import shuffle\nfrom src.dataloader import IMDB\nfrom sklearn.model_selection import StratifiedShuffleSplit\nfrom sklearn.metrics import confusion_matrix\nfrom src.model import get_model\nfrom src.embedding import *",
"_____no_output_____"
]
],
[
[
"# load data and embedding ",
"_____no_output_____"
]
],
[
[
"# load confing\nimport yaml\nimport pickle\nwith open('config.yaml', 'r') as f:\n conf = yaml.load(f)\nBATCH_SIZE = conf[\"MODEL\"][\"BATCH_SIZE\"]\nMAX_EPOCHS = conf[\"MODEL\"][\"MAX_EPOCHS\"]\nWORD2VEC_MODEL = conf[\"EMBEDDING\"][\"WORD2VEC_MODEL\"]",
"_____no_output_____"
],
[
"print('load data ...')\nX_train = np.load('data/X_train.npy')\ny_train = np.load('data/y_train.npy')\nX_test = np.load('data/X_test.npy')\ny_test = np.load('data/y_test.npy')\nX_val = np.load('data/X_val.npy')\ny_val = np.load('data/y_val.npy')\n\nwith open('data/word_index.pkl', 'rb') as f:\n word_index = pickle.load(f)",
"_____no_output_____"
],
[
"# load embedding\nfrom gensim import models\nword2vec_model = models.KeyedVectors.load_word2vec_format(WORD2VEC_MODEL, binary=True)\nembeddings_index, embedding_dim = get_embeddings_index(word2vec_model)\nembedding_layer = get_embedding_layer(word_index, embeddings_index, embedding_dim, True)\nword2vec_model = None",
"_____no_output_____"
]
],
[
[
"# train model",
"_____no_output_____"
]
],
[
[
"lstm = get_model(2, embedding_layer)",
"_____no_output_____"
],
[
"callback = tf.keras.callbacks.EarlyStopping(monitor='val_loss', patience=2)\nlstm.compile(\n loss='binary_crossentropy',\n optimizer=tf.keras.optimizers.Adam(0.0001),\n metrics=['accuracy'])",
"_____no_output_____"
],
[
"lstm.summary()",
"_____no_output_____"
],
[
"# train model\nhistory = lstm.fit(\n X_train, y_train,\n batch_size=BATCH_SIZE,\n epochs=MAX_EPOCHS,\n callbacks=[callback],\n validation_data=(X_test, y_test)\n)",
"_____no_output_____"
]
],
[
[
"# history",
"_____no_output_____"
]
],
[
[
"from matplotlib import pyplot as plt\n# \"Accuracy\"\nplt.plot(history.history['acc'])\nplt.plot(history.history['val_acc'])\nplt.title('model accuracy')\nplt.ylabel('accuracy')\nplt.xlabel('epoch')\nplt.legend(['train', 'validation'], loc='upper left')\nplt.show()\n# \"Loss\"\nplt.plot(history.history['loss'])\nplt.plot(history.history['val_loss'])\nplt.title('model loss')\nplt.ylabel('loss')\nplt.xlabel('epoch')\nplt.legend(['train', 'validation'], loc='upper left')\nplt.show()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0922fd5869ef568e651f6ea63e4fcf38e6edc16 | 20,997 | ipynb | Jupyter Notebook | hourly_model/XGB.ipynb | NREL/TrafficVolumeEstimation | 419b008b0da4809fbb13d65b4b1afa5572bda24e | [
"BSD-3-Clause"
] | null | null | null | hourly_model/XGB.ipynb | NREL/TrafficVolumeEstimation | 419b008b0da4809fbb13d65b4b1afa5572bda24e | [
"BSD-3-Clause"
] | null | null | null | hourly_model/XGB.ipynb | NREL/TrafficVolumeEstimation | 419b008b0da4809fbb13d65b4b1afa5572bda24e | [
"BSD-3-Clause"
] | 1 | 2021-08-04T19:13:30.000Z | 2021-08-04T19:13:30.000Z | 35.348485 | 139 | 0.536077 | [
[
[
"%matplotlib inline\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport math\nimport os\nimport random\nimport pickle\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.ensemble import RandomForestRegressor\nfrom sklearn.ensemble import GradientBoostingRegressor\nfrom sklearn.ensemble import GradientBoostingClassifier\nfrom sklearn.ensemble import AdaBoostRegressor\nfrom sklearn.ensemble import AdaBoostClassifier\nfrom sklearn.tree import DecisionTreeRegressor\nfrom sklearn.metrics import mean_squared_error, r2_score\nfrom sklearn.metrics import confusion_matrix\nfrom sklearn.metrics import roc_curve, auc\nfrom sklearn import preprocessing \nfrom xgboost import XGBRegressor\nfrom datetime import datetime\nfrom bayes_opt import BayesianOptimization\nrandom.seed(1234)",
"_____no_output_____"
]
],
[
[
"# Define Functions",
"_____no_output_____"
]
],
[
[
"# Performce cross validation using xgboost\ndef xgboostcv(X, y, fold, n_estimators, lr, depth, n_jobs, gamma, min_cw, subsample, colsample):\n uid = np.unique(fold)\n model_pred = np.zeros(X.shape[0])\n model_valid_loss = np.zeros(len(uid))\n model_train_loss = np.zeros(len(uid))\n for i in uid:\n x_valid = X[fold==i]\n x_train = X[fold!=i]\n y_valid = y[fold==i]\n y_train = y[fold!=i]\n model = XGBRegressor(n_estimators=n_estimators, learning_rate=lr, \n max_depth = depth, n_jobs = n_jobs, \n gamma = gamma, min_child_weight = min_cw,\n subsample = subsample, colsample_bytree = colsample, random_state=1234)\n model.fit(x_train, y_train)\n\n pred = model.predict(x_valid)\n model_pred[fold==i] = pred\n model_valid_loss[uid==i] = mean_squared_error(y_valid, pred)\n model_train_loss[uid==i] = mean_squared_error(y_train, model.predict(x_train))\n return {'pred':model_pred, 'valid_loss':model_valid_loss, 'train_loss':model_train_loss}\n\n# Compute MSE for xgboost cross validation\ndef xgboostcv_mse(n, p, depth, g, min_cw, subsample, colsample):\n model_cv = xgboostcv(X_train, y_train, fold_train, \n int(n)*100, 10**p, int(depth), n_nodes, \n 10**g, min_cw, subsample, colsample)\n MSE = mean_squared_error(y_train, model_cv['pred'])\n return -MSE\n\n# Display model performance metrics for each cv iteration\ndef cv_performance(model, y, fold):\n uid = np.unique(fold)\n pred = np.round(model['pred'])\n y = y.reshape(-1)\n model_valid_mse = np.zeros(len(uid))\n model_valid_mae = np.zeros(len(uid))\n model_valid_r2 = np.zeros(len(uid))\n for i in uid:\n pred_i = pred[fold==i]\n y_i = y[fold==i]\n model_valid_mse[uid==i] = mean_squared_error(y_i, pred_i)\n model_valid_mae[uid==i] = np.abs(pred_i-y_i).mean()\n model_valid_r2[uid==i] = r2_score(y_i, pred_i)\n \n results = pd.DataFrame(0, index=uid, \n columns=['valid_mse', 'valid_mae', 'valid_r2', \n 'valid_loss', 'train_loss'])\n results['valid_mse'] = model_valid_mse\n results['valid_mae'] = model_valid_mae\n results['valid_r2'] = model_valid_r2\n results['valid_loss'] = model['valid_loss']\n results['train_loss'] = model['train_loss']\n print(results)\n\n# Display overall model performance metrics\ndef cv_overall_performance(y, y_pred):\n overall_MSE = mean_squared_error(y, y_pred)\n overall_MAE = (np.abs(y_pred-y)).mean()\n overall_RMSE = np.sqrt(np.square(y_pred-y).mean())\n overall_R2 = r2_score(y, y_pred)\n print(\"XGB overall MSE: %0.4f\" %overall_MSE)\n print(\"XGB overall MAE: %0.4f\" %overall_MAE)\n print(\"XGB overall RMSE: %0.4f\" %overall_RMSE)\n print(\"XGB overall R^2: %0.4f\" %overall_R2) \n\n# Plot variable importance\ndef plot_importance(model, columns):\n importances = pd.Series(model.feature_importances_, index = columns).sort_values(ascending=False)\n n = len(columns)\n plt.figure(figsize=(10,15))\n plt.barh(np.arange(n)+0.5, importances)\n plt.yticks(np.arange(0.5,n+0.5), importances.index)\n plt.tick_params(axis='both', which='major', labelsize=22)\n plt.ylim([0,n])\n plt.gca().invert_yaxis()\n plt.savefig('variable_importance.png', dpi = 150)\n\n# Save xgboost model\ndef save(obj, path):\n pkl_fl = open(path, 'wb')\n pickle.dump(obj, pkl_fl)\n pkl_fl.close()\n\n# Load xgboost model\ndef load(path):\n f = open(path, 'rb')\n obj = pickle.load(f)\n f.close()\n return(obj)",
"_____no_output_____"
]
],
[
[
"# Parameter Values",
"_____no_output_____"
]
],
[
[
"# Set a few values\nvalidation_only = False # Whether test model with test data\nn_nodes = 96 # Number of computing nodes used for hyperparamter tunning\ntrained = False # If a trained model exits\ncols_drop = ['StationId', 'Date', 'PenRate', 'NumberOfLanes', 'Dir', 'FC', 'Month'] # Columns to be dropped\nif trained:\n params = load('params.dat')\n xgb_cv = load('xgb_cv.dat')\n xgb = load('xgb.dat')",
"_____no_output_____"
]
],
[
[
"# Read Data",
"_____no_output_____"
]
],
[
[
"if validation_only:\n raw_data_train = pd.read_csv(\"final_train_data.csv\")\n data = raw_data_train.drop(cols_drop, axis=1)\n if 'Dir' in data.columns:\n data[['Dir']] = data[['Dir']].astype('category')\n one_hot = pd.get_dummies(data[['Dir']])\n data = data.drop(['Dir'], axis = 1)\n data = data.join(one_hot)\n if 'FC' in data.columns:\n data[['FC']] = data[['FC']].astype('category')\n one_hot = pd.get_dummies(data[['FC']])\n data = data.drop(['FC'], axis = 1)\n data = data.join(one_hot)\n week_dict = {\"DayOfWeek\": {'Monday': 1, 'Tuesday': 2, 'Wednesday': 3, 'Thursday': 4, \n 'Friday': 5, 'Saturday': 6, 'Sunday': 7}}\n data = data.replace(week_dict)\n\n X = data.drop(['Volume', 'fold'], axis=1)\n X_col = X.columns\n y = data[['Volume']]\n\n fold_train = data[['fold']].values.reshape(-1)\n X_train = X.values\n y_train = y.values\n \nelse:\n raw_data_train = pd.read_csv(\"final_train_data.csv\")\n raw_data_test = pd.read_csv(\"final_test_data.csv\")\n raw_data_test1 = pd.DataFrame(np.concatenate((raw_data_test.values, np.zeros(raw_data_test.shape[0]).reshape(-1, 1)), axis=1),\n columns = raw_data_test.columns.append(pd.Index(['fold'])))\n raw_data = pd.DataFrame(np.concatenate((raw_data_train.values, raw_data_test1.values), axis=0), \n columns = raw_data_train.columns)\n\n data = raw_data.drop(cols_drop, axis=1)\n if 'Dir' in data.columns:\n data[['Dir']] = data[['Dir']].astype('category')\n one_hot = pd.get_dummies(data[['Dir']])\n data = data.drop(['Dir'], axis = 1)\n data = data.join(one_hot)\n if 'FC' in data.columns:\n data[['FC']] = data[['FC']].astype('category')\n one_hot = pd.get_dummies(data[['FC']])\n data = data.drop(['FC'], axis = 1)\n data = data.join(one_hot)\n week_dict = {\"DayOfWeek\": {'Monday': 1, 'Tuesday': 2, 'Wednesday': 3, 'Thursday': 4, \n 'Friday': 5, 'Saturday': 6, 'Sunday': 7}}\n data = data.replace(week_dict)\n\n X = data.drop(['Volume'], axis=1)\n y = data[['Volume']]\n\n X_train = X.loc[X.fold!=0, :]\n fold_train = X_train[['fold']].values.reshape(-1)\n X_col = X_train.drop(['fold'], axis = 1).columns\n X_train = X_train.drop(['fold'], axis = 1).values\n y_train = y.loc[X.fold!=0, :].values\n\n X_test = X.loc[X.fold==0, :]\n X_test = X_test.drop(['fold'], axis = 1).values\n y_test = y.loc[X.fold==0, :].values",
"_____no_output_____"
],
[
"X_col",
"_____no_output_____"
],
[
"# Explain variable names\nX_name_dict = {'Temp': 'Temperature', 'WindSp': 'Wind Speed', 'Precip': 'Precipitation', 'Snow': 'Snow', \n 'Long': 'Longitude', 'Lat': 'Latitude', 'NumberOfLanes': 'Number of Lanes', 'SpeedLimit': 'Speed Limit', \n 'FRC': 'TomTom FRC', 'DayOfWeek': 'Day of Week', 'Month': 'Month', 'Hour': 'Hour', \n 'AvgSp': 'Average Speed', 'ProbeCount': 'Probe Count', 'Dir_E': 'Direction(East)', \n 'Dir_N': 'Direction(North)', 'Dir_S': 'Direction(South)', 'Dir_W': 'Direction(West)', \n 'FC_3R': 'FHWA FC(3R)', 'FC_3U': 'FHWA FC(3U)', 'FC_4R': 'FHWA FC(4R)', 'FC_4U': 'FHWA FC(4U)', \n 'FC_5R': 'FHWA FC(5R)', 'FC_5U': 'FHWA FC(5U)', 'FC_7R': 'FHWA FC(7R)', 'FC_7U': 'FHWA FC(7U)'}",
"_____no_output_____"
],
[
"data.head()",
"_____no_output_____"
],
[
"X_train.shape",
"_____no_output_____"
],
[
"if validation_only == False:\n print(X_test.shape)",
"_____no_output_____"
]
],
[
[
"# Cross Validation & Hyperparameter Optimization",
"_____no_output_____"
]
],
[
[
"# Set hyperparameter ranges for Bayesian optimization \nxgboostBO = BayesianOptimization(xgboostcv_mse,\n {'n': (1, 10),\n 'p': (-4, 0),\n 'depth': (2, 10),\n 'g': (-3, 0),\n 'min_cw': (1, 10), \n 'subsample': (0.5, 1), \n 'colsample': (0.5, 1)\n })",
"_____no_output_____"
],
[
"# Use Bayesian optimization to tune hyperparameters\nimport time\nstart_time = time.time()\nxgboostBO.maximize(init_points=10, n_iter = 50)\nprint('-'*53)\nprint('Final Results')\nprint('XGBOOST: %f' % xgboostBO.max['target'])\nprint(\"--- %s seconds ---\" % (time.time() - start_time))",
"_____no_output_____"
],
[
"# Save the hyperparameters the yield the highest model performance\nparams = xgboostBO.max['params']\nsave(params, 'params.dat')\nparams",
"_____no_output_____"
],
[
"# Perform cross validation using the optimal hyperparameters\nxgb_cv = xgboostcv(X_train, y_train, fold_train, int(params['n'])*100, \n 10**params['p'], int(params['depth']), n_nodes, \n 10**params['g'], params['min_cw'], params['subsample'], params['colsample'])",
"_____no_output_____"
],
[
"# Display cv results for each iteration\ncv_performance(xgb_cv, y_train, fold_train)",
"_____no_output_____"
],
[
"# Display overall cv results\ncv_pred = xgb_cv['pred']\ncv_pred[cv_pred<0] = 0\ncv_overall_performance(y_train.reshape(-1), cv_pred)",
"_____no_output_____"
],
[
"# Save the cv results\nsave(xgb_cv, 'xgb_cv.dat')",
"_____no_output_____"
]
],
[
[
"# Model Test",
"_____no_output_____"
]
],
[
[
"# Build a xgboost using all the training data with the optimal hyperparameter\nxgb = XGBRegressor(n_estimators=int(params['n'])*100, learning_rate=10**params['p'], max_depth = int(params['depth']), \n n_jobs = n_nodes, gamma = 10**params['g'], min_child_weight = params['min_cw'], \n subsample = params['subsample'], colsample_bytree = params['colsample'], random_state=1234)\nxgb.fit(X_train, y_train)",
"_____no_output_____"
],
[
"# Test the trained model with test data\nif validation_only == False:\n y_pred = xgb.predict(X_test)\n y_pred[y_pred<0] = 0\n cv_overall_performance(y_test.reshape(-1), y_pred)",
"_____no_output_____"
],
[
"# Plot variable importance\ncol_names = [X_name_dict[i] for i in X_col]\nplot_importance(xgb, col_names)",
"_____no_output_____"
],
[
"# Save the trained xgboost model\nsave(xgb, 'xgb.dat')",
"_____no_output_____"
],
[
"# Produce cross validation estimates or estimates for test data\ntrain_data_pred = pd.DataFrame(np.concatenate((raw_data_train.values, cv_pred.reshape(-1, 1)), axis=1),\n columns = raw_data_train.columns.append(pd.Index(['PredVolume'])))\ntrain_data_pred.to_csv('train_data_pred.csv', index = False)\n\nif validation_only == False:\n test_data_pred = pd.DataFrame(np.concatenate((raw_data_test.values, y_pred.reshape(-1, 1)), axis=1),\n columns = raw_data_test.columns.append(pd.Index(['PredVolume'])))\n test_data_pred.to_csv('test_data_pred.csv', index = False)\n",
"_____no_output_____"
]
],
[
[
"# Plot Estimations vs. Observations",
"_____no_output_____"
]
],
[
[
"# Prepare data to plot estimated and observed values\nif validation_only:\n if trained:\n plot_df = pd.read_csv(\"train_data_pred.csv\")\n else:\n plot_df = train_data_pred\nelse:\n if trained:\n plot_df = pd.read_csv(\"test_data_pred.csv\")\n else:\n plot_df = test_data_pred \nplot_df = plot_df.sort_values(by=['StationId', 'Date', 'Dir', 'Hour'])\nplot_df = plot_df.set_index(pd.Index(range(plot_df.shape[0])))",
"_____no_output_____"
],
[
"# Define a function to plot estimated and observed values for a day\ndef plot_daily_estimate(frc):\n indices = plot_df.index[(plot_df.FRC == frc) & (plot_df.Hour == 0)].tolist()\n from_index = np.random.choice(indices, 1)[0]\n to_index = from_index + 23\n plot_df_sub = plot_df.loc[from_index:to_index, :]\n time = pd.date_range(plot_df_sub.Date.iloc[0] + ' 00:00:00', periods=24, freq='H')\n plt.figure(figsize=(20,10))\n plt.plot(time, plot_df_sub.PredVolume, 'b-', label='XGBoost', lw=2)\n plt.plot(time, plot_df_sub.Volume, 'r--', label='Observed', lw=3)\n plt.tick_params(axis='both', which='major', labelsize=24)\n plt.ylabel('Volume (vehs/hr)', fontsize=24)\n plt.xlabel(\"Time\", fontsize=24)\n plt.legend(loc='upper left', shadow=True, fontsize=24)\n plt.title('Station ID: {0}, MAE={1}, FRC = {2}'.format(\n plot_df_sub.StationId.iloc[0],\n round(np.abs(plot_df_sub.PredVolume-plot_df_sub.Volume).mean()),\n plot_df_sub.FRC.iloc[0]), fontsize=40)\n plt.savefig('frc_{0}.png'.format(frc), dpi = 150)\n return(plot_df_sub)",
"_____no_output_____"
],
[
"# Define a function to plot estimated and observed values for a week\ndef plot_weekly_estimate(frc):\n indices = plot_df.index[(plot_df.FRC == frc) & (plot_df.Hour == 0) & (plot_df.DayOfWeek == 'Monday')].tolist()\n from_index = np.random.choice(indices, 1)[0]\n to_index = from_index + 24*7-1\n plot_df_sub = plot_df.loc[from_index:to_index, :]\n time = pd.date_range(plot_df_sub.Date.iloc[0] + ' 00:00:00', periods=24*7, freq='H')\n plt.figure(figsize=(20,10))\n plt.plot(time, plot_df_sub.PredVolume, 'b-', label='XGBoost', lw=2)\n plt.plot(time, plot_df_sub.Volume, 'r--', label='Observed', lw=3)\n plt.tick_params(axis='both', which='major', labelsize=24)\n plt.ylabel('Volume (vehs/hr)', fontsize=24)\n plt.xlabel(\"Time\", fontsize=24)\n plt.legend(loc='upper left', shadow=True, fontsize=24)\n plt.title('Station ID: {0}, MAE={1}, FRC = {2}'.format(\n plot_df_sub.StationId.iloc[0],\n round(np.abs(plot_df_sub.PredVolume-plot_df_sub.Volume).mean()),\n plot_df_sub.FRC.iloc[0]), fontsize=40)\n plt.savefig('frc_{0}.png'.format(frc), dpi = 150)\n return(plot_df_sub)",
"_____no_output_____"
],
[
"# Plot estimated and observed values for a day\nfrc2_daily_plot = plot_daily_estimate(2)\nsave(frc2_daily_plot, 'frc2_daily_plot.dat')",
"_____no_output_____"
],
[
"# Plot estimated and observed values for a week\nfrc3_weekly_plot = plot_weekly_estimate(3)\nsave(frc3_weekly_plot, 'frc3_weekly_plot.dat')",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
d0923bbf12908606078f64bf4977d117a41cbe1d | 64,587 | ipynb | Jupyter Notebook | examples/Unit Tests.ipynb | Genes-N-Risks/genocode | 769b9486b1f3012c9ad3df81161840490c736e94 | [
"MIT"
] | null | null | null | examples/Unit Tests.ipynb | Genes-N-Risks/genocode | 769b9486b1f3012c9ad3df81161840490c736e94 | [
"MIT"
] | 1 | 2020-06-17T18:34:54.000Z | 2020-06-17T18:34:54.000Z | examples/Unit Tests.ipynb | Genes-N-Risks/genocode | 769b9486b1f3012c9ad3df81161840490c736e94 | [
"MIT"
] | 2 | 2020-04-25T20:17:24.000Z | 2021-01-10T00:48:59.000Z | 54.78117 | 94 | 0.583647 | [
[
[
"First import the \"datavis\" module",
"_____no_output_____"
]
],
[
[
"import sys\nsys.path.append('..')\nimport numpy as np\nimport datavis \nimport vectorized_datavis",
"_____no_output_____"
],
[
"def test_se_to_sd():\n \"\"\"\n Test that the value returned is a float value\n \"\"\"\n sdev = datavis.se_to_sd(0.5, 1000)\n assert isinstance(sdev, float),\\\n \"Returned data type is not a float number\"",
"_____no_output_____"
],
[
"test_se_to_sd() ",
"_____no_output_____"
],
[
"def test_ci_to_sd():\n \"\"\"\n Test that the value returned is a float value\n \"\"\"\n sdev = datavis.ci_to_sd(0.2, 0.4) \n assert isinstance(sdev, float),\\\n \"Returned data type is not a float number\"",
"_____no_output_____"
],
[
"test_ci_to_sd() ",
"_____no_output_____"
],
[
"def test_datagen():\n \"\"\"\n Test that the data returned is a numpy.ndarray\n \"\"\"\n randdata = datavis.datagen(25, 0.2, 0.4)\n assert isinstance(randdata, np.ndarray),\\\n \"Returned data type is not a numpy.ndarray\"",
"_____no_output_____"
],
[
"test_datagen() ",
"_____no_output_____"
],
[
"def test_correctdatatype():\n \"\"\"\n Test that the statistical parameters returned are float numbers\n \"\"\"\n fmean, fsdev, fserror, fuci, flci = datavis.correctdatatype(0.2, 0.4)\n assert isinstance(fmean, float),\\\n \"Returned data type is not a float number\"",
"_____no_output_____"
],
[
"test_correctdatatype() ",
"_____no_output_____"
],
[
"def test_compounddata():\n \"\"\"\n Test that the data returned are numpy.ndarrays\n \"\"\"\n datagenerated1, datagenerated2, datagenerated3 = \\\n datavis.compounddata\\\n (mean1=24.12,sdev1=3.87,mean2=24.43,sdev2=3.94,mean3=24.82,sdev3=3.95,size=1000) \n assert isinstance(datagenerated1, np.ndarray),\\\n \"Returned data are not numpy.ndarrays\"",
"_____no_output_____"
],
[
"test_compounddata() ",
"_____no_output_____"
],
[
"def test_databinning():\n \"\"\"\n Test that the data returned are numpy.ndarrays\n \"\"\"\n datagenerated1, datagenerated2, datagenerated3 = \\\n datavis.compounddata\\\n (mean1=24.12,sdev1=3.87,mean2=24.43,sdev2=3.94,mean3=24.82,sdev3=3.95,size=1000)\n bins = np.linspace(10,40,num=30)\n yhist1, yhist2, yhist3 = \\\n datavis.databinning\\\n (datagenerated1, datagenerated2, datagenerated3,bins_list=bins) \n assert isinstance(yhist1, np.ndarray),\\\n \"Returned data are not numpy.ndarrays\"",
"_____no_output_____"
],
[
"test_databinning() ",
"_____no_output_____"
],
[
"def test_pdfgen():\n \"\"\"\n Test that the data returned are numpy.ndarrays\n \"\"\"\n bins = np.linspace(10,40,num=30)\n mean1 = 24.12\n sdev1 = 3.87\n mean2 = 24.43\n sdev2 = 3.94\n mean3 = 24.82\n sdev3 = 3.95 \n pdf1, pdf2, pdf3 = datavis.pdfgen\\\n (mean1, sdev1, mean2, sdev2, mean3, sdev3, bins_list=bins)\n assert isinstance(pdf1, np.ndarray),\\\n \"Returned data are not numpy.ndarrays\"",
"_____no_output_____"
],
[
"test_pdfgen() ",
"_____no_output_____"
],
[
"def test_percent_overlap():\n \"\"\"\n Test that the data returned is a tuple\n \"\"\"\n mean1 = 24.12\n sdev1 = 3.87\n mean2 = 24.43\n sdev2 = 3.94\n mean3 = 24.82\n sdev3 = 3.95\n overlap_11_perc, overlap_12_perc, overlap_13_perc = \\\n datavis.percent_overlap\\\n (mean1, sdev1, mean2, sdev2, mean3, sdev3)\n assert isinstance\\\n (datavis.percent_overlap\\\n (mean1, sdev1, mean2, sdev2, mean3, sdev3), tuple),\\\n \"Returned data is not numpy.float64 type\"",
"_____no_output_____"
],
[
"test_percent_overlap() ",
"_____no_output_____"
],
[
"mean = np.array([24.12, 24.43, 24.82])\nsdev = np.array([3.87, 3.94, 3.95]) ",
"_____no_output_____"
],
[
"vectorized_datavis.compounddata(mean, sdev) ",
"_____no_output_____"
],
[
"def test_compounddata():\n \"\"\"\n Test that the data returned are numpy.ndarrays\n \"\"\"\n mean = np.array([24.12, 24.43, 24.82])\n sdev = np.array([3.87, 3.94, 3.95]) \n datagenerated = vectorized_datavis.compounddata(mean, sdev) \n assert isinstance(datagenerated, np.ndarray),\\\n \"Returned data are not numpy.ndarrays\"",
"_____no_output_____"
],
[
"test_compounddata() ",
"_____no_output_____"
],
[
"def test_databinning():\n \"\"\"\n Test that the data returned are numpy.ndarrays\n \"\"\"\n mean = np.array([24.12, 24.43, 24.82])\n sdev = np.array([3.87, 3.94, 3.95]) \n datagenerated = vectorized_datavis.compounddata(mean, sdev)\n bins = np.linspace(10, 40, num=30)\n yhist = vectorized_datavis.databinning(datagenerated, bins_list=bins) \n assert isinstance(yhist, np.ndarray),\\\n \"Returned data are not numpy.ndarrays\" ",
"_____no_output_____"
],
[
"test_databinning()",
"_____no_output_____"
],
[
"def test_pdfgen():\n \"\"\"\n Test that the data returned are numpy.ndarrays\n \"\"\"\n bins = np.linspace(10,40,num=30)\n mean = np.array([24.12, 24.43, 24.82])\n sdev = np.array([3.87, 3.94, 3.95]) \n pdf = vectorized_datavis.pdfgen(mean, sdev, bins_list=bins)\n assert isinstance(pdf, np.ndarray),\\\n \"Returned data are not numpy.ndarrays\"",
"_____no_output_____"
],
[
"test_pdfgen() ",
"_____no_output_____"
],
[
"def test_percent_overlap():\n \"\"\"\n Test that the data returned is a numpy.ndarray\n \"\"\"\n mean = np.array([24.12, 24.43, 24.82])\n sdev = np.array([3.87, 3.94, 3.95]) \n overlap_perc_1w = vectorized_datavis.percent_overlap\\\n (mean, sdev)\n assert isinstance(overlap_perc_1w, np.ndarray),\\\n \"Returned data is not a numpy.ndarray\"",
"_____no_output_____"
],
[
"test_percent_overlap() ",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0923d3497e83ea656bc88b6de4572763ef01382 | 56,447 | ipynb | Jupyter Notebook | IDS_2020/RegressionSVM/Instruction4-RegressionSVM-without solutions.ipynb | pmnatos/DataScience | a90cafe0f94316b7808e73903a0ce8873c4cf780 | [
"Apache-2.0"
] | null | null | null | IDS_2020/RegressionSVM/Instruction4-RegressionSVM-without solutions.ipynb | pmnatos/DataScience | a90cafe0f94316b7808e73903a0ce8873c4cf780 | [
"Apache-2.0"
] | null | null | null | IDS_2020/RegressionSVM/Instruction4-RegressionSVM-without solutions.ipynb | pmnatos/DataScience | a90cafe0f94316b7808e73903a0ce8873c4cf780 | [
"Apache-2.0"
] | null | null | null | 67.359189 | 15,260 | 0.771892 | [
[
[
"# IDS Instruction: Regression\n(Lisa Mannel)",
"_____no_output_____"
],
[
"## Simple linear regression",
"_____no_output_____"
],
[
"First we import the packages necessary fo this instruction:",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport pandas as pd\nfrom sklearn.linear_model import LinearRegression\nfrom sklearn.metrics import mean_squared_error, mean_absolute_error",
"_____no_output_____"
]
],
[
[
"Consider the data set \"df\" with feature variables \"x\" and \"y\" given below.",
"_____no_output_____"
]
],
[
[
"df1 = pd.DataFrame({'x': [0, 1, 2, 3, 4, 5, 6, 7, 8, 9], 'y': [1, 3, 2, 5, 7, 8, 8, 9, 10, 12]})\nprint(df1)",
" x y\n0 0 1\n1 1 3\n2 2 2\n3 3 5\n4 4 7\n5 5 8\n6 6 8\n7 7 9\n8 8 10\n9 9 12\n"
]
],
[
[
"To get a first impression of the given data, let's have a look at its scatter plot:",
"_____no_output_____"
]
],
[
[
"plt.scatter(df1.x, df1.y, color = \"y\", marker = \"o\", s = 40)\nplt.xlabel('x') \nplt.ylabel('y')\nplt.title('first overview of the data')\nplt.show()",
"_____no_output_____"
]
],
[
[
"We can already see a linear correlation between x and y. Assume the feature x to be descriptive, while y is our target feature. We want a linear function, y=ax+b, that predicts y as accurately as possible based on x. To achieve this goal we use linear regression from the sklearn package.",
"_____no_output_____"
]
],
[
[
"#define the set of descriptive features (in this case only 'x' is in that set) and the target feature (in this case 'y')\ndescriptiveFeatures1=df1[['x']]\ntargetFeature1=df1['y']\n\n#define the classifier\nclassifier = LinearRegression()\n#train the classifier\nmodel1 = classifier.fit(descriptiveFeatures1, targetFeature1)",
"_____no_output_____"
]
],
[
[
"Now we can use the classifier to predict y. We print the predictions as well as the coefficient and bias (*intercept*) of the linear function.",
"_____no_output_____"
]
],
[
[
"#use the classifier to make prediction\ntargetFeature1_predict = classifier.predict(descriptiveFeatures1)\nprint(targetFeature1_predict)\n#print coefficient and intercept\nprint('Coefficients: \\n', classifier.coef_)\nprint('Intercept: \\n', classifier.intercept_)",
"[ 1.23636364 2.40606061 3.57575758 4.74545455 5.91515152 7.08484848\n 8.25454545 9.42424242 10.59393939 11.76363636]\nCoefficients: \n [1.16969697]\nIntercept: \n 1.2363636363636399\n"
]
],
[
[
"Let's visualize our regression function with the scatterplot showing the original data set. Herefore, we use the predicted values.",
"_____no_output_____"
]
],
[
[
"#visualize data points\nplt.scatter(df1.x, df1.y, color = \"y\", marker = \"o\", s = 40) \n#visualize regression function\nplt.plot(descriptiveFeatures1, targetFeature1_predict, color = \"g\") \nplt.xlabel('x') \nplt.ylabel('y') \nplt.title('the data and the regression function')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### <span style=\"color:green\"> Now it is your turn. </span> Build a simple linear regression for the data below. Use col1 as descriptive feature and col2 as target feature. Also plot your results.",
"_____no_output_____"
]
],
[
[
"df2 = pd.DataFrame({'col1': [770, 677, 428, 410, 371, 504, 1136, 695, 551, 550], 'col2': [54, 47, 28, 38, 29, 38, 80, 52, 45, 40]})\n#Your turn\n",
"_____no_output_____"
]
],
[
[
"### Evaluation\n\nUsually, the model and its predictions is not sufficient. In the following we want to evaluate our classifiers. \n\nLet's start by computing their error. The sklearn.metrics package contains several errors such as\n\n* Mean squared error\n* Mean absolute error\n* Mean squared log error\n* Median absolute error\n",
"_____no_output_____"
]
],
[
[
"#computing the squared error of the first model\nprint(\"Mean squared error model 1: %.2f\" % mean_squared_error(targetFeature1, targetFeature1_predict))",
"Mean squared error model 1: 0.56\n"
]
],
[
[
"We can also visualize the errors:",
"_____no_output_____"
]
],
[
[
"plt.scatter(targetFeature1_predict, (targetFeature1 - targetFeature1_predict) ** 2, color = \"blue\", s = 10,) \n \n## plotting line to visualize zero error \nplt.hlines(y = 0, xmin = 0, xmax = 15, linewidth = 2) \n \n## plot title \nplt.title(\"Squared errors Model 1\") \n \n## function to show plot \nplt.show() ",
"_____no_output_____"
]
],
[
[
"### <span style=\"color:green\"> Now it is your turn. </span> Compute the mean squared error and visualize the squared errors. Play around using different error metrics.",
"_____no_output_____"
]
],
[
[
"#Your turn\n",
"_____no_output_____"
]
],
[
[
"## Handling multiple descriptive features at once - Multiple linear regression\nIn most cases, we will have more than one descriptive feature . As an example we use an example data set of the scikit package. The dataset describes housing prices in Boston based on several attributes. Note, in this format the data is already split into descriptive features and a target feature.",
"_____no_output_____"
]
],
[
[
"from sklearn import datasets ## imports datasets from scikit-learn\ndf3 = datasets.load_boston()\n\n#The sklearn package provides the data splitted into a set of descriptive features and a target feature.\n#We can easily transform this format into the pandas data frame as used above.\ndescriptiveFeatures3 = pd.DataFrame(df3.data, columns=df3.feature_names)\ntargetFeature3 = pd.DataFrame(df3.target, columns=['target'])\nprint('Descriptive features:')\nprint(descriptiveFeatures3.head())\nprint('Target feature:')\nprint(targetFeature3.head())",
"Descriptive features:\n CRIM ZN INDUS CHAS NOX RM AGE DIS RAD TAX \\\n0 0.00632 18.0 2.31 0.0 0.538 6.575 65.2 4.0900 1.0 296.0 \n1 0.02731 0.0 7.07 0.0 0.469 6.421 78.9 4.9671 2.0 242.0 \n2 0.02729 0.0 7.07 0.0 0.469 7.185 61.1 4.9671 2.0 242.0 \n3 0.03237 0.0 2.18 0.0 0.458 6.998 45.8 6.0622 3.0 222.0 \n4 0.06905 0.0 2.18 0.0 0.458 7.147 54.2 6.0622 3.0 222.0 \n\n PTRATIO B LSTAT \n0 15.3 396.90 4.98 \n1 17.8 396.90 9.14 \n2 17.8 392.83 4.03 \n3 18.7 394.63 2.94 \n4 18.7 396.90 5.33 \nTarget feature:\n target\n0 24.0\n1 21.6\n2 34.7\n3 33.4\n4 36.2\n"
]
],
[
[
"To predict the housing price we will use a Multiple Linear Regression model. In Python this is very straightforward: we use the same function as for simple linear regression, but our set of descriptive features now contains more than one element (see above).",
"_____no_output_____"
]
],
[
[
"classifier = LinearRegression()\nmodel3 = classifier.fit(descriptiveFeatures3,targetFeature3)\n\ntargetFeature3_predict = classifier.predict(descriptiveFeatures3)\nprint('Coefficients: \\n', classifier.coef_)\nprint('Intercept: \\n', classifier.intercept_)\nprint(\"Mean squared error: %.2f\" % mean_squared_error(targetFeature3, targetFeature3_predict))",
"Coefficients: \n [[-1.08011358e-01 4.64204584e-02 2.05586264e-02 2.68673382e+00\n -1.77666112e+01 3.80986521e+00 6.92224640e-04 -1.47556685e+00\n 3.06049479e-01 -1.23345939e-02 -9.52747232e-01 9.31168327e-03\n -5.24758378e-01]]\nIntercept: \n [36.45948839]\nMean squared error: 21.89\n"
]
],
[
[
"As you can see above, we have a coefficient for each descriptive feature.",
"_____no_output_____"
],
[
"## Handling categorical descriptive features\nSo far we always encountered numerical dscriptive features, but data sets can also contain categorical attributes. The regression function can only handle numerical input. There are several ways to tranform our categorical data to numerical data (for example using one-hot encoding as explained in the lecture: we introduce a 0/1 feature for every possible value of our categorical attribute). For adequate data, another possibility is to replace each categorical value by a numerical value and adding an ordering with it. \n\nPopular possibilities to achieve this transformation are\n\n* the get_dummies function of pandas\n* the OneHotEncoder of scikit\n* the LabelEncoder of scikit\n\nAfter encoding the attributes we can apply our regular regression function.",
"_____no_output_____"
]
],
[
[
"#example using pandas\ndf4 = pd.DataFrame({'A':['a','b','c'],'B':['c','b','a'] })\none_hot_pd = pd.get_dummies(df4)\none_hot_pd",
"_____no_output_____"
],
[
"#example using scikit\nfrom sklearn.preprocessing import LabelEncoder, OneHotEncoder\n\n#apply the one hot encoder\nencoder = OneHotEncoder(categories='auto')\nencoder.fit(df4)\ndf4_OneHot = encoder.transform(df4).toarray()\nprint('Transformed by One-hot Encoding: ')\nprint(df4_OneHot)\n\n# encode labels with value between 0 and n_classes-1\nencoder = LabelEncoder()\ndf4_LE = df4.apply(encoder.fit_transform)\nprint('Replacing categories by numerical labels: ')\nprint(df4_LE.head())",
"Transformed by One-hot Encoding: \n[[1. 0. 0. 0. 0. 1.]\n [0. 1. 0. 0. 1. 0.]\n [0. 0. 1. 1. 0. 0.]]\nReplacing categories by numerical labels: \n A B\n0 0 2\n1 1 1\n2 2 0\n"
]
],
[
[
"### <span style=\"color:green\"> Now it is your turn. </span> Perform linear regression using the data set given below. Don't forget to transform your categorical descriptive features. The rental price attribute represents the target variable. ",
"_____no_output_____"
]
],
[
[
"df5 = pd.DataFrame({'Size':[500,550,620,630,665],'Floor':[4,7,9,5,8], 'Energy rating':['C', 'A', 'A', 'B', 'C'], 'Rental price': [320,380,400,390,385] })\n#Your turn\n",
"_____no_output_____"
]
],
[
[
"## Predicting a categorical target value - Logistic regression",
"_____no_output_____"
],
[
"We might also encounter data sets where our target feature is categorical. Here we don't transform them into numerical values, but insetad we use a logistic regression function. Luckily, sklearn provides us with a suitable function that is similar to the linear equivalent. Similar to linear regression, we can compute logistic regression on a single descriptive variable as well as on multiple variables.",
"_____no_output_____"
]
],
[
[
"# Importing the dataset\niris = pd.read_csv('iris.csv')\n\nprint('First look at the data set: ')\nprint(iris.head())\n\n#defining the descriptive and target features\ndescriptiveFeatures_iris = iris[['sepal_length']] #we only use the attribute 'sepal_length' in this example\ntargetFeature_iris = iris['species'] #we want to predict the 'species' of iris\n\nfrom sklearn.linear_model import LogisticRegression\nclassifier = LogisticRegression(solver = 'liblinear', multi_class = 'ovr')\nclassifier.fit(descriptiveFeatures_iris, targetFeature_iris)\n\ntargetFeature_iris_pred = classifier.predict(descriptiveFeatures_iris)\n\nprint('Coefficients: \\n', classifier.coef_)\nprint('Intercept: \\n', classifier.intercept_)",
"First look at the data set: \n sepal_length sepal_width petal_length petal_width species\n0 5.1 3.5 1.4 0.2 setosa\n1 4.9 3.0 1.4 0.2 setosa\n2 4.7 3.2 1.3 0.2 setosa\n3 4.6 3.1 1.5 0.2 setosa\n4 5.0 3.6 1.4 0.2 setosa\nCoefficients: \n [[-0.86959145]\n [ 0.01223362]\n [ 0.57972675]]\nIntercept: \n [ 4.16186636 -0.74244291 -3.9921824 ]\n"
]
],
[
[
"### <span style=\"color:green\"> Now it is your turn. </span> In the example above we only used the first attribute as descriptive variable. Change the example such that all available attributes are used.",
"_____no_output_____"
]
],
[
[
"#Your turn",
"_____no_output_____"
]
],
[
[
"Note, that the regression classifier (both logistic and non-logistic) can be tweaked using several parameters. This includes, but is not limited to, non-linear regression. Check out the documentation for details and feel free to play around!",
"_____no_output_____"
],
[
"# Support Vector Machines",
"_____no_output_____"
],
[
"Aside from regression models, the sklearn package also provides us with a function for training support vector machines. Looking at the example below we see that they can be trained in similar ways. We still use the iris data set for illustration.",
"_____no_output_____"
]
],
[
[
"from sklearn.svm import SVC\n\n#define descriptive and target features as before\ndescriptiveFeatures_iris = iris[['sepal_length', 'sepal_width', 'petal_length', 'petal_width']]\ntargetFeature_iris = iris['species']\n\n#this time, we train an SVM classifier\nclassifier = SVC(C=1, kernel='linear', gamma = 'auto')\nclassifier.fit(descriptiveFeatures_iris, targetFeature_iris)\n\ntargetFeature_iris_predict = classifier.predict(descriptiveFeatures_iris)\ntargetFeature_iris_predict[0:5] #show the first 5 predicted values",
"_____no_output_____"
]
],
[
[
"As explained in the lecture, a support vector machine is defined by its support vectors. In the sklearn package we can access them and their properties very easily:\n\n* support_: indicies of support vectors\n* support_vectors_: the support vectors\n* n_support_: the number of support vectors for each class",
"_____no_output_____"
]
],
[
[
"print('Indicies of support vectors:')\nprint(classifier.support_)\n\nprint('The support vectors:')\nprint(classifier.support_vectors_)\n\nprint('The number of support vectors for each class:')\nprint(classifier.n_support_)",
"Indicies of support vectors:\n[ 23 24 41 52 56 63 66 68 70 72 76 77 83 84 98 106 110 119\n 123 126 127 129 133 138 146 147 149]\nThe support vectors:\n[[5.1 3.3 1.7 0.5]\n [4.8 3.4 1.9 0.2]\n [4.5 2.3 1.3 0.3]\n [6.9 3.1 4.9 1.5]\n [6.3 3.3 4.7 1.6]\n [6.1 2.9 4.7 1.4]\n [5.6 3. 4.5 1.5]\n [6.2 2.2 4.5 1.5]\n [5.9 3.2 4.8 1.8]\n [6.3 2.5 4.9 1.5]\n [6.8 2.8 4.8 1.4]\n [6.7 3. 5. 1.7]\n [6. 2.7 5.1 1.6]\n [5.4 3. 4.5 1.5]\n [5.1 2.5 3. 1.1]\n [4.9 2.5 4.5 1.7]\n [6.5 3.2 5.1 2. ]\n [6. 2.2 5. 1.5]\n [6.3 2.7 4.9 1.8]\n [6.2 2.8 4.8 1.8]\n [6.1 3. 4.9 1.8]\n [7.2 3. 5.8 1.6]\n [6.3 2.8 5.1 1.5]\n [6. 3. 4.8 1.8]\n [6.3 2.5 5. 1.9]\n [6.5 3. 5.2 2. ]\n [5.9 3. 5.1 1.8]]\nThe number of support vectors for each class:\n[ 3 12 12]\n"
]
],
[
[
"We can also calculate the distance of the data points to the separating hyperplane by using the decision_function(X) method. Score(X,y) calculates the mean accuracy of the classification. The classification report shows metrics such as precision, recall, f1-score and support. You will learn more about these quality metrics in a few lectures.",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import classification_report\nclassifier.decision_function(descriptiveFeatures_iris)\nprint('Accuracy: \\n', classifier.score(descriptiveFeatures_iris,targetFeature_iris))\nprint('Classification report: \\n')\nprint(classification_report(targetFeature_iris, targetFeature_iris_predict)) ",
"Accuracy: \n 0.9933333333333333\nClassification report: \n\n precision recall f1-score support\n\n setosa 1.00 1.00 1.00 50\n versicolor 1.00 0.98 0.99 50\n virginica 0.98 1.00 0.99 50\n\n accuracy 0.99 150\n macro avg 0.99 0.99 0.99 150\nweighted avg 0.99 0.99 0.99 150\n\n"
]
],
[
[
"The SVC has many parameters. In the lecture you learned about the concept of kernels. Scikit gives you the opportunity to try different kernel functions.\nFurthermore, the parameter C tells the SVM optimization problem how much you want to avoid misclassifying each training example. ",
"_____no_output_____"
],
[
"On the scikit website you can find more information about the available kernels etc. http://scikit-learn.org/stable/modules/generated/sklearn.svm.SVC.html",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d0924730c94276d78f689c138cd87917704b7115 | 36,579 | ipynb | Jupyter Notebook | customer_churn/brazilian-ecommerce/Notes1.ipynb | jzhang1031/pandas_exercises | 4000e4350c610bbdfedd1fa8f1836a6a0f7def9c | [
"BSD-3-Clause"
] | null | null | null | customer_churn/brazilian-ecommerce/Notes1.ipynb | jzhang1031/pandas_exercises | 4000e4350c610bbdfedd1fa8f1836a6a0f7def9c | [
"BSD-3-Clause"
] | null | null | null | customer_churn/brazilian-ecommerce/Notes1.ipynb | jzhang1031/pandas_exercises | 4000e4350c610bbdfedd1fa8f1836a6a0f7def9c | [
"BSD-3-Clause"
] | null | null | null | 33.375 | 536 | 0.42631 | [
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"products = pd.read_csv('olist_products_dataset.csv')\nproducts.head()",
"_____no_output_____"
],
[
"# find the most common product color\nproducts.product_category_name.value_counts().idxmax()",
"_____no_output_____"
],
[
"orders = pd.read_csv('olist_orders_dataset.csv')\norders.head()",
"_____no_output_____"
],
[
"total = pd.merge(transacitions, product, on='product_id')",
"_____no_output_____"
],
[
"orders['date'] = pd.to_datetime(orders.order_purchase_timestamp)\n\n# red or not\ndef status(x):\n if x == 'delivered':\n return 1\n else:\n return 0\n\norders['status'] = orders.order_status.apply(status)\n\norders.head()",
"_____no_output_____"
],
[
"print(orders.date.max())\nprint(orders.date.min())\nprint(orders.date.apply(['max','min']))",
"2018-10-17 17:30:18\n2016-09-04 21:15:19\nmax 2018-10-17 17:30:18\nmin 2016-09-04 21:15:19\nName: date, dtype: datetime64[ns]\n"
],
[
"#aggregate the data by customer and filter it to the pre-Oct-2012 time period\nagg = orders[orders['date'] < '2018-01-01 00:00:00'].groupby('customer_id')",
"_____no_output_____"
],
[
"# use this data to create as many predictor features\n\n# number of transcactins\nnumber_of_transactions = agg.count()['order_id']\n\n# total money spent\n## aggregate on the original transactions_data\n## transactions['total_money'] = transactions['purchased_quantity'] * transactions['unit_product_price'] \n## agg.total_money.sum()\n\n# red product\nred_or_not = agg.sum()\n\n# total purchased quantity\n\n# average price for product\n## total money / total quantity\n\n# transaction frequency",
"_____no_output_____"
],
[
"feature = pd.concat([number_of_transactions, red_or_not],axis = 1)\nfeature.head()",
"_____no_output_____"
],
[
"# create response feature\nimport numpy as np\norder2018 = orders[orders['date'] >= '2018-01-01 00:00:00']\norder2018['target'] = np.ones(order2018.shape[0],dtype=int)\ntarget = order2018.groupby('customer_id').sum().drop('status',axis = 1)\ntarget.head()",
"//anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:4: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n after removing the cwd from sys.path.\n"
],
[
"features = pd.merge(feature, target, how = 'left',on = 'customer_id')\nfeatures.head()",
"_____no_output_____"
],
[
"# change NaN to 0\nfeatures = features.fillna(0)\nfeatures.index.is_unique\nfeatures.head()",
"_____no_output_____"
],
[
"#save output data as a csv file\n\nfeatures.to_csv('features_data.csv')",
"_____no_output_____"
],
[
"data = pd.read_csv('features_data.csv',index_col = 'customer_id')\ndata.head()",
"_____no_output_____"
],
[
"X = data.drop('target', axis = 1)\ny = data['target']\n\n# train_test_split\nfrom sklearn.model_selection import train_test_split\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=42)\n\nfrom sklearn.linear_model import LogisticRegression\nlogmodel = LogisticRegression()\nlogmodel.fit(X_train,y_train)\npredictions = logmodel.predict(X_test)",
"//anaconda3/lib/python3.7/site-packages/sklearn/linear_model/logistic.py:432: FutureWarning: Default solver will be changed to 'lbfgs' in 0.22. Specify a solver to silence this warning.\n FutureWarning)\n"
],
[
"#evaluation\nfrom sklearn.metrics import classification_report\nprint(classification_report(y_test,predictions))",
" precision recall f1-score support\n\n 0.0 1.00 1.00 1.00 15042\n 1.0 1.00 1.00 1.00 17774\n\n accuracy 1.00 32816\n macro avg 1.00 1.00 1.00 32816\nweighted avg 1.00 1.00 1.00 32816\n\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d092487ffa70d34aa83034bee176f79710d59627 | 19,477 | ipynb | Jupyter Notebook | Exam project/Jacob legemappe/ANALYSIS_JENS_CLEAN.ipynb | tnv875/Group18-NoTeeth | f5861d58e880d99f8d6ce0beaaf5e93059d00929 | [
"MIT"
] | null | null | null | Exam project/Jacob legemappe/ANALYSIS_JENS_CLEAN.ipynb | tnv875/Group18-NoTeeth | f5861d58e880d99f8d6ce0beaaf5e93059d00929 | [
"MIT"
] | null | null | null | Exam project/Jacob legemappe/ANALYSIS_JENS_CLEAN.ipynb | tnv875/Group18-NoTeeth | f5861d58e880d99f8d6ce0beaaf5e93059d00929 | [
"MIT"
] | null | null | null | 33.523236 | 194 | 0.560969 | [
[
[
"import scraping_class\nlogfile = 'log.txt'## name your log file.\nconnector = scraping_class.Connector(logfile)\nimport requests\nfrom bs4 import BeautifulSoup\nfrom tqdm import tqdm_notebook",
"_____no_output_____"
],
[
"import pandas as pd\nimport numpy as np\nimport html5lib\nimport sys\nimport pickle\nfrom tqdm import tqdm_notebook\nimport seaborn as sns",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nfrom matplotlib import pyplot",
"_____no_output_____"
],
[
"with open('df_final.pkl', 'rb') as f:\n df = pickle.load(f)",
"_____no_output_____"
]
],
[
[
"### Grade distrubtion by Faculties (used in the appendix)",
"_____no_output_____"
]
],
[
[
"# note: no NaN's\nprint(df['Fakultet'].unique())",
"_____no_output_____"
],
[
"# Number of courses by faculty\nprint(sum(df['Fakultet'] == 'Det Natur- og Biovidenskabelige Fakultet'))\nprint(sum(df['Fakultet'] == 'Det Samfundsvidenskabelige Fakultet'))\nprint(sum(df['Fakultet'] == 'Det Humanistiske Fakultet'))\nprint(sum(df['Fakultet'] == 'Det Sundhedsvidenskabelige Fakultet'))\nprint(sum(df['Fakultet'] == 'Det Juridiske Fakultet'))\nprint(sum(df['Fakultet'] == 'Det Teologiske Fakultet'))",
"_____no_output_____"
],
[
"#Number of grades given for each faculty. \nlist_number_of_grades_faculties = []\nfor i in tqdm_notebook(df['Fakultet'].unique()):\n df_number_grades = df[df['Fakultet'] == i]\n list_number_of_grades_faculties.append(int(sum(df_number_grades[[12, 10, 7, 4, 2, 0, -3]].sum(skipna = True))))\n\nlist_number_of_grades_faculties;",
"_____no_output_____"
],
[
"#Number of passing grades given for each faculty. \nlist_no_fail_number_of_grades_faculties = []\nfor i in tqdm_notebook(df['Fakultet'].unique()):\n df_number_grades = df[df['Fakultet'] == i]\n list_no_fail_number_of_grades_faculties.append(int(sum(df_number_grades[[12, 10, 7, 4, 2]].sum(skipna = True))))\n\nlist_no_fail_number_of_grades_faculties",
"_____no_output_____"
]
],
[
[
"### Grade distrubtion by Faculties, weighted against ECTS",
"_____no_output_____"
]
],
[
[
"# We need to weight the grades according to ECTS points. If we do not small courses will have the same weight as\n# bigger courses. \ndf['Weigthed_m3'] = df['Credit_edit'] * df[-3]\ndf['Weigthed_00'] = df['Credit_edit'] * df[0]\ndf['Weigthed_02'] = df['Credit_edit'] * df[2]\ndf['Weigthed_4'] = df['Credit_edit'] * df[4]\ndf['Weigthed_7'] = df['Credit_edit'] * df[7]\ndf['Weigthed_10'] = df['Credit_edit'] * df[10]\ndf['Weigthed_12'] = df['Credit_edit'] * df[12]\ndf[['Credit_edit',-3,'Weigthed_m3',0,'Weigthed_00',2,'Weigthed_02',4,'Weigthed_4',7,'Weigthed_7',10,'Weigthed_10',12,'Weigthed_12']];",
"_____no_output_____"
],
[
"y_ects_inner = []\ny_ects = []\nx = ['-3','00','02','4','7','10','12']\n\n# Looking at each faculty \nfor i in tqdm_notebook(df['Fakultet'].unique()):\n df_faculty = df[df['Fakultet']==i]\n \n # Using the weighted grades this time.\n for k in ['Weigthed_m3','Weigthed_00','Weigthed_02','Weigthed_4','Weigthed_7','Weigthed_10','Weigthed_12']:\n y_ects_inner.append(df_faculty[k].sum(skipna = True)) \n \n y_ects.append(y_ects_inner)\n y_ects_inner=[]",
"_____no_output_____"
],
[
"# calc frequencies \ny_ects_freq_inner = []\ny_ects_freq = []\n\n# running through each faculty\nfor i in range(len(y_ects)):\n \n # calc frequencies \n for q in range(len(y_ects[i])):\n y_ects_freq_inner.append(y_ects[i][q]/sum(y_ects[i]))\n\n y_ects_freq.append(y_ects_freq_inner)\n y_ects_freq_inner = []",
"_____no_output_____"
],
[
"# This figure is used in the analysis. It uses WEIGHTED grades\nf, ax = plt.subplots(figsize=(15,10))\n\nplt.subplot(2, 3, 1)\nplt.title(Faculty_names[0], fontsize = 16, weight = 'bold')\nplt.ylim([0,0.30])\nplt.grid(axis ='y',zorder=0)\nplt.ylabel('Frequency',fontsize=14)\nplt.annotate('Number of grades th=0.93, edgecolor='black',zorder=3)\ngiven: '+ str(list_number_of_grades_faculties[0]), (0,0), (0, -20),fontsize= 13, xycoords='axes fraction', textcoords='offset points', va='top')\nplt.bar(x, y_ects_freq[0], wid\nplt.subplot(2, 3, 2)\nplt.title(Faculty_names[1], fontsize = 16, weight = 'bold')\nplt.ylim([0,0.30])\nplt.grid(axis ='y',zorder=0)\nplt.annotate('Number of grades given: '+ str(list_number_of_grades_faculties[1]), (0,0), (0, -20),fontsize= 13, xycoords='axes fraction', textcoords='offset points', va='top')\nplt.bar(x, y_ects_freq[1], width=0.93, edgecolor='black',zorder=3)\n\nplt.subplot(2, 3, 3)\nplt.title(Faculty_names[2], fontsize = 16, weight = 'bold')\nplt.ylim([0,0.30])\nplt.grid(axis ='y',zorder=0)\nplt.annotate('Number of grades given: '+ str(list_number_of_grades_faculties[2]), (0,0), (0, -20),fontsize= 13, xycoords='axes fraction', textcoords='offset points', va='top')\nplt.bar(x, y_ects_freq[2], width=0.93, edgecolor='black',zorder=3)\n\nplt.subplot(2, 3, 4)\nplt.title(Faculty_names[3], fontsize = 16, weight = 'bold')\nplt.ylim([0,0.30])\nplt.grid(axis ='y',zorder=0)\nplt.ylabel('Frequency',fontsize=14)\nplt.annotate('Number of grades given: '+ str(list_number_of_grades_faculties[3]), (0,0), (0, -20),fontsize= 13, xycoords='axes fraction', textcoords='offset points', va='top')\nplt.bar(x, y_ects_freq[3], width=0.93, edgecolor='black',zorder=3)\n\nplt.subplot(2, 3, 5)\nplt.title(Faculty_names[4], fontsize = 16, weight = 'bold')\nplt.ylim([0,0.30])\nplt.grid(axis ='y',zorder=0)\nplt.annotate('Number of grades given: '+ str(list_number_of_grades_faculties[4]), (0,0), (0, -20),fontsize= 13, xycoords='axes fraction', textcoords='offset points', va='top')\nplt.bar(x, y_ects_freq[4], width=0.93, edgecolor='black',zorder=3)\n\nplt.subplot(2, 3, 6)\nplt.title(Faculty_names[5], fontsize = 16, weight = 'bold')\nplt.ylim([0,0.30])\nplt.grid(axis ='y',zorder=0)\nplt.annotate('Number of grades given: '+ str(list_number_of_grades_faculties[5]), (0,0), (0, -20),fontsize= 13, xycoords='axes fraction', textcoords='offset points', va='top')\nplt.bar(x, y_ects_freq[5], width=0.93, edgecolor='black',zorder=3)\n\nf.savefig('histogram_gades_split_faculty_ECTS_weight.png')",
"_____no_output_____"
]
],
[
[
"### Faculties, weighted against ECTS - droppping -3 and 00",
"_____no_output_____"
]
],
[
[
"y_no_fail_ects = []\nx_no_fail = ['02','4','7','10','12']\n\n# Looking at each faculty \nfor i in tqdm_notebook(df['Fakultet'].unique()):\n df_faculty = df[df['Fakultet']==i]\n \n y_no_fail_ects_inner=[]\n \n for k in ['Weigthed_02','Weigthed_4','Weigthed_7','Weigthed_10','Weigthed_12']:\n y_no_fail_ects_inner.append(df_faculty[k].sum(skipna = True)) \n \n y_no_fail_ects.append(y_no_fail_ects_inner)\n \ny_no_fail_ects",
"_____no_output_____"
],
[
"# calc frequencies \ny_no_fail_ects_freq = []\n\n# running through each faculty\nfor i in range(len(y_ects)):\n \n y_no_fail_ects_freq_inner = []\n \n # calc frequencies \n for q in range(len(y_no_fail_ects[i])):\n y_no_fail_ects_freq_inner.append(y_no_fail_ects[i][q]/sum(y_no_fail_ects[i]))\n\n y_no_fail_ects_freq.append(y_no_fail_ects_freq_inner)\n\n\ny_no_fail_ects_freq",
"_____no_output_____"
],
[
"# This figure is used in the analysis\nf, ax = plt.subplots(figsize=(15,10))\n\nplt.subplot(2, 3, 1)\nplt.title(Faculty_names[0], fontsize = 16, weight = 'bold')\nplt.ylim([0,0.35])\nplt.grid(axis ='y',zorder=0)\nplt.ylabel('Frequency',fontsize=14)\nplt.annotate('Number of passing grades: '+ str(list_no_fail_number_of_grades_faculties[0]), (0,0), (0, -20),fontsize= 13, xycoords='axes fraction', textcoords='offset points', va='top')\nplt.bar(x_no_fail, y_no_fail_ects_freq[0], width=0.93, edgecolor='black',zorder=3)\n\nplt.subplot(2, 3, 2)\nplt.title(Faculty_names[1], fontsize = 16, weight = 'bold')\nplt.ylim([0,0.35])\nplt.grid(axis ='y',zorder=0)\nplt.annotate('Number of passing grades: '+ str(list_no_fail_number_of_grades_faculties[1]), (0,0), (0, -20),fontsize= 13, xycoords='axes fraction', textcoords='offset points', va='top')\nplt.bar(x_no_fail, y_no_fail_ects_freq[1], width=0.93, edgecolor='black',zorder=3)\n\nplt.subplot(2, 3, 3)\nplt.title(Faculty_names[2], fontsize = 16, weight = 'bold')\nplt.ylim([0,0.35])\nplt.grid(axis ='y',zorder=0)\nplt.annotate('Number of passing grades: '+ str(list_no_fail_number_of_grades_faculties[2]), (0,0), (0, -20),fontsize= 13, xycoords='axes fraction', textcoords='offset points', va='top')\nplt.bar(x_no_fail, y_no_fail_ects_freq[2], width=0.93, edgecolor='black',zorder=3)\n\nplt.subplot(2, 3, 4)\nplt.title(Faculty_names[3], fontsize = 16, weight = 'bold')\nplt.ylim([0,0.35])\nplt.grid(axis ='y',zorder=0)\nplt.ylabel('Frequency',fontsize=14)\nplt.annotate('Number of passing grades: '+ str(list_no_fail_number_of_grades_faculties[3]), (0,0), (0, -20),fontsize= 13, xycoords='axes fraction', textcoords='offset points', va='top')\nplt.bar(x_no_fail, y_no_fail_ects_freq[3], width=0.93, edgecolor='black',zorder=3)\n\nplt.subplot(2, 3, 5)\nplt.title(Faculty_names[4], fontsize = 16, weight = 'bold')\nplt.ylim([0,0.35])\nplt.grid(axis ='y',zorder=0)\nplt.annotate('Number of passing grades: '+ str(list_no_fail_number_of_grades_faculties[4]), (0,0), (0, -20),fontsize= 13, xycoords='axes fraction', textcoords='offset points', va='top')\nplt.bar(x_no_fail, y_no_fail_ects_freq[4], width=0.93, edgecolor='black',zorder=3)\n\nplt.subplot(2, 3, 6)\nplt.title(Faculty_names[5], fontsize = 16, weight = 'bold')\nplt.ylim([0,0.35])\nplt.grid(axis ='y',zorder=0)\nplt.annotate('Number of passing grades: '+ str(list_no_fail_number_of_grades_faculties[5]), (0,0), (0, -20),fontsize= 13, xycoords='axes fraction', textcoords='offset points', va='top')\nplt.bar(x_no_fail, y_no_fail_ects_freq[5], width=0.93, edgecolor='black',zorder=3)\n\nf.savefig('histogram_gades_split_faculty_ECTS_weight_NO_FAIL.png')",
"_____no_output_____"
]
],
[
[
"### Calculating GPA by faculty",
"_____no_output_____"
]
],
[
[
"from math import isnan\nimport math",
"_____no_output_____"
],
[
"#Calculate gpa when ONLY PASSED exams are counted\nsnit = []\nfor i in range(0,len(df)):\n x_02 = df[2][i]\n x_04 = df[4][i]\n x_07 = df[7][i]\n x_10 = df[10][i]\n x_12 = df[12][i]\n \n number = (x_12,x_10,x_07,x_04,x_02)\n grades = [12,10,7,4,2]\n mydick = dict(zip(grades,number))\n cleandick = {k: mydick[k] for k in mydick if not isnan(mydick[k])}\n \n num = sum([x * y for x,y in mydick.items()])\n den = sum(mydick.values())\n \n snit.append(num/den)\n \ndf[\"Snit\"] = snit",
"_____no_output_____"
],
[
"# Here I calculate the GPA of some form of assessment\ndef gpa(df,string):\n \n x_gpa = []\n x_sho = []\n x_ect = []\n\n for i in range(0,len(df)):\n if df[\"Fakultet\"][i] == string:\n if math.isnan(df[\"Snit\"][i]) == False:\n x_gpa.append(float(df[\"Snit\"][i]))\n x_sho.append(float(df[\"Fremmødte\"][i]))\n x_ect.append(float(df[\"Credit_edit\"][i]))\n\n den = 0\n num = 0\n for i in range(0,len(x_gpa)):\n den = x_sho[i]*x_ect[i] + den\n num = x_gpa[i]*x_sho[i]*x_ect[i] + num\n out = num/den\n return out",
"_____no_output_____"
],
[
"# Looping through each faculty\n\nfor i in df['Fakultet'].unique():\n print(gpa(df,i))",
"_____no_output_____"
]
],
[
[
"### Type of assessments broken down by faculties",
"_____no_output_____"
]
],
[
[
"list_type_ass=list(df['Type of assessmet_edit'].unique())",
"_____no_output_____"
],
[
"y_type_ass_inner = []\ny_type_ass = []\n\n# Looking at each faculty \nfor i in tqdm_notebook(df['Fakultet'].unique()):\n df_faculty = df[df['Fakultet']==i]\n \n # Running through each type of assessment.\n for k in list_type_ass:\n # Summing number of passed people for all courses (in each faculty) broken down on each tyep of assessment\n y_type_ass_inner.append(df_faculty[df_faculty['Type of assessmet_edit']==k]['Antal bestået'].sum(skipna = True)) \n \n y_type_ass.append(y_type_ass_inner)\n y_type_ass_inner=[]",
"_____no_output_____"
],
[
"# Creating the categories which we want for plot.\ncategories = []\n\nfor i in range(len(df['Fakultet'].unique())):\n \n categories_inner = []\n \n # Oral\n categories_inner.append(y_type_ass[i][1])\n \n # Written not under invigilation\n categories_inner.append(y_type_ass[i][2])\n \n # Written under invigilation\n categories_inner.append(y_type_ass[i][4])\n \n # Rest\n categories_inner.append(y_type_ass[i][0]+y_type_ass[i][3]+y_type_ass[i][5]+y_type_ass[i][6]+y_type_ass[i][7]\\\n +y_type_ass[i][8]+y_type_ass[i][9])\n \n categories.append(categories_inner)",
"_____no_output_____"
],
[
"#calc share.\n\nlist_categories_share = []\n\n# Running through each faculty\nfor i in range(len(categories)):\n \n categories_share_inner = []\n \n # For each faculty calc type of ass shares. \n for k in range(len(categories[i])):\n categories_share= categories[i][k]/sum(categories[i])*100 # times a 100 for %\n categories_share_inner.append(categories_share)\n \n list_categories_share.append(categories_share_inner) ",
"_____no_output_____"
],
[
"# Converting list to DataFrame \ndfcat= pd.DataFrame(list_categories_share) \ndfcat\ndfcat=dfcat.T\ndfcat.columns = ['Science','Social Sciences','Humanities','Health & Medical Sciences','Law','Theology']\ndfcat.rename(index={0:'Oral',1:'Written not under invigilation',2:'Written under invigilation',3:'Rest'})\n#dfcat.index.name = 'type_ass'\ndfcat=dfcat.T\ndfcat.columns = ['Oral','Written not invigilation','Written invigilation','Rest']",
"_____no_output_____"
],
[
"colors = [\"#011f4b\",\"#005b96\",\"#6497b1\",'#b3cde0']\n\n\ndfcat.plot(kind='bar', stacked=True, color = colors, fontsize = 12)\n#plt.legend(bbox_to_anchor=(0, 1), loc='upper left', ncol=1)\nplt.rcParams[\"figure.figsize\"] = [15,15]\nplt.legend(bbox_to_anchor=(0,1.02,1,0.2), loc=\"lower left\", mode=\"expand\", borderaxespad=0, ncol=2, fontsize = 12) #,weight = 'bold')\nplt.tight_layout()\nplt.savefig('stacked_bar_share_ass.png')",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d09251832d8f89802bf35936f11fa73e19ef0ee2 | 195,918 | ipynb | Jupyter Notebook | SentimentalAnalysisWithGPTNeo.ipynb | bhadreshpsavani/ExploringSentimentalAnalysis | a48a8b44850cb0df7ae5fd19943fa26a3a66142c | [
"Apache-2.0"
] | 1 | 2022-01-14T04:47:43.000Z | 2022-01-14T04:47:43.000Z | SentimentalAnalysisWithGPTNeo.ipynb | bhadreshpsavani/ExploringSentimentalAnalysis | a48a8b44850cb0df7ae5fd19943fa26a3a66142c | [
"Apache-2.0"
] | 2 | 2020-10-04T09:21:28.000Z | 2020-10-04T09:23:36.000Z | SentimentalAnalysisWithGPTNeo.ipynb | bhadreshpsavani/ExploringSentimentalAnalysis | a48a8b44850cb0df7ae5fd19943fa26a3a66142c | [
"Apache-2.0"
] | 3 | 2021-11-30T20:23:40.000Z | 2022-02-03T18:11:32.000Z | 37.953894 | 1,505 | 0.506661 | [
[
[
"<a href=\"https://colab.research.google.com/github/bhadreshpsavani/ExploringSentimentalAnalysis/blob/main/SentimentalAnalysisWithGPTNeo.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"## Step1. Import and Load Data",
"_____no_output_____"
]
],
[
[
"!pip install -q pip install git+https://github.com/huggingface/transformers.git\n!pip install -q datasets",
" Installing build dependencies ... \u001b[?25l\u001b[?25hdone\n Getting requirements to build wheel ... \u001b[?25l\u001b[?25hdone\n Preparing wheel metadata ... \u001b[?25l\u001b[?25hdone\n\u001b[K |████████████████████████████████| 901kB 6.5MB/s \n\u001b[K |████████████████████████████████| 3.3MB 16.3MB/s \n\u001b[?25h Building wheel for transformers (PEP 517) ... \u001b[?25l\u001b[?25hdone\n\u001b[K |████████████████████████████████| 235kB 5.2MB/s \n\u001b[K |████████████████████████████████| 112kB 8.9MB/s \n\u001b[K |████████████████████████████████| 245kB 9.9MB/s \n\u001b[?25h"
],
[
"from datasets import load_dataset\nemotions = load_dataset(\"emotion\")",
"_____no_output_____"
],
[
"import torch\ndevice = torch.device(\"cuda\" if torch.cuda.is_available() else \"cpu\")",
"_____no_output_____"
]
],
[
[
"## Step2. Preprocess Data",
"_____no_output_____"
]
],
[
[
"from transformers import AutoTokenizer\n\nmodel_name = \"EleutherAI/gpt-neo-125M\"\ntokenizer = AutoTokenizer.from_pretrained(model_name)\n\ndef tokenize(batch):\n return tokenizer(batch[\"text\"], padding=True, truncation=True)",
"_____no_output_____"
],
[
"tokenizer",
"_____no_output_____"
],
[
"tokenizer.add_special_tokens({'pad_token': '<|pad|>'})\ntokenizer",
"_____no_output_____"
],
[
"emotions_encoded = emotions.map(tokenize, batched=True, batch_size=None)",
"_____no_output_____"
],
[
"from transformers import AutoModelForSequenceClassification\nnum_labels = 6\nmodel = (AutoModelForSequenceClassification.from_pretrained(model_name, num_labels=num_labels).to(device))",
"_____no_output_____"
],
[
"emotions_encoded[\"train\"].features",
"_____no_output_____"
],
[
"emotions_encoded.set_format(\"torch\", columns=[\"input_ids\", \"attention_mask\", \"label\"])\nemotions_encoded[\"train\"].features",
"_____no_output_____"
],
[
"from sklearn.metrics import accuracy_score, f1_score\n\ndef compute_metrics(pred):\n labels = pred.label_ids\n preds = pred.predictions.argmax(-1)\n f1 = f1_score(labels, preds, average=\"weighted\")\n acc = accuracy_score(labels, preds)\n return {\"accuracy\": acc, \"f1\": f1}",
"_____no_output_____"
],
[
"from transformers import Trainer, TrainingArguments\n\nbatch_size = 2\nlogging_steps = len(emotions_encoded[\"train\"]) // batch_size\ntraining_args = TrainingArguments(output_dir=\"results\",\n num_train_epochs=2,\n learning_rate=2e-5,\n per_device_train_batch_size=batch_size,\n per_device_eval_batch_size=batch_size,\n load_best_model_at_end=True,\n metric_for_best_model=\"f1\",\n weight_decay=0.01,\n evaluation_strategy=\"epoch\",\n disable_tqdm=False,\n logging_steps=logging_steps,)",
"_____no_output_____"
],
[
"from transformers import Trainer\n\ntrainer = Trainer(model=model, args=training_args,\n compute_metrics=compute_metrics,\n train_dataset=emotions_encoded[\"train\"],\n eval_dataset=emotions_encoded[\"validation\"])\ntrainer.train();",
"_____no_output_____"
],
[
"results = trainer.evaluate()\nresults",
"_____no_output_____"
],
[
"preds_output = trainer.predict(emotions_encoded[\"validation\"])\npreds_output.metrics",
"_____no_output_____"
],
[
"import numpy as np\nfrom sklearn.metrics import plot_confusion_matrix\ny_valid = np.array(emotions_encoded[\"validation\"][\"label\"])\ny_preds = np.argmax(preds_output.predictions, axis=1)\nlabels = ['sadness', 'joy', 'love', 'anger', 'fear', 'surprise']\nplot_confusion_matrix(y_preds, y_valid, labels)",
"_____no_output_____"
],
[
"model.save_pretrained('./model')\ntokenizer.save_pretrained('./model')",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d09259602e20222af42d20b16843170972c22f07 | 1,871 | ipynb | Jupyter Notebook | examples/00 - adapter trimming.ipynb | britm92/NET_analysis | 42bcbaafa10e284b86ea8f71bdf488d88f8c1b43 | [
"MIT"
] | null | null | null | examples/00 - adapter trimming.ipynb | britm92/NET_analysis | 42bcbaafa10e284b86ea8f71bdf488d88f8c1b43 | [
"MIT"
] | null | null | null | examples/00 - adapter trimming.ipynb | britm92/NET_analysis | 42bcbaafa10e284b86ea8f71bdf488d88f8c1b43 | [
"MIT"
] | 1 | 2020-07-22T14:35:13.000Z | 2020-07-22T14:35:13.000Z | 23.987179 | 88 | 0.445751 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d092644b84debe134ce58ce5f93c2c0d8997565d | 278,692 | ipynb | Jupyter Notebook | Cluster/kmeans-kmedoids/KMedoids-blobs-3-0.3-varypts.ipynb | bcottman/photon_experiments | e2097dc809bd73482936b25b7504b1b9211512b2 | [
"MIT"
] | null | null | null | Cluster/kmeans-kmedoids/KMedoids-blobs-3-0.3-varypts.ipynb | bcottman/photon_experiments | e2097dc809bd73482936b25b7504b1b9211512b2 | [
"MIT"
] | null | null | null | Cluster/kmeans-kmedoids/KMedoids-blobs-3-0.3-varypts.ipynb | bcottman/photon_experiments | e2097dc809bd73482936b25b7504b1b9211512b2 | [
"MIT"
] | null | null | null | 180.734112 | 97,076 | 0.790238 | [
[
[
"## parameters",
"_____no_output_____"
]
],
[
[
"CLUSTER_ALGO = 'KMedoids'\nC_SHAPE ='circle'\n#C_SHAPE ='CIRCLE'\n#C_SHAPE ='ellipse'\n#N_CLUSTERS = [50,300, 1000]\nN_CLUSTERS = [3]\nCLUSTERS_STD = 0.3\nN_P_CLUSTERS = [3, 30, 300, 3000]\nN_CLUSTERS_S = N_CLUSTERS[0]\nINNER_FOLDS = 3\nOUTER_FOLDS = 3",
"_____no_output_____"
]
],
[
[
"## includes",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport matplotlib\n\nimport matplotlib.pyplot as plt\nfrom sklearn.datasets import load_breast_cancer\nfrom sklearn.datasets import load_iris\nfrom sklearn.datasets import make_blobs\nfrom sklearn.datasets import make_moons\n",
"_____no_output_____"
],
[
"%load_ext autoreload\n%autoreload 2\npackages = !conda list\npackages",
"_____no_output_____"
]
],
[
[
"## Output registry",
"_____no_output_____"
]
],
[
[
"from __future__ import print_function\nimport sys, os\n\nold__file__ = !pwd\n__file__ = !cd ../../../photon ;pwd\n#__file__ = !pwd\n__file__ = __file__[0]\n__file__\nsys.path.append(__file__)\nprint(sys.path)\nos.chdir(old__file__[0])\n!pwd\nold__file__[0]",
"['/docker/photon_experiments/Cluster/kmeans-kmedoids', '/opt/conda/lib/python37.zip', '/opt/conda/lib/python3.7', '/opt/conda/lib/python3.7/lib-dynload', '', '/opt/conda/lib/python3.7/site-packages', '/opt/conda/lib/python3.7/site-packages/IPython/extensions', '/home/jovyan/.ipython', '/docker/photon']\n/docker/photon_experiments/Cluster/kmeans-kmedoids\n"
],
[
"\nimport seaborn as sns; sns.set() # for plot styling\nimport numpy as np\nimport pandas as pd\nfrom math import floor,ceil\nfrom sklearn.model_selection import KFold\nfrom sklearn.manifold import TSNE\nimport itertools\nimport seaborn as sns\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\n#set font size of labels on matplotlib plots\nplt.rc('font', size=16)\n\n#set style of plots\nsns.set_style('white')\n\n#define a custom palette\nPALLET = ['#40111D', '#DCD5E4', '#E7CC74'\n ,'#39C8C6', '#AC5583', '#D3500C'\n ,'#FFB139', '#98ADA7', '#AD989E'\n ,'#708090','#6C8570','#3E534D'\n ,'#0B8FD3','#0B47D3','#96D30B' \n ,'#630C3A','#F1D0AF','#64788B' \n ,'#8B7764','#7A3C5D','#77648B'\n ,'#eaff39','#39ff4e','#4e39ff'\n ,'#ff4e39','#87ff39','#ff3987', ]\nN_PALLET = len(PALLET)\nsns.set_palette(PALLET)\nsns.palplot(PALLET)\n\n\nfrom clusim.clustering import Clustering, remap2match\nimport clusim.sim as sim\n\nfrom photonai.base import Hyperpipe, PipelineElement, Preprocessing, OutputSettings\nfrom photonai.optimization import FloatRange, Categorical, IntegerRange\nfrom photonai.base.photon_elements import PhotonRegistry\nfrom photonai.visual.graphics import plot_cm\nfrom photonai.photonlogger.logger import logger\n#from photonai.base.registry.registry import PhotonRegistry",
"/opt/conda/lib/python3.7/site-packages/sklearn/externals/joblib/__init__.py:15: DeprecationWarning: sklearn.externals.joblib is deprecated in 0.21 and will be removed in 0.23. Please import this functionality directly from joblib, which can be installed with: pip install joblib. If this warning is raised when loading pickled models, you may need to re-serialize those models with scikit-learn 0.21+.\n warnings.warn(msg, category=DeprecationWarning)\n"
]
],
[
[
"## function defintions",
"_____no_output_____"
]
],
[
[
"def yield_parameters_ellipse(n_p_clusters):\n cluster_std = CLUSTERS_STD\n for n_p_cluster in n_p_clusters:\n for n_cluster in N_CLUSTERS:\n print('ncluster:', n_cluster)\n n_cluster_std = [cluster_std for k in range(n_cluster)]\n n_samples = [n_p_cluster for k in range(n_cluster)]\n data_X, data_y = make_blobs(n_samples=n_samples,\n cluster_std=n_cluster_std, random_state=0)\n transformation = [[0.6, -0.6], [-0.4, 0.8]]\n X_ellipse = np.dot(data_X, transformation)\n yield [X_ellipse, data_y, n_cluster]",
"_____no_output_____"
],
[
"def yield_parameters(n_p_clusters):\n cluster_std = CLUSTERS_STD\n for n_p_cluster in n_p_clusters:\n for n_cluster in N_CLUSTERS:\n n_cluster_std = [cluster_std for k in range(n_cluster)]\n n_samples = [n_p_cluster for k in range(n_cluster)]\n data_X, data_y = make_blobs(n_samples=n_samples,\n cluster_std=n_cluster_std, random_state=0)\n yield [data_X, data_y, n_cluster]",
"_____no_output_____"
],
[
"def results_to_df(results):\n ll = []\n for obj in results:\n ll.append([obj.operation,\n obj.value,\n obj.metric_name])\n _results=pd.DataFrame(ll).pivot(index=2, columns=0, values=1)\n _results.columns=['Mean','STD']\n \n return(_results)",
"_____no_output_____"
],
[
"def cluster_plot(my_pipe, data_X, n_cluster, PALLET):\n y_pred= my_pipe.predict(data_X)\n data = pd.DataFrame(data_X[:, 0],columns=['x'])\n data['y'] = data_X[:, 1]\n data['labels'] = y_pred\n facet = sns.lmplot(data=data, x='x', y='y', hue='labels', \n aspect= 1.0, height=10,\n fit_reg=False, legend=True, legend_out=True)\n\n customPalette = PALLET #*ceil((n_cluster/N_PALLET +2))\n for i, label in enumerate( np.sort(data['labels'].unique())):\n plt.annotate(label, \n data.loc[data['labels']==label,['x','y']].mean(),\n horizontalalignment='center',\n verticalalignment='center',\n size=5, weight='bold',\n color='white',\n backgroundcolor=customPalette[i]) \n\n plt.show()\n return y_pred",
"_____no_output_____"
],
[
"def simple_output(string: str, number: int) -> None:\n print(string, number)\n logger.info(string, number )",
"_____no_output_____"
],
[
"__file__ = \"exp1.log\"\nbase_folder = os.path.dirname(os.path.abspath(''))\ncustom_elements_folder = os.path.join(base_folder, 'custom_elements')\ncustom_elements_folder",
"_____no_output_____"
],
[
"registry = PhotonRegistry(custom_elements_folder=custom_elements_folder)\nregistry.activate()\nregistry.PHOTON_REGISTRIES,PhotonRegistry.PHOTON_REGISTRIES",
"_____no_output_____"
],
[
"registry.activate()\nregistry.list_available_elements()\n# take off last name",
"\nPhotonCore\nARDRegression sklearn.linear_model.ARDRegression Estimator\nAdaBoostClassifier sklearn.ensemble.AdaBoostClassifier Estimator\nAdaBoostRegressor sklearn.ensemble.AdaBoostRegressor Estimator\nBaggingClassifier sklearn.ensemble.BaggingClassifier Estimator\nBaggingRegressor sklearn.ensemble.BaggingRegressor Estimator\nBayesianGaussianMixture sklearn.mixture.BayesianGaussianMixture Estimator\nBayesianRidge sklearn.linear_model.BayesianRidge Estimator\nBernoulliNB sklearn.naive_bayes.BernoulliNB Estimator\nBernoulliRBM sklearn.neural_network.BernoulliRBM Estimator\nBinarizer sklearn.preprocessing.Binarizer Transformer\nCCA sklearn.cross_decomposition.CCA Transformer\nConfounderRemoval photonai.modelwrapper.ConfounderRemoval.ConfounderRemoval Transformer\nDecisionTreeClassifier sklearn.tree.DecisionTreeClassifier Estimator\nDecisionTreeRegressor sklearn.tree.DecisionTreeRegressor Estimator\nDictionaryLearning sklearn.decomposition.DictionaryLearning Transformer\nDummyClassifier sklearn.dummy.DummyClassifier Estimator\nDummyRegressor sklearn.dummy.DummyRegressor Estimator\nElasticNet sklearn.linear_model.ElasticNet Estimator\nExtraDecisionTreeClassifier sklearn.tree.ExtraDecisionTreeClassifier Estimator\nExtraDecisionTreeRegressor sklearn.tree.ExtraDecisionTreeRegressor Estimator\nExtraTreesClassifier sklearn.ensemble.ExtraTreesClassifier Estimator\nExtraTreesRegressor sklearn.ensemble.ExtraTreesRegressor Estimator\nFClassifSelectPercentile photonai.modelwrapper.FeatureSelection.FClassifSelectPercentile Transformer\nFRegressionFilterPValue photonai.modelwrapper.FeatureSelection.FRegressionFilterPValue Transformer\nFRegressionSelectPercentile photonai.modelwrapper.FeatureSelection.FRegressionSelectPercentile Transformer\nFactorAnalysis sklearn.decomposition.FactorAnalysis Transformer\nFastICA sklearn.decomposition.FastICA Transformer\nFeatureEncoder photonai.modelwrapper.OrdinalEncoder.FeatureEncoder Transformer\nFunctionTransformer sklearn.preprocessing.FunctionTransformer Transformer\nGaussianMixture sklearn.mixture.GaussianMixture Estimator\nGaussianNB sklearn.naive_bayes.GaussianNB Estimator\nGaussianProcessClassifier sklearn.gaussian_process.GaussianProcessClassifier Estimator\nGaussianProcessRegressor sklearn.gaussian_process.GaussianProcessRegressor Estimator\nGenericUnivariateSelect sklearn.feature_selection.GenericUnivariateSelect Transformer\nGradientBoostingClassifier sklearn.ensemble.GradientBoostingClassifier Estimator\nGradientBoostingRegressor sklearn.ensemble.GradientBoostingRegressor Estimator\nHuberRegressor sklearn.linear_model.HuberRegressor Estimator\nImbalancedDataTransformer photonai.modelwrapper.imbalanced_data_transformer.ImbalancedDataTransformer Transformer\nIncrementalPCA sklearn.decomposition.IncrementalPCA Transformer\nKNeighborsClassifier sklearn.neighbors.KNeighborsClassifier Estimator\nKNeighborsRegressor sklearn.neighbors.KNeighborsRegressor Estimator\nKerasBaseClassifier photonai.modelwrapper.keras_base_models.KerasBaseClassifier Estimator\nKerasBaseRegression photonai.modelwrapper.keras_base_models.KerasBaseRegression Estimator\nKerasDnnClassifier photonai.modelwrapper.keras_dnn_classifier.KerasDnnClassifier Estimator\nKerasDnnRegressor photonai.modelwrapper.keras_dnn_regressor.KerasDnnRegressor Estimator\nKernelCenterer sklearn.preprocessing.KernelCenterer Transformer\nKernelPCA sklearn.decomposition.KernelPCA Transformer\nKernelRidge sklearn.kernel_ridge.KernelRidge Estimator\nLabelEncoder photonai.modelwrapper.LabelEncoder.LabelEncoder Transformer\nLars sklearn.linear_model.Lars Estimator\nLasso sklearn.linear_model.Lasso Estimator\nLassoFeatureSelection photonai.modelwrapper.FeatureSelection.LassoFeatureSelection Transformer\nLassoLars sklearn.linear_model.LassoLars Estimator\nLatentDirichletAllocation sklearn.decomposition.LatentDirichletAllocation Transformer\nLinearRegression sklearn.linear_model.LinearRegression Estimator\nLinearSVC sklearn.svm.LinearSVC Estimator\nLinearSVR sklearn.svm.LinearSVR Estimator\nLogisticRegression sklearn.linear_model.LogisticRegression Estimator\nMLPClassifier sklearn.neural_network.MLPClassifier Estimator\nMLPRegressor sklearn.neural_network.MLPRegressor Estimator\nMaxAbsScaler sklearn.preprocessing.MaxAbsScaler Transformer\nMinMaxScaler sklearn.preprocessing.MinMaxScaler Transformer\nMiniBatchDictionaryLearning sklearn.decomposition.MiniBatchDictionaryLearning Transformer\nMiniBatchSparsePCA sklearn.decomposition.MiniBatchSparsePCA Transformer\nMultinomialNB sklearn.naive_bayes.MultinomialNB Estimator\nNMF sklearn.decompositcion.NMF Transformer\nNearestCentroid sklearn.neighbors.NearestCentroid Estimator\nNormalizer sklearn.preprocessing.Normalizer Transformer\nNuSVC sklearn.svm.NuSVC Estimator\nNuSVR sklearn.svm.NuSVR Estimator\nOneClassSVM sklearn.svm.OneClassSVM Estimator\nPCA sklearn.decomposition.PCA Transformer\nPLSCanonical sklearn.cross_decomposition.PLSCanonical Transformer\nPLSRegression sklearn.cross_decomposition.PLSRegression Transformer\nPLSSVD sklearn.cross_decomposition.PLSSVD Transformer\nPassiveAggressiveClassifier sklearn.linear_model.PassiveAggressiveClassifier Estimator\nPassiveAggressiveRegressor sklearn.linear_model.PassiveAggressiveRegressor Estimator\nPerceptron sklearn.linear_model.Perceptron Estimator\nPhotonMLPClassifier photonai.modelwrapper.PhotonMLPClassifier.PhotonMLPClassifier Estimator\nPhotonOneClassSVM photonai.modelwrapper.PhotonOneClassSVM.PhotonOneClassSVM Estimator\nPhotonTestXPredictor photonai.test.processing_tests.results_tests.XPredictor Estimator\nPhotonVotingClassifier photonai.modelwrapper.Voting.PhotonVotingClassifier Estimator\nPhotonVotingRegressor photonai.modelwrapper.Voting.PhotonVotingRegressor Estimator\nPolynomialFeatures sklearn.preprocessing.PolynomialFeatures Transformer\nPowerTransformer sklearn.preprocessing.PowerTransformer Transformer\nQuantileTransformer sklearn.preprocessing.QuantileTransformer Transformer\nRANSACRegressor sklearn.linear_model.RANSACRegressor Estimator\nRFE sklearn.feature_selection.RFE Transformer\nRFECV sklearn.feature_selection.RFECV Transformer\nRadiusNeighborsClassifier sklearn.neighbors.RadiusNeighborsClassifier Estimator\nRadiusNeighborsRegressor sklearn.neighbors.RadiusNeighborsRegressor Estimator\nRandomForestClassifier sklearn.ensemble.RandomForestClassifier Estimator\nRandomForestRegressor sklearn.ensemble.RandomForestRegressor Estimator\nRandomTreesEmbedding sklearn.ensemble.RandomTreesEmbedding Transformer\nRangeRestrictor photonai.modelwrapper.RangeRestrictor.RangeRestrictor Estimator\nRidge sklearn.linear_model.Ridge Estimator\nRidgeClassifier sklearn.linear_model.RidgeClassifier Estimator\nRobustScaler sklearn.preprocessing.RobustScaler Transformer\nSGDClassifier sklearn.linear_model.SGDClassifier Estimator\nSGDRegressor sklearn.linear_model.SGDRegressor Estimator\nSVC sklearn.svm.SVC Estimator\nSVR sklearn.svm.SVR Estimator\nSamplePairingClassification photonai.modelwrapper.SamplePairing.SamplePairingClassification Transformer\nSamplePairingRegression photonai.modelwrapper.SamplePairing.SamplePairingRegression Transformer\nSelectFdr sklearn.feature_selection.SelectFdr Transformer\nSelectFpr sklearn.feature_selection.SelectFpr Transformer\nSelectFromModel sklearn.feature_selection.SelectFromModel Transformer\nSelectFwe sklearn.feature_selection.SelectFwe Transformer\nSelectKBest sklearn.feature_selection.SelectKBest Transformer\nSelectPercentile sklearn.feature_selection.SelectPercentile Transformer\nSimpleImputer sklearn.impute.SimpleImputer Transformer\nSourceSplitter photonai.modelwrapper.source_splitter.SourceSplitter Transformer\nSparseCoder sklearn.decomposition.SparseCoder Transformer\nSparsePCA sklearn.decomposition.SparsePCA Transformer\nStandardScaler sklearn.preprocessing.StandardScaler Transformer\nTheilSenRegressor sklearn.linear_model.TheilSenRegressor Estimator\nTruncatedSVD sklearn.decomposition.TruncatedSVD Transformer\nVarianceThreshold sklearn.feature_selection.VarianceThreshold Transformer\ndict_learning sklearn.decomposition.dict_learning Transformer\ndict_learning_online sklearn.decomposition.dict_learning_online Transformer\nfastica sklearn.decomposition.fastica Transformer\nsparse_encode sklearn.decomposition.sparse_encode Transformer\n\nPhotonCluster\nKMeans sklearn.cluster.KMeans Estimator\nKMedoids sklearn_extra.cluster.KMedoids Estimator\n\nPhotonNeuro\nBrainAtlas photonai.neuro.brain_atlas.BrainAtlas Transformer\nBrainMask photonai.neuro.brain_atlas.BrainMask Transformer\nPatchImages photonai.neuro.nifti_transformations.PatchImages Transformer\nResampleImages photonai.neuro.nifti_transformations.ResampleImages Transformer\nSmoothImages photonai.neuro.nifti_transformations.SmoothImages Transformer\n"
]
],
[
[
"## KMeans blobs",
"_____no_output_____"
]
],
[
[
"\nregistry.info(CLUSTER_ALGO)",
"----------------------------------\nName: KMedoids\nNamespace: sklearn_extra.cluster\n----------------------------------\nPossible Hyperparameters as derived from constructor:\nn_clusters n_clusters=8 \nmetric metric='euclidean' \ninit init='heuristic' \nmax_iter max_iter=300 \nrandom_state random_state=None \n----------------------------------\n"
],
[
"def hyper_cluster(cluster_name):\n if C_SHAPE == 'ellipse' :\n yield_cluster = yield_parameters_ellipse\n else: \n yield_cluster = yield_parameters\n \n n_p_clusters = N_P_CLUSTERS\n for data_X, data_y,n_cluster in yield_cluster(n_p_clusters):\n simple_output('CLUSTER_ALGO:', CLUSTER_ALGO)\n simple_output('C_SHAPE:',C_SHAPE)\n simple_output('n_cluster:', n_cluster)\n simple_output('CLUSTERS_STD:', CLUSTERS_STD)\n \n simple_output('INNER_FOLDS:', INNER_FOLDS)\n simple_output('OUTER_FOLDS:', OUTER_FOLDS) \n simple_output('n_points:', len(data_y))\n\n X = data_X.copy(); y = data_y.copy()\n # DESIGN YOUR PIPELINE\n settings = OutputSettings(project_folder='./tmp/')\n \n my_pipe = Hyperpipe('batching',\n optimizer='sk_opt',\n # optimizer_params={'n_configurations': 25},\n metrics=['ARI', 'MI', 'HCV', 'FM'],\n best_config_metric='ARI',\n outer_cv=KFold(n_splits=OUTER_FOLDS),\n inner_cv=KFold(n_splits=INNER_FOLDS),\n verbosity=0,\n output_settings=settings)\n\n\n my_pipe += PipelineElement(cluster_name, hyperparameters={\n 'n_clusters': IntegerRange(floor(n_cluster*.7)\n , ceil(n_cluster*1.2)),\n },random_state=777)\n\n logger.info('Cluster optimization range:', floor(n_cluster*.7), ceil(n_cluster*1.2))\n print('Cluster optimization range:', floor(n_cluster*.7), ceil(n_cluster*1.2)) \n\n # NOW TRAIN YOUR PIPELINE\n my_pipe.fit(X, y)\n\n debug = True\n #------------------------------plot\n y_pred=cluster_plot(my_pipe, X, n_cluster, PALLET)\n #--------------------------------- best\n print(pd.DataFrame(my_pipe.best_config.items()\n ,columns=['n_clusters', 'k']))\n #------------------------------\n print('train','\\n'\n ,results_to_df(my_pipe.results.metrics_train))\n print('test','\\n'\n ,results_to_df(my_pipe.results.metrics_test))\n #------------------------------ \n # turn the ground-truth labels into a clusim Clustering\n true_clustering = Clustering().from_membership_list(y) \n kmeans_clustering = Clustering().from_membership_list(y_pred) # lets see how similar the predicted k-means clustering is to the true clustering\n #------------------------------\n # using all available similar measures!\n row_format2 =\"{:>25}\" * (2)\n for simfunc in sim.available_similarity_measures:\n print(row_format2.format(simfunc, eval('sim.' + simfunc+'(true_clustering, kmeans_clustering)')))\n #------------------------------# The element-centric similarity is particularly useful for understanding\n # how a clustering method performed\n\n # Let's start with the single similarity value:\n elsim = sim.element_sim(true_clustering, kmeans_clustering)\n print(\"Element-centric similarity: {}\".format(elsim))",
"_____no_output_____"
],
[
"hyper_cluster(CLUSTER_ALGO)",
"CLUSTER_ALGO: KMedoids\nC_SHAPE: circle\nn_cluster: 3\nINNER_FOLDS: 3\nOUTER_FOLDS: 3\nn_points: 9\nCluster optimization range: 2 4\n***************************************************************************************************************\nPHOTON ANALYSIS: batching\n***************************************************************************************************************\n\n********************************************************\nOuter Cross validation Fold 1\n********************************************************\n---------------------------------------------------------------------------------------------------------------\nBEST_CONFIG \n---------------------------------------------------------------------------------------------------------------\n{\n \"KMedoids\": [\n \"n_clusters=4\"\n ]\n}\n+--------+-------------------+------------------+\n| METRIC | PERFORMANCE TRAIN | PERFORMANCE TEST |\n+--------+-------------------+------------------+\n| ARI | 0.7619 | 1.0000 |\n| MI | 0.6154 | 1.0000 |\n| HCV | 1.0000 | 1.0000 |\n| FM | 0.8165 | 0.0000 |\n+--------+-------------------+------------------+\n\n********************************************************\nOuter Cross validation Fold 2\n********************************************************\n---------------------------------------------------------------------------------------------------------------\nBEST_CONFIG \n---------------------------------------------------------------------------------------------------------------\n{\n \"KMedoids\": [\n \"n_clusters=4\"\n ]\n}\n+--------+-------------------+------------------+\n| METRIC | PERFORMANCE TRAIN | PERFORMANCE TEST |\n+--------+-------------------+------------------+\n| ARI | 0.7619 | 1.0000 |\n| MI | 0.6154 | 1.0000 |\n| HCV | 1.0000 | 1.0000 |\n| FM | 0.8165 | 0.0000 |\n+--------+-------------------+------------------+\n\n********************************************************\nOuter Cross validation Fold 3\n********************************************************\n---------------------------------------------------------------------------------------------------------------\nBEST_CONFIG \n---------------------------------------------------------------------------------------------------------------\n{\n \"KMedoids\": [\n \"n_clusters=4\"\n ]\n}\n+--------+-------------------+------------------+\n| METRIC | PERFORMANCE TRAIN | PERFORMANCE TEST |\n+--------+-------------------+------------------+\n| ARI | 0.7619 | 1.0000 |\n| MI | 0.6154 | 1.0000 |\n| HCV | 1.0000 | 1.0000 |\n| FM | 0.8165 | 0.0000 |\n+--------+-------------------+------------------+\n\n===============================================================================================================\nOVERALL BEST CONFIGURATION\n===============================================================================================================\n{\n \"KMedoids\": [\n \"n_clusters=4\"\n ]\n}\n\nAnalysis batching done in 0:00:24.506777\nYour results are stored in ./tmp/batching_results_2020-06-04_16-20-14\n***************************************************************************************************************\nPHOTON 1.0.0b - www.photon-ai.com \n n_clusters k\n0 KMedoids__n_clusters 4\ntrain \n Mean STD\n2 \nARI 0.761905 0.000000e+00\nFM 0.816497 1.110223e-16\nHCV 1.000000 0.000000e+00\nMI 0.615385 0.000000e+00\ntest \n Mean STD\n2 \nARI 1.0 0.0\nFM 0.0 0.0\nHCV 1.0 0.0\nMI 1.0 0.0\n jaccard_index 0.7777777777777778\n rand_index 0.9444444444444444\n adjrand_index 0.8399999999999999\n fowlkes_mallows_index 0.8819171036881969\n fmeasure 0.875\n purity_index 0.8888888888888888\n classification_error 0.11111111111111116\n czekanowski_index 0.875\n dice_index 0.875\n sorensen_index 0.875\n rogers_tanimoto_index 0.8947368421052632\n southwood_index 3.5\n pearson_correlation 0.0038314176245210726\n corrected_chance 0.7446541086468912\n sample_expected_sim 0.0\n nmi 0.9119400080526283\n mi 1.5849625007211563\n adj_mi 0.8546410755834939\n rmi 0.4545024595933558\n vi 0.30609861135149674\n geometric_accuracy 0.9428090415820634\n overlap_quality -0.0\n onmi 0.8869847631644618\n omega_index 0.84\nElement-centric similarity: 0.8518518518518519\nCLUSTER_ALGO: KMedoids\nC_SHAPE: circle\nn_cluster: 3\nINNER_FOLDS: 3\nOUTER_FOLDS: 3\nn_points: 90\nCluster optimization range: 2 4\n***************************************************************************************************************\nPHOTON ANALYSIS: batching\n***************************************************************************************************************\n\n********************************************************\nOuter Cross validation Fold 1\n********************************************************\n---------------------------------------------------------------------------------------------------------------\nBEST_CONFIG \n---------------------------------------------------------------------------------------------------------------\n{\n \"KMedoids\": [\n \"n_clusters=3\"\n ]\n}\n+--------+-------------------+------------------+\n| METRIC | PERFORMANCE TRAIN | PERFORMANCE TEST |\n+--------+-------------------+------------------+\n| ARI | 1.0000 | 1.0000 |\n| MI | 1.0000 | 1.0000 |\n| HCV | 1.0000 | 1.0000 |\n| FM | 1.0000 | 1.0000 |\n+--------+-------------------+------------------+\n\n********************************************************\nOuter Cross validation Fold 2\n********************************************************\n---------------------------------------------------------------------------------------------------------------\nBEST_CONFIG \n---------------------------------------------------------------------------------------------------------------\n{\n \"KMedoids\": [\n \"n_clusters=2\"\n ]\n}\n+--------+-------------------+------------------+\n| METRIC | PERFORMANCE TRAIN | PERFORMANCE TEST |\n+--------+-------------------+------------------+\n| ARI | 0.6397 | 0.3800 |\n| MI | 0.5713 | 0.4221 |\n| HCV | 0.5782 | 0.4455 |\n| FM | 0.8002 | 0.7184 |\n+--------+-------------------+------------------+\n\n********************************************************\nOuter Cross validation Fold 3\n********************************************************\n---------------------------------------------------------------------------------------------------------------\nBEST_CONFIG \n---------------------------------------------------------------------------------------------------------------\n{\n \"KMedoids\": [\n \"n_clusters=3\"\n ]\n}\n+--------+-------------------+------------------+\n| METRIC | PERFORMANCE TRAIN | PERFORMANCE TEST |\n+--------+-------------------+------------------+\n| ARI | 1.0000 | 1.0000 |\n| MI | 1.0000 | 1.0000 |\n| HCV | 1.0000 | 1.0000 |\n| FM | 1.0000 | 1.0000 |\n+--------+-------------------+------------------+\n\n===============================================================================================================\nOVERALL BEST CONFIGURATION\n===============================================================================================================\n{\n \"KMedoids\": [\n \"n_clusters=3\"\n ]\n}\n"
],
[
"Cottman\n*-+\n*+-=END",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d0926a5c77a737dd5a93b9b17edad14978154fec | 41,664 | ipynb | Jupyter Notebook | notebooks/MultiProcessParamSweepLambdaMart.ipynb | skallumadi/BachPropagate | 0bb0054d182c954bad6f002189285991fa62daa2 | [
"Apache-2.0"
] | 5 | 2018-07-21T12:06:33.000Z | 2018-12-16T17:40:20.000Z | notebooks/MultiProcessParamSweepLambdaMart.ipynb | skallumadi/BachPropagate | 0bb0054d182c954bad6f002189285991fa62daa2 | [
"Apache-2.0"
] | 10 | 2020-03-24T15:52:37.000Z | 2022-03-11T23:24:36.000Z | notebooks/MultiProcessParamSweepLambdaMart.ipynb | skallumadi/BachPropagate | 0bb0054d182c954bad6f002189285991fa62daa2 | [
"Apache-2.0"
] | 3 | 2018-10-18T14:40:26.000Z | 2021-07-06T19:30:09.000Z | 62.464768 | 1,282 | 0.648377 | [
[
[
"from collections import OrderedDict\nfrom collections import namedtuple\n\nimport numpy as np\nfrom scipy import stats\n\n\n# R precision\ndef r_precision(targets, predictions, max_n_predictions=500):\n # Assumes predictions are sorted by relevance\n # First, cap the number of predictions\n predictions = predictions[:max_n_predictions]\n\n # Calculate metric\n target_set = set(targets)\n target_count = len(target_set)\n return float(len(set(predictions[:target_count]).intersection(target_set))) / target_count\n\ndef dcg(relevant_elements, retrieved_elements, k, *args, **kwargs):\n \"\"\"Compute the Discounted Cumulative Gain.\n Rewards elements being retrieved in descending order of relevance.\n \\[ DCG = rel_1 + \\sum_{i=2}^{|R|} \\frac{rel_i}{\\log_2(i + 1)} \\]\n Args:\n retrieved_elements (list): List of retrieved elements\n relevant_elements (list): List of relevant elements\n k (int): 1-based index of the maximum element in retrieved_elements\n taken in the computation\n Note: The vector `retrieved_elements` is truncated at first, THEN\n deduplication is done, keeping only the first occurence of each element.\n Returns:\n DCG value\n \"\"\"\n retrieved_elements = __get_unique(retrieved_elements[:k])\n relevant_elements = __get_unique(relevant_elements)\n if len(retrieved_elements) == 0 or len(relevant_elements) == 0:\n return 0.0\n # Computes an ordered vector of 1.0 and 0.0\n score = [float(el in relevant_elements) for el in retrieved_elements]\n # return score[0] + np.sum(score[1:] / np.log2(\n # 1 + np.arange(2, len(score) + 1)))\n return np.sum(score / np.log2(1 + np.arange(1, len(score) + 1)))\n\n\ndef ndcg(relevant_elements, retrieved_elements, k, *args, **kwargs):\n \"\"\"Compute the Normalized Discounted Cumulative Gain.\n Rewards elements being retrieved in descending order of relevance.\n The metric is determined by calculating the DCG and dividing it by the\n ideal or optimal DCG in the case that all recommended tracks are relevant.\n Note:\n The ideal DCG or IDCG is on our case equal to:\n \\[ IDCG = 1+\\sum_{i=2}^{min(\\left| G \\right|, k)}\\frac{1}{\\log_2(i +1)}\\]\n If the size of the set intersection of \\( G \\) and \\( R \\), is empty, then\n the IDCG is equal to 0. The NDCG metric is now calculated as:\n \\[ NDCG = \\frac{DCG}{IDCG + \\delta} \\]\n with \\( \\delta \\) a (very) small constant.\n The vector `retrieved_elements` is truncated at first, THEN\n deduplication is done, keeping only the first occurence of each element.\n Args:\n retrieved_elements (list): List of retrieved elements\n relevant_elements (list): List of relevant elements\n k (int): 1-based index of the maximum element in retrieved_elements\n taken in the computation\n Returns:\n NDCG value\n \"\"\"\n\n # TODO: When https://github.com/scikit-learn/scikit-learn/pull/9951 is\n # merged...\n idcg = dcg(\n relevant_elements, relevant_elements, min(k, len(relevant_elements)))\n if idcg == 0:\n raise ValueError(\"relevent_elements is empty, the metric is\"\n \"not defined\")\n true_dcg = dcg(relevant_elements, retrieved_elements, k)\n return true_dcg / idcg\n\n\ndef __get_unique(original_list):\n \"\"\"Get only unique values of a list but keep the order of the first\n occurence of each element\n \"\"\"\n return list(OrderedDict.fromkeys(original_list))\n\n\nMetrics = namedtuple('Metrics', ['r_precision', 'ndcg', 'plex_clicks'])\n\n\n# playlist extender clicks\ndef playlist_extender_clicks(targets, predictions, max_n_predictions=500):\n # Assumes predictions are sorted by relevance\n # First, cap the number of predictions\n predictions = predictions[:max_n_predictions]\n\n # Calculate metric\n i = set(predictions).intersection(set(targets))\n for index, t in enumerate(predictions):\n for track in i:\n if t == track:\n return float(int(index / 10))\n return float(max_n_predictions / 10.0 + 1)\n\n\n# def compute all metrics\ndef get_all_metrics(targets, predictions, k):\n return Metrics(r_precision(targets, predictions, k),\n ndcg(targets, predictions, k),\n playlist_extender_clicks(targets, predictions, k))\n\n\nMetricsSummary = namedtuple('MetricsSummary', ['mean_r_precision',\n 'mean_ndcg',\n 'mean_plex_clicks',\n 'coverage'])\n",
"_____no_output_____"
],
[
"#java -jar RankLib-2.10.jar -train BigRecall-TrainingFile750-2080.txt -ranker 6 -metric2t NDCG@20 -save BigRecallTrain750-2080Model-1Trees-NDCG20-tc1-lr05-leaf5.txt -tree 1 -tc 1 -shrinkage 0.5 -leaf 5\n#java -jar RankLib-2.10.jar -load BigRecallTrain750-2080Model-500Trees-NDCG20-tc1-lr05-leaf50.txt -rank BigRecallTestingFile750.txt -score BRScores750-2080Model-500Trees-NDCG20-tc1-lr05-leaf50.txt\n\n#java -jar RankLib-2.10.jar -train BigRecall-TrainingFile750-2080.txt -ranker 6 -metric2t NDCG@20 -save BigRecallTrain750-2080Model-500Trees-NDCG20-tc1-lr05-leaf50.txt -tree 500 -tc 1 -shrinkage 0.5 -leaf 50\n\n\nimport os\nTrainingFile='./Training/BigRecall-TrainingFile750-2080.txt'\nTestingFile='./Training/BigRecallTestingFile750.txt'\ntrees=[500]\ntcVals=[-1]\nshrinkages=[0.5]\nleaves= [50]\n\n\ndef createCommand(tree,tc,lr,leaf):\n opModelFile= './Training/ModelsAndScores/BigRecallTrain750-2080Model-'+str(tree)+'Trees-NDCG20-tc'+str(tc)+'-lr'+str(lr).replace('.','')+'-leaf'+str(leaf)+'.txt'\n trainCommand= 'java -jar ./Training/RankLib-2.10.jar -train ./Training/BigRecall-TrainingFile750-2080.txt -ranker 6 -metric2t NDCG@20 -silent -save '+ opModelFile+ ' -tree '+str(tree)+' -tc '+str(tc)+ ' -shrinkage '+str(lr)+ ' -leaf '+ str(leaf) +' -missingzero'\n #BRScores750-2080Model-1Trees-NDCG20-tc1-lr05-leaf5.txt\n opScoresFile='./Training/ModelsAndScores/BRScores750-2080Model-'+opModelFile.split('Model-')[1]\n testCommand= 'java -jar ./Training/RankLib-2.10.jar -load '+opModelFile+' -rank ./Training/BigRecallTestingFile750.txt -score '+opScoresFile\n return (opModelFile,trainCommand, opScoresFile, testCommand)\nparamSweep=[]\nfor tree in trees:\n for tc in tcVals:\n for lr in shrinkages:\n for leaf in leaves:\n paramSweep.append(createCommand(tree,tc,lr,leaf))",
"_____no_output_____"
],
[
"import multiprocessing as mp\nimport codecs\nimport os\nimport subprocess\n\ndef ExecuteRanklib(execTuples):\n try:\n trainCommand=execTuples[1]\n train= subprocess.check_output(trainCommand.split())\n print train\n print '----------'\n scoreCommand=execTuples[3]\n test=subprocess.check_output(scoreCommand.split())\n print test\n print '----------'\n \n except:\n print execTuples\n\n\npool = mp.Pool(processes=10)\npool.map(ExecuteRanklib, paramSweep)",
"Process PoolWorker-30:\nProcess PoolWorker-23:\nProcess PoolWorker-24:\nProcess PoolWorker-27:\nProcess PoolWorker-26:\nProcess PoolWorker-25:\nProcess PoolWorker-28:\n"
],
[
"TestFile='./Training/BigRecallTestingFile750.txt'\n\nwith open(TestFile) as f:\n test = f.readlines()\n\nPidTestTracks={}\nfor l in test:\n pid=l.split()[1].split(':')[1].strip()\n track=l.split('#')[1].strip()\n PidTestTracks.setdefault(pid,[]).append(track)\n \n###skip\nimport os\n\nMeta1Resultspath='/home/ubuntu/SpotifyChallenge/notebooks/Reranking/TestingQueryResults/Meta1/'\nMeta2Resultspath='/home/ubuntu/SpotifyChallenge/notebooks/Reranking/TestingQueryResults/Meta2/'\nQEPRFResultspath='/home/ubuntu/SpotifyChallenge/notebooks/Reranking/TestingQueryResults/QEPRF750/'\n\nMeta1Files=[Meta1Resultspath+x for x in os.listdir(Meta1Resultspath)]\nMeta2Files=[Meta2Resultspath+x for x in os.listdir(Meta2Resultspath)]\nQEPRFFiles=[QEPRFResultspath+x for x in os.listdir(QEPRFResultspath)]\n\n###skip\nimport codecs\ndef parseMetaFiles(path):\n playlistId=path.split('/')[-1].split('.op')[0]\n with codecs.open(path, 'r', encoding='utf-8') as f:\n lines = f.read().splitlines()\n rank=0\n resultSet=[]\n for result in lines[1:]:\n try:\n rank=rank+1\n splits=result.split('\\t')\n score = splits[0]\n trackid= splits[1]\n resultSet.append((rank,trackid,score))\n except:\n print result\n return \"QueryError\"\n return(playlistId,resultSet)\n\n####skip\nMeta1Op=[]\nerr1=[]\nMeta2Op=[]\nerr2=[]\nfor f in Meta1Files:\n res=parseMetaFiles(f)\n if res !=\"QueryError\":\n Meta1Op.append(res)\n else:\n err1.append(f)\nfor f in Meta2Files:\n res=parseMetaFiles(f)\n if res !=\"QueryError\":\n Meta2Op.append(res)\n else:\n err2.append(f)\n \n####skip\nimport codecs\ndef QEPRFParse(path):\n playlistId=path.split('/')[-1].split('.op')[0]\n with codecs.open(path, 'r', encoding='utf-8') as f:\n lines = f.read().splitlines()\n inputQueries=lines[0].split('# query: ')[1].split()\n resultSet=[]\n pairResults= lines[1].split(' #weight(')[2].split(') )')[0].split('\" ')\n rank=0\n for result in pairResults[:-1]:\n try:\n rank=rank+1\n splits=result.split('\"')\n score = splits[0].strip()\n trackid= splits[1].strip()\n resultSet.append((rank,trackid,score))\n except:\n print result\n return \"QueryError\"\n return(playlistId,inputQueries,resultSet)\n\n###skip\nQEPRFOp=[]\nerr3=[]\nfor f in QEPRFFiles:\n res=QEPRFParse(f)\n if res !=\"QueryError\":\n QEPRFOp.append(res)\n else:\n err3.append(f)\n \n###skip\nimport pickle\npidTrackMapping=pickle.load(open('./BiPartites/AllDataPidTrackListBipartite.pkl','rb'))\n\n####skip\nimport pickle\nimport os\nimport codecs\nfrom random import shuffle\n\npkl = os.listdir('./SplitsInformation/')\n\ncount=0\nDS={}\nfor fpkl in pkl:\n if fpkl in ['testing25RandPid.pkl', 'testing25Pid.pkl', 'testing1Pid.pkl', 'testing100Pid.pkl', 'testing10Pid.pkl', 'testing5Pid.pkl', 'testing100RandPid.pkl']:\n testType=fpkl.replace('.pkl','')\n if 'Rand' in fpkl:\n listLen=int(fpkl.split('testing')[1].split('Rand')[0])\n qtype='Rand'\n else :\n listLen=int(fpkl.split('testing')[1].split('Pid')[0])\n qtype='Normal'\n testingPids=pickle.load(open('./SplitsInformation/'+fpkl,'rb'))\n for pid in testingPids:\n pid=str(pid)\n referenceSet=[x.replace('spotify:track:','') for x in pidTrackMapping[pid]]\n DS[pid]=(testType,qtype,listLen,referenceSet)\n\n####skip\nimport pickle\nimport os\nimport codecs\nfrom random import shuffle\n\npkl = os.listdir('./SplitsInformation/')\ntestingTitleonlyPids=[]\nfor fpkl in pkl:\n if fpkl =='testingOnlyTitlePid.pkl':\n testType=fpkl.replace('.pkl','')\n listLen=0\n qtype='Normal'\n testingPids=pickle.load(open('./SplitsInformation/'+fpkl,'rb'))\n for pid in testingPids:\n pid=str(pid)\n referenceSet=[x.replace('spotify:track:','') for x in pidTrackMapping[pid]]\n DS[pid]=(testType,qtype,listLen,referenceSet)\ntestingTitleonlyPids=[str(x) for x in testingPids]\n\n",
"# EXCEPTION in query 759505: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n# EXCEPTION in query 864483: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n# EXCEPTION in query 110994: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n# EXCEPTION in query 591668: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n# EXCEPTION in query 754459: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n# EXCEPTION in query 479140: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n# EXCEPTION in query 221635: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n# EXCEPTION in query 841212: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n# EXCEPTION in query 943553: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n# EXCEPTION in query 512541: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n# EXCEPTION in query 841351: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n# EXCEPTION in query 609540: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n# EXCEPTION in query 603765: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n# EXCEPTION in query 593650: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n# EXCEPTION in query 759505: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n# EXCEPTION in query 864483: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n# EXCEPTION in query 110994: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n# EXCEPTION in query 591668: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n# EXCEPTION in query 754459: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n# EXCEPTION in query 479140: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n# EXCEPTION in query 221635: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n# EXCEPTION in query 841212: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n# EXCEPTION in query 943553: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n# EXCEPTION in query 512541: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n# EXCEPTION in query 841351: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n# EXCEPTION in query 609540: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n# EXCEPTION in query 603765: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n# EXCEPTION in query 593650: IndriRunQuery.cpp(410): QueryThread::_runQuery Exception\n"
],
[
"from collections import defaultdict\nfrom random import shuffle\n\nfor comb in paramSweep:\n scoresfile= comb[2]\n with open(scoresfile) as f:\n scores = f.readlines()\n \n PidTracksScores={}\n for l in scores:\n pid=l.split()[0].strip()\n trackScore=l.split()[2].strip()\n PidTracksScores.setdefault(pid,[]).append(float(trackScore))\n \n \n rerankedCandidates={}\n for pid,tracksList in PidTestTracks.items():\n scoresList=PidTracksScores[pid]\n zippedPairs=zip(tracksList,scoresList)\n shuffle(zippedPairs)\n rerankedCandidates[pid]=[x[0] for x in sorted(zippedPairs, key=lambda x: x[1], reverse=True)]\n \n ####continue here\n evalSets=[]\n for pl in QEPRFOp:\n plId=pl[0]\n exposed=pl[1]\n candidates=rerankedCandidates[plId]\n candidates=[x for x in candidates if x not in exposed]\n refVals= DS[plId]\n testtype=refVals[0]\n orderType=refVals[1]\n exposedLen=refVals[2]\n playlist=refVals[3]\n if orderType=='Normal':\n groundTruth=playlist[exposedLen:]\n else:\n groundTruth=[x for x in playlist if x not in exposed]\n evalSets.append((groundTruth, candidates[:500], testtype, exposedLen))\n\n for pl in Meta2Op:\n plId=pl[0]\n if plId in testingTitleonlyPids and plId in rerankedCandidates:\n exposed=[]\n candidates=rerankedCandidates[plId]\n refVals= DS[plId]\n testtype=refVals[0]\n orderType=refVals[1]\n exposedLen=refVals[2]\n playlist=refVals[3]\n groundTruth=playlist[exposedLen:]\n evalSets.append((groundTruth, candidates[:500], testtype, exposedLen))\n ####continue here\n\n '''\n r_precision(targets, predictions, k),\n ndcg(targets, predictions, k),\n playlist_extender_clicks(targets, predictions, k)\n '''\n\n indivSumsCounts= defaultdict(int)\n indivSumsRecall = defaultdict(int)\n indivSumsNdcg = defaultdict(int)\n indivSumsRprec = defaultdict(int)\n indivSumsClicks = defaultdict(int)\n globalNdcg=0\n globalRprec=0\n globalClicks=0\n globalRecall=0\n \n count=0\n for evalTuple in evalSets:\n targets=evalTuple[0]\n predictions=evalTuple[1]\n testType=evalTuple[2]\n tupNdcg=ndcg(targets,predictions,500)\n tuprprec=r_precision(targets,predictions,500)\n tupClicks=playlist_extender_clicks(targets,predictions,500)\n globalNdcg+=tupNdcg\n indivSumsNdcg[testType]+=tupNdcg\n globalRprec+=tuprprec\n indivSumsRprec[testType]+=tuprprec\n globalClicks+=tupClicks\n indivSumsClicks[testType]+=tupClicks\n\n indivSumsCounts[testType]+=1\n\n recallSetSize= len(set(predictions)&set(targets))\n refSetSize=len(targets)\n recall=recallSetSize*1.0/refSetSize\n globalRecall+=recall\n indivSumsRecall[testType]+=recall\n\n count+=1\n for k, v in indivSumsCounts.items():\n indivSumsRecall[k]=indivSumsRecall[k]/v\n indivSumsNdcg[k]=indivSumsNdcg[k]/v\n indivSumsRprec[k]=indivSumsRprec[k]/v\n indivSumsClicks[k]=indivSumsClicks[k]/v\n \n print scoresfile , 'Recall:' , globalRecall/count,'NDCG:', globalNdcg/count, 'RPrec:', globalRprec/count,'Clicks:', globalClicks/count\n ",
"./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr09-leaf5.txt Recall: 0.496151328292 NDCG: 0.301458189779 RPrec: 0.128395610746 Clicks: 4.9145316253\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr09-leaf10.txt Recall: 0.500125251481 NDCG: 0.303755406247 RPrec: 0.129254491629 Clicks: 4.83206565252\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr09-leaf50.txt Recall: 0.504065314042 NDCG: 0.305168030632 RPrec: 0.128284880919 Clicks: 4.72037630104\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr09-leaf100.txt Recall: 0.504055750427 NDCG: 0.303409031112 RPrec: 0.126503903426 Clicks: 4.77862289832\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr09-leaf250.txt Recall: 0.499622453926 NDCG: 0.293808130357 RPrec: 0.117779772983 Clicks: 4.95756605284\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr09-leaf500.txt Recall: 0.49332748546 NDCG: 0.283637203164 RPrec: 0.110198804203 Clicks: 5.10168134508\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr05-leaf5.txt Recall: 0.496164453175 NDCG: 0.301517925208 RPrec: 0.128266662223 Clicks: 4.91713370697\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr05-leaf10.txt Recall: 0.500127634339 NDCG: 0.303780567203 RPrec: 0.129224492888 Clicks: 4.83186549239\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr05-leaf50.txt Recall: 0.504065314042 NDCG: 0.30515435263 RPrec: 0.128284880919 Clicks: 4.72017614091\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr05-leaf100.txt Recall: 0.504055750427 NDCG: 0.303422657114 RPrec: 0.126503903426 Clicks: 4.77862289832\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr05-leaf250.txt Recall: 0.499622453926 NDCG: 0.293801992519 RPrec: 0.117771432978 Clicks: 4.95756605284\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr05-leaf500.txt Recall: 0.49332748546 NDCG: 0.283652023598 RPrec: 0.110198804203 Clicks: 5.10168134508\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr01-leaf5.txt Recall: 0.496165407699 NDCG: 0.301369844886 RPrec: 0.12817000256 Clicks: 4.91993594876\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr01-leaf10.txt Recall: 0.500127634339 NDCG: 0.303673536871 RPrec: 0.12900945099 Clicks: 4.83206565252\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr01-leaf50.txt Recall: 0.504065314042 NDCG: 0.305162989172 RPrec: 0.128292579385 Clicks: 4.72037630104\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr01-leaf100.txt Recall: 0.504055750427 NDCG: 0.303382816495 RPrec: 0.126503903426 Clicks: 4.77862289832\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr01-leaf250.txt Recall: 0.499622453926 NDCG: 0.293783396179 RPrec: 0.117779772983 Clicks: 4.95756605284\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr01-leaf500.txt Recall: 0.49332748546 NDCG: 0.283655799509 RPrec: 0.110198804203 Clicks: 5.10168134508\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr001-leaf5.txt Recall: 0.496152935437 NDCG: 0.301572391142 RPrec: 0.128464317926 Clicks: 4.91533226581\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr001-leaf10.txt Recall: 0.500141931491 NDCG: 0.3037733597 RPrec: 0.129199724959 Clicks: 4.83086469175\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr001-leaf50.txt Recall: 0.504065314042 NDCG: 0.305154036099 RPrec: 0.128292579385 Clicks: 4.72037630104\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr001-leaf100.txt Recall: 0.504055750427 NDCG: 0.30341355168 RPrec: 0.126503903426 Clicks: 4.77842273819\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr001-leaf250.txt Recall: 0.499622453926 NDCG: 0.293802539574 RPrec: 0.117771432978 Clicks: 4.95756605284\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr001-leaf500.txt Recall: 0.49332748546 NDCG: 0.283656416129 RPrec: 0.110198804203 Clicks: 5.10168134508\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr0001-leaf5.txt Recall: 0.496165066787 NDCG: 0.301400850531 RPrec: 0.128153690613 Clicks: 4.91653322658\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr0001-leaf10.txt Recall: 0.500120485763 NDCG: 0.30376808345 RPrec: 0.128972121528 Clicks: 4.83106485188\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr0001-leaf50.txt Recall: 0.504065314042 NDCG: 0.305138898946 RPrec: 0.128284880919 Clicks: 4.72017614091\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr0001-leaf100.txt Recall: 0.504055750427 NDCG: 0.303350668028 RPrec: 0.126503903426 Clicks: 4.77862289832\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr0001-leaf250.txt Recall: 0.499622453926 NDCG: 0.293790149638 RPrec: 0.117771432978 Clicks: 4.95756605284\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc-1-lr0001-leaf500.txt Recall: 0.49332748546 NDCG: 0.283655797676 RPrec: 0.110198804203 Clicks: 5.10168134508\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc1-lr09-leaf5.txt Recall: 0.289335874105 NDCG: 0.157248885563 RPrec: 0.0373704395215 Clicks: 10.1407125701\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc1-lr09-leaf10.txt Recall: 0.291616788317 NDCG: 0.15700164328 RPrec: 0.0371544095338 Clicks: 10.1214971978\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc1-lr09-leaf50.txt Recall: 0.290032866751 NDCG: 0.157043067529 RPrec: 0.0375357093035 Clicks: 10.1435148118\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc1-lr09-leaf100.txt Recall: 0.293505634966 NDCG: 0.158466253 RPrec: 0.0372050045938 Clicks: 9.98899119295\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc1-lr09-leaf250.txt Recall: 0.291211243576 NDCG: 0.156863875924 RPrec: 0.0368054582285 Clicks: 10.1777421938\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc1-lr09-leaf500.txt Recall: 0.290273764549 NDCG: 0.157702522635 RPrec: 0.0380846257769 Clicks: 10.2063650921\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc1-lr05-leaf5.txt Recall: 0.288963565608 NDCG: 0.156078859737 RPrec: 0.0369598227219 Clicks: 10.2135708567\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc1-lr05-leaf10.txt Recall: 0.292462374914 NDCG: 0.157396341281 RPrec: 0.0371254143317 Clicks: 10.1989591673\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc1-lr05-leaf50.txt Recall: 0.289984230325 NDCG: 0.157202004015 RPrec: 0.0372611621779 Clicks: 10.2189751801\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc1-lr05-leaf100.txt Recall: 0.290166535871 NDCG: 0.157728661532 RPrec: 0.0374146897118 Clicks: 10.2357886309\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc1-lr05-leaf250.txt Recall: 0.290899587463 NDCG: 0.157710404275 RPrec: 0.0372930430812 Clicks: 10.15632506\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc1-lr05-leaf500.txt Recall: 0.292045907732 NDCG: 0.157762590374 RPrec: 0.0373490364783 Clicks: 9.92373899119\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc1-lr01-leaf5.txt Recall: 0.289518977162 NDCG: 0.15662697437 RPrec: 0.0376159443714 Clicks: 10.1353082466\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc1-lr01-leaf10.txt Recall: 0.290570467606 NDCG: 0.156576601465 RPrec: 0.0370202337618 Clicks: 10.2415932746\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc1-lr01-leaf50.txt Recall: 0.289711990082 NDCG: 0.156976433957 RPrec: 0.0369661264375 Clicks: 10.0258206565\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc1-lr01-leaf100.txt Recall: 0.289126374553 NDCG: 0.157181360169 RPrec: 0.0373717403651 Clicks: 10.0930744596\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc1-lr01-leaf250.txt Recall: 0.291311056084 NDCG: 0.156940280153 RPrec: 0.0370540913273 Clicks: 10.0924739792\n./Training/ModelsAndScores/BRScores750-2080Model-100Trees-NDCG20-tc1-lr01-leaf500.txt Recall: 0.290188286261 NDCG: 0.157513402744 RPrec: 0.0374607698482 Clicks: 10.0040032026\n"
],
[
" Recall 0.5964542020518547,\n NDCG 0.30332032798678032,\n RPrec 0.12934009424035461,\n Clicks 5.1286",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0928c376aad98853888a51491be9369822ba32b | 371,922 | ipynb | Jupyter Notebook | Assignments/1_network_science.ipynb | carparel/NTDS | 6bb07de2888b23f4b7fd429b9adbae77629b3e95 | [
"Apache-2.0"
] | null | null | null | Assignments/1_network_science.ipynb | carparel/NTDS | 6bb07de2888b23f4b7fd429b9adbae77629b3e95 | [
"Apache-2.0"
] | null | null | null | Assignments/1_network_science.ipynb | carparel/NTDS | 6bb07de2888b23f4b7fd429b9adbae77629b3e95 | [
"Apache-2.0"
] | 2 | 2020-01-21T14:49:05.000Z | 2020-05-18T14:23:16.000Z | 179.153179 | 127,916 | 0.894647 | [
[
[
"# [NTDS'19] assignment 1: network science\n[ntds'19]: https://github.com/mdeff/ntds_2019\n\n[Eda Bayram](https://lts4.epfl.ch/bayram), [EPFL LTS4](https://lts4.epfl.ch) and\n[Nikolaos Karalias](https://people.epfl.ch/nikolaos.karalias), [EPFL LTS2](https://lts2.epfl.ch).",
"_____no_output_____"
],
[
"## Students\n\n* Team: `<5>`\n* `<Alice Bizeul, Gaia Carparelli, Antoine Spahr and Hugues Vinzant`",
"_____no_output_____"
],
[
"## Rules\n\nGrading:\n* The first deadline is for individual submissions. The second deadline is for the team submission.\n* All team members will receive the same grade based on the team solution submitted on the second deadline.\n* As a fallback, a team can ask for individual grading. In that case, solutions submitted on the first deadline are graded.\n* Collaboration between team members is encouraged. No collaboration between teams is allowed.\n\nSubmission:\n* Textual answers shall be short. Typically one to two sentences.\n* Code has to be clean.\n* You cannot import any other library than we imported.\n Note that Networkx is imported in the second section and cannot be used in the first.\n* When submitting, the notebook is executed and the results are stored. I.e., if you open the notebook again it should show numerical results and plots. We won't be able to execute your notebooks.\n* The notebook is re-executed from a blank state before submission. That is to be sure it is reproducible. You can click \"Kernel\" then \"Restart Kernel and Run All Cells\" in Jupyter.",
"_____no_output_____"
],
[
"## Objective\n\nThe purpose of this milestone is to explore a given dataset, represent it by network by constructing different graphs. In the first section, you will analyze the network properties. In the second section, you will explore various network models and find out the network model fitting the ones you construct from the dataset.",
"_____no_output_____"
],
[
"## Cora Dataset\n\nThe [Cora dataset](https://linqs.soe.ucsc.edu/node/236) consists of scientific publications classified into one of seven research fields. \n\n* **Citation graph:** the citation network can be constructed from the connections given in the `cora.cites` file.\n* **Feature graph:** each publication in the dataset is described by a 0/1-valued word vector indicating the absence/presence of the corresponding word from the dictionary and its research field, given in the `cora.content` file. The dictionary consists of 1433 unique words. A feature graph can be constructed using the Euclidean distance between the feature vector of the publications.\n\nThe [`README`](data/cora/README) provides details about the content of [`cora.cites`](data/cora/cora.cites) and [`cora.content`](data/cora/cora.content).",
"_____no_output_____"
],
[
"## Section 1: Network Properties",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nfrom matplotlib import pyplot as plt\n\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"### Question 1: Construct a Citation Graph and a Feature Graph",
"_____no_output_____"
],
[
"Read the `cora.content` file into a Pandas DataFrame by setting a header for the column names. Check the `README` file.",
"_____no_output_____"
]
],
[
[
"column_list = ['paper_id'] + [str(i) for i in range(1,1434)] + ['class_label']\npd_content = pd.read_csv('data/cora/cora.content', delimiter='\\t', names=column_list) \npd_content.head()",
"_____no_output_____"
]
],
[
[
"Print out the number of papers contained in each of the reasearch fields.\n\n**Hint:** You can use the `value_counts()` function.",
"_____no_output_____"
]
],
[
[
"pd_content['class_label'].value_counts()",
"_____no_output_____"
]
],
[
[
"Select all papers from a field of your choice and store their feature vectors into a NumPy array.\nCheck its shape.",
"_____no_output_____"
]
],
[
[
"my_field = 'Neural_Networks'\nfeatures = pd_content[pd_content['class_label'] == my_field].drop(columns=['paper_id','class_label']).to_numpy()\nfeatures.shape",
"_____no_output_____"
]
],
[
[
"Let $D$ be the Euclidean distance matrix whose $(i,j)$ entry corresponds to the Euclidean distance between feature vectors $i$ and $j$.\nUsing the feature vectors of the papers from the field which you have selected, construct $D$ as a Numpy array. ",
"_____no_output_____"
]
],
[
[
"distance = np.zeros([features.shape[0],features.shape[0]])\n\nfor i in range(features.shape[0]):\n distance[i] = np.sqrt(np.sum((features[i,:] - features)**2, axis=1))",
"_____no_output_____"
],
[
"distance.shape",
"_____no_output_____"
]
],
[
[
"Check the mean pairwise distance $\\mathbb{E}[D]$.",
"_____no_output_____"
]
],
[
[
"# Mean on the upper triangle as the matrix is symetric (we also excluded the diagonal)\nmean_distance = distance[np.triu_indices(distance.shape[1],1)].mean()\nprint('Mean euclidian distance between feature vectors of papers on Neural Networks: {}'.format(mean_distance))",
"Mean euclidian distance between feature vectors of papers on Neural Networks: 5.696602496555962\n"
]
],
[
[
"Plot an histogram of the euclidean distances.",
"_____no_output_____"
]
],
[
[
"fig,ax = plt.subplots(1,1,figsize=(8, 8))\nax.hist(distance.flatten(), density=True, bins=20, color='salmon', edgecolor='black', linewidth=1);\nax.set_title(\"Histogram of Euclidean distances between Neural-networks papers\")\nax.set_xlabel(\"Euclidian Distances\")\nax.set_ylabel(\"Frequency\")\nax.grid(True, which='major', axis='y')\nax.set_axisbelow(True)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Now create an adjacency matrix for the papers by thresholding the Euclidean distance matrix.\nThe resulting (unweighted) adjacency matrix should have entries\n$$ A_{ij} = \\begin{cases} 1, \\; \\text{if} \\; d(i,j)< \\mathbb{E}[D], \\; i \\neq j, \\\\ 0, \\; \\text{otherwise.} \\end{cases} $$\n\nFirst, let us choose the mean distance as the threshold.",
"_____no_output_____"
]
],
[
[
"threshold = mean_distance\nA_feature = np.where(distance < threshold, 1, 0)\nnp.fill_diagonal(A_feature,0)",
"_____no_output_____"
]
],
[
[
"Now read the `cora.cites` file and construct the citation graph by converting the given citation connections into an adjacency matrix.",
"_____no_output_____"
]
],
[
[
"cora_cites = np.genfromtxt('data/cora/cora.cites', delimiter='\\t')",
"_____no_output_____"
],
[
"papers = np.unique(cora_cites)\nA_citation = np.zeros([papers.size, papers.size])\nfor i in range(cora_cites.shape[0]):\n A_citation[np.where(papers==cora_cites[i,1]),np.where(papers==cora_cites[i,0])] = 1\nA_citation.shape",
"_____no_output_____"
]
],
[
[
"Get the adjacency matrix of the citation graph for the field that you chose.\nYou have to appropriately reduce the adjacency matrix of the citation graph.",
"_____no_output_____"
]
],
[
[
"# get the paper id from the chosen field\nfield_id = pd_content[pd_content['class_label'] == my_field][\"paper_id\"].unique()\n\n# get the index of those paper in the A_citation matrix (similar to index on the vector 'papers')\nfield_citation_id = np.empty(field_id.shape[0]).astype(int)\nfor i in range(field_id.shape[0]):\n field_citation_id[i] = np.where(papers == field_id[i])[0]\n\n# get the A_citation matrix only at the index of the paper in the field \nA_citation = A_citation[field_citation_id][:,field_citation_id] \nA_citation.shape",
"_____no_output_____"
]
],
[
[
"Check if your adjacency matrix is symmetric. Symmetrize your final adjacency matrix if it's not already symmetric.",
"_____no_output_____"
]
],
[
[
"# a matrix is symetric if it's the same as its transpose\nprint('The citation adjency matrix for papers on Neural Networks is symmetric: {}'.format(np.all(A_citation == A_citation.transpose())))\n# symetrize it by taking the maximum between A and A.transposed\nA_citation = np.maximum(A_citation, A_citation.transpose())\n\n# To verify if the matrix is symetric\nprint('After modifiying the matrix, it is symmetric: {}'.format(np.count_nonzero(A_citation - A_citation.transpose())==0))",
"The citation adjency matrix for papers on Neural Networks is symmetric: False\nAfter modifiying the matrix, it is symmetric: True\n"
]
],
[
[
"Check the shape of your adjacency matrix again.",
"_____no_output_____"
]
],
[
[
"A_citation.shape",
"_____no_output_____"
]
],
[
[
"### Question 2: Degree Distribution and Moments",
"_____no_output_____"
],
[
"What is the total number of edges in each graph?",
"_____no_output_____"
]
],
[
[
"num_edges_feature = int(np.sum(A_feature)/2) # only half of the matrix\nnum_edges_citation = int(np.sum(A_citation)/2)\nprint(f\"Number of edges in the feature graph: {num_edges_feature}\")\nprint(f\"Number of edges in the citation graph: {num_edges_citation}\")",
"Number of edges in the feature graph: 136771\nNumber of edges in the citation graph: 1175\n"
]
],
[
[
"Plot the degree distribution histogram for each of the graphs.",
"_____no_output_____"
]
],
[
[
"degrees_citation = A_citation.sum(axis=1) # degree = nbr of connections --> sum of ones over columns (axis=1)\ndegrees_feature = A_feature.sum(axis=1) \n\ndeg_hist_normalization = np.ones(degrees_citation.shape[0]) / degrees_citation.shape[0]\n\nfig, axes = plt.subplots(1, 2, figsize=(16, 8))\naxes[0].set_title('Citation graph degree distribution')\naxes[0].hist(degrees_citation, weights=deg_hist_normalization, bins=20, color='salmon', edgecolor='black', linewidth=1);\naxes[1].set_title('Feature graph degree distribution')\naxes[1].hist(degrees_feature, weights=deg_hist_normalization, bins=20, color='salmon', edgecolor='black', linewidth=1);",
"_____no_output_____"
]
],
[
[
"Calculate the first and second moments of the degree distribution of each graph.",
"_____no_output_____"
]
],
[
[
"cit_moment_1 = np.mean(degrees_citation)\ncit_moment_2 = np.var(degrees_citation)\n\nfeat_moment_1 = np.mean(degrees_feature)\nfeat_moment_2 = np.var(degrees_feature)\n\nprint(f\"1st moment of citation graph: {cit_moment_1:.3f}\")\nprint(f\"2nd moment of citation graph: {cit_moment_2:.3f}\")\nprint(f\"1st moment of feature graph: {feat_moment_1:.3f}\")\nprint(f\"2nd moment of feature graph: {feat_moment_2:.3f}\")",
"1st moment of citation graph: 2.873\n2nd moment of citation graph: 15.512\n1st moment of feature graph: 334.403\n2nd moment of feature graph: 55375.549\n"
]
],
[
[
"What information do the moments provide you about the graphs?\nExplain the differences in moments between graphs by comparing their degree distributions.",
"_____no_output_____"
],
[
"### Answer :\n**<br>The moments provide an idea of the sparsity of the graphs and the way the data is distributed using numerical values. The first moment is associated with the average value, the second to the variance of the distribution. A large 1st moment would mean a large number of edges per node on average, whereas the 2nd moment give information about the spread of the node's degree around the average value (variance).\n<br> Citation degree distribution 1st moment lays around 2.8, and the second one is higher (around 15.5) with a large number of nodes (818). It thus means that there are many nodes with a small degree but there are also larger hubs, the nework is likely to be sparse. The feature degree distribution moments are larger, meaning a rather dense graph. There are many nodes with a degree of above 800 (15%), and since the network contains 818 nodes, it means that many nodes are almost saturated. The high variance shows that the degree distribution is more diffuse around the average value than for the citation graph.**",
"_____no_output_____"
],
[
"Select the 20 largest hubs for each of the graphs and remove them. Observe the sparsity pattern of the adjacency matrices of the citation and feature graphs before and after such a reduction.",
"_____no_output_____"
]
],
[
[
"smallest_feat_hub_idx = np.argpartition(degrees_feature, degrees_feature.shape[0]-20)[:-20]\nsmallest_feat_hub_idx.sort()\nreduced_A_feature = A_feature[smallest_feat_hub_idx][:,smallest_feat_hub_idx]\n\nsmallest_cit_hub_idx = np.argpartition(degrees_citation, degrees_citation.shape[0]-20)[:-20]\nsmallest_cit_hub_idx.sort()\nreduced_A_citation = A_citation[smallest_cit_hub_idx][:,smallest_cit_hub_idx]\n\nfig, axes = plt.subplots(2, 2, figsize=(16, 16))\naxes[0, 0].set_title('Feature graph: adjacency matrix sparsity pattern')\naxes[0, 0].spy(A_feature);\naxes[0, 1].set_title('Feature graph without top 20 hubs: adjacency matrix sparsity pattern')\naxes[0, 1].spy(reduced_A_feature);\naxes[1, 0].set_title('Citation graph: adjacency matrix sparsity pattern')\naxes[1, 0].spy(A_citation);\naxes[1, 1].set_title('Citation graph without top 20 hubs: adjacency matrix sparsity pattern')\naxes[1, 1].spy(reduced_A_citation);",
"_____no_output_____"
]
],
[
[
"Plot the new degree distribution histograms.",
"_____no_output_____"
]
],
[
[
"reduced_degrees_feat = reduced_A_feature.sum(axis=1)\nreduced_degrees_cit = reduced_A_citation.sum(axis=1)\n\ndeg_hist_normalization = np.ones(reduced_degrees_feat.shape[0])/reduced_degrees_feat.shape[0]\nfig, axes = plt.subplots(1, 2, figsize=(16, 8))\naxes[0].set_title('Citation graph degree distribution')\naxes[0].hist(reduced_degrees_cit, weights=deg_hist_normalization, bins=8, color='salmon', edgecolor='black', linewidth=1);\naxes[1].set_title('Feature graph degree distribution')\naxes[1].hist(reduced_degrees_feat, weights=deg_hist_normalization, bins=20, color='salmon', edgecolor='black', linewidth=1);",
"_____no_output_____"
]
],
[
[
"Compute the first and second moments for the new graphs.",
"_____no_output_____"
]
],
[
[
"reduced_cit_moment_1 = np.mean(reduced_degrees_cit)\nreduced_cit_moment_2 = np.var(reduced_degrees_cit)\n\nreduced_feat_moment_1 = np.mean(reduced_degrees_feat)\nreduced_feat_moment_2 = np.var(reduced_degrees_feat)\n\nprint(f\"Citation graph first moment: {reduced_cit_moment_1:.3f}\")\nprint(f\"Citation graph second moment: {reduced_cit_moment_2:.3f}\")\nprint(f\"Feature graph first moment: {reduced_feat_moment_1:.3f}\")\nprint(f\"Feature graph second moment: {reduced_feat_moment_2:.3f}\")",
"Citation graph first moment: 1.972\nCitation graph second moment: 2.380\nFeature graph first moment: 302.308\nFeature graph second moment: 50780.035\n"
]
],
[
[
"Print the number of edges in the reduced graphs.",
"_____no_output_____"
]
],
[
[
"num_edges_reduced_feature = int(np.sum(reduced_A_feature)/2)\nnum_edges_reduced_citation = int(np.sum(reduced_A_citation)/2)\nprint(f\"Number of edges in the reduced feature graph: {num_edges_reduced_feature}\")\nprint(f\"Number of edges in the reduced citation graph: {num_edges_reduced_citation}\")",
"Number of edges in the reduced feature graph: 120621\nNumber of edges in the reduced citation graph: 787\n"
]
],
[
[
"Is the effect of removing the hubs the same for both networks? Look at the percentage changes for each moment. Which of the moments is affected the most and in which graph? Explain why. \n\n**Hint:** Examine the degree distributions.",
"_____no_output_____"
]
],
[
[
"change_cit_moment_1 = (reduced_cit_moment_1-cit_moment_1)/cit_moment_1\nchange_cit_moment_2 = (reduced_cit_moment_2-cit_moment_2)/cit_moment_2\nchange_feat_moment_1 = (reduced_feat_moment_1-feat_moment_1)/feat_moment_1\nchange_feat_moment_2 = (reduced_feat_moment_2-feat_moment_2)/feat_moment_2\n\nprint(f\"% Percentage of change for citation 1st moment: {change_cit_moment_1*100:.3f}\")\nprint(f\"% Percentage of change for citation 2nd moment: {change_cit_moment_2*100:.3f}\")\nprint(f\"% Percentage of change for feature 1st moment: {change_feat_moment_1*100:.3f}\")\nprint(f\"% Percentage of change for feature 2nd moment: {change_feat_moment_2*100:.3f}\")",
"% Percentage of change for citation 1st moment: -31.343\n% Percentage of change for citation 2nd moment: -84.656\n% Percentage of change for feature 1st moment: -9.598\n% Percentage of change for feature 2nd moment: -8.299\n"
]
],
[
[
"### Answer : \n**After looking of the percentage of change of moments, we can notice that the removal of the 20 largest hubs affects way more the citation degree distribution than the feature degree distribution. The 2nd moment of the citation degree distribution is reduced by almost 85%, this can be due to the fact that the percentage of nodes with a high degree was lower for the citation than for the feature network and they were thus all removed as part of the 20 largest hubs, resulting in a much lower variance (less spread distribution).**\n\n**In conclusion, the new citation distribution is more condensed around its mean value, the degree landscape is hence more uniform. Regarding the feature degree distribution, a consistent number of nodes remain hotspots.**",
"_____no_output_____"
],
[
"### Question 3: Pruning, sparsity, paths",
"_____no_output_____"
],
[
"By adjusting the threshold of the euclidean distance matrix, prune the feature graph so that its number of edges is roughly close (within a hundred edges) to the number of edges in the citation graph.",
"_____no_output_____"
]
],
[
[
"threshold = np.max(distance)\ndiagonal = distance.shape[0]\nthreshold_flag = False\nepsilon = 0.01*threshold \ntolerance = 250\n\nwhile threshold > 0 and not threshold_flag:\n threshold -= epsilon # steps of 1% of maximum\n n_edge = int((np.count_nonzero(np.where(distance < threshold, 1, 0)) - diagonal)/2)\n \n # within a hundred edges\n if abs(num_edges_citation - n_edge) < tolerance:\n threshold_flag = True\n print(f'Found a threshold : {threshold:.3f}')\n\nA_feature_pruned = np.where(distance < threshold, 1, 0)\nnp.fill_diagonal(A_feature_pruned, 0)\nnum_edges_feature_pruned = int(np.count_nonzero(A_feature_pruned)/2)\n\nprint(f\"Number of edges in the feature graph: {num_edges_feature}\")\nprint(f\"Number of edges in the feature graph after pruning: {num_edges_feature_pruned}\")\nprint(f\"Number of edges in the citation graph: {num_edges_citation}\")",
"Found a threshold : 2.957\nNumber of edges in the feature graph: 136771\nNumber of edges in the feature graph after pruning: 1386\nNumber of edges in the citation graph: 1175\n"
]
],
[
[
"### Remark:\n**The distribution of distances (which is a distribution of integers) for this particular field (Neural Networks) doesn't allow a configuration where the number of edges is roughly close (whithin a hundred of edges) to the citation distribution. This is independant of the chosen epsilon . The closest match is 250 edges apart.**",
"_____no_output_____"
],
[
"Check your results by comparing the sparsity patterns and total number of edges between the graphs.",
"_____no_output_____"
]
],
[
[
"fig, axes = plt.subplots(1, 2, figsize=(12, 6))\naxes[0].set_title('Citation graph sparsity')\naxes[0].spy(A_citation);\naxes[1].set_title('Feature graph sparsity')\naxes[1].spy(A_feature_pruned);",
"_____no_output_____"
]
],
[
[
"Let $C_{k}(i,j)$ denote the number of paths of length $k$ from node $i$ to node $j$. \n\nWe define the path matrix $P$, with entries\n$ P_{ij} = \\displaystyle\\sum_{k=0}^{N}C_{k}(i,j). $",
"_____no_output_____"
],
[
"Calculate the path matrices for both the citation and the unpruned feature graphs for $N =10$. \n\n**Hint:** Use [powers of the adjacency matrix](https://en.wikipedia.org/wiki/Adjacency_matrix#Matrix_powers).",
"_____no_output_____"
]
],
[
[
"def path_matrix(A, N=10):\n \"\"\"Compute the path matrix for matrix A for N power \"\"\"\n power_A = [A]\n for i in range(N-1):\n power_A.append(np.matmul(power_A[-1], A))\n\n return np.stack(power_A, axis=2).sum(axis=2)",
"_____no_output_____"
],
[
"path_matrix_citation = path_matrix(A_citation)\npath_matrix_feature = path_matrix(A_feature)",
"_____no_output_____"
]
],
[
[
"Check the sparsity pattern for both of path matrices.",
"_____no_output_____"
]
],
[
[
"fig, axes = plt.subplots(1, 2, figsize=(16, 9))\naxes[0].set_title('Citation Path matrix sparsity')\naxes[0].spy(path_matrix_citation);\naxes[1].set_title('Feature Path matrix sparsity')\naxes[1].spy(path_matrix_feature, vmin=0, vmax=1); #scaling the color bar",
"_____no_output_____"
]
],
[
[
"Now calculate the path matrix of the pruned feature graph for $N=10$. Plot the corresponding sparsity pattern. Is there any difference?",
"_____no_output_____"
]
],
[
[
"path_matrix_pruned = path_matrix(A_feature_pruned)\n\nplt.figure(figsize=(12, 6))\nplt.title('Feature Path matrix sparsity')\nplt.spy(path_matrix_pruned);",
"_____no_output_____"
]
],
[
[
"### Your answer here:\n<br> **Many combinations of nodes have a path matrix value of zero now, meaning that they are not within the reach of N = 10 nodes from another node. This makes sense as many edges were removed in the pruning procedure (from 136000 to 1400). Hence, the number of possible paths from i to j was reduced, reducing at the same time the amount of paths of size N. The increase of the sparsity of the adjency matrix increases the diameter of a network.**",
"_____no_output_____"
],
[
"Describe how you can use the above process of counting paths to determine whether a graph is connected or not. Is the original (unpruned) feature graph connected?",
"_____no_output_____"
],
[
"### Answer:\n<br> **The graph is connected if all points are within the reach of others. In others words, if when increasing $N$, we are able to reach a point where the path matrix doesn't contain any null value, it means that the graph is connected. Therefore, even if the path matrix has some null values it can be connected, this depends on the chosen N-value.\n<br> For example, if 20 nodes are aligned and linked then we know that all point are reachable. Even though, the number of paths of length 10 between the first and the last node remain 0.**",
"_____no_output_____"
],
[
"If the graph is connected, how can you guess its diameter using the path matrix?",
"_____no_output_____"
],
[
"### Answer :\n<br> **The diameter coresponds to the minimum $N$ ($N$ being a non negative integer) for which the path matrix does not contain any null value.**",
"_____no_output_____"
],
[
"If any of your graphs is connected, calculate the diameter using that process.",
"_____no_output_____"
]
],
[
[
"N=0\ndiameter = None\nd_found = False \nwhile not d_found:\n N += 1\n P = path_matrix(A_feature, N)\n if np.count_nonzero(P == 0) == 0: # if there are no zero in P\n d_found = True\n diameter = N\n\nprint(f\"The diameter of the feature graph (which is connected) is: {diameter}\")",
"The diameter of the feature graph (which is connected) is: 2\n"
]
],
[
[
"Check if your guess was correct using [NetworkX](https://networkx.github.io/documentation/stable/reference/algorithms/generated/networkx.algorithms.distance_measures.diameter.html).\nNote: usage of NetworkX is only allowed in this part of Section 1.",
"_____no_output_____"
]
],
[
[
"import networkx as nx\nfeature_graph = nx.from_numpy_matrix(A_feature)\nprint(f\"Diameter of feature graph according to networkx: {nx.diameter(feature_graph)}\")",
"Diameter of feature graph according to networkx: 2\n"
]
],
[
[
"## Section 2: Network Models",
"_____no_output_____"
],
[
"In this section, you will analyze the feature and citation graphs you constructed in the previous section in terms of the network model types.\nFor this purpose, you can use the NetworkX libary imported below.",
"_____no_output_____"
]
],
[
[
"import networkx as nx",
"_____no_output_____"
]
],
[
[
"Let us create NetworkX graph objects from the adjacency matrices computed in the previous section.",
"_____no_output_____"
]
],
[
[
"G_citation = nx.from_numpy_matrix(A_citation)\nprint('Number of nodes: {}, Number of edges: {}'. format(G_citation.number_of_nodes(), G_citation.number_of_edges()))\nprint('Number of self-loops: {}, Number of connected components: {}'. format(G_citation.number_of_selfloops(), nx.number_connected_components(G_citation)))",
"Number of nodes: 818, Number of edges: 1175\nNumber of self-loops: 0, Number of connected components: 104\n"
]
],
[
[
"In the rest of this assignment, we will consider the pruned feature graph as the feature network.",
"_____no_output_____"
]
],
[
[
"G_feature = nx.from_numpy_matrix(A_feature_pruned)\nprint('Number of nodes: {}, Number of edges: {}'. format(G_feature.number_of_nodes(), G_feature.number_of_edges()))\nprint('Number of self-loops: {}, Number of connected components: {}'. format(G_feature.number_of_selfloops(), nx.number_connected_components(G_feature)))",
"Number of nodes: 818, Number of edges: 1386\nNumber of self-loops: 0, Number of connected components: 684\n"
]
],
[
[
"### Question 4: Simulation with Erdős–Rényi and Barabási–Albert models",
"_____no_output_____"
],
[
"Create an Erdős–Rényi and a Barabási–Albert graph using NetworkX to simulate the citation graph and the feature graph you have. When choosing parameters for the networks, take into account the number of vertices and edges of the original networks.",
"_____no_output_____"
],
[
"The number of nodes should exactly match the number of nodes in the original citation and feature graphs.",
"_____no_output_____"
]
],
[
[
"assert len(G_citation.nodes()) == len(G_feature.nodes())\nn = len(G_citation.nodes())\nprint('The number of nodes ({}) matches the original number of nodes: {}'.format(n,n==A_citation.shape[0]))",
"The number of nodes (818) matches the original number of nodes: True\n"
]
],
[
[
"The number of match shall fit the average of the number of edges in the citation and the feature graph.",
"_____no_output_____"
]
],
[
[
"m = np.round((G_citation.size() + G_feature.size()) / 2)\nprint('The number of match ({}) fits the average number of edges: {}'.format(m,m==np.round(np.mean([num_edges_citation,num_edges_feature_pruned]))))",
"The number of match (1280.0) fits the average number of edges: True\n"
]
],
[
[
"How do you determine the probability parameter for the Erdős–Rényi graph?",
"_____no_output_____"
],
[
"### Answer:\n**<br>Based on the principles ruling random networks (no preferential attachment but a random attachment), the expected number of edges is given by : $\\langle L \\rangle = p\\frac{N(N-1)}{2}$ , where $\\langle L \\rangle$ is the average number of edges, $N$, the number of nodes and $p$, the probability parameter.\n<br> Therefore we can get $p$ from the number of edges we want and the number of nodes we have : $ p = \\langle L \\rangle\\frac{2}{N(N-1)}$ \n<br> The number of expected edges is given by $m$ in our case (defined as being the average of edges between the two original networks). $N$ is the same as in the original graphs**",
"_____no_output_____"
]
],
[
[
"p = m*2/(n*(n-1))\nG_er = nx.erdos_renyi_graph(n, p)",
"_____no_output_____"
]
],
[
[
"Check the number of edges in the Erdős–Rényi graph.",
"_____no_output_____"
]
],
[
[
"print('My Erdos-Rényi network that simulates the citation graph has {} edges.'.format(G_er.size()))",
"My Erdos-Rényi network that simulates the citation graph has 1238 edges.\n"
]
],
[
[
"How do you determine the preferential attachment parameter for Barabási–Albert graphs?",
"_____no_output_____"
],
[
"### Answer :\n<br>**The Barabasi-Albert model uses growth and preferential attachement to build a scale-free network. The network is constructed by progressivly adding nodes to the network and adding a fixed number of edges, q, to each node added. Those edges are preferentially drawn towards already existing nodes with a high degree (preferential attachment). \n<br> By the end of the process, the network contains n nodes and hence $n * q$ edges. Knowing that the final number of edges is defined by $m$, the parameter $q = m/n$**",
"_____no_output_____"
]
],
[
[
"q = int(m/n)\nG_ba = nx.barabasi_albert_graph(n, q)",
"_____no_output_____"
]
],
[
[
"Check the number of edges in the Barabási–Albert graph.",
"_____no_output_____"
]
],
[
[
"print('My Barabási-Albert network that simulates the citation graph has {} edges.'.format(G_ba.size()))",
"My Barabási-Albert network that simulates the citation graph has 817 edges.\n"
]
],
[
[
"### Question 5: Giant Component",
"_____no_output_____"
],
[
"Check the size of the largest connected component in the citation and feature graphs.",
"_____no_output_____"
]
],
[
[
"giant_citation = max(nx.connected_component_subgraphs(G_citation), key=len)\nprint('The giant component of the citation graph has {} nodes and {} edges.'.format(giant_citation.number_of_nodes(), giant_citation.size()))",
"The giant component of the citation graph has 636 nodes and 1079 edges.\n"
],
[
"giant_feature = max(nx.connected_component_subgraphs(G_feature), key=len)\nprint('The giant component of the feature graph has {} nodes and {} edges.'.format(giant_feature.number_of_nodes(), giant_feature.size()))",
"The giant component of the feature graph has 117 nodes and 1364 edges.\n"
]
],
[
[
"Check the size of the giant components in the generated Erdős–Rényi graph.",
"_____no_output_____"
]
],
[
[
"giant_er = max(nx.connected_component_subgraphs(G_er), key=len)\nprint('The giant component of the Erdos-Rényi network has {} nodes and {} edges.'.format(giant_er.number_of_nodes(), giant_er.size()))",
"The giant component of the Erdos-Rényi network has 771 nodes and 1234 edges.\n"
]
],
[
[
"Let us match the number of nodes in the giant component of the feature graph by simulating a new Erdős–Rényi network.\nHow do you choose the probability parameter this time? \n\n**Hint:** Recall the expected giant component size from the lectures.",
"_____no_output_____"
],
[
"### Answer :\n**<br> We can see the average degree of a network/each node as being the probability p multiplied by the amount of nodes to which it can connect ($N-1$, because the network is not recursive, N the number of nodes,): $\\langle k \\rangle = p . (N-1)$\n<br>We can establish that, $S$, the portion of nodes in the Giant Component is given by $S = \\frac{N_{GC}}{N}$ ($N_{GC}$, the number of nodes in the giant component) and $u$, the probability that $i$ is not linked to the GC via any other node $j$. U is also the portion of nodes not in the GC : $u = 1 - S$.\n<br>Knowing that for one $j$ among the $N-1$ nodes, this probability can be seen as the probability to have no link with $j$ if $j$ is in the GC or having a link with $j$ if $j$ is not being in the GC ($p . u$), we can establish that: $u = (1 - p - p.u)^{N-1}$\n<br> Using the relationship mentionned above : $p = \\frac{<k>}{(N-1)}$, applying a log on both sides of the relationship and taking the Taylor expansion : \n<br>$S = 1-e^{-\\langle k \\rangle S}$\n<br> => $e^{-\\langle k \\rangle S} =1-S$\n<br> => $-\\langle k \\rangle S = ln(1-S)$\n<br> => $\\langle k \\rangle = -\\frac{1}{S}ln(1-S)$\n<br> This expression of the average degree is then used to define $p$ : $p = \\frac{\\langle k \\rangle}{N-1} = \\frac{-\\frac{1}{S}ln(1-S)}{N-1}$**",
"_____no_output_____"
]
],
[
[
"GC_node = giant_feature.number_of_nodes()\nS = GC_node/n\navg_k = -1/S*np.log(1-S)",
"_____no_output_____"
],
[
"p_new = avg_k/(n-1)\nG_er_new = nx.erdos_renyi_graph(n, p_new)",
"_____no_output_____"
]
],
[
[
"Check the size of the new Erdős–Rényi network and its giant component.",
"_____no_output_____"
]
],
[
[
"print('My new Erdos Renyi network that simulates the citation graph has {} edges.'.format(G_er_new.size()))\ngiant_er_new = max(nx.connected_component_subgraphs(G_er_new), key=len)\nprint('The giant component of the new Erdos-Rényi network has {} nodes and {} edges.'.format(giant_er_new.number_of_nodes(), giant_er_new.size()))",
"My new Erdos Renyi network that simulates the citation graph has 437 edges.\nThe giant component of the new Erdos-Rényi network has 208 nodes and 210 edges.\n"
]
],
[
[
"### Question 6: Degree Distributions",
"_____no_output_____"
],
[
"Recall the degree distribution of the citation and the feature graph.",
"_____no_output_____"
]
],
[
[
"fig, axes = plt.subplots(1, 2, figsize=(15, 6),sharex = True)\naxes[0].set_title('Citation graph')\ncitation_degrees = [deg for (node, deg) in G_citation.degree()]\naxes[0].hist(citation_degrees, bins=20, color='salmon', edgecolor='black', linewidth=1);\naxes[1].set_title('Feature graph')\nfeature_degrees = [deg for (node, deg) in G_feature.degree()]\naxes[1].hist(feature_degrees, bins=20, color='salmon', edgecolor='black', linewidth=1);",
"_____no_output_____"
]
],
[
[
"What does the degree distribution tell us about a network? Can you make a prediction on the network model type of the citation and the feature graph by looking at their degree distributions?",
"_____no_output_____"
],
[
"### Answer :\n\n<br> **The degree distribution tell us about the sparsity of a network.\nBoth show a power law degree distribution (many nodes with few edges but a couple of big components with a lot of edges). Hence they should fall in the scale-free network category which have a similar degree distribution. Therefore the Barabasi-Albert model which is a random scale free model is probably the best match.\n<br> Those distributions are indeed power laws as it could be seen by the linear behavior of the distribution using a log scale.**",
"_____no_output_____"
],
[
"Now, plot the degree distribution historgrams for the simulated networks.",
"_____no_output_____"
]
],
[
[
"fig, axes = plt.subplots(1, 3, figsize=(20, 8))\naxes[0].set_title('Erdos-Rényi network')\ner_degrees = [deg for (node, deg) in G_er.degree()]\naxes[0].hist(er_degrees, bins=10, color='salmon', edgecolor='black', linewidth=1)\naxes[1].set_title('Barabási-Albert network')\nba_degrees = [deg for (node, deg) in G_ba.degree()]\naxes[1].hist(ba_degrees, bins=10, color='salmon', edgecolor='black', linewidth=1)\naxes[2].set_title('new Erdos-Rényi network')\ner_new_degrees = [deg for (node, deg) in G_er_new.degree()]\naxes[2].hist(er_new_degrees, bins=6, color='salmon', edgecolor='black', linewidth=1)\nplt.show()",
"_____no_output_____"
]
],
[
[
"In terms of the degree distribution, is there a good match between the citation and feature graphs and the simulated networks?\nFor the citation graph, choose one of the simulated networks above that match its degree distribution best. Indicate your preference below.",
"_____no_output_____"
],
[
"### Answer :\n<br> **Regarding the feature network, none of the distributions above matche the range of degrees of the feature network. Also none of the above distributions, model the large portion of hotspots seen in the feature graph. <br>Regarding the citation network, the Barabasi-Albert network seem to be a good match. Indeed, the range of values as well as the power-law shape of the model is close to the distribution of the citation graph showed earlier. Hence, a scale free model seems to be the best match to model the citation network for the Neural Networks field.**",
"_____no_output_____"
],
[
"You can also simulate a network using the configuration model to match its degree disctribution exactly. Refer to [Configuration model](https://networkx.github.io/documentation/stable/reference/generated/networkx.generators.degree_seq.configuration_model.html#networkx.generators.degree_seq.configuration_model).\n\nLet us create another network to match the degree distribution of the feature graph. ",
"_____no_output_____"
]
],
[
[
"G_config = nx.configuration_model(feature_degrees) \nprint('Configuration model has {} nodes and {} edges.'.format(G_config.number_of_nodes(), G_config.size()))",
"Configuration model has 818 nodes and 1386 edges.\n"
]
],
[
[
"Does it mean that we create the same graph with the feature graph by the configuration model? If not, how do you understand that they are not the same?",
"_____no_output_____"
],
[
"### Answer :\n<br> **No we don't create the same graph, the number of edges, nodes and degree distribution can be the same but the links can be different. For example, in a group of three papers, various configurations are possible using only 2 links.\n<br> Also the function used to create this model considers self loops and paralell edges which is not the case for the real feature graph. Hence the network resulting from this modelisation will most probably not be identical to the original graph.**",
"_____no_output_____"
],
[
"### Question 7: Clustering Coefficient",
"_____no_output_____"
],
[
"Let us check the average clustering coefficient of the original citation and feature graphs. ",
"_____no_output_____"
]
],
[
[
"nx.average_clustering(G_citation)",
"_____no_output_____"
],
[
"nx.average_clustering(G_feature)",
"_____no_output_____"
]
],
[
[
"What does the clustering coefficient tell us about a network? Comment on the values you obtain for the citation and feature graph.",
"_____no_output_____"
],
[
"### Answer :\n**<br>Clustering coefficient is linked to the presence of subgroups (or clusters) in the network. A high clustering coefficient means that a node is very likely to be part of a subgroup. Here we can observe that the clustering coefficient of the citation graph is higher (almost double) than the one of the feature graph, this can highlight the fact that citations are more likely to form subgroups than feature.**\n",
"_____no_output_____"
],
[
"Now, let us check the average clustering coefficient for the simulated networks.",
"_____no_output_____"
]
],
[
[
"nx.average_clustering(G_er)",
"_____no_output_____"
],
[
"nx.average_clustering(G_ba)",
"_____no_output_____"
],
[
"nx.average_clustering(nx.Graph(G_config))",
"_____no_output_____"
]
],
[
[
"Comment on the values you obtain for the simulated networks. Is there any good match to the citation or feature graph in terms of clustering coefficient?",
"_____no_output_____"
],
[
"### Answer :\n<br> **No, there is not any match. The clustering coefficients are rather small compared to the ones for feature and citation graphs. Random networks have generally small clustering coefficients because they don't tend to form subgroups as the pairing is random.**",
"_____no_output_____"
],
[
"Check the other [network model generators](https://networkx.github.io/documentation/networkx-1.10/reference/generators.html) provided by NetworkX. Which one do you predict to have a better match to the citation graph or the feature graph in terms of degree distribution and clustering coefficient at the same time? Justify your answer.",
"_____no_output_____"
],
[
"### Answer :\n<br> **Based on the course notes about Watts Strogatz model which is a extension of the random network model generating small world properties and high clustering, we tested the watts_strogatz_graph function provided by NetworkX. We used the average degree ($k = m*2/n$) as an initial guess of the number of nearest neighbours to which we connect each node. We then modulated the rewiring probability to find a good match. Results did not show any satisfying match for the clustering coefficient (it was always rather low compared to the original networks). We then tuned parameter k by increasing it for a fixed p of 0.5 (corresponding to the small-world property). k was originally very low and we wanted to increase the occurence of clusters. At k =100, the clustering coefficient matched our expectations (being close to the clustering coeffients of the two distributions) but the distribtion did not match a powerlaw. In conclusion, watts-strogratz was left aside as no combination of parameters enabled to match the clustering coefficent as well as the shape of the distribution.\n<br>After scrolling through the documention of NetworkX, we came across the power_law_cluster function. According to the documentation, parameter n is the number of nodes, (n = 818 in our case). The second parameter, k, is the _number of random edges to add to each new node_ which we chose to be the average degree of the original graph as an initial guess. Parameter p, the probability of connecting two nodes which already share a common neighbour (forming a triangle) is chosen to be the average of average clustering coefficient across the original distributions. This yield a clustering coefficient that was a bit low compared with our expectations. We therefore tuned this parameter to better match the coefficients, results showed that a good comprise was reached at p = 0.27.** ",
"_____no_output_____"
],
[
"If you find a better fit, create a graph object below for that network model. Print the number of edges and the average clustering coefficient. Plot the histogram of the degree distribution.",
"_____no_output_____"
]
],
[
[
"k = m*2/n\np = (nx.average_clustering(G_citation) + nx.average_clustering(G_feature))*0.8\nG_pwc = nx.powerlaw_cluster_graph(n, int(k), p)\n \nprint('Power law cluster model has {} edges.'.format(G_pwc.size()))\nprint('Power law cluster model has a clustering coefficient of {}'.format(nx.average_clustering(G_pwc)))\nprint('Citation model has {} edges.'.format(G_citation.size()))\nprint('Citation model has a clustering coefficient of {}'.format(nx.average_clustering(G_citation)))\nprint('Feature model has {} edges.'.format(G_feature.size()))\nprint('Feature model has a clustering coefficient of {}'.format(nx.average_clustering(G_feature)))\n\nfig, axs = plt.subplots(1, 3, figsize=(15, 5), sharey=True, sharex=True)\naxs[0].set_title('PWC graph')\nws_degrees = [deg for (node, deg) in G_pwc.degree()]\naxs[0].hist(ws_degrees, bins=20, color='salmon', edgecolor='black', linewidth=1)\naxs[1].set_title('Citation graph')\ncitation_degrees = [deg for (node, deg) in G_citation.degree()]\naxs[1].hist(citation_degrees, bins=20, color='salmon', edgecolor='black', linewidth=1)\naxs[2].set_title('Feature graph')\nfeature_degree = [deg for (node, deg) in G_feature.degree()]\naxs[2].hist(feature_degree, bins=20, color='salmon', edgecolor='black', linewidth=1)\nplt.show()",
"Power law cluster model has 2440 edges.\nPower law cluster model has a clustering coefficient of 0.16384898371644135\nCitation model has 1175 edges.\nCitation model has a clustering coefficient of 0.21693567980632222\nFeature model has 1386 edges.\nFeature model has a clustering coefficient of 0.1220744470334593\n"
]
],
[
[
"Comment on the similarities of your match.",
"_____no_output_____"
],
[
"### Answer :\n<br> **At this point, the decay of the power law distribution (PWC) had an intermediate behavior between the citation and the feature graph and the clustering coefficient which fall in between the original graphs (~0.17). The degree range is roughly equilvalent in all three above distributions. \n<br> We hence found a model that satisfies both the clustering coefficients and the degree distribution. Also the final model raises distributions which have the expected intermediate behavior in comparison with the original distributions.**",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d0928ed2f553ce4c59bd78ded9291136812b4037 | 2,879 | ipynb | Jupyter Notebook | [CODE] GECODING/GEOCODING of Dataset for Amenities.ipynb | wca91/MSBA_DBA5106_Project_Group_30 | 9eebf3912184d33d4f9e6446b719fc756ea3507c | [
"MIT"
] | null | null | null | [CODE] GECODING/GEOCODING of Dataset for Amenities.ipynb | wca91/MSBA_DBA5106_Project_Group_30 | 9eebf3912184d33d4f9e6446b719fc756ea3507c | [
"MIT"
] | null | null | null | [CODE] GECODING/GEOCODING of Dataset for Amenities.ipynb | wca91/MSBA_DBA5106_Project_Group_30 | 9eebf3912184d33d4f9e6446b719fc756ea3507c | [
"MIT"
] | null | null | null | 27.160377 | 126 | 0.557138 | [
[
[
"import json\nimport requests\nimport pandas as pd\nfrom geopy.distance import geodesic",
"_____no_output_____"
],
[
"def find_postal(lst, filename):\n \n for index,add in enumerate(lst):\n url= \"https://developers.onemap.sg/commonapi/search?returnGeom=Y&getAddrDetails=Y&pageNum=1&searchVal=\"+add\n #print(index,url)\n response = requests.get(url)\n data = json.loads(response.text) \n \n temp_df = pd.DataFrame.from_dict(data[\"results\"])\n temp_df[\"address\"] = add\n \n if index == 0:\n file = temp_df\n else:\n file = file.append(temp_df)\n file.to_csv(filename + '.csv')",
"_____no_output_____"
],
[
"cc = pd.read_csv('.\\Amenities\\CC.csv')\nsupermarket = pd.read_csv('.\\Amenities\\supermarket_cleaned.csv')\nhawker = pd.read_csv('.\\Amenities\\list-of-government-markets-hawker-centres.csv')\nmrt = pd.read_csv('.\\Amenities\\mrtsg.csv')\nnpc = pd.read_csv('.\\Amenities\\PoliceCentre.csv')\n",
"_____no_output_____"
],
[
"unique_address_cc = cc['CC'].unique()\nunique_address_supermarket = supermarket['address'].unique()\nunique_address_hawker = hawker['name_of_centre'].unique()\nunique_address_mrt = mrt['STN_NAME'].unique()\nunique_address_npc = npc['NPC'].unique()\n",
"_____no_output_____"
],
[
"find_postal(unique_address_supermarket, \"supermarket_geoloc\")\nfind_postal(unique_address_hawker, \"hawker_geoloc\")\nfind_postal(unique_address_mrt , \"mrt_geoloc\")\nfind_postal(unique_address_cc, \"cc_geoloc\")\nfind_postal(unique_address_supermarket, \"npc_geoloc\")",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
d0929c71563d62399baec57c388a7f3a941f3e25 | 672 | ipynb | Jupyter Notebook | adventofcode/adventofcode.ipynb | headwinds/python-notebooks | d478de27f07d5f2164a1bedd4ca313dd88ab3446 | [
"MIT"
] | 2 | 2019-11-04T17:41:44.000Z | 2020-01-12T01:56:40.000Z | adventofcode/adventofcode.ipynb | headwinds/python-notebooks | d478de27f07d5f2164a1bedd4ca313dd88ab3446 | [
"MIT"
] | null | null | null | adventofcode/adventofcode.ipynb | headwinds/python-notebooks | d478de27f07d5f2164a1bedd4ca313dd88ab3446 | [
"MIT"
] | 1 | 2021-09-06T07:16:25.000Z | 2021-09-06T07:16:25.000Z | 17.684211 | 64 | 0.516369 | [
[
[
"import pandas as pd\ndata = pd.read_csv('input.txt', sep=\" \", header=None)\n\n\n",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code"
]
] |
d092a35e7c2d53580efa3b6db265ae60ada4bfef | 1,002,342 | ipynb | Jupyter Notebook | notebooks/reports/iris.ipynb | Atrus619/CSDGAN | 712be213e59b32a79a4970684d726af63616edaf | [
"MIT"
] | 1 | 2019-06-27T12:14:39.000Z | 2019-06-27T12:14:39.000Z | notebooks/reports/iris.ipynb | Atrus619/CSDGAN | 712be213e59b32a79a4970684d726af63616edaf | [
"MIT"
] | 21 | 2019-08-06T12:44:14.000Z | 2019-11-03T14:28:46.000Z | notebooks/reports/iris.ipynb | Atrus619/Synthetic_Data_GAN_Capstone | 712be213e59b32a79a4970684d726af63616edaf | [
"MIT"
] | 1 | 2020-07-25T13:14:31.000Z | 2020-07-25T13:14:31.000Z | 1,017.606091 | 236,065 | 0.953785 | [
[
[
"# Implementing a CGAN for the Iris data set to generate synthetic data\n### Import necessary modules and packages",
"_____no_output_____"
]
],
[
[
"import os\nwhile os.path.basename(os.getcwd()) != 'Synthetic_Data_GAN_Capstone':\n os.chdir('..')\nfrom utils.utils import *\nsafe_mkdir('experiments')\nfrom utils.data_loading import load_raw_dataset\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport torch\nimport torch.nn as nn\nimport torch.optim as optim\nfrom torch.autograd import Variable\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.preprocessing import StandardScaler\nfrom models.VGAN import VGAN_Generator, VGAN_Discriminator\nfrom models.CGAN_iris import CGAN_Generator, CGAN_Discriminator\nimport random",
"_____no_output_____"
]
],
[
[
"### Set random seed for reproducibility",
"_____no_output_____"
]
],
[
[
"manualSeed = 999\nprint(\"Random Seed: \", manualSeed)\nrandom.seed(manualSeed)\ntorch.manual_seed(manualSeed)",
"Random Seed: 999\n"
]
],
[
[
"### Import and briefly inspect data",
"_____no_output_____"
]
],
[
[
"iris = load_raw_dataset('iris')\niris.head()",
"_____no_output_____"
]
],
[
[
"### Preprocessing data\nSplit 50-50 so we can demonstrate the effectiveness of additional data",
"_____no_output_____"
]
],
[
[
"x_train, x_test, y_train, y_test = train_test_split(iris.drop(columns='species'), iris.species, test_size=0.5, stratify=iris.species, random_state=manualSeed)\nprint(\"x_train:\", x_train.shape)\nprint(\"x_test:\", x_test.shape)",
"x_train: (75, 4)\nx_test: (75, 4)\n"
]
],
[
[
"### Model parameters (feel free to play with these)",
"_____no_output_____"
]
],
[
[
"nz = 32 # Size of generator noise input\nH = 16 # Size of hidden network layer\nout_dim = x_train.shape[1] # Size of output\nbs = x_train.shape[0] # Full data set\nnc = 3 # 3 different types of label in this problem\nnum_batches = 1\nnum_epochs = 10000\nexp_name = 'experiments/iris_1x16'\nsafe_mkdir(exp_name)",
"_____no_output_____"
]
],
[
[
"### Adam optimizer hyperparameters\nI set these based on the original paper, but feel free to play with them as well.",
"_____no_output_____"
]
],
[
[
"lr = 2e-4\nbeta1 = 0.5\nbeta2 = 0.999",
"_____no_output_____"
]
],
[
[
"### Set the device",
"_____no_output_____"
]
],
[
[
"device = torch.device(\"cuda:0\" if (torch.cuda.is_available()) else \"cpu\")",
"_____no_output_____"
]
],
[
[
"### Scale continuous inputs for neural networks",
"_____no_output_____"
]
],
[
[
"scaler = StandardScaler()\nx_train = scaler.fit_transform(x_train)\nx_train_tensor = torch.tensor(x_train, dtype=torch.float)\ny_train_dummies = pd.get_dummies(y_train)\ny_train_dummies_tensor = torch.tensor(y_train_dummies.values, dtype=torch.float)",
"_____no_output_____"
]
],
[
[
"### Instantiate nets",
"_____no_output_____"
]
],
[
[
"netG = CGAN_Generator(nz=nz, H=H, out_dim=out_dim, nc=nc, bs=bs, lr=lr, beta1=beta1, beta2=beta2).to(device)\nnetD = CGAN_Discriminator(H=H, out_dim=out_dim, nc=nc, lr=lr, beta1=beta1, beta2=beta2).to(device)",
"_____no_output_____"
]
],
[
[
"### Print models\nI chose to avoid using sequential mode in case I wanted to create non-sequential networks, it is more flexible in my opinion, but does not print out as nicely",
"_____no_output_____"
]
],
[
[
"print(netG)\nprint(netD)",
"CGAN_Generator(\n (fc1): Linear(in_features=35, out_features=16, bias=True)\n (output): Linear(in_features=16, out_features=4, bias=True)\n (act): LeakyReLU(negative_slope=0.2)\n (loss_fn): BCELoss()\n)\nCGAN_Discriminator(\n (fc1): Linear(in_features=7, out_features=16, bias=True)\n (output): Linear(in_features=16, out_features=1, bias=True)\n (act): LeakyReLU(negative_slope=0.2)\n (m): Sigmoid()\n (loss_fn): BCELoss()\n)\n"
]
],
[
[
"### Define labels",
"_____no_output_____"
]
],
[
[
"real_label = 1\nfake_label = 0",
"_____no_output_____"
]
],
[
[
"### Training Loop\nLook through the comments to better understand the steps that are taking place",
"_____no_output_____"
]
],
[
[
"print(\"Starting Training Loop...\")\nfor epoch in range(num_epochs):\n for i in range(num_batches): # Only one batch per epoch since our data is horrifically small\n # Update Discriminator\n # All real batch first\n real_data = x_train_tensor.to(device) # Format batch (entire data set in this case)\n real_classes = y_train_dummies_tensor.to(device)\n label = torch.full((bs,), real_label, device=device) # All real labels\n\n output = netD(real_data, real_classes).view(-1) # Forward pass with real data through Discriminator\n netD.train_one_step_real(output, label)\n\n # All fake batch next\n noise = torch.randn(bs, nz, device=device) # Generate batch of latent vectors\n fake = netG(noise, real_classes) # Fake image batch with netG\n label.fill_(fake_label)\n output = netD(fake.detach(), real_classes).view(-1)\n netD.train_one_step_fake(output, label)\n netD.combine_and_update_opt()\n netD.update_history()\n\n # Update Generator\n label.fill_(real_label) # Reverse labels, fakes are real for generator cost\n output = netD(fake, real_classes).view(-1) # Since D has been updated, perform another forward pass of all-fakes through D\n netG.train_one_step(output, label)\n netG.update_history()\n\n # Output training stats\n if epoch % 1000 == 0 or (epoch == num_epochs-1):\n print('[%d/%d]\\tLoss_D: %.4f\\tLoss_G: %.4f\\tD(x): %.4f\\tD(G(z)): %.4f / %.4f'\n % (epoch+1, num_epochs, netD.loss.item(), netG.loss.item(), netD.D_x, netD.D_G_z1, netG.D_G_z2))\n with torch.no_grad():\n fake = netG(netG.fixed_noise, real_classes).detach().cpu()\n netG.fixed_noise_outputs.append(scaler.inverse_transform(fake))\nprint(\"Training Complete\")",
"Starting Training Loop...\n[1/10000]\tLoss_D: 1.3938\tLoss_G: 0.7742\tD(x): 0.4603\tD(G(z)): 0.4609 / 0.4611\n[1001/10000]\tLoss_D: 1.3487\tLoss_G: 0.7206\tD(x): 0.5089\tD(G(z)): 0.4883 / 0.4882\n[2001/10000]\tLoss_D: 1.3557\tLoss_G: 0.7212\tD(x): 0.5025\tD(G(z)): 0.4864 / 0.4864\n[3001/10000]\tLoss_D: 1.4118\tLoss_G: 0.6679\tD(x): 0.5029\tD(G(z)): 0.5138 / 0.5135\n[4001/10000]\tLoss_D: 1.3725\tLoss_G: 0.7053\tD(x): 0.5021\tD(G(z)): 0.4943 / 0.4942\n[5001/10000]\tLoss_D: 1.3690\tLoss_G: 0.7011\tD(x): 0.5064\tD(G(z)): 0.4967 / 0.4966\n[6001/10000]\tLoss_D: 1.3813\tLoss_G: 0.6954\tD(x): 0.5028\tD(G(z)): 0.4993 / 0.4994\n[7001/10000]\tLoss_D: 1.3799\tLoss_G: 0.6996\tD(x): 0.5006\tD(G(z)): 0.4970 / 0.4969\n[8001/10000]\tLoss_D: 1.3810\tLoss_G: 0.6978\tD(x): 0.5006\tD(G(z)): 0.4976 / 0.4978\n[9001/10000]\tLoss_D: 1.3871\tLoss_G: 0.6914\tD(x): 0.5008\tD(G(z)): 0.5010 / 0.5010\n[10000/10000]\tLoss_D: 1.3838\tLoss_G: 0.6955\tD(x): 0.5003\tD(G(z)): 0.4989 / 0.4989\nTraining Complete\n"
]
],
[
[
"### Output diagnostic plots tracking training progress and statistics",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\ntraining_plots(netD=netD, netG=netG, num_epochs=num_epochs, save=exp_name)",
"/home/aj/.local/lib/python3.7/site-packages/matplotlib/figure.py:445: UserWarning: Matplotlib is currently using module://ipykernel.pylab.backend_inline, which is a non-GUI backend, so cannot show the figure.\n % get_backend())\n"
],
[
"plot_layer_scatters(netG, title=\"Generator\", save=exp_name)",
"/home/aj/.local/lib/python3.7/site-packages/matplotlib/figure.py:445: UserWarning: Matplotlib is currently using module://ipykernel.pylab.backend_inline, which is a non-GUI backend, so cannot show the figure.\n % get_backend())\n"
],
[
"plot_layer_scatters(netD, title=\"Discriminator\", save=exp_name)",
"/home/aj/.local/lib/python3.7/site-packages/matplotlib/figure.py:445: UserWarning: Matplotlib is currently using module://ipykernel.pylab.backend_inline, which is a non-GUI backend, so cannot show the figure.\n % get_backend())\n"
]
],
[
[
"It looks like training stabilized fairly quickly, after only a few thousand iterations. The fact that the weight norm increased over time probably means that this network would benefit from some regularization.",
"_____no_output_____"
],
[
"### Compare performance of training on fake data versus real data\nIn this next section, we will lightly tune two models via cross-validation. The first model will be trained on the 75 real training data examples and tested on the remaining 75 testing data examples, whereas the second set of models will be trained on different amounts of generated data (no real data involved whatsoever). We will then compare performance and plot some graphs to evaluate our CGAN.",
"_____no_output_____"
]
],
[
[
"y_test_dummies = pd.get_dummies(y_test)\nprint(\"Dummy columns match?\", all(y_train_dummies.columns == y_test_dummies.columns))\nx_test = scaler.transform(x_test)\nlabels_list = [x for x in y_train_dummies.columns]\nparam_grid = {'tol': [1e-9, 1e-8, 1e-7, 1e-6, 1e-5],\n 'C': [0.5, 0.75, 1, 1.25],\n 'l1_ratio': [0, 0.25, 0.5, 0.75, 1]}",
"Dummy columns match? True\n"
]
],
[
[
"### Train on real data",
"_____no_output_____"
]
],
[
[
"model_real, score_real = train_test_logistic_reg(x_train, y_train, x_test, y_test, param_grid=param_grid, cv=5, random_state=manualSeed, labels=labels_list)",
"Accuracy: 0.92\nBest Parameters: {'C': 1.25, 'l1_ratio': 0.5, 'tol': 1e-09}\n precision recall f1-score support\n\n Iris-setosa 1.00 1.00 1.00 25\nIris-versicolor 0.85 0.92 0.88 25\n Iris-virginica 0.91 0.84 0.87 25\n\n accuracy 0.92 75\n macro avg 0.92 0.92 0.92 75\n weighted avg 0.92 0.92 0.92 75\n\n[[25 0 0]\n [ 0 23 2]\n [ 0 4 21]]\n"
]
],
[
[
"### Train on various levels of fake data",
"_____no_output_____"
]
],
[
[
"test_range = [75, 150, 300, 600, 1200]\nfake_bs = bs\nfake_models = []\nfake_scores = []\nfor size in test_range:\n num_batches = size // fake_bs + 1\n genned_data = np.empty((0, out_dim))\n genned_labels = np.empty(0)\n rem = size\n while rem > 0:\n curr_size = min(fake_bs, rem)\n noise = torch.randn(curr_size, nz, device=device)\n fake_labels, output_labels = gen_labels(size=curr_size, num_classes=nc, labels_list=labels_list)\n fake_labels = fake_labels.to(device)\n rem -= curr_size\n fake_data = netG(noise, fake_labels).cpu().detach().numpy()\n genned_data = np.concatenate((genned_data, fake_data))\n genned_labels = np.concatenate((genned_labels, output_labels))\n print(\"For size of:\", size)\n model_fake_tmp, score_fake_tmp = train_test_logistic_reg(genned_data, genned_labels, x_test, y_test,\n param_grid=param_grid, cv=5, random_state=manualSeed, labels=labels_list)\n fake_models.append(model_fake_tmp)\n fake_scores.append(score_fake_tmp)",
"For size of: 75\nAccuracy: 0.92\nBest Parameters: {'C': 0.5, 'l1_ratio': 0.25, 'tol': 1e-09}\n precision recall f1-score support\n\n Iris-setosa 1.00 1.00 1.00 25\nIris-versicolor 0.88 0.88 0.88 25\n Iris-virginica 0.88 0.88 0.88 25\n\n accuracy 0.92 75\n macro avg 0.92 0.92 0.92 75\n weighted avg 0.92 0.92 0.92 75\n\n[[25 0 0]\n [ 0 22 3]\n [ 0 3 22]]\nFor size of: 150\nAccuracy: 0.92\nBest Parameters: {'C': 0.5, 'l1_ratio': 1, 'tol': 1e-09}\n precision recall f1-score support\n\n Iris-setosa 1.00 1.00 1.00 25\nIris-versicolor 0.88 0.88 0.88 25\n Iris-virginica 0.88 0.88 0.88 25\n\n accuracy 0.92 75\n macro avg 0.92 0.92 0.92 75\n weighted avg 0.92 0.92 0.92 75\n\n[[25 0 0]\n [ 0 22 3]\n [ 0 3 22]]\nFor size of: 300\nAccuracy: 0.92\nBest Parameters: {'C': 0.5, 'l1_ratio': 0, 'tol': 1e-09}\n precision recall f1-score support\n\n Iris-setosa 1.00 1.00 1.00 25\nIris-versicolor 0.88 0.88 0.88 25\n Iris-virginica 0.88 0.88 0.88 25\n\n accuracy 0.92 75\n macro avg 0.92 0.92 0.92 75\n weighted avg 0.92 0.92 0.92 75\n\n[[25 0 0]\n [ 0 22 3]\n [ 0 3 22]]\nFor size of: 600\nAccuracy: 0.92\nBest Parameters: {'C': 0.5, 'l1_ratio': 0.75, 'tol': 1e-09}\n precision recall f1-score support\n\n Iris-setosa 1.00 1.00 1.00 25\nIris-versicolor 0.88 0.88 0.88 25\n Iris-virginica 0.88 0.88 0.88 25\n\n accuracy 0.92 75\n macro avg 0.92 0.92 0.92 75\n weighted avg 0.92 0.92 0.92 75\n\n[[25 0 0]\n [ 0 22 3]\n [ 0 3 22]]\nFor size of: 1200\nAccuracy: 0.9466666666666667\nBest Parameters: {'C': 0.5, 'l1_ratio': 0, 'tol': 1e-09}\n precision recall f1-score support\n\n Iris-setosa 1.00 1.00 1.00 25\nIris-versicolor 0.89 0.96 0.92 25\n Iris-virginica 0.96 0.88 0.92 25\n\n accuracy 0.95 75\n macro avg 0.95 0.95 0.95 75\n weighted avg 0.95 0.95 0.95 75\n\n[[25 0 0]\n [ 0 24 1]\n [ 0 3 22]]\n"
]
],
[
[
"Well, it looks like this experiment was a success. The models trained on fake data were actually able to outperform models trained on real data, which supports the belief that the CGAN is able to understand the distribution of the data it was trained on and generate meaningful examples that can be used to add additional information to the model.",
"_____no_output_____"
],
[
"Let's visualize some of the distributions of outputs to get a better idea of what took place",
"_____no_output_____"
]
],
[
[
"iris_plot_scatters(genned_data, genned_labels, \"Fake Data\", scaler, alpha=0.5, save=exp_name) # Fake data",
"_____no_output_____"
],
[
"iris_plot_scatters(iris.drop(columns='species'), np.array(iris.species), \"Full Real Data Set\", alpha=0.5, save=exp_name) # All real data",
"_____no_output_____"
],
[
"iris_plot_densities(genned_data, genned_labels, \"Fake Data\", scaler, save=exp_name) # Fake data",
"/home/aj/.local/lib/python3.7/site-packages/matplotlib/figure.py:445: UserWarning: Matplotlib is currently using module://ipykernel.pylab.backend_inline, which is a non-GUI backend, so cannot show the figure.\n % get_backend())\n"
],
[
"iris_plot_densities(iris.drop(columns='species'), np.array(iris.species), \"Full Real Data Set\", save=exp_name) # All real data",
"/home/aj/.local/lib/python3.7/site-packages/matplotlib/figure.py:445: UserWarning: Matplotlib is currently using module://ipykernel.pylab.backend_inline, which is a non-GUI backend, so cannot show the figure.\n % get_backend())\n"
],
[
"plot_scatter_matrix(genned_data, \"Fake Data\", iris.drop(columns='species'), scaler=scaler, save=exp_name)",
"_____no_output_____"
],
[
"plot_scatter_matrix(iris.drop(columns='species'), \"Real Data\", iris.drop(columns='species'), scaler=None, save=exp_name)",
"_____no_output_____"
]
],
[
[
"Finally, I present a summary of the test results ran above",
"_____no_output_____"
]
],
[
[
"fake_data_training_plots(real_range=bs, score_real=score_real, test_range=test_range, fake_scores=fake_scores, save=exp_name)",
"/home/aj/.local/lib/python3.7/site-packages/matplotlib/figure.py:445: UserWarning: Matplotlib is currently using module://ipykernel.pylab.backend_inline, which is a non-GUI backend, so cannot show the figure.\n % get_backend())\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d092a928138fe7cafafa0f84a83c9e93e2f3e25e | 39,707 | ipynb | Jupyter Notebook | codes/compare_optimizer/Haarinitstate_nlayer_4qubit.ipynb | vutuanhai237/QuantumTomography | 52916096482d7e7cd29782c049478bbba901d9bd | [
"MIT"
] | 2 | 2021-12-11T07:49:46.000Z | 2022-03-04T07:11:30.000Z | codes/compare_optimizer/Haarinitstate_nlayer_4qubit.ipynb | vutuanhai237/QuantumTomography | 52916096482d7e7cd29782c049478bbba901d9bd | [
"MIT"
] | null | null | null | codes/compare_optimizer/Haarinitstate_nlayer_4qubit.ipynb | vutuanhai237/QuantumTomography | 52916096482d7e7cd29782c049478bbba901d9bd | [
"MIT"
] | null | null | null | 177.263393 | 33,958 | 0.901705 | [
[
[
"import qiskit\nimport numpy as np, matplotlib.pyplot as plt\nimport sys\nsys.path.insert(1, '../')\nimport qtm.base, qtm.constant, qtm.nqubit, qtm.onequbit, qtm.fubini_study",
"_____no_output_____"
],
[
"num_qubits = 3\nnum_layers = 2\npsi = 2*np.random.rand(2**num_qubits)-1\npsi = psi / np.linalg.norm(psi)\nqc_origin = qiskit.QuantumCircuit(num_qubits, num_qubits)\nqc_origin.initialize(psi, range(0, num_qubits))\nthetas_origin = np.ones((num_layers*num_qubits*3))",
"_____no_output_____"
],
[
"qc = qc_origin.copy()\nthetas = thetas_origin.copy()\nthetas, loss_values_sgd = qtm.base.fit(\n qc, num_steps = 100, thetas = thetas, \n create_circuit_func = qtm.nqubit.u_cluster_nlayer_nqubit, \n grad_func = qtm.base.grad_loss,\n loss_func = qtm.base.loss_basis,\n optimizer = qtm.base.sgd,\n verbose = 1,\n num_layers = num_layers\n)",
"Step: 100%|██████████| 100/100 [14:02<00:00, 8.43s/it]\n"
],
[
"qc = qc_origin.copy()\nthetas = thetas_origin.copy()\nthetas, loss_values_adam = qtm.base.fit(\n qc, num_steps = 100, thetas = thetas, \n create_circuit_func = qtm.nqubit.u_cluster_nlayer_nqubit, \n grad_func = qtm.base.grad_loss,\n loss_func = qtm.base.loss_basis,\n optimizer = qtm.base.adam,\n verbose = 1,\n num_layers = num_layers\n)",
"Step: 100%|██████████| 100/100 [11:04<00:00, 6.65s/it]\n"
],
[
"qc = qc_origin.copy()\nthetas = thetas_origin.copy()\nthetas, loss_values_qfsm = qtm.base.fit(\n qc, num_steps = 100, thetas = thetas, \n create_circuit_func = qtm.nqubit.u_cluster_nlayer_nqubit, \n grad_func = qtm.base.grad_loss,\n loss_func = qtm.base.loss_basis,\n optimizer = qtm.base.qng_fubini_study,\n verbose = 1,\n num_layers = num_layers\n)",
"Step: 100%|██████████| 100/100 [37:54<00:00, 22.75s/it]\n"
],
[
"qc = qc_origin.copy()\nthetas = thetas_origin.copy()\nthetas, loss_values_qfim = qtm.base.fit(\n qc, num_steps = 100, thetas = thetas, \n create_circuit_func = qtm.nqubit.u_cluster_nlayer_nqubit, \n grad_func = qtm.base.grad_loss,\n loss_func = qtm.base.loss_basis,\n optimizer = qtm.base.qng_qfim,\n verbose = 1,\n num_layers = num_layers\n)",
"Step: 100%|██████████| 100/100 [29:50<00:00, 17.90s/it]\n"
],
[
"qc = qc_origin.copy()\nthetas = thetas_origin.copy()\nthetas, loss_values_adam_qfim = qtm.base.fit(\n qc, num_steps = 100, thetas = thetas, \n create_circuit_func = qtm.nqubit.u_cluster_nlayer_nqubit, \n grad_func = qtm.base.grad_loss,\n loss_func = qtm.base.loss_basis,\n optimizer = qtm.base.qng_adam,\n verbose = 1,\n num_layers = num_layers\n)",
"Step: 100%|██████████| 100/100 [29:22<00:00, 17.62s/it]\n"
],
[
"plt.plot(loss_values_sgd, label=\"SGD\")\nplt.plot(loss_values_adam, label=\"Adam\")\nplt.plot(loss_values_qfsm, label=\"QNG-QFSM\")\nplt.plot(loss_values_qfim, label=\"QNG-QFIM\")\nplt.plot(loss_values_adam_qfim, label=\"Adam QNG-QFIM\")\nplt.xlabel(\"Step\")\nplt.ylabel(\"Cost value\")\nplt.ylim(-0.05, 1.05)\nplt.legend(prop={'size': 8}, loc=1)\nplt.savefig(str(num_qubits) + '.svg', format='svg')\nplt.show()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d092abb69c5156ef413ed19c1fe5c5370af88aa8 | 286,310 | ipynb | Jupyter Notebook | nasa-turbofan-rul-lstm/notebooks/3 - Data preparation.ipynb | michaelhoarau/sagemaker-predictive-maintenance | 5f35d75d12d1e398a9d77508d11a2ffbe0c413ae | [
"MIT-0",
"MIT"
] | null | null | null | nasa-turbofan-rul-lstm/notebooks/3 - Data preparation.ipynb | michaelhoarau/sagemaker-predictive-maintenance | 5f35d75d12d1e398a9d77508d11a2ffbe0c413ae | [
"MIT-0",
"MIT"
] | null | null | null | nasa-turbofan-rul-lstm/notebooks/3 - Data preparation.ipynb | michaelhoarau/sagemaker-predictive-maintenance | 5f35d75d12d1e398a9d77508d11a2ffbe0c413ae | [
"MIT-0",
"MIT"
] | null | null | null | 169.213948 | 221,820 | 0.830443 | [
[
[
"# Predictive Maintenance using Machine Learning on Sagemaker\n*Part 3 - Timeseries data preparation*",
"_____no_output_____"
],
[
"## Initialization\n---\nDirectory structure to run this notebook:\n```\nnasa-turbofan-rul-lstm\n|\n+--- data\n| |\n| +--- interim: intermediate data we can manipulate and process\n| |\n| \\--- raw: *immutable* data downloaded from the source website\n|\n+--- notebooks: all the notebooks are positionned here\n|\n+--- src: utility python modules are stored here\n```\n### Imports",
"_____no_output_____"
]
],
[
[
"%load_ext autoreload\n\nimport matplotlib.pyplot as plt\nimport sagemaker\nimport boto3\nimport os\nimport errno\nimport pandas as pd\nimport numpy as np\nimport seaborn as sns\nimport json\nimport sys\nimport s3fs\nimport mxnet as mx\nimport joblib",
"_____no_output_____"
],
[
"%matplotlib inline\n%autoreload 2\n\nsns.set_style('darkgrid')\nsys.path.append('../src')\n\nfigures = []\n\nINTERIM_DATA = '../data/interim'\nPROCESSED_DATA = '../data/processed'",
"_____no_output_____"
]
],
[
[
"### Loading data from the previous notebook",
"_____no_output_____"
]
],
[
[
"# Load data from the notebook local storage:\n%store -r reduced_train_data\n%store -r reduced_test_data\n\n# If the data are not present in the notebook local storage, we need to load them from disk:\nsuccess_msg = 'Loaded \"reduced_train_data\"'\nif 'reduced_train_data' not in locals():\n print('Nothing in notebook store, trying to load from disk.')\n try:\n local_path = '../data/interim'\n reduced_train_data = pd.read_csv(os.path.join(local_path, 'reduced_train_data.csv'))\n reduced_train_data = reduced_train_data.set_index(['unit_number', 'time'])\n \n print(success_msg)\n \n except Exception as e:\n if (e.errno == errno.ENOENT):\n print('Files not found to load train data from: you need to execute the previous notebook.')\n \nelse:\n print('Train data found in notebook environment.')\n print(success_msg)\n \nsuccess_msg = 'Loaded \"reduced_test_data\"'\nif 'reduced_test_data' not in locals():\n print('Nothing in notebook store, trying to load from disk.')\n try:\n local_path = '../data/interim'\n reduced_test_data = pd.read_csv(os.path.join(local_path, 'reduced_test_data.csv'))\n reduced_test_data = reduced_test_data.set_index(['unit_number', 'time'])\n \n print(success_msg)\n \n except Exception as e:\n if (e.errno == errno.ENOENT):\n print('Files not found to load test data from: you need to execute the previous notebook.')\n \nelse:\n print('Test data found in notebook environment.')\n print(success_msg)",
"no stored variable reduced_train_data\nno stored variable reduced_test_data\nNothing in notebook store, trying to load from disk.\nLoaded \"reduced_train_data\"\nNothing in notebook store, trying to load from disk.\nLoaded \"reduced_test_data\"\n"
],
[
"print(reduced_train_data.shape)\nreduced_train_data.head()",
"(20631, 19)\n"
],
[
"print(reduced_test_data.shape)\nreduced_test_data.head()",
"(13096, 26)\n"
]
],
[
[
"### Study parameters",
"_____no_output_____"
]
],
[
[
"sequence_length = 20",
"_____no_output_____"
]
],
[
[
"## Normalization\n---\n### Normalizing the training data\nFirst, we build some scalers based on the training data:",
"_____no_output_____"
]
],
[
[
"from sklearn import preprocessing\n\n# Isolate the columns to normalize:\nnormalized_cols = reduced_train_data.columns.difference(['true_rul', 'piecewise_rul'])\n\n# Build MinMax scalers for the features and the labels:\nfeatures_scaler = preprocessing.MinMaxScaler()\nlabels_scaler = preprocessing.MinMaxScaler()\n\n# Normalize the operational settings and sensor measurements data (our features):\nreduced_train_data['sensor_measurement_17'] = reduced_train_data['sensor_measurement_17'].astype(np.float64)\nnormalized_data = pd.DataFrame(\n features_scaler.fit_transform(reduced_train_data[normalized_cols]),\n columns=normalized_cols, \n index=reduced_train_data.index\n)\n\n# Normalizing the labels data:\nreduced_train_data['piecewise_rul'] = reduced_train_data['piecewise_rul'].astype(np.float64)\nnormalized_training_labels = pd.DataFrame(\n labels_scaler.fit_transform(reduced_train_data[['piecewise_rul']]),\n columns=['piecewise_rul'], \n index=reduced_train_data.index\n)\n\n# Join the normalized features with the RUL (label) data:\njoined_data = normalized_training_labels.join(normalized_data)\nnormalized_train_data = joined_data.reindex(columns=reduced_train_data.columns)\nnormalized_train_data['true_rul'] = reduced_train_data['true_rul']\nprint(normalized_train_data.shape)\nnormalized_train_data.head()",
"(20631, 19)\n"
]
],
[
[
"### Normalizing the testing data\nNext, we apply these normalizers to the testing data:",
"_____no_output_____"
]
],
[
[
"normalized_test_data = pd.DataFrame(\n features_scaler.transform(reduced_test_data[normalized_cols]),\n columns=normalized_cols,\n index=reduced_test_data.index\n)\n\nreduced_test_data['piecewise_rul'] = reduced_test_data['piecewise_rul'].astype(np.float64)\n\nnormalized_test_labels = pd.DataFrame(\n labels_scaler.transform(reduced_test_data[['piecewise_rul']]),\n columns=['piecewise_rul'], \n index=reduced_test_data.index\n)\n\n# Join the normalize data with the RUL data:\njoined_data = normalized_test_labels.join(normalized_test_data)\nnormalized_test_data = joined_data.reindex(columns=reduced_test_data.columns)\nnormalized_test_data['true_rul'] = reduced_test_data['true_rul']\nprint(normalized_test_data.shape)\nnormalized_test_data.head()",
"(13096, 26)\n"
]
],
[
[
"## Sequences generation\n---",
"_____no_output_____"
]
],
[
[
"from lstm_utils import generate_sequences, generate_labels, generate_training_sequences, generate_testing_sequences\n\n# Building features, target and engine unit lists:\nfeatures = normalized_train_data.columns.difference(['true_rul', 'piecewise_rul'])\ntarget = ['piecewise_rul']\nunit_list = list(set(normalized_train_data.index.get_level_values(level=0).tolist()))\n\n# Generating features and labels for the training sequences:\ntrain_sequences = generate_training_sequences(normalized_train_data, sequence_length, features, unit_list)\ntrain_labels = generate_training_sequences(normalized_train_data, sequence_length, target, unit_list)\ntest_sequences, test_labels, unit_span = generate_testing_sequences(normalized_test_data, sequence_length, features, target, unit_list)\n\n# Checking sequences shapes:\nprint('train_sequences:', train_sequences.shape)\nprint('train_labels:', train_labels.shape)\nprint('test_sequences:', test_sequences.shape)\nprint('test_labels:', test_labels.shape)",
"Unit 1 test sequence ignored, not enough data points.\nUnit 22 test sequence ignored, not enough data points.\nUnit 39 test sequence ignored, not enough data points.\nUnit 85 test sequence ignored, not enough data points.\ntrain_sequences: (18631, 20, 17)\ntrain_labels: (18631, 1)\ntest_sequences: (11035, 20, 17)\ntest_labels: (11035, 1)\n"
]
],
[
[
"### Visualizing the sequences\nLet's visualize the sequences we built for an engine (e.g. unit 3 in the example below) to understand what will be fed to the LSTM model:",
"_____no_output_____"
]
],
[
[
"from lstm_utils import plot_timestep, plot_text\n\n# We use the normalized sequences for the plot but the original data for the label (RUL) for understanding purpose:\ncurrent_unit = 1\ntmp_sequences = generate_training_sequences(normalized_train_data, sequence_length, features, [current_unit])\ntmp_labels = generate_training_sequences(reduced_train_data, sequence_length, target, [current_unit])\n\n# Initialize the graphics:\nprint('Sequences generated for unit {}:\\n'.format(current_unit))\nsns.set_style('white')\nfig = plt.figure(figsize=(35,6))\n\n# Initialize the loop:\nnb_signals = min(12, len(tmp_sequences[0][0]))\nplots_per_row = nb_signals + 3\nnb_rows = 7\nnb_cols = plots_per_row\ncurrent_row = 0\nprevious_index = -1\ntimesteps = [\n # We will plot the first 3 sequences (first timesteps fed to the LSTM model):\n 0, 1, 2, \n\n # And the last 3 ones:\n len(tmp_sequences) - 3, len(tmp_sequences) - 2, len(tmp_sequences) - 1\n]\n\n# Loops through all the timesteps we want to plot:\nfor i in timesteps:\n # We draw a vertical ellispsis for the hole in the timesteps:\n if (i - previous_index > 1):\n plot_text(fig, nb_rows, nb_cols, nb_signals, current_row, '. . .', 1, no_axis=True, main_title='', plots_per_row=plots_per_row, options={'fontsize': 32, 'rotation': 270})\n current_row += 1\n\n # Timestep column:\n previous_index = i\n plot_text(fig, nb_rows, nb_cols, nb_signals, current_row, 'T{}'.format(i), 1, no_axis=True, main_title='Timestep', plots_per_row=plots_per_row, options={'fontsize': 16})\n\n # For a given timestep, we want to loop through all the signals to plot:\n plot_timestep(nb_rows, nb_cols, nb_signals, current_row, 2, tmp_sequences[i].T, features.tolist(), plots_per_row=plots_per_row)\n\n # Then we draw an ellipsis:\n plot_text(fig, nb_rows, nb_cols, nb_signals, current_row, '. . .', nb_signals + 2, no_axis=True, main_title='', plots_per_row=plots_per_row, options={'fontsize': 32})\n\n # Finally, we show the remaining useful life at the end of the row for this timestep:\n plot_text(fig, nb_rows, nb_cols, nb_signals, current_row, int(tmp_labels[i][0]), nb_signals + 3, no_axis=False, main_title='RUL', plots_per_row=plots_per_row, options={'fontsize': 16, 'color': '#CC0000'})\n\n current_row += 1",
"Sequences generated for unit 1:\n\n"
]
],
[
[
"## Cleanup\n---",
"_____no_output_____"
],
[
"### Storing data for the next notebook",
"_____no_output_____"
]
],
[
[
"%store labels_scaler\n%store test_sequences\n%store test_labels\n%store unit_span\n\n#columns = normalized_train_data.columns.tolist()\n#%store columns\n#%store train_sequences\n#%store train_labels\n#%store normalized_train_data\n#%store normalized_test_data\n#%store sequence_length",
"Stored 'labels_scaler' (MinMaxScaler)\nStored 'test_sequences' (ndarray)\nStored 'test_labels' (ndarray)\nStored 'unit_span' (list)\n"
]
],
[
[
"### Persisting these data to disk\nThis is useful in case you want to be able to execute each notebook independantly (from one session to another) and don't want to reexecute every notebooks whenever you want to focus on a particular step. Let's start by persisting train and test sequences and the associated labels:",
"_____no_output_____"
]
],
[
[
"import h5py as h5\n\ntrain_data = os.path.join(PROCESSED_DATA, 'train.h5')\nwith h5.File(train_data, 'w') as ftrain:\n ftrain.create_dataset('train_sequences', data=train_sequences)\n ftrain.create_dataset('train_labels', data=train_labels)\n ftrain.close()\n\ntest_data = os.path.join(PROCESSED_DATA, 'test.h5')\nwith h5.File(test_data, 'w') as ftest:\n ftest.create_dataset('test_sequences', data=test_sequences)\n ftest.create_dataset('test_labels', data=test_labels)\n ftest.close()",
"_____no_output_____"
]
],
[
[
"Pushing these files to S3:",
"_____no_output_____"
]
],
[
[
"sagemaker_session = sagemaker.Session()\nbucket = sagemaker_session.default_bucket()\nprefix = 'nasa-rul-lstm/data'\ntrain_data_location = 's3://{}/{}'.format(bucket, prefix)\n\ns3_resource = boto3.Session().resource('s3')\ns3_resource.Bucket(bucket).Object('{}/train/train.h5'.format(prefix)).upload_file(train_data)\ns3_resource.Bucket(bucket).Object('{}/test/test.h5'.format(prefix)).upload_file(test_data)\n\n# Build the data channel and write it to disk:\ndata_channels = {'train': 's3://{}/{}/train/train.h5'.format(bucket, prefix)}\nwith open(os.path.join(PROCESSED_DATA, 'data_channels.txt'), 'w') as f:\n f.write(str(data_channels))\n\n%store data_channels",
"Stored 'data_channels' (dict)\n"
]
],
[
[
"Storing the other elements on disk:",
"_____no_output_____"
]
],
[
[
"with open(os.path.join(PROCESSED_DATA, 'unit_span.lst'), 'w') as f:\n f.write(str(unit_span))\n\n_ = joblib.dump(labels_scaler, os.path.join(PROCESSED_DATA, 'labels_scaler.joblib'))",
"_____no_output_____"
]
],
[
[
"### Memory cleanup",
"_____no_output_____"
]
],
[
[
"fig.clear()\nplt.close(fig)\n\nimport gc\n_ = gc.collect()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d092b71bad89ace8e511938e461a01aba882215f | 357,372 | ipynb | Jupyter Notebook | 01-Square Boards(RUS).ipynb | esimonov/chess_prob | ed438803dbde7d5a981b6e38b0d0ebb73e20339f | [
"MIT"
] | null | null | null | 01-Square Boards(RUS).ipynb | esimonov/chess_prob | ed438803dbde7d5a981b6e38b0d0ebb73e20339f | [
"MIT"
] | null | null | null | 01-Square Boards(RUS).ipynb | esimonov/chess_prob | ed438803dbde7d5a981b6e38b0d0ebb73e20339f | [
"MIT"
] | null | null | null | 306.493997 | 73,766 | 0.909847 | [
[
[
"# О вероятности попасть под удар фигуры, поставленной случайным образом на шахматную доску\n",
"_____no_output_____"
],
[
"На шахматную доску случайным образом поставлены две фигуры. С какой вероятностью первая фигура бьёт вторую? В данном ноутбуке представлен расчёт этой вероятности для каждой шахматной фигуры как функции от размера доски. Рассмотрены только квадратные доски. Фигуры полагаются поставленными одновременно (обязательно стоят на разных клетках), выбор каждого из полей равновероятен.",
"_____no_output_____"
],
[
"Степень (валентность) вершины $v$ графа $G$ - количество рёбер графа $G$, инцидентных вершине $v$.",
"_____no_output_____"
],
[
"Граф ходов шахматной фигуры (далее Граф) - граф, изображающий все возможные ходы фигуры на шахматной доске - каждая вершина соответствует клетке на доске, а рёбра соответствуют возможным ходам.\nТогда валентность вершины Графа - количество полей, которые бьёт фигура, будучи поставленной на соответствующую этой вершине клетку. В целях упрощения речи далее в тексте используется формулировка \"клетка имеет валентность\", однако понятно, что валентность имеет не клетка, а соответствующая ей вершина в Графе.",
"_____no_output_____"
],
[
"Если событие $X$ может произойти только при выполнении одного из событий $H_1, H_2,..., H_n$, которые образуют полную группу несовместных событий, то вероятность $P(X)$ вычисляется по формуле: $$P(X) = P(H_1) \\cdot P(X|H_1) + P(H_2) \\cdot P(X|H_2) + ... + P(H_n) \\cdot P(X|H_n),$$ которая называется формулой полной вероятности.",
"_____no_output_____"
],
[
"Пусть событие $X_{piece}$ = «Первая *фигура (piece)* бьёт вторую на доске размера $n\\times n$», $a$ - некоторое значение валентности, $b$ - количество клеток, имеющих валентность $a$, каждая из гипотез $H_i$ = «Первая фигура стоит на клетке с валентностью $a_i$». Тогда $P(H_i) = \\frac{b_i}{n^{2}}$ в силу классического определения вероятности - это отношение количества исходов, благоприятствующих событию $H_i$, к количеству всех равновозможных исходов. $P(X_{piece}|H_i) = \\frac{a_i}{n^{2}-1}$ по той же причине - событию $X_{piece}$ при условии $H_i$ благоприятствует $a_i$ исходов (вторая фигура стоит под ударом клетки с валентностью $a_i$),а количество всех равновозможных исходов уменьшилось на единицу, так как одна из клеток уже занята первой фигурой.",
"_____no_output_____"
],
[
"### Импорты",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom string import ascii_uppercase as alphabet",
"_____no_output_____"
]
],
[
[
"### Функция создания доски",
"_____no_output_____"
],
[
"Создание доски как пустого массива заданной формы. Также нам понадобится массив такой же формы для хранения цвета надписи на клетках.\nЗаполнение доски: 0 - чёрные клетки, 1 - белые.",
"_____no_output_____"
]
],
[
[
"def get_board(board_size):\n\n x, y = np.meshgrid(range(board_size), range(board_size))\n\n board = np.empty(shape=(board_size, board_size), dtype='uint8')\n text_colors = np.empty_like(board, dtype='<U5')\n\n # force left bottom corner cell to be black\n if board_size % 2 == 0:\n extra_term = 1\n else:\n extra_term = 0\n\n for i, j in zip(x.flatten(), y.flatten()):\n board[i, j] = (i + j + extra_term) % 2\n # text color should be the opposite to a cell color\n text_colors[i, j] = 'black' if board[i, j] else 'white'\n\n return board, text_colors",
"_____no_output_____"
],
[
"def get_valencies(piece, board):\n # Get valencies for the given piece on the given board\n valencies = np.empty_like(board)\n\n if piece == 'Pawn':\n valencies = pawn(valencies)\n\n elif piece == 'Knight':\n valencies = knight(valencies)\n\n elif piece == 'Rook':\n valencies = rook(valencies)\n\n elif piece == 'King':\n valencies = king(valencies)\n\n else:\n valencies = bishop_or_queen(piece, valencies)\n\n return valencies",
"_____no_output_____"
]
],
[
[
"### Функция рисования доски",
"_____no_output_____"
],
[
"Функция создаёт изображение шахматной доски, каждая клетка которой подписана соответствующим значением валентности.",
"_____no_output_____"
]
],
[
[
"def plot_board(board, text_colors, piece):\n\n board_size = np.shape(board)[0]\n\n x, y = np.meshgrid(range(board_size), range(board_size))\n\n # let figure size be dependent on the board size\n plt.figure(figsize=(3*board_size/4, 3*board_size/4))\n ax = plt.subplot(111)\n\n ax.imshow(board, cmap='gray', interpolation='none')\n\n # Display valency (degree) values\n val_board = get_valencies(piece, board)\n\n for i, j, valency, text_col in zip(x.flatten(), y.flatten(),\n val_board.flatten(),\n text_colors.flatten()):\n ax.text(i, j, str(valency), color=text_col,\n va='center', ha='center', fontsize=20)\n\n ax.set_xticks(np.arange(board_size+1)) # one tick per cell\n ax.set_xticklabels(alphabet[:board_size]) # set letters as ticklabels\n # one tick per cell\n ax.set_yticks(np.arange(board_size+1))\n # set numbers as ticklabels (upside down)\n ax.set_yticklabels(np.arange(board_size, 0, -1))\n\n ax.axis('tight') # get rid of the white spaces on the edges\n ax.set_title(piece, fontsize=30)\n plt.show()",
"_____no_output_____"
]
],
[
[
"## Пешка",
"_____no_output_____"
],
[
"### Функция, возвращающая массив валентностей пешечного Графа",
"_____no_output_____"
],
[
"Иллюстрирует изложенные ниже соображения.",
"_____no_output_____"
]
],
[
[
"def pawn(valencies):\n\n valencies[0, :] = 0 # empty horizontal line\n valencies[1:, 0] = valencies[1:, -1] = 1 # vertical edges\n valencies[1:, 1:-1] = 2\n\n return valencies",
"_____no_output_____"
]
],
[
[
"Рассмотрим несколько частных случаев в поисках закономерности.",
"_____no_output_____"
]
],
[
[
"def special_cases(piece, board_sizes):\n ''' Plot boards of every board_size,\n contained in board_sizes list for given piece.\n '''\n for board_size in board_sizes:\n board, text_colors = get_board(board_size=board_size)\n plot_board(board, text_colors, piece=piece)",
"_____no_output_____"
],
[
"special_cases(piece='Pawn', board_sizes=range(4,6))",
"_____no_output_____"
]
],
[
[
"Закономерность очевидна - всегда присутствует горизонталь (верхняя или нижняя - в зависимости от цвета фигуры), с которой пешка не бьёт ни одну клетку - все поля этой горизонтали $0$-валентны. Их количество равно $n$. \nНа крайних вертикалях расположены $1$-валентные клетки, которых $2(n-1)$ штук.\nВсе остальные поля - $2$-валентны, и расположены они прямоугольником размера $(n-1)\\times(n-2)$.",
"_____no_output_____"
],
[
"Тогда $$ P(X_{pawn}) = \\frac{n\\cdot 0}{n^{2}(n^{2}-1)} + \\frac{2(n-1)\\cdot 1}{n^{2}(n^{2}-1)} + \\frac{(n-1)(n-2)\\cdot 2}{n^{2}(n^{2}-1)}= \\frac{2(n-1)({\\color{Green}1}+n-{\\color{Green}2})}{n^{2}(n^{2}-1)} = \\frac{2(n-1)^{2}}{n^{2}(n^{2}-1)}. $$ Так как $(n^{2}-1) = (n+1)(n-1)$, $$ P(X_{pawn}) = \\frac{2(n-1)}{n^{2}(n+1)}. $$",
"_____no_output_____"
],
[
"## Конь",
"_____no_output_____"
],
[
"### Функция, возвращающая массив валентностей Графа коня",
"_____no_output_____"
]
],
[
[
"def knight(valencies):\n\n board_size = valencies.shape[0]\n\n if board_size > 3:\n # Four points in each corner are the same for any board size > 3.\n \n # corner cells\n valencies[0, 0] = valencies[0, -1] = \\\n valencies[-1, 0] = valencies[-1, -1] = 2\n\n # cells horizontally/vertically adjacent to the corners\n valencies[0, 1] = valencies[1, 0] = \\\n valencies[0, -2] = valencies[1, -1] = \\\n valencies[-2, 0] = valencies[-1, 1] = \\\n valencies[-2, -1] = valencies[-1, -2] = 3\n\n # cells diagonally adjacent\n valencies[1, 1] = valencies[1, -2] = \\\n valencies[-2, 1] = valencies[-2, -2] = 4\n\n if board_size > 4:\n\n valencies[0, 2:-2] = valencies[2:-2, 0] = \\\n valencies[2:-2, -1] = valencies[-1, 2:-2] = 4\n\n valencies[1, 2:-2] = valencies[2:-2, 1] = \\\n valencies[2:-2, -2] = valencies[-2, 2:-2] = 6\n \n valencies[2:-2, 2:-2] = 8\n\n # Patholigical cases\n elif board_size == 3:\n valencies = 2 * np.ones((board_size, board_size), dtype='uint8')\n valencies[1, 1] = 0\n\n else:\n valencies = np.zeros((board_size, board_size), dtype='uint8')\n\n return valencies",
"_____no_output_____"
],
[
"special_cases(piece='Knight', board_sizes=[4,5,6])",
"_____no_output_____"
]
],
[
[
"Количество $2$- и $3$-валентных клеток фиксировано при любом $n\\geq 4$. Первые расположены в углах, а вторые прилегают к ним по вертикали и горизонтали. Стало быть, количество $2$-валентных клеток равно $4$, а $3$-валентных - $8$, вдвое больше. $4$-валентные клетки образуют арифметическую прогрессию с начальным элементом $4$ и шагом $4$ для всех $n\\geq 4$ (при увеличении $n$ на единицу с каждой стороны появляется одна $4$-валентная клетка). Легко видеть, что рост количества $6$-валентных клеток устроен аналогично, однако существуют они только при $n\\geq 5$. Таким образом, $4$-валентных клеток $4(n-3)$, а $6$-валентных клеток - $4(n-4)$ штук. Количество $8$-валентных клеток растёт квадратично, к тому же, они существуют только при $n\\geq 5$. То есть, их количество - $(n-4)^2$. Итого имеем:",
"_____no_output_____"
],
[
"$$ P(X_{knight}) = \\frac{4\\cdot 2}{n^{2}(n^{2}-1)} + \\frac{8\\cdot 3}{n^{2}(n^{2}-1)} + \\frac{4(n-3)\\cdot 4}{n^{2}(n^{2}-1)} +$$ $$ + \\frac{4(n-4)\\cdot 6}{n^{2}(n^{2}-1)} + \\frac{(n-4)^2\\cdot 8}{n^{2}(n^{2}-1)} = \\frac{32 + 24(n-4) + 16(n-3) + 8(n-4)^{2}}{n^{2}(n^{2}-1)} = $$ \n$$ \\frac{8(4+3(n-4)+2(n-3)+(n-4)^{2})}{n^{2}(n^{2}-1)} = \\frac{8({\\color{Green} 4}+{\\color{Red} {3n}}-{\\color{Green} {12}}+{\\color{Red} {2n}} - {\\color{Green} 6}+ n^{2}-{\\color{Red} {8n}}+{\\color{Green} {16}})}{n^{2}(n^{2}-1)} = $$ \n$$= \\frac{8(n^{2}-3n+2)}{n^{2}(n^{2}-1)} = \\frac{8(n-1)(n-2)}{n^{2}(n^{2}-1)} = \\frac{8(n-2)}{n^{2}(n+1)}. $$",
"_____no_output_____"
],
[
"## Офицер",
"_____no_output_____"
],
[
"### Функция, возвращающая массив валентностей Графа офицера (и ферзя)",
"_____no_output_____"
],
[
"Расположение валентностей для офицера и ферзя практически идентично, за исключением того, что наименьшее значение валентности для ферзя в три раза превышает оное для офицера.",
"_____no_output_____"
]
],
[
[
"def bishop_or_queen(piece, valencies):\n \n board_size = np.shape(valencies)[0]\n \n if piece == 'Bishop':\n smallest_val = board_size-1\n\n else:\n smallest_val = 3*(board_size-1)\n \n # external square\n valencies[0, :] = valencies[:, 0] = \\\n valencies[:, -1] = valencies[-1, :] = smallest_val\n \n # internal sqares\n for i in range (1, int(board_size/2)+1):\n # top, left\n # right, bottom\n valencies[i, i:-i] = valencies[i:-i, i] = \\\n valencies[i:-i, -(i+1)] = valencies[-(i+1), i:-i] = \\\n smallest_val + 2*i\n \n return valencies",
"_____no_output_____"
],
[
"special_cases(piece='Bishop', board_sizes=range(4,8))",
"_____no_output_____"
]
],
[
[
"Видно, что эквивалентные клетки располагаются по периметрам образованных клетками концентрических квадратов. Поскольку при чётных $n$ в центре доски расположены $4$ поля с максимальной валентностью, а при нечётных - одно, случаи чётных и нечётных $n$ представляется удобным рассмотреть раздельно.",
"_____no_output_____"
],
[
"### Чётные $n$",
"_____no_output_____"
],
[
"Каково количество различных значений валентности, а также их величина? Наименьшее значение равно $(n-1)$, так как это количество клеток на диагонали минус клетка, на которой стоит сама фигура. Наибольшее значение - $(n-1) + (n-2) = (2n-3)$, так как оно больше наименьшего значения на количество клеток, расположенных на диагонали квадрата со стороной $(n-1)$ минус клетка, на которой стоит сама фигура. ",
"_____no_output_____"
],
[
"Пусть $s$ - количество шагов размера $2$, которое требуется совершить для перехода от значения $(n-1)$ к значению $(2n-3)$. Тогда\n$$ n-1 + 2s = 2n-3, $$ $$ 2s = {\\color{Red} {2n}} - {\\color{Green} 3} - {\\color{Red} n} + {\\color{Green} 1} = n - 2 \\Rightarrow s = \\frac{n-2}{2}. $$\nТак как $n$ - чётное, $s$ $\\in \\mathbb{Z}$.",
"_____no_output_____"
],
[
"Однако ввиду того, что *один* шаг совершается между *двумя* разными значениями, количество различных значений валентности на единицу больше количества шагов, требующихся для перехода от минимального до максимального. В таком случае имеем $\\frac{n-2}{2} + 1 = \\frac{n}{2} - {\\color{Green} 1} +{\\color{Green} 1} = \\frac{n}{2}.$ Итого, на доске со стороной $n$ содержится $\\frac{n}{2}$ клеток с различными значениями валентности - $\\frac{n}{2}$ концентрических квадратов.",
"_____no_output_____"
],
[
"В каком количестве представлено каждое из значений? Количество элементов, расположенных по периметру образованного клетками квадрата со стороной $\\lambda$, равно учетверённой стороне минус четыре угловые клетки, которые оказываются учтёнными дважды. Тогда количество клеток с одноимённым значением валентности равно $4\\lambda-4 = 4(\\lambda-1)$, где $\\lambda$ изменяется с шагом $2$ в пределах от $2$ (центральный квадрат) до $n$ (внешний).",
"_____no_output_____"
],
[
"При этом от $\\lambda$ зависит не только количество значений валентности, но и их величина - она равна сумме $\\lambda$ и наименьшего из значений валентности, встречающихся на доске. Таким образом, имея наименьшее значение валентности, а также количество концентрических квадратов, нетрудно составить зависимую от $\\lambda$ сумму $P(X^{even}_{bishop}) = \\sum_{}P(H_i) \\cdot P(X|H_i)$. Однако удобнее суммировать по индексу, который изменяется с шагом $1$, потому заменим $k = \\frac{\\lambda}{2}.$ Теперь можно записать:",
"_____no_output_____"
],
[
"$$ P(X^{even}_{bishop}) = \\sum_{k = 1}^{\\frac{n}{2}} \\frac{4(n+1-2k)\\cdot(n-3+2k)} {n^{2}(n^{2}-1)} = \\frac{4}{n^{2}(n^{2}-1)} \\sum_{k = 1}^{\\frac{n}{2}} n^{2} - {\\color{Red} {3n}} + {\\color{Blue} {2kn}} + {\\color{Red} {n}} - 3 + {\\color{Cyan} {2k}} - {\\color{Blue} {2kn}} + {\\color{Cyan} {6k}} - 4k^{2} = $$ \n\n$$ =\\frac{4}{n^{2}(n^{2}-1)} \\sum_{k = 1}^{\\frac{n}{2}} n^{2} - 2n - 3 + 8k - 4k^{2}. $$ \nВынесем первые три слагаемых за знак суммы, так как они не зависят от $k$, умножив их на $\\frac{n}{2}$ - количество раз, которое они встречаются в сумме: \n\n$$ P(X^{even}_{bishop}) = \\frac{4}{n^{2}(n^{2}-1)}\\ [\\frac{n}{2}(n^{2} - 2n - 3) + \\sum_{k = 1}^{\\frac{n}{2}}8k - 4k^{2}] $$",
"_____no_output_____"
],
[
"Рассмотрим отдельно выражение под знаком суммы.",
"_____no_output_____"
],
[
"$$ \\sum_{k = 1}^{\\frac{n}{2}}8k - 4k^{2} = 8\\sum_{k = 1}^{\\frac{n}{2}}k - 4\\sum_{k = 1}^{\\frac{n}{2}} k^{2}. $$",
"_____no_output_____"
],
[
"Обозначим $ S_1 = 8\\sum_{k = 1}^{\\frac{n}{2}}k$, $ S_2 = 4\\sum_{k = 1}^{\\frac{n}{2}} k^{2}. $",
"_____no_output_____"
],
[
"$S_1$ - это умноженная на $8$ сумма первых $\\frac{n}{2}$ натуральных чисел, которая есть сумма первых $\\frac{n}{2}$ членов арифметической прогрессии, поэтому ",
"_____no_output_____"
],
[
"$$ S_1 = 8\\frac{\\frac{n}{2}(\\frac{n}{2}+1)}{2} = 4\\frac{n}{2}(\\frac{n}{2}+1) = 2n(\\frac{n}{2}+1) = \\frac{2n^2}{2}+2n = n^2 + 2n = n(n+2). $$",
"_____no_output_____"
],
[
"$S_2$ - это умноженная на 4 сумма квадратов первых $\\frac{n}{2}$ натуральных чисел, поэтому",
"_____no_output_____"
],
[
"$$ S_2 = 4\\frac{\\frac{n}{2}(\\frac{n}{2}+1)(2\\frac{n}{2}+1)}{6} = \\frac{n(n+2)(n+1)}{6}. $$",
"_____no_output_____"
],
[
"$$ S_1 - S_2 = n(n+2) - \\frac{n(n+2)(n+1)}{6} = n(n+2) (1 - \\frac{(n+1)}{6}) = $$ $$ = \\frac{n(n+2)({\\color{Green} 6}-n-{\\color{Green} 1})}{6} = \\frac{n(n+2)(-n + 5)}{6} = -\\frac{n(n+2)(n-5)}{6}.$$ ",
"_____no_output_____"
],
[
"Тогда",
"_____no_output_____"
],
[
"$$ P(X^{even}_{bishop}) = \\frac{4}{n^{2}(n^{2}-1)}\\ [\\frac{n}{2}(n^{2} - 2n - 3) - \\frac{n(n+2)(n-5)}{6} ] = $$ $$ = \\frac{4}{n^{2}(n^{2}-1)}\\ [\\frac{n(3n^{2} - 6n - 9)}{6} - \\frac{n(n+2)(n-5)}{6} ] = $$ $$ = \\frac{4n}{6n^{2}(n^{2}-1)}({\\color{Orange} {3n^{2}}} - {\\color{Red} {6n}} - {\\color{Green} 9} - {\\color{Orange} {n^2}} + {\\color{Red} {5n}} - {\\color{Red} {2n}} + {\\color{Green} {10}}) = $$ $$ =\\frac{2}{3n(n^{2}-1)}(2n^2 - 3n + 1) = \\frac{2(2n-1)(n-1)}{3n(n^{2}-1)} = \\frac{2(2n-1)}{3n(n+1)}. $$",
"_____no_output_____"
],
[
"### Нечётные $n$",
"_____no_output_____"
],
[
"Каково количество различных значений валентности? Наименьшее значение равно $(n-1)$ из тех же рассуждений, что и для чётных $n$. Наибольшее значение, очевидно, равно удвоенному наименьшему - $(n-1) + (n-1) = 2(n-1)$. ",
"_____no_output_____"
],
[
"Пусть $s$ - количество шагов размера $2$, которое требуется совершить для перехода от значения $(n-1)$ к значению $2(n-1)$. Тогда\n$$n-1 + 2s = 2n-2,$$ $$2s = {\\color{Red} {2n}} - {\\color{Green} 2} - {\\color{Red} n} + {\\color{Green} 1} = n - 1 \\Rightarrow s = \\frac{n-1}{2}.$$\nТак как $n$ - нечётное, $s$ $\\in \\mathbb{Z}$. Итого имеем $\\frac{n-1}{2} + 1 = \\frac{n}{2} - {\\color{Green} {\\frac{1}{2}}} +{\\color{Green} 1} = \\frac{n}{2} + \\frac{1}{2} = \\frac{n+1}{2}$ клеток с различными значениями валентности.",
"_____no_output_____"
],
[
"В каком количестве представлено каждое из значений? Рассуждения для чётных и нечётных $n$ идентичны, за исключением того, что выражение $4(\\lambda-1)$ равно нулю при $\\lambda = 1$ (в центральной клетке доски). По этой причине слагаемое $P(H_{\\frac{n+1}{2}}) \\cdot P(X|H_{\\frac{n+1}{2}})$ должно быть вынесено за знак общей суммы, а индекс суммирования будет принимать на единицу меньше значений: $\\frac{n+1}{2} - 1 = \\frac{n}{2} + \\frac{1}{2} - 1 = \\frac{n}{2} + {\\color{Green} {\\frac{1}{2}}} - {\\color{Green} 1} = \\frac{n}{2} - \\frac{1}{2} = \\frac{n-1}{2}.$",
"_____no_output_____"
],
[
"Тогда можно записать:",
"_____no_output_____"
],
[
"$$ P(X^{odd}_{bishop}) = \\frac{1\\cdot 2(n-1)}{n^{2}(n^{2}-1)} + \\sum_{k = 1}^{\\frac{n-1}{2}} \\frac{4(n+1-2k)\\cdot(n-3+2k)} {n^{2}(n^{2}-1)}. $$",
"_____no_output_____"
],
[
"Легко видеть, что выражение под знаком суммы отличается от $P(X^{even}_{bishop})$ только верхней границей суммирования. Следовательно, аналогично предыдущим выкладкам можно обозначить: $ S_1 = 8\\sum_{k = 1}^{\\frac{n-1}{2}}k$, $ S_2 = 4\\sum_{k = 1}^{\\frac{n-1}{2}} k^{2}. $",
"_____no_output_____"
],
[
"$$ S_1 = 8\\frac{\\frac{n-1}{2}(\\frac{n-1}{2}+1)}{2} = 4\\frac{n-1}{2}(\\frac{n+1}{2}) = (n-1)(n+1). $$",
"_____no_output_____"
],
[
"$$ S_2 = 4\\frac{\\frac{n-1}{2}(\\frac{n-1}{2}+1)(2\\frac{n-1}{2}+1)}{6} = 4\\frac{\\frac{n-1}{2}(\\frac{n-1}{2}+1)(2\\frac{n-1}{2}+1)}{6} = \\frac{(n-1)(\\frac{n+1}{2})n}{3} = \\frac{(n-1)(n+1)n}{6}. $$",
"_____no_output_____"
],
[
"$$ S_1 - S_2 = (n-1)(n+1) - \\frac{(n-1)(n+1)n}{6} = (n-1)(n+1)(1 - \\frac{n}{6}) = \\frac{(n-1)(n+1)(6 - n)}{6} = -\\frac{(n-1)(n+1)(n-6)}{6}. $$",
"_____no_output_____"
],
[
"Тогда",
"_____no_output_____"
],
[
"$$ P(X^{odd}_{bishop}) = \\frac{2(n-1)}{n^{2}(n^{2}-1)} + \\frac{4}{n^{2}(n^{2}-1)}\\ [\\frac{n-1}{2}(n^{2} - 2n - 3) -\\frac{(n-1)(n+1)(n-6)}{6}] = $$ $$ =\\frac{2}{n^{2}(n+1)} + \\frac{4(n-1)}{n^{2}(n^{2}-1)} [\\frac{3n^2 - 6n - 9}{6} -\\frac{(n+1)(n-6)}{6}] = $$ $$ =\\frac{2}{n^{2}(n+1)} + \\frac{4}{6n^{2}(n+1)}({\\color{Orange} {3n^2}} - {\\color{Red} {6n}} - {\\color{Green} 9} - {\\color{Orange} {n^2}} + {\\color{Red} {6n}} - {\\color{Red} n} + {\\color{Green} 6}) = $$ $$ =\\frac{2}{n^{2}(n+1)} + \\frac{4}{6n^{2}(n+1)}(2n^2 - n - 3) = \\frac{{\\color{Green} {12}} + 8n^2 - 4n - {\\color{Green} {12}}}{6n^{2}(n+1)} = \\frac{4n(2n-1)}{6n^{2}(n+1)} = \\frac{2(2n-1)}{3n(n+1)}. $$",
"_____no_output_____"
],
[
"Как видно, чётность доски не влияет на рассматриваемую вероятность: $P(X^{even}_{bishop}) = P(X^{odd}_{bishop}) = P(X_{bishop}) = \\frac{2(2n-1)}{3n(n+1)}$.",
"_____no_output_____"
],
[
"## Ладья",
"_____no_output_____"
],
[
"### Функция, возвращающая массив валентностей ладейного Графа",
"_____no_output_____"
]
],
[
[
"def rook(valencies):\n \n board_size = np.shape(valencies)[0]\n \n x, y = np.meshgrid(range(board_size), range(board_size))\n \n for i, j in zip(x.flatten(), y.flatten()):\n valencies[i, j] = 2*(board_size-1)\n \n return valencies ",
"_____no_output_____"
],
[
"special_cases(piece='Rook', board_sizes=range(4,6))",
"_____no_output_____"
]
],
[
[
"Известная особенность ладьи - независимо от расположения на доске, она всегда контролирует постоянное количество полей, а именно $2(n-1)$ - это сумма полей по горизонтали и вертикали минус поле, на котором стоит сама ладья.\n$$P(X_{rook}) = \\frac{n^{2}\\cdot 2(n-1)}{n^{2}(n^{2}-1)} = \\frac{2}{(n+1)}.$$",
"_____no_output_____"
],
[
"## Ферзь",
"_____no_output_____"
]
],
[
[
"special_cases(piece='Queen', board_sizes=range(4,8))",
"_____no_output_____"
]
],
[
[
"Поскольку ферзь сочетает в себе возможности офицера и ладьи, выражение для него может быть получено как сумма выражений для этих фигур:",
"_____no_output_____"
],
[
"$$ P(X_{queen}) = \\frac{2(2n-1)}{3n(n+1)} + \\frac{2}{n+1} = \\frac{2(2n-1) + 6n}{3n(n+1)} = \\frac{{\\color{Red} {4n}} - 2 + {\\color{Red} {6n}}}{3n(n+1)} = \\frac{10n - 2}{3n(n+1)} = \\frac{2(5n-1)}{3n(n+1)}. $$",
"_____no_output_____"
],
[
"## Король",
"_____no_output_____"
],
[
"### Функция, возвращающая массив валентностей Графа короля",
"_____no_output_____"
]
],
[
[
"def king(valencies):\n \n # corners : top left = top right = \\\n # bottom left = bottom right\n valencies[0, 0] = valencies[0, -1] = \\\n valencies[-1, 0] = valencies[-1, -1] = 3\n \n # edges : top, left, right, bottom\n valencies[0, 1:-1] = valencies[1:-1, 0] = \\\n valencies[1:-1, -1] = valencies[-1, 1:-1] = 5\n \n # center\n valencies[1:-1, 1:-1] = 8\n \n return valencies",
"_____no_output_____"
],
[
"special_cases(piece='King', board_sizes=range(4,6))",
"_____no_output_____"
]
],
[
[
"Видно, что края доски, за исключением $3$-валентных углов, $5$-валентны, а всё оставшееся пространство $8$-валентно. Ввиду того, что краёв $4$, а $5$-валентных клеток на одном краю $(n-2)$ штук, имеем: \n$$ P(X_{king}) = \\frac{4\\cdot 3}{n^{2}(n^{2}-1)} +\\frac{4(n-2)\\cdot 5}{n^{2}(n^{2}-1)} +\\frac{(n-2)^2\\cdot 8}{n^{2}(n^{2}-1)} = \\frac{12 + 20(n-2) + 8(n-2)^2}{n^{2}(n^{2}-1)} = $$\n$$ = \\frac{4(3 + 5(n-2)+2(n-2)^2)}{n^{2}(n^{2}-1)} = \\frac{4(3 + 5n-10+2(n^2 - 4n + 4))}{n^{2}(n^{2}-1)} = \\frac{4({\\color{Green} 3} + {\\color{Red} {5n}}-{\\color{Green} {10}}+2n^2 - {\\color{Red} {8n}} + {\\color{Green} {8}} )}{n^{2}(n^{2}-1)} = $$ \n$$ =\\frac{4(2n^2 - 3n + 1)}{n^{2}(n^{2}-1)} = \\frac{4(2n-1)(n-1)}{n^{2}(n^{2}-1)} = \\frac{4(2n-1)}{n^{2}(n+1)}. $$",
"_____no_output_____"
],
[
"### Функция, возвращающая значение $P(X_{piece})$",
"_____no_output_____"
]
],
[
[
"def get_probabilities(piece, n):\n \n # NOTE: Results can be incorrect for large n because of dividing by\n # the huge denominator!\n if piece == 'Pawn':\n return 2*(n-1)/((n**2)*(n+1)) \n \n elif piece == 'Knight':\n return 8*(n-2)/((n**2)*(n+1)) \n\n elif piece == 'Bishop':\n return 2*(2*n-1)/(3*n*(n+1)) \n \n elif piece == 'Rook':\n return 2/(n+1) \n\n elif piece == 'Queen':\n return 2*(5*n-1)/(3*n*(n+1)) \n\n elif piece == 'King':\n return 4*(2*n-1)/(n**2*(n+1))\n",
"_____no_output_____"
]
],
[
[
"Для проверки аналитических результатов используем метод вычисления вероятности «в лоб» - напрямую из массива валентностей.",
"_____no_output_____"
]
],
[
[
"def straightforward_prob(piece, board_size):\n # Get probability directly from the board of valencies\n board, _ = get_board(board_size)\n val_board = get_valencies(piece, board)\n \n unique, counts = np.unique(val_board, return_counts=True)\n prob = np.dot(unique, counts)/((board_size)**2 * (board_size**2 - 1))\n \n return prob ",
"_____no_output_____"
]
],
[
[
"График, отображающий зависимость вероятности от размера доски, представлен как функция действительного переменного в целях наглядности. ",
"_____no_output_____"
]
],
[
[
"start = 2\nend = 16\nstep = 0.02\nx = np.arange(start, end)\nnames_list = ['Pawn', 'Knight', 'Bishop', 'Rook', 'Queen', 'King']\n\n# Check if analytical results match straightforward calculations\nfor name in names_list:\n for board_size in x:\n y = get_probabilities(name, board_size)\n if not y == straightforward_prob(name, board_size):\n print('Mistake in equation for %s' % name)\nprint('Analytical results approved')\n\n# Let's expand the range from Z to R for the sake of visual clarity\nx = np.arange(start, end, step)\nfig, ax = plt.subplots(figsize=(10, 8)) \n\nfor name in names_list:\n y = get_probabilities(name, x) \n plt.plot(x, y, label=name, linewidth=3.0)\n \nlegend = plt.legend(loc='upper right')\n\nfor label in legend.get_lines():\n label.set_linewidth(3) \n\nfor label in legend.get_texts(): \n label.set_fontsize(26)\n\nplt.xlabel(\"Size of a board\", fontsize=20) \nplt.ylabel(\"Probability\", fontsize=20) \nplt.show() ",
"Analytical results approved\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d092b93d2bfafe5167397f020ad1d7fb4240d0bd | 246,047 | ipynb | Jupyter Notebook | courses/machine_learning/deepdive2/recommendation_systems/solutions/wals.ipynb | ssensalo/training-data-analyst | ddd0c422880634df43c3fab5dae1d2e3edc9d3d2 | [
"Apache-2.0"
] | 1 | 2016-04-18T10:36:49.000Z | 2016-04-18T10:36:49.000Z | courses/machine_learning/deepdive2/recommendation_systems/solutions/wals.ipynb | GoogleCloudPlatform/data-analyst-training | d18cc31961312af3520b134f05ed92012ea5b16a | [
"Apache-2.0"
] | null | null | null | courses/machine_learning/deepdive2/recommendation_systems/solutions/wals.ipynb | GoogleCloudPlatform/data-analyst-training | d18cc31961312af3520b134f05ed92012ea5b16a | [
"Apache-2.0"
] | null | null | null | 108.534186 | 121,824 | 0.802729 | [
[
[
"# Collaborative Filtering on Google Analytics Data\n",
"_____no_output_____"
],
[
"### Learning objectives\n1. Prepare the user-item matrix and use it with WALS.\n2. Train a `WALSMatrixFactorization` within TensorFlow locally and on AI Platform.\n3. Visualize the embedding vectors with principal components analysis.\n",
"_____no_output_____"
],
[
"## Overview\nThis notebook demonstrates how to implement a WALS matrix refactorization approach to do collaborative filtering.\n\nEach learning objective will correspond to a __#TODO__ in the [student lab notebook](../labs/wals.ipynb) -- try to complete that notebook first before reviewing this solution notebook.\n",
"_____no_output_____"
]
],
[
[
"import os\nPROJECT = \"cloud-training-demos\" # REPLACE WITH YOUR PROJECT ID\nBUCKET = \"cloud-training-demos-ml\" # REPLACE WITH YOUR BUCKET NAME\nREGION = \"us-central1\" # REPLACE WITH YOUR BUCKET REGION e.g. us-central1\n\n# Do not change these\nos.environ[\"PROJECT\"] = PROJECT\nos.environ[\"BUCKET\"] = BUCKET\nos.environ[\"REGION\"] = REGION\nos.environ[\"TFVERSION\"] = \"1.15\"",
"_____no_output_____"
],
[
"%%bash\ngcloud config set project $PROJECT\ngcloud config set compute/region $REGION",
"Updated property [core/project].\nUpdated property [compute/region].\n"
],
[
"import tensorflow as tf\nprint(tf.__version__)",
"1.15.5\n"
]
],
[
[
"## Create raw dataset\n<p>\nFor collaborative filtering, you don't need to know anything about either the users or the content. Essentially, all you need to know is userId, itemId, and rating that the particular user gave the particular item.\n<p>\nIn this case, you are working with newspaper articles. The company doesn't ask their users to rate the articles. However, you can use the time-spent on the page as a proxy for rating.\n<p>\nNormally, you would also add a time filter to this (\"latest 7 days\"), but your dataset is itself limited to a few days.",
"_____no_output_____"
]
],
[
[
"from google.cloud import bigquery\nbq = bigquery.Client(project = PROJECT)\n\nsql = \"\"\"\nWITH CTE_visitor_page_content AS (\n SELECT\n # Schema: https://support.google.com/analytics/answer/3437719?hl=en\n # For a completely unique visit-session ID, you combine combination of fullVisitorId and visitNumber:\n CONCAT(fullVisitorID,'-',CAST(visitNumber AS STRING)) AS visitorId,\n (SELECT MAX(IF(index=10, value, NULL)) FROM UNNEST(hits.customDimensions)) AS latestContentId, \n (LEAD(hits.time, 1) OVER (PARTITION BY fullVisitorId ORDER BY hits.time ASC) - hits.time) AS session_duration \n FROM\n `cloud-training-demos.GA360_test.ga_sessions_sample`, \n UNNEST(hits) AS hits\n WHERE \n # only include hits on pages\n hits.type = \"PAGE\"\nGROUP BY \n fullVisitorId,\n visitNumber,\n latestContentId,\n hits.time )\n-- Aggregate web stats\nSELECT \n visitorId,\n latestContentId as contentId,\n SUM(session_duration) AS session_duration\nFROM\n CTE_visitor_page_content\nWHERE\n latestContentId IS NOT NULL \nGROUP BY\n visitorId, \n latestContentId\nHAVING \n session_duration > 0\n\"\"\"\n\ndf = bq.query(sql).to_dataframe()\ndf.head()",
"_____no_output_____"
],
[
"stats = df.describe()\nstats",
"_____no_output_____"
],
[
"df[[\"session_duration\"]].plot(kind=\"hist\", logy=True, bins=100, figsize=[8,5])",
"_____no_output_____"
],
[
"# The rating is the session_duration scaled to be in the range 0-1. This will help with training.\nmedian = stats.loc[\"50%\", \"session_duration\"]\ndf[\"rating\"] = 0.3 * df[\"session_duration\"] / median\ndf.loc[df[\"rating\"] > 1, \"rating\"] = 1\ndf[[\"rating\"]].plot(kind=\"hist\", logy=True, bins=100, figsize=[8,5])",
"_____no_output_____"
],
[
"del df[\"session_duration\"]",
"_____no_output_____"
],
[
"%%bash\nrm -rf data\nmkdir data",
"_____no_output_____"
],
[
"# TODO 1: Write object to a comma-separated values (csv) file.\ndf.to_csv(path_or_buf = \"data/collab_raw.csv\", index = False, header = False)",
"_____no_output_____"
],
[
"!head data/collab_raw.csv",
"1012012094517511217-1,299949290,1.0\n1614877438160481626-349,29455408,1.0\n1815317450115839569-235,299807267,0.8569622728727375\n1950053456137072413-10,243386345,0.5474309284215766\n2179122255490468209-180,299925086,0.03222095793478647\n2477455524323251766-1,299936493,0.3514756604914127\n277149937495438413-384,299777707,0.1444742737261316\n2912361176772133138-147,299796840,1.0\n3868650583032521586-577,299814183,0.5796732212068414\n4086929260756256102-380,299946732,1.0\n"
]
],
[
[
"## Create dataset for WALS\n<p>\nThe raw dataset (above) won't work for WALS:\n<ol>\n<li> The userId and itemId have to be 0,1,2 ... so you need to create a mapping from visitorId (in the raw data) to userId and contentId (in the raw data) to itemId.\n<li> You will need to save the above mapping to a file because at prediction time, you'll need to know how to map the contentId in the table above to the itemId.\n<li> You'll need two files: a \"rows\" dataset where all the items for a particular user are listed; and a \"columns\" dataset where all the users for a particular item are listed.\n</ol>\n\n<p>\n\n### Mapping",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\ndef create_mapping(values, filename):\n with open(filename, 'w') as ofp:\n value_to_id = {value:idx for idx, value in enumerate(values.unique())}\n for value, idx in value_to_id.items():\n ofp.write(\"{},{}\\n\".format(value, idx))\n return value_to_id\n\ndf = pd.read_csv(filepath_or_buffer = \"data/collab_raw.csv\",\n header = None,\n names = [\"visitorId\", \"contentId\", \"rating\"],\n dtype = {\"visitorId\": str, \"contentId\": str, \"rating\": np.float})\ndf.to_csv(path_or_buf = \"data/collab_raw.csv\", index = False, header = False)\nuser_mapping = create_mapping(df[\"visitorId\"], \"data/users.csv\")\nitem_mapping = create_mapping(df[\"contentId\"], \"data/items.csv\")",
"_____no_output_____"
],
[
"!head -3 data/*.csv",
"==> data/collab_raw.csv <==\n1012012094517511217-1,299949290,1.0\n1614877438160481626-349,29455408,1.0\n1815317450115839569-235,299807267,0.8569622728727375\n\n==> data/items.csv <==\n299949290,0\n29455408,1\n299807267,2\n\n==> data/users.csv <==\n1012012094517511217-1,0\n1614877438160481626-349,1\n1815317450115839569-235,2\n"
],
[
"df[\"userId\"] = df[\"visitorId\"].map(user_mapping.get)\ndf[\"itemId\"] = df[\"contentId\"].map(item_mapping.get)",
"_____no_output_____"
],
[
"mapped_df = df[[\"userId\", \"itemId\", \"rating\"]]\nmapped_df.to_csv(path_or_buf = \"data/collab_mapped.csv\", index = False, header = False)\nmapped_df.head()",
"_____no_output_____"
]
],
[
[
"### Creating rows and columns datasets",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nmapped_df = pd.read_csv(filepath_or_buffer = \"data/collab_mapped.csv\", header = None, names = [\"userId\", \"itemId\", \"rating\"])\nmapped_df.head()",
"_____no_output_____"
],
[
"NITEMS = np.max(mapped_df[\"itemId\"]) + 1\nNUSERS = np.max(mapped_df[\"userId\"]) + 1\nmapped_df[\"rating\"] = np.round(mapped_df[\"rating\"].values, 2)\nprint(\"{} items, {} users, {} interactions\".format( NITEMS, NUSERS, len(mapped_df) ))",
"5662 items, 120983 users, 284343 interactions\n"
],
[
"grouped_by_items = mapped_df.groupby(\"itemId\")\niter = 0\nfor item, grouped in grouped_by_items:\n print(item, grouped[\"userId\"].values, grouped[\"rating\"].values)\n iter = iter + 1\n if iter > 5:\n break",
"0 [ 0 357 561 608 746 1119 1159 1207 1315 1348 1370 1396\n 1940 2035 2143 2254 3169 3313 176 3692 611 4003 4528 1432\n 4625 4862 4879 5110 5324 5409 5506 5693 5732 5743 5838 491\n 7167 7349 7908 7919 8064 8217 5758 8764 8829 9050 9140 9311\n 9417 9820 10368 10418 10809 10822 10911 11351 11761 9110 11982 3345\n 12292 12599 12841 12863 13342 13758 13986 14038 14153 14357 15069 15074\n 15106 15346 15424 15519 13030 16265 16363 5322 16416 16560 16749 17042\n 17077 17403 17442 6664 17575 17674 17735 17916 17990 18152 18181 13354\n 18383 18532 16206 13652 18643 18905 11473 19213 19655 19685 19699 17437\n 19817 3948 20195 15545 20285 20733 20792 16549 21222 5923 21627 21796\n 12084 14759 22053 22116 22691 1287 22717 20511 23252 23446 23479 5683\n 23796 17283 24177 24348 810 10004 24800 25200 13567 1899 8200 25455\n 25570 25629 25806 25822 25870 25904 11926 26330 26598 26610 26751 26914\n 27307 28187 6666 579 28556 28604 28708 28731 28756 24762 28916 28966\n 29452 29521 11392 11503 29771 29829 29935 12012 30161 30496 3827 26662\n 30686 30746 17931 30809 13231 30980 20602 31324 31418 31621 31898 3034\n 31997 389 32253 32413 32502 12924 32719 32744 32817 32890 32929 5062\n 29483 33477 33528 14164 33617 33642 33703 33771 33834 34045 34066 34101\n 3455 28351 9561 34206 34522 4378 34789 34857 34994 35004 2160 35210\n 35447 35588 35605 35725 35815 35818 36006 36111 26557 24649 36384 26845\n 4605 22817 22916 37058 37093 37389 19517 9461 37818 6890 22314 37945\n 38436 33298 21167 38896 25831 19582 39375 39554 28739 32642 39832 40055\n 4750 40269 31276 40332 21443 11597 350 41247 41345 41441 918 4525\n 18200 25024 15950 40285 8029 42042 2197 11288 8735 30551 890 32677\n 43227 34610 43447 27111 1548 31044 1807 43717 21284 44031 11636 2802\n 44168 44245 21948 44417 12241 6776 45088 45172 38633 45347 2248 45488\n 45576 8867 45720 37552 35866 45944 46118 46227 41742 46690 46730 1973\n 46791 46843 46892 46937 5483 47006 29697 14330 47309 24170 47509 47611\n 47882 7307 4978 38715 43861 37051 48606 19146 35573 3092 30196 48951\n 49159 49214 4064 39936 41673 49432 49461 49488 10504 49776 11221 49973\n 50009 5664 6297 42753 22220 34315 15468 50756 38125 38223 50883 50957\n 25159 51131 51187 18763 5550 51564 17084 51793 39490 51911 44702 52123\n 52484 8354 8701 11873 46 53227 53297 15206 18026 15900 53759 53760\n 7810 53840 10923 53895 20950 5332 43946 54472 54477 54496 6715 4022\n 7296 54877 54980 30992 27101 55045 55085 41871 55100 48350 2392 8750\n 55727 55746 6746 55976 12697 12787 56230 56266 53648 31410 42156 56638\n 23987 57145 57182 41351 24499 4302 7263 18016 55019 40434 2871 28104\n 40941] [1. 0.19 0.93 0.55 0.4 0.54 0. 0.09 0.57 1. 0.36 0.43 0.37 0.44\n 0.57 0.03 0.08 0.02 0.16 0.36 0.07 0.72 0.87 0.04 0.64 0.69 0.05 0.4\n 0.14 0.03 0.06 0.36 0.6 0.03 0.07 0.01 0.6 0.56 0.36 0.75 0.36 1.\n 1. 0.16 0.58 1. 0.98 0.37 1. 0.92 0.45 0.49 0.47 1. 1. 0.19\n 0.01 0.09 0.07 0.27 0.34 0.21 0.01 0.01 0.91 0.57 0.28 0.92 0.02 0.65\n 0.25 0.87 0.46 1. 0. 1. 0.37 1. 0.53 1. 1. 0.42 0.1 1.\n 0.47 1. 1. 0.41 0.04 0.44 1. 0.34 0.81 0.56 0.32 0.04 0.25 0.03\n 0.73 1. 0.03 0.21 0.05 0.11 0.26 1. 0.67 1. 0.4 0.69 0.19 0.1\n 0.14 0.98 0.46 0.46 0.11 0.94 0.46 0.92 0.1 1. 0.39 1. 0.67 0.23\n 0.76 1. 1. 0.68 0.68 0.25 0.41 0.79 0.81 0.05 0.72 0.04 0.07 0.34\n 1. 0.61 0.32 1. 0.37 1. 0.81 0.41 0.78 0.39 0.02 0.27 0.91 1.\n 0.21 1. 0.19 1. 0.33 0.66 0.56 0.04 0.43 0.78 0.47 0.93 0.52 0.15\n 0.61 0.2 1. 0.45 0.03 0.48 0.09 0.44 0.14 0.14 0.01 0.05 0.01 0.52\n 0.36 0.07 1. 0.13 0.57 1. 1. 1. 0.74 0.63 0.02 0.03 0.17 0.63\n 0.54 0.02 0.72 0.07 0.43 1. 1. 0.01 0.62 0.05 1. 0.03 0.26 0.48\n 0.5 1. 0.46 0.3 0.14 0.57 0.51 0.75 0.8 0.01 0.02 0.6 1. 1.\n 0.08 0.48 1. 0.18 0.91 0.54 0.3 0.21 0.52 0.03 0.03 1. 0.06 0.46\n 0.05 0.47 0.42 0.32 0.13 0.25 0.95 0.04 0.92 1. 0.5 0.45 0.32 0.99\n 0.6 0.78 0.17 0.32 0.24 0.77 0.46 0.66 0.15 0.78 0.27 0.31 0.36 0.25\n 0.76 0.04 0.43 1. 0.43 0.26 0.63 0.78 0.62 0.48 0.48 1. 0.08 0.01\n 0.48 0.41 0.17 0.49 0.45 0.69 0.14 0.76 1. 1. 0.58 0.02 0.52 1.\n 0.87 0.03 0.08 0.73 0.03 0. 0.78 0.28 0.27 0.99 0.62 0.25 0.37 0.95\n 0.37 0.96 0.45 0.16 0.08 1. 0.24 0. 0.03 0.04 0.5 0.21 0.31 0.09\n 0.85 0.03 0.4 0.07 0.47 0.41 0.62 0.01 0.89 0.07 0.29 0.04 0.92 1.\n 0.16 0.04 0.6 0.27 0.57 0.15 0.56 0.93 0.28 1. 0.69 0.61 0.32 0.01\n 0.49 1. 0.07 0.72 0.28 1. 0.68 0.04 0.26 0.28 0.09 0.03 1. 0.78\n 1. 0.4 0.55 0.36 0.04 0.62 1. 0.5 0.12 1. 0.33 1. 0.46 0.05\n 0.09 0.67 0.72 0.49 1. 0.47 0.57 0.38 0.91 1. 0.01 0.41 0.04 1.\n 0.19 0.06 0.68 0.77 0.11 0.31 0.15 0.33 0.47 0.91 0.24 0.11 0.4 1.\n 0.64 0.11 1. 0.08 1. 0.6 0.04 0.34 0.38 0.26 1. 0.64 0.87 0.24\n 0.45 0.01 1. 1. 0.45 0.09 1. 0.43 0.03 0.47 0.8 0.41 0.44 0.14\n 0.49 0.31 0.54 0.02 0.09 0.68 0.12 0.14 0.63 0.3 0.76]\n1 [ 1 2468] [1. 0.7]\n2 [ 2 1102 1529 2209 2413 2581 3155 3185 3365 4460 4792 5042\n 5789 6980 6992 4111 7403 8052 8308 8340 8556 8895 8900 9868\n 10187 10376 7594 10775 8029 11270 11376 11448 11476 11694 12310 506\n 12741 13139 13329 7864 5291 14267 14268 252 14961 15235 7685 10716\n 17004 11878 17652 17680 18180 18217 18310 3300 19828 20221 20732 15562\n 1973 23535 14400 23824 23971 24335 3162 26143 26206 26495 26980 27012\n 2311 21946 28358 29211 29313 23310 11251 29759 8261 32172 32538 32573\n 32609 32652 33063 19200 33716 34004 34027 26366 34225 7478 34974 1822\n 29483 36061 36252 36515 37262 31746 37485 37525 38397 38712 39130 39311\n 39542 17502 40027 5166 37229 21481 11594 16836 21615 40891 9510 28679\n 41525 41646 32836 31141 41886 13820 21732 41299 10152 7790 43557 43733\n 45115 42408 14421 9141 9847 36258 46677 29794 25806 33827 21989 47656\n 34544 47983 48065 48113 36663 8812 48672 21872 4671 16359 49926 11663\n 3563 32327 50606 1237 50914 18245 51523 25907 12246 51978 935 52065\n 52407 48171 31205 19055 39118 2893 11765 576 7143 53514 43349 53634\n 39075 24216 54609 14269 2859 55690 56129 13214 2195 8700 40892 3061\n 57183 34328 57368 57392 57453 57577 24940 15854 51041 58076 44054] [0.86 0.41 0.47 0.05 0.51 0.66 0.38 0.35 0.34 1. 0.63 0.03 0.12 1.\n 0.08 0.14 0.09 0.4 0.29 0.12 0.58 0.36 0.29 1. 0.77 0.6 0.13 0.16\n 0.25 0.57 0.28 1. 0.67 0.36 0.65 1. 1. 0.66 1. 1. 0.83 0.02\n 0.04 0.53 0.35 0.5 1. 0.31 0.53 0.12 1. 1. 0. 0.21 0.37 0.07\n 0.26 0.05 1. 0.27 0.36 0.45 0.62 0.4 0.17 0.06 0.01 0.17 1. 0.15\n 0.06 0.49 0.02 0.08 0.87 0.7 1. 0.12 0.28 0.27 0.37 1. 1. 1.\n 1. 0.65 0.1 1. 1. 1. 0.09 0.26 0.34 0.64 0.63 0.23 0.41 0.17\n 0.02 0.43 0.15 0.23 0.3 0.77 1. 0.03 0.3 0.29 1. 0.03 0.03 0.02\n 0.63 0.02 0.47 0.88 0.17 1. 0.01 0.55 1. 1. 0.3 1. 0.18 0.08\n 1. 0.14 1. 0.26 1. 1. 0.3 0.12 0.05 0.46 0.44 0.72 0.01 0.09\n 0.12 0.08 0.96 0.29 1. 0.23 1. 0.28 0.13 0.7 1. 0.34 0.41 0.23\n 1. 0.17 0.46 0.1 0.33 0.21 1. 0.22 0.31 0.05 0.94 0.85 0.27 0.4\n 1. 0.63 1. 0.86 0.19 0.05 0.42 0.1 0.39 1. 1. 1. 0.26 0.11\n 0.25 0.66 0.85 0.22 1. 0.57 0.42 0.36 1. 0.01 1. 0.03 1. 0.21\n 0.5 1. 0.02 0.27 0.97 0.22 0.23]\n3 [ 3 229 900 1536 1981 2145 3838 4953 5330 5621 5691 5812\n 2819 5912 4244 8307 8393 9131 9233 9502 10069 1583 11109 2172\n 11343 11523 11678 5900 11742 12195 12458 13794 14493 11849 15437 15626\n 16240 16306 17400 17487 943 19053 19166 19696 20219 20813 21088 14139\n 21438 21452 2769 14680 22111 1871 23358 18875 23458 23747 277 12870\n 15755 25332 25413 25429 23586 25680 25784 25950 26765 26787 25493 27777\n 27784 8781 28011 17464 17581 3867 29041 29193 30621 30648 31975 9890\n 15607 10167 34736 20722 35220 35542 9065 6254 35804 24425 1571 5514\n 37175 37571 38077 38505 38513 38626 38696 2380 38866 35728 26277 26306\n 39862 20435 40303 40561 9149 9816 20673 42241 23790 43064 28689 26975\n 17580 4789 29170 33382 45426 45521 19523 534 41264 46245 30931 40558\n 8763 45661 39787 17924 22876 48134 2196 48646 46138 4013 10587 49717\n 50057 50196 50482 50773 51551 9846 52286 52329 52691 11639 2848 52949\n 45879 39435 53681 16032 54144 54364 3390 54672 9912 22638 42022 8134\n 23284 21698 6364 14871 1254 34824 25050 56524 56637 14496 57294 7553\n 57764 16540 58108] [0.55 0.75 0.2 0. 1. 0.45 0.08 1. 0.08 0.2 0.04 0.49 0.1 0.34\n 0.27 0.35 0.09 0.89 0.13 0.16 0.18 0.38 0.46 0.6 0.28 0.57 0.8 0.12\n 0.27 0.75 0.53 0.68 0.06 0.29 1. 0.15 0.68 0.63 0.44 1. 0.33 0.16\n 0.29 0.5 0.02 0.2 0.04 0.38 0.22 0.02 0.67 0.41 0.28 1. 1. 0.28\n 0.35 1. 0.61 0.54 0.15 0.24 0.5 0.37 0.38 0.4 1. 0.39 0.12 0.6\n 0.38 0.03 0.08 0.89 1. 0.37 0.23 0.39 0.02 0.18 0.33 0.24 0.85 0.73\n 1. 0.7 0.33 0.37 1. 0.6 1. 0.59 1. 1. 0.41 0.16 0.1 0.35\n 0.67 0.51 0.28 0.56 0.5 0.39 0.02 0.56 0.53 0.63 0.59 0.16 0.05 0.19\n 0.2 1. 0.32 0.7 0.78 0.12 0.05 0.43 0.51 0.37 0.5 0.14 1. 0.27\n 0.2 1. 0.92 0.07 0.04 0.41 0.03 0.55 0.31 0.07 0.47 1. 0.07 0.34\n 0.02 0.76 0.6 0.43 0.48 0.31 0.41 0.13 0.7 0.49 0.03 0.09 0.08 0.43\n 0.34 0.26 0.15 1. 0.13 0.41 1. 0.4 0.14 0.25 0.62 0.71 0.6 0.32\n 0.72 0.24 0.33 0.45 0.48 0.75 0.1 0.38 0.3 0.09 0.22 0.23 0.26 0.51\n 0.18]\n4 [ 4 11 115 135 211 214 272 401 537 701 849 854\n 869 1427 1580 1589 1901 1993 2020 2054 2177 2488 2553 2583\n 2643 2674 2714 2969 3286 3641 3866 3875 3923 4016 4042 4379\n 4565 4708 4728 4836 4861 4943 5155 2113 5346 5438 5500 5526\n 5615 5645 5652 5837 5904 5910 6152 6242 6367 6714 6861 6886\n 6887 6967 6977 7086 7236 7399 7418 7448 7453 4791 4799 7841\n 7869 7880 7884 8073 8115 8255 8434 8509 8534 8581 8832 9135\n 6250 9251 9438 9559 6705 9806 9907 9980 10011 10150 10433 10520\n 10553 10667 10754 7999 10915 10966 11101 11110 11277 11303 11400 5622\n 11508 11683 11901 9207 12085 12144 12328 12436 12501 12518 12723 4094\n 1053 7192 12958 12976 13075 13079 13184 4610 13288 13394 13455 13527\n 13626 13637 13665 13718 14316 14329 14389 14395 14496 14565 14625 14691\n 14899 15044 15061 15134 15153 15208 15473 10234 15613 15696 15717 15737\n 15800 15811 15929 16227 16668 14256 17233 17308 17341 12287 17397 17413\n 17489 17667 12769 17890 17901 17972 15673 10383 18184 18229 18347 4851\n 18476 4928 18504 18818 8423 18961 19004 2694 8931 19323 19449 3197\n 148 19903 19932 20081 20169 20283 20325 20563 20650 13477 20791 20806\n 20863 21101 21127 21196 21366 21433 21460 21549 21612 21619 21849 67\n 22025 22059 22066 22085 22122 22138 6947 22427 22445 22607 22838 23093\n 16290 23375 23379 23614 23658 23687 5803 23739 24060 24150 12271 24229\n 24323 913 942 24783 24858 25041 25151 23081 25189 25247 25264 25340\n 25381 25452 25577 8711 25803 25838 14328 25846 25877 3069 26022 12009\n 9219 12066 145 26194 26202 26215 14814 12247 26338 26435 26489 26563\n 26597 26697 17807 26756 26813 26884 1468 18422 27243 18454 27293 27326\n 27409 18843 2486 27746 5698 27841 5757 11591 5938 28139 28293 28309\n 26286 14975 28469 12554 28546 28562 28595 28725 28788 28829 29026 29156\n 29217 27262 10875 29342 29407 29511 29605 29635 18938 29641 29654 29678\n 19058 29787 21486 29854 16939 29943 29955 29965 30246 6717 26496 847\n 30710 30752 30788 30914 5109 31306 11053 23383 31475 31480 31506 8460\n 2542 31653 5736 31697 31758 31816 31837 31893 17038 32028 26204 19643\n 32203 32324 26492 32406 32408 3941 26608 4020 32643 32793 15739 4711\n 10563 32971 32977 20602 33010 1611 33022 1674 33069 33095 33237 1975\n 23286 33365 33372 33511 33540 14140 29736 33600 33671 33708 33734 33745\n 33856 33934 33950 32081 3398 34078 34108 9509 34265 6885 28612 34385\n 34757 34801 1573 34894 1732 34983 10794 35006 31206 1879 35055 35113\n 27415 8146 35199 23409 35299 35415 35430 35464 35507 8881 35670 35693\n 35710 11918 35811 28226 35966 36155 36198 36243 10036 36318 28771 36331\n 36349 4406 10302 32805 36593 32980 36665 36679 34971 36768 33146 23172\n 36859 5148 10999 36892 5233 37025 37028 37046 37141 37186 2685 37337\n 37360 5905 19239 2912 37712 37763 19910 12556 37887 37899 37914 37923\n 37939 17724 978 38140 38236 38270 38351 38379 38385 34838 38491 33246\n 38709 11160 38803 16525 38942 38972 14234 39149 9073 39421 12282 39470\n 692 39643 38006 39730 39819 39884 15761 40024 40042 40128 4892 40251\n 40273 27460 40487 40693 40768 40771 40880 40917 40957 26185 3451 24322\n 41207 41210 41451 41461 12835 41615 41665 41775 41857 41879 41928 41965\n 27413 13747 42096 42211 27667 42492 42515 14447 42735 42792 9414 42853\n 42896 24473 43016 43125 12859 43275 43350 43401 10491 43866 43887 43952\n 44094 16871 44156 44185 40906 44341 39555 605 44619 44655 24577 44921\n 1466 43422 32979 1578 7923 45198 45216 45289 45373 8290 45637 45678\n 19258 45746 6060 45779 24010 45866 21997 17371 46297 46331 10232 46433\n 46473 46494 46495 46506 29102 22967 7777 46700 16404 46903 18946 47022\n 38954 47049 47080 47156 37401 19369 47429 47674 47761 47812 47864 7314\n 1280 48052 29125 31068 7806 48221 48270 13813 48393 11225 8328 48453\n 48471 48528 48578 48584 2658 9057 6440 48950 48966 32321 19919 49224\n 15438 49348 49364 49375 10342 49422 49474 49477 49485 15861 49516 49542\n 49556 31065 49741 49752 49892 49910 49914 18921 49986 50041 50312 100\n 193 209 3781 20160 983 50852 50956 50995 51129 51234 51275 51359\n 51424 35606 51568 11750 28162 51844 51845 51909 34248 51975 52018 52034\n 43088 26660 52110 52117 52156 15568 52262 52327 4740 10580 52415 52437\n 52453 52457 16263 52534 52607 25613 11392 27783 52753 14152 2790 2796\n 42529 28042 52924 50297 19541 12130 19633 53120 3411 374 53196 53208\n 53238 30422 53285 3996 53447 53522 18015 1584 4882 40246 1805 53832\n 43622 49727 18660 8116 2196 54082 54187 5988 54298 33831 24346 12547\n 17615 37948 54899 15688 54930 55076 36713 29373 55238 55247 36905 29553\n 13905 55366 55423 55700 48851 55741 123 51800 55883 55891 562 3728\n 26619 4000 46230 56050 15380 4289 56157 56170 7316 56250 10454 22850\n 22861 56339 56392 56411 7861 56487 56621 38748 56647 56659 56756 56758\n 44295 56996 57040 35819 19557 53138 47486 12504 57297 57315 57389 4124\n 17865 57528 4723 55069 57701 57727 56437 49790 25403 23572 42360 58134\n 58189 28004 21649 58217 48758 58265] [0.03 1. 0.62 0.29 1. 0.12 0.05 1. 0.18 0.19 0.02 0.01 0.05 0.27\n 0.43 0.42 0.03 0.43 0.11 1. 0.33 0.24 0.01 1. 0.36 0.04 1. 0.02\n 0.07 0.41 0.12 0.25 0.05 0.03 0.31 0.59 0.01 0.22 0.13 0.38 1. 0.02\n 0.14 1. 0. 0.01 0.04 0.08 0.23 0.22 0.12 0.2 0.57 0.26 0.05 0.21\n 0.88 0.22 0.71 0.57 0. 0.15 0.13 0.07 0.5 0.28 0.03 0.06 0.32 0.18\n 0.28 0.42 1. 0.13 0.14 0.04 0.14 0.22 1. 0.03 0.22 0.03 0.04 0.29\n 0.02 0. 0.41 0.19 0.49 0.15 0.35 0.11 0.23 0.26 0.55 0.01 0.33 0.05\n 0.23 0.19 0.46 0.61 0.35 0.39 0.33 0.2 0.83 0.17 0.07 0.02 0.15 0.05\n 0.4 0.11 0.61 0.15 0.2 0.54 0.01 0.1 0.13 0.01 0.05 0. 0.52 0.06\n 0.26 0.22 0.09 0.04 0.09 0.01 0. 0.01 0.04 0.35 0.24 0.13 0.28 0.78\n 0.23 0. 1. 0.04 0.22 0.47 0.57 0.19 1. 0.1 0.26 0.03 0.29 1.\n 0.18 0.1 0.35 0.04 0.09 0.08 0.02 0.29 0.18 0.09 0.02 0.41 1. 0.4\n 0.71 0.03 0.06 0.01 0.11 0.05 0.15 0.53 0.14 0.25 0.04 0.05 0.5 0.21\n 0.03 0.06 1. 0.03 0.03 0.16 0.59 0.08 0.34 0.41 0.11 0.12 0.18 0.1\n 0.39 0.05 0.34 0.02 0.19 0.18 0. 0.23 0.33 0.16 1. 0.68 0.06 0.13\n 1. 0.32 0.02 0.26 0.31 0.2 0.36 0.49 0. 0.02 0.1 0.05 1. 0.52\n 0.05 0.26 0.27 0.85 0.16 0.05 0.11 0.05 0.1 0.72 0.05 0.13 0.2 0.04\n 0.13 0.19 0.05 0.08 0.36 0.01 0.86 0.22 0.03 0.55 0.05 0.28 0.25 0.55\n 0.41 0.43 0.31 0.39 0.19 0.11 0.37 0.2 0.56 0.42 1. 0.59 0.27 0.11\n 0.25 0.02 0.2 0.4 0.01 0.33 0.03 0.02 0.12 0.32 0.07 0.08 0.04 0.13\n 0.05 0.43 0.01 1. 0.41 0.74 0.5 0.01 0.14 0.07 0.5 1. 0.7 0.35\n 0.15 0.32 1. 0.07 0.14 1. 0.06 0.62 0.04 0.06 0.47 0.04 0.24 0.03\n 0.26 0.01 1. 0.02 0. 0.36 0.29 0.02 0.16 0.15 0.1 0.04 0.24 0.41\n 0.4 0.44 0.04 0.13 0.21 0.31 0.34 0.25 0.37 0.02 0.03 0.8 0.03 0.18\n 1. 0.24 0.18 0.1 0.4 0.24 0.03 0.35 0.71 1. 0.2 0.36 0.66 0.\n 1. 0.64 0.26 0.24 0.56 0.5 0.18 0.03 0.24 0.65 0.04 0.02 0.26 0.09\n 1. 0.03 0.29 0.12 1. 0.16 0.01 0.62 0.25 0. 0.41 0.4 0.18 0.01\n 0.65 0.22 0.04 0.07 0.31 0.53 0.43 0.26 0.52 0.22 0.01 0.58 0.03 0.23\n 0.42 0.13 0.32 0.06 0.17 0.19 0.04 0.37 0.12 0.58 0.08 0.27 0. 0.35\n 0.34 0.48 0.33 0.1 1. 0.14 0.07 0.3 0.21 0. 0.08 0.24 0.38 0.52\n 0.05 1. 0.09 0.17 0.24 0.44 0.47 0.34 0.33 0.18 0.28 0.01 0.34 0.08\n 0.29 0.37 0.6 0.05 0.5 0.15 0.27 0.08 0.2 0.63 0.45 0.23 0.03 0.1\n 0.01 0.05 0.71 0.2 0.1 0.38 0.43 0.42 0.09 0. 0.1 0.02 0.64 0.5\n 1. 0.09 0.23 0.34 0.04 0.3 0.07 0.1 0.44 1. 0.04 0.02 1. 0.37\n 0.34 0.22 0.14 0.02 0.38 1. 0.07 0.39 0.31 0.41 0.15 0.02 0.87 0.09\n 0.04 0.09 0.15 0.67 0.01 0.31 0.33 0.56 0.53 0.07 0.29 0.85 0.41 0.07\n 0.27 0.01 0.1 0.12 0.03 0.69 0.28 0.04 0.09 0.03 0.39 0.29 0.6 1.\n 0.17 0.01 0.06 0.88 0.17 0.02 0.37 0.47 1. 1. 0.36 0.06 0.4 0.01\n 0.35 0.12 0.44 0.02 0.05 0.56 0.17 0.2 0.04 1. 0.14 0.26 0.59 0.14\n 0.28 0.04 0.25 0.19 0.69 0.43 0.01 0.3 0.09 0.34 0.14 0.21 0.27 0.05\n 0.26 0.43 0.8 0.51 0.42 0.05 0.1 0.03 0.25 0.05 0.19 0.36 0.08 0.27\n 1. 0.01 0.08 0.16 0.17 0.11 0.08 0.11 0.04 0.51 0.09 0.24 0.6 0.02\n 0.03 0.02 0.21 0.03 0.29 0.68 0.19 0.38 0.05 0.03 1. 0.39 0.01 0.\n 1. 0.34 0.77 0.46 0.88 0.06 0.01 0.09 0.32 0.22 0.55 0.16 0.25 0.25\n 0.91 0.6 0.25 0.14 0.25 0.37 0.24 0.12 0.03 0.28 0.32 0.42 0.39 0.16\n 0.27 0.09 0.16 0.19 0.43 0.06 0.08 0.07 0.03 0.39 0.64 0.11 0.28 0.04\n 0.22 0.23 0.05 0.08 0.01 0.16 0.08 0.39 0.26 0.1 0.09 1. 0.05 0.36\n 0.04 0.14 0.35 0.35 0.31 0.54 0. 1. 0.16 0.17 0.11 0.03 0.13 0.23\n 0.2 0.68 0.09 1. 0.08 0.33 1. 0.06 0.03 0.59 0.02 0.35 0.26 0.29\n 0.46 1. 0.71 0.41 0.01 0.22 0.64 0.06 0.23 0.15 0.32 0.38 0.38 0.07\n 0.04 0.97 0.19 0.21 0.94 0.4 0.14 0.05 0.02 0.26 0.29 0.06 0.04 0.02\n 0.36 0.13 0.58 0.18 0.91 0.75 0.01 0.51 0.23 0.01 0.3 0.03 0.34 0.02\n 0.09 0.2 0.25 0.77 0.72 0.24 0.01 0.19 0.62 0.11 0.05 0.36 0.3 0.89\n 0.44 0.23 0.81 0.77 0.46 0.01 0.01 0.39 0.15 0.02 0.07 0.11 0.18 0.3\n 0.6 1. 0.32 0.32 0.14 0.33 0.38 0.39 0.01 0.37 0.45 0.14 0.06 0.15\n 0.03 0.64 0.02 0.41 0.18 0.46 0.48 1. 0.02 0.04 0.05 0.42 0.03 0.57\n 0.03 0.89 0.08 0.6 0.29 0. 0.28 0.55 0. 0.74 1. 0.15 0.12 0.06\n 0.08 1. 0.01 1. 0.06 0.27 0.08 0.1 0.03 0.21 0.06 0.2 0.01 0.04\n 0.28 0.1 1. 0.3 0.13 0.02 0.13 0.77 1. 0.15 0.03 0.7 0.45 0.12\n 0.06 0.32 0.43 0.53 0.06 0.3 0. 0.03]\n5 [ 5 32 35 148 363 442 504 609 712 716 1332 1341\n 1378 1449 2196 2588 3374 3386 430 4312 4409 4562 4605 4678\n 5004 5150 5344 5808 5960 6131 6307 7093 1039 7298 4441 7523\n 7585 7591 7787 7809 7810 8081 8096 8642 8867 8968 9073 9148\n 9252 9364 3641 9612 9744 9908 10014 10046 10168 10360 4568 10552\n 5101 11053 11139 11625 11864 84 12479 12668 13220 13295 13588 13704\n 13840 11230 14441 14520 14992 15136 15159 12687 15293 15407 15495 15499\n 15563 15696 15962 16071 5173 16292 16331 16429 11331 16591 16628 16766\n 5850 11831 17033 17187 17325 17371 397 12373 491 17514 17810 4147\n 17834 17862 17864 17919 18652 18672 18827 18853 18967 19247 14446 19400\n 19513 6608 19884 12660 20025 20143 20158 20359 20567 20757 2042 21115\n 8513 21446 11764 21884 17283 22307 22333 22457 1040 4510 22916 4787\n 2211 23507 23555 23703 21781 24170 24396 6879 10286 25120 25138 16197\n 25333 25336 25445 18806 25485 25499 25916 26238 26331 26387 503 26555\n 20075 4299 26821 26826 4485 22845 27098 27164 13475 27304 13540 27350\n 27568 27714 27884 2749 27910 28063 28399 3595 28593 28644 15673 18378\n 11392 29697 29701 29948 29994 9157 352 22155 30461 7134 30769 10180\n 15560 10216 10256 24823 30874 30904 24985 31491 19097 31743 8868 32274\n 9807 32553 32691 32758 32779 4480 23107 33177 33503 5852 33819 33894\n 34146 15031 34318 34340 9795 32636 15769 32894 35058 35167 35200 8210\n 35295 5486 35355 25614 35471 21661 6118 35745 36015 36020 26531 10030\n 36919 5264 21147 37207 37314 2847 37395 11747 332 7170 7238 15557\n 4570 4623 15936 38428 36713 13507 1843 38615 2070 14139 39040 39349\n 3327 30307 356 34198 39500 39509 38022 39980 39994 40543 8566 41389\n 15848 34880 42376 42506 5994 42529 42567 17077 3254 30141 14945 42874\n 42924 43068 43073 7018 17816 32767 26924 43336 32929 2113 21094 44187\n 3301 44513 44604 37866 9920 44807 28792 1317 45141 45456 12276 15046\n 7013 46329 46352 26844 10675 46685 10934 5164 45537 47046 5741 39072\n 33703 11683 16994 47639 47704 47768 870 47774 47883 47908 47951 48031\n 29253 16191 2279 48494 48615 48617 48763 48907 365 20236 28824 43298\n 49510 49636 29630 3378 50913 18170 15794 51086 38645 21319 19197 51573\n 17109 48818 51733 51736 22144 3969 52020 46235 41543 52237 49460 32903\n 32939 38357 4966 5074 52642 11624 6084 52983 28209 53054 51717 53100\n 53159 53202 26522 39777 15720 39987 53605 53982 54013 2305 5679 5923\n 3528 30606 28809 4669 15967 27425 55390 30030 55740 26330 42950 52107\n 49331 7708 4874 16290 23251 33270 13942 23468 56677 27686 56716 56845\n 28011 56936 56989 17149 19800 7473 54960 57780 51142 57892 57907 57916\n 48467 19009 25706 58104 37435] [0.35 0.02 0.04 1. 0.36 0.2 0.47 0.02 1. 0.12 0.25 0.02 0.31 0.03\n 0.2 0.22 0.14 0.13 0.38 1. 0.31 0.15 0.42 0.44 0.28 0.02 0.58 0.01\n 0.14 0.02 0.02 0.59 0.46 0.48 0.34 0.35 0.28 0.13 0.08 0.09 0.4 0.24\n 0.41 0.04 0.23 0.26 1. 0.32 0.67 0.33 0.24 0.13 0.04 0.24 0.34 0.29\n 0.15 0.63 0.54 0.33 0.59 0.06 0.09 1. 0.44 1. 0.46 0.02 0.14 0.26\n 0.29 0.64 0.19 0.37 0.23 0.12 0.16 0.36 0.2 0.25 0.21 1. 0. 0.25\n 0.12 0.19 0.01 1. 0.34 0.09 0.74 1. 0.02 0.38 0.19 0.21 0.24 0.45\n 0.27 0.17 0.43 0.23 0.21 0.3 0.04 0.16 0.25 0.04 0.22 0.34 0.2 0.07\n 0.35 0.46 0.07 0.26 0.11 0.08 0.11 0.28 1. 0.05 0.2 0.64 0.25 0.44\n 0.04 0.11 0.08 0.26 0.37 1. 0.71 1. 0.33 0.66 0.41 0.38 0.23 0.11\n 0.65 0.24 0.4 0.02 0.53 0.34 1. 1. 0.23 1. 0.54 0.59 0.29 0.07\n 0.59 0.15 0.24 0.3 1. 0.32 0.01 0.14 0.49 0.14 0.17 0.15 0.02 0.75\n 0.5 0.01 0.19 0.14 0.2 0.35 0.31 0.06 0.23 0.21 0.27 0.56 0.05 0.2\n 0.22 0.08 0.34 0.41 0.46 0.46 0.2 0.51 0.41 0.39 0.33 0.29 0.77 0.21\n 0.2 0.25 1. 0.23 0.25 0.01 0.67 1. 0.06 0.38 0.24 0.42 0.21 0.1\n 0.03 0.19 0.03 0.19 1. 0.11 0.89 0.36 0.02 0.17 1. 0.28 0.18 0.29\n 0.12 0. 0.21 0.36 0.08 0.02 0.43 0.03 1. 0.15 0.2 0.22 0.23 0.32\n 0.06 0.18 1. 0.41 1. 0.27 0.03 0.52 1. 0.69 0.02 0.05 0.36 0.15\n 0.06 0.2 0.22 0.03 0.36 0.57 0.44 0.04 0.5 0.37 0.02 0.11 0.08 0.29\n 0.18 0.12 0.01 0. 0.24 0.12 0.14 0.46 0.01 0.26 0.01 0.25 0.29 0.98\n 0.21 0.18 0.08 0.11 0.71 0.23 0.49 0.35 0.24 0.02 0.4 0.14 0.02 0.01\n 0.33 0.26 0.05 0.3 0.01 0.27 0.14 0.12 1. 0.09 0.22 0.23 0.28 0.02\n 0.6 0.01 0.94 0.21 0.19 0.05 0.02 0.6 0.23 0.33 0.16 0.36 0.3 0.\n 0.03 0.23 0.01 1. 0.06 0.05 0.34 0.01 0.5 0.74 0.18 0.22 0.33 0.03\n 0.33 0.14 0.85 0.13 0.29 0.21 0.32 0.35 0.42 0.22 0.1 1. 0.27 0.12\n 0.38 0.4 0.46 0.06 0.25 1. 0.04 0.18 0.32 0.03 0.32 0.16 0.4 0.25\n 0.37 1. 0.02 1. 0.03 0.16 0.11 0.01 0.04 0.16 0.13 0.12 0.31 0.2\n 0.02 0.06 0.02 0.31 0.12 0.25 0.12 0.12 0.39 0.48 0.1 0.01 0.21 0.2\n 0.05 0.79 0.22 0.39 0.96 0. 0.5 0.06 0.12 0.32 0.12 0.77 0.46 0.15\n 0.3 0.94 0.58 0.01 0.04 0.35 0.89 1. 0.16 0.01 1. 0.48 0.68 0.68\n 0.62 0.32 0.13 0.21 0.24 0.45 0.81 1. 0.5 0.29 0.39 1. 1. 0.61\n 1. 0.21 0.29 0.17 0.07 0.17 0.81 0.04 0.03 0.01 0.04 0.39 0.04 0.44\n 0.49]\n"
],
[
"import tensorflow as tf\ngrouped_by_items = mapped_df.groupby(\"itemId\")\nwith tf.python_io.TFRecordWriter(\"data/users_for_item\") as ofp:\n for item, grouped in grouped_by_items:\n example = tf.train.Example(features = tf.train.Features(feature = {\n \"key\": tf.train.Feature(int64_list = tf.train.Int64List(value = [item])),\n \"indices\": tf.train.Feature(int64_list = tf.train.Int64List(value = grouped[\"userId\"].values)),\n \"values\": tf.train.Feature(float_list = tf.train.FloatList(value = grouped[\"rating\"].values))\n }))\n ofp.write(example.SerializeToString())",
"WARNING:tensorflow:From /tmp/ipykernel_13523/1646577164.py:3: The name tf.python_io.TFRecordWriter is deprecated. Please use tf.io.TFRecordWriter instead.\n\n"
],
[
"grouped_by_users = mapped_df.groupby(\"userId\")\nwith tf.python_io.TFRecordWriter(\"data/items_for_user\") as ofp:\n for user, grouped in grouped_by_users:\n example = tf.train.Example(features = tf.train.Features(feature = {\n \"key\": tf.train.Feature(int64_list = tf.train.Int64List(value = [user])),\n \"indices\": tf.train.Feature(int64_list = tf.train.Int64List(value = grouped[\"itemId\"].values)),\n \"values\": tf.train.Feature(float_list = tf.train.FloatList(value = grouped[\"rating\"].values))\n }))\n ofp.write(example.SerializeToString())",
"_____no_output_____"
],
[
"!ls -lrt data",
"total 36580\n-rw-r--r-- 1 jupyter jupyter 14124035 May 16 17:32 collab_raw.csv\n-rw-r--r-- 1 jupyter jupyter 3529114 May 16 17:32 users.csv\n-rw-r--r-- 1 jupyter jupyter 82083 May 16 17:32 items.csv\n-rw-r--r-- 1 jupyter jupyter 7667828 May 16 17:32 collab_mapped.csv\n-rw-r--r-- 1 jupyter jupyter 2296476 May 16 17:33 users_for_item\n-rw-r--r-- 1 jupyter jupyter 9743438 May 16 17:33 items_for_user\n"
]
],
[
[
"To summarize, you created the following data files from collab_raw.csv:\n<ol>\n<li> ```collab_mapped.csv``` is essentially the same data as in ```collab_raw.csv``` except that ```visitorId``` and ```contentId``` which are business-specific have been mapped to ```userId``` and ```itemId``` which are enumerated in 0,1,2,.... The mappings themselves are stored in ```items.csv``` and ```users.csv``` so that they can be used during inference.\n<li> ```users_for_item``` contains all the users/ratings for each item in TFExample format\n<li> ```items_for_user``` contains all the items/ratings for each user in TFExample format\n</ol>",
"_____no_output_____"
],
[
"## Train with WALS\n\nOnce you have the dataset, do matrix factorization with WALS using the [WALSMatrixFactorization](https://www.tensorflow.org/versions/r1.15/api_docs/python/tf/contrib/factorization/WALSMatrixFactorization) in the contrib directory.\nThis is an estimator model, so it should be relatively familiar.\n<p>\nAs usual, you write an input_fn to provide the data to the model, and then create the Estimator to do train_and_evaluate.\nBecause it is in contrib and hasn't moved over to tf.estimator yet, you use tf.contrib.learn.Experiment to handle the training loop.<p>\n",
"_____no_output_____"
]
],
[
[
"import os\nimport tensorflow as tf\nfrom tensorflow.python.lib.io import file_io\nfrom tensorflow.contrib.factorization import WALSMatrixFactorization\n \ndef read_dataset(mode, args):\n# TODO 2: Decode the example\n def decode_example(protos, vocab_size):\n features = {\n \"key\": tf.FixedLenFeature(shape = [1], dtype = tf.int64),\n \"indices\": tf.VarLenFeature(dtype = tf.int64),\n \"values\": tf.VarLenFeature(dtype = tf.float32)}\n parsed_features = tf.parse_single_example(serialized = protos, features = features)\n values = tf.sparse_merge(sp_ids = parsed_features[\"indices\"], sp_values = parsed_features[\"values\"], vocab_size = vocab_size)\n # Save key to remap after batching\n # This is a temporary workaround to assign correct row numbers in each batch.\n # You can ignore details of this part and remap_keys().\n key = parsed_features[\"key\"]\n decoded_sparse_tensor = tf.SparseTensor(indices = tf.concat(values = [values.indices, [key]], axis = 0), \n values = tf.concat(values = [values.values, [0.0]], axis = 0), \n dense_shape = values.dense_shape)\n return decoded_sparse_tensor\n \n \n def remap_keys(sparse_tensor):\n # Current indices of your SparseTensor that you need to fix\n bad_indices = sparse_tensor.indices # shape = (current_batch_size * (number_of_items/users[i] + 1), 2)\n # Current values of your SparseTensor that you need to fix\n bad_values = sparse_tensor.values # shape = (current_batch_size * (number_of_items/users[i] + 1),)\n\n # Since batch is ordered, the last value for a batch index is the user\n # Find where the batch index chages to extract the user rows\n # 1 where user, else 0\n user_mask = tf.concat(values = [bad_indices[1:,0] - bad_indices[:-1,0], tf.constant(value = [1], dtype = tf.int64)], axis = 0) # shape = (current_batch_size * (number_of_items/users[i] + 1), 2)\n\n # Mask out the user rows from the values\n good_values = tf.boolean_mask(tensor = bad_values, mask = tf.equal(x = user_mask, y = 0)) # shape = (current_batch_size * number_of_items/users[i],)\n item_indices = tf.boolean_mask(tensor = bad_indices, mask = tf.equal(x = user_mask, y = 0)) # shape = (current_batch_size * number_of_items/users[i],)\n user_indices = tf.boolean_mask(tensor = bad_indices, mask = tf.equal(x = user_mask, y = 1))[:, 1] # shape = (current_batch_size,)\n\n good_user_indices = tf.gather(params = user_indices, indices = item_indices[:,0]) # shape = (current_batch_size * number_of_items/users[i],)\n\n # User and item indices are rank 1, need to make rank 1 to concat\n good_user_indices_expanded = tf.expand_dims(input = good_user_indices, axis = -1) # shape = (current_batch_size * number_of_items/users[i], 1)\n good_item_indices_expanded = tf.expand_dims(input = item_indices[:, 1], axis = -1) # shape = (current_batch_size * number_of_items/users[i], 1)\n good_indices = tf.concat(values = [good_user_indices_expanded, good_item_indices_expanded], axis = 1) # shape = (current_batch_size * number_of_items/users[i], 2)\n\n remapped_sparse_tensor = tf.SparseTensor(indices = good_indices, values = good_values, dense_shape = sparse_tensor.dense_shape)\n return remapped_sparse_tensor\n\n \n def parse_tfrecords(filename, vocab_size):\n if mode == tf.estimator.ModeKeys.TRAIN:\n num_epochs = None # indefinitely\n else:\n num_epochs = 1 # end-of-input after this\n\n files = tf.gfile.Glob(filename = os.path.join(args[\"input_path\"], filename))\n\n # Create dataset from file list\n dataset = tf.data.TFRecordDataset(files)\n dataset = dataset.map(map_func = lambda x: decode_example(x, vocab_size))\n dataset = dataset.repeat(count = num_epochs)\n dataset = dataset.batch(batch_size = args[\"batch_size\"])\n dataset = dataset.map(map_func = lambda x: remap_keys(x))\n return dataset.make_one_shot_iterator().get_next()\n \n def _input_fn():\n features = {\n WALSMatrixFactorization.INPUT_ROWS: parse_tfrecords(\"items_for_user\", args[\"nitems\"]),\n WALSMatrixFactorization.INPUT_COLS: parse_tfrecords(\"users_for_item\", args[\"nusers\"]),\n WALSMatrixFactorization.PROJECT_ROW: tf.constant(True)\n }\n return features, None\n\n return _input_fn",
"_____no_output_____"
]
],
[
[
"This code is helpful in developing the input function. You don't need it in production.",
"_____no_output_____"
]
],
[
[
"def try_out():\n with tf.Session() as sess:\n fn = read_dataset(\n mode = tf.estimator.ModeKeys.EVAL, \n args = {\"input_path\": \"data\", \"batch_size\": 4, \"nitems\": NITEMS, \"nusers\": NUSERS})\n feats, _ = fn()\n \n print(feats[\"input_rows\"].eval())\n print(feats[\"input_rows\"].eval())\n\ntry_out()",
"WARNING:tensorflow:From /tmp/ipykernel_13523/171886433.py:2: The name tf.Session is deprecated. Please use tf.compat.v1.Session instead.\n\nWARNING:tensorflow:From /tmp/ipykernel_13523/3925013467.py:57: The name tf.gfile.Glob is deprecated. Please use tf.io.gfile.glob instead.\n\nWARNING:tensorflow:From /opt/conda/lib/python3.7/site-packages/tensorflow_core/python/autograph/converters/directives.py:119: The name tf.FixedLenFeature is deprecated. Please use tf.io.FixedLenFeature instead.\n\nWARNING:tensorflow:From /opt/conda/lib/python3.7/site-packages/tensorflow_core/python/autograph/converters/directives.py:119: The name tf.VarLenFeature is deprecated. Please use tf.io.VarLenFeature instead.\n\nWARNING:tensorflow:From /opt/conda/lib/python3.7/site-packages/tensorflow_core/python/autograph/converters/directives.py:119: The name tf.parse_single_example is deprecated. Please use tf.io.parse_single_example instead.\n\n"
],
[
"def find_top_k(user, item_factors, k):\n all_items = tf.matmul(a = tf.expand_dims(input = user, axis = 0), b = tf.transpose(a = item_factors))\n topk = tf.nn.top_k(input = all_items, k = k)\n return tf.cast(x = topk.indices, dtype = tf.int64)\n \ndef batch_predict(args):\n import numpy as np\n with tf.Session() as sess:\n estimator = tf.contrib.factorization.WALSMatrixFactorization(\n num_rows = args[\"nusers\"], \n num_cols = args[\"nitems\"],\n embedding_dimension = args[\"n_embeds\"],\n model_dir = args[\"output_dir\"])\n \n # This is how you would get the row factors for out-of-vocab user data\n # row_factors = list(estimator.get_projections(input_fn=read_dataset(tf.estimator.ModeKeys.EVAL, args)))\n # user_factors = tf.convert_to_tensor(np.array(row_factors))\n\n # But for in-vocab data, the row factors are already in the checkpoint\n user_factors = tf.convert_to_tensor(value = estimator.get_row_factors()[0]) # (nusers, nembeds)\n # In either case, you have to assume catalog doesn\"t change, so col_factors are read in\n item_factors = tf.convert_to_tensor(value = estimator.get_col_factors()[0])# (nitems, nembeds)\n\n # For each user, find the top K items\n topk = tf.squeeze(input = tf.map_fn(fn = lambda user: find_top_k(user, item_factors, args[\"topk\"]), elems = user_factors, dtype = tf.int64))\n with file_io.FileIO(os.path.join(args[\"output_dir\"], \"batch_pred.txt\"), mode = 'w') as f:\n for best_items_for_user in topk.eval():\n f.write(\",\".join(str(x) for x in best_items_for_user) + '\\n')\n\ndef train_and_evaluate(args):\n train_steps = int(0.5 + (1.0 * args[\"num_epochs\"] * args[\"nusers\"]) / args[\"batch_size\"])\n steps_in_epoch = int(0.5 + args[\"nusers\"] / args[\"batch_size\"])\n print(\"Will train for {} steps, evaluating once every {} steps\".format(train_steps, steps_in_epoch))\n def experiment_fn(output_dir):\n return tf.contrib.learn.Experiment(\n tf.contrib.factorization.WALSMatrixFactorization(\n num_rows = args[\"nusers\"], \n num_cols = args[\"nitems\"],\n embedding_dimension = args[\"n_embeds\"],\n model_dir = args[\"output_dir\"]),\n train_input_fn = read_dataset(tf.estimator.ModeKeys.TRAIN, args),\n eval_input_fn = read_dataset(tf.estimator.ModeKeys.EVAL, args),\n train_steps = train_steps,\n eval_steps = 1,\n min_eval_frequency = steps_in_epoch\n )\n\n from tensorflow.contrib.learn.python.learn import learn_runner\n learn_runner.run(experiment_fn = experiment_fn, output_dir = args[\"output_dir\"])\n \n batch_predict(args)",
"_____no_output_____"
],
[
"import shutil\nshutil.rmtree(path = \"wals_trained\", ignore_errors=True)\ntrain_and_evaluate({\n \"output_dir\": \"wals_trained\",\n \"input_path\": \"data/\",\n \"num_epochs\": 0.05,\n \"nitems\": NITEMS,\n \"nusers\": NUSERS,\n\n \"batch_size\": 512,\n \"n_embeds\": 10,\n \"topk\": 3\n })",
"Will train for 12 steps, evaluating once every 236 steps\nWARNING:tensorflow:From /tmp/ipykernel_13523/1819520735.py:49: run (from tensorflow.contrib.learn.python.learn.learn_runner) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse tf.estimator.train_and_evaluate.\nWARNING:tensorflow:\nThe TensorFlow contrib module will not be included in TensorFlow 2.0.\nFor more information, please see:\n * https://github.com/tensorflow/community/blob/master/rfcs/20180907-contrib-sunset.md\n * https://github.com/tensorflow/addons\n * https://github.com/tensorflow/io (for I/O related ops)\nIf you depend on functionality not listed there, please file an issue.\n\nWARNING:tensorflow:From /opt/conda/lib/python3.7/site-packages/tensorflow_core/contrib/learn/python/learn/estimators/estimator.py:1180: BaseEstimator.__init__ (from tensorflow.contrib.learn.python.learn.estimators.estimator) is deprecated and will be removed in a future version.\nInstructions for updating:\nPlease replace uses of any Estimator from tf.contrib.learn with an Estimator from tf.estimator.*\nWARNING:tensorflow:From /opt/conda/lib/python3.7/site-packages/tensorflow_core/contrib/learn/python/learn/estimators/estimator.py:427: RunConfig.__init__ (from tensorflow.contrib.learn.python.learn.estimators.run_config) is deprecated and will be removed in a future version.\nInstructions for updating:\nWhen switching to tf.estimator.Estimator, use tf.estimator.RunConfig instead.\nINFO:tensorflow:Using default config.\nINFO:tensorflow:Using config: {'_task_type': None, '_task_id': 0, '_cluster_spec': <tensorflow.python.training.server_lib.ClusterSpec object at 0x7fc3e838dfd0>, '_master': '', '_num_ps_replicas': 0, '_num_worker_replicas': 0, '_environment': 'local', '_is_chief': True, '_evaluation_master': '', '_train_distribute': None, '_eval_distribute': None, '_experimental_max_worker_delay_secs': None, '_device_fn': None, '_tf_config': gpu_options {\n per_process_gpu_memory_fraction: 1.0\n}\n, '_tf_random_seed': None, '_save_summary_steps': 100, '_save_checkpoints_secs': 600, '_log_step_count_steps': 100, '_protocol': None, '_session_config': None, '_save_checkpoints_steps': None, '_keep_checkpoint_max': 5, '_keep_checkpoint_every_n_hours': 10000, '_model_dir': 'wals_trained', '_session_creation_timeout_secs': 7200}\nWARNING:tensorflow:From /tmp/ipykernel_13523/1819520735.py:45: Experiment.__init__ (from tensorflow.contrib.learn.python.learn.experiment) is deprecated and will be removed in a future version.\nInstructions for updating:\nPlease switch to tf.estimator.train_and_evaluate. You will also have to convert to a tf.estimator.Estimator.\nWARNING:tensorflow:From /opt/conda/lib/python3.7/site-packages/tensorflow_core/contrib/learn/python/learn/monitors.py:279: BaseMonitor.__init__ (from tensorflow.contrib.learn.python.learn.monitors) is deprecated and will be removed after 2016-12-05.\nInstructions for updating:\nMonitors are deprecated. Please use tf.train.SessionRunHook.\nWARNING:tensorflow:From /opt/conda/lib/python3.7/site-packages/tensorflow_core/python/training/training_util.py:236: Variable.initialized_value (from tensorflow.python.ops.variables) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse Variable.read_value. Variables in 2.X are initialized automatically both in eager and graph (inside tf.defun) contexts.\nWARNING:tensorflow:From /opt/conda/lib/python3.7/site-packages/tensorflow_core/contrib/factorization/python/ops/wals.py:315: ModelFnOps.__new__ (from tensorflow.contrib.learn.python.learn.estimators.model_fn) is deprecated and will be removed in a future version.\nInstructions for updating:\nWhen switching to tf.estimator.Estimator, use tf.estimator.EstimatorSpec. You can use the `estimator_spec` method to create an equivalent one.\nINFO:tensorflow:Create CheckpointSaverHook.\nINFO:tensorflow:Graph was finalized.\nINFO:tensorflow:Running local_init_op.\nINFO:tensorflow:Done running local_init_op.\nINFO:tensorflow:Saving checkpoints for 0 into wals_trained/model.ckpt.\nINFO:tensorflow:SweepHook running init op.\nINFO:tensorflow:SweepHook running prep ops for the row sweep.\nINFO:tensorflow:Next fit step starting.\nINFO:tensorflow:loss = 193699.97, step = 1\nINFO:tensorflow:Next fit step starting.\nINFO:tensorflow:Next fit step starting.\nINFO:tensorflow:Next fit step starting.\nINFO:tensorflow:Next fit step starting.\nINFO:tensorflow:Next fit step starting.\nINFO:tensorflow:Next fit step starting.\nINFO:tensorflow:Next fit step starting.\nINFO:tensorflow:Next fit step starting.\nINFO:tensorflow:Next fit step starting.\nINFO:tensorflow:Next fit step starting.\nINFO:tensorflow:Next fit step starting.\nINFO:tensorflow:Saving checkpoints for 12 into wals_trained/model.ckpt.\nINFO:tensorflow:Loss for final step: 162428.39.\nINFO:tensorflow:Starting evaluation at 2022-05-16T17:40:42Z\nINFO:tensorflow:Graph was finalized.\nINFO:tensorflow:Restoring parameters from wals_trained/model.ckpt-12\nINFO:tensorflow:Running local_init_op.\nINFO:tensorflow:Done running local_init_op.\nINFO:tensorflow:Evaluation [1/1]\nINFO:tensorflow:Finished evaluation at 2022-05-16-17:40:43\nINFO:tensorflow:Saving dict for global step 12: global_step = 12, loss = 193699.97\nINFO:tensorflow:Using default config.\nINFO:tensorflow:Using config: {'_task_type': None, '_task_id': 0, '_cluster_spec': <tensorflow.python.training.server_lib.ClusterSpec object at 0x7fc3e838dfd0>, '_master': '', '_num_ps_replicas': 0, '_num_worker_replicas': 0, '_environment': 'local', '_is_chief': True, '_evaluation_master': '', '_train_distribute': None, '_eval_distribute': None, '_experimental_max_worker_delay_secs': None, '_device_fn': None, '_tf_config': gpu_options {\n per_process_gpu_memory_fraction: 1.0\n}\n, '_tf_random_seed': None, '_save_summary_steps': 100, '_save_checkpoints_secs': 600, '_log_step_count_steps': 100, '_protocol': None, '_session_config': None, '_save_checkpoints_steps': None, '_keep_checkpoint_max': 5, '_keep_checkpoint_every_n_hours': 10000, '_model_dir': 'wals_trained', '_session_creation_timeout_secs': 7200}\n"
],
[
"!ls wals_trained",
"batch_pred.txt\ncheckpoint\neval\nevents.out.tfevents.1652722839.tensorflow-1-15-20220516-225105\ngraph.pbtxt\nmodel.ckpt-0.data-00000-of-00001\nmodel.ckpt-0.index\nmodel.ckpt-0.meta\nmodel.ckpt-12.data-00000-of-00001\nmodel.ckpt-12.index\nmodel.ckpt-12.meta\n"
],
[
"!head wals_trained/batch_pred.txt",
"5526,129,885\n4712,2402,2620\n2710,139,1001\n885,5526,3450\n202,370,2839\n4972,1085,4505\n4896,1687,2833\n3418,5095,2702\n3537,479,4896\n1001,4516,2347\n"
]
],
[
[
"## Run as a Python module\n\nLet's run it as Python module for just a few steps.",
"_____no_output_____"
]
],
[
[
"os.environ[\"NITEMS\"] = str(NITEMS)\nos.environ[\"NUSERS\"] = str(NUSERS)",
"_____no_output_____"
],
[
"%%bash\nrm -rf wals.tar.gz wals_trained\ngcloud ai-platform local train \\\n --module-name=walsmodel.task \\\n --package-path=${PWD}/walsmodel \\\n -- \\\n --output_dir=${PWD}/wals_trained \\\n --input_path=${PWD}/data \\\n --num_epochs=0.01 --nitems=${NITEMS} --nusers=${NUSERS} \\\n --job-dir=./tmp",
"Will train for 2 steps, evaluating once every 236 steps\n"
]
],
[
[
"## Run on Cloud",
"_____no_output_____"
]
],
[
[
"%%bash\ngsutil -m cp data/* gs://${BUCKET}/wals/data",
"Copying file://data/collab_mapped.csv [Content-Type=text/csv]...\nCopying file://data/collab_raw.csv [Content-Type=text/csv]... \nCopying file://data/items.csv [Content-Type=text/csv]... \nCopying file://data/items_for_user [Content-Type=application/octet-stream]... \nCopying file://data/users_for_item [Content-Type=application/octet-stream]... \nCopying file://data/users.csv [Content-Type=text/csv]... \n- [6/6 files][ 35.7 MiB/ 35.7 MiB] 100% Done \nOperation completed over 6 objects/35.7 MiB. \n"
],
[
"%%bash\nOUTDIR=gs://${BUCKET}/wals/model_trained\nJOBNAME=wals_$(date -u +%y%m%d_%H%M%S)\necho $OUTDIR $REGION $JOBNAME\ngsutil -m rm -rf $OUTDIR\ngcloud ai-platform jobs submit training $JOBNAME \\\n --region=$REGION \\\n --module-name=walsmodel.task \\\n --package-path=${PWD}/walsmodel \\\n --job-dir=$OUTDIR \\\n --staging-bucket=gs://$BUCKET \\\n --scale-tier=BASIC_GPU \\\n --runtime-version=$TFVERSION \\\n -- \\\n --output_dir=$OUTDIR \\\n --input_path=gs://${BUCKET}/wals/data \\\n --num_epochs=10 --nitems=${NITEMS} --nusers=${NUSERS} ",
"gs://qwiklabs-gcp-01-7f6e984e70dc/wals/model_trained us-central1 wals_220516_174236\njobId: wals_220516_174236\nstate: QUEUED\n"
]
],
[
[
"This will take <b>10 minutes</b> to complete. Rerun the above command until the jobs gets submitted.",
"_____no_output_____"
],
[
"## Get row and column factors\n\nOnce you have a trained WALS model, you can get row and column factors (user and item embeddings) from the checkpoint file. You'll look at how to use these in the section on building a recommendation system using deep neural networks.",
"_____no_output_____"
]
],
[
[
"def get_factors(args):\n with tf.Session() as sess:\n estimator = tf.contrib.factorization.WALSMatrixFactorization(\n num_rows = args[\"nusers\"], \n num_cols = args[\"nitems\"],\n embedding_dimension = args[\"n_embeds\"],\n model_dir = args[\"output_dir\"])\n \n row_factors = estimator.get_row_factors()[0]\n col_factors = estimator.get_col_factors()[0]\n return row_factors, col_factors",
"_____no_output_____"
],
[
"args = {\n \"output_dir\": \"gs://{}/wals/model_trained\".format(BUCKET),\n \"nitems\": NITEMS,\n \"nusers\": NUSERS,\n \"n_embeds\": 10\n }\n\nuser_embeddings, item_embeddings = get_factors(args)\nprint(user_embeddings[:3])\nprint(item_embeddings[:3])",
"INFO:tensorflow:Using default config.\nINFO:tensorflow:Using config: {'_task_type': None, '_task_id': 0, '_cluster_spec': <tensorflow.python.training.server_lib.ClusterSpec object at 0x7fc3e9a79a10>, '_master': '', '_num_ps_replicas': 0, '_num_worker_replicas': 0, '_environment': 'local', '_is_chief': True, '_evaluation_master': '', '_train_distribute': None, '_eval_distribute': None, '_experimental_max_worker_delay_secs': None, '_device_fn': None, '_tf_config': gpu_options {\n per_process_gpu_memory_fraction: 1.0\n}\n, '_tf_random_seed': None, '_save_summary_steps': 100, '_save_checkpoints_secs': 600, '_log_step_count_steps': 100, '_protocol': None, '_session_config': None, '_save_checkpoints_steps': None, '_keep_checkpoint_max': 5, '_keep_checkpoint_every_n_hours': 10000, '_model_dir': 'gs://qwiklabs-gcp-01-7f6e984e70dc/wals/model_trained', '_session_creation_timeout_secs': 7200}\n[[ 3.6948849e-03 -8.5906551e-04 4.8303331e-04 -1.5890118e-03\n 3.2951573e-03 1.8742392e-03 -4.0969993e-03 6.1582108e-03\n -2.8773610e-04 -1.3332820e-03]\n [ 1.3797252e-03 -6.1344338e-04 1.8084486e-03 -1.3052947e-03\n -5.8687606e-04 1.2303573e-03 4.5222897e-04 1.5270878e-03\n -3.6727260e-03 -3.0614049e-04]\n [-1.8781899e-04 -3.4702761e-04 -2.0322317e-05 -3.4224652e-04\n -4.4325300e-04 -1.1415767e-03 1.1274662e-04 5.7519245e-04\n 1.1694311e-04 7.0931134e-04]]\n[[ 1.55777712e+01 -1.05091429e+00 9.74276304e-01 -1.42321882e+01\n 5.99148512e+00 2.02531338e+00 -1.14337492e+01 1.73737488e+01\n -6.27120638e+00 -3.14125824e+00]\n [ 1.13881841e-01 -1.24881916e-01 1.47017851e-01 3.90907042e-02\n -2.54981425e-02 -2.25878507e-02 -8.38336051e-02 -3.10666151e-02\n -1.92982569e-01 5.66481380e-03]\n [-2.36634836e-01 -1.71446967e+00 -2.04065472e-01 -2.04369950e+00\n -1.53748310e+00 -4.64286995e+00 1.42189160e-01 2.47772884e+00\n -9.40964222e-01 3.68809652e+00]]\n"
]
],
[
[
"You can visualize the embedding vectors using dimensional reduction techniques such as PCA.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nfrom mpl_toolkits.mplot3d import Axes3D\nfrom sklearn.decomposition import PCA\n\npca = PCA(n_components = 3)\npca.fit(user_embeddings)\n# TODO 3: Apply the mapping (transform) to user embeddings\nuser_embeddings_pca = pca.transform(user_embeddings)\n\nfig = plt.figure(figsize = (8,8))\nax = fig.add_subplot(111, projection = \"3d\")\nxs, ys, zs = user_embeddings_pca[::150].T\nax.scatter(xs, ys, zs)",
"_____no_output_____"
]
],
[
[
"<pre>\n# Copyright 2022 Google Inc. All Rights Reserved.\n#\n# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# http://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.\n</pre>",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d092bb10a2d398e399aa9cccfc2a28bf0e9843fd | 23,326 | ipynb | Jupyter Notebook | content/materials/notebooks/2020/l13_arbitrary_inputs.ipynb | moorepants/eng122 | 7bcd502eae4ab0ec9d463389acca2290b263ba64 | [
"CC-BY-4.0"
] | 2 | 2020-05-11T05:56:54.000Z | 2020-12-20T13:44:12.000Z | content/materials/notebooks/2020/l13_arbitrary_inputs.ipynb | moorepants/eng122 | 7bcd502eae4ab0ec9d463389acca2290b263ba64 | [
"CC-BY-4.0"
] | 12 | 2016-09-20T01:26:33.000Z | 2020-01-22T02:10:33.000Z | content/materials/notebooks/2020/l13_arbitrary_inputs.ipynb | moorepants/eng122 | 7bcd502eae4ab0ec9d463389acca2290b263ba64 | [
"CC-BY-4.0"
] | 4 | 2017-06-09T01:28:45.000Z | 2020-02-29T02:36:18.000Z | 35.449848 | 1,457 | 0.557618 | [
[
[
"import numpy as np\nfrom resonance.nonlinear_systems import SingleDoFNonLinearSystem",
"_____no_output_____"
]
],
[
[
"To apply arbitrary forcing to a single degree of freedom linear or nonlinear system, you can do so with `SingleDoFNonLinearSystem` (`SingleDoFLinearSystem` does not support arbitrary forcing...yet).\n\nAdd constants, a generalized coordinate, and a generalized speed to the system.",
"_____no_output_____"
]
],
[
[
"sys = SingleDoFNonLinearSystem()",
"_____no_output_____"
],
[
"sys.constants['m'] = 100 # kg\nsys.constants['c'] = 1.1*1.2*0.5/2\nsys.constants['k'] = 10\nsys.constants['Fo'] = 1000 # N\nsys.constants['Ft'] = 100 # N/s\nsys.constants['to'] = 3.0 # s\n\nsys.coordinates['x'] = 0.0\nsys.speeds['v'] = 0.0",
"_____no_output_____"
]
],
[
[
"Create a function that evaluates the first order form of the non-linear equations of motion. In this case:\n\n$$\n\\dot{x} = v \\\\\nm\\dot{v} + c \\textrm{sgn}(v)v^2 + k \\textrm{sgn}(x)x^2 = F(t)\n$$\n\nMake the arbitrary forcing term, $F$, an input to this function.",
"_____no_output_____"
]
],
[
[
"def eval_eom(x, v, m, c, k, F):\n xdot = v\n vdot = (F - np.sign(v)*c*v**2 - np.sign(x)*k*x**2) / m\n return xdot, vdot",
"_____no_output_____"
]
],
[
[
"Note that you cannot add this to the system because `F` has not been defined.",
"_____no_output_____"
]
],
[
[
"sys.diff_eq_func = eval_eom",
"_____no_output_____"
]
],
[
[
"To rememdy this, create a function that returns the input value given the appropriate constants and time.",
"_____no_output_____"
]
],
[
[
"def eval_step_input(Fo, to, time):\n if time < to:\n return 0.0\n else:\n return Fo",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n%matplotlib widget",
"_____no_output_____"
],
[
"ts = np.linspace(0, 10)\nplt.plot(ts, eval_step_input(5.0, 3.0, ts))",
"_____no_output_____"
],
[
"ts < 3.0",
"_____no_output_____"
],
[
"def eval_step_input(Fo, to, time):\n F = np.empty_like(time)\n for i, ti in enumerate(time):\n if ti < to:\n F[i] = 0.0\n else:\n F[i] = Fo\n return F",
"_____no_output_____"
],
[
"plt.plot(ts, eval_step_input(5.0, 3.0, ts))",
"_____no_output_____"
],
[
"eval_step_input(5.0, 3.0, ts)",
"_____no_output_____"
],
[
"eval_step_input(5.0, 3.0, 7.0)",
"_____no_output_____"
],
[
"def eval_step_input(Fo, to, time):\n if np.isscalar(time):\n if time < to:\n return 0.0\n else:\n return Fo\n else:\n F = np.empty_like(time)\n for i, ti in enumerate(time):\n if ti < to:\n F[i] = 0.0\n else:\n F[i] = Fo\n return F",
"_____no_output_____"
],
[
"eval_step_input(5.0, 3.0, 7.0)",
"_____no_output_____"
],
[
"eval_step_input(5.0, 3.0, ts)",
"_____no_output_____"
],
[
"True * 5.0",
"_____no_output_____"
],
[
"False * 5.0",
"_____no_output_____"
],
[
"(ts >= 3.0)*5.0",
"_____no_output_____"
],
[
"(5.0 >= 3.0)*5.0",
"_____no_output_____"
],
[
"def eval_step_input(Fo, to, time):\n return (time >=to)*Fo",
"_____no_output_____"
],
[
"eval_step_input(5.0, 3.0, ts)",
"_____no_output_____"
],
[
"eval_step_input(5.0, 3.0, 7.0)",
"_____no_output_____"
],
[
"sys.add_measurement('F', eval_step_input)",
"_____no_output_____"
],
[
"sys.diff_eq_func = eval_eom",
"_____no_output_____"
],
[
"traj = sys.free_response(20.0)",
"_____no_output_____"
],
[
"traj.plot(subplots=True)",
"_____no_output_____"
],
[
"def eval_ramp_input(Ft, to, time):\n return (time >= to)*(Ft*time - Ft*to)",
"_____no_output_____"
],
[
"del sys.measurements['F']",
"_____no_output_____"
],
[
"sys.add_measurement('F', eval_ramp_input)",
"_____no_output_____"
],
[
"sys.measurements",
"_____no_output_____"
],
[
"traj = sys.free_response(20.0)",
"_____no_output_____"
],
[
"traj.plot(subplots=True)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d092cde0c4d9b0aca71abd8960b235fc5a5462c0 | 5,392 | ipynb | Jupyter Notebook | examples/SGTPy's paper notebooks/Fit Equilibrium Ethanol + CPME.ipynb | MatKie/SGTPy | 8e98d92fedd2b07d834e547e5154ec8f70d80728 | [
"MIT"
] | 12 | 2020-12-27T17:04:33.000Z | 2021-07-19T06:28:28.000Z | examples/SGTPy's paper notebooks/Fit Equilibrium Ethanol + CPME.ipynb | MatKie/SGTPy | 8e98d92fedd2b07d834e547e5154ec8f70d80728 | [
"MIT"
] | 2 | 2021-05-15T14:27:57.000Z | 2021-08-19T15:42:24.000Z | examples/SGTPy's paper notebooks/Fit Equilibrium Ethanol + CPME.ipynb | MatKie/SGTPy | 8e98d92fedd2b07d834e547e5154ec8f70d80728 | [
"MIT"
] | 5 | 2021-02-21T01:33:29.000Z | 2021-07-26T15:11:08.000Z | 31.717647 | 241 | 0.523739 | [
[
[
"# Fit $k_{ij}$ and $r_c^{ABij}$ interactions parameter of Ethanol and CPME\n\nThis notebook has te purpose of showing how to optimize the $k_{ij}$ and $r_c^{ABij}$ for a mixture with induced association.\n\nFirst it's needed to import the necessary modules",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nfrom sgtpy import component, mixture, saftvrmie\nfrom sgtpy.fit import fit_cross",
"_____no_output_____"
]
],
[
[
"Now that the functions are available it is necessary to create the mixture.",
"_____no_output_____"
]
],
[
[
"ethanol = component('ethanol2C', ms = 1.7728, sigma = 3.5592 , eps = 224.50,\n lambda_r = 11.319, lambda_a = 6., eAB = 3018.05, rcAB = 0.3547,\n rdAB = 0.4, sites = [1,0,1], cii= 5.3141080872882285e-20)\n\ncpme = component('cpme', ms = 2.32521144, sigma = 4.13606074, eps = 343.91193798, lambda_r = 14.15484877, \n lambda_a = 6.0, npol = 1.91990385,mupol = 1.27, sites =[0,0,1], cii = 3.5213681817448466e-19)\n\nmix = mixture(ethanol, cpme)",
"_____no_output_____"
]
],
[
[
"Now the experimental equilibria data is read and a tuple is created. It includes the experimental liquid composition, vapor composition, equilibrium temperature and pressure. This is done with ```datavle = (Xexp, Yexp, Texp, Pexp)```\n",
"_____no_output_____"
]
],
[
[
"# Experimental data obtained from Mejia, Cartes, J. Chem. Eng. Data, vol. 64, no. 5, pp. 1970–1977, 2019\n\n# Experimental temperature saturation in K\nTexp = np.array([355.77, 346.42, 342.82, 340.41, 338.95, 337.78, 336.95, 336.29,\n 335.72, 335.3 , 334.92, 334.61, 334.35, 334.09, 333.92, 333.79,\n 333.72, 333.72, 333.81, 334.06, 334.58])\n\n# Experimental pressure in Pa\nPexp = np.array([50000., 50000., 50000., 50000., 50000., 50000., 50000., 50000.,\n 50000., 50000., 50000., 50000., 50000., 50000., 50000., 50000.,\n 50000., 50000., 50000., 50000., 50000.])\n\n# Experimental liquid composition\nXexp = np.array([[0. , 0.065, 0.11 , 0.161, 0.203, 0.253, 0.301, 0.351, 0.402,\n 0.446, 0.497, 0.541, 0.588, 0.643, 0.689, 0.743, 0.785, 0.837,\n 0.893, 0.947, 1. ],\n [1. , 0.935, 0.89 , 0.839, 0.797, 0.747, 0.699, 0.649, 0.598,\n 0.554, 0.503, 0.459, 0.412, 0.357, 0.311, 0.257, 0.215, 0.163,\n 0.107, 0.053, 0. ]])\n\n# Experimental vapor composition\nYexp = np.array([[0. , 0.302, 0.411, 0.48 , 0.527, 0.567, 0.592, 0.614, 0.642,\n 0.657, 0.678, 0.694, 0.71 , 0.737, 0.753, 0.781, 0.801, 0.837,\n 0.883, 0.929, 1. ],\n [1. , 0.698, 0.589, 0.52 , 0.473, 0.433, 0.408, 0.386, 0.358,\n 0.343, 0.322, 0.306, 0.29 , 0.263, 0.247, 0.219, 0.199, 0.163,\n 0.117, 0.071, 0. ]])\n\ndatavle = (Xexp, Yexp, Texp, Pexp)",
"_____no_output_____"
]
],
[
[
"The function ```fit_cross``` optimize the $k_{ij}$ correction and $r_c^{ABij}$ distance. An initial guess is needed, as well as the mixture object, the index of the self-associating component and the equilibria data.",
"_____no_output_____"
]
],
[
[
"#initial guesses for kij and rcij\nx0 = [0.01015194, 2.23153033]\nfit_cross(x0, mix, assoc=0, datavle=datavle)",
"_____no_output_____"
]
],
[
[
"For more information just run:\n```fit_cross?```",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d092d384f32019d2124226ae9f7f1fbb7c9d288a | 23,119 | ipynb | Jupyter Notebook | ML SERIES - NAIVE BAYES/MACHINE LEARNING TOP ALGORITHMS - NAIVE BAYES CLASSIFIER.ipynb | acadena-repo/MACHINE-LEARNING-TOP-ALGORITHMS-SERIES | 9396b927194bf50225b33c5cf3263f956d7972ab | [
"MIT"
] | null | null | null | ML SERIES - NAIVE BAYES/MACHINE LEARNING TOP ALGORITHMS - NAIVE BAYES CLASSIFIER.ipynb | acadena-repo/MACHINE-LEARNING-TOP-ALGORITHMS-SERIES | 9396b927194bf50225b33c5cf3263f956d7972ab | [
"MIT"
] | null | null | null | ML SERIES - NAIVE BAYES/MACHINE LEARNING TOP ALGORITHMS - NAIVE BAYES CLASSIFIER.ipynb | acadena-repo/MACHINE-LEARNING-TOP-ALGORITHMS-SERIES | 9396b927194bf50225b33c5cf3263f956d7972ab | [
"MIT"
] | null | null | null | 30.300131 | 456 | 0.470695 | [
[
[
"# BAYES CLASSIFIERS\n\nFor any classifier $f:{X \\to Y}$, it's prediction error is:\n\n$P(f(x) \\ne Y) = \\mathbb{E}[ \\mathbb{1}(f(X) \\ne Y)] = \\mathbb{E}[\\mathbb{E}[ \\mathbb{1}(f(X) \\ne Y)|X]]$\n\nFor each $x \\in X$,\n\n$$\\mathbb{E}[ \\mathbb{1}(f(X) \\ne Y)|X = x] = \\sum\\limits_{y \\in Y} P(Y = y|X = x) \\cdot \\mathbb{1}(f(x) \\ne y)$$\n\nThe above quantity is minimized for this particular $x \\in X$ when,\n\n$$f(x) = \\underset{y \\in Y}{argmax} \\space P(Y = y|X = x) \\space \\star$$\n\nA classifier $f$ with property $ \\star$ for all $x \\in X$ is called the `Bayes Classifier`\n",
"_____no_output_____"
],
[
"Under the assumption $(X,Y) \\overset{iid}{\\sim} P$, the optimal classifier is:\n$$f^{\\star}(x) = \\underset{y \\in Y}{argmax} \\space P(Y = y|X = x)$$\n\nAnd from _Bayes Rule_ we equivalently have:\n\n$$f^{\\star}(x) = \\underset{y \\in Y}{argmax} \\space P(Y = y) \\space P(X = x|Y = y)$$\n\nWhere\n- $P(Y =y)$ is called _the class prior_\n- $P(X = x|Y= y)$ is called _the class conditional distribution_ of $X$\n\nAssuming $X = \\mathbb{R}, Y = \\{ 0,1 \\}$, and the distribution of $P \\space \\text{of} \\space (X,Y)$ is as follows:\n- _Class prior_: $P(Y = y) = \\pi_y, y \\in \\{ 0,1 \\}$\n- _Class conditional density_ for class $y \\in \\{ 0,1 \\}: p_y (x) = N(x|\\mu_y,\\sigma^2_y)$\n\n$$f^{\\star}(x) = \\underset{y \\in \\{ 0,1 \\}}{argmax} \\space P(Y = y) \\space P(X = x|Y = y) = \n \\begin{cases}\n 1 & \\text{if} \\space \\frac{\\pi_1}{\\sigma_1}\\space exp[- \\frac{(x - \\mu_1)^2}{2 \\sigma^2_1}] > \\frac{\\pi_0}{\\sigma_0}\\space exp[- \\frac{(x - \\mu_0)^2}{2 \\sigma^2_0}]\\\\\n 0 & \\text{otherwise}\n \\end{cases}$$",
"_____no_output_____"
],
[
"### _Bayes Classifier_\n",
"_____no_output_____"
],
[
"The `Bayes Classifier` has the smallest prediction error of all classifiers. The problem is that we need to know the distribution of $P$ in order to construct the `Bayes Classifier`",
"_____no_output_____"
],
[
"# NAIVE BAYES CLASSIFIER\n\nA simplifying assumtion that the features values are conditionally independent given the label, the probability of observing the conjunction $x_1, x_2, x_3, ..., x_d$ is the product of the probabilities for the individual features:\n\n$$ p(x_1, x_2, x_3, ..., x_d|y) = \\prod \\limits_j \\space p(x_j|y)$$\n\nThen the `Naive Bayes Classifier` is defined as:\n\n$$f^{\\star}(x) = \\underset{y \\in Y}{argmax} \\space p(y) \\space \\prod \\limits_j \\space p(x_j|y)$$\n\nWe can estimate these two terms based on the **frequency counts** in the dataset. If the features are real-valued, Naive Bayes can be extended assuming that features follow a Gaussian distribution. This extension is called `Gaussian Naive Bayes`. Other functions can be used to estimate the distribution but the Gaussian distribution is the easiest to work with due to we only need to estimate the mean and the standard deviation from the dataset.\n\nOk, let's start with the implementation of `Gaussian Naive Bayes` from scratch.",
"_____no_output_____"
]
],
[
[
"##IMPORTING ALL NECESSARY SUPPORT LIBRARIES\nimport math as mt\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"def separate_by_label(dataset):\n separate = dict()\n for i in range(len(dataset)):\n row = dataset[i]\n label = row[-1]\n if (label not in separate):\n separate[label] = list()\n separate[label].append(row)\n \n return separate",
"_____no_output_____"
],
[
"def mean(list_num):\n return sum(list_num)/len(list_num)",
"_____no_output_____"
],
[
"def stdv(list_num):\n mu = mean(list_num)\n var = sum([(x - mu)**2 for x in list_num])/(len(list_num) - 1)\n \n return mt.sqrt(var)",
"_____no_output_____"
],
[
"def stats_per_feature(ds):\n '''\n argument:\n > ds: 1-D Array with the all data separated by class\n returns:\n > stats: 1-D Array with statistics summary for each feature\n '''\n stats = [(mean(col), stdv(col), len(col)) for col in zip(*ds)]\n del(stats[-1])\n \n return stats",
"_____no_output_____"
],
[
"def summary_by_class(dataset):\n sep_label = separate_by_label(dataset)\n summary = dict()\n for label, rows in sep_label.items():\n summary[label] = stats_per_feature(rows)\n \n return summary",
"_____no_output_____"
],
[
"def gaussian_pdf(mean, stdv, x):\n _exp = mt.exp(-1*((x - mean)**2/(2*stdv**2)))\n return (1/(mt.sqrt(2 * mt.pi)*stdv)) * _exp",
"_____no_output_____"
]
],
[
[
"Now it is time to use the statistics calculated from the data to calculate probabilities for new data.\n\nProbabilities are calculated separately for each class, so we calculate the probability that a new piece of data belongs to the first class, then calculate the probability that it belongs to the second class, and so on for all the classes.\n\nFor example, if we have two inputs $x_1 and \\space x_2$ the calculation of the probability that those belong to class = _y_ is:\n\n$$P(class = y|x_1,x_2) = P(x_1|class = y) \\cdot P(x_2|class = y) \\cdot P(class = y)$$",
"_____no_output_____"
]
],
[
[
"def class_probabilities(summary, row):\n total = sum([summary[label][0][2] for label in summary])\n probabilities = dict()\n \n for class_, class_summary in summary.items():\n probabilities[class_] = summary[class_][0][2]/total\n for i in range(len(class_summary)):\n mean, stdev, count = class_summary[i]\n probabilities[class_] *= gaussian_pdf(row[i], mean, stdev)\n \n return probabilities",
"_____no_output_____"
],
[
"def predict(summary, row):\n cls_prob = class_probabilities(summary, row)\n _label, _prob = None, -1.0\n for class_, probability in cls_prob.items():\n if _label is None or probability > _prob:\n _prob = probability\n _label = class_\n \n return _label ",
"_____no_output_____"
]
],
[
[
"In order to verify proper implementation a **toy dataset** is used to evaluate the algorithm.",
"_____no_output_____"
]
],
[
[
"dataset = [[3.393533211,2.331273381,0],\n[3.110073483,1.781539638,0],\n[1.343808831,3.368360954,0],\n[3.582294042,4.67917911,0],\n[2.280362439,2.866990263,0],\n[7.423436942,4.696522875,1],\n[5.745051997,3.533989803,1],\n[9.172168622,2.511101045,1],\n[7.792783481,3.424088941,1],\n[7.939820817,0.791637231,1]]\n\nsummaries = summary_by_class(dataset)\nfor row in dataset:\n y_pred = predict(summaries, row)\n y_real = row[-1]\n print(\"Expected={0}, Predicted={1}\".format(y_real, y_pred))",
"Expected=0, Predicted=0\nExpected=0, Predicted=0\nExpected=0, Predicted=0\nExpected=0, Predicted=0\nExpected=0, Predicted=0\nExpected=1, Predicted=1\nExpected=1, Predicted=1\nExpected=1, Predicted=1\nExpected=1, Predicted=1\nExpected=1, Predicted=1\n"
]
],
[
[
"# _GAUSSIAN NAIVE BAYES APPLICATION_\n\nFrom the `UCI Machine Learning Repository` which contains Iris dataset, we will train our `Gaussian Naive Bayes` model. The Iris dataset is perhaps the best known database to be found in the pattern recognition literature. Fisher's paper is a classic in the field and is referenced frequently to this day.\n\nThe dataset contains 3 classes of 50 instances each, where each class refers to a type of iris plant. One class is linearly separable from the other 2; the latter are not linearly separable from each other.\n\nThe dataset have 150 instances and the following attributes:\n\n 1. sepal length in cm\n 2. sepal width in cm\n 3. petal length in cm\n 4. petal width in cm\n 5. class: \n -- Iris Setosa\n -- Iris Versicolour\n -- Iris Virginica",
"_____no_output_____"
],
[
"To compare the performance of our _Classifier_ on the **Iris** dataset, a Gaussian Naive Bayes model from `sklearn` will be fit on the dataset and classification report for both models is generated.",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\nfrom sklearn.naive_bayes import GaussianNB\nfrom sklearn.metrics import classification_report",
"_____no_output_____"
],
[
"##LOADING 'IRIS' DATASET\ncolumns = ['sepal-len','sepal-wid','petal-len','petal-wid','class']\ndf = pd.read_csv('./data/Iris.csv', names = columns)\ndf.head()",
"_____no_output_____"
],
[
"df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 150 entries, 0 to 149\nData columns (total 5 columns):\nsepal-len 150 non-null float64\nsepal-wid 150 non-null float64\npetal-len 150 non-null float64\npetal-wid 150 non-null float64\nclass 150 non-null object\ndtypes: float64(4), object(1)\nmemory usage: 5.9+ KB\n"
]
],
[
[
"Due to the class variable type is `categorical` we need first to encode it as numeric type in order to be feed it into our models.",
"_____no_output_____"
]
],
[
[
"def encoder(df, class_value_pair):\n for class_name, value in class_value_pair.items():\n df['class'] = df['class'].replace(class_name, value)\n \n return df\nclass_encoder = {'Iris-setosa':0, 'Iris-versicolor':1, 'Iris-virginica':2}\ndf = encoder(df, class_encoder)\ndf.head()",
"_____no_output_____"
],
[
"df['class'].value_counts().sort_index()",
"_____no_output_____"
]
],
[
[
"Once the preprocessing is complete the dataset will be split into a `Training` & `Test` dataset.",
"_____no_output_____"
]
],
[
[
"X_ = df.drop(['class'],axis = 1)\ny = df['class']\nX_train, X_test, y_train, y_test = train_test_split(X_, y, test_size = 0.30, random_state = 5)",
"_____no_output_____"
]
],
[
[
"Now, we can `train` our customized model. Noticed that our _Gaussian Naive Bayes_ model expects a complete dataset (attributes and labels) in order to calculate the summaries.",
"_____no_output_____"
]
],
[
[
"ds_train = pd.concat([X_train, y_train], axis = 1)\nGNB_custom = summary_by_class(ds_train.values.tolist())",
"_____no_output_____"
],
[
"ds_test = pd.concat([X_test, y_test], axis = 1)\ncust_pred = [predict(GNB_custom, row) for row in ds_test.values.tolist()]\ncust_pred = np.array(cust_pred, dtype = 'int64')",
"_____no_output_____"
],
[
"cust_pred",
"_____no_output_____"
]
],
[
[
"Now an instance of `sklearn` _Gaussian Naive Bayes_ model is created and fit it with the training data and an array of predictions is obtained in order to get out performance comparation",
"_____no_output_____"
]
],
[
[
"##GET AND INSTANCE OF GAUSSIAN NAIVE BAYES MODEL\nGNB_skln = GaussianNB()\nGNB_skln.fit(X_train, y_train)\n\n##CREATE SKLEARN PREDICTIONS ARRAY\nsk_pred = GNB_skln.predict(X_test)",
"_____no_output_____"
],
[
"sk_pred",
"_____no_output_____"
]
],
[
[
"By last, a comparison on both models is performed thru a _Classification Report_",
"_____no_output_____"
]
],
[
[
"print(\"Sklearn:\")\nprint(classification_report(y_test, sk_pred))\nprint(\"Custom:\")\nprint(classification_report(y_test, cust_pred))",
"Sklearn:\n precision recall f1-score support\n\n 0 1.00 1.00 1.00 15\n 1 0.88 0.94 0.91 16\n 2 0.92 0.86 0.89 14\n\n micro avg 0.93 0.93 0.93 45\n macro avg 0.94 0.93 0.93 45\nweighted avg 0.93 0.93 0.93 45\n\nCustom:\n precision recall f1-score support\n\n 0 1.00 1.00 1.00 15\n 1 0.88 0.94 0.91 16\n 2 0.92 0.86 0.89 14\n\n micro avg 0.93 0.93 0.93 45\n macro avg 0.94 0.93 0.93 45\nweighted avg 0.93 0.93 0.93 45\n\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d092d4685763cc9357c5b3e903adfa0b41f87ecb | 269,695 | ipynb | Jupyter Notebook | ReinforcementLearning/TemporalDifference/Temporal_Difference_Solution.ipynb | xuebai1990/DeepLearningNanodegree | 2bf06d24ae5eb9dfabeb764949704bfb8867351f | [
"MIT"
] | 7 | 2019-03-18T01:02:18.000Z | 2020-01-04T18:17:52.000Z | ReinforcementLearning/TemporalDifference/Temporal_Difference_Solution.ipynb | xuebai1990/DeepLearningNanodegree | 2bf06d24ae5eb9dfabeb764949704bfb8867351f | [
"MIT"
] | null | null | null | ReinforcementLearning/TemporalDifference/Temporal_Difference_Solution.ipynb | xuebai1990/DeepLearningNanodegree | 2bf06d24ae5eb9dfabeb764949704bfb8867351f | [
"MIT"
] | 2 | 2019-03-18T08:22:09.000Z | 2019-03-19T05:14:40.000Z | 316.915394 | 47,736 | 0.915475 | [
[
[
"# Mini Project: Temporal-Difference Methods\n\nIn this notebook, you will write your own implementations of many Temporal-Difference (TD) methods.\n\nWhile we have provided some starter code, you are welcome to erase these hints and write your code from scratch.",
"_____no_output_____"
],
[
"### Part 0: Explore CliffWalkingEnv\n\nUse the code cell below to create an instance of the [CliffWalking](https://github.com/openai/gym/blob/master/gym/envs/toy_text/cliffwalking.py) environment.",
"_____no_output_____"
]
],
[
[
"import gym\nenv = gym.make('CliffWalking-v0')",
"_____no_output_____"
]
],
[
[
"The agent moves through a $4\\times 12$ gridworld, with states numbered as follows:\n```\n[[ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11],\n [12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23],\n [24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35],\n [36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47]]\n```\nAt the start of any episode, state `36` is the initial state. State `47` is the only terminal state, and the cliff corresponds to states `37` through `46`.\n\nThe agent has 4 potential actions:\n```\nUP = 0\nRIGHT = 1\nDOWN = 2\nLEFT = 3\n```\n\nThus, $\\mathcal{S}^+=\\{0, 1, \\ldots, 47\\}$, and $\\mathcal{A} =\\{0, 1, 2, 3\\}$. Verify this by running the code cell below.",
"_____no_output_____"
]
],
[
[
"print(env.action_space)\nprint(env.observation_space)",
"Discrete(4)\nDiscrete(48)\n"
]
],
[
[
"In this mini-project, we will build towards finding the optimal policy for the CliffWalking environment. The optimal state-value function is visualized below. Please take the time now to make sure that you understand _why_ this is the optimal state-value function.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom plot_utils import plot_values\n\n# define the optimal state-value function\nV_opt = np.zeros((4,12))\nV_opt[0:13][0] = -np.arange(3, 15)[::-1]\nV_opt[0:13][1] = -np.arange(3, 15)[::-1] + 1\nV_opt[0:13][2] = -np.arange(3, 15)[::-1] + 2\nV_opt[3][0] = -13\n\nplot_values(V_opt)",
"_____no_output_____"
]
],
[
[
"### Part 1: TD Prediction: State Values\n\nIn this section, you will write your own implementation of TD prediction (for estimating the state-value function).\n\nWe will begin by investigating a policy where the agent moves:\n- `RIGHT` in states `0` through `10`, inclusive, \n- `DOWN` in states `11`, `23`, and `35`, and\n- `UP` in states `12` through `22`, inclusive, states `24` through `34`, inclusive, and state `36`.\n\nThe policy is specified and printed below. Note that states where the agent does not choose an action have been marked with `-1`.",
"_____no_output_____"
]
],
[
[
"policy = np.hstack([1*np.ones(11), 2, 0, np.zeros(10), 2, 0, np.zeros(10), 2, 0, -1*np.ones(11)])\nprint(\"\\nPolicy (UP = 0, RIGHT = 1, DOWN = 2, LEFT = 3, N/A = -1):\")\nprint(policy.reshape(4,12))",
"\nPolicy (UP = 0, RIGHT = 1, DOWN = 2, LEFT = 3, N/A = -1):\n[[ 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 2.]\n [ 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 2.]\n [ 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 2.]\n [ 0. -1. -1. -1. -1. -1. -1. -1. -1. -1. -1. -1.]]\n"
]
],
[
[
"Run the next cell to visualize the state-value function that corresponds to this policy. Make sure that you take the time to understand why this is the corresponding value function!",
"_____no_output_____"
]
],
[
[
"V_true = np.zeros((4,12))\nfor i in range(3):\n V_true[0:12][i] = -np.arange(3, 15)[::-1] - i\nV_true[1][11] = -2\nV_true[2][11] = -1\nV_true[3][0] = -17\n\nplot_values(V_true)",
"_____no_output_____"
]
],
[
[
"The above figure is what you will try to approximate through the TD prediction algorithm.\n\nYour algorithm for TD prediction has five arguments:\n- `env`: This is an instance of an OpenAI Gym environment.\n- `num_episodes`: This is the number of episodes that are generated through agent-environment interaction.\n- `policy`: This is a 1D numpy array with `policy.shape` equal to the number of states (`env.nS`). `policy[s]` returns the action that the agent chooses when in state `s`.\n- `alpha`: This is the step-size parameter for the update step.\n- `gamma`: This is the discount rate. It must be a value between 0 and 1, inclusive (default value: `1`).\n\nThe algorithm returns as output:\n- `V`: This is a dictionary where `V[s]` is the estimated value of state `s`.\n\nPlease complete the function in the code cell below.",
"_____no_output_____"
]
],
[
[
"from collections import defaultdict, deque\nimport sys\n\ndef td_prediction(env, num_episodes, policy, alpha, gamma=1.0):\n # initialize empty dictionaries of floats\n V = defaultdict(float)\n # loop over episodes\n for i_episode in range(1, num_episodes+1):\n # monitor progress\n if i_episode % 100 == 0:\n print(\"\\rEpisode {}/{}\".format(i_episode, num_episodes), end=\"\")\n sys.stdout.flush()\n # begin an episode, observe S\n state = env.reset()\n while True:\n # choose action A\n action = policy[state]\n # take action A, observe R, S'\n next_state, reward, done, info = env.step(action)\n # perform updates\n V[state] = V[state] + (alpha * (reward + (gamma * V[next_state]) - V[state])) \n # S <- S'\n state = next_state\n # end episode if reached terminal state\n if done:\n break \n return V",
"_____no_output_____"
]
],
[
[
"Run the code cell below to test your implementation and visualize the estimated state-value function. If the code cell returns **PASSED**, then you have implemented the function correctly! Feel free to change the `num_episodes` and `alpha` parameters that are supplied to the function. However, if you'd like to ensure the accuracy of the unit test, please do not change the value of `gamma` from the default.",
"_____no_output_____"
]
],
[
[
"import check_test\n\n# evaluate the policy and reshape the state-value function\nV_pred = td_prediction(env, 5000, policy, .01)\n\n# please do not change the code below this line\nV_pred_plot = np.reshape([V_pred[key] if key in V_pred else 0 for key in np.arange(48)], (4,12)) \ncheck_test.run_check('td_prediction_check', V_pred_plot)\nplot_values(V_pred_plot)",
"Episode 5000/5000"
]
],
[
[
"How close is your estimated state-value function to the true state-value function corresponding to the policy? \n\nYou might notice that some of the state values are not estimated by the agent. This is because under this policy, the agent will not visit all of the states. In the TD prediction algorithm, the agent can only estimate the values corresponding to states that are visited.",
"_____no_output_____"
],
[
"### Part 2: TD Control: Sarsa\n\nIn this section, you will write your own implementation of the Sarsa control algorithm.\n\nYour algorithm has four arguments:\n- `env`: This is an instance of an OpenAI Gym environment.\n- `num_episodes`: This is the number of episodes that are generated through agent-environment interaction.\n- `alpha`: This is the step-size parameter for the update step.\n- `gamma`: This is the discount rate. It must be a value between 0 and 1, inclusive (default value: `1`).\n\nThe algorithm returns as output:\n- `Q`: This is a dictionary (of one-dimensional arrays) where `Q[s][a]` is the estimated action value corresponding to state `s` and action `a`.\n\nPlease complete the function in the code cell below.\n\n(_Feel free to define additional functions to help you to organize your code._)",
"_____no_output_____"
]
],
[
[
"def update_Q(Qsa, Qsa_next, reward, alpha, gamma):\n \"\"\" updates the action-value function estimate using the most recent time step \"\"\"\n return Qsa + (alpha * (reward + (gamma * Qsa_next) - Qsa))\n\ndef epsilon_greedy_probs(env, Q_s, i_episode, eps=None):\n \"\"\" obtains the action probabilities corresponding to epsilon-greedy policy \"\"\"\n epsilon = 1.0 / i_episode\n if eps is not None:\n epsilon = eps\n policy_s = np.ones(env.nA) * epsilon / env.nA\n policy_s[np.argmax(Q_s)] = 1 - epsilon + (epsilon / env.nA)\n return policy_s",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n%matplotlib inline\n\ndef sarsa(env, num_episodes, alpha, gamma=1.0):\n # initialize action-value function (empty dictionary of arrays)\n Q = defaultdict(lambda: np.zeros(env.nA))\n # initialize performance monitor\n plot_every = 100\n tmp_scores = deque(maxlen=plot_every)\n scores = deque(maxlen=num_episodes)\n # loop over episodes\n for i_episode in range(1, num_episodes+1):\n # monitor progress\n if i_episode % 100 == 0:\n print(\"\\rEpisode {}/{}\".format(i_episode, num_episodes), end=\"\")\n sys.stdout.flush() \n # initialize score\n score = 0\n # begin an episode, observe S\n state = env.reset() \n # get epsilon-greedy action probabilities\n policy_s = epsilon_greedy_probs(env, Q[state], i_episode)\n # pick action A\n action = np.random.choice(np.arange(env.nA), p=policy_s)\n # limit number of time steps per episode\n for t_step in np.arange(300):\n # take action A, observe R, S'\n next_state, reward, done, info = env.step(action)\n # add reward to score\n score += reward\n if not done:\n # get epsilon-greedy action probabilities\n policy_s = epsilon_greedy_probs(env, Q[next_state], i_episode)\n # pick next action A'\n next_action = np.random.choice(np.arange(env.nA), p=policy_s)\n # update TD estimate of Q\n Q[state][action] = update_Q(Q[state][action], Q[next_state][next_action], \n reward, alpha, gamma)\n # S <- S'\n state = next_state\n # A <- A'\n action = next_action\n if done:\n # update TD estimate of Q\n Q[state][action] = update_Q(Q[state][action], 0, reward, alpha, gamma)\n # append score\n tmp_scores.append(score)\n break\n if (i_episode % plot_every == 0):\n scores.append(np.mean(tmp_scores))\n # plot performance\n plt.plot(np.linspace(0,num_episodes,len(scores),endpoint=False),np.asarray(scores))\n plt.xlabel('Episode Number')\n plt.ylabel('Average Reward (Over Next %d Episodes)' % plot_every)\n plt.show()\n # print best 100-episode performance\n print(('Best Average Reward over %d Episodes: ' % plot_every), np.max(scores))\n return Q",
"_____no_output_____"
]
],
[
[
"Use the next code cell to visualize the **_estimated_** optimal policy and the corresponding state-value function. \n\nIf the code cell returns **PASSED**, then you have implemented the function correctly! Feel free to change the `num_episodes` and `alpha` parameters that are supplied to the function. However, if you'd like to ensure the accuracy of the unit test, please do not change the value of `gamma` from the default.",
"_____no_output_____"
]
],
[
[
"# obtain the estimated optimal policy and corresponding action-value function\nQ_sarsa = sarsa(env, 5000, .01)\n\n# print the estimated optimal policy\npolicy_sarsa = np.array([np.argmax(Q_sarsa[key]) if key in Q_sarsa else -1 for key in np.arange(48)]).reshape(4,12)\ncheck_test.run_check('td_control_check', policy_sarsa)\nprint(\"\\nEstimated Optimal Policy (UP = 0, RIGHT = 1, DOWN = 2, LEFT = 3, N/A = -1):\")\nprint(policy_sarsa)\n\n# plot the estimated optimal state-value function\nV_sarsa = ([np.max(Q_sarsa[key]) if key in Q_sarsa else 0 for key in np.arange(48)])\nplot_values(V_sarsa)",
"Episode 5000/5000"
]
],
[
[
"### Part 3: TD Control: Q-learning\n\nIn this section, you will write your own implementation of the Q-learning control algorithm.\n\nYour algorithm has four arguments:\n- `env`: This is an instance of an OpenAI Gym environment.\n- `num_episodes`: This is the number of episodes that are generated through agent-environment interaction.\n- `alpha`: This is the step-size parameter for the update step.\n- `gamma`: This is the discount rate. It must be a value between 0 and 1, inclusive (default value: `1`).\n\nThe algorithm returns as output:\n- `Q`: This is a dictionary (of one-dimensional arrays) where `Q[s][a]` is the estimated action value corresponding to state `s` and action `a`.\n\nPlease complete the function in the code cell below.\n\n(_Feel free to define additional functions to help you to organize your code._)",
"_____no_output_____"
]
],
[
[
"def q_learning(env, num_episodes, alpha, gamma=1.0):\n # initialize action-value function (empty dictionary of arrays)\n Q = defaultdict(lambda: np.zeros(env.nA))\n # initialize performance monitor\n plot_every = 100\n tmp_scores = deque(maxlen=plot_every)\n scores = deque(maxlen=num_episodes)\n # loop over episodes\n for i_episode in range(1, num_episodes+1):\n # monitor progress\n if i_episode % 100 == 0:\n print(\"\\rEpisode {}/{}\".format(i_episode, num_episodes), end=\"\")\n sys.stdout.flush()\n # initialize score\n score = 0\n # begin an episode, observe S\n state = env.reset()\n while True:\n # get epsilon-greedy action probabilities\n policy_s = epsilon_greedy_probs(env, Q[state], i_episode)\n # pick next action A\n action = np.random.choice(np.arange(env.nA), p=policy_s)\n # take action A, observe R, S'\n next_state, reward, done, info = env.step(action)\n # add reward to score\n score += reward\n # update Q\n Q[state][action] = update_Q(Q[state][action], np.max(Q[next_state]), \\\n reward, alpha, gamma) \n # S <- S'\n state = next_state\n # until S is terminal\n if done:\n # append score\n tmp_scores.append(score)\n break\n if (i_episode % plot_every == 0):\n scores.append(np.mean(tmp_scores))\n # plot performance\n plt.plot(np.linspace(0,num_episodes,len(scores),endpoint=False),np.asarray(scores))\n plt.xlabel('Episode Number')\n plt.ylabel('Average Reward (Over Next %d Episodes)' % plot_every)\n plt.show()\n # print best 100-episode performance\n print(('Best Average Reward over %d Episodes: ' % plot_every), np.max(scores))\n return Q",
"_____no_output_____"
]
],
[
[
"Use the next code cell to visualize the **_estimated_** optimal policy and the corresponding state-value function. \n\nIf the code cell returns **PASSED**, then you have implemented the function correctly! Feel free to change the `num_episodes` and `alpha` parameters that are supplied to the function. However, if you'd like to ensure the accuracy of the unit test, please do not change the value of `gamma` from the default.",
"_____no_output_____"
]
],
[
[
"# obtain the estimated optimal policy and corresponding action-value function\nQ_sarsamax = q_learning(env, 5000, .01)\n\n# print the estimated optimal policy\npolicy_sarsamax = np.array([np.argmax(Q_sarsamax[key]) if key in Q_sarsamax else -1 for key in np.arange(48)]).reshape((4,12))\ncheck_test.run_check('td_control_check', policy_sarsamax)\nprint(\"\\nEstimated Optimal Policy (UP = 0, RIGHT = 1, DOWN = 2, LEFT = 3, N/A = -1):\")\nprint(policy_sarsamax)\n\n# plot the estimated optimal state-value function\nplot_values([np.max(Q_sarsamax[key]) if key in Q_sarsamax else 0 for key in np.arange(48)])",
"Episode 5000/5000"
]
],
[
[
"### Part 4: TD Control: Expected Sarsa\n\nIn this section, you will write your own implementation of the Expected Sarsa control algorithm.\n\nYour algorithm has four arguments:\n- `env`: This is an instance of an OpenAI Gym environment.\n- `num_episodes`: This is the number of episodes that are generated through agent-environment interaction.\n- `alpha`: This is the step-size parameter for the update step.\n- `gamma`: This is the discount rate. It must be a value between 0 and 1, inclusive (default value: `1`).\n\nThe algorithm returns as output:\n- `Q`: This is a dictionary (of one-dimensional arrays) where `Q[s][a]` is the estimated action value corresponding to state `s` and action `a`.\n\nPlease complete the function in the code cell below.\n\n(_Feel free to define additional functions to help you to organize your code._)",
"_____no_output_____"
]
],
[
[
"def expected_sarsa(env, num_episodes, alpha, gamma=1.0):\n # initialize action-value function (empty dictionary of arrays)\n Q = defaultdict(lambda: np.zeros(env.nA))\n # initialize performance monitor\n plot_every = 100\n tmp_scores = deque(maxlen=plot_every)\n scores = deque(maxlen=num_episodes)\n # loop over episodes\n for i_episode in range(1, num_episodes+1):\n # monitor progress\n if i_episode % 100 == 0:\n print(\"\\rEpisode {}/{}\".format(i_episode, num_episodes), end=\"\")\n sys.stdout.flush()\n # initialize score\n score = 0\n # begin an episode\n state = env.reset()\n # get epsilon-greedy action probabilities\n policy_s = epsilon_greedy_probs(env, Q[state], i_episode, 0.005)\n while True:\n # pick next action\n action = np.random.choice(np.arange(env.nA), p=policy_s)\n # take action A, observe R, S'\n next_state, reward, done, info = env.step(action)\n # add reward to score\n score += reward\n # get epsilon-greedy action probabilities (for S')\n policy_s = epsilon_greedy_probs(env, Q[next_state], i_episode, 0.005)\n # update Q\n Q[state][action] = update_Q(Q[state][action], np.dot(Q[next_state], policy_s), \\\n reward, alpha, gamma) \n # S <- S'\n state = next_state\n # until S is terminal\n if done:\n # append score\n tmp_scores.append(score)\n break\n if (i_episode % plot_every == 0):\n scores.append(np.mean(tmp_scores))\n # plot performance\n plt.plot(np.linspace(0,num_episodes,len(scores),endpoint=False),np.asarray(scores))\n plt.xlabel('Episode Number')\n plt.ylabel('Average Reward (Over Next %d Episodes)' % plot_every)\n plt.show()\n # print best 100-episode performance\n print(('Best Average Reward over %d Episodes: ' % plot_every), np.max(scores))\n return Q",
"_____no_output_____"
]
],
[
[
"Use the next code cell to visualize the **_estimated_** optimal policy and the corresponding state-value function. \n\nIf the code cell returns **PASSED**, then you have implemented the function correctly! Feel free to change the `num_episodes` and `alpha` parameters that are supplied to the function. However, if you'd like to ensure the accuracy of the unit test, please do not change the value of `gamma` from the default.",
"_____no_output_____"
]
],
[
[
"# obtain the estimated optimal policy and corresponding action-value function\nQ_expsarsa = expected_sarsa(env, 10000, 1)\n\n# print the estimated optimal policy\npolicy_expsarsa = np.array([np.argmax(Q_expsarsa[key]) if key in Q_expsarsa else -1 for key in np.arange(48)]).reshape(4,12)\ncheck_test.run_check('td_control_check', policy_expsarsa)\nprint(\"\\nEstimated Optimal Policy (UP = 0, RIGHT = 1, DOWN = 2, LEFT = 3, N/A = -1):\")\nprint(policy_expsarsa)\n\n# plot the estimated optimal state-value function\nplot_values([np.max(Q_expsarsa[key]) if key in Q_expsarsa else 0 for key in np.arange(48)])",
"Episode 10000/10000"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d092d4c3f4114df6fe22e86420f65b5ad97a08e1 | 640,195 | ipynb | Jupyter Notebook | LS_DS_114_Making_Data_backed_Assertions.ipynb | elizabethts/DS-Unit-1-Sprint-1-Dealing-With-Data | c84688b0cad643b90c6daa8943c11996f6366a88 | [
"MIT"
] | null | null | null | LS_DS_114_Making_Data_backed_Assertions.ipynb | elizabethts/DS-Unit-1-Sprint-1-Dealing-With-Data | c84688b0cad643b90c6daa8943c11996f6366a88 | [
"MIT"
] | null | null | null | LS_DS_114_Making_Data_backed_Assertions.ipynb | elizabethts/DS-Unit-1-Sprint-1-Dealing-With-Data | c84688b0cad643b90c6daa8943c11996f6366a88 | [
"MIT"
] | null | null | null | 81.325584 | 98,334 | 0.630713 | [
[
[
"<a href=\"https://colab.research.google.com/github/elizabethts/DS-Unit-1-Sprint-1-Dealing-With-Data/blob/master/LS_DS_114_Making_Data_backed_Assertions.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Lambda School Data Science - Making Data-backed Assertions\n\nThis is, for many, the main point of data science - to create and support reasoned arguments based on evidence. It's not a topic to master in a day, but it is worth some focused time thinking about and structuring your approach to it.",
"_____no_output_____"
],
[
"## Lecture - generating a confounding variable\n\nThe prewatch material told a story about a hypothetical health condition where both the drug usage and overall health outcome were related to gender - thus making gender a confounding variable, obfuscating the possible relationship between the drug and the outcome.\n\nLet's use Python to generate data that actually behaves in this fashion!",
"_____no_output_____"
]
],
[
[
"import random\ndir(random) # Reminding ourselves what we can do here\nrandom.seed(10) # Sets Random Seed for Reproducibility",
"_____no_output_____"
],
[
"# Let's think of another scenario:\n# We work for a company that sells accessories for mobile phones.\n# They have an ecommerce site, and we are supposed to analyze logs\n# to determine what sort of usage is related to purchases, and thus guide\n# website development to encourage higher conversion.\n\n# The hypothesis - users who spend longer on the site tend\n# to spend more. Seems reasonable, no?\n\n# But there's a confounding variable! If they're on a phone, they:\n# a) Spend less time on the site, but\n# b) Are more likely to be interested in the actual products!\n\n# Let's use namedtuple to represent our data\n\nfrom collections import namedtuple\n# purchased and mobile are bools, time_on_site in seconds\nUser = namedtuple('User', ['purchased','time_on_site', 'mobile'])\n\nexample_user = User(False, 12, False)\nprint(example_user)",
"User(purchased=False, time_on_site=12, mobile=False)\n"
],
[
"# And now let's generate 1000 example users\n# 750 mobile, 250 not (i.e. desktop)\n# A desktop user has a base conversion likelihood of 10%\n# And it goes up by 1% for each 15 seconds they spend on the site\n# And they spend anywhere from 10 seconds to 10 minutes on the site (uniform)\n# Mobile users spend on average half as much time on the site as desktop\n# But have three times as much base likelihood of buying something\n\nusers = []\n\nfor _ in range(250):\n # Desktop users\n time_on_site = random.uniform(10, 600)\n purchased = random.random() < 0.1 + (time_on_site / 1500)\n users.append(User(purchased, time_on_site, False))\n \nfor _ in range(750):\n # Mobile users\n time_on_site = random.uniform(5, 300)\n purchased = random.random() < 0.3 + (time_on_site / 1500)\n users.append(User(purchased, time_on_site, True))\n \nrandom.shuffle(users)\nprint(users[:10])",
"[User(purchased=False, time_on_site=172.07500125969045, mobile=False), User(purchased=False, time_on_site=242.1604565076447, mobile=True), User(purchased=True, time_on_site=172.4562884302345, mobile=True), User(purchased=False, time_on_site=134.30741730988564, mobile=True), User(purchased=False, time_on_site=176.6659151415657, mobile=False), User(purchased=False, time_on_site=98.57704667574383, mobile=True), User(purchased=False, time_on_site=141.90635886960914, mobile=True), User(purchased=False, time_on_site=46.30954508769639, mobile=True), User(purchased=True, time_on_site=568.9570603645093, mobile=False), User(purchased=False, time_on_site=64.57737234489078, mobile=True)]\n"
],
[
"# Let's put this in a dataframe so we can look at it more easily\nimport pandas as pd\nuser_data = pd.DataFrame(users)\nuser_data.head()",
"_____no_output_____"
],
[
"user_data.dtypes",
"_____no_output_____"
],
[
"user_data.isnull().sum()",
"_____no_output_____"
],
[
"user_data.describe()",
"_____no_output_____"
],
[
"import numpy as np\nuser_data.describe(exclude=[np.number])",
"_____no_output_____"
],
[
"# Let's use crosstabulation to try to see what's going on\npd.crosstab(user_data['purchased'], user_data['time_on_site'])",
"_____no_output_____"
],
[
"# !pip freeze # 0.24.2\n!pip install pandas==0.23.4",
"Requirement already satisfied: pandas==0.23.4 in /usr/local/lib/python3.6/dist-packages (0.23.4)\nRequirement already satisfied: pytz>=2011k in /usr/local/lib/python3.6/dist-packages (from pandas==0.23.4) (2018.9)\nRequirement already satisfied: numpy>=1.9.0 in /usr/local/lib/python3.6/dist-packages (from pandas==0.23.4) (1.16.3)\nRequirement already satisfied: python-dateutil>=2.5.0 in /usr/local/lib/python3.6/dist-packages (from pandas==0.23.4) (2.5.3)\nRequirement already satisfied: six>=1.5 in /usr/local/lib/python3.6/dist-packages (from python-dateutil>=2.5.0->pandas==0.23.4) (1.12.0)\n"
],
[
"!pip freeze",
"absl-py==0.7.1\nalabaster==0.7.12\nalbumentations==0.1.12\naltair==3.0.1\nastor==0.8.0\nastropy==3.0.5\natari-py==0.1.15\natomicwrites==1.3.0\nattrs==19.1.0\naudioread==2.1.7\nautograd==1.2\nBabel==2.6.0\nbackcall==0.1.0\nbackports.tempfile==1.0\nbackports.weakref==1.0.post1\nbeautifulsoup4==4.6.3\nbleach==3.1.0\nblis==0.2.4\nbokeh==1.0.4\nboto==2.49.0\nboto3==1.9.156\nbotocore==1.12.156\nBottleneck==1.2.1\nbranca==0.3.1\nbs4==0.0.1\nbz2file==0.98\ncachetools==3.1.1\ncertifi==2019.3.9\ncffi==1.12.3\nchainer==5.4.0\nchardet==3.0.4\nClick==7.0\ncloudpickle==0.6.1\ncmake==3.12.0\ncolorlover==0.3.0\ncommunity==1.0.0b1\ncontextlib2==0.5.5\nconvertdate==2.1.3\ncoverage==3.7.1\ncoveralls==0.5\ncrcmod==1.7\ncufflinks==0.14.6\ncvxopt==1.2.3\ncvxpy==1.0.15\ncycler==0.10.0\ncymem==2.0.2\nCython==0.29.7\ncytoolz==0.9.0.1\ndaft==0.0.4\ndask==1.1.5\ndataclasses==0.6\ndatascience==0.10.6\ndecorator==4.4.0\ndefusedxml==0.6.0\ndill==0.2.9\ndistributed==1.25.3\nDjango==2.2.1\ndlib==19.16.0\ndm-sonnet==1.32\ndocopt==0.6.2\ndocutils==0.14\ndopamine-rl==1.0.5\neasydict==1.9\necos==2.0.7.post1\neditdistance==0.5.3\nen-core-web-sm==2.0.0\nentrypoints==0.3\nenum34==1.1.6\nephem==3.7.6.0\net-xmlfile==1.0.1\nfa2==0.3.5\nfancyimpute==0.4.3\nfastai==1.0.52\nfastcache==1.1.0\nfastdtw==0.3.2\nfastprogress==0.1.21\nfastrlock==0.4\nfbprophet==0.5\nfeather-format==0.4.0\nfeaturetools==0.4.1\nfilelock==3.0.12\nfix-yahoo-finance==0.0.22\nFlask==1.0.3\nfolium==0.8.3\nfuture==0.16.0\ngast==0.2.2\nGDAL==2.2.2\ngdown==3.6.4\ngensim==3.6.0\ngeographiclib==1.49\ngeopy==1.17.0\ngevent==1.4.0\ngin-config==0.1.4\nglob2==0.6\ngoogle==2.0.2\ngoogle-api-core==1.11.0\ngoogle-api-python-client==1.6.7\ngoogle-auth==1.4.2\ngoogle-auth-httplib2==0.0.3\ngoogle-auth-oauthlib==0.3.0\ngoogle-cloud-bigquery==1.8.1\ngoogle-cloud-core==0.29.1\ngoogle-cloud-language==1.0.2\ngoogle-cloud-storage==1.13.2\ngoogle-cloud-translate==1.3.3\ngoogle-colab==1.0.0\ngoogle-resumable-media==0.3.2\ngoogleapis-common-protos==1.6.0\ngoogledrivedownloader==0.4\ngraph-nets==1.0.4\ngraphviz==0.10.1\ngreenlet==0.4.15\ngrpcio==1.15.0\ngspread==3.0.1\ngspread-dataframe==3.0.2\ngunicorn==19.9.0\ngym==0.10.11\nh5py==2.8.0\nHeapDict==1.0.0\nholidays==0.9.10\nhtml5lib==1.0.1\nhttpimport==0.5.16\nhttplib2==0.11.3\nhumanize==0.5.1\nhyperopt==0.1.2\nideep4py==2.0.0.post3\nidna==2.8\nimage==1.5.27\nimageio==2.4.1\nimagesize==1.1.0\nimbalanced-learn==0.4.3\nimblearn==0.0\nimgaug==0.2.9\nimutils==0.5.2\ninflect==2.1.0\nintel-openmp==2019.0\nintervaltree==2.1.0\nipykernel==4.6.1\nipython==5.5.0\nipython-genutils==0.2.0\nipython-sql==0.3.9\nipywidgets==7.4.2\nitsdangerous==1.1.0\njdcal==1.4.1\njedi==0.13.3\njieba==0.39\nJinja2==2.10.1\njmespath==0.9.4\njoblib==0.12.5\njpeg4py==0.1.4\njsonschema==2.6.0\njupyter==1.0.0\njupyter-client==5.2.4\njupyter-console==6.0.0\njupyter-core==4.4.0\nkaggle==1.5.3\nkapre==0.1.3.1\nKeras==2.2.4\nKeras-Applications==1.0.7\nKeras-Preprocessing==1.0.9\nkeras-vis==0.4.1\nkiwisolver==1.1.0\nknnimpute==0.1.0\nlibrosa==0.6.3\nlightgbm==2.2.3\nllvmlite==0.28.0\nlmdb==0.94\nlucid==0.3.8\nlunardate==0.2.0\nlxml==4.2.6\nmagenta==0.3.19\nMarkdown==3.1.1\nMarkupSafe==1.1.1\nmatplotlib==3.0.3\nmatplotlib-venn==0.11.5\nmesh-tensorflow==0.0.5\nmido==1.2.6\nmir-eval==0.5\nmissingno==0.4.1\nmistune==0.8.4\nmkl==2019.0\nmlxtend==0.14.0\nmock==3.0.5\nmore-itertools==7.0.0\nmoviepy==0.2.3.5\nmpi4py==3.0.1\nmpmath==1.1.0\nmsgpack==0.5.6\nmsgpack-numpy==0.4.3.2\nmultiprocess==0.70.7\nmultitasking==0.0.9\nmurmurhash==1.0.2\nmusic21==5.5.0\nnatsort==5.5.0\nnbconvert==5.5.0\nnbformat==4.4.0\nnetworkx==2.3\nnibabel==2.3.3\nnltk==3.2.5\nnose==1.3.7\nnotebook==5.2.2\nnp-utils==0.5.10.0\nnumba==0.40.1\nnumexpr==2.6.9\nnumpy==1.16.3\nnvidia-ml-py3==7.352.0\noauth2client==4.1.3\noauthlib==3.0.1\nokgrade==0.4.3\nolefile==0.46\nopencv-contrib-python==3.4.3.18\nopencv-python==3.4.5.20\nopenpyxl==2.5.9\nosqp==0.5.0\npackaging==19.0\npandas==0.23.4\npandas-datareader==0.7.0\npandas-gbq==0.4.1\npandas-profiling==1.4.1\npandocfilters==1.4.2\nparso==0.4.0\npathlib==1.0.1\npatsy==0.5.1\npexpect==4.7.0\npickleshare==0.7.5\nPillow==4.3.0\npip-tools==3.6.1\nplac==0.9.6\nplotly==3.6.1\npluggy==0.7.1\nportpicker==1.2.0\nprefetch-generator==1.0.1\npreshed==2.0.1\npretty-midi==0.2.8\nprettytable==0.7.2\nprogressbar2==3.38.0\nprometheus-client==0.6.0\npromise==2.2.1\nprompt-toolkit==1.0.16\nprotobuf==3.7.1\npsutil==5.4.8\npsycopg2==2.7.6.1\nptyprocess==0.6.0\npy==1.8.0\npyarrow==0.13.0\npyasn1==0.4.5\npyasn1-modules==0.2.5\npycocotools==2.0.0\npycparser==2.19\npydot==1.3.0\npydot-ng==2.0.0\npydotplus==2.0.2\npyemd==0.5.1\npyglet==1.3.2\nPygments==2.1.3\npygobject==3.26.1\npymc3==3.6\npymongo==3.8.0\npymystem3==0.2.0\nPyOpenGL==3.1.0\npyparsing==2.4.0\npyrsistent==0.15.2\npysndfile==1.3.2\nPySocks==1.7.0\npystan==2.19.0.0\npytest==3.6.4\npython-apt==1.6.4\npython-chess==0.23.11\npython-dateutil==2.5.3\npython-louvain==0.13\npython-rtmidi==1.3.0\npython-slugify==3.0.2\npython-utils==2.3.0\npytz==2018.9\nPyWavelets==1.0.3\nPyYAML==3.13\npyzmq==17.0.0\nqtconsole==4.5.1\nregex==2018.1.10\nrequests==2.21.0\nrequests-oauthlib==1.2.0\nresampy==0.2.1\nretrying==1.3.3\nrpy2==2.9.5\nrsa==4.0\ns3fs==0.2.1\ns3transfer==0.2.0\nscikit-image==0.15.0\nscikit-learn==0.21.2\nscipy==1.3.0\nscreen-resolution-extra==0.0.0\nscs==2.1.0\nseaborn==0.9.0\nsemantic-version==2.6.0\nSend2Trash==1.5.0\nsetuptools-git==1.2\nShapely==1.6.4.post2\nsimplegeneric==0.8.1\nsix==1.12.0\nsklearn==0.0\nsklearn-pandas==1.8.0\nsmart-open==1.8.3\nsnowballstemmer==1.2.1\nsortedcontainers==2.1.0\nsoupsieve==1.9.1\nspacy==2.0.18\nSphinx==1.8.5\nsphinxcontrib-websupport==1.1.2\nSQLAlchemy==1.3.3\nsqlparse==0.3.0\nsrsly==0.0.5\nstable-baselines==2.2.1\nstatsmodels==0.9.0\nsympy==1.1.1\ntables==3.4.4\ntabulate==0.8.3\ntblib==1.4.0\ntensor2tensor==1.11.0\ntensorboard==1.13.1\ntensorboardcolab==0.0.22\ntensorflow==1.13.1\ntensorflow-estimator==1.13.0\ntensorflow-hub==0.4.0\ntensorflow-metadata==0.13.0\ntensorflow-probability==0.6.0\ntermcolor==1.1.0\nterminado==0.8.2\ntestpath==0.4.2\ntext-unidecode==1.2\ntextblob==0.15.3\ntextgenrnn==1.4.1\ntfds-nightly==1.0.2.dev201905240105\ntflearn==0.3.2\nTheano==1.0.4\nthinc==6.12.1\ntoolz==0.9.0\ntorch==1.1.0\ntorchsummary==1.5.1\ntorchtext==0.3.1\ntorchvision==0.3.0\ntornado==4.5.3\ntqdm==4.28.1\ntraitlets==4.3.2\ntweepy==3.6.0\ntyping==3.6.6\ntzlocal==1.5.1\nujson==1.35\numap-learn==0.3.8\nuritemplate==3.0.0\nurllib3==1.24.3\nvega-datasets==0.7.0\nwasabi==0.2.2\nwcwidth==0.1.7\nwebencodings==0.5.1\nWerkzeug==0.15.4\nwidgetsnbextension==3.4.2\nwordcloud==1.5.0\nwrapt==1.10.11\nxarray==0.11.3\nxgboost==0.90\nxkit==0.0.0\nxlrd==1.1.0\nxlwt==1.3.0\nyellowbrick==0.9.1\nzict==0.1.4\nzmq==0.0.0\n"
],
[
"# OK, that's not quite what we want\n# Time is continuous! We need to put it in discrete buckets\n# Pandas calls these bins, and pandas.cut helps make them\n\ntime_bins = pd.cut(user_data['time_on_site'], 6) # 6 equal-sized bins\npd.crosstab(user_data['purchased'], time_bins)",
"_____no_output_____"
],
[
"# We can make this a bit clearer by normalizing (getting %)\npd.crosstab(user_data['purchased'], time_bins, normalize='columns')",
"_____no_output_____"
],
[
"# That seems counter to our hypothesis\n# More time on the site can actually have fewer purchases\n\n# But we know why, since we generated the data!\n# Let's look at mobile and purchased\npd.crosstab(user_data['purchased'], user_data['mobile'], normalize='columns')",
"_____no_output_____"
],
[
"# Yep, mobile users are more likely to buy things\n# But we're still not seeing the *whole* story until we look at all 3 at once\n\n# Live/stretch goal - how can we do that?\n\nct = pd.crosstab(user_data['mobile'], [user_data['purchased'], time_bins],\n rownames = ['device'],\n colnames = ['purchased','time_on_site'],\n normalize = 'index'\n )\nct\n",
"_____no_output_____"
],
[
"type(ct)",
"_____no_output_____"
],
[
"pt = pd.pivot_table(user_data, values = 'purchased', index = time_bins)\npt",
"_____no_output_____"
],
[
"pt.plot.bar();",
"_____no_output_____"
],
[
"ct = pd.crosstab(time_bins, [user_data['purchased'], user_data['mobile']], \n normalize = 'columns')\nct",
"_____no_output_____"
],
[
"ct_final = ct.iloc[:, [2,3]]\nct_final.plot(kind = 'bar', stacked = True);",
"_____no_output_____"
]
],
[
[
"## Assignment - what's going on here?\n\nConsider the data in `persons.csv` (already prepared for you, in the repo for the week). It has four columns - a unique id, followed by age (in years), weight (in lbs), and exercise time (in minutes/week) of 1200 (hypothetical) people.\n\nTry to figure out which variables are possibly related to each other, and which may be confounding relationships.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"# TODO - your code here\n# Use what we did live in lecture as an example\n\n# HINT - you can find the raw URL on GitHub and potentially use that\n# to load the data with read_csv, or you can upload it yourself\n\n\n#load data\npersons_url = 'https://raw.githubusercontent.com/LambdaSchool/DS-Unit-1-Sprint-1-Dealing-With-Data/master/module3-databackedassertions/persons.csv'\npersons_df = pd.read_csv(persons_url)\npersons_df.head()",
"_____no_output_____"
],
[
"persons_df.shape ",
"_____no_output_____"
],
[
"time_range = pd.cut(persons_df['exercise_time'],30) #cut time into 5 groups\nage_range = pd.cut(persons_df['age'],3) #cut time into 5 groups\nweight_range = pd.cut(persons_df['weight'],30)\npersons_ct = pd.crosstab(persons_df['weight'], \n [age_range, persons_df['exercise_time']])\npersons_ct",
"_____no_output_____"
],
[
"group1 = persons_df[persons_df.age <30]\ngroup2 = persons_df[(persons_df.age >=30) & (persons_df.age<50)]\ngroup3 = persons_df[persons_df.age >=50]",
"_____no_output_____"
],
[
"fig, ax1 = plt.subplots()\nax1.set_axisbelow(True)\nax1.set_xlabel('Exercise Time')\nax1.set_ylabel('Weight')\nax1.set_title('Weight vs Exercise Time (Age <30)')\nplt.gca().invert_yaxis()\nplt.scatter(group1.exercise_time, group1.weight, color ='red', alpha = .4)\n\nfig, ax2 = plt.subplots()\nax2.set_axisbelow(True)\nax2.set_xlabel('Exercise Time')\nax2.set_ylabel('Weight')\nax2.set_title('Weight vs Exercise Time (Age 30-50)')\nplt.gca().invert_yaxis()\nplt.scatter(group2.exercise_time, group2.weight, color = 'teal', alpha = .4)\n\nfig, ax3 = plt.subplots()\nax3.set_axisbelow(True)\nax3.set_xlabel('Exercise Time')\nax3.set_ylabel('Weight')\nax3.set_title('Weight vs Exercise Time (Age > 50)')\nplt.gca().invert_yaxis()\nplt.scatter(group3.exercise_time, group3.weight, color = 'yellow', alpha = 0.4);\n\n\nfig, ax4 = plt.subplots()\nax4.set_axisbelow(True)\nax4.set_xlabel('Exercise Time')\nax4.set_ylabel('Weight')\nax4.set_title('Weight vs Exercise Time for All Ages')\nplt.scatter(group2.exercise_time, group2.weight, color = 'teal', alpha = .6)\nplt.scatter(group3.exercise_time, group3.weight,color = 'yellow', alpha = 0.6);\nplt.scatter(group1.exercise_time, group1.weight, color ='red', alpha = .6)\nplt.gca().invert_yaxis()\n\n\nfig, ax5 = plt.subplots()\nax5.set_axisbelow(True)\nax5.set_xlabel('Exercise Time')\nax5.set_ylabel('Age')\nax5.set_title('Age vs Exercise Time (All Ages)')\nplt.scatter(group2.exercise_time, group2.age, color = 'teal', alpha = .6)\nplt.scatter(group3.exercise_time, group3.age,color = 'yellow', alpha = 0.6);\nplt.scatter(group1.exercise_time, group1.age, color ='red', alpha = .6)\n\nfig, ax6 = plt.subplots()\nax6.set_axisbelow(True)\nax6.set_xlabel('Weight')\nax6.set_ylabel('Age')\nax6.set_title('Age vs Weight (All Ages)')\nplt.scatter(group2.weight, group2.age, color = 'teal', alpha = .6)\nplt.scatter(group3.weight, group3.age,color = 'yellow', alpha = 0.6);\nplt.scatter(group1.weight, group1.age, color ='red', alpha = .6);\n",
"_____no_output_____"
],
[
"#Conclusions: \n# -People who exercise more weight less\n# -The group age >=50 tends to exercise for a shorter amount of time ",
"_____no_output_____"
]
],
[
[
"### Assignment questions\n\nAfter you've worked on some code, answer the following questions in this text block:\n\n1. What are the variable types in the data?\n \n \n age - ordinal and continuous\n weight - ordinal and continuous\n exercise time - ordinal and continuous\n\n2. What are the relationships between the variables?\n \n \n Weight decreases as exercise time increases so they are inversely related. Weight increases as age increases so they are directly related\n\n3. Which relationships are \"real\", and which spurious?\n\n\n The relationship between weight and exercise time is real and the relationship between age and weight is spurious (older people tend to exercise less)\n",
"_____no_output_____"
],
[
"## Stretch goals and resources\n\nFollowing are *optional* things for you to take a look at. Focus on the above assignment first, and make sure to commit and push your changes to GitHub.\n\n- [Spurious Correlations](http://tylervigen.com/spurious-correlations)\n- [NIH on controlling for confounding variables](https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4017459/)\n\nStretch goals:\n\n- Produce your own plot inspired by the Spurious Correlation visualizations (and consider writing a blog post about it - both the content and how you made it)\n- Pick one of the techniques that NIH highlights for confounding variables - we'll be going into many of them later, but see if you can find which Python modules may help (hint - check scikit-learn)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
d092e422a885737ab72785e45a1a9bf51432da6c | 99,041 | ipynb | Jupyter Notebook | .ipynb_checkpoints/assignment 2 and 3-Copy1-checkpoint.ipynb | clintonmaiko/ADS-Assignment-2-3 | 12588590c8c8ea5b1579cf53798706e664ef0325 | [
"MIT"
] | null | null | null | .ipynb_checkpoints/assignment 2 and 3-Copy1-checkpoint.ipynb | clintonmaiko/ADS-Assignment-2-3 | 12588590c8c8ea5b1579cf53798706e664ef0325 | [
"MIT"
] | null | null | null | .ipynb_checkpoints/assignment 2 and 3-Copy1-checkpoint.ipynb | clintonmaiko/ADS-Assignment-2-3 | 12588590c8c8ea5b1579cf53798706e664ef0325 | [
"MIT"
] | null | null | null | 93.346843 | 61,744 | 0.723953 | [
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n",
"_____no_output_____"
],
[
"#data importation\ndf = pd.read_csv(\"HR-Employee-Attrition.csv\")\n\ndf.head()",
"_____no_output_____"
],
[
"#data analysis\ndf.dtypes",
"_____no_output_____"
],
[
"#data analysis\ndf.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1470 entries, 0 to 1469\nData columns (total 35 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Age 1470 non-null int64 \n 1 Attrition 1470 non-null object\n 2 BusinessTravel 1470 non-null object\n 3 DailyRate 1470 non-null int64 \n 4 Department 1470 non-null object\n 5 DistanceFromHome 1470 non-null int64 \n 6 Education 1470 non-null int64 \n 7 EducationField 1470 non-null object\n 8 EmployeeCount 1470 non-null int64 \n 9 EmployeeNumber 1470 non-null int64 \n 10 EnvironmentSatisfaction 1470 non-null int64 \n 11 Gender 1470 non-null object\n 12 HourlyRate 1470 non-null int64 \n 13 JobInvolvement 1470 non-null int64 \n 14 JobLevel 1470 non-null int64 \n 15 JobRole 1470 non-null object\n 16 JobSatisfaction 1470 non-null int64 \n 17 MaritalStatus 1470 non-null object\n 18 MonthlyIncome 1470 non-null int64 \n 19 MonthlyRate 1470 non-null int64 \n 20 NumCompaniesWorked 1470 non-null int64 \n 21 Over18 1470 non-null object\n 22 OverTime 1470 non-null object\n 23 PercentSalaryHike 1470 non-null int64 \n 24 PerformanceRating 1470 non-null int64 \n 25 RelationshipSatisfaction 1470 non-null int64 \n 26 StandardHours 1470 non-null int64 \n 27 StockOptionLevel 1470 non-null int64 \n 28 TotalWorkingYears 1470 non-null int64 \n 29 TrainingTimesLastYear 1470 non-null int64 \n 30 WorkLifeBalance 1470 non-null int64 \n 31 YearsAtCompany 1470 non-null int64 \n 32 YearsInCurrentRole 1470 non-null int64 \n 33 YearsSinceLastPromotion 1470 non-null int64 \n 34 YearsWithCurrManager 1470 non-null int64 \ndtypes: int64(26), object(9)\nmemory usage: 402.1+ KB\n"
],
[
"#Showing a breakdown of distance from home by job role and attrition.\ndf.groupby([\"DistanceFromHome\", \"JobRole\"])[\"Attrition\"].count()\n\n\n",
"_____no_output_____"
],
[
"#Comparing average monthly income by education and attrition.\ndf.groupby([\"MonthlyIncome\", \"Education\"])[\"Attrition\"].count()\n\n\n\n\n",
"_____no_output_____"
],
[
"#getting average montly income income and creating column\nav_Monthlyincome = df[\"MonthlyIncome\"].mean()\n\ndf = pd.DataFrame(df)\ndf['av_Monthlyincome'] = av_Monthlyincome\n\ndf.head()\n\n",
"_____no_output_____"
],
[
"# comparing the three columns\ndf_comparison = pd.DataFrame(df, columns = ['av_Monthlyincome','Education', 'Attrition'] )\n\ndf_comparison.head(10)\n",
"_____no_output_____"
],
[
"#ASSIGHNMENT 3\n#Based on your analysis in Assignment 2, express your results in the\n#form of the most appropriate visualization using any visualization library of your choice.",
"_____no_output_____"
],
[
"# visualisation on breakdown of distance from home by job role and attrition.\n\nsns.catplot(x=\"DistanceFromHome\",y=\"JobRole\",hue=\"Attrition\",data=df,palette=\"Set2\")\nplt.title(\"How Distance from home and job role effect the attrition\");\n\n\n",
"_____no_output_____"
],
[
"# visualisation of Comparing average monthly income by education and attrition\n\n#fig, ax = plt.subplots(figsize = (8,8))\n#sns.barplot(y = 'av_Monthlyincome',x = 'Education',hue = 'Attrition',data = df,ax = ax)\n",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d092f1ae25c71756518906b0d736ddac5fcc6c69 | 4,746 | ipynb | Jupyter Notebook | notebooks/cifar10/01_train_cifar.ipynb | Yu-Group/adaptive-wavelets | e67f726e741d83c94c3aee3ed97a772db4ce0bb3 | [
"MIT"
] | 22 | 2021-02-13T05:22:13.000Z | 2022-03-07T09:55:55.000Z | notebooks/cifar10/01_train_cifar.ipynb | Yu-Group/adaptive-wavelets | e67f726e741d83c94c3aee3ed97a772db4ce0bb3 | [
"MIT"
] | null | null | null | notebooks/cifar10/01_train_cifar.ipynb | Yu-Group/adaptive-wavelets | e67f726e741d83c94c3aee3ed97a772db4ce0bb3 | [
"MIT"
] | 5 | 2021-12-11T13:43:19.000Z | 2022-03-19T07:07:37.000Z | 28.590361 | 98 | 0.527391 | [
[
[
"%load_ext autoreload\n%autoreload 2\n%matplotlib inline\nimport numpy as np\nimport torch\nimport matplotlib.pyplot as plt\nfrom tqdm import tqdm\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport torch.optim as optim\nimport util\nimport sys\nsys.path.append(\"../..\")",
"The autoreload extension is already loaded. To reload it, use:\n %reload_ext autoreload\n"
],
[
"import cifar10\ndevice = 'cuda' if torch.cuda.is_available() else 'cpu'\nprint(device)\n\n# load data \ndata_path = \"../../data/cifar10\"\ntrain_loader, test_loader = cifar10.get_dataloader(data_path, batch_size=100, download=True)",
"cuda\nFiles already downloaded and verified\nFiles already downloaded and verified\n"
],
[
"class Net(nn.Module):\n def __init__(self):\n super().__init__()\n self.conv1 = nn.Conv2d(3, 6, 5)\n self.pool = nn.MaxPool2d(2, 2)\n self.conv2 = nn.Conv2d(6, 16, 5)\n self.fc1 = nn.Linear(16 * 5 * 5, 120)\n self.fc2 = nn.Linear(120, 84)\n self.fc3 = nn.Linear(84, 10)\n\n def forward(self, x):\n x = self.pool(F.relu(self.conv1(x)))\n x = self.pool(F.relu(self.conv2(x)))\n x = torch.flatten(x, 1) # flatten all dimensions except batch\n x = F.relu(self.fc1(x))\n x = F.relu(self.fc2(x))\n x = self.fc3(x)\n return x\n \n# define model and hyperparms\ncriterion = nn.CrossEntropyLoss()\noptimizer = optim.Adam(net.parameters(), lr=0.001)\nnum_epochs = 10\nnet = Net().to(device) ",
"_____no_output_____"
]
],
[
[
"# train cnn",
"_____no_output_____"
]
],
[
[
"util.train(model, device, train_loader, test_loader, optimizer, num_epochs, criterion)",
"Train Epoch: 1 [45000/50000 (90%)]\tLoss: 1.500776\nTest set: Average loss: 0.0002, Accuracy: 4550/10000 (45.50%)\n\nTrain Epoch: 2 [45000/50000 (90%)]\tLoss: 1.379729\nTest set: Average loss: 0.0001, Accuracy: 5390/10000 (53.90%)\n\nTrain Epoch: 3 [45000/50000 (90%)]\tLoss: 1.330050\nTest set: Average loss: 0.0001, Accuracy: 5640/10000 (56.40%)\n\nTrain Epoch: 4 [45000/50000 (90%)]\tLoss: 1.189881\nTest set: Average loss: 0.0001, Accuracy: 5753/10000 (57.53%)\n\nTrain Epoch: 5 [45000/50000 (90%)]\tLoss: 1.287006\nTest set: Average loss: 0.0001, Accuracy: 6042/10000 (60.42%)\n\nTrain Epoch: 6 [45000/50000 (90%)]\tLoss: 1.345478\nTest set: Average loss: 0.0001, Accuracy: 5962/10000 (59.62%)\n\nTrain Epoch: 7 [45000/50000 (90%)]\tLoss: 1.042157\nTest set: Average loss: 0.0001, Accuracy: 6065/10000 (60.65%)\n\nTrain Epoch: 8 [45000/50000 (90%)]\tLoss: 1.040688\nTest set: Average loss: 0.0001, Accuracy: 6236/10000 (62.36%)\n\nTrain Epoch: 9 [45000/50000 (90%)]\tLoss: 1.040002\nTest set: Average loss: 0.0001, Accuracy: 6186/10000 (61.86%)\n\nTrain Epoch: 10 [45000/50000 (90%)]\tLoss: 0.792405\nTest set: Average loss: 0.0001, Accuracy: 6311/10000 (63.11%)\n\n"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d093033c03f2a1e529678c6424fd1e4fc5c717e2 | 728,152 | ipynb | Jupyter Notebook | pairs_trading.ipynb | snowr/statarb_rw | 9b69d4cad71bf784bc01abacadb449ef7659124f | [
"Apache-2.0"
] | null | null | null | pairs_trading.ipynb | snowr/statarb_rw | 9b69d4cad71bf784bc01abacadb449ef7659124f | [
"Apache-2.0"
] | null | null | null | pairs_trading.ipynb | snowr/statarb_rw | 9b69d4cad71bf784bc01abacadb449ef7659124f | [
"Apache-2.0"
] | null | null | null | 305.946218 | 212,664 | 0.897745 | [
[
[
"import numpy as np\nimport pandas_datareader as pdr\nimport datetime as dt\nimport pandas as pd\nimport matplotlib.pyplot as plt",
"/usr/lib/python3/dist-packages/requests/__init__.py:89: RequestsDependencyWarning: urllib3 (1.26.7) or chardet (3.0.4) doesn't match a supported version!\n warnings.warn(\"urllib3 ({}) or chardet ({}) doesn't match a supported \"\n"
],
[
"start = dt.datetime(2012, 6, 1)\nend = dt.datetime(2022, 3, 9)\n\nstock = ['fb']\nstock_data = pdr.get_data_yahoo(stock, start, end)\n\npair = ['snap']\npair_data = pdr.get_data_yahoo(pair, start, end)\n\n",
"_____no_output_____"
],
[
"main_df = pd.DataFrame()\npair_df = pd.DataFrame()\n\nmain_df[\"Close\"] = stock_data[[\"Close\"]]\nmain_df[\"Open\"] = stock_data[[\"Open\"]]\nmain_df[\"High\"] = stock_data[[\"High\"]]\nmain_df[\"Low\"] = stock_data[[\"Low\"]]\nmain_df[\"Volume\"] = stock_data[[\"Volume\"]]\nmain_df[\"stock_prev_close\"] = main_df[\"Close\"].shift(1)\nmain_df['Date'] = main_df.index\n\npair_df[\"Close\"] = pair_data[[\"Close\"]]\npair_df[\"Open\"] = pair_data[[\"Open\"]]\npair_df[\"High\"] = pair_data[[\"High\"]]\npair_df[\"Low\"] = pair_data[[\"Low\"]]\npair_df[\"Volume\"] = pair_data[[\"Volume\"]]\npair_df[\"pair_prev_close\"] = pair_df[\"Close\"].shift(1)\npair_df['Date'] = pair_df.index\n",
"_____no_output_____"
],
[
"merged = pd.merge(main_df.reset_index(drop=True), pair_df.reset_index(drop=True), on=[\"Date\"], how=\"left\")",
"_____no_output_____"
],
[
"merged",
"_____no_output_____"
],
[
"merged['ratio_spread'] = merged['Close_x'] / merged['Close_y']\nmerged['prev_ratio_spread'] = merged['ratio_spread'].shift(1)\nmerged['close_spread'] = np.log(merged['Close_x']) - np.log(merged['Close_y'])\nmerged['close_spread_sma20'] = merged['close_spread'].rolling(20).mean()\nmerged['close_spread_std20'] = merged['close_spread'].rolling(20).std()\nmerged['zscore'] = (merged['close_spread'] - merged['close_spread_sma20']) / merged['close_spread_std20']\nmerged['prev_zscore'] = merged['zscore'].shift(1)\nmerged['lsr'] = np.log(merged['ratio_spread']) - np.log(merged['prev_ratio_spread'])\nmerged['strat_return'] = merged['prev_zscore'] * -1 * (merged['lsr'] - np.log(merged['ratio_spread']))",
"_____no_output_____"
],
[
"merged",
"_____no_output_____"
],
[
"figsize=(10,5)\nax = merged.plot(x=\"Date\", y=\"zscore\", legend=False,figsize=figsize)",
"_____no_output_____"
],
[
"figsize=(10,5)\nax = merged.plot(x=\"Date\", y=\"lsr\", legend=False,figsize=figsize)",
"_____no_output_____"
],
[
"figsize=(10,5)\nax = merged.plot(x=\"Date\", y=\"strat_return\", legend=False,figsize=figsize)",
"_____no_output_____"
],
[
"figsize=(30,10)\nax = merged[1200:].plot(y=\"strat_return\", legend=False,figsize=figsize)\n",
"_____no_output_____"
]
],
[
[
"# Monthly Aggregates - Work in Progress - I forgot to set price to 0 - Need to rewatch video!",
"_____no_output_____"
]
],
[
[
"merged['date_trunc_month'] = merged.apply(lambda x: dt.datetime.strftime(x['Date'],'%Y-%m'), axis=1)",
"_____no_output_____"
],
[
"merged['date_trunc_month']",
"_____no_output_____"
],
[
"merged['is_start_of_month'] = merged.apply(lambda x: x.name == min(merged[merged['date_trunc_month'] == x['date_trunc_month']].index), axis=1)\nmerged['is_end_of_month'] = merged.apply(lambda x: x.name == max(merged[merged['date_trunc_month'] == x['date_trunc_month']].index), axis=1)",
"_____no_output_____"
],
[
"merged[merged['is_start_of_month'] == True]",
"_____no_output_____"
]
],
[
[
"```sql\nSELECT\n startofmonth,\n stock1,\n stock2,\n SUM(lsr) as lsr\nFROM dailyreturns\nGROUP BY\n startofmonth,\n stock1,\n stock2\n```",
"_____no_output_____"
]
],
[
[
"merged[(merged['is_start_of_month'] == True)][['date_trunc_month','lsr']]",
"_____no_output_____"
],
[
"merged[(merged['is_start_of_month'] == True) | (merged['is_end_of_month'] == True)][['date_trunc_month','lsr']].plot()",
"_____no_output_____"
],
[
"merged['lsr'] = merged.apply(lambda x: 0 if x['is_end_of_month'] is True else x['lsr'], axis=1)",
"_____no_output_____"
],
[
"merged[merged['is_end_of_month'] == True]",
"_____no_output_____"
],
[
"monthly_agg = merged[(merged['is_start_of_month'] == True) | (merged['is_end_of_month'] == True)].copy()",
"_____no_output_____"
],
[
"monthly_agg.reset_index(drop=True)\nmonthly_agg = monthly_agg.dropna()",
"_____no_output_____"
],
[
"monthly_agg",
"_____no_output_____"
],
[
"figsize=(30,10)\nax = monthly_agg.plot(y=\"lsr\", x=\"date_trunc_month\", legend=False,figsize=figsize)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d093095046e756cd8a19653513f5978c5f1f90aa | 978,382 | ipynb | Jupyter Notebook | convolutional-neural-networks/cifar-cnn/cifar10_cnn_augmentation.ipynb | gaoxiao/deep-learning-v2-pytorch | 3ce54645575093fa1dd98a83b71fcc750b50ef8a | [
"MIT"
] | null | null | null | convolutional-neural-networks/cifar-cnn/cifar10_cnn_augmentation.ipynb | gaoxiao/deep-learning-v2-pytorch | 3ce54645575093fa1dd98a83b71fcc750b50ef8a | [
"MIT"
] | null | null | null | convolutional-neural-networks/cifar-cnn/cifar10_cnn_augmentation.ipynb | gaoxiao/deep-learning-v2-pytorch | 3ce54645575093fa1dd98a83b71fcc750b50ef8a | [
"MIT"
] | null | null | null | 1,455.925595 | 721,036 | 0.954532 | [
[
[
"# Convolutional Neural Networks\n---\nIn this notebook, we train a **CNN** to classify images from the CIFAR-10 database.\n\nThe images in this database are small color images that fall into one of ten classes; some example images are pictured below.\n\n<img src='notebook_ims/cifar_data.png' width=70% height=70% />",
"_____no_output_____"
],
[
"### Test for [CUDA](http://pytorch.org/docs/stable/cuda.html)\n\nSince these are larger (32x32x3) images, it may prove useful to speed up your training time by using a GPU. CUDA is a parallel computing platform and CUDA Tensors are the same as typical Tensors, only they utilize GPU's for computation.",
"_____no_output_____"
]
],
[
[
"import torch\nimport numpy as np\n\n# check if CUDA is available\ntrain_on_gpu = torch.cuda.is_available()\n\nif not train_on_gpu:\n print('CUDA is not available. Training on CPU ...')\nelse:\n print('CUDA is available! Training on GPU ...')",
"CUDA is available! Training on GPU ...\n"
]
],
[
[
"---\n## Load and Augment the [Data](http://pytorch.org/docs/stable/torchvision/datasets.html)\n\nDownloading may take a minute. We load in the training and test data, split the training data into a training and validation set, then create DataLoaders for each of these sets of data.\n\n#### Augmentation\n\nIn this cell, we perform some simple [data augmentation](https://medium.com/nanonets/how-to-use-deep-learning-when-you-have-limited-data-part-2-data-augmentation-c26971dc8ced) by randomly flipping and rotating the given image data. We do this by defining a torchvision `transform`, and you can learn about all the transforms that are used to pre-process and augment data, [here](https://pytorch.org/docs/stable/torchvision/transforms.html).\n\n#### TODO: Look at the [transformation documentation](https://pytorch.org/docs/stable/torchvision/transforms.html); add more augmentation transforms, and see how your model performs.\n\nThis type of data augmentation should add some positional variety to these images, so that when we train a model on this data, it will be robust in the face of geometric changes (i.e. it will recognize a ship, no matter which direction it is facing). It's recommended that you choose one or two transforms.",
"_____no_output_____"
]
],
[
[
"from torchvision import datasets\nimport torchvision.transforms as transforms\nfrom torch.utils.data.sampler import SubsetRandomSampler\n\n# number of subprocesses to use for data loading\nnum_workers = 0\n# how many samples per batch to load\nbatch_size = 20\n# percentage of training set to use as validation\nvalid_size = 0.2\n\n# convert data to a normalized torch.FloatTensor\ntransform = transforms.Compose([\n transforms.RandomHorizontalFlip(), # randomly flip and rotate\n transforms.RandomRotation(10),\n transforms.ToTensor(),\n transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))\n ])\n\n# choose the training and test datasets\ntrain_data = datasets.CIFAR10('data', train=True,\n download=True, transform=transform)\ntest_data = datasets.CIFAR10('data', train=False,\n download=True, transform=transform)\n\n# obtain training indices that will be used for validation\nnum_train = len(train_data)\nindices = list(range(num_train))\nnp.random.shuffle(indices)\nsplit = int(np.floor(valid_size * num_train))\ntrain_idx, valid_idx = indices[split:], indices[:split]\n\n# define samplers for obtaining training and validation batches\ntrain_sampler = SubsetRandomSampler(train_idx)\nvalid_sampler = SubsetRandomSampler(valid_idx)\n\n# prepare data loaders (combine dataset and sampler)\ntrain_loader = torch.utils.data.DataLoader(train_data, batch_size=batch_size,\n sampler=train_sampler, num_workers=num_workers)\nvalid_loader = torch.utils.data.DataLoader(train_data, batch_size=batch_size, \n sampler=valid_sampler, num_workers=num_workers)\ntest_loader = torch.utils.data.DataLoader(test_data, batch_size=batch_size, \n num_workers=num_workers)\n\n# specify the image classes\nclasses = ['airplane', 'automobile', 'bird', 'cat', 'deer',\n 'dog', 'frog', 'horse', 'ship', 'truck']",
"Files already downloaded and verified\nFiles already downloaded and verified\n"
]
],
[
[
"### Visualize a Batch of Training Data",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n%matplotlib inline\n\n# helper function to un-normalize and display an image\ndef imshow(img):\n img = img / 2 + 0.5 # unnormalize\n plt.imshow(np.transpose(img, (1, 2, 0))) # convert from Tensor image",
"_____no_output_____"
],
[
"# obtain one batch of training images\ndataiter = iter(train_loader)\nimages, labels = dataiter.next()\nimages = images.numpy() # convert images to numpy for display\n\n# plot the images in the batch, along with the corresponding labels\nfig = plt.figure(figsize=(25, 4))\n# display 20 images\nfor idx in np.arange(20):\n ax = fig.add_subplot(2, 20/2, idx+1, xticks=[], yticks=[])\n imshow(images[idx])\n ax.set_title(classes[labels[idx]])",
"_____no_output_____"
]
],
[
[
"### View an Image in More Detail\n\nHere, we look at the normalized red, green, and blue (RGB) color channels as three separate, grayscale intensity images.",
"_____no_output_____"
]
],
[
[
"rgb_img = np.squeeze(images[3])\nchannels = ['red channel', 'green channel', 'blue channel']\n\nfig = plt.figure(figsize = (36, 36)) \nfor idx in np.arange(rgb_img.shape[0]):\n ax = fig.add_subplot(1, 3, idx + 1)\n img = rgb_img[idx]\n ax.imshow(img, cmap='gray')\n ax.set_title(channels[idx])\n width, height = img.shape\n thresh = img.max()/2.5\n for x in range(width):\n for y in range(height):\n val = round(img[x][y],2) if img[x][y] !=0 else 0\n ax.annotate(str(val), xy=(y,x),\n horizontalalignment='center',\n verticalalignment='center', size=8,\n color='white' if img[x][y]<thresh else 'black')",
"_____no_output_____"
]
],
[
[
"---\n## Define the Network [Architecture](http://pytorch.org/docs/stable/nn.html)\n\nThis time, you'll define a CNN architecture. Instead of an MLP, which used linear, fully-connected layers, you'll use the following:\n* [Convolutional layers](https://pytorch.org/docs/stable/nn.html#conv2d), which can be thought of as stack of filtered images.\n* [Maxpooling layers](https://pytorch.org/docs/stable/nn.html#maxpool2d), which reduce the x-y size of an input, keeping only the most _active_ pixels from the previous layer.\n* The usual Linear + Dropout layers to avoid overfitting and produce a 10-dim output.\n\nA network with 2 convolutional layers is shown in the image below and in the code, and you've been given starter code with one convolutional and one maxpooling layer.\n\n<img src='notebook_ims/2_layer_conv.png' height=50% width=50% />\n\n#### TODO: Define a model with multiple convolutional layers, and define the feedforward metwork behavior.\n\nThe more convolutional layers you include, the more complex patterns in color and shape a model can detect. It's suggested that your final model include 2 or 3 convolutional layers as well as linear layers + dropout in between to avoid overfitting. \n\nIt's good practice to look at existing research and implementations of related models as a starting point for defining your own models. You may find it useful to look at [this PyTorch classification example](https://github.com/pytorch/tutorials/blob/master/beginner_source/blitz/cifar10_tutorial.py) or [this, more complex Keras example](https://github.com/keras-team/keras/blob/master/examples/cifar10_cnn.py) to help decide on a final structure.\n\n#### Output volume for a convolutional layer\n\nTo compute the output size of a given convolutional layer we can perform the following calculation (taken from [Stanford's cs231n course](http://cs231n.github.io/convolutional-networks/#layers)):\n> We can compute the spatial size of the output volume as a function of the input volume size (W), the kernel/filter size (F), the stride with which they are applied (S), and the amount of zero padding used (P) on the border. The correct formula for calculating how many neurons define the output_W is given by `(W−F+2P)/S+1`. \n\nFor example for a 7x7 input and a 3x3 filter with stride 1 and pad 0 we would get a 5x5 output. With stride 2 we would get a 3x3 output.",
"_____no_output_____"
]
],
[
[
"import torch.nn as nn\nimport torch.nn.functional as F\n\n# define the CNN architecture\nclass Net(nn.Module):\n def __init__(self):\n super(Net, self).__init__()\n # convolutional layer (sees 32x32x3 image tensor)\n self.conv1 = nn.Conv2d(3, 16, 3, padding=1)\n # convolutional layer (sees 16x16x16 tensor)\n self.conv2 = nn.Conv2d(16, 32, 3, padding=1)\n # convolutional layer (sees 8x8x32 tensor)\n self.conv3 = nn.Conv2d(32, 64, 3, padding=1)\n # max pooling layer\n self.pool = nn.MaxPool2d(2, 2)\n # linear layer (64 * 4 * 4 -> 500)\n self.fc1 = nn.Linear(64 * 4 * 4, 500)\n # linear layer (500 -> 10)\n self.fc2 = nn.Linear(500, 10)\n # dropout layer (p=0.25)\n self.dropout = nn.Dropout(0.25)\n\n def forward(self, x):\n # add sequence of convolutional and max pooling layers\n x = self.pool(F.relu(self.conv1(x)))\n x = self.pool(F.relu(self.conv2(x)))\n x = self.pool(F.relu(self.conv3(x)))\n # flatten image input\n x = x.view(-1, 64 * 4 * 4)\n # add dropout layer\n x = self.dropout(x)\n # add 1st hidden layer, with relu activation function\n x = F.relu(self.fc1(x))\n # add dropout layer\n x = self.dropout(x)\n # add 2nd hidden layer, with relu activation function\n x = self.fc2(x)\n return x\n\n# create a complete CNN\nmodel = Net()\nprint(model)\n\n# move tensors to GPU if CUDA is available\nif train_on_gpu:\n model.cuda()",
"Net(\n (conv1): Conv2d(3, 16, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (conv2): Conv2d(16, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (conv3): Conv2d(32, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (pool): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)\n (fc1): Linear(in_features=1024, out_features=500, bias=True)\n (fc2): Linear(in_features=500, out_features=10, bias=True)\n (dropout): Dropout(p=0.25)\n)\n"
]
],
[
[
"### Specify [Loss Function](http://pytorch.org/docs/stable/nn.html#loss-functions) and [Optimizer](http://pytorch.org/docs/stable/optim.html)\n\nDecide on a loss and optimization function that is best suited for this classification task. The linked code examples from above, may be a good starting point; [this PyTorch classification example](https://github.com/pytorch/tutorials/blob/master/beginner_source/blitz/cifar10_tutorial.py) or [this, more complex Keras example](https://github.com/keras-team/keras/blob/master/examples/cifar10_cnn.py). Pay close attention to the value for **learning rate** as this value determines how your model converges to a small error.\n\n#### TODO: Define the loss and optimizer and see how these choices change the loss over time.",
"_____no_output_____"
]
],
[
[
"import torch.optim as optim\n\n# specify loss function (categorical cross-entropy)\ncriterion = nn.CrossEntropyLoss()\n\n# specify optimizer\noptimizer = optim.SGD(model.parameters(), lr=0.01)",
"_____no_output_____"
]
],
[
[
"---\n## Train the Network\n\nRemember to look at how the training and validation loss decreases over time; if the validation loss ever increases it indicates possible overfitting.",
"_____no_output_____"
]
],
[
[
"# number of epochs to train the model\nn_epochs = 30\n\nvalid_loss_min = np.Inf # track change in validation loss\n\nfor epoch in range(1, n_epochs+1):\n\n # keep track of training and validation loss\n train_loss = 0.0\n valid_loss = 0.0\n \n ###################\n # train the model #\n ###################\n model.train()\n for batch_idx, (data, target) in enumerate(train_loader):\n # move tensors to GPU if CUDA is available\n if train_on_gpu:\n data, target = data.cuda(), target.cuda()\n # clear the gradients of all optimized variables\n optimizer.zero_grad()\n # forward pass: compute predicted outputs by passing inputs to the model\n output = model(data)\n # calculate the batch loss\n loss = criterion(output, target)\n # backward pass: compute gradient of the loss with respect to model parameters\n loss.backward()\n # perform a single optimization step (parameter update)\n optimizer.step()\n # update training loss\n train_loss += loss.item()*data.size(0)\n \n ###################### \n # validate the model #\n ######################\n model.eval()\n for batch_idx, (data, target) in enumerate(valid_loader):\n # move tensors to GPU if CUDA is available\n if train_on_gpu:\n data, target = data.cuda(), target.cuda()\n # forward pass: compute predicted outputs by passing inputs to the model\n output = model(data)\n # calculate the batch loss\n loss = criterion(output, target)\n # update average validation loss \n valid_loss += loss.item()*data.size(0)\n \n # calculate average losses\n train_loss = train_loss/len(train_loader.dataset)\n valid_loss = valid_loss/len(valid_loader.dataset)\n \n # print training/validation statistics \n print('Epoch: {} \\tTraining Loss: {:.6f} \\tValidation Loss: {:.6f}'.format(\n epoch, train_loss, valid_loss))\n \n # save model if validation loss has decreased\n if valid_loss <= valid_loss_min:\n print('Validation loss decreased ({:.6f} --> {:.6f}). Saving model ...'.format(\n valid_loss_min,\n valid_loss))\n torch.save(model.state_dict(), 'model_augmented.pt')\n valid_loss_min = valid_loss",
"Epoch: 1 \tTraining Loss: 1.483400 \tValidation Loss: 0.357015\nValidation loss decreased (inf --> 0.357015). Saving model ...\nEpoch: 2 \tTraining Loss: 1.462634 \tValidation Loss: 0.355032\nValidation loss decreased (0.357015 --> 0.355032). Saving model ...\nEpoch: 3 \tTraining Loss: 1.454201 \tValidation Loss: 0.351629\nValidation loss decreased (0.355032 --> 0.351629). Saving model ...\nEpoch: 4 \tTraining Loss: 1.446663 \tValidation Loss: 0.349965\nValidation loss decreased (0.351629 --> 0.349965). Saving model ...\nEpoch: 5 \tTraining Loss: 1.444058 \tValidation Loss: 0.348746\nValidation loss decreased (0.349965 --> 0.348746). Saving model ...\nEpoch: 6 \tTraining Loss: 1.443467 \tValidation Loss: 0.350312\nEpoch: 7 \tTraining Loss: 1.435970 \tValidation Loss: 0.347319\nValidation loss decreased (0.348746 --> 0.347319). Saving model ...\nEpoch: 8 \tTraining Loss: 1.430920 \tValidation Loss: 0.347215\nValidation loss decreased (0.347319 --> 0.347215). Saving model ...\nEpoch: 9 \tTraining Loss: 1.428707 \tValidation Loss: 0.347475\nEpoch: 10 \tTraining Loss: 1.426082 \tValidation Loss: 0.344052\nValidation loss decreased (0.347215 --> 0.344052). Saving model ...\nEpoch: 11 \tTraining Loss: 1.421921 \tValidation Loss: 0.343847\nValidation loss decreased (0.344052 --> 0.343847). Saving model ...\nEpoch: 12 \tTraining Loss: 1.419442 \tValidation Loss: 0.344311\nEpoch: 13 \tTraining Loss: 1.418585 \tValidation Loss: 0.345510\nEpoch: 14 \tTraining Loss: 1.415068 \tValidation Loss: 0.342422\nValidation loss decreased (0.343847 --> 0.342422). Saving model ...\nEpoch: 15 \tTraining Loss: 1.410706 \tValidation Loss: 0.340904\nValidation loss decreased (0.342422 --> 0.340904). Saving model ...\nEpoch: 16 \tTraining Loss: 1.415896 \tValidation Loss: 0.342239\nEpoch: 17 \tTraining Loss: 1.408537 \tValidation Loss: 0.339082\nValidation loss decreased (0.340904 --> 0.339082). Saving model ...\nEpoch: 18 \tTraining Loss: 1.406269 \tValidation Loss: 0.343943\nEpoch: 19 \tTraining Loss: 1.404933 \tValidation Loss: 0.338068\nValidation loss decreased (0.339082 --> 0.338068). Saving model ...\nEpoch: 20 \tTraining Loss: 1.404783 \tValidation Loss: 0.341946\nEpoch: 21 \tTraining Loss: 1.401647 \tValidation Loss: 0.340137\nEpoch: 22 \tTraining Loss: 1.402606 \tValidation Loss: 0.339872\nEpoch: 23 \tTraining Loss: 1.398441 \tValidation Loss: 0.337148\nValidation loss decreased (0.338068 --> 0.337148). Saving model ...\nEpoch: 24 \tTraining Loss: 1.401196 \tValidation Loss: 0.335357\nValidation loss decreased (0.337148 --> 0.335357). Saving model ...\nEpoch: 25 \tTraining Loss: 1.392405 \tValidation Loss: 0.339850\nEpoch: 26 \tTraining Loss: 1.392132 \tValidation Loss: 0.339279\nEpoch: 27 \tTraining Loss: 1.387730 \tValidation Loss: 0.331665\nValidation loss decreased (0.335357 --> 0.331665). Saving model ...\nEpoch: 28 \tTraining Loss: 1.380706 \tValidation Loss: 0.340137\nEpoch: 29 \tTraining Loss: 1.374192 \tValidation Loss: 0.332150\nEpoch: 30 \tTraining Loss: 1.372476 \tValidation Loss: 0.332638\n"
]
],
[
[
"### Load the Model with the Lowest Validation Loss",
"_____no_output_____"
]
],
[
[
"model.load_state_dict(torch.load('model_augmented.pt'))",
"_____no_output_____"
]
],
[
[
"---\n## Test the Trained Network\n\nTest your trained model on previously unseen data! A \"good\" result will be a CNN that gets around 70% (or more, try your best!) accuracy on these test images.",
"_____no_output_____"
]
],
[
[
"# track test loss\ntest_loss = 0.0\nclass_correct = list(0. for i in range(10))\nclass_total = list(0. for i in range(10))\n\nmodel.eval()\n# iterate over test data\nfor batch_idx, (data, target) in enumerate(test_loader):\n # move tensors to GPU if CUDA is available\n if train_on_gpu:\n data, target = data.cuda(), target.cuda()\n # forward pass: compute predicted outputs by passing inputs to the model\n output = model(data)\n # calculate the batch loss\n loss = criterion(output, target)\n # update test loss \n test_loss += loss.item()*data.size(0)\n # convert output probabilities to predicted class\n _, pred = torch.max(output, 1) \n # compare predictions to true label\n correct_tensor = pred.eq(target.data.view_as(pred))\n correct = np.squeeze(correct_tensor.numpy()) if not train_on_gpu else np.squeeze(correct_tensor.cpu().numpy())\n # calculate test accuracy for each object class\n for i in range(batch_size):\n label = target.data[i]\n class_correct[label] += correct[i].item()\n class_total[label] += 1\n\n# average test loss\ntest_loss = test_loss/len(test_loader.dataset)\nprint('Test Loss: {:.6f}\\n'.format(test_loss))\n\nfor i in range(10):\n if class_total[i] > 0:\n print('Test Accuracy of %5s: %2d%% (%2d/%2d)' % (\n classes[i], 100 * class_correct[i] / class_total[i],\n np.sum(class_correct[i]), np.sum(class_total[i])))\n else:\n print('Test Accuracy of %5s: N/A (no training examples)' % (classes[i]))\n\nprint('\\nTest Accuracy (Overall): %2d%% (%2d/%2d)' % (\n 100. * np.sum(class_correct) / np.sum(class_total),\n np.sum(class_correct), np.sum(class_total)))",
"Test Loss: 1.650751\n\nTest Accuracy of airplane: 37% (374/1000)\nTest Accuracy of automobile: 47% (471/1000)\nTest Accuracy of bird: 8% (80/1000)\nTest Accuracy of cat: 20% (203/1000)\nTest Accuracy of deer: 39% (390/1000)\nTest Accuracy of dog: 27% (274/1000)\nTest Accuracy of frog: 37% (372/1000)\nTest Accuracy of horse: 48% (483/1000)\nTest Accuracy of ship: 47% (473/1000)\nTest Accuracy of truck: 58% (581/1000)\n\nTest Accuracy (Overall): 37% (3701/10000)\n"
]
],
[
[
"### Visualize Sample Test Results",
"_____no_output_____"
]
],
[
[
"# obtain one batch of test images\ndataiter = iter(test_loader)\nimages, labels = dataiter.next()\nimages.numpy()\n\n# move model inputs to cuda, if GPU available\nif train_on_gpu:\n images = images.cuda()\n\n# get sample outputs\noutput = model(images)\n# convert output probabilities to predicted class\n_, preds_tensor = torch.max(output, 1)\npreds = np.squeeze(preds_tensor.numpy()) if not train_on_gpu else np.squeeze(preds_tensor.cpu().numpy())\n\n# plot the images in the batch, along with predicted and true labels\nfig = plt.figure(figsize=(25, 4))\nfor idx in np.arange(20):\n ax = fig.add_subplot(2, 20/2, idx+1, xticks=[], yticks=[])\n imshow(images[idx])\n ax.set_title(\"{} ({})\".format(classes[preds[idx]], classes[labels[idx]]),\n color=(\"green\" if preds[idx]==labels[idx].item() else \"red\"))",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0931034e7af3b1eb9927dcad49b06263e02add9 | 67,527 | ipynb | Jupyter Notebook | nbs/03_data.core.ipynb | aaminggo/fastai | fbc60a2bbfc13a92a71af657e0052761116ab447 | [
"Apache-2.0"
] | 1 | 2021-01-24T23:17:40.000Z | 2021-01-24T23:17:40.000Z | nbs/03_data.core.ipynb | aaminggo/fastai | fbc60a2bbfc13a92a71af657e0052761116ab447 | [
"Apache-2.0"
] | null | null | null | nbs/03_data.core.ipynb | aaminggo/fastai | fbc60a2bbfc13a92a71af657e0052761116ab447 | [
"Apache-2.0"
] | 1 | 2020-11-28T19:12:10.000Z | 2020-11-28T19:12:10.000Z | 32.621739 | 828 | 0.535623 | [
[
[
"#hide\n#skip\n! [ -e /content ] && pip install -Uqq fastai # upgrade fastai on colab",
"_____no_output_____"
],
[
"#default_exp data.core",
"_____no_output_____"
],
[
"#export\nfrom fastai.torch_basics import *\nfrom fastai.data.load import *",
"_____no_output_____"
],
[
"#hide\nfrom nbdev.showdoc import *",
"_____no_output_____"
]
],
[
[
"# Data core\n\n> Core functionality for gathering data",
"_____no_output_____"
],
[
"The classes here provide functionality for applying a list of transforms to a set of items (`TfmdLists`, `Datasets`) or a `DataLoader` (`TfmdDl`) as well as the base class used to gather the data for model training: `DataLoaders`.",
"_____no_output_____"
],
[
"## TfmdDL -",
"_____no_output_____"
]
],
[
[
"#export\n@typedispatch\ndef show_batch(x, y, samples, ctxs=None, max_n=9, **kwargs):\n if ctxs is None: ctxs = Inf.nones\n if hasattr(samples[0], 'show'):\n ctxs = [s.show(ctx=c, **kwargs) for s,c,_ in zip(samples,ctxs,range(max_n))]\n else:\n for i in range_of(samples[0]):\n ctxs = [b.show(ctx=c, **kwargs) for b,c,_ in zip(samples.itemgot(i),ctxs,range(max_n))]\n return ctxs",
"_____no_output_____"
]
],
[
[
"`show_batch` is a type-dispatched function that is responsible for showing decoded `samples`. `x` and `y` are the input and the target in the batch to be shown, and are passed along to dispatch on their types. There is a different implementation of `show_batch` if `x` is a `TensorImage` or a `TensorText` for instance (see vision.core or text.data for more details). `ctxs` can be passed but the function is responsible to create them if necessary. `kwargs` depend on the specific implementation.",
"_____no_output_____"
]
],
[
[
"#export\n@typedispatch\ndef show_results(x, y, samples, outs, ctxs=None, max_n=9, **kwargs):\n if ctxs is None: ctxs = Inf.nones\n for i in range(len(samples[0])):\n ctxs = [b.show(ctx=c, **kwargs) for b,c,_ in zip(samples.itemgot(i),ctxs,range(max_n))]\n for i in range(len(outs[0])):\n ctxs = [b.show(ctx=c, **kwargs) for b,c,_ in zip(outs.itemgot(i),ctxs,range(max_n))]\n return ctxs",
"_____no_output_____"
]
],
[
[
"`show_results` is a type-dispatched function that is responsible for showing decoded `samples` and their corresponding `outs`. Like in `show_batch`, `x` and `y` are the input and the target in the batch to be shown, and are passed along to dispatch on their types. `ctxs` can be passed but the function is responsible to create them if necessary. `kwargs` depend on the specific implementation.",
"_____no_output_____"
]
],
[
[
"#export\n_all_ = [\"show_batch\", \"show_results\"]",
"_____no_output_____"
],
[
"#export\n_batch_tfms = ('after_item','before_batch','after_batch')",
"_____no_output_____"
],
[
"#export\n@log_args(but_as=DataLoader.__init__)\n@delegates()\nclass TfmdDL(DataLoader):\n \"Transformed `DataLoader`\"\n def __init__(self, dataset, bs=64, shuffle=False, num_workers=None, verbose=False, do_setup=True, **kwargs):\n if num_workers is None: num_workers = min(16, defaults.cpus)\n for nm in _batch_tfms: kwargs[nm] = Pipeline(kwargs.get(nm,None))\n super().__init__(dataset, bs=bs, shuffle=shuffle, num_workers=num_workers, **kwargs)\n if do_setup:\n for nm in _batch_tfms:\n pv(f\"Setting up {nm}: {kwargs[nm]}\", verbose)\n kwargs[nm].setup(self)\n\n def _one_pass(self):\n b = self.do_batch([self.do_item(0)])\n if self.device is not None: b = to_device(b, self.device)\n its = self.after_batch(b)\n self._n_inp = 1 if not isinstance(its, (list,tuple)) or len(its)==1 else len(its)-1\n self._types = explode_types(its)\n\n def _retain_dl(self,b):\n if not getattr(self, '_types', None): self._one_pass()\n return retain_types(b, typs=self._types)\n\n @delegates(DataLoader.new)\n def new(self, dataset=None, cls=None, **kwargs):\n res = super().new(dataset, cls, do_setup=False, **kwargs)\n if not hasattr(self, '_n_inp') or not hasattr(self, '_types'):\n try:\n self._one_pass()\n res._n_inp,res._types = self._n_inp,self._types\n except: print(\"Could not do one pass in your dataloader, there is something wrong in it\")\n else: res._n_inp,res._types = self._n_inp,self._types\n return res\n\n def before_iter(self):\n super().before_iter()\n split_idx = getattr(self.dataset, 'split_idx', None)\n for nm in _batch_tfms:\n f = getattr(self,nm)\n if isinstance(f,Pipeline): f.split_idx=split_idx\n\n def decode(self, b): return self.before_batch.decode(to_cpu(self.after_batch.decode(self._retain_dl(b))))\n def decode_batch(self, b, max_n=9, full=True): return self._decode_batch(self.decode(b), max_n, full)\n\n def _decode_batch(self, b, max_n=9, full=True):\n f = self.after_item.decode\n f = compose(f, partial(getattr(self.dataset,'decode',noop), full = full))\n return L(batch_to_samples(b, max_n=max_n)).map(f)\n\n def _pre_show_batch(self, b, max_n=9):\n \"Decode `b` to be ready for `show_batch`\"\n b = self.decode(b)\n if hasattr(b, 'show'): return b,None,None\n its = self._decode_batch(b, max_n, full=False)\n if not is_listy(b): b,its = [b],L((o,) for o in its)\n return detuplify(b[:self.n_inp]),detuplify(b[self.n_inp:]),its\n\n def show_batch(self, b=None, max_n=9, ctxs=None, show=True, unique=False, **kwargs):\n if unique:\n old_get_idxs = self.get_idxs\n self.get_idxs = lambda: Inf.zeros\n if b is None: b = self.one_batch()\n if not show: return self._pre_show_batch(b, max_n=max_n)\n show_batch(*self._pre_show_batch(b, max_n=max_n), ctxs=ctxs, max_n=max_n, **kwargs)\n if unique: self.get_idxs = old_get_idxs\n\n def show_results(self, b, out, max_n=9, ctxs=None, show=True, **kwargs):\n x,y,its = self.show_batch(b, max_n=max_n, show=False)\n b_out = type(b)(b[:self.n_inp] + (tuple(out) if is_listy(out) else (out,)))\n x1,y1,outs = self.show_batch(b_out, max_n=max_n, show=False)\n res = (x,x1,None,None) if its is None else (x, y, its, outs.itemgot(slice(self.n_inp,None)))\n if not show: return res\n show_results(*res, ctxs=ctxs, max_n=max_n, **kwargs)\n\n @property\n def n_inp(self):\n if hasattr(self.dataset, 'n_inp'): return self.dataset.n_inp\n if not hasattr(self, '_n_inp'): self._one_pass()\n return self._n_inp\n\n def to(self, device):\n self.device = device\n for tfm in self.after_batch.fs:\n for a in L(getattr(tfm, 'parameters', None)): setattr(tfm, a, getattr(tfm, a).to(device))\n return self",
"_____no_output_____"
]
],
[
[
"A `TfmdDL` is a `DataLoader` that creates `Pipeline` from a list of `Transform`s for the callbacks `after_item`, `before_batch` and `after_batch`. As a result, it can decode or show a processed `batch`.",
"_____no_output_____"
]
],
[
[
"#export\nadd_docs(TfmdDL,\n decode=\"Decode `b` using `tfms`\",\n decode_batch=\"Decode `b` entirely\",\n new=\"Create a new version of self with a few changed attributes\",\n show_batch=\"Show `b` (defaults to `one_batch`), a list of lists of pipeline outputs (i.e. output of a `DataLoader`)\",\n show_results=\"Show each item of `b` and `out`\",\n before_iter=\"override\",\n to=\"Put self and its transforms state on `device`\")",
"_____no_output_____"
],
[
"class _Category(int, ShowTitle): pass",
"_____no_output_____"
],
[
"#Test retain type\nclass NegTfm(Transform):\n def encodes(self, x): return torch.neg(x)\n def decodes(self, x): return torch.neg(x)\n \ntdl = TfmdDL([(TensorImage([1]),)] * 4, after_batch=NegTfm(), bs=4, num_workers=4)\nb = tdl.one_batch()\ntest_eq(type(b[0]), TensorImage)\nb = (tensor([1.,1.,1.,1.]),)\ntest_eq(type(tdl.decode_batch(b)[0][0]), TensorImage)",
"_____no_output_____"
],
[
"class A(Transform): \n def encodes(self, x): return x \n def decodes(self, x): return TitledInt(x) \n\n@Transform\ndef f(x)->None: return fastuple((x,x))\n\nstart = torch.arange(50)\ntest_eq_type(f(2), fastuple((2,2)))",
"_____no_output_____"
],
[
"a = A()\ntdl = TfmdDL(start, after_item=lambda x: (a(x), f(x)), bs=4)\nx,y = tdl.one_batch()\ntest_eq(type(y), fastuple)\n\ns = tdl.decode_batch((x,y))\ntest_eq(type(s[0][1]), fastuple)",
"_____no_output_____"
],
[
"tdl = TfmdDL(torch.arange(0,50), after_item=A(), after_batch=NegTfm(), bs=4)\ntest_eq(tdl.dataset[0], start[0])\ntest_eq(len(tdl), (50-1)//4+1)\ntest_eq(tdl.bs, 4)\ntest_stdout(tdl.show_batch, '0\\n1\\n2\\n3')\ntest_stdout(partial(tdl.show_batch, unique=True), '0\\n0\\n0\\n0')",
"_____no_output_____"
],
[
"class B(Transform):\n parameters = 'a'\n def __init__(self): self.a = torch.tensor(0.)\n def encodes(self, x): x\n \ntdl = TfmdDL([(TensorImage([1]),)] * 4, after_batch=B(), bs=4)\ntest_eq(tdl.after_batch.fs[0].a.device, torch.device('cpu'))\ntdl.to(default_device())\ntest_eq(tdl.after_batch.fs[0].a.device, default_device())",
"_____no_output_____"
]
],
[
[
"### Methods",
"_____no_output_____"
]
],
[
[
"show_doc(TfmdDL.one_batch)",
"_____no_output_____"
],
[
"tfm = NegTfm()\ntdl = TfmdDL(start, after_batch=tfm, bs=4)",
"_____no_output_____"
],
[
"b = tdl.one_batch()\ntest_eq(tensor([0,-1,-2,-3]), b)",
"_____no_output_____"
],
[
"show_doc(TfmdDL.decode)",
"_____no_output_____"
],
[
"test_eq(tdl.decode(b), tensor(0,1,2,3))",
"_____no_output_____"
],
[
"show_doc(TfmdDL.decode_batch)",
"_____no_output_____"
],
[
"test_eq(tdl.decode_batch(b), [0,1,2,3])",
"_____no_output_____"
],
[
"show_doc(TfmdDL.show_batch)",
"_____no_output_____"
],
[
"show_doc(TfmdDL.to)",
"_____no_output_____"
]
],
[
[
"## DataLoaders -",
"_____no_output_____"
]
],
[
[
"# export\n@docs\nclass DataLoaders(GetAttr):\n \"Basic wrapper around several `DataLoader`s.\"\n _default='train'\n def __init__(self, *loaders, path='.', device=None):\n self.loaders,self.path = list(loaders),Path(path)\n if device is not None or hasattr(loaders[0],'to'): self.device = device\n\n def __getitem__(self, i): return self.loaders[i]\n def new_empty(self):\n loaders = [dl.new(dl.dataset.new_empty()) for dl in self.loaders]\n return type(self)(*loaders, path=self.path, device=self.device)\n\n def _set(i, self, v): self.loaders[i] = v\n train ,valid = add_props(lambda i,x: x[i], _set)\n train_ds,valid_ds = add_props(lambda i,x: x[i].dataset)\n\n @property\n def device(self): return self._device\n\n @device.setter\n def device(self, d):\n for dl in self.loaders: dl.to(d)\n self._device = d\n\n def to(self, device):\n self.device = device\n return self\n\n def cuda(self): return self.to(device=default_device())\n def cpu(self): return self.to(device=torch.device('cpu'))\n\n @classmethod\n def from_dsets(cls, *ds, path='.', bs=64, device=None, dl_type=TfmdDL, **kwargs):\n default = (True,) + (False,) * (len(ds)-1)\n defaults = {'shuffle': default, 'drop_last': default}\n for nm in _batch_tfms:\n if nm in kwargs: kwargs[nm] = Pipeline(kwargs[nm])\n kwargs = merge(defaults, {k: tuplify(v, match=ds) for k,v in kwargs.items()})\n kwargs = [{k: v[i] for k,v in kwargs.items()} for i in range_of(ds)]\n return cls(*[dl_type(d, bs=bs, **k) for d,k in zip(ds, kwargs)], path=path, device=device)\n\n @classmethod\n def from_dblock(cls, dblock, source, path='.', bs=64, val_bs=None, shuffle_train=True, device=None, **kwargs):\n return dblock.dataloaders(source, path=path, bs=bs, val_bs=val_bs, shuffle_train=shuffle_train, device=device, **kwargs)\n\n _docs=dict(__getitem__=\"Retrieve `DataLoader` at `i` (`0` is training, `1` is validation)\",\n train=\"Training `DataLoader`\",\n valid=\"Validation `DataLoader`\",\n train_ds=\"Training `Dataset`\",\n valid_ds=\"Validation `Dataset`\",\n to=\"Use `device`\",\n cuda=\"Use the gpu if available\",\n cpu=\"Use the cpu\",\n new_empty=\"Create a new empty version of `self` with the same transforms\",\n from_dblock=\"Create a dataloaders from a given `dblock`\")",
"_____no_output_____"
],
[
"dls = DataLoaders(tdl,tdl)\nx = dls.train.one_batch()\nx2 = first(tdl)\ntest_eq(x,x2)\nx2 = dls.one_batch()\ntest_eq(x,x2)",
"_____no_output_____"
],
[
"#hide\n#test assignment works\ndls.train = dls.train.new(bs=4)",
"_____no_output_____"
]
],
[
[
"### Methods",
"_____no_output_____"
]
],
[
[
"show_doc(DataLoaders.__getitem__)",
"_____no_output_____"
],
[
"x2 = dls[0].one_batch()\ntest_eq(x,x2)",
"_____no_output_____"
],
[
"show_doc(DataLoaders.train, name=\"DataLoaders.train\")",
"_____no_output_____"
],
[
"show_doc(DataLoaders.valid, name=\"DataLoaders.valid\")",
"_____no_output_____"
],
[
"show_doc(DataLoaders.train_ds, name=\"DataLoaders.train_ds\")",
"_____no_output_____"
],
[
"show_doc(DataLoaders.valid_ds, name=\"DataLoaders.valid_ds\")",
"_____no_output_____"
]
],
[
[
"## TfmdLists -",
"_____no_output_____"
]
],
[
[
"#export\nclass FilteredBase:\n \"Base class for lists with subsets\"\n _dl_type,_dbunch_type = TfmdDL,DataLoaders\n def __init__(self, *args, dl_type=None, **kwargs):\n if dl_type is not None: self._dl_type = dl_type\n self.dataloaders = delegates(self._dl_type.__init__)(self.dataloaders)\n super().__init__(*args, **kwargs)\n\n @property\n def n_subsets(self): return len(self.splits)\n def _new(self, items, **kwargs): return super()._new(items, splits=self.splits, **kwargs)\n def subset(self): raise NotImplemented\n\n def dataloaders(self, bs=64, val_bs=None, shuffle_train=True, n=None, path='.', dl_type=None, dl_kwargs=None,\n device=None, **kwargs):\n if device is None: device=default_device()\n if dl_kwargs is None: dl_kwargs = [{}] * self.n_subsets\n if dl_type is None: dl_type = self._dl_type\n drop_last = kwargs.pop('drop_last', shuffle_train)\n dl = dl_type(self.subset(0), bs=bs, shuffle=shuffle_train, drop_last=drop_last, n=n, device=device,\n **merge(kwargs, dl_kwargs[0]))\n dls = [dl] + [dl.new(self.subset(i), bs=(bs if val_bs is None else val_bs), shuffle=False, drop_last=False,\n n=None, **dl_kwargs[i]) for i in range(1, self.n_subsets)]\n return self._dbunch_type(*dls, path=path, device=device)\n\nFilteredBase.train,FilteredBase.valid = add_props(lambda i,x: x.subset(i))",
"_____no_output_____"
],
[
"#export\nclass TfmdLists(FilteredBase, L, GetAttr):\n \"A `Pipeline` of `tfms` applied to a collection of `items`\"\n _default='tfms'\n def __init__(self, items, tfms, use_list=None, do_setup=True, split_idx=None, train_setup=True,\n splits=None, types=None, verbose=False, dl_type=None):\n super().__init__(items, use_list=use_list)\n if dl_type is not None: self._dl_type = dl_type\n self.splits = L([slice(None),[]] if splits is None else splits).map(mask2idxs)\n if isinstance(tfms,TfmdLists): tfms = tfms.tfms\n if isinstance(tfms,Pipeline): do_setup=False\n self.tfms = Pipeline(tfms, split_idx=split_idx)\n store_attr('types,split_idx')\n if do_setup:\n pv(f\"Setting up {self.tfms}\", verbose)\n self.setup(train_setup=train_setup)\n\n def _new(self, items, split_idx=None, **kwargs):\n split_idx = ifnone(split_idx,self.split_idx)\n return super()._new(items, tfms=self.tfms, do_setup=False, types=self.types, split_idx=split_idx, **kwargs)\n def subset(self, i): return self._new(self._get(self.splits[i]), split_idx=i)\n def _after_item(self, o): return self.tfms(o)\n def __repr__(self): return f\"{self.__class__.__name__}: {self.items}\\ntfms - {self.tfms.fs}\"\n def __iter__(self): return (self[i] for i in range(len(self)))\n def show(self, o, **kwargs): return self.tfms.show(o, **kwargs)\n def decode(self, o, **kwargs): return self.tfms.decode(o, **kwargs)\n def __call__(self, o, **kwargs): return self.tfms.__call__(o, **kwargs)\n def overlapping_splits(self): return L(Counter(self.splits.concat()).values()).filter(gt(1))\n def new_empty(self): return self._new([])\n\n def setup(self, train_setup=True):\n self.tfms.setup(self, train_setup)\n if len(self) != 0:\n x = super().__getitem__(0) if self.splits is None else super().__getitem__(self.splits[0])[0]\n self.types = []\n for f in self.tfms.fs:\n self.types.append(getattr(f, 'input_types', type(x)))\n x = f(x)\n self.types.append(type(x))\n types = L(t if is_listy(t) else [t] for t in self.types).concat().unique()\n self.pretty_types = '\\n'.join([f' - {t}' for t in types])\n\n def infer_idx(self, x):\n # TODO: check if we really need this, or can simplify\n idx = 0\n for t in self.types:\n if isinstance(x, t): break\n idx += 1\n types = L(t if is_listy(t) else [t] for t in self.types).concat().unique()\n pretty_types = '\\n'.join([f' - {t}' for t in types])\n assert idx < len(self.types), f\"Expected an input of type in \\n{pretty_types}\\n but got {type(x)}\"\n return idx\n\n def infer(self, x):\n return compose_tfms(x, tfms=self.tfms.fs[self.infer_idx(x):], split_idx=self.split_idx)\n\n def __getitem__(self, idx):\n res = super().__getitem__(idx)\n if self._after_item is None: return res\n return self._after_item(res) if is_indexer(idx) else res.map(self._after_item)",
"_____no_output_____"
],
[
"#export\nadd_docs(TfmdLists,\n setup=\"Transform setup with self\",\n decode=\"From `Pipeline`\",\n show=\"From `Pipeline`\",\n overlapping_splits=\"All splits that are in more than one split\",\n subset=\"New `TfmdLists` with same tfms that only includes items in `i`th split\",\n infer_idx=\"Finds the index where `self.tfms` can be applied to `x`, depending on the type of `x`\",\n infer=\"Apply `self.tfms` to `x` starting at the right tfm depending on the type of `x`\",\n new_empty=\"A new version of `self` but with no items\")",
"_____no_output_____"
],
[
"#exports\ndef decode_at(o, idx):\n \"Decoded item at `idx`\"\n return o.decode(o[idx])",
"_____no_output_____"
],
[
"#exports\ndef show_at(o, idx, **kwargs):\n \"Show item at `idx`\",\n return o.show(o[idx], **kwargs)",
"_____no_output_____"
]
],
[
[
"A `TfmdLists` combines a collection of object with a `Pipeline`. `tfms` can either be a `Pipeline` or a list of transforms, in which case, it will wrap them in a `Pipeline`. `use_list` is passed along to `L` with the `items` and `split_idx` are passed to each transform of the `Pipeline`. `do_setup` indicates if the `Pipeline.setup` method should be called during initialization.",
"_____no_output_____"
]
],
[
[
"class _IntFloatTfm(Transform):\n def encodes(self, o): return TitledInt(o)\n def decodes(self, o): return TitledFloat(o)\nint2f_tfm=_IntFloatTfm()\n\ndef _neg(o): return -o\nneg_tfm = Transform(_neg, _neg)",
"_____no_output_____"
],
[
"items = L([1.,2.,3.]); tfms = [neg_tfm, int2f_tfm]\ntl = TfmdLists(items, tfms=tfms)\ntest_eq_type(tl[0], TitledInt(-1))\ntest_eq_type(tl[1], TitledInt(-2))\ntest_eq_type(tl.decode(tl[2]), TitledFloat(3.))\ntest_stdout(lambda: show_at(tl, 2), '-3')\ntest_eq(tl.types, [float, float, TitledInt])\ntl",
"_____no_output_____"
],
[
"# add splits to TfmdLists\nsplits = [[0,2],[1]]\ntl = TfmdLists(items, tfms=tfms, splits=splits)\ntest_eq(tl.n_subsets, 2)\ntest_eq(tl.train, tl.subset(0))\ntest_eq(tl.valid, tl.subset(1))\ntest_eq(tl.train.items, items[splits[0]])\ntest_eq(tl.valid.items, items[splits[1]])\ntest_eq(tl.train.tfms.split_idx, 0)\ntest_eq(tl.valid.tfms.split_idx, 1)\ntest_eq(tl.train.new_empty().split_idx, 0)\ntest_eq(tl.valid.new_empty().split_idx, 1)\ntest_eq_type(tl.splits, L(splits))\nassert not tl.overlapping_splits()",
"_____no_output_____"
],
[
"df = pd.DataFrame(dict(a=[1,2,3],b=[2,3,4]))\ntl = TfmdLists(df, lambda o: o.a+1, splits=[[0],[1,2]])\ntest_eq(tl[1,2], [3,4])\ntr = tl.subset(0)\ntest_eq(tr[:], [2])\nval = tl.subset(1)\ntest_eq(val[:], [3,4])",
"_____no_output_____"
],
[
"class _B(Transform):\n def __init__(self): self.m = 0\n def encodes(self, o): return o+self.m\n def decodes(self, o): return o-self.m\n def setups(self, items): \n print(items)\n self.m = tensor(items).float().mean().item()\n\n# test for setup, which updates `self.m`\ntl = TfmdLists(items, _B())\ntest_eq(tl.m, 2)",
"TfmdLists: [1.0, 2.0, 3.0]\ntfms - (#0) []\n"
]
],
[
[
"Here's how we can use `TfmdLists.setup` to implement a simple category list, getting labels from a mock file list:",
"_____no_output_____"
]
],
[
[
"class _Cat(Transform):\n order = 1\n def encodes(self, o): return int(self.o2i[o])\n def decodes(self, o): return TitledStr(self.vocab[o])\n def setups(self, items): self.vocab,self.o2i = uniqueify(L(items), sort=True, bidir=True)\ntcat = _Cat()\n\ndef _lbl(o): return TitledStr(o.split('_')[0])\n\n# Check that tfms are sorted by `order` & `_lbl` is called first\nfns = ['dog_0.jpg','cat_0.jpg','cat_2.jpg','cat_1.jpg','dog_1.jpg']\ntl = TfmdLists(fns, [tcat,_lbl])\nexp_voc = ['cat','dog']\ntest_eq(tcat.vocab, exp_voc)\ntest_eq(tl.tfms.vocab, exp_voc)\ntest_eq(tl.vocab, exp_voc)\ntest_eq(tl, (1,0,0,0,1))\ntest_eq([tl.decode(o) for o in tl], ('dog','cat','cat','cat','dog'))",
"_____no_output_____"
],
[
"#Check only the training set is taken into account for setup\ntl = TfmdLists(fns, [tcat,_lbl], splits=[[0,4], [1,2,3]])\ntest_eq(tcat.vocab, ['dog'])",
"_____no_output_____"
],
[
"tfm = NegTfm(split_idx=1)\ntds = TfmdLists(start, A())\ntdl = TfmdDL(tds, after_batch=tfm, bs=4)\nx = tdl.one_batch()\ntest_eq(x, torch.arange(4))\ntds.split_idx = 1\nx = tdl.one_batch()\ntest_eq(x, -torch.arange(4))\ntds.split_idx = 0\nx = tdl.one_batch()\ntest_eq(x, torch.arange(4))",
"_____no_output_____"
],
[
"tds = TfmdLists(start, A())\ntdl = TfmdDL(tds, after_batch=NegTfm(), bs=4)\ntest_eq(tdl.dataset[0], start[0])\ntest_eq(len(tdl), (len(tds)-1)//4+1)\ntest_eq(tdl.bs, 4)\ntest_stdout(tdl.show_batch, '0\\n1\\n2\\n3')",
"_____no_output_____"
],
[
"show_doc(TfmdLists.subset)",
"_____no_output_____"
],
[
"show_doc(TfmdLists.infer_idx)",
"_____no_output_____"
],
[
"show_doc(TfmdLists.infer)",
"_____no_output_____"
],
[
"def mult(x): return x*2\nmult.order = 2\n\nfns = ['dog_0.jpg','cat_0.jpg','cat_2.jpg','cat_1.jpg','dog_1.jpg']\ntl = TfmdLists(fns, [_lbl,_Cat(),mult])\n\ntest_eq(tl.infer_idx('dog_45.jpg'), 0)\ntest_eq(tl.infer('dog_45.jpg'), 2)\n\ntest_eq(tl.infer_idx(4), 2)\ntest_eq(tl.infer(4), 8)\n\ntest_fail(lambda: tl.infer_idx(2.0))\ntest_fail(lambda: tl.infer(2.0))",
"_____no_output_____"
],
[
"#hide\n#Test input_types works on a Transform\ncat = _Cat()\ncat.input_types = (str, float)\ntl = TfmdLists(fns, [_lbl,cat,mult])\ntest_eq(tl.infer_idx(2.0), 1)\n\n#Test type annotations work on a function\ndef mult(x:(int,float)): return x*2\nmult.order = 2\ntl = TfmdLists(fns, [_lbl,_Cat(),mult])\ntest_eq(tl.infer_idx(2.0), 2)",
"_____no_output_____"
]
],
[
[
"## Datasets -",
"_____no_output_____"
]
],
[
[
"#export\n@docs\n@delegates(TfmdLists)\nclass Datasets(FilteredBase):\n \"A dataset that creates a tuple from each `tfms`, passed through `item_tfms`\"\n def __init__(self, items=None, tfms=None, tls=None, n_inp=None, dl_type=None, **kwargs):\n super().__init__(dl_type=dl_type)\n self.tls = L(tls if tls else [TfmdLists(items, t, **kwargs) for t in L(ifnone(tfms,[None]))])\n self.n_inp = ifnone(n_inp, max(1, len(self.tls)-1))\n\n def __getitem__(self, it):\n res = tuple([tl[it] for tl in self.tls])\n return res if is_indexer(it) else list(zip(*res))\n\n def __getattr__(self,k): return gather_attrs(self, k, 'tls')\n def __dir__(self): return super().__dir__() + gather_attr_names(self, 'tls')\n def __len__(self): return len(self.tls[0])\n def __iter__(self): return (self[i] for i in range(len(self)))\n def __repr__(self): return coll_repr(self)\n def decode(self, o, full=True): return tuple(tl.decode(o_, full=full) for o_,tl in zip(o,tuplify(self.tls, match=o)))\n def subset(self, i): return type(self)(tls=L(tl.subset(i) for tl in self.tls), n_inp=self.n_inp)\n def _new(self, items, *args, **kwargs): return super()._new(items, tfms=self.tfms, do_setup=False, **kwargs)\n def overlapping_splits(self): return self.tls[0].overlapping_splits()\n def new_empty(self): return type(self)(tls=[tl.new_empty() for tl in self.tls], n_inp=self.n_inp)\n @property\n def splits(self): return self.tls[0].splits\n @property\n def split_idx(self): return self.tls[0].tfms.split_idx\n @property\n def items(self): return self.tls[0].items\n @items.setter\n def items(self, v):\n for tl in self.tls: tl.items = v\n\n def show(self, o, ctx=None, **kwargs):\n for o_,tl in zip(o,self.tls): ctx = tl.show(o_, ctx=ctx, **kwargs)\n return ctx\n\n @contextmanager\n def set_split_idx(self, i):\n old_split_idx = self.split_idx\n for tl in self.tls: tl.tfms.split_idx = i\n try: yield self\n finally:\n for tl in self.tls: tl.tfms.split_idx = old_split_idx\n\n _docs=dict(\n decode=\"Compose `decode` of all `tuple_tfms` then all `tfms` on `i`\",\n show=\"Show item `o` in `ctx`\",\n dataloaders=\"Get a `DataLoaders`\",\n overlapping_splits=\"All splits that are in more than one split\",\n subset=\"New `Datasets` that only includes subset `i`\",\n new_empty=\"Create a new empty version of the `self`, keeping only the transforms\",\n set_split_idx=\"Contextmanager to use the same `Datasets` with another `split_idx`\"\n )",
"_____no_output_____"
]
],
[
[
"A `Datasets` creates a tuple from `items` (typically input,target) by applying to them each list of `Transform` (or `Pipeline`) in `tfms`. Note that if `tfms` contains only one list of `tfms`, the items given by `Datasets` will be tuples of one element. \n\n`n_inp` is the number of elements in the tuples that should be considered part of the input and will default to 1 if `tfms` consists of one set of transforms, `len(tfms)-1` otherwise. In most cases, the number of elements in the tuples spit out by `Datasets` will be 2 (for input,target) but it can happen that there is 3 (Siamese networks or tabular data) in which case we need to be able to determine when the inputs end and the targets begin.",
"_____no_output_____"
]
],
[
[
"items = [1,2,3,4]\ndsets = Datasets(items, [[neg_tfm,int2f_tfm], [add(1)]])\nt = dsets[0]\ntest_eq(t, (-1,2))\ntest_eq(dsets[0,1,2], [(-1,2),(-2,3),(-3,4)])\ntest_eq(dsets.n_inp, 1)\ndsets.decode(t)",
"_____no_output_____"
],
[
"class Norm(Transform):\n def encodes(self, o): return (o-self.m)/self.s\n def decodes(self, o): return (o*self.s)+self.m\n def setups(self, items):\n its = tensor(items).float()\n self.m,self.s = its.mean(),its.std()",
"_____no_output_____"
],
[
"items = [1,2,3,4]\nnrm = Norm()\ndsets = Datasets(items, [[neg_tfm,int2f_tfm], [neg_tfm,nrm]])\n\nx,y = zip(*dsets)\ntest_close(tensor(y).mean(), 0)\ntest_close(tensor(y).std(), 1)\ntest_eq(x, (-1,-2,-3,-4,))\ntest_eq(nrm.m, -2.5)\ntest_stdout(lambda:show_at(dsets, 1), '-2')\n\ntest_eq(dsets.m, nrm.m)\ntest_eq(dsets.norm.m, nrm.m)\ntest_eq(dsets.train.norm.m, nrm.m)",
"_____no_output_____"
],
[
"#hide\n#Check filtering is properly applied\nclass B(Transform):\n def encodes(self, x)->None: return int(x+1)\n def decodes(self, x): return TitledInt(x-1)\nadd1 = B(split_idx=1)\n\ndsets = Datasets(items, [neg_tfm, [neg_tfm,int2f_tfm,add1]], splits=[[3],[0,1,2]])\ntest_eq(dsets[1], [-2,-2])\ntest_eq(dsets.valid[1], [-2,-1])\ntest_eq(dsets.valid[[1,1]], [[-2,-1], [-2,-1]])\ntest_eq(dsets.train[0], [-4,-4])",
"_____no_output_____"
],
[
"test_fns = ['dog_0.jpg','cat_0.jpg','cat_2.jpg','cat_1.jpg','kid_1.jpg']\ntcat = _Cat()\ndsets = Datasets(test_fns, [[tcat,_lbl]], splits=[[0,1,2], [3,4]])\ntest_eq(tcat.vocab, ['cat','dog'])\ntest_eq(dsets.train, [(1,),(0,),(0,)])\ntest_eq(dsets.valid[0], (0,))\ntest_stdout(lambda: show_at(dsets.train, 0), \"dog\")",
"_____no_output_____"
],
[
"inp = [0,1,2,3,4]\ndsets = Datasets(inp, tfms=[None])\n\ntest_eq(*dsets[2], 2) # Retrieve one item (subset 0 is the default)\ntest_eq(dsets[1,2], [(1,),(2,)]) # Retrieve two items by index\nmask = [True,False,False,True,False]\ntest_eq(dsets[mask], [(0,),(3,)]) # Retrieve two items by mask",
"_____no_output_____"
],
[
"inp = pd.DataFrame(dict(a=[5,1,2,3,4]))\ndsets = Datasets(inp, tfms=attrgetter('a')).subset(0)\ntest_eq(*dsets[2], 2) # Retrieve one item (subset 0 is the default)\ntest_eq(dsets[1,2], [(1,),(2,)]) # Retrieve two items by index\nmask = [True,False,False,True,False]\ntest_eq(dsets[mask], [(5,),(3,)]) # Retrieve two items by mask",
"_____no_output_____"
],
[
"#test n_inp\ninp = [0,1,2,3,4]\ndsets = Datasets(inp, tfms=[None])\ntest_eq(dsets.n_inp, 1)\ndsets = Datasets(inp, tfms=[[None],[None],[None]])\ntest_eq(dsets.n_inp, 2)\ndsets = Datasets(inp, tfms=[[None],[None],[None]], n_inp=1)\ntest_eq(dsets.n_inp, 1)",
"_____no_output_____"
],
[
"# splits can be indices\ndsets = Datasets(range(5), tfms=[None], splits=[tensor([0,2]), [1,3,4]])\n\ntest_eq(dsets.subset(0), [(0,),(2,)])\ntest_eq(dsets.train, [(0,),(2,)]) # Subset 0 is aliased to `train`\ntest_eq(dsets.subset(1), [(1,),(3,),(4,)])\ntest_eq(dsets.valid, [(1,),(3,),(4,)]) # Subset 1 is aliased to `valid`\ntest_eq(*dsets.valid[2], 4)\n#assert '[(1,),(3,),(4,)]' in str(dsets) and '[(0,),(2,)]' in str(dsets)\ndsets",
"_____no_output_____"
],
[
"# splits can be boolean masks (they don't have to cover all items, but must be disjoint)\nsplits = [[False,True,True,False,True], [True,False,False,False,False]]\ndsets = Datasets(range(5), tfms=[None], splits=splits)\n\ntest_eq(dsets.train, [(1,),(2,),(4,)])\ntest_eq(dsets.valid, [(0,)])",
"_____no_output_____"
],
[
"# apply transforms to all items\ntfm = [[lambda x: x*2,lambda x: x+1]]\nsplits = [[1,2],[0,3,4]]\ndsets = Datasets(range(5), tfm, splits=splits)\ntest_eq(dsets.train,[(3,),(5,)])\ntest_eq(dsets.valid,[(1,),(7,),(9,)])\ntest_eq(dsets.train[False,True], [(5,)])",
"_____no_output_____"
],
[
"# only transform subset 1\nclass _Tfm(Transform):\n split_idx=1\n def encodes(self, x): return x*2\n def decodes(self, x): return TitledStr(x//2)",
"_____no_output_____"
],
[
"dsets = Datasets(range(5), [_Tfm()], splits=[[1,2],[0,3,4]])\ntest_eq(dsets.train,[(1,),(2,)])\ntest_eq(dsets.valid,[(0,),(6,),(8,)])\ntest_eq(dsets.train[False,True], [(2,)])\ndsets",
"_____no_output_____"
],
[
"#A context manager to change the split_idx and apply the validation transform on the training set\nds = dsets.train\nwith ds.set_split_idx(1):\n test_eq(ds,[(2,),(4,)])\ntest_eq(dsets.train,[(1,),(2,)])",
"_____no_output_____"
],
[
"#hide\n#Test Datasets pickles\ndsrc1 = pickle.loads(pickle.dumps(dsets))\ntest_eq(dsets.train, dsrc1.train)\ntest_eq(dsets.valid, dsrc1.valid)",
"_____no_output_____"
],
[
"dsets = Datasets(range(5), [_Tfm(),noop], splits=[[1,2],[0,3,4]])\ntest_eq(dsets.train,[(1,1),(2,2)])\ntest_eq(dsets.valid,[(0,0),(6,3),(8,4)])",
"_____no_output_____"
],
[
"start = torch.arange(0,50)\ntds = Datasets(start, [A()])\ntdl = TfmdDL(tds, after_item=NegTfm(), bs=4)\nb = tdl.one_batch()\ntest_eq(tdl.decode_batch(b), ((0,),(1,),(2,),(3,)))\ntest_stdout(tdl.show_batch, \"0\\n1\\n2\\n3\")",
"_____no_output_____"
],
[
"# only transform subset 1\nclass _Tfm(Transform):\n split_idx=1\n def encodes(self, x): return x*2\n\ndsets = Datasets(range(8), [None], splits=[[1,2,5,7],[0,3,4,6]])",
"_____no_output_____"
],
[
"# only transform subset 1\nclass _Tfm(Transform):\n split_idx=1\n def encodes(self, x): return x*2\n\ndsets = Datasets(range(8), [None], splits=[[1,2,5,7],[0,3,4,6]])\ndls = dsets.dataloaders(bs=4, after_batch=_Tfm(), shuffle_train=False, device=torch.device('cpu'))\ntest_eq(dls.train, [(tensor([1,2,5, 7]),)])\ntest_eq(dls.valid, [(tensor([0,6,8,12]),)])\ntest_eq(dls.n_inp, 1)",
"_____no_output_____"
]
],
[
[
"### Methods",
"_____no_output_____"
]
],
[
[
"items = [1,2,3,4]\ndsets = Datasets(items, [[neg_tfm,int2f_tfm]])",
"_____no_output_____"
],
[
"#hide_input\n_dsrc = Datasets([1,2])\nshow_doc(_dsrc.dataloaders, name=\"Datasets.dataloaders\")",
"_____no_output_____"
],
[
"show_doc(Datasets.decode)",
"_____no_output_____"
],
[
"test_eq(*dsets[0], -1)\ntest_eq(*dsets.decode((-1,)), 1)",
"_____no_output_____"
],
[
"show_doc(Datasets.show)",
"_____no_output_____"
],
[
"test_stdout(lambda:dsets.show(dsets[1]), '-2')",
"_____no_output_____"
],
[
"show_doc(Datasets.new_empty)",
"_____no_output_____"
],
[
"items = [1,2,3,4]\nnrm = Norm()\ndsets = Datasets(items, [[neg_tfm,int2f_tfm], [neg_tfm]])\nempty = dsets.new_empty()\ntest_eq(empty.items, [])",
"_____no_output_____"
],
[
"#hide\n#test it works for dataframes too\ndf = pd.DataFrame({'a':[1,2,3,4,5], 'b':[6,7,8,9,10]})\ndsets = Datasets(df, [[attrgetter('a')], [attrgetter('b')]])\nempty = dsets.new_empty()",
"_____no_output_____"
]
],
[
[
"## Add test set for inference",
"_____no_output_____"
]
],
[
[
"# only transform subset 1\nclass _Tfm1(Transform):\n split_idx=0\n def encodes(self, x): return x*3\n\ndsets = Datasets(range(8), [[_Tfm(),_Tfm1()]], splits=[[1,2,5,7],[0,3,4,6]])\ntest_eq(dsets.train, [(3,),(6,),(15,),(21,)])\ntest_eq(dsets.valid, [(0,),(6,),(8,),(12,)])",
"_____no_output_____"
],
[
"#export\ndef test_set(dsets, test_items, rm_tfms=None, with_labels=False):\n \"Create a test set from `test_items` using validation transforms of `dsets`\"\n if isinstance(dsets, Datasets):\n tls = dsets.tls if with_labels else dsets.tls[:dsets.n_inp]\n test_tls = [tl._new(test_items, split_idx=1) for tl in tls]\n if rm_tfms is None: rm_tfms = [tl.infer_idx(get_first(test_items)) for tl in test_tls]\n else: rm_tfms = tuplify(rm_tfms, match=test_tls)\n for i,j in enumerate(rm_tfms): test_tls[i].tfms.fs = test_tls[i].tfms.fs[j:]\n return Datasets(tls=test_tls)\n elif isinstance(dsets, TfmdLists):\n test_tl = dsets._new(test_items, split_idx=1)\n if rm_tfms is None: rm_tfms = dsets.infer_idx(get_first(test_items))\n test_tl.tfms.fs = test_tl.tfms.fs[rm_tfms:]\n return test_tl\n else: raise Exception(f\"This method requires using the fastai library to assemble your data. Expected a `Datasets` or a `TfmdLists` but got {dsets.__class__.__name__}\")",
"_____no_output_____"
],
[
"class _Tfm1(Transform):\n split_idx=0\n def encodes(self, x): return x*3\n\ndsets = Datasets(range(8), [[_Tfm(),_Tfm1()]], splits=[[1,2,5,7],[0,3,4,6]])\ntest_eq(dsets.train, [(3,),(6,),(15,),(21,)])\ntest_eq(dsets.valid, [(0,),(6,),(8,),(12,)])\n\n#Tranform of the validation set are applied\ntst = test_set(dsets, [1,2,3])\ntest_eq(tst, [(2,),(4,),(6,)])",
"_____no_output_____"
],
[
"#hide\n#Test with different types\ntfm = _Tfm1()\ntfm.split_idx,tfm.order = None,2\ndsets = Datasets(['dog', 'cat', 'cat', 'dog'], [[_Cat(),tfm]])\n\n#With strings\ntest_eq(test_set(dsets, ['dog', 'cat', 'cat']), [(3,), (0,), (0,)])\n#With ints\ntest_eq(test_set(dsets, [1,2]), [(3,), (6,)])",
"_____no_output_____"
],
[
"#hide\n#Test with various input lengths\ndsets = Datasets(range(8), [[_Tfm(),_Tfm1()],[_Tfm(),_Tfm1()],[_Tfm(),_Tfm1()]], splits=[[1,2,5,7],[0,3,4,6]])\ntst = test_set(dsets, [1,2,3])\ntest_eq(tst, [(2,2),(4,4),(6,6)])\n\ndsets = Datasets(range(8), [[_Tfm(),_Tfm1()],[_Tfm(),_Tfm1()],[_Tfm(),_Tfm1()]], splits=[[1,2,5,7],[0,3,4,6]], n_inp=1)\ntst = test_set(dsets, [1,2,3])\ntest_eq(tst, [(2,),(4,),(6,)])",
"_____no_output_____"
],
[
"#hide\n#Test with rm_tfms\ndsets = Datasets(range(8), [[_Tfm(),_Tfm()]], splits=[[1,2,5,7],[0,3,4,6]])\ntst = test_set(dsets, [1,2,3])\ntest_eq(tst, [(4,),(8,),(12,)])\n\ndsets = Datasets(range(8), [[_Tfm(),_Tfm()]], splits=[[1,2,5,7],[0,3,4,6]])\ntst = test_set(dsets, [1,2,3], rm_tfms=1)\ntest_eq(tst, [(2,),(4,),(6,)])\n\ndsets = Datasets(range(8), [[_Tfm(),_Tfm()], [_Tfm(),_Tfm()]], splits=[[1,2,5,7],[0,3,4,6]], n_inp=2)\ntst = test_set(dsets, [1,2,3], rm_tfms=(1,0))\ntest_eq(tst, [(2,4),(4,8),(6,12)])",
"_____no_output_____"
],
[
"#export\n@delegates(TfmdDL.__init__)\n@patch\ndef test_dl(self:DataLoaders, test_items, rm_type_tfms=None, with_labels=False, **kwargs):\n \"Create a test dataloader from `test_items` using validation transforms of `dls`\"\n test_ds = test_set(self.valid_ds, test_items, rm_tfms=rm_type_tfms, with_labels=with_labels\n ) if isinstance(self.valid_ds, (Datasets, TfmdLists)) else test_items\n return self.valid.new(test_ds, **kwargs)",
"_____no_output_____"
],
[
"dsets = Datasets(range(8), [[_Tfm(),_Tfm1()]], splits=[[1,2,5,7],[0,3,4,6]])\ndls = dsets.dataloaders(bs=4, device=torch.device('cpu'))",
"_____no_output_____"
],
[
"dsets = Datasets(range(8), [[_Tfm(),_Tfm1()]], splits=[[1,2,5,7],[0,3,4,6]])\ndls = dsets.dataloaders(bs=4, device=torch.device('cpu'))\ntst_dl = dls.test_dl([2,3,4,5])\ntest_eq(tst_dl._n_inp, 1)\ntest_eq(list(tst_dl), [(tensor([ 4, 6, 8, 10]),)])\n#Test you can change transforms\ntst_dl = dls.test_dl([2,3,4,5], after_item=add1)\ntest_eq(list(tst_dl), [(tensor([ 5, 7, 9, 11]),)])",
"_____no_output_____"
]
],
[
[
"## Export -",
"_____no_output_____"
]
],
[
[
"#hide\nfrom nbdev.export import notebook2script\nnotebook2script()",
"Converted 00_torch_core.ipynb.\nConverted 01_layers.ipynb.\nConverted 01a_losses.ipynb.\nConverted 02_data.load.ipynb.\nConverted 03_data.core.ipynb.\nConverted 04_data.external.ipynb.\nConverted 05_data.transforms.ipynb.\nConverted 06_data.block.ipynb.\nConverted 07_vision.core.ipynb.\nConverted 08_vision.data.ipynb.\nConverted 09_vision.augment.ipynb.\nConverted 09b_vision.utils.ipynb.\nConverted 09c_vision.widgets.ipynb.\nConverted 10_tutorial.pets.ipynb.\nConverted 10b_tutorial.albumentations.ipynb.\nConverted 11_vision.models.xresnet.ipynb.\nConverted 12_optimizer.ipynb.\nConverted 13_callback.core.ipynb.\nConverted 13a_learner.ipynb.\nConverted 13b_metrics.ipynb.\nConverted 14_callback.schedule.ipynb.\nConverted 14a_callback.data.ipynb.\nConverted 15_callback.hook.ipynb.\nConverted 15a_vision.models.unet.ipynb.\nConverted 16_callback.progress.ipynb.\nConverted 17_callback.tracker.ipynb.\nConverted 18_callback.fp16.ipynb.\nConverted 18a_callback.training.ipynb.\nConverted 18b_callback.preds.ipynb.\nConverted 19_callback.mixup.ipynb.\nConverted 20_interpret.ipynb.\nConverted 20a_distributed.ipynb.\nConverted 21_vision.learner.ipynb.\nConverted 22_tutorial.imagenette.ipynb.\nConverted 23_tutorial.vision.ipynb.\nConverted 24_tutorial.siamese.ipynb.\nConverted 24_vision.gan.ipynb.\nConverted 30_text.core.ipynb.\nConverted 31_text.data.ipynb.\nConverted 32_text.models.awdlstm.ipynb.\nConverted 33_text.models.core.ipynb.\nConverted 34_callback.rnn.ipynb.\nConverted 35_tutorial.wikitext.ipynb.\nConverted 36_text.models.qrnn.ipynb.\nConverted 37_text.learner.ipynb.\nConverted 38_tutorial.text.ipynb.\nConverted 39_tutorial.transformers.ipynb.\nConverted 40_tabular.core.ipynb.\nConverted 41_tabular.data.ipynb.\nConverted 42_tabular.model.ipynb.\nConverted 43_tabular.learner.ipynb.\nConverted 44_tutorial.tabular.ipynb.\nConverted 45_collab.ipynb.\nConverted 46_tutorial.collab.ipynb.\nConverted 50_tutorial.datablock.ipynb.\nConverted 60_medical.imaging.ipynb.\nConverted 61_tutorial.medical_imaging.ipynb.\nConverted 65_medical.text.ipynb.\nConverted 70_callback.wandb.ipynb.\nConverted 71_callback.tensorboard.ipynb.\nConverted 72_callback.neptune.ipynb.\nConverted 73_callback.captum.ipynb.\nConverted 74_callback.cutmix.ipynb.\nConverted 97_test_utils.ipynb.\nConverted 99_pytorch_doc.ipynb.\nConverted dev-setup.ipynb.\nConverted index.ipynb.\nConverted quick_start.ipynb.\nConverted tutorial.ipynb.\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d09317f88e08098cfee0c36cbeeefe55876b3ff3 | 35,796 | ipynb | Jupyter Notebook | week6/2_Functional_Behavior.ipynb | BrandMan2299/Notebooks | 73145c2f87ff773307175fef77d5cd9fee57159f | [
"CC-BY-4.0"
] | 1,061 | 2019-04-01T19:51:40.000Z | 2022-03-28T19:18:59.000Z | week6/2_Functional_Behavior.ipynb | BrandMan2299/Notebooks | 73145c2f87ff773307175fef77d5cd9fee57159f | [
"CC-BY-4.0"
] | 21 | 2019-04-14T20:25:38.000Z | 2022-02-07T20:45:11.000Z | week6/2_Functional_Behavior.ipynb | zoharsacks/Notebooks | cbe8430ce9fd541e2513a3283f53a5aa0464085b | [
"CC-BY-4.0"
] | 180 | 2017-04-22T21:49:42.000Z | 2022-03-24T05:17:32.000Z | 32.336043 | 438 | 0.553917 | [
[
[
"<img src=\"images/logo.jpg\" style=\"display: block; margin-left: auto; margin-right: auto;\" alt=\"לוגו של מיזם לימוד הפייתון. נחש מצויר בצבעי צהוב וכחול, הנע בין האותיות של שם הקורס: לומדים פייתון. הסלוגן המופיע מעל לשם הקורס הוא מיזם חינמי ללימוד תכנות בעברית.\">",
"_____no_output_____"
],
[
"# <span style=\"text-align: right; direction: rtl; float: right;\">התנהגות של פונקציות</span>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n בפסקאות הקרובות נבחן פונקציות מזווית ראייה מעט שונה מהרגיל.<br>\n בואו נקפוץ ישירות למים!\n</p>",
"_____no_output_____"
],
[
"### <span style=\"text-align: right; direction: rtl; float: right; clear: both;\">שם של פונקציה</span>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n תכונה מעניינת שמתקיימת בפייתון היא שפונקציה היא ערך, בדיוק כמו כל ערך אחר.<br>\n נגדיר פונקציה שמעלה מספר בריבוע:\n</p>",
"_____no_output_____"
]
],
[
[
"def square(x):\n return x ** 2",
"_____no_output_____"
]
],
[
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n נוכל לבדוק מאיזה טיפוס הפונקציה (אנחנו לא קוראים לה עם סוגריים אחרי שמה – רק מציינים את שמה):\n</p>",
"_____no_output_____"
]
],
[
[
"type(square)",
"_____no_output_____"
]
],
[
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n ואפילו לבצע השמה שלה למשתנה, כך ששם המשתנה החדש יצביע עליה:\n</p>",
"_____no_output_____"
]
],
[
[
"ribua = square\n\nprint(square(5))\nprint(ribua(5))",
"_____no_output_____"
]
],
[
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n מה מתרחש בתא למעלה?<br>\n כשהגדרנו את הפונקציה <var>square</var>, יצרנו לייזר עם התווית <var>square</var> שמצביע לפונקציה שמעלה מספר בריבוע.<br>\n בהשמה שביצענו בשורה הראשונה בתא שלמעלה, הלייזר שעליו מודבקת התווית <var>ribua</var> כוון אל אותה הפונקציה שעליה מצביע הלייזר <var>square</var>.<br>\n כעת <var>square</var> ו־<var>ribua</var> מצביעים לאותה פונקציה. אפשר לבדוק זאת כך:\n</p>",
"_____no_output_____"
]
],
[
[
"ribua is square",
"_____no_output_____"
]
],
[
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n בשלב הזה אצטרך לבקש מכם לחגור חגורות, כי זה לא הולך להיות טיול רגיל הפעם.\n</p>",
"_____no_output_____"
],
[
"### <span style=\"text-align: right; direction: rtl; float: right; clear: both;\">פונקציות במבנים מורכבים</span>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n אם פונקציה היא בסך הכול ערך, ואם אפשר להתייחס לשם שלה בכל מקום, אין סיבה שלא נוכל ליצור רשימה של פונקציות!<br>\n ננסה לממש את הרעיון:\n</p>",
"_____no_output_____"
]
],
[
[
"def add(num1, num2):\n return num1 + num2\n\n\ndef subtract(num1, num2):\n return num1 - num2\n\n\ndef multiply(num1, num2):\n return num1 * num2\n\n\ndef divide(num1, num2):\n return num1 / num2\n\n\nfunctions = [add, subtract, multiply, divide]",
"_____no_output_____"
]
],
[
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n כעת יש לנו רשימה בעלת 4 איברים, שכל אחד מהם מצביע לפונקציה שונה.<br>\n אם נרצה לבצע פעולת חיבור, נוכל לקרוא ישירות ל־<var>add</var> או (בשביל התרגול) לנסות לאחזר אותה מהרשימה שיצרנו:\n</p>",
"_____no_output_____"
]
],
[
[
"# Option 1\nprint(add(5, 2))\n\n# Option 2\nmath_function = functions[0]\nprint(math_function(5, 2))\n\n# Option 3 (ugly, but works!)\nprint(functions[0](5, 2))",
"_____no_output_____"
]
],
[
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n אם נרצה, נוכל אפילו לעבור על רשימת הפונקציות בעזרת לולאה ולהפעיל את כולן, זו אחר זו:\n</p>",
"_____no_output_____"
]
],
[
[
"for function in functions:\n print(function(5, 2))",
"_____no_output_____"
]
],
[
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n בכל איטרציה של לולאת ה־<code>for</code>, המשתנה <var>function</var> עבר להצביע על הפונקציה הבאה מתוך רשימת <var>functions</var>.<br>\n בשורה הבאה קראנו לאותה הפונקציה ש־<var>function</var> מצביע עליה, והדפסנו את הערך שהיא החזירה. \n</p>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n כיוון שרשימה היא מבנה ששומר על סדר האיברים שבו, התוצאות מודפסות בסדר שבו הפונקציות שמורות ברשימה.<br>\n התוצאה הראשונה שאנחנו רואים היא תוצאת פונקציית החיבור, השנייה היא תוצאת פונקציית החיסור וכן הלאה.\n</p>",
"_____no_output_____"
],
[
"#### <span style=\"text-align: right; direction: rtl; float: right; clear: both;\">תרגיל ביניים: סוגרים חשבון</span>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n כתבו פונקציה בשם <var>calc</var> שמקבלת כפרמטר שני מספרים וסימן של פעולה חשבונית.<br>\n הסימן יכול להיות אחד מאלה: <code>+</code>, <code>-</code>, <code>*</code> או <code>/</code>.<br>\n מטרת הפונקציה היא להחזיר את תוצאת הביטוי החשבוני שהופעל על שני המספרים.<br>\n בפתרונכם, השתמשו בהגדרת הפונקציות מלמעלה ובמילון.\n</p>",
"_____no_output_____"
],
[
"### <span style=\"text-align: right; direction: rtl; float: right; clear: both;\">העברת פונקציה כפרמטר</span>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n נמשיך ללהטט בפונקציות.<br>\n</p>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n פונקציה נקראת \"<dfn>פונקציה מסדר גבוה</dfn>\" (<dfn>higher order function</dfn>) אם היא מקבלת כפרמטר פונקציה.<br>\n ניקח לדוגמה את הפונקציה <var>calculate</var>:\n</p>",
"_____no_output_____"
]
],
[
[
"def calculate(function, num1, num2):\n return function(num1, num2)",
"_____no_output_____"
]
],
[
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n בקריאה ל־<var>calculate</var>, נצטרך להעביר פונקציה ושני מספרים.<br>\n נעביר לדוגמה את הפונקציה <var>divide</var> שהגדרנו קודם לכן:\n</p>",
"_____no_output_____"
]
],
[
[
"calculate(divide, 5, 2)",
"_____no_output_____"
]
],
[
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n מה שמתרחש במקרה הזה הוא שהעברנו את הפונקציה <var>divide</var> כארגומנט ראשון.<br>\n הפרמטר <var>function</var> בפונקציה <var>calculate</var> מצביע כעת על פונקציית החילוק שהגדרנו למעלה.<br>\n מכאן, שהפונקציה תחזיר את התוצאה של <code>divide(5, 2)</code> – הרי היא 2.5.\n</p>",
"_____no_output_____"
],
[
"#### <span style=\"text-align: right; direction: rtl; float: right; clear: both;\">תרגיל ביניים: מפה לפה</span>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n כתבו generator בשם <var>apply</var> שמקבל כפרמטר ראשון פונקציה (<var>func</var>), וכפרמטר שני iterable (<var dir=\"rtl\">iter</var>).<br>\n עבור כל איבר ב־iterable, ה־generator יניב את האיבר אחרי שהופעלה עליו הפונקציה <var>func</var>, דהיינו – <code>func(item)</code>.<br>\n</p>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n ודאו שהרצת התא הבא מחזירה <code>True</code> עבור הקוד שלכם:\n</p>",
"_____no_output_____"
]
],
[
[
"def square(number):\n return number ** 2\n\n\nsquare_check = apply(square, [5, -1, 6, -8, 0])\ntuple(square_check) == (25, 1, 36, 64, 0)",
"_____no_output_____"
]
],
[
[
"### <span style=\"text-align: right; direction: rtl; float: right; clear: both;\">סיכום ביניים</span>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n וואו. זה היה די משוגע.\n</p>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n אז למעשה, פונקציות בפייתון הן ערך לכל דבר, כמו מחרוזות ומספרים!<br>\n אפשר לאחסן אותן במשתנים, לשלוח אותן כארגומנטים ולכלול אותם בתוך מבני נתונים מורכבים יותר.<br>\n אנשי התיאוריה של מדעי המחשב נתנו להתנהגות כזו שם: \"<dfn>אזרח ממדרגה ראשונה</dfn>\" (<dfn>first class citizen</dfn>).<br>\n אם כך, אפשר להגיד על פונקציות בפייתון שהן אזרחיות ממדרגה ראשונה.\n</p>",
"_____no_output_____"
],
[
"## <span style=\"text-align: right; direction: rtl; float: right; clear: both;\">פונקציות מסדר גבוה בפייתון</span>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n החדשות הטובות הן שכבר עשינו היכרות קלה עם המונח פונקציות מסדר גבוה.<br>\n עכשיו, כשאנחנו יודעים שמדובר בפונקציות שמקבלות פונקציה כפרמטר, נתחיל ללכלך קצת את הידיים.<br>\n נציג כמה פונקציות פייתוניות מעניינות שכאלו:\n</p>",
"_____no_output_____"
],
[
"### <span style=\"text-align: right; direction: rtl; float: right; clear: both;\">הפונקציה map</span>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n הפונקציה <var>map</var> מקבלת פונקציה כפרמטר הראשון, ו־iterable כפרמטר השני.<br>\n <var>map</var> מפעילה את הפונקציה מהפרמטר הראשון על כל אחד מהאיברים שהועברו ב־iterable.<br>\n היא מחזירה iterator שמורכב מהערכים שחזרו מהפעלת הפונקציה.<br>\n</p>",
"_____no_output_____"
],
[
"\n<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n במילים אחרות, <var>map</var> יוצרת iterable חדש.<br>\n ה־iterable כולל את הערך שהוחזר מהפונקציה עבור כל איבר ב־<code>iterable</code> שהועבר.\n</p>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n לדוגמה:\n</p>",
"_____no_output_____"
]
],
[
[
"squared_items = map(square, [1, 6, -1, 8, 0, 3, -3, 9, -8, 8, -7])\nprint(tuple(squared_items))",
"_____no_output_____"
]
],
[
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n הפונקציה קיבלה כארגומנט ראשון את הפונקציה <var>square</var> שהגדרנו למעלה, שמטרתה העלאת מספר בריבוע.<br>\n כארגומנט שני היא קיבלה את רשימת כל המספרים שאנחנו רוצים שהפונקציה תרוץ עליהם.<br>\n כשהעברנו ל־<var>map</var> את הארגומנטים הללו, <var>map</var> החזירה לנו ב־iterator (מבנה שאפשר לעבור עליו איבר־איבר) את התוצאה:<br>\n הריבוע, קרי החזקה השנייה, של כל אחד מהאיברים ברשימה שהועברה כארגומנט השני.\n</p>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n למעשה, אפשר להגיד ש־<code>map</code> שקולה לפונקציה הבאה:\n</p>",
"_____no_output_____"
]
],
[
[
"def my_map(function, iterable):\n for item in iterable:\n yield function(item)",
"_____no_output_____"
]
],
[
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n הנה דוגמה נוספת לשימוש ב־<var>map</var>:\n</p>",
"_____no_output_____"
]
],
[
[
"numbers = [(2, 4), (1, 4, 2), (1, 3, 5, 6, 2), (3, )]\nsums = map(sum, numbers)\nprint(tuple(sums))",
"_____no_output_____"
]
],
[
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n במקרה הזה, בכל מעבר, קיבלה הפונקציה <var>sum</var> איבר אחד מתוך הרשימה – tuple.<br>\n היא סכמה את האיברים של כל tuple שקיבלה, וכך החזירה לנו את הסכומים של כל ה־tuple־ים – זה אחרי זה.\n</p>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n ודוגמה אחרונה:\n</p>",
"_____no_output_____"
]
],
[
[
"def add_one(number):\n return number + 1\n\n\nincremented = map(add_one, (1, 2, 3))\nprint(tuple(incremented))",
"_____no_output_____"
]
],
[
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n בדוגמה הזו יצרנו פונקציה משל עצמנו, ואותה העברנו ל־map.<br>\n מטרת דוגמה זו היא להדגיש שאין שוני בין העברת פונקציה שקיימת בפייתון לבין פונקציה שאנחנו יצרנו.\n</p>",
"_____no_output_____"
],
[
"<div class=\"align-center\" style=\"display: flex; text-align: right; direction: rtl; clear: both;\">\n <div style=\"display: flex; width: 10%; float: right; clear: both;\">\n <img src=\"images/exercise.svg\" style=\"height: 50px !important;\" alt=\"תרגול\"> \n </div>\n <div style=\"width: 70%\">\n <p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n כתבו פונקציה שמקבלת רשימת מחרוזות של שתי מילים: שם פרטי ושם משפחה.<br>\n הפונקציה תשתמש ב־map כדי להחזיר מכולן רק את השם הפרטי.\n </p>\n </div>\n <div style=\"display: flex; width: 20%; border-right: 0.1rem solid #A5A5A5; padding: 1rem 2rem;\">\n <p style=\"text-align: center; direction: rtl; justify-content: center; align-items: center; clear: both;\">\n <strong>חשוב!</strong><br>\n פתרו לפני שתמשיכו!\n </p>\n </div>\n</div>",
"_____no_output_____"
],
[
"### <span style=\"text-align: right; direction: rtl; float: right; clear: both;\">הפונקציה filter</span>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n הפונקציה <var>filter</var> מקבלת פונקציה כפרמטר ראשון, ו־iterable כפרמטר שני.<br>\n <var>filter</var> מפעילה על כל אחד מאיברי ה־iterable את הפונקציה, ומחזירה את האיבר אך ורק אם הערך שחזר מהפונקציה שקול ל־<code>True</code>.<br>\n אם ערך ההחזרה שקול ל־<code>False</code> – הערך \"יבלע\" ב־<var>filter</var> ולא יחזור ממנה.\n</p>",
"_____no_output_____"
],
[
"\n<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n במילים אחרות, <var>filter</var> יוצרת iterable חדש ומחזירה אותו.<br>\n ה־iterable כולל רק את האיברים שעבורם הפונקציה שהועברה החזירה ערך השקול ל־<code>True</code>.\n</p>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n נבנה, לדוגמה, פונקציה שמחזירה אם אדם הוא בגיר.<br>\n הפונקציה תקבל כפרמטר גיל, ותחזיר <code>True</code> כאשר הגיל שהועבר לה הוא לפחות 18, ו־<code>False</code> אחרת.\n</p>",
"_____no_output_____"
]
],
[
[
"def is_mature(age):\n return age >= 18",
"_____no_output_____"
]
],
[
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n נגדיר רשימת גילים, ונבקש מ־<var>filter</var> לסנן אותם לפי הפונקציה שהגדרנו:\n</p>",
"_____no_output_____"
]
],
[
[
"ages = [0, 1, 4, 10, 20, 35, 56, 84, 120]\nmature_ages = filter(is_mature, ages)\nprint(tuple(mature_ages))",
"_____no_output_____"
]
],
[
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n כפי שלמדנו, <var>filter</var> מחזירה לנו רק גילים השווים ל־18 או גדולים ממנו.\n</p>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n נחדד שהפונקציה שאנחנו מעבירים ל־<var>filter</var> לא חייבת להחזיר בהכרח <code>True</code> או <code>False</code>.<br>\n הערך 0, לדוגמה, שקול ל־<code>False</code>, ולכן <var>filter</var> תסנן כל ערך שעבורו הפונקציה תחזיר 0: \n</p>",
"_____no_output_____"
]
],
[
[
"to_sum = [(1, -1), (2, 5), (5, -3, -2), (1, 2, 3)]\nsum_is_not_zero = filter(sum, to_sum)\nprint(tuple(sum_is_not_zero))",
"_____no_output_____"
]
],
[
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n בתא האחרון העברנו ל־<var>filter</var> את sum כפונקציה שאותה אנחנו רוצים להפעיל, ואת <var>to_sum</var> כאיברים שעליהם אנחנו רוצים לפעול.<br>\n ה־tuple־ים שסכום איבריהם היה 0 סוננו, וקיבלנו חזרה iterator שהאיברים בו הם אך ורק אלו שסכומם שונה מ־0.\n</p>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n כטריק אחרון, נלמד ש־<var>filter</var> יכולה לקבל גם <code>None</code> בתור הפרמטר הראשון, במקום פונקציה.<br>\n זה יגרום ל־<var>filter</var> לא להפעיל פונקציה על האיברים שהועברו, כלומר לסנן אותם כמו שהם.<br>\n איברים השקולים ל־<code>True</code> יוחזרו, ואיברים השקולים ל־<code>False</code> לא יוחזרו:\n</p>",
"_____no_output_____"
]
],
[
[
"to_sum = [0, \"\", None, 0.0, True, False, \"Hello\"]\nequivalent_to_true = filter(None, to_sum)\nprint(tuple(equivalent_to_true))",
"_____no_output_____"
]
],
[
[
"<div class=\"align-center\" style=\"display: flex; text-align: right; direction: rtl; clear: both;\">\n <div style=\"display: flex; width: 10%; float: right; clear: both;\">\n <img src=\"images/exercise.svg\" style=\"height: 50px !important;\" alt=\"תרגול\"> \n </div>\n <div style=\"width: 70%\">\n <p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n כתבו פונקציה שמקבלת רשימת מחרוזות, ומחזירה רק את המחרוזות הפלינדרומיות שבה.<br>\n מחרוזת נחשבת פלינדרום אם קריאתה מימין לשמאל ומשמאל לימין יוצרת אותו ביטוי.<br>\n השתמשו ב־<var>filter</var>.\n </p>\n </div>\n <div style=\"display: flex; width: 20%; border-right: 0.1rem solid #A5A5A5; padding: 1rem 2rem;\">\n <p style=\"text-align: center; direction: rtl; justify-content: center; align-items: center; clear: both;\">\n <strong>חשוב!</strong><br>\n פתרו לפני שתמשיכו!\n </p>\n </div>\n</div>",
"_____no_output_____"
],
[
"## <span style=\"text-align: right; direction: rtl; float: right; clear: both;\">פונקציות אנונימיות</span>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n תעלול נוסף שנוסיף לארגז הכלים שלנו הוא <dfn>פונקציות אנונימיות</dfn> (<dfn>anonymous functions</dfn>).<br>\n אל תיבהלו מהשם המאיים – בסך הכול פירושו הוא \"פונקציות שאין להן שם\".<br>\n</p>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n לפני שאתם מרימים גבה ושואלים את עצמכם למה הן שימושיות, בואו נבחן כמה דוגמאות.<br>\n ניזכר בהגדרת פונקציית החיבור שיצרנו לא מזמן:\n</p>",
"_____no_output_____"
]
],
[
[
"def add(num1, num2):\n return num1 + num2",
"_____no_output_____"
]
],
[
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n ונגדיר את אותה הפונקציה בדיוק בצורה אנונימית:\n</p>",
"_____no_output_____"
]
],
[
[
"add = lambda num1, num2: num1 + num2\n\nprint(add(5, 2))",
"_____no_output_____"
]
],
[
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n לפני שנסביר איפה החלק של ה\"פונקציה בלי שם\" נתמקד בצד ימין של ההשמה.<br>\n כיצד מנוסחת הגדרת פונקציה אנונימית?\n</p>",
"_____no_output_____"
],
[
"<ol style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n <li>הצהרנו שברצוננו ליצור פונקציה אנונימית בעזרת מילת המפתח <code>lambda</code>.</li>\n <li>מייד אחריה, ציינו את שמות כל הפרמטרים שהפונקציה תקבל, כשהם מופרדים בפסיק זה מזה.</li>\n <li>כדי להפריד בין רשימת הפרמטרים לערך ההחזרה של הפונקציה, השתמשנו בנקודתיים.</li>\n <li>אחרי הנקודתיים, כתבנו את הביטוי שאנחנו רוצים שהפונקציה תחזיר.</li>\n</ol>",
"_____no_output_____"
],
[
"<figure>\n <img src=\"images/lambda.png\" style=\"max-width: 500px; margin-right: auto; margin-left: auto; text-align: center;\" alt=\"בתמונה מופיעה הגדרת ה־lambda שביצענו קודם לכן. מעל המילה lambda המודגשת בירוק ישנו פס מקווקו, ומעליו רשום 'הצהרה'. מימין למילה lambda כתוב num1 (פסיק) num2, מעליהם קו מקווקו ומעליו המילה 'פרמטרים'. מימין לפרמטרים יש נקודתיים, ואז num1 (הסימן פלוס) num2. מעליהם קו מקווקו, ומעליו המילה 'ערך החזרה'.\"/>\n <figcaption style=\"margin-top: 2rem; text-align: center; direction: rtl;\">חלקי ההגדרה של פונקציה אנונימית בעזרת מילת המפתח <code>lambda</code><br><span style=\"color: white;\">A girl has no name</span></figcaption>\n</figure>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n במה שונה ההגדרה של פונקציה זו מההגדרה של פונקציה רגילה?<br>\n היא לא באמת שונה.<br>\n המטרה היא לאפשר תחביר שיקל על חיינו כשאנחנו רוצים לכתוב פונקציה קצרצרה שאורכה שורה אחת.\n</p>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n נראה, לדוגמה, שימוש ב־<var>filter</var> כדי לסנן את כל האיברים שאינם חיוביים:\n</p>",
"_____no_output_____"
]
],
[
[
"def is_positive(number):\n return number > 0\n\n\nnumbers = [-2, -1, 0, 1, 2]\npositive_numbers = filter(is_positive, numbers)\nprint(tuple(positive_numbers))",
"_____no_output_____"
]
],
[
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n במקום להגדיר פונקציה חדשה שנקראת <var>is_positive</var>, נוכל להשתמש בפונקציה אנונימית:\n</p>",
"_____no_output_____"
]
],
[
[
"numbers = [-2, -1, 0, 1, 2]\npositive_numbers = filter(lambda n: n > 0, numbers)\nprint(tuple(positive_numbers))",
"_____no_output_____"
]
],
[
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n איך זה עובד?<br>\n במקום להעביר ל־<var>filter</var> פונקציה שיצרנו מבעוד מועד, השתמשנו ב־<code>lambda</code> כדי ליצור פונקציה ממש באותה השורה.<br>\n הפונקציה שהגדרנו מקבלת מספר (<var>n</var>), ומחזירה <code>True</code> אם הוא חיובי, או <code>False</code> אחרת.<br>\n שימו לב שבצורה זו באמת לא היינו צריכים לתת שם לפונקציה שהגדרנו.\n</p>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n השימוש בפונקציות אנונימיות לא מוגבל ל־<var>map</var> ול־<var>filter</var>, כמובן.<br>\n מקובל להשתמש ב־<code>lambda</code> גם עבור פונקציות כמו <var>sorted</var>, שמקבלות פונקציה בתור ארגומנט.\n</p>",
"_____no_output_____"
],
[
"<div class=\"align-center\" style=\"display: flex; text-align: right; direction: rtl;\">\n <div style=\"display: flex; width: 10%; float: right; \">\n <img src=\"images/recall.svg\" style=\"height: 50px !important;\" alt=\"תזכורת\" title=\"תזכורת\"> \n </div>\n <div style=\"width: 90%\">\n <p style=\"text-align: right; direction: rtl;\">\n הפונקציה <code>sorted</code> מאפשרת לנו לסדר ערכים, ואפילו להגדיר עבורה לפי מה לסדר אותם.<br>\n לרענון בנוגע לשימוש בפונקציה גשו למחברת בנושא פונקציות מובנות בשבוע 4.\n </p>\n </div>\n</div>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n נסדר, למשל, את הדמויות ברשימה הבאה, לפי תאריך הולדתן:\n</p>",
"_____no_output_____"
]
],
[
[
"closet = [\n {'name': 'Peter', 'year_of_birth': 1927, 'gender': 'Male'},\n {'name': 'Edmund', 'year_of_birth': 1930, 'gender': 'Male'},\n {'name': 'Lucy', 'year_of_birth': 1932, 'gender': 'Female'},\n {'name': 'Susan', 'year_of_birth': 1928, 'gender': 'Female'},\n {'name': 'Jadis', 'year_of_birth': 0, 'gender': 'Female'},\n]",
"_____no_output_____"
]
],
[
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n נרצה שסידור הרשימה יתבצע לפי המפתח <var>year_of_birth</var>.<br>\n כלומר, בהינתן מילון שמייצג דמות בשם <var>d</var>, יש להשיג את <code dir=\"ltr\">d['year_of_birth']</code>, ולפיו לבצע את סידור הרשימה.<br>\n ניגש למלאכה:\n</p>",
"_____no_output_____"
]
],
[
[
"sorted(closet, key=lambda d: d['year_of_birth'])",
"_____no_output_____"
]
],
[
[
"<div class=\"align-center\" style=\"display: flex; text-align: right; direction: rtl;\">\n <div style=\"display: flex; width: 10%; float: right; \">\n <img src=\"images/warning.png\" style=\"height: 50px !important;\" alt=\"אזהרה!\"> \n </div>\n <div style=\"width: 90%\">\n <p style=\"text-align: right; direction: rtl;\">\n פונקציות אנונימיות הן יכולת חביבה שאמורה לסייע לכם לכתוב קוד נאה וקריא.<br>\n כלל אצבע טוב לחיים הוא להימנע משימוש בהן כאשר הן מסרבלות את הקוד.\n </p>\n </div>\n</div>",
"_____no_output_____"
],
[
"<div class=\"align-center\" style=\"display: flex; text-align: right; direction: rtl; clear: both;\">\n <div style=\"display: flex; width: 10%; float: right; clear: both;\">\n <img src=\"images/exercise.svg\" style=\"height: 50px !important;\" alt=\"תרגיל\" title=\"תרגיל\"> \n </div>\n <div style=\"width: 70%\">\n <p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n סדרו את הדמויות ב־<var>closet</var> לפי האות האחרונה בשמם. \n </p>\n </div>\n</div>",
"_____no_output_____"
],
[
"## <span style=\"align: right; direction: rtl; float: right; clear: both;\">מונחים</span>",
"_____no_output_____"
],
[
"<dl style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n <dt>פונקציה מסדר גבוה</dt>\n <dd>פונקציה שמקבלת פונקציה כאחד הארגומנטים, או שמחזירה פונקציה כערך ההחזרה שלה.</dd>\n <dt>אזרח ממדרגה ראשונה</dt>\n <dd>ישות תכנותית המתנהגת בשפת התכנות כערך לכל דבר. בפייתון, פונקציות הן אזרחיות ממדרגה ראשונה.<dd>\n <dt>פונקציה אנונימית, פונקציית <code>lambda</code></dt>\n <dd>פונקציה ללא שם המיועדת להגדרת פונקציה בשורה אחת, לרוב לשימוש חד־פעמי. בעברית: פונקציית למדא.</dd>\n</dl>",
"_____no_output_____"
],
[
"## <span style=\"align: right; direction: rtl; float: right; clear: both;\">תרגילים</span>",
"_____no_output_____"
],
[
"### <span style=\"align: right; direction: rtl; float: right; clear: both;\">פילטר מותאם אישית</span>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n כתבו פונקציה בשם <var>my_filter</var> שמתנהגת בדיוק כמו הפונקציה <var>filter</var>.<br>\n בפתירת התרגיל, המנעו משימוש ב־<var>filter</var> או במודולים.\n</p>",
"_____no_output_____"
],
[
"### <span style=\"align: right; direction: rtl; float: right; clear: both;\">נשאר? חיובי</span>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\nכתבו פונקציה בשם <var>get_positive_numbers</var> שמקבלת מהמשתמש קלט בעזרת <var>input</var>.<br>\nהמשתמש יתבקש להזין סדרה של מספרים המופרדים בפסיק זה מזה.<br>\nהפונקציה תחזיר את כל המספרים החיוביים שהמשתמש הזין, כרשימה של מספרים מסוג <code>int</code>.<br>\nאפשר להניח שהקלט מהמשתמש תקין.\n</p>",
"_____no_output_____"
],
[
"### <span style=\"align: right; direction: rtl; float: right; clear: both;\">ריצת 2,000</span>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n כתבו פונקציה בשם <var>timer</var> שמקבלת כפרמטר פונקציה (נקרא לה <var>f</var>) ופרמטרים נוספים.<br>\n הפונקציה <var>timer</var> תמדוד כמה זמן רצה פונקציה <var>f</var> כשמועברים אליה אותם פרמטרים. <br>\n</p>",
"_____no_output_____"
],
[
"<p style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n לדוגמה:\n</p>",
"_____no_output_____"
],
[
"<ol style=\"text-align: right; direction: rtl; float: right; clear: both;\">\n <li>עבור הקריאה <code dir=\"ltr\">timer(print, \"Hello\")</code>, תחזיר הפונקציה את משך זמן הביצוע של <code dir=\"ltr\">print(\"Hello\")</code>.</li>\n <li>עבור הקריאה <code dir=\"ltr\">timer(zip, [1, 2, 3], [4, 5, 6])</code>, תחזיר הפונקציה את משך זמן הביצוע של <code dir=\"ltr\">zip([1, 2, 3], [4, 5, 6])</code>.</li>\n <li>עבור הקריאה <code dir=\"ltr\">timer(\"Hi {name}\".format, name=\"Bug\")</code>, תחזיר הפונקציה את משך זמן הביצוע של <code dir=\"ltr\">\"Hi {name}\".format(name=\"Bug\")</code></li>\n</ol>",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d0932943f8b6d53e9bce19567c41e056a02ccf59 | 18,513 | ipynb | Jupyter Notebook | model_notebooks/UBC/model.ipynb | indralab/adeft_indra | 6f039b58b6dea5eefa529cf15afaffff2d485513 | [
"BSD-2-Clause"
] | null | null | null | model_notebooks/UBC/model.ipynb | indralab/adeft_indra | 6f039b58b6dea5eefa529cf15afaffff2d485513 | [
"BSD-2-Clause"
] | null | null | null | model_notebooks/UBC/model.ipynb | indralab/adeft_indra | 6f039b58b6dea5eefa529cf15afaffff2d485513 | [
"BSD-2-Clause"
] | null | null | null | 30.752492 | 1,456 | 0.556042 | [
[
[
"import os\nimport json\nimport pickle\nimport random\nfrom collections import defaultdict, Counter\n\nfrom indra.literature.adeft_tools import universal_extract_text\nfrom indra.databases.hgnc_client import get_hgnc_name, get_hgnc_id\n\nfrom adeft.discover import AdeftMiner\nfrom adeft.gui import ground_with_gui\nfrom adeft.modeling.label import AdeftLabeler\nfrom adeft.modeling.classify import AdeftClassifier\nfrom adeft.disambiguate import AdeftDisambiguator, load_disambiguator\n\n\nfrom adeft_indra.ground.ground import AdeftGrounder\nfrom adeft_indra.model_building.s3 import model_to_s3\nfrom adeft_indra.model_building.escape import escape_filename\nfrom adeft_indra.db.content import get_pmids_for_agent_text, get_pmids_for_entity, \\\n get_plaintexts_for_pmids",
"_____no_output_____"
],
[
"adeft_grounder = AdeftGrounder()",
"_____no_output_____"
],
[
"shortforms = ['UBC']\nmodel_name = ':'.join(sorted(escape_filename(shortform) for shortform in shortforms))\nresults_path = os.path.abspath(os.path.join('../..', 'results', model_name))",
"_____no_output_____"
],
[
"miners = dict()\nall_texts = {}\nfor shortform in shortforms:\n pmids = get_pmids_for_agent_text(shortform)\n text_dict = get_plaintexts_for_pmids(pmids, contains=shortforms)\n text_dict = {pmid: text for pmid, text in text_dict.items() if len(text) > 5}\n miners[shortform] = AdeftMiner(shortform)\n miners[shortform].process_texts(text_dict.values())\n all_texts.update(text_dict)\n\nlongform_dict = {}\nfor shortform in shortforms:\n longforms = miners[shortform].get_longforms()\n longforms = [(longform, count, score) for longform, count, score in longforms\n if count*score > 2]\n longform_dict[shortform] = longforms\n \ncombined_longforms = Counter()\nfor longform_rows in longform_dict.values():\n combined_longforms.update({longform: count for longform, count, score\n in longform_rows})\ngrounding_map = {}\nnames = {}\nfor longform in combined_longforms:\n groundings = adeft_grounder.ground(longform)\n if groundings:\n grounding = groundings[0]['grounding']\n grounding_map[longform] = grounding\n names[grounding] = groundings[0]['name']\nlongforms, counts = zip(*combined_longforms.most_common())\npos_labels = []",
"_____no_output_____"
],
[
"list(zip(longforms, counts))",
"_____no_output_____"
],
[
"try:\n disamb = load_disambiguator(shortforms[0])\n for shortform, gm in disamb.grounding_dict.items():\n for longform, grounding in gm.items():\n grounding_map[longform] = grounding\n for grounding, name in disamb.names.items():\n names[grounding] = name\n pos_labels = disamb.pos_labels\nexcept Exception:\n pass",
"_____no_output_____"
],
[
"names",
"_____no_output_____"
],
[
"grounding_map, names, pos_labels = ground_with_gui(longforms, counts, \n grounding_map=grounding_map,\n names=names, pos_labels=pos_labels, no_browser=True, port=8891)",
"INFO: [2020-11-10 03:01:29] indra.ontology.bio.ontology - Loading INDRA bio ontology from cache at /home/ubuntu/.indra/bio_ontology/1.4/bio_ontology.pkl\n"
],
[
"result = [grounding_map, names, pos_labels]",
"_____no_output_____"
],
[
"result",
"_____no_output_____"
],
[
"grounding_map, names, pos_labels = [{'ubiquitin c': 'HGNC:12468',\n 'ubiquitin conjugating': 'FPLX:UBE2',\n 'ubiquitin conjugating enzyme': 'FPLX:UBE2',\n 'unipolar brush cells': 'ungrounded',\n 'urinary bladder cancer': 'MESH:D001749',\n 'urothelial bladder cancer': 'MESH:D001749',\n 'urothelial bladder carcinoma': 'MESH:D001749'},\n {'HGNC:12468': 'UBC',\n 'FPLX:UBE2': 'UBE2',\n 'MESH:D001749': 'Urinary Bladder Neoplasms'},\n ['FPLX:UBE2', 'HGNC:12468', 'MESH:D001749']]",
"_____no_output_____"
],
[
"excluded_longforms = []",
"_____no_output_____"
],
[
"grounding_dict = {shortform: {longform: grounding_map[longform] \n for longform, _, _ in longforms if longform in grounding_map\n and longform not in excluded_longforms}\n for shortform, longforms in longform_dict.items()}\nresult = [grounding_dict, names, pos_labels]\n\nif not os.path.exists(results_path):\n os.mkdir(results_path)\nwith open(os.path.join(results_path, f'{model_name}_preliminary_grounding_info.json'), 'w') as f:\n json.dump(result, f)",
"_____no_output_____"
],
[
"additional_entities = {}",
"_____no_output_____"
],
[
"unambiguous_agent_texts = {}",
"_____no_output_____"
],
[
"labeler = AdeftLabeler(grounding_dict)\ncorpus = labeler.build_from_texts((text, pmid) for pmid, text in all_texts.items())\nagent_text_pmid_map = defaultdict(list)\nfor text, label, id_ in corpus:\n agent_text_pmid_map[label].append(id_)\n\nentity_pmid_map = {entity: set(get_pmids_for_entity(*entity.split(':', maxsplit=1),\n major_topic=True))for entity in additional_entities}",
"_____no_output_____"
],
[
"intersection1 = []\nfor entity1, pmids1 in entity_pmid_map.items():\n for entity2, pmids2 in entity_pmid_map.items():\n intersection1.append((entity1, entity2, len(pmids1 & pmids2)))",
"_____no_output_____"
],
[
"intersection2 = []\nfor entity1, pmids1 in agent_text_pmid_map.items():\n for entity2, pmids2 in entity_pmid_map.items():\n intersection2.append((entity1, entity2, len(set(pmids1) & pmids2)))",
"_____no_output_____"
],
[
"intersection1",
"_____no_output_____"
],
[
"intersection2",
"_____no_output_____"
],
[
"all_used_pmids = set()\nfor entity, agent_texts in unambiguous_agent_texts.items():\n used_pmids = set()\n for agent_text in agent_texts[1]:\n pmids = set(get_pmids_for_agent_text(agent_text))\n new_pmids = list(pmids - all_texts.keys() - used_pmids)\n text_dict = get_plaintexts_for_pmids(new_pmids, contains=agent_texts)\n corpus.extend([(text, entity, pmid) for pmid, text in text_dict.items() if len(text) >= 5])\n used_pmids.update(new_pmids)\n all_used_pmids.update(used_pmids)\n \nfor entity, pmids in entity_pmid_map.items():\n new_pmids = list(set(pmids) - all_texts.keys() - all_used_pmids)\n if len(new_pmids) > 10000:\n new_pmids = random.choices(new_pmids, k=10000)\n _, contains = additional_entities[entity]\n text_dict = get_plaintexts_for_pmids(new_pmids, contains=contains)\n corpus.extend([(text, entity, pmid) for pmid, text in text_dict.items() if len(text) >= 5])",
"_____no_output_____"
],
[
"names.update({key: value[0] for key, value in additional_entities.items()})\nnames.update({key: value[0] for key, value in unambiguous_agent_texts.items()})\npos_labels = list(set(pos_labels) | additional_entities.keys() |\n unambiguous_agent_texts.keys())",
"_____no_output_____"
],
[
"%%capture\n\nclassifier = AdeftClassifier(shortforms, pos_labels=pos_labels, random_state=1729)\nparam_grid = {'C': [100.0], 'max_features': [10000]}\ntexts, labels, pmids = zip(*corpus)\nclassifier.cv(texts, labels, param_grid, cv=5, n_jobs=5)",
"INFO: [2020-11-10 03:02:57] /adeft/PP/adeft/adeft/modeling/classify.py - Beginning grid search in parameter space:\n{'C': [100.0], 'max_features': [10000]}\nINFO: [2020-11-10 03:02:59] /adeft/PP/adeft/adeft/modeling/classify.py - Best f1 score of 0.8145351127630539 found for parameter values:\n{'logit__C': 100.0, 'tfidf__max_features': 10000}\n"
],
[
"classifier.stats",
"_____no_output_____"
],
[
"disamb = AdeftDisambiguator(classifier, grounding_dict, names)",
"_____no_output_____"
],
[
"disamb.dump(model_name, results_path)",
"_____no_output_____"
],
[
"print(disamb.info())",
"Disambiguation model for UBC\n\nProduces the disambiguations:\n\tUBC*\tHGNC:12468\n\tUBE2*\tFPLX:UBE2\n\tUrinary Bladder Neoplasms*\tMESH:D001749\n\nClass level metrics:\n--------------------\nGrounding \tCount\tF1 \nUrinary Bladder Neoplasms*\t57\t0.91538\n UBC*\t16\t0.55778\n UBE2*\t10\t0.62667\n Ungrounded\t 2\t 0.0\n\nWeighted Metrics:\n-----------------\n\tF1 score:\t0.81454\n\tPrecision:\t0.8339\n\tRecall:\t\t0.85515\n\n* Positive labels\nSee Docstring for explanation\n\n"
],
[
"model_to_s3(disamb)",
"_____no_output_____"
],
[
"preds = [disamb.disambiguate(text) for text in all_texts.values()]",
"_____no_output_____"
],
[
"texts = [text for pred, text in zip(preds, all_texts.values()) if pred[0] == 'HGNC:10967']",
"_____no_output_____"
],
[
"texts[3]",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d09336fdcbf19804e260054e91e1c192454f8b71 | 1,717 | ipynb | Jupyter Notebook | lectures/1_TeoriaProbabilidades/parte5.ipynb | magister-informatica-uach/INFO337 | 45d7faabbd4ed5b25a575ee065551b87b097f92e | [
"Unlicense"
] | 4 | 2021-06-12T04:07:26.000Z | 2022-03-27T23:22:59.000Z | lectures/1_TeoriaProbabilidades/parte5.ipynb | magister-informatica-uach/INFO337 | 45d7faabbd4ed5b25a575ee065551b87b097f92e | [
"Unlicense"
] | null | null | null | lectures/1_TeoriaProbabilidades/parte5.ipynb | magister-informatica-uach/INFO337 | 45d7faabbd4ed5b25a575ee065551b87b097f92e | [
"Unlicense"
] | 1 | 2019-11-07T14:49:09.000Z | 2019-11-07T14:49:09.000Z | 28.147541 | 201 | 0.506115 | [
[
[
"# Teoremas Asintóticos\n\n## Ley débil de los grandes números\nSea $\\{X_i\\}_{i=1}^N$ un conjunto de $N$ v.a. independientes idénticamente distribuidas (iid) de media $\\mu$ y varianza finita $\\sigma^2$. Entonces, para cualquier $\\epsilon >0$ se tiene:\n\n$$\\lim_{N \\to \\infty} P\\left\\{\\middle | \\frac{\\sum_i X_i}{N} - \\mu \\,\\middle | > \\epsilon \\right\\} = 0$$\n",
"_____no_output_____"
],
[
"## Teorema del Límite Central\nSean $X_1,...X_N$ v.a. iid de media $\\mu$ y varianza $\\sigma^2$ entonces\n$S_N=X_1+...X_N$ cumple:\n\n$$ \\lim_{N \\to \\infty}P\\left( \\frac{S_N - N\\mu}{\\sigma\\sqrt N} \\leq z\\right) = F_Z(z) \n\\qquad Z \\sim \\cal{N}(0,1)$$\n\n\nEn particular $m= \\frac{S_N}{N}= \\frac{\\sum_{i=1}^N \\limits X_i}{N}$ cumple:\n\n$$ \\lim_{N \\to \\infty}P\\left( \\frac{m - \\mu}{\\sigma/\\sqrt N} \\leq z\\right) = F_Z(z) $$\n\nEs decir que es posible aproximar la distribución de $m$ por ${\\cal{N}}(\\mu,\\frac{\\sigma^2}{N})$\n",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown"
]
] |
d09341726c3d450868ffd7025915eba35380a548 | 195,920 | ipynb | Jupyter Notebook | Script/kmodes-cluster-perfil-consumo-cliente.ipynb | helenacypreste/cluster-cliente-perfil-consumo | fbdb3bc5ddc17e1932b6910926d35639cf13db32 | [
"MIT"
] | null | null | null | Script/kmodes-cluster-perfil-consumo-cliente.ipynb | helenacypreste/cluster-cliente-perfil-consumo | fbdb3bc5ddc17e1932b6910926d35639cf13db32 | [
"MIT"
] | null | null | null | Script/kmodes-cluster-perfil-consumo-cliente.ipynb | helenacypreste/cluster-cliente-perfil-consumo | fbdb3bc5ddc17e1932b6910926d35639cf13db32 | [
"MIT"
] | null | null | null | 105.050938 | 129,764 | 0.751501 | [
[
[
"# Identificar Perfil de Consumo de Clientes de uma Instituição Financeira",
"_____no_output_____"
],
[
"## Uma breve introdução",
"_____no_output_____"
],
[
"Uma Instituição Financeira X tem o interesse em identificar o perfil de gastos dos seus clientes. Identificando os clientes certos, eles podem melhorar a comunicação dos ativos promocionais, utilizar com mais eficiência os canais de comunicação e aumentar o engajamento dos clientes com o uso do seu produto.",
"_____no_output_____"
],
[
"## Sobre o estudo",
"_____no_output_____"
],
[
"Os dados estão anonimizados por questões de segurança. No dataset, temos o valor gasto de 121818 clientes, no ano de 2019, em cada ramo de atividade. A base já está limpa. O intuito aqui é apresentar uma possibilidade de fazer a clusterização de clientes com base em seu consumo. Os dados são de 1 ano a fim de diminuir o efeito da sazonalidade.",
"_____no_output_____"
],
[
"## Importando a biblioteca",
"_____no_output_____"
],
[
"Será necessário instalar o pacote kmodes, caso não tenha. ",
"_____no_output_____"
]
],
[
[
"# pip install --upgrade kmodes",
"_____no_output_____"
],
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport os\nimport random\nimport plotly.express as px\n# pacote do modelo\nfrom kmodes.kmodes import KModes\n# from sklearn.model_selection import train_test_split\n\nimport warnings\nwarnings.filterwarnings(\"ignore\")\n\nrandom.seed(2020)\n\npd.options.display.float_format = '{:,.2f}'.format",
"_____no_output_____"
]
],
[
[
"## Leitura dos dados",
"_____no_output_____"
]
],
[
[
"# Carregar os dados\ndados = pd.read_csv('../../dados_gasto_rmat.csv', sep=',')",
"_____no_output_____"
],
[
"dados.head(10)",
"_____no_output_____"
],
[
"print('O arquivo possui ' + str(dados.shape[0]) + ' linhas e ' + str(dados.shape[1]) + ' colunas.')",
"O arquivo possui 3898614 linhas e 3 colunas.\n"
],
[
"# Tipos dos dados\ndados.dtypes",
"_____no_output_____"
],
[
"# Verificando valores nulos\ndados.isnull().values.any()",
"_____no_output_____"
]
],
[
[
"A base não possui valores nulos.",
"_____no_output_____"
],
[
"## Visualização dos dados",
"_____no_output_____"
]
],
[
[
"# Total de clientes\nlen(dados['CLIENTE'].unique())",
"_____no_output_____"
]
],
[
[
"Soma do valor gasto em cada ramo de atividade.",
"_____no_output_____"
]
],
[
[
"# Dados agregados por RMAT e o percentual gasto\ndados_agreg = dados.groupby(['RMAT'])['VALOR_GASTO'].sum().reset_index()\ndados_agreg['percentual'] = round(dados_agreg['VALOR_GASTO']/sum(dados_agreg['VALOR_GASTO'])*100,2)\ndados_agreg.head()",
"_____no_output_____"
],
[
"# Traz o 30 RMATs que tiveram maior gasto dos clientes\ntop_rotulos = dados_agreg.sort_values(by = 'percentual', ascending = False)[:30]",
"_____no_output_____"
],
[
"# Gráfico dos RMATs\nax = top_rotulos.plot.barh(x='RMAT', y='percentual', rot=0, figsize = (20, 15), fontsize=20, color='violet')\nplt.title('Percentual do valor gasto por Ramo de Atividade', fontsize=22)\nplt.xlabel('')\nplt.ylabel('')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Supermercados, postos de combustível e drogarias são os ramos com maior consumo dos clientes, representando 13.48%, 7.07% e 5.75%, respectivamente. ",
"_____no_output_____"
],
[
"### Construção da base para o modelo",
"_____no_output_____"
],
[
"Vamos construir uma base que seja a nível cliente e as colunas serão os percentuais de consumo em cada ramo de atividade. Aqui, temos muitos percentuais 0 em determinados ramos de atividade e isso tem impacto negativo na construção do modelo. Portanto, iremos categorizar as variáveis de acordo com a variação do percentual para cada atributo.",
"_____no_output_____"
]
],
[
[
"dados.head()",
"_____no_output_____"
]
],
[
[
"A função pivot faz a transposição dos dados, transformando o valor de \"RMAT\" em colunas.",
"_____no_output_____"
]
],
[
[
"# Inverter o data frame (colocar nos rmats como coluna)\ncli_pivot = dados.pivot(index='CLIENTE', columns='RMAT', values='VALOR_GASTO')\ncli_pivot.fillna(0, inplace = True)",
"_____no_output_____"
],
[
"# Calcular o percentual de volume transacionado de cada cliente por rmat \ncli_pivot = cli_pivot.apply(lambda x: x.apply(lambda y: 100*y/sum(x)),axis = 1)\ncli_pivot.head()",
"_____no_output_____"
]
],
[
[
"Abaixo, uma função que quebra os valores de cada coluna em quantidade de categorias escolhido. Nesse caso, utilizamos 8 quebras.",
"_____no_output_____"
]
],
[
[
"# Funnção para fazer categorização das variaveis\ndef hcut(df, colunas, nlevs, prefixo=''):\n x = df.copy()\n for c in colunas:\n \n x[prefixo+c] = pd.cut(x[c] , bins=nlevs, include_lowest = False, precision=0)\n \n return x",
"_____no_output_____"
],
[
"base_cluster = hcut(cli_pivot, cli_pivot\n .columns, 8, 'esc_')",
"_____no_output_____"
],
[
"base_cluster.head()",
"_____no_output_____"
]
],
[
[
"Agora, filtraremos as variáveis necessárias para a modelagem. ",
"_____no_output_____"
]
],
[
[
"# Selecionar somentes as colunas categorizadas que serão utilizadas no modelo\nfilter_col = [col for col in base_cluster if\n col.startswith('esc_')]\ndf1 = base_cluster.loc[:,filter_col].reset_index() \ndf1.head()",
"_____no_output_____"
]
],
[
[
"### Criação do modelo ",
"_____no_output_____"
],
[
"Para nosso caso, utilizaremos o método de clusterização chamado K-modes. Esse método é uma extensão do K-means. Em vez de distâncias, ele usa dissimilaridade (isto é, quantificação das incompatibilidades totais entre dois objetos: quanto menor esse número, masi semelhantes são os dois objetos). Além disso, ele utiliza modas. Cada vetor de elementos é criado de forma a minimizar as diferenças entre o próprio vetor e cada objetos dos dados. Assim, teremos tantos vetores de modas quanto o número de clusters necessários, pois eles atuam como centróides. \n",
"_____no_output_____"
],
[
"Aqui, iremos fazer a divisão dos clientes em 7 clusters. ",
"_____no_output_____"
]
],
[
[
"km_huang = KModes(n_clusters=7, init = \"Huang\", n_init = 5, verbose=1, random_state=2020)\nfitClusters = km_huang.fit_predict(df1)",
"Init: initializing centroids\nInit: initializing clusters\nStarting iterations...\nRun 1, iteration: 1/100, moves: 37911, cost: 285526.0\nRun 1, iteration: 2/100, moves: 6829, cost: 285526.0\nInit: initializing centroids\nInit: initializing clusters\nStarting iterations...\nRun 2, iteration: 1/100, moves: 24674, cost: 296153.0\nInit: initializing centroids\nInit: initializing clusters\nStarting iterations...\nRun 3, iteration: 1/100, moves: 38186, cost: 311798.0\nRun 3, iteration: 2/100, moves: 1175, cost: 311797.0\nRun 3, iteration: 3/100, moves: 0, cost: 311797.0\nInit: initializing centroids\nInit: initializing clusters\nStarting iterations...\nRun 4, iteration: 1/100, moves: 49330, cost: 291921.0\nRun 4, iteration: 2/100, moves: 5319, cost: 291920.0\nRun 4, iteration: 3/100, moves: 0, cost: 291920.0\nInit: initializing centroids\nInit: initializing clusters\nStarting iterations...\nRun 5, iteration: 1/100, moves: 35294, cost: 289461.0\nRun 5, iteration: 2/100, moves: 14295, cost: 289460.0\nRun 5, iteration: 3/100, moves: 0, cost: 289460.0\nBest run was number 1\n"
],
[
"# Adiciona os clusters na base\ndf1['cluster'] = fitClusters\nbase_cluster['cluster'] = fitClusters",
"_____no_output_____"
]
],
[
[
"#### Percentual de clientes por cluster. ",
"_____no_output_____"
]
],
[
[
"df1['cluster'].value_counts()/len(df1)*100",
"_____no_output_____"
],
[
"df2 = base_cluster.drop(columns=filter_col)",
"_____no_output_____"
],
[
"df2.head()",
"_____no_output_____"
],
[
"# Para visualização dos clusters\n\n# from sklearn.decomposition import PCA\n# pca_2 = PCA(2)\n# plot_columns = pca_2.fit_transform(base_cluster.iloc[:,0:65])\n# plt.scatter(x=plot_columns[:,0], y=plot_columns[:,1], c=fitClusters,)\n# plt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d09345fb0cb362ce01e8b9baa1fb54fceca1e0f5 | 4,170 | ipynb | Jupyter Notebook | Code/9.generadores/02-Iterators and Generators Homework.ipynb | davidMartinVergues/PYTHON | dd39d3aabfc43b3cb09aadb2919e51d03364117d | [
"DOC"
] | null | null | null | Code/9.generadores/02-Iterators and Generators Homework.ipynb | davidMartinVergues/PYTHON | dd39d3aabfc43b3cb09aadb2919e51d03364117d | [
"DOC"
] | null | null | null | Code/9.generadores/02-Iterators and Generators Homework.ipynb | davidMartinVergues/PYTHON | dd39d3aabfc43b3cb09aadb2919e51d03364117d | [
"DOC"
] | null | null | null | 19.952153 | 166 | 0.489928 | [
[
[
"# Iterators and Generators Homework \n\n### Problem 1\n\nCreate a generator that generates the squares of numbers up to some number N.",
"_____no_output_____"
]
],
[
[
"# option 1\ndef gensquares(N):\n end = N\n start = 0\n while start<end:\n yield start**2\n start += 1\n\n# option 2\ndef gensquares2(N):\n\n for num in range(N):\n yield num**2\n\n\n ",
"_____no_output_____"
],
[
"for x in gensquares2(10):\n print(x)",
"0\n1\n4\n9\n16\n25\n36\n49\n64\n81\n"
]
],
[
[
"### Problem 2\n\nCreate a generator that yields \"n\" random numbers between a low and high number (that are inputs). <br>Note: Use the random library. For example:",
"_____no_output_____"
]
],
[
[
"import random\n\nrandom.randint(1,10)",
"_____no_output_____"
],
[
"def rand_num(low,high,n):\n for times in range(n):\n yield random.randint(low,high)\n\n ",
"_____no_output_____"
],
[
"for num in rand_num(1,10,12):\n print(num)",
"1\n5\n4\n6\n9\n5\n5\n4\n2\n7\n9\n6\n"
]
],
[
[
"### Problem 3\n\nUse the iter() function to convert the string below into an iterator:\n",
"_____no_output_____"
]
],
[
[
"s = 'hello'\n\n#code here\ns_iter = iter()",
"_____no_output_____"
]
],
[
[
"### Problem 4\nExplain a use case for a generator using a yield statement where you would not want to use a normal function with a return statement.<br><br><br><br><br><br>\n\n",
"_____no_output_____"
],
[
"### Extra Credit!\nCan you explain what *gencomp* is in the code below? (Note: We never covered this in lecture! You will have to do some Googling/Stack Overflowing!)",
"_____no_output_____"
]
],
[
[
"my_list = [1,2,3,4,5]\n\ngencomp = (item for item in my_list if item > 3)\n\nfor item in gencomp:\n print(item)",
"4\n5\n"
]
],
[
[
"Hint: Google *generator comprehension*!\n\n# Great Job!",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0935bdcaa66591a029c6a0ae86e4b636cc89d11 | 42,608 | ipynb | Jupyter Notebook | Improving_Computer_Vision_Using_CNN.ipynb | snalahi/Introduction-to-TensorFlow-for-Artificial-Intelligence-Machine-Learning-and-Deep-Learning | 390b113b03c3d1c15366a281a8a0078d9ccef326 | [
"MIT"
] | 1 | 2021-05-29T21:04:01.000Z | 2021-05-29T21:04:01.000Z | Improving_Computer_Vision_Using_CNN.ipynb | snalahi/Introduction-to-TensorFlow-for-Artificial-Intelligence-Machine-Learning-and-Deep-Learning | 390b113b03c3d1c15366a281a8a0078d9ccef326 | [
"MIT"
] | null | null | null | Improving_Computer_Vision_Using_CNN.ipynb | snalahi/Introduction-to-TensorFlow-for-Artificial-Intelligence-Machine-Learning-and-Deep-Learning | 390b113b03c3d1c15366a281a8a0078d9ccef326 | [
"MIT"
] | null | null | null | 79.492537 | 19,268 | 0.76542 | [
[
[
"##### Copyright 2019 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"#Improving Computer Vision Accuracy using Convolutions\n\nIn the previous lessons you saw how to do fashion recognition using a Deep Neural Network (DNN) containing three layers -- the input layer (in the shape of the data), the output layer (in the shape of the desired output) and a hidden layer. You experimented with the impact of different sizes of hidden layer, number of training epochs etc on the final accuracy.\n\nFor convenience, here's the entire code again. Run it and take a note of the test accuracy that is printed out at the end. ",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nmnist = tf.keras.datasets.fashion_mnist\n(training_images, training_labels), (test_images, test_labels) = mnist.load_data()\ntraining_images=training_images / 255.0\ntest_images=test_images / 255.0\nmodel = tf.keras.models.Sequential([\n tf.keras.layers.Flatten(),\n tf.keras.layers.Dense(128, activation=tf.nn.relu),\n tf.keras.layers.Dense(10, activation=tf.nn.softmax)\n])\nmodel.compile(optimizer='adam', loss='sparse_categorical_crossentropy', metrics=['accuracy'])\nmodel.fit(training_images, training_labels, epochs=5)\n\ntest_loss = model.evaluate(test_images, test_labels)",
"Downloading data from https://storage.googleapis.com/tensorflow/tf-keras-datasets/train-labels-idx1-ubyte.gz\n32768/29515 [=================================] - 0s 0us/step\nDownloading data from https://storage.googleapis.com/tensorflow/tf-keras-datasets/train-images-idx3-ubyte.gz\n26427392/26421880 [==============================] - 0s 0us/step\nDownloading data from https://storage.googleapis.com/tensorflow/tf-keras-datasets/t10k-labels-idx1-ubyte.gz\n8192/5148 [===============================================] - 0s 0us/step\nDownloading data from https://storage.googleapis.com/tensorflow/tf-keras-datasets/t10k-images-idx3-ubyte.gz\n4423680/4422102 [==============================] - 0s 0us/step\nEpoch 1/5\n1875/1875 [==============================] - 4s 2ms/step - loss: 0.4968 - accuracy: 0.8242\nEpoch 2/5\n1875/1875 [==============================] - 4s 2ms/step - loss: 0.3721 - accuracy: 0.8649\nEpoch 3/5\n1875/1875 [==============================] - 4s 2ms/step - loss: 0.3326 - accuracy: 0.8787\nEpoch 4/5\n1875/1875 [==============================] - 4s 2ms/step - loss: 0.3107 - accuracy: 0.8857\nEpoch 5/5\n1875/1875 [==============================] - 4s 2ms/step - loss: 0.2946 - accuracy: 0.8911\n313/313 [==============================] - 1s 1ms/step - loss: 0.3476 - accuracy: 0.8750\n"
]
],
[
[
"Your accuracy is probably about 89% on training and 87% on validation...not bad...But how do you make that even better? One way is to use something called Convolutions. I'm not going to details on Convolutions here, but the ultimate concept is that they narrow down the content of the image to focus on specific, distinct, details. \n\nIf you've ever done image processing using a filter (like this: https://en.wikipedia.org/wiki/Kernel_(image_processing)) then convolutions will look very familiar.\n\nIn short, you take an array (usually 3x3 or 5x5) and pass it over the image. By changing the underlying pixels based on the formula within that matrix, you can do things like edge detection. So, for example, if you look at the above link, you'll see a 3x3 that is defined for edge detection where the middle cell is 8, and all of its neighbors are -1. In this case, for each pixel, you would multiply its value by 8, then subtract the value of each neighbor. Do this for every pixel, and you'll end up with a new image that has the edges enhanced.\n\nThis is perfect for computer vision, because often it's features that can get highlighted like this that distinguish one item for another, and the amount of information needed is then much less...because you'll just train on the highlighted features.\n\nThat's the concept of Convolutional Neural Networks. Add some layers to do convolution before you have the dense layers, and then the information going to the dense layers is more focussed, and possibly more accurate.\n\nRun the below code -- this is the same neural network as earlier, but this time with Convolutional layers added first. It will take longer, but look at the impact on the accuracy:",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nprint(tf.__version__)\nmnist = tf.keras.datasets.fashion_mnist\n(training_images, training_labels), (test_images, test_labels) = mnist.load_data()\n# Reshaping the images to tell the convolutional layers that the images are in greyscale by adding an extra dimension of 1\ntraining_images=training_images.reshape(60000, 28, 28, 1)\ntraining_images=training_images / 255.0\ntest_images = test_images.reshape(10000, 28, 28, 1)\ntest_images=test_images/255.0\nmodel = tf.keras.models.Sequential([\n # Mind it, Convolutions and MaxPooling are always applied before the Deep Neural Network Layers\n # Why 2D? because, applied convolutions and maxpoolings are 2D array in nature (having rows and columns)\n # Here 64 is the total number of convolutional filters of size (3, 3) applied\n ### Be careful about the shapes!!! You obviously need to mention the input_shape at the first Conv2D(), otherwise it will turn\n ### into an error!!! \n tf.keras.layers.Conv2D(64, (3, 3), activation='relu', input_shape=(28, 28, 1)),\n tf.keras.layers.MaxPooling2D(2, 2),\n tf.keras.layers.Conv2D(64, (3, 3), activation='relu'),\n tf.keras.layers.MaxPooling2D(2, 2),\n tf.keras.layers.Flatten(),\n tf.keras.layers.Dense(128, activation='relu'),\n tf.keras.layers.Dense(10, activation='softmax')\n])\nmodel.compile(optimizer='adam', loss='sparse_categorical_crossentropy', metrics=['accuracy'])\n# Mind it, model.summary() is a way of cross-verification that the DNN with Convolutions is applied correctly with accurate\n# shape retention\nmodel.summary()\nmodel.fit(training_images, training_labels, epochs=5)\ntest_loss = model.evaluate(test_images, test_labels)\n",
"2.5.0\nModel: \"sequential_3\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nconv2d_4 (Conv2D) (None, 26, 26, 64) 640 \n_________________________________________________________________\nmax_pooling2d_4 (MaxPooling2 (None, 13, 13, 64) 0 \n_________________________________________________________________\nconv2d_5 (Conv2D) (None, 11, 11, 64) 36928 \n_________________________________________________________________\nmax_pooling2d_5 (MaxPooling2 (None, 5, 5, 64) 0 \n_________________________________________________________________\nflatten_3 (Flatten) (None, 1600) 0 \n_________________________________________________________________\ndense_6 (Dense) (None, 128) 204928 \n_________________________________________________________________\ndense_7 (Dense) (None, 10) 1290 \n=================================================================\nTotal params: 243,786\nTrainable params: 243,786\nNon-trainable params: 0\n_________________________________________________________________\nEpoch 1/5\n1875/1875 [==============================] - 81s 43ms/step - loss: 0.4397 - accuracy: 0.8383\nEpoch 2/5\n1875/1875 [==============================] - 79s 42ms/step - loss: 0.2954 - accuracy: 0.8918\nEpoch 3/5\n1875/1875 [==============================] - 79s 42ms/step - loss: 0.2518 - accuracy: 0.9076\nEpoch 4/5\n1875/1875 [==============================] - 78s 42ms/step - loss: 0.2214 - accuracy: 0.9164\nEpoch 5/5\n1875/1875 [==============================] - 78s 42ms/step - loss: 0.1924 - accuracy: 0.9273\n313/313 [==============================] - 4s 12ms/step - loss: 0.2676 - accuracy: 0.9057\n"
]
],
[
[
"It's likely gone up to about 93% on the training data and 91% on the validation data. \n\nThat's significant, and a step in the right direction!\n\nTry running it for more epochs -- say about 20, and explore the results! But while the results might seem really good, the validation results may actually go down, due to something called 'overfitting' which will be discussed later. \n\n(In a nutshell, 'overfitting' occurs when the network learns the data from the training set really well, but it's too specialised to only that data, and as a result is less effective at seeing *other* data. For example, if all your life you only saw red shoes, then when you see a red shoe you would be very good at identifying it, but blue suade shoes might confuse you...and you know you should never mess with my blue suede shoes.)\n\nThen, look at the code again, and see, step by step how the Convolutions were built:",
"_____no_output_____"
],
[
"Step 1 is to gather the data. You'll notice that there's a bit of a change here in that the training data needed to be reshaped. That's because the first convolution expects a single tensor containing everything, so instead of 60,000 28x28x1 items in a list, we have a single 4D list that is 60,000x28x28x1, and the same for the test images. If you don't do this, you'll get an error when training as the Convolutions do not recognize the shape. \n\n\n\n```\nimport tensorflow as tf\nmnist = tf.keras.datasets.fashion_mnist\n(training_images, training_labels), (test_images, test_labels) = mnist.load_data()\ntraining_images=training_images.reshape(60000, 28, 28, 1)\ntraining_images=training_images / 255.0\ntest_images = test_images.reshape(10000, 28, 28, 1)\ntest_images=test_images/255.0\n```\n",
"_____no_output_____"
],
[
"Next is to define your model. Now instead of the input layer at the top, you're going to add a Convolution. The parameters are:\n\n1. The number of convolutions you want to generate. Purely arbitrary, but good to start with something in the order of 32\n2. The size of the Convolution, in this case a 3x3 grid\n3. The activation function to use -- in this case we'll use relu, which you might recall is the equivalent of returning x when x>0, else returning 0\n4. In the first layer, the shape of the input data.\n\nYou'll follow the Convolution with a MaxPooling layer which is then designed to compress the image, while maintaining the content of the features that were highlighted by the convlution. By specifying (2,2) for the MaxPooling, the effect is to quarter the size of the image. Without going into too much detail here, the idea is that it creates a 2x2 array of pixels, and picks the biggest one, thus turning 4 pixels into 1. It repeats this across the image, and in so doing halves the number of horizontal, and halves the number of vertical pixels, effectively reducing the image by 25%.\n\nYou can call model.summary() to see the size and shape of the network, and you'll notice that after every MaxPooling layer, the image size is reduced in this way. \n\n\n```\nmodel = tf.keras.models.Sequential([\n tf.keras.layers.Conv2D(32, (3,3), activation='relu', input_shape=(28, 28, 1)),\n tf.keras.layers.MaxPooling2D(2, 2),\n```\n",
"_____no_output_____"
],
[
"Add another convolution\n\n\n\n```\n tf.keras.layers.Conv2D(64, (3,3), activation='relu'),\n tf.keras.layers.MaxPooling2D(2,2)\n```\n",
"_____no_output_____"
],
[
"Now flatten the output. After this you'll just have the same DNN structure as the non convolutional version\n\n```\n tf.keras.layers.Flatten(),\n```\n",
"_____no_output_____"
],
[
"The same 128 dense layers, and 10 output layers as in the pre-convolution example:\n\n\n\n```\n tf.keras.layers.Dense(128, activation='relu'),\n tf.keras.layers.Dense(10, activation='softmax')\n])\n```\n",
"_____no_output_____"
],
[
"Now compile the model, call the fit method to do the training, and evaluate the loss and accuracy from the test set.\n\n\n\n```\nmodel.compile(optimizer='adam', loss='sparse_categorical_crossentropy', metrics=['accuracy'])\nmodel.fit(training_images, training_labels, epochs=5)\ntest_loss, test_acc = model.evaluate(test_images, test_labels)\nprint(test_acc)\n```\n",
"_____no_output_____"
],
[
"# Visualizing the Convolutions and Pooling\n\nThis code will show us the convolutions graphically. The print (test_labels[;100]) shows us the first 100 labels in the test set, and you can see that the ones at index 0, index 23 and index 28 are all the same value (9). They're all shoes. Let's take a look at the result of running the convolution on each, and you'll begin to see common features between them emerge. Now, when the DNN is training on that data, it's working with a lot less, and it's perhaps finding a commonality between shoes based on this convolution/pooling combination.",
"_____no_output_____"
]
],
[
[
"print(test_labels[:100])",
"[9 2 1 1 6 1 4 6 5 7 4 5 7 3 4 1 2 4 8 0 2 5 7 9 1 4 6 0 9 3 8 8 3 3 8 0 7\n 5 7 9 6 1 3 7 6 7 2 1 2 2 4 4 5 8 2 2 8 4 8 0 7 7 8 5 1 1 2 3 9 8 7 0 2 6\n 2 3 1 2 8 4 1 8 5 9 5 0 3 2 0 6 5 3 6 7 1 8 0 1 4 2]\n"
],
[
"import matplotlib.pyplot as plt\nf, axarr = plt.subplots(3,4)\nFIRST_IMAGE=0\nSECOND_IMAGE=7\nTHIRD_IMAGE=26\nCONVOLUTION_NUMBER = 1\nfrom tensorflow.keras import models\nlayer_outputs = [layer.output for layer in model.layers]\nactivation_model = tf.keras.models.Model(inputs = model.input, outputs = layer_outputs)\nfor x in range(0,4):\n f1 = activation_model.predict(test_images[FIRST_IMAGE].reshape(1, 28, 28, 1))[x]\n axarr[0,x].imshow(f1[0, : , :, CONVOLUTION_NUMBER], cmap='inferno')\n axarr[0,x].grid(False)\n f2 = activation_model.predict(test_images[SECOND_IMAGE].reshape(1, 28, 28, 1))[x]\n axarr[1,x].imshow(f2[0, : , :, CONVOLUTION_NUMBER], cmap='inferno')\n axarr[1,x].grid(False)\n f3 = activation_model.predict(test_images[THIRD_IMAGE].reshape(1, 28, 28, 1))[x]\n axarr[2,x].imshow(f3[0, : , :, CONVOLUTION_NUMBER], cmap='inferno')\n axarr[2,x].grid(False)",
"_____no_output_____"
]
],
[
[
"EXERCISES\n\n1. Try editing the convolutions. Change the 32s to either 16 or 64. What impact will this have on accuracy and/or training time.\n\n2. Remove the final Convolution. What impact will this have on accuracy or training time?\n\n3. How about adding more Convolutions? What impact do you think this will have? Experiment with it.\n\n4. Remove all Convolutions but the first. What impact do you think this will have? Experiment with it. \n\n5. In the previous lesson you implemented a callback to check on the loss function and to cancel training once it hit a certain amount. See if you can implement that here!",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nprint(tf.__version__)\nmnist = tf.keras.datasets.mnist\n(training_images, training_labels), (test_images, test_labels) = mnist.load_data()\ntraining_images=training_images.reshape(60000, 28, 28, 1)\ntraining_images=training_images / 255.0\ntest_images = test_images.reshape(10000, 28, 28, 1)\ntest_images=test_images/255.0\nmodel = tf.keras.models.Sequential([\n tf.keras.layers.Conv2D(32, (3,3), activation='relu', input_shape=(28, 28, 1)),\n tf.keras.layers.MaxPooling2D(2, 2),\n tf.keras.layers.Flatten(),\n tf.keras.layers.Dense(128, activation='relu'),\n tf.keras.layers.Dense(10, activation='softmax')\n])\nmodel.compile(optimizer='adam', loss='sparse_categorical_crossentropy', metrics=['accuracy'])\nmodel.fit(training_images, training_labels, epochs=10)\ntest_loss, test_acc = model.evaluate(test_images, test_labels)\nprint(test_acc)",
"2.5.0\nDownloading data from https://storage.googleapis.com/tensorflow/tf-keras-datasets/mnist.npz\n11493376/11490434 [==============================] - 0s 0us/step\nEpoch 1/10\n1875/1875 [==============================] - 34s 18ms/step - loss: 0.1424 - accuracy: 0.9569\nEpoch 2/10\n1875/1875 [==============================] - 34s 18ms/step - loss: 0.0502 - accuracy: 0.9842\nEpoch 3/10\n1875/1875 [==============================] - 34s 18ms/step - loss: 0.0311 - accuracy: 0.9901\nEpoch 4/10\n1875/1875 [==============================] - 34s 18ms/step - loss: 0.0217 - accuracy: 0.9932\nEpoch 5/10\n1875/1875 [==============================] - 34s 18ms/step - loss: 0.0138 - accuracy: 0.9955\nEpoch 6/10\n1875/1875 [==============================] - 34s 18ms/step - loss: 0.0100 - accuracy: 0.9969\nEpoch 7/10\n1875/1875 [==============================] - 34s 18ms/step - loss: 0.0078 - accuracy: 0.9974\nEpoch 8/10\n1875/1875 [==============================] - 34s 18ms/step - loss: 0.0063 - accuracy: 0.9980\nEpoch 9/10\n1875/1875 [==============================] - 34s 18ms/step - loss: 0.0046 - accuracy: 0.9986\nEpoch 10/10\n1875/1875 [==============================] - 34s 18ms/step - loss: 0.0055 - accuracy: 0.9982\n313/313 [==============================] - 2s 6ms/step - loss: 0.0584 - accuracy: 0.9866\n0.9865999817848206\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0936e61dca151b3b924633e2cc139a8864f8b33 | 38,154 | ipynb | Jupyter Notebook | examples/fundamentals/fundamentals.ipynb | yo252yo/ml4a | a5c37265460f037d2b0dae7c022e5944b9a7b8a6 | [
"MIT"
] | 1,110 | 2016-06-02T23:58:41.000Z | 2020-11-29T07:24:20.000Z | examples/fundamentals/fundamentals.ipynb | yo252yo/ml4a | a5c37265460f037d2b0dae7c022e5944b9a7b8a6 | [
"MIT"
] | 49 | 2016-08-14T22:58:41.000Z | 2020-07-17T17:59:56.000Z | examples/fundamentals/fundamentals.ipynb | yo252yo/ml4a | a5c37265460f037d2b0dae7c022e5944b9a7b8a6 | [
"MIT"
] | 300 | 2016-06-13T23:06:55.000Z | 2020-11-18T22:42:55.000Z | 44.624561 | 820 | 0.622949 | [
[
[
"## Fundamentals, introduction to machine learning\n\n\nThe purpose of these guides is to go a bit deeper into the details behind common machine learning methods, assuming little math background, and teach you how to use popular machine learning Python packages. In particular, we'll focus on the Numpy and PyTorch libraries.\n\nI'll assume you have some experience programming with Python -- if not, check out the initial [fundamentals of Python guide](https://github.com/ml4a/ml4a-guides/blob/master/notebooks/intro_python.ipynb) or for a longer, more comprehensive resource: [Learn Python the Hard Way](http://learnpythonthehardway.org/book/). It will really help to illustrate the concepts introduced here.\n\nNumpy underlies most Python machine learning packages and is great for performing quick sketches or working through calculations. PyTorch rivals alternative libraries, such as TensorFlow, for its flexibility and ease of use. Despite the high level appearance of PyTorch, it can be quite low-level, which is great for experimenting with novel algorithms. PyTorch can seamlessly be integrated with distributed computation libraries, like Ray, to make the Kessel Run in less than 12 parsecs (citation needed). \n\nThese guides will present the formal math for concepts alongside Python code examples since this often (for me at least) is a lot easier to develop an intuition for. Each guide is also available as an iPython notebook for your own experimentation.\n\nThe guides are not meant to exhaustively cover the field of machine learning but I hope they will instill you with the confidence and knowledge to explore further on your own.\n\nIf you do want more details, you might enjoy my [artificial intelligence notes](http://frnsys.com/ai_notes).\n\n\n### Modeling the world\n\nYou've probably seen various machine learning algorithms pop up -- linear regression, SVMs, neural networks, random forests, etc. How are they all related? What do they have in common? What is machine learning for anyways?\n\nFirst, let's consider the general, fundamental problem all machine learning is concerned with, leaving aside the algorithm name soup for now. The primary concern of machine learning is _modeling the world_.\n\nWe can model phenomena or systems -- both natural and artificial, if you want to make that distinction -- with mathematical functions. We see something out in the world and want to describe it in some way, we want to formalize how two or more things are related, and we can do that with a function. The problem is, for a given phenomenon, how do we figure out what function to use? There are infinitely many to choose from!\n\nBefore this gets too abstract, let's use an example to make things more concrete.\n\nSay we have a bunch of data about the heights and weights of a species of deer. We want to understand how these two variables are related -- in particular, given the weight of a deer, can we predict its height?\n\nYou might see where this is going. The data looks like a line, and lines in general are described by functions of the form $y = mx + b$.\n\nRemember that lines vary depending on what the values of $m$ and $b$ are:\n\n\n\nThus $m$ and $b$ uniquely define a function -- thus they are called the _parameters_ of the function -- and when it comes to machine learning, these parameters are what we ultimately want to learn. So when I say there are infinitely many functions to choose from, it is because $m$ and $b$ can pretty much take on any value. Machine learning techniques essentially search through these possible functions to find parameters that best fit the data you have. One way machine learning algorithms are differentiated is by how exactly they conduct this search (i.e. how they learn parameters).\n\nIn this case we've (reasonably) assumed the function takes the form $y = mx + b$, but conceivably you may have data that doesn't take the form of a line. Real world data is typically a lot more convoluted-looking. Maybe the true function has a $sin$ in it, for example.\n\nThis is where another main distinction between machine learning algorithms comes in -- certain algorithms can model only certain forms of functions. _Linear regression_, for example, can only model linear functions, as indicated by its name. Neural networks, on the other hand, are _universal function approximators_, which mean they can (in theory) approximate _any_ function, no matter how exotic. This doesn't necessarily make them a better method, just better suited for certain circumstances (there are many other considerations when choosing an algorithm).\n\nFor now, let's return to the line function. Now that we've looked at the $m$ and $b$ variables, let's consider the input variable $x$. A function takes a numerical input; that is $x$ must be a number of some kind. That's pretty straightforward here since the deer weights are already numbers. But this is not always the case! What if we want to predict the sales price of a house. A house is not a number. We have to find a way to _represent_ it as a number (or as several numbers, i.e. a vector, which will be detailed in a moment), e.g. by its square footage. This challenge of representation is a major part of machine learning; the practice of building representations is known as _feature engineering_ since each variable (e.g. square footage or zip code) used for the representation is called a _feature_.\n\nIf you think about it, representation is a practice we regularly engage in. The word \"house\" is not a house any more than an image of a house is -- there is no true \"house\" anyways, it is always a constellation of various physical and nonphysical components.\n\nThat's about it -- broadly speaking, machine learning is basically a bunch of algorithms that learn you a function, which is to say they learn the parameters that uniquely define a function.\n\n### Vectors\n\nIn the line example before I mentioned that we might have multiple numbers representing an input. For example, a house probably can't be solely represented by its square footage -- perhaps we also want to consider how many bedrooms it has, or how high the ceilings are, or its distance from local transportation. How do we group these numbers together?\n\nThat's what _vectors_ are for (they come up for many other reasons too, but we'll focus on representation for now). Vectors, along with matrices and other tensors (which will be explained a bit further down), could be considered the \"primitives\" of machine learning.\n\nThe Numpy library is best for dealing with vectors (and other tensors) in Python. A more complete introduction to Numpy is provided in the [numpy and basic mathematics guide](https://github.com/ml4a/ml4a-guides/blob/master/notebooks/math_review_numpy.ipynb).\n\nLet's import `numpy` with the alias `nf`:",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
]
],
[
[
"You may have encountered vectors before in high school or college -- to use Python terms, a vector is like a list of numbers. The mathematical notation is quite similar to Python code, e.g. `[5,4]`, but `numpy` has its own way of instantiating a vector:",
"_____no_output_____"
]
],
[
[
"v = np.array([5, 4])",
"_____no_output_____"
]
],
[
[
"$$\nv = \\begin{bmatrix} 5 \\\\ 4 \\end{bmatrix}\n$$\n\nVectors are usually represented with lowercase variables.\n\nNote that we never specified how _many_ numbers (also called _components_) a vector has - because it can have any amount. The amount of components a vector has is called its _dimensionality_. The example vector above has two dimensions. The vector `x = [8,1,3]` has three dimensions, and so on. Components are usually indicated by their index (usually using 1-indexing), e.g. in the previous vector, $x_1$ refers to the value $8$.\n\n\"Dimensions\" in the context of vectors is just like the spatial dimensions you spend every day in. These dimensions define a __space__, so a two-dimensional vector, e.g. `[5,4]`, can describe a point in 2D space and a three-dimensional vector, e.g. `[8,1,3]`, can describe a point in 3D space. As mentioned before, there is no limit to the amount of dimensions a vector may have (technically, there must be one or more dimensions), so we could conceivably have space consisting of thousands or tens of thousands of dimensions. At that point we can't rely on the same human intuitions about space as we could when working with just two or three dimensions. In practice, most interesting applications of machine learning deal with many, many dimensions.\n\nWe can get a better sense of this by plotting a vector out. For instance, a 2D vector `[5,0]` would look like:\n\n\n\nSo in a sense vectors can be thought of lines that \"point\" to the position they specify - here the vector is a line \"pointing\" to `[5,0]`. If the vector were 3D, e.g. `[8,1,3]`, then we would have to visualize it in 3D space, and so on.\n\nSo vectors are great - they allow us to form logical groupings of numbers. For instance, if we're talking about cities on a map we would want to group their latitude and longitude together. We'd represent Lagos with `[6.455027, 3.384082]` and Beijing separately with `[39.9042, 116.4074]`. If we have an inventory of books for sale, we could represent each book with its own vector consisting of its price, number of pages, and remaining stock.\n\nTo use vectors in functions, there are a few mathematical operations you need to know.\n\n### Basic vector operations\n\nVectors can be added (and subtracted) easily:",
"_____no_output_____"
]
],
[
[
"np.array([6, 2]) + np.array([-4, 4])",
"_____no_output_____"
]
],
[
[
"$$\n\\begin{bmatrix} 6 \\\\ 2 \\end{bmatrix} + \\begin{bmatrix} -4 \\\\ 4 \\end{bmatrix} = \\begin{bmatrix} 6 + -4 \\\\ 2 + 4 \\end{bmatrix} = \\begin{bmatrix} 2 \\\\ 6 \\end{bmatrix}\n$$\n\nHowever, when it comes to vector multiplication there are many different kinds.\n\nThe simplest is _vector-scalar_ multiplication:",
"_____no_output_____"
]
],
[
[
"3 * np.array([2, 1])",
"_____no_output_____"
]
],
[
[
"$$\n3\\begin{bmatrix} 2 \\\\ 1 \\end{bmatrix} = \\begin{bmatrix} 3 \\times 2 \\\\ 3 \\times 1\n\\end{bmatrix} = \\begin{bmatrix} 6 \\\\ 3 \\end{bmatrix}\n$$\n\nBut when you multiply two vectors together you have a few options. I'll cover the two most important ones here.\n\nThe one you might have thought of is the _element-wise product_, also called the _pointwise product_, _component-wise product_, or the _Hadamard product_, typically notated with $\\odot$. This just involves multiplying the corresponding elements of each vector together, resulting in another vector:",
"_____no_output_____"
]
],
[
[
"np.array([6, 2]) * np.array([-4, 4])",
"_____no_output_____"
]
],
[
[
"$$\n\\begin{bmatrix} 6 \\\\ 2 \\end{bmatrix} \\odot \\begin{bmatrix} -4 \\\\ 4 \\end{bmatrix} = \\begin{bmatrix} 6 \\times -4 \\\\ 2 \\times 4 \\end{bmatrix} = \\begin{bmatrix} -24 \\\\ 8 \\end{bmatrix}\n$$\n\nThe other vector product, which you'll encounter a lot, is the _dot product_, also called _inner product_, usually notated with $\\cdot$ (though when vectors are placed side-by-side this often implies dot multiplication). This involves multiplying corresponding elements of each vector and then summing the resulting vector's components (so this results in a scalar rather than another vector).",
"_____no_output_____"
]
],
[
[
"np.dot(np.array([6, 2]), np.array([-4, 4]))",
"_____no_output_____"
]
],
[
[
"$$\n\\begin{bmatrix} 6 \\\\ 2 \\end{bmatrix} \\cdot \\begin{bmatrix} -4 \\\\ 4 \\end{bmatrix} = (6 \\times -4) + (2 \\times 4) = -16\n$$\n\nThe more general formulation is:",
"_____no_output_____"
]
],
[
[
"# a slow pure-Python dot product\ndef dot(a, b):\n assert len(a) == len(b)\n return sum(a_i * b_i for a_i, b_i in zip(a,b))",
"_____no_output_____"
]
],
[
[
"$$\n\\begin{aligned}\n\\vec{a} \\cdot \\vec{b} &= \\begin{bmatrix} a_1 \\\\ a_2 \\\\ \\vdots \\\\ a_n \\end{bmatrix} \\cdot \\begin{bmatrix} b_1 \\\\ b_2 \\\\ \\vdots \\\\ b_n \\end{bmatrix} = a_1b_1 + a_2b_2 + \\dots + a_nb_n \\\\\n&= \\sum^n_{i=1} a_i b_i\n\\end{aligned}\n$$\n\nNote that the vectors in these operations must have the same dimensions!\n\nPerhaps the most important vector operation mentioned here is the dot product. We'll return to the house example to see why. Let's say want to represent a house with three variables: square footage, number of bedrooms, and the number of bathrooms. For convenience we'll notate the variables $x_1, x_2, x_3$, respectively. We're working in three dimensions now so instead of learning a line we're learning a _hyperplane_ (if we were working with two dimensions we'd be learning a plane, \"hyperplane\" is the term for the equivalent of a plane in higher dimensions).\n\nAside from the different name, the function we're learning is essentially of the same form as before, just with more variables and thus more parameters. We'll notate each parameter as $\\theta_i$ as is the convention (you may see $\\beta_i$ used elsewhere), and for the intercept (what was the $b$ term in the original line), we'll add in a dummy variable $x_0 = 1$ as is the typical practice (thus $\\theta_0$ is equivalent to $b$):",
"_____no_output_____"
]
],
[
[
"# this is so clumsy in python;\n# this will become more concise in a bit\ndef f(x0, x1, x2, x3, theta0, theta1, theta2, theta3):\n return theta0 * x0\\\n + theta1 * x1\\\n + theta2 * x2\\\n + theta3 * x3",
"_____no_output_____"
]
],
[
[
"$$\ny = \\theta_0 x_0 + \\theta_1 x_1 + \\theta_2 x_2 + \\theta_3 x_3\n$$\n\nThis kind of looks like the dot product, doesn't it? In fact, we can re-write this entire function as a dot product. We define our feature vector $x = [x_0, x_1, x_2, x_3]$ and our parameter vector $\\theta = [\\theta_0, \\theta_1, \\theta_2, \\theta_3]$, then re-write the function:",
"_____no_output_____"
]
],
[
[
"def f(x, theta):\n return x.dot(theta)",
"_____no_output_____"
]
],
[
[
"$$\ny = \\theta x\n$$\n\nSo that's how we incorporate multiple features in a representation.\n\nThere's a whole lot more to vectors than what's presented here, but this is the ground-level knowledge you should have of them. Other aspects of vectors will be explained as they come up.\n\n## Learning\n\nSo machine learning algorithms learn parameters - how do they do it?\n\nHere we're focusing on the most common kind of machine learning - _supervised_ learning. In supervised learning, the algorithm learns parameters from data which includes both the inputs and the true outputs. This data is called _training_ data.\n\nAlthough they vary on specifics, there is a general approach that supervised machine learning algorithms use to learn parameters. The idea is that the algorithm takes an input example, inputs it into the current guess at the function (called the _hypothesis_, notate $h_{\\theta}$), and then checks how wrong its output is against the true output. The algorithm then updates its hypothesis (that is, its guesses for the parameters), accordingly.\n\n\"How wrong\" an algorithm is, can vary depending on the _loss function_ it is using. The loss function takes the algorithm's current guess for the output, $\\hat y$, and the true output, $y$, and returns some value quantifying its wrongness. Certain loss functions are more appropriate for certain tasks, which we'll get into later.\n\nWe'll get into the specifies of how the algorithm determines what kind of update to perform (i.e. how much each parameter changes), but before we do that we should consider how we manage batches of training examples (i.e. multiple training vectors) simultaneously.\n\n\n## Matrices\n\n__Matrices__ are in a sense a \"vector\" of vectors. That is, where a vector can be thought of as a logical grouping of numbers, a matrix can be thought of as a logical grouping of vectors. So if a vector represents a book in our catalog (id, price, number in stock), a matrix could represent the entire catalog (each row refers to a book). Or if we want to represent a grayscale image, the matrix can represent the brightness values of the pixels in the image.",
"_____no_output_____"
]
],
[
[
"A = np.array([\n [6, 8, 0],\n [8, 2, 7],\n [3, 3, 9],\n [3, 8, 6]\n])",
"_____no_output_____"
]
],
[
[
"$$\n\\mathbf A =\n\\begin{bmatrix}\n6 & 8 & 0 \\\\\n8 & 2 & 7 \\\\\n3 & 3 & 9 \\\\\n3 & 8 & 6\n\\end{bmatrix}\n$$\n\nMatrices are usually represented with uppercase variables.\n\nNote that the \"vectors\" in the matrix must have the same dimension. The matrix's dimensions are expressed in the form $m \\times n$, meaning that there are $m$ rows and $n$ columns. So the example matrix has dimensions of $4 \\times 3$. Numpy calls these dimensions a matrix's \"shape\".\n\nWe can access a particular element, $A_{i,j}$, in a matrix by its indices. Say we want to refer to the element in the 2nd row and the 3rd column (remember that python uses 0-indexing):",
"_____no_output_____"
]
],
[
[
"A[1,2]",
"_____no_output_____"
]
],
[
[
"### Basic matrix operations\n\nLike vectors, matrix addition and subtraction is straightforward (again, they must be of the same dimensions):",
"_____no_output_____"
]
],
[
[
"B = np.array([\n [8, 3, 7],\n [2, 9, 6],\n [2, 5, 6],\n [5, 0, 6]\n])\n\nA + B",
"_____no_output_____"
]
],
[
[
"$$\n\\begin{aligned}\n\\mathbf B &=\n\\begin{bmatrix}\n8 & 3 & 7 \\\\\n2 & 9 & 6 \\\\\n2 & 5 & 6 \\\\\n5 & 0 & 6\n\\end{bmatrix} \\\\\nA + B &=\n\\begin{bmatrix}\n8+6 & 3+8 & 7+0 \\\\\n2+8 & 9+2 & 6+7 \\\\\n2+3 & 5+3 & 6+9 \\\\\n5+3 & 0+8 & 6+6\n\\end{bmatrix} \\\\\n&=\n\\begin{bmatrix}\n14 & 11 & 7 \\\\\n10 & 11 & 13 \\\\\n5 & 8 & 15 \\\\\n8 & 8 & 12\n\\end{bmatrix} \\\\\n\\end{aligned}\n$$\n\nMatrices also have a few different multiplication operations, like vectors.\n\n_Matrix-scalar multiplication_ is similar to vector-scalar multiplication - you just distribute the scalar, multiplying it with each element in the matrix.\n\n_Matrix-vector products_ require that the vector has the same dimension as the matrix has columns, i.e. for an $m \\times n$ matrix, the vector must be $n$-dimensional. The operation basically involves taking the dot product of each matrix row with the vector:",
"_____no_output_____"
]
],
[
[
"# a slow pure-Python matrix-vector product,\n# using our previous dot product implementation\ndef matrix_vector_product(M, v):\n return [np.dot(row, v) for row in M]\n\n# or, with numpy, you could use np.matmul(A,v)",
"_____no_output_____"
]
],
[
[
"$$\n\\mathbf M v =\n\\begin{bmatrix}\nM_{1} \\cdot v \\\\\n\\vdots \\\\\nM_{m} \\cdot v \\\\\n\\end{bmatrix}\n$$\n\nWe have a few options when it comes to multiplying matrices with matrices.\n\nHowever, before we go any further we should talk about the _tranpose_ operation - this just involves switching the columns and rows of a matrix. The transpose of a matrix $A$ is notated $A^T$:",
"_____no_output_____"
]
],
[
[
"A = np.array([\n [1,2,3],\n [4,5,6]\n ])\n\nnp.transpose(A)",
"_____no_output_____"
]
],
[
[
"$$\n\\begin{aligned}\n\\mathbf A &=\n\\begin{bmatrix}\n1 & 2 & 3 \\\\\n4 & 5 & 6\n\\end{bmatrix} \\\\\n\\mathbf A^T &=\n\\begin{bmatrix}\n1 & 4 \\\\\n2 & 5 \\\\\n3 & 6\n\\end{bmatrix}\n\\end{aligned}\n$$\n\n\nFor matrix-matrix products, the matrix on the lefthand must have the same number of columns as the righthand's rows. To be more concrete, we'll represent a matrix-matrix product as $A B$ and we'll say that $A$ has $m \\times n$ dimensions. For this operation to work, $B$ must have $n \\times p$ dimensions. The resulting product will have $m \\times p$ dimensions.",
"_____no_output_____"
]
],
[
[
"# a slow pure-Python matrix Hadamard product\ndef matrix_matrix_product(A, B):\n _, a_cols = np.shape(A)\n b_rows, _ = np.shape(B)\n assert a_cols == b_rows\n\n result = []\n # tranpose B so we can iterate over its columns\n for col in np.tranpose(B):\n # using our previous implementation\n result.append(\n matrix_vector_product(A, col))\n return np.transpose(result)",
"_____no_output_____"
]
],
[
[
"$$\n\\mathbf AB =\n\\begin{bmatrix}\nA B^T_1 \\\\\n\\vdots \\\\\nA B^T_p\n\\end{bmatrix}^T\n$$\n\nFinally, like with vectors, we also have Hadamard (element-wise) products:",
"_____no_output_____"
]
],
[
[
"# a slow pure-Python matrix-matrix product\n# or, with numpy, you can use A * B\ndef matrix_matrix_hadamard(A, B):\n result = []\n for a_row, b_row in zip(A, B):\n result.append(\n zip(a_i * b_i for a_i, b_i in zip(a_row, b_row)))",
"_____no_output_____"
]
],
[
[
"$$\n\\mathbf A \\odot B =\n\\begin{bmatrix}\nA_{1,1} B_{1,1} & \\dots & A_{1,n} B_{1,n} \\\\\n\\vdots & \\dots & \\vdots \\\\\nA_{m,1} B_{m,1} & \\dots & A_{m,n} B_{m,n}\n\\end{bmatrix}\n$$\n\nLike vector Hadamard products, this requires that the two matrices share the same dimensions.\n\n## Tensors\n\nWe've seen vectors, which is like a list of numbers, and matrices, which is like a list of a list of numbers. We can generalize this concept even further, for instance, with a list of a list of a list of numbers and so on. What all of these structures are called are _tensors_ (i.e. the \"tensor\" in \"TensorFlow\"). They are distinguished by their _rank_, which, if you're thinking in the \"list of lists\" way, refers to the number of nestings. So a vector has a rank of one (just a list of numbers) and a matrix has a rank of two (a list of a list of numbers).\n\nAnother way to think of rank is by number of indices necessary to access an element in the tensor. An element in a vector is accessed by one index, e.g. `v[i]`, so it is of rank one. An element in a matrix is accessed by two indices, e.g. `M[i,j]`, so it is of rank two.\n\nWhy is the concept of a tensor useful? Before we referred to vectors as a logical grouping of numbers and matrices as a logical grouping of vectors. What if we need a logical grouping of matrices? That's what 3rd-rank tensors are! A matrix can represent a grayscale image, but what about a color image with three color channels (red, green, blue)? With a 3rd-rank tensor, we could represent each channel as its own matrix and group them together.\n\n## Learning continued\n\nWhen the current hypothesis is wrong, how does the algorithm know how to adjust the parameters?\n\nLet's take a step back and look at it another way. The loss function measures the wrongness of the hypothesis $h_{\\theta}$ - another way of saying this is the loss function is a function of the parameters $\\theta$. So we could notate it as $L(\\theta)$.\n\nThe minimum of $L(\\theta)$ is the point where the parameters guess $\\theta$ is least wrong (at best, $L(\\theta) = 0$, i.e. a perfect score, though this is not always good, as will be explained later); i.e. the best guess for the parameters.\n\nSo the algorithm learns the best-fitting function by minimizing its loss function. That is, we can frame this as an optimization problem.\n\nThere are many techniques to solve an optimization problem - sometimes they can be solved analytically (i.e. by moving around variables and isolating the one you want to solve for), but more often than not we must solve them numerically, i.e. by guessing a lot of different values - but not randomly!\n\nThe prevailing technique now is called _gradient descent_, and to understand how it works, we have to understand derivatives.\n\n## Derivatives\n\nDerivatives are everywhere in machine learning, so it's worthwhile become a bit familiar with them. I won't go into specifics on differentiation (how to calculate derivatives) because now we're spoiled with automatic differentiation, but it's still good to have a solid intuition about derivatives themselves.\n\nA derivative expresses a rate of (instantaneous) change - they are always about how one variable quantity changes with respect to another variable quantity. That's basically all there is to it. For instance, velocity is a derivative which expresses how position changes with respect to time. Another interpretation, which is more relevant to machine learning, is that a derivative tells us how to change one variable to achieve a desired change in the other variable. Velocity, for instance, tells us how to change position by \"changing\" time.\n\nTo get a better understanding of _instantaneous_ change, consider a cyclist, cycling on a line. We have data about their position over time. We could calculate an average velocity over the data's entire time period, but we typically prefer to know the velocity at any given _moment_ (i.e. at any _instant_).\n\nLet's get more concrete first. Let's say we have data for $n$ seconds, i.e. from $t_0$ to $t_n$ seconds, and the position at any given second $i$ is $p_i$. If we wanted to get the rate of change in position over the entire time interval, we'd just do:",
"_____no_output_____"
]
],
[
[
"positions = [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, # moving forward\n 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, # pausing\n 9, 8, 7, 6, 5, 4, 3, 2, 1, 0] # moving backwards\nt_0 = 0\nt_n = 29\n(positions[t_n] - positions[t_0])/t_n",
"_____no_output_____"
]
],
[
[
"$$\nv = \\frac{p_n - p_0}{n}\n$$\n\nThis kind of makes it look like the cyclist didn't move at all. It would probably be more useful to identify the velocity at a given second $t$. Thus we want to come up with some function $v(t)$ which gives us the velocity at some second $t$. We can apply the same approach we just used to get the velocity over the entire time interval, but we focus on a shorter time interval instead. To get the _instantaneous_ change at $t$ we just keep reducing the interval we look at until it is basically 0.\n\nDerivatives have a special notation. A derivative of a function $f(x)$ with respect to a variable $x$ is notated:\n\n$$\n\\frac{\\delta f(x)}{\\delta x}\n$$\n\nSo if position is a function of time, e.g. $p = f(t)$, then velocity can be represented as $\\frac{\\delta p}{\\delta t}$. To drive the point home, this derivative is also a function of time (derivatives are functions of what their \"with respect to\" variable is).\n\nSince we are often computing derivatives of a function with respect to its input, a shorthand for the derivative of a function $f(x)$ with respect to $x$ can also be notated $f'(x)$.\n\n### The Chain Rule\n\nA very important property of derivatives is the _chain rule_ (there are other \"chain rules\" throughout mathematics, if we want to be specific, this is the \"chain rule of derivatives\"). The chain rule is important because it allows us to take complicated nested functions and more manageably differentiate them.\n\nLet's look at an example to make this concrete:",
"_____no_output_____"
]
],
[
[
"def g(x):\n return x**2\n\ndef h(x):\n return x**3\n\ndef f(x):\n return g(h(x))\n\n# derivatives\ndef g_(x):\n return 2*x\n\ndef h_(x):\n return 3*(x**2)",
"_____no_output_____"
]
],
[
[
"$$\n\\begin{aligned}\ng(x) &= x^2 \\\\\nh(x) &= x^3 \\\\\nf(x) &= g(h(x)) \\\\\ng'(x) &= 2x \\\\\nh'(x) &= 3x^2\n\\end{aligned}\n$$\n\nWe're interested in understanding how $f(x)$ changes with respect to $x$, so we want to compute the derivative of $f(x)$. The chain rule allows us to individually differentiate the component functions of $f(x)$ and multiply those to get $f'(x)$:",
"_____no_output_____"
]
],
[
[
"def f_(x):\n return g_(x) * h_(x)",
"_____no_output_____"
]
],
[
[
"$$\n\\frac{df}{dx} = \\frac{dg}{dh} \\frac{dh}{dx}\n$$\n\nThis example is a bit contrived (there is a very easy way to differentiate this particular example that doesn't involve the chain rule) but if $g(x)$ and $h(x)$ were really nasty functions, the chain rule makes them quite a lot easier to deal with.\n\nThe chain rule can be applied to nested functions ad nauseaum! You can apply it to something crazy like $f(g(h(u(q(p(x))))))$. In fact, with deep neural networks, you are typically dealing with function compositions even more gnarly than this, so the chain rule is cornerstone there.\n\n\n### Partial derivatives and gradients\n\nThe functions we've looked at so far just have a single input, but you can imagine many scenarios where you'd want to work with functions with some arbitrary number of inputs (i.e. a _multivariable_ function), like $f(x,y,z)$.\n\nHere's where _partial deriatives_ come into play. Partial derivatives are just like regular derivatives except we use them for multivariable functions; it just means we only differentiate with respect to one variable at a time. So for $f(x,y,z)$, we'd have a partial derivative with respect to $x$, i.e. $\\frac{\\partial f}{\\partial x}$ (note the slightly different notation), one with respect to $y$, i.e. $\\frac{\\partial f}{\\partial y}$, and one with respect to $z$, i.e. $\\frac{\\partial f}{\\partial z}$.\n\nThat's pretty simple! But it would be useful to group these partial derivatives together in some way. If we put these partial derivatives together in a vector, the resulting vector is the _gradient_ of $f$, notated $\\nabla f$ (the symbol is called \"nabla\").\n\n\n### Higher-order derivatives\n\nWe saw that velocity is the derivative of position because it describes how position changes over time. Acceleration similarly describes how _velocity_ changes over time, so we'd say that acceleration is the derivative of velocity. We can also say that acceleration is the _second-order_ derivative of position (that is, it is the derivative of its derivative).\n\nThis is the general idea behind higher-order derivatives.\n\n## Gradient descent\n\nOnce you understand derivatives, gradient descent is really, really simple. The basic idea is that we use the derivative of the loss $L(\\theta)$ with respect to $\\theta$ and figure out which way the loss is decreasing, then \"move\" the parameter guess in that direction.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0937f4cdf22afde272ae2957f3741e2b5396c52 | 6,917 | ipynb | Jupyter Notebook | course/0_Check_Environment.ipynb | tetsu/zero-deep-learning | 5626f668679a72e129af887630ea629a503a70e9 | [
"MIT"
] | null | null | null | course/0_Check_Environment.ipynb | tetsu/zero-deep-learning | 5626f668679a72e129af887630ea629a503a70e9 | [
"MIT"
] | null | null | null | course/0_Check_Environment.ipynb | tetsu/zero-deep-learning | 5626f668679a72e129af887630ea629a503a70e9 | [
"MIT"
] | null | null | null | 26.706564 | 149 | 0.521324 | [
[
[
"# Check Environment\nThis notebook checks that you have correctly created the environment and that all packages needed are installed.",
"_____no_output_____"
],
[
"## Environment\n\nThe next command should return a line like (Mac/Linux):\n\n /<YOUR-HOME-FOLDER>/anaconda/envs/ztdl/bin/python\n\nor like (Windows 10):\n\n C:\\\\<YOUR-HOME-FOLDER>\\\\Anaconda3\\\\envs\\\\ztdl\\\\python.exe\n\nIn particular you should make sure that you are using the python executable from within the course environment.\n\nIf that's not the case do this:\n\n1. close this notebook\n2. go to the terminal and stop jupyer notebook\n3. make sure that you have activated the environment, you should see a prompt like:\n\n (ztdl) $\n4. (optional) if you don't see that prompt activate the environment:\n - mac/linux:\n \n conda activate ztdl\n\n - windows:\n\n activate ztdl\n5. restart jupyter notebook",
"_____no_output_____"
]
],
[
[
"import os\nimport sys\nsys.executable",
"_____no_output_____"
]
],
[
[
"## Python 3.6\n\nThe next line should say that you're using Python 3.6.x from Continuum Analytics. At the time of publication it looks like this (Mac/Linux):\n\n Python 3.6.7 |Anaconda, Inc.| (default, Oct 23 2018, 14:01:38)\n [GCC 4.2.1 Compatible Clang 4.0.1 (tags/RELEASE_401/final)] on darwin\n Type \"help\", \"copyright\", \"credits\" or \"license\" for more information.\n\nor like this (Windows 10):\n\n Python 3.6.7 |Anaconda, Inc.| (default, Oct 28 2018, 19:44:12) [MSC v.1915 64 bit (AMD64)] on win32\n Type \"help\", \"copyright\", \"credits\" or \"license\" for more information.\n\nbut date and exact version of GCC may change in the future.\n\nIf you see a different version of python, go back to the previous step and make sure you created and activated the environment correctly.",
"_____no_output_____"
]
],
[
[
"import sys\nsys.version",
"_____no_output_____"
]
],
[
[
"## Jupyter\n\nCheck that Jupyter is running from within the environment. The next line should look like (Mac/Linux):\n\n /<YOUR-HOME-FOLDER>/anaconda/envs/ztdl/lib/python3.6/site-packages/jupyter.py'\n\nor like this (Windows 10):\n\n C:\\\\Users\\\\paperspace\\\\Anaconda3\\\\envs\\\\ztdl\\\\lib\\\\site-packages\\\\jupyter.py",
"_____no_output_____"
]
],
[
[
"import jupyter\njupyter.__file__",
"_____no_output_____"
]
],
[
[
"## Other packages\n\nHere we will check that all the packages are installed and have the correct versions. If everything is ok you should see:\n \n Using TensorFlow backend.\n \n Houston we are go!\n\nIf there's any issue here please make sure you have checked the previous steps and if it's all good please send us a question in the Q&A forum.",
"_____no_output_____"
]
],
[
[
"import pip\nimport numpy\nimport jupyter\nimport matplotlib\nimport sklearn\nimport scipy\nimport pandas\nimport PIL\nimport seaborn\nimport h5py\nimport tensorflow\nimport keras\n\n\ndef check_version(pkg, version):\n actual = pkg.__version__.split('.')\n if len(actual) == 3:\n actual_major = '.'.join(actual[:2])\n elif len(actual) == 2:\n actual_major = '.'.join(actual)\n else:\n raise NotImplementedError(pkg.__name__ +\n \"actual version :\"+\n pkg.__version__)\n try:\n assert(actual_major == version)\n except Exception as ex:\n print(\"{} {}\\t=> {}\".format(pkg.__name__,\n version,\n pkg.__version__))\n raise ex\n\ncheck_version(pip, '10.0')\ncheck_version(numpy, '1.15')\ncheck_version(matplotlib, '3.0')\ncheck_version(sklearn, '0.20')\ncheck_version(scipy, '1.1')\ncheck_version(pandas, '0.23')\ncheck_version(PIL, '5.3')\ncheck_version(seaborn, '0.9')\ncheck_version(h5py, '2.8')\ncheck_version(tensorflow, '1.11')\ncheck_version(keras, '2.2')\n\nprint(\"Houston we are go!\")",
"Houston we are go!\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d093893a523a3416db84382f03afadcf4abf5a93 | 164,723 | ipynb | Jupyter Notebook | Alphabet_Soup_Charity_Final.ipynb | serastva/Deep_learning_challenge | b7ca9b3d561f78da3d298fd649929ef6c1debf60 | [
"ADSL"
] | null | null | null | Alphabet_Soup_Charity_Final.ipynb | serastva/Deep_learning_challenge | b7ca9b3d561f78da3d298fd649929ef6c1debf60 | [
"ADSL"
] | null | null | null | Alphabet_Soup_Charity_Final.ipynb | serastva/Deep_learning_challenge | b7ca9b3d561f78da3d298fd649929ef6c1debf60 | [
"ADSL"
] | null | null | null | 49.931191 | 509 | 0.351038 | [
[
[
"## Preprocessing",
"_____no_output_____"
]
],
[
[
"# Import our dependencies\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.preprocessing import StandardScaler\nimport pandas as pd\nimport tensorflow as tf\n\n# Import and read the charity_data.csv.\nimport pandas as pd \napplication_df = pd.read_csv(\"charity_data.csv\")\napplication_df.head()",
"_____no_output_____"
],
[
"# Drop the non-beneficial ID columns, 'EIN' and 'NAME'.\napplication_df = application_df.drop(['EIN', 'NAME'], axis=1)",
"_____no_output_____"
],
[
"# Determine the number of unique values in each column.\napplication_df.nunique()",
"_____no_output_____"
],
[
"# Look at APPLICATION_TYPE value counts for binning\nval_count = application_df['APPLICATION_TYPE'].value_counts()\nval_count",
"_____no_output_____"
],
[
"# Choose a cutoff value and create a list of application types to be replaced\n# use the variable name `application_types_to_replace`\napplication_types_to_replace = list(val_count[val_count<200].index)\n\n# Replace in dataframe\nfor app in application_types_to_replace:\n application_df['APPLICATION_TYPE'] = application_df['APPLICATION_TYPE'].replace(app,\"Other\")\n\n# Check to make sure binning was successful\napplication_df['APPLICATION_TYPE'].value_counts()",
"_____no_output_____"
],
[
"# Look at CLASSIFICATION value counts for binning\nclass_val_count = application_df['CLASSIFICATION'].value_counts()\nclass_val_count",
"_____no_output_____"
],
[
"# You may find it helpful to look at CLASSIFICATION value counts >1\nclass_val_count2 = class_val_count[class_val_count>1]\nclass_val_count2",
"_____no_output_____"
],
[
"# Choose a cutoff value and create a list of classifications to be replaced\n# use the variable name `classifications_to_replace`\nclassifications_to_replace =class_val_count.loc[class_val_count<1000].index\n\n# Replace in dataframe\nfor cls in classifications_to_replace:\n application_df['CLASSIFICATION'] = application_df['CLASSIFICATION'].replace(cls,\"Other\")\n \n# Check to make sure binning was successful\napplication_df['CLASSIFICATION'].value_counts()",
"_____no_output_____"
],
[
"# Convert categorical data to numeric with `pd.get_dummies`\ndummies_df = pd.get_dummies(application_df)\ndummies_df",
"_____no_output_____"
],
[
"# Split our preprocessed data into our features and target arrays\nX = dummies_df.drop(['IS_SUCCESSFUL'],1).values\ny = dummies_df['IS_SUCCESSFUL'].values\n\n# Split the preprocessed data into a training and testing dataset\nX_train, X_test, y_train, y_test = train_test_split(X,y,random_state= 42)",
"_____no_output_____"
],
[
"print(X.shape)\nprint(y.shape)",
"(34299, 43)\n(34299,)\n"
],
[
"# Create a StandardScaler instances\nscaler = StandardScaler()\n\n# Fit the StandardScaler\nX_scaler = scaler.fit(X_train)\n\n# Scale the data\nX_train_scaled = X_scaler.transform(X_train)\nX_test_scaled = X_scaler.transform(X_test)",
"_____no_output_____"
]
],
[
[
"## Compile, Train and Evaluate the Model",
"_____no_output_____"
]
],
[
[
"# Define the model - deep neural net, i.e., the number of input features and hidden nodes for each layer.\ninput_features = len(X_train_scaled[0])\nhidden_nodes_layer1 = 80\nhidden_nodes_layer2 = 30\n#hidden_nodes_layer3 = 60\n\nnn = tf.keras.models.Sequential()\n\n# First hidden layer\nnn.add(tf.keras.layers.Dense(units=hidden_nodes_layer1, input_dim=input_features, activation='relu'))\n\n# Second hidden layer\nnn.add(tf.keras.layers.Dense(units=hidden_nodes_layer2, activation='relu'))\n\n# Third hidden layer\n#nn.add(tf.keras.layers.Dense(units=hidden_nodes_layer3, activation='relu'))\n\n\n# Output layer\nnn.add(tf.keras.layers.Dense(units=1, activation='sigmoid'))\n\n# Check the structure of the model\nnn.summary()",
"Model: \"sequential\"\n_________________________________________________________________\n Layer (type) Output Shape Param # \n=================================================================\n dense (Dense) (None, 80) 3520 \n \n dense_1 (Dense) (None, 30) 2430 \n \n dense_2 (Dense) (None, 1) 31 \n \n=================================================================\nTotal params: 5,981\nTrainable params: 5,981\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"# Compile the model\nnn.compile(loss=\"binary_crossentropy\", optimizer=\"adam\", metrics=[\"accuracy\"])",
"_____no_output_____"
],
[
"# Train the model\nfit_model = nn.fit(X_train_scaled, y_train, epochs=100)",
"Epoch 1/100\n804/804 [==============================] - 6s 4ms/step - loss: 0.5712 - accuracy: 0.7208\nEpoch 2/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5569 - accuracy: 0.7266\nEpoch 3/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5536 - accuracy: 0.7293\nEpoch 4/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5510 - accuracy: 0.7303\nEpoch 5/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5499 - accuracy: 0.7304\nEpoch 6/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5480 - accuracy: 0.7320\nEpoch 7/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5478 - accuracy: 0.7309\nEpoch 8/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5474 - accuracy: 0.7331\nEpoch 9/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5469 - accuracy: 0.7323\nEpoch 10/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5458 - accuracy: 0.7325\nEpoch 11/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5453 - accuracy: 0.7330\nEpoch 12/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5449 - accuracy: 0.7341\nEpoch 13/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5452 - accuracy: 0.7334\nEpoch 14/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5439 - accuracy: 0.7335\nEpoch 15/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5440 - accuracy: 0.7343\nEpoch 16/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5434 - accuracy: 0.7356\nEpoch 17/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5433 - accuracy: 0.7352\nEpoch 18/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5432 - accuracy: 0.7352\nEpoch 19/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5426 - accuracy: 0.7358\nEpoch 20/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5422 - accuracy: 0.7352\nEpoch 21/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5421 - accuracy: 0.7355\nEpoch 22/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5419 - accuracy: 0.7353\nEpoch 23/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5420 - accuracy: 0.7350\nEpoch 24/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5418 - accuracy: 0.7355\nEpoch 25/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5412 - accuracy: 0.7357\nEpoch 26/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5410 - accuracy: 0.7351\nEpoch 27/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5410 - accuracy: 0.7360\nEpoch 28/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5410 - accuracy: 0.7352\nEpoch 29/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5408 - accuracy: 0.7356\nEpoch 30/100\n804/804 [==============================] - 4s 5ms/step - loss: 0.5399 - accuracy: 0.7365\nEpoch 31/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5402 - accuracy: 0.7359\nEpoch 32/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5400 - accuracy: 0.7365\nEpoch 33/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5399 - accuracy: 0.7357\nEpoch 34/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5396 - accuracy: 0.7373\nEpoch 35/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5398 - accuracy: 0.7365\nEpoch 36/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5396 - accuracy: 0.7376\nEpoch 37/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5394 - accuracy: 0.7365\nEpoch 38/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5391 - accuracy: 0.7371\nEpoch 39/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5391 - accuracy: 0.7362\nEpoch 40/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5392 - accuracy: 0.7369\nEpoch 41/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5389 - accuracy: 0.7372\nEpoch 42/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5386 - accuracy: 0.7381\nEpoch 43/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5381 - accuracy: 0.7374\nEpoch 44/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5384 - accuracy: 0.7375\nEpoch 45/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5381 - accuracy: 0.7382\nEpoch 46/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5383 - accuracy: 0.7387\nEpoch 47/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5381 - accuracy: 0.7379\nEpoch 48/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5378 - accuracy: 0.7370\nEpoch 49/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5374 - accuracy: 0.7376\nEpoch 50/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5376 - accuracy: 0.7380\nEpoch 51/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5373 - accuracy: 0.7378\nEpoch 52/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5377 - accuracy: 0.7378\nEpoch 53/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5371 - accuracy: 0.7385\nEpoch 54/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5370 - accuracy: 0.7377\nEpoch 55/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5371 - accuracy: 0.7388\nEpoch 56/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5369 - accuracy: 0.7391\nEpoch 57/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5371 - accuracy: 0.7386\nEpoch 58/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5368 - accuracy: 0.7392\nEpoch 59/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5364 - accuracy: 0.7386\nEpoch 60/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5367 - accuracy: 0.7389\nEpoch 61/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5362 - accuracy: 0.7396\nEpoch 62/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5364 - accuracy: 0.7386\nEpoch 63/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5363 - accuracy: 0.7391\nEpoch 64/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5363 - accuracy: 0.7387\nEpoch 65/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5362 - accuracy: 0.7396\nEpoch 66/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5361 - accuracy: 0.7388\nEpoch 67/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5360 - accuracy: 0.7391\nEpoch 68/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5356 - accuracy: 0.7392\nEpoch 69/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5360 - accuracy: 0.7385\nEpoch 70/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5355 - accuracy: 0.7391\nEpoch 71/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5355 - accuracy: 0.7397\nEpoch 72/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5358 - accuracy: 0.7391\nEpoch 73/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5357 - accuracy: 0.7401\nEpoch 74/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5356 - accuracy: 0.7388\nEpoch 75/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5356 - accuracy: 0.7394\nEpoch 76/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5356 - accuracy: 0.7393\nEpoch 77/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5352 - accuracy: 0.7394\nEpoch 78/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5352 - accuracy: 0.7400\nEpoch 79/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5353 - accuracy: 0.7398\nEpoch 80/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5352 - accuracy: 0.7393\nEpoch 81/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5351 - accuracy: 0.7396\nEpoch 82/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5349 - accuracy: 0.7391\nEpoch 83/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5348 - accuracy: 0.7395\nEpoch 84/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5346 - accuracy: 0.7397\nEpoch 85/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5349 - accuracy: 0.7394\nEpoch 86/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5347 - accuracy: 0.7397\nEpoch 87/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5347 - accuracy: 0.7397\nEpoch 88/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5348 - accuracy: 0.7400\nEpoch 89/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5348 - accuracy: 0.7408\nEpoch 90/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5345 - accuracy: 0.7397\nEpoch 91/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5345 - accuracy: 0.7397\nEpoch 92/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5341 - accuracy: 0.7402\nEpoch 93/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5344 - accuracy: 0.7406\nEpoch 94/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5344 - accuracy: 0.7398\nEpoch 95/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5343 - accuracy: 0.7411\nEpoch 96/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5346 - accuracy: 0.7396\nEpoch 97/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5339 - accuracy: 0.7392\nEpoch 98/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5344 - accuracy: 0.7401\nEpoch 99/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5337 - accuracy: 0.7396\nEpoch 100/100\n804/804 [==============================] - 3s 4ms/step - loss: 0.5339 - accuracy: 0.7398\n"
],
[
"# Evaluate the model using the test data\nmodel_loss, model_accuracy = nn.evaluate(X_test_scaled,y_test,verbose=2)\nprint(f\"Loss: {model_loss}, Accuracy: {model_accuracy}\")",
"268/268 - 1s - loss: 0.5639 - accuracy: 0.7277 - 735ms/epoch - 3ms/step\nLoss: 0.5639024972915649, Accuracy: 0.7276967763900757\n"
],
[
"# Export our model to HDF5 file\nnn.save(\"AlphabetSoupCharity.h5\") ",
"_____no_output_____"
]
],
[
[
"**Optimization 1**\n",
"_____no_output_____"
]
],
[
[
"# Define the model - deep neural net, i.e., the number of input features and hidden nodes for each layer.\ninput_features = len(X_train_scaled[0])\nhidden_nodes_layer1 = 43\nhidden_nodes_layer2 = 25\n#hidden_nodes_layer3 = 15\n\nnn2 = tf.keras.models.Sequential()\n\n# First hidden layer\nnn2.add(tf.keras.layers.Dense(units=hidden_nodes_layer1, input_dim=input_features, activation='relu'))\n\n# Second hidden layer\nnn2.add(tf.keras.layers.Dense(units=hidden_nodes_layer2, activation='relu'))\n\n# Third hidden layer\n#nn.add(tf.keras.layers.Dense(units=hidden_nodes_layer3, activation='relu'))\n\n\n# Output layer\nnn2.add(tf.keras.layers.Dense(units=1, activation='sigmoid'))\n\n# Check the structure of the model\nnn2.summary()",
"Model: \"sequential_1\"\n_________________________________________________________________\n Layer (type) Output Shape Param # \n=================================================================\n dense_3 (Dense) (None, 43) 1892 \n \n dense_4 (Dense) (None, 25) 1100 \n \n dense_5 (Dense) (None, 1) 26 \n \n=================================================================\nTotal params: 3,018\nTrainable params: 3,018\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"# Compile the model\nnn2.compile(loss=\"binary_crossentropy\", optimizer=\"adam\", metrics=[\"accuracy\"])",
"_____no_output_____"
],
[
"# Train the model\nfit_model = nn2.fit(X_train_scaled, y_train, epochs=200)",
"Epoch 1/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5765 - accuracy: 0.7133\nEpoch 2/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5556 - accuracy: 0.7291\nEpoch 3/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5529 - accuracy: 0.7300\nEpoch 4/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5502 - accuracy: 0.7303\nEpoch 5/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5501 - accuracy: 0.7304\nEpoch 6/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5483 - accuracy: 0.7320\nEpoch 7/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5479 - accuracy: 0.7312\nEpoch 8/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5479 - accuracy: 0.7320\nEpoch 9/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5471 - accuracy: 0.7326\nEpoch 10/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5465 - accuracy: 0.7324\nEpoch 11/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5452 - accuracy: 0.7329\nEpoch 12/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5456 - accuracy: 0.7337\nEpoch 13/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5450 - accuracy: 0.7342\nEpoch 14/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5449 - accuracy: 0.7331\nEpoch 15/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5444 - accuracy: 0.7332\nEpoch 16/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5445 - accuracy: 0.7333\nEpoch 17/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5435 - accuracy: 0.7346\nEpoch 18/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5434 - accuracy: 0.7346\nEpoch 19/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5432 - accuracy: 0.7338\nEpoch 20/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5431 - accuracy: 0.7353\nEpoch 21/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5428 - accuracy: 0.7347\nEpoch 22/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5428 - accuracy: 0.7352\nEpoch 23/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5422 - accuracy: 0.7347\nEpoch 24/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5426 - accuracy: 0.7349\nEpoch 25/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5424 - accuracy: 0.7362\nEpoch 26/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5423 - accuracy: 0.7345\nEpoch 27/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5417 - accuracy: 0.7363\nEpoch 28/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5419 - accuracy: 0.7358\nEpoch 29/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5418 - accuracy: 0.7355\nEpoch 30/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5414 - accuracy: 0.7350\nEpoch 31/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5411 - accuracy: 0.7355\nEpoch 32/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5407 - accuracy: 0.7353\nEpoch 33/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5406 - accuracy: 0.7371\nEpoch 34/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5409 - accuracy: 0.7364\nEpoch 35/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5406 - accuracy: 0.7362\nEpoch 36/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5403 - accuracy: 0.7364\nEpoch 37/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5402 - accuracy: 0.7360\nEpoch 38/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5405 - accuracy: 0.7365\nEpoch 39/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5401 - accuracy: 0.7369\nEpoch 40/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5401 - accuracy: 0.7364\nEpoch 41/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5403 - accuracy: 0.7366\nEpoch 42/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5396 - accuracy: 0.7362\nEpoch 43/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5397 - accuracy: 0.7358\nEpoch 44/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5392 - accuracy: 0.7365\nEpoch 45/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5395 - accuracy: 0.7370\nEpoch 46/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5396 - accuracy: 0.7374\nEpoch 47/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5392 - accuracy: 0.7359\nEpoch 48/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5391 - accuracy: 0.7369\nEpoch 49/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5391 - accuracy: 0.7369\nEpoch 50/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5389 - accuracy: 0.7381\nEpoch 51/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5390 - accuracy: 0.7371\nEpoch 52/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5392 - accuracy: 0.7378\nEpoch 53/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5390 - accuracy: 0.7373\nEpoch 54/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5388 - accuracy: 0.7374\nEpoch 55/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5387 - accuracy: 0.7372\nEpoch 56/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5388 - accuracy: 0.7363\nEpoch 57/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5389 - accuracy: 0.7354\nEpoch 58/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5386 - accuracy: 0.7375\nEpoch 59/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5384 - accuracy: 0.7376\nEpoch 60/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5379 - accuracy: 0.7375\nEpoch 61/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5381 - accuracy: 0.7382\nEpoch 62/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5380 - accuracy: 0.7381\nEpoch 63/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5379 - accuracy: 0.7371\nEpoch 64/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5381 - accuracy: 0.7380\nEpoch 65/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5380 - accuracy: 0.7383\nEpoch 66/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5381 - accuracy: 0.7378\nEpoch 67/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5378 - accuracy: 0.7379\nEpoch 68/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5380 - accuracy: 0.7382\nEpoch 69/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5378 - accuracy: 0.7382\nEpoch 70/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5379 - accuracy: 0.7381\nEpoch 71/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5375 - accuracy: 0.7378\nEpoch 72/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5374 - accuracy: 0.7385\nEpoch 73/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5375 - accuracy: 0.7372\nEpoch 74/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5374 - accuracy: 0.7385\nEpoch 75/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5373 - accuracy: 0.7374\nEpoch 76/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5370 - accuracy: 0.7387\nEpoch 77/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5372 - accuracy: 0.7391\nEpoch 78/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5372 - accuracy: 0.7388\nEpoch 79/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5372 - accuracy: 0.7390\nEpoch 80/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5369 - accuracy: 0.7386\nEpoch 81/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5371 - accuracy: 0.7389\nEpoch 82/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5369 - accuracy: 0.7382\nEpoch 83/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5368 - accuracy: 0.7391\nEpoch 84/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5371 - accuracy: 0.7379\nEpoch 85/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5366 - accuracy: 0.7374\nEpoch 86/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5370 - accuracy: 0.7371\nEpoch 87/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5364 - accuracy: 0.7390\nEpoch 88/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5368 - accuracy: 0.7386\nEpoch 89/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5366 - accuracy: 0.7390\nEpoch 90/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5363 - accuracy: 0.7394\nEpoch 91/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5361 - accuracy: 0.7385\nEpoch 92/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5366 - accuracy: 0.7393\nEpoch 93/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5366 - accuracy: 0.7389\nEpoch 94/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5362 - accuracy: 0.7390\nEpoch 95/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5362 - accuracy: 0.7391\nEpoch 96/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5362 - accuracy: 0.7386\nEpoch 97/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5362 - accuracy: 0.7386\nEpoch 98/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5359 - accuracy: 0.7388\nEpoch 99/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5365 - accuracy: 0.7385\nEpoch 100/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5364 - accuracy: 0.7388\nEpoch 101/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5363 - accuracy: 0.7390\nEpoch 102/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5359 - accuracy: 0.7375\nEpoch 103/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5361 - accuracy: 0.7385\nEpoch 104/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5360 - accuracy: 0.7389\nEpoch 105/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5356 - accuracy: 0.7393\nEpoch 106/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5358 - accuracy: 0.7392\nEpoch 107/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5357 - accuracy: 0.7386\nEpoch 108/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5361 - accuracy: 0.7393\nEpoch 109/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5359 - accuracy: 0.7388\nEpoch 110/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5354 - accuracy: 0.7387\nEpoch 111/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5358 - accuracy: 0.7396\nEpoch 112/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5355 - accuracy: 0.7398\nEpoch 113/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5360 - accuracy: 0.7397\nEpoch 114/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5356 - accuracy: 0.7381\nEpoch 115/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5357 - accuracy: 0.7390\nEpoch 116/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5352 - accuracy: 0.7388\nEpoch 117/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5353 - accuracy: 0.7391\nEpoch 118/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5356 - accuracy: 0.7393\nEpoch 119/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5358 - accuracy: 0.7394\nEpoch 120/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5350 - accuracy: 0.7397\nEpoch 121/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5354 - accuracy: 0.7384\nEpoch 122/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5353 - accuracy: 0.7401\nEpoch 123/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5352 - accuracy: 0.7393\nEpoch 124/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5352 - accuracy: 0.7404\nEpoch 125/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5349 - accuracy: 0.7392\nEpoch 126/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5349 - accuracy: 0.7404\nEpoch 127/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5346 - accuracy: 0.7402\nEpoch 128/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5352 - accuracy: 0.7403\nEpoch 129/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5350 - accuracy: 0.7398\nEpoch 130/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5350 - accuracy: 0.7398\nEpoch 131/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5344 - accuracy: 0.7394\nEpoch 132/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5349 - accuracy: 0.7402\nEpoch 133/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5348 - accuracy: 0.7397\nEpoch 134/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5352 - accuracy: 0.7392\nEpoch 135/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5349 - accuracy: 0.7408\nEpoch 136/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5348 - accuracy: 0.7403\nEpoch 137/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5349 - accuracy: 0.7401\nEpoch 138/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5350 - accuracy: 0.7404\nEpoch 139/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5345 - accuracy: 0.7397\nEpoch 140/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5350 - accuracy: 0.7396\nEpoch 141/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5342 - accuracy: 0.7399\nEpoch 142/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5347 - accuracy: 0.7405\nEpoch 143/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5347 - accuracy: 0.7389\nEpoch 144/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5347 - accuracy: 0.7405\nEpoch 145/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5348 - accuracy: 0.7396\nEpoch 146/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5344 - accuracy: 0.7407\nEpoch 147/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5347 - accuracy: 0.7402\nEpoch 148/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5346 - accuracy: 0.7401\nEpoch 149/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5344 - accuracy: 0.7399\nEpoch 150/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5344 - accuracy: 0.7398\nEpoch 151/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5346 - accuracy: 0.7400\nEpoch 152/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5346 - accuracy: 0.7414\nEpoch 153/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5344 - accuracy: 0.7408\nEpoch 154/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5345 - accuracy: 0.7395\nEpoch 155/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5340 - accuracy: 0.7400\nEpoch 156/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5341 - accuracy: 0.7400\nEpoch 157/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5346 - accuracy: 0.7404\nEpoch 158/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5342 - accuracy: 0.7407\nEpoch 159/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5344 - accuracy: 0.7406\nEpoch 160/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5344 - accuracy: 0.7408\nEpoch 161/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5341 - accuracy: 0.7414\nEpoch 162/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5341 - accuracy: 0.7405\nEpoch 163/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5341 - accuracy: 0.7398\nEpoch 164/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5345 - accuracy: 0.7401\nEpoch 165/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5338 - accuracy: 0.7411\nEpoch 166/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5339 - accuracy: 0.7408\nEpoch 167/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5340 - accuracy: 0.7404\nEpoch 168/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5341 - accuracy: 0.7392\nEpoch 169/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5340 - accuracy: 0.7400\nEpoch 170/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5338 - accuracy: 0.7400\nEpoch 171/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5340 - accuracy: 0.7409\nEpoch 172/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5342 - accuracy: 0.7396\nEpoch 173/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5342 - accuracy: 0.7405\nEpoch 174/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5339 - accuracy: 0.7404\nEpoch 175/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5339 - accuracy: 0.7404\nEpoch 176/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5338 - accuracy: 0.7408\nEpoch 177/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5343 - accuracy: 0.7395\nEpoch 178/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5338 - accuracy: 0.7408\nEpoch 179/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5340 - accuracy: 0.7402\nEpoch 180/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5338 - accuracy: 0.7406\nEpoch 181/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5337 - accuracy: 0.7410\nEpoch 182/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5337 - accuracy: 0.7401\nEpoch 183/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5339 - accuracy: 0.7399\nEpoch 184/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5336 - accuracy: 0.7407\nEpoch 185/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5338 - accuracy: 0.7408\nEpoch 186/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5338 - accuracy: 0.7412\nEpoch 187/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5334 - accuracy: 0.7405\nEpoch 188/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5336 - accuracy: 0.7407\nEpoch 189/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5337 - accuracy: 0.7416\nEpoch 190/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5337 - accuracy: 0.7409\nEpoch 191/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5338 - accuracy: 0.7408\nEpoch 192/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5336 - accuracy: 0.7405\nEpoch 193/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5332 - accuracy: 0.7399\nEpoch 194/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5334 - accuracy: 0.7404\nEpoch 195/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5335 - accuracy: 0.7408\nEpoch 196/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5337 - accuracy: 0.7407\nEpoch 197/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5337 - accuracy: 0.7414\nEpoch 198/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5338 - accuracy: 0.7406\nEpoch 199/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5333 - accuracy: 0.7411\nEpoch 200/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5334 - accuracy: 0.7413\n"
],
[
"# Evaluate the model using the test data\nmodel_loss, model_accuracy = nn2.evaluate(X_test_scaled,y_test,verbose=2)\nprint(f\"Loss: {model_loss}, Accuracy: {model_accuracy}\")",
"268/268 - 1s - loss: 0.5651 - accuracy: 0.7263 - 711ms/epoch - 3ms/step\nLoss: 0.5651004910469055, Accuracy: 0.7262973785400391\n"
],
[
"# Export our model to HDF5 file\nnn2.save(\"AlphabetSoupCharity_optimize_1.h5\") ",
"_____no_output_____"
]
],
[
[
"**Optimization 2**",
"_____no_output_____"
]
],
[
[
"# Define the model - deep neural net, i.e., the number of input features and hidden nodes for each layer.\ninput_features = len(X_train_scaled[0])\nhidden_nodes_layer1 = 20\nhidden_nodes_layer2 = 40\nhidden_nodes_layer3 = 80\n\nnn3 = tf.keras.models.Sequential()\n\n# First hidden layer\nnn3.add(tf.keras.layers.Dense(units=hidden_nodes_layer1, input_dim=input_features, activation='relu'))\n\n# Second hidden layer\nnn3.add(tf.keras.layers.Dense(units=hidden_nodes_layer2, activation='relu'))\n\n# Third hidden layer\nnn3.add(tf.keras.layers.Dense(units=hidden_nodes_layer3, activation='relu'))\n\n\n# Output layer\nnn3.add(tf.keras.layers.Dense(units=1, activation='sigmoid'))\n\n# Check the structure of the model\nnn3.summary()",
"Model: \"sequential_2\"\n_________________________________________________________________\n Layer (type) Output Shape Param # \n=================================================================\n dense_6 (Dense) (None, 20) 880 \n \n dense_7 (Dense) (None, 40) 840 \n \n dense_8 (Dense) (None, 80) 3280 \n \n dense_9 (Dense) (None, 1) 81 \n \n=================================================================\nTotal params: 5,081\nTrainable params: 5,081\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"# Compile the model\nnn3.compile(loss=\"binary_crossentropy\", optimizer=\"adam\", metrics=[\"accuracy\"])",
"_____no_output_____"
],
[
"# Train the model\nfit_model = nn3.fit(X_train_scaled, y_train, epochs=200)",
"Epoch 1/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5725 - accuracy: 0.7208\nEpoch 2/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5562 - accuracy: 0.7286\nEpoch 3/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5533 - accuracy: 0.7282\nEpoch 4/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5513 - accuracy: 0.7301\nEpoch 5/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5496 - accuracy: 0.7310\nEpoch 6/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5487 - accuracy: 0.7313\nEpoch 7/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5481 - accuracy: 0.7322\nEpoch 8/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5475 - accuracy: 0.7318\nEpoch 9/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5466 - accuracy: 0.7338\nEpoch 10/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5458 - accuracy: 0.7333\nEpoch 11/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5454 - accuracy: 0.7338\nEpoch 12/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5449 - accuracy: 0.7329\nEpoch 13/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5450 - accuracy: 0.7344\nEpoch 14/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5440 - accuracy: 0.7359\nEpoch 15/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5445 - accuracy: 0.7355\nEpoch 16/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5440 - accuracy: 0.7336\nEpoch 17/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5435 - accuracy: 0.7348\nEpoch 18/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5432 - accuracy: 0.7350\nEpoch 19/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5430 - accuracy: 0.7345\nEpoch 20/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5425 - accuracy: 0.7348\nEpoch 21/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5424 - accuracy: 0.7357\nEpoch 22/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5422 - accuracy: 0.7357\nEpoch 23/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5421 - accuracy: 0.7358\nEpoch 24/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5422 - accuracy: 0.7355\nEpoch 25/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5416 - accuracy: 0.7355\nEpoch 26/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5413 - accuracy: 0.7360\nEpoch 27/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5411 - accuracy: 0.7364\nEpoch 28/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5413 - accuracy: 0.7354\nEpoch 29/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5411 - accuracy: 0.7364\nEpoch 30/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5409 - accuracy: 0.7360\nEpoch 31/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5408 - accuracy: 0.7365\nEpoch 32/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5402 - accuracy: 0.7362\nEpoch 33/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5400 - accuracy: 0.7365\nEpoch 34/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5404 - accuracy: 0.7358\nEpoch 35/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5397 - accuracy: 0.7385\nEpoch 36/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5399 - accuracy: 0.7366\nEpoch 37/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5398 - accuracy: 0.7373\nEpoch 38/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5392 - accuracy: 0.7375\nEpoch 39/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5390 - accuracy: 0.7381\nEpoch 40/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5388 - accuracy: 0.7381\nEpoch 41/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5388 - accuracy: 0.7387\nEpoch 42/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5385 - accuracy: 0.7371\nEpoch 43/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5386 - accuracy: 0.7370\nEpoch 44/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5387 - accuracy: 0.7381\nEpoch 45/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5385 - accuracy: 0.7369\nEpoch 46/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5380 - accuracy: 0.7388\nEpoch 47/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5386 - accuracy: 0.7378\nEpoch 48/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5382 - accuracy: 0.7378\nEpoch 49/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5383 - accuracy: 0.7373\nEpoch 50/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5378 - accuracy: 0.7378\nEpoch 51/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5374 - accuracy: 0.7384\nEpoch 52/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5374 - accuracy: 0.7378\nEpoch 53/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5383 - accuracy: 0.7377\nEpoch 54/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5376 - accuracy: 0.7383\nEpoch 55/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5375 - accuracy: 0.7379\nEpoch 56/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5371 - accuracy: 0.7383\nEpoch 57/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5368 - accuracy: 0.7398\nEpoch 58/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5369 - accuracy: 0.7389\nEpoch 59/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5372 - accuracy: 0.7373\nEpoch 60/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5368 - accuracy: 0.7392\nEpoch 61/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5364 - accuracy: 0.7394\nEpoch 62/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5367 - accuracy: 0.7384\nEpoch 63/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5364 - accuracy: 0.7392\nEpoch 64/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5364 - accuracy: 0.7392\nEpoch 65/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5363 - accuracy: 0.7396\nEpoch 66/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5365 - accuracy: 0.7389\nEpoch 67/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5361 - accuracy: 0.7392\nEpoch 68/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5361 - accuracy: 0.7396\nEpoch 69/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5361 - accuracy: 0.7387\nEpoch 70/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5359 - accuracy: 0.7388\nEpoch 71/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5355 - accuracy: 0.7399\nEpoch 72/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5360 - accuracy: 0.7387\nEpoch 73/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5361 - accuracy: 0.7396\nEpoch 74/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5359 - accuracy: 0.7393\nEpoch 75/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5354 - accuracy: 0.7394\nEpoch 76/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5354 - accuracy: 0.7399\nEpoch 77/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5354 - accuracy: 0.7388\nEpoch 78/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5360 - accuracy: 0.7395\nEpoch 79/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5353 - accuracy: 0.7400\nEpoch 80/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5350 - accuracy: 0.7395\nEpoch 81/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5353 - accuracy: 0.7394\nEpoch 82/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5351 - accuracy: 0.7392\nEpoch 83/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5347 - accuracy: 0.7406\nEpoch 84/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5348 - accuracy: 0.7406\nEpoch 85/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5350 - accuracy: 0.7392\nEpoch 86/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5344 - accuracy: 0.7406\nEpoch 87/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5348 - accuracy: 0.7406\nEpoch 88/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5348 - accuracy: 0.7396\nEpoch 89/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5349 - accuracy: 0.7395\nEpoch 90/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5344 - accuracy: 0.7397\nEpoch 91/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5344 - accuracy: 0.7401\nEpoch 92/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5344 - accuracy: 0.7406\nEpoch 93/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5347 - accuracy: 0.7402\nEpoch 94/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5345 - accuracy: 0.7390\nEpoch 95/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5346 - accuracy: 0.7399\nEpoch 96/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5345 - accuracy: 0.7397\nEpoch 97/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5343 - accuracy: 0.7404\nEpoch 98/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5340 - accuracy: 0.7404\nEpoch 99/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5338 - accuracy: 0.7403\nEpoch 100/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5345 - accuracy: 0.7407\nEpoch 101/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5343 - accuracy: 0.7411\nEpoch 102/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5340 - accuracy: 0.7400\nEpoch 103/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5339 - accuracy: 0.7404\nEpoch 104/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5342 - accuracy: 0.7402\nEpoch 105/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5338 - accuracy: 0.7405\nEpoch 106/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5340 - accuracy: 0.7404\nEpoch 107/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5345 - accuracy: 0.7402\nEpoch 108/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5337 - accuracy: 0.7400\nEpoch 109/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5335 - accuracy: 0.7406\nEpoch 110/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5335 - accuracy: 0.7407\nEpoch 111/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5338 - accuracy: 0.7400\nEpoch 112/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5338 - accuracy: 0.7396\nEpoch 113/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5332 - accuracy: 0.7409\nEpoch 114/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5332 - accuracy: 0.7404\nEpoch 115/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5334 - accuracy: 0.7409\nEpoch 116/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5334 - accuracy: 0.7404\nEpoch 117/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5332 - accuracy: 0.7407\nEpoch 118/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5336 - accuracy: 0.7400\nEpoch 119/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5333 - accuracy: 0.7411\nEpoch 120/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5338 - accuracy: 0.7402\nEpoch 121/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5340 - accuracy: 0.7397\nEpoch 122/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5329 - accuracy: 0.7405\nEpoch 123/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5332 - accuracy: 0.7413\nEpoch 124/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5331 - accuracy: 0.7410\nEpoch 125/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5332 - accuracy: 0.7407\nEpoch 126/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5333 - accuracy: 0.7410\nEpoch 127/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5333 - accuracy: 0.7409\nEpoch 128/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5333 - accuracy: 0.7403\nEpoch 129/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5330 - accuracy: 0.7410\nEpoch 130/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5338 - accuracy: 0.7405\nEpoch 131/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5331 - accuracy: 0.7407\nEpoch 132/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5330 - accuracy: 0.7410\nEpoch 133/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5331 - accuracy: 0.7400\nEpoch 134/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5328 - accuracy: 0.7413\nEpoch 135/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5330 - accuracy: 0.7405\nEpoch 136/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5332 - accuracy: 0.7407\nEpoch 137/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5330 - accuracy: 0.7405\nEpoch 138/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5329 - accuracy: 0.7406\nEpoch 139/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5327 - accuracy: 0.7406\nEpoch 140/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5328 - accuracy: 0.7407\nEpoch 141/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5329 - accuracy: 0.7405\nEpoch 142/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5333 - accuracy: 0.7402\nEpoch 143/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5332 - accuracy: 0.7413\nEpoch 144/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5324 - accuracy: 0.7412\nEpoch 145/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5327 - accuracy: 0.7404\nEpoch 146/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5325 - accuracy: 0.7411\nEpoch 147/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5326 - accuracy: 0.7408\nEpoch 148/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5325 - accuracy: 0.7410\nEpoch 149/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5330 - accuracy: 0.7406\nEpoch 150/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5327 - accuracy: 0.7409\nEpoch 151/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5327 - accuracy: 0.7411\nEpoch 152/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5328 - accuracy: 0.7402\nEpoch 153/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5323 - accuracy: 0.7409\nEpoch 154/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5318 - accuracy: 0.7419\nEpoch 155/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5323 - accuracy: 0.7416\nEpoch 156/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5325 - accuracy: 0.7413\nEpoch 157/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5323 - accuracy: 0.7414\nEpoch 158/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5327 - accuracy: 0.7406\nEpoch 159/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5321 - accuracy: 0.7409\nEpoch 160/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5324 - accuracy: 0.7406\nEpoch 161/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5324 - accuracy: 0.7409\nEpoch 162/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5323 - accuracy: 0.7412\nEpoch 163/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5321 - accuracy: 0.7404\nEpoch 164/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5323 - accuracy: 0.7411\nEpoch 165/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5323 - accuracy: 0.7415\nEpoch 166/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5322 - accuracy: 0.7405\nEpoch 167/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5318 - accuracy: 0.7413\nEpoch 168/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5323 - accuracy: 0.7415\nEpoch 169/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5318 - accuracy: 0.7405\nEpoch 170/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5319 - accuracy: 0.7409\nEpoch 171/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5320 - accuracy: 0.7414\nEpoch 172/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5327 - accuracy: 0.7408\nEpoch 173/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5317 - accuracy: 0.7411\nEpoch 174/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5320 - accuracy: 0.7412\nEpoch 175/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5315 - accuracy: 0.7411\nEpoch 176/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5318 - accuracy: 0.7409\nEpoch 177/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5323 - accuracy: 0.7404\nEpoch 178/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5321 - accuracy: 0.7405\nEpoch 179/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5321 - accuracy: 0.7403\nEpoch 180/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5320 - accuracy: 0.7415\nEpoch 181/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5319 - accuracy: 0.7414\nEpoch 182/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5318 - accuracy: 0.7411\nEpoch 183/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5318 - accuracy: 0.7409\nEpoch 184/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5322 - accuracy: 0.7413\nEpoch 185/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5314 - accuracy: 0.7421\nEpoch 186/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5319 - accuracy: 0.7406\nEpoch 187/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5316 - accuracy: 0.7407\nEpoch 188/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5329 - accuracy: 0.7408\nEpoch 189/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5319 - accuracy: 0.7419\nEpoch 190/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5316 - accuracy: 0.7412\nEpoch 191/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5314 - accuracy: 0.7413\nEpoch 192/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5317 - accuracy: 0.7422\nEpoch 193/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5321 - accuracy: 0.7412\nEpoch 194/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5318 - accuracy: 0.7406\nEpoch 195/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5313 - accuracy: 0.7405\nEpoch 196/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5315 - accuracy: 0.7416\nEpoch 197/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5313 - accuracy: 0.7410\nEpoch 198/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5313 - accuracy: 0.7413\nEpoch 199/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5317 - accuracy: 0.7414\nEpoch 200/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5322 - accuracy: 0.7413\n"
],
[
"# Evaluate the model using the test data\nmodel_loss, model_accuracy = nn3.evaluate(X_test_scaled,y_test,verbose=2)\nprint(f\"Loss: {model_loss}, Accuracy: {model_accuracy}\")",
"268/268 - 1s - loss: 0.5761 - accuracy: 0.7322 - 742ms/epoch - 3ms/step\nLoss: 0.576053261756897, Accuracy: 0.7322449088096619\n"
],
[
"# Export our model to HDF5 file\nnn3.save(\"AlphabetSoupCharity_optimize_2.h5\") ",
"_____no_output_____"
],
[
"# Define the model - deep neural net, i.e., the number of input features and hidden nodes for each layer.\ninput_features = len(X_train_scaled[0])\nhidden_nodes_layer1 = 60\nhidden_nodes_layer2 = 60\nhidden_nodes_layer3 = 40\nhidden_nodes_layer4 = 20\n\nnn4 = tf.keras.models.Sequential()\n\n# First hidden layer\nnn4.add(tf.keras.layers.Dense(units=hidden_nodes_layer1, input_dim=input_features, activation='relu'))\n\n# Second hidden layer\nnn4.add(tf.keras.layers.Dense(units=hidden_nodes_layer2, activation='relu'))\n\n# Third hidden layer\nnn4.add(tf.keras.layers.Dense(units=hidden_nodes_layer3, activation='relu'))\n\n# Forth hidden layer\nnn4.add(tf.keras.layers.Dense(units=hidden_nodes_layer4, activation='relu'))\n\n# Output layer\nnn4.add(tf.keras.layers.Dense(units=1, activation='sigmoid'))\n\n# Check the structure of the model\nnn4.summary()",
"Model: \"sequential_3\"\n_________________________________________________________________\n Layer (type) Output Shape Param # \n=================================================================\n dense_10 (Dense) (None, 60) 2640 \n \n dense_11 (Dense) (None, 60) 3660 \n \n dense_12 (Dense) (None, 40) 2440 \n \n dense_13 (Dense) (None, 20) 820 \n \n dense_14 (Dense) (None, 1) 21 \n \n=================================================================\nTotal params: 9,581\nTrainable params: 9,581\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"# Compile the model\nnn4.compile(loss=\"binary_crossentropy\", optimizer=\"adam\", metrics=[\"accuracy\"])",
"_____no_output_____"
],
[
"# Train the model\nfit_model = nn4.fit(X_train_scaled, y_train, epochs=200)",
"Epoch 1/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5713 - accuracy: 0.7197\nEpoch 2/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5558 - accuracy: 0.7264\nEpoch 3/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5527 - accuracy: 0.7309\nEpoch 4/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5503 - accuracy: 0.7327\nEpoch 5/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5492 - accuracy: 0.7311\nEpoch 6/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5477 - accuracy: 0.7318\nEpoch 7/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5467 - accuracy: 0.7329\nEpoch 8/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5464 - accuracy: 0.7341\nEpoch 9/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5451 - accuracy: 0.7333\nEpoch 10/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5449 - accuracy: 0.7344\nEpoch 11/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5445 - accuracy: 0.7340\nEpoch 12/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5445 - accuracy: 0.7341\nEpoch 13/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5434 - accuracy: 0.7356\nEpoch 14/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5433 - accuracy: 0.7345\nEpoch 15/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5424 - accuracy: 0.7358\nEpoch 16/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5425 - accuracy: 0.7345\nEpoch 17/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5421 - accuracy: 0.7348\nEpoch 18/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5413 - accuracy: 0.7360\nEpoch 19/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5418 - accuracy: 0.7361\nEpoch 20/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5409 - accuracy: 0.7369\nEpoch 21/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5404 - accuracy: 0.7359\nEpoch 22/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5405 - accuracy: 0.7367\nEpoch 23/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5406 - accuracy: 0.7365\nEpoch 24/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5404 - accuracy: 0.7367\nEpoch 25/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5400 - accuracy: 0.7362\nEpoch 26/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5394 - accuracy: 0.7367\nEpoch 27/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5395 - accuracy: 0.7362\nEpoch 28/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5388 - accuracy: 0.7372\nEpoch 29/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5393 - accuracy: 0.7373\nEpoch 30/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5388 - accuracy: 0.7372\nEpoch 31/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5386 - accuracy: 0.7379\nEpoch 32/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5382 - accuracy: 0.7374\nEpoch 33/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5383 - accuracy: 0.7385\nEpoch 34/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5380 - accuracy: 0.7381\nEpoch 35/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5373 - accuracy: 0.7378\nEpoch 36/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5374 - accuracy: 0.7385\nEpoch 37/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5382 - accuracy: 0.7376\nEpoch 38/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5371 - accuracy: 0.7384\nEpoch 39/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5368 - accuracy: 0.7386\nEpoch 40/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5366 - accuracy: 0.7382\nEpoch 41/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5367 - accuracy: 0.7381\nEpoch 42/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5365 - accuracy: 0.7390\nEpoch 43/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5361 - accuracy: 0.7388\nEpoch 44/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5364 - accuracy: 0.7386\nEpoch 45/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5366 - accuracy: 0.7387\nEpoch 46/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5362 - accuracy: 0.7388\nEpoch 47/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5357 - accuracy: 0.7383\nEpoch 48/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5359 - accuracy: 0.7384\nEpoch 49/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5360 - accuracy: 0.7391\nEpoch 50/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5361 - accuracy: 0.7383\nEpoch 51/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5354 - accuracy: 0.7395\nEpoch 52/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5350 - accuracy: 0.7369\nEpoch 53/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5351 - accuracy: 0.7400\nEpoch 54/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5349 - accuracy: 0.7390\nEpoch 55/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5347 - accuracy: 0.7383\nEpoch 56/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5349 - accuracy: 0.7394\nEpoch 57/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5346 - accuracy: 0.7392\nEpoch 58/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5350 - accuracy: 0.7393\nEpoch 59/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5346 - accuracy: 0.7399\nEpoch 60/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5344 - accuracy: 0.7396\nEpoch 61/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5354 - accuracy: 0.7391\nEpoch 62/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5346 - accuracy: 0.7385\nEpoch 63/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5340 - accuracy: 0.7401\nEpoch 64/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5343 - accuracy: 0.7396\nEpoch 65/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5336 - accuracy: 0.7402\nEpoch 66/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5339 - accuracy: 0.7397\nEpoch 67/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5338 - accuracy: 0.7391\nEpoch 68/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5342 - accuracy: 0.7405\nEpoch 69/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5340 - accuracy: 0.7395\nEpoch 70/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5339 - accuracy: 0.7398\nEpoch 71/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5338 - accuracy: 0.7402\nEpoch 72/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5338 - accuracy: 0.7403\nEpoch 73/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5340 - accuracy: 0.7405\nEpoch 74/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5331 - accuracy: 0.7405\nEpoch 75/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5336 - accuracy: 0.7409\nEpoch 76/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5335 - accuracy: 0.7397\nEpoch 77/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5331 - accuracy: 0.7400\nEpoch 78/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5334 - accuracy: 0.7397\nEpoch 79/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5330 - accuracy: 0.7403\nEpoch 80/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5330 - accuracy: 0.7398\nEpoch 81/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5327 - accuracy: 0.7406\nEpoch 82/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5328 - accuracy: 0.7401\nEpoch 83/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5331 - accuracy: 0.7401\nEpoch 84/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5321 - accuracy: 0.7399\nEpoch 85/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5328 - accuracy: 0.7410\nEpoch 86/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5329 - accuracy: 0.7403\nEpoch 87/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5342 - accuracy: 0.7405\nEpoch 88/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5320 - accuracy: 0.7407\nEpoch 89/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5342 - accuracy: 0.7401\nEpoch 90/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5322 - accuracy: 0.7394\nEpoch 91/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5325 - accuracy: 0.7401\nEpoch 92/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5329 - accuracy: 0.7402\nEpoch 93/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5328 - accuracy: 0.7407\nEpoch 94/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5324 - accuracy: 0.7404\nEpoch 95/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5317 - accuracy: 0.7410\nEpoch 96/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5321 - accuracy: 0.7408\nEpoch 97/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5325 - accuracy: 0.7405\nEpoch 98/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5323 - accuracy: 0.7408\nEpoch 99/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5325 - accuracy: 0.7415\nEpoch 100/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5320 - accuracy: 0.7408\nEpoch 101/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5323 - accuracy: 0.7402\nEpoch 102/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5318 - accuracy: 0.7409\nEpoch 103/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5317 - accuracy: 0.7411\nEpoch 104/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5329 - accuracy: 0.7411\nEpoch 105/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5314 - accuracy: 0.7411\nEpoch 106/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5310 - accuracy: 0.7411\nEpoch 107/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5316 - accuracy: 0.7407\nEpoch 108/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5320 - accuracy: 0.7407\nEpoch 109/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5319 - accuracy: 0.7409\nEpoch 110/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5315 - accuracy: 0.7401\nEpoch 111/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5316 - accuracy: 0.7403\nEpoch 112/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5314 - accuracy: 0.7405\nEpoch 113/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5315 - accuracy: 0.7413\nEpoch 114/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5315 - accuracy: 0.7406\nEpoch 115/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5318 - accuracy: 0.7399\nEpoch 116/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5321 - accuracy: 0.7406\nEpoch 117/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5319 - accuracy: 0.7416\nEpoch 118/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5320 - accuracy: 0.7402\nEpoch 119/200\n804/804 [==============================] - 3s 4ms/step - loss: 0.5310 - accuracy: 0.7407\nEpoch 120/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5310 - accuracy: 0.7414\nEpoch 121/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5363 - accuracy: 0.7405\nEpoch 122/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5319 - accuracy: 0.7409\nEpoch 123/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5320 - accuracy: 0.7413\nEpoch 124/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5312 - accuracy: 0.7405\nEpoch 125/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5311 - accuracy: 0.7409\nEpoch 126/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5321 - accuracy: 0.7423\nEpoch 127/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5308 - accuracy: 0.7414\nEpoch 128/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5314 - accuracy: 0.7408\nEpoch 129/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5304 - accuracy: 0.7407\nEpoch 130/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5317 - accuracy: 0.7411\nEpoch 131/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5320 - accuracy: 0.7421\nEpoch 132/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5310 - accuracy: 0.7409\nEpoch 133/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5311 - accuracy: 0.7404\nEpoch 134/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5302 - accuracy: 0.7419\nEpoch 135/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5303 - accuracy: 0.7418\nEpoch 136/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5308 - accuracy: 0.7408\nEpoch 137/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5320 - accuracy: 0.7410\nEpoch 138/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5311 - accuracy: 0.7413\nEpoch 139/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5308 - accuracy: 0.7407\nEpoch 140/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5307 - accuracy: 0.7412\nEpoch 141/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5306 - accuracy: 0.7410\nEpoch 142/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5310 - accuracy: 0.7406\nEpoch 143/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5310 - accuracy: 0.7417\nEpoch 144/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5307 - accuracy: 0.7415\nEpoch 145/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5307 - accuracy: 0.7411\nEpoch 146/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5307 - accuracy: 0.7414\nEpoch 147/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5306 - accuracy: 0.7410\nEpoch 148/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5304 - accuracy: 0.7413\nEpoch 149/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5356 - accuracy: 0.7408\nEpoch 150/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5323 - accuracy: 0.7413\nEpoch 151/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5305 - accuracy: 0.7417\nEpoch 152/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5303 - accuracy: 0.7422\nEpoch 153/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5302 - accuracy: 0.7415\nEpoch 154/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5306 - accuracy: 0.7411\nEpoch 155/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5307 - accuracy: 0.7410\nEpoch 156/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5304 - accuracy: 0.7411\nEpoch 157/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5308 - accuracy: 0.7413\nEpoch 158/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5307 - accuracy: 0.7417\nEpoch 159/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5303 - accuracy: 0.7418\nEpoch 160/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5302 - accuracy: 0.7413\nEpoch 161/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5319 - accuracy: 0.7419\nEpoch 162/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5309 - accuracy: 0.7408\nEpoch 163/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5317 - accuracy: 0.7412\nEpoch 164/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5300 - accuracy: 0.7416\nEpoch 165/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5306 - accuracy: 0.7408\nEpoch 166/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5305 - accuracy: 0.7414\nEpoch 167/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5303 - accuracy: 0.7413\nEpoch 168/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5302 - accuracy: 0.7425\nEpoch 169/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5308 - accuracy: 0.7410\nEpoch 170/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5310 - accuracy: 0.7408\nEpoch 171/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5309 - accuracy: 0.7414\nEpoch 172/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5301 - accuracy: 0.7416\nEpoch 173/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5301 - accuracy: 0.7418\nEpoch 174/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5300 - accuracy: 0.7421\nEpoch 175/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5305 - accuracy: 0.7413\nEpoch 176/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5299 - accuracy: 0.7411\nEpoch 177/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5304 - accuracy: 0.7421\nEpoch 178/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5321 - accuracy: 0.7406\nEpoch 179/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5306 - accuracy: 0.7412\nEpoch 180/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5302 - accuracy: 0.7415\nEpoch 181/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5303 - accuracy: 0.7418\nEpoch 182/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5298 - accuracy: 0.7416\nEpoch 183/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5304 - accuracy: 0.7419\nEpoch 184/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5299 - accuracy: 0.7420\nEpoch 185/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5302 - accuracy: 0.7412\nEpoch 186/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5309 - accuracy: 0.7416\nEpoch 187/200\n804/804 [==============================] - 4s 4ms/step - loss: 0.5302 - accuracy: 0.7416\nEpoch 188/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5298 - accuracy: 0.7407\nEpoch 189/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5297 - accuracy: 0.7411\nEpoch 190/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5307 - accuracy: 0.7413\nEpoch 191/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5302 - accuracy: 0.7416\nEpoch 192/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5296 - accuracy: 0.7416\nEpoch 193/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5300 - accuracy: 0.7416\nEpoch 194/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5301 - accuracy: 0.7420\nEpoch 195/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5303 - accuracy: 0.7409\nEpoch 196/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5303 - accuracy: 0.7416\nEpoch 197/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5300 - accuracy: 0.7414\nEpoch 198/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5296 - accuracy: 0.7414\nEpoch 199/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5295 - accuracy: 0.7413\nEpoch 200/200\n804/804 [==============================] - 4s 5ms/step - loss: 0.5300 - accuracy: 0.7407\n"
],
[
"# Evaluate the model using the test data\nmodel_loss, model_accuracy = nn4.evaluate(X_test_scaled,y_test,verbose=2)\nprint(f\"Loss: {model_loss}, Accuracy: {model_accuracy}\")",
"268/268 - 1s - loss: 0.6118 - accuracy: 0.7266 - 769ms/epoch - 3ms/step\nLoss: 0.6118495464324951, Accuracy: 0.7266472578048706\n"
],
[
"# Export our model to HDF5 file\nnn4.save(\"AlphabetSoupCharity_optimize_3.h5\") ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0938dd09eda707152a4f90b09dd20d4b840e9a3 | 10,056 | ipynb | Jupyter Notebook | notebooks/PE_Numpy.ipynb | NickBaynham/aimldl | 163bfd3fb834ae6811da4b0fadccfcb228ed2e79 | [
"MIT"
] | null | null | null | notebooks/PE_Numpy.ipynb | NickBaynham/aimldl | 163bfd3fb834ae6811da4b0fadccfcb228ed2e79 | [
"MIT"
] | null | null | null | notebooks/PE_Numpy.ipynb | NickBaynham/aimldl | 163bfd3fb834ae6811da4b0fadccfcb228ed2e79 | [
"MIT"
] | null | null | null | 22.198675 | 262 | 0.412589 | [
[
[
"# Numpy",
"_____no_output_____"
],
[
"We have seen python basic data structures in our last section. They are great but lack specialized features for data analysis. Like, adding rows, columns, operating on 2d matrices aren't readily available. So, we will use *numpy* for such functions.\n\n",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
]
],
[
[
"Numpy operates on *nd* arrays. These are similar to lists but contains homogenous elements but easier to store 2-d data.",
"_____no_output_____"
]
],
[
[
"l1 = [1,2,3,4]\nnd1 = np.array(l1)\nprint(nd1)\n\nl2 = [5,6,7,8]\nnd2 = np.array([l1,l2])\nprint(nd2)",
"[1 2 3 4]\n[[1 2 3 4]\n [5 6 7 8]]\n"
]
],
[
[
"Sum functions on np.array()",
"_____no_output_____"
]
],
[
[
"print(nd2.shape)\n\nprint(nd2.size)\n\nprint(nd2.dtype)",
"(2, 4)\n8\nint64\n"
]
],
[
[
"### Question 1\n\nCreate an identity 2d-array or matrix (with ones across the diagonal).\n\n[ **Hint:** You can also use **np.identity()** function ]",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"### Question 2\n\nCreate a 2d-array or matrix of order 3x3 with values = 9,8,7,6,5,4,3,2,1 arranged in the same order.\n\nUse: **np.array()** function\n\n",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"### Question 3\n\nInterchange both the rows and columns of the given matrix.\n\nHint: You can use the transpose **.T**)",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"### Question 4\nAdd + 1 to all the elements in the given matrix.",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"Similarly you can do operations like scalar substraction, division, multiplication (operating on each element in the matrix)",
"_____no_output_____"
],
[
"### Question 5\n\nFind the mean of all elements in the given matrix nd6.\nnd6 = [[ 1 4 9 121 144 169]\n [ 16 25 36 196 225 256]\n [ 49 64 81 289 324 361]]\n \n Use: **.mean()** function\n",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"### Question 7\n\nFind the dot product of two given matrices.\n\n[**Hint:** Use **np.dot()**]",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"### Array Slicing/Indexing:\n\n- Now we'll learn to access multiple elements or a range of elements from an array.",
"_____no_output_____"
]
],
[
[
"x = np.arange(20)\nx",
"_____no_output_____"
]
],
[
[
"### Question 8\n\nPrint the array elements from start to 4th position\n",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"### Question 9\n\n- Return elements from first position with step size 2. (Difference between consecutive elements is 2)",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"### Question 10\n\nReverse the array using array indexing:",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d0939ee7cd60f635905ade3c10e94cfc8cfb21c6 | 62,865 | ipynb | Jupyter Notebook | notebooks/figure-occurence-boxplot.ipynb | bo1929/ethologger | 0d00a42d856b82d15ed0898b0895c25bfb11aa5d | [
"MIT"
] | 2 | 2021-08-11T20:27:41.000Z | 2021-08-24T18:10:34.000Z | notebooks/figure-occurence-boxplot.ipynb | bo1929/ethologger | 0d00a42d856b82d15ed0898b0895c25bfb11aa5d | [
"MIT"
] | null | null | null | notebooks/figure-occurence-boxplot.ipynb | bo1929/ethologger | 0d00a42d856b82d15ed0898b0895c25bfb11aa5d | [
"MIT"
] | 1 | 2021-08-15T18:59:30.000Z | 2021-08-15T18:59:30.000Z | 312.761194 | 57,376 | 0.933095 | [
[
[
"%load_ext autoreload\n%autoreload 2\nimport yaml\nimport time\nfrom pathlib import Path",
"_____no_output_____"
],
[
"import matplotlib as mpl\nimport seaborn as sns\nimport numpy as np\nplt = mpl.pyplot",
"_____no_output_____"
],
[
"FPS = 30",
"_____no_output_____"
],
[
"main_cfg_path = \"/home/bo/tmp/presentation-project-attempt4/main_cfg.yaml\"\nwith open(main_cfg_path, \"r\") as stream:\n try:\n main_cfg = yaml.safe_load(stream)\n except yaml.YAMLError as exc:\n raise exc",
"_____no_output_____"
],
[
"epxt_names = main_cfg[\"experiment_data_paths\"].keys()\nprint(epxt_names)",
"dict_keys(['FlyF1-03082020164520', 'FlyF14-08172021175459', 'FlyF15-08182021173222', 'FlyF17-08192021173437', 'FlyF2-03092020175006', 'FlyF20-08202021171828', 'FlyF3-08092021173846', 'FlyF5-08102021175053', 'FlyM12-08162021175307', 'FlyM16-08182021173236', 'FlyM19-08202021171805', 'FlyM21-08262021165434', 'FlyM4-03062020153616', 'FlyM5-03072020150452', 'FlyM8-08112021174107', 'FlyM9-08122021171549'])\n"
],
[
"cluster_labels_dict = {}\nfor expt_name in epxt_names:\n cluster_labels_dict[expt_name] = np.load(Path(main_cfg[\"project_path\"]) / expt_name / \"cluster_labels.npy\")",
"_____no_output_____"
],
[
"# Cluster you're interested in.\ncluster_lbls = [1,2,23,10]\nlbl_name = \"Haltere-switch\"\n\ncluster_occurence_dict = {}\nfor expt_name in epxt_names:\n cluster_labels = cluster_labels_dict[expt_name]\n mask_lbl = np.full(cluster_labels.shape[0], False)\n for lbl in cluster_lbls:\n mask_lbl[cluster_labels == lbl] = True\n print(expt_name,len(np.nonzero(mask_lbl)[0]))\n cluster_occurence_dict[expt_name] = np.nonzero(mask_lbl)[0]",
"FlyF1-03082020164520 2856\nFlyF14-08172021175459 1121\nFlyF15-08182021173222 4714\nFlyF17-08192021173437 2877\nFlyF2-03092020175006 2326\nFlyF20-08202021171828 2639\nFlyF3-08092021173846 2259\nFlyF5-08102021175053 3564\nFlyM12-08162021175307 7137\nFlyM16-08182021173236 2139\nFlyM19-08202021171805 3517\nFlyM21-08262021165434 1179\nFlyM4-03062020153616 3107\nFlyM5-03072020150452 2002\nFlyM8-08112021174107 8112\nFlyM9-08122021171549 14888\n"
],
[
"cluster_occurence_times = np.array(list(cluster_occurence_dict.values()), dtype=object) / FPS\nfly_expt_names = list(cluster_occurence_dict.keys())\nfig, ax = plt.subplots(figsize=(15,10))\nax = sns.boxplot(data=cluster_occurence_times, orient=\"h\", showfliers=False, palette=\"tab20\", ax=ax)\nax.set_yticklabels(fly_expt_names,)\nformatter = mpl.ticker.FuncFormatter(lambda sec, x: time.strftime(\"%H:%M\", time.gmtime(sec)))\nax.xaxis.set_major_formatter(formatter)\nax.set_title(f\"{lbl_name} Occurence Time Boxplot\", fontsize=40)",
"_____no_output_____"
]
],
[
[
"## ",
"_____no_output_____"
]
]
] | [
"code",
"markdown"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
d0939fb35eff4d7c8fe8d98e640ab11734e59459 | 9,251 | ipynb | Jupyter Notebook | notebooks/analysis/TheGraphDataAnalysis.ipynb | trangnv/geb-simulations-h20 | df86e1ad1ff8e98cf2c3f6025d1626d260a3b125 | [
"MIT"
] | 7 | 2021-08-31T13:11:51.000Z | 2022-02-10T09:05:16.000Z | notebooks/analysis/TheGraphDataAnalysis.ipynb | trangnv/geb-simulations-h20 | df86e1ad1ff8e98cf2c3f6025d1626d260a3b125 | [
"MIT"
] | null | null | null | notebooks/analysis/TheGraphDataAnalysis.ipynb | trangnv/geb-simulations-h20 | df86e1ad1ff8e98cf2c3f6025d1626d260a3b125 | [
"MIT"
] | 8 | 2021-09-03T08:29:09.000Z | 2021-12-04T04:20:49.000Z | 27.780781 | 311 | 0.600476 | [
[
[
"# The Graph Data Access\n\nIn this notebook, we read in the data that was generated and saved as a csv from the [TheGraphDataSetCreation](TheGraphDataSetCreation.ipynb) notebook. \n\n\nGoals of this notebook are to obtain:\n\n* Signals, states, event and sequences\n* Volatility metrics\n* ID perceived shocks (correlated with announcements)\n* Signal for target price\n* Signal for market price\n* Error plot\n\nAs a starting point for moving to a decision support system.",
"_____no_output_____"
]
],
[
[
"# import libraries\nimport pandas as pd\nimport seaborn as sns\nimport matplotlib.pyplot as plt\nimport scipy as sp\nfrom statsmodels.distributions.empirical_distribution import ECDF\nimport scipy.stats as stats",
"_____no_output_____"
]
],
[
[
"## Import data and add additional attributes",
"_____no_output_____"
]
],
[
[
"graphData = pd.read_csv('saved_results/RaiLiveGraphData.csv')\ndel graphData['Unnamed: 0']",
"_____no_output_____"
],
[
"graphData.head()",
"_____no_output_____"
],
[
"graphData.describe()",
"_____no_output_____"
],
[
"graphData.plot(x='blockNumber',y='redemptionPriceActual',kind='line',title='redemptionPriceActual')",
"_____no_output_____"
],
[
"graphData.plot(x='blockNumber',y='redemptionRateActual',kind='line',title='redemptionRateActual')",
"_____no_output_____"
],
[
"graphData['error'] = graphData['redemptionPriceActual'] - graphData['marketPriceUsd']\ngraphData['error_integral'] = graphData['error'].cumsum()",
"_____no_output_____"
],
[
"graphData.plot(x='blockNumber',y='error',kind='line',title='error')",
"_____no_output_____"
],
[
"graphData.plot(x='blockNumber',y='error_integral',kind='line',title='Steady state error')",
"_____no_output_____"
]
],
[
[
"## Error experimentation\n\n#### Note: not taking into account control period",
"_____no_output_____"
]
],
[
[
"kp = 2e-7\n#ki = (-kp * error)/(integral_error)\n# computing at each time, what would the value of ki need to be such that the redemption price would be constant\ngraphData['equilibriation_ki'] = (-kp * graphData.error)/graphData.error_integral",
"_____no_output_____"
],
[
"# todo iterate through labels and append negative\ngraphData['equilibriation_ki'].apply(lambda x: -x).plot(logy = True,title='Actual equilibriation_ki - flipped sign for log plotting')\nplt.hlines(5e-9, 0, 450, linestyles='solid', label='Recommended ki - flipped sign', color='r')\nplt.hlines(-(graphData['equilibriation_ki'].median()), 0, 450, linestyles='solid', label='median actual ki - flipped', color='g')\nlocs,labels = plt.yticks() # Get the current locations and labelsyticks\nnew_locs = []\nfor i in locs:\n new_locs.append('-'+str(i))\nplt.yticks(locs, new_locs)\nplt.legend(loc=\"upper right\")",
"_____no_output_____"
],
[
"graphData['equilibriation_ki'].median()",
"_____no_output_____"
]
],
[
[
"### Counterfactual if intergral control rate had been median the whole time",
"_____no_output_____"
]
],
[
[
"graphData['counterfactual_redemption_rate'] = (kp * graphData['error'] + graphData['equilibriation_ki'].median())/ graphData['error_integral']",
"_____no_output_____"
],
[
"subsetGraph = graphData.iloc[50:]\nsns.lineplot(data=subsetGraph,x=\"blockNumber\", y=\"counterfactual_redemption_rate\",label='Counterfactual')\nax2 = plt.twinx()\n# let reflexer know this is wrong\nsns.lineplot(data=subsetGraph,x=\"blockNumber\", y=\"redemptionRateActual\",ax=ax2,color='r',label='Actual')\nplt.title('Actual redemption rate vs counterfactual')\nplt.legend(loc=\"upper left\")\n",
"_____no_output_____"
]
],
[
[
"## Goodness of fit tests\nWhether or not counterfactual is far enough from actual to reject null that they are from the same distributions.",
"_____no_output_____"
]
],
[
[
"# fit a cdf\necdf = ECDF(subsetGraph.redemptionRateActual.values)\necdf2 = ECDF(subsetGraph.counterfactual_redemption_rate.values)\n\nplt.plot(ecdf.x,ecdf.y,color='r')\nplt.title('redemptionRateActual ECDF')\nplt.show()\n\nplt.plot(ecdf2.x,ecdf2.y,color='b')\nplt.title('counterfactual_redemption_rate ECDF')\nplt.show()\n\nalpha = 0.05\n\nstatistic, p_value = stats.ks_2samp(subsetGraph.redemptionRateActual.values, subsetGraph.counterfactual_redemption_rate.values) # two sided\nif p_value > alpha:\n decision = \"Sample is from the distribution\"\nelif p_value <= alpha:\n decision = \"Sample is not from the distribution\"\n\nprint(p_value)\nprint(decision)",
"_____no_output_____"
]
],
[
[
"Based on our analysis using the Kolmogorov-Smirnov Goodness-of-Fit Test, the distributions are very different. As can be seen above from their EDCF plots, you can see a different in their distributions, however pay close attention to the x axis and you can see the distribution difference is significant. ",
"_____no_output_____"
]
],
[
[
"# scatterplot of linear regressoin residuals\nsns.residplot(x='blockNumber', y='redemptionRateActual', data=subsetGraph, label='redemptionRateActual')\nplt.title('redemptionRateActual regression residuals')",
"_____no_output_____"
],
[
"sns.residplot(x='blockNumber', y='counterfactual_redemption_rate', data=subsetGraph,label='counterfactual_redemption_rate')\nplt.title('counterfactual_redemption_rate regression residuals')",
"_____no_output_____"
],
[
"graphData.plot(x='blockNumber',y='globalDebt',kind='line',title='globalDebt')",
"_____no_output_____"
],
[
"graphData.plot(x='blockNumber',y='erc20CoinTotalSupply',kind='line',title='erc20CoinTotalSupply')",
"_____no_output_____"
],
[
"graphData.plot(x='blockNumber',y='marketPriceEth',kind='line',title='marketPriceEth')",
"_____no_output_____"
],
[
"graphData.plot(x='blockNumber',y='marketPriceUsd',kind='line',title='marketPriceUsd')",
"_____no_output_____"
]
],
[
[
"## Conclusion\n\nUsing The Graph, a lot of data about the Rai system can be obtained for analyzing the health of the system. With some data manipulation, these data streams could be intergrated into the Rai cadCAD model to turn it into a true decision support system.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
d093a6071aa1e970a92aab38a29e6ddb2a3fece9 | 13,017 | ipynb | Jupyter Notebook | src/notebooks/mimic_data_preparation_no_discharge_notes_4.ipynb | jplasser/CNEP | 849be943035b0c73a0002475403b046201691a92 | [
"MIT"
] | 2 | 2021-12-12T13:19:55.000Z | 2022-02-04T15:31:35.000Z | src/notebooks/mimic_data_preparation_no_discharge_notes_4.ipynb | jplasser/CNEP | 849be943035b0c73a0002475403b046201691a92 | [
"MIT"
] | null | null | null | src/notebooks/mimic_data_preparation_no_discharge_notes_4.ipynb | jplasser/CNEP | 849be943035b0c73a0002475403b046201691a92 | [
"MIT"
] | null | null | null | 35.086253 | 340 | 0.519551 | [
[
[
"import pandas as pd\nimport numpy as np\nimport pickle\n\nBASEDIR_MIMIC = '/Volumes/MyData/MIMIC_data/mimiciii/1.4'",
"_____no_output_____"
],
[
"def get_note_events():\n n_rows = 100000\n\n icd9_code = pd.read_csv(f\"{BASEDIR_MIMIC}/DIAGNOSES_ICD.csv\", index_col = None)\n # create the iterator\n noteevents_iterator = pd.read_csv(\n f\"{BASEDIR_MIMIC}/NOTEEVENTS.csv\",\n iterator=True,\n chunksize=n_rows)\n\n events_list = ['Discharge summary',\n 'Echo',\n 'ECG',\n 'Nursing',\n 'Physician ',\n 'Rehab Services',\n 'Case Management ',\n 'Respiratory ',\n 'Nutrition',\n 'General',\n 'Social Work',\n 'Pharmacy',\n 'Consult',\n 'Radiology',\n 'Nursing/other']\n\n # concatenate according to a filter to get our noteevents data\n noteevents = pd.concat(\n [noteevents_chunk[\n np.logical_and(\n noteevents_chunk.CATEGORY.isin(events_list[1:]),\n noteevents_chunk.DESCRIPTION.isin([\"Report\"])\n )\n ]\n for noteevents_chunk in noteevents_iterator])\n\n # drop all nan in column HADM_ID\n noteevents = noteevents.dropna(subset=[\"HADM_ID\"])\n noteevents.HADM_ID = noteevents.HADM_ID.astype(int)\n try:\n assert len(noteevents.drop_duplicates([\"SUBJECT_ID\",\"HADM_ID\"])) == len(noteevents)\n except AssertionError as e:\n print(\"There are duplicates on Primary Key Set\")\n \n noteevents.CHARTDATE = pd.to_datetime(noteevents.CHARTDATE , format = '%Y-%m-%d %H:%M:%S', errors = 'coerce')\n pd.set_option('display.max_colwidth',50)\n noteevents.sort_values([\"SUBJECT_ID\",\"HADM_ID\",\"CHARTDATE\"], inplace =True)\n #noteevents.drop_duplicates([\"SUBJECT_ID\",\"HADM_ID\"], inplace = True)\n\n noteevents.reset_index(drop = True, inplace = True)\n \n top_values = (icd9_code.groupby('ICD9_CODE').\n agg({\"SUBJECT_ID\": \"nunique\"}).\n reset_index().sort_values(['SUBJECT_ID'], ascending = False).ICD9_CODE.tolist()[:15])\n\n # icd9_code = icd9_code[icd9_code.ICD9_CODE.isin(top_values)]\n icd9_code = icd9_code[icd9_code.ICD9_CODE.isin(top_values)]\n \n import re\n import itertools\n\n def clean_text(text):\n return [x for x in list(itertools.chain.from_iterable([t.split(\"<>\") for t in text.replace(\"\\n\",\" \").split(\" \")])) if len(x) > 0]\n\n\n# irrelevant_tags = [\"Admission Date:\", \"Date of Birth:\", \"Service:\", \"Attending:\", \"Facility:\", \"Medications on Admission:\", \"Discharge Medications:\", \"Completed by:\",\n# \"Dictated By:\" , \"Department:\" , \"Provider:\"]\n\n updated_text = [\"<>\".join([\" \".join(re.split(\"\\n\\d|\\n\\s+\",re.sub(\"^(.*?):\",\"\",x).strip()))\n for x in text.split(\"\\n\\n\")]) for text in noteevents.TEXT]\n updated_text = [re.sub(\"(\\[.*?\\])\", \"\", text) for text in updated_text]\n\n updated_text = [\" \".join(clean_text(x)) for x in updated_text]\n noteevents[\"CLEAN_TEXT\"] = updated_text\n \n return noteevents\n\nnoteevents = get_note_events()",
"/var/folders/6p/bxnlvss50bs19f3950g4dfw00000gn/T/ipykernel_36653/2260032821.py:78: DtypeWarning: Columns (5) have mixed types.Specify dtype option on import or set low_memory=False.\n noteevents = get_note_events()\n"
],
[
"def mapNotes(dataset):\n print(f\"Mapping notes on {dataset}.\")\n df = pickle.load(open(f'../data/mimic3/train_data_mimic3/{dataset}', 'rb'))\n \n BASEDIR_MIMIC = '/Volumes/MyData/MIMIC_data/mimiciii/1.4'\n icustays = pd.read_csv(f\"{BASEDIR_MIMIC}/ICUSTAYS.csv\", index_col = None)\n \n # SUBJECT_ID \"_\" ICUSTAY_ID \"_episode\" episode \"_timeseries_readmission.csv\"\n\n import re\n \n episodes = df['names']\n \n regex = r\"(\\d+)_(\\d+)_episode(\\d+)_timeseries_readmission\\.csv\"\n\n sid = []\n hadmids = []\n icustayid = [] # ICUSTAYS.csv ICUSTAY_ID\n episode = []\n notestexts = []\n notextepis = []\n for epi in episodes:\n match = re.findall(regex, epi) #, re.MULTILINE)\n sid.append(int(match[0][0]))\n icustayid.append(int(match[0][1]))\n episode.append(int(match[0][2]))\n hadmid = icustays[icustays['ICUSTAY_ID']==int(match[0][1])]['HADM_ID']\n hadmids.append(int(hadmid))\n try:\n #text = noteevents[noteevents['HADM_ID']==int(hadmid)]['TEXT'].iloc[0]\n #text = noteevents[noteevents['HADM_ID']==int(hadmid)]['CLEAN_TEXT'].iloc[0]\n text = \"\\n\\n\".join([t for t in noteevents[noteevents['HADM_ID']==int(hadmid)]['CLEAN_TEXT']])\n except:\n notextepis.append(int(hadmid))\n text = ''\n notestexts.append(text)\n\n print(len(episodes), len(notextepis), len(set(notextepis)))\n print(len(sid), len(hadmids), len(df['names']))\n \n notesfull = pd.DataFrame({'SUBJECT_ID':sid, 'HADM_ID':hadmids, 'ICUSTAY_ID':icustayid, 'EPISODE':episode, 'CLEAN_TEXT':notestexts})\n \n # save full data\n filename = f'./events_notes_{dataset}'\n\n with open(filename + '.pickle', 'wb') as handle:\n pickle.dump(notesfull, handle, protocol=pickle.HIGHEST_PROTOCOL)\n \n print(f\"Finished mapping notes on {dataset}.\\n\")",
"_____no_output_____"
],
[
"def combineData(dataset):\n print(f\"Combining data for all {dataset}.\")\n df = pickle.load(open(f'../data/mimic3/train_data_mimic3/{dataset}', 'rb'))\n print(df.keys(), len(df['data']),len(df['names']), df['data'][0].shape, len(df['data'][1]), len(df['names']))\n\n notes = pickle.load(open(f'clinical_notes_{dataset}.pickle', 'rb'))\n eventsnotes = pickle.load(open(f'events_notes_{dataset}.pickle', 'rb'))\n\n # how many empty text rows\n # np.where(notes.applymap(lambda x: x == ''))\n\n # how many empty text rows\n print(f\"There are {len(list(notes[notes['CLEAN_TEXT'] == ''].index))} empty rows in notes.\")\n print(f\"There are {len(list(eventsnotes[eventsnotes['CLEAN_TEXT'] == ''].index))} empty rows in eventsnotes.\")\n \n X = df['data'][0]\n y = np.array(df['data'][1])\n N = list(notes.CLEAN_TEXT)\n EN = list(eventsnotes.CLEAN_TEXT)\n\n # check if all three data sets have the same size/length\n assert len(X) == len(y) == len(N) == len(EN)\n\n empty_ind_N = list(notes[notes['CLEAN_TEXT'] == ''].index)\n empty_ind_EN = list(notes[eventsnotes['CLEAN_TEXT'] == ''].index)\n N_ = np.array(N)\n EN_ = np.array(EN)\n\n mask = np.ones(len(notes), np.bool)\n mask[empty_ind_N] = 0\n mask[empty_ind_EN] = 0\n \n good_notes = N_[mask]\n good_eventsnotes = EN_[mask]\n good_X = X[mask]\n good_y = y[mask]\n\n print(f\"Final shapes = {good_X.shape, good_y.shape, good_notes.shape}\")\n\n data = {'inputs': good_X,\n 'labels': good_y,\n 'eventsnotes': good_eventsnotes,\n 'notes': good_notes}\n\n # save full data\n filename = f'./new_{dataset}_CNEP'\n #full_data.to_csv(filename + '.csv', index = None)\n\n with open(filename + '.pickle', 'wb') as handle:\n pickle.dump(data, handle, protocol=pickle.HIGHEST_PROTOCOL)\n \n print(\"finished.\\n\")",
"_____no_output_____"
],
[
"all_datasets = ['train_data', 'test_data', 'val_data']\n\nfor dataset in all_datasets:\n print(f\"\\n\\nProcessing dataset {dataset}.\")\n mapNotes(dataset)\n combineData(dataset)\n ",
"\n\nProcessing dataset train_data.\nMapping notes on train_data.\n15392 0 0\n15392 15392 15392\nFinished mapping notes on train_data.\n\nCombining data for all train_data.\ndict_keys(['data', 'names']) 2 15392 (15392, 48, 390) 15392 15392\nThere are 374 empty rows in notes.\nThere are 680 empty rows in eventsnotes.\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
d093abbb549f6522458c844a9e63573c58c28a16 | 268,468 | ipynb | Jupyter Notebook | course/.ipynb_checkpoints/8 Recurrent Neural Networks-checkpoint.ipynb | ResitKadir1/aws-DL | 280e26daff56fe3350dbf47674200ecbf41c6dd3 | [
"MIT"
] | null | null | null | course/.ipynb_checkpoints/8 Recurrent Neural Networks-checkpoint.ipynb | ResitKadir1/aws-DL | 280e26daff56fe3350dbf47674200ecbf41c6dd3 | [
"MIT"
] | null | null | null | course/.ipynb_checkpoints/8 Recurrent Neural Networks-checkpoint.ipynb | ResitKadir1/aws-DL | 280e26daff56fe3350dbf47674200ecbf41c6dd3 | [
"MIT"
] | null | null | null | 149.481069 | 39,416 | 0.85523 | [
[
[
"# Recurrent Neural Networks",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\n%matplotlib inline\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"## Time series forecasting",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv('../data/cansim-0800020-eng-6674700030567901031.csv',\n skiprows=6, skipfooter=9,\n engine='python')\ndf.head()",
"_____no_output_____"
],
[
"from pandas.tseries.offsets import MonthEnd",
"_____no_output_____"
],
[
"df['Adjustments'] = pd.to_datetime(df['Adjustments']) + MonthEnd(1)\ndf = df.set_index('Adjustments')\ndf.head()",
"_____no_output_____"
],
[
"df.plot()",
"_____no_output_____"
],
[
"split_date = pd.Timestamp('01-01-2011')",
"_____no_output_____"
],
[
"train = df.loc[:split_date, ['Unadjusted']]\ntest = df.loc[split_date:, ['Unadjusted']]",
"_____no_output_____"
],
[
"ax = train.plot()\ntest.plot(ax=ax)\nplt.legend(['train', 'test'])",
"_____no_output_____"
],
[
"from sklearn.preprocessing import MinMaxScaler\n\nsc = MinMaxScaler()\n#not fitting our test (important)\ntrain_sc = sc.fit_transform(train)\ntest_sc = sc.transform(test)",
"_____no_output_____"
],
[
"train_sc[:4]",
"_____no_output_____"
],
[
"#model learn from previous value such as 0. will learn from 0.01402033\nX_train = train_sc[:-1]\ny_train = train_sc[1:]\n\nX_test = test_sc[:-1]\ny_test = test_sc[1:]",
"_____no_output_____"
]
],
[
[
"### Fully connected predictor",
"_____no_output_____"
]
],
[
[
"from tensorflow.keras.models import Sequential\nfrom tensorflow.keras.layers import Dense\nimport tensorflow.keras.backend as K\nfrom tensorflow.keras.callbacks import EarlyStopping",
"_____no_output_____"
],
[
"K.clear_session()\n\nmodel = Sequential()\nmodel.add(Dense(12, #12 nodes\n input_dim=1, #one input \n activation='relu'))\nmodel.add(Dense(1)) #one output ,no activation function this is regression problem\nmodel.compile(loss='mean_squared_error', optimizer='adam')\nmodel.summary()",
"Model: \"sequential\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense (Dense) (None, 12) 24 \n_________________________________________________________________\ndense_1 (Dense) (None, 1) 13 \n=================================================================\nTotal params: 37\nTrainable params: 37\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"early_stop = EarlyStopping(monitor='loss', patience=1, verbose=1)",
"_____no_output_____"
],
[
"model.fit(X_train, y_train, epochs=200,\n batch_size=2, verbose=1,\n callbacks=[early_stop])",
"Epoch 1/200\n120/120 [==============================] - 0s 2ms/step - loss: 0.0100\nEpoch 2/200\n120/120 [==============================] - 0s 2ms/step - loss: 0.0101\nEpoch 3/200\n120/120 [==============================] - 0s 2ms/step - loss: 0.0100\nEpoch 4/200\n120/120 [==============================] - 0s 2ms/step - loss: 0.0102\nEpoch 5/200\n120/120 [==============================] - 0s 2ms/step - loss: 0.0100\nEpoch 6/200\n120/120 [==============================] - 1s 5ms/step - loss: 0.0099\nEpoch 7/200\n120/120 [==============================] - 0s 3ms/step - loss: 0.0102\nEpoch 8/200\n120/120 [==============================] - 0s 3ms/step - loss: 0.0101\nEpoch 9/200\n120/120 [==============================] - 0s 4ms/step - loss: 0.0099\nEpoch 10/200\n120/120 [==============================] - 0s 3ms/step - loss: 0.0099\nEpoch 11/200\n120/120 [==============================] - 1s 5ms/step - loss: 0.0100\nEpoch 12/200\n120/120 [==============================] - 0s 2ms/step - loss: 0.0100\nEpoch 13/200\n120/120 [==============================] - 0s 2ms/step - loss: 0.0101\nEpoch 14/200\n120/120 [==============================] - 0s 2ms/step - loss: 0.0100\nEpoch 15/200\n120/120 [==============================] - 0s 1ms/step - loss: 0.0101\nEpoch 16/200\n120/120 [==============================] - 0s 1ms/step - loss: 0.0099\nEpoch 17/200\n120/120 [==============================] - 0s 1ms/step - loss: 0.0101\nEpoch 18/200\n120/120 [==============================] - 0s 1ms/step - loss: 0.0100\nEpoch 19/200\n120/120 [==============================] - 0s 1ms/step - loss: 0.0097\nEpoch 20/200\n120/120 [==============================] - 0s 1ms/step - loss: 0.0101\nEpoch 21/200\n120/120 [==============================] - 0s 1ms/step - loss: 0.0100\nEpoch 22/200\n120/120 [==============================] - 0s 1ms/step - loss: 0.0099\nEpoch 23/200\n120/120 [==============================] - 0s 1ms/step - loss: 0.0102\nEpoch 24/200\n120/120 [==============================] - 0s 1ms/step - loss: 0.0102\nEpoch 25/200\n120/120 [==============================] - 0s 1ms/step - loss: 0.0099\nEpoch 26/200\n120/120 [==============================] - 0s 1ms/step - loss: 0.0100\nEpoch 27/200\n120/120 [==============================] - 0s 1ms/step - loss: 0.0101\nEpoch 28/200\n120/120 [==============================] - 0s 1ms/step - loss: 0.0100\nEpoch 29/200\n120/120 [==============================] - 0s 2ms/step - loss: 0.0099\nEpoch 00029: early stopping\n"
],
[
"y_pred = model.predict(X_test)",
"_____no_output_____"
],
[
"plt.plot(y_test)\nplt.plot(y_pred)\n#fully connected\n#very bad performance , our model just learnt to mirroring",
"_____no_output_____"
]
],
[
[
"### Recurrent predictor",
"_____no_output_____"
]
],
[
[
"from tensorflow.keras.layers import LSTM",
"_____no_output_____"
],
[
"X_train.shape",
"_____no_output_____"
],
[
"#3D tensor with shape (batch_size, timesteps, input_dim)\nX_train[:, None].shape",
"_____no_output_____"
],
[
"X_train_t = X_train[:, None]\nX_test_t = X_test[:, None]",
"_____no_output_____"
],
[
"K.clear_session()\nmodel = Sequential()\n\nmodel.add(LSTM(6, \n input_shape=(1, 1)#1 timestep ,1 number\n ))\n\nmodel.add(Dense(1))\n\nmodel.compile(loss='mean_squared_error', optimizer='adam')",
"_____no_output_____"
],
[
"model.fit(X_train_t, y_train,\n epochs=100, \n batch_size=1 #training each data point\n , verbose=1,\n callbacks=[early_stop])",
"Epoch 1/100\n239/239 [==============================] - 2s 4ms/step - loss: 0.0736A: 0s - loss:\nEpoch 2/100\n239/239 [==============================] - 1s 2ms/step - loss: 0.0263\nEpoch 3/100\n239/239 [==============================] - 1s 3ms/step - loss: 0.0200\nEpoch 4/100\n239/239 [==============================] - 1s 2ms/step - loss: 0.0153\nEpoch 5/100\n239/239 [==============================] - 1s 2ms/step - loss: 0.0125\nEpoch 6/100\n239/239 [==============================] - 1s 2ms/step - loss: 0.0110\nEpoch 7/100\n239/239 [==============================] - 1s 3ms/step - loss: 0.0104\nEpoch 8/100\n239/239 [==============================] - 1s 4ms/step - loss: 0.0101A: 0s - loss: 0.\nEpoch 9/100\n239/239 [==============================] - 1s 4ms/step - loss: 0.0102A: 0s - loss: 0.\nEpoch 10/100\n239/239 [==============================] - 1s 2ms/step - loss: 0.0101\nEpoch 11/100\n239/239 [==============================] - 1s 2ms/step - loss: 0.0101\nEpoch 12/100\n239/239 [==============================] - 1s 2ms/step - loss: 0.0101\nEpoch 13/100\n239/239 [==============================] - 1s 3ms/step - loss: 0.0101\nEpoch 14/100\n239/239 [==============================] - 1s 3ms/step - loss: 0.0099\nEpoch 15/100\n239/239 [==============================] - 1s 4ms/step - loss: 0.0099\nEpoch 16/100\n239/239 [==============================] - 1s 3ms/step - loss: 0.0100\nEpoch 17/100\n239/239 [==============================] - 1s 5ms/step - loss: 0.0099\nEpoch 18/100\n239/239 [==============================] - 1s 3ms/step - loss: 0.0101\nEpoch 19/100\n239/239 [==============================] - 1s 3ms/step - loss: 0.0099\nEpoch 20/100\n239/239 [==============================] - 1s 3ms/step - loss: 0.0102\nEpoch 21/100\n239/239 [==============================] - 1s 3ms/step - loss: 0.0101\nEpoch 22/100\n239/239 [==============================] - 1s 3ms/step - loss: 0.0100\nEpoch 23/100\n239/239 [==============================] - 1s 4ms/step - loss: 0.0100\nEpoch 24/100\n239/239 [==============================] - 1s 4ms/step - loss: 0.0100\nEpoch 00024: early stopping\n"
],
[
"y_pred = model.predict(X_test_t)\nplt.plot(y_test)\nplt.plot(y_pred)\n#unfortunately LSTM didnt improve model performance\n#now we try Windows",
"_____no_output_____"
]
],
[
[
"## Windows",
"_____no_output_____"
]
],
[
[
"train_sc.shape",
"_____no_output_____"
],
[
"train_sc_df = pd.DataFrame(train_sc, columns=['Scaled'], index=train.index)\ntest_sc_df = pd.DataFrame(test_sc, columns=['Scaled'], index=test.index)\ntrain_sc_df.head()",
"_____no_output_____"
],
[
"#12 months back shifts\nfor s in range(1, 13):\n train_sc_df['shift_{}'.format(s)] = train_sc_df['Scaled'].shift(s)\n test_sc_df['shift_{}'.format(s)] = test_sc_df['Scaled'].shift(s)",
"_____no_output_____"
],
[
"train_sc_df.head(13)",
"_____no_output_____"
],
[
"#drop null data which mean drop first year\nX_train = train_sc_df.dropna().drop('Scaled', axis=1)\ny_train = train_sc_df.dropna()[['Scaled']]\n\nX_test = test_sc_df.dropna().drop('Scaled', axis=1)\ny_test = test_sc_df.dropna()[['Scaled']]",
"_____no_output_____"
],
[
"X_train.head()",
"_____no_output_____"
],
[
"X_train.shape",
"_____no_output_____"
],
[
"X_train = X_train.values\nX_test= X_test.values\n\ny_train = y_train.values\ny_test = y_test.values",
"_____no_output_____"
]
],
[
[
"### Fully Connected on Windows",
"_____no_output_____"
]
],
[
[
"K.clear_session()\n\nmodel = Sequential()\nmodel.add(Dense(12, input_dim=12, activation='relu'))\nmodel.add(Dense(1))\nmodel.compile(loss='mean_squared_error', optimizer='adam')\nmodel.summary()",
"Model: \"sequential\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense (Dense) (None, 12) 156 \n_________________________________________________________________\ndense_1 (Dense) (None, 1) 13 \n=================================================================\nTotal params: 169\nTrainable params: 169\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"model.fit(X_train, y_train, epochs=200,\n batch_size=1, verbose=1, callbacks=[early_stop])",
"Epoch 1/200\n228/228 [==============================] - 1s 2ms/step - loss: 0.0329\nEpoch 2/200\n228/228 [==============================] - 0s 2ms/step - loss: 0.0110\nEpoch 3/200\n228/228 [==============================] - 0s 1ms/step - loss: 0.0087\nEpoch 4/200\n228/228 [==============================] - 0s 1ms/step - loss: 0.0071\nEpoch 5/200\n228/228 [==============================] - 0s 1ms/step - loss: 0.0060\nEpoch 6/200\n228/228 [==============================] - 0s 1ms/step - loss: 0.0053\nEpoch 7/200\n228/228 [==============================] - 0s 1ms/step - loss: 0.0046\nEpoch 8/200\n228/228 [==============================] - 0s 1ms/step - loss: 0.0039\nEpoch 9/200\n228/228 [==============================] - 1s 3ms/step - loss: 0.0034\nEpoch 10/200\n228/228 [==============================] - 1s 2ms/step - loss: 0.0028\nEpoch 11/200\n228/228 [==============================] - 0s 1ms/step - loss: 0.0024\nEpoch 12/200\n228/228 [==============================] - 0s 1ms/step - loss: 0.0023\nEpoch 13/200\n228/228 [==============================] - 0s 1ms/step - loss: 0.0020\nEpoch 14/200\n228/228 [==============================] - 0s 2ms/step - loss: 0.0020\nEpoch 00014: early stopping\n"
],
[
"y_pred = model.predict(X_test)\nplt.plot(y_test)\nplt.plot(y_pred)\n\n#this model way more better than precious models\n#our expectataion lines overlapping",
"WARNING:tensorflow:5 out of the last 13 calls to <function Model.make_predict_function.<locals>.predict_function at 0x7fde7bc085f0> triggered tf.function retracing. Tracing is expensive and the excessive number of tracings could be due to (1) creating @tf.function repeatedly in a loop, (2) passing tensors with different shapes, (3) passing Python objects instead of tensors. For (1), please define your @tf.function outside of the loop. For (2), @tf.function has experimental_relax_shapes=True option that relaxes argument shapes that can avoid unnecessary retracing. For (3), please refer to https://www.tensorflow.org/guide/function#controlling_retracing and https://www.tensorflow.org/api_docs/python/tf/function for more details.\n"
]
],
[
[
"### LSTM on Windows",
"_____no_output_____"
]
],
[
[
"X_train_t = X_train.reshape(X_train.shape[0], 1, 12)\nX_test_t = X_test.reshape(X_test.shape[0], 1, 12)",
"_____no_output_____"
],
[
"X_train_t.shape #one time instance with 12 vector coordinates",
"_____no_output_____"
],
[
"K.clear_session()\nmodel = Sequential()\n\nmodel.add(LSTM(6, input_shape=(1, 12)#our shaped dimensions,parameter 6*12*3?\n ))\n\nmodel.add(Dense(1))\n\nmodel.compile(loss='mean_squared_error', optimizer='adam')",
"_____no_output_____"
],
[
"model.summary()",
"Model: \"sequential\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nlstm (LSTM) (None, 6) 456 \n_________________________________________________________________\ndense (Dense) (None, 1) 7 \n=================================================================\nTotal params: 463\nTrainable params: 463\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"model.fit(X_train_t, y_train, epochs=100,\n batch_size=1, verbose=1, callbacks=[early_stop])",
"Epoch 1/100\n228/228 [==============================] - 2s 2ms/step - loss: 0.0190\nEpoch 2/100\n228/228 [==============================] - 0s 2ms/step - loss: 0.0075\nEpoch 3/100\n228/228 [==============================] - 0s 2ms/step - loss: 0.0066\nEpoch 4/100\n228/228 [==============================] - 0s 2ms/step - loss: 0.0057\nEpoch 5/100\n228/228 [==============================] - 0s 2ms/step - loss: 0.0046\nEpoch 6/100\n228/228 [==============================] - 1s 2ms/step - loss: 0.0035\nEpoch 7/100\n228/228 [==============================] - 1s 4ms/step - loss: 0.0029- ETA: 1s - \nEpoch 8/100\n228/228 [==============================] - 1s 3ms/step - loss: 0.0023\nEpoch 9/100\n228/228 [==============================] - 0s 2ms/step - loss: 0.0021\nEpoch 10/100\n228/228 [==============================] - 1s 2ms/step - loss: 0.0018\nEpoch 11/100\n228/228 [==============================] - 1s 2ms/step - loss: 0.0019\nEpoch 00011: early stopping\n"
],
[
"y_pred = model.predict(X_test_t)\nplt.plot(y_test)\nplt.plot(y_pred)\n#best model",
"WARNING:tensorflow:5 out of the last 12 calls to <function Model.make_predict_function.<locals>.predict_function at 0x7fde7ca8ad40> triggered tf.function retracing. Tracing is expensive and the excessive number of tracings could be due to (1) creating @tf.function repeatedly in a loop, (2) passing tensors with different shapes, (3) passing Python objects instead of tensors. For (1), please define your @tf.function outside of the loop. For (2), @tf.function has experimental_relax_shapes=True option that relaxes argument shapes that can avoid unnecessary retracing. For (3), please refer to https://www.tensorflow.org/guide/function#controlling_retracing and https://www.tensorflow.org/api_docs/python/tf/function for more details.\n"
]
],
[
[
"## Exercise 1\n\nIn the model above we reshaped the input shape to: `(num_samples, 1, 12)`, i.e. we treated a window of 12 months as a vector of 12 coordinates that we simultaneously passed to all the LSTM nodes. An alternative way to look at the problem is to reshape the input to `(num_samples, 12, 1)`. This means we consider each input window as a sequence of 12 values that we will pass in sequence to the LSTM. In principle this looks like a more accurate description of our situation. But does it yield better predictions? Let's check it.\n\n- Reshape `X_train` and `X_test` so that they represent a set of univariate sequences\n- retrain the same LSTM(6) model, you'll have to adapt the `input_shape`\n- check the performance of this new model, is it better at predicting the test data?",
"_____no_output_____"
],
[
"## Exercise 2\n\nRNN models can be applied to images too. In general we can apply them to any data where there's a connnection between nearby units. Let's see how we can easily build a model that works with images.\n\n- Load the MNIST data, by now you should be able to do it blindfolded :)\n- reshape it so that an image looks like a long sequence of pixels\n- create a recurrent model and train it on the training data\n- how does it perform compared to a fully connected? How does it compare to Convolutional Neural Networks?\n\n(feel free to run this exercise on a cloud GPU if it's too slow on your laptop)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
d093b4a98f514ace0fa5876b5be313b768f304b3 | 99,533 | ipynb | Jupyter Notebook | notebooks/handwritten_digits_tunning.ipynb | dgavieira/handwritten-digits-recognition-app | f4d59eac8ee36545e9bdfbe92ba3e63b67f2e4af | [
"Apache-2.0"
] | null | null | null | notebooks/handwritten_digits_tunning.ipynb | dgavieira/handwritten-digits-recognition-app | f4d59eac8ee36545e9bdfbe92ba3e63b67f2e4af | [
"Apache-2.0"
] | null | null | null | notebooks/handwritten_digits_tunning.ipynb | dgavieira/handwritten-digits-recognition-app | f4d59eac8ee36545e9bdfbe92ba3e63b67f2e4af | [
"Apache-2.0"
] | null | null | null | 149.224888 | 38,378 | 0.854169 | [
[
[
"# Handwritten Digits Classifier with Improved Accuracy using Data Augmentation",
"_____no_output_____"
],
[
"In previous steps, we trained a model that could recognize handwritten digits using the MNIST dataset. We were able to achieve above 98% accuracy on our validation dataset. However, when you deploy the model in an Android app and test it, you probably noticed some accuracy issue. Although the app was able to recognize digits that you drew, the accuracy is probably way lower than 98%.\n\nIn this notebook we will explore the couse of the accuracy drop and use data augmentation to improve deployment accuracy.",
"_____no_output_____"
],
[
"## Preparation\n\nLet's start by importing TensorFlow and other supporting libraries that are used for data processing and visualization.",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nfrom tensorflow import keras\n\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\nimport random\n\nprint(tf.__version__)",
"2.6.0\n"
]
],
[
[
"Import the MNIST dataset",
"_____no_output_____"
]
],
[
[
"mnist = keras.datasets.mnist\n(train_images, train_labels), (test_images, test_labels) = mnist.load_data()\n\n# Normalize the input image so that each pixel value is between 0 to 1.\ntrain_images = train_images / 255.0\ntest_images = test_images / 255.0\n\n# Add a color dimension to the images in \"train\" and \"validate\" dataset to\n# leverage Keras's data augmentation utilities later.\ntrain_images = np.expand_dims(train_images, axis=3)\ntest_images = np.expand_dims(test_images, axis=3)",
"Downloading data from https://storage.googleapis.com/tensorflow/tf-keras-datasets/mnist.npz\n11493376/11490434 [==============================] - 0s 0us/step\n11501568/11490434 [==============================] - 0s 0us/step\n"
]
],
[
[
"Define an utility function so that we can create quickly create multiple models with the same model architecture for comparison.\n",
"_____no_output_____"
]
],
[
[
"def create_model():\n model = keras.Sequential(\n [\n keras.layers.InputLayer(input_shape=(28,28,1)),\n keras.layers.Conv2D(filters=32, kernel_size=(3,3), activation=tf.nn.relu),\n keras.layers.Conv2D(filters=64, kernel_size=(3,3), activation=tf.nn.relu),\n keras.layers.MaxPooling2D(pool_size=(2,2)),\n keras.layers.Dropout(0.25),\n keras.layers.Flatten(),\n keras.layers.Dense(10, activation=tf.nn.softmax)\n ]\n )\n model.compile(optimizer='adam',\n loss='sparse_categorical_crossentropy',\n metrics=['accuracy'])\n return model",
"_____no_output_____"
]
],
[
[
"Confirm that our model can achieve above 98% accuracy on MNIST Dataset.",
"_____no_output_____"
]
],
[
[
"base_model = create_model()\nbase_model.fit(\n train_images,\n train_labels,\n epochs=5,\n validation_data=(test_images, test_labels)\n)",
"Epoch 1/5\n1875/1875 [==============================] - 43s 7ms/step - loss: 0.1418 - accuracy: 0.9569 - val_loss: 0.0580 - val_accuracy: 0.9815\nEpoch 2/5\n1875/1875 [==============================] - 12s 7ms/step - loss: 0.0544 - accuracy: 0.9836 - val_loss: 0.0515 - val_accuracy: 0.9829\nEpoch 3/5\n1875/1875 [==============================] - 13s 7ms/step - loss: 0.0414 - accuracy: 0.9869 - val_loss: 0.0385 - val_accuracy: 0.9875\nEpoch 4/5\n1875/1875 [==============================] - 12s 7ms/step - loss: 0.0324 - accuracy: 0.9900 - val_loss: 0.0362 - val_accuracy: 0.9891\nEpoch 5/5\n1875/1875 [==============================] - 12s 7ms/step - loss: 0.0267 - accuracy: 0.9913 - val_loss: 0.0373 - val_accuracy: 0.9884\n"
]
],
[
[
"## Troubleshoot the accuracy drop\n\nLet's see the digit images in MNIST again and guess the cause of the accuracy drop we experienced in deployment.",
"_____no_output_____"
]
],
[
[
"# Show the first 25 images in the training dataset.\nplt.figure(figsize=(10,10))\nfor i in range(25):\n plt.subplot(5,5,i+1)\n plt.xticks([])\n plt.yticks([])\n plt.grid(False)\n plt.imshow(np.squeeze(train_images[i], axis=2), cmap=plt.cm.gray)\n plt.xlabel(train_labels[i])\nplt.show()",
"_____no_output_____"
]
],
[
[
"We can see from the 25 images above that the digits are about the same size, and they are in the center of the images. Let's verify if this assumption is true across the MNIST dataset.",
"_____no_output_____"
]
],
[
[
"# An utility function that returns where the digit is in the image.\ndef digit_area(mnist_image):\n # Remove the color axes\n mnist_image = np.squeeze(mnist_image, axis=2) \n\n # Extract the list of columns that contain at least 1 pixel from the digit\n x_nonzero = np.nonzero(np.amax(mnist_image, 0))\n x_min = np.min(x_nonzero)\n x_max = np.max(x_nonzero)\n\n # Extract the list of rows that contain at least 1 pixel from the digit\n y_nonzero = np.nonzero(np.amax(mnist_image, 1))\n y_min = np.min(y_nonzero)\n y_max = np.max(y_nonzero)\n\n return [x_min, x_max, y_min, y_max]\n\n# Calculate the area containing the digit across MNIST dataset\ndigit_area_rows = []\nfor image in train_images:\n digit_area_row = digit_area(image)\n digit_area_rows.append(digit_area_row)\ndigit_area_df = pd.DataFrame(\n digit_area_rows,\n columns=['x_min', 'x_max', 'y_min', 'y_max']\n)\ndigit_area_df.hist()",
"_____no_output_____"
]
],
[
[
"Now for the histogram, you can confirm that the digit in MNIST images are fitted nicely in an certain area at the center of the images.\n\n[MNIST Range](https://download.tensorflow.org/models/tflite/digit_classifier/mnist_range.png)\n\nHowever, when you wrote digits in your Android app, you probably did not pay attention to make sure your digit fit in the virtual area that the digits appear in MNIST dataset. The Machine Learning Model have not seen such data before so it performed poorly, especially when you wrote a a digit that was off the center of the drawing pad.\n\nLet's add some data augmentation to the MNIST dataset to verify if our assumption is true. We will distort our MNIST dataset by adding:\n* Rotation\n* Width and height shift\n* Shear\n* Zoom",
"_____no_output_____"
]
],
[
[
"# Define data augmentation\n\ndatagen = keras.preprocessing.image.ImageDataGenerator(\n rotation_range=30,\n width_shift_range=0.25,\n height_shift_range=0.25,\n shear_range=0.25,\n zoom_range=0.2\n)\n\n# Generate augmented data from MNIST dataset\ntrain_generator = datagen.flow(train_images, train_labels)\ntest_generator = datagen.flow(test_images, test_labels)",
"_____no_output_____"
]
],
[
[
"Let's see what our digit images look like after augmentation. You can see that we now clearly have much more variation on how the digits are placed in the images.",
"_____no_output_____"
]
],
[
[
"augmented_images, augmented_labels = next(train_generator)\nplt.figure(figsize=(10,10))\nfor i in range(25):\n plt.subplot(5, 5, i+1)\n plt.xticks([])\n plt.yticks([])\n plt.grid(False)\n plt.imshow(np.squeeze(augmented_images[i], axis=2), cmap=plt.cm.gray)\n plt.xlabel('Label: %d' % augmented_labels[i])\nplt.show()",
"_____no_output_____"
]
],
[
[
"Let's evaluate the digit classifier model that we trained earlier on this augmented test dataset and see if it makes accuracy drop.",
"_____no_output_____"
]
],
[
[
"base_model.evaluate(test_generator)",
"313/313 [==============================] - 6s 18ms/step - loss: 5.6356 - accuracy: 0.3272\n"
]
],
[
[
"You can see that accuracy significantly dropped to below 40% in augmented test dataset.",
"_____no_output_____"
],
[
"## Improve accuracy with data augmentation\n\nNow let's train our model using augmented dataset to make it perform better in deployment.",
"_____no_output_____"
]
],
[
[
"improved_model = create_model()\nimproved_model.fit(train_generator, epochs=5, validation_data=test_generator)",
"Epoch 1/5\n1875/1875 [==============================] - 41s 22ms/step - loss: 0.9860 - accuracy: 0.6849 - val_loss: 0.5181 - val_accuracy: 0.8516\nEpoch 2/5\n1875/1875 [==============================] - 40s 22ms/step - loss: 0.5070 - accuracy: 0.8491 - val_loss: 0.3765 - val_accuracy: 0.8877\nEpoch 3/5\n1875/1875 [==============================] - 41s 22ms/step - loss: 0.4145 - accuracy: 0.8758 - val_loss: 0.3064 - val_accuracy: 0.9102\nEpoch 4/5\n1875/1875 [==============================] - 41s 22ms/step - loss: 0.3671 - accuracy: 0.8896 - val_loss: 0.2749 - val_accuracy: 0.9219\nEpoch 5/5\n1875/1875 [==============================] - 41s 22ms/step - loss: 0.3275 - accuracy: 0.9017 - val_loss: 0.2593 - val_accuracy: 0.9249\n"
]
],
[
[
"We can see that as the models saw more distorted digit images during training, its accuracy evaluated distorted test digit images were significantly improved to about 90%.",
"_____no_output_____"
],
[
"## Convert to TensorFlow Lite\n\nLet's convert the improved model to TensorFlow Lite and redeploy the Android app.",
"_____no_output_____"
]
],
[
[
"# Convert Keras model to TF Lite format and quantize.\n\nconverter = tf.lite.TFLiteConverter.from_keras_model(improved_model)\nconverter.optimizations = [tf.lite.Optimize.DEFAULT]\ntflite_quantized_model = converter.convert()\n\n# Save the Quantized Model to file to the Downloads Directory\nf = open('mnist-improved.tflite', \"wb\")\nf.write(tflite_quantized_model)\nf.close()\n\n# Download the digit classification model\nfrom google.colab import files\nfiles.download('mnist-improved.tflite')",
"INFO:tensorflow:Assets written to: /tmp/tmpm1ypiyxg/assets\n"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
d093b7ce620ae99233856eec6c3565d60d57f972 | 4,218 | ipynb | Jupyter Notebook | 1-1.NNLM/NNLM_Tensor.ipynb | wurikiji/nlp-tutorial | 05706149dbf2056a74bbd7b983eb7610e4ba70bd | [
"MIT"
] | 6 | 2019-06-13T17:50:33.000Z | 2021-08-20T03:13:53.000Z | 1-1.NNLM/NNLM_Tensor.ipynb | Matafight/nlp-tutorial | f53f08079c2b7b1a897592ef1569df7ca240c07e | [
"MIT"
] | null | null | null | 1-1.NNLM/NNLM_Tensor.ipynb | Matafight/nlp-tutorial | f53f08079c2b7b1a897592ef1569df7ca240c07e | [
"MIT"
] | 4 | 2019-07-24T01:57:41.000Z | 2021-01-08T03:42:06.000Z | 36.678261 | 122 | 0.486486 | [
[
[
"# code by Tae Hwan Jung @graykode\nimport tensorflow as tf\nimport numpy as np\n\ntf.reset_default_graph()\n\nsentences = [ \"i like dog\", \"i love coffee\", \"i hate milk\"]\n\nword_list = \" \".join(sentences).split()\nword_list = list(set(word_list))\nword_dict = {w: i for i, w in enumerate(word_list)}\nnumber_dict = {i: w for i, w in enumerate(word_list)}\nn_class = len(word_dict) # number of Vocabulary\n\n# NNLM Parameter\nn_step = 2 # number of steps ['i like', 'i love', 'i hate']\nn_hidden = 2 # number of hidden units\n\ndef make_batch(sentences):\n input_batch = []\n target_batch = []\n\n for sen in sentences:\n word = sen.split()\n input = [word_dict[n] for n in word[:-1]]\n target = word_dict[word[-1]]\n\n input_batch.append(np.eye(n_class)[input])\n target_batch.append(np.eye(n_class)[target])\n\n return input_batch, target_batch\n\n# Model\nX = tf.placeholder(tf.float32, [None, n_step, n_class]) # [batch_size, number of steps, number of Vocabulary]\nY = tf.placeholder(tf.float32, [None, n_class])\n\ninput = tf.reshape(X, shape=[-1, n_step * n_class]) # [batch_size, n_step * n_class]\nH = tf.Variable(tf.random_normal([n_step * n_class, n_hidden]))\nd = tf.Variable(tf.random_normal([n_hidden]))\nU = tf.Variable(tf.random_normal([n_hidden, n_class]))\nb = tf.Variable(tf.random_normal([n_class]))\n\ntanh = tf.nn.tanh(d + tf.matmul(input, H)) # [batch_size, n_hidden]\nmodel = tf.matmul(tanh, U) + b # [batch_size, n_class]\n\ncost = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits_v2(logits=model, labels=Y))\noptimizer = tf.train.AdamOptimizer(0.001).minimize(cost)\nprediction =tf.argmax(model, 1)\n\n# Training\ninit = tf.global_variables_initializer()\nsess = tf.Session()\nsess.run(init)\n\ninput_batch, target_batch = make_batch(sentences)\n\nfor epoch in range(5000):\n _, loss = sess.run([optimizer, cost], feed_dict={X: input_batch, Y: target_batch})\n if (epoch + 1)%1000 == 0:\n print('Epoch:', '%04d' % (epoch + 1), 'cost =', '{:.6f}'.format(loss))\n\n# Predict\npredict = sess.run([prediction], feed_dict={X: input_batch})\n\n# Test\ninput = [sen.split()[:2] for sen in sentences]\nprint([sen.split()[:2] for sen in sentences], '->', [number_dict[n] for n in predict[0]])",
"Epoch: 1000 cost = 0.468996\nEpoch: 2000 cost = 0.125292\nEpoch: 3000 cost = 0.045533\nEpoch: 4000 cost = 0.020888\nEpoch: 5000 cost = 0.010938\n[['i', 'like'], ['i', 'love'], ['i', 'hate']] -> ['dog', 'coffee', 'milk']\n"
]
]
] | [
"code"
] | [
[
"code"
]
] |
d093b85d925aacefee767942cae3e90116335a5e | 7,790 | ipynb | Jupyter Notebook | nltk_treinar_etiquetador.ipynb | ilexistools/ebralc2021 | 76a63117554e3df4f9b3204223d62f23fb107650 | [
"MIT"
] | null | null | null | nltk_treinar_etiquetador.ipynb | ilexistools/ebralc2021 | 76a63117554e3df4f9b3204223d62f23fb107650 | [
"MIT"
] | null | null | null | nltk_treinar_etiquetador.ipynb | ilexistools/ebralc2021 | 76a63117554e3df4f9b3204223d62f23fb107650 | [
"MIT"
] | null | null | null | 35.248869 | 643 | 0.556226 | [
[
[
"<a href=\"https://colab.research.google.com/github/ilexistools/ebralc2021/blob/main/nltk_treinar_etiquetador.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"# Treinar um etiquetador morfossintático\n\nPara realizar a etiquetagem morfossintática de textos, é preciso obter um etiquetador. O NLTK possui diversas opções para a criação de classificadores e etiquetadores de palavras: DefaultTagger, RegexpTagger, UnigramTagger, BigramTagger, TrigramTagger, BrillTagger, além de outros classificadores.\n\nA criação de etiquetadores requer dados de treinamento, textos previamente etiquetados, no formato de sentenças etiquetadas, especificamente em listas de tuplas para o NLTK. A partir dos dados e algoritmos de treinamento, cria-se um objeto (etiquetador) que pode ser armazenado para usos futuros, uma vez que o treinamento leva um tempo considerável.",
"_____no_output_____"
],
[
"# Recursos\n\nPara realizar o teste de utilização, precisamos carregar o corpus mc-morpho e o tokenizador 'punkt' da biblioteca do nltk:\n\n",
"_____no_output_____"
]
],
[
[
"import nltk\n\nnltk.download('mac_morpho')\nnltk.download('punkt')",
"[nltk_data] Downloading package mac_morpho to /root/nltk_data...\n[nltk_data] Package mac_morpho is already up-to-date!\n[nltk_data] Downloading package punkt to /root/nltk_data...\n[nltk_data] Unzipping tokenizers/punkt.zip.\n"
],
[
"",
"_____no_output_____"
],
[
"import nltk\nimport pickle\nfrom nltk.corpus import mac_morpho\n\n# prepara dados de treinamento e teste\nsents = mac_morpho.tagged_sents()\ntrein = sents[0:30000]\nteste = sents[13000:]\n\n# treina um etiquetador sequencial\netq1 = nltk.DefaultTagger('N')\netq2 = nltk.UnigramTagger(trein,backoff=etq1)\netq3 = nltk.BigramTagger(trein,backoff=etq2)\n\n# imprime a acurácia dos etiquetadores\nprint('DefaultTagger', etq1.evaluate(teste))\nprint('UnigramTagger', etq2.evaluate(teste))\nprint('BigramTagger', etq3.evaluate(teste))\n\n# armazena o etiquetador treinado\nwith open('etq.pickle','wb') as fh:\n pickle.dump(etq3,fh)",
"DefaultTagger 0.20087708474599295\nUnigramTagger 0.8237940367746194\nBigramTagger 0.842816406510894\n"
]
],
[
[
"No exemplo, carregamos os dados etiquetados para treinamento e teste do etiquetador. Separamos uma quantidade maior de sentenças para treino (70%) e outra menor para teste (30%).\n\nEm seguida, iniciamos o processo de treinamento de um etiquetador sequencial a partir de três modelos diferentes combinados. O etiquetador 'DefaultTagger' atribui uma etiqueta padrão ('N') para todas as palavras. O etiquetador 'UnigramTagger', treinado com as sentenças, atribui a etiqueta mais provável para a palavra a partir de um dicionário criado internamente. O etiquetador 'BigramTagger', também treinado com as sentenças etiquetadas, atribui a etiqueta mais provável para a palavra com base na etiqueta anterior (hipótese de Markov). A combinação dos etiquetadores é feita sequencialmente por meio do argumento 'backoff''.\n\nTendo realizado o treinamento do etiquetador, avaliamos a precisão de cada etapa por meio da função ‘evaluate()’, passando como argumento a variável ‘teste’, que armazena parte das sentenças etiquetadas do corpus MacMorpho. No processo, obtemos a impressão do desempenho de cada um dos etiquetadores em separado. O etiquetador final, resultado da combinação de todos, obtém uma precisão de 84% com os dados de teste.\n\nPor fim, armazenamos o etiquetador treinado para usos posteriores por meio da função ‘dump’, do módulo ‘pickle’. \n\nPara verificar a funcionalidade do etiquetador, realizamos o seguinte teste:",
"_____no_output_____"
]
],
[
[
"import nltk\nimport pickle\n\n# carrega o etiquetador treinado\nwith open('etq.pickle','rb') as fh:\n etiquetador = pickle.load(fh)\n\n# armazena um texto a ser etiquetado como teste\ntexto = 'Estamos realizando um teste agora.'\n\n# itemiza o texto\nitens = nltk.word_tokenize(texto,language='portuguese')\n\n# etiqueta os itens\nitens_etiquetados = etiquetador.tag(itens)\n\n# imprime o resultado\nprint(itens_etiquetados)\n",
"[('Estamos', 'V'), ('realizando', 'V'), ('um', 'ART'), ('teste', 'N'), ('agora', 'ADV'), ('.', '.')]\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d093bacb37c6ca0dac5e87f0838006f2f6ec9ee7 | 30,447 | ipynb | Jupyter Notebook | Python_Graph/A simple graph with optimal pruning based routing.ipynb | phoophoo187/Privacy_SDN_Edge_IoT | 3ee6e0fb36c6d86cf8caf4599a35c04b0ade9a8c | [
"MIT"
] | null | null | null | Python_Graph/A simple graph with optimal pruning based routing.ipynb | phoophoo187/Privacy_SDN_Edge_IoT | 3ee6e0fb36c6d86cf8caf4599a35c04b0ade9a8c | [
"MIT"
] | null | null | null | Python_Graph/A simple graph with optimal pruning based routing.ipynb | phoophoo187/Privacy_SDN_Edge_IoT | 3ee6e0fb36c6d86cf8caf4599a35c04b0ade9a8c | [
"MIT"
] | null | null | null | 85.047486 | 20,884 | 0.805104 | [
[
[
"# An example of an optimal pruning based routing algorithm\n- based on a simple graph and Dijkstra's algorithm with concave cost function\n- Create a simple graph with multiple edge's attributes¶\n - weight = w_ij\n - concave = c_ij where i,j is nodes",
"_____no_output_____"
]
],
[
[
"import networkx as nx\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"## Define functions",
"_____no_output_____"
]
],
[
[
"def add_multi_link_attributes(G,attr1,attr2):\n \"\"\"\n This funtion is to add the multiple link attributes to graph G\n input: G : graph\n attr1 : link attribute 1\n attr2 : link attribute 2\n output : G\n \"\"\"\n i = 0\n for (u, v) in G.edges():\n G.add_edge(u,v,w=attr1[i],c=attr2[i])\n i = i+1 \n return G\n",
"_____no_output_____"
],
[
"def draw_graph(G,pos):\n \"\"\"\n This function is to draw a graph with the fixed position\n input : G : graph\n pos: postions of all nodes with the dictionary of coordinates (x,y)\n \"\"\"\n edge_labels = {} ## add edge lables from edge attribute\n for u, v, data in G.edges(data=True):\n edge_labels[u, v] = data\n\n nx.draw_networkx(G,pos)\n nx.draw_networkx_edge_labels(G,pos,edge_labels=edge_labels)",
"_____no_output_____"
],
[
"def remove_Edge(G,rm_edge_list):\n \"\"\"\n This function is to remove edges in the rm_edge_list from G\n \"\"\"\n G.remove_edges_from(rm_edge_list)\n G.edges()\n return G\n ",
"_____no_output_____"
],
[
"def compare_path(path1,path2):\n \n if collections.Counter(path1) == collections.Counter(path2):\n print (\"The lists l1 and l2 are the same\") \n flag = True\n else: \n print (\"The lists l1 and l2 are not the same\") \n flag = False\n return flag",
"_____no_output_____"
],
[
"def additive_path_cost(G, path, attr):\n \"\"\"\n This function is to find the path cost based on the additive costs\n : Path_Cost = sum_{edges in the path}attr[edge]\n Input : G : graph\n path : path is a list of nodes in the path\n attr : attribute of edges\n output : path_cost\n \"\"\"\n return sum([G[path[i]][path[i+1]][attr] for i in range(len(path)-1)])",
"_____no_output_____"
],
[
"## Calculate concave path cost from attr\ndef max_path_cost(G, path, attr):\n \"\"\"\n This function is to find the path cost based on the Concave costs\n : Path_Cost = max{edges in the path}attr[edge]\n Input : G : graph\n path : path is a list of nodes in the path\n attr : attribute of edges\n output : path_cost\n \"\"\"\n return max([G[path[i]][path[i+1]][attr] for i in range(len(path)-1)])",
"_____no_output_____"
],
[
"def rm_edge_constraint(G,Cons):\n rm_edge_list = []\n for u, v, data in G.edges(data=True):\n e = (u,v)\n cost = G.get_edge_data(*e)\n print(cost)\n if cost['c'] >= Cons:\n rm_edge_list.append(e)\n print(rm_edge_list)\n \n remove_Edge(G,rm_edge_list)\n\n return G",
"_____no_output_____"
],
[
"def has_path(G, source, target):\n \"\"\"Return True if G has a path from source to target, False otherwise.\n\n Parameters\n ----------\n G : NetworkX graph\n\n source : node\n Starting node for path\n\n target : node\n Ending node for path\n \"\"\"\n try:\n sp = nx.shortest_path(G,source, target)\n except nx.NetworkXNoPath:\n return False\n return True\n",
"_____no_output_____"
],
[
"def Optimum_prun_based_routing(G,S,D,L):\n \"\"\"\n This function is to find the optimal path from S to D with constraint L \n Input : G : graph\n S : Source\n D : Destination\n L : constraint\n \"\"\"\n if has_path(G, S, D):\n \n Shortest_path = nx.dijkstra_path(G, S, D, weight='w')\n Opt_path = Shortest_path\n while len(Shortest_path) != 0:\n path_cost = additive_path_cost(G, Shortest_path, 'w') \n print(path_cost)\n if path_cost <= L:\n \"\"\"go to concave cost\"\"\"\n PathConcave_cost = max_path_cost(G, Shortest_path, 'c')\n G = rm_edge_constraint(G,PathConcave_cost) # remove all links where the concave link is greater than PathConcave_cost\n \n Opt_path = Shortest_path\n if has_path(G, S, D):\n Shortest_path = nx.dijkstra_path(G, S, D, weight='w')\n else:\n Shortest_path = []\n \n else:\n pass \n else:\n print('No path from', S, ' to ', D)\n Opt_path = []\n return Opt_path\n \n \n ",
"_____no_output_____"
]
],
[
[
"## Create a graph",
"_____no_output_____"
]
],
[
[
"G = nx.Graph()\nedge_list = [('S', 'B'), ('S', 'A'), ('S','E'), ('B','A'), ('B','D'), ('A','D'), ('E','D')]\nWeight_edge_list = [2, 2, 3, 2, 1, 2, 2]\nConcave_edge_list = [1, 3, 3, 1, 4, 3, 1]\npos = { 'S': (0,50), 'B': (50, 100), 'A': (50, 50), 'E': (50, 0), 'D': (100, 50)} # draw by position\n\nG.add_edges_from(edge_list)\nG = add_multi_link_attributes(G,Weight_edge_list,Concave_edge_list)\ndraw_graph(G,pos)\n",
"_____no_output_____"
]
],
[
[
"## Run the optimum-pruning-based-routing algorithm",
"_____no_output_____"
]
],
[
[
"Optimum_prun_based_routing(G,'S','D',5)",
"3\n{'w': 2, 'c': 1}\n{'w': 2, 'c': 3}\n{'w': 3, 'c': 3}\n{'w': 2, 'c': 1}\n{'w': 1, 'c': 4}\n[('B', 'D')]\n{'w': 2, 'c': 3}\n{'w': 2, 'c': 1}\n4\n{'w': 2, 'c': 1}\n{'w': 2, 'c': 3}\n[('S', 'A')]\n{'w': 3, 'c': 3}\n[('S', 'A'), ('S', 'E')]\n{'w': 2, 'c': 1}\n{'w': 2, 'c': 3}\n[('S', 'A'), ('S', 'E'), ('A', 'D')]\n{'w': 2, 'c': 1}\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d093c58ff17e9900f092f1409bc5ad5c8de00493 | 57,330 | ipynb | Jupyter Notebook | User Independence Analysis/ipynb/Basic Checking.ipynb | ramanathanmurugappan/User-Independent-Human-Stress-Detection | 56d8fc52ecc3e8751ef07304eb349f88ab5d08df | [
"MIT"
] | null | null | null | User Independence Analysis/ipynb/Basic Checking.ipynb | ramanathanmurugappan/User-Independent-Human-Stress-Detection | 56d8fc52ecc3e8751ef07304eb349f88ab5d08df | [
"MIT"
] | null | null | null | User Independence Analysis/ipynb/Basic Checking.ipynb | ramanathanmurugappan/User-Independent-Human-Stress-Detection | 56d8fc52ecc3e8751ef07304eb349f88ab5d08df | [
"MIT"
] | 3 | 2020-09-29T17:48:39.000Z | 2021-05-06T17:55:53.000Z | 33.25406 | 94 | 0.359759 | [
[
[
"%%time\nimport pandas as pd\nimport numpy as np\nfrom sklearn.ensemble import ExtraTreesClassifier\nfrom sklearn.metrics import classification_report\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.linear_model import LogisticRegression\nfrom imblearn.over_sampling import SMOTE\nimport matplotlib.pyplot as plt\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.preprocessing import MinMaxScaler\nfrom sklearn.preprocessing import RobustScaler\nfrom sklearn.preprocessing import Normalizer",
"CPU times: user 71 µs, sys: 0 ns, total: 71 µs\nWall time: 74.6 µs\n"
],
[
"df=pd.read_csv('60s_window_merged.csv',index_col=0)\ndf",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df.columns.tolist()",
"_____no_output_____"
],
[
"df.describe()",
"_____no_output_____"
],
[
"df.shape",
"_____no_output_____"
],
[
"df['gender_ male'].unique()",
"_____no_output_____"
],
[
"df['subject'].unique()",
"_____no_output_____"
],
[
"df['subject'].value_counts()",
"_____no_output_____"
],
[
"df['label'].unique()",
"_____no_output_____"
],
[
"df['label'].value_counts()",
"_____no_output_____"
],
[
"df.shape",
"_____no_output_____"
],
[
"features=df.columns.tolist()\nfeatures\n\nremoved = ['label']\nfor rem in removed:\n features.remove(rem)\n\nfeatures\n",
"_____no_output_____"
],
[
"train=df[df['subject']<=9]\ntest=df[df['subject']>9]",
"_____no_output_____"
],
[
"print(train.shape)\nprint(test.shape)",
"(413, 58)\n(372, 58)\n"
],
[
"from sklearn.preprocessing import MinMaxScaler\nscaler = MinMaxScaler(feature_range = (0, 1))\n\ntraining_scaled = scaler.fit_transform(train)",
"_____no_output_____"
],
[
"training_scaled.shape",
"_____no_output_____"
],
[
"features_set = []\nlabels = []\nfor i in range(5, 785):\n features_set.append(training_scaled[i-5:i, 0])\n labels.append(training_scaled[i, 0])",
"_____no_output_____"
],
[
"features_set, labels = np.array(features_set), np.array(labels)",
"_____no_output_____"
],
[
"features_set.shape",
"_____no_output_____"
],
[
"features_set = np.reshape(features_set, (780,5,1))",
"_____no_output_____"
],
[
"from keras.models import Sequential\nfrom keras.layers import Dense\nfrom keras.layers import LSTM\nfrom keras.layers import Dropout",
"_____no_output_____"
],
[
"model = Sequential()",
"_____no_output_____"
],
[
"model.add(LSTM(units=50, return_sequences=True, input_shape=(features_set.shape[1], 1)))",
"_____no_output_____"
],
[
"model.add(Dropout(0.2))",
"_____no_output_____"
],
[
"model.add(LSTM(units=50, return_sequences=True))\nmodel.add(Dropout(0.2))\n\nmodel.add(LSTM(units=50, return_sequences=True))\nmodel.add(Dropout(0.2))\n\nmodel.add(LSTM(units=50))\nmodel.add(Dropout(0.2))",
"_____no_output_____"
],
[
"model.add(Dense(units = 1))",
"_____no_output_____"
],
[
"model.compile(optimizer = 'adam', loss = 'mean_squared_error')",
"_____no_output_____"
],
[
"model.fit(features_set, labels, epochs = 100, batch_size = 32)",
"Epoch 1/100\n780/780 [==============================] - 4s 5ms/step - loss: 0.2330\nEpoch 2/100\n780/780 [==============================] - 0s 615us/step - loss: 0.0576\nEpoch 3/100\n780/780 [==============================] - 0s 622us/step - loss: 0.0496\nEpoch 4/100\n780/780 [==============================] - 0s 611us/step - loss: 0.0498\nEpoch 5/100\n780/780 [==============================] - 0s 624us/step - loss: 0.0499\nEpoch 6/100\n780/780 [==============================] - 0s 614us/step - loss: 0.0477\nEpoch 7/100\n780/780 [==============================] - 0s 615us/step - loss: 0.0457\nEpoch 8/100\n780/780 [==============================] - 0s 619us/step - loss: 0.0460\nEpoch 9/100\n780/780 [==============================] - 0s 622us/step - loss: 0.0464\nEpoch 10/100\n780/780 [==============================] - 0s 620us/step - loss: 0.0461\nEpoch 11/100\n780/780 [==============================] - 0s 625us/step - loss: 0.0464\nEpoch 12/100\n780/780 [==============================] - 0s 613us/step - loss: 0.0446\nEpoch 13/100\n780/780 [==============================] - 0s 624us/step - loss: 0.0460\nEpoch 14/100\n780/780 [==============================] - 0s 617us/step - loss: 0.0443\nEpoch 15/100\n780/780 [==============================] - 0s 619us/step - loss: 0.0450\nEpoch 16/100\n780/780 [==============================] - 0s 616us/step - loss: 0.0454\nEpoch 17/100\n780/780 [==============================] - 0s 625us/step - loss: 0.0444\nEpoch 18/100\n780/780 [==============================] - 0s 613us/step - loss: 0.0421\nEpoch 19/100\n780/780 [==============================] - 0s 613us/step - loss: 0.0458\nEpoch 20/100\n780/780 [==============================] - 0s 624us/step - loss: 0.0423\nEpoch 21/100\n780/780 [==============================] - 0s 622us/step - loss: 0.0447\nEpoch 22/100\n780/780 [==============================] - 0s 613us/step - loss: 0.0419\nEpoch 23/100\n780/780 [==============================] - 0s 616us/step - loss: 0.0412\nEpoch 24/100\n780/780 [==============================] - 0s 607us/step - loss: 0.0396\nEpoch 25/100\n780/780 [==============================] - 0s 615us/step - loss: 0.0403\nEpoch 26/100\n780/780 [==============================] - 0s 616us/step - loss: 0.0381\nEpoch 27/100\n780/780 [==============================] - 0s 620us/step - loss: 0.0375\nEpoch 28/100\n780/780 [==============================] - 0s 628us/step - loss: 0.0366\nEpoch 29/100\n780/780 [==============================] - 0s 618us/step - loss: 0.0360\nEpoch 30/100\n780/780 [==============================] - 0s 612us/step - loss: 0.0364\nEpoch 31/100\n780/780 [==============================] - 0s 621us/step - loss: 0.0361\nEpoch 32/100\n780/780 [==============================] - 0s 613us/step - loss: 0.0366\nEpoch 33/100\n780/780 [==============================] - 0s 619us/step - loss: 0.0362\nEpoch 34/100\n780/780 [==============================] - 0s 627us/step - loss: 0.0332\nEpoch 35/100\n780/780 [==============================] - 0s 625us/step - loss: 0.0355\nEpoch 36/100\n780/780 [==============================] - 0s 631us/step - loss: 0.0350\nEpoch 37/100\n780/780 [==============================] - 0s 626us/step - loss: 0.0317\nEpoch 38/100\n780/780 [==============================] - 0s 630us/step - loss: 0.0313\nEpoch 39/100\n780/780 [==============================] - 0s 631us/step - loss: 0.0310\nEpoch 40/100\n780/780 [==============================] - 1s 644us/step - loss: 0.0311\nEpoch 41/100\n780/780 [==============================] - 0s 633us/step - loss: 0.0304\nEpoch 42/100\n780/780 [==============================] - 0s 632us/step - loss: 0.0304\nEpoch 43/100\n780/780 [==============================] - 0s 629us/step - loss: 0.0285\nEpoch 44/100\n780/780 [==============================] - 0s 624us/step - loss: 0.0285\nEpoch 45/100\n780/780 [==============================] - 0s 632us/step - loss: 0.0297\nEpoch 46/100\n780/780 [==============================] - 0s 618us/step - loss: 0.0293\nEpoch 47/100\n780/780 [==============================] - 0s 619us/step - loss: 0.0289\nEpoch 48/100\n780/780 [==============================] - 0s 628us/step - loss: 0.0273\nEpoch 49/100\n780/780 [==============================] - 0s 625us/step - loss: 0.0298\nEpoch 50/100\n780/780 [==============================] - 0s 627us/step - loss: 0.0276\nEpoch 51/100\n780/780 [==============================] - 0s 631us/step - loss: 0.0277\nEpoch 52/100\n780/780 [==============================] - 0s 633us/step - loss: 0.0268\nEpoch 53/100\n780/780 [==============================] - 0s 626us/step - loss: 0.0284\nEpoch 54/100\n780/780 [==============================] - 0s 633us/step - loss: 0.0282\nEpoch 55/100\n780/780 [==============================] - 0s 627us/step - loss: 0.0281\nEpoch 56/100\n780/780 [==============================] - 0s 622us/step - loss: 0.0271\nEpoch 57/100\n780/780 [==============================] - 0s 633us/step - loss: 0.0294\nEpoch 58/100\n780/780 [==============================] - 0s 629us/step - loss: 0.0281\nEpoch 59/100\n780/780 [==============================] - 0s 637us/step - loss: 0.0284\nEpoch 60/100\n780/780 [==============================] - 0s 635us/step - loss: 0.0273\nEpoch 61/100\n780/780 [==============================] - 0s 623us/step - loss: 0.0263\nEpoch 62/100\n780/780 [==============================] - 1s 642us/step - loss: 0.0300\nEpoch 63/100\n780/780 [==============================] - 0s 628us/step - loss: 0.0256\nEpoch 64/100\n780/780 [==============================] - 0s 635us/step - loss: 0.0260\nEpoch 65/100\n780/780 [==============================] - 0s 620us/step - loss: 0.0266\nEpoch 66/100\n780/780 [==============================] - 0s 631us/step - loss: 0.0273\nEpoch 67/100\n780/780 [==============================] - 0s 630us/step - loss: 0.0262\nEpoch 68/100\n780/780 [==============================] - 0s 623us/step - loss: 0.0262\nEpoch 69/100\n780/780 [==============================] - 0s 621us/step - loss: 0.0261\nEpoch 70/100\n780/780 [==============================] - 0s 623us/step - loss: 0.0249\nEpoch 71/100\n780/780 [==============================] - 0s 617us/step - loss: 0.0273\nEpoch 72/100\n780/780 [==============================] - 0s 620us/step - loss: 0.0266\nEpoch 73/100\n780/780 [==============================] - 0s 621us/step - loss: 0.0266\nEpoch 74/100\n780/780 [==============================] - 0s 622us/step - loss: 0.0262\nEpoch 75/100\n780/780 [==============================] - 0s 623us/step - loss: 0.0253\nEpoch 76/100\n780/780 [==============================] - 0s 620us/step - loss: 0.0260\nEpoch 77/100\n780/780 [==============================] - 0s 628us/step - loss: 0.0245\nEpoch 78/100\n780/780 [==============================] - 0s 627us/step - loss: 0.0264\nEpoch 79/100\n780/780 [==============================] - 0s 629us/step - loss: 0.0249\nEpoch 80/100\n780/780 [==============================] - 0s 618us/step - loss: 0.0253\nEpoch 81/100\n780/780 [==============================] - 0s 615us/step - loss: 0.0259\nEpoch 82/100\n780/780 [==============================] - 0s 618us/step - loss: 0.0264\nEpoch 83/100\n780/780 [==============================] - 0s 621us/step - loss: 0.0254\nEpoch 84/100\n780/780 [==============================] - 0s 620us/step - loss: 0.0255\nEpoch 85/100\n780/780 [==============================] - 0s 613us/step - loss: 0.0251\nEpoch 86/100\n780/780 [==============================] - 0s 617us/step - loss: 0.0249\nEpoch 87/100\n780/780 [==============================] - 0s 619us/step - loss: 0.0265\nEpoch 88/100\n780/780 [==============================] - 0s 612us/step - loss: 0.0270\nEpoch 89/100\n780/780 [==============================] - 0s 611us/step - loss: 0.0249\nEpoch 90/100\n780/780 [==============================] - 0s 602us/step - loss: 0.0242\nEpoch 91/100\n780/780 [==============================] - 0s 613us/step - loss: 0.0254\nEpoch 92/100\n780/780 [==============================] - 0s 614us/step - loss: 0.0242\nEpoch 93/100\n780/780 [==============================] - 0s 612us/step - loss: 0.0255\nEpoch 94/100\n780/780 [==============================] - 0s 622us/step - loss: 0.0255\nEpoch 95/100\n780/780 [==============================] - 0s 608us/step - loss: 0.0257\nEpoch 96/100\n780/780 [==============================] - 0s 614us/step - loss: 0.0253\nEpoch 97/100\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d093cf1806dc88634ffbff5ea8f5c93045dcdeaf | 96,642 | ipynb | Jupyter Notebook | titanic_survival_exploration.ipynb | numanyilmaz/titanic_survival_exploration | 713705f1b1d438488d00577aec606a82f99193a4 | [
"MIT"
] | null | null | null | titanic_survival_exploration.ipynb | numanyilmaz/titanic_survival_exploration | 713705f1b1d438488d00577aec606a82f99193a4 | [
"MIT"
] | null | null | null | titanic_survival_exploration.ipynb | numanyilmaz/titanic_survival_exploration | 713705f1b1d438488d00577aec606a82f99193a4 | [
"MIT"
] | null | null | null | 112.243902 | 16,706 | 0.809265 | [
[
[
"# Machine Learning Engineer Nanodegree\n## Introduction and Foundations\n## Project: Titanic Survival Exploration\n\nIn 1912, the ship RMS Titanic struck an iceberg on its maiden voyage and sank, resulting in the deaths of most of its passengers and crew. In this introductory project, we will explore a subset of the RMS Titanic passenger manifest to determine which features best predict whether someone survived or did not survive. To complete this project, you will need to implement several conditional predictions and answer the questions below. Your project submission will be evaluated based on the completion of the code and your responses to the questions.\n> **Tip:** Quoted sections like this will provide helpful instructions on how to navigate and use an iPython notebook. ",
"_____no_output_____"
],
[
"# Getting Started\nTo begin working with the RMS Titanic passenger data, we'll first need to `import` the functionality we need, and load our data into a `pandas` DataFrame. \nRun the code cell below to load our data and display the first few entries (passengers) for examination using the `.head()` function.\n> **Tip:** You can run a code cell by clicking on the cell and using the keyboard shortcut **Shift + Enter** or **Shift + Return**. Alternatively, a code cell can be executed using the **Play** button in the hotbar after selecting it. Markdown cells (text cells like this one) can be edited by double-clicking, and saved using these same shortcuts. [Markdown](http://daringfireball.net/projects/markdown/syntax) allows you to write easy-to-read plain text that can be converted to HTML.",
"_____no_output_____"
]
],
[
[
"# Import libraries necessary for this project\nimport numpy as np\nimport pandas as pd\nfrom IPython.display import display # Allows the use of display() for DataFrames\n\n# Import supplementary visualizations code visuals.py\nimport visuals as vs\n\n# Pretty display for notebooks\n%matplotlib inline\n\n# Load the dataset\nin_file = 'titanic_data.csv'\nfull_data = pd.read_csv(in_file)\n\n# Print the first few entries of the RMS Titanic data\ndisplay(full_data.head())",
"_____no_output_____"
]
],
[
[
"From a sample of the RMS Titanic data, we can see the various features present for each passenger on the ship:\n- **Survived**: Outcome of survival (0 = No; 1 = Yes)\n- **Pclass**: Socio-economic class (1 = Upper class; 2 = Middle class; 3 = Lower class)\n- **Name**: Name of passenger\n- **Sex**: Sex of the passenger\n- **Age**: Age of the passenger (Some entries contain `NaN`)\n- **SibSp**: Number of siblings and spouses of the passenger aboard\n- **Parch**: Number of parents and children of the passenger aboard\n- **Ticket**: Ticket number of the passenger\n- **Fare**: Fare paid by the passenger\n- **Cabin** Cabin number of the passenger (Some entries contain `NaN`)\n- **Embarked**: Port of embarkation of the passenger (C = Cherbourg; Q = Queenstown; S = Southampton)\n\nSince we're interested in the outcome of survival for each passenger or crew member, we can remove the **Survived** feature from this dataset and store it as its own separate variable `outcomes`. We will use these outcomes as our prediction targets. \nRun the code cell below to remove **Survived** as a feature of the dataset and store it in `outcomes`.",
"_____no_output_____"
]
],
[
[
"# Store the 'Survived' feature in a new variable and remove it from the dataset\noutcomes = full_data['Survived']\ndata = full_data.drop('Survived', axis = 1)\n\n# Show the new dataset with 'Survived' removed\ndisplay(data.head())",
"_____no_output_____"
]
],
[
[
"The very same sample of the RMS Titanic data now shows the **Survived** feature removed from the DataFrame. Note that `data` (the passenger data) and `outcomes` (the outcomes of survival) are now *paired*. That means for any passenger `data.loc[i]`, they have the survival outcome `outcomes[i]`.\n\nTo measure the performance of our predictions, we need a metric to score our predictions against the true outcomes of survival. Since we are interested in how *accurate* our predictions are, we will calculate the proportion of passengers where our prediction of their survival is correct. Run the code cell below to create our `accuracy_score` function and test a prediction on the first five passengers. \n\n**Think:** *Out of the first five passengers, if we predict that all of them survived, what would you expect the accuracy of our predictions to be?*",
"_____no_output_____"
]
],
[
[
"def accuracy_score(truth, pred):\n \"\"\" Returns accuracy score for input truth and predictions. \"\"\"\n \n # Ensure that the number of predictions matches number of outcomes\n if len(truth) == len(pred): \n \n # Calculate and return the accuracy as a percent\n return \"Predictions have an accuracy of {:.2f}%.\".format((truth == pred).mean()*100)\n \n else:\n return \"Number of predictions does not match number of outcomes!\"\n \n# Test the 'accuracy_score' function\npredictions = pd.Series(np.ones(5, dtype = int))\nprint accuracy_score(outcomes[:5], predictions)",
"Predictions have an accuracy of 60.00%.\n"
]
],
[
[
"> **Tip:** If you save an iPython Notebook, the output from running code blocks will also be saved. However, the state of your workspace will be reset once a new session is started. Make sure that you run all of the code blocks from your previous session to reestablish variables and functions before picking up where you last left off.\n\n# Making Predictions\n\nIf we were asked to make a prediction about any passenger aboard the RMS Titanic whom we knew nothing about, then the best prediction we could make would be that they did not survive. This is because we can assume that a majority of the passengers (more than 50%) did not survive the ship sinking. \nThe `predictions_0` function below will always predict that a passenger did not survive.",
"_____no_output_____"
]
],
[
[
"def predictions_0(data):\n \"\"\" Model with no features. Always predicts a passenger did not survive. \"\"\"\n\n predictions = []\n for _, passenger in data.iterrows():\n \n # Predict the survival of 'passenger'\n predictions.append(0)\n \n # Return our predictions\n return pd.Series(predictions)\n\n# Make the predictions\npredictions = predictions_0(data)",
"_____no_output_____"
]
],
[
[
"### Question 1\n*Using the RMS Titanic data, how accurate would a prediction be that none of the passengers survived?* \n**Hint:** Run the code cell below to see the accuracy of this prediction.",
"_____no_output_____"
]
],
[
[
"print accuracy_score(outcomes, predictions)",
"Predictions have an accuracy of 61.62%.\n"
]
],
[
[
"**Answer:** 61.62%",
"_____no_output_____"
],
[
"***\nLet's take a look at whether the feature **Sex** has any indication of survival rates among passengers using the `survival_stats` function. This function is defined in the `visuals.py` Python script included with this project. The first two parameters passed to the function are the RMS Titanic data and passenger survival outcomes, respectively. The third parameter indicates which feature we want to plot survival statistics across. \nRun the code cell below to plot the survival outcomes of passengers based on their sex.",
"_____no_output_____"
]
],
[
[
"vs.survival_stats(data, outcomes, 'Sex')",
"_____no_output_____"
]
],
[
[
"Examining the survival statistics, a large majority of males did not survive the ship sinking. However, a majority of females *did* survive the ship sinking. Let's build on our previous prediction: If a passenger was female, then we will predict that they survived. Otherwise, we will predict the passenger did not survive. \nFill in the missing code below so that the function will make this prediction. \n**Hint:** You can access the values of each feature for a passenger like a dictionary. For example, `passenger['Sex']` is the sex of the passenger.",
"_____no_output_____"
]
],
[
[
"def predictions_1(data):\n \"\"\" Model with one feature: \n - Predict a passenger survived if they are female. \"\"\"\n \n predictions = []\n for _, passenger in data.iterrows():\n \n if passenger['Sex'] == 'female':\n predictions.append(1)\n else:\n predictions.append(0)\n \n # Return our predictions\n return pd.Series(predictions)\n\n# Make the predictions\npredictions = predictions_1(data)",
"_____no_output_____"
]
],
[
[
"### Question 2\n*How accurate would a prediction be that all female passengers survived and the remaining passengers did not survive?* \n**Hint:** Run the code cell below to see the accuracy of this prediction.",
"_____no_output_____"
]
],
[
[
"print accuracy_score(outcomes, predictions)",
"Predictions have an accuracy of 78.68%.\n"
]
],
[
[
"**Answer**: 78.68%",
"_____no_output_____"
],
[
"***\nUsing just the **Sex** feature for each passenger, we are able to increase the accuracy of our predictions by a significant margin. Now, let's consider using an additional feature to see if we can further improve our predictions. For example, consider all of the male passengers aboard the RMS Titanic: Can we find a subset of those passengers that had a higher rate of survival? Let's start by looking at the **Age** of each male, by again using the `survival_stats` function. This time, we'll use a fourth parameter to filter out the data so that only passengers with the **Sex** 'male' will be included. \nRun the code cell below to plot the survival outcomes of male passengers based on their age.",
"_____no_output_____"
]
],
[
[
"vs.survival_stats(data, outcomes, 'Age', [\"Sex == 'male'\"])",
"_____no_output_____"
]
],
[
[
"Examining the survival statistics, the majority of males younger than 10 survived the ship sinking, whereas most males age 10 or older *did not survive* the ship sinking. Let's continue to build on our previous prediction: If a passenger was female, then we will predict they survive. If a passenger was male and younger than 10, then we will also predict they survive. Otherwise, we will predict they do not survive. \nFill in the missing code below so that the function will make this prediction. \n**Hint:** You can start your implementation of this function using the prediction code you wrote earlier from `predictions_1`.",
"_____no_output_____"
]
],
[
[
"def predictions_2(data):\n \"\"\" Model with two features: \n - Predict a passenger survived if they are female.\n - Predict a passenger survived if they are male and younger than 10. \"\"\"\n \n predictions = []\n for _, passenger in data.iterrows():\n \n if passenger['Sex'] == 'female':\n predictions.append(1)\n elif passenger['Sex'] == 'male' and passenger['Age'] < 10:\n predictions.append(1)\n else:\n predictions.append(0)\n \n # Return our predictions\n return pd.Series(predictions)\n\n# Make the predictions\npredictions = predictions_2(data)",
"_____no_output_____"
]
],
[
[
"### Question 3\n*How accurate would a prediction be that all female passengers and all male passengers younger than 10 survived?* \n**Hint:** Run the code cell below to see the accuracy of this prediction.",
"_____no_output_____"
]
],
[
[
"print accuracy_score(outcomes, predictions)",
"Predictions have an accuracy of 79.35%.\n"
]
],
[
[
"**Answer**: 79.35%",
"_____no_output_____"
],
[
"***\nAdding the feature **Age** as a condition in conjunction with **Sex** improves the accuracy by a small margin more than with simply using the feature **Sex** alone. Now it's your turn: Find a series of features and conditions to split the data on to obtain an outcome prediction accuracy of at least 80%. This may require multiple features and multiple levels of conditional statements to succeed. You can use the same feature multiple times with different conditions. \n**Pclass**, **Sex**, **Age**, **SibSp**, and **Parch** are some suggested features to try.\n\nUse the `survival_stats` function below to to examine various survival statistics. \n**Hint:** To use mulitple filter conditions, put each condition in the list passed as the last argument. Example: `[\"Sex == 'male'\", \"Age < 18\"]`",
"_____no_output_____"
]
],
[
[
"vs.survival_stats(data, outcomes, 'SibSp', [\"Sex == 'female'\"])",
"_____no_output_____"
]
],
[
[
"Adding number of siblings and spouses of the passenger for females increased accuracy. Having less than 3 siblings and spouses increasing chance of survive, 3 or 4 siblings and spouses reduce survive and there is no survival chance for females who had more than 4 siblings and spouses.",
"_____no_output_____"
]
],
[
[
"vs.survival_stats(data, outcomes, 'Age', [\"Sex == 'male'\", \"Pclass == 1\"])",
"_____no_output_____"
]
],
[
[
"For male who was in first class with the age between 30 and 40 has high chance of survive. ",
"_____no_output_____"
],
[
"After exploring the survival statistics visualization, fill in the missing code below so that the function will make your prediction. \nMake sure to keep track of the various features and conditions you tried before arriving at your final prediction model. \n**Hint:** You can start your implementation of this function using the prediction code you wrote earlier from `predictions_2`.",
"_____no_output_____"
]
],
[
[
"def predictions_3(data):\n \"\"\" Model with multiple features. Makes a prediction with an accuracy of at least 80%. \"\"\"\n \n predictions = []\n for _, passenger in data.iterrows():\n \n if passenger['Sex'] == 'female' and passenger['SibSp'] < 3:\n predictions.append(1)\n \n elif passenger['Sex'] == 'female' and passenger['SibSp'] < 5:\n predictions.append(0)\n \n elif passenger['Sex'] == 'female' and passenger['SibSp'] >=5 :\n predictions.append(0) \n # code above incrased the accuracy up to 80.36% \n \n elif passenger['Sex'] == 'male' and passenger['Age'] < 10:\n predictions.append(1)\n \n #vs.survival_stats(data, outcomes, 'Age', [\"Sex == 'male'\", \"Pclass == 1\"]) \n #elif statement below increases accuracy to 80.58\n elif passenger['Sex'] == 'male' and passenger['Age'] < 40 and passenger['Age'] > 30 and passenger['Pclass'] == 1:\n predictions.append(1)\n \n else:\n predictions.append(0)\n \n \n # Return our predictions\n return pd.Series(predictions)\n\n# Make the predictions\npredictions = predictions_3(data)",
"_____no_output_____"
]
],
[
[
"### Question 4\n*Describe the steps you took to implement the final prediction model so that it got an accuracy of at least 80%. What features did you look at? Were certain features more informative than others? Which conditions did you use to split the survival outcomes in the data? How accurate are your predictions?* \n**Hint:** Run the code cell below to see the accuracy of your predictions.",
"_____no_output_____"
]
],
[
[
"print accuracy_score(outcomes, predictions)",
"Predictions have an accuracy of 80.58%.\n"
]
],
[
[
"**Answer**: 80.58%",
"_____no_output_____"
],
[
"# Conclusion\n\nAfter several iterations of exploring and conditioning on the data, you have built a useful algorithm for predicting the survival of each passenger aboard the RMS Titanic. The technique applied in this project is a manual implementation of a simple machine learning model, the *decision tree*. A decision tree splits a set of data into smaller and smaller groups (called *nodes*), by one feature at a time. Each time a subset of the data is split, our predictions become more accurate if each of the resulting subgroups are more homogeneous (contain similar labels) than before. The advantage of having a computer do things for us is that it will be more exhaustive and more precise than our manual exploration above. [This link](http://www.r2d3.us/visual-intro-to-machine-learning-part-1/) provides another introduction into machine learning using a decision tree.\n\nA decision tree is just one of many models that come from *supervised learning*. In supervised learning, we attempt to use features of the data to predict or model things with objective outcome labels. That is to say, each of our data points has a known outcome value, such as a categorical, discrete label like `'Survived'`, or a numerical, continuous value like predicting the price of a house.\n\n### Question 5\n*Think of a real-world scenario where supervised learning could be applied. What would be the outcome variable that you are trying to predict? Name two features about the data used in this scenario that might be helpful for making the predictions.* ",
"_____no_output_____"
],
[
"**Answer**: Weather forecast would be a good example. We could predict the next days temperature based on previous years data. In this case, Humidity, wind could be the features and temparature would be the target.",
"_____no_output_____"
],
[
"> **Note**: Once you have completed all of the code implementations and successfully answered each question above, you may finalize your work by exporting the iPython Notebook as an HTML document. You can do this by using the menu above and navigating to \n**File -> Download as -> HTML (.html)**. Include the finished document along with this notebook as your submission.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d093d3a1ba4e0f276442db47bb010f9b8248f233 | 6,297 | ipynb | Jupyter Notebook | quantum_circuits/examples.ipynb | JinLi711/quantum_circuits | b2483afcd6b8976f74486b302cb6a1ab7c8114a9 | [
"MIT"
] | 1 | 2021-05-15T08:51:56.000Z | 2021-05-15T08:51:56.000Z | quantum_circuits/examples.ipynb | JinLi711/quantum_circuits | b2483afcd6b8976f74486b302cb6a1ab7c8114a9 | [
"MIT"
] | null | null | null | quantum_circuits/examples.ipynb | JinLi711/quantum_circuits | b2483afcd6b8976f74486b302cb6a1ab7c8114a9 | [
"MIT"
] | null | null | null | 21.940767 | 439 | 0.480387 | [
[
[
"Examples on how to use this package:",
"_____no_output_____"
]
],
[
[
"from circuit import Circuit",
"_____no_output_____"
]
],
[
[
"# Circuit Creation",
"_____no_output_____"
]
],
[
[
"circ = Circuit(3,5)\ncirc.X(2)\ncirc.H()\ncirc.barrier()",
"_____no_output_____"
]
],
[
[
"# Getting the Current State",
"_____no_output_____"
]
],
[
[
"circ.get_state()",
"_____no_output_____"
],
[
"circ.get_state(output='tensor')",
"_____no_output_____"
]
],
[
[
"# Execute Simulation",
"_____no_output_____"
]
],
[
[
"circ.execute(num_instances=800)",
"_____no_output_____"
]
],
[
[
"# Code To OpenQASM",
"_____no_output_____"
]
],
[
[
"print(circ.compile())",
"OPENQASM 2.0;\ninclude \"qelib1.inc\";\n\nqreg q[3]\ncreg c[5]\n\nx q[2];\nh q[None];\nbarrier q;\n\n"
]
],
[
[
"# Measurement",
"_____no_output_____"
]
],
[
[
"circ = Circuit(2, 3)\ncirc.X(1)\n\n# measure qubit one and write the result to classical bit 1\ncirc.measure(0, 1)\ncirc.bits[1]",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d093ee4e9ed3878dd7d048d2a7a7288ec7596f76 | 73,646 | ipynb | Jupyter Notebook | notebooks/to_check/Restitch_to_test_angle.ipynb | linnarsson-lab/pysmFISH_auto | d963f1319099c36873c2c5afd41bd15e7361a00e | [
"MIT"
] | 3 | 2022-01-15T07:49:15.000Z | 2022-01-18T08:18:38.000Z | notebooks/to_check/Restitch_to_test_angle.ipynb | linnarsson-lab/pysmFISH_auto | d963f1319099c36873c2c5afd41bd15e7361a00e | [
"MIT"
] | null | null | null | notebooks/to_check/Restitch_to_test_angle.ipynb | linnarsson-lab/pysmFISH_auto | d963f1319099c36873c2c5afd41bd15e7361a00e | [
"MIT"
] | 3 | 2022-01-14T15:47:54.000Z | 2022-01-24T14:58:24.000Z | 57.089922 | 195 | 0.689732 | [
[
[
"%load_ext autoreload\n%autoreload 2",
"The autoreload extension is already loaded. To reload it, use:\n %reload_ext autoreload\n"
],
[
"import numpy as np\nfrom pathlib import Path\n",
"_____no_output_____"
],
[
"from pysmFISH import stitching\nfrom pysmFISH import io\nfrom pysmFISH import data_models",
"_____no_output_____"
],
[
"from dask import dataframe as dd\nfrom dask.distributed import Client",
"_____no_output_____"
],
[
"client = Client()",
"_____no_output_____"
],
[
"client",
"_____no_output_____"
],
[
"experiment_fpath = Path('/Users/simone.codeluppi/Documents/data_analysis_jlabs_sc/eel_adult_mouse_brain_atlas_project/LBEXP20200925_EEL_Mouse_-140um/data/LBEXP20200925_EEL_Mouse_-140um')\ndataset_fpath = experiment_fpath / '211028_17_17_59_LBEXP20200925_EEL_Mouse_-140um_img_data_dataset.parquet'\nmicroscope_coords_pxl_fpath = experiment_fpath / 'results' / 'microscope_tile_corners_coords_pxl.npy'",
"_____no_output_____"
],
[
"dataset = data_models.Dataset()",
"_____no_output_____"
],
[
"dataset.load_dataset(dataset_fpath)",
"_____no_output_____"
],
[
"metadata = dataset.collect_metadata(dataset.dataset)",
"_____no_output_____"
],
[
"tile_corners_coords_pxl = np.load(microscope_coords_pxl_fpath)",
"_____no_output_____"
],
[
"reference_corner_fov_position = 'old-robofish2'",
"_____no_output_____"
],
[
"for fpath in (experiment_fpath / 'results').glob('*decoded_fov*'):\n print(f\"processing {fpath.stem}\")\n stitching.stitch_using_coords_general(fpath,tile_corners_coords_pxl,reference_corner_fov_position,metadata,'microscope_stitched')",
"processing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_253\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_243\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_406\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_416\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_618\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_608\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_135\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_125\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_15\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_392\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_382\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_288\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_298\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_67\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_77\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_157\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_147\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_702\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_712\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_464\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_474\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_349\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_231\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_359\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_221\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_480\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_490\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_627\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_83\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_637\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_93\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_314\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_304\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_541\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_439\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_551\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_429\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_523\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_533\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_376\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_366\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_168\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_178\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_645\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_655\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_6\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_58\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_48\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_291\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_281\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_663\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_673\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_505\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_515\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_350\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_228\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_340\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_238\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_332\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_322\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_567\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_577\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_601\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_611\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_195\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_185\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_442\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_452\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_217\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_207\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_171\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_161\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_724\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_734\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_41\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_51\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_583\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_593\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_499\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_489\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_687\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_23\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_697\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_33\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_746\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_756\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_113\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_103\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_275\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_265\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_558\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_420\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_548\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_430\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_672\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_662\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_239\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_341\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_229\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_351\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_514\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_504\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_280\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_290\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_184\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_194\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_576\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_566\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_323\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_333\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_610\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_600\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_50\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_40\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_592\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_582\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_206\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_216\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_453\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_443\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_735\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_725\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_160\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_170\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_102\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_112\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_757\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_747\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_431\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_549\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_421\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_559\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_264\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_274\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_488\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_498\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_32\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_696\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_22\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_686\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_14\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_383\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_393\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_417\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_407\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_242\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_252\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_124\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_134\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_609\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_619\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_713\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_703\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_146\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_156\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_220\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_358\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_230\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_348\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_475\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_465\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_299\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_289\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_76\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_66\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_92\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_636\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_82\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_626\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_428\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_550\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_438\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_540\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_305\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_315\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_491\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_481\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_49\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_59\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_7\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_367\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_377\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_532\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_522\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_654\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_644\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_179\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_169\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_388\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_398\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_196\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_186\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_602\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_612\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_249\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_331\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_259\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_321\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_564\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_574\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_506\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_516\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_353\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_343\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_718\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_660\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_708\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_670\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_292\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_282\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_276\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_266\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_423\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_433\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_745\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_99\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_89\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_755\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_110\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_100\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_20\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_684\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_30\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_694\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_580\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_590\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_42\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_52\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_172\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_162\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_727\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_737\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_441\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_539\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_451\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_529\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_214\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_204\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_467\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_477\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_232\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_222\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_154\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_144\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_701\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_679\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_711\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_669\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_64\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_74\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_391\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_381\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_16\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_136\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_126\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_250\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_328\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_240\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_338\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_405\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_415\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_599\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_589\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_5\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_646\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_656\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_458\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_520\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_448\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_530\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_375\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_365\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_317\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_307\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_542\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_552\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_80\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_624\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_90\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_634\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_109\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_119\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_39\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_29\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_483\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_493\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_75\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_65\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_223\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_233\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_476\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_466\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_668\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_710\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_678\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_700\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_145\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_155\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_127\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_137\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_414\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_404\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_339\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_241\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_329\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_251\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_380\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_390\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_17\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_657\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_647\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_364\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_374\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_531\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_449\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_521\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_459\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_588\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_598\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_4\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_28\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_38\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_492\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_482\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_553\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_543\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_306\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_316\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_118\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_108\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_635\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_91\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_625\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_81\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_613\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_603\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_575\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_565\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_320\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_258\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_330\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_248\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_399\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_389\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_187\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_197\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_283\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_293\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_342\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_352\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_517\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_507\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_671\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_709\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_661\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_719\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_695\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_31\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_685\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_21\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_432\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_422\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_267\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_277\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_101\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_111\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_88\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_754\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_744\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_98\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_736\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_726\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_163\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_173\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_205\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_215\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_528\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_450\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_538\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_440\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_591\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_581\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_53\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_43\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_680\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_24\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_690\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_34\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_114\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_104\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_639\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_741\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_629\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_751\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_427\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_437\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_272\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_262\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_368\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_210\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_378\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_200\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_445\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_455\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_723\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_733\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_176\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_166\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_46\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_56\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_8\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_584\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_594\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_560\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_418\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_570\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_408\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_335\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_325\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_606\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_616\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_192\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_182\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_296\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_286\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_79\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_69\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_664\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_674\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_149\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_159\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_357\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_347\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_502\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_512\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_371\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_209\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_361\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_219\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_524\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_534\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_642\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_652\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_1\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_487\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_497\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_699\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_689\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_620\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_84\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_630\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_94\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_748\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_546\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_556\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_313\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_303\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_60\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_70\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_705\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_715\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_150\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_140\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_236\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_226\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_463\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_473\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_579\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_401\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_569\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_411\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_254\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_244\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_132\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_122\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_12\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_395\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_385\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_0\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_535\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_525\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_218\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_360\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_208\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_370\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_653\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_643\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_95\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_749\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_631\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_85\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_621\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_302\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_312\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_557\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_547\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_496\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_486\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_688\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_698\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_141\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_151\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_714\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_704\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_472\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_462\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_227\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_237\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_71\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_61\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_13\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_384\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_394\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_245\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_255\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_410\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_568\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_400\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_578\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_123\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_133\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_750\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_628\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_740\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_638\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_105\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_115\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_263\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_273\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_436\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_426\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_35\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_691\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_25\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_681\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_9\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_57\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_47\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_595\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_585\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_454\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_444\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_201\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_379\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_211\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_369\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_167\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_177\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_732\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_722\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_183\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_193\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_324\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_334\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_409\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_571\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_419\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_561\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_617\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_607\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_158\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_148\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_675\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_665\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_513\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_503\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_346\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_356\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_287\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_297\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_68\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_78\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_545\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_555\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_268\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_310\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_278\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_300\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_87\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_623\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_97\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_633\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_484\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_494\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_2\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_739\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_641\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_729\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_651\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_372\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_362\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_527\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_537\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_396\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_386\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_188\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_198\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_11\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_131\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_121\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_402\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_412\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_257\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_247\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_235\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_225\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_460\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_518\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_470\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_508\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_706\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_716\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_153\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_143\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_63\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_73\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_587\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_597\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_45\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_55\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_720\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_658\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_730\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_648\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_175\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_165\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_213\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_203\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_446\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_456\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_424\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_434\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_271\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_309\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_261\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_319\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_117\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_107\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_742\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_752\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_27\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_683\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_37\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_693\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_354\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_344\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_479\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_501\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_469\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_511\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_667\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_677\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_295\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_285\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_191\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_181\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_18\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_128\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_138\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_605\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_615\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_563\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_573\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_336\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_326\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_164\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_174\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_649\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_731\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_659\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_721\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_457\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_447\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_202\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_212\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_596\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_586\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_54\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_44\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_692\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_36\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_682\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_26\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_318\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_260\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_308\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_270\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_435\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_425\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_753\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_743\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_106\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_116\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_284\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_294\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_510\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_468\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_500\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_478\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_345\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_355\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_676\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_666\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_614\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_604\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_139\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_129\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_327\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_337\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_572\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_562\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_19\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_180\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_190\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_495\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_485\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_301\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_279\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_311\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_269\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_554\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_544\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_632\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_96\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_622\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_86\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_650\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_728\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_640\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_738\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_536\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_526\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_363\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_373\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_3\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_120\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_130\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_246\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_256\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_413\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_403\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_387\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_397\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_10\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_199\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_189\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_72\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_62\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_509\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_471\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_519\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_461\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_224\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_234\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_142\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_152\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_717\nprocessing LBEXP20200925_EEL_Mouse_-140um_decoded_fov_707\n"
],
[
"stitching_selected = 'microscope_stitched'\ninput_file_tag = 'decoded_fov'\nfile_tag = 'old-robofish2'\nselected_Hdistance = 3/16",
"_____no_output_____"
],
[
"io.simple_output_plotting(experiment_fpath, stitching_selected, \n selected_Hdistance, client, input_file_tag, file_tag)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d093f816feeb34938d19753900f009e32248a3ad | 530,934 | ipynb | Jupyter Notebook | figures/Results-mcmc-corner-plot.ipynb | benjaminrose/SNIa-Local-Environments | 92713be96a89da991fe53bffcc596a5c0942fc37 | [
"MIT"
] | 1 | 2020-09-19T22:08:51.000Z | 2020-09-19T22:08:51.000Z | figures/Results-mcmc-corner-plot.ipynb | benjaminrose/SNIa-Local-Environments | 92713be96a89da991fe53bffcc596a5c0942fc37 | [
"MIT"
] | 9 | 2017-12-11T19:15:33.000Z | 2018-04-18T19:08:34.000Z | figures/Results-mcmc-corner-plot.ipynb | benjaminrose/SNIa-Local-Environments | 92713be96a89da991fe53bffcc596a5c0942fc37 | [
"MIT"
] | 3 | 2020-08-13T03:45:09.000Z | 2020-08-19T22:31:00.000Z | 886.367279 | 513,332 | 0.951467 | [
[
[
"# Make Corner Plots of Posterior Distributions\n\nThis file allows me to quickly and repeatedly make the cornor plot to examin the results of the MCMC analsys",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport matplotlib\nimport pandas as pd\nfrom astropy.table import Table\nimport corner\n# import seaborn",
"_____no_output_____"
],
[
"matplotlib.rcParams.update({'font.size': 11})",
"_____no_output_____"
]
],
[
[
"This function is the general function that is repeated called throught the file. One benifite to this system, is that I only need to update to higher quality labels in one place. ",
"_____no_output_____"
]
],
[
[
"def corner_plot(file_, saved_file, truths=None, third=0):\n data = Table.read(file_, format='ascii.commented_header', delimiter='\\t')\n if third !=0:\n size = len(data)\n data = data[(third-1)*size//3:(third)*size//3]\n data = data.to_pandas()\n data.dropna(inplace=True)\n \n \n # look at corner.hist2d(levels) to not have too many conturs on a plot\n # http://corner.readthedocs.io/en/latest/api.html\n fig = corner.corner(data, show_titles=True, use_math_text=True,\n bins=25, quantiles=[0.16, 0.84], smooth=1, \n plot_datapoints=False,\n labels=[r\"$\\log(z/z_{\\odot})$\", r\"$\\tau_2$\", r\"$\\tau$\", \n r\"$t_{0}$\", r\"$t_{i}$\", r'$\\phi$', \n '$\\delta$', 'age'],\n truths=truths, range=[0.99]*8\n )\n\n \n fig.savefig(saved_file)",
"_____no_output_____"
]
],
[
[
"## One Object",
"_____no_output_____"
]
],
[
[
"#run one object\nSN = 16185\nfile_ = f'../resources/SN{SN}_campbell_chain.tsv'\nsaved_file = f'SN{SN}-mcmc-2018-12-21.pdf'\ncorner_plot(file_, saved_file)",
"_____no_output_____"
]
],
[
[
"## Messier Objects",
"_____no_output_____"
]
],
[
[
"# run all Messier objects \nfor id in [63, 82, 87, 89, 91, 101, 105, 108]:\n file_ = f'../resources/SN{id}_messier_chain.tsv'\n saved_file = f'messierTests/12-29-M{id}.pdf'\n print(f'\\nMaking {saved_file}')\n corner_plot(file_, saved_file)",
"\nMaking messierTests/12-29-M63.pdf\n"
],
[
"# One Messier Object\nID = 63\nfile_ = f'../resources/SN{ID}_messier_chain.tsv'\nsaved_file = f'messierTests/12-22-M{ID}.pdf'\nprint(f'\\nMaking {saved_file}')\ncorner_plot(file_, saved_file)",
"\nMaking messierTests/12-22-M63.pdf\n"
]
],
[
[
"## Circle Test -- old",
"_____no_output_____"
]
],
[
[
"# run on circle test \nfor id in [1, 2, 3, 4, 5, 6, 7]:\n file_ = f'../resources/SN{id}_chain.tsv'\n saved_file = f'circleTests/12-19-C{id}.pdf'\n print(f'\\nMaking {saved_file}')\n corner_plot(file_, saved_file)",
"\nMaking circleTests/07-31-C1.pdf\n"
],
[
"# run on circle test 3 with truths\nfile_ = f'../resources/SN3_chain.tsv'\nsaved_file = f'circleTests/07-31-C3-truths.pdf'\n\ndata = Table.read(file_, format='ascii.commented_header', delimiter='\\t')\ndata = data.to_pandas()\ndata.dropna(inplace=True)\n\nfig = corner.corner(data, show_titles=True, use_math_text=True,\n quantiles=[0.16, 0.5, 0.84], smooth=0.5, \n plot_datapoints=False,\n labels=[\"$logZ_{sol}$\", \"$dust_2$\", r\"$\\tau$\", \n \"$t_{start}$\", \"$t_{trans}$\", 'sf slope', \n 'c', 'Age'],\n truths=[-0.5, 0.1, 7.0, 3.0, 10, 15.0, -25, None]\n )\n\nfig.savefig(saved_file)",
"_____no_output_____"
],
[
"# run on circle test 1 with truths\nfile_ = f'../resources/SN1_chain_2017-09-11.tsv'\nsaved_file = f'circleTests/09-11-C1-truths.pdf'\ntruths=[-0.5, 0.1, 0.5, 1.5, 9.0, -1.0, -25, None]\n\ncorner_plot(file_, saved_file, truths)\n\n# data = Table.read(file_, format='ascii.commented_header', delimiter='\\t')\n# data = data.to_pandas()\n# data.dropna(inplace=True)\n#\n# fig = corner.corner(data, show_titles=True, use_math_text=True,\n# quantiles=[0.16, 0.5, 0.84], smooth=0.5, \n# plot_datapoints=False,\n# labels=[\"$logZ_{sol}$\", \"$dust_2$\", r\"$\\tau$\", \n# \"$t_{start}$\", \"$t_{trans}$\", 'sf slope', \n# 'c', 'Age'],\n# truths=[-0.5, 0.1, 0.5, 1.5, 9.0, -1.0, -25, None]\n# )\n\n# fig.savefig(saved_file)",
"WARNING:root:Too few points to create valid contours\n"
]
],
[
[
"## Test all Circle Tests",
"_____no_output_____"
]
],
[
[
"# for slope\n# truths = {\n# 1 : [-0.5, 0.1, 0.5, 1.5, 9.0, -1.0, -25, 10.68],\n# 2 : [-0.5, 0.1, 0.5, 1.5, 9.0, 15.0, -25, 1.41],\n# 3 : [-0.5, 0.1, 7.0, 3.0, 10, 15.0, -25, 1.75],\n# 4 : [-0.5, 0.1, 7.0, 3.0, 13.0, 0.0, -25, 4.28],\n# 5 : [-1.5, 0.1, 0.5, 1.5, 9.0, -1.0, -25, 10.68],\n# 6 : [-0.5, 0.8, 7.0, 3.0, 10.0, 15.0, -25, 1.75],\n# 7 : [-0.5, 0.1, 0.5, 1.5, 6.0, 15.0, -25, ] \n# }\n\n# for phi\ntruths = {\n1 : [-0.5, 0.1, 0.5, 1.5, 9.0, -0.785, -25, 10.68],\n2 : [-0.5, 0.1, 0.5, 1.5, 9.0, 1.504, -25, 1.41],\n3 : [-0.5, 0.1, 7.0, 3.0, 10, 1.504, -25, 1.75],\n4 : [-0.5, 0.1, 7.0, 3.0, 13.0, 0.0, -25, 4.28],\n5 : [-1.5, 0.1, 0.5, 1.5, 9.0, -0.785, -25, 10.68],\n6 : [-0.5, 0.8, 7.0, 3.0, 10.0, 1.504, -25, 1.75],\n7 : [-0.5, 0.1, 0.5, 1.5, 6.0, 1.504, -25, 2.40],\n8 : [-0.5, 0.1, 0.1, 8.0, 12.0, 1.52, -25, 0.437]\n}",
"_____no_output_____"
],
[
"for id_ in np.arange(8) + 1:\n file_ = f'../resources/SN{id_}_circle_chain.tsv'\n saved_file = f'circleTests/C{id_}-truths-0717.pdf'\n print(f'\\nMaking {saved_file}')\n corner_plot(file_, saved_file, truths[id_])",
"\nMaking circleTests/C1-truths-0717.pdf\n"
],
[
"# just one cirlce test\nid_ = 8\nfile_ = f'../resources/SN{id_}_circle_chain.tsv'\nsaved_file = f'circleTests/C{id_}-truths-0717_1.pdf'\ncorner_plot(file_, saved_file, truths[id_])",
"/usr/local/lib/python3.6/site-packages/matplotlib/pyplot.py:524: RuntimeWarning: More than 20 figures have been opened. Figures created through the pyplot interface (`matplotlib.pyplot.figure`) are retained until explicitly closed and may consume too much memory. (To control this warning, see the rcParam `figure.max_open_warning`).\n max_open_warning, RuntimeWarning)\n"
]
],
[
[
"# Check sections of chain",
"_____no_output_____"
]
],
[
[
"file_ = f'../resources/SN2_chain.tsv'\nsaved_file = f'circleTests/C2-3.pdf'\nprint(f'\\nMaking {saved_file}')\ncorner_plot(file_, saved_file, truths[2], third=3)",
"\nMaking circleTests/C2-3.pdf\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d093fdbd144916355b3446cc135eee051ed832f5 | 5,746 | ipynb | Jupyter Notebook | lectures/07_lecture/2d_lp_example.ipynb | edmund735/companion | 95ef489b6f79f8c375c4ac060c3c1a1506332f8c | [
"MIT"
] | null | null | null | lectures/07_lecture/2d_lp_example.ipynb | edmund735/companion | 95ef489b6f79f8c375c4ac060c3c1a1506332f8c | [
"MIT"
] | null | null | null | lectures/07_lecture/2d_lp_example.ipynb | edmund735/companion | 95ef489b6f79f8c375c4ac060c3c1a1506332f8c | [
"MIT"
] | null | null | null | 31.922222 | 142 | 0.434041 | [
[
[
"import cvxpy as cp\n\n#make decision variable\nx = cp.Variable(2)\n\n#define objective\nobjective = x[0]\n\n#define constraints\nconstraints = [ -1 <= x[0], x[0] <= 1, #inequalities\n -1 <= x[1], x[1] <= 1, #inequalities\n x[0] + x[1] == -1] #equalities\n\n#make problem and solve\nprob = cp.Problem(cp.Minimize(objective),constraints)\nprob.solve()",
"_____no_output_____"
],
[
"#define objective\nobjective = x[0]\n\n#define constraints\nconstraints = [ cp.abs(x[0]) <= 1, #inequalities\n cp.abs(x[1]) <= 1, #inequalities\n x[0] + x[1] == -1] #equalities\n\n#make problem and solve\nprob = cp.Problem(cp.Minimize(objective),constraints)\nprob.solve()",
"_____no_output_____"
],
[
"#define objective\nobjective = x[0]\n\n#define constraints\nconstraints = [ cp.norm(x, 'inf') <= 1, #inequalities\n cp.sum(x) == -1] #equalities\n\n#make problem and solve\nprob = cp.Problem(cp.Minimize(objective),constraints)\nprob.solve()\nprob.solve(solver='ECOS', verbose=True) ",
"===============================================================================\n CVXPY \n v1.1.18 \n===============================================================================\n(CVXPY) Feb 13 05:00:50 PM: Your problem has 2 variables, 2 constraints, and 0 parameters.\n(CVXPY) Feb 13 05:00:50 PM: It is compliant with the following grammars: DCP, DQCP\n(CVXPY) Feb 13 05:00:50 PM: (If you need to solve this problem multiple times, but with different data, consider using parameters.)\n(CVXPY) Feb 13 05:00:50 PM: CVXPY will first compile your problem; then, it will invoke a numerical solver to obtain a solution.\n-------------------------------------------------------------------------------\n Compilation \n-------------------------------------------------------------------------------\n(CVXPY) Feb 13 05:00:50 PM: Compiling problem (target solver=ECOS).\n(CVXPY) Feb 13 05:00:50 PM: Reduction chain: Dcp2Cone -> CvxAttr2Constr -> ConeMatrixStuffing -> ECOS\n(CVXPY) Feb 13 05:00:50 PM: Applying reduction Dcp2Cone\n(CVXPY) Feb 13 05:00:50 PM: Applying reduction CvxAttr2Constr\n(CVXPY) Feb 13 05:00:50 PM: Applying reduction ConeMatrixStuffing\n(CVXPY) Feb 13 05:00:50 PM: Applying reduction ECOS\n(CVXPY) Feb 13 05:00:50 PM: Finished problem compilation (took 1.140e-02 seconds).\n-------------------------------------------------------------------------------\n Numerical solver \n-------------------------------------------------------------------------------\n(CVXPY) Feb 13 05:00:50 PM: Invoking solver ECOS to obtain a solution.\n-------------------------------------------------------------------------------\n Summary \n-------------------------------------------------------------------------------\n(CVXPY) Feb 13 05:00:50 PM: Problem status: optimal\n(CVXPY) Feb 13 05:00:50 PM: Optimal value: -1.000e+00\n(CVXPY) Feb 13 05:00:50 PM: Compilation took 1.140e-02 seconds\n(CVXPY) Feb 13 05:00:50 PM: Solver (including time spent in interface) took 3.099e-04 seconds\n"
],
[
"print(\"x = \", x.value)",
"x = [-1.00000000e+00 -8.24139516e-12]\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code"
]
] |
d09403f15f8d5084937d53a3ac45fa7b0737dfab | 27,910 | ipynb | Jupyter Notebook | Soccer_SQLite.ipynb | gequitz/Issuing-Credit-Cards-and-SQL-Assignment | 53fa1a59b38f5779a31f48efcad6f306febecae2 | [
"MIT"
] | null | null | null | Soccer_SQLite.ipynb | gequitz/Issuing-Credit-Cards-and-SQL-Assignment | 53fa1a59b38f5779a31f48efcad6f306febecae2 | [
"MIT"
] | null | null | null | Soccer_SQLite.ipynb | gequitz/Issuing-Credit-Cards-and-SQL-Assignment | 53fa1a59b38f5779a31f48efcad6f306febecae2 | [
"MIT"
] | null | null | null | 33.789346 | 339 | 0.460122 | [
[
[
"import pandas as pd\nimport sqlite3\n\nconn = sqlite3.connect('database.sqlite')\n\nquery = \"SELECT * FROM sqlite_master\"\n\ndf_schema = pd.read_sql_query(query, conn)\n\ndf_schema.tbl_name.unique()",
"_____no_output_____"
],
[
"df_schema.head(20)",
"_____no_output_____"
],
[
"cur = conn.cursor()\ncur.execute('''SELECT * from pragma_table_info(\"Player_Attributes\")''' )\nrows = cur.fetchall()\n \nfor row in rows[:]:\n print(row)",
"(0, 'id', 'INTEGER', 0, None, 1)\n(1, 'player_fifa_api_id', 'INTEGER', 0, None, 0)\n(2, 'player_api_id', 'INTEGER', 0, None, 0)\n(3, 'date', 'TEXT', 0, None, 0)\n(4, 'overall_rating', 'INTEGER', 0, None, 0)\n(5, 'potential', 'INTEGER', 0, None, 0)\n(6, 'preferred_foot', 'TEXT', 0, None, 0)\n(7, 'attacking_work_rate', 'TEXT', 0, None, 0)\n(8, 'defensive_work_rate', 'TEXT', 0, None, 0)\n(9, 'crossing', 'INTEGER', 0, None, 0)\n(10, 'finishing', 'INTEGER', 0, None, 0)\n(11, 'heading_accuracy', 'INTEGER', 0, None, 0)\n(12, 'short_passing', 'INTEGER', 0, None, 0)\n(13, 'volleys', 'INTEGER', 0, None, 0)\n(14, 'dribbling', 'INTEGER', 0, None, 0)\n(15, 'curve', 'INTEGER', 0, None, 0)\n(16, 'free_kick_accuracy', 'INTEGER', 0, None, 0)\n(17, 'long_passing', 'INTEGER', 0, None, 0)\n(18, 'ball_control', 'INTEGER', 0, None, 0)\n(19, 'acceleration', 'INTEGER', 0, None, 0)\n(20, 'sprint_speed', 'INTEGER', 0, None, 0)\n(21, 'agility', 'INTEGER', 0, None, 0)\n(22, 'reactions', 'INTEGER', 0, None, 0)\n(23, 'balance', 'INTEGER', 0, None, 0)\n(24, 'shot_power', 'INTEGER', 0, None, 0)\n(25, 'jumping', 'INTEGER', 0, None, 0)\n(26, 'stamina', 'INTEGER', 0, None, 0)\n(27, 'strength', 'INTEGER', 0, None, 0)\n(28, 'long_shots', 'INTEGER', 0, None, 0)\n(29, 'aggression', 'INTEGER', 0, None, 0)\n(30, 'interceptions', 'INTEGER', 0, None, 0)\n(31, 'positioning', 'INTEGER', 0, None, 0)\n(32, 'vision', 'INTEGER', 0, None, 0)\n(33, 'penalties', 'INTEGER', 0, None, 0)\n(34, 'marking', 'INTEGER', 0, None, 0)\n(35, 'standing_tackle', 'INTEGER', 0, None, 0)\n(36, 'sliding_tackle', 'INTEGER', 0, None, 0)\n(37, 'gk_diving', 'INTEGER', 0, None, 0)\n(38, 'gk_handling', 'INTEGER', 0, None, 0)\n(39, 'gk_kicking', 'INTEGER', 0, None, 0)\n(40, 'gk_positioning', 'INTEGER', 0, None, 0)\n(41, 'gk_reflexes', 'INTEGER', 0, None, 0)\n"
],
[
"#df_schema.to_csv(\"soccer_schema.csv\")",
"_____no_output_____"
],
[
"cur = conn.cursor()\ncur.execute('''SELECT * from pragma_table_info(\"Player\")''' )\nrows = cur.fetchall()\n \nfor row in rows[:]:\n print(row)",
"(0, 'id', 'INTEGER', 0, None, 1)\n(1, 'player_api_id', 'INTEGER', 0, None, 0)\n(2, 'player_name', 'TEXT', 0, None, 0)\n(3, 'player_fifa_api_id', 'INTEGER', 0, None, 0)\n(4, 'birthday', 'TEXT', 0, None, 0)\n(5, 'height', 'INTEGER', 0, None, 0)\n(6, 'weight', 'INTEGER', 0, None, 0)\n"
],
[
"cur = conn.cursor()\ncur.execute('''SELECT * from pragma_table_info(\"Match\")''' )\nrows = cur.fetchall()\n \nfor row in rows[:]:\n print(row)",
"(0, 'id', 'INTEGER', 0, None, 1)\n(1, 'country_id', 'INTEGER', 0, None, 0)\n(2, 'league_id', 'INTEGER', 0, None, 0)\n(3, 'season', 'TEXT', 0, None, 0)\n(4, 'stage', 'INTEGER', 0, None, 0)\n(5, 'date', 'TEXT', 0, None, 0)\n(6, 'match_api_id', 'INTEGER', 0, None, 0)\n(7, 'home_team_api_id', 'INTEGER', 0, None, 0)\n(8, 'away_team_api_id', 'INTEGER', 0, None, 0)\n(9, 'home_team_goal', 'INTEGER', 0, None, 0)\n(10, 'away_team_goal', 'INTEGER', 0, None, 0)\n(11, 'home_player_X1', 'INTEGER', 0, None, 0)\n(12, 'home_player_X2', 'INTEGER', 0, None, 0)\n(13, 'home_player_X3', 'INTEGER', 0, None, 0)\n(14, 'home_player_X4', 'INTEGER', 0, None, 0)\n(15, 'home_player_X5', 'INTEGER', 0, None, 0)\n(16, 'home_player_X6', 'INTEGER', 0, None, 0)\n(17, 'home_player_X7', 'INTEGER', 0, None, 0)\n(18, 'home_player_X8', 'INTEGER', 0, None, 0)\n(19, 'home_player_X9', 'INTEGER', 0, None, 0)\n(20, 'home_player_X10', 'INTEGER', 0, None, 0)\n(21, 'home_player_X11', 'INTEGER', 0, None, 0)\n(22, 'away_player_X1', 'INTEGER', 0, None, 0)\n(23, 'away_player_X2', 'INTEGER', 0, None, 0)\n(24, 'away_player_X3', 'INTEGER', 0, None, 0)\n(25, 'away_player_X4', 'INTEGER', 0, None, 0)\n(26, 'away_player_X5', 'INTEGER', 0, None, 0)\n(27, 'away_player_X6', 'INTEGER', 0, None, 0)\n(28, 'away_player_X7', 'INTEGER', 0, None, 0)\n(29, 'away_player_X8', 'INTEGER', 0, None, 0)\n(30, 'away_player_X9', 'INTEGER', 0, None, 0)\n(31, 'away_player_X10', 'INTEGER', 0, None, 0)\n(32, 'away_player_X11', 'INTEGER', 0, None, 0)\n(33, 'home_player_Y1', 'INTEGER', 0, None, 0)\n(34, 'home_player_Y2', 'INTEGER', 0, None, 0)\n(35, 'home_player_Y3', 'INTEGER', 0, None, 0)\n(36, 'home_player_Y4', 'INTEGER', 0, None, 0)\n(37, 'home_player_Y5', 'INTEGER', 0, None, 0)\n(38, 'home_player_Y6', 'INTEGER', 0, None, 0)\n(39, 'home_player_Y7', 'INTEGER', 0, None, 0)\n(40, 'home_player_Y8', 'INTEGER', 0, None, 0)\n(41, 'home_player_Y9', 'INTEGER', 0, None, 0)\n(42, 'home_player_Y10', 'INTEGER', 0, None, 0)\n(43, 'home_player_Y11', 'INTEGER', 0, None, 0)\n(44, 'away_player_Y1', 'INTEGER', 0, None, 0)\n(45, 'away_player_Y2', 'INTEGER', 0, None, 0)\n(46, 'away_player_Y3', 'INTEGER', 0, None, 0)\n(47, 'away_player_Y4', 'INTEGER', 0, None, 0)\n(48, 'away_player_Y5', 'INTEGER', 0, None, 0)\n(49, 'away_player_Y6', 'INTEGER', 0, None, 0)\n(50, 'away_player_Y7', 'INTEGER', 0, None, 0)\n(51, 'away_player_Y8', 'INTEGER', 0, None, 0)\n(52, 'away_player_Y9', 'INTEGER', 0, None, 0)\n(53, 'away_player_Y10', 'INTEGER', 0, None, 0)\n(54, 'away_player_Y11', 'INTEGER', 0, None, 0)\n(55, 'home_player_1', 'INTEGER', 0, None, 0)\n(56, 'home_player_2', 'INTEGER', 0, None, 0)\n(57, 'home_player_3', 'INTEGER', 0, None, 0)\n(58, 'home_player_4', 'INTEGER', 0, None, 0)\n(59, 'home_player_5', 'INTEGER', 0, None, 0)\n(60, 'home_player_6', 'INTEGER', 0, None, 0)\n(61, 'home_player_7', 'INTEGER', 0, None, 0)\n(62, 'home_player_8', 'INTEGER', 0, None, 0)\n(63, 'home_player_9', 'INTEGER', 0, None, 0)\n(64, 'home_player_10', 'INTEGER', 0, None, 0)\n(65, 'home_player_11', 'INTEGER', 0, None, 0)\n(66, 'away_player_1', 'INTEGER', 0, None, 0)\n(67, 'away_player_2', 'INTEGER', 0, None, 0)\n(68, 'away_player_3', 'INTEGER', 0, None, 0)\n(69, 'away_player_4', 'INTEGER', 0, None, 0)\n(70, 'away_player_5', 'INTEGER', 0, None, 0)\n(71, 'away_player_6', 'INTEGER', 0, None, 0)\n(72, 'away_player_7', 'INTEGER', 0, None, 0)\n(73, 'away_player_8', 'INTEGER', 0, None, 0)\n(74, 'away_player_9', 'INTEGER', 0, None, 0)\n(75, 'away_player_10', 'INTEGER', 0, None, 0)\n(76, 'away_player_11', 'INTEGER', 0, None, 0)\n(77, 'goal', 'TEXT', 0, None, 0)\n(78, 'shoton', 'TEXT', 0, None, 0)\n(79, 'shotoff', 'TEXT', 0, None, 0)\n(80, 'foulcommit', 'TEXT', 0, None, 0)\n(81, 'card', 'TEXT', 0, None, 0)\n(82, 'cross', 'TEXT', 0, None, 0)\n(83, 'corner', 'TEXT', 0, None, 0)\n(84, 'possession', 'TEXT', 0, None, 0)\n(85, 'B365H', 'NUMERIC', 0, None, 0)\n(86, 'B365D', 'NUMERIC', 0, None, 0)\n(87, 'B365A', 'NUMERIC', 0, None, 0)\n(88, 'BWH', 'NUMERIC', 0, None, 0)\n(89, 'BWD', 'NUMERIC', 0, None, 0)\n(90, 'BWA', 'NUMERIC', 0, None, 0)\n(91, 'IWH', 'NUMERIC', 0, None, 0)\n(92, 'IWD', 'NUMERIC', 0, None, 0)\n(93, 'IWA', 'NUMERIC', 0, None, 0)\n(94, 'LBH', 'NUMERIC', 0, None, 0)\n(95, 'LBD', 'NUMERIC', 0, None, 0)\n(96, 'LBA', 'NUMERIC', 0, None, 0)\n(97, 'PSH', 'NUMERIC', 0, None, 0)\n(98, 'PSD', 'NUMERIC', 0, None, 0)\n(99, 'PSA', 'NUMERIC', 0, None, 0)\n(100, 'WHH', 'NUMERIC', 0, None, 0)\n(101, 'WHD', 'NUMERIC', 0, None, 0)\n(102, 'WHA', 'NUMERIC', 0, None, 0)\n(103, 'SJH', 'NUMERIC', 0, None, 0)\n(104, 'SJD', 'NUMERIC', 0, None, 0)\n(105, 'SJA', 'NUMERIC', 0, None, 0)\n(106, 'VCH', 'NUMERIC', 0, None, 0)\n(107, 'VCD', 'NUMERIC', 0, None, 0)\n(108, 'VCA', 'NUMERIC', 0, None, 0)\n(109, 'GBH', 'NUMERIC', 0, None, 0)\n(110, 'GBD', 'NUMERIC', 0, None, 0)\n(111, 'GBA', 'NUMERIC', 0, None, 0)\n(112, 'BSH', 'NUMERIC', 0, None, 0)\n(113, 'BSD', 'NUMERIC', 0, None, 0)\n(114, 'BSA', 'NUMERIC', 0, None, 0)\n"
],
[
"cur = conn.cursor()\ncur.execute('''SELECT * from pragma_table_info(\"League\")''' )\nrows = cur.fetchall()\n \nfor row in rows[:]:\n print(row)",
"(0, 'id', 'INTEGER', 0, None, 1)\n(1, 'country_id', 'INTEGER', 0, None, 0)\n(2, 'name', 'TEXT', 0, None, 0)\n"
],
[
"cur = conn.cursor()\ncur.execute('''SELECT * from pragma_table_info(\"Country\")''' )\nrows = cur.fetchall()\n \nfor row in rows[:]:\n print(row)",
"(0, 'id', 'INTEGER', 0, None, 1)\n(1, 'name', 'TEXT', 0, None, 0)\n"
],
[
"cur = conn.cursor()\ncur.execute('''SELECT * from pragma_table_info(\"Team\")''' )\nrows = cur.fetchall()\n \nfor row in rows[:]:\n print(row)",
"(0, 'id', 'INTEGER', 0, None, 1)\n(1, 'team_api_id', 'INTEGER', 0, None, 0)\n(2, 'team_fifa_api_id', 'INTEGER', 0, None, 0)\n(3, 'team_long_name', 'TEXT', 0, None, 0)\n(4, 'team_short_name', 'TEXT', 0, None, 0)\n"
],
[
"cur = conn.cursor()\ncur.execute('''SELECT * from pragma_table_info(\"Team_Attributes\")''' )\nrows = cur.fetchall()\n \nfor row in rows[:]:\n print(row)",
"(0, 'id', 'INTEGER', 0, None, 1)\n(1, 'team_fifa_api_id', 'INTEGER', 0, None, 0)\n(2, 'team_api_id', 'INTEGER', 0, None, 0)\n(3, 'date', 'TEXT', 0, None, 0)\n(4, 'buildUpPlaySpeed', 'INTEGER', 0, None, 0)\n(5, 'buildUpPlaySpeedClass', 'TEXT', 0, None, 0)\n(6, 'buildUpPlayDribbling', 'INTEGER', 0, None, 0)\n(7, 'buildUpPlayDribblingClass', 'TEXT', 0, None, 0)\n(8, 'buildUpPlayPassing', 'INTEGER', 0, None, 0)\n(9, 'buildUpPlayPassingClass', 'TEXT', 0, None, 0)\n(10, 'buildUpPlayPositioningClass', 'TEXT', 0, None, 0)\n(11, 'chanceCreationPassing', 'INTEGER', 0, None, 0)\n(12, 'chanceCreationPassingClass', 'TEXT', 0, None, 0)\n(13, 'chanceCreationCrossing', 'INTEGER', 0, None, 0)\n(14, 'chanceCreationCrossingClass', 'TEXT', 0, None, 0)\n(15, 'chanceCreationShooting', 'INTEGER', 0, None, 0)\n(16, 'chanceCreationShootingClass', 'TEXT', 0, None, 0)\n(17, 'chanceCreationPositioningClass', 'TEXT', 0, None, 0)\n(18, 'defencePressure', 'INTEGER', 0, None, 0)\n(19, 'defencePressureClass', 'TEXT', 0, None, 0)\n(20, 'defenceAggression', 'INTEGER', 0, None, 0)\n(21, 'defenceAggressionClass', 'TEXT', 0, None, 0)\n(22, 'defenceTeamWidth', 'INTEGER', 0, None, 0)\n(23, 'defenceTeamWidthClass', 'TEXT', 0, None, 0)\n(24, 'defenceDefenderLineClass', 'TEXT', 0, None, 0)\n"
]
],
[
[
"## Question 1: Which team scored the most points when playing at home?",
"_____no_output_____"
]
],
[
[
"cur = conn.cursor()\ncur.execute(\"SELECT sum(a.home_team_goal) as sum_goals, b.team_api_id, b.team_long_name FROM match a, team b WHERE a.home_team_api_id = b.team_api_id group by b.team_api_id order by sum_goals desc limit 1\" )\nrows = cur.fetchall()\n \nfor row in rows[:]:\n print(row)\n",
"(505, 8633, 'Real Madrid CF')\n"
]
],
[
[
"## Question 2: Did this team also score the most points when playing away?",
"_____no_output_____"
]
],
[
[
"cur = conn.cursor()\n\ncur.execute(\"SELECT sum(a.away_team_goal) as sum_goals, b.team_api_id, b.team_long_name FROM match a, team b WHERE a.away_team_api_id = b.team_api_id group by b.team_api_id order by sum_goals desc limit 1\" )\nrows = cur.fetchall()\n \nfor row in rows[:]:\n print(row)",
"(354, 8634, 'FC Barcelona')\n"
]
],
[
[
"## Question 3: How many matches resulted in a tie?",
"_____no_output_____"
]
],
[
[
"cur = conn.cursor()\ncur.execute(\"SELECT count(match_api_id) FROM match where home_team_goal = away_team_goal\" )\nrows = cur.fetchall()\n \nfor row in rows[:]:\n print(row)",
"(6596,)\n"
]
],
[
[
"## Question 4: How many players have Smith for their last name? How many have 'smith' anywhere in their name?",
"_____no_output_____"
]
],
[
[
"cur = conn.cursor()\ncur.execute(\"SELECT COUNT(player_name) FROM Player where player_name LIKE '% smith' \" )\nrows = cur.fetchall()\n \nfor row in rows[:]:\n print(row)\n\n",
"(15,)\n"
],
[
"cur = conn.cursor()\ncur.execute(\"SELECT COUNT(player_name) FROM Player where player_name LIKE '%smith%' \" )\nrows = cur.fetchall()\n \nfor row in rows[:]:\n print(row)",
"(18,)\n"
]
],
[
[
"## Question 5: What was the median tie score? Use the value determined in the previous question for the number of tie games. Hint: PostgreSQL does not have a median function. Instead, think about the steps required to calculate a median and use the WITH command to store stepwise results as a table and then operate on these results.",
"_____no_output_____"
]
],
[
[
"cur = conn.cursor()\n#cur.execute(\"WITH goal_list AS (SELECT home_team_goal FROM match where home_team_goal = away_team_goal order \\\n# by home_team_goal desc) select home_team_goal from goal_list limit 1 offset 6596/2\" )\n\ncur.execute(\"WITH goal_list AS (SELECT home_team_goal FROM match where home_team_goal = away_team_goal order \\\n by home_team_goal desc) select home_team_goal from goal_list limit 1 offset (select count(*) from goal_list)/2\" )\n\n\nrows = cur.fetchall()\n \nfor row in rows[:20]:\n print(row)\n \n ",
"(1,)\n"
]
],
[
[
"## Question 6: What percentage of players prefer their left or right foot? Hint: Calculate either the right or left foot, whichever is easier based on how you setup the problem.",
"_____no_output_____"
]
],
[
[
"cur = conn.cursor()\n\n\ncur.execute(\"SELECT (COUNT(DISTINCT(player_api_id)) * 100.0 / (SELECT COUNT(DISTINCT(player_api_id)) FROM Player_Attributes)) \\\n FROM Player_Attributes WHERE preferred_foot LIKE '%right%' \" )\n\n\nrows = cur.fetchall()\n \nfor row in rows[:20]:\n print(row)\n \n#SELECT (COUNT(DISTINCT(player_api_id)) * 100.0 / (SELECT COUNT(DISTINCT(player_api_id)) FROM Player_Attributes)) as percentage\n#FROM Player_Attributes\n#WHERE preferred_foot LIKE '%left%'",
"(81.18444846292948,)\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0940e2fb4ebaf25a5c72ee00b4bc7dbbe4a5e69 | 418,730 | ipynb | Jupyter Notebook | example.ipynb | dalekreitler-bnl/mxscreen | 9eae13654839875c8b6fe3ed3bdafd6bd3b3094c | [
"MIT"
] | null | null | null | example.ipynb | dalekreitler-bnl/mxscreen | 9eae13654839875c8b6fe3ed3bdafd6bd3b3094c | [
"MIT"
] | null | null | null | example.ipynb | dalekreitler-bnl/mxscreen | 9eae13654839875c8b6fe3ed3bdafd6bd3b3094c | [
"MIT"
] | null | null | null | 803.704415 | 20,124 | 0.949875 | [
[
[
"from mxscreen.burn import burner\nimport matplotlib.pyplot as plt\n\nf1 = \"/Users/dkreitler/xray/mxscreen_test_data/proteinaseK_static_burn_center_24539_master.h5\"\nf2 = \"/Users/dkreitler/xray/mxscreen_test_data/SPOT_BURN_PK_DIALS.XDS\"\nf3 = \"/Users/dkreitler/xray/mxscreen_test_data/SPOT_BURN_PK_DOZOR.XDS\"\n \nburnExperimentDials = burner.BurnExperiment(f2,\n f1,\n nResShells=10,\n resRangeAll=(1,5),\n nPsiWedges=1,\n psiRangeAll=(0,180),\n frameRangeAll=(2,175),\n decayStrategy=\"doubleExponential\")\n\nburnExperimentDials.psiBounds()\ndialsBounds=burnExperimentDials.resBounds()\nprint('dialsBounds',dialsBounds)\nboundsdict={'resBounds': dialsBounds}\nburnExperimentDozor = burner.BurnExperiment(f3,\n f1,\n nResShells=10,\n resRangeAll=(1,5),\n nPsiWedges=1,\n psiRangeAll=(270,360),\n frameRangeAll=(2,175),\n decayStrategy=\"doubleExponential\",\n **boundsdict)\nburnExperimentDozor.psiBounds()\n#burnExperimentDozor.resBounds()\nburnExperimentDials.rangesDictList()\nburnExperimentDozor.rangesDictList()\nburnExperimentDials.spotGroupList()\nburnExperimentDozor.spotGroupList()\nburnExperimentDials.fitSpotGroups()\nburnExperimentDozor.fitSpotGroups()\nresultsDials = burnExperimentDials.experimentPlot()\nresultsDozor = burnExperimentDozor.experimentPlot()\n \nprint('dials results\\n',resultsDials)\nprint('dozor results\\n',resultsDozor)\n \nplt.plot(resultsDials[:,0],resultsDials[:,1],'b-o',label='dials')\nplt.plot(resultsDozor[:,0],resultsDozor[:,1],'g-o',label='dozor')\nplt.legend()\nplt.ylabel('Half life as Frame no.')\nplt.xlabel('resolution bin number')\nplt.show()",
"254518\nDIMSIZE 25451\nRESBOUNDS [1.11504301 1.97165096 2.22320969 2.46486892 2.69921323 2.92650869\n 3.20759107 3.48046812 3.85988894 4.27777565 4.99837116]\ndialsBounds [1.11504301 1.97165096 2.22320969 2.46486892 2.69921323 2.92650869\n 3.20759107 3.48046812 3.85988894 4.27777565 4.99837116]\nresRangeBounds [1.11504301 1.97165096 2.22320969 2.46486892 2.69921323 2.92650869\n 3.20759107 3.48046812 3.85988894 4.27777565 4.99837116]\nresRangeBounds [1.11504301 1.97165096 2.22320969 2.46486892 2.69921323 2.92650869\n 3.20759107 3.48046812 3.85988894 4.27777565 4.99837116]\nRANGESTEST {'resBin': 0, 'psiWedge': 0, 'resRange': (1.1150430064108419, 1.9716509636401365), 'psiRange': (0.0, 180.0), 'frameRange': (2, 175)}\n"
]
]
] | [
"code"
] | [
[
"code"
]
] |
d09458dc146d61d7a28bccb28c40b4b9dd86fc31 | 14,282 | ipynb | Jupyter Notebook | Heart Disease Risk Prediction/Heart Disease Risk Prediction.ipynb | somyasriv16/AN319_H4CK3RS_SIH2020 | 5a6e15ec76530425b4e611f663bb3d322ff3e8e8 | [
"MIT"
] | null | null | null | Heart Disease Risk Prediction/Heart Disease Risk Prediction.ipynb | somyasriv16/AN319_H4CK3RS_SIH2020 | 5a6e15ec76530425b4e611f663bb3d322ff3e8e8 | [
"MIT"
] | null | null | null | Heart Disease Risk Prediction/Heart Disease Risk Prediction.ipynb | somyasriv16/AN319_H4CK3RS_SIH2020 | 5a6e15ec76530425b4e611f663bb3d322ff3e8e8 | [
"MIT"
] | null | null | null | 14,282 | 14,282 | 0.696541 | [
[
[
"# Importing the libraries",
"_____no_output_____"
]
],
[
[
"%pip install tensorflow",
"_____no_output_____"
],
[
"import pandas as pd\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport tensorflow as tf\nimport seaborn as sns",
"_____no_output_____"
]
],
[
[
"# Importing The Dataset",
"_____no_output_____"
]
],
[
[
"dataset = pd.read_csv(\"../input/framingham-heart-study-dataset/framingham.csv\")",
"_____no_output_____"
]
],
[
[
"# Analysing The Data",
"_____no_output_____"
]
],
[
[
"dataset.shape",
"_____no_output_____"
],
[
"dataset.dtypes",
"_____no_output_____"
],
[
"dataset.info",
"_____no_output_____"
]
],
[
[
"# Visualizing the data",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize = (8,8))\nax = fig.gca()\ndataset.hist(ax=ax)\nplt.show()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\nax.hist(dataset[\"TenYearCHD\"],color = \"yellow\")\nax.set_title(' To predict heart disease')\nax.set_xlabel('TenYearCHD')\nax.set_ylabel('Frequency')",
"_____no_output_____"
],
[
"data = np.random.random([100,4])\nsns.violinplot(data=data, palette=['r','g','b','m'])",
"_____no_output_____"
]
],
[
[
"# Separating the dependent and independent variables",
"_____no_output_____"
]
],
[
[
"X = dataset.iloc[:,:-1].values\ny = dataset.iloc[:,-1].values",
"_____no_output_____"
],
[
"np.isnan(X).sum()",
"_____no_output_____"
],
[
"np.isnan(y).sum()",
"_____no_output_____"
]
],
[
[
"# Taking Care of Missing Values",
"_____no_output_____"
]
],
[
[
"from sklearn.impute import SimpleImputer\nsi = SimpleImputer(missing_values = np.nan, strategy = 'mean')\nX = si.fit_transform(X)",
"_____no_output_____"
],
[
"y.shape",
"_____no_output_____"
],
[
"np.isnan(X).sum()",
"_____no_output_____"
],
[
"np.isnan(y).sum()",
"_____no_output_____"
],
[
"dataset.isna().sum()",
"_____no_output_____"
]
],
[
[
"# Splitting into Training and test Data",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\nX_train,X_test,y_train,y_test = train_test_split(X,y,test_size = 0.3,random_state = 0)",
"_____no_output_____"
]
],
[
[
"# Normalising The data",
"_____no_output_____"
]
],
[
[
"from sklearn.preprocessing import StandardScaler\nsc = StandardScaler()\nX_train = sc.fit_transform(X_train)\nX_test = sc.transform(X_test)",
"_____no_output_____"
],
[
"X_train",
"_____no_output_____"
],
[
"y_train",
"_____no_output_____"
],
[
"np.isnan(X_train).sum()",
"_____no_output_____"
],
[
"np.isnan(y_train).sum()",
"_____no_output_____"
]
],
[
[
"# Preparing ANN Model with two layers",
"_____no_output_____"
]
],
[
[
"ann = tf.keras.models.Sequential()",
"_____no_output_____"
],
[
"ann.add(tf.keras.layers.Dense(units = 6, activation = 'relu'))",
"_____no_output_____"
],
[
"ann.add(tf.keras.layers.Dense(units = 6, activation='relu'))",
"_____no_output_____"
],
[
"ann.add(tf.keras.layers.Dense(units = 1,activation='sigmoid'))",
"_____no_output_____"
],
[
"ann.compile(optimizer = 'adam', loss = 'binary_crossentropy', metrics = ['accuracy'])",
"_____no_output_____"
],
[
"model = ann.fit(X_train,y_train,validation_data=(X_test,y_test), batch_size = 32,epochs=100)",
"_____no_output_____"
],
[
"y_pred = ann.predict(X_test)\ny_pred = (y_pred > 0.5)\nprint(np.concatenate((y_pred.reshape(len(y_pred),1), y_test.reshape(len(y_test),1)),1))",
"_____no_output_____"
],
[
"from sklearn.metrics import confusion_matrix, accuracy_score, classification_report\ncm = confusion_matrix(y_test, y_pred)\nprint(cm)\naccuracy_score(y_test, y_pred)",
"_____no_output_____"
]
],
[
[
"# Model Accuracy Visualisation",
"_____no_output_____"
]
],
[
[
"plt.plot(model.history['accuracy'])\nplt.plot(model.history['val_accuracy'])\nplt.title('Model Accuracy')\nplt.ylabel('Accuracy')\nplt.xlabel('Epoch')\nplt.legend(['Train', 'Validation'], loc='lower right')\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Model Loss Visualisation",
"_____no_output_____"
]
],
[
[
"plt.plot(model.history['loss'])\nplt.plot(model.history['val_loss'])\nplt.title('Model Loss')\nplt.ylabel('loss')\nplt.xlabel('Epoch')\nplt.legend(['Train', 'Validation'], loc='upper right')\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Calculating Different Metrics",
"_____no_output_____"
]
],
[
[
"print(classification_report(y_test, y_pred))",
"_____no_output_____"
]
],
[
[
"# Using MLP Classifier for Prediction",
"_____no_output_____"
]
],
[
[
"from sklearn.neural_network import MLPClassifier\nclassifier = MLPClassifier(hidden_layer_sizes=(150,100,50), max_iter=300,activation = 'relu',solver='adam',random_state=1)",
"_____no_output_____"
],
[
"classifier.fit(X_train, y_train)",
"_____no_output_____"
],
[
"y_pred = classifier.predict(X_test)\ny_pred = (y_pred > 0.5)\nprint(np.concatenate((y_pred.reshape(len(y_pred),1), y_test.reshape(len(y_test),1)),1))",
"_____no_output_____"
],
[
"from sklearn.metrics import confusion_matrix, accuracy_score, classification_report\ncm = confusion_matrix(y_test, y_pred)\nprint(cm)\naccuracy_score(y_test, y_pred)",
"_____no_output_____"
],
[
"print(classification_report(y_test, y_pred))",
"_____no_output_____"
]
],
[
[
"# Visualising The MLP Model After Applying the PCA method",
"_____no_output_____"
]
],
[
[
"from sklearn.decomposition import PCA\npca = PCA(n_components=2)\n\nX_train = pca.fit_transform(X_train)\nX_test = pca.transform(X_test)",
"_____no_output_____"
],
[
"classifier.fit(X_train, y_train)",
"_____no_output_____"
],
[
"def visualization_train(model):\n sns.set_context(context='notebook',font_scale=2)\n plt.figure(figsize=(16,9))\n from matplotlib.colors import ListedColormap\n X_set, y_set = X_train, y_train\n X1, X2 = np.meshgrid(np.arange(start = X_set[:, 0].min() - 1, stop = X_set[:, 0].max() + 1, step = 0.01),\n np.arange(start = X_set[:, 1].min() - 1, stop = X_set[:, 1].max() + 1, step = 0.01))\n plt.contourf(X1, X2, classifier.predict(np.array([X1.ravel(), X2.ravel()]).T).reshape(X1.shape),\n alpha = 0.6, cmap = ListedColormap(('red', 'green')))\n plt.xlim(X1.min(), X1.max())\n plt.ylim(X2.min(), X2.max())\n for i, j in enumerate(np.unique(y_set)):\n plt.scatter(X_set[y_set == j, 0], X_set[y_set == j, 1],\n c = ListedColormap(('red', 'green'))(i), label = j)\n plt.title(\"%s Model on training data\" %(model))\n plt.xlabel('PC 1')\n plt.ylabel('PC 2')\n plt.legend()\ndef visualization_test(model):\n sns.set_context(context='notebook',font_scale=2)\n plt.figure(figsize=(16,9))\n from matplotlib.colors import ListedColormap\n X_set, y_set = X_test, y_test\n X1, X2 = np.meshgrid(np.arange(start = X_set[:, 0].min() - 1, stop = X_set[:, 0].max() + 1, step = 0.01),\n np.arange(start = X_set[:, 1].min() - 1, stop = X_set[:, 1].max() + 1, step = 0.01))\n plt.contourf(X1, X2, classifier.predict(np.array([X1.ravel(), X2.ravel()]).T).reshape(X1.shape),\n alpha = 0.6, cmap = ListedColormap(('red', 'green')))\n plt.xlim(X1.min(), X1.max())\n plt.ylim(X2.min(), X2.max())\n for i, j in enumerate(np.unique(y_set)):\n plt.scatter(X_set[y_set == j, 0], X_set[y_set == j, 1],\n c = ListedColormap(('red', 'green'))(i), label = j)\n plt.title(\"%s Test Set\" %(model))\n plt.xlabel('PC 1')\n plt.ylabel('PC 2')\n plt.legend()",
"_____no_output_____"
],
[
"visualization_train(model= 'MLP')",
"_____no_output_____"
]
],
[
[
"# Saving a machine learning Model",
"_____no_output_____"
]
],
[
[
"#import joblib\n#joblib.dump(ann, 'ann_model.pkl') \n#joblib.dump(sc, 'sc_model.pkl') ",
"_____no_output_____"
],
[
"#knn_from_joblib = joblib.load('mlp_model.pkl')\n#sc_model = joblib.load('sc_model.pkl') ",
"_____no_output_____"
]
],
[
[
"# Saving a tensorflow model",
"_____no_output_____"
]
],
[
[
"#!pip install h5py",
"_____no_output_____"
],
[
"#ann.save('ann_model.h5')",
"_____no_output_____"
],
[
"#model = tf.keras.models.load_model('ann_model.h5')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d09463b9a3106e4273176817073e181032aaa38b | 68,203 | ipynb | Jupyter Notebook | lessons/13_Step_10.ipynb | fayazr/CFDPython | 83d175d4940322ec4e0f3b251543cd40d387913d | [
"CC-BY-3.0"
] | 1 | 2018-02-05T09:03:55.000Z | 2018-02-05T09:03:55.000Z | lessons/13_Step_10.ipynb | abahman/CFDPython | 83d175d4940322ec4e0f3b251543cd40d387913d | [
"CC-BY-3.0"
] | null | null | null | lessons/13_Step_10.ipynb | abahman/CFDPython | 83d175d4940322ec4e0f3b251543cd40d387913d | [
"CC-BY-3.0"
] | 1 | 2019-08-09T23:12:06.000Z | 2019-08-09T23:12:06.000Z | 171.364322 | 54,141 | 0.859669 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d0946abbcb737c6eb40e2ed03b8e92a66f3b0a88 | 346,549 | ipynb | Jupyter Notebook | 16. Reinforcement Learning.ipynb | MRD-Git/Hands-On-Machine-Learning | 41bb0adcd788d11922546e4f391e7604c78176cc | [
"Apache-2.0"
] | null | null | null | 16. Reinforcement Learning.ipynb | MRD-Git/Hands-On-Machine-Learning | 41bb0adcd788d11922546e4f391e7604c78176cc | [
"Apache-2.0"
] | null | null | null | 16. Reinforcement Learning.ipynb | MRD-Git/Hands-On-Machine-Learning | 41bb0adcd788d11922546e4f391e7604c78176cc | [
"Apache-2.0"
] | null | null | null | 62.239404 | 34,211 | 0.617688 | [
[
[
"# Reinforcement Learning\npage 441<br>\nFor details, see\n- https://github.com/ageron/handson-ml/blob/master/16_reinforcement_learning.ipynb,\n- https://gym.openai.com,\n- http://www0.cs.ucl.ac.uk/staff/d.silver/web/Teaching.html,\n- https://www.jstor.org/stable/24900506?seq=1#metadata_info_tab_contents,\n- http://book.pythontips.com/en/latest/enumerate.html, and\n- https://docs.python.org/2.3/whatsnew/section-slices.html (on slices).\n\nReinforcement Learning is one of the most excititng fields of Machine Learning today. It has been around since the 1950s but flying below the radar most of the time. In 2013, *DeepMind* (at that time still an independent start-up in London) produced a reinforcement learning algorithm that could achieve superhuman performance of playing about any Atari game without having prior domain knowledge. Notably, *AlphaGo*, also a reinforcement learning algorithm by Deepmind, beat world champion *Lee Sedol* in the board game *Go* in March 2016. This has become possible by applying the power of Deep Learning to the field of Reinforcement Learning.<br><br>\nTwo very important concepts for Reinforcement Learning are **policy gradients** and **deep Q-networks** (DQN).\n## Learning to Optimize Rewards\npage 442<br>\nReinforcement learning utilizes a software **agent** that makes **observations** and takes **actions** in an **environment** by which it can earn **rewards**. Its objective is to learn to act in a way that is expected to maximise long-term rewards. This can be linked to human behaviour as humans seem to try to minimize pain (negative reward / penalty) and maximise pleasure (positive reward). The setting of Reinforcement Learning is quite broad. The following list shows some typical applications.\n- The agent can be the program controlling a walking robot. In this case, the environment is the real world, the agent obersrves the environment through a set of *sensors* such as cameras and touch sensors, and its actions consist of sending signals to activate motors. It may be programmed to get positive rewards whenever it approaches the target destination, and negative rewards whenever it wastes time, goes in the wrong direction, or falls down.\n- The agent can be the program controlling Ms. Pac-Man. In this case, the environment is a simulation of the Atari game, the actions are the nine possible joystick positions (upper left, down, center, and so on), the observations are screenshots, and the rewards are just the game points.\n- Similarly, the agent can be the program playing a board game such as the game of *Go*.\n- The agent does not have to control a physically (or virtually ) moving thing. For example, it can be a smart thermostat, getting rewards whenever it is close to the target temperature and saves energy, and negative rewards when humans need to tweak the temperature, so the agent must learn to anticipate human needs.\n- The agent can observe stock market prices and decide how much to buy or sell every second. Rewards are obviously the monetary gains and losses.\n\nNote that it is not necessary to have both positive and negative rewards. For example, using only negative rewards can be useful when a task shall be finished as soon as possible. There are many more applications of Reinforcement Learning, e.g., self-driving cars or placing ads on a webpage (maximizing clicks and/or buys).\n## Policy Search\npage 444<br>\nThe algorithm that the software agent uses to determine its action is called its **policy**. For example, the policy could be a neural network taking inputs (observations) and producing outputs (actions). The policy can be pretty much any algorithm. If that algorithm is not deterministic, it is called a *stochastic policy*.<br><br>\n\nA simple, stochastic policy for a vacuum cleaner could be to go straight for a specified time and then turn with probability $p$ by a random angle $r$. In that example, the algorithm's *policy parameters* are $p$ and $r$. One example of *policy search* would be to try out all possible parameter values and then settle for those values that perform best (e.g., most dust picked up per time). However, if the *policy space* is too large, as is often the case, a near-optimal policy will likely be missed by such brute force approach.<br><br>\n\n*Genetic algorithms* are another way to explore the policy space. For example, one may randomly create 100 policies, rank their performance on the task, and then filter out the bad performers. This could be done by strictly filtering out the 80 lowest performers, or filter out a policy with a probability that is high if the performance is low (this works often better). To restore the population of 100 policies, each of the 20 survivors may produce 4 offsprings, i.e., copies of the survivor[s] (parent algorithm[s]) with slight random modifications of its [their] policy parameters. With a single parent [two or more parents], this is referred to as asexual [sexual] reproduction. The surviving policies together with their offsprins constitute the next generation. This scheme can be continued until a satisfactory policy is found.<br><br>\nYet another approach is to use optimization techniques, e.g., *gradient ascent* (similar to gradient descent) to maximise the reward (similar to minimising the cost).\n## Introduction to OpenAI Gym\npage 445<br>\nIn order to train an agent, that agent needs an *environment* to operate in. The environment evolves subject to the agent's actions and the agent observes the environment. For example, an Atari game simulator is the environment for an agent that tries to learn to play an Atari game. If the agent tries to learn how to walk a humanoid robot in the real world, then the real world is the environment. Yet, the real world may have limitations when compared to software environments: if the real-world robot gets damaged, just clicking \"undo\" or \"restart\" will not suffice. It is also not possible to speed up training by having the time run faster (which would be the equivalent of increasing the clock rate or using a faster processor). And having thousands or millions of real-world robots train in parallel will be much more expensive than running simulations in parallel.<br><br>\nIn short, one generally wants to use a *simulated environment* at least for getting started. Here, we use the *OpenAI gym* (link at the top). A minimal installation only requires to run \"pip3 install --upgrade gym\" in a terminal. We shall try it out!",
"_____no_output_____"
]
],
[
[
"import gym # import gym from OpenAI\nenv = gym.make(\"CartPole-v0\") # make CartPole environment\nprint(env) # print the cartpole environment\nobs = env.reset() # reset observations (in case this is necessary)\nprint(obs) # print the observations",
"\u001b[33mWARN: gym.spaces.Box autodetected dtype as <class 'numpy.float32'>. Please provide explicit dtype.\u001b[0m\n<TimeLimit<CartPoleEnv<CartPole-v0>>>\n[ 0.01911763 -0.04981526 0.02235611 0.01423974]\n"
]
],
[
[
"The \"make()\" function creates an environment (here, the CartPole environment). After creation, the environment must be reset via the \"reset()\" method. This returns the first observation, in the form of four floats in a $1\\times4$ NumPy array: (i) the horizontal position (0=center), (ii) the velocity, (iii) the angle of the pole (0=vertical), and (iv) its angular velocity.\n<br>\nSo far so good. Some environments including the CartPole environment demand access to the screen to visualize the environment. As explained in the Github link (linked above), this may be problematic when Jupyter is used. However, one can work around this issue by using the following function (taken from the Github link) that will render the CartPole environment within a Jupyter notebook.",
"_____no_output_____"
]
],
[
[
"# further imports and plot specifications (taken from Github link above)\nimport numpy as np\n%matplotlib nbagg\nimport matplotlib\nimport matplotlib.animation as animation\nimport matplotlib.pyplot as plt\nplt.rcParams[\"axes.labelsize\"] = 14\nplt.rcParams[\"xtick.labelsize\"] = 12\nplt.rcParams[\"ytick.labelsize\"] = 12\n# functions for rendering of the CartPole environment within the Jupyter notebook (taken from Github-link)\nfrom PIL import Image, ImageDraw\ntry:\n from pyglet.gl import gl_info\n openai_cart_pole_rendering = True\nexcept Exception:\n openai_cart_pole_rendering = False\ndef render_cart_pole(env, obs):\n if openai_cart_pole_rendering:\n return env.render(mode=\"rgb_array\")\n else:\n img_w = 600\n img_h = 400\n cart_w = img_w // 12\n cart_h = img_h // 15\n pole_len = img_h // 3.5\n pole_w = img_w // 80 + 1\n x_width = 2\n max_ang = 0.2\n bg_col = (255, 255, 255)\n cart_col = 0x000000\n pole_col = 0x669acc\n pos, vel, ang, ang_vel = obs\n img = Image.new(\"RGB\", (img_w, img_h), bg_col)\n draw = ImageDraw.Draw(img)\n cart_x = pos * img_w // x_width + img_w // x_width\n cart_y = img_h * 95 // 100\n top_pole_x = cart_x + pole_len * np.sin(ang)\n top_pole_y = cart_y - cart_h // 2 - pole_len * np.cos(ang)\n draw.line((0, cart_y, img_w, cart_y), fill=0)\n draw.rectangle((cart_x - cart_w // 2, cart_y - cart_h // 2,\n cart_x + cart_w // 2, cart_y + cart_h // 2), fill=cart_col)\n draw.line((cart_x, cart_y - cart_h // 2, top_pole_x, top_pole_y), fill=pole_col, width=pole_w)\n return np.array(img)\ndef plot_cart_pole(env, obs):\n plt.close()\n img = render_cart_pole(env, obs)\n plt.imshow(img)\n plt.axis(\"off\")\n plt.show()\n# now, employ the above code\nopenai_cart_pole_rendering = False # here, we do not even try to display the CartPole environment outside Jupyter\nplot_cart_pole(env, obs)",
"_____no_output_____"
]
],
[
[
"Nice. So this is a visualization OpenAI's CartPole environment. After the visualization is finished (it may be a movie), it is best to \"close\" the visualization by tapping the off-button at its top-right corner.<br><br>\n**Suggestion or Tip**<br>\nUnfortunately, the CartPole (and a few other environments) renders the image to the screen even if you set the mode to \"rgb_array\". The only way to avoid this is to use a fake X server such as Xvfb or Xdummy. For example, you can install Xvfb and start Python using the following [shell] command: 'xcfb-run -s \"-screen 0 1400x900x24\" python'. Or use the xvfb wrapper package (https://goo.gl/wR1oJl). [With the code from the Github link, we do not have to bother about all this.]<br><br>\nTo continue, we ought to know what actions are possible. This can be checked via the environment's \"action_space\" method.",
"_____no_output_____"
]
],
[
[
"env.action_space # check what actions are possible",
"_____no_output_____"
]
],
[
[
"There are two possible actions: accelerating left (represented by the integer 0) or right (integer 1). Other environments are going to have other action spaces. We accelerate the cart in the direction that it is leaning.",
"_____no_output_____"
]
],
[
[
"obs = env.reset() # reset again to get different first observations when rerunning this cell\nprint(obs)\nangle = obs[2] # get the angle\nprint(angle) \naction = int((1 + np.sign(angle)) / 2) # compute the action\nprint(action)\nobs, reward, done, info = env.step(action) # submit the action to the environment so the environment can evlolve\nprint(obs, reward, done, info)",
"[-0.01435942 0.0420023 0.00417418 0.0271142 ]\n0.004174176652464501\n1\n[-0.01351937 0.23706414 0.00471646 -0.26424881] 1.0 False {}\n"
]
],
[
[
"The \"step()\" method executes the chosen action and returns 4 values:\n1. \"obs\"<br>\nThis is the next observation.\n2. \"reward\"<br>\nThis environment gives a reward for every time step (not more and not less). The only goal is to keep balancing the cart as long as possble.\n3. \"done\"<br>\nThe value of this dictionary is \"False\" while the episode is running and \"True\" when the episode is finished. It will be finished when the cart moves out of its boundaries or the pole tilts too much.\n4. info<br>\nThis dictionary may provide extra useful information (but not in the CartPole environment). This information can be useful for understanding the problem but shall not be used for adapting the policy. That would be cheating.\n\nWe now hardcode a simple policy that always accelerates the cart in the direction it is leaning.",
"_____no_output_____"
]
],
[
[
"# define a basic policy\ndef basic_policy(obs):\n angle = obs[2] # angle\n return 0 if angle < 0 else 1 # return action\ntotals = [] # container for total rewards of different episodes\n# run 500 episodes of up to 1000 steps each\nfor episode in range(500): # run 500 episodes\n episode_rewards = 0 # for each episode, start with 0 rewards ...\n obs = env.reset() # ... and initialize the environment\n for step in range(1000): # attempt 1000 steps for each episode (the policy might fail way before)\n action = basic_policy(obs) # use our basic policy to infer the action from the observation\n obs, reward, done, info = env.step(action) # apply the action and make a step in the environment\n episode_rewards += reward # add the earned reward\n if done: # if the environment says that it has finished ...\n break # ... this shall be the final step within the current episode\n totals.append(episode_rewards) # put the final episode rewards into the container for all the total rewards\n# print statistics on the earned rewards\nprint(\"mean:\", np.mean(totals),\", std:\", np.std(totals),\", min:\", np.min(totals),\", max:\", np.max(totals))",
"mean: 41.938 , std: 8.855854334845397 , min: 24.0 , max: 68.0\n"
]
],
[
[
"Even for 500 episodes, this policy did not manage to reach only 100 steps, not to speak of 1000. As can be seen from the additional code shown under the above Github link, the reason for this is that the pole quickly begins strongly oscillating back and forth. Certainly, the policy is just too simple: also taking the angular velocity of the pole into account should help avoiding wild angular oscillations. A more sophisticated policy might employ a neural network.\n## Neural Network Policies\npage 448<br>\nLet's not just talk the talk but also walk the walk by actually implementing a neural network policy for the CartPole environment. This environment accepts only 2 possible actions: 0 (left) and 1 (right). So one output layer will be sufficient: it returns a value $p$ between 0 and 1. We will accelerate right with probability $p$ and – obviously – left with probability $1-p$.<br>\nWhy do we not go right with probability ${\\rm heaviside}(p-0.5)$? This would hamper the algorithm from exploring further, i.e., it would slow down (or even stop) the learning progress, as can be illustrated with the following analogue. Consider going to a foreign restaurant for the first time. All items on the menu seem equally OK. You choose absolutely randomly one dish. If you happen to like it, you might from now on order this dish every time you visit the restaurant. Fair enough. But there might be even better dishes that you will never find out about if you stick to that policy. By just choosing this menu with a probability in the interval $(0.5,1)$, you will continue to explore the menu, and develop a better understanding of what menu is the best.<br><br>\nNote that the CartPole environment returns on every instance the complete necessary information: (i) it contains the velocities (position and angle), so previous observations are not necessary to deduce them, and (ii) it is noise free, so previous observations are not necessary to deduce the actual values from an average. In that sense, the CartPole environment is really as simple as can be.<br><br>\nThe environment returns 4 observations so we will employ 4 input neurons. We have already mentioned that we only need one single output neuron. To keep things simple, this shall be it: 4 input neurons in the input and in the only hidden layer and 1 output neuron in the output layer. Now, let's cast this in code!",
"_____no_output_____"
]
],
[
[
"# code partially taken from Github link above\nimport tensorflow as tf\ndef reset_graph(seed=42):\n tf.reset_default_graph()\n tf.set_random_seed(seed)\n np.random.seed(seed)\nreset_graph()\n# 1. specify the neural network architecture\nn_inputs = 4\nn_hidden = 4\nn_outputs = 1\ninitializer = tf.variance_scaling_initializer()\n# 2. build the neural network\nX = tf.placeholder(tf.float32, shape=[None, n_inputs])\nhidden = tf.layers.dense(X, n_hidden, activation=tf.nn.elu, kernel_initializer=initializer)\nlogits = tf.layers.dense(hidden, n_outputs, kernel_initializer=initializer)\noutputs = tf.nn.sigmoid(logits)\n# 3. select a random action based on the estimated probabilities\np_left_and_right = tf.concat(axis=1, values=[outputs, 1 - outputs])\naction = tf.multinomial(tf.log(p_left_and_right), num_samples=1)\ninit = tf.global_variables_initializer()\n# maximal number of steps and container for video frames\nn_max_steps = 1000\nframes = []\n# start a session, run the graph, and close the environment (see Github link)\nwith tf.Session() as sess:\n init.run()\n obs = env.reset()\n for step in range(n_max_steps):\n img = render_cart_pole(env, obs)\n frames.append(img)\n action_val = action.eval(feed_dict={X: obs.reshape(1, n_inputs)})\n obs, reward, done, info = env.step(action_val[0][0])\n if done:\n break\nenv.close()\n# use the below functions and commands to show an animation of the frames\ndef update_scene(num, frames, patch):\n patch.set_data(frames[num])\n return patch,\ndef plot_animation(frames, repeat=False, interval=40):\n plt.close()\n fig = plt.figure()\n patch = plt.imshow(frames[0])\n plt.axis(\"off\")\n return animation.FuncAnimation(fig, update_scene, fargs=(frames, patch),\n frames=len(frames), repeat=repeat, interval=interval)\nvideo = plot_animation(frames)\nplt.show()",
"/anaconda3/lib/python3.6/site-packages/tensorflow/python/framework/dtypes.py:493: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint8 = np.dtype([(\"qint8\", np.int8, 1)])\n/anaconda3/lib/python3.6/site-packages/tensorflow/python/framework/dtypes.py:494: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint8 = np.dtype([(\"quint8\", np.uint8, 1)])\n/anaconda3/lib/python3.6/site-packages/tensorflow/python/framework/dtypes.py:495: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint16 = np.dtype([(\"qint16\", np.int16, 1)])\n/anaconda3/lib/python3.6/site-packages/tensorflow/python/framework/dtypes.py:496: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint16 = np.dtype([(\"quint16\", np.uint16, 1)])\n/anaconda3/lib/python3.6/site-packages/tensorflow/python/framework/dtypes.py:497: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint32 = np.dtype([(\"qint32\", np.int32, 1)])\n/anaconda3/lib/python3.6/site-packages/tensorflow/python/framework/dtypes.py:502: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n np_resource = np.dtype([(\"resource\", np.ubyte, 1)])\n"
]
],
[
[
"Above, the numbered comments indicate what is happening in the subsequent code. Here are some details.\n1. Definition of the Neural Network architecture (c.f. Figure 16-5 on page 449 of the book).\n2. Building the Neural Network. Obviously, it is a vanilla (most basic) version. Via the sigmoid activation function, the output lies in the interval [0,1]. If more than two actions were possible, this scheme should use one output neuron per action and a softmax activation function instead of a sigmoid (refer to Chapter 4).\n3. Use a multinomial probability distribution to map the probability to one single action. See https://en.wikipedia.org/wiki/Multinomial_distribution and the book for details.\n\n## Evaluating Actions: The Credit Assignment Problem\npage 451<br>\nIf we knew the optimal (target) probability for taking a certain action, we could just do regular machine learning by trying to minimize the cross entropy between that target probability and the estimated probability that we obtain from our policy (see above and below \"Neural Network Policies\"). However, we do not know the optimal probability for taking a certain action in a certain situation. We have to guess it based on subsequent rewards and penalties (negative rewards). However, rewards are often rare and usually delayed. So it is not clear which previous actions have contributed to earning the subsequent reward. This is the *Credit Assignment Problem*.<br>\nOne way to tackle the Credit Assignment Problem is to introduce a *discount rate* $r\\in[0,1]$. A reward $x$ that occurs $n$ steps after an action - with $n=0$ if the reward is earned directly after the action - adds a score of $r^nx$ to the rewards assigned to that action. For exmaple, if $r=0.8$ and an action is immediately followed by a reward of $+10$, then $0$ after the next action, and $-50$ after the second next action, then that initial action is assigned a score of\n$$+10+0-50\\times 0.8^2=10-50\\times0.64=10-32=-22\\,.$$\nTypical discount rates are 0.95 and 0.99. When actions have rather immediate consequences, $r=0.95$ is expected to be more suitable than $r=0.99$.<br>\nOf course it can happen that in one specific run of the program, a good action happens to be followed by an observation with negative reward. However, on average, a good action will be followed by positive rewards (otherwise it is not a good action). So we need to make many runs to smoothen out the effect of untypical evolutions. Then the scores need to be normalized. In the CartPole environment for example, one could discretize the continuous observations into a finite number of intervals, such that different observations may lie within the same (or different) margins. Then, the scores within one interval should be added up and divided by the number of times the according action was taken, thus giving a representative score. Such scores shall be calculated for all possible actions given that observation. The mean score over all actions needs to be subtracted from each action's score. The resulting number should probably be normalized by the standard deviation and then mapped to probabilities assigned to taking these actions.\n## Policy Gradients\npage 452<br>\nThe training procedure that has been outlined above shall now be implemented using TensorFlow.\n- First, let the neural network policy play the game several times and **at each step compute the gradients** that would make the chosen action even more likely, but don't apply thes gradients yet. Note that **the gradients involve the observation / state** of the system!\n- Once you have **run several episodes (to reduce noise from odd evolutions)**, compute each action's score (using the method described in the previous paragraph).\n- If an action's score is positive, it means that the action was good and you want to apply the grdients computed earler to make the action even more likel to be chosen in the future. However, if the score is negative, it means the action was bad and you want to apply the opposite gradients to make this action slightly *less* likely in the future. The solution is simply to **multiply each gradient vector by the corresponding action's score**.\n- Finally, compute the mean of all the resulting gradient vectors, and us it to perform a **Gradient Descent** step.\n\nWe start with two functions. The first function calculated receives a list of rewards corresponding that occur one step after another and returns a list of accumulated and discounted rewards. The second function receives a list of such lists as well as a discount rate and returns the centered (subtraction of the total mean) and normalized (division by the total standard deviation) lists of rewards. Both functions are tested with simple inputs.",
"_____no_output_____"
]
],
[
[
"# build a function that calculates the accumulated and discounted rewards for all steps\ndef discount_rewards(rewards, discount_rate):\n discounted_rewards = np.empty(len(rewards))\n cumulative_rewards = 0\n for step in reversed(range(len(rewards))): # go from end to start\n cumulative_rewards = rewards[step] + cumulative_rewards * discount_rate # the current reward has no discount\n discounted_rewards[step] = cumulative_rewards\n return discounted_rewards\n# after rewards from 10 episodes, center and normalize the rewards in order to calculate the gradient (below)\ndef discount_and_normalize_rewards(all_rewards, discount_rate):\n all_discounted_rewards = [discount_rewards(rewards, discount_rate) # make a list of the lists with ...\n for rewards in all_rewards] # ... the rewards (i.e., a matrix)\n flat_rewards = np.concatenate(all_discounted_rewards) # flatten this list to calculate ...\n reward_mean = flat_rewards.mean() # ... the mean and ...\n reward_std = flat_rewards.std() # ... the standard deviation\n return [(discounted_rewards - reward_mean)/reward_std # return a list of centered and normalized ...\n for discounted_rewards in all_discounted_rewards] # ... lists of rewards\n# check whether these functions work\ndiscount_rewards([10,0,-50], 0.8)\ndiscount_and_normalize_rewards([[10, 0, -50], [10, 20]], 0.8)",
"_____no_output_____"
]
],
[
[
"The following cell is a combination of code from the book and from the github link above. It implements the entire algorithm so it is a bit complicated. Comments shall assist in understanding the code. The default value of \"n_iterations\" (\"n_max_steps\") [\"discount_rate\"] is 250 (1000) [0.95] but can be increased for better performance.",
"_____no_output_____"
]
],
[
[
"# see also \"Visualizing the Graph and Training Curves Using TensorBoard\" in chapter / notebook 9 to review ...\n# ... how to save a graph and display it in tensorboard; important ingredients: ...\n# ... (i) reset_graph(), (ii) file_writer, and (iii) file_writer.close(); possible the \"saver\" is also important\n# create a directory with a time stamp\nfrom datetime import datetime # import datetime\nnow = datetime.utcnow().strftime(\"%Y%m%d%H%M%S\") # get current UTC time as a string with specified format\nroot_logdir = \"./tf_logs/16_RL/Cartpole\" # directory in the same folder as this .ipynb notebook\nlogdir = \"{}/policy_gradients-{}/\".format(root_logdir, now) # folder with timestamp (inside the folder \"tf_logs\")\nprint(logdir) # print the total path relative to this notebook\n# reset the graph and setup the environment (cartpole)\nreset_graph()\nenv = gym.make(\"CartPole-v0\")\n# 1. specify the neural network architecture\nn_inputs = 4\nn_hidden = 4\nn_outputs = 1\ninitializer = tf.contrib.layers.variance_scaling_initializer()\nlearning_rate = 0.01\n# 2. build the neural network\nX = tf.placeholder(tf.float32, shape=[None, n_inputs])\nhidden = tf.layers.dense(X, n_hidden, activation=tf.nn.elu, kernel_initializer=initializer)\nlogits = tf.layers.dense(hidden, n_outputs, kernel_initializer=initializer)\noutputs = tf.nn.sigmoid(logits)\np_left_and_right = tf.concat(axis=1, values=[outputs, 1 - outputs])\naction = tf.multinomial(tf.log(p_left_and_right), num_samples=1)\ny = 1. - tf.to_float(action)\n# 3. cost function (cross entropy), optimizer, and calculation of gradients\ncross_entropy = tf.nn.sigmoid_cross_entropy_with_logits(labels=y, logits=logits)\noptimizer = tf.train.AdamOptimizer(learning_rate)\ngrads_and_vars = optimizer.compute_gradients(cross_entropy)\n# 4. rearrange gradients and variables\ngradients = [grad for grad, variable in grads_and_vars]\ngradient_placeholders = []\ngrads_and_vars_feed = []\nfor grad, variable in grads_and_vars:\n gradient_placeholder = tf.placeholder(tf.float32, shape=grad.get_shape())\n gradient_placeholders.append(gradient_placeholder)\n grads_and_vars_feed.append((gradient_placeholder, variable))\ntraining_op = optimizer.apply_gradients(grads_and_vars_feed)\n# 5. file writer, global initializer, and saver\nfile_writer = tf.summary.FileWriter(logdir, tf.get_default_graph()) # save the graph graph under \"logdir\"\ninit = tf.global_variables_initializer()\nsaver = tf.train.Saver()\n# 6. details for the iterations that will be run below\nn_iterations = 250 # default is 250\nn_max_steps = 1000 # default is 1000\nn_games_per_update = 10\nsave_iterations = 10\ndiscount_rate = 0.95 # default is 0.95\n# 7. start a session and run it\nwith tf.Session() as sess:\n init.run()\n # 7.1. loop through iterations\n for iteration in range(n_iterations):\n all_rewards = [] # container for rewards lists\n all_gradients = [] # container for gradients lists\n # 7.2. loop through games\n for game in range(n_games_per_update):\n current_rewards = [] # container for unnormalized rewards from current episode\n current_gradients = [] # container for gradients from current episode\n obs = env.reset() # get first observation\n # 7.3 make steps in the current game\n for step in range(n_max_steps):\n action_val, gradients_val = sess.run([action, gradients], # feed the observation to the model and ...\n feed_dict={X: obs.reshape(1, n_inputs)}) # ... receive the ...\n # ... action and the gradient (see 4. above)\n obs, reward, done, info = env.step(action_val[0][0]) # feed action and receive next observation\n current_rewards.append(reward) # store reward in container\n current_gradients.append(gradients_val) # store gradients in container\n if done: # stop when done\n break\n all_rewards.append(current_rewards) # update list of rewards\n all_gradients.append(current_gradients) # update list of gradients\n # 7.4. we have now run the current policy for \"n_games_per_update\" times and it shall be updated\n all_rewards = discount_and_normalize_rewards(all_rewards, discount_rate) # use the function(s) defined above\n # 7.5. fill place holders with actual values\n feed_dict = {}\n for var_index, grad_placeholder in enumerate(gradient_placeholders): # loop through placeholders\n mean_gradients = np.mean([reward * all_gradients[game_index][step][var_index] # calculate ...\n for game_index, rewards in enumerate(all_rewards) # ... the ...\n for step, reward in enumerate(rewards)], # ... mean ...\n axis=0) # ... gradients and ...\n feed_dict[grad_placeholder] = mean_gradients # ... feed them\n # 7.6. run the training operation\n sess.run(training_op, feed_dict=feed_dict)\n # 7.7. save every now and then\n if iteration % save_iterations == 0:\n saver.save(sess, \"./tf_logs/16_RL/Cartpole/my_policy_net_pg.ckpt\")\n# 8. close the file writer and the environment (cartpole)\nfile_writer.close()\nenv.close()",
"./tf_logs/16_RL/Cartpole/policy_gradients-20190919190805/\n\u001b[33mWARN: gym.spaces.Box autodetected dtype as <class 'numpy.float32'>. Please provide explicit dtype.\u001b[0m\n"
]
],
[
[
"Using another function from the github link, we apply the trained model and visualize the performance of the agent in its environment. The performance differs from run to run, presumably due to random initialization of the environment.",
"_____no_output_____"
]
],
[
[
"# use code from the github link above to apply the trained model to the cartpole environment and show the results\nmodel = 1\ndef render_policy_net(model_path, action, X, n_max_steps = 1000):\n frames = []\n env = gym.make(\"CartPole-v0\")\n obs = env.reset()\n with tf.Session() as sess:\n saver.restore(sess, model_path)\n for step in range(n_max_steps):\n img = render_cart_pole(env, obs)\n frames.append(img)\n action_val = action.eval(feed_dict={X: obs.reshape(1, n_inputs)})\n obs, reward, done, info = env.step(action_val[0][0])\n if done:\n break\n env.close()\n return frames\nif model == 1:\n frames = render_policy_net(\"./tf_logs/16_RL/Cartpole/my_policy_net_pg.ckpt\", action, X, n_max_steps=1000)\nelse:\n frames = render_policy_net(\"./tf_logs/16_RL/Cartpole/my_good_policy_net_pg.ckpt\",\n action, X, n_max_steps=1000)\nvideo = plot_animation(frames)\nplt.show()",
"\u001b[33mWARN: gym.spaces.Box autodetected dtype as <class 'numpy.float32'>. Please provide explicit dtype.\u001b[0m\nINFO:tensorflow:Restoring parameters from ./tf_logs/16_RL/Cartpole/my_policy_net_pg.ckpt\n"
]
],
[
[
"This looks much better than without updating the gradients. Almost skillful, at least witn \"n_iterations = 1000\", \"n_max_steps = 1000\", and \"discount_rate = 0.999\". The according model is saved under \"./tf_logs/Cartpole/my_1000_1000_0999_policy_net_pg.ckpt\". In fact, **this algorithm is really powerful**. It can be used for much harder problems. **AlphaGo** was also based on it (and on *Monte Carlo Tree Search*, which is beyond the scope of this book).<br><br>\n**Suggestion or Tip**<br>\nResearchers try to find algorithms that work well even when the agent initially knows nothing about the environment. However, unless you are writing a paper, you should inject as much prior knowledge as possible into the agent, as it will speed up training dramatically. For example, you could add negative rewards proportional to the distance from the center of the screen, and to the pole's angle. Also, if you already have a reasonably good policy (e.g., hardcoded), you may want to train the nerual network to imitate it before using policy gradients to imporve it.<br><br>\nBut next, we go a different path: the agent shall calculate what future rewards (possibly discounted) it expects after an action in a certain situation. The action shall be chosen based on that expectation. To this end, some more theory is necessary.\n## Markov Decision Processes\npage 457<br>\nA *Markov chain* is a stochastic process where a system changes between a finite number of states and the probability to transition from state $s$ to state $s'$ is only determined by the pair $(s,s')$, not on previous states (the system has no memory). Andrey Markov studied these processes in the early 20th century. If the transition $(s,s)$ has probability $1$, then the state $s$ is a *terminal state*: the system cannot leave it (see also Figure 16-7 on page 458 of the book). There can be any integer $n\\geq0$ number of terminal states. Markov chains are heavily used in thermodynamics, chemistry, statistics, and much more.<br>\nMarkov decision processes were first decribed by Richard Bellmann in the 1950s (see the jstor-link above). They are Markov chains where at each state $s$, there is one or more action $a$ that an *agent* can choose from and the transition probilitiy between state $s$ and $s'$ depends on action $a$. Moreover, some transitions $(s,a,s')$ return a reward (positive or negative). The agent's goal is to find a policy that maximizes rewards over time (see also Figure 16-8 on page 459).<br><br>\nIf an agent acts optimally, then the *Bellman Optimality Equation* applies. This recursive equation states that the optimal value of the current state is equal to the reward it will get on average after taking one optimal action, plus the expected optimal vlaue of all possible next states that this action can lead to:<br><br>\n$$V^*(s)={\\rm max}_a\\sum_{s'}T(s,a,s')[R(s,a,s')+\\gamma*V^*(s')]\\quad\\text{for all $s$.}$$\nHere,\n- $V^*(s)$ is the *optimal state value* of state $s$,\n- $T(s,a,s')$ is the transition probability from state $s$ to state $s'$, given that the agent chose action $a$,\n- $R(s,a,s')$ is the reward that the agent gets when it goes from state $s$ to state$s'$, given that the agent chose action $a$, and\n- $\\gamma$ is the discount rate.\n\nThe **Value Iteration** algorithm will have the values $V(s)$ - initialized to 0 - converge towards their optima $V^*(s)$ by iteratively updating them:<br><br>\n$$V_{k+1}(s)\\leftarrow{\\rm max}_a\\sum_{s'}T(s,a,s')[R(s,a,s')+\\gamma V_k(s')]\\quad\\text{for all $s$.}$$\nHere, $V_k(s)$ is the estimated value of state $s$ in the $k$-th iteration and $\\lim_{k\\to\\infty}V_k(s)=V^*(s)$.\n<br><br>\n**General note**<br>\nThis algorithm is an example of *Dynamic Programming*, which breaks down a complex problem (in this case estimating a potentially infinite sum of discounted future rewards) into tractable sub-problems that can be tackled iteratively (in this case finding the action that maximizes the average reward plus the discounted next state value).<br><br>\nYet even when $V^*(s)$ is known, it is still unclear what the agent shall do. Bellman thus extended the concept of state values $V(s)$ to *state-action values*, called *Q-values*. The optimal Q-value of the state-action pair $(s,a)$, noted $Q^*(s,a)$, is the sum of discounted future rewards the agent will earn on average when it applies to correct policy, i.e., choosing the right action $a$ when in state $s$. The according **Q-value iteration** algorithm is<br><br>\n$$Q_{k+1}(s,a)\\leftarrow\\sum_{s'}T(s,a,s')[R(s,a,s')+\\gamma\\,{\\rm max}_{a'}Q_k(s',a')],\\quad\\text{for all $(s,a)$.}$$<br>\nThis is very similar to the value iteration algorithm. Now, the optimal policy $\\pi^*(s)$ is clear: when in state $s$, apply that action $a$ for which the Q-value is highest: $\\pi^*(s)={\\rm argmax}_aQ^*(s,a)$. We will practice this on the Markov decision process shown in Figure 16-8 on page 459 of the book.",
"_____no_output_____"
]
],
[
[
"nan = np.nan # for transitions that do not exist (and thus have probability 0 but shall not be updated)\n# transition probabilities\nT = np.array([\n [[0.7, 0.3, 0.0], [1.0, 0.0, 0.0], [0.8, 0.2, 0.0]], # at state s_0: actions a_0, a_1, a_2 to states s_0, s_1, s_2\n [[0.0, 1.0, 0.0], [nan, nan, nan], [0.0, 0.0, 1.0]], # at state s_1\n [[nan, nan, nan], [0.8, 0.1, 0.1], [nan, nan, nan]] # at state s_2\n])\n# rewards associated with the transitions above\nR = np.array([\n [[10., 0.0, 0.0], [0.0, 0.0, 0.0], [0.0, 0.0, 0.0]],\n [[10., 0.0, 0.0], [nan, nan, nan], [0.0, 0.0, -50]],\n [[nan, nan, nan], [40., 0.0, 0.0], [nan, nan, nan]]\n])\n# possible actions\npossible_actions = [\n [0, 1, 2], # in state s0\n [0, 2], # in state s1\n [1]] # in state s2\n# Q value initialization\nQ = np.full((3, 3), -np.inf) # -infinity for impossible actions\nfor state, actions in enumerate(possible_actions):\n print(state, actions)\n Q[state, actions] = 0.0 # 0.0 for possible actions\nprint(Q)",
"0 [0, 1, 2]\n1 [0, 2]\n2 [1]\n[[ 0. 0. 0.]\n [ 0. -inf 0.]\n [-inf 0. -inf]]\n"
]
],
[
[
"Above, we have specified the model (lists of transition probabilities T, rewards R, and possible actions) and initialized the Q-values. Using $-\\infty$ for impossible actions ensures that those actions will not be chosen by the policy. The enumerate command can be quite helpful and is therefore shortly introduced below.",
"_____no_output_____"
]
],
[
[
"# see the python tips link above\nfor counter, value in enumerate([\"apple\", \"banana\", \"grapes\", \"pear\"]):\n print(counter, value)\nprint()\nfor counter, value in enumerate([\"apple\", \"banana\", \"grapes\", \"pear\"], 1):\n print(counter, value)\nprint()\ncounter_list = list(enumerate([\"apple\", \"banana\", \"grapes\", \"pear\"], 1))\nprint(counter_list)",
"0 apple\n1 banana\n2 grapes\n3 pear\n\n1 apple\n2 banana\n3 grapes\n4 pear\n\n[(1, 'apple'), (2, 'banana'), (3, 'grapes'), (4, 'pear')]\n"
]
],
[
[
"Now, define residual parameters and run the Q-value iteration algorithm. The results are very similar to the ones in the book.",
"_____no_output_____"
]
],
[
[
"# learning parameters\nlearning_rate = 0.01\ndiscount_rate = 0.95 # try 0.95 and 0.9 (they give different results)\nn_iterations = 100\n# Q-value iteration\nfor iteration in range(n_iterations): # loop over iterations\n Q_prev = Q.copy() # previous Q\n for s in range(3): # loop over states s\n for a in possible_actions[s]: # loop over available actions a\n # update Q[s, a]\n Q[s, a] = np.sum([T[s, a, sp] * (R[s, a, sp] + # transition probability to sp times ...\n discount_rate*np.max(Q_prev[sp])) # ... ( reward + max Q(sp) )\n for sp in range(3)]) # sum over sp (sp = s prime)\nprint(Q) # print final Q\nnp.argmax(Q, axis=1) # best action for a given state (i.e., in a given row)",
"[[21.88646117 20.79149867 16.854807 ]\n [ 1.10804034 -inf 1.16703135]\n [ -inf 53.8607061 -inf]]\n"
]
],
[
[
"The Q-value iteration algorithm gives different results for different discount rates $\\gamma$. For $\\gamma=0.95$, the optimal policy is to choose action 0 (2) [1] in state 0 (1) [2]. But this changes to actions 0 (0) [1] if $\\gamma=0.9$. And it makes sense: when someone appreciates the present more than possible future rewards, there is less motivation to go through the fire.\n## Temporal Difference Learning and Q-Learning\npage 461<br>\nReinforcement Learning problems can often be modelled with Markov Decision Processes, yet initially, there is no knowledge on transition probabilities $T(s,a,s')$ and rewards $R(s,a,s')$. The algorithm must experience every connection at least once to obtain the rewards and multiple times to get a glimplse of the probabilities.<br>\n**Temporal Difference Learning** (TD Learning) is similar to *Value Iteration* (see above) but adapted to the fact that the agent has only partial knowledge of the MDP. In fact, it often only knows the possible states and actions and thus must *explore* the MDP via an *exploration policy* in order to recursively update the estimates for the state values based on observed transitions and rewards. This is achieved via the **Temporal Difference Learning algorithm** (Equation 16-4 in the book),\n$$V_{k+1}\\leftarrow(1-\\alpha)V_k(s)+\\alpha(r+\\gamma V_k(s'))\\,,$$\nwhere $\\alpha$ is the learning rate (e.g., 0.01).<br>\nFor each state $s$, this algorithm simply keeps track of a running average of the next reward (for the transition to the state it will end up at next) and future rewards (via the state value of the next state, since the agent is assumed to act optimally). This algorithm is stochastic: the next state $s'$ and the reward gained by going to it are not known at state $s$: this transition only happens with a certain probability and thus will – in general – be different from time to time.<br><br>\n**Suggestion or Tip**<br>\nTD Learning has many similarities with Stochasitc Gradient Descent, in particular the fact that it handles one sample at a time. Just like SGD, it can only truly converge if you gradually reduce the learning rate (otherwise it will keep bouncing around the optimum).<br><br>\nThe **Q-Learning algorithm** resembles the *Temporal Difference Learning algorithm* just like the *Q-Value Iteration algorithm* resembles the *Value Iteration algorithm* (see Equation 16-5 in the book):\n$$Q_{k+1}(s,a)\\leftarrow(1-\\alpha)Q_k(s,a)+\\alpha(r+\\gamma\\,{\\rm max}_{a'}Q_k(s',a'))\\,.$$\n<br>\nIn comparison with the *TD Learning algorithm* above, there is another degree of freedom: the action $a$. It is assumed that the agent will act optimally. As a consequence, the maximum (w.r.t. actions a') Q-Value is chosen for the next state-action pair. Now to the implementation!",
"_____no_output_____"
]
],
[
[
"n_states = 3 # number of MDP states\nn_actions = 3 # number of actions\nn_steps = 20000 # number of steps (=iterations)\nalpha = 0.01 # learning rate\ngamma = 0.99 # discount rate\n# class for a Markov Decision Process (taken from the github link above)\nclass MDPEnvironment(object):\n def __init__(self, start_state=0): # necessary __init__\n self.start_state=start_state\n self.reset()\n def reset(self): # reset the environment\n self.total_rewards = 0\n self.state = self.start_state\n def step(self, action): # make a step\n next_state =np.random.choice(range(3), # use the transition probabilities T above and the ...\n p=T[self.state][action]) # ... state and action to infer the next state\n reward = R[self.state][action][next_state] # in this fashion, also calculate the obtained reward\n self.state = next_state # update the state ...\n self.total_rewards += reward # ... and the total rewards\n return self.state, reward\n# function that returns a random action that is in line the given state (taken from github link above)\ndef policy_random(state):\n return np.random.choice(possible_actions[state]) # see definition of \"possible_actions\" above\nexploration_policy = policy_random # use that function as an exploration policy\n# reinitialize the Q values (they had been updated above)\nQ = np.full((3, 3), -np.inf) # -infinity for impossible actions\nfor state, actions in enumerate(possible_actions): # see (unchanged) defintion of possible_actions above\n Q[state, actions] = 0.0 # 0.0 for possible actions\nprint(Q)\n# use the class defined above\nenv = MDPEnvironment()\n# loop through all steps (=iterations)\nfor step in range(n_steps):\n action = exploration_policy(env.state) # give our \"exploration_policy = policy_random\" the ...\n # ... current state and obtain an action in return\n state = env.state # obtain the state from the environment\n next_state, reward = env.step(action) # apply the action to obtain the next state and reward\n next_value = np.max(Q[next_state]) # assuming optimal behavior, use the highest Q-value ...\n # ... that is in line with the next state (maximise ...\n # ... w.r.t. the actions)\n Q[state, action] = (1-alpha)*Q[state, action] + alpha*(reward + gamma * next_value) # update Q value\nQ # return the final Q values",
"[[ 0. 0. 0.]\n [ 0. -inf 0.]\n [-inf 0. -inf]]\n"
]
],
[
[
"The above implementation is different from the book (where prior knowledge on the transition probabilities is used). Accordingly, the resulting Q-values are different from those in the book and closer to the ones listed in the github link above.<br>\nWith enough iterations, this algorithm will converge towards theo optimal Q-values. It is called an **off-policy** algorithm since the policy that is used for training is different from the one that will be applied. Surprisingly, this algorithm converges towards the optimal policy by stumbling around: it's a stochastic process. So although this works, there should be a more promising approach. This shall be studied next.\n### Exploration Policies\npage 463<br>\nAbove, the transitions probabilites $T(s,a,s')$ are used to explore the MDP and obtain a policy. However, this might take a long time. An alternative approach is to use an **$\\epsilon$-greedy policy** (greedy = gierig): in each state, it chooses the state-action value Q(s,a) with the highest score with probability $1-\\epsilon$ or a random action (i.e., Q(s,...)) with probability $\\epsilon$. This way, the policy will explorte the interesting parts of the network more intensely while still getting in touch (eventually) with all other parts. It is common to start with $\\epsilon=1$ (totally random) and transition to $\\epsilon=0.05$ (choose highest Q-value most of the time).<br>\nAnother way to incite the agent to explore the entire network is to artificially increase the value of state action pairs that have not been visited frequently. This can be done like so (Equation 16-6 in the book):<br><br>\n$$Q(s,a)\\leftarrow(1-\\alpha)Q(s,a)+\\alpha\\left(r+\\gamma\\,{\\rm max}_{\\alpha'}f(Q(s',a'),N(s',a')\\right)\\,,$$<br>\nwhere $\\alpha$ is the learning rate, $\\gamma$ is the discount rate, and $f(q,n)$ is a function of the Q-value $q$ and the additional reward $n$, e.g. $f(q,n)=q+K/(1+n)$, where $K$ is a **curiosity hyperparameter** and $n$ is the number of times the Q-value $q$ has been visited.\n### Approximate Q-Learning\npage 464<br>\nUnfortunately, Q-Learning as described above dose not scale well. For example, in Ms. Pac-Man there exist more than 250 pellets that can be already eaten or not. This alone gives rise to $2^{250}\\approx10^{75}$ states, i.e., more than the number of atoms in the universe. And this is just the pellets! There is no way, a policy could completely explore the according MDP.<br>\nThe solution is **Approximate Q-Learning** where estimates for the Q-values are inferred from a managable number of parameters. For a long time, the way to go had been to hand-craft features, e.g., the distances between Ms. Pac-Man and the ghosts. But DeepMind showed that deep neural networks can perform much better. On top, they do not require feature engineering. A DNN that is used to estimate Q-Values is called a **Deep Q-Network (DQN)**, and using a DQN for Approximate Q-Learning (and subsequently inferring a policy from the Q-Values) is called **Deep Q-Learning**.\n\nThe rest of this chapter is about using Deep Q-Learning to train a computer on Ms. Pac-Man, much like DeepMind did in 2013. This code is quite versatile. It works for many Atari games but tends to have problems with games with long-running storylines.\n## Learning to Play Ms. Pac-Man Using Deep Q-Learning\npage 464<br>\nSome installations shall be made:\n- Homebrew (see http://brew.sh/). For macOS, run '/usr/bin/ruby -e \"$(curl -fsSL https://raw.githubusercontent.com/Homebrew/install/master/install)\"' in the terminal and for Ubuntu, use 'apg-get install -y python3-numpy python3-dev cmake zlib1g-dev libjpeg-dev\\xvfb libav-tools xorg-dev python3-opengl libboost-all-dev libsdl2-dev swig'.\n- Extra python modules for the gym of OpenAI, via \"pip3 install --upgrade 'gym[all]'\".\n\nWith these, we should manage to create a Ms. Pac-Man environment.",
"_____no_output_____"
]
],
[
[
"env = gym.make(\"MsPacman-v0\")\nobs = env.reset()\nprint(obs.shape) # [height, width, channels]\nprint(env.action_space) # possible actions",
"(210, 160, 3)\nDiscrete(9)\n"
]
],
[
[
"So the agent observes a picture (screenshot) of 210 pixels height and 160 pixels width, with each pixel having three color values: the intensity of red, green, and blue. The agent may react by returning an action to the environment. There are nine discrete actions, corresponding to the nine joystick positions ([up, middle, down]x[left, middle, right]). The screenshots are a bit too large and grayscale should suffice. So we preprocess the data by cropping the image to the relevant part and converting the color image to grayscale. This will speed up training.",
"_____no_output_____"
]
],
[
[
"# see https://docs.python.org/2.3/whatsnew/section-slices.html for details on slices\ntrial_list = [[0.0,0.1,0.2,0.3,0.4],\n [0,1,2,3,4],\n [0,10,20,30,40]]\nprint(trial_list[:2]) # print list elements until before element 2, i.e., 0 and 1\nprint(trial_list[::2]) # print all list elements that are reached by step size 2, i.e., 0 and 2, not 1\nprint(trial_list[:3]) # print list elements until before element 3, i.e., 0, 1, and 2 (this means all, here)\nprint(trial_list[::3]) # print all list elements that are reached by step size 3, i.e., only 0, not 1 and 2\n# back to the book\nmspacman_color = np.array([210, 164, 74]).mean()\ndef preprocess_observation(obs):\n img = obs[1:176:2, ::2] # use only every second line from line 1 to line 176 (top towards bottom) and in ...\n # ... each line, use only every second row\n img = img.mean(axis=2) # go to grayscale by replacing the 3 rgb values by their average\n img[img==mspacman_color] = 0 # draw Ms. Pac-Man in black\n img = (img-128)/128 - 1 # normalize grayscale\n return img.reshape(88, 80, 1) # reshape to 88 rows and 80 columns; each scalar entry is the color in grayscale ...\n # ... and thus is a bit different from the book\n#print(preprocess_observation(obs))\nprint(preprocess_observation(obs).shape)\n# slightly modified from the github link above\nplt.figure(figsize=(9, 5))\nplt.subplot(121)\nplt.title(\"Original observation (160×210 RGB)\")\nplt.imshow(obs)\nplt.axis(\"off\")\nplt.subplot(122)\nplt.title(\"Preprocessed observation (88×80 greyscale)\")\n# the output of \"preprocess_observation()\" is reshaped as to be in line with the placeholder \"X_state\", to which ...\n# ... it will be fed; below, another reshape operation is appropriate to conform the (possbily updated w.r.t. the ...\n# ... book) \"plt.imshow()\" command, https://matplotlib.org/api/_as_gen/matplotlib.pyplot.imshow.html\nplt.imshow(preprocess_observation(obs).reshape(88,80), interpolation=\"nearest\", cmap=\"gray\")\nplt.axis(\"off\")\nplt.show()",
"[[0.0, 0.1, 0.2, 0.3, 0.4], [0, 1, 2, 3, 4]]\n[[0.0, 0.1, 0.2, 0.3, 0.4], [0, 10, 20, 30, 40]]\n[[0.0, 0.1, 0.2, 0.3, 0.4], [0, 1, 2, 3, 4], [0, 10, 20, 30, 40]]\n[[0.0, 0.1, 0.2, 0.3, 0.4]]\n(88, 80, 1)\n"
]
],
[
[
"The next task will be to build the DQN (*Deep Q-Network*). In principle, one could just feed it a state-action pair (s, a) and have it estimate the Q-Value Q(s, a). But since the actions are discrete and only 9 actions are possible, one can also feed the DQN the state s and have it estimate Q(s, a) for all 9 possible actions. The DQN shall have 3 convolutional layers followed by 2 fully connected layers, the last of which is the output layer. An actor-critic approach is used. They share the same architecture but have different tasks: the actor plays and the critic watches the play and tries to identify and fix shortcomings. After each few iterations, the critic network is copied to the actor network. So while they share the same architecture their parameters are different. Next, we define a function to build these DQNs.",
"_____no_output_____"
]
],
[
[
"### construction phase\n# resetting the graph seems to always be a good idea\nreset_graph()\n# DQN architecture / hyperparameters\ninput_height = 88\ninput_width = 80\ninput_channels = 1\nconv_n_maps = [32, 64, 64]\nconv_kernel_sizes = [(8,8), (4,4), (3,3)]\nconv_strides = [4, 2, 1]\nconv_paddings = [\"SAME\"] * 3\nconv_activation =[tf.nn.relu] * 3\nn_hidden_in = 64 * 11 * 10 # conv3 has 64 maps of 11x10 each\nn_hidden = 512\nhidden_activation = tf.nn.relu # corrected from \"[tf.nn.relu]*3\", according to the code on github (see link above)\nn_outputs = env.action_space.n # 9 discrete actions are available\ninitializer = tf.contrib.layers.variance_scaling_initializer()\n# function to build networks with the above architecture; the function receives a state (observation) and a name\ndef q_network(X_state, name):\n prev_layer = X_state # input from the input layer\n conv_layers = [] # container for all the layers of the convolutional network (input layer is not part of that)\n with tf.variable_scope(name) as scope:\n # loop through tuples, see \"https://docs.python.org/3.3/library/functions.html#zip\" for details;\n # in the first (second) [next] step take the first (second) [next] elements of the lists conv_n_maps, ...\n # ... conv_kernel_sizes, etc.; this continues until one of these lists has arrived at the end;\n # here, all these lists have length 3 (see above)\n for n_maps, kernel_size, stride, padding, activation in zip(conv_n_maps, conv_kernel_sizes, conv_strides,\n conv_paddings, conv_activation):\n # in the next step (of the for-loop), this layer will be the \"previous\" one\n # [\"stride\" -> \"strides\", see https://www.tensorflow.org/api_docs/python/tf/layers/conv2d]\n prev_layer = tf.layers.conv2d(prev_layer, filters=n_maps, kernel_size=kernel_size, strides=stride,\n padding=padding, activation=activation, kernel_initializer=initializer)\n # put the current layer into the container for all the layers of the convolutional neural network\n conv_layers.append(prev_layer)\n # make the last output a vector so it it can be passed easily to the upcoming fully connected layer\n last_conv_layer_flat = tf.reshape(prev_layer, shape=[-1, n_hidden_in])\n # first hidden layer\n hidden = tf.layers.dense(last_conv_layer_flat, n_hidden, activation=hidden_activation,\n kernel_initializer=initializer)\n # second hidden layer = output layer (these are the results!)\n outputs = tf.layers.dense(hidden, n_outputs, kernel_initializer=initializer)\n # let tensorflow figure out what variables shall be trained ...\n trainable_vars = tf.get_collection(tf.GraphKeys.TRAINABLE_VARIABLES, scope=scope.name)\n # ... and give these variables names so they can be distinguished in the graph\n trainable_vars_by_name = {var.name[len(scope.name):]: var for var in trainable_vars}\n return outputs, trainable_vars_by_name # return the outputs and the dictionary of trainable variables\n# input, actor, critic, and model copying\nX_state = tf.placeholder(tf.float32, shape=[None, input_height, input_width, input_channels]) # input placeholder\nactor_q_values, actor_vars = q_network(X_state, name=\"q_networks/actor\") # actor model\n#q_values = actor_q_values.eval(feed_dict={X_state: [state]})\ncritic_q_values, critic_vars = q_network(X_state, name=\"q_networks/critic\") # critic model\ncopy_ops = [actor_var.assign(critic_vars[var_name]) for var_name, actor_var in actor_vars.items()] # copy critic ... \ncopy_critic_to_actor = tf.group(*copy_ops) # ... to actor\n# some more theory is due\nprint(\"next, some text...\")",
"next, some text...\n"
]
],
[
[
"The somehow initialized Q-values will have the actor network play the game (initially somehow random), update the encountered Q-values (via the default discounted rewards), and leave the remaining Q-values unchanged. Every now and then, the critic network is tasked with predicting all Q-values by fitting the weights and biases. The Q-values resulting from the fitted weights and biases will be slightly different. This is supervised learning. The cost function that the critic network shall minimize is<br><br>\n$$J(\\theta_{\\rm critic})=\\frac{1}{m}\\sum_{i=1}^m\\left(y^{(i)}-Q(s^{(i)},a^{(i)},\\theta_{\\rm critic})\\right)^2{\\rm , with }\\quad y^{(i)}=r^{(i)}+\\gamma {\\rm max}_{a'}Q\\left(s'^{(i)},a^{(i)},\\theta_{\\rm actor}\\right)\\,,$$\nwhere\n- $s^{(i)},\\,a^{(i)},\\,r^{(i)},\\,s'^{(i)}$ are respectively the state, action, reward, and the next state of the $i^{\\rm th}$ memory sampled from the replay memory,\n- $m$ is the size of the memory batch,\n- $\\theta_{\\rm critic}$ and $\\theta_{\\rm actor}$ are the critic and the actor's parameters,\n- $Q(s^{(i)},a^{(i)},\\theta_{\\rm critic})$ is the critic DQN's prediction of the $i^{\\rm th}$ memorized state-action's Q-value,\n- $Q(s^{(i)},a^{(i)},\\theta_{\\rm actor})$ is the actor DQN's prediction of Q-value it can expect from the next state $s'^{\\rm(i)}$ if it chooses action $a'$,\n- $y^{(i)}$ is the target Q-value for the $i^{\\rm th}$ memory. Note that it is equal to the reward actually observed by the actor, plus the actor's *prediction* of what future rewards it should expect if it were to play optimally (as far as it knows), and\n- $J(\\theta_{\\rm critic})$ is the cost function used to train the critic DQN. As you can see, it is just the mean squared error between the target Q-values $y^{(i)}$ as estimated by the actor DQN, and the critic DQN's prediction of these Q-values.\n\n**General note**<br>\nThe replay memory [see code and/or text in the book] is optional but highly recommended. Without it , you would train the critic DQN using consecutive experiences that may be very correlated. This would introduce a lot of bias and slow down the training algorithm's convergence. By using a replay memory, we ensure that the memories fed to the training algorithm can be fairly uncorrelated.",
"_____no_output_____"
]
],
[
[
"# the actor DQN computes 9 Q-values: 1 for each possible action; use one-hot encoding to select only the one that ...\n# ... is actually chosen (https://www.tensorflow.org/api_docs/python/tf/one_hot)\nX_action = tf.placeholder(tf.int32, shape=[None])\nq_value = tf.reduce_sum(critic_q_values * tf.one_hot(X_action, n_outputs), axis=1, keep_dims=True)\n# feed the Q-values for the critic network through a placeholder y and do all the rest for training operations\ny = tf.placeholder(tf.float32, shape=[None, 1])\ncost = tf.reduce_mean(tf.square(y - q_value))\nglobal_step = tf.Variable(0, trainable=False, name=\"global_step\")\noptimizer = tf.train.AdamOptimizer(learning_rate)\ntraining_op = optimizer.minimize(cost, global_step=global_step)\n# initializer and saver nodes\ninit = tf.global_variables_initializer()\nsaver = tf.train.Saver()",
"WARNING:tensorflow:From <ipython-input-17-483165648def>:4: calling reduce_sum (from tensorflow.python.ops.math_ops) with keep_dims is deprecated and will be removed in a future version.\nInstructions for updating:\nkeep_dims is deprecated, use keepdims instead\n"
]
],
[
[
"For the execution phase, we will need the following tools.",
"_____no_output_____"
]
],
[
[
"# time keeping; for details, see the contribution of user \"daviewales\" on ...\n# ... https://codereview.stackexchange.com/questions/26534/is-there-a-better-way-to-count-seconds-in-python\nimport time\ntime_start = time.time()\n# another import, see https://docs.python.org/3/library/collections.html#collections.deque\nfrom collections import deque # deque or \"double-ended queue\" is similar to a python list but more performant\nreplay_memory_size = 10000 # name says it\nreplay_memory = deque([], maxlen=replay_memory_size) # deque container for the replay memory\n# function that samples random memories\ndef sample_memories(batch_size):\n # use \"np.rand\" instead of \"rnd\", see ...\n # ... \"https://docs.scipy.org/doc/numpy-1.15.0/reference/generated/numpy.random.permutation.html\"\n indices = np.random.permutation(len(replay_memory))[:batch_size] # take \"batch_size\" random indices\n cols = [[], [], [], [], []] # state, action, reward, next_state, continue\n # loop over indices\n for idx in indices:\n memory = replay_memory[idx] # specifice memory\n for col, value in zip(cols, memory): # fancy way of moving the values in the ...\n col.append(value) # ... memory into col\n cols = [np.array(col) for col in cols] # make entries numpy arrays\n return (cols[0], cols[1], cols[2].reshape(-1,1), cols[3], cols[4].reshape(-1,1))\n# use an epsilon-greedy policy\neps_min = 0.05 # next action is random with probability of 5%\neps_max = 1.0 # next action will be random for sure\neps_decay_steps = 50000 # decay schedule for epsilon\n# receive the Q values for the current state and the current step\ndef epsilon_greedy(q_values, step):\n epsilon = max(eps_min, eps_max - (eps_max-eps_min) * step/eps_decay_steps) # calculate the current epsilon\n if np.random.rand() < epsilon: # choice of random or optimal action\n return np.random.randint(n_outputs) # random action\n else:\n return np.argmax(q_values) # optimal action\n### execution phase\nn_steps = 10 # total number of training steps (default: 100000)\ntraining_start = 1000 # start training after 1000 game iterations\ntraining_interval = 3 # run a training step every few game iterations (default: 3)\nsave_steps = 50 # save the model every 50 training steps\ncopy_steps = 25 # copy the critic to the actor every few training steps (default: 25)\ndiscount_rate = 0.95 # discount rate (default: 0.95)\nskip_start = 90 # skip the start of every game (it's just waiting time)\nbatch_size = 50 # batch size (default: 50)\niteration = 0 # initialize the game iterations counter\ncheckpoint_path = \"./tf_logs/16_RL/MsPacMan/my_dqn.ckpt\" # checkpoint path\ndone = True # env needs to be reset\n# remaining import\nimport os\n# start the session\nwith tf.Session() as sess:\n # restore a session if one has been stored\n if os.path.isfile(checkpoint_path):\n saver.restore(sess, checkpoint_path)\n # otherwise start from scratch\n else:\n init.run()\n while True: # continue training ...\n step = global_step.eval() # ... until step ...\n if step >= n_steps: # ... has reached n_steps ...\n break # ... (stop in that latter case)\n iteration += 1 # count the iterations\n if done: # game over: start again ...\n # restart 1/3\n obs = env.reset() # ... and reset the environment\n # restart 2/3\n for skip in range(skip_start): # skip the start of each game\n obs, reward, done, info = env.step(0) # get data from environment (Ms. Pacman)\n # restart 3/3\n state = preprocess_observation(obs) # preprocess the state of the game (screenshot)\n # actor evaluates what to do\n q_values = actor_q_values.eval(feed_dict={X_state: [state]}) # get available Q-values for the current state\n action = epsilon_greedy(q_values, step) # apply the epsilon-greedy policy\n # actor plays\n obs, reward, done, info = env.step(action) # apply the action and get new data from env\n next_state = preprocess_observation(obs) # preprocess the next state (screenshot)\n # let's memorize what just happened\n replay_memory.append((state, action, reward, next_state, 1.0 - done))\n state = next_state\n # go to the next iteration of the while loop (i) before training starts and (ii) if learning is not scheduled\n if iteration < training_start or iteration % training_interval != 0:\n continue\n # if learning is scheduled for the current iteration, get samples from the memory, update the Q-values ...\n # ... that the actor learned as well as the rewards, and run a training operation\n X_state_val, X_action_val, rewards, X_next_state_val, continues = (sample_memories(batch_size))\n next_q_values = actor_q_values.eval(feed_dict={X_state: X_next_state_val})\n max_next_q_values = np.max(next_q_values, axis=1, keepdims=True)\n y_val = rewards + continues * discount_rate * max_next_q_values\n training_op.run(feed_dict={X_state: X_state_val, X_action: X_action_val, y: y_val})\n # regularly copy critic to actor and ... \n if step % copy_steps == 0:\n copy_critic_to_actor.run()\n # ... save the session so it could be restored\n if step % save_steps == 0:\n saver.save(sess, checkpoint_path)\n# the output of this time keeping (see top of this cell) will be in seconds\ntime_finish = time.time()\nprint(time_finish - time_start)",
"14.940124988555908\n"
]
],
[
[
"With a function from the github link above, we can visualize the play of the trained model.",
"_____no_output_____"
]
],
[
[
"frames = []\nn_max_steps = 10000\nwith tf.Session() as sess:\n saver.restore(sess, checkpoint_path)\n obs = env.reset()\n for step in range(n_max_steps):\n state = preprocess_observation(obs)\n q_values = actor_q_values.eval(feed_dict={X_state: [state]}) # from \"online_q_values\" to \"actor_q_values\"\n action = np.argmax(q_values)\n obs, reward, done, info = env.step(action)\n img = env.render(mode=\"rgb_array\")\n frames.append(img)\n if done:\n break\nplot_animation(frames)",
"INFO:tensorflow:Restoring parameters from ./tf_logs/16_RL/MsPacMan/my_dqn.ckpt\n"
]
],
[
[
"**Suggestion or Tip**<br>\nUnfortunately, training is very slow: if you use your laptop for training, it will take days before Ms. Pac-Man gets any good, and if you look at the learning curve, measuring the average rewards per episode, you will notice that it is extremely noisy. At some points there may be no apparent progress for a very long time until suddenly the agent learns to survive a reasonable amount of time. As mentioned earlier, one solution is to inject as much prior knowledge as possible into the model (e.g., through preprocessing, rewards, and so on), and you can also try to bootstrap the model by first training it to imitate a basic strategy. In any case, RL still requires quite a lot of patience and tweaking but the end result is very exciting.\n## Exercises\npage 473<br>\n### 1.-7.\nSolutions are shown in Appendix A of the book and in the separate notebook *ExercisesWithoutCode*.\n### 8.\nUse Deep Q-Learning to tackle OpenAI gym's \"BypedalWalker-v2\". The Q-networks do not need to be very deep for this task.",
"_____no_output_____"
]
],
[
[
"# everything below is mainly from the github link above\n# the rendering command (see github link) does not work here; the according code is adapted; frames are not ...\n# ... displayed, here\nenv = gym.make(\"BipedalWalker-v2\")\nprint(env)\nobs = env.reset()\nprint(obs)\nprint(env.action_space)\nprint(env.action_space.low)\nprint(env.action_space.high)",
"\u001b[33mWARN: gym.spaces.Box autodetected dtype as <class 'numpy.float32'>. Please provide explicit dtype.\u001b[0m\n\u001b[33mWARN: gym.spaces.Box autodetected dtype as <class 'numpy.float32'>. Please provide explicit dtype.\u001b[0m\n<TimeLimit<BipedalWalker<BipedalWalker-v2>>>\n[ 2.74743512e-03 -1.15991896e-05 9.02299415e-04 -1.59999228e-02\n 9.19910073e-02 -1.19072467e-03 8.60251635e-01 2.28293116e-03\n 1.00000000e+00 3.23961042e-02 -1.19064655e-03 8.53801474e-01\n 8.42094344e-04 1.00000000e+00 4.40814018e-01 4.45820123e-01\n 4.61422771e-01 4.89550203e-01 5.34102798e-01 6.02461040e-01\n 7.09148884e-01 8.85931849e-01 1.00000000e+00 1.00000000e+00]\nBox(4,)\n[-1. -1. -1. -1.]\n[1. 1. 1. 1.]\n"
]
],
[
[
"Build all possible combinations of actions (cf. above).",
"_____no_output_____"
]
],
[
[
"# see https://docs.python.org/3.7/library/itertools.html#itertools.product\nfrom itertools import product\npossible_torques = np.array([-1.0, 0.0, 1.0])\npossible_actions = np.array(list(product(possible_torques, repeat=4)))\npossible_actions.shape\nprint(possible_actions)",
"[[-1. -1. -1. -1.]\n [-1. -1. -1. 0.]\n [-1. -1. -1. 1.]\n [-1. -1. 0. -1.]\n [-1. -1. 0. 0.]\n [-1. -1. 0. 1.]\n [-1. -1. 1. -1.]\n [-1. -1. 1. 0.]\n [-1. -1. 1. 1.]\n [-1. 0. -1. -1.]\n [-1. 0. -1. 0.]\n [-1. 0. -1. 1.]\n [-1. 0. 0. -1.]\n [-1. 0. 0. 0.]\n [-1. 0. 0. 1.]\n [-1. 0. 1. -1.]\n [-1. 0. 1. 0.]\n [-1. 0. 1. 1.]\n [-1. 1. -1. -1.]\n [-1. 1. -1. 0.]\n [-1. 1. -1. 1.]\n [-1. 1. 0. -1.]\n [-1. 1. 0. 0.]\n [-1. 1. 0. 1.]\n [-1. 1. 1. -1.]\n [-1. 1. 1. 0.]\n [-1. 1. 1. 1.]\n [ 0. -1. -1. -1.]\n [ 0. -1. -1. 0.]\n [ 0. -1. -1. 1.]\n [ 0. -1. 0. -1.]\n [ 0. -1. 0. 0.]\n [ 0. -1. 0. 1.]\n [ 0. -1. 1. -1.]\n [ 0. -1. 1. 0.]\n [ 0. -1. 1. 1.]\n [ 0. 0. -1. -1.]\n [ 0. 0. -1. 0.]\n [ 0. 0. -1. 1.]\n [ 0. 0. 0. -1.]\n [ 0. 0. 0. 0.]\n [ 0. 0. 0. 1.]\n [ 0. 0. 1. -1.]\n [ 0. 0. 1. 0.]\n [ 0. 0. 1. 1.]\n [ 0. 1. -1. -1.]\n [ 0. 1. -1. 0.]\n [ 0. 1. -1. 1.]\n [ 0. 1. 0. -1.]\n [ 0. 1. 0. 0.]\n [ 0. 1. 0. 1.]\n [ 0. 1. 1. -1.]\n [ 0. 1. 1. 0.]\n [ 0. 1. 1. 1.]\n [ 1. -1. -1. -1.]\n [ 1. -1. -1. 0.]\n [ 1. -1. -1. 1.]\n [ 1. -1. 0. -1.]\n [ 1. -1. 0. 0.]\n [ 1. -1. 0. 1.]\n [ 1. -1. 1. -1.]\n [ 1. -1. 1. 0.]\n [ 1. -1. 1. 1.]\n [ 1. 0. -1. -1.]\n [ 1. 0. -1. 0.]\n [ 1. 0. -1. 1.]\n [ 1. 0. 0. -1.]\n [ 1. 0. 0. 0.]\n [ 1. 0. 0. 1.]\n [ 1. 0. 1. -1.]\n [ 1. 0. 1. 0.]\n [ 1. 0. 1. 1.]\n [ 1. 1. -1. -1.]\n [ 1. 1. -1. 0.]\n [ 1. 1. -1. 1.]\n [ 1. 1. 0. -1.]\n [ 1. 1. 0. 0.]\n [ 1. 1. 0. 1.]\n [ 1. 1. 1. -1.]\n [ 1. 1. 1. 0.]\n [ 1. 1. 1. 1.]]\n"
]
],
[
[
"Build a network and see how it performs.",
"_____no_output_____"
]
],
[
[
"tf.reset_default_graph()\n# 1. Specify the network architecture\nn_inputs = env.observation_space.shape[0] # == 24\nn_hidden = 10\nn_outputs = len(possible_actions) # == 625\ninitializer = tf.variance_scaling_initializer()\n# 2. Build the neural network\nX = tf.placeholder(tf.float32, shape=[None, n_inputs])\nhidden = tf.layers.dense(X, n_hidden, activation=tf.nn.selu, kernel_initializer=initializer)\nlogits = tf.layers.dense(hidden, n_outputs, kernel_initializer=initializer)\noutputs = tf.nn.softmax(logits)\n# 3. Select a random action based on the estimated probabilities\naction_index = tf.squeeze(tf.multinomial(logits, num_samples=1), axis=-1)\n# 4. Training\nlearning_rate = 0.01\ny = tf.one_hot(action_index, depth=len(possible_actions))\ncross_entropy = tf.nn.softmax_cross_entropy_with_logits_v2(labels=y, logits=logits)\noptimizer = tf.train.AdamOptimizer(learning_rate)\ngrads_and_vars = optimizer.compute_gradients(cross_entropy)\ngradients = [grad for grad, variable in grads_and_vars]\ngradient_placeholders = []\ngrads_and_vars_feed = []\nfor grad, variable in grads_and_vars:\n gradient_placeholder = tf.placeholder(tf.float32, shape=grad.get_shape())\n gradient_placeholders.append(gradient_placeholder)\n grads_and_vars_feed.append((gradient_placeholder, variable))\ntraining_op = optimizer.apply_gradients(grads_and_vars_feed)\ninit = tf.global_variables_initializer()\nsaver = tf.train.Saver()\n# 5. execute it, count the rewards, and show them\ndef run_bipedal_walker(model_path=None, n_max_steps =1000): # function from github but adapted to counting rewards ...\n env = gym.make(\"BipedalWalker-v2\") # ... and showing them instead of showing rendered frames\n with tf.Session() as sess:\n if model_path is None:\n init.run()\n else:\n saver.restore(sess, model_path)\n obs = env.reset()\n rewards = 0\n for step in range(n_max_steps):\n action_index_val = action_index.eval(feed_dict={X: obs.reshape(1, n_inputs)})\n action = possible_actions[action_index_val]\n obs, reward, done, info = env.step(action[0])\n rewards += reward\n if done:\n break\n env.close()\n return rewards\nrun_bipedal_walker()",
"\u001b[33mWARN: gym.spaces.Box autodetected dtype as <class 'numpy.float32'>. Please provide explicit dtype.\u001b[0m\n\u001b[33mWARN: gym.spaces.Box autodetected dtype as <class 'numpy.float32'>. Please provide explicit dtype.\u001b[0m\n"
]
],
[
[
"The model does not perform well because it has not been trained, yet. This shall be done!",
"_____no_output_____"
]
],
[
[
"n_games_per_update = 10 # default is 10\nn_max_steps = 10 # default is 1000\nn_iterations = 100 # default is 1000\nsave_iterations = 10 # default is 10\ndiscount_rate = 0.95 # default is 0.95\nwith tf.Session() as sess:\n init.run()\n for iteration in range(n_iterations):\n print(\"\\rIteration: {}/{}\".format(iteration + 1, n_iterations), end=\"\")\n all_rewards = []\n all_gradients = []\n for game in range(n_games_per_update):\n current_rewards = []\n current_gradients = []\n obs = env.reset()\n for step in range(n_max_steps):\n action_index_val, gradients_val = sess.run([action_index, gradients],\n feed_dict={X: obs.reshape(1, n_inputs)})\n action = possible_actions[action_index_val]\n obs, reward, done, info = env.step(action[0])\n current_rewards.append(reward)\n current_gradients.append(gradients_val)\n if done:\n break\n all_rewards.append(current_rewards)\n all_gradients.append(current_gradients)\n all_rewards = discount_and_normalize_rewards(all_rewards, discount_rate=discount_rate)\n feed_dict = {}\n for var_index, gradient_placeholder in enumerate(gradient_placeholders):\n mean_gradients = np.mean([reward * all_gradients[game_index][step][var_index]\n for game_index, rewards in enumerate(all_rewards)\n for step, reward in enumerate(rewards)], axis=0)\n feed_dict[gradient_placeholder] = mean_gradients\n sess.run(training_op, feed_dict=feed_dict)\n if iteration % save_iterations == 0:\n saver.save(sess, \"./tf_logs/16_RL/BiPedal/my_bipedal_walker_pg.ckpt\")\nrun_bipedal_walker(\"./tf_logs/16_RL/BiPedal/my_bipedal_walker_pg.ckpt\")",
"Iteration: 100/100\u001b[33mWARN: gym.spaces.Box autodetected dtype as <class 'numpy.float32'>. Please provide explicit dtype.\u001b[0m\n\u001b[33mWARN: gym.spaces.Box autodetected dtype as <class 'numpy.float32'>. Please provide explicit dtype.\u001b[0m\nINFO:tensorflow:Restoring parameters from ./tf_logs/16_RL/BiPedal/my_bipedal_walker_pg.ckpt\n"
]
],
[
[
"With enough training, the network does indeed better.\n### 9. Use policy gradients to train an agent to play *Pong*, the famous Atari game (\"Pong-v0\" in the OpenAI gym). Beware: an individual observation is insufficient to tell the direction and speed of the ball. One solution is to pass two observations at a time to the neural network policy. To reduce dimensionality and speed up training, you should definitely preprocess these images (crop, resize, and convert them to black and white), and possibly merge them into a single image (e.g., by overlaying them).\n\nA solution may follow later.\n### 10. If you have about \\$100 to spare, you can purchase a Raspberry Pi 3 plus some cheap robotics components, install TensorFlow on the Pi, and go wild! For an example, check out this fun post (https://goo.gl/Eu5u28) by Lukas Biewald, or take a look at GoPiGo or BrickPi. Why not try to build a real-life cartpole by training the robot using policy gradients? Or build a robotic spider that learns to walk; give it rewards any time it gets closer to some objective (you will need sensors to measure the distance to the objective). The only limit is your imagination.\n\nA solution for this may follow even later.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d09473686cb44a21cdde369c00b4175438ec4edc | 14,977 | ipynb | Jupyter Notebook | dynamic_rnn.ipynb | abhihirekhan/-TensorFlow-basic-models-and-neural-networks-Tableau | 30215ddb8198bd8cc2b80b38475066f1da874025 | [
"MIT"
] | null | null | null | dynamic_rnn.ipynb | abhihirekhan/-TensorFlow-basic-models-and-neural-networks-Tableau | 30215ddb8198bd8cc2b80b38475066f1da874025 | [
"MIT"
] | null | null | null | dynamic_rnn.ipynb | abhihirekhan/-TensorFlow-basic-models-and-neural-networks-Tableau | 30215ddb8198bd8cc2b80b38475066f1da874025 | [
"MIT"
] | null | null | null | 42.91404 | 254 | 0.570475 | [
[
[
"# Dynamic Recurrent Neural Network.\n\nTensorFlow implementation of a Recurrent Neural Network (LSTM) that performs dynamic computation over sequences with variable length. This example is using a toy dataset to classify linear sequences. The generated sequences have variable length.\n\n ",
"_____no_output_____"
],
[
"## RNN Overview\n\n<img src=\"http://colah.github.io/posts/2015-08-Understanding-LSTMs/img/RNN-unrolled.png\" alt=\"nn\" style=\"width: 600px;\"/>\n\nReferences:\n- [Long Short Term Memory](http://deeplearning.cs.cmu.edu/pdfs/Hochreiter97_lstm.pdf), Sepp Hochreiter & Jurgen Schmidhuber, Neural Computation 9(8): 1735-1780, 1997.",
"_____no_output_____"
]
],
[
[
"from __future__ import print_function\n\nimport tensorflow as tf\nimport random",
"_____no_output_____"
],
[
"# ====================\n# TOY DATA GENERATOR\n# ====================\n\nclass ToySequenceData(object):\n \"\"\" Generate sequence of data with dynamic length.\n This class generate samples for training:\n - Class 0: linear sequences (i.e. [0, 1, 2, 3,...])\n - Class 1: random sequences (i.e. [1, 3, 10, 7,...])\n\n NOTICE:\n We have to pad each sequence to reach 'max_seq_len' for TensorFlow\n consistency (we cannot feed a numpy array with inconsistent\n dimensions). The dynamic calculation will then be perform thanks to\n 'seqlen' attribute that records every actual sequence length.\n \"\"\"\n def __init__(self, n_samples=1000, max_seq_len=20, min_seq_len=3,\n max_value=1000):\n self.data = []\n self.labels = []\n self.seqlen = []\n for i in range(n_samples):\n # Random sequence length\n len = random.randint(min_seq_len, max_seq_len)\n # Monitor sequence length for TensorFlow dynamic calculation\n self.seqlen.append(len)\n # Add a random or linear int sequence (50% prob)\n if random.random() < .5:\n # Generate a linear sequence\n rand_start = random.randint(0, max_value - len)\n s = [[float(i)/max_value] for i in\n range(rand_start, rand_start + len)]\n # Pad sequence for dimension consistency\n s += [[0.] for i in range(max_seq_len - len)]\n self.data.append(s)\n self.labels.append([1., 0.])\n else:\n # Generate a random sequence\n s = [[float(random.randint(0, max_value))/max_value]\n for i in range(len)]\n # Pad sequence for dimension consistency\n s += [[0.] for i in range(max_seq_len - len)]\n self.data.append(s)\n self.labels.append([0., 1.])\n self.batch_id = 0\n\n def next(self, batch_size):\n \"\"\" Return a batch of data. When dataset end is reached, start over.\n \"\"\"\n if self.batch_id == len(self.data):\n self.batch_id = 0\n batch_data = (self.data[self.batch_id:min(self.batch_id +\n batch_size, len(self.data))])\n batch_labels = (self.labels[self.batch_id:min(self.batch_id +\n batch_size, len(self.data))])\n batch_seqlen = (self.seqlen[self.batch_id:min(self.batch_id +\n batch_size, len(self.data))])\n self.batch_id = min(self.batch_id + batch_size, len(self.data))\n return batch_data, batch_labels, batch_seqlen",
"_____no_output_____"
],
[
"# ==========\n# MODEL\n# ==========\n\n# Parameters\nlearning_rate = 0.01\ntraining_steps = 10000\nbatch_size = 128\ndisplay_step = 200\n\n# Network Parameters\nseq_max_len = 20 # Sequence max length\nn_hidden = 64 # hidden layer num of features\nn_classes = 2 # linear sequence or not\n\ntrainset = ToySequenceData(n_samples=1000, max_seq_len=seq_max_len)\ntestset = ToySequenceData(n_samples=500, max_seq_len=seq_max_len)\n\n# tf Graph input\nx = tf.placeholder(\"float\", [None, seq_max_len, 1])\ny = tf.placeholder(\"float\", [None, n_classes])\n# A placeholder for indicating each sequence length\nseqlen = tf.placeholder(tf.int32, [None])\n\n# Define weights\nweights = {\n 'out': tf.Variable(tf.random_normal([n_hidden, n_classes]))\n}\nbiases = {\n 'out': tf.Variable(tf.random_normal([n_classes]))\n}",
"_____no_output_____"
],
[
"def dynamicRNN(x, seqlen, weights, biases):\n\n # Prepare data shape to match `rnn` function requirements\n # Current data input shape: (batch_size, n_steps, n_input)\n # Required shape: 'n_steps' tensors list of shape (batch_size, n_input)\n \n # Unstack to get a list of 'n_steps' tensors of shape (batch_size, n_input)\n x = tf.unstack(x, seq_max_len, 1)\n\n # Define a lstm cell with tensorflow\n lstm_cell = tf.contrib.rnn.BasicLSTMCell(n_hidden)\n\n # Get lstm cell output, providing 'sequence_length' will perform dynamic\n # calculation.\n outputs, states = tf.contrib.rnn.static_rnn(lstm_cell, x, dtype=tf.float32,\n sequence_length=seqlen)\n\n # When performing dynamic calculation, we must retrieve the last\n # dynamically computed output, i.e., if a sequence length is 10, we need\n # to retrieve the 10th output.\n # However TensorFlow doesn't support advanced indexing yet, so we build\n # a custom op that for each sample in batch size, get its length and\n # get the corresponding relevant output.\n\n # 'outputs' is a list of output at every timestep, we pack them in a Tensor\n # and change back dimension to [batch_size, n_step, n_input]\n outputs = tf.stack(outputs)\n outputs = tf.transpose(outputs, [1, 0, 2])\n\n # Hack to build the indexing and retrieve the right output.\n batch_size = tf.shape(outputs)[0]\n # Start indices for each sample\n index = tf.range(0, batch_size) * seq_max_len + (seqlen - 1)\n # Indexing\n outputs = tf.gather(tf.reshape(outputs, [-1, n_hidden]), index)\n\n # Linear activation, using outputs computed above\n return tf.matmul(outputs, weights['out']) + biases['out']",
"_____no_output_____"
],
[
"pred = dynamicRNN(x, seqlen, weights, biases)\n\n# Define loss and optimizer\ncost = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(logits=pred, labels=y))\noptimizer = tf.train.GradientDescentOptimizer(learning_rate=learning_rate).minimize(cost)\n\n# Evaluate model\ncorrect_pred = tf.equal(tf.argmax(pred,1), tf.argmax(y,1))\naccuracy = tf.reduce_mean(tf.cast(correct_pred, tf.float32))\n\n# Initialize the variables (i.e. assign their default value)\ninit = tf.global_variables_initializer()",
"/Users/aymeric.damien/anaconda2/lib/python2.7/site-packages/tensorflow/python/ops/gradients_impl.py:93: UserWarning: Converting sparse IndexedSlices to a dense Tensor of unknown shape. This may consume a large amount of memory.\n \"Converting sparse IndexedSlices to a dense Tensor of unknown shape. \"\n"
],
[
"# Start training\nwith tf.Session() as sess:\n\n # Run the initializer\n sess.run(init)\n\n for step in range(1, training_steps+1):\n batch_x, batch_y, batch_seqlen = trainset.next(batch_size)\n # Run optimization op (backprop)\n sess.run(optimizer, feed_dict={x: batch_x, y: batch_y,\n seqlen: batch_seqlen})\n if step % display_step == 0 or step == 1:\n # Calculate batch accuracy & loss\n acc, loss = sess.run([accuracy, cost], feed_dict={x: batch_x, y: batch_y,\n seqlen: batch_seqlen})\n print(\"Step \" + str(step) + \", Minibatch Loss= \" + \\\n \"{:.6f}\".format(loss) + \", Training Accuracy= \" + \\\n \"{:.5f}\".format(acc))\n\n print(\"Optimization Finished!\")\n\n # Calculate accuracy\n test_data = testset.data\n test_label = testset.labels\n test_seqlen = testset.seqlen\n print(\"Testing Accuracy:\", \\\n sess.run(accuracy, feed_dict={x: test_data, y: test_label,\n seqlen: test_seqlen}))",
"Step 1, Minibatch Loss= 0.864517, Training Accuracy= 0.42188\nStep 200, Minibatch Loss= 0.686012, Training Accuracy= 0.43269\nStep 400, Minibatch Loss= 0.682970, Training Accuracy= 0.48077\nStep 600, Minibatch Loss= 0.679640, Training Accuracy= 0.50962\nStep 800, Minibatch Loss= 0.675208, Training Accuracy= 0.53846\nStep 1000, Minibatch Loss= 0.668636, Training Accuracy= 0.56731\nStep 1200, Minibatch Loss= 0.657525, Training Accuracy= 0.62500\nStep 1400, Minibatch Loss= 0.635423, Training Accuracy= 0.67308\nStep 1600, Minibatch Loss= 0.580433, Training Accuracy= 0.75962\nStep 1800, Minibatch Loss= 0.475599, Training Accuracy= 0.81731\nStep 2000, Minibatch Loss= 0.434865, Training Accuracy= 0.83654\nStep 2200, Minibatch Loss= 0.423690, Training Accuracy= 0.85577\nStep 2400, Minibatch Loss= 0.417472, Training Accuracy= 0.85577\nStep 2600, Minibatch Loss= 0.412906, Training Accuracy= 0.85577\nStep 2800, Minibatch Loss= 0.409193, Training Accuracy= 0.85577\nStep 3000, Minibatch Loss= 0.406035, Training Accuracy= 0.86538\nStep 3200, Minibatch Loss= 0.403287, Training Accuracy= 0.87500\nStep 3400, Minibatch Loss= 0.400862, Training Accuracy= 0.87500\nStep 3600, Minibatch Loss= 0.398704, Training Accuracy= 0.86538\nStep 3800, Minibatch Loss= 0.396768, Training Accuracy= 0.86538\nStep 4000, Minibatch Loss= 0.395017, Training Accuracy= 0.86538\nStep 4200, Minibatch Loss= 0.393422, Training Accuracy= 0.86538\nStep 4400, Minibatch Loss= 0.391957, Training Accuracy= 0.85577\nStep 4600, Minibatch Loss= 0.390600, Training Accuracy= 0.85577\nStep 4800, Minibatch Loss= 0.389334, Training Accuracy= 0.86538\nStep 5000, Minibatch Loss= 0.388143, Training Accuracy= 0.86538\nStep 5200, Minibatch Loss= 0.387015, Training Accuracy= 0.86538\nStep 5400, Minibatch Loss= 0.385940, Training Accuracy= 0.86538\nStep 5600, Minibatch Loss= 0.384907, Training Accuracy= 0.86538\nStep 5800, Minibatch Loss= 0.383904, Training Accuracy= 0.85577\nStep 6000, Minibatch Loss= 0.382921, Training Accuracy= 0.86538\nStep 6200, Minibatch Loss= 0.381941, Training Accuracy= 0.86538\nStep 6400, Minibatch Loss= 0.380947, Training Accuracy= 0.86538\nStep 6600, Minibatch Loss= 0.379912, Training Accuracy= 0.86538\nStep 6800, Minibatch Loss= 0.378796, Training Accuracy= 0.86538\nStep 7000, Minibatch Loss= 0.377540, Training Accuracy= 0.86538\nStep 7200, Minibatch Loss= 0.376041, Training Accuracy= 0.86538\nStep 7400, Minibatch Loss= 0.374130, Training Accuracy= 0.85577\nStep 7600, Minibatch Loss= 0.371514, Training Accuracy= 0.85577\nStep 7800, Minibatch Loss= 0.367723, Training Accuracy= 0.85577\nStep 8000, Minibatch Loss= 0.362049, Training Accuracy= 0.85577\nStep 8200, Minibatch Loss= 0.353558, Training Accuracy= 0.85577\nStep 8400, Minibatch Loss= 0.341072, Training Accuracy= 0.86538\nStep 8600, Minibatch Loss= 0.323062, Training Accuracy= 0.87500\nStep 8800, Minibatch Loss= 0.299278, Training Accuracy= 0.89423\nStep 9000, Minibatch Loss= 0.273857, Training Accuracy= 0.90385\nStep 9200, Minibatch Loss= 0.248392, Training Accuracy= 0.91346\nStep 9400, Minibatch Loss= 0.221348, Training Accuracy= 0.92308\nStep 9600, Minibatch Loss= 0.191947, Training Accuracy= 0.92308\nStep 9800, Minibatch Loss= 0.159308, Training Accuracy= 0.93269\nStep 10000, Minibatch Loss= 0.136938, Training Accuracy= 0.96154\nOptimization Finished!\nTesting Accuracy: 0.952\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d09476661ac3256ab054fc3b010d0ef75197db37 | 6,080 | ipynb | Jupyter Notebook | experiments/breakout/mc_random_query_efficiency.ipynb | Tony-Cheng/Active-Reinforcement-Learning | 50bb65106ae1f957d8cb6cb5706ce1285519e6b4 | [
"MIT"
] | null | null | null | experiments/breakout/mc_random_query_efficiency.ipynb | Tony-Cheng/Active-Reinforcement-Learning | 50bb65106ae1f957d8cb6cb5706ce1285519e6b4 | [
"MIT"
] | null | null | null | experiments/breakout/mc_random_query_efficiency.ipynb | Tony-Cheng/Active-Reinforcement-Learning | 50bb65106ae1f957d8cb6cb5706ce1285519e6b4 | [
"MIT"
] | null | null | null | 29.658537 | 141 | 0.583553 | [
[
[
"import sys, os\nsys.path.append(os.path.abspath('../..'))",
"_____no_output_____"
],
[
"from tqdm.notebook import tqdm\nimport math\nimport gym\nimport torch\nimport torch.optim as optim \nfrom torch.utils.tensorboard import SummaryWriter\nfrom collections import deque\n\nfrom networks.dqn_atari import MC_DQN\nfrom utils.memory import RankedReplayMemory, LabeledReplayMemory\nfrom utils.optimization import AMN_perc_optimization, AMN_optimization_epochs\nfrom environments.atari_wrappers import make_atari, wrap_deepmind\nfrom utils.atari_utils import fp, ActionSelector, evaluate\nfrom utils.acquisition_functions import mc_random\n\nimport imageio\nfrom utils.visualization import visualize_AMN",
"_____no_output_____"
],
[
"env_name = 'Breakout'\nenv_raw = make_atari('{}NoFrameskip-v4'.format(env_name))\nenv = wrap_deepmind(env_raw, frame_stack=False, episode_life=False, clip_rewards=True)\nc,h,w = c,h,w = fp(env.reset()).shape\nn_actions = env.action_space.n",
"_____no_output_____"
],
[
"BATCH_SIZE = 128\nLR = 0.0000625\nGAMMA = 0.99\nEPS = 0.05\nNUM_STEPS = 10000000\nNOT_LABELLED_CAPACITY = 10000\nLABELLED_CAPACITY = 100000\nINITIAL_STEPS=NOT_LABELLED_CAPACITY\nEPOCHS=20\nPERCENTAGE = 0.01\n\nNAME = 'mc_random_1_large_buffer_many_epochs'",
"_____no_output_____"
],
[
"device = torch.device(\"cuda\" if torch.cuda.is_available() else \"cpu\") # if gpu is to be used\nAMN_net = MC_DQN(n_actions).to(device)\n# AMN_net = ENS_DQN(n_actions).to(device)\nexpert_net = torch.load(\"models/dqn_expert_breakout_model\").to(device)\nAMN_net.apply(AMN_net.init_weights)\nexpert_net.eval()\noptimizer = optim.Adam(AMN_net.parameters(), lr=LR, eps=1.5e-4)\n# optimizer = optim.Adam(AMN_net.parameters(), lr=LR, eps=1.5e-4)",
"_____no_output_____"
],
[
"memory = LabeledReplayMemory(NOT_LABELLED_CAPACITY, LABELLED_CAPACITY, [5,h,w], n_actions, mc_random, AMN_net, device=device)\naction_selector = ActionSelector(EPS, EPS, AMN_net, 1, n_actions, device)",
"_____no_output_____"
],
[
"steps_done = 0\nwriter = SummaryWriter(f'runs/{NAME}')",
"_____no_output_____"
],
[
"q = deque(maxlen=5)\ndone=True\neps = 0\nepisode_len = 0\nnum_labels = 0",
"_____no_output_____"
],
[
"progressive = tqdm(range(NUM_STEPS), total=NUM_STEPS, ncols=400, leave=False, unit='b')\nfor step in progressive:\n if done:\n env.reset()\n sum_reward = 0\n episode_len = 0\n img, _, _, _ = env.step(1) # BREAKOUT specific !!!\n for i in range(10): # no-op\n n_frame, _, _, _ = env.step(0)\n n_frame = fp(n_frame)\n q.append(n_frame)\n \n # Select and perform an action\n state = torch.cat(list(q))[1:].unsqueeze(0)\n action, eps = action_selector.select_action(state)\n n_frame, reward, done, info = env.step(action)\n n_frame = fp(n_frame)\n\n # 5 frame as memory\n q.append(n_frame)\n memory.push(torch.cat(list(q)).unsqueeze(0), action, reward, done) # here the n_frame means next frame from the previous time step\n episode_len += 1\n\n # Perform one step of the optimization (on the target network)\n if step % NOT_LABELLED_CAPACITY == 0 and step > INITIAL_STEPS:\n num_labels += memory.label_sample(percentage=PERCENTAGE)\n loss = AMN_optimization_epochs(AMN_net, expert_net, optimizer, memory, EPOCHS,\n batch_size=BATCH_SIZE, device=device)\n if loss is not None:\n writer.add_scalar('loss_vs_#labels', loss, num_labels)\n evaluated_reward = evaluate(step, AMN_net, device, env_raw, n_actions, eps=0.05, num_episode=15)\n writer.add_scalar('reward_vs_#labels', evaluated_reward, num_labels)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d094a9a4a89f17a45c8b297e2721931da673637a | 964,183 | ipynb | Jupyter Notebook | NY Permits.ipynb | parasgulati8/Data-Analysis | 4ff049fc31b9265b02c3de505df306eb0ce14797 | [
"MIT"
] | null | null | null | NY Permits.ipynb | parasgulati8/Data-Analysis | 4ff049fc31b9265b02c3de505df306eb0ce14797 | [
"MIT"
] | null | null | null | NY Permits.ipynb | parasgulati8/Data-Analysis | 4ff049fc31b9265b02c3de505df306eb0ce14797 | [
"MIT"
] | null | null | null | 413.812446 | 311,272 | 0.911161 | [
[
[
"# **Exploratory Analysis**",
"_____no_output_____"
],
[
"First of all, let's import some useful libraries that will be used in the analysis.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"Now, the dataset stored in drive needs to be retieved. I am using google colab for this exploration with TPU hardware accelerator for faster computation. To get the data from drive the drive needs to be mounted first. ",
"_____no_output_____"
]
],
[
[
"from google.colab import drive\ndrive.mount('/content/drive/')",
"Go to this URL in a browser: https://accounts.google.com/o/oauth2/auth?client_id=947318989803-6bn6qk8qdgf4n4g3pfee6491hc0brc4i.apps.googleusercontent.com&redirect_uri=urn%3Aietf%3Awg%3Aoauth%3A2.0%3Aoob&scope=email%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fdocs.test%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fdrive%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fdrive.photos.readonly%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fpeopleapi.readonly&response_type=code\n\nEnter your authorization code:\n··········\nMounted at /content/drive/\n"
]
],
[
[
"Once the drive is successfully mounted, I fetched the data and stored it in a pandas dataframe.",
"_____no_output_____"
]
],
[
[
"dataset = pd.read_csv(\"/content/drive/My Drive/Colab Notebooks/DOB_Permit_Issuance.csv\")",
"/usr/local/lib/python3.6/dist-packages/IPython/core/interactiveshell.py:2718: DtypeWarning: Columns (1,8,9,10,15,25,31,33,34,35,36,51,52) have mixed types. Specify dtype option on import or set low_memory=False.\n interactivity=interactivity, compiler=compiler, result=result)\n"
]
],
[
[
"To get the gist of the dataset, I used pandas describe function that gives a very broad understanding of the data. ",
"_____no_output_____"
]
],
[
[
"pd.set_option('display.max_colwidth', 1000)\npd.set_option('display.max_rows', 1000)\npd.set_option('display.max_columns', 500)\npd.set_option('display.width', 1000)\ndataset.describe()",
"_____no_output_____"
]
],
[
[
"From the describe function, we now know that the dataset has almost 3.5 M data. Let's take a look at the dataset now.",
"_____no_output_____"
]
],
[
[
"dataset.head()",
"_____no_output_____"
]
],
[
[
"I can see there are lot's of NaNs in many columns. To better analyse the data, the NaNs needs to be removed or dealt with. But first, let's see hoe many NaNs are there is each column.",
"_____no_output_____"
]
],
[
[
"dataset.isna().sum()",
"_____no_output_____"
]
],
[
[
"\n\n---\n\n\nThe information above is very useful in feature selection. Observing the columns with very high number of NaNs, such as :\n\n\nColumn | NaNs \n------------|--------\n Special District 1 | 3121182\n Special District 2 | 3439516\n Permittee's Other Title | 3236862\n HIC License | 3477843\n Site Safety Mgr's Last Name | 3481861\n Site Safety Mgr's First Name | 3481885\n Site Safety Mgr Business Name |3490529\n Residential | 2139591\n Superintendent First & Last Name | 1814931\n Superintendent Business Name | 1847714\n Self_Cert | 1274022\n Permit Subtype | 1393411\n Oil Gas | 3470104\n\n\n---\n\n\nFrom the column_info sheet of file 'DD_DOB_Permit_Issuance_2018_11_02', I know that some of the column has a meaning related to blanks. For example, for the Residential column, there are either 'Yes' or 'Blanks'. So it's safe to assume that the blanks are associated with 'No'. \n\nSimilarly, to fill the blanks based on relevant information from column_info, I am using below mappings for some columns:\n\n* Residential : No\n* Site Fill : None\n* Oil Gas : None\n* Self_Cert : N\n* Act as Superintendent : N\n* Non-Profit : N\n\n\n \n\n\n\n\n\n\n",
"_____no_output_____"
]
],
[
[
"values = {'Residential': 'No','Site Fill':'NONE', 'Oil Gas':'None', 'Self_Cert':'N', 'Act as Superintendent':'N',\n 'Non-Profit':'N' }\ndataset = dataset.fillna(value= values)",
"_____no_output_____"
]
],
[
[
"Since there are many columns with blank spaces and we cannot fill the blanks with appropriate information, it's better to drop these column as they do not add value to the analysis.\n\nI will drop the following columns :\n\n* Special District 1\n* Special District 2\n* Work Type\n* Permit Subtype\n* Permittee's First Name\n* Permittee's Last Name\n* Permittee's Business Name\n* Permittee's Phone #\n* Permittee's Other Title\n* HIC License\n* Site Safety Mgr's First Name\n* Site Safety Mgr's Last Name\n* Site Safety Mgr Business Name\n* Superintendent First & Last Name\n* Superintendent Business Name\n* Owner's Business Name\n* Owner's First Name\n* Owner's Last Name\n* Owner's House #\n* Owner's House Street Name\n* Owner's Phone #\n* DOBRunDate",
"_____no_output_____"
]
],
[
[
"dataset.drop(\"Special District 1\", axis=1, inplace=True)\ndataset.drop(\"Special District 2\", axis=1, inplace=True)\ndataset.drop(\"Work Type\", axis=1, inplace=True) #since work type and permit type give same information\ndataset.drop(\"Permit Subtype\", axis=1, inplace=True)\ndataset.drop(\"Permittee's First Name\", axis=1, inplace=True)\ndataset.drop(\"Permittee's Last Name\", axis=1, inplace=True)\ndataset.drop(\"Permittee's Business Name\", axis=1, inplace=True)\ndataset.drop(\"Permittee's Phone #\", axis=1, inplace=True)\ndataset.drop(\"Permittee's Other Title\", axis=1, inplace=True) #Permit Subtype\ndataset.drop(\"HIC License\", axis=1, inplace=True)\ndataset.drop(\"Site Safety Mgr's First Name\", axis=1, inplace=True)\ndataset.drop(\"Site Safety Mgr's Last Name\", axis=1, inplace=True)\ndataset.drop(\"Site Safety Mgr Business Name\", axis=1, inplace=True)\ndataset.drop(\"Superintendent First & Last Name\", axis=1, inplace=True)\ndataset.drop(\"Superintendent Business Name\", axis=1, inplace=True)\ndataset.drop(\"Owner's Business Name\", axis=1, inplace=True)\ndataset.drop(\"Owner's First Name\", axis=1, inplace=True)\ndataset.drop(\"Owner's Last Name\", axis=1, inplace=True)\ndataset.drop(\"Owner's House #\", axis=1, inplace=True)\ndataset.drop(\"Owner's House Street Name\", axis=1, inplace=True)\ndataset.drop(\"Owner's Phone #\", axis=1, inplace=True)\ndataset.drop(\"DOBRunDate\", axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"Let's take a look at the remaining columns and their number of blanks again.",
"_____no_output_____"
]
],
[
[
"dataset.isna().sum()",
"_____no_output_____"
]
],
[
[
"We still have blanks in few columns left. One way to deal with them is to replace them with mean of that column or the most frequent entry of that column. Mean can onlly be applied to numerical columns and even for numerical columns such as Longitude and Lattitude, this mean might not be right to replace all the missing value with single longitude or lattitude. This will skew the column and would not result in fair analysis of data.\n\nSimilarly, if we use the most frequently used entry to replace all the blanks in that column, it will either skew the matrix or the data itself would not make sense. For example, the for a particular location, there is a state, city, zip code and street name associated with it. If we replace the missing entries in zip code with most frequent entry, then it might result in a data that is having a different state, city and a zip code of another location. \n\nTherefore, to clean the data I will drop the rows with NaNs. We will still have enough data for exploration.",
"_____no_output_____"
]
],
[
[
"dataset.dropna(inplace=True)\ndataset.isna().sum()",
"_____no_output_____"
]
],
[
[
"\n---\n\nNow the dataset looks clean and we can proceed with analysis. I will try find the correlation between columns using the correlation matrix.",
"_____no_output_____"
]
],
[
[
"# Correlation matrix\ndef plotCorrelationMatrix(df, graphWidth):\n filename = 'DOB_Permit_Issuance.csv'\n df = df[[col for col in df if df[col].nunique() > 1]] # keep columns where there are more than 1 unique values\n corr = df.corr()\n plt.figure(num=None, figsize=(graphWidth, graphWidth), dpi=80, facecolor='w', edgecolor='k')\n corrMat = plt.matshow(corr, fignum = 1)\n plt.xticks(range(len(corr.columns)), corr.columns, rotation=90)\n plt.yticks(range(len(corr.columns)), corr.columns)\n plt.gca().xaxis.tick_bottom()\n plt.colorbar(corrMat)\n plt.title(f'Correlation Matrix for {filename}', fontsize=15)\n plt.show()",
"_____no_output_____"
],
[
"plotCorrelationMatrix(dataset, 15)",
"_____no_output_____"
]
],
[
[
"We can see that there is strong positive relationship between :\n\n* Zip Code and Job #\n* Job # and Council_District\n* Zip Code and Council_District\n",
"_____no_output_____"
],
[
"---\n\n\nTo get more insight of the data and its column-wise data distribution, I will plot the columns using bar graphs. For displaying purposes, I will pick columns that have between 1 and 50 unique values.",
"_____no_output_____"
]
],
[
[
"def plotColumns(dataframe, nGraphShown, nGraphPerRow):\n nunique = dataframe.nunique()\n dataframe = dataframe[[col for col in dataframe if nunique[col] > 1 and nunique[col] < 50]]\n nRow, nCol = dataframe.shape\n columnNames = list(dataframe)\n nGraphRow = (nCol + nGraphPerRow - 1) / nGraphPerRow\n plt.figure(num = None, figsize = (6 * nGraphPerRow, 8 * nGraphRow), dpi = 80, facecolor = 'w', edgecolor = 'k')\n for i in range(min(nCol, nGraphShown)):\n plt.subplot(nGraphRow, nGraphPerRow, i + 1)\n columndataframe = dataframe.iloc[:, i]\n if (not np.issubdtype(type(columndataframe.iloc[0]), np.number)):\n valueCounts = columndataframe.value_counts()\n valueCounts.plot.bar()\n else:\n columndataframe.hist()\n plt.ylabel('counts')\n plt.xticks(rotation = 90)\n plt.title(f'{columnNames[i]} (column {i})')\n plt.tight_layout(pad = 1.0, w_pad = 1.0, h_pad = 1.0)\n plt.show()",
"_____no_output_____"
],
[
"plotColumns(dataset, 38, 3)",
"_____no_output_____"
]
],
[
[
"From the Borough graph, it's evident that the Manhattan has highest number of filing for the permit. Then the second popuar borough after manhattan is Brooklyn with huge margin. Then comes the Queens with almost same number of permits as Brooklyn. We see another plunge in permit numbers with Bronx and Staten Island.\n\n---\n\nJob Document number is the number of documents that were added with the file during the application. Mostly all the filings required a single document. There are some permits that had two documents and then higher number of documents are negligible.\n\n---\n\nThere is a pattern in Job Type as well. We can see that the most populat Job Type or Work Type is A2 with more than 1.75 M permits. The second most popular work type is NB (new building) with around 400,000 permits. The number of permits decreases with A3, A1, DM and SG where DM and SG are significantly less than other types.\n\n---\n\nSelf_Cert\tindicates whether or not the application was submitted as Professionally Certified. A Professional Engineer (PE) or Registered Architect (RA) can certify compliance with applicable laws and codes on applications filed by him/her as applicant. Plot shows mostly were not filed by Professional Engineer or Registered Architect. \n\n---\n\nBldg Type indicates\tlegal occupancy classification. The most popular type of building type is '2' with more than 2M permits.\n\n---\n\nMost of the buildings are non Residential and only about 1.3M buildinngs were residential.\n\n---\n\nPermit Status\tindicates the current status of the permit application. Corresponding plot for the column suggests that most of the permits are 'Issued' and very small number were 'Reissued'. The 'In-Progress' and 'Revoked' are negligible. \n\n---\nFiling Status\tindicates if this is the first time the permit is being applied for or if the permit is being renewed. A large amount of permits were in 'Initial' status and less than half of that were in 'Renewal' state.\n\n---\n\nPermit Type\tThe specific type of work covered by the permit.\tThis is a two character code to indicate the type of work. There are 7 types of permits where EW has the highest number. The number indicates decreasing trend with PL, EQ, AL, NB, SG, and FO. \n\n---\n\nA sequential number assigned to each issuance of the particular permit from initial issuance to each subsequent renewal. Every initial permit should have a 01 sequence number. Every additional renewal receives a number that increases by 1 (ex: 02, 03, 04). Most of the permits have less than 5 sequence number.\n\n---\n\nIf the permit is for work on fuel burning equipment, this indicates whether it burns oil or gas. Most of the permits are for neither Oil nor Gas. A very small fraction of permits is for Oil and there is negligible number of permits for Gas.\n\n---\n\nSite Fill indicates the source of any fill dirt that will be used on the construction site. When over 300 cubic yards of fill is being used, the Department is required to inform Sanitation of where the fill is coming from and the amount. About 1.1M entries didn't mention any Site Fill type indicating that the less than 300 cubic yards of fill is being used. Almost permits are not applicable. About 300,000 permits were for on-site fill and less than 100,000 were for off-site.\n\n\n---\nProfessional license type of the person that the permit was issued to. In most of the cases the person was holding GC type. Then the number showcased a decreasing trend with MP, FS, OB, SI, NW, OW, PE, RA, and HI where NW, OW, PE, RA, and HI are negligible in number.\n\n---\n\nAct as Superintendent indicates if the permittee acts as the Construction Superintendent for the work site. Only about 1.1M people responded 'Yes' to this and majority respondded with 'No'\n\n\n---\n\nOwner's Business Type\tindicates the type of entity that owns the building where the work will be performed. Mostly the entities owning the builing were 'Corporations'. With slightly less than 'Corporation', 'Individual' type stands at the second position and 'Partnership' holds the third position. Other business types like are less significant in number.\n\n\n\n---\n\nNon-Profit indicates if the building is owned by a non-profit. Less than 250,000 buildings were owned by 'Non-Profit' and more than 2.75M were not 'Non-Profit'.\n\n\n\n\n",
"_____no_output_____"
],
[
"# Borough-wise analysis\nLet's now dive deeper in the data and take a closer look. Is the trend we see above is same across all Boroughs ?. Does every borough have same type of building? Is 'EW' the most popular permit type across all cities? What about the 'owner's business type' and 'Job Type' ? I will try to find the answers to all these by exploring the pattern in each bouough.",
"_____no_output_____"
],
[
"### Bldg Type in each borough",
"_____no_output_____"
]
],
[
[
"manhattan_bldg_type = dataset[dataset.BOROUGH == 'MANHATTAN'][['Bldg Type']]\nmanhattan_bldg_type.reset_index(drop=True, inplace=True)\n\nbrooklyn_bldg_type = dataset[dataset.BOROUGH == 'BROOKLYN'][['Bldg Type']]\nbrooklyn_bldg_type.reset_index(drop=True, inplace=True)\n\nbronx_bldg_type = dataset[dataset.BOROUGH == 'BRONX'][['Bldg Type']]\nbronx_bldg_type.reset_index(drop=True, inplace=True)\n\nqueens_bldg_type = dataset[dataset.BOROUGH == 'QUEENS'][['Bldg Type']]\nqueens_bldg_type.reset_index(drop=True, inplace=True)\n\nstaten_island_bldg_type = dataset[dataset.BOROUGH == 'STATEN ISLAND'][['Bldg Type']]\nstaten_island_bldg_type.reset_index(drop=True, inplace=True)\n\nbuilding_type = pd.DataFrame()\nbuilding_type['manhattan_bldg_type'] = manhattan_bldg_type #brooklyn_bldg_type\nbuilding_type['brooklyn_bldg_type'] = brooklyn_bldg_type \nbuilding_type['bronx_bldg_type'] = bronx_bldg_type \nbuilding_type['queens_bldg_type'] = queens_bldg_type \nbuilding_type['staten_island_bldg_type'] = staten_island_bldg_type ",
"_____no_output_____"
],
[
"plotColumns(building_type, 5, 3)",
"_____no_output_____"
]
],
[
[
"**Analysis**\n\nThe builing type is either '1' or '2' and we earlier discovered that type '2' were significantly popular as compared to type '1'. However, this is not true for all the Boroughs. For manhattan, this trend still holds true. But for other locations, the type '1' buildings are comparable in number with that of type '2'. More interestingly, in case of Staten Island, the type '1' is significantly popular beating type '2' with a good margin. ",
"_____no_output_____"
],
[
"### Permit Type in Each Borough",
"_____no_output_____"
]
],
[
[
"manhattan_permit_type = dataset[dataset.BOROUGH == 'MANHATTAN'][['Permit Type']]\nmanhattan_permit_type.reset_index(drop=True, inplace=True)\n\nbrooklyn_permit_type = dataset[dataset.BOROUGH == 'BROOKLYN'][['Permit Type']]\nbrooklyn_permit_type.reset_index(drop=True, inplace=True)\n\nbronx_permit_type = dataset[dataset.BOROUGH == 'BRONX'][['Permit Type']]\nbronx_permit_type.reset_index(drop=True, inplace=True)\n\nqueens_permit_type = dataset[dataset.BOROUGH == 'QUEENS'][['Permit Type']]\nqueens_permit_type.reset_index(drop=True, inplace=True)\n\nstaten_island_permit_type = dataset[dataset.BOROUGH == 'STATEN ISLAND'][['Permit Type']]\nstaten_island_permit_type.reset_index(drop=True, inplace=True)\n\npermit_type = pd.DataFrame()\npermit_type['manhattan_permit_type'] = manhattan_permit_type #brooklyn_permit_type\npermit_type['brooklyn_permit_type'] = brooklyn_permit_type \npermit_type['bronx_permit_type'] = bronx_permit_type \npermit_type['queens_permit_type'] = queens_permit_type \npermit_type['staten_island_permit_type'] = staten_island_permit_type ",
"_____no_output_____"
],
[
"plotColumns(permit_type, 5, 3)",
"_____no_output_____"
]
],
[
[
"**Analysis**\n\nIn Permit Type we earlier discovered that type 'EW' was the most popular and significantly higher in number as compared to other types. However, this is true for most of the Boroughs except Staten Island. However, other types of Permit are shuffled in each Borough. Below is the type of permits in decreasing order for each borough.\n\nManhattan | Brooklyn | Queens |Bronx | Staten Island \n------------|--------|--------------|-------------\nEW | EW | EW| EW | PL \nPL | PL | PL | PL |NB \nEQ | EQ | EQ |EQ |EW \nAL | AL | NB | AL | EQ\nSQ | NB | AL | SQ | AL\nNB| FO| SG |FO | DM\nFO|DM|FO| DM| SG\nDM|SG|DM|SG|FO",
"_____no_output_____"
],
[
"### Owner's Business Type in Each Borough",
"_____no_output_____"
]
],
[
[
"manhattan_owners_business_type = dataset[dataset.BOROUGH == 'MANHATTAN'][['Owner\\'s Business Type']]\nmanhattan_owners_business_type.reset_index(drop=True, inplace=True)\n\nbrooklyn_owners_business_type = dataset[dataset.BOROUGH == 'BROOKLYN'][['Owner\\'s Business Type']]\nbrooklyn_owners_business_type.reset_index(drop=True, inplace=True)\n\nbronx_owners_business_type = dataset[dataset.BOROUGH == 'BRONX'][['Owner\\'s Business Type']]\nbronx_owners_business_type.reset_index(drop=True, inplace=True)\n\nqueens_owners_business_type = dataset[dataset.BOROUGH == 'QUEENS'][['Owner\\'s Business Type']]\nqueens_owners_business_type.reset_index(drop=True, inplace=True)\n\nstaten_island_owners_business_type = dataset[dataset.BOROUGH == 'STATEN ISLAND'][['Owner\\'s Business Type']]\nstaten_island_owners_business_type.reset_index(drop=True, inplace=True)\n\nowners_business_type = pd.DataFrame()\nowners_business_type['manhattan_owners_business_type'] = manhattan_owners_business_type #brooklyn_owners_business_type\nowners_business_type['brooklyn_owners_business_type'] = brooklyn_owners_business_type \nowners_business_type['bronx_owners_business_type'] = bronx_owners_business_type \nowners_business_type['queens_owners_business_type'] = queens_owners_business_type \nowners_business_type['staten_island_owners_business_type'] = staten_island_owners_business_type ",
"_____no_output_____"
],
[
"plotColumns(owners_business_type, 5, 3)",
"_____no_output_____"
]
],
[
[
"**Analysis**\n\nWe earlier discovered that the 'Corporation' was the most popular 'Owner's Business type' and 'Individual' type was closely competing with it. Taking a closer look at each borough reveals that the trend highly varies in all the Boroughs. In Manhattan, the 'Corporation' is still the highest but the 'Individual' is substituted by 'Partnership'. In Brooklyn, 'Individual' holds the top place and 'Corporation' and 'Partnership' are on second and third place respectively.\n\nIn Bronx, the 'Corporation' and 'Individual' are closely competing while 'Corporation' holds the highest number and 'Partnership' is on third place.\n\nFor Queens and Staten Island, the 'Individual' holds the top place and 'Corporation' and 'Partnership' are on second and third place respectively. This is consistent with the trend observed in 'Brooklyn'.",
"_____no_output_____"
],
[
"### Job Type in Each Borough",
"_____no_output_____"
]
],
[
[
"manhattan_job_type = dataset[dataset.BOROUGH == 'MANHATTAN'][['Job Type']]\nmanhattan_job_type.reset_index(drop=True, inplace=True)\n\nbrooklyn_job_type = dataset[dataset.BOROUGH == 'BROOKLYN'][['Job Type']]\nbrooklyn_job_type.reset_index(drop=True, inplace=True)\n\nbronx_job_type = dataset[dataset.BOROUGH == 'BRONX'][['Job Type']]\nbronx_job_type.reset_index(drop=True, inplace=True)\n\nqueens_job_type = dataset[dataset.BOROUGH == 'QUEENS'][['Job Type']]\nqueens_job_type.reset_index(drop=True, inplace=True)\n\nstaten_island_job_type = dataset[dataset.BOROUGH == 'STATEN ISLAND'][['Job Type']]\nstaten_island_job_type.reset_index(drop=True, inplace=True)\n\njob_type = pd.DataFrame()\njob_type['manhattan_job_type'] = manhattan_job_type #brooklyn_job_type\njob_type['brooklyn_job_type'] = brooklyn_job_type \njob_type['bronx_job_type'] = bronx_job_type \njob_type['queens_job_type'] = queens_job_type \njob_type['staten_island_job_type'] = staten_island_job_type ",
"_____no_output_____"
],
[
"plotColumns(job_type, 5, 3)",
"_____no_output_____"
]
],
[
[
"**Analysis**\n\nWe earlier discovered that the 'A2' was the most popular 'Job type' and numbers showed a decreasing trend with 'NB', 'A3', 'A1', 'DM', and 'SG'. Taking a closer look at each borough revealsa slightly different trend. For example, in Manhattan, the 'A2' is still the highest but the 'NB' is pushed beyond 'A1' to the fourth place while in Staten Island 'NB' holds the first place. Below is the type of Jobs in decreasing order for each borough. \n\nOverall | Manhattan | Brooklyn | Queens |Bronx | Staten Island \n------------|--------|--------------|-------------\nA2|A2|A2|A2|A2|NB\nNB|A3|NB|NB|NB|A2\nA3|A1|A1|A3|A1|A1\nA1|NB|A3|A1|A3|A3\nDM|SG|DM|SG|DM|DM\nSG|DM|SG|DM|SG|SG",
"_____no_output_____"
],
[
"# Permits Per Years",
"_____no_output_____"
],
[
"### Is there is trend in the number of permits issued each year? Let's find out!",
"_____no_output_____"
],
[
"First, the date format of 'Issuance Date' needs to be converted to python Datetime format and then only the year needs to be extracted from the date.",
"_____no_output_____"
]
],
[
[
"dataset['Issuance Date'] = pd.to_datetime(dataset['Issuance Date'])\ndataset['Issuance Date'] =dataset['Issuance Date'].dt.year",
"_____no_output_____"
]
],
[
[
"Once the dates are replaced by the corresponding years, we can plot the graph.",
"_____no_output_____"
]
],
[
[
"timeline = dataset['Issuance Date'].value_counts(ascending=True)\ntimeline = timeline.sort_index()",
"_____no_output_____"
],
[
"timeline.to_frame()",
"_____no_output_____"
],
[
"timeline.plot.bar()",
"_____no_output_____"
]
],
[
[
"**Analysis**\n\nWe can observe that the number of permits has been consistently increasing each year. The number got stagnant from 1993 to 1996 and then it increased exponentially from 1997 until 2007. The applications decreasedfor a couple of years and then rose exponentially from 2010 to 2017.",
"_____no_output_____"
],
[
"### Borough-Wise Analysis of Timeline",
"_____no_output_____"
],
[
"Whether the trend is consistent across all borough? Is there a time when construction slowed down in a city and surged in other ?",
"_____no_output_____"
]
],
[
[
"manhattan_permits_issued = dataset[dataset.BOROUGH == 'MANHATTAN'][['Issuance Date']]\nmanhattan_permits_issued.reset_index(drop=True, inplace=True)\n\nbrooklyn_permits_issued = dataset[dataset.BOROUGH == 'BROOKLYN'][['Issuance Date']]\nbrooklyn_permits_issued.reset_index(drop=True, inplace=True)\n\nbronx_permits_issued = dataset[dataset.BOROUGH == 'BRONX'][['Issuance Date']]\nbronx_permits_issued.reset_index(drop=True, inplace=True)\n\nqueens_permits_issued = dataset[dataset.BOROUGH == 'QUEENS'][['Issuance Date']]\nqueens_permits_issued.reset_index(drop=True, inplace=True)\n\nstaten_island_permits_issued = dataset[dataset.BOROUGH == 'STATEN ISLAND'][['Issuance Date']]\nstaten_island_permits_issued.reset_index(drop=True, inplace=True)\n\npermits_issued = pd.DataFrame()\npermits_issued['manhattan_permits_issued'] = manhattan_permits_issued #brooklyn_permits_issued\npermits_issued['brooklyn_permits_issued'] = brooklyn_permits_issued \npermits_issued['bronx_permits_issued'] = bronx_permits_issued \npermits_issued['queens_permits_issued'] = queens_permits_issued \npermits_issued['staten_island_permits_issued'] = staten_island_permits_issued ",
"_____no_output_____"
],
[
"plotColumns(permits_issued, 5, 3)",
"_____no_output_____"
]
],
[
[
"In Manhattan and Brooklyn, most number of applications were filed during 2010 and 2015 period.\n\nBronx and Queens observed the highest number of filings in recent years from 2015 to 2019. \n\nStaten Island had most applications during 2000 to 2015 and then there is again a surge in applications around 2015 but the number of applications is very less compared to its counterparts.",
"_____no_output_____"
],
[
"# Permits per month",
"_____no_output_____"
],
[
"### Is there is trend in the number of permits filed every month? Let's explore!",
"_____no_output_____"
],
[
"First, the date format of 'Filing Date' needs to be converted to python Datetime format and then only the month needs to be extracted from the date.",
"_____no_output_____"
]
],
[
[
"dataset['Filing Date'] = pd.to_datetime(dataset['Filing Date'])\ndataset['Filing Date'] =dataset['Filing Date'].dt.month",
"_____no_output_____"
],
[
"months = dataset['Filing Date'].value_counts()\nmonths.to_frame()\nmonths = months.sort_index()\nplotColumns(months, 1, 1)",
"_____no_output_____"
]
],
[
[
"Mostly all the months gets equal number of permit applications. February has less days, so that justifies the less number. \n\nOverall, Month of 'March' has highest permit filings. ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
d094b08c24514a2d6772bf6fd1de8a829cc7b804 | 85,227 | ipynb | Jupyter Notebook | labs/lab07.ipynb | Dhannai/MAT281_portfolio | 3ac902e00f8fc519cafc762b214c8e89a3741c6f | [
"MIT"
] | null | null | null | labs/lab07.ipynb | Dhannai/MAT281_portfolio | 3ac902e00f8fc519cafc762b214c8e89a3741c6f | [
"MIT"
] | null | null | null | labs/lab07.ipynb | Dhannai/MAT281_portfolio | 3ac902e00f8fc519cafc762b214c8e89a3741c6f | [
"MIT"
] | null | null | null | 149.783831 | 68,248 | 0.876307 | [
[
[
"# Laboratorio 7",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport altair as alt\n\nfrom sklearn import datasets, linear_model\nfrom sklearn.metrics import mean_squared_error, r2_score\n\nalt.themes.enable('opaque')\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"En este laboratorio utilizaremos los mismos datos de diabetes vistos en la clase",
"_____no_output_____"
]
],
[
[
"diabetes_X, diabetes_y = datasets.load_diabetes(return_X_y=True, as_frame=True)\ndiabetes = pd.concat([diabetes_X, diabetes_y], axis=1)\ndiabetes.head()",
"_____no_output_____"
]
],
[
[
"## Pregunta 1\n\n(1 pto)\n\n* ¿Por qué la columna de sexo tiene esos valores?\n* ¿Cuál es la columna a predecir?\n* ¿Crees que es necesario escalar o transformar los datos antes de comenzar el modelamiento?",
"_____no_output_____"
],
[
"__Respuesta:__\n\n* Porque los valores se encuentran escalados y centrados por su desviación estandar multiplicada por la cantidad de muestras, entonces tienen una norma unitaria.\n* La columna `target`\n* Sí, para que el algoritmo los pueda reconocer más fácil y el usuario pueda comparar de manera más simple, ya que los datos vienen con escalas y magnitudes distintas.",
"_____no_output_____"
],
[
"## Pregunta 2\n\n(1 pto)\n\nRealiza dos regresiones lineales con todas las _features_, el primer caso incluyendo intercepto y el segundo sin intercepto. Luego obtén la predicción para así calcular el error cuadrático medio y coeficiente de determinación de cada uno de ellos.",
"_____no_output_____"
]
],
[
[
"regr_with_incerpet = linear_model.LinearRegression(fit_intercept=True)\nregr_with_incerpet.fit(diabetes_X, diabetes_y)",
"_____no_output_____"
],
[
"diabetes_y_pred_with_intercept = regr_with_incerpet.predict(diabetes_X)",
"_____no_output_____"
],
[
"# Coeficientes\nprint(f\"Coefficients: \\n{regr_with_incerpet.coef_}\\n\")\n# Intercepto\nprint(f\"Intercept: \\n{regr_with_incerpet.intercept_}\\n\")\n# Error cuadrático medio\nprint(f\"Mean squared error: {mean_squared_error(diabetes_y, diabetes_y_pred_with_intercept):.2f}\\n\")\n# Coeficiente de determinación\nprint(f\"Coefficient of determination: {r2_score(diabetes_y, diabetes_y_pred_with_intercept):.2f}\")",
"Coefficients: \n[ -10.01219782 -239.81908937 519.83978679 324.39042769 -792.18416163\n 476.74583782 101.04457032 177.06417623 751.27932109 67.62538639]\n\nIntercept: \n152.1334841628965\n\nMean squared error: 2859.69\n\nCoefficient of determination: 0.52\n"
],
[
"regr_without_incerpet = linear_model.LinearRegression(fit_intercept=False)\nregr_without_incerpet.fit(diabetes_X, diabetes_y)",
"_____no_output_____"
],
[
"diabetes_y_pred_without_intercept = regr_without_incerpet.predict(diabetes_X)",
"_____no_output_____"
],
[
"# Coeficientes\nprint(f\"Coefficients: \\n{regr_without_incerpet.coef_}\\n\")\n# Error cuadrático medio\nprint(f\"Mean squared error: {mean_squared_error(diabetes_y, diabetes_y_pred_without_intercept):.2f}\\n\")\n# Coeficiente de determinación\nprint(f\"Coefficient of determination: {r2_score(diabetes_y, diabetes_y_pred_without_intercept):.2f}\")",
"Coefficients: \n[ -10.01219782 -239.81908937 519.83978679 324.39042769 -792.18416163\n 476.74583782 101.04457032 177.06417623 751.27932109 67.62538639]\n\nMean squared error: 26004.29\n\nCoefficient of determination: -3.39\n"
]
],
[
[
"**Pregunta: ¿Qué tan bueno fue el ajuste del modelo?**",
"_____no_output_____"
],
[
"__Respuesta:__\nNo es muy bueno, ya que el coeficiente de determinación es 0.52 para el caso con intercepto, y para que sea una buena aproximación debería ser cercano a 1. Para el caso sin intercepto el R2 es negativo por lo que es mejor utilizar el promedio de los datos, ya que estos entregarían un mejor ajuste, además se debe considerar que el error es mucho mayor que para el caso con intercepto, entonces tampoco es un buen ajuste.",
"_____no_output_____"
],
[
"## Pregunta 3\n\n(1 pto)\n\nRealizar multiples regresiones lineales utilizando una sola _feature_ a la vez. \n\nEn cada iteración:\n\n- Crea un arreglo `X`con solo una feature filtrando `X`.\n- Crea un modelo de regresión lineal con intercepto.\n- Ajusta el modelo anterior.\n- Genera una predicción con el modelo.\n- Calcula e imprime las métricas de la pregunta anterior.",
"_____no_output_____"
]
],
[
[
"for col in diabetes_X.columns:\n X_i = diabetes_X[col].to_frame()\n regr_i = linear_model.LinearRegression(fit_intercept=True)\n regr_i.fit(X_i,diabetes_y)\n diabetes_y_pred_i = regr_i.predict(X_i)\n print(f\"Feature: {col}\")\n print(f\"\\tCoefficients: {regr_i.coef_}\")\n print(f\"\\tIntercept: {regr_i.intercept_}\")\n print(f\"\\tMean squared error: {mean_squared_error(diabetes_y, diabetes_y_pred_i):.2f}\")\n print(f\"\\tCoefficient of determination: {r2_score(diabetes_y, diabetes_y_pred_i):.2f}\\n\")",
"Feature: age\n\tCoefficients: [304.18307453]\n\tIntercept: 152.13348416289605\n\tMean squared error: 5720.55\n\tCoefficient of determination: 0.04\n\nFeature: sex\n\tCoefficients: [69.71535568]\n\tIntercept: 152.13348416289594\n\tMean squared error: 5918.89\n\tCoefficient of determination: 0.00\n\nFeature: bmi\n\tCoefficients: [949.43526038]\n\tIntercept: 152.1334841628967\n\tMean squared error: 3890.46\n\tCoefficient of determination: 0.34\n\nFeature: bp\n\tCoefficients: [714.7416437]\n\tIntercept: 152.13348416289585\n\tMean squared error: 4774.10\n\tCoefficient of determination: 0.19\n\nFeature: s1\n\tCoefficients: [343.25445189]\n\tIntercept: 152.13348416289597\n\tMean squared error: 5663.32\n\tCoefficient of determination: 0.04\n\nFeature: s2\n\tCoefficients: [281.78459335]\n\tIntercept: 152.1334841628959\n\tMean squared error: 5750.24\n\tCoefficient of determination: 0.03\n\nFeature: s3\n\tCoefficients: [-639.14527932]\n\tIntercept: 152.13348416289566\n\tMean squared error: 5005.66\n\tCoefficient of determination: 0.16\n\nFeature: s4\n\tCoefficients: [696.88303009]\n\tIntercept: 152.13348416289568\n\tMean squared error: 4831.14\n\tCoefficient of determination: 0.19\n\nFeature: s5\n\tCoefficients: [916.13872282]\n\tIntercept: 152.13348416289628\n\tMean squared error: 4030.99\n\tCoefficient of determination: 0.32\n\nFeature: s6\n\tCoefficients: [619.22282068]\n\tIntercept: 152.13348416289614\n\tMean squared error: 5062.38\n\tCoefficient of determination: 0.15\n\n"
]
],
[
[
"**Pregunta: Si tuvieras que escoger una sola _feauture_, ¿Cuál sería? ¿Por qué?**",
"_____no_output_____"
],
[
"**Respuesta:** Escogería la Feature bmi, porque es la que tiene le menor error y el coeficiente de determinación más cercano a 1.",
"_____no_output_____"
],
[
"## Ejercicio 4\n\n(1 pto)\n\nCon la feature escogida en el ejercicio 3 realiza el siguiente gráfico:\n\n- Scatter Plot\n- Eje X: Valores de la feature escogida.\n- Eje Y: Valores de la columna a predecir (target).\n- En color rojo dibuja la recta correspondiente a la regresión lineal (utilizando `intercept_`y `coefs_`).\n- Coloca un título adecuado, nombre de los ejes, etc.\n\nPuedes utilizar `matplotlib` o `altair`, el que prefieras.",
"_____no_output_____"
]
],
[
[
"regr = linear_model.LinearRegression(fit_intercept=True).fit(diabetes_X['bmi'].to_frame(), diabetes_y)\ndiabetes_y_pred_bmi = regr.predict(diabetes_X['bmi'].to_frame()) \n",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n%matplotlib inline\nfrom matplotlib import rcParams\nrcParams['font.family'] = 'serif'\nrcParams['font.size'] = 16",
"_____no_output_____"
],
[
"x = diabetes_X['bmi'].values\ny = diabetes_y\nx_reg = np.arange(-0.1, 0.2, 0.01)\ny_reg = regr.intercept_ + regr.coef_ * x_reg\n\nfig = plt.figure(figsize=(20,8))\nfig.suptitle('Regresión lineal body mass index (bmi) versus progresión de la enfermedad (target)')\nax = fig.add_subplot()\nax.figsize=(20,8)\nax.scatter(x,y,c='k');\nax.plot(x_reg,y_reg,c='r');\nax.legend([\"Regresión\", \"Datos \"]);\nax.set_xlabel('Body Mass Index')\nax.set_ylabel('target');\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
]
] |
d094b8809270ab324de585c88a5ec60d1b011678 | 85,400 | ipynb | Jupyter Notebook | Geolocation/Notebooks/Des1Des2OverheadComp.ipynb | radical-experiments/iceberg_escience | e5c230a23395a71a4adf554730ea3d77f923166c | [
"MIT"
] | 1 | 2019-05-24T02:19:29.000Z | 2019-05-24T02:19:29.000Z | Geolocation/Notebooks/Des1Des2OverheadComp.ipynb | radical-experiments/iceberg_seals | e5c230a23395a71a4adf554730ea3d77f923166c | [
"MIT"
] | null | null | null | Geolocation/Notebooks/Des1Des2OverheadComp.ipynb | radical-experiments/iceberg_seals | e5c230a23395a71a4adf554730ea3d77f923166c | [
"MIT"
] | null | null | null | 148.780488 | 30,680 | 0.861698 | [
[
[
"import radical.analytics as ra\nimport radical.pilot as rp\nimport radical.utils as ru\nimport radical.entk as re\nimport os\nfrom glob import glob\nimport numpy as np\nfrom matplotlib import pyplot as plt\nfrom matplotlib import cm\nimport csv\nimport pandas as pd\nimport json\nimport matplotlib as mpl\nmpl.rcParams['text.usetex'] = True\nmpl.rcParams['text.latex.unicode'] = True\n\nblues = cm.get_cmap(plt.get_cmap('Blues'))\ngreens = cm.get_cmap(plt.get_cmap('Greens'))\nreds = cm.get_cmap(plt.get_cmap('Reds'))\noranges = cm.get_cmap(plt.get_cmap('Oranges'))\npurples = cm.get_cmap(plt.get_cmap('Purples'))\ngreys = cm.get_cmap(plt.get_cmap('Greys'))\n\nfrom IPython.core.display import display, HTML\ndisplay(HTML(\"<style>.container { width:100% !important; }</style>\"))\nimport warnings\nwarnings.filterwarnings('ignore')\n!radical-stack",
"_____no_output_____"
]
],
[
[
"## Design 1",
"_____no_output_____"
]
],
[
[
"!tar -xzvf ../Data/Design1/entk.session-design1-54875/entk.session-design1-54875.tar.gz -C ../Data/Design1/entk.session-design1-54875/\n!tar -xzvf ../Data/Design1/entk.session-design1-54875/entk_workflow.tar.gz -C ../Data/Design1/entk.session-design1-54875/",
"x entk.session-design1-54875.json\nx entk_workflow.json\n"
],
[
"des1DF = pd.DataFrame(columns=['TTX','AgentOverhead','ClientOverhead','EnTKOverhead'])\nwork_file = open('../Data/Design1/entk.session-design1-54875/entk_workflow.json')\nwork_json = json.load(work_file)\nwork_file.close()\n\nworkflow = work_json['workflows'][1]\nunit_ids = list()\nfor pipe in workflow['pipes']:\n unit_path = pipe['stages'][1]['tasks'][0]['path']\n unit_id = unit_path.split('/')[-2]\n if unit_id != 'unit.000000':\n unit_ids.append(unit_id)\n\nsids=['entk.session-design1-54875']\nfor sid in sids:\n re_session = ra.Session(stype='radical.entk',src='../Data/Design1',sid=sid)\n rp_session = ra.Session(stype='radical.pilot',src='../Data/Design1/'+sid)\n units = rp_session.filter(etype='unit', inplace=False, uid=unit_ids)\n pilot = rp_session.filter(etype='pilot', inplace=False)\n units_duration = units.duration(event=[{ru.EVENT: 'exec_start'},{ru.EVENT: 'exec_stop'}])\n units_agent = units.duration (event=[{ru.EVENT: 'state',ru.STATE: rp.AGENT_STAGING_INPUT},{ru.EVENT: 'staging_uprof_stop'}])\n all_units = rp_session.filter(etype='unit', inplace=False)\n disc_unit = rp_session.filter(etype='unit', inplace=False, uid='unit.000000')\n disc_time = disc_unit.duration([rp.NEW, rp.DONE])\n units_client = units.duration([rp.NEW, rp.DONE])\n appmanager = re_session.filter(etype='appmanager',inplace=False)\n t_p2 = pilot.duration(event=[{ru.EVENT: 'bootstrap_0_start'}, {ru.EVENT: 'cmd'}])\n resource_manager = re_session.filter(etype='resource_manager',inplace=False)\n app_duration = appmanager.duration(event=[{ru.EVENT:\"amgr run started\"},{ru.EVENT:\"termination done\"}])\n res_duration = resource_manager.duration(event=[{ru.EVENT:\"rreq submitted\"},{ru.EVENT:\"resource active\"}])\n ttx = units_duration\n agent_overhead = abs(units_agent - units_duration)\n client_overhead = units_client - units_agent\n entk_overhead = app_duration - units_client - res_duration - all_units.duration(event=[{ru.EVENT: 'exec_start'},{ru.EVENT: 'exec_stop'}]) + ttx\n des1DF.loc[len(des1DF)] = [ttx, agent_overhead, client_overhead, entk_overhead]",
"_____no_output_____"
],
[
"print(des1DF)",
" TTX AgentOverhead ClientOverhead EnTKOverhead\n0 11717.529445 978.254925 45306.359832 12857.508548\n"
]
],
[
[
"## Design 2",
"_____no_output_____"
]
],
[
[
"des2DF = pd.DataFrame(columns=['TTX','SetupOverhead','SetupOverhead2','AgentOverhead','ClientOverhead'])\nsids = ['design2_11K_run5'] \nfor sid in sids:\n Node1 = pd.DataFrame(columns=['Start','End','Type'])\n Node1Tilling = pd.read_csv('../Data/Design2/'+sid+'/pilot.0000/unit.000002/geolocate1.csv')\n for index,row in Node1Tilling.iterrows():\n Node1.loc[len(Node1)] = [row['Start'],row['End'],'Geo1']\n Node1Tilling = pd.read_csv('../Data/Design2/'+sid+'/pilot.0000/unit.000002/geolocate2.csv')\n for index,row in Node1Tilling.iterrows():\n Node1.loc[len(Node1)] = [row['Start'],row['End'],'Geo2']\n Node1Tilling = pd.read_csv('../Data/Design2/'+sid+'/pilot.0000/unit.000002/ransac1.csv')\n for index,row in Node1Tilling.iterrows():\n Node1.loc[len(Node1)] = [row['Start'],row['End'],'Ransac1']\n Node2 = pd.DataFrame(columns=['Start','End','Type'])\n Node2Tilling = pd.read_csv('../Data/Design2/'+sid+'/pilot.0000/unit.000003/geolocate3.csv')\n for index,row in Node2Tilling.iterrows():\n Node2.loc[len(Node2)] = [row['Start'],row['End'],'Geo3']\n Node2Tilling = pd.read_csv('../Data/Design2/'+sid+'/pilot.0000/unit.000003/geolocate4.csv')\n for index,row in Node2Tilling.iterrows():\n Node2.loc[len(Node2)] = [row['Start'],row['End'],'Geo4']\n Node2Tilling = pd.read_csv('../Data/Design2/'+sid+'/pilot.0000/unit.000003/ransac2.csv')\n for index,row in Node2Tilling.iterrows():\n Node2.loc[len(Node2)] = [row['Start'],row['End'],'Ransac2']\n Node3 = pd.DataFrame(columns=['Start','End','Type'])\n Node3Tilling = pd.read_csv('../Data/Design2/'+sid+'/pilot.0000/unit.000004/geolocate5.csv')\n for index,row in Node3Tilling.iterrows():\n Node3.loc[len(Node3)] = [row['Start'],row['End'],'Geo5']\n Node3Tilling = pd.read_csv('../Data/Design2/'+sid+'/pilot.0000/unit.000004/geolocate6.csv')\n for index,row in Node3Tilling.iterrows():\n Node3.loc[len(Node3)] = [row['Start'],row['End'],'Geo6']\n Node3Tilling = pd.read_csv('../Data/Design2/'+sid+'/pilot.0000/unit.000004/ransac3.csv')\n for index,row in Node3Tilling.iterrows():\n Node3.loc[len(Node3)] = [row['Start'],row['End'],'Ransac3']\n Node4 = pd.DataFrame(columns=['Start','End','Type'])\n Node4Tilling = pd.read_csv('../Data/Design2/'+sid+'/pilot.0000/unit.000005/geolocate7.csv')\n for index,row in Node4Tilling.iterrows():\n Node4.loc[len(Node4)] = [row['Start'],row['End'],'Geo7']\n Node4Tilling = pd.read_csv('../Data/Design2/'+sid+'/pilot.0000/unit.000005/geolocate8.csv')\n for index,row in Node4Tilling.iterrows():\n Node4.loc[len(Node4)] = [row['Start'],row['End'],'Geo8']\n Node4Tilling = pd.read_csv('../Data/Design2/'+sid+'/pilot.0000/unit.000005/ransac4.csv')\n for index,row in Node4Tilling.iterrows():\n Node4.loc[len(Node4)] = [row['Start'],row['End'],'Ransac4']\n AllNodes = pd.DataFrame(columns=['Start','End','Type'])\n AllNodes = AllNodes.append(Node1)\n AllNodes = AllNodes.append(Node2)\n AllNodes = AllNodes.append(Node3)\n AllNodes = AllNodes.append(Node4)\n AllNodes.reset_index(inplace=True,drop='index')\n rp_sessionDes2 = ra.Session(stype='radical.pilot',src='../Data/Design2/'+sid)\n unitsDes2 = rp_sessionDes2.filter(etype='unit', inplace=False)\n execUnits = unitsDes2.filter(uid=['unit.000002','unit.000003','unit.000004','unit.000005'],inplace=False)\n exec_units_setup_des2 = execUnits.duration(event=[{ru.EVENT: 'exec_start'},{ru.EVENT: 'exec_stop'}])\n exec_units_agent_des2 = execUnits.duration([rp.AGENT_STAGING_INPUT, rp.UMGR_STAGING_OUTPUT_PENDING])\n exec_units_clientDes2 = execUnits.duration([rp.NEW, rp.DONE])\n SetupUnit = unitsDes2.filter(uid=['unit.000000'],inplace=False)\n setup_units_clientDes2 = SetupUnit.duration(event=[{ru.STATE: rp.NEW},{ru.EVENT: 'exec_start'}])\n pilotDes2 = rp_sessionDes2.filter(etype='pilot', inplace=False)\n pilot_duration = pilotDes2.duration([rp.PMGR_ACTIVE,rp.FINAL])\n des2_duration = AllNodes['End'].max() - AllNodes['Start'].min()\n setupDes2_overhead = exec_units_setup_des2 - des2_duration\n agentDes2_overhead = exec_units_agent_des2 - exec_units_setup_des2\n clientDes2_overhead = exec_units_clientDes2 - exec_units_agent_des2\n des2DF.loc[len(des2DF)] = [des2_duration, setup_units_clientDes2, setupDes2_overhead, agentDes2_overhead, clientDes2_overhead]",
"_____no_output_____"
],
[
"print(des2DF)",
" TTX SetupOverhead SetupOverhead2 AgentOverhead ClientOverhead\n0 6703.852195 81.679263 7.051244 0.18611 22.274512\n"
]
],
[
[
"## Design 2A",
"_____no_output_____"
]
],
[
[
"sid='../Data/Design2a/design2a_11k_test5/'\nrp_sessionDes2 = ra.Session(stype='radical.pilot',src=sid)\nunitsDes2 = rp_sessionDes2.filter(etype='unit', inplace=False)\nexecUnits = unitsDes2.filter(uid=['unit.000002','unit.000003','unit.000004','unit.000001'],inplace=False)\nexec_units_setup_des2 = execUnits.duration(event=[{ru.EVENT: 'exec_start'},{ru.EVENT: 'exec_stop'}])\nexec_units_agent_des2 = execUnits.duration([rp.AGENT_STAGING_INPUT, rp.UMGR_STAGING_OUTPUT_PENDING])\nexec_units_clientDes2 = execUnits.duration([rp.NEW, rp.DONE])\nSetupUnit = unitsDes2.filter(uid=['unit.000000'],inplace=False)\nsetup_units_clientDes2 = SetupUnit.duration(event=[{ru.STATE: rp.NEW},{ru.EVENT: 'exec_start'}])\npilotDes2 = rp_sessionDes2.filter(etype='pilot', inplace=False)",
"WARNING: profile \"../Data/Design2a/design2a_11k_test5/pmgr.0000.launching.0.child.prof\" not correctly closed.\nWARNING: profile \"../Data/Design2a/design2a_11k_test5/pmgr.0000.launching.0.child.prof\" not correctly closed.\nWARNING: profile \"../Data/Design2a/design2a_11k_test5/pmgr.0000.launching.0.child.prof\" not correctly closed.\nWARNING: profile \"../Data/Design2a/design2a_11k_test5/pmgr.0000.launching.0.child.prof\" not correctly closed.\nWARNING: profile \"../Data/Design2a/design2a_11k_test5/pmgr.0000.launching.0.child.prof\" not correctly closed.\n"
],
[
"Node1 = pd.DataFrame(columns=['Start','End','Type'])\nNode1Tilling = pd.read_csv('../Data/Design2a/design2a_11k_test5/pilot.0000/unit.000000/geolocate1.csv')\nfor index,row in Node1Tilling.iterrows():\n Node1.loc[len(Node1)] = [row['Start'],row['End'],'Geo1']\nNode1Tilling = pd.read_csv('../Data/Design2a/design2a_11k_test5/pilot.0000/unit.000000/geolocate2.csv')\nfor index,row in Node1Tilling.iterrows():\n Node1.loc[len(Node1)] = [row['Start'],row['End'],'Geo2']\n\nNode1Tilling = pd.read_csv('../Data/Design2a/design2a_11k_test5/pilot.0000/unit.000000/ransac1.csv')\nfor index,row in Node1Tilling.iterrows():\n Node1.loc[len(Node1)] = [row['Start'],row['End'],'Ransac1']\n",
"_____no_output_____"
],
[
"Node2 = pd.DataFrame(columns=['Start','End','Type'])\nNode2Tilling = pd.read_csv('../Data/Design2a/design2a_11k_test5/pilot.0000/unit.000001/geolocate3.csv')\nfor index,row in Node2Tilling.iterrows():\n Node2.loc[len(Node2)] = [row['Start'],row['End'],'Geo3']\nNode2Tilling = pd.read_csv('../Data/Design2a/design2a_11k_test5/pilot.0000/unit.000001/geolocate4.csv')\nfor index,row in Node2Tilling.iterrows():\n Node2.loc[len(Node2)] = [row['Start'],row['End'],'Geo4']\n\nNode2Tilling = pd.read_csv('../Data/Design2a/design2a_11k_test5/pilot.0000/unit.000001/ransac2.csv')\nfor index,row in Node2Tilling.iterrows():\n Node2.loc[len(Node2)] = [row['Start'],row['End'],'Ransac2']",
"_____no_output_____"
],
[
"Node3 = pd.DataFrame(columns=['Start','End','Type'])\nNode3Tilling = pd.read_csv('../Data/Design2a/design2a_11k_test5/pilot.0000/unit.000002/geolocate5.csv')\nfor index,row in Node3Tilling.iterrows():\n Node3.loc[len(Node3)] = [row['Start'],row['End'],'Geo5']\nNode3Tilling = pd.read_csv('../Data/Design2a/design2a_11k_test5/pilot.0000/unit.000002/geolocate6.csv')\nfor index,row in Node3Tilling.iterrows():\n Node3.loc[len(Node3)] = [row['Start'],row['End'],'Geo6']\n\nNode3Tilling = pd.read_csv('../Data/Design2a/design2a_11k_test5/pilot.0000/unit.000002/ransac3.csv')\nfor index,row in Node3Tilling.iterrows():\n Node3.loc[len(Node3)] = [row['Start'],row['End'],'Ransac3']\n",
"_____no_output_____"
],
[
"Node4 = pd.DataFrame(columns=['Start','End','Type'])\nNode4Tilling = pd.read_csv('../Data/Design2a/design2a_11k_test5/pilot.0000/unit.000003/geolocate7.csv')\nfor index,row in Node4Tilling.iterrows():\n Node4.loc[len(Node4)] = [row['Start'],row['End'],'Geo7']\nNode4Tilling = pd.read_csv('../Data/Design2a/design2a_11k_test5/pilot.0000/unit.000003/geolocate8.csv')\nfor index,row in Node4Tilling.iterrows():\n Node4.loc[len(Node4)] = [row['Start'],row['End'],'Geo8']\n\nNode4Tilling = pd.read_csv('../Data/Design2a/design2a_11k_test5/pilot.0000/unit.000003/ransac4.csv')\nfor index,row in Node4Tilling.iterrows():\n Node4.loc[len(Node4)] = [row['Start'],row['End'],'Ransac4']\n",
"_____no_output_____"
],
[
"des2ADF = pd.DataFrame(columns=['TTX','SetupOverhead','AgentOverhead','ClientOverhead'])\nAllNodes = pd.DataFrame(columns=['Start','End','Type'])\nAllNodes = AllNodes.append(Node1)\nAllNodes = AllNodes.append(Node2)\nAllNodes = AllNodes.append(Node3)\nAllNodes = AllNodes.append(Node4)\nAllNodes.reset_index(inplace=True,drop='index')\nrp_sessionDes2 = ra.Session(stype='radical.pilot',src=sid)\nunitsDes2 = rp_sessionDes2.filter(etype='unit', inplace=False)\nexecUnits = unitsDes2.filter(uid=['unit.000000','unit.000001','unit.000002','unit.000003'],inplace=False)\nexec_units_setup_des2 = unitsDes2.duration(event=[{ru.EVENT: 'exec_start'},{ru.EVENT: 'exec_stop'}])\nexec_units_agent_des2 = unitsDes2.duration([rp.AGENT_STAGING_INPUT, rp.UMGR_STAGING_OUTPUT_PENDING])\nexec_units_clientDes2 = execUnits.duration([rp.NEW, rp.DONE])\npilotDes2 = rp_sessionDes2.filter(etype='pilot', inplace=False)\npilot_duration = pilotDes2.duration([rp.PMGR_ACTIVE,rp.FINAL])\ndes2_duration = AllNodes['End'].max() - AllNodes['Start'].min()\nsetupDes2_overhead = exec_units_setup_des2 - des2_duration\nagentDes2_overhead = exec_units_agent_des2 - exec_units_setup_des2\nclientDes2_overhead = exec_units_clientDes2 - exec_units_agent_des2\nqueue_time = max(pilotDes2.timestamps(event=[{ru.STATE: rp.PMGR_ACTIVE}]))- max(execUnits.timestamps(event=[{ru.STATE: rp.AGENT_STAGING_INPUT_PENDING}]))\ndes2ADF.loc[len(des2ADF)] = [des2_duration, setupDes2_overhead, agentDes2_overhead, clientDes2_overhead-queue_time]",
"WARNING: profile \"../Data/Design2a/design2a_11k_test5/pmgr.0000.launching.0.child.prof\" not correctly closed.\nWARNING: profile \"../Data/Design2a/design2a_11k_test5/pmgr.0000.launching.0.child.prof\" not correctly closed.\nWARNING: profile \"../Data/Design2a/design2a_11k_test5/pmgr.0000.launching.0.child.prof\" not correctly closed.\nWARNING: profile \"../Data/Design2a/design2a_11k_test5/pmgr.0000.launching.0.child.prof\" not correctly closed.\n"
],
[
"print(des2ADF)",
" TTX SetupOverhead AgentOverhead ClientOverhead\n0 6172.103771 22.297043 0.246021 193.781951\n"
],
[
"fig, axis = plt.subplots(nrows=1,ncols=1, figsize=(15,7.5))\nx1 = np.arange(3)\n_ = axis.bar(x1[0], des1DF['TTX'].mean(), width=0.5, color=blues(300), label='Design 1 TTX')\n_ = axis.bar(x1[1], des2DF['TTX'].mean(), width=0.5, color=blues(200), label='Design 2 TTX')\n_ = axis.bar(x1[2], des2ADF['TTX'].mean(), width=0.5, color=blues(100), label='Design 2A TTX')\n_ = axis.set_xticks([0,1,2])\n_ = axis.grid(which='both', linestyle=':', linewidth=1)\n_ = axis.set_xticklabels(['Design 1', 'Design 2','Design 2A'], fontsize=36)\n_ = axis.set_ylabel('Time in seconds', fontsize=26)\n_ = axis.set_yticklabels(axis.get_yticks().astype('int').tolist(),fontsize=24)\n#fig.savefig('geo_ttx.pdf',dpi=800,bbox='tight')",
"_____no_output_____"
],
[
"dist_overhead = np.load('../Data/dist_dataset.npy')",
"_____no_output_____"
],
[
"DiscDurations = [1861.404363739,\n1872.631383787,\n1870.355146581,\n1852.347904858,\n1857.771844937,\n1868.644424397,\n1873.176510421,\n1851.527881958,\n1870.128898667,\n1856.676059379]",
"_____no_output_____"
],
[
"fig, axis = plt.subplots(nrows=1, ncols=1, figsize=(9,7.5))\nx1 = np.arange(3)\n\n_ = axis.bar(x1[0], des1DF['AgentOverhead'].mean(),width=0.5, color=reds(200),label='RP Agent Overhead Design 1')\n_ = axis.bar(x1[0], des1DF['ClientOverhead'].mean(), bottom=des1DF['AgentOverhead'].mean(),width=0.5, color=reds(150),label='RP Client Overhead Design 1')\n_ = axis.bar(x1[0], des1DF['EnTKOverhead'].mean(),bottom=des1DF['ClientOverhead'].mean()+des1DF['AgentOverhead'].mean(),width=0.5, color=reds(100),label='EnTK Overheads Design 1')\n_ = axis.bar(x1[0], np.mean(DiscDurations), yerr=np.std(DiscDurations), bottom=des1DF['ClientOverhead'].mean()+des1DF['AgentOverhead'].mean() + des1DF['EnTKOverhead'].mean(), \n width=0.5, color=reds(50),label='Design 1 Dataset Discovery')\n\n_ = axis.bar(x1[1],des2DF['AgentOverhead'].mean(),width=0.5, color=greens(200),label='RP Agent Overhead Design 2')\n_ = axis.bar(x1[1],des2DF['ClientOverhead'].mean(),bottom=des2DF['AgentOverhead'].mean(),width=0.5, color=greens(150),label='RP Client Overhead Design 2')\n_ = axis.bar(x1[1],(des2DF['SetupOverhead'] + des2DF['SetupOverhead2']).mean(),bottom=des2DF['ClientOverhead'].mean()+des2DF['AgentOverhead'].mean(),width=0.5, color=greens(100),label='Design 2 Setup Overhead')\n_ = axis.bar(x1[1],np.mean(DiscDurations), yerr=np.std(DiscDurations), bottom=des2DF['ClientOverhead'].mean()+des2DF['AgentOverhead'].mean() + (des2DF['SetupOverhead']+des2DF['SetupOverhead2']).mean(),\n width=0.5, color=greens(50),label='Design 2 Dataset Discovery')\n\n\n_ = axis.bar(x1[2],des2ADF['AgentOverhead'].mean(),#yerr=des2ADF['AgentOverhead'].std(),\n width=0.5, color=purples(250),label='RP Agent Overhead Design 2A',log=1)\n_ = axis.bar(x1[2],des2ADF['ClientOverhead'].mean(),#yerr=des2ADF['ClientOverhead'].std(),\n bottom=des2ADF['AgentOverhead'].mean(),width=0.5, color=purples(200),label='RP Client Overhead Design 2A')\n_ = axis.bar(x1[2],des2ADF['SetupOverhead'].mean(),#yerr=des2ADF['SetupOverhead'].std(),\n bottom=des2ADF['ClientOverhead'].mean()+des2ADF['AgentOverhead'].mean(),width=0.5, color=purples(150),label='Design 2A Setup Overhead')\n_ = axis.bar(x1[2],dist_overhead.mean(),yerr=dist_overhead.std(),\n bottom=des2ADF['ClientOverhead'].mean()+des2ADF['AgentOverhead'].mean()+des2ADF['SetupOverhead'].mean(),width=0.5, color=purples(100),label='Design 2A Distributing Overhead')\n_ = axis.bar(x1[2],np.mean(DiscDurations), yerr=np.std(DiscDurations),\n bottom=des2ADF['ClientOverhead'].mean()+des2ADF['AgentOverhead'].mean()+des2ADF['SetupOverhead'].mean() + dist_overhead.mean(),\n width=0.5, color=purples(50),label='Design 2A Dataset Discovery')\n\n\n_ = axis.set_xticks([0,1,2])\n_ = axis.grid(which='both', linestyle=':', linewidth=1)\n_ = axis.set_ylabel('Time in seconds', fontsize=26)\n_ = axis.set_xticklabels(['Design 1', 'Design 2','Design 2A'], fontsize=26)\n_ = axis.set_yticks([1,10,100,1000,10000,100000])\n_ = axis.set_yticklabels(axis.get_yticks().astype('int').tolist(),fontsize=24)\n#_ = axis.legend(fontsize=22,loc = 'lower center', bbox_to_anchor = (0,-.55,1,1), ncol=2)\n#_ = fig.subplots_adjust(bottom=.205)\nfig.savefig('geo_overheads.pdf',dpi=800,pad_inches=0)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d094c6c30f7998fc404771618902234c61369e3f | 5,866 | ipynb | Jupyter Notebook | 1_RunExtension.ipynb | kevcunnane/msbuild_ads_demo | f9a587aa64fbc543bc5b927520a78e5d45dd7f6a | [
"MIT"
] | 3 | 2019-05-30T10:07:23.000Z | 2019-09-24T17:30:22.000Z | 1_RunExtension.ipynb | kevcunnane/msbuild_ads_demo | f9a587aa64fbc543bc5b927520a78e5d45dd7f6a | [
"MIT"
] | null | null | null | 1_RunExtension.ipynb | kevcunnane/msbuild_ads_demo | f9a587aa64fbc543bc5b927520a78e5d45dd7f6a | [
"MIT"
] | 2 | 2019-05-21T13:44:58.000Z | 2020-08-07T01:11:25.000Z | 72.419753 | 661 | 0.600068 | [
[
[
"# Running, Debugging, Testing & Packaging\n",
"_____no_output_____"
]
],
[
[
"!code ./1-helloconnectedworld",
"_____no_output_____"
]
],
[
[
"\nLet's look at the key parts of our app:\n\n**package.json**\nThis defines all contributions: commands, context menus, UI, everything! \n\n```json\n \"activationEvents\": [\n // Use \"*\" to start on application start. If contributing commands, only start on command to speed up user experience\n \"onCommand:extension.showCurrentConnection\"\n ],\n \"main\": \"./out/extension\",\n \"contributes\": {\n \"commands\": [\n {\n \"command\": \"extension.showCurrentConnection\",\n \"title\": \"Show Current Connection\"\n }\n ]\n },\n\n```",
"_____no_output_____"
],
[
"**extension.ts** is our extension control center. Your extension always starts here by registering your extension points, and using built-in APIs to query connections, show messages, and much more \n\n```ts\n context.subscriptions.push(vscode.commands.registerCommand('extension.showCurrentConnection', () => {\n sqlops.connection.getCurrentConnection().then(connection => {\n let connectionId = connection ? connection.connectionId : 'No connection found!';\n vscode.window.showInformationMessage(connectionId);\n }, error => {\n console.info(error);\n });\n }));\n\n```",
"_____no_output_____"
],
[
"### VSCode APIs\n\nAll* VSCode APIs are defined in Azure Data Studio meaning VSCode extensions just work. These include common workspace, window and language services features\n> *Debugger APIs are defined but the debugger is not implemented",
"_____no_output_____"
],
[
"### sqlops / azdata APIs**\n\nAzure Data Studio APIs are in the sqlops namespace. These cover Connection, Query, advanced UI (dialogs, wizards, and other standardized UI controls]), and the Data Management Protocol (DMP).\n> These are moving to a new **azdata** namespace. We will cover the types of changes being made to simplify development as part of this demo",
"_____no_output_____"
],
[
"# Run your code\n\n* In VSCode, you should have the \"Azure Data Studio Debug\" extension installed\n\n\n\n* Hit F5 or go to the debugger section and click the Run button\n* Azure Data Studio will launch\n* Hit `Ctrl+Shift+P` and choose **Show Current Connection**\n* It will show **No Connection Found**. How do we find out what's wrong? Let's go and debug it!",
"_____no_output_____"
],
[
"# Debug your code\n\n* As for any app, click inside the code and set a breakdpoint on the line\n\n```\n let connectionId = connection ? connection.connectionId : 'No connection found!';\n```\n\n* Run the command again\n* We will see that the connection is not getting returned. Why might that be? It's because nobody connected to one!\n* Open a connection and try again. This time you will see all the available information about this connection.",
"_____no_output_____"
],
[
"# Testing your code\n\nIf you like to write tests, you have a template built-into your extension. You can even debug using the **Extension Tests** option in the debugger dropdown. This uses standard Javascript test frameworks (Mocha) and is able to integrate with all the usual actions.",
"_____no_output_____"
],
[
"# Packaging your extension\n\nPackaging is as easy as running `vsce package` from the root of the extension.\n* The first time you run this, you'll see errors if you didn't edit your Readme and other key files\n* Update Readme.md so it's not a default value\n* Similarly delete Changelog contents or update as needed\n* Delete the **vsc-extension-quickstart.md` file\n\nNow if you re-run, you'll get a .vsix file\n\n## Installing your extension for testing\n* In Azure Data Studio, hit `Ctrl+Shift+P` and choose **Extensions: Install from VSIX...**\n* Pick your file and click OK\n* It'll install and be available - no reload necessary!",
"_____no_output_____"
],
[
"## Publishing your extension\n\nFollow our [Extension Authoring](https://github.com/Microsoft/azuredatastudio/wiki/Extension-Authoring) guide which has details on publishing to the extension gallery. If you have any issues reach out to us on Twitter [@AzureDataStudio](https://twitter.com/azuredatastudio)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d094d3175d4494df357c077c919c9c3fe985358e | 50,274 | ipynb | Jupyter Notebook | ix-1-lab-intro/4-networks.ipynb | emlg/Internet-Analytics | f26df7e736571bdb7cfad2c0043617761101b798 | [
"MIT"
] | null | null | null | ix-1-lab-intro/4-networks.ipynb | emlg/Internet-Analytics | f26df7e736571bdb7cfad2c0043617761101b798 | [
"MIT"
] | null | null | null | ix-1-lab-intro/4-networks.ipynb | emlg/Internet-Analytics | f26df7e736571bdb7cfad2c0043617761101b798 | [
"MIT"
] | null | null | null | 212.126582 | 37,722 | 0.899332 | [
[
[
"# Networks\n\nIn this last exercise, you will start to put into practice some of the concepts learned in class. Start by importing the libraries that you will need.",
"_____no_output_____"
]
],
[
[
"import networkx as nx\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"We give you the network of the 34 members of [karate clubs](https://en.wikipedia.org/wiki/Zachary's_karate_club) on the campus. Each node is a member of one of two clubs (\"Mr. Hi\" or \"Officer\") and there is an edge between two nodes if the two are friends outside of the club.",
"_____no_output_____"
]
],
[
[
"karate_club = nx.karate_club_graph()\n",
"_____no_output_____"
]
],
[
[
"## Visualization\n\n1. Visualize the network by\n - painting every 'Mr. Hi' member in **blue** and every 'Officer' member in **red**, and\n - drawing the label corresponding to the member index (between 1 and 34) on each node.\n2. What can you tell about the nodes and their connections? Do you see some nodes with more/less links than others? Who can they be?",
"_____no_output_____"
]
],
[
[
"# Your code goes here\ncolor_map = []\nfor i in range(0, karate_club.order()):\n if karate_club.node[i]['club'] == \"Mr. Hi\":\n color_map.append('blue')\n else:\n color_map.append('red')\n\nnx.draw_networkx(karate_club, node_color = color_map, with_labels = True)\nplt.axis('off');",
"_____no_output_____"
]
],
[
[
"## Degree Distribution\n\n1. Plot the degree distribution of the network.\n2. Does it confirm your answer to the previous question?",
"_____no_output_____"
]
],
[
[
"# Your code goes here\ndegre = list()\nfor v in karate_club:\n degre.append(karate_club.degree(v))\n \nplt.plot(degre, '*')\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d094d3d2299f17e8ad23903907c2bdffdf5c43bf | 110,344 | ipynb | Jupyter Notebook | autoencoder/Convolutional_Autoencoder_Solution.ipynb | vinidixit/deep-learning-repo | 6b207e326cc937930a0512a8c599e86e48c297b2 | [
"MIT"
] | 7 | 2019-03-07T10:05:03.000Z | 2020-07-01T06:40:15.000Z | autoencoder/Convolutional_Autoencoder_Solution.ipynb | mazharul-miraz/deep-learning-repo | 6b207e326cc937930a0512a8c599e86e48c297b2 | [
"MIT"
] | 120 | 2020-01-28T21:35:50.000Z | 2022-03-11T23:25:52.000Z | autoencoder/Convolutional_Autoencoder_Solution.ipynb | mazharul-miraz/deep-learning-repo | 6b207e326cc937930a0512a8c599e86e48c297b2 | [
"MIT"
] | 9 | 2018-03-21T04:48:53.000Z | 2020-05-05T00:38:46.000Z | 153.468707 | 48,642 | 0.843018 | [
[
[
"# Convolutional Autoencoder\n\nSticking with the MNIST dataset, let's improve our autoencoder's performance using convolutional layers. Again, loading modules and the data.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport numpy as np\nimport tensorflow as tf\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"from tensorflow.examples.tutorials.mnist import input_data\nmnist = input_data.read_data_sets('MNIST_data', validation_size=0)",
"Extracting MNIST_data/train-images-idx3-ubyte.gz\nExtracting MNIST_data/train-labels-idx1-ubyte.gz\nExtracting MNIST_data/t10k-images-idx3-ubyte.gz\nExtracting MNIST_data/t10k-labels-idx1-ubyte.gz\n"
],
[
"img = mnist.train.images[2]\nplt.imshow(img.reshape((28, 28)), cmap='Greys_r')",
"_____no_output_____"
]
],
[
[
"## Network Architecture\n\nThe encoder part of the network will be a typical convolutional pyramid. Each convolutional layer will be followed by a max-pooling layer to reduce the dimensions of the layers. The decoder though might be something new to you. The decoder needs to convert from a narrow representation to a wide reconstructed image. For example, the representation could be a 4x4x8 max-pool layer. This is the output of the encoder, but also the input to the decoder. We want to get a 28x28x1 image out from the decoder so we need to work our way back up from the narrow decoder input layer. A schematic of the network is shown below.\n\n<img src='assets/convolutional_autoencoder.png' width=500px>\n\nHere our final encoder layer has size 4x4x8 = 128. The original images have size 28x28 = 784, so the encoded vector is roughly 16% the size of the original image. These are just suggested sizes for each of the layers. Feel free to change the depths and sizes, but remember our goal here is to find a small representation of the input data.\n\n### What's going on with the decoder\n\nOkay, so the decoder has these \"Upsample\" layers that you might not have seen before. First off, I'll discuss a bit what these layers *aren't*. Usually, you'll see **transposed convolution** layers used to increase the width and height of the layers. They work almost exactly the same as convolutional layers, but in reverse. A stride in the input layer results in a larger stride in the transposed convolution layer. For example, if you have a 3x3 kernel, a 3x3 patch in the input layer will be reduced to one unit in a convolutional layer. Comparatively, one unit in the input layer will be expanded to a 3x3 path in a transposed convolution layer. The TensorFlow API provides us with an easy way to create the layers, [`tf.nn.conv2d_transpose`](https://www.tensorflow.org/api_docs/python/tf/nn/conv2d_transpose). \n\nHowever, transposed convolution layers can lead to artifacts in the final images, such as checkerboard patterns. This is due to overlap in the kernels which can be avoided by setting the stride and kernel size equal. In [this Distill article](http://distill.pub/2016/deconv-checkerboard/) from Augustus Odena, *et al*, the authors show that these checkerboard artifacts can be avoided by resizing the layers using nearest neighbor or bilinear interpolation (upsampling) followed by a convolutional layer. In TensorFlow, this is easily done with [`tf.image.resize_images`](https://www.tensorflow.org/versions/r1.1/api_docs/python/tf/image/resize_images), followed by a convolution. Be sure to read the Distill article to get a better understanding of deconvolutional layers and why we're using upsampling.\n\n> **Exercise:** Build the network shown above. Remember that a convolutional layer with strides of 1 and 'same' padding won't reduce the height and width. That is, if the input is 28x28 and the convolution layer has stride = 1 and 'same' padding, the convolutional layer will also be 28x28. The max-pool layers are used the reduce the width and height. A stride of 2 will reduce the size by a factor of 2. Odena *et al* claim that nearest neighbor interpolation works best for the upsampling, so make sure to include that as a parameter in `tf.image.resize_images` or use [`tf.image.resize_nearest_neighbor`]( `https://www.tensorflow.org/api_docs/python/tf/image/resize_nearest_neighbor). For convolutional layers, use [`tf.layers.conv2d`](https://www.tensorflow.org/api_docs/python/tf/layers/conv2d). For example, you would write `conv1 = tf.layers.conv2d(inputs, 32, (5,5), padding='same', activation=tf.nn.relu)` for a layer with a depth of 32, a 5x5 kernel, stride of (1,1), padding is 'same', and a ReLU activation. Similarly, for the max-pool layers, use [`tf.layers.max_pooling2d`](https://www.tensorflow.org/api_docs/python/tf/layers/max_pooling2d).",
"_____no_output_____"
]
],
[
[
"inputs_ = tf.placeholder(tf.float32, (None, 28, 28, 1), name='inputs')\ntargets_ = tf.placeholder(tf.float32, (None, 28, 28, 1), name='targets')\n\n### Encoder\nconv1 = tf.layers.conv2d(inputs_, 16, (3,3), padding='same', activation=tf.nn.relu)\n# Now 28x28x16\nmaxpool1 = tf.layers.max_pooling2d(conv1, (2,2), (2,2), padding='same')\n# Now 14x14x16\nconv2 = tf.layers.conv2d(maxpool1, 8, (3,3), padding='same', activation=tf.nn.relu)\n# Now 14x14x8\nmaxpool2 = tf.layers.max_pooling2d(conv2, (2,2), (2,2), padding='same')\n# Now 7x7x8\nconv3 = tf.layers.conv2d(maxpool2, 8, (3,3), padding='same', activation=tf.nn.relu)\n# Now 7x7x8\nencoded = tf.layers.max_pooling2d(conv3, (2,2), (2,2), padding='same')\n# Now 4x4x8\n\n### Decoder\nupsample1 = tf.image.resize_nearest_neighbor(encoded, (7,7))\n# Now 7x7x8\nconv4 = tf.layers.conv2d(upsample1, 8, (3,3), padding='same', activation=tf.nn.relu)\n# Now 7x7x8\nupsample2 = tf.image.resize_nearest_neighbor(conv4, (14,14))\n# Now 14x14x8\nconv5 = tf.layers.conv2d(upsample2, 8, (3,3), padding='same', activation=tf.nn.relu)\n# Now 14x14x8\nupsample3 = tf.image.resize_nearest_neighbor(conv5, (28,28))\n# Now 28x28x8\nconv6 = tf.layers.conv2d(upsample3, 16, (3,3), padding='same', activation=tf.nn.relu)\n# Now 28x28x16\n\nlogits = tf.layers.conv2d(conv6, 1, (3,3), padding='same', activation=None)\n#Now 28x28x1\n\ndecoded = tf.nn.sigmoid(logits, name='decoded')\n\nloss = tf.nn.sigmoid_cross_entropy_with_logits(labels=targets_, logits=logits)\ncost = tf.reduce_mean(loss)\nopt = tf.train.AdamOptimizer(0.001).minimize(cost)",
"_____no_output_____"
]
],
[
[
"## Training\n\nAs before, here wi'll train the network. Instead of flattening the images though, we can pass them in as 28x28x1 arrays.",
"_____no_output_____"
]
],
[
[
"sess = tf.Session()",
"_____no_output_____"
],
[
"epochs = 1\nbatch_size = 200\nsess.run(tf.global_variables_initializer())\nfor e in range(epochs):\n for ii in range(mnist.train.num_examples//batch_size):\n batch = mnist.train.next_batch(batch_size)\n imgs = batch[0].reshape((-1, 28, 28, 1))\n batch_cost, _ = sess.run([cost, opt], feed_dict={inputs_: imgs,\n targets_: imgs})\n\n print(\"Epoch: {}/{}...\".format(e+1, epochs),\n \"Training loss: {:.4f}\".format(batch_cost))",
"Epoch: 1/1... Training loss: 0.6928\nEpoch: 1/1... Training loss: 0.6904\nEpoch: 1/1... Training loss: 0.6882\nEpoch: 1/1... Training loss: 0.6855\nEpoch: 1/1... Training loss: 0.6820\nEpoch: 1/1... Training loss: 0.6781\nEpoch: 1/1... Training loss: 0.6737\nEpoch: 1/1... Training loss: 0.6678\nEpoch: 1/1... Training loss: 0.6595\nEpoch: 1/1... Training loss: 0.6511\nEpoch: 1/1... Training loss: 0.6411\nEpoch: 1/1... Training loss: 0.6301\nEpoch: 1/1... Training loss: 0.6194\nEpoch: 1/1... Training loss: 0.6014\nEpoch: 1/1... Training loss: 0.5887\nEpoch: 1/1... Training loss: 0.5697\nEpoch: 1/1... Training loss: 0.5519\nEpoch: 1/1... Training loss: 0.5418\nEpoch: 1/1... Training loss: 0.5345\nEpoch: 1/1... Training loss: 0.5279\nEpoch: 1/1... Training loss: 0.5188\nEpoch: 1/1... Training loss: 0.5222\nEpoch: 1/1... Training loss: 0.5193\nEpoch: 1/1... Training loss: 0.5131\nEpoch: 1/1... Training loss: 0.4972\nEpoch: 1/1... Training loss: 0.4846\nEpoch: 1/1... Training loss: 0.4719\nEpoch: 1/1... Training loss: 0.4638\nEpoch: 1/1... Training loss: 0.4571\nEpoch: 1/1... Training loss: 0.4516\nEpoch: 1/1... Training loss: 0.4379\nEpoch: 1/1... Training loss: 0.4336\nEpoch: 1/1... Training loss: 0.4269\nEpoch: 1/1... Training loss: 0.4233\nEpoch: 1/1... Training loss: 0.4084\nEpoch: 1/1... Training loss: 0.4051\nEpoch: 1/1... Training loss: 0.3949\nEpoch: 1/1... Training loss: 0.3842\nEpoch: 1/1... Training loss: 0.3678\nEpoch: 1/1... Training loss: 0.3623\nEpoch: 1/1... Training loss: 0.3531\nEpoch: 1/1... Training loss: 0.3350\nEpoch: 1/1... Training loss: 0.3347\nEpoch: 1/1... Training loss: 0.3238\nEpoch: 1/1... Training loss: 0.3218\nEpoch: 1/1... Training loss: 0.3118\nEpoch: 1/1... Training loss: 0.3039\nEpoch: 1/1... Training loss: 0.2953\nEpoch: 1/1... Training loss: 0.2906\nEpoch: 1/1... Training loss: 0.2925\nEpoch: 1/1... Training loss: 0.2761\nEpoch: 1/1... Training loss: 0.2733\nEpoch: 1/1... Training loss: 0.2694\nEpoch: 1/1... Training loss: 0.2667\nEpoch: 1/1... Training loss: 0.2571\nEpoch: 1/1... Training loss: 0.2525\nEpoch: 1/1... Training loss: 0.2478\nEpoch: 1/1... Training loss: 0.2507\nEpoch: 1/1... Training loss: 0.2313\nEpoch: 1/1... Training loss: 0.2328\nEpoch: 1/1... Training loss: 0.2367\nEpoch: 1/1... Training loss: 0.2315\nEpoch: 1/1... Training loss: 0.2283\nEpoch: 1/1... Training loss: 0.2218\nEpoch: 1/1... Training loss: 0.2229\nEpoch: 1/1... Training loss: 0.2180\nEpoch: 1/1... Training loss: 0.2174\nEpoch: 1/1... Training loss: 0.2179\nEpoch: 1/1... Training loss: 0.2124\nEpoch: 1/1... Training loss: 0.2118\nEpoch: 1/1... Training loss: 0.2176\nEpoch: 1/1... Training loss: 0.2255\nEpoch: 1/1... Training loss: 0.2171\nEpoch: 1/1... Training loss: 0.2096\nEpoch: 1/1... Training loss: 0.2090\nEpoch: 1/1... Training loss: 0.2093\nEpoch: 1/1... Training loss: 0.2001\nEpoch: 1/1... Training loss: 0.2020\nEpoch: 1/1... Training loss: 0.2018\nEpoch: 1/1... Training loss: 0.2014\nEpoch: 1/1... Training loss: 0.2029\nEpoch: 1/1... Training loss: 0.2051\nEpoch: 1/1... Training loss: 0.2041\nEpoch: 1/1... Training loss: 0.2014\nEpoch: 1/1... Training loss: 0.2027\nEpoch: 1/1... Training loss: 0.1952\nEpoch: 1/1... Training loss: 0.2021\nEpoch: 1/1... Training loss: 0.1974\nEpoch: 1/1... Training loss: 0.1986\nEpoch: 1/1... Training loss: 0.1966\nEpoch: 1/1... Training loss: 0.1989\nEpoch: 1/1... Training loss: 0.1925\nEpoch: 1/1... Training loss: 0.1927\nEpoch: 1/1... Training loss: 0.1929\nEpoch: 1/1... Training loss: 0.1983\nEpoch: 1/1... Training loss: 0.1964\nEpoch: 1/1... Training loss: 0.1968\nEpoch: 1/1... Training loss: 0.1883\nEpoch: 1/1... Training loss: 0.1912\nEpoch: 1/1... Training loss: 0.1927\nEpoch: 1/1... Training loss: 0.1858\nEpoch: 1/1... Training loss: 0.1927\nEpoch: 1/1... Training loss: 0.1879\nEpoch: 1/1... Training loss: 0.1849\nEpoch: 1/1... Training loss: 0.1868\nEpoch: 1/1... Training loss: 0.1919\nEpoch: 1/1... Training loss: 0.1855\nEpoch: 1/1... Training loss: 0.1920\nEpoch: 1/1... Training loss: 0.1841\nEpoch: 1/1... Training loss: 0.1948\nEpoch: 1/1... Training loss: 0.1864\nEpoch: 1/1... Training loss: 0.1823\nEpoch: 1/1... Training loss: 0.1907\nEpoch: 1/1... Training loss: 0.1804\nEpoch: 1/1... Training loss: 0.1835\nEpoch: 1/1... Training loss: 0.1883\nEpoch: 1/1... Training loss: 0.1908\nEpoch: 1/1... Training loss: 0.1845\nEpoch: 1/1... Training loss: 0.1823\nEpoch: 1/1... Training loss: 0.1810\nEpoch: 1/1... Training loss: 0.1742\nEpoch: 1/1... Training loss: 0.1863\nEpoch: 1/1... Training loss: 0.1846\nEpoch: 1/1... Training loss: 0.1814\nEpoch: 1/1... Training loss: 0.1779\nEpoch: 1/1... Training loss: 0.1745\nEpoch: 1/1... Training loss: 0.1820\nEpoch: 1/1... Training loss: 0.1773\nEpoch: 1/1... Training loss: 0.1747\nEpoch: 1/1... Training loss: 0.1831\nEpoch: 1/1... Training loss: 0.1806\nEpoch: 1/1... Training loss: 0.1752\nEpoch: 1/1... Training loss: 0.1780\nEpoch: 1/1... Training loss: 0.1746\nEpoch: 1/1... Training loss: 0.1796\nEpoch: 1/1... Training loss: 0.1796\nEpoch: 1/1... Training loss: 0.1789\nEpoch: 1/1... Training loss: 0.1795\nEpoch: 1/1... Training loss: 0.1839\nEpoch: 1/1... Training loss: 0.1829\nEpoch: 1/1... Training loss: 0.1836\nEpoch: 1/1... Training loss: 0.1761\nEpoch: 1/1... Training loss: 0.1781\nEpoch: 1/1... Training loss: 0.1799\nEpoch: 1/1... Training loss: 0.1823\nEpoch: 1/1... Training loss: 0.1694\nEpoch: 1/1... Training loss: 0.1761\nEpoch: 1/1... Training loss: 0.1769\nEpoch: 1/1... Training loss: 0.1761\nEpoch: 1/1... Training loss: 0.1726\nEpoch: 1/1... Training loss: 0.1762\nEpoch: 1/1... Training loss: 0.1758\nEpoch: 1/1... Training loss: 0.1752\nEpoch: 1/1... Training loss: 0.1764\nEpoch: 1/1... Training loss: 0.1787\nEpoch: 1/1... Training loss: 0.1755\nEpoch: 1/1... Training loss: 0.1769\nEpoch: 1/1... Training loss: 0.1748\nEpoch: 1/1... Training loss: 0.1758\nEpoch: 1/1... Training loss: 0.1709\nEpoch: 1/1... Training loss: 0.1685\nEpoch: 1/1... Training loss: 0.1653\nEpoch: 1/1... Training loss: 0.1713\nEpoch: 1/1... Training loss: 0.1697\nEpoch: 1/1... Training loss: 0.1742\nEpoch: 1/1... Training loss: 0.1747\nEpoch: 1/1... Training loss: 0.1692\nEpoch: 1/1... Training loss: 0.1755\nEpoch: 1/1... Training loss: 0.1721\nEpoch: 1/1... Training loss: 0.1757\nEpoch: 1/1... Training loss: 0.1682\nEpoch: 1/1... Training loss: 0.1666\nEpoch: 1/1... Training loss: 0.1705\nEpoch: 1/1... Training loss: 0.1687\nEpoch: 1/1... Training loss: 0.1729\nEpoch: 1/1... Training loss: 0.1671\nEpoch: 1/1... Training loss: 0.1702\nEpoch: 1/1... Training loss: 0.1686\nEpoch: 1/1... Training loss: 0.1639\nEpoch: 1/1... Training loss: 0.1642\nEpoch: 1/1... Training loss: 0.1701\nEpoch: 1/1... Training loss: 0.1653\nEpoch: 1/1... Training loss: 0.1717\nEpoch: 1/1... Training loss: 0.1670\nEpoch: 1/1... Training loss: 0.1693\nEpoch: 1/1... Training loss: 0.1684\nEpoch: 1/1... Training loss: 0.1633\nEpoch: 1/1... Training loss: 0.1683\nEpoch: 1/1... Training loss: 0.1631\nEpoch: 1/1... Training loss: 0.1649\nEpoch: 1/1... Training loss: 0.1647\nEpoch: 1/1... Training loss: 0.1619\nEpoch: 1/1... Training loss: 0.1692\nEpoch: 1/1... Training loss: 0.1682\nEpoch: 1/1... Training loss: 0.1650\nEpoch: 1/1... Training loss: 0.1637\nEpoch: 1/1... Training loss: 0.1603\nEpoch: 1/1... Training loss: 0.1661\nEpoch: 1/1... Training loss: 0.1598\nEpoch: 1/1... Training loss: 0.1623\nEpoch: 1/1... Training loss: 0.1640\nEpoch: 1/1... Training loss: 0.1716\nEpoch: 1/1... Training loss: 0.1661\nEpoch: 1/1... Training loss: 0.1607\nEpoch: 1/1... Training loss: 0.1660\nEpoch: 1/1... Training loss: 0.1619\nEpoch: 1/1... Training loss: 0.1584\nEpoch: 1/1... Training loss: 0.1719\nEpoch: 1/1... Training loss: 0.1593\nEpoch: 1/1... Training loss: 0.1601\nEpoch: 1/1... Training loss: 0.1651\nEpoch: 1/1... Training loss: 0.1625\nEpoch: 1/1... Training loss: 0.1679\nEpoch: 1/1... Training loss: 0.1633\nEpoch: 1/1... Training loss: 0.1610\nEpoch: 1/1... Training loss: 0.1583\nEpoch: 1/1... Training loss: 0.1611\nEpoch: 1/1... Training loss: 0.1611\nEpoch: 1/1... Training loss: 0.1621\nEpoch: 1/1... Training loss: 0.1597\nEpoch: 1/1... Training loss: 0.1638\nEpoch: 1/1... Training loss: 0.1663\nEpoch: 1/1... Training loss: 0.1640\nEpoch: 1/1... Training loss: 0.1550\nEpoch: 1/1... Training loss: 0.1608\nEpoch: 1/1... Training loss: 0.1604\nEpoch: 1/1... Training loss: 0.1610\nEpoch: 1/1... Training loss: 0.1547\n"
],
[
"fig, axes = plt.subplots(nrows=2, ncols=10, sharex=True, sharey=True, figsize=(20,4))\nin_imgs = mnist.test.images[:10]\nreconstructed = sess.run(decoded, feed_dict={inputs_: in_imgs.reshape((10, 28, 28, 1))})\n\nfor images, row in zip([in_imgs, reconstructed], axes):\n for img, ax in zip(images, row):\n ax.imshow(img.reshape((28, 28)), cmap='Greys_r')\n ax.get_xaxis().set_visible(False)\n ax.get_yaxis().set_visible(False)\n\n\nfig.tight_layout(pad=0.1)",
"_____no_output_____"
],
[
"sess.close()",
"_____no_output_____"
]
],
[
[
"## Denoising\n\nAs I've mentioned before, autoencoders like the ones you've built so far aren't too useful in practive. However, they can be used to denoise images quite successfully just by training the network on noisy images. We can create the noisy images ourselves by adding Gaussian noise to the training images, then clipping the values to be between 0 and 1. We'll use noisy images as input and the original, clean images as targets. Here's an example of the noisy images I generated and the denoised images.\n\n\n\n\nSince this is a harder problem for the network, we'll want to use deeper convolutional layers here, more feature maps. I suggest something like 32-32-16 for the depths of the convolutional layers in the encoder, and the same depths going backward through the decoder. Otherwise the architecture is the same as before.\n\n> **Exercise:** Build the network for the denoising autoencoder. It's the same as before, but with deeper layers. I suggest 32-32-16 for the depths, but you can play with these numbers, or add more layers.",
"_____no_output_____"
]
],
[
[
"inputs_ = tf.placeholder(tf.float32, (None, 28, 28, 1), name='inputs')\ntargets_ = tf.placeholder(tf.float32, (None, 28, 28, 1), name='targets')\n\n### Encoder\nconv1 = tf.layers.conv2d(inputs_, 32, (3,3), padding='same', activation=tf.nn.relu)\n# Now 28x28x32\nmaxpool1 = tf.layers.max_pooling2d(conv1, (2,2), (2,2), padding='same')\n# Now 14x14x32\nconv2 = tf.layers.conv2d(maxpool1, 32, (3,3), padding='same', activation=tf.nn.relu)\n# Now 14x14x32\nmaxpool2 = tf.layers.max_pooling2d(conv2, (2,2), (2,2), padding='same')\n# Now 7x7x32\nconv3 = tf.layers.conv2d(maxpool2, 16, (3,3), padding='same', activation=tf.nn.relu)\n# Now 7x7x16\nencoded = tf.layers.max_pooling2d(conv3, (2,2), (2,2), padding='same')\n# Now 4x4x16\n\n### Decoder\nupsample1 = tf.image.resize_nearest_neighbor(encoded, (7,7))\n# Now 7x7x16\nconv4 = tf.layers.conv2d(upsample1, 16, (3,3), padding='same', activation=tf.nn.relu)\n# Now 7x7x16\nupsample2 = tf.image.resize_nearest_neighbor(conv4, (14,14))\n# Now 14x14x16\nconv5 = tf.layers.conv2d(upsample2, 32, (3,3), padding='same', activation=tf.nn.relu)\n# Now 14x14x32\nupsample3 = tf.image.resize_nearest_neighbor(conv5, (28,28))\n# Now 28x28x32\nconv6 = tf.layers.conv2d(upsample3, 32, (3,3), padding='same', activation=tf.nn.relu)\n# Now 28x28x32\n\nlogits = tf.layers.conv2d(conv6, 1, (3,3), padding='same', activation=None)\n#Now 28x28x1\n\ndecoded = tf.nn.sigmoid(logits, name='decoded')\n\nloss = tf.nn.sigmoid_cross_entropy_with_logits(labels=targets_, logits=logits)\ncost = tf.reduce_mean(loss)\nopt = tf.train.AdamOptimizer(0.001).minimize(cost)",
"_____no_output_____"
],
[
"sess = tf.Session()",
"_____no_output_____"
],
[
"epochs = 100\nbatch_size = 200\n# Set's how much noise we're adding to the MNIST images\nnoise_factor = 0.5\nsess.run(tf.global_variables_initializer())\nfor e in range(epochs):\n for ii in range(mnist.train.num_examples//batch_size):\n batch = mnist.train.next_batch(batch_size)\n # Get images from the batch\n imgs = batch[0].reshape((-1, 28, 28, 1))\n \n # Add random noise to the input images\n noisy_imgs = imgs + noise_factor * np.random.randn(*imgs.shape)\n # Clip the images to be between 0 and 1\n noisy_imgs = np.clip(noisy_imgs, 0., 1.)\n \n # Noisy images as inputs, original images as targets\n batch_cost, _ = sess.run([cost, opt], feed_dict={inputs_: noisy_imgs,\n targets_: imgs})\n\n print(\"Epoch: {}/{}...\".format(e+1, epochs),\n \"Training loss: {:.4f}\".format(batch_cost))",
"_____no_output_____"
]
],
[
[
"## Checking out the performance\n\nHere I'm adding noise to the test images and passing them through the autoencoder. It does a suprising great job of removing the noise, even though it's sometimes difficult to tell what the original number is.",
"_____no_output_____"
]
],
[
[
"fig, axes = plt.subplots(nrows=2, ncols=10, sharex=True, sharey=True, figsize=(20,4))\nin_imgs = mnist.test.images[:10]\nnoisy_imgs = in_imgs + noise_factor * np.random.randn(*in_imgs.shape)\nnoisy_imgs = np.clip(noisy_imgs, 0., 1.)\n\nreconstructed = sess.run(decoded, feed_dict={inputs_: noisy_imgs.reshape((10, 28, 28, 1))})\n\nfor images, row in zip([noisy_imgs, reconstructed], axes):\n for img, ax in zip(images, row):\n ax.imshow(img.reshape((28, 28)), cmap='Greys_r')\n ax.get_xaxis().set_visible(False)\n ax.get_yaxis().set_visible(False)\n\nfig.tight_layout(pad=0.1)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d094d6656cf53fda4e7ce9982905291c9b04a15f | 8,509 | ipynb | Jupyter Notebook | 1.1. Create_Data_Augmentation_With_Augmentor.ipynb | christianquicano/tflite_classification_banknotes | eebac0a4128832b7ebb891f533ddabbeec23e361 | [
"MIT"
] | null | null | null | 1.1. Create_Data_Augmentation_With_Augmentor.ipynb | christianquicano/tflite_classification_banknotes | eebac0a4128832b7ebb891f533ddabbeec23e361 | [
"MIT"
] | null | null | null | 1.1. Create_Data_Augmentation_With_Augmentor.ipynb | christianquicano/tflite_classification_banknotes | eebac0a4128832b7ebb891f533ddabbeec23e361 | [
"MIT"
] | null | null | null | 25.476048 | 142 | 0.543542 | [
[
[
"# 1.1. Create_Data_Augmentation_With_Augmentor\nhttps://github.com/mdbloice/Augmentor",
"_____no_output_____"
]
],
[
[
"# install the package\n# !pip install Augmentor",
"_____no_output_____"
],
[
"import Augmentor",
"_____no_output_____"
],
[
"label = '200_frente'\npath_img = 'data_augmentation/source/' + label + '/'\nsample_per_configurator = 50",
"_____no_output_____"
],
[
"# random distortion + rotate\np1 = Augmentor.Pipeline(path_img)\np1.rotate(probability=1.0, max_left_rotation=20, max_right_rotation=20)\np1.random_distortion(probability=1, grid_width=30, grid_height=30, magnitude=8)\n#p1.sample(sample_per_configurator)\np1.process()",
"\rExecuting Pipeline: 0%| | 0/25 [00:00<?, ? Samples/s]"
],
[
"# random distortion + rotate\np1 = Augmentor.Pipeline(path_img)\np1.rotate(probability=1.0, max_left_rotation=20, max_right_rotation=20)\np1.random_distortion(probability=1, grid_width=30, grid_height=30, magnitude=8)\np1.process()",
"\rExecuting Pipeline: 0%| | 0/25 [00:00<?, ? Samples/s]"
],
[
"# random distortion + rotate\np3_1 = Augmentor.Pipeline(path_img)\np3_1.rotate(probability=1, max_left_rotation=25, max_right_rotation=25)\np3_1.random_distortion(probability=1, grid_width=30, grid_height=30, magnitude=8)\np3_1.process()",
"\rExecuting Pipeline: 0%| | 0/25 [00:00<?, ? Samples/s]"
],
[
"# random skew + rotate + random distortion\np4 = Augmentor.Pipeline(path_img)\np4.rotate(probability=1, max_left_rotation=25, max_right_rotation=25)\np4.skew(probability=1, magnitude=0.6)\np4.random_distortion(probability=1, grid_width=30, grid_height=30, magnitude=8)\np4.process()",
"\rExecuting Pipeline: 0%| | 0/25 [00:00<?, ? Samples/s]"
],
[
"# random skew + rotate\np4 = Augmentor.Pipeline(path_img)\np4.rotate(probability=1, max_left_rotation=25, max_right_rotation=25)\np4.skew(probability=1, magnitude=0.8)\np4.process()",
"\rExecuting Pipeline: 0%| | 0/25 [00:00<?, ? Samples/s]"
],
[
"# shear + rotate\np5 = Augmentor.Pipeline(path_img)\np5.shear(probability=1, max_shear_left=25, max_shear_right=25)\np5.rotate(probability=1, max_left_rotation=25, max_right_rotation=25)\n#p5.random_distortion(probability=0.6, grid_width=40, grid_height=40, magnitude=8)\np5.process()",
"\rExecuting Pipeline: 0%| | 0/25 [00:00<?, ? Samples/s]"
],
[
"# shear + rotate + random distortion\np5 = Augmentor.Pipeline(path_img)\np5.shear(probability=1, max_shear_left=25, max_shear_right=25)\np5.rotate(probability=1, max_left_rotation=25, max_right_rotation=25)\np5.random_distortion(probability=1, grid_width=30, grid_height=30, magnitude=8)\np5.process()",
"\rExecuting Pipeline: 0%| | 0/25 [00:00<?, ? Samples/s]"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d094da275464c7e190fa732af847bf2dcef827aa | 146,035 | ipynb | Jupyter Notebook | notebooks/Area_within_Historic_District_Exploratory.ipynb | NYCPlanning/db-equitable-development-tool | b24d83dc4092489995cabcdcb611642c1c8ee3b2 | [
"MIT"
] | 1 | 2021-12-30T21:03:56.000Z | 2021-12-30T21:03:56.000Z | notebooks/Area_within_Historic_District_Exploratory.ipynb | NYCPlanning/db-equitable-development-tool | b24d83dc4092489995cabcdcb611642c1c8ee3b2 | [
"MIT"
] | 209 | 2021-10-20T19:03:04.000Z | 2022-03-31T21:02:37.000Z | notebooks/Area_within_Historic_District_Exploratory.ipynb | NYCPlanning/db-equitable-development-tool | b24d83dc4092489995cabcdcb611642c1c8ee3b2 | [
"MIT"
] | null | null | null | 103.865576 | 33,388 | 0.772376 | [
[
[
"Practice geospatial aggregations in geopandas before writing them to .py files",
"_____no_output_____"
]
],
[
[
"%load_ext autoreload\n%autoreload 2\n",
"_____no_output_____"
],
[
"import sys\nsys.path.append('../utils')\nimport wd_management\nwd_management.set_wd_root()",
"called set_wd_root. current working directory is /Users/alexanderweinstein/Documents/Data_Products/db-equitable-development-tool/notebooks\n"
],
[
"import geopandas as gp\nimport pandas as pd\nimport requests",
"_____no_output_____"
],
[
"res = requests.get('https://services5.arcgis.com/GfwWNkhOj9bNBqoJ/arcgis/rest/services/NYC_Public_Use_Microdata_Areas_PUMAs_2010/FeatureServer/0/query?where=1=1&outFields=*&outSR=4326&f=pgeojson')\nres_json = res.json()",
"_____no_output_____"
],
[
"NYC_PUMAs = gp.GeoDataFrame.from_features(res_json['features'])\nNYC_PUMAs.set_crs('EPSG:4326',inplace=True)\nNYC_PUMAs.set_index('PUMA', inplace=True)\nNYC_PUMAs.head(5)",
"_____no_output_____"
],
[
"NYC_PUMAs.plot()",
"_____no_output_____"
]
],
[
[
"Ok looks good. Load in historic districts. [This stackoverflow post](https://gis.stackexchange.com/questions/327197/typeerror-input-geometry-column-must-contain-valid-geometry-objects) was helpful ",
"_____no_output_____"
]
],
[
[
"from shapely import wkt",
"_____no_output_____"
],
[
"hd= gp.read_file('.library/lpc_historic_district_areas.csv')\nhd['the_geom'] = hd['the_geom'].apply(wkt.loads)\nhd.set_geometry(col='the_geom', inplace=True, crs='EPSG:4326')",
"_____no_output_____"
],
[
"hd= hd.explode(column='the_geom')\nhd.set_geometry('the_geom',inplace=True)\nhd = hd.to_crs('EPSG:2263')\nhd = hd.reset_index()\nhd.plot()",
"/var/folders/tg/01b257jj0dzg5fzzc3p3gj0w0000gq/T/ipykernel_26102/3499734924.py:1: FutureWarning: Currently, index_parts defaults to True, but in the future, it will default to False to be consistent with Pandas. Use `index_parts=True` to keep the current behavior and True/False to silence the warning.\n hd= hd.explode(column='the_geom')\n"
]
],
[
[
"Ok great next do some geospatial analysis. Start only with PUMA 3807 as it has a lot of historic area",
"_____no_output_____"
]
],
[
[
"def fraction_area_historic(PUMA, hd):\n try:\n gdf = gp.GeoDataFrame(geometry = [PUMA.geometry], crs = 'EPSG:4326')\n gdf = gdf.to_crs('EPSG:2263')\n overlay = gp.overlay(hd, gdf, 'intersection')\n if overlay.empty:\n return 0, 0\n else:\n fraction = overlay.area.sum()/gdf.geometry.area.sum()\n return fraction, overlay.area.sum()/(5280**2)\n except Exception as e:\n print(f'broke on {PUMA}')\n print(e)\n",
"_____no_output_____"
],
[
"NYC_PUMAs[['fraction_area_historic', 'total_area_historic']] = NYC_PUMAs.apply(fraction_area_historic, axis=1, args=(hd,), result_type='expand')",
"_____no_output_____"
],
[
"NYC_PUMAs.sort_values('fraction_area_historic', ascending=False)",
"_____no_output_____"
]
],
[
[
"Superimpose PUMA 3801's historic districts on it to see if 38% looks right",
"_____no_output_____"
]
],
[
[
"def visualize_overlay(PUMA):\n test_PUMA = NYC_PUMAs.loc[[PUMA]].to_crs('EPSG:2263')\n base = test_PUMA.plot(color='green', edgecolor='black')\n overlay = gp.overlay(hd, test_PUMA, 'intersection')\n overlay.plot(ax=base, color='red');",
"_____no_output_____"
],
[
"visualize_overlay('3810')",
"_____no_output_____"
]
],
[
[
"Ok great that looks like about a third to me",
"_____no_output_____"
],
[
"From eyeballing map, more than 20% of PUMA 3806 on UWS looks to be historic",
"_____no_output_____"
]
],
[
[
"visualize_overlay('3806')",
"_____no_output_____"
]
],
[
[
"Ah ok the PUMA geography from includes central park. Worth flagging ",
"_____no_output_____"
],
[
"### Question from Renae:\nRenae points out that description of historic districts says \"including items that may have been denied designation or overturned.\"\nLook at dataset to see if columns point to this clearly",
"_____no_output_____"
]
],
[
[
"hd.head(5)",
"_____no_output_____"
],
[
"hd.groupby('status_of_').size()",
"_____no_output_____"
],
[
"hd.groupby('current_').size()",
"_____no_output_____"
],
[
"hd.groupby('last_actio').size()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d094dda4c67366ec220a6713366b1030f8d51d31 | 72,046 | ipynb | Jupyter Notebook | Fine_Tuning_BERT_for_Spam_Classification.ipynb | lamiathu/Fine-Tuning-BERT | 371909ad22aa7110e3b47f2c5d195ef8550acd0d | [
"Apache-2.0"
] | 81 | 2020-08-12T18:31:02.000Z | 2022-03-11T12:00:21.000Z | Fine_Tuning_BERT_for_Spam_Classification.ipynb | C-Ritam98/Fine-Tuning-BERT | 371909ad22aa7110e3b47f2c5d195ef8550acd0d | [
"Apache-2.0"
] | 13 | 2020-09-28T10:05:17.000Z | 2022-01-28T05:29:03.000Z | Fine_Tuning_BERT_for_Spam_Classification.ipynb | C-Ritam98/Fine-Tuning-BERT | 371909ad22aa7110e3b47f2c5d195ef8550acd0d | [
"Apache-2.0"
] | 81 | 2020-07-23T22:35:49.000Z | 2022-03-27T16:39:34.000Z | 35.351325 | 7,254 | 0.499139 | [
[
[
"<a href=\"https://colab.research.google.com/github/prateekjoshi565/Fine-Tuning-BERT/blob/master/Fine_Tuning_BERT_for_Spam_Classification.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Install Transformers Library",
"_____no_output_____"
]
],
[
[
"!pip install transformers",
"_____no_output_____"
],
[
"import numpy as np\nimport pandas as pd\nimport torch\nimport torch.nn as nn\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import classification_report\nimport transformers\nfrom transformers import AutoModel, BertTokenizerFast\n\n# specify GPU\ndevice = torch.device(\"cuda\")",
"_____no_output_____"
]
],
[
[
"# Load Dataset",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv(\"spamdata_v2.csv\")\ndf.head()",
"_____no_output_____"
],
[
"df.shape",
"_____no_output_____"
],
[
"# check class distribution\ndf['label'].value_counts(normalize = True)",
"_____no_output_____"
]
],
[
[
"# Split train dataset into train, validation and test sets",
"_____no_output_____"
]
],
[
[
"train_text, temp_text, train_labels, temp_labels = train_test_split(df['text'], df['label'], \n random_state=2018, \n test_size=0.3, \n stratify=df['label'])\n\n# we will use temp_text and temp_labels to create validation and test set\nval_text, test_text, val_labels, test_labels = train_test_split(temp_text, temp_labels, \n random_state=2018, \n test_size=0.5, \n stratify=temp_labels)",
"_____no_output_____"
]
],
[
[
"# Import BERT Model and BERT Tokenizer",
"_____no_output_____"
]
],
[
[
"# import BERT-base pretrained model\nbert = AutoModel.from_pretrained('bert-base-uncased')\n\n# Load the BERT tokenizer\ntokenizer = BertTokenizerFast.from_pretrained('bert-base-uncased')",
"_____no_output_____"
],
[
"# sample data\ntext = [\"this is a bert model tutorial\", \"we will fine-tune a bert model\"]\n\n# encode text\nsent_id = tokenizer.batch_encode_plus(text, padding=True, return_token_type_ids=False)",
"_____no_output_____"
],
[
"# output\nprint(sent_id)",
"{'input_ids': [[101, 2023, 2003, 1037, 14324, 2944, 14924, 4818, 102, 0], [101, 2057, 2097, 2986, 1011, 8694, 1037, 14324, 2944, 102]], 'attention_mask': [[1, 1, 1, 1, 1, 1, 1, 1, 1, 0], [1, 1, 1, 1, 1, 1, 1, 1, 1, 1]]}\n"
]
],
[
[
"# Tokenization",
"_____no_output_____"
]
],
[
[
"# get length of all the messages in the train set\nseq_len = [len(i.split()) for i in train_text]\n\npd.Series(seq_len).hist(bins = 30)",
"_____no_output_____"
],
[
"max_seq_len = 25",
"_____no_output_____"
],
[
"# tokenize and encode sequences in the training set\ntokens_train = tokenizer.batch_encode_plus(\n train_text.tolist(),\n max_length = max_seq_len,\n pad_to_max_length=True,\n truncation=True,\n return_token_type_ids=False\n)\n\n# tokenize and encode sequences in the validation set\ntokens_val = tokenizer.batch_encode_plus(\n val_text.tolist(),\n max_length = max_seq_len,\n pad_to_max_length=True,\n truncation=True,\n return_token_type_ids=False\n)\n\n# tokenize and encode sequences in the test set\ntokens_test = tokenizer.batch_encode_plus(\n test_text.tolist(),\n max_length = max_seq_len,\n pad_to_max_length=True,\n truncation=True,\n return_token_type_ids=False\n)",
"_____no_output_____"
]
],
[
[
"# Convert Integer Sequences to Tensors",
"_____no_output_____"
]
],
[
[
"# for train set\ntrain_seq = torch.tensor(tokens_train['input_ids'])\ntrain_mask = torch.tensor(tokens_train['attention_mask'])\ntrain_y = torch.tensor(train_labels.tolist())\n\n# for validation set\nval_seq = torch.tensor(tokens_val['input_ids'])\nval_mask = torch.tensor(tokens_val['attention_mask'])\nval_y = torch.tensor(val_labels.tolist())\n\n# for test set\ntest_seq = torch.tensor(tokens_test['input_ids'])\ntest_mask = torch.tensor(tokens_test['attention_mask'])\ntest_y = torch.tensor(test_labels.tolist())",
"_____no_output_____"
]
],
[
[
"# Create DataLoaders",
"_____no_output_____"
]
],
[
[
"from torch.utils.data import TensorDataset, DataLoader, RandomSampler, SequentialSampler\n\n#define a batch size\nbatch_size = 32\n\n# wrap tensors\ntrain_data = TensorDataset(train_seq, train_mask, train_y)\n\n# sampler for sampling the data during training\ntrain_sampler = RandomSampler(train_data)\n\n# dataLoader for train set\ntrain_dataloader = DataLoader(train_data, sampler=train_sampler, batch_size=batch_size)\n\n# wrap tensors\nval_data = TensorDataset(val_seq, val_mask, val_y)\n\n# sampler for sampling the data during training\nval_sampler = SequentialSampler(val_data)\n\n# dataLoader for validation set\nval_dataloader = DataLoader(val_data, sampler = val_sampler, batch_size=batch_size)",
"_____no_output_____"
]
],
[
[
"# Freeze BERT Parameters",
"_____no_output_____"
]
],
[
[
"# freeze all the parameters\nfor param in bert.parameters():\n param.requires_grad = False",
"_____no_output_____"
]
],
[
[
"# Define Model Architecture",
"_____no_output_____"
]
],
[
[
"class BERT_Arch(nn.Module):\n\n def __init__(self, bert):\n \n super(BERT_Arch, self).__init__()\n\n self.bert = bert \n \n # dropout layer\n self.dropout = nn.Dropout(0.1)\n \n # relu activation function\n self.relu = nn.ReLU()\n\n # dense layer 1\n self.fc1 = nn.Linear(768,512)\n \n # dense layer 2 (Output layer)\n self.fc2 = nn.Linear(512,2)\n\n #softmax activation function\n self.softmax = nn.LogSoftmax(dim=1)\n\n #define the forward pass\n def forward(self, sent_id, mask):\n\n #pass the inputs to the model \n _, cls_hs = self.bert(sent_id, attention_mask=mask)\n \n x = self.fc1(cls_hs)\n\n x = self.relu(x)\n\n x = self.dropout(x)\n\n # output layer\n x = self.fc2(x)\n \n # apply softmax activation\n x = self.softmax(x)\n\n return x",
"_____no_output_____"
],
[
"# pass the pre-trained BERT to our define architecture\nmodel = BERT_Arch(bert)\n\n# push the model to GPU\nmodel = model.to(device)",
"_____no_output_____"
],
[
"# optimizer from hugging face transformers\nfrom transformers import AdamW\n\n# define the optimizer\noptimizer = AdamW(model.parameters(), lr = 1e-3)",
"_____no_output_____"
]
],
[
[
"# Find Class Weights",
"_____no_output_____"
]
],
[
[
"from sklearn.utils.class_weight import compute_class_weight\n\n#compute the class weights\nclass_wts = compute_class_weight('balanced', np.unique(train_labels), train_labels)\n\nprint(class_wts)",
"[0.57743559 3.72848948]\n"
],
[
"# convert class weights to tensor\nweights= torch.tensor(class_wts,dtype=torch.float)\nweights = weights.to(device)\n\n# loss function\ncross_entropy = nn.NLLLoss(weight=weights) \n\n# number of training epochs\nepochs = 10",
"_____no_output_____"
]
],
[
[
"# Fine-Tune BERT",
"_____no_output_____"
]
],
[
[
"# function to train the model\ndef train():\n \n model.train()\n\n total_loss, total_accuracy = 0, 0\n \n # empty list to save model predictions\n total_preds=[]\n \n # iterate over batches\n for step,batch in enumerate(train_dataloader):\n \n # progress update after every 50 batches.\n if step % 50 == 0 and not step == 0:\n print(' Batch {:>5,} of {:>5,}.'.format(step, len(train_dataloader)))\n\n # push the batch to gpu\n batch = [r.to(device) for r in batch]\n \n sent_id, mask, labels = batch\n\n # clear previously calculated gradients \n model.zero_grad() \n\n # get model predictions for the current batch\n preds = model(sent_id, mask)\n\n # compute the loss between actual and predicted values\n loss = cross_entropy(preds, labels)\n\n # add on to the total loss\n total_loss = total_loss + loss.item()\n\n # backward pass to calculate the gradients\n loss.backward()\n\n # clip the the gradients to 1.0. It helps in preventing the exploding gradient problem\n torch.nn.utils.clip_grad_norm_(model.parameters(), 1.0)\n\n # update parameters\n optimizer.step()\n\n # model predictions are stored on GPU. So, push it to CPU\n preds=preds.detach().cpu().numpy()\n\n # append the model predictions\n total_preds.append(preds)\n\n # compute the training loss of the epoch\n avg_loss = total_loss / len(train_dataloader)\n \n # predictions are in the form of (no. of batches, size of batch, no. of classes).\n # reshape the predictions in form of (number of samples, no. of classes)\n total_preds = np.concatenate(total_preds, axis=0)\n\n #returns the loss and predictions\n return avg_loss, total_preds",
"_____no_output_____"
],
[
"# function for evaluating the model\ndef evaluate():\n \n print(\"\\nEvaluating...\")\n \n # deactivate dropout layers\n model.eval()\n\n total_loss, total_accuracy = 0, 0\n \n # empty list to save the model predictions\n total_preds = []\n\n # iterate over batches\n for step,batch in enumerate(val_dataloader):\n \n # Progress update every 50 batches.\n if step % 50 == 0 and not step == 0:\n \n # Calculate elapsed time in minutes.\n elapsed = format_time(time.time() - t0)\n \n # Report progress.\n print(' Batch {:>5,} of {:>5,}.'.format(step, len(val_dataloader)))\n\n # push the batch to gpu\n batch = [t.to(device) for t in batch]\n\n sent_id, mask, labels = batch\n\n # deactivate autograd\n with torch.no_grad():\n \n # model predictions\n preds = model(sent_id, mask)\n\n # compute the validation loss between actual and predicted values\n loss = cross_entropy(preds,labels)\n\n total_loss = total_loss + loss.item()\n\n preds = preds.detach().cpu().numpy()\n\n total_preds.append(preds)\n\n # compute the validation loss of the epoch\n avg_loss = total_loss / len(val_dataloader) \n\n # reshape the predictions in form of (number of samples, no. of classes)\n total_preds = np.concatenate(total_preds, axis=0)\n\n return avg_loss, total_preds",
"_____no_output_____"
]
],
[
[
"# Start Model Training",
"_____no_output_____"
]
],
[
[
"# set initial loss to infinite\nbest_valid_loss = float('inf')\n\n# empty lists to store training and validation loss of each epoch\ntrain_losses=[]\nvalid_losses=[]\n\n#for each epoch\nfor epoch in range(epochs):\n \n print('\\n Epoch {:} / {:}'.format(epoch + 1, epochs))\n \n #train model\n train_loss, _ = train()\n \n #evaluate model\n valid_loss, _ = evaluate()\n \n #save the best model\n if valid_loss < best_valid_loss:\n best_valid_loss = valid_loss\n torch.save(model.state_dict(), 'saved_weights.pt')\n \n # append training and validation loss\n train_losses.append(train_loss)\n valid_losses.append(valid_loss)\n \n print(f'\\nTraining Loss: {train_loss:.3f}')\n print(f'Validation Loss: {valid_loss:.3f}')",
"\n Epoch 1 / 10\n Batch 50 of 122.\n Batch 100 of 122.\n\nEvaluating...\n\nTraining Loss: 0.526\nValidation Loss: 0.656\n\n Epoch 2 / 10\n Batch 50 of 122.\n Batch 100 of 122.\n\nEvaluating...\n\nTraining Loss: 0.345\nValidation Loss: 0.231\n\n Epoch 3 / 10\n Batch 50 of 122.\n Batch 100 of 122.\n\nEvaluating...\n\nTraining Loss: 0.344\nValidation Loss: 0.194\n\n Epoch 4 / 10\n Batch 50 of 122.\n Batch 100 of 122.\n\nEvaluating...\n\nTraining Loss: 0.223\nValidation Loss: 0.171\n\n Epoch 5 / 10\n Batch 50 of 122.\n Batch 100 of 122.\n\nEvaluating...\n\nTraining Loss: 0.219\nValidation Loss: 0.178\n\n Epoch 6 / 10\n Batch 50 of 122.\n Batch 100 of 122.\n\nEvaluating...\n\nTraining Loss: 0.215\nValidation Loss: 0.180\n\n Epoch 7 / 10\n Batch 50 of 122.\n Batch 100 of 122.\n\nEvaluating...\n\nTraining Loss: 0.247\nValidation Loss: 0.262\n\n Epoch 8 / 10\n Batch 50 of 122.\n Batch 100 of 122.\n\nEvaluating...\n\nTraining Loss: 0.224\nValidation Loss: 0.217\n\n Epoch 9 / 10\n Batch 50 of 122.\n Batch 100 of 122.\n\nEvaluating...\n\nTraining Loss: 0.217\nValidation Loss: 0.148\n\n Epoch 10 / 10\n Batch 50 of 122.\n Batch 100 of 122.\n\nEvaluating...\n\nTraining Loss: 0.231\nValidation Loss: 0.639\n"
]
],
[
[
"# Load Saved Model",
"_____no_output_____"
]
],
[
[
"#load weights of best model\npath = 'saved_weights.pt'\nmodel.load_state_dict(torch.load(path))",
"_____no_output_____"
]
],
[
[
"# Get Predictions for Test Data",
"_____no_output_____"
]
],
[
[
"# get predictions for test data\nwith torch.no_grad():\n preds = model(test_seq.to(device), test_mask.to(device))\n preds = preds.detach().cpu().numpy()",
"_____no_output_____"
],
[
"# model's performance\npreds = np.argmax(preds, axis = 1)\nprint(classification_report(test_y, preds))",
" precision recall f1-score support\n\n 0 0.99 0.98 0.98 724\n 1 0.88 0.92 0.90 112\n\n accuracy 0.97 836\n macro avg 0.93 0.95 0.94 836\nweighted avg 0.97 0.97 0.97 836\n\n"
],
[
"# confusion matrix\npd.crosstab(test_y, preds)",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d094e23268e267b51e4ce90602268376b058e1ce | 391,550 | ipynb | Jupyter Notebook | 06-Python-APIs_HW/Instructions/starter_code/WeatherPy.ipynb | jmbruner37/6_python_api | 1be67d412dc440c0ebd3f149b387eb4fe31baf27 | [
"ADSL"
] | null | null | null | 06-Python-APIs_HW/Instructions/starter_code/WeatherPy.ipynb | jmbruner37/6_python_api | 1be67d412dc440c0ebd3f149b387eb4fe31baf27 | [
"ADSL"
] | null | null | null | 06-Python-APIs_HW/Instructions/starter_code/WeatherPy.ipynb | jmbruner37/6_python_api | 1be67d412dc440c0ebd3f149b387eb4fe31baf27 | [
"ADSL"
] | null | null | null | 217.406996 | 43,072 | 0.898654 | [
[
[
"# WeatherPy\n----\n\n#### Note\n* Instructions have been included for each segment. You do not have to follow them exactly, but they are included to help you think through the steps.",
"_____no_output_____"
]
],
[
[
"# Dependencies and Setup\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport numpy as np\nimport requests\nimport time\nimport random\nimport json\nimport scipy.stats as st\nfrom scipy.stats import linregress\n\n# Import API key\nfrom api_keys import weather_api_key\nfrom api_keys import g_key\n\n# Incorporated citipy to determine city based on latitude and longitude\n\nfrom citipy import citipy\n\n# Output File (CSV)\noutput_data_file = \"output_data/cities.csv\"\n\n# Range of latitudes and longitudes\nlat_range = (-90, 90)\nlng_range = (-180, 180)",
"_____no_output_____"
]
],
[
[
"## Generate Cities List",
"_____no_output_____"
]
],
[
[
"# List for holding lat_lngs and cities\nlat_lngs = []\ncities = []\n\n# Create a set of random lat and lng combinations\nlats = np.random.uniform(lat_range[0], lat_range[1], size=1500)\nlngs = np.random.uniform(lng_range[0], lng_range[1], size=1500)\nlat_lngs = zip(lats, lngs)\n\n# Identify nearest city for each lat, lng combination\nfor lat_lng in lat_lngs:\n city = citipy.nearest_city(lat_lng[0], lat_lng[1]).city_name\n \n # If the city is unique, then add it to a our cities list\n if city not in cities:\n cities.append(city)\n\n# Print the city count to confirm sufficient count\nlen(cities)",
"_____no_output_____"
],
[
"city_names_ls = []\ncloudiness_ls = []\ncountry_ls = []\ndate_ls = []\nhumidity_ls = []\nlat_ls = []\nlng_ls = []\nmax_temp_ls = []\nwind_speed_ls = []\nindex_counter = 0\nset_counter = 1",
"_____no_output_____"
]
],
[
[
"### Perform API Calls\n* Perform a weather check on each city using a series of successive API calls.\n* Include a print log of each city as it'sbeing processed (with the city number and city name).\n",
"_____no_output_____"
]
],
[
[
"print(\"Beginning Data Retrieval\")\nprint(\"-------------------------------\")\n\nbase_url = \"http://api.openweathermap.org/data/2.5/weather?\"\nunits = \"imperial\"\nurl = f\"{base_url}appid={weather_api_key}&units={units}&q=\"\n\nfor index, city in enumerate(cities, start = 1):\n try:\n response = requests.get(url + city).json()\n city_names_ls.append(response[\"name\"])\n cloudiness_ls.append(response[\"clouds\"][\"all\"])\n country_ls.append(response[\"sys\"][\"country\"])\n date_ls.append(response[\"dt\"])\n humidity_ls.append(response[\"main\"][\"humidity\"])\n lat_ls.append(response[\"coord\"][\"lat\"])\n lng_ls.append(response[\"coord\"][\"lon\"])\n max_temp_ls.append(response[\"main\"][\"temp_max\"])\n wind_speed_ls.append(response[\"wind\"][\"speed\"])\n if index_counter > 49:\n index_counter = 0\n set_counter = set_counter + 1\n \n else:\n index_counter = index_counter + 1\n \n print(f\"Processing Record {index_counter} of Set {set_counter} : {city}\") \n \n except(KeyError, IndexError):\n print(\"City not found. Skipping...\")\n \n \n\nprint(\"-------------------------------\")\nprint(\"Data Retrieval Complete\")\nprint(\"-------------------------------\")",
"Beginning Data Retrieval\n-------------------------------\nProcessing Record 1 of Set 1 : albany\nProcessing Record 2 of Set 1 : terney\nCity not found. Skipping...\nProcessing Record 3 of Set 1 : pedasi\nProcessing Record 4 of Set 1 : wakkanai\nProcessing Record 5 of Set 1 : talnakh\nCity not found. Skipping...\nProcessing Record 6 of Set 1 : mataura\nProcessing Record 7 of Set 1 : bilma\nCity not found. Skipping...\nProcessing Record 8 of Set 1 : makakilo city\nProcessing Record 9 of Set 1 : hobart\nProcessing Record 10 of Set 1 : brae\nProcessing Record 11 of Set 1 : busselton\nProcessing Record 12 of Set 1 : yar-sale\nProcessing Record 13 of Set 1 : bluff\nProcessing Record 14 of Set 1 : norman wells\nProcessing Record 15 of Set 1 : qasigiannguit\nProcessing Record 16 of Set 1 : claresholm\nProcessing Record 17 of Set 1 : ribeira grande\nProcessing Record 18 of Set 1 : hermanus\nProcessing Record 19 of Set 1 : korem\nCity not found. Skipping...\nProcessing Record 20 of Set 1 : balabac\nProcessing Record 21 of Set 1 : moose factory\nProcessing Record 22 of Set 1 : butaritari\nProcessing Record 23 of Set 1 : kapaa\nProcessing Record 24 of Set 1 : longyearbyen\nProcessing Record 25 of Set 1 : oda\nProcessing Record 26 of Set 1 : uri\nProcessing Record 27 of Set 1 : eucaliptus\nProcessing Record 28 of Set 1 : chitradurga\nProcessing Record 29 of Set 1 : rikitea\nProcessing Record 30 of Set 1 : nikolskoye\nProcessing Record 31 of Set 1 : hithadhoo\nProcessing Record 32 of Set 1 : la ronge\nProcessing Record 33 of Set 1 : rocha\nProcessing Record 34 of Set 1 : general roca\nProcessing Record 35 of Set 1 : cape town\nProcessing Record 36 of Set 1 : mar del plata\nProcessing Record 37 of Set 1 : almaznyy\nProcessing Record 38 of Set 1 : dunedin\nProcessing Record 39 of Set 1 : salalah\nProcessing Record 40 of Set 1 : kavaratti\nProcessing Record 41 of Set 1 : teguldet\nProcessing Record 42 of Set 1 : tuatapere\nProcessing Record 43 of Set 1 : ancud\nCity not found. Skipping...\nCity not found. Skipping...\nProcessing Record 44 of Set 1 : port alfred\nProcessing Record 45 of Set 1 : hilo\nProcessing Record 46 of Set 1 : lebu\nCity not found. Skipping...\nProcessing Record 47 of Set 1 : hommelvik\nCity not found. Skipping...\nProcessing Record 48 of Set 1 : mundo novo\nProcessing Record 49 of Set 1 : abnub\nProcessing Record 50 of Set 1 : airai\nProcessing Record 0 of Set 2 : qaanaaq\nProcessing Record 1 of Set 2 : chuy\nProcessing Record 2 of Set 2 : akureyri\nProcessing Record 3 of Set 2 : ponta do sol\nProcessing Record 4 of Set 2 : thompson\nProcessing Record 5 of Set 2 : muqui\nProcessing Record 6 of Set 2 : port elizabeth\nProcessing Record 7 of Set 2 : caucaia\nProcessing Record 8 of Set 2 : chokurdakh\nProcessing Record 9 of Set 2 : nanortalik\nProcessing Record 10 of Set 2 : castro\nProcessing Record 11 of Set 2 : atuona\nProcessing Record 12 of Set 2 : coquimbo\nProcessing Record 13 of Set 2 : alofi\nProcessing Record 14 of Set 2 : punta arenas\nProcessing Record 15 of Set 2 : erzin\nProcessing Record 16 of Set 2 : jalu\nProcessing Record 17 of Set 2 : ushuaia\nProcessing Record 18 of Set 2 : torbay\nProcessing Record 19 of Set 2 : roma\nProcessing Record 20 of Set 2 : parys\nProcessing Record 21 of Set 2 : aitape\nProcessing Record 22 of Set 2 : shache\nProcessing Record 23 of Set 2 : langsa\nProcessing Record 24 of Set 2 : asau\nProcessing Record 25 of Set 2 : ahipara\nProcessing Record 26 of Set 2 : loandjili\nProcessing Record 27 of Set 2 : carnarvon\nProcessing Record 28 of Set 2 : pierre\nProcessing Record 29 of Set 2 : marsh harbour\nProcessing Record 30 of Set 2 : masumbwe\nProcessing Record 31 of Set 2 : puerto ayora\nProcessing Record 32 of Set 2 : pitimbu\nProcessing Record 33 of Set 2 : saldanha\nProcessing Record 34 of Set 2 : igrim\nProcessing Record 35 of Set 2 : yumen\nProcessing Record 36 of Set 2 : tasiilaq\nCity not found. Skipping...\nProcessing Record 37 of Set 2 : new norfolk\nProcessing Record 38 of Set 2 : deputatskiy\nProcessing Record 39 of Set 2 : vao\nProcessing Record 40 of Set 2 : taltal\nProcessing Record 41 of Set 2 : el estor\nProcessing Record 42 of Set 2 : emporia\nProcessing Record 43 of Set 2 : vila velha\nProcessing Record 44 of Set 2 : nelson bay\nProcessing Record 45 of Set 2 : kavieng\nCity not found. Skipping...\nProcessing Record 46 of Set 2 : moate\nProcessing Record 47 of Set 2 : yellowknife\nProcessing Record 48 of Set 2 : ahuimanu\nProcessing Record 49 of Set 2 : jamestown\nProcessing Record 50 of Set 2 : east london\nProcessing Record 0 of Set 3 : deep river\nProcessing Record 1 of Set 3 : hede\nProcessing Record 2 of Set 3 : la orilla\nProcessing Record 3 of Set 3 : hovd\nCity not found. Skipping...\nProcessing Record 4 of Set 3 : atar\nProcessing Record 5 of Set 3 : pevek\nProcessing Record 6 of Set 3 : fortuna\nProcessing Record 7 of Set 3 : constitucion\nCity not found. Skipping...\nProcessing Record 8 of Set 3 : west lake stevens\nProcessing Record 9 of Set 3 : kyaikto\nProcessing Record 10 of Set 3 : constantine\nProcessing Record 11 of Set 3 : monzon\nProcessing Record 12 of Set 3 : bethel\nProcessing Record 13 of Set 3 : san miguel\nProcessing Record 14 of Set 3 : pangnirtung\nCity not found. Skipping...\nProcessing Record 15 of Set 3 : nalut\nCity not found. Skipping...\nProcessing Record 16 of Set 3 : arraial do cabo\nProcessing Record 17 of Set 3 : nedjo\nProcessing Record 18 of Set 3 : bambous virieux\nProcessing Record 19 of Set 3 : birjand\nProcessing Record 20 of Set 3 : sorland\nProcessing Record 21 of Set 3 : victoria\nProcessing Record 22 of Set 3 : nandyal\nProcessing Record 23 of Set 3 : dakar\nProcessing Record 24 of Set 3 : paengaroa\nProcessing Record 25 of Set 3 : vaini\nProcessing Record 26 of Set 3 : port hardy\nProcessing Record 27 of Set 3 : klaksvik\nProcessing Record 28 of Set 3 : culebra\nProcessing Record 29 of Set 3 : mehamn\nProcessing Record 30 of Set 3 : guisa\nProcessing Record 31 of Set 3 : male\nCity not found. Skipping...\nProcessing Record 32 of Set 3 : salym\nProcessing Record 33 of Set 3 : barrow\nProcessing Record 34 of Set 3 : jining\nProcessing Record 35 of Set 3 : patiya\nProcessing Record 36 of Set 3 : itoman\nCity not found. Skipping...\nProcessing Record 37 of Set 3 : korla\nProcessing Record 38 of Set 3 : saint george\nProcessing Record 39 of Set 3 : caravelas\nProcessing Record 40 of Set 3 : touros\nCity not found. Skipping...\nProcessing Record 41 of Set 3 : saint-augustin\nProcessing Record 42 of Set 3 : college\nProcessing Record 43 of Set 3 : hami\nProcessing Record 44 of Set 3 : guerrero negro\nCity not found. Skipping...\nProcessing Record 45 of Set 3 : verkh-usugli\nProcessing Record 46 of Set 3 : krasnoarmeysk\nProcessing Record 47 of Set 3 : kirkwall\nProcessing Record 48 of Set 3 : bredasdorp\nProcessing Record 49 of Set 3 : huilong\nProcessing Record 50 of Set 3 : paita\nProcessing Record 0 of Set 4 : berdigestyakh\nProcessing Record 1 of Set 4 : tiksi\nProcessing Record 2 of Set 4 : hasaki\nProcessing Record 3 of Set 4 : kodiak\nProcessing Record 4 of Set 4 : dikson\nProcessing Record 5 of Set 4 : mahebourg\nProcessing Record 6 of Set 4 : kaseda\nProcessing Record 7 of Set 4 : margate\nProcessing Record 8 of Set 4 : hay river\nProcessing Record 9 of Set 4 : dharchula\nProcessing Record 10 of Set 4 : daru\nProcessing Record 11 of Set 4 : codrington\nProcessing Record 12 of Set 4 : pasni\nCity not found. Skipping...\nProcessing Record 13 of Set 4 : evensk\nProcessing Record 14 of Set 4 : shiyan\nProcessing Record 15 of Set 4 : lexington park\nProcessing Record 16 of Set 4 : buraydah\nProcessing Record 17 of Set 4 : port-gentil\nProcessing Record 18 of Set 4 : maralal\nProcessing Record 19 of Set 4 : agua dulce\nProcessing Record 20 of Set 4 : jian\nProcessing Record 21 of Set 4 : mafinga\nProcessing Record 22 of Set 4 : pisco\nProcessing Record 23 of Set 4 : preobrazheniye\nProcessing Record 24 of Set 4 : ramshir\nProcessing Record 25 of Set 4 : puerto narino\nProcessing Record 26 of Set 4 : keetmanshoop\nProcessing Record 27 of Set 4 : waipawa\nProcessing Record 28 of Set 4 : winneba\nCity not found. Skipping...\nProcessing Record 29 of Set 4 : bitung\nProcessing Record 30 of Set 4 : sept-iles\nProcessing Record 31 of Set 4 : provideniya\nProcessing Record 32 of Set 4 : chapais\nProcessing Record 33 of Set 4 : xushan\nProcessing Record 34 of Set 4 : mount isa\n"
]
],
[
[
"### Convert Raw Data to DataFrame\n* Export the city data into a .csv.\n* Display the DataFrame",
"_____no_output_____"
]
],
[
[
"weather_df = pd.DataFrame({ \n \"City\" : city_names_ls,\n \"Cloudiness\" : cloudiness_ls,\n \"Country\" : country_ls,\n \"Date\" : date_ls,\n \"Humidity\" : humidity_ls,\n \"Lat\" : lat_ls,\n \"Lng\" : lng_ls,\n \"Max Temp\" : max_temp_ls,\n \"Wind Speed\" : wind_speed_ls\n})\nweather_df.count()",
"_____no_output_____"
],
[
"weather_df",
"_____no_output_____"
],
[
"weather_df.to_csv('../output_data/cities.csv')",
"_____no_output_____"
]
],
[
[
"## Inspect the data and remove the cities where the humidity > 100%.\n----\nSkip this step if there are no cities that have humidity > 100%. ",
"_____no_output_____"
]
],
[
[
"# Get the indices of cities that have humidity over 100%.\n",
"_____no_output_____"
],
[
"# Make a new DataFrame equal to the city data to drop all humidity outliers by index.\n# Passing \"inplace=False\" will make a copy of the city_data DataFrame, which we call \"clean_city_data\".\n",
"_____no_output_____"
],
[
"# Extract relevant fields from the data frame\n\n\n# Export the City_Data into a csv\n",
"_____no_output_____"
]
],
[
[
"## Plotting the Data\n* Use proper labeling of the plots using plot titles (including date of analysis) and axes labels.\n* Save the plotted figures as .pngs.",
"_____no_output_____"
],
[
"## Latitude vs. Temperature Plot",
"_____no_output_____"
]
],
[
[
"plt.scatter(weather_df[\"Lat\"], weather_df[\"Max Temp\"], facecolor = \"darkblue\", edgecolor = \"darkgrey\")\n\nplt.title(\"City Latitude vs. Max Temperature (07/13/20)\")\n\nplt.ylabel(\"Max Temperature (F)\")\nplt.xlabel(\"Latitude\")\n\nplt.grid(True)\n\nplt.savefig(\"../Images/City Latitude vs Max Temperature.png\")\n\nplt.show()\nprint(\"This plot shows that as cities get further away from equator that they are cooler\")",
"_____no_output_____"
]
],
[
[
"## Latitude vs. Humidity Plot",
"_____no_output_____"
]
],
[
[
"plt.scatter(weather_df[\"Lat\"], weather_df[\"Humidity\"], facecolor = \"darkblue\", edgecolor = \"darkgrey\")\n\nplt.title(\"City Latitude vs. Humidity (07/13/20)\")\n\nplt.ylabel(\"Humidity (%)\")\nplt.xlabel(\"Latitude\")\n\nplt.grid(True)\n\nplt.savefig(\"../Images/City Latitude vs Humidity.png\")\n\nplt.show()\nprint(\"This plot shows that humidity is well spread throughout cities despite location\")",
"_____no_output_____"
]
],
[
[
"## Latitude vs. Cloudiness Plot",
"_____no_output_____"
]
],
[
[
"plt.scatter(weather_df[\"Lat\"], weather_df[\"Cloudiness\"], facecolor = \"darkblue\", edgecolor = \"darkgrey\")\n\nplt.title(\"City Latitude vs. Cloudiness (07/13/20)\")\n\nplt.ylabel(\"Cloudiness (%)\")\nplt.xlabel(\"Latitude\")\n\nplt.grid(True)\n\nplt.savefig(\"../Images/City Latitude vs Cloudiness.png\")\n\nplt.show()\nprint(\"This plot shows that cloudiness is well spread throughout cities despite location\")",
"_____no_output_____"
]
],
[
[
"## Latitude vs. Wind Speed Plot",
"_____no_output_____"
]
],
[
[
"plt.scatter(weather_df[\"Lat\"], weather_df[\"Wind Speed\"], facecolor = \"darkblue\", edgecolor = \"darkgrey\")\n\nplt.title(\"City Latitude vs. Windspeed (07/13/20)\")\n\nplt.ylabel(\"Windspeed (mph)\")\nplt.xlabel(\"Latitude\")\n\nplt.grid(True)\n\nplt.savefig(\"../Images/City Latitude vs Windspeed.png\")\n\nplt.show()\nprint(\"This plot shows that windspeed is well spread throughout cities despite location\")",
"_____no_output_____"
]
],
[
[
"## Linear Regression",
"_____no_output_____"
]
],
[
[
"# OPTIONAL: Create a function to create Linear Regression plots\nnor_hemi = weather_df.loc[weather_df[\"Lat\"] >= 0]\nsou_hemi = weather_df.loc[weather_df[\"Lat\"] < 0]",
"_____no_output_____"
],
[
"def linear_agression(x,y):\n print(f\"The r-squared is : {round(st.pearsonr(x, y)[0],2)}\")\n (slope, intercept, rvalue, pvalue, stderr) = linregress(x, y)\n regress_values = x * slope + intercept\n line_eq = \"y = \" + str(round(slope,2)) + \"x + \" + str(round(intercept,2))\n plt.scatter(x, y)\n plt.plot(x,regress_values,\"r-\")\n return line_eq\n\ndef annotate(line_eq, a, b):\n plt.annotate(line_eq,(a,b),fontsize=15,color=\"red\")",
"_____no_output_____"
]
],
[
[
"#### Northern Hemisphere - Max Temp vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"equation = linear_agression(nor_hemi[\"Lat\"], nor_hemi[\"Max Temp\"])\n\n\nannotate(equation, 10, 40)\n\n\nplt.ylabel(\"Max Temp (F)\")\nplt.xlabel(\"Latitude\")\n\nplt.savefig(\"../Images/NorHemi Max Temp vs. Latitude Linear Regression.png\")\nprint(\"This linear regression shows that cities gets hotter as get closer to equator in northern hemisphere\")",
"The r-squared is : -0.65\nThis linear regression shows that cities gets hotter as get closer to equator in northern hemisphere\n"
]
],
[
[
"#### Southern Hemisphere - Max Temp vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"equation = linear_agression(sou_hemi[\"Lat\"], sou_hemi[\"Max Temp\"])\n\nannotate(equation, -50, 80)\n\nplt.ylabel(\"Max Temperature (F)\")\nplt.xlabel(\"Latitude\")\n\nplt.savefig(\"../Images/SouHemi Max Temp vs. Latitude Linear Regression.png\")\nprint(\"This linear regression shows that cities gets colder as get away to equator in southern hemisphere\")",
"The r-squared is : 0.74\n"
]
],
[
[
"#### Northern Hemisphere - Humidity (%) vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"equation = linear_agression(nor_hemi[\"Lat\"], nor_hemi[\"Humidity\"])\n\n\nannotate(equation, 1, 5)\n\n\nplt.ylabel(\"Humidity\")\nplt.xlabel(\"Latitude\")\n\n\nplt.savefig(\"../Images/NorHemi Humidity vs. Latitude Linear Regression.png\")\nprint(\"This linear regression shows that cities' humidity doesn't change much as get closer to equator in northern hemisphere\")",
"The r-squared is : -0.08\n"
]
],
[
[
"#### Southern Hemisphere - Humidity (%) vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"equation = linear_agression(sou_hemi[\"Lat\"], sou_hemi[\"Humidity\"])\n\nannotate(equation, -50, 20)\n\nplt.ylabel(\"Humidity\")\nplt.xlabel(\"Latitude\")\n\nplt.savefig(\"../Images/SouHemi Humidity vs. Latitude Linear Regression.png\")\nprint(\"This linear regression shows that cities' humidity doesn't change much as get closer to equator in southern hemisphere\")",
"The r-squared is : 0.09\n"
]
],
[
[
"#### Northern Hemisphere - Cloudiness (%) vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"equation = linear_agression(nor_hemi[\"Lat\"], nor_hemi[\"Cloudiness\"])\n\n\nannotate(equation, 0, 0)\n\n\nplt.ylabel(\"Cloudiness\")\nplt.xlabel(\"Latitude\")\n\n\nplt.savefig(\"../Images/NorHemi Cloudiness vs. Latitude Linear Regression.png\")\nprint(\"This linear regression shows that cities' cloudiness doesn't change much as get closer to equator in northern hemisphere\")",
"The r-squared is : -0.01\n"
]
],
[
[
"#### Southern Hemisphere - Cloudiness (%) vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"equation = linear_agression(sou_hemi[\"Lat\"], sou_hemi[\"Cloudiness\"])\n\nannotate(equation, -50, 50)\n\nplt.ylabel(\"Cloudiness\")\nplt.xlabel(\"Latitude\")\n\nplt.savefig(\"../Images/SouHemi Cloudiness vs. Latitude Linear Regression.png\")\nprint(\"This linear regression shows that cities' cloudiness doesn't change much as get closer to equator in southern hemisphere\")",
"The r-squared is : 0.1\n"
]
],
[
[
"#### Northern Hemisphere - Wind Speed (mph) vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"equation = linear_agression(nor_hemi[\"Lat\"], nor_hemi[\"Wind Speed\"])\n\n\nannotate(equation, 40, 25)\n\n\nplt.ylabel(\"Wind Speed\")\nplt.xlabel(\"Latitude\")\n\n\nplt.savefig(\"../Images/NorHemi Wind Speed vs. Latitude Linear Regression.png\")\nprint(\"This linear regression shows that cities' windiness doesn't change much as get closer to equator in northern hemisphere\")",
"The r-squared is : -0.0\n"
]
],
[
[
"#### Southern Hemisphere - Wind Speed (mph) vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"equation = linear_agression(sou_hemi[\"Lat\"], sou_hemi[\"Wind Speed\"])\n\nannotate(equation, -50, 25)\n\nplt.ylabel(\"Wind Speed\")\nplt.xlabel(\"Latitude\")\n\nplt.savefig(\"../Images/SouHemi Wind Speed vs. Latitude Linear Regression.png\")\nprint(\"This linear regression shows that cities' windiness doesn't change much as get closer to equator in northern hemisphere\")",
"The r-squared is : -0.24\n"
],
[
"#Three Observable Trends\n\n#1- Out of the cities analyzed, tends to be hotter by the equatator\n#2- This time of year there tends to be more wind in cities away from equator in southern hemisphere\n#3- Humidty, cloudiness, and wind speeds did not any obvious trends in the nothern hemisphere.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d094e5648dd98e86aa0e563087967c60546f6cff | 48,242 | ipynb | Jupyter Notebook | class3_null_and_conditions/identify_null.ipynb | techsparksguru/data_analysis_pandas_spark_koalas | 384801961f84d94f4c68901332bf1874d0ac5929 | [
"Apache-2.0"
] | 3 | 2020-07-17T16:46:43.000Z | 2022-02-09T10:49:50.000Z | class3_null_and_conditions/identify_null.ipynb | techsparksguru/pandas_basic_training | 3caacf9e8b166d5bc19a508795a06a9024ca4bf9 | [
"Apache-2.0"
] | null | null | null | class3_null_and_conditions/identify_null.ipynb | techsparksguru/pandas_basic_training | 3caacf9e8b166d5bc19a508795a06a9024ca4bf9 | [
"Apache-2.0"
] | 2 | 2020-04-08T10:53:04.000Z | 2021-03-10T03:26:06.000Z | 26.608935 | 112 | 0.299656 | [
[
[
"import pandas as pd\ndf = pd.read_csv('../height_weight.csv')",
"_____no_output_____"
],
[
"# Observe the count of values in each column\ndf.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 200 entries, 0 to 199\nData columns (total 5 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 sex 200 non-null object \n 1 weight 200 non-null int64 \n 2 height 200 non-null int64 \n 3 repwt 183 non-null float64\n 4 repht 183 non-null float64\ndtypes: float64(2), int64(2), object(1)\nmemory usage: 7.9+ KB\n"
],
[
"# Are there any null/NaN values in the dataframe\ndf.isnull()",
"_____no_output_____"
],
[
"# Which columns have at least one null value ?\ndf.isnull().any()",
"_____no_output_____"
],
[
"# Print and see \ndf[['height','weight','repwt']].head(20)",
"_____no_output_____"
],
[
"# Which columns have all null values ?\ndf.isnull().all()",
"_____no_output_____"
],
[
"# Which rows have at least one null value?\ndf.isnull().any(axis=1)",
"_____no_output_____"
],
[
"#which rows have ALL null values ?\ndf.isnull().all(axis=1)",
"_____no_output_____"
],
[
"# Counting is difficult, so chain any() again. This tells us there is every row with at least one value\ndf.isnull().all(axis=1).any()",
"_____no_output_____"
],
[
"# Get all rows where at least one value in the row is null\ndf[df.isnull().any(axis=1)]",
"_____no_output_____"
],
[
"# Which columns do NOT have null value ?\ndf.notnull().any()",
"_____no_output_____"
],
[
"df['repwt'].head(20)",
"_____no_output_____"
],
[
"# Get rows that have MIN_TEMP_GROUND null\ndf[df['repwt'].isnull()]",
"_____no_output_____"
],
[
"# Get rows that have MIN_TEMP_GROUND not null\ndf[df['repwt'].notnull()]",
"_____no_output_____"
],
[
"list_of_not_null_indices = df[df['repwt'].notnull()].index.values.tolist()",
"_____no_output_____"
],
[
"# Define a range starting from 5 until 200 with steps of 2, convert to list\nx = list(range(5,200,2))\nfor i in x:\n print(i)",
"5\n7\n9\n11\n13\n15\n17\n19\n21\n23\n25\n27\n29\n31\n33\n35\n37\n39\n41\n43\n45\n47\n49\n51\n53\n55\n57\n59\n61\n63\n65\n67\n69\n71\n73\n75\n77\n79\n81\n83\n85\n87\n89\n91\n93\n95\n97\n99\n101\n103\n105\n107\n109\n111\n113\n115\n117\n119\n121\n123\n125\n127\n129\n131\n133\n135\n137\n139\n141\n143\n145\n147\n149\n151\n153\n155\n157\n159\n161\n163\n165\n167\n169\n171\n173\n175\n177\n179\n181\n183\n185\n187\n189\n191\n193\n195\n197\n199\n"
],
[
"if x == list_of_not_null_indices:\n print(\"equal\")",
"_____no_output_____"
],
[
"# Drop all rows that have MIN_TEMP_GROUND null \ndf.drop(df[df['repwt'].isnull()].index.values.tolist())",
"_____no_output_____"
],
[
"# Observe that the original dataframe is still the same. So how do you change original dataframe ?\ndf",
"_____no_output_____"
],
[
"# Drop all rows that have MIN_TEMP_GROUND null with inplace=True\ndf.drop(df[df['repwt'].isnull()].index.values.tolist(),inplace=True)",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"# Maybe you want to sent this data to someone, save it to file \ndf.to_xlsx('blah.xlsx')",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0950351c1b11269acd58183cb7d43411bfb01c4 | 8,743 | ipynb | Jupyter Notebook | 4_diciembre1358.ipynb | jorge23amury/daa_2021_1 | e41fb096cc6c26839f4f10af4b5a4abcc450a12d | [
"MIT"
] | null | null | null | 4_diciembre1358.ipynb | jorge23amury/daa_2021_1 | e41fb096cc6c26839f4f10af4b5a4abcc450a12d | [
"MIT"
] | null | null | null | 4_diciembre1358.ipynb | jorge23amury/daa_2021_1 | e41fb096cc6c26839f4f10af4b5a4abcc450a12d | [
"MIT"
] | null | null | null | 36.128099 | 570 | 0.385451 | [
[
[
"<a href=\"https://colab.research.google.com/github/jorge23amury/daa_2021_1/blob/master/4_diciembre1358.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"\"\"\"\nArray2D\n\"\"\"\n\nclass Array2D:\n\n def __init__(self,rows, cols, value):\n self.__cols = cols\n self.__rows = rows\n self.__array=[[value for x in range(self.__cols)] for y in range(self.__rows)]\n\n def to_string(self):\n [print(\"---\",end=\"\") for x in range(self.__cols)]\n print(\"\")\n for ren in self.__array:\n print(ren)\n [print(\"---\",end=\"\") for x in range(self.__cols)]\n print(\"\")\n\n def get_num_rows(self):\n return self.__rows\n\n def get_num_cols(self):\n return self.__cols\n\n def get_item(self,row,col):\n return self.__array[row][col]\n\n def set_item( self , row , col , valor ):\n self.__array[row][col]=valor\n\n def clearing(self, valor=0):\n for ren in range(self.__rows):\n for col in range(self.__cols):\n self.__array[ren][col]=valor",
"_____no_output_____"
],
[
"class Stack:\n def __init__(self):\n self.__data = []\n self.__size = 0\n\n def pop(self):\n return self.__data.pop()\n\n def get_size(self):\n return self.__size\n\n def peak(self):\n if len(self.__data) > 0:\n return self.__data[-1]\n else:\n return None\n\n def push(self,value):\n self.__data.append(value)\n self.__size += 1\n\n def to_string(self):\n print(\"-\"*6)\n for dato in self.__data[::-1]:\n print(f\"| {dato} |\")\n print(\"/\" * 6)\n print(\"\")\n",
"_____no_output_____"
],
[
"class Laberinto_ADT:\n def __init__(self, archivo):\n self.__laberinto = Array2D(0, 0, 0)\n self.__camino = Stack()\n self.__rens = 0\n self.__cols = 0\n self.__entrada = (0, 0)\n entrada = open(archivo, 'rt')\n datos = entrada.readlines()\n #print(datos)\n self.__rens = int(datos.pop(0).strip())\n self.__cols = int(datos.pop(0).strip())\n self.__entrada = list(datos[0].strip().split(','))\n self.__entrada[0] = int(self.__entrada[0])\n self.__entrada[1] = int(self.__entrada[1])\n self.__camino.push((self.__entrada[1],[0]))\n\n\n datos.pop(0) #eleminamos la tupla\n\n print(self.__rens, self.__cols, self.__entrada)\n print(datos)\n\n self.__laberinto = Array2D(self.__rens, self.__cols, '1')\n for renglon in range(self.__rens):\n info_ren = datos[renglon].strip().split(',')\n\n for columna in range(self.__cols):\n self.__laberinto.set_item(renglon, columna, info_ren[columna])\n\n self.__laberinto.to_string()\n \n def resolver(self):\n actual = self.__camino.peek()\n\n\n def imprime_camino(self):\n self.__camino.to_string()\n\n \n def mostrar(self):\n self.__laberinto.to_string()\n\n if self.__laberinto.get_item( actual[0], actual[1] - 1 ) == '0' and self.__laberinto.get_item( actual[0], actual[1] - 1 ) != 'X' and self.__previa ! = actual[0], actual[1] - 1:\n self.__previa = actual\n self.__camino.push(actual[0], actual[1]-1)\n\n\n elif self.__laberinto.get_item(actual[0]-1 , actual[1]) == '0' and self.__\n \n \n elif l ==2 :\n pass\n elif l== 2:\n pass\n else:\n self.__laberinto.set_item(actual[0])\n def otros():\n pass\n",
"_____no_output_____"
],
[
"#main\nlaberinto = Laberinto_ADT(\"entrada.txt\")\nlaberinto.mostrar()\nlaberinto.imprime_camino()",
"7 7 [6, 2]\n['1,1,1,1,1,1,1\\n', '1,1,1,1,1,1,1\\n', '1,1,1,1,1,1,1\\n', '1,1,1,1,1,1,1\\n', '1,0,0,0,0,0,S\\n', '1,1,0,1,1,1,1\\n', '1,1,E,1,1,1,1']\n---------------------\n['1', '1', '1', '1', '1', '1', '1']\n['1', '1', '1', '1', '1', '1', '1']\n['1', '1', '1', '1', '1', '1', '1']\n['1', '1', '1', '1', '1', '1', '1']\n['1', '0', '0', '0', '0', '0', 'S']\n['1', '1', '0', '1', '1', '1', '1']\n['1', '1', 'E', '1', '1', '1', '1']\n---------------------\n---------------------\n['1', '1', '1', '1', '1', '1', '1']\n['1', '1', '1', '1', '1', '1', '1']\n['1', '1', '1', '1', '1', '1', '1']\n['1', '1', '1', '1', '1', '1', '1']\n['1', '0', '0', '0', '0', '0', 'S']\n['1', '1', '0', '1', '1', '1', '1']\n['1', '1', 'E', '1', '1', '1', '1']\n---------------------\n------\n| (2, [0]) |\n//////\n\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d0952dc807de69587c259d488e2dd52a9acdd004 | 13,141 | ipynb | Jupyter Notebook | projects/crispy_shifties/03_fold_bound_states.ipynb | proleu/crispy_shifty | 87393581a67cbeda287f858a8860145b6ccb5768 | [
"MIT"
] | 4 | 2022-01-11T23:40:12.000Z | 2022-03-03T00:42:57.000Z | projects/crispy_shifties/03_fold_bound_states.ipynb | proleu/crispy_shifty | 87393581a67cbeda287f858a8860145b6ccb5768 | [
"MIT"
] | null | null | null | projects/crispy_shifties/03_fold_bound_states.ipynb | proleu/crispy_shifty | 87393581a67cbeda287f858a8860145b6ccb5768 | [
"MIT"
] | null | null | null | 26.818367 | 299 | 0.556655 | [
[
[
"# Run hacked AlphaFold2 on the designed bound states",
"_____no_output_____"
],
[
"### Imports",
"_____no_output_____"
]
],
[
[
"%load_ext lab_black\n# Python standard library\nfrom glob import glob\nimport os\nimport socket\nimport sys\n\n# 3rd party library imports\nimport dask\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport pyrosetta\nimport numpy as np\nimport scipy\nimport seaborn as sns\nfrom tqdm.auto import tqdm # jupyter compatible progress bar\n\ntqdm.pandas() # link tqdm to pandas\n# Notebook magic\n# save plots in the notebook\n%matplotlib inline\n# reloads modules automatically before executing cells\n%load_ext autoreload\n%autoreload 2\nprint(f\"running in directory: {os.getcwd()}\") # where are we?\nprint(f\"running on node: {socket.gethostname()}\") # what node are we on?",
"running in directory: /mnt/home/pleung/projects/crispy_shifty/projects/crispy_shifties\nrunning on node: dig154\n"
]
],
[
[
"### Set working directory to the root of the crispy_shifty repo\nTODO set to projects dir",
"_____no_output_____"
]
],
[
[
"os.chdir(\"/home/pleung/projects/crispy_shifty\")\n# os.chdir(\"/projects/crispy_shifty\")",
"_____no_output_____"
]
],
[
[
"### Run AF2 on the designed bound states\nTODO",
"_____no_output_____"
]
],
[
[
"from crispy_shifty.utils.io import gen_array_tasks\n\nsimulation_name = \"03_fold_bound_states\"\ndesign_list_file = os.path.join(\n os.getcwd(),\n \"projects/crispy_shifties/02_mpnn_bound_states/test_mpnn_states.pair\", # TODO\n)\noutput_path = os.path.join(os.getcwd(), f\"projects/crispy_shifties/{simulation_name}\")\n\noptions = \" \".join(\n [\n \"out:level 200\",\n ]\n)\nextra_kwargs = {\"models\": \"1\"}\n\ngen_array_tasks(\n distribute_func=\"crispy_shifty.protocols.folding.fold_bound_state\",\n design_list_file=design_list_file,\n output_path=output_path,\n queue=\"gpu\", # TODO\n cores=2,\n memory=\"16G\", # TODO\n gres=\"--gres=gpu:rtx2080:1\", # TODO\n # TODO perlmutter_mode=True,\n nstruct=1,\n nstruct_per_task=1,\n options=options,\n extra_kwargs=extra_kwargs,\n simulation_name=simulation_name,\n)",
"/projects/crispy_shifty/envs/crispy/lib/python3.8/site-packages/dask_jobqueue/core.py:20: FutureWarning: tmpfile is deprecated and will be removed in a future release. Please use dask.utils.tmpfile instead.\n from distributed.utils import tmpfile\n"
],
[
"# !sbatch -a 1-$(cat /mnt/home/pleung/projects/crispy_shifty/projects/crispy_shifties/02_mpnn_bound_states/tasks.cmds | wc -l) /mnt/home/pleung/projects/crispy_shifty/projects/crispy_shifties/02_mpnn_bound_states/run.sh",
"_____no_output_____"
]
],
[
[
"### Collect scorefiles of designed bound states and concatenate\nTODO change to projects dir",
"_____no_output_____"
]
],
[
[
"sys.path.insert(0, \"~/projects/crispy_shifty\") # TODO\nfrom crispy_shifty.utils.io import collect_score_file\n\nsimulation_name = \"03_fold_bound_states\"\noutput_path = os.path.join(os.getcwd(), f\"projects/crispy_shifties/{simulation_name}\")\n\nif not os.path.exists(os.path.join(output_path, \"scores.json\")):\n collect_score_file(output_path, \"scores\")",
"/projects/crispy_shifty/envs/crispy/lib/python3.8/site-packages/dask_jobqueue/core.py:20: FutureWarning: tmpfile is deprecated and will be removed in a future release. Please use dask.utils.tmpfile instead.\n from distributed.utils import tmpfile\n"
]
],
[
[
"### Load resulting concatenated scorefile\nTODO change to projects dir",
"_____no_output_____"
]
],
[
[
"sys.path.insert(0, \"~/projects/crispy_shifty\") # TODO\nfrom crispy_shifty.utils.io import parse_scorefile_linear\n\noutput_path = os.path.join(os.getcwd(), f\"projects/crispy_shifties/{simulation_name}\")\n\nscores_df = parse_scorefile_linear(os.path.join(output_path, \"scores.json\"))\nscores_df = scores_df.convert_dtypes()",
"_____no_output_____"
]
],
[
[
"### Setup for plotting",
"_____no_output_____"
]
],
[
[
"sns.set(\n context=\"talk\",\n font_scale=1, # make the font larger; default is pretty small\n style=\"ticks\", # make the background white with black lines\n palette=\"colorblind\", # a color palette that is colorblind friendly!\n)",
"_____no_output_____"
]
],
[
[
"### Data exploration\nGonna remove the Rosetta sfxn scoreterms for now",
"_____no_output_____"
]
],
[
[
"from crispy_shifty.protocols.design import beta_nov16_terms\n\nscores_df = scores_df[\n [term for term in scores_df.columns if term not in beta_nov16_terms]\n]\nprint(len(scores_df))",
"1621\n"
],
[
"j = 0\nfor i, r in scores_df.iterrows():\n if (r[\"designed_by\"]) == \"rosetta\":\n j += 1\n\nprint(j)",
"10\n"
]
],
[
[
"### Save a list of outputs",
"_____no_output_____"
]
],
[
[
"# simulation_name = \"03_fold_bound_states\"\n# output_path = os.path.join(os.getcwd(), f\"projects/crispy_shifties/{simulation_name}\")\n\n# with open(os.path.join(output_path, \"folded_states.list\"), \"w\") as f:\n# for path in tqdm(scores_df.index):\n# print(path, file=f)",
"_____no_output_____"
]
],
[
[
"### Prototyping blocks",
"_____no_output_____"
],
[
"test `fold_bound_state`",
"_____no_output_____"
]
],
[
[
"%%time \nfrom operator import gt, lt\nimport pyrosetta\n\nfilter_dict = {\n \"mean_plddt\": (gt, 85.0),\n \"rmsd_to_reference\": (lt, 2.2),\n \"mean_pae_interaction\": (lt, 10.0),\n}\n\nrank_on = \"mean_plddt\"\nprefix = \"mpnn_seq\"\n\npyrosetta.init()\n\n\nsys.path.insert(0, \"~/projects/crispy_shifty/\") # TODO projects\nfrom crispy_shifty.protocols.folding import fold_bound_state\n\nt = fold_bound_state(\n None,\n **{\n 'fasta_path': '/mnt/home/pleung/projects/crispy_shifty/projects/crispy_shifties/02_mpnn_bound_states/fastas/0000/02_mpnn_bound_states_25a76fae39514121922e2b477b5b9813.fa',\n \"filter_dict\": filter_dict,\n \"models\": [1], # TODO\n 'pdb_path': '/mnt/home/pleung/projects/crispy_shifty/projects/crispy_shifties/02_mpnn_bound_states/decoys/0000/02_mpnn_bound_states_25a76fae39514121922e2b477b5b9813.pdb.bz2',\n 'prefix': prefix,\n 'rank_on': rank_on,\n# 'fasta_path': 'bar.fa',\n# \"models\": [1, 2], # TODO\n# 'pdb_path': 'foo.pdb.bz2',\n \n }\n)\nfor i, tppose in enumerate(t):\n tppose.pose.dump_pdb(f\"{i}.pdb\")",
"_____no_output_____"
],
[
"tppose.pose.scores",
"_____no_output_____"
]
],
[
[
"test `generate_decoys_from_pose`",
"_____no_output_____"
]
],
[
[
"from operator import gt, lt\nfrom crispy_shifty.protocols.folding import generate_decoys_from_pose\n\nfilter_dict = {\n \"mean_plddt\": (gt, 85.0),\n \"rmsd_to_reference\": (lt, 2.2),\n \"mean_pae_interaction\": (lt, 10.0),\n}\n\nrank_on = \"mean_plddt\"\nprefix = \"mpnn_seq\"\n\ntpose = tppose.pose.clone()\n\ngenr = generate_decoys_from_pose(\n tpose, prefix=prefix, rank_on=rank_on, filter_dict=filter_dict\n)\nfor d in genr:\n print(d.sequence())",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0952dc941869268542cd94a83ae22fe611fac21 | 144,195 | ipynb | Jupyter Notebook | Train_and_Test_Notebooks/ResNet101_RoadTest.ipynb | parshwa1999/Map-Segmentation | 0c45c887fb2363125771bccafc27a953ad05ce5a | [
"MIT"
] | 5 | 2020-03-23T18:43:00.000Z | 2021-12-14T09:52:12.000Z | Train_and_Test_Notebooks/ResNet101_RoadTest.ipynb | parshwa1999/Map-Segmentation | 0c45c887fb2363125771bccafc27a953ad05ce5a | [
"MIT"
] | null | null | null | Train_and_Test_Notebooks/ResNet101_RoadTest.ipynb | parshwa1999/Map-Segmentation | 0c45c887fb2363125771bccafc27a953ad05ce5a | [
"MIT"
] | 1 | 2020-09-08T12:34:39.000Z | 2020-09-08T12:34:39.000Z | 77.943243 | 6,832 | 0.636194 | [
[
[
"<a href=\"https://colab.research.google.com/github/parshwa1999/Map-Segmentation/blob/master/ResNet_RoadTest.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Segmentation of Road from Satellite imagery",
"_____no_output_____"
],
[
"## Importing Libraries",
"_____no_output_____"
]
],
[
[
"import warnings\nwarnings.filterwarnings('ignore')\n\nimport os\nimport cv2\n#from google.colab.patches import cv2_imshow\nimport numpy as np\nimport tensorflow as tf\nimport pandas as pd\nfrom keras.models import Model, load_model\nfrom skimage.morphology import label\nimport pickle\nfrom keras import backend as K\n\nfrom matplotlib import pyplot as plt\nfrom tqdm import tqdm_notebook\nimport random\nfrom skimage.io import imread, imshow, imread_collection, concatenate_images\nfrom matplotlib import pyplot as plt\nimport h5py\n\nseed = 56",
"_____no_output_____"
],
[
"from google.colab import drive\ndrive.mount('/content/gdrive/')\nbase_path = \"gdrive/My\\ Drive/MapSegClean/\"\n%cd gdrive/My\\ Drive/MapSegClean/",
"Drive already mounted at /content/gdrive/; to attempt to forcibly remount, call drive.mount(\"/content/gdrive/\", force_remount=True).\n/content/gdrive/My Drive/MapSegClean\n"
]
],
[
[
"## Defining Custom Loss functions and accuracy Metric.",
"_____no_output_____"
]
],
[
[
"#Source: https://towardsdatascience.com/metrics-to-evaluate-your-semantic-segmentation-model-6bcb99639aa2\nfrom keras import backend as K\ndef iou_coef(y_true, y_pred, smooth=1):\n intersection = K.sum(K.abs(y_true * y_pred), axis=[1,2,3])\n union = K.sum(y_true,[1,2,3])+K.sum(y_pred,[1,2,3])-intersection\n iou = K.mean((intersection + smooth) / (union + smooth), axis=0)\n \n return iou",
"_____no_output_____"
],
[
"def dice_coef(y_true, y_pred, smooth = 1):\n y_true_f = K.flatten(y_true)\n y_pred_f = K.flatten(y_pred)\n intersection = K.sum(y_true_f * y_pred_f)\n return (2. * intersection + smooth) / (K.sum(y_true_f) + K.sum(y_pred_f) + smooth)\n\ndef soft_dice_loss(y_true, y_pred):\n return 1-dice_coef(y_true, y_pred)",
"_____no_output_____"
]
],
[
[
"## Defining Our Model",
"_____no_output_____"
]
],
[
[
"pip install -U segmentation-models",
"Requirement already up-to-date: segmentation-models in /usr/local/lib/python3.6/dist-packages (1.0.1)\nRequirement already satisfied, skipping upgrade: image-classifiers==1.0.0 in /usr/local/lib/python3.6/dist-packages (from segmentation-models) (1.0.0)\nRequirement already satisfied, skipping upgrade: efficientnet==1.0.0 in /usr/local/lib/python3.6/dist-packages (from segmentation-models) (1.0.0)\nRequirement already satisfied, skipping upgrade: keras-applications<=1.0.8,>=1.0.7 in /usr/local/lib/python3.6/dist-packages (from segmentation-models) (1.0.8)\nRequirement already satisfied, skipping upgrade: scikit-image in /usr/local/lib/python3.6/dist-packages (from efficientnet==1.0.0->segmentation-models) (0.16.2)\nRequirement already satisfied, skipping upgrade: h5py in /usr/local/lib/python3.6/dist-packages (from keras-applications<=1.0.8,>=1.0.7->segmentation-models) (2.8.0)\nRequirement already satisfied, skipping upgrade: numpy>=1.9.1 in /usr/local/lib/python3.6/dist-packages (from keras-applications<=1.0.8,>=1.0.7->segmentation-models) (1.17.5)\nRequirement already satisfied, skipping upgrade: imageio>=2.3.0 in /usr/local/lib/python3.6/dist-packages (from scikit-image->efficientnet==1.0.0->segmentation-models) (2.4.1)\nRequirement already satisfied, skipping upgrade: matplotlib!=3.0.0,>=2.0.0 in /usr/local/lib/python3.6/dist-packages (from scikit-image->efficientnet==1.0.0->segmentation-models) (3.1.3)\nRequirement already satisfied, skipping upgrade: PyWavelets>=0.4.0 in /usr/local/lib/python3.6/dist-packages (from scikit-image->efficientnet==1.0.0->segmentation-models) (1.1.1)\nRequirement already satisfied, skipping upgrade: scipy>=0.19.0 in /usr/local/lib/python3.6/dist-packages (from scikit-image->efficientnet==1.0.0->segmentation-models) (1.4.1)\nRequirement already satisfied, skipping upgrade: pillow>=4.3.0 in /usr/local/lib/python3.6/dist-packages (from scikit-image->efficientnet==1.0.0->segmentation-models) (6.2.2)\nRequirement already satisfied, skipping upgrade: networkx>=2.0 in /usr/local/lib/python3.6/dist-packages (from scikit-image->efficientnet==1.0.0->segmentation-models) (2.4)\nRequirement already satisfied, skipping upgrade: six in /usr/local/lib/python3.6/dist-packages (from h5py->keras-applications<=1.0.8,>=1.0.7->segmentation-models) (1.12.0)\nRequirement already satisfied, skipping upgrade: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in /usr/local/lib/python3.6/dist-packages (from matplotlib!=3.0.0,>=2.0.0->scikit-image->efficientnet==1.0.0->segmentation-models) (2.4.6)\nRequirement already satisfied, skipping upgrade: cycler>=0.10 in /usr/local/lib/python3.6/dist-packages (from matplotlib!=3.0.0,>=2.0.0->scikit-image->efficientnet==1.0.0->segmentation-models) (0.10.0)\nRequirement already satisfied, skipping upgrade: python-dateutil>=2.1 in /usr/local/lib/python3.6/dist-packages (from matplotlib!=3.0.0,>=2.0.0->scikit-image->efficientnet==1.0.0->segmentation-models) (2.6.1)\nRequirement already satisfied, skipping upgrade: kiwisolver>=1.0.1 in /usr/local/lib/python3.6/dist-packages (from matplotlib!=3.0.0,>=2.0.0->scikit-image->efficientnet==1.0.0->segmentation-models) (1.1.0)\nRequirement already satisfied, skipping upgrade: decorator>=4.3.0 in /usr/local/lib/python3.6/dist-packages (from networkx>=2.0->scikit-image->efficientnet==1.0.0->segmentation-models) (4.4.1)\nRequirement already satisfied, skipping upgrade: setuptools in /usr/local/lib/python3.6/dist-packages (from kiwisolver>=1.0.1->matplotlib!=3.0.0,>=2.0.0->scikit-image->efficientnet==1.0.0->segmentation-models) (45.1.0)\n"
],
[
"from keras.models import Model, load_model\nimport tensorflow as tf\nfrom keras.layers import Input\nfrom keras.layers.core import Dropout, Lambda\nfrom keras.layers.convolutional import Conv2D, Conv2DTranspose\nfrom keras.layers.pooling import MaxPooling2D\nfrom keras.layers.merge import concatenate\nfrom keras import optimizers\nfrom keras.layers import BatchNormalization\nimport keras\n\nfrom segmentation_models import Unet\nfrom segmentation_models import get_preprocessing\nfrom segmentation_models.losses import bce_jaccard_loss\nfrom segmentation_models.metrics import iou_score",
"Segmentation Models: using `keras` framework.\n"
],
[
"model = Unet('resnet101', input_shape=(256, 256, 3), encoder_weights=None)\n#model = Unet(input_shape=(256, 256, 3), weights=None, activation='elu')\nmodel.summary()\n\n# fit model\n",
"WARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:541: The name tf.placeholder is deprecated. Please use tf.compat.v1.placeholder instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:66: The name tf.get_default_graph is deprecated. Please use tf.compat.v1.get_default_graph instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:190: The name tf.get_default_session is deprecated. Please use tf.compat.v1.get_default_session instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:197: The name tf.ConfigProto is deprecated. Please use tf.compat.v1.ConfigProto instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:203: The name tf.Session is deprecated. Please use tf.compat.v1.Session instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:207: The name tf.global_variables is deprecated. Please use tf.compat.v1.global_variables instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:216: The name tf.is_variable_initialized is deprecated. Please use tf.compat.v1.is_variable_initialized instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:223: The name tf.variables_initializer is deprecated. Please use tf.compat.v1.variables_initializer instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:2041: The name tf.nn.fused_batch_norm is deprecated. Please use tf.compat.v1.nn.fused_batch_norm instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:148: The name tf.placeholder_with_default is deprecated. Please use tf.compat.v1.placeholder_with_default instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:4432: The name tf.random_uniform is deprecated. Please use tf.random.uniform instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:4267: The name tf.nn.max_pool is deprecated. Please use tf.nn.max_pool2d instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:2239: The name tf.image.resize_nearest_neighbor is deprecated. Please use tf.compat.v1.image.resize_nearest_neighbor instead.\n\nModel: \"model_2\"\n__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ndata (InputLayer) (None, 256, 256, 3) 0 \n__________________________________________________________________________________________________\nbn_data (BatchNormalization) (None, 256, 256, 3) 9 data[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_1 (ZeroPadding2D (None, 262, 262, 3) 0 bn_data[0][0] \n__________________________________________________________________________________________________\nconv0 (Conv2D) (None, 128, 128, 64) 9408 zero_padding2d_1[0][0] \n__________________________________________________________________________________________________\nbn0 (BatchNormalization) (None, 128, 128, 64) 256 conv0[0][0] \n__________________________________________________________________________________________________\nrelu0 (Activation) (None, 128, 128, 64) 0 bn0[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_2 (ZeroPadding2D (None, 130, 130, 64) 0 relu0[0][0] \n__________________________________________________________________________________________________\npooling0 (MaxPooling2D) (None, 64, 64, 64) 0 zero_padding2d_2[0][0] \n__________________________________________________________________________________________________\nstage1_unit1_bn1 (BatchNormaliz (None, 64, 64, 64) 256 pooling0[0][0] \n__________________________________________________________________________________________________\nstage1_unit1_relu1 (Activation) (None, 64, 64, 64) 0 stage1_unit1_bn1[0][0] \n__________________________________________________________________________________________________\nstage1_unit1_conv1 (Conv2D) (None, 64, 64, 64) 4096 stage1_unit1_relu1[0][0] \n__________________________________________________________________________________________________\nstage1_unit1_bn2 (BatchNormaliz (None, 64, 64, 64) 256 stage1_unit1_conv1[0][0] \n__________________________________________________________________________________________________\nstage1_unit1_relu2 (Activation) (None, 64, 64, 64) 0 stage1_unit1_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_3 (ZeroPadding2D (None, 66, 66, 64) 0 stage1_unit1_relu2[0][0] \n__________________________________________________________________________________________________\nstage1_unit1_conv2 (Conv2D) (None, 64, 64, 64) 36864 zero_padding2d_3[0][0] \n__________________________________________________________________________________________________\nstage1_unit1_bn3 (BatchNormaliz (None, 64, 64, 64) 256 stage1_unit1_conv2[0][0] \n__________________________________________________________________________________________________\nstage1_unit1_relu3 (Activation) (None, 64, 64, 64) 0 stage1_unit1_bn3[0][0] \n__________________________________________________________________________________________________\nstage1_unit1_conv3 (Conv2D) (None, 64, 64, 256) 16384 stage1_unit1_relu3[0][0] \n__________________________________________________________________________________________________\nstage1_unit1_sc (Conv2D) (None, 64, 64, 256) 16384 stage1_unit1_relu1[0][0] \n__________________________________________________________________________________________________\nadd_1 (Add) (None, 64, 64, 256) 0 stage1_unit1_conv3[0][0] \n stage1_unit1_sc[0][0] \n__________________________________________________________________________________________________\nstage1_unit2_bn1 (BatchNormaliz (None, 64, 64, 256) 1024 add_1[0][0] \n__________________________________________________________________________________________________\nstage1_unit2_relu1 (Activation) (None, 64, 64, 256) 0 stage1_unit2_bn1[0][0] \n__________________________________________________________________________________________________\nstage1_unit2_conv1 (Conv2D) (None, 64, 64, 64) 16384 stage1_unit2_relu1[0][0] \n__________________________________________________________________________________________________\nstage1_unit2_bn2 (BatchNormaliz (None, 64, 64, 64) 256 stage1_unit2_conv1[0][0] \n__________________________________________________________________________________________________\nstage1_unit2_relu2 (Activation) (None, 64, 64, 64) 0 stage1_unit2_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_4 (ZeroPadding2D (None, 66, 66, 64) 0 stage1_unit2_relu2[0][0] \n__________________________________________________________________________________________________\nstage1_unit2_conv2 (Conv2D) (None, 64, 64, 64) 36864 zero_padding2d_4[0][0] \n__________________________________________________________________________________________________\nstage1_unit2_bn3 (BatchNormaliz (None, 64, 64, 64) 256 stage1_unit2_conv2[0][0] \n__________________________________________________________________________________________________\nstage1_unit2_relu3 (Activation) (None, 64, 64, 64) 0 stage1_unit2_bn3[0][0] \n__________________________________________________________________________________________________\nstage1_unit2_conv3 (Conv2D) (None, 64, 64, 256) 16384 stage1_unit2_relu3[0][0] \n__________________________________________________________________________________________________\nadd_2 (Add) (None, 64, 64, 256) 0 stage1_unit2_conv3[0][0] \n add_1[0][0] \n__________________________________________________________________________________________________\nstage1_unit3_bn1 (BatchNormaliz (None, 64, 64, 256) 1024 add_2[0][0] \n__________________________________________________________________________________________________\nstage1_unit3_relu1 (Activation) (None, 64, 64, 256) 0 stage1_unit3_bn1[0][0] \n__________________________________________________________________________________________________\nstage1_unit3_conv1 (Conv2D) (None, 64, 64, 64) 16384 stage1_unit3_relu1[0][0] \n__________________________________________________________________________________________________\nstage1_unit3_bn2 (BatchNormaliz (None, 64, 64, 64) 256 stage1_unit3_conv1[0][0] \n__________________________________________________________________________________________________\nstage1_unit3_relu2 (Activation) (None, 64, 64, 64) 0 stage1_unit3_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_5 (ZeroPadding2D (None, 66, 66, 64) 0 stage1_unit3_relu2[0][0] \n__________________________________________________________________________________________________\nstage1_unit3_conv2 (Conv2D) (None, 64, 64, 64) 36864 zero_padding2d_5[0][0] \n__________________________________________________________________________________________________\nstage1_unit3_bn3 (BatchNormaliz (None, 64, 64, 64) 256 stage1_unit3_conv2[0][0] \n__________________________________________________________________________________________________\nstage1_unit3_relu3 (Activation) (None, 64, 64, 64) 0 stage1_unit3_bn3[0][0] \n__________________________________________________________________________________________________\nstage1_unit3_conv3 (Conv2D) (None, 64, 64, 256) 16384 stage1_unit3_relu3[0][0] \n__________________________________________________________________________________________________\nadd_3 (Add) (None, 64, 64, 256) 0 stage1_unit3_conv3[0][0] \n add_2[0][0] \n__________________________________________________________________________________________________\nstage2_unit1_bn1 (BatchNormaliz (None, 64, 64, 256) 1024 add_3[0][0] \n__________________________________________________________________________________________________\nstage2_unit1_relu1 (Activation) (None, 64, 64, 256) 0 stage2_unit1_bn1[0][0] \n__________________________________________________________________________________________________\nstage2_unit1_conv1 (Conv2D) (None, 64, 64, 128) 32768 stage2_unit1_relu1[0][0] \n__________________________________________________________________________________________________\nstage2_unit1_bn2 (BatchNormaliz (None, 64, 64, 128) 512 stage2_unit1_conv1[0][0] \n__________________________________________________________________________________________________\nstage2_unit1_relu2 (Activation) (None, 64, 64, 128) 0 stage2_unit1_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_6 (ZeroPadding2D (None, 66, 66, 128) 0 stage2_unit1_relu2[0][0] \n__________________________________________________________________________________________________\nstage2_unit1_conv2 (Conv2D) (None, 32, 32, 128) 147456 zero_padding2d_6[0][0] \n__________________________________________________________________________________________________\nstage2_unit1_bn3 (BatchNormaliz (None, 32, 32, 128) 512 stage2_unit1_conv2[0][0] \n__________________________________________________________________________________________________\nstage2_unit1_relu3 (Activation) (None, 32, 32, 128) 0 stage2_unit1_bn3[0][0] \n__________________________________________________________________________________________________\nstage2_unit1_conv3 (Conv2D) (None, 32, 32, 512) 65536 stage2_unit1_relu3[0][0] \n__________________________________________________________________________________________________\nstage2_unit1_sc (Conv2D) (None, 32, 32, 512) 131072 stage2_unit1_relu1[0][0] \n__________________________________________________________________________________________________\nadd_4 (Add) (None, 32, 32, 512) 0 stage2_unit1_conv3[0][0] \n stage2_unit1_sc[0][0] \n__________________________________________________________________________________________________\nstage2_unit2_bn1 (BatchNormaliz (None, 32, 32, 512) 2048 add_4[0][0] \n__________________________________________________________________________________________________\nstage2_unit2_relu1 (Activation) (None, 32, 32, 512) 0 stage2_unit2_bn1[0][0] \n__________________________________________________________________________________________________\nstage2_unit2_conv1 (Conv2D) (None, 32, 32, 128) 65536 stage2_unit2_relu1[0][0] \n__________________________________________________________________________________________________\nstage2_unit2_bn2 (BatchNormaliz (None, 32, 32, 128) 512 stage2_unit2_conv1[0][0] \n__________________________________________________________________________________________________\nstage2_unit2_relu2 (Activation) (None, 32, 32, 128) 0 stage2_unit2_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_7 (ZeroPadding2D (None, 34, 34, 128) 0 stage2_unit2_relu2[0][0] \n__________________________________________________________________________________________________\nstage2_unit2_conv2 (Conv2D) (None, 32, 32, 128) 147456 zero_padding2d_7[0][0] \n__________________________________________________________________________________________________\nstage2_unit2_bn3 (BatchNormaliz (None, 32, 32, 128) 512 stage2_unit2_conv2[0][0] \n__________________________________________________________________________________________________\nstage2_unit2_relu3 (Activation) (None, 32, 32, 128) 0 stage2_unit2_bn3[0][0] \n__________________________________________________________________________________________________\nstage2_unit2_conv3 (Conv2D) (None, 32, 32, 512) 65536 stage2_unit2_relu3[0][0] \n__________________________________________________________________________________________________\nadd_5 (Add) (None, 32, 32, 512) 0 stage2_unit2_conv3[0][0] \n add_4[0][0] \n__________________________________________________________________________________________________\nstage2_unit3_bn1 (BatchNormaliz (None, 32, 32, 512) 2048 add_5[0][0] \n__________________________________________________________________________________________________\nstage2_unit3_relu1 (Activation) (None, 32, 32, 512) 0 stage2_unit3_bn1[0][0] \n__________________________________________________________________________________________________\nstage2_unit3_conv1 (Conv2D) (None, 32, 32, 128) 65536 stage2_unit3_relu1[0][0] \n__________________________________________________________________________________________________\nstage2_unit3_bn2 (BatchNormaliz (None, 32, 32, 128) 512 stage2_unit3_conv1[0][0] \n__________________________________________________________________________________________________\nstage2_unit3_relu2 (Activation) (None, 32, 32, 128) 0 stage2_unit3_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_8 (ZeroPadding2D (None, 34, 34, 128) 0 stage2_unit3_relu2[0][0] \n__________________________________________________________________________________________________\nstage2_unit3_conv2 (Conv2D) (None, 32, 32, 128) 147456 zero_padding2d_8[0][0] \n__________________________________________________________________________________________________\nstage2_unit3_bn3 (BatchNormaliz (None, 32, 32, 128) 512 stage2_unit3_conv2[0][0] \n__________________________________________________________________________________________________\nstage2_unit3_relu3 (Activation) (None, 32, 32, 128) 0 stage2_unit3_bn3[0][0] \n__________________________________________________________________________________________________\nstage2_unit3_conv3 (Conv2D) (None, 32, 32, 512) 65536 stage2_unit3_relu3[0][0] \n__________________________________________________________________________________________________\nadd_6 (Add) (None, 32, 32, 512) 0 stage2_unit3_conv3[0][0] \n add_5[0][0] \n__________________________________________________________________________________________________\nstage2_unit4_bn1 (BatchNormaliz (None, 32, 32, 512) 2048 add_6[0][0] \n__________________________________________________________________________________________________\nstage2_unit4_relu1 (Activation) (None, 32, 32, 512) 0 stage2_unit4_bn1[0][0] \n__________________________________________________________________________________________________\nstage2_unit4_conv1 (Conv2D) (None, 32, 32, 128) 65536 stage2_unit4_relu1[0][0] \n__________________________________________________________________________________________________\nstage2_unit4_bn2 (BatchNormaliz (None, 32, 32, 128) 512 stage2_unit4_conv1[0][0] \n__________________________________________________________________________________________________\nstage2_unit4_relu2 (Activation) (None, 32, 32, 128) 0 stage2_unit4_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_9 (ZeroPadding2D (None, 34, 34, 128) 0 stage2_unit4_relu2[0][0] \n__________________________________________________________________________________________________\nstage2_unit4_conv2 (Conv2D) (None, 32, 32, 128) 147456 zero_padding2d_9[0][0] \n__________________________________________________________________________________________________\nstage2_unit4_bn3 (BatchNormaliz (None, 32, 32, 128) 512 stage2_unit4_conv2[0][0] \n__________________________________________________________________________________________________\nstage2_unit4_relu3 (Activation) (None, 32, 32, 128) 0 stage2_unit4_bn3[0][0] \n__________________________________________________________________________________________________\nstage2_unit4_conv3 (Conv2D) (None, 32, 32, 512) 65536 stage2_unit4_relu3[0][0] \n__________________________________________________________________________________________________\nadd_7 (Add) (None, 32, 32, 512) 0 stage2_unit4_conv3[0][0] \n add_6[0][0] \n__________________________________________________________________________________________________\nstage3_unit1_bn1 (BatchNormaliz (None, 32, 32, 512) 2048 add_7[0][0] \n__________________________________________________________________________________________________\nstage3_unit1_relu1 (Activation) (None, 32, 32, 512) 0 stage3_unit1_bn1[0][0] \n__________________________________________________________________________________________________\nstage3_unit1_conv1 (Conv2D) (None, 32, 32, 256) 131072 stage3_unit1_relu1[0][0] \n__________________________________________________________________________________________________\nstage3_unit1_bn2 (BatchNormaliz (None, 32, 32, 256) 1024 stage3_unit1_conv1[0][0] \n__________________________________________________________________________________________________\nstage3_unit1_relu2 (Activation) (None, 32, 32, 256) 0 stage3_unit1_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_10 (ZeroPadding2 (None, 34, 34, 256) 0 stage3_unit1_relu2[0][0] \n__________________________________________________________________________________________________\nstage3_unit1_conv2 (Conv2D) (None, 16, 16, 256) 589824 zero_padding2d_10[0][0] \n__________________________________________________________________________________________________\nstage3_unit1_bn3 (BatchNormaliz (None, 16, 16, 256) 1024 stage3_unit1_conv2[0][0] \n__________________________________________________________________________________________________\nstage3_unit1_relu3 (Activation) (None, 16, 16, 256) 0 stage3_unit1_bn3[0][0] \n__________________________________________________________________________________________________\nstage3_unit1_conv3 (Conv2D) (None, 16, 16, 1024) 262144 stage3_unit1_relu3[0][0] \n__________________________________________________________________________________________________\nstage3_unit1_sc (Conv2D) (None, 16, 16, 1024) 524288 stage3_unit1_relu1[0][0] \n__________________________________________________________________________________________________\nadd_8 (Add) (None, 16, 16, 1024) 0 stage3_unit1_conv3[0][0] \n stage3_unit1_sc[0][0] \n__________________________________________________________________________________________________\nstage3_unit2_bn1 (BatchNormaliz (None, 16, 16, 1024) 4096 add_8[0][0] \n__________________________________________________________________________________________________\nstage3_unit2_relu1 (Activation) (None, 16, 16, 1024) 0 stage3_unit2_bn1[0][0] \n__________________________________________________________________________________________________\nstage3_unit2_conv1 (Conv2D) (None, 16, 16, 256) 262144 stage3_unit2_relu1[0][0] \n__________________________________________________________________________________________________\nstage3_unit2_bn2 (BatchNormaliz (None, 16, 16, 256) 1024 stage3_unit2_conv1[0][0] \n__________________________________________________________________________________________________\nstage3_unit2_relu2 (Activation) (None, 16, 16, 256) 0 stage3_unit2_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_11 (ZeroPadding2 (None, 18, 18, 256) 0 stage3_unit2_relu2[0][0] \n__________________________________________________________________________________________________\nstage3_unit2_conv2 (Conv2D) (None, 16, 16, 256) 589824 zero_padding2d_11[0][0] \n__________________________________________________________________________________________________\nstage3_unit2_bn3 (BatchNormaliz (None, 16, 16, 256) 1024 stage3_unit2_conv2[0][0] \n__________________________________________________________________________________________________\nstage3_unit2_relu3 (Activation) (None, 16, 16, 256) 0 stage3_unit2_bn3[0][0] \n__________________________________________________________________________________________________\nstage3_unit2_conv3 (Conv2D) (None, 16, 16, 1024) 262144 stage3_unit2_relu3[0][0] \n__________________________________________________________________________________________________\nadd_9 (Add) (None, 16, 16, 1024) 0 stage3_unit2_conv3[0][0] \n add_8[0][0] \n__________________________________________________________________________________________________\nstage3_unit3_bn1 (BatchNormaliz (None, 16, 16, 1024) 4096 add_9[0][0] \n__________________________________________________________________________________________________\nstage3_unit3_relu1 (Activation) (None, 16, 16, 1024) 0 stage3_unit3_bn1[0][0] \n__________________________________________________________________________________________________\nstage3_unit3_conv1 (Conv2D) (None, 16, 16, 256) 262144 stage3_unit3_relu1[0][0] \n__________________________________________________________________________________________________\nstage3_unit3_bn2 (BatchNormaliz (None, 16, 16, 256) 1024 stage3_unit3_conv1[0][0] \n__________________________________________________________________________________________________\nstage3_unit3_relu2 (Activation) (None, 16, 16, 256) 0 stage3_unit3_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_12 (ZeroPadding2 (None, 18, 18, 256) 0 stage3_unit3_relu2[0][0] \n__________________________________________________________________________________________________\nstage3_unit3_conv2 (Conv2D) (None, 16, 16, 256) 589824 zero_padding2d_12[0][0] \n__________________________________________________________________________________________________\nstage3_unit3_bn3 (BatchNormaliz (None, 16, 16, 256) 1024 stage3_unit3_conv2[0][0] \n__________________________________________________________________________________________________\nstage3_unit3_relu3 (Activation) (None, 16, 16, 256) 0 stage3_unit3_bn3[0][0] \n__________________________________________________________________________________________________\nstage3_unit3_conv3 (Conv2D) (None, 16, 16, 1024) 262144 stage3_unit3_relu3[0][0] \n__________________________________________________________________________________________________\nadd_10 (Add) (None, 16, 16, 1024) 0 stage3_unit3_conv3[0][0] \n add_9[0][0] \n__________________________________________________________________________________________________\nstage3_unit4_bn1 (BatchNormaliz (None, 16, 16, 1024) 4096 add_10[0][0] \n__________________________________________________________________________________________________\nstage3_unit4_relu1 (Activation) (None, 16, 16, 1024) 0 stage3_unit4_bn1[0][0] \n__________________________________________________________________________________________________\nstage3_unit4_conv1 (Conv2D) (None, 16, 16, 256) 262144 stage3_unit4_relu1[0][0] \n__________________________________________________________________________________________________\nstage3_unit4_bn2 (BatchNormaliz (None, 16, 16, 256) 1024 stage3_unit4_conv1[0][0] \n__________________________________________________________________________________________________\nstage3_unit4_relu2 (Activation) (None, 16, 16, 256) 0 stage3_unit4_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_13 (ZeroPadding2 (None, 18, 18, 256) 0 stage3_unit4_relu2[0][0] \n__________________________________________________________________________________________________\nstage3_unit4_conv2 (Conv2D) (None, 16, 16, 256) 589824 zero_padding2d_13[0][0] \n__________________________________________________________________________________________________\nstage3_unit4_bn3 (BatchNormaliz (None, 16, 16, 256) 1024 stage3_unit4_conv2[0][0] \n__________________________________________________________________________________________________\nstage3_unit4_relu3 (Activation) (None, 16, 16, 256) 0 stage3_unit4_bn3[0][0] \n__________________________________________________________________________________________________\nstage3_unit4_conv3 (Conv2D) (None, 16, 16, 1024) 262144 stage3_unit4_relu3[0][0] \n__________________________________________________________________________________________________\nadd_11 (Add) (None, 16, 16, 1024) 0 stage3_unit4_conv3[0][0] \n add_10[0][0] \n__________________________________________________________________________________________________\nstage3_unit5_bn1 (BatchNormaliz (None, 16, 16, 1024) 4096 add_11[0][0] \n__________________________________________________________________________________________________\nstage3_unit5_relu1 (Activation) (None, 16, 16, 1024) 0 stage3_unit5_bn1[0][0] \n__________________________________________________________________________________________________\nstage3_unit5_conv1 (Conv2D) (None, 16, 16, 256) 262144 stage3_unit5_relu1[0][0] \n__________________________________________________________________________________________________\nstage3_unit5_bn2 (BatchNormaliz (None, 16, 16, 256) 1024 stage3_unit5_conv1[0][0] \n__________________________________________________________________________________________________\nstage3_unit5_relu2 (Activation) (None, 16, 16, 256) 0 stage3_unit5_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_14 (ZeroPadding2 (None, 18, 18, 256) 0 stage3_unit5_relu2[0][0] \n__________________________________________________________________________________________________\nstage3_unit5_conv2 (Conv2D) (None, 16, 16, 256) 589824 zero_padding2d_14[0][0] \n__________________________________________________________________________________________________\nstage3_unit5_bn3 (BatchNormaliz (None, 16, 16, 256) 1024 stage3_unit5_conv2[0][0] \n__________________________________________________________________________________________________\nstage3_unit5_relu3 (Activation) (None, 16, 16, 256) 0 stage3_unit5_bn3[0][0] \n__________________________________________________________________________________________________\nstage3_unit5_conv3 (Conv2D) (None, 16, 16, 1024) 262144 stage3_unit5_relu3[0][0] \n__________________________________________________________________________________________________\nadd_12 (Add) (None, 16, 16, 1024) 0 stage3_unit5_conv3[0][0] \n add_11[0][0] \n__________________________________________________________________________________________________\nstage3_unit6_bn1 (BatchNormaliz (None, 16, 16, 1024) 4096 add_12[0][0] \n__________________________________________________________________________________________________\nstage3_unit6_relu1 (Activation) (None, 16, 16, 1024) 0 stage3_unit6_bn1[0][0] \n__________________________________________________________________________________________________\nstage3_unit6_conv1 (Conv2D) (None, 16, 16, 256) 262144 stage3_unit6_relu1[0][0] \n__________________________________________________________________________________________________\nstage3_unit6_bn2 (BatchNormaliz (None, 16, 16, 256) 1024 stage3_unit6_conv1[0][0] \n__________________________________________________________________________________________________\nstage3_unit6_relu2 (Activation) (None, 16, 16, 256) 0 stage3_unit6_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_15 (ZeroPadding2 (None, 18, 18, 256) 0 stage3_unit6_relu2[0][0] \n__________________________________________________________________________________________________\nstage3_unit6_conv2 (Conv2D) (None, 16, 16, 256) 589824 zero_padding2d_15[0][0] \n__________________________________________________________________________________________________\nstage3_unit6_bn3 (BatchNormaliz (None, 16, 16, 256) 1024 stage3_unit6_conv2[0][0] \n__________________________________________________________________________________________________\nstage3_unit6_relu3 (Activation) (None, 16, 16, 256) 0 stage3_unit6_bn3[0][0] \n__________________________________________________________________________________________________\nstage3_unit6_conv3 (Conv2D) (None, 16, 16, 1024) 262144 stage3_unit6_relu3[0][0] \n__________________________________________________________________________________________________\nadd_13 (Add) (None, 16, 16, 1024) 0 stage3_unit6_conv3[0][0] \n add_12[0][0] \n__________________________________________________________________________________________________\nstage3_unit7_bn1 (BatchNormaliz (None, 16, 16, 1024) 4096 add_13[0][0] \n__________________________________________________________________________________________________\nstage3_unit7_relu1 (Activation) (None, 16, 16, 1024) 0 stage3_unit7_bn1[0][0] \n__________________________________________________________________________________________________\nstage3_unit7_conv1 (Conv2D) (None, 16, 16, 256) 262144 stage3_unit7_relu1[0][0] \n__________________________________________________________________________________________________\nstage3_unit7_bn2 (BatchNormaliz (None, 16, 16, 256) 1024 stage3_unit7_conv1[0][0] \n__________________________________________________________________________________________________\nstage3_unit7_relu2 (Activation) (None, 16, 16, 256) 0 stage3_unit7_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_16 (ZeroPadding2 (None, 18, 18, 256) 0 stage3_unit7_relu2[0][0] \n__________________________________________________________________________________________________\nstage3_unit7_conv2 (Conv2D) (None, 16, 16, 256) 589824 zero_padding2d_16[0][0] \n__________________________________________________________________________________________________\nstage3_unit7_bn3 (BatchNormaliz (None, 16, 16, 256) 1024 stage3_unit7_conv2[0][0] \n__________________________________________________________________________________________________\nstage3_unit7_relu3 (Activation) (None, 16, 16, 256) 0 stage3_unit7_bn3[0][0] \n__________________________________________________________________________________________________\nstage3_unit7_conv3 (Conv2D) (None, 16, 16, 1024) 262144 stage3_unit7_relu3[0][0] \n__________________________________________________________________________________________________\nadd_14 (Add) (None, 16, 16, 1024) 0 stage3_unit7_conv3[0][0] \n add_13[0][0] \n__________________________________________________________________________________________________\nstage3_unit8_bn1 (BatchNormaliz (None, 16, 16, 1024) 4096 add_14[0][0] \n__________________________________________________________________________________________________\nstage3_unit8_relu1 (Activation) (None, 16, 16, 1024) 0 stage3_unit8_bn1[0][0] \n__________________________________________________________________________________________________\nstage3_unit8_conv1 (Conv2D) (None, 16, 16, 256) 262144 stage3_unit8_relu1[0][0] \n__________________________________________________________________________________________________\nstage3_unit8_bn2 (BatchNormaliz (None, 16, 16, 256) 1024 stage3_unit8_conv1[0][0] \n__________________________________________________________________________________________________\nstage3_unit8_relu2 (Activation) (None, 16, 16, 256) 0 stage3_unit8_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_17 (ZeroPadding2 (None, 18, 18, 256) 0 stage3_unit8_relu2[0][0] \n__________________________________________________________________________________________________\nstage3_unit8_conv2 (Conv2D) (None, 16, 16, 256) 589824 zero_padding2d_17[0][0] \n__________________________________________________________________________________________________\nstage3_unit8_bn3 (BatchNormaliz (None, 16, 16, 256) 1024 stage3_unit8_conv2[0][0] \n__________________________________________________________________________________________________\nstage3_unit8_relu3 (Activation) (None, 16, 16, 256) 0 stage3_unit8_bn3[0][0] \n__________________________________________________________________________________________________\nstage3_unit8_conv3 (Conv2D) (None, 16, 16, 1024) 262144 stage3_unit8_relu3[0][0] \n__________________________________________________________________________________________________\nadd_15 (Add) (None, 16, 16, 1024) 0 stage3_unit8_conv3[0][0] \n add_14[0][0] \n__________________________________________________________________________________________________\nstage3_unit9_bn1 (BatchNormaliz (None, 16, 16, 1024) 4096 add_15[0][0] \n__________________________________________________________________________________________________\nstage3_unit9_relu1 (Activation) (None, 16, 16, 1024) 0 stage3_unit9_bn1[0][0] \n__________________________________________________________________________________________________\nstage3_unit9_conv1 (Conv2D) (None, 16, 16, 256) 262144 stage3_unit9_relu1[0][0] \n__________________________________________________________________________________________________\nstage3_unit9_bn2 (BatchNormaliz (None, 16, 16, 256) 1024 stage3_unit9_conv1[0][0] \n__________________________________________________________________________________________________\nstage3_unit9_relu2 (Activation) (None, 16, 16, 256) 0 stage3_unit9_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_18 (ZeroPadding2 (None, 18, 18, 256) 0 stage3_unit9_relu2[0][0] \n__________________________________________________________________________________________________\nstage3_unit9_conv2 (Conv2D) (None, 16, 16, 256) 589824 zero_padding2d_18[0][0] \n__________________________________________________________________________________________________\nstage3_unit9_bn3 (BatchNormaliz (None, 16, 16, 256) 1024 stage3_unit9_conv2[0][0] \n__________________________________________________________________________________________________\nstage3_unit9_relu3 (Activation) (None, 16, 16, 256) 0 stage3_unit9_bn3[0][0] \n__________________________________________________________________________________________________\nstage3_unit9_conv3 (Conv2D) (None, 16, 16, 1024) 262144 stage3_unit9_relu3[0][0] \n__________________________________________________________________________________________________\nadd_16 (Add) (None, 16, 16, 1024) 0 stage3_unit9_conv3[0][0] \n add_15[0][0] \n__________________________________________________________________________________________________\nstage3_unit10_bn1 (BatchNormali (None, 16, 16, 1024) 4096 add_16[0][0] \n__________________________________________________________________________________________________\nstage3_unit10_relu1 (Activation (None, 16, 16, 1024) 0 stage3_unit10_bn1[0][0] \n__________________________________________________________________________________________________\nstage3_unit10_conv1 (Conv2D) (None, 16, 16, 256) 262144 stage3_unit10_relu1[0][0] \n__________________________________________________________________________________________________\nstage3_unit10_bn2 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit10_conv1[0][0] \n__________________________________________________________________________________________________\nstage3_unit10_relu2 (Activation (None, 16, 16, 256) 0 stage3_unit10_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_19 (ZeroPadding2 (None, 18, 18, 256) 0 stage3_unit10_relu2[0][0] \n__________________________________________________________________________________________________\nstage3_unit10_conv2 (Conv2D) (None, 16, 16, 256) 589824 zero_padding2d_19[0][0] \n__________________________________________________________________________________________________\nstage3_unit10_bn3 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit10_conv2[0][0] \n__________________________________________________________________________________________________\nstage3_unit10_relu3 (Activation (None, 16, 16, 256) 0 stage3_unit10_bn3[0][0] \n__________________________________________________________________________________________________\nstage3_unit10_conv3 (Conv2D) (None, 16, 16, 1024) 262144 stage3_unit10_relu3[0][0] \n__________________________________________________________________________________________________\nadd_17 (Add) (None, 16, 16, 1024) 0 stage3_unit10_conv3[0][0] \n add_16[0][0] \n__________________________________________________________________________________________________\nstage3_unit11_bn1 (BatchNormali (None, 16, 16, 1024) 4096 add_17[0][0] \n__________________________________________________________________________________________________\nstage3_unit11_relu1 (Activation (None, 16, 16, 1024) 0 stage3_unit11_bn1[0][0] \n__________________________________________________________________________________________________\nstage3_unit11_conv1 (Conv2D) (None, 16, 16, 256) 262144 stage3_unit11_relu1[0][0] \n__________________________________________________________________________________________________\nstage3_unit11_bn2 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit11_conv1[0][0] \n__________________________________________________________________________________________________\nstage3_unit11_relu2 (Activation (None, 16, 16, 256) 0 stage3_unit11_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_20 (ZeroPadding2 (None, 18, 18, 256) 0 stage3_unit11_relu2[0][0] \n__________________________________________________________________________________________________\nstage3_unit11_conv2 (Conv2D) (None, 16, 16, 256) 589824 zero_padding2d_20[0][0] \n__________________________________________________________________________________________________\nstage3_unit11_bn3 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit11_conv2[0][0] \n__________________________________________________________________________________________________\nstage3_unit11_relu3 (Activation (None, 16, 16, 256) 0 stage3_unit11_bn3[0][0] \n__________________________________________________________________________________________________\nstage3_unit11_conv3 (Conv2D) (None, 16, 16, 1024) 262144 stage3_unit11_relu3[0][0] \n__________________________________________________________________________________________________\nadd_18 (Add) (None, 16, 16, 1024) 0 stage3_unit11_conv3[0][0] \n add_17[0][0] \n__________________________________________________________________________________________________\nstage3_unit12_bn1 (BatchNormali (None, 16, 16, 1024) 4096 add_18[0][0] \n__________________________________________________________________________________________________\nstage3_unit12_relu1 (Activation (None, 16, 16, 1024) 0 stage3_unit12_bn1[0][0] \n__________________________________________________________________________________________________\nstage3_unit12_conv1 (Conv2D) (None, 16, 16, 256) 262144 stage3_unit12_relu1[0][0] \n__________________________________________________________________________________________________\nstage3_unit12_bn2 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit12_conv1[0][0] \n__________________________________________________________________________________________________\nstage3_unit12_relu2 (Activation (None, 16, 16, 256) 0 stage3_unit12_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_21 (ZeroPadding2 (None, 18, 18, 256) 0 stage3_unit12_relu2[0][0] \n__________________________________________________________________________________________________\nstage3_unit12_conv2 (Conv2D) (None, 16, 16, 256) 589824 zero_padding2d_21[0][0] \n__________________________________________________________________________________________________\nstage3_unit12_bn3 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit12_conv2[0][0] \n__________________________________________________________________________________________________\nstage3_unit12_relu3 (Activation (None, 16, 16, 256) 0 stage3_unit12_bn3[0][0] \n__________________________________________________________________________________________________\nstage3_unit12_conv3 (Conv2D) (None, 16, 16, 1024) 262144 stage3_unit12_relu3[0][0] \n__________________________________________________________________________________________________\nadd_19 (Add) (None, 16, 16, 1024) 0 stage3_unit12_conv3[0][0] \n add_18[0][0] \n__________________________________________________________________________________________________\nstage3_unit13_bn1 (BatchNormali (None, 16, 16, 1024) 4096 add_19[0][0] \n__________________________________________________________________________________________________\nstage3_unit13_relu1 (Activation (None, 16, 16, 1024) 0 stage3_unit13_bn1[0][0] \n__________________________________________________________________________________________________\nstage3_unit13_conv1 (Conv2D) (None, 16, 16, 256) 262144 stage3_unit13_relu1[0][0] \n__________________________________________________________________________________________________\nstage3_unit13_bn2 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit13_conv1[0][0] \n__________________________________________________________________________________________________\nstage3_unit13_relu2 (Activation (None, 16, 16, 256) 0 stage3_unit13_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_22 (ZeroPadding2 (None, 18, 18, 256) 0 stage3_unit13_relu2[0][0] \n__________________________________________________________________________________________________\nstage3_unit13_conv2 (Conv2D) (None, 16, 16, 256) 589824 zero_padding2d_22[0][0] \n__________________________________________________________________________________________________\nstage3_unit13_bn3 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit13_conv2[0][0] \n__________________________________________________________________________________________________\nstage3_unit13_relu3 (Activation (None, 16, 16, 256) 0 stage3_unit13_bn3[0][0] \n__________________________________________________________________________________________________\nstage3_unit13_conv3 (Conv2D) (None, 16, 16, 1024) 262144 stage3_unit13_relu3[0][0] \n__________________________________________________________________________________________________\nadd_20 (Add) (None, 16, 16, 1024) 0 stage3_unit13_conv3[0][0] \n add_19[0][0] \n__________________________________________________________________________________________________\nstage3_unit14_bn1 (BatchNormali (None, 16, 16, 1024) 4096 add_20[0][0] \n__________________________________________________________________________________________________\nstage3_unit14_relu1 (Activation (None, 16, 16, 1024) 0 stage3_unit14_bn1[0][0] \n__________________________________________________________________________________________________\nstage3_unit14_conv1 (Conv2D) (None, 16, 16, 256) 262144 stage3_unit14_relu1[0][0] \n__________________________________________________________________________________________________\nstage3_unit14_bn2 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit14_conv1[0][0] \n__________________________________________________________________________________________________\nstage3_unit14_relu2 (Activation (None, 16, 16, 256) 0 stage3_unit14_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_23 (ZeroPadding2 (None, 18, 18, 256) 0 stage3_unit14_relu2[0][0] \n__________________________________________________________________________________________________\nstage3_unit14_conv2 (Conv2D) (None, 16, 16, 256) 589824 zero_padding2d_23[0][0] \n__________________________________________________________________________________________________\nstage3_unit14_bn3 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit14_conv2[0][0] \n__________________________________________________________________________________________________\nstage3_unit14_relu3 (Activation (None, 16, 16, 256) 0 stage3_unit14_bn3[0][0] \n__________________________________________________________________________________________________\nstage3_unit14_conv3 (Conv2D) (None, 16, 16, 1024) 262144 stage3_unit14_relu3[0][0] \n__________________________________________________________________________________________________\nadd_21 (Add) (None, 16, 16, 1024) 0 stage3_unit14_conv3[0][0] \n add_20[0][0] \n__________________________________________________________________________________________________\nstage3_unit15_bn1 (BatchNormali (None, 16, 16, 1024) 4096 add_21[0][0] \n__________________________________________________________________________________________________\nstage3_unit15_relu1 (Activation (None, 16, 16, 1024) 0 stage3_unit15_bn1[0][0] \n__________________________________________________________________________________________________\nstage3_unit15_conv1 (Conv2D) (None, 16, 16, 256) 262144 stage3_unit15_relu1[0][0] \n__________________________________________________________________________________________________\nstage3_unit15_bn2 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit15_conv1[0][0] \n__________________________________________________________________________________________________\nstage3_unit15_relu2 (Activation (None, 16, 16, 256) 0 stage3_unit15_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_24 (ZeroPadding2 (None, 18, 18, 256) 0 stage3_unit15_relu2[0][0] \n__________________________________________________________________________________________________\nstage3_unit15_conv2 (Conv2D) (None, 16, 16, 256) 589824 zero_padding2d_24[0][0] \n__________________________________________________________________________________________________\nstage3_unit15_bn3 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit15_conv2[0][0] \n__________________________________________________________________________________________________\nstage3_unit15_relu3 (Activation (None, 16, 16, 256) 0 stage3_unit15_bn3[0][0] \n__________________________________________________________________________________________________\nstage3_unit15_conv3 (Conv2D) (None, 16, 16, 1024) 262144 stage3_unit15_relu3[0][0] \n__________________________________________________________________________________________________\nadd_22 (Add) (None, 16, 16, 1024) 0 stage3_unit15_conv3[0][0] \n add_21[0][0] \n__________________________________________________________________________________________________\nstage3_unit16_bn1 (BatchNormali (None, 16, 16, 1024) 4096 add_22[0][0] \n__________________________________________________________________________________________________\nstage3_unit16_relu1 (Activation (None, 16, 16, 1024) 0 stage3_unit16_bn1[0][0] \n__________________________________________________________________________________________________\nstage3_unit16_conv1 (Conv2D) (None, 16, 16, 256) 262144 stage3_unit16_relu1[0][0] \n__________________________________________________________________________________________________\nstage3_unit16_bn2 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit16_conv1[0][0] \n__________________________________________________________________________________________________\nstage3_unit16_relu2 (Activation (None, 16, 16, 256) 0 stage3_unit16_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_25 (ZeroPadding2 (None, 18, 18, 256) 0 stage3_unit16_relu2[0][0] \n__________________________________________________________________________________________________\nstage3_unit16_conv2 (Conv2D) (None, 16, 16, 256) 589824 zero_padding2d_25[0][0] \n__________________________________________________________________________________________________\nstage3_unit16_bn3 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit16_conv2[0][0] \n__________________________________________________________________________________________________\nstage3_unit16_relu3 (Activation (None, 16, 16, 256) 0 stage3_unit16_bn3[0][0] \n__________________________________________________________________________________________________\nstage3_unit16_conv3 (Conv2D) (None, 16, 16, 1024) 262144 stage3_unit16_relu3[0][0] \n__________________________________________________________________________________________________\nadd_23 (Add) (None, 16, 16, 1024) 0 stage3_unit16_conv3[0][0] \n add_22[0][0] \n__________________________________________________________________________________________________\nstage3_unit17_bn1 (BatchNormali (None, 16, 16, 1024) 4096 add_23[0][0] \n__________________________________________________________________________________________________\nstage3_unit17_relu1 (Activation (None, 16, 16, 1024) 0 stage3_unit17_bn1[0][0] \n__________________________________________________________________________________________________\nstage3_unit17_conv1 (Conv2D) (None, 16, 16, 256) 262144 stage3_unit17_relu1[0][0] \n__________________________________________________________________________________________________\nstage3_unit17_bn2 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit17_conv1[0][0] \n__________________________________________________________________________________________________\nstage3_unit17_relu2 (Activation (None, 16, 16, 256) 0 stage3_unit17_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_26 (ZeroPadding2 (None, 18, 18, 256) 0 stage3_unit17_relu2[0][0] \n__________________________________________________________________________________________________\nstage3_unit17_conv2 (Conv2D) (None, 16, 16, 256) 589824 zero_padding2d_26[0][0] \n__________________________________________________________________________________________________\nstage3_unit17_bn3 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit17_conv2[0][0] \n__________________________________________________________________________________________________\nstage3_unit17_relu3 (Activation (None, 16, 16, 256) 0 stage3_unit17_bn3[0][0] \n__________________________________________________________________________________________________\nstage3_unit17_conv3 (Conv2D) (None, 16, 16, 1024) 262144 stage3_unit17_relu3[0][0] \n__________________________________________________________________________________________________\nadd_24 (Add) (None, 16, 16, 1024) 0 stage3_unit17_conv3[0][0] \n add_23[0][0] \n__________________________________________________________________________________________________\nstage3_unit18_bn1 (BatchNormali (None, 16, 16, 1024) 4096 add_24[0][0] \n__________________________________________________________________________________________________\nstage3_unit18_relu1 (Activation (None, 16, 16, 1024) 0 stage3_unit18_bn1[0][0] \n__________________________________________________________________________________________________\nstage3_unit18_conv1 (Conv2D) (None, 16, 16, 256) 262144 stage3_unit18_relu1[0][0] \n__________________________________________________________________________________________________\nstage3_unit18_bn2 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit18_conv1[0][0] \n__________________________________________________________________________________________________\nstage3_unit18_relu2 (Activation (None, 16, 16, 256) 0 stage3_unit18_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_27 (ZeroPadding2 (None, 18, 18, 256) 0 stage3_unit18_relu2[0][0] \n__________________________________________________________________________________________________\nstage3_unit18_conv2 (Conv2D) (None, 16, 16, 256) 589824 zero_padding2d_27[0][0] \n__________________________________________________________________________________________________\nstage3_unit18_bn3 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit18_conv2[0][0] \n__________________________________________________________________________________________________\nstage3_unit18_relu3 (Activation (None, 16, 16, 256) 0 stage3_unit18_bn3[0][0] \n__________________________________________________________________________________________________\nstage3_unit18_conv3 (Conv2D) (None, 16, 16, 1024) 262144 stage3_unit18_relu3[0][0] \n__________________________________________________________________________________________________\nadd_25 (Add) (None, 16, 16, 1024) 0 stage3_unit18_conv3[0][0] \n add_24[0][0] \n__________________________________________________________________________________________________\nstage3_unit19_bn1 (BatchNormali (None, 16, 16, 1024) 4096 add_25[0][0] \n__________________________________________________________________________________________________\nstage3_unit19_relu1 (Activation (None, 16, 16, 1024) 0 stage3_unit19_bn1[0][0] \n__________________________________________________________________________________________________\nstage3_unit19_conv1 (Conv2D) (None, 16, 16, 256) 262144 stage3_unit19_relu1[0][0] \n__________________________________________________________________________________________________\nstage3_unit19_bn2 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit19_conv1[0][0] \n__________________________________________________________________________________________________\nstage3_unit19_relu2 (Activation (None, 16, 16, 256) 0 stage3_unit19_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_28 (ZeroPadding2 (None, 18, 18, 256) 0 stage3_unit19_relu2[0][0] \n__________________________________________________________________________________________________\nstage3_unit19_conv2 (Conv2D) (None, 16, 16, 256) 589824 zero_padding2d_28[0][0] \n__________________________________________________________________________________________________\nstage3_unit19_bn3 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit19_conv2[0][0] \n__________________________________________________________________________________________________\nstage3_unit19_relu3 (Activation (None, 16, 16, 256) 0 stage3_unit19_bn3[0][0] \n__________________________________________________________________________________________________\nstage3_unit19_conv3 (Conv2D) (None, 16, 16, 1024) 262144 stage3_unit19_relu3[0][0] \n__________________________________________________________________________________________________\nadd_26 (Add) (None, 16, 16, 1024) 0 stage3_unit19_conv3[0][0] \n add_25[0][0] \n__________________________________________________________________________________________________\nstage3_unit20_bn1 (BatchNormali (None, 16, 16, 1024) 4096 add_26[0][0] \n__________________________________________________________________________________________________\nstage3_unit20_relu1 (Activation (None, 16, 16, 1024) 0 stage3_unit20_bn1[0][0] \n__________________________________________________________________________________________________\nstage3_unit20_conv1 (Conv2D) (None, 16, 16, 256) 262144 stage3_unit20_relu1[0][0] \n__________________________________________________________________________________________________\nstage3_unit20_bn2 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit20_conv1[0][0] \n__________________________________________________________________________________________________\nstage3_unit20_relu2 (Activation (None, 16, 16, 256) 0 stage3_unit20_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_29 (ZeroPadding2 (None, 18, 18, 256) 0 stage3_unit20_relu2[0][0] \n__________________________________________________________________________________________________\nstage3_unit20_conv2 (Conv2D) (None, 16, 16, 256) 589824 zero_padding2d_29[0][0] \n__________________________________________________________________________________________________\nstage3_unit20_bn3 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit20_conv2[0][0] \n__________________________________________________________________________________________________\nstage3_unit20_relu3 (Activation (None, 16, 16, 256) 0 stage3_unit20_bn3[0][0] \n__________________________________________________________________________________________________\nstage3_unit20_conv3 (Conv2D) (None, 16, 16, 1024) 262144 stage3_unit20_relu3[0][0] \n__________________________________________________________________________________________________\nadd_27 (Add) (None, 16, 16, 1024) 0 stage3_unit20_conv3[0][0] \n add_26[0][0] \n__________________________________________________________________________________________________\nstage3_unit21_bn1 (BatchNormali (None, 16, 16, 1024) 4096 add_27[0][0] \n__________________________________________________________________________________________________\nstage3_unit21_relu1 (Activation (None, 16, 16, 1024) 0 stage3_unit21_bn1[0][0] \n__________________________________________________________________________________________________\nstage3_unit21_conv1 (Conv2D) (None, 16, 16, 256) 262144 stage3_unit21_relu1[0][0] \n__________________________________________________________________________________________________\nstage3_unit21_bn2 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit21_conv1[0][0] \n__________________________________________________________________________________________________\nstage3_unit21_relu2 (Activation (None, 16, 16, 256) 0 stage3_unit21_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_30 (ZeroPadding2 (None, 18, 18, 256) 0 stage3_unit21_relu2[0][0] \n__________________________________________________________________________________________________\nstage3_unit21_conv2 (Conv2D) (None, 16, 16, 256) 589824 zero_padding2d_30[0][0] \n__________________________________________________________________________________________________\nstage3_unit21_bn3 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit21_conv2[0][0] \n__________________________________________________________________________________________________\nstage3_unit21_relu3 (Activation (None, 16, 16, 256) 0 stage3_unit21_bn3[0][0] \n__________________________________________________________________________________________________\nstage3_unit21_conv3 (Conv2D) (None, 16, 16, 1024) 262144 stage3_unit21_relu3[0][0] \n__________________________________________________________________________________________________\nadd_28 (Add) (None, 16, 16, 1024) 0 stage3_unit21_conv3[0][0] \n add_27[0][0] \n__________________________________________________________________________________________________\nstage3_unit22_bn1 (BatchNormali (None, 16, 16, 1024) 4096 add_28[0][0] \n__________________________________________________________________________________________________\nstage3_unit22_relu1 (Activation (None, 16, 16, 1024) 0 stage3_unit22_bn1[0][0] \n__________________________________________________________________________________________________\nstage3_unit22_conv1 (Conv2D) (None, 16, 16, 256) 262144 stage3_unit22_relu1[0][0] \n__________________________________________________________________________________________________\nstage3_unit22_bn2 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit22_conv1[0][0] \n__________________________________________________________________________________________________\nstage3_unit22_relu2 (Activation (None, 16, 16, 256) 0 stage3_unit22_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_31 (ZeroPadding2 (None, 18, 18, 256) 0 stage3_unit22_relu2[0][0] \n__________________________________________________________________________________________________\nstage3_unit22_conv2 (Conv2D) (None, 16, 16, 256) 589824 zero_padding2d_31[0][0] \n__________________________________________________________________________________________________\nstage3_unit22_bn3 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit22_conv2[0][0] \n__________________________________________________________________________________________________\nstage3_unit22_relu3 (Activation (None, 16, 16, 256) 0 stage3_unit22_bn3[0][0] \n__________________________________________________________________________________________________\nstage3_unit22_conv3 (Conv2D) (None, 16, 16, 1024) 262144 stage3_unit22_relu3[0][0] \n__________________________________________________________________________________________________\nadd_29 (Add) (None, 16, 16, 1024) 0 stage3_unit22_conv3[0][0] \n add_28[0][0] \n__________________________________________________________________________________________________\nstage3_unit23_bn1 (BatchNormali (None, 16, 16, 1024) 4096 add_29[0][0] \n__________________________________________________________________________________________________\nstage3_unit23_relu1 (Activation (None, 16, 16, 1024) 0 stage3_unit23_bn1[0][0] \n__________________________________________________________________________________________________\nstage3_unit23_conv1 (Conv2D) (None, 16, 16, 256) 262144 stage3_unit23_relu1[0][0] \n__________________________________________________________________________________________________\nstage3_unit23_bn2 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit23_conv1[0][0] \n__________________________________________________________________________________________________\nstage3_unit23_relu2 (Activation (None, 16, 16, 256) 0 stage3_unit23_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_32 (ZeroPadding2 (None, 18, 18, 256) 0 stage3_unit23_relu2[0][0] \n__________________________________________________________________________________________________\nstage3_unit23_conv2 (Conv2D) (None, 16, 16, 256) 589824 zero_padding2d_32[0][0] \n__________________________________________________________________________________________________\nstage3_unit23_bn3 (BatchNormali (None, 16, 16, 256) 1024 stage3_unit23_conv2[0][0] \n__________________________________________________________________________________________________\nstage3_unit23_relu3 (Activation (None, 16, 16, 256) 0 stage3_unit23_bn3[0][0] \n__________________________________________________________________________________________________\nstage3_unit23_conv3 (Conv2D) (None, 16, 16, 1024) 262144 stage3_unit23_relu3[0][0] \n__________________________________________________________________________________________________\nadd_30 (Add) (None, 16, 16, 1024) 0 stage3_unit23_conv3[0][0] \n add_29[0][0] \n__________________________________________________________________________________________________\nstage4_unit1_bn1 (BatchNormaliz (None, 16, 16, 1024) 4096 add_30[0][0] \n__________________________________________________________________________________________________\nstage4_unit1_relu1 (Activation) (None, 16, 16, 1024) 0 stage4_unit1_bn1[0][0] \n__________________________________________________________________________________________________\nstage4_unit1_conv1 (Conv2D) (None, 16, 16, 512) 524288 stage4_unit1_relu1[0][0] \n__________________________________________________________________________________________________\nstage4_unit1_bn2 (BatchNormaliz (None, 16, 16, 512) 2048 stage4_unit1_conv1[0][0] \n__________________________________________________________________________________________________\nstage4_unit1_relu2 (Activation) (None, 16, 16, 512) 0 stage4_unit1_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_33 (ZeroPadding2 (None, 18, 18, 512) 0 stage4_unit1_relu2[0][0] \n__________________________________________________________________________________________________\nstage4_unit1_conv2 (Conv2D) (None, 8, 8, 512) 2359296 zero_padding2d_33[0][0] \n__________________________________________________________________________________________________\nstage4_unit1_bn3 (BatchNormaliz (None, 8, 8, 512) 2048 stage4_unit1_conv2[0][0] \n__________________________________________________________________________________________________\nstage4_unit1_relu3 (Activation) (None, 8, 8, 512) 0 stage4_unit1_bn3[0][0] \n__________________________________________________________________________________________________\nstage4_unit1_conv3 (Conv2D) (None, 8, 8, 2048) 1048576 stage4_unit1_relu3[0][0] \n__________________________________________________________________________________________________\nstage4_unit1_sc (Conv2D) (None, 8, 8, 2048) 2097152 stage4_unit1_relu1[0][0] \n__________________________________________________________________________________________________\nadd_31 (Add) (None, 8, 8, 2048) 0 stage4_unit1_conv3[0][0] \n stage4_unit1_sc[0][0] \n__________________________________________________________________________________________________\nstage4_unit2_bn1 (BatchNormaliz (None, 8, 8, 2048) 8192 add_31[0][0] \n__________________________________________________________________________________________________\nstage4_unit2_relu1 (Activation) (None, 8, 8, 2048) 0 stage4_unit2_bn1[0][0] \n__________________________________________________________________________________________________\nstage4_unit2_conv1 (Conv2D) (None, 8, 8, 512) 1048576 stage4_unit2_relu1[0][0] \n__________________________________________________________________________________________________\nstage4_unit2_bn2 (BatchNormaliz (None, 8, 8, 512) 2048 stage4_unit2_conv1[0][0] \n__________________________________________________________________________________________________\nstage4_unit2_relu2 (Activation) (None, 8, 8, 512) 0 stage4_unit2_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_34 (ZeroPadding2 (None, 10, 10, 512) 0 stage4_unit2_relu2[0][0] \n__________________________________________________________________________________________________\nstage4_unit2_conv2 (Conv2D) (None, 8, 8, 512) 2359296 zero_padding2d_34[0][0] \n__________________________________________________________________________________________________\nstage4_unit2_bn3 (BatchNormaliz (None, 8, 8, 512) 2048 stage4_unit2_conv2[0][0] \n__________________________________________________________________________________________________\nstage4_unit2_relu3 (Activation) (None, 8, 8, 512) 0 stage4_unit2_bn3[0][0] \n__________________________________________________________________________________________________\nstage4_unit2_conv3 (Conv2D) (None, 8, 8, 2048) 1048576 stage4_unit2_relu3[0][0] \n__________________________________________________________________________________________________\nadd_32 (Add) (None, 8, 8, 2048) 0 stage4_unit2_conv3[0][0] \n add_31[0][0] \n__________________________________________________________________________________________________\nstage4_unit3_bn1 (BatchNormaliz (None, 8, 8, 2048) 8192 add_32[0][0] \n__________________________________________________________________________________________________\nstage4_unit3_relu1 (Activation) (None, 8, 8, 2048) 0 stage4_unit3_bn1[0][0] \n__________________________________________________________________________________________________\nstage4_unit3_conv1 (Conv2D) (None, 8, 8, 512) 1048576 stage4_unit3_relu1[0][0] \n__________________________________________________________________________________________________\nstage4_unit3_bn2 (BatchNormaliz (None, 8, 8, 512) 2048 stage4_unit3_conv1[0][0] \n__________________________________________________________________________________________________\nstage4_unit3_relu2 (Activation) (None, 8, 8, 512) 0 stage4_unit3_bn2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_35 (ZeroPadding2 (None, 10, 10, 512) 0 stage4_unit3_relu2[0][0] \n__________________________________________________________________________________________________\nstage4_unit3_conv2 (Conv2D) (None, 8, 8, 512) 2359296 zero_padding2d_35[0][0] \n__________________________________________________________________________________________________\nstage4_unit3_bn3 (BatchNormaliz (None, 8, 8, 512) 2048 stage4_unit3_conv2[0][0] \n__________________________________________________________________________________________________\nstage4_unit3_relu3 (Activation) (None, 8, 8, 512) 0 stage4_unit3_bn3[0][0] \n__________________________________________________________________________________________________\nstage4_unit3_conv3 (Conv2D) (None, 8, 8, 2048) 1048576 stage4_unit3_relu3[0][0] \n__________________________________________________________________________________________________\nadd_33 (Add) (None, 8, 8, 2048) 0 stage4_unit3_conv3[0][0] \n add_32[0][0] \n__________________________________________________________________________________________________\nbn1 (BatchNormalization) (None, 8, 8, 2048) 8192 add_33[0][0] \n__________________________________________________________________________________________________\nrelu1 (Activation) (None, 8, 8, 2048) 0 bn1[0][0] \n__________________________________________________________________________________________________\ndecoder_stage0_upsampling (UpSa (None, 16, 16, 2048) 0 relu1[0][0] \n__________________________________________________________________________________________________\ndecoder_stage0_concat (Concaten (None, 16, 16, 3072) 0 decoder_stage0_upsampling[0][0] \n stage4_unit1_relu1[0][0] \n__________________________________________________________________________________________________\ndecoder_stage0a_conv (Conv2D) (None, 16, 16, 256) 7077888 decoder_stage0_concat[0][0] \n__________________________________________________________________________________________________\ndecoder_stage0a_bn (BatchNormal (None, 16, 16, 256) 1024 decoder_stage0a_conv[0][0] \n__________________________________________________________________________________________________\ndecoder_stage0a_relu (Activatio (None, 16, 16, 256) 0 decoder_stage0a_bn[0][0] \n__________________________________________________________________________________________________\ndecoder_stage0b_conv (Conv2D) (None, 16, 16, 256) 589824 decoder_stage0a_relu[0][0] \n__________________________________________________________________________________________________\ndecoder_stage0b_bn (BatchNormal (None, 16, 16, 256) 1024 decoder_stage0b_conv[0][0] \n__________________________________________________________________________________________________\ndecoder_stage0b_relu (Activatio (None, 16, 16, 256) 0 decoder_stage0b_bn[0][0] \n__________________________________________________________________________________________________\ndecoder_stage1_upsampling (UpSa (None, 32, 32, 256) 0 decoder_stage0b_relu[0][0] \n__________________________________________________________________________________________________\ndecoder_stage1_concat (Concaten (None, 32, 32, 768) 0 decoder_stage1_upsampling[0][0] \n stage3_unit1_relu1[0][0] \n__________________________________________________________________________________________________\ndecoder_stage1a_conv (Conv2D) (None, 32, 32, 128) 884736 decoder_stage1_concat[0][0] \n__________________________________________________________________________________________________\ndecoder_stage1a_bn (BatchNormal (None, 32, 32, 128) 512 decoder_stage1a_conv[0][0] \n__________________________________________________________________________________________________\ndecoder_stage1a_relu (Activatio (None, 32, 32, 128) 0 decoder_stage1a_bn[0][0] \n__________________________________________________________________________________________________\ndecoder_stage1b_conv (Conv2D) (None, 32, 32, 128) 147456 decoder_stage1a_relu[0][0] \n__________________________________________________________________________________________________\ndecoder_stage1b_bn (BatchNormal (None, 32, 32, 128) 512 decoder_stage1b_conv[0][0] \n__________________________________________________________________________________________________\ndecoder_stage1b_relu (Activatio (None, 32, 32, 128) 0 decoder_stage1b_bn[0][0] \n__________________________________________________________________________________________________\ndecoder_stage2_upsampling (UpSa (None, 64, 64, 128) 0 decoder_stage1b_relu[0][0] \n__________________________________________________________________________________________________\ndecoder_stage2_concat (Concaten (None, 64, 64, 384) 0 decoder_stage2_upsampling[0][0] \n stage2_unit1_relu1[0][0] \n__________________________________________________________________________________________________\ndecoder_stage2a_conv (Conv2D) (None, 64, 64, 64) 221184 decoder_stage2_concat[0][0] \n__________________________________________________________________________________________________\ndecoder_stage2a_bn (BatchNormal (None, 64, 64, 64) 256 decoder_stage2a_conv[0][0] \n__________________________________________________________________________________________________\ndecoder_stage2a_relu (Activatio (None, 64, 64, 64) 0 decoder_stage2a_bn[0][0] \n__________________________________________________________________________________________________\ndecoder_stage2b_conv (Conv2D) (None, 64, 64, 64) 36864 decoder_stage2a_relu[0][0] \n__________________________________________________________________________________________________\ndecoder_stage2b_bn (BatchNormal (None, 64, 64, 64) 256 decoder_stage2b_conv[0][0] \n__________________________________________________________________________________________________\ndecoder_stage2b_relu (Activatio (None, 64, 64, 64) 0 decoder_stage2b_bn[0][0] \n__________________________________________________________________________________________________\ndecoder_stage3_upsampling (UpSa (None, 128, 128, 64) 0 decoder_stage2b_relu[0][0] \n__________________________________________________________________________________________________\ndecoder_stage3_concat (Concaten (None, 128, 128, 128 0 decoder_stage3_upsampling[0][0] \n relu0[0][0] \n__________________________________________________________________________________________________\ndecoder_stage3a_conv (Conv2D) (None, 128, 128, 32) 36864 decoder_stage3_concat[0][0] \n__________________________________________________________________________________________________\ndecoder_stage3a_bn (BatchNormal (None, 128, 128, 32) 128 decoder_stage3a_conv[0][0] \n__________________________________________________________________________________________________\ndecoder_stage3a_relu (Activatio (None, 128, 128, 32) 0 decoder_stage3a_bn[0][0] \n__________________________________________________________________________________________________\ndecoder_stage3b_conv (Conv2D) (None, 128, 128, 32) 9216 decoder_stage3a_relu[0][0] \n__________________________________________________________________________________________________\ndecoder_stage3b_bn (BatchNormal (None, 128, 128, 32) 128 decoder_stage3b_conv[0][0] \n__________________________________________________________________________________________________\ndecoder_stage3b_relu (Activatio (None, 128, 128, 32) 0 decoder_stage3b_bn[0][0] \n__________________________________________________________________________________________________\ndecoder_stage4_upsampling (UpSa (None, 256, 256, 32) 0 decoder_stage3b_relu[0][0] \n__________________________________________________________________________________________________\ndecoder_stage4a_conv (Conv2D) (None, 256, 256, 16) 4608 decoder_stage4_upsampling[0][0] \n__________________________________________________________________________________________________\ndecoder_stage4a_bn (BatchNormal (None, 256, 256, 16) 64 decoder_stage4a_conv[0][0] \n__________________________________________________________________________________________________\ndecoder_stage4a_relu (Activatio (None, 256, 256, 16) 0 decoder_stage4a_bn[0][0] \n__________________________________________________________________________________________________\ndecoder_stage4b_conv (Conv2D) (None, 256, 256, 16) 2304 decoder_stage4a_relu[0][0] \n__________________________________________________________________________________________________\ndecoder_stage4b_bn (BatchNormal (None, 256, 256, 16) 64 decoder_stage4b_conv[0][0] \n__________________________________________________________________________________________________\ndecoder_stage4b_relu (Activatio (None, 256, 256, 16) 0 decoder_stage4b_bn[0][0] \n__________________________________________________________________________________________________\nfinal_conv (Conv2D) (None, 256, 256, 1) 145 decoder_stage4b_relu[0][0] \n__________________________________________________________________________________________________\nsigmoid (Activation) (None, 256, 256, 1) 0 final_conv[0][0] \n==================================================================================================\nTotal params: 51,605,466\nTrainable params: 51,505,684\nNon-trainable params: 99,782\n__________________________________________________________________________________________________\n"
]
],
[
[
"### HYPER_PARAMETERS",
"_____no_output_____"
]
],
[
[
"LEARNING_RATE = 0.0001",
"_____no_output_____"
]
],
[
[
"### Initializing Callbacks",
"_____no_output_____"
]
],
[
[
"#from tensorboardcolab import TensorBoardColab, TensorBoardColabCallback\nfrom keras.callbacks import EarlyStopping, ModelCheckpoint, ReduceLROnPlateau\nfrom datetime import datetime",
"_____no_output_____"
],
[
"model_path = \"./Models/Resnet_road_weights.h5\"\ncheckpointer = ModelCheckpoint(model_path,\n monitor=\"val_loss\",\n mode=\"min\",\n save_best_only = True,\n verbose=1)\n\nearlystopper = EarlyStopping(monitor = 'val_loss', \n min_delta = 0, \n patience = 5,\n verbose = 1,\n restore_best_weights = True)\n\nlr_reducer = ReduceLROnPlateau(monitor='val_loss',\n factor=0.1,\n patience=4,\n verbose=1,\n epsilon=1e-4)",
"_____no_output_____"
]
],
[
[
"### Compiling the model",
"_____no_output_____"
]
],
[
[
"opt = keras.optimizers.adam(LEARNING_RATE)\nmodel.compile(\n optimizer=opt,\n loss=soft_dice_loss,\n metrics=[iou_coef])",
"WARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/optimizers.py:793: The name tf.train.Optimizer is deprecated. Please use tf.compat.v1.train.Optimizer instead.\n\n"
]
],
[
[
"## Testing our Model",
"_____no_output_____"
],
[
"### On Test Images",
"_____no_output_____"
]
],
[
[
"model.load_weights(\"Models/Resnet_road_weights.h5\")",
"_____no_output_____"
],
[
"import cv2\nimport glob\nimport numpy as np\nimport h5py\n\n#test_images = np.array([cv2.imread(file) for file in glob.glob(\"/home/bisag/Desktop/Road-Segmentation/I/\")])\n#test_masks = np.array([cv2.imread(file) for file in glob.glob(\"/home/bisag/Desktop/Road-Segmentation/M/\")])\ntest_masks = []\ntest_images = []\nfiles = glob.glob (\"TestI/*.png\")\nfor myFile in files:\n print(myFile)\n image = cv2.imread (myFile)\n test_images.append (image)\n myFile = 'TestM' + myFile[5:len(myFile)]\n image = cv2.cvtColor(cv2.imread (myFile), cv2.COLOR_BGR2GRAY)\n test_masks.append (image)\n\n\n#files = glob.glob (\"TestM/*.png\")\n#for myFile in files:\n# print(myFile)\n\n \n#test_images = cv2.imread(\"/home/bisag/Desktop/Road-Segmentation/I/1.png\")\n#test_masks = cv2.imread(\"/home/bisag/Desktop/Road-Segmentation/M/1.png\")\n\ntest_images = np.array(test_images)\ntest_masks = np.array(test_masks)\n\ntest_masks = np.expand_dims(test_masks, -1)\nprint(\"Unique elements in the train mask:\", np.unique(test_masks))\n\nprint(test_images.shape)\nprint(test_masks.shape)\n\ntest_images = test_images.astype(np.float16)/255\ntest_masks = test_masks.astype(np.float16)/255\n",
"TestI/10378690_15_24.png\nTestI/10378690_15_4.png\nTestI/10228705_15_22.png\nTestI/10378675_15_14.png\nTestI/10078690_15_18.png\nTestI/10228660_15_20.png\nTestI/10378675_15_4.png\nTestI/10078735_15_25.png\nTestI/10228675_15_22.png\nTestI/10078690_15_16.png\nTestI/10228735_15_20.png\nTestI/10078720_15_14.png\nTestI/10378675_15_24.png\nTestI/10378690_15_5.png\nTestI/10078660_15_4.png\nTestI/10228705_15_24.png\nTestI/10228735_15_13.png\nTestI/10228720_15_2.png\nTestI/10378660_15_19.png\nTestI/10378675_15_13.png\nTestI/10228705_15_7.png\nTestI/10228765_15_25.png\nTestI/10228675_15_6.png\nTestI/10228675_15_9.png\nTestI/10228720_15_16.png\nTestI/10228705_15_19.png\nTestI/10228750_15_12.png\nTestI/10078720_15_18.png\nTestI/10228675_15_14.png\nTestI/10228750_15_5.png\nUnique elements in the train mask: [ 0 255]\n(30, 256, 256, 3)\n(30, 256, 256, 1)\n"
],
[
"import sys\ndef sizeof_fmt(num, suffix='B'):\n ''' by Fred Cirera, https://stackoverflow.com/a/1094933/1870254, modified'''\n for unit in ['','Ki','Mi','Gi','Ti','Pi','Ei','Zi']:\n if abs(num) < 1024.0:\n return \"%3.1f %s%s\" % (num, unit, suffix)\n num /= 1024.0\n return \"%.1f %s%s\" % (num, 'Yi', suffix)\n\nfor name, size in sorted(((name, sys.getsizeof(value)) for name, value in locals().items()),\n key= lambda x: -x[1])[:10]:\n print(\"{:>30}: {:>8}\".format(name, sizeof_fmt(size)))",
" test_images: 11.3 MiB\n test_masks: 3.8 MiB\n image: 64.1 KiB\n Conv2D: 2.0 KiB\n BatchNormalization: 2.0 KiB\n MaxPooling2D: 1.4 KiB\n EarlyStopping: 1.4 KiB\n ModelCheckpoint: 1.4 KiB\n ReduceLROnPlateau: 1.4 KiB\n _i: 1.2 KiB\n"
],
[
"test_masks_tmp = []\nfor i in test_masks:\n image = cv2.cvtColor(i, cv2.COLOR_BGR2GRAY)\n test_masks_tmp.append (image)\n\ntest_images = np.array(test_images)\ntest_masks = np.array(test_masks_tmp)\n\ntest_masks = np.expand_dims(test_masks, -1)\n\n#print(np.unique(test_masks))\nprint(test_images.shape)\nprint(test_masks.shape)\ndel test_masks_tmp",
"_____no_output_____"
],
[
"model.evaluate(test_images, test_masks)",
"\r30/30 [==============================] - 18s 584ms/step\n"
],
[
"predictions = model.predict(test_images, verbose=1)",
"\r30/30 [==============================] - 17s 555ms/step\n"
],
[
"thresh_val = 0.1\npredicton_threshold = (predictions > thresh_val).astype(np.uint8)",
"_____no_output_____"
],
[
"plt.figure()\n#plt.subplot(2, 1, 1)\nplt.imshow(np.squeeze(predictions[19][:,:,0]))\nplt.show()\n\n",
"_____no_output_____"
],
[
"import matplotlib\n\nfor i in range(len(predictions)):\n #print(\"Results/\" + str(i) + \"Image.png\")\n matplotlib.image.imsave( \"Results/\" + str(i) + \"Image.png\" , np.squeeze(test_images[i][:,:,0]))\n matplotlib.image.imsave( \"Results/\" + str(i) + \"GroundTruth.png\" , np.squeeze(test_masks[i][:,:,0]))\n #cv2.imwrite( \"/home/bisag/Desktop/Road-Segmentation/Results/\" + str(i) + \"Prediction.png\" , np.squeeze(predictions[i][:,:,0]))\n #cv2.imwrite( \"/home/bisag/Desktop/Road-Segmentation/Results/\" + str(i) + \"Prediction_Threshold.png\" , np.squeeze(predicton_threshold[i][:,:,0]))\n #matplotlib.image.imsave('/home/bisag/Desktop/Road-Segmentation/Results/000.png', np.squeeze(predicton_threshold[0][:,:,0]))\n matplotlib.image.imsave(\"Results/\" + str(i) + \"Prediction.png\" , np.squeeze(predictions[i][:,:,0]))\n matplotlib.image.imsave( \"Results/\" + str(i) + \"Prediction_Threshold.png\" , np.squeeze(predicton_threshold[i][:,:,0]))\n\n #imshow(np.squeeze(predictions[0][:,:,0]))\n\n\n#import scipy.misc\n#scipy.misc.imsave('/home/bisag/Desktop/Road-Segmentation/Results/00.png', np.squeeze(predictions[0][:,:,0]))",
"_____no_output_____"
],
[
"model.load_weights(\"/home/parshwa/Desktop/Road-Segmentation/Models/weights.h5\")",
"_____no_output_____"
]
],
[
[
"### Just Test",
"_____no_output_____"
]
],
[
[
"\"\"\"Test\"\"\"\nimport cv2\nimport glob\nimport numpy as np\nimport h5py\n\n#test_images = np.array([cv2.imread(file) for file in glob.glob(\"/home/bisag/Desktop/Road-Segmentation/I/\")])\n#test_masks = np.array([cv2.imread(file) for file in glob.glob(\"/home/bisag/Desktop/Road-Segmentation/M/\")])\n\ntest_images = []\nfiles = glob.glob (\"/home/parshwa/Desktop/Road-Segmentation/Test/*.png\")\nfor myFile in files:\n print(myFile)\n image = cv2.imread (myFile)\n test_images.append (image)\n\n\n \n#test_images = cv2.imread(\"/home/bisag/Desktop/Road-Segmentation/I/1.png\")\n#test_masks = cv2.imread(\"/home/bisag/Desktop/Road-Segmentation/M/1.png\")\n\ntest_images = np.array(test_images)\n\nprint(test_images.shape)\n\n",
"_____no_output_____"
],
[
"predictions = model.predict(test_images, verbose=1)",
"_____no_output_____"
],
[
"thresh_val = 0.1\npredicton_threshold = (predictions > thresh_val).astype(np.uint8)",
"_____no_output_____"
],
[
"import matplotlib\n\nfor i in range(len(predictions)):\n cv2.imwrite( \"/home/parshwa/Desktop/Road-Segmentation/Results/\" + str(i) + \"Image.png\" , np.squeeze(test_images[i][:,:,0]))\n #cv2.imwrite( \"/home/bisag/Desktop/Road-Segmentation/Results/\" + str(i) + \"Prediction.png\" , np.squeeze(predictions[i][:,:,0]))\n #cv2.imwrite( \"/home/bisag/Desktop/Road-Segmentation/Results/\" + str(i) + \"Prediction_Threshold.png\" , np.squeeze(predicton_threshold[i][:,:,0]))\n #matplotlib.image.imsave('/home/bisag/Desktop/Road-Segmentation/Results/000.png', np.squeeze(predicton_threshold[0][:,:,0]))\n matplotlib.image.imsave(\"/home/parshwa/Desktop/Road-Segmentation/Results/\" + str(i) + \"Prediction.png\" , np.squeeze(predictions[i][:,:,0]))\n matplotlib.image.imsave( \"/home/parshwa/Desktop/Road-Segmentation/Results/\" + str(i) + \"Prediction_Threshold.png\" , np.squeeze(predicton_threshold[i][:,:,0]))\n\n #imshow(np.squeeze(predictions[0][:,:,0]))\n\nimshow(np.squeeze(predictions[0][:,:,0]))\n\n#import scipy.misc\n#scipy.misc.imsave('/home/bisag/Desktop/Road-Segmentation/Results/00.png', np.squeeze(predictions[0][:,:,0]))",
"_____no_output_____"
],
[
"\"\"\"Visualise\"\"\"\ndef layer_to_visualize(layer):\n inputs = [K.learning_phase()] + model.inputs\n\n _convout1_f = K.function(inputs, [layer.output])\n def convout1_f(X):\n # The [0] is to disable the training phase flag\n return _convout1_f([0] + [X])\n\n convolutions = convout1_f(img_to_visualize)\n convolutions = np.squeeze(convolutions)\n\n print ('Shape of conv:', convolutions.shape)\n\n n = convolutions.shape[0]\n n = int(np.ceil(np.sqrt(n)))\n\n # Visualization of each filter of the layer\n fig = plt.figure(figsize=(12,8))\n for i in range(len(convolutions)):\n ax = fig.add_subplot(n,n,i+1)\n ax.imshow(convolutions[i], cmap='gray')\n",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0953577bf25aabc837ac00cbe163ecdf41b4e2c | 76,096 | ipynb | Jupyter Notebook | coursework/03-CNN-Convolutional-Neural-Networks/MNIST - CNN.ipynb | saint1729/deep-learning-pytorch | 7157c32d58a8d053cf56811eb03af9adb37597d1 | [
"MIT"
] | null | null | null | coursework/03-CNN-Convolutional-Neural-Networks/MNIST - CNN.ipynb | saint1729/deep-learning-pytorch | 7157c32d58a8d053cf56811eb03af9adb37597d1 | [
"MIT"
] | null | null | null | coursework/03-CNN-Convolutional-Neural-Networks/MNIST - CNN.ipynb | saint1729/deep-learning-pytorch | 7157c32d58a8d053cf56811eb03af9adb37597d1 | [
"MIT"
] | null | null | null | 77.412004 | 18,000 | 0.733573 | [
[
[
"import torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nfrom torch.utils.data import DataLoader\nfrom torchvision import datasets, transforms\nfrom torchvision.utils import make_grid\n\nimport numpy as np\nimport pandas as pd\nfrom sklearn.metrics import confusion_matrix\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\nimport time",
"_____no_output_____"
],
[
"transform = transforms.ToTensor()",
"_____no_output_____"
],
[
"train_data = datasets.MNIST(root='../Data', train=True, download=True, transform=transform)",
"_____no_output_____"
],
[
"test_data = datasets.MNIST(root='../Data', train=False, download=True, transform=transform)",
"_____no_output_____"
],
[
"train_data",
"_____no_output_____"
],
[
"test_data",
"_____no_output_____"
],
[
"train_loader = DataLoader(train_data, batch_size=10, shuffle=True)\ntest_loader = DataLoader(test_data, batch_size=10, shuffle=False)",
"_____no_output_____"
],
[
"conv1 = nn.Conv2d(1, 6, 3, 1)\nconv2 = nn.Conv2d(6, 16, 3, 1)",
"_____no_output_____"
],
[
"for i, (X_train, y_train) in enumerate(train_data):\n break",
"_____no_output_____"
],
[
"x = X_train.view(1, 1, 28, 28)",
"_____no_output_____"
],
[
"x = F.relu(conv1(x))",
"_____no_output_____"
],
[
"x.shape",
"_____no_output_____"
],
[
"x = F.max_pool2d(x, 2, 2)",
"_____no_output_____"
],
[
"x.shape",
"_____no_output_____"
],
[
"x = F.relu(conv2(x))",
"_____no_output_____"
],
[
"x.shape",
"_____no_output_____"
],
[
"x = F.max_pool2d(x, 2, 2)",
"_____no_output_____"
],
[
"x.shape",
"_____no_output_____"
],
[
"x.view(-1, 16 * 5 * 5).shape",
"_____no_output_____"
],
[
"class ConvolutionalNetwork(nn.Module):\n def __init__(self):\n super().__init__()\n self.conv1 = nn.Conv2d(1, 6, 3, 1)\n self.conv2 = nn.Conv2d(6, 16, 3, 1)\n self.fc1 = nn.Linear(5 * 5 * 16, 120)\n self.fc2 = nn.Linear(120, 84)\n self.fc3 = nn.Linear(84, 10)\n \n def forward(self, X):\n X = F.relu(self.conv1(X))\n X = F.max_pool2d(X, 2, 2)\n X = F.relu(self.conv2(X))\n X = F.max_pool2d(X, 2, 2)\n X = X.view(-1, 16 * 5 * 5)\n X = F.relu(self.fc1(X))\n X = F.relu(self.fc2(X))\n X = F.log_softmax(self.fc3(X), dim=1)\n return X",
"_____no_output_____"
],
[
"torch.manual_seed(42)\nmodel = ConvolutionalNetwork()\nmodel",
"_____no_output_____"
],
[
"for param in model.parameters():\n print(param.numel())",
"54\n6\n864\n16\n48000\n120\n10080\n84\n840\n10\n"
],
[
"criterion = nn.CrossEntropyLoss()\noptimizer = torch.optim.Adam(model.parameters(), lr=0.001)",
"_____no_output_____"
],
[
"start_time = time.time()\n\n\nepochs = 5\ntrain_losses = []\ntest_losses = []\ntrain_correct = []\ntest_correct = []\n\nfor i in range(epochs):\n trn_corr = 0\n tst_corr = 0\n \n for b, (X_train, y_train) in enumerate(train_loader):\n b += 1\n \n y_pred = model(X_train)\n loss = criterion(y_pred, y_train)\n \n predicted = torch.max(y_pred.data, 1)[1]\n batch_corr = (predicted == y_train).sum()\n trn_corr += batch_corr\n \n optimizer.zero_grad()\n loss.backward()\n optimizer.step()\n \n if b % 600 == 0:\n print(f\"Epoch: {i} BATCH: {b} LOSS: {loss.item()}\")\n \n train_losses.append(loss)\n train_correct.append(trn_corr)\n \n with torch.no_grad():\n for b, (X_test, y_test) in enumerate(test_loader):\n y_val = model(X_test)\n predicted = torch.max(y_val.data, 1)[1]\n tst_corr += (predicted == y_test).sum()\n \n loss = criterion(y_val, y_test)\n test_losses.append(loss)\n test_correct.append(tst_corr)\n\n\ncurrent_time = time.time()\ntotal = current_time - start_time\nprint(f\"Training took {total / 60} minutes\")",
"Epoch: 0 BATCH: 600 LOSS: 0.4816969037055969\nEpoch: 0 BATCH: 1200 LOSS: 0.10990709066390991\nEpoch: 0 BATCH: 1800 LOSS: 0.018104346469044685\nEpoch: 0 BATCH: 2400 LOSS: 0.10995674133300781\nEpoch: 0 BATCH: 3000 LOSS: 0.0026193559169769287\nEpoch: 0 BATCH: 3600 LOSS: 0.05913165211677551\nEpoch: 0 BATCH: 4200 LOSS: 0.11463085561990738\nEpoch: 0 BATCH: 4800 LOSS: 0.0028016851283609867\nEpoch: 0 BATCH: 5400 LOSS: 0.005568644963204861\nEpoch: 0 BATCH: 6000 LOSS: 0.0599198155105114\n1\ntensor([[-1.2366e+01, -1.0606e+01, -8.0966e+00, -7.9539e+00, -1.1628e+01,\n -1.3640e+01, -2.1273e+01, -7.1607e-04, -1.2043e+01, -1.1117e+01],\n [-1.3008e+01, -1.0265e+01, -4.8040e-05, -1.1553e+01, -1.5649e+01,\n -1.9043e+01, -1.8359e+01, -1.4002e+01, -1.4960e+01, -2.0766e+01],\n [-1.2239e+01, -6.1326e-04, -1.0169e+01, -1.1805e+01, -8.2123e+00,\n -1.1868e+01, -9.4521e+00, -9.3454e+00, -9.1483e+00, -1.1348e+01],\n [-2.9480e-03, -1.2429e+01, -7.2616e+00, -8.2204e+00, -1.0337e+01,\n -7.9653e+00, -8.3047e+00, -9.6860e+00, -8.4244e+00, -6.8496e+00],\n [-1.0381e+01, -9.6773e+00, -9.5559e+00, -9.9186e+00, -1.7862e-02,\n -8.6334e+00, -7.7808e+00, -1.1773e+01, -7.8653e+00, -4.1043e+00],\n [-1.6763e+01, -4.6133e-05, -1.3613e+01, -1.5910e+01, -1.0585e+01,\n -1.6703e+01, -1.4645e+01, -1.1517e+01, -1.1675e+01, -1.4628e+01],\n [-1.7681e+01, -7.4023e+00, -1.2501e+01, -1.3932e+01, -3.5442e-02,\n -8.9475e+00, -1.1434e+01, -1.0010e+01, -3.3923e+00, -7.8493e+00],\n [-1.1723e+01, -6.5215e+00, -8.9891e+00, -3.8989e+00, -4.4738e+00,\n -6.3457e+00, -1.2474e+01, -9.6107e+00, -5.6009e+00, -3.9566e-02],\n [-1.0328e+01, -1.0709e+01, -1.4352e+01, -8.9741e+00, -1.5124e+01,\n -1.6314e-02, -4.1450e+00, -1.6266e+01, -8.8490e+00, -1.1356e+01],\n [-1.6031e+01, -1.3104e+01, -1.5597e+01, -1.2535e+01, -5.8834e+00,\n -1.2290e+01, -1.8808e+01, -1.0926e+01, -8.3061e+00, -3.0654e-03]])\n2\ntensor([[-1.2366e+01, -1.0606e+01, -8.0966e+00, -7.9539e+00, -1.1628e+01,\n -1.3640e+01, -2.1273e+01, -7.1607e-04, -1.2043e+01, -1.1117e+01],\n [-1.3008e+01, -1.0265e+01, -4.8040e-05, -1.1553e+01, -1.5649e+01,\n -1.9043e+01, -1.8359e+01, -1.4002e+01, -1.4960e+01, -2.0766e+01],\n [-1.2239e+01, -6.1326e-04, -1.0169e+01, -1.1805e+01, -8.2123e+00,\n -1.1868e+01, -9.4521e+00, -9.3454e+00, -9.1483e+00, -1.1348e+01],\n [-2.9480e-03, -1.2429e+01, -7.2616e+00, -8.2204e+00, -1.0337e+01,\n -7.9653e+00, -8.3047e+00, -9.6860e+00, -8.4244e+00, -6.8496e+00],\n [-1.0381e+01, -9.6773e+00, -9.5559e+00, -9.9186e+00, -1.7862e-02,\n -8.6334e+00, -7.7808e+00, -1.1773e+01, -7.8653e+00, -4.1043e+00],\n [-1.6763e+01, -4.6133e-05, -1.3613e+01, -1.5910e+01, -1.0585e+01,\n -1.6703e+01, -1.4645e+01, -1.1517e+01, -1.1675e+01, -1.4628e+01],\n [-1.7681e+01, -7.4023e+00, -1.2501e+01, -1.3932e+01, -3.5442e-02,\n -8.9475e+00, -1.1434e+01, -1.0010e+01, -3.3923e+00, -7.8493e+00],\n [-1.1723e+01, -6.5215e+00, -8.9891e+00, -3.8989e+00, -4.4738e+00,\n -6.3457e+00, -1.2474e+01, -9.6107e+00, -5.6009e+00, -3.9566e-02],\n [-1.0328e+01, -1.0709e+01, -1.4352e+01, -8.9741e+00, -1.5124e+01,\n -1.6314e-02, -4.1450e+00, -1.6266e+01, -8.8490e+00, -1.1356e+01],\n [-1.6031e+01, -1.3104e+01, -1.5597e+01, -1.2535e+01, -5.8834e+00,\n -1.2290e+01, -1.8808e+01, -1.0926e+01, -8.3061e+00, -3.0654e-03]])\n3\ntorch.return_types.max(\nvalues=tensor([-7.1607e-04, -4.8040e-05, -6.1326e-04, -2.9480e-03, -1.7862e-02,\n -4.6133e-05, -3.5442e-02, -3.9566e-02, -1.6314e-02, -3.0654e-03]),\nindices=tensor([7, 2, 1, 0, 4, 1, 4, 9, 5, 9]))\n4\ntensor([7, 2, 1, 0, 4, 1, 4, 9, 5, 9])\nEpoch: 1 BATCH: 600 LOSS: 0.053106147795915604\nEpoch: 1 BATCH: 1200 LOSS: 0.004053960554301739\nEpoch: 1 BATCH: 1800 LOSS: 0.03800049051642418\nEpoch: 1 BATCH: 2400 LOSS: 0.015968404710292816\nEpoch: 1 BATCH: 3000 LOSS: 6.104153726482764e-05\nEpoch: 1 BATCH: 3600 LOSS: 0.1219397634267807\nEpoch: 1 BATCH: 4200 LOSS: 0.014603614807128906\nEpoch: 1 BATCH: 4800 LOSS: 0.18480448424816132\nEpoch: 1 BATCH: 5400 LOSS: 0.07728765904903412\nEpoch: 1 BATCH: 6000 LOSS: 0.34217339754104614\n1\ntensor([[-2.0286e+01, -1.3063e+01, -1.4802e+01, -1.0365e+01, -1.3617e+01,\n -1.7535e+01, -2.9164e+01, -6.7351e-05, -1.6291e+01, -1.0350e+01],\n [-1.8556e+01, -1.0925e+01, -1.8000e-05, -1.9779e+01, -2.0506e+01,\n -2.3138e+01, -1.9619e+01, -2.0428e+01, -1.9775e+01, -2.2517e+01],\n [-1.8916e+01, -1.9073e-06, -1.5206e+01, -2.0411e+01, -1.5486e+01,\n -1.7805e+01, -1.3694e+01, -1.5155e+01, -1.6003e+01, -1.8355e+01],\n [-2.2937e-02, -1.1946e+01, -1.0117e+01, -1.3681e+01, -1.0582e+01,\n -9.2423e+00, -3.8017e+00, -1.3006e+01, -9.7555e+00, -9.0820e+00],\n [-1.6076e+01, -1.5480e+01, -1.2990e+01, -1.6069e+01, -8.1893e-05,\n -1.2965e+01, -1.0986e+01, -1.5674e+01, -1.5135e+01, -9.7299e+00],\n [-2.2853e+01, 0.0000e+00, -1.9285e+01, -2.5378e+01, -1.8172e+01,\n -2.3402e+01, -2.0000e+01, -1.7685e+01, -1.9226e+01, -2.1037e+01],\n [-1.9746e+01, -1.1097e+01, -1.5131e+01, -1.8213e+01, -3.7344e-03,\n -1.0519e+01, -1.0793e+01, -1.3119e+01, -5.6752e+00, -8.3661e+00],\n [-1.4230e+01, -1.0998e+01, -1.3725e+01, -1.0768e+01, -5.6841e+00,\n -9.6870e+00, -1.5299e+01, -1.4647e+01, -8.3078e+00, -3.7557e-03],\n [-1.2204e+01, -1.0382e+01, -1.6367e+01, -1.2602e+01, -1.1860e+01,\n -4.7523e-03, -5.5372e+00, -1.3765e+01, -7.2255e+00, -1.0482e+01],\n [-1.8447e+01, -1.6706e+01, -1.9218e+01, -1.5037e+01, -8.4691e+00,\n -1.4987e+01, -2.1147e+01, -1.2826e+01, -1.0301e+01, -2.4685e-04]])\n2\ntensor([[-2.0286e+01, -1.3063e+01, -1.4802e+01, -1.0365e+01, -1.3617e+01,\n -1.7535e+01, -2.9164e+01, -6.7351e-05, -1.6291e+01, -1.0350e+01],\n [-1.8556e+01, -1.0925e+01, -1.8000e-05, -1.9779e+01, -2.0506e+01,\n -2.3138e+01, -1.9619e+01, -2.0428e+01, -1.9775e+01, -2.2517e+01],\n [-1.8916e+01, -1.9073e-06, -1.5206e+01, -2.0411e+01, -1.5486e+01,\n -1.7805e+01, -1.3694e+01, -1.5155e+01, -1.6003e+01, -1.8355e+01],\n [-2.2937e-02, -1.1946e+01, -1.0117e+01, -1.3681e+01, -1.0582e+01,\n -9.2423e+00, -3.8017e+00, -1.3006e+01, -9.7555e+00, -9.0820e+00],\n [-1.6076e+01, -1.5480e+01, -1.2990e+01, -1.6069e+01, -8.1893e-05,\n -1.2965e+01, -1.0986e+01, -1.5674e+01, -1.5135e+01, -9.7299e+00],\n [-2.2853e+01, 0.0000e+00, -1.9285e+01, -2.5378e+01, -1.8172e+01,\n -2.3402e+01, -2.0000e+01, -1.7685e+01, -1.9226e+01, -2.1037e+01],\n [-1.9746e+01, -1.1097e+01, -1.5131e+01, -1.8213e+01, -3.7344e-03,\n -1.0519e+01, -1.0793e+01, -1.3119e+01, -5.6752e+00, -8.3661e+00],\n [-1.4230e+01, -1.0998e+01, -1.3725e+01, -1.0768e+01, -5.6841e+00,\n -9.6870e+00, -1.5299e+01, -1.4647e+01, -8.3078e+00, -3.7557e-03],\n [-1.2204e+01, -1.0382e+01, -1.6367e+01, -1.2602e+01, -1.1860e+01,\n -4.7523e-03, -5.5372e+00, -1.3765e+01, -7.2255e+00, -1.0482e+01],\n [-1.8447e+01, -1.6706e+01, -1.9218e+01, -1.5037e+01, -8.4691e+00,\n -1.4987e+01, -2.1147e+01, -1.2826e+01, -1.0301e+01, -2.4685e-04]])\n3\ntorch.return_types.max(\nvalues=tensor([-6.7351e-05, -1.8000e-05, -1.9073e-06, -2.2937e-02, -8.1893e-05,\n 0.0000e+00, -3.7344e-03, -3.7557e-03, -4.7523e-03, -2.4685e-04]),\nindices=tensor([7, 2, 1, 0, 4, 1, 4, 9, 5, 9]))\n4\ntensor([7, 2, 1, 0, 4, 1, 4, 9, 5, 9])\nEpoch: 2 BATCH: 600 LOSS: 0.0053770639933645725\nEpoch: 2 BATCH: 1200 LOSS: 0.006665552966296673\nEpoch: 2 BATCH: 1800 LOSS: 0.011250527575612068\nEpoch: 2 BATCH: 2400 LOSS: 0.005573090631514788\nEpoch: 2 BATCH: 3000 LOSS: 0.0022761584259569645\nEpoch: 2 BATCH: 3600 LOSS: 0.03163021057844162\nEpoch: 2 BATCH: 4200 LOSS: 0.0023643814492970705\nEpoch: 2 BATCH: 4800 LOSS: 0.0005138001288287342\nEpoch: 2 BATCH: 5400 LOSS: 0.0036241114139556885\nEpoch: 2 BATCH: 6000 LOSS: 0.00020174476958345622\n1\ntensor([[-2.2738e+01, -1.7007e+01, -1.5693e+01, -1.4687e+01, -1.5451e+01,\n -1.8895e+01, -2.9453e+01, -1.5497e-06, -1.7160e+01, -1.4190e+01],\n [-1.3311e+01, -9.1252e+00, -1.1134e-04, -1.7959e+01, -1.6399e+01,\n -1.7465e+01, -1.5193e+01, -1.5884e+01, -1.4886e+01, -1.9549e+01],\n [-1.8627e+01, -1.0311e-04, -1.2386e+01, -1.9073e+01, -1.0729e+01,\n -1.4375e+01, -1.1519e+01, -1.2524e+01, -9.6815e+00, -1.5025e+01],\n [-3.8146e-05, -1.5521e+01, -1.4249e+01, -1.6109e+01, -1.8881e+01,\n -1.3486e+01, -1.0393e+01, -1.7400e+01, -1.2196e+01, -1.5480e+01],\n [-1.6919e+01, -1.4237e+01, -1.2351e+01, -1.7684e+01, -1.3076e-04,\n -1.5466e+01, -1.1082e+01, -1.4038e+01, -1.2894e+01, -9.1448e+00],\n [-2.1768e+01, -1.7166e-05, -1.5405e+01, -2.3151e+01, -1.2385e+01,\n -1.8221e+01, -1.6042e+01, -1.4886e+01, -1.1304e+01, -1.6864e+01],\n [-2.2937e+01, -1.1746e+01, -1.6224e+01, -2.2977e+01, -6.1719e-04,\n -1.3826e+01, -1.2442e+01, -1.2570e+01, -7.6997e+00, -8.8200e+00],\n [-1.4564e+01, -9.9941e+00, -9.3720e+00, -1.3149e+01, -4.6236e+00,\n -1.2857e+01, -1.6203e+01, -1.3221e+01, -7.8410e+00, -1.0403e-02],\n [-6.9392e+00, -9.3670e+00, -1.1382e+01, -8.2846e+00, -1.0247e+01,\n -9.9605e-02, -3.0977e+00, -9.3800e+00, -3.1124e+00, -5.5940e+00],\n [-2.1361e+01, -1.9765e+01, -2.0151e+01, -1.8553e+01, -9.2187e+00,\n -1.6997e+01, -2.3103e+01, -1.2750e+01, -9.3910e+00, -1.8547e-04]])\n2\ntensor([[-2.2738e+01, -1.7007e+01, -1.5693e+01, -1.4687e+01, -1.5451e+01,\n -1.8895e+01, -2.9453e+01, -1.5497e-06, -1.7160e+01, -1.4190e+01],\n [-1.3311e+01, -9.1252e+00, -1.1134e-04, -1.7959e+01, -1.6399e+01,\n -1.7465e+01, -1.5193e+01, -1.5884e+01, -1.4886e+01, -1.9549e+01],\n [-1.8627e+01, -1.0311e-04, -1.2386e+01, -1.9073e+01, -1.0729e+01,\n -1.4375e+01, -1.1519e+01, -1.2524e+01, -9.6815e+00, -1.5025e+01],\n [-3.8146e-05, -1.5521e+01, -1.4249e+01, -1.6109e+01, -1.8881e+01,\n -1.3486e+01, -1.0393e+01, -1.7400e+01, -1.2196e+01, -1.5480e+01],\n [-1.6919e+01, -1.4237e+01, -1.2351e+01, -1.7684e+01, -1.3076e-04,\n -1.5466e+01, -1.1082e+01, -1.4038e+01, -1.2894e+01, -9.1448e+00],\n [-2.1768e+01, -1.7166e-05, -1.5405e+01, -2.3151e+01, -1.2385e+01,\n -1.8221e+01, -1.6042e+01, -1.4886e+01, -1.1304e+01, -1.6864e+01],\n [-2.2937e+01, -1.1746e+01, -1.6224e+01, -2.2977e+01, -6.1719e-04,\n -1.3826e+01, -1.2442e+01, -1.2570e+01, -7.6997e+00, -8.8200e+00],\n [-1.4564e+01, -9.9941e+00, -9.3720e+00, -1.3149e+01, -4.6236e+00,\n -1.2857e+01, -1.6203e+01, -1.3221e+01, -7.8410e+00, -1.0403e-02],\n [-6.9392e+00, -9.3670e+00, -1.1382e+01, -8.2846e+00, -1.0247e+01,\n -9.9605e-02, -3.0977e+00, -9.3800e+00, -3.1124e+00, -5.5940e+00],\n [-2.1361e+01, -1.9765e+01, -2.0151e+01, -1.8553e+01, -9.2187e+00,\n -1.6997e+01, -2.3103e+01, -1.2750e+01, -9.3910e+00, -1.8547e-04]])\n3\ntorch.return_types.max(\nvalues=tensor([-1.5497e-06, -1.1134e-04, -1.0311e-04, -3.8146e-05, -1.3076e-04,\n -1.7166e-05, -6.1719e-04, -1.0403e-02, -9.9605e-02, -1.8547e-04]),\nindices=tensor([7, 2, 1, 0, 4, 1, 4, 9, 5, 9]))\n4\ntensor([7, 2, 1, 0, 4, 1, 4, 9, 5, 9])\n"
],
[
"with torch.no_grad():\n plt.plot(train_losses, label='train loss')\n plt.plot(test_losses, label='validation loss')\n plt.title('LOSS AT EPOCH')\n plt.legend()",
"_____no_output_____"
],
[
"plt.plot([t / 600 for t in train_correct], label='training accuracy')\nplt.plot([t / 100 for t in test_correct], label='validation accuracy')\nplt.title('Accuracy at the end of each epoch')\nplt.legend()",
"_____no_output_____"
],
[
"test_load_all = DataLoader(test_data, batch_size=10000, shuffle=False)",
"_____no_output_____"
],
[
"with torch.no_grad():\n correct = 0\n for X_test, y_test in test_load_all:\n y_val = model(X_test)\n predicted = torch.max(y_val, 1)[1]\n correct += (predicted == y_test).sum()",
"_____no_output_____"
],
[
"correct.item() * 100 / len(test_data)",
"_____no_output_____"
],
[
"np.set_printoptions(formatter=dict(int=lambda x: f'{x:4}'))\nprint(np.arange(10).reshape(1, 10))\nprint()\n\nprint(confusion_matrix(predicted.view(-1), y_test.view(-1)))",
"[[ 0 1 2 3 4 5 6 7 8 9]]\n\n[[ 977 0 3 0 0 4 5 1 8 1]\n [ 0 1129 2 0 1 0 3 9 1 3]\n [ 0 1 1015 1 0 1 1 4 3 0]\n [ 0 0 1 1006 0 10 0 1 1 1]\n [ 0 0 1 0 978 0 1 1 3 11]\n [ 0 2 0 1 0 870 0 0 0 2]\n [ 1 2 1 0 2 4 947 0 0 0]\n [ 2 1 7 0 0 1 0 1008 1 6]\n [ 0 0 2 1 0 0 1 3 950 8]\n [ 0 0 0 1 1 2 0 1 7 977]]\n"
],
[
"plt.imshow(test_data[333][0].reshape(28, 28))",
"_____no_output_____"
],
[
"model.eval()\nwith torch.no_grad():\n new_prediction = model(test_data[333][0].view(1, 1, 28, 28))",
"_____no_output_____"
],
[
"new_prediction",
"_____no_output_____"
],
[
"new_prediction.argmax()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0953b8c1d209f262f038f9111db0e6fdf4d2fab | 970,143 | ipynb | Jupyter Notebook | Session_2_ and_3_Seaborn.ipynb | reeha-parkar/applied-statistics | 6700ccdfdb6ed84e238467c900b6c1e4771d1dd5 | [
"MIT"
] | null | null | null | Session_2_ and_3_Seaborn.ipynb | reeha-parkar/applied-statistics | 6700ccdfdb6ed84e238467c900b6c1e4771d1dd5 | [
"MIT"
] | null | null | null | Session_2_ and_3_Seaborn.ipynb | reeha-parkar/applied-statistics | 6700ccdfdb6ed84e238467c900b6c1e4771d1dd5 | [
"MIT"
] | null | null | null | 1,959.884848 | 785,420 | 0.957421 | [
[
[
"### Seaborn (Working on the diabetes dataset)",
"_____no_output_____"
]
],
[
[
"import seaborn as sns\nimport matplotlib.pyplot as plt\nimport pandas as pd",
"_____no_output_____"
],
[
"df = pd.read_csv('diabetes.csv')",
"_____no_output_____"
],
[
"sns.lmplot(x='Age', y='BloodPressure', data=df)\nplt.show()",
"_____no_output_____"
],
[
"sns.lmplot(x='Age', y='Glucose', data=df, \n fit_reg=False, # No regression line\n hue='Outcome' # color by evaluation stage\n )\nplt.show()",
"_____no_output_____"
],
[
"sns.swarmplot(x='Outcome', y='Glucose', data=df)\nplt.show()",
"_____no_output_____"
],
[
"sns.lmplot(x='Outcome', y='Glucose', data=df)\nplt.show()",
"_____no_output_____"
],
[
"sns.boxplot(data=df.loc[df['Outcome']==0, ['Glucose']])\nplt.show()",
"_____no_output_____"
],
[
"sns.boxplot(data=df.loc[df['Outcome']==1, ['Glucose']])\nplt.show()",
"_____no_output_____"
],
[
"df.corr",
"_____no_output_____"
],
[
"sns.set(style='ticks', color_codes=True)\nsns.pairplot(df)\nplt.show()",
"_____no_output_____"
],
[
"y = df['Outcome']\ny",
"_____no_output_____"
],
[
"x = df.iloc[:, 0:8]\nx",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d09541ac44540930fa0f44d2132e92301b2c2e31 | 23,676 | ipynb | Jupyter Notebook | week04_approx_rl/homework_lasagne.ipynb | Kirili4ik/Practical_RL | b49d639163a84fccfce9cccb8fb7b1b7fc502713 | [
"Unlicense"
] | 5,304 | 2017-01-23T16:26:50.000Z | 2022-03-31T21:21:45.000Z | week04_approx_rl/homework_lasagne.ipynb | Kirili4ik/Practical_RL | b49d639163a84fccfce9cccb8fb7b1b7fc502713 | [
"Unlicense"
] | 451 | 2017-01-23T20:56:34.000Z | 2022-03-21T19:52:19.000Z | week04_approx_rl/homework_lasagne.ipynb | Kirili4ik/Practical_RL | b49d639163a84fccfce9cccb8fb7b1b7fc502713 | [
"Unlicense"
] | 1,731 | 2017-01-23T16:23:18.000Z | 2022-03-31T20:44:33.000Z | 30.828125 | 248 | 0.569353 | [
[
[
"## Homework 4\n\nToday we'll start by reproducing the DQN and then try improving it with the tricks we learned on the lecture:\n* Target networks\n* Double q-learning\n* Prioritized experience replay\n* Dueling DQN\n* Bootstrap DQN",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\n%matplotlib inline\n\n\n# If you are running on a server, launch xvfb to record game videos\n# Please make sure you have xvfb installed\nimport os\nif type(os.environ.get(\"DISPLAY\")) is not str or len(os.environ.get(\"DISPLAY\")) == 0:\n !bash ../xvfb start\n os.environ['DISPLAY'] = ':1'",
"_____no_output_____"
]
],
[
[
"# Processing game image (2 pts)\n\nRaw Atari images are large, 210x160x3 by default. However, we don't need that level of detail in order to learn them.\n\nWe can thus save a lot of time by preprocessing game image, including\n* Resizing to a smaller shape\n* Converting to grayscale\n* Cropping irrelevant image parts",
"_____no_output_____"
]
],
[
[
"from gym.core import ObservationWrapper\nfrom gym.spaces import Box\n\nfrom scipy.misc import imresize\n\n\nclass PreprocessAtari(ObservationWrapper):\n def __init__(self, env):\n \"\"\"A gym wrapper that crops, scales image into the desired shapes and optionally grayscales it.\"\"\"\n ObservationWrapper.__init__(self, env)\n\n self.img_size = (64, 64)\n self.observation_space = Box(0.0, 1.0, self.img_size)\n\n def observation(self, img):\n \"\"\"what happens to each observation\"\"\"\n\n # Here's what you need to do:\n # * crop image, remove irrelevant parts\n # * resize image to self.img_size\n # (use imresize imported above or any library you want,\n # e.g. opencv, skimage, PIL, keras)\n # * cast image to grayscale\n # * convert image pixels to (0,1) range, float32 type\n\n <Your code here>\n return <YOUR CODE>",
"_____no_output_____"
],
[
"import gym\n\n\ndef make_env():\n env = gym.make(\"KungFuMasterDeterministic-v0\") # create raw env\n return PreprocessAtari(env) # apply your wrapper\n\n\n# spawn game instance for tests\nenv = make_env()\n\nobservation_shape = env.observation_space.shape\nn_actions = env.action_space.n\n\nobs = env.reset()",
"_____no_output_____"
],
[
"# test observation\nassert obs.shape == observation_shape\nassert obs.dtype == 'float32'\nassert len(np.unique(obs)) > 2, \"your image must not be binary\"\nassert 0 <= np.min(obs) and np.max(\n obs) <= 1, \"convert image pixels to (0,1) range\"\n\nprint \"Formal tests seem fine. Here's an example of what you'll get.\"\n\nplt.title(\"what your network gonna see\")\nplt.imshow(obs, interpolation='none', cmap='gray')",
"_____no_output_____"
],
[
"plt.figure(figsize=[12, 12])\nenv.reset()\nfor i in range(16):\n for _ in range(10):\n new_obs = env.step(env.action_space.sample())[0]\n plt.subplot(4, 4, i+1)\n plt.imshow(new_obs, interpolation='none', cmap='gray')",
"_____no_output_____"
],
[
"# dispose of the game instance\ndel env",
"_____no_output_____"
]
],
[
[
"# Building a DQN (2 pts)\nHere we define a simple agent that maps game images into Qvalues using simple convolutional neural network.\n\n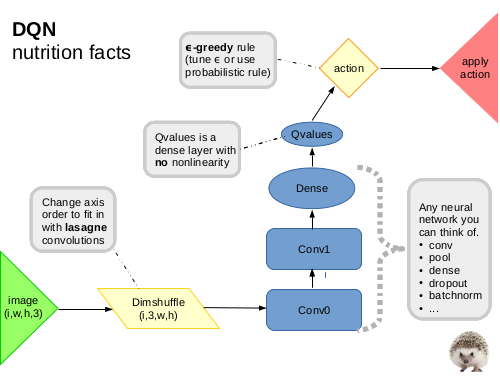",
"_____no_output_____"
]
],
[
[
"# setup theano/lasagne. Prefer GPU. Fallback to CPU (will print warning)\n%env THEANO_FLAGS = floatX = float32\n\nimport theano\nimport lasagne\nfrom lasagne.layers import *\nfrom theano import tensor as T",
"_____no_output_____"
],
[
"# observation\nobservation_layer = InputLayer(\n (None,)+observation_shape) # game image, [batch,64,64]",
"_____no_output_____"
],
[
"# 4-tick window over images\nfrom agentnet.memory import WindowAugmentation\n\n# window size [batch,4,64,64]\nprev_wnd = InputLayer((None, 4)+observation_shape)\n\nnew_wnd = WindowAugmentation( < current observation layer> , prev_wnd)\n\n# if you changed img size, remove assert\nassert new_wnd.output_shape == (None, 4, 64, 64)",
"_____no_output_____"
],
[
"from lasagne.nonlinearities import elu, tanh, softmax, rectify\n\n<network body, growing from new_wnd. several conv layers or something similar would do>\n\ndense = <final dense layer with 256 neurons>",
"_____no_output_____"
],
[
"# qvalues layer\nqvalues_layer = <a dense layer that predicts q-values>\n\nassert qvalues_layer.nonlinearity is not rectify",
"_____no_output_____"
],
[
"# sample actions proportionally to policy_layer\nfrom agentnet.resolver import EpsilonGreedyResolver\naction_layer = EpsilonGreedyResolver(qvalues_layer)",
"_____no_output_____"
]
],
[
[
"### Define agent\nHere you will need to declare how your agent works\n\n* `observation_layers` and `action_layers` are the input and output of agent in MDP.\n* `policy_estimators` must contain whatever you need for training\n * In our case, that's `qvalues_layer`, but you'll need to add more when implementing target network.\n* agent_states contains our frame buffer. \n * The code `{new_wnd:prev_wnd}` reads as \"`new_wnd becomes prev_wnd next turn`\"",
"_____no_output_____"
]
],
[
[
"from agentnet.agent import Agent\n# agent\nagent = Agent(observation_layers=<YOUR CODE>,\n policy_estimators=<YOUR CODE>,\n action_layers=<YOUR CODE>,\n agent_states={new_wnd: prev_wnd},)",
"_____no_output_____"
]
],
[
[
"# Create and manage a pool of Atari sessions to play with\n\n* To make training more stable, we shall have an entire batch of game sessions each happening independent of others\n* Why several parallel agents help training: http://arxiv.org/pdf/1602.01783v1.pdf\n* Alternative approach: store more sessions: https://www.cs.toronto.edu/~vmnih/docs/dqn.pdf",
"_____no_output_____"
]
],
[
[
"from agentnet.experiments.openai_gym.pool import EnvPool\n\npool = EnvPool(agent, make_env, n_games=16) # 16 parallel game sessions",
"_____no_output_____"
],
[
"%%time\n# interact for 7 ticks\n_, action_log, reward_log, _, _, _ = pool.interact(5)\n\nprint('actions:')\nprint(action_log[0])\nprint(\"rewards\")\nprint(reward_log[0])",
"_____no_output_____"
],
[
"# load first sessions (this function calls interact and remembers sessions)\nSEQ_LENGTH = 10 # sub-session length\npool.update(SEQ_LENGTH)",
"_____no_output_____"
]
],
[
[
"# Q-learning\n\nWe train our agent based on sessions it has played in `pool.update(SEQ_LENGTH)`\n\nTo do so, we first obtain sequences of observations, rewards, actions, q-values, etc.\n\nActions and rewards have shape `[n_games,seq_length]`, q-values are `[n_games,seq_length,n_actions]`",
"_____no_output_____"
]
],
[
[
"# get agent's Qvalues obtained via experience replay\nreplay = pool.experience_replay\n\nactions, rewards, is_alive = replay.actions[0], replay.rewards, replay.is_alive\n\n_, _, _, _, qvalues = agent.get_sessions(\n replay,\n session_length=SEQ_LENGTH,\n experience_replay=True,\n)\n\nassert actions.ndim == rewards.ndim == is_alive.ndim == 2, \"actions, rewards and is_alive must have shape [batch,time]\"\nassert qvalues.ndim == 3, \"q-values must have shape [batch,time,n_actions]\"",
"_____no_output_____"
],
[
"# compute V(s) as Qvalues of best actions.\n# For homework assignment, you will need to use target net\n# or special double q-learning objective here\n\nstate_values_target = <YOUR CODE: compute V(s) 2d tensor by taking T.argmax of qvalues over correct axis>\n\nassert state_values_target.eval().shape = qvalues.eval().shape[:2]",
"_____no_output_____"
],
[
"from agentnet.learning.generic import get_n_step_value_reference\n\n# get reference Q-values via Q-learning algorithm\nreference_qvalues = get_n_step_value_reference(\n state_values=state_values_target,\n rewards=rewards/100.,\n is_alive=is_alive,\n n_steps=10,\n gamma_or_gammas=0.99,\n)\n\n# consider it constant\nfrom theano.gradient import disconnected_grad\nreference_qvalues = disconnected_grad(reference_qvalues)",
"_____no_output_____"
],
[
"# get predicted Q-values for committed actions by both current and target networks\nfrom agentnet.learning.generic import get_values_for_actions\naction_qvalues = get_values_for_actions(qvalues, actions)",
"_____no_output_____"
],
[
"# loss for Qlearning =\n# (Q(s,a) - (r+ gamma*r' + gamma^2*r'' + ... +gamma^10*Q(s_{t+10},a_max)))^2\n\nelwise_mse_loss = <mean squared error between action qvalues and reference qvalues>\n\n# mean over all batches and time ticks\nloss = (elwise_mse_loss*is_alive).mean()",
"_____no_output_____"
],
[
"# Since it's a single lasagne network, one can get it's weights, output, etc\nweights = <YOUR CODE: get all trainable params>\nweights",
"_____no_output_____"
],
[
"# Compute weight updates\nupdates = <your favorite optimizer>\n\n# compile train function\ntrain_step = theano.function([], loss, updates=updates)",
"_____no_output_____"
]
],
[
[
"# Demo run\nas usual...",
"_____no_output_____"
]
],
[
[
"action_layer.epsilon.set_value(0.05)\nuntrained_reward = np.mean(pool.evaluate(save_path=\"./records\",\n record_video=True))",
"_____no_output_____"
],
[
"# show video\nfrom IPython.display import HTML\nimport os\n\nvideo_names = list(\n filter(lambda s: s.endswith(\".mp4\"), os.listdir(\"./records/\")))\n\nHTML(\"\"\"\n<video width=\"640\" height=\"480\" controls>\n <source src=\"{}\" type=\"video/mp4\">\n</video>\n\"\"\".format(\"./records/\" + video_names[-1])) # this may or may not be _last_ video. Try other indices",
"_____no_output_____"
]
],
[
[
"# Training loop",
"_____no_output_____"
]
],
[
[
"# starting epoch\nepoch_counter = 1\n\n# full game rewards\nrewards = {}\nloss, reward_per_tick, reward = 0, 0, 0",
"_____no_output_____"
],
[
"from tqdm import trange\nfrom IPython.display import clear_output\n\n\nfor i in trange(150000):\n\n # update agent's epsilon (in e-greedy policy)\n current_epsilon = 0.05 + 0.45*np.exp(-epoch_counter/20000.)\n action_layer.epsilon.set_value(np.float32(current_epsilon))\n\n # play\n pool.update(SEQ_LENGTH)\n\n # train\n loss = 0.95*loss + 0.05*train_step()\n\n if epoch_counter % 10 == 0:\n # average reward per game tick in current experience replay pool\n reward_per_tick = 0.95*reward_per_tick + 0.05 * \\\n pool.experience_replay.rewards.get_value().mean()\n print(\"iter=%i\\tepsilon=%.3f\\tloss=%.3f\\treward/tick=%.3f\" % (epoch_counter,\n current_epsilon,\n loss,\n reward_per_tick))\n\n # record current learning progress and show learning curves\n if epoch_counter % 100 == 0:\n action_layer.epsilon.set_value(0.05)\n reward = 0.95*reward + 0.05*np.mean(pool.evaluate(record_video=False))\n action_layer.epsilon.set_value(np.float32(current_epsilon))\n\n rewards[epoch_counter] = reward\n\n clear_output(True)\n plt.plot(*zip(*sorted(rewards.items(), key=lambda (t, r): t)))\n plt.show()\n\n epoch_counter += 1\n\n\n# Time to drink some coffee!",
"_____no_output_____"
]
],
[
[
"# Evaluating results\n * Here we plot learning curves and sample testimonials",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nplt.plot(*zip(*sorted(rewards.items(), key=lambda k: k[0])))",
"_____no_output_____"
],
[
"from agentnet.utils.persistence import save, load\nsave(action_layer, \"pacman.pcl\")",
"_____no_output_____"
],
[
"action_layer.epsilon.set_value(0.05)\nrw = pool.evaluate(n_games=20, save_path=\"./records\", record_video=False)\nprint(\"mean session score=%f.5\" % np.mean(rw))",
"_____no_output_____"
],
[
"# show video\nfrom IPython.display import HTML\nimport os\n\nvideo_names = list(\n filter(lambda s: s.endswith(\".mp4\"), os.listdir(\"./records/\")))\n\nHTML(\"\"\"\n<video width=\"640\" height=\"480\" controls>\n <source src=\"{}\" type=\"video/mp4\">\n</video>\n\"\"\".format(\"./videos/\" + video_names[-1])) # this may or may not be _last_ video. Try other indices",
"_____no_output_____"
]
],
[
[
"## Assignment part I (5 pts)\n\nWe'll start by implementing target network to stabilize training.\n\nThere are two ways to do so: \n\n\n__1)__ Manually write lasagne network, or clone it via [one of those methods](https://github.com/Lasagne/Lasagne/issues/720).\n\nYou will need to implement loading weights from original network to target network.\n\nWe recommend thoroughly debugging your code on simple tests before applying it in Atari dqn.\n\n__2)__ Use pre-build functionality from [here](http://agentnet.readthedocs.io/en/master/modules/target_network.html)\n\n```\nfrom agentnet.target_network import TargetNetwork\ntarget_net = TargetNetwork(qvalues_layer)\nold_qvalues = target_net.output_layers\n\n#agent's policy_estimators must now become (qvalues,old_qvalues)\n\n_,_,_,_,(qvalues,old_qvalues) = agent.get_sessions(...) #replaying experience\n\n\ntarget_net.load_weights()#loads weights, so target network is now exactly same as main network\n\ntarget_net.load_weights(0.01)# w_target = 0.99*w_target + 0.01*w_new\n```",
"_____no_output_____"
],
[
"## Bonus I (2+ pts)\n\nImplement and train double q-learning.\n\nThis task contains of\n* Implementing __double q-learning__ or __dueling q-learning__ or both (see tips below)\n* Training a network till convergence\n * Full points will be awwarded if your network gets average score of >=10 (see \"evaluating results\")\n * Higher score = more points as usual\n * If you're running out of time, it's okay to submit a solution that hasn't converged yet and updating it when it converges. _Lateness penalty will not increase for second submission_, so submitting first one in time gets you no penalty.\n\n\n#### Tips:\n* Implementing __double q-learning__ shouldn't be a problem if you've already have target networks in place.\n * As one option, use `get_values_for_actions(<some q-values tensor3>,<some indices>)`.\n * You will probably need `T.argmax` to select best actions\n * Here's an original [article](https://arxiv.org/abs/1509.06461)\n\n* __Dueling__ architecture is also quite straightforward if you have standard DQN.\n * You will need to change network architecture, namely the q-values layer\n * It must now contain two heads: V(s) and A(s,a), both dense layers\n * You should then add them up via elemwise sum layer or a [custom](http://lasagne.readthedocs.io/en/latest/user/custom_layers.html) layer.\n * Here's an [article](https://arxiv.org/pdf/1511.06581.pdf)\n \nHere's a template for your convenience:",
"_____no_output_____"
]
],
[
[
"from lasagne.layers import *\n\n\nclass DuelingQvaluesLayer(MergeLayer):\n def get_output_for(self, inputs, **tags):\n V, A = inputs\n return <YOUR CODE: add them up :)>\n\n def get_output_shape_for(self, input_shapes, **tags):\n V_shape, A_shape=input_shapes\n assert len(\n V_shape) == 2 and V_shape[-1] == 1, \"V layer (first param) shape must be [batch,tick,1]\"\n return A_shape # shape of q-values is same as predicted advantages",
"_____no_output_____"
],
[
"# mock-up tests\nimport theano.tensor as T\nv_tensor = -T.arange(10).reshape((10, 1))\nV = InputLayer((None, 1), v_tensor)\n\na_tensor = T.arange(30).reshape((10, 3))\nA = InputLayer((None, 1), a_tensor)\n\nQ = DuelingQvaluesLayer([V, A])",
"_____no_output_____"
],
[
"import numpy as np\nassert np.allclose(get_output(Q).eval(), (v_tensor+a_tensor).eval())\nprint(\"looks good\")",
"_____no_output_____"
]
],
[
[
"## Bonus II (5+ pts): Prioritized experience replay\n\nIn this section, you're invited to implement prioritized experience replay\n\n* You will probably need to provide a custom data structure\n* Once pool.update is called, collect the pool.experience_replay.observations, actions, rewards and is_alive and store them in your data structure\n* You can now sample such transitions in proportion to the error (see [article](https://arxiv.org/abs/1511.05952)) for training.\n\nIt's probably more convenient to explicitly declare inputs for \"sample observations\", \"sample actions\" and so on to plug them into q-learning.\n\nPrioritized (and even normal) experience replay should greatly reduce amount of game sessions you need to play in order to achieve good performance. \n\nWhile it's effect on runtime is limited for atari, more complicated envs (further in the course) will certainly benefit for it.\n\nPrioritized experience replay only supports off-policy algorithms, so pls enforce `n_steps=1` in your q-learning reference computation (default is 10).",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
d095504e957e4d09ab052d13e0266cf1db8f27e5 | 42,073 | ipynb | Jupyter Notebook | Real Time Face Emotion Detection.ipynb | datascientist1234/qwer | 972ae3d5f724d326c817934a249f611000ce4692 | [
"MIT"
] | null | null | null | Real Time Face Emotion Detection.ipynb | datascientist1234/qwer | 972ae3d5f724d326c817934a249f611000ce4692 | [
"MIT"
] | null | null | null | Real Time Face Emotion Detection.ipynb | datascientist1234/qwer | 972ae3d5f724d326c817934a249f611000ce4692 | [
"MIT"
] | null | null | null | 170.336032 | 752 | 0.751076 | [
[
[
"# installing keras\n!pip install keras",
"Requirement already satisfied: keras in c:\\users\\apoorva\\appdata\\local\\programs\\python\\python38\\lib\\site-packages (2.4.3)\nRequirement already satisfied: scipy>=0.14 in c:\\users\\apoorva\\appdata\\local\\programs\\python\\python38\\lib\\site-packages (from keras) (1.6.3)\nRequirement already satisfied: h5py in c:\\users\\apoorva\\appdata\\local\\programs\\python\\python38\\lib\\site-packages (from keras) (2.10.0)\nRequirement already satisfied: pyyaml in c:\\users\\apoorva\\appdata\\local\\programs\\python\\python38\\lib\\site-packages (from keras) (5.4.1)\nRequirement already satisfied: numpy>=1.9.1 in c:\\users\\apoorva\\appdata\\local\\programs\\python\\python38\\lib\\site-packages (from keras) (1.19.5)\nRequirement already satisfied: six in c:\\users\\apoorva\\appdata\\local\\programs\\python\\python38\\lib\\site-packages (from h5py->keras) (1.15.0)\n"
],
[
"# installing opencv\n!pip install opencv-python",
"Requirement already satisfied: opencv-python in c:\\users\\apoorva\\appdata\\local\\programs\\python\\python38\\lib\\site-packages (4.5.1.48)\nRequirement already satisfied: numpy>=1.17.3 in c:\\users\\apoorva\\appdata\\local\\programs\\python\\python38\\lib\\site-packages (from opencv-python) (1.19.5)\n"
],
[
"# installing opencv full package\n!pip install opencv-contrib-python",
"Requirement already satisfied: opencv-contrib-python in c:\\users\\apoorva\\appdata\\local\\programs\\python\\python38\\lib\\site-packages (4.5.1.48)\nRequirement already satisfied: numpy>=1.17.3 in c:\\users\\apoorva\\appdata\\local\\programs\\python\\python38\\lib\\site-packages (from opencv-contrib-python) (1.19.5)\n"
],
[
"import cv2\nfrom keras.models import load_model\nimport numpy as np\nface_detect = cv2.CascadeClassifier('haarcascade_frontalface_default.xml') \n\n \n\ndef face_detection(img,size=0.5):\n img_gray = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY) # converting image into grayscale image\n face_roi = face_detect.detectMultiScale(img_gray, 1.3,1) # ROI (region of interest of detected face) \n class_labels = ['Angry','Happy','Neutral','Fear']\n \n \n if face_roi is (): # checking if face_roi is empty that is if no face detected\n return img\n\n for(x,y,w,h) in face_roi: # iterating through faces and draw rectangle over each face\n x = x - 5\n w = w + 10\n y = y + 7\n h = h + 2\n cv2.rectangle(img, (x,y),(x+w,y+h),(125,125,10), 1) # (x,y)- top left point ; (x+w,y+h)-bottom right point ; (125,125,10)-colour of rectangle ; 1- thickness \n img_gray_crop = img_gray[y:y+h,x:x+w] # croping gray scale image \n img_color_crop = img[y:y+h,x:x+w] # croping color image\n \n model=load_model('model.h5')\n final_image = cv2.resize(img_color_crop, (48,48)) # size of colured image is resized to 48,48\n final_image = np.expand_dims(final_image, axis = 0) # array is expanded by inserting axis at position 0\n final_image = final_image/255.0 # feature scaling of final image\n \n prediction = model.predict(final_image) # predicting emotion of captured image from the trained model\n label=class_labels[prediction.argmax()] # finding the label of class which has maximaum probalility \n cv2.putText(frame,label, (50,60), cv2.FONT_HERSHEY_SCRIPT_COMPLEX,2, (120,10,200),3) \n # putText is used to draw a detected emotion on image\n # (50,60)-top left coordinate FONT_HERSHEY_SCRIPT_COMPLEX-font type\n # 2-fontscale (120,10,200)-font colour 3-font thickness\n\n\n img_color_crop = cv2.flip(img_color_crop, 1) # fliping the image\n return img\n\ncap = cv2.VideoCapture(0) # capturing the video that is live webcam\n\nwhile True:\n ret, frame = cap.read()\n cv2.imshow('LIVE', face_detection(frame)) # captured frame will be sent to face_detection function for emotion detection\n if cv2.waitKey(1) & 0xFF == ord('q'):\n break\n\ncap.release()\ncv2.destroyAllWindows()",
"<>:14: SyntaxWarning: \"is\" with a literal. Did you mean \"==\"?\n<>:14: SyntaxWarning: \"is\" with a literal. Did you mean \"==\"?\n<ipython-input-4-94db6c585d1c>:14: SyntaxWarning: \"is\" with a literal. Did you mean \"==\"?\n if face_roi is (): # checking if face_roi is empty that is if no face detected\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code"
]
] |
d095546b7001e8abf0f5bd4219c4fb95b1ba7c73 | 8,353 | ipynb | Jupyter Notebook | GearboxCreditAccounts-YearnLPs.ipynb | apeir99n/gearbox-analytics | 09a747a74b90925ac850353334d28ed1b8b0e502 | [
"MIT"
] | null | null | null | GearboxCreditAccounts-YearnLPs.ipynb | apeir99n/gearbox-analytics | 09a747a74b90925ac850353334d28ed1b8b0e502 | [
"MIT"
] | null | null | null | GearboxCreditAccounts-YearnLPs.ipynb | apeir99n/gearbox-analytics | 09a747a74b90925ac850353334d28ed1b8b0e502 | [
"MIT"
] | null | null | null | 32.885827 | 134 | 0.580749 | [
[
[
"# (c) amantay from https://github.com/AmantayAbdurakhmanov/misc/blob/master/Geabox-Yearn.ipynb",
"_____no_output_____"
],
[
"import json \nimport os\nfrom dotenv import load_dotenv \nfrom web3 import Web3\nfrom multicall import Call, Multicall\n\nload_dotenv() # add this line\nRPC_Endpoint = os.getenv('RPC_NODE')\n\n",
"_____no_output_____"
],
[
"GearboxAddressProvider = Web3.toChecksumAddress('0xcF64698AFF7E5f27A11dff868AF228653ba53be0') #gearbox address provider\nyVaultUSDC = Web3.toChecksumAddress('0xa354F35829Ae975e850e23e9615b11Da1B3dC4DE') #Yearn Vault Address\n\nABI = \"\"\"[{\"name\":\"getAccountFactory\",\n \"inputs\":[],\n \"outputs\":[{\"internalType\":\"address\",\"name\":\"\",\"type\":\"address\"}],\n \"stateMutability\":\"view\",\"type\":\"function\"},\n {\"name\":\"countCreditAccounts\",\n \"inputs\":[],\n \"outputs\":[{\"internalType\":\"uint256\",\"name\":\"\",\"type\":\"uint256\"}],\n \"stateMutability\":\"view\",\"type\":\"function\"},\n {\"name\":\"decimals\",\n \"inputs\":[],\n \"outputs\":[{\"name\":\"\",\"type\":\"uint256\"}],\n \"stateMutability\":\"view\",\"type\":\"function\"\n }\n ]\n \"\"\"\n\ndef chunks(lst, n):\n \"\"\"Yield successive n-sized chunks from lst.\"\"\"\n for i in range(0, len(lst), n):\n yield lst[i:i + n]\n\nw3_eth = Web3(Web3.HTTPProvider(RPC_Endpoint, request_kwargs={'timeout': 20}))\nprint ('Ethereum connected:', w3_eth.isConnected())",
"Ethereum connected: True\n"
],
[
"AccountFactory = w3_eth.eth.contract(address=GearboxAddressProvider, abi=ABI).functions.getAccountFactory().call()\nprint('AccountFactory:', AccountFactory)\n\ncountCreditAccounts = w3_eth.eth.contract(address=AccountFactory, abi=ABI).functions.countCreditAccounts().call()\nprint('countCreditAccounts:', countCreditAccounts)\n\nyVaultUSDC_decimals = w3_eth.eth.contract(address=yVaultUSDC, abi=ABI).functions.decimals().call()\nprint('yVaultUSDC_decimals:', yVaultUSDC_decimals)\n",
"AccountFactory: 0x444CD42BaEdDEB707eeD823f7177b9ABcC779C04\ncountCreditAccounts: 5001\nyVaultUSDC_decimals: 6\n"
],
[
"idList = list(range(countCreditAccounts))\nmulti_idCA = {}\n\nfor ids in list(chunks(idList, 400)): #chunk size for multicall = 400\n #d_ca = get_data_multicall(df.loc[id_range], 'creditAccounts', df_abi, AccountFactory)\n multi_result = Multicall([\n Call(AccountFactory, ['creditAccounts(uint256)(address)', x], [[x, Web3.toChecksumAddress]]) for x in ids\n ]\n ,_w3 = w3_eth)\n \n multi_result = multi_result()\n multi_idCA.update(multi_result)\n \n",
"_____no_output_____"
],
[
"multi_idBalance = {}\nfor ids in list(chunks(list(multi_idCA), 400)): #chunk size for multicall = 400\n multi_result = Multicall([\n Call(yVaultUSDC, ['balanceOf(address)(uint256)', multi_idCA[x]], [[x, None]]) for x in ids\n ]\n ,_w3 = w3_eth)\n \n multi_result = multi_result()\n multi_idBalance.update(multi_result)\n",
"_____no_output_____"
],
[
"multi_CA = {multi_idCA[x]:multi_idBalance[x] for x in multi_idCA}",
"_____no_output_____"
],
[
"#only with existing balance\n{key:value for key, value in multi_CA.items() if value > 0}",
"_____no_output_____"
],
[
"#decimal values\n{key:value*10**(-yVaultUSDC_decimals) for key, value in multi_CA.items() if value > 0}",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d095568a5f04c66507a7ed2e450aec56b2b2ce3e | 45,749 | ipynb | Jupyter Notebook | Jupyter_notebook/ISER2021/Path cover/Chlorophyll/.ipynb_checkpoints/20201006-Sabattus-path-cover-chloro-checkpoint.ipynb | dartmouthrobotics/epscor_asv_data_analysis | 438205fc899eca67b33fb43d51bf538db6c734b4 | [
"MIT"
] | null | null | null | Jupyter_notebook/ISER2021/Path cover/Chlorophyll/.ipynb_checkpoints/20201006-Sabattus-path-cover-chloro-checkpoint.ipynb | dartmouthrobotics/epscor_asv_data_analysis | 438205fc899eca67b33fb43d51bf538db6c734b4 | [
"MIT"
] | null | null | null | Jupyter_notebook/ISER2021/Path cover/Chlorophyll/.ipynb_checkpoints/20201006-Sabattus-path-cover-chloro-checkpoint.ipynb | dartmouthrobotics/epscor_asv_data_analysis | 438205fc899eca67b33fb43d51bf538db6c734b4 | [
"MIT"
] | null | null | null | 156.674658 | 38,044 | 0.885899 | [
[
[
"#!/usr/bin/env python\nimport matplotlib.pyplot as plt\nimport matplotlib.colors as colors\nimport matplotlib.tri as tri\nfrom mpl_toolkits.axes_grid1.anchored_artists import AnchoredSizeBar\nfrom matplotlib import ticker, cm\nimport numpy as np\nfrom numpy import ma\nimport csv",
"_____no_output_____"
],
[
"degree_sign= u'\\N{DEGREE SIGN}'\n\nCSV_FILE_PATH = '../../../Data/ISER2021/Sabattus-catabot-20201006.csv'\n#CSV_FILE_PATH2 = '../../../Data/ISER2021/Sunapee-20200715-path-2.csv'\n#CSV_FILE_PATH3 = '../../../Data/ISER2021/Sunapee-20200715-path-3.csv'\n\nwith open(CSV_FILE_PATH, 'r') as csv_file:\n reader = csv.reader(csv_file)\n path1_list = np.array(list(reader))\n \n\"\"\" \nwith open(CSV_FILE_PATH2, 'r') as csv_file:\n reader = csv.reader(csv_file)\n path2_list = np.array(list(reader))\n \nwith open(CSV_FILE_PATH3, 'r') as csv_file:\n reader = csv.reader(csv_file)\n path3_list = np.array(list(reader))\n\"\"\"\n#=============================== 07/15 ===============================\n\n# one independent\n\n# temp\n#z = path1_list[0:1833,23]\n#z = z.astype('float32')\n\n# DO\nz = path1_list[0:1833,30]\nz = z.astype('float32')\n\n# gps x,y\nx = path1_list[0:1833,2]\nx = x.astype('float32')\ny = path1_list[0:1833,1]\ny = y.astype('float32')\n\n\"\"\"\n\n# PATH 1\n# temp\nz1 = path1_list[0:2126,23]\nz1 = z1.astype('float32')\n\n# gps x,y\nx1 = path1_list[0:2126,2]\nx1 = x1.astype('float32')\ny1 = path1_list[0:2126,1]\ny1 = y1.astype('float32')\n\n## PATH 2\n# temp\nz2 = path2_list[0:998,23]\nz2 = z2.astype('float32')\n\n# gps x,y\nx2 = path2_list[0:998,2]\nx2 = x2.astype('float32')\ny2 = path2_list[0:998,1]\ny2 = y2.astype('float32')\n\n## PATH 3\n# temp\nz3 = path3_list[0:597,23]\nz3 = z3.astype('float32')\n\n# gps x,y\nx3 = path3_list[0:597,2]\nx3 = x3.astype('float32')\ny3 = path3_list[0:597,1]\ny3 = y3.astype('float32')\n\nx = np.concatenate([x1, x2, x3])\ny = np.concatenate([y1, y2, y3])\nz = np.concatenate([z1, z2, z3])\n\"\"\"\n#=====================================================================\n\nf, ax = plt.subplots()\n#ax.set_title('Catabot 10/06 Lake Sabattus: Temperature (' + degree_sign + 'C)')\nax.set_title('Catabot 10/06 Lake Sabattus: DO (%sat)')\n\nvmax=96.4\nvmin=82.4\nlevels = np.linspace(vmin,vmax, 100)\n\ncs = ax.tricontourf(x,y,z, 10, norm=colors.SymLogNorm(linthresh=0.03, linscale=0.03), levels=levels,vmax=vmax,vmin=vmin)\n#cs = ax.tricontourf(x,y,z, 20, vmin=24.35, vmax=26.94)\n#cs = ax.tricontourf(x,y,z, 20)\n\ncb_ticklabel = np.linspace(82.4, 96.4, 10)\n#cb = f.colorbar(cs, ticks=cb_ticklabel, orientation='horizontal', format='%.1f')\n\n\nax.set_xlabel('Longitude')\nplt.xlim([-70.1070, -70.1040])\nax.set_xticks(np.arange(-70.1070, -70.1039, 0.001))\nf.canvas.draw()\nax.set_xticklabels(['-70.1070', '-70.1060', '-70.1050', '-70.1040'])\n\nax.set_ylabel('Latitude')\nplt.ylim([44.1230, 44.1255])\nax.set_yticks(np.arange(44.1230, 44.1256, 0.0005))\nf.canvas.draw()\nax.set_yticklabels(['44.1230', '44.1235', '44.1240', '44.1245', '44.1250', '44.1255'])\n\n# path 1,2,3\nax.plot(x,y,marker='o', color='k', markersize=0.1)\n#ax.plot(x2,y2,marker='o', color='b', markersize=0.1)\n#ax.plot(x3,y3,marker='o', color='r', markersize=0.1)\n\nax.set_aspect('equal')\nplt.grid(True)\n\n# boathouse\nax.plot(np.array([-70.1060711]), np.array([44.1251027]), color='k', marker=(5,1), markersize=16)\nax.plot(np.array([-70.1060711]), np.array([44.1251027]), color='#FF4500', marker=(5,1), markersize=8)\n\n\n\"\"\"\n# summer buoy\nax.plot(np.array([-72.033128]), np.array([43.4096079]), color='k', marker='o', markersize=13)\nax.plot(np.array([-72.033128]), np.array([43.4096079]), color='yellow', marker='o', markersize=8)\n\n# boathouse\nax.plot(np.array([-72.0369625]), np.array([43.4100466]), color='k', marker=(5,1), markersize=16)\nax.plot(np.array([-72.0369625]), np.array([43.4100466]), color='#FF4500', marker=(5,1), markersize=8)\n\n# winter buoy\nax.plot(np.array([-72.0365116]), np.array([43.410345]), color='k', marker='o', markersize=13)\nax.plot(np.array([-72.0365116]), np.array([43.410345]), color='m', marker='o', markersize=8)\n\"\"\"\n\nbar = AnchoredSizeBar(ax.transData, 0.00046, '40 m', 'upper right', pad=0.6, frameon=False)\nax.add_artist(bar)\n\n\nplt.show()\n#f.savefig('1006-Sabattus-DO.pdf', bbox_inches ='tight')",
"_____no_output_____"
],
[
"min(x)",
"_____no_output_____"
],
[
"max(x)",
"_____no_output_____"
],
[
"min(y)",
"_____no_output_____"
],
[
"max(y)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d095771fc43850951f1356e547fde79233548604 | 140,206 | ipynb | Jupyter Notebook | notebooks/mlmachine_part_2.ipynb | klahrich/mlmachine | 6a3bc4eac93e7ddead0736404a228abbb2fc27b3 | [
"MIT"
] | null | null | null | notebooks/mlmachine_part_2.ipynb | klahrich/mlmachine | 6a3bc4eac93e7ddead0736404a228abbb2fc27b3 | [
"MIT"
] | null | null | null | notebooks/mlmachine_part_2.ipynb | klahrich/mlmachine | 6a3bc4eac93e7ddead0736404a228abbb2fc27b3 | [
"MIT"
] | null | null | null | 75.910125 | 42,016 | 0.736673 | [
[
[
"__mlmachine - GroupbyImputer, KFoldEncoder, and Skew Correction__\n<br><br>\nWelcome to Example Notebook 2. If you're new to mlmachine, check out [Example Notebook 1](https://github.com/petersontylerd/mlmachine/blob/master/notebooks/mlmachine_part_1.ipynb).\n<br><br>\nCheck out the [GitHub repository](https://github.com/petersontylerd/mlmachine).\n<br><br>\n\n1. [Missing Values - Assessment & GroupbyImputer](#Missing-Values-Assessment-&-GroupbyImputer)\n 1. [Assessment](#Assessment)\n 1. [GroupbyImputer](#GroupbyImputer)\n 1. [Imputation](#Imputation)\n1. [KFold Encoding - Exotic Encoding Without the Leakage](#KFold-Encoding-Exotic-Encoding-Without-the-Leakage)\n 1. [KFoldEncoder](#KFoldEncoder)\n1. [Box, Cox, Yeo & Johnson - Skew Correctors](#Box,-Cox,-Yeo-&-Johnson-Skew-Correctors)\n 1. [Assessment](#Assessment-1)\n 1. [Skew correction](#Skew-correction)",
"_____no_output_____"
],
[
"---\n# Missing Values - Assessment & GroupbyImputer\n---\n<br><br>\nLet's start by instantiating a couple `Machine()` objects, one for our training data and a second for our validation data:\n<br><br>",
"_____no_output_____"
],
[
"<a id = 'Missing-Values-Assessment-&-GroupbyImputer'></a>",
"_____no_output_____"
]
],
[
[
"# import libraries\nimport numpy as np\nimport pandas as pd\n\n# import mlmachine tools\nimport mlmachine as mlm\nfrom mlmachine.data import titanic\n\n# use titanic() function to create DataFrames for training and validation datasets\ndf_train, df_valid = titanic()\n\n# ordinal encoding hierarchy\nordinal_encodings = {\"Pclass\": [1, 2, 3]}\n\n# instantiate a Machine object for the training data\nmlmachine_titanic_train = mlm.Machine(\n data=df_train,\n target=\"Survived\",\n remove_features=[\"PassengerId\",\"Ticket\",\"Name\"],\n identify_as_continuous=[\"Age\",\"Fare\"],\n identify_as_count=[\"Parch\",\"SibSp\"],\n identify_as_nominal=[\"Embarked\"],\n identify_as_ordinal=[\"Pclass\"],\n ordinal_encodings=ordinal_encodings,\n is_classification=True,\n)\n\n# instantiate a Machine object for the validation data\nmlmachine_titanic_valid = mlm.Machine(\n data=df_valid,\n remove_features=[\"PassengerId\",\"Ticket\",\"Name\"],\n identify_as_continuous=[\"Age\",\"Fare\"],\n identify_as_count=[\"Parch\",\"SibSp\"],\n identify_as_nominal=[\"Embarked\"],\n identify_as_ordinal=[\"Pclass\"],\n ordinal_encodings=ordinal_encodings,\n is_classification=True,\n)\n",
"~/.pyenv/versions/main37/lib/python3.7/site-packages/sklearn/externals/joblib/__init__.py:15: FutureWarning: sklearn.externals.joblib is deprecated in 0.21 and will be removed in 0.23. Please import this functionality directly from joblib, which can be installed with: pip install joblib. If this warning is raised when loading pickled models, you may need to re-serialize those models with scikit-learn 0.21+.\n warnings.warn(msg, category=FutureWarning)\n"
]
],
[
[
"---\n## Assessment\n---\n<br><br>\nEach `Machine()` object contains a method for summarizing missingness in tabular form and in graphical form:\n<br><br>",
"_____no_output_____"
],
[
"<a id = 'Assessment'></a>",
"_____no_output_____"
]
],
[
[
"# generate missingness summary for training data\nmlmachine_titanic_train.eda_missing_summary(display_df=True)",
"_____no_output_____"
]
],
[
[
"---\n<br><br>\nBy default, this method acts on the `data` attribute associated with `mlmachine_train`. Let's do the same for the validation dataset:\n<br><br>",
"_____no_output_____"
]
],
[
[
"# generate missingness summary for validation data\nmlmachine_titanic_valid.eda_missing_summary(display_df=True)",
"_____no_output_____"
]
],
[
[
"---\n<br><br>\nNext, we need to determine if there are features with missing values in the training data, but not the validation data, and vice versa. This informs how we should set up our transformation pipeline. For example, if a feature has missing values in the validation dataset, but not the training dataset, we will still want to `fit_transform()` this feature on the training data to learn imputation values to apply on the nulls in the validation dataset.\n<br><br>\n\nWe could eyeball the tables and visuals above to compare the state of missingness in the two datasets, but this can be tedious, particularly with large datasets. Instead, we will leverage a method within our `Machine()` object. We simply pass the validation dataset to `mlmachine_titanic_train`'s method `missing_col_compare`, which returns a bidirectional missingness summary.\n<br><br>",
"_____no_output_____"
]
],
[
[
"# generate missingness comparison summary\nmlmachine_titanic_train.missing_column_compare(\n validation_data=mlmachine_titanic_valid.data,\n)",
"Feature has missing values in validation data, not training data.\n{'Fare'}\n\nFeature has missing values in training data, not validation data.\n{'Embarked'}\n"
]
],
[
[
"---\n<br><br>\nThe key observation here is that \"Fare\" is fully populated in the training data, but not the validation data. We need to make sure our pipeline learns how to impute these missing values based on the training data, despite the fact that the training data is not missing any values in this feature.\n<br><br>",
"_____no_output_____"
],
[
"---\n## GroupbyImputer\n---\n<br><br>\nmlmachine includes a transformer called `GroupbyImputer()`, which makes it easy to perform the same basic imputation techniques provided by Scikit-learn's `SimpleImputer()`, but with the added ability to group by another feature in the dataset. Let's see an example:\n<br><br>",
"_____no_output_____"
],
[
"<a id = 'GroupbyImputer'></a>",
"_____no_output_____"
]
],
[
[
"# import mlmachine tools\nfrom mlmachine.features.preprocessing import GroupbyImputer\n\n# instantiate GroupbyImputer to fill \"Age\" mean, grouped by \"SibSp\"\nimpute = GroupbyImputer(null_column=\"Age\", groupby_column=\"SibSp\", strategy=\"mean\")\nimpute.fit_transform(mlmachine_titanic_train.data[[\"Age\",\"SibSp\"]])\ndisplay(impute.train_value)",
"_____no_output_____"
]
],
[
[
"---\n<br><br>\nIn the code snippet above, we mean impute \"Age\", grouped by \"SibSp\". We pass \"Age\" to the `null_column` parameter to indicate which column contains the nulls, and pass \"SibSp\" to the `groupby_column` parameter. The strategy parameter receives the same instructions as Scikit-learn's `SimpleImputer()` - \"mean\", \"median\" and \"most_frequent\".\n<br><br>\n\nTo inspect the learned values, we can display the object's `train_value` attribute, which is a `DataFrame` containing the category/value pairs\n<br><br>\n\n`GroupbyImputer` uses these pairs to impute the missing values in \"Age\". If, in the unlikely circumstance, a level in `groupby_column` has only null values in `null_column`, then the missing values associated with that level will be imputed with the mean, median or mode of the entire feature.\n<br><br>",
"_____no_output_____"
],
[
"---\n## Imputation\n---\n<br><br>\nNow we're going to use `GroupbyImputer()` within `PandasFeatureUnion()` to impute nulls in both the training and validation datasets.\n<br><br>",
"_____no_output_____"
],
[
"<a id = 'Imputation'></a>",
"_____no_output_____"
]
],
[
[
"# import libraries\nfrom sklearn.impute import SimpleImputer\nfrom sklearn.pipeline import make_pipeline\n\n# import mlmachine tools\nfrom mlmachine.features.preprocessing import (\n DataFrameSelector,\n PandasTransformer,\n PandasFeatureUnion,\n)\n\n# create imputation PandasFeatureUnion pipeline\nimpute_pipe = PandasFeatureUnion([\n (\"age\", make_pipeline(\n DataFrameSelector(include_columns=[\"Age\",\"SibSp\"]),\n GroupbyImputer(null_column=\"Age\", groupby_column=\"SibSp\", strategy=\"mean\")\n )),\n (\"fare\", make_pipeline(\n DataFrameSelector(include_columns=[\"Fare\",\"Pclass\"]),\n GroupbyImputer(null_column=\"Fare\", groupby_column=\"Pclass\", strategy=\"mean\")\n )),\n (\"embarked\", make_pipeline(\n DataFrameSelector(include_columns=[\"Embarked\"]),\n PandasTransformer(SimpleImputer(strategy=\"most_frequent\"))\n )),\n (\"cabin\", make_pipeline(\n DataFrameSelector(include_columns=[\"Cabin\"]),\n PandasTransformer(SimpleImputer(strategy=\"constant\", fill_value=\"X\"))\n )),\n (\"diff\", make_pipeline(\n DataFrameSelector(exclude_columns=[\"Age\",\"Fare\",\"Embarked\",\"Cabin\"])\n )),\n])\n\n# fit and transform training data, transform validation data\nmlmachine_titanic_train.data = impute_pipe.fit_transform(mlmachine_titanic_train.data)\nmlmachine_titanic_valid.data = impute_pipe.transform(mlmachine_titanic_valid.data)",
"_____no_output_____"
],
[
"mlmachine_titanic_train.data[:20]",
"_____no_output_____"
]
],
[
[
"---\n<br><br>\n`GroupbyImputer()` makes two appearances in this `PandasFeatureUnion()` operation. On line 4, we groupby the feature \"SibSp\" to impute the mean \"Age\" value, and on line 8 we groupby the feature \"Pclass\" to impute the mean \"Fare\" value. \n<br><br>\n\nImputations for \"Embarked\" and \"Cabin\" are completed in straightforward fashion - \"Embarked\" is simply imputed with the mode, and \"Cabin\" is imputed with the constant value of \"X\".\n<br><br>\n\nLastly, we `fit_transform()` the `PandasFeatureUnion()` on `mlmachine_titanic_train.data` and finish filling our nulls by calling `transform()` on `mlmachine_titanic_valid.data`.\n<br><br>",
"_____no_output_____"
],
[
"---\n# KFold Encoding - Exotic Encoding Without the Leakage\n---\n<br><br>\nTarget value-based encoding techniques such as mean encoding, CatBoost Encoding, and Weight of Evidence encoding are often discussed in the context of Kaggle competitions. The primary advantage of these techniques is that they use the target variable to inform the encoded feature's values. However, this comes with the risk of leaking target information into the encoded values. \n<br><br>\n\nKFold cross-validation assists in avoiding this problem. The key is to apply the encoded values to the out-of-fold observations only. This visualization illustrates the general pattern:\n<br><br>\n\n<br><br>\n\n<br><br>\n\n- Separate a validation subset from the training dataset.\n- Learn the encoded values from the training data and the associated target values.\n- Apply the learned values to the validation observations only.\n- Repeat the process on the K-1 remaining folds.",
"_____no_output_____"
],
[
"<a id = 'KFold-Encoding-Exotic-Encoding-Without-the-Leakage'></a>",
"_____no_output_____"
],
[
"---\n## KFoldEncoder\n---\n<br><br>\nmlmachine has a class called `KFoldEncoder` that facilitates KFold encoding with an encoder of choice. Let's use a small subset of our features to see how this works. \n<br><br>\n\nWe want to target encode two features: \"Pclass\" and \"Age\". Since \"Age\" is a continuous feature, we first need to map the values to bins, which is effectively an ordinal categorical column. We handle all of this in the simple `PandasFeatureUnion` below:\n<br><br>",
"_____no_output_____"
],
[
"<a id = 'KFoldEncoder'></a>",
"_____no_output_____"
]
],
[
[
"# import libraries\nfrom sklearn.preprocessing import KBinsDiscretizer\n\n# create simple encoding PandasFeatureUnion pipeline\nencode_pipe = PandasFeatureUnion([\n (\"bin\", make_pipeline(\n DataFrameSelector(include_columns=[\"Age\"]),\n PandasTransformer(KBinsDiscretizer(encode=\"ordinal\"))\n )),\n (\"select\", make_pipeline(\n DataFrameSelector(include_columns=[\"Age\",\"Pclass\"])\n )),\n])\n\n# fit and transform training data, transform validation data\nmlmachine_titanic_train.data = encode_pipe.fit_transform(mlmachine_titanic_train.data)\nmlmachine_titanic_valid.data = encode_pipe.fit_transform(mlmachine_titanic_valid.data)\n\n# update mlm_dtypes\nmlmachine_titanic_train.update_dtypes()\nmlmachine_titanic_valid.update_dtypes()",
"_____no_output_____"
]
],
[
[
"---\n<br><br>\nThis operation returns a binned version of \"Age\", as well as the original \"Age\" and \"Pclass\" features.\n<br><br>",
"_____no_output_____"
]
],
[
[
"mlmachine_titanic_train.data[:10]",
"_____no_output_____"
]
],
[
[
"---\n<br><br>\nNext, we target encode both \"Pclass\" and \"Age_binned_5\" using mean encoding, CatBoost encoding and Weight of Evidence encoding as provided by the package category_encoders. \n<br><br>",
"_____no_output_____"
]
],
[
[
"# import libraries\nfrom sklearn.model_selection import KFold\nfrom category_encoders import WOEEncoder, TargetEncoder, CatBoostEncoder\n\n# import mlmachine tools\nfrom mlmachine.features.preprocessing import KFoldEncoder\n\n# create KFold encoding PandasFeatureUnion pipeline\ntarget_encode_pipe = PandasFeatureUnion([\n (\"target\", make_pipeline(\n DataFrameSelector(include_mlm_dtypes=[\"category\"], exclude_columns=[\"Cabin\"]), \n KFoldEncoder(\n target=mlmachine_titanic_train.target,\n cv=KFold(n_splits=5, shuffle=True, random_state=0),\n encoder=TargetEncoder,\n ),\n )),\n (\"woe\", make_pipeline(\n DataFrameSelector(include_mlm_dtypes=[\"category\"]),\n KFoldEncoder(\n target=mlmachine_titanic_train.target,\n cv=KFold(n_splits=5, shuffle=False),\n encoder=WOEEncoder,\n ),\n )),\n (\"catboost\", make_pipeline(\n DataFrameSelector(include_mlm_dtypes=[\"category\"]),\n KFoldEncoder(\n target=mlmachine_titanic_train.target,\n cv=KFold(n_splits=5, shuffle=False),\n encoder=CatBoostEncoder,\n ),\n )),\n (\"diff\", make_pipeline(\n DataFrameSelector(exclude_mlm_dtypes=[\"category\"]),\n )),\n])\n\n# fit and transform training data, transform validation data\nmlmachine_titanic_train.data = target_encode_pipe.fit_transform(mlmachine_titanic_train.data)\nmlmachine_titanic_valid.data = target_encode_pipe.transform(mlmachine_titanic_valid.data)\n\n# update mlm_dtypes\nmlmachine_titanic_train.update_dtypes()\nmlmachine_titanic_valid.update_dtypes()",
"_____no_output_____"
],
[
"mlmachine_titanic_train.data[:10]",
"_____no_output_____"
]
],
[
[
"---\n<br><br>\nLet's review the key `KFoldEncoder()` parameters:\n- `target`: the target attribute of our mlmachine_titanic_train object\n- `cv`: a cross-validation object\n- `encoder`: a target encoder class\n<br><br>\n\n`KFoldEncoder()` learns the encoded values on the training data, and applies the values to the out-of-fold observations. \n<br><br>\n\nOn the validation data, the process is simpler: we calculate the average out-of-fold encodings applied to the training data and apply these values to all validation observations.\n<br><br>",
"_____no_output_____"
],
[
"---\n# Box, Cox, Yeo & Johnson - Skew Correctors\n---",
"_____no_output_____"
],
[
"<a id = 'Box,-Cox,-Yeo-&-Johnson-Skew-Correctors'></a>",
"_____no_output_____"
],
[
"---\n## Assessment\n---\n<br><br>\nJust as we have a quick method for evaluating missingness, we have a quick method for evaluating skew.\n<br><br>",
"_____no_output_____"
],
[
"<a id = 'Assessment-1'></a>",
"_____no_output_____"
]
],
[
[
"# generate skewness summary\nmlmachine_titanic_train.skew_summary()",
"_____no_output_____"
]
],
[
[
"---\n<br><br>\nThe `skew_summary()` method returns a `DataFrame` that summarizes the skew for each feature, along with a \"Percent zero\" column, which informs us of the percentage of values in the feature that are zero.\n<br><br>",
"_____no_output_____"
],
[
"---\n## Skew correction\n---\n<br><br>\nmlmachine contains a class called `DualTransformer()`, which, by default, applies both Yeo-Johnson and Box-Cox transformations to the specified features with the intent of correcting skew. The Box-Cox transformation automatically seeks the lambda value which maximizes the log-likelihood function. \n<br><br>\n\nSince Box-Cox transformation requires all values in a feature to be greater than zero, `DualTransformer()` applies one of two simple feature adjustments when this rule is violated:\n<br><br>\n- If the minimum value in a feature is zero, each value in that feature is increased by a value of 1 prior to transformation. \n- If the minimum value is less than zero, then each feature value is increased by the absolute value of the minimum value in the feature plus 1 prior to transformation.\n<br><br>\n\nLet's use `DualTransformer()` to see if we can minimize the skew in the original \"Age\" feature:\n<br><br>",
"_____no_output_____"
],
[
"<a id = 'Skew-correction'></a>",
"_____no_output_____"
]
],
[
[
"# import mlmachine tools\nfrom mlmachine.features.preprocessing import DualTransformer\n\n# create skew correction PandasFeatureUnion pipeline\nskew_pipe = PandasFeatureUnion([\n (\"skew\", make_pipeline(\n DataFrameSelector(include_columns=[\"Age\"]),\n DualTransformer(),\n )), \n])\n\n# fit and transform training data, transform validation data\nmlmachine_titanic_train.data = skew_pipe.fit_transform(mlmachine_titanic_train.data)\nmlmachine_titanic_valid.data = skew_pipe.transform(mlmachine_titanic_valid.data)\n\n# update mlm_dtypes\nmlmachine_titanic_train.update_dtypes()\nmlmachine_titanic_valid.update_dtypes()",
"_____no_output_____"
],
[
"mlmachine_titanic_train.data[:10]",
"_____no_output_____"
]
],
[
[
"---\n<br><br>\n`DualTransformer()` adds the features \"Age_BoxCox\" and \"Age_YeoJohnson\". Let's execute `skew_summary()` again to see if `DualTransformer()` addressed the skew in our original feature:\n<br><br>\n\"Age_BoxCox\" and \"Age_YeoJohnson\" have a skew of 0.0286 and 0.0483, respectively.\n<br><br>",
"_____no_output_____"
]
],
[
[
"# generate skewness summary\nmlmachine_titanic_train.skew_summary()",
"_____no_output_____"
]
],
[
[
"---\n<br><br>\nStar the [GitHub repository](https://github.com/petersontylerd/mlmachine), and stay tuned for additional notebooks.\n<br><br>",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0957e49e8348405617e5668a35e41fbe1b0c269 | 135,344 | ipynb | Jupyter Notebook | nbs/01_analysis.ipynb | dmitryhits/ProtonBeamTherapy | 1ae7a40229631b3f25c0f9cefcfc3b2c2aa5111d | [
"Apache-2.0"
] | null | null | null | nbs/01_analysis.ipynb | dmitryhits/ProtonBeamTherapy | 1ae7a40229631b3f25c0f9cefcfc3b2c2aa5111d | [
"Apache-2.0"
] | null | null | null | nbs/01_analysis.ipynb | dmitryhits/ProtonBeamTherapy | 1ae7a40229631b3f25c0f9cefcfc3b2c2aa5111d | [
"Apache-2.0"
] | 1 | 2021-07-13T09:12:33.000Z | 2021-07-13T09:12:33.000Z | 108.2752 | 20,808 | 0.836661 | [
[
[
"# default_exp analysis",
"_____no_output_____"
]
],
[
[
"# Tools to analyze the results of Gate simulations",
"_____no_output_____"
]
],
[
[
"#hide\nfrom nbdev.showdoc import *",
"_____no_output_____"
]
],
[
[
"## Dependencies",
"_____no_output_____"
]
],
[
[
"#export\nimport pandas as pd\nimport uproot as rt\nimport awkward as ak\n\nfrom scipy.stats import moyal\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport math\nfrom scipy import stats\nfrom scipy.stats import rv_continuous\n# import pylandau\nfrom matplotlib.pyplot import hist2d\nimport matplotlib.colors as mcolors\nimport glob",
"_____no_output_____"
],
[
"#export\ndef find_max_nonzero(array_hist):\n \"\"\"returns an upper boundary of the continuos non-zero bins\n \n input a histogram array output from plt.hist\n \"\"\"\n previous = -1\n preprevious = -1\n p_b = -1\n pp_b = -1\n for v, b in zip(array_hist[0],array_hist[1]):\n if preprevious != 0 and previous == 0 and v == 0:\n return math.ceil(p_b)\n pp_b = p_b\n p_b = b\n preprevious = previous\n previous = v\n ",
"_____no_output_____"
],
[
"show_doc(find_max_nonzero)",
"_____no_output_____"
],
[
"#export\ndef find_range(param):\n \"\"\"removes a tail in the upper range of the histogram\"\"\"\n array_hist = plt.hist(param, bins=100)\n upper_limit = find_max_nonzero(array_hist)\n ret = -1\n for _ in range(10):\n print(f'upper limit: {upper_limit}')\n ret = upper_limit\n array_hist = plt.hist(param[param < upper_limit], bins=100)\n upper_limit = find_max_nonzero(array_hist)\n if ret == upper_limit:\n break\n return ret",
"_____no_output_____"
],
[
"show_doc(find_range)",
"_____no_output_____"
],
[
"#export\ndef get_edep_data(df, sensor=-1):\n \"\"\"returns an array of energies deposited in each event (keV)\"\"\"\n \n \n \n # sum all energy deposited in each event and convert the result to keV\n if sensor == -1:\n edep = df.groupby(['eventID'])['edep'].sum()*1000\n else:\n edep = (df[df['volumeID'] == sensor].groupby(['eventID']))['edep'].sum()*1000\n return edep",
"_____no_output_____"
],
[
"show_doc(get_edep_data)",
"_____no_output_____"
],
[
"#export\ndef get_df_subentry2(root_file_name):\n \"\"\"returns a dataframe that contains only subentry 2 data\n \n This subentry seems to contain all the relevant information\"\"\"\n \n df = pd.DataFrame()\n with rt.open(f'{root_file_name}:Hits') as tree:\n df = ak.to_pandas(tree.arrays())\n return df.xs(2, level='subentry')",
"_____no_output_____"
],
[
"show_doc(get_df_subentry2)",
"_____no_output_____"
],
[
"#export\ndef get_phasespace_df(timestamp, layer):\n root_file = f\"../results/tracker_{timestamp}_{layer}.root:PhaseSpace\"\n df = pd.DataFrame()\n with rt.open(root_file) as tree:\n df = ak.to_pandas(tree.arrays())\n return df",
"_____no_output_____"
],
[
"root_file = \"../results/dose_2021May10_181812_1-Dose.root\"\nfile = rt.open(root_file)",
"_____no_output_____"
],
[
"file.keys()",
"_____no_output_____"
],
[
"#export\ndef get_Ekin(df, particle='proton'):\n return df[df['ParticleName'] == particle]['Ekine']",
"_____no_output_____"
],
[
"#export\ndef extract_dose(timestamp):\n '''return numpy array of the dose for all phantom layers\n \n '''\n # get all the Dose files for a give timestamp\n files = glob.glob(f'../results/dose_{timestamp}_*-Dose.txt')\n # sort them by the layer number\n files.sort(key=lambda x: int(x.split('_')[-1].rstrip('-Dose.txt')))\n dose = []\n for file in files:\n d = []\n with open(file) as f:\n for line in f:\n # ignore the lines starting with #\n if not line.startswith('#'):\n d.append(float(line))\n # The beam is in the negative 'y' direction\n # so is the numbering of layers\n # while the data in the files is in positive direction\n # so it needs to be reversed\n dose += reversed(d)\n\n return np.array(dose)",
"_____no_output_____"
]
],
[
[
"ph0 = get_phasespace_df('2021May03_141349', 0)\nph5 = get_phasespace_df('2021May03_141349', 5)\nph10 = get_phasespace_df('2021May03_141349', 10)\nph15 = get_phasespace_df('2021May03_141349', 15)\nph19 = get_phasespace_df('2021May03_141349', 19)",
"_____no_output_____"
]
],
[
[
"get_Ekin(ph0).hist(bins=50)\nget_Ekin(ph19).hist(bins=50)",
"_____no_output_____"
],
[
"print(round(stats.tstd(get_Ekin(ph0), (175, 200)), 2), 'MeV')\nprint(round(stats.tstd(get_Ekin(ph19), (55, 90)), 2), 'MeV')",
"1.22 MeV\n3.75 MeV\n"
],
[
"from matplotlib import colors\nx0 = ph19[(ph19[\"ParticleName\"]=='proton') & (ph19['CreatorProcess'] == 0) & (ph19['NuclearProcess'] == 0)]['X']\nz0 = ph19[(ph19[\"ParticleName\"]=='proton') & (ph19['CreatorProcess'] == 0) & (ph19['NuclearProcess'] == 0)]['Z']\nx1 = ph19[(ph19[\"ParticleName\"]=='proton')]['X']\nz1 = ph19[(ph19[\"ParticleName\"]=='proton')]['Z']\n\nfig, (ax_all, ax_prim) = plt.subplots(1,2, figsize = (14, 5))\n__ =ax_all.hist2d(x1, z1, bins=50, range=[[-50, 50],[-50,50]], norm=colors.LogNorm())\nax_all.set_title(\"lateral position of all protons in the last phantom layer\")\nax_all.set_xlabel('X (mm)')\nax_all.set_ylabel('Z (mm)')\n__ = ax_prim.hist2d(x0, z0, bins=50, range=[[-50, 50],[-50,50]], norm=colors.LogNorm())\nax_prim.set_xlabel('X (mm)')\nax_prim.set_ylabel('Z (mm)')\n_ =ax_prim.set_title(\"lateral position of primary protons that only experienced Coulomb scattering\")",
"_____no_output_____"
]
],
[
[
"Lateral positions of protons in the last phantom layer. On the left are all protons, on the right are only the primary protons that did not experience nuclear scattering.",
"_____no_output_____"
]
],
[
[
"x0 = ph19[(ph19[\"ParticleName\"]=='proton') & (ph19['CreatorProcess'] == 0) & (ph19['NuclearProcess'] == 0)]['X']\nekin0 = ph19[(ph19[\"ParticleName\"]=='proton') & (ph19['CreatorProcess'] == 0) & (ph19['NuclearProcess'] == 0)]['Ekine']\nx1 = ph19[(ph19[\"ParticleName\"]=='proton')]['X']\nekin1 = ph19[(ph19[\"ParticleName\"]=='proton')]['Ekine']\nfig, (ax_all, ax_prim) = plt.subplots(1,2, figsize = (14, 5), sharey=True)\n__ =ax_all.hist2d(x1, ekin1, bins=50, norm=colors.LogNorm(), range=[[-70, 70],[0, 90]])\nax_all.set_ylabel('E_kin (MeV)')\nax_all.set_xlabel('X (mm)')\n__ = ax_prim.hist2d(x0, ekin0, bins=50, norm=colors.LogNorm(), range=[[-70, 70],[0, 90]])\nax_prim.set_xlabel('X (mm)')",
"_____no_output_____"
]
],
[
[
"Kinetic energy deposited by particle versus the position of the hit (left) all protons (right) protons from the primary beam that did not experience nuclear scattering",
"_____no_output_____"
]
],
[
[
"ph = pd.merge(ph0, ph5, on=\"EventID\", suffixes=(\"\", \"_5\"))\nph = pd.merge(ph, ph10, on=\"EventID\", suffixes=(\"\", \"_10\"))\nph = pd.merge(ph, ph15, on=\"EventID\", suffixes=(\"\", \"_15\"))\nph = pd.merge(ph, ph19, on=\"EventID\", suffixes=(\"\", \"_19\"))",
"_____no_output_____"
],
[
"def select(ph):\n result = (ph[f\"CreatorProcess\"] == 0) & (ph[f\"ParticleName\"] == \"proton\")\n for x in [ \"_5\", \"_10\", \"_15\", \"_19\"]:\n result = result & (ph[f\"CreatorProcess{x}\"] == 0) & (ph[f\"ParticleName{x}\"] == \"proton\")\n return result",
"_____no_output_____"
],
[
"ph100 = ph[(ph[\"EventID\"]<100) & select(ph)]",
"_____no_output_____"
],
[
"x = np.array(ph100[ph100[\"EventID\"] == 1][[\"X\", \"X_5\", \"X_10\", \"X_15\", \"X_19\"]]).flatten()\ny = np.array(ph100[ph100[\"EventID\"] == 1][[\"Y\", \"Y_5\", \"Y_10\", \"Y_15\", \"Y_19\"]]).flatten()",
"_____no_output_____"
],
[
"y = np.array(y).flatten()\nx = np.array(x).flatten()",
"_____no_output_____"
],
[
"plt.plot(y,x, 'o')",
"_____no_output_____"
],
[
"ph100[\"EventID\"].head()",
"_____no_output_____"
],
[
"from mpl_toolkits.mplot3d import Axes3D",
"_____no_output_____"
]
],
[
[
"## Example showing energy deposition with 3 sensors",
"_____no_output_____"
]
],
[
[
"df2 = get_df_subentry2('results/TrackerHits.root')\nedep = get_edep_data(df2, sensor=0)\n_ = plt.hist(edep, bins=100, range=(0,1000))\n_ = plt.hist(get_edep_data(df2, sensor=1), bins=100, range=(0,1000))\n_ = plt.hist(get_edep_data(df2, sensor=2), bins=100, range=(0,1000))",
"_____no_output_____"
],
[
"null_columns = [col for col in df2.columns if df2[col].max() == 0 and df2[col].min() == 0]\ndf2.drop(columns=null_columns, inplace=True)",
"_____no_output_____"
],
[
"single_value_columns = [col for col in df2.columns if df2[col].max() == df2[col].min()]\ndf2.drop(columns=single_value_columns, inplace=True)\ndf2.head()",
"_____no_output_____"
],
[
"_ = plt.hist2d(df2['posX']-df2['sourcePosX'], df2['posY'], bins=(100, 80), norm=mcolors.LogNorm())",
"_____no_output_____"
],
[
"df2_sensor0 = df2[df2.volumeID == 0]",
"_____no_output_____"
],
[
"_= plt.hist((df2_sensor0[(df2_sensor0['processName']=='Transportation') & (df2_sensor0['posY']==-47.25)]).edep,log=True, density=True, bins = 100)\n_= plt.hist((df2_sensor0[(df2_sensor0['processName']=='Transportation') & (df2_sensor0['posY']==-47.75)]).edep,log=True, density=True,bins = 100)",
"_____no_output_____"
],
[
"_= hist2d(df2.volumeID, df2.posY, bins=(12,100), norm=mcolors.LogNorm())",
"_____no_output_____"
],
[
"_ = hist2d(df2.trackLength, df2.volumeID, bins=(100, 12), norm=mcolors.LogNorm())",
"_____no_output_____"
],
[
"import pylandau\nclass landau_gen(rv_continuous):\n r\"\"\"A Landau continuous random variable.\n %(before_notes)s\n Notes\n -----\n The probability density function for `Landau` is:\n \n for a real number :math:`x`.\n %(after_notes)s\n This distribution has utility in high-energy physics and radiation\n detection. It describes the energy loss of a charged relativistic\n particle due to ionization of the medium . \n \"\"\"\n \n def _pdf(self, x):\n return pylandau.landau_pdf(np.float64(x))\n\n\nlandau = landau_gen(name=\"landau\")",
"_____no_output_____"
],
[
"#hide\nfrom nbdev.export import notebook2script; notebook2script()",
"Converted 00_macrotools.ipynb.\nConverted 01_analysis.ipynb.\nConverted 02_scanners.ipynb.\nConverted 20_analysis0.ipynb.\nConverted 21_analysis1.ipynb.\nConverted index.ipynb.\n"
],
[
"loc,scale = moyal.fit(edep)\nprint(loc, scale)",
"383.5713661065298 215.44240570369095\n"
],
[
"fig1, ax1 = plt.subplots(figsize=(7, 3))\nx = np.linspace(0, 100, 200)\nax1.plot(x, moyal.pdf(x, loc, scale), label = 'Moyal MLE fit')\n_ = ax1.hist(edep[edep < 100], bins = 100, histtype='step', density= True, label = 'sim data')\nax1.plot(x, landau.pdf(x, 23.973851592546183, 2.921658875656049), label='Landau MLE fit')\nax1.plot(x, landau.pdf(x, 24.13, 2.629), label='Meine Landau mit deine fit Parametern')\n#ax1.scatter(GeV8_data.energy, GeV8_data.counts/4400, label = 'data', marker='o', c = 'green', alpha = 0.5)\nplt.xlabel('keV')\nax1.legend()",
"<ipython-input-18-afb1cad1840f>:17: DeprecationWarning: `np.float` is a deprecated alias for the builtin `float`. To silence this warning, use `float` by itself. Doing this will not modify any behavior and is safe. If you specifically wanted the numpy scalar type, use `np.float64` here.\nDeprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations\n return pylandau.landau_pdf(np.float64(x))\n<ipython-input-18-afb1cad1840f>:17: DeprecationWarning: `np.float` is a deprecated alias for the builtin `float`. To silence this warning, use `float` by itself. Doing this will not modify any behavior and is safe. If you specifically wanted the numpy scalar type, use `np.float64` here.\nDeprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations\n return pylandau.landau_pdf(np.float64(x))\n"
],
[
"loc,scale = moyal.fit(edep[edep < 50])\nprint(loc, scale)",
"4.607171285784132 3.5588357359363867\n"
],
[
"m = np.mean(edep)\nem = stats.sem(edep)\ntm = stats.tmean(edep, limits=(edep.min(),np.mean(edep) + 1 * np.std(edep) + 2))\netm = stats.tsem(edep, limits=(edep.min(),np.mean(edep) + 1 * np.std(edep) + 2))\nprint(f'Mean: {m}, Error on mean: {em}, SNR: {m/em}')\nprint(f'Trimmed mean {tm}, Error on trimmed mean: {etm}, SNR: {tm/etm}')\n#print(stats.mode(np.round(edep, 0)))\n",
"Mean: 465.60009765625, Error on mean: 8.538391660737398, SNR: 54.53018743532778\nTrimmed mean 400.85728265348166, Error on trimmed mean: 1.1922625847147752, SNR: 336.21560199289377\n"
],
[
"## edep.to_csv('simdata.csv', sep =',', mode='w')",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"raw",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"raw"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d09594d4146bdac738e7faeb75fe6fc1e057907f | 56,001 | ipynb | Jupyter Notebook | 09_recurrent_neural_networks/tf_seq2seq.ipynb | JiaheXu/Machine-Learning-Codebase | 0280fbc0a048133d71fa1f8129c147d267fefc3e | [
"MIT"
] | null | null | null | 09_recurrent_neural_networks/tf_seq2seq.ipynb | JiaheXu/Machine-Learning-Codebase | 0280fbc0a048133d71fa1f8129c147d267fefc3e | [
"MIT"
] | null | null | null | 09_recurrent_neural_networks/tf_seq2seq.ipynb | JiaheXu/Machine-Learning-Codebase | 0280fbc0a048133d71fa1f8129c147d267fefc3e | [
"MIT"
] | null | null | null | 36.530333 | 198 | 0.497563 | [
[
[
"# 序列到序列学习(seq2seq)\n\n在`seq2seq`中,\n特定的“<eos>”表示序列结束词元。\n一旦输出序列生成此词元,模型就会停止预测。\n在循环神经网络解码器的初始化时间步,有两个特定的设计决定:\n首先,特定的“<bos>”表示序列开始词元,它是解码器的输入序列的第一个词元。\n其次,使用循环神经网络编码器最终的隐状态来初始化解码器的隐状态。\n这种设计将输入序列的编码信息送入到解码器中来生成输出序列的。\n在其他一些设计中 :cite:`Cho.Van-Merrienboer.Gulcehre.ea.2014`,\n编码器最终的隐状态在每一个时间步都作为解码器的输入序列的一部分。\n类似于 :`language_model`中语言模型的训练,\n可以允许标签成为原始的输出序列,\n从源序列词元“<bos>”、“Ils”、“regardent”、“.”\n到新序列词元\n“Ils”、“regardent”、“.”、“<eos>”来移动预测的位置。\n\n下面,我们动手构建 :`seq2seq`的设计,\n并将基于 :`machine_translation`中\n介绍的“英-法”数据集来训练这个机器翻译模型。\n",
"_____no_output_____"
]
],
[
[
"import collections\nimport math\nimport tensorflow as tf\nfrom d2l import tensorflow as d2l",
"_____no_output_____"
]
],
[
[
"## 编码器\n\n从技术上讲,编码器将长度可变的输入序列转换成\n形状固定的上下文变量$\\mathbf{c}$,\n并且将输入序列的信息在该上下文变量中进行编码。\n如 :numref:`fig_seq2seq`所示,可以使用循环神经网络来设计编码器。\n\n考虑由一个序列组成的样本(批量大小是$1$)。\n假设输入序列是$x_1, \\ldots, x_T$,\n其中$x_t$是输入文本序列中的第$t$个词元。\n在时间步$t$,循环神经网络将词元$x_t$的输入特征向量\n$\\mathbf{x}_t$和$\\mathbf{h} _{t-1}$(即上一时间步的隐状态)\n转换为$\\mathbf{h}_t$(即当前步的隐状态)。\n使用一个函数$f$来描述循环神经网络的循环层所做的变换:\n\n$$\\mathbf{h}_t = f(\\mathbf{x}_t, \\mathbf{h}_{t-1}). $$\n\n总之,编码器通过选定的函数$q$,\n将所有时间步的隐状态转换为上下文变量:\n\n$$\\mathbf{c} = q(\\mathbf{h}_1, \\ldots, \\mathbf{h}_T).$$\n\n比如,当选择$q(\\mathbf{h}_1, \\ldots, \\mathbf{h}_T) = \\mathbf{h}_T$时\n(就像 :numref:`fig_seq2seq`中一样),\n上下文变量仅仅是输入序列在最后时间步的隐状态$\\mathbf{h}_T$。\n\n到目前为止,我们使用的是一个单向循环神经网络来设计编码器,\n其中隐状态只依赖于输入子序列,\n这个子序列是由输入序列的开始位置到隐状态所在的时间步的位置\n(包括隐状态所在的时间步)组成。\n我们也可以使用双向循环神经网络构造编码器,\n其中隐状态依赖于两个输入子序列,\n两个子序列是由隐状态所在的时间步的位置之前的序列和之后的序列\n(包括隐状态所在的时间步),\n因此隐状态对整个序列的信息都进行了编码。\n\n现在,让我们[**实现循环神经网络编码器**]。\n注意,我们使用了*嵌入层*(embedding layer)\n来获得输入序列中每个词元的特征向量。\n嵌入层的权重是一个矩阵,\n其行数等于输入词表的大小(`vocab_size`),\n其列数等于特征向量的维度(`embed_size`)。\n对于任意输入词元的索引$i$,\n嵌入层获取权重矩阵的第$i$行(从$0$开始)以返回其特征向量。\n另外,本文选择了一个多层门控循环单元来实现编码器。\n\n",
"_____no_output_____"
]
],
[
[
"class Encoder(tf.keras.layers.Layer):\n \"\"\"编码器-解码器架构的基本编码器接口\"\"\"\n def __init__(self, **kwargs):\n super(Encoder, self).__init__(**kwargs)\n\n def call(self, X, *args, **kwargs):\n raise NotImplementedError",
"_____no_output_____"
],
[
"#@save\nclass Seq2SeqEncoder(d2l.Encoder):\n \"\"\"用于序列到序列学习的循环神经网络编码器\"\"\"\n def __init__(self, vocab_size, embed_size, num_hiddens, num_layers, dropout=0, **kwargs):\n super().__init__(*kwargs)\n # 嵌入层\n self.embedding = tf.keras.layers.Embedding(vocab_size, embed_size)\n self.rnn = tf.keras.layers.RNN(tf.keras.layers.StackedRNNCells(\n [tf.keras.layers.GRUCell(num_hiddens, dropout=dropout)\n for _ in range(num_layers)]), return_sequences=True,\n return_state=True)\n\n def call(self, X, *args, **kwargs):\n # 输入'X'的形状:(`batch_size`, `num_steps`)\n # 输出'X'的形状:(`batch_size`, `num_steps`, `embed_size`)\n X = self.embedding(X)\n output = self.rnn(X, **kwargs)\n state = output[1:]\n return output[0], state",
"_____no_output_____"
]
],
[
[
"循环层返回变量的说明可以参考 :numref:`sec_rnn-concise`。\n\n下面,我们实例化[**上述编码器的实现**]:\n我们使用一个两层门控循环单元编码器,其隐藏单元数为$16$。\n给定一小批量的输入序列`X`(批量大小为$4$,时间步为$7$)。\n在完成所有时间步后,\n最后一层的隐状态的输出是一个张量(`output`由编码器的循环层返回),\n其形状为(时间步数,批量大小,隐藏单元数)。\n",
"_____no_output_____"
]
],
[
[
"encoder = Seq2SeqEncoder(vocab_size=10, embed_size=8, num_hiddens=16, num_layers=2)\nX = tf.zeros((4, 7))\noutput, state = encoder(X, training=False)\noutput.shape",
"_____no_output_____"
]
],
[
[
"由于这里使用的是门控循环单元,\n所以在最后一个时间步的多层隐状态的形状是\n(隐藏层的数量,批量大小,隐藏单元的数量)。\n如果使用长短期记忆网络,`state`中还将包含记忆单元信息。\n",
"_____no_output_____"
]
],
[
[
"len(state), [element.shape for element in state]",
"_____no_output_____"
]
],
[
[
"## [**解码器**]\n:label:`sec_seq2seq_decoder`\n\n正如上文提到的,编码器输出的上下文变量$\\mathbf{c}$\n对整个输入序列$x_1, \\ldots, x_T$进行编码。\n来自训练数据集的输出序列$y_1, y_2, \\ldots, y_{T'}$,\n对于每个时间步$t'$(与输入序列或编码器的时间步$t$不同),\n解码器输出$y_{t'}$的概率取决于先前的输出子序列\n$y_1, \\ldots, y_{t'-1}$和上下文变量$\\mathbf{c}$,\n即$P(y_{t'} \\mid y_1, \\ldots, y_{t'-1}, \\mathbf{c})$。\n\n为了在序列上模型化这种条件概率,\n我们可以使用另一个循环神经网络作为解码器。\n在输出序列上的任意时间步$t^\\prime$,\n循环神经网络将来自上一时间步的输出$y_{t^\\prime-1}$\n和上下文变量$\\mathbf{c}$作为其输入,\n然后在当前时间步将它们和上一隐状态\n$\\mathbf{s}_{t^\\prime-1}$转换为\n隐状态$\\mathbf{s}_{t^\\prime}$。\n因此,可以使用函数$g$来表示解码器的隐藏层的变换:\n\n$$\\mathbf{s}_{t^\\prime} = g(y_{t^\\prime-1}, \\mathbf{c}, \\mathbf{s}_{t^\\prime-1}).$$\n:eqlabel:`eq_seq2seq_s_t`\n\n在获得解码器的隐状态之后,\n我们可以使用输出层和softmax操作\n来计算在时间步$t^\\prime$时输出$y_{t^\\prime}$的条件概率分布\n$P(y_{t^\\prime} \\mid y_1, \\ldots, y_{t^\\prime-1}, \\mathbf{c})$。\n\n根据 :numref:`fig_seq2seq`,当实现解码器时,\n我们直接使用编码器最后一个时间步的隐状态来初始化解码器的隐状态。\n这就要求使用循环神经网络实现的编码器和解码器具有相同数量的层和隐藏单元。\n为了进一步包含经过编码的输入序列的信息,\n上下文变量在所有的时间步与解码器的输入进行拼接(concatenate)。\n为了预测输出词元的概率分布,\n在循环神经网络解码器的最后一层使用全连接层来变换隐状态。\n",
"_____no_output_____"
],
[
"### 解码器输入: hidden state ‘c’, 当前词向量 ‘x’, 输出:目标语言词向量 ‘output’\n### 解码器要得到解码是的hidden state需要c和x, 所以解码器的RNNcell的输入是embed_size + num_hiddens 维\n### 解码器最后要根据hidden state得到目标语言,所以最后要加一个nn.Linear(num_hiddens, vocab_size)",
"_____no_output_____"
]
],
[
[
"class Decoder(tf.keras.layers.Layer):\n \"\"\"编码器-解码器架构的基本解码器接口\"\"\"\n def __init__(self, **kwargs):\n super(Decoder, self).__init__(**kwargs)\n\n def init_state(self, enc_outputs, *args):\n raise NotImplementedError\n\n def call(self, X, state, **kwargs):\n raise NotImplementedError",
"_____no_output_____"
],
[
"class Seq2SeqDecoder(d2l.Decoder):\n \"\"\"用于序列到序列学习的循环神经网络解码器\"\"\"\n def __init__(self, vocab_size, embed_size, num_hiddens, num_layers,\n dropout=0, **kwargs):\n super().__init__(**kwargs)\n self.embedding = tf.keras.layers.Embedding(vocab_size, embed_size)\n self.rnn = tf.keras.layers.RNN(tf.keras.layers.StackedRNNCells(\n [tf.keras.layers.GRUCell(num_hiddens, dropout=dropout)\n for _ in range(num_layers)]), return_sequences=True,\n return_state=True)\n self.dense = tf.keras.layers.Dense(vocab_size)\n\n def init_state(self, enc_outputs, *args):\n return enc_outputs[1]\n\n def call(self, X, state, **kwargs):\n # 输出'X'的形状:(`batch_size`, `num_steps`, `embed_size`)\n X = self.embedding(X)\n # 广播`context`,使其具有与`X`相同的`num_steps`\n context = tf.repeat(tf.expand_dims(state[-1], axis=1), repeats=X.shape[1], axis=1)\n X_and_context = tf.concat((X, context), axis=2)\n rnn_output = self.rnn(X_and_context, state, **kwargs)\n output = self.dense(rnn_output[0])\n # `output`的形状: (`batch_size`, `num_steps`, `vocab_size`)\n # `state`是一个包含`num_layers`个元素的列表,每个元素的形状: (`batch_size`, `num_hiddens`)\n return output, rnn_output[1:]",
"_____no_output_____"
]
],
[
[
"下面,我们用与前面提到的编码器中相同的超参数来[**实例化解码器**]。\n如我们所见,解码器的输出形状变为(批量大小,时间步数,词表大小),\n其中张量的最后一个维度存储预测的词元分布。\n",
"_____no_output_____"
]
],
[
[
"decoder = Seq2SeqDecoder(vocab_size=10, embed_size=8, num_hiddens=16, num_layers=2)\nstate = decoder.init_state(encoder(X))\noutput, state = decoder(X, state, training=False)\noutput.shape, len(state), state[0].shape",
"_____no_output_____"
]
],
[
[
"## 损失函数\n\n在每个时间步,解码器预测了输出词元的概率分布。\n类似于语言模型,可以使用softmax来获得分布,\n并通过计算交叉熵损失函数来进行优化。\n回想一下 :`machine_translation`中,\n特定的填充词元被添加到序列的末尾,\n因此不同长度的序列可以以相同形状的小批量加载。\n但是,我们应该将填充词元的预测排除在损失函数的计算之外。\n\n为此,我们可以使用下面的`sequence_mask`函数\n[**通过零值化屏蔽不相关的项**],\n以便后面任何不相关预测的计算都是与零的乘积,结果都等于零。\n例如,如果两个序列的有效长度(不包括填充词元)分别为$1$和$2$,\n则第一个序列的第一项和第二个序列的前两项之后的剩余项将被清除为零。\n\n",
"_____no_output_____"
]
],
[
[
"#@save\ndef sequence_mask(X, valid_len, value=0):\n \"\"\"在序列中屏蔽不相关的项\"\"\"\n maxlen = X.shape[1]\n mask = tf.range(start=0, limit=maxlen, dtype=tf.float32)[\n None, :] < tf.cast(valid_len[:, None], dtype=tf.float32)\n\n if len(X.shape) == 3:\n return tf.where(tf.expand_dims(mask, axis=-1), X, value)\n else:\n return tf.where(mask, X, value)\n\nX = tf.constant([[1, 2, 3], [4, 5, 6]])\nsequence_mask(X, tf.constant([1, 2]))",
"_____no_output_____"
]
],
[
[
"(**我们还可以使用此函数屏蔽最后几个轴上的所有项。**)如果愿意,也可以使用指定的非零值来替换这些项。\n",
"_____no_output_____"
]
],
[
[
"X = tf.ones((2,3,4))\nsequence_mask(X, tf.constant([1, 2]), value=-1)",
"_____no_output_____"
]
],
[
[
"现在,我们可以[**通过扩展softmax交叉熵损失函数来遮蔽不相关的预测**]。\n最初,所有预测词元的掩码都设置为1。\n一旦给定了有效长度,与填充词元对应的掩码将被设置为0。\n最后,将所有词元的损失乘以掩码,以过滤掉损失中填充词元产生的不相关预测。\n",
"_____no_output_____"
]
],
[
[
"#@save\nclass MaskedSoftmaxCELoss(tf.keras.losses.Loss):\n \"\"\"带遮蔽的softmax交叉熵损失函数\"\"\"\n def __init__(self, valid_len):\n super().__init__(reduction='none')\n self.valid_len = valid_len\n\n # `pred` 的形状:(`batch_size`, `num_steps`, `vocab_size`)\n # `label` 的形状:(`batch_size`, `num_steps`)\n # `valid_len` 的形状:(`batch_size`,)\n def call(self, label, pred):\n weights = tf.ones_like(label, dtype=tf.float32)\n weights = sequence_mask(weights, self.valid_len)\n label_one_hot = tf.one_hot(label, depth=pred.shape[-1])\n unweighted_loss = tf.keras.losses.CategoricalCrossentropy(\n from_logits=True, reduction='none')(label_one_hot, pred)\n weighted_loss = tf.reduce_mean((unweighted_loss*weights), axis=1)\n return weighted_loss",
"_____no_output_____"
]
],
[
[
"我们可以创建三个相同的序列来进行[**代码健全性检查**],\n然后分别指定这些序列的有效长度为$4$、$2$和$0$。\n结果就是,第一个序列的损失应为第二个序列的两倍,而第三个序列的损失应为零。\n",
"_____no_output_____"
]
],
[
[
"loss = MaskedSoftmaxCELoss(tf.constant([4, 2, 0]))\nloss(tf.ones((3,4), dtype = tf.int32), tf.ones((3, 4, 10))).numpy()",
"_____no_output_____"
]
],
[
[
"## [**训练**]\n:label:`sec_seq2seq_training`\n\n在下面的循环训练过程中,如 :numref:`fig_seq2seq`所示,\n特定的序列开始词元(“<bos>”)和\n原始的输出序列(不包括序列结束词元“<eos>”)\n拼接在一起作为解码器的输入。\n这被称为*强制教学*(teacher forcing),\n因为原始的输出序列(词元的标签)被送入解码器。\n或者,将来自上一个时间步的*预测*得到的词元作为解码器的当前输入。\n",
"_____no_output_____"
]
],
[
[
"#@save\ndef train_seq2seq(net, data_iter, lr, num_epochs, tgt_vocab, device):\n \"\"\"训练序列到序列模型\"\"\"\n optimizer = tf.keras.optimizers.Adam(learning_rate=lr)\n animator = d2l.Animator(xlabel=\"epoch\", ylabel=\"loss\",\n xlim=[10, num_epochs])\n for epoch in range(num_epochs):\n timer = d2l.Timer()\n metric = d2l.Accumulator(2) # 训练损失总和,词元数量\n for batch in data_iter:\n X, X_valid_len, Y, Y_valid_len = [x for x in batch]\n bos = tf.reshape(tf.constant([tgt_vocab['<bos>']] * Y.shape[0]),\n shape=(-1, 1))\n dec_input = tf.concat([bos, Y[:, :-1]], 1) # 强制教学\n with tf.GradientTape() as tape:\n Y_hat, _ = net(X, dec_input, X_valid_len, training=True)\n l = MaskedSoftmaxCELoss(Y_valid_len)(Y, Y_hat)\n gradients = tape.gradient(l, net.trainable_variables)\n gradients = d2l.grad_clipping(gradients, 1)\n optimizer.apply_gradients(zip(gradients, net.trainable_variables))\n num_tokens = tf.reduce_sum(Y_valid_len).numpy()\n metric.add(tf.reduce_sum(l), num_tokens)\n if (epoch + 1) % 10 == 0:\n animator.add(epoch + 1, (metric[0] / metric[1],))\n print(f'loss {metric[0] / metric[1]:.3f}, {metric[1] / timer.stop():.1f} '\n f'tokens/sec on {str(device)}')",
"_____no_output_____"
]
],
[
[
"现在,在机器翻译数据集上,我们可以\n[**创建和训练一个循环神经网络“编码器-解码器”模型**]用于序列到序列的学习。\n",
"_____no_output_____"
]
],
[
[
"class EncoderDecoder(tf.keras.Model):\n \"\"\"编码器-解码器架构的基类\"\"\"\n def __init__(self, encoder, decoder, **kwargs):\n super(EncoderDecoder, self).__init__(**kwargs)\n self.encoder = encoder\n self.decoder = decoder\n\n def call(self, enc_X, dec_X, *args, **kwargs):\n enc_outputs = self.encoder(enc_X, *args, **kwargs)\n dec_state = self.decoder.init_state(enc_outputs, *args)\n return self.decoder(dec_X, dec_state, **kwargs)",
"_____no_output_____"
],
[
"embed_size, num_hiddens, num_layers, dropout = 32, 32, 2, 0.1\nbatch_size, num_steps = 64, 10\nlr, num_epochs, device = 0.005, 300, d2l.try_gpu()\n\ntrain_iter, src_vocab, tgt_vocab = d2l.load_data_nmt(batch_size, num_steps)\nencoder = Seq2SeqEncoder(len(src_vocab), embed_size, num_hiddens, num_layers,\n dropout)\ndecoder = Seq2SeqDecoder(len(tgt_vocab), embed_size, num_hiddens, num_layers,\n dropout)\nnet = EncoderDecoder(encoder, decoder)\ntrain_seq2seq(net, train_iter, lr, num_epochs, tgt_vocab, device)",
"loss 0.027, 2120.5 tokens/sec on <tensorflow.python.eager.context._EagerDeviceContext object at 0x000001E3A0071D40>\n"
]
],
[
[
"## [**预测**]\n\n为了采用一个接着一个词元的方式预测输出序列,\n每个解码器当前时间步的输入都将来自于前一时间步的预测词元。\n与训练类似,序列开始词元(“<bos>”)\n在初始时间步被输入到解码器中。\n该预测过程如 :`seq2seq_predict`所示,\n当输出序列的预测遇到序列结束词元(“<eos>”)时,预测就结束了。\n\n我们将在 :`beam-search`中介绍不同的序列生成策略。\n",
"_____no_output_____"
]
],
[
[
"#@save\ndef predict_seq2seq(net, src_sentence, src_vocab, tgt_vocab, num_steps,\n save_attention_weights=False):\n \"\"\"序列到序列模型的预测\"\"\"\n src_tokens = src_vocab[src_sentence.lower().split(' ')] + [\n src_vocab['<eos>']]\n enc_valid_len = tf.constant([len(src_tokens)])\n src_tokens = d2l.truncate_pad(src_tokens, num_steps, src_vocab['<pad>'])\n # 添加批量轴\n enc_X = tf.expand_dims(src_tokens, axis=0)\n enc_outputs = net.encoder(enc_X, enc_valid_len, training=False)\n dec_state = net.decoder.init_state(enc_outputs, enc_valid_len)\n # 添加批量轴\n dec_X = tf.expand_dims(tf.constant([tgt_vocab['<bos>']]), axis=0)\n output_seq, attention_weight_seq = [], []\n for _ in range(num_steps):\n Y, dec_state = net.decoder(dec_X, dec_state, training=False)\n # 我们使用具有预测最高可能性的词元,作为解码器在下一时间步的输入\n dec_X = tf.argmax(Y, axis=2)\n pred = tf.squeeze(dec_X, axis=0)\n # 保存注意力权重\n if save_attention_weights:\n attention_weight_seq.append(net.decoder.attention_weights)\n # 一旦序列结束词元被预测,输出序列的生成就完成了\n if pred == tgt_vocab['<eos>']:\n break\n output_seq.append(pred.numpy())\n return ' '.join(tgt_vocab.to_tokens(tf.reshape(output_seq,\n shape = -1).numpy().tolist())), attention_weight_seq",
"_____no_output_____"
]
],
[
[
"## 预测序列的评估\n\n我们可以通过与真实的标签序列进行比较来评估预测序列。\n虽然 :cite:`Papineni.Roukos.Ward.ea.2002`\n提出的BLEU(bilingual evaluation understudy)\n最先是用于评估机器翻译的结果,\n但现在它已经被广泛用于测量许多应用的输出序列的质量。\n原则上说,对于预测序列中的任意$n$元语法(n-grams),\nBLEU的评估都是这个$n$元语法是否出现在标签序列中。\n\n我们将BLEU定义为:\n\n$$ \\exp\\left(\\min\\left(0, 1 - \\frac{\\mathrm{len}_{\\text{label}}}{\\mathrm{len}_{\\text{pred}}}\\right)\\right) \\prod_{n=1}^k p_n^{1/2^n},$$\n:eqlabel:`eq_bleu`\n\n其中$\\mathrm{len}_{\\text{label}}$表示标签序列中的词元数和\n$\\mathrm{len}_{\\text{pred}}$表示预测序列中的词元数,\n$k$是用于匹配的最长的$n$元语法。\n另外,用$p_n$表示$n$元语法的精确度,它是两个数量的比值:\n第一个是预测序列与标签序列中匹配的$n$元语法的数量,\n第二个是预测序列中$n$元语法的数量的比率。\n具体地说,给定标签序列$A$、$B$、$C$、$D$、$E$、$F$\n和预测序列$A$、$B$、$B$、$C$、$D$,\n我们有$p_1 = 4/5$、$p_2 = 3/4$、$p_3 = 1/3$和$p_4 = 0$。\n\n根据 :eqref:`eq_bleu`中BLEU的定义,\n当预测序列与标签序列完全相同时,BLEU为$1$。\n此外,由于$n$元语法越长则匹配难度越大,\n所以BLEU为更长的$n$元语法的精确度分配更大的权重。\n具体来说,当$p_n$固定时,$p_n^{1/2^n}$\n会随着$n$的增长而增加(原始论文使用$p_n^{1/n}$)。\n而且,由于预测的序列越短获得的$p_n$值越高,\n所以 :eqref:`eq_bleu`中乘法项之前的系数用于惩罚较短的预测序列。\n例如,当$k=2$时,给定标签序列$A$、$B$、$C$、$D$、$E$、$F$\n和预测序列$A$、$B$,尽管$p_1 = p_2 = 1$,\n惩罚因子$\\exp(1-6/2) \\approx 0.14$会降低BLEU。\n\n[**BLEU的代码实现**]如下。\n",
"_____no_output_____"
]
],
[
[
"def bleu(pred_seq, label_seq, k): #@save\n \"\"\"计算BLEU\"\"\"\n pred_tokens, label_tokens = pred_seq.split(' '), label_seq.split(' ')\n len_pred, len_label = len(pred_tokens), len(label_tokens)\n score = math.exp(min(0, 1 - len_label / len_pred))\n for n in range(1, k + 1):\n num_matches, label_subs = 0, collections.defaultdict(int)\n for i in range(len_label - n + 1):\n label_subs[''.join(label_tokens[i: i + n])] += 1\n for i in range(len_pred - n + 1):\n if label_subs[''.join(pred_tokens[i: i + n])] > 0:\n num_matches += 1\n label_subs[''.join(pred_tokens[i: i + n])] -= 1\n score *= math.pow(num_matches / (len_pred - n + 1), math.pow(0.5, n))\n return score",
"_____no_output_____"
]
],
[
[
"最后,利用训练好的循环神经网络“编码器-解码器”模型,\n[**将几个英语句子翻译成法语**],并计算BLEU的最终结果。\n",
"_____no_output_____"
]
],
[
[
"engs = ['go .', \"i lost .\", 'he\\'s calm .', 'i\\'m home .']\nfras = ['va !', 'j\\'ai perdu .', 'il est calme .', 'je suis chez moi .']\nfor eng, fra in zip(engs, fras):\n translation, attention_weight_seq = predict_seq2seq(\n net, eng, src_vocab, tgt_vocab, num_steps)\n print(f'{eng} => {translation}, bleu {bleu(translation, fra, k=2):.3f}')",
"go . => va !, bleu 1.000\ni lost . => j'ai perdu ., bleu 1.000\nhe's calm . => il est bon ?, bleu 0.537\ni'm home . => je suis chez moi ., bleu 1.000\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.