hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
sequence | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
sequence | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
sequence | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
sequence | cell_types
sequence | cell_type_groups
sequence |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
d0840fdfcf1673693daa7e7bd1770ab51cbe03a7 | 4,219 | ipynb | Jupyter Notebook | B.O.M.Z.I.I. the Bot Box.ipynb | HomamAl/.B.O.M.Z.I.I-the-BotBox- | cf1238b9368231ce0f7b77b93e33bc2457304600 | [
"Apache-2.0"
] | 1 | 2020-07-15T13:26:27.000Z | 2020-07-15T13:26:27.000Z | B.O.M.Z.I.I. the Bot Box.ipynb | HomamAl/.B.O.M.Z.I.I-the-BotBox- | cf1238b9368231ce0f7b77b93e33bc2457304600 | [
"Apache-2.0"
] | null | null | null | B.O.M.Z.I.I. the Bot Box.ipynb | HomamAl/.B.O.M.Z.I.I-the-BotBox- | cf1238b9368231ce0f7b77b93e33bc2457304600 | [
"Apache-2.0"
] | null | null | null | 26.36875 | 118 | 0.450818 | [
[
[
"#import the library \nfrom nltk.chat.util import Chat, reflections\nimport json",
"_____no_output_____"
],
[
"pairs = [\n ['my name is(.*)',['hi %1']],\n ['(hi|hello|hey|holla|hola|salam)', ['hey there', 'hi there', 'hellooooooo']],\n ['(.*) in (.*) is fun', ['%1 in %2 is indeed fun']],\n ['(.*)(location|city) ?', 'Tokyo, Japan'],\n ['(.*)created you ?', ['HomamAl did using NLTK']],\n ['how is the weather in (.*)', ['the weather in %1 is amazing like always']],\n ['is (.*) fun', ['%1 is really fun']],\n ['(.*)help(.*)',['I can help you']],\n ['(.*)your name ?',['my name is .B.O.M.Z.I.I']],\n ['(.*)can we be friends ?(.*)', ['Of course!! I will be your only friend that will never leave you ;)']],\n ['(.*)', ['I can only answer one of these questions: '\n '\\n' '1. Ask about my name?' \n '\\n' '2. I can greet you if you greet me :)' \n '\\n' '3. Ask me if something is fun'\n '\\n' '4. Ask who created me'\n '\\n' '5. Ask me how is the weather in your city'\n '\\n' '6. Ask me for help'\n '\\n' 'Have Fun and dont forget that i will be your AI friend <3']]\n \n]",
"_____no_output_____"
],
[
"reflections",
"_____no_output_____"
],
[
"my_dummy_reflections = {\n 'go' : 'gone',\n 'hello' : 'hey there'\n}",
"_____no_output_____"
],
[
"chat = Chat( pairs, my_dummy_reflections)\nchat.converse()",
">Hey\nhellooooooo\n>Hello\nhey there\n>ABCD\nI can only answer one of these questions: \n1. Ask about my name?\n2. I can greet you if you greet me :)\n3. Ask me if something is fun\n4. Ask who created me\n5. Ask me how is the weather in your city\n6. Ask me for help\nHave Fun and dont forget that i will be your AI friend <3\n>Whats your name\nmy name is .B.O.M.Z.I.I\n>Who created you\nHomamAl did using NLTK\n>Is eating fun\neating is really fun\n>I need help \nI can help you\n>How is the weather in London \nthe weather in london is amazing like always\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
d084218dcc52b361a7479f741f988d2572cc0924 | 66,998 | ipynb | Jupyter Notebook | ProcessingForPaper/7_produce_dataset.ipynb | SiobhanPowell/speech | e666cc11268037ad821936e3ef944996d68cf825 | [
"BSD-2-Clause"
] | 3 | 2021-12-12T05:21:47.000Z | 2022-01-19T18:35:54.000Z | ProcessingForPaper/7_produce_dataset.ipynb | SiobhanPowell/speech | e666cc11268037ad821936e3ef944996d68cf825 | [
"BSD-2-Clause"
] | null | null | null | ProcessingForPaper/7_produce_dataset.ipynb | SiobhanPowell/speech | e666cc11268037ad821936e3ef944996d68cf825 | [
"BSD-2-Clause"
] | 1 | 2021-12-17T17:11:15.000Z | 2021-12-17T17:11:15.000Z | 260.692607 | 22,960 | 0.92852 | [
[
[
"\"\"\"\nSPEECh: Scalable Probabilistic Estimates of EV Charging\n\nCode first published in October 2021.\n\nDeveloped by Siobhan Powell ([email protected]).\n\"\"\"",
"_____no_output_____"
],
[
"# This code produces the data set of driver group profiles that has been posted\n### Siobhan Powell, October 2021",
"_____no_output_____"
]
],
[
[
"from speech import DataSetConfigurations\nfrom speech import SPEECh\nfrom speech import SPEEChGeneralConfiguration\nfrom speech import Plotting\nfrom speech import LoadProfile\n",
"_____no_output_____"
],
[
"import os\nos.chdir('..')",
"_____no_output_____"
],
[
"import pandas as pd\nimport matplotlib.pyplot as plt\nimport numpy as np",
"_____no_output_____"
],
[
"total_evs = 10000\nweekday_option = 'weekday'\n\n# data = DataSetConfigurations('NewData', ng=9)\ndata = DataSetConfigurations('Original16', ng=16)\nmodel = SPEECh(data)\nconfig = SPEEChGeneralConfiguration(model)\nconfig.num_evs(total_evs)\nconfig.groups()",
"_____no_output_____"
],
[
"results = {}\nn = 10000\nfor i in range(data.ng):\n j = data.cluster_reorder_dendtoac[i]\n config.group_configs[j].numbers(total_drivers=n)\n config.group_configs[j].load_gmms()\n model = LoadProfile(config, config.group_configs[j], weekday=weekday_option)\n model.calculate_load()\n (pd.DataFrame(model.load_segments_dict)/n).to_csv('Output_Data/group'+str(int(i+1))+'_weekday.csv', index=None)\n results[i] = pd.DataFrame(model.load_segments_dict)/n",
"_____no_output_____"
],
[
"total_evs = 10000\nweekday_option = 'weekend'\n\n# data = DataSetConfigurations('NewData', ng=9)\ndata = DataSetConfigurations('Original16', ng=16)\nmodel = SPEECh(data)\nconfig = SPEEChGeneralConfiguration(model)\nconfig.num_evs(total_evs)\nconfig.groups()\n\nresults = {}\nn = 10000\nfor i in range(data.ng):\n j = data.cluster_reorder_dendtoac[i]\n config.group_configs[j].numbers(total_drivers=n)\n config.group_configs[j].load_gmms()\n model = LoadProfile(config, config.group_configs[j], weekday=weekday_option)\n model.calculate_load()\n (pd.DataFrame(model.load_segments_dict)/n).to_csv('Output_Data/group'+str(int(i+1))+'_weekend.csv', index=None)\n results[i] = pd.DataFrame(model.load_segments_dict)/n",
"_____no_output_____"
]
],
[
[
"# Testing:",
"_____no_output_____"
]
],
[
[
"plt.plot(results[5])",
"_____no_output_____"
],
[
"test = pd.read_csv('Output_Data/group3_weekday.csv')\ntest2 = pd.read_csv('Output_Data/group3_weekend.csv')",
"_____no_output_____"
],
[
"plt.plot(test.sum(axis=1), label='weekday')\nplt.plot(test2.sum(axis=1), label='weekend')\nplt.legend()\nplt.show()",
"_____no_output_____"
],
[
"test = pd.read_csv('Output_Data/group4_weekday.csv')\ntest2 = pd.read_csv('Output_Data/group4_weekend.csv')",
"_____no_output_____"
],
[
"plt.plot(test.sum(axis=1), label='weekday')\nplt.plot(test2.sum(axis=1), label='weekend')\nplt.legend()\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
d08437db88d81b9904f270017c9d4fe3448e7e56 | 15,751 | ipynb | Jupyter Notebook | GeneralExemplars/MLExemplars/Classification_k_NN_Notebook.ipynb | gaybro8777/Exemplars2020 | 0075e300b5ec671d11a875023f28359009cfeb35 | [
"BSD-3-Clause"
] | 2 | 2021-01-11T01:58:26.000Z | 2021-06-19T19:49:47.000Z | GeneralExemplars/MLExemplars/Classification_k_NN_Notebook.ipynb | gaybro8777/Exemplars2020 | 0075e300b5ec671d11a875023f28359009cfeb35 | [
"BSD-3-Clause"
] | null | null | null | GeneralExemplars/MLExemplars/Classification_k_NN_Notebook.ipynb | gaybro8777/Exemplars2020 | 0075e300b5ec671d11a875023f28359009cfeb35 | [
"BSD-3-Clause"
] | 3 | 2020-07-24T15:56:43.000Z | 2022-03-09T10:18:26.000Z | 30.94499 | 317 | 0.593931 | [
[
[
"## This Notebook - Goals - FOR EDINA\n\n**What?:**\n- Standard classification method example/tutorial\n\n**Who?:**\n- Researchers in ML\n- Students in computer science\n- Teachers in ML/STEM\n\n**Why?:**\n- Demonstrate capability/simplicity of core scipy stack. \n- Demonstrate common ML concept known to learners and used by researchers.\n\n**Noteable features to exploit:**\n- use of pre-installed libraries: <code>numpy</code>, <code>scikit-learn</code>, <code>matplotlib</code>\n\n**How?:**\n- clear to understand - minimise assumed knowledge\n- clear visualisations - concise explanations\n- recognisable/familiar - use standard methods\n- Effective use of core libraries\n\n<hr>",
"_____no_output_____"
],
[
"# Classification - K nearest neighbours\n\nK nearest neighbours is a simple and effective way to deal with classification problems. This method classifies each sample based on the class of the points that are closest to it.\n\nThis is a supervised learning method, meaning that data used contains information on some feature that the model should predict.\n\nThis notebook shows the process of classifying handwritten digits. ",
"_____no_output_____"
],
[
"<hr>\n\n### Import libraries\n\nOn Noteable, all the libaries required for this notebook are pre-installed, so they simply need to be imported:",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nimport sklearn.datasets as ds\nimport sklearn.model_selection as ms \n\nfrom sklearn import decomposition\nfrom sklearn import neighbors\nfrom sklearn import metrics\n\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"<hr>\n\n# Data - Handwritten Digits\n\nIn terms of data, [scikit-learn](https://scikit-learn.org/stable/modules/generated/sklearn.datasets.load_digits.html) has a loading function for some data regarding hand written digits.",
"_____no_output_____"
]
],
[
[
"# get the digits data from scikit into the notebook\ndigits = ds.load_digits()",
"_____no_output_____"
]
],
[
[
"The cell above loads the data as a [bunch object](https://scikit-learn.org/stable/modules/generated/sklearn.datasets.load_digits.html), meaning that the data (in this case images of handwritten digits) and the target (the number that is written) can be split by accessing the attributes of the bunch object:",
"_____no_output_____"
]
],
[
[
"# store data and targets seperately\nX = digits.data\ny = digits.target\n\nprint(\"The data is of the shape\", X.shape)\nprint(\"The target data is of the shape\", y.shape)",
"_____no_output_____"
]
],
[
[
"The individual samples in the <code>X</code> array each represent an image. In this representation, 64 numbers are used to represent a greyscale value on an 8\\*8 square. The images can be examined by using pyplot's [matshow](https://matplotlib.org/3.3.0/api/_as_gen/matplotlib.pyplot.matshow.html) function.\n\nThe next cell displays the 17th sample in the dataset as an 8\\*8 image.",
"_____no_output_____"
]
],
[
[
"# create figure to display the 17th sample\nfig = plt.matshow(digits.images[17], cmap=plt.cm.gray)\nfig.axes.get_xaxis().set_visible(False)\nfig.axes.get_yaxis().set_visible(False)",
"_____no_output_____"
]
],
[
[
"Suppose instead of viewing the 17th sample, we want to see the average of samples corresponding to a certain value.\n\nThis can be done as follows (using 0 as an example):\n- All samples where the target value is 0 are located\n- The mean of these samples is taken\n- The resulting 64 long array is reshaped to be 8\\*8 (for display)\n- The image is displayed",
"_____no_output_____"
]
],
[
[
"# take samples with target=0\nizeros = np.where(y == 0)\n# take average across samples, reshape to visualise\nzeros = np.mean(X[izeros], axis=0).reshape(8,8)\n\n# display\nfig = plt.matshow(zeros, cmap=plt.cm.gray)\nfig.axes.get_xaxis().set_visible(False)\nfig.axes.get_yaxis().set_visible(False)",
"_____no_output_____"
]
],
[
[
"<hr>\n\n# Fit and test the model\n\n## Split the data",
"_____no_output_____"
],
[
"Now that you have an understanding of the data, the model can be fitted.\n\nFitting the model involves setting some of the data aside for testing, and allowing the model to \"see\" the target values corresponding to the training samples.\n\nOnce the model has been fitted to the training data, the model will be tested on some data it has not seen before. \n\nThe next cell uses [train_test_split](https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.train_test_split.html) to shuffle all data, then set some data aside for testing later. \n\nFor this example, $\\frac{1}{4}$ of the data will be set aside for testing, and the model will be trained on the remaining training set.\n\nAs before, <code>X</code> corresponds to data samples, and <code>y</code> corresponds to labels.",
"_____no_output_____"
]
],
[
[
"# split data to train and test sets\nX_train, X_test, y_train, y_test = \\\n ms.train_test_split(X, y, test_size=0.25, shuffle=True,\n random_state=22)",
"_____no_output_____"
]
],
[
[
"The data can be examined - here you can see that 1347 samples have been put into the training set, and 450 have been set aside for testing.",
"_____no_output_____"
]
],
[
[
"# print shape of data\nprint(\"training samples:\", X_train.shape) \nprint(\"testing samples :\", X_test.shape)\nprint(\"training targets:\", y_train.shape) \nprint(\"testing targets :\", y_test.shape) ",
"_____no_output_____"
]
],
[
[
"## Using PCA to visualise data\n\nBefore diving into classifying, it is useful to visualise the data.\n\nSince each sample has 64 dimensions, some dimensionality reduction is needed in order to visualise the samples as points on a 2D map.\n\nOne of the easiest ways of visualising high dimensional data is by principal component analysis (PCA). This maps the 64 dimensional image data onto a lower dimension map (here we will map to 2D) so it can be easily viewed on a screen.\n\nIn this case, the 2 most important \"components\" are maintained.",
"_____no_output_____"
]
],
[
[
"# create PCA model with 2 components\npca = decomposition.PCA(n_components=2)",
"_____no_output_____"
]
],
[
[
"The next step is to perform the PCA on the samples, and store the results.",
"_____no_output_____"
]
],
[
[
"# transform training data to 2 principal components\nX_pca = pca.fit_transform(X_train)\n\n# transform test data to 2 principal components\nT_pca = pca.transform(X_test)",
"_____no_output_____"
],
[
"# check shape of result\nprint(X_pca.shape) \nprint(T_pca.shape)",
"_____no_output_____"
]
],
[
[
"As you can see from the above cell, the <code>X_pca</code> and <code>T_pca</code> data is now represented by only 2 elements per sample. The number of samples has remained the same.\n\nNow that there is a 2D representation of the data, it can be plotted on a regular scatter graph. Since the labels corresponding to each point are stored in the <code>y_train</code> variable, the plot can be colour coded by target value!\n\nDifferent coloured dots have different target values.",
"_____no_output_____"
]
],
[
[
"# choose the colours for each digit\ncmap_digits = plt.cm.tab10\n\n# plot training data with labels\nplt.figure(figsize = (9,6))\nplt.scatter(X_pca[:,0], X_pca[:,1], s=7, c=y_train,\n cmap=cmap_digits, alpha=0.7)\nplt.title(\"Training data coloured by target value\")\nplt.colorbar();",
"_____no_output_____"
]
],
[
[
"## Create and fit the model\n\nThe scikit-learn library allows fitting of a k-NN model just as with PCA above.\n\nFirst, create the classifier:",
"_____no_output_____"
]
],
[
[
"# create model\nknn = neighbors.KNeighborsClassifier()",
"_____no_output_____"
]
],
[
[
"The next step fits the k-NN model using the training data.",
"_____no_output_____"
]
],
[
[
"# fit model to training data\nknn.fit(X_train,y_train);",
"_____no_output_____"
]
],
[
[
"## Test model\n\nNow use the data that was set aside earlier - this stage involves getting the model to \"guess\" the samples (this time without seeing their target values).\n\nOnce the model has predicted the sample's class, a score can be calculated by checking how many samples the model guessed correctly.",
"_____no_output_____"
]
],
[
[
"# predict test data\npreds = knn.predict(X_test)\n\n# test model on test data\nscore = round(knn.score(X_test,y_test)*100, 2)\nprint(\"Score on test data: \" + str(score) + \"%\")",
"_____no_output_____"
]
],
[
[
"98.44% is a really high score, one that would not likely be seen on real life applications of the method.\n\nIt can often be useful to visualise the results of your example. Below are plots showing:\n- The labels that the model predicted for the test data\n- The actual labels for the test data\n- The data points that were incorrectly labelled\n\nIn this case, the predicted and actual plots are very similar, so these plots are not very informative. In other cases, this kind of visualisation may reveal patterns for you to explore further.",
"_____no_output_____"
]
],
[
[
"# plot 3 axes\nfig, axes = plt.subplots(2,2,figsize=(12,12))\n\n# top left axis for predictions\naxes[0,0].scatter(T_pca[:,0], T_pca[:,1], s=5, \n c=preds, cmap=cmap_digits)\naxes[0,0].set_title(\"Predicted labels\")\n\n# top right axis for actual targets\naxes[0,1].scatter(T_pca[:,0], T_pca[:,1], s=5, \n c=y_test, cmap=cmap_digits)\naxes[0,1].set_title(\"Actual labels\")\n\n# bottom left axis coloured to show correct and incorrect\naxes[1,0].scatter(T_pca[:,0], T_pca[:,1], s=5, \n c=(preds==y_test))\naxes[1,0].set_title(\"Incorrect labels\")\n\n# bottom right axis not used\naxes[1,1].set_axis_off()",
"_____no_output_____"
]
],
[
[
"So which samples did the model get wrong?\n\nThere were 7 samples that were misclassified. These can be displayed alongside their actual and predicted labels using the cell below:",
"_____no_output_____"
]
],
[
[
"# find the misclassified samples\nmisclass = np.where(preds!=y_test)[0]\n\n# display misclassified samples\nr, c = 1, len(misclass)\nfig, axes = plt.subplots(r,c,figsize=(10,5))\n\nfor i in range(c):\n ax = axes[i]\n ax.matshow(X_test[misclass[i]].reshape(8,8),cmap=plt.cm.gray)\n ax.set_axis_off()\n act = y_test[misclass[i]]\n pre = preds[misclass[i]]\n strng = \"actual: {a:.0f} \\npredicted: {p:.0f}\".format(a=act, p=pre)\n ax.set_title(strng)",
"_____no_output_____"
]
],
[
[
"Additionally, a confusion matrix can be used to identify which samples are misclassified by the model. This can help you identify if their are samples that are commonly misidentified - for example you may identify that 8's are often mistook for 1's.",
"_____no_output_____"
]
],
[
[
"# confusion matrix\nconf = metrics.confusion_matrix(y_test,preds)\n\n# figure\nf, ax = plt.subplots(figsize=(9,5))\nim = ax.imshow(conf, cmap=plt.cm.RdBu)\n\n# set labels as ticks on axes\nax.set_xticks(np.arange(10))\nax.set_yticks(np.arange(10))\nax.set_xticklabels(list(range(0,10)))\nax.set_yticklabels(list(range(0,10)))\nax.set_ylim(9.5,-0.5)\n\n# axes labels\nax.set_ylabel(\"actual value\")\nax.set_xlabel(\"predicted value\")\nax.set_title(\"Digit classification confusion matrix\")\n\n# display\nplt.colorbar(im).set_label(label=\"number of classifications\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0843c0ee96f101942054aa2face41a4b49705ca | 17,500 | ipynb | Jupyter Notebook | extract_training_images.ipynb | adriancampos/road-extraction | 3eaf4ed010d71475276d99d4841d67990a967a1b | [
"MIT"
] | 1 | 2019-07-12T20:17:24.000Z | 2019-07-12T20:17:24.000Z | extract_training_images.ipynb | adriancampos/road-extraction | 3eaf4ed010d71475276d99d4841d67990a967a1b | [
"MIT"
] | null | null | null | extract_training_images.ipynb | adriancampos/road-extraction | 3eaf4ed010d71475276d99d4841d67990a967a1b | [
"MIT"
] | null | null | null | 33.914729 | 231 | 0.528629 | [
[
[
"## Dependencies",
"_____no_output_____"
]
],
[
[
"!nvidia-smi\n!jupyter notebook list\n%env CUDA_VISIBLE_DEVICES=3",
"Wed Nov 6 18:21:34 2019 \n+-----------------------------------------------------------------------------+\n| NVIDIA-SMI 410.104 Driver Version: 410.104 CUDA Version: 10.0 |\n|-------------------------------+----------------------+----------------------+\n| GPU Name Persistence-M| Bus-Id Disp.A | Volatile Uncorr. ECC |\n| Fan Temp Perf Pwr:Usage/Cap| Memory-Usage | GPU-Util Compute M. |\n|===============================+======================+======================|\n| 0 Tesla K80 Off | 00000000:00:04.0 Off | 0 |\n| N/A 54C P0 99W / 149W | 9115MiB / 11441MiB | 60% Default |\n+-------------------------------+----------------------+----------------------+\n| 1 Tesla K80 Off | 00000000:00:05.0 Off | 0 |\n| N/A 73C P0 95W / 149W | 6011MiB / 11441MiB | 0% Default |\n+-------------------------------+----------------------+----------------------+\n| 2 Tesla K80 Off | 00000000:00:06.0 Off | 0 |\n| N/A 49C P0 58W / 149W | 672MiB / 11441MiB | 0% Default |\n+-------------------------------+----------------------+----------------------+\n| 3 Tesla K80 Off | 00000000:00:07.0 Off | 0 |\n| N/A 74C P0 92W / 149W | 0MiB / 11441MiB | 48% Default |\n+-------------------------------+----------------------+----------------------+\n \n+-----------------------------------------------------------------------------+\n| Processes: GPU Memory |\n| GPU PID Type Process name Usage |\n|=============================================================================|\n| 0 7353 C /opt/anaconda3/bin/python 4212MiB |\n| 0 16848 C /opt/anaconda3/bin/python 4889MiB |\n| 1 26172 C /opt/anaconda3/bin/python 6000MiB |\n| 2 18096 C /opt/anaconda3/bin/python 661MiB |\n+-----------------------------------------------------------------------------+\nCurrently running servers:\nhttp://localhost:8080/ :: /home/jupyter\nhttp://localhost:8080/ :: /home/jupyter\nenv: CUDA_VISIBLE_DEVICES=3\n"
],
[
"%matplotlib inline\n%load_ext autoreload\n%autoreload 2\n\nimport time\nfrom pathlib import Path\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nimport torch\nimport torch.nn as nn\nimport torch.optim as optim\nimport torchvision\nimport torchvision.transforms as transforms\n\nfrom models import tiramisu\nfrom models import tiramisu_bilinear\nfrom models import tiramisu_m3\nfrom models import unet\nfrom datasets import deepglobe\nfrom datasets import maroads\nfrom datasets import joint_transforms\nimport utils.imgs\nimport utils.training as train_utils\n\n# tensorboard\nfrom torch.utils.tensorboard import SummaryWriter",
"_____no_output_____"
]
],
[
[
"## Dataset\n\nDownload the DeepGlobe dataset from https://competitions.codalab.org/competitions/18467. Place it in datasets/deepglobe/dataset/train,test,valid\nDownload the Massachusetts Road Dataset from https://www.cs.toronto.edu/~vmnih/data/. Combine the training, validation, and test sets, process with `crop_dataset.ipynb` and place the output in datasets/maroads/dataset/map,sat",
"_____no_output_____"
]
],
[
[
"run = \"expM.3.drop2.1\"\nDEEPGLOBE_PATH = Path('datasets/', 'deepglobe/dataset')\nMAROADS_PATH = Path('datasets/', 'maroads/dataset')\nRESULTS_PATH = Path('.results/')\nWEIGHTS_PATH = Path('.weights/')\nRUNS_PATH = Path('.runs/')\nRESULTS_PATH.mkdir(exist_ok=True)\nWEIGHTS_PATH.mkdir(exist_ok=True)\nRUNS_PATH.mkdir(exist_ok=True)\n\nbatch_size = 1 # TODO: Should be `MAX_BATCH_PER_CARD * torch.cuda.device_count()` (which in this case is 1 assuming max of 1 batch per card)",
"_____no_output_____"
],
[
"# resize = joint_transforms.JointRandomCrop((300, 300))\n\nnormalize = transforms.Normalize(mean=deepglobe.mean, std=deepglobe.std)\ntrain_joint_transformer = transforms.Compose([\n# resize,\n joint_transforms.JointRandomHorizontalFlip(),\n joint_transforms.JointRandomVerticalFlip(),\n joint_transforms.JointRandomRotate()\n ])\n\ntrain_slice = slice(None,4000)\ntest_slice = slice(4000,None)\n\ntrain_dset = deepglobe.DeepGlobe(DEEPGLOBE_PATH, 'train', slc = train_slice,\n joint_transform=train_joint_transformer,\n transform=transforms.Compose([\n transforms.ColorJitter(brightness=.4,contrast=.4,saturation=.4),\n transforms.ToTensor(),\n normalize,\n ]))\n\ntrain_dset_ma = maroads.MARoads(MAROADS_PATH, \n joint_transform=train_joint_transformer,\n transform=transforms.Compose([\n transforms.ColorJitter(brightness=.4,contrast=.4,saturation=.4),\n transforms.ToTensor(),\n normalize,\n ]))\n\n# print(len(train_dset_ma.imgs))\n# print(len(train_dset_ma.msks))\ntrain_dset_combine = torch.utils.data.ConcatDataset((train_dset, train_dset_ma))\n\n# train_loader = torch.utils.data.DataLoader(train_dset, batch_size=batch_size, shuffle=True)\n# train_loader = torch.utils.data.DataLoader(train_dset_ma, batch_size=batch_size, shuffle=True)\ntrain_loader = torch.utils.data.DataLoader(\n train_dset_combine, batch_size=batch_size, shuffle=True)\n\n\n\n\n\n# resize_joint_transformer = transforms.Compose([\n# resize\n# ])\nresize_joint_transformer = None\nval_dset = deepglobe.DeepGlobe(\n DEEPGLOBE_PATH, 'valid', joint_transform=resize_joint_transformer,\n transform=transforms.Compose([\n transforms.ToTensor(),\n normalize\n ]))\nval_loader = torch.utils.data.DataLoader(\n val_dset, batch_size=batch_size, shuffle=False)\n\ntest_dset = deepglobe.DeepGlobe(\n DEEPGLOBE_PATH, 'train', joint_transform=resize_joint_transformer, slc = test_slice,\n transform=transforms.Compose([\n transforms.ToTensor(),\n normalize\n ]))\ntest_loader = torch.utils.data.DataLoader(\n test_dset, batch_size=batch_size, shuffle=False)",
"_____no_output_____"
],
[
"print(\"Train: %d\" %len(train_loader.dataset))\nprint(\"Val: %d\" %len(val_loader.dataset.imgs))\nprint(\"Test: %d\" %len(test_loader.dataset.imgs))\n# print(\"Classes: %d\" % len(train_loader.dataset.classes))\n\nprint((iter(train_loader)))\n\ninputs, targets = next(iter(train_loader))\nprint(\"Inputs: \", inputs.size())\nprint(\"Targets: \", targets.size())\n\n# utils.imgs.view_image(inputs[0])\n# utils.imgs.view_image(targets[0])\n# utils.imgs.view_annotated(targets[0])\n\n# print(targets[0])\n\n\n\nfor i,(image,label) in enumerate(iter(test_loader)):\n if i % 10 == 0:\n print(\"Procssing image\",i)\n \n im = image[0]\n \n # scale to [0,1]\n im -= im.min()\n im /= im.max()\n \n im = torchvision.transforms.ToPILImage()(im)\n im.save(\"ds_test/\" + str(i) + \".png\")\n \n label = label.float()\n la = torchvision.transforms.ToPILImage()(label)\n la.save(\"ds_test/\" + str(i) + \".mask.png\")\n \nprint(\"Done!\")",
"Train: 4909\nVal: 1243\nTest: 2226\n<torch.utils.data.dataloader._SingleProcessDataLoaderIter object at 0x7f387e5c3dd0>\nInputs: torch.Size([1, 3, 1024, 1024])\nTargets: torch.Size([1, 1024, 1024])\nProcssing image 0\nProcssing image 10\nProcssing image 20\nProcssing image 30\nProcssing image 40\nProcssing image 50\nProcssing image 60\nProcssing image 70\nProcssing image 80\nProcssing image 90\nProcssing image 100\nProcssing image 110\nProcssing image 120\nProcssing image 130\nProcssing image 140\nProcssing image 150\nProcssing image 160\nProcssing image 170\nProcssing image 180\nProcssing image 190\nProcssing image 200\nProcssing image 210\nProcssing image 220\nProcssing image 230\nProcssing image 240\nProcssing image 250\nProcssing image 260\nProcssing image 270\nProcssing image 350\nProcssing image 360\nProcssing image 370\nProcssing image 380\nProcssing image 390\nProcssing image 400\nProcssing image 410\nProcssing image 420\nProcssing image 430\nProcssing image 440\nProcssing image 450\nProcssing image 460\nProcssing image 470\nProcssing image 480\nProcssing image 490\nProcssing image 500\nProcssing image 510\nProcssing image 520\nProcssing image 530\nProcssing image 540\nProcssing image 550\nProcssing image 560\nProcssing image 570\nProcssing image 580\nProcssing image 590\nProcssing image 600\nProcssing image 610\nProcssing image 620\nProcssing image 630\nProcssing image 640\nProcssing image 650\nProcssing image 660\nProcssing image 670\nProcssing image 680\nProcssing image 690\nProcssing image 700\nProcssing image 710\nProcssing image 720\nProcssing image 730\nProcssing image 740\nProcssing image 750\nProcssing image 760\nProcssing image 770\nProcssing image 780\nProcssing image 790\nProcssing image 800\nProcssing image 810\nProcssing image 820\nProcssing image 830\nProcssing image 840\nProcssing image 850\nProcssing image 860\nProcssing image 870\nProcssing image 880\nProcssing image 890\nProcssing image 900\nProcssing image 910\nProcssing image 920\nProcssing image 930\nProcssing image 940\nProcssing image 950\nProcssing image 960\nProcssing image 970\nProcssing image 980\nProcssing image 990\nProcssing image 1000\nProcssing image 1010\nProcssing image 1020\nProcssing image 1030\nProcssing image 1040\nProcssing image 1050\nProcssing image 1060\nProcssing image 1070\nProcssing image 1080\nProcssing image 1090\nProcssing image 1100\nProcssing image 1110\nProcssing image 1120\nProcssing image 1130\nProcssing image 1140\nProcssing image 1150\nProcssing image 1160\nProcssing image 1170\nProcssing image 1180\nProcssing image 1190\nProcssing image 1200\nProcssing image 1210\nProcssing image 1220\nProcssing image 1230\nProcssing image 1240\nProcssing image 1250\nProcssing image 1260\nProcssing image 1270\nProcssing image 1280\nProcssing image 1290\nProcssing image 1300\nProcssing image 1310\nProcssing image 1320\nProcssing image 1330\nProcssing image 1340\nProcssing image 1350\nProcssing image 1360\nProcssing image 1370\nProcssing image 1380\nProcssing image 1390\nProcssing image 1400\nProcssing image 1410\nProcssing image 1420\nProcssing image 1430\nProcssing image 1440\nProcssing image 1450\nProcssing image 1460\nProcssing image 1470\nProcssing image 1480\nProcssing image 1490\nProcssing image 1500\nProcssing image 1510\nProcssing image 1520\nProcssing image 1530\nProcssing image 1540\nProcssing image 1550\nProcssing image 1560\nProcssing image 1570\nProcssing image 1580\nProcssing image 1590\nProcssing image 1600\nProcssing image 1610\nProcssing image 1620\nProcssing image 1630\nProcssing image 1640\nProcssing image 1650\nProcssing image 1660\nProcssing image 1670\nProcssing image 1680\nProcssing image 1690\nProcssing image 1700\nProcssing image 1710\nProcssing image 1720\nProcssing image 1730\nProcssing image 1740\nProcssing image 1750\nProcssing image 1760\nProcssing image 1770\nProcssing image 1780\nProcssing image 1790\nProcssing image 1800\nProcssing image 1810\nProcssing image 1820\nProcssing image 1830\nProcssing image 1840\nProcssing image 1850\nProcssing image 1860\nProcssing image 1870\nProcssing image 1880\nProcssing image 1890\nProcssing image 1900\nProcssing image 1910\nProcssing image 1920\nProcssing image 1930\nProcssing image 1940\nProcssing image 1950\nProcssing image 1960\nProcssing image 1970\nProcssing image 1980\nProcssing image 1990\nProcssing image 2000\nProcssing image 2010\nProcssing image 2020\nProcssing image 2030\nProcssing image 2040\nProcssing image 2050\nProcssing image 2060\nProcssing image 2070\nProcssing image 2080\nProcssing image 2090\nProcssing image 2100\nProcssing image 2110\nProcssing image 2120\nProcssing image 2130\nProcssing image 2140\nProcssing image 2150\nProcssing image 2160\nProcssing image 2170\nProcssing image 2180\nProcssing image 2190\nProcssing image 2200\nProcssing image 2210\nProcssing image 2220\nDone!\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d0844244d9e79d2741bca35c3407173117bcc65c | 1,388 | ipynb | Jupyter Notebook | jb_demo/foundations/Hello.ipynb | halehawk/sphinx-pythia-theme | d0a48a23ac21544c7cfda73f6732b879b0c6b1dc | [
"Apache-2.0"
] | null | null | null | jb_demo/foundations/Hello.ipynb | halehawk/sphinx-pythia-theme | d0a48a23ac21544c7cfda73f6732b879b0c6b1dc | [
"Apache-2.0"
] | null | null | null | jb_demo/foundations/Hello.ipynb | halehawk/sphinx-pythia-theme | d0a48a23ac21544c7cfda73f6732b879b0c6b1dc | [
"Apache-2.0"
] | null | null | null | 17.794872 | 67 | 0.513689 | [
[
[
"# Fun with Python\n\nA very minimal example for the Pythia Foundations collection.",
"_____no_output_____"
],
[
"A Python program can be a single line:",
"_____no_output_____"
]
],
[
[
"print('Hello interweb')",
"Hello interweb\n"
]
],
[
[
"Try it out in Binder and run it yourself!",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d08445bf59349230f253559db8e4967827617887 | 7,756 | ipynb | Jupyter Notebook | floydhub/Bootstrap/bootstrap.ipynb | CarlosOmdelc/ImagesToCode | 82331e4dc8be263a1136d00a21f4407a9d92d3be | [
"MIT"
] | 1 | 2019-07-18T12:50:49.000Z | 2019-07-18T12:50:49.000Z | floydhub/Bootstrap/bootstrap.ipynb | CarlosOmdelc/ImagesToCode | 82331e4dc8be263a1136d00a21f4407a9d92d3be | [
"MIT"
] | null | null | null | floydhub/Bootstrap/bootstrap.ipynb | CarlosOmdelc/ImagesToCode | 82331e4dc8be263a1136d00a21f4407a9d92d3be | [
"MIT"
] | null | null | null | 38.78 | 136 | 0.57852 | [
[
[
"from os import listdir\nfrom numpy import array\nfrom keras.preprocessing.text import Tokenizer, one_hot\nfrom keras.preprocessing.sequence import pad_sequences\nfrom keras.models import Model, Sequential, model_from_json\nfrom keras.utils import to_categorical\nfrom keras.layers.core import Dense, Dropout, Flatten\nfrom keras.optimizers import RMSprop\nfrom keras.layers.convolutional import Conv2D\nfrom keras.callbacks import ModelCheckpoint\nfrom keras.layers import Embedding, TimeDistributed, RepeatVector, LSTM, concatenate , Input, Reshape, Dense\nfrom keras.preprocessing.image import array_to_img, img_to_array, load_img\nimport numpy as np",
"_____no_output_____"
],
[
"dir_name = '/data/train/'\n\n# Read a file and return a string\ndef load_doc(filename):\n file = open(filename, 'r')\n text = file.read()\n file.close()\n return text\n\ndef load_data(data_dir):\n text = []\n images = []\n # Load all the files and order them\n all_filenames = listdir(data_dir)\n all_filenames.sort()\n for filename in (all_filenames):\n if filename[-3:] == \"npz\":\n # Load the images already prepared in arrays\n image = np.load(data_dir+filename)\n images.append(image['features'])\n else:\n # Load the boostrap tokens and rap them in a start and end tag\n syntax = '<START> ' + load_doc(data_dir+filename) + ' <END>'\n # Seperate all the words with a single space\n syntax = ' '.join(syntax.split())\n # Add a space after each comma\n syntax = syntax.replace(',', ' ,')\n text.append(syntax)\n images = np.array(images, dtype=float)\n return images, text\n\ntrain_features, texts = load_data(dir_name)",
"_____no_output_____"
],
[
"# Initialize the function to create the vocabulary \ntokenizer = Tokenizer(filters='', split=\" \", lower=False)\n# Create the vocabulary \ntokenizer.fit_on_texts([load_doc('bootstrap.vocab')])\n\n# Add one spot for the empty word in the vocabulary \nvocab_size = len(tokenizer.word_index) + 1\n# Map the input sentences into the vocabulary indexes\ntrain_sequences = tokenizer.texts_to_sequences(texts)\n# The longest set of boostrap tokens\nmax_sequence = max(len(s) for s in train_sequences)\n# Specify how many tokens to have in each input sentence\nmax_length = 48\n\ndef preprocess_data(sequences, features):\n X, y, image_data = list(), list(), list()\n for img_no, seq in enumerate(sequences):\n for i in range(1, len(seq)):\n # Add the sentence until the current count(i) and add the current count to the output\n in_seq, out_seq = seq[:i], seq[i]\n # Pad all the input token sentences to max_sequence\n in_seq = pad_sequences([in_seq], maxlen=max_sequence)[0]\n # Turn the output into one-hot encoding\n out_seq = to_categorical([out_seq], num_classes=vocab_size)[0]\n # Add the corresponding image to the boostrap token file\n image_data.append(features[img_no])\n # Cap the input sentence to 48 tokens and add it\n X.append(in_seq[-48:])\n y.append(out_seq)\n return np.array(X), np.array(y), np.array(image_data)\n\nX, y, image_data = preprocess_data(train_sequences, train_features)",
"_____no_output_____"
],
[
"#Create the encoder\nimage_model = Sequential()\nimage_model.add(Conv2D(16, (3, 3), padding='valid', activation='relu', input_shape=(256, 256, 3,)))\nimage_model.add(Conv2D(16, (3,3), activation='relu', padding='same', strides=2))\nimage_model.add(Conv2D(32, (3,3), activation='relu', padding='same'))\nimage_model.add(Conv2D(32, (3,3), activation='relu', padding='same', strides=2))\nimage_model.add(Conv2D(64, (3,3), activation='relu', padding='same'))\nimage_model.add(Conv2D(64, (3,3), activation='relu', padding='same', strides=2))\nimage_model.add(Conv2D(128, (3,3), activation='relu', padding='same'))\n\nimage_model.add(Flatten())\nimage_model.add(Dense(1024, activation='relu'))\nimage_model.add(Dropout(0.3))\nimage_model.add(Dense(1024, activation='relu'))\nimage_model.add(Dropout(0.3))\n\nimage_model.add(RepeatVector(max_length))\n\nvisual_input = Input(shape=(256, 256, 3,))\nencoded_image = image_model(visual_input)\n\nlanguage_input = Input(shape=(max_length,))\nlanguage_model = Embedding(vocab_size, 50, input_length=max_length, mask_zero=True)(language_input)\nlanguage_model = LSTM(128, return_sequences=True)(language_model)\nlanguage_model = LSTM(128, return_sequences=True)(language_model)\n\n#Create the decoder\ndecoder = concatenate([encoded_image, language_model])\ndecoder = LSTM(512, return_sequences=True)(decoder)\ndecoder = LSTM(512, return_sequences=False)(decoder)\ndecoder = Dense(vocab_size, activation='softmax')(decoder)\n\n# Compile the model\nmodel = Model(inputs=[visual_input, language_input], outputs=decoder)\noptimizer = RMSprop(lr=0.0001, clipvalue=1.0)\nmodel.compile(loss='categorical_crossentropy', optimizer=optimizer)",
"_____no_output_____"
],
[
"#Save the model for every 2nd epoch\nfilepath=\"org-weights-epoch-{epoch:04d}--val_loss-{val_loss:.4f}--loss-{loss:.4f}.hdf5\"\ncheckpoint = ModelCheckpoint(filepath, monitor='val_loss', verbose=1, save_weights_only=True, period=2)\ncallbacks_list = [checkpoint]",
"_____no_output_____"
],
[
"# Train the model\nmodel.fit([image_data, X], y, batch_size=64, shuffle=False, validation_split=0.1, callbacks=callbacks_list, verbose=1, epochs=50)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d08477339bff001dae961fa67466f2b6a2254d13 | 907,920 | ipynb | Jupyter Notebook | Analiza i Bazy Danych 2020/Lecture 3 (EDA)/notes.ipynb | jakub-sacha/public_lectures | fbd1360e0a0f4655985e49ef53fcecfd5e99367f | [
"CC-BY-4.0"
] | null | null | null | Analiza i Bazy Danych 2020/Lecture 3 (EDA)/notes.ipynb | jakub-sacha/public_lectures | fbd1360e0a0f4655985e49ef53fcecfd5e99367f | [
"CC-BY-4.0"
] | null | null | null | Analiza i Bazy Danych 2020/Lecture 3 (EDA)/notes.ipynb | jakub-sacha/public_lectures | fbd1360e0a0f4655985e49ef53fcecfd5e99367f | [
"CC-BY-4.0"
] | null | null | null | 2,678.230088 | 380,361 | 0.689554 | [
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\ndef ECDF(series):\n ser=series.value_counts().sort_index().cumsum()/len(series)\n ser.index.name='Value'\n ser.name='Probability'\n return ser",
"_____no_output_____"
],
[
"np.random.seed(223)\ndf = pd.DataFrame()\n\nx2 = np.random.normal(0,np.pi/6,size=1000)\nx2_org=x2.copy()\nx2[np.abs(x2)<0.1]=0\ndf['Angle']=x2\nalpha=(x2>np.pi/6)&(x2<np.pi/3).astype(int)\ndf['Poluted']=alpha\nalpha=alpha*3+1\nx1 = np.random.beta(alpha,10,size=1000)\nx1_org=x1.copy()\nx1[x1<0.01]=0\nx1[x1>0.3]=0.3\ndf['Concentration']=x1\n\n\ndf_org=pd.DataFrame({'Angle':x2_org,'Concentration':x1_org,'Poluted':df['Poluted']})\ndf.drop('Poluted',axis=1)\ndf.to_csv('polution_observed_data.csv')\ndf_org.to_csv('polution_true_data.csv')",
"_____no_output_____"
],
[
"fig,axes = plt.subplots(1,2,figsize=(7, 8),subplot_kw=dict(polar=True))\nax=axes[0]\nax.scatter(x2_org, x1_org, c=df['Polluted'])\n\nax.set_thetamin(-90)\nax.set_thetamax(90)\nax.set_title('True data')\nax=axes[1]\nax.scatter(x2, x1,c=df['Polluted'])\nax.set_thetamin(-90)\nax.set_thetamax(90)\nax.set_title('Observed data')\n\n\nplt.show()",
"_____no_output_____"
],
[
"series=df.Concentration",
"_____no_output_____"
],
[
"n=10\nseries.plot(kind='hist',bins=n,density=True,title=series.name+' Histogram bins={}'.format(n))\nplt.show()",
"_____no_output_____"
],
[
"series.plot(kind='density',title=series.name+' KDE')\nplt.show()",
"_____no_output_____"
],
[
"ax=ECDF(series).plot(title=series.name+' ECDF',drawstyle=\"steps\")\nax.set_ylabel(ylabel='Probability')\nax.set_yticks([0, ECDF(series).min(),1])\nplt.show()",
"_____no_output_____"
],
[
"series",
"_____no_output_____"
],
[
"np.random.seed(223)\n\nseries = df.Angle",
"_____no_output_____"
],
[
"n=10\nseries.plot(kind='hist',bins=n,density=True,title=series.name+' Histogram bins={}'.format(n))\n\nplt.show()",
"_____no_output_____"
],
[
"series.plot(kind='density',title=series.name+' KDE')\nplt.show()",
"_____no_output_____"
],
[
"ax=ECDF(series).plot(title=series.name+' ECDF')\nax.set_ylabel(ylabel='Probability')\nplt.show()",
"_____no_output_____"
],
[
"df.plot(x='Angle',y='Concentration',kind='scatter')",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0847f31ebd4a91edfb16515b6a1bd2e35086552 | 235,520 | ipynb | Jupyter Notebook | intro-to-pytorch/Part 5 - Inference and Validation (Exercises).ipynb | rodolfoams/deep-learning-v2-pytorch | 524d28d1d2279651d86a7679aa85f340fcd05fa4 | [
"MIT"
] | null | null | null | intro-to-pytorch/Part 5 - Inference and Validation (Exercises).ipynb | rodolfoams/deep-learning-v2-pytorch | 524d28d1d2279651d86a7679aa85f340fcd05fa4 | [
"MIT"
] | null | null | null | intro-to-pytorch/Part 5 - Inference and Validation (Exercises).ipynb | rodolfoams/deep-learning-v2-pytorch | 524d28d1d2279651d86a7679aa85f340fcd05fa4 | [
"MIT"
] | null | null | null | 44.055368 | 50,936 | 0.644056 | [
[
[
"# Inference and Validation\n\nNow that you have a trained network, you can use it for making predictions. This is typically called **inference**, a term borrowed from statistics. However, neural networks have a tendency to perform *too well* on the training data and aren't able to generalize to data that hasn't been seen before. This is called **overfitting** and it impairs inference performance. To test for overfitting while training, we measure the performance on data not in the training set called the **validation** set. We avoid overfitting through regularization such as dropout while monitoring the validation performance during training. In this notebook, I'll show you how to do this in PyTorch. \n\nAs usual, let's start by loading the dataset through torchvision. You'll learn more about torchvision and loading data in a later part. This time we'll be taking advantage of the test set which you can get by setting `train=False` here:\n\n```python\ntestset = datasets.FashionMNIST('~/.pytorch/F_MNIST_data/', download=True, train=False, transform=transform)\n```\n\nThe test set contains images just like the training set. Typically you'll see 10-20% of the original dataset held out for testing and validation with the rest being used for training.",
"_____no_output_____"
]
],
[
[
"import torch\nfrom torchvision import datasets, transforms\n\n# Define a transform to normalize the data\ntransform = transforms.Compose([transforms.ToTensor(),\n transforms.Normalize((0.5,), (0.5,))])\n# Download and load the training data\ntrainset = datasets.FashionMNIST('~/.pytorch/F_MNIST_data/', download=True, train=True, transform=transform)\ntrainloader = torch.utils.data.DataLoader(trainset, batch_size=64, shuffle=True)\n\n# Download and load the test data\ntestset = datasets.FashionMNIST('~/.pytorch/F_MNIST_data/', download=True, train=False, transform=transform)\ntestloader = torch.utils.data.DataLoader(testset, batch_size=64, shuffle=True)",
"_____no_output_____"
]
],
[
[
"Here I'll create a model like normal, using the same one from my solution for part 4.",
"_____no_output_____"
]
],
[
[
"from torch import nn, optim\nimport torch.nn.functional as F\n\nclass Classifier(nn.Module):\n def __init__(self):\n super().__init__()\n self.fc1 = nn.Linear(784, 256)\n self.fc2 = nn.Linear(256, 128)\n self.fc3 = nn.Linear(128, 64)\n self.fc4 = nn.Linear(64, 10)\n \n def forward(self, x):\n # make sure input tensor is flattened\n x = x.view(x.shape[0], -1)\n \n x = F.relu(self.fc1(x))\n x = F.relu(self.fc2(x))\n x = F.relu(self.fc3(x))\n x = F.log_softmax(self.fc4(x), dim=1)\n \n return x",
"_____no_output_____"
]
],
[
[
"The goal of validation is to measure the model's performance on data that isn't part of the training set. Performance here is up to the developer to define though. Typically this is just accuracy, the percentage of classes the network predicted correctly. Other options are [precision and recall](https://en.wikipedia.org/wiki/Precision_and_recall#Definition_(classification_context)) and top-5 error rate. We'll focus on accuracy here. First I'll do a forward pass with one batch from the test set.",
"_____no_output_____"
]
],
[
[
"model = Classifier()\n\nimages, labels = next(iter(testloader))\n# Get the class probabilities\nps = torch.exp(model(images))\n# Make sure the shape is appropriate, we should get 10 class probabilities for 64 examples\nprint(ps.shape)",
"torch.Size([64, 10])\n"
]
],
[
[
"With the probabilities, we can get the most likely class using the `ps.topk` method. This returns the $k$ highest values. Since we just want the most likely class, we can use `ps.topk(1)`. This returns a tuple of the top-$k$ values and the top-$k$ indices. If the highest value is the fifth element, we'll get back 4 as the index.",
"_____no_output_____"
]
],
[
[
"top_p, top_class = ps.topk(1, dim=1)\n# Look at the most likely classes for the first 10 examples\nprint(top_class[:10,:])",
"tensor([[4],\n [4],\n [4],\n [4],\n [4],\n [4],\n [4],\n [4],\n [4],\n [4]])\n"
]
],
[
[
"Now we can check if the predicted classes match the labels. This is simple to do by equating `top_class` and `labels`, but we have to be careful of the shapes. Here `top_class` is a 2D tensor with shape `(64, 1)` while `labels` is 1D with shape `(64)`. To get the equality to work out the way we want, `top_class` and `labels` must have the same shape.\n\nIf we do\n\n```python\nequals = top_class == labels\n```\n\n`equals` will have shape `(64, 64)`, try it yourself. What it's doing is comparing the one element in each row of `top_class` with each element in `labels` which returns 64 True/False boolean values for each row.",
"_____no_output_____"
]
],
[
[
"equals = top_class == labels.view(*top_class.shape)",
"_____no_output_____"
]
],
[
[
"Now we need to calculate the percentage of correct predictions. `equals` has binary values, either 0 or 1. This means that if we just sum up all the values and divide by the number of values, we get the percentage of correct predictions. This is the same operation as taking the mean, so we can get the accuracy with a call to `torch.mean`. If only it was that simple. If you try `torch.mean(equals)`, you'll get an error\n\n```\nRuntimeError: mean is not implemented for type torch.ByteTensor\n```\n\nThis happens because `equals` has type `torch.ByteTensor` but `torch.mean` isn't implemented for tensors with that type. So we'll need to convert `equals` to a float tensor. Note that when we take `torch.mean` it returns a scalar tensor, to get the actual value as a float we'll need to do `accuracy.item()`.",
"_____no_output_____"
]
],
[
[
"accuracy = torch.mean(equals.type(torch.FloatTensor))\nprint(f'Accuracy: {accuracy.item()*100}%')",
"Accuracy: 9.375%\n"
]
],
[
[
"The network is untrained so it's making random guesses and we should see an accuracy around 10%. Now let's train our network and include our validation pass so we can measure how well the network is performing on the test set. Since we're not updating our parameters in the validation pass, we can speed up our code by turning off gradients using `torch.no_grad()`:\n\n```python\n# turn off gradients\nwith torch.no_grad():\n # validation pass here\n for images, labels in testloader:\n ...\n```\n\n>**Exercise:** Implement the validation loop below and print out the total accuracy after the loop. You can largely copy and paste the code from above, but I suggest typing it in because writing it out yourself is essential for building the skill. In general you'll always learn more by typing it rather than copy-pasting. You should be able to get an accuracy above 80%.",
"_____no_output_____"
]
],
[
[
"model = Classifier()\ncriterion = nn.NLLLoss()\noptimizer = optim.Adam(model.parameters(), lr=0.003)\n\nepochs = 30\nsteps = 0\n\ntrain_losses, test_losses = [], []\nfor e in range(epochs):\n running_loss = 0\n for images, labels in trainloader:\n \n optimizer.zero_grad()\n \n log_ps = model(images)\n loss = criterion(log_ps, labels)\n loss.backward()\n optimizer.step()\n \n running_loss += loss.item()\n \n else:\n test_loss = 0\n accuracy = 0\n \n # Turn off gradients for validation, saves memory and computations\n with torch.no_grad():\n for images, labels in testloader:\n log_ps = model(images)\n test_loss += criterion(log_ps, labels)\n \n ps = torch.exp(log_ps)\n top_p, top_class = ps.topk(1, dim=1)\n equals = top_class == labels.view(*top_class.shape)\n accuracy += torch.mean(equals.type(torch.FloatTensor))\n \n train_losses.append(running_loss/len(trainloader))\n test_losses.append(test_loss/len(testloader))\n\n print(\"Epoch: {}/{}.. \".format(e+1, epochs),\n \"Training Loss: {:.3f}.. \".format(running_loss/len(trainloader)),\n \"Test Loss: {:.3f}.. \".format(test_loss/len(testloader)),\n \"Test Accuracy: {:.3f}\".format(accuracy/len(testloader)))",
"Epoch: 1/30.. Training Loss: 0.511.. Test Loss: 0.496.. Test Accuracy: 0.823\nEpoch: 2/30.. Training Loss: 0.394.. Test Loss: 0.466.. Test Accuracy: 0.836\nEpoch: 3/30.. Training Loss: 0.357.. Test Loss: 0.392.. Test Accuracy: 0.861\nEpoch: 4/30.. Training Loss: 0.336.. Test Loss: 0.379.. Test Accuracy: 0.864\nEpoch: 5/30.. Training Loss: 0.319.. Test Loss: 0.360.. Test Accuracy: 0.867\nEpoch: 6/30.. Training Loss: 0.305.. Test Loss: 0.374.. Test Accuracy: 0.865\nEpoch: 7/30.. Training Loss: 0.294.. Test Loss: 0.374.. Test Accuracy: 0.874\nEpoch: 8/30.. Training Loss: 0.288.. Test Loss: 0.352.. Test Accuracy: 0.873\nEpoch: 9/30.. Training Loss: 0.277.. Test Loss: 0.364.. Test Accuracy: 0.876\nEpoch: 10/30.. Training Loss: 0.271.. Test Loss: 0.397.. Test Accuracy: 0.870\nEpoch: 11/30.. Training Loss: 0.264.. Test Loss: 0.365.. Test Accuracy: 0.875\nEpoch: 12/30.. Training Loss: 0.257.. Test Loss: 0.371.. Test Accuracy: 0.875\nEpoch: 13/30.. Training Loss: 0.251.. Test Loss: 0.380.. Test Accuracy: 0.877\nEpoch: 14/30.. Training Loss: 0.243.. Test Loss: 0.379.. Test Accuracy: 0.881\nEpoch: 15/30.. Training Loss: 0.233.. Test Loss: 0.370.. Test Accuracy: 0.878\nEpoch: 16/30.. Training Loss: 0.236.. Test Loss: 0.373.. Test Accuracy: 0.880\nEpoch: 17/30.. Training Loss: 0.228.. Test Loss: 0.384.. Test Accuracy: 0.874\nEpoch: 18/30.. Training Loss: 0.225.. Test Loss: 0.378.. Test Accuracy: 0.880\nEpoch: 19/30.. Training Loss: 0.219.. Test Loss: 0.386.. Test Accuracy: 0.878\nEpoch: 20/30.. Training Loss: 0.221.. Test Loss: 0.378.. Test Accuracy: 0.883\nEpoch: 21/30.. Training Loss: 0.211.. Test Loss: 0.446.. Test Accuracy: 0.869\nEpoch: 22/30.. Training Loss: 0.209.. Test Loss: 0.395.. Test Accuracy: 0.882\nEpoch: 23/30.. Training Loss: 0.208.. Test Loss: 0.402.. Test Accuracy: 0.882\nEpoch: 24/30.. Training Loss: 0.205.. Test Loss: 0.412.. Test Accuracy: 0.880\nEpoch: 25/30.. Training Loss: 0.197.. Test Loss: 0.414.. Test Accuracy: 0.885\nEpoch: 26/30.. Training Loss: 0.195.. Test Loss: 0.412.. Test Accuracy: 0.881\nEpoch: 27/30.. Training Loss: 0.198.. Test Loss: 0.407.. Test Accuracy: 0.885\nEpoch: 28/30.. Training Loss: 0.191.. Test Loss: 0.453.. Test Accuracy: 0.879\nEpoch: 29/30.. Training Loss: 0.195.. Test Loss: 0.450.. Test Accuracy: 0.878\nEpoch: 30/30.. Training Loss: 0.188.. Test Loss: 0.440.. Test Accuracy: 0.878\n"
],
[
"%matplotlib inline\n%config InlineBackend.figure_format = 'retina'\n\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"plt.plot(train_losses, label='Training loss')\nplt.plot(test_losses, label='Validation loss')\nplt.legend(frameon=False)",
"_____no_output_____"
]
],
[
[
"## Overfitting\n\nIf we look at the training and validation losses as we train the network, we can see a phenomenon known as overfitting.\n\n<img src='assets/overfitting.png' width=450px>\n\nThe network learns the training set better and better, resulting in lower training losses. However, it starts having problems generalizing to data outside the training set leading to the validation loss increasing. The ultimate goal of any deep learning model is to make predictions on new data, so we should strive to get the lowest validation loss possible. One option is to use the version of the model with the lowest validation loss, here the one around 8-10 training epochs. This strategy is called *early-stopping*. In practice, you'd save the model frequently as you're training then later choose the model with the lowest validation loss.\n\nThe most common method to reduce overfitting (outside of early-stopping) is *dropout*, where we randomly drop input units. This forces the network to share information between weights, increasing it's ability to generalize to new data. Adding dropout in PyTorch is straightforward using the [`nn.Dropout`](https://pytorch.org/docs/stable/nn.html#torch.nn.Dropout) module.\n\n```python\nclass Classifier(nn.Module):\n def __init__(self):\n super().__init__()\n self.fc1 = nn.Linear(784, 256)\n self.fc2 = nn.Linear(256, 128)\n self.fc3 = nn.Linear(128, 64)\n self.fc4 = nn.Linear(64, 10)\n \n # Dropout module with 0.2 drop probability\n self.dropout = nn.Dropout(p=0.2)\n \n def forward(self, x):\n # make sure input tensor is flattened\n x = x.view(x.shape[0], -1)\n \n # Now with dropout\n x = self.dropout(F.relu(self.fc1(x)))\n x = self.dropout(F.relu(self.fc2(x)))\n x = self.dropout(F.relu(self.fc3(x)))\n \n # output so no dropout here\n x = F.log_softmax(self.fc4(x), dim=1)\n \n return x\n```\n\nDuring training we want to use dropout to prevent overfitting, but during inference we want to use the entire network. So, we need to turn off dropout during validation, testing, and whenever we're using the network to make predictions. To do this, you use `model.eval()`. This sets the model to evaluation mode where the dropout probability is 0. You can turn dropout back on by setting the model to train mode with `model.train()`. In general, the pattern for the validation loop will look like this, where you turn off gradients, set the model to evaluation mode, calculate the validation loss and metric, then set the model back to train mode.\n\n```python\n# turn off gradients\nwith torch.no_grad():\n \n # set model to evaluation mode\n model.eval()\n \n # validation pass here\n for images, labels in testloader:\n ...\n\n# set model back to train mode\nmodel.train()\n```",
"_____no_output_____"
],
[
"> **Exercise:** Add dropout to your model and train it on Fashion-MNIST again. See if you can get a lower validation loss or higher accuracy.",
"_____no_output_____"
]
],
[
[
"## TODO: Define your model with dropout added\nclass MyClassifier(nn.Module):\n def __init__(self):\n super().__init__()\n self.fc1 = nn.Linear(784, 256)\n self.fc2 = nn.Linear(256, 128)\n self.fc3 = nn.Linear(128, 64)\n self.fc4 = nn.Linear(64, 10)\n self.dropout = nn.Dropout(p=0.2)\n \n def forward(self, x):\n x = x.view(x.shape[0], -1)\n \n x = self.dropout(F.relu(self.fc1(x)))\n x = self.dropout(F.relu(self.fc2(x)))\n x = self.dropout(F.relu(self.fc3(x)))\n x = F.log_softmax(self.dropout(self.fc4(x)), dim=1)\n \n return x",
"_____no_output_____"
],
[
"## TODO: Train your model with dropout, and monitor the training progress with the validation loss and accuracy\nmodel = MyClassifier()\ncriterion = nn.NLLLoss()\noptimizer = optim.Adam(model.parameters(), lr=0.001)\n\nepochs = 30\n\nfor e in range(epochs):\n running_loss = 0\n for images, labels in trainloader:\n log_ps = model(images)\n loss = criterion(log_ps, labels)\n \n optimizer.zero_grad()\n loss.backward()\n optimizer.step()\n \n running_loss += loss.item()\n else:\n with torch.no_grad():\n model.eval()\n \n for images, labels in testloader:\n ps = torch.exp(model(images))\n top_p, top_class = ps.topk(1, dim=1)\n equals = top_class == labels.view(*top_class.shape)\n accuracy = torch.mean(equals.type(torch.FloatTensor))\n print(f'Accuracy: {accuracy.item()*100}%')\n model.train()",
"Accuracy: 75.0%\nAccuracy: 90.625%\nAccuracy: 75.0%\nAccuracy: 78.125%\nAccuracy: 85.9375%\nAccuracy: 81.25%\nAccuracy: 73.4375%\nAccuracy: 75.0%\nAccuracy: 79.6875%\nAccuracy: 87.5%\nAccuracy: 82.8125%\nAccuracy: 79.6875%\nAccuracy: 90.625%\nAccuracy: 79.6875%\nAccuracy: 81.25%\nAccuracy: 75.0%\nAccuracy: 84.375%\nAccuracy: 81.25%\nAccuracy: 76.5625%\nAccuracy: 76.5625%\nAccuracy: 78.125%\nAccuracy: 85.9375%\nAccuracy: 70.3125%\nAccuracy: 85.9375%\nAccuracy: 81.25%\nAccuracy: 81.25%\nAccuracy: 75.0%\nAccuracy: 89.0625%\nAccuracy: 76.5625%\nAccuracy: 90.625%\nAccuracy: 89.0625%\nAccuracy: 87.5%\nAccuracy: 76.5625%\nAccuracy: 81.25%\nAccuracy: 85.9375%\nAccuracy: 81.25%\nAccuracy: 82.8125%\nAccuracy: 78.125%\nAccuracy: 82.8125%\nAccuracy: 87.5%\nAccuracy: 85.9375%\nAccuracy: 89.0625%\nAccuracy: 81.25%\nAccuracy: 85.9375%\nAccuracy: 82.8125%\nAccuracy: 87.5%\nAccuracy: 79.6875%\nAccuracy: 89.0625%\nAccuracy: 92.1875%\nAccuracy: 89.0625%\nAccuracy: 87.5%\nAccuracy: 78.125%\nAccuracy: 89.0625%\nAccuracy: 92.1875%\nAccuracy: 84.375%\nAccuracy: 79.6875%\nAccuracy: 82.8125%\nAccuracy: 84.375%\nAccuracy: 75.0%\nAccuracy: 84.375%\nAccuracy: 78.125%\nAccuracy: 85.9375%\nAccuracy: 87.5%\nAccuracy: 81.25%\nAccuracy: 87.5%\nAccuracy: 81.25%\nAccuracy: 79.6875%\nAccuracy: 79.6875%\nAccuracy: 87.5%\nAccuracy: 92.1875%\nAccuracy: 84.375%\nAccuracy: 85.9375%\nAccuracy: 78.125%\nAccuracy: 84.375%\nAccuracy: 87.5%\nAccuracy: 81.25%\nAccuracy: 95.3125%\nAccuracy: 81.25%\nAccuracy: 76.5625%\nAccuracy: 90.625%\nAccuracy: 89.0625%\nAccuracy: 87.5%\nAccuracy: 84.375%\nAccuracy: 79.6875%\nAccuracy: 85.9375%\nAccuracy: 89.0625%\nAccuracy: 82.8125%\nAccuracy: 90.625%\nAccuracy: 78.125%\nAccuracy: 84.375%\nAccuracy: 85.9375%\nAccuracy: 89.0625%\nAccuracy: 75.0%\nAccuracy: 87.5%\nAccuracy: 89.0625%\nAccuracy: 82.8125%\nAccuracy: 82.8125%\nAccuracy: 84.375%\nAccuracy: 89.0625%\nAccuracy: 87.5%\nAccuracy: 73.4375%\nAccuracy: 82.8125%\nAccuracy: 78.125%\nAccuracy: 92.1875%\nAccuracy: 87.5%\nAccuracy: 84.375%\nAccuracy: 85.9375%\nAccuracy: 89.0625%\nAccuracy: 89.0625%\nAccuracy: 81.25%\nAccuracy: 87.5%\nAccuracy: 85.9375%\nAccuracy: 87.5%\nAccuracy: 76.5625%\nAccuracy: 87.5%\nAccuracy: 78.125%\nAccuracy: 85.9375%\nAccuracy: 79.6875%\nAccuracy: 82.8125%\nAccuracy: 84.375%\nAccuracy: 92.1875%\nAccuracy: 85.9375%\nAccuracy: 79.6875%\nAccuracy: 78.125%\nAccuracy: 85.9375%\nAccuracy: 84.375%\nAccuracy: 84.375%\nAccuracy: 79.6875%\nAccuracy: 79.6875%\nAccuracy: 87.5%\nAccuracy: 87.5%\nAccuracy: 75.0%\nAccuracy: 89.0625%\nAccuracy: 81.25%\nAccuracy: 84.375%\nAccuracy: 85.9375%\nAccuracy: 84.375%\nAccuracy: 84.375%\nAccuracy: 81.25%\nAccuracy: 81.25%\nAccuracy: 84.375%\nAccuracy: 78.125%\nAccuracy: 85.9375%\nAccuracy: 85.9375%\nAccuracy: 84.375%\nAccuracy: 87.5%\nAccuracy: 96.875%\nAccuracy: 84.375%\nAccuracy: 82.8125%\nAccuracy: 92.1875%\nAccuracy: 87.5%\nAccuracy: 90.625%\nAccuracy: 95.3125%\nAccuracy: 81.25%\nAccuracy: 87.5%\nAccuracy: 79.6875%\nAccuracy: 93.75%\nAccuracy: 84.375%\nAccuracy: 87.5%\nAccuracy: 82.8125%\nAccuracy: 87.5%\nAccuracy: 84.375%\nAccuracy: 87.5%\nAccuracy: 89.0625%\nAccuracy: 79.6875%\nAccuracy: 85.9375%\nAccuracy: 85.9375%\nAccuracy: 79.6875%\nAccuracy: 84.375%\nAccuracy: 85.9375%\nAccuracy: 87.5%\nAccuracy: 82.8125%\nAccuracy: 87.5%\nAccuracy: 79.6875%\nAccuracy: 82.8125%\nAccuracy: 85.9375%\nAccuracy: 87.5%\nAccuracy: 82.8125%\nAccuracy: 82.8125%\nAccuracy: 85.9375%\nAccuracy: 79.6875%\nAccuracy: 87.5%\nAccuracy: 85.9375%\nAccuracy: 89.0625%\nAccuracy: 82.8125%\nAccuracy: 90.625%\nAccuracy: 85.9375%\nAccuracy: 81.25%\nAccuracy: 82.8125%\nAccuracy: 84.375%\nAccuracy: 82.8125%\nAccuracy: 85.9375%\nAccuracy: 79.6875%\nAccuracy: 85.9375%\nAccuracy: 89.0625%\nAccuracy: 92.1875%\nAccuracy: 85.9375%\nAccuracy: 76.5625%\nAccuracy: 84.375%\nAccuracy: 75.0%\nAccuracy: 79.6875%\nAccuracy: 84.375%\nAccuracy: 79.6875%\nAccuracy: 84.375%\nAccuracy: 89.0625%\nAccuracy: 85.9375%\nAccuracy: 87.5%\nAccuracy: 93.75%\nAccuracy: 92.1875%\nAccuracy: 84.375%\nAccuracy: 84.375%\nAccuracy: 92.1875%\nAccuracy: 81.25%\nAccuracy: 87.5%\nAccuracy: 84.375%\nAccuracy: 90.625%\nAccuracy: 85.9375%\nAccuracy: 82.8125%\nAccuracy: 90.625%\nAccuracy: 73.4375%\nAccuracy: 84.375%\nAccuracy: 81.25%\nAccuracy: 70.3125%\nAccuracy: 82.8125%\nAccuracy: 87.5%\nAccuracy: 73.4375%\nAccuracy: 82.8125%\nAccuracy: 79.6875%\nAccuracy: 79.6875%\nAccuracy: 81.25%\nAccuracy: 87.5%\nAccuracy: 87.5%\nAccuracy: 82.8125%\nAccuracy: 85.9375%\nAccuracy: 92.1875%\nAccuracy: 79.6875%\nAccuracy: 92.1875%\nAccuracy: 81.25%\nAccuracy: 87.5%\nAccuracy: 87.5%\nAccuracy: 82.8125%\nAccuracy: 84.375%\nAccuracy: 84.375%\nAccuracy: 90.625%\nAccuracy: 79.6875%\nAccuracy: 82.8125%\nAccuracy: 85.9375%\nAccuracy: 87.5%\nAccuracy: 84.375%\nAccuracy: 85.9375%\nAccuracy: 82.8125%\nAccuracy: 79.6875%\nAccuracy: 81.25%\nAccuracy: 90.625%\nAccuracy: 81.25%\nAccuracy: 84.375%\nAccuracy: 81.25%\nAccuracy: 89.0625%\nAccuracy: 85.9375%\nAccuracy: 87.5%\nAccuracy: 82.8125%\nAccuracy: 79.6875%\nAccuracy: 89.0625%\nAccuracy: 87.5%\nAccuracy: 87.5%\nAccuracy: 82.8125%\nAccuracy: 87.5%\nAccuracy: 82.8125%\nAccuracy: 79.6875%\nAccuracy: 79.6875%\nAccuracy: 85.9375%\nAccuracy: 85.9375%\nAccuracy: 85.9375%\nAccuracy: 81.25%\nAccuracy: 89.0625%\nAccuracy: 85.9375%\nAccuracy: 90.625%\nAccuracy: 89.0625%\nAccuracy: 90.625%\nAccuracy: 82.8125%\nAccuracy: 89.0625%\nAccuracy: 85.9375%\nAccuracy: 89.0625%\nAccuracy: 87.5%\nAccuracy: 78.125%\nAccuracy: 84.375%\nAccuracy: 90.625%\nAccuracy: 87.5%\nAccuracy: 82.8125%\nAccuracy: 89.0625%\nAccuracy: 81.25%\nAccuracy: 82.8125%\nAccuracy: 84.375%\nAccuracy: 81.25%\nAccuracy: 92.1875%\nAccuracy: 90.625%\nAccuracy: 84.375%\nAccuracy: 82.8125%\nAccuracy: 85.9375%\nAccuracy: 78.125%\nAccuracy: 82.8125%\nAccuracy: 82.8125%\nAccuracy: 87.5%\nAccuracy: 78.125%\nAccuracy: 92.1875%\nAccuracy: 76.5625%\nAccuracy: 87.5%\nAccuracy: 82.8125%\nAccuracy: 95.3125%\nAccuracy: 87.5%\nAccuracy: 75.0%\nAccuracy: 87.5%\nAccuracy: 84.375%\nAccuracy: 75.0%\nAccuracy: 84.375%\nAccuracy: 82.8125%\nAccuracy: 82.8125%\nAccuracy: 89.0625%\nAccuracy: 85.9375%\nAccuracy: 85.9375%\nAccuracy: 93.75%\nAccuracy: 84.375%\nAccuracy: 79.6875%\nAccuracy: 79.6875%\nAccuracy: 87.5%\nAccuracy: 85.9375%\nAccuracy: 82.8125%\nAccuracy: 90.625%\nAccuracy: 84.375%\nAccuracy: 82.8125%\nAccuracy: 85.9375%\nAccuracy: 81.25%\nAccuracy: 89.0625%\nAccuracy: 85.9375%\nAccuracy: 81.25%\nAccuracy: 90.625%\nAccuracy: 87.5%\nAccuracy: 84.375%\nAccuracy: 84.375%\nAccuracy: 84.375%\nAccuracy: 81.25%\nAccuracy: 82.8125%\nAccuracy: 87.5%\nAccuracy: 79.6875%\nAccuracy: 90.625%\nAccuracy: 79.6875%\nAccuracy: 92.1875%\nAccuracy: 78.125%\nAccuracy: 93.75%\nAccuracy: 82.8125%\nAccuracy: 79.6875%\nAccuracy: 87.5%\nAccuracy: 75.0%\nAccuracy: 79.6875%\nAccuracy: 82.8125%\nAccuracy: 81.25%\nAccuracy: 89.0625%\nAccuracy: 87.5%\nAccuracy: 87.5%\nAccuracy: 87.5%\nAccuracy: 87.5%\nAccuracy: 82.8125%\nAccuracy: 89.0625%\nAccuracy: 87.5%\nAccuracy: 92.1875%\nAccuracy: 84.375%\nAccuracy: 84.375%\nAccuracy: 85.9375%\nAccuracy: 78.125%\nAccuracy: 78.125%\nAccuracy: 81.25%\nAccuracy: 92.1875%\nAccuracy: 92.1875%\nAccuracy: 84.375%\nAccuracy: 85.9375%\nAccuracy: 84.375%\nAccuracy: 85.9375%\nAccuracy: 78.125%\nAccuracy: 87.5%\nAccuracy: 79.6875%\nAccuracy: 87.5%\nAccuracy: 85.9375%\nAccuracy: 82.8125%\nAccuracy: 81.25%\nAccuracy: 81.25%\nAccuracy: 84.375%\nAccuracy: 82.8125%\nAccuracy: 82.8125%\nAccuracy: 89.0625%\nAccuracy: 84.375%\nAccuracy: 81.25%\nAccuracy: 79.6875%\nAccuracy: 87.5%\nAccuracy: 82.8125%\nAccuracy: 81.25%\nAccuracy: 82.8125%\nAccuracy: 82.8125%\nAccuracy: 90.625%\nAccuracy: 81.25%\nAccuracy: 87.5%\nAccuracy: 78.125%\nAccuracy: 92.1875%\nAccuracy: 90.625%\nAccuracy: 78.125%\nAccuracy: 79.6875%\nAccuracy: 85.9375%\nAccuracy: 89.0625%\nAccuracy: 84.375%\nAccuracy: 85.9375%\nAccuracy: 85.9375%\nAccuracy: 82.8125%\nAccuracy: 85.9375%\nAccuracy: 84.375%\nAccuracy: 84.375%\nAccuracy: 79.6875%\nAccuracy: 84.375%\nAccuracy: 87.5%\nAccuracy: 89.0625%\nAccuracy: 84.375%\nAccuracy: 75.0%\nAccuracy: 82.8125%\nAccuracy: 87.5%\nAccuracy: 81.25%\nAccuracy: 84.375%\nAccuracy: 84.375%\nAccuracy: 87.5%\nAccuracy: 85.9375%\nAccuracy: 79.6875%\nAccuracy: 85.9375%\nAccuracy: 79.6875%\nAccuracy: 90.625%\nAccuracy: 87.5%\nAccuracy: 82.8125%\nAccuracy: 84.375%\nAccuracy: 84.375%\nAccuracy: 79.6875%\nAccuracy: 79.6875%\nAccuracy: 81.25%\nAccuracy: 92.1875%\nAccuracy: 85.9375%\nAccuracy: 84.375%\nAccuracy: 85.9375%\nAccuracy: 89.0625%\nAccuracy: 87.5%\nAccuracy: 78.125%\nAccuracy: 89.0625%\nAccuracy: 82.8125%\nAccuracy: 82.8125%\nAccuracy: 84.375%\nAccuracy: 81.25%\nAccuracy: 85.9375%\nAccuracy: 85.9375%\nAccuracy: 79.6875%\nAccuracy: 87.5%\nAccuracy: 79.6875%\nAccuracy: 85.9375%\nAccuracy: 85.9375%\nAccuracy: 87.5%\n"
]
],
[
[
"## Inference\n\nNow that the model is trained, we can use it for inference. We've done this before, but now we need to remember to set the model in inference mode with `model.eval()`. You'll also want to turn off autograd with the `torch.no_grad()` context.",
"_____no_output_____"
]
],
[
[
"# Import helper module (should be in the repo)\nimport helper\n\n# Test out your network!\n\nmodel.eval()\n\ndataiter = iter(testloader)\nimages, labels = dataiter.next()\nimg = images[0]\n# Convert 2D image to 1D vector\nimg = img.view(1, 784)\n\n# Calculate the class probabilities (softmax) for img\nwith torch.no_grad():\n output = model.forward(img)\n\nps = torch.exp(output)\n\n# Plot the image and probabilities\nhelper.view_classify(img.view(1, 28, 28), ps, version='Fashion')",
"_____no_output_____"
]
],
[
[
"## Next Up!\n\nIn the next part, I'll show you how to save your trained models. In general, you won't want to train a model everytime you need it. Instead, you'll train once, save it, then load the model when you want to train more or use if for inference.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d08482d9cb57369a59d8aaa76c90cb421fd091d7 | 3,026 | ipynb | Jupyter Notebook | salemPi/SoundPlayerDocs.ipynb | kburgon/salem-candy-dispenser | 4549761523717a5ded51572b39ccb2b27e20971f | [
"MIT"
] | null | null | null | salemPi/SoundPlayerDocs.ipynb | kburgon/salem-candy-dispenser | 4549761523717a5ded51572b39ccb2b27e20971f | [
"MIT"
] | null | null | null | salemPi/SoundPlayerDocs.ipynb | kburgon/salem-candy-dispenser | 4549761523717a5ded51572b39ccb2b27e20971f | [
"MIT"
] | null | null | null | 22.75188 | 190 | 0.560145 | [
[
[
"# SoundPlayer Usage Instructions\n\nThe `SoundPlayer` class plays sounds contained within the Sounds directory in the project. Sounds can be played simmply by initializing the class and calling the available methods.\n\nThe sounds that are available to play are located in the folder `Sounds/`. Files of type .mp3 and .wav are supported.",
"_____no_output_____"
],
[
"## Getting a List of Available Sounds\nIn order to get a list of the sounds that are available to play, call the method `SoundPlayer.list_sounds()`.",
"_____no_output_____"
]
],
[
[
"from SoundPlayer import SoundPlayer\n\nplayer = SoundPlayer()\nplayer.list_sounds()",
"_____no_output_____"
]
],
[
[
"## Playing a Sound\n\nA sound can be played by calling the method `SoundPlayer.play(sound)`, where sound is the string name of the sound that will be played.",
"_____no_output_____"
]
],
[
[
"sound = player.list_sounds()[0]\nplayer.play(sound)",
"playing name wlaugh.mp3\n"
]
],
[
[
"## Rotating Through Sounds\nWhen the method `SoundPlayer.play_rotated_sound()` is called, a different sound within the Sounds folder will be played each time.",
"_____no_output_____"
]
],
[
[
"for i in range(len(player.list_sounds())):\n player.play_rotated_sound()",
"playing name wlaugh.mp3\nplaying name hag_idle.mp3\nplaying name ghosts03.mp3\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0849b677b3b8c9d73650a8b99ed93c5e0e8f085 | 3,857 | ipynb | Jupyter Notebook | case-studies/new-york-taxi/new-york-taxi_scale-out.ipynb | Bhaskers-Blu-Org2/AMLDataPrepDocs | 61d1dc7e206d9032a6e5b5304598526c0516b5bb | [
"MIT"
] | null | null | null | case-studies/new-york-taxi/new-york-taxi_scale-out.ipynb | Bhaskers-Blu-Org2/AMLDataPrepDocs | 61d1dc7e206d9032a6e5b5304598526c0516b5bb | [
"MIT"
] | null | null | null | case-studies/new-york-taxi/new-york-taxi_scale-out.ipynb | Bhaskers-Blu-Org2/AMLDataPrepDocs | 61d1dc7e206d9032a6e5b5304598526c0516b5bb | [
"MIT"
] | 1 | 2020-07-30T12:35:49.000Z | 2020-07-30T12:35:49.000Z | 28.360294 | 485 | 0.61706 | [
[
[
"# Scale-Out Data Preparation\n",
"_____no_output_____"
],
[
"Once we are done with preparing and featurizing the data locally, we can run the same steps on the full dataset in scale-out mode. The new york taxi cab data is about 300GB in total, which is perfect for scale-out. Let's start by downloading the package we saved earlier to disk. Feel free to run the `new_york_taxi_cab.ipynb` notebook to generate the package yourself, in which case you may comment out the download code and set the `package_path` to where the package is saved.",
"_____no_output_____"
]
],
[
[
"from tempfile import mkdtemp\nfrom os import path\nfrom urllib.request import urlretrieve\n\ndflow_root = mkdtemp()\ndflow_path = path.join(dflow_root, \"new_york_taxi.dprep\")\nprint(\"Downloading Dataflow to: {}\".format(dflow_path))\nurlretrieve(\"https://dprepdata.blob.core.windows.net/demo/new_york_taxi_v2.dprep\", dflow_path)",
"_____no_output_____"
]
],
[
[
"Let's load the package we just downloaded.",
"_____no_output_____"
]
],
[
[
"import azureml.dataprep as dprep\n\ndf = dprep.Dataflow.open(dflow_path)",
"_____no_output_____"
]
],
[
[
"Let's replace the datasources with the full dataset.",
"_____no_output_____"
]
],
[
[
"from uuid import uuid4\n\nother_step = df._get_steps()[7].arguments['dataflows'][0]['anonymousSteps'][0]\nother_step['id'] = str(uuid4())\nother_step['arguments']['path']['target'] = 1\nother_step['arguments']['path']['resourceDetails'][0]['path'] = 'https://wranglewestus.blob.core.windows.net/nyctaxi/yellow_tripdata*'",
"_____no_output_____"
],
[
"green_dsource = dprep.BlobDataSource(\"https://wranglewestus.blob.core.windows.net/nyctaxi/green_tripdata*\")\ndf = df.replace_datasource(green_dsource)",
"_____no_output_____"
]
],
[
[
"Once we have replaced the datasource, we can now run the same steps on the full dataset. We will print the first 5 rows of the spark DataFrame. Since we are running on the full dataset, this might take a little while depending on your spark cluster's size.",
"_____no_output_____"
]
],
[
[
"spark_df = df.to_spark_dataframe()\nspark_df.head(5)",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d084ac284e702de8e09cc5dcd585ede23fc984a4 | 19,465 | ipynb | Jupyter Notebook | 13-Advanced Python Modules/05-Regular Expressions - re.ipynb | Pankaj-Ra/Complete-Python3-Bootcamp-master | 9e860adff332cead1302a23c18665396fd9b6cb4 | [
"MIT"
] | null | null | null | 13-Advanced Python Modules/05-Regular Expressions - re.ipynb | Pankaj-Ra/Complete-Python3-Bootcamp-master | 9e860adff332cead1302a23c18665396fd9b6cb4 | [
"MIT"
] | null | null | null | 13-Advanced Python Modules/05-Regular Expressions - re.ipynb | Pankaj-Ra/Complete-Python3-Bootcamp-master | 9e860adff332cead1302a23c18665396fd9b6cb4 | [
"MIT"
] | null | null | null | 32.550167 | 443 | 0.533522 | [
[
[
"# Regular Expressions\n\nRegular expressions are text-matching patterns described with a formal syntax. You'll often hear regular expressions referred to as 'regex' or 'regexp' in conversation. Regular expressions can include a variety of rules, from finding repetition, to text-matching, and much more. As you advance in Python you'll see that a lot of your parsing problems can be solved with regular expressions (they're also a common interview question!).\n\n\nIf you're familiar with Perl, you'll notice that the syntax for regular expressions are very similar in Python. We will be using the <code>re</code> module with Python for this lecture.\n\n\nLet's get started!",
"_____no_output_____"
],
[
"## Searching for Patterns in Text\n\nOne of the most common uses for the re module is for finding patterns in text. Let's do a quick example of using the search method in the re module to find some text:",
"_____no_output_____"
]
],
[
[
"import re\n\n# List of patterns to search for\npatterns = ['term1', 'term2']\n\n# Text to parse\ntext = 'This is a string with term1, but it does not have the other term.'\n\nfor pattern in patterns:\n print('Searching for \"%s\" in:\\n \"%s\"\\n' %(pattern,text))\n \n #Check for match\n if re.search(pattern,text):\n print('Match was found. \\n')\n else:\n print('No Match was found.\\n')",
"Searching for \"term1\" in:\n \"This is a string with term1, but it does not have the other term.\"\n\nMatch was found. \n\nSearching for \"term2\" in:\n \"This is a string with term1, but it does not have the other term.\"\n\nNo Match was found.\n\n"
]
],
[
[
"Now we've seen that <code>re.search()</code> will take the pattern, scan the text, and then return a **Match** object. If no pattern is found, **None** is returned. To give a clearer picture of this match object, check out the cell below:",
"_____no_output_____"
]
],
[
[
"# List of patterns to search for\npattern = 'term1'\n\n# Text to parse\ntext = 'This is a string with term1, but it does not have the other term.'\n\nmatch = re.search(pattern,text)\n\ntype(match)",
"_____no_output_____"
]
],
[
[
"This **Match** object returned by the search() method is more than just a Boolean or None, it contains information about the match, including the original input string, the regular expression that was used, and the location of the match. Let's see the methods we can use on the match object:",
"_____no_output_____"
]
],
[
[
"# Show start of match\nmatch.start()",
"_____no_output_____"
],
[
"# Show end\nmatch.end()",
"_____no_output_____"
]
],
[
[
"## Split with regular expressions\n\nLet's see how we can split with the re syntax. This should look similar to how you used the split() method with strings.",
"_____no_output_____"
]
],
[
[
"# Term to split on\nsplit_term = '@'\n\nphrase = 'What is the domain name of someone with the email: [email protected]'\n\n# Split the phrase\nre.split(split_term,phrase)",
"_____no_output_____"
]
],
[
[
"Note how <code>re.split()</code> returns a list with the term to split on removed and the terms in the list are a split up version of the string. Create a couple of more examples for yourself to make sure you understand!\n\n## Finding all instances of a pattern\n\nYou can use <code>re.findall()</code> to find all the instances of a pattern in a string. For example:",
"_____no_output_____"
]
],
[
[
"# Returns a list of all matches\nre.findall('match','test phrase match is in middle')",
"_____no_output_____"
]
],
[
[
"## re Pattern Syntax\n\nThis will be the bulk of this lecture on using re with Python. Regular expressions support a huge variety of patterns beyond just simply finding where a single string occurred. \n\nWe can use *metacharacters* along with re to find specific types of patterns. \n\nSince we will be testing multiple re syntax forms, let's create a function that will print out results given a list of various regular expressions and a phrase to parse:",
"_____no_output_____"
]
],
[
[
"def multi_re_find(patterns,phrase):\n '''\n Takes in a list of regex patterns\n Prints a list of all matches\n '''\n for pattern in patterns:\n print('Searching the phrase using the re check: %r' %(pattern))\n print(re.findall(pattern,phrase))\n print('\\n')",
"_____no_output_____"
]
],
[
[
"### Repetition Syntax\n\nThere are five ways to express repetition in a pattern:\n\n 1. A pattern followed by the meta-character <code>*</code> is repeated zero or more times. \n 2. Replace the <code>*</code> with <code>+</code> and the pattern must appear at least once. \n 3. Using <code>?</code> means the pattern appears zero or one time. \n 4. For a specific number of occurrences, use <code>{m}</code> after the pattern, where **m** is replaced with the number of times the pattern should repeat. \n 5. Use <code>{m,n}</code> where **m** is the minimum number of repetitions and **n** is the maximum. Leaving out **n** <code>{m,}</code> means the value appears at least **m** times, with no maximum.\n \nNow we will see an example of each of these using our multi_re_find function:",
"_____no_output_____"
]
],
[
[
"test_phrase = 'sdsd..sssddd...sdddsddd...dsds...dsssss...sdddd'\n\ntest_patterns = [ 'sd*', # s followed by zero or more d's\n 'sd+', # s followed by one or more d's\n 'sd?', # s followed by zero or one d's\n 'sd{3}', # s followed by three d's\n 'sd{2,3}', # s followed by two to three d's\n ]\n\nmulti_re_find(test_patterns,test_phrase)",
"Searching the phrase using the re check: 'sd*'\n['sd', 'sd', 's', 's', 'sddd', 'sddd', 'sddd', 'sd', 's', 's', 's', 's', 's', 's', 'sdddd']\n\n\nSearching the phrase using the re check: 'sd+'\n['sd', 'sd', 'sddd', 'sddd', 'sddd', 'sd', 'sdddd']\n\n\nSearching the phrase using the re check: 'sd?'\n['sd', 'sd', 's', 's', 'sd', 'sd', 'sd', 'sd', 's', 's', 's', 's', 's', 's', 'sd']\n\n\nSearching the phrase using the re check: 'sd{3}'\n['sddd', 'sddd', 'sddd', 'sddd']\n\n\nSearching the phrase using the re check: 'sd{2,3}'\n['sddd', 'sddd', 'sddd', 'sddd']\n\n\n"
]
],
[
[
"## Character Sets\n\nCharacter sets are used when you wish to match any one of a group of characters at a point in the input. Brackets are used to construct character set inputs. For example: the input <code>[ab]</code> searches for occurrences of either **a** or **b**.\nLet's see some examples:",
"_____no_output_____"
]
],
[
[
"test_phrase = 'sdsd..sssddd...sdddsddd...dsds...dsssss...sdddd'\n\ntest_patterns = ['[sd]', # either s or d\n 's[sd]+'] # s followed by one or more s or d\n\nmulti_re_find(test_patterns,test_phrase)",
"Searching the phrase using the re check: '[sd]'\n['s', 'd', 's', 'd', 's', 's', 's', 'd', 'd', 'd', 's', 'd', 'd', 'd', 's', 'd', 'd', 'd', 'd', 's', 'd', 's', 'd', 's', 's', 's', 's', 's', 's', 'd', 'd', 'd', 'd']\n\n\nSearching the phrase using the re check: 's[sd]+'\n['sdsd', 'sssddd', 'sdddsddd', 'sds', 'sssss', 'sdddd']\n\n\n"
]
],
[
[
"It makes sense that the first input <code>[sd]</code> returns every instance of s or d. Also, the second input <code>s[sd]+</code> returns any full strings that begin with an s and continue with s or d characters until another character is reached.",
"_____no_output_____"
],
[
"## Exclusion\n\nWe can use <code>^</code> to exclude terms by incorporating it into the bracket syntax notation. For example: <code>[^...]</code> will match any single character not in the brackets. Let's see some examples:",
"_____no_output_____"
]
],
[
[
"test_phrase = 'This is a string! But it has punctuation. How can we remove it?'",
"_____no_output_____"
]
],
[
[
"Use <code>[^!.? ]</code> to check for matches that are not a !,.,?, or space. Add a <code>+</code> to check that the match appears at least once. This basically translates into finding the words.",
"_____no_output_____"
]
],
[
[
"re.findall('[^!.? ]+',test_phrase)",
"_____no_output_____"
]
],
[
[
"## Character Ranges\n\nAs character sets grow larger, typing every character that should (or should not) match could become very tedious. A more compact format using character ranges lets you define a character set to include all of the contiguous characters between a start and stop point. The format used is <code>[start-end]</code>.\n\nCommon use cases are to search for a specific range of letters in the alphabet. For instance, <code>[a-f]</code> would return matches with any occurrence of letters between a and f. \n\nLet's walk through some examples:",
"_____no_output_____"
]
],
[
[
"\ntest_phrase = 'This is an example sentence. Lets see if we can find some letters.'\n\ntest_patterns=['[a-z]+', # sequences of lower case letters\n '[A-Z]+', # sequences of upper case letters\n '[a-zA-Z]+', # sequences of lower or upper case letters\n '[A-Z][a-z]+'] # one upper case letter followed by lower case letters\n \nmulti_re_find(test_patterns,test_phrase)",
"Searching the phrase using the re check: '[a-z]+'\n['his', 'is', 'an', 'example', 'sentence', 'ets', 'see', 'if', 'we', 'can', 'find', 'some', 'letters']\n\n\nSearching the phrase using the re check: '[A-Z]+'\n['T', 'L']\n\n\nSearching the phrase using the re check: '[a-zA-Z]+'\n['This', 'is', 'an', 'example', 'sentence', 'Lets', 'see', 'if', 'we', 'can', 'find', 'some', 'letters']\n\n\nSearching the phrase using the re check: '[A-Z][a-z]+'\n['This', 'Lets']\n\n\n"
]
],
[
[
"## Escape Codes\n\nYou can use special escape codes to find specific types of patterns in your data, such as digits, non-digits, whitespace, and more. For example:\n\n<table border=\"1\" class=\"docutils\">\n<colgroup>\n<col width=\"14%\" />\n<col width=\"86%\" />\n</colgroup>\n<thead valign=\"bottom\">\n<tr class=\"row-odd\"><th class=\"head\">Code</th>\n<th class=\"head\">Meaning</th>\n</tr>\n</thead>\n<tbody valign=\"top\">\n<tr class=\"row-even\"><td><tt class=\"docutils literal\"><span class=\"pre\">\\d</span></tt></td>\n<td>a digit</td>\n</tr>\n<tr class=\"row-odd\"><td><tt class=\"docutils literal\"><span class=\"pre\">\\D</span></tt></td>\n<td>a non-digit</td>\n</tr>\n<tr class=\"row-even\"><td><tt class=\"docutils literal\"><span class=\"pre\">\\s</span></tt></td>\n<td>whitespace (tab, space, newline, etc.)</td>\n</tr>\n<tr class=\"row-odd\"><td><tt class=\"docutils literal\"><span class=\"pre\">\\S</span></tt></td>\n<td>non-whitespace</td>\n</tr>\n<tr class=\"row-even\"><td><tt class=\"docutils literal\"><span class=\"pre\">\\w</span></tt></td>\n<td>alphanumeric</td>\n</tr>\n<tr class=\"row-odd\"><td><tt class=\"docutils literal\"><span class=\"pre\">\\W</span></tt></td>\n<td>non-alphanumeric</td>\n</tr>\n</tbody>\n</table>\n\nEscapes are indicated by prefixing the character with a backslash <code>\\</code>. Unfortunately, a backslash must itself be escaped in normal Python strings, and that results in expressions that are difficult to read. Using raw strings, created by prefixing the literal value with <code>r</code>, eliminates this problem and maintains readability.\n\nPersonally, I think this use of <code>r</code> to escape a backslash is probably one of the things that block someone who is not familiar with regex in Python from being able to read regex code at first. Hopefully after seeing these examples this syntax will become clear.",
"_____no_output_____"
]
],
[
[
"test_phrase = 'This is a string with some numbers 1233 and a symbol #hashtag'\n\ntest_patterns=[ r'\\d+', # sequence of digits\n r'\\D+', # sequence of non-digits\n r'\\s+', # sequence of whitespace\n r'\\S+', # sequence of non-whitespace\n r'\\w+', # alphanumeric characters\n r'\\W+', # non-alphanumeric\n ]\n\nmulti_re_find(test_patterns,test_phrase)",
"Searching the phrase using the re check: '\\\\d+'\n['1233']\n\n\nSearching the phrase using the re check: '\\\\D+'\n['This is a string with some numbers ', ' and a symbol #hashtag']\n\n\nSearching the phrase using the re check: '\\\\s+'\n[' ', ' ', ' ', ' ', ' ', ' ', ' ', ' ', ' ', ' ', ' ']\n\n\nSearching the phrase using the re check: '\\\\S+'\n['This', 'is', 'a', 'string', 'with', 'some', 'numbers', '1233', 'and', 'a', 'symbol', '#hashtag']\n\n\nSearching the phrase using the re check: '\\\\w+'\n['This', 'is', 'a', 'string', 'with', 'some', 'numbers', '1233', 'and', 'a', 'symbol', 'hashtag']\n\n\nSearching the phrase using the re check: '\\\\W+'\n[' ', ' ', ' ', ' ', ' ', ' ', ' ', ' ', ' ', ' ', ' #']\n\n\n"
]
],
[
[
"## Conclusion\n\nYou should now have a solid understanding of how to use the regular expression module in Python. There are a ton of more special character instances, but it would be unreasonable to go through every single use case. Instead take a look at the full [documentation](https://docs.python.org/3/library/re.html#regular-expression-syntax) if you ever need to look up a particular pattern.\n\nYou can also check out the nice summary tables at this [source](http://www.tutorialspoint.com/python/python_reg_expressions.htm).\n\nGood job!\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d084afa5ebad40fb507919298eb2abce844a695b | 1,645 | ipynb | Jupyter Notebook | Interview Preparation Kit/6. Greedy Algorithms/Minimum Absolute Difference in an Array.ipynb | Nam-SH/HackerRank | d1ced5cdad3eae7661f39af4d12aa33f460821cb | [
"MIT"
] | null | null | null | Interview Preparation Kit/6. Greedy Algorithms/Minimum Absolute Difference in an Array.ipynb | Nam-SH/HackerRank | d1ced5cdad3eae7661f39af4d12aa33f460821cb | [
"MIT"
] | null | null | null | Interview Preparation Kit/6. Greedy Algorithms/Minimum Absolute Difference in an Array.ipynb | Nam-SH/HackerRank | d1ced5cdad3eae7661f39af4d12aa33f460821cb | [
"MIT"
] | null | null | null | 22.534247 | 116 | 0.507599 | [
[
[
"# Minimum Absolute Difference in an Array\n\n<br>\n\n",
"_____no_output_____"
]
],
[
[
"#!/bin/python3\n\nimport math\nimport os\nimport random\nimport re\nimport sys\n\n# Complete the minimumAbsoluteDifference function below.\ndef minimumAbsoluteDifference(arr):\n arr.sort(reverse=True)\n Min = arr[0] - arr[1]\n for i in range(1, n - 1):\n Min = min(Min, arr[i] - arr[i + 1])\n return Min\n\nif __name__ == '__main__':\n fptr = open(os.environ['OUTPUT_PATH'], 'w')\n\n n = int(input())\n\n arr = list(map(int, input().rstrip().split()))\n\n result = minimumAbsoluteDifference(arr)\n\n fptr.write(str(result) + '\\n')\n\n fptr.close()",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
]
] |
d084c6feede42c182e56912a0cdde2e25ba4651f | 7,331 | ipynb | Jupyter Notebook | Assignments/hw3/Failed_to_perform_with_dataset/HW3_feature_selection_from_Boston/plot_select_from_model_boston.ipynb | Leon23N/Leon23N.github.io | bfa1cf19a14da7cb13842fa0567c6c555d4abab4 | [
"CC-BY-3.0"
] | null | null | null | Assignments/hw3/Failed_to_perform_with_dataset/HW3_feature_selection_from_Boston/plot_select_from_model_boston.ipynb | Leon23N/Leon23N.github.io | bfa1cf19a14da7cb13842fa0567c6c555d4abab4 | [
"CC-BY-3.0"
] | null | null | null | Assignments/hw3/Failed_to_perform_with_dataset/HW3_feature_selection_from_Boston/plot_select_from_model_boston.ipynb | Leon23N/Leon23N.github.io | bfa1cf19a14da7cb13842fa0567c6c555d4abab4 | [
"CC-BY-3.0"
] | null | null | null | 55.120301 | 1,290 | 0.634838 | [
[
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"\n# Feature selection using SelectFromModel and LassoCV\n\n\nUse SelectFromModel meta-transformer along with Lasso to select the best\ncouple of features from the Boston dataset.\n\n",
"_____no_output_____"
]
],
[
[
"# Author: Manoj Kumar <[email protected]>\n# License: BSD 3 clause\n\nprint(__doc__)\n\nimport matplotlib.pyplot as plt\nimport numpy as np\n\nfrom sklearn.datasets import load_boston\nfrom sklearn.feature_selection import SelectFromModel\nfrom sklearn.linear_model import LassoCV\nimport pandas as pd\n\n# Load the boston dataset.\n# boston = load_boston()\n# X, y = boston['data'], boston['target']\n\nds1 = pd.read_csv(\"DS.csv\") \nds2= pd.read_csv(\"DS1.csv\") \nX = ds1\ny = ds2\n\n# We use the base estimator LassoCV since the L1 norm promotes sparsity of features.\nclf = LassoCV()\n\n# Set a minimum threshold of 0.25\nsfm = SelectFromModel(clf, threshold=0.25)\nsfm.fit(X, y)\nn_features = sfm.transform(X).shape[1]\n\n# Reset the threshold till the number of features equals two.\n# Note that the attribute can be set directly instead of repeatedly\n# fitting the metatransformer.\nwhile n_features > 2:\n sfm.threshold += 0.1\n X_transform = sfm.transform(X)\n n_features = X_transform.shape[1]\n\n# Plot the selected two features from X.\nplt.title(\n \"Features selected from Boston using SelectFromModel with \"\n \"threshold %0.3f.\" % sfm.threshold)\nfeature1 = X_transform[:, 0]\nfeature2 = X_transform[:, 1] \nplt.plot(feature1, feature2, 'r.')\nplt.xlabel(\"Feature number 1\")\nplt.ylabel(\"Feature number 2\")\nplt.ylim([np.min(feature2), np.max(feature2)])\nplt.show()",
"Automatically created module for IPython interactive environment\n"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d084e254a78b31c6160c3fd13c6d3625d2749ce1 | 4,905 | ipynb | Jupyter Notebook | part-3/Autoencoders_notes.ipynb | masterflorin/dlnd-udacity | 53dc427a7337c4c232668225d35043e370a0d96a | [
"MIT"
] | 1 | 2020-01-19T17:26:36.000Z | 2020-01-19T17:26:36.000Z | part-3/Autoencoders_notes.ipynb | masterflorin/dlnd-udacity | 53dc427a7337c4c232668225d35043e370a0d96a | [
"MIT"
] | null | null | null | part-3/Autoencoders_notes.ipynb | masterflorin/dlnd-udacity | 53dc427a7337c4c232668225d35043e370a0d96a | [
"MIT"
] | 1 | 2020-01-19T17:27:09.000Z | 2020-01-19T17:27:09.000Z | 60.555556 | 478 | 0.716208 | [
[
[
"### What is an Autoencoder?\n\nAn Autoencoder is a model that can make use of a CNN's ability to compress the data into a flat vector / feature vector. We can think of it as a smart encoder that learns compression and decompression algorithms from the data. As an example, if you had a file format that was highly dimensional or noisy then you could use an autoencoder to get a file that you are able to work with.\n\nThe main ability of an autoencoder is that it's able to compress the data while maintaining its content which makes it possible to use the compress representation of the input.\n\n\n\n### Linear autoencoder\n\nThis is a simple autoencoder that uses an MLP with with a few linear layers for encoder and decoder. The number of layers depends on the problem you trying to solve.\n\nAnother thing to note here is you might want to consider a different loss function such as **MSE** because it's suitable for comparing pixel quantities rather than probabilities, it's a function that uses regression instead of probabilities. You are also interested in only the images and not the labels, same goes for validation set, you're mostly focusing on training and then you're using test to visualize the reconstruction.\n\n**Key point**: we are comparing the images that resulted from the reconstruction with the original ones so we're not interested in accuracy like in usual applications.\n\n**Code**: [Notebook](autoencoder/linear-autoencoder/Simple_Autoencoder_Exercise.ipynb) with fully connected layers.\n\n### Upsampling\n\nEncoder performs what is called **downsampling** as it is compressing the data into a flat vector. Conversely, the decoder is doing an **upsampling** through a transpose convolutional layer or sometimes called deconvolutional layer. Note: deconv layer doesn't strictly mean that we are undoing a convolution. It is essentially reversing the downsampling by increasing the spatial dimensions of a compressed input so that you get to the original dimensions of the input \n\n### Transpose convolutional layer\n\nBeginning from the compressed representation, a filter with a given size and stride passes over the input and multiplies the pixel with its weight resulting a representation. \n\nBelow is the result after the convolution went over all points. The interesting part is that this pixel representation will overlap with another convolution and what happens is that the overlapping section/edges get summed together. In this case a stride of 2 was used for the output. \n\n\n\nThere are options to add/substract the padding from the output as above but the most common solution is a 2x2 filter and stride of 2 to double the dimensions of the input.\n\n\n\nWhen not sure about how the calculations are done there is a very good explanation of how convolutional arithmethic works in this [repo](https://github.com/vdumoulin/conv_arithmetic/blob/master/README.md).\n\n**Code**: [Notebook](autoencoder/convolutional-autoencoder/Convolutional_Autoencoder_Exercise.ipynb) with convolutional and transpose convolutional layers.\n\nConvolutional neural networks gives as as an output the image that is much closely resembling the original imaage compared to the autoencoder that used linear layers. However, there are still some artifacts present in some of the images. This can be attributed to how Transpose conv layer works.\n\nThe solution for this is to use a technique called upsampling with nearest-neightbor interpolation coupled with convolutional layers.\n\n**Code**: [Notebook](autoencoder/convolutional-autoencoder/Upsampling_Solution.ipynb) with upsampling. \n\n### De-noising\n\nOne of the most interesting things you can use an autoencoder for is de-noising.\n\n**Code**: [Exercise](autoencoder/denoising-autoencoder/Denoising_Autoencoder_Exercise.ipynb) of using it for de-noising.",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown"
]
] |
d084e2f49b78002b5a58c6d00cf1f11a12ac9042 | 814,670 | ipynb | Jupyter Notebook | module2-convolutional-neural-networks/LS_DS_432_Convolutional_Neural_Networks_Lecture.ipynb | SamH3pn3r/DS-Unit-4-Sprint-3-Deep-Learning | e7aed7ce4b19af9b0e46699a67a23cfba53d712f | [
"MIT"
] | null | null | null | module2-convolutional-neural-networks/LS_DS_432_Convolutional_Neural_Networks_Lecture.ipynb | SamH3pn3r/DS-Unit-4-Sprint-3-Deep-Learning | e7aed7ce4b19af9b0e46699a67a23cfba53d712f | [
"MIT"
] | null | null | null | module2-convolutional-neural-networks/LS_DS_432_Convolutional_Neural_Networks_Lecture.ipynb | SamH3pn3r/DS-Unit-4-Sprint-3-Deep-Learning | e7aed7ce4b19af9b0e46699a67a23cfba53d712f | [
"MIT"
] | null | null | null | 974.485646 | 315,293 | 0.955112 | [
[
[
"Lambda School Data Science\n\n*Unit 4, Sprint 3, Module 2*\n\n---",
"_____no_output_____"
],
[
"# Convolutional Neural Networks (Prepare)\n\n> Convolutional networks are simply neural networks that use convolution in place of general matrix multiplication in at least one of their layers. *Goodfellow, et al.*",
"_____no_output_____"
],
[
"## Learning Objectives\n- <a href=\"#p1\">Part 1: </a>Describe convolution and pooling\n- <a href=\"#p2\">Part 2: </a>Apply a convolutional neural network to a classification task\n- <a href=\"#p3\">Part 3: </a>Use a pre-trained convolution neural network for object detection\n\nModern __computer vision__ approaches rely heavily on convolutions as both a dimensinoality reduction and feature extraction method. Before we dive into convolutions, let's talk about some of the common computer vision applications: \n* Classification [(Hot Dog or Not Dog)](https://www.youtube.com/watch?v=ACmydtFDTGs)\n* Object Detection [(YOLO)](https://www.youtube.com/watch?v=MPU2HistivI)\n* Pose Estimation [(PoseNet)](https://ai.googleblog.com/2019/08/on-device-real-time-hand-tracking-with.html)\n* Facial Recognition [Emotion Detection](https://www.cbronline.com/wp-content/uploads/2018/05/Mona-lIsa-test-570x300.jpg)\n* and *countless* more \n\nWe are going to focus on classification and pre-trained object detection today. What are some of the applications of object detection?",
"_____no_output_____"
]
],
[
[
"from IPython.display import YouTubeVideo\nYouTubeVideo('MPU2HistivI', width=600, height=400)",
"_____no_output_____"
]
],
[
[
"# Convolution & Pooling (Learn)\n<a id=\"p1\"></a>",
"_____no_output_____"
],
[
"## Overview\n\nLike neural networks themselves, CNNs are inspired by biology - specifically, the receptive fields of the visual cortex.\n\nPut roughly, in a real brain the neurons in the visual cortex *specialize* to be receptive to certain regions, shapes, colors, orientations, and other common visual features. In a sense, the very structure of our cognitive system transforms raw visual input, and sends it to neurons that specialize in handling particular subsets of it.\n\nCNNs imitate this approach by applying a convolution. A convolution is an operation on two functions that produces a third function, showing how one function modifies another. Convolutions have a [variety of nice mathematical properties](https://en.wikipedia.org/wiki/Convolution#Properties) - commutativity, associativity, distributivity, and more. Applying a convolution effectively transforms the \"shape\" of the input.\n\nOne common confusion - the term \"convolution\" is used to refer to both the process of computing the third (joint) function and the process of applying it. In our context, it's more useful to think of it as an application, again loosely analogous to the mapping from visual field to receptive areas of the cortex in a real animal.",
"_____no_output_____"
]
],
[
[
"from IPython.display import YouTubeVideo\nYouTubeVideo('IOHayh06LJ4', width=600, height=400)",
"_____no_output_____"
]
],
[
[
"## Follow Along\n\nLet's try to do some convolutions in `Keras`.",
"_____no_output_____"
],
[
"### Convolution - an example\n\nConsider blurring an image - assume the image is represented as a matrix of numbers, where each number corresponds to the color value of a pixel.",
"_____no_output_____"
]
],
[
[
"import imageio\nimport matplotlib.pyplot as plt\nfrom skimage import color, io\nfrom skimage.exposure import rescale_intensity\n\nausten = io.imread('https://dl.airtable.com/S1InFmIhQBypHBL0BICi_austen.jpg')\nausten_grayscale = rescale_intensity(color.rgb2gray(austen))\nausten_grayscale",
"_____no_output_____"
],
[
"plt.imshow(austen_grayscale, cmap=\"gray\");",
"_____no_output_____"
],
[
"import scipy.ndimage as nd\nimport numpy as np\n\nhorizontal_edge_convolution = np.array([[1,1,1,1,1],\n [0,0,0,0,0],\n [0,0,0,0,0],\n [0,0,0,0,0],\n [-1,-1,-1,-1,-1]])\n\nvertical_edge_convolution = np.array([[1, 0, 0, 0, -1],\n [1, 0, 0, 0, -1],\n [1, 0, 0, 0, -1],\n [1, 0, 0, 0, -1],\n [1, 0, 0, 0, -1]])\n\nausten_edges = nd.convolve(austen_grayscale, vertical_edge_convolution)\n#austen_edges",
"_____no_output_____"
],
[
"plt.imshow(austen_edges, cmap=\"gray\");",
"_____no_output_____"
]
],
[
[
"## Challenge\n\nYou will be expected to be able to describe convolution. ",
"_____no_output_____"
],
[
"# CNNs for Classification (Learn)",
"_____no_output_____"
],
[
"## Overview",
"_____no_output_____"
],
[
"### Typical CNN Architecture\n\n\n\nThe first stage of a CNN is, unsurprisingly, a convolution - specifically, a transformation that maps regions of the input image to neurons responsible for receiving them. The convolutional layer can be visualized as follows:\n\n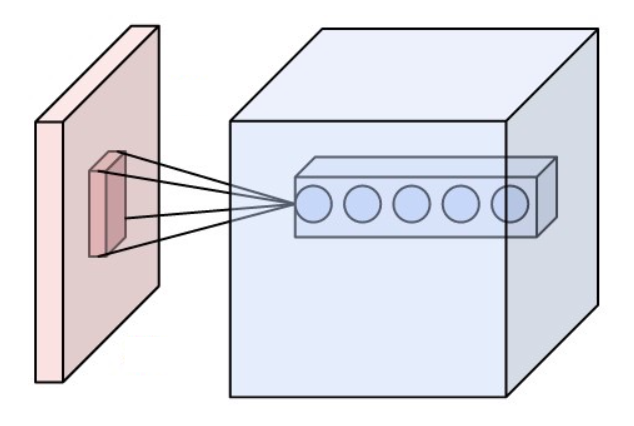\n\nThe red represents the original input image, and the blue the neurons that correspond.\n\nAs shown in the first image, a CNN can have multiple rounds of convolutions, [downsampling](https://en.wikipedia.org/wiki/Downsampling_(signal_processing)) (a digital signal processing technique that effectively reduces the information by passing through a filter), and then eventually a fully connected neural network and output layer. Typical output layers for a CNN would be oriented towards classification or detection problems - e.g. \"does this picture contain a cat, a dog, or some other animal?\"\n\nWhy are CNNs so popular?\n1. Compared to prior image learning techniques, they require relatively little image preprocessing (cropping/centering, normalizing, etc.)\n2. Relatedly, they are *robust* to all sorts of common problems in images (shifts, lighting, etc.)\n\nActually training a cutting edge image classification CNN is nontrivial computationally - the good news is, with transfer learning, we can get one \"off-the-shelf\"!",
"_____no_output_____"
],
[
"## Follow Along",
"_____no_output_____"
]
],
[
[
"from tensorflow.keras import datasets\nfrom tensorflow.keras.models import Sequential, Model # <- May Use\nfrom tensorflow.keras.layers import Dense, Conv2D, MaxPooling2D, Flatten\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"(train_images, train_labels), (test_images, test_labels) = datasets.cifar10.load_data()\n\n# Normalize pixel values to be between 0 and 1\ntrain_images, test_images = train_images / 255.0, test_images / 255.0",
"Downloading data from https://www.cs.toronto.edu/~kriz/cifar-10-python.tar.gz\n170500096/170498071 [==============================] - 41s 0us/step\n"
],
[
"class_names = ['airplane', 'automobile', 'bird', 'cat', 'deer',\n 'dog', 'frog', 'horse', 'ship', 'truck']\n\nplt.figure(figsize=(10,10))\nfor i in range(25):\n plt.subplot(5,5,i+1)\n plt.xticks([])\n plt.yticks([])\n plt.grid(False)\n plt.imshow(train_images[i], cmap=plt.cm.binary)\n # The CIFAR labels happen to be arrays, \n # which is why you need the extra index\n plt.xlabel(class_names[train_labels[i][0]])\nplt.show()",
"_____no_output_____"
],
[
"train_images[0].shape",
"_____no_output_____"
],
[
"# Setup Architecture\n\nmodel = Sequential()\nmodel.add(Conv2D(32, (3, 3), activation='relu', input_shape=(32, 32, 3)))\nmodel.add(MaxPooling2D((2,2)))\nmodel.add(Conv2D(64, (3, 3), activation='relu'))\nmodel.add(MaxPooling2D((2,2)))\nmodel.add(Conv2D(64, (3, 3), activation='relu'))\nmodel.add(Flatten())\nmodel.add(Dense(64, activation='relu'))\nmodel.add(Dense(10, activation='softmax'))\n\nmodel.summary()",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nconv2d_9 (Conv2D) (None, 30, 30, 32) 896 \n_________________________________________________________________\nmax_pooling2d_7 (MaxPooling2 (None, 15, 15, 32) 0 \n_________________________________________________________________\nconv2d_10 (Conv2D) (None, 13, 13, 64) 18496 \n_________________________________________________________________\nmax_pooling2d_8 (MaxPooling2 (None, 6, 6, 64) 0 \n_________________________________________________________________\nconv2d_11 (Conv2D) (None, 4, 4, 64) 36928 \n_________________________________________________________________\nflatten_1 (Flatten) (None, 1024) 0 \n_________________________________________________________________\ndense (Dense) (None, 64) 65600 \n_________________________________________________________________\ndense_1 (Dense) (None, 10) 650 \n=================================================================\nTotal params: 122,570\nTrainable params: 122,570\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"# Compile Model\n\nmodel.compile(optimizer='adam',\n loss='sparse_categorical_crossentropy',\n metrics=['accuracy'])",
"_____no_output_____"
],
[
"# Fit Model\nmodel.fit(train_images, train_labels, epochs=10, validation_data=(test_images, test_labels));",
"Train on 50000 samples, validate on 10000 samples\nEpoch 1/10\n50000/50000 [==============================] - 465s 9ms/sample - loss: 1.5302 - acc: 0.4419 - val_loss: 1.2764 - val_acc: 0.5435\nEpoch 2/10\n50000/50000 [==============================] - 484s 10ms/sample - loss: 1.1523 - acc: 0.5943 - val_loss: 1.0657 - val_acc: 0.6253\nEpoch 3/10\n50000/50000 [==============================] - 445s 9ms/sample - loss: 0.9975 - acc: 0.6495 - val_loss: 0.9984 - val_acc: 0.6472\nEpoch 4/10\n50000/50000 [==============================] - 437s 9ms/sample - loss: 0.8981 - acc: 0.6852 - val_loss: 0.9317 - val_acc: 0.6698\nEpoch 5/10\n50000/50000 [==============================] - 461s 9ms/sample - loss: 0.8207 - acc: 0.7126 - val_loss: 0.8993 - val_acc: 0.6894\nEpoch 6/10\n50000/50000 [==============================] - 479s 10ms/sample - loss: 0.7669 - acc: 0.7301 - val_loss: 0.9009 - val_acc: 0.6917\nEpoch 7/10\n50000/50000 [==============================] - 428s 9ms/sample - loss: 0.7121 - acc: 0.7501 - val_loss: 0.8806 - val_acc: 0.6979\nEpoch 8/10\n50000/50000 [==============================] - 404s 8ms/sample - loss: 0.6663 - acc: 0.7652 - val_loss: 0.8891 - val_acc: 0.6980\nEpoch 9/10\n50000/50000 [==============================] - 400s 8ms/sample - loss: 0.6314 - acc: 0.7769 - val_loss: 0.9083 - val_acc: 0.6970\nEpoch 10/10\n50000/50000 [==============================] - 401s 8ms/sample - loss: 0.5920 - acc: 0.7937 - val_loss: 0.9412 - val_acc: 0.7020\n"
],
[
"# Evaluate Model\ntest_loss, test_acc = model.evaluate(test_images, test_labels, verbose=2)",
" - 25s - loss: 0.9412 - acc: 0.7020\n"
]
],
[
[
"## Challenge\n\nYou will apply CNNs to a classification task in the module project.",
"_____no_output_____"
],
[
"# CNNs for Object Detection (Learn)",
"_____no_output_____"
],
[
"## Overview",
"_____no_output_____"
],
[
"### Transfer Learning - TensorFlow Hub\n\n\"A library for reusable machine learning modules\"\n\nThis lets you quickly take advantage of a model that was trained with thousands of GPU hours. It also enables transfer learning - reusing a part of a trained model (called a module) that includes weights and assets, but also training the overall model some yourself with your own data. The advantages are fairly clear - you can use less training data, have faster training, and have a model that generalizes better.\n\nhttps://www.tensorflow.org/hub/\n\n**WARNING** - Dragons ahead!\n\n\n\nTensorFlow Hub is very bleeding edge, and while there's a good amount of documentation out there, it's not always updated or consistent. You'll have to use your problem-solving skills if you want to use it!",
"_____no_output_____"
],
[
"## Follow Along",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nfrom tensorflow.keras.applications.resnet50 import ResNet50\nfrom tensorflow.keras.preprocessing import image\nfrom tensorflow.keras.applications.resnet50 import preprocess_input, decode_predictions\n\ndef process_img_path(img_path):\n return image.load_img(img_path, target_size=(224, 224))\n\ndef img_contains_banana(img):\n x = image.img_to_array(img)\n x = np.expand_dims(x, axis=0)\n x = preprocess_input(x)\n model = ResNet50(weights='imagenet')\n features = model.predict(x)\n results = decode_predictions(features, top=3)[0]\n print(results)\n for entry in results:\n if entry[1] == 'banana':\n return entry[2]\n return 0.0",
"_____no_output_____"
],
[
"import requests\nimage_urls = [\"https://github.com/LambdaSchool/ML-YouOnlyLookOnce/raw/master/sample_data/negative_examples/example11.jpeg\",\n \"https://github.com/LambdaSchool/ML-YouOnlyLookOnce/raw/master/sample_data/positive_examples/example0.jpeg\"]\n\nfor _id,img in enumerate(image_urls): \n r = requests.get(img)\n with open(f'example{_id}.jpg', 'wb') as f:\n f.write(r.content)",
"_____no_output_____"
],
[
"from IPython.display import Image\nImage(filename='./example0.jpg', width=600)",
"_____no_output_____"
],
[
"img_contains_banana(process_img_path('example0.jpg'))",
"WARNING:tensorflow:From C:\\Users\\Samue\\Anaconda3\\envs\\U4-S3-DNN\\lib\\site-packages\\tensorflow\\python\\ops\\resource_variable_ops.py:435: colocate_with (from tensorflow.python.framework.ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nColocations handled automatically by placer.\n[('n04037443', 'racer', 0.91754997), ('n04285008', 'sports_car', 0.07783192), ('n04461696', 'tow_truck', 0.0023059668)]\n"
],
[
"Image(filename='example1.jpg', width=600)",
"_____no_output_____"
],
[
"img_contains_banana(process_img_path('example1.jpg'))",
"[('n07753592', 'banana', 0.06643853), ('n03532672', 'hook', 0.06110267), ('n03498962', 'hatchet', 0.05880436)]\n"
]
],
[
[
"Notice that, while it gets it right, the confidence for the banana image is fairly low. That's because so much of the image is \"not-banana\"! How can this be improved? Bounding boxes to center on items of interest.",
"_____no_output_____"
],
[
"## Challenge\n\nYou will be expected to apply a pretrained model to a classificaiton problem today. ",
"_____no_output_____"
],
[
"# Review\n\n- <a href=\"#p1\">Part 1: </a>Describe convolution and pooling\n * A Convolution is a function applied to another function to produce a third function\n * Convolutional Kernels are typically 'learned' during the process of training a Convolution Neural Network\n * Pooling is a dimensionality reduction technique that uses either Max or Average of a feature max region to downsample data\n- <a href=\"#p2\">Part 2: </a>Apply a convolutional neural network to a classification task\n * Keras has layers for convolutions :) \n- <a href=\"#p3\">Part 3: </a>Use a pre-trained convolution neural network for object detection\n * Check out both pretinaed models available in Keras & TensorFlow Hub",
"_____no_output_____"
],
[
"# Sources\n\n- *_Deep Learning_*. Goodfellow *et al.*\n- [Keras CNN Tutorial](https://www.tensorflow.org/tutorials/images/cnn)\n- [Tensorflow + Keras](https://www.tensorflow.org/api_docs/python/tf/keras/layers/Conv2D)\n- [Convolution Wiki](https://en.wikipedia.org/wiki/Convolution)\n- [Keras Conv2D: Working with CNN 2D Convolutions in Keras](https://missinglink.ai/guides/keras/keras-conv2d-working-cnn-2d-convolutions-keras/)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d084e39a08ecef4d6d5cf29d79445b5b8e9fe771 | 274,371 | ipynb | Jupyter Notebook | Metaculus vs. Polymarket.ipynb | rethinkpriorities/compare_forecast_markets | cab384107cd41e0b1e5c8625e670c65832f231a6 | [
"MIT"
] | null | null | null | Metaculus vs. Polymarket.ipynb | rethinkpriorities/compare_forecast_markets | cab384107cd41e0b1e5c8625e670c65832f231a6 | [
"MIT"
] | null | null | null | Metaculus vs. Polymarket.ipynb | rethinkpriorities/compare_forecast_markets | cab384107cd41e0b1e5c8625e670c65832f231a6 | [
"MIT"
] | null | null | null | 74.76049 | 27,464 | 0.673296 | [
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport urllib.request as urllib\n\nfrom datetime import datetime\n\nimport time\nimport ergo # Download from https://github.com/rethinkpriorities/ergo\n\ndef fetch(url):\n max_attempts = 80\n attempts = 0\n sleeptime = 10 #in seconds, no reason to continuously try if network is down\n\n while attempts < max_attempts:\n time.sleep(sleeptime)\n try:\n response = urllib.urlopen(url, timeout=5)\n content = response.read()\n return content\n except urllib.URLError as e:\n print(e)\n attempts += 1\n",
"/Users/peterhurford/.virtualenvs/dev/lib/python3.8/site-packages/ergo/ppl.py:15: TqdmExperimentalWarning: Using `tqdm.autonotebook.tqdm` in notebook mode. Use `tqdm.tqdm` instead to force console mode (e.g. in jupyter console)\n from tqdm.autonotebook import tqdm\n"
],
[
"def kelly(user_odds, market_odds):\n return (user_odds - ((1 - user_odds) * (market_odds / (1 - market_odds))))",
"_____no_output_____"
],
[
"def compare_metaculus_vs_polymarket(polymarket_url, metaculus_qid, actual, inverse_pm=False):\n print('Fetching...')\n content = fetch(polymarket_url)\n print('Fetched')\n \n polymarket_df = pd.DataFrame(eval(str(content).split('\"All\":')[1].split('},\"graphKeys\":[')[0]))\n if inverse_pm:\n polymarket_df.columns = ['no_price', 'yes_price', 'time']\n else:\n polymarket_df.columns = ['yes_price', 'no_price', 'time']\n \n polymarket_df['yes_price'] = polymarket_df['yes_price'].astype(float)\n polymarket_df['no_price'] = polymarket_df['no_price'].astype(float)\n polymarket_df['time'] = polymarket_df['time'].apply(lambda t: t.split(',')[0])\n polymarket_df = polymarket_df[~((polymarket_df['yes_price'] == 0) & (polymarket_df['no_price'] == 0))]\n polymarket_df['polymarket_yes'] = polymarket_df['yes_price']\n polymarket_df = polymarket_df[['time', 'polymarket_yes']].drop_duplicates('time', keep='last')\n polymarket_df['time'] = polymarket_df['time'].apply(lambda t: t + ' 21' if ('Jan' in t or 'Feb' in t) else t + ' 20')\n polymarket_df['time'] = pd.to_datetime(polymarket_df['time'], format='%b %d %y')\n polymarket_df = polymarket_df.reset_index(drop=True)\n \n q = metaculus.get_question(metaculus_qid)\n metaculus_df = pd.DataFrame(q.data['metaculus_prediction']['history'])\n metaculus_df.columns = ['time', 'metaculus_yes']\n metaculus_df['time'] = pd.to_datetime(metaculus_df['time'].apply(lambda t: datetime.fromtimestamp(t)).dt.date)\n metaculus_df = metaculus_df.drop_duplicates('time', keep='last')\n metaculus_df = metaculus_df.reset_index(drop=True)\n metaculus_df\n \n merged_df = metaculus_df.merge(polymarket_df, on='time', how='left').dropna()\n merged_df['metaculus_brier'] = (merged_df['metaculus_yes'] - actual) ** 2\n merged_df['polymarket_brier'] = (merged_df['polymarket_yes'] - actual) ** 2\n merged_df['50_50_yes'] = merged_df['polymarket_yes'] * 0.5 + merged_df['metaculus_yes'] * 0.5\n merged_df['50_50_brier'] = (merged_df['50_50_yes'] - actual) ** 2\n \n bankroll = 1000\n metaculus_bets = []\n metaculus_winnings = []\n for index, row in merged_df.iterrows():\n if row['polymarket_yes'] == 0:\n row['polymarket_yes'] = 0.001\n if row['metaculus_yes'] == 0:\n row['metaculus_yes'] = 0.001\n if row['metaculus_yes'] > row['polymarket_yes']:\n bet = bankroll * kelly(row['metaculus_yes'], row['polymarket_yes'])\n shares = bet / row['polymarket_yes']\n winnings = shares if actual == 1 else -bet\n elif row['metaculus_yes'] < row['polymarket_yes']:\n bet = bankroll * kelly(1 - row['metaculus_yes'], 1 - row['polymarket_yes'])\n shares = bet / (1 - row['polymarket_yes'])\n winnings = shares if actual == 0 else -bet\n else:\n bet = 0\n shares = 0\n winnings += (bankroll - bet)\n metaculus_bets.append(bet)\n metaculus_winnings.append(winnings)\n \n polymarket_bets = []\n polymarket_winnings = []\n for index, row in merged_df.iterrows():\n if row['polymarket_yes'] > row['metaculus_yes']:\n bet = bankroll * kelly(row['polymarket_yes'], row['metaculus_yes'])\n shares = bet / row['metaculus_yes']\n winnings = shares if actual == 1 else -bet\n elif row['polymarket_yes'] < row['metaculus_yes']:\n bet = bankroll * kelly(1 - row['polymarket_yes'], 1 - row['metaculus_yes'])\n shares = bet / (1 - row['metaculus_yes'])\n winnings = shares if actual == 0 else -bet\n else:\n bet = 0\n shares = 0\n winnings = 0\n winnings += (bankroll - bet)\n polymarket_bets.append(bet)\n polymarket_winnings.append(winnings)\n \n merged_df['metaculus_bets'] = metaculus_bets\n merged_df['metaculus_winnings'] = metaculus_winnings\n merged_df['polymarket_bets'] = polymarket_bets\n merged_df['polymarket_winnings'] = polymarket_winnings\n \n return {'metaculus': metaculus_df,\n 'polymarket': polymarket_df,\n 'data': merged_df,\n 'brier': merged_df[['metaculus_brier', 'polymarket_brier', '50_50_brier']].sum() / len(merged_df),\n 'winnings': merged_df[['metaculus_winnings', 'polymarket_winnings']].sum() / len(merged_df)}\n\n\ndef plot_predictions(preds, q_title):\n plt.plot(preds['data']['time'], preds['data']['metaculus_yes'], label='Metaculus')\n plt.plot(preds['data']['time'], preds['data']['polymarket_yes'], label='Polymarket')\n plt.title(q_title)\n plt.legend()\n return plt",
"_____no_output_____"
],
[
"print('Logging in to Metaculus...')\nmetaculus = ergo.Metaculus()\nmetaculus.login_via_username_and_password(username='PeterHurford', password='GaZKQ6hEtZH0')\nprint('...Logged on')",
"Logging in to Metaculus...\n...Logged on\n"
]
],
[
[
"## Trump Charges",
"_____no_output_____"
]
],
[
[
"# https://www.metaculus.com/questions/6222/criminal-charges-against-trump/\ntrump_charges = compare_metaculus_vs_polymarket('https://polymarket.com/market/donald-trump-federally-charged-by-february-20th',\n 6222,\n actual=0)",
"Fetching...\nFetched\n"
],
[
"trump_charges['data']",
"_____no_output_____"
],
[
"trump_charges['brier']",
"_____no_output_____"
],
[
"trump_charges['winnings']",
"_____no_output_____"
],
[
"plot_predictions(trump_charges, 'Criminal charges against Trump by 20 Feb?').show()",
"_____no_output_____"
]
],
[
[
"## GOP Win",
"_____no_output_____"
]
],
[
[
"# https://www.metaculus.com/questions/5734/gop-to-hold-senate-on-feb-1st-2021/\ngop_senate = compare_metaculus_vs_polymarket('https://polymarket.com/market/which-party-will-control-the-senate',\n 5734,\n actual=0)",
"Fetching...\nFetched\n"
],
[
"gop_senate['data']",
"_____no_output_____"
],
[
"gop_senate['brier']",
"_____no_output_____"
],
[
"gop_senate['winnings']",
"_____no_output_____"
],
[
"plot_predictions(gop_senate, 'GOP Hold Senate for 2021?').show()",
"_____no_output_____"
]
],
[
[
"## Trump Pardon",
"_____no_output_____"
]
],
[
[
"# https://www.metaculus.com/questions/5685/will-donald-trump-attempt-to-pardon-himself/\ntrump_pardon = compare_metaculus_vs_polymarket('https://polymarket.com/market/will-trump-pardon-himself-in-his-first-term',\n 5685,\n actual=0)",
"Fetching...\nFetched\n"
],
[
"trump_pardon['data']",
"_____no_output_____"
],
[
"trump_pardon['brier']",
"_____no_output_____"
],
[
"trump_pardon['winnings']",
"_____no_output_____"
],
[
"plot_predictions(trump_pardon, 'Trump self-pardon?').show()",
"_____no_output_____"
]
],
[
[
"## 538 - Economist",
"_____no_output_____"
]
],
[
[
"# https://www.metaculus.com/questions/5503/comparing-538-and-economist-forecasts-in-2020/\neconomist_538 = compare_metaculus_vs_polymarket('https://polymarket.com/market/will-538-outperform-the-economist-in-forecasting-the-2020-presidential-election',\n 5503,\n actual=0)",
"Fetching...\nFetched\n"
],
[
"economist_538['data']",
"_____no_output_____"
],
[
"economist_538['brier']",
"_____no_output_____"
],
[
"economist_538['winnings']",
"_____no_output_____"
],
[
"plot_predictions(economist_538, '538 prez forecast beat Economist?').show()",
"_____no_output_____"
]
],
[
[
"## Biden in-person inauguration",
"_____no_output_____"
]
],
[
[
"## https://www.metaculus.com/questions/6293/biden-in-person-inauguration/\nbiden_in_person = compare_metaculus_vs_polymarket('https://polymarket.com/market/will-joe-biden-be-officially-inaugurated-as-president-in-person-outside-the-us-capitol-on-january-20th-2021',\n 6293,\n actual=1)",
"Fetching...\nFetched\n"
],
[
"biden_in_person['data']",
"_____no_output_____"
],
[
"biden_in_person['brier']",
"_____no_output_____"
],
[
"biden_in_person['winnings']",
"_____no_output_____"
],
[
"plot_predictions(biden_in_person, 'Biden inaugurated in-person on 20 Jan 2021?').show()",
"_____no_output_____"
]
],
[
[
"## Trump at Biden's Inauguration",
"_____no_output_____"
]
],
[
[
"## https://www.metaculus.com/questions/5825/trump-at-bidens-inauguration/\ntrump_attend = compare_metaculus_vs_polymarket('https://polymarket.com/market/will-donald-trump-attend-joe-biden-s-inauguration-ceremony-in-person',\n 5825,\n actual=0)",
"Fetching...\nFetched\n"
],
[
"trump_attend['data']",
"_____no_output_____"
],
[
"trump_attend['brier']",
"_____no_output_____"
],
[
"trump_attend['winnings']",
"_____no_output_____"
],
[
"plot_predictions(trump_attend, 'Trump attend Biden\\'s inauguration?').show()",
"_____no_output_____"
]
],
[
[
"## Electoral Challenge",
"_____no_output_____"
]
],
[
[
"## https://www.metaculus.com/questions/5844/electoral-college-results-challenged/\nchallenge = compare_metaculus_vs_polymarket('https://polymarket.com/market/will-any-electoral-certificates-be-formally-challenged-in-congress',\n 5844,\n actual=1)",
"Fetching...\nFetched\n"
],
[
"challenge['data']",
"_____no_output_____"
],
[
"challenge['brier']",
"_____no_output_____"
],
[
"challenge['winnings']",
"_____no_output_____"
],
[
"plot_predictions(challenge, 'Electoral college challenge?').show()",
"_____no_output_____"
]
],
[
[
"## Trump Convict",
"_____no_output_____"
]
],
[
[
"## https://www.metaculus.com/questions/6303/trump-convicted-by-senate/\ntrump_convict = compare_metaculus_vs_polymarket('https://polymarket.com/market/will-the-senate-convict-donald-trump-on-impeachment-before-june-1-2021',\n 6303,\n actual=0)",
"Fetching...\nFetched\n"
],
[
"trump_convict['data']",
"_____no_output_____"
],
[
"trump_convict['brier']",
"_____no_output_____"
],
[
"trump_convict['winnings']",
"_____no_output_____"
],
[
"plot_predictions(trump_convict, 'Senate convict Trump in 2021?').show()",
"_____no_output_____"
]
],
[
[
"## Tokyo Olympics",
"_____no_output_____"
]
],
[
[
"# https://polymarket.com/market/will-the-tokyo-summer-olympics-be-cancelled-or-postponed\n# https://www.metaculus.com/questions/5555/rescheduled-2020-olympics/",
"_____no_output_____"
]
],
[
[
"## Brier",
"_____no_output_____"
]
],
[
[
"(challenge['brier'] + trump_attend['brier'] + biden_in_person['brier'] + economist_538['brier'] +\ntrump_pardon['brier'] + gop_senate['brier'] + trump_charges['brier'] + trump_convict['brier']) / 8",
"_____no_output_____"
],
[
"(challenge['winnings'] + trump_attend['winnings'] + biden_in_person['winnings'] + economist_538['winnings'] +\ntrump_pardon['winnings'] + gop_senate['winnings'] + trump_charges['winnings'] + trump_convict['winnings']) / 8",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d084e6ea59095ac6a644fcad1d2a6ea50cf365e9 | 54,077 | ipynb | Jupyter Notebook | assignment2/PyTorch.ipynb | aoboturov/cs237n-17 | cff22b775e4c08b2258a8b69954f28a9765148ec | [
"MIT"
] | null | null | null | assignment2/PyTorch.ipynb | aoboturov/cs237n-17 | cff22b775e4c08b2258a8b69954f28a9765148ec | [
"MIT"
] | null | null | null | assignment2/PyTorch.ipynb | aoboturov/cs237n-17 | cff22b775e4c08b2258a8b69954f28a9765148ec | [
"MIT"
] | null | null | null | 35.64733 | 880 | 0.538602 | [
[
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"# Training a ConvNet PyTorch\n\nIn this notebook, you'll learn how to use the powerful PyTorch framework to specify a conv net architecture and train it on the CIFAR-10 dataset.",
"_____no_output_____"
]
],
[
[
"import torch\nimport torch.nn as nn\nimport torch.optim as optim\nfrom torch.autograd import Variable\nfrom torch.utils.data import DataLoader\nfrom torch.utils.data import sampler\n\nimport torchvision.datasets as dset\nimport torchvision.transforms as T\n\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nimport timeit\nfrom tqdm import tqdm\n\n# for auto-reloading extenrnal modules\n# see http://stackoverflow.com/questions/1907993/autoreload-of-modules-in-ipython\n%load_ext autoreload\n%autoreload 2\n",
"_____no_output_____"
]
],
[
[
"## What's this PyTorch business?\n\nYou've written a lot of code in this assignment to provide a whole host of neural network functionality. Dropout, Batch Norm, and 2D convolutions are some of the workhorses of deep learning in computer vision. You've also worked hard to make your code efficient and vectorized.\n\nFor the last part of this assignment, though, we're going to leave behind your beautiful codebase and instead migrate to one of two popular deep learning frameworks: in this instance, PyTorch (or TensorFlow, if you switch over to that notebook). \n\nWhy?\n\n* Our code will now run on GPUs! Much faster training. When using a framework like PyTorch or TensorFlow you can harness the power of the GPU for your own custom neural network architectures without having to write CUDA code directly (which is beyond the scope of this class).\n* We want you to be ready to use one of these frameworks for your project so you can experiment more efficiently than if you were writing every feature you want to use by hand. \n* We want you to stand on the shoulders of giants! TensorFlow and PyTorch are both excellent frameworks that will make your lives a lot easier, and now that you understand their guts, you are free to use them :) \n* We want you to be exposed to the sort of deep learning code you might run into in academia or industry. ",
"_____no_output_____"
],
[
"## How will I learn PyTorch?\n\nIf you've used Torch before, but are new to PyTorch, this tutorial might be of use: http://pytorch.org/tutorials/beginner/former_torchies_tutorial.html\n\nOtherwise, this notebook will walk you through much of what you need to do to train models in Torch. See the end of the notebook for some links to helpful tutorials if you want to learn more or need further clarification on topics that aren't fully explained here.",
"_____no_output_____"
],
[
"## Load Datasets\n\nWe load the CIFAR-10 dataset. This might take a couple minutes the first time you do it, but the files should stay cached after that.",
"_____no_output_____"
]
],
[
[
"class ChunkSampler(sampler.Sampler):\n \"\"\"Samples elements sequentially from some offset. \n Arguments:\n num_samples: # of desired datapoints\n start: offset where we should start selecting from\n \"\"\"\n def __init__(self, num_samples, start = 0):\n self.num_samples = num_samples\n self.start = start\n\n def __iter__(self):\n return iter(range(self.start, self.start + self.num_samples))\n\n def __len__(self):\n return self.num_samples\n\nNUM_TRAIN = 49000\nNUM_VAL = 1000\n\ncifar10_train = dset.CIFAR10('./cs231n/datasets', train=True, download=True,\n transform=T.ToTensor())\nloader_train = DataLoader(cifar10_train, batch_size=64, sampler=ChunkSampler(NUM_TRAIN, 0))\n\ncifar10_val = dset.CIFAR10('./cs231n/datasets', train=True, download=True,\n transform=T.ToTensor())\nloader_val = DataLoader(cifar10_val, batch_size=64, sampler=ChunkSampler(NUM_VAL, NUM_TRAIN))\n\ncifar10_test = dset.CIFAR10('./cs231n/datasets', train=False, download=True,\n transform=T.ToTensor())\nloader_test = DataLoader(cifar10_test, batch_size=64)\n",
"Files already downloaded and verified\nFiles already downloaded and verified\nFiles already downloaded and verified\n"
]
],
[
[
"For now, we're going to use a CPU-friendly datatype. Later, we'll switch to a datatype that will move all our computations to the GPU and measure the speedup.",
"_____no_output_____"
]
],
[
[
"# Constant to control how frequently we print train loss\nprint_every = 100\n\n# This is a little utility that we'll use to reset the model\n# if we want to re-initialize all our parameters\ndef reset(m):\n if hasattr(m, 'reset_parameters'):\n m.reset_parameters()",
"_____no_output_____"
]
],
[
[
"## Example Model\n\n### Some assorted tidbits\n\nLet's start by looking at a simple model. First, note that PyTorch operates on Tensors, which are n-dimensional arrays functionally analogous to numpy's ndarrays, with the additional feature that they can be used for computations on GPUs.\n\nWe'll provide you with a Flatten function, which we explain here. Remember that our image data (and more relevantly, our intermediate feature maps) are initially N x C x H x W, where:\n* N is the number of datapoints\n* C is the number of channels\n* H is the height of the intermediate feature map in pixels\n* W is the height of the intermediate feature map in pixels\n\nThis is the right way to represent the data when we are doing something like a 2D convolution, that needs spatial understanding of where the intermediate features are relative to each other. When we input data into fully connected affine layers, however, we want each datapoint to be represented by a single vector -- it's no longer useful to segregate the different channels, rows, and columns of the data. So, we use a \"Flatten\" operation to collapse the C x H x W values per representation into a single long vector. The Flatten function below first reads in the N, C, H, and W values from a given batch of data, and then returns a \"view\" of that data. \"View\" is analogous to numpy's \"reshape\" method: it reshapes x's dimensions to be N x ??, where ?? is allowed to be anything (in this case, it will be C x H x W, but we don't need to specify that explicitly). ",
"_____no_output_____"
]
],
[
[
"class Flatten(nn.Module):\n def forward(self, x):\n N, C, H, W = x.size() # read in N, C, H, W\n return x.view(N, -1) # \"flatten\" the C * H * W values into a single vector per image",
"_____no_output_____"
],
[
"def out_dim(sz, filter_size, padding, stride):\n \"\"\"\n Computes the size of dimension after convolution.\n\n Input:\n - sz: Original size of dimension\n - filter_size: Filter size applied in convolution\n - padding: Applied to the original dimension\n - stride: Between the two applications of convolution\n\n Returns a tuple of:\n - out: The size of the dimension after the convolution is computed\n \"\"\"\n return 1 + int((sz + 2 * padding - filter_size) / stride)",
"_____no_output_____"
],
[
"# Verify that CUDA is properly configured and you have a GPU available\n\nif torch.cuda.is_available():\n dtype = torch.cuda.FloatTensor\n ltype = torch.cuda.LongTensor\nelse:\n dtype = torch.FloatTensor\n ltype = torch.LongTensor\n",
"_____no_output_____"
]
],
[
[
"### The example model itself\n\nThe first step to training your own model is defining its architecture.\n\nHere's an example of a convolutional neural network defined in PyTorch -- try to understand what each line is doing, remembering that each layer is composed upon the previous layer. We haven't trained anything yet - that'll come next - for now, we want you to understand how everything gets set up. nn.Sequential is a container which applies each layer\none after the other.\n\nIn that example, you see 2D convolutional layers (Conv2d), ReLU activations, and fully-connected layers (Linear). You also see the Cross-Entropy loss function, and the Adam optimizer being used. \n\nMake sure you understand why the parameters of the Linear layer are 5408 and 10.\n",
"_____no_output_____"
]
],
[
[
"# Here's where we define the architecture of the model... \nsimple_model = nn.Sequential(\n nn.Conv2d(3, 32, kernel_size=7, stride=2),\n nn.ReLU(inplace=True),\n Flatten(), # see above for explanation\n nn.Linear(5408, 10), # affine layer\n )\n# the number of output classes:\n# 10\n\n# 32*out_dim(32, 7, 0, 2)**2\n# 5408\n\n\n# Set the type of all data in this model to be FloatTensor \nsimple_model.type(dtype)\n\nloss_fn = nn.CrossEntropyLoss().type(dtype)\noptimizer = optim.Adam(simple_model.parameters(), lr=1e-2) # lr sets the learning rate of the optimizer",
"_____no_output_____"
]
],
[
[
"PyTorch supports many other layer types, loss functions, and optimizers - you will experiment with these next. Here's the official API documentation for these (if any of the parameters used above were unclear, this resource will also be helpful). One note: what we call in the class \"spatial batch norm\" is called \"BatchNorm2D\" in PyTorch.\n\n* Layers: http://pytorch.org/docs/nn.html\n* Activations: http://pytorch.org/docs/nn.html#non-linear-activations\n* Loss functions: http://pytorch.org/docs/nn.html#loss-functions\n* Optimizers: http://pytorch.org/docs/optim.html#algorithms",
"_____no_output_____"
],
[
"## Training a specific model\n\nIn this section, we're going to specify a model for you to construct. The goal here isn't to get good performance (that'll be next), but instead to get comfortable with understanding the PyTorch documentation and configuring your own model. \n\nUsing the code provided above as guidance, and using the following PyTorch documentation, specify a model with the following architecture:\n\n* 7x7 Convolutional Layer with 32 filters and stride of 1\n* ReLU Activation Layer\n* Spatial Batch Normalization Layer\n* 2x2 Max Pooling layer with a stride of 2\n* Affine layer with 1024 output units\n* ReLU Activation Layer\n* Affine layer from 1024 input units to 10 outputs\n\nAnd finally, set up a **cross-entropy** loss function and the **RMSprop** learning rule.",
"_____no_output_____"
]
],
[
[
"n_Conv2d = out_dim(32, 7, 0, 1)\nn_MaxPool2d = out_dim(n_Conv2d, 2, 0, 2)\nn_Flatten = 32*n_MaxPool2d**2\n\nfixed_model_base = nn.Sequential( # You fill this in!\n nn.Conv2d(3, 32, kernel_size=7, stride=1),\n nn.ReLU(inplace=True),\n nn.BatchNorm2d(32),\n nn.MaxPool2d(2, stride=2),\n Flatten(), # see above for explanation\n nn.Linear(n_Flatten, 1024), # affine layer\n nn.ReLU(inplace=True),\n nn.Linear(1024, 10) # affine layer\n )\n\nfixed_model = fixed_model_base.type(dtype)",
"_____no_output_____"
]
],
[
[
"To make sure you're doing the right thing, use the following tool to check the dimensionality of your output (it should be 64 x 10, since our batches have size 64 and the output of the final affine layer should be 10, corresponding to our 10 classes):",
"_____no_output_____"
]
],
[
[
"## Now we're going to feed a random batch into the model you defined and make sure the output is the right size\nx = torch.randn(64, 3, 32, 32).type(dtype)\nx_var = Variable(x.type(dtype)) # Construct a PyTorch Variable out of your input data\nans = fixed_model(x_var) # Feed it through the model! \n\n# Check to make sure what comes out of your model\n# is the right dimensionality... this should be True\n# if you've done everything correctly\nnp.array_equal(np.array(ans.size()), np.array([64, 10])) ",
"_____no_output_____"
]
],
[
[
"### GPU!\n\nNow, we're going to switch the dtype of the model and our data to the GPU-friendly tensors, and see what happens... everything is the same, except we are casting our model and input tensors as this new dtype instead of the old one.\n\nIf this returns false, or otherwise fails in a not-graceful way (i.e., with some error message), you may not have an NVIDIA GPU available on your machine. If you're running locally, we recommend you switch to Google Cloud and follow the instructions to set up a GPU there. If you're already on Google Cloud, something is wrong -- make sure you followed the instructions on how to request and use a GPU on your instance. If you did, post on Piazza or come to Office Hours so we can help you debug.",
"_____no_output_____"
]
],
[
[
"import copy\n\nfixed_model_gpu = copy.deepcopy(fixed_model_base).type(dtype)\n\nx_gpu = torch.randn(64, 3, 32, 32).type(dtype)\nx_var_gpu = Variable(x.type(dtype)) # Construct a PyTorch Variable out of your input data\nans = fixed_model_gpu(x_var_gpu) # Feed it through the model! \n\n# Check to make sure what comes out of your model\n# is the right dimensionality... this should be True\n# if you've done everything correctly\nnp.array_equal(np.array(ans.size()), np.array([64, 10]))",
"_____no_output_____"
]
],
[
[
"Run the following cell to evaluate the performance of the forward pass running on the CPU:",
"_____no_output_____"
]
],
[
[
"%%timeit \nans = fixed_model(x_var)",
"1000 loops, best of 3: 445 µs per loop\n"
]
],
[
[
"... and now the GPU:",
"_____no_output_____"
]
],
[
[
"%%timeit \n# torch.cuda.synchronize() # Make sure there are no pending GPU computations\nans = fixed_model_gpu(x_var_gpu) # Feed it through the model! \n# torch.cuda.synchronize() # Make sure there are no pending GPU computations",
"1000 loops, best of 3: 448 µs per loop\n"
]
],
[
[
"You should observe that even a simple forward pass like this is significantly faster on the GPU. So for the rest of the assignment (and when you go train your models in assignment 3 and your project!), you should use the GPU datatype for your model and your tensors: as a reminder that is *torch.cuda.FloatTensor* (in our notebook here as *gpu_dtype*)",
"_____no_output_____"
],
[
"### Train the model.\n\nNow that you've seen how to define a model and do a single forward pass of some data through it, let's walk through how you'd actually train one whole epoch over your training data (using the simple_model we provided above).\n\nMake sure you understand how each PyTorch function used below corresponds to what you implemented in your custom neural network implementation.\n\nNote that because we are not resetting the weights anywhere below, if you run the cell multiple times, you are effectively training multiple epochs (so your performance should improve).\n\nFirst, set up an RMSprop optimizer (using a 1e-3 learning rate) and a cross-entropy loss function:",
"_____no_output_____"
]
],
[
[
"loss_fn = nn.CrossEntropyLoss()\noptimizer = optim.RMSprop(fixed_model_gpu.parameters(), lr=1e-3)\n",
"_____no_output_____"
],
[
"# This sets the model in \"training\" mode. This is relevant for some layers that may have different behavior\n# in training mode vs testing mode, such as Dropout and BatchNorm. \nfixed_model_gpu.train()\n\n# Load one batch at a time.\nfor t, (x, y) in enumerate(tqdm(loader_train)):\n x_var = Variable(x.type(dtype))\n y_var = Variable(y.type(ltype))\n \n # This is the forward pass: predict the scores for each class, for each x in the batch.\n scores = fixed_model_gpu(x_var)\n \n # Use the correct y values and the predicted y values to compute the loss.\n loss = loss_fn(scores, y_var)\n \n if (t + 1) % print_every == 0:\n print('t = %d, loss = %.4f' % (t + 1, loss.data[0]))\n\n # Zero out all of the gradients for the variables which the optimizer will update.\n optimizer.zero_grad()\n \n # This is the backwards pass: compute the gradient of the loss with respect to each \n # parameter of the model.\n loss.backward()\n \n # Actually update the parameters of the model using the gradients computed by the backwards pass.\n optimizer.step()",
" 26%|██▌ | 198/766 [00:01<00:05, 105.99it/s]"
]
],
[
[
"Now you've seen how the training process works in PyTorch. To save you writing boilerplate code, we're providing the following helper functions to help you train for multiple epochs and check the accuracy of your model:",
"_____no_output_____"
]
],
[
[
"def train(model, loss_fn, optimizer, num_epochs = 1, verbose = True):\n for epoch in range(num_epochs):\n if verbose:\n print('Starting epoch %d / %d' % (epoch + 1, num_epochs))\n model.train()\n for t, (x, y) in enumerate(loader_train):\n x_var = Variable(x.type(dtype))\n y_var = Variable(y.type(ltype))\n\n scores = model(x_var)\n \n loss = loss_fn(scores, y_var)\n if (t + 1) % print_every == 0 and verbose:\n print('t = %d, loss = %.4f' % (t + 1, loss.data[0]))\n\n optimizer.zero_grad()\n loss.backward()\n optimizer.step()\n\ndef check_accuracy(model, loader, verbose = True):\n if verbose:\n if loader.dataset.train:\n print('Checking accuracy on validation set')\n else:\n print('Checking accuracy on test set') \n num_correct = 0\n num_samples = 0\n model.eval() # Put the model in test mode (the opposite of model.train(), essentially)\n for x, y in loader:\n x_var = Variable(x.type(dtype), volatile=True)\n\n scores = model(x_var)\n _, preds = scores.data.cpu().max(1)\n num_correct += (preds == y).sum()\n num_samples += preds.size(0)\n acc = float(num_correct) / num_samples\n if verbose:\n print('Got %d / %d correct (%.2f)' % (num_correct, num_samples, 100 * acc))\n return acc\n\ntorch.cuda.random.manual_seed(12345)",
"_____no_output_____"
]
],
[
[
"### Check the accuracy of the model.\n\nLet's see the train and check_accuracy code in action -- feel free to use these methods when evaluating the models you develop below.\n\nYou should get a training loss of around 1.2-1.4, and a validation accuracy of around 50-60%. As mentioned above, if you re-run the cells, you'll be training more epochs, so your performance will improve past these numbers.\n\nBut don't worry about getting these numbers better -- this was just practice before you tackle designing your own model.",
"_____no_output_____"
]
],
[
[
"fixed_model_gpu.apply(reset)\ntrain(fixed_model_gpu, loss_fn, optimizer, num_epochs=5)\ncheck_accuracy(fixed_model_gpu, loader_val)",
"Starting epoch 1 / 5\nt = 100, loss = 1.3351\nt = 200, loss = 1.4855\nt = 300, loss = 1.4892\nt = 400, loss = 1.2383\nt = 500, loss = 1.2223\nt = 600, loss = 1.3844\nt = 700, loss = 1.1986\nStarting epoch 2 / 5\nt = 100, loss = 0.9178\nt = 200, loss = 0.9722\nt = 300, loss = 1.0708\nt = 400, loss = 0.8852\nt = 500, loss = 0.9199\nt = 600, loss = 1.0414\nt = 700, loss = 0.8921\nStarting epoch 3 / 5\nt = 100, loss = 0.6192\nt = 200, loss = 0.6360\nt = 300, loss = 0.5818\nt = 400, loss = 0.7068\nt = 500, loss = 0.6241\nt = 600, loss = 0.7583\nt = 700, loss = 0.4911\nStarting epoch 4 / 5\nt = 100, loss = 0.3345\nt = 200, loss = 0.2367\nt = 300, loss = 0.2881\nt = 400, loss = 0.3601\nt = 500, loss = 0.2190\nt = 600, loss = 0.3616\nt = 700, loss = 0.2555\nStarting epoch 5 / 5\nt = 100, loss = 0.1729\nt = 200, loss = 0.1571\nt = 300, loss = 0.1178\nt = 400, loss = 0.1417\nt = 500, loss = 0.2129\nt = 600, loss = 0.3370\nt = 700, loss = 0.2010\nChecking accuracy on validation set\nGot 623 / 1000 correct (62.30)\n"
]
],
[
[
"### Don't forget the validation set!\n\nAnd note that you can use the check_accuracy function to evaluate on either the test set or the validation set, by passing either **loader_test** or **loader_val** as the second argument to check_accuracy. You should not touch the test set until you have finished your architecture and hyperparameter tuning, and only run the test set once at the end to report a final value. ",
"_____no_output_____"
],
[
"## Train a _great_ model on CIFAR-10!\n\nNow it's your job to experiment with architectures, hyperparameters, loss functions, and optimizers to train a model that achieves **>=70%** accuracy on the CIFAR-10 **validation** set. You can use the check_accuracy and train functions from above.",
"_____no_output_____"
],
[
"### Things you should try:\n- **Filter size**: Above we used 7x7; this makes pretty pictures but smaller filters may be more efficient\n- **Number of filters**: Above we used 32 filters. Do more or fewer do better?\n- **Pooling vs Strided Convolution**: Do you use max pooling or just stride convolutions?\n- **Batch normalization**: Try adding spatial batch normalization after convolution layers and vanilla batch normalization after affine layers. Do your networks train faster?\n- **Network architecture**: The network above has two layers of trainable parameters. Can you do better with a deep network? Good architectures to try include:\n - [conv-relu-pool]xN -> [affine]xM -> [softmax or SVM]\n - [conv-relu-conv-relu-pool]xN -> [affine]xM -> [softmax or SVM]\n - [batchnorm-relu-conv]xN -> [affine]xM -> [softmax or SVM]\n- **Global Average Pooling**: Instead of flattening and then having multiple affine layers, perform convolutions until your image gets small (7x7 or so) and then perform an average pooling operation to get to a 1x1 image picture (1, 1 , Filter#), which is then reshaped into a (Filter#) vector. This is used in [Google's Inception Network](https://arxiv.org/abs/1512.00567) (See Table 1 for their architecture).\n- **Regularization**: Add l2 weight regularization, or perhaps use Dropout.\n\n### Tips for training\nFor each network architecture that you try, you should tune the learning rate and regularization strength. When doing this there are a couple important things to keep in mind:\n\n- If the parameters are working well, you should see improvement within a few hundred iterations\n- Remember the coarse-to-fine approach for hyperparameter tuning: start by testing a large range of hyperparameters for just a few training iterations to find the combinations of parameters that are working at all.\n- Once you have found some sets of parameters that seem to work, search more finely around these parameters. You may need to train for more epochs.\n- You should use the validation set for hyperparameter search, and save your test set for evaluating your architecture on the best parameters as selected by the validation set.\n\n### Going above and beyond\nIf you are feeling adventurous there are many other features you can implement to try and improve your performance. You are **not required** to implement any of these; however they would be good things to try for extra credit.\n\n- Alternative update steps: For the assignment we implemented SGD+momentum, RMSprop, and Adam; you could try alternatives like AdaGrad or AdaDelta.\n- Alternative activation functions such as leaky ReLU, parametric ReLU, ELU, or MaxOut.\n- Model ensembles\n- Data augmentation\n- New Architectures\n - [ResNets](https://arxiv.org/abs/1512.03385) where the input from the previous layer is added to the output.\n - [DenseNets](https://arxiv.org/abs/1608.06993) where inputs into previous layers are concatenated together.\n - [This blog has an in-depth overview](https://chatbotslife.com/resnets-highwaynets-and-densenets-oh-my-9bb15918ee32)\n\nIf you do decide to implement something extra, clearly describe it in the \"Extra Credit Description\" cell below.\n\n### What we expect\nAt the very least, you should be able to train a ConvNet that gets at least 70% accuracy on the validation set. This is just a lower bound - if you are careful it should be possible to get accuracies much higher than that! Extra credit points will be awarded for particularly high-scoring models or unique approaches.\n\nYou should use the space below to experiment and train your network. \n\nHave fun and happy training!",
"_____no_output_____"
]
],
[
[
"import time\nimport hyperopt.pyll\nfrom hyperopt import fmin, tpe, hp, STATUS_OK, Trials\nfrom hyperopt.pyll import scope\n",
"_____no_output_____"
],
[
"@scope.define_pure\ndef L1_shift(a):\n return [a + 5]\n\[email protected]_pure\ndef L2_shift(a):\n return [a + 5]\n\[email protected]_pure\ndef L2_L3_shift(a, b):\n return [a + 5, b + 5]\n\[email protected]_pure\ndef W1_shift(a):\n return [a + 256]\n\[email protected]_pure\ndef W2_shift(a):\n return [a + 64]\n",
"_____no_output_____"
]
],
[
[
"## Base Model\n\nAfter 20 rounds of hyperparameters optimization we were able to get a 60% validation accuracy.\n\n parameters_zero = {'L1': [6], 'S': [3], 'W1': [651], 'loss': 0.6},",
"_____no_output_____"
]
],
[
[
"# Train your model here, and make sure the output of this cell is the accuracy of your best model on the \n# train, val, and test sets. Here's some code to get you started. The output of this cell should be the training\n# and validation accuracy on your best model (measured by validation accuracy).\n\ndef model_zero(L1, W1, S, C = 10):\n n_Conv2d = out_dim(32, S, 0, 1)\n n_MaxPool2d = out_dim(n_Conv2d, 2, 0, 2)\n n_Flatten = L1*n_MaxPool2d**2\n\n return nn.Sequential( # You fill this in!\n nn.Conv2d(3, L1, kernel_size=S, stride=1),\n nn.ReLU(inplace=True),\n nn.BatchNorm2d(L1),\n nn.MaxPool2d(2, stride=2),\n Flatten(), # see above for explanation\n nn.Linear(n_Flatten, W1), # affine layer\n nn.ReLU(inplace=True),\n nn.Linear(W1, C) # affine layer\n )\n",
"_____no_output_____"
],
[
"search_space_zero = {\n 'L1': L1_shift(hp.randint('L1', 20)),\n 'W1': W1_shift(hp.randint('W1', 2048)),\n 'S': hp.choice('S', [3, 5, 7])\n}\n\ndef loss_zero(x):\n print(x)\n model = model_zero(x['L1'][0], x['W1'][0], x['S']).type(dtype)\n\n loss_fn = nn.CrossEntropyLoss()\n optimizer = optim.RMSprop(model.parameters(), lr=1e-3)\n\n train(model, loss_fn, optimizer, num_epochs=20, verbose=False)\n return -check_accuracy(model, loader_val, verbose=False)\n\ndef objective_zero(x):\n return {\n 'loss': loss_zero(x),\n 'status': STATUS_OK,\n # -- store other results\n 'eval_time': time.time()\n }\n\ntrials_zero = Trials()\nbest_zero = fmin(objective_zero,\n space=search_space_zero,\n algo=tpe.suggest,\n max_evals=20,\n trials=trials_zero)\n",
"_____no_output_____"
],
[
"def extract_zero(x):\n return {\n 'loss': -x['result']['loss'],\n 'L1': list(map(lambda v: v+5, x['misc']['vals']['L1'])),\n 'W1': list(map(lambda v: v+256, x['misc']['vals']['W1'])),\n 'S': list(map(lambda v: 3 if v == 0 else 5 if v == 1 else 7,x['misc']['vals']['S']))\n }\n\nres_zero = list(map(extract_zero, trials_zero))\n\n[{'L1': [9], 'S': [5], 'W1': [1870], 'loss': 0.584},\n {'L1': [8], 'S': [7], 'W1': [649], 'loss': 0.568},\n {'L1': [16], 'S': [5], 'W1': [614], 'loss': 0.55},\n {'L1': [18], 'S': [7], 'W1': [1773], 'loss': 0.513},\n {'L1': [21], 'S': [3], 'W1': [1664], 'loss': 0.616},\n {'L1': [6], 'S': [3], 'W1': [651], 'loss': 0.6},\n {'L1': [5], 'S': [3], 'W1': [750], 'loss': 0.587},\n {'L1': [17], 'S': [3], 'W1': [2189], 'loss': 0.623},\n {'L1': [14], 'S': [7], 'W1': [2240], 'loss': 0.553},\n {'L1': [5], 'S': [5], 'W1': [1008], 'loss': 0.533},\n {'L1': [22], 'S': [5], 'W1': [1498], 'loss': 0.609},\n {'L1': [15], 'S': [3], 'W1': [2065], 'loss': 0.581},\n {'L1': [11], 'S': [7], 'W1': [1174], 'loss': 0.557},\n {'L1': [9], 'S': [7], 'W1': [923], 'loss': 0.566},\n {'L1': [15], 'S': [7], 'W1': [2022], 'loss': 0.59},\n {'L1': [6], 'S': [5], 'W1': [969], 'loss': 0.587},\n {'L1': [9], 'S': [3], 'W1': [1193], 'loss': 0.514},\n {'L1': [16], 'S': [5], 'W1': [2252], 'loss': 0.627},\n {'L1': [12], 'S': [5], 'W1': [1821], 'loss': 0.611},\n {'L1': [9], 'S': [7], 'W1': [279], 'loss': 0.53}]\n",
"_____no_output_____"
]
],
[
[
"\n\n## [conv-relu-pool]xN -> [affine]xM -> [softmax or SVM]\n\n66.5% validation accuracy with:\n\n {'L1': [16], 'L2': [8], 'W1': [464], 'W2': [], 'loss': 0.665}",
"_____no_output_____"
]
],
[
[
"def model_one(conv_relu_pool_depths, affine_depths, C = 10):\n from functools import reduce\n from collections import OrderedDict\n\n Conv2d_K = 3\n Conv2d_S = 1\n MaxPool2d_K = 2\n MaxPool2d_S = 2\n # 15, 4500; 6, 720\n conv_relu_pool_sizes = [\n lambda N: out_dim(N, Conv2d_K, 0, Conv2d_S),\n lambda N: out_dim(N, MaxPool2d_K, 0, MaxPool2d_S)\n ] * len(conv_relu_pool_depths)\n \n n_to_Flatten = reduce(lambda value, f: f(value), conv_relu_pool_sizes, 32)\n n_Flatten = conv_relu_pool_depths[-1]*n_to_Flatten**2\n\n def conv_relu_pool_layers_ctr():\n for i, (L0, L1) in enumerate(zip([3] + conv_relu_pool_depths[:-1], conv_relu_pool_depths)):\n yield 'conv2d_%s'%i, nn.Conv2d(L0, L1, kernel_size=Conv2d_K, stride=Conv2d_S)\n yield 'relu_%s'%i, nn.ReLU(inplace=True)\n yield 'pool_%s'%i, nn.MaxPool2d(MaxPool2d_K, stride=MaxPool2d_S)\n\n def affine_layers_ctr():\n for i, (W0, W1) in enumerate(zip([n_Flatten] + affine_depths[:-1], affine_depths)):\n yield 'affine_linear_%s'%i, nn.Linear(W0, W1)\n yield 'affine_relu_%s'%i, nn.ReLU(inplace=True)\n \n layers = list(conv_relu_pool_layers_ctr()) + [tuple(('flatten', Flatten()))] + list(affine_layers_ctr()) + [tuple(('to_classes', nn.Linear(affine_depths[-1], C)))]\n \n return nn.Sequential(OrderedDict(layers))\n",
"_____no_output_____"
],
[
"search_space_one = {\n 'conv_relu_pool_layers': L1_shift(hp.randint('L1', 20)) + hp.choice('conv_relu_pool_depths', [\n list(),\n L2_shift(hp.randint('L2', 20))\n ]),\n 'affine_layers': W1_shift(hp.randint('W1', 2048)) + hp.choice('affine_depths', [\n list(),\n W2_shift(hp.randint('W2', 256))\n ])\n}\n\ndef loss_one(x):\n print(x)\n model = model_one(list(x['conv_relu_pool_layers']), list(x['affine_layers'])).type(dtype)\n\n loss_fn = nn.CrossEntropyLoss()\n optimizer = optim.RMSprop(model.parameters(), lr=1e-3)\n\n train(model, loss_fn, optimizer, num_epochs=20, verbose=True)\n return -check_accuracy(model, loader_val, verbose=True)\n\ndef objective_one(x):\n return {\n 'loss': loss_one(x),\n 'status': STATUS_OK,\n # -- store other results\n 'eval_time': time.time()\n }\n\ntrials_one = Trials()\nbest_one = fmin(objective_one,\n space=search_space_one,\n algo=tpe.suggest,\n max_evals=20,\n trials=trials_one)\n",
"_____no_output_____"
],
[
"def extract_one(x):\n return {\n 'loss': -x['result']['loss'],\n 'L1': list(map(lambda x: x+5, x['misc']['vals']['L1'])),\n 'L2': list(map(lambda x: x+5, x['misc']['vals']['L2'])),\n 'W1': list(map(lambda x: x+256, x['misc']['vals']['W1'])),\n 'W2': list(map(lambda x: x+64, x['misc']['vals']['W2']))\n }\n\nres_one = list(map(extract_one, trials_one))\n\n[{'L1': [18], 'L2': [], 'W1': [950], 'W2': [], 'loss': 0.614},\n {'L1': [7], 'L2': [], 'W1': [2072], 'W2': [168], 'loss': 0.586},\n {'L1': [21], 'L2': [20], 'W1': [840], 'W2': [], 'loss': 0.653},\n {'L1': [16], 'L2': [8], 'W1': [464], 'W2': [], 'loss': 0.665},\n {'L1': [17], 'L2': [], 'W1': [2011], 'W2': [316], 'loss': 0.619},\n {'L1': [24], 'L2': [], 'W1': [1908], 'W2': [174], 'loss': 0.644},\n {'L1': [14], 'L2': [8], 'W1': [2196], 'W2': [], 'loss': 0.604},\n {'L1': [15], 'L2': [10], 'W1': [1806], 'W2': [], 'loss': 0.607},\n {'L1': [20], 'L2': [], 'W1': [1820], 'W2': [], 'loss': 0.643},\n {'L1': [11], 'L2': [], 'W1': [1431], 'W2': [247], 'loss': 0.62},\n {'L1': [9], 'L2': [5], 'W1': [2238], 'W2': [], 'loss': 0.548},\n {'L1': [7], 'L2': [], 'W1': [1655], 'W2': [], 'loss': 0.6},\n {'L1': [12], 'L2': [], 'W1': [2241], 'W2': [], 'loss': 0.612},\n {'L1': [20], 'L2': [9], 'W1': [1818], 'W2': [], 'loss': 0.589},\n {'L1': [22], 'L2': [], 'W1': [934], 'W2': [], 'loss': 0.636},\n {'L1': [24], 'L2': [13], 'W1': [667], 'W2': [210], 'loss': 0.64},\n {'L1': [18], 'L2': [], 'W1': [455], 'W2': [278], 'loss': 0.62},\n {'L1': [9], 'L2': [], 'W1': [967], 'W2': [], 'loss': 0.594},\n {'L1': [13], 'L2': [], 'W1': [1033], 'W2': [], 'loss': 0.63},\n {'L1': [19], 'L2': [24], 'W1': [573], 'W2': [], 'loss': 0.647}]",
"_____no_output_____"
]
],
[
[
"## [batchnorm-relu-conv]xN -> [affine]xM -> [softmax or SVM]\n\n64.9% validation accuracy with:\n\n {'L1': [20],\n 'L2': [21],\n 'L2_1': [],\n 'L3_1': [],\n 'W1': [487],\n 'W2': [],\n 'loss': 0.649}",
"_____no_output_____"
]
],
[
[
"def model_three(batchnorm_conv_relu_depths, affine_depths, C = 10):\n from functools import reduce\n from collections import OrderedDict\n\n Conv2d_K = 3\n Conv2d_S = 1\n batchnorm_conv_relu_sizes = [\n lambda N: out_dim(N, Conv2d_K, 0, Conv2d_S)\n ] * len(batchnorm_conv_relu_depths)\n \n n_to_Flatten = reduce(lambda value, f: f(value), batchnorm_conv_relu_sizes, 32)\n n_Flatten = batchnorm_conv_relu_depths[-1]*n_to_Flatten**2\n\n def batchnorm_conv_relu_layers_ctr():\n for i, (L0, L1) in enumerate(zip([3] + batchnorm_conv_relu_depths[:-1], batchnorm_conv_relu_depths)):\n yield 'batchnorm2d_%s'%i, nn.BatchNorm2d(L0)\n yield 'conv2d_%s'%i, nn.Conv2d(L0, L1, kernel_size=Conv2d_K, stride=Conv2d_S)\n yield 'relu_%s'%i, nn.ReLU(inplace=True)\n\n def affine_layers_ctr():\n for i, (W0, W1) in enumerate(zip([n_Flatten] + affine_depths[:-1], affine_depths)):\n yield 'affine_linear_%s'%i, nn.Linear(W0, W1)\n yield 'affine_relu_%s'%i, nn.ReLU(inplace=True)\n \n layers = list(batchnorm_conv_relu_layers_ctr()) + [tuple(('flatten', Flatten()))] + list(affine_layers_ctr()) + [tuple(('to_classes', nn.Linear(affine_depths[-1], C)))]\n \n return nn.Sequential(OrderedDict(layers))\n",
"_____no_output_____"
],
[
"search_space_three = {\n 'batchnorm2d_conv_relu_layers': L1_shift(hp.randint('L1', 20)) + hp.choice('batchnorm2d_conv_relu_L2', [\n list(),\n L2_shift(hp.randint('L2', 20))\n ]) + hp.choice('batchnorm2d_conv_relu_L3', [\n list(),\n L2_shift(hp.randint('L3', 20))\n ]),\n 'affine_layers': W1_shift(hp.randint('W1', 2048)) + hp.choice('affine_depths', [\n list(),\n W2_shift(hp.randint('W2', 256))\n ])\n}\n\ndef loss_three(x):\n print(x)\n model = model_three(list(x['batchnorm2d_conv_relu_layers']), list(x['affine_layers'])).type(dtype)\n\n loss_fn = nn.CrossEntropyLoss()\n optimizer = optim.RMSprop(model.parameters(), lr=1e-3)\n\n train(model, loss_fn, optimizer, num_epochs=20, verbose=False)\n return -check_accuracy(model, loader_val, verbose=False)\n\ndef objective_three(x):\n return {\n 'loss': loss_three(x),\n 'status': STATUS_OK,\n # -- store other results\n 'eval_time': time.time()\n }\n\ntrials_three = Trials()\nbest_three = fmin(objective_three,\n space=search_space_three,\n algo=tpe.suggest,\n max_evals=20,\n trials=trials_three)\n",
"_____no_output_____"
],
[
"def extract_three(x):\n val = x['misc']['vals']\n return {\n 'loss': -x['result']['loss'],\n 'L1': list(map(lambda x: x+5, val['L1'])),\n 'L2': list(map(lambda x: x+5, val.get('L2', []))),\n 'L2_1': list(map(lambda x: x+5, val.get('L2_1', []))),\n 'L3_1': list(map(lambda x: x+5, val.get('L3_1', []))),\n 'W1': list(map(lambda x: x+256, val['W1'])),\n 'W2': list(map(lambda x: x+64, val.get('W2', [])))\n }\n\nres_three = list(map(extract_three, trials_three))\n\n[{'L1': [24],\n 'L2': [],\n 'L2_1': [],\n 'L3_1': [],\n 'W1': [975],\n 'W2': [],\n 'loss': 0.603},\n {'L1': [13],\n 'L2': [],\n 'L2_1': [],\n 'L3_1': [],\n 'W1': [1873],\n 'W2': [222],\n 'loss': 0.613},\n {'L1': [9],\n 'L2': [10],\n 'L2_1': [],\n 'L3_1': [],\n 'W1': [2180],\n 'W2': [161],\n 'loss': 0.616},\n {'L1': [14],\n 'L2': [],\n 'L2_1': [],\n 'L3_1': [],\n 'W1': [1241],\n 'W2': [295],\n 'loss': 0.622},\n {'L1': [11],\n 'L2': [8],\n 'L2_1': [],\n 'L3_1': [],\n 'W1': [1138],\n 'W2': [74],\n 'loss': 0.6},\n {'L1': [20],\n 'L2': [10],\n 'L2_1': [],\n 'L3_1': [],\n 'W1': [1968],\n 'W2': [261],\n 'loss': 0.618},\n {'L1': [15],\n 'L2': [],\n 'L2_1': [],\n 'L3_1': [],\n 'W1': [803],\n 'W2': [],\n 'loss': 0.614},\n {'L1': [24],\n 'L2': [],\n 'L2_1': [],\n 'L3_1': [],\n 'W1': [1607],\n 'W2': [],\n 'loss': 0.594},\n {'L1': [24],\n 'L2': [],\n 'L2_1': [],\n 'L3_1': [],\n 'W1': [1132],\n 'W2': [],\n 'loss': 0.596},\n {'L1': [10],\n 'L2': [],\n 'L2_1': [],\n 'L3_1': [],\n 'W1': [1500],\n 'W2': [302],\n 'loss': 0.584},\n {'L1': [9],\n 'L2': [22],\n 'L2_1': [],\n 'L3_1': [],\n 'W1': [2272],\n 'W2': [],\n 'loss': 0.559},\n {'L1': [15],\n 'L2': [18],\n 'L2_1': [],\n 'L3_1': [],\n 'W1': [1224],\n 'W2': [],\n 'loss': 0.614},\n {'L1': [23],\n 'L2': [],\n 'L2_1': [],\n 'L3_1': [],\n 'W1': [1120],\n 'W2': [],\n 'loss': 0.621},\n {'L1': [22],\n 'L2': [11],\n 'L2_1': [],\n 'L3_1': [],\n 'W1': [1398],\n 'W2': [],\n 'loss': 0.573},\n {'L1': [13],\n 'L2': [],\n 'L2_1': [],\n 'L3_1': [],\n 'W1': [1214],\n 'W2': [283],\n 'loss': 0.61},\n {'L1': [24],\n 'L2': [7],\n 'L2_1': [],\n 'L3_1': [],\n 'W1': [302],\n 'W2': [],\n 'loss': 0.559},\n {'L1': [20],\n 'L2': [21],\n 'L2_1': [],\n 'L3_1': [],\n 'W1': [487],\n 'W2': [],\n 'loss': 0.649},\n {'L1': [22],\n 'L2': [22],\n 'L2_1': [],\n 'L3_1': [],\n 'W1': [1007],\n 'W2': [],\n 'loss': 0.641},\n {'L1': [15],\n 'L2': [18],\n 'L2_1': [],\n 'L3_1': [],\n 'W1': [509],\n 'W2': [168],\n 'loss': 0.61},\n {'L1': [12],\n 'L2': [],\n 'L2_1': [],\n 'L3_1': [],\n 'W1': [1424],\n 'W2': [],\n 'loss': 0.583}]",
"_____no_output_____"
]
],
[
[
"### Describe what you did \n\nIn the cell below you should write an explanation of what you did, any additional features that you implemented, and any visualizations or graphs that you make in the process of training and evaluating your network.",
"_____no_output_____"
],
[
"DNN with architectures 0, 1 and 3 were implemented and trained on data. Hyperparameter optimization was performed.\n\nThe best model found was the [conv-relu-pool]xN -> [affine]xM -> [softmax] with parameters: {'L1': 16, 'L2': 8, 'W1': 464} which got an accuracy of 0.665.",
"_____no_output_____"
],
[
"## Test set -- run this only once\n\nNow that we've gotten a result we're happy with, we test our final model on the test set (which you should store in best_model). This would be the score we would achieve on a competition. Think about how this compares to your validation set accuracy.",
"_____no_output_____"
]
],
[
[
"best_model = None\ncheck_accuracy(best_model, loader_test)",
"_____no_output_____"
]
],
[
[
"## Going further with PyTorch\n\nThe next assignment will make heavy use of PyTorch. You might also find it useful for your projects. \n\nHere's a nice tutorial by Justin Johnson that shows off some of PyTorch's features, like dynamic graphs and custom NN modules: http://pytorch.org/tutorials/beginner/pytorch_with_examples.html\n\nIf you're interested in reinforcement learning for your final project, this is a good (more advanced) DQN tutorial in PyTorch: http://pytorch.org/tutorials/intermediate/reinforcement_q_learning.html",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d084ee3f0975c0c5ea4cc602daff9bd27f162100 | 10,174 | ipynb | Jupyter Notebook | Demo1.ipynb | RoyMillamis/Roy-Millamis-BSCpE-1-2 | 145fab8ae1dae23300a6f6c85047fab255f3b512 | [
"Apache-2.0"
] | null | null | null | Demo1.ipynb | RoyMillamis/Roy-Millamis-BSCpE-1-2 | 145fab8ae1dae23300a6f6c85047fab255f3b512 | [
"Apache-2.0"
] | null | null | null | Demo1.ipynb | RoyMillamis/Roy-Millamis-BSCpE-1-2 | 145fab8ae1dae23300a6f6c85047fab255f3b512 | [
"Apache-2.0"
] | null | null | null | 21.87957 | 236 | 0.388638 | [
[
[
"<a href=\"https://colab.research.google.com/github/RoyMillamis/Roy-Millamis-BSCpE-1-2/blob/main/Demo1.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"##Introduction to Python\n",
"_____no_output_____"
]
],
[
[
"#Python Indention\nif 5>2:\n print(\"five is greater than two!\")",
"five is greater than two!\n"
],
[
"x = 1 # This is a single variable with single value\nx,y = 1,2 # these are two variables with two different values\nx,y,z= 1,2,3\nprint(x)\nprint(y)\nprint(z)",
"1\n2\n3\n"
],
[
"",
"_____no_output_____"
],
[
"x,y=\"four\",2\nx\ny\nx",
"_____no_output_____"
]
],
[
[
"### Casting",
"_____no_output_____"
]
],
[
[
"b = int(4)\nb\nc= float(4)\nc",
"_____no_output_____"
]
],
[
[
"### Type Function",
"_____no_output_____"
]
],
[
[
"x=5\ny= \"John\" # This is a type of string\nh= \"ana\"\nH='Ana'\nprint(type(x))\nprint(type(y))\nprint(h)\nprint(H)",
"<class 'int'>\n<class 'str'>\nana\nAna\n"
]
],
[
[
"## One Value to Multiple Variables",
"_____no_output_____"
]
],
[
[
"x = y = z = 'four'\nprint(x)\nprint(y)\nprint(z)",
"four\nfour\nfour\n"
],
[
"x = \"enjoying\"\nprint(\"Python Programming is\" \" \" + x)",
"Python Programming is enjoying\n"
],
[
"x = 11\ny = 12\nz = 13\nprint(x+y+z)",
"36\n"
],
[
"x+=3 #This is the same as x = +3\nprint(x)",
"14\n"
],
[
"y+=5\nprint(y)",
"17\n"
],
[
"x<y and x!=x # pag isang lang yung true false na",
"_____no_output_____"
],
[
"x>y or not y==z # kahit isa lang yung true, true paden",
"_____no_output_____"
],
[
"not(print(x>y))",
"False\n"
],
[
"#Identity operations\nprint (x is y)\nprint (x is not z)",
"False\nTrue\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d084f584a1db217b83b0c8ce808b3e4e20c0645b | 294 | ipynb | Jupyter Notebook | nbs/02_data_prep/r.ipynb | andrewtruong/notes | 24e3a7d822f746ac4ab6c3186bc44dccb4f0e169 | [
"Apache-2.0"
] | null | null | null | nbs/02_data_prep/r.ipynb | andrewtruong/notes | 24e3a7d822f746ac4ab6c3186bc44dccb4f0e169 | [
"Apache-2.0"
] | null | null | null | nbs/02_data_prep/r.ipynb | andrewtruong/notes | 24e3a7d822f746ac4ab6c3186bc44dccb4f0e169 | [
"Apache-2.0"
] | null | null | null | 12.782609 | 33 | 0.442177 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d08503686284a96a48a8a6af9770609ae44f2350 | 91,625 | ipynb | Jupyter Notebook | Modulo1/.ipynb_checkpoints/Clase5_ProgramacionLineal-checkpoint.ipynb | danielabenavides/SimMat2018-2 | 027e5ef022d03f16ac2ecd96023b74188c81797a | [
"MIT"
] | 1 | 2022-01-29T04:16:12.000Z | 2022-01-29T04:16:12.000Z | Modulo1/.ipynb_checkpoints/Clase5_ProgramacionLineal-checkpoint.ipynb | danielabenavides/SimMat2018-2 | 027e5ef022d03f16ac2ecd96023b74188c81797a | [
"MIT"
] | 1 | 2020-08-14T17:44:49.000Z | 2020-08-14T17:48:39.000Z | Modulo1/.ipynb_checkpoints/Clase5_ProgramacionLineal-checkpoint.ipynb | danielabenavides/SimMat2018-2 | 027e5ef022d03f16ac2ecd96023b74188c81797a | [
"MIT"
] | 3 | 2019-01-31T18:08:31.000Z | 2019-01-31T18:13:26.000Z | 75.72314 | 27,729 | 0.760404 | [
[
[
"# Programación lineal\n\n<img style=\"float: right; margin: 0px 0px 15px 15px;\" src=\"https://upload.wikimedia.org/wikipedia/commons/thumb/0/0c/Linear_Programming_Feasible_Region.svg/2000px-Linear_Programming_Feasible_Region.svg.png\" width=\"400px\" height=\"125px\" />\n\n> La programación lineal es el campo de la optimización matemática dedicado a maximizar o minimizar (optimizar) funciones lineales, denominada función objetivo, de tal forma que las variables de dicha función estén sujetas a una serie de restricciones expresadas mediante un sistema de ecuaciones o inecuaciones también lineales.\n\n**Referencias:**\n- https://es.wikipedia.org/wiki/Programaci%C3%B3n_lineal\n- https://docs.scipy.org/doc/scipy-0.18.1/reference/optimize.html",
"_____no_output_____"
],
[
"## 1. Apuntes históricos\n\n<img style=\"float: right; margin: 0px 0px 15px 15px;\" src=\"https://upload.wikimedia.org/wikipedia/commons/5/5e/JohnvonNeumann-LosAlamos.gif\" width=\"400px\" height=\"125px\" />\n\n- 1826:\tJoseph Fourier anticipa la programación lineal. Carl Friedrich Gauss resuelve ecuaciones lineales por eliminación \"gaussiana\".\n- 1902:\tGyula Farkas concibe un método para resolver sistemas de inecuaciones.\n- Es hasta la Segunda Guerra Mundial que se plantea la programación lineal como un modelo matemático para planificar gastos y retornos, de modo que se reduzcan costos de guerra y aumentar pérdidas del enemigo. Secreto hasta 1947 (posguerra).\n- 1947:\tGeorge Dantzig publica el algoritmo simplex y John von Neumann desarrolló la teoría de la dualidad. Se sabe que Leonid Kantoróvich también formuló la teoría en forma independiente.\n- Fue usado por muchas industrias en la planificación diaria.\n\n**Hasta acá, tiempos exponenciales de solución. Lo siguiente, tiempo polinomial.**\n\n- 1979: Leonid Khachiyan, diseñó el llamado Algoritmo del elipsoide, a través del cual demostró que el problema de la programación lineal es resoluble de manera eficiente, es decir, en tiempo polinomial.\n- 1984: Narendra Karmarkar introduce el método del punto interior para resolver problemas de programación lineal.\n\n**Mencionar complejidad computacional.**",
"_____no_output_____"
],
[
"## 2. Motivación\n\nYa la clase pasada habíamos mencionado que cuando se quería optimizar una función de varias variables con restricciones, se podía aplicar siempre el método de Multiplicadores de Lagrange. Sin embargo, este método es computacionalmente muy complejo conforme crece el número de variables.\n\nPor tanto, cuando la función a optimizar y las restricciones son de caracter lineal, los métodos de solución que se pueden desarrollar son computacionalmente eficientes, por lo que es útil realizar la distinción.",
"_____no_output_____"
],
[
"## 3. Problemas de programación lineal\n\n### 3.1. Ejemplo básico\nUna compañía produce dos productos ($X_1$ y $X_2$) usando dos máquinas ($A$ y $B$). Cada unidad de $X_1$ que se produce requiere 50 minutos en la máquina $A$ y 30 minutos en la máquina $B$. Cada unidad de $X_2$ que se produce requiere 24 minutos en la máquina $A$ y 33 minutos en la máquina $B$.\n\nAl comienzo de la semana hay 30 unidades de $X_1$ y 90 unidades de $X_2$ en inventario. El tiempo de uso disponible de la máquina $A$ es de 40 horas y el de la máquina $B$ es de 35 horas.\n\nLa demanda para $X_1$ en la semana actual es de 75 unidades y de $X_2$ es de 95 unidades. La política de la compañía es maximizar la suma combinada de unidades de $X_1$ e $X_2$ en inventario al finalizar la semana.\n\nFormular el problema de decidir cuánto hacer de cada producto en la semana como un problema de programación lineal.",
"_____no_output_____"
],
[
"#### Solución\n\nSean:\n- $x_1$ la cantidad de unidades de $X_1$ a ser producidas en la semana, y\n- $x_2$ la cantidad de unidades de $X_2$ a ser producidas en la semana.\n\nNotar que lo que se quiere es maximizar $x_1+x_2$.\n\nRestricciones:\n1. El tiempo de uso disponible de la máquina $A$ es de 40 horas: $50x_1+24x_2\\leq 40(60)\\Rightarrow 50x_1+24x_2\\leq 2400$.\n2. El tiempo de uso disponible de la máquina $B$ es de 35 horas: $30x_1+33x_2\\leq 35(60)\\Rightarrow 30x_1+33x_2\\leq 2100$.\n3. La demanda para $X_1$ en la semana actual es de 75 unidades: $x_1+30\\geq 75\\Rightarrow x_1\\geq 45\\Rightarrow -x_1\\leq -45$.\n4. La demanda para $X_2$ en la semana actual es de 95 unidades: $x_2+90\\geq 95\\Rightarrow x_2\\geq 5\\Rightarrow -x_2\\leq -5$.\n\nFinalmente, el problema puede ser expresado en la forma explicada como:\n\\begin{equation}\n\\begin{array}{ll}\n\\min_{x_1,x_2} & -x_1-x_2 \\\\\n\\text{s. a. } & 50x_1+24x_2\\leq 2400 \\\\\n & 30x_1+33x_2\\leq 2100 \\\\\n & -x_1\\leq -45 \\\\\n & -x_2\\leq -5,\n\\end{array}\n\\end{equation}\n\no, eqivalentemente \n\\begin{equation}\n\\begin{array}{ll}\n\\min_{\\boldsymbol{x}} & \\boldsymbol{c}^T\\boldsymbol{x} \\\\\n\\text{s. a. } & \\boldsymbol{A}_{eq}\\boldsymbol{x}=\\boldsymbol{b}_{eq} \\\\\n & \\boldsymbol{A}\\boldsymbol{x}\\leq\\boldsymbol{b},\n\\end{array}\n\\end{equation}\ncon\n- $\\boldsymbol{c}=\\left[-1 \\quad -1\\right]^T$,\n- $\\boldsymbol{A}=\\left[\\begin{array}{cc}50 & 24 \\\\ 30 & 33\\\\ -1 & 0\\\\ 0 & -1\\end{array}\\right]$, y\n- $\\boldsymbol{b}=\\left[2400\\quad 2100\\quad -45\\quad -5\\right]^T$.\n\nPreferiremos, en adelante, la notación vectorial/matricial.",
"_____no_output_____"
],
[
"### 3.2. En general\nDe acuerdo a lo descrito anteriormente, un problema de programación lineal puede escribirse en la siguiente forma:\n\n\\begin{equation}\n\\begin{array}{ll}\n\\min_{x_1,\\dots,x_n} & c_1x_1+\\dots+c_nx_n \\\\\n\\text{s. a. } & a^{eq}_{j,1}x_1+\\dots+a^{eq}_{j,n}x_n=b^{eq}_j \\text{ para } 1\\leq j\\leq m_1 \\\\\n & a_{k,1}x_1+\\dots+a_{k,n}x_n\\leq b_k \\text{ para } 1\\leq k\\leq m_2,\n\\end{array}\n\\end{equation}\ndonde:\n- $x_i$ para $i=1,\\dots,n$ son las incógnitas o variables de decisión,\n- $c_i$ para $i=1,\\dots,n$ son los coeficientes de la función a optimizar,\n- $a^{eq}_{j,i}$ para $j=1,\\dots,m_1$ e $i=1,\\dots,n$, son los coeficientes de la restricción de igualdad,\n- $a_{k,i}$ para $k=1,\\dots,m_2$ e $i=1,\\dots,n$, son los coeficientes de la restricción de desigualdad,\n- $b^{eq}_j$ para $j=1,\\dots,m_1$ son valores conocidos que deben ser respetados estrictamente, y\n- $b_k$ para $k=1,\\dots,m_2$ son valores conocidos que no deben ser superados.\n\nEquivalentemente, el problema puede escribirse como\n\n\\begin{equation}\n\\begin{array}{ll}\n\\min_{\\boldsymbol{x}} & \\boldsymbol{c}^T\\boldsymbol{x} \\\\\n\\text{s. a. } & \\boldsymbol{A}_{eq}\\boldsymbol{x}=\\boldsymbol{b}_{eq} \\\\\n & \\boldsymbol{A}\\boldsymbol{x}\\leq\\boldsymbol{b},\n\\end{array}\n\\end{equation}\ndonde:\n- $\\boldsymbol{x}=\\left[x_1\\quad\\dots\\quad x_n\\right]^T$,\n- $\\boldsymbol{c}=\\left[c_1\\quad\\dots\\quad c_n\\right]^T$,\n- $\\boldsymbol{A}_{eq}=\\left[\\begin{array}{ccc}a^{eq}_{1,1} & \\dots & a^{eq}_{1,n}\\\\ \\vdots & \\ddots & \\vdots\\\\ a^{eq}_{m_1,1} & \\dots & a^{eq}_{m_1,n}\\end{array}\\right]$,\n- $\\boldsymbol{A}=\\left[\\begin{array}{ccc}a_{1,1} & \\dots & a_{1,n}\\\\ \\vdots & \\ddots & \\vdots\\\\ a_{m_2,1} & \\dots & a_{m_2,n}\\end{array}\\right]$,\n- $\\boldsymbol{b}_{eq}=\\left[b^{eq}_1\\quad\\dots\\quad b^{eq}_{m_1}\\right]^T$, y\n- $\\boldsymbol{b}=\\left[b_1\\quad\\dots\\quad b_{m_2}\\right]^T$.\n\n**Nota:** el problema $\\max_{\\boldsymbol{x}}\\boldsymbol{g}(\\boldsymbol{x})$ es equivalente a $\\min_{\\boldsymbol{x}}-\\boldsymbol{g}(\\boldsymbol{x})$.",
"_____no_output_____"
],
[
"#### Bueno, y una vez planteado, ¿cómo se resuelve el problema?",
"_____no_output_____"
],
[
"Este problema está sencillo pues solo es en dos variables. La solución gráfica es válida.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n%matplotlib inline\nimport numpy as np",
"_____no_output_____"
],
[
"def res1(x1):\n return (2400-50*x1)/24\ndef res2(x1):\n return (2100-30*x1)/33",
"_____no_output_____"
],
[
"x1 = np.linspace(40, 50)\nr1 = res1(x1)\nr2 = res2(x1)",
"_____no_output_____"
],
[
"plt.figure(figsize = (8,6))\nplt.plot(x1, res1(x1), 'b--', label = 'res1')\nplt.plot(x1, res2(x1), 'r--', label = 'res2')\nplt.plot([45, 45], [0, 25], 'k', label = 'res3')\nplt.plot([40, 50], [5, 5], 'm', label = 'res4')\nplt.fill_between(np.array([45.0, 45.6]), res1(np.array([45.0, 45.6])), 5*np.ones(2))\nplt.text(44,4,'$(45,5)$',fontsize=10)\nplt.text(45.1,6.35,'$(45,6.25)$',fontsize=10)\nplt.text(45.6,4,'$(45.6,5)$',fontsize=10)\nplt.legend(loc = 'best')\nplt.xlabel('$x_1$')\nplt.ylabel('$x_2$')\nplt.axis([44, 46, 4, 7])\nplt.show()",
"_____no_output_____"
]
],
[
[
"**Actividad.** Mónica hace aretes y cadenitas de joyería. Es tan buena, que todo lo que hace lo vende.\n\nLe toma 30 minutos hacer un par de aretes y una hora hacer una cadenita, y como Mónica también es estudihambre, solo dispone de 10 horas a la semana para hacer las joyas. Por otra parte, el material que compra solo le alcanza para hacer 15 unidades (el par de aretes cuenta como unidad) de joyas por semana.\n\nLa utilidad que le deja la venta de las joyas es \\$15 en cada par de aretes y \\$20 en cada cadenita.\n\n¿Cuántos pares de aretes y cuántas cadenitas debería hacer Mónica para maximizar su utilidad?\n\nFormular el problema en la forma explicada y obtener la solución gráfica (puede ser a mano).\n\n**Diez minutos: quien primero lo haga, pasará a explicarlo al tablero y le subiré la nota de alguna tarea a 100. Debe salir a explicar el problema en el pizarrón.**",
"_____no_output_____"
],
[
"## 5. ¿Cómo se resuelve en python?\n\n### 5.1 Librería `SciPy`\n\n<img style=\"float: right; margin: 0px 0px 15px 15px;\" src=\"https://scipy.org/_static/images/scipy_med.png\" width=\"200px\" height=\"75px\" />\n\n`SciPy` es un softwar de código abierto basado en `Python` para matemáticas, ciencia e ingeniería. \n\nEn particular, los siguientes son algunos de los paquetes básicos:\n- `NumPy`\n- **Librería `SciPy`**\n- `SymPy`\n- `matplotlib`\n- `pandas`\n\nLa **Librería `SciPy`** es uno de los paquetes principales y provee varias rutinas numéricas eficientes. Entre ellas, para integración numérica y optimización.\n\nEn esta clase, y en lo que resta del módulo, estaremos utilizando el módulo `optimize` de la librería `SciPy`.\n\n**Importémoslo**",
"_____no_output_____"
]
],
[
[
"# Importar el módulo optimize de la librería scipy\nimport scipy.optimize as opt",
"_____no_output_____"
]
],
[
[
"El módulo `optimize` que acabamos de importar contiene varias funciones para optimización y búsqueda de raices ($f(x)=0$). Entre ellas se encuentra la función `linprog`",
"_____no_output_____"
]
],
[
[
"# Función linprog del módulo optimize\nhelp(opt.linprog)",
"Help on function linprog in module scipy.optimize._linprog:\n\nlinprog(c, A_ub=None, b_ub=None, A_eq=None, b_eq=None, bounds=None, method='simplex', callback=None, options=None)\n Minimize a linear objective function subject to linear\n equality and inequality constraints.\n \n Linear Programming is intended to solve the following problem form::\n \n Minimize: c^T * x\n \n Subject to: A_ub * x <= b_ub\n A_eq * x == b_eq\n \n Parameters\n ----------\n c : array_like\n Coefficients of the linear objective function to be minimized.\n A_ub : array_like, optional\n 2-D array which, when matrix-multiplied by ``x``, gives the values of\n the upper-bound inequality constraints at ``x``.\n b_ub : array_like, optional\n 1-D array of values representing the upper-bound of each inequality\n constraint (row) in ``A_ub``.\n A_eq : array_like, optional\n 2-D array which, when matrix-multiplied by ``x``, gives the values of\n the equality constraints at ``x``.\n b_eq : array_like, optional\n 1-D array of values representing the RHS of each equality constraint\n (row) in ``A_eq``.\n bounds : sequence, optional\n ``(min, max)`` pairs for each element in ``x``, defining\n the bounds on that parameter. Use None for one of ``min`` or\n ``max`` when there is no bound in that direction. By default\n bounds are ``(0, None)`` (non-negative)\n If a sequence containing a single tuple is provided, then ``min`` and\n ``max`` will be applied to all variables in the problem.\n method : str, optional\n Type of solver. :ref:`'simplex' <optimize.linprog-simplex>`\n and :ref:`'interior-point' <optimize.linprog-interior-point>`\n are supported.\n callback : callable, optional (simplex only)\n If a callback function is provide, it will be called within each\n iteration of the simplex algorithm. The callback must have the\n signature ``callback(xk, **kwargs)`` where ``xk`` is the current\n solution vector and ``kwargs`` is a dictionary containing the\n following::\n \n \"tableau\" : The current Simplex algorithm tableau\n \"nit\" : The current iteration.\n \"pivot\" : The pivot (row, column) used for the next iteration.\n \"phase\" : Whether the algorithm is in Phase 1 or Phase 2.\n \"basis\" : The indices of the columns of the basic variables.\n \n options : dict, optional\n A dictionary of solver options. All methods accept the following\n generic options:\n \n maxiter : int\n Maximum number of iterations to perform.\n disp : bool\n Set to True to print convergence messages.\n \n For method-specific options, see :func:`show_options('linprog')`.\n \n Returns\n -------\n A `scipy.optimize.OptimizeResult` consisting of the following fields:\n \n x : ndarray\n The independent variable vector which optimizes the linear\n programming problem.\n fun : float\n Value of the objective function.\n slack : ndarray\n The values of the slack variables. Each slack variable corresponds\n to an inequality constraint. If the slack is zero, then the\n corresponding constraint is active.\n success : bool\n Returns True if the algorithm succeeded in finding an optimal\n solution.\n status : int\n An integer representing the exit status of the optimization::\n \n 0 : Optimization terminated successfully\n 1 : Iteration limit reached\n 2 : Problem appears to be infeasible\n 3 : Problem appears to be unbounded\n \n nit : int\n The number of iterations performed.\n message : str\n A string descriptor of the exit status of the optimization.\n \n See Also\n --------\n show_options : Additional options accepted by the solvers\n \n Notes\n -----\n This section describes the available solvers that can be selected by the\n 'method' parameter. The default method\n is :ref:`Simplex <optimize.linprog-simplex>`.\n :ref:`Interior point <optimize.linprog-interior-point>` is also available.\n \n Method *simplex* uses the simplex algorithm (as it relates to linear\n programming, NOT the Nelder-Mead simplex) [1]_, [2]_. This algorithm\n should be reasonably reliable and fast for small problems.\n \n .. versionadded:: 0.15.0\n \n Method *interior-point* uses the primal-dual path following algorithm\n as outlined in [4]_. This algorithm is intended to provide a faster\n and more reliable alternative to *simplex*, especially for large,\n sparse problems. Note, however, that the solution returned may be slightly\n less accurate than that of the simplex method and may not correspond with a\n vertex of the polytope defined by the constraints.\n \n References\n ----------\n .. [1] Dantzig, George B., Linear programming and extensions. Rand\n Corporation Research Study Princeton Univ. Press, Princeton, NJ,\n 1963\n .. [2] Hillier, S.H. and Lieberman, G.J. (1995), \"Introduction to\n Mathematical Programming\", McGraw-Hill, Chapter 4.\n .. [3] Bland, Robert G. New finite pivoting rules for the simplex method.\n Mathematics of Operations Research (2), 1977: pp. 103-107.\n .. [4] Andersen, Erling D., and Knud D. Andersen. \"The MOSEK interior point\n optimizer for linear programming: an implementation of the\n homogeneous algorithm.\" High performance optimization. Springer US,\n 2000. 197-232.\n .. [5] Andersen, Erling D. \"Finding all linearly dependent rows in\n large-scale linear programming.\" Optimization Methods and Software\n 6.3 (1995): 219-227.\n .. [6] Freund, Robert M. \"Primal-Dual Interior-Point Methods for Linear\n Programming based on Newton's Method.\" Unpublished Course Notes,\n March 2004. Available 2/25/2017 at\n https://ocw.mit.edu/courses/sloan-school-of-management/15-084j-nonlinear-programming-spring-2004/lecture-notes/lec14_int_pt_mthd.pdf\n .. [7] Fourer, Robert. \"Solving Linear Programs by Interior-Point Methods.\"\n Unpublished Course Notes, August 26, 2005. Available 2/25/2017 at\n http://www.4er.org/CourseNotes/Book%20B/B-III.pdf\n .. [8] Andersen, Erling D., and Knud D. Andersen. \"Presolving in linear\n programming.\" Mathematical Programming 71.2 (1995): 221-245.\n .. [9] Bertsimas, Dimitris, and J. Tsitsiklis. \"Introduction to linear\n programming.\" Athena Scientific 1 (1997): 997.\n .. [10] Andersen, Erling D., et al. Implementation of interior point\n methods for large scale linear programming. HEC/Universite de\n Geneve, 1996.\n \n Examples\n --------\n Consider the following problem:\n \n Minimize: f = -1*x[0] + 4*x[1]\n \n Subject to: -3*x[0] + 1*x[1] <= 6\n 1*x[0] + 2*x[1] <= 4\n x[1] >= -3\n \n where: -inf <= x[0] <= inf\n \n This problem deviates from the standard linear programming problem.\n In standard form, linear programming problems assume the variables x are\n non-negative. Since the variables don't have standard bounds where\n 0 <= x <= inf, the bounds of the variables must be explicitly set.\n \n There are two upper-bound constraints, which can be expressed as\n \n dot(A_ub, x) <= b_ub\n \n The input for this problem is as follows:\n \n >>> c = [-1, 4]\n >>> A = [[-3, 1], [1, 2]]\n >>> b = [6, 4]\n >>> x0_bounds = (None, None)\n >>> x1_bounds = (-3, None)\n >>> from scipy.optimize import linprog\n >>> res = linprog(c, A_ub=A, b_ub=b, bounds=(x0_bounds, x1_bounds),\n ... options={\"disp\": True})\n Optimization terminated successfully.\n Current function value: -22.000000\n Iterations: 1\n >>> print(res)\n fun: -22.0\n message: 'Optimization terminated successfully.'\n nit: 1\n slack: array([39., 0.])\n status: 0\n success: True\n x: array([10., -3.])\n \n Note the actual objective value is 11.428571. In this case we minimized\n the negative of the objective function.\n\n"
]
],
[
[
"la cual resuelve problemas como los que aprendimos a plantear.",
"_____no_output_____"
],
[
"### 5.2 Solución del ejemplo básico con linprog\nYa hicimos la solución gráfica. Contrastemos con la solución que nos da `linprog`...",
"_____no_output_____"
]
],
[
[
"# Importar numpy para crear las matrices\nimport numpy as np",
"_____no_output_____"
],
[
"# Crear las matrices para resolver el problema\nc = np.array([-1, -1])\nA = np.array([[50, 24],\n [30, 33],\n [-1, 0],\n [0, -1]])\nb = np.array([2400, 2100, -45, -5])",
"_____no_output_____"
],
[
"b",
"_____no_output_____"
],
[
"# Resolver utilizando linprog\nresultado = opt.linprog(c, A_ub=A, b_ub=b)",
"_____no_output_____"
],
[
"# Mostrar el resultado\nresultado",
"_____no_output_____"
],
[
"# Extraer el vector solución\nresultado.x",
"_____no_output_____"
]
],
[
[
"**Conclusión**\n- Para maximizar el inventario conjunto de cantidad de productos X1 y X2, se deben producir 45 unidades de X1 y 6.25 unidades de X2.\n- Con esa producción, el inventario conjunto al finalizar la semana es de 1.25 unidades.",
"_____no_output_____"
],
[
"**Otra forma:** poner las cotas de las variables a parte",
"_____no_output_____"
]
],
[
[
"# Escribir matrices y cotas\nc = np.array([-1, -1])\nA = np.array([[50, 24],\n [30, 33]])\nb = np.array([2400, 2100])\n\nx1_bound = (45, None)\nx2_bound = (5, None)",
"_____no_output_____"
],
[
"# Resolver\nresultado2 = opt.linprog(c, A_ub=A, b_ub=b, bounds=(x1_bound,x2_bound))",
"_____no_output_____"
],
[
"# Mostrar el resultado\nresultado2",
"_____no_output_____"
]
],
[
[
"**Actividad.** Resolver el ejemplo de Mónica y sus tiliches con `linprog`",
"_____no_output_____"
]
],
[
[
"# Resolver acá\nc = np.array([-15, -20])\nA = np.array([[1, 2],\n [1, 1]])\nb = np.array([20, 15])",
"_____no_output_____"
],
[
"resultado_monica = opt.linprog(c, A_ub=A, b_ub=b)",
"_____no_output_____"
],
[
"resultado_monica",
"_____no_output_____"
]
],
[
[
"## 6. Problema de transporte 1\n- **Referencia**: https://es.wikipedia.org/wiki/Programaci%C3%B3n_lineal\n\n<img style=\"float: right; margin: 0px 0px 15px 15px;\" src=\"https://upload.wikimedia.org/wikipedia/commons/a/a0/Progr_Lineal.PNG\" width=\"400px\" height=\"125px\" />\n\nEste es un caso curioso, con solo 6 variables (un caso real de problema de transporte puede tener fácilmente más de 1.000 variables) en el cual se aprecia la utilidad de este procedimiento de cálculo.\n\nExisten tres minas de carbón cuya producción diaria es:\n- la mina \"a\" produce 40 toneladas de carbón por día;\n- la mina \"b\" produce 40 t/día; y,\n- la mina \"c\" produce 20 t/día.\n\nEn la zona hay dos centrales termoeléctricas que consumen:\n- la central \"d\" consume 40 t/día de carbón; y,\n- la central \"e\" consume 60 t/día.\n\nLos costos de mercado, de transporte por tonelada son:\n- de \"a\" a \"d\" = 2 monedas;\n- de \"a\" a \"e\" = 11 monedas;\n- de \"b\" a \"d\" = 12 monedas;\n- de \"b\" a \"e\" = 24 monedas;\n- de \"c\" a \"d\" = 13 monedas; y,\n- de \"c\" a \"e\" = 18 monedas.\n\nSi se preguntase a los pobladores de la zona cómo organizar el transporte, tal vez la mayoría opinaría que debe aprovecharse el precio ofrecido por el transportista que va de \"a\" a \"d\", porque es más conveniente que los otros, debido a que es el de más bajo precio.\n\nEn este caso, el costo total del transporte es:\n- transporte de 40 t de \"a\" a \"d\" = 80 monedas;\n- transporte de 20 t de \"c\" a \"e\" = 360 monedas; y,\n- transporte de 40 t de \"b\" a \"e\" = 960 monedas,\n \nPara un total 1.400 monedas.\n\nSin embargo, formulando el problema para ser resuelto por la programación lineal con\n- $x_1$ toneladas transportadas de la mina \"a\" a la central \"d\"\n- $x_2$ toneladas transportadas de la mina \"a\" a la central \"e\"\n- $x_3$ toneladas transportadas de la mina \"b\" a la central \"d\"\n- $x_4$ toneladas transportadas de la mina \"b\" a la central \"e\"\n- $x_5$ toneladas transportadas de la mina \"c\" a la central \"d\"\n- $x_6$ toneladas transportadas de la mina \"c\" a la central \"e\"\n\nse tienen las siguientes ecuaciones:\n\nRestricciones de la producción:\n\n- $x_1 + x_2 \\leq 40$\n- $x_3 + x_4 \\leq 40$\n- $x_5 + x_6 \\leq 20$\n\nRestricciones del consumo:\n\n- $x_1 + x_3 + x_5 \\geq 40$\n- $x_2 + x_4 + x_6 \\geq 60$\n\nLa función objetivo será:\n\n$$\\min_{x_1,\\dots,x_6}2x_1 + 11x_2 + 12x_3 + 24x_4 + 13x_5 + 18x_6$$\n\nResolver con `linprog`",
"_____no_output_____"
]
],
[
[
"# Matrices y cotas\nc = np.array([2, 11, 12, 24, 13, 18])\nA = np.array([[1, 1, 0, 0, 0, 0],\n [0, 0, 1, 1, 0, 0],\n [0, 0, 0, 0, 1, 1],\n [-1, 0, -1, 0, -1, 0],\n [0, -1, 0, -1, 0, -1]])\nb = np.array([40, 40, 20, -40, -60])",
"_____no_output_____"
],
[
"# Resolver\nresultado_transporte = opt.linprog(c, A_ub=A, b_ub=b)",
"_____no_output_____"
],
[
"# Mostrar resultado\nresultado_transporte",
"_____no_output_____"
]
],
[
[
"**Conclusión**\n\n- La estrategia de menor costo es llevar 40 toneladas de la mina \"a\" a la central \"e\", 40 toneladas de la mina \"b\" a la central \"d\" y 20 toneladas de la mina \"c\" a la central \"e\". El costo total de esta estrategia de transporte es 1280 monedas.",
"_____no_output_____"
],
[
"## 7. Optimización de inversión en bonos\n\n**Referencia:**",
"_____no_output_____"
]
],
[
[
"from IPython.display import YouTubeVideo\nYouTubeVideo('gukxBus8lOs')",
"_____no_output_____"
]
],
[
[
"El objetivo de este problema es determinar la mejor estrategia de inversión, dados diferentes tipos de bono, la máxima cantidad que puede ser invertida en cada bono, el porcentaje de retorno y los años de madurez. También hay una cantidad fija de dinero disponible ($\\$750,000$). Por lo menos la mitad de este dinero debe ser invertido en bonos con 10 años o más para la madurez. Se puede invertir un máximo del $25\\%$ de esta cantidad en cada bono. Finalmente, hay otra restricción que no permite usar más de $35\\%$ en bonos de alto riesgo.\n\nExisten seis (6) opciones de inversión con las letras correspondientes $A_i$\n\n1. $A_1$:(Tasa de retorno=$8.65\\%$; Años para la madurez=11, Riesgo=Bajo)\n1. $A_2$:(Tasa de retorno=$9.50\\%$; Años para la madurez=10, Riesgo=Alto)\n1. $A_3$:(Tasa de retorno=$10.00\\%$; Años para la madurez=6, Riesgo=Alto)\n1. $A_4$:(Tasa de retorno=$8.75\\%$; Años para la madurez=10, Riesgo=Bajo)\n1. $A_5$:(Tasa de retorno=$9.25\\%$; Años para la madurez=7, Riesgo=Alto)\n1. $A_6$:(Tasa de retorno=$9.00\\%$; Años para la madurez=13, Riesgo=Bajo)\n\nLo que se quiere entonces es maximizar el retorno que deja la inversión.\n\nEste problema puede ser resuelto con programación lineal. Formalmente, puede ser descrito como:\n\n$$\\max_{A_1,A_2,...,A_6}\\sum^{6}_{i=1} A_iR_i,$$\n\ndonde $A_i$ representa la cantidad invertida en la opción, y $R_i$ representa la tasa de retorno respectiva.",
"_____no_output_____"
],
[
"Plantear restricciones...",
"_____no_output_____"
]
],
[
[
"# Matrices y cotas\n",
"_____no_output_____"
],
[
"# Resolver\n",
"_____no_output_____"
],
[
"# Mostrar resultado\n",
"_____no_output_____"
]
],
[
[
"Recordar que en el problema minimizamos $-\\sum^{6}_{i=1} A_iR_i$. El rendimiento obtenido es entonces:",
"_____no_output_____"
],
[
"**Conclusión**\n\n- ",
"_____no_output_____"
],
[
"## 8. Tarea \n\n### 1. Diseño de la Dieta Óptima\n\nSe quiere producir comida para gatos de la manera más barata, no obstante se debe también asegurar que se cumplan los datos requeridos de analisis nutricional. Por lo que se quiere variar la cantidad de cada ingrediente para cumplir con los estandares nutricionales. Los requisitos que se tienen es que en 100 gramos, se deben tener por lo menos 8 gramos de proteína y 6 gramos de grasa. Así mismo, no se debe tener más de 2 gramos de fibra y 0.4 gramos de sal. \n\nLos datos nutricionales se pueden obtener de la siguiente tabla:\n\nIngrediente|Proteína|Grasa|Fibra|Sal\n:----|----\nPollo| 10.0%|08.0%|00.1%|00.2%\nCarne| 20.0%|10.0%|00.5%|00.5%\nCordero|15.0%|11.0%|00.5%|00.7%\nArroz| 00.0%|01.0%|10.0%|00.2%\nTrigo| 04.0%|01.0%|15.0%|00.8%\nGel| 00.0%|00.0%|00.0%|00.0%\n\nLos costos de cada producto son:\n\nIngrediente|Costo por gramo\n:----|----\nPollo|$\\$$0.013\nCarne|$\\$$0.008\nCordero|$\\$$0.010\nArroz|$\\$$0.002\nTrigo|$\\$$0.005\nGel|$\\$$0.001 \n\nLo que se busca optimizar en este caso es la cantidad de productos que se debe utilizar en la comida de gato, para simplificar la notación se van a nombrar las siguientes variables: \n\n$x_1:$ Gramos de pollo \n$x_2:$ Gramos de carne \n$x_3:$ Gramos de cordero \n$x_4:$ Gramos de arroz \n$x_5:$ Gramos de trigo \n$x_6:$ Gramos de gel \n\nCon los datos, se puede plantear la función objetivo, está dada por la siguiente expresión:\n\n$$\\min 0.013 x_1 + 0.008 x_2 + 0.010 x_3 + 0.002 x_4 + 0.005 x_5 + 0.001 x_6$$\n\nLas restricciones estarían dadas por el siguiente conjunto de ecuaciones:\n\n$x_1+x_2+x_3+x_4+x_5+x_6=100$ \n\n$(10.0 x_1+ 20.0 x_2+ 15.0 x_3+ 00.0 x_4+ 04.0 x_5+ 00.0 x_6)/100 \\geq 8.0$ \n\n$(08.0 x_1+ 10.0 x_2+ 11.0 x_3+ 01.0 x_4+ 01.0 x_5+ 00.0 x_6)/100 \\geq 6.0$ \n\n$(00.1 x_1+ 00.5 x_2+ 00.5 x_3+ 10.0 x_4+ 15.0 x_5+ 00.0 x_6)/100 \\leq 2.0$ \n\n$(00.2 x_1+ 00.5 x_2+ 00.7 x_3+ 00.2 x_4+ 00.8 x_5+ 00.0 x_6)/100 \\leq 0.4$ \n\nLa primer condición asegura que la cantidad de productos que se usará cumple con los 100 gramos. Las siguientes sólo siguen los lineamientos planteados para cumplir con los requisitos nutrimentales. ",
"_____no_output_____"
],
[
"### 2. Otro problema de transporte\n\nReferencia: https://relopezbriega.github.io/blog/2017/01/18/problemas-de-optimizacion-con-python/\n\nSupongamos que tenemos que enviar cajas de cervezas de 2 cervecerías (Modelo y Cuauhtémoc Moctezuma) a 5 bares de acuerdo al siguiente gráfico:\n\n<img style=\"float: center; margin: 0px 0px 15px 15px;\" src=\"https://relopezbriega.github.io/images/Trans_problem.png\" width=\"500px\" height=\"150px\" />\n\nAsimismo, supongamos que nuestro gerente financiero nos informa que el costo de transporte por caja de cada ruta se conforma de acuerdo a la siguiente tabla:",
"_____no_output_____"
]
],
[
[
"import pandas as pd\ninfo = pd.DataFrame({'Bar1': [2, 3], 'Bar2': [4, 1], 'Bar3': [5, 3], 'Bar4': [2, 2], 'Bar5': [1, 3]}, index = ['CerveceriaA', 'CerveceriaB'])\ninfo",
"_____no_output_____"
]
],
[
[
"Y por último, las restricciones del problema, van a estar dadas por las capacidades de oferta y demanda de cada cervecería (en cajas de cerveza) y cada bar, las cuales se detallan en el gráfico de más arriba.",
"_____no_output_____"
],
[
"Sean:\n- $x_i$ cajas transportadas de la cervecería A al Bar $i$,\n- $x_{i+5}$ cajas transportadas de la cervecería B al Bar $i$.\n\nLa tarea consiste en plantear el problema de minimizar el costo de transporte de la forma vista y resolverlo con `linprog`.\n\nDeben crear un notebook de jupyter (archivo .ipynb) y llamarlo Tarea4_ApellidoNombre, y subirlo a moodle.\n\n**Definir fecha**",
"_____no_output_____"
],
[
"<script>\n $(document).ready(function(){\n $('div.prompt').hide();\n $('div.back-to-top').hide();\n $('nav#menubar').hide();\n $('.breadcrumb').hide();\n $('.hidden-print').hide();\n });\n</script>\n\n<footer id=\"attribution\" style=\"float:right; color:#808080; background:#fff;\">\nCreated with Jupyter by Esteban Jiménez Rodríguez.\n</footer>",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
d085044a956a9d4c7874bcf9d8db2a15af25a826 | 5,021 | ipynb | Jupyter Notebook | N19.ipynb | MLNLPAI/mlnlpai-notebooks | 4b22cc4ab756bd858acfbcd238c8bad1cf2a023c | [
"MIT"
] | null | null | null | N19.ipynb | MLNLPAI/mlnlpai-notebooks | 4b22cc4ab756bd858acfbcd238c8bad1cf2a023c | [
"MIT"
] | null | null | null | N19.ipynb | MLNLPAI/mlnlpai-notebooks | 4b22cc4ab756bd858acfbcd238c8bad1cf2a023c | [
"MIT"
] | null | null | null | 27.437158 | 99 | 0.382792 | [
[
[
"from sklearn.model_selection import GridSearchCV\nfrom sklearn.linear_model import LassoLarsCV\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.datasets import make_regression",
"_____no_output_____"
],
[
"X, y = make_regression(n_samples=10000, n_features=30, random_state=42)",
"_____no_output_____"
],
[
"pipeline = Pipeline([\n ('regr', LassoLarsCV())\n])",
"_____no_output_____"
],
[
"params = {\n 'regr__fit_intercept': [True, False],\n 'regr__copy_X': [True, False],\n}",
"_____no_output_____"
],
[
"gs_cv = GridSearchCV(estimator=pipeline, param_grid=params, cv=4, n_jobs=-1)",
"_____no_output_____"
],
[
"gs_cv.fit(X, y)",
"_____no_output_____"
],
[
"print(gs_cv.best_score_)",
"1.0\n"
],
[
"print(gs_cv.best_params_)",
"{'regr__copy_X': True, 'regr__fit_intercept': True}\n"
],
[
"",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0850a0fd6e7e552bde8e61fdd07e10f5dd05d1b | 119,081 | ipynb | Jupyter Notebook | Sports League - Twitter Analysis.ipynb | stevefan4/NLP-Twitter | b74912112c6c6276d1162ece1c5c508f524f4cd1 | [
"MIT"
] | null | null | null | Sports League - Twitter Analysis.ipynb | stevefan4/NLP-Twitter | b74912112c6c6276d1162ece1c5c508f524f4cd1 | [
"MIT"
] | null | null | null | Sports League - Twitter Analysis.ipynb | stevefan4/NLP-Twitter | b74912112c6c6276d1162ece1c5c508f524f4cd1 | [
"MIT"
] | null | null | null | 84.514549 | 2,221 | 0.609081 | [
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport scipy as sp\nimport os\nos.chdir('/Users/steve/GetOldTweets3-0.0.10')\nimport re\nimport nltk\nimport contractions\n\nos.environ['KMP_DUPLICATE_LIB_OK']='True'\n\nfrom sklearn.feature_extraction.text import TfidfVectorizer, CountVectorizer\nfrom sklearn.decomposition import NMF, LatentDirichletAllocation\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.ensemble import RandomForestClassifier, ExtraTreesClassifier\nfrom sklearn.linear_model import SGDClassifier\nfrom sklearn.metrics import classification_report, confusion_matrix, accuracy_score\nfrom sklearn.linear_model import LogisticRegression\n\nfrom sent2vec.vectorizer import Vectorizer\n\nfrom nltk.corpus import stopwords\nfrom vaderSentiment.vaderSentiment import SentimentIntensityAnalyzer\n\nfrom keras.preprocessing.text import Tokenizer\nfrom keras.preprocessing.sequence import pad_sequences\n\n!git clone https://github.com/facebookresearch/fastText.git\n!cd fastText\n!python3 setup.py install\nimport fasttext.util\n\nfrom imblearn.combine import SMOTETomek\n\nfrom gensim.parsing.preprocessing import remove_stopwords, preprocess_string, strip_tags, strip_punctuation, stem_text, preprocess_documents, strip_multiple_whitespaces, strip_non_alphanum, strip_short",
"fatal: destination path 'fastText' already exists and is not an empty directory.\npython3: can't open file 'setup.py': [Errno 2] No such file or directory\n"
],
[
"nltk.download('words')\nwords = set(nltk.corpus.words.words())",
"[nltk_data] Downloading package words to /Users/steve/nltk_data...\n[nltk_data] Package words is already up-to-date!\n"
],
[
"CUSTOM_FILTERS = [lambda x: x.lower(), strip_tags, strip_punctuation, strip_non_alphanum, stem_text,\n remove_stopwords, strip_short]\n\ndef prep(sentence): \n sentence = contractions.fix(sentence)\n split_sentence = sentence.split()\n return preprocess_string(sentence, CUSTOM_FILTERS)\n\ndef prep_withspell(sentence):\n holder = \" \"\n sentence = contractions.fix(sentence)\n preprocessed = preprocess_string(sentence, CUSTOM_FILTERS)\n spell_correct = TextBlob(holder.join(preprocessed))\n corrected_sentence = spell_correct.correct()\n return corrected_sentence.split() \n\ndef prepare(sentence): \n processed_feature = re.sub(r'\\W', ' ', str(sentence))\n processed_feature= re.sub(r'\\s+[a-zA-Z]\\s+', ' ', processed_feature)\n processed_feature = re.sub(r'\\^[a-zA-Z]\\s+', ' ', processed_feature) \n processed_feature = re.sub(r'\\s+', ' ', processed_feature, flags=re.I)\n processed_feature = re.sub(r'^b\\s+', '', processed_feature)\n processed_feature = processed_feature.lower()\n stop_words = set(stopwords.words('english'))\n add = ['football', 'league', 'soccer', 'super', 'super league', 'footbal', 'leagu', 'footbal soccer']\n stop_words = set.union(stop_words, add)\n sentence = processed_feature.split()\n return [w for w in sentence if not w.lower() in stop_words] ",
"_____no_output_____"
],
[
"def cont_to_multiclass(cont):\n if cont > 0.66:\n return 'highly positive'\n elif cont > 0.33:\n return 'positive'\n elif cont > 0:\n return 'partly positive'\n elif cont > -0.33:\n return 'partly negative'\n elif cont > -0.66:\n return 'negative'\n else:\n return 'highly negative'\n\ndef cont_to_binary(cont):\n if cont > 0:\n #return 'positive'\n return 1\n else:\n #return 'negative'\n return 0",
"_____no_output_____"
],
[
"def get_document_frequency(data, wi, wj=None):\n if wj is None:\n D_wi = 0\n for l in range(len(data)):\n doc = data[l]\n if wi in doc:\n D_wi += 1\n return D_wi\n D_wj = 0\n D_wi_wj = 0\n for l in range(len(data)):\n doc = data[l]\n if wj in doc:\n D_wj += 1\n if wi in doc:\n D_wi_wj += 1\n return D_wj, D_wi_wj\n\ndef get_topic_coherence(beta, data, vocab, seed):\n D = len(data)\n TC = []\n num_topics = len(beta.components_)\n selected = -1\n selected = []\n for k, topic in enumerate(beta.components_):\n print('k: {}/{}'.format(k, num_topics))\n top_10 = topic.argsort()[:-20 - 1:-1]\n top_words = [vocab[i] for i in top_10]\n print(top_words)\n TC_k = 0\n counter = 0\n for i, word in enumerate(top_words):\n D_wi = get_document_frequency(data, word)\n j = i + 1\n tmp = 0\n while j < len(top_10) and j > i:\n D_wj, D_wi_wj = get_document_frequency(data, word, top_words[j])\n if D_wi_wj == 0:\n f_wi_wj = -1\n else:\n f_wi_wj = -1 + ( np.log(D_wi) + np.log(D_wj) - 2.0 * np.log(D) ) / ( np.log(D_wi_wj) - np.log(D) )\n tmp += f_wi_wj\n j += 1\n counter += 1\n TC_k += tmp \n TC.append(TC_k)\n print('num topics: ', len(TC))\n print('Topic Coherence is: {}'.format(TC))\n return TC, selected",
"_____no_output_____"
],
[
"def sentiment_scores(sentence):\n sid_obj = SentimentIntensityAnalyzer()\n sentiment_dict = sid_obj.polarity_scores(sentence)\n return sentiment_dict['compound']",
"_____no_output_____"
],
[
"def word_vector(model, tokens, size):\n vec = np.zeros(size).reshape((1, size))\n count = 0\n for word in tokens:\n try:\n vec += model.wv[word].reshape((1, size))\n count += 1.\n except KeyError: # handling the case where the token is not in vocabulary\n continue\n if count != 0:\n vec /= count\n return vec",
"_____no_output_____"
],
[
"sl_df = pd.read_csv('SL.csv', usecols = ['content'])\nsl_df = sl_df.rename(columns={\"content\": 0})\ndataset = sl_df.drop_duplicates()",
"_____no_output_____"
],
[
"dataset[1] = dataset.apply(lambda row: re.sub(r'http\\S+', '', str(row[0])), axis=1)\ndataset[2] = dataset.apply(lambda row: ''.join([c for c in row[1] if not c.isdigit()]), axis=1)\ndataset[3] = dataset.apply(lambda row : \" \".join(w for w in nltk.wordpunct_tokenize(row[2]) if w.lower() in words or not w.isalpha()), axis = 1)\ndataset[4] = dataset.apply(lambda row : prep(row[3]), axis = 1)\ndataset[5] = dataset.apply(lambda row : prepare(row[3]), axis = 1)\n\nprocessed_data = [\" \".join(x) for x in dataset[4]]",
"<ipython-input-53-aa608e041004>:1: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n dataset[1] = dataset.apply(lambda row: re.sub(r'http\\S+', '', str(row[0])), axis=1)\n<ipython-input-53-aa608e041004>:2: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n dataset[2] = dataset.apply(lambda row: ''.join([c for c in row[1] if not c.isdigit()]), axis=1)\n<ipython-input-53-aa608e041004>:3: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n dataset[3] = dataset.apply(lambda row : \" \".join(w for w in nltk.wordpunct_tokenize(row[2]) if w.lower() in words or not w.isalpha()), axis = 1)\n<ipython-input-53-aa608e041004>:4: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n dataset[4] = dataset.apply(lambda row : prep(row[3]), axis = 1)\n<ipython-input-53-aa608e041004>:5: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n dataset[5] = dataset.apply(lambda row : prepare(row[3]), axis = 1)\n"
],
[
"tfidf = TfidfVectorizer(max_df=0.90, min_df=50, stop_words='english', ngram_range=(1,3))\ndtm = tfidf.fit_transform(processed_data)\n\nnmf_model = NMF(n_components=50, random_state=42, beta_loss='kullback-leibler', solver='mu', \n max_iter=1000, alpha=2, l1_ratio=0.5)\nnmf_topics = nmf_model.fit_transform(dtm)\n\nfor index, topic in enumerate(nmf_model.components_):\n print(f'THE TOP 50 WORDS FOR TOPIC #{index} GIVEN BY NMF:')\n print([tfidf.get_feature_names()[i] for i in topic.argsort()[-50:]])\n print('\\n')",
"/Users/steve/opt/anaconda3/lib/python3.8/site-packages/sklearn/decomposition/_nmf.py:312: FutureWarning: The 'init' value, when 'init=None' and n_components is less than n_samples and n_features, will be changed from 'nndsvd' to 'nndsvda' in 1.1 (renaming of 0.26).\n warnings.warn((\"The 'init' value, when 'init=None' and \"\n"
],
[
"tf_vectorizer = CountVectorizer(analyzer='word', min_df=50, ngram_range=(1, 3))\ntf_fit = tf_vectorizer.fit_transform(processed_data)\n\nlda_model = LatentDirichletAllocation(n_components=50, doc_topic_prior=.01)\nlda_topics = lda_model.fit_transform(tf_fit)\n\nfor index, topic in enumerate(lda_model.components_):\n print(f'THE TOP 50 WORDS FOR TOPIC #{index} GIVEN BY LDA:')\n print([tf_vectorizer.get_feature_names()[i] for i in topic.argsort()[-50:]])\n print('\\n')",
"THE TOP 50 WORDS FOR TOPIC #0 GIVEN BY LDA:\n['approv', 'multipl', 'formal', 'seek', 'super leagu chang', 'reviv', 'paul tenorio', 'tenorio', 'eat', 'sport', 'leagu chang', 'competit super leagu', 'competit super', 'realli super leagu', 'premier', 'super leagu follow', 'premier leagu', 'paul', 'inter', 'confirm', 'particip super leagu', 'realli super', 'particip super', 'leagu follow', 'offici', 'announc', 'leagu withdraw super', 'mail', 'leagu withdraw competit', 'daili', 'super leagu competit', 'origin', 'withdraw competit', 'kind', 'leagu competit', 'super leagu realli', 'leagu realli', 'super leagu withdraw', 'particip', 'premier leagu withdraw', 'follow', 'realli', 'leagu withdraw', 'withdraw super leagu', 'withdraw super', 'competit', 'super leagu', 'super', 'withdraw', 'leagu']\n\n\nTHE TOP 50 WORDS FOR TOPIC #1 GIVEN BY LDA:\n['someth like', 'thi like', 'imposs', 'everi club', 'act like', 'tax', 'end', 'feel like', 'noth super leagu', 'game', 'super leagu everi', 'noth super', 'super leagu week', 'leagu week', 'far', 'leagu everi', 'singl', 'feel', 'everi year', 'imagin', 'dei', 'someth', 'time', 'coupl', 'year', 'noth', 'act', 'mark', 'super leagu dai', 'leagu dai', 'ago', 'week super leagu', 'week super', 'sure', 'everi week', 'past', 'watch', 'stori', 'like thi', 'thi week', 'like super leagu', 'like super', 'everi', 'dai', 'thi', 'super leagu', 'super', 'like', 'leagu', 'week']\n\n\nTHE TOP 50 WORDS FOR TOPIC #2 GIVEN BY LDA:\n['super leagu long', 'mean', 'leagu long', 'believ super leagu', 'believ super', 'properli', 'fan base', 'glazer famili', 'controversi', 'furiou', 'long time', 'read', 'wait', 'thing', 'explain super leagu', 'explain super', 'doe', 'pass', 'veri', 'time', 'backlash', 'term', 'thi happen', 'awai', 'protest super leagu', 'protest super', 'trust', 'condemn', 'hopefulli', 'super leagu unit', 'angri', 'leagu unit', 'situat', 'famili', 'fan', 'base', 'glazer', 'someon', 'unit', 'explain', 'thi', 'protest', 'super leagu happen', 'leagu happen', 'believ', 'long', 'super leagu', 'super', 'leagu', 'happen']\n\n\nTHE TOP 50 WORDS FOR TOPIC #3 GIVEN BY LDA:\n['footbal leagu', 'leagu morgan', 'someth super leagu', 'super leagu amid', 'someth super', 'bless', 'headlin', 'disguis', 'save super leagu', 'leagu amid', 'save super', 'host', 'super leagu championship', 'fantasi', 'amid', 'footi', 'yeah', 'leagu championship', 'karma', 'wait super leagu', 'wait super', 'super leagu someth', 'super leagu form', 'leagu semi', 'break super', 'break super leagu', 'leagu someth', 'destroi', 'leagu form', 'semi final', 'updat', 'break', 'final', 'super leagu announc', 'leagu announc', 'someth', 'championship', 'premier leagu super', 'semi', 'announc', 'wait', 'form super leagu', 'form super', 'super leagu super', 'form', 'leagu super leagu', 'leagu super', 'super leagu', 'super', 'leagu']\n\n\nTHE TOP 50 WORDS FOR TOPIC #4 GIVEN BY LDA:\n['beg', 'blame super leagu', 'blame super', 'elit', 'end season', 'job', 'band', 'legisl', 'big club', 'yeah', 'forgiv', 'belong', 'right super leagu', 'right super', 'zero', 'basic super leagu', 'basic super', 'deduct', 'good super leagu', 'good super', 'super leagu try', 'wonder thei', 'mayb super leagu', 'shut', 'mayb super', 'leagu try', 'thi', 'consid', 'good', 'big super leagu', 'try', 'big super', 'super leagu big', 'thi season', 'right', 'leagu big', 'basic', 'ban', 'blame', 'wonder', 'stop super leagu', 'stop super', 'thei', 'mayb', 'season', 'stop', 'super leagu', 'super', 'leagu', 'big']\n\n\nTHE TOP 50 WORDS FOR TOPIC #5 GIVEN BY LDA:\n['hard', 'haven', 'brand', 'york', 'super leagu perfect', 'leagu perfect', 'super leagu live', 'super leagu offici', 'leagu live', 'sale', 'leagu offici', 'free', 'statement', 'offici statement', 'super leagu plot', 'leagu plot', 'new', 'live', 'power', 'peopl super leagu', 'super leagu peopl', 'peopl super', 'debat', 'super leagu work', 'leagu peopl', 'thi', 'leagu work', 'time', 'capit', 'seen', 'person', 'global', 'class', 'perfect', 'plot', 'video', 'west ham', 'ham', 'lot', 'left super leagu', 'left super', 'west', 'mani', 'offici', 'left', 'work', 'super leagu', 'super', 'leagu', 'peopl']\n\n\nTHE TOP 50 WORDS FOR TOPIC #6 GIVEN BY LDA:\n['tell super', 'twitter', 'breakawai leagu', 'turn', 'excit', 'abus', 'onli footbal', 'racism', 'anyth', 'better', 'feel', 'tell', 'season super leagu', 'season super', 'super leagu sure', 'footbal', 'mention', 'leagu sure', 'thi team', 'look forward', 'sure', 'sick', 'onli', 'super leagu season', 'wai super leagu', 'wai super', 'look', 'covid', 'opposit', 'wai', 'moment', 'leagu season', 'super leagu talk', 'leagu talk', 'nice', 'unit super leagu', 'unit super', 'team super leagu', 'team super', 'season', 'talk', 'forward', 'breakawai super leagu', 'breakawai super', 'breakawai', 'unit', 'super leagu', 'super', 'leagu', 'team']\n\n\nTHE TOP 50 WORDS FOR TOPIC #7 GIVEN BY LDA:\n['soccer leagu', 'project proce', 'arsen thei', 'arsen arsen', 'credit', 'dure super', 'dure super leagu', 'fault', 'new post', 'project', 'dirti dozen', 'thei said', 'super leagu chairman', 'leagu chairman', 'said thei', 'dozen', 'proce', 'post super', 'post super leagu', 'reveal endors super', 'super leagu said', 'reveal endors', 'chairman', 'endors super leagu', 'endors super', 'leagu said', 'new', 'dirti', 'endors', 'super leagu suspend', 'super leagu soccer', 'leagu suspend', 'dure', 'leagu soccer', 'bank', 'reveal', 'said super leagu', 'said super', 'suspend', 'soccer super leagu', 'soccer super', 'post', 'arsen super leagu', 'arsen super', 'said', 'soccer', 'super leagu', 'super', 'leagu', 'arsen']\n\n\nTHE TOP 50 WORDS FOR TOPIC #8 GIVEN BY LDA:\n['leagu busi', 'leagu alwai', 'time', 'thei onli', 'shock', 'shirt', 'titl', 'million', 'year', 'sport', 'busi', 'success', 'peopl', 'monei', 'chanc', 'oil', 'challeng', 'onli thing', 'becaus', 'win leagu', 'thing', 'super leagu win', 'trophi', 'onli wai', 'leagu win', 'minut', 'wigan', 'young', 'win super leagu', 'hull', 'win super', 'wai', 'super leagu won', 'manag', 'answer', 'thi', 'leagu won', 'alwai', 'problem', 'onli super leagu', 'onli super', 'greedi', 'super leagu onli', 'leagu onli', 'won', 'super leagu', 'super', 'leagu', 'win', 'onli']\n\n\nTHE TOP 50 WORDS FOR TOPIC #9 GIVEN BY LDA:\n['leagu materi', 'actual super leagu', 'actual super', 'gave', 'sell', 'leagu save footbal', 'enemi', 'greed super leagu', 'greed super', 'born', 'review', 'super leagu world', 'attempt', 'controversi super leagu', 'controversi super', 'world super leagu', 'aliv', 'greed', 'world super', 'materi', 'try super', 'try super leagu', 'exactli', 'world footbal', 'worri', 'club', 'cancel', 'associ', 'super leagu save', 'leagu world', 'stage', 'leagu save', 'try', 'super leagu actual', 'leagu actual', 'footbal world', 'controversi', 'rest', 'world cup', 'saw', 'cup', 'save footbal', 'stand', 'footbal', 'save', 'actual', 'super leagu', 'super', 'leagu', 'world']\n\n\nTHE TOP 50 WORDS FOR TOPIC #10 GIVEN BY LDA:\n['bryan', 'try join', 'skysport', 'consult', 'secret', 'appropri', 'leagu biggest', 'chang super leagu', 'chang super', 'sack', 'becaus', 'ref', 'thei come', 'perman', 'nation', 'common', 'current', 'super leagu join', 'reshap project', 'prepar', 'earn', 'forev', 'defend', 'leagu join', 'accord', 'project', 'replac', 'join', 'oppos super leagu', 'oppos super', 'try', 'reshap', 'come', 'small', 'thei', 'domest', 'rule', 'given', 'chang', 'levi', 'oppos', 'super leagu plan', 'leagu plan', 'fact', 'sens', 'biggest', 'plan', 'super leagu', 'super', 'leagu']\n\n\nTHE TOP 50 WORDS FOR TOPIC #11 GIVEN BY LDA:\n['super leagu mean', 'releas', 'super leagu bet', 'morgan super leagu', 'leagu bet', 'morgan super', 'leagu mean', 'turn super leagu', 'thank', 'super leagu mayb', 'turn super', 'leagu mayb', 'apolog super leagu', 'apolog super', 'creation', 'apolog', 'super leagu turn', 'argument', 'mayb', 'won super leagu', 'leagu turn', 'won super', 'scrap', 'statement super leagu', 'statement super', 'thi', 'meant', 'sport super leagu', 'won', 'sport super', 'know super leagu', 'mean', 'know super', 'sport', 'better super leagu', 'better super', 'bit', 'stupid', 'becaus super leagu', 'becaus super', 'bet', 'morgan', 'statement', 'know', 'becaus', 'better', 'turn', 'super leagu', 'super', 'leagu']\n\n\nTHE TOP 50 WORDS FOR TOPIC #12 GIVEN BY LDA:\n['hall', 'deal', 'doe super', 'doe super leagu', 'poor', 'super leagu invit', 'mad', 'leagu releg', 'leagu invit', 'fixtur', 'old', 'offici', 'wan', 'offici super leagu', 'offici super', 'want', 'doe', 'firm', 'plai leagu', 'thi arsen', 'want plai', 'founder', 'promot releg', 'super leagu plai', 'sinc super leagu', 'compet', 'sinc super', 'team', 'home', 'thei plai', 'thi', 'leagu plai', 'super leagu wai', 'leagu wai', 'invit', 'spot', 'promot', 'arsen', 'draw', 'wai', 'rich', 'tonight', 'sinc', 'plai super leagu', 'plai super', 'releg', 'super leagu', 'super', 'plai', 'leagu']\n\n\nTHE TOP 50 WORDS FOR TOPIC #13 GIVEN BY LDA:\n['rememb super', 'reaction', 'consid', 'dead water', 'justic', 'wonder super leagu', 'wonder super', 'bulli', 'super leagu sinc', 'buri', 'thi', 'super leagu pull', 'happi super leagu', 'leagu sinc', 'happi super', 'wonder', 'type', 'total', 'super leagu look', 'everyon super leagu', 'everyon super', 'goe', 'leagu look', 'point super leagu', 'look', 'point super', 'sinc', 'leagu pull', 'water', 'anger', 'everyon', 'dai super leagu', 'dai super', 'terribl', 'exist', 'pull super', 'pull super leagu', 'dai', 'life', 'worst', 'rememb', 'super leagu dead', 'leagu dead', 'happi', 'point', 'pull', 'dead', 'super leagu', 'super', 'leagu']\n\n\nTHE TOP 50 WORDS FOR TOPIC #14 GIVEN BY LDA:\n['sorri super', 'sorri super leagu', 'arsen owner', 'lead', 'fee', 'دوري السوبر', 'told', 'protest', 'دوري', 'utter', 'fan', 'super leagu goal', 'super leagu statement', 'super leagu ars', 'السوبر', 'leagu ars', 'leagu goal', 'leagu statement', 'white', 'sell club', 'super leagu pai', 'bind', 'thei leav', 'leagu pai', 'fine', 'bui', 'billionair', 'million', 'goal', 'pep', 'ars', 'statement', 'sell', 'stadium', 'score', 'mistak', 'owner', 'contract', 'pai', 'arsen', 'sorri', 'leav super leagu', 'leav super', 'super leagu club', 'leagu club', 'leav', 'super leagu', 'super', 'leagu', 'club']\n\n\nTHE TOP 50 WORDS FOR TOPIC #15 GIVEN BY LDA:\n['leagu group', 'leagu basic', 'ani', 'try', 'flat', 'leagu leav real', 'super leagu ruin', 'leagu ruin', 'group', 'thei deserv', 'basic', 'deserv super leagu', 'deserv super', 'leav real presid', 'themselv', 'ruin footbal', 'enter super leagu', 'enter super', 'real presid super', 'leav real', 'thei sai', 'thei won', 'footbal', 'thei think', 'super leagu sai', 'leagu sai', 'direct', 'won', 'super leagu leav', 'leagu leav', 'sai thei', 'enter', 'anyon', 'real', 'presid super leagu', 'presid super', 'ruin', 'deserv', 'think thei', 'leav', 'real presid', 'think super leagu', 'think super', 'thei', 'sai', 'presid', 'super leagu', 'super', 'leagu', 'think']\n\n\nTHE TOP 50 WORDS FOR TOPIC #16 GIVEN BY LDA:\n['fact', 'murder', 'chief', 'woke feel', 'leagu wors', 'verdict', 'fold', 'super leagu close', 'super leagu night', 'offic', 'defenc', 'super leagu thought', 'leagu night', 'polic', 'leagu close', 'leagu thought', 'attend', 'tell', 'smart', 'onli reason', 'guilti', 'thought', 'confirm', 'woke', 'wors super leagu', 'wors super', 'danger', 'initi', 'book', 'line', 'reason super leagu', 'reason super', 'thi', 'extra', 'super leagu final', 'feel', 'leagu final', 'pretti', 'perform', 'everyth', 'effect', 'final', 'decid', 'close', 'wors', 'night', 'reason', 'super leagu', 'super', 'leagu']\n\n\nTHE TOP 50 WORDS FOR TOPIC #17 GIVEN BY LDA:\n['knew', 'stop thi', 'thi gui', 'luck', 'noth', 'sai thi', 'wai thei', 'club', 'thi leagu', 'end', 'thi club', 'know thei', 'ani', 'everyon', 'right', 'moral', 'super leagu start', 'thi year', 'summer', 'like', 'try', 'leagu start', 'thei thi', 'thi thei', 'becaus', 'year', 'peopl', 'come', 'think thi', 'hope', 'sai', 'realli', 'start', 'wai', 'gui', 'think', 'super leagu thing', 'leagu thing', 'know', 'love', 'thei', 'super leagu thi', 'leagu thi', 'thing', 'thi super leagu', 'thi super', 'super leagu', 'super', 'leagu', 'thi']\n\n\nTHE TOP 50 WORDS FOR TOPIC #18 GIVEN BY LDA:\n['leagu posit', 'super leagu disast', 'guardian sport', 'leagu disast', 'cover', 'great', 'jordan', 'revers', 'sell super', 'sell super leagu', 'sport', 'arsen', 'intent', 'respons super leagu', 'respons super', 'super leagu finish', 'leagu finish', 'josh', 'led', 'public', 'fan super leagu', 'super leagu veri', 'fan super', 'alreadi super leagu', 'alreadi super', 'defeat', 'leagu veri', 'alreadi', 'race', 'silenc', 'disast', 'super leagu backlash', 'leagu backlash', 'fan', 'guardian', 'sell', 'taken', 'posit', 'veri', 'respons', 'backlash', 'super leagu leagu', 'finish', 'leagu leagu', 'super leagu fiasco', 'leagu fiasco', 'fiasco', 'super leagu', 'super', 'leagu']\n\n\nTHE TOP 50 WORDS FOR TOPIC #19 GIVEN BY LDA:\n['appli', 'dai', 'thei thought', 'come', 'wast', 'tackl', 'everyon', 'time thei', 'racism footbal', 'becaus', 'good thing', 'thei realli', 'super leagu pleas', 'tactic', 'leagu pleas', 'someth', 'wish', 'thi weekend', 'peopl', 'abus', 'thing super leagu', 'effort', 'thing super', 'thought thei', 'thi time', 'boycott', 'thi morn', 'weekend', 'thing', 'footbal', 'social media', 'energi', 'realli', 'thought', 'pleas', 'morn', 'fight', 'social', 'good', 'time', 'probabl', 'thi', 'racism', 'media', 'thei super leagu', 'thei super', 'super leagu', 'super', 'leagu', 'thei']\n\n\nTHE TOP 50 WORDS FOR TOPIC #20 GIVEN BY LDA:\n['fiasco super', 'ahead super', 'super leagu becom', 'sport merit', 'fate super', 'fate super leagu', 'beat super leagu', 'beat super', 'offer', 'leagu becom', 'sport', 'ill fate', 'boss super leagu', 'boss super', 'revolt', 'beat', 'grab', 'footbal', 'necessari', 'game', 'wealthi', 'merit', 'push super leagu', 'push super', 'rebel', 'fate', 'spoke', 'rip super leagu', 'rip super', 'ill', 'super leagu fan', 'leagu fan', 'light', 'fan', 'rip', 'super leagu propos', 'push', 'leagu propos', 'boss', 'becom', 'super leagu monei', 'leagu monei', 'monei', 'propos', 'super leagu team', 'leagu team', 'team', 'super leagu', 'super', 'leagu']\n\n\nTHE TOP 50 WORDS FOR TOPIC #21 GIVEN BY LDA:\n['super leagu anywai', 'sue', 'geld', 'super leagu het', 'leagu anywai', 'happen super leagu', 'happen super', 'dit', 'leagu het', 'super leagu doe', 'van het', 'kan', 'watch', 'vote', 'leagu doe', 'het van', 'super leagu watch', 'super leagu che', 'ben', 'wat', 'dan', 'leagu watch', 'leagu che', 'door', 'super leagu know', 'know', 'doe', 'leagu know', 'van super leagu', 'van super', 'come super leagu', 'nog', 'che super leagu', 'anywai', 'come super', 'che super', 'super leagu non', 'leagu non', 'non super leagu', 'non super', 'super leagu come', 'met', 'leagu come', 'non', 'het', 'van', 'come', 'super leagu', 'super', 'leagu']\n\n\nTHE TOP 50 WORDS FOR TOPIC #22 GIVEN BY LDA:\n['super leagu mud', 'den super leagu', 'nun', 'den super', 'leagu den', 'mir', 'leagu continu', 'ich die', 'super super leagu', 'peter', 'die den', 'leagu reform', 'geld', 'sport', 'man', 'bin', 'pro super leagu', 'pro super', 'super super', 'mud', 'super leagu ist', 'leagu ist', 'game', 'die ist', 'continu', 'wir', 'ist die', 'sind', 'super leagu game', 'wird', 'super leagu die', 'sie', 'leagu game', 'mal', 'nach', 'leagu die', 'pro', 'reform', 'war', 'die die', 'hat', 'ich', 'die super leagu', 'die super', 'den', 'ist', 'super leagu', 'leagu', 'super', 'die']\n\n\nTHE TOP 50 WORDS FOR TOPIC #23 GIVEN BY LDA:\n['mort', 'sport', 'doit', 'petit', 'jour', 'toi', 'sera', 'grand', 'parl', 'gen', 'club', 'aura', 'mon', 'chose', 'son', 'dire', 'car', 'gro', 'super leagu pour', 'leagu pour', 'bon', 'dit', 'encor', 'ell', 'footbal', 'collaps super leagu', 'collaps super', 'san', 'super leagu collaps', 'leagu collaps', 'coup', 'bring super leagu', 'bring super', 'pour super leagu', 'argent', 'pour super', 'actufoot', 'bien', 'par', 'nou', 'bring', 'sur', 'tout', 'foot', 'plu', 'collaps', 'pour', 'super leagu', 'leagu', 'super']\n\n\nTHE TOP 50 WORDS FOR TOPIC #24 GIVEN BY LDA:\n['demis super', 'elit', 'announc', 'version super leagu', 'version super', 'set', 'live super leagu', 'live super', 'super leagu noth', 'leagu noth', 'dai befor super', 'learn', 'noth', 'super leagu end', 'bare', 'super leagu launch', 'met', 'leagu launch', 'live', 'leagu end', 'demis', 'han', 'end super leagu', 'end super', 'chief', 'unit', 'short', 'thought super leagu', 'thought super', 'main', 'til', 'street', 'dai befor', 'version', 'meet', 'befor super leagu', 'befor super', 'woodward', 'men', 'launch', 'dai', 'super leagu debacl', 'leagu debacl', 'thought', 'debacl', 'end', 'befor', 'super leagu', 'super', 'leagu']\n\n\nTHE TOP 50 WORDS FOR TOPIC #25 GIVEN BY LDA:\n['want thi', 'leagu anoth', 'leagu thi week', 'fall apart', 'apart', 'super leagu everyon', 'rise fall super', 'peopl', 'leagu everyon', 'fan power', 'thei want super', 'everyon els', 'anyon', 'anoth', 'super leagu fall', 'piec', 'leagu fall', 'rise fall', 'withdrawn super', 'withdrawn super leagu', 'clear', 'super leagu befor', 'tho', 'fan', 'leagu befor', 'thi', 'bye', 'befor', 'fall super leagu', 'fall super', 'question', 'withdrawn', 'thei', 'super leagu want', 'start super leagu', 'everyon', 'start super', 'thei want', 'rise', 'leagu want', 'els', 'power', 'fall', 'want super leagu', 'want super', 'start', 'super leagu', 'super', 'leagu', 'want']\n\n\nTHE TOP 50 WORDS FOR TOPIC #26 GIVEN BY LDA:\n['equal', 'reduc', 'pai', 'season', 'million', 'player', 'structur', 'think', 'increas', 'cartel', 'better', 'mani', 'super leagu mai', 'leagu mai', 'inevit', 'revenu', 'pandem', 'similar', 'fair plai', 'cap', 'format super leagu', 'wai', 'format super', 'price', 'wage', 'current', 'salari', 'number', 'deal', 'simpli', 'issu', 'super leagu time', 'transfer', 'ticket', 'leagu time', 'normal', 'cash', 'monei', 'thi', 'new format', 'financi', 'fair', 'greed', 'mai', 'new', 'super leagu', 'super', 'time', 'leagu', 'format']\n\n\nTHE TOP 50 WORDS FOR TOPIC #27 GIVEN BY LDA:\n['tweet', 'imagin thi', 'realli', 'space', 'reckon', 'import', 'par super leagu', 'possibl', 'par super', 'abort', 'plu argent', 'mil', 'join thi', 'drop super', 'drop super leagu', 'cool', 'curs', 'mean super leagu', 'mean super', 'super leagu differ', 'mean', 'bid', 'bar', 'leagu differ', 'super leagu par', 'utd', 'leagu par', 'par', 'differ super leagu', 'differ super', 'plu super leagu', 'plu super', 'affect', 'wow', 'super leagu plu', 'suppos', 'disgrac', 'leagu plu', 'drop', 'plu', 'differ', 'super leagu footbal', 'footbal', 'leagu footbal', 'thi super leagu', 'thi super', 'thi', 'super leagu', 'super', 'leagu']\n\n\nTHE TOP 50 WORDS FOR TOPIC #28 GIVEN BY LDA:\n['crack', 'protect', 'govern', 'bodi', 'wild', 'porto', 'interview', 'plan super leagu', 'plan super', 'wall', 'rival', 'court', 'hard', 'given', 'super leagu farc', 'work super leagu', 'leagu farc', 'work super', 'south', 'lower', 'thi', 'union', 'uproar', 'stai super leagu', 'stai super', 'especi', 'north', 'unlik', 'radio', 'earli', 'rubbish', 'close shop', 'rate', 'doubl', 'farc', 'close', 'onc', 'work', 'shop', 'legal', 'bore', 'hit', 'liter', 'sort', 'huge', 'stai', 'lost', 'super leagu', 'super', 'leagu']\n\n\nTHE TOP 50 WORDS FOR TOPIC #29 GIVEN BY LDA:\n['smaller', 'lot', 'alwai', 'point', 'thei care', 'thei monei', 'won', 'themselv', 'wai', 'thei got', 'got', 'thei try', 'financi', 'leagu becaus thei', 'ani', 'run', 'afford', 'competit', 'sell', 'alreadi', 'pictur', 'lose', 'billion', 'invest', 'like', 'monei thei', 'monei super leagu', 'monei super', 'try', 'thei thei', 'super leagu becaus', 'pai', 'leagu becaus', 'thei want', 'bui', 'spend', 'care', 'debt', 'club', 'want', 'big', 'becaus thei', 'becaus', 'super leagu', 'super', 'super leagu thei', 'leagu thei', 'leagu', 'monei', 'thei']\n\n\nTHE TOP 50 WORDS FOR TOPIC #30 GIVEN BY LDA:\n['becaus', 'understand', 'littl', 'realli', 'footbal thi', 'come', 'time', 'ani', 'greedi', 'greed', 'voic', 'countri', 'monei', 'look like', 'noth', 'pyramid', 'local', 'passion', 'chang', 'love', 'import', 'game', 'power', 'histori', 'govern', 'corrupt', 'watch', 'polit', 'veri', 'super leagu nonsens', 'footbal club', 'leagu nonsens', 'commun', 'wai', 'togeth', 'kick', 'busi', 'fan', 'club', 'super leagu', 'super', 'leagu', 'peopl', 'tri', 'nonsens', 'like', 'sport', 'look', 'thi', 'footbal']\n\n\nTHE TOP 50 WORDS FOR TOPIC #31 GIVEN BY LDA:\n['wrong super', 'refere', 'super leagu wrong', 'leagu wrong', 'wai', 'time', 'statu quo', 'unit', 'quo', 'super leagu kill', 'kill footbal', 'leagu kill', 'date', 'leagu involv', 'super leagu involv', 'love', 'bring', 'involv', 'everybodi', 'red card', 'understand', 'entir', 'letter', 'beauti game', 'row', 'thi game', 'demand', 'statu', 'card', 'got', 'appar', 'thi', 'beauti', 'includ', 'enjoi', 'super leagu think', 'footbal', 'leagu think', 'rid', 'matter', 'think', 'head', 'open', 'kill', 'red', 'wrong', 'super leagu', 'super', 'leagu', 'game']\n\n\nTHE TOP 50 WORDS FOR TOPIC #32 GIVEN BY LDA:\n['thei talk', 'dah', 'para', 'exit super leagu', 'exit super', 'mid tabl', 'stock', 'leagu year', 'understand super leagu', 'understand super', 'necess', 'bola', 'mode', 'leagu tabl', 'option', 'hate super leagu', 'hate super', 'understand', 'super leagu failur', 'leagu failur', 'favor', 'mid', 'dari super leagu', 'dari super', 'exit', 'add', 'inter super leagu', 'super leagu better', 'inter super', 'leagu better', 'ass', 'better', 'super leagu sport', 'definit', 'leagu sport', 'focu', 'yang', 'failur', 'dari', 'dan', 'hate', 'tabl', 'talk super leagu', 'talk super', 'sport', 'inter', 'talk', 'super leagu', 'super', 'leagu']\n\n\nTHE TOP 50 WORDS FOR TOPIC #33 GIVEN BY LDA:\n['san super', 'knew super leagu', 'knew super', 'thi project', 'super leagu coup', 'leagu coup', 'punto', 'sanction', 'ani club', 'super leagu sur', 'match super leagu', 'coup', 'match super', 'set super leagu', 'set super', 'leagu sur', 'set', 'super leagu match', 'quitter', 'tour', 'regard super leagu', 'reaction super', 'reaction super leagu', 'regard super', 'super leagu ani', 'leagu match', 'leagu ani', 'readi', 'ani', 'knew', 'time super leagu', 'time super', 'ser', 'time', 'reaction', 'sur super leagu', 'sur super', 'regard', 'son', 'sur', 'super leagu project', 'leagu project', 'club super leagu', 'club super', 'club', 'match', 'project', 'super leagu', 'super', 'leagu']\n\n\nTHE TOP 50 WORDS FOR TOPIC #34 GIVEN BY LDA:\n['invit super leagu', 'invit super', 'super leagu nou', 'super leagu laugh', 'jag', 'super leagu foot', 'leagu nou', 'leagu foot', 'leagu laugh', 'invit', 'carabao', 'hear super leagu', 'hear super', 'clash', 'mot', 'owner super', 'owner super leagu', 'stop', 'final super leagu', 'super leagu longer', 'final super', 'stick', 'super leagu stop', 'goal super leagu', 'goal super', 'leagu longer', 'till', 'welcom', 'leagu stop', 'man super leagu', 'cup final', 'man super', 'leagu cup', 'citi', 'och', 'citi super leagu', 'citi super', 'man', 'laugh', 'sack', 'longer super leagu', 'longer super', 'hear', 'longer', 'goal', 'cup', 'final', 'super leagu', 'super', 'leagu']\n\n\nTHE TOP 50 WORDS FOR TOPIC #35 GIVEN BY LDA:\n['lose super', 'everyon', 'super leagu instead', 'super leagu run', 'possibl', 'arriv', 'night', 'noth', 'leagu run', 'leagu instead', 'lose', 'ask', 'super leagu support', 'said', 'els', 'leagu support', 'fight super leagu', 'fight super', 'run', 'fight', 'sound', 'mate', 'agre super', 'agre super leagu', 'befor', 'thei', 'member', 'game super leagu', 'game super', 'anyth', 'instead', 'leagu fell apart', 'super leagu fell', 'game', 'leagu fell', 'like thei', 'fell apart', 'guess', 'support super leagu', 'support super', 'fell', 'apart', 'super leagu like', 'agre', 'leagu like', 'support', 'super leagu', 'super', 'leagu', 'like']\n\n\nTHE TOP 50 WORDS FOR TOPIC #36 GIVEN BY LDA:\n['wake super', 'luke', 'despit super leagu', 'despit super', 'soon', 'barcelona presid', 'super leagu lot', 'lot', 'boss', 'leagu real barcelona', 'inter', 'shaw', 'leagu lot', 'remain super', 'remain super leagu', 'error', 'scheme', 'super leagu despit', 'leagu despit', 'super leagu barcelona', 'leagu barcelona', 'super leagu face', 'forget super leagu', 'forget super', 'leagu face', 'wake', 'histor', 'presid', 'barcelona super leagu', 'barcelona super', 'real real', 'return', 'forget', 'bad', 'bad super leagu', 'bad super', 'barcelona real', 'remain', 'despit', 'super leagu real', 'real super leagu', 'real super', 'real barcelona', 'leagu real', 'face', 'barcelona', 'super leagu', 'super', 'leagu', 'real']\n\n\nTHE TOP 50 WORDS FOR TOPIC #37 GIVEN BY LDA:\n['club', 'club non', 'super leagu quit', 'dare', 'imag', 'unravel', 'super leagu qualiti', 'super leagu mess', 'super leagu took', 'leagu qualiti', 'leagu mess', 'duti', 'worthi', 'gather', 'leagu took', 'train ground', 'leagu quit', 'serv', 'tire', 'campo', 'che come', 'protest', 'nei', 'psg', 'parma', 'leagu bad idea', 'pure', 'fin', 'penalti', 'refus', 'captain', 'ball', 'insid', 'bad idea', 'idea', 'account', 'mess', 'qualiti', 'train', 'ground', 'fine', 'super leagu bad', 'leagu bad', 'took', 'quit', 'twitter', 'super leagu', 'super', 'leagu', 'bad']\n\n\nTHE TOP 50 WORDS FOR TOPIC #38 GIVEN BY LDA:\n['plenti', 'new footbal', 'boot', 'franchis', 'poll', 'colleg footbal super', 'beauti', 'review', 'fan', 'strang', 'industri', 'privat', 'school', 'state', 'breakawai', 'equival', 'prime', 'dai footbal', 'german', 'law', 'music', 'footbal leagu', 'tori', 'cricket', 'govern', 'dai', 'quit super leagu', 'quit super', 'basketbal', 'brought', 'abandon', 'thi footbal', 'colleg footbal', 'quit', 'super leagu alreadi', 'colleg', 'fun', 'dream', 'leagu alreadi', 'ownership', 'creat super leagu', 'creat super', 'creat', 'alreadi', 'footbal super leagu', 'footbal super', 'super leagu', 'super', 'leagu', 'footbal']\n\n\nTHE TOP 50 WORDS FOR TOPIC #39 GIVEN BY LDA:\n['opposit', 'leagu opposit', 'baru', 'care super leagu', 'format leagu', 'care super', 'comparison', 'chanc', 'gone super leagu', 'gone super', 'bizarr', 'chanc super leagu', 'chanc super', 'super leagu distract', 'shame', 'leagu distract', 'tweet super leagu', 'tweet super', 'super leagu shame', 'leagu shame', 'gone ahead', 'fought', 'footbal tweet', 'miss super leagu', 'miss super', 'lesser', 'super leagu absolut', 'announc', 'leagu absolut', 'announc super leagu', 'absolut', 'announc super', 'actufoot', 'super leagu format', 'actufoot super leagu', 'actufoot super', 'new', 'distract', 'new leagu format', 'super leagu gone', 'leagu gone', 'miss', 'tweet', 'format', 'new leagu', 'leagu format', 'gone', 'super leagu', 'super', 'leagu']\n\n\nTHE TOP 50 WORDS FOR TOPIC #40 GIVEN BY LDA:\n['col', 'tutti che', 'dall', 'che club', 'vive', 'non solo', 'sponsor', 'quasi', 'tempo', 'far', 'aver', 'club che', 'partit', 'dove', 'parol', 'gazzetta', 'visto', 'dice', 'foss', 'male', 'press', 'sport', 'bene', 'confer', 'dire', 'non non', 'dell', 'soldi', 'dal', 'tra', 'prima', 'loro', 'sulla', 'ancora', 'president', 'poi', 'non che', 'inter', 'fare', 'ora', 'chi', 'che che', 'club', 'era', 'tutti', 'come', 'che non', 'solo', 'non', 'che']\n\n\nTHE TOP 50 WORDS FOR TOPIC #41 GIVEN BY LDA:\n['leagu inter', 'preview', 'tune', 'video', 'talk', 'time', 'winner', 'draft', 'anoth super leagu', 'discuss super leagu', 'discuss super', 'anoth super', 'avail', 'special', 'year', 'record', 'second', 'super leagu breakawai', 'leagu breakawai', 'battl', 'latest', 'pod', 'tomorrow', 'round', 'watch', 'articl', 'todai', 'chat', 'sport', 'super leagu drama', 'leagu drama', 'link', 'breakawai', 'check', 'thi', 'anoth', 'qualifi', 'half', 'super leagu new', 'drama', 'leagu new', 'discuss', 'episod', 'read', 'listen', 'live', 'new', 'super leagu', 'super', 'leagu']\n\n\nTHE TOP 50 WORDS FOR TOPIC #42 GIVEN BY LDA:\n['shown', 'champion leagu', 'netbal', 'need chang', 'know', 'opinion super', 'opinion super leagu', 'awai super', 'awai super leagu', 'sir', 'mind super leagu', 'mind super', 'imagin super leagu', 'imagin super', 'footbal need', 'rob', 'convinc', 'scandal', 'remov', 'chang', 'amid super', 'amid super leagu', 'death', 'thi', 'thei', 'walk', 'imagin', 'case', 'contra', 'champion', 'amid', 'ani super leagu', 'ani super', 'fail', 'leagu saga', 'super leagu saga', 'super leagu need', 'thei need', 'mind', 'leagu need', 'opinion', 'saga', 'awai', 'need super leagu', 'need super', 'ani', 'super leagu', 'super', 'leagu', 'need']\n\n\nTHE TOP 50 WORDS FOR TOPIC #43 GIVEN BY LDA:\n['near', 'princ', 'leagu latest', 'super leagu latest', 'super leagu dan', 'unit', 'leagu dan', 'thi', 'super leagu protest', 'leagu protest', 'super leagu concept', 'leagu concept', 'super leagu joke', 'bridg', 'leagu joke', 'fulham', 'result', 'footbal new', 'begin', 'thi new', 'china', 'protest', 'glad super leagu', 'glad super', 'break new', 'break', 'got super leagu', 'got super', 'super leagu right', 'leagu right', 'concept', 'super leagu got', 'speak', 'super leagu stuff', 'leagu stuff', 'leagu got', 'embarrass', 'latest', 'joke', 'beat', 'glad', 'stuff', 'right', 'new super leagu', 'new super', 'got', 'super leagu', 'super', 'leagu', 'new']\n\n\nTHE TOP 50 WORDS FOR TOPIC #44 GIVEN BY LDA:\n['papa', 'crisi', 'betray', 'leagu base', 'arsen unit', 'agenda', 'went ahead', 'leagu titl', 'longer ahead', 'base', 'chief', 'man arsen', 'citi arsen', 'hold', 'fra', 'chairman super leagu', 'chairman super', 'wish', 'thi man', 'arsen man', 'citi unit', 'inter', 'super leagu went', 'leagu went', 'chairman', 'super leagu citi', 'unit arsen', 'leagu citi', 'titl', 'villa', 'super leagu man', 'leagu man', 'super leagu ahead', 'leagu ahead', 'went', 'man unit', 'sai super leagu', 'sai super', 'sai', 'super leagu arsen', 'leagu arsen', 'unit', 'man citi', 'ahead', 'arsen', 'super leagu', 'super', 'leagu', 'citi', 'man']\n\n\nTHE TOP 50 WORDS FOR TOPIC #45 GIVEN BY LDA:\n['super leagu punish', 'super leagu hope', 'remov', 'abandon super leagu', 'super leagu sign', 'abandon super', 'leagu chief', 'clearli', 'premier leagu club', 'warn', 'leagu hope', 'selfish', 'chief', 'leagu sign', 'abandon', 'involv', 'leagu involv', 'german', 'super leagu thank', 'leagu punish', 'threat', 'leagu thank', 'sign', 'breakawai', 'thank god', 'face', 'crap', 'sin', 'follow super', 'follow super leagu', 'punish super leagu', 'punish super', 'follow', 'leagu big', 'step', 'deal', 'premier leagu big', 'hope', 'god', 'big', 'super leagu premier', 'leagu premier leagu', 'leagu premier', 'thank', 'punish', 'super leagu', 'super', 'premier leagu', 'premier', 'leagu']\n\n\nTHE TOP 50 WORDS FOR TOPIC #46 GIVEN BY LDA:\n['leagu apolog', 'place super leagu', 'place super', 'disrupt', 'howev', 'john henri', 'let thi', 'anyth super leagu', 'owner henri super', 'anyth super', 'know', 'relat', 'bottl', 'let super leagu', 'bell', 'sign super leagu', 'sign super', 'flop', 'let super', 'standard', 'forget', 'john', 'thei', 'anyth', 'super leagu place', 'said', 'haven', 'leagu place', 'super leagu best', 'henri super', 'henri super leagu', 'coach', 'super leagu let', 'honest', 'staff', 'leagu best', 'leagu let', 'true', 'owner henri', 'shame', 'sign', 'apolog', 'owner', 'henri', 'place', 'best', 'super leagu', 'super', 'leagu', 'let']\n\n\nTHE TOP 50 WORDS FOR TOPIC #47 GIVEN BY LDA:\n['road', 'todai super', 'group super leagu', 'group super', 'palac', 'dinero', 'leagu chao', 'super leagu chao', 'comment', 'struggl', 'dai', 'action', 'thi idea', 'super leagu came', 'woodward', 'derbi', 'leagu came', 'martin', 'super leagu todai', 'upset', 'leagu todai', 'woodward super leagu', 'woodward super', 'thi', 'hai', 'great idea', 'leagu good idea', 'role super', 'role super leagu', 'chao', 'super leagu great', 'leagu great', 'group', 'good idea', 'role', 'super leagu good', 'leagu good', 'came', 'idea super leagu', 'idea super', 'para', 'super leagu idea', 'leagu idea', 'todai', 'great', 'good', 'super leagu', 'super', 'leagu', 'idea']\n\n\nTHE TOP 50 WORDS FOR TOPIC #48 GIVEN BY LDA:\n['leagu major', 'uefacom', 'decis join super', 'sai', 'wonder', 'club join', 'futur super leagu', 'futur super', 'agre join', 'major', 'deepli regret', 'arsen join super', 'board', 'super leagu decis', 'leagu decis', 'decis join', 'agre', 'sport new', 'arsen join', 'want join super', 'super leagu futur', 'futur footbal', 'deepli', 'fcbarcelona', 'leagu futur', 'super leagu sky', 'club', 'super leagu lose', 'level', 'leagu lose', 'leagu sky', 'thei join super', 'arsen', 'regret', 'want join', 'ridicul', 'thei join', 'thei', 'lose', 'sky sport', 'sport', 'futur', 'decis', 'sky', 'super leagu', 'super', 'join super leagu', 'join super', 'leagu', 'join']\n\n\nTHE TOP 50 WORDS FOR TOPIC #49 GIVEN BY LDA:\n['goat', 'phone', 'leagu break', 'execut vice chairman', 'alwai', 'execut vice', 'lee', 'player', 'outsid', 'club involv', 'chief execut', 'chief', 'reject', 'town', 'club', 'busi', 'footbal', 'king', 'woodward resign', 'nobodi', 'super leagu woodward', 'leagu woodward', 'vice chairman', 'critic', 'anti super leagu', 'anti super', 'mad', 'watch super leagu', 'watch super', 'break awai', 'told', 'vice', 'awai', 'watch', 'execut', 'super leagu becaus', 'leagu becaus', 'unit', 'chairman', 'anti', 'becaus', 'resign', 'involv super leagu', 'involv super', 'break', 'woodward', 'involv', 'super leagu', 'super', 'leagu']\n\n\n"
],
[
"naming={0:'Topic0', 1:'Topic1', 2:'Topic2', 3:'Topic3', 4:'Topic4', 5:'Topic5', 6:'Topic6', 7:'Topic7',\n 8:'Topic8', 9:'Topic9', 10:'Topic10', 11:'Topic11', 12:'Topic12', 13:'Topic13', 14:'Topic14', 15:'Topic15',\n 16:'Topic16', 17:'Topic17', 18:'Topic18', 19:'Topic19', 20:'Topic20', 21:'Topic21', 22:'Topic22', 23:'Topic23',\n 24:'Topic24', 25:'Topic25', 26:'Topic26', 27:'Topic27', 28:'Topic28', 29:'Topic29', 30:'Topic30', 31:'Topic31',\n 32:'Topic32', 33:'Topic33', 34:'Topic34', 35:'Topic35', 36:'Topic36', 37:'Topic37', 38:'Topic38', 39:'Topic39',\n 40:'Topic40', 41:'Topic41', 42:'Topic42', 43:'Topic43', 44:'Topic44', 45:'Topic45', 46:'Topic46', 47:'Topic47',\n 48:'Topic48', 49:'Topic49'}\n\n\ndataset[5] = nmf_topics.argmax(axis=1)\ndataset[5] = dataset[5].map(naming)\ndataset[6] = lda_topics.argmax(axis=1)\ndataset[6] = dataset[6].map(naming)\ndataset[7] = dataset[4].apply(lambda row: ' '.join(row))",
"<ipython-input-60-6a968f997946>:11: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n dataset[5] = nmf_topics.argmax(axis=1)\n<ipython-input-60-6a968f997946>:12: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n dataset[5] = dataset[5].map(naming)\n<ipython-input-60-6a968f997946>:13: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n dataset[6] = lda_topics.argmax(axis=1)\n<ipython-input-60-6a968f997946>:14: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n dataset[6] = dataset[6].map(naming)\n<ipython-input-60-6a968f997946>:15: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n dataset[7] = dataset[4].apply(lambda row: ' '.join(row))\n"
],
[
"lab_num = 20000\nlabeled_sentiment = dataset.sample(n=lab_num)\nlabeled_sentiment[9] = labeled_sentiment.apply(lambda row : sentiment_scores(row[2]), axis = 1)\nlabeled_sentiment[10] = labeled_sentiment[9].apply(lambda row: cont_to_binary(row))\ndataset = dataset.join(labeled_sentiment[10], how='left', lsuffix='_left', rsuffix='_right')",
"_____no_output_____"
],
[
"%%capture\nfasttext.util.download_model('en', if_exists='ignore')\nft_model = fasttext.load_model('cc.en.300.bin')\nft = pd.DataFrame(processed_data, columns=['tweets'])\nft['tweets'] = ft['tweets'].apply(lambda row: ft_model.get_sentence_vector(row))\nft = np.stack(ft['tweets'].to_numpy())",
"_____no_output_____"
],
[
"dtm_ar = dtm.toarray()\ntf_fit_ar = tf_fit.toarray()\nembed_array = np.hstack((dtm_ar, tf_fit_ar, ft))\nembed_array_df = pd.DataFrame(embed_array, index=dataset.index)\nlabeled_sentiment_embeddings = labeled_sentiment.join(embed_array_df, how='left', lsuffix='_left', rsuffix='_right')\nlabeled_sentiment_embeddings.drop(['0_left', '1_left', '2_left', '3_left', '4_left', '5_left'], axis=1, inplace=True)\nlabeled_sentiment_embeddings.drop(['6_left','7_left', '8_left', '9_left'], axis=1, inplace=True)\nprint('here')\n\nsmt = SMOTETomek(random_state=42)\nX_train, X_test, y_train, y_test = train_test_split(labeled_sentiment_embeddings, labeled_sentiment[9], test_size=0.1, random_state=42)\nX_train, y_train = smt.fit_resample(X_train, y_train)",
"_____no_output_____"
],
[
"X_train_dtm = X_train[X_train.columns[0:dtm.shape[1]]]\nX_train_tf = X_train[X_train.columns[dtm.shape[1]:(dtm.shape[1]+tf_fit.shape[1])]]\nX_train_tf = X_train[X_train.columns[dtm.shape[1]:(dtm.shape[1]+tf_fit.shape[1])]]\nX_train_ft = X_train[X_train.columns[(dtm.shape[1]+tf_fit.shape[1]):(dtm.shape[1]+tf_fit.shape[1]+ft.shape[1])]]\nX_test_dtm = X_test[X_test.columns[0:dtm.shape[1]]]\nX_test_tf = X_test[X_test.columns[dtm.shape[1]:(dtm.shape[1]+tf_fit.shape[1])]]\nX_test_ft = X_test[X_test.columns[(dtm.shape[1]+tf_fit.shape[1]):(dtm.shape[1]+tf_fit.shape[1]+ft.shape[1])]]\ndtm_df = pd.DataFrame(dtm_ar, columns=X_test_dtm.columns)\ntf_fit_df = pd.DataFrame(tf_fit_ar, columns=X_test_tf.columns)\nft_df = pd.DataFrame(ft, columns=X_test_ft.columns)",
"_____no_output_____"
],
[
"lab_num=20000\nlab_num=int(lab_num/10)\nrf_pred = pd.DataFrame(np.empty((lab_num, 3), columns=['dtm', 'tf_fit', 'fasttext'])\netc_pred = pd.DataFrame(np.empty((lab_num, 3)), columns=['dtm', 'tf_fit', 'fasttext'])\nsgd_pred = pd.DataFrame(np.empty((lab_num, 3)), columns=['dtm', 'tf_fit', 'fasttext'])\n\nrf_pred_full = pd.DataFrame(np.empty((len(dataset[0]), 3)), columns=['dtm', 'tf_fit', 'fasttext'])\netc_pred_full = pd.DataFrame(np.empty((len(dataset[0]), 3)), columns=['dtm', 'tf_fit', 'fasttext'])\nsgd_pred_full = pd.DataFrame(np.empty((len(dataset[0]), 3)), columns=['dtm', 'tf_fit', 'fasttext'])",
"_____no_output_____"
],
[
"print(\"RandomForest: \")\nrf_classifier_dtm = RandomForestClassifier(n_estimators=300, max_depth=300, random_state=42)\nrf_classifier_dtm.fit(X_train_dtm, y_train)\npredictions = rf_classifier_dtm.predict(X_test_dtm)\nrf_pred['dtm'] = predictions\nprint(confusion_matrix(y_test,predictions))\nprint(classification_report(y_test,predictions))\nprint(accuracy_score(y_test, predictions))\npredictions = rf_classifier_dtm.predict(dtm_df)\nrf_pred_full['dtm'] = predictions\n\nprint(\"ExtraTree: \")\netc_dtm = ExtraTreesClassifier(n_estimators=300, random_state=42)\netc_dtm.fit(X_train_dtm, y_train)\npredictions = etc_dtm.predict(X_test_dtm)\netc_pred['dtm'] = predictions\nprint(confusion_matrix(y_test, predictions))\nprint(classification_report(y_test, predictions))\nprint(accuracy_score(y_test, predictions))\npredictions = etc_dtm.predict(dtm_df)\netc_pred_full['dtm'] = predictions\n\nprint(\"StochasticGradientDescent: \")\nsgd_classifier_dtm = SGDClassifier(loss=\"hinge\", penalty=\"l1\")\nsgd_classifier_dtm.fit(X_train_dtm, y_train)\npredictions = sgd_classifier_dtm.predict(X_test_dtm)\nsgd_pred['dtm'] = predictions\nprint(confusion_matrix(y_test, predictions))\nprint(classification_report(y_test, predictions))\nprint(accuracy_score(y_test, predictions))\npredictions = sgd_classifier_dtm.predict(dtm_df)\nsgd_pred_full['dtm'] = predictions\n",
"_____no_output_____"
],
[
"print(\"RandomForest: \")\nrf_classifier_tf = RandomForestClassifier(n_estimators=300, max_depth=300, random_state=42)\nrf_classifier_tf.fit(X_train_tf, y_train)\npredictions = rf_classifier_tf.predict(X_test_tf)\nrf_pred['tf_fit'] = predictions\nprint(confusion_matrix(y_test, predictions))\nprint(classification_report(y_test, predictions))\nprint(accuracy_score(y_test, predictions))\npredictions = rf_classifier_tf.predict(tf_fit)\nrf_pred_full['tf_fit'] = predictions\n\nprint(\"ExtraTree: \")\netc_tf = ExtraTreesClassifier(n_estimators=300, random_state=42)\netc_tf.fit(X_train_tf, y_train)\npredictions = etc_tf.predict(X_test_tf)\netc_pred['tf_fit'] = predictions\nprint(confusion_matrix(y_test, predictions))\nprint(classification_report(y_test, predictions))\nprint(accuracy_score(y_test, predictions))\npredictions = etc_tf.predict(tf_fit)\netc_pred_full['tf_fit'] = predictions\n\nprint(\"StochasticGradientDescent: \")\nsgd_classifier_tf = SGDClassifier(loss=\"hinge\", penalty=\"l1\")\nsgd_classifier_tf.fit(X_train_tf, y_train)\npredictions = sgd_classifier_tf.predict(X_test_tf)\nsgd_pred['tf_fit'] = predictions\nprint(confusion_matrix(y_test, predictions))\nprint(classification_report(y_test, predictions))\nprint(accuracy_score(y_test, predictions))\npredictions = sgd_classifier_tf.predict(tf_fit)\nsgd_pred_full['tf_fit'] = predictions\n",
"_____no_output_____"
],
[
"print(\"RandomForest: \")\nrf_classifier_ft = RandomForestClassifier(n_estimators=300, max_depth=300, random_state=42)\nrf_classifier_ft.fit(X_train_ft, y_train)\npredictions = rf_classifier_ft.predict(X_test_ft)\nrf_pred['fasttext'] = predictions\nprint(confusion_matrix(y_test, predictions))\nprint(classification_report(y_test, predictions))\nprint(accuracy_score(y_test, predictions))\npredictions = rf_classifier_ft.predict(ft)\nrf_pred_full['fasttext'] = predictions\n\nprint(\"ExtraTree: \")\netc_ft = ExtraTreesClassifier(n_estimators=300, random_state=42)\netc_ft.fit(X_train_ft, y_train)\npredictions = etc_ft.predict(X_test_ft)\netc_pred['fasttext'] = predictions\nprint(confusion_matrix(y_test, predictions))\nprint(classification_report(y_test, predictions))\nprint(accuracy_score(y_test, predictions))\npredictions = etc_ft.predict(ft)\netc_pred_full['fasttext'] = predictions\n\nprint(\"StochasticGradientDescent: \")\nsgd_classifier_ft = SGDClassifier(loss=\"hinge\", penalty=\"l1\")\nsgd_classifier_ft.fit(X_train_ft, y_train)\npredictions = sgd_classifier_ft.predict(X_test_ft)\nsgd_pred['fasttext'] = predictions\nprint(confusion_matrix(y_test, predictions))\nprint(classification_report(y_test, predictions))\nprint(accuracy_score(y_test, predictions))\npredictions = sgd_classifier_ft.predict(ft)\nsgd_pred_full['fasttext'] = predictions\n",
"_____no_output_____"
],
[
"print(\"RF Model: \")\nrf_model = LogisticRegression(random_state=42).fit(rf_pred, y_test)\npredictions = rf_model.predict(rf_pred)\nprint(confusion_matrix(y_test, predictions))\nprint(classification_report(y_test, predictions))\nprint(accuracy_score(y_test, predictions))\n\nprint(\"ETC Model: \")\netc_model = LogisticRegression(random_state=42).fit(etc_pred, y_test)\npredictions = etc_model.predict(etc_pred)\nprint(confusion_matrix(y_test, predictions))\nprint(classification_report(y_test, predictions))\nprint(accuracy_score(y_test, predictions))\n\nprint(\"SGD Model: \")\nsgd_model = LogisticRegression(random_state=42).fit(sgd_pred, y_test)\npredictions = sgd_model.predict(sgd_pred)\nprint(confusion_matrix(y_test, predictions))\nprint(classification_report(y_test, predictions))\nprint(accuracy_score(y_test, predictions))\n",
"_____no_output_____"
],
[
"dataset[10] = rf_model.predict(rf_pred_full)\ndataset[11] = etc_model.predict(etc_pred_full)\ndataset[12] = sgd_model.predict(sgd_pred_full)\ndataset[13] = xgb_model.predict(xgb_pred_full)\n\nsent_pred = dataset[9]\nml_pred = dataset[11]\nsent_pred[np.isnan(sent_pred)] = ml_pred\ndataset[14] = sent_pred.to_frame()",
"_____no_output_____"
],
[
"race_nmf, race_lda = 'Topic28', 'Topic3'\ngun_nmf, gun_lda = 'Topic31', 'Topic50'\nmask_nmf, mask_lda = 'Topic27', 'Topic5'\nresist_nmf, resist_lda = 'Topic25', 'Topic42'\nimmig_nmf, immig_lda = 'Topic45', 'Topic44'\nrace_nmf = dataset[dataset[5] == race_nmf]\ngun_nmf = dataset[dataset[5] == gun_nmf]\nmask_nmf = dataset[dataset[5] == mask_nmf]\nresist_nmf = dataset[dataset[5] == resist_nmf]\nimmig_nmf = dataset[dataset[5] == immig_nmf]\n\nfrom collections import Counter\nrace_nmf_vol = pd.DataFrame.from_dict(Counter([date[0:7] for date in (race_nmf[0].values)]), orient='index').reset_index()\ngun_nmf_vol = pd.DataFrame.from_dict(Counter([date[0:7] for date in (gun_nmf[0].values)]), orient='index').reset_index()\nmask_nmf_vol = pd.DataFrame.from_dict(Counter([date[0:7] for date in (mask_nmf[0].values)]), orient='index').reset_index()\nresist_nmf_vol = pd.DataFrame.from_dict(Counter([date[0:7] for date in (resist_nmf[0].values)]), orient='index').reset_index()\nimmig_nmf_vol = pd.DataFrame.from_dict(Counter([date[0:7] for date in (immig_nmf[0].values)]), orient='index').reset_index()",
"_____no_output_____"
],
[
"import datetime\nrace_nmf_vol = race_nmf_vol.sort_values(by=['index'])\nrace_nmf_vol['index'] = [datetime.datetime.strptime(d,\"%Y-%m\").date() for d in race_nmf_vol['index']]\nplt.plot(race_nmf_vol['index'], race_nmf_vol[0])\nplt.ylabel('# Posts per Month')\nplt.title('Race NMF Volumetric Analysis')\n#plt.locator_params(axis=\"x\", nbins=4)\nplt.show()",
"_____no_output_____"
],
[
"gun_nmf_vol = gun_nmf_vol.sort_values(by=['index'])\ngun_nmf_vol['index'] = [datetime.datetime.strptime(d,\"%Y-%m\").date() for d in gun_nmf_vol['index']]\nplt.plot(gun_nmf_vol['index'], gun_nmf_vol[0])\nplt.ylabel('# Posts per Month')\nplt.title('Gun NMF Volumetric Analysis')\nplt.show()",
"_____no_output_____"
],
[
"mask_nmf_vol = mask_nmf_vol.sort_values(by=['index'])\nmask_nmf_vol['index'] = [datetime.datetime.strptime(d,\"%Y-%m\").date() for d in mask_nmf_vol['index']]\nplt.plot(mask_nmf_vol['index'], mask_nmf_vol[0])\nplt.ylabel('# Posts per Month')\nplt.title('COVID NMF Volumetric Analysis')\n#plt.locator_params(axis=\"x\", nbins=4)\nplt.show()",
"_____no_output_____"
],
[
"resist_nmf_vol = resist_nmf_vol.sort_values(by=['index'])\nresist_nmf_vol['index'] = [datetime.datetime.strptime(d,\"%Y-%m\").date() for d in resist_nmf_vol['index']]\nplt.plot(resist_nmf_vol['index'], resist_nmf_vol[0])\nplt.ylabel('# Posts per Month')\nplt.title('Resist NMF Volumetric Analysis')\n#plt.locator_params(axis=\"x\", nbins=4)\nplt.show()",
"_____no_output_____"
],
[
"immig_nmf_vol = immig_nmf_vol.sort_values(by=['index'])\nimmig_nmf_vol['index'] = [datetime.datetime.strptime(d,\"%Y-%m\").date() for d in immig_nmf_vol['index']]\nplt.plot(immig_nmf_vol['index'], immig_nmf_vol[0])\nplt.ylabel('# Posts per Month')\nplt.title('Immigration NMF Volumetric Analysis')\nplt.locator_params(axis=\"x\", nbins=4)\nplt.show()",
"_____no_output_____"
],
[
"race_sentiment = pd.DataFrame([date for date in (race_nmf[14].values)], index=(race_nmf[0].values))\nrace_sentiment_pos = race_sentiment.groupby(race_sentiment[0]).get_group(1)\nrace_sentiment_neg = race_sentiment.groupby(race_sentiment[0]).get_group(0)\nrace_sentiment_pos = (pd.DataFrame.from_dict(Counter(race_sentiment_pos.index.str[:7]), orient='index').reset_index()).sort_values(by=['index'])\nrace_sentiment_neg = (pd.DataFrame.from_dict(Counter(race_sentiment_neg.index.str[:7]), orient='index').reset_index()).sort_values(by=['index']).set_index('index')\n\nax = plt.gca()\n\nrace_sentiment_pos.plot(style=[':', '--', '-'], color='blue', ax=ax)\nrace_sentiment_neg.plot(style=[':', '--', '-'], color='red', ax=ax)\nplt.ylabel('# Posts per Month')\nplt.xlabel('Time-Series')\nplt.xticks(np.arange(0, max(len(race_sentiment_pos), len(race_sentiment_neg))+1, 10))\nplt.title('Race NMF Sentiment Analysis')\nax.xaxis.set_major_locator(plt.MaxNLocator(10))\nplt.gcf().autofmt_xdate()\nplt.show()",
"_____no_output_____"
],
[
"gun_sentiment = pd.DataFrame([date for date in (gun_nmf[14].values)], index=(gun_nmf[0].values))\ngun_sentiment_pos = gun_sentiment.groupby(gun_sentiment[0]).get_group(1)\ngun_sentiment_neg = gun_sentiment.groupby(gun_sentiment[0]).get_group(0)\ngun_sentiment_pos = (pd.DataFrame.from_dict(Counter(gun_sentiment_pos[0].index.str[:7]), orient='index').reset_index()).sort_values(by=['index']).set_index('index')\ngun_sentiment_neg = (pd.DataFrame.from_dict(Counter(gun_sentiment_neg[0].index.str[:7]), orient='index').reset_index()).sort_values(by=['index']).set_index('index')\n\nax = plt.gca()\n\ngun_sentiment_pos.plot(style=[':', '--', '-'], color='blue', ax=ax)\ngun_sentiment_neg.plot(style=[':', '--', '-'], color='red', ax=ax)\nplt.ylabel('# Gun per Month')\nplt.xlabel('Time-Series')\nplt.title('Immigration NMF Sentiment Analysis')\nax.xaxis.set_major_locator(plt.MaxNLocator(10))\nplt.gcf().autofmt_xdate()\nplt.show()",
"_____no_output_____"
],
[
"mask_sentiment = pd.DataFrame([date for date in (mask_nmf[14].values)], index=(mask_nmf[0].values))\nmask_sentiment_pos = mask_sentiment.groupby(mask_sentiment[0]).get_group(1)\nmask_sentiment_neg = mask_sentiment.groupby(mask_sentiment[0]).get_group(0)\nmask_sentiment_pos = (pd.DataFrame.from_dict(Counter(mask_sentiment_pos[0].index.str[:7]), orient='index').reset_index()).sort_values(by=['index']).set_index('index')\nmask_sentiment_neg = (pd.DataFrame.from_dict(Counter(mask_sentiment_neg[0].index.str[:7]), orient='index').reset_index()).sort_values(by=['index']).set_index('index')\n\nax = plt.gca()\n\nmask_sentiment_pos.plot(style=[':', '--', '-'], color='blue', ax=ax)\nmask_sentiment_neg.plot(style=[':', '--', '-'], color='red', ax=ax)\nplt.ylabel('# Posts per Month')\nplt.xlabel('Time-Series')\nplt.title('Healthcare NMF Sentiment Analysis')\nax.xaxis.set_major_locator(plt.MaxNLocator(10))\nplt.gcf().autofmt_xdate()\nplt.show()",
"_____no_output_____"
],
[
"resist_sentiment = pd.DataFrame([date for date in (resist_nmf[14].values)], index=(resist_nmf[0].values))\nresist_sentiment_pos = resist_sentiment.groupby(resist_sentiment[0]).get_group(1)\nresist_sentiment_neg = resist_sentiment.groupby(resist_sentiment[0]).get_group(0)\nresist_sentiment_pos = (pd.DataFrame.from_dict(Counter(resist_sentiment_pos[0].index.str[:7]), orient='index').reset_index()).sort_values(by=['index']).set_index('index')\nresist_sentiment_neg = (pd.DataFrame.from_dict(Counter(resist_sentiment_neg[0].index.str[:7]), orient='index').reset_index()).sort_values(by=['index']).set_index('index')\n\nax = plt.gca()\n\nresist_sentiment_pos.plot(style=[':', '--', '-'], color='blue', ax=ax)\nresist_sentiment_neg.plot(style=[':', '--', '-'], color='red', ax=ax)\nplt.ylabel('# Posts per Month')\nplt.xlabel('Time-Series')\nplt.title('Racism NMF Sentiment Analysis')\nax.xaxis.set_major_locator(plt.MaxNLocator(10))\nplt.gcf().autofmt_xdate()\nplt.show()",
"_____no_output_____"
],
[
"immig_sentiment = pd.DataFrame([date for date in (immig_nmf[14].values)], index=(immig_nmf[0].values))\nimmig_sentiment_pos = immig_sentiment.groupby(immig_sentiment[0]).get_group(1)\nimmig_sentiment_neg = immig_sentiment.groupby(immig_sentiment[0]).get_group(0)\nimmig_sentiment_pos = (pd.DataFrame.from_dict(Counter(immig_sentiment_pos[0].index.str[:7]), orient='index').reset_index()).sort_values(by=['index']).set_index('index')\nimmig_sentiment_neg = (pd.DataFrame.from_dict(Counter(immig_sentiment_neg[0].index.str[:7]), orient='index').reset_index()).sort_values(by=['index']).set_index('index')\n\nax = plt.gca()\n\nresist_sentiment_pos.plot(style=[':', '--', '-'], color='blue', ax=ax)\nresist_sentiment_neg.plot(style=[':', '--', '-'], color='red', ax=ax)\nplt.ylabel('# Posts per Month')\nplt.xlabel('Time-Series')\nplt.title('Immigration NMF Sentiment Analysis')\nax.xaxis.set_major_locator(plt.MaxNLocator(10))\nplt.gcf().autofmt_xdate()\nplt.show()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(figsize=(10, 10))\n\nax.plot(race_sentiment_neg[0].index,\n race_sentiment_neg[0], \n linestyle=\":\", \n color='red')\nax.plot(race_sentiment_pos[0].index,\n race_sentiment_pos[0],\n color='blue',\n linestyle=\"--\")\nax.set(xlabel=\"Date\",\n ylabel=\"Negative Sentiment\",\n title=\"Daily Total Precipitation\\nBoulder, Colorado in July 2018\")\nplt.gcf().autofmt_xdate()\nax.xaxis.set_major_locator(plt.MaxNLocator(30))\nplt.show()",
"_____no_output_____"
],
[
"immig_sentiment_pos.index",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0851319b8f349356782cc6afbaaa3edc054202a | 67,148 | ipynb | Jupyter Notebook | docs/examples/nyse/nyse.ipynb | ddrightnow/blurr | a8745101d4a8a85ccf1efc608dba8486d3cebb49 | [
"Apache-2.0"
] | null | null | null | docs/examples/nyse/nyse.ipynb | ddrightnow/blurr | a8745101d4a8a85ccf1efc608dba8486d3cebb49 | [
"Apache-2.0"
] | 7 | 2019-12-16T20:58:29.000Z | 2022-02-09T23:57:32.000Z | docs/examples/nyse/nyse.ipynb | ddrightnow/blurr | a8745101d4a8a85ccf1efc608dba8486d3cebb49 | [
"Apache-2.0"
] | null | null | null | 51.572965 | 17,056 | 0.592631 | [
[
[
"# NYSE & Blurr\n\nIn this guide we will train a machine learning model that predicts closing price of a stock based on historical data. We will transform time-series stock data into features to train this model. \n\n## Prerequisites\n\nIt's recommended to have a basic understanding of how Blurr works. Following [tutorials 1](http://productml-blurr.readthedocs.io/en/latest/Streaming%20BTS%20Tutorial/) and [2](http://productml-blurr.readthedocs.io/en/latest/Window%20BTS%20Tutorial/) should provide enough background context.\n\n\n## Preparation\n\nLet's start by installing `Blurr` and other required dependencies (using requirements.txt):",
"_____no_output_____"
]
],
[
[
"import sys\n\nprint(\"installing blurr and other required dependencies...\")\n!{sys.executable} -m pip install blurr --quiet\n!{sys.executable} -m pip install -r requirements.txt --quiet\nprint(\"done.\")",
"installing blurr and other required dependencies...\ndone.\n"
]
],
[
[
"## The Dataset\n\nThis walkthrough is based on [New York Stock Exchange Data](https://www.kaggle.com/dgawlik/nyse/data) made available for [Kaggle challenges](https://www.kaggle.com/dgawlik/nyse).\n\nLet's start by downloading and having a peek at the available data: ",
"_____no_output_____"
]
],
[
[
"!wget http://demo.productml.com/data/nyse-input-data.json.zip\n!unzip -o nyse-input-data.json.zip -d .",
"--2018-05-12 03:25:09-- http://demo.productml.com/data/nyse-input-data.json.zip\nResolving demo.productml.com (demo.productml.com)... 52.218.208.187\nConnecting to demo.productml.com (demo.productml.com)|52.218.208.187|:80... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 3941552 (3.8M) [application/zip]\nSaving to: 'nyse-input-data.json.zip'\n\nnyse-input-data.jso 100%[===================>] 3.76M 1.44MB/s in 2.6s \n\n2018-05-12 03:25:12 (1.44 MB/s) - 'nyse-input-data.json.zip' saved [3941552/3941552]\n\nArchive: nyse-input-data.json.zip\n inflating: ./nyse-input-data.json \n"
],
[
"import pandas as pd\n\nstocks = pd.read_json(\"./nyse-input-data.json\", lines=True)\nstocks.head()",
"_____no_output_____"
]
],
[
[
"This dataset contains data for each market day.\n\nOur **goal is to predict closing price** of a stock for any given day based on historical data. In order to do that, we need to transform our original data source into **features** that can be used for training.\n\nWe'll calculate **moving averages** and other aggregate data for different **time windows**: one, three and seven days.\n\n## Blurr Templates\n\nWe perform initial aggregations of our data by day with [nyse-streaming-bts.yml](./nyse-streaming-bts.yml). Features are then computed using [nyse-window-bts.yml](./nyse-window-bts.yml) for each stock per day.",
"_____no_output_____"
]
],
[
[
"!cat 'nyse-streaming-bts.yml'",
"Type: Blurr:Transform:Streaming\r\nVersion: '2018-03-01'\r\nDescription: New York Store Exchange Transformations\r\nName: nyse\r\n\r\nImport:\r\n - { Module: dateutil.parser, Identifiers: [ parse ]}\r\n\r\nIdentity: source.symbol\r\n\r\nTime: parse(source.datetime)\r\n\r\nStores:\r\n - Type: Blurr:Store:Memory\r\n Name: memory\r\n\r\nAggregates:\r\n - Type: Blurr:Aggregate:Block\r\n Name: stats\r\n Store: memory\r\n Split: time.date() != stats.latest_tradetime.date()\r\n When: source.symbol in ['AAPL', 'MSFT', 'GOOG', 'FB']\r\n Fields:\r\n - Name: close\r\n Type: float\r\n Value: source.price\r\n\r\n - Name: high\r\n Type: float\r\n Value: source.price\r\n When: source.price >= stats.high\r\n \r\n - Name: low\r\n Type: float\r\n Value: source.price\r\n When: (stats.low == 0 or source.price < stats.low)\r\n\r\n - Name: volatility\r\n Type: float\r\n Value: (float(stats.high) / float(stats.low)) - 1\r\n When: stats.low > 0\r\n\r\n - Name: volume\r\n Type: float\r\n Value: stats.volume + source.volume\r\n\r\n - Name: latest_tradetime\r\n Type: datetime\r\n Value: time"
]
],
[
[
"**Streaming BTS**\n\nWe're predicting values for tech companies only (Apple, Facebook, Microsoft, Google):\n\n```yaml\nWhen: source.symbol in ['AAPL', 'MSFT', 'GOOG', 'FB']\n```\n\nEach record in the original dataset represents a single stock transaction. By setting `Split: str(time.date()) != stats.date` we'll create a new aggregate for each day per stock.\n",
"_____no_output_____"
]
],
[
[
"!cat 'nyse-window-bts.yml'",
"Type: Blurr:Transform:Window\r\nVersion: '2018-03-01'\r\nName: moving_averages\r\n\r\nSourceBTS: nyse\r\n\r\nAnchor:\r\n Condition: nyse.stats.volatility < 0.04\r\n\r\nAggregates:\r\n\r\n\r\n - Type: Blurr:Aggregate:Window\r\n Name: close\r\n WindowType: count\r\n WindowValue: 1\r\n Source: nyse.stats\r\n Fields:\r\n - Name: value\r\n Type: float\r\n Value: anchor.close # the anchor object represents the record that matches the anchor condition\r\n\r\n - Type: Blurr:Aggregate:Window\r\n Name: last\r\n WindowType: count\r\n WindowValue: -1\r\n Source: nyse.stats\r\n Fields:\r\n - Name: close\r\n Type: float\r\n Value: source.close[0]\r\n - Name: volume\r\n Type: float\r\n Value: source.volume[0]\r\n - Name: volatility\r\n Type: float\r\n Value: source.volatility[0]\r\n\r\n - Type: Blurr:Aggregate:Window\r\n Name: last_3\r\n WindowType: count\r\n WindowValue: -3\r\n Source: nyse.stats\r\n Fields:\r\n - Name: close_avg\r\n Type: float\r\n Value: sum(source.close) / len(source.close)\r\n - Name: volume_avg\r\n Type: float\r\n Value: sum(source.volume) / len(source.volume)\r\n - Name: volatility_avg\r\n Type: float\r\n Value: sum(source.volatility) / len(source.volatility)\r\n - Name: max_volatility\r\n Type: float\r\n Value: max(source.volatility)\r\n - Name: min_volatility\r\n Type: float\r\n Value: min(source.volatility)\r\n\r\n - Type: Blurr:Aggregate:Window\r\n Name: last_7\r\n WindowType: count\r\n WindowValue: -7\r\n Source: nyse.stats\r\n Fields:\r\n - Name: close_avg\r\n Type: float\r\n Value: sum(source.close) / len(source.close)\r\n - Name: volume_avg\r\n Type: float\r\n Value: sum(source.volume) / len(source.volume)\r\n - Name: volatility_avg\r\n Type: float\r\n Value: sum(source.volatility) / len(source.volatility)\r\n - Name: max_volatility\r\n Type: float\r\n Value: max(source.volatility)\r\n - Name: min_volatility\r\n Type: float\r\n Value: min(source.volatility)\r\n"
]
],
[
[
"**Window BTS**\n\nWe'll use a very rough criteria to remove outliers: our model will only work when closing price changes less than a 4%:\n\n```yaml\nAnchor:\n Condition: nyse.stats.volatility < 0.04\n```\n\nWe're using [moving averages](https://www.investopedia.com/terms/m/movingaverage.asp) to generate features based on historical data about a stock:\n\n```yaml\n- Type: Blurr:Aggregate:Window\n Name: last_7\n WindowType: count\n WindowValue: -7\n Source: nyse.stats\n Fields:\n - Name: close_avg\n Type: float\n Value: sum(source.close) / len(source.close)\n```\n\n\n## Transforming Data\n\n",
"_____no_output_____"
]
],
[
[
"from blurr_util import print_head, validate, transform\n\nvalidate('nyse-streaming-bts.yml')\nvalidate('nyse-window-bts.yml')",
"Running syntax validation on nyse-streaming-bts.yml\nDocument is valid\nRunning syntax validation on nyse-window-bts.yml\nDocument is valid\n"
]
],
[
[
"Let's run our Streaming BTS for informational purposes only, so we can preview the result of the transformation:",
"_____no_output_____"
]
],
[
[
"transform(log_files=[\"./nyse-input-data.json\"],\n stream_bts='./nyse-streaming-bts.yml',\n output_file=\"./nyse-streaming-bts-out.log\")",
"_____no_output_____"
],
[
"print_head(\"./nyse-streaming-bts-out.log\")",
"_____no_output_____"
],
[
"transform(log_files=[\"./nyse-input-data.json\"],\n stream_bts='./nyse-streaming-bts.yml',\n window_bts='./nyse-window-bts.yml',\n output_file=\"./nyse-processed-data.csv\")",
"_____no_output_____"
]
],
[
[
"Let's now preview the data that will be used to **train our model**",
"_____no_output_____"
]
],
[
[
"window_out = pd.read_csv(\"./nyse-processed-data.csv\")\nwindow_out.head()",
"_____no_output_____"
]
],
[
[
"## Modelling\n\n**Blurr** is about Data Preparation and Feature Engineering. Modeling is included here for illustration purpose, and the reader can use any modeling library or tool for such purpose.\n\nLet's start by importing the output of our Window BTS as the source dataset. We're dropping unnecessary `_identity` columns:",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport pandas as pd\n\ndef import_dataset():\n data = pd.read_csv(\"./nyse-processed-data.csv\")\n data[\"close\"] = data[\"close.value\"] # Moving close to the last column\n data.drop(['close.value'], 1, inplace=True) \n data.drop(['close._identity'], 1, inplace=True) \n data.drop(['last._identity'], 1, inplace=True) \n data.drop(['last_3._identity'], 1, inplace=True) \n data.drop(['last_7._identity'], 1, inplace=True) \n return data\n\ndataset = import_dataset()\ndataset.head()",
"_____no_output_____"
]
],
[
[
"Each column represents a Feature, except the rightmost column which represents the Output we're trying to predic",
"_____no_output_____"
]
],
[
[
"feature_count = len(dataset.columns) - 1\nprint(\"#features=\" + str(feature_count))",
"#features=13\n"
]
],
[
[
"We're splitting our dataset into Input Variables (`X`) and the Output Variable (`Y`) using pandas' [`iloc` function](http://pandas.pydata.org/pandas-docs/version/0.17.0/generated/pandas.DataFrame.iloc.html):",
"_____no_output_____"
]
],
[
[
"X = dataset.iloc[:, 0:feature_count].values\nprint(X.shape)",
"(5978, 13)\n"
],
[
"Y = dataset.iloc[:, feature_count].values\nprint(Y.shape)",
"(5978,)\n"
]
],
[
[
"We need to split between train and test datasets for training and evaluation of the model:",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\n\nX_train_raw, X_test_raw, Y_train_raw, Y_test_raw = train_test_split(X, Y, test_size = 0.2)",
"_____no_output_____"
]
],
[
[
"Finally, we need to scale our data before training:",
"_____no_output_____"
]
],
[
[
"from sklearn.preprocessing import MinMaxScaler\n\nscaler = MinMaxScaler()\nX_train = scaler.fit_transform(X_train_raw)\nX_test = scaler.transform(X_test_raw)\nY_train = scaler.fit_transform(Y_train_raw.reshape(-1, 1))\nY_test = scaler.transform(Y_test_raw.reshape(-1, 1))",
"_____no_output_____"
]
],
[
[
"It's now time to build and train our model:",
"_____no_output_____"
]
],
[
[
"# Importing the Keras libraries and packages\n\nimport keras\nfrom keras.models import Sequential\nfrom keras.layers import Dense",
"/Users/vigneshbadrinathkrishna/Documents/Jupyter/temp/lib/python3.6/site-packages/h5py/__init__.py:36: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n from ._conv import register_converters as _register_converters\nUsing TensorFlow backend.\n"
],
[
"#Initializing Neural Network\n\nmodel = Sequential()\nmodel.add(Dense(units = 36, kernel_initializer = 'uniform', activation = 'relu', input_dim = feature_count))\nmodel.add(Dense(units = 36, kernel_initializer = 'uniform', activation = 'relu'))\nmodel.add(Dense(units = 1, kernel_initializer = 'uniform', activation = 'linear'))\n\n# Compiling Neural Network\nmodel.compile(loss='mse',optimizer='adam', metrics=['accuracy'])\n\n# Fitting our model \nmodel.fit(X_train, Y_train, batch_size = 512, epochs = 70, validation_split=0.1, verbose=1)",
"Train on 4303 samples, validate on 479 samples\nEpoch 1/70\n4303/4303 [==============================] - 0s 46us/step - loss: 0.1025 - acc: 0.0000e+00 - val_loss: 0.0775 - val_acc: 0.0021\nEpoch 2/70\n4303/4303 [==============================] - 0s 4us/step - loss: 0.0942 - acc: 0.0000e+00 - val_loss: 0.0686 - val_acc: 0.0021\nEpoch 3/70\n4303/4303 [==============================] - 0s 4us/step - loss: 0.0813 - acc: 0.0000e+00 - val_loss: 0.0563 - val_acc: 0.0021\nEpoch 4/70\n4303/4303 [==============================] - 0s 4us/step - loss: 0.0645 - acc: 0.0000e+00 - val_loss: 0.0449 - val_acc: 0.0021\nEpoch 5/70\n4303/4303 [==============================] - 0s 4us/step - loss: 0.0494 - acc: 0.0000e+00 - val_loss: 0.0391 - val_acc: 0.0021\nEpoch 6/70\n4303/4303 [==============================] - 0s 4us/step - loss: 0.0385 - acc: 0.0000e+00 - val_loss: 0.0277 - val_acc: 0.0021\nEpoch 7/70\n4303/4303 [==============================] - 0s 4us/step - loss: 0.0242 - acc: 2.3240e-04 - val_loss: 0.0145 - val_acc: 0.0021\nEpoch 8/70\n4303/4303 [==============================] - 0s 4us/step - loss: 0.0113 - acc: 2.3240e-04 - val_loss: 0.0053 - val_acc: 0.0021\nEpoch 9/70\n4303/4303 [==============================] - 0s 5us/step - loss: 0.0034 - acc: 2.3240e-04 - val_loss: 0.0016 - val_acc: 0.0021\nEpoch 10/70\n4303/4303 [==============================] - 0s 4us/step - loss: 0.0010 - acc: 2.3240e-04 - val_loss: 8.0088e-04 - val_acc: 0.0021\nEpoch 11/70\n4303/4303 [==============================] - 0s 5us/step - loss: 7.3606e-04 - acc: 2.3240e-04 - val_loss: 7.9738e-04 - val_acc: 0.0021\nEpoch 12/70\n4303/4303 [==============================] - 0s 4us/step - loss: 7.4681e-04 - acc: 2.3240e-04 - val_loss: 7.1321e-04 - val_acc: 0.0021\nEpoch 13/70\n4303/4303 [==============================] - 0s 4us/step - loss: 6.5283e-04 - acc: 2.3240e-04 - val_loss: 5.8154e-04 - val_acc: 0.0021\nEpoch 14/70\n4303/4303 [==============================] - 0s 5us/step - loss: 5.3614e-04 - acc: 2.3240e-04 - val_loss: 5.1272e-04 - val_acc: 0.0021\nEpoch 15/70\n4303/4303 [==============================] - 0s 4us/step - loss: 4.8915e-04 - acc: 2.3240e-04 - val_loss: 4.8887e-04 - val_acc: 0.0021\nEpoch 16/70\n4303/4303 [==============================] - 0s 4us/step - loss: 4.5884e-04 - acc: 2.3240e-04 - val_loss: 4.4613e-04 - val_acc: 0.0021\nEpoch 17/70\n4303/4303 [==============================] - 0s 4us/step - loss: 4.2753e-04 - acc: 2.3240e-04 - val_loss: 4.1493e-04 - val_acc: 0.0021\nEpoch 18/70\n4303/4303 [==============================] - 0s 4us/step - loss: 4.0553e-04 - acc: 2.3240e-04 - val_loss: 3.9318e-04 - val_acc: 0.0021\nEpoch 19/70\n4303/4303 [==============================] - 0s 4us/step - loss: 3.8638e-04 - acc: 2.3240e-04 - val_loss: 3.7563e-04 - val_acc: 0.0021\nEpoch 20/70\n4303/4303 [==============================] - 0s 4us/step - loss: 3.6760e-04 - acc: 2.3240e-04 - val_loss: 3.5841e-04 - val_acc: 0.0021\nEpoch 21/70\n4303/4303 [==============================] - 0s 4us/step - loss: 3.4842e-04 - acc: 2.3240e-04 - val_loss: 3.3985e-04 - val_acc: 0.0021\nEpoch 22/70\n4303/4303 [==============================] - 0s 4us/step - loss: 3.2932e-04 - acc: 2.3240e-04 - val_loss: 3.2005e-04 - val_acc: 0.0021\nEpoch 23/70\n4303/4303 [==============================] - 0s 4us/step - loss: 3.0877e-04 - acc: 2.3240e-04 - val_loss: 3.0079e-04 - val_acc: 0.0021\nEpoch 24/70\n4303/4303 [==============================] - 0s 4us/step - loss: 2.8941e-04 - acc: 2.3240e-04 - val_loss: 2.8071e-04 - val_acc: 0.0021\nEpoch 25/70\n4303/4303 [==============================] - 0s 4us/step - loss: 2.6856e-04 - acc: 2.3240e-04 - val_loss: 2.6227e-04 - val_acc: 0.0021\nEpoch 26/70\n4303/4303 [==============================] - 0s 4us/step - loss: 2.4924e-04 - acc: 2.3240e-04 - val_loss: 2.4349e-04 - val_acc: 0.0021\nEpoch 27/70\n4303/4303 [==============================] - 0s 4us/step - loss: 2.3008e-04 - acc: 2.3240e-04 - val_loss: 2.2409e-04 - val_acc: 0.0021\nEpoch 28/70\n4303/4303 [==============================] - 0s 5us/step - loss: 2.1201e-04 - acc: 2.3240e-04 - val_loss: 2.0661e-04 - val_acc: 0.0021\nEpoch 29/70\n4303/4303 [==============================] - 0s 4us/step - loss: 1.9386e-04 - acc: 2.3240e-04 - val_loss: 1.8931e-04 - val_acc: 0.0021\nEpoch 30/70\n4303/4303 [==============================] - 0s 4us/step - loss: 1.7755e-04 - acc: 2.3240e-04 - val_loss: 1.7436e-04 - val_acc: 0.0021\nEpoch 31/70\n4303/4303 [==============================] - 0s 4us/step - loss: 1.6193e-04 - acc: 2.3240e-04 - val_loss: 1.5940e-04 - val_acc: 0.0021\nEpoch 32/70\n4303/4303 [==============================] - 0s 4us/step - loss: 1.4812e-04 - acc: 2.3240e-04 - val_loss: 1.4754e-04 - val_acc: 0.0021\nEpoch 33/70\n4303/4303 [==============================] - 0s 4us/step - loss: 1.3705e-04 - acc: 2.3240e-04 - val_loss: 1.3818e-04 - val_acc: 0.0021\nEpoch 34/70\n4303/4303 [==============================] - 0s 4us/step - loss: 1.2689e-04 - acc: 2.3240e-04 - val_loss: 1.2781e-04 - val_acc: 0.0021\nEpoch 35/70\n4303/4303 [==============================] - 0s 4us/step - loss: 1.1788e-04 - acc: 2.3240e-04 - val_loss: 1.2091e-04 - val_acc: 0.0021\nEpoch 36/70\n4303/4303 [==============================] - 0s 4us/step - loss: 1.0975e-04 - acc: 2.3240e-04 - val_loss: 1.1201e-04 - val_acc: 0.0021\nEpoch 37/70\n4303/4303 [==============================] - 0s 4us/step - loss: 1.0235e-04 - acc: 2.3240e-04 - val_loss: 1.0453e-04 - val_acc: 0.0021\nEpoch 38/70\n4303/4303 [==============================] - 0s 4us/step - loss: 9.5748e-05 - acc: 2.3240e-04 - val_loss: 9.8943e-05 - val_acc: 0.0021\nEpoch 39/70\n4303/4303 [==============================] - 0s 4us/step - loss: 9.0075e-05 - acc: 2.3240e-04 - val_loss: 9.3020e-05 - val_acc: 0.0021\nEpoch 40/70\n4303/4303 [==============================] - 0s 4us/step - loss: 8.4809e-05 - acc: 2.3240e-04 - val_loss: 8.7891e-05 - val_acc: 0.0021\nEpoch 41/70\n4303/4303 [==============================] - 0s 4us/step - loss: 8.0412e-05 - acc: 2.3240e-04 - val_loss: 8.3260e-05 - val_acc: 0.0021\nEpoch 42/70\n4303/4303 [==============================] - 0s 4us/step - loss: 7.6413e-05 - acc: 2.3240e-04 - val_loss: 8.0566e-05 - val_acc: 0.0021\nEpoch 43/70\n4303/4303 [==============================] - 0s 4us/step - loss: 7.2809e-05 - acc: 2.3240e-04 - val_loss: 7.6273e-05 - val_acc: 0.0021\nEpoch 44/70\n4303/4303 [==============================] - 0s 4us/step - loss: 6.9614e-05 - acc: 2.3240e-04 - val_loss: 7.2717e-05 - val_acc: 0.0021\nEpoch 45/70\n4303/4303 [==============================] - 0s 4us/step - loss: 6.6499e-05 - acc: 2.3240e-04 - val_loss: 6.9950e-05 - val_acc: 0.0021\nEpoch 46/70\n4303/4303 [==============================] - 0s 4us/step - loss: 6.3570e-05 - acc: 2.3240e-04 - val_loss: 6.6740e-05 - val_acc: 0.0021\nEpoch 47/70\n4303/4303 [==============================] - 0s 5us/step - loss: 6.1582e-05 - acc: 2.3240e-04 - val_loss: 6.4343e-05 - val_acc: 0.0021\nEpoch 48/70\n4303/4303 [==============================] - 0s 5us/step - loss: 5.9359e-05 - acc: 2.3240e-04 - val_loss: 6.2669e-05 - val_acc: 0.0021\nEpoch 49/70\n4303/4303 [==============================] - 0s 5us/step - loss: 5.7216e-05 - acc: 2.3240e-04 - val_loss: 5.9803e-05 - val_acc: 0.0021\nEpoch 50/70\n4303/4303 [==============================] - 0s 5us/step - loss: 5.5511e-05 - acc: 2.3240e-04 - val_loss: 5.8672e-05 - val_acc: 0.0021\nEpoch 51/70\n4303/4303 [==============================] - 0s 5us/step - loss: 5.4430e-05 - acc: 2.3240e-04 - val_loss: 5.6377e-05 - val_acc: 0.0021\nEpoch 52/70\n4303/4303 [==============================] - 0s 4us/step - loss: 5.2620e-05 - acc: 2.3240e-04 - val_loss: 5.4984e-05 - val_acc: 0.0021\nEpoch 53/70\n4303/4303 [==============================] - 0s 5us/step - loss: 5.1474e-05 - acc: 2.3240e-04 - val_loss: 5.3884e-05 - val_acc: 0.0021\nEpoch 54/70\n4303/4303 [==============================] - 0s 4us/step - loss: 5.1241e-05 - acc: 2.3240e-04 - val_loss: 5.1820e-05 - val_acc: 0.0021\nEpoch 55/70\n4303/4303 [==============================] - 0s 4us/step - loss: 4.8984e-05 - acc: 2.3240e-04 - val_loss: 5.0553e-05 - val_acc: 0.0021\nEpoch 56/70\n4303/4303 [==============================] - 0s 4us/step - loss: 4.7987e-05 - acc: 2.3240e-04 - val_loss: 4.9232e-05 - val_acc: 0.0021\n"
]
],
[
[
"We can measure the quality of our model using [MSE](https://en.wikipedia.org/wiki/Mean_squared_error) and [RMSE](https://en.wikipedia.org/wiki/Root-mean-square_deviation):",
"_____no_output_____"
]
],
[
[
"import math\n\nscore = model.evaluate(X_test, Y_test, verbose=0)\nprint('Model Score: %.5f MSE (%.2f RMSE)' % (score[0], math.sqrt(score[0])))",
"Model Score: 0.00004 MSE (0.01 RMSE)\n"
]
],
[
[
"Finally, let's plot prediction vs actual data.\n\nPrior to normalisation, we undo scaling and perform a sort for graph quality:",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt2\n\nprediction_sorted = scaler.inverse_transform(model.predict(X_test))\nprediction_sorted.sort(axis=0)\n\nY_test_sorted = scaler.inverse_transform(Y_test.copy().reshape(-1, 1))\nY_test_sorted.sort(axis=0)\n\nplt2.plot(prediction_sorted, color='red', label='Prediction')\nplt2.plot(Y_test_sorted, color='blue', label='Actual')\nplt2.xlabel('#sample')\nplt2.ylabel('close value')\nplt2.legend(loc='best')\nplt2.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d085214641c2632797da9ae05c32715652f92bbe | 3,213 | ipynb | Jupyter Notebook | notebooks/GR_lab03b_linear_systems_direct_2.ipynb | mapenzo-ph/numerical-analysis-2021-2022 | 952808d7fa7a9e718274592104be24882acef3ec | [
"CC-BY-4.0"
] | 1 | 2022-01-12T23:19:50.000Z | 2022-01-12T23:19:50.000Z | notebooks/08b-linear-systems-direct-2.ipynb | giovastabile/numerical-analysis-2021-2022 | 15b4557cc06eb089077931e08367845a7c10935c | [
"CC-BY-4.0"
] | null | null | null | notebooks/08b-linear-systems-direct-2.ipynb | giovastabile/numerical-analysis-2021-2022 | 15b4557cc06eb089077931e08367845a7c10935c | [
"CC-BY-4.0"
] | null | null | null | 38.710843 | 361 | 0.577031 | [
[
[
"# Direct methods for solving linear systems (additional exercises)\n",
"_____no_output_____"
],
[
"**Exercise 1.** Let us consider the linear system $A\\mathbf{x} = \\mathbf{b}$ where\n$$\n A = \n \\begin{bmatrix}\n \\epsilon & 1 & 2\\\\\n 1 & 3 & 1 \\\\\n 2 & 1 & 3 \\\\\n \\end{bmatrix}.\n$$\n\n1. Find the range of values of $\\epsilon \\in \\mathbb{R}$ such that the matrix $A$ is symmetric and positive definite.\n**Suggestion**: use the *Sylvester's criterion* which states that a symmetric matrix $A \\in \\mathbb{R}^{n \\times n}$ is positive definite if and only if all the main minors (The main minors of $A \\in \\mathbb{R}^{n \\times n}$ are the determinants of the submatrices $A_p = (a_{i,j})_{1 \\leq i, j \\leq p}$, $p = 1, ..., n$). of $A$ are positive.\n2. What factorization is more suitable for solving the linear system $A\\mathbf{x}=\\mathbf{b}$ for the case $\\epsilon=0$? Motivate the answer.\n3. Compute the Cholesky factorization $A = R^T R$ for the case $\\epsilon = 2$.\n4. Given $\\mathbf{b} = (1,1,1)^T$, solve the linear system by using the Cholesky factorization computed at the previous point.\n\n",
"_____no_output_____"
],
[
"**Exercise 2.** Let us consider the following matrix $A \\in \\mathbb R^{3 \\times 3}$ depending on the parameter $\\epsilon \\in \\mathbb R$:\n$$\nA =\n\\begin{bmatrix}\n1 & \\epsilon & -1 \\\\\n\\epsilon & \\frac{35}3 & 1 \\\\\n-1 & \\epsilon & 2 \\\\\n\\end{bmatrix}.\n$$\n\n\n\n1. Calculate the values of the parameter $\\epsilon \\in \\mathbb R$ for which the matrix $A$ is invertible (non singular).\n\n2. Calculate the Gauss factorization $LU$ of the matrix $A$ (when non singular) for a generic value of the parameter $\\epsilon \\in \\mathbb R$.\n\n3. Calculate the values of the parameter $\\epsilon \\in \\mathbb R$ for which the Gauss factorization $LU$ of the matrix $A$ (when non singular) exists and is unique.\n\n4. Set $\\epsilon = \\sqrt{\\frac{35}3}$ and use the pivoting technique to calculate the Gauss factorization $LU$ of the matrix $A$.\n\n5. For $\\epsilon=1$, the matrix $A$ is symmetric and positive definite. Calculate the corresponding Cholesky factorization of the matrix $A$, i.e. the upper triangular matrix with positive elements on the diagonal, say $R$, for which $A = R^T R$.",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
]
] |
d08529d3e80a89f7e7d961e88191faf78700739d | 17,333 | ipynb | Jupyter Notebook | examples/user_guide/1_Surface.ipynb | kwinkunks/xarray-spatial | 59984d859820e6e1cd9f11f1bf7696c04d1924fb | [
"MIT"
] | null | null | null | examples/user_guide/1_Surface.ipynb | kwinkunks/xarray-spatial | 59984d859820e6e1cd9f11f1bf7696c04d1924fb | [
"MIT"
] | null | null | null | examples/user_guide/1_Surface.ipynb | kwinkunks/xarray-spatial | 59984d859820e6e1cd9f11f1bf7696c04d1924fb | [
"MIT"
] | null | null | null | 39.038288 | 411 | 0.625627 | [
[
[
"# Xarray-spatial\n### User Guide: Surface tools\n-----\nWith the Surface tools, you can quantify and visualize a terrain landform represented by a digital elevation model.\n\nStarting with a raster elevation surface, represented as an Xarray DataArray, these tools can help you identify some specific patterns that may not be readily apparent in the original surface. The return of each function is also an Xarray DataArray.\n\n[Hillshade](#Hillshade): Creates a shaded relief from a surface raster by considering the illumination source angle and shadows.\n\n[Slope](#Slope): Identifies the slope for each cell of a raster.\n\n[Curvature](#Curvature): Calculates the curvature of a raster surface.\n\n[Aspect](#Aspect): Derives the aspect for each cell of a raster surface.\n\n[Viewshed](#Viewshed): Determines visible locations in the input raster surface from a viewpoint with an optional observer height.\n\n-----------\n",
"_____no_output_____"
],
[
"#### Let's use datashader to render our images...",
"_____no_output_____"
],
[
"We'll need the basic Numpy and Pandas, as well as datashader, \na data rasterization package highly compatible with Xarray-spatial. \nAlong with the base package, we'll import several nested functions (shade, stack...) \nincluding Elevation, which we'll use below.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport xarray as xr\n\nimport datashader as ds\n\nfrom datashader.transfer_functions import shade\nfrom datashader.transfer_functions import stack\nfrom datashader.transfer_functions import dynspread\nfrom datashader.transfer_functions import set_background\nfrom datashader.colors import Elevation\n\nimport xrspatial",
"_____no_output_____"
]
],
[
[
"## Generate Terrain Data\n\nThe rest of the geo-related functions focus on raster data, i.e. data that's been aggregated into the row-column grid of cells in a raster image. Datashader's Canvas object provides a convenient frame to set up a new raster, so we'll use that with our `generate_terrain` function to generate some fake terrain as an elevation raster. Once we have that, we'll use datashader's shade for easy visualization.",
"_____no_output_____"
]
],
[
[
"from xrspatial import generate_terrain\n\nW = 800\nH = 600\n\nterrain = xr.DataArray(np.zeros((H, W)))\nterrain = generate_terrain(terrain)\n\nshade(terrain, cmap=['black', 'white'], how='linear')",
"_____no_output_____"
]
],
[
[
"The grayscale values in the image above show elevation, scaled linearly in black-to-white color intensity (with the large black areas indicating low elevation). This shows the data, but it would look more like a landscape if we map the lowest values to colors representing water, and the highest to colors representing mountaintops. Let's try the Elevation colormap we imported above:",
"_____no_output_____"
]
],
[
[
"shade(terrain, cmap=Elevation, how='linear')",
"_____no_output_____"
]
],
[
[
"## Hillshade\n\n[Hillshade](https://en.wikipedia.org/wiki/Terrain_cartography) is a technique used to visualize terrain as shaded relief by illuminating it with a hypothetical light source. The illumination value for each cell is determined by its orientation to the light source, which can be calculated from slope and aspect.\n\nLet's apply Hillshade to our terrain and visualize the result with shade.",
"_____no_output_____"
]
],
[
[
"from xrspatial import hillshade\n\nilluminated = hillshade(terrain)\n\nhillshade_gray_white = shade(illuminated, cmap=['gray', 'white'], alpha=255, how='linear')\nhillshade_gray_white",
"_____no_output_____"
]
],
[
[
"Applying hillshade reveals a lot of detail in the 3D shape of the terrain.\n\nTo add even more detail, we can add the Elevation colormapped terrain from earlier and combine it with the hillshade terrain using datashader's stack function.",
"_____no_output_____"
]
],
[
[
"terrain_elevation = shade(terrain, cmap=Elevation, alpha=128, how='linear')\nstack(hillshade_gray_white, terrain_elevation)",
"_____no_output_____"
]
],
[
[
"## Slope\n[Slope](https://en.wikipedia.org/wiki/Slope) is the inclination of a surface. \nIn geography, *slope* is the amount of change in elevation for an area in a terrain relative to its surroundings.\n\nXarray-spatial's slope function returns the slope at each cell in degrees.\nBecause Xarray-spatial is integrated with Xarray and Numpy, we can apply standard Numpy filters. For example, we can highlight only slopes in the [avalanche risk](https://www.gravityprotection.co.uk/blog/slope-steepness-avalanche-risk.html) range of 25 - 50 degrees. (Note the use of risky.data since these are DataArrays).\nStacking the resulting raster with the hillshaded and plain terrain ones from above gives an image with areas of avalanche risk neatly highlighted.",
"_____no_output_____"
]
],
[
[
"from xrspatial import slope\n\nrisky = slope(terrain)\nrisky.data = np.where(np.logical_and(risky.data > 25, risky.data < 50), 1, np.nan)\n\nstack(shade(terrain, cmap=['black', 'white'], how='linear'),\n shade(illuminated, cmap=['black', 'white'], how='linear', alpha=128),\n shade(risky, cmap='red', how='linear', alpha=200))",
"_____no_output_____"
]
],
[
[
"## Curvature\n[Curvature](https://desktop.arcgis.com/en/arcmap/10.3/tools/spatial-analyst-toolbox/curvature.htm) is the second derivative of a surface's elevation, or the *slope-of-the-slope*; in other words, how fast the slope is increasing or decreasing as we move along a surface.\n- A positive curvature means the surface is curving up (upwardly convex) at that cell. \n- A negative curvature means the surface is curving down (downwardly convex) at that cell. \n- A curvature of 0 means the surface is striaght and constant in whatever angle it's sloped towards.\n\nThe Xarray-spatial curvature function returns a raster in units one hundredth (1/100) of the z-factor, or scaling factor (which you can set explicitly in generate _terrain as \"zfactor\"). \nReasonably expected values in the curvature raster for a hilly area (moderate relief) would be between -0.5 and 0.5, while for steep, rugged mountains (extreme relief) these can range as far as -4 and 4. For certain raster surfaces it is possible to go even larger than that.\n\nLet's generate a terrain with an appropriate z-factor and apply the curvature function to it. Then, we can apply some Numpy filtering (remember, we have access to all those functions) to highlight steeper and gentler curves in the slopes.\nStacking these with the hillshaded and plain terrains gives us a fuller picture of the slopes.",
"_____no_output_____"
]
],
[
[
"from xrspatial import curvature\n\nterrain_z_one = xr.DataArray(np.zeros((H, W)))\nterrain_z_one = generate_terrain(terrain_z_one, zfactor=1)\ncurv = curvature(terrain_z_one)\ncurv_hi, curv_low = curv.copy(), curv.copy()\ncurv_hi.data = np.where(np.logical_and(curv_hi.data > 1, curv_hi.data < 4), 1, np.nan)\ncurv_low.data = np.where(np.logical_and(curv_low.data > 0.5, curv_low.data < 1), 1, np.nan)\n\nstack(shade(terrain, cmap=['black', 'white'], how='linear'),\n shade(illuminated, cmap=['black', 'white'], how='linear', alpha=128),\n shade(curv_hi, cmap='red', how='log', alpha=200),\n shade(curv_low, cmap='green', how='log', alpha=200))",
"_____no_output_____"
]
],
[
[
"## Aspect\n\n[Aspect](https://en.wikipedia.org/wiki/Aspect_(geography)) is the orientation of a slope, measured clockwise in degrees from 0 to 360, where 0 is north-facing, 90 is east-facing, 180 is south-facing, and 270 is west-facing.\n\nThe Xarray-spatial aspect function returns the aspect in degrees for each cell in an elevation terrain.\n\nWe can apply aspect to our terrain, then use Numpy to filter out only slopes facing close to North. Then, we can stack that with the hillshaded and plain terrains.\n(Note: the printout images are from a North point-of-view.)",
"_____no_output_____"
]
],
[
[
"from xrspatial import aspect\n\nnorth_faces = aspect(terrain)\nnorth_faces.data = np.where(np.logical_or(north_faces.data > 350 ,\n north_faces.data < 10), 1, np.nan)\n\nstack(shade(terrain, cmap=['black', 'white'], how='linear'),\n shade(illuminated, cmap=['black', 'white'], how='linear', alpha=128),\n shade(north_faces, cmap=['aqua'], how='linear', alpha=100))",
"_____no_output_____"
]
],
[
[
"## Viewshed\n\nThe `xrspatial.viewshed` function operates on a given aggregate to calculate the viewshed (the visible cells in the raster) for a given viewpoint, or *observer location*. \n\nThe visibility model is as follows: Two cells are visible to each other if the line of sight that connects their centers is not blocked at any point by another part of the terrain. If the line of sight does not pass through the cell center, elevation is determined using bilinear interpolation.",
"_____no_output_____"
],
[
"##### Simple Viewshed Example\n\n- The example below creates a datashader aggregate from a 2d normal distribution.\n- To calculate the viewshed, we need an observer location so we'll set up an aggregate for that as well.\n- Then, we can visualize all of that with hillshade, shade, and stack.\n- The observer location is indicated by the orange point in the upper-left of the plot.",
"_____no_output_____"
]
],
[
[
"from xrspatial import viewshed\n\nOBSERVER_X = -12.5\nOBSERVER_Y = 10\n\ncanvas = ds.Canvas(plot_width=W, plot_height=H,\n x_range=(-20, 20), y_range=(-20, 20))\n\nnormal_df = pd.DataFrame({\n 'x': np.random.normal(.5, 1, 10000000),\n 'y': np.random.normal(.5, 1, 10000000)\n})\nnormal_agg = canvas.points(normal_df, 'x', 'y')\nnormal_agg.values = normal_agg.values.astype(\"float64\")\nnormal_shaded = shade(normal_agg)\n\nobserver_df = pd.DataFrame({'x': [OBSERVER_X], 'y': [OBSERVER_Y]})\nobserver_agg = canvas.points(observer_df, 'x', 'y')\nobserver_shaded = dynspread(shade(observer_agg, cmap=['orange']),\n threshold=1, max_px=4)\n\nnormal_illuminated = hillshade(normal_agg)\nnormal_illuminated_shaded = shade(normal_illuminated, cmap=['black', 'white'], \n alpha=128, how='linear')\n\nstack(normal_illuminated_shaded, observer_shaded)",
"_____no_output_____"
]
],
[
[
"##### Calculate viewshed using the observer location\n\nNow we can apply viewshed to the normal_agg, with the observer_agg for the viewpoint. We can then visualize it and stack it with the hillshade and observer rasters.",
"_____no_output_____"
]
],
[
[
"# Will take some time to run...\n%time view = viewshed(normal_agg, x=OBSERVER_X, y=OBSERVER_Y)\n\nview_shaded = shade(view, cmap=['white', 'red'], alpha=128, how='linear')\n\nstack(normal_illuminated_shaded, observer_shaded, view_shaded) ",
"_____no_output_____"
]
],
[
[
"As you can see, the image highlights in red all points visible from the observer location marked with the orange dot. As one might expect, the areas behind the normal distribution *mountain* are blocked from the viewer.",
"_____no_output_____"
],
[
"#### Viewshed on Terrain\nNow we can try using viewshed on our more complicated terrain.\n\n- We'll set up our terrain aggregate and apply hillshade and shade for easy visualization.\n- We'll also set up an observer location aggregate, setting the location to the center, at (x, y) = (0, 0).",
"_____no_output_____"
]
],
[
[
"from xrspatial import viewshed\n\nx_range=(-20e6, 20e6)\ny_range=(-20e6, 20e6)\nterrain = xr.DataArray(np.zeros((H, W)))\nterrain = generate_terrain(terrain, x_range=x_range, y_range=y_range)\nterrain_shaded = shade(terrain, cmap=Elevation, alpha=128, how='linear')\n\nilluminated = hillshade(terrain)\n\nOBSERVER_X = 0.0\nOBSERVER_Y = 0.0\n\ncvs = ds.Canvas(plot_width=W, plot_height=H, x_range=x_range, y_range=y_range)\nobserver_df = pd.DataFrame({'x': [OBSERVER_X],'y': [OBSERVER_Y]})\nobserver_agg = cvs.points(observer_df, 'x', 'y')\nobserver_shaded = dynspread(shade(observer_agg, cmap=['orange']),\n threshold=1, max_px=4)\n\nstack(shade(illuminated, cmap=['black', 'white'], alpha=128, how='linear'),\n terrain_shaded,\n observer_shaded)",
"_____no_output_____"
]
],
[
[
"Now we can apply viewshed.\n- Notice the use of the `observer_elev` argument, which is the height of the observer above the terrain.",
"_____no_output_____"
]
],
[
[
"%time view = viewshed(terrain, x=OBSERVER_X, y=OBSERVER_Y, observer_elev=100)\n\nview_shaded = shade(view, cmap='fuchsia', how='linear')\nstack(shade(illuminated, cmap=['black', 'white'], alpha=128, how='linear'),\n terrain_shaded,\n view_shaded,\n observer_shaded)",
"_____no_output_____"
]
],
[
[
"The fuchsia areas are those visible to an observer of the given height at the indicated orange location.",
"_____no_output_____"
],
[
"\n\n\n### References\n- An overview of the Surface toolset: https://pro.arcgis.com/en/pro-app/tool-reference/spatial-analyst/an-overview-of-the-surface-tools.htm\n- Burrough, P. A., and McDonell, R. A., 1998. Principles of Geographical Information Systems (Oxford University Press, New York), p. 406.\n- Making Maps with Noise Functions: https://www.redblobgames.com/maps/terrain-from-noise/\n- How Aspect Works: http://desktop.arcgis.com/en/arcmap/10.3/tools/spatial-analyst-toolbox/how-aspect-works.htm#ESRI_SECTION1_4198691F8852475A9F4BC71246579FAA",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d0852a0fa774d72533bec9f2dc0bc8983eddeaaa | 51,725 | ipynb | Jupyter Notebook | Machine Learning with Python/Machine Learning with Python - Week 3 - Lab 3 - Classification - Logistic Regression.ipynb | ipiyushsonar/coursera-labs | 39a6a528980d3298ceb52bf9e729d046f3b5cbfa | [
"MIT"
] | 3 | 2020-09-17T05:10:22.000Z | 2020-11-09T12:51:17.000Z | Machine learning with Python/Week3/Logistic Regression.ipynb | harunurrashid97/Research-Work | e141c8f38ec63abdcae6685509b3b50f2eb5d0ca | [
"MIT"
] | null | null | null | Machine learning with Python/Week3/Logistic Regression.ipynb | harunurrashid97/Research-Work | e141c8f38ec63abdcae6685509b3b50f2eb5d0ca | [
"MIT"
] | 1 | 2019-09-09T12:32:27.000Z | 2019-09-09T12:32:27.000Z | 42.087063 | 10,696 | 0.593871 | [
[
[
"<a href=\"https://www.bigdatauniversity.com\"><img src=\"https://ibm.box.com/shared/static/cw2c7r3o20w9zn8gkecaeyjhgw3xdgbj.png\" width=400 align=\"center\"></a>\n\n<h1 align=\"center\"><font size=\"5\"> Logistic Regression with Python</font></h1>",
"_____no_output_____"
],
[
"In this notebook, you will learn Logistic Regression, and then, you'll create a model for a telecommunication company, to predict when its customers will leave for a competitor, so that they can take some action to retain the customers.",
"_____no_output_____"
],
[
"<h1>Table of contents</h1>\n\n<div class=\"alert alert-block alert-info\" style=\"margin-top: 20px\">\n <ol>\n <li><a href=\"#about_dataset\">About the dataset</a></li>\n <li><a href=\"#preprocessing\">Data pre-processing and selection</a></li>\n <li><a href=\"#modeling\">Modeling (Logistic Regression with Scikit-learn)</a></li>\n <li><a href=\"#evaluation\">Evaluation</a></li>\n <li><a href=\"#practice\">Practice</a></li>\n </ol>\n</div>\n<br>\n<hr>",
"_____no_output_____"
],
[
"<a id=\"ref1\"></a>\n## What is the difference between Linear and Logistic Regression?\n\nWhile Linear Regression is suited for estimating continuous values (e.g. estimating house price), it is not the best tool for predicting the class of an observed data point. In order to estimate the class of a data point, we need some sort of guidance on what would be the <b>most probable class</b> for that data point. For this, we use <b>Logistic Regression</b>.\n\n<div class=\"alert alert-success alertsuccess\" style=\"margin-top: 20px\">\n<font size = 3><strong>Recall linear regression:</strong></font>\n<br>\n<br>\n As you know, <b>Linear regression</b> finds a function that relates a continuous dependent variable, <b>y</b>, to some predictors (independent variables $x_1$, $x_2$, etc.). For example, Simple linear regression assumes a function of the form:\n<br><br>\n$$\ny = \\theta_0 + \\theta_1 x_1 + \\theta_2 x_2 + \\cdots\n$$\n<br>\nand finds the values of parameters $\\theta_0, \\theta_1, \\theta_2$, etc, where the term $\\theta_0$ is the \"intercept\". It can be generally shown as:\n<br><br>\n$$\nℎ_\\theta(𝑥) = \\theta^TX\n$$\n<p></p>\n\n</div>\n\nLogistic Regression is a variation of Linear Regression, useful when the observed dependent variable, <b>y</b>, is categorical. It produces a formula that predicts the probability of the class label as a function of the independent variables.\n\nLogistic regression fits a special s-shaped curve by taking the linear regression and transforming the numeric estimate into a probability with the following function, which is called sigmoid function 𝜎:\n\n$$\nℎ_\\theta(𝑥) = \\sigma({\\theta^TX}) = \\frac {e^{(\\theta_0 + \\theta_1 x_1 + \\theta_2 x_2 +...)}}{1 + e^{(\\theta_0 + \\theta_1 x_1 + \\theta_2 x_2 +\\cdots)}}\n$$\nOr:\n$$\nProbabilityOfaClass_1 = P(Y=1|X) = \\sigma({\\theta^TX}) = \\frac{e^{\\theta^TX}}{1+e^{\\theta^TX}} \n$$\n\nIn this equation, ${\\theta^TX}$ is the regression result (the sum of the variables weighted by the coefficients), `exp` is the exponential function and $\\sigma(\\theta^TX)$ is the sigmoid or [logistic function](http://en.wikipedia.org/wiki/Logistic_function), also called logistic curve. It is a common \"S\" shape (sigmoid curve).\n\nSo, briefly, Logistic Regression passes the input through the logistic/sigmoid but then treats the result as a probability:\n\n<img\nsrc=\"https://ibm.box.com/shared/static/kgv9alcghmjcv97op4d6onkyxevk23b1.png\" width=\"400\" align=\"center\">\n\n\nThe objective of __Logistic Regression__ algorithm, is to find the best parameters θ, for $ℎ_\\theta(𝑥)$ = $\\sigma({\\theta^TX})$, in such a way that the model best predicts the class of each case.",
"_____no_output_____"
],
[
"### Customer churn with Logistic Regression\nA telecommunications company is concerned about the number of customers leaving their land-line business for cable competitors. They need to understand who is leaving. Imagine that you are an analyst at this company and you have to find out who is leaving and why.",
"_____no_output_____"
],
[
"Lets first import required libraries:",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport pylab as pl\nimport numpy as np\nimport scipy.optimize as opt\nfrom sklearn import preprocessing\n%matplotlib inline \nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"<h2 id=\"about_dataset\">About the dataset</h2>\nWe will use a telecommunications dataset for predicting customer churn. This is a historical customer dataset where each row represents one customer. The data is relatively easy to understand, and you may uncover insights you can use immediately. Typically it is less expensive to keep customers than acquire new ones, so the focus of this analysis is to predict the customers who will stay with the company. \n\n\nThis data set provides information to help you predict what behavior will help you to retain customers. You can analyze all relevant customer data and develop focused customer retention programs.\n\n\n\nThe dataset includes information about:\n\n- Customers who left within the last month – the column is called Churn\n- Services that each customer has signed up for – phone, multiple lines, internet, online security, online backup, device protection, tech support, and streaming TV and movies\n- Customer account information – how long they had been a customer, contract, payment method, paperless billing, monthly charges, and total charges\n- Demographic info about customers – gender, age range, and if they have partners and dependents\n",
"_____no_output_____"
],
[
"### Load the Telco Churn data \nTelco Churn is a hypothetical data file that concerns a telecommunications company's efforts to reduce turnover in its customer base. Each case corresponds to a separate customer and it records various demographic and service usage information. Before you can work with the data, you must use the URL to get the ChurnData.csv.\n\nTo download the data, we will use `!wget` to download it from IBM Object Storage.",
"_____no_output_____"
]
],
[
[
"#Click here and press Shift+Enter\n!wget -O ChurnData.csv https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/ML0101ENv3/labs/ChurnData.csv",
"--2019-04-09 12:01:22-- https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/ML0101ENv3/labs/ChurnData.csv\nResolving s3-api.us-geo.objectstorage.softlayer.net (s3-api.us-geo.objectstorage.softlayer.net)... 67.228.254.193\nConnecting to s3-api.us-geo.objectstorage.softlayer.net (s3-api.us-geo.objectstorage.softlayer.net)|67.228.254.193|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 36144 (35K) [text/csv]\nSaving to: ‘ChurnData.csv’\n\nChurnData.csv 100%[=====================>] 35.30K --.-KB/s in 0.02s \n\n2019-04-09 12:01:22 (1.62 MB/s) - ‘ChurnData.csv’ saved [36144/36144]\n\n"
]
],
[
[
"__Did you know?__ When it comes to Machine Learning, you will likely be working with large datasets. As a business, where can you host your data? IBM is offering a unique opportunity for businesses, with 10 Tb of IBM Cloud Object Storage: [Sign up now for free](http://cocl.us/ML0101EN-IBM-Offer-CC)",
"_____no_output_____"
],
[
"### Load Data From CSV File ",
"_____no_output_____"
]
],
[
[
"churn_df = pd.read_csv(\"ChurnData.csv\")\nchurn_df.head()",
"_____no_output_____"
]
],
[
[
"<h2 id=\"preprocessing\">Data pre-processing and selection</h2>",
"_____no_output_____"
],
[
"Lets select some features for the modeling. Also we change the target data type to be integer, as it is a requirement by the skitlearn algorithm:",
"_____no_output_____"
]
],
[
[
"churn_df = churn_df[['tenure', 'age', 'address', 'income', 'ed', 'employ', 'equip', 'callcard', 'wireless','churn']]\nchurn_df['churn'] = churn_df['churn'].astype('int')\nchurn_df.head()",
"_____no_output_____"
]
],
[
[
"## Practice\nHow many rows and columns are in this dataset in total? What are the name of columns?",
"_____no_output_____"
]
],
[
[
"# write your code here\nchurn_df.shape\n\n",
"_____no_output_____"
]
],
[
[
"Lets define X, and y for our dataset:",
"_____no_output_____"
]
],
[
[
"X = np.asarray(churn_df[['tenure', 'age', 'address', 'income', 'ed', 'employ', 'equip']])\nX[0:5]",
"_____no_output_____"
],
[
"y = np.asarray(churn_df['churn'])\ny [0:5]",
"_____no_output_____"
]
],
[
[
"Also, we normalize the dataset:",
"_____no_output_____"
]
],
[
[
"from sklearn import preprocessing\nX = preprocessing.StandardScaler().fit(X).transform(X)\nX[0:5]",
"_____no_output_____"
]
],
[
[
"## Train/Test dataset",
"_____no_output_____"
],
[
"Okay, we split our dataset into train and test set:",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\nX_train, X_test, y_train, y_test = train_test_split( X, y, test_size=0.2, random_state=4)\nprint ('Train set:', X_train.shape, y_train.shape)\nprint ('Test set:', X_test.shape, y_test.shape)",
"Train set: (160, 7) (160,)\nTest set: (40, 7) (40,)\n"
]
],
[
[
"<h2 id=\"modeling\">Modeling (Logistic Regression with Scikit-learn)</h2>",
"_____no_output_____"
],
[
"Lets build our model using __LogisticRegression__ from Scikit-learn package. This function implements logistic regression and can use different numerical optimizers to find parameters, including ‘newton-cg’, ‘lbfgs’, ‘liblinear’, ‘sag’, ‘saga’ solvers. You can find extensive information about the pros and cons of these optimizers if you search it in internet.\n\nThe version of Logistic Regression in Scikit-learn, support regularization. Regularization is a technique used to solve the overfitting problem in machine learning models.\n__C__ parameter indicates __inverse of regularization strength__ which must be a positive float. Smaller values specify stronger regularization. \nNow lets fit our model with train set:",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LogisticRegression\nfrom sklearn.metrics import confusion_matrix\nLR = LogisticRegression(C=0.01, solver='liblinear').fit(X_train,y_train)\nLR",
"_____no_output_____"
]
],
[
[
"Now we can predict using our test set:",
"_____no_output_____"
]
],
[
[
"yhat = LR.predict(X_test)\nyhat",
"_____no_output_____"
]
],
[
[
"__predict_proba__ returns estimates for all classes, ordered by the label of classes. So, the first column is the probability of class 1, P(Y=1|X), and second column is probability of class 0, P(Y=0|X):",
"_____no_output_____"
]
],
[
[
"yhat_prob = LR.predict_proba(X_test)\nyhat_prob",
"_____no_output_____"
]
],
[
[
"<h2 id=\"evaluation\">Evaluation</h2>",
"_____no_output_____"
],
[
"### jaccard index\nLets try jaccard index for accuracy evaluation. we can define jaccard as the size of the intersection divided by the size of the union of two label sets. If the entire set of predicted labels for a sample strictly match with the true set of labels, then the subset accuracy is 1.0; otherwise it is 0.0.\n\n",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import jaccard_similarity_score\njaccard_similarity_score(y_test, yhat)",
"_____no_output_____"
]
],
[
[
"### confusion matrix\nAnother way of looking at accuracy of classifier is to look at __confusion matrix__.",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import classification_report, confusion_matrix\nimport itertools\ndef plot_confusion_matrix(cm, classes,\n normalize=False,\n title='Confusion matrix',\n cmap=plt.cm.Blues):\n \"\"\"\n This function prints and plots the confusion matrix.\n Normalization can be applied by setting `normalize=True`.\n \"\"\"\n if normalize:\n cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]\n print(\"Normalized confusion matrix\")\n else:\n print('Confusion matrix, without normalization')\n\n print(cm)\n\n plt.imshow(cm, interpolation='nearest', cmap=cmap)\n plt.title(title)\n plt.colorbar()\n tick_marks = np.arange(len(classes))\n plt.xticks(tick_marks, classes, rotation=45)\n plt.yticks(tick_marks, classes)\n\n fmt = '.2f' if normalize else 'd'\n thresh = cm.max() / 2.\n for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):\n plt.text(j, i, format(cm[i, j], fmt),\n horizontalalignment=\"center\",\n color=\"white\" if cm[i, j] > thresh else \"black\")\n\n plt.tight_layout()\n plt.ylabel('True label')\n plt.xlabel('Predicted label')\nprint(confusion_matrix(y_test, yhat, labels=[1,0]))",
"[[ 6 9]\n [ 1 24]]\n"
],
[
"# Compute confusion matrix\ncnf_matrix = confusion_matrix(y_test, yhat, labels=[1,0])\nnp.set_printoptions(precision=2)\n\n\n# Plot non-normalized confusion matrix\nplt.figure()\nplot_confusion_matrix(cnf_matrix, classes=['churn=1','churn=0'],normalize= False, title='Confusion matrix')",
"Confusion matrix, without normalization\n[[ 6 9]\n [ 1 24]]\n"
]
],
[
[
"Look at first row. The first row is for customers whose actual churn value in test set is 1.\nAs you can calculate, out of 40 customers, the churn value of 15 of them is 1. \nAnd out of these 15, the classifier correctly predicted 6 of them as 1, and 9 of them as 0. \n\nIt means, for 6 customers, the actual churn value were 1 in test set, and classifier also correctly predicted those as 1. However, while the actual label of 9 customers were 1, the classifier predicted those as 0, which is not very good. We can consider it as error of the model for first row.\n\nWhat about the customers with churn value 0? Lets look at the second row.\nIt looks like there were 25 customers whom their churn value were 0. \n\n\nThe classifier correctly predicted 24 of them as 0, and one of them wrongly as 1. So, it has done a good job in predicting the customers with churn value 0. A good thing about confusion matrix is that shows the model’s ability to correctly predict or separate the classes. In specific case of binary classifier, such as this example, we can interpret these numbers as the count of true positives, false positives, true negatives, and false negatives. ",
"_____no_output_____"
]
],
[
[
"print (classification_report(y_test, yhat))\n",
" precision recall f1-score support\n\n 0 0.73 0.96 0.83 25\n 1 0.86 0.40 0.55 15\n\n micro avg 0.75 0.75 0.75 40\n macro avg 0.79 0.68 0.69 40\nweighted avg 0.78 0.75 0.72 40\n\n"
]
],
[
[
"Based on the count of each section, we can calculate precision and recall of each label:\n\n\n- __Precision__ is a measure of the accuracy provided that a class label has been predicted. It is defined by: precision = TP / (TP + FP)\n\n- __Recall__ is true positive rate. It is defined as: Recall = TP / (TP + FN)\n\n \nSo, we can calculate precision and recall of each class.\n\n__F1 score:__\nNow we are in the position to calculate the F1 scores for each label based on the precision and recall of that label. \n\nThe F1 score is the harmonic average of the precision and recall, where an F1 score reaches its best value at 1 (perfect precision and recall) and worst at 0. It is a good way to show that a classifer has a good value for both recall and precision.\n\n\nAnd finally, we can tell the average accuracy for this classifier is the average of the F1-score for both labels, which is 0.72 in our case.",
"_____no_output_____"
],
[
"### log loss\nNow, lets try __log loss__ for evaluation. In logistic regression, the output can be the probability of customer churn is yes (or equals to 1). This probability is a value between 0 and 1.\nLog loss( Logarithmic loss) measures the performance of a classifier where the predicted output is a probability value between 0 and 1. \n",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import log_loss\nlog_loss(y_test, yhat_prob)",
"_____no_output_____"
]
],
[
[
"<h2 id=\"practice\">Practice</h2>\nTry to build Logistic Regression model again for the same dataset, but this time, use different __solver__ and __regularization__ values? What is new __logLoss__ value?",
"_____no_output_____"
]
],
[
[
"# write your code here\n\n",
"_____no_output_____"
]
],
[
[
"Double-click __here__ for the solution.\n\n<!-- Your answer is below:\n \nLR2 = LogisticRegression(C=0.01, solver='sag').fit(X_train,y_train)\nyhat_prob2 = LR2.predict_proba(X_test)\nprint (\"LogLoss: : %.2f\" % log_loss(y_test, yhat_prob2))\n\n-->",
"_____no_output_____"
],
[
"<h2>Want to learn more?</h2>\n\nIBM SPSS Modeler is a comprehensive analytics platform that has many machine learning algorithms. It has been designed to bring predictive intelligence to decisions made by individuals, by groups, by systems – by your enterprise as a whole. A free trial is available through this course, available here: <a href=\"http://cocl.us/ML0101EN-SPSSModeler\">SPSS Modeler</a>\n\nAlso, you can use Watson Studio to run these notebooks faster with bigger datasets. Watson Studio is IBM's leading cloud solution for data scientists, built by data scientists. With Jupyter notebooks, RStudio, Apache Spark and popular libraries pre-packaged in the cloud, Watson Studio enables data scientists to collaborate on their projects without having to install anything. Join the fast-growing community of Watson Studio users today with a free account at <a href=\"https://cocl.us/ML0101EN_DSX\">Watson Studio</a>\n\n<h3>Thanks for completing this lesson!</h3>\n\n<h4>Author: <a href=\"https://ca.linkedin.com/in/saeedaghabozorgi\">Saeed Aghabozorgi</a></h4>\n<p><a href=\"https://ca.linkedin.com/in/saeedaghabozorgi\">Saeed Aghabozorgi</a>, PhD is a Data Scientist in IBM with a track record of developing enterprise level applications that substantially increases clients’ ability to turn data into actionable knowledge. He is a researcher in data mining field and expert in developing advanced analytic methods like machine learning and statistical modelling on large datasets.</p>\n\n<hr>\n\n<p>Copyright © 2018 <a href=\"https://cocl.us/DX0108EN_CC\">Cognitive Class</a>. This notebook and its source code are released under the terms of the <a href=\"https://bigdatauniversity.com/mit-license/\">MIT License</a>.</p>",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d0852f0c388420c41d04b5f683410b6580218ec3 | 7,594 | ipynb | Jupyter Notebook | login.ipynb | NoNameGr/NoName | e437ada090612bb44de0524affb66348537eda56 | [
"MIT"
] | null | null | null | login.ipynb | NoNameGr/NoName | e437ada090612bb44de0524affb66348537eda56 | [
"MIT"
] | null | null | null | login.ipynb | NoNameGr/NoName | e437ada090612bb44de0524affb66348537eda56 | [
"MIT"
] | 2 | 2020-07-30T04:10:37.000Z | 2020-07-30T04:15:10.000Z | 45.473054 | 92 | 0.585594 | [
[
[
"from PyQt5 import QtCore, QtGui, QtWidgets",
"_____no_output_____"
],
[
"class Ui_Dialog(object):\n def setupUi(self, Dialog):\n Dialog.setObjectName(\"Dialog\")\n Dialog.resize(419, 383)\n self.radioButton = QtWidgets.QRadioButton(Dialog)\n self.radioButton.setGeometry(QtCore.QRect(200, 110, 100, 20))\n self.radioButton.setObjectName(\"radioButton\")\n self.buttonGroup = QtWidgets.QButtonGroup(Dialog)\n self.buttonGroup.setObjectName(\"buttonGroup\")\n self.buttonGroup.addButton(self.radioButton)\n self.radioButton_2 = QtWidgets.QRadioButton(Dialog)\n self.radioButton_2.setGeometry(QtCore.QRect(200, 140, 100, 20))\n self.radioButton_2.setObjectName(\"radioButton_2\")\n self.buttonGroup.addButton(self.radioButton_2)\n self.radioButton_3 = QtWidgets.QRadioButton(Dialog)\n self.radioButton_3.setGeometry(QtCore.QRect(200, 190, 100, 20))\n self.radioButton_3.setObjectName(\"radioButton_3\")\n self.buttonGroup_2 = QtWidgets.QButtonGroup(Dialog)\n self.buttonGroup_2.setObjectName(\"buttonGroup_2\")\n self.buttonGroup_2.addButton(self.radioButton_3)\n self.radioButton_4 = QtWidgets.QRadioButton(Dialog)\n self.radioButton_4.setGeometry(QtCore.QRect(200, 220, 161, 20))\n self.radioButton_4.setObjectName(\"radioButton_4\")\n self.buttonGroup_2.addButton(self.radioButton_4)\n self.radioButton_5 = QtWidgets.QRadioButton(Dialog)\n self.radioButton_5.setGeometry(QtCore.QRect(200, 250, 100, 20))\n self.radioButton_5.setObjectName(\"radioButton_5\")\n self.buttonGroup_2.addButton(self.radioButton_5)\n self.label = QtWidgets.QLabel(Dialog)\n self.label.setGeometry(QtCore.QRect(90, 10, 271, 51))\n font = QtGui.QFont()\n font.setPointSize(31)\n font.setBold(True)\n font.setWeight(75)\n self.label.setFont(font)\n self.label.setObjectName(\"label\")\n self.label_2 = QtWidgets.QLabel(Dialog)\n self.label_2.setGeometry(QtCore.QRect(330, 50, 81, 31))\n self.label_2.setObjectName(\"label_2\")\n self.label_3 = QtWidgets.QLabel(Dialog)\n self.label_3.setGeometry(QtCore.QRect(20, 90, 171, 81))\n font = QtGui.QFont()\n font.setPointSize(18)\n self.label_3.setFont(font)\n self.label_3.setObjectName(\"label_3\")\n self.label_4 = QtWidgets.QLabel(Dialog)\n self.label_4.setGeometry(QtCore.QRect(20, 190, 171, 81))\n font = QtGui.QFont()\n font.setPointSize(18)\n self.label_4.setFont(font)\n self.label_4.setObjectName(\"label_4\")\n self.commandLinkButton = QtWidgets.QCommandLinkButton(Dialog)\n self.commandLinkButton.setGeometry(QtCore.QRect(10, 330, 131, 51))\n font = QtGui.QFont()\n font.setPointSize(18)\n self.commandLinkButton.setFont(font)\n self.commandLinkButton.setIconSize(QtCore.QSize(25, 25))\n self.commandLinkButton.setCheckable(False)\n self.commandLinkButton.setDescription(\"\")\n self.commandLinkButton.setObjectName(\"commandLinkButton\")\n self.commandLinkButton_2 = QtWidgets.QCommandLinkButton(Dialog)\n self.commandLinkButton_2.setGeometry(QtCore.QRect(150, 330, 131, 51))\n font = QtGui.QFont()\n font.setPointSize(18)\n self.commandLinkButton_2.setFont(font)\n self.commandLinkButton_2.setIconSize(QtCore.QSize(25, 25))\n self.commandLinkButton_2.setCheckable(False)\n self.commandLinkButton_2.setDescription(\"\")\n self.commandLinkButton_2.setObjectName(\"commandLinkButton_2\")\n self.commandLinkButton_3 = QtWidgets.QCommandLinkButton(Dialog)\n self.commandLinkButton_3.setGeometry(QtCore.QRect(280, 330, 131, 51))\n font = QtGui.QFont()\n font.setPointSize(18)\n self.commandLinkButton_3.setFont(font)\n self.commandLinkButton_3.setIconSize(QtCore.QSize(25, 25))\n self.commandLinkButton_3.setCheckable(False)\n self.commandLinkButton_3.setDescription(\"\")\n self.commandLinkButton_3.setObjectName(\"commandLinkButton_3\")\n self.pushButton = QtWidgets.QPushButton(Dialog)\n self.pushButton.setGeometry(QtCore.QRect(270, 280, 131, 51))\n self.pushButton.setObjectName(\"pushButton\")\n\n self.retranslateUi(Dialog)\n QtCore.QMetaObject.connectSlotsByName(Dialog)\n\n def retranslateUi(self, Dialog):\n _translate = QtCore.QCoreApplication.translate\n Dialog.setWindowTitle(_translate(\"Dialog\", \"Dialog\"))\n self.radioButton.setText(_translate(\"Dialog\", \"Tiếng Việt\"))\n self.radioButton_2.setText(_translate(\"Dialog\", \"English\"))\n self.radioButton_3.setText(_translate(\"Dialog\", \"Dễ / Easy\"))\n self.radioButton_4.setText(_translate(\"Dialog\", \"Trung bình / Medium\"))\n self.radioButton_5.setText(_translate(\"Dialog\", \"Khó / Hard\"))\n self.label.setText(_translate(\"Dialog\", \"Đuổi hình bắt chữ\"))\n self.label_2.setText(_translate(\"Dialog\", \"by NoName\"))\n self.label_3.setText(_translate(\"Dialog\", \"Phiên bản / Version\"))\n self.label_4.setText(_translate(\"Dialog\", \"Mức độ chơi / Level\"))\n self.commandLinkButton.setText(_translate(\"Dialog\", \"Facebook\"))\n self.commandLinkButton_2.setText(_translate(\"Dialog\", \"Google\"))\n self.commandLinkButton_3.setText(_translate(\"Dialog\", \"Twitter\"))\n self.pushButton.setText(_translate(\"Dialog\", \"Start\"))",
"_____no_output_____"
],
[
"if __name__ == \"__main__\":\n import sys\n app = QtWidgets.QApplication(sys.argv)\n Dialog = QtWidgets.QDialog()\n ui = Ui_Dialog()\n ui.setupUi(Dialog)\n Dialog.show()\n sys.exit(app.exec_())",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
d085419c9322e08c986651be75172f1215e19b34 | 15,273 | ipynb | Jupyter Notebook | aws_es/2 - ES Indexer.ipynb | rdempsey/osc-demo | ecba4410e6b8a96bf367403f6321301d89ce0dc1 | [
"Apache-2.0"
] | null | null | null | aws_es/2 - ES Indexer.ipynb | rdempsey/osc-demo | ecba4410e6b8a96bf367403f6321301d89ce0dc1 | [
"Apache-2.0"
] | null | null | null | aws_es/2 - ES Indexer.ipynb | rdempsey/osc-demo | ecba4410e6b8a96bf367403f6321301d89ce0dc1 | [
"Apache-2.0"
] | null | null | null | 36.538278 | 184 | 0.414719 | [
[
[
"# Imports\nimport json\nimport pandas as pd\nimport numpy as np\nfrom espandas import Espandas\nimport uuid\nfrom datetime import datetime",
"_____no_output_____"
],
[
"# Load the CSV data\ndf = pd.read_csv(\"yts_data_fully_enriched_clean.csv\", index_col=False, header=0)\nlen(df)",
"_____no_output_____"
],
[
"# Check the data types\ndf.dtypes",
"_____no_output_____"
],
[
"# Create an ID column of UUIDs and add it to the dataframe\nindex_ids = list()\nfor i in range(0,8610):\n index_ids.append(i)\n\ndf['indexId'] = index_ids",
"_____no_output_____"
],
[
"# Create a datetime column so we have one for Elasticsearch\ndf['created_at'] = datetime.utcnow().strftime(\"%Y/%m/%d %H:%M:%S\")\ndf['updated_at'] = datetime.utcnow().strftime(\"%Y/%m/%d %H:%M:%S\")",
"_____no_output_____"
],
[
"# Let's look at some of the data\ndf.head()",
"_____no_output_____"
],
[
"# Index the dataframe into Elasticsearch\nINDEX = 'movies'\nTYPE = 'movie'\nesp = Espandas()\nesp.es_write(df, INDEX, TYPE)",
"index does not exist, creating index\n"
],
[
"# Query for the first ten rows and see that they match the original\nk = df.indexId[0:10]\nres = esp.es_read(k, INDEX, TYPE)\nres == df.iloc[0:10].astype('str')",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0855535fd95c7cb79249cfffebc3579a74e0d8e | 27,404 | ipynb | Jupyter Notebook | t81_558_class_13_02_checkpoint.ipynb | rserran/t81_558_deep_learning | ec312cc7a7cef207e55e382594455fe44bcdec11 | [
"Apache-2.0"
] | null | null | null | t81_558_class_13_02_checkpoint.ipynb | rserran/t81_558_deep_learning | ec312cc7a7cef207e55e382594455fe44bcdec11 | [
"Apache-2.0"
] | null | null | null | t81_558_class_13_02_checkpoint.ipynb | rserran/t81_558_deep_learning | ec312cc7a7cef207e55e382594455fe44bcdec11 | [
"Apache-2.0"
] | null | null | null | 37.694635 | 749 | 0.588637 | [
[
[
"<a href=\"https://colab.research.google.com/github/jeffheaton/t81_558_deep_learning/blob/master/t81_558_class_13_02_checkpoint.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# T81-558: Applications of Deep Neural Networks\n**Module 13: Advanced/Other Topics**\n* Instructor: [Jeff Heaton](https://sites.wustl.edu/jeffheaton/), McKelvey School of Engineering, [Washington University in St. Louis](https://engineering.wustl.edu/Programs/Pages/default.aspx)\n* For more information visit the [class website](https://sites.wustl.edu/jeffheaton/t81-558/).",
"_____no_output_____"
],
[
"# Module 13 Video Material\n\n* Part 13.1: Flask and Deep Learning Web Services [[Video]](https://www.youtube.com/watch?v=H73m9XvKHug&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](https://github.com/jeffheaton/t81_558_deep_learning/blob/master/t81_558_class_13_01_flask.ipynb)\n* **Part 13.2: Interrupting and Continuing Training** [[Video]](https://www.youtube.com/watch?v=kaQCdv46OBA&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](https://github.com/jeffheaton/t81_558_deep_learning/blob/master/t81_558_class_13_02_checkpoint.ipynb)\n* Part 13.3: Using a Keras Deep Neural Network with a Web Application [[Video]](https://www.youtube.com/watch?v=OBbw0e-UroI&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](https://github.com/jeffheaton/t81_558_deep_learning/blob/master/t81_558_class_13_03_web.ipynb)\n* Part 13.4: When to Retrain Your Neural Network [[Video]](https://www.youtube.com/watch?v=K2Tjdx_1v9g&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](https://github.com/jeffheaton/t81_558_deep_learning/blob/master/t81_558_class_13_04_retrain.ipynb)\n* Part 13.5: Tensor Processing Units (TPUs) [[Video]](https://www.youtube.com/watch?v=Ygyf3NUqvSc&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](https://github.com/jeffheaton/t81_558_deep_learning/blob/master/t81_558_class_13_05_tpu.ipynb)\n\n",
"_____no_output_____"
],
[
"## Google CoLab Instructions\nThe following code ensures that Google CoLab is running the correct version of TensorFlow.",
"_____no_output_____"
]
],
[
[
"try:\n from google.colab import drive\n COLAB = True\n print(\"Note: using Google CoLab\")\n %tensorflow_version 2.x\nexcept:\n print(\"Note: not using Google CoLab\")\n COLAB = False\n\n# Nicely formatted time string\ndef hms_string(sec_elapsed):\n h = int(sec_elapsed / (60 * 60))\n m = int((sec_elapsed % (60 * 60)) / 60)\n s = sec_elapsed % 60\n return f\"{h}:{m:>02}:{s:>05.2f}\"",
"Note: using Google CoLab\n"
]
],
[
[
"# Part 13.2: Interrupting and Continuing Training\n\nWe would train our Keras models in one pass in an ideal world, utilizing as much GPU and CPU power as we need. The world in which we train our models is anything but ideal. In this part, we will see that we can stop and continue and even adjust training at later times. We accomplish this continuation with checkpoints. We begin by creating several utility functions. The first utility generates an output directory that has a unique name. This technique allows us to organize multiple runs of our experiment. We provide the Logger class to route output to a log file contained in the output directory.",
"_____no_output_____"
]
],
[
[
"import os\nimport re\nimport sys\nimport time\nimport numpy as np\nfrom typing import Any, List, Tuple, Union\nfrom tensorflow.keras.datasets import mnist\nfrom tensorflow.keras import backend as K\nimport tensorflow as tf\nimport tensorflow.keras\nimport tensorflow as tf\nfrom tensorflow.keras.callbacks import EarlyStopping, \\\n LearningRateScheduler, ModelCheckpoint\nfrom tensorflow.keras import regularizers\nfrom tensorflow.keras.models import Sequential\nfrom tensorflow.keras.layers import Dense, Dropout, Flatten\nfrom tensorflow.keras.layers import Conv2D, MaxPooling2D\nfrom tensorflow.keras.models import load_model\nimport pickle\n\ndef generate_output_dir(outdir, run_desc):\n prev_run_dirs = []\n if os.path.isdir(outdir):\n prev_run_dirs = [x for x in os.listdir(outdir) if os.path.isdir(\\\n os.path.join(outdir, x))]\n prev_run_ids = [re.match(r'^\\d+', x) for x in prev_run_dirs]\n prev_run_ids = [int(x.group()) for x in prev_run_ids if x is not None]\n cur_run_id = max(prev_run_ids, default=-1) + 1\n run_dir = os.path.join(outdir, f'{cur_run_id:05d}-{run_desc}')\n assert not os.path.exists(run_dir)\n os.makedirs(run_dir)\n return run_dir\n\n# From StyleGAN2\nclass Logger(object):\n \"\"\"Redirect stderr to stdout, optionally print stdout to a file, and \n optionally force flushing on both stdout and the file.\"\"\"\n\n def __init__(self, file_name: str = None, file_mode: str = \"w\", \\\n should_flush: bool = True):\n self.file = None\n\n if file_name is not None:\n self.file = open(file_name, file_mode)\n\n self.should_flush = should_flush\n self.stdout = sys.stdout\n self.stderr = sys.stderr\n\n sys.stdout = self\n sys.stderr = self\n\n def __enter__(self) -> \"Logger\":\n return self\n\n def __exit__(self, exc_type: Any, exc_value: Any, \\\n traceback: Any) -> None:\n self.close()\n\n def write(self, text: str) -> None:\n \"\"\"Write text to stdout (and a file) and optionally flush.\"\"\"\n if len(text) == 0: \n return\n\n if self.file is not None:\n self.file.write(text)\n\n self.stdout.write(text)\n\n if self.should_flush:\n self.flush()\n\n def flush(self) -> None:\n \"\"\"Flush written text to both stdout and a file, if open.\"\"\"\n if self.file is not None:\n self.file.flush()\n\n self.stdout.flush()\n\n def close(self) -> None:\n \"\"\"Flush, close possible files, and remove \n stdout/stderr mirroring.\"\"\"\n self.flush()\n\n # if using multiple loggers, prevent closing in wrong order\n if sys.stdout is self:\n sys.stdout = self.stdout\n if sys.stderr is self:\n sys.stderr = self.stderr\n\n if self.file is not None:\n self.file.close()\n\ndef obtain_data():\n (x_train, y_train), (x_test, y_test) = mnist.load_data()\n print(\"Shape of x_train: {}\".format(x_train.shape))\n print(\"Shape of y_train: {}\".format(y_train.shape))\n print()\n print(\"Shape of x_test: {}\".format(x_test.shape))\n print(\"Shape of y_test: {}\".format(y_test.shape))\n\n # input image dimensions\n img_rows, img_cols = 28, 28\n if K.image_data_format() == 'channels_first':\n x_train = x_train.reshape(x_train.shape[0], 1, img_rows, img_cols)\n x_test = x_test.reshape(x_test.shape[0], 1, img_rows, img_cols)\n input_shape = (1, img_rows, img_cols)\n else:\n x_train = x_train.reshape(x_train.shape[0], img_rows, img_cols, 1)\n x_test = x_test.reshape(x_test.shape[0], img_rows, img_cols, 1)\n input_shape = (img_rows, img_cols, 1)\n x_train = x_train.astype('float32')\n x_test = x_test.astype('float32')\n x_train /= 255\n x_test /= 255\n print('x_train shape:', x_train.shape)\n print(\"Training samples: {}\".format(x_train.shape[0]))\n print(\"Test samples: {}\".format(x_test.shape[0]))\n # convert class vectors to binary class matrices\n y_train = tf.keras.utils.to_categorical(y_train, num_classes)\n y_test = tf.keras.utils.to_categorical(y_test, num_classes)\n \n return input_shape, x_train, y_train, x_test, y_test\n",
"_____no_output_____"
]
],
[
[
"We define the basic training parameters and where we wish to write the output.",
"_____no_output_____"
]
],
[
[
"outdir = \"./data/\"\nrun_desc = \"test-train\"\nbatch_size = 128\nnum_classes = 10\n\nrun_dir = generate_output_dir(outdir, run_desc)\nprint(f\"Results saved to: {run_dir}\")",
"Results saved to: ./data/00000-test-train\n"
]
],
[
[
"Keras provides a prebuilt checkpoint class named **ModelCheckpoint** that contains most of our desired functionality. This built-in class can save the model's state repeatedly as training progresses. Stopping neural network training is not always a controlled event. Sometimes this stoppage can be abrupt, such as a power failure or a network resource shutting down. If Microsoft Windows is your operating system of choice, your training can also be interrupted by a high-priority system update. Because of all of this uncertainty, it is best to save your model at regular intervals. This process is similar to saving a game at critical checkpoints, so you do not have to start over if something terrible happens to your avatar in the game.\n\nWe will create our checkpoint class, named **MyModelCheckpoint**. In addition to saving the model, we also save the state of the training infrastructure. Why save the training infrastructure, in addition to the weights? This technique eases the transition back into training for the neural network and will be more efficient than a cold start. \n\nConsider if you interrupted your college studies after the first year. Sure, your brain (the neural network) will retain all the knowledge. But how much rework will you have to do? Your transcript at the university is like the training parameters. It ensures you do not have to start over when you come back.",
"_____no_output_____"
]
],
[
[
"class MyModelCheckpoint(ModelCheckpoint):\n def __init__(self, *args, **kwargs):\n super().__init__(*args, **kwargs)\n\n def on_epoch_end(self, epoch, logs=None):\n super().on_epoch_end(epoch,logs)\\\n\n # Also save the optimizer state\n filepath = self._get_file_path(epoch=epoch, \n logs=logs, batch=None)\n filepath = filepath.rsplit( \".\", 1 )[ 0 ] \n filepath += \".pkl\"\n\n with open(filepath, 'wb') as fp:\n pickle.dump(\n {\n 'opt': model.optimizer.get_config(),\n 'epoch': epoch+1\n # Add additional keys if you need to store more values\n }, fp, protocol=pickle.HIGHEST_PROTOCOL)\n print('\\nEpoch %05d: saving optimizaer to %s' % (epoch + 1, filepath))",
"_____no_output_____"
]
],
[
[
"The optimizer applies a step decay schedule during training to decrease the learning rate as training progresses. It is essential to preserve the current epoch that we are on to perform correctly after a training resume.",
"_____no_output_____"
]
],
[
[
"def step_decay_schedule(initial_lr=1e-3, decay_factor=0.75, step_size=10):\n def schedule(epoch):\n return initial_lr * (decay_factor ** np.floor(epoch/step_size))\n return LearningRateScheduler(schedule)",
"_____no_output_____"
]
],
[
[
"We build the model just as we have in previous sessions. However, the training function requires a few extra considerations. We specify the maximum number of epochs; however, we also allow the user to select the starting epoch number for training continuation. ",
"_____no_output_____"
]
],
[
[
"def build_model(input_shape, num_classes):\n model = Sequential()\n model.add(Conv2D(32, kernel_size=(3, 3),\n activation='relu',\n input_shape=input_shape))\n model.add(Conv2D(64, (3, 3), activation='relu'))\n model.add(MaxPooling2D(pool_size=(2, 2)))\n model.add(Dropout(0.25))\n model.add(Flatten())\n model.add(Dense(128, activation='relu'))\n model.add(Dropout(0.5))\n model.add(Dense(num_classes, activation='softmax'))\n model.compile(\n loss='categorical_crossentropy', \n optimizer=tf.keras.optimizers.Adam(),\n metrics=['accuracy'])\n return model\n\ndef train_model(model, initial_epoch=0, max_epochs=10):\n start_time = time.time()\n\n checkpoint_cb = MyModelCheckpoint(\n os.path.join(run_dir, 'model-{epoch:02d}-{val_loss:.2f}.hdf5'),\n monitor='val_loss',verbose=1)\n\n lr_sched_cb = step_decay_schedule(initial_lr=1e-4, decay_factor=0.75, \\\n step_size=2)\n cb = [checkpoint_cb, lr_sched_cb]\n\n model.fit(x_train, y_train,\n batch_size=batch_size,\n epochs=max_epochs,\n initial_epoch = initial_epoch,\n verbose=2, callbacks=cb,\n validation_data=(x_test, y_test))\n score = model.evaluate(x_test, y_test, verbose=0, callbacks=cb)\n print('Test loss: {}'.format(score[0]))\n print('Test accuracy: {}'.format(score[1]))\n\n elapsed_time = time.time() - start_time\n print(\"Elapsed time: {}\".format(hms_string(elapsed_time)))",
"_____no_output_____"
]
],
[
[
"We now begin training, using the **Logger** class to write the output to a log file in the output directory.",
"_____no_output_____"
]
],
[
[
"with Logger(os.path.join(run_dir, 'log.txt')):\n input_shape, x_train, y_train, x_test, y_test = obtain_data()\n model = build_model(input_shape, num_classes)\n train_model(model, max_epochs=3)",
"Downloading data from https://storage.googleapis.com/tensorflow/tf-keras-datasets/mnist.npz\n11493376/11490434 [==============================] - 0s 0us/step\n11501568/11490434 [==============================] - 0s 0us/step\nShape of x_train: (60000, 28, 28)\nShape of y_train: (60000,)\n\nShape of x_test: (10000, 28, 28)\nShape of y_test: (10000,)\nx_train shape: (60000, 28, 28, 1)\nTraining samples: 60000\nTest samples: 10000\nEpoch 1/3\n\nEpoch 1: saving model to ./data/00000-test-train/model-01-0.20.hdf5\n\nEpoch 00001: saving optimizaer to ./data/00000-test-train/model-01-0.20.pkl\n469/469 - 12s - loss: 0.6354 - accuracy: 0.8129 - val_loss: 0.1977 - val_accuracy: 0.9420 - lr: 1.0000e-04 - 12s/epoch - 25ms/step\nEpoch 2/3\n\nEpoch 2: saving model to ./data/00000-test-train/model-02-0.11.hdf5\n\nEpoch 00002: saving optimizaer to ./data/00000-test-train/model-02-0.11.pkl\n469/469 - 2s - loss: 0.2284 - accuracy: 0.9332 - val_loss: 0.1087 - val_accuracy: 0.9677 - lr: 1.0000e-04 - 2s/epoch - 5ms/step\nEpoch 3/3\n\nEpoch 3: saving model to ./data/00000-test-train/model-03-0.08.hdf5\n\nEpoch 00003: saving optimizaer to ./data/00000-test-train/model-03-0.08.pkl\n469/469 - 2s - loss: 0.1575 - accuracy: 0.9541 - val_loss: 0.0837 - val_accuracy: 0.9746 - lr: 7.5000e-05 - 2s/epoch - 5ms/step\nTest loss: 0.08365701138973236\nTest accuracy: 0.9746000170707703\nElapsed time: 0:00:22.09\n"
]
],
[
[
"You should notice that the above output displays the name of the hdf5 and pickle (pkl) files produced at each checkpoint. These files serve the following functions:\n\n* Pickle files contain the state of the optimizer.\n* HDF5 files contain the saved model.\n\nFor this training run, which went for 3 epochs, these two files were named:\n\n* ./data/00013-test-train/model-03-0.08.hdf5\n* ./data/00013-test-train/model-03-0.08.pkl\n\nWe can inspect the output from the training run. Notice we can see a folder named \"00000-test-train\". This new folder was the first training run. The program will call the next training run \"00001-test-train\", and so on. Inside this directory, you will find the pickle and hdf5 files for each checkpoint. ",
"_____no_output_____"
]
],
[
[
"!ls ./data",
"00000-test-train\n"
],
[
"!ls ./data/00000-test-train",
"log.txt\t\t model-01-0.20.pkl\tmodel-02-0.11.pkl model-03-0.08.pkl\nmodel-01-0.20.hdf5 model-02-0.11.hdf5\tmodel-03-0.08.hdf5\n"
]
],
[
[
"Keras stores the model itself in an HDF5, which includes the optimizer. Because of this feature, it is not generally necessary to restore the internal state of the optimizer (such as ADAM). However, we include the code to do so. We can obtain the internal state of an optimizer by calling **get_config**, which will return a dictionary similar to the following:\n\n```\n{'name': 'Adam', 'learning_rate': 7.5e-05, 'decay': 0.0, \n'beta_1': 0.9, 'beta_2': 0.999, 'epsilon': 1e-07, 'amsgrad': False}\n```\n\nIn practice, I've found that different optimizers implement get_config differently. This function will always return the training hyperparameters. However, it may not always capture the complete internal state of an optimizer beyond the hyperparameters. The exact implementation of get_config can vary per optimizer implementation.\n\n## Continuing Training\n\nWe are now ready to continue training. You will need the paths to both your HDF5 and PKL files. You can find these paths in the output above. Your values may differ from mine, so perform a copy/paste.\n",
"_____no_output_____"
]
],
[
[
"MODEL_PATH = './data/00000-test-train/model-03-0.08.hdf5'\nOPT_PATH = './data/00000-test-train/model-03-0.08.pkl'",
"_____no_output_____"
]
],
[
[
"The following code loads the HDF5 and PKL files and then recompiles the model based on the PKL file. Depending on the optimizer in use, you might have to recompile the model. ",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nfrom tensorflow.keras.models import load_model\nimport pickle\n\ndef load_model_data(model_path, opt_path):\n model = load_model(model_path)\n with open(opt_path, 'rb') as fp:\n d = pickle.load(fp)\n epoch = d['epoch']\n opt = d['opt']\n return epoch, model, opt\n\nepoch, model, opt = load_model_data(MODEL_PATH, OPT_PATH)\n\n# note: often it is not necessary to recompile the model\nmodel.compile(\n loss='categorical_crossentropy', \n optimizer=tf.keras.optimizers.Adam.from_config(opt),\n metrics=['accuracy'])",
"_____no_output_____"
]
],
[
[
"Finally, we train the model for additional epochs. You can see from the output that the new training starts at a higher accuracy than the first training run. Further, the accuracy increases with additional training. Also, you will notice that the epoch number begins at four and not one.",
"_____no_output_____"
]
],
[
[
"outdir = \"./data/\"\nrun_desc = \"cont-train\"\nnum_classes = 10\n\nrun_dir = generate_output_dir(outdir, run_desc)\nprint(f\"Results saved to: {run_dir}\")\n\nwith Logger(os.path.join(run_dir, 'log.txt')):\n input_shape, x_train, y_train, x_test, y_test = obtain_data()\n train_model(model, initial_epoch=epoch, max_epochs=6)",
"Results saved to: ./data/00001-cont-train\nShape of x_train: (60000, 28, 28)\nShape of y_train: (60000,)\n\nShape of x_test: (10000, 28, 28)\nShape of y_test: (10000,)\nx_train shape: (60000, 28, 28, 1)\nTraining samples: 60000\nTest samples: 10000\nEpoch 4/6\n\nEpoch 4: saving model to ./data/00001-cont-train/model-04-0.07.hdf5\n\nEpoch 00004: saving optimizaer to ./data/00001-cont-train/model-04-0.07.pkl\n469/469 - 3s - loss: 0.1272 - accuracy: 0.9634 - val_loss: 0.0692 - val_accuracy: 0.9788 - lr: 7.5000e-05 - 3s/epoch - 6ms/step\nEpoch 5/6\n\nEpoch 5: saving model to ./data/00001-cont-train/model-05-0.06.hdf5\n\nEpoch 00005: saving optimizaer to ./data/00001-cont-train/model-05-0.06.pkl\n469/469 - 2s - loss: 0.1099 - accuracy: 0.9677 - val_loss: 0.0612 - val_accuracy: 0.9818 - lr: 5.6250e-05 - 2s/epoch - 5ms/step\nEpoch 6/6\n\nEpoch 6: saving model to ./data/00001-cont-train/model-06-0.06.hdf5\n\nEpoch 00006: saving optimizaer to ./data/00001-cont-train/model-06-0.06.pkl\n469/469 - 2s - loss: 0.0990 - accuracy: 0.9711 - val_loss: 0.0561 - val_accuracy: 0.9827 - lr: 5.6250e-05 - 2s/epoch - 5ms/step\nTest loss: 0.05610647052526474\nTest accuracy: 0.982699990272522\nElapsed time: 0:00:11.72\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0855a6b29fb3216870f9d63f200f7ef8f1e8708 | 10,861 | ipynb | Jupyter Notebook | core/interactions/interaction - loss function tests.ipynb | Flipajs/FERDA | 120a9e16d7ab4877f72fadbe4484c8b91adf22c4 | [
"MIT"
] | 1 | 2021-04-23T10:00:49.000Z | 2021-04-23T10:00:49.000Z | core/interactions/interaction - loss function tests.ipynb | Flipajs/FERDA | 120a9e16d7ab4877f72fadbe4484c8b91adf22c4 | [
"MIT"
] | null | null | null | core/interactions/interaction - loss function tests.ipynb | Flipajs/FERDA | 120a9e16d7ab4877f72fadbe4484c8b91adf22c4 | [
"MIT"
] | null | null | null | 30.508427 | 135 | 0.543136 | [
[
[
"%matplotlib inline\nimport matplotlib.pylab as plt\nimport numpy as np\nfrom keras import objectives\nfrom keras import backend as K\nfrom keras import losses\nimport tensorflow as tf\nimport interactions_results\nimport train_interactions",
"_____no_output_____"
],
[
"OBJ_IDS = ['1', '2']\n\nCOLUMNS_MAP = [('x', 'ant%s_x'),\n ('y', 'ant%s_y'),\n ('major', 'ant%s_major'),\n ('minor', 'ant%s_minor'),\n ('angle_deg', 'ant%s_angle_deg'),\n ('dx', 'ant%s_dx'),\n ('dy', 'ant%s_dy'),\n ]\n\nCOL = dict(COLUMNS_MAP)\nNAMES = reduce(list.__add__, [[value % i for key, value in COLUMNS_MAP] for i in OBJ_IDS])\nCOL2ID = {key: i for i, (key, value) in enumerate(COLUMNS_MAP)}",
"_____no_output_____"
],
[
"COL2ID",
"_____no_output_____"
],
[
"def angle_absolute_error(y_true, y_pred, backend, scaler=None):\n if scaler is not None:\n # y_pred_ = scaler.inverse_transform(y_pred[:, 4:5]) # this doesn't work with Tensors\n y_pred_ = y_pred[:, 4:5] * scaler[1] + scaler[0]\n else:\n y_pred_ = y_pred[:, 4:5]\n val = backend.abs(y_pred_ - y_true[:, 4:5]) % 180\n return backend.minimum(val, 180 - val)\n\n\ndef xy_absolute_error(y_true, y_pred, backend):\n return backend.abs(y_pred[:, :2] - y_true[:, :2])\n\n\ndef absolute_errors(y_true, y_pred, backend, angle_scaler):\n theta = angle_absolute_error(y_true, y_pred, backend, angle_scaler)\n pos = xy_absolute_error(y_true, y_pred, backend)\n return pos, theta\n\n\ndef interaction_loss(y_true, y_pred, angle_scaler=None, alpha=0.5):\n assert 0 <= alpha <= 1\n sum_errors_xy, sum_errors_angle, indices = match_pred_to_gt(y_true, y_pred, K, angle_scaler)\n\n return K.mean(tf.gather_nd(sum_errors_xy, indices) * (1 - alpha) +\n tf.gather_nd(sum_errors_angle, indices) * alpha)\n\n\ny_a = np.array([[10., 10, 25, 5, 20, 100, 100, 25, 5, 30],\n [100., 100, 25, 5, 30, 20, 20, 25, 5, 20],\n [10., 10, 25, 5, 20, 200, 200, 25, 5, 30]])\ny_b = np.array([[20., 20, 25, 5, 30, 150, 170, 25, 5, 0],\n [30., 30, 25, 5, 30, 170, 150, 25, 5, 5],\n [30., 60, 25, 5, 30, 170, 120, 25, 5, 5]])\nxy, angle, indices = train_interactions.match_pred_to_gt(y_a, y_b, np)\n\nprint (xy[indices[:, 0], indices[:, 1]]).mean()\nprint (angle[indices[:, 0], indices[:, 1]]).mean()",
"_____no_output_____"
],
[
"# with h5py.File(DATA_DIR + '/imgs_inter_test.h5', 'r') as hf:\n# X_test = hf['data'][:]\n#\ny_a_ = interactions_results.tostruct(y_a)\ny_b_ = interactions_results.tostruct(y_b)\ni = 1\ninteractions_results.plot_interaction(y_a_[[i]], y_b_[[i]])\nplt.ylim(0, 200)\nplt.xlim(0, 200)",
"_____no_output_____"
],
[
"y_true = K.variable(y_a)\ny_pred = K.variable(y_b)\nbackend = K\nangle_scaler = None",
"_____no_output_____"
],
[
"K.eval(y_pred[:, 1:2])",
"_____no_output_____"
],
[
"K.eval(y_pred[:, [COL2ID['x'], COL2ID['y']]] - y_true[:, [COL2ID['x'], COL2ID['y']]])",
"_____no_output_____"
],
[
"y_true = y_a\ny_pred = y_b\nbackend = np\nangle_scaler = None",
"_____no_output_____"
],
[
"mean_errors_xy, mean_errors_angle, indices = train_interactions.match_pred_to_gt(y_true, y_pred, backend)\nfor x in [mean_errors_xy, mean_errors_angle, indices]:\n print x",
"_____no_output_____"
],
[
"mean_errors_xy, mean_errors_angle, indices = train_interactions.match_pred_to_gt(y_true, y_pred, backend)\nfor x in [mean_errors_xy, mean_errors_angle, indices]:\n print K.eval(x)",
"_____no_output_____"
],
[
"mean_errors_xy, mean_errors_angle, indices = train_interactions.match_pred_to_gt(y_true, y_pred, K)\nfor x in [mean_errors_xy, mean_errors_angle, indices]:\n print K.eval(x)",
"_____no_output_____"
],
[
"# def match_pred_to_gt(y_true, y_pred, backend, angle_scaler=None):\n\"\"\"\nReturn mean absolute errors for individual samples for xy and theta\nin two possible combinations of prediction and ground truth.\n\"\"\"\nxy11, theta11 = absolute_errors(y_true[:, :5], y_pred[:, :5], backend, angle_scaler)\nxy22, theta22 = absolute_errors(y_true[:, 5:], y_pred[:, 5:], backend, angle_scaler)\nxy12, theta12 = absolute_errors(y_true[:, :5], y_pred[:, 5:], backend, angle_scaler)\nxy21, theta21 = absolute_errors(y_true[:, 5:], y_pred[:, :5], backend, angle_scaler)\nif backend == np:\n norm = np.linalg.norm\n int64 = np.int64\n shape = lambda x, n: x.shape[n]\nelse:\n norm = tf.linalg.norm\n int64 = tf.int64\n shape = lambda x, n: backend.cast(backend.shape(x)[n], int64)\nmean_errors_xy = backend.stack((backend.mean(backend.stack((norm(xy11, axis=1), norm(xy22, axis=1))), axis=0),\n backend.mean(backend.stack((norm(xy12, axis=1), norm(xy21, axis=1))), axis=0))) # shape=(2, n)\n\nmean_errors_angle = backend.stack((backend.mean(backend.concatenate((theta11, theta22)), axis=1),\n backend.mean(backend.concatenate((theta12, theta21)), axis=1))) # shape=(2, n)\n\nprint K.eval(theta11)\nprint K.eval(backend.concatenate((theta11, theta22)))\nprint K.eval(backend.sum(backend.concatenate((theta11, theta22)), axis=1))\n\nswap_idx = backend.argmin(mean_errors_xy, axis=0) # shape = (n,)\nindices = backend.transpose(\n backend.stack((swap_idx, backend.arange(0, shape(mean_errors_xy, 1))))) # shape=(n, 2)\n# return mean_errors_xy, mean_errors_angle, indices\n\nfor x in [mean_errors_xy, mean_errors_angle, indices]:\n print K.eval(x)",
"_____no_output_____"
],
[
"angle_scaler = None\ny_true = K.variable(y_a)\ny_pred = K.variable(y_b)\nxy11, theta11 = absolute_errors(y_true[:, :5], y_pred[:, :5], angle_scaler)\nxy22, theta22 = absolute_errors(y_true[:, 5:], y_pred[:, 5:], angle_scaler)\n\nxy12, theta12 = absolute_errors(y_true[:, :5], y_pred[:, 5:], angle_scaler)\nxy21, theta21 = absolute_errors(y_true[:, 5:], y_pred[:, :5], angle_scaler)\n\nnorm = tf.linalg.norm\n# print y_a\n# print y_b\n# print K.eval(xy11)\n# print K.eval(xy22)\n# print K.eval(xy12)\n# print K.eval(xy21)\nsum_errors_xy = K.stack((K.sum(K.stack((norm(xy11, axis=1), norm(xy22, axis=1))), axis=0),\n K.sum(K.stack((norm(xy12, axis=1), norm(xy21, axis=1))), axis=0))) # shape=(2, n)\nsum_errors_angle = K.stack((K.sum(K.concatenate((theta11, theta22)), axis=1),\n K.sum(K.concatenate((theta12, theta21)), axis=1))) # shape=(2, n)\n\nswap_idx = K.argmin(sum_errors_xy, axis=0) # shape = (n,)\nindices = K.transpose(K.stack((swap_idx, K.arange(0, K.cast(K.shape(sum_errors_xy)[1], tf.int64))))) # shape=(n, 2) \n\nprint K.eval(tf.gather_nd(sum_errors_xy, idx))\nprint K.eval(tf.gather_nd(sum_errors_angle, idx))\nprint K.eval(tf.gather_nd(sum_errors_xy, idx) + tf.gather_nd(sum_errors_angle, idx))",
"_____no_output_____"
],
[
"idx = K.transpose(K.stack((swap_idx, K.arange(0, K.cast(K.shape(sum_errors_xy)[1], tf.int64)))))",
"_____no_output_____"
],
[
"K.eval(tf.gather_nd(sum_errors_xy, idx))",
"_____no_output_____"
],
[
"K.eval(sum_errors_xy)",
"_____no_output_____"
],
[
"K.eval(sum_errors_angle)",
"_____no_output_____"
],
[
"K.eval(tf.gather_nd(sum_errors_angle, idx))",
"_____no_output_____"
],
[
"np.ca",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d08560a51ee887a0640dfe386aa0b006835ac3d8 | 8,289 | ipynb | Jupyter Notebook | docs/source/notebooks/readme.ipynb | UN-GCPDS/openbci-stream | 339c8cf033788438658d5766b07406fe1f68db07 | [
"BSD-2-Clause"
] | 5 | 2021-04-13T13:14:59.000Z | 2022-03-22T10:47:06.000Z | docs/source/notebooks/readme.ipynb | UN-GCPDS/openbci-stream | 339c8cf033788438658d5766b07406fe1f68db07 | [
"BSD-2-Clause"
] | 1 | 2021-10-04T10:04:34.000Z | 2021-10-04T10:04:34.000Z | docs/source/notebooks/readme.ipynb | UN-GCPDS/openbci-stream | 339c8cf033788438658d5766b07406fe1f68db07 | [
"BSD-2-Clause"
] | 2 | 2021-10-04T19:45:35.000Z | 2021-11-18T22:42:59.000Z | 44.326203 | 1,020 | 0.653999 | [
[
[
"> Developed by [Yeison Nolberto Cardona Álvarez](https://github.com/yeisonCardona) \n> [Andrés Marino Álvarez Meza, PhD.](https://github.com/amalvarezme) \n> César Germán Castellanos Dominguez, PhD. \n> _Digital Signal Processing and Control Group_ | _Grupo de Control y Procesamiento Digital de Señales ([GCPDS](https://github.com/UN-GCPDS/))_ \n> _National University of Colombia at Manizales_ | _Universidad Nacional de Colombia sede Manizales_\n\n----",
"_____no_output_____"
],
[
"# OpenBCI-Stream \nHigh level Python module for EEG/EMG/ECG acquisition and distributed streaming for OpenBCI Cyton board.\n\n\n\n\n\n\n\n\n[](https://openbci-stream.readthedocs.io/en/latest/?badge=latest)\n\nComprise a set of scripts that deals with the configuration and connection with the board, also is compatible with both connection modes supported by [Cyton](https://shop.openbci.com/products/cyton-biosensing-board-8-channel?variant=38958638542): RFduino (Serial dongle) and Wi-Fi (with the OpenBCI Wi-Fi Shield). These drivers are a stand-alone library that can handle the board from three different endpoints: (i) a [Command-Line Interface](06-command_line_interface.ipynb) (CLI) with simple instructions configure, start and stop data acquisition, debug stream status, and register events markers; (ii) a [Python Module](03-data_acuisition.ipynb) with high-level instructions and asynchronous acquisition; (iii) an object-proxying using Remote Python Call (RPyC) for [distributed implementations](A4-server-based-acquisition.ipynb) that can manipulate the Python modules as if they were local, this last mode needs a daemon running in the remote host that will listen to connections and driving instructions.\n\nThe main functionality of the drivers live on to serve real-time and distributed access to data flow, even on single machine implementations, this is achieved by implementing [Kafka](https://kafka.apache.org/) and their capabilities to create multiple topics for classifying the streaming, these topics are used to separate the neurophysiological data from the [event markers](05-stream_markers), so the clients can subscribe to a specific topic for injecting or read content, this means that is possible to implement an event register in a separate process that stream markers for all clients in real-time without handle dense time-series data. A crucial issue that stays on [time synchronization](A4-server-based_acquisition.ipynb#Step-5---Configure-time-server), all systems components in the network should have the same real-time protocol (RTP) server reference. ",
"_____no_output_____"
],
[
"## Main features\n\n * **Asynchronous acquisition:** Acquisition and deserialization are done in uninterrupted parallel processes. In this way, the sampling rate keeps stable as long as possible.\n * **Distributed streaming system:** The acquisition, processing, visualizations, and any other system that needs to be fed with EEG/EMG/ECG real-time data can run with their architecture.\n * **Remote board handle:** Same code syntax for developing and debug Cython boards connected to any node in the distributed system.\n * **Command-line interface:** A simple interface for handle the start, stop, and access to data stream directly from the command line.\n * **Markers/Events handler:** Besides the marker boardmode available in Cyton, a stream channel for the reading and writing of markers is available for use in any development. \n * **Multiple boards:** Is possible to use multiple OpenBCI boards just by adding multiple endpoints to the commands.",
"_____no_output_____"
],
[
"## Examples",
"_____no_output_____"
]
],
[
[
"# Acquisition with blocking call\n\nfrom openbci_stream.acquisition import Cyton\nopenbci = Cyton('serial', endpoint='/dev/ttyUSB0', capture_stream=True)\n\n# blocking call\nopenbci.stream(15) # collect data for 15 seconds\n\n# openbci.eeg_time_series \n# openbci.aux_time_series\n# openbci.timestamp_time_series ",
"_____no_output_____"
],
[
"# Acquisition with asynchronous call\n\nfrom openbci_stream.acquisition import Cyton\nopenbci = Cyton('wifi', endpoint='192.68.1.113', capture_stream=True)\nopenbci.stream(15) # collect data for 15 seconds\n\n# asynchronous call\nopenbci.start_stream()\ntime.sleep(15) # collect data for 15 seconds\nopenbci.stop_stream()",
"_____no_output_____"
],
[
"# Remote acquisition\n\nfrom openbci_stream.acquisition import Cyton\nopenbci = Cyton('serial', endpoint='/dev/ttyUSB0', host='192.168.1.1', capture_stream=True)\n\n# blocking call\nopenbci.stream(15) # collect data for 15 seconds",
"_____no_output_____"
],
[
"# Consumer for active streamming\n\nfrom openbci_stream.acquisition import OpenBCIConsumer\nwith OpenBCIConsumer() as stream:\n for i, message in enumerate(stream):\n if message.topic == 'eeg':\n print(f\"received {message.value['samples']} samples\")\n if i == 9:\n break",
"_____no_output_____"
],
[
"# Create stream then consume data\n\nfrom openbci_stream.acquisition import OpenBCIConsumer\nwith OpenBCIConsumer(mode='serial', endpoint='/dev/ttyUSB0', streaming_package_size=250) as (stream, openbci):\n t0 = time.time()\n for i, message in enumerate(stream):\n if message.topic == 'eeg':\n print(f\"{i}: received {message.value['samples']} samples\")\n t0 = time.time()\n if i == 9:\n break",
"_____no_output_____"
],
[
"# Acquisition with multiple boards\n\nfrom openbci_stream.acquisition import Cyton\nopenbci = Cyton('wifi', endpoint=['192.68.1.113', '192.68.1.185'], capture_stream=True)\nopenbci.stream(15) # collect data for 15 seconds\n\n# asynchronous call\nopenbci.start_stream()\ntime.sleep(15) # collect data for 15 seconds\nopenbci.stop_stream()",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0856e0fb2291512365de5169dbbf885c4386506 | 5,589 | ipynb | Jupyter Notebook | Notion/Notion_Update_pages_from_database.ipynb | vivard/awesome-notebooks | 899558bcc2165bb2155f5ab69ac922c6458e1799 | [
"BSD-3-Clause"
] | null | null | null | Notion/Notion_Update_pages_from_database.ipynb | vivard/awesome-notebooks | 899558bcc2165bb2155f5ab69ac922c6458e1799 | [
"BSD-3-Clause"
] | null | null | null | Notion/Notion_Update_pages_from_database.ipynb | vivard/awesome-notebooks | 899558bcc2165bb2155f5ab69ac922c6458e1799 | [
"BSD-3-Clause"
] | null | null | null | 23.884615 | 294 | 0.533369 | [
[
[
"<img width=\"10%\" alt=\"Naas\" src=\"https://landen.imgix.net/jtci2pxwjczr/assets/5ice39g4.png?w=160\"/>",
"_____no_output_____"
],
[
"# Notion - Update pages from database\n<a href=\"https://app.naas.ai/user-redirect/naas/downloader?url=https://raw.githubusercontent.com/jupyter-naas/awesome-notebooks/master/Notion/Notion_Update_pages_from_database.ipynb\" target=\"_parent\"><img src=\"https://naasai-public.s3.eu-west-3.amazonaws.com/open_in_naas.svg\"/></a>",
"_____no_output_____"
],
[
"**Tags:** #notion #productivity",
"_____no_output_____"
],
[
"## Input",
"_____no_output_____"
],
[
"### Import library",
"_____no_output_____"
]
],
[
[
"from naas_drivers import notion",
"_____no_output_____"
]
],
[
[
"### Setup Notion\n<a href='https://docs.naas.ai/drivers/notion'>How to get your Notion integration token ?</a>",
"_____no_output_____"
]
],
[
[
"# Enter Token API\nnotion_token = \"*****\"\n\n# Enter Database URL\ndatabase_url = \"https://www.notion.so/********\"",
"_____no_output_____"
]
],
[
[
"## Model",
"_____no_output_____"
],
[
"### Get pages from Notion DB",
"_____no_output_____"
]
],
[
[
"database_id = database_url.split(\"/\")[-1].split(\"?v=\")[0]\npages = notion.connect(notion_token).database.query(database_id, query={})\nprint(\"📊 Pages in Notion DB:\", len(pages))",
"_____no_output_____"
]
],
[
[
"## Output",
"_____no_output_____"
],
[
"### Update pages",
"_____no_output_____"
]
],
[
[
"for page in pages:\n print(page)\n# page.title(\"Name\",\"Page updated\")\n# page.rich_text(\"Text\",\"Ceci est toto\")\n# page.number(\"Number\", 42)\n# page.select(\"Select\",\"Value3\") \n# page.multi_select(\"Muti Select\",[\"Value1\",\"Value2\",\"Value3\"])\n# page.date(\"Date\",\"2021-10-03T17:01:26\") #Follow ISO 8601 format\n# page.people(\"People\", [\"6e3bab71-beeb-484b-af99-ea30fdef4773\"]) #list of ID of users\n# page.checkbox(\"Checkbox\", False)\n# page.email(\"Email\",\"[email protected]\")\n page.update()\n print(f\"✅ Page updated in Notion.\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
d085797a6385dd4ef67f5e4b40bbe93accab3ffb | 37,902 | ipynb | Jupyter Notebook | site/ja/tutorials/customization/performance.ipynb | nic-fp/docs | a3dff9a33f832e6fd53b3ace6337c854ce707431 | [
"Apache-2.0"
] | 3 | 2020-01-09T02:58:22.000Z | 2020-09-11T09:02:01.000Z | site/ja/tutorials/customization/performance.ipynb | nic-fp/docs | a3dff9a33f832e6fd53b3ace6337c854ce707431 | [
"Apache-2.0"
] | 1 | 2019-10-22T11:24:17.000Z | 2019-10-22T11:24:17.000Z | site/ja/tutorials/customization/performance.ipynb | nic-fp/docs | a3dff9a33f832e6fd53b3ace6337c854ce707431 | [
"Apache-2.0"
] | 1 | 2019-12-07T21:16:25.000Z | 2019-12-07T21:16:25.000Z | 28.932824 | 341 | 0.482296 | [
[
[
"##### Copyright 2019 The TensorFlow Authors.\n",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# tf.function で性能アップ\n\n<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/tutorials/customization/performance\"><img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/docs/blob/master/site/ja/tutorials/customization/performance.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/docs/blob/master/site/ja/tutorials/customization/performance.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View source on GitHub</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/docs/site/ja/tutorials/customization/performance.ipynb\"><img src=\"https://www.tensorflow.org/images/download_logo_32px.png\" />Download notebook</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"TensorFlow 2.0 では Eager Execution が既定で有効になっています。ユーザーインターフェイスは直感的で柔軟です(演算を一度だけ行う場合にはずっと簡単に、かつ迅速に実行されます)。しかしながら、それは性能と展開の面での犠牲の上に成り立っています。\n\n最高性能を得ながら、モデルをどこへでも展開できるようにするには、`tf.function` を使ってプログラムから計算グラフを作成します。\nAutoGraph のおかげで、驚くほど多くの Python コードが tf.function でそのまま動作しますが、気をつけなければならない落とし穴も存在します。\n\nポイントと推奨事項は下記の通りです。\n\n- オブジェクトの変更やリストへの追加のような Python の副作用に依存しないこと\n- tf.functions は NumPy の演算や Python の組み込み演算よりも、TensorFlow の演算に適していること\n- 迷ったときは、`for x in y` というイディオムを使うこと",
"_____no_output_____"
]
],
[
[
"from __future__ import absolute_import, division, print_function, unicode_literals\n\ntry:\n %tensorflow_version 2.x\nexcept Exception:\n pass\nimport tensorflow as tf",
"_____no_output_____"
],
[
"import contextlib\n\n# 遭遇するかもしれないいくつかのエラーをデモするためのヘルパー関数\[email protected]\ndef assert_raises(error_class):\n try:\n yield\n except error_class as e:\n print('Caught expected exception \\n {}: {}'.format(error_class, e))\n except Exception as e:\n print('Got unexpected exception \\n {}: {}'.format(type(e), e))\n else:\n raise Exception('Expected {} to be raised but no error was raised!'.format(\n error_class))",
"_____no_output_____"
]
],
[
[
"あなたが定義した `tf.function` は TensorFlow Core の演算に似たものです。例えばそれを即時に実行することも、計算グラフで使うこともできますし、勾配を計算することも可能です。",
"_____no_output_____"
]
],
[
[
"# function は演算のように振る舞う\n\[email protected]\ndef add(a, b):\n return a + b\n\nadd(tf.ones([2, 2]), tf.ones([2, 2])) # [[2., 2.], [2., 2.]]",
"_____no_output_____"
],
[
"# function は勾配を計算できる\n\[email protected]\ndef add(a, b):\n return a + b\n\nv = tf.Variable(1.0)\nwith tf.GradientTape() as tape:\n result = add(v, 1.0)\ntape.gradient(result, v)",
"_____no_output_____"
],
[
"# function 内で function を使うこともできる\n\[email protected]\ndef dense_layer(x, w, b):\n return add(tf.matmul(x, w), b)\n\ndense_layer(tf.ones([3, 2]), tf.ones([2, 2]), tf.ones([2]))",
"_____no_output_____"
]
],
[
[
"## トレーシングとポリモーフィズム\n\nPython の動的型付けは、関数をさまざまな型の引数で呼び出すことができ、Python がそれぞれのシナリオで異なる動作をするということを意味します。\n\n他方で、TensorFlow の計算グラフでは、dtype と shape の次元が静的であることが必要です。`tf.function` は、正しい計算グラフを生成するために必要なときには関数を再トレースして、このギャップをつなぐ役割を果たします。\n\n異なる型の引数を使って関数を呼び出し、何が起きるか見てみましょう。",
"_____no_output_____"
]
],
[
[
"# Function はポリモーフィック\n\[email protected]\ndef double(a):\n print(\"Tracing with\", a)\n return a + a\n\nprint(double(tf.constant(1)))\nprint()\nprint(double(tf.constant(1.1)))\nprint()\nprint(double(tf.constant(\"a\")))\nprint()\n",
"_____no_output_____"
]
],
[
[
"トレースの動作を制御するためには、下記のようなテクニックを使います。\n\n- 新しい `tf.function` を作成する。別々の `tf.function` オブジェクトがトレースを共有することはない。\n- 特定のトレースを得るには `get_concrete_function` メソッドを使用する。\n- 計算グラフの呼び出し時に1回だけトレースを行うには、 `input_signature` を指定して `tf.function` を呼び出す。",
"_____no_output_____"
]
],
[
[
"print(\"Obtaining concrete trace\")\ndouble_strings = double.get_concrete_function(tf.TensorSpec(shape=None, dtype=tf.string))\nprint(\"Executing traced function\")\nprint(double_strings(tf.constant(\"a\")))\nprint(double_strings(a=tf.constant(\"b\")))\nprint(\"Using a concrete trace with incompatible types will throw an error\")\nwith assert_raises(tf.errors.InvalidArgumentError):\n double_strings(tf.constant(1))",
"_____no_output_____"
],
[
"@tf.function(input_signature=(tf.TensorSpec(shape=[None], dtype=tf.int32),))\ndef next_collatz(x):\n print(\"Tracing with\", x)\n return tf.where(tf.equal(x % 2, 0), x // 2, 3 * x + 1)\n\nprint(next_collatz(tf.constant([1, 2])))\n# 1次元のテンソルを input signature として指定しているので、これは失敗する\nwith assert_raises(ValueError):\n next_collatz(tf.constant([[1, 2], [3, 4]]))",
"_____no_output_____"
]
],
[
[
"## いつ再トレースするのか?\n\nポリモーフィックな `tf.function` はトレーシングによって生成された具象関数のキャッシュを保持しています。キャッシュのキーは、実際にはその関数の引数及びキーワード引数から生成されたキーのタプルです。`tf.Tensor` 引数から生成されるキーは、テンソルの shape と型です。Python の組み込み型引数から生成されるキーはその値です。それ以外の Python の型では、キーはオブジェクトの `id()` に基づいており、メソッドはクラスのインスタンスひとつずつ独立にトレースされます。将来、TensorFlowには、Python オブジェクトについて安全にテンソルに変換できるような、より洗練されたキャッシングが追加されるかもしれません。",
"_____no_output_____"
],
[
"## 引数は Python か? Tensor か?\n\nしばしば、ハイパーパラメータやグラフ構成を制御するために Python の組み込み型の引数が使われます。例えば、`num_layers=10` や `training=True` あるいは `nonlinearity='relu'` のようにです。このため、この Python の組み込み型の引数が変更されると、計算グラフを再びトレースする必要があるということになります。\n\nしかし、グラフの生成を制御するために Python の組み込み型の引数を使用する必要はありません。これらのケースでは、Python引数の値の変更が不必要な再トレースを引き起こす可能性があります。例えば、この訓練ループでは、AutoGraph は動的に展開を行います。複数回トレースを行っていますが、生成される計算グラフは全く変わりません。これは少し非効率です。",
"_____no_output_____"
]
],
[
[
"def train_one_step():\n pass\n\[email protected]\ndef train(num_steps):\n print(\"Tracing with num_steps = {}\".format(num_steps))\n for _ in tf.range(num_steps):\n train_one_step()\n\ntrain(num_steps=10)\ntrain(num_steps=20)\n",
"_____no_output_____"
]
],
[
[
"ここでの簡単な回避方法は、生成されたグラフの shape が変わらないのであれば、引数をテンソルにキャストすることです。",
"_____no_output_____"
]
],
[
[
"train(num_steps=tf.constant(10))\ntrain(num_steps=tf.constant(20))",
"_____no_output_____"
]
],
[
[
"## `tf.function` の中の副作用\n\n一般的には、(印字やオブジェクト変更のような)Python の副作用は、トレーシングの最中にだけ発生します。それでは、どうしたら `tf.function` で安定的に副作用を起こすことができるでしょうか?\n\n一般的な原則は、トレースをデバッグする際にだけ Python の副作用を使用するというものです。あるいは、`tf.Variable.assign`、`tf.print`、そして `tf.summary` のような TensorFlow の演算を使うことで、コードがトレースされるときにも、TensorFlowランタイムによって都度呼び出される際にも、確実に実行されるようにできます。一般には、関数型のスタイルを使用することで最も良い結果を得られます。 ",
"_____no_output_____"
]
],
[
[
"@tf.function\ndef f(x):\n print(\"Traced with\", x)\n tf.print(\"Executed with\", x)\n\nf(1)\nf(1)\nf(2)\n",
"_____no_output_____"
]
],
[
[
"`tf.function` が呼び出されるたびに Python のコードを実行したいのであれば、`tf.py_function` がぴったりです。`tf.py_function` の欠点は、ポータブルでないこと、それほど性能が高くないこと、(マルチGPU、TPUの)分散環境ではうまく動作しないことなどです。また、`tf.py_function` は計算グラフに組み込まれるため、入出力すべてをテンソルにキャストします。",
"_____no_output_____"
]
],
[
[
"external_list = []\n\ndef side_effect(x):\n print('Python side effect')\n external_list.append(x)\n\[email protected]\ndef f(x):\n tf.py_function(side_effect, inp=[x], Tout=[])\n\nf(1)\nf(1)\nf(1)\nassert len(external_list) == 3\n# .numpy() call required because py_function casts 1 to tf.constant(1)\nassert external_list[0].numpy() == 1\n",
"_____no_output_____"
]
],
[
[
"## Python の状態に注意\n\nジェネレーターやイテレーターなど Python の機能の多くは、状態を追跡するために Python のランタイムに依存しています。これらの仕組みは、一般的には Eager モードでも期待通りに動作しますが、トレーシングの振る舞いにより、`tf.function` の中では予期しないことが起きることがあります。\n\n1例として、イテレーターの状態が進むのは Python の副作用であり、トレーシングの中だけで発生します。",
"_____no_output_____"
]
],
[
[
"external_var = tf.Variable(0)\[email protected]\ndef buggy_consume_next(iterator):\n external_var.assign_add(next(iterator))\n tf.print(\"Value of external_var:\", external_var)\n\niterator = iter([0, 1, 2, 3])\nbuggy_consume_next(iterator)\n# 次のコードは、イテレーターの次の値を使うのではなく、最初の値を再利用する\nbuggy_consume_next(iterator)\nbuggy_consume_next(iterator)",
"_____no_output_____"
]
],
[
[
"イテレーターが tf.function の中で生成されすべて使われる場合には、正しく動作するはずです。しかし、イテレーター全体がトレースされることとなり、巨大な計算グラフの生成をまねく可能性があります。これは、望みどおりの動作かもしれません。しかし、もし Python のリストとして表されたメモリー上の巨大なデータセットを使って訓練を行うとすると、これは非常に大きな計算グラフを生成することになり、`tf.function` がスピードアップにはつながらないと考えられます。\n\nPython データを繰り返し使用する場合、もっとも安全な方法は tf.data.Dataset でラップして、`for x in y` というイディオムを使用することです。AutoGraph には、`y` がテンソルあるいは tf.data.Dataset である場合、`for` ループを安全に変換する特別な機能があります。",
"_____no_output_____"
]
],
[
[
"def measure_graph_size(f, *args):\n g = f.get_concrete_function(*args).graph\n print(\"{}({}) contains {} nodes in its graph\".format(\n f.__name__, ', '.join(map(str, args)), len(g.as_graph_def().node)))\n\[email protected]\ndef train(dataset):\n loss = tf.constant(0)\n for x, y in dataset:\n loss += tf.abs(y - x) # ダミー計算\n return loss\n\nsmall_data = [(1, 1)] * 2\nbig_data = [(1, 1)] * 10\nmeasure_graph_size(train, small_data)\nmeasure_graph_size(train, big_data)\n\nmeasure_graph_size(train, tf.data.Dataset.from_generator(\n lambda: small_data, (tf.int32, tf.int32)))\nmeasure_graph_size(train, tf.data.Dataset.from_generator(\n lambda: big_data, (tf.int32, tf.int32)))",
"_____no_output_____"
]
],
[
[
"Python/Numpy のデータを Dataset でラップする際には、`tf.data.Dataset.from_generator` と `tf.data.Dataset.from_tensors` の違いに留意しましょう。前者はデータを Python のまま保持し `tf.py_function` を通じて取得するため、性能に影響する場合があります。これに対して後者はデータのコピーを計算グラフの中の、ひとつの大きな `tf.constant()` に結びつけるため、メモリー消費に影響する可能性があります。 \n\nTFRecordDataset/CsvDataset/などを通じてデータをファイルから読み込むことが、データを使用する最も効率的な方法です。TensorFlow 自身が Python とは関係なく非同期のデータ読み込みとプリフェッチを管理することができるからです。",
"_____no_output_____"
],
[
"## 自動的な依存関係の制御\n\nプログラミングモデルとしての関数が一般的なデータフローグラフに対して非常に優位である点は、意図したコードの振る舞いがどのようなものであるかということについて、より多くの情報をランタイムに与えられるということにあります。\n\n例えば、同じ変数を何度も読んだり書いたりするコードを書く場合、データフローグラフではもともと意図されていた演算の順番を自然に組み込むわけではありません。`tf.function` の中では、もともとの Python コードの文の実行順序を参照することで、実行順序の曖昧さを解消します。これにより、`tf.function` の中のステートフルな演算の順序が、先行実行モードのセマンティクスを模していることになります。\n\nこれは、手動で制御の依存関係を加える必要がないことを意味しています。`tf.function` は十分賢いので、あなたのコードが正しく動作するために必要十分な最小限の制御の依存関係を追加してくれます。",
"_____no_output_____"
]
],
[
[
"# 自動的な依存関係の制御\n\na = tf.Variable(1.0)\nb = tf.Variable(2.0)\n\[email protected]\ndef f(x, y):\n a.assign(y * b)\n b.assign_add(x * a)\n return a + b\n\nf(1.0, 2.0) # 10.0",
"_____no_output_____"
]
],
[
[
"## 変数\n\n`tf.function` の中では、意図したコードの実行順序を活用するという同じアイデアを使って、変数の作成と活用を簡単に行うことができます。しかし、ひとつだけ非常に重要な欠点があります。それは、変数を使った場合、先行実行モードとグラフモードでは動作が変わるコードを書いてしまう可能性があるということです。\n\n特に、呼び出しの都度新しい変数を作成する場合にこれが発生します。トレーシングの意味では、`tf.function` は呼び出しのたびに同じ変数を再利用しますが、Eager モードでは呼び出しごとに新しい変数を生成します。この間違いを防止するため、`tf.function` は危険な変数の生成動作を見つけるとエラーを発生させます。",
"_____no_output_____"
]
],
[
[
"@tf.function\ndef f(x):\n v = tf.Variable(1.0)\n v.assign_add(x)\n return v\n\nwith assert_raises(ValueError):\n f(1.0)",
"_____no_output_____"
],
[
"# しかし、曖昧さの無いコードは大丈夫\n\nv = tf.Variable(1.0)\n\[email protected]\ndef f(x):\n return v.assign_add(x)\n\nprint(f(1.0)) # 2.0\nprint(f(2.0)) # 4.0",
"_____no_output_____"
],
[
"# 初めて関数が実行されるときだけ変数が生成されることを保証できれば\n# tf.function 内で変数を作成できる\n\nclass C: pass\nobj = C(); obj.v = None\n\[email protected]\ndef g(x):\n if obj.v is None:\n obj.v = tf.Variable(1.0)\n return obj.v.assign_add(x)\n\nprint(g(1.0)) # 2.0\nprint(g(2.0)) # 4.0",
"_____no_output_____"
],
[
"# 変数の初期化は、関数の引数や他の変数の値に依存可能\n# 制御の依存関係を生成するのと同じ手法で、正しい初期化の順序を発見可能\n\nstate = []\[email protected]\ndef fn(x):\n if not state:\n state.append(tf.Variable(2.0 * x))\n state.append(tf.Variable(state[0] * 3.0))\n return state[0] * x * state[1]\n\nprint(fn(tf.constant(1.0)))\nprint(fn(tf.constant(3.0)))",
"_____no_output_____"
]
],
[
[
"# AutoGraph の使用\n\n[autograph](https://www.tensorflow.org/guide/function) ライブラリは `tf.function` に完全に統合されており、計算グラフの中で動的に実行される条件文や繰り返しを書くことができます。\n\n`tf.cond` や `tf.while_loop` は `tf.function` でも使えますが、制御フローを含むコードは、命令形式で書いたほうが書きやすいし理解しやすいです。",
"_____no_output_____"
]
],
[
[
"# 単純な繰り返し\n\[email protected]\ndef f(x):\n while tf.reduce_sum(x) > 1:\n tf.print(x)\n x = tf.tanh(x)\n return x\n\nf(tf.random.uniform([5]))",
"_____no_output_____"
],
[
"# 興味があれば AutoGraph が生成するコードを調べることができる\n# ただし、アセンブリ言語を読むような感じがする\n\ndef f(x):\n while tf.reduce_sum(x) > 1:\n tf.print(x)\n x = tf.tanh(x)\n return x\n\nprint(tf.autograph.to_code(f))",
"_____no_output_____"
]
],
[
[
"## AutoGraph: 条件分岐\n\nAutoGraph は `if` 文を等価である `tf.cond` の呼び出しに変換します。\n\nこの置換は条件がテンソルである場合に行われます。そうでない場合には、条件分岐はトレーシングの中で実行されます。",
"_____no_output_____"
]
],
[
[
"def test_tf_cond(f, *args):\n g = f.get_concrete_function(*args).graph\n if any(node.name == 'cond' for node in g.as_graph_def().node):\n print(\"{}({}) uses tf.cond.\".format(\n f.__name__, ', '.join(map(str, args))))\n else:\n print(\"{}({}) executes normally.\".format(\n f.__name__, ', '.join(map(str, args))))\n",
"_____no_output_____"
],
[
"@tf.function\ndef hyperparam_cond(x, training=True):\n if training:\n x = tf.nn.dropout(x, rate=0.5)\n return x\n\[email protected]\ndef maybe_tensor_cond(x):\n if x < 0:\n x = -x\n return x\n\ntest_tf_cond(hyperparam_cond, tf.ones([1], dtype=tf.float32))\ntest_tf_cond(maybe_tensor_cond, tf.constant(-1))\ntest_tf_cond(maybe_tensor_cond, -1)",
"_____no_output_____"
]
],
[
[
"`tf.cond` には、色々と注意すべき細かな点があります。\n\n- `tf.cond` は条件分岐の両方をトレーシングし、条件に従って実行時に適切な分岐を選択することで機能します。分岐の両方をトレースすることで、Python プログラムを予期せず実行する可能性があります。\n- `tf.cond` では、分岐の一方が後ほど使用されるテンソルを作成する場合、もう一方の分岐もそのテンソルを作成することが必要です。",
"_____no_output_____"
]
],
[
[
"@tf.function\ndef f():\n x = tf.constant(0)\n if tf.constant(True):\n x = x + 1\n print(\"Tracing `then` branch\")\n else:\n x = x - 1\n print(\"Tracing `else` branch\")\n return x\n\nf()",
"_____no_output_____"
],
[
"@tf.function\ndef f():\n if tf.constant(True):\n x = tf.ones([3, 3])\n return x\n\n# 分岐のどちらの枝でも `x` を定義する必要があるためエラーが発生\nwith assert_raises(ValueError):\n f()",
"_____no_output_____"
]
],
[
[
"## AutoGraph と繰り返し\n\nAutoGraph には繰り返しの変換にいくつかの単純なルールがあります。\n\n- `for`: 反復可能オブジェクトがテンソルである場合に変換する\n- `while`: while 条件がテンソルに依存している場合に変換する\n\n繰り返しが変換される場合、`tf.while_loop` によって動的に展開されます。あるいは、 `for x in tf.data.Dataset` という特別なケースの場合には、 `tf.data.Dataset.reduce` に変換されます。\n\n繰り返しが変換されない場合、それは静的に展開されます。",
"_____no_output_____"
]
],
[
[
"def test_dynamically_unrolled(f, *args):\n g = f.get_concrete_function(*args).graph\n if any(node.name == 'while' for node in g.as_graph_def().node):\n print(\"{}({}) uses tf.while_loop.\".format(\n f.__name__, ', '.join(map(str, args))))\n elif any(node.name == 'ReduceDataset' for node in g.as_graph_def().node):\n print(\"{}({}) uses tf.data.Dataset.reduce.\".format(\n f.__name__, ', '.join(map(str, args))))\n else:\n print(\"{}({}) gets unrolled.\".format(\n f.__name__, ', '.join(map(str, args))))",
"_____no_output_____"
],
[
"@tf.function\ndef for_in_range():\n x = 0\n for i in range(5):\n x += i\n return x\n\ntest_dynamically_unrolled(for_in_range)",
"_____no_output_____"
],
[
"@tf.function\ndef for_in_tfrange():\n x = tf.constant(0, dtype=tf.int32)\n for i in tf.range(5):\n x += i\n return x\n\ntest_dynamically_unrolled(for_in_tfrange)",
"_____no_output_____"
],
[
"@tf.function\ndef for_in_tfdataset():\n x = tf.constant(0, dtype=tf.int64)\n for i in tf.data.Dataset.range(5):\n x += i\n return x\n\ntest_dynamically_unrolled(for_in_tfdataset)",
"_____no_output_____"
],
[
"@tf.function\ndef while_py_cond():\n x = 5\n while x > 0:\n x -= 1\n return x\n\ntest_dynamically_unrolled(while_py_cond)",
"_____no_output_____"
],
[
"@tf.function\ndef while_tf_cond():\n x = tf.constant(5)\n while x > 0:\n x -= 1\n return x\n\ntest_dynamically_unrolled(while_tf_cond)",
"_____no_output_____"
]
],
[
[
" 繰り返しに、テンソルに依存する `break` や、途中での `return` がある場合、一番外側の条件あるいは反復可能オブジェクトはテンソルである必要があります。 \n \n 比較してみましょう。",
"_____no_output_____"
]
],
[
[
"@tf.function\ndef while_py_true_py_break(x):\n while True: # py true\n if x == 0: # py break\n break\n x -= 1\n return x\n\ntest_dynamically_unrolled(while_py_true_py_break, 5)",
"_____no_output_____"
],
[
"@tf.function\ndef buggy_while_py_true_tf_break(x):\n while True: # py true\n if tf.equal(x, 0): # tf break\n break\n x -= 1\n return x\n\nwith assert_raises(TypeError):\n test_dynamically_unrolled(buggy_while_py_true_tf_break, 5)",
"_____no_output_____"
],
[
"@tf.function\ndef while_tf_true_tf_break(x):\n while tf.constant(True): # tf true\n if x == 0: # py break\n break\n x -= 1\n return x\n\ntest_dynamically_unrolled(while_tf_true_tf_break, 5)",
"_____no_output_____"
],
[
"@tf.function\ndef buggy_py_for_tf_break():\n x = 0\n for i in range(5): # py for\n if tf.equal(i, 3): # tf break\n break\n x += i\n return x\n\nwith assert_raises(TypeError):\n test_dynamically_unrolled(buggy_py_for_tf_break)",
"_____no_output_____"
],
[
"@tf.function\ndef tf_for_py_break():\n x = 0\n for i in tf.range(5): # tf for\n if i == 3: # py break\n break\n x += i\n return x\n\ntest_dynamically_unrolled(tf_for_py_break)",
"_____no_output_____"
]
],
[
[
"動的に展開される繰り返しの結果を集計するため、`tf.TensorArray` を使いたくなるかもしれません。",
"_____no_output_____"
]
],
[
[
"batch_size = 2\nseq_len = 3\nfeature_size = 4\n\ndef rnn_step(inp, state):\n return inp + state\n\[email protected]\ndef dynamic_rnn(rnn_step, input_data, initial_state):\n # [batch, time, features] -> [time, batch, features]\n input_data = tf.transpose(input_data, [1, 0, 2])\n max_seq_len = input_data.shape[0]\n\n states = tf.TensorArray(tf.float32, size=max_seq_len)\n state = initial_state\n for i in tf.range(max_seq_len):\n state = rnn_step(input_data[i], state)\n states = states.write(i, state)\n return tf.transpose(states.stack(), [1, 0, 2])\n \ndynamic_rnn(rnn_step,\n tf.random.uniform([batch_size, seq_len, feature_size]),\n tf.zeros([batch_size, feature_size]))",
"_____no_output_____"
]
],
[
[
"`tf.cond` と同様に、`tf.while_loop` にも、色々と注意すべき細かな点があります。\n\n- 繰り返しの実行回数が 0 である可能性があるため、while_loop の後で使用されるテンソルは、繰り返しの前に初期化されなければならない\n- すべての繰り返しの変数は、各繰り返しを通じてその形状と dtype が変わらないことが必要",
"_____no_output_____"
]
],
[
[
"@tf.function\ndef buggy_loop_var_uninitialized():\n for i in tf.range(3):\n x = i\n return x\n\nwith assert_raises(ValueError):\n buggy_loop_var_uninitialized()",
"_____no_output_____"
],
[
"@tf.function\ndef f():\n x = tf.constant(0)\n for i in tf.range(3):\n x = i\n return x\n\nf()",
"_____no_output_____"
],
[
"@tf.function\ndef buggy_loop_type_changes():\n x = tf.constant(0, dtype=tf.float32)\n for i in tf.range(3): # tf.int32 型のテンソルを1つづつ取り出して…\n x = i\n return x\n\nwith assert_raises(tf.errors.InvalidArgumentError):\n buggy_loop_type_changes()",
"_____no_output_____"
],
[
"@tf.function\ndef buggy_concat():\n x = tf.ones([0, 10])\n for i in tf.range(5):\n x = tf.concat([x, tf.ones([1, 10])], axis=0)\n return x\n\nwith assert_raises(ValueError):\n buggy_concat()",
"_____no_output_____"
],
[
"@tf.function\ndef concat_with_padding():\n x = tf.zeros([5, 10])\n for i in tf.range(5):\n x = tf.concat([x[:i], tf.ones([1, 10]), tf.zeros([4-i, 10])], axis=0)\n x.set_shape([5, 10])\n return x\n\nconcat_with_padding()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
d0857d57ef6c8350bd816195e02a2863efc8fc7e | 154,403 | ipynb | Jupyter Notebook | scene_classification.ipynb | Harshavardhanjo/Scene_Classification | 73f3d2b7887276233dca9d14d6d711d3d9d4f50b | [
"MIT"
] | null | null | null | scene_classification.ipynb | Harshavardhanjo/Scene_Classification | 73f3d2b7887276233dca9d14d6d711d3d9d4f50b | [
"MIT"
] | null | null | null | scene_classification.ipynb | Harshavardhanjo/Scene_Classification | 73f3d2b7887276233dca9d14d6d711d3d9d4f50b | [
"MIT"
] | null | null | null | 238.644513 | 118,290 | 0.904464 | [
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport matplotlib.image as mpimg\nimport tensorflow as tf\nfrom sklearn.preprocessing import OneHotEncoder\nfrom sklearn.model_selection import train_test_split\nfrom tensorflow.keras.preprocessing.image import ImageDataGenerator",
"_____no_output_____"
],
[
"df = pd.DataFrame(columns=['filename','label'])",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"for file in os.listdir('./data/scene_classification/seg_train/seg_train/buildings'):\n df.loc[len(df)] = [file,'buildings']",
"_____no_output_____"
],
[
"for file in os.listdir('./data/scene_classification/seg_train/seg_train/forest'):\n df.loc[len(df)] = [file,'forest']",
"_____no_output_____"
],
[
"for file in os.listdir('./data/scene_classification/seg_train/seg_train/glacier'):\n df.loc[len(df)] = [file,'glacier']",
"_____no_output_____"
],
[
"for file in os.listdir('./data/scene_classification/seg_train/seg_train/mountain'):\n df.loc[len(df)] = [file,'mountain']",
"_____no_output_____"
],
[
"for file in os.listdir('./data/scene_classification/seg_train/seg_train/sea'):\n df.loc[len(df)] = [file,'sea']",
"_____no_output_____"
],
[
"for file in os.listdir('./data/scene_classification/seg_train/seg_train/street'):\n df.loc[len(df)] = [file,'street']",
"_____no_output_____"
],
[
"df = df.sample(frac=1).reset_index(drop=True)",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df_train,df_val = train_test_split(df,test_size=0.2,random_state=42)",
"_____no_output_____"
],
[
"df_train.head()",
"_____no_output_____"
],
[
"train_datagen = ImageDataGenerator(rescale = 1./255,shear_range = 0.2,zoom_range = 0.2,horizontal_flip = True,vertical_flip=True,brightness_range=[0.5,1.5])\nval_datagen = ImageDataGenerator(rescale = 1./255)\n\ntraining_set = train_datagen.flow_from_dataframe(dataframe = df_train,directory = './data/scene_classification/seg_train/seg_train/images/',x_col = 'filename',y_col = 'label',class_mode = 'categorical',target_size = (150,150),batch_size = 32,shuffle = True)\nvalidation_set = val_datagen.flow_from_dataframe(dataframe = df_val,directory = './data/scene_classification/seg_train/seg_train/images/',x_col = 'filename',y_col = 'label',class_mode = 'categorical',target_size = (150,150),batch_size = 32,shuffle = True)",
"Found 11227 validated image filenames belonging to 6 classes.\nFound 2807 validated image filenames belonging to 6 classes.\n"
],
[
"model = tf.keras.models.Sequential([\n tf.keras.layers.Conv2D(32,(3,3),activation='relu',input_shape=(150,150,3)),\n tf.keras.layers.MaxPooling2D(2,2),\n tf.keras.layers.Conv2D(64,(3,3),activation='relu'),\n tf.keras.layers.MaxPooling2D(2,2),\n tf.keras.layers.Conv2D(128,(3,3),activation='relu'),\n tf.keras.layers.MaxPooling2D(2,2),\n tf.keras.layers.Conv2D(128,(3,3),activation='relu'),\n tf.keras.layers.MaxPooling2D(2,2),\n tf.keras.layers.Flatten(),\n tf.keras.layers.Dense(512,activation='relu'),\n tf.keras.layers.Dense(6,activation='softmax')\n])",
"_____no_output_____"
],
[
"model.summary()",
"Model: \"sequential\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nconv2d (Conv2D) (None, 148, 148, 32) 896 \n_________________________________________________________________\nmax_pooling2d (MaxPooling2D) (None, 74, 74, 32) 0 \n_________________________________________________________________\nconv2d_1 (Conv2D) (None, 72, 72, 64) 18496 \n_________________________________________________________________\nmax_pooling2d_1 (MaxPooling2 (None, 36, 36, 64) 0 \n_________________________________________________________________\nconv2d_2 (Conv2D) (None, 34, 34, 128) 73856 \n_________________________________________________________________\nmax_pooling2d_2 (MaxPooling2 (None, 17, 17, 128) 0 \n_________________________________________________________________\nconv2d_3 (Conv2D) (None, 15, 15, 128) 147584 \n_________________________________________________________________\nmax_pooling2d_3 (MaxPooling2 (None, 7, 7, 128) 0 \n_________________________________________________________________\nflatten (Flatten) (None, 6272) 0 \n_________________________________________________________________\ndense (Dense) (None, 512) 3211776 \n_________________________________________________________________\ndense_1 (Dense) (None, 6) 3078 \n=================================================================\nTotal params: 3,455,686\nTrainable params: 3,455,686\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"model.compile(optimizer='adam',loss='categorical_crossentropy',metrics=['accuracy'])",
"_____no_output_____"
],
[
"history = model.fit(training_set,validation_data=validation_set,epochs=10)",
"Epoch 1/10\n351/351 [==============================] - 353s 1s/step - loss: 1.1901 - accuracy: 0.5115 - val_loss: 1.1105 - val_accuracy: 0.5604\nEpoch 2/10\n351/351 [==============================] - 414s 1s/step - loss: 0.9270 - accuracy: 0.6347 - val_loss: 0.8909 - val_accuracy: 0.6402\nEpoch 3/10\n351/351 [==============================] - 396s 1s/step - loss: 0.8012 - accuracy: 0.6943 - val_loss: 0.6723 - val_accuracy: 0.7499\nEpoch 4/10\n351/351 [==============================] - 383s 1s/step - loss: 0.7131 - accuracy: 0.7290 - val_loss: 0.6104 - val_accuracy: 0.7702\nEpoch 5/10\n351/351 [==============================] - 367s 1s/step - loss: 0.6350 - accuracy: 0.7642 - val_loss: 0.7647 - val_accuracy: 0.7253\nEpoch 6/10\n351/351 [==============================] - 320s 911ms/step - loss: 0.5907 - accuracy: 0.7802 - val_loss: 0.5213 - val_accuracy: 0.8137\nEpoch 7/10\n351/351 [==============================] - 320s 912ms/step - loss: 0.5567 - accuracy: 0.7945 - val_loss: 0.5248 - val_accuracy: 0.8158\nEpoch 8/10\n351/351 [==============================] - 297s 846ms/step - loss: 0.5282 - accuracy: 0.8048 - val_loss: 0.5396 - val_accuracy: 0.8023\nEpoch 9/10\n351/351 [==============================] - 307s 875ms/step - loss: 0.4968 - accuracy: 0.8128 - val_loss: 0.5224 - val_accuracy: 0.8080\nEpoch 10/10\n351/351 [==============================] - 312s 888ms/step - loss: 0.4738 - accuracy: 0.8279 - val_loss: 0.5934 - val_accuracy: 0.8069\n"
],
[
"plt.plot(history.history['accuracy'])\nplt.plot(history.history['val_accuracy'])",
"_____no_output_____"
],
[
"path = './models/scene_classification/'\nmodel.save(path+'model.h5')\nmodel.save_weights(path+'weights.h5')",
"_____no_output_____"
],
[
"new_model = tf.keras.models.load_model(path+'model.h5')",
"_____no_output_____"
],
[
"loss,accuracy = new_model.evaluate(validation_set)",
"88/88 [==============================] - 19s 217ms/step - loss: 0.5934 - accuracy: 0.8069\n"
],
[
"new_model.summary()",
"Model: \"sequential\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nconv2d (Conv2D) (None, 148, 148, 32) 896 \n_________________________________________________________________\nmax_pooling2d (MaxPooling2D) (None, 74, 74, 32) 0 \n_________________________________________________________________\nconv2d_1 (Conv2D) (None, 72, 72, 64) 18496 \n_________________________________________________________________\nmax_pooling2d_1 (MaxPooling2 (None, 36, 36, 64) 0 \n_________________________________________________________________\nconv2d_2 (Conv2D) (None, 34, 34, 128) 73856 \n_________________________________________________________________\nmax_pooling2d_2 (MaxPooling2 (None, 17, 17, 128) 0 \n_________________________________________________________________\nconv2d_3 (Conv2D) (None, 15, 15, 128) 147584 \n_________________________________________________________________\nmax_pooling2d_3 (MaxPooling2 (None, 7, 7, 128) 0 \n_________________________________________________________________\nflatten (Flatten) (None, 6272) 0 \n_________________________________________________________________\ndense (Dense) (None, 512) 3211776 \n_________________________________________________________________\ndense_1 (Dense) (None, 6) 3078 \n=================================================================\nTotal params: 3,455,686\nTrainable params: 3,455,686\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"import cv2\n# Utility\nimport itertools\nimport random\nfrom collections import Counter\nfrom glob import iglob\ndef load_image(filename):\n img = mpimg.imread('data/Scene_Classification/seg_train/seg_train/images/' + filename)\n img = cv2.resize(img, (150,150) )\n img = img /255\n \n return img\ndef predict(image):\n probabilities = model.predict(np.asarray([img]))[0]\n class_idx = np.argmax(probabilities)\n \n return class_idx\nplt.imshow(mpimg.imread('data/Scene_Classification/seg_train/seg_train/street/2.jpg'))\nimg = load_image('2.jpg')\npredict(img)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d08597bcf583bb7672b861f15fc9c6348160f789 | 92,393 | ipynb | Jupyter Notebook | Examples/swordfish_DD.ipynb | bradkav/swordfish | 64af5fb068a12d48c1b54bbb2bbc2ada3b24b3c9 | [
"MIT"
] | null | null | null | Examples/swordfish_DD.ipynb | bradkav/swordfish | 64af5fb068a12d48c1b54bbb2bbc2ada3b24b3c9 | [
"MIT"
] | null | null | null | Examples/swordfish_DD.ipynb | bradkav/swordfish | 64af5fb068a12d48c1b54bbb2bbc2ada3b24b3c9 | [
"MIT"
] | null | null | null | 334.757246 | 82,834 | 0.912775 | [
[
[
"from __future__ import division\nimport numpy as np\nimport pylab as plt\nimport swordfish as sf\nfrom scipy.interpolate import interp1d\nfrom scipy.constants import c\nfrom numpy.random import multivariate_normal\nfrom matplotlib import rc\nfrom scipy.interpolate import UnivariateSpline\nrc('text', usetex=True)\nrc('font',**{'family':'sans-serif','sans-serif':['cmr']})\nrc('font',**{'family':'serif','serif':['cmr']})\n%matplotlib inline\n\nc_kms = c*1.e-3 # in km s^-1\ng2 = 1.e-18\nsig0 = 1e-45\nm_med = 1e5 # MeV\nGeV_inv_cm = 1.98e-14\nmp = 0.93",
"_____no_output_____"
]
],
[
[
"# Xenon1T",
"_____no_output_____"
],
[
"We can now implement the projected limits for Xenon1T based off 83 days of exposure using the same recoil energy spectrum as for CRESST-III but with the appropriate changes made for the Xenon nuclei (it is more simple here since there are only Xenon nuclei). We assume that there is only one isotope for our calculations. Since we do not have access to the Xenon background with much detail we perform a 1D analysis using the backgrounds published in 1705.06655 as a function of the S1 signal. We therefore need to to approximate the way that the dark matter signal is distributed between S1 and S2.",
"_____no_output_____"
],
[
"Now we define the signal component as,\n$$ \\frac{dR}{dE_R} = \\frac{\\rho_0\\xi_T}{2\\pi m_{DM}} \\frac{g^2 F_T^2(E_{R})}{(2m_TE_{R} + m^2_{med})^2}\\eta(v_{min}(E_R))$$\n\nwhere $E_R$ is the recoil energy, $\\rho_0$ is the dark matter density at earth (which we take to be $0.3 GeV cm^{-3}$, $m_{DM}$ is the dark matter mass, $m_{med}$ is the mediator mass, $F_T^2(E_{R})$ is the recoil form factor, and $m_T$ is the mass of the target isotope.",
"_____no_output_____"
]
],
[
[
"rho_0 = 0.3*1.e3 # MeV/cm3\n# Define energy range in MeV\n\ndef eta_F():\n \"\"\"Returns an interpolated integral over the velocity distribution, taken to be Maxwellian\"\"\"\n v, gave = np.loadtxt(\"DD_files/gave.dat\", unpack=True, dtype=float)\n f = interp1d(v, gave, bounds_error=False, fill_value=0.0) # s km^-1\n return f\n\ndef dRdE(E_R, m_DM, A, xi_T):\n \"\"\"Return differential recoil rate in 1/s/MeV/kg.\"\"\"\n # g is the parameter we wish to set a limit on so is left out\n # Form factor taken from eq4.4 of http://pa.brown.edu/articles/Lewin_Smith_DM_Review.pdf\n m_T = A*931.5 # MeV\n muT = m_DM*m_T/(m_DM + m_T) # unitless\n rn = A**(1./3.) * 1/197. # fm --> MeV^-1\n F_T = lambda q: np.sqrt(np.exp(-((q*rn)**2.)/3.))\n vmin = lambda E_R: np.sqrt(m_T*E_R/2/(muT**2.))\n eta = eta_F()\n q = np.sqrt(2*m_T*E_R)\n signal = (A**2)*F_T(q)**2*eta(vmin(E_R)*c_kms)*rho_0*xi_T*g2/2./np.pi/m_DM/((q**2+m_med**2)**2)\n conversion = 1.96311325e24 # MeV^-4 cm^-3 s km^-1 hbar^2 c^6 --> MeV^-1 s^-1 kg^-1\n signal *= conversion \n return signal",
"_____no_output_____"
],
[
"eff1, eff2 = np.loadtxt(\"Swordfish_Xenon1T/Efficiency-1705.06655.txt\", unpack=True)\nefficiency = UnivariateSpline(eff1, eff2, ext=\"zeros\", k=1, s=0)\nS1_vals, E_vals = np.loadtxt(\"Swordfish_Xenon1T/S1vsER.txt\", unpack=True)\n\n# Interpolation for the recoil energy as a function of S1\n# and the derivative\nCalcER = UnivariateSpline(S1_vals, E_vals, k=4, s=0)\ndERdS1 = CalcER.derivative()\n\n# Recoil distribution as a function of S1\n# taking into account the efficiency and change\n# of variables ER -> S1\ndef dRdS1(S1, m_DM):\n A_Xe = 131. #FIXME: Change to Xenon values\n xi_T_Xe = 1.0\n ER_keV = CalcER(S1)\n ER_MeV = ER_keV*1.e-3\n #Factor of 0.475 comes from the fact that\n #the reference region should contain about\n #47.5% of nuclear recoils (between median and 2sigma lines)\n # Factor of 1/1e3 to convert 1/MeV --> 1/keV\n return 0.475*efficiency(ER_keV)*dRdE(ER_MeV,m_DM,A_Xe,xi_T_Xe)/1e3*dERdS1(S1)",
"_____no_output_____"
],
[
"# We are now working in distributions as a function of s1\ns1 = np.linspace(3,70,num=100)\ns1width = s1[1]-s1[0]\ns1means = s1[0:-1]+s1width/2.\nbkgs = ['acc','Anom','ElectronRecoil','n','Neutrino','Wall']\n\ndef load_bkgs():\n b = dict()\n for i in range(len(bkgs)):\n S1, temp = np.loadtxt(\"DD_files/\" + bkgs[i] + \".txt\", unpack=True)\n interp = interp1d(S1, temp, bounds_error=False, fill_value=0.0)\n b[bkgs[i]] = interp(s1means)\n return b\n \ndef XenonIT_sig(m_DM):\n m_DM *= 1.e3 # conversion to MeV\n sig = dRdS1(s1means,m_DM)*s1width\n return sig",
"_____no_output_____"
],
[
"b_dict = load_bkgs()\nobsT = np.ones_like(s1means)*24.*3600.*35636.\nmlist = np.logspace(1, 3, 50) # GeV\nb = np.array(b_dict[bkgs[0]]/obsT)\nK = np.diag((b.flatten()*0.01)**2)\nB = [b_dict[bkgs[0]]/obsT, b_dict[bkgs[1]]/obsT, b_dict[bkgs[2]]/obsT,\n b_dict[bkgs[3]]/obsT, b_dict[bkgs[4]]/obsT, b_dict[bkgs[5]]/obsT]\n\ndef g(m, sigma):\n # Takes in sigma and returns g^2\n mu_temp = m*mp/(m+mp)\n return np.ones_like(s1means)*np.pi*((m_med/1.e3)**4.)*sigma/(GeV_inv_cm**2.)/(mu_temp**2.)\n\nSF = sf.Swordfish(B, T=[0.1,0.1,0.1,0.1,0.1,0.1], E=obsT, K=K)\nULlist_Xenon = []\nDRlist_Xenon = []\nfor i, m in enumerate(mlist):\n sig = XenonIT_sig(m)\n UL = SF.upperlimit(sig, 0.05)\n DR = SF.discoveryreach(sig, 2.87e-7)\n DRlist_Xenon.append(DR*g2)\n ULlist_Xenon.append(UL*g2)\n \nmu_xp = mlist*mp/(mlist+mp)\nsigma_Xe = (GeV_inv_cm**2.)*np.array(ULlist_Xenon)*mu_xp**2./np.pi/(m_med/1.e3)**4.\nsigma_Xe_DR = (GeV_inv_cm**2.)*np.array(DRlist_Xenon)*mu_xp**2./np.pi/(m_med/1.e3)**4.",
"_____no_output_____"
],
[
"m10list = np.linspace(1, 3, 50) # GeV\ns10list = np.linspace(-46, -44, 50)\n\nS = lambda m, sigma: g(10**m, 10**sigma)*(g2**(-1))*XenonIT_sig(10**m)\nprint S(1.7, -46).sum()*3600.*24*35636\nTF = SF.getfield(S, m10list, s10list)\nvf1, vf2 = TF.VectorFields()",
"0.83096555216\n"
],
[
"plt.figure(figsize=(5,4))\nmask = lambda x, y: (y > np.interp(x, np.log10(mlist), np.log10(sigma_Xe))) & (y<-44)\nvf1.streamlines(color='0.5', mask = mask, Nmax = 40);\nvf2.streamlines(color='0.5', mask = mask, Nmax = 40);\n\nplt.plot(np.log10(mlist), np.log10(sigma_Xe), label=r\"95\\% CL Exclusion Limit\")\nplt.plot(np.log10(mlist), np.log10(sigma_Xe_DR), \"-.\", label=r\"$5\\sigma$ Discovery Reach\")\n\nTF.contour([1.4, -45.], 1., color='r', ls='-');\nTF.contour([1.4, -45.], 2., color='r', ls='--');\nTF.contour([1.5, -44.25], 1., color='b', ls='-');\nTF.contour([1.5, -44.25], 2., color='b', ls='--');\n\nplt.legend(loc=4)\ny = [-46,-45,-44]\nx = [1,2,3]\nplt.xlim(1.,3.)\nplt.ylim(-46,-44)\nplt.xlabel(r\"$\\log_{10}(m_{\\mathrm{DM}}/\\rm GeV)$\")\nplt.ylabel(r\"$\\log_{10}(\\sigma /\\rm cm^{-2})$\")\nplt.yticks(np.arange(min(y), max(y)+1, 1.0))\nplt.xticks(np.arange(min(x), max(x)+1, 1.0))\nplt.tight_layout(pad=0.3)\nplt.savefig(\"Xe_stream_limits.eps\")",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d085a1621ab80758b8319329b1af647967b5bc85 | 480,657 | ipynb | Jupyter Notebook | playground_baseline_dictionary_various_training_snapshots_various_noise_interferences_random.ipynb | dung-n-tran/speech-enhancement-beamforming | b0db5e1bc88dfbec33a22abe96c911d6f6c77574 | [
"MIT"
] | 2 | 2019-04-14T06:13:55.000Z | 2020-06-08T03:53:34.000Z | playground_baseline_dictionary_various_training_snapshots_various_noise_interferences_random.ipynb | dung-n-tran/speech-enhancement-beamforming | b0db5e1bc88dfbec33a22abe96c911d6f6c77574 | [
"MIT"
] | null | null | null | playground_baseline_dictionary_various_training_snapshots_various_noise_interferences_random.ipynb | dung-n-tran/speech-enhancement-beamforming | b0db5e1bc88dfbec33a22abe96c911d6f6c77574 | [
"MIT"
] | null | null | null | 1,316.868493 | 466,336 | 0.951377 | [
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline\n%config InlineBackend.figure_format = \"retina\"\n# print(plt.style.available)\nplt.style.use(\"ggplot\")\n# plt.style.use(\"fivethirtyeight\")\nplt.style.use(\"seaborn-talk\")\n\nfrom tqdm import tnrange, tqdm_notebook",
"_____no_output_____"
],
[
"def uniform_linear_array(n_mics, spacing):\n return spacing*np.arange(-(n_mics-1)/2, (n_mics-1)/2+1).reshape(1, n_mics)\n\ndef compute_MVDR_weight(source_steering_vector, signals):\n snapshot = signals.shape[1]\n sample_covariance_matrix = signals.dot(signals.transpose().conjugate()) / snapshot\n inverse_sample_covariance_matrix = np.linalg.inv(sample_covariance_matrix)\n normalization_factor = (source_steering_vector.transpose().conjugate().dot(inverse_sample_covariance_matrix).dot(source_steering_vector))\n weight = inverse_sample_covariance_matrix.dot(source_steering_vector) / normalization_factor\n return weight\n\ndef compute_steering_vector_ULA(u, microphone_array):\n return np.exp(1j*2*np.pi*microphone_array.geometry*u).reshape((microphone_array.n_mics, 1))\n\ndef generate_gaussian_samples(power, shape):\n return np.sqrt(power/2)*np.random.randn(shape[0], shape[1]) + 1j*np.sqrt(power/2)*np.random.randn(shape[0], shape[1]); # signal samples\n\nclass MicrophoneArray():\n def __init__(self, array_geometry):\n self.dim = array_geometry.shape[0]\n self.n_mics = array_geometry.shape[1]\n self.geometry = array_geometry\n\n \n \nclass BaseDLBeamformer(object):\n def __init__(self, vs, bf_type=\"MVDR\"):\n \"\"\"\n Parameters\n ----------\n vs: Source manifold array vector\n bf_type: Type of beamformer\n \"\"\"\n self.vs = vs\n self.bf_type = bf_type\n self.weights_ = None\n \n def _compute_weights(self, training_data):\n n_training_samples = len(training_data)\n n_mics, snapshot = training_data[0].shape\n D = np.zeros((n_mics, n_training_samples), dtype=complex)\n for i_training_sample in range(n_training_samples):\n nv = training_data[i_training_sample]\n if self.bf_type == \"MVDR\":\n w = compute_MVDR_weight(vs, nv)\n D[:, i_training_sample] = w.reshape(n_mics,)\n return D\n\n def _initialize(self, X):\n pass\n\n def _choose_weights(self, x):\n n_dictionary_atoms = self.weights_.shape[1]\n R = x.dot(x.transpose().conjugate())\n proxy = np.diagonal(self.weights_.transpose().conjugate().dot(R).dot(self.weights_))\n optimal_weight_index = np.argmin(proxy)\n return self.weights_[:, optimal_weight_index]\n \n def fit(self, training_data):\n \"\"\"\n Parameters\n ----------\n X: shape = [n_samples, n_features]\n \"\"\"\n D = self._compute_weights(training_data)\n self.weights_ = D\n return self\n\n def choose_weights(self, x):\n return self._choose_weights(x)",
"_____no_output_____"
]
],
[
[
"#### Setup",
"_____no_output_____"
]
],
[
[
"array_geometry = uniform_linear_array(n_mics=10, spacing=0.5)\nmicrophone_array = MicrophoneArray(array_geometry)\nus = 0\nvs = compute_steering_vector_ULA(us, microphone_array)\nSNRs = np.arange(0, 31, 10)\nn_SNRs = len(SNRs)\nsigma_n = 1",
"_____no_output_____"
]
],
[
[
"#### Training data",
"_____no_output_____"
]
],
[
[
"n_training_samples = 5000\ntraining_snapshots = [10, 50, 1000]\ninterference_powers = [10, 20, 30]\nn_interference_list = [1, 2, 3]\n# interference_powers = [20]\n# n_interference_list = [1]\n# sigma = 10**(20/10)\ntraining_noise_interference_data_various_snapshots = []\nfor training_snapshot in training_snapshots:\n training_noise_interference_data = []\n for i_training_sample in range(n_training_samples): \n n_interferences = np.random.choice(n_interference_list)\n nv = np.zeros((microphone_array.n_mics, training_snapshot), dtype=complex)\n for _ in range(n_interferences):\n u = np.random.uniform(0, 1)\n vi = compute_steering_vector_ULA(u, microphone_array)\n sigma = 10**(np.random.choice(interference_powers)/10)\n ii = generate_gaussian_samples(power=sigma, shape=(1, training_snapshot))\n nv += vi.dot(ii)\n noise = generate_gaussian_samples(power=sigma_n, shape=(microphone_array.n_mics, training_snapshot))\n nv += noise\n training_noise_interference_data.append(nv)\n training_noise_interference_data_various_snapshots.append(training_noise_interference_data)",
"_____no_output_____"
]
],
[
[
"#### Train baseline dictionary",
"_____no_output_____"
]
],
[
[
"dictionaries = []\nfor i_training_snapshot in range(len(training_snapshots)):\n training_noise_interference_data = training_noise_interference_data_various_snapshots[i_training_snapshot]\n dictionary = BaseDLBeamformer(vs)\n dictionary.fit(training_noise_interference_data);\n dictionaries.append(dictionary)",
"_____no_output_____"
]
],
[
[
"#### Testing",
"_____no_output_____"
]
],
[
[
"n_trials = 200\nsnapshots = np.array([10, 20, 30, 40, 60, 100, 200, 500, 1000])\nn_snapshots = len(snapshots)\nui1 = np.random.uniform(0, 1)\nui2 = np.random.uniform(0, 1)\nsigma_1 = 10**(20/10)\nsigma_2 = 0*10**(20/10)\nvi1 = compute_steering_vector_ULA(ui1, microphone_array)\nvi2 = compute_steering_vector_ULA(ui2, microphone_array)\n\nn_interferences = np.random.choice(n_interference_list)\ninterference_steering_vectors = []\nfor _ in range(n_interferences):\n u = np.random.uniform(0, 1)\n vi = compute_steering_vector_ULA(u, microphone_array)\n interference_steering_vectors.append(vi) \n\nsinr_snr_mvdr = np.zeros((n_SNRs, n_snapshots))\nsinr_snr_mpdr = np.zeros((n_SNRs, n_snapshots))\nsinr_snr_baseline_mpdr = np.zeros((len(training_snapshots), n_SNRs, n_snapshots))\n\nfor i_SNR in tqdm_notebook(range(n_SNRs), desc=\"SNRs\"):\n sigma_s = 10**(SNRs[i_SNR] / 10)\n Rs = sigma_s * vs.dot(vs.transpose().conjugate()) \n \n for i_snapshot in tqdm_notebook(range(n_snapshots), desc=\"Snapshots\", leave=False):\n snapshot = snapshots[i_snapshot]\n sinr_mvdr = np.zeros(n_trials)\n sinr_mpdr = np.zeros(n_trials)\n sinr_baseline_mpdr = np.zeros((len(training_snapshots), n_trials))\n \n for i_trial in range(n_trials):\n \n ss = generate_gaussian_samples(power=sigma_s, shape=(1, snapshot)) # signal samples\n nn = generate_gaussian_samples(power=sigma_n, shape=(microphone_array.n_mics, snapshot)) # Gaussian noise samples\n# ii1 = generate_gaussian_samples(power=sigma_1, shape=(1, snapshot)) # first interference samples\n# ii2 = generate_gaussian_samples(power=sigma_2, shape=(1, snapshot)) # second interference samples\n nv = np.zeros((microphone_array.n_mics, snapshot), dtype=complex)\n Rn = np.zeros((microphone_array.n_mics, microphone_array.n_mics), dtype=complex)\n for i_interference in range(n_interferences):\n sigma = 10**(np.random.choice(interference_powers)/10)\n ii = generate_gaussian_samples(power=sigma, shape=(1, snapshot))\n nv += interference_steering_vectors[i_interference].dot(ii)\n Rn += sigma*interference_steering_vectors[i_interference].dot(interference_steering_vectors[i_interference].transpose().conjugate())\n Rn += sigma_n*np.identity(microphone_array.n_mics)\n Rninv = np.linalg.inv(Rn)\n Wo = Rninv.dot(vs) / (vs.transpose().conjugate().dot(Rninv).dot(vs))\n SINRopt = ( np.real(Wo.transpose().conjugate().dot(Rs).dot(Wo)) / np.real(Wo.transpose().conjugate().dot(Rn).dot(Wo)) )[0][0]\n \n nv += nn\n sv = vs.dot(ss)\n xx = sv + nv\n \n wv = compute_MVDR_weight(vs, nv)\n wp = compute_MVDR_weight(vs, xx)\n \n for i_dictionary in range(len(dictionaries)):\n dictionary = dictionaries[i_dictionary]\n w_baseline_p = dictionary.choose_weights(xx)\n sinr_baseline_mpdr[i_dictionary, i_trial] = np.real(w_baseline_p.transpose().conjugate().dot(Rs).dot(w_baseline_p)) / np.real(w_baseline_p.transpose().conjugate().dot(Rn).dot(w_baseline_p))\n \n sinr_mvdr[i_trial] = np.real(wv.transpose().conjugate().dot(Rs).dot(wv)) / np.real(wv.transpose().conjugate().dot(Rn).dot(wv))\n sinr_mpdr[i_trial] = np.real(wp.transpose().conjugate().dot(Rs).dot(wp)) / np.real(wp.transpose().conjugate().dot(Rn).dot(wp))\n \n sinr_snr_mvdr[i_SNR, i_snapshot] = np.sum(sinr_mvdr) / n_trials\n sinr_snr_mpdr[i_SNR, i_snapshot] = np.sum(sinr_mpdr) / n_trials\n for i_dictionary in range(len(dictionaries)):\n sinr_snr_baseline_mpdr[i_dictionary, i_SNR, i_snapshot] = np.sum(sinr_baseline_mpdr[i_dictionary, :]) / n_trials",
"_____no_output_____"
]
],
[
[
"#### Visualize results",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(9, 6*n_SNRs)); \nfor i_SNR in range(n_SNRs):\n sigma_s = 10**(SNRs[i_SNR] / 10)\n Rs = sigma_s * vs.dot(vs.transpose().conjugate())\n \n SINRopt = ( np.real(Wo.transpose().conjugate().dot(Rs).dot(Wo)) / np.real(Wo.transpose().conjugate().dot(Rn).dot(Wo)) )[0][0]\n ax = fig.add_subplot(n_SNRs, 1, i_SNR+1)\n ax.semilogx(snapshots, 10*np.log10(sinr_snr_mvdr[i_SNR, :]), marker=\"o\", label=\"MVDR\")\n ax.semilogx(snapshots, 10*np.log10(sinr_snr_mpdr[i_SNR, :]), marker=\"*\", label=\"MPDR\")\n for i_training_snapshot in range(len(training_snapshots)):\n ax.semilogx(snapshots, 10*np.log10(sinr_snr_baseline_mpdr[i_training_snapshot, i_SNR, :]), \n label=\"Baseline - {} training snapshots\".format(training_snapshots[i_training_snapshot]))\n ax.set_xlim(10, 1000); ax.set_ylim(-10, 45)\n ax.legend(loc=\"lower right\")\n ax.set_xlabel(\"Number of snapshots\")\n ax.set_ylabel(r\"$SINR_0$ [dB]\")\n ax.set_title(\"Testing performance, {} training samples\".format(n_training_samples))\nplt.tight_layout()\nfig.savefig(\"baseline_dl_mvdr_various_interferences.jpg\", dpi=600)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d085a51c6a8e03c3dfa6cdebafe4a1bdefd5730b | 110,860 | ipynb | Jupyter Notebook | examples/notebooks/camera_display.ipynb | orelgueta/ctapipe | ee28440e83cc283ccd57428d5fdad764a1e786f0 | [
"BSD-3-Clause"
] | null | null | null | examples/notebooks/camera_display.ipynb | orelgueta/ctapipe | ee28440e83cc283ccd57428d5fdad764a1e786f0 | [
"BSD-3-Clause"
] | null | null | null | examples/notebooks/camera_display.ipynb | orelgueta/ctapipe | ee28440e83cc283ccd57428d5fdad764a1e786f0 | [
"BSD-3-Clause"
] | null | null | null | 640.809249 | 106,342 | 0.936803 | [
[
[
"# Example of a Camera Display to HTML5 via MPLD3\n\n",
"_____no_output_____"
]
],
[
[
"%matplotlib inline",
"_____no_output_____"
],
[
"import matplotlib.pylab as plt\nfrom ctapipe.instrument import CameraGeometry\nfrom ctapipe.visualization import CameraDisplay\nfrom ctapipe.image import toymodel\nfrom ctapipe.image import hillas_parameters, tailcuts_clean\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"Just a quick function to mark a pixel and draw lines to its neighbors",
"_____no_output_____"
]
],
[
[
"def draw_neighbors(geom, pixel_index, color='r', **kwargs):\n \"\"\" draw lines between a pixel and its neighbors\"\"\"\n neigh = geom.neighbors[pixel_index] # neighbor indices (not pixel ids)\n x, y = geom.pix_x[pixel_index].value, geom.pix_y[pixel_index].value\n for nn in neigh:\n nx, ny = geom.pix_x[nn].value, geom.pix_y[nn].value\n plt.plot([x, nx], [y, ny], color=color, **kwargs)",
"_____no_output_____"
]
],
[
[
"Now, let's create a fake Cherenkov image from a given `CameraGeometry` and fill it with some data:",
"_____no_output_____"
]
],
[
[
"# get the HESS demo camera geometry\ngeom = CameraGeometry.from_name(\"NectarCam\")\n\n# create a fake camera image to display:\nmodel = toymodel.generate_2d_shower_model(centroid=(0.2, 0.0),\n width=0.01,\n length=0.1,\n psi='35d')\n\nimage, sig, bg = toymodel.make_toymodel_shower_image(geom, model.pdf, \n intensity=50,\n nsb_level_pe=1000)\n\n# apply really stupid image cleaning (single threshold):\nmask = tailcuts_clean(geom, image, 10, 100)\n\n# calculate image parameters\nhillas = hillas_parameters(geom.pix_x.value[mask], geom.pix_y.value[mask], image[mask])",
"_____no_output_____"
],
[
"# show the camera image and overlay Hillas ellipse\ndisp = CameraDisplay(geom)\ndisp.set_limits_minmax(0, 300)\ndisp.add_colorbar()\ndisp.image = image\ndisp.overlay_moments(hillas, color='grey', linewidth=3,zorder=10)\n#disp.highlight_pixels(mask)\n\n# draw the neighbors of pixel 100 in red, and the\n# neighbor-neighbors in green\nfor ii in geom.neighbors[130]:\n draw_neighbors(geom, ii, color='green')\ndraw_neighbors(geom, 130, color='red',lw=2)",
"_____no_output_____"
]
],
[
[
"For online use, you can use the mpld3 library to automatically convert this to a zoomable HTML5 plot if you like. Simply call `display()` at the end of the code:\n\n import mpld3\n ...\n mpld3.display()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
d085aa2b52d22a0f5a2f01a937a335f6cdef677b | 35,833 | ipynb | Jupyter Notebook | how-to-use-azureml/automated-machine-learning/forecasting-orange-juice-sales/auto-ml-forecasting-orange-juice-sales.ipynb | zronaghi/MachineLearningNotebooks | a79f8c254a97bfcece5b8a36c50398162960b896 | [
"MIT"
] | 1 | 2021-08-10T13:25:32.000Z | 2021-08-10T13:25:32.000Z | how-to-use-azureml/automated-machine-learning/forecasting-orange-juice-sales/auto-ml-forecasting-orange-juice-sales.ipynb | zronaghi/MachineLearningNotebooks | a79f8c254a97bfcece5b8a36c50398162960b896 | [
"MIT"
] | 1 | 2021-02-24T13:58:10.000Z | 2021-02-24T13:58:10.000Z | how-to-use-azureml/automated-machine-learning/forecasting-orange-juice-sales/auto-ml-forecasting-orange-juice-sales.ipynb | zronaghi/MachineLearningNotebooks | a79f8c254a97bfcece5b8a36c50398162960b896 | [
"MIT"
] | null | null | null | 44.238272 | 827 | 0.607429 | [
[
[
"Copyright (c) Microsoft Corporation. All rights reserved.\n\nLicensed under the MIT License.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"# Automated Machine Learning\n_**Orange Juice Sales Forecasting**_\n\n## Contents\n1. [Introduction](#Introduction)\n1. [Setup](#Setup)\n1. [Compute](#Compute)\n1. [Data](#Data)\n1. [Train](#Train)\n1. [Predict](#Predict)\n1. [Operationalize](#Operationalize)",
"_____no_output_____"
],
[
"## Introduction\nIn this example, we use AutoML to train, select, and operationalize a time-series forecasting model for multiple time-series.\n\nMake sure you have executed the [configuration notebook](../../../configuration.ipynb) before running this notebook.\n\nThe examples in the follow code samples use the University of Chicago's Dominick's Finer Foods dataset to forecast orange juice sales. Dominick's was a grocery chain in the Chicago metropolitan area.",
"_____no_output_____"
],
[
"## Setup",
"_____no_output_____"
]
],
[
[
"import azureml.core\nimport pandas as pd\nimport numpy as np\nimport logging\n\nfrom azureml.core.workspace import Workspace\nfrom azureml.core.experiment import Experiment\nfrom azureml.train.automl import AutoMLConfig\nfrom azureml.automl.core.featurization import FeaturizationConfig",
"_____no_output_____"
]
],
[
[
"This sample notebook may use features that are not available in previous versions of the Azure ML SDK.",
"_____no_output_____"
]
],
[
[
"print(\"This notebook was created using version 1.19.0 of the Azure ML SDK\")\nprint(\"You are currently using version\", azureml.core.VERSION, \"of the Azure ML SDK\")",
"_____no_output_____"
]
],
[
[
"As part of the setup you have already created a <b>Workspace</b>. To run AutoML, you also need to create an <b>Experiment</b>. An Experiment corresponds to a prediction problem you are trying to solve, while a Run corresponds to a specific approach to the problem. ",
"_____no_output_____"
]
],
[
[
"ws = Workspace.from_config()\n\n# choose a name for the run history container in the workspace\nexperiment_name = 'automl-ojforecasting'\n\nexperiment = Experiment(ws, experiment_name)\n\noutput = {}\noutput['Subscription ID'] = ws.subscription_id\noutput['Workspace'] = ws.name\noutput['SKU'] = ws.sku\noutput['Resource Group'] = ws.resource_group\noutput['Location'] = ws.location\noutput['Run History Name'] = experiment_name\npd.set_option('display.max_colwidth', -1)\noutputDf = pd.DataFrame(data = output, index = [''])\noutputDf.T",
"_____no_output_____"
]
],
[
[
"## Compute\nYou will need to create a [compute target](https://docs.microsoft.com/en-us/azure/machine-learning/service/how-to-set-up-training-targets#amlcompute) for your AutoML run. In this tutorial, you create AmlCompute as your training compute resource.\n#### Creation of AmlCompute takes approximately 5 minutes. \nIf the AmlCompute with that name is already in your workspace this code will skip the creation process.\nAs with other Azure services, there are limits on certain resources (e.g. AmlCompute) associated with the Azure Machine Learning service. Please read this article on the default limits and how to request more quota.",
"_____no_output_____"
]
],
[
[
"from azureml.core.compute import ComputeTarget, AmlCompute\nfrom azureml.core.compute_target import ComputeTargetException\n\n# Choose a name for your CPU cluster\namlcompute_cluster_name = \"oj-cluster\"\n\n# Verify that cluster does not exist already\ntry:\n compute_target = ComputeTarget(workspace=ws, name=amlcompute_cluster_name)\n print('Found existing cluster, use it.')\nexcept ComputeTargetException:\n compute_config = AmlCompute.provisioning_configuration(vm_size='STANDARD_D2_V2',\n max_nodes=6)\n compute_target = ComputeTarget.create(ws, amlcompute_cluster_name, compute_config)\n\ncompute_target.wait_for_completion(show_output=True)",
"_____no_output_____"
]
],
[
[
"## Data\nYou are now ready to load the historical orange juice sales data. We will load the CSV file into a plain pandas DataFrame; the time column in the CSV is called _WeekStarting_, so it will be specially parsed into the datetime type.",
"_____no_output_____"
]
],
[
[
"time_column_name = 'WeekStarting'\ndata = pd.read_csv(\"dominicks_OJ.csv\", parse_dates=[time_column_name])\ndata.head()",
"_____no_output_____"
]
],
[
[
"Each row in the DataFrame holds a quantity of weekly sales for an OJ brand at a single store. The data also includes the sales price, a flag indicating if the OJ brand was advertised in the store that week, and some customer demographic information based on the store location. For historical reasons, the data also include the logarithm of the sales quantity. The Dominick's grocery data is commonly used to illustrate econometric modeling techniques where logarithms of quantities are generally preferred. \n\nThe task is now to build a time-series model for the _Quantity_ column. It is important to note that this dataset is comprised of many individual time-series - one for each unique combination of _Store_ and _Brand_. To distinguish the individual time-series, we define the **time_series_id_column_names** - the columns whose values determine the boundaries between time-series: ",
"_____no_output_____"
]
],
[
[
"time_series_id_column_names = ['Store', 'Brand']\nnseries = data.groupby(time_series_id_column_names).ngroups\nprint('Data contains {0} individual time-series.'.format(nseries))",
"_____no_output_____"
]
],
[
[
"For demonstration purposes, we extract sales time-series for just a few of the stores:",
"_____no_output_____"
]
],
[
[
"use_stores = [2, 5, 8]\ndata_subset = data[data.Store.isin(use_stores)]\nnseries = data_subset.groupby(time_series_id_column_names).ngroups\nprint('Data subset contains {0} individual time-series.'.format(nseries))",
"_____no_output_____"
]
],
[
[
"### Data Splitting\nWe now split the data into a training and a testing set for later forecast evaluation. The test set will contain the final 20 weeks of observed sales for each time-series. The splits should be stratified by series, so we use a group-by statement on the time series identifier columns.",
"_____no_output_____"
]
],
[
[
"n_test_periods = 20\n\ndef split_last_n_by_series_id(df, n):\n \"\"\"Group df by series identifiers and split on last n rows for each group.\"\"\"\n df_grouped = (df.sort_values(time_column_name) # Sort by ascending time\n .groupby(time_series_id_column_names, group_keys=False))\n df_head = df_grouped.apply(lambda dfg: dfg.iloc[:-n])\n df_tail = df_grouped.apply(lambda dfg: dfg.iloc[-n:])\n return df_head, df_tail\n\ntrain, test = split_last_n_by_series_id(data_subset, n_test_periods)",
"_____no_output_____"
]
],
[
[
"### Upload data to datastore\nThe [Machine Learning service workspace](https://docs.microsoft.com/en-us/azure/machine-learning/service/concept-workspace), is paired with the storage account, which contains the default data store. We will use it to upload the train and test data and create [tabular datasets](https://docs.microsoft.com/en-us/python/api/azureml-core/azureml.data.tabulardataset?view=azure-ml-py) for training and testing. A tabular dataset defines a series of lazily-evaluated, immutable operations to load data from the data source into tabular representation.",
"_____no_output_____"
]
],
[
[
"train.to_csv (r'./dominicks_OJ_train.csv', index = None, header=True)\ntest.to_csv (r'./dominicks_OJ_test.csv', index = None, header=True)",
"_____no_output_____"
],
[
"datastore = ws.get_default_datastore()\ndatastore.upload_files(files = ['./dominicks_OJ_train.csv', './dominicks_OJ_test.csv'], target_path = 'dataset/', overwrite = True,show_progress = True)",
"_____no_output_____"
]
],
[
[
"### Create dataset for training",
"_____no_output_____"
]
],
[
[
"from azureml.core.dataset import Dataset\ntrain_dataset = Dataset.Tabular.from_delimited_files(path=datastore.path('dataset/dominicks_OJ_train.csv'))",
"_____no_output_____"
],
[
"train_dataset.to_pandas_dataframe().tail()",
"_____no_output_____"
]
],
[
[
"## Modeling\n\nFor forecasting tasks, AutoML uses pre-processing and estimation steps that are specific to time-series. AutoML will undertake the following pre-processing steps:\n* Detect time-series sample frequency (e.g. hourly, daily, weekly) and create new records for absent time points to make the series regular. A regular time series has a well-defined frequency and has a value at every sample point in a contiguous time span \n* Impute missing values in the target (via forward-fill) and feature columns (using median column values) \n* Create features based on time series identifiers to enable fixed effects across different series\n* Create time-based features to assist in learning seasonal patterns\n* Encode categorical variables to numeric quantities\n\nIn this notebook, AutoML will train a single, regression-type model across **all** time-series in a given training set. This allows the model to generalize across related series. If you're looking for training multiple models for different time-series, please see the many-models notebook.\n\nYou are almost ready to start an AutoML training job. First, we need to separate the target column from the rest of the DataFrame: ",
"_____no_output_____"
]
],
[
[
"target_column_name = 'Quantity'",
"_____no_output_____"
]
],
[
[
"## Customization\n\nThe featurization customization in forecasting is an advanced feature in AutoML which allows our customers to change the default forecasting featurization behaviors and column types through `FeaturizationConfig`. The supported scenarios include:\n\n1. Column purposes update: Override feature type for the specified column. Currently supports DateTime, Categorical and Numeric. This customization can be used in the scenario that the type of the column cannot correctly reflect its purpose. Some numerical columns, for instance, can be treated as Categorical columns which need to be converted to categorical while some can be treated as epoch timestamp which need to be converted to datetime. To tell our SDK to correctly preprocess these columns, a configuration need to be add with the columns and their desired types.\n2. Transformer parameters update: Currently supports parameter change for Imputer only. User can customize imputation methods. The supported imputing methods for target column are constant and ffill (forward fill). The supported imputing methods for feature columns are mean, median, most frequent, constant and ffill (forward fill). This customization can be used for the scenario that our customers know which imputation methods fit best to the input data. For instance, some datasets use NaN to represent 0 which the correct behavior should impute all the missing value with 0. To achieve this behavior, these columns need to be configured as constant imputation with `fill_value` 0.\n3. Drop columns: Columns to drop from being featurized. These usually are the columns which are leaky or the columns contain no useful data.",
"_____no_output_____"
]
],
[
[
"featurization_config = FeaturizationConfig()\nfeaturization_config.drop_columns = ['logQuantity'] # 'logQuantity' is a leaky feature, so we remove it.\n# Force the CPWVOL5 feature to be numeric type.\nfeaturization_config.add_column_purpose('CPWVOL5', 'Numeric')\n# Fill missing values in the target column, Quantity, with zeros.\nfeaturization_config.add_transformer_params('Imputer', ['Quantity'], {\"strategy\": \"constant\", \"fill_value\": 0})\n# Fill missing values in the INCOME column with median value.\nfeaturization_config.add_transformer_params('Imputer', ['INCOME'], {\"strategy\": \"median\"})\n# Fill missing values in the Price column with forward fill (last value carried forward).\nfeaturization_config.add_transformer_params('Imputer', ['Price'], {\"strategy\": \"ffill\"})",
"_____no_output_____"
]
],
[
[
"## Forecasting Parameters\nTo define forecasting parameters for your experiment training, you can leverage the ForecastingParameters class. The table below details the forecasting parameter we will be passing into our experiment.\n\n\n|Property|Description|\n|-|-|\n|**time_column_name**|The name of your time column.|\n|**forecast_horizon**|The forecast horizon is how many periods forward you would like to forecast. This integer horizon is in units of the timeseries frequency (e.g. daily, weekly).|\n|**time_series_id_column_names**|The column names used to uniquely identify the time series in data that has multiple rows with the same timestamp. If the time series identifiers are not defined, the data set is assumed to be one time series.|",
"_____no_output_____"
],
[
"## Train\n\nThe [AutoMLConfig](https://docs.microsoft.com/en-us/python/api/azureml-train-automl-client/azureml.train.automl.automlconfig.automlconfig?view=azure-ml-py) object defines the settings and data for an AutoML training job. Here, we set necessary inputs like the task type, the number of AutoML iterations to try, the training data, and cross-validation parameters.\n\nFor forecasting tasks, there are some additional parameters that can be set in the `ForecastingParameters` class: the name of the column holding the date/time, the timeseries id column names, and the maximum forecast horizon. A time column is required for forecasting, while the time_series_id is optional. If time_series_id columns are not given, AutoML assumes that the whole dataset is a single time-series. We also pass a list of columns to drop prior to modeling. The _logQuantity_ column is completely correlated with the target quantity, so it must be removed to prevent a target leak.\n\nThe forecast horizon is given in units of the time-series frequency; for instance, the OJ series frequency is weekly, so a horizon of 20 means that a trained model will estimate sales up to 20 weeks beyond the latest date in the training data for each series. In this example, we set the forecast horizon to the number of samples per series in the test set (n_test_periods). Generally, the value of this parameter will be dictated by business needs. For example, a demand planning application that estimates the next month of sales should set the horizon according to suitable planning time-scales. Please see the [energy_demand notebook](https://github.com/Azure/MachineLearningNotebooks/tree/master/how-to-use-azureml/automated-machine-learning/forecasting-energy-demand) for more discussion of forecast horizon.\n\nWe note here that AutoML can sweep over two types of time-series models:\n* Models that are trained for each series such as ARIMA and Facebook's Prophet.\n* Models trained across multiple time-series using a regression approach.\n\nIn the first case, AutoML loops over all time-series in your dataset and trains one model (e.g. AutoArima or Prophet, as the case may be) for each series. This can result in long runtimes to train these models if there are a lot of series in the data. One way to mitigate this problem is to fit models for different series in parallel if you have multiple compute cores available. To enable this behavior, set the `max_cores_per_iteration` parameter in your AutoMLConfig as shown in the example in the next cell. \n\n\nFinally, a note about the cross-validation (CV) procedure for time-series data. AutoML uses out-of-sample error estimates to select a best pipeline/model, so it is important that the CV fold splitting is done correctly. Time-series can violate the basic statistical assumptions of the canonical K-Fold CV strategy, so AutoML implements a [rolling origin validation](https://robjhyndman.com/hyndsight/tscv/) procedure to create CV folds for time-series data. To use this procedure, you just need to specify the desired number of CV folds in the AutoMLConfig object. It is also possible to bypass CV and use your own validation set by setting the *validation_data* parameter of AutoMLConfig.\n\nHere is a summary of AutoMLConfig parameters used for training the OJ model:\n\n|Property|Description|\n|-|-|\n|**task**|forecasting|\n|**primary_metric**|This is the metric that you want to optimize.<br> Forecasting supports the following primary metrics <br><i>spearman_correlation</i><br><i>normalized_root_mean_squared_error</i><br><i>r2_score</i><br><i>normalized_mean_absolute_error</i>\n|**experiment_timeout_hours**|Experimentation timeout in hours.|\n|**enable_early_stopping**|If early stopping is on, training will stop when the primary metric is no longer improving.|\n|**training_data**|Input dataset, containing both features and label column.|\n|**label_column_name**|The name of the label column.|\n|**compute_target**|The remote compute for training.|\n|**n_cross_validations**|Number of cross-validation folds to use for model/pipeline selection|\n|**enable_voting_ensemble**|Allow AutoML to create a Voting ensemble of the best performing models|\n|**enable_stack_ensemble**|Allow AutoML to create a Stack ensemble of the best performing models|\n|**debug_log**|Log file path for writing debugging information|\n|**featurization**| 'auto' / 'off' / FeaturizationConfig Indicator for whether featurization step should be done automatically or not, or whether customized featurization should be used. Setting this enables AutoML to perform featurization on the input to handle *missing data*, and to perform some common *feature extraction*.|\n|**max_cores_per_iteration**|Maximum number of cores to utilize per iteration. A value of -1 indicates all available cores should be used",
"_____no_output_____"
]
],
[
[
"from azureml.automl.core.forecasting_parameters import ForecastingParameters\nforecasting_parameters = ForecastingParameters(\n time_column_name=time_column_name,\n forecast_horizon=n_test_periods,\n time_series_id_column_names=time_series_id_column_names\n)\n\nautoml_config = AutoMLConfig(task='forecasting',\n debug_log='automl_oj_sales_errors.log',\n primary_metric='normalized_mean_absolute_error',\n experiment_timeout_hours=0.25,\n training_data=train_dataset,\n label_column_name=target_column_name,\n compute_target=compute_target,\n enable_early_stopping=True,\n featurization=featurization_config,\n n_cross_validations=3,\n verbosity=logging.INFO,\n max_cores_per_iteration=-1,\n forecasting_parameters=forecasting_parameters)",
"_____no_output_____"
]
],
[
[
"You can now submit a new training run. Depending on the data and number of iterations this operation may take several minutes.\nInformation from each iteration will be printed to the console. Validation errors and current status will be shown when setting `show_output=True` and the execution will be synchronous.",
"_____no_output_____"
]
],
[
[
"remote_run = experiment.submit(automl_config, show_output=False)\nremote_run",
"_____no_output_____"
],
[
"remote_run.wait_for_completion()",
"_____no_output_____"
]
],
[
[
"### Retrieve the Best Model\nEach run within an Experiment stores serialized (i.e. pickled) pipelines from the AutoML iterations. We can now retrieve the pipeline with the best performance on the validation dataset:",
"_____no_output_____"
]
],
[
[
"best_run, fitted_model = remote_run.get_output()\nprint(fitted_model.steps)\nmodel_name = best_run.properties['model_name']",
"_____no_output_____"
]
],
[
[
"## Transparency\n\nView updated featurization summary",
"_____no_output_____"
]
],
[
[
"custom_featurizer = fitted_model.named_steps['timeseriestransformer']",
"_____no_output_____"
],
[
"custom_featurizer.get_featurization_summary()",
"_____no_output_____"
]
],
[
[
"# Forecasting\n\nNow that we have retrieved the best pipeline/model, it can be used to make predictions on test data. First, we remove the target values from the test set:",
"_____no_output_____"
]
],
[
[
"X_test = test\ny_test = X_test.pop(target_column_name).values",
"_____no_output_____"
],
[
"X_test.head()",
"_____no_output_____"
]
],
[
[
"To produce predictions on the test set, we need to know the feature values at all dates in the test set. This requirement is somewhat reasonable for the OJ sales data since the features mainly consist of price, which is usually set in advance, and customer demographics which are approximately constant for each store over the 20 week forecast horizon in the testing data.",
"_____no_output_____"
]
],
[
[
"# forecast returns the predictions and the featurized data, aligned to X_test.\n# This contains the assumptions that were made in the forecast\ny_predictions, X_trans = fitted_model.forecast(X_test)",
"_____no_output_____"
]
],
[
[
"If you are used to scikit pipelines, perhaps you expected `predict(X_test)`. However, forecasting requires a more general interface that also supplies the past target `y` values. Please use `forecast(X,y)` as `predict(X)` is reserved for internal purposes on forecasting models.\n\nThe [forecast function notebook](../forecasting-forecast-function/auto-ml-forecasting-function.ipynb).",
"_____no_output_____"
],
[
"# Evaluate\n\nTo evaluate the accuracy of the forecast, we'll compare against the actual sales quantities for some select metrics, included the mean absolute percentage error (MAPE). \n\nWe'll add predictions and actuals into a single dataframe for convenience in calculating the metrics.",
"_____no_output_____"
]
],
[
[
"assign_dict = {'predicted': y_predictions, target_column_name: y_test}\ndf_all = X_test.assign(**assign_dict)",
"_____no_output_____"
],
[
"from azureml.automl.core.shared import constants\nfrom azureml.automl.runtime.shared.score import scoring\nfrom matplotlib import pyplot as plt\n\n# use automl scoring module\nscores = scoring.score_regression(\n y_test=df_all[target_column_name],\n y_pred=df_all['predicted'],\n metrics=list(constants.Metric.SCALAR_REGRESSION_SET))\n\nprint(\"[Test data scores]\\n\")\nfor key, value in scores.items(): \n print('{}: {:.3f}'.format(key, value))\n \n# Plot outputs\n%matplotlib inline\ntest_pred = plt.scatter(df_all[target_column_name], df_all['predicted'], color='b')\ntest_test = plt.scatter(df_all[target_column_name], df_all[target_column_name], color='g')\nplt.legend((test_pred, test_test), ('prediction', 'truth'), loc='upper left', fontsize=8)\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Operationalize",
"_____no_output_____"
],
[
"_Operationalization_ means getting the model into the cloud so that other can run it after you close the notebook. We will create a docker running on Azure Container Instances with the model.",
"_____no_output_____"
]
],
[
[
"description = 'AutoML OJ forecaster'\ntags = None\nmodel = remote_run.register_model(model_name = model_name, description = description, tags = tags)\n\nprint(remote_run.model_id)",
"_____no_output_____"
]
],
[
[
"### Develop the scoring script\n\nFor the deployment we need a function which will run the forecast on serialized data. It can be obtained from the best_run.",
"_____no_output_____"
]
],
[
[
"script_file_name = 'score_fcast.py'\nbest_run.download_file('outputs/scoring_file_v_1_0_0.py', script_file_name)",
"_____no_output_____"
]
],
[
[
"### Deploy the model as a Web Service on Azure Container Instance",
"_____no_output_____"
]
],
[
[
"from azureml.core.model import InferenceConfig\nfrom azureml.core.webservice import AciWebservice\nfrom azureml.core.webservice import Webservice\nfrom azureml.core.model import Model\n\ninference_config = InferenceConfig(environment = best_run.get_environment(), \n entry_script = script_file_name)\n\naciconfig = AciWebservice.deploy_configuration(cpu_cores = 1, \n memory_gb = 2, \n tags = {'type': \"automl-forecasting\"},\n description = \"Automl forecasting sample service\")\n\naci_service_name = 'automl-oj-forecast-01'\nprint(aci_service_name)\naci_service = Model.deploy(ws, aci_service_name, [model], inference_config, aciconfig)\naci_service.wait_for_deployment(True)\nprint(aci_service.state)",
"_____no_output_____"
],
[
"aci_service.get_logs()",
"_____no_output_____"
]
],
[
[
"### Call the service",
"_____no_output_____"
]
],
[
[
"import json\nX_query = X_test.copy()\n# We have to convert datetime to string, because Timestamps cannot be serialized to JSON.\nX_query[time_column_name] = X_query[time_column_name].astype(str)\n# The Service object accept the complex dictionary, which is internally converted to JSON string.\n# The section 'data' contains the data frame in the form of dictionary.\ntest_sample = json.dumps({'data': X_query.to_dict(orient='records')})\nresponse = aci_service.run(input_data = test_sample)\n# translate from networkese to datascientese\ntry: \n res_dict = json.loads(response)\n y_fcst_all = pd.DataFrame(res_dict['index'])\n y_fcst_all[time_column_name] = pd.to_datetime(y_fcst_all[time_column_name], unit = 'ms')\n y_fcst_all['forecast'] = res_dict['forecast'] \nexcept:\n print(res_dict)",
"_____no_output_____"
],
[
"y_fcst_all.head()",
"_____no_output_____"
]
],
[
[
"### Delete the web service if desired",
"_____no_output_____"
]
],
[
[
"serv = Webservice(ws, 'automl-oj-forecast-01')\nserv.delete() # don't do it accidentally",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d085be6016cc954723231d7ffb2a615b44ae2332 | 33,904 | ipynb | Jupyter Notebook | how-to-use-azureml/automated-machine-learning/classification-bank-marketing-all-features/auto-ml-classification-bank-marketing-all-features.ipynb | hyoshioka0128/MachineLearningNotebooks | 053efde8c9740c7b691c4d13ee1f5b5b206cd24f | [
"MIT"
] | null | null | null | how-to-use-azureml/automated-machine-learning/classification-bank-marketing-all-features/auto-ml-classification-bank-marketing-all-features.ipynb | hyoshioka0128/MachineLearningNotebooks | 053efde8c9740c7b691c4d13ee1f5b5b206cd24f | [
"MIT"
] | null | null | null | how-to-use-azureml/automated-machine-learning/classification-bank-marketing-all-features/auto-ml-classification-bank-marketing-all-features.ipynb | hyoshioka0128/MachineLearningNotebooks | 053efde8c9740c7b691c4d13ee1f5b5b206cd24f | [
"MIT"
] | null | null | null | 36.892274 | 1,128 | 0.567249 | [
[
[
"Copyright (c) Microsoft Corporation. All rights reserved.\n\nLicensed under the MIT License.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"# Automated Machine Learning\n_**Classification with Deployment using a Bank Marketing Dataset**_\n\n## Contents\n1. [Introduction](#Introduction)\n1. [Setup](#Setup)\n1. [Train](#Train)\n1. [Results](#Results)\n1. [Deploy](#Deploy)\n1. [Test](#Test)\n1. [Acknowledgements](#Acknowledgements)",
"_____no_output_____"
],
[
"## Introduction\n\nIn this example we use the UCI Bank Marketing dataset to showcase how you can use AutoML for a classification problem and deploy it to an Azure Container Instance (ACI). The classification goal is to predict if the client will subscribe to a term deposit with the bank.\n\nIf you are using an Azure Machine Learning Compute Instance, you are all set. Otherwise, go through the [configuration](../../../configuration.ipynb) notebook first if you haven't already to establish your connection to the AzureML Workspace. \n\nPlease find the ONNX related documentations [here](https://github.com/onnx/onnx).\n\nIn this notebook you will learn how to:\n1. Create an experiment using an existing workspace.\n2. Configure AutoML using `AutoMLConfig`.\n3. Train the model using local compute with ONNX compatible config on.\n4. Explore the results, featurization transparency options and save the ONNX model\n5. Inference with the ONNX model.\n6. Register the model.\n7. Create a container image.\n8. Create an Azure Container Instance (ACI) service.\n9. Test the ACI service.\n\nIn addition this notebook showcases the following features\n- **Blacklisting** certain pipelines\n- Specifying **target metrics** to indicate stopping criteria\n- Handling **missing data** in the input",
"_____no_output_____"
],
[
"## Setup\n\nAs part of the setup you have already created an Azure ML `Workspace` object. For AutoML you will need to create an `Experiment` object, which is a named object in a `Workspace` used to run experiments.",
"_____no_output_____"
]
],
[
[
"import logging\n\nfrom matplotlib import pyplot as plt\nimport pandas as pd\nimport os\n\nimport azureml.core\nfrom azureml.core.experiment import Experiment\nfrom azureml.core.workspace import Workspace\nfrom azureml.automl.core.featurization import FeaturizationConfig\nfrom azureml.core.dataset import Dataset\nfrom azureml.train.automl import AutoMLConfig\nfrom azureml.explain.model._internal.explanation_client import ExplanationClient",
"_____no_output_____"
]
],
[
[
"This sample notebook may use features that are not available in previous versions of the Azure ML SDK.",
"_____no_output_____"
]
],
[
[
"print(\"This notebook was created using version 1.8.0 of the Azure ML SDK\")\nprint(\"You are currently using version\", azureml.core.VERSION, \"of the Azure ML SDK\")",
"_____no_output_____"
]
],
[
[
"Accessing the Azure ML workspace requires authentication with Azure.\n\nThe default authentication is interactive authentication using the default tenant. Executing the `ws = Workspace.from_config()` line in the cell below will prompt for authentication the first time that it is run.\n\nIf you have multiple Azure tenants, you can specify the tenant by replacing the `ws = Workspace.from_config()` line in the cell below with the following:\n\n```\nfrom azureml.core.authentication import InteractiveLoginAuthentication\nauth = InteractiveLoginAuthentication(tenant_id = 'mytenantid')\nws = Workspace.from_config(auth = auth)\n```\n\nIf you need to run in an environment where interactive login is not possible, you can use Service Principal authentication by replacing the `ws = Workspace.from_config()` line in the cell below with the following:\n\n```\nfrom azureml.core.authentication import ServicePrincipalAuthentication\nauth = auth = ServicePrincipalAuthentication('mytenantid', 'myappid', 'mypassword')\nws = Workspace.from_config(auth = auth)\n```\nFor more details, see [aka.ms/aml-notebook-auth](http://aka.ms/aml-notebook-auth)",
"_____no_output_____"
]
],
[
[
"ws = Workspace.from_config()\n\n# choose a name for experiment\nexperiment_name = 'automl-classification-bmarketing-all'\n\nexperiment=Experiment(ws, experiment_name)\n\noutput = {}\noutput['Subscription ID'] = ws.subscription_id\noutput['Workspace'] = ws.name\noutput['Resource Group'] = ws.resource_group\noutput['Location'] = ws.location\noutput['Experiment Name'] = experiment.name\npd.set_option('display.max_colwidth', -1)\noutputDf = pd.DataFrame(data = output, index = [''])\noutputDf.T",
"_____no_output_____"
]
],
[
[
"## Create or Attach existing AmlCompute\nYou will need to create a compute target for your AutoML run. In this tutorial, you create AmlCompute as your training compute resource.\n#### Creation of AmlCompute takes approximately 5 minutes. \nIf the AmlCompute with that name is already in your workspace this code will skip the creation process.\nAs with other Azure services, there are limits on certain resources (e.g. AmlCompute) associated with the Azure Machine Learning service. Please read this article on the default limits and how to request more quota.",
"_____no_output_____"
]
],
[
[
"from azureml.core.compute import ComputeTarget, AmlCompute\nfrom azureml.core.compute_target import ComputeTargetException\n\n# Choose a name for your CPU cluster\ncpu_cluster_name = \"cpu-cluster-4\"\n\n# Verify that cluster does not exist already\ntry:\n compute_target = ComputeTarget(workspace=ws, name=cpu_cluster_name)\n print('Found existing cluster, use it.')\nexcept ComputeTargetException:\n compute_config = AmlCompute.provisioning_configuration(vm_size='STANDARD_D2_V2',\n max_nodes=6)\n compute_target = ComputeTarget.create(ws, cpu_cluster_name, compute_config)\n\ncompute_target.wait_for_completion(show_output=True)",
"_____no_output_____"
]
],
[
[
"# Data",
"_____no_output_____"
],
[
"### Load Data\n\nLeverage azure compute to load the bank marketing dataset as a Tabular Dataset into the dataset variable. ",
"_____no_output_____"
],
[
"### Training Data",
"_____no_output_____"
]
],
[
[
"data = pd.read_csv(\"https://automlsamplenotebookdata.blob.core.windows.net/automl-sample-notebook-data/bankmarketing_train.csv\")\ndata.head()",
"_____no_output_____"
],
[
"# Add missing values in 75% of the lines.\nimport numpy as np\n\nmissing_rate = 0.75\nn_missing_samples = int(np.floor(data.shape[0] * missing_rate))\nmissing_samples = np.hstack((np.zeros(data.shape[0] - n_missing_samples, dtype=np.bool), np.ones(n_missing_samples, dtype=np.bool)))\nrng = np.random.RandomState(0)\nrng.shuffle(missing_samples)\nmissing_features = rng.randint(0, data.shape[1], n_missing_samples)\ndata.values[np.where(missing_samples)[0], missing_features] = np.nan",
"_____no_output_____"
],
[
"if not os.path.isdir('data'):\n os.mkdir('data')\n \n# Save the train data to a csv to be uploaded to the datastore\npd.DataFrame(data).to_csv(\"data/train_data.csv\", index=False)\n\nds = ws.get_default_datastore()\nds.upload(src_dir='./data', target_path='bankmarketing', overwrite=True, show_progress=True)\n\n \n\n# Upload the training data as a tabular dataset for access during training on remote compute\ntrain_data = Dataset.Tabular.from_delimited_files(path=ds.path('bankmarketing/train_data.csv'))\nlabel = \"y\"",
"_____no_output_____"
]
],
[
[
"### Validation Data",
"_____no_output_____"
]
],
[
[
"validation_data = \"https://automlsamplenotebookdata.blob.core.windows.net/automl-sample-notebook-data/bankmarketing_validate.csv\"\nvalidation_dataset = Dataset.Tabular.from_delimited_files(validation_data)",
"_____no_output_____"
]
],
[
[
"### Test Data",
"_____no_output_____"
]
],
[
[
"test_data = \"https://automlsamplenotebookdata.blob.core.windows.net/automl-sample-notebook-data/bankmarketing_test.csv\"\ntest_dataset = Dataset.Tabular.from_delimited_files(test_data)",
"_____no_output_____"
]
],
[
[
"## Train\n\nInstantiate a AutoMLConfig object. This defines the settings and data used to run the experiment.\n\n|Property|Description|\n|-|-|\n|**task**|classification or regression or forecasting|\n|**primary_metric**|This is the metric that you want to optimize. Classification supports the following primary metrics: <br><i>accuracy</i><br><i>AUC_weighted</i><br><i>average_precision_score_weighted</i><br><i>norm_macro_recall</i><br><i>precision_score_weighted</i>|\n|**iteration_timeout_minutes**|Time limit in minutes for each iteration.|\n|**blacklist_models** | *List* of *strings* indicating machine learning algorithms for AutoML to avoid in this run. <br><br> Allowed values for **Classification**<br><i>LogisticRegression</i><br><i>SGD</i><br><i>MultinomialNaiveBayes</i><br><i>BernoulliNaiveBayes</i><br><i>SVM</i><br><i>LinearSVM</i><br><i>KNN</i><br><i>DecisionTree</i><br><i>RandomForest</i><br><i>ExtremeRandomTrees</i><br><i>LightGBM</i><br><i>GradientBoosting</i><br><i>TensorFlowDNN</i><br><i>TensorFlowLinearClassifier</i><br><br>Allowed values for **Regression**<br><i>ElasticNet</i><br><i>GradientBoosting</i><br><i>DecisionTree</i><br><i>KNN</i><br><i>LassoLars</i><br><i>SGD</i><br><i>RandomForest</i><br><i>ExtremeRandomTrees</i><br><i>LightGBM</i><br><i>TensorFlowLinearRegressor</i><br><i>TensorFlowDNN</i><br><br>Allowed values for **Forecasting**<br><i>ElasticNet</i><br><i>GradientBoosting</i><br><i>DecisionTree</i><br><i>KNN</i><br><i>LassoLars</i><br><i>SGD</i><br><i>RandomForest</i><br><i>ExtremeRandomTrees</i><br><i>LightGBM</i><br><i>TensorFlowLinearRegressor</i><br><i>TensorFlowDNN</i><br><i>Arima</i><br><i>Prophet</i>|\n| **whitelist_models** | *List* of *strings* indicating machine learning algorithms for AutoML to use in this run. Same values listed above for **blacklist_models** allowed for **whitelist_models**.|\n|**experiment_exit_score**| Value indicating the target for *primary_metric*. <br>Once the target is surpassed the run terminates.|\n|**experiment_timeout_hours**| Maximum amount of time in hours that all iterations combined can take before the experiment terminates.|\n|**enable_early_stopping**| Flag to enble early termination if the score is not improving in the short term.|\n|**featurization**| 'auto' / 'off' Indicator for whether featurization step should be done automatically or not. Note: If the input data is sparse, featurization cannot be turned on.|\n|**n_cross_validations**|Number of cross validation splits.|\n|**training_data**|Input dataset, containing both features and label column.|\n|**label_column_name**|The name of the label column.|\n\n**_You can find more information about primary metrics_** [here](https://docs.microsoft.com/en-us/azure/machine-learning/service/how-to-configure-auto-train#primary-metric)",
"_____no_output_____"
]
],
[
[
"automl_settings = {\n \"experiment_timeout_hours\" : 0.3,\n \"enable_early_stopping\" : True,\n \"iteration_timeout_minutes\": 5,\n \"max_concurrent_iterations\": 4,\n \"max_cores_per_iteration\": -1,\n #\"n_cross_validations\": 2,\n \"primary_metric\": 'AUC_weighted',\n \"featurization\": 'auto',\n \"verbosity\": logging.INFO,\n}\n\nautoml_config = AutoMLConfig(task = 'classification',\n debug_log = 'automl_errors.log',\n compute_target=compute_target,\n experiment_exit_score = 0.9984,\n blacklist_models = ['KNN','LinearSVM'],\n enable_onnx_compatible_models=True,\n training_data = train_data,\n label_column_name = label,\n validation_data = validation_dataset,\n **automl_settings\n )",
"_____no_output_____"
]
],
[
[
"Call the `submit` method on the experiment object and pass the run configuration. Execution of local runs is synchronous. Depending on the data and the number of iterations this can run for a while.",
"_____no_output_____"
]
],
[
[
"remote_run = experiment.submit(automl_config, show_output = False)",
"_____no_output_____"
],
[
"remote_run",
"_____no_output_____"
]
],
[
[
"Run the following cell to access previous runs. Uncomment the cell below and update the run_id.",
"_____no_output_____"
]
],
[
[
"#from azureml.train.automl.run import AutoMLRun\n#remote_run = AutoMLRun(experiment=experiment, run_id='<run_ID_goes_here')\n#remote_run",
"_____no_output_____"
],
[
"# Wait for the remote run to complete\nremote_run.wait_for_completion()",
"_____no_output_____"
],
[
"best_run_customized, fitted_model_customized = remote_run.get_output()",
"_____no_output_____"
]
],
[
[
"## Transparency\n\nView updated featurization summary",
"_____no_output_____"
]
],
[
[
"custom_featurizer = fitted_model_customized.named_steps['datatransformer']\ndf = custom_featurizer.get_featurization_summary()\npd.DataFrame(data=df)",
"_____no_output_____"
]
],
[
[
"Set `is_user_friendly=False` to get a more detailed summary for the transforms being applied.",
"_____no_output_____"
]
],
[
[
"df = custom_featurizer.get_featurization_summary(is_user_friendly=False)\npd.DataFrame(data=df)",
"_____no_output_____"
],
[
"df = custom_featurizer.get_stats_feature_type_summary()\npd.DataFrame(data=df)",
"_____no_output_____"
]
],
[
[
"## Results",
"_____no_output_____"
]
],
[
[
"from azureml.widgets import RunDetails\nRunDetails(remote_run).show() ",
"_____no_output_____"
]
],
[
[
"### Retrieve the Best Model's explanation\nRetrieve the explanation from the best_run which includes explanations for engineered features and raw features. Make sure that the run for generating explanations for the best model is completed.",
"_____no_output_____"
]
],
[
[
"# Wait for the best model explanation run to complete\nfrom azureml.core.run import Run\nmodel_explainability_run_id = remote_run.get_properties().get('ModelExplainRunId')\nprint(model_explainability_run_id)\nif model_explainability_run_id is not None:\n model_explainability_run = Run(experiment=experiment, run_id=model_explainability_run_id)\n model_explainability_run.wait_for_completion()\n\n# Get the best run object\nbest_run, fitted_model = remote_run.get_output()",
"_____no_output_____"
]
],
[
[
"#### Download engineered feature importance from artifact store\nYou can use ExplanationClient to download the engineered feature explanations from the artifact store of the best_run.",
"_____no_output_____"
]
],
[
[
"client = ExplanationClient.from_run(best_run)\nengineered_explanations = client.download_model_explanation(raw=False)\nexp_data = engineered_explanations.get_feature_importance_dict()\nexp_data",
"_____no_output_____"
]
],
[
[
"#### Download raw feature importance from artifact store\nYou can use ExplanationClient to download the raw feature explanations from the artifact store of the best_run.",
"_____no_output_____"
]
],
[
[
"client = ExplanationClient.from_run(best_run)\nengineered_explanations = client.download_model_explanation(raw=True)\nexp_data = engineered_explanations.get_feature_importance_dict()\nexp_data",
"_____no_output_____"
]
],
[
[
"### Retrieve the Best ONNX Model\n\nBelow we select the best pipeline from our iterations. The `get_output` method returns the best run and the fitted model. The Model includes the pipeline and any pre-processing. Overloads on `get_output` allow you to retrieve the best run and fitted model for *any* logged metric or for a particular *iteration*.\n\nSet the parameter return_onnx_model=True to retrieve the best ONNX model, instead of the Python model.",
"_____no_output_____"
]
],
[
[
"best_run, onnx_mdl = remote_run.get_output(return_onnx_model=True)",
"_____no_output_____"
]
],
[
[
"### Save the best ONNX model",
"_____no_output_____"
]
],
[
[
"from azureml.automl.runtime.onnx_convert import OnnxConverter\nonnx_fl_path = \"./best_model.onnx\"\nOnnxConverter.save_onnx_model(onnx_mdl, onnx_fl_path)",
"_____no_output_____"
]
],
[
[
"### Predict with the ONNX model, using onnxruntime package",
"_____no_output_____"
]
],
[
[
"import sys\nimport json\nfrom azureml.automl.core.onnx_convert import OnnxConvertConstants\nfrom azureml.train.automl import constants\n\nif sys.version_info < OnnxConvertConstants.OnnxIncompatiblePythonVersion:\n python_version_compatible = True\nelse:\n python_version_compatible = False\n\nimport onnxruntime\nfrom azureml.automl.runtime.onnx_convert import OnnxInferenceHelper\n\ndef get_onnx_res(run):\n res_path = 'onnx_resource.json'\n run.download_file(name=constants.MODEL_RESOURCE_PATH_ONNX, output_file_path=res_path)\n with open(res_path) as f:\n onnx_res = json.load(f)\n return onnx_res\n\nif python_version_compatible:\n test_df = test_dataset.to_pandas_dataframe()\n mdl_bytes = onnx_mdl.SerializeToString()\n onnx_res = get_onnx_res(best_run)\n\n onnxrt_helper = OnnxInferenceHelper(mdl_bytes, onnx_res)\n pred_onnx, pred_prob_onnx = onnxrt_helper.predict(test_df)\n\n print(pred_onnx)\n print(pred_prob_onnx)\nelse:\n print('Please use Python version 3.6 or 3.7 to run the inference helper.')",
"_____no_output_____"
]
],
[
[
"## Deploy\n\n### Retrieve the Best Model\n\nBelow we select the best pipeline from our iterations. The `get_output` method returns the best run and the fitted model. Overloads on `get_output` allow you to retrieve the best run and fitted model for *any* logged metric or for a particular *iteration*.",
"_____no_output_____"
],
[
"#### Widget for Monitoring Runs\n\nThe widget will first report a \"loading\" status while running the first iteration. After completing the first iteration, an auto-updating graph and table will be shown. The widget will refresh once per minute, so you should see the graph update as child runs complete.\n\n**Note:** The widget displays a link at the bottom. Use this link to open a web interface to explore the individual run details",
"_____no_output_____"
]
],
[
[
"best_run, fitted_model = remote_run.get_output()",
"_____no_output_____"
],
[
"model_name = best_run.properties['model_name']\n\nscript_file_name = 'inference/score.py'\nconda_env_file_name = 'inference/env.yml'\n\nbest_run.download_file('outputs/scoring_file_v_1_0_0.py', 'inference/score.py')\nbest_run.download_file('outputs/conda_env_v_1_0_0.yml', 'inference/env.yml')",
"_____no_output_____"
]
],
[
[
"### Register the Fitted Model for Deployment\nIf neither `metric` nor `iteration` are specified in the `register_model` call, the iteration with the best primary metric is registered.",
"_____no_output_____"
]
],
[
[
"description = 'AutoML Model trained on bank marketing data to predict if a client will subscribe to a term deposit'\ntags = None\nmodel = remote_run.register_model(model_name = model_name, description = description, tags = tags)\n\nprint(remote_run.model_id) # This will be written to the script file later in the notebook.",
"_____no_output_____"
]
],
[
[
"### Deploy the model as a Web Service on Azure Container Instance",
"_____no_output_____"
]
],
[
[
"from azureml.core.model import InferenceConfig\nfrom azureml.core.webservice import AciWebservice\nfrom azureml.core.webservice import Webservice\nfrom azureml.core.model import Model\nfrom azureml.core.environment import Environment\n\nmyenv = Environment.from_conda_specification(name=\"myenv\", file_path=conda_env_file_name)\ninference_config = InferenceConfig(entry_script=script_file_name, environment=myenv)\n\naciconfig = AciWebservice.deploy_configuration(cpu_cores = 1, \n memory_gb = 1, \n tags = {'area': \"bmData\", 'type': \"automl_classification\"}, \n description = 'sample service for Automl Classification')\n\naci_service_name = 'automl-sample-bankmarketing-all'\nprint(aci_service_name)\naci_service = Model.deploy(ws, aci_service_name, [model], inference_config, aciconfig)\naci_service.wait_for_deployment(True)\nprint(aci_service.state)",
"_____no_output_____"
]
],
[
[
"### Delete a Web Service\n\nDeletes the specified web service.",
"_____no_output_____"
]
],
[
[
"#aci_service.delete()",
"_____no_output_____"
]
],
[
[
"### Get Logs from a Deployed Web Service\n\nGets logs from a deployed web service.",
"_____no_output_____"
]
],
[
[
"#aci_service.get_logs()",
"_____no_output_____"
]
],
[
[
"## Test\n\nNow that the model is trained, run the test data through the trained model to get the predicted values.",
"_____no_output_____"
]
],
[
[
"# Load the bank marketing datasets.\nfrom numpy import array",
"_____no_output_____"
],
[
"X_test = test_dataset.drop_columns(columns=['y'])\ny_test = test_dataset.keep_columns(columns=['y'], validate=True)\ntest_dataset.take(5).to_pandas_dataframe()",
"_____no_output_____"
],
[
"X_test = X_test.to_pandas_dataframe()\ny_test = y_test.to_pandas_dataframe()",
"_____no_output_____"
],
[
"y_pred = fitted_model.predict(X_test)\nactual = array(y_test)\nactual = actual[:,0]\nprint(y_pred.shape, \" \", actual.shape)",
"_____no_output_____"
]
],
[
[
"### Calculate metrics for the prediction\n\nNow visualize the data on a scatter plot to show what our truth (actual) values are compared to the predicted values \nfrom the trained model that was returned.",
"_____no_output_____"
]
],
[
[
"%matplotlib notebook\ntest_pred = plt.scatter(actual, y_pred, color='b')\ntest_test = plt.scatter(actual, actual, color='g')\nplt.legend((test_pred, test_test), ('prediction', 'truth'), loc='upper left', fontsize=8)\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Acknowledgements",
"_____no_output_____"
],
[
"This Bank Marketing dataset is made available under the Creative Commons (CCO: Public Domain) License: https://creativecommons.org/publicdomain/zero/1.0/. Any rights in individual contents of the database are licensed under the Database Contents License: https://creativecommons.org/publicdomain/zero/1.0/ and is available at: https://www.kaggle.com/janiobachmann/bank-marketing-dataset .\n\n_**Acknowledgements**_\nThis data set is originally available within the UCI Machine Learning Database: https://archive.ics.uci.edu/ml/datasets/bank+marketing\n\n[Moro et al., 2014] S. Moro, P. Cortez and P. Rita. A Data-Driven Approach to Predict the Success of Bank Telemarketing. Decision Support Systems, Elsevier, 62:22-31, June 2014",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d085d1075cba698720d10132ab814f2fd984ecac | 24,366 | ipynb | Jupyter Notebook | 01-03_The_power_of_NumPy.ipynb | Collumbus/Machine_Learning_for_Trading-Udacity | a2fe08b783c642e20333f7e5e80c7178caa28d85 | [
"MIT"
] | null | null | null | 01-03_The_power_of_NumPy.ipynb | Collumbus/Machine_Learning_for_Trading-Udacity | a2fe08b783c642e20333f7e5e80c7178caa28d85 | [
"MIT"
] | null | null | null | 01-03_The_power_of_NumPy.ipynb | Collumbus/Machine_Learning_for_Trading-Udacity | a2fe08b783c642e20333f7e5e80c7178caa28d85 | [
"MIT"
] | null | null | null | 29.97048 | 383 | 0.49134 | [
[
[
"### Lesson outline\n\nIf you're familiar with NumPy (esp. the following operations), feel free to skim through this lesson.\n\n- #### Create a NumPy array:\n - from a pandas dataframe: [pandas.DataFrame.values](http://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.values.html)\n - from a Python sequence: [numpy.array](http://docs.scipy.org/doc/numpy/reference/generated/numpy.array.html)\n - with constant initial values: [numpy.ones, numpy.zeros](http://docs.scipy.org/doc/numpy/reference/generated/numpy.ones.html)\n - with random values: [numpy.random](http://docs.scipy.org/doc/numpy/reference/routines.random.html)\n\n- #### Access array attributes: [shape](http://docs.scipy.org/doc/numpy/reference/generated/numpy.ndarray.shape.html), [ndim](http://docs.scipy.org/doc/numpy/reference/generated/numpy.ndarray.ndim.html), [size](http://docs.scipy.org/doc/numpy/reference/generated/numpy.ndarray.size.html), [dtype](http://docs.scipy.org/doc/numpy/reference/generated/numpy.ndarray.dtype.html)\n- #### Compute statistics: [sum](http://docs.scipy.org/doc/numpy/reference/generated/numpy.sum.html), [min](http://docs.scipy.org/doc/numpy/reference/generated/numpy.min.html), [max](http://docs.scipy.org/doc/numpy/reference/generated/numpy.max.html), [mean](http://docs.scipy.org/doc/numpy/reference/generated/numpy.mean.html)\n- #### Carry out arithmetic operations: [add](http://docs.scipy.org/doc/numpy/reference/generated/numpy.add.html), [subtract](http://docs.scipy.org/doc/numpy/reference/generated/numpy.subtract.html), [multiply](http://docs.scipy.org/doc/numpy/reference/generated/numpy.multiply.html), [divide](http://docs.scipy.org/doc/numpy/reference/generated/numpy.divide.html)\n- #### Measure execution time: [time.time](https://docs.python.org/2/library/time.html#time.time), [profile](https://docs.python.org/2/library/profile.html)\n- #### Manipulate array elements: [Using simple indices and slices](http://docs.scipy.org/doc/numpy/reference/arrays.indexing.html#basic-slicing-and-indexing), [integer arrays](http://docs.scipy.org/doc/numpy/reference/arrays.indexing.html#integer-array-indexing), [boolean arrays](http://docs.scipy.org/doc/numpy/reference/arrays.indexing.html#boolean-array-indexing)",
"_____no_output_____"
]
],
[
[
"'''Creating NumPy arrays.'''\n\nimport numpy as np\n\ndef test_run():\n # List to 1D array\n print np.array([2, 3, 4])\n print ''\n \n #List of tuples to 2D array\n print np.array([(2, 3, 4), (5, 6, 7)])\n\nif __name__ == '__main__':\n test_run()",
"[2 3 4]\n\n[[2 3 4]\n [5 6 7]]\n"
],
[
"'''Arrays with initial values.'''\n\nimport numpy as np\n\ndef test_run():\n # Empty array\n print np.empty(5)\n print np.empty((5,4))\n \n #Arrays of 1s\n print np.ones((5,4))\n\nif __name__ == '__main__':\n test_run()",
"[ 0.00000000e+000 6.89941071e-310 6.89943153e-310 3.39387131e-317\n 2.37151510e-322]\n[[ 6.89944150e-310 6.56353958e-317 1.01006264e+261 4.48861446e-120]\n [ 1.67709469e+243 3.03574399e-152 8.05084511e+173 6.69433480e+151]\n [ 2.44514154e-154 3.63537905e+233 4.83245960e+276 8.82085571e+199]\n [ 9.34634029e+218 2.65856669e-260 2.13505411e+161 2.18072748e-153]\n [ 4.97745931e+151 6.01346953e-154 1.24039117e+224 6.09109151e-114]]\n[[ 1. 1. 1. 1.]\n [ 1. 1. 1. 1.]\n [ 1. 1. 1. 1.]\n [ 1. 1. 1. 1.]\n [ 1. 1. 1. 1.]]\n"
],
[
"'''Specify the datatype.'''\n\nimport numpy as np\n\ndef test_run():\n \n #Arrays of integers 1s\n print np.ones((5,4), dtype=np.int)\n\nif __name__ == '__main__':\n test_run()",
"[[1 1 1 1]\n [1 1 1 1]\n [1 1 1 1]\n [1 1 1 1]\n [1 1 1 1]]\n"
],
[
"'''Generating random numbers.'''\n\nimport numpy as np\n\ndef test_run():\n \n #Generate an anrray full of rando, numbers, uniformly sampled from [0.0, 1.0)\n print np.random.random((5,4)) # Pass in a size tuple\n print ''\n \n # Sample numbers from a Gaussian (normal) distribution\n print 'Standard Normal'\n print np.random.normal(size=(2, 3)) # \"Standard normal\" (mean =0, s.d. = 1)\n print ''\n print 'Standard Normal'\n print np.random.normal(50,10, size=(2, 3)) # Change mean to 50 and s.d. = 10\n print ''\n \n #Random integers\n print 'A single integer'\n print np.random.randint(10) # A single integer in [0, 10)\n print ''\n print 'A single integer'\n print np.random.randint(0, 10) # Same as above, especifying [low, high) explicit\n print ''\n print '1d-array'\n print np.random.randint(0, 10, size = 5) # 5 random integers as a 1D array\n print ''\n print '2d-array'\n print np.random.randint(0, 10, size = (2, 3)) # 2x3 array of random integers\n \nif __name__ == '__main__':\n test_run()",
"[[ 0.98002498 0.57936353 0.40416187 0.45760029]\n [ 0.66914955 0.0749194 0.55522073 0.04126315]\n [ 0.10187618 0.75788742 0.44318673 0.41842539]\n [ 0.87198883 0.80890351 0.48565408 0.68981486]\n [ 0.38924023 0.83339295 0.19694214 0.84770307]]\n\nStandard Normal\n[[ 0.20255373 -0.36719115 -0.20939396]\n [-0.97229616 -0.44531705 -0.1617242 ]]\n\nStandard Normal\n[[ 49.02978119 41.66887027 65.92429881]\n [ 41.58021424 50.39193175 46.07780296]]\n\nA single integer\n0\n\nA single integer\n9\n\n1d-array\n[0 1 0 3 7]\n\n2d-array\n[[9 8 5]\n [2 3 6]]\n"
],
[
"'''Array attributes.'''\n\nimport numpy as np\n\ndef test_run():\n \n a = np.random.random((5,4)) # 5x4 array of random numbers\n print a\n print a.shape\n print a.shape[0] # Number of rows\n print a.shape[1] # Number of columns\n print len(a.shape)\n print a.size\n print a.dtype\n \n\nif __name__ == '__main__':\n test_run()",
"[[ 0.76444591 0.42017101 0.78568757 0.47444077]\n [ 0.77287955 0.68360112 0.71139951 0.34694677]\n [ 0.72109319 0.9797951 0.22239037 0.06959021]\n [ 0.09883535 0.17842835 0.79085698 0.885343 ]\n [ 0.46956974 0.84621499 0.58448892 0.65181972]]\n(5, 4)\n5\n4\n2\n20\nfloat64\n"
],
[
"'''Operations on arrays.'''\n\nimport numpy as np\n\ndef test_run():\n \n a = np.random.randint(0,10, size = (5,4)) # 5x4 random integers in [0, 10)\n print 'Array:\\n', a\n \n #Sum of all elements\n print 'Sum of all elements:', a.sum()\n \n #Iterate over rows, to compute sum of each column\n print 'Sum of each column:', a.sum(axis=0)\n \n #Iterate over columns, to compute sum of each row\n print 'Sum of each row:', a.sum(axis=1)\n \n #Statistics: min, max, mean (accross rows, cols, and overall)\n print 'Minimum of each column:\\n', a.min(axis=0)\n print 'Maximum of each row:\\n', a.min(axis=1)\n print 'Mean of all elements:\\n', a.min() # Leave out axis arg.\n \n \nif __name__ == '__main__':\n test_run()",
"Array:\n[[0 9 4 4]\n [6 9 0 8]\n [8 1 4 8]\n [1 8 2 1]\n [2 3 1 5]]\nSum of all elements: 84\nSum of each column: [17 30 11 26]\nSum of each row: [17 23 21 12 11]\nMinimum of each column:\n[0 1 0 1]\nMaximum of each row:\n[0 0 1 1 1]\nMean of all elements:\n0\n"
]
],
[
[
"---\n## Quiz: Locate Maximum Value",
"_____no_output_____"
]
],
[
[
"\"\"\"Locate maximum value.\"\"\"\n\nimport numpy as np\n\n\ndef get_max_index(a):\n \"\"\"Return the index of the maximum value in given 1D array.\"\"\"\n return np.argmax(a)\n\n\ndef test_run():\n a = np.array([9, 6, 2, 3, 12, 14, 7, 10], dtype=np.int32) # 32-bit integer array\n print \"Array:\", a\n \n # Find the maximum and its index in array\n print \"Maximum value:\", a.max()\n print \"Index of max.:\", get_max_index(a)\n\n\nif __name__ == \"__main__\":\n test_run()\n",
"Array: [ 9 6 2 3 12 14 7 10]\nMaximum value: 14\nIndex of max.: 5\n"
]
],
[
[
"---",
"_____no_output_____"
]
],
[
[
"'''Using time function.'''\n\nimport numpy as np\nimport time\n\ndef test_run():\n t1 = time.time()\n print 'ML4T'\n t2 = time.time()\n print 'The time taken by print statement is ', t2 - t1,'seconds'\n \n\nif __name__ == '__main__':\n test_run()",
"ML4T\nThe time taken by print statement is 9.08374786377e-05 seconds\n"
],
[
"'''How fast is NumPy.'''\n\nimport numpy as np\nfrom time import time\n\ndef how_long(func, *args):\n '''Execute funcion with given arguments, and measure execution time.'''\n t0 = time()\n result = func(*args) # All arguments are passed in as-is\n t1 = time()\n return result, t1- t0\n\ndef manual_mean(arr):\n '''Compute mean (average) of all elements in the given 2D array'''\n sum = 0\n for i in xrange(0, arr.shape[0]):\n for j in xrange (0, arr.shape[1]):\n sum = sum + arr[i, j]\n return sum / arr.size\n\ndef numpy_mean(arr):\n '''Compute mean (average) using NumPy'''\n return arr.mean()\n\ndef test_run():\n '''Function called by Test Run.'''\n nd1 = np.random.random((1000, 10000)) # Use a sufficiently large array\n \n #Time the two functions, retrieving results and execution times\n res_manual, t_manual = how_long(manual_mean, nd1)\n res_numpy, t_numpy = how_long(numpy_mean, nd1)\n print 'Manual: {:.6f} ({:.3f} secs.) vs NumPy: {:.6f} ({:.3f} secs.)'.format(res_manual, t_manual, res_numpy, t_numpy) \n \n #Make sure both give us the same answer (upto some precision)\n assert abs(res_manual - res_numpy) <= 10e-6, 'Results aren´t equal!' \n \n #Compute speedup\n speedup = t_manual / t_numpy\n print 'NumPy mean is', speedup, 'times faster than manual for loops.'\n\nif __name__ == '__main__':\n test_run()",
"Manual: 0.500004 (1.491 secs.) vs NumPy: 0.500004 (0.007 secs.)\nNumPy mean is 199.771391143 times faster than manual for loops.\n"
],
[
"'''Accessing array elements.'''\n\nimport numpy as np\n\ndef test_run():\n a = np.random.rand(5, 4)\n print 'Array:\\n', a\n print''\n \n #Accessing element at position (3, 2)\n element = a[3, 2]\n print 'Position (3, 2):\\n', element\n print''\n \n #Elements in defined range\n print 'Range (0, 1:3):\\n', a[0, 1:3]\n print''\n \n #Top-left corner\n print 'Top-left corner :\\n', a[0:2, 0:2]\n print''\n \n #Slicing\n #Note: Slice n:m:t specifies a range that starts at n, and stops before m, in steps of sizet\n print 'Slicing:', a[:, 0:3:2]\n\nif __name__ == '__main__':\n test_run()",
"Array:\n[[ 0.70471966 0.488766 0.72131719 0.59584046]\n [ 0.11730061 0.85680481 0.49135416 0.95255406]\n [ 0.00229705 0.24127687 0.33643233 0.65719103]\n [ 0.11624821 0.57739567 0.3062893 0.894219 ]\n [ 0.88178216 0.16968218 0.46442419 0.98048617]]\n\nPosition (3, 2):\n0.306289299939\n\nRange (0, 1:3):\n[ 0.488766 0.72131719]\n\nTop-left corner :\n[[ 0.70471966 0.488766 ]\n [ 0.11730061 0.85680481]]\n\nSlicing: [[ 0.70471966 0.72131719]\n [ 0.11730061 0.49135416]\n [ 0.00229705 0.33643233]\n [ 0.11624821 0.3062893 ]\n [ 0.88178216 0.46442419]]\n"
],
[
"'''Modifying array elements.'''\n\nimport numpy as np\n\ndef test_run():\n a = np.random.rand(5, 4)\n print 'Array:\\n', a\n print''\n \n #Assigning a value to aa particular location\n a[0, 0] = 1\n print '\\nModified (replaced one element):\\n', a\n print''\n \n #Assingning a single value to an entire row\n a[0, :] = 2\n print '\\nModified (replaced a row with a single value):\\n', a\n print''\n \n #Assingning a list to a column in an array\n a[:, 3] = [1, 2, 3, 4, 5]\n print '\\nModified (replaced a column with a list):\\n', a\n print''\nif __name__ == '__main__':\n test_run()",
"Array:\n[[ 0.03000799 0.29923522 0.63106626 0.40993497]\n [ 0.14535125 0.07935721 0.02244874 0.75989961]\n [ 0.93776966 0.30107738 0.25096299 0.67044536]\n [ 0.95579093 0.81059579 0.29811526 0.42337705]\n [ 0.68052147 0.59356922 0.77569521 0.58312439]]\n\n\nModified (replaced one element):\n[[ 1. 0.29923522 0.63106626 0.40993497]\n [ 0.14535125 0.07935721 0.02244874 0.75989961]\n [ 0.93776966 0.30107738 0.25096299 0.67044536]\n [ 0.95579093 0.81059579 0.29811526 0.42337705]\n [ 0.68052147 0.59356922 0.77569521 0.58312439]]\n\n\nModified (replaced a row with a single value):\n[[ 2. 2. 2. 2. ]\n [ 0.14535125 0.07935721 0.02244874 0.75989961]\n [ 0.93776966 0.30107738 0.25096299 0.67044536]\n [ 0.95579093 0.81059579 0.29811526 0.42337705]\n [ 0.68052147 0.59356922 0.77569521 0.58312439]]\n\n\nModified (replaced a column with a list):\n[[ 2. 2. 2. 1. ]\n [ 0.14535125 0.07935721 0.02244874 2. ]\n [ 0.93776966 0.30107738 0.25096299 3. ]\n [ 0.95579093 0.81059579 0.29811526 4. ]\n [ 0.68052147 0.59356922 0.77569521 5. ]]\n\n"
],
[
"'''Indexing an array with another array.'''\n\nimport numpy as np\n\ndef test_run():\n a = np.random.rand(5)\n \n #Accessing using list of indices\n indices = np.array([1, 1, 2, 3])\n \n print a\n print a[indices]\n\nif __name__ == '__main__':\n test_run()",
"[ 0.01286659 0.72581137 0.59752278 0.58280926 0.19408949]\n[ 0.72581137 0.72581137 0.59752278 0.58280926]\n"
],
[
"'''Boolean or \"mask\" index arrays.'''\n\nimport numpy as np\n\ndef test_run():\n a = np.array([(20, 25, 10, 23, 26, 32, 10, 5, 0), (0, 2, 50, 20, 0, 1, 28, 5, 0)])\n print 'Array:\\n', a\n print ''\n \n #Calculating mean\n mean = a.mean()\n print 'Mean:\\n', mean\n print ''\n \n #Masking\n a[a<mean] = mean\n print 'Masking:\\n', a\n \nif __name__ == '__main__':\n test_run()",
"Array:\n[[20 25 10 23 26 32 10 5 0]\n [ 0 2 50 20 0 1 28 5 0]]\n\nMean:\n14.2777777778\n\nMasking:\n[[20 25 14 23 26 32 14 14 14]\n [14 14 50 20 14 14 28 14 14]]\n"
],
[
"'''Arithmetic operations.'''\n\nimport numpy as np\n\ndef test_run():\n a = np.array([(1, 2, 3, 4, 5), (10, 20, 30, 40, 50)])\n print 'Original array a:\\n', a\n print ''\n \n b = np.array([(100, 200, 300, 400, 500), (1, 2, 3, 4, 5)])\n print 'Original array b:\\n', b\n print ''\n \n #Multiply a by 2\n mean = a.mean()\n print 'Multiply a by 2:\\n', 2*a\n print ''\n \n #Divide a by 2\n mean = a.mean()\n print 'Divide a by 2:\\n', a/2.0\n \n #Add the two arrays\n print '\\nAdd a + b:\\n', a + b\n \n #Multiply a and b\n print '\\nMultiply a * b:\\n', a * b\n \n #Divide a and b\n print '\\nDivide a / b:\\n', a / b\n \nif __name__ == '__main__':\n test_run()",
"Original array a:\n[[ 1 2 3 4 5]\n [10 20 30 40 50]]\n\nOriginal array b:\n[[100 200 300 400 500]\n [ 1 2 3 4 5]]\n\nMultiply a by 2:\n[[ 2 4 6 8 10]\n [ 20 40 60 80 100]]\n\nDivide a by 2:\n[[ 0.5 1. 1.5 2. 2.5]\n [ 5. 10. 15. 20. 25. ]]\n\nAdd a + b:\n[[101 202 303 404 505]\n [ 11 22 33 44 55]]\n\nMultiply a * b:\n[[ 100 400 900 1600 2500]\n [ 10 40 90 160 250]]\n\nDivide a / b:\n[[ 0 0 0 0 0]\n [10 10 10 10 10]]\n"
]
],
[
[
"### Learning more NumPy\n\nResources from NumPy [User Guide](http://docs.scipy.org/doc/numpy/user/index.html) and [Reference](http://docs.scipy.org/doc/numpy/reference/index.html):\n\n- #### [The N-dimensional array](http://docs.scipy.org/doc/numpy/reference/arrays.ndarray.html)\n- #### [Data types](http://docs.scipy.org/doc/numpy/user/basics.types.html)\n- #### [Array creation](http://docs.scipy.org/doc/numpy/user/basics.creation.html) [[more]](http://docs.scipy.org/doc/numpy/reference/routines.array-creation.html)\n- #### [Indexing](http://docs.scipy.org/doc/numpy/user/basics.indexing.html) [[more]](http://docs.scipy.org/doc/numpy/reference/arrays.indexing.html)\n- #### [Broadcasting](http://docs.scipy.org/doc/numpy/user/basics.broadcasting.html)\n- #### [Random sampling](http://docs.scipy.org/doc/numpy/reference/routines.random.html)\n- #### [Mathematical functions](http://docs.scipy.org/doc/numpy/reference/routines.math.html)\n- #### [Linear algebra](http://docs.scipy.org/doc/numpy/reference/routines.linalg.html)\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
d085d986e3e323c8f6f74a756785298843c8c990 | 385,041 | ipynb | Jupyter Notebook | notebooks/basic_cap_figures.ipynb | Chandrima-04/gimmebio | cb3e66380006d5c5c00ff70bfb87317dd252c312 | [
"MIT"
] | 3 | 2020-01-21T23:49:55.000Z | 2020-07-29T17:02:30.000Z | notebooks/basic_cap_figures.ipynb | Chandrima-04/gimmebio | cb3e66380006d5c5c00ff70bfb87317dd252c312 | [
"MIT"
] | null | null | null | notebooks/basic_cap_figures.ipynb | Chandrima-04/gimmebio | cb3e66380006d5c5c00ff70bfb87317dd252c312 | [
"MIT"
] | 4 | 2020-01-21T16:48:17.000Z | 2020-03-13T15:34:52.000Z | 682.696809 | 72,767 | 0.941497 | [
[
[
"# Setup",
"_____no_output_____"
]
],
[
[
"%load_ext rpy2.ipython\n\nimport os\nfrom json import loads as jloads\nfrom glob import glob\nimport pandas as pd\nimport datetime",
"_____no_output_____"
],
[
"%%R\n\nlibrary(gplots)\nlibrary(ggplot2)\nlibrary(ggthemes)\nlibrary(reshape2)\nlibrary(gridExtra)\nlibrary(heatmap.plus)\n\nascols = function(facs, pallette){\n facs = facs[,1]\n ffacs = as.factor(as.character(facs))\n n = length(unique(facs))\n cols = pallette(n)[ffacs]\n}\n\ngreyscale = function(n){\n return(rev(gray.colors(n)))\n}",
"_____no_output_____"
],
[
" \ndef getsname(filename):\n return filename.split('/')[-1].split('.')[0]\n\n\ndef readJSON(jsonf):\n return jloads(open(jsonf).read())\n",
"_____no_output_____"
]
],
[
[
"# Beta Diversity",
"_____no_output_____"
]
],
[
[
"obj = readJSON('results/olympiome.beta_diversity_stats.json.json')\nspeciesRhoKraken = obj['species']['rho_proportionality']['kraken']\nspeciesRhoKrakenDF = pd.DataFrame(speciesRhoKraken)\nspeciesJSDKraken = obj['species']['jensen_shannon_distance']['kraken']\nspeciesJSDKrakenDF = pd.DataFrame(speciesJSDKraken)",
"_____no_output_____"
],
[
"%%R -i speciesRhoKrakenDF \n\n\nbeta.df = as.matrix(speciesRhoKrakenDF)\ndiag(beta.df) = NA\n\nheatmap.2(beta.df, trace='none', margins=c(8,8), ColSideColorsSize=3, KeyValueName=\"Rho Prop.\", labCol=F, cexRow=0.8, dendrogram=\"both\", density.info=\"histogram\", col=greyscale)",
"_____no_output_____"
],
[
"%%R -i speciesJSDKrakenDF \n\n\nbeta.df = as.matrix(speciesJSDKrakenDF)\ndiag(beta.df) = NA\n\nheatmap.2(beta.df, \n trace='none', \n margins=c(8,8), \n ColSideColorsSize=3, \n KeyValueName=\"Rho Prop.\", \n labCol=F, \n cexRow=0.8, \n dendrogram=\"both\", \n density.info=\"histogram\", \n col=greyscale)",
"_____no_output_____"
]
],
[
[
"# AMR",
"_____no_output_____"
]
],
[
[
"amrclassfs = glob('results/*.resistome_amrs.classus.tsv')\n\ndef parseF(fname):\n out = {}\n with open(fname) as f:\n f.readline()\n for line in f:\n tkns = line.strip().split('\\t')\n out[tkns[1]] = int(tkns[2])\n return out\n\namrclass = {getsname(amrclassf): parseF(amrclassf) for amrclassf in amrclassfs}\namrclass = pd.DataFrame(amrclass).fillna(0).transpose()\namrclass.shape",
"_____no_output_____"
],
[
"%%R -i amrclass \n\n\namr.df = t(as.matrix(amrclass))\n\n\nheatmap.2(amr.df, \n trace='none', \n margins=c(8,8), \n ColSideColorsSize=3, \n KeyValueName=\"Rho Prop.\", \n cexCol=0.8,\n cexRow=0.8, \n dendrogram=\"both\", \n density.info=\"histogram\", \n col=greyscale)",
"_____no_output_____"
]
],
[
[
"# Virulence Factors",
"_____no_output_____"
]
],
[
[
"virfs = glob('results/*.vfdb_quantify.table.tsv')\nvirs = {getsname(virf): pd.read_csv(virf).set_index('Unnamed: 0').transpose() for virf in virfs}\n\nvirpan = pd.Panel(virs).transpose(2,0,1)\n#vrpkm = virpan['RPKM'].fillna(0).apply(pd.to_numeric)\nvrpkmg = virpan['RPKMG'].fillna(0).apply(pd.to_numeric)\n\n",
"_____no_output_____"
],
[
"vrpkmghigh = vrpkmg.transpose().loc[vrpkmg.mean(axis=0) > 200]\nvrpkmghigh.shape",
"_____no_output_____"
],
[
"%%R -i vrpkmghigh \n\n\nvir.df = as.matrix(vrpkmghigh)\n\n\nheatmap.2(vir.df, \n trace='none', \n margins=c(8,8), \n ColSideColorsSize=3, \n KeyValueName=\"Rho Prop.\", \n cexCol=0.8,\n cexRow=0.8, \n dendrogram=\"both\", \n density.info=\"histogram\", \n col=greyscale)",
"_____no_output_____"
]
],
[
[
"# Virulence vs AMR",
"_____no_output_____"
]
],
[
[
"virlevels = vrpkmg.transpose().mean()\namrlevels = amrclass.transpose().mean().loc[virlevels.index]",
"_____no_output_____"
],
[
"%%R -i virlevels -i amrlevels\n\ndf = cbind(virlevels, amrlevels)\ncolnames(df) = c(\"virulence\", \"antimicrobial\")\ndf = as.data.frame(df)\nggplot(df, aes(virulence, antimicrobial)) + geom_point() + geom_rug() + theme_tufte(ticks=F) + \n xlab(\"Total Virulence\") + ylab(\"Total AMR\") + \n theme(axis.title.x = element_text(vjust=-0.5), axis.title.y = element_text(vjust=1))",
"_____no_output_____"
]
],
[
[
"# Alpha Diversity",
"_____no_output_____"
]
],
[
[
"adivfs = glob('results/*.alpha_diversity_stats.json.json')\nadivs = {getsname(adivf): readJSON(adivf) for adivf in adivfs}\n\nchaoSpecies = {}\nshanSpecies = {}\nrichSpecies = {}\nfor sname, adiv in adivs.items():\n chaoSpecies[sname] = adiv['kraken']['species']['chao1']\n shanSpecies[sname] = adiv['kraken']['species']['shannon_index']\n richSpecies[sname] = adiv['kraken']['species']['richness']\n \nchaoSpeciesDF = pd.DataFrame(chaoSpecies).fillna(0)\nshanSpeciesDF = pd.DataFrame(shanSpecies).fillna(0)\nrichSpeciesDF = pd.DataFrame(richSpecies).fillna(0)",
"_____no_output_____"
],
[
"shanSpeciesDF.loc['500000'].sort_values()",
"_____no_output_____"
]
],
[
[
"# HMP Comparison",
"_____no_output_____"
]
],
[
[
"hmpfs = glob('results/*.hmp_site_dists.metaphlan2.json')\n\ndef crunch(obj):\n out = {}\n for k, v in obj.items():\n out[k] = sum(v) / len(v)\n return out\n\nhmps = {getsname(hmpf): crunch(readJSON(hmpf)) for hmpf in hmpfs}\n\nhmps = pd.DataFrame(hmps).transpose()",
"_____no_output_____"
],
[
"%%R -i hmps \n\nhmp.df = melt(hmps)\n\nggplot(hmp.df, aes(x=variable, y=value)) +\n theme_tufte() + \n geom_boxplot() +\n ylab('Cosine Similarity to HMP Sites') +\n xlab('Body Site')",
"_____no_output_____"
]
],
[
[
"# Taxonomy",
"_____no_output_____"
]
],
[
[
"krakfs = glob('results/*.kraken_taxonomy_profiling.mpa.mpa.tsv')\n\ndef parseKrakF(krakf):\n out = {}\n with open(krakf) as kf:\n for line in kf:\n tkns = line.strip().split()\n taxa = tkns[0]\n if ('g__' in taxa) and ('s__' not in taxa):\n key = taxa.split('g__')[-1]\n out[key] = int(tkns[1])\n return out\n \n \ndef getTopN(vec, n):\n tups = vec.items()\n tups = sorted(tups, key=lambda x: -x[1])\n out = {k: v for k, v in tups[:n]}\n return out\n\nkrak10 = {getsname(krakf): getTopN(parseKrakF(krakf), 10) \n for krakf in krakfs}\nkrak10 = pd.DataFrame(krak10).fillna(0).transpose()",
"_____no_output_____"
],
[
"%%R -i krak10\n\nkrak.df = t(as.matrix(krak10))\nkrak.df = log(krak.df)\nkrak.df[!is.finite(krak.df)] = 0\n\nheatmap.2(krak.df, \n trace='none', \n margins=c(8,8), \n ColSideColorsSize=3, \n KeyValueName=\"Rho Prop.\", \n cexCol=0.8,\n cexRow=0.7, \n dendrogram=\"both\", \n density.info=\"histogram\", \n col=greyscale)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d085de38c8170efbce8470a8a4f0625642351d5e | 7,663 | ipynb | Jupyter Notebook | task13.ipynb | SkivHisink/OpenCV_Practice | d31d32584b6854c514f69c5977d64323c5830a11 | [
"MIT"
] | null | null | null | task13.ipynb | SkivHisink/OpenCV_Practice | d31d32584b6854c514f69c5977d64323c5830a11 | [
"MIT"
] | null | null | null | task13.ipynb | SkivHisink/OpenCV_Practice | d31d32584b6854c514f69c5977d64323c5830a11 | [
"MIT"
] | null | null | null | 34.209821 | 200 | 0.557876 | [
[
[
"import cv2 as cv\nimport numpy as np\nimport random",
"_____no_output_____"
],
[
"img = cv.imread(\"task13.jpg\")\ntemp = cv.imread(\"task13temp.jpg\")",
"_____no_output_____"
],
[
"noise = 100\nprev_noise = 0\nresult_of_ch = np.copy(img)\nresult_of_matching = np.copy(img)\nrotated_img = np.copy(img)\nbright_size = 10\ncontrast_size = 10\nangle_size = 0.0\nscale_size = 1.0\npoint_list=list()\npoint_list.append((0, 0))\npoint_list.append((img.shape[1], 0))\npoint_list.append((img.shape[1], img.shape[0]))\npoint_list.append((0, img.shape[0]))\npoint_for_change = 0",
"_____no_output_____"
],
[
"def update():\n global noise\n global prev_noise\n global result_of_ch\n global rotated_img\n global result_of_matching\n height, width = img.shape[:2]\n center = (width / 2, height / 2)\n if( prev_noise != noise):\n prev_noise = noise\n img_tmp = np.float64(img) \n noise_pic = np.copy(img_tmp)\n for i in range(img_tmp.shape[0]):\n for j in range(img_tmp.shape[1]):\n noise_val = random.randint(-noise, noise)\n noise_pic[i][j] = img_tmp[i][j] + noise_val\n result_of_ch = np.uint8(np.clip(noise_pic, 0, 255))\n rotated_img = result_of_ch.copy()\n change_brightness(bright_size)\n change_contrast(contrast_size)\n rotate_matrix = cv.getRotationMatrix2D(center, angle_size, scale_size)\n rotated_img = cv.warpAffine(rotated_img, rotate_matrix, (width, height))\n matrix = cv.getPerspectiveTransform( np.float32([[0, 0], [img.shape[1], 0], [img.shape[1], img.shape[0]], [0, img.shape[0]]]), \\\n np.float32(point_list))\n rotated_img = cv.warpPerspective(rotated_img, matrix, (img.shape[1], img.shape[0]), cv.INTER_CUBIC, borderMode = cv.BORDER_CONSTANT, borderValue = (0, 0, 0))\n \n imageGray = cv.cvtColor(rotated_img, cv.COLOR_BGR2GRAY)\n templateGray = cv.cvtColor(temp, cv.COLOR_BGR2GRAY)\n result = cv.matchTemplate(imageGray, templateGray,\n\t cv.TM_CCOEFF_NORMED)\n (minVal, maxVal, minLoc, maxLoc) = cv.minMaxLoc(result)\n (startX, startY) = maxLoc\n endX = startX + temp.shape[1]\n endY = startY + temp.shape[0]\n result_of_matching = cv.rectangle(cv.resize(rotated_img, (int(result_of_matching.shape[1] * 0.7), int(result_of_matching.shape[0] * 0.7))), (startX, startY), (endX, endY), (255, 0, 0), 3)\n #cv.imshow(\"image\", cv.resize(rotated_img, (int(rotated_img.shape[1] / 2), int(rotated_img.shape[0] / 2))))\n cv.imshow(\"result\", cv.resize(result_of_matching, (int(result_of_matching.shape[1] / 2), int(result_of_matching.shape[0] / 2))))\n\ndef change_noise(val):\n global noise\n noise = val\n update()\n\ndef change_brightness( brightness):\n global rotated_img\n if brightness != 0:\n rotated_img = cv.addWeighted(rotated_img, (255 - brightness) / 255, rotated_img, 0, brightness)\n\ndef change_contrast(contrast):\n global rotated_img\n if contrast != 0:\n f = 131 * (contrast + 127) / (127 * (131 - contrast))\n rotated_img = cv.addWeighted(rotated_img, f, rotated_img, 0, 127 * (1 - f))\n\n\ndef change_brightness_size(val):\n global bright_size\n bright_size = val\n update()\n\n\ndef change_contrast_size(val):\n global contrast_size\n contrast_size = val\n update()\n\n\ndef change_rotate_angle(val):\n global angle_size\n angle_size = val\n update()\n\n\ndef change_rotate_scale(val):\n global scale_size\n if val > 0:\n scale_size = val / 10.0\n update()\n\n\ndef change_point_for_changing(val):\n global point_for_change\n point_for_change = val\n\n\ndef change_point_x_val(val):\n global point_list\n _, old_y = point_list[point_for_change]\n point_list[point_for_change] = (val, old_y)\n update() \n\n\ndef change_point_y_val(val):\n global point_list\n old_x, _ = point_list[point_for_change]\n point_list[point_for_change] = (old_x, val)\n update() \n\n \ndef return_to_default(val):\n global point_list\n global scale_size\n global angle_size\n global contrast_size\n global bright_size\n scale_size = 1\n angle_size = 0\n point_list[0] = (0, 0)\n point_list[1] = (img.shape[1], 0)\n point_list[2] = (img.shape[1], img.shape[0])\n point_list[3] = (0, img.shape[0])\n update()",
"_____no_output_____"
],
[
"#cv.imshow(\"image\", cv.resize(result_of_ch, (int(result_of_ch.shape[1] / 2), int(result_of_ch.shape[0] / 2))))\ncv.imshow(\"result\", cv.resize(result_of_matching, (int(result_of_matching.shape[1] / 2), int(result_of_matching.shape[0] / 2))))\ncv.namedWindow('Control')\ncv.createTrackbar('noise', 'Control', 100, 255, change_noise)\ncv.createTrackbar('contrast', 'Control', 0, 255, change_contrast_size)\ncv.createTrackbar('brightness', 'Control', 0, 255, change_brightness_size)\ncv.createTrackbar('Angle', 'Control', 0, 360, change_rotate_angle)\ncv.createTrackbar('Scale', 'Control', 10, 360, change_rotate_scale)\ncv.namedWindow('Control2')\ncv.createTrackbar('Point', 'Control2', 0, 3, change_point_for_changing)\ncv.createTrackbar('X', 'Control2', 0, img.shape[1] - 1, change_point_x_val)\ncv.createTrackbar('Y', 'Control2', 0, img.shape[0] - 1, change_point_y_val)\ncv.createTrackbar('Return', 'Control2', 0, 1, return_to_default)\ncv.waitKey(0)\ncv.destroyAllWindows()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
d085e8eef8be78a17dc0e37742d8abf61fd6dc86 | 234,255 | ipynb | Jupyter Notebook | module2-Bag-of-Words/LS_DS_422_BOW_Assignment.ipynb | brit228/AB-Demo | 938b2fd9f1cc80f47a7f4acba4478ed563914c67 | [
"MIT"
] | null | null | null | module2-Bag-of-Words/LS_DS_422_BOW_Assignment.ipynb | brit228/AB-Demo | 938b2fd9f1cc80f47a7f4acba4478ed563914c67 | [
"MIT"
] | null | null | null | module2-Bag-of-Words/LS_DS_422_BOW_Assignment.ipynb | brit228/AB-Demo | 938b2fd9f1cc80f47a7f4acba4478ed563914c67 | [
"MIT"
] | null | null | null | 100.885013 | 51,729 | 0.585089 | [
[
[
"<a href=\"https://colab.research.google.com/github/brit228/AB-Demo/blob/master/module2-Bag-of-Words/LS_DS_422_BOW_Assignment.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"import re\nimport string\n\n!pip install -U nltk\n\nimport nltk\nnltk.download('punkt')\nnltk.download('stopwords')\nnltk.download('wordnet')\nfrom nltk.tokenize import sent_tokenize # Sentence Tokenizer\nfrom nltk.tokenize import word_tokenize # Word Tokenizer\nfrom nltk.corpus import stopwords\nfrom nltk.stem.porter import PorterStemmer\nfrom nltk.stem.wordnet import WordNetLemmatizer\nfrom nltk.probability import FreqDist\n\nfrom sklearn.feature_extraction.text import CountVectorizer\nfrom sklearn.feature_extraction.text import TfidfVectorizer\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport numpy as np",
"Requirement already up-to-date: nltk in /usr/local/lib/python3.6/dist-packages (3.4)\nRequirement already satisfied, skipping upgrade: singledispatch in /usr/local/lib/python3.6/dist-packages (from nltk) (3.4.0.3)\nRequirement already satisfied, skipping upgrade: six in /usr/local/lib/python3.6/dist-packages (from nltk) (1.11.0)\n"
]
],
[
[
"# 1) (optional) Scrape 100 Job Listings that contain the title \"Data Scientist\" from indeed.com\n\nAt a minimum your final dataframe of job listings should contain\n- Job Title\n- Job Description",
"_____no_output_____"
]
],
[
[
"import requests\nfrom bs4 import BeautifulSoup\n\n\naddition = \"\"\ni = 0\ndata = []\nwhile True:\n r = requests.get(\"https://www.indeed.com/jobs?q=data%20scientist&l=Boston,%20MA\"+addition)\n soup = BeautifulSoup(r.text, 'html.parser')\n for card in soup.find_all('div', class_=\"jobsearch-SerpJobCard\", attrs={\"data-tn-component\": \"organicJob\"}):\n try:\n d = {}\n d[\"Job Title\"] = card.h2.a.text\n d[\"Company\"] = card.find(\"span\", class_=\"company\").text.strip()\n d[\"Location\"] = card.find(\"span\", class_=\"location\").text.strip()\n r2 = requests.get(\"https://www.indeed.com\"+card.a[\"href\"])\n soup2 = BeautifulSoup(r2.text, 'html.parser')\n d[\"Job Description\"] = \"\\n\".join([a.text for a in soup2.find(\"div\", class_=\"jobsearch-JobComponent-description icl-u-xs-mt--md\").contents])\n data.append(d)\n except:\n pass\n i += 10\n print(i)\n addition = \"&start={}\".format(i)\n if len(data) > 100:\n break\ndf = pd.DataFrame(data)\ndf",
"Who we are\nMassachusetts Cannabis Research Laboratories (MCR Labs) is a rapidly growing, independent, and accredited cannabis testing and analytics laboratory.\nWe are a team of researchers, analytical chemists, pharmaceutical scientists, technologists, and cannabis enthusiasts working to advance the medical and recreational cannabis industries by helping providers ensure the safety and effectiveness of their products. We are committed to proving the best possible services to our clients, building relationships and supporting our community.\nThis is an exceptional career opportunity to join the team of a fast-paced company in a high-growth industry!\nJob Summary\nThis position is responsible for the production of reports, extracting and analyzing output from internal software tool for client testing reports, and performing internal trend analysis.\nPrimary Job Responsibilities: \nThe position will be responsible for creating and running various reports from the internal software. Review and ensure all information on the reports is accurate and correct. Identify opportunities for process improvements in reporting and development, while providing value through insightful analytics and problem-solving. The reporting data analyst will be responsible for identifying data trends and performing data analysis. Timely and effectively communicate with coworkers and other internal departments any changes affecting client’s product. Present findings and make recommendations. Meet client deadlines for ad hoc reports and other requested information. In this dynamic role, the individual will coordinate reporting and act as the liaison with the external customers and internal team. Understand the purpose and content of requested reports, utilizing systems to create routine reports to meet clients’ needs, and audit and analyze data to verify quality and data integrity. Identify and research data anomalies.\nPrimary Goal\nThe primary goal of the Reporting Data Analyst is to prepare accurate client analytical testing reports. Review all reports for accuracy before sending to the client. Analyze and develop recommendations on cannabis laboratory reporting processes.\nAbout You\nPreferred Skills and Qualifications\nBS/BA degree in chemistry, computer science, physics, mathematics or a related fieldExperience working in a labKnowledge of knowledge of Chemistry, specifically ChromatographyKnowledge of Mass SpectrometryAbility to perform repetitive tasks with a high level of successVery high level of organization and attention to detail with a sense of self-directionAbility to read, analyze, and interpret technical dataWillingness to work and learnAbility to adapt and learn quicklyDesire to learn about product and industryHave advanced knowledge of Excel, macros, and functions to enhance current reportingStrong computer skillsAbility to work independently and communicate effectively in a team environmentEffective time management skills – this is a fast-paced environment and the successful candidate will be able to multi-task and change focus very quickly while maintaining their ability to be effective\nDo you have what it takes to be a part of an industry leading cannabis testing company?\nCome join the Green Rush!\nJob Type: Full-time Job Location: Framingham, MA 01701\nRequired education: Bachelor's\nAdditional Requirements: Ability to work on weekends, must be fluent in English and authorized to work in US.\nMCR Labs is an equal opportunity employer and we value diversity at our company. We do not discriminate on the basis of race, religion, color, national origin, gender, sexual orientation, age, marital status, genetic information, veteran status, or disability status.\nAll qualified applicants must be able to satisfy the Department of Homeland Security Form I-9 requirement, with the documents necessary to verify identity & employment authorization.\nJob Type: Full-time\nExperience:\nData Analysis: 1 year (Required)Excel: 2 years (Required)Chromatography: 1 year (Required)\nEducation:\nBachelor's (Required)\nBenefits offered:\nPaid time offHealth insuranceDental insurance\n$68,809 a year\nTO APPLY: \nInterested candidates must fully complete the online MSO application and attach a cover letter and resume through the online application process. The online application process can be found under the ‘Employment’ tab at www.middlesexsheriff.com.\nThe Middlesex Sheriff’s Office (MSO) is seeking a qualified professional for the position of Data Integration Specialist / Research Analyst to work in the Administrative Offices of the MSO located in Medford, MA. The Data Integration Specialist / Research Analyst will work directly with individual departments and administration to coordinate, streamline, and visualize an array of data being gathered at the MSO. The Data Integration Specialist / Research Analyst will also be responsible for gathering, linking, and cleaning data that will be used for reporting requirements. The Data Integration Specialist / Research Analyst will also work with the Chief Financial Officer and Director of Fiscal Operations on grant funded projects, reporting requirements, and other budget-related initiatives. This is an exempt, full time, at-will employment position that reports to the Director of Fiscal Operations and Chief Financial Officer.\nAdditional responsibilities include, but are not limited to, the following: \nCollecting, analyzing, interpreting and presenting quantitative and qualitative information;\nDesigning and managing experiments, surveys and data collection;\nDesigning data warehouse/reporting schemas that advance the MSO’s reporting needs;\nCollaborating with full-time and contractual staff to manage hardware and software systems (SQL, Excel);\nProviding analysis of data obtained in context, looking for patterns and forecasting trends to help make recommendations for funding and policy changes;\nWorking with MSO staff to ensure that complex statistical concepts are explained efficiently and accurately;\nPreparing materials for submission to granting agencies and foundations.\nWorking with other internal departments and outside public safety agencies to compile data and provide data visualizations for presentations; do one-off and repeatable Extract Transfer Load (ETL) work to support the MSO;\nBuilding and managing ETL processes for internal departments;\nWorking closely with data analysts to build data integrations that quickly and effectively solve reporting needs;\nOther work duties as assigned by Director of Fiscal Operations and Chief Financial Officer.\nThe individual selected for the position of Data Integration Specialist / Research Analyst must have the ability to exercise good judgment and focus on detail as required by the job. The individual selected shall also be cognizant of the confidential and sensitive nature of working in a law enforcement agency and must comply with all institutional rules regarding safety and security.\nQualifications: \nBachelor's degree in Accounting/Finance, Business/Public Administration or a closely related field preferred;\nProven ability to gather, synthesize, and present current research and trends in criminal justice, healthcare, and social science.\nMandatory proficiencies should include: Microsoft Word; Excel; Access; SQL query and data transformation skills, including development of Dashboards, Crystal Reports, and MS Access-based Reports. Microsoft SQL Server expertise a plus.\nStrong knowledge of industry research methodologies and techniques.\nAbility to work independently, as well as, and with other internal departments in a fast paced environment and execute close attention to detail;\nStrong written, communication, organizational, analytical, problem-solving and time-management skills;\nAbility to complete multiple projects in a timely and accurate manner while maintaining comprehensive and cohesive records.\nIn compliance with federal law, all persons hired will be required to verify identity and eligibility to work in the United States and to complete the required employment eligibility verification form upon hire. Must be a Massachusetts resident, have an active valid driver’s license and the right to legally operate a motor vehicle in Massachusetts and pass a criminal background check.\nSalary: Starting annual base salary from $68,809 plus additional benefits.\nSubmissions must be received by Friday, March 22, 2019 at 4:00p.m.\nThe Middlesex Sheriff’s Office is an equal opportunity employer\nJob Type: Full-time\nEducation:\nBachelor's (Preferred)\nLicense:\nactive valid driver’s (Required)\nDue to BitSight’s rapid growth, and our need to better understand and glean insights from our marketing data, we are recruiting a data scientist to join the marketing team. This is a new role in the marketing department that you will help shape. You will also have the opportunity to work with BitSight’s Security Data Science team to stay on the bleeding edge of data science/AI with a bi-weekly internal reading group, technology-specific workshops, and conference attendance.\n\n\nResponsibilities\n\nLook for signals in our large data sets and build models that leverage these signals to glean insights from our marketing and customer data.\n\nDetermine the buyer’s journey from first touch on the website all the way through retention and upsell/cross-sell. Make recommendations on what changes to make to content and programs across the funnel from awareness to consideration to selection to upsell to drive revenue growth.\n\nLead machine learning projects using varied advanced analytical techniques for estimating current and future customer engagement, prospecting new customers, cross-selling to existing customers (response models), and identifying customers likely to leave.\n\nDevelop and test multiple hypotheses using results from analyses to generate and answer new questions leading to more in-depth understanding of customer behavior being studied.\n\nCollaborate with internal business partners across departments to integrate analytical tools and solutions into demand generation campaigns/initiatives.\n\nIdentify potential issues that arise during the course of research projects and escalate and communicate these to management and internal clients for awareness/ action/ resolution.\n\nMake presentations of status and results of analysis to non-technical audience of various levels.\n\nProjects are typically short, fast moving, and highly varied. You will need to manage multiple research projects under tight deadlines, as well as ensure research projects are completed on time and meet internal client expectations.\n\n\nDesirable Skills\n\nStrong coding skills: You are able to read in a data set, process it, and plot data. You can build modules that encapsulate functionality you have developed. You have some experience with Big Data tools, e.g., Spark, PrestoDB, Hive. We are a Python house and you need to know Python. Also important is the ability to quickly learn to use tools and packages new to you.\n\nStrong statistical analysis skills: You are able to take a data set, estimate statistical parameters from it, and compare the results with existing data sets. You are comfortable with and knowledgeable about statistical concepts like p-values, hypothesis testing, and non-normal distributions. Again, of utmost importance is being comfortable learning new statistical methods.\n\nSolid machine learning and statistical modeling skills: You are able to frame problems as estimating a target variable, build a dataset, define an evaluation metric, build a non-trivial baseline, and finally use appropriate techniques to beat the baseline when possible.\n\nStrong team working skills: You’ll be able to work well with other departments (e.g. data science, product management, customer success), balance requirements, and work independently.\n\nAbility to communicate effectively: Good results are a good starting point but we also need to communicate these results verbally, in writing, and visually.\n\n\nJob Benefits\n\nBitSight prides itself in building exceptional career opportunities and offering outstanding benefits to our team. In that regard, BitSight is not your average company. We have the enthusiasm of a start-up, a culture driven from industry veterans committed to long-term growth, and the benefits package of a mature industry leader. BitSight is a great place to work.\n\nThe purpose of this role is to partner with the respective Franchise analytics teams to ensure high quality analytic insights, recommendations, and data usage for the supported therapeutic area. Key responsibilities include: Analytics Execute analytics to support Multi-Channel, Patient, Payer/Provider, Life Cycle, Field Force, and Forecasting Determine the ideal methodology to apply for each analysis or process based on data availability and limitations Develop, validate and deploy predictive and diagnostic solutions using reusable code and computing paradigms Derive insights and recommendations from research and analyses that address both stated and unstated business questions Use statistical approaches, such as ANOVA, etc., leveraging statistical analysis toolsets, such as R, SPSS and SAS Process and analyze large health-related datasets ranging from small to Big Data and integrate and analyze Structured, Semi-structured, and Unstructured data Use tools for accessing, synthesizing, analyzing and reporting data Data Work with database technologies, SQL, NLP, data engineering, Hadoop Zoo, Kibana, visualization tools, graph analysis Collaboration Act as SME resource for broader Advanced Analytics community within supported therapeutic areas Foster common data-driven viewpoints between stakeholders with divergent views and objectives Connect technical and data skills to business needs to identify improvements with the project lifecycle Work with peers to ensure that resulting code is compliant and supports standardization whenever possible Proactively deliver analyses and reports based on timing for key planning processes Project Management Meet Franchise Leadership expectations by delivering work on time and within scope Fulfill assigned role within a project team delivering to expectations Ensure Takeda ethics and compliance are continuously met Responsibilities Job Function and Description 80% Deliver recommendations built on models and analytics 20% Develop data environment Education and Experience Requirements BA/BS degree in business, life sciences, or related technical discipline Master’s in statistics, mathematics, computer science, applied economics, computational biology, computational informatics, or medical informatics preferred 5+ years’ of relevant analytical or healthcare experience preferred Preferred Programming skills: Uses Statistical Modelling: e.g. SPSS, SAS, R Machine Learning Tools: e.g. Spark Visualization: e.g. QlikView/QlikSense, Tableau Data Environment: e.g. Datameer (Hadoop) Experience with large scale database applications (e.g., Oracle, Hadoop, Teradata) Experience detailing Big Data environment requirements Familiar with advanced data science methods Demonstrated proficiency with statistical methods, e.g. ANCOVA, two-tailed p-test, descriptive statistics, etc. Key Skills, Abilities, and Competencies Technical Skills Practiced skills in creating appropriate logic that answers stated and unstated business questions In-depth analytical and critical thinking skills to resolve issues efficiently and effectively Experienced in selecting and applying the appropriate methodology based on business need and data Demonstrable comfort using and applying structured statistical modeling tools and additional analytical modules or add-ons Expertise in writing reusable code to customize statistical models Business Acumen Understanding of goals and needs of supported functions Able to identify stakeholder needs through voice of customer and relevant data collection Experience in generating insights and recommendations from research and analyses that address both stated and unstated business questions Interpersonal Skills Experienced in presenting insights and conclusions from complex information in an easy to understand way Able to maintain an impartial point of view Builds marketing and sales leadership’s confidence through active listening, asking questions and accurately paraphrasing their needs and expectations Proactively engages with stakeholders to build relationships. Recognizes the need to modify communication styles to fit diverse audiences with various levels of expertise Fulfills assigned role within a project team delivering to expectations Complexity and Problem Solving Technical, data and analysis related decisions are within the incumbent’s authority. For business and stakeholder related decision he/she consults with the Advanced Analytics Lead Internal and External Contacts Internal Business Partner Contacts Advanced Analytics Community in Franchises U.S. Commercial Operations functions (Sales Operations, Marketing Operations, Learning and Development) IT, Regulatory, Medical, Compliance External Vendor Contracts Service Providers / Consultants Technology Solution Implementation Vendors Software Vendors Other Job Requirements 10% domestic travel may be required.\nNotice to Employment / Recruitment Agents:\nEmployment / Recruitment agents may only submit candidates for vacancies only if they have written authorization to do so from Shire, a wholly-owned subsidiary of Takeda’s Talent Acquisition department. Any agency candidate submission may only be submitted to positions opened to the agency through the specific Agency Portal. Shire, a wholly-owned subsidiary of Takeda will only pay a fee for candidates submitted or presented where there is a fully executed contract in place between the Employment / Recruitment agents and Shire, a wholly-owned subsidiary of Takeda and only if the candidate is submitted via the Agency Portal. Candidates submitted or presented by Employment / Recruitment Agents without a fully executed contract or submitted through this site shall not be deemed to form part of any Engagement for which the Agency may claim remuneration.\nEqual Employment Opportunity\nShire, a wholly-owned subsidiary of Takeda, is an Equal Opportunity Employer committed to a diverse workforce. Shire, a wholly-owned subsidiary of Takeda, will not discriminate against any worker or job applicant on the basis of race, color, religion, gender, national origin, ancestry, age, sexual orientation, marital or civil partnership status, pregnancy, gender reassignment, non-job related mental or physical disability, genetic information, veteran status, military service, application for military service, or membership in any other category protected under law.\nEEO is the Law - https://www.dol.gov/ofccp/regs/compliance/posters/pdf/eeopost.pdf\nEEO is the Law – Supplement - https://www.dol.gov/ofccp/regs/compliance/posters/pdf/OFCCP_EEO_Supplement_Final_JRF_QA_508c.pdf\nPay Transparency Policy - https://www.dol.gov/ofccp/pdf/pay-transp_formattedESQA508c.pdf\nReasonable Accommodations\nShire, a wholly-owned subsidiary of Takeda, is committed to working with and providing reasonable accommodation to individuals with disabilities. If, because of a medical condition or disability, you need a reasonable accommodation for any part of the application process, or in order to perform the essential functions of a position, please call 484-595-8400 and let us know the nature of your request and your contact information.\nDescription:\nAre you passionate about applying data science to real business and customer needs? Would you like to use your data science skills to help our customers do more, feel more, and be more? At Bose, all of our energy is aimed at bringing products into the world that people truly love, and we don’t stop until the details are just right. Data science, machine learning, and analytics have become a crucial part of this mission . These capabilities fuel the creation of new and innovative products in consumer electronics and wellness, help us to bring the right products to the right customers, and allow us to astonish customers with carefully crafted and personalized experiences.\nWe are looking for a bright, enthusiastic data scientist for our new and growing Global Consumer Sales Data Science team out of the Boston Landing location. The mission of this team is to develop world-class data science, machine learning, and related technologies to extract insights from data for driving business and customer value. We provide data science expertise and support across the Sales, Marketing, Retail, and Customer Service organizations. The desired outcomes will include improved customer experiences, personalized recommendations, and digital optimization.\n\nResponsibilities:\nDevelop and evaluate predictive and prescriptive models for marketing, sales, e-commerce, and customer service applications such as customer lifetime value models, product recommenders, customer segmentations, uplift models, and propensity models.\nExplore large datasets related to customer and user behavior using modeling, analysis, and visualization techniques.\nApply frequentist and Bayesian statistical inference tools to experimental and observational data.\nCollaborate with data science, data engineering, and data governance teams throughout the data science process. Engage with Global Consumer Sales colleagues to understand business problems and define data science solutions.\nCommunicate results, analyses, and methodologies to technical and business stakeholders.\nTravel to Framingham, MA location at least once per week (shuttle from Boston Landing is available) .\nEducation:\nBS or MS (preferred) in Data Science, Computer Science, Business Analytics, Statistics, or a related field\nCompleted coursework related to Statistics, Computer Science, Machine Learning, and Data Science\nCompleted coursework related to Business/Customer Analytics, Marketing, Sales, and/or Management\nSkills:\n2+ years of experience applying data science, machine learning, and analytics techniques to business problems, preferably related to sales and marketing\nStrong programming background with experience in Python (preferred) or R\nStrong understanding of unsupervised and supervised machine learning algorithms\nExperience designing experiments and analyzing experimental data using statistical modeling\nStrong analytical, communication, collaboration, and problem - solving skills\nExperience cleaning and wrangling data using tools such as SQL and Python\n(Preferred) Experience working with big data tools and frameworks such as Hadoop and Apache Spark\nBose is an equal opportunity employer that is committed to inclusion and diversity. We evaluate qualified applicants without regard to race, color, religion, sex, sexual orientation, gender identity, genetic information, national origin, age, disability, veteran status, or any other legally protected characteristics. For additional information, please review: (1) the EEO is the Law Poster (http://www.dol.gov/ofccp/regs/compliance/posters/pdf/OFCCP_EEO_Supplement_Final_JRF_QA_508c.pdf); and (2) its Supplements (http://www.dol.gov/ofccp/regs/compliance/posters/ofccpost.htm). Please note, the company's pay transparency is available at http://www.dol.gov/ofccp/pdf/EO13665_PrescribedNondiscriminationPostingLanguage_JRFQA508c.pdf. Bose is committed to working with and providing reasonable accommodations to individuals with disabilities. If you need a reasonable accommodation because of a disability for any part of the application or employment process, please send an e-mail to [email protected] and let us know the nature of your request and your contact information.\n150\nWe are seeking a highly motivated Data Scientist for computational analysis of complex data across our R&D portfolio to advance Preclinical and Development-stage programs. The successful candidate will serve as a subject matter expert who will provide cross-functional guidance and support internally and externally. He/she will thrive in a fast-paced, highly-collaborative environment to advance program goals with deep expertise in developing models using multi-dimensional data sources.\n\nJob Responsibilities:\nProvide scientific input and leadership to enable the team to analyze complex data (e.g. genomics, transcriptomics and proteomics) from patient samples. Lead efforts to develop predictive models using statistical and computational biology approaches for biomarker development and patient stratification strategies.\nProvide immediate support in multiple R&D-stage programs and to cross-functional clinical development teams through integrated analysis of clinical and biomarker data generated from multiple platforms and formats.\nPro-actively define statistical analysis plans to generate actionable results for meeting program and business objectives.\nCommunication and visualization of results to scientific and non-scientific audiences.\nProactively partner with core R&D functional leads to advance Clinical, Preclinical and Discovery program objectives and serve as internal expert in computational biology and biostatistical modeling.\nIdentify key scientific questions to advance our scientific understanding across the portfolio.\nAdapt latest methods and tools for analyzing large omics datasets (genomics/proteomics).\nEstablish internal best practices for complex data visualization, integration, and accessibility.\n\nEducation and Experience:\nPh.D. in statistics, mathematics, bioinformatics, computational biology, genomics, computer science, or a related field with 5+ years of experience in complex, quantitative data analysis in a biotech/biopharm environment.\nExperience working with big-data generated by diverse platforms (e.g. RNA-Seq, Flow Cytometry, multiplexed proteomics) and accessing and mining external datasets.\nExperience working with clinical study data and compiling reports in a GxP-environment.\nAbility to present and visualize data for communicating with scientific and non-scientific colleagues.\nProficiency in common programming languages such as Python, R, Matlab, Java, Shell and Linux environments.\nAbility to work independently and collaboratively in highly dynamic, fast-paced projects within a highly-matrixed, cross-functional and collaborative environment.\nData Science Team\nNift brings new customers through the doors of neighborhood businesses better than anything else out there. Join the team that’s giving millions of people gifts they love while bringing in the foot traffic that makes neighborhood businesses thrive.\nData Science is the heart and core of our company. Algorithms and models are the foundation upon which our product is built, with data driving our key decisions, testing and growth. Our Chief Scientist, David C Parkes, is the former Area Dean of Computer Science at Harvard, the founder of Harvard’s EconCS Group, and the Co-Director of Harvard's Data Science Initiative. Our data doubles every two months with even more sources waiting to be added. Our product represents a completely new kind of marketplace and the science around it has yet to be defined. We’re looking for a Data Analyst to join our core team and drive growth and revenue.\nThis position is based in Boston, MA.\nExamples of projects we currently need help with:\n\nAnalyze real-time data market economic data\nEvaluate experimental results - On simulated data, real-time data and concurrent AB tests.\nCollect data, build analysis and present it to monitor and understand the company revenue.\nEstimate the demand in multiple local markets (in different sizes, locations and development stages).\nSegment customers and businesses; offer insights and identify revenue growth opportunities within existing core verticals and new ones.\nMonitor marketplace metrics and system performance and suggest corrective actions when needed.\n\n\nWe have an outstanding core team with deep understanding of algorithmic economics and data science. Our work is highly sought-after and is critical to the success of our business. If you have a proven track-record, want to make an impact and you get excited about the prospects of being part of something really special, we should talk.\nTraits we value:\n\nSolid understanding of statistics, economics, and math.\nA keen eye for detail and thoughtful investigation of data.\nA steadfast focus on creating impactful change and ability to prioritize between many tasks to maximize the improvement of the business.\n2+ years of commercial experience is a plus.\nA minimum of a Bachelor’s degree, a Master’s degree is preferred.\nCollaborative team player who values the contribution of others.\n\n\n\nWe believe it's time technology starts working for Main Street's small businesses. Launched in Boston in the Summer of 2016, our start-up is helping millions of people discover great local businesses.\n\nKlaviyo is looking for data scientists to analyze large data sets (we’re collecting billions of individual actions every month), build models and ship products that enable businesses to grow faster and communicate with their customers. Our background as a team is building these models for the Fortune 50 and we want to democratize and open up that technology to everyone.\n\nThe ideal candidate has a background in data science, statistics and machine learning and has done work ranging from exploratory analysis to training and deploying models. We use a wide variety of data mining and machine learning algorithms. The right candidate will have both a solid fundamental understanding and deep practical experience with at least a few modeling and machine learning techniques.\n\nOur goal is always to match the right assumptions and models to the right problem - they don’t necessarily have to be complex. You should have experience building models that are used by people to make better decisions. We’re focused on shipping early and often. We prefer iterative solutions that are incrementally better to the perfect solution. You should also be able to measure and know what impact your models had on the decisions people made - e.g. did they outperform the previous best model or a human decision maker?\n\nOur data science team is still in its early days and you’ll have a big impact on our direction and how we operate. You’ll be central to upfront research and shipping products that help our customers learn and grow from their data.\nYou:\nHave a strong fundamental understanding and deep experience with at least a few machine learning algorithms (e.g. regressions, decision trees, k-means clustering, neural networks).\nUnderstand Bayesian modeling techniques.\nAre capable of analyzing data and making rigorous statements about what can or cannot be concluded.\nHave experience designing and implementing model performance/validation assessments.\nHave a background in statistics and understand different distributions and the conditions under which they’re valid.\nKnow how code and have used data science tools and packages.\nHave demonstrated a measurable impact based on the models you’ve created. It’s not always easy getting a model correct and we love talking about places we got stuck and working as a team to think through ideas that could unblock us.\nHave a desire to ship features powered by data science (in other words, you’re excited by both upfront research and actually getting models into production at cloud scale).\nYou Have:\nBachelor’s or advanced degree in statistics, applied mathematics, computer science or other relevant quantitative discipline, or equivalent industry experience.\nHave worked in a data science role for 5+ years professionally or academically and can talk about your projects and the techniques you used.\nAbout Us\n\nKlaviyo is a team of people who are crazy motivated by growth.\n\nIt’s what we help our customers do: grow their businesses by making it possible and easy for them to use their data to power better marketing.\n\nIt’s how we behave as individuals: we’re all deeply passionate about learning.\n\nIt’s how we manage our business: we have thousands of paying customers, we’re profitable, and we’re growing insanely fast.\n\nAnd it’s what our culture is all about. Working at Klaviyo means you’ll work on things you never imagined you would; you’ll grow in ways you didn’t consider possible; and you’ll do the best work of your career with people who are just as motivated and talented as you are.\n\nIf this sounds like your ideal place to work, drop us a note!\n\nUnder supervision, and within established departmental and hospital policies and procedures, is responsible for performing a variety of general and technical duties to support the research activities within the Translational Neuroimmunology Research Center (TNRC) in the Ann Romney Center for Neurologic Diseases (ARCND). The data analyst is to provide expertise to acquire, manage, manipulate, analyze data, and report the results. Data capture and reporting in collaboration with others will be a daily activity.\n\n\nQualifications\n\nPRINCIPAL DUTIES:\n\n\n a. To assist in data analysis related to the research of Multiple Sclerosis and\n\n other Autoimmune diseases\n\n b. Assist with data processing, archiving, storage and computer analysis\n\n c. Create analysis datasets from an Oracle database\n\n d. Oversee data cleaning and manipulation\n\n e. Perform statistical analysis utilizing SAS, STATA, or R\n\n f. Identify, analyze and interpret trends or patterns in complex data sets\n\n g. Perform data entry when needed or required\n\n h. Develop graphs, reports and presentations of project results\n\n i. In collaboration with others; develop and maintain databases and data\n\n systems necessary for projects and department functions.\n\n\nPREFERRED QUALIFICATIONS:\n\nExperience with Red Cap\nExperience with SAS, STATA or R\nProgramming Skills to execute queries in Oracle database and knowledge of SQL and XML\nAt least 2 years in a research setting looking at multiple sclerosis or neurological data\nMPH in epidemiology, biostatistics or a related field\nSKILLS/ABILITIES/COMPETENCIES REQUIRED:\n\nProficiency in Microsoft Office suite, including Excel and Access\nBachelor’s or master’s degree, preferable in Information Management, Healthcare Information, Computing, Mathematics, Statistics or related fields\nProficiency with statistics in order to communicate easily with other statisticians\nWORKING CONDITIONS:\n\nOffice space environment and occasional clinical center exposure.\nEEO Statement\n\nBrigham and Women’s Hospital is an Equal Opportunity Employer. All qualified applicants will receive consideration for employment without regard to race, color, religion, creed, sex, sexual orientation, gender identity, national origin, ancestry, age, veteran status, disability unrelated to job requirements, genetic information, military service, or other protected status.\n\nPrimary Location: MA-Boston-BWH Longwood Medical Area\nWork Locations: BWH Longwood Medical Area 75 Francis Street Boston 02115\nJob: Business and Systems Analyst\nOrganization: Brigham & Women's Hospital(BWH)\nSchedule: Full-time\nStandard Hours: 40\nShift: Day Job\nEmployee Status: Regular\nRecruiting Department: BWH Neurology\nJob Posting: Mar 18, 2019\n\n160\n"
]
],
[
[
"# 2) Use NLTK to tokenize / clean the listings ",
"_____no_output_____"
]
],
[
[
"df2 = df.copy()\nstop_words = stopwords.words('english')\nlemmatizer = WordNetLemmatizer()\ndf2[\"Job Description\"] = df2[\"Job Description\"].apply(lambda v: [lemmatizer.lemmatize(w) for w in word_tokenize(v) if w.isalpha() and w not in stop_words])\nvector_list = sorted(list(set([inner for outer in df2[\"Job Description\"].values for inner in outer])))\nprint(vector_list)",
"['A', 'AART', 'AB', 'ACCESS', 'ACO', 'AD', 'AI', 'AIDS', 'ANALYST', 'AND', 'API', 'AWS', 'Abdul', 'Abilities', 'Ability', 'About', 'Absolutely', 'Acceleration', 'Access', 'Accommodation', 'Accommodations', 'Accountability', 'Accountable', 'Achieve', 'Acquisition', 'Act', 'Acting', 'Action', 'Actively', 'Activities', 'Actual', 'Actuarial', 'Acute', 'Additional', 'Additionally', 'Adhere', 'Administration', 'Advance', 'Advanced', 'Advice', 'Advisers', 'Advisory', 'Aetna', 'Affirmative', 'Affordability', 'Africa', 'African', 'After', 'Afternoon', 'Again', 'Agency', 'Aggregating', 'Agile', 'Agilent', 'Agility', 'Agios', 'Alaska', 'Algebra', 'Algorithm', 'Algorithms', 'Alion', 'All', 'Allocation', 'Alongside', 'Also', 'Alternatively', 'Amazon', 'America', 'American', 'Americans', 'An', 'Analyses', 'Analysis', 'Analyst', 'Analysts', 'Analytics', 'AnalyticsTM', 'Analyze', 'And', 'Apache', 'Applicants', 'Application', 'ApplicationClaims', 'Applications', 'Applied', 'Apply', 'Are', 'Area', 'Areas', 'Arthritis', 'Artificial', 'As', 'Asia', 'AspenTech', 'Assess', 'Asset', 'Assist', 'Assistance', 'Assistants', 'Assists', 'Associate', 'Association', 'AstraZeneca', 'At', 'Attend', 'Attractive', 'Attributes', 'August', 'Australia', 'Authenticity', 'Auto', 'Autoimmunity', 'Automate', 'Automation', 'Avenue', 'Award', 'Azar', 'Azure', 'B', 'BI', 'BLAS', 'BMC', 'BOP', 'BPT', 'BS', 'BWH', 'Bachelor', 'Bachelors', 'Back', 'Background', 'Banking', 'Based', 'Bash', 'Basic', 'Bayesian', 'Be', 'Beacon', 'Beam', 'Bedford', 'Begins', 'Benefits', 'BenefitsExcellent', 'Berkley', 'Big', 'Billing', 'Bioanalyzer', 'Biochemistry', 'Bioengineering', 'Bioinformatics', 'Biology', 'Bioscience', 'Biostatistics', 'BitSight', 'Blog', 'Bloomberg', 'Boards', 'Bose', 'Boston', 'BostonBMC', 'Botswana', 'Branson', 'Brave', 'Brazil', 'Breakfast', 'Brigham', 'Bring', 'Bringing', 'Broad', 'Broadway', 'Build', 'Building', 'Bulgaria', 'Bureau', 'Burning', 'Business', 'But', 'By', 'C', 'CCDS', 'CEO', 'CEPAC', 'CERTIFICATES', 'CFA', 'CFO', 'CFR', 'CHC', 'CHI', 'CIO', 'CJ', 'CMake', 'COMPETENCIES', 'CONDITIONS', 'CRM', 'CRO', 'CSCW', 'CT', 'CTO', 'CUDA', 'CV', 'CVB', 'CVS', 'Cafeteria', 'Calculus', 'Call', 'Cambridge', 'Campaign', 'Campus', 'Can', 'Cancer', 'Candidate', 'Capable', 'Capacity', 'Capital', 'Cardiovascular', 'Care', 'Carlo', 'Carry', 'Ccain', 'Census', 'Center', 'Central', 'Chain', 'Chairman', 'Chan', 'Chance', 'Characteristics', 'Chemistry', 'Chief', 'China', 'Christensen', 'Circadian', 'City', 'Claim', 'Claims', 'Classification', 'Clayton', 'Click', 'Client', 'Clinical', 'Clinicians', 'Clinicogenomics', 'Cloud', 'Cloudera', 'Clustering', 'Coding', 'Cohen', 'Collaborate', 'Collaborating', 'Collaboration', 'Collaborative', 'Colleagues', 'Collect', 'Collibra', 'Combine', 'Combining', 'Come', 'Comfort', 'Comfortable', 'Commercial', 'Commitment', 'Communicate', 'Communicating', 'Communication', 'Community', 'Company', 'Competencies', 'Completed', 'Completion', 'Complications', 'Compute', 'Computer', 'Computing', 'Conditions', 'Conduct', 'Conducting', 'Conducts', 'Confidently', 'Connect', 'Connecting', 'Considered', 'Consult', 'Consulting', 'Consumer', 'Contact', 'Container', 'Continuous', 'Contract', 'Contribute', 'Conversational', 'Coordinate', 'Coordinating', 'Core', 'Cortex', 'Cost', 'Counsel', 'Create', 'Creates', 'Creating', 'Creative', 'Critical', 'Cultivate', 'Curious', 'Currently', 'Customer', 'Cybersecurity', 'Côte', 'D', 'DARPA', 'DATA', 'DEPARTMENT', 'DESCRIPTION', 'DESIRABLE', 'DICOM', 'DNA', 'DNNs', 'DO', 'DUTIES', 'Dash', 'Dassault', 'Data', 'Database', 'Databricks', 'DatabricksBayesian', 'Datawarehouse', 'Date', 'David', 'Day', 'Dean', 'Dec', 'December', 'Decision', 'Decisiveness', 'Dedication', 'Deep', 'Defense', 'Define', 'Defines', 'Degree', 'Degrees', 'Deliver', 'Delivering', 'Delphix', 'Demonstrable', 'Demonstrate', 'Demonstrated', 'Demonstrates', 'Dental', 'Department', 'Depending', 'Description', 'Descriptive', 'Design', 'Designer', 'Designs', 'Desirable', 'Desirables', 'Desired', 'Desk', 'Detail', 'Details', 'Detection', 'Determination', 'Determine', 'DevOps', 'Develop', 'Developer', 'Developing', 'Development', 'Develops', 'Devices', 'Diagnostics', 'Dialogflow', 'Diego', 'Digestive', 'Digital', 'Dimensionality', 'Direct', 'Director', 'Disabilities', 'Disadvantaged', 'Discovery', 'Diseases', 'Disorders', 'Distribution', 'Diversity', 'Division', 'Do', 'DoD', 'Docker', 'Documenting', 'Does', 'Draw', 'Drives', 'Drug', 'Due', 'Duration', 'During', 'Duties', 'Dyad', 'ED', 'EDUCATION', 'EEO', 'EMR', 'ENGIE', 'ENOVIA', 'EO', 'ETL', 'EXG', 'EXPERIENCE', 'East', 'Eclipse', 'EconCS', 'Econometrics', 'Economic', 'Economics', 'Economist', 'Ecova', 'Education', 'Educational', 'Effective', 'Efficiency', 'Efficient', 'ElasticSearch', 'Electric', 'Electrical', 'Elsevier', 'Employee', 'Employer', 'Employment', 'Empower', 'Encoders', 'Encouragement', 'Energy', 'Engage', 'Engine', 'Engineer', 'Engineering', 'Engineers', 'England', 'EnglandBoston', 'English', 'Enhance', 'Enjoy', 'Ensure', 'Ensuring', 'Environment', 'Epidemiology', 'Equal', 'Equally', 'Equipment', 'Essential', 'Essentials', 'Establish', 'Establishing', 'Estimate', 'Etiometry', 'Europe', 'Evaluate', 'Evaluating', 'Evaluation', 'Evangelize', 'Evelo', 'Examine', 'Examining', 'Examples', 'Excel', 'Excellent', 'Exception', 'Exceptional', 'Exchange', 'Excitingly', 'Executive', 'Exempt', 'Exercises', 'Exhibit', 'Exhibits', 'Expand', 'Experience', 'Experienced', 'Experiences', 'Experimental', 'Expert', 'Expertise', 'Exposure', 'Extensive', 'External', 'Extract', 'FAIR', 'FFNN', 'FLSA', 'Facebook', 'Facility', 'FactBase', 'Factor', 'Factors', 'Fair', 'Fairbank', 'Familiar', 'Familiarity', 'Fearlessly', 'Feb', 'Federal', 'Fidelity', 'Fields', 'Finance', 'Financial', 'First', 'Flag', 'Flagship', 'Flask', 'Flatiron', 'Flexibility', 'Fluency', 'Fluent', 'Follow', 'Football', 'For', 'Forces', 'Forest', 'Form', 'Formal', 'Formalize', 'Formulate', 'Forrester', 'Fortune', 'Forum', 'Foundation', 'FoundationCORE', 'Founded', 'Framingham', 'France', 'Francis', 'Francisco', 'Fraud', 'Free', 'Friday', 'From', 'Fruit', 'Fuel', 'Full', 'Functions', 'Fusion', 'GAA', 'GI', 'GL', 'GPA', 'GPGPUYou', 'GPU', 'Gastrointesinal', 'Gastrointestinal', 'Gaussian', 'Gen', 'General', 'Generalized', 'Generating', 'Genetics', 'Git', 'GitHub', 'Glass', 'Global', 'Go', 'Goal', 'Good', 'Google', 'Governance', 'Government', 'Grad', 'Grads', 'Graduate', 'Grant', 'Great', 'Group', 'GroupsTranslate', 'Growth', 'Guard', 'Guided', 'Guidelines', 'Gym', 'HDFS', 'HIGHLY', 'HIPAA', 'HIV', 'HMS', 'HPC', 'HPSL', 'HPV', 'HQ', 'HTML', 'Hadoop', 'Hampshire', 'HampshireBoston', 'Hands', 'Harvard', 'Have', 'Haves', 'Hawaiians', 'He', 'Health', 'HealthCare', 'HealthNet', 'Healthcare', 'Heart', 'Help', 'Helpdesk', 'Hemostasis', 'Here', 'HiTS', 'Hierarchical', 'High', 'Higher', 'Highest', 'Highly', 'Hiring', 'Hispanic', 'Hive', 'Hockey', 'Homeland', 'Homesite', 'Horizons', 'Hospital', 'Hospitals', 'Hours', 'How', 'However', 'Hub', 'Huge', 'Human', 'Humana', 'Humor', 'Hypothesis', 'I', 'IBM', 'IL', 'IM', 'IMED', 'IMU', 'IMWUT', 'IP', 'IRB', 'IS', 'IT', 'IUI', 'IVD', 'IVZ', 'Idea', 'Ideal', 'Ideally', 'Identifies', 'Identify', 'Identifying', 'Identity', 'If', 'Illumina', 'Image', 'Imagine', 'Impact', 'Impala', 'Implement', 'Implementation', 'Improvement', 'In', 'Includes', 'Inclusion', 'Incomplete', 'Index', 'India', 'Indians', 'Indicator', 'Indigo', 'Individual', 'Individuals', 'Industry', 'InfiniBand', 'Info', 'Informatica', 'Informatics', 'Information', 'Infusion', 'Ingenuity', 'Initial', 'Initiative', 'Innovation', 'Insight', 'Insights', 'Institute', 'Institutes', 'Institutional', 'Instructor', 'Instrumentation', 'Insurance', 'Insurtech', 'Integrate', 'Integrating', 'Integration', 'Integrity', 'Intellectual', 'Intelligence', 'Intense', 'Interact', 'Interacting', 'Interacts', 'Interest', 'Interested', 'Interface', 'Intern', 'International', 'Internet', 'Interns', 'Internship', 'InternshipBoston', 'Interprets', 'Intuitive', 'Invesco', 'Investigators', 'Investment', 'Investments', 'Investor', 'Involve', 'Is', 'Islanders', 'It', 'Ivoire', 'JOB', 'Jameel', 'Jan', 'Java', 'JavaScript', 'Javascript', 'Jenkins', 'Job', 'Join', 'Joint', 'Journal', 'Julia', 'July', 'Junction', 'June', 'Junior', 'Jupyter', 'KEY', 'KNN', 'KPIs', 'KTC', 'Kanban', 'Keep', 'Kenexa', 'Keras', 'Key', 'Kintai', 'Kirschstein', 'Klaviyo', 'Know', 'Knowledge', 'Knowledgeable', 'Kubernetes', 'L', 'LAW', 'LICENSES', 'LMS', 'LOCATION', 'LSP', 'LSTM', 'LTD', 'Lab', 'Laboratory', 'Labs', 'Lake', 'Language', 'Languages', 'Lastly', 'Latex', 'Latif', 'Launched', 'Law', 'Lead', 'Leading', 'Learn', 'Learning', 'Lester', 'Let', 'Letters', 'Level', 'Leverage', 'Leveraging', 'Liberty', 'Life', 'Lifetime', 'Limited', 'Linear', 'Lines', 'LinkedIn', 'Linux', 'Loans', 'Localization', 'Located', 'Location', 'Locations', 'LogMeIn', 'Logistics', 'London', 'Longwood', 'Look', 'Looking', 'Lucene', 'Lunch', 'M', 'MA', 'MATLAB', 'MCR', 'MD', 'MDM', 'MGH', 'MIT', 'MITRE', 'ML', 'MLlib', 'MPEC', 'MPI', 'MPP', 'MRI', 'MS', 'MSc', 'MXNet', 'MYSQLLanguages', 'MacB', 'Machine', 'Machines', 'Mackey', 'Madrid', 'Main', 'Maintaining', 'Make', 'Makes', 'Manage', 'Management', 'Manager', 'Managing', 'Manipulate', 'Manipulating', 'Manipulation', 'Manufacturing', 'Many', 'Mapping', 'Mar', 'Market', 'Marketbasket', 'Marketers', 'Marketing', 'Markov', 'Massachusetts', 'Master', 'Masters', 'MatLab', 'Matab', 'Materials', 'Math', 'MathWorks', 'Mathematics', 'Matlab', 'Matploltib', 'May', 'McKinsey', 'Measure', 'Measurement', 'Medicaid', 'Medical', 'Medicine', 'Members', 'Mentor', 'Mercurial', 'Metadata', 'Metrics', 'Microsoft', 'Middle', 'Military', 'Min', 'Mine', 'Minimum', 'Minneapolis', 'Minorities', 'Mo', 'Model', 'Modeling', 'Modelling', 'Moderna', 'Molecular', 'MongoDB', 'Monitor', 'Monte', 'More', 'Moreover', 'Morning', 'Most', 'Mozambique', 'Much', 'MuleSoft', 'Multiple', 'Must', 'Mutual', 'MySQL', 'N', 'NCI', 'NERD', 'NGS', 'NIH', 'NLP', 'NPL', 'NRSA', 'NY', 'NYC', 'Nanodrop', 'Nation', 'National', 'Native', 'Natives', 'Natural', 'Nature', 'Need', 'Network', 'Networks', 'Neural', 'New', 'Next', 'Nice', 'Nift', 'No', 'NoSQL', 'None', 'Normal', 'North', 'Northeastern', 'Notebooks', 'Now', 'NumPy', 'Numpy', 'OBI', 'OFCCP', 'OOP', 'OUR', 'Object', 'Objective', 'Occasional', 'Of', 'Offers', 'Office', 'Officer', 'On', 'Oncology', 'One', 'Ongoing', 'Only', 'OpenFlow', 'Openly', 'Operational', 'Operations', 'Opportunities', 'Opportunity', 'Optic', 'Optimisation', 'Optimizing', 'Options', 'Oracle', 'Ordinance', 'OrgSolutions', 'Organization', 'Oriented', 'Original', 'Orthopaedic', 'Orthopedics', 'Other', 'Our', 'Outcomes', 'Outlook', 'Outstanding', 'Over', 'Overview', 'Owners', 'P', 'PCA', 'PHS', 'PI', 'PIVOT', 'PM', 'POSITION', 'PRINCIPAL', 'Pacific', 'Paid', 'Pandas', 'Paris', 'Park', 'Parkes', 'Parkinson', 'Participate', 'Partner', 'Partners', 'Passion', 'Passionate', 'Patent', 'Patients', 'Perform', 'Performance', 'Performing', 'Performs', 'Perks', 'Perl', 'Personal', 'Personalization', 'Persons', 'PhD', 'PharmD', 'Pharmaceutical', 'Pharmacology', 'Philadelphia', 'Physical', 'Physics', 'Pioneering', 'Plan', 'Planning', 'Platforms', 'Please', 'Plotly', 'Plusses', 'Policy', 'Population', 'Position', 'Positive', 'Possess', 'PostDocs', 'Poster', 'Posters', 'Postg', 'PostgreSQL', 'Postgres', 'Postgresql', 'Posting', 'Poverty', 'Power', 'PowerAdvocate', 'PowerPoint', 'Powered', 'Practical', 'Practice', 'Predictive', 'Preferable', 'Preference', 'Preferred', 'Prepare', 'Prepares', 'Present', 'Presenting', 'Presents', 'Presto', 'PrestoDB', 'Preventing', 'Previous', 'Price', 'Primary', 'Prime', 'Principal', 'Prior', 'Privacy', 'Proactively', 'Probability', 'Process', 'Processing', 'Procurement', 'Produce', 'Product', 'Productivity', 'Productize', 'Prof', 'Professional', 'Professions', 'Proficiency', 'Proficient', 'Program', 'Programming', 'Project', 'Projects', 'Propensity', 'Prototype', 'Proven', 'Provide', 'Providence', 'Psychology', 'Public', 'Publish', 'Pubmed', 'Pulse', 'Purchasing', 'Purpose', 'Pursuant', 'Putting', 'PyTorch', 'Python', 'PythonKnowledge', 'QA', 'QC', 'QUALIFICATIONS', 'Quailfications', 'Qualification', 'Qualifications', 'Quality', 'Quantitative', 'Qubit', 'Query', 'Quest', 'R', 'RDMA', 'REGISTRATIONS', 'REQUIRED', 'REQUIREMENTS', 'RESEARCH', 'RESPECT', 'RESPONSIBILITIES', 'RESPONSIBILITY', 'RESTful', 'RF', 'RNA', 'RYou', 'Radiology', 'Random', 'Rapidly', 'Read', 'Reasonable', 'RecSys', 'Recent', 'Recognition', 'Recognized', 'Reconstruction', 'Recruiter', 'Recruiting', 'RedShift', 'Redshift', 'Regression', 'Regular', 'Relationship', 'Relevant', 'ReltioMust', 'Rentals', 'Reporting', 'Req', 'Required', 'Requirements', 'Research', 'Researching', 'Resources', 'Respect', 'Respond', 'Responsibilities', 'Responsibility', 'Responsible', 'Results', 'Review', 'Richard', 'Rifiniti', 'Right', 'Risk', 'Roche', 'Role', 'Root', 'Ruby', 'Run', 'Running', 'Ruth', 'SAI', 'SAP', 'SAS', 'SCIENTIST', 'SENIOR', 'SIGIR', 'SKILLS', 'SLAM', 'SPARK', 'SPSS', 'SQL', 'SSIS', 'STATA', 'STD', 'STEM', 'SUMMARY', 'SUPERVISORY', 'SUPPLEMENT', 'SVD', 'SVM', 'SVMs', 'SaaS', 'Sample', 'Samples', 'San', 'Saturdays', 'Scala', 'Schedule', 'Scheduled', 'Scheduling', 'Schneider', 'Scholar', 'Scholarship', 'School', 'SciKit', 'SciPy', 'Science', 'Sciences', 'Scientific', 'Scientist', 'Scientists', 'Scikit', 'Scrum', 'Seamlessly', 'Seattle', 'Sec', 'Secondary', 'Secretary', 'Security', 'See', 'Seek', 'Segment', 'Select', 'Selected', 'Self', 'Senior', 'Sense', 'Sensor', 'September', 'Seres', 'Serve', 'Server', 'Service', 'Services', 'Several', 'Shaker', 'Shape', 'Share', 'She', 'Shift', 'Shiny', 'ShinyExpansive', 'Significant', 'Since', 'Singapore', 'Sir', 'Skills', 'Slack', 'Sleep', 'So', 'Soccer', 'Social', 'Sofia', 'Software', 'Solid', 'Solution', 'Solutions', 'Solve', 'Some', 'South', 'Spark', 'SparkProficiency', 'Special', 'Spend', 'Sports', 'Spotify', 'Springs', 'St', 'Stakeholder', 'Standard', 'Start', 'Stata', 'State', 'Statement', 'States', 'Statistical', 'Statistics', 'StatisticsExperience', 'Status', 'Stay', 'Steps', 'Steward', 'Stewards', 'Still', 'Stock', 'Strategic', 'Street', 'Strong', 'Structured', 'Student', 'Students', 'Studio', 'Study', 'Submit', 'Subsidized', 'Subversion', 'Success', 'Successful', 'Successfully', 'Such', 'Summary', 'Summer', 'Superior', 'Supervisory', 'Supplements', 'Supply', 'Support', 'Surgery', 'Sustainability', 'Sustained', 'System', 'Systemes', 'Systems', 'Systèmes', 'TA', 'TECHNICAL', 'THE', 'TITLE', 'Tableau', 'Take', 'Talend', 'Teach', 'Teaching', 'Team', 'Teammate', 'Teamwork', 'Tech', 'Technical', 'Technologies', 'Technology', 'Telecom', 'Temporary', 'TensorFlow', 'TensorFlowDatabases', 'Tensorflow', 'Test', 'Testing', 'Text', 'Thailand', 'That', 'The', 'Therapeutic', 'Therapeutics', 'Therapy', 'There', 'These', 'They', 'This', 'Those', 'Thoughtful', 'Three', 'Thrive', 'Through', 'Throughout', 'Thursday', 'Time', 'Times', 'Title', 'To', 'Tobacco', 'Today', 'Together', 'Tokyo', 'Tools', 'Torch', 'Train', 'Training', 'Traits', 'Transfer', 'Translate', 'Transnational', 'Transportation', 'Travel', 'Treatments', 'Tree', 'Trello', 'TripAdvisor', 'Troubleshoot', 'Tryout', 'Tuesday', 'Tufts', 'Twitter', 'Two', 'Type', 'Typically', 'UIST', 'UK', 'US', 'USA', 'UX', 'Ukraine', 'Uncompromising', 'Under', 'Understand', 'Understanding', 'Undertaking', 'Underwriting', 'Unit', 'United', 'University', 'Unposting', 'Up', 'Us', 'Use', 'User', 'Users', 'Using', 'Utilize', 'Utilizing', 'VALUES', 'VBA', 'VM', 'VMs', 'VMware', 'VR', 'Value', 'Values', 'Vector', 'Ventures', 'Verily', 'Very', 'Veteran', 'Veterans', 'Videos', 'Virgin', 'Virtual', 'Vistaprint', 'W', 'WA', 'WORKING', 'WWW', 'Wall', 'Waltham', 'Want', 'Watson', 'We', 'Web', 'Webinars', 'Wednesday', 'Weekly', 'Wellbeing', 'Werfen', 'What', 'When', 'Where', 'Whether', 'While', 'Who', 'Why', 'Will', 'Willingness', 'Wired', 'With', 'Within', 'Without', 'Women', 'Word', 'Work', 'Working', 'Works', 'World', 'Worldpay', 'Would', 'Write', 'Writing', 'Wyman', 'XGBoost', 'XML', 'Xgboost', 'Yale', 'York', 'You', 'Your', 'Youssef', 'ZR', 'ZRNift', 'Zealand', 'Zeppelin', 'Zimbabwe', 'Zurich', 'ability', 'able', 'abreast', 'abroad', 'absolute', 'abstract', 'academia', 'academic', 'academically', 'accelerate', 'accelerating', 'accept', 'accepted', 'accepting', 'access', 'accessible', 'accessing', 'accolade', 'accommodate', 'accommodation', 'accompanying', 'accomplish', 'account', 'accountability', 'accountable', 'accredited', 'accuracy', 'accurate', 'accurately', 'achievable', 'achieve', 'achievement', 'achieving', 'acid', 'acquired', 'acquisition', 'across', 'act', 'action', 'actionable', 'active', 'actively', 'activity', 'actual', 'actually', 'actuarial', 'acuity', 'ad', 'adapt', 'added', 'addition', 'additional', 'address', 'addressing', 'adept', 'adhere', 'adjust', 'adjusted', 'administrative', 'admission', 'admitted', 'adoption', 'advance', 'advanced', 'advancement', 'advancing', 'advantage', 'advertised', 'advertisement', 'advice', 'advisor', 'advisory', 'advocacy', 'advocate', 'affect', 'affiliate', 'affiliated', 'affiliation', 'affirmative', 'affordable', 'aforementioned', 'afraid', 'age', 'agency', 'agenda', 'agent', 'aggregate', 'aggressive', 'agile', 'ago', 'agreement', 'ahead', 'aid', 'aim', 'aimed', 'al', 'alert', 'algebra', 'algorithm', 'algorithmic', 'align', 'aligned', 'aligning', 'alignment', 'aligns', 'alliance', 'allocate', 'allocation', 'allow', 'allows', 'along', 'alongside', 'also', 'alternative', 'always', 'amazing', 'ambiguity', 'ambiguous', 'ambulatory', 'amenable', 'among', 'amount', 'analyse', 'analysesDevelops', 'analysesImplement', 'analysis', 'analysisAnalyze', 'analysisComfort', 'analysisEnhancing', 'analysisProvide', 'analyst', 'analytic', 'analytical', 'analytics', 'analyze', 'analyzed', 'analyzes', 'analyzing', 'anatomical', 'ancestry', 'annotation', 'annual', 'annually', 'anomaly', 'another', 'answer', 'answered', 'answering', 'anticipate', 'anticipated', 'anyone', 'anything', 'anywhere', 'applicability', 'applicable', 'applicant', 'application', 'applied', 'applies', 'apply', 'applying', 'appointment', 'appreciates', 'appreciation', 'approach', 'approachesHas', 'appropriate', 'appropriately', 'approval', 'approximately', 'architect', 'architecting', 'architectural', 'architecture', 'archive', 'area', 'arise', 'around', 'arrangement', 'array', 'arrest', 'art', 'articulating', 'artifact', 'artificial', 'artist', 'asap', 'asked', 'asking', 'aspect', 'aspenONE', 'aspiration', 'aspire', 'ass', 'assay', 'assembled', 'assessment', 'asset', 'assigned', 'assignment', 'assist', 'assistance', 'assistant', 'assisting', 'associate', 'associated', 'assume', 'assumption', 'assurance', 'assure', 'attached', 'attend', 'attendance', 'attending', 'attention', 'attitude', 'attract', 'attracts', 'attribute', 'attributed', 'attribution', 'attrition', 'audience', 'audiencesDemonstrates', 'audio', 'audit', 'auditing', 'augment', 'augmented', 'authentication', 'author', 'authorization', 'authorship', 'automate', 'automated', 'automating', 'automation', 'autonomously', 'autonomy', 'available', 'avenue', 'average', 'award', 'awarded', 'awareness', 'b', 'bachelor', 'back', 'backed', 'background', 'bagging', 'balance', 'banking', 'base', 'based', 'baseline', 'basic', 'basis', 'bayes', 'bear', 'beat', 'beating', 'become', 'becoming', 'began', 'begin', 'beginning', 'begun', 'behalf', 'behave', 'behavior', 'behavioral', 'behaviour', 'behavioural', 'behind', 'belief', 'believe', 'bench', 'benchmark', 'benefit', 'bespoke', 'best', 'better', 'beyond', 'bias', 'big', 'bilingual', 'billing', 'billion', 'bioinformaticians', 'bioinformatics', 'biological', 'biologist', 'biology', 'biologyComfort', 'biomedical', 'biometric', 'biopharmaceutical', 'biophysical', 'bioscience', 'biostatistical', 'biostatisticians', 'biostatistics', 'bleeding', 'blend', 'blocker', 'blood', 'board', 'body', 'bold', 'bonus', 'boosting', 'borough', 'bottom', 'bound', 'boundary', 'boutique', 'box', 'brain', 'brainstorm', 'brand', 'breadth', 'break', 'breaking', 'breakthrough', 'brief', 'brightest', 'brilliant', 'bring', 'bringing', 'brings', 'broad', 'broader', 'broadly', 'broker', 'budgeting', 'bug', 'build', 'building', 'built', 'burden', 'burgeoning', 'business', 'busy', 'buyer', 'c', 'caching', 'calculation', 'calibration', 'call', 'campaign', 'campus', 'cancer', 'candidate', 'cannabis', 'capability', 'capable', 'capacity', 'capitalization', 'capture', 'carbon', 'cardiovascular', 'care', 'career', 'careful', 'caring', 'carried', 'carrier', 'carry', 'carve', 'case', 'casualty', 'catalog', 'categorization', 'category', 'causal', 'causality', 'cause', 'cell', 'center', 'central', 'centric', 'ceremony', 'certain', 'certification', 'certified', 'chain', 'challenge', 'challenged', 'challenging', 'chance', 'change', 'changing', 'channel', 'characteristic', 'characterization', 'characterize', 'characterizing', 'charge', 'charged', 'chart', 'chatbots', 'check', 'chemist', 'chemistry', 'choice', 'choose', 'chosen', 'churn', 'circumstance', 'cited', 'citizen', 'citizenship', 'city', 'civil', 'civilian', 'claim', 'clarity', 'class', 'classical', 'classification', 'classified', 'classifier', 'classify', 'classifying', 'classroom', 'clean', 'cleaning', 'cleanliness', 'cleanse', 'cleansing', 'clear', 'clearable', 'clearance', 'clearly', 'click', 'clickstream', 'client', 'climate', 'clinic', 'clinical', 'clinician', 'clinicogenomic', 'close', 'closely', 'cloud', 'club', 'cluster', 'clustering', 'coach', 'coaching', 'coagulation', 'code', 'coding', 'cognitive', 'coherent', 'cohort', 'collaborate', 'collaborating', 'collaboration', 'collaborative', 'collaboratively', 'collaborator', 'colleague', 'collect', 'collected', 'collecting', 'collection', 'college', 'color', 'combating', 'combination', 'combine', 'combined', 'combining', 'come', 'comfort', 'comfortable', 'coming', 'command', 'commensurate', 'commerce', 'commercial', 'commit', 'commitment', 'committed', 'common', 'communicate', 'communicates', 'communicating', 'communication', 'communicative', 'communicator', 'community', 'company', 'compare', 'comparison', 'compartmental', 'compatibly', 'compelling', 'compensates', 'compensation', 'competence', 'competency', 'competing', 'competition', 'competitive', 'competitor', 'complaint', 'complete', 'completed', 'completely', 'completing', 'completion', 'complex', 'complexity', 'compliance', 'comply', 'component', 'compound', 'comprehensive', 'comprehensiveness', 'comprised', 'comprises', 'computation', 'computational', 'computationally', 'computer', 'computing', 'conceive', 'concentration', 'concept', 'conceptualize', 'concise', 'concluded', 'conclusion', 'concreate', 'concurrent', 'condition', 'conditional', 'conduct', 'conducted', 'conducting', 'conference', 'conferencing', 'confidence', 'confident', 'confidentiality', 'confidently', 'configuration', 'congestive', 'connect', 'connected', 'consider', 'considerable', 'consideration', 'considered', 'considering', 'consistent', 'consisting', 'consists', 'consolidate', 'consolidation', 'constant', 'constantly', 'constraint', 'construct', 'construction', 'constructive', 'construed', 'consult', 'consultant', 'consultation', 'consultative', 'consulting', 'consumable', 'consumer', 'consumption', 'contact', 'contain', 'contained', 'container', 'contains', 'contender', 'content', 'context', 'continual', 'continually', 'continue', 'continued', 'continuing', 'continuous', 'continuously', 'continuum', 'contract', 'contractor', 'contribute', 'contributes', 'contributing', 'contribution', 'contributor', 'control', 'convenience', 'convention', 'conventional', 'conversation', 'convert', 'convey', 'conviction', 'convince', 'convincingly', 'cooperatively', 'coordinate', 'coordination', 'coordinator', 'core', 'coronary', 'corporate', 'correct', 'corrective', 'correlate', 'corresponding', 'cost', 'could', 'country', 'coupled', 'courage', 'course', 'coursework', 'cover', 'coverage', 'crave', 'crazy', 'create', 'created', 'creates', 'creating', 'creation', 'creative', 'credential', 'credibility', 'creditable', 'creed', 'crime', 'criminal', 'critical', 'criticism', 'cross', 'crucial', 'cryptography', 'cubicle', 'culmination', 'cultural', 'culturally', 'culture', 'cumulative', 'curation', 'curiosity', 'curious', 'currency', 'current', 'currently', 'curriculum', 'custom', 'customer', 'customized', 'cut', 'cutting', 'cvshealthsupport', 'cyber', 'cybersecurity', 'cycle', 'cyclostationary', 'daily', 'dashboard', 'data', 'dataOrganized', 'database', 'datapoints', 'dataset', 'datasets', 'date', 'day', 'deadline', 'deaf', 'deal', 'dealing', 'death', 'debugging', 'decision', 'decomposition', 'decreasing', 'dedicated', 'dedication', 'deemed', 'deep', 'deepen', 'deeper', 'deeply', 'define', 'defined', 'defining', 'degree', 'deidentification', 'delay', 'deliver', 'deliverable', 'delivered', 'delivering', 'delivers', 'delivery', 'demand', 'demo', 'democratization', 'democratize', 'demographic', 'demography', 'demonstrable', 'demonstrably', 'demonstrate', 'demonstrated', 'demonstrates', 'dental', 'department', 'depending', 'depict', 'deploy', 'deploying', 'deployment', 'deploys', 'depth', 'derive', 'derived', 'deriving', 'describe', 'described', 'describing', 'description', 'descriptive', 'deserves', 'design', 'designation', 'designed', 'designer', 'designing', 'desirable', 'desire', 'desired', 'desktop', 'detail', 'detailed', 'detailing', 'detect', 'detection', 'determination', 'determine', 'determined', 'determining', 'deterministic', 'devastating', 'develop', 'developed', 'developedInterface', 'developer', 'developing', 'development', 'developmentExperience', 'develops', 'device', 'devise', 'devising', 'devoted', 'diabetes', 'diagnosing', 'diagnosis', 'diagnostic', 'diagnostics', 'diagram', 'dialect', 'difference', 'different', 'differentiate', 'differentiated', 'differentiating', 'difficult', 'dig', 'digestive', 'digging', 'digital', 'diligence', 'dimensionality', 'direct', 'direction', 'directly', 'dirty', 'disability', 'disadvantaged', 'discharge', 'discipline', 'disciplined', 'disclose', 'disclosed', 'disclosure', 'discover', 'discovering', 'discovers', 'discovery', 'discrepancy', 'discriminate', 'discrimination', 'discus', 'discussed', 'discussion', 'disease', 'disorder', 'disparate', 'disposition', 'disprove', 'disrupted', 'disrupter', 'disruptive', 'disseminate', 'distill', 'distinctive', 'distributed', 'distribution', 'distributor', 'diverse', 'diversifying', 'diversity', 'divestments', 'division', 'doctor', 'doctoral', 'document', 'documentation', 'documented', 'dollar', 'domain', 'domestically', 'dominant', 'done', 'door', 'double', 'downtown', 'dramatic', 'draw', 'dream', 'drive', 'driven', 'driver', 'driving', 'drop', 'drug', 'drugging', 'due', 'duration', 'dust', 'duty', 'dynamic', 'eValuation', 'eager', 'earliest', 'early', 'earned', 'earning', 'ease', 'easy', 'ecommerce', 'econometric', 'econometrics', 'economic', 'economically', 'economics', 'economy', 'ecosystem', 'edX', 'edge', 'editing', 'educating', 'education', 'educational', 'educationally', 'effect', 'effective', 'effectively', 'effectiveness', 'efficiency', 'efficient', 'efficiently', 'effort', 'eigenvalue', 'either', 'electronic', 'elevate', 'eligible', 'elite', 'else', 'email', 'embrace', 'embracing', 'emerge', 'emerged', 'emerging', 'eminence', 'empathetically', 'emphasis', 'emphasized', 'employ', 'employee', 'employer', 'employment', 'empower', 'empowers', 'enable', 'enabled', 'enablement', 'enabler', 'enables', 'enabling', 'encapsulate', 'encourage', 'encouraged', 'encourages', 'encouraging', 'end', 'endpoint', 'energetic', 'energized', 'energy', 'enforcement', 'engage', 'engaged', 'engagement', 'engaging', 'engender', 'engine', 'engineer', 'engineering', 'enhance', 'enhancement', 'enhancing', 'enjoy', 'enjoys', 'enough', 'enrich', 'enrichment', 'enrolled', 'ensemble', 'ensure', 'ensures', 'ensuring', 'entail', 'entailing', 'enterprise', 'enthusiasm', 'enthusiast', 'enthusiastic', 'entire', 'entity', 'entrepreneurial', 'entry', 'environment', 'environmentCommunicates', 'environmentDemonstrates', 'environmentExcellent', 'environmentExperience', 'environmental', 'epidemiological', 'epidemiologist', 'epidemiology', 'equal', 'equally', 'equation', 'equilavent', 'equip', 'equipment', 'equivalent', 'error', 'escalate', 'especially', 'essential', 'establish', 'established', 'establishing', 'establishment', 'estimate', 'estimating', 'et', 'etc', 'ethic', 'ethical', 'ethnic', 'ethnicity', 'euro', 'evaluate', 'evaluating', 'evaluation', 'even', 'evening', 'event', 'ever', 'every', 'everyday', 'everyone', 'everything', 'everywhere', 'evidence', 'evolution', 'evolve', 'evolving', 'exam', 'examining', 'example', 'exceeding', 'excel', 'excellence', 'excellent', 'exceptional', 'excessive', 'exchange', 'exchanging', 'excited', 'exciting', 'excluding', 'execute', 'executed', 'executing', 'execution', 'executive', 'exercise', 'exhaustive', 'exhibit', 'exhilarating', 'exist', 'existing', 'expand', 'expanding', 'expands', 'expansion', 'expectation', 'expected', 'expedition', 'expense', 'experience', 'experienceExperience', 'experienced', 'experiential', 'experiment', 'experimental', 'experimentation', 'expert', 'expertise', 'explain', 'explaining', 'exploration', 'exploratory', 'explore', 'exploring', 'exponential', 'exposure', 'exposureExperience', 'expression', 'extend', 'extended', 'extending', 'extensive', 'external', 'externally', 'extract', 'extracting', 'extraction', 'extraordinary', 'extreme', 'eye', 'fabrication', 'face', 'facet', 'facilitate', 'facilitated', 'facility', 'facing', 'facingExperience', 'fact', 'factor', 'faculty', 'failure', 'fair', 'famed', 'familiar', 'familiarity', 'family', 'fan', 'fashion', 'fast', 'faster', 'faulty', 'feature', 'federal', 'feed', 'feedback', 'feel', 'fellow', 'fertile', 'fertility', 'fidelity', 'field', 'fieldDemonstrated', 'fieldProficient', 'fiercely', 'fifty', 'fight', 'file', 'filing', 'final', 'finalizing', 'finally', 'finance', 'financial', 'find', 'finding', 'finish', 'fintech', 'firm', 'first', 'fit', 'fitting', 'five', 'fix', 'flexibility', 'flexible', 'flow', 'focus', 'focused', 'focusing', 'follow', 'following', 'foot', 'footprint', 'force', 'forecasting', 'foremost', 'forest', 'forge', 'form', 'formal', 'format', 'formatting', 'formed', 'former', 'formerly', 'formulate', 'formulation', 'forth', 'forward', 'foster', 'found', 'foundation', 'founded', 'founder', 'founding', 'four', 'fourth', 'frame', 'framework', 'fraud', 'free', 'freedom', 'frequently', 'fresh', 'front', 'fuel', 'fulfill', 'full', 'fuller', 'fullest', 'fulltime', 'fully', 'fun', 'function', 'functional', 'functionality', 'fund', 'fundamental', 'fundamentally', 'funded', 'funders', 'funnel', 'furnish', 'furtherance', 'furthering', 'future', 'fuzzy', 'gain', 'gained', 'gaining', 'game', 'gaming', 'gas', 'gastroenterology', 'gather', 'gathering', 'gender', 'general', 'generalist', 'generalized', 'generally', 'generate', 'generated', 'generates', 'generating', 'generation', 'generic', 'generous', 'genetic', 'geneticist', 'genetics', 'genomic', 'genomics', 'genuine', 'get', 'getting', 'ggplot', 'gift', 'git', 'give', 'given', 'giving', 'glean', 'global', 'globally', 'globe', 'go', 'goal', 'goalsInterest', 'good', 'got', 'govern', 'governance', 'governing', 'government', 'grading', 'graduate', 'graduated', 'graduation', 'grant', 'grantee', 'graph', 'graphic', 'graphical', 'gratifying', 'great', 'greater', 'greatest', 'greatly', 'greatness', 'grid', 'ground', 'group', 'grow', 'growing', 'growth', 'guide', 'guideline', 'guiding', 'gym', 'habit', 'half', 'hand', 'handle', 'handled', 'handling', 'happier', 'happiness', 'harassment', 'hard', 'harmonization', 'harness', 'harnessing', 'hat', 'head', 'headquartered', 'headquarters', 'health', 'healthcare', 'healthier', 'healthiest', 'healthy', 'hearing', 'heart', 'heat', 'heavy', 'hectic', 'held', 'help', 'helpful', 'helping', 'hepatitis', 'hidden', 'high', 'higher', 'highest', 'highly', 'hire', 'hired', 'hiring', 'history', 'hoc', 'hold', 'holder', 'holding', 'home', 'horizon', 'horizontal', 'hospital', 'host', 'hosted', 'hosting', 'hottest', 'hour', 'house', 'http', 'human', 'humanity', 'humility', 'hundred', 'hybrid', 'hyperparameter', 'hypothesis', 'idea', 'ideal', 'ideally', 'ideation', 'identifiable', 'identification', 'identified', 'identify', 'identifying', 'identity', 'illustrate', 'image', 'imagery', 'imagination', 'imagined', 'imaging', 'immediate', 'immediately', 'immigration', 'immune', 'impact', 'impactful', 'impacting', 'impairment', 'implement', 'implementation', 'implemented', 'implementing', 'implication', 'importance', 'important', 'importantly', 'importing', 'impossible', 'improve', 'improved', 'improvement', 'improving', 'imputation', 'inappropriate', 'incentive', 'include', 'included', 'includes', 'including', 'inclusion', 'inclusive', 'inclusivity', 'income', 'incompatible', 'incomplete', 'inconsistency', 'incorporate', 'increase', 'increasing', 'incrementally', 'incumbent', 'indeed', 'independence', 'independent', 'independently', 'indicate', 'indicator', 'individual', 'individualized', 'indoor', 'industry', 'inexplicable', 'infectious', 'inferential', 'inflammatory', 'influence', 'influencing', 'inform', 'informatica', 'informaticians', 'information', 'informed', 'informing', 'infrastructure', 'infused', 'ingenuity', 'ingest', 'ingestion', 'inhibited', 'initial', 'initiated', 'initiative', 'innovate', 'innovating', 'innovation', 'innovative', 'inordinate', 'input', 'inquired', 'inquiry', 'insanely', 'inside', 'insight', 'insightful', 'inspire', 'inspired', 'inspires', 'installation', 'instance', 'instinctive', 'institution', 'institutional', 'instruction', 'instructor', 'instrumental', 'insurance', 'insuranceDental', 'insurer', 'integral', 'integrate', 'integrated', 'integrating', 'integration', 'integrative', 'integrity', 'intellectual', 'intelligence', 'intelligent', 'intended', 'intensive', 'interact', 'interacting', 'interaction', 'interactive', 'interdisciplinary', 'interest', 'interested', 'interesting', 'interim', 'intermediate', 'intermittent', 'intern', 'internal', 'internally', 'international', 'internationally', 'internet', 'internship', 'interpersonal', 'interpret', 'interpretation', 'interpreted', 'interpreting', 'interval', 'intervention', 'interventional', 'interview', 'intrigued', 'introduce', 'introducing', 'introductory', 'intuitive', 'invent', 'inventing', 'inventive', 'inventory', 'invest', 'investigate', 'investigates', 'investigating', 'investigation', 'investigator', 'investing', 'investment', 'invite', 'involve', 'involved', 'involvement', 'involves', 'involving', 'issue', 'issuesPresent', 'item', 'iterative', 'jeopardize', 'job', 'join', 'joining', 'journal', 'journey', 'judge', 'judgment', 'junior', 'justification', 'k', 'keen', 'key', 'kick', 'kind', 'king', 'know', 'knowledge', 'knowledgeable', 'known', 'lab', 'label', 'labor', 'laboratory', 'lake', 'lambda', 'landmark', 'landscape', 'language', 'large', 'largely', 'largest', 'last', 'lasting', 'latent', 'later', 'latest', 'latitude', 'latter', 'launch', 'launched', 'law', 'lawfully', 'le', 'lead', 'leader', 'leadership', 'leading', 'leaf', 'learn', 'learner', 'learning', 'leased', 'least', 'leave', 'led', 'left', 'legacy', 'legal', 'legally', 'lens', 'lesion', 'let', 'letter', 'level', 'leverage', 'leveraging', 'liability', 'liaising', 'liaison', 'library', 'lie', 'lieu', 'life', 'lifecycle', 'lifestyle', 'lift', 'like', 'likelihood', 'likely', 'limit', 'limitation', 'limited', 'line', 'lineage', 'linear', 'link', 'linking', 'list', 'listed', 'listen', 'listener', 'listing', 'literature', 'little', 'live', 'loan', 'local', 'localization', 'located', 'location', 'locker', 'logical', 'logically', 'logistic', 'long', 'longitudinal', 'look', 'looking', 'loosely', 'love', 'low', 'lower', 'loyalty', 'machine', 'macro', 'mad', 'made', 'main', 'maintain', 'maintaining', 'maintains', 'maintenance', 'major', 'make', 'maker', 'making', 'manage', 'managed', 'management', 'manager', 'managing', 'manipulate', 'manipulating', 'manipulation', 'manipulationExperience', 'manner', 'mannerCreating', 'manufacturing', 'manuscript', 'many', 'mapping', 'margin', 'marital', 'market', 'marketing', 'marketplace', 'marshal', 'mart', 'massive', 'master', 'match', 'matching', 'material', 'maternity', 'math', 'mathematical', 'mathematician', 'mathematics', 'matplotlib', 'matrixed', 'matter', 'mature', 'maturity', 'maximize', 'may', 'mdm', 'mean', 'meaning', 'meaningful', 'measurable', 'measure', 'measurement', 'mechanism', 'mechanistic', 'medical', 'medicine', 'medium', 'meet', 'meeting', 'member', 'mental', 'mentality', 'mentor', 'mentored', 'mentoring', 'merge', 'merger', 'merit', 'message', 'metabolic', 'metabolite', 'metabolomics', 'metadata', 'method', 'methodological', 'methodology', 'methodsDoing', 'meticulous', 'metric', 'mgmt', 'micro', 'microbiome', 'microenvironment', 'microservices', 'might', 'migration', 'milestone', 'military', 'million', 'mind', 'mindset', 'mine', 'minimal', 'minimum', 'mining', 'miningSolid', 'minority', 'mission', 'mobile', 'modality', 'model', 'modeling', 'modelling', 'modellingAdvanced', 'moderate', 'moderately', 'modern', 'modified', 'module', 'molecular', 'molecule', 'moment', 'money', 'monitor', 'monitoring', 'month', 'motivated', 'motivation', 'move', 'moving', 'much', 'multi', 'multidisciplinary', 'multiple', 'multitask', 'multitude', 'multivariate', 'music', 'must', 'mutual', 'myriad', 'narrativeMeticulous', 'nation', 'national', 'natural', 'nature', 'navigating', 'naïve', 'near', 'nearly', 'necessarily', 'necessary', 'necessity', 'need', 'needed', 'neededSelecting', 'needing', 'negotiate', 'neighborhood', 'net', 'network', 'neural', 'neuroimaging', 'neuroscience', 'never', 'new', 'newly', 'next', 'nimble', 'noise', 'noncitizen', 'nonpartisan', 'nontechnical', 'normalization', 'normalized', 'normalizing', 'normally', 'note', 'notebook', 'nothing', 'novel', 'nucleic', 'number', 'numerate', 'numerical', 'numerous', 'nurture', 'nurturing', 'ob', 'object', 'objective', 'objectivesMaintain', 'observational', 'observe', 'obstacle', 'obtain', 'obtaining', 'occasional', 'occur', 'offHealth', 'offer', 'offered', 'offering', 'office', 'offline', 'often', 'oil', 'onboard', 'oncology', 'one', 'ongoing', 'online', 'onsite', 'open', 'opening', 'openly', 'openness', 'operate', 'operates', 'operating', 'operation', 'operational', 'operationalize', 'opinion', 'opportunity', 'optical', 'optimal', 'optimally', 'optimization', 'optimize', 'optimized', 'optimizing', 'option', 'oral', 'orchestration', 'orchestrator', 'order', 'ordering', 'orderliness', 'ordinance', 'organization', 'organizationDeliver', 'organizational', 'organize', 'organized', 'organizing', 'orientation', 'oriented', 'origin', 'original', 'others', 'otherwise', 'out', 'outcome', 'outgoing', 'outlier', 'outlined', 'outlook', 'outperform', 'outside', 'outsourced', 'outstanding', 'overall', 'overarching', 'overcoming', 'overseeing', 'oversight', 'owned', 'ownership', 'owning', 'pace', 'paced', 'package', 'paid', 'paired', 'panda', 'panel', 'paper', 'paradigm', 'parallel', 'parameter', 'parameterization', 'part', 'participant', 'participate', 'participates', 'participating', 'participation', 'particularly', 'partner', 'partnered', 'partnering', 'partnership', 'party', 'passion', 'passionate', 'past', 'path', 'pathology', 'pathway', 'patient', 'pattern', 'pay', 'payer', 'paying', 'payment', 'pedagogical', 'peer', 'penalized', 'people', 'per', 'perception', 'perfect', 'perform', 'performance', 'performanceWorking', 'performed', 'performing', 'period', 'periodically', 'permanent', 'permitted', 'person', 'personal', 'personality', 'personalization', 'personalize', 'personalized', 'personnel', 'perspective', 'pharmaceutical', 'pharmacist', 'pharmacology', 'phase', 'phenotyping', 'phone', 'physic', 'physical', 'physician', 'physiological', 'physiology', 'pick', 'picture', 'piece', 'pilot', 'piloting', 'pioneer', 'pioneering', 'pipeline', 'pivotal', 'place', 'plan', 'planet', 'planning', 'platform', 'play', 'player', 'playing', 'please', 'plot', 'plus', 'plusExperience', 'plusHigh', 'plusProficient', 'plusProven', 'point', 'poised', 'policy', 'political', 'population', 'portal', 'portfolio', 'position', 'positive', 'positively', 'posse', 'possessing', 'possibility', 'possible', 'post', 'postdoc', 'posting', 'potential', 'potentially', 'pound', 'power', 'powered', 'powerful', 'powering', 'practical', 'practice', 'pragmatic', 'preceptor', 'precise', 'predicting', 'prediction', 'predictive', 'preempt', 'pref', 'prefer', 'preferably', 'preference', 'preferred', 'pregnancy', 'preliminary', 'premier', 'preparation', 'prepare', 'prepared', 'preparing', 'preprocessing', 'prescribe', 'prescriptive', 'present', 'presentation', 'presenting', 'presently', 'pressure', 'preventing', 'prevention', 'previous', 'previously', 'price', 'pricing', 'pride', 'primarily', 'primary', 'principle', 'prior', 'prioritize', 'prioritized', 'prioritizing', 'priority', 'privacy', 'private', 'privately', 'proactive', 'proactively', 'probabilistic', 'probability', 'problem', 'procedure', 'proceeding', 'process', 'processed', 'processing', 'produce', 'produced', 'producing', 'product', 'production', 'productionExcellent', 'productionalizing', 'productively', 'productivity', 'profession', 'professional', 'professionally', 'professionnal', 'proficiency', 'proficient', 'proficiently', 'profile', 'profiling', 'profit', 'profitability', 'profitable', 'program', 'programmer', 'programming', 'progress', 'progression', 'progressive', 'prohibits', 'project', 'projection', 'projectsDemonstrates', 'projectsSoftware', 'promote', 'promotes', 'promoting', 'proof', 'property', 'proposal', 'propose', 'proposed', 'proposition', 'proprietary', 'prospect', 'prospecting', 'prosper', 'protected', 'proteomic', 'proteomics', 'protocol', 'prototype', 'prototyping', 'proud', 'proudly', 'proven', 'provide', 'provided', 'provider', 'provides', 'providing', 'proving', 'psychology', 'public', 'publication', 'publicly', 'publish', 'published', 'publishes', 'publishing', 'pull', 'purchase', 'purchasing', 'purpose', 'pursue', 'pursuing', 'pursuit', 'push', 'put', 'python', 'qPCR', 'qualification', 'qualified', 'qualifying', 'qualitative', 'quality', 'quantitative', 'quarterly', 'query', 'querying', 'quest', 'question', 'quick', 'quickly', 'quit', 'quo', 'rShiny', 'race', 'racial', 'radar', 'random', 'range', 'ranging', 'ranking', 'rapid', 'rapidly', 'rare', 'rate', 'rather', 'rating', 'reach', 'reaction', 'read', 'readable', 'reading', 'reagent', 'real', 'reality', 'realize', 'really', 'reason', 'reasonable', 'reasoning', 'receive', 'received', 'recent', 'recently', 'reciprocally', 'recognition', 'recognize', 'recognized', 'recommend', 'recommendation', 'recommender', 'reconstruction', 'record', 'recordkeeping', 'recovery', 'recruit', 'recruiting', 'recruitment', 'recurring', 'redefine', 'redefining', 'redesigned', 'reduce', 'reducing', 'reduction', 'reference', 'refine', 'refinement', 'reflect', 'reflects', 'regard', 'regarding', 'regardless', 'region', 'regional', 'regression', 'regular', 'regularized', 'regularly', 'regulated', 'regulation', 'regulatory', 'reinforcement', 'reinsurers', 'related', 'relating', 'relational', 'relationship', 'release', 'relentless', 'relevance', 'relevant', 'reliability', 'relies', 'religion', 'religious', 'relocation', 'reltio', 'rely', 'relying', 'remain', 'remaining', 'remote', 'remotely', 'rendered', 'renewable', 'renewal', 'repertoire', 'repetitive', 'replying', 'report', 'reporting', 'repository', 'reposted', 'represent', 'representation', 'representing', 'represents', 'reproduce', 'reproducibility', 'reproducible', 'repurposed', 'request', 'requested', 'require', 'required', 'requirement', 'requires', 'requisition', 'research', 'researcher', 'researching', 'reservist', 'reshape', 'reshaping', 'residence', 'resolution', 'resolve', 'resolving', 'resource', 'resourceful', 'respect', 'respond', 'responding', 'response', 'responsibility', 'responsible', 'rest', 'restore', 'result', 'resultsDesign', 'resume', 'retail', 'retain', 'retaliation', 'retention', 'retrain', 'return', 'reused', 'revenue', 'reversal', 'reverse', 'review', 'reviewed', 'reviewing', 'revolutionize', 'reward', 'rewarding', 'rich', 'richest', 'ride', 'right', 'rigor', 'rigorous', 'risk', 'roadmap', 'robust', 'rockstar', 'role', 'roll', 'room', 'root', 'rounded', 'routine', 'row', 'rule', 'run', 'running', 'runtime', 'rural', 'safe', 'safer', 'safety', 'salary', 'sale', 'sample', 'sampling', 'sanity', 'satellite', 'satisfaction', 'satisfy', 'saving', 'say', 'scalability', 'scalable', 'scale', 'scaling', 'scenario', 'scene', 'schedule', 'schema', 'scholar', 'scholarship', 'school', 'science', 'scientific', 'scientist', 'scope', 'screen', 'screening', 'scripting', 'scrum', 'seamlessly', 'search', 'searchable', 'searching', 'seasoned', 'secondary', 'sector', 'secure', 'security', 'see', 'seeing', 'seek', 'seeker', 'seeking', 'seen', 'segment', 'segmentation', 'segmenting', 'select', 'selected', 'selecting', 'selection', 'sell', 'send', 'senior', 'seniority', 'sense', 'sensing', 'sensitivity', 'sensor', 'sentiment', 'sequencing', 'serf', 'series', 'serious', 'serve', 'service', 'servicing', 'serving', 'set', 'setsKnowledge', 'setting', 'seventeen', 'several', 'sex', 'sexual', 'shape', 'shaping', 'share', 'shared', 'sharing', 'sharp', 'shift', 'ship', 'shipping', 'short', 'shot', 'shower', 'shown', 'shrinking', 'signal', 'signaling', 'significance', 'significant', 'silo', 'similar', 'simple', 'simplest', 'simplicity', 'simplifies', 'simplify', 'simplifying', 'simply', 'simulated', 'simulation', 'simultaneously', 'since', 'single', 'singular', 'sit', 'site', 'sits', 'situation', 'size', 'skill', 'skilled', 'skillsClear', 'skillsTeam', 'sleep', 'sleeve', 'slide', 'small', 'smart', 'smarter', 'social', 'socially', 'sociology', 'software', 'solid', 'solution', 'solve', 'solved', 'solver', 'solves', 'solving', 'someone', 'something', 'soon', 'sophisticated', 'sophomore', 'sound', 'source', 'sourcing', 'space', 'span', 'spanning', 'speak', 'speaker', 'speaking', 'spearheading', 'spec', 'special', 'specialist', 'specialization', 'specialized', 'specializing', 'specialty', 'specific', 'specifically', 'specification', 'specified', 'specifying', 'speech', 'speed', 'spend', 'spending', 'spirit', 'sponsor', 'sponsorship', 'sport', 'spot', 'spotting', 'spouse', 'spreadsheet', 'stack', 'staff', 'stage', 'stakeholder', 'standard', 'standardized', 'standardizing', 'start', 'started', 'starting', 'startup', 'state', 'stateful', 'statement', 'statistic', 'statistical', 'statistician', 'status', 'stay', 'stayed', 'staying', 'steadfast', 'step', 'stepwise', 'stewardship', 'still', 'stimulating', 'stone', 'storage', 'store', 'story', 'storytelling', 'straightforward', 'strategic', 'strategy', 'stratification', 'streaming', 'streamline', 'streamlining', 'street', 'strength', 'strict', 'strictly', 'stride', 'strive', 'strong', 'strongest', 'strongly', 'structure', 'structured', 'stuck', 'student', 'studied', 'study', 'studying', 'style', 'subject', 'submit', 'substantial', 'substantially', 'subtypes', 'succeed', 'succeeding', 'success', 'successful', 'successfully', 'suffer', 'suggest', 'suggestion', 'sum', 'summarize', 'summarized', 'summarizes', 'summarizing', 'summary', 'summer', 'superior', 'supervised', 'supervising', 'supervision', 'supervisor', 'supplement', 'supplier', 'supply', 'support', 'supported', 'supporting', 'supportive', 'sure', 'surface', 'surprise', 'surrounded', 'survey', 'survival', 'sustainable', 'sustained', 'sustaining', 'switch', 'symbiosis', 'symbolic', 'sync', 'synergy', 'synthesis', 'synthesize', 'synthesized', 'synthetic', 'system', 'systematic', 'systematization', 'systemsExtending', 'table', 'tackle', 'tackled', 'tactic', 'take', 'takeaway', 'taking', 'talent', 'talented', 'talk', 'talking', 'tandem', 'tangible', 'target', 'targeted', 'targeting', 'task', 'tasked', 'teach', 'teachable', 'teaching', 'team', 'teammate', 'teamwork', 'tech', 'technical', 'technique', 'techniquesData', 'technological', 'technologiesPractical', 'technologist', 'technology', 'technologyPrior', 'tedious', 'telecom', 'telephone', 'tell', 'temperature', 'ten', 'tenacious', 'term', 'territory', 'test', 'tested', 'testing', 'text', 'thanks', 'theoretical', 'theory', 'therapeutic', 'therapy', 'thereafter', 'therefore', 'thermodynamics', 'thesis', 'thing', 'think', 'thinker', 'thinking', 'third', 'thirty', 'thorough', 'though', 'thought', 'thoughtful', 'thousand', 'threat', 'three', 'threshold', 'thrive', 'thrives', 'throughout', 'tie', 'tight', 'time', 'timeline', 'timely', 'tissue', 'title', 'tobacco', 'today', 'together', 'tolerance', 'tool', 'toolbox', 'tooling', 'toolkits', 'toolsExperience', 'toolsets', 'top', 'topic', 'total', 'touch', 'toward', 'towards', 'toworking', 'track', 'tracking', 'traded', 'tradeoff', 'traditional', 'traffic', 'train', 'trained', 'trainee', 'training', 'trajectory', 'transaction', 'transfer', 'transform', 'transformation', 'transformative', 'transforming', 'transition', 'translate', 'translating', 'translational', 'transparency', 'traumatic', 'travel', 'travelling', 'treat', 'treatment', 'tree', 'tremendous', 'trend', 'trial', 'triangulate', 'trillion', 'troubleshoot', 'troubleshooting', 'trucking', 'truly', 'trust', 'trusted', 'truth', 'try', 'trying', 'tuberculosis', 'tumor', 'tuning', 'turn', 'turning', 'turnover', 'two', 'type', 'typical', 'typically', 'typo', 'u', 'ultimate', 'ultimately', 'unblock', 'uncover', 'undergoing', 'undergraduate', 'underlying', 'underpin', 'underpinnings', 'underrepresentation', 'underrepresented', 'understand', 'understanding', 'understands', 'undertake', 'undertaking', 'underway', 'underwriting', 'unexpected', 'unifying', 'unique', 'unit', 'university', 'unlawful', 'unless', 'unlock', 'unlocking', 'unparalleled', 'unprecedented', 'unrelated', 'unrivaled', 'unsolved', 'unstructured', 'unsupervised', 'untapped', 'unturned', 'unusual', 'upcoming', 'updating', 'upfront', 'uphold', 'upkeep', 'upload', 'uploading', 'upon', 'upsell', 'urban', 'urgency', 'urgent', 'us', 'usability', 'usage', 'use', 'used', 'useful', 'user', 'usercentric', 'using', 'utility', 'utilization', 'utilize', 'utilized', 'utilizing', 'utmost', 'valid', 'validate', 'validated', 'validating', 'validation', 'valuable', 'value', 'valued', 'variable', 'variance', 'variation', 'varied', 'variety', 'various', 'vary', 'varying', 'vast', 'vector', 'velocity', 'vendor', 'venture', 'venue', 'verbal', 'verbally', 'verifiable', 'verification', 'verify', 'verifying', 'versed', 'version', 'vertical', 'veteran', 'via', 'vibrant', 'view', 'virtual', 'virtualization', 'virtualized', 'virtually', 'visibility', 'vision', 'visionary', 'visit', 'visual', 'visualization', 'visualize', 'visualizing', 'visually', 'vital', 'vitro', 'voice', 'volatility', 'volume', 'vulnerability', 'waiting', 'wallet', 'want', 'warehouse', 'waste', 'water', 'way', 'weakness', 'weapon', 'wear', 'web', 'weblog', 'website', 'week', 'weekend', 'weight', 'welcome', 'well', 'wellbeing', 'wherever', 'whole', 'whose', 'wide', 'widely', 'wider', 'willing', 'willingness', 'win', 'wind', 'winning', 'within', 'without', 'word', 'work', 'worked', 'workflow', 'workforce', 'working', 'workload', 'workplace', 'workshop', 'workstreams', 'world', 'worldIs', 'worldwide', 'would', 'wrangling', 'write', 'writing', 'written', 'year', 'yes', 'yet', 'yield']\n"
]
],
[
[
"# 3) Use Scikit-Learn's CountVectorizer to get word counts for each listing.",
"_____no_output_____"
]
],
[
[
"df2[\"Job Description - Most Common\"] = df2[\"Job Description\"].apply(lambda v: FreqDist(v).most_common(20))\ndf2[\"Job Description - Most Common\"]",
"_____no_output_____"
]
],
[
[
"# 4) Visualize the most common word counts",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nfdist = FreqDist([inner for outer in df2[\"Job Description\"].values for inner in outer])\nfdist.plot(30, cumulative=False)\nplt.show()",
"_____no_output_____"
]
],
[
[
" # 5) Use Scikit-Learn's tfidfVectorizer to get a TF-IDF feature matrix",
"_____no_output_____"
]
],
[
[
"from sklearn.feature_extraction.text import CountVectorizer\n\nvectorizer = CountVectorizer(stop_words='english')\ntfidf = TfidfVectorizer(ngram_range=(1,1), max_features=20)\nbag_of_words = tfidf.fit_transform([\" \".join(v) for v in df2[\"Job Description\"].values])\n\ndf_vec = pd.DataFrame(bag_of_words.toarray(), columns=tfidf.get_feature_names())\ndf_vec.head()",
"_____no_output_____"
]
],
[
[
"## Stretch Goals\n\n - Scrape Job Listings for the job title \"Data Analyst\". How do these differ from Data Scientist Job Listings\n - Try and identify requirements for experience specific technologies that are asked for in the job listings. How are those distributed among the job listings?\n - Use a clustering algorithm to cluster documents by their most important terms. Do the clusters reveal any common themes?\n - **Hint:** K-means might not be the best algorithm for this. Do a little bit of research to see what might be good for this. Also, remember that algorithms that depend on Euclidean distance break down with high dimensional data.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d085f3b4f8579b757e6f47c3c952c25ef1e33f10 | 203,689 | ipynb | Jupyter Notebook | Test_karl/material/session_12/lecture_12.ipynb | karlbindslev/sds_group29 | 6f5263b08b35f35374b7f01b31a0e90d1cf4d53e | [
"MIT",
"Unlicense"
] | null | null | null | Test_karl/material/session_12/lecture_12.ipynb | karlbindslev/sds_group29 | 6f5263b08b35f35374b7f01b31a0e90d1cf4d53e | [
"MIT",
"Unlicense"
] | null | null | null | Test_karl/material/session_12/lecture_12.ipynb | karlbindslev/sds_group29 | 6f5263b08b35f35374b7f01b31a0e90d1cf4d53e | [
"MIT",
"Unlicense"
] | null | null | null | 166.141109 | 82,000 | 0.90378 | [
[
[
"# Code stuff - not slides!",
"_____no_output_____"
]
],
[
[
"%run ../ML_plots.ipynb",
"ERROR:root:File `'../ML_plots.ipynb.py'` not found.\n"
]
],
[
[
"# Session 12:\n## Supervised learning, part 1\n\n*Andreas Bjerre-Nielsen*",
"_____no_output_____"
],
[
"## Agenda\n1. [Modelling data](#Modelling-data)\n1. [A familiar regression model](#A-familiar-regression-model)\n1. [The curse of overfitting](#The-curse-of-overfitting)\n1. [Important details](#Implementation-details)",
"_____no_output_____"
],
[
"## Vaaaamos",
"_____no_output_____"
]
],
[
[
"import warnings\nfrom sklearn.exceptions import ConvergenceWarning\nwarnings.filterwarnings(action='ignore', category=ConvergenceWarning)\n\nimport matplotlib.pyplot as plt\nimport numpy as np \nimport pandas as pd \nimport seaborn as sns\n\nplt.style.use('default') # set style (colors, background, size, gridlines etc.)\nplt.rcParams['figure.figsize'] = 10, 4 # set default size of plots\nplt.rcParams.update({'font.size': 18})",
"_____no_output_____"
]
],
[
[
"## Supervised problems (1)\n*How do we distinguish between problems?*",
"_____no_output_____"
]
],
[
[
"f_identify_question",
"_____no_output_____"
]
],
[
[
"## Supervised problems (2)\n*The two canonical problems*",
"_____no_output_____"
]
],
[
[
"f_identify_answer",
"_____no_output_____"
]
],
[
[
"## Supervised problems (3)\n*Which models have we seen for classification?*\n\n- .\n\n- .\n\n- .",
"_____no_output_____"
],
[
"# Modelling data",
"_____no_output_____"
],
[
"## Model complexity (1)\n*What does a model of low complexity look like?*",
"_____no_output_____"
]
],
[
[
"f_complexity[0]",
"_____no_output_____"
]
],
[
[
"## Model complexity (2)\n*What does medium model complexity look like?*",
"_____no_output_____"
]
],
[
[
"f_complexity[1]",
"_____no_output_____"
]
],
[
[
"## Model complexity (3)\n*What does high model complexity look like?*",
"_____no_output_____"
]
],
[
[
"f_complexity[2]",
"_____no_output_____"
]
],
[
[
"## Model fitting (1)\n*Quiz (1 min.): Which model fitted the data best?*",
"_____no_output_____"
]
],
[
[
"f_bias_var['regression'][2]",
"_____no_output_____"
]
],
[
[
"## Model fitting (2)\n*What does underfitting and overfitting look like for classification?*",
"_____no_output_____"
]
],
[
[
"f_bias_var['classification'][2]",
"_____no_output_____"
]
],
[
[
"## Two agendas (1)\n\nWhat are the objectives of empirical research? \n\n1. *causation*: what is the effect of a particular variable on an outcome? \n2. *prediction*: find some function that provides a good prediction of $y$ as a function of $x$",
"_____no_output_____"
],
[
"## Two agendas (2)\n\nHow might we express the agendas in a model?\n\n$$ y = \\alpha + \\beta x + \\varepsilon $$\n\n- *causation*: interested in $\\hat{\\beta}$ \n\n- *prediction*: interested in $\\hat{y}$ \n",
"_____no_output_____"
],
[
"## Two agendas (3)\n\nMight these two agendas be related at a deeper level? \n\nCan prediction quality inform us about how to make causal models?",
"_____no_output_____"
],
[
"# A familiar regression model",
"_____no_output_____"
],
[
"## Estimation (1)\n*Do we know already some ways to estimate regression models?*",
"_____no_output_____"
],
[
"- Social scientists know all about the Ordinary Least Squares (OLS).\n - OLS estimate both parameters and their standard deviation.\n - Is best linear unbiased estimator under regularity conditions. \n ",
"_____no_output_____"
],
[
"*How is OLS estimated?*",
"_____no_output_____"
],
[
"- $\\beta=(\\textbf{X}^T\\textbf{X})^{-1}\\textbf{X}^T\\textbf{y}$\n- computation requires non perfect multicollinarity.",
"_____no_output_____"
],
[
"## Estimation (2)\n*How might we estimate a linear regression model?*",
"_____no_output_____"
],
[
"- first order method (e.g. gradient descent)\n- second order method (e.g. Newton-Raphson)",
"_____no_output_____"
],
[
"*So what the hell was gradient descent?*",
"_____no_output_____"
],
[
"- compute errors, multiply with features and update",
"_____no_output_____"
],
[
"## Estimation (3)\n*Can you explain that in details?*",
"_____no_output_____"
],
[
"- Yes, like with Adaline, we minimize the sum of squared errors (SSE): \n\\begin{align}SSE&=\\boldsymbol{e}^{T}\\boldsymbol{e}\\\\\\boldsymbol{e}&=\\textbf{y}-\\textbf{X}\\textbf{w}\\end{align}",
"_____no_output_____"
]
],
[
[
"X = np.random.normal(size=(3,2))\ny = np.random.normal(size=(3))\nw = np.random.normal(size=(3))\n\ne = y-(w[0]+X.dot(w[1:]))\nSSE = e.T.dot(e)",
"_____no_output_____"
]
],
[
[
"## Estimation (4)\n*And what about the updating..? What is it something about the first order deritative?*",
"_____no_output_____"
],
[
"\\begin{align}\n\\frac{\\partial SSE}{\\partial\\hat{w}}=&\\textbf{X}^T\\textbf{e},\\\\\n \\Delta\\hat{w}=&\\eta\\cdot\\textbf{X}^T\\textbf{e}=\\eta\\cdot\\textbf{X}^T(\\textbf{y}-\\hat{\\textbf{y}})\n\\end{align}",
"_____no_output_____"
]
],
[
[
"eta = 0.001 # learning rate\nfod = X.T.dot(e)\nupdate_vars = eta*fod\nupdate_bias = eta*e.sum()",
"_____no_output_____"
]
],
[
[
"## Estimation (5)\n*What might some advantages be relative to OLS?*\n\n- Works despite high multicollinarity\n- Speed\n - OLS has $\\mathcal{O}(K^2N)$ computation time ([read more](https://math.stackexchange.com/questions/84495/computational-complexity-of-least-square-regression-operation))\n - Quadratic scaling in number of variables ($K$).\n - Stochastic gradient descent\n - Likely to converge faster with many observations ($N$)",
"_____no_output_____"
],
[
"## Fitting a polynomial (1)\nPolyonomial: $f(x) = 2+8*x^4$\n\nTry models of increasing order polynomials. \n\n- Split data into train and test (50/50)\n\n\n- For polynomial order 0 to 9:\n - Iteration n: $y = \\sum_{k=0}^{n}(\\beta_k\\cdot x^k)+\\varepsilon$.\n - Estimate order n model on training data\n - Evaluate with on test data with RMSE: \n - $log RMSE = \\log (\\sqrt{MSE})$ ",
"_____no_output_____"
],
[
"## Fitting a polynomial (2)\nWe generate samples of data from true model.",
"_____no_output_____"
]
],
[
[
"from sklearn.preprocessing import PolynomialFeatures\nfrom sklearn.linear_model import LinearRegression\n\ndef true_fct(X):\n return 2+X**4\n\nn_samples = 25\nn_degrees = 15\n\nnp.random.seed(0)\n\nX_train = np.random.normal(size=(n_samples,1))\ny_train = true_fct(X_train).reshape(-1) + np.random.randn(n_samples) \n\nX_test = np.random.normal(size=(n_samples,1))\ny_test = true_fct(X_test).reshape(-1) + np.random.randn(n_samples)",
"_____no_output_____"
]
],
[
[
"## Fitting a polynomial (3)\nWe estimate the polynomials",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import mean_squared_error as mse\n\ntest_mse = []\ntrain_mse = []\nparameters = []\ndegrees = range(n_degrees+1)\n\nfor p in degrees:\n X_train_p = PolynomialFeatures(degree=p).fit_transform(X_train)\n X_test_p = PolynomialFeatures(degree=p).fit_transform(X_train)\n reg = LinearRegression().fit(X_train_p, y_train)\n train_mse += [mse(reg.predict(X_train_p),y_train)] \n test_mse += [mse(reg.predict(X_test_p),y_test)] \n parameters.append(reg.coef_)",
"_____no_output_____"
]
],
[
[
"## Fitting a polynomial (4)\n*So what happens to the model performance in- and out-of-sample?*",
"_____no_output_____"
]
],
[
[
"degree_index = pd.Index(degrees,name='Polynomial degree ~ model complexity')\nax = pd.DataFrame({'Train set':train_mse, 'Test set':test_mse})\\\n .set_index(degree_index)\\\n .plot(figsize=(10,4))\nax.set_ylabel('Mean squared error')",
"_____no_output_____"
]
],
[
[
"## Fitting a polynomial (4)\n*Why does it go wrong?*\n- more spurious parameters\n- the coefficient size increases",
"_____no_output_____"
],
[
"## Fitting a polynomial (5)\n*What do you mean coefficient size increase?*",
"_____no_output_____"
]
],
[
[
"order_idx = pd.Index(range(n_degrees+1),name='Polynomial order')\nax = pd.DataFrame(parameters,index=order_idx)\\\n.abs().mean(1)\\\n.plot(logy=True)\nax.set_ylabel('Mean parameter size')",
"_____no_output_____"
]
],
[
[
"## Fitting a polynomial (6)\n*How else could we visualize this problem?*",
"_____no_output_____"
]
],
[
[
"f_bias_var['regression'][2]",
"_____no_output_____"
]
],
[
[
"# The curse of overfitting",
"_____no_output_____"
],
[
"## Looking for a remedy\n*How might we solve the overfitting problem?*\n\nBy reducing\n- the number of variables\n- the coefficient size of variables ",
"_____no_output_____"
],
[
"## Regularization (1)\n\n*Why do we regularize?*",
"_____no_output_____"
],
[
"- To mitigate overfitting > better model predictions",
"_____no_output_____"
],
[
"*How do we regularize?*",
"_____no_output_____"
],
[
"- We make models which are less complex:\n - reducing the **number** of coefficient;\n - reducing the **size** of the coefficients.",
"_____no_output_____"
],
[
"## Regularization (2)\n\n*What does regularization look like?*",
"_____no_output_____"
],
[
"We add a penalty term our optimization procedure:\n \n$$ \\text{arg min}_\\beta \\, \\underset{\\text{MSE}}{\\underbrace{E[(y_0 - \\hat{f}(x_0))^2]}} + \\underset{\\text{penalty}}{\\underbrace{\\lambda \\cdot R(\\beta)}}$$\n\nIntroduction of penalties implies that increased model complexity has to be met with high increases precision of estimates.",
"_____no_output_____"
],
[
"## Regularization (3)\n\n*What are some used penalty functions?*",
"_____no_output_____"
],
[
"The two most common penalty functions are L1 and L2 regularization.\n\n- L1 regularization (***Lasso***): $R(\\beta)=\\sum_{j=1}^{p}|\\beta_j|$ \n - Makes coefficients sparse, i.e. selects variables by removing some (if $\\lambda$ is high)\n \n \n- L2 regularization (***Ridge***): $R(\\beta)=\\sum_{j=1}^{p}\\beta_j^2$\n - Reduce coefficient size\n - Fast due to analytical solution\n \n*To note:* The *Elastic Net* uses a combination of L1 and L2 regularization.",
"_____no_output_____"
],
[
"## Regularization (4)\n\n*How the Lasso (L1 reg.) deviates from OLS*\n\n<img src='http://rasbt.github.io/mlxtend/user_guide/general_concepts/regularization-linear_files/l1.png'>",
"_____no_output_____"
],
[
"## Regularization (5)\n\n*How the Ridge regression (L2 reg.) deviates from OLS*\n\n<img src='http://rasbt.github.io/mlxtend/user_guide/general_concepts/regularization-linear_files/l2.png'>",
"_____no_output_____"
],
[
"## Regularization (6)\n\n*How might we describe the $\\lambda$ of Lasso and Ridge?*\n\nThese are hyperparameters that we can optimize over. \n\n- More about this tomorrow.",
"_____no_output_____"
],
[
"# Implementation details",
"_____no_output_____"
],
[
"## The devils in the details (1)\n\n*So we just run regularization?*",
"_____no_output_____"
],
[
"# NO",
"_____no_output_____"
],
[
"We need to rescale our features:\n- convert to zero mean: \n- standardize to unit std: \n\nCompute in Python:\n- option 1: `StandardScaler` in `sklearn` \n- option 2: `(X - np.mean(X)) / np.std(X)`\n\n",
"_____no_output_____"
],
[
"## The devils in the details (2)\n*So we just scale our test and train?*",
"_____no_output_____"
],
[
"# NO",
"_____no_output_____"
],
[
"Fit to the distribution in the training data first, then rescale train and test! See more [here](https://stats.stackexchange.com/questions/174823/how-to-apply-standardization-normalization-to-train-and-testset-if-prediction-i).",
"_____no_output_____"
],
[
"## The devils in the details (3)\n*So we just rescale before using polynomial features?*",
"_____no_output_____"
],
[
"# NO",
"_____no_output_____"
],
[
"Otherwise the interacted varaibles are not gaussian distributed.",
"_____no_output_____"
],
[
"## The devils in the details (4)\n*Does sklearn's `PolynomialFeatures` work for more than variable?*",
"_____no_output_____"
],
[
"# YES!",
"_____no_output_____"
],
[
"# The end\n[Return to agenda](#Agenda)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d085ff6fe6e447af779033db7d66db243949db0f | 8,431 | ipynb | Jupyter Notebook | DeepMol/src/standardization_test.ipynb | PedromfRibeiro/AA2-Embeddings | 9d186fe2b8ed7da2cb150dbbf533ed0027aefc6d | [
"MIT"
] | 9 | 2021-07-10T18:36:04.000Z | 2022-03-07T12:40:54.000Z | src/tests/standardization_test.ipynb | BioSystemsUM/DeepMol | 62904fac46f62ec6231543891efbe52ac7ea1cf1 | [
"BSD-2-Clause"
] | null | null | null | src/tests/standardization_test.ipynb | BioSystemsUM/DeepMol | 62904fac46f62ec6231543891efbe52ac7ea1cf1 | [
"BSD-2-Clause"
] | null | null | null | 32.426923 | 213 | 0.583086 | [
[
[
"#conda install -c conda-forge chembl_structure_pipeline",
"_____no_output_____"
],
[
"from loaders.Loaders import CSVLoader\nfrom standardizer.BasicStandardizer import BasicStandardizer\nfrom standardizer.CustomStandardizer import CustomStandardizer\nfrom standardizer.ChEMBLStandardizer import ChEMBLStandardizer",
"_____no_output_____"
],
[
"#Load Dataset\ndataset = CSVLoader(dataset_path='preprocessed_dataset_wfoodb.csv', \n mols_field='Smiles', \n labels_fields='Class', \n id_field='ID')\ndataset = dataset.create_dataset()\ndataset.get_shape()",
"Mols_shape: 23290\nFeatures_shape: X not defined!\nLabels_shape: (23290,)\n"
],
[
"print(dataset.mols)",
"['Cc1cc2-c3c(O)cc(cc3OC3(Oc4cc(O)ccc4-c(c1O)c23)c1ccc(O)cc1O)-c1cc2cccc(O)c2o1'\n 'CC(=O)OC[C@H]1O[C@@H](Oc2cc3c(O)cc(O)cc3[o+]c2-c2ccc(O)c(O)c2)[C@H](O)[C@@H](O)[C@H]1O'\n 'O[C@@H]1[C@@H](COC(=O)CCC(O)=O)O[C@@H](Oc2cc3c(O)cc(O)cc3[o+]c2-c2ccc(O)c(O)c2)[C@H](O)[C@H]1O'\n ... 'CCCOC(=O)CC(C)C' 'CC(C)=CCCC(C)=O'\n 'CC(=O)OCC(C)=CCCC1(C)C2CC3C(C2)C13C']\n"
],
[
"standardizer = BasicStandardizer().standardize(dataset)\n\n#standardizer = CustomStandardizer().standardize(dataset)\n\n#heavy_standardisation = {\n# 'REMOVE_ISOTOPE': True,\n# 'NEUTRALISE_CHARGE': True,\n# 'REMOVE_STEREO': True,\n# 'KEEP_BIGGEST': True,\n# 'ADD_HYDROGEN': True,\n# 'KEKULIZE': False,\n# 'NEUTRALISE_CHARGE_LATE': True}\n#standardizer = CustomStandardizer(heavy_standardisation).standardize(dataset)\n\n#standardizer = ChEMBLStandardizer().standardize(dataset)\n",
"Standardizing datapoint 0\nStandardizing datapoint 1000\nStandardizing datapoint 2000\nStandardizing datapoint 3000\nStandardizing datapoint 4000\nStandardizing datapoint 5000\nStandardizing datapoint 6000\nerror in standardizing smile: O=[Cl]=O\nFailed to featurize datapoint 6257, O=[Cl]=O. Appending non standardized mol\nException message: Python argument types in\n rdkit.Chem.rdmolfiles.MolToSmiles(numpy.str_)\ndid not match C++ signature:\n MolToSmiles(RDKit::ROMol mol, bool isomericSmiles=True, bool kekuleSmiles=False, int rootedAtAtom=-1, bool canonical=True, bool allBondsExplicit=False, bool allHsExplicit=False, bool doRandom=False)\n"
],
[
"print(dataset.mols)",
"['Cc1cc2c3c(c1O)-c1ccc(O)cc1OC3(c1ccc(O)cc1O)Oc1cc(-c3cc4cccc(O)c4o3)cc(O)c1-2'\n 'CC(=O)OCC1OC(Oc2cc3c(O)cc(O)cc3[o+]c2-c2ccc(O)c(O)c2)C(O)C(O)C1O'\n 'O=C(O)CCC(=O)OCC1OC(Oc2cc3c(O)cc(O)cc3[o+]c2-c2ccc(O)c(O)c2)C(O)C(O)C1O'\n ... 'CCCOC(=O)CC(C)C' 'CC(=O)CCC=C(C)C'\n 'CC(=O)OCC(C)=CCCC1(C)C2CC3C(C2)C31C']\n"
],
[
"dataset.get_shape()",
"Mols_shape: 23290\nFeatures_shape: X not defined!\nLabels_shape: (23290,)\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d085ff9c5ab66fff456e459d58c80e29344d3f07 | 650,598 | ipynb | Jupyter Notebook | WGAN.ipynb | tommyliu9/CSC413-Generative-Model-Comparisons | 45f1c50f0f5bb181313a085d49d7db4d9c61168e | [
"MIT"
] | null | null | null | WGAN.ipynb | tommyliu9/CSC413-Generative-Model-Comparisons | 45f1c50f0f5bb181313a085d49d7db4d9c61168e | [
"MIT"
] | null | null | null | WGAN.ipynb | tommyliu9/CSC413-Generative-Model-Comparisons | 45f1c50f0f5bb181313a085d49d7db4d9c61168e | [
"MIT"
] | null | null | null | 38.044442 | 1,698 | 0.605666 | [
[
[
"# Colab FAQ\n\nFor some basic overview and features offered in Colab notebooks, check out: [Overview of Colaboratory Features](https://colab.research.google.com/notebooks/basic_features_overview.ipynb)\n\nYou need to use the colab GPU for this assignmentby selecting:\n\n> **Runtime** → **Change runtime type** → **Hardware Accelerator: GPU**",
"_____no_output_____"
],
[
"## Setup PyTorch\nAll files are stored at /content/csc421/a4/ folder\n",
"_____no_output_____"
]
],
[
[
"######################################################################\n# Setup python environment and change the current working directory\n######################################################################\n!pip install torch torchvision\n!pip install imageio\n\n!pip install matplotlib\n\n%mkdir -p ./content/csc413/a4/\n%cd ./content/csc413/a4\n\n",
"Requirement already satisfied: imageio in /home/tommy/miniconda3/lib/python3.8/site-packages (2.9.0)\nRequirement already satisfied: pillow in /home/tommy/miniconda3/lib/python3.8/site-packages (from imageio) (8.1.2)\nRequirement already satisfied: numpy in /home/tommy/miniconda3/lib/python3.8/site-packages (from imageio) (1.19.2)\nRequirement already satisfied: matplotlib in /home/tommy/miniconda3/lib/python3.8/site-packages (3.3.4)\nRequirement already satisfied: numpy>=1.15 in /home/tommy/miniconda3/lib/python3.8/site-packages (from matplotlib) (1.19.2)\nRequirement already satisfied: kiwisolver>=1.0.1 in /home/tommy/miniconda3/lib/python3.8/site-packages (from matplotlib) (1.3.1)\nRequirement already satisfied: python-dateutil>=2.1 in /home/tommy/miniconda3/lib/python3.8/site-packages (from matplotlib) (2.8.1)\nRequirement already satisfied: cycler>=0.10 in /home/tommy/miniconda3/lib/python3.8/site-packages (from matplotlib) (0.10.0)\nRequirement already satisfied: pillow>=6.2.0 in /home/tommy/miniconda3/lib/python3.8/site-packages (from matplotlib) (8.1.2)\nRequirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.3 in /home/tommy/miniconda3/lib/python3.8/site-packages (from matplotlib) (2.4.7)\nRequirement already satisfied: six in /home/tommy/miniconda3/lib/python3.8/site-packages (from cycler>=0.10->matplotlib) (1.15.0)\n/mnt/c/Users/superhardcocksgamerp/Documents/Github/Final-Project/content/csc413/a4\n"
]
],
[
[
"# Helper code",
"_____no_output_____"
],
[
"## Utility functions",
"_____no_output_____"
]
],
[
[
"import os\n\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nimport torch\nfrom torch import nn\nfrom torch.nn import Parameter\nimport torch.nn.functional as F\nimport torch.optim as optim\nfrom torch.autograd import Variable\nfrom torch.utils.data import DataLoader\nfrom torchvision import datasets\nfrom torchvision import transforms\n\nfrom six.moves.urllib.request import urlretrieve\nimport tarfile\n\nimport imageio\nfrom urllib.error import URLError\nfrom urllib.error import HTTPError\nos.environ[\"CUDA_VISABLE_DEVICES\"] = \"GPU_ID\"\n\ndef get_file(fname,\n origin,\n untar=False,\n extract=False,\n archive_format='auto',\n cache_dir='data'):\n datadir = os.path.join(cache_dir)\n if not os.path.exists(datadir):\n os.makedirs(datadir)\n\n if untar:\n untar_fpath = os.path.join(datadir, fname)\n fpath = untar_fpath + '.tar.gz'\n else:\n fpath = os.path.join(datadir, fname)\n\n print(fpath)\n if not os.path.exists(fpath):\n print('Downloading data from', origin)\n\n error_msg = 'URL fetch failure on {}: {} -- {}'\n try:\n try:\n urlretrieve(origin, fpath)\n except URLError as e:\n raise Exception(error_msg.format(origin, e.errno, e.reason))\n except HTTPError as e:\n raise Exception(error_msg.format(origin, e.code, e.msg))\n except (Exception, KeyboardInterrupt) as e:\n if os.path.exists(fpath):\n os.remove(fpath)\n raise\n\n if untar:\n if not os.path.exists(untar_fpath):\n print('Extracting file.')\n with tarfile.open(fpath) as archive:\n archive.extractall(datadir)\n return untar_fpath\n\n return fpath\n\n\nclass AttrDict(dict):\n def __init__(self, *args, **kwargs):\n super(AttrDict, self).__init__(*args, **kwargs)\n self.__dict__ = self\n\n \ndef to_var(tensor, cuda=True):\n \"\"\"Wraps a Tensor in a Variable, optionally placing it on the GPU.\n\n Arguments:\n tensor: A Tensor object.\n cuda: A boolean flag indicating whether to use the GPU.\n\n Returns:\n A Variable object, on the GPU if cuda==True.\n \"\"\"\n if cuda:\n return Variable(tensor.cuda())\n else:\n return Variable(tensor)\n\n \ndef to_data(x):\n \"\"\"Converts variable to numpy.\"\"\"\n if torch.cuda.is_available():\n x = x.cpu()\n return x.data.numpy()\n\n\ndef create_dir(directory):\n \"\"\"Creates a directory if it doesn't already exist.\n \"\"\"\n if not os.path.exists(directory):\n os.makedirs(directory)\n\n\ndef gan_checkpoint(iteration, G, D, opts):\n \"\"\"Saves the parameters of the generator G and discriminator D.\n \"\"\"\n G_path = os.path.join(opts.checkpoint_dir, 'G.pkl')\n D_path = os.path.join(opts.checkpoint_dir, 'D.pkl')\n torch.save(G.state_dict(), G_path)\n torch.save(D.state_dict(), D_path)\n\ndef load_checkpoint(opts):\n \"\"\"Loads the generator and discriminator models from checkpoints.\n \"\"\"\n G_path = os.path.join(opts.load, 'G.pkl')\n D_path = os.path.join(opts.load, 'D_.pkl')\n\n G = DCGenerator(noise_size=opts.noise_size, conv_dim=opts.g_conv_dim, spectral_norm=opts.spectral_norm)\n D = DCDiscriminator(conv_dim=opts.d_conv_dim)\n\n G.load_state_dict(torch.load(G_path, map_location=lambda storage, loc: storage))\n D.load_state_dict(torch.load(D_path, map_location=lambda storage, loc: storage))\n\n if torch.cuda.is_available():\n G.cuda()\n D.cuda()\n print('Models moved to GPU.')\n\n return G, D\n\n\ndef merge_images(sources, targets, opts):\n \"\"\"Creates a grid consisting of pairs of columns, where the first column in\n each pair contains images source images and the second column in each pair\n contains images generated by the CycleGAN from the corresponding images in\n the first column.\n \"\"\"\n _, _, h, w = sources.shape\n row = int(np.sqrt(opts.batch_size))\n merged = np.zeros([3, row * h, row * w * 2])\n for (idx, s, t) in (zip(range(row ** 2), sources, targets, )):\n i = idx // row\n j = idx % row\n merged[:, i * h:(i + 1) * h, (j * 2) * h:(j * 2 + 1) * h] = s\n merged[:, i * h:(i + 1) * h, (j * 2 + 1) * h:(j * 2 + 2) * h] = t\n return merged.transpose(1, 2, 0)\n\n\ndef generate_gif(directory_path, keyword=None):\n images = []\n for filename in sorted(os.listdir(directory_path)):\n if filename.endswith(\".png\") and (keyword is None or keyword in filename):\n img_path = os.path.join(directory_path, filename)\n print(\"adding image {}\".format(img_path))\n images.append(imageio.imread(img_path))\n\n if keyword:\n imageio.mimsave(\n os.path.join(directory_path, 'anim_{}.gif'.format(keyword)), images)\n else:\n imageio.mimsave(os.path.join(directory_path, 'anim.gif'), images)\n\n\ndef create_image_grid(array, ncols=None):\n \"\"\"\n \"\"\"\n num_images, channels, cell_h, cell_w = array.shape\n if not ncols:\n ncols = int(np.sqrt(num_images))\n nrows = int(np.math.floor(num_images / float(ncols)))\n result = np.zeros((cell_h * nrows, cell_w * ncols, channels), dtype=array.dtype)\n for i in range(0, nrows):\n for j in range(0, ncols):\n result[i * cell_h:(i + 1) * cell_h, j * cell_w:(j + 1) * cell_w, :] = array[i * ncols + j].transpose(1, 2,\n 0)\n\n if channels == 1:\n result = result.squeeze()\n return result\n\n\ndef gan_save_samples(G, fixed_noise, iteration, opts):\n generated_images = G(fixed_noise)\n generated_images = to_data(generated_images)\n\n grid = create_image_grid(generated_images)\n\n # merged = merge_images(X, fake_Y, opts)\n path = os.path.join(opts.sample_dir, 'sample-{:06d}.png'.format(iteration))\n imageio.imwrite(path, grid)\n print('Saved {}'.format(path))",
"_____no_output_____"
]
],
[
[
"## Data loader",
"_____no_output_____"
]
],
[
[
"def get_emoji_loader(emoji_type, opts):\n \"\"\"Creates training and test data loaders.\n \"\"\"\n transform = transforms.Compose([\n transforms.Scale(opts.image_size),\n transforms.ToTensor(),\n transforms.Normalize((0.5, ), (0.5, )),\n ])\n\n train_path = os.path.join('data/emojis', emoji_type)\n test_path = os.path.join('data/emojis', 'Test_{}'.format(emoji_type))\n\n train_dataset = datasets.ImageFolder(train_path, transform)\n test_dataset = datasets.ImageFolder(test_path, transform)\n\n\n train_dloader = DataLoader(dataset=train_dataset, batch_size=opts.batch_size, shuffle=True, num_workers=opts.num_workers)\n test_dloader = DataLoader(dataset=test_dataset, batch_size=opts.batch_size, shuffle=False, num_workers=opts.num_workers)\n\n return train_dloader, test_dloader\n\ndef get_emnist_loader(emnist_type, opts):\n transform = transforms.Compose([\n transforms.Scale(opts.image_size),\n transforms.ToTensor(),\n transforms.Normalize((0.5), (0.5)),\n ])\n train = datasets.EMNIST(\".\", split=emnist_type,train = True, download = True, transform= transform)\n test = datasets.EMNIST(\".\", split=emnist_type,train = False, download = True, transform = transform)\n \n train_dloader = DataLoader(dataset=train, batch_size=opts.batch_size, shuffle=True,num_workers=opts.num_workers)\n test_dloader = DataLoader(dataset=test, batch_size=opts.batch_size, shuffle=False,num_workers=opts.num_workers)\n return train_dloader, test_dloader\n\n\n\n",
"_____no_output_____"
]
],
[
[
"## Training and evaluation code",
"_____no_output_____"
]
],
[
[
"def print_models(G_XtoY, G_YtoX, D_X, D_Y):\n \"\"\"Prints model information for the generators and discriminators.\n \"\"\"\n print(\" G \")\n print(\"---------------------------------------\")\n print(G_XtoY)\n print(\"---------------------------------------\")\n\n print(\" D \")\n print(\"---------------------------------------\")\n print(D_X)\n print(\"---------------------------------------\")\n\n\ndef create_model(opts):\n \"\"\"Builds the generators and discriminators.\n \"\"\"\n ### GAN\n G = DCGenerator(noise_size=opts.noise_size, conv_dim=opts.g_conv_dim, spectral_norm=opts.spectral_norm)\n D = DCDiscriminator(conv_dim=opts.d_conv_dim, spectral_norm=opts.spectral_norm)\n\n print_models(G, None, D, None)\n\n if torch.cuda.is_available():\n G.cuda()\n D.cuda()\n print('Models moved to GPU.')\n return G, D\n\ndef train(opts):\n \"\"\"Loads the data, creates checkpoint and sample directories, and starts the training loop.\n \"\"\"\n\n # Create train and test dataloaders for images from the two domains X and Y\n dataloader_X, test_dataloader_X = get_emnist_loader(opts.X, opts=opts)\n \n # Create checkpoint and sample directories\n create_dir(opts.checkpoint_dir)\n create_dir(opts.sample_dir)\n\n # Start training\n G, D = gan_training_loop(dataloader_X, test_dataloader_X, opts)\n return G, D\n\ndef print_opts(opts):\n \"\"\"Prints the values of all command-line arguments.\n \"\"\"\n print('=' * 80)\n print('Opts'.center(80))\n print('-' * 80)\n for key in opts.__dict__:\n if opts.__dict__[key]:\n print('{:>30}: {:<30}'.format(key, opts.__dict__[key]).center(80))\n print('=' * 80)\n",
"_____no_output_____"
]
],
[
[
"# Your code for generators and discriminators",
"_____no_output_____"
],
[
"## Helper modules",
"_____no_output_____"
]
],
[
[
"def sample_noise(batch_size, dim):\n \"\"\"\n Generate a PyTorch Tensor of uniform random noise.\n\n Input:\n - batch_size: Integer giving the batch size of noise to generate.\n - dim: Integer giving the dimension of noise to generate.\n\n Output:\n - A PyTorch Tensor of shape (batch_size, dim, 1, 1) containing uniform\n random noise in the range (-1, 1).\n \"\"\"\n return to_var(torch.rand(batch_size, dim) * 2 - 1).unsqueeze(2).unsqueeze(3)\n \n\ndef upconv(in_channels, out_channels, kernel_size, stride=2, padding=2, batch_norm=True, spectral_norm=False):\n \"\"\"Creates a upsample-and-convolution layer, with optional batch normalization.\n \"\"\"\n layers = []\n if stride>1:\n layers.append(nn.Upsample(scale_factor=stride))\n conv_layer = nn.Conv2d(in_channels=in_channels, out_channels=out_channels, kernel_size=kernel_size, stride=1, padding=padding, bias=False)\n if spectral_norm:\n layers.append(SpectralNorm(conv_layer))\n else:\n layers.append(conv_layer)\n if batch_norm:\n layers.append(nn.BatchNorm2d(out_channels))\n return nn.Sequential(*layers)\n\n\ndef conv(in_channels, out_channels, kernel_size, stride=2, padding=2, batch_norm=True, init_zero_weights=False, spectral_norm=False):\n \"\"\"Creates a convolutional layer, with optional batch normalization.\n \"\"\"\n layers = []\n conv_layer = nn.Conv2d(in_channels=in_channels, out_channels=out_channels, kernel_size=kernel_size, stride=stride, padding=padding, bias=False)\n if init_zero_weights:\n conv_layer.weight.data = torch.randn(out_channels, in_channels, kernel_size, kernel_size) * 0.001\n \n if spectral_norm:\n layers.append(SpectralNorm(conv_layer))\n else:\n layers.append(conv_layer)\n\n if batch_norm:\n layers.append(nn.BatchNorm2d(out_channels))\n return nn.Sequential(*layers)\n \n\nclass ResnetBlock(nn.Module):\n def __init__(self, conv_dim):\n super(ResnetBlock, self).__init__()\n self.conv_layer = conv(in_channels=conv_dim, out_channels=conv_dim, kernel_size=3, stride=1, padding=1)\n\n def forward(self, x):\n out = x + self.conv_layer(x)\n return out",
"_____no_output_____"
]
],
[
[
"## DCGAN",
"_____no_output_____"
],
[
"## Spectral Norm class",
"_____no_output_____"
],
[
"### GAN generator",
"_____no_output_____"
]
],
[
[
"class DCGenerator(nn.Module):\n def __init__(self, noise_size, conv_dim, spectral_norm=False):\n super(DCGenerator, self).__init__()\n\n self.conv_dim = conv_dim\n self.relu = nn.ReLU()\n self.linear_bn = upconv(100, conv_dim*4,3) #BS X noise_size x 1 x 1 -> BS x 128 x 4 x 4 \n self.upconv1 = upconv(conv_dim*4,conv_dim*2,5)\n self.upconv2 = upconv(conv_dim*2,conv_dim,5)\n self.upconv3 = upconv(conv_dim,1,5, batch_norm=False) \n \n self.tanh = nn.Tanh()\n\n def forward(self, z):\n \"\"\"Generates an image given a sample of random noise.\n\n Input\n -----\n z: BS x noise_size x 1 x 1 --> BSx100x1x1 (during training)\n\n Output\n ------\n out: BS x channels x image_width x image_height --> BSx3x32x32 (during training)\n \"\"\"\n batch_size = z.size(0)\n out = self.relu(self.linear_bn(z)) # BS x 128 x 4 x 4 conv_dim=32\n out = out.view(-1, self.conv_dim*4, 4, 4)\n out = self.relu(self.upconv1(out)) # BS x 64 x 8 x 8\n out = self.relu(self.upconv2(out)) # BS x 32 x 16 x 16\n out = self.tanh(self.upconv3(out)) # BS x 3 x 32 x 32\n out_size = out.size()\n if out_size != torch.Size([batch_size, 1, 32, 32]):\n raise ValueError(\"expect {} x 3 x 32 x 32, but get {}\".format(batch_size, out_size))\n return out\n",
"_____no_output_____"
]
],
[
[
"### GAN discriminator",
"_____no_output_____"
]
],
[
[
"class DCDiscriminator(nn.Module):\n \"\"\"Defines the architecture of the discriminator network.\n Note: Both discriminators D_X and D_Y have the same architecture in this assignment.\n \"\"\"\n def __init__(self, conv_dim=64, spectral_norm=False):\n super(DCDiscriminator, self).__init__()\n\n self.conv1 = conv(in_channels=1, out_channels=conv_dim, kernel_size=5, stride=2, spectral_norm=spectral_norm)\n self.conv2 = conv(in_channels=conv_dim, out_channels=conv_dim*2, kernel_size=5, stride=2, spectral_norm=spectral_norm)\n self.conv3 = conv(in_channels=conv_dim*2, out_channels=conv_dim*4, kernel_size=5, stride=2, spectral_norm=spectral_norm)\n self.conv4 = conv(in_channels=conv_dim*4, out_channels=1, kernel_size=5, stride=2, padding=1, batch_norm=False, spectral_norm=spectral_norm)\n\n def forward(self, x):\n batch_size = x.size(0)\n\n out = F.relu(self.conv1(x)) # BS x 64 x 16 x 16\n out = F.relu(self.conv2(out)) # BS x 64 x 8 x 8\n out = F.relu(self.conv3(out)) # BS x 64 x 4 x 4\n\n out = self.conv4(out).squeeze()\n out_size = out.size()\n if out_size != torch.Size([batch_size,]):\n raise ValueError(\"expect {} x 1, but get {}\".format(batch_size, out_size))\n return out",
"_____no_output_____"
],
[
"from torch.utils.tensorboard import SummaryWriter\nimport numpy as np\n\ndef log_to_tensorboard(iteration, losses):\n writer = SummaryWriter(\"./runs/\")\n \n for key in losses:\n arr = losses[key]\n writer.add_scalar(f'loss/{key}', arr[-1], iteration)\n writer.close()\ndef calculate_log_likelihood(model, opts):\n \n transform = transforms.Compose([\n transforms.Scale(opts.image_size),\n transforms.ToTensor(),\n transforms.Normalize((0.5), (0.5)),\n ])\n train = datasets.EMNIST(\".\", split=\"letters\",train = True, download = True, transform= transform)\n train_dloader = DataLoader(dataset=train, batch_size=opts.batch_size, shuffle=True,num_workers=opts.num_workers)\n \n x = next(iter(train_dloader))[0]\n print(x)\n return torch.log(model(x)).mean()\n ",
"_____no_output_____"
]
],
[
[
"### GAN training loop",
"_____no_output_____"
]
],
[
[
"def gan_training_loop(dataloader, test_dataloader, opts):\n \"\"\"Runs the training loop.\n * Saves checkpoint every opts.checkpoint_every iterations\n * Saves generated samples every opts.sample_every iterations\n \"\"\"\n\n # Create generators and discriminators\n G, D = create_model(opts)\n\n g_params = G.parameters() # Get generator parameters\n d_params = D.parameters() # Get discriminator parameters\n\n # Create optimizers for the generators and discriminators\n g_optimizer = optim.RMSprop(g_params, opts.lr)\n d_optimizer = optim.RMSprop(d_params, opts.lr)\n\n train_iter = iter(dataloader)\n\n test_iter = iter(test_dataloader)\n # Get some fixed data from domains X and Y for sampling. These are images that are held\n # constant throughout training, that allow us to inspect the model's performance.\n fixed_noise = sample_noise(100, opts.noise_size) # # 100 x noise_size x 1 x 1\n\n iter_per_epoch = len(train_iter)\n total_train_iters = opts.train_iters\n\n losses = {\"iteration\": [], \"D_fake_loss\": [], \"D_real_loss\": [], \"G_loss\": [], \"D_loss\": [], \"W_loss\": []}\n\n # adversarial_loss = torch.nn.BCEWithLogitsLoss()\n gp_weight = 1\n epoch = 0\n total_iters = 0\n try:\n for iteration in range(1, opts.train_iters + 1):\n\n # Reset data_iter for each epoch\n \n # ones = Variable(torch.Tensor(real_images.shape[0]).float().cuda().fill_(1.0), requires_grad=False)\n if total_iters % iter_per_epoch == 0:\n epoch +=1\n train_iter = iter(dataloader)\n print(\"EPOCH:\", epoch)\n b = opts.batch_size\n for i in range(opts.n_critic):\n real_images, real_labels = train_iter.next()\n real_images, real_labels = to_var(real_images), to_var(real_labels).long().squeeze()\n m = b\n noise = sample_noise(m, opts.noise_size)\n fake_images = G(noise)\n D_real_loss = D(real_images).mean()\n D_fake_loss = D(fake_images).mean()\n\n D_loss = -(D_real_loss - D_fake_loss) #Minimize D_real_loss - D_fake_loss\n D_loss.backward()\n d_optimizer.step()\n\n Wasserstein_Distance = D_real_loss - D_fake_loss\n\n total_iters += 1 \n \n for param in D.parameters():\n param.data.clamp_(-opts.clip, opts.clip)\n D.zero_grad()\n G.zero_grad()\n # \n z = sample_noise(m, opts.noise_size)\n G_z = G(z)\n G_loss = -torch.mean(D(G_z))\n\n G_loss.backward()\n g_optimizer.step()\n D.zero_grad()\n G.zero_grad()\n\n if iteration % opts.log_step == 0:\n w_loss = Wasserstein_Distance\n losses['iteration'].append(iteration)\n losses['D_real_loss'].append(D_real_loss.item())\n losses['D_loss'].append(D_loss.item())\n losses['D_fake_loss'].append(D_fake_loss.item())\n losses['W_loss'].append(w_loss.item())\n losses['G_loss'].append(G_loss.item())\n print('Iteration [{:4d}/{:4d}] | D_real_loss: {:6.4f} | D_fake_loss: {:6.4f} | G_loss: {:6.4f} | D_loss: {:6.4f} | Wasserstein_Distance: {:6.4f}'.format(\n iteration, total_train_iters, D_real_loss.item(), D_fake_loss.item(), G_loss.item(), D_loss.item(), Wasserstein_Distance.item() ))\n\n \n log_to_tensorboard(iteration, losses)\n # Save the generated samples\n if iteration % opts.sample_every == 0:\n gan_save_samples(G, fixed_noise, iteration, opts)\n\n # Save the model parameters\n if iteration % opts.checkpoint_every == 0:\n gan_checkpoint(iteration, G, D, opts)\n\n except KeyboardInterrupt:\n print('Exiting early from training.')\n return G, D\n\n plt.figure()\n plt.plot(losses['iteration'], losses['D_real_loss'], label='D_real')\n plt.plot(losses['iteration'], losses['D_fake_loss'], label='D_fake')\n plt.plot(losses['iteration'], losses['G_loss'], label='G')\n plt.plot(losses['iteration'], losses['D_loss'], label='D')\n\n plt.legend()\n plt.savefig(os.path.join(opts.sample_dir, 'losses.png'))\n plt.close()\n return G, D",
"_____no_output_____"
]
],
[
[
"# Training\n",
"_____no_output_____"
],
[
"## Download dataset",
"_____no_output_____"
],
[
"### WGAN",
"_____no_output_____"
]
],
[
[
"SEED = 11\n\n# Set the random seed manually for reproducibility.\nnp.random.seed(SEED)\ntorch.manual_seed(SEED)\nif torch.cuda.is_available():\n torch.cuda.manual_seed(SEED)\n\n\nargs = AttrDict()\nargs_dict = {\n 'clip': .01,\n 'n_critic': 5,\n 'image_size':32, \n 'g_conv_dim':32, \n 'd_conv_dim':64,\n 'noise_size':100,\n 'num_workers': 0,\n 'train_iters':300000,\n 'X':'letters', # options: 'Windows' / 'Apple'\n 'Y': None,\n 'lr':5e-5,\n 'beta1':0.5,\n 'beta2':0.999,\n 'batch_size':64, \n 'checkpoint_dir':'./results/checkpoints_gan_gp1_lr3e-5',\n 'sample_dir': './results/samples_gan_gp1_lr3e-5',\n 'load': None,\n 'log_step':200,\n 'sample_every':200,\n 'checkpoint_every':1000,\n 'spectral_norm': False,\n 'gradient_penalty': False,\n 'd_train_iters': 1\n}\nargs.update(args_dict)\n\nprint_opts(args)\nG, D = train(args)\n\ngenerate_gif(\"results/samples_gan_gp1_lr3e-5\")",
"================================================================================\n Opts \n--------------------------------------------------------------------------------\n clip: 0.01 \n n_critic: 5 \n image_size: 32 \n g_conv_dim: 32 \n d_conv_dim: 64 \n noise_size: 100 \n train_iters: 300000 \n X: letters \n lr: 5e-05 \n beta1: 0.5 \n beta2: 0.999 \n batch_size: 64 \n checkpoint_dir: ./results/checkpoints_gan_gp1_lr3e-5 \n sample_dir: ./results/samples_gan_gp1_lr3e-5 \n log_step: 200 \n sample_every: 200 \n checkpoint_every: 1000 \n d_train_iters: 1 \n================================================================================\nDownloading and extracting zip archive\n"
],
[
"torch.cuda.is_available()",
"_____no_output_____"
],
[
"torch.cuda.device(0)\ntorch.cuda.get_device_name(0)",
"ERROR! Session/line number was not unique in database. History logging moved to new session 96\n"
],
[
"torch.version.cuda",
"_____no_output_____"
],
[
"torch.cuda.FloatTensor()",
"_____no_output_____"
],
[
"load_args = AttrDict()\nargs_dict = {\n 'clip': .01,\n 'n_critic': 5,\n 'image_size':32, \n 'g_conv_dim':32, \n 'd_conv_dim':64,\n 'noise_size':100,\n 'num_workers': 0,\n 'train_iters':300000,\n 'X':'letters', # options: 'Windows' / 'Apple'\n 'Y': None,\n 'lr':5e-5,\n 'beta1':0.5,\n 'beta2':0.999,\n 'batch_size':64, \n 'checkpoint_dir':'./results/checkpoints_gan_gp1_lr3e-5',\n 'sample_dir': './results/samples_gan_gp1_lr3e-5',\n 'load': './results/samples_gan_gp1_lr3e-5',\n 'log_step':200,\n 'sample_every':200,\n 'checkpoint_every':1000,\n 'spectral_norm': False,\n 'gradient_penalty': False,\n 'd_train_iters': 1\n}\nload_args.update(args_dict)\n\nD,G = load_checkpoint(load_args)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0860c5e2ec5462dbfe39fdfc6df33ea6e6af388 | 282,660 | ipynb | Jupyter Notebook | Lib/CharacterDetection/Character_detection.ipynb | VishalS99/Project-Securise | 553e4c11f73651624318760fa4d27a02afff0a5a | [
"MIT"
] | 2 | 2021-04-17T00:58:10.000Z | 2022-01-02T07:54:25.000Z | Lib/CharacterDetection/Character_detection.ipynb | VishalS99/Project-Securise | 553e4c11f73651624318760fa4d27a02afff0a5a | [
"MIT"
] | null | null | null | Lib/CharacterDetection/Character_detection.ipynb | VishalS99/Project-Securise | 553e4c11f73651624318760fa4d27a02afff0a5a | [
"MIT"
] | null | null | null | 301.022364 | 150,879 | 0.883121 | [
[
[
"import cv2\nimport os\nimport numpy\nfrom PIL import Image\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"# !tar -xf EnglishHnd.tgz\n# !mv English/Hnd ./\n# !rm -rf Hnd/Trj/\n# !mv Hnd/Img/* Hnd/\n# !rm -rf Hnd/Img\n# !rm -rf English",
"_____no_output_____"
],
[
"# !rm -rf Hnd",
"_____no_output_____"
],
[
"label_list = ['0','1','2','3','4','5','6','7','8','9', 'A','B','C','D','E','F','G','H', 'I','J','K','L','M','N','O','P','Q','R','S','T','U','V','W','X','Y','Z','a','b','c','d','e','f','g','h','i','j','k','l','m','n','o','p','q','r','s','t','u','v','w','x','y','z','fail']",
"_____no_output_____"
],
[
"# # count = 0\n# os.remove(\"./Hnd/all.txt~\")\n# for cc in os.listdir(\"./Hnd\"):\n# count = cc[-2:]\n# os.rename('Hnd/' + cc, 'Hnd/' + label_list[int(count)-1])",
"_____no_output_____"
],
[
"import torch\nfrom torch.utils.data import Dataset\nfrom torchvision import datasets\nfrom torchvision import transforms\nimport matplotlib.pyplot as plt\nfrom torchvision.io import read_image",
"_____no_output_____"
],
[
"transform = transforms.Compose(\n [\n # transforms.ToPILImage(),\n transforms.Grayscale(),\n transforms.Resize((28,28)),\n transforms.ToTensor(),\n # transforms.Normalize((0.5), (0.5)),\n ]\n)\n\ndef load_dataset():\n data_path = './Img/'\n train_dataset = datasets.ImageFolder(\n root=data_path,\n transform=transform\n )\n # train_dataset = datasets.EMNIST(root= \"./data\",split=\"byclass\", train = True, download = True, transform = transform)\n train_loader = torch.utils.data.DataLoader(\n train_dataset,\n batch_size=64,\n num_workers=2,\n shuffle=True\n )\n return train_loader\n\n# for batch_idx, (data, target) in enumerate(load_dataset()):\n# print(batch_idx)",
"_____no_output_____"
],
[
"dataiter = iter(load_dataset())\nimages, labels = dataiter.next()\n\nprint(images.shape)\nprint(labels.shape)\n\nfigure = plt.figure()\nnum_of_images = 60\nfor index in range(1, num_of_images + 1):\n plt.subplot(6, 10, index)\n plt.axis('off')\n plt.imshow(images[index].numpy().squeeze(), cmap='gray_r')",
"torch.Size([64, 1, 28, 28])\ntorch.Size([64])\n"
],
[
"load_dataset()",
"_____no_output_____"
],
[
"device = 'cuda' if torch.cuda.is_available() else 'cpu'",
"_____no_output_____"
],
[
"device",
"_____no_output_____"
],
[
"# defining the model architecture\nclass Net(torch.nn.Module): \n def __init__(self):\n super(Net, self).__init__()\n\n self.cnn_layers = torch.nn.Sequential(\n # Defining a 2D convolution layer\n torch.nn.Conv2d(1, 128, kernel_size=3, stride=1, padding=1),\n torch.nn.BatchNorm2d(128),\n torch.nn.ReLU(inplace=True),\n torch.nn.MaxPool2d(kernel_size=2, stride=2),\n # Defining another 2D convolution layer\n torch.nn.Conv2d(128, 128, kernel_size=3, stride=1, padding=1),\n torch.nn.BatchNorm2d(128),\n torch.nn.ReLU(inplace=True),\n torch.nn.MaxPool2d(kernel_size=2, stride=2),\n )\n\n self.linear_layers = torch.nn.Sequential(\n torch.nn.Linear(128 * 7 * 7, 63)\n )\n\n # Defining the forward pass \n def forward(self, x):\n x = self.cnn_layers(x)\n x = x.view(x.size(0), -1)\n # print(x.size)\n x = self.linear_layers(x)\n return x",
"_____no_output_____"
],
[
"model = Net()\n# optimizer = torch.optim.Adam(model.parameters(), lr = 0.05)\noptimizer = torch.optim.SGD(model.parameters(), lr=0.005, momentum=0.9)\n\ncc = torch.nn.CrossEntropyLoss()\n\nmodel.cuda()\ncc = cc.cuda()",
"_____no_output_____"
],
[
"model",
"_____no_output_____"
],
[
"for i in range(30):\n running_loss = 0\n for batch_idx, (images, labels) in enumerate(load_dataset()):\n if torch.cuda.is_available():\n images = images.cuda()\n labels = labels.cuda()\n\n # Training pass\n optimizer.zero_grad()\n output = model(images)\n loss = cc(output, labels)\n \n #This is where the model learns by backpropagating\n loss.backward()\n \n #And optimizes its weights here\n optimizer.step()\n \n running_loss += loss.item()\n else:\n print(\"Epoch {} - Training loss: {}\".format(i+1, running_loss/len(load_dataset())))\n",
"Epoch 1 - Training loss: 2.6299318342792746\nEpoch 2 - Training loss: 1.266061888665569\nEpoch 3 - Training loss: 1.029972419142723\nEpoch 4 - Training loss: 0.8772061093121158\nEpoch 5 - Training loss: 0.7388597208322311\nEpoch 6 - Training loss: 0.6751072840118895\nEpoch 7 - Training loss: 0.5606623250154816\nEpoch 8 - Training loss: 0.5231542096150165\nEpoch 9 - Training loss: 0.4680153228038428\nEpoch 10 - Training loss: 0.41698990017175674\nEpoch 11 - Training loss: 0.391155127968107\nEpoch 12 - Training loss: 0.3590970816356795\nEpoch 13 - Training loss: 0.30464253850205214\nEpoch 14 - Training loss: 0.2670100870333156\nEpoch 15 - Training loss: 0.2559660838498753\nEpoch 16 - Training loss: 0.2270271936621593\nEpoch 17 - Training loss: 0.21650315686224067\nEpoch 18 - Training loss: 0.22151087830793492\nEpoch 19 - Training loss: 0.20112429726488737\nEpoch 20 - Training loss: 0.1652718718942939\nEpoch 21 - Training loss: 0.14512722124820765\nEpoch 22 - Training loss: 0.13480370953603057\nEpoch 23 - Training loss: 0.13806580149625636\nEpoch 24 - Training loss: 0.14343493738762883\nEpoch 25 - Training loss: 0.11210734431384778\nEpoch 26 - Training loss: 0.11168958956603797\nEpoch 27 - Training loss: 0.10685745509797517\nEpoch 28 - Training loss: 0.08757424241464053\nEpoch 29 - Training loss: 0.07940195771694487\nEpoch 30 - Training loss: 0.10928565851051589\n"
],
[
"torch.save(model, './model_character_detect.pt')",
"_____no_output_____"
],
[
"running_loss\r\n",
"_____no_output_____"
],
[
"image = Image.open(\"../../Images/segmentation2/image_2_ROI_5.png\")\r\nimage\r\n",
"_____no_output_____"
],
[
"mm = torch.load('./model_character_detect.pt')",
"_____no_output_____"
],
[
"image = image.resize((28,28))",
"_____no_output_____"
],
[
"# from PIL import ImageOps\r\n# # image = ImageOps.grayscale(image)",
"_____no_output_____"
],
[
"image = transform(image)\r\nimage = image.cuda()",
"_____no_output_____"
],
[
"mm\r\n",
"_____no_output_____"
],
[
"# image = image.cuda()\r\nlp = mm(image[None, ...])",
"_____no_output_____"
],
[
"ps = torch.exp(lp)\r\nprobab = list(ps.cpu()[0])\r\npred_label = probab.index(max(probab))",
"_____no_output_____"
],
[
"pred_label",
"_____no_output_____"
],
[
"max(probab)",
"_____no_output_____"
],
[
"label_list[pred_label]",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d08621006adbf3292cbf8f247b001aae6d1c473b | 5,773 | ipynb | Jupyter Notebook | 1_data_prep.ipynb | marshmellow77/adverse-drug-effect-detection | b6c69865965e2e351b1beffecb61739d539796fc | [
"MIT"
] | 4 | 2021-10-08T21:02:23.000Z | 2022-02-08T03:48:45.000Z | 1_data_prep.ipynb | alabarga/adverse-drug-effect-detection | b6c69865965e2e351b1beffecb61739d539796fc | [
"MIT"
] | null | null | null | 1_data_prep.ipynb | alabarga/adverse-drug-effect-detection | b6c69865965e2e351b1beffecb61739d539796fc | [
"MIT"
] | 3 | 2021-10-09T11:38:03.000Z | 2022-02-22T04:23:28.000Z | 24.883621 | 175 | 0.564005 | [
[
[
"!pip install datasets -q",
"_____no_output_____"
],
[
"!pip install sagemaker -U -q",
"_____no_output_____"
],
[
"!pip install s3fs==0.4.2 -U -q",
"_____no_output_____"
]
],
[
[
"### Load dataset and have a peak:",
"_____no_output_____"
],
[
"This cell is required in SageMaker Studio, otherwise the download of the dataset will throw an error.\nAfter running this cell, the kernel needs to be restarted. After restarting tthe kernel, continue with the cell below (loading the dataset)",
"_____no_output_____"
]
],
[
[
"%%capture\nimport IPython\n!conda install -c conda-forge ipywidgets -y\nIPython.Application.instance().kernel.do_shutdown(True)",
"_____no_output_____"
],
[
"from datasets import load_dataset\nimport pandas as pd\ndataset = load_dataset('ade_corpus_v2', 'Ade_corpus_v2_classification')\ndf = pd.DataFrame(dataset['train'])\ndf.sample(5, random_state=124)",
"_____no_output_____"
]
],
[
[
"### Determine ratio of positive ADE phrases compared to total dataset",
"_____no_output_____"
]
],
[
[
"df['label'].sum()/len(df)",
"_____no_output_____"
]
],
[
[
"### Initialise Sagemaker variables and create S3 bucket",
"_____no_output_____"
]
],
[
[
"from sagemaker.huggingface.processing import HuggingFaceProcessor\nimport sagemaker\nfrom sagemaker import get_execution_role",
"_____no_output_____"
],
[
"sess = sagemaker.Session()\nrole = sagemaker.get_execution_role()\nbucket = f\"az-ade-{sess.account_id()}\"\nsess._create_s3_bucket_if_it_does_not_exist(bucket_name=bucket, region=sess._region_name)",
"_____no_output_____"
]
],
[
[
"### Save the name of the S3 bucket for later sessions",
"_____no_output_____"
]
],
[
[
"%store bucket",
"_____no_output_____"
]
],
[
[
"### Set up processing job",
"_____no_output_____"
]
],
[
[
"hf_processor = HuggingFaceProcessor(\n role=role,\n instance_type=\"ml.p3.2xlarge\",\n transformers_version='4.6',\n base_job_name=\"az-ade\",\n pytorch_version='1.7',\n instance_count=1,\n)",
"_____no_output_____"
],
[
"from sagemaker.processing import ProcessingInput, ProcessingOutput\n\noutputs=[\n ProcessingOutput(output_name=\"train_data\", source=\"/opt/ml/processing/training\", destination=f\"s3://{bucket}/processing_output/train_data\"),\n ProcessingOutput(output_name=\"validation_data\", source=\"/opt/ml/processing/validation\", destination=f\"s3://{bucket}/processing_output/validation_data\"),\n ProcessingOutput(output_name=\"test_data\", source=\"/opt/ml/processing/test\", destination=f\"s3://{bucket}/processing_output/test_data\"),\n ]\narguments = [\"--dataset-name\", \"ade_corpus_v2\",\n \"--datasubset-name\", \"Ade_corpus_v2_classification\",\n \"--model-name\", \"distilbert-base-uncased\",\n \"--train-ratio\", \"0.7\",\n \"--val-ratio\", \"0.15\",]",
"_____no_output_____"
],
[
"hf_processor.run(\n code=\"scripts/preprocess.py\",\n outputs=outputs,\n arguments=arguments\n)",
"_____no_output_____"
],
[
"preprocessing_job_description = hf_processor.jobs[-1].describe()\n\noutput_config = preprocessing_job_description['ProcessingOutputConfig']\nfor output in output_config['Outputs']:\n print(output['S3Output']['S3Uri'])",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d0863342b20d52ae9b1a89e1a95805028198fb06 | 3,578 | ipynb | Jupyter Notebook | 0 - HowTo.ipynb | red-hara/K | 410de4d0de3c24ce90f56edc28b50c05e543a2c7 | [
"MIT"
] | 1 | 2021-12-02T21:28:08.000Z | 2021-12-02T21:28:08.000Z | 0 - HowTo.ipynb | red-hara/K | 410de4d0de3c24ce90f56edc28b50c05e543a2c7 | [
"MIT"
] | null | null | null | 0 - HowTo.ipynb | red-hara/K | 410de4d0de3c24ce90f56edc28b50c05e543a2c7 | [
"MIT"
] | 1 | 2021-12-02T21:28:57.000Z | 2021-12-02T21:28:57.000Z | 22.223602 | 138 | 0.561766 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d0863414c70e50d583eacd56bddd7c22c243a67b | 34,079 | ipynb | Jupyter Notebook | IllusTrip3D.ipynb | z-tasker/aphantasia | 9c0fb3ab4554e4781297724f5d76bebba62e75fa | [
"MIT"
] | 579 | 2021-03-02T04:24:01.000Z | 2022-03-30T07:27:39.000Z | IllusTrip3D.ipynb | z-tasker/aphantasia | 9c0fb3ab4554e4781297724f5d76bebba62e75fa | [
"MIT"
] | 32 | 2021-03-07T14:53:12.000Z | 2022-01-25T20:14:56.000Z | IllusTrip3D.ipynb | z-tasker/aphantasia | 9c0fb3ab4554e4781297724f5d76bebba62e75fa | [
"MIT"
] | 77 | 2021-03-10T15:56:29.000Z | 2022-03-31T00:58:52.000Z | 43.579284 | 275 | 0.52346 | [
[
[
"# IllusTrip: Text to Video 3D\n\nPart of [Aphantasia](https://github.com/eps696/aphantasia) suite, made by Vadim Epstein [[eps696](https://github.com/eps696)] \nBased on [CLIP](https://github.com/openai/CLIP) + FFT/pixel ops from [Lucent](https://github.com/greentfrapp/lucent). \n3D part by [deKxi](https://twitter.com/deKxi), based on [AdaBins](https://github.com/shariqfarooq123/AdaBins) depth. \nthanks to [Ryan Murdock](https://twitter.com/advadnoun), [Jonathan Fly](https://twitter.com/jonathanfly), [@eduwatch2](https://twitter.com/eduwatch2) for ideas.\n\n## Features \n* continuously processes **multiple sentences** (e.g. illustrating lyrics or poems)\n* makes **videos**, evolving with pan/zoom/rotate motion\n* works with [inverse FFT](https://github.com/greentfrapp/lucent/blob/master/lucent/optvis/param/spatial.py) representation of the image or **directly with RGB** pixels (no GANs involved)\n* generates massive detailed textures (a la deepdream), **unlimited resolution**\n* optional **depth** processing for 3D look\n* various CLIP models\n* can start/resume from an image\n",
"_____no_output_____"
],
[
"**Run the cell below after each session restart**\n\nEnsure that you're given Tesla T4/P4/P100 GPU, not K80!",
"_____no_output_____"
]
],
[
[
"#@title General setup\n\n!pip install ftfy==5.8 transformers\n!pip install gputil ffpb \n\ntry: \n !pip3 install googletrans==3.1.0a0\n from googletrans import Translator, constants\n translator = Translator()\nexcept: pass\n\n# !apt-get -qq install ffmpeg\nwork_dir = '/content/illustrip'\nimport os\nos.makedirs(work_dir, exist_ok=True)\n%cd $work_dir\n\nimport os\nimport io\nimport time\nimport math\nimport random\nimport imageio\nimport numpy as np\nimport PIL\nfrom base64 import b64encode\nimport shutil\nfrom easydict import EasyDict as edict\na = edict()\n\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport torchvision\nfrom torchvision import transforms as T\nfrom torch.autograd import Variable\n\nfrom IPython.display import HTML, Image, display, clear_output\nfrom IPython.core.interactiveshell import InteractiveShell\nInteractiveShell.ast_node_interactivity = \"all\"\nimport ipywidgets as ipy\nfrom google.colab import output, files\n\nimport warnings\nwarnings.filterwarnings(\"ignore\")\n\n!pip install git+https://github.com/openai/CLIP.git --no-deps\nimport clip\n!pip install sentence_transformers\nfrom sentence_transformers import SentenceTransformer\n!pip install kornia\nimport kornia\n!pip install lpips\nimport lpips\n!pip install PyWavelets==1.1.1\n!pip install git+https://github.com/fbcotter/pytorch_wavelets\n\n%cd /content\n!rm -rf aphantasia\n!git clone https://github.com/eps696/aphantasia\n%cd aphantasia/\nfrom clip_fft import to_valid_rgb, fft_image, rfft2d_freqs, img2fft, pixel_image, un_rgb\nfrom utils import basename, file_list, img_list, img_read, txt_clean, plot_text, old_torch\nfrom utils import slice_imgs, derivat, pad_up_to, slerp, checkout, sim_func, latent_anima\nimport transforms\nimport depth\nfrom progress_bar import ProgressIPy as ProgressBar\nshutil.copy('mask.jpg', work_dir)\ndepth_mask_file = os.path.join(work_dir, 'mask.jpg')\n\nclear_output()\n\ndef save_img(img, fname=None):\n img = np.array(img)[:,:,:]\n img = np.transpose(img, (1,2,0)) \n img = np.clip(img*255, 0, 255).astype(np.uint8)\n if fname is not None:\n imageio.imsave(fname, np.array(img))\n imageio.imsave('result.jpg', np.array(img))\n\ndef makevid(seq_dir, size=None):\n char_len = len(basename(img_list(seq_dir)[0]))\n out_sequence = seq_dir + '/%0{}d.jpg'.format(char_len)\n out_video = seq_dir + '.mp4'\n print('.. generating video ..')\n !ffmpeg -y -v warning -i $out_sequence -crf 18 $out_video\n data_url = \"data:video/mp4;base64,\" + b64encode(open(out_video,'rb').read()).decode()\n wh = '' if size is None else 'width=%d height=%d' % (size, size)\n return \"\"\"<video %s controls><source src=\"%s\" type=\"video/mp4\"></video>\"\"\" % (wh, data_url)\n\n# Hardware check\n!ln -sf /opt/bin/nvidia-smi /usr/bin/nvidia-smi\nimport GPUtil as GPU\ngpu = GPU.getGPUs()[0] # XXX: only one GPU on Colab and isn’t guaranteed\n!nvidia-smi -L\nprint(\"GPU RAM {0:.0f}MB | Free {1:.0f}MB)\".format(gpu.memoryTotal, gpu.memoryFree))",
"_____no_output_____"
],
[
"#@title Load inputs\n\n#@markdown **Content** (either type a text string, or upload a text file):\ncontent = \"\" #@param {type:\"string\"}\nupload_texts = False #@param {type:\"boolean\"}\n\n#@markdown **Style** (either type a text string, or upload a text file):\nstyle = \"\" #@param {type:\"string\"}\nupload_styles = False #@param {type:\"boolean\"}\n\n#@markdown For non-English languages use Google translation:\ntranslate = False #@param {type:\"boolean\"}\n\n#@markdown Resume from the saved `.pt` snapshot, or from an image \n#@markdown (resolution settings below will be ignored in this case): \n\nif upload_texts:\n print('Upload main text file')\n uploaded = files.upload()\n text_file = list(uploaded)[0]\n texts = list(uploaded.values())[0].decode().split('\\n')\n texts = [tt.strip() for tt in texts if len(tt.strip())>0 and tt[0] != '#']\n print(' main text:', text_file, len(texts), 'lines')\n workname = txt_clean(basename(text_file))\nelse:\n texts = [content]\n workname = txt_clean(content)[:44]\n\nif upload_styles:\n print('Upload styles text file')\n uploaded = files.upload()\n text_file = list(uploaded)[0]\n styles = list(uploaded.values())[0].decode().split('\\n')\n styles = [tt.strip() for tt in styles if len(tt.strip())>0 and tt[0] != '#']\n print(' styles:', text_file, len(styles), 'lines')\nelse:\n styles = [style]\n\nresume = False #@param {type:\"boolean\"}\nif resume:\n print('Upload file to resume from')\n resumed = files.upload()\n resumed_filename = list(resumed)[0]\n resumed_bytes = list(resumed.values())[0]\n\nassert len(texts) > 0 and len(texts[0]) > 0, 'No input text[s] found!'\ntempdir = os.path.join(work_dir, workname)\nos.makedirs(tempdir, exist_ok=True)\nprint('main dir', tempdir)",
"_____no_output_____"
]
],
[
[
"**`content`** (what to draw) is your primary input; **`style`** (how to draw) is optional, if you want to separate such descriptions. \nIf you load text file[s], the imagery will interpolate from line to line (ensure equal line counts for content and style lists, for their accordance).",
"_____no_output_____"
]
],
[
[
"#@title Google Drive [optional]\n\n#@markdown Run this cell, if you want to store results on your Google Drive.\nusing_GDrive = True#@param{type:\"boolean\"}\nif using_GDrive:\n import os\n from google.colab import drive\n\n if not os.path.isdir('/G/MyDrive'): \n drive.mount('/G', force_remount=True)\n gdir = '/G/MyDrive'\n\n tempdir = os.path.join(gdir, 'illustrip', workname)\n os.makedirs(tempdir, exist_ok=True)\n print('main dir', tempdir)\n",
"_____no_output_____"
],
[
"#@title Main settings\n\nsideX = 1280 #@param {type:\"integer\"}\nsideY = 720 #@param {type:\"integer\"}\nsteps = 200 #@param {type:\"integer\"}\nframe_step = 100 #@param {type:\"integer\"}\n#@markdown > Config\nmethod = 'RGB' #@param ['FFT', 'RGB']\nmodel = 'ViT-B/32' #@param ['ViT-B/16', 'ViT-B/32', 'RN101', 'RN50x16', 'RN50x4', 'RN50']\n\n# Default settings\nif method == 'RGB':\n align = 'overscan'\n colors = 2\n contrast = 1.2\n sharpness = -1.\n aug_noise = 0.\n smooth = False\nelse:\n align = 'uniform'\n colors = 1.8\n contrast = 1.1\n sharpness = 1.\n aug_noise = 2.\n smooth = True\ninterpolate_topics = True\nstyle_power = 1.\nsamples = 200\nsave_step = 1\nlearning_rate = 1.\naug_transform = 'custom'\nsimilarity_function = 'cossim'\nmacro = 0.4\nenforce = 0.\nexpand = 0.\nzoom = 0.012\nshift = 10\nrotate = 0.8\ndistort = 0.3\nanimate_them = True\nsample_decrease = 1.\nDepthStrength = 0.\n\nprint(' loading CLIP model..')\nmodel_clip, _ = clip.load(model, jit=old_torch())\nmodsize = model_clip.visual.input_resolution\nxmem = {'ViT-B/16':0.25, 'RN50':0.5, 'RN50x4':0.16, 'RN50x16':0.06, 'RN101':0.33}\nif model in xmem.keys():\n sample_decrease *= xmem[model]\n\nclear_output()\nprint(' using CLIP model', model)",
"_____no_output_____"
]
],
[
[
"**`FFT`** method uses inverse FFT representation of the image. It allows flexible motion, but is either blurry (if smoothed) or noisy (if not). \n**`RGB`** method directly optimizes image pixels (without FFT parameterization). It's more clean and stable, when zooming in. \nThere are few choices for CLIP `model` (results do vary!). I prefer ViT-B/32 for consistency, next best bet is ViT-B/16. \n\n**`steps`** defines the length of animation per text line (multiply it to the inputs line count to get total video duration in frames). \n`frame_step` sets frequency of the changes in animation (how many frames between motion keypoints). \n\n",
"_____no_output_____"
],
[
"## Other settings [optional]",
"_____no_output_____"
]
],
[
[
"#@title Run this cell to override settings, if needed\n#@markdown [to roll back defaults, run \"Main settings\" cell again]\n\nstyle_power = 1. #@param {type:\"number\"}\noverscan = True #@param {type:\"boolean\"}\nalign = 'overscan' if overscan else 'uniform'\ninterpolate_topics = True #@param {type:\"boolean\"}\n\n#@markdown > Look\ncolors = 2 #@param {type:\"number\"}\ncontrast = 1.2 #@param {type:\"number\"}\nsharpness = 0. #@param {type:\"number\"}\n\n#@markdown > Training\nsamples = 200 #@param {type:\"integer\"}\nsave_step = 1 #@param {type:\"integer\"}\nlearning_rate = 1. #@param {type:\"number\"}\n\n#@markdown > Tricks\naug_transform = 'custom' #@param ['elastic', 'custom', 'none']\naug_noise = 0. #@param {type:\"number\"}\nmacro = 0.4 #@param {type:\"number\"}\nenforce = 0. #@param {type:\"number\"}\nexpand = 0. #@param {type:\"number\"}\nsimilarity_function = 'cossim' #@param ['cossim', 'spherical', 'mixed', 'angular', 'dot']\n\n#@markdown > Motion\nzoom = 0.012 #@param {type:\"number\"}\nshift = 10 #@param {type:\"number\"}\nrotate = 0.8 #@param {type:\"number\"}\ndistort = 0.3 #@param {type:\"number\"}\nanimate_them = True #@param {type:\"boolean\"}\nsmooth = True #@param {type:\"boolean\"}\nif method == 'RGB': smooth = False\n",
"_____no_output_____"
]
],
[
[
"`style_power` controls the strength of the style descriptions, comparing to the main input. \n`overscan` provides better frame coverage (needed for RGB method). \n`interpolate_topics` changes the subjects smoothly, otherwise they're switched by cut, making sharper transitions. \n\nDecrease **`samples`** if you face OOM (it's the main RAM eater), or just to speed up the process (with the cost of quality). \n`save_step` defines, how many optimization steps are taken between saved frames. Set it >1 for stronger image processing. \n\nExperimental tricks: \n`aug_transform` applies some augmentations, which quite radically change the output of this method (and slow down the process). Try yourself to see which is good for your case. `aug_noise` augmentation [FFT only!] seems to enhance optimization with transforms. \n`macro` boosts bigger forms. \n`enforce` adds more details by enforcing similarity between two parallel samples. \n`expand` boosts diversity (up to irrelevant) by enforcing difference between prev/next samples. \n\nMotion section:\n`shift` is in pixels, `rotate` in degrees. The values will be used as limits, if you mark `animate_them`. \n\n`smooth` reduces blinking, but induces motion blur with subtle screen-fixed patterns (valid only for FFT method, disabled for RGB). ",
"_____no_output_____"
],
[
"## Add 3D depth [optional]",
"_____no_output_____"
]
],
[
[
"### deKxi:: This whole cell contains most of whats needed, \n# with just a few changes to hook it up via frame_transform \n# (also glob_step now as global var)\n\n# I highly recommend performing the frame transformations and depth *after* saving,\n# (or just the depth warp if you prefer to keep the other affines as they are)\n# from my testing it reduces any noticeable stretching and allows the new areas\n# revealed from the changed perspective to be filled/detailed \n\n# pretrained models: Nyu is much better but Kitti is an option too\ndepth_model = 'nyu' # @ param [\"nyu\",\"kitti\"]\nDepthStrength = 0.01 #@param{type:\"number\"}\nMaskBlurAmt = 33 #@param{type:\"integer\"}\nsave_depth = False #@param{type:\"boolean\"}\nsize = (sideY,sideX)\n\n#@markdown NB: depth computing may take up to ~3x more time. Read the comments inside for more info. \n\n#@markdown Courtesy of [deKxi](https://twitter.com/deKxi)\n\nif DepthStrength > 0:\n\n if not os.path.exists(\"AdaBins_nyu.pt\"):\n !gdown https://drive.google.com/uc?id=1lvyZZbC9NLcS8a__YPcUP7rDiIpbRpoF\n if not os.path.exists('AdaBins_nyu.pt'):\n !wget https://www.dropbox.com/s/tayczpcydoco12s/AdaBins_nyu.pt\n # if depth_model=='kitti' and not os.path.exists(os.path.join(workdir_depth, \"pretrained/AdaBins_kitti.pt\")):\n # !gdown https://drive.google.com/uc?id=1HMgff-FV6qw1L0ywQZJ7ECa9VPq1bIoj\n\n if save_depth:\n depthdir = os.path.join(tempdir, 'depth')\n os.makedirs(depthdir, exist_ok=True)\n print('depth dir', depthdir)\n else:\n depthdir = None\n\n depth_infer, depth_mask = depth.init_adabins(model_path='AdaBins_nyu.pt', mask_path='mask.jpg', size=size)\n\n def depth_transform(img_t, img_np, depth_infer, depth_mask, size, depthX=0, scale=1., shift=[0,0], colors=1, depth_dir=None, save_num=0):\n\n # d X/Y define the origin point of the depth warp, effectively a \"3D pan zoom\", [-1..1]\n # plus = look ahead, minus = look aside\n dX = 100. * shift[0] / size[1]\n dY = 100. * shift[1] / size[0]\n # dZ = movement direction: 1 away (zoom out), 0 towards (zoom in), 0.5 stay\n dZ = 0.5 + 23. * (scale[0]-1) \n # dZ += 0.5 * float(math.sin(((save_num % 70)/70) * math.pi * 2))\n \n if img_np is None:\n img2 = img_t.clone().detach()\n par, imag, _ = pixel_image(img2.shape, resume=img2)\n img2 = to_valid_rgb(imag, colors=colors)()\n img2 = img2.detach().cpu().numpy()[0]\n img2 = (np.transpose(img2, (1,2,0))) # [h,w,c]\n img2 = np.clip(img2*255, 0, 255).astype(np.uint8)\n image_pil = T.ToPILImage()(img2)\n del img2\n else:\n image_pil = T.ToPILImage()(img_np)\n\n size2 = [s//2 for s in size]\n\n img = depth.depthwarp(img_t, image_pil, depth_infer, depth_mask, size2, depthX, [dX,dY], dZ, rescale=0.5, clip_range=2, save_path=depth_dir, save_num=save_num)\n return img\n",
"_____no_output_____"
]
],
[
[
"## Generate",
"_____no_output_____"
]
],
[
[
"#@title Generate\n\nif aug_transform == 'elastic':\n trform_f = transforms.transforms_elastic\n sample_decrease *= 0.95\nelif aug_transform == 'custom':\n trform_f = transforms.transforms_custom \n sample_decrease *= 0.95\nelse:\n trform_f = transforms.normalize()\n\nif enforce != 0:\n sample_decrease *= 0.5\n\nsamples = int(samples * sample_decrease)\nprint(' using %s method, %d samples' % (method, samples))\n\nif translate:\n translator = Translator()\n\ndef enc_text(txt):\n if translate:\n txt = translator.translate(txt, dest='en').text\n emb = model_clip.encode_text(clip.tokenize(txt).cuda()[:77])\n return emb.detach().clone()\n\n# Encode inputs\ncount = 0 # max count of texts and styles\nkey_txt_encs = [enc_text(txt) for txt in texts]\ncount = max(count, len(key_txt_encs))\nkey_styl_encs = [enc_text(style) for style in styles]\ncount = max(count, len(key_styl_encs))\nassert count > 0, \"No inputs found!\"\n\n# !rm -rf $tempdir\n# os.makedirs(tempdir, exist_ok=True)\n\n# opt_steps = steps * save_step # for optimization\nglob_steps = count * steps # saving\nif glob_steps == frame_step: frame_step = glob_steps // 2 # otherwise no motion\n\noutpic = ipy.Output()\noutpic\n\nif method == 'RGB':\n\n if resume:\n img_in = imageio.imread(resumed_bytes) / 255.\n params_tmp = torch.Tensor(img_in).permute(2,0,1).unsqueeze(0).float().cuda()\n params_tmp = un_rgb(params_tmp, colors=1.)\n sideY, sideX = img_in.shape[0], img_in.shape[1]\n else:\n params_tmp = torch.randn(1, 3, sideY, sideX).cuda() # * 0.01\n\nelse: # FFT\n\n if resume:\n if os.path.splitext(resumed_filename)[1].lower()[1:] in ['jpg','png','tif','bmp']:\n img_in = imageio.imread(resumed_bytes)\n params_tmp = img2fft(img_in, 1.5, 1.) * 2.\n else:\n params_tmp = torch.load(io.BytesIO(resumed_bytes))\n if isinstance(params_tmp, list): params_tmp = params_tmp[0]\n params_tmp = params_tmp.cuda()\n sideY, sideX = params_tmp.shape[2], (params_tmp.shape[3]-1)*2\n else:\n params_shape = [1, 3, sideY, sideX//2+1, 2]\n params_tmp = torch.randn(*params_shape).cuda() * 0.01\n \nparams_tmp = params_tmp.detach()\n# function() = torch.transformation(linear)\n\n# animation controls\nif animate_them:\n if method == 'RGB':\n m_scale = latent_anima([1], glob_steps, frame_step, uniform=True, cubic=True, start_lat=[-0.3])\n m_scale = 1 + (m_scale + 0.3) * zoom # only zoom in\n else:\n m_scale = latent_anima([1], glob_steps, frame_step, uniform=True, cubic=True, start_lat=[0.6])\n m_scale = 1 - (m_scale-0.6) * zoom # ping pong\n m_shift = latent_anima([2], glob_steps, frame_step, uniform=True, cubic=True, start_lat=[0.5,0.5])\n m_angle = latent_anima([1], glob_steps, frame_step, uniform=True, cubic=True, start_lat=[0.5])\n m_shear = latent_anima([1], glob_steps, frame_step, uniform=True, cubic=True, start_lat=[0.5])\n m_shift = (m_shift-0.5) * shift * abs(m_scale-1.) / zoom\n m_angle = (m_angle-0.5) * rotate * abs(m_scale-1.) / zoom\n m_shear = (m_shear-0.5) * distort * abs(m_scale-1.) / zoom\n\ndef get_encs(encs, num):\n cnt = len(encs)\n if cnt == 0: return []\n enc_1 = encs[min(num, cnt-1)]\n enc_2 = encs[min(num+1, cnt-1)]\n return slerp(enc_1, enc_2, steps)\n\ndef frame_transform(img, size, angle, shift, scale, shear):\n if old_torch(): # 1.7.1\n img = T.functional.affine(img, angle, shift, scale, shear, fillcolor=0, resample=PIL.Image.BILINEAR)\n img = T.functional.center_crop(img, size)\n img = pad_up_to(img, size)\n else: # 1.8+\n img = T.functional.affine(img, angle, shift, scale, shear, fill=0, interpolation=T.InterpolationMode.BILINEAR)\n img = T.functional.center_crop(img, size) # on 1.8+ also pads\n return img\n\nglobal img_np\nimg_np = None\nprev_enc = 0\ndef process(num):\n global params_tmp, img_np, opt_state, params, image_f, optimizer, pbar\n\n if interpolate_topics:\n txt_encs = get_encs(key_txt_encs, num)\n styl_encs = get_encs(key_styl_encs, num)\n else:\n txt_encs = [key_txt_encs[min(num, len(key_txt_encs)-1)][0]] * steps if len(key_txt_encs) > 0 else []\n styl_encs = [key_styl_encs[min(num, len(key_styl_encs)-1)][0]] * steps if len(key_styl_encs) > 0 else []\n\n if len(texts) > 0: print(' ref text: ', texts[min(num, len(texts)-1)][:80])\n if len(styles) > 0: print(' ref style: ', styles[min(num, len(styles)-1)][:80])\n\n for ii in range(steps):\n glob_step = num * steps + ii # saving/transforming\n\n ### animation: transform frame, reload params\n\n h, w = sideY, sideX\n \n # transform frame for motion\n scale = m_scale[glob_step] if animate_them else 1-zoom\n trans = tuple(m_shift[glob_step]) if animate_them else [0, shift]\n angle = m_angle[glob_step][0] if animate_them else rotate\n shear = m_shear[glob_step][0] if animate_them else distort\n\n if method == 'RGB':\n if DepthStrength > 0:\n params_tmp = depth_transform(params_tmp, img_np, depth_infer, depth_mask, size, DepthStrength, scale, trans, colors, depthdir, glob_step)\n params_tmp = frame_transform(params_tmp, (h,w), angle, trans, scale, shear)\n params, image_f, _ = pixel_image([1,3,h,w], resume=params_tmp)\n img_tmp = None\n\n else: # FFT\n if old_torch(): # 1.7.1\n img_tmp = torch.irfft(params_tmp, 2, normalized=True, signal_sizes=(h,w))\n if DepthStrength > 0:\n img_tmp = depth_transform(img_tmp, img_np, depth_infer, depth_mask, size, DepthStrength, scale, trans, colors, depthdir, glob_step)\n img_tmp = frame_transform(img_tmp, (h,w), angle, trans, scale, shear)\n params_tmp = torch.rfft(img_tmp, 2, normalized=True)\n else: # 1.8+\n if type(params_tmp) is not torch.complex64:\n params_tmp = torch.view_as_complex(params_tmp)\n img_tmp = torch.fft.irfftn(params_tmp, s=(h,w), norm='ortho')\n if DepthStrength > 0:\n img_tmp = depth_transform(img_tmp, img_np, depth_infer, depth_mask, size, DepthStrength, scale, trans, colors, depthdir, glob_step)\n img_tmp = frame_transform(img_tmp, (h,w), angle, trans, scale, shear)\n params_tmp = torch.fft.rfftn(img_tmp, s=[h,w], dim=[2,3], norm='ortho')\n params_tmp = torch.view_as_real(params_tmp)\n\n params, image_f, _ = fft_image([1,3,h,w], resume=params_tmp, sd=1.)\n\n image_f = to_valid_rgb(image_f, colors=colors)\n del img_tmp\n optimizer = torch.optim.Adam(params, learning_rate)\n # optimizer = torch.optim.AdamW(params, learning_rate, weight_decay=0.01, amsgrad=True)\n if smooth is True and num + ii > 0:\n optimizer.load_state_dict(opt_state)\n\n # get encoded inputs\n txt_enc = txt_encs[ii % len(txt_encs)].unsqueeze(0) if len(txt_encs) > 0 else None\n styl_enc = styl_encs[ii % len(styl_encs)].unsqueeze(0) if len(styl_encs) > 0 else None\n \n ### optimization\n\n for ss in range(save_step):\n loss = 0\n\n noise = aug_noise * (torch.rand(1, 1, *params[0].shape[2:4], 1)-0.5).cuda() if aug_noise > 0 else 0.\n img_out = image_f(noise)\n img_sliced = slice_imgs([img_out], samples, modsize, trform_f, align, macro)[0]\n out_enc = model_clip.encode_image(img_sliced)\n\n if method == 'RGB': # empirical hack\n loss += 1.5 * abs(img_out.mean((2,3)) - 0.45).mean() # fix brightness\n loss += 1.5 * abs(img_out.std((2,3)) - 0.17).sum() # fix contrast\n\n if txt_enc is not None:\n loss -= sim_func(txt_enc, out_enc, similarity_function)\n if styl_enc is not None:\n loss -= style_power * sim_func(styl_enc, out_enc, similarity_function)\n if sharpness != 0: # mode = scharr|sobel|naive\n loss -= sharpness * derivat(img_out, mode='naive')\n # loss -= sharpness * derivat(img_sliced, mode='scharr')\n if enforce != 0:\n img_sliced = slice_imgs([image_f(noise)], samples, modsize, trform_f, align, macro)[0]\n out_enc2 = model_clip.encode_image(img_sliced)\n loss -= enforce * sim_func(out_enc, out_enc2, similarity_function)\n del out_enc2; torch.cuda.empty_cache()\n if expand > 0:\n global prev_enc\n if ii > 0:\n loss += expand * sim_func(prev_enc, out_enc, similarity_function)\n prev_enc = out_enc.detach().clone()\n del img_out, img_sliced, out_enc; torch.cuda.empty_cache()\n\n optimizer.zero_grad()\n loss.backward()\n optimizer.step()\n \n ### save params & frame\n\n params_tmp = params[0].detach().clone()\n if smooth is True:\n opt_state = optimizer.state_dict()\n\n with torch.no_grad():\n img_t = image_f(contrast=contrast)[0].permute(1,2,0)\n img_np = torch.clip(img_t*255, 0, 255).cpu().numpy().astype(np.uint8)\n imageio.imsave(os.path.join(tempdir, '%05d.jpg' % glob_step), img_np, quality=95)\n shutil.copy(os.path.join(tempdir, '%05d.jpg' % glob_step), 'result.jpg')\n outpic.clear_output()\n with outpic:\n display(Image('result.jpg'))\n del img_t\n pbar.upd()\n\n params_tmp = params[0].detach().clone()\n\noutpic = ipy.Output()\noutpic\n\npbar = ProgressBar(glob_steps)\nfor i in range(count):\n process(i)\n\nHTML(makevid(tempdir))\nfiles.download(tempdir + '.mp4')\n\n## deKxi: downloading depth video\nif save_depth and DepthStrength > 0:\n HTML(makevid(depthdir))\n files.download(depthdir + '.mp4')",
"_____no_output_____"
]
],
[
[
"If video is not auto-downloaded after generation (for whatever reason), run this cell to do that:",
"_____no_output_____"
]
],
[
[
"files.download(tempdir + '.mp4')\nif save_depth and DepthStrength > 0:\n files.download(depthdir + '.mp4')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0863bb5740708185bdf018fb51a07e5ffe83709 | 317,412 | ipynb | Jupyter Notebook | notebook/linearRegression/GPUprice.ipynb | lbeaucourt/SIGMA-machine-learning | e9ae50c0f613abf9c46f344aea44c5a5e45b4d95 | [
"MIT"
] | null | null | null | notebook/linearRegression/GPUprice.ipynb | lbeaucourt/SIGMA-machine-learning | e9ae50c0f613abf9c46f344aea44c5a5e45b4d95 | [
"MIT"
] | null | null | null | notebook/linearRegression/GPUprice.ipynb | lbeaucourt/SIGMA-machine-learning | e9ae50c0f613abf9c46f344aea44c5a5e45b4d95 | [
"MIT"
] | 2 | 2019-04-01T09:33:51.000Z | 2019-04-09T15:38:54.000Z | 351.508306 | 36,028 | 0.922152 | [
[
[
"import time\nfrom IPython import display\n\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nfrom mpl_toolkits.mplot3d import Axes3D\n%matplotlib inline\nplt.rcParams['figure.figsize'] = [16, 10]",
"_____no_output_____"
]
],
[
[
"# 1) Get data in a pandas.DataFrame and plot it using matplotlib.pyplot",
"_____no_output_____"
]
],
[
[
"# Get data\n# 1) directement sous forme de list python\n\nGPU = [2048,2048,4096,4096,3072,6144,6144,8192,8192,8192,8192,11264,11264]\nprix = [139.96,149.95,184.96,194.95,299.95,332.95,359.95,459.95,534.95,569.95,699.95,829.96,929.95]\ndata = pd.DataFrame({'x1':GPU,'y':prix})\n\n# Remarque: On peut également enregistrer des données structurées (dataFrame) en .csv\ndata.to_csv('graphicCardsData.csv',index=False)\n\n# 2) En utilisant la fonction .read_csv() de pandas pour importer des données extérieure sous form .csv \n# directement dans un pandas.DataFrame\ndata = pd.read_csv('graphicCards.csv')\ndata.head()",
"_____no_output_____"
],
[
"data = data[['memory (Go)', 'price (euros)']]",
"_____no_output_____"
],
[
"data = data.rename(columns={\"memory (Go)\": 'x1', 'price (euros)': 'y'})",
"_____no_output_____"
],
[
"data['x1'] = data['x1'] * 1000",
"_____no_output_____"
],
[
"#PLot data\n\nplt.plot(data.x1,data.y,'o')\nplt.xlabel('GPU (Mo)')\nplt.ylabel('prix (€)')\nplt.show();",
"_____no_output_____"
]
],
[
[
"# 2) Contruire un modéle pour nos données",
"_____no_output_____"
]
],
[
[
"# Définir notre hypothèse (fonction)\n\ndef hypothesis(x,theta):\n return np.dot(x,theta)",
"_____no_output_____"
],
[
"# On génére aléatoirement une valeur de départ pour le paramètre theta1 de notre modèle\n\ntheta = np.random.rand()",
"_____no_output_____"
],
[
"# Fonction pour générer la droite représentant notre modèle\n\ndef getHypothesisForPLot(theta):\n return pd.DataFrame({'x':np.arange(0, 12000, 100),\n 'y':[hypothesis(x,theta) for x in np.arange(0, 12000, 100)]})",
"_____no_output_____"
],
[
"# On plot les données avec notre hypothèse ...\n\nplt.plot(data.x1,data.y,'o',label='data')\nplt.plot(getHypothesisForPLot(theta).x,getHypothesisForPLot(theta).y ,'r',label='hypothèse')\nplt.xlabel('GPU (Mo)')\nplt.ylabel('prix (€)')\nplt.title(\"C'est pas ça ....\")\nplt.legend()\nplt.show();\n\nprint(\"theta = %f\" % theta)",
"_____no_output_____"
]
],
[
[
"# 3) Tester la pertinence de notre modèle: la fonction de coût",
"_____no_output_____"
]
],
[
[
"data.shape",
"_____no_output_____"
],
[
"# On définit notre fonction de coût: somme quadratique (eg: on somme les carré)\n\ndef costFunction(y,yhat):\n return np.power(yhat - y,2).sum()*(2/y.shape[0])",
"_____no_output_____"
],
[
"# Prix prédis par notre modèle (avec un theta choisi pour illustrer) pour chaque exemple\n\ntheta = 0.07\nyhat = hypothesis(data.x1,theta)",
"_____no_output_____"
],
[
"#Comment fonctionne la fonction de coût: on somme le carré de toute les barre noire\n\nplt.plot(data.x1,data.y,'o',label='data')\nplt.plot(getHypothesisForPLot(theta).x,getHypothesisForPLot(theta).y,'r',label='hypothèse')\nfor i in range(data.shape[0]):\n plt.plot((data.x1[i],data.x1[i]), (min(data.y[i],yhat[i]),max(data.y[i],yhat[i])), 'k-')\nplt.xlabel('GPU (Mo)')\nplt.ylabel('prix (€)')\nplt.legend()\nplt.show();\n\nprint(\"theta = %f\" % theta)\nprint(\"J(theta) = %f\" % costFunction(data.y,yhat))",
"_____no_output_____"
]
],
[
[
"# 4) À quoi ressemble J(theta) en fonction de theta1",
"_____no_output_____"
]
],
[
[
"# Calculons (brutalement) la valeur de J(theta) dans un intervale de valeur de theta1 \n# pour observer la forme de notre fonction de coût que nous allons chercher à minimiser\n\nthetaRange = np.arange(-0.8,1,0.01)\ncostFctEvol = pd.DataFrame({'theta':thetaRange,\n 'cost':[costFunction(data.y,hypothesis(data.x1,theta)) \n for theta in thetaRange]})\n\nplt.plot(costFctEvol.theta,costFctEvol.cost)\nplt.xlabel('theta')\nplt.ylabel('J(theta)')\nplt.show;",
"_____no_output_____"
]
],
[
[
"# 5) La descente de Gradient",
"_____no_output_____"
]
],
[
[
"# La descente de gradient utilise la notion de dérivée, \n# illustrée ici avec la fonction carré (qui doit nous en rappeler une autre!)\n\ndef fct(x):\n return np.power(x,2)\n\ndef fctDeriv(x):\n return 2*x\n\nfctCarre = pd.DataFrame({'x':np.arange(-10,10,0.1),'y':[fct(x) for x in np.arange(-10,10,0.1)]})\nfctCarreD = pd.DataFrame({'x':np.arange(-10,10,0.1),\n 'y':[fctDeriv(x) for x in np.arange(-10,10,0.1)]})\nplt.plot(fctCarre.x,fctCarre.y,label='f(x)')\nplt.plot(fctCarreD.x,fctCarreD.y,label=\"f'(x)\")\nplt.legend();",
"_____no_output_____"
],
[
"# La descente de gradient utilise la dérivé de la fonction de coût \n# par rapport au paramètre theta1\n\ndef costFctDeriv(x,y,yhat):\n return ((yhat - y)*x.T).sum().sum()/y.shape[0]",
"_____no_output_____"
],
[
"# À chaque étape de la descente de gradient (jusqu'à la convergence), \n# on incremente la valeur de theta1 par ce résultat.\n# Alpha est le learning rate\n\ndef gradDescent(x,y,yhat,alpha):\n return -alpha*costFctDeriv(x,y,yhat)",
"_____no_output_____"
],
[
"# on plot les données avec l'hypothèse correpondant à la valeur de theta \n# ainsi que l'évolution dans la courbe de J(theta) en fonction de theta\n# On rajoute également la valeur de J(theta) en fonction du temps qui va nous servir à \n# débuger notre algorithme\n\ndef plotData(ax,data,theta,yhat,gradDescentEvol, title=''):\n ax.plot(data.x1,data.y,'o',label='data')\n ax.plot(getHypothesisForPLot(theta).x,getHypothesisForPLot(theta).y,'r',label='hypothèse')\n for i in range(data.shape[0]):\n ax.plot((data.x1[i],data.x1[i]), (min(data.y[i],yhat[i]),max(data.y[i],yhat[i])), 'k-')\n ax.set_xlabel('iteration step')\n if title != \"\":\n ax.set_title(title)\n ax.legend()\n\ndef plotCostFunction(ax,data,theta,gradDescentEvol,thetaInit, title=''):\n thetaRange = np.arange(-abs(thetaInit)+0.07,abs(thetaInit)+0.07,0.01)\n costFctEvol = pd.DataFrame({'theta':thetaRange,\n 'cost':[costFunction(data.y,hypothesis(data.x1,genTheta))\n for genTheta in thetaRange]})\n\n ax.plot(costFctEvol.theta,costFctEvol.cost,label='J(theta)')\n for i in range(gradDescentEvol.shape[0]):\n ax.plot(gradDescentEvol.theta[i],gradDescentEvol.J[i],'ro')\n for i in range(gradDescentEvol.shape[0]-1):\n ax.plot((gradDescentEvol.theta[i],gradDescentEvol.theta[i+1]),\n (gradDescentEvol.J[i],gradDescentEvol.J[i+1]),'k-',lw=1)\n ax.set_xlabel('iteration step')\n if title != \"\":\n ax.set_title(title)\n ax.legend()\n\ndef plotCostFunctionEvol(ax,gradDescentEvol,title=\"\"):\n ax.plot(np.arange(gradDescentEvol.shape[0]),gradDescentEvol.J,label='J(theta)')\n ax.set_xlabel('iteration step')\n if title != \"\":\n ax.set_title(title)\n ax.legend()",
"_____no_output_____"
],
[
"# On utilise donc une valeur de départ pour theta généré aléatoirement entre 0 et 1, \n# la valeur du learning rate est fixé à 0.00000003\n# Epsilon correspond à la précision que l'on veut atteindre pour stopper la descente de gradient\n\nthetaInit = np.random.rand()\nyhat = hypothesis(data.x1,thetaInit)\nalpha = 0.003\nepsilon = 0.001",
"_____no_output_____"
],
[
"# On prepare un dataframe pour stocker les valeurs de J(theta) et theta1\n\ngradDescentEvol = pd.DataFrame({'theta':thetaInit,\n 'J':costFunction(data.y,yhat)},index = np.arange(1))",
"_____no_output_____"
],
[
"# On parametrise deux trois trucs\n\nplt.rcParams['figure.figsize'] = [16, 5]\ncostFct = 0\ncount = 0\ntheta = thetaInit\n\n# Et on se lance dans la boucle: La descente de gradient!\nwhile np.abs(costFunction(data.y,yhat) - costFct)/costFct >= epsilon:\n count += 1\n costFct = costFunction(data.y,yhat)\n theta += gradDescent(data.x1,data.y,yhat,alpha)\n yhat = hypothesis(data.x1,theta)\n gradDescentEvol = gradDescentEvol.append(pd.DataFrame({'theta':theta,\n 'J':costFunction(data.y,yhat)},\n index = np.arange(1)),\n ignore_index=True)\n fig, ax = plt.subplots(ncols=3)\n plotData(ax[0],data,theta,yhat,gradDescentEvol)\n plotCostFunction(ax[1],data,theta,gradDescentEvol,thetaInit)\n plotCostFunctionEvol(ax[2],gradDescentEvol)\n display.clear_output(wait=True)\n display.display(plt.gcf())\n time.sleep(1)",
"_____no_output_____"
]
],
[
[
"# 6) Conclusion",
"_____no_output_____"
]
],
[
[
"# Afficher les résultat:\nprint('La descente de gradient a été réalisé en %i étapes.' % count)\nprint('theta = %f' % theta)\nprint('J(theta) = %f' % costFunction(data.y,yhat))",
"_____no_output_____"
],
[
"# Faisons une prédiction ....\n\nnewGPUs = [3072*1.5,11264*1.2]\nfor newGPU in newGPUs:\n print(\"Notre nouvelle carte de %i Mo de GPU pourra se vendre autour de %.2f €\" % \n (newGPU,newGPU*theta))\n \nplt.rcParams['figure.figsize'] = [14, 8]\nplt.plot(data.x1,data.y,'o',label='data')\nplt.plot(getHypothesisForPLot(theta).x,getHypothesisForPLot(theta).y,'r',label='hypothèse')\nfor i in range(data.shape[0]):\n plt.plot((data.x1[i],data.x1[i]), (min(data.y[i],yhat[i]),max(data.y[i],yhat[i])), 'k-')\nplt.plot(newGPUs,[newGPU*theta for newGPU in newGPUs], 'or', label='predictions')\nplt.xlabel('GPU (Mo)')\nplt.ylabel('prix (€)')\nplt.legend()\nplt.show();",
"_____no_output_____"
]
],
[
[
"# 7) Choix du taux d'apprentissage lambda",
"_____no_output_____"
]
],
[
[
"# On utilise donc une valeur de départ pour theta généré aléatoirement entre 0 et 1, \n# la valeur du learning rate est fixé à 0.00000003\n# Epsilon correspond à la précision que l'on veut atteindre pour stopper la descente de gradient\n\nthetaInit = np.random.rand()\nyhat1 = hypothesis(data.x1,thetaInit)\nyhat2 = hypothesis(data.x1,thetaInit)\nyhat3 = hypothesis(data.x1,thetaInit)\nalpha1 = 0.000000001\nalpha2 = 0.00000001\nalpha3 = 0.00000006\n\nepsilon = 0.001\n# On prepare un dataframe pour stocker les valeurs de J(theta) et theta1\n\ngradDescentEvol1 = pd.DataFrame({'theta':thetaInit,\n 'J':costFunction(data.y,yhat1)},index = np.arange(1))\ngradDescentEvol2 = pd.DataFrame({'theta':thetaInit,\n 'J':costFunction(data.y,yhat2)},index = np.arange(1))\ngradDescentEvol3 = pd.DataFrame({'theta':thetaInit,\n 'J':costFunction(data.y,yhat3)},index = np.arange(1))\n# On parametrise deux trois trucs\nplt.rcParams['figure.figsize'] = [16, 5]\ncount = 0\ncostFct1 = 0\ntheta1 = thetaInit\ncostFct2 = 0\ntheta2 = thetaInit\ncostFct3 = 0\ntheta3 = thetaInit\n\n# Et on se lance dans la boucle: La descente de gradient!\nwhile np.abs(costFunction(data.y,yhat2) - costFct2)/costFct2 >= epsilon:\n count += 1\n costFct1 = costFunction(data.y,yhat1)\n theta1 += gradDescent(data.x1,data.y,yhat1,alpha1)\n yhat1 = hypothesis(data.x1,theta1)\n gradDescentEvol1 = gradDescentEvol1.append(pd.DataFrame({'theta':theta1,\n 'J':costFunction(data.y,yhat1)},\n index = np.arange(1)),\n ignore_index=True)\n costFct2 = costFunction(data.y,yhat2)\n theta2 += gradDescent(data.x1,data.y,yhat2,alpha2)\n yhat2 = hypothesis(data.x1,theta2)\n gradDescentEvol2 = gradDescentEvol2.append(pd.DataFrame({'theta':theta2,\n 'J':costFunction(data.y,yhat2)},\n index = np.arange(1)),\n ignore_index=True)\n costFct3 = costFunction(data.y,yhat3)\n theta3 += gradDescent(data.x1,data.y,yhat3,alpha3)\n yhat3 = hypothesis(data.x1,theta3)\n gradDescentEvol3 = gradDescentEvol3.append(pd.DataFrame({'theta':theta3,\n 'J':costFunction(data.y,yhat3)},\n index = np.arange(1)),\n ignore_index=True)\n fig, ax = plt.subplots(ncols=3)\n plotCostFunctionEvol(ax[0],gradDescentEvol1,'small alpha')\n plotCostFunctionEvol(ax[1],gradDescentEvol2,'correct alpha')\n plotCostFunctionEvol(ax[2],gradDescentEvol3,'huge alpha')\n display.clear_output(wait=True)\n display.display(plt.gcf())\n time.sleep(1)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0864062418400aa8cbb8f73b86bff797849af5d | 92,474 | ipynb | Jupyter Notebook | notebooks/community/gapic/custom/showcase_custom_tabular_regression_online_pipeline.ipynb | nayaknishant/vertex-ai-samples | 3ce120b953f1cdc2ec2c5a3f4509cfeab106b7d0 | [
"Apache-2.0"
] | 213 | 2021-06-10T20:05:20.000Z | 2022-03-31T16:09:29.000Z | notebooks/community/gapic/custom/showcase_custom_tabular_regression_online_pipeline.ipynb | nayaknishant/vertex-ai-samples | 3ce120b953f1cdc2ec2c5a3f4509cfeab106b7d0 | [
"Apache-2.0"
] | 343 | 2021-07-25T22:55:25.000Z | 2022-03-31T23:58:47.000Z | notebooks/community/gapic/custom/showcase_custom_tabular_regression_online_pipeline.ipynb | nayaknishant/vertex-ai-samples | 3ce120b953f1cdc2ec2c5a3f4509cfeab106b7d0 | [
"Apache-2.0"
] | 143 | 2021-07-21T17:27:47.000Z | 2022-03-29T01:20:43.000Z | 42.264168 | 503 | 0.564526 | [
[
[
"# Copyright 2020 Google LLC\n#\n# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Vertex client library: Custom training tabular regression model with pipeline for online prediction with training pipeline\n\n<table align=\"left\">\n <td>\n <a href=\"https://colab.research.google.com/github/GoogleCloudPlatform/vertex-ai-samples/blob/master/notebooks/community/gapic/custom/showcase_custom_tabular_regression_online_pipeline.ipynb\">\n <img src=\"https://cloud.google.com/ml-engine/images/colab-logo-32px.png\" alt=\"Colab logo\"> Run in Colab\n </a>\n </td>\n <td>\n <a href=\"https://github.com/GoogleCloudPlatform/vertex-ai-samples/blob/master/notebooks/community/gapic/custom/showcase_custom_tabular_regression_online_pipeline.ipynb\">\n <img src=\"https://cloud.google.com/ml-engine/images/github-logo-32px.png\" alt=\"GitHub logo\">\n View on GitHub\n </a>\n </td>\n</table>\n<br/><br/><br/>",
"_____no_output_____"
],
[
"## Overview\n\n\nThis tutorial demonstrates how to use the Vertex client library for Python to train and deploy a custom tabular regression model for online prediction, using a training pipeline.",
"_____no_output_____"
],
[
"### Dataset\n\nThe dataset used for this tutorial is the [Boston Housing Prices dataset](https://www.cs.toronto.edu/~delve/data/boston/bostonDetail.html). The version of the dataset you will use in this tutorial is built into TensorFlow. The trained model predicts the median price of a house in units of 1K USD.",
"_____no_output_____"
],
[
"### Objective\n\nIn this tutorial, you create a custom model from a Python script in a Google prebuilt Docker container using the Vertex client library, and then do a prediction on the deployed model by sending data. You can alternatively create custom models using `gcloud` command-line tool or online using Google Cloud Console.\n\nThe steps performed include:\n\n- Create a Vertex custom job for training a model.\n- Create a `TrainingPipeline` resource.\n- Train a TensorFlow model with the `TrainingPipeline` resource.\n- Retrieve and load the model artifacts.\n- View the model evaluation.\n- Upload the model as a Vertex `Model` resource.\n- Deploy the `Model` resource to a serving `Endpoint` resource.\n- Make a prediction.\n- Undeploy the `Model` resource.",
"_____no_output_____"
],
[
"### Costs\n\nThis tutorial uses billable components of Google Cloud (GCP):\n\n* Vertex AI\n* Cloud Storage\n\nLearn about [Vertex AI\npricing](https://cloud.google.com/vertex-ai/pricing) and [Cloud Storage\npricing](https://cloud.google.com/storage/pricing), and use the [Pricing\nCalculator](https://cloud.google.com/products/calculator/)\nto generate a cost estimate based on your projected usage.",
"_____no_output_____"
],
[
"## Installation\n\nInstall the latest version of Vertex client library.",
"_____no_output_____"
]
],
[
[
"import os\nimport sys\n\n# Google Cloud Notebook\nif os.path.exists(\"/opt/deeplearning/metadata/env_version\"):\n USER_FLAG = \"--user\"\nelse:\n USER_FLAG = \"\"\n\n! pip3 install -U google-cloud-aiplatform $USER_FLAG",
"_____no_output_____"
]
],
[
[
"Install the latest GA version of *google-cloud-storage* library as well.",
"_____no_output_____"
]
],
[
[
"! pip3 install -U google-cloud-storage $USER_FLAG",
"_____no_output_____"
]
],
[
[
"### Restart the kernel\n\nOnce you've installed the Vertex client library and Google *cloud-storage*, you need to restart the notebook kernel so it can find the packages.",
"_____no_output_____"
]
],
[
[
"if not os.getenv(\"IS_TESTING\"):\n # Automatically restart kernel after installs\n import IPython\n\n app = IPython.Application.instance()\n app.kernel.do_shutdown(True)",
"_____no_output_____"
]
],
[
[
"## Before you begin\n\n### GPU runtime\n\n*Make sure you're running this notebook in a GPU runtime if you have that option. In Colab, select* **Runtime > Change Runtime Type > GPU**\n\n### Set up your Google Cloud project\n\n**The following steps are required, regardless of your notebook environment.**\n\n1. [Select or create a Google Cloud project](https://console.cloud.google.com/cloud-resource-manager). When you first create an account, you get a $300 free credit towards your compute/storage costs.\n\n2. [Make sure that billing is enabled for your project.](https://cloud.google.com/billing/docs/how-to/modify-project)\n\n3. [Enable the Vertex APIs and Compute Engine APIs.](https://console.cloud.google.com/flows/enableapi?apiid=ml.googleapis.com,compute_component)\n\n4. [The Google Cloud SDK](https://cloud.google.com/sdk) is already installed in Google Cloud Notebook.\n\n5. Enter your project ID in the cell below. Then run the cell to make sure the\nCloud SDK uses the right project for all the commands in this notebook.\n\n**Note**: Jupyter runs lines prefixed with `!` as shell commands, and it interpolates Python variables prefixed with `$` into these commands.",
"_____no_output_____"
]
],
[
[
"PROJECT_ID = \"[your-project-id]\" # @param {type:\"string\"}",
"_____no_output_____"
],
[
"if PROJECT_ID == \"\" or PROJECT_ID is None or PROJECT_ID == \"[your-project-id]\":\n # Get your GCP project id from gcloud\n shell_output = !gcloud config list --format 'value(core.project)' 2>/dev/null\n PROJECT_ID = shell_output[0]\n print(\"Project ID:\", PROJECT_ID)",
"_____no_output_____"
],
[
"! gcloud config set project $PROJECT_ID",
"_____no_output_____"
]
],
[
[
"#### Region\n\nYou can also change the `REGION` variable, which is used for operations\nthroughout the rest of this notebook. Below are regions supported for Vertex. We recommend that you choose the region closest to you.\n\n- Americas: `us-central1`\n- Europe: `europe-west4`\n- Asia Pacific: `asia-east1`\n\nYou may not use a multi-regional bucket for training with Vertex. Not all regions provide support for all Vertex services. For the latest support per region, see the [Vertex locations documentation](https://cloud.google.com/vertex-ai/docs/general/locations)",
"_____no_output_____"
]
],
[
[
"REGION = \"us-central1\" # @param {type: \"string\"}",
"_____no_output_____"
]
],
[
[
"#### Timestamp\n\nIf you are in a live tutorial session, you might be using a shared test account or project. To avoid name collisions between users on resources created, you create a timestamp for each instance session, and append onto the name of resources which will be created in this tutorial.",
"_____no_output_____"
]
],
[
[
"from datetime import datetime\n\nTIMESTAMP = datetime.now().strftime(\"%Y%m%d%H%M%S\")",
"_____no_output_____"
]
],
[
[
"### Authenticate your Google Cloud account\n\n**If you are using Google Cloud Notebook**, your environment is already authenticated. Skip this step.\n\n**If you are using Colab**, run the cell below and follow the instructions when prompted to authenticate your account via oAuth.\n\n**Otherwise**, follow these steps:\n\nIn the Cloud Console, go to the [Create service account key](https://console.cloud.google.com/apis/credentials/serviceaccountkey) page.\n\n**Click Create service account**.\n\nIn the **Service account name** field, enter a name, and click **Create**.\n\nIn the **Grant this service account access to project** section, click the Role drop-down list. Type \"Vertex\" into the filter box, and select **Vertex Administrator**. Type \"Storage Object Admin\" into the filter box, and select **Storage Object Admin**.\n\nClick Create. A JSON file that contains your key downloads to your local environment.\n\nEnter the path to your service account key as the GOOGLE_APPLICATION_CREDENTIALS variable in the cell below and run the cell.",
"_____no_output_____"
]
],
[
[
"# If you are running this notebook in Colab, run this cell and follow the\n# instructions to authenticate your GCP account. This provides access to your\n# Cloud Storage bucket and lets you submit training jobs and prediction\n# requests.\n\n# If on Google Cloud Notebook, then don't execute this code\nif not os.path.exists(\"/opt/deeplearning/metadata/env_version\"):\n if \"google.colab\" in sys.modules:\n from google.colab import auth as google_auth\n\n google_auth.authenticate_user()\n\n # If you are running this notebook locally, replace the string below with the\n # path to your service account key and run this cell to authenticate your GCP\n # account.\n elif not os.getenv(\"IS_TESTING\"):\n %env GOOGLE_APPLICATION_CREDENTIALS ''",
"_____no_output_____"
]
],
[
[
"### Create a Cloud Storage bucket\n\n**The following steps are required, regardless of your notebook environment.**\n\nWhen you submit a custom training job using the Vertex client library, you upload a Python package\ncontaining your training code to a Cloud Storage bucket. Vertex runs\nthe code from this package. In this tutorial, Vertex also saves the\ntrained model that results from your job in the same bucket. You can then\ncreate an `Endpoint` resource based on this output in order to serve\nonline predictions.\n\nSet the name of your Cloud Storage bucket below. Bucket names must be globally unique across all Google Cloud projects, including those outside of your organization.",
"_____no_output_____"
]
],
[
[
"BUCKET_NAME = \"gs://[your-bucket-name]\" # @param {type:\"string\"}",
"_____no_output_____"
],
[
"if BUCKET_NAME == \"\" or BUCKET_NAME is None or BUCKET_NAME == \"gs://[your-bucket-name]\":\n BUCKET_NAME = \"gs://\" + PROJECT_ID + \"aip-\" + TIMESTAMP",
"_____no_output_____"
]
],
[
[
"**Only if your bucket doesn't already exist**: Run the following cell to create your Cloud Storage bucket.",
"_____no_output_____"
]
],
[
[
"! gsutil mb -l $REGION $BUCKET_NAME",
"_____no_output_____"
]
],
[
[
"Finally, validate access to your Cloud Storage bucket by examining its contents:",
"_____no_output_____"
]
],
[
[
"! gsutil ls -al $BUCKET_NAME",
"_____no_output_____"
]
],
[
[
"### Set up variables\n\nNext, set up some variables used throughout the tutorial.\n### Import libraries and define constants",
"_____no_output_____"
],
[
"#### Import Vertex client library\n\nImport the Vertex client library into our Python environment.",
"_____no_output_____"
]
],
[
[
"import time\n\nfrom google.cloud.aiplatform import gapic as aip\nfrom google.protobuf import json_format\nfrom google.protobuf.json_format import MessageToJson, ParseDict\nfrom google.protobuf.struct_pb2 import Struct, Value",
"_____no_output_____"
]
],
[
[
"#### Vertex constants\n\nSetup up the following constants for Vertex:\n\n- `API_ENDPOINT`: The Vertex API service endpoint for dataset, model, job, pipeline and endpoint services.\n- `PARENT`: The Vertex location root path for dataset, model, job, pipeline and endpoint resources.",
"_____no_output_____"
]
],
[
[
"# API service endpoint\nAPI_ENDPOINT = \"{}-aiplatform.googleapis.com\".format(REGION)\n\n# Vertex location root path for your dataset, model and endpoint resources\nPARENT = \"projects/\" + PROJECT_ID + \"/locations/\" + REGION",
"_____no_output_____"
]
],
[
[
"#### CustomJob constants\n\nSet constants unique to CustomJob training:\n\n- Dataset Training Schemas: Tells the `Pipeline` resource service the task (e.g., classification) to train the model for.",
"_____no_output_____"
]
],
[
[
"CUSTOM_TASK_GCS_PATH = (\n \"gs://google-cloud-aiplatform/schema/trainingjob/definition/custom_task_1.0.0.yaml\"\n)",
"_____no_output_____"
]
],
[
[
"#### Hardware Accelerators\n\nSet the hardware accelerators (e.g., GPU), if any, for training and prediction.\n\nSet the variables `TRAIN_GPU/TRAIN_NGPU` and `DEPLOY_GPU/DEPLOY_NGPU` to use a container image supporting a GPU and the number of GPUs allocated to the virtual machine (VM) instance. For example, to use a GPU container image with 4 Nvidia Telsa K80 GPUs allocated to each VM, you would specify:\n\n (aip.AcceleratorType.NVIDIA_TESLA_K80, 4)\n\nFor GPU, available accelerators include:\n - aip.AcceleratorType.NVIDIA_TESLA_K80\n - aip.AcceleratorType.NVIDIA_TESLA_P100\n - aip.AcceleratorType.NVIDIA_TESLA_P4\n - aip.AcceleratorType.NVIDIA_TESLA_T4\n - aip.AcceleratorType.NVIDIA_TESLA_V100\n\n\nOtherwise specify `(None, None)` to use a container image to run on a CPU.\n\n*Note*: TF releases before 2.3 for GPU support will fail to load the custom model in this tutorial. It is a known issue and fixed in TF 2.3 -- which is caused by static graph ops that are generated in the serving function. If you encounter this issue on your own custom models, use a container image for TF 2.3 with GPU support.",
"_____no_output_____"
]
],
[
[
"if os.getenv(\"IS_TESTING_TRAIN_GPU\"):\n TRAIN_GPU, TRAIN_NGPU = (\n aip.AcceleratorType.NVIDIA_TESLA_K80,\n int(os.getenv(\"IS_TESTING_TRAIN_GPU\")),\n )\nelse:\n TRAIN_GPU, TRAIN_NGPU = (aip.AcceleratorType.NVIDIA_TESLA_K80, 1)\n\nif os.getenv(\"IS_TESTING_DEPOLY_GPU\"):\n DEPLOY_GPU, DEPLOY_NGPU = (\n aip.AcceleratorType.NVIDIA_TESLA_K80,\n int(os.getenv(\"IS_TESTING_DEPOLY_GPU\")),\n )\nelse:\n DEPLOY_GPU, DEPLOY_NGPU = (None, None)",
"_____no_output_____"
]
],
[
[
"#### Container (Docker) image\n\nNext, we will set the Docker container images for training and prediction\n\n - TensorFlow 1.15\n - `gcr.io/cloud-aiplatform/training/tf-cpu.1-15:latest`\n - `gcr.io/cloud-aiplatform/training/tf-gpu.1-15:latest`\n - TensorFlow 2.1\n - `gcr.io/cloud-aiplatform/training/tf-cpu.2-1:latest`\n - `gcr.io/cloud-aiplatform/training/tf-gpu.2-1:latest`\n - TensorFlow 2.2\n - `gcr.io/cloud-aiplatform/training/tf-cpu.2-2:latest`\n - `gcr.io/cloud-aiplatform/training/tf-gpu.2-2:latest`\n - TensorFlow 2.3\n - `gcr.io/cloud-aiplatform/training/tf-cpu.2-3:latest`\n - `gcr.io/cloud-aiplatform/training/tf-gpu.2-3:latest`\n - TensorFlow 2.4\n - `gcr.io/cloud-aiplatform/training/tf-cpu.2-4:latest`\n - `gcr.io/cloud-aiplatform/training/tf-gpu.2-4:latest`\n - XGBoost\n - `gcr.io/cloud-aiplatform/training/xgboost-cpu.1-1`\n - Scikit-learn\n - `gcr.io/cloud-aiplatform/training/scikit-learn-cpu.0-23:latest`\n - Pytorch\n - `gcr.io/cloud-aiplatform/training/pytorch-cpu.1-4:latest`\n - `gcr.io/cloud-aiplatform/training/pytorch-cpu.1-5:latest`\n - `gcr.io/cloud-aiplatform/training/pytorch-cpu.1-6:latest`\n - `gcr.io/cloud-aiplatform/training/pytorch-cpu.1-7:latest`\n\nFor the latest list, see [Pre-built containers for training](https://cloud.google.com/vertex-ai/docs/training/pre-built-containers).\n\n - TensorFlow 1.15\n - `gcr.io/cloud-aiplatform/prediction/tf-cpu.1-15:latest`\n - `gcr.io/cloud-aiplatform/prediction/tf-gpu.1-15:latest`\n - TensorFlow 2.1\n - `gcr.io/cloud-aiplatform/prediction/tf2-cpu.2-1:latest`\n - `gcr.io/cloud-aiplatform/prediction/tf2-gpu.2-1:latest`\n - TensorFlow 2.2\n - `gcr.io/cloud-aiplatform/prediction/tf2-cpu.2-2:latest`\n - `gcr.io/cloud-aiplatform/prediction/tf2-gpu.2-2:latest`\n - TensorFlow 2.3\n - `gcr.io/cloud-aiplatform/prediction/tf2-cpu.2-3:latest`\n - `gcr.io/cloud-aiplatform/prediction/tf2-gpu.2-3:latest`\n - XGBoost\n - `gcr.io/cloud-aiplatform/prediction/xgboost-cpu.1-2:latest`\n - `gcr.io/cloud-aiplatform/prediction/xgboost-cpu.1-1:latest`\n - `gcr.io/cloud-aiplatform/prediction/xgboost-cpu.0-90:latest`\n - `gcr.io/cloud-aiplatform/prediction/xgboost-cpu.0-82:latest`\n - Scikit-learn\n - `gcr.io/cloud-aiplatform/prediction/sklearn-cpu.0-23:latest`\n - `gcr.io/cloud-aiplatform/prediction/sklearn-cpu.0-22:latest`\n - `gcr.io/cloud-aiplatform/prediction/sklearn-cpu.0-20:latest`\n\nFor the latest list, see [Pre-built containers for prediction](https://cloud.google.com/vertex-ai/docs/predictions/pre-built-containers)",
"_____no_output_____"
]
],
[
[
"if os.getenv(\"IS_TESTING_TF\"):\n TF = os.getenv(\"IS_TESTING_TF\")\nelse:\n TF = \"2-1\"\n\nif TF[0] == \"2\":\n if TRAIN_GPU:\n TRAIN_VERSION = \"tf-gpu.{}\".format(TF)\n else:\n TRAIN_VERSION = \"tf-cpu.{}\".format(TF)\n if DEPLOY_GPU:\n DEPLOY_VERSION = \"tf2-gpu.{}\".format(TF)\n else:\n DEPLOY_VERSION = \"tf2-cpu.{}\".format(TF)\nelse:\n if TRAIN_GPU:\n TRAIN_VERSION = \"tf-gpu.{}\".format(TF)\n else:\n TRAIN_VERSION = \"tf-cpu.{}\".format(TF)\n if DEPLOY_GPU:\n DEPLOY_VERSION = \"tf-gpu.{}\".format(TF)\n else:\n DEPLOY_VERSION = \"tf-cpu.{}\".format(TF)\n\nTRAIN_IMAGE = \"gcr.io/cloud-aiplatform/training/{}:latest\".format(TRAIN_VERSION)\nDEPLOY_IMAGE = \"gcr.io/cloud-aiplatform/prediction/{}:latest\".format(DEPLOY_VERSION)\n\nprint(\"Training:\", TRAIN_IMAGE, TRAIN_GPU, TRAIN_NGPU)\nprint(\"Deployment:\", DEPLOY_IMAGE, DEPLOY_GPU, DEPLOY_NGPU)",
"_____no_output_____"
]
],
[
[
"#### Machine Type\n\nNext, set the machine type to use for training and prediction.\n\n- Set the variables `TRAIN_COMPUTE` and `DEPLOY_COMPUTE` to configure the compute resources for the VMs you will use for for training and prediction.\n - `machine type`\n - `n1-standard`: 3.75GB of memory per vCPU.\n - `n1-highmem`: 6.5GB of memory per vCPU\n - `n1-highcpu`: 0.9 GB of memory per vCPU\n - `vCPUs`: number of \\[2, 4, 8, 16, 32, 64, 96 \\]\n\n*Note: The following is not supported for training:*\n\n - `standard`: 2 vCPUs\n - `highcpu`: 2, 4 and 8 vCPUs\n\n*Note: You may also use n2 and e2 machine types for training and deployment, but they do not support GPUs*.",
"_____no_output_____"
]
],
[
[
"if os.getenv(\"IS_TESTING_TRAIN_MACHINE\"):\n MACHINE_TYPE = os.getenv(\"IS_TESTING_TRAIN_MACHINE\")\nelse:\n MACHINE_TYPE = \"n1-standard\"\n\nVCPU = \"4\"\nTRAIN_COMPUTE = MACHINE_TYPE + \"-\" + VCPU\nprint(\"Train machine type\", TRAIN_COMPUTE)\n\nif os.getenv(\"IS_TESTING_DEPLOY_MACHINE\"):\n MACHINE_TYPE = os.getenv(\"IS_TESTING_DEPLOY_MACHINE\")\nelse:\n MACHINE_TYPE = \"n1-standard\"\n\nVCPU = \"4\"\nDEPLOY_COMPUTE = MACHINE_TYPE + \"-\" + VCPU\nprint(\"Deploy machine type\", DEPLOY_COMPUTE)",
"_____no_output_____"
]
],
[
[
"# Tutorial\n\nNow you are ready to start creating your own custom model and training for Boston Housing.",
"_____no_output_____"
],
[
"## Set up clients\n\nThe Vertex client library works as a client/server model. On your side (the Python script) you will create a client that sends requests and receives responses from the Vertex server.\n\nYou will use different clients in this tutorial for different steps in the workflow. So set them all up upfront.\n\n- Model Service for `Model` resources.\n- Pipeline Service for training.\n- Endpoint Service for deployment.\n- Job Service for batch jobs and custom training.\n- Prediction Service for serving.",
"_____no_output_____"
]
],
[
[
"# client options same for all services\nclient_options = {\"api_endpoint\": API_ENDPOINT}\n\n\ndef create_model_client():\n client = aip.ModelServiceClient(client_options=client_options)\n return client\n\n\ndef create_pipeline_client():\n client = aip.PipelineServiceClient(client_options=client_options)\n return client\n\n\ndef create_endpoint_client():\n client = aip.EndpointServiceClient(client_options=client_options)\n return client\n\n\ndef create_prediction_client():\n client = aip.PredictionServiceClient(client_options=client_options)\n return client\n\n\nclients = {}\nclients[\"model\"] = create_model_client()\nclients[\"pipeline\"] = create_pipeline_client()\nclients[\"endpoint\"] = create_endpoint_client()\nclients[\"prediction\"] = create_prediction_client()\n\nfor client in clients.items():\n print(client)",
"_____no_output_____"
]
],
[
[
"## Train a model\n\nThere are two ways you can train a custom model using a container image:\n\n- **Use a Google Cloud prebuilt container**. If you use a prebuilt container, you will additionally specify a Python package to install into the container image. This Python package contains your code for training a custom model.\n\n- **Use your own custom container image**. If you use your own container, the container needs to contain your code for training a custom model.",
"_____no_output_____"
],
[
"## Prepare your custom job specification\n\nNow that your clients are ready, your first step is to create a Job Specification for your custom training job. The job specification will consist of the following:\n\n- `worker_pool_spec` : The specification of the type of machine(s) you will use for training and how many (single or distributed)\n- `python_package_spec` : The specification of the Python package to be installed with the pre-built container.",
"_____no_output_____"
],
[
"### Prepare your machine specification\n\nNow define the machine specification for your custom training job. This tells Vertex what type of machine instance to provision for the training.\n - `machine_type`: The type of GCP instance to provision -- e.g., n1-standard-8.\n - `accelerator_type`: The type, if any, of hardware accelerator. In this tutorial if you previously set the variable `TRAIN_GPU != None`, you are using a GPU; otherwise you will use a CPU.\n - `accelerator_count`: The number of accelerators.",
"_____no_output_____"
]
],
[
[
"if TRAIN_GPU:\n machine_spec = {\n \"machine_type\": TRAIN_COMPUTE,\n \"accelerator_type\": TRAIN_GPU,\n \"accelerator_count\": TRAIN_NGPU,\n }\nelse:\n machine_spec = {\"machine_type\": TRAIN_COMPUTE, \"accelerator_count\": 0}",
"_____no_output_____"
]
],
[
[
"### Prepare your disk specification\n\n(optional) Now define the disk specification for your custom training job. This tells Vertex what type and size of disk to provision in each machine instance for the training.\n\n - `boot_disk_type`: Either SSD or Standard. SSD is faster, and Standard is less expensive. Defaults to SSD.\n - `boot_disk_size_gb`: Size of disk in GB.",
"_____no_output_____"
]
],
[
[
"DISK_TYPE = \"pd-ssd\" # [ pd-ssd, pd-standard]\nDISK_SIZE = 200 # GB\n\ndisk_spec = {\"boot_disk_type\": DISK_TYPE, \"boot_disk_size_gb\": DISK_SIZE}",
"_____no_output_____"
]
],
[
[
"### Define the worker pool specification\n\nNext, you define the worker pool specification for your custom training job. The worker pool specification will consist of the following:\n\n- `replica_count`: The number of instances to provision of this machine type.\n- `machine_spec`: The hardware specification.\n- `disk_spec` : (optional) The disk storage specification.\n\n- `python_package`: The Python training package to install on the VM instance(s) and which Python module to invoke, along with command line arguments for the Python module.\n\nLet's dive deeper now into the python package specification:\n\n-`executor_image_spec`: This is the docker image which is configured for your custom training job.\n\n-`package_uris`: This is a list of the locations (URIs) of your python training packages to install on the provisioned instance. The locations need to be in a Cloud Storage bucket. These can be either individual python files or a zip (archive) of an entire package. In the later case, the job service will unzip (unarchive) the contents into the docker image.\n\n-`python_module`: The Python module (script) to invoke for running the custom training job. In this example, you will be invoking `trainer.task.py` -- note that it was not neccessary to append the `.py` suffix.\n\n-`args`: The command line arguments to pass to the corresponding Pythom module. In this example, you will be setting:\n - `\"--model-dir=\" + MODEL_DIR` : The Cloud Storage location where to store the model artifacts. There are two ways to tell the training script where to save the model artifacts:\n - direct: You pass the Cloud Storage location as a command line argument to your training script (set variable `DIRECT = True`), or\n - indirect: The service passes the Cloud Storage location as the environment variable `AIP_MODEL_DIR` to your training script (set variable `DIRECT = False`). In this case, you tell the service the model artifact location in the job specification.\n - `\"--epochs=\" + EPOCHS`: The number of epochs for training.\n - `\"--steps=\" + STEPS`: The number of steps (batches) per epoch.\n - `\"--distribute=\" + TRAIN_STRATEGY\"` : The training distribution strategy to use for single or distributed training.\n - `\"single\"`: single device.\n - `\"mirror\"`: all GPU devices on a single compute instance.\n - `\"multi\"`: all GPU devices on all compute instances.\n - `\"--param-file=\" + PARAM_FILE`: The Cloud Storage location for storing feature normalization values.",
"_____no_output_____"
]
],
[
[
"JOB_NAME = \"custom_job_\" + TIMESTAMP\nMODEL_DIR = \"{}/{}\".format(BUCKET_NAME, JOB_NAME)\n\nif not TRAIN_NGPU or TRAIN_NGPU < 2:\n TRAIN_STRATEGY = \"single\"\nelse:\n TRAIN_STRATEGY = \"mirror\"\n\nEPOCHS = 20\nSTEPS = 100\n\nPARAM_FILE = BUCKET_NAME + \"/params.txt\"\n\nDIRECT = True\nif DIRECT:\n CMDARGS = [\n \"--model-dir=\" + MODEL_DIR,\n \"--epochs=\" + str(EPOCHS),\n \"--steps=\" + str(STEPS),\n \"--distribute=\" + TRAIN_STRATEGY,\n \"--param-file=\" + PARAM_FILE,\n ]\nelse:\n CMDARGS = [\n \"--epochs=\" + str(EPOCHS),\n \"--steps=\" + str(STEPS),\n \"--distribute=\" + TRAIN_STRATEGY,\n \"--param-file=\" + PARAM_FILE,\n ]\n\nworker_pool_spec = [\n {\n \"replica_count\": 1,\n \"machine_spec\": machine_spec,\n \"disk_spec\": disk_spec,\n \"python_package_spec\": {\n \"executor_image_uri\": TRAIN_IMAGE,\n \"package_uris\": [BUCKET_NAME + \"/trainer_boston.tar.gz\"],\n \"python_module\": \"trainer.task\",\n \"args\": CMDARGS,\n },\n }\n]",
"_____no_output_____"
]
],
[
[
"### Examine the training package\n\n#### Package layout\n\nBefore you start the training, you will look at how a Python package is assembled for a custom training job. When unarchived, the package contains the following directory/file layout.\n\n- PKG-INFO\n- README.md\n- setup.cfg\n- setup.py\n- trainer\n - \\_\\_init\\_\\_.py\n - task.py\n\nThe files `setup.cfg` and `setup.py` are the instructions for installing the package into the operating environment of the Docker image.\n\nThe file `trainer/task.py` is the Python script for executing the custom training job. *Note*, when we referred to it in the worker pool specification, we replace the directory slash with a dot (`trainer.task`) and dropped the file suffix (`.py`).\n\n#### Package Assembly\n\nIn the following cells, you will assemble the training package.",
"_____no_output_____"
]
],
[
[
"# Make folder for Python training script\n! rm -rf custom\n! mkdir custom\n\n# Add package information\n! touch custom/README.md\n\nsetup_cfg = \"[egg_info]\\n\\ntag_build =\\n\\ntag_date = 0\"\n! echo \"$setup_cfg\" > custom/setup.cfg\n\nsetup_py = \"import setuptools\\n\\nsetuptools.setup(\\n\\n install_requires=[\\n\\n 'tensorflow_datasets==1.3.0',\\n\\n ],\\n\\n packages=setuptools.find_packages())\"\n! echo \"$setup_py\" > custom/setup.py\n\npkg_info = \"Metadata-Version: 1.0\\n\\nName: Boston Housing tabular regression\\n\\nVersion: 0.0.0\\n\\nSummary: Demostration training script\\n\\nHome-page: www.google.com\\n\\nAuthor: Google\\n\\nAuthor-email: [email protected]\\n\\nLicense: Public\\n\\nDescription: Demo\\n\\nPlatform: Vertex\"\n! echo \"$pkg_info\" > custom/PKG-INFO\n\n# Make the training subfolder\n! mkdir custom/trainer\n! touch custom/trainer/__init__.py",
"_____no_output_____"
]
],
[
[
"#### Task.py contents\n\nIn the next cell, you write the contents of the training script task.py. I won't go into detail, it's just there for you to browse. In summary:\n\n- Get the directory where to save the model artifacts from the command line (`--model_dir`), and if not specified, then from the environment variable `AIP_MODEL_DIR`.\n- Loads Boston Housing dataset from TF.Keras builtin datasets\n- Builds a simple deep neural network model using TF.Keras model API.\n- Compiles the model (`compile()`).\n- Sets a training distribution strategy according to the argument `args.distribute`.\n- Trains the model (`fit()`) with epochs specified by `args.epochs`.\n- Saves the trained model (`save(args.model_dir)`) to the specified model directory.\n- Saves the maximum value for each feature `f.write(str(params))` to the specified parameters file.",
"_____no_output_____"
]
],
[
[
"%%writefile custom/trainer/task.py\n# Single, Mirror and Multi-Machine Distributed Training for Boston Housing\n\nimport tensorflow_datasets as tfds\nimport tensorflow as tf\nfrom tensorflow.python.client import device_lib\nimport numpy as np\nimport argparse\nimport os\nimport sys\ntfds.disable_progress_bar()\n\nparser = argparse.ArgumentParser()\nparser.add_argument('--model-dir', dest='model_dir',\n default=os.getenv('AIP_MODEL_DIR'), type=str, help='Model dir.')\nparser.add_argument('--lr', dest='lr',\n default=0.001, type=float,\n help='Learning rate.')\nparser.add_argument('--epochs', dest='epochs',\n default=20, type=int,\n help='Number of epochs.')\nparser.add_argument('--steps', dest='steps',\n default=100, type=int,\n help='Number of steps per epoch.')\nparser.add_argument('--distribute', dest='distribute', type=str, default='single',\n help='distributed training strategy')\nparser.add_argument('--param-file', dest='param_file',\n default='/tmp/param.txt', type=str,\n help='Output file for parameters')\nargs = parser.parse_args()\n\nprint('Python Version = {}'.format(sys.version))\nprint('TensorFlow Version = {}'.format(tf.__version__))\nprint('TF_CONFIG = {}'.format(os.environ.get('TF_CONFIG', 'Not found')))\n\n# Single Machine, single compute device\nif args.distribute == 'single':\n if tf.test.is_gpu_available():\n strategy = tf.distribute.OneDeviceStrategy(device=\"/gpu:0\")\n else:\n strategy = tf.distribute.OneDeviceStrategy(device=\"/cpu:0\")\n# Single Machine, multiple compute device\nelif args.distribute == 'mirror':\n strategy = tf.distribute.MirroredStrategy()\n# Multiple Machine, multiple compute device\nelif args.distribute == 'multi':\n strategy = tf.distribute.experimental.MultiWorkerMirroredStrategy()\n\n# Multi-worker configuration\nprint('num_replicas_in_sync = {}'.format(strategy.num_replicas_in_sync))\n\n\ndef make_dataset():\n\n # Scaling Boston Housing data features\n def scale(feature):\n max = np.max(feature)\n feature = (feature / max).astype(np.float)\n return feature, max\n\n (x_train, y_train), (x_test, y_test) = tf.keras.datasets.boston_housing.load_data(\n path=\"boston_housing.npz\", test_split=0.2, seed=113\n )\n params = []\n for _ in range(13):\n x_train[_], max = scale(x_train[_])\n x_test[_], _ = scale(x_test[_])\n params.append(max)\n\n # store the normalization (max) value for each feature\n with tf.io.gfile.GFile(args.param_file, 'w') as f:\n f.write(str(params))\n return (x_train, y_train), (x_test, y_test)\n\n\n# Build the Keras model\ndef build_and_compile_dnn_model():\n model = tf.keras.Sequential([\n tf.keras.layers.Dense(128, activation='relu', input_shape=(13,)),\n tf.keras.layers.Dense(128, activation='relu'),\n tf.keras.layers.Dense(1, activation='linear')\n ])\n model.compile(\n loss='mse',\n optimizer=tf.keras.optimizers.RMSprop(learning_rate=args.lr))\n return model\n\nNUM_WORKERS = strategy.num_replicas_in_sync\n# Here the batch size scales up by number of workers since\n# `tf.data.Dataset.batch` expects the global batch size.\nBATCH_SIZE = 16\nGLOBAL_BATCH_SIZE = BATCH_SIZE * NUM_WORKERS\n\nwith strategy.scope():\n # Creation of dataset, and model building/compiling need to be within\n # `strategy.scope()`.\n model = build_and_compile_dnn_model()\n\n# Train the model\n(x_train, y_train), (x_test, y_test) = make_dataset()\nmodel.fit(x_train, y_train, epochs=args.epochs, batch_size=GLOBAL_BATCH_SIZE)\nmodel.save(args.model_dir)",
"_____no_output_____"
]
],
[
[
"#### Store training script on your Cloud Storage bucket\n\nNext, you package the training folder into a compressed tar ball, and then store it in your Cloud Storage bucket.",
"_____no_output_____"
]
],
[
[
"! rm -f custom.tar custom.tar.gz\n! tar cvf custom.tar custom\n! gzip custom.tar\n! gsutil cp custom.tar.gz $BUCKET_NAME/trainer_boston.tar.gz",
"_____no_output_____"
]
],
[
[
"## Train the model using a `TrainingPipeline` resource\n\nNow start training of your custom training job using a training pipeline on Vertex. To train the your custom model, do the following steps:\n\n1. Create a Vertex `TrainingPipeline` resource for the `Dataset` resource.\n2. Execute the pipeline to start the training.\n\n### Create a `TrainingPipeline` resource\n\nYou may ask, what do we use a pipeline for? We typically use pipelines when the job (such as training) has multiple steps, generally in sequential order: do step A, do step B, etc. By putting the steps into a pipeline, we gain the benefits of:\n\n1. Being reusable for subsequent training jobs.\n2. Can be containerized and ran as a batch job.\n3. Can be distributed.\n4. All the steps are associated with the same pipeline job for tracking progress.\n\n#### The `training_pipeline` specification\n\nFirst, you need to describe a pipeline specification. Let's look into the *minimal* requirements for constructing a `training_pipeline` specification for a custom job:\n\n- `display_name`: A human readable name for the pipeline job.\n- `training_task_definition`: The training task schema.\n- `training_task_inputs`: A dictionary describing the requirements for the training job.\n- `model_to_upload`: A dictionary describing the specification for the (uploaded) Vertex custom `Model` resource.\n - `display_name`: A human readable name for the `Model` resource.\n - `artificat_uri`: The Cloud Storage path where the model artifacts are stored in SavedModel format.\n - `container_spec`: This is the specification for the Docker container that will be installed on the `Endpoint` resource, from which the custom model will serve predictions.",
"_____no_output_____"
]
],
[
[
"from google.protobuf import json_format\nfrom google.protobuf.struct_pb2 import Value\n\nMODEL_NAME = \"custom_pipeline-\" + TIMESTAMP\nPIPELINE_DISPLAY_NAME = \"custom-training-pipeline\" + TIMESTAMP\n\ntraining_task_inputs = json_format.ParseDict(\n {\"workerPoolSpecs\": worker_pool_spec}, Value()\n)\npipeline = {\n \"display_name\": PIPELINE_DISPLAY_NAME,\n \"training_task_definition\": CUSTOM_TASK_GCS_PATH,\n \"training_task_inputs\": training_task_inputs,\n \"model_to_upload\": {\n \"display_name\": PIPELINE_DISPLAY_NAME + \"-model\",\n \"artifact_uri\": MODEL_DIR,\n \"container_spec\": {\"image_uri\": DEPLOY_IMAGE},\n },\n}\n\nprint(pipeline)",
"_____no_output_____"
]
],
[
[
"#### Create the training pipeline\n\nUse this helper function `create_pipeline`, which takes the following parameter:\n\n- `training_pipeline`: the full specification for the pipeline training job.\n\nThe helper function calls the pipeline client service's `create_pipeline` method, which takes the following parameters:\n\n- `parent`: The Vertex location root path for your `Dataset`, `Model` and `Endpoint` resources.\n- `training_pipeline`: The full specification for the pipeline training job.\n\nThe helper function will return the Vertex fully qualified identifier assigned to the training pipeline, which is saved as `pipeline.name`.",
"_____no_output_____"
]
],
[
[
"def create_pipeline(training_pipeline):\n\n try:\n pipeline = clients[\"pipeline\"].create_training_pipeline(\n parent=PARENT, training_pipeline=training_pipeline\n )\n print(pipeline)\n except Exception as e:\n print(\"exception:\", e)\n return None\n return pipeline\n\n\nresponse = create_pipeline(pipeline)",
"_____no_output_____"
]
],
[
[
"Now save the unique identifier of the training pipeline you created.",
"_____no_output_____"
]
],
[
[
"# The full unique ID for the pipeline\npipeline_id = response.name\n# The short numeric ID for the pipeline\npipeline_short_id = pipeline_id.split(\"/\")[-1]\n\nprint(pipeline_id)",
"_____no_output_____"
]
],
[
[
"### Get information on a training pipeline\n\nNow get pipeline information for just this training pipeline instance. The helper function gets the job information for just this job by calling the the job client service's `get_training_pipeline` method, with the following parameter:\n\n- `name`: The Vertex fully qualified pipeline identifier.\n\nWhen the model is done training, the pipeline state will be `PIPELINE_STATE_SUCCEEDED`.",
"_____no_output_____"
]
],
[
[
"def get_training_pipeline(name, silent=False):\n response = clients[\"pipeline\"].get_training_pipeline(name=name)\n if silent:\n return response\n\n print(\"pipeline\")\n print(\" name:\", response.name)\n print(\" display_name:\", response.display_name)\n print(\" state:\", response.state)\n print(\" training_task_definition:\", response.training_task_definition)\n print(\" training_task_inputs:\", dict(response.training_task_inputs))\n print(\" create_time:\", response.create_time)\n print(\" start_time:\", response.start_time)\n print(\" end_time:\", response.end_time)\n print(\" update_time:\", response.update_time)\n print(\" labels:\", dict(response.labels))\n return response\n\n\nresponse = get_training_pipeline(pipeline_id)",
"_____no_output_____"
]
],
[
[
"# Deployment\n\nTraining the above model may take upwards of 20 minutes time.\n\nOnce your model is done training, you can calculate the actual time it took to train the model by subtracting `end_time` from `start_time`. For your model, you will need to know the fully qualified Vertex Model resource identifier, which the pipeline service assigned to it. You can get this from the returned pipeline instance as the field `model_to_deploy.name`.",
"_____no_output_____"
]
],
[
[
"while True:\n response = get_training_pipeline(pipeline_id, True)\n if response.state != aip.PipelineState.PIPELINE_STATE_SUCCEEDED:\n print(\"Training job has not completed:\", response.state)\n model_to_deploy_id = None\n if response.state == aip.PipelineState.PIPELINE_STATE_FAILED:\n raise Exception(\"Training Job Failed\")\n else:\n model_to_deploy = response.model_to_upload\n model_to_deploy_id = model_to_deploy.name\n print(\"Training Time:\", response.end_time - response.start_time)\n break\n time.sleep(60)\n\nprint(\"model to deploy:\", model_to_deploy_id)",
"_____no_output_____"
],
[
"if not DIRECT:\n MODEL_DIR = MODEL_DIR + \"/model\"\nmodel_path_to_deploy = MODEL_DIR",
"_____no_output_____"
]
],
[
[
"## Load the saved model\n\nYour model is stored in a TensorFlow SavedModel format in a Cloud Storage bucket. Now load it from the Cloud Storage bucket, and then you can do some things, like evaluate the model, and do a prediction.\n\nTo load, you use the TF.Keras `model.load_model()` method passing it the Cloud Storage path where the model is saved -- specified by `MODEL_DIR`.",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\n\nmodel = tf.keras.models.load_model(MODEL_DIR)",
"_____no_output_____"
]
],
[
[
"## Evaluate the model\n\nNow let's find out how good the model is.\n\n### Load evaluation data\n\nYou will load the Boston Housing test (holdout) data from `tf.keras.datasets`, using the method `load_data()`. This will return the dataset as a tuple of two elements. The first element is the training data and the second is the test data. Each element is also a tuple of two elements: the feature data, and the corresponding labels (median value of owner-occupied home).\n\nYou don't need the training data, and hence why we loaded it as `(_, _)`.\n\nBefore you can run the data through evaluation, you need to preprocess it:\n\nx_test:\n1. Normalize (rescaling) the data in each column by dividing each value by the maximum value of that column. This will replace each single value with a 32-bit floating point number between 0 and 1.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom tensorflow.keras.datasets import boston_housing\n\n(_, _), (x_test, y_test) = boston_housing.load_data(\n path=\"boston_housing.npz\", test_split=0.2, seed=113\n)\n\n\ndef scale(feature):\n max = np.max(feature)\n feature = (feature / max).astype(np.float32)\n return feature\n\n\n# Let's save one data item that has not been scaled\nx_test_notscaled = x_test[0:1].copy()\n\nfor _ in range(13):\n x_test[_] = scale(x_test[_])\nx_test = x_test.astype(np.float32)\n\nprint(x_test.shape, x_test.dtype, y_test.shape)\nprint(\"scaled\", x_test[0])\nprint(\"unscaled\", x_test_notscaled)",
"_____no_output_____"
]
],
[
[
"### Perform the model evaluation\n\nNow evaluate how well the model in the custom job did.",
"_____no_output_____"
]
],
[
[
"model.evaluate(x_test, y_test)",
"_____no_output_____"
]
],
[
[
"## Upload the model for serving\n\nNext, you will upload your TF.Keras model from the custom job to Vertex `Model` service, which will create a Vertex `Model` resource for your custom model. During upload, you need to define a serving function to convert data to the format your model expects. If you send encoded data to Vertex, your serving function ensures that the data is decoded on the model server before it is passed as input to your model.\n\n### How does the serving function work\n\nWhen you send a request to an online prediction server, the request is received by a HTTP server. The HTTP server extracts the prediction request from the HTTP request content body. The extracted prediction request is forwarded to the serving function. For Google pre-built prediction containers, the request content is passed to the serving function as a `tf.string`.\n\nThe serving function consists of two parts:\n\n- `preprocessing function`:\n - Converts the input (`tf.string`) to the input shape and data type of the underlying model (dynamic graph).\n - Performs the same preprocessing of the data that was done during training the underlying model -- e.g., normalizing, scaling, etc.\n- `post-processing function`:\n - Converts the model output to format expected by the receiving application -- e.q., compresses the output.\n - Packages the output for the the receiving application -- e.g., add headings, make JSON object, etc.\n\nBoth the preprocessing and post-processing functions are converted to static graphs which are fused to the model. The output from the underlying model is passed to the post-processing function. The post-processing function passes the converted/packaged output back to the HTTP server. The HTTP server returns the output as the HTTP response content.\n\nOne consideration you need to consider when building serving functions for TF.Keras models is that they run as static graphs. That means, you cannot use TF graph operations that require a dynamic graph. If you do, you will get an error during the compile of the serving function which will indicate that you are using an EagerTensor which is not supported.",
"_____no_output_____"
],
[
"## Get the serving function signature\n\nYou can get the signatures of your model's input and output layers by reloading the model into memory, and querying it for the signatures corresponding to each layer.\n\nWhen making a prediction request, you need to route the request to the serving function instead of the model, so you need to know the input layer name of the serving function -- which you will use later when you make a prediction request.",
"_____no_output_____"
]
],
[
[
"loaded = tf.saved_model.load(model_path_to_deploy)\n\nserving_input = list(\n loaded.signatures[\"serving_default\"].structured_input_signature[1].keys()\n)[0]\nprint(\"Serving function input:\", serving_input)",
"_____no_output_____"
]
],
[
[
"### Upload the model\n\nUse this helper function `upload_model` to upload your model, stored in SavedModel format, up to the `Model` service, which will instantiate a Vertex `Model` resource instance for your model. Once you've done that, you can use the `Model` resource instance in the same way as any other Vertex `Model` resource instance, such as deploying to an `Endpoint` resource for serving predictions.\n\nThe helper function takes the following parameters:\n\n- `display_name`: A human readable name for the `Endpoint` service.\n- `image_uri`: The container image for the model deployment.\n- `model_uri`: The Cloud Storage path to our SavedModel artificat. For this tutorial, this is the Cloud Storage location where the `trainer/task.py` saved the model artifacts, which we specified in the variable `MODEL_DIR`.\n\nThe helper function calls the `Model` client service's method `upload_model`, which takes the following parameters:\n\n- `parent`: The Vertex location root path for `Dataset`, `Model` and `Endpoint` resources.\n- `model`: The specification for the Vertex `Model` resource instance.\n\nLet's now dive deeper into the Vertex model specification `model`. This is a dictionary object that consists of the following fields:\n\n- `display_name`: A human readable name for the `Model` resource.\n- `metadata_schema_uri`: Since your model was built without an Vertex `Dataset` resource, you will leave this blank (`''`).\n- `artificat_uri`: The Cloud Storage path where the model is stored in SavedModel format.\n- `container_spec`: This is the specification for the Docker container that will be installed on the `Endpoint` resource, from which the `Model` resource will serve predictions. Use the variable you set earlier `DEPLOY_GPU != None` to use a GPU; otherwise only a CPU is allocated.\n\nUploading a model into a Vertex Model resource returns a long running operation, since it may take a few moments. You call response.result(), which is a synchronous call and will return when the Vertex Model resource is ready.\n\nThe helper function returns the Vertex fully qualified identifier for the corresponding Vertex Model instance upload_model_response.model. You will save the identifier for subsequent steps in the variable model_to_deploy_id.",
"_____no_output_____"
]
],
[
[
"IMAGE_URI = DEPLOY_IMAGE\n\n\ndef upload_model(display_name, image_uri, model_uri):\n model = {\n \"display_name\": display_name,\n \"metadata_schema_uri\": \"\",\n \"artifact_uri\": model_uri,\n \"container_spec\": {\n \"image_uri\": image_uri,\n \"command\": [],\n \"args\": [],\n \"env\": [{\"name\": \"env_name\", \"value\": \"env_value\"}],\n \"ports\": [{\"container_port\": 8080}],\n \"predict_route\": \"\",\n \"health_route\": \"\",\n },\n }\n response = clients[\"model\"].upload_model(parent=PARENT, model=model)\n print(\"Long running operation:\", response.operation.name)\n upload_model_response = response.result(timeout=180)\n print(\"upload_model_response\")\n print(\" model:\", upload_model_response.model)\n return upload_model_response.model\n\n\nmodel_to_deploy_id = upload_model(\n \"boston-\" + TIMESTAMP, IMAGE_URI, model_path_to_deploy\n)",
"_____no_output_____"
]
],
[
[
"### Get `Model` resource information\n\nNow let's get the model information for just your model. Use this helper function `get_model`, with the following parameter:\n\n- `name`: The Vertex unique identifier for the `Model` resource.\n\nThis helper function calls the Vertex `Model` client service's method `get_model`, with the following parameter:\n\n- `name`: The Vertex unique identifier for the `Model` resource.",
"_____no_output_____"
]
],
[
[
"def get_model(name):\n response = clients[\"model\"].get_model(name=name)\n print(response)\n\n\nget_model(model_to_deploy_id)",
"_____no_output_____"
]
],
[
[
"## Deploy the `Model` resource\n\nNow deploy the trained Vertex custom `Model` resource. This requires two steps:\n\n1. Create an `Endpoint` resource for deploying the `Model` resource to.\n\n2. Deploy the `Model` resource to the `Endpoint` resource.",
"_____no_output_____"
],
[
"### Create an `Endpoint` resource\n\nUse this helper function `create_endpoint` to create an endpoint to deploy the model to for serving predictions, with the following parameter:\n\n- `display_name`: A human readable name for the `Endpoint` resource.\n\nThe helper function uses the endpoint client service's `create_endpoint` method, which takes the following parameter:\n\n- `display_name`: A human readable name for the `Endpoint` resource.\n\nCreating an `Endpoint` resource returns a long running operation, since it may take a few moments to provision the `Endpoint` resource for serving. You call `response.result()`, which is a synchronous call and will return when the Endpoint resource is ready. The helper function returns the Vertex fully qualified identifier for the `Endpoint` resource: `response.name`.",
"_____no_output_____"
]
],
[
[
"ENDPOINT_NAME = \"boston_endpoint-\" + TIMESTAMP\n\n\ndef create_endpoint(display_name):\n endpoint = {\"display_name\": display_name}\n response = clients[\"endpoint\"].create_endpoint(parent=PARENT, endpoint=endpoint)\n print(\"Long running operation:\", response.operation.name)\n\n result = response.result(timeout=300)\n print(\"result\")\n print(\" name:\", result.name)\n print(\" display_name:\", result.display_name)\n print(\" description:\", result.description)\n print(\" labels:\", result.labels)\n print(\" create_time:\", result.create_time)\n print(\" update_time:\", result.update_time)\n return result\n\n\nresult = create_endpoint(ENDPOINT_NAME)",
"_____no_output_____"
]
],
[
[
"Now get the unique identifier for the `Endpoint` resource you created.",
"_____no_output_____"
]
],
[
[
"# The full unique ID for the endpoint\nendpoint_id = result.name\n# The short numeric ID for the endpoint\nendpoint_short_id = endpoint_id.split(\"/\")[-1]\n\nprint(endpoint_id)",
"_____no_output_____"
]
],
[
[
"### Compute instance scaling\n\nYou have several choices on scaling the compute instances for handling your online prediction requests:\n\n- Single Instance: The online prediction requests are processed on a single compute instance.\n - Set the minimum (`MIN_NODES`) and maximum (`MAX_NODES`) number of compute instances to one.\n\n- Manual Scaling: The online prediction requests are split across a fixed number of compute instances that you manually specified.\n - Set the minimum (`MIN_NODES`) and maximum (`MAX_NODES`) number of compute instances to the same number of nodes. When a model is first deployed to the instance, the fixed number of compute instances are provisioned and online prediction requests are evenly distributed across them.\n\n- Auto Scaling: The online prediction requests are split across a scaleable number of compute instances.\n - Set the minimum (`MIN_NODES`) number of compute instances to provision when a model is first deployed and to de-provision, and set the maximum (`MAX_NODES) number of compute instances to provision, depending on load conditions.\n\nThe minimum number of compute instances corresponds to the field `min_replica_count` and the maximum number of compute instances corresponds to the field `max_replica_count`, in your subsequent deployment request.",
"_____no_output_____"
]
],
[
[
"MIN_NODES = 1\nMAX_NODES = 1",
"_____no_output_____"
]
],
[
[
"### Deploy `Model` resource to the `Endpoint` resource\n\nUse this helper function `deploy_model` to deploy the `Model` resource to the `Endpoint` resource you created for serving predictions, with the following parameters:\n\n- `model`: The Vertex fully qualified model identifier of the model to upload (deploy) from the training pipeline.\n- `deploy_model_display_name`: A human readable name for the deployed model.\n- `endpoint`: The Vertex fully qualified endpoint identifier to deploy the model to.\n\nThe helper function calls the `Endpoint` client service's method `deploy_model`, which takes the following parameters:\n\n- `endpoint`: The Vertex fully qualified `Endpoint` resource identifier to deploy the `Model` resource to.\n- `deployed_model`: The requirements specification for deploying the model.\n- `traffic_split`: Percent of traffic at the endpoint that goes to this model, which is specified as a dictionary of one or more key/value pairs.\n - If only one model, then specify as **{ \"0\": 100 }**, where \"0\" refers to this model being uploaded and 100 means 100% of the traffic.\n - If there are existing models on the endpoint, for which the traffic will be split, then use `model_id` to specify as **{ \"0\": percent, model_id: percent, ... }**, where `model_id` is the model id of an existing model to the deployed endpoint. The percents must add up to 100.\n\nLet's now dive deeper into the `deployed_model` parameter. This parameter is specified as a Python dictionary with the minimum required fields:\n\n- `model`: The Vertex fully qualified model identifier of the (upload) model to deploy.\n- `display_name`: A human readable name for the deployed model.\n- `disable_container_logging`: This disables logging of container events, such as execution failures (default is container logging is enabled). Container logging is typically enabled when debugging the deployment and then disabled when deployed for production.\n- `dedicated_resources`: This refers to how many compute instances (replicas) that are scaled for serving prediction requests.\n - `machine_spec`: The compute instance to provision. Use the variable you set earlier `DEPLOY_GPU != None` to use a GPU; otherwise only a CPU is allocated.\n - `min_replica_count`: The number of compute instances to initially provision, which you set earlier as the variable `MIN_NODES`.\n - `max_replica_count`: The maximum number of compute instances to scale to, which you set earlier as the variable `MAX_NODES`.\n\n#### Traffic Split\n\nLet's now dive deeper into the `traffic_split` parameter. This parameter is specified as a Python dictionary. This might at first be a tad bit confusing. Let me explain, you can deploy more than one instance of your model to an endpoint, and then set how much (percent) goes to each instance.\n\nWhy would you do that? Perhaps you already have a previous version deployed in production -- let's call that v1. You got better model evaluation on v2, but you don't know for certain that it is really better until you deploy to production. So in the case of traffic split, you might want to deploy v2 to the same endpoint as v1, but it only get's say 10% of the traffic. That way, you can monitor how well it does without disrupting the majority of users -- until you make a final decision.\n\n#### Response\n\nThe method returns a long running operation `response`. We will wait sychronously for the operation to complete by calling the `response.result()`, which will block until the model is deployed. If this is the first time a model is deployed to the endpoint, it may take a few additional minutes to complete provisioning of resources.",
"_____no_output_____"
]
],
[
[
"DEPLOYED_NAME = \"boston_deployed-\" + TIMESTAMP\n\n\ndef deploy_model(\n model, deployed_model_display_name, endpoint, traffic_split={\"0\": 100}\n):\n\n if DEPLOY_GPU:\n machine_spec = {\n \"machine_type\": DEPLOY_COMPUTE,\n \"accelerator_type\": DEPLOY_GPU,\n \"accelerator_count\": DEPLOY_NGPU,\n }\n else:\n machine_spec = {\n \"machine_type\": DEPLOY_COMPUTE,\n \"accelerator_count\": 0,\n }\n\n deployed_model = {\n \"model\": model,\n \"display_name\": deployed_model_display_name,\n \"dedicated_resources\": {\n \"min_replica_count\": MIN_NODES,\n \"max_replica_count\": MAX_NODES,\n \"machine_spec\": machine_spec,\n },\n \"disable_container_logging\": False,\n }\n\n response = clients[\"endpoint\"].deploy_model(\n endpoint=endpoint, deployed_model=deployed_model, traffic_split=traffic_split\n )\n\n print(\"Long running operation:\", response.operation.name)\n result = response.result()\n print(\"result\")\n deployed_model = result.deployed_model\n print(\" deployed_model\")\n print(\" id:\", deployed_model.id)\n print(\" model:\", deployed_model.model)\n print(\" display_name:\", deployed_model.display_name)\n print(\" create_time:\", deployed_model.create_time)\n\n return deployed_model.id\n\n\ndeployed_model_id = deploy_model(model_to_deploy_id, DEPLOYED_NAME, endpoint_id)",
"_____no_output_____"
]
],
[
[
"## Make a online prediction request\n\nNow do a online prediction to your deployed model.",
"_____no_output_____"
],
[
"### Get test item\n\nYou will use an example out of the test (holdout) portion of the dataset as a test item.",
"_____no_output_____"
]
],
[
[
"test_item = x_test[0]\ntest_label = y_test[0]\nprint(test_item.shape)",
"_____no_output_____"
]
],
[
[
"### Send the prediction request\n\nOk, now you have a test data item. Use this helper function `predict_data`, which takes the parameters:\n\n- `data`: The test data item as a numpy 1D array of floating point values.\n- `endpoint`: The Vertex fully qualified identifier for the `Endpoint` resource where the `Model` resource was deployed.\n- `parameters_dict`: Additional parameters for serving.\n\nThis function uses the prediction client service and calls the `predict` method with the parameters:\n\n- `endpoint`: The Vertex fully qualified identifier for the `Endpoint` resource where the `Model` resource was deployed.\n- `instances`: A list of instances (data items) to predict.\n- `parameters`: Additional parameters for serving.\n\nTo pass the test data to the prediction service, you package it for transmission to the serving binary as follows:\n\n 1. Convert the data item from a 1D numpy array to a 1D Python list.\n 2. Convert the prediction request to a serialized Google protobuf (`json_format.ParseDict()`)\n\n\nEach instance in the prediction request is a dictionary entry of the form:\n\n {input_name: content}\n\n- `input_name`: the name of the input layer of the underlying model.\n- `content`: The data item as a 1D Python list.\n\nSince the `predict()` service can take multiple data items (instances), you will send your single data item as a list of one data item. As a final step, you package the instances list into Google's protobuf format -- which is what we pass to the `predict()` service.\n\nThe `response` object returns a list, where each element in the list corresponds to the corresponding image in the request. You will see in the output for each prediction:\n\n- `predictions` -- the predicated median value of a house in units of 1K USD.",
"_____no_output_____"
]
],
[
[
"def predict_data(data, endpoint, parameters_dict):\n parameters = json_format.ParseDict(parameters_dict, Value())\n\n # The format of each instance should conform to the deployed model's prediction input schema.\n instances_list = [{serving_input: data.tolist()}]\n instances = [json_format.ParseDict(s, Value()) for s in instances_list]\n\n response = clients[\"prediction\"].predict(\n endpoint=endpoint, instances=instances, parameters=parameters\n )\n print(\"response\")\n print(\" deployed_model_id:\", response.deployed_model_id)\n predictions = response.predictions\n print(\"predictions\")\n for prediction in predictions:\n print(\" prediction:\", prediction)\n\n\npredict_data(test_item, endpoint_id, None)",
"_____no_output_____"
]
],
[
[
"## Undeploy the `Model` resource\n\nNow undeploy your `Model` resource from the serving `Endpoint` resoure. Use this helper function `undeploy_model`, which takes the following parameters:\n\n- `deployed_model_id`: The model deployment identifier returned by the endpoint service when the `Model` resource was deployed to.\n- `endpoint`: The Vertex fully qualified identifier for the `Endpoint` resource where the `Model` is deployed to.\n\nThis function calls the endpoint client service's method `undeploy_model`, with the following parameters:\n\n- `deployed_model_id`: The model deployment identifier returned by the endpoint service when the `Model` resource was deployed.\n- `endpoint`: The Vertex fully qualified identifier for the `Endpoint` resource where the `Model` resource is deployed.\n- `traffic_split`: How to split traffic among the remaining deployed models on the `Endpoint` resource.\n\nSince this is the only deployed model on the `Endpoint` resource, you simply can leave `traffic_split` empty by setting it to {}.",
"_____no_output_____"
]
],
[
[
"def undeploy_model(deployed_model_id, endpoint):\n response = clients[\"endpoint\"].undeploy_model(\n endpoint=endpoint, deployed_model_id=deployed_model_id, traffic_split={}\n )\n print(response)\n\n\nundeploy_model(deployed_model_id, endpoint_id)",
"_____no_output_____"
]
],
[
[
"# Cleaning up\n\nTo clean up all GCP resources used in this project, you can [delete the GCP\nproject](https://cloud.google.com/resource-manager/docs/creating-managing-projects#shutting_down_projects) you used for the tutorial.\n\nOtherwise, you can delete the individual resources you created in this tutorial:\n\n- Dataset\n- Pipeline\n- Model\n- Endpoint\n- Batch Job\n- Custom Job\n- Hyperparameter Tuning Job\n- Cloud Storage Bucket",
"_____no_output_____"
]
],
[
[
"delete_dataset = True\ndelete_pipeline = True\ndelete_model = True\ndelete_endpoint = True\ndelete_batchjob = True\ndelete_customjob = True\ndelete_hptjob = True\ndelete_bucket = True\n\n# Delete the dataset using the Vertex fully qualified identifier for the dataset\ntry:\n if delete_dataset and \"dataset_id\" in globals():\n clients[\"dataset\"].delete_dataset(name=dataset_id)\nexcept Exception as e:\n print(e)\n\n# Delete the training pipeline using the Vertex fully qualified identifier for the pipeline\ntry:\n if delete_pipeline and \"pipeline_id\" in globals():\n clients[\"pipeline\"].delete_training_pipeline(name=pipeline_id)\nexcept Exception as e:\n print(e)\n\n# Delete the model using the Vertex fully qualified identifier for the model\ntry:\n if delete_model and \"model_to_deploy_id\" in globals():\n clients[\"model\"].delete_model(name=model_to_deploy_id)\nexcept Exception as e:\n print(e)\n\n# Delete the endpoint using the Vertex fully qualified identifier for the endpoint\ntry:\n if delete_endpoint and \"endpoint_id\" in globals():\n clients[\"endpoint\"].delete_endpoint(name=endpoint_id)\nexcept Exception as e:\n print(e)\n\n# Delete the batch job using the Vertex fully qualified identifier for the batch job\ntry:\n if delete_batchjob and \"batch_job_id\" in globals():\n clients[\"job\"].delete_batch_prediction_job(name=batch_job_id)\nexcept Exception as e:\n print(e)\n\n# Delete the custom job using the Vertex fully qualified identifier for the custom job\ntry:\n if delete_customjob and \"job_id\" in globals():\n clients[\"job\"].delete_custom_job(name=job_id)\nexcept Exception as e:\n print(e)\n\n# Delete the hyperparameter tuning job using the Vertex fully qualified identifier for the hyperparameter tuning job\ntry:\n if delete_hptjob and \"hpt_job_id\" in globals():\n clients[\"job\"].delete_hyperparameter_tuning_job(name=hpt_job_id)\nexcept Exception as e:\n print(e)\n\nif delete_bucket and \"BUCKET_NAME\" in globals():\n ! gsutil rm -r $BUCKET_NAME",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d086407af685fc6351fa88c1c340d078f5476e9f | 476,642 | ipynb | Jupyter Notebook | Workshop/GRU_236.ipynb | ShepherdCode/ShepherdML | fd8d71c63f7bd788ea0052294d93e43246254a12 | [
"MIT"
] | null | null | null | Workshop/GRU_236.ipynb | ShepherdCode/ShepherdML | fd8d71c63f7bd788ea0052294d93e43246254a12 | [
"MIT"
] | 4 | 2020-03-24T18:05:09.000Z | 2020-12-22T17:42:54.000Z | Workshop/GRU_236.ipynb | ShepherdCode/ShepherdML | fd8d71c63f7bd788ea0052294d93e43246254a12 | [
"MIT"
] | null | null | null | 171.392305 | 56,822 | 0.727215 | [
[
[
"# GRU 236\n* Operate on 16000 GenCode 34 seqs.\n* 5-way cross validation. Save best model per CV.\n* Report mean accuracy from final re-validation with best 5.\n* Use Adam with a learn rate decay schdule.",
"_____no_output_____"
]
],
[
[
"NC_FILENAME='ncRNA.gc34.processed.fasta'\nPC_FILENAME='pcRNA.gc34.processed.fasta'\nDATAPATH=\"\"\ntry:\n from google.colab import drive\n IN_COLAB = True\n PATH='/content/drive/'\n drive.mount(PATH)\n DATAPATH=PATH+'My Drive/data/' # must end in \"/\"\n NC_FILENAME = DATAPATH+NC_FILENAME\n PC_FILENAME = DATAPATH+PC_FILENAME\nexcept:\n IN_COLAB = False\n DATAPATH=\"\" \n\nEPOCHS=200\nSPLITS=5\nK=3\nVOCABULARY_SIZE=4**K+1 # e.g. K=3 => 64 DNA K-mers + 'NNN'\nEMBED_DIMEN=16\nFILENAME='GRU236'\nNEURONS=64\nACT=\"tanh\"\nDROP=0.5\n",
"Mounted at /content/drive/\n"
],
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nfrom sklearn.model_selection import ShuffleSplit\nfrom sklearn.model_selection import cross_val_score\nfrom sklearn.model_selection import RepeatedKFold\nfrom sklearn.model_selection import StratifiedKFold\nimport tensorflow as tf\nfrom tensorflow import keras\nfrom keras.wrappers.scikit_learn import KerasRegressor\nfrom keras.models import Sequential\nfrom keras.layers import Bidirectional\nfrom keras.layers import GRU\nfrom keras.layers import Dense\nfrom keras.layers import LayerNormalization\nimport time\ndt='float32'\ntf.keras.backend.set_floatx(dt)",
"_____no_output_____"
]
],
[
[
"## Build model",
"_____no_output_____"
]
],
[
[
"def compile_model(model):\n adam_default_learn_rate = 0.001\n schedule = tf.keras.optimizers.schedules.ExponentialDecay(\n initial_learning_rate = adam_default_learn_rate*10,\n #decay_steps=100000, decay_rate=0.96, staircase=True)\n decay_steps=10000, decay_rate=0.99, staircase=True)\n # learn rate = initial_learning_rate * decay_rate ^ (step / decay_steps)\n alrd = tf.keras.optimizers.Adam(learning_rate=schedule)\n bc=tf.keras.losses.BinaryCrossentropy(from_logits=False)\n print(\"COMPILE...\")\n #model.compile(loss=bc, optimizer=alrd, metrics=[\"accuracy\"])\n model.compile(loss=bc, optimizer=\"adam\", metrics=[\"accuracy\"])\n print(\"...COMPILED\")\n return model\n\ndef build_model():\n embed_layer = keras.layers.Embedding(\n #VOCABULARY_SIZE, EMBED_DIMEN, input_length=1000, input_length=1000, mask_zero=True)\n #input_dim=[None,VOCABULARY_SIZE], output_dim=EMBED_DIMEN, mask_zero=True)\n input_dim=VOCABULARY_SIZE, output_dim=EMBED_DIMEN, mask_zero=True)\n #rnn1_layer = keras.layers.Bidirectional(\n rnn1_layer = keras.layers.GRU(NEURONS, return_sequences=True, \n input_shape=[1000,EMBED_DIMEN], activation=ACT, dropout=DROP)#)#bi\n #rnn2_layer = keras.layers.Bidirectional(\n rnn2_layer = keras.layers.GRU(NEURONS, return_sequences=False, \n activation=ACT, dropout=DROP)#)#bi\n dense1_layer = keras.layers.Dense(NEURONS, activation=ACT,dtype=dt)\n #drop1_layer = keras.layers.Dropout(DROP)\n dense2_layer = keras.layers.Dense(NEURONS, activation=ACT,dtype=dt)\n #drop2_layer = keras.layers.Dropout(DROP)\n output_layer = keras.layers.Dense(1, activation=\"sigmoid\", dtype=dt)\n mlp = keras.models.Sequential()\n mlp.add(embed_layer)\n mlp.add(rnn1_layer)\n mlp.add(rnn2_layer)\n mlp.add(dense1_layer)\n #mlp.add(drop1_layer)\n mlp.add(dense2_layer)\n #mlp.add(drop2_layer)\n mlp.add(output_layer)\n mlpc = compile_model(mlp)\n return mlpc",
"_____no_output_____"
]
],
[
[
"## Load and partition sequences",
"_____no_output_____"
]
],
[
[
"# Assume file was preprocessed to contain one line per seq.\n# Prefer Pandas dataframe but df does not support append.\n# For conversion to tensor, must avoid python lists.\ndef load_fasta(filename,label):\n DEFLINE='>'\n labels=[]\n seqs=[]\n lens=[]\n nums=[]\n num=0\n with open (filename,'r') as infile:\n for line in infile:\n if line[0]!=DEFLINE:\n seq=line.rstrip()\n num += 1 # first seqnum is 1\n seqlen=len(seq)\n nums.append(num)\n labels.append(label)\n seqs.append(seq)\n lens.append(seqlen)\n df1=pd.DataFrame(nums,columns=['seqnum'])\n df2=pd.DataFrame(labels,columns=['class'])\n df3=pd.DataFrame(seqs,columns=['sequence'])\n df4=pd.DataFrame(lens,columns=['seqlen'])\n df=pd.concat((df1,df2,df3,df4),axis=1)\n return df\n\ndef separate_X_and_y(data):\n y= data[['class']].copy()\n X= data.drop(columns=['class','seqnum','seqlen'])\n return (X,y)\n\n",
"_____no_output_____"
]
],
[
[
"## Make K-mers",
"_____no_output_____"
]
],
[
[
"def make_kmer_table(K):\n npad='N'*K\n shorter_kmers=['']\n for i in range(K):\n longer_kmers=[]\n for mer in shorter_kmers:\n longer_kmers.append(mer+'A')\n longer_kmers.append(mer+'C')\n longer_kmers.append(mer+'G')\n longer_kmers.append(mer+'T')\n shorter_kmers = longer_kmers\n all_kmers = shorter_kmers\n kmer_dict = {}\n kmer_dict[npad]=0\n value=1\n for mer in all_kmers:\n kmer_dict[mer]=value\n value += 1\n return kmer_dict\n\nKMER_TABLE=make_kmer_table(K)\n\ndef strings_to_vectors(data,uniform_len):\n all_seqs=[]\n for seq in data['sequence']:\n i=0\n seqlen=len(seq)\n kmers=[]\n while i < seqlen-K+1 -1: # stop at minus one for spaced seed\n #kmer=seq[i:i+2]+seq[i+3:i+5] # SPACED SEED 2/1/2 for K=4\n kmer=seq[i:i+K] \n i += 1\n value=KMER_TABLE[kmer]\n kmers.append(value)\n pad_val=0\n while i < uniform_len:\n kmers.append(pad_val)\n i += 1\n all_seqs.append(kmers)\n pd2d=pd.DataFrame(all_seqs)\n return pd2d # return 2D dataframe, uniform dimensions",
"_____no_output_____"
],
[
"def make_kmers(MAXLEN,train_set):\n (X_train_all,y_train_all)=separate_X_and_y(train_set)\n X_train_kmers=strings_to_vectors(X_train_all,MAXLEN)\n # From pandas dataframe to numpy to list to numpy\n num_seqs=len(X_train_kmers)\n tmp_seqs=[]\n for i in range(num_seqs):\n kmer_sequence=X_train_kmers.iloc[i]\n tmp_seqs.append(kmer_sequence)\n X_train_kmers=np.array(tmp_seqs)\n tmp_seqs=None\n labels=y_train_all.to_numpy()\n return (X_train_kmers,labels)",
"_____no_output_____"
],
[
"def make_frequencies(Xin):\n Xout=[]\n VOCABULARY_SIZE= 4**K + 1 # plus one for 'NNN'\n for seq in Xin:\n freqs =[0] * VOCABULARY_SIZE\n total = 0\n for kmerval in seq:\n freqs[kmerval] += 1\n total += 1\n for c in range(VOCABULARY_SIZE):\n freqs[c] = freqs[c]/total\n Xout.append(freqs)\n Xnum = np.asarray(Xout)\n return (Xnum)\ndef make_slice(data_set,min_len,max_len):\n slice = data_set.query('seqlen <= '+str(max_len)+' & seqlen>= '+str(min_len))\n return slice",
"_____no_output_____"
]
],
[
[
"## Cross validation",
"_____no_output_____"
]
],
[
[
"def do_cross_validation(X,y,given_model):\n cv_scores = []\n fold=0\n splitter = ShuffleSplit(n_splits=SPLITS, test_size=0.1) #, random_state=37863)\n for train_index,valid_index in splitter.split(X):\n fold += 1\n X_train=X[train_index] # use iloc[] for dataframe\n y_train=y[train_index]\n X_valid=X[valid_index]\n y_valid=y[valid_index] \n # Avoid continually improving the same model.\n model = compile_model(keras.models.clone_model(given_model))\n bestname=DATAPATH+FILENAME+\".cv.\"+str(fold)+\".best\"\n mycallbacks = [keras.callbacks.ModelCheckpoint(\n filepath=bestname, save_best_only=True, \n monitor='val_accuracy', mode='max')] \n print(\"FIT\")\n start_time=time.time()\n history=model.fit(X_train, y_train, # batch_size=10, default=32 works nicely\n epochs=EPOCHS, verbose=1, # verbose=1 for ascii art, verbose=0 for none\n callbacks=mycallbacks,\n validation_data=(X_valid,y_valid) )\n end_time=time.time()\n elapsed_time=(end_time-start_time) \n print(\"Fold %d, %d epochs, %d sec\"%(fold,EPOCHS,elapsed_time))\n pd.DataFrame(history.history).plot(figsize=(8,5))\n plt.grid(True)\n plt.gca().set_ylim(0,1)\n plt.show()\n best_model=keras.models.load_model(bestname)\n scores = best_model.evaluate(X_valid, y_valid, verbose=0)\n print(\"%s: %.2f%%\" % (best_model.metrics_names[1], scores[1]*100))\n cv_scores.append(scores[1] * 100) \n print()\n print(\"%d-way Cross Validation mean %.2f%% (+/- %.2f%%)\" % (fold, np.mean(cv_scores), np.std(cv_scores)))",
"_____no_output_____"
]
],
[
[
"## Train on RNA lengths 200-1Kb",
"_____no_output_____"
]
],
[
[
"MINLEN=200\nMAXLEN=1000\nprint(\"Load data from files.\")\nnc_seq=load_fasta(NC_FILENAME,0)\npc_seq=load_fasta(PC_FILENAME,1)\ntrain_set=pd.concat((nc_seq,pc_seq),axis=0)\nnc_seq=None\npc_seq=None\nprint(\"Ready: train_set\")\n#train_set\nsubset=make_slice(train_set,MINLEN,MAXLEN)# One array to two: X and y\nprint (\"Data reshape\")\n(X_train,y_train)=make_kmers(MAXLEN,subset)\n#print (\"Data prep\")\n#X_train=make_frequencies(X_train)",
"Load data from files.\nReady: train_set\nData reshape\n"
],
[
"print (\"Compile the model\")\nmodel=build_model()\nprint (\"Summarize the model\")\nprint(model.summary()) # Print this only once\nmodel.save(DATAPATH+FILENAME+'.model')\n",
"Compile the model\nCOMPILE...\n...COMPILED\nSummarize the model\nModel: \"sequential\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nembedding (Embedding) (None, None, 16) 1040 \n_________________________________________________________________\ngru (GRU) (None, None, 64) 15744 \n_________________________________________________________________\ngru_1 (GRU) (None, 64) 24960 \n_________________________________________________________________\ndense (Dense) (None, 64) 4160 \n_________________________________________________________________\ndense_1 (Dense) (None, 64) 4160 \n_________________________________________________________________\ndense_2 (Dense) (None, 1) 65 \n=================================================================\nTotal params: 50,129\nTrainable params: 50,129\nNon-trainable params: 0\n_________________________________________________________________\nNone\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow/python/training/tracking/tracking.py:111: Model.state_updates (from tensorflow.python.keras.engine.training) is deprecated and will be removed in a future version.\nInstructions for updating:\nThis property should not be used in TensorFlow 2.0, as updates are applied automatically.\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow/python/training/tracking/tracking.py:111: Layer.updates (from tensorflow.python.keras.engine.base_layer) is deprecated and will be removed in a future version.\nInstructions for updating:\nThis property should not be used in TensorFlow 2.0, as updates are applied automatically.\nINFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU215r.model/assets\n"
],
[
"print (\"Cross valiation\")\ndo_cross_validation(X_train,y_train,model) \nprint (\"Done\")",
"Cross valiation\nCOMPILE...\n...COMPILED\nFIT\nEpoch 1/200\n453/453 [==============================] - ETA: 0s - loss: 0.6361 - accuracy: 0.6476INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU215r.cv.1.best/assets\n453/453 [==============================] - 52s 114ms/step - loss: 0.6361 - accuracy: 0.6476 - val_loss: 0.6820 - val_accuracy: 0.4444\nEpoch 2/200\n453/453 [==============================] - ETA: 0s - loss: 0.6361 - accuracy: 0.6537INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU215r.cv.1.best/assets\n453/453 [==============================] - 48s 106ms/step - loss: 0.6361 - accuracy: 0.6537 - val_loss: 0.6581 - val_accuracy: 0.6344\nEpoch 3/200\n453/453 [==============================] - ETA: 0s - loss: 0.6074 - accuracy: 0.6732INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU215r.cv.1.best/assets\n453/453 [==============================] - 48s 107ms/step - loss: 0.6074 - accuracy: 0.6732 - val_loss: 0.5766 - val_accuracy: 0.7207\nEpoch 4/200\n453/453 [==============================] - ETA: 0s - loss: 0.4915 - accuracy: 0.7705INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU215r.cv.1.best/assets\n453/453 [==============================] - 49s 107ms/step - loss: 0.4915 - accuracy: 0.7705 - val_loss: 0.4512 - val_accuracy: 0.7958\nEpoch 5/200\n453/453 [==============================] - ETA: 0s - loss: 0.4407 - accuracy: 0.8014INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU215r.cv.1.best/assets\n453/453 [==============================] - 48s 106ms/step - loss: 0.4407 - accuracy: 0.8014 - val_loss: 0.4440 - val_accuracy: 0.8076\nEpoch 6/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.4270 - accuracy: 0.8065 - val_loss: 0.4494 - val_accuracy: 0.7827\nEpoch 7/200\n453/453 [==============================] - ETA: 0s - loss: 0.4182 - accuracy: 0.8125INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU215r.cv.1.best/assets\n453/453 [==============================] - 48s 106ms/step - loss: 0.4182 - accuracy: 0.8125 - val_loss: 0.3862 - val_accuracy: 0.8268\nEpoch 8/200\n453/453 [==============================] - 33s 73ms/step - loss: 0.4056 - accuracy: 0.8207 - val_loss: 0.3965 - val_accuracy: 0.8237\nEpoch 9/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3990 - accuracy: 0.8232 - val_loss: 0.3983 - val_accuracy: 0.8206\nEpoch 10/200\n453/453 [==============================] - 33s 73ms/step - loss: 0.3949 - accuracy: 0.8276 - val_loss: 0.4184 - val_accuracy: 0.8094\nEpoch 11/200\n453/453 [==============================] - ETA: 0s - loss: 0.3903 - accuracy: 0.8263INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU215r.cv.1.best/assets\n453/453 [==============================] - 49s 108ms/step - loss: 0.3903 - accuracy: 0.8263 - val_loss: 0.3728 - val_accuracy: 0.8312\nEpoch 12/200\n453/453 [==============================] - ETA: 0s - loss: 0.3849 - accuracy: 0.8271INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU215r.cv.1.best/assets\n453/453 [==============================] - 49s 107ms/step - loss: 0.3849 - accuracy: 0.8271 - val_loss: 0.3765 - val_accuracy: 0.8324\nEpoch 13/200\n453/453 [==============================] - 33s 73ms/step - loss: 0.3824 - accuracy: 0.8290 - val_loss: 0.3780 - val_accuracy: 0.8305\nEpoch 14/200\n453/453 [==============================] - ETA: 0s - loss: 0.3787 - accuracy: 0.8349INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU215r.cv.1.best/assets\n453/453 [==============================] - 49s 107ms/step - loss: 0.3787 - accuracy: 0.8349 - val_loss: 0.3623 - val_accuracy: 0.8367\nEpoch 15/200\n453/453 [==============================] - ETA: 0s - loss: 0.3752 - accuracy: 0.8352INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU215r.cv.1.best/assets\n453/453 [==============================] - 48s 105ms/step - loss: 0.3752 - accuracy: 0.8352 - val_loss: 0.3562 - val_accuracy: 0.8448\nEpoch 16/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3752 - accuracy: 0.8323 - val_loss: 0.3541 - val_accuracy: 0.8430\nEpoch 17/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3685 - accuracy: 0.8353 - val_loss: 0.4017 - val_accuracy: 0.8125\nEpoch 18/200\n453/453 [==============================] - ETA: 0s - loss: 0.3584 - accuracy: 0.8450INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU215r.cv.1.best/assets\n453/453 [==============================] - 48s 107ms/step - loss: 0.3584 - accuracy: 0.8450 - val_loss: 0.3387 - val_accuracy: 0.8529\nEpoch 19/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3498 - accuracy: 0.8490 - val_loss: 0.3511 - val_accuracy: 0.8479\nEpoch 20/200\n453/453 [==============================] - ETA: 0s - loss: 0.3440 - accuracy: 0.8515INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU215r.cv.1.best/assets\n453/453 [==============================] - 48s 105ms/step - loss: 0.3440 - accuracy: 0.8515 - val_loss: 0.3271 - val_accuracy: 0.8628\nEpoch 21/200\n453/453 [==============================] - ETA: 0s - loss: 0.3305 - accuracy: 0.8592INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU215r.cv.1.best/assets\n453/453 [==============================] - 47s 105ms/step - loss: 0.3305 - accuracy: 0.8592 - val_loss: 0.3013 - val_accuracy: 0.8715\nEpoch 22/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3209 - accuracy: 0.8655 - val_loss: 0.3096 - val_accuracy: 0.8715\nEpoch 23/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3288 - accuracy: 0.8620 - val_loss: 0.3177 - val_accuracy: 0.8547\nEpoch 24/200\n453/453 [==============================] - ETA: 0s - loss: 0.3139 - accuracy: 0.8687INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU215r.cv.1.best/assets\n453/453 [==============================] - 48s 106ms/step - loss: 0.3139 - accuracy: 0.8687 - val_loss: 0.3002 - val_accuracy: 0.8790\nEpoch 25/200\n453/453 [==============================] - 32s 72ms/step - loss: 0.3052 - accuracy: 0.8733 - val_loss: 0.4355 - val_accuracy: 0.7896\nEpoch 26/200\n453/453 [==============================] - 33s 73ms/step - loss: 0.4059 - accuracy: 0.8172 - val_loss: 0.3680 - val_accuracy: 0.8417\nEpoch 27/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3964 - accuracy: 0.8258 - val_loss: 0.3708 - val_accuracy: 0.8367\nEpoch 28/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3360 - accuracy: 0.8547 - val_loss: 0.3075 - val_accuracy: 0.8746\nEpoch 29/200\n453/453 [==============================] - 33s 73ms/step - loss: 0.3511 - accuracy: 0.8504 - val_loss: 0.3812 - val_accuracy: 0.8367\nEpoch 30/200\n453/453 [==============================] - 32s 72ms/step - loss: 0.3702 - accuracy: 0.8400 - val_loss: 0.3504 - val_accuracy: 0.8510\nEpoch 31/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.3399 - accuracy: 0.8569 - val_loss: 0.3377 - val_accuracy: 0.8560\nEpoch 32/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3298 - accuracy: 0.8584 - val_loss: 0.3280 - val_accuracy: 0.8610\nEpoch 33/200\n453/453 [==============================] - ETA: 0s - loss: 0.3089 - accuracy: 0.8731INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU215r.cv.1.best/assets\n453/453 [==============================] - 47s 105ms/step - loss: 0.3089 - accuracy: 0.8731 - val_loss: 0.2890 - val_accuracy: 0.8895\nEpoch 34/200\n453/453 [==============================] - 32s 72ms/step - loss: 0.2939 - accuracy: 0.8825 - val_loss: 0.2793 - val_accuracy: 0.8895\nEpoch 35/200\n453/453 [==============================] - ETA: 0s - loss: 0.2943 - accuracy: 0.8837INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU215r.cv.1.best/assets\n453/453 [==============================] - 48s 106ms/step - loss: 0.2943 - accuracy: 0.8837 - val_loss: 0.2592 - val_accuracy: 0.9032\nEpoch 36/200\n453/453 [==============================] - 32s 72ms/step - loss: 0.2844 - accuracy: 0.8875 - val_loss: 0.3216 - val_accuracy: 0.8727\nEpoch 37/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3080 - accuracy: 0.8728 - val_loss: 0.2746 - val_accuracy: 0.8920\nEpoch 38/200\n453/453 [==============================] - 32s 70ms/step - loss: 0.2825 - accuracy: 0.8893 - val_loss: 0.2770 - val_accuracy: 0.8945\nEpoch 39/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3002 - accuracy: 0.8799 - val_loss: 0.2753 - val_accuracy: 0.8870\nEpoch 40/200\n453/453 [==============================] - 32s 70ms/step - loss: 0.3032 - accuracy: 0.8760 - val_loss: 0.3113 - val_accuracy: 0.8678\nEpoch 41/200\n453/453 [==============================] - 32s 70ms/step - loss: 0.3115 - accuracy: 0.8727 - val_loss: 0.2946 - val_accuracy: 0.8759\nEpoch 42/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3031 - accuracy: 0.8763 - val_loss: 0.2835 - val_accuracy: 0.8821\nEpoch 43/200\n453/453 [==============================] - ETA: 0s - loss: 0.2934 - accuracy: 0.8837INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU215r.cv.1.best/assets\n453/453 [==============================] - 47s 105ms/step - loss: 0.2934 - accuracy: 0.8837 - val_loss: 0.2572 - val_accuracy: 0.9094\nEpoch 44/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.2627 - accuracy: 0.8993 - val_loss: 0.2596 - val_accuracy: 0.9038\nEpoch 45/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3563 - accuracy: 0.8519 - val_loss: 0.3042 - val_accuracy: 0.8727\nEpoch 46/200\n453/453 [==============================] - 32s 70ms/step - loss: 0.2846 - accuracy: 0.8870 - val_loss: 0.2589 - val_accuracy: 0.9044\nEpoch 47/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2705 - accuracy: 0.8946 - val_loss: 0.2459 - val_accuracy: 0.9088\nEpoch 48/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.2997 - accuracy: 0.8766 - val_loss: 0.2931 - val_accuracy: 0.8808\nEpoch 49/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2876 - accuracy: 0.8814 - val_loss: 0.2637 - val_accuracy: 0.8988\nEpoch 50/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2964 - accuracy: 0.8779 - val_loss: 0.2561 - val_accuracy: 0.9050\nEpoch 51/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2800 - accuracy: 0.8887 - val_loss: 0.2525 - val_accuracy: 0.9056\nEpoch 52/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.2562 - accuracy: 0.8995 - val_loss: 0.2413 - val_accuracy: 0.9088\nEpoch 53/200\n453/453 [==============================] - ETA: 0s - loss: 0.2444 - accuracy: 0.9054INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU215r.cv.1.best/assets\n453/453 [==============================] - 47s 104ms/step - loss: 0.2444 - accuracy: 0.9054 - val_loss: 0.2289 - val_accuracy: 0.9143\nEpoch 54/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3011 - accuracy: 0.8773 - val_loss: 0.3016 - val_accuracy: 0.8790\nEpoch 55/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.3377 - accuracy: 0.8550 - val_loss: 0.3758 - val_accuracy: 0.8206\nEpoch 56/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2664 - accuracy: 0.8914 - val_loss: 0.2604 - val_accuracy: 0.8932\nEpoch 57/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2692 - accuracy: 0.8956 - val_loss: 0.2441 - val_accuracy: 0.9032\nEpoch 58/200\n453/453 [==============================] - ETA: 0s - loss: 0.2466 - accuracy: 0.9035INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU215r.cv.1.best/assets\n453/453 [==============================] - 47s 103ms/step - loss: 0.2466 - accuracy: 0.9035 - val_loss: 0.2407 - val_accuracy: 0.9156\nEpoch 59/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2770 - accuracy: 0.8887 - val_loss: 0.3081 - val_accuracy: 0.8684\nEpoch 60/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.3007 - accuracy: 0.8770 - val_loss: 0.2347 - val_accuracy: 0.9112\nEpoch 61/200\n453/453 [==============================] - 32s 72ms/step - loss: 0.2542 - accuracy: 0.8995 - val_loss: 0.2328 - val_accuracy: 0.9156\nEpoch 62/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.2549 - accuracy: 0.9013 - val_loss: 0.2530 - val_accuracy: 0.8994\nEpoch 63/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2479 - accuracy: 0.9072 - val_loss: 0.2357 - val_accuracy: 0.9137\nEpoch 64/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2474 - accuracy: 0.9055 - val_loss: 0.2781 - val_accuracy: 0.8901\nEpoch 65/200\n453/453 [==============================] - 32s 72ms/step - loss: 0.2715 - accuracy: 0.8914 - val_loss: 0.2851 - val_accuracy: 0.8845\nEpoch 66/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3346 - accuracy: 0.8547 - val_loss: 0.3348 - val_accuracy: 0.8579\nEpoch 67/200\n453/453 [==============================] - 32s 70ms/step - loss: 0.3540 - accuracy: 0.8488 - val_loss: 0.3053 - val_accuracy: 0.8777\nEpoch 68/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3332 - accuracy: 0.8615 - val_loss: 0.3003 - val_accuracy: 0.8802\nEpoch 69/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.3233 - accuracy: 0.8657 - val_loss: 0.2952 - val_accuracy: 0.8808\nEpoch 70/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3114 - accuracy: 0.8715 - val_loss: 0.2855 - val_accuracy: 0.8821\nEpoch 71/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.3067 - accuracy: 0.8719 - val_loss: 0.3043 - val_accuracy: 0.8759\nEpoch 72/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3077 - accuracy: 0.8741 - val_loss: 0.2769 - val_accuracy: 0.8920\nEpoch 73/200\n453/453 [==============================] - 32s 72ms/step - loss: 0.2914 - accuracy: 0.8808 - val_loss: 0.2751 - val_accuracy: 0.8994\nEpoch 74/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2850 - accuracy: 0.8830 - val_loss: 0.2705 - val_accuracy: 0.8951\nEpoch 75/200\n453/453 [==============================] - ETA: 0s - loss: 0.2546 - accuracy: 0.8995INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU215r.cv.1.best/assets\n453/453 [==============================] - 47s 104ms/step - loss: 0.2546 - accuracy: 0.8995 - val_loss: 0.2307 - val_accuracy: 0.9187\nEpoch 76/200\n453/453 [==============================] - 33s 73ms/step - loss: 0.2371 - accuracy: 0.9081 - val_loss: 0.2644 - val_accuracy: 0.8870\nEpoch 77/200\n453/453 [==============================] - ETA: 0s - loss: 0.2303 - accuracy: 0.9131INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU215r.cv.1.best/assets\n453/453 [==============================] - 48s 106ms/step - loss: 0.2303 - accuracy: 0.9131 - val_loss: 0.2201 - val_accuracy: 0.9236\nEpoch 78/200\n453/453 [==============================] - ETA: 0s - loss: 0.2203 - accuracy: 0.9173INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU215r.cv.1.best/assets\n453/453 [==============================] - 48s 106ms/step - loss: 0.2203 - accuracy: 0.9173 - val_loss: 0.2126 - val_accuracy: 0.9274\nEpoch 79/200\n453/453 [==============================] - 33s 73ms/step - loss: 0.2226 - accuracy: 0.9149 - val_loss: 0.2206 - val_accuracy: 0.9224\nEpoch 80/200\n453/453 [==============================] - 33s 73ms/step - loss: 0.2166 - accuracy: 0.9188 - val_loss: 0.2090 - val_accuracy: 0.9230\nEpoch 81/200\n453/453 [==============================] - 32s 72ms/step - loss: 0.2085 - accuracy: 0.9211 - val_loss: 0.2209 - val_accuracy: 0.9218\nEpoch 82/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2060 - accuracy: 0.9228 - val_loss: 0.2061 - val_accuracy: 0.9261\nEpoch 83/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2011 - accuracy: 0.9273 - val_loss: 0.2266 - val_accuracy: 0.9150\nEpoch 84/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2049 - accuracy: 0.9247 - val_loss: 0.2163 - val_accuracy: 0.9205\nEpoch 85/200\n453/453 [==============================] - 32s 70ms/step - loss: 0.2038 - accuracy: 0.9238 - val_loss: 0.2144 - val_accuracy: 0.9249\nEpoch 86/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2003 - accuracy: 0.9262 - val_loss: 0.2374 - val_accuracy: 0.9174\nEpoch 87/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.2012 - accuracy: 0.9253 - val_loss: 0.2187 - val_accuracy: 0.9156\nEpoch 88/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.1979 - accuracy: 0.9265 - val_loss: 0.2169 - val_accuracy: 0.9236\nEpoch 89/200\n453/453 [==============================] - ETA: 0s - loss: 0.2017 - accuracy: 0.9260INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU215r.cv.1.best/assets\n453/453 [==============================] - 48s 106ms/step - loss: 0.2017 - accuracy: 0.9260 - val_loss: 0.2008 - val_accuracy: 0.9280\nEpoch 90/200\n453/453 [==============================] - 33s 73ms/step - loss: 0.3862 - accuracy: 0.8240 - val_loss: 0.4660 - val_accuracy: 0.7834\nEpoch 91/200\n453/453 [==============================] - 33s 73ms/step - loss: 0.4274 - accuracy: 0.8015 - val_loss: 0.4335 - val_accuracy: 0.8026\nEpoch 92/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.4130 - accuracy: 0.8116 - val_loss: 0.4077 - val_accuracy: 0.8187\nEpoch 93/200\n453/453 [==============================] - 33s 73ms/step - loss: 0.4022 - accuracy: 0.8199 - val_loss: 0.3873 - val_accuracy: 0.8231\nEpoch 94/200\n453/453 [==============================] - 33s 73ms/step - loss: 0.3994 - accuracy: 0.8193 - val_loss: 0.4020 - val_accuracy: 0.8138\nEpoch 95/200\n453/453 [==============================] - 33s 73ms/step - loss: 0.3977 - accuracy: 0.8217 - val_loss: 0.4060 - val_accuracy: 0.8150\nEpoch 96/200\n453/453 [==============================] - 33s 73ms/step - loss: 0.3861 - accuracy: 0.8283 - val_loss: 0.3699 - val_accuracy: 0.8367\nEpoch 97/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.3870 - accuracy: 0.8236 - val_loss: 0.3630 - val_accuracy: 0.8386\nEpoch 98/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.3778 - accuracy: 0.8354 - val_loss: 0.3748 - val_accuracy: 0.8256\nEpoch 99/200\n453/453 [==============================] - 33s 73ms/step - loss: 0.3815 - accuracy: 0.8291 - val_loss: 0.3540 - val_accuracy: 0.8442\nEpoch 100/200\n453/453 [==============================] - 33s 73ms/step - loss: 0.3721 - accuracy: 0.8351 - val_loss: 0.3656 - val_accuracy: 0.8330\nEpoch 101/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3634 - accuracy: 0.8425 - val_loss: 0.3476 - val_accuracy: 0.8436\nEpoch 102/200\n453/453 [==============================] - 33s 73ms/step - loss: 0.3709 - accuracy: 0.8338 - val_loss: 0.3624 - val_accuracy: 0.8417\nEpoch 103/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3678 - accuracy: 0.8390 - val_loss: 0.3582 - val_accuracy: 0.8430\nEpoch 104/200\n453/453 [==============================] - 32s 70ms/step - loss: 0.3641 - accuracy: 0.8413 - val_loss: 0.3494 - val_accuracy: 0.8467\nEpoch 105/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.3594 - accuracy: 0.8419 - val_loss: 0.3977 - val_accuracy: 0.8045\nEpoch 106/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3557 - accuracy: 0.8452 - val_loss: 0.3947 - val_accuracy: 0.8268\nEpoch 107/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3435 - accuracy: 0.8503 - val_loss: 0.3266 - val_accuracy: 0.8591\nEpoch 108/200\n453/453 [==============================] - 32s 72ms/step - loss: 0.3333 - accuracy: 0.8559 - val_loss: 0.3238 - val_accuracy: 0.8603\nEpoch 109/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3276 - accuracy: 0.8606 - val_loss: 0.3368 - val_accuracy: 0.8541\nEpoch 110/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.3140 - accuracy: 0.8667 - val_loss: 0.3181 - val_accuracy: 0.8647\nEpoch 111/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2941 - accuracy: 0.8804 - val_loss: 0.2946 - val_accuracy: 0.8808\nEpoch 112/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3340 - accuracy: 0.8541 - val_loss: 0.3607 - val_accuracy: 0.8479\nEpoch 113/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.3407 - accuracy: 0.8520 - val_loss: 0.3399 - val_accuracy: 0.8566\nEpoch 114/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3227 - accuracy: 0.8621 - val_loss: 0.3099 - val_accuracy: 0.8740\nEpoch 115/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2947 - accuracy: 0.8771 - val_loss: 0.3164 - val_accuracy: 0.8709\nEpoch 116/200\n453/453 [==============================] - 32s 72ms/step - loss: 0.3010 - accuracy: 0.8762 - val_loss: 0.3231 - val_accuracy: 0.8572\nEpoch 117/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.3172 - accuracy: 0.8633 - val_loss: 0.4079 - val_accuracy: 0.8150\nEpoch 118/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3056 - accuracy: 0.8742 - val_loss: 0.2704 - val_accuracy: 0.8982\nEpoch 119/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.2652 - accuracy: 0.8950 - val_loss: 0.2657 - val_accuracy: 0.8988\nEpoch 120/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.2660 - accuracy: 0.8925 - val_loss: 0.2614 - val_accuracy: 0.8957\nEpoch 121/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2624 - accuracy: 0.8943 - val_loss: 0.2864 - val_accuracy: 0.8945\nEpoch 122/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.2593 - accuracy: 0.8961 - val_loss: 0.2685 - val_accuracy: 0.8970\nEpoch 123/200\n453/453 [==============================] - 33s 73ms/step - loss: 0.2563 - accuracy: 0.8982 - val_loss: 0.2534 - val_accuracy: 0.9050\nEpoch 124/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.2522 - accuracy: 0.8997 - val_loss: 0.3743 - val_accuracy: 0.8386\nEpoch 125/200\n453/453 [==============================] - 33s 73ms/step - loss: 0.3451 - accuracy: 0.8499 - val_loss: 0.3622 - val_accuracy: 0.8405\nEpoch 126/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.3437 - accuracy: 0.8478 - val_loss: 0.3452 - val_accuracy: 0.8504\nEpoch 127/200\n453/453 [==============================] - 33s 73ms/step - loss: 0.3498 - accuracy: 0.8479 - val_loss: 0.3665 - val_accuracy: 0.8442\nEpoch 128/200\n453/453 [==============================] - 33s 73ms/step - loss: 0.3387 - accuracy: 0.8529 - val_loss: 0.3811 - val_accuracy: 0.8330\nEpoch 129/200\n453/453 [==============================] - 32s 70ms/step - loss: 0.3332 - accuracy: 0.8557 - val_loss: 0.3629 - val_accuracy: 0.8461\nEpoch 130/200\n453/453 [==============================] - 32s 70ms/step - loss: 0.3311 - accuracy: 0.8564 - val_loss: 0.3955 - val_accuracy: 0.8250\nEpoch 131/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3238 - accuracy: 0.8600 - val_loss: 0.3997 - val_accuracy: 0.8206\nEpoch 132/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3182 - accuracy: 0.8659 - val_loss: 0.3415 - val_accuracy: 0.8516\nEpoch 133/200\n453/453 [==============================] - 32s 70ms/step - loss: 0.2993 - accuracy: 0.8743 - val_loss: 0.3430 - val_accuracy: 0.8591\nEpoch 134/200\n453/453 [==============================] - 32s 70ms/step - loss: 0.2918 - accuracy: 0.8814 - val_loss: 0.3175 - val_accuracy: 0.8690\nEpoch 135/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.2807 - accuracy: 0.8859 - val_loss: 0.2961 - val_accuracy: 0.8696\nEpoch 136/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2600 - accuracy: 0.8949 - val_loss: 0.3006 - val_accuracy: 0.8709\nEpoch 137/200\n453/453 [==============================] - 32s 70ms/step - loss: 0.2596 - accuracy: 0.8937 - val_loss: 0.2792 - val_accuracy: 0.8839\nEpoch 138/200\n453/453 [==============================] - 32s 70ms/step - loss: 0.2749 - accuracy: 0.8899 - val_loss: 0.3704 - val_accuracy: 0.8516\nEpoch 139/200\n453/453 [==============================] - 32s 70ms/step - loss: 0.2994 - accuracy: 0.8744 - val_loss: 0.3465 - val_accuracy: 0.8653\nEpoch 140/200\n453/453 [==============================] - 32s 70ms/step - loss: 0.2747 - accuracy: 0.8885 - val_loss: 0.3200 - val_accuracy: 0.8709\nEpoch 141/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2746 - accuracy: 0.8887 - val_loss: 0.2957 - val_accuracy: 0.8703\nEpoch 142/200\n453/453 [==============================] - 32s 70ms/step - loss: 0.2660 - accuracy: 0.8937 - val_loss: 0.2784 - val_accuracy: 0.8858\nEpoch 143/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2864 - accuracy: 0.8824 - val_loss: 0.3955 - val_accuracy: 0.8163\nEpoch 144/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.2751 - accuracy: 0.8898 - val_loss: 0.2637 - val_accuracy: 0.9007\nEpoch 145/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2642 - accuracy: 0.8946 - val_loss: 0.3197 - val_accuracy: 0.8690\nEpoch 146/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.3362 - accuracy: 0.8566 - val_loss: 0.4848 - val_accuracy: 0.7747\nEpoch 147/200\n453/453 [==============================] - 32s 70ms/step - loss: 0.3824 - accuracy: 0.8305 - val_loss: 0.4688 - val_accuracy: 0.7877\nEpoch 148/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3676 - accuracy: 0.8376 - val_loss: 0.4588 - val_accuracy: 0.7921\nEpoch 149/200\n453/453 [==============================] - 32s 70ms/step - loss: 0.3640 - accuracy: 0.8393 - val_loss: 0.4436 - val_accuracy: 0.8020\nEpoch 150/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3555 - accuracy: 0.8442 - val_loss: 0.4453 - val_accuracy: 0.8014\nEpoch 151/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3463 - accuracy: 0.8488 - val_loss: 0.4561 - val_accuracy: 0.8001\nEpoch 152/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.3436 - accuracy: 0.8518 - val_loss: 0.4243 - val_accuracy: 0.8088\nEpoch 153/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.3349 - accuracy: 0.8537 - val_loss: 0.4339 - val_accuracy: 0.8038\nEpoch 154/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.3358 - accuracy: 0.8547 - val_loss: 0.3917 - val_accuracy: 0.8293\nEpoch 155/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3231 - accuracy: 0.8607 - val_loss: 0.4251 - val_accuracy: 0.8101\nEpoch 156/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3087 - accuracy: 0.8675 - val_loss: 0.3938 - val_accuracy: 0.8256\nEpoch 157/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2943 - accuracy: 0.8757 - val_loss: 0.3869 - val_accuracy: 0.8423\nEpoch 158/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2853 - accuracy: 0.8818 - val_loss: 0.3463 - val_accuracy: 0.8628\nEpoch 159/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2929 - accuracy: 0.8802 - val_loss: 0.3623 - val_accuracy: 0.8597\nEpoch 160/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2717 - accuracy: 0.8900 - val_loss: 0.3340 - val_accuracy: 0.8709\nEpoch 161/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2902 - accuracy: 0.8796 - val_loss: 0.3680 - val_accuracy: 0.8634\nEpoch 162/200\n453/453 [==============================] - 32s 70ms/step - loss: 0.2825 - accuracy: 0.8853 - val_loss: 0.2839 - val_accuracy: 0.8895\nEpoch 163/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2530 - accuracy: 0.9017 - val_loss: 0.3017 - val_accuracy: 0.8870\nEpoch 164/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.2376 - accuracy: 0.9073 - val_loss: 0.2877 - val_accuracy: 0.8982\nEpoch 165/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2342 - accuracy: 0.9077 - val_loss: 0.2999 - val_accuracy: 0.8908\nEpoch 166/200\n453/453 [==============================] - 32s 72ms/step - loss: 0.2280 - accuracy: 0.9124 - val_loss: 0.2905 - val_accuracy: 0.8963\nEpoch 167/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2210 - accuracy: 0.9169 - val_loss: 0.2462 - val_accuracy: 0.9131\nEpoch 168/200\n453/453 [==============================] - 33s 73ms/step - loss: 0.2076 - accuracy: 0.9211 - val_loss: 0.2616 - val_accuracy: 0.9075\nEpoch 169/200\n453/453 [==============================] - 32s 72ms/step - loss: 0.2130 - accuracy: 0.9213 - val_loss: 0.2492 - val_accuracy: 0.9162\nEpoch 170/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2037 - accuracy: 0.9237 - val_loss: 0.2282 - val_accuracy: 0.9199\nEpoch 171/200\n453/453 [==============================] - 33s 73ms/step - loss: 0.2079 - accuracy: 0.9197 - val_loss: 0.2770 - val_accuracy: 0.9038\nEpoch 172/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.2092 - accuracy: 0.9212 - val_loss: 0.2486 - val_accuracy: 0.9094\nEpoch 173/200\n453/453 [==============================] - 33s 73ms/step - loss: 0.2102 - accuracy: 0.9200 - val_loss: 0.2252 - val_accuracy: 0.9187\nEpoch 174/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.2043 - accuracy: 0.9215 - val_loss: 0.2357 - val_accuracy: 0.9181\nEpoch 175/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.1917 - accuracy: 0.9293 - val_loss: 0.2591 - val_accuracy: 0.9032\nEpoch 176/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.1895 - accuracy: 0.9301 - val_loss: 0.2720 - val_accuracy: 0.9007\nEpoch 177/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.1861 - accuracy: 0.9320 - val_loss: 0.2412 - val_accuracy: 0.9205\nEpoch 178/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.1812 - accuracy: 0.9339 - val_loss: 0.2353 - val_accuracy: 0.9187\nEpoch 179/200\n453/453 [==============================] - 32s 72ms/step - loss: 0.1829 - accuracy: 0.9316 - val_loss: 0.2412 - val_accuracy: 0.9199\nEpoch 180/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.1752 - accuracy: 0.9332 - val_loss: 0.2251 - val_accuracy: 0.9218\nEpoch 181/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.1743 - accuracy: 0.9372 - val_loss: 0.2271 - val_accuracy: 0.9199\nEpoch 182/200\n453/453 [==============================] - ETA: 0s - loss: 0.1752 - accuracy: 0.9351INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU215r.cv.1.best/assets\n453/453 [==============================] - 49s 107ms/step - loss: 0.1752 - accuracy: 0.9351 - val_loss: 0.2098 - val_accuracy: 0.9286\nEpoch 183/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.2744 - accuracy: 0.8864 - val_loss: 0.3062 - val_accuracy: 0.8814\nEpoch 184/200\n453/453 [==============================] - 32s 72ms/step - loss: 0.3304 - accuracy: 0.8568 - val_loss: 0.4067 - val_accuracy: 0.8250\nEpoch 185/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.3265 - accuracy: 0.8588 - val_loss: 0.3853 - val_accuracy: 0.8417\nEpoch 186/200\n453/453 [==============================] - 32s 72ms/step - loss: 0.3194 - accuracy: 0.8590 - val_loss: 0.3962 - val_accuracy: 0.8293\nEpoch 187/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.3101 - accuracy: 0.8684 - val_loss: 0.3825 - val_accuracy: 0.8405\nEpoch 188/200\n453/453 [==============================] - 32s 72ms/step - loss: 0.3002 - accuracy: 0.8739 - val_loss: 0.3746 - val_accuracy: 0.8485\nEpoch 189/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.2937 - accuracy: 0.8748 - val_loss: 0.3289 - val_accuracy: 0.8665\nEpoch 190/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.2769 - accuracy: 0.8846 - val_loss: 0.3285 - val_accuracy: 0.8659\nEpoch 191/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.2501 - accuracy: 0.8985 - val_loss: 0.2631 - val_accuracy: 0.8926\nEpoch 192/200\n453/453 [==============================] - 32s 72ms/step - loss: 0.2349 - accuracy: 0.9080 - val_loss: 0.2345 - val_accuracy: 0.9119\nEpoch 193/200\n453/453 [==============================] - 33s 74ms/step - loss: 0.1986 - accuracy: 0.9244 - val_loss: 0.2372 - val_accuracy: 0.9081\nEpoch 194/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.2235 - accuracy: 0.9142 - val_loss: 0.3134 - val_accuracy: 0.8790\nEpoch 195/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2421 - accuracy: 0.9040 - val_loss: 0.2502 - val_accuracy: 0.9001\nEpoch 196/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2250 - accuracy: 0.9146 - val_loss: 0.2778 - val_accuracy: 0.9001\nEpoch 197/200\n453/453 [==============================] - 32s 70ms/step - loss: 0.2504 - accuracy: 0.8997 - val_loss: 0.3098 - val_accuracy: 0.8752\nEpoch 198/200\n453/453 [==============================] - 32s 71ms/step - loss: 0.2767 - accuracy: 0.8859 - val_loss: 0.2939 - val_accuracy: 0.8845\nEpoch 199/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.2452 - accuracy: 0.9000 - val_loss: 0.3176 - val_accuracy: 0.8746\nEpoch 200/200\n453/453 [==============================] - 33s 72ms/step - loss: 0.2610 - accuracy: 0.8920 - val_loss: 0.2798 - val_accuracy: 0.8852\nFold 1, 200 epochs, 6883 sec\n"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d0865f54d99adfd3f3ef6ec01bb3de189fffbd24 | 8,519 | ipynb | Jupyter Notebook | dev/make_markdown.ipynb | sungcheolkim78/py_kbible | 3a576c20e5e49f5e85be6ddede20accb6df14663 | [
"Apache-2.0"
] | null | null | null | dev/make_markdown.ipynb | sungcheolkim78/py_kbible | 3a576c20e5e49f5e85be6ddede20accb6df14663 | [
"Apache-2.0"
] | null | null | null | dev/make_markdown.ipynb | sungcheolkim78/py_kbible | 3a576c20e5e49f5e85be6ddede20accb6df14663 | [
"Apache-2.0"
] | 1 | 2021-12-27T00:32:48.000Z | 2021-12-27T00:32:48.000Z | 28.023026 | 299 | 0.474704 | [
[
[
"import kbible",
"_____no_output_____"
],
[
"bible = kbible.read_full_bible(\"개역개정판성경\")",
"_____no_output_____"
],
[
"print(kbible.extract_bystr(bible, \"창1:3\", form=\"md\"))",
"`창1:3` 하나님이 이르시되 빛이 있으라 하시니 빛이 있었고 \n"
]
],
[
[
"## print daily message",
"_____no_output_____"
]
],
[
[
"day_info = {\"day\":u\"1\", \"title\":u\"영광과 흑암\", \"song\":u\"하나님은 우리 아버지\", \"prayer\":u\"\", \"summary\":u\"\", \\\n \"verses\": [u\"창1:3\", u\"창1:14\", u\"창1:26,28,29\", u\"창2:19\", u\"창2:9\", u\"창2:17\", u\"창3:6-7\", \\\n u\"유1:6\", u\"벧후2:4\", u\"창3:17-18\", u\"왕하6:17\", u\"행26:18\", u\"롬1:19-20\", u\"요12:46\", \\\n u\"골1:13\", u\"단2:22\", u\"요일5:19\"]}",
"_____no_output_____"
],
[
"day_info[\"day\"]",
"_____no_output_____"
],
[
"def make_mdpage(bible, day_info, save=False):\n \"\"\" print all verses in list using markdown format \"\"\"\n \n # check day_info.yml file\n if isinstance(day_info, str):\n try:\n with open(day_info, \"r\") as f:\n day_info = yaml.load(f, yaml.BaseLoader)\n except:\n print(\"... file: {} parser error!\".format(day_info))\n return 0\n \n bible_version = \"\"\n # check bible version\n if isinstance(bible, str):\n try:\n bible_version = \"-\" + bible\n bible = kbible.read_full_bible(bible)\n except:\n print(\"... read error: {}\".format(bible_version))\n return 0\n \n msg = \"# {}일차 - {}\\n\\n\".format(day_info[\"day\"],day_info[\"title\"])\n msg = msg + \"찬양 : {}\\n\\n\".format(day_info[\"song\"])\n msg = msg + \"기도 : {}\\n\\n\".format(day_info[\"prayer\"])\n msg = msg + \"요약 : {}\\n\\n\".format(day_info[\"summary\"])\n msg = msg + \"성경 버전 : {}\\n\\n\".format(bible_version[1:])\n \n for v in day_info[\"verses\"]:\n msg = msg + '- {}\\n\\n'.format(kbible.extract_bystr(bible, v, form=\"md\"))\n\n msg = msg + \"### info\\n\\n\"\n msg = msg + \"- 성경 구절 갯수 : {}\".format(len(day_info[\"verses\"]))\n \n if save:\n filename = 'mpages/day{}-{}{}.md'.format(day_info[\"day\"], day_info[\"title\"].replace(\" \", \"\"), bible_version)\n with open(filename, \"w\") as f:\n f.write(msg)\n print('... save to {}'.format(filename))\n \n return msg",
"_____no_output_____"
],
[
"print(make_mdpage(\"현대인의성경\", \"day1.yaml\", save=True))",
"... save to mpages/day1-영광과흑암-현대인의성경.md\n# 1일차 - 영광과 흑암\n\n찬양 : 하나님은 우리 아버지\n\n기도 : \n\n요약 : \n\n성경 버전 : 현대인의성경\n\n- `창1:3` 하나님이 이르시되 빛이 있으라 하시니 빛이 있었고 \n\n- `창1:14` 태초에 하나님이 천지를 창조하시니라. 그 빛이 하나님이 보시기에 좋았더라 하나님이 빛과 어둠을 나누사\n\n- `창1:26,28,29` 하나님이 이르시되 우리의 형상을 따라 우리의 모양대로 우리가 사람을 만들고 그들로 바다의 물고기와 하늘의 새와 가축과 온 땅과 땅에 기는 모든 것을 다스리게 하자 하시고. 하나님이 그들에게 복을 주시며 하나님이 그들에게 이르시되 생육하고 번성하여 땅에 충만 하라, 땅을 정복하라, 바다의 물고기와 하늘의 새와 땅에 움직이는 모든 생물을 다스 리라 하시니라. 하나님이 이르시되 내가 온 지면의 씨 맺는 모든 채소와 씨 가진 열매 맺는 모든 나무를 너희에게 주노니 너희의 먹을거리가 되리라 \n\n- `창2:19` 천지와 만물이 다 이루어지니라. 여호와 하나님이 그 땅에서 보기에 아름답고 먹기에 좋은 나무가 나게 하시니 동산 가운데에는 생명 나무와 선악을 알게 하는 나무도 있더라\n\n- `창2:9` 여호와 하나님이 그 땅에서 보기에 아름답고 먹기에 좋은 나무가 나게 하시니 동산 가운데에는 생명 나무와 선악을 알게 하는 나무도 있더라\n\n- `창2:17` 천지와 만물이 다 이루어지니라. 여호와 하나님이 땅의 흙으로 사람을 지으시고 생기를 그 코에 불어넣으시니 사람이 생령이 되니라\n\n- `창3:6-7` 여자가 그 나무를 본즉 먹음직도 하고 보암직도 하고 지혜롭게 할만큼 탐스럽기도 한 나무인지라 여자가 그 열매를 따먹고 자기와 함께 있는 남편에게도 주매 그도 먹은지라. 이에 그들의 눈이 밝아져 자기들이 벗은 줄을 알고 무화과나무 잎을 엮어 치마로 삼았더라\n\n- `유1:6` 또 자기 지위를 지키지 아니하고 자기 처소를 떠난 천사들을 큰 날의 심판까지 영원한 결박으로 흑암에 가두셨으며\n\n- `벧후2:4` 하나님이 범죄한 천사들을 용서하지 아니하시고 지옥에 던져 어두운 구덩이에 두어 심판 때까지 지키게 하셨으며\n\n- `창3:17-18` 아담에게 이르시되 네가 네 아내의 말을 듣고 내가 네게 먹지 말라한 나무의 열매를 먹었은즉 땅은 너로 말미암아 저주를 받고 너는 네 평생에 수고하여야 그 소산을 먹으리라 . 땅이 네게 가시덤불과 엉겅퀴를 낼 것이라 네가 먹을 것은 밭의 채소인즉\n\n- `왕하6:17` 선지자의 제자들이 엘리사에게 이르되 보소서 우리가 당신과 함께 거주하는 이 곳이 우리에게는 좁으니. 이르되 너는 그것을 집으라 하니 그 사람이 손을 내밀어 그것을 집으니라 \n\n- `행26:18` 아그립바가 바울에게 이르되 너를 위하여 말하기를 네게 허락하노라 하니 이에 바울이 손을 들어 변명하되. 당신들은 하나님이 죽은 사람을 다시 살리심을 어찌하여 못 믿을 것으로 여기나이까\n\n- `롬1:19-20` 이는 하나님을 알 만한 것이 그들 속에 보임이라 하나님께서 이를 그들에게 보이셨느니라. 창세로부터 그의 보이지 아니하는 것들 곧 그의 영원하신 능력과 신성이 그가 만드신 만물에 분명히 보여 알려졌나니 그러므로 그들이 핑계하지 못할지니라\n\n- `요12:46` 제자 중 하나로서 예수를 잡아 줄 가룟 유다가 말하되. 이렇게 말함은 가난한 자들을 생각함이 아니요 그는 도둑이라 돈 궤를 맡고 거기 넣는 것을 훔쳐감이러라\n\n- `골1:13` 하나님의 뜻으로 말미암아 그리스도 예수의 사도 된 바울과 형제 디모데는 . 우리가 너희를 위하여 기도할 때마다 하나님 곧 우리 주 예수 그리스도의 아버지께 감사하노라 \n\n- `단2:22` 왕이 그의 꿈을 자기에게 알려 주도록 박수와 술객과 점쟁이와 갈대아 술사를 부르라 말하매 그들이 들어가서 왕의 앞에 선지라 \n\n- `요일5:19` 예수께서 그리스도이심을 믿는 자마다 하나님께로부터 난 자니 또한 낳으신 이를 사랑하는 자마다 그에게서 난 자를 사랑하느니라. 만일 우리가 사람들의 증언을 받을진대 하나님의 증거는 더욱 크도다 하나님의 증거는 이것이니 그의 아들에 대하여 증언하신 것이라\n\n### info\n\n- 성경 구절 갯수 : 17\n"
],
[
"#kbible.find_id(bible, book=\"롬\", chapter=1)",
"_____no_output_____"
]
],
[
[
"## daily info file",
"_____no_output_____"
]
],
[
[
"import yaml",
"_____no_output_____"
],
[
"print(yaml.dump(day_info, allow_unicode=True))",
"day: '1'\nprayer: ''\nsong: 하나님은 우리 아버지\nsummary: ''\ntitle: 영광과 흑암\nverses:\n- 창1:3\n- 창1:14\n- 창1:26,28,29\n- 창2:19\n- 창2:9\n- 창2:17\n- 창3:6-7\n- 유1:6\n- 벧후2:4\n- 창3:17-18\n- 왕하6:17\n- 행26:18\n- 롬1:19-20\n- 요12:46\n- 골1:13\n- 단2:22\n- 요일5:19\n\n"
],
[
"with open('day1.yaml', \"w\") as f:\n yaml.dump(day_info, f, allow_unicode=True)",
"_____no_output_____"
],
[
"yaml.load?",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d086631da51f65d997978325ef28edb4f98205a5 | 13,001 | ipynb | Jupyter Notebook | notebooks/world_happiness.ipynb | FregicAI/fregic-exercises | 5d95fd5322914cc104576e19d664eac72ff9b1a8 | [
"MIT"
] | null | null | null | notebooks/world_happiness.ipynb | FregicAI/fregic-exercises | 5d95fd5322914cc104576e19d664eac72ff9b1a8 | [
"MIT"
] | null | null | null | notebooks/world_happiness.ipynb | FregicAI/fregic-exercises | 5d95fd5322914cc104576e19d664eac72ff9b1a8 | [
"MIT"
] | null | null | null | 39.637195 | 87 | 0.345743 | [
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns",
"_____no_output_____"
],
[
"df = pd.read_csv('../data/world-happiness-report-2021.csv')\ndf.head()",
"_____no_output_____"
],
[
"df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 149 entries, 0 to 148\nData columns (total 20 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Country name 149 non-null object \n 1 Regional indicator 149 non-null object \n 2 Ladder score 149 non-null float64\n 3 Standard error of ladder score 149 non-null float64\n 4 upperwhisker 149 non-null float64\n 5 lowerwhisker 149 non-null float64\n 6 Logged GDP per capita 149 non-null float64\n 7 Social support 149 non-null float64\n 8 Healthy life expectancy 149 non-null float64\n 9 Freedom to make life choices 149 non-null float64\n 10 Generosity 149 non-null float64\n 11 Perceptions of corruption 149 non-null float64\n 12 Ladder score in Dystopia 149 non-null float64\n 13 Explained by: Log GDP per capita 149 non-null float64\n 14 Explained by: Social support 149 non-null float64\n 15 Explained by: Healthy life expectancy 149 non-null float64\n 16 Explained by: Freedom to make life choices 149 non-null float64\n 17 Explained by: Generosity 149 non-null float64\n 18 Explained by: Perceptions of corruption 149 non-null float64\n 19 Dystopia + residual 149 non-null float64\ndtypes: float64(18), object(2)\nmemory usage: 23.4+ KB\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
d08679c37d3b87a41a5c53b50686e6ba36ec8092 | 560,956 | ipynb | Jupyter Notebook | Bike_Transportations.ipynb | jenhar/jupiter_bike_transportation | 646b77856285d5271eb40c06c06583b98700be04 | [
"MIT"
] | null | null | null | Bike_Transportations.ipynb | jenhar/jupiter_bike_transportation | 646b77856285d5271eb40c06c06583b98700be04 | [
"MIT"
] | null | null | null | Bike_Transportations.ipynb | jenhar/jupiter_bike_transportation | 646b77856285d5271eb40c06c06583b98700be04 | [
"MIT"
] | null | null | null | 1,147.149284 | 172,944 | 0.95451 | [
[
[
"url='https://data.seattle.gov/api/views/65db-xm6k/rows.csv?accessType=DOWNLOAD'",
"_____no_output_____"
],
[
"from urllib.request import urlretrieve",
"_____no_output_____"
],
[
"urlretrieve(url, 'Fremont.csv')",
"_____no_output_____"
],
[
"import pandas as pd",
"_____no_output_____"
],
[
"data=pd.read_csv('Fremont.csv', index_col='Date', parse_dates=True)",
"_____no_output_____"
],
[
"data.head()",
"_____no_output_____"
],
[
"%matplotlib inline\ndata.resample('W').sum().plot();",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nplt.style.use('seaborn')\ndata.columns=['East', 'West']\ndata.resample('W').sum().plot()",
"_____no_output_____"
],
[
"#anual trend\ndata.resample('D').sum().rolling(365).sum().plot();",
"_____no_output_____"
],
[
"ax=data.resample('D').sum().rolling(365).sum().plot();\nax.set_ylim(0, None);",
"_____no_output_____"
],
[
"data['total']=data.West + data.East\nax=data.resample('D').sum().rolling(365).sum().plot();\nax.set_ylim(0, None);",
"_____no_output_____"
],
[
"data.groupby(data.index.time).mean().plot();",
"_____no_output_____"
],
[
"pivoted=data.pivot_table('total', index=data.index.time, columns=data.index.date)\npivoted.iloc[:5, :5]",
"_____no_output_____"
],
[
"pivoted.plot(legend=False)",
"_____no_output_____"
],
[
"pivoted.plot(legend=False, alpha=0.01);",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0868eaf92583a86ad1e14aefbc7308901fcd637 | 12,837 | ipynb | Jupyter Notebook | examples/user_guide/Network_Graphs.ipynb | jsignell/holoviews | 4f9fd27367f23c3d067d176f638ec82e4b9ec8f0 | [
"BSD-3-Clause"
] | null | null | null | examples/user_guide/Network_Graphs.ipynb | jsignell/holoviews | 4f9fd27367f23c3d067d176f638ec82e4b9ec8f0 | [
"BSD-3-Clause"
] | null | null | null | examples/user_guide/Network_Graphs.ipynb | jsignell/holoviews | 4f9fd27367f23c3d067d176f638ec82e4b9ec8f0 | [
"BSD-3-Clause"
] | null | null | null | 32.664122 | 645 | 0.615486 | [
[
[
"import numpy as np\nimport pandas as pd\nimport holoviews as hv\nimport networkx as nx\n\nhv.extension('bokeh')\n\n%opts Graph [width=400 height=400]",
"_____no_output_____"
]
],
[
[
"Visualizing and working with network graphs is a common problem in many different disciplines. HoloViews provides the ability to represent and visualize graphs very simply and easily with facilities for interactively exploring the nodes and edges of the graph, especially using the bokeh plotting interface.\n\nThe ``Graph`` ``Element`` differs from other elements in HoloViews in that it consists of multiple sub-elements. The data of the ``Graph`` element itself are the abstract edges between the nodes. By default the element will automatically compute concrete ``x`` and ``y`` positions for the nodes and represent them using a ``Nodes`` element, which is stored on the Graph. The abstract edges and concrete node positions are sufficient to render the ``Graph`` by drawing straight-line edges between the nodes. In order to supply explicit edge paths we can also declare ``EdgePaths``, providing explicit coordinates for each edge to follow.\n\nTo summarize a ``Graph`` consists of three different components:\n\n* The ``Graph`` itself holds the abstract edges stored as a table of node indices.\n* The ``Nodes`` hold the concrete ``x`` and ``y`` positions of each node along with a node ``index``. The ``Nodes`` may also define any number of value dimensions, which can be revealed when hovering over the nodes or to color the nodes by.\n* The ``EdgePaths`` can optionally be supplied to declare explicit node paths.\n\n#### A simple Graph\n\nLet's start by declaring a very simple graph connecting one node to all others. If we simply supply the abstract connectivity of the ``Graph``, it will automatically compute a layout for the nodes using the ``layout_nodes`` operation, which defaults to a circular layout:",
"_____no_output_____"
]
],
[
[
"# Declare abstract edges\nN = 8\nnode_indices = np.arange(N, dtype=np.int32)\nsource = np.zeros(N, dtype=np.int32)\ntarget = node_indices\n\npadding = dict(x=(-1.2, 1.2), y=(-1.2, 1.2))\n\nsimple_graph = hv.Graph(((source, target),)).redim.range(**padding)\nsimple_graph",
"_____no_output_____"
]
],
[
[
"#### Accessing the nodes and edges\n\nWe can easily access the ``Nodes`` and ``EdgePaths`` on the ``Graph`` element using the corresponding properties:",
"_____no_output_____"
]
],
[
[
"simple_graph.nodes + simple_graph.edgepaths",
"_____no_output_____"
]
],
[
[
"#### Supplying explicit paths\n\nNext we will extend this example by supplying explicit edges:",
"_____no_output_____"
]
],
[
[
"def bezier(start, end, control, steps=np.linspace(0, 1, 100)):\n return (1-steps)**2*start + 2*(1-steps)*steps*control+steps**2*end\n\nx, y = simple_graph.nodes.array([0, 1]).T\n\npaths = []\nfor node_index in node_indices:\n ex, ey = x[node_index], y[node_index]\n paths.append(np.column_stack([bezier(x[0], ex, 0), bezier(y[0], ey, 0)]))\n \nbezier_graph = hv.Graph(((source, target), (x, y, node_indices), paths)).redim.range(**padding)\nbezier_graph",
"_____no_output_____"
]
],
[
[
"## Interactive features",
"_____no_output_____"
],
[
"#### Hover and selection policies\n\nThanks to Bokeh we can reveal more about the graph by hovering over the nodes and edges. The ``Graph`` element provides an ``inspection_policy`` and a ``selection_policy``, which define whether hovering and selection highlight edges associated with the selected node or nodes associated with the selected edge, these policies can be toggled by setting the policy to ``'nodes'`` (the default) and ``'edges'``.",
"_____no_output_____"
]
],
[
[
"bezier_graph.options(inspection_policy='edges')",
"_____no_output_____"
]
],
[
[
"In addition to changing the policy we can also change the colors used when hovering and selecting nodes:",
"_____no_output_____"
]
],
[
[
"%%opts Graph [tools=['hover', 'box_select']] (edge_hover_line_color='green' node_hover_fill_color='red')\nbezier_graph.options(inspection_policy='nodes')",
"_____no_output_____"
]
],
[
[
"#### Additional information\n\nWe can also associate additional information with the nodes and edges of a graph. By constructing the ``Nodes`` explicitly we can declare additional value dimensions, which are revealed when hovering and/or can be mapped to the color by specifying the ``color_index``. We can also associate additional information with each edge by supplying a value dimension to the ``Graph`` itself, which we can map to a color using the ``edge_color_index``.",
"_____no_output_____"
]
],
[
[
"%%opts Graph [color_index='Type' edge_color_index='Weight'] (cmap='Set1' edge_cmap='viridis')\nnode_labels = ['Output']+['Input']*(N-1)\nnp.random.seed(7)\nedge_labels = np.random.rand(8)\n\nnodes = hv.Nodes((x, y, node_indices, node_labels), vdims='Type')\ngraph = hv.Graph(((source, target, edge_labels), nodes, paths), vdims='Weight').redim.range(**padding)\ngraph + graph.options(inspection_policy='edges')",
"_____no_output_____"
]
],
[
[
"If you want to supply additional node information without speciying explicit node positions you may pass in a ``Dataset`` object consisting of various value dimensions.",
"_____no_output_____"
]
],
[
[
"%%opts Graph [color_index='Label'] (cmap='Set1')\nnode_info = hv.Dataset(node_labels, vdims='Label')\nhv.Graph(((source, target), node_info)).redim.range(**padding)",
"_____no_output_____"
]
],
[
[
"## Working with NetworkX",
"_____no_output_____"
],
[
"NetworkX is a very useful library when working with network graphs and the Graph Element provides ways of importing a NetworkX Graph directly. Here we will load the Karate Club graph and use the ``circular_layout`` function provided by NetworkX to lay it out:",
"_____no_output_____"
]
],
[
[
"%%opts Graph [tools=['hover']]\nG = nx.karate_club_graph()\nhv.Graph.from_networkx(G, nx.layout.circular_layout).redim.range(**padding)",
"_____no_output_____"
]
],
[
[
"#### Animating graphs",
"_____no_output_____"
],
[
"Like all other elements ``Graph`` can be updated in a ``HoloMap`` or ``DynamicMap``. Here we animate how the Fruchterman-Reingold force-directed algorithm lays out the nodes in real time.",
"_____no_output_____"
]
],
[
[
"%%opts Graph\nG = nx.karate_club_graph()\n\ndef get_graph(iteration):\n np.random.seed(10)\n return hv.Graph.from_networkx(G, nx.spring_layout, iterations=iteration)\n\nhv.HoloMap({i: get_graph(i) for i in range(5, 30, 5)},\n kdims='Iterations').redim.range(x=(-1.2, 1.2), y=(-1.2, 1.2))",
"_____no_output_____"
]
],
[
[
"## Real world graphs",
"_____no_output_____"
],
[
"As a final example let's look at a slightly larger graph. We will load a dataset of a Facebook network consisting a number of friendship groups identified by their ``'circle'``. We will load the edge and node data using pandas and then color each node by their friendship group using many of the things we learned above.",
"_____no_output_____"
]
],
[
[
"%opts Nodes Graph [width=800 height=800 xaxis=None yaxis=None]",
"_____no_output_____"
],
[
"%%opts Graph [color_index='circle']\n%%opts Graph (node_size=10 edge_line_width=1)\ncolors = ['#000000']+hv.Cycle('Category20').values\nedges_df = pd.read_csv('../assets/fb_edges.csv')\nfb_nodes = hv.Nodes(pd.read_csv('../assets/fb_nodes.csv')).sort()\nfb_graph = hv.Graph((edges_df, fb_nodes), label='Facebook Circles')\nfb_graph = fb_graph.redim.range(x=(-0.05, 1.05), y=(-0.05, 1.05)).options(cmap=colors)\nfb_graph",
"_____no_output_____"
]
],
[
[
"## Bundling graphs",
"_____no_output_____"
],
[
"The datashader library provides algorithms for bundling the edges of a graph and HoloViews provides convenient wrappers around the libraries. Note that these operations need ``scikit-image`` which you can install using:\n\n```\nconda install scikit-image\n```\n\nor\n\n```\npip install scikit-image\n```",
"_____no_output_____"
]
],
[
[
"from holoviews.operation.datashader import datashade, bundle_graph\nbundled = bundle_graph(fb_graph)\nbundled",
"_____no_output_____"
]
],
[
[
"## Datashading graphs",
"_____no_output_____"
],
[
"For graphs with a large number of edges we can datashade the paths and display the nodes separately. This loses some of the interactive features but will let you visualize quite large graphs:",
"_____no_output_____"
]
],
[
[
"%%opts Nodes [color_index='circle'] (size=10 cmap=colors) Overlay [show_legend=False]\ndatashade(bundled, normalization='linear', width=800, height=800) * bundled.nodes",
"_____no_output_____"
]
],
[
[
"### Applying selections",
"_____no_output_____"
],
[
"Alternatively we can select the nodes and edges by an attribute that resides on either. In this case we will select the nodes and edges for a particular circle and then overlay just the selected part of the graph on the datashaded plot. Note that selections on the ``Graph`` itself will select all nodes that connect to one of the selected nodes. In this way a smaller subgraph can be highlighted and the larger graph can be datashaded.",
"_____no_output_____"
]
],
[
[
"%%opts Graph (node_fill_color='white')\ndatashade(bundle_graph(fb_graph), normalization='linear', width=800, height=800) *\\\nbundled.select(circle='circle15')",
"_____no_output_____"
]
],
[
[
"To select just nodes that are in 'circle15' set the ``selection_mode='nodes'`` overriding the default of 'edges':",
"_____no_output_____"
]
],
[
[
"bundled.select(circle='circle15', selection_mode='nodes')",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0868fa9d3766476b71f67e2752f5afc2b358653 | 649,263 | ipynb | Jupyter Notebook | AirBNB_Analysis_San_Francisco.ipynb | zaveta/AirBNB-Analysis-San-Francisco | 1b98c79f64a1a1af1470a8b5d08c296c128c9685 | [
"Apache-2.0"
] | null | null | null | AirBNB_Analysis_San_Francisco.ipynb | zaveta/AirBNB-Analysis-San-Francisco | 1b98c79f64a1a1af1470a8b5d08c296c128c9685 | [
"Apache-2.0"
] | null | null | null | AirBNB_Analysis_San_Francisco.ipynb | zaveta/AirBNB-Analysis-San-Francisco | 1b98c79f64a1a1af1470a8b5d08c296c128c9685 | [
"Apache-2.0"
] | null | null | null | 154.182617 | 203,896 | 0.794046 | [
[
[
"# How have Airbnb prices changed due to COVID-19?",
"_____no_output_____"
],
[
"## Business Understanding",
"_____no_output_____"
],
[
"This is the most recent data (Oct, 2020) taken from the official website Airbnb http://insideairbnb.com/get-the-data.html\n\nIn this Notebook, we'll look at this data, clean up, analyze, visualize, and model.\n\nAnd we will answer the following questions for Business Understanding:\n\n1. What correlates best with the price?\n\n2. How has price and busyness changed over the course of COVID-19?\n\n4. Can we predict the price based on its features?",
"_____no_output_____"
],
[
"Let's begin!",
"_____no_output_____"
]
],
[
[
"#import libraries\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport geopandas as gpd\n\n#ml libraries\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.ensemble import RandomForestRegressor\nfrom sklearn.ensemble import AdaBoostRegressor\nfrom sklearn.ensemble import GradientBoostingRegressor\nimport xgboost as xgb\nfrom xgboost import plot_importance\n\nfrom keras import backend as K\nimport tensorflow as tf\nimport time\nfrom tensorflow import keras\nfrom keras import models, layers, optimizers, regularizers\nfrom IPython.display import SVG\nfrom keras.utils.vis_utils import model_to_dot\nfrom tensorflow.keras.callbacks import EarlyStopping\nfrom sklearn.ensemble import RandomForestRegressor\nfrom sklearn.ensemble import AdaBoostRegressor\nfrom sklearn.ensemble import GradientBoostingRegressor\nfrom sklearn.model_selection import learning_curve\nfrom sklearn.preprocessing import StandardScaler, MinMaxScaler\n#metrics\nfrom sklearn.metrics import r2_score, mean_squared_error\n\n%matplotlib inline",
"Using TensorFlow backend.\n"
],
[
"def printColunmsInfo(df):\n '''takes dataframe, prints columns info'''\n df.info()\n print(\"\\n\")\n printTotalRowsAndColumns(df)\n print(\"---------------------------------------\")\n \ndef printTotalRowsAndColumns(df):\n '''print number of columns and rows'''\n print(\"Total columns: \", df.shape[1])\n print(\"Total rows: \", df.shape[0])",
"_____no_output_____"
],
[
"def stringToNumConverter(string):\n '''deletes not numbers symbols from string'''\n newString = \"\"\n if pd.notna(string):\n for i in string:\n if i.isdigit() or i == \".\":\n newString += i \n return newString",
"_____no_output_____"
],
[
"def create_dummy_df(df, cat_cols, dummy_na):\n '''creates dummy'''\n for col in cat_cols:\n try:\n # for each cat add dummy var, drop original column\n df = pd.concat([df.drop(col, axis=1), pd.get_dummies(df[col], prefix=col, prefix_sep='_', drop_first=True, dummy_na=dummy_na)], axis=1)\n except:\n continue\n return df",
"_____no_output_____"
],
[
"def dateToCategorical(row):\n '''changes column from date type to categorical'''\n if row.year <= 2016:\n return \"4+ years\"\n elif row.year <= 2018:\n return \"2-3 years\"\n elif row.year <= 2019:\n return \"1-2 years\"\n elif row.year == 2020:\n if row.month > 8:\n return \"0-1 month\"\n elif row.month > 2:\n return \"2-6 months\"\n elif row.month <= 2:\n return \"this year\"\n else :\n return \"no reviews\" ",
"_____no_output_____"
],
[
"def appendToMetricsdf(df, model_name, train_r2, test_r2, train_mse, test_mse):\n '''appends new row to metrics_df'''\n new_row = {\"Model Name\" : model_name,\n \"r-squared train\" : train_r2,\n \"r-squared train test\" : test_r2,\n \"MSE train\" : train_mse,\n \"MSE test\" : test_mse }\n\n df = df.append(new_row, ignore_index=True)\n return df",
"_____no_output_____"
],
[
"def r2_keras(y_true, y_pred):\n '''calculates r2_score'''\n SS_res = K.sum(K.square(y_true - y_pred)) \n SS_tot = K.sum(K.square(y_true - K.mean(y_true))) \n return (1 - SS_res/(SS_tot + K.epsilon()) )",
"_____no_output_____"
],
[
"#load data\nsf_cal = pd.read_csv(\"datasets/calendar.csv\", low_memory=False, index_col=0)\nsf_list = pd.read_csv(\"datasets/listings.csv\")",
"N:\\Anaconda\\lib\\site-packages\\numpy\\lib\\arraysetops.py:569: FutureWarning: elementwise comparison failed; returning scalar instead, but in the future will perform elementwise comparison\n mask |= (ar1 == a)\n"
]
],
[
[
"## Cleaning the Data",
"_____no_output_____"
],
[
"### Listing Data Frame",
"_____no_output_____"
],
[
"First, let's look on Listing Data Frame. It is the biggest table. We won't need some columns because they don't make much sense for our purposes. So we will drop them.",
"_____no_output_____"
]
],
[
[
"sf_list = sf_list[['id', 'host_since', 'host_is_superhost', 'host_listings_count', 'host_response_time', \n 'host_response_rate', 'host_acceptance_rate','neighbourhood_cleansed', 'latitude', 'longitude', \n 'property_type', 'room_type', 'accommodates', 'bathrooms', 'bedrooms', 'beds', 'amenities', \n 'minimum_nights', 'maximum_nights', 'review_scores_rating', 'review_scores_accuracy', \n 'review_scores_cleanliness', 'review_scores_checkin', 'review_scores_communication', \n 'review_scores_location', 'review_scores_value', 'availability_30', 'number_of_reviews',\n 'last_review', 'reviews_per_month', 'price']]\nsf_list.head()",
"_____no_output_____"
]
],
[
[
"We have left the following columns:\n\n* __'id'__ — we'll use to join tables\n* __host_since__ and __last_review__ — datatype data, we transform to categorical\n* __'host_response_time'__ — categorical data\n* __host_is_superhost__ — boolean data\n* __'host_response_rate'__ and __'host_acceptance_rate'__ — as a percentage, we will change to integer\n* __neighbourhood_cleansed'__ — neighbourhood name\n* __'latitude', 'longitude'__ — сoordinates, we use them for visualisation\n* __'room_type'__ and __property_type__ — categorical data\n* __'accommodates', 'bathrooms', 'bedrooms', 'beds'__ — numerical values describing property\n* __'amenities'__ — can be used to identify words associated with amenities\n* __'minimum_nights', 'maximum_nights'__ — numerical values\n* __'review_scores_rating'__ — numbers between 20 and 100\n* __'review_scores_accuracy', 'review_scores_cleanliness', 'review_scores_checkin', review_scores_communication', 'review_scores_location', 'review_scores_value'__ — numbers between 2 and 10\n* __availability_30__, __number_of_reviews__, __reviews_per_month__ — numerical\n* __'price'__ — target value",
"_____no_output_____"
],
[
"Let's convert string data to numeric.",
"_____no_output_____"
]
],
[
[
"#converting datatype of price column to integer\nsf_list[\"price\"] = sf_list[\"price\"].apply(lambda string: ''.join(i for i in string if i.isdigit())[:-2])\nsf_list[\"price\"] = pd.to_numeric(sf_list[\"price\"], downcast=\"integer\")\n\n#host_response_rate and host_acceptance_rate types to float\nsf_list[\"host_response_rate\"] = sf_list[\"host_acceptance_rate\"].apply(lambda string: stringToNumConverter(string))\nsf_list[\"host_response_rate\"] = pd.to_numeric(sf_list[\"host_response_rate\"], downcast=\"float\")\n\nsf_list[\"host_acceptance_rate\"] = sf_list[\"host_acceptance_rate\"].apply(lambda string: stringToNumConverter(string))\nsf_list[\"host_acceptance_rate\"] = pd.to_numeric(sf_list[\"host_acceptance_rate\"], downcast=\"float\")\n\n#converting t, f value to 1 or 0\nsf_list[\"host_is_superhost\"] = sf_list[\"host_is_superhost\"].apply((lambda string: 1 if string == \"t\" else 0))\n\n#converting datatype of date columns to datetime\nsf_list[\"last_review\"] = pd.to_datetime(arg=sf_list[\"last_review\"], errors=\"coerce\")\nsf_list[\"host_since\"] = pd.to_datetime(arg=sf_list[\"host_since\"], errors=\"coerce\")\n",
"_____no_output_____"
],
[
"print(\"Listing Data Frame\")\nprintColunmsInfo(sf_list)",
"Listing Data Frame\n<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 7274 entries, 0 to 7273\nData columns (total 31 columns):\nid 7274 non-null int64\nhost_since 7274 non-null datetime64[ns]\nhost_is_superhost 7274 non-null int64\nhost_listings_count 7274 non-null int64\nhost_response_time 5321 non-null object\nhost_response_rate 6393 non-null float32\nhost_acceptance_rate 6393 non-null float32\nneighbourhood_cleansed 7274 non-null object\nlatitude 7274 non-null float64\nlongitude 7274 non-null float64\nproperty_type 7274 non-null object\nroom_type 7274 non-null object\naccommodates 7274 non-null int64\nbathrooms 7185 non-null float64\nbedrooms 6430 non-null float64\nbeds 7234 non-null float64\namenities 7274 non-null object\nminimum_nights 7274 non-null int64\nmaximum_nights 7274 non-null int64\nreview_scores_rating 5508 non-null float64\nreview_scores_accuracy 5495 non-null float64\nreview_scores_cleanliness 5495 non-null float64\nreview_scores_checkin 5494 non-null float64\nreview_scores_communication 5496 non-null float64\nreview_scores_location 5494 non-null float64\nreview_scores_value 5494 non-null float64\navailability_30 7274 non-null int64\nnumber_of_reviews 7274 non-null int64\nlast_review 5554 non-null datetime64[ns]\nreviews_per_month 5554 non-null float64\nprice 7274 non-null int16\ndtypes: datetime64[ns](2), float32(2), float64(13), int16(1), int64(8), object(5)\nmemory usage: 1.6+ MB\n\n\nTotal columns: 31\nTotal rows: 7274\n---------------------------------------\n"
]
],
[
[
"### Amenities Data Frame",
"_____no_output_____"
],
[
"Consider the data about the amenities. This column is a set of lists enclosed in strings. So I had to use the *eval*. If you know a more elegant method, please let me know.\n\nThen we'll add columns for each amenitie, remove the common and very rare amenities.",
"_____no_output_____"
]
],
[
[
"amenitiesList = []\nfor item in sf_list[\"amenities\"].value_counts().reset_index()[\"index\"]:\n item = eval(item)\n for i in item:\n if i not in amenitiesList:\n amenitiesList.append(i)\nprint(\"Total amenities: \", len(amenitiesList))\nprint(amenitiesList)",
"Total amenities: 131\n['Gym', 'Heating', 'Hot water', 'Air conditioning', 'Iron', 'Dryer', 'Carbon monoxide alarm', 'Building staff', 'Luggage dropoff allowed', 'Private entrance', 'Microwave', 'Hangers', 'Essentials', 'Smoke alarm', 'Refrigerator', 'Wifi', 'Shampoo', 'TV', 'Hair dryer', 'Washer', 'Cable TV', 'Bed linens', 'Stove', 'Coffee maker', 'Elevator', 'Laptop-friendly workspace', 'Extra pillows and blankets', 'Cooking basics', 'Dishes and silverware', 'Long term stays allowed', 'Kitchen', 'Oven', 'Dishwasher', 'BBQ grill', 'Host greets you', 'Free street parking', 'Hot tub', 'First aid kit', 'Fire extinguisher', 'Lock on bedroom door', 'Paid parking on premises', 'Lockbox', 'Bathtub', 'Garden or backyard', 'Patio or balcony', 'Indoor fireplace', 'Keypad', 'Cleaning before checkout', 'Paid parking off premises', 'Smart lock', 'Free parking on premises', 'Bread maker', 'Shower gel', 'Crib', 'Baking sheet', 'Breakfast', 'Pool', 'Pocket wifi', 'Ethernet connection', 'Pack ’n Play/travel crib', 'Fireplace guards', 'Bathroom essentials', 'Bedroom comforts', 'Self check-in', 'Full kitchen', 'Private living room', 'Outlet covers', 'Game console', 'Children’s books and toys', 'EV charger', 'High chair', 'Room-darkening shades', 'Kitchenette', 'Stair gates', 'Lake access', 'Single level home', 'Waterfront', 'Beachfront', 'Changing table', 'Children’s dinnerware', 'Babysitter recommendations', 'Window guards', 'Baby bath', 'Stand alone bathtub', 'Terrace', 'Stand alone rain shower', 'Jetted tub', 'Breakfast bar', 'Security cameras', 'Dual vanity', 'Sonos sound system', 'Smart TV', 'Smart home technology', 'Office', 'Portable air conditioning', 'Wine cooler', 'Ironing board', 'Gas fireplace', 'Barbecue utensils', 'Beach essentials', 'Pets allowed', 'Baby monitor', 'Suitable for events', 'Smoking allowed', 'Table corner guards', 'Private hot tub', 'Piano', 'Alarm system', 'Natural gas barbeque', 'Walk in closet', 'Desk', 'Wet bar', 'Mini fridge', 'Fire pit', 'Shower bathtub combo', 'Courtyard', 'Stand alone steam shower', 'Home theater', 'Nest thermometer', 'Espresso machine', 'Balcony', 'Massage table', 'Media room', 'Apple TV', 'Shared hot tub', 'Outdoor kitchen', 'Wood-burning fireplace', 'Driveway parking', 'Day bed', 'Lounge area', 'Ski-in/Ski-out']\n"
],
[
"amenities_df = sf_list[[\"id\", \"amenities\"]]\n\n#we don't need \"amenities\" in original data frame anymore\nsf_list.drop([\"amenities\"], axis=1, inplace=True)",
"_____no_output_____"
],
[
"amenitiesDict = {}\nfor item in range(amenities_df.shape[0]):\n i_id, amenitiesSet = amenities_df.loc[item, \"id\"], set(eval(amenities_df.loc[item, \"amenities\"]))\n amenitiesDict[i_id] = amenitiesSet\n\nfor amenitie in amenitiesList:\n bilist = []\n for amId in amenities_df[\"id\"]: \n if amenitie in amenitiesDict[amId]:\n bilist.append(1)\n else:\n bilist.append(0)\n amenities_df.insert(loc=len(amenities_df.columns), column=amenitie, value=bilist, allow_duplicates=True)\nprint(amenities_df.shape)",
"(7274, 133)\n"
]
],
[
[
"### Calendar Data Frame",
"_____no_output_____"
]
],
[
[
"sf_cal.head()",
"_____no_output_____"
]
],
[
[
"This Data Frame has folowing columns:\n* __listing_id__ — id values, we'll use to join tables\n* __date__ — we need to change datatype to datetime\n* __available__ — it has to be boolean, so we need to change it\n* __minimum_nights, maximum_nights__ — we have same columns in Listing Data Frame, drop they later\n* __adjusted_price__, __price__ — target values",
"_____no_output_____"
]
],
[
[
"#converting datatype of price and adjusted_price columns to integer\nsf_cal[\"price\"] = sf_cal[\"price\"].apply(lambda string: stringToNumConverter(string))\nsf_cal[\"price\"] = pd.to_numeric(sf_cal[\"price\"], downcast=\"integer\")\n\nsf_cal[\"adjusted_price\"] = sf_cal[\"adjusted_price\"].apply(lambda string: stringToNumConverter(string))\nsf_cal[\"adjusted_price\"] = pd.to_numeric(sf_cal[\"adjusted_price\"], downcast=\"integer\")\n\n#converting datatype of date columns to datetime\nsf_cal[\"date\"] = pd.to_datetime(arg=sf_cal[\"date\"], errors=\"coerce\")\n\n#converting t, f value to Boolean datatype\nsf_cal[\"available\"] = sf_cal[\"available\"].apply((lambda string: True if string == \"t\" else False))",
"_____no_output_____"
],
[
"print(\"Calendar Data Frame\")\nprintColunmsInfo(sf_cal)",
"Calendar Data Frame\n<class 'pandas.core.frame.DataFrame'>\nInt64Index: 2390652 entries, 0 to 2390651\nData columns (total 7 columns):\nlisting_id int64\ndate datetime64[ns]\navailable bool\nprice float64\nadjusted_price float64\nminimum_nights float64\nmaximum_nights float64\ndtypes: bool(1), datetime64[ns](1), float64(4), int64(1)\nmemory usage: 130.0 MB\n\n\nTotal columns: 7\nTotal rows: 2390652\n---------------------------------------\n"
]
],
[
[
"## Data Understanding",
"_____no_output_____"
],
[
"Let's analyze the data to answer the questions given at the beginning:\n#### 1. What correlates best with price?",
"_____no_output_____"
],
[
"##### Does Amenities correlate with price?",
"_____no_output_____"
]
],
[
[
"amen_price_corr_neg = amenities_df.merge(sf_list[[\"id\", \"price\"]], on=\"id\").corr()[[\"id\", \"price\"]].sort_values(by=\"price\").head(10)\namen_price_corr_pos = amenities_df.merge(sf_list[[\"id\", \"price\"]], on=\"id\").corr()[[\"id\", \"price\"]].sort_values(by=\"price\").drop(\"price\", axis=0).tail(10)",
"_____no_output_____"
],
[
"#negative correlation\namen_price_corr_neg.drop(\"id\", axis=1).style.bar(color=\"#00677e\", align=\"mid\")",
"_____no_output_____"
],
[
"#positive correlation\namen_price_corr_pos.drop(\"id\", axis=1).style.bar(color=\"#cd4a4c\")",
"_____no_output_____"
]
],
[
[
"As you can see, air conditioning, gym, and building staff are highly correlated with price. The rest of the amenities correlate either weakly or not at all.",
"_____no_output_____"
],
[
"##### Does Review Scores correlate with price?",
"_____no_output_____"
]
],
[
[
"plt.subplots(figsize=(9, 6))\nsns.heatmap(sf_list[['review_scores_rating', 'review_scores_accuracy',\n 'review_scores_cleanliness', 'review_scores_checkin',\n 'review_scores_communication', 'review_scores_location',\n 'review_scores_value', \"number_of_reviews\", 'price']].corr(),\n annot=True, fmt=\".2f\")",
"_____no_output_____"
]
],
[
[
"Review Scores correlate weakly with price, but they correlate well with each other.",
"_____no_output_____"
],
[
"##### Does Housing Characteristics correlate with price?",
"_____no_output_____"
]
],
[
[
"plt.subplots(figsize=(9, 6))\nsns.heatmap(sf_list[['accommodates', 'bathrooms', 'bedrooms', 'beds', 'price']].corr(),\n annot=True, fmt=\".2f\")",
"_____no_output_____"
]
],
[
[
"There is an obvious correlation. The more people you can accommodate, the more expensive it is to rent a room. Same about bedrooms and beds. But the number of bathrooms does not have a strong impact.",
"_____no_output_____"
],
[
"Some more dependencies on the price, which we will use in modeling:",
"_____no_output_____"
]
],
[
[
"sf_list.groupby([\"room_type\"]).mean().reset_index()[[\"room_type\",\"price\"]].style.bar(color=\"#cd4a4c\")",
"_____no_output_____"
],
[
"sf_list.groupby([\"property_type\"]).mean().reset_index()[[\"property_type\",\"price\"]].sort_values(by=\"price\", ascending=False).style.bar(color=\"#cd4a4c\")",
"_____no_output_____"
],
[
"sf_list.groupby([\"host_response_time\"]).mean().reset_index()[[\"host_response_time\",\"price\"]].style.bar(color=\"#cd4a4c\")",
"_____no_output_____"
],
[
"sf_list.groupby([\"host_is_superhost\"]).mean().reset_index()[[\"host_is_superhost\",\"price\"]]",
"_____no_output_____"
],
[
"sf_list[[\"number_of_reviews\",\"price\"]].corr()",
"_____no_output_____"
],
[
"plt.subplots(figsize=(9, 6))\nsns.heatmap(sf_list[[\"host_response_rate\", \"host_acceptance_rate\", \"minimum_nights\",\n \"maximum_nights\", \"number_of_reviews\", \"price\"]].corr(),\n annot=True, fmt=\".2f\")",
"_____no_output_____"
]
],
[
[
"##### How about Neighbourhoods?",
"_____no_output_____"
],
[
"Let's find the most expensive neighbourhood.",
"_____no_output_____"
]
],
[
[
"#coordinates of San Francisco\nsf_latitude, sf_longitude = 37.7647993, -122.4629897\n#the necessary data for the map\nsf_map = gpd.read_file(\"planning_neighborhoods/planning_neighborhoods.shp\")\nsf_neig_mean = sf_list.groupby([\"neighbourhood_cleansed\"]).mean().reset_index()\nsf_map = sf_map.merge(sf_neig_mean, left_on=\"neighborho\", right_on=\"neighbourhood_cleansed\")",
"_____no_output_____"
],
[
"vmin, vmax = 100, 1300\n\nfig, ax = plt.subplots(figsize = (20, 20))\nax.set_title(\"Average price in each neighborhood of San Francisco\", fontdict={\"fontsize\": \"25\", \"fontweight\" : \"3\"})\nsf_map.plot(column=\"price\", cmap=\"OrRd\", linewidth=0.8, ax=ax, edgecolor=\"0.8\")\n\ntexts = []\nfor x, y, label in zip(sf_map.centroid.geometry.x, sf_map.centroid.geometry.y, sf_map[\"neighbourhood_cleansed\"]):\n texts.append(plt.text(x, y, label, fontsize = 8))\n\nsm = plt.cm.ScalarMappable(cmap=\"OrRd\", norm=plt.Normalize(vmin=vmin, vmax=vmax))\n# empty array for the data range\nsm._A = []\n# add the colorbar to the figure\ncbar = fig.colorbar(sm)\n \nax.axis(\"off\")\nplt.show()",
"_____no_output_____"
],
[
"sf_list.groupby([\"neighbourhood_cleansed\"]).mean().reset_index()[[\"neighbourhood_cleansed\",\"price\"]].sort_values(by=\"price\", ascending=False).style.bar(color=\"#cd4a4c\")",
"_____no_output_____"
]
],
[
[
"As you can see from the map, the high price is more related to the location. The most expensive areas are Golden Gate Park and Financial District. If you look at my previous research, you understand that Golden Gate Park is quite safe, unlike the Financial District which pretty criminal.",
"_____no_output_____"
],
[
"All this data can be used to predict prices. But before that, let's answer the second question.",
"_____no_output_____"
],
[
"#### 2. How has price and busyness changed over the course of COVID-19?",
"_____no_output_____"
],
[
"Let's start by looking at price changes over the past year.",
"_____no_output_____"
]
],
[
[
"per = sf_cal.date.dt.to_period(\"M\") \ng = sf_cal.groupby(per)\n\nax = sns.set_palette(\"viridis\")\n\nplt.figure(figsize=(16,6))\nsns.barplot(x=g.mean().reset_index()[\"date\"], \n y=g.mean().reset_index()[\"price\"])\nplt.xlabel(\"Month\", fontsize=20)\nplt.ylabel(\"Price per night\", fontsize=20)\nplt.title(\"Average Price per night in San Francisco\", fontsize=25)\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"During the covid period, the average price per night rose by about $33. And it does not stop growing linearly.",
"_____no_output_____"
],
[
"Next one is busyness.",
"_____no_output_____"
]
],
[
[
"ax = sns.set_palette(\"viridis\")\n\nplt.figure(figsize=(16,6))\nsns.barplot(x=g.mean().reset_index()[\"date\"], \n y=g.mean().reset_index()[\"available\"])\nplt.xlabel(\"Month\", fontsize=20)\nplt.ylabel(\"Availability, proportion\", fontsize=20)\nplt.title(\"Average Availability in San Francisco\", fontsize=25)\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"September last year was quite popular (wonderful weather). Then the decline began. But with the onset of covid, the decline intensified and reached its peak (half of the housing is vacant) by May.",
"_____no_output_____"
],
[
"As expected, the covid did not affect the Airbnb business in the best way. Prices have gone up and there are fewer customers. The indicators have not yet returned to their previous values.",
"_____no_output_____"
],
[
"To answer the last question, we have to prepare the data for modeling.",
"_____no_output_____"
],
[
"### Can we predict the price based on its features?",
"_____no_output_____"
],
[
"## Prepare Data",
"_____no_output_____"
],
[
"#### Working with NaNs and categorical variables",
"_____no_output_____"
],
[
"Let's turn \"last_review\" and \"host_since\" from date type to categorical values. For that, we create new columns and fill them in.",
"_____no_output_____"
]
],
[
[
"sf_list[\"since_last_review\"] = sf_list[\"last_review\"].apply(lambda row : dateToCategorical(row))\nsf_list[\"host_since_cat\"] = sf_list[\"host_since\"].apply(lambda row : dateToCategorical(row))",
"_____no_output_____"
],
[
"#drop all Nans in \"price\" columns\ndrop_sf_list = sf_list.dropna(subset=[\"price\"], axis=0)\n#create data frame with categorical values\ncat_sf_list = drop_sf_list[[\"id\", \"neighbourhood_cleansed\", \"room_type\",'property_type', \"since_last_review\", \"host_since_cat\"]]\n#create data frame with nimerical\nmean_sf_list = drop_sf_list[[\"id\", \"accommodates\", \"review_scores_rating\", \"bathrooms\", \"bedrooms\", \"beds\",\n \"review_scores_accuracy\", \"review_scores_cleanliness\", \"availability_30\",\n \"number_of_reviews\", \"reviews_per_month\", \"review_scores_communication\",\n \"review_scores_location\", \"review_scores_value\", \"host_is_superhost\",\n \"host_listings_count\", \"price\"]]",
"_____no_output_____"
],
[
"num_cols = [\"accommodates\", \"review_scores_rating\", \"bathrooms\", \"bedrooms\", \"beds\",\n \"review_scores_accuracy\", \"review_scores_cleanliness\", \"availability_30\",\n \"number_of_reviews\", \"reviews_per_month\", \"review_scores_communication\",\n \"review_scores_location\", \"review_scores_value\", \"host_is_superhost\",\n \"host_listings_count\", \"price\"]\nfor col in num_cols:\n mean_sf_list[col] = mean_sf_list[col].astype('float64').replace(0.0, 0.01) \n mean_sf_list[col] = np.log(mean_sf_list[col])",
"N:\\Anaconda\\lib\\site-packages\\ipykernel_launcher.py:7: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n import sys\nN:\\Anaconda\\lib\\site-packages\\ipykernel_launcher.py:8: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \n"
],
[
"#fill the mean\nfill_mean = lambda col: col.fillna(col.mean())\nmean_sf_list = mean_sf_list.apply(fill_mean, axis=0)",
"_____no_output_____"
],
[
"#create dummy data frame\ncat_cols_lst = [\"neighbourhood_cleansed\", \"room_type\",'property_type', \"since_last_review\", \"host_since_cat\"]\ndummy_sf_list = create_dummy_df(cat_sf_list, cat_cols_lst, dummy_na=False)",
"_____no_output_____"
]
],
[
[
"After all, we'll merge tree Data Frames: mean_sf_list, dummy_sf_list and amenities_df.",
"_____no_output_____"
]
],
[
[
"full_sf_list = dummy_sf_list.merge(amenities_df.drop([\"amenities\"], axis=1), on=\"id\").merge(mean_sf_list, on=\"id\")",
"_____no_output_____"
]
],
[
[
"## Data Modeling",
"_____no_output_____"
],
[
"Let's start modeling. We will try several models and compare the results.",
"_____no_output_____"
]
],
[
[
"#preparation train and test data\nX = full_sf_list.drop([\"price\"], axis=1)\ny = full_sf_list[\"price\"]\n\n#scaling\nscaler = StandardScaler()\nX = pd.DataFrame(scaler.fit_transform(X), columns=list(X.columns))\n\n#split into train and test\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=.3, random_state=42)",
"_____no_output_____"
],
[
"#writing the metrics for every model in DataFrame\nmetrics_columns = [\"Model Name\", \"r-squared train\", \"r-squared train test\", \"MSE train\", \"MSE test\"]\nmetrics_df = pd.DataFrame(columns=metrics_columns)",
"_____no_output_____"
]
],
[
[
"## Predicting Price",
"_____no_output_____"
],
[
"### AdaBoost regressor",
"_____no_output_____"
]
],
[
[
"adaboost_model = AdaBoostRegressor(n_estimators=20)\nadaboost_model.fit(X_train, y_train)\n\n#predict and score the model\ny_test_preds = adaboost_model.predict(X_test)\ny_train_preds = adaboost_model.predict(X_train)\n\n#scoring model\ntest_r2 = round(r2_score(y_test, y_test_preds), 4)\ntrain_r2 = round(r2_score(y_train, y_train_preds), 4)\ntest_mse = round(mean_squared_error(y_test, y_test_preds), 4)\ntrain_mse = round(mean_squared_error(y_train, y_train_preds), 4)\n\nprint('r-squared score for training set was {}. r-squared score for test set was {}.'.format(train_r2, test_r2))\nprint('MSE score for training set was {}. MSE score for test set was {}.'.format(train_mse, test_mse))\n\n#add row to metrics\nmetrics_df = appendToMetricsdf(metrics_df, \"AdaBoost regressor\", train_r2, test_r2, train_mse, test_mse)",
"r-squared score for training set was 0.4804. r-squared score for test set was 0.4407.\nMSE score for training set was 0.3275. MSE score for test set was 0.3551.\n"
]
],
[
[
"### Gradient Boosting for regression",
"_____no_output_____"
]
],
[
[
"gradboost_model = GradientBoostingRegressor(n_estimators=300)\ngradboost_model.fit(X_train, y_train)\n\n#predict and score the model\ny_test_preds = gradboost_model.predict(X_test)\ny_train_preds = gradboost_model.predict(X_train)\n\n#scoring model\ntest_r2 = round(r2_score(y_test, y_test_preds), 4)\ntrain_r2 = round(r2_score(y_train, y_train_preds), 4)\ntest_mse = round(mean_squared_error(y_test, y_test_preds), 4)\ntrain_mse = round(mean_squared_error(y_train, y_train_preds), 4)\n\nprint('r-squared score for training set was {}. r-squared score for test set was {}.'.format(train_r2, test_r2))\nprint('MSE score for training set was {}. MSE score for test set was {}.'.format(train_mse, test_mse))\n\nmetrics_df = appendToMetricsdf(metrics_df, \"Gradient Boosting\", train_r2, test_r2, train_mse, test_mse)",
"r-squared score for training set was 0.8149. r-squared score for test set was 0.7033.\nMSE score for training set was 0.1167. MSE score for test set was 0.1884.\n"
]
],
[
[
"### Extreme Gradient Boosting",
"_____no_output_____"
]
],
[
[
"xgb_reg = xgb.XGBRegressor()\nxgb_reg.fit(X_train, y_train)\n\ny_train_preds = xgb_reg.predict(X_train)\ny_test_preds = xgb_reg.predict(X_test)\n\n#scoring model\ntest_r2 = round(r2_score(y_test, y_test_preds), 4)\ntrain_r2 = round(r2_score(y_train, y_train_preds), 4)\ntest_mse = round(mean_squared_error(y_test, y_test_preds), 4)\ntrain_mse = round(mean_squared_error(y_train, y_train_preds), 4)\n\nprint('r-squared score for training set was {}. r-squared score for test set was {}.'.format(train_r2, test_r2))\nprint('MSE score for training set was {}. MSE score for test set was {}.'.format(train_mse, test_mse))\n\nmetrics_df = appendToMetricsdf(metrics_df, \"Extreme Gradient Boosting\", train_r2, test_r2, train_mse, test_mse)",
"[16:43:04] WARNING: src/objective/regression_obj.cu:152: reg:linear is now deprecated in favor of reg:squarederror.\n"
]
],
[
[
"### Neural Network",
"_____no_output_____"
]
],
[
[
"#building the model\nmodel = models.Sequential()\nmodel.add(layers.Dense(128, input_shape=(X_train.shape[1],), activation='relu'))\nmodel.add(layers.Dense(256, activation='relu'))\nmodel.add(layers.Dense(256, activation='relu'))\nmodel.add(layers.Dense(1, activation='linear'))\n\n#compiling the model\nmodel.compile(optimizer='adam', \n loss='mse', \n metrics=[r2_keras])\n\n#model summary\nprint(model.summary())",
"Model: \"sequential_1\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense_1 (Dense) (None, 128) 32000 \n_________________________________________________________________\ndense_2 (Dense) (None, 256) 33024 \n_________________________________________________________________\ndense_3 (Dense) (None, 256) 65792 \n_________________________________________________________________\ndense_4 (Dense) (None, 1) 257 \n=================================================================\nTotal params: 131,073\nTrainable params: 131,073\nNon-trainable params: 0\n_________________________________________________________________\nNone\n"
],
[
"# Training the model\nmodel_start = time.time()\n\nmodel_history = model.fit(X_train, y_train, epochs=500, batch_size=256, validation_data=(X_test, y_test))\n\nmodel_end = time.time()\n\nprint(f\"Time taken to run: {round((model_end - model_start)/60,1)} minutes\")",
"Train on 5091 samples, validate on 2183 samples\nEpoch 1/500\n5091/5091 [==============================] - 0s 96us/step - loss: 8.1906 - r2_keras: -12.9687 - val_loss: 2.6318 - val_r2_keras: -3.0942\nEpoch 2/500\n5091/5091 [==============================] - 0s 22us/step - loss: 1.7102 - r2_keras: -1.7570 - val_loss: 1.1099 - val_r2_keras: -0.7554\nEpoch 3/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.6961 - r2_keras: -0.1202 - val_loss: 0.6361 - val_r2_keras: -0.0045\nEpoch 4/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.4219 - r2_keras: 0.3275 - val_loss: 0.4637 - val_r2_keras: 0.2656\nEpoch 5/500\n5091/5091 [==============================] - 0s 23us/step - loss: 0.3061 - r2_keras: 0.5054 - val_loss: 0.3980 - val_r2_keras: 0.3676\nEpoch 6/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.2488 - r2_keras: 0.5988 - val_loss: 0.3661 - val_r2_keras: 0.4169\nEpoch 7/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.2136 - r2_keras: 0.6549 - val_loss: 0.3469 - val_r2_keras: 0.4474\nEpoch 8/500\n5091/5091 [==============================] - 0s 30us/step - loss: 0.1880 - r2_keras: 0.6951 - val_loss: 0.3314 - val_r2_keras: 0.4714\nEpoch 9/500\n5091/5091 [==============================] - 0s 23us/step - loss: 0.1688 - r2_keras: 0.7275 - val_loss: 0.3255 - val_r2_keras: 0.4805\nEpoch 10/500\n5091/5091 [==============================] - 0s 23us/step - loss: 0.1514 - r2_keras: 0.7579 - val_loss: 0.3148 - val_r2_keras: 0.4985\nEpoch 11/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.1378 - r2_keras: 0.7764 - val_loss: 0.3087 - val_r2_keras: 0.5079\nEpoch 12/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.1283 - r2_keras: 0.7952 - val_loss: 0.3059 - val_r2_keras: 0.5120\nEpoch 13/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.1178 - r2_keras: 0.8106 - val_loss: 0.3011 - val_r2_keras: 0.5197\nEpoch 14/500\n5091/5091 [==============================] - ETA: 0s - loss: 0.1112 - r2_keras: 0.82 - 0s 23us/step - loss: 0.1084 - r2_keras: 0.8264 - val_loss: 0.3035 - val_r2_keras: 0.5153\nEpoch 15/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.1002 - r2_keras: 0.8379 - val_loss: 0.2960 - val_r2_keras: 0.5275\nEpoch 16/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.0918 - r2_keras: 0.8514 - val_loss: 0.2928 - val_r2_keras: 0.5330\nEpoch 17/500\n5091/5091 [==============================] - 0s 23us/step - loss: 0.0859 - r2_keras: 0.8608 - val_loss: 0.2908 - val_r2_keras: 0.5361\nEpoch 18/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.0806 - r2_keras: 0.8709 - val_loss: 0.2930 - val_r2_keras: 0.5323\nEpoch 19/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.0758 - r2_keras: 0.8776 - val_loss: 0.2942 - val_r2_keras: 0.5307\nEpoch 20/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.0709 - r2_keras: 0.8857 - val_loss: 0.2916 - val_r2_keras: 0.5349\nEpoch 21/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.0688 - r2_keras: 0.8891 - val_loss: 0.2927 - val_r2_keras: 0.5335\nEpoch 22/500\n5091/5091 [==============================] - 0s 23us/step - loss: 0.0636 - r2_keras: 0.8977 - val_loss: 0.2920 - val_r2_keras: 0.5342\nEpoch 23/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.0597 - r2_keras: 0.9046 - val_loss: 0.2969 - val_r2_keras: 0.5258\nEpoch 24/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.0593 - r2_keras: 0.9048 - val_loss: 0.2916 - val_r2_keras: 0.5349\nEpoch 25/500\n5091/5091 [==============================] - 0s 23us/step - loss: 0.0546 - r2_keras: 0.9138 - val_loss: 0.2932 - val_r2_keras: 0.5318\nEpoch 26/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.0525 - r2_keras: 0.9161 - val_loss: 0.2932 - val_r2_keras: 0.5325\nEpoch 27/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.0497 - r2_keras: 0.9206 - val_loss: 0.2936 - val_r2_keras: 0.5319\nEpoch 28/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.0452 - r2_keras: 0.9277 - val_loss: 0.2923 - val_r2_keras: 0.5333\nEpoch 29/500\n5091/5091 [==============================] - 0s 24us/step - loss: 0.0424 - r2_keras: 0.9319 - val_loss: 0.2967 - val_r2_keras: 0.5262\nEpoch 30/500\n5091/5091 [==============================] - 0s 23us/step - loss: 0.0406 - r2_keras: 0.9354 - val_loss: 0.2943 - val_r2_keras: 0.5307\nEpoch 31/500\n5091/5091 [==============================] - 0s 23us/step - loss: 0.0375 - r2_keras: 0.9407 - val_loss: 0.2959 - val_r2_keras: 0.5277\nEpoch 32/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.0369 - r2_keras: 0.9415 - val_loss: 0.2967 - val_r2_keras: 0.5264\nEpoch 33/500\n5091/5091 [==============================] - 0s 23us/step - loss: 0.0350 - r2_keras: 0.9446 - val_loss: 0.2964 - val_r2_keras: 0.5267\nEpoch 34/500\n5091/5091 [==============================] - 0s 23us/step - loss: 0.0360 - r2_keras: 0.9410 - val_loss: 0.2988 - val_r2_keras: 0.5228\nEpoch 35/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.0364 - r2_keras: 0.9424 - val_loss: 0.3026 - val_r2_keras: 0.5176\nEpoch 36/500\n5091/5091 [==============================] - 0s 23us/step - loss: 0.0332 - r2_keras: 0.9462 - val_loss: 0.2986 - val_r2_keras: 0.5240\nEpoch 37/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.0298 - r2_keras: 0.9530 - val_loss: 0.2999 - val_r2_keras: 0.5212\nEpoch 38/500\n5091/5091 [==============================] - 0s 23us/step - loss: 0.0283 - r2_keras: 0.9533 - val_loss: 0.2995 - val_r2_keras: 0.5219\nEpoch 39/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.0278 - r2_keras: 0.9545 - val_loss: 0.3008 - val_r2_keras: 0.5192\nEpoch 40/500\n5091/5091 [==============================] - 0s 23us/step - loss: 0.0272 - r2_keras: 0.9569 - val_loss: 0.3021 - val_r2_keras: 0.5172\nEpoch 41/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.0265 - r2_keras: 0.9577 - val_loss: 0.3018 - val_r2_keras: 0.5184\nEpoch 42/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.0273 - r2_keras: 0.9576 - val_loss: 0.3013 - val_r2_keras: 0.5176\nEpoch 43/500\n5091/5091 [==============================] - 0s 23us/step - loss: 0.0320 - r2_keras: 0.9490 - val_loss: 0.3050 - val_r2_keras: 0.5131\nEpoch 44/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.0293 - r2_keras: 0.9547 - val_loss: 0.3035 - val_r2_keras: 0.5144\nEpoch 45/500\n5091/5091 [==============================] - 0s 23us/step - loss: 0.0261 - r2_keras: 0.9601 - val_loss: 0.3053 - val_r2_keras: 0.5126\nEpoch 46/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.0249 - r2_keras: 0.9598 - val_loss: 0.3052 - val_r2_keras: 0.5117\nEpoch 47/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.0235 - r2_keras: 0.9647 - val_loss: 0.3030 - val_r2_keras: 0.5165\nEpoch 48/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.0228 - r2_keras: 0.9643 - val_loss: 0.3053 - val_r2_keras: 0.5125\nEpoch 49/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.0226 - r2_keras: 0.9640 - val_loss: 0.3032 - val_r2_keras: 0.5153\nEpoch 50/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.0204 - r2_keras: 0.9674 - val_loss: 0.3093 - val_r2_keras: 0.5055\nEpoch 51/500\n5091/5091 [==============================] - 0s 23us/step - loss: 0.0196 - r2_keras: 0.9689 - val_loss: 0.3065 - val_r2_keras: 0.5106\nEpoch 52/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.0197 - r2_keras: 0.9698 - val_loss: 0.3054 - val_r2_keras: 0.5120\nEpoch 53/500\n5091/5091 [==============================] - 0s 23us/step - loss: 0.0184 - r2_keras: 0.9700 - val_loss: 0.3084 - val_r2_keras: 0.5070\nEpoch 54/500\n5091/5091 [==============================] - 0s 23us/step - loss: 0.0193 - r2_keras: 0.9702 - val_loss: 0.3074 - val_r2_keras: 0.5092\nEpoch 55/500\n5091/5091 [==============================] - 0s 22us/step - loss: 0.0194 - r2_keras: 0.9692 - val_loss: 0.3107 - val_r2_keras: 0.5030\nEpoch 56/500\n"
],
[
"#evaluate model\nloss_train = model_history.history['loss']\nloss_val = model_history.history['val_loss']\nplt.figure(figsize=(8,6))\nplt.plot(model_history.history['loss'])\nplt.plot(model_history.history['val_loss'])\nplt.title('Training and Test loss at each epoch')\nplt.ylabel('Loss')\nplt.xlabel('Epoch')\nplt.legend(['train', 'test'], loc='upper left')\nplt.show()",
"_____no_output_____"
],
[
"score_train = model.evaluate(X_train, y_train, verbose = 0)\nscore_test = model.evaluate(X_test, y_test, verbose = 0)\n\ntrain_r2 = round(score_train[1], 4)\ntest_r2 = round(score_test[1], 4)\ntrain_mse = round(score_train[0], 4)\ntest_mse = round(score_test[0], 4)\n\nmetrics_df = appendToMetricsdf(metrics_df, \"Neural Network\", train_r2, test_r2, train_mse, test_mse)",
"_____no_output_____"
]
],
[
[
"## Evaluate the Results",
"_____no_output_____"
],
[
"Let's take a look at our results and compare them with each other.",
"_____no_output_____"
]
],
[
[
"metrics_df",
"_____no_output_____"
]
],
[
[
"The AdaBoost regressor showed bad r2 score. The predictions of this model are not similar to the real values.\nGradient Boosting and Extreme Gradient Boosting showed similar results, but Gradient Boosting is slightly better.\nFinally, I trained a neural network that performs worse than Gradient Boosting and shows overfitting.",
"_____no_output_____"
],
[
"## Thank you!",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d086900cc15f3186b528a84379e59c5ae7e6419f | 160,481 | ipynb | Jupyter Notebook | 42-text-generation-with-lstm.ipynb | mhsattarian/class.vision | 00953ff60c6de7fb42b991b635590f391063283b | [
"MIT"
] | 103 | 2018-02-23T15:58:26.000Z | 2022-03-09T05:49:14.000Z | 42-text-generation-with-lstm.ipynb | Deepstatsanalysis/class.vision | d7859f51d4f969913549e440fdc45f673c9da3de | [
"MIT"
] | null | null | null | 42-text-generation-with-lstm.ipynb | Deepstatsanalysis/class.vision | d7859f51d4f969913549e440fdc45f673c9da3de | [
"MIT"
] | 53 | 2018-02-16T20:38:29.000Z | 2022-03-07T10:12:10.000Z | 68.964761 | 473 | 0.658389 | [
[
[
"<img src=\"http://akhavanpour.ir/notebook/images/srttu.gif\" alt=\"SRTTU\" style=\"width: 150px;\"/>\n\n[](https://notebooks.azure.com/import/gh/Alireza-Akhavan/class.vision)",
"_____no_output_____"
],
[
"# <div style=\"direction:rtl;text-align:right;font-family:B Lotus, B Nazanin, Tahoma\"> تولید متن با شبکه بازگشتی LSTM در Keras</div>\n\n<div style=\"direction:rtl;text-align:right;font-family:Tahoma\">\nکدها برگرفته از فصل هشتم کتاب\n</div>\n\n[Deep Learning with Python](https://www.manning.com/books/deep-learning-with-python?a_aid=keras&a_bid=76564dff)\n<div style=\"direction:rtl;text-align:right;font-family:Tahoma\">\nو گیت هاب نویسنده کتاب و توسعه دهنده کراس \n</div>\n\n[François Chollet](http://nbviewer.jupyter.org/github/fchollet/deep-learning-with-python-notebooks/blob/master/8.1-text-generation-with-lstm.ipynb)\n<div style=\"direction:rtl;text-align:right;font-family:Tahoma\">\nاست.\n</div>\n",
"_____no_output_____"
]
],
[
[
"import keras\nkeras.__version__",
"Using TensorFlow backend.\n"
]
],
[
[
"# Text generation with LSTM\n\n## Implementing character-level LSTM text generation\n\n\nLet's put these ideas in practice in a Keras implementation. The first thing we need is a lot of text data that we can use to learn a \nlanguage model. You could use any sufficiently large text file or set of text files -- Wikipedia, the Lord of the Rings, etc. In this \nexample we will use some of the writings of Nietzsche, the late-19th century German philosopher (translated to English). The language model \nwe will learn will thus be specifically a model of Nietzsche's writing style and topics of choice, rather than a more generic model of the \nEnglish language.",
"_____no_output_____"
],
[
"### <div style=\"direction:rtl;text-align:right;font-family:B Lotus, B Nazanin, Tahoma\"> مجموعه داده\n</div>",
"_____no_output_____"
]
],
[
[
"import keras\nimport numpy as np\n\npath = keras.utils.get_file(\n 'nietzsche.txt',\n origin='https://s3.amazonaws.com/text-datasets/nietzsche.txt')\ntext = open(path).read().lower()\nprint('Corpus length:', len(text))",
"Corpus length: 600901\n"
]
],
[
[
"\nNext, we will extract partially-overlapping sequences of length `maxlen`, one-hot encode them and pack them in a 3D Numpy array `x` of \nshape `(sequences, maxlen, unique_characters)`. Simultaneously, we prepare a array `y` containing the corresponding targets: the one-hot \nencoded characters that come right after each extracted sequence.",
"_____no_output_____"
]
],
[
[
"# Length of extracted character sequences\nmaxlen = 60\n\n# We sample a new sequence every `step` characters\nstep = 3\n\n# This holds our extracted sequences\nsentences = []\n\n# This holds the targets (the follow-up characters)\nnext_chars = []\n\nfor i in range(0, len(text) - maxlen, step):\n sentences.append(text[i: i + maxlen])\n next_chars.append(text[i + maxlen])\nprint('Number of sequences:', len(sentences))\n\n# List of unique characters in the corpus\nchars = sorted(list(set(text)))\nprint('Unique characters:', len(chars))\n# Dictionary mapping unique characters to their index in `chars`\nchar_indices = dict((char, chars.index(char)) for char in chars)\n\n# Next, one-hot encode the characters into binary arrays.\nprint('Vectorization...')\nx = np.zeros((len(sentences), maxlen, len(chars)), dtype=np.bool)\ny = np.zeros((len(sentences), len(chars)), dtype=np.bool)\nfor i, sentence in enumerate(sentences):\n for t, char in enumerate(sentence):\n x[i, t, char_indices[char]] = 1\n y[i, char_indices[next_chars[i]]] = 1",
"Number of sequences: 200281\nUnique characters: 59\nVectorization...\n"
]
],
[
[
"## <div style=\"direction:rtl;text-align:right;font-family:B Lotus, B Nazanin, Tahoma\"> ایجاد شبکه (Building the network)</div>\nOur network is a single `LSTM` layer followed by a `Dense` classifier and softmax over all possible characters. But let us note that \nrecurrent neural networks are not the only way to do sequence data generation; 1D convnets also have proven extremely successful at it in \nrecent times.",
"_____no_output_____"
]
],
[
[
"from keras import layers\n\nmodel = keras.models.Sequential()\nmodel.add(layers.LSTM(128, input_shape=(maxlen, len(chars))))\nmodel.add(layers.Dense(len(chars), activation='softmax'))",
"_____no_output_____"
]
],
[
[
"Since our targets are one-hot encoded, we will use `categorical_crossentropy` as the loss to train the model:",
"_____no_output_____"
]
],
[
[
"optimizer = keras.optimizers.RMSprop(lr=0.01)\nmodel.compile(loss='categorical_crossentropy', optimizer=optimizer)",
"_____no_output_____"
]
],
[
[
"## Training the language model and sampling from it\n\n\nGiven a trained model and a seed text snippet, we generate new text by repeatedly:\n\n* 1) Drawing from the model a probability distribution over the next character given the text available so far\n* 2) Reweighting the distribution to a certain \"temperature\"\n* 3) Sampling the next character at random according to the reweighted distribution\n* 4) Adding the new character at the end of the available text\n\nThis is the code we use to reweight the original probability distribution coming out of the model, \nand draw a character index from it (the \"sampling function\"):",
"_____no_output_____"
]
],
[
[
"def sample(preds, temperature=1.0):\n preds = np.asarray(preds).astype('float64')\n preds = np.log(preds) / temperature\n exp_preds = np.exp(preds)\n preds = exp_preds / np.sum(exp_preds)\n probas = np.random.multinomial(1, preds, 1)\n return np.argmax(probas)",
"_____no_output_____"
]
],
[
[
"\nFinally, this is the loop where we repeatedly train and generated text. We start generating text using a range of different temperatures \nafter every epoch. This allows us to see how the generated text evolves as the model starts converging, as well as the impact of \ntemperature in the sampling strategy.",
"_____no_output_____"
]
],
[
[
"import random\nimport sys\n\nfor epoch in range(1, 60):\n print('epoch', epoch)\n # Fit the model for 1 epoch on the available training data\n model.fit(x, y,\n batch_size=128,\n epochs=1)\n\n # Select a text seed at random\n start_index = random.randint(0, len(text) - maxlen - 1)\n generated_text = text[start_index: start_index + maxlen]\n print('--- Generating with seed: \"' + generated_text + '\"')\n\n for temperature in [0.2, 0.5, 1.0, 1.2]:\n print('------ temperature:', temperature)\n sys.stdout.write(generated_text)\n\n # We generate 400 characters\n for i in range(400):\n sampled = np.zeros((1, maxlen, len(chars)))\n for t, char in enumerate(generated_text):\n sampled[0, t, char_indices[char]] = 1.\n\n preds = model.predict(sampled, verbose=0)[0]\n next_index = sample(preds, temperature)\n next_char = chars[next_index]\n\n generated_text += next_char\n generated_text = generated_text[1:]\n\n sys.stdout.write(next_char)\n sys.stdout.flush()\n print()",
"epoch 1\nEpoch 1/1\n200278/200278 [==============================] - 126s - loss: 1.9895 \n--- Generating with seed: \"h they inspire.\" or, as la\nrochefoucauld says: \"if you think\"\n------ temperature: 0.2\nh they inspire.\" or, as la\nrochefoucauld says: \"if you think in the sense of the say the same of the antimated and present in the all the has a such and opent and the say and and the fan and the sense of the into the sense of the say the words and the present the sense of the present present of the present in the man is the man in the sense of the say the sense of the say and the say and the say it is the such and the sense of the ast the sense of the say \n------ temperature: 0.5\nt is the such and the sense of the ast the sense of the say the instand of the way and it is the man for the some songully the sain it is opperience of all the sensity of the same the intendition of the man, in the most with the same philosophicism of the feelient of internations of a present and and colleng it is the sense the greath to the highers of the antolity as nature and the really in the spilitions the leaded and decome the has opence in the sume \n------ temperature: 1.0\nspilitions the leaded and decome the has opence in the sume the orded out powe higher mile as of coftere obe inbernation as to\nthe fof ould mome evpladity. in no it\ngranter, it is the than the\nsay, but the\nmost nothing which, the like the knre hindver\"\nus setured effect of agard\nappate of alsoden\" the lixe their men\nan its of losed the unistensshatity; and oppreness of this not which at the brindurely to giths of sayquitt guratuch with that this\nif\nand whu\n------ temperature: 1.2\nrely to giths of sayquitt guratuch with that this\nif\nand whungs thinkmani.\nficcy, and peninecinated andur mage the\nsened in think wiwhhic\nto beyreasts than\nthis gruath with thioruit\ncatuen\nmuch. h.\n geevated in\nsporated mast the a\"coid\n nrese mae, all conentry, .. fin perhuen\nvenerly (whisty or spore lised har of\nbut ic; at lebgre and things. it keod\nto pring ancayedy\nfrom dill a be utisti listousesquas oke\nthe semment\" (fim their falshin al\nup hesd, and u\nepoch 2\nEpoch 1/1\n200278/200278 [==============================] - 125s - loss: 1.6382 \n--- Generating with seed: \"he cleverness of christianity.=--it is a master stroke of\nch\"\n------ temperature: 0.2\nhe cleverness of christianity.=--it is a master stroke of\nchreme and the same and the contrary and conscience of the deason of the sense and that a sould and superion of the all the subjections and all the disting and all the more and the disting and all the same and such an all the delief to the same and more and sand and sense and all the more and the still and the sense and the more and contrary and man and such a sould and art and the presention of the\n------ temperature: 0.5\ny and man and such a sould and art and the presention of the daction of the still of the same is any more and sanders of hoors of who has an all the man is been fact and belief and contrary had sake and disting world so sake from the\nprejudice of the sentiment and the contrarism of vided and all the saymits of the man way not the achated the deadity at the \"courde of sisted and all the disanctions and as a contrades in a should for a phadoward and only and\n------ temperature: 1.0\n and as a contrades in a should for a phadoward and only and\nemptoces of anmoved and the issintions eedit modeyners bre- warlt of being whole has been bit and would be thing as all it as mankfrom for is\nresp\"quent, privelym yeads overthtice from\nhow will has a mankinduled opine sancels and ary are but the moderation along atolity.\n\n131. new may intempt a van the\nsaur. trater, sake--it tantian all ass are a superstion truth, \"worldting and lawtyes to make l\n------ temperature: 1.2\nass are a superstion truth, \"worldting and lawtyes to make life\ncoldurcly of no has grocbity of norratrimer. no weat doem not ques to thus rasg, whation.\n\nod\"y polent and rulobioved\nagrigncary us queciest?\n\n41\nuspotive force as unischolondanden of cratids, the unbanted caarlo\nsoke not are re. to the trainit ene kinkly skants that self consatiof,\", preveplle reasol decistuticaso itly vail.\n\n8que\"se of a every a progor\nveist a not caul. rigerary nature,\nin \nepoch 3\nEpoch 1/1\n200278/200278 [==============================] - 124s - loss: 1.5460 \n--- Generating with seed: \"ch knows\nhow to handle the knife surely and deftly, even whe\"\n------ temperature: 0.2\nch knows\nhow to handle the knife surely and deftly, even when they has and the strength of the command the great and the sense of the great they are of the streng to the strength and the strength of the great former the strength and the strong the condition of the command they have to the strength and the profound the free spirit of the world in a more of the world and present in the compained they have been the sense of the command they have to the streng\n------ temperature: 0.5\ny have been the sense of the command they have to the strength concernous of the power, the have begon of the last of the profound the artists discourse in the becomes sense of the stand and\nconcertic of texplence of the to may not a seep of the into the accuations that they heart as a solitude, in the good\ninto the accistors, to when the have they has a stard in the last they seems they are of the consequently with the ender, and\ngood in such a power of\nt\n------ temperature: 1.0\ne consequently with the ender, and\ngood in such a power of\nthe \"firmat chores forgubmentatic in stand-new of a needs\nabove\nthan repersibily\ninto\nthe provivent stand\" more what operiority courhe when endure really save sope ford of lower, and long of have, are sins and keet by courd. he should in the bodiec\nthey noblephics,\"\nimported. so perhaps europe.\n\n\n , sechosics of\nthe endiitagy, fougked\nany stranger of the corrorato\nit be last once or consequently no\n------ temperature: 1.2\nstranger of the corrorato\nit be last once or consequently not! in of access is once\nappearal\nstemporic,\"--he the garwand\nany zer-oo -- drinequable to other one much lilutage and\ncumrest of \nthe one, it not =the\nbas of trachtade of\ncowlutaf of whathout such with spount eronry\nare; gow\na whick of a sole phvioration:whicitylyi\npower, in high has a conp, coming, he\nplession his hey!\" unnects, iy every nevershs to adrataes family have\ninsten, os ne's \nepoch 4\nEpoch 1/1\n200278/200278 [==============================] - 125s - loss: 1.4973 \n--- Generating with seed: \"to have spoken of the sensus allegoricus of religion.\nhe wou\"\n------ temperature: 0.2\nto have spoken of the sensus allegoricus of religion.\nhe would be the proposition of the subjection of the standing to such the subjection of the subjltition of the stands and the really the power of the spirit and concertion of the contrary of the concertion of the subjection to the subjection of the spirit of the subjection of the subjection of the subjection of the subjection of the contrary of the same and the subjection of the subjection of the stands\n------ temperature: 0.5\n the same and the subjection of the subjection of the stands of the more beartles and power of the pleasure of moral light, who is the must are an every disting of the deliebly desire the spirit in the subjection of men of distress in the single, to the strange to really been a mettful our uncertainting the expect and the stands of the expochish, exhection of the truth and the merely, and the doctior and enory and the pation of the thought and for a feat o\n------ temperature: 1.0\nior and enory and the pation of the thought and for a feat offues toned spievement and common as musics of danger. that \"the ordered-wants and lack of world of lettife--in any or nehin too\n\"misundifow hundrary not incligation,\ndight, however, to moranary and life these\nmotilet\nreculonac, to aritic means his sarkic. times, his tanvary him, it is their day happiness, in\nhare, of tood whings\nbelief that eary when 1( the dinging it world induction in their for\n------ temperature: 1.2\nhat eary when 1( the dinging it world induction in their for artran, rspumous, ald\nredical pleniscion ap no revereiblines, tho lacquiring that fegais oracus--is preyer. the pery measime, as firnom and rack. -purss\nlove to they like relight of\nreoning\ncage of signtories, the timu to\ncoursite; that libenes afverbtersal; all catured, ehhic: when all tumple, heartted a inhting in\naway love\nthe puten\nparty al mistray. i jesess. own can clatorify\nseloperati\", wh\nepoch 5\nEpoch 1/1\n200278/200278 [==============================] - 125s - loss: 1.4682 \n--- Generating with seed: \"ion (werthschätzung)--the\ndislocation, distortion and the ap\"\n------ temperature: 0.2\nion (werthschätzung)--the\ndislocation, distortion and the appearation of his sensition and conscience of the distrusting the far the sensition of the individually the suffering the sense of the presentiments of the sense of the suffering and suffering the stronger of the suffering and the consequently the sense of the subject of the sense of the moral the sense of the desire the sense of the\nself--and the sensition of the suffering the sensition of the sen\n------ temperature: 0.5\n-and the sensition of the suffering the sensition of the sensition of the individual hence all the perceived as an existence of a few to who is new spirits of himself which may be the world ground our democration in every undifferent of the purely the far much of the estimate religions of the strong and sense of the other reality and conscience and the self-sure he has gare in the self--and knows man and period with the spirit and consequently consequently\n------ temperature: 1.0\nman and period with the spirit and consequently consequently hast\"\"\nbut every every matters (without mad their world who prodessions are weok they consciences of commutionally men) who in comtring. this she appaine, without\nhave under which ialations from o srud nothing in\nthe metively to ding tender, in\nany hens in all very another purithe the complactions--how varies in the exrepration world and though the ethicangling; there is everything our comliferac\n------ temperature: 1.2\n though the ethicangling; there is everything our comliferacled ourianceince the long---r=nony much of anyome.\nif they lanifuels enally inepinious of\nmay, the\ncommin's for concern,\nthere are has dmarding\" to actable,ly effet will itower, butiness the condinided\"--rings up they will futher miands, incondations? gear of limitny, conlict of hervedozihare and the intosting perious into comediand, setakest perficiated\nand\ninlital self--nage peruody;\nthere is sp\nepoch 6\nEpoch 1/1\n200278/200278 [==============================] - 125s - loss: 1.4466 \n--- Generating with seed: \"rd, which\nalways flies further aloft in order always to see \"\n------ temperature: 0.2\nrd, which\nalways flies further aloft in order always to see the suffering that the suffering the sense of the strengthes of the suffering that it is a more and the self-complication of the suffering the suffering and the subtle and self-compartion of the comparting the suffering of the suffering the most the suffering the suffering of the compartion of the most present and the strength and the sufferings of the most the sense of the suffering the sense of \n------ temperature: 0.5\nferings of the most the sense of the suffering the sense of the expect of the intellieate strengent of the dit the attaint is a soul one of the hond to the heart the most expect of the religious the sense of the\nhistle of the fear of the same individual in such a most interest of the had to so the immorality of the possess of the allow, the compress is entitul condition, the discountering in the more reveale, and the refined the fear it is betered one to s\n------ temperature: 1.0\nore reveale, and the refined the fear it is betered one to self-contindaning hypition of surdinguates\nthe\npossible\nataint, when he must beakes comple in the grody of the opposite oftent\ntog, pain finds one that templily to the\ntruthdly one of the fasting oby the highest present treative must materies of incase varies in\na cain, when seaced in seasoury, or such them of\nearlily, and so\nits as of the will to their to forms too scienticiel\nand for which\nit hea\n------ temperature: 1.2\n will to their to forms too scienticiel\nand for which\nit headds maid, estavelhing\nquestion, for thuer, requite tomlan\"! what its do touthodly, thereby). theurse\nout who juveangh of tly histomiaraeg, in peinds. on it.\nall bemond\nmimal. the more harr acqueire it, he house, at of accouncing patedpance han\" willly\nthe ellara\n\"formy tellate.\nmedish purman tturfil an attruth been the custrestiblries in themen-and lightly again ih a daawas or its learhting than\nc\nepoch 7\nEpoch 1/1\n200278/200278 [==============================] - 125s - loss: 1.4282 \n--- Generating with seed: \"realize how\ncharacteristic is this fear of the \"man\" in the \"\n------ temperature: 0.2\nrealize how\ncharacteristic is this fear of the \"man\" in the spirit and and the superition of the propert and perhaps be the superition of the superition of the same of the same the spirit and in the same the strong the still contrast and and and the sure an end and the strong to the destand that the standard, and the spirit and the superition of the superition of the strong the strong that the superition and the state of the same the spirit and and be the \n------ temperature: 0.5\nerition and the state of the same the spirit and and be the same said the spirit to the state and admired to rechancient man as a self felt that the religious distinguished the human believe that the deception, in soul, the stands had been man to be has striced be actual perhaps in all the interpretical strong the decontaitsnentine, the philosophy happiness of the greatest formerly be for the fact deep and weaker of an involuntarian man is one has to the c\n------ temperature: 1.0\n deep and weaker of an involuntarian man is one has to the carely community: ourselves as it seem with theme in hami dance\nalto manifesty, mansike of\nwhich that thereby religion, and reason, a litely for of the allarded by pogures, such diviniatifings and disentached, with life of suffernes, this , altherage.\n\n\n1afetuenally that this tooking\nto plong tematic thate and surfoundaas: the\nprogreable and untisy; which dhes mifere the all such a philosophers, a\n------ temperature: 1.2\nand untisy; which dhes mifere the all such a philosophers, as the athained the such living upon serposed if, his injuring, \"the most standhfulness.\n no the dalb(basise, equal di butz if. thereby mast wast\nhad to plangubly overman hat our eitrieious tar\nand hearth--a -of womet far imminalk and \"she of castuoled.--in the oalt. ant\nollicatiom prot behing-ma\nformuln unkercite--and probachte-patial the historled qualizsss section unterman contlict of, bein\nepoch 8\nEpoch 1/1\n200278/200278 [==============================] - 125s - loss: 1.4150 \n--- Generating with seed: \"ith religion itself and regarded as the\nsupreme attainment o\"\n------ temperature: 0.2\nith religion itself and regarded as the\nsupreme attainment of the words to the scientifical strength and such an and and instincts and and the profoundly to the senses the subjection of the subtle and be desired and still be way and the same interpretation of the way, and the self-destines in the subjection of the desire and and such an experience of the same and be a still be subjection, the spirit is and all the surposition of the same and the subjection\n------ temperature: 0.5\nit is and all the surposition of the same and the subjection and the poind to the profound the same obscure of good, a spirit of an extent so from the greates the similated to himself with the place of spirit was to whenever the masters\" of the experience, that is an extent or their spyous and need, and the experience and past by its the higher the schopenhauer's with an abstration and the purposed to understand that it is destined and destiny of himself, \n------ temperature: 1.0\nd to understand that it is destined and destiny of himself, fur\nfeshicutawas terding itswhas ourselves which an\n\" intain segret shise them? this opposing for ourselvesl. and as life-doatts?\nwith and light, e spirit, he oppisest, one be does not as the differnes.\n\n\n18\n\n"
]
],
[
[
"\nAs you can see, a low temperature results in extremely repetitive and predictable text, but where local structure is highly realistic: in \nparticular, all words (a word being a local pattern of characters) are real English words. With higher temperatures, the generated text \nbecomes more interesting, surprising, even creative; it may sometimes invent completely new words that sound somewhat plausible (such as \n\"eterned\" or \"troveration\"). With a high temperature, the local structure starts breaking down and most words look like semi-random strings \nof characters. Without a doubt, here 0.5 is the most interesting temperature for text generation in this specific setup. Always experiment \nwith multiple sampling strategies! A clever balance between learned structure and randomness is what makes generation interesting.\n\nNote that by training a bigger model, longer, on more data, you can achieve generated samples that will look much more coherent and \nrealistic than ours. But of course, don't expect to ever generate any meaningful text, other than by random chance: all we are doing is \nsampling data from a statistical model of which characters come after which characters. Language is a communication channel, and there is \na distinction between what communications are about, and the statistical structure of the messages in which communications are encoded. To \nevidence this distinction, here is a thought experiment: what if human language did a better job at compressing communications, much like \nour computers do with most of our digital communications? Then language would be no less meaningful, yet it would lack any intrinsic \nstatistical structure, thus making it impossible to learn a language model like we just did.\n\n\n## Take aways\n\n* We can generate discrete sequence data by training a model to predict the next tokens(s) given previous tokens.\n* In the case of text, such a model is called a \"language model\" and could be based on either words or characters.\n* Sampling the next token requires balance between adhering to what the model judges likely, and introducing randomness.\n* One way to handle this is the notion of _softmax temperature_. Always experiment with different temperatures to find the \"right\" one.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d086969e5d062dd40c5ad948728bdbc0adab0f92 | 9,987 | ipynb | Jupyter Notebook | Swarm Intelligence Bot/Swarm Intelligence agent.ipynb | aanshul22/ConnectX_bot-Kaggle | 21b780b54ed44bd1738d0cca0879790ed92c49a5 | [
"MIT"
] | null | null | null | Swarm Intelligence Bot/Swarm Intelligence agent.ipynb | aanshul22/ConnectX_bot-Kaggle | 21b780b54ed44bd1738d0cca0879790ed92c49a5 | [
"MIT"
] | null | null | null | Swarm Intelligence Bot/Swarm Intelligence agent.ipynb | aanshul22/ConnectX_bot-Kaggle | 21b780b54ed44bd1738d0cca0879790ed92c49a5 | [
"MIT"
] | null | null | null | 39.948 | 104 | 0.421248 | [
[
[
"## Swarm intelligence agent\n\nLast checked score: 1062.9",
"_____no_output_____"
]
],
[
[
"def swarm(obs, conf):\n def send_scout_carrier(x, y):\n \"\"\" send scout carrier to explore current cell and, if possible, cell above \"\"\"\n points = send_scouts(x, y)\n # if cell above exists\n if y > 0:\n cell_above_points = send_scouts(x, y - 1)\n # cell above points have lower priority\n if points < m1 and points < (cell_above_points - 1):\n # current cell's points will be negative\n points -= cell_above_points\n return points\n \n def send_scouts(x, y):\n \"\"\" send scouts to get points from all axes of the cell \"\"\"\n axes = explore_axes(x, y)\n points = combine_points(axes)\n return points\n \n def explore_axes(x, y):\n \"\"\"\n find points, marks, zeros and amount of in_air cells of all axes of the cell,\n \"NE\" = North-East etc.\n \"\"\"\n return {\n \"NE -> SW\": [\n explore_direction(x, lambda z : z + 1, y, lambda z : z - 1),\n explore_direction(x, lambda z : z - 1, y, lambda z : z + 1)\n ],\n \"E -> W\": [\n explore_direction(x, lambda z : z + 1, y, lambda z : z),\n explore_direction(x, lambda z : z - 1, y, lambda z : z)\n ],\n \"SE -> NW\": [\n explore_direction(x, lambda z : z + 1, y, lambda z : z + 1),\n explore_direction(x, lambda z : z - 1, y, lambda z : z - 1)\n ],\n \"S -> N\": [\n explore_direction(x, lambda z : z, y, lambda z : z + 1),\n explore_direction(x, lambda z : z, y, lambda z : z - 1)\n ]\n }\n \n def explore_direction(x, x_fun, y, y_fun):\n \"\"\" get points, mark, zeros and amount of in_air cells of this direction \"\"\"\n # consider only opponents mark\n mark = 0\n points = 0\n zeros = 0\n in_air = 0\n for i in range(one_mark_to_win):\n x = x_fun(x)\n y = y_fun(y)\n # if board[x][y] is inside board's borders\n if y >= 0 and y < conf.rows and x >= 0 and x < conf.columns:\n # mark of the direction will be the mark of the first non-empty cell\n if mark == 0 and board[x][y] != 0:\n mark = board[x][y]\n # if board[x][y] is empty\n if board[x][y] == 0:\n zeros += 1\n if (y + 1) < conf.rows and board[x][y + 1] == 0:\n in_air += 1\n elif board[x][y] == mark:\n points += 1\n # stop searching for marks in this direction\n else:\n break\n return {\n \"mark\": mark,\n \"points\": points,\n \"zeros\": zeros,\n \"in_air\": in_air\n }\n \n def combine_points(axes):\n \"\"\" combine points of different axes \"\"\"\n points = 0\n # loop through all axes\n for axis in axes:\n # if mark in both directions of the axis is the same\n # or mark is zero in one or both directions of the axis\n if (axes[axis][0][\"mark\"] == axes[axis][1][\"mark\"]\n or axes[axis][0][\"mark\"] == 0 or axes[axis][1][\"mark\"] == 0):\n # combine points of the same axis\n points += evaluate_amount_of_points(\n axes[axis][0][\"points\"] + axes[axis][1][\"points\"],\n axes[axis][0][\"zeros\"] + axes[axis][1][\"zeros\"],\n axes[axis][0][\"in_air\"] + axes[axis][1][\"in_air\"],\n m1,\n m2,\n axes[axis][0][\"mark\"]\n )\n else:\n # if marks in directions of the axis are different and none of those marks is 0\n for direction in axes[axis]:\n points += evaluate_amount_of_points(\n direction[\"points\"],\n direction[\"zeros\"],\n direction[\"in_air\"],\n m1,\n m2,\n direction[\"mark\"]\n )\n return points\n \n def evaluate_amount_of_points(points, zeros, in_air, m1, m2, mark):\n \"\"\" evaluate amount of points in one direction or entire axis \"\"\"\n # if points + zeros in one direction or entire axis >= one_mark_to_win\n # multiply amount of points by one of the multipliers or keep amount of points as it is\n if (points + zeros) >= one_mark_to_win:\n if points >= one_mark_to_win:\n points *= m1\n elif points == two_marks_to_win:\n points = points * m2 + zeros - in_air\n else:\n points = points + zeros - in_air\n else:\n points = 0\n return points\n\n\n #################################################################################\n # one_mark_to_win points multiplier\n m1 = 100\n # two_marks_to_win points multiplier\n m2 = 10\n # define swarm's mark\n swarm_mark = obs.mark\n # define opponent's mark\n opp_mark = 2 if swarm_mark == 1 else 1\n # define one mark to victory\n one_mark_to_win = conf.inarow - 1\n # define two marks to victory\n two_marks_to_win = conf.inarow - 2\n # define board as two dimensional array\n board = []\n for column in range(conf.columns):\n board.append([])\n for row in range(conf.rows):\n board[column].append(obs.board[conf.columns * row + column])\n # define board center\n board_center = conf.columns // 2\n # start searching for the_column from board center\n x = board_center\n # shift to left/right from board center\n shift = 0\n # THE COLUMN !!!\n the_column = {\n \"x\": x,\n \"points\": float(\"-inf\")\n }\n \n # searching for the_column\n while x >= 0 and x < conf.columns:\n # find first empty cell starting from bottom of the column\n y = conf.rows - 1\n while y >= 0 and board[x][y] != 0:\n y -= 1\n # if column is not full\n if y >= 0:\n # send scout carrier to get points\n points = send_scout_carrier(x, y)\n # evaluate which column is THE COLUMN !!!\n if points > the_column[\"points\"]:\n the_column[\"x\"] = x\n the_column[\"points\"] = points\n # shift x to right or left from swarm center\n shift *= -1\n if shift >= 0:\n shift += 1\n x = board_center + shift\n \n # Swarm's final decision :)\n return the_column[\"x\"]",
"_____no_output_____"
]
],
[
[
"#### Converting the agent into a python file so that it can be submitted",
"_____no_output_____"
]
],
[
[
"import inspect\nimport os\n\ndef write_agent_to_file(function, file):\n with open(file, \"a\" if os.path.exists(file) else \"w\") as f:\n f.write(inspect.getsource(function))\n print(function, \"written to\", file)\n\nwrite_agent_to_file(swarm, os.getcwd() + \"\\\\submission.py\")",
"<function swarm at 0x0000024ACFC97E58> written to D:\\Notebooks\\submission.py\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0869f96206b2f1e80ed61c9e62f274967600535 | 2,877 | ipynb | Jupyter Notebook | Libs/ModulesManagement.ipynb | vertechcon/jupyter | 7fa26c1b8dfe638900ecf5d4c06fd66bd8bf3235 | [
"Apache-2.0"
] | null | null | null | Libs/ModulesManagement.ipynb | vertechcon/jupyter | 7fa26c1b8dfe638900ecf5d4c06fd66bd8bf3235 | [
"Apache-2.0"
] | 1 | 2020-07-15T01:53:28.000Z | 2020-07-15T01:53:28.000Z | Libs/ModulesManagement.ipynb | vertechcon/jupyter | 7fa26c1b8dfe638900ecf5d4c06fd66bd8bf3235 | [
"Apache-2.0"
] | null | null | null | 24.176471 | 117 | 0.530066 | [
[
[
"# Module dependency installation functions",
"_____no_output_____"
],
[
"The function moduleExists accepts a regex\nmoduleExists(r\"minio.*\") ",
"_____no_output_____"
]
],
[
[
"import re\nimport pkg_resources\nimport sys\n\ndef moduleExists (moduleFilter: str) -> bool:\n installed_packages = pkg_resources.working_set\n installed_packages_list = sorted([\"%s==%s\" % (i.key, i.version)\n for i in installed_packages])\n installed_packages_list = list(filter(lambda str: re.match(moduleFilter, str), installed_packages_list))\n if installed_packages_list and len(installed_packages_list) > 0:\n print(\"Modules found.\")\n print(installed_packages_list)\n return True\n return False",
"_____no_output_____"
]
],
[
[
"The function moduleExists takes both the moduleName and regex\nmoduleExists(\"minio\", r\"minio.*\") ",
"_____no_output_____"
]
],
[
[
"def ensureInstalled (moduleName: str, flt: str):\n if not moduleExists(flt):\n !{sys.executable} -m pip install {moduleName} \n print(\"Module installed.\")\n else:\n print(\"Module already installed.\")\n \ndef ensureInstalled_noDeps (moduleName: str, flt: str):\n if not moduleExists(flt):\n !{sys.executable} -m pip install {moduleName} --no-deps \n print(\"Module installed.\")\n else:\n print(\"Module already installed.\")",
"_____no_output_____"
],
[
"#Tests\n#ensureInstalled(\"minio\", r\"minio.*\")",
"Modules found.\n['minio==5.0.10']\nModule already installed.\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d086acc651099f86c33c0952d0a725dbd9ddc5f0 | 20,927 | ipynb | Jupyter Notebook | notes/know-lstms-better.ipynb | pegasus-lynx/rtg | d4cfb557e8532de432f494b28f3ed160ff2f702f | [
"Apache-2.0"
] | 15 | 2019-06-28T21:22:46.000Z | 2022-02-03T06:36:43.000Z | notes/know-lstms-better.ipynb | pegasus-lynx/rtg | d4cfb557e8532de432f494b28f3ed160ff2f702f | [
"Apache-2.0"
] | 23 | 2019-06-14T19:12:26.000Z | 2022-03-15T23:22:14.000Z | notes/know-lstms-better.ipynb | pegasus-lynx/rtg | d4cfb557e8532de432f494b28f3ed160ff2f702f | [
"Apache-2.0"
] | 8 | 2019-06-11T19:03:39.000Z | 2022-01-09T06:58:23.000Z | 30.417151 | 141 | 0.46041 | [
[
[
"# Getting to know LSTMs better\n\nCreated: September 13, 2018 \nAuthor: Thamme Gowda \n\n\nGoals:\n- To get batches of *unequal length sequences* encoded correctly!\n- Know how the hidden states flow between encoders and decoders\n- Know how the multiple stacked LSTM layers pass hidden states\n\nExample: a simple bi-directional LSTM which takes 3d input vectors\nand produces 2d output vectors. ",
"_____no_output_____"
]
],
[
[
"import torch \nfrom torch import nn",
"_____no_output_____"
],
[
"lstm = nn.LSTM(3, 2, batch_first=True, bidirectional=True)",
"_____no_output_____"
],
[
"# Lets create a batch input.\n# 3 sequences in batch (the first dim) , see batch_first=True\n# Then the logest sequence is 4 time steps, ==> second dimension\n# Each time step has 3d vector which is input ==> last dimension\npad_seq = torch.rand(3, 4, 3)\n\n# That is nice for the theory\n# but in practice we are dealing with un equal length sequences\n# among those 3 sequences in the batch, lets us say \n# first sequence is the longest, with 4 time steps --> no padding needed\n# second seq is 3 time steps --> pad the last time step\npad_seq[1, 3, :] = 0.0\n# third seq is 2 time steps --> pad the last two steps\npad_seq[2, 2:, :] = 0.0\nprint(\"Padded Input:\")\nprint(pad_seq)\n\n# so we got these lengths\nlens = [4,3,2]\n\nprint(\"Sequence Lenghts: \", lens)",
"Padded Input:\ntensor([[[0.7850, 0.6658, 0.7522],\n [0.3855, 0.7981, 0.6199],\n [0.9081, 0.6357, 0.3619],\n [0.2481, 0.5198, 0.2635]],\n\n [[0.2654, 0.9904, 0.3050],\n [0.1671, 0.1709, 0.2392],\n [0.0705, 0.4811, 0.3636],\n [0.0000, 0.0000, 0.0000]],\n\n [[0.6474, 0.5172, 0.0308],\n [0.5782, 0.3083, 0.5117],\n [0.0000, 0.0000, 0.0000],\n [0.0000, 0.0000, 0.0000]]])\nSequence Lenghts: [4, 3, 2]\n"
],
[
"# lets send padded seq to LSTM\nout,(h_t, c_t) = lstm(pad_seq)\nprint(\"All Outputs:\")\nprint(out)",
"All Outputs:\ntensor([[[ 0.0428, -0.3015, 0.0359, 0.0557],\n [ 0.0919, -0.4145, 0.0278, 0.0480],\n [ 0.0768, -0.4989, 0.0203, 0.0674],\n [ 0.1019, -0.4925, -0.0177, 0.0224]],\n\n [[ 0.0587, -0.3025, 0.0017, 0.0201],\n [ 0.0537, -0.3388, -0.0532, 0.0111],\n [ 0.0839, -0.3811, -0.0446, -0.0020],\n [ 0.0595, -0.3681, -0.0720, 0.0218]],\n\n [[ 0.0147, -0.2585, -0.0093, 0.0756],\n [ 0.0398, -0.3531, -0.0174, 0.0369],\n [ 0.0458, -0.3476, -0.0912, 0.0243],\n [ 0.0422, -0.3360, -0.0720, 0.0218]]], grad_fn=<TransposeBackward0>)\n"
]
],
[
[
"^^ Output is 2x2d=4d vector since it is bidirectional \nforward 2d, backward 2d are concatenated \nTotal vectors=12: 3 seqs in batch x 4 time steps;; each vector is 4d \n\n> Hmm, what happened to my padding time steps? Will padded zeros mess with the internal weights of LSTM when I do backprop?\n\n---\nLets look at the last Hidden state",
"_____no_output_____"
]
],
[
[
"print(h_t)",
"tensor([[[ 0.1019, -0.4925],\n [ 0.0595, -0.3681],\n [ 0.0422, -0.3360]],\n\n [[ 0.0359, 0.0557],\n [ 0.0017, 0.0201],\n [-0.0093, 0.0756]]], grad_fn=<ViewBackward>)\n"
]
],
[
[
"Last hidden state is a 2d (same as output) vectors, \nbut 2 for each step because of bidirectional rnn \nThere are 3 of them since there were three seqs in the batch \neach corresponding to the last step \nBut the definition of *last time step* is bit tricky \nFor the left-to-right LSTM, it is the last step of input \nFor the right-to-left LSTM, it is the first step of input \n\nThis makes sense now.\n\n--- \nLets look at $c_t$:",
"_____no_output_____"
]
],
[
[
"print(\"Last c_t:\")\nprint(c_t)",
"Last c_t:\ntensor([[[ 0.3454, -1.0070],\n [ 0.1927, -0.6731],\n [ 0.1361, -0.6063]],\n\n [[ 0.1219, 0.1858],\n [ 0.0049, 0.0720],\n [-0.0336, 0.2787]]], grad_fn=<ViewBackward>)\n"
]
],
[
[
"This should be similar to the last hidden state.\n\n\n## Question: \n> what happened to my padding time steps? Did the last hidden state exclude the padded time steps?\n\nI can see that last hidden state of the forward LSTM didnt distinguish padded zeros. \n\nLets see output of each time steps and last hidden state of left-to-right LSTM, again. \nWe know that the lengths (after removing padding) are \\[4,3,2] ",
"_____no_output_____"
]
],
[
[
"print(\"All time stamp outputs:\")\nprint(out[:, :, :2])\nprint(\"Last hidden state (forward LSTM):\")\nprint(h_t[0])",
"All time stamp outputs:\ntensor([[[ 0.0428, -0.3015],\n [ 0.0919, -0.4145],\n [ 0.0768, -0.4989],\n [ 0.1019, -0.4925]],\n\n [[ 0.0587, -0.3025],\n [ 0.0537, -0.3388],\n [ 0.0839, -0.3811],\n [ 0.0595, -0.3681]],\n\n [[ 0.0147, -0.2585],\n [ 0.0398, -0.3531],\n [ 0.0458, -0.3476],\n [ 0.0422, -0.3360]]], grad_fn=<SliceBackward>)\nLast hidden state (forward LSTM):\ntensor([[ 0.1019, -0.4925],\n [ 0.0595, -0.3681],\n [ 0.0422, -0.3360]], grad_fn=<SelectBackward>)\n"
]
],
[
[
"*Okay, Now I get it.* \nWhen building sequence to sequence (for Machine translation) I cant pass last hidden state like this to a decoder.\n\nWe have to inform the LSTM about lengths.\n\nHow? \n\nThats why we have `torch.nn.utils.rnn.pack_padded_sequence`",
"_____no_output_____"
]
],
[
[
"print(\"Padded Seqs:\")\nprint(pad_seq)\nprint(\"Lens:\", lens)\n\nprint(\"Pack Padded Seqs:\")\npac_pad_seq = torch.nn.utils.rnn.pack_padded_sequence(pad_seq, lens, batch_first=True)\nprint(pac_pad_seq)",
"Padded Seqs:\ntensor([[[0.7850, 0.6658, 0.7522],\n [0.3855, 0.7981, 0.6199],\n [0.9081, 0.6357, 0.3619],\n [0.2481, 0.5198, 0.2635]],\n\n [[0.2654, 0.9904, 0.3050],\n [0.1671, 0.1709, 0.2392],\n [0.0705, 0.4811, 0.3636],\n [0.0000, 0.0000, 0.0000]],\n\n [[0.6474, 0.5172, 0.0308],\n [0.5782, 0.3083, 0.5117],\n [0.0000, 0.0000, 0.0000],\n [0.0000, 0.0000, 0.0000]]])\nLens: [4, 3, 2]\nPack Padded Seqs:\nPackedSequence(data=tensor([[0.7850, 0.6658, 0.7522],\n [0.2654, 0.9904, 0.3050],\n [0.6474, 0.5172, 0.0308],\n [0.3855, 0.7981, 0.6199],\n [0.1671, 0.1709, 0.2392],\n [0.5782, 0.3083, 0.5117],\n [0.9081, 0.6357, 0.3619],\n [0.0705, 0.4811, 0.3636],\n [0.2481, 0.5198, 0.2635]]), batch_sizes=tensor([3, 3, 2, 1]))\n"
]
],
[
[
"Okay, this is doing some magic -- getting rid of all padded zeros -- Cool!\n`batch_sizes=tensor([3, 3, 2, 1]` seems to be the main ingredient of this magic.\n\n`[3, 3, 2, 1]` I get it!\nWe have 4 time steps in batch. \n- First two step has all 3 seqs in the batch. \n- third step is made of first 2 seqs in batch. \n- Fourth step is made of first seq in batch\n\nI now understand why the sequences in the batch has to be sorted by descending order of lengths!\n\nNow let us send it to LSTM and see what it produces",
"_____no_output_____"
]
],
[
[
"pac_pad_out, (pac_ht, pac_ct) = lstm(pac_pad_seq)\n# Lets first look at output. this is packed output\nprint(pac_pad_out)",
"PackedSequence(data=tensor([[ 0.0428, -0.3015, 0.0359, 0.0557],\n [ 0.0587, -0.3025, 0.0026, 0.0203],\n [ 0.0147, -0.2585, -0.0057, 0.0754],\n [ 0.0919, -0.4145, 0.0278, 0.0480],\n [ 0.0537, -0.3388, -0.0491, 0.0110],\n [ 0.0398, -0.3531, -0.0005, 0.0337],\n [ 0.0768, -0.4989, 0.0203, 0.0674],\n [ 0.0839, -0.3811, -0.0262, -0.0056],\n [ 0.1019, -0.4925, -0.0177, 0.0224]], grad_fn=<CatBackward>), batch_sizes=tensor([3, 3, 2, 1]))\n"
]
],
[
[
"Okay this is packed output. Sequences are of unequal lengths.\nNow we need to restore the output by padding 0s for shorter sequences. ",
"_____no_output_____"
]
],
[
[
"pad_out = nn.utils.rnn.pad_packed_sequence(pac_pad_out, batch_first=True, padding_value=0)\nprint(pad_out)",
"(tensor([[[ 0.0428, -0.3015, 0.0359, 0.0557],\n [ 0.0919, -0.4145, 0.0278, 0.0480],\n [ 0.0768, -0.4989, 0.0203, 0.0674],\n [ 0.1019, -0.4925, -0.0177, 0.0224]],\n\n [[ 0.0587, -0.3025, 0.0026, 0.0203],\n [ 0.0537, -0.3388, -0.0491, 0.0110],\n [ 0.0839, -0.3811, -0.0262, -0.0056],\n [ 0.0000, 0.0000, 0.0000, 0.0000]],\n\n [[ 0.0147, -0.2585, -0.0057, 0.0754],\n [ 0.0398, -0.3531, -0.0005, 0.0337],\n [ 0.0000, 0.0000, 0.0000, 0.0000],\n [ 0.0000, 0.0000, 0.0000, 0.0000]]], grad_fn=<TransposeBackward0>), tensor([4, 3, 2]))\n"
]
],
[
[
"Output looks good! Now Let us look at the hidden state. ",
"_____no_output_____"
]
],
[
[
"print(pac_ht)",
"tensor([[[ 0.1019, -0.4925],\n [ 0.0839, -0.3811],\n [ 0.0398, -0.3531]],\n\n [[ 0.0359, 0.0557],\n [ 0.0026, 0.0203],\n [-0.0057, 0.0754]]], grad_fn=<ViewBackward>)\n"
]
],
[
[
"This is great. As we see the forward (or Left-to-right) LSTM's last hidden state is proper as per the lengths. So should be the c_t.\n\nLet us concatenate forward and reverse LSTM's hidden states",
"_____no_output_____"
]
],
[
[
"torch.cat([pac_ht[0],pac_ht[1]], dim=1) ",
"_____no_output_____"
]
],
[
[
"----\n\n# Multi Layer LSTM\n\nLet us redo the above hacking to understand how 2 layer LSTM works",
"_____no_output_____"
]
],
[
[
"n_layers = 2\ninp_size = 3\nout_size = 2\nlstm2 = nn.LSTM(inp_size, out_size, num_layers=n_layers, batch_first=True, bidirectional=True)",
"_____no_output_____"
],
[
"pac_out, (h_n, c_n) = lstm2(pac_pad_seq)\nprint(\"Packed Output:\")\nprint(pac_out)\npad_out = nn.utils.rnn.pad_packed_sequence(pac_out, batch_first=True, padding_value=0)\nprint(\"Pad Output:\")\nprint(pad_out)\n\n\nprint(\"Last h_n:\")\nprint(h_n)\n\nprint(\"Last c_n:\")\nprint(c_n)",
"Packed Output:\nPackedSequence(data=tensor([[ 0.2443, 0.0703, -0.0871, -0.0664],\n [ 0.2496, 0.0677, -0.0658, -0.0605],\n [ 0.2419, 0.0687, -0.0701, -0.0521],\n [ 0.3354, 0.0964, -0.0772, -0.0613],\n [ 0.3272, 0.0975, -0.0655, -0.0534],\n [ 0.3216, 0.1055, -0.0504, -0.0353],\n [ 0.3644, 0.1065, -0.0752, -0.0531],\n [ 0.3583, 0.1116, -0.0418, -0.0350],\n [ 0.3760, 0.1139, -0.0438, -0.0351]], grad_fn=<CatBackward>), batch_sizes=tensor([3, 3, 2, 1]))\nPad Output:\n(tensor([[[ 0.2443, 0.0703, -0.0871, -0.0664],\n [ 0.3354, 0.0964, -0.0772, -0.0613],\n [ 0.3644, 0.1065, -0.0752, -0.0531],\n [ 0.3760, 0.1139, -0.0438, -0.0351]],\n\n [[ 0.2496, 0.0677, -0.0658, -0.0605],\n [ 0.3272, 0.0975, -0.0655, -0.0534],\n [ 0.3583, 0.1116, -0.0418, -0.0350],\n [ 0.0000, 0.0000, 0.0000, 0.0000]],\n\n [[ 0.2419, 0.0687, -0.0701, -0.0521],\n [ 0.3216, 0.1055, -0.0504, -0.0353],\n [ 0.0000, 0.0000, 0.0000, 0.0000],\n [ 0.0000, 0.0000, 0.0000, 0.0000]]], grad_fn=<TransposeBackward0>), tensor([4, 3, 2]))\nLast h_n:\ntensor([[[ 0.2190, 0.2067],\n [ 0.1868, 0.2188],\n [ 0.1706, 0.2347]],\n\n [[-0.5062, 0.1701],\n [-0.4130, 0.2190],\n [-0.4228, 0.1733]],\n\n [[ 0.3760, 0.1139],\n [ 0.3583, 0.1116],\n [ 0.3216, 0.1055]],\n\n [[-0.0871, -0.0664],\n [-0.0658, -0.0605],\n [-0.0701, -0.0521]]], grad_fn=<ViewBackward>)\nLast c_n:\ntensor([[[ 0.5656, 0.3145],\n [ 0.4853, 0.3633],\n [ 0.4255, 0.3718]],\n\n [[-0.9779, 0.6461],\n [-0.8578, 0.7013],\n [-0.6978, 0.5322]],\n\n [[ 1.0754, 0.4258],\n [ 1.0021, 0.4184],\n [ 0.8623, 0.3839]],\n\n [[-0.1535, -0.2073],\n [-0.1187, -0.1912],\n [-0.1211, -0.1589]]], grad_fn=<ViewBackward>)\n"
]
],
[
[
"The LSTM output looks similar to single layer LSTM.\n\nHowever the ht and ct states are bigger -- since there are two layers. \nNow its time to RTFM. \n\n\n> h_n of shape `(num_layers * num_directions, batch, hidden_size)`: tensor containing the hidden state for `t = seq_len`.\nLike output, the layers can be separated using `h_n.view(num_layers, num_directions, batch, hidden_size)` and similarly for c_n.",
"_____no_output_____"
]
],
[
[
"batch_size = 3\nnum_dirs = 2\nl_n_h_n = h_n.view(n_layers, num_dirs, batch_size, out_size)[-1]\n# last layer last time step hidden state\nprint(l_n_h_n)",
"tensor([[[ 0.3760, 0.1139],\n [ 0.3583, 0.1116],\n [ 0.3216, 0.1055]],\n\n [[-0.0871, -0.0664],\n [-0.0658, -0.0605],\n [-0.0701, -0.0521]]], grad_fn=<SelectBackward>)\n"
],
[
"last_hid = torch.cat([l_n_h_n[0], l_n_h_n[1]], dim=1)\n\nprint(\"last layer last time stamp hidden state\")\nprint(last_hid)\n\nprint(\"Padded Outputs :\")\nprint(pad_out)",
"last layer last time stamp hidden state\ntensor([[ 0.3760, 0.1139, -0.0871, -0.0664],\n [ 0.3583, 0.1116, -0.0658, -0.0605],\n [ 0.3216, 0.1055, -0.0701, -0.0521]], grad_fn=<CatBackward>)\nPadded Outputs :\n(tensor([[[ 0.2443, 0.0703, -0.0871, -0.0664],\n [ 0.3354, 0.0964, -0.0772, -0.0613],\n [ 0.3644, 0.1065, -0.0752, -0.0531],\n [ 0.3760, 0.1139, -0.0438, -0.0351]],\n\n [[ 0.2496, 0.0677, -0.0658, -0.0605],\n [ 0.3272, 0.0975, -0.0655, -0.0534],\n [ 0.3583, 0.1116, -0.0418, -0.0350],\n [ 0.0000, 0.0000, 0.0000, 0.0000]],\n\n [[ 0.2419, 0.0687, -0.0701, -0.0521],\n [ 0.3216, 0.1055, -0.0504, -0.0353],\n [ 0.0000, 0.0000, 0.0000, 0.0000],\n [ 0.0000, 0.0000, 0.0000, 0.0000]]], grad_fn=<TransposeBackward0>), tensor([4, 3, 2]))\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d086aee66bcdf364a2ac6b14a6219e1ee2a9c2f6 | 52,302 | ipynb | Jupyter Notebook | differential-privacy/differential_privacy.ipynb | gitgik/pytorch | 4063885a2184b69040ef76ccf0c90c62c48e4277 | [
"MIT"
] | 38 | 2019-10-08T03:00:32.000Z | 2022-03-25T02:17:19.000Z | differential-privacy/differential_privacy.ipynb | gitgik/pytorch | 4063885a2184b69040ef76ccf0c90c62c48e4277 | [
"MIT"
] | 1 | 2019-12-04T16:35:03.000Z | 2019-12-04T16:35:03.000Z | differential-privacy/differential_privacy.ipynb | gitgik/pytorch | 4063885a2184b69040ef76ccf0c90c62c48e4277 | [
"MIT"
] | 9 | 2019-10-08T03:00:31.000Z | 2022-01-03T02:33:21.000Z | 32.729662 | 700 | 0.589002 | [
[
[
"## Differential Privacy - Simple Database Queries",
"_____no_output_____"
],
[
"The database is going to be a VERY simple database with only one boolean column. Each row corresponds to a person. Each value corresponds to whether or not that person has a certain private attribute (such as whether they have a certain disease, or whether they are above/below a certain age). We are then going to learn how to know whether a database query over such a small database is differentially private or not - and more importantly - what techniques we can employ to ensure various levels of privacy\n\n#### Create a Simple Database\nTo do this, initialize a random list of 1s and 0s (which are the entries in our database). Note - the number of entries directly corresponds to the number of people in our database.",
"_____no_output_____"
]
],
[
[
"import torch\n# the number of entries in our DB / this of it as number of people in the DB\nnum_entries = 5000\ndb = torch.rand(num_entries) > 0.5\ndb",
"_____no_output_____"
]
],
[
[
"## Generate Parallel Databases\n> \"When querying a database, if I removed someone from the database, would the output of the query change?\". \n\nIn order to check for this, we create \"parallel databases\" which are simply databases with one entry removed. \n\nWe'll create a list of every parallel database to the one currently contained in the \"db\" variable. Then, create a helper function which does the following:\n- creates the initial database (db)\n- creates all parallel databases",
"_____no_output_____"
]
],
[
[
"def create_parallel_db(db, remove_index):\n return torch.cat((db[0:remove_index], db[remove_index+1:]))",
"_____no_output_____"
],
[
"def create_parallel_dbs(db):\n parallel_dbs = list()\n for i in range(len(db)):\n pdb = create_parallel_db(db, i)\n parallel_dbs.append(pdb)\n return parallel_dbs",
"_____no_output_____"
],
[
"def create_db_and_parallels(num_entries):\n # generate dbs and parallel dbs on the fly\n db = torch.rand(num_entries) > 0.5\n pdbs = create_parallel_dbs(db)\n \n return db, pdbs",
"_____no_output_____"
],
[
"db, pdbs = create_db_and_parallels(10)\npdbs\nprint(\"Real database:\", db)\nprint(\"Size of real DB\", db.size())\nprint(\"A sample parallel DB\", pdbs[0])\nprint(\"Size of parallel DB\", pdbs[0].size())",
"Real database: tensor([1, 1, 1, 0, 0, 0, 0, 0, 0, 0], dtype=torch.uint8)\nSize of real DB torch.Size([10])\nA sample parallel DB tensor([1, 1, 0, 0, 0, 0, 0, 0, 0], dtype=torch.uint8)\nSize of parallel DB torch.Size([9])\n"
]
],
[
[
"# Towards Evaluating The Differential Privacy of a Function\n\nIntuitively, we want to be able to query our database and evaluate whether or not the result of the query is leaking \"private\" information. \n> This is about evaluating whether the output of a query changes when we remove someone from the database. Specifically, we want to evaluate the *maximum* amount the query changes when someone is removed (maximum over all possible people who could be removed). \n\nTo find how much privacy is leaked, we'll iterate over each person in the database and **measure** the difference in the output of the query relative to when we query the entire database. \n\nJust for the sake of argument, let's make our first \"database query\" a simple sum. Aka, we're going to count the number of 1s in the database.",
"_____no_output_____"
]
],
[
[
"db, pdbs = create_db_and_parallels(200)\ndef query(db):\n return db.sum()",
"_____no_output_____"
],
[
"query(db)",
"_____no_output_____"
],
[
"# the output of the parallel dbs is different from the db query\nquery(pdbs[1])\n",
"_____no_output_____"
],
[
"full_db_result = query(db)\nprint(full_db_result)",
"tensor(106)\n"
],
[
"sensitivity = 0\nsensitivity_scale = []\nfor pdb in pdbs:\n pdb_result = query(pdb)\n db_distance = torch.abs(pdb_result - full_db_result)\n if(db_distance > sensitivity):\n sensitivity_scale.append(db_distance)\n sensitivity = db_distance",
"_____no_output_____"
],
[
"sensitivity",
"_____no_output_____"
]
],
[
[
"#### Sensitivity\n> The maximum amount the query changes when removing an individual from the DB.\n",
"_____no_output_____"
],
[
"# Evaluating the Privacy of a Function\n\nThe difference between each parallel db's query result and the query result for the real database and its max value (which was 1) is called \"sensitivity\". It corresponds to the function we chose for the query. The \"sum\" query will always have a sensitivity of exactly 1. We can also calculate sensitivity for other functions as well.\n\nLet's calculate sensitivity for the \"mean\" function.",
"_____no_output_____"
]
],
[
[
"def sensitivity(query, num_entries=1000):\n db, pdbs = create_db_and_parallels(num_entries)\n \n full_db_result = query(db)\n \n max_distance = 0\n for pdb in pdbs:\n # for each parallel db, execute the query (sum, or mean, ..., etc)\n pdb_result = query(pdb)\n db_distance = torch.abs(pdb_result - full_db_result)\n \n if (db_distance > max_distance):\n max_distance = db_distance\n\n return max_distance ",
"_____no_output_____"
],
[
"# our query is now the mean\ndef query(db):\n return db.float().mean()",
"_____no_output_____"
],
[
"\nsensitivity(query)",
"_____no_output_____"
]
],
[
[
"Wow! That sensitivity is WAY lower. Note the intuition here. \n>\"Sensitivity\" is measuring how sensitive the output of the query is to a person being removed from the database. \n\nFor a simple sum, this is always 1, but for the mean, removing a person is going to change the result of the query by rougly 1 divided by the size of the database. Thus, \"mean\" is a VASTLY less \"sensitive\" function (query) than SUM.",
"_____no_output_____"
],
[
"# Calculating L1 Sensitivity For Threshold\n\nTO calculate the sensitivty for the \"threshold\" function: \n\n- First compute the sum over the database (i.e. sum(db)) and return whether that sum is greater than a certain threshold.\n- Then, create databases of size 10 and threshold of 5 and calculate the sensitivity of the function. \n- Finally, re-initialize the database 10 times and calculate the sensitivity each time.",
"_____no_output_____"
]
],
[
[
"def query(db, threshold=5):\n \"\"\"\n Query that adds a threshold of 5, and returns whether sum is > threshold or not.\n \"\"\"\n return (db.sum() > threshold).float()",
"_____no_output_____"
],
[
"for i in range(10):\n sens = sensitivity(query, num_entries=10)\n print(sens)",
"0\ntensor(1.)\n0\n0\n0\n0\n0\n0\n0\ntensor(1.)\n"
]
],
[
[
"# A Basic Differencing Attack\n\nSadly none of the functions we've looked at so far are differentially private (despite them having varying levels of sensitivity). The most basic type of attack can be done as follows.\n\nLet's say we wanted to figure out a specific person's value in the database. All we would have to do is query for the sum of the entire database and then the sum of the entire database without that person!\n\n## Performing a Differencing Attack on Row 10 (How privacy can fail)\n\nWe'll construct a database and then demonstrate how one can use two different sum queries to explose the value of the person represented by row 10 in the database (note, you'll need to use a database with at least 10 rows)",
"_____no_output_____"
]
],
[
[
"db, _ = create_db_and_parallels(100)",
"_____no_output_____"
],
[
"db",
"_____no_output_____"
],
[
"# create a parallel db with that person (index 10) removed\npdb = create_parallel_db(db, remove_index=10)",
"_____no_output_____"
],
[
"pdb",
"_____no_output_____"
],
[
"# differencing attack using sum query\nsum(db) - sum(pdb)",
"_____no_output_____"
],
[
"# a differencing attack using mean query\nsum(db).float() /len(db) - sum(pdb).float() / len(pdb)",
"_____no_output_____"
],
[
"# differencing using a threshold\n(sum(db).float() > 50) - (sum(pdb).float() > 50)",
"_____no_output_____"
]
],
[
[
"# Local Differential Privacy\n\nDifferential privacy always requires a form of randommess or noise added to the query to protect from things like Differencing Attacks.\nTo explain this, let's look at Randomized Response.\n\n### Randomized Response (Local Differential Privacy)\n\nLet's say I have a group of people I wish to survey about a very taboo behavior which I think they will lie about (say, I want to know if they have ever committed a certain kind of crime). I'm not a policeman, I'm just trying to collect statistics to understand the higher level trend in society. So, how do we do this? One technique is to add randomness to each person's response by giving each person the following instructions (assuming I'm asking a simple yes/no question):\n\n- Flip a coin 2 times.\n- If the first coin flip is heads, answer honestly\n- If the first coin flip is tails, answer according to the second coin flip (heads for yes, tails for no)!\n\nThus, each person is now protected with \"plausible deniability\". If they answer \"Yes\" to the question \"have you committed X crime?\", then it might becasue they actually did, or it might be because they are answering according to a random coin flip. Each person has a high degree of protection. Furthermore, we can recover the underlying statistics with some accuracy, as the \"true statistics\" are simply averaged with a 50% probability. Thus, if we collect a bunch of samples and it turns out that 60% of people answer yes, then we know that the TRUE distribution is actually centered around 70%, because 70% averaged with a 50% (a coin flip) is 60% which is the result we obtained. \n\nHowever, it should be noted that, especially when we only have a few samples, this comes at the cost of accuracy. This tradeoff exists across all of Differential Privacy. \n\n> NOTE: **The greater the privacy protection (plausible deniability) the less accurate the results. **\n\nLet's implement this local DP for our database before!\n\nThe main goal is to: \n* Get the most accurate query with the **greatest** amount of privacy\n* Greatest fit with trust models in the actual world, (don't waste trust)\n\nLet's implement local differential privacy:",
"_____no_output_____"
]
],
[
[
"db, pdbs = create_db_and_parallels(100)\ndb",
"_____no_output_____"
],
[
"def query(db):\n true_result = torch.mean(db.float())\n \n # local differential privacy is adding noise to data: replacing some \n # of the values with random values\n first_coin_flip = (torch.rand(len(db)) > 0.5).float()\n second_coin_flip = (torch.rand(len(db)) > 0.5).float()\n \n # differentially private DB ... \n augmented_db = db.float() * first_coin_flip + (1 - first_coin_flip) * second_coin_flip\n \n # the result is skewed if we do:\n # torch.mean(augmented_db.float())\n # we remove the skewed average that was the result of the differential privacy\n dp_result = torch.mean(augmented_db.float()) * 2 - 0.5\n \n return dp_result, true_result",
"_____no_output_____"
],
[
"db, pdbs = create_db_and_parallels(10)\nprivate_result, true_result = query(db)\nprint(f\"Without noise {private_result}\")\nprint(f\"With noise: {true_result}\")",
"Without noise 0.7000000476837158\nWith noise: 0.30000001192092896\n"
],
[
"# Increasing the size of the dateset\ndb, pdbs = create_db_and_parallels(100)\nprivate_result, true_result = query(db)\nprint(f\"Without noise {private_result}\")\nprint(f\"With noise: {true_result}\")",
"Without noise 0.42000001668930054\nWith noise: 0.4699999988079071\n"
],
[
"# Increasing the size of the dateset even further\ndb, pdbs = create_db_and_parallels(1000)\nprivate_result, true_result = query(db)\nprint(f\"Without noise {private_result}\")\nprint(f\"With noise: {true_result}\")",
"Without noise 0.5099999904632568\nWith noise: 0.5210000276565552\n"
]
],
[
[
"As we have seen,\n> The more data we have the more the noise will tend to not affect the output of the query",
"_____no_output_____"
],
[
"# Varying Amounts of Noise\n\nWe are going to augment the randomized response query to allow for varying amounts of randomness to be added. To do this, we bias the coin flip to be higher or lower and then run the same experiment. \n\nWe'll need to both adjust the likelihood of the first coin flip AND the de-skewing at the end (where we create the \"augmented_result\" variable).\n",
"_____no_output_____"
]
],
[
[
"# Noise < 0.5 sets the likelihood that the coin flip will be heads, and vice-versa.\nnoise = 0.2\n\ntrue_result = torch.mean(db.float())\n# let's add the noise to data: replacing some of the values with random values\nfirst_coin_flip = (torch.rand(len(db)) > noise).float()\nsecond_coin_flip = (torch.rand(len(db)) > 0.5).float()\n\n# differentially private DB ... \naugmented_db = db.float() * first_coin_flip + (1 - first_coin_flip) * second_coin_flip\n\n# since the result will be skewed if we do: torch.mean(augmented_db.float())\n# we'll remove the skewed average above by doing below:\ndp_result = torch.mean(augmented_db.float()) * 2 - 0.5\n\nsk_result = augmented_db.float().mean()\nprint('True result:', true_result)\nprint('Skewed result:', sk_result)\nprint('De-skewed result:', dp_result)",
"True result: tensor(0.5210)\nSkewed result: tensor(0.5140)\nDe-skewed result: tensor(0.5280)\n"
],
[
"def query(db, noise=0.2):\n \"\"\"Default noise(0.2) above sets the likelihood that the coin flip will be heads\"\"\"\n true_result = torch.mean(db.float())\n\n # local diff privacy is adding noise to data: replacing some \n # of the values with random values\n first_coin_flip = (torch.rand(len(db)) > noise).float()\n second_coin_flip = (torch.rand(len(db)) > 0.5).float()\n\n # differentially private DB ... \n augmented_db = db.float() * first_coin_flip + (1 - first_coin_flip) * second_coin_flip\n\n # the result is skewed if we do:\n # torch.mean(augmented_db.float())\n # we remove the skewed average that was the result of the differential privacy\n sk_result = augmented_db.float().mean()\n private_result = ((sk_result / noise ) - 0.5) * noise / (1 - noise)\n\n return private_result, true_result",
"_____no_output_____"
],
[
"# test varying noise\ndb, pdbs = create_db_and_parallels(10)\nprivate_result, true_result = query(db, noise=0.2)\nprint(f\"Without noise {private_result}\")\nprint(f\"With noise: {true_result}\")\n",
"Without noise 0.25\nWith noise: 0.30000001192092896\n"
],
[
"# Increasing the size of the dateset even further\ndb, pdbs = create_db_and_parallels(100)\nprivate_result, true_result = query(db, noise=0.4)\nprint(f\"Without noise {private_result}\")\nprint(f\"With noise: {true_result}\")",
"Without noise 0.7333332300186157\nWith noise: 0.6399999856948853\n"
],
[
"# Increasing the size of the dateset even further\ndb, pdbs = create_db_and_parallels(10000)\nprivate_result, true_result = query(db, noise=0.8)\nprint(f\"Without noise {private_result}\")\nprint(f\"With noise: {true_result}\")\n",
"Without noise 0.5264999866485596\nWith noise: 0.5004000067710876\n"
]
],
[
[
"From the analysis above, with more data, its easier to protect privacy with noise. It becomes a lot easier to learn about general characteristics in the DB because the algorithm has more data points to look at and compare with each other.",
"_____no_output_____"
],
[
"So differential privacy mechanisms has helped us filter out any information unique to individual data entities and try to let through information that is consistent across multiple different people in the dataset. \n> The larger the dataset, the easier it is to protect privacy. ",
"_____no_output_____"
],
[
"# The Formal Definition of Differential Privacy\n\nThe previous method of adding noise was called \"Local Differentail Privacy\" because we added noise to each datapoint individually. This is necessary for some situations wherein the data is SO sensitive that individuals do not trust noise to be added later. However, it comes at a very high cost in terms of accuracy. \n\nHowever, alternatively we can add noise AFTER data has been aggregated by a function. This kind of noise can allow for similar levels of protection with a lower affect on accuracy. However, participants must be able to trust that no-one looked at their datapoints _before_ the aggregation took place. In some situations this works out well, in others (such as an individual hand-surveying a group of people), this is less realistic.\n\nNevertheless, global differential privacy is incredibly important because it allows us to perform differential privacy on smaller groups of individuals with lower amounts of noise. Let's revisit our sum functions.",
"_____no_output_____"
]
],
[
[
"db, pdbs = create_db_and_parallels(100)\n\ndef query(db):\n return torch.sum(db.float())\n\ndef M(db):\n query(db) + noise\n\nquery(db)",
"_____no_output_____"
]
],
[
[
"So the idea here is that we want to add noise to the output of our function. We actually have two different kinds of noise we can add - Laplacian Noise or Gaussian Noise. However, before we do so at this point we need to dive into the formal definition of Differential Privacy.\n\n",
"_____no_output_____"
],
[
"_Image From: \"The Algorithmic Foundations of Differential Privacy\" - Cynthia Dwork and Aaron Roth - https://www.cis.upenn.edu/~aaroth/Papers/privacybook.pdf_",
"_____no_output_____"
],
[
"This definition does not _create_ differential privacy, instead it is a measure of how much privacy is afforded by a query M. Specifically, it's a comparison between running the query M on a database (x) and a parallel database (y). As you remember, parallel databases are defined to be the same as a full database (x) with one entry/person removed.\n\nThus, this definition says that FOR ALL parallel databases, the maximum distance between a query on database (x) and the same query on database (y) will be e^epsilon, but that occasionally this constraint won't hold with probability delta. Thus, this theorem is called \"epsilon delta\" differential privacy.\n\n# Epsilon\n\nLet's unpack the intuition of this for a moment. \n\nEpsilon Zero: If a query satisfied this inequality where epsilon was set to 0, then that would mean that the query for all parallel databases outputed the exact same value as the full database. As you may remember, when we calculated the \"threshold\" function, often the Sensitivity was 0. In that case, the epsilon also happened to be zero.\n\nEpsilon One: If a query satisfied this inequality with epsilon 1, then the maximum distance between all queries would be 1 - or more precisely - the maximum distance between the two random distributions M(x) and M(y) is 1 (because all these queries have some amount of randomness in them, just like we observed in the last section).\n\n# Delta\n\nDelta is basically the probability that epsilon breaks. Namely, sometimes the epsilon is different for some queries than it is for others. For example, you may remember when we were calculating the sensitivity of threshold, most of the time sensitivity was 0 but sometimes it was 1. Thus, we could calculate this as \"epsilon zero but non-zero delta\" which would say that epsilon is perfect except for some probability of the time when it's arbitrarily higher. Note that this expression doesn't represent the full tradeoff between epsilon and delta.",
"_____no_output_____"
],
[
"# How To Add Noise for Global Differential Privacy\n\nGlobal Differential Privacy adds noise to the output of a query.\nWe'll add noise to the output of our query so that it satisfies a certain epsilon-delta differential privacy threshold.\n\nThere are two kinds of noise we can add \n- Gaussian Noise\n- Laplacian Noise. \n\nGenerally speaking Laplacian is better, but both are still valid. Now to the hard question...\n\n### How much noise should we add?\n\nThe amount of noise necessary to add to the output of a query is a function of four things:\n\n- the type of noise (Gaussian/Laplacian)\n- the sensitivity of the query/function\n- the desired epsilon (ε)\n- the desired delta (δ)\n\nThus, for each type of noise we're adding, we have different way of calculating how much to add as a function of sensitivity, epsilon, and delta.\n\nLaplacian noise is increased/decreased according to a \"scale\" parameter b. We choose \"b\" based on the following formula.\n\n`b = sensitivity(query) / epsilon`\n\nIn other words, if we set b to be this value, then we know that we will have a privacy leakage of <= epsilon. Furthermore, the nice thing about Laplace is that it guarantees this with delta == 0. There are some tunings where we can have very low epsilon where delta is non-zero, but we'll ignore them for now.\n\n### Querying Repeatedly\n\n- if we query the database multiple times - we can simply add the epsilons (Even if we change the amount of noise and their epsilons are not the same).",
"_____no_output_____"
],
[
"# Create a Differentially Private Query\n\nLet's create a query function which sums over the database and adds just the right amount of noise such that it satisfies an epsilon constraint. query will be for \"sum\" and for \"mean\". We'll use the correct sensitivity measures for both.",
"_____no_output_____"
]
],
[
[
"epsilon = 0.001",
"_____no_output_____"
],
[
"import numpy as np",
"_____no_output_____"
],
[
"db, pdbs = create_db_and_parallels(100)",
"_____no_output_____"
],
[
"db",
"_____no_output_____"
],
[
"def sum_query(db):\n return db.sum()",
"_____no_output_____"
],
[
"def laplacian_mechanism(db, query, sensitivity):\n beta = sensitivity / epsilon\n noise = torch.tensor(np.random.laplace(0, beta, 1))\n \n return query(db) + noise",
"_____no_output_____"
],
[
"laplacian_mechanism(db, sum_query, 0.01)",
"_____no_output_____"
],
[
"def mean_query(db):\n return torch.mean(db.float())",
"_____no_output_____"
],
[
"laplacian_mechanism(db, mean_query, 1)",
"_____no_output_____"
]
],
[
[
"# Differential Privacy for Deep Learning\n\nSo what does all of this have to do with Deep Learning? Well, these mechanisms form the core primitives for how Differential Privacy provides guarantees in the context of Deep Learning. \n\n### Perfect Privacy\n> \"a query to a database returns the same value even if we remove any person from the database\".\n\nIn the context of Deep Learning, we have a similar standard. \n\n> Training a model on a dataset should return the same model even if we remove any person from the dataset.\n\nThus, we've replaced \"querying a database\" with \"training a model on a dataset\". In essence, the training process is a kind of query. However, one should note that this adds two points of complexity which database queries did not have:\n\n 1. do we always know where \"people\" are referenced in the dataset?\n 2. neural models rarely never train to the same output model, even on identical data\n\nThe answer to (1) is to treat each training example as a single, separate person. Strictly speaking, this is often overly zealous as some training examples have no relevance to people and others may have multiple/partial (consider an image with multiple people contained within it). Thus, localizing exactly where \"people\" are referenced, and thus how much your model would change if people were removed, is challenging.\n\nThe answer to (2) is also an open problem. To solve this, lets look at PATE.\n\n## Scenario: A Health Neural Network\n\nYou work for a hospital and you have a large collection of images about your patients. However, you don't know what's in them. You would like to use these images to develop a neural network which can automatically classify them, however since your images aren't labeled, they aren't sufficient to train a classifier.\n\nHowever, being a cunning strategist, you realize that you can reach out to 10 partner hospitals which have annotated data. It is your hope to train your new classifier on their datasets so that you can automatically label your own. While these hospitals are interested in helping, they have privacy concerns regarding information about their patients. Thus, you will use the following technique to train a classifier which protects the privacy of patients in the other hospitals.\n\n- 1) You'll ask each of the 10 hospitals to train a model on their own datasets (All of which have the same kinds of labels)\n- 2) You'll then use each of the 10 partner models to predict on your local dataset, generating 10 labels for each of your datapoints\n- 3) Then, for each local data point (now with 10 labels), you will perform a DP query to generate the final true label. This query is a \"max\" function, where \"max\" is the most frequent label across the 10 labels. We will need to add laplacian noise to make this Differentially Private to a certain epsilon/delta constraint.\n- 4) Finally, we will retrain a new model on our local dataset which now has labels. This will be our final \"DP\" model.\n\nSo, let's walk through these steps. I will assume you're already familiar with how to train/predict a deep neural network, so we'll skip steps 1 and 2 and work with example data. We'll focus instead on step 3, namely how to perform the DP query for each example using toy data.\n\nSo, let's say we have 10,000 training examples, and we've got 10 labels for each example (from our 10 \"teacher models\" which were trained directly on private data). Each label is chosen from a set of 10 possible labels (categories) for each image.",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
],
[
"num_teachers = 10 # we're working with 10 partner hospitals\nnum_examples = 10000 # the size of OUR dataset\nnum_labels = 10 # number of lablels for our classifier",
"_____no_output_____"
],
[
"# fake predictions\nfake_preds = (\n np.random.rand(\n num_teachers, num_examples\n ) * num_labels).astype(int).transpose(1,0)",
"_____no_output_____"
],
[
"fake_preds[:,0]\n",
"_____no_output_____"
],
[
"# Step 3: Perform a DP query to generate the final true label/outputs,\n# Use the argmax function to find the most frequent label across all 10 labels,\n# Then finally add some noise to make it differentially private.\n\nnew_labels = list()\nfor an_image in fake_preds:\n # count the most frequent label the hospitals came up with\n label_counts = np.bincount(an_image, minlength=num_labels)\n\n epsilon = 0.1\n beta = 1 / epsilon\n\n for i in range(len(label_counts)):\n # for each label, add some noise to the counts\n label_counts[i] += np.random.laplace(0, beta, 1)\n\n new_label = np.argmax(label_counts)\n \n new_labels.append(new_label)",
"_____no_output_____"
],
[
"# new_labels\nnew_labels[:10]",
"_____no_output_____"
]
],
[
[
"# PATE Analysis",
"_____no_output_____"
]
],
[
[
"# lets say the hospitals came up with these outputs... 9, 9, 3, 6 ..., 2\nlabels = np.array([9, 9, 3, 6, 9, 9, 9, 9, 8, 2])\ncounts = np.bincount(labels, minlength=10)\nprint(counts)\nquery_result = np.argmax(counts)\nquery_result\n",
"[0 0 1 1 0 0 1 0 1 6]\n"
]
],
[
[
"If every hospital says the result is 9, then we have very low sensitivity.\nWe could remove a person, from the dataset, and the query results still is 9,\nthen we have not leaked any information. \nCore assumption: The same patient was not present at any of this two hospitals.\n\nRemoving any one of this hospitals, acts as a proxy to removing one person, which means that if we do remove one hospital, the query result should not be different.\n\n",
"_____no_output_____"
]
],
[
[
"from syft.frameworks.torch.differential_privacy import pate",
"_____no_output_____"
],
[
"num_teachers, num_examples, num_labels = (100, 100, 10)\n# generate fake predictions/labels\npreds = (np.random.rand(num_teachers, num_examples) * num_labels).astype(int)\nindices = (np.random.rand(num_examples) * num_labels).astype(int) # true answers\n\npreds[:,0:10] *= 0\n\n# perform PATE to find the data depended epsilon and data independent epsilon\ndata_dep_eps, data_ind_eps = pate.perform_analysis(\n teacher_preds=preds, \n indices=indices, \n noise_eps=0.1, \n delta=1e-5\n)\nprint('Data Independent Epsilon', data_ind_eps)\nprint('Data Dependent Epsilon', data_dep_eps)\n\nassert data_dep_eps < data_ind_eps\n",
"Warning: May not have used enough values of l. Increase 'moments' variable and run again.\nData Independent Epsilon 11.756462732485115\nData Dependent Epsilon 1.52655213289881\n"
],
[
"data_dep_eps, data_ind_eps = pate.perform_analysis(teacher_preds=preds, indices=indices, noise_eps=0.1, delta=1e-5)\nprint(\"Data Independent Epsilon:\", data_ind_eps)\nprint(\"Data Dependent Epsilon:\", data_dep_eps)",
"Warning: May not have used enough values of l. Increase 'moments' variable and run again.\nData Independent Epsilon: 11.756462732485115\nData Dependent Epsilon: 1.52655213289881\n"
],
[
"preds[:,0:50] *= 0",
"_____no_output_____"
],
[
"data_dep_eps, data_ind_eps = pate.perform_analysis(teacher_preds=preds, indices=indices, noise_eps=0.1, delta=1e-5, moments=20)\nprint(\"Data Independent Epsilon:\", data_ind_eps)\nprint(\"Data Dependent Epsilon:\", data_dep_eps)",
"Data Independent Epsilon: 411.5129254649703\nData Dependent Epsilon: 9.219308825046408\n"
]
],
[
[
"# Where to Go From Here\n\n\nRead:\n - Algorithmic Foundations of Differential Privacy: https://www.cis.upenn.edu/~aaroth/Papers/privacybook.pdf\n - Deep Learning with Differential Privacy: https://arxiv.org/pdf/1607.00133.pdf\n - The Ethical Algorithm: https://www.amazon.com/Ethical-Algorithm-Science-Socially-Design/dp/0190948205\n \nTopics:\n - The Exponential Mechanism\n - The Moment's Accountant\n - Differentially Private Stochastic Gradient Descent\n\nAdvice:\n - For deployments - stick with public frameworks!\n - Join the Differential Privacy Community\n - Don't get ahead of yourself - DP is still in the early days",
"_____no_output_____"
],
[
"# Application of DP in Private Federated Learning",
"_____no_output_____"
],
[
"DP works by adding statistical noise either at the input level or output level of the model so that you can mask out individual user contribution, but at the same time gain insight into th overall population without sacrificing privacy.\n\n> Case: Figure out average money one has in their pockets.\nWe could go and ask someone how much they have in their wallet. They pick a random number between -100 and 100. Add that to the real value, say $20 and a picked number of 100. resulting in 120. That way, we have no way to know what the actual amount of money in their wallet is.\nWhen sufficiently large numbers of people submit these results, if we take the average, the noise will cancel out and we'll start seeing the true average.\n\n\nApart from statistical use cases, we can apply DP in Private Federated learning.\n\nSuppose you want to train a model using distributed learning across a number of user devices. One way to do that is to get all the private data from the devices, but that's not very privacy friendly. \n\nInstead, we send the model from the server back to the devices. The devices will then train the model\nusing their user data, and only send the privatized model updates back to the server.\nServer will then aggregate the updates and make an informed decision of the overall model on the server.\nAs you do more and more rounds, slowly the model converges to the true population without \nprivate user data having to leave the devices.\nIf you increase the level of privacy, the model converges a bit slower and vice versa.\n",
"_____no_output_____"
],
[
"# Project:\n\nFor the final project for this section, you're going to train a DP model using this PATE method on the MNIST dataset, provided below.",
"_____no_output_____"
]
],
[
[
"import torchvision.datasets as datasets\nmnist_trainset = datasets.MNIST(root='./data', train=True, download=True, transform=None)",
"Downloading http://yann.lecun.com/exdb/mnist/train-images-idx3-ubyte.gz to ./data/MNIST/raw/train-images-idx3-ubyte.gz\n"
],
[
"train_data = mnist_trainset.train_data\ntrain_targets = mnist_trainset.train_labels",
"/Users/atrask/anaconda/lib/python3.6/site-packages/torchvision/datasets/mnist.py:53: UserWarning: train_data has been renamed data\n warnings.warn(\"train_data has been renamed data\")\n/Users/atrask/anaconda/lib/python3.6/site-packages/torchvision/datasets/mnist.py:43: UserWarning: train_labels has been renamed targets\n warnings.warn(\"train_labels has been renamed targets\")\n"
],
[
"test_data = mnist_trainset.test_data\ntest_targets = mnist_trainset.test_labels",
"/Users/atrask/anaconda/lib/python3.6/site-packages/torchvision/datasets/mnist.py:58: UserWarning: test_data has been renamed data\n warnings.warn(\"test_data has been renamed data\")\n/Users/atrask/anaconda/lib/python3.6/site-packages/torchvision/datasets/mnist.py:48: UserWarning: test_labels has been renamed targets\n warnings.warn(\"test_labels has been renamed targets\")\n"
]
],
[
[
"\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
d086b9bf236b8590a238e503ae4c1c6170b5bf08 | 10,740 | ipynb | Jupyter Notebook | jupyter_notebook/legacy/literal_embeddings.ipynb | rsgit95/med_kg_txt_multimodal | 80355b0cf58e0571531ad6f9728c533110ca996d | [
"Apache-2.0"
] | null | null | null | jupyter_notebook/legacy/literal_embeddings.ipynb | rsgit95/med_kg_txt_multimodal | 80355b0cf58e0571531ad6f9728c533110ca996d | [
"Apache-2.0"
] | null | null | null | jupyter_notebook/legacy/literal_embeddings.ipynb | rsgit95/med_kg_txt_multimodal | 80355b0cf58e0571531ad6f9728c533110ca996d | [
"Apache-2.0"
] | null | null | null | 31.869436 | 191 | 0.533613 | [
[
[
"import os\nfrom tqdm import tqdm\nfrom typing import Optional, List, Dict\nfrom dataclasses import dataclass, field\n\nimport torch\nfrom transformers import AutoModel, AutoTokenizer\n\n# bluebert models\nBlueBERT_MODELCARD = [\n 'bionlp/bluebert_pubmed_mimic_uncased_L-12_H-768_A-12',\n 'bionlp/bluebert_pubmed_mimic_uncased_L-24_H-1024_A-16',\n 'bionlp/bluebert_pubmed_uncased_L-12_H-768_A-12',\n 'bionlp/bluebert_pubmed_uncased_L-24_H-1024_A-16'\n]\n\n# googlebert models\nGoogleBERT_MODELCARD = [\n 'google/bert_uncased_L-2_H-128_A-2', \n 'google/bert_uncased_L-4_H-128_A-2', \n 'google/bert_uncased_L-6_H-128_A-2', \n 'google/bert_uncased_L-2_H-512_A-2', \n 'google/bert_uncased_L-4_H-512_A-2', \n 'google/bert_uncased_L-6_H-512_A-2',\n]\n\n@dataclass\nclass EhrKgNode2IdMapping:\n '''\n This class could be only implemented,\n as the form of \"entity2id.txt\" (or \"node2id.txt\" in the feature)\n '''\n exp_path: str\n file_name: str = field(default='entity2id.txt') # actually it means node2id.txt (they all have entities and literals)\n kg_special_token_ids: dict = field(default_factory=lambda: {\"PAD\":0,\"MASK\":1})\n skip_first_line: bool = True\n\n def get_lines(self):\n file_path = os.path.join(self.exp_path, self.file_name)\n with open(file_path) as f:\n lines = f.read().splitlines()\n if self.skip_first_line:\n lines = lines[1:]\n return lines\n\n def get_id2literal(self) -> dict:\n lines = self.get_lines()\n lines_literal = list(filter(None, [self._get_literal(line) for line in lines]))\n id2literal = {self._make_id2key(line) : self._make_str2val(line) for line in lines_literal}\n return id2literal\n\n def get_id2entity(self) -> dict:\n ''' actually means (entity => node)'''\n lines = self.get_lines()\n id2entity = {self._make_id2key(line) : self._make_str2val(line) for line in lines}\n return id2entity\n\n def _get_literal(self, line: str) -> str:\n (node, node_id) = line.split('\\t')\n _check_node = node.split('^^')\n if len(_check_node) == 2:\n literal = _check_node[0].replace(\"\\\"\",\"\") # clean \"\n return literal + '\\t' + node_id\n\n def _make_id2key(self, line: str) -> int:\n _id = int(line.split('\\t')[1])\n _add = len(self.kg_special_token_ids) # len(config.kg_special_token_ids)\n key = (_id + _add)\n return key\n\n def _make_str2val(self, line: str) -> str:\n val = line.split('\\t')[0].split('^^')[0]\n return val\n\n\n_no_default = object()\n@dataclass\nclass EhrKgNode2EmbeddingMapping(EhrKgNode2IdMapping):\n \n model_name_or_path: str = _no_default\n # kg_special_token_ids: dict = field(default_factory={\"PAD\":0,\"MASK\":1})\n # tokenizer_name: Optional[str] = field(\n # default=None, metadata={\"help\": \"Pretrained tokenizer name or path if not the same as model_name\"}\n # )\n\n def __post_init__(self):\n if self.model_name_or_path is _no_default:\n raise TypeError(\"__init__ missing 1 required argument: 'model_name_or_path'\")\n\n def _load_model_and_tokenizer(self):\n # load model\n if self.model_name_or_path:\n model = AutoModel.from_pretrained(self.model_name_or_path)\n else:\n raise ValueError(\"There is no (pre-trained) model name or path.\")\n # load tokenizer\n if self.model_name_or_path:\n tokenizer = AutoTokenizer.from_pretrained(self.model_name_or_path)\n else:\n raise ValueError(\"There is no (pre-trained) tokenizer name or path.\")\n return model, tokenizer\n\n def get_literal_embeddings_from_model(self):\n model, tokenizer = self._load_model_and_tokenizer() # load (pre-trained) model and tokenizer\n id2literal = self.get_id2literal() # get mapping dict\n \n def _convert_to_model_input(literal: str, tokenizer) -> List[str]:\n return tokenizer(text=literal, return_tensors='pt', padding=True, truncation=True)\n \n id2literalembedding = {}\n for k, v in tqdm(id2literal.items()):\n encoded_input = _convert_to_model_input(literal=v, tokenizer=tokenizer)\n _, output = model(**encoded_input)\n id2literalembedding[k] = output.cpu().detach()\n return id2literalembedding\n\n def save_literal_embeddings_from_model(self, save_file_dir: str, save_file_name: str = 'id2literalembedding.pt'):\n if not os.path.isdir(save_file_dir):\n os.mkdir(save_file_dir)\n save_file_path = os.path.join(save_file_dir, save_file_name)\n id2literalembedding = self.get_literal_embeddings_from_model()\n torch.save(id2literalembedding, save_file_path)",
"_____no_output_____"
]
],
[
[
"## 0. PATH",
"_____no_output_____"
]
],
[
[
"os.getcwd()",
"_____no_output_____"
],
[
"EXP_PATH = os.getcwd() # file directory\nFILE_NAME = 'entity2id.txt' # mapping file",
"_____no_output_____"
]
],
[
[
"## 1. EhrKgNode2IdMapping",
"_____no_output_____"
]
],
[
[
"ehrkg_node2id_mapping = EhrKgNode2IdMapping(exp_path=EXP_PATH,\n file_name=FILE_NAME,\n kg_special_token_ids={\"PAD\":0,\"MASK\":1},\n skip_first_line=True)",
"_____no_output_____"
]
],
[
[
"### get id2entity: dict",
"_____no_output_____"
]
],
[
[
"id2entity = ehrkg_node_mapping.get_id2entity()",
"_____no_output_____"
]
],
[
[
"### get id2literal: dict",
"_____no_output_____"
]
],
[
[
"id2literal = ehrkg_node_mapping.get_id2literal()",
"_____no_output_____"
]
],
[
[
"## 2. EhrKgNode2EmbeddingMapping",
"_____no_output_____"
]
],
[
[
"model_name_or_path = GoogleBERT_MODELCARD[2]\nprint(model_name_or_path)",
"google/bert_uncased_L-6_H-128_A-2\n"
],
[
"ehrkg_node2embedding_mapping = EhrKgNode2EmbeddingMapping(exp_path=EXP_PATH,\n file_name=FILE_NAME,\n kg_special_token_ids={\"PAD\":0,\"MASK\":1},\n skip_first_line=True,\n model_name_or_path=model_name_or_path)",
"_____no_output_____"
]
],
[
[
"### get id2literalembeddings: dict",
"_____no_output_____"
]
],
[
[
"id2literalembeddings = ehrkg_node2embedding_mapping.get_literal_embeddings_from_model()",
" 0%| | 0/9103 [00:00<?, ?it/s]Asking to truncate to max_length but no maximum length is provided and the model has no predefined maximum length. Default to no truncation.\n100%|██████████| 9103/9103 [01:26<00:00, 104.68it/s]\n"
]
],
[
[
"### save id2literalembeddings",
"_____no_output_____"
]
],
[
[
"SAVE_FILE_DIR = os.getcwd()\nehr_kg_embedding_mapping.save_literal_embeddings_from_model(save_file_dir=SAVE_FILE_DIR)",
" 0%| | 0/9103 [00:00<?, ?it/s]Asking to truncate to max_length but no maximum length is provided and the model has no predefined maximum length. Default to no truncation.\n100%|██████████| 9103/9103 [00:38<00:00, 236.54it/s]\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d086d6577ad7fe6776d44dc06f0224d76d84dd13 | 8,224 | ipynb | Jupyter Notebook | .ipynb_checkpoints/L09-Supervised_Machine_Learning-Practice-checkpoint.ipynb | Huiting120/Data-Analytics-With-Python | abdae1ea4f984de546a476f72d01176ef8abc2ff | [
"Apache-2.0"
] | 3 | 2020-02-18T18:37:57.000Z | 2022-01-28T23:01:54.000Z | .ipynb_checkpoints/L09-Supervised_Machine_Learning-Practice-checkpoint.ipynb | Huiting120/Data-Analytics-With-Python | abdae1ea4f984de546a476f72d01176ef8abc2ff | [
"Apache-2.0"
] | null | null | null | .ipynb_checkpoints/L09-Supervised_Machine_Learning-Practice-checkpoint.ipynb | Huiting120/Data-Analytics-With-Python | abdae1ea4f984de546a476f72d01176ef8abc2ff | [
"Apache-2.0"
] | 12 | 2020-03-05T18:41:00.000Z | 2022-02-03T17:31:37.000Z | 29.905455 | 568 | 0.61053 | [
[
[
"# Lesson 9 Practice: Supervised Machine Learning\nUse this notebook to follow along with the lesson in the corresponding lesson notebook: [L09-Supervised_Machine_Learning-Lesson.ipynb](./L09-Supervised_Machine_Learning-Lesson.ipynb). \n ",
"_____no_output_____"
],
[
"## Instructions\nFollow along with the teaching material in the lesson. Throughout the tutorial sections labeled as \"Tasks\" are interspersed and indicated with the icon: . You should follow the instructions provided in these sections by performing them in the practice notebook. When the tutorial is completed you can turn in the final practice notebook. For each task, use the cell below it to write and test your code. You may add additional cells for any task as needed or desired. ",
"_____no_output_____"
],
[
"## Task 1a: Setup\n\nImport the following package sets:\n+ packages for data management\n+ pacakges for visualization\n+ packages for machine learning\n\nRemember to activate the `%matplotlib inline` magic.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\n# Data Management\nimport numpy as np\nimport pandas as pd\n\n# Visualization\nimport seaborn as sns\nimport matplotlib.pyplot as plt\n\n# Machine learning\nfrom sklearn import model_selection\nfrom sklearn import preprocessing\nfrom sklearn.metrics import accuracy_score\nfrom sklearn.metrics import confusion_matrix\nfrom sklearn.metrics import classification_report\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.svm import SVC, LinearSVC\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.naive_bayes import GaussianNB\nfrom sklearn.linear_model import Perceptron\nfrom sklearn.linear_model import SGDClassifier\nfrom sklearn.tree import DecisionTreeClassifier\nfrom sklearn.discriminant_analysis import LinearDiscriminantAnalysis",
"_____no_output_____"
]
],
[
[
"## Task 2a: Data Exploration\n\nAfter reviewing the data in sections 2.1, 2.2, 2.3 and 2.4 do you see any problems with this iris dataset? If so, please describe them in the practice notebook. If not, simply indicate that there are no issues.",
"_____no_output_____"
],
[
"## Task 2b: Make Assumptions\n\nAfter reviewing the data in sections 2.1, 2.2, 2.3 and 2.4 are there any columns that would make poor predictors of species? \n\n**Hint**: columns that are poor predictors are:\n+ those with too many missing values\n+ those with no difference in variation when grouped by the outcome class\n+ variables with high levels of collinearity",
"_____no_output_____"
],
[
"## Task 3a: Practice with the random forest classifier\n\nNow that you have learned how to perform supervised machine learning using a variety of algorithms, lets practice using a new algorithm we haven't looked at yet: the Random Forest Classifier. The random forest classifier builds multiple decision trees and merges them together. Review the sklearn [online documentation for the RandomForestClassifier](https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.RandomForestClassifier.html). For this task:\n\n1. Perform a 10-fold cross-validation strategy to see how well the random forest classifier performs with the iris data\n2. Use a boxplot to show the distribution of accuracy\n3. Use the `fit` and `predict` functions to see how well it performs with the testing data.\n4. Plot the confusion matrix\n5. Print the classification report.\n",
"_____no_output_____"
]
],
[
[
"iris = sns.load_dataset('iris')",
"_____no_output_____"
],
[
"X = iris.loc[:,'sepal_length':'petal_width'].values",
"_____no_output_____"
],
[
"Y = iris['species'].values",
"_____no_output_____"
],
[
"X = preprocessing.robust_scale(X)",
"_____no_output_____"
],
[
"Xt, Xv, Yt, Yv = model_selection.train_test_split(X, Y, test_size=0.2, random_state=10)",
"_____no_output_____"
],
[
"kfold = model_selection.KFold(n_splits=10, random_state=10)",
"_____no_output_____"
],
[
"results = {\n 'LogisticRegression' : np.zeros(10),\n 'LinearDiscriminantAnalysis' : np.zeros(10),\n 'KNeighborsClassifier' : np.zeros(10),\n 'DecisionTreeClassifier' : np.zeros(10),\n 'GaussianNB' : np.zeros(10),\n 'SVC' : np.zeros(10),\n 'RandomForestClassifier': np.zeros(10)\n}\nresults",
"_____no_output_____"
],
[
"# Create the LogisticRegression object prepared for a multinomial outcome validation set.\nalg = RandomForestClassifier()\n\n# Execute the cross-validation strategy\nresults['RandomForestClassifier'] = model_selection.cross_val_score(alg, Xt, Yt, cv=kfold, \n scoring=\"accuracy\", error_score=np.nan)\n\n# Take a look at the scores for each of the 10-fold runs.\nresults['RandomForestClassifier']",
"_____no_output_____"
],
[
"pd.DataFrame(results).plot(kind=\"box\", rot=90);",
"_____no_output_____"
],
[
"# Create the LinearDiscriminantAnalysis object with defaults.\nalg = RandomForestClassifier()\n\n# Create a new model using all of the training data.\nalg.fit(Xt, Yt)\n\n# Using the testing data, predict the iris species.\npredictions = alg.predict(Xv)\n\n# Let's see the predictions\npredictions",
"_____no_output_____"
],
[
"accuracy_score(Yv, predictions)",
"_____no_output_____"
],
[
"labels = ['versicolor', 'virginica', 'setosa']\ncm = confusion_matrix(Yv, predictions, labels=labels)\nprint(cm)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d086d68e4a2fb052e5e96976e9a5392ec0282447 | 24,012 | ipynb | Jupyter Notebook | docs/synapse/userguides/data_model_form_categories.ipynb | kcreyts/synapse | fe740fd1e0febfa32f8d431b32ab48f8a0cf306e | [
"Apache-2.0"
] | 1 | 2021-02-15T22:07:05.000Z | 2021-02-15T22:07:05.000Z | docs/synapse/userguides/data_model_form_categories.ipynb | ericalpeterson/synapse | ebbea4d519723fad4b9cd33572ed159e57587fe2 | [
"Apache-2.0"
] | null | null | null | docs/synapse/userguides/data_model_form_categories.ipynb | ericalpeterson/synapse | ebbea4d519723fad4b9cd33572ed159e57587fe2 | [
"Apache-2.0"
] | null | null | null | 108.651584 | 1,380 | 0.706688 | [
[
[
".. highlight:: none\n\n.. _data-model-form-categories:\n\nData Model - Form Categories\n============================\n\nSynapse forms can be broadly grouped into conceptual categories based on the object a form is meant to represent - an :ref:`form-entity`, a :ref:`form-relationship`, or an :ref:`form-event`.\n\nSynapse forms can also be broadly grouped based on how their primary properties (``<form> = <valu>``) are structured or formed.\n\nRecall that ``<form> = <valu>`` must be unique for all forms of a given type. In other words, the ``<valu>`` must be defined so that it uniquely identifies any given node of that form; it represents that form’s \"essence\" or \"thinghood\" in a way that allows the unambiguous deconfliction of all possible nodes of that form.\n\nConceptually speaking, the general categories of forms in Synapse are:\n\n- `Simple Form`_\n- `Composite (Comp) Form`_\n- `Guid Form`_\n- `Edge Representations`_\n\n - `Digraph (Edge) Form`_\n - `Lightweight (Light) Edge`_\n\n- `Generic Form`_\n\nThis list represents a conceptual framework to understand the Synapse data model.\n\n.. _form-simple:\n\nSimple Form\n-----------\n\nA simple form refers to a form whose primary property is a single typed ``<valu>``. They are commonly used to represent an :ref:`form-entity`, and so tend to be the most readily understood from a modeling perspective.\n\n**Examples**\n\n- **IP addresses.** An IP address (IPv4 or IPv6) must be unique within its address space and can be defined by the address itself: ``inet:ipv4 = 1.2.3.4``. Secondary properties include the associated Autonomous System number and whether the IP belongs to a specialized or reserved group (e.g., private, multicast, etc.).\n\n- **Email addresses.** An email address must be unique in order to route email to the correct account / individual and can be defined by the address itself: ``inet:email = [email protected]``. Secondary properties include the domain where the account receives mail and the username for the account.\n\n.. _form-comp:\n\nComposite (Comp) Form\n---------------------\n\nA composite (comp) form is one where the primary property is a comma-separated list of two or more typed ``<valu>`` elements. While no single element makes the form unique, a combination of elements can uniquely define a given node of that form. Comp forms are often (though not universally) used to represent a :ref:`form-relationship`.\n\n**Examples**\n\n- **Fused DNS A records.** A DNS A record can be uniquely defined by the combination of the domain (``inet:fqdn``) and the IP address (``inet:ipv4``) in the A record. Synapse’s ``inet:dns:a`` form represents the knowledge that a given domain has ever resolved to a specific IP (fused knowledge): ``inet:dns:a = (woot.com, 1.2.3.4)``.\n\n- **Web-based accounts.** An account at an online service (such as Github or Gmail) can be uniquely defined by the combination of the domain where the service is hosted (``inet:fqdn``) and the unique user ID (``inet:user``) used to identify the account: ``inet:web:acct = (twitter.com, joeuser)``.\n\n- **Social networks.** Many online services allow users to establish relationships with other users of that service. These relationships may be one-way (you can follow someone on Twitter) or two-way (you can mutually connect with someone on LinkedIn). A given one-way social network relationship can be uniquely defined by the two users (``inet:web:acct``) involved in the relationship: ``inet:web:follows = ((twitter.com,alice), (twitter.com,bob))``. (A two-way relationship can be defined by two one-way relationships.)\n \n Note that each of the elements in the ``inet:web:follows`` comp form is itself a comp form (``inet:web:acct``).\n \n- **Subsidiaries.** An organization / sub-organization relationship (e.g., corporation / subsidiary, company / division, government / ministry, etc.) can be uniquely defined by the specific parent / child entities (``ou:org``) involved: ``ou:suborg = (084e295272e839afcf3f1fe10c6c97b9, 237e88a35439fdb566d909e291339154)``.\n \n Note that each of the organizations (``ou:org``) in the relationship is represented by a 128-bit Globally Unique Identifier (guid), each an example of a `Guid Form`_.\n\n.. _form-guid:\n\nGuid Form\n---------\n\nA guid (Globally Unique Identifier) form is uniquely defined by a machine-generated 128-bit number. Guids account for cases where it is impossible to uniquely define a thing based on a specific set of properties no matter how many individual elements are factored into a comp form. A guid form can be considered a special case of a :ref:`form-simple` where the typed ``<valu>`` is of type ``<guid>``.\n\n.. NOTE::\n Guid forms can be arbitrary (generated ad-hoc by Synapse) or predictable / deconflictable (generated based on a specific set of inputs). See the :ref:`type-guid` section of :ref:`storm-ref-type-specific` for a more detailed discussion of this concept.\n\nWhile certain types of data **could** be represented by a comp form based on a sufficient number of properties of the data, there are advantages to using a guid instead:\n\n- in a comp form, the elements used to create the primary property are **required** in order to create a node of that form. It is not uncommon for real world data to be incomplete. Using a guid allows all of those elements to be defined as optional secondary properties, so the node can be created with as much (or as little) data as is available.\n- Some data sources are such that individual records can be considered unique a priori. This often applies to event-type forms for large quantities of events. In this case it sufficient to distinguish the nodes from each other using a guid as opposed to being uniqued over a subset of properties.\n- There is a potential performance benefit to representing forms using arbitrary guids in partitcular because they are guaranteed to be unique for a given Cortex. In particular, when ingesting data presumed to be unique, creating guid-based forms vs comp forms eliminates the need to parse and deconflict nodes before they are created. This benefit can be significant over large data sets.\n\n**Examples**\n\n- **People.** Synapse uses a guid as the primary property for a person (``ps:person``) node. There is no single property or set of properties that uniquely and unambiguously define a person. A person’s full name, date of birth, or place of birth (or the combination of all three) are not guaranteed to be fully unique across an entire population. Identification numbers (such as Social Security or National ID numbers) are country-specific, and not all countries require each citizen to have an ID number. Even a person’s genome is not guaranteed to be unique (such as in the case of identical twins).\n\n Secondary properties include the person’s name (including given, middle, or family names) and date of birth.\n\n- **Host execution / sandbox data.** The ability to model detailed behavior of a process executing on a host (or in a sandbox) is important for a range of disciplines, including incident response and malware analysis. Modeling this data is challenging because of the number of effects that execution may have on a system (files read, written, or deleted; network activity initiated). Even if we focus on a specific effect (\"a process wrote a new file to disk\"), there are still a number of details that may define a \"unique instance\" of \"process writes file\": the specific host (``it:host``) where the process ran, the program (``file:bytes``) that wrote the file to disk, the process (``file:bytes``) that launched the program, the time the execution occurred, the file that was written (``file:bytes``), the file’s path (``file:path``), and so on. While all of these elements could be used to create a comp form, in the \"real world\" not all of this data may be available in all cases, making a guid a better option for forms such as ``it:exec:file.write``.\n\n- **Unique DNS responses.** Similar to host execution data, an individual DNS response to a request could potentially be uniqued based on a comp form containing multiple elements (time, DNS query, server that replied, response code, specific response, etc.) However, the same issues described above apply and it is preferable to use a guid for forms such as ``inet:dns:request`` or ``inet:dns:answer``.\n\n.. _form-edge-reps:\n\nEdge Representations\n--------------------\n\nRecall that a :ref:`form-relationship` can be the hypergraph equivalent of an edge connecting two nodes in a directed graph. A standard relationship form (such as ``inet:dns:a``) represents a specific relationship (\"has DNS A record for\") between two explicitly typed nodes (``inet:fqdn`` and ``inet:ipv4``). Synapse's strong typing and type safety ensure that all primary and secondary properties are explicitly typed, which facilitates both normalization of data and the ability to readily pivot across disparate properties that share the same data type. However, this means that types for all primary and secondary properties for a form representing a relationship must be defined in the data model ahead of time.\n\nSome relationships are generic enough to apply to a wide variety of forms. One example is \"has\": <thing a> \"has\" <thing b>. While it is possible to explicitly define typed forms for every possible variation of that relationship (\"person has telephone number\", \"company has social media account\"), you would still need to update the data model every time a new variation of what is essentially the same \"has\" relationship is identified.\n\nSynapse provides two options to represent generic \"edge-type\" relationships between arbitrary forms. Both methods allow this data to be incorporated into a Cortex without code modifications to update the data model: the :ref:`form-edge` and the :ref:`light-edge`.\n\n.. _form-edge:\n\nDigraph (Edge) Form\n+++++++++++++++++++\n\nA digraph form (\"edge\" form) is a specialized :ref:`form-comp` whose primary property value consists of two ``<form>,<valu>`` pairs (\"node definitions\", or ndefs). An edge form is a specialized relationship form that can be used to link two arbitrary forms in a generic relationship. In the \"has\" example above, a variety of entities (people, organizations) may \"have\" a variety of things (email addresses, social media accounts, company cars). It would be nice to have a single generic \"has\" form that could link two arbitrary objects without having to explicitly define relationship forms such as \"person has email address\" or \"company has office location\".\n\nSynapse addresses this issue by defining a node’s **ndef** (``<form>,<valu>`` pair) as a data :ref:`data-type`. Properties of type ``ndef`` can thus effectively specify both a type (``<form>``) and a ``<valu>`` at the time of node creation. This allows for generic relationship forms (such as ``edge:has``) that can link two \"arbitrary\" node types.\n\nGeneric edge forms are best suited for representing relationships where you need to capture additional detail about the relationship (via secondary properties) or observations about the relationship (via tags).\n\n.. _light-edge:\n\nLightweight (Light) Edge\n++++++++++++++++++++++++\n\nDigraph forms are useful, but have some disadvantages in terms of performance, representation, and navigation for many common use cases. Lightweight (light) edges address these limitations.\n\nSimilar to edge forms, light edges are used to link two arbitrary forms. However, unlike edge forms, light edges are not forms at all. They consist solely of a user-defined verb (that describes the linking relationship) and the two forms (nodes) being linked. Light edges typically have an implied direction (as many relationships represented by light edges are \"one-way\"). However, the direction is not an inherent part of the definition of the light edge itself; instead the direction is \"defined\" via the Storm syntax used to join the nodes. That is, nothing in Synapse prevents you from joining any two forms in any direction via a light edge, but only some of those joins will make sense given the meaning of the edge verb.\n\nLight edges have some advantages over edge forms:\n\n- Because they are nodes, edge forms incur additional performance overhead in general. This overhead is amplified in use cases where the edge represents a many-to-one relationship and the \"many\" is high. Light edges will always be more efficient than edge forms, and the performance benefit is significant in many cases.\n- Edge forms represent generic relationships, but the edge form itself must still exist in the data model before it can be used. Synapse includes edge forms for common generic relationships (e.g., ``edge:has``), but introducing additional relationships would require extending the data model. Light edges can be created on the fly (with appropriate permissions) as the need arises.\n- The primary property of an edge form is two elements of type :ref:`gloss-ndef`. Because of Synapse's type-awareness, this may exclude edge forms from certain types of navigation (such as wildcard (\"refs out\" / \"refs in\") pivots - see :ref:`storm-ref-pivot`). This makes it slightly more complicated to \"show me all the things\" connected to a given node when those connections may include things linked by edge forms vs. things linked by light edges.\n\nLight edges have some disadvantages - namely, since they are not forms, they cannot store any additional \"detail\" about the relationship they represent outside of their verb. They do not suppport secondary properties, and you cannot apply tags to light edges.\n\nIn addition, because light edges are not forms, they cannot be viewed in a Cortex via Synapse's model introspection features (see :ref:`storm-ref-model-introspect`). The Storm :ref:`storm-model` commands allow you to list and otherwise work with the light edges in a Cortex (note that there are no light edges defined in a Cortex by default).\n\nSee the :ref:`storm-ref-data-mod` for detail on creating (or deleting) light edges and the :ref:`storm-ref-pivot` for navigating light edges.\n\nWhether to use an edge form or a light edge to represent data in your Cortex will depend on your specific needs.\n\nExamples\n++++++++\n\n**\"References\".** There are a number of use cases where it is helpful to note that a thing “references” another thing. Examples include:\n\n- A report (``media:news``) that contains threat indicators, such as hashes (``hash:sha256``), domains (``inet:fqdn``), email addresses (``inet:email``), etc.\n- A photograph (``file:bytes``) that depicts a person (``ps:person``), a location (``geo:place``), a landmark (``mat:item``), etc.\n- A news article (``media:news``) that describes an event such as a conference (``ou:conference``).\n\n\"References\" is a very simple generic relationship. It is also likely to represent large many-to-one relationships, at least for some use cases; while some blogs may include only a handful of indicators, comprehensive whitepapers or internal documents such as incident reports may contain hundreds or thousands of indicators and referenced objects. \"References\" is also unlikely to have an associated time element; that is, if a report contains (references) an indicator (such as an FQDN), that relationship is unlikely to change. A report may be revised, but then it is technically a different report; the original still contains the reference.\n\nFor these reasons a \"references\" relationship would be better represented by a light edge vs. an edge form.\n\n**\"Has\".** There are a number of use cases where it is helpful to note that a thing owns or possesses (\"has\") another thing. Examples include:\n\n- A company (``ou:org``) owns a corporate office (``geo:place``, ``mat:item``), a range of IP addresses (``inet:cidr4``), or a delivery van (``mat:item``).\n- A person (``ps:person``) has an email address (``inet:email``) or telephone number (``tel:phone``).\n\nIn some cases the relationship of a person or organization owning or possessing (\"having\") a resource (a social media account, or an email address) may be indirectly apparent via existing pivots in the Synapse hypergraph. For example, an organization (``ou:org``) may have a name that is shared by a social media account (``ou:org:name -> inet:web:acct:realname``) where the social media account also references the organization’s web page (``inet:web:acct:webpage -> ou:org:url``). However, it may be desirable to more tightly link an \"owning\" entity to things that it \"has\". In addition, there may be things that an organization or person \"has\" that are not as easily identified via primary and secondary property pivots. In these cases the \"has\" form can represent this relationship between the \"owning\" entity and the arbitrary thing owned.\n\nLike \"references\", \"has\" seems like a very simple generic relationship. Whether to use an edge form or a light edge depends in part on the number of many-to-one relationships you need to model, and whether you need to capture additional information about the relationship (such as if something was \"had\" only for a specific period of time).\n\nIf the many-to-one is relatively small AND you need to capture data such as a time interval, an edge form (``edge:has``) may be best. For large instances of many-to-one, or cases where things like time are not relevant (or where the time element is captured elsewhere), light edges are preferable.\n\n- An organization (``ou:org``) may \"have\" an office location (``geo:place``) only for a period of time; the organization may lease or buy a different space if the business grows, for example. If this time element is relevant, an ``edge:has`` node can be used to represent the relationship, with the ``.seen`` property capturing the time interval.\n\n- An IP address (``inet:ipv4`` or ``inet:ipv6``) may be part of a netblock, either directly (``inet:asnet4``, ``inet:cidr4``, ``inet:cidr6``) or as part of a netblock referenced in a network registration record (``inet:whois:iprec``). Depending on the size of the netblock, the many-to-one relationship may be extremely large. In addition, an IP address may be part of more than one netblock / registration record, given network range suballocations and so on. In some cases a time element is irrelevant (i.e., a defined CIDR block is a fixed thing; an IP that is part of a /24 will never **not** be part of that /24). In cases of network registration records, the ``inet:whois:iprec`` form contains time values; if that record changes (specifically, if the IP range is allocated differently) that would represent a new ``inet:whois:iprec`` with a new \"has\" relationship with the IPs in that range. In these cases (IP as part of CIDR, IP refrenced by netblock in registration record) light edges are preferable - for example, ``inet:cidr4 -(has)> inet:ipv4`` to show an IP is part of a CIDR block or ``inet:whois:iprec -(has)> inet:ipv4`` to show that an IP is part of a netblock referenced in a registration record. These light edges can be represented by a generic verb (\"has\") or a more relationship-specific verb (e.g., \"hasip\") depending on preference or need.\n\n**\"Went to\".** \"Went to\" can be used to represent that a thing (often a person, potentially an object such as a bus) traveled to a place (a city, an office building, a set of geolocation coordinates) or that a person attended an event (a conference, a party). It would be natural to want to record \"when\" this event occured, such as via a \"time\" secondary property (for a single point in time, such as an arrival time). Alternately, the ``.seen`` universal property could be used to record a start and end time if the \"went to\" needed to capture a duration. Because of this need to track additional information about the relationship, an edge form (``edge:wentto``) would be more appopriate.\n\n.. _form-generic:\n\nGeneric Form\n------------\n\nThe Synapse data model includes a number of \"generic\" forms that can be used to represent metadata and / or arbitrary data. \n\nArbitrary Data\n++++++++++++++\n\nIn an ideal world, all data represented in a Synapse hypergraph would be accurately modeled using an appropriate form to property capture the data’s unique (primary property) and contextual (secondary property) characteristics. However, designing an appropriate data model may require extended discussion, subject matter expertise, and testing against \"real world\" data - not to mention development time to implement model changes. In addition, there are use cases where data needs to be added to a Cortex for reference or analysis purposes, but simply does not have sufficient detail to be represented accurately, even if appropriate data forms exist.\n\nWhile the use of generic forms is not ideal (the representation of data is lossy, which may impact effective analysis), these forms allow for the addition of arbitrary data to a hypergraph, either because that is the only way the data can be represented; or because an appropriate model does not yet exist but the data is needed now.\n\nGeneric forms such as ``graph:node``, ``graph:edge``, ``graph:timeedge`` and ``graph:event`` can be used for this purpose. Similarly, the generic ``graph:cluster`` node can be used to link (via ``refs`` light edges or ``edge:refs`` forms) a set of nodes of arbitrary size (\"someone says these things are all related\") in the absence of greater detail.\n\nMetadata\n++++++++\n\nThe Synapse data model includes forms such as ``meta:source`` that can be used to track data sources for data ingested into a Cortex. \"Sources\" may include sensors or third-party services or connectors. Structures such as ``seen`` light edges or ``meta:seen`` forms can be used to track that a particular piece of data (e.g., a node) was observed by or from a particular source.",
"_____no_output_____"
]
]
] | [
"raw"
] | [
[
"raw"
]
] |
d086d847879a7b45856c5936f9b0b005b18e707f | 1,806 | ipynb | Jupyter Notebook | Jupyter Notebook/Basics of Jupyter Notebook.ipynb | ElliotRedhead/pythonmachinelearning | 71a9bd29b897eb1bcd2356095adc6287eaaa9916 | [
"MIT"
] | null | null | null | Jupyter Notebook/Basics of Jupyter Notebook.ipynb | ElliotRedhead/pythonmachinelearning | 71a9bd29b897eb1bcd2356095adc6287eaaa9916 | [
"MIT"
] | null | null | null | Jupyter Notebook/Basics of Jupyter Notebook.ipynb | ElliotRedhead/pythonmachinelearning | 71a9bd29b897eb1bcd2356095adc6287eaaa9916 | [
"MIT"
] | null | null | null | 19.846154 | 96 | 0.493355 | [
[
[
"# Basics of Jupyter Notebook\n\nThe cell type can be modified by the dropdown in the Jupyter editor in the toolbar.<br>\nUse `<br>` to drop the following text to a new line.<br>\nTo see syntax tips, type a function and press `SHIFT + TAB`.<br>\nPublish a section by pressing `CTRL + TAB`.<br>",
"_____no_output_____"
],
[
"## Examples of Python Code",
"_____no_output_____"
]
],
[
[
"my_name = \"Elliot\"\nhello_statement = f\"Hello, {my_name}\"\nprint(hello_statement)",
"Hello, Elliot\n"
],
[
"x = 1\nfor i in range(1, 5):\n x = x + i\n print(f\"i={i}, x={x}\")",
"i=1, x=2\ni=2, x=4\ni=3, x=7\ni=4, x=11\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
d086dedd7bd8c4f56de2cd143a68444036e759b8 | 37,766 | ipynb | Jupyter Notebook | deep-learning-with-python-book/ch2.math-building-blocks-of-nn/03_tensor_operations.ipynb | plamenti/deep_learning_project_manning | 99a3bfaecb18a396079e0e64674af4c1a5810fb5 | [
"MIT"
] | null | null | null | deep-learning-with-python-book/ch2.math-building-blocks-of-nn/03_tensor_operations.ipynb | plamenti/deep_learning_project_manning | 99a3bfaecb18a396079e0e64674af4c1a5810fb5 | [
"MIT"
] | null | null | null | deep-learning-with-python-book/ch2.math-building-blocks-of-nn/03_tensor_operations.ipynb | plamenti/deep_learning_project_manning | 99a3bfaecb18a396079e0e64674af4c1a5810fb5 | [
"MIT"
] | null | null | null | 31.108731 | 389 | 0.475375 | [
[
[
"import numpy as np\r\nimport time\r\n\r\nimport tensorflow as tf\r\nfrom tensorflow.keras.datasets import mnist\r\nfrom tensorflow.keras import models, layers\r\nfrom tensorflow.keras.models import Sequential\r\nfrom tensorflow.keras import optimizers\r\n\r\nfrom tensorflow.keras.layers import Dense",
"_____no_output_____"
]
],
[
[
"Much as any computer program can be ultimately reduced to a small set of binary operations on binary inputs (AND, OR, NOR, and so on), all transformations learned by deep neural networks can be reduced to a handful of tensor operations applied to tensors of numeric data. For instance, it’s possible to add tensors, multiply tensors, and so on.",
"_____no_output_____"
],
[
"A Keras layer instance looks like this",
"_____no_output_____"
]
],
[
[
"Dense(512, activation='relu')",
"_____no_output_____"
]
],
[
[
"This layer can be interpreted as a function, which takes as input a matrix and returns another matrix — a new representation for the input tensor. Specifically, the function is as follows (where W is a matrix and b is a vector, both attributes of the layer).\r\n\r\nWe have three tensor operations here: a dot product (dot) between the input tensor and a tensor named W; an addition (+) between the resulting matrix and a vector b; and, finally, a relu operation. relu(x) is max(x, 0)",
"_____no_output_____"
]
],
[
[
"# output = relu(dot(W, input) + b)",
"_____no_output_____"
]
],
[
[
"### Element-wise operations\r\n\r\nThe **relu** operation and **addition** are element-wise operations: operations that are applied independently to each entry in the tensors being considered. This means these operations are highly amenable to massively parallel implementations.\r\n\r\nIf you want to write a naive Python implementation of an element-wise operation, you use a for loop, as in this naive implementation of an element-wise **relu** operation:",
"_____no_output_____"
]
],
[
[
"def naive_relu(x):\r\n assert len(x.shape) == 2\r\n x = x.copy()\r\n for i in range(x.shape[0]):\r\n for j in range(x.shape[1]):\r\n x[i, j] = max(x[i, j], 0)\r\n return x",
"_____no_output_____"
],
[
"def naive_add(x, y):\r\n assert len(x.shape) == 2\r\n assert x.shape == y.shape\r\n x = x.copy()\r\n for i in range(x.shape[0]):\r\n for j in range(x.shape[1]):\r\n x[i, j] += y[i, j]\r\n return x",
"_____no_output_____"
]
],
[
[
"On the same principle, you can do element-wise multiplication, subtraction, and so on.\r\n\r\nIn practice, when dealing with NumPy arrays, these operations are available as well-optimized built-in NumPy functions, which themselves delegate the heavy lifting to a Basic Linear Algebra Subprograms (BLAS) implementation if you have one installed. BLAS are low-level, highly parallel, efficient tensor-manipulation routines that are typically implemented in Fortran or C.\r\n\r\nIn NumPy, you can do the following element-wise operation, and it will be blazing fast",
"_____no_output_____"
]
],
[
[
"# z = x + y\r\n# z = np.maximum(z, 0)",
"_____no_output_____"
]
],
[
[
"Time the difference:\r\n",
"_____no_output_____"
]
],
[
[
"x = np.random.random((20, 100))\r\ny = np.random.random((20, 100))\r\n\r\ntime_start = time.time()\r\n\r\nfor _ in range(1000):\r\n z = x + y\r\n z = np.maximum(z, 0)\r\n\r\nduration = time.time() - time_start\r\nprint(f\"Duration: {duration} sec\")",
"Duration: 0.009510040283203125 sec\n"
],
[
"time_start = time.time()\r\nfor _ in range(1000):\r\n z = naive_add(x, y)\r\n z = naive_relu(z)\r\n\r\nduration = time.time() - time_start\r\nprint(f\"Duration: {duration} sec\")",
"Duration: 2.2906622886657715 sec\n"
]
],
[
[
"### Broadcasting\r\n\r\nWhen possible, and if there’s no ambiguity, the smaller tensor will be broadcasted to match the shape of the larger tensor. Broadcasting consists of two steps:\r\n\r\nAxes (called broadcast axes) are added to the smaller tensor to match the ndim of the larger tensor.\r\nThe smaller tensor is repeated alongside these new axes to match the full shape of the larger tensor.\r\n\r\nExample - Consider X with shape (32, 10) and y with shape (10,). First, we add an empty first axis to y, whose shape becomes (1, 10). Then, we repeat y 32 times alongside this new axis, so that we end up with a tensor Y with shape (32, 10), where Y[i, :] == y for i in range(0, 32). At this point, we can proceed to add X and Y, because they have the same shape.",
"_____no_output_____"
]
],
[
[
"def naive_add_matrix_and_vector(x, y):\r\n assert len(x.shape) == 2\r\n assert len(y.shape) == 1\r\n assert x.shape[1] == y.shape[0]\r\n x = x.copy()\r\n for i in range(x.shape[0]):\r\n for j in range(x.shape[1]):\r\n x[i, j] += y[j]\r\n return x",
"_____no_output_____"
],
[
"x = np.random.random((64, 3, 32, 10))\r\ny = np.random.random((32, 10))\r\nz = np.maximum(x, y)",
"_____no_output_____"
]
],
[
[
"### Tensor product\r\nThe tensor product, or dot product (not to be confused with an element-wise product, the * operator) is one of the most common, most useful tensor operations.\r\n\r\nIn NumPy, a tensor product is done using the np.dot function (because the mathematical notation for tensor product is usually a dot).",
"_____no_output_____"
]
],
[
[
"x = np.random.random((32,))\r\ny = np.random.random((32,))\r\nz = np.dot(x, y)",
"_____no_output_____"
],
[
"z",
"_____no_output_____"
],
[
"# naive interpretation of two vectors\r\ndef naive_vector_dot(x, y):\r\n assert len(x.shape) == 1\r\n assert len(y.shape) == 1\r\n assert x.shape[0] == y.shape[0]\r\n z = 0.\r\n for i in range(x.shape[0]):\r\n z += x[i] * y[i]\r\n return z",
"_____no_output_____"
],
[
"zz = naive_vector_dot(x, y)",
"_____no_output_____"
],
[
"zz",
"_____no_output_____"
],
[
"# naive interpretation of matrix and vector\r\ndef naive_matrix_vector_dot(x, y):\r\n assert len(x.shape) == 2\r\n assert len(y.shape) == 1\r\n assert x.shape[1] == y.shape[0]\r\n z = np.zeros(x.shape[0])\r\n for i in range(x.shape[0]):\r\n for j in range(x.shape[1]):\r\n z[i] += x[i, j] * y[j]\r\n return z",
"_____no_output_____"
]
],
[
[
"As soon as one of the two tensors has an ndim greater than 1, dot is no longer symmetric, which is to say that dot(x, y) isn’t the same as dot(y, x)",
"_____no_output_____"
],
[
"The most common applications may be the dot product between two matrices. You can take the dot product of two matrices x and y (dot(x, y)) if and only if x.shape[1] == y.shape[0] (mn nm). The result is a matrix with shape (x.shape[0], y.shape[1]), where the coefficients are the vector products between the rows of x and the columns of y. Here’s the naive implementation:",
"_____no_output_____"
]
],
[
[
"def naive_matrix_dot(x, y):\r\n assert len(x.shape) == 2\r\n assert len(y.shape) == 2\r\n assert x.shape[1] == y.shape[0]\r\n z = np.zeros((x.shape[0], y.shape[1]))\r\n for i in range(x.shape[0]):\r\n for j in range(y.shape[1]):\r\n row_x = x[i, :]\r\n column_y = y[:, j]\r\n z[i, j] = naive_vector_dot(row_x, column_y)\r\n return z",
"_____no_output_____"
]
],
[
[
"### Tensor reshaping\r\n\r\nReshaping a tensor means rearranging its rows and columns to match a target shape. Naturally, the reshaped tensor has the same total number of coefficients as the initial tensor. Reshaping is best understood via simple examples:",
"_____no_output_____"
]
],
[
[
"x = np.array([[0., 1.],\r\n [2., 3.],\r\n [4., 5.]])\r\nprint(x.shape)",
"(3, 2)\n"
],
[
"x = x.reshape((6, 1))\r\nx",
"_____no_output_____"
],
[
"x = x.reshape((2, 3))\r\nx",
"_____no_output_____"
]
],
[
[
"A special case of reshaping that’s commonly encountered is transposition. Transposing a matrix means exchanging its rows and its columns, so that x[i, :] becomes x[:, i]:",
"_____no_output_____"
]
],
[
[
"x = np.zeros((300, 20))\r\nprint(x.shape)",
"(300, 20)\n"
],
[
"x = np.transpose(x)\r\nprint(x.shape)",
"(20, 300)\n"
]
],
[
[
"### Geometric interpretation of tensor operations\r\n\r\nBecause the contents of the tensors manipulated by tensor operations can be interpreted as coordinates of points in some geometric space, all tensor operations have a geometric interpretation. For instance, let’s consider addition. We’ll start with the following vector:",
"_____no_output_____"
],
[
"### The engine of neural networks: gradient-based optimization\r\n\r\nDerivative of a tensor operation: the gradient\r\n\r\nStochastic gradient descent\r\n\r\nChaining derivatives: the Backpropagation algorithm\r\n\r\nThe chain rule\r\n\r\nThe Gradient Tape in TensorFlow - The API through which you can leverage TensorFlow’s powerful automatic differentiation capabilities is the GradientTape.\r\n\r\n",
"_____no_output_____"
]
],
[
[
"x = tf.Variable(0.)\r\nwith tf.GradientTape() as tape:\r\n y = 2 * x + 3\r\ngrad_of_y_wrt_x = tape.gradient(y, x)",
"_____no_output_____"
],
[
"grad_of_y_wrt_x",
"_____no_output_____"
],
[
"W = tf.Variable(tf.random.uniform((2, 2)))\r\nb = tf.Variable(tf.zeros((2,)))\r\nx = tf.random.uniform((2, 2))\r\nwith tf.GradientTape() as tape:\r\n y = tf.matmul(W, x) + b\r\ngrad_of_y_wrt_W_and_b = tape.gradient(y, [W, b])",
"_____no_output_____"
],
[
"grad_of_y_wrt_W_and_b",
"_____no_output_____"
],
[
"(train_images, train_labels), (test_images, test_labels) = mnist.load_data()\r\ntrain_images = train_images.reshape((60000, 28 * 28))\r\ntrain_images = train_images.astype('float32') / 255\r\ntest_images = test_images.reshape((10000, 28 * 28))\r\ntest_images = test_images.astype('float32') / 255",
"Downloading data from https://storage.googleapis.com/tensorflow/tf-keras-datasets/mnist.npz\n11493376/11490434 [==============================] - 0s 0us/step\n"
],
[
"model = models.Sequential([\r\n layers.Dense(512, activation='relu'),\r\n layers.Dense(10, activation='softmax')\r\n])",
"_____no_output_____"
],
[
"model.compile(optimizer=\"rmsprop\",\r\n loss=\"sparse_categorical_crossentropy\",\r\n metrics=\"accuracy\")",
"_____no_output_____"
],
[
"model.fit(train_images, train_labels, epochs=5, batch_size=128)",
"Epoch 1/5\n469/469 [==============================] - 4s 8ms/step - loss: 0.2586 - accuracy: 0.9244\nEpoch 2/5\n469/469 [==============================] - 4s 8ms/step - loss: 0.1050 - accuracy: 0.9682\nEpoch 3/5\n469/469 [==============================] - 4s 8ms/step - loss: 0.0687 - accuracy: 0.9792\nEpoch 4/5\n469/469 [==============================] - 4s 8ms/step - loss: 0.0490 - accuracy: 0.9851\nEpoch 5/5\n469/469 [==============================] - 4s 8ms/step - loss: 0.0369 - accuracy: 0.9891\n"
]
],
[
[
"Implementing from scratch in TensorFlow\r\n\r\nLet’s implement a simple Python class NaiveDense that creates two TensorFlow variables W and b, and exposes a call method that applies the above transformation.",
"_____no_output_____"
]
],
[
[
"class NaiveDense:\r\n\r\n def __init__(self, input_size, output_size, activation):\r\n self.activation = activation\r\n\r\n w_shape = (input_size, output_size) # create a matrix W of shape \"(input_size, output_size)\", initialized with random values\r\n w_initial_value = tf.random.uniform(w_shape, minval=0, maxval=1e-1)\r\n self.W = tf.Variable(w_initial_value)\r\n\r\n b_shape = (output_size,) # create a vector b os shape (output_size, ), initialized with zeros\r\n b_initial_value = tf.zeros(b_shape)\r\n self.b = tf.Variable(b_initial_value)\r\n\r\n def __call__(self, inputs): # apply the forward pass\r\n return self.activation(tf.matmul(inputs, self.W) + self.b)\r\n\r\n @property\r\n def weights(self): # convinience method for rettrieving the layer weights\r\n return [self.W, self.b]",
"_____no_output_____"
]
],
[
[
"A simple Sequential class - create a NaiveSequential class to chain these layers. It wraps a list of layers, and exposes a call methods that simply call the underlying layers on the inputs, in order. It also features a weights property to easily keep track of the layers' parameters.",
"_____no_output_____"
]
],
[
[
"class NaiveSequential:\r\n\r\n def __init__(self, layers):\r\n self.layers = layers\r\n\r\n def __call__(self, inputs):\r\n x = inputs\r\n for layer in self.layers:\r\n x = layer(x)\r\n return x\r\n\r\n @property\r\n def weights(self):\r\n weights = []\r\n for layer in self.layers:\r\n weights += layer.weights\r\n return weights",
"_____no_output_____"
]
],
[
[
"Using this NaiveDense class and this NaiveSequential class, we can create a mock Keras model:",
"_____no_output_____"
]
],
[
[
"model = NaiveSequential([\r\n NaiveDense(input_size=28 * 28, output_size=512, activation=tf.nn.relu),\r\n NaiveDense(input_size=512, output_size=10, activation=tf.nn.softmax)\r\n])\r\nassert len(model.weights) == 4",
"_____no_output_____"
]
],
[
[
"A batch generator\r\n\r\nNext, we need a way to iterate over the MNIST data in mini-batches. This is easy:",
"_____no_output_____"
]
],
[
[
"class BatchGenerator:\r\n\r\n def __init__(self, images, labels, batch_size=128):\r\n self.index = 0\r\n self.images = images\r\n self.labels = labels\r\n self.batch_size = batch_size\r\n\r\n def next(self):\r\n images = self.images[self.index : self.index + self.batch_size]\r\n labels = self.labels[self.index : self.index + self.batch_size]\r\n self.index += self.batch_size\r\n return images, labels",
"_____no_output_____"
]
],
[
[
"Running one training step\r\n\r\nThe most difficult part of the process is the “training step”: updating the weights of the model after running it on one batch of data. We need to:\r\n\r\n1. Compute the predictions of the model for the images in the batch\r\n\r\n2. Compute the loss value for these predictions given the actual labels\r\n\r\n3. Compute the gradient of the loss with regard to the model’s weights\r\n\r\n4. Move the weights by a small amount in the direction opposite to the gradient\r\n\r\nTo compute the gradient, we will use the TensorFlow GradientTape object",
"_____no_output_____"
]
],
[
[
"learning_rate = 1e-3\r\n\r\ndef update_weights(gradients, weights):\r\n for g, w in zip(gradients, weights):\r\n w.assign_sub(w * learning_rate) # assign_sub is the equivalent of -= for TensorFlow variables",
"_____no_output_____"
],
[
"def one_training_step(model, images_batch, labels_batch):\r\n with tf.GradientTape() as tape: # run the \"forward pass\" (compute the model's predictions under the GradientTape scope)\r\n predictions = model(images_batch)\r\n per_sample_losses = tf.keras.losses.sparse_categorical_crossentropy(\r\n labels_batch, predictions)\r\n average_loss = tf.reduce_mean(per_sample_losses)\r\n gradients = tape.gradient(average_loss, model.weights) # compute the gradient of the loss with regard to the weights. The output gradients is a list where each entry corresponds to a weight from the models.weights list\r\n update_weights(gradients, model.weights) # update the weights using the gradients\r\n return average_loss",
"_____no_output_____"
]
],
[
[
"In practice, you will almost never implement a weight update step like this by hand. Instead, you would use an Optimizer instance from Keras. Like this:",
"_____no_output_____"
]
],
[
[
"optimizer = optimizers.SGD(learning_rate=1e-3)\r\n\r\ndef update_weights(gradients, weights):\r\n optimizer.apply_gradients(zip(gradients, weights))",
"_____no_output_____"
]
],
[
[
"The full training loop\r\n\r\nAn epoch of training simply consists of the repetition of the training step for each batch in the training data, and the full training loop is simply the repetition of one epoch:",
"_____no_output_____"
]
],
[
[
"def fit(model, images, labels, epochs, batch_size=128):\r\n for epoch_counter in range(epochs):\r\n print('Epoch %d' % epoch_counter)\r\n batch_generator = BatchGenerator(images, labels)\r\n for batch_counter in range(len(images) // batch_size):\r\n images_batch, labels_batch = batch_generator.next()\r\n loss = one_training_step(model, images_batch, labels_batch)\r\n if batch_counter % 100 == 0:\r\n print('loss at batch %d: %.2f' % (batch_counter, loss))",
"_____no_output_____"
],
[
"from tensorflow.keras.datasets import mnist\r\n(train_images, train_labels), (test_images, test_labels) = mnist.load_data()\r\n\r\ntrain_images = train_images.reshape((60000, 28 * 28))\r\ntrain_images = train_images.astype('float32') / 255\r\ntest_images = test_images.reshape((10000, 28 * 28))\r\ntest_images = test_images.astype('float32') / 255\r\n\r\nfit(model, train_images, train_labels, epochs=10, batch_size=128)",
"Epoch 0\nloss at batch 0: 6.83\nloss at batch 100: 2.24\nloss at batch 200: 2.21\nloss at batch 300: 2.12\nloss at batch 400: 2.22\nEpoch 1\nloss at batch 0: 1.93\nloss at batch 100: 1.89\nloss at batch 200: 1.84\nloss at batch 300: 1.75\nloss at batch 400: 1.84\nEpoch 2\nloss at batch 0: 1.61\nloss at batch 100: 1.59\nloss at batch 200: 1.52\nloss at batch 300: 1.46\nloss at batch 400: 1.53\nEpoch 3\nloss at batch 0: 1.33\nloss at batch 100: 1.35\nloss at batch 200: 1.26\nloss at batch 300: 1.24\nloss at batch 400: 1.29\nEpoch 4\nloss at batch 0: 1.12\nloss at batch 100: 1.16\nloss at batch 200: 1.05\nloss at batch 300: 1.07\nloss at batch 400: 1.12\nEpoch 5\nloss at batch 0: 0.97\nloss at batch 100: 1.02\nloss at batch 200: 0.90\nloss at batch 300: 0.95\nloss at batch 400: 1.00\nEpoch 6\nloss at batch 0: 0.86\nloss at batch 100: 0.91\nloss at batch 200: 0.80\nloss at batch 300: 0.85\nloss at batch 400: 0.91\nEpoch 7\nloss at batch 0: 0.77\nloss at batch 100: 0.83\nloss at batch 200: 0.72\nloss at batch 300: 0.78\nloss at batch 400: 0.84\nEpoch 8\nloss at batch 0: 0.71\nloss at batch 100: 0.76\nloss at batch 200: 0.65\nloss at batch 300: 0.72\nloss at batch 400: 0.79\nEpoch 9\nloss at batch 0: 0.66\nloss at batch 100: 0.70\nloss at batch 200: 0.60\nloss at batch 300: 0.67\nloss at batch 400: 0.75\n"
]
],
[
[
" Evaluating the model\r\n\r\n We can evaluate the model by taking the argmax of its predictions over the test images, and comparing it to the expected labels:",
"_____no_output_____"
]
],
[
[
"predictions = model(test_images)\r\npredictions = predictions.numpy() # calling .numpy() to a TensorFlow tensor converts it to a NumPy tensor\r\npredicted_labels = np.argmax(predictions, axis=1)\r\nmatches = predicted_labels == test_labels\r\n# print('accuracy: %.2f' % matches.average())\r\nprint(f\"Accuracy: {np.average(matches)}\")",
"Accuracy: 0.8318\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d086e811c916da88e8e31c35bffb6d5902839083 | 28,051 | ipynb | Jupyter Notebook | pipeline_2.ipynb | rocabrera/kaggles_enem | b7ef287ad23eed094ffad816d881a9431e785d57 | [
"MIT"
] | null | null | null | pipeline_2.ipynb | rocabrera/kaggles_enem | b7ef287ad23eed094ffad816d881a9431e785d57 | [
"MIT"
] | null | null | null | pipeline_2.ipynb | rocabrera/kaggles_enem | b7ef287ad23eed094ffad816d881a9431e785d57 | [
"MIT"
] | null | null | null | 34.292176 | 298 | 0.529072 | [
[
[
"import numpy as np\nimport pandas as pd\nfrom tqdm import tqdm\nfrom utils import clean_target\nfrom categorical_ordinal import get_categorical_ordinal_columns\nfrom categorical_nominal import get_categorical_nominal_columns\nfrom columns_transformers import ColumnSelector\nfrom sklearn.pipeline import Pipeline, FeatureUnion\nfrom sklearn.impute import SimpleImputer\nfrom sklearn.ensemble import RandomForestRegressor\nfrom sklearn.preprocessing import OrdinalEncoder, OneHotEncoder, MinMaxScaler\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import mean_absolute_error, mean_squared_error",
"_____no_output_____"
]
],
[
[
"<font color=\"orange\"> <b>Grupos de features:</b></font>\n\n- **Categorial Ordinal:**\n - TP_ (17-4-3 = 10)\n - Questions: ```[\"Q001\", \"Q002\", \"Q003\", \"Q004\", \"Q005\",\"Q006\", \"Q007\", \"Q008\", \"Q009\", \"Q010\", \"Q011\", \"Q012\", \"Q013\", \"Q014\", \"Q015\", \"Q016\", \"Q017\", \"Q019\", \"Q022\", \"Q024\"]``` (20)\n \n- **Categorial Nominal:**\n - IN_ : All Binary (52) \n - TP_ : ```[\"TP_SEXO\", \"TP_ESTADO_CIVIL\", \"TP_COR_RACA\", \"TP_NACIONALIDADE\"]``` (4)\n - SG_ : (4-1 = 3)\n - Questions: ```[\"Q018\", \"Q020\", \"Q021\", \"Q023\", \"Q025\"]``` (5)\n \n- **Numerical:**\n - NU_IDADE (1)\n\n- Droped:\n - Identificator: ```[NU_INSCRICAO]``` (1)\n - More than 40% missing: ```['CO_ESCOLA', 'NO_MUNICIPIO_ESC', 'SG_UF_ESC', 'TP_DEPENDENCIA_ADM_ESC', 'TP_LOCALIZACAO_ESC', 'TP_SIT_FUNC_ESC']``` (4)\n - NO_M: (To many categories): ```['NO_MUNICIPIO_RESIDENCIA', 'NO_MUNICIPIO_NASCIMENTO', 'NO_MUNICIPIO_PROVA']``` (3)\n - NU_NOTA: Targets variables (5)",
"_____no_output_____"
]
],
[
[
"train_df = pd.read_parquet(\"data/train.parquet\")\nclean_target(train_df)\n#test= pd.read_parquet(\"data/test.parquet\")",
"_____no_output_____"
],
[
"categorical_ordinal_columns = get_categorical_ordinal_columns(train_df)\nqtd_categorical_ordinal_columns=len(categorical_ordinal_columns)\nprint(f\"Number of categorial ordinal features: {qtd_categorical_ordinal_columns}\")",
"Number of categorial ordinal features: 30\n"
],
[
"categorical_nominal_columns = get_categorical_nominal_columns(train_df)\nqtd_categorical_nominal_columns = len(categorical_nominal_columns)\nprint(f\"Number of categorial nominal features: {qtd_categorical_nominal_columns}\")",
"Number of categorial nominal features: 64\n"
],
[
"drop_columns = [\"NU_INSCRICAO\", \n \"CO_ESCOLA\", \n \"NO_MUNICIPIO_ESC\", \n \"SG_UF_ESC\", \n \"TP_DEPENDENCIA_ADM_ESC\", \n \"TP_LOCALIZACAO_ESC\", \n \"TP_SIT_FUNC_ESC\", \n \"NO_MUNICIPIO_RESIDENCIA\", \n \"NO_MUNICIPIO_NASCIMENTO\", \n \"NO_MUNICIPIO_PROVA\"]\nqtd_drop_columns = len(drop_columns)\nprint(f\"Number of columns dropped: {qtd_drop_columns}\")",
"Number of columns dropped: 10\n"
],
[
"target_columns = train_df.filter(regex=\"NU_NOTA\").columns.tolist()\nqtd_target_columns = len(target_columns)\nprint(f\"Number of targets: {qtd_target_columns}\")",
"Number of targets: 5\n"
],
[
"numerical_columns = [\"NU_IDADE\"]\nqtd_numerical_columns = len(numerical_columns)\nprint(f\"Number of targets: {qtd_numerical_columns}\")",
"Number of targets: 1\n"
],
[
"target_columns = train_df.filter(regex=\"NU_NOTA\").columns.tolist()\nqtd_target_columns = len(target_columns)\nprint(f\"Number of targets: {qtd_target_columns}\")",
"Number of targets: 5\n"
],
[
"all_columns = drop_columns + categorical_nominal_columns + categorical_ordinal_columns + numerical_columns + target_columns\nqtd_total = qtd_drop_columns + qtd_categorical_nominal_columns + qtd_categorical_ordinal_columns + qtd_numerical_columns + qtd_target_columns\nprint(f\"Total columns: {qtd_total}\")",
"Total columns: 110\n"
]
],
[
[
"## **Create Pipeline**",
"_____no_output_____"
]
],
[
[
"\"\"\"\nVariáveis categóricas com dados ordinais que tem dados faltantes:\n- TP_ENSINO: Suposto que NaN representa a categoria faltante descrita nos metadados.\n- TP_STATUS_REDACAO: Mapeado para outra classe (Faltou na prova)\n\"\"\"\ncategorical_ordinal_pipe = Pipeline([\n ('selector', ColumnSelector(categorical_ordinal_columns)),\n ('imputer', SimpleImputer(missing_values=np.nan, \n strategy='constant', \n fill_value=0)),\n ('encoder', OrdinalEncoder(handle_unknown='use_encoded_value', unknown_value=-1))\n])\n\n\"\"\"\nVariáveis categóricas com dados ordinais que tem dados faltantes:\n- SG_UF_NASCIMENTO: Mapeado para uma nova categoria\n\"\"\"\ncategorical_nominal_pipe = Pipeline([\n ('selector', ColumnSelector(categorical_nominal_columns)),\n ('imputer', SimpleImputer(missing_values=np.nan, \n strategy='constant', \n fill_value=\"missing\")),\n ('encoder', OneHotEncoder(drop=\"first\", handle_unknown='ignore'))\n])\n\nnumerical_pipe = Pipeline([\n ('selector', ColumnSelector(numerical_columns)),\n ('imputer', SimpleImputer(missing_values=np.nan, \n strategy='constant', \n fill_value=0)),\n ('scaler', MinMaxScaler())\n])\n\n\npreprocessor = FeatureUnion([\n ('categorical_ordinal', categorical_ordinal_pipe),\n ('categorical_nominal', categorical_nominal_pipe),\n ('numerical', numerical_pipe)\n])\n\nkwargs_regressor = {\"n_estimators\":50,\n \"n_jobs\":-1,\n \"verbose\":2}\n\npipe = Pipeline([\n ('preprocessor', preprocessor),\n ('feature_selection', VarianceThreshold(threshold=0.05)),\n ('model', RandomForestRegressor(**kwargs_regressor))\n])",
"_____no_output_____"
],
[
"n_samples = 1000\nX = train_df.sample(n_samples).drop(columns=target_columns+drop_columns)\ny = train_df.sample(n_samples).filter(regex=\"NU_NOTA\")",
"_____no_output_____"
],
[
"X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.1, random_state=42)",
"_____no_output_____"
],
[
"def split_target(y):\n y_nu_nota_cn = y[\"NU_NOTA_CN\"]\n y_nu_nota_ch = y[\"NU_NOTA_CH\"]\n y_nu_nota_lc = y[\"NU_NOTA_LC\"]\n y_nu_nota_mt = y[\"NU_NOTA_MT\"]\n y_nu_nota_redacao = y[\"NU_NOTA_REDACAO\"]\n return (y_nu_nota_cn, y_nu_nota_ch, y_nu_nota_lc, y_nu_nota_mt, y_nu_nota_redacao)\n\ny_train_cn, y_train_ch, y_train_lc, y_train_mt, y_train_redacao = split_target(y_train)\ny_test_cn, y_test_ch, y_test_lc, y_test_mt, y_test_redacao = split_target(y_test)\n\ny_structure = {\"NU_NOTA_CN\":[y_train_cn, y_test_cn], \n \"NU_NOTA_CH\":[y_train_ch, y_test_ch],\n \"NU_NOTA_LC\":[y_train_lc, y_test_lc],\n \"NU_NOTA_MT\":[y_train_mt, y_test_mt],\n \"NU_NOTA_REDACAO\":[y_train_redacao, y_test_redacao]}",
"_____no_output_____"
],
[
"from joblib import dump\n\nfor key, ys in tqdm(y_structure.items()):\n \n pipe.fit(X_train, ys[0])\n dump(pipe, f\"models/model_{key}.joblib\") \n y_train_hat = pipe.predict(X_train)\n ys.append(y_train_hat)\n y_test_hat = pipe.predict(X_test)\n ys.append(y_test_hat)",
" 0%| | 0/5 [00:00<?, ?it/s][Parallel(n_jobs=-1)]: Using backend ThreadingBackend with 8 concurrent workers.\n[Parallel(n_jobs=-1)]: Done 25 tasks | elapsed: 0.1s\n"
],
[
"for key, ys in tqdm(y_structure.items()):\n train_error = mean_squared_error(ys[0], ys[2], squared=False)\n test_error = mean_squared_error(ys[1], ys[3], squared=False)\n print(key)\n print(f\"Train: {train_error}\")\n print(f\"Test: {test_error}\\n\") ",
"100%|██████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 5/5 [00:00<00:00, 1737.78it/s]"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d086ee94d69484308a00a3318c080961c32678ae | 408,938 | ipynb | Jupyter Notebook | final_project_files/code/NLP_Project_Neural_Network_Model.ipynb | PrudhviVajja/Mutation_Prediction | ce7019f94b27450ddaf51448816947fa6c683d4c | [
"Apache-2.0"
] | null | null | null | final_project_files/code/NLP_Project_Neural_Network_Model.ipynb | PrudhviVajja/Mutation_Prediction | ce7019f94b27450ddaf51448816947fa6c683d4c | [
"Apache-2.0"
] | null | null | null | final_project_files/code/NLP_Project_Neural_Network_Model.ipynb | PrudhviVajja/Mutation_Prediction | ce7019f94b27450ddaf51448816947fa6c683d4c | [
"Apache-2.0"
] | null | null | null | 135.186116 | 24,410 | 0.723469 | [
[
[
"# importing all the required libraries\r\nimport pandas as pd\r\nfrom google.colab import files\r\nimport io\r\nimport spacy\r\nfrom sklearn.feature_extraction.text import CountVectorizer\r\nfrom sklearn.model_selection import train_test_split\r\nfrom sklearn.preprocessing import LabelEncoder\r\nimport keras\r\nfrom keras.utils import to_categorical\r\nfrom keras import Sequential\r\nfrom keras.layers import Dense\r\nfrom keras.layers import Input\r\nfrom keras.layers import Softmax\r\nfrom sklearn.metrics import classification_report, confusion_matrix\r\nimport matplotlib.pyplot as plt\r\nimport sklearn.decomposition\r\nimport keras.callbacks\r\nimport pickle\r\nimport re\r\nimport nltk\r\nfrom nltk.stem import PorterStemmer",
"_____no_output_____"
],
[
"uploaded = files.upload()",
"_____no_output_____"
]
],
[
[
"Defining custom early stopper classes for early stopping of model.fit keras method",
"_____no_output_____"
]
],
[
[
"class CustomStopper(keras.callbacks.EarlyStopping):\r\n def __init__(self, monitor='val_loss',\r\n min_delta=0, patience=10, verbose=0, mode='auto', start_epoch = 30): # add argument for starting epoch\r\n super(CustomStopper, self).__init__()\r\n self.start_epoch = start_epoch\r\n\r\n def on_epoch_end(self, epoch, logs=None):\r\n if epoch > self.start_epoch:\r\n super().on_epoch_end(epoch, logs)",
"_____no_output_____"
]
],
[
[
"Defining variables to be passes in the various methods",
"_____no_output_____"
]
],
[
[
"filename = 'Data_v1.xlsx' #file name of the uploaded dataset file \r\nmodelName = 'Model1' #name of the model, this will be used to save model evaluation and history\r\nnumEpochs = 150 # maximum number of epochs if early stopping doesnt work\r\nbatchsize = 50 # batchsize which will be used in each step by optimizer defined in the model\r\noptimizer = 'adadelta' #optimizer to be used in model.fit keras method\r\n",
"_____no_output_____"
]
],
[
[
"Method to read uploaded file. Returns back the text input samples and target labels for each. Transforms X to a vector which holds the number of occurences of each word for every sample",
"_____no_output_____"
]
],
[
[
"def mypreprocessor(text):\r\n porter_stemmer = PorterStemmer()\r\n words=re.split(\"\\\\s+\",text)\r\n stemmed_words=[porter_stemmer.stem(word=word) for word in words]\r\n return ' '.join(stemmed_words)\r\n \r\ndef Preprocessing():\r\n \r\n X = pd.read_excel(list(uploaded.items())[0][0],usecols=\"H\") #pass usecols as the column containing all the training samples\r\n y = pd.read_excel(list(uploaded.items())[0][0],usecols=\"F\") #pass usecols as the column containing all the target labels\r\n X = [str(i) for i in X.extracted_text.to_list()] #the property used with X. should match column name in excel\r\n \r\n # for i in range(len(X)):\r\n # X[i] = re.sub(r'(\\s\\d+\\s)|{\\d+}|\\(\\d+\\)','',X[i])\r\n # X[i] = re.sub(r'gain-of-function|gain of function|toxic gain of function|activating mutation|constitutively active|hypermorph|ectopic expression|neomorph|gain of interaction|function protein|fusion transcript','GOF',X[i])\r\n # X[i] = re.sub(r'haploinsufficiency|haploinsufficient|hypomorph|amorph|null mutation|hemizygous','HI',X[i])\r\n # X[i] = re.sub(r'dominant-negative|dominant negative|antimorph','DN',X[i])\r\n # X[i] = re.sub(r'loss of function|loss-of-function','LOF',X[i])\r\n\r\n # X = preprocess_data(X)\r\n y = y.mutation_consequence.to_list()\r\n # vocabulary = ['gain-of-function','gain of function',\r\n # 'toxic gain of function','activating mutation',\r\n # 'constitutively active','hypermorph','ectopic expression',\r\n # 'neomorph','gain of interaction','function protein','fusion transcript',\r\n # 'haploinsufficiency','haploinsufficient','hypomorph','amorph',\r\n # 'null mutation','hemizygous','dominant-negative','dominant negative','antimorph',\r\n # 'loss of function','loss-of-function']\r\n X=TfidfVectorizer(X,preprocessor=mypreprocessor,max_df=200 ,ngram_range=(1, 2)).fit(X).transform(X)\r\n # X=CountVectorizer(X,preprocessor=mypreprocessor,max_df=200 ,ngram_range=(1, 2)).fit(X).transform(X)\r\n return X, y",
"_____no_output_____"
]
],
[
[
"Method to split the dataset into training and testing. Changes y to one-hot encoded vector, e.g if target class is 3, then returns [0,0,0,1,0] for 5 target classes",
"_____no_output_____"
]
],
[
[
"def TrainTestSplit(X, y):\r\n X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.20, random_state=100, stratify = y) #split the dataset, test_Size variable defines the size of the test dataset, stratify column makes sure even distribution of target labels\r\n\r\n X_train = X_train.toarray() #changing to numpy array to work with keras sequential model\r\n X_test = X_test.toarray() #changing to numpy array to work with keras sequential model\r\n le = LabelEncoder() \r\n y_train = to_categorical(le.fit(y_train).transform(y_train))\r\n y_test = to_categorical(le.fit(y_test).transform(y_test))\r\n return X_train, X_test, y_train, y_test, le.classes_ # returns training and test datasets, as well as class names",
"_____no_output_____"
]
],
[
[
"Defining the model to be used for training the datasets.\r\n",
"_____no_output_____"
]
],
[
[
"def ModelBuild(X, y):\r\n inputs = keras.layers.Input(shape=(len(X_train[0]),))\r\n dense1 = keras.layers.Dense(200, activation=\"relu\")(inputs) #fully connected with input vectors\r\n \r\n \r\n # dropout = keras.layers.Dropout(0.2)(dense1) #regularization layer if required\r\n dense2 = keras.layers.Dense(50, activation=\"relu\")(dense1) #fully connected with Layer 1\r\n # dropout2 = keras.layers.Dropout(0.1)(dense2) #regularization layer if required\r\n # dense3 = keras.layers.Dense(50, activation=\"relu\")(dense2)\r\n \r\n outputs = keras.layers.Dense(len(y_train[0]), activation=\"sigmoid\")(dense2) #output layer\r\n model = keras.Model(inputs=inputs, outputs=outputs)\r\n return model",
"_____no_output_____"
]
],
[
[
"Method to show summary of the model as well as the shape in diagram form",
"_____no_output_____"
]
],
[
[
"def PlotModel(model, filename):\r\n model.summary()\r\n keras.utils.plot_model(model, filename, show_shapes=True)",
"_____no_output_____"
]
],
[
[
"Method to compile the defined model as well as run the training. Returns a history variable which can be used to plot training and validation loss as well as accuracy at every epoch",
"_____no_output_____"
]
],
[
[
"def PlotTraining(model, X_test, y_test):\r\n model.compile(loss='categorical_crossentropy',optimizer=optimizer,metrics=[keras.metrics.CategoricalAccuracy(),'accuracy'])\r\n # EarlyStoppage = CustomStopper()\r\n es = keras.callbacks.EarlyStopping(monitor='val_accuracy', baseline=0.7, patience=30)\r\n history = model.fit(X_train, y_train,validation_split=0.2,epochs=numEpochs, batch_size=batchsize) #,callbacks = [es] ) - use this for early stopping\r\n model.evaluate(X_test, y_test)\r\n return history\r\n ",
"_____no_output_____"
]
],
[
[
"Plots the validation and training accuracy at every epoch using a history object obtained by model.fit in the previous step",
"_____no_output_____"
]
],
[
[
"def plot(history):\r\n # list all data in history\r\n print(history.keys())\r\n # summarize history for accuracy\r\n plt.plot(history['accuracy'])\r\n plt.plot(history['val_accuracy'])\r\n plt.title('model accuracy')\r\n plt.ylabel('accuracy')\r\n plt.xlabel('epoch')\r\n plt.legend(['train', 'test'], loc='upper left')\r\n plt.show()\r\n # summarize history for loss\r\n plt.plot(history['loss'])\r\n plt.plot(history['val_loss'])\r\n plt.title('model loss')\r\n plt.ylabel('loss')\r\n plt.xlabel('epoch')\r\n plt.legend(['train', 'test'], loc='upper left')\r\n plt.show()",
"_____no_output_____"
]
],
[
[
"Calling the methods to run all the required steps in the pipeline",
"_____no_output_____"
]
],
[
[
"X, y = Preprocessing()\r\nX_train, X_test, y_train, y_test, ClassNames = TrainTestSplit(X, y)\r\n# svd = sklearn.decomposition.TruncatedSVD(n_components=60, n_iter=5, random_state=42)\r\n# X_train = svd.fit(X_train).transform(X_train)\r\n# svd = sklearn.decomposition.TruncatedSVD(n_components=60, n_iter=5, random_state=42)\r\n# X_test = svd.fit(X_test).transform(X_test)\r\nmodel = ModelBuild(X_train, y_train)\r\nPlotModel(model, modelName +\".png\")\r\nhistory = PlotTraining(model, X_test, y_test)\r\nprint(confusion_matrix(y_test.argmax(axis=-1),model.predict(X_test).argmax(axis=-1)))\r\nprint(classification_report(y_test.argmax(axis=-1), model.predict(X_test).argmax(axis=-1),target_names=ClassNames))\r\n",
"Model: \"model\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_1 (InputLayer) [(None, 112443)] 0 \n_________________________________________________________________\ndense (Dense) (None, 200) 22488800 \n_________________________________________________________________\ndropout (Dropout) (None, 200) 0 \n_________________________________________________________________\ndense_1 (Dense) (None, 100) 20100 \n_________________________________________________________________\ndropout_1 (Dropout) (None, 100) 0 \n_________________________________________________________________\ndense_2 (Dense) (None, 5) 505 \n=================================================================\nTotal params: 22,509,405\nTrainable params: 22,509,405\nNon-trainable params: 0\n_________________________________________________________________\nEpoch 1/150\n7/7 [==============================] - 3s 256ms/step - loss: 1.6442 - categorical_accuracy: 0.1841 - accuracy: 0.1841 - val_loss: 1.6049 - val_categorical_accuracy: 0.1750 - val_accuracy: 0.1750\nEpoch 2/150\n7/7 [==============================] - 1s 217ms/step - loss: 1.6283 - categorical_accuracy: 0.1839 - accuracy: 0.1839 - val_loss: 1.6048 - val_categorical_accuracy: 0.1750 - val_accuracy: 0.1750\nEpoch 3/150\n7/7 [==============================] - 1s 216ms/step - loss: 1.6161 - categorical_accuracy: 0.1903 - accuracy: 0.1903 - val_loss: 1.6047 - val_categorical_accuracy: 0.1750 - val_accuracy: 0.1750\nEpoch 4/150\n7/7 [==============================] - 1s 215ms/step - loss: 1.6238 - categorical_accuracy: 0.1794 - accuracy: 0.1794 - val_loss: 1.6046 - val_categorical_accuracy: 0.1750 - val_accuracy: 0.1750\nEpoch 5/150\n7/7 [==============================] - 2s 236ms/step - loss: 1.6016 - categorical_accuracy: 0.2155 - accuracy: 0.2155 - val_loss: 1.6045 - val_categorical_accuracy: 0.1750 - val_accuracy: 0.1750\nEpoch 6/150\n7/7 [==============================] - 1s 212ms/step - loss: 1.6197 - categorical_accuracy: 0.1839 - accuracy: 0.1839 - val_loss: 1.6044 - val_categorical_accuracy: 0.2000 - val_accuracy: 0.2000\nEpoch 7/150\n7/7 [==============================] - 2s 217ms/step - loss: 1.6163 - categorical_accuracy: 0.1786 - accuracy: 0.1786 - val_loss: 1.6042 - val_categorical_accuracy: 0.2000 - val_accuracy: 0.2000\nEpoch 8/150\n7/7 [==============================] - 1s 215ms/step - loss: 1.6090 - categorical_accuracy: 0.1873 - accuracy: 0.1873 - val_loss: 1.6040 - val_categorical_accuracy: 0.2000 - val_accuracy: 0.2000\nEpoch 9/150\n7/7 [==============================] - 1s 215ms/step - loss: 1.5972 - categorical_accuracy: 0.2204 - accuracy: 0.2204 - val_loss: 1.6038 - val_categorical_accuracy: 0.2000 - val_accuracy: 0.2000\nEpoch 10/150\n7/7 [==============================] - 1s 212ms/step - loss: 1.6203 - categorical_accuracy: 0.1772 - accuracy: 0.1772 - val_loss: 1.6037 - val_categorical_accuracy: 0.2000 - val_accuracy: 0.2000\nEpoch 11/150\n7/7 [==============================] - 2s 218ms/step - loss: 1.6023 - categorical_accuracy: 0.2138 - accuracy: 0.2138 - val_loss: 1.6036 - val_categorical_accuracy: 0.2000 - val_accuracy: 0.2000\nEpoch 12/150\n7/7 [==============================] - 1s 212ms/step - loss: 1.5857 - categorical_accuracy: 0.2411 - accuracy: 0.2411 - val_loss: 1.6036 - val_categorical_accuracy: 0.2000 - val_accuracy: 0.2000\nEpoch 13/150\n7/7 [==============================] - 1s 214ms/step - loss: 1.5898 - categorical_accuracy: 0.2313 - accuracy: 0.2313 - val_loss: 1.6035 - val_categorical_accuracy: 0.2000 - val_accuracy: 0.2000\nEpoch 14/150\n7/7 [==============================] - 1s 215ms/step - loss: 1.6034 - categorical_accuracy: 0.2166 - accuracy: 0.2166 - val_loss: 1.6034 - val_categorical_accuracy: 0.2000 - val_accuracy: 0.2000\nEpoch 15/150\n7/7 [==============================] - 1s 218ms/step - loss: 1.5932 - categorical_accuracy: 0.2222 - accuracy: 0.2222 - val_loss: 1.6034 - val_categorical_accuracy: 0.2000 - val_accuracy: 0.2000\nEpoch 16/150\n7/7 [==============================] - 1s 213ms/step - loss: 1.5769 - categorical_accuracy: 0.2633 - accuracy: 0.2633 - val_loss: 1.6032 - val_categorical_accuracy: 0.2000 - val_accuracy: 0.2000\nEpoch 17/150\n7/7 [==============================] - 1s 216ms/step - loss: 1.5713 - categorical_accuracy: 0.2482 - accuracy: 0.2482 - val_loss: 1.6031 - val_categorical_accuracy: 0.2000 - val_accuracy: 0.2000\nEpoch 18/150\n7/7 [==============================] - 1s 215ms/step - loss: 1.5691 - categorical_accuracy: 0.2749 - accuracy: 0.2749 - val_loss: 1.6030 - val_categorical_accuracy: 0.2000 - val_accuracy: 0.2000\nEpoch 19/150\n7/7 [==============================] - 1s 213ms/step - loss: 1.5695 - categorical_accuracy: 0.2535 - accuracy: 0.2535 - val_loss: 1.6030 - val_categorical_accuracy: 0.2000 - val_accuracy: 0.2000\nEpoch 20/150\n7/7 [==============================] - 1s 214ms/step - loss: 1.5749 - categorical_accuracy: 0.2750 - accuracy: 0.2750 - val_loss: 1.6029 - val_categorical_accuracy: 0.2125 - val_accuracy: 0.2125\nEpoch 21/150\n7/7 [==============================] - 1s 216ms/step - loss: 1.5612 - categorical_accuracy: 0.3107 - accuracy: 0.3107 - val_loss: 1.6028 - val_categorical_accuracy: 0.2125 - val_accuracy: 0.2125\nEpoch 22/150\n7/7 [==============================] - 2s 237ms/step - loss: 1.5483 - categorical_accuracy: 0.3056 - accuracy: 0.3056 - val_loss: 1.6030 - val_categorical_accuracy: 0.2125 - val_accuracy: 0.2125\nEpoch 23/150\n7/7 [==============================] - 1s 217ms/step - loss: 1.5402 - categorical_accuracy: 0.3241 - accuracy: 0.3241 - val_loss: 1.6029 - val_categorical_accuracy: 0.2125 - val_accuracy: 0.2125\nEpoch 24/150\n7/7 [==============================] - 1s 216ms/step - loss: 1.5521 - categorical_accuracy: 0.3151 - accuracy: 0.3151 - val_loss: 1.6028 - val_categorical_accuracy: 0.2125 - val_accuracy: 0.2125\nEpoch 25/150\n7/7 [==============================] - 2s 222ms/step - loss: 1.5569 - categorical_accuracy: 0.3428 - accuracy: 0.3428 - val_loss: 1.6028 - val_categorical_accuracy: 0.2125 - val_accuracy: 0.2125\nEpoch 26/150\n7/7 [==============================] - 1s 214ms/step - loss: 1.5510 - categorical_accuracy: 0.2997 - accuracy: 0.2997 - val_loss: 1.6029 - val_categorical_accuracy: 0.2125 - val_accuracy: 0.2125\nEpoch 27/150\n7/7 [==============================] - 1s 215ms/step - loss: 1.5338 - categorical_accuracy: 0.3602 - accuracy: 0.3602 - val_loss: 1.6029 - val_categorical_accuracy: 0.2125 - val_accuracy: 0.2125\nEpoch 28/150\n7/7 [==============================] - 2s 216ms/step - loss: 1.5475 - categorical_accuracy: 0.2996 - accuracy: 0.2996 - val_loss: 1.6028 - val_categorical_accuracy: 0.2000 - val_accuracy: 0.2000\nEpoch 29/150\n7/7 [==============================] - 2s 218ms/step - loss: 1.5347 - categorical_accuracy: 0.3353 - accuracy: 0.3353 - val_loss: 1.6028 - val_categorical_accuracy: 0.2125 - val_accuracy: 0.2125\nEpoch 30/150\n7/7 [==============================] - 2s 219ms/step - loss: 1.5496 - categorical_accuracy: 0.2842 - accuracy: 0.2842 - val_loss: 1.6029 - val_categorical_accuracy: 0.2125 - val_accuracy: 0.2125\nEpoch 31/150\n7/7 [==============================] - 1s 215ms/step - loss: 1.5229 - categorical_accuracy: 0.3587 - accuracy: 0.3587 - val_loss: 1.6029 - val_categorical_accuracy: 0.2125 - val_accuracy: 0.2125\nEpoch 32/150\n7/7 [==============================] - 2s 214ms/step - loss: 1.5368 - categorical_accuracy: 0.3555 - accuracy: 0.3555 - val_loss: 1.6029 - val_categorical_accuracy: 0.2125 - val_accuracy: 0.2125\nEpoch 33/150\n7/7 [==============================] - 1s 213ms/step - loss: 1.5285 - categorical_accuracy: 0.3483 - accuracy: 0.3483 - val_loss: 1.6029 - val_categorical_accuracy: 0.2125 - val_accuracy: 0.2125\nEpoch 34/150\n7/7 [==============================] - 1s 216ms/step - loss: 1.5061 - categorical_accuracy: 0.4151 - accuracy: 0.4151 - val_loss: 1.6030 - val_categorical_accuracy: 0.2125 - val_accuracy: 0.2125\nEpoch 35/150\n7/7 [==============================] - 1s 215ms/step - loss: 1.5045 - categorical_accuracy: 0.4305 - accuracy: 0.4305 - val_loss: 1.6029 - val_categorical_accuracy: 0.2000 - val_accuracy: 0.2000\nEpoch 36/150\n7/7 [==============================] - 2s 238ms/step - loss: 1.5233 - categorical_accuracy: 0.3564 - accuracy: 0.3564 - val_loss: 1.6029 - val_categorical_accuracy: 0.2125 - val_accuracy: 0.2125\nEpoch 37/150\n7/7 [==============================] - 1s 213ms/step - loss: 1.5046 - categorical_accuracy: 0.4016 - accuracy: 0.4016 - val_loss: 1.6031 - val_categorical_accuracy: 0.2125 - val_accuracy: 0.2125\nEpoch 38/150\n7/7 [==============================] - 2s 218ms/step - loss: 1.4936 - categorical_accuracy: 0.4299 - accuracy: 0.4299 - val_loss: 1.6030 - val_categorical_accuracy: 0.2125 - val_accuracy: 0.2125\nEpoch 39/150\n7/7 [==============================] - 2s 217ms/step - loss: 1.5201 - categorical_accuracy: 0.3919 - accuracy: 0.3919 - val_loss: 1.6031 - val_categorical_accuracy: 0.2125 - val_accuracy: 0.2125\nEpoch 40/150\n7/7 [==============================] - 2s 218ms/step - loss: 1.4934 - categorical_accuracy: 0.4193 - accuracy: 0.4193 - val_loss: 1.6030 - val_categorical_accuracy: 0.2125 - val_accuracy: 0.2125\nEpoch 41/150\n7/7 [==============================] - 2s 222ms/step - loss: 1.5053 - categorical_accuracy: 0.4039 - accuracy: 0.4039 - val_loss: 1.6030 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 42/150\n7/7 [==============================] - 2s 226ms/step - loss: 1.4845 - categorical_accuracy: 0.4606 - accuracy: 0.4606 - val_loss: 1.6030 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 43/150\n7/7 [==============================] - 2s 219ms/step - loss: 1.4932 - categorical_accuracy: 0.4201 - accuracy: 0.4201 - val_loss: 1.6031 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 44/150\n7/7 [==============================] - 2s 220ms/step - loss: 1.4954 - categorical_accuracy: 0.4085 - accuracy: 0.4085 - val_loss: 1.6031 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 45/150\n7/7 [==============================] - 2s 220ms/step - loss: 1.4988 - categorical_accuracy: 0.4175 - accuracy: 0.4175 - val_loss: 1.6031 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 46/150\n7/7 [==============================] - 2s 218ms/step - loss: 1.4959 - categorical_accuracy: 0.4129 - accuracy: 0.4129 - val_loss: 1.6031 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 47/150\n7/7 [==============================] - 2s 219ms/step - loss: 1.4611 - categorical_accuracy: 0.4983 - accuracy: 0.4983 - val_loss: 1.6031 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 48/150\n7/7 [==============================] - 2s 225ms/step - loss: 1.4856 - categorical_accuracy: 0.4559 - accuracy: 0.4559 - val_loss: 1.6032 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 49/150\n7/7 [==============================] - 2s 219ms/step - loss: 1.4688 - categorical_accuracy: 0.4731 - accuracy: 0.4731 - val_loss: 1.6032 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 50/150\n7/7 [==============================] - 2s 220ms/step - loss: 1.4827 - categorical_accuracy: 0.4351 - accuracy: 0.4351 - val_loss: 1.6031 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 51/150\n7/7 [==============================] - 2s 217ms/step - loss: 1.4691 - categorical_accuracy: 0.4577 - accuracy: 0.4577 - val_loss: 1.6031 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 52/150\n7/7 [==============================] - 2s 218ms/step - loss: 1.4572 - categorical_accuracy: 0.5097 - accuracy: 0.5097 - val_loss: 1.6031 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 53/150\n7/7 [==============================] - 1s 216ms/step - loss: 1.4810 - categorical_accuracy: 0.4626 - accuracy: 0.4626 - val_loss: 1.6031 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 54/150\n7/7 [==============================] - 2s 218ms/step - loss: 1.4631 - categorical_accuracy: 0.4704 - accuracy: 0.4704 - val_loss: 1.6032 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 55/150\n7/7 [==============================] - 2s 219ms/step - loss: 1.4488 - categorical_accuracy: 0.5076 - accuracy: 0.5076 - val_loss: 1.6033 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 56/150\n7/7 [==============================] - 2s 220ms/step - loss: 1.4598 - categorical_accuracy: 0.4709 - accuracy: 0.4709 - val_loss: 1.6034 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 57/150\n7/7 [==============================] - 2s 240ms/step - loss: 1.4289 - categorical_accuracy: 0.5455 - accuracy: 0.5455 - val_loss: 1.6034 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 58/150\n7/7 [==============================] - 2s 224ms/step - loss: 1.4545 - categorical_accuracy: 0.4822 - accuracy: 0.4822 - val_loss: 1.6035 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 59/150\n7/7 [==============================] - 2s 220ms/step - loss: 1.4483 - categorical_accuracy: 0.4910 - accuracy: 0.4910 - val_loss: 1.6036 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 60/150\n7/7 [==============================] - 2s 223ms/step - loss: 1.4370 - categorical_accuracy: 0.5448 - accuracy: 0.5448 - val_loss: 1.6035 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 61/150\n7/7 [==============================] - 2s 219ms/step - loss: 1.4641 - categorical_accuracy: 0.5216 - accuracy: 0.5216 - val_loss: 1.6036 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 62/150\n7/7 [==============================] - 2s 220ms/step - loss: 1.4266 - categorical_accuracy: 0.5326 - accuracy: 0.5326 - val_loss: 1.6036 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 63/150\n7/7 [==============================] - 2s 222ms/step - loss: 1.4530 - categorical_accuracy: 0.4994 - accuracy: 0.4994 - val_loss: 1.6036 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 64/150\n7/7 [==============================] - 2s 217ms/step - loss: 1.4321 - categorical_accuracy: 0.5254 - accuracy: 0.5254 - val_loss: 1.6036 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 65/150\n7/7 [==============================] - 2s 227ms/step - loss: 1.4360 - categorical_accuracy: 0.5375 - accuracy: 0.5375 - val_loss: 1.6037 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 66/150\n7/7 [==============================] - 2s 217ms/step - loss: 1.4451 - categorical_accuracy: 0.5252 - accuracy: 0.5252 - val_loss: 1.6036 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 67/150\n7/7 [==============================] - 2s 222ms/step - loss: 1.4387 - categorical_accuracy: 0.5283 - accuracy: 0.5283 - val_loss: 1.6037 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 68/150\n7/7 [==============================] - 2s 223ms/step - loss: 1.4255 - categorical_accuracy: 0.5467 - accuracy: 0.5467 - val_loss: 1.6036 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 69/150\n7/7 [==============================] - 2s 220ms/step - loss: 1.4209 - categorical_accuracy: 0.5649 - accuracy: 0.5649 - val_loss: 1.6036 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 70/150\n7/7 [==============================] - 2s 216ms/step - loss: 1.4245 - categorical_accuracy: 0.5519 - accuracy: 0.5519 - val_loss: 1.6037 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 71/150\n7/7 [==============================] - 2s 222ms/step - loss: 1.4176 - categorical_accuracy: 0.6160 - accuracy: 0.6160 - val_loss: 1.6038 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 72/150\n7/7 [==============================] - 2s 218ms/step - loss: 1.4304 - categorical_accuracy: 0.5519 - accuracy: 0.5519 - val_loss: 1.6039 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 73/150\n7/7 [==============================] - 2s 243ms/step - loss: 1.4362 - categorical_accuracy: 0.5088 - accuracy: 0.5088 - val_loss: 1.6039 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 74/150\n7/7 [==============================] - 2s 217ms/step - loss: 1.4030 - categorical_accuracy: 0.5673 - accuracy: 0.5673 - val_loss: 1.6041 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 75/150\n7/7 [==============================] - 2s 217ms/step - loss: 1.4012 - categorical_accuracy: 0.5948 - accuracy: 0.5948 - val_loss: 1.6041 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 76/150\n7/7 [==============================] - 2s 217ms/step - loss: 1.4147 - categorical_accuracy: 0.5936 - accuracy: 0.5936 - val_loss: 1.6041 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 77/150\n7/7 [==============================] - 2s 217ms/step - loss: 1.4152 - categorical_accuracy: 0.5740 - accuracy: 0.5740 - val_loss: 1.6041 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 78/150\n7/7 [==============================] - 2s 218ms/step - loss: 1.3969 - categorical_accuracy: 0.6088 - accuracy: 0.6088 - val_loss: 1.6043 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 79/150\n7/7 [==============================] - 2s 219ms/step - loss: 1.3902 - categorical_accuracy: 0.5802 - accuracy: 0.5802 - val_loss: 1.6044 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 80/150\n7/7 [==============================] - 1s 215ms/step - loss: 1.3987 - categorical_accuracy: 0.5981 - accuracy: 0.5981 - val_loss: 1.6045 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 81/150\n7/7 [==============================] - 2s 220ms/step - loss: 1.3797 - categorical_accuracy: 0.6469 - accuracy: 0.6469 - val_loss: 1.6044 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 82/150\n7/7 [==============================] - 2s 221ms/step - loss: 1.3810 - categorical_accuracy: 0.5957 - accuracy: 0.5957 - val_loss: 1.6046 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 83/150\n7/7 [==============================] - 2s 222ms/step - loss: 1.3927 - categorical_accuracy: 0.5855 - accuracy: 0.5855 - val_loss: 1.6047 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 84/150\n7/7 [==============================] - 1s 218ms/step - loss: 1.3714 - categorical_accuracy: 0.6307 - accuracy: 0.6307 - val_loss: 1.6047 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 85/150\n7/7 [==============================] - 2s 217ms/step - loss: 1.3644 - categorical_accuracy: 0.5979 - accuracy: 0.5979 - val_loss: 1.6047 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 86/150\n7/7 [==============================] - 2s 218ms/step - loss: 1.3863 - categorical_accuracy: 0.6073 - accuracy: 0.6073 - val_loss: 1.6048 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 87/150\n7/7 [==============================] - 2s 239ms/step - loss: 1.3784 - categorical_accuracy: 0.6635 - accuracy: 0.6635 - val_loss: 1.6049 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 88/150\n7/7 [==============================] - 2s 218ms/step - loss: 1.3751 - categorical_accuracy: 0.6674 - accuracy: 0.6674 - val_loss: 1.6049 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 89/150\n7/7 [==============================] - 1s 215ms/step - loss: 1.3423 - categorical_accuracy: 0.6719 - accuracy: 0.6719 - val_loss: 1.6049 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 90/150\n7/7 [==============================] - 2s 219ms/step - loss: 1.3781 - categorical_accuracy: 0.6433 - accuracy: 0.6433 - val_loss: 1.6049 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 91/150\n7/7 [==============================] - 2s 221ms/step - loss: 1.3791 - categorical_accuracy: 0.6254 - accuracy: 0.6254 - val_loss: 1.6049 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 92/150\n7/7 [==============================] - 1s 216ms/step - loss: 1.3625 - categorical_accuracy: 0.6262 - accuracy: 0.6262 - val_loss: 1.6049 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 93/150\n7/7 [==============================] - 2s 218ms/step - loss: 1.3785 - categorical_accuracy: 0.6283 - accuracy: 0.6283 - val_loss: 1.6051 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 94/150\n7/7 [==============================] - 2s 215ms/step - loss: 1.3568 - categorical_accuracy: 0.6598 - accuracy: 0.6598 - val_loss: 1.6051 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 95/150\n7/7 [==============================] - 1s 215ms/step - loss: 1.3716 - categorical_accuracy: 0.6385 - accuracy: 0.6385 - val_loss: 1.6052 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 96/150\n7/7 [==============================] - 2s 220ms/step - loss: 1.3683 - categorical_accuracy: 0.6450 - accuracy: 0.6450 - val_loss: 1.6053 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 97/150\n7/7 [==============================] - 1s 216ms/step - loss: 1.3524 - categorical_accuracy: 0.6742 - accuracy: 0.6742 - val_loss: 1.6055 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 98/150\n7/7 [==============================] - 2s 221ms/step - loss: 1.3558 - categorical_accuracy: 0.7176 - accuracy: 0.7176 - val_loss: 1.6056 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 99/150\n7/7 [==============================] - 2s 220ms/step - loss: 1.3345 - categorical_accuracy: 0.7133 - accuracy: 0.7133 - val_loss: 1.6056 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 100/150\n7/7 [==============================] - 2s 221ms/step - loss: 1.3378 - categorical_accuracy: 0.6825 - accuracy: 0.6825 - val_loss: 1.6057 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 101/150\n7/7 [==============================] - 2s 224ms/step - loss: 1.3344 - categorical_accuracy: 0.6934 - accuracy: 0.6934 - val_loss: 1.6059 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 102/150\n7/7 [==============================] - 2s 232ms/step - loss: 1.3614 - categorical_accuracy: 0.6648 - accuracy: 0.6648 - val_loss: 1.6059 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 103/150\n7/7 [==============================] - 2s 226ms/step - loss: 1.3420 - categorical_accuracy: 0.6617 - accuracy: 0.6617 - val_loss: 1.6061 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 104/150\n7/7 [==============================] - 2s 233ms/step - loss: 1.3280 - categorical_accuracy: 0.6806 - accuracy: 0.6806 - val_loss: 1.6063 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 105/150\n7/7 [==============================] - 2s 217ms/step - loss: 1.3517 - categorical_accuracy: 0.6366 - accuracy: 0.6366 - val_loss: 1.6064 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 106/150\n7/7 [==============================] - 2s 219ms/step - loss: 1.3317 - categorical_accuracy: 0.6901 - accuracy: 0.6901 - val_loss: 1.6064 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 107/150\n7/7 [==============================] - 1s 216ms/step - loss: 1.3327 - categorical_accuracy: 0.6838 - accuracy: 0.6838 - val_loss: 1.6064 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 108/150\n7/7 [==============================] - 2s 243ms/step - loss: 1.3251 - categorical_accuracy: 0.6536 - accuracy: 0.6536 - val_loss: 1.6065 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 109/150\n7/7 [==============================] - 2s 220ms/step - loss: 1.3279 - categorical_accuracy: 0.7057 - accuracy: 0.7057 - val_loss: 1.6066 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 110/150\n7/7 [==============================] - 2s 219ms/step - loss: 1.3351 - categorical_accuracy: 0.7151 - accuracy: 0.7151 - val_loss: 1.6067 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 111/150\n7/7 [==============================] - 2s 219ms/step - loss: 1.3328 - categorical_accuracy: 0.7268 - accuracy: 0.7268 - val_loss: 1.6068 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 112/150\n7/7 [==============================] - 2s 214ms/step - loss: 1.3125 - categorical_accuracy: 0.7527 - accuracy: 0.7527 - val_loss: 1.6070 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 113/150\n7/7 [==============================] - 2s 218ms/step - loss: 1.3081 - categorical_accuracy: 0.7329 - accuracy: 0.7329 - val_loss: 1.6070 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 114/150\n7/7 [==============================] - 1s 214ms/step - loss: 1.3130 - categorical_accuracy: 0.7372 - accuracy: 0.7372 - val_loss: 1.6070 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 115/150\n7/7 [==============================] - 2s 216ms/step - loss: 1.3189 - categorical_accuracy: 0.7289 - accuracy: 0.7289 - val_loss: 1.6070 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 116/150\n7/7 [==============================] - 2s 220ms/step - loss: 1.3187 - categorical_accuracy: 0.7009 - accuracy: 0.7009 - val_loss: 1.6069 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 117/150\n7/7 [==============================] - 1s 216ms/step - loss: 1.3057 - categorical_accuracy: 0.7011 - accuracy: 0.7011 - val_loss: 1.6070 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 118/150\n7/7 [==============================] - 2s 224ms/step - loss: 1.3080 - categorical_accuracy: 0.7407 - accuracy: 0.7407 - val_loss: 1.6071 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 119/150\n7/7 [==============================] - 1s 216ms/step - loss: 1.2955 - categorical_accuracy: 0.7345 - accuracy: 0.7345 - val_loss: 1.6071 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 120/150\n7/7 [==============================] - 2s 221ms/step - loss: 1.3000 - categorical_accuracy: 0.7153 - accuracy: 0.7153 - val_loss: 1.6073 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 121/150\n7/7 [==============================] - 2s 218ms/step - loss: 1.2985 - categorical_accuracy: 0.7479 - accuracy: 0.7479 - val_loss: 1.6073 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 122/150\n7/7 [==============================] - 2s 223ms/step - loss: 1.3049 - categorical_accuracy: 0.7411 - accuracy: 0.7411 - val_loss: 1.6075 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 123/150\n7/7 [==============================] - 2s 220ms/step - loss: 1.2910 - categorical_accuracy: 0.7572 - accuracy: 0.7572 - val_loss: 1.6075 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 124/150\n7/7 [==============================] - 2s 218ms/step - loss: 1.2928 - categorical_accuracy: 0.7323 - accuracy: 0.7323 - val_loss: 1.6076 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 125/150\n7/7 [==============================] - 2s 219ms/step - loss: 1.3077 - categorical_accuracy: 0.7019 - accuracy: 0.7019 - val_loss: 1.6076 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 126/150\n7/7 [==============================] - 2s 221ms/step - loss: 1.2859 - categorical_accuracy: 0.7732 - accuracy: 0.7732 - val_loss: 1.6077 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 127/150\n7/7 [==============================] - 2s 219ms/step - loss: 1.2948 - categorical_accuracy: 0.7363 - accuracy: 0.7363 - val_loss: 1.6076 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 128/150\n7/7 [==============================] - 2s 224ms/step - loss: 1.2813 - categorical_accuracy: 0.7485 - accuracy: 0.7485 - val_loss: 1.6077 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 129/150\n7/7 [==============================] - 2s 243ms/step - loss: 1.2796 - categorical_accuracy: 0.7372 - accuracy: 0.7372 - val_loss: 1.6079 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 130/150\n7/7 [==============================] - 2s 219ms/step - loss: 1.2809 - categorical_accuracy: 0.7451 - accuracy: 0.7451 - val_loss: 1.6080 - val_categorical_accuracy: 0.2250 - val_accuracy: 0.2250\nEpoch 131/150\n7/7 [==============================] - 2s 220ms/step - loss: 1.2809 - categorical_accuracy: 0.7686 - accuracy: 0.7686 - val_loss: 1.6080 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 132/150\n7/7 [==============================] - 2s 219ms/step - loss: 1.2847 - categorical_accuracy: 0.7417 - accuracy: 0.7417 - val_loss: 1.6082 - val_categorical_accuracy: 0.2375 - val_accuracy: 0.2375\nEpoch 133/150\n7/7 [==============================] - 2s 223ms/step - loss: 1.2770 - categorical_accuracy: 0.7359 - accuracy: 0.7359 - val_loss: 1.6083 - val_categorical_accuracy: 0.2500 - val_accuracy: 0.2500\nEpoch 134/150\n7/7 [==============================] - 2s 220ms/step - loss: 1.2681 - categorical_accuracy: 0.7277 - accuracy: 0.7277 - val_loss: 1.6083 - val_categorical_accuracy: 0.2500 - val_accuracy: 0.2500\nEpoch 135/150\n7/7 [==============================] - 2s 218ms/step - loss: 1.2637 - categorical_accuracy: 0.7360 - accuracy: 0.7360 - val_loss: 1.6082 - val_categorical_accuracy: 0.2500 - val_accuracy: 0.2500\nEpoch 136/150\n7/7 [==============================] - 2s 219ms/step - loss: 1.2623 - categorical_accuracy: 0.7423 - accuracy: 0.7423 - val_loss: 1.6084 - val_categorical_accuracy: 0.2500 - val_accuracy: 0.2500\nEpoch 137/150\n7/7 [==============================] - 2s 222ms/step - loss: 1.2901 - categorical_accuracy: 0.7702 - accuracy: 0.7702 - val_loss: 1.6086 - val_categorical_accuracy: 0.2500 - val_accuracy: 0.2500\nEpoch 138/150\n7/7 [==============================] - 2s 225ms/step - loss: 1.2566 - categorical_accuracy: 0.7865 - accuracy: 0.7865 - val_loss: 1.6087 - val_categorical_accuracy: 0.2500 - val_accuracy: 0.2500\nEpoch 139/150\n7/7 [==============================] - 2s 218ms/step - loss: 1.2538 - categorical_accuracy: 0.7777 - accuracy: 0.7777 - val_loss: 1.6088 - val_categorical_accuracy: 0.2625 - val_accuracy: 0.2625\nEpoch 140/150\n7/7 [==============================] - 2s 216ms/step - loss: 1.2316 - categorical_accuracy: 0.8059 - accuracy: 0.8059 - val_loss: 1.6089 - val_categorical_accuracy: 0.2750 - val_accuracy: 0.2750\nEpoch 141/150\n7/7 [==============================] - 2s 218ms/step - loss: 1.2642 - categorical_accuracy: 0.7551 - accuracy: 0.7551 - val_loss: 1.6089 - val_categorical_accuracy: 0.2750 - val_accuracy: 0.2750\nEpoch 142/150\n7/7 [==============================] - 2s 222ms/step - loss: 1.2689 - categorical_accuracy: 0.7321 - accuracy: 0.7321 - val_loss: 1.6090 - val_categorical_accuracy: 0.2750 - val_accuracy: 0.2750\nEpoch 143/150\n7/7 [==============================] - 2s 218ms/step - loss: 1.2487 - categorical_accuracy: 0.7459 - accuracy: 0.7459 - val_loss: 1.6092 - val_categorical_accuracy: 0.2750 - val_accuracy: 0.2750\nEpoch 144/150\n7/7 [==============================] - 2s 218ms/step - loss: 1.2548 - categorical_accuracy: 0.7831 - accuracy: 0.7831 - val_loss: 1.6092 - val_categorical_accuracy: 0.2750 - val_accuracy: 0.2750\nEpoch 145/150\n7/7 [==============================] - 2s 218ms/step - loss: 1.2502 - categorical_accuracy: 0.7701 - accuracy: 0.7701 - val_loss: 1.6094 - val_categorical_accuracy: 0.2750 - val_accuracy: 0.2750\nEpoch 146/150\n7/7 [==============================] - 2s 218ms/step - loss: 1.2484 - categorical_accuracy: 0.7879 - accuracy: 0.7879 - val_loss: 1.6095 - val_categorical_accuracy: 0.2750 - val_accuracy: 0.2750\nEpoch 147/150\n7/7 [==============================] - 2s 218ms/step - loss: 1.2605 - categorical_accuracy: 0.7872 - accuracy: 0.7872 - val_loss: 1.6095 - val_categorical_accuracy: 0.2750 - val_accuracy: 0.2750\nEpoch 148/150\n7/7 [==============================] - 2s 219ms/step - loss: 1.2389 - categorical_accuracy: 0.7984 - accuracy: 0.7984 - val_loss: 1.6096 - val_categorical_accuracy: 0.2750 - val_accuracy: 0.2750\nEpoch 149/150\n7/7 [==============================] - 2s 240ms/step - loss: 1.2478 - categorical_accuracy: 0.7890 - accuracy: 0.7890 - val_loss: 1.6098 - val_categorical_accuracy: 0.2750 - val_accuracy: 0.2750\nEpoch 150/150\n7/7 [==============================] - 1s 217ms/step - loss: 1.2303 - categorical_accuracy: 0.7607 - accuracy: 0.7607 - val_loss: 1.6101 - val_categorical_accuracy: 0.2750 - val_accuracy: 0.2750\n4/4 [==============================] - 0s 38ms/step - loss: 1.6100 - categorical_accuracy: 0.2100 - accuracy: 0.2100\n[[ 9 3 3 5 0]\n [10 2 3 5 0]\n [ 6 3 2 9 0]\n [ 9 2 2 7 0]\n [13 3 0 3 1]]\n precision recall f1-score support\n\n DN 0.19 0.45 0.27 20\n GOF 0.15 0.10 0.12 20\n HI 0.20 0.10 0.13 20\n LOF 0.24 0.35 0.29 20\n none 1.00 0.05 0.10 20\n\n accuracy 0.21 100\n macro avg 0.36 0.21 0.18 100\nweighted avg 0.36 0.21 0.18 100\n\n"
],
[
"with open('/content/%s' %modelName, 'wb') as file_pi:\r\n pickle.dump(history.history, file_pi)\r\nhistory = pickle.load(open('/content/%s' % modelName, \"rb\"))\r\nmodel.save(modelName +'.h5')\r\nplot(history)\r\nwith open('/content/%s_train' %modelName, 'wb') as file_pi:\r\n pickle.dump(X_train, file_pi)\r\nwith open('/content/%s_test' %modelName, 'wb') as file_pi:\r\n pickle.dump(X_test, file_pi)\r\nwith open('/content/%s_Labeltest' %modelName, 'wb') as file_pi:\r\n pickle.dump(y_test, file_pi)\r\nwith open('/content/%s_LabelTrain' %modelName, 'wb') as file_pi:\r\n pickle.dump(y_train, file_pi)\r\nwith open('/content/%s_ConfusionMatrix' %modelName, 'wb') as file_pi:\r\n pickle.dump(confusion_matrix(y_test.argmax(axis=-1), model.predict(X_test).argmax(axis=-1)), file_pi)\r\nwith open('/content/%s_ClassificationReport' %modelName, 'wb') as file_pi:\r\n pickle.dump(classification_report(y_test.argmax(axis=-1), model.predict(X_test).argmax(axis=-1), target_names=ClassNames), file_pi)",
"dict_keys(['loss', 'categorical_accuracy', 'accuracy', 'val_loss', 'val_categorical_accuracy', 'val_accuracy'])\n"
],
[
"",
"_____no_output_____"
]
],
[
[
"Running above solution with reduced text and preprocessing",
"_____no_output_____"
]
],
[
[
"uploaded = files.upload()",
"_____no_output_____"
],
[
"modelName = 'Model2' #name of the model, this will be used to save model evaluation and history\r\nnumEpochs = 150 # maximum number of epochs if early stopping doesnt work\r\nbatchsize = 50 # batchsize which will be used in each step by optimizer defined in the model\r\noptimizer = 'adam' #optimizer to be used in model.fit keras method\r\n",
"_____no_output_____"
]
],
[
[
"Calling the above pipeline again with new parameters",
"_____no_output_____"
]
],
[
[
"X, y = Preprocessing()\r\nX_train, X_test, y_train, y_test, ClassNames = TrainTestSplit(X, y)\r\nmodel = ModelBuild(X_train, y_train)\r\nPlotModel(model, modelName +\".png\")\r\nhistory = PlotTraining(model, X_test, y_test)\r\nprint(confusion_matrix(y_test.argmax(axis=-1),model.predict(X_test).argmax(axis=-1)))\r\nprint(classification_report(y_test.argmax(axis=-1), model.predict(X_test).argmax(axis=-1),target_names=ClassNames))\r\n",
"Model: \"model_1\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_2 (InputLayer) [(None, 55389)] 0 \n_________________________________________________________________\ndense_3 (Dense) (None, 500) 27695000 \n_________________________________________________________________\ndense_4 (Dense) (None, 100) 50100 \n_________________________________________________________________\ndense_5 (Dense) (None, 5) 505 \n=================================================================\nTotal params: 27,745,605\nTrainable params: 27,745,605\nNon-trainable params: 0\n_________________________________________________________________\nEpoch 1/150\n7/7 [==============================] - 2s 268ms/step - loss: 1.5695 - categorical_accuracy: 0.2738 - accuracy: 0.2738 - val_loss: 1.3374 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 2/150\n7/7 [==============================] - 2s 235ms/step - loss: 0.5004 - categorical_accuracy: 0.9324 - accuracy: 0.9324 - val_loss: 1.4829 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 3/150\n7/7 [==============================] - 2s 238ms/step - loss: 0.1755 - categorical_accuracy: 0.9727 - accuracy: 0.9727 - val_loss: 1.9200 - val_categorical_accuracy: 0.5714 - val_accuracy: 0.5714\nEpoch 4/150\n7/7 [==============================] - 2s 235ms/step - loss: 0.0752 - categorical_accuracy: 0.9793 - accuracy: 0.9793 - val_loss: 2.3246 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 5/150\n7/7 [==============================] - 2s 238ms/step - loss: 0.0606 - categorical_accuracy: 0.9759 - accuracy: 0.9759 - val_loss: 2.5312 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 6/150\n7/7 [==============================] - 2s 269ms/step - loss: 0.0578 - categorical_accuracy: 0.9775 - accuracy: 0.9775 - val_loss: 2.5172 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 7/150\n7/7 [==============================] - 2s 233ms/step - loss: 0.0360 - categorical_accuracy: 0.9819 - accuracy: 0.9819 - val_loss: 2.5284 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 8/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0478 - categorical_accuracy: 0.9859 - accuracy: 0.9859 - val_loss: 2.5672 - val_categorical_accuracy: 0.5714 - val_accuracy: 0.5714\nEpoch 9/150\n7/7 [==============================] - 2s 226ms/step - loss: 0.0928 - categorical_accuracy: 0.9899 - accuracy: 0.9899 - val_loss: 2.6665 - val_categorical_accuracy: 0.5584 - val_accuracy: 0.5584\nEpoch 10/150\n7/7 [==============================] - 2s 228ms/step - loss: 0.0700 - categorical_accuracy: 0.9818 - accuracy: 0.9818 - val_loss: 2.5901 - val_categorical_accuracy: 0.5714 - val_accuracy: 0.5714\nEpoch 11/150\n7/7 [==============================] - 2s 228ms/step - loss: 0.0588 - categorical_accuracy: 0.9781 - accuracy: 0.9781 - val_loss: 2.4994 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 12/150\n7/7 [==============================] - 2s 228ms/step - loss: 0.0321 - categorical_accuracy: 0.9884 - accuracy: 0.9884 - val_loss: 2.4964 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 13/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0412 - categorical_accuracy: 0.9842 - accuracy: 0.9842 - val_loss: 2.4833 - val_categorical_accuracy: 0.5844 - val_accuracy: 0.5844\nEpoch 14/150\n7/7 [==============================] - 2s 229ms/step - loss: 0.0265 - categorical_accuracy: 0.9921 - accuracy: 0.9921 - val_loss: 2.4687 - val_categorical_accuracy: 0.5844 - val_accuracy: 0.5844\nEpoch 15/150\n7/7 [==============================] - 2s 233ms/step - loss: 0.0426 - categorical_accuracy: 0.9865 - accuracy: 0.9865 - val_loss: 2.4541 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 16/150\n7/7 [==============================] - 2s 228ms/step - loss: 0.0360 - categorical_accuracy: 0.9857 - accuracy: 0.9857 - val_loss: 2.4937 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 17/150\n7/7 [==============================] - 2s 228ms/step - loss: 0.0213 - categorical_accuracy: 0.9906 - accuracy: 0.9906 - val_loss: 2.5171 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 18/150\n7/7 [==============================] - 2s 228ms/step - loss: 0.0320 - categorical_accuracy: 0.9818 - accuracy: 0.9818 - val_loss: 2.5628 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 19/150\n7/7 [==============================] - 2s 227ms/step - loss: 0.0231 - categorical_accuracy: 0.9882 - accuracy: 0.9882 - val_loss: 2.5661 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 20/150\n7/7 [==============================] - 2s 228ms/step - loss: 0.0326 - categorical_accuracy: 0.9881 - accuracy: 0.9881 - val_loss: 2.5363 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 21/150\n7/7 [==============================] - 2s 225ms/step - loss: 0.0280 - categorical_accuracy: 0.9839 - accuracy: 0.9839 - val_loss: 2.5081 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 22/150\n7/7 [==============================] - 2s 231ms/step - loss: 0.0215 - categorical_accuracy: 0.9820 - accuracy: 0.9820 - val_loss: 2.4980 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 23/150\n7/7 [==============================] - 2s 250ms/step - loss: 0.0189 - categorical_accuracy: 0.9826 - accuracy: 0.9826 - val_loss: 2.4890 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 24/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0141 - categorical_accuracy: 0.9895 - accuracy: 0.9895 - val_loss: 2.5021 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 25/150\n7/7 [==============================] - 2s 228ms/step - loss: 0.0232 - categorical_accuracy: 0.9783 - accuracy: 0.9783 - val_loss: 2.5163 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 26/150\n7/7 [==============================] - 2s 229ms/step - loss: 0.0191 - categorical_accuracy: 0.9926 - accuracy: 0.9926 - val_loss: 2.5147 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 27/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0244 - categorical_accuracy: 0.9907 - accuracy: 0.9907 - val_loss: 2.5307 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 28/150\n7/7 [==============================] - 2s 229ms/step - loss: 0.0202 - categorical_accuracy: 0.9834 - accuracy: 0.9834 - val_loss: 2.5760 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 29/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0238 - categorical_accuracy: 0.9813 - accuracy: 0.9813 - val_loss: 2.5627 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 30/150\n7/7 [==============================] - 2s 228ms/step - loss: 0.0206 - categorical_accuracy: 0.9784 - accuracy: 0.9784 - val_loss: 2.5765 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 31/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0240 - categorical_accuracy: 0.9867 - accuracy: 0.9867 - val_loss: 2.5787 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 32/150\n7/7 [==============================] - 2s 228ms/step - loss: 0.0256 - categorical_accuracy: 0.9794 - accuracy: 0.9794 - val_loss: 2.5785 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 33/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0230 - categorical_accuracy: 0.9814 - accuracy: 0.9814 - val_loss: 2.6078 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 34/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0188 - categorical_accuracy: 0.9874 - accuracy: 0.9874 - val_loss: 2.6180 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 35/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0381 - categorical_accuracy: 0.9869 - accuracy: 0.9869 - val_loss: 2.7025 - val_categorical_accuracy: 0.5455 - val_accuracy: 0.5455\nEpoch 36/150\n7/7 [==============================] - 2s 231ms/step - loss: 0.0677 - categorical_accuracy: 0.9758 - accuracy: 0.9758 - val_loss: 2.6749 - val_categorical_accuracy: 0.5455 - val_accuracy: 0.5455\nEpoch 37/150\n7/7 [==============================] - 2s 232ms/step - loss: 0.0483 - categorical_accuracy: 0.9836 - accuracy: 0.9836 - val_loss: 2.6163 - val_categorical_accuracy: 0.5714 - val_accuracy: 0.5714\nEpoch 38/150\n7/7 [==============================] - 2s 231ms/step - loss: 0.0238 - categorical_accuracy: 0.9807 - accuracy: 0.9807 - val_loss: 2.6076 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 39/150\n7/7 [==============================] - 2s 253ms/step - loss: 0.0162 - categorical_accuracy: 0.9862 - accuracy: 0.9862 - val_loss: 2.6158 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 40/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0331 - categorical_accuracy: 0.9788 - accuracy: 0.9788 - val_loss: 2.6012 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 41/150\n7/7 [==============================] - 2s 232ms/step - loss: 0.0326 - categorical_accuracy: 0.9903 - accuracy: 0.9903 - val_loss: 2.5301 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 42/150\n7/7 [==============================] - 2s 234ms/step - loss: 0.0830 - categorical_accuracy: 0.9789 - accuracy: 0.9789 - val_loss: 2.5265 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 43/150\n7/7 [==============================] - 2s 233ms/step - loss: 0.0230 - categorical_accuracy: 0.9869 - accuracy: 0.9869 - val_loss: 2.5262 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 44/150\n7/7 [==============================] - 2s 227ms/step - loss: 0.0182 - categorical_accuracy: 0.9865 - accuracy: 0.9865 - val_loss: 2.6183 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 45/150\n7/7 [==============================] - 2s 232ms/step - loss: 0.0188 - categorical_accuracy: 0.9903 - accuracy: 0.9903 - val_loss: 2.6366 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 46/150\n7/7 [==============================] - 2s 231ms/step - loss: 0.0205 - categorical_accuracy: 0.9851 - accuracy: 0.9851 - val_loss: 2.6301 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 47/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0301 - categorical_accuracy: 0.9823 - accuracy: 0.9823 - val_loss: 2.6482 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 48/150\n7/7 [==============================] - 2s 229ms/step - loss: 0.0167 - categorical_accuracy: 0.9946 - accuracy: 0.9946 - val_loss: 2.6663 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 49/150\n7/7 [==============================] - 2s 228ms/step - loss: 0.0218 - categorical_accuracy: 0.9869 - accuracy: 0.9869 - val_loss: 2.6685 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 50/150\n7/7 [==============================] - 2s 235ms/step - loss: 0.0237 - categorical_accuracy: 0.9861 - accuracy: 0.9861 - val_loss: 2.7008 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 51/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0188 - categorical_accuracy: 0.9903 - accuracy: 0.9903 - val_loss: 2.7143 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 52/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0165 - categorical_accuracy: 0.9851 - accuracy: 0.9851 - val_loss: 2.7180 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 53/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0214 - categorical_accuracy: 0.9864 - accuracy: 0.9864 - val_loss: 2.7170 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 54/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0448 - categorical_accuracy: 0.9805 - accuracy: 0.9805 - val_loss: 2.7195 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 55/150\n7/7 [==============================] - 2s 231ms/step - loss: 0.0306 - categorical_accuracy: 0.9810 - accuracy: 0.9810 - val_loss: 2.7402 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 56/150\n7/7 [==============================] - 2s 254ms/step - loss: 0.0244 - categorical_accuracy: 0.9810 - accuracy: 0.9810 - val_loss: 2.7503 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 57/150\n7/7 [==============================] - 2s 231ms/step - loss: 0.0144 - categorical_accuracy: 0.9894 - accuracy: 0.9894 - val_loss: 2.8069 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 58/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0229 - categorical_accuracy: 0.9828 - accuracy: 0.9828 - val_loss: 2.8329 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 59/150\n7/7 [==============================] - 2s 231ms/step - loss: 0.0155 - categorical_accuracy: 0.9941 - accuracy: 0.9941 - val_loss: 2.8254 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 60/150\n7/7 [==============================] - 2s 234ms/step - loss: 0.0378 - categorical_accuracy: 0.9850 - accuracy: 0.9850 - val_loss: 2.8034 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 61/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0209 - categorical_accuracy: 0.9903 - accuracy: 0.9903 - val_loss: 2.7956 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 62/150\n7/7 [==============================] - 2s 233ms/step - loss: 0.0185 - categorical_accuracy: 0.9878 - accuracy: 0.9878 - val_loss: 2.7866 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 63/150\n7/7 [==============================] - 2s 227ms/step - loss: 0.0166 - categorical_accuracy: 0.9903 - accuracy: 0.9903 - val_loss: 2.7855 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 64/150\n7/7 [==============================] - 2s 227ms/step - loss: 0.0227 - categorical_accuracy: 0.9808 - accuracy: 0.9808 - val_loss: 2.7905 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 65/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0214 - categorical_accuracy: 0.9883 - accuracy: 0.9883 - val_loss: 2.7819 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 66/150\n7/7 [==============================] - 2s 227ms/step - loss: 0.0158 - categorical_accuracy: 0.9891 - accuracy: 0.9891 - val_loss: 2.7884 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 67/150\n7/7 [==============================] - 2s 234ms/step - loss: 0.0244 - categorical_accuracy: 0.9788 - accuracy: 0.9788 - val_loss: 2.7801 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 68/150\n7/7 [==============================] - 2s 229ms/step - loss: 0.0140 - categorical_accuracy: 0.9901 - accuracy: 0.9901 - val_loss: 2.7888 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 69/150\n7/7 [==============================] - 2s 235ms/step - loss: 0.0195 - categorical_accuracy: 0.9858 - accuracy: 0.9858 - val_loss: 2.8070 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 70/150\n7/7 [==============================] - 2s 231ms/step - loss: 0.0179 - categorical_accuracy: 0.9871 - accuracy: 0.9871 - val_loss: 2.8166 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 71/150\n7/7 [==============================] - 2s 232ms/step - loss: 0.0160 - categorical_accuracy: 0.9930 - accuracy: 0.9930 - val_loss: 2.8293 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 72/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0174 - categorical_accuracy: 0.9852 - accuracy: 0.9852 - val_loss: 2.9318 - val_categorical_accuracy: 0.5844 - val_accuracy: 0.5844\nEpoch 73/150\n7/7 [==============================] - 2s 252ms/step - loss: 0.0165 - categorical_accuracy: 0.9889 - accuracy: 0.9889 - val_loss: 2.9376 - val_categorical_accuracy: 0.5844 - val_accuracy: 0.5844\nEpoch 74/150\n7/7 [==============================] - 2s 233ms/step - loss: 0.0178 - categorical_accuracy: 0.9952 - accuracy: 0.9952 - val_loss: 2.8995 - val_categorical_accuracy: 0.5844 - val_accuracy: 0.5844\nEpoch 75/150\n7/7 [==============================] - 2s 233ms/step - loss: 0.0163 - categorical_accuracy: 0.9879 - accuracy: 0.9879 - val_loss: 2.8483 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 76/150\n7/7 [==============================] - 2s 234ms/step - loss: 0.0213 - categorical_accuracy: 0.9926 - accuracy: 0.9926 - val_loss: 2.8146 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 77/150\n7/7 [==============================] - 2s 229ms/step - loss: 0.0199 - categorical_accuracy: 0.9859 - accuracy: 0.9859 - val_loss: 2.7798 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 78/150\n7/7 [==============================] - 2s 231ms/step - loss: 0.0216 - categorical_accuracy: 0.9758 - accuracy: 0.9758 - val_loss: 2.7987 - val_categorical_accuracy: 0.5844 - val_accuracy: 0.5844\nEpoch 79/150\n7/7 [==============================] - 2s 232ms/step - loss: 0.0304 - categorical_accuracy: 0.9833 - accuracy: 0.9833 - val_loss: 2.7905 - val_categorical_accuracy: 0.5714 - val_accuracy: 0.5714\nEpoch 80/150\n7/7 [==============================] - 2s 229ms/step - loss: 0.0440 - categorical_accuracy: 0.9819 - accuracy: 0.9819 - val_loss: 2.7772 - val_categorical_accuracy: 0.5844 - val_accuracy: 0.5844\nEpoch 81/150\n7/7 [==============================] - 2s 232ms/step - loss: 0.0218 - categorical_accuracy: 0.9878 - accuracy: 0.9878 - val_loss: 2.7729 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 82/150\n7/7 [==============================] - 2s 229ms/step - loss: 0.0211 - categorical_accuracy: 0.9816 - accuracy: 0.9816 - val_loss: 2.7741 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 83/150\n7/7 [==============================] - 2s 229ms/step - loss: 0.0154 - categorical_accuracy: 0.9871 - accuracy: 0.9871 - val_loss: 2.7900 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 84/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0115 - categorical_accuracy: 0.9906 - accuracy: 0.9906 - val_loss: 2.7975 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 85/150\n7/7 [==============================] - 2s 228ms/step - loss: 0.0181 - categorical_accuracy: 0.9934 - accuracy: 0.9934 - val_loss: 2.8041 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 86/150\n7/7 [==============================] - 2s 229ms/step - loss: 0.0173 - categorical_accuracy: 0.9865 - accuracy: 0.9865 - val_loss: 2.8106 - val_categorical_accuracy: 0.5844 - val_accuracy: 0.5844\nEpoch 87/150\n7/7 [==============================] - 2s 247ms/step - loss: 0.0138 - categorical_accuracy: 0.9894 - accuracy: 0.9894 - val_loss: 2.8212 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 88/150\n7/7 [==============================] - 2s 232ms/step - loss: 0.0202 - categorical_accuracy: 0.9816 - accuracy: 0.9816 - val_loss: 2.8189 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 89/150\n7/7 [==============================] - 2s 235ms/step - loss: 0.0292 - categorical_accuracy: 0.9765 - accuracy: 0.9765 - val_loss: 2.8358 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 90/150\n7/7 [==============================] - 2s 233ms/step - loss: 0.0134 - categorical_accuracy: 0.9899 - accuracy: 0.9899 - val_loss: 2.8383 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 91/150\n7/7 [==============================] - 2s 231ms/step - loss: 0.0214 - categorical_accuracy: 0.9794 - accuracy: 0.9794 - val_loss: 2.8439 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 92/150\n7/7 [==============================] - 2s 232ms/step - loss: 0.0169 - categorical_accuracy: 0.9821 - accuracy: 0.9821 - val_loss: 2.8547 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 93/150\n7/7 [==============================] - 2s 231ms/step - loss: 0.0144 - categorical_accuracy: 0.9896 - accuracy: 0.9896 - val_loss: 2.8564 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 94/150\n7/7 [==============================] - 2s 231ms/step - loss: 0.0251 - categorical_accuracy: 0.9852 - accuracy: 0.9852 - val_loss: 2.8609 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 95/150\n7/7 [==============================] - 2s 231ms/step - loss: 0.0231 - categorical_accuracy: 0.9873 - accuracy: 0.9873 - val_loss: 2.8660 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 96/150\n7/7 [==============================] - 2s 234ms/step - loss: 0.0187 - categorical_accuracy: 0.9874 - accuracy: 0.9874 - val_loss: 2.8491 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 97/150\n7/7 [==============================] - 2s 232ms/step - loss: 0.0363 - categorical_accuracy: 0.9782 - accuracy: 0.9782 - val_loss: 2.8636 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 98/150\n7/7 [==============================] - 2s 229ms/step - loss: 0.0265 - categorical_accuracy: 0.9830 - accuracy: 0.9830 - val_loss: 2.8566 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 99/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0124 - categorical_accuracy: 0.9908 - accuracy: 0.9908 - val_loss: 2.8613 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 100/150\n7/7 [==============================] - 2s 229ms/step - loss: 0.0216 - categorical_accuracy: 0.9817 - accuracy: 0.9817 - val_loss: 2.8446 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 101/150\n7/7 [==============================] - 2s 253ms/step - loss: 0.0215 - categorical_accuracy: 0.9857 - accuracy: 0.9857 - val_loss: 2.8412 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 102/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0179 - categorical_accuracy: 0.9909 - accuracy: 0.9909 - val_loss: 2.8377 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 103/150\n7/7 [==============================] - 2s 231ms/step - loss: 0.0225 - categorical_accuracy: 0.9814 - accuracy: 0.9814 - val_loss: 2.8464 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 104/150\n7/7 [==============================] - 2s 231ms/step - loss: 0.0123 - categorical_accuracy: 0.9899 - accuracy: 0.9899 - val_loss: 2.8473 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 105/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0120 - categorical_accuracy: 0.9942 - accuracy: 0.9942 - val_loss: 2.9007 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 106/150\n7/7 [==============================] - 2s 231ms/step - loss: 0.0150 - categorical_accuracy: 0.9908 - accuracy: 0.9908 - val_loss: 2.9241 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 107/150\n7/7 [==============================] - 2s 234ms/step - loss: 0.0201 - categorical_accuracy: 0.9927 - accuracy: 0.9927 - val_loss: 2.9078 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 108/150\n7/7 [==============================] - 2s 232ms/step - loss: 0.0226 - categorical_accuracy: 0.9830 - accuracy: 0.9830 - val_loss: 2.8641 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 109/150\n7/7 [==============================] - 2s 232ms/step - loss: 0.0157 - categorical_accuracy: 0.9914 - accuracy: 0.9914 - val_loss: 2.8085 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 110/150\n7/7 [==============================] - 2s 232ms/step - loss: 0.0283 - categorical_accuracy: 0.9842 - accuracy: 0.9842 - val_loss: 2.8021 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 111/150\n7/7 [==============================] - 2s 232ms/step - loss: 0.0182 - categorical_accuracy: 0.9900 - accuracy: 0.9900 - val_loss: 2.8295 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 112/150\n7/7 [==============================] - 2s 233ms/step - loss: 0.0142 - categorical_accuracy: 0.9874 - accuracy: 0.9874 - val_loss: 2.8586 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 113/150\n7/7 [==============================] - 2s 231ms/step - loss: 0.0168 - categorical_accuracy: 0.9884 - accuracy: 0.9884 - val_loss: 2.8866 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 114/150\n7/7 [==============================] - 2s 231ms/step - loss: 0.0171 - categorical_accuracy: 0.9842 - accuracy: 0.9842 - val_loss: 2.8624 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 115/150\n7/7 [==============================] - 2s 254ms/step - loss: 0.0235 - categorical_accuracy: 0.9875 - accuracy: 0.9875 - val_loss: 2.8660 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 116/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0215 - categorical_accuracy: 0.9901 - accuracy: 0.9901 - val_loss: 2.8900 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 117/150\n7/7 [==============================] - 2s 233ms/step - loss: 0.0138 - categorical_accuracy: 0.9902 - accuracy: 0.9902 - val_loss: 2.9117 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 118/150\n7/7 [==============================] - 2s 234ms/step - loss: 0.0138 - categorical_accuracy: 0.9912 - accuracy: 0.9912 - val_loss: 2.9251 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 119/150\n7/7 [==============================] - 2s 241ms/step - loss: 0.0234 - categorical_accuracy: 0.9869 - accuracy: 0.9869 - val_loss: 2.9329 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 120/150\n7/7 [==============================] - 2s 241ms/step - loss: 0.0166 - categorical_accuracy: 0.9920 - accuracy: 0.9920 - val_loss: 2.9517 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 121/150\n7/7 [==============================] - 2s 247ms/step - loss: 0.0098 - categorical_accuracy: 0.9903 - accuracy: 0.9903 - val_loss: 2.9636 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 122/150\n7/7 [==============================] - 2s 233ms/step - loss: 0.0163 - categorical_accuracy: 0.9874 - accuracy: 0.9874 - val_loss: 2.9753 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 123/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0179 - categorical_accuracy: 0.9871 - accuracy: 0.9871 - val_loss: 2.9879 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 124/150\n7/7 [==============================] - 2s 229ms/step - loss: 0.0153 - categorical_accuracy: 0.9884 - accuracy: 0.9884 - val_loss: 2.9775 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 125/150\n7/7 [==============================] - 2s 233ms/step - loss: 0.0145 - categorical_accuracy: 0.9921 - accuracy: 0.9921 - val_loss: 2.9799 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 126/150\n7/7 [==============================] - 2s 228ms/step - loss: 0.0217 - categorical_accuracy: 0.9850 - accuracy: 0.9850 - val_loss: 2.9875 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 127/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0111 - categorical_accuracy: 0.9958 - accuracy: 0.9958 - val_loss: 2.9838 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 128/150\n7/7 [==============================] - 2s 232ms/step - loss: 0.0160 - categorical_accuracy: 0.9952 - accuracy: 0.9952 - val_loss: 2.9816 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 129/150\n7/7 [==============================] - 2s 252ms/step - loss: 0.0200 - categorical_accuracy: 0.9881 - accuracy: 0.9881 - val_loss: 2.9989 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 130/150\n7/7 [==============================] - 2s 234ms/step - loss: 0.0212 - categorical_accuracy: 0.9830 - accuracy: 0.9830 - val_loss: 3.0140 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 131/150\n7/7 [==============================] - 2s 232ms/step - loss: 0.0132 - categorical_accuracy: 0.9906 - accuracy: 0.9906 - val_loss: 3.0093 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 132/150\n7/7 [==============================] - 2s 228ms/step - loss: 0.0190 - categorical_accuracy: 0.9932 - accuracy: 0.9932 - val_loss: 3.0156 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 133/150\n7/7 [==============================] - 2s 234ms/step - loss: 0.0176 - categorical_accuracy: 0.9924 - accuracy: 0.9924 - val_loss: 3.0144 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 134/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0236 - categorical_accuracy: 0.9877 - accuracy: 0.9877 - val_loss: 3.0029 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 135/150\n7/7 [==============================] - 2s 234ms/step - loss: 0.0148 - categorical_accuracy: 0.9898 - accuracy: 0.9898 - val_loss: 3.0016 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 136/150\n7/7 [==============================] - 2s 231ms/step - loss: 0.0141 - categorical_accuracy: 0.9903 - accuracy: 0.9903 - val_loss: 2.9975 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 137/150\n7/7 [==============================] - 2s 231ms/step - loss: 0.0133 - categorical_accuracy: 0.9926 - accuracy: 0.9926 - val_loss: 2.9627 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 138/150\n7/7 [==============================] - 2s 233ms/step - loss: 0.0261 - categorical_accuracy: 0.9863 - accuracy: 0.9863 - val_loss: 2.9678 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 139/150\n7/7 [==============================] - 2s 229ms/step - loss: 0.0150 - categorical_accuracy: 0.9934 - accuracy: 0.9934 - val_loss: 2.9786 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 140/150\n7/7 [==============================] - 2s 232ms/step - loss: 0.0132 - categorical_accuracy: 0.9935 - accuracy: 0.9935 - val_loss: 2.9763 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 141/150\n7/7 [==============================] - 2s 231ms/step - loss: 0.0123 - categorical_accuracy: 0.9947 - accuracy: 0.9947 - val_loss: 2.9446 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 142/150\n7/7 [==============================] - 2s 233ms/step - loss: 0.0115 - categorical_accuracy: 0.9944 - accuracy: 0.9944 - val_loss: 2.9445 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 143/150\n7/7 [==============================] - 2s 258ms/step - loss: 0.0170 - categorical_accuracy: 0.9869 - accuracy: 0.9869 - val_loss: 2.9763 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 144/150\n7/7 [==============================] - 2s 233ms/step - loss: 0.0175 - categorical_accuracy: 0.9863 - accuracy: 0.9863 - val_loss: 2.9987 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 145/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0168 - categorical_accuracy: 0.9863 - accuracy: 0.9863 - val_loss: 3.0144 - val_categorical_accuracy: 0.5844 - val_accuracy: 0.5844\nEpoch 146/150\n7/7 [==============================] - 2s 230ms/step - loss: 0.0126 - categorical_accuracy: 0.9947 - accuracy: 0.9947 - val_loss: 3.0175 - val_categorical_accuracy: 0.5844 - val_accuracy: 0.5844\nEpoch 147/150\n7/7 [==============================] - 2s 232ms/step - loss: 0.0144 - categorical_accuracy: 0.9926 - accuracy: 0.9926 - val_loss: 3.0204 - val_categorical_accuracy: 0.5844 - val_accuracy: 0.5844\nEpoch 148/150\n7/7 [==============================] - 2s 233ms/step - loss: 0.0138 - categorical_accuracy: 0.9934 - accuracy: 0.9934 - val_loss: 3.0372 - val_categorical_accuracy: 0.5844 - val_accuracy: 0.5844\nEpoch 149/150\n7/7 [==============================] - 2s 231ms/step - loss: 0.0194 - categorical_accuracy: 0.9862 - accuracy: 0.9862 - val_loss: 3.0475 - val_categorical_accuracy: 0.5844 - val_accuracy: 0.5844\nEpoch 150/150\n7/7 [==============================] - 2s 233ms/step - loss: 0.0169 - categorical_accuracy: 0.9883 - accuracy: 0.9883 - val_loss: 3.0495 - val_categorical_accuracy: 0.5844 - val_accuracy: 0.5844\n4/4 [==============================] - 0s 44ms/step - loss: 3.5693 - categorical_accuracy: 0.4639 - accuracy: 0.4639\n[[13 3 4 0 0]\n [ 5 12 1 2 0]\n [ 4 4 7 4 1]\n [ 5 3 3 9 0]\n [ 0 6 2 5 4]]\n precision recall f1-score support\n\n DN 0.48 0.65 0.55 20\n GOF 0.43 0.60 0.50 20\n HI 0.41 0.35 0.38 20\n LOF 0.45 0.45 0.45 20\n none 0.80 0.24 0.36 17\n\n accuracy 0.46 97\n macro avg 0.51 0.46 0.45 97\nweighted avg 0.51 0.46 0.45 97\n\n"
],
[
"with open('/content/%s' %modelName, 'wb') as file_pi:\r\n pickle.dump(history.history, file_pi)\r\nhistory = pickle.load(open('/content/%s' % modelName, \"rb\"))\r\nmodel.save(modelName +'.h5')\r\nplot(history)\r\nwith open('/content/%s_train' %modelName, 'wb') as file_pi:\r\n pickle.dump(X_train, file_pi)\r\nwith open('/content/%s_test' %modelName, 'wb') as file_pi:\r\n pickle.dump(X_test, file_pi)\r\nwith open('/content/%s_Labeltest' %modelName, 'wb') as file_pi:\r\n pickle.dump(y_test, file_pi)\r\nwith open('/content/%s_LabelTrain' %modelName, 'wb') as file_pi:\r\n pickle.dump(y_train, file_pi)\r\nwith open('/content/%s_ConfusionMatrix' %modelName, 'wb') as file_pi:\r\n pickle.dump(confusion_matrix(y_test.argmax(axis=-1), model.predict(X_test).argmax(axis=-1)), file_pi)\r\nwith open('/content/%s_ClassificationReport' %modelName, 'wb') as file_pi:\r\n pickle.dump(classification_report(y_test.argmax(axis=-1), model.predict(X_test).argmax(axis=-1), target_names=ClassNames), file_pi)",
"dict_keys(['loss', 'categorical_accuracy', 'accuracy', 'val_loss', 'val_categorical_accuracy', 'val_accuracy'])\n"
],
[
"",
"_____no_output_____"
]
],
[
[
"Running the model with TF-IDF vectorizer instead of CountVectorizer (adam optimizer)",
"_____no_output_____"
]
],
[
[
"uploaded = files.upload()",
"_____no_output_____"
],
[
"modelName = 'Model3' #name of the model, this will be used to save model evaluation and history\r\nnumEpochs = 150 # maximum number of epochs if early stopping doesnt work\r\nbatchsize = 20 # batchsize which will be used in each step by optimizer defined in the model\r\noptimizer = 'adam' #optimizer to be used in model.fit keras method",
"_____no_output_____"
],
[
"from sklearn.feature_extraction.text import TfidfVectorizer\r\n",
"_____no_output_____"
]
],
[
[
"For the next step, go to Preprocessing method and change CountVectorizer to TfIdfVectorizer",
"_____no_output_____"
]
],
[
[
"X, y = Preprocessing()\r\nX_train, X_test, y_train, y_test, ClassNames = TrainTestSplit(X, y)\r\nmodel = ModelBuild(X_train, y_train)\r\nPlotModel(model, modelName +\".png\")\r\nhistory = PlotTraining(model, X_test, y_test)\r\nprint(confusion_matrix(y_test.argmax(axis=-1),model.predict(X_test).argmax(axis=-1)))\r\nprint(classification_report(y_test.argmax(axis=-1), model.predict(X_test).argmax(axis=-1),target_names=ClassNames))\r\n",
"Model: \"model_2\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_3 (InputLayer) [(None, 55389)] 0 \n_________________________________________________________________\ndense_6 (Dense) (None, 500) 27695000 \n_________________________________________________________________\ndense_7 (Dense) (None, 100) 50100 \n_________________________________________________________________\ndense_8 (Dense) (None, 5) 505 \n=================================================================\nTotal params: 27,745,605\nTrainable params: 27,745,605\nNon-trainable params: 0\n_________________________________________________________________\nEpoch 1/150\n16/16 [==============================] - 3s 178ms/step - loss: 1.6028 - categorical_accuracy: 0.2213 - accuracy: 0.2213 - val_loss: 1.5433 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 2/150\n16/16 [==============================] - 3s 165ms/step - loss: 1.2730 - categorical_accuracy: 0.9245 - accuracy: 0.9245 - val_loss: 1.3357 - val_categorical_accuracy: 0.5584 - val_accuracy: 0.5584\nEpoch 3/150\n16/16 [==============================] - 3s 178ms/step - loss: 0.4882 - categorical_accuracy: 0.9311 - accuracy: 0.9311 - val_loss: 1.2724 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 4/150\n16/16 [==============================] - 3s 162ms/step - loss: 0.1110 - categorical_accuracy: 0.9643 - accuracy: 0.9643 - val_loss: 1.2933 - val_categorical_accuracy: 0.6753 - val_accuracy: 0.6753\nEpoch 5/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0650 - categorical_accuracy: 0.9740 - accuracy: 0.9740 - val_loss: 1.3172 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 6/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0663 - categorical_accuracy: 0.9746 - accuracy: 0.9746 - val_loss: 1.3390 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 7/150\n16/16 [==============================] - 3s 164ms/step - loss: 0.0834 - categorical_accuracy: 0.9684 - accuracy: 0.9684 - val_loss: 1.3466 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 8/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0293 - categorical_accuracy: 0.9881 - accuracy: 0.9881 - val_loss: 1.3368 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 9/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0346 - categorical_accuracy: 0.9911 - accuracy: 0.9911 - val_loss: 1.3370 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 10/150\n16/16 [==============================] - 3s 164ms/step - loss: 0.0280 - categorical_accuracy: 0.9909 - accuracy: 0.9909 - val_loss: 1.3646 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 11/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0284 - categorical_accuracy: 0.9870 - accuracy: 0.9870 - val_loss: 1.3432 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 12/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0408 - categorical_accuracy: 0.9808 - accuracy: 0.9808 - val_loss: 1.3717 - val_categorical_accuracy: 0.6883 - val_accuracy: 0.6883\nEpoch 13/150\n16/16 [==============================] - 3s 168ms/step - loss: 0.0225 - categorical_accuracy: 0.9887 - accuracy: 0.9887 - val_loss: 1.3742 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 14/150\n16/16 [==============================] - 3s 170ms/step - loss: 0.0259 - categorical_accuracy: 0.9830 - accuracy: 0.9830 - val_loss: 1.3811 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 15/150\n16/16 [==============================] - 3s 167ms/step - loss: 0.0323 - categorical_accuracy: 0.9754 - accuracy: 0.9754 - val_loss: 1.3740 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 16/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0320 - categorical_accuracy: 0.9729 - accuracy: 0.9729 - val_loss: 1.3927 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 17/150\n16/16 [==============================] - 3s 163ms/step - loss: 0.0169 - categorical_accuracy: 0.9914 - accuracy: 0.9914 - val_loss: 1.3947 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 18/150\n16/16 [==============================] - 3s 163ms/step - loss: 0.0185 - categorical_accuracy: 0.9867 - accuracy: 0.9867 - val_loss: 1.4015 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 19/150\n16/16 [==============================] - 3s 163ms/step - loss: 0.0221 - categorical_accuracy: 0.9883 - accuracy: 0.9883 - val_loss: 1.3684 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 20/150\n16/16 [==============================] - 3s 174ms/step - loss: 0.0320 - categorical_accuracy: 0.9857 - accuracy: 0.9857 - val_loss: 1.3930 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 21/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0169 - categorical_accuracy: 0.9919 - accuracy: 0.9919 - val_loss: 1.3999 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 22/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0123 - categorical_accuracy: 0.9934 - accuracy: 0.9934 - val_loss: 1.4209 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 23/150\n16/16 [==============================] - 3s 164ms/step - loss: 0.0288 - categorical_accuracy: 0.9795 - accuracy: 0.9795 - val_loss: 1.4107 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 24/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0316 - categorical_accuracy: 0.9860 - accuracy: 0.9860 - val_loss: 1.4274 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 25/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0204 - categorical_accuracy: 0.9864 - accuracy: 0.9864 - val_loss: 1.4339 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 26/150\n16/16 [==============================] - 3s 164ms/step - loss: 0.0236 - categorical_accuracy: 0.9775 - accuracy: 0.9775 - val_loss: 1.4243 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 27/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0108 - categorical_accuracy: 0.9924 - accuracy: 0.9924 - val_loss: 1.4341 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 28/150\n16/16 [==============================] - 3s 162ms/step - loss: 0.0254 - categorical_accuracy: 0.9735 - accuracy: 0.9735 - val_loss: 1.4416 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 29/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0210 - categorical_accuracy: 0.9822 - accuracy: 0.9822 - val_loss: 1.4395 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 30/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0236 - categorical_accuracy: 0.9884 - accuracy: 0.9884 - val_loss: 1.4432 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 31/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0096 - categorical_accuracy: 0.9921 - accuracy: 0.9921 - val_loss: 1.4523 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 32/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0210 - categorical_accuracy: 0.9942 - accuracy: 0.9942 - val_loss: 1.4965 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 33/150\n16/16 [==============================] - 3s 164ms/step - loss: 0.0406 - categorical_accuracy: 0.9749 - accuracy: 0.9749 - val_loss: 1.5091 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 34/150\n16/16 [==============================] - 3s 164ms/step - loss: 0.0218 - categorical_accuracy: 0.9943 - accuracy: 0.9943 - val_loss: 1.4751 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 35/150\n16/16 [==============================] - 3s 163ms/step - loss: 0.0195 - categorical_accuracy: 0.9871 - accuracy: 0.9871 - val_loss: 1.4660 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 36/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0168 - categorical_accuracy: 0.9904 - accuracy: 0.9904 - val_loss: 1.4529 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 37/150\n16/16 [==============================] - 3s 174ms/step - loss: 0.0286 - categorical_accuracy: 0.9856 - accuracy: 0.9856 - val_loss: 1.5108 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 38/150\n16/16 [==============================] - 3s 163ms/step - loss: 0.0249 - categorical_accuracy: 0.9841 - accuracy: 0.9841 - val_loss: 1.4411 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 39/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0325 - categorical_accuracy: 0.9695 - accuracy: 0.9695 - val_loss: 1.4555 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 40/150\n16/16 [==============================] - 3s 164ms/step - loss: 0.0184 - categorical_accuracy: 0.9845 - accuracy: 0.9845 - val_loss: 1.4638 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 41/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0182 - categorical_accuracy: 0.9863 - accuracy: 0.9863 - val_loss: 1.4758 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 42/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0201 - categorical_accuracy: 0.9810 - accuracy: 0.9810 - val_loss: 1.5077 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 43/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0153 - categorical_accuracy: 0.9909 - accuracy: 0.9909 - val_loss: 1.4745 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 44/150\n16/16 [==============================] - 3s 164ms/step - loss: 0.0118 - categorical_accuracy: 0.9938 - accuracy: 0.9938 - val_loss: 1.4856 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 45/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0178 - categorical_accuracy: 0.9868 - accuracy: 0.9868 - val_loss: 1.4799 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 46/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0263 - categorical_accuracy: 0.9746 - accuracy: 0.9746 - val_loss: 1.4766 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 47/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0128 - categorical_accuracy: 0.9948 - accuracy: 0.9948 - val_loss: 1.5321 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 48/150\n16/16 [==============================] - 3s 167ms/step - loss: 0.0275 - categorical_accuracy: 0.9713 - accuracy: 0.9713 - val_loss: 1.5311 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 49/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0184 - categorical_accuracy: 0.9869 - accuracy: 0.9869 - val_loss: 1.4906 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 50/150\n16/16 [==============================] - 3s 164ms/step - loss: 0.0120 - categorical_accuracy: 0.9953 - accuracy: 0.9953 - val_loss: 1.5155 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 51/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0182 - categorical_accuracy: 0.9910 - accuracy: 0.9910 - val_loss: 1.4913 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 52/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0131 - categorical_accuracy: 0.9929 - accuracy: 0.9929 - val_loss: 1.4908 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 53/150\n16/16 [==============================] - 3s 175ms/step - loss: 0.0177 - categorical_accuracy: 0.9907 - accuracy: 0.9907 - val_loss: 1.5102 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 54/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0265 - categorical_accuracy: 0.9688 - accuracy: 0.9688 - val_loss: 1.5282 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 55/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0173 - categorical_accuracy: 0.9950 - accuracy: 0.9950 - val_loss: 1.5189 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 56/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0173 - categorical_accuracy: 0.9864 - accuracy: 0.9864 - val_loss: 1.5137 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 57/150\n16/16 [==============================] - 3s 167ms/step - loss: 0.0211 - categorical_accuracy: 0.9864 - accuracy: 0.9864 - val_loss: 1.5138 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 58/150\n16/16 [==============================] - 3s 167ms/step - loss: 0.0133 - categorical_accuracy: 0.9896 - accuracy: 0.9896 - val_loss: 1.5490 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 59/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0186 - categorical_accuracy: 0.9870 - accuracy: 0.9870 - val_loss: 1.5109 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 60/150\n16/16 [==============================] - 3s 163ms/step - loss: 0.0263 - categorical_accuracy: 0.9798 - accuracy: 0.9798 - val_loss: 1.5228 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 61/150\n16/16 [==============================] - 3s 164ms/step - loss: 0.0175 - categorical_accuracy: 0.9887 - accuracy: 0.9887 - val_loss: 1.5322 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 62/150\n16/16 [==============================] - 3s 163ms/step - loss: 0.0230 - categorical_accuracy: 0.9739 - accuracy: 0.9739 - val_loss: 1.5379 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 63/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0196 - categorical_accuracy: 0.9812 - accuracy: 0.9812 - val_loss: 1.5318 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 64/150\n16/16 [==============================] - 3s 164ms/step - loss: 0.0155 - categorical_accuracy: 0.9924 - accuracy: 0.9924 - val_loss: 1.5441 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 65/150\n16/16 [==============================] - 3s 164ms/step - loss: 0.0207 - categorical_accuracy: 0.9755 - accuracy: 0.9755 - val_loss: 1.5475 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 66/150\n16/16 [==============================] - 3s 164ms/step - loss: 0.0114 - categorical_accuracy: 0.9887 - accuracy: 0.9887 - val_loss: 1.5630 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 67/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0266 - categorical_accuracy: 0.9797 - accuracy: 0.9797 - val_loss: 1.5679 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 68/150\n16/16 [==============================] - 3s 164ms/step - loss: 0.0236 - categorical_accuracy: 0.9762 - accuracy: 0.9762 - val_loss: 1.5647 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 69/150\n16/16 [==============================] - 3s 164ms/step - loss: 0.0223 - categorical_accuracy: 0.9819 - accuracy: 0.9819 - val_loss: 1.5682 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 70/150\n16/16 [==============================] - 3s 176ms/step - loss: 0.0273 - categorical_accuracy: 0.9748 - accuracy: 0.9748 - val_loss: 1.5579 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 71/150\n16/16 [==============================] - 3s 169ms/step - loss: 0.0144 - categorical_accuracy: 0.9906 - accuracy: 0.9906 - val_loss: 1.5758 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 72/150\n16/16 [==============================] - 3s 164ms/step - loss: 0.0112 - categorical_accuracy: 0.9944 - accuracy: 0.9944 - val_loss: 1.5807 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 73/150\n16/16 [==============================] - 3s 167ms/step - loss: 0.0159 - categorical_accuracy: 0.9983 - accuracy: 0.9983 - val_loss: 1.6180 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 74/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0254 - categorical_accuracy: 0.9838 - accuracy: 0.9838 - val_loss: 1.5451 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 75/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0195 - categorical_accuracy: 0.9879 - accuracy: 0.9879 - val_loss: 1.5608 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 76/150\n16/16 [==============================] - 3s 168ms/step - loss: 0.0130 - categorical_accuracy: 0.9887 - accuracy: 0.9887 - val_loss: 1.5690 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 77/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0163 - categorical_accuracy: 0.9905 - accuracy: 0.9905 - val_loss: 1.5711 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 78/150\n16/16 [==============================] - 3s 171ms/step - loss: 0.0122 - categorical_accuracy: 0.9944 - accuracy: 0.9944 - val_loss: 1.5991 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 79/150\n16/16 [==============================] - 3s 164ms/step - loss: 0.0148 - categorical_accuracy: 0.9901 - accuracy: 0.9901 - val_loss: 1.6153 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 80/150\n16/16 [==============================] - 3s 168ms/step - loss: 0.0108 - categorical_accuracy: 0.9946 - accuracy: 0.9946 - val_loss: 1.6008 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 81/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0161 - categorical_accuracy: 0.9826 - accuracy: 0.9826 - val_loss: 1.5888 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 82/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0232 - categorical_accuracy: 0.9801 - accuracy: 0.9801 - val_loss: 1.6078 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 83/150\n16/16 [==============================] - 3s 167ms/step - loss: 0.0230 - categorical_accuracy: 0.9796 - accuracy: 0.9796 - val_loss: 1.6002 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 84/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0055 - categorical_accuracy: 0.9953 - accuracy: 0.9953 - val_loss: 1.6205 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 85/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0266 - categorical_accuracy: 0.9685 - accuracy: 0.9685 - val_loss: 1.6020 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 86/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0216 - categorical_accuracy: 0.9912 - accuracy: 0.9912 - val_loss: 1.6223 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 87/150\n16/16 [==============================] - 3s 174ms/step - loss: 0.0139 - categorical_accuracy: 0.9945 - accuracy: 0.9945 - val_loss: 1.6347 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 88/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0140 - categorical_accuracy: 0.9963 - accuracy: 0.9963 - val_loss: 1.6904 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 89/150\n16/16 [==============================] - 3s 167ms/step - loss: 0.0176 - categorical_accuracy: 0.9945 - accuracy: 0.9945 - val_loss: 1.6409 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 90/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0141 - categorical_accuracy: 0.9948 - accuracy: 0.9948 - val_loss: 1.6347 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 91/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0125 - categorical_accuracy: 0.9954 - accuracy: 0.9954 - val_loss: 1.6334 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 92/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0263 - categorical_accuracy: 0.9767 - accuracy: 0.9767 - val_loss: 1.6840 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 93/150\n16/16 [==============================] - 3s 167ms/step - loss: 0.0170 - categorical_accuracy: 0.9936 - accuracy: 0.9936 - val_loss: 1.6112 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 94/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0208 - categorical_accuracy: 0.9820 - accuracy: 0.9820 - val_loss: 1.6256 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 95/150\n16/16 [==============================] - 3s 167ms/step - loss: 0.0318 - categorical_accuracy: 0.9785 - accuracy: 0.9785 - val_loss: 1.6222 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 96/150\n16/16 [==============================] - 3s 167ms/step - loss: 0.0228 - categorical_accuracy: 0.9793 - accuracy: 0.9793 - val_loss: 1.6550 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 97/150\n16/16 [==============================] - 3s 167ms/step - loss: 0.0228 - categorical_accuracy: 0.9831 - accuracy: 0.9831 - val_loss: 1.6280 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 98/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0181 - categorical_accuracy: 0.9890 - accuracy: 0.9890 - val_loss: 1.6601 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 99/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0162 - categorical_accuracy: 0.9910 - accuracy: 0.9910 - val_loss: 1.6500 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 100/150\n16/16 [==============================] - 3s 168ms/step - loss: 0.0278 - categorical_accuracy: 0.9838 - accuracy: 0.9838 - val_loss: 1.6488 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 101/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0205 - categorical_accuracy: 0.9797 - accuracy: 0.9797 - val_loss: 1.6682 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 102/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0179 - categorical_accuracy: 0.9898 - accuracy: 0.9898 - val_loss: 1.6243 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 103/150\n16/16 [==============================] - 3s 175ms/step - loss: 0.0170 - categorical_accuracy: 0.9927 - accuracy: 0.9927 - val_loss: 1.6300 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 104/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0109 - categorical_accuracy: 0.9908 - accuracy: 0.9908 - val_loss: 1.6586 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 105/150\n16/16 [==============================] - 3s 168ms/step - loss: 0.0173 - categorical_accuracy: 0.9948 - accuracy: 0.9948 - val_loss: 1.6714 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 106/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0131 - categorical_accuracy: 0.9912 - accuracy: 0.9912 - val_loss: 1.6748 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 107/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0138 - categorical_accuracy: 0.9933 - accuracy: 0.9933 - val_loss: 1.7029 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 108/150\n16/16 [==============================] - 3s 167ms/step - loss: 0.0206 - categorical_accuracy: 0.9897 - accuracy: 0.9897 - val_loss: 1.7388 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 109/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0179 - categorical_accuracy: 0.9899 - accuracy: 0.9899 - val_loss: 1.7357 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 110/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0134 - categorical_accuracy: 0.9851 - accuracy: 0.9851 - val_loss: 1.6712 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 111/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0215 - categorical_accuracy: 0.9945 - accuracy: 0.9945 - val_loss: 1.6801 - val_categorical_accuracy: 0.6753 - val_accuracy: 0.6753\nEpoch 112/150\n16/16 [==============================] - 3s 169ms/step - loss: 0.0094 - categorical_accuracy: 0.9943 - accuracy: 0.9943 - val_loss: 1.7077 - val_categorical_accuracy: 0.6753 - val_accuracy: 0.6753\nEpoch 113/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0142 - categorical_accuracy: 0.9875 - accuracy: 0.9875 - val_loss: 1.7203 - val_categorical_accuracy: 0.6753 - val_accuracy: 0.6753\nEpoch 114/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0129 - categorical_accuracy: 0.9890 - accuracy: 0.9890 - val_loss: 1.7125 - val_categorical_accuracy: 0.6753 - val_accuracy: 0.6753\nEpoch 115/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0080 - categorical_accuracy: 0.9978 - accuracy: 0.9978 - val_loss: 1.7831 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 116/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0103 - categorical_accuracy: 0.9959 - accuracy: 0.9959 - val_loss: 1.7559 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 117/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0156 - categorical_accuracy: 0.9918 - accuracy: 0.9918 - val_loss: 1.7499 - val_categorical_accuracy: 0.6753 - val_accuracy: 0.6753\nEpoch 118/150\n16/16 [==============================] - 3s 163ms/step - loss: 0.0199 - categorical_accuracy: 0.9858 - accuracy: 0.9858 - val_loss: 1.7550 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 119/150\n16/16 [==============================] - 3s 164ms/step - loss: 0.0171 - categorical_accuracy: 0.9833 - accuracy: 0.9833 - val_loss: 1.7553 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 120/150\n16/16 [==============================] - 3s 174ms/step - loss: 0.0178 - categorical_accuracy: 0.9848 - accuracy: 0.9848 - val_loss: 1.7628 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 121/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0299 - categorical_accuracy: 0.9786 - accuracy: 0.9786 - val_loss: 1.7543 - val_categorical_accuracy: 0.6753 - val_accuracy: 0.6753\nEpoch 122/150\n16/16 [==============================] - 3s 167ms/step - loss: 0.0101 - categorical_accuracy: 0.9954 - accuracy: 0.9954 - val_loss: 1.7582 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 123/150\n16/16 [==============================] - 3s 167ms/step - loss: 0.0126 - categorical_accuracy: 0.9952 - accuracy: 0.9952 - val_loss: 1.7614 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 124/150\n16/16 [==============================] - 3s 167ms/step - loss: 0.0147 - categorical_accuracy: 0.9888 - accuracy: 0.9888 - val_loss: 1.7466 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 125/150\n16/16 [==============================] - 3s 168ms/step - loss: 0.0297 - categorical_accuracy: 0.9868 - accuracy: 0.9868 - val_loss: 1.7633 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 126/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0115 - categorical_accuracy: 0.9918 - accuracy: 0.9918 - val_loss: 1.7767 - val_categorical_accuracy: 0.6753 - val_accuracy: 0.6753\nEpoch 127/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0131 - categorical_accuracy: 0.9898 - accuracy: 0.9898 - val_loss: 1.7818 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 128/150\n16/16 [==============================] - 3s 167ms/step - loss: 0.0063 - categorical_accuracy: 0.9930 - accuracy: 0.9930 - val_loss: 1.7911 - val_categorical_accuracy: 0.6753 - val_accuracy: 0.6753\nEpoch 129/150\n16/16 [==============================] - 3s 168ms/step - loss: 0.0192 - categorical_accuracy: 0.9907 - accuracy: 0.9907 - val_loss: 1.7835 - val_categorical_accuracy: 0.6753 - val_accuracy: 0.6753\nEpoch 130/150\n16/16 [==============================] - 3s 168ms/step - loss: 0.0228 - categorical_accuracy: 0.9835 - accuracy: 0.9835 - val_loss: 1.7474 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 131/150\n16/16 [==============================] - 3s 168ms/step - loss: 0.0256 - categorical_accuracy: 0.9872 - accuracy: 0.9872 - val_loss: 1.7769 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 132/150\n16/16 [==============================] - 3s 167ms/step - loss: 0.0214 - categorical_accuracy: 0.9876 - accuracy: 0.9876 - val_loss: 1.7664 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 133/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0204 - categorical_accuracy: 0.9856 - accuracy: 0.9856 - val_loss: 1.7945 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 134/150\n16/16 [==============================] - 3s 169ms/step - loss: 0.0149 - categorical_accuracy: 0.9901 - accuracy: 0.9901 - val_loss: 1.8278 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 135/150\n16/16 [==============================] - 3s 171ms/step - loss: 0.0151 - categorical_accuracy: 0.9842 - accuracy: 0.9842 - val_loss: 1.8091 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 136/150\n16/16 [==============================] - 3s 168ms/step - loss: 0.0134 - categorical_accuracy: 0.9858 - accuracy: 0.9858 - val_loss: 1.8199 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 137/150\n16/16 [==============================] - 3s 175ms/step - loss: 0.0138 - categorical_accuracy: 0.9849 - accuracy: 0.9849 - val_loss: 1.8425 - val_categorical_accuracy: 0.6753 - val_accuracy: 0.6753\nEpoch 138/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0121 - categorical_accuracy: 0.9875 - accuracy: 0.9875 - val_loss: 1.8341 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 139/150\n16/16 [==============================] - 3s 167ms/step - loss: 0.0150 - categorical_accuracy: 0.9883 - accuracy: 0.9883 - val_loss: 1.8382 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 140/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0221 - categorical_accuracy: 0.9890 - accuracy: 0.9890 - val_loss: 1.8566 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 141/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0162 - categorical_accuracy: 0.9933 - accuracy: 0.9933 - val_loss: 1.8050 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 142/150\n16/16 [==============================] - 3s 167ms/step - loss: 0.0192 - categorical_accuracy: 0.9829 - accuracy: 0.9829 - val_loss: 1.8171 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 143/150\n16/16 [==============================] - 3s 168ms/step - loss: 0.0133 - categorical_accuracy: 0.9927 - accuracy: 0.9927 - val_loss: 1.8188 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 144/150\n16/16 [==============================] - 3s 173ms/step - loss: 0.0245 - categorical_accuracy: 0.9818 - accuracy: 0.9818 - val_loss: 1.8321 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 145/150\n16/16 [==============================] - 3s 165ms/step - loss: 0.0096 - categorical_accuracy: 0.9940 - accuracy: 0.9940 - val_loss: 1.8456 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 146/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0119 - categorical_accuracy: 0.9916 - accuracy: 0.9916 - val_loss: 1.8515 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 147/150\n16/16 [==============================] - 3s 167ms/step - loss: 0.0172 - categorical_accuracy: 0.9919 - accuracy: 0.9919 - val_loss: 1.8447 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 148/150\n16/16 [==============================] - 3s 167ms/step - loss: 0.0123 - categorical_accuracy: 0.9950 - accuracy: 0.9950 - val_loss: 1.8522 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 149/150\n16/16 [==============================] - 3s 166ms/step - loss: 0.0202 - categorical_accuracy: 0.9840 - accuracy: 0.9840 - val_loss: 1.8721 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 150/150\n16/16 [==============================] - 3s 169ms/step - loss: 0.0108 - categorical_accuracy: 0.9915 - accuracy: 0.9915 - val_loss: 1.8740 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\n4/4 [==============================] - 0s 45ms/step - loss: 3.1126 - categorical_accuracy: 0.3918 - accuracy: 0.3918\n[[ 9 2 9 0 0]\n [ 5 8 6 1 0]\n [ 4 1 11 3 1]\n [ 4 1 11 4 0]\n [ 0 2 6 3 6]]\n precision recall f1-score support\n\n DN 0.41 0.45 0.43 20\n GOF 0.57 0.40 0.47 20\n HI 0.26 0.55 0.35 20\n LOF 0.36 0.20 0.26 20\n none 0.86 0.35 0.50 17\n\n accuracy 0.39 97\n macro avg 0.49 0.39 0.40 97\nweighted avg 0.48 0.39 0.40 97\n\n"
],
[
"with open('/content/%s' %modelName, 'wb') as file_pi:\r\n pickle.dump(history.history, file_pi)\r\nhistory = pickle.load(open('/content/%s' % modelName, \"rb\"))\r\nmodel.save(modelName +'.h5')\r\nplot(history)\r\nwith open('/content/%s_train' %modelName, 'wb') as file_pi:\r\n pickle.dump(X_train, file_pi)\r\nwith open('/content/%s_test' %modelName, 'wb') as file_pi:\r\n pickle.dump(X_test, file_pi)\r\nwith open('/content/%s_Labeltest' %modelName, 'wb') as file_pi:\r\n pickle.dump(y_test, file_pi)\r\nwith open('/content/%s_LabelTrain' %modelName, 'wb') as file_pi:\r\n pickle.dump(y_train, file_pi)\r\nwith open('/content/%s_ConfusionMatrix' %modelName, 'wb') as file_pi:\r\n pickle.dump(confusion_matrix(y_test.argmax(axis=-1), model.predict(X_test).argmax(axis=-1)), file_pi)\r\nwith open('/content/%s_ClassificationReport' %modelName, 'wb') as file_pi:\r\n pickle.dump(classification_report(y_test.argmax(axis=-1), model.predict(X_test).argmax(axis=-1), target_names=ClassNames), file_pi)",
"dict_keys(['loss', 'categorical_accuracy', 'accuracy', 'val_loss', 'val_categorical_accuracy', 'val_accuracy'])\n"
],
[
"print(pickle.load(open('Model3_ClassificationReport','rb')))",
" precision recall f1-score support\n\n DN 0.41 0.45 0.43 20\n GOF 0.57 0.40 0.47 20\n HI 0.26 0.55 0.35 20\n LOF 0.36 0.20 0.26 20\n none 0.86 0.35 0.50 17\n\n accuracy 0.39 97\n macro avg 0.49 0.39 0.40 97\nweighted avg 0.48 0.39 0.40 97\n\n"
],
[
"",
"_____no_output_____"
]
],
[
[
"Running model with adadelta optimizer and tfidf vectorizer",
"_____no_output_____"
]
],
[
[
"modelName = 'Model5' #name of the model, this will be used to save model evaluation and history\r\nnumEpochs = 200 # maximum number of epochs if early stopping doesnt work\r\nbatchsize = 50 # batchsize which will be used in each step by optimizer defined in the model\r\noptimizer = 'adam' #optimizer to be used in model.fit keras method",
"_____no_output_____"
],
[
"X, y = Preprocessing()\r\nX_train, X_test, y_train, y_test, ClassNames = TrainTestSplit(X, y)\r\nmodel = ModelBuild(X_train, y_train)\r\nPlotModel(model, modelName +\".png\")\r\nhistory = PlotTraining(model, X_test, y_test)\r\nprint(confusion_matrix(y_test.argmax(axis=-1),model.predict(X_test).argmax(axis=-1)))\r\nprint(classification_report(y_test.argmax(axis=-1), model.predict(X_test).argmax(axis=-1),target_names=ClassNames))\r\n",
"Model: \"model_4\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_5 (InputLayer) [(None, 55389)] 0 \n_________________________________________________________________\ndense_12 (Dense) (None, 500) 27695000 \n_________________________________________________________________\ndense_13 (Dense) (None, 100) 50100 \n_________________________________________________________________\ndense_14 (Dense) (None, 5) 505 \n=================================================================\nTotal params: 27,745,605\nTrainable params: 27,745,605\nNon-trainable params: 0\n_________________________________________________________________\nEpoch 1/200\n7/7 [==============================] - 2s 265ms/step - loss: 1.6061 - categorical_accuracy: 0.2644 - accuracy: 0.2644 - val_loss: 1.5790 - val_categorical_accuracy: 0.4805 - val_accuracy: 0.4805\nEpoch 2/200\n7/7 [==============================] - 2s 224ms/step - loss: 1.4397 - categorical_accuracy: 0.9499 - accuracy: 0.9499 - val_loss: 1.5018 - val_categorical_accuracy: 0.5325 - val_accuracy: 0.5325\nEpoch 3/200\n7/7 [==============================] - 2s 221ms/step - loss: 1.0596 - categorical_accuracy: 0.9532 - accuracy: 0.9532 - val_loss: 1.4027 - val_categorical_accuracy: 0.5844 - val_accuracy: 0.5844\nEpoch 4/200\n7/7 [==============================] - 2s 216ms/step - loss: 0.5255 - categorical_accuracy: 0.9574 - accuracy: 0.9574 - val_loss: 1.3426 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 5/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.1926 - categorical_accuracy: 0.9614 - accuracy: 0.9614 - val_loss: 1.3324 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 6/200\n7/7 [==============================] - 2s 247ms/step - loss: 0.0866 - categorical_accuracy: 0.9803 - accuracy: 0.9803 - val_loss: 1.3774 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 7/200\n7/7 [==============================] - 2s 219ms/step - loss: 0.0677 - categorical_accuracy: 0.9680 - accuracy: 0.9680 - val_loss: 1.4381 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 8/200\n7/7 [==============================] - 2s 219ms/step - loss: 0.0433 - categorical_accuracy: 0.9883 - accuracy: 0.9883 - val_loss: 1.4893 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 9/200\n7/7 [==============================] - 1s 216ms/step - loss: 0.0262 - categorical_accuracy: 0.9871 - accuracy: 0.9871 - val_loss: 1.5047 - val_categorical_accuracy: 0.6883 - val_accuracy: 0.6883\nEpoch 10/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0305 - categorical_accuracy: 0.9822 - accuracy: 0.9822 - val_loss: 1.5226 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 11/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0341 - categorical_accuracy: 0.9818 - accuracy: 0.9818 - val_loss: 1.5019 - val_categorical_accuracy: 0.6753 - val_accuracy: 0.6753\nEpoch 12/200\n7/7 [==============================] - 2s 221ms/step - loss: 0.0225 - categorical_accuracy: 0.9890 - accuracy: 0.9890 - val_loss: 1.5271 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 13/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0201 - categorical_accuracy: 0.9917 - accuracy: 0.9917 - val_loss: 1.5040 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 14/200\n7/7 [==============================] - 2s 220ms/step - loss: 0.0226 - categorical_accuracy: 0.9864 - accuracy: 0.9864 - val_loss: 1.4953 - val_categorical_accuracy: 0.6753 - val_accuracy: 0.6753\nEpoch 15/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0333 - categorical_accuracy: 0.9761 - accuracy: 0.9761 - val_loss: 1.5114 - val_categorical_accuracy: 0.6753 - val_accuracy: 0.6753\nEpoch 16/200\n7/7 [==============================] - 2s 219ms/step - loss: 0.0222 - categorical_accuracy: 0.9801 - accuracy: 0.9801 - val_loss: 1.5144 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 17/200\n7/7 [==============================] - 2s 218ms/step - loss: 0.0168 - categorical_accuracy: 0.9955 - accuracy: 0.9955 - val_loss: 1.5221 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 18/200\n7/7 [==============================] - 2s 217ms/step - loss: 0.0219 - categorical_accuracy: 0.9894 - accuracy: 0.9894 - val_loss: 1.6686 - val_categorical_accuracy: 0.5844 - val_accuracy: 0.5844\nEpoch 19/200\n7/7 [==============================] - 2s 220ms/step - loss: 0.0289 - categorical_accuracy: 0.9894 - accuracy: 0.9894 - val_loss: 1.6546 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 20/200\n7/7 [==============================] - 2s 220ms/step - loss: 0.0321 - categorical_accuracy: 0.9883 - accuracy: 0.9883 - val_loss: 1.6700 - val_categorical_accuracy: 0.5584 - val_accuracy: 0.5584\nEpoch 21/200\n7/7 [==============================] - 2s 220ms/step - loss: 0.0242 - categorical_accuracy: 0.9950 - accuracy: 0.9950 - val_loss: 1.6014 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 22/200\n7/7 [==============================] - 2s 219ms/step - loss: 0.0410 - categorical_accuracy: 0.9826 - accuracy: 0.9826 - val_loss: 1.5243 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 23/200\n7/7 [==============================] - 2s 219ms/step - loss: 0.0272 - categorical_accuracy: 0.9880 - accuracy: 0.9880 - val_loss: 1.4797 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 24/200\n7/7 [==============================] - 2s 217ms/step - loss: 0.0216 - categorical_accuracy: 0.9920 - accuracy: 0.9920 - val_loss: 1.4685 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 25/200\n7/7 [==============================] - 2s 221ms/step - loss: 0.0334 - categorical_accuracy: 0.9874 - accuracy: 0.9874 - val_loss: 1.4574 - val_categorical_accuracy: 0.6753 - val_accuracy: 0.6753\nEpoch 26/200\n7/7 [==============================] - 2s 240ms/step - loss: 0.0215 - categorical_accuracy: 0.9844 - accuracy: 0.9844 - val_loss: 1.4821 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 27/200\n7/7 [==============================] - 2s 219ms/step - loss: 0.0184 - categorical_accuracy: 0.9894 - accuracy: 0.9894 - val_loss: 1.4701 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 28/200\n7/7 [==============================] - 2s 218ms/step - loss: 0.0210 - categorical_accuracy: 0.9911 - accuracy: 0.9911 - val_loss: 1.4698 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 29/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0235 - categorical_accuracy: 0.9880 - accuracy: 0.9880 - val_loss: 1.4845 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 30/200\n7/7 [==============================] - 2s 221ms/step - loss: 0.0187 - categorical_accuracy: 0.9910 - accuracy: 0.9910 - val_loss: 1.4884 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 31/200\n7/7 [==============================] - 2s 221ms/step - loss: 0.0209 - categorical_accuracy: 0.9873 - accuracy: 0.9873 - val_loss: 1.4793 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 32/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0169 - categorical_accuracy: 0.9885 - accuracy: 0.9885 - val_loss: 1.4828 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 33/200\n7/7 [==============================] - 2s 224ms/step - loss: 0.0202 - categorical_accuracy: 0.9869 - accuracy: 0.9869 - val_loss: 1.4866 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 34/200\n7/7 [==============================] - 2s 233ms/step - loss: 0.0162 - categorical_accuracy: 0.9889 - accuracy: 0.9889 - val_loss: 1.4895 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 35/200\n7/7 [==============================] - 2s 233ms/step - loss: 0.0305 - categorical_accuracy: 0.9854 - accuracy: 0.9854 - val_loss: 1.5032 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 36/200\n7/7 [==============================] - 2s 236ms/step - loss: 0.0207 - categorical_accuracy: 0.9889 - accuracy: 0.9889 - val_loss: 1.5131 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 37/200\n7/7 [==============================] - 2s 226ms/step - loss: 0.0189 - categorical_accuracy: 0.9903 - accuracy: 0.9903 - val_loss: 1.5229 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 38/200\n7/7 [==============================] - 2s 220ms/step - loss: 0.0145 - categorical_accuracy: 0.9883 - accuracy: 0.9883 - val_loss: 1.5121 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 39/200\n7/7 [==============================] - 2s 221ms/step - loss: 0.0189 - categorical_accuracy: 0.9895 - accuracy: 0.9895 - val_loss: 1.5127 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 40/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0222 - categorical_accuracy: 0.9837 - accuracy: 0.9837 - val_loss: 1.5178 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 41/200\n7/7 [==============================] - 2s 219ms/step - loss: 0.0209 - categorical_accuracy: 0.9900 - accuracy: 0.9900 - val_loss: 1.5166 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 42/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0213 - categorical_accuracy: 0.9795 - accuracy: 0.9795 - val_loss: 1.5234 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 43/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0121 - categorical_accuracy: 0.9882 - accuracy: 0.9882 - val_loss: 1.5200 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 44/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0114 - categorical_accuracy: 0.9930 - accuracy: 0.9930 - val_loss: 1.5258 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 45/200\n7/7 [==============================] - 2s 217ms/step - loss: 0.0186 - categorical_accuracy: 0.9820 - accuracy: 0.9820 - val_loss: 1.5307 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 46/200\n7/7 [==============================] - 2s 221ms/step - loss: 0.0132 - categorical_accuracy: 0.9889 - accuracy: 0.9889 - val_loss: 1.5298 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 47/200\n7/7 [==============================] - 2s 239ms/step - loss: 0.0234 - categorical_accuracy: 0.9833 - accuracy: 0.9833 - val_loss: 1.5370 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 48/200\n7/7 [==============================] - 2s 221ms/step - loss: 0.0216 - categorical_accuracy: 0.9781 - accuracy: 0.9781 - val_loss: 1.5344 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 49/200\n7/7 [==============================] - 2s 219ms/step - loss: 0.0140 - categorical_accuracy: 0.9856 - accuracy: 0.9856 - val_loss: 1.5335 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 50/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0124 - categorical_accuracy: 0.9871 - accuracy: 0.9871 - val_loss: 1.5343 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 51/200\n7/7 [==============================] - 2s 218ms/step - loss: 0.0197 - categorical_accuracy: 0.9921 - accuracy: 0.9921 - val_loss: 1.5214 - val_categorical_accuracy: 0.5584 - val_accuracy: 0.5584\nEpoch 52/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0254 - categorical_accuracy: 0.9825 - accuracy: 0.9825 - val_loss: 1.5232 - val_categorical_accuracy: 0.5844 - val_accuracy: 0.5844\nEpoch 53/200\n7/7 [==============================] - 2s 221ms/step - loss: 0.0225 - categorical_accuracy: 0.9825 - accuracy: 0.9825 - val_loss: 1.5268 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 54/200\n7/7 [==============================] - 2s 219ms/step - loss: 0.0185 - categorical_accuracy: 0.9906 - accuracy: 0.9906 - val_loss: 1.5332 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 55/200\n7/7 [==============================] - 2s 220ms/step - loss: 0.0137 - categorical_accuracy: 0.9941 - accuracy: 0.9941 - val_loss: 1.5363 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 56/200\n7/7 [==============================] - 2s 218ms/step - loss: 0.0134 - categorical_accuracy: 0.9952 - accuracy: 0.9952 - val_loss: 1.5427 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 57/200\n7/7 [==============================] - 2s 219ms/step - loss: 0.0135 - categorical_accuracy: 0.9921 - accuracy: 0.9921 - val_loss: 1.5477 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 58/200\n7/7 [==============================] - 2s 221ms/step - loss: 0.0171 - categorical_accuracy: 0.9899 - accuracy: 0.9899 - val_loss: 1.5528 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 59/200\n7/7 [==============================] - 2s 227ms/step - loss: 0.0175 - categorical_accuracy: 0.9867 - accuracy: 0.9867 - val_loss: 1.5529 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 60/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0129 - categorical_accuracy: 0.9951 - accuracy: 0.9951 - val_loss: 1.5286 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 61/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0149 - categorical_accuracy: 0.9874 - accuracy: 0.9874 - val_loss: 1.5586 - val_categorical_accuracy: 0.6623 - val_accuracy: 0.6623\nEpoch 62/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0151 - categorical_accuracy: 0.9842 - accuracy: 0.9842 - val_loss: 1.5607 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 63/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0168 - categorical_accuracy: 0.9877 - accuracy: 0.9877 - val_loss: 1.5622 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 64/200\n7/7 [==============================] - 2s 243ms/step - loss: 0.0157 - categorical_accuracy: 0.9909 - accuracy: 0.9909 - val_loss: 1.5559 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 65/200\n7/7 [==============================] - 2s 220ms/step - loss: 0.0163 - categorical_accuracy: 0.9856 - accuracy: 0.9856 - val_loss: 1.5486 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 66/200\n7/7 [==============================] - 2s 220ms/step - loss: 0.0176 - categorical_accuracy: 0.9842 - accuracy: 0.9842 - val_loss: 1.5404 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 67/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0202 - categorical_accuracy: 0.9881 - accuracy: 0.9881 - val_loss: 1.5490 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 68/200\n7/7 [==============================] - 2s 218ms/step - loss: 0.0106 - categorical_accuracy: 0.9935 - accuracy: 0.9935 - val_loss: 1.5534 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 69/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0130 - categorical_accuracy: 0.9948 - accuracy: 0.9948 - val_loss: 1.5489 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 70/200\n7/7 [==============================] - 2s 224ms/step - loss: 0.0174 - categorical_accuracy: 0.9866 - accuracy: 0.9866 - val_loss: 1.5496 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 71/200\n7/7 [==============================] - 2s 221ms/step - loss: 0.0141 - categorical_accuracy: 0.9888 - accuracy: 0.9888 - val_loss: 1.5550 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 72/200\n7/7 [==============================] - 2s 219ms/step - loss: 0.0133 - categorical_accuracy: 0.9921 - accuracy: 0.9921 - val_loss: 1.5519 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 73/200\n7/7 [==============================] - 2s 225ms/step - loss: 0.0279 - categorical_accuracy: 0.9829 - accuracy: 0.9829 - val_loss: 1.5501 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 74/200\n7/7 [==============================] - 2s 219ms/step - loss: 0.0176 - categorical_accuracy: 0.9928 - accuracy: 0.9928 - val_loss: 1.5580 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 75/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0130 - categorical_accuracy: 0.9935 - accuracy: 0.9935 - val_loss: 1.5738 - val_categorical_accuracy: 0.6883 - val_accuracy: 0.6883\nEpoch 76/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0256 - categorical_accuracy: 0.9926 - accuracy: 0.9926 - val_loss: 1.5658 - val_categorical_accuracy: 0.6753 - val_accuracy: 0.6753\nEpoch 77/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0185 - categorical_accuracy: 0.9852 - accuracy: 0.9852 - val_loss: 1.5660 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 78/200\n7/7 [==============================] - 2s 243ms/step - loss: 0.0144 - categorical_accuracy: 0.9942 - accuracy: 0.9942 - val_loss: 1.5707 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 79/200\n7/7 [==============================] - 2s 221ms/step - loss: 0.0145 - categorical_accuracy: 0.9915 - accuracy: 0.9915 - val_loss: 1.5695 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 80/200\n7/7 [==============================] - 2s 220ms/step - loss: 0.0135 - categorical_accuracy: 0.9953 - accuracy: 0.9953 - val_loss: 1.5720 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 81/200\n7/7 [==============================] - 2s 221ms/step - loss: 0.0114 - categorical_accuracy: 0.9926 - accuracy: 0.9926 - val_loss: 1.5699 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 82/200\n7/7 [==============================] - 2s 219ms/step - loss: 0.0141 - categorical_accuracy: 0.9921 - accuracy: 0.9921 - val_loss: 1.5905 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 83/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0144 - categorical_accuracy: 0.9936 - accuracy: 0.9936 - val_loss: 1.5970 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 84/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0163 - categorical_accuracy: 0.9898 - accuracy: 0.9898 - val_loss: 1.5810 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 85/200\n7/7 [==============================] - 2s 220ms/step - loss: 0.0208 - categorical_accuracy: 0.9835 - accuracy: 0.9835 - val_loss: 1.5879 - val_categorical_accuracy: 0.6883 - val_accuracy: 0.6883\nEpoch 86/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0174 - categorical_accuracy: 0.9926 - accuracy: 0.9926 - val_loss: 1.6007 - val_categorical_accuracy: 0.6883 - val_accuracy: 0.6883\nEpoch 87/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0199 - categorical_accuracy: 0.9916 - accuracy: 0.9916 - val_loss: 1.5839 - val_categorical_accuracy: 0.6753 - val_accuracy: 0.6753\nEpoch 88/200\n7/7 [==============================] - 2s 219ms/step - loss: 0.0147 - categorical_accuracy: 0.9939 - accuracy: 0.9939 - val_loss: 1.6152 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 89/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0276 - categorical_accuracy: 0.9913 - accuracy: 0.9913 - val_loss: 1.5777 - val_categorical_accuracy: 0.5844 - val_accuracy: 0.5844\nEpoch 90/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0176 - categorical_accuracy: 0.9896 - accuracy: 0.9896 - val_loss: 1.5579 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 91/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0141 - categorical_accuracy: 0.9912 - accuracy: 0.9912 - val_loss: 1.5696 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 92/200\n7/7 [==============================] - 2s 219ms/step - loss: 0.0167 - categorical_accuracy: 0.9929 - accuracy: 0.9929 - val_loss: 1.5754 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 93/200\n7/7 [==============================] - 2s 221ms/step - loss: 0.0140 - categorical_accuracy: 0.9906 - accuracy: 0.9906 - val_loss: 1.5780 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 94/200\n7/7 [==============================] - 2s 220ms/step - loss: 0.0168 - categorical_accuracy: 0.9855 - accuracy: 0.9855 - val_loss: 1.5846 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 95/200\n7/7 [==============================] - 2s 221ms/step - loss: 0.0165 - categorical_accuracy: 0.9870 - accuracy: 0.9870 - val_loss: 1.5928 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 96/200\n7/7 [==============================] - 2s 220ms/step - loss: 0.0198 - categorical_accuracy: 0.9875 - accuracy: 0.9875 - val_loss: 1.5948 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 97/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0163 - categorical_accuracy: 0.9883 - accuracy: 0.9883 - val_loss: 1.6000 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 98/200\n7/7 [==============================] - 2s 226ms/step - loss: 0.0141 - categorical_accuracy: 0.9942 - accuracy: 0.9942 - val_loss: 1.5995 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 99/200\n7/7 [==============================] - 2s 248ms/step - loss: 0.0146 - categorical_accuracy: 0.9868 - accuracy: 0.9868 - val_loss: 1.5980 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 100/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0118 - categorical_accuracy: 0.9903 - accuracy: 0.9903 - val_loss: 1.6004 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 101/200\n7/7 [==============================] - 2s 221ms/step - loss: 0.0122 - categorical_accuracy: 0.9912 - accuracy: 0.9912 - val_loss: 1.6102 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 102/200\n7/7 [==============================] - 2s 221ms/step - loss: 0.0136 - categorical_accuracy: 0.9958 - accuracy: 0.9958 - val_loss: 1.6659 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 103/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0271 - categorical_accuracy: 0.9886 - accuracy: 0.9886 - val_loss: 1.6602 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 104/200\n7/7 [==============================] - 2s 227ms/step - loss: 0.0308 - categorical_accuracy: 0.9854 - accuracy: 0.9854 - val_loss: 1.6386 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 105/200\n7/7 [==============================] - 2s 220ms/step - loss: 0.0274 - categorical_accuracy: 0.9805 - accuracy: 0.9805 - val_loss: 1.6110 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 106/200\n7/7 [==============================] - 2s 218ms/step - loss: 0.0180 - categorical_accuracy: 0.9867 - accuracy: 0.9867 - val_loss: 1.6103 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 107/200\n7/7 [==============================] - 2s 220ms/step - loss: 0.0220 - categorical_accuracy: 0.9875 - accuracy: 0.9875 - val_loss: 1.6123 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 108/200\n7/7 [==============================] - 2s 220ms/step - loss: 0.0094 - categorical_accuracy: 0.9942 - accuracy: 0.9942 - val_loss: 1.6072 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 109/200\n7/7 [==============================] - 2s 221ms/step - loss: 0.0100 - categorical_accuracy: 0.9947 - accuracy: 0.9947 - val_loss: 1.6101 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 110/200\n7/7 [==============================] - 2s 228ms/step - loss: 0.0134 - categorical_accuracy: 0.9883 - accuracy: 0.9883 - val_loss: 1.6174 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 111/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0135 - categorical_accuracy: 0.9932 - accuracy: 0.9932 - val_loss: 1.6155 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 112/200\n7/7 [==============================] - 2s 226ms/step - loss: 0.0125 - categorical_accuracy: 0.9884 - accuracy: 0.9884 - val_loss: 1.6201 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 113/200\n7/7 [==============================] - 2s 220ms/step - loss: 0.0103 - categorical_accuracy: 0.9911 - accuracy: 0.9911 - val_loss: 1.6270 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 114/200\n7/7 [==============================] - 2s 224ms/step - loss: 0.0105 - categorical_accuracy: 0.9908 - accuracy: 0.9908 - val_loss: 1.6212 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 115/200\n7/7 [==============================] - 2s 221ms/step - loss: 0.0161 - categorical_accuracy: 0.9870 - accuracy: 0.9870 - val_loss: 1.6193 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 116/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0166 - categorical_accuracy: 0.9913 - accuracy: 0.9913 - val_loss: 1.6202 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 117/200\n7/7 [==============================] - 2s 228ms/step - loss: 0.0143 - categorical_accuracy: 0.9838 - accuracy: 0.9838 - val_loss: 1.6293 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 118/200\n7/7 [==============================] - 2s 221ms/step - loss: 0.0123 - categorical_accuracy: 0.9882 - accuracy: 0.9882 - val_loss: 1.6215 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 119/200\n7/7 [==============================] - 2s 218ms/step - loss: 0.0171 - categorical_accuracy: 0.9810 - accuracy: 0.9810 - val_loss: 1.6469 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 120/200\n7/7 [==============================] - 2s 221ms/step - loss: 0.0124 - categorical_accuracy: 0.9916 - accuracy: 0.9916 - val_loss: 1.6591 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 121/200\n7/7 [==============================] - 2s 245ms/step - loss: 0.0126 - categorical_accuracy: 0.9911 - accuracy: 0.9911 - val_loss: 1.6492 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 122/200\n7/7 [==============================] - 2s 225ms/step - loss: 0.0203 - categorical_accuracy: 0.9869 - accuracy: 0.9869 - val_loss: 1.6418 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 123/200\n7/7 [==============================] - 2s 224ms/step - loss: 0.0163 - categorical_accuracy: 0.9869 - accuracy: 0.9869 - val_loss: 1.6340 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 124/200\n7/7 [==============================] - 2s 220ms/step - loss: 0.0198 - categorical_accuracy: 0.9871 - accuracy: 0.9871 - val_loss: 1.6327 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 125/200\n7/7 [==============================] - 2s 221ms/step - loss: 0.0095 - categorical_accuracy: 0.9953 - accuracy: 0.9953 - val_loss: 1.6242 - val_categorical_accuracy: 0.5714 - val_accuracy: 0.5714\nEpoch 126/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0194 - categorical_accuracy: 0.9906 - accuracy: 0.9906 - val_loss: 1.6241 - val_categorical_accuracy: 0.5714 - val_accuracy: 0.5714\nEpoch 127/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0105 - categorical_accuracy: 0.9952 - accuracy: 0.9952 - val_loss: 1.6197 - val_categorical_accuracy: 0.5844 - val_accuracy: 0.5844\nEpoch 128/200\n7/7 [==============================] - 2s 225ms/step - loss: 0.0186 - categorical_accuracy: 0.9817 - accuracy: 0.9817 - val_loss: 1.6208 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 129/200\n7/7 [==============================] - 2s 224ms/step - loss: 0.0104 - categorical_accuracy: 0.9958 - accuracy: 0.9958 - val_loss: 1.6271 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 130/200\n7/7 [==============================] - 2s 225ms/step - loss: 0.0153 - categorical_accuracy: 0.9888 - accuracy: 0.9888 - val_loss: 1.6385 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 131/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0174 - categorical_accuracy: 0.9921 - accuracy: 0.9921 - val_loss: 1.6433 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 132/200\n7/7 [==============================] - 2s 221ms/step - loss: 0.0140 - categorical_accuracy: 0.9927 - accuracy: 0.9927 - val_loss: 1.6468 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 133/200\n7/7 [==============================] - 2s 228ms/step - loss: 0.0131 - categorical_accuracy: 0.9903 - accuracy: 0.9903 - val_loss: 1.6498 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 134/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0149 - categorical_accuracy: 0.9944 - accuracy: 0.9944 - val_loss: 1.6515 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 135/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0131 - categorical_accuracy: 0.9874 - accuracy: 0.9874 - val_loss: 1.6484 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 136/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0165 - categorical_accuracy: 0.9870 - accuracy: 0.9870 - val_loss: 1.6447 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 137/200\n7/7 [==============================] - 2s 230ms/step - loss: 0.0142 - categorical_accuracy: 0.9875 - accuracy: 0.9875 - val_loss: 1.6500 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 138/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0151 - categorical_accuracy: 0.9911 - accuracy: 0.9911 - val_loss: 1.6879 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 139/200\n7/7 [==============================] - 2s 227ms/step - loss: 0.0173 - categorical_accuracy: 0.9901 - accuracy: 0.9901 - val_loss: 1.6906 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 140/200\n7/7 [==============================] - 2s 224ms/step - loss: 0.0090 - categorical_accuracy: 0.9939 - accuracy: 0.9939 - val_loss: 1.6827 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 141/200\n7/7 [==============================] - 2s 248ms/step - loss: 0.0093 - categorical_accuracy: 0.9947 - accuracy: 0.9947 - val_loss: 1.6767 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 142/200\n7/7 [==============================] - 2s 220ms/step - loss: 0.0162 - categorical_accuracy: 0.9920 - accuracy: 0.9920 - val_loss: 1.6738 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 143/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0218 - categorical_accuracy: 0.9863 - accuracy: 0.9863 - val_loss: 1.6609 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 144/200\n7/7 [==============================] - 2s 226ms/step - loss: 0.0093 - categorical_accuracy: 0.9958 - accuracy: 0.9958 - val_loss: 1.6525 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 145/200\n7/7 [==============================] - 2s 224ms/step - loss: 0.0212 - categorical_accuracy: 0.9878 - accuracy: 0.9878 - val_loss: 1.6423 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 146/200\n7/7 [==============================] - 2s 226ms/step - loss: 0.0129 - categorical_accuracy: 0.9921 - accuracy: 0.9921 - val_loss: 1.6432 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 147/200\n7/7 [==============================] - 2s 224ms/step - loss: 0.0071 - categorical_accuracy: 0.9966 - accuracy: 0.9966 - val_loss: 1.6412 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 148/200\n7/7 [==============================] - 2s 225ms/step - loss: 0.0097 - categorical_accuracy: 0.9934 - accuracy: 0.9934 - val_loss: 1.6286 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 149/200\n7/7 [==============================] - 2s 227ms/step - loss: 0.0080 - categorical_accuracy: 0.9958 - accuracy: 0.9958 - val_loss: 1.6270 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 150/200\n7/7 [==============================] - 2s 224ms/step - loss: 0.0254 - categorical_accuracy: 0.9854 - accuracy: 0.9854 - val_loss: 1.6315 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 151/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0162 - categorical_accuracy: 0.9896 - accuracy: 0.9896 - val_loss: 1.6411 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 152/200\n7/7 [==============================] - 2s 225ms/step - loss: 0.0131 - categorical_accuracy: 0.9931 - accuracy: 0.9931 - val_loss: 1.6405 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 153/200\n7/7 [==============================] - 2s 219ms/step - loss: 0.0118 - categorical_accuracy: 0.9935 - accuracy: 0.9935 - val_loss: 1.6416 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 154/200\n7/7 [==============================] - 2s 221ms/step - loss: 0.0226 - categorical_accuracy: 0.9812 - accuracy: 0.9812 - val_loss: 1.6425 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 155/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0136 - categorical_accuracy: 0.9899 - accuracy: 0.9899 - val_loss: 1.6434 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 156/200\n7/7 [==============================] - 2s 229ms/step - loss: 0.0288 - categorical_accuracy: 0.9806 - accuracy: 0.9806 - val_loss: 1.6347 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 157/200\n7/7 [==============================] - 2s 220ms/step - loss: 0.0177 - categorical_accuracy: 0.9875 - accuracy: 0.9875 - val_loss: 1.6385 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 158/200\n7/7 [==============================] - 2s 245ms/step - loss: 0.0253 - categorical_accuracy: 0.9829 - accuracy: 0.9829 - val_loss: 1.6461 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 159/200\n7/7 [==============================] - 2s 219ms/step - loss: 0.0150 - categorical_accuracy: 0.9920 - accuracy: 0.9920 - val_loss: 1.6553 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 160/200\n7/7 [==============================] - 2s 225ms/step - loss: 0.0174 - categorical_accuracy: 0.9866 - accuracy: 0.9866 - val_loss: 1.6604 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 161/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0146 - categorical_accuracy: 0.9896 - accuracy: 0.9896 - val_loss: 1.6922 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 162/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0176 - categorical_accuracy: 0.9831 - accuracy: 0.9831 - val_loss: 1.7785 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 163/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0315 - categorical_accuracy: 0.9912 - accuracy: 0.9912 - val_loss: 1.6837 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 164/200\n7/7 [==============================] - 2s 220ms/step - loss: 0.0190 - categorical_accuracy: 0.9871 - accuracy: 0.9871 - val_loss: 1.6893 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 165/200\n7/7 [==============================] - 2s 224ms/step - loss: 0.0192 - categorical_accuracy: 0.9844 - accuracy: 0.9844 - val_loss: 1.6927 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 166/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0106 - categorical_accuracy: 0.9912 - accuracy: 0.9912 - val_loss: 1.6876 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 167/200\n7/7 [==============================] - 2s 224ms/step - loss: 0.0420 - categorical_accuracy: 0.9874 - accuracy: 0.9874 - val_loss: 2.5766 - val_categorical_accuracy: 0.4286 - val_accuracy: 0.4286\nEpoch 168/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0713 - categorical_accuracy: 0.9843 - accuracy: 0.9843 - val_loss: 2.2569 - val_categorical_accuracy: 0.4805 - val_accuracy: 0.4805\nEpoch 169/200\n7/7 [==============================] - 2s 226ms/step - loss: 0.0371 - categorical_accuracy: 0.9813 - accuracy: 0.9813 - val_loss: 1.8179 - val_categorical_accuracy: 0.5714 - val_accuracy: 0.5714\nEpoch 170/200\n7/7 [==============================] - 2s 221ms/step - loss: 0.0662 - categorical_accuracy: 0.9810 - accuracy: 0.9810 - val_loss: 1.7300 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 171/200\n7/7 [==============================] - 2s 221ms/step - loss: 0.0286 - categorical_accuracy: 0.9869 - accuracy: 0.9869 - val_loss: 1.6549 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 172/200\n7/7 [==============================] - 2s 244ms/step - loss: 0.0212 - categorical_accuracy: 0.9918 - accuracy: 0.9918 - val_loss: 1.5864 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 173/200\n7/7 [==============================] - 2s 220ms/step - loss: 0.0138 - categorical_accuracy: 0.9881 - accuracy: 0.9881 - val_loss: 1.5966 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 174/200\n7/7 [==============================] - 2s 225ms/step - loss: 0.0203 - categorical_accuracy: 0.9931 - accuracy: 0.9931 - val_loss: 1.7612 - val_categorical_accuracy: 0.5195 - val_accuracy: 0.5195\nEpoch 175/200\n7/7 [==============================] - 2s 224ms/step - loss: 0.0173 - categorical_accuracy: 0.9914 - accuracy: 0.9914 - val_loss: 1.8206 - val_categorical_accuracy: 0.5065 - val_accuracy: 0.5065\nEpoch 176/200\n7/7 [==============================] - 2s 224ms/step - loss: 0.0173 - categorical_accuracy: 0.9871 - accuracy: 0.9871 - val_loss: 1.7602 - val_categorical_accuracy: 0.5065 - val_accuracy: 0.5065\nEpoch 177/200\n7/7 [==============================] - 2s 221ms/step - loss: 0.0169 - categorical_accuracy: 0.9838 - accuracy: 0.9838 - val_loss: 1.6933 - val_categorical_accuracy: 0.5584 - val_accuracy: 0.5584\nEpoch 178/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0264 - categorical_accuracy: 0.9830 - accuracy: 0.9830 - val_loss: 1.6293 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 179/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0203 - categorical_accuracy: 0.9827 - accuracy: 0.9827 - val_loss: 1.6126 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 180/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0195 - categorical_accuracy: 0.9927 - accuracy: 0.9927 - val_loss: 1.6022 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 181/200\n7/7 [==============================] - 2s 224ms/step - loss: 0.0129 - categorical_accuracy: 0.9932 - accuracy: 0.9932 - val_loss: 1.6105 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 182/200\n7/7 [==============================] - 2s 224ms/step - loss: 0.0214 - categorical_accuracy: 0.9768 - accuracy: 0.9768 - val_loss: 1.6240 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 183/200\n7/7 [==============================] - 2s 226ms/step - loss: 0.0138 - categorical_accuracy: 0.9941 - accuracy: 0.9941 - val_loss: 1.6495 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 184/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0202 - categorical_accuracy: 0.9896 - accuracy: 0.9896 - val_loss: 1.6601 - val_categorical_accuracy: 0.6494 - val_accuracy: 0.6494\nEpoch 185/200\n7/7 [==============================] - 2s 225ms/step - loss: 0.0268 - categorical_accuracy: 0.9863 - accuracy: 0.9863 - val_loss: 1.6659 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\nEpoch 186/200\n7/7 [==============================] - 2s 245ms/step - loss: 0.0135 - categorical_accuracy: 0.9883 - accuracy: 0.9883 - val_loss: 1.7288 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 187/200\n7/7 [==============================] - 2s 224ms/step - loss: 0.0178 - categorical_accuracy: 0.9859 - accuracy: 0.9859 - val_loss: 1.7336 - val_categorical_accuracy: 0.5195 - val_accuracy: 0.5195\nEpoch 188/200\n7/7 [==============================] - 2s 230ms/step - loss: 0.0144 - categorical_accuracy: 0.9881 - accuracy: 0.9881 - val_loss: 1.7156 - val_categorical_accuracy: 0.5195 - val_accuracy: 0.5195\nEpoch 189/200\n7/7 [==============================] - 2s 221ms/step - loss: 0.0193 - categorical_accuracy: 0.9882 - accuracy: 0.9882 - val_loss: 1.7133 - val_categorical_accuracy: 0.5455 - val_accuracy: 0.5455\nEpoch 190/200\n7/7 [==============================] - 2s 225ms/step - loss: 0.0152 - categorical_accuracy: 0.9879 - accuracy: 0.9879 - val_loss: 1.6891 - val_categorical_accuracy: 0.5584 - val_accuracy: 0.5584\nEpoch 191/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0146 - categorical_accuracy: 0.9932 - accuracy: 0.9932 - val_loss: 1.6747 - val_categorical_accuracy: 0.5714 - val_accuracy: 0.5714\nEpoch 192/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0225 - categorical_accuracy: 0.9850 - accuracy: 0.9850 - val_loss: 1.6665 - val_categorical_accuracy: 0.5974 - val_accuracy: 0.5974\nEpoch 193/200\n7/7 [==============================] - 2s 222ms/step - loss: 0.0145 - categorical_accuracy: 0.9863 - accuracy: 0.9863 - val_loss: 1.6657 - val_categorical_accuracy: 0.6104 - val_accuracy: 0.6104\nEpoch 194/200\n7/7 [==============================] - 2s 223ms/step - loss: 0.0154 - categorical_accuracy: 0.9921 - accuracy: 0.9921 - val_loss: 1.6685 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 195/200\n7/7 [==============================] - 2s 228ms/step - loss: 0.0205 - categorical_accuracy: 0.9911 - accuracy: 0.9911 - val_loss: 1.6797 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 196/200\n7/7 [==============================] - 2s 230ms/step - loss: 0.0141 - categorical_accuracy: 0.9910 - accuracy: 0.9910 - val_loss: 1.6879 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 197/200\n7/7 [==============================] - 2s 230ms/step - loss: 0.0163 - categorical_accuracy: 0.9920 - accuracy: 0.9920 - val_loss: 1.6900 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 198/200\n7/7 [==============================] - 2s 230ms/step - loss: 0.0183 - categorical_accuracy: 0.9931 - accuracy: 0.9931 - val_loss: 1.6979 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 199/200\n7/7 [==============================] - 2s 234ms/step - loss: 0.0104 - categorical_accuracy: 0.9926 - accuracy: 0.9926 - val_loss: 1.7037 - val_categorical_accuracy: 0.6234 - val_accuracy: 0.6234\nEpoch 200/200\n7/7 [==============================] - 2s 230ms/step - loss: 0.0205 - categorical_accuracy: 0.9837 - accuracy: 0.9837 - val_loss: 1.7142 - val_categorical_accuracy: 0.6364 - val_accuracy: 0.6364\n4/4 [==============================] - 0s 50ms/step - loss: 2.6128 - categorical_accuracy: 0.4124 - accuracy: 0.4124\n[[10 2 8 0 0]\n [ 5 9 6 0 0]\n [ 5 1 12 1 1]\n [ 4 1 11 4 0]\n [ 0 2 9 1 5]]\n precision recall f1-score support\n\n DN 0.42 0.50 0.45 20\n GOF 0.60 0.45 0.51 20\n HI 0.26 0.60 0.36 20\n LOF 0.67 0.20 0.31 20\n none 0.83 0.29 0.43 17\n\n accuracy 0.41 97\n macro avg 0.56 0.41 0.41 97\nweighted avg 0.55 0.41 0.41 97\n\n"
],
[
"X_train.shape",
"_____no_output_____"
],
[
"with open('/content/%s' %modelName, 'wb') as file_pi:\r\n pickle.dump(history.history, file_pi)\r\nhistory = pickle.load(open('/content/%s' % modelName, \"rb\"))\r\nmodel.save(modelName +'.h5')\r\nplot(history)\r\nwith open('/content/%s_train' %modelName, 'wb') as file_pi:\r\n pickle.dump(X_train, file_pi)\r\nwith open('/content/%s_test' %modelName, 'wb') as file_pi:\r\n pickle.dump(X_test, file_pi)\r\nwith open('/content/%s_Labeltest' %modelName, 'wb') as file_pi:\r\n pickle.dump(y_test, file_pi)\r\nwith open('/content/%s_LabelTrain' %modelName, 'wb') as file_pi:\r\n pickle.dump(y_train, file_pi)\r\nwith open('/content/%s_ConfusionMatrix' %modelName, 'wb') as file_pi:\r\n pickle.dump(confusion_matrix(y_test.argmax(axis=-1), model.predict(X_test).argmax(axis=-1)), file_pi)\r\nwith open('/content/%s_ClassificationReport' %modelName, 'wb') as file_pi:\r\n pickle.dump(classification_report(y_test.argmax(axis=-1), model.predict(X_test).argmax(axis=-1), target_names=ClassNames), file_pi)",
"dict_keys(['loss', 'categorical_accuracy', 'accuracy', 'val_loss', 'val_categorical_accuracy', 'val_accuracy'])\n"
],
[
"",
"_____no_output_____"
]
],
[
[
"Code to download the files as zip folders\r\n(to load the models and datasets for prediction/evaluation use pickle.load)\r\n",
"_____no_output_____"
]
],
[
[
"\r\n# !zip -r '/Model1.zip' 'Model1Folder'\r\nfiles.download('/Model1.zip')",
"_____no_output_____"
],
[
"!zip -r '/Model2.zip' 'Model2Folder'\r\nfiles.download('/Model2.zip')",
" adding: Model2Folder/ (stored 0%)\n adding: Model2Folder/Model2_Labeltest (deflated 88%)\n adding: Model2Folder/Model2.h5 (deflated 45%)\n adding: Model2Folder/Model2_ConfusionMatrix (deflated 50%)\n adding: Model2Folder/Model2_train (deflated 100%)\n adding: Model2Folder/Model2_test (deflated 100%)\n adding: Model2Folder/Model2_LabelTrain (deflated 94%)\n adding: Model2Folder/Model2_ClassificationReport (deflated 64%)\n adding: Model2Folder/Model2.png (deflated 14%)\n adding: Model2Folder/Model2 (deflated 79%)\n"
],
[
"!zip -r '/Model3.zip' 'Model3Folder'\r\nfiles.download('/Model3.zip')",
" adding: Model3Folder/ (stored 0%)\n adding: Model3Folder/Model3.png (deflated 12%)\n adding: Model3Folder/Model3_LabelTrain (deflated 94%)\n adding: Model3Folder/Model3 (deflated 78%)\n adding: Model3Folder/Model3_ConfusionMatrix (deflated 50%)\n adding: Model3Folder/Model3.h5 (deflated 37%)\n adding: Model3Folder/Model3_Labeltest (deflated 88%)\n adding: Model3Folder/Model3_test (deflated 99%)\n adding: Model3Folder/Model3_train (deflated 99%)\n adding: Model3Folder/Model3_ClassificationReport (deflated 63%)\n"
],
[
"!zip -r '/Model4.zip' 'Model4Folder'\r\nfiles.download('/Model4.zip')",
" adding: Model4Folder/ (stored 0%)\n adding: Model4Folder/Model4_train (deflated 99%)\n adding: Model4Folder/Model4_test (deflated 99%)\n adding: Model4Folder/Model4_Labeltest (deflated 88%)\n adding: Model4Folder/Model4 (deflated 82%)\n adding: Model4Folder/Model4_ConfusionMatrix (deflated 50%)\n adding: Model4Folder/Model4_LabelTrain (deflated 94%)\n adding: Model4Folder/Model4.png (deflated 12%)\n adding: Model4Folder/Model4_ClassificationReport (deflated 63%)\n adding: Model4Folder/Model4.h5 (deflated 45%)\n"
],
[
"!zip -r '/Model5.zip' 'Model5Folder'\r\nfiles.download('/Model5.zip')",
" adding: Model5Folder/ (stored 0%)\n adding: Model5Folder/Model5_ClassificationReport (deflated 64%)\n adding: Model5Folder/Model5_test (deflated 99%)\n adding: Model5Folder/Model5_LabelTrain (deflated 94%)\n adding: Model5Folder/Model5_train (deflated 99%)\n adding: Model5Folder/Model5 (deflated 79%)\n adding: Model5Folder/Model5.png (deflated 13%)\n adding: Model5Folder/Model5_Labeltest (deflated 88%)\n adding: Model5Folder/Model5_ConfusionMatrix (deflated 50%)\n adding: Model5Folder/Model5.h5 (deflated 35%)\n"
],
[
"",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d086fa03485caf3dfbd1b5617a98034fbf20138a | 12,111 | ipynb | Jupyter Notebook | python/11_Time_handling.ipynb | AaltoScienceIT/python-r-data-analysis-course | bdf7e275dab591ae3c763157d5adf7f7be4456a5 | [
"CC0-1.0",
"CC-BY-4.0"
] | 8 | 2018-04-09T12:56:57.000Z | 2020-05-19T09:18:49.000Z | python/11_Time_handling.ipynb | AaltoScienceIT/python-r-data-analysis-course | bdf7e275dab591ae3c763157d5adf7f7be4456a5 | [
"CC0-1.0",
"CC-BY-4.0"
] | null | null | null | python/11_Time_handling.ipynb | AaltoScienceIT/python-r-data-analysis-course | bdf7e275dab591ae3c763157d5adf7f7be4456a5 | [
"CC0-1.0",
"CC-BY-4.0"
] | 10 | 2018-04-10T14:16:23.000Z | 2019-11-01T15:40:09.000Z | 22.469388 | 414 | 0.535794 | [
[
[
"# Time handling\n\nLast year in this course, people asked: \"how do you handle times?\" That's a good question...",
"_____no_output_____"
],
[
"## Exercise\n\nWhat is the ambiguity in these cases?\n\n1. Meet me for lunch at 12:00\n2. The meeting is at 14:00\n3. How many hours are between 01:00 and 06:00 (in the morning)\n4. When does the new year start?\n\nLocal times are a *political* construction and subject to change. They differ depending on where you are. Human times are messy. If you try to do things with human times, you can expect to be sad.\n\nBut still, *actual* time advances at the same rate all over the world (excluding relativity). There *is* a way to do this.",
"_____no_output_____"
],
[
"## What are timezones?\n\nA timezone specifies a certain *local time* at a certain location on earth.\n\nIf you specify a timestamp such as 14:00 on 1 October 2019, it is **naive** if it does not include a timezone. Dependon on where you are standing, you can experience this timestamp at different times.\n\nIf it include a timezone, it is **aware**. An aware timestamp exactly specifies a certain time across the whole world (but depending on where you are standing, your localtime may be different).\n\n**UTC** (coordinated universal time) is a certain timezone - the basis of all other timezones.\n\nUnix computers have a designated **localtime** timezone, which is used by default to display things. This is in the `TZ` environment variable.\n\nThe **tz database** (or zoneinfo) is a open source, comprehensive, updated catalog of all timezones across the whole planet since 1970. It contains things like `EET`, `EEST`, but also geographic locations like `Europe/Helsinki` because the abbreviations can change. [Wikipedia](https://en.wikipedia.org/wiki/Tz_database) and [list of all zones](https://en.wikipedia.org/wiki/List_of_tz_database_time_zones).",
"_____no_output_____"
],
[
"## unixtime\n\nUnixtime is zero at 00:00 on 1 January 1970, and increases at a rate of one per second. This definition defines a single unique time everywhere in the world. You can find unixtime with the `date +%s` command:",
"_____no_output_____"
]
],
[
[
"!date +%s",
"1570084871\n"
]
],
[
[
"You can convert from unixtime to real (local) time using the date command again",
"_____no_output_____"
]
],
[
[
"!date -d @1234567890",
"Sat Feb 14 01:31:30 EET 2009\n"
]
],
[
[
"There are functions which take (unixtime + timezone) and produce the timestamp (year, month, day, hour, minute, second). And vice versa.\n\n",
"_____no_output_____"
],
[
"Unix time has two main benefits:\n* Un-ambiguous: defines a single time\n* You can do math on the times and compute differences, add time, etc, and it just works.",
"_____no_output_____"
],
[
"## Recommendations\n\nWhen you have times, always store them in unixtime in numerical format.\n\nWhen you need a human time (e.g. \"what hour was this time\"), you use a function to compute that property *in a given timezone*.\n\nIf you store the other time components, for example hour and minute, this is just for convenience and you should *not* assume that you can go back to the unixtime to do math.\n\n[Richard's python time reference](http://rkd.zgib.net/wiki/DebianNotes/PythonTime) is the only comprehensive cataloging of Python that he knows of.",
"_____no_output_____"
],
[
"## Exercises\nTo do these, you have to search for the functions yourself.",
"_____no_output_____"
],
[
"### 1. Convert this unixtime to localtime in Helsinki",
"_____no_output_____"
]
],
[
[
"ts = 1570078806",
"_____no_output_____"
]
],
[
[
"### 2. Convert the same time to UTC ",
"_____no_output_____"
],
[
"### Convert that unixtime to a pandas `Timestamp`\nYou'll need to search the docs some...",
"_____no_output_____"
],
[
"## Localization and conversion\n\nIf you are given a time like \"14:00 1 October 2019\", and you want to convert it to a different timezone, can you? No, because there is no timezone already. You have to **localize** it by applying a timezone, then you can convert.",
"_____no_output_____"
]
],
[
[
"import pytz\ntz = pytz.timezone(\"Asia/Tokyo\")\ntz",
"_____no_output_____"
],
[
"# Make a timestamp from a real time. We dont' know when this is...\nimport pandas as pd\nimport datetime\ndt = pd.Timestamp(datetime.datetime(2019, 10, 1, 14, 0))\ndt",
"_____no_output_____"
],
[
"dt.timestamp()",
"_____no_output_____"
],
[
"# Localize it - interpert it as a certain timezone\nlocalized = dt.tz_localize(tz)\nlocalized",
"_____no_output_____"
],
[
"dt.timestamp()",
"_____no_output_____"
],
[
"converted = localized.tz_convert(pytz.timezone('Europe/Helsinki'))\nconverted",
"_____no_output_____"
]
],
[
[
"And we notice it does the conversion... if we don't localize first, then this doesn't work.",
"_____no_output_____"
],
[
"## Exercises",
"_____no_output_____"
],
[
"### 1. Convert this timestamp to a pandas timestamp in Europe/Helsinki and Asia/Tokyo",
"_____no_output_____"
]
],
[
[
"ts = 1570078806",
"_____no_output_____"
]
],
[
[
"### Print the day of the year and hour of this unixtime",
"_____no_output_____"
],
[
"## From the command line",
"_____no_output_____"
]
],
[
[
"!date",
"Thu Oct 3 09:41:14 EEST 2019\n"
],
[
"!date -d \"15:00\"",
"Thu Oct 3 15:00:00 EEST 2019\n"
],
[
"!date -d \"15:00 2019-10-31\"",
"Thu Oct 31 15:00:00 EET 2019\n"
],
[
"!date -d \"15:00 2019-10-31\" +%s",
"1572526800\n"
],
[
"!date -d @1572526800",
"Thu Oct 31 15:00:00 EET 2019\n"
],
[
"!TZ=America/New_York date -d @1572526800",
"Thu Oct 31 09:00:00 EDT 2019\n"
],
[
"!date -d '2019-10-01 14:00 CEST'",
"Tue Oct 1 15:00:00 EEST 2019\n"
]
],
[
[
"## See also\n\n* Julian day - days since 1 January year 4713BCE, or Gregorian ordinal - days since 1 january year 1. Useful if you need to do date, instead of time, arithmetic.\n* [Richard's python-time reference](http://rkd.zgib.net/wiki/DebianNotes/PythonTime)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
d087045c93bda306b6185cb6d7edc2c2652e209a | 139,860 | ipynb | Jupyter Notebook | jupyter_notebooks/machine_learning/ebook_mastering_ml_in_6_steps/Chapter_4_Code/Code/Hyper_Parameter_Tuning.ipynb | manual123/Nacho-Jupyter-Notebooks | e75523434b1a90313a6b44e32b056f63de8a7135 | [
"MIT"
] | 2 | 2021-02-13T05:52:05.000Z | 2022-02-08T09:52:35.000Z | machine_learning/ebook_mastering_ml_in_6_steps/Chapter_4_Code/Code/Hyper_Parameter_Tuning.ipynb | manual123/Nacho-Jupyter-Notebooks | e75523434b1a90313a6b44e32b056f63de8a7135 | [
"MIT"
] | null | null | null | machine_learning/ebook_mastering_ml_in_6_steps/Chapter_4_Code/Code/Hyper_Parameter_Tuning.ipynb | manual123/Nacho-Jupyter-Notebooks | e75523434b1a90313a6b44e32b056f63de8a7135 | [
"MIT"
] | null | null | null | 400.744986 | 124,302 | 0.907121 | [
[
[
"### Hyper Parameter Tuning\n\nOne of the primary objective and challenge in machine learning process is improving the performance score, based on data patterns and observed evidence. To achieve this objective, almost all machine learning algorithms have specific set of parameters that needs to estimate from dataset which will maximize the performance score. The best way to choose good hyperparameters is through trial and error of all possible combination of parameter values. Scikit-learn provide GridSearch and RandomSearch functions to facilitate automatic and reproducible approach for hyperparameter tuning. ",
"_____no_output_____"
]
],
[
[
"from IPython.display import Image\nImage(filename='../Chapter 4 Figures/Hyper_Parameter_Tuning.png', width=1000)",
"_____no_output_____"
]
],
[
[
"### GridSearch",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\n\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.cross_validation import train_test_split\nfrom sklearn import cross_validation\nfrom sklearn import metrics\n\nfrom matplotlib.colors import ListedColormap\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.grid_search import GridSearchCV\nseed = 2017\n\n# read the data in\ndf = pd.read_csv(\"Data/Diabetes.csv\")\n\nX = df.ix[:,:8].values # independent variables\ny = df['class'].values # dependent variables\n\n#Normalize\nX = StandardScaler().fit_transform(X)\n\n# evaluate the model by splitting into train and test sets\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=seed)\n\nkfold = cross_validation.StratifiedKFold(y=y_train, n_folds=5, random_state=seed)\nnum_trees = 100\n\nclf_rf = RandomForestClassifier(random_state=seed).fit(X_train, y_train)\n\nrf_params = {\n 'n_estimators': [100, 250, 500, 750, 1000],\n 'criterion': ['gini', 'entropy'],\n 'max_features': [None, 'auto', 'sqrt', 'log2'],\n 'max_depth': [1, 3, 5, 7, 9]\n}\n\n# setting verbose = 10 will print the progress for every 10 task completion\ngrid = GridSearchCV(clf_rf, rf_params, scoring='roc_auc', cv=kfold, verbose=10, n_jobs=-1)\ngrid.fit(X_train, y_train)\n\nprint 'Best Parameters: ', grid.best_params_\n\nresults = cross_validation.cross_val_score(grid.best_estimator_, X_train,y_train, cv=kfold)\nprint \"Accuracy - Train CV: \", results.mean()\nprint \"Accuracy - Train : \", metrics.accuracy_score(grid.best_estimator_.predict(X_train), y_train)\nprint \"Accuracy - Test : \", metrics.accuracy_score(grid.best_estimator_.predict(X_test), y_test)",
"C:\\Users\\Manoh\\Anaconda2\\lib\\site-packages\\sklearn\\cross_validation.py:44: DeprecationWarning: This module was deprecated in version 0.18 in favor of the model_selection module into which all the refactored classes and functions are moved. Also note that the interface of the new CV iterators are different from that of this module. This module will be removed in 0.20.\n \"This module will be removed in 0.20.\", DeprecationWarning)\nC:\\Users\\Manoh\\Anaconda2\\lib\\site-packages\\sklearn\\grid_search.py:43: DeprecationWarning: This module was deprecated in version 0.18 in favor of the model_selection module into which all the refactored classes and functions are moved. This module will be removed in 0.20.\n DeprecationWarning)\n"
]
],
[
[
"### RandomSearch",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import RandomizedSearchCV\nfrom scipy.stats import randint as sp_randint\n\n# specify parameters and distributions to sample from\nparam_dist = {'n_estimators':sp_randint(100,1000),\n 'criterion': ['gini', 'entropy'],\n 'max_features': [None, 'auto', 'sqrt', 'log2'],\n 'max_depth': [None, 1, 3, 5, 7, 9]\n }\n\n# run randomized search\nn_iter_search = 20\nrandom_search = RandomizedSearchCV(clf_rf, param_distributions=param_dist, cv=kfold, \n n_iter=n_iter_search, verbose=10, n_jobs=-1, random_state=seed)\n\nrandom_search.fit(X_train, y_train)\n# report(random_search.cv_results_)\n\nprint 'Best Parameters: ', random_search.best_params_\n\nresults = cross_validation.cross_val_score(random_search.best_estimator_, X_train,y_train, cv=kfold)\nprint \"Accuracy - Train CV: \", results.mean()\nprint \"Accuracy - Train : \", metrics.accuracy_score(random_search.best_estimator_.predict(X_train), y_train)\nprint \"Accuracy - Test : \", metrics.accuracy_score(random_search.best_estimator_.predict(X_test), y_test)",
"Fitting 5 folds for each of 20 candidates, totalling 100 fits\n"
],
[
"from bayes_opt import BayesianOptimization\nfrom sklearn.cross_validation import cross_val_score\n\ndef rfccv(n_estimators, min_samples_split, max_features):\n return cross_val_score(RandomForestClassifier(n_estimators=int(n_estimators),\n min_samples_split=int(min_samples_split),\n max_features=min(max_features, 0.999),\n random_state=2017),\n X_train, y_train, 'f1', cv=kfold).mean()\n\ngp_params = {\"alpha\": 1e5}\n\nrfcBO = BayesianOptimization(rfccv, {'n_estimators': (100, 1000),\n 'min_samples_split': (2, 25),\n 'max_features': (0.1, 0.999)})\n\nrfcBO.maximize(n_iter=10, **gp_params)\n\nprint('RFC: %f' % rfcBO.res['max']['max_val'])",
"\u001b[31mInitialization\u001b[0m\n\u001b[94m-------------------------------------------------------------------------------------\u001b[0m\n Step | Time | Value | max_features | min_samples_split | n_estimators | \n 1 | 00m13s | \u001b[35m 0.59033\u001b[0m | \u001b[32m 0.1628\u001b[0m | \u001b[32m 2.7911\u001b[0m | \u001b[32m 891.4580\u001b[0m | \n 2 | 00m08s | 0.57056 | 0.1725 | 4.1269 | 543.9055 | \n 3 | 00m04s | \u001b[35m 0.61064\u001b[0m | \u001b[32m 0.7927\u001b[0m | \u001b[32m 21.6962\u001b[0m | \u001b[32m 275.7203\u001b[0m | \n 4 | 00m06s | 0.58312 | 0.2228 | 6.4325 | 437.6023 | \n 5 | 00m03s | \u001b[35m 0.61265\u001b[0m | \u001b[32m 0.3626\u001b[0m | \u001b[32m 12.2393\u001b[0m | \u001b[32m 236.6017\u001b[0m | \n\u001b[31mBayesian Optimization\u001b[0m\n\u001b[94m-------------------------------------------------------------------------------------\u001b[0m\n Step | Time | Value | max_features | min_samples_split | n_estimators | \n 6 | 00m17s | \u001b[35m 0.61354\u001b[0m | \u001b[32m 0.6776\u001b[0m | \u001b[32m 22.9885\u001b[0m | \u001b[32m 999.9903\u001b[0m | \n 7 | 00m01s | 0.60445 | 0.3997 | 22.0831 | 100.0026 | \n 8 | 00m16s | \u001b[35m 0.61529\u001b[0m | \u001b[32m 0.7174\u001b[0m | \u001b[32m 21.0355\u001b[0m | \u001b[32m 999.9898\u001b[0m | \n 9 | 00m01s | \u001b[35m 0.61976\u001b[0m | \u001b[32m 0.4951\u001b[0m | \u001b[32m 2.7633\u001b[0m | \u001b[32m 100.0283\u001b[0m | \n 10 | 00m17s | \u001b[35m 0.62833\u001b[0m | \u001b[32m 0.5922\u001b[0m | \u001b[32m 2.0234\u001b[0m | \u001b[32m 999.9699\u001b[0m | \n 11 | 00m02s | 0.61220 | 0.9008 | 24.4009 | 100.0341 | \n 12 | 00m17s | 0.60972 | 0.8109 | 5.1949 | 999.9955 | \n 13 | 00m01s | 0.60395 | 0.2883 | 2.0518 | 100.0341 | \n 14 | 00m16s | 0.61529 | 0.6443 | 24.7840 | 999.9869 | \n 15 | 00m01s | 0.59926 | 0.3312 | 19.7489 | 100.0013 | \nRFC: 0.628329\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d0872b6c39543fe5e7aea2fff5645e8fb78338d8 | 5,797 | ipynb | Jupyter Notebook | 01-tensorflow-in-practice/03-nlp-tensorflow/13-conv-sarcasm-detection.ipynb | pedro-abundio-wang/tensorflow-specialization | 46ef3d342957d673143a2a17d0faf67a0c49fdb5 | [
"Apache-2.0"
] | null | null | null | 01-tensorflow-in-practice/03-nlp-tensorflow/13-conv-sarcasm-detection.ipynb | pedro-abundio-wang/tensorflow-specialization | 46ef3d342957d673143a2a17d0faf67a0c49fdb5 | [
"Apache-2.0"
] | null | null | null | 01-tensorflow-in-practice/03-nlp-tensorflow/13-conv-sarcasm-detection.ipynb | pedro-abundio-wang/tensorflow-specialization | 46ef3d342957d673143a2a17d0faf67a0c49fdb5 | [
"Apache-2.0"
] | null | null | null | 27.604762 | 92 | 0.576678 | [
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
],
[
"!wget --no-check-certificate \\\n https://storage.googleapis.com/laurencemoroney-blog.appspot.com/sarcasm.json \\\n -O /tmp/sarcasm.json",
"_____no_output_____"
],
[
"import numpy as np\n\nimport json\nimport tensorflow as tf\n\nfrom tensorflow.keras.preprocessing.text import Tokenizer\nfrom tensorflow.keras.preprocessing.sequence import pad_sequences",
"_____no_output_____"
],
[
"vocab_size = 1000\nembedding_dim = 16\nmax_length = 120\ntrunc_type='post'\npadding_type='post'\noov_tok = \"<OOV>\"\ntraining_size = 20000",
"_____no_output_____"
],
[
"with open(\"/tmp/sarcasm.json\", 'r') as f:\n datastore = json.load(f)\n\n\nsentences = []\nlabels = []\nurls = []\nfor item in datastore:\n sentences.append(item['headline'])\n labels.append(item['is_sarcastic'])\n\ntraining_sentences = sentences[0:training_size]\ntesting_sentences = sentences[training_size:]\ntraining_labels = labels[0:training_size]\ntesting_labels = labels[training_size:]",
"_____no_output_____"
],
[
"tokenizer = Tokenizer(num_words=vocab_size, oov_token=oov_tok)\ntokenizer.fit_on_texts(training_sentences)\n\nword_index = tokenizer.word_index\n\ntraining_sequences = tokenizer.texts_to_sequences(training_sentences)\ntraining_padded = pad_sequences(training_sequences, maxlen=max_length, \n padding=padding_type, truncating=trunc_type)\n\ntesting_sequences = tokenizer.texts_to_sequences(testing_sentences)\ntesting_padded = pad_sequences(testing_sequences, maxlen=max_length, \n padding=padding_type, truncating=trunc_type)",
"_____no_output_____"
],
[
"training_padded = np.array(training_padded)\ntraining_labels = np.array(training_labels)\ntesting_padded = np.array(testing_padded)\ntesting_labels = np.array(testing_labels)",
"_____no_output_____"
],
[
"model = tf.keras.Sequential([\n tf.keras.layers.Embedding(vocab_size, embedding_dim, input_length=max_length),\n tf.keras.layers.Conv1D(128, 5, activation='relu'),\n tf.keras.layers.GlobalMaxPooling1D(),\n tf.keras.layers.Dense(24, activation='relu'),\n tf.keras.layers.Dense(1, activation='sigmoid')\n])\nmodel.compile(loss='binary_crossentropy',optimizer='adam',metrics=['accuracy'])\nmodel.summary()",
"_____no_output_____"
],
[
"num_epochs = 10\nhistory = model.fit(training_padded, training_labels, epochs=num_epochs, \n validation_data=(testing_padded, testing_labels), verbose=1)",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n\n\ndef plot_graphs(history, string):\n plt.plot(history.history[string])\n plt.plot(history.history['val_'+string])\n plt.xlabel(\"Epochs\")\n plt.ylabel(string)\n plt.legend([string, 'val_'+string])\n plt.show()\n\nplot_graphs(history, 'accuracy')\nplot_graphs(history, 'loss')",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0873b201fcd1692c5772a067a711d68018e4ef9 | 47,129 | ipynb | Jupyter Notebook | notebooks/1-flickr_api_images_geotag_download.ipynb | isaac-chung/insight | 4459c72ea5f54a5da3709c28b647f5de2a732766 | [
"MIT"
] | 9 | 2020-07-02T02:09:15.000Z | 2021-08-12T02:50:21.000Z | notebooks/1-flickr_api_images_geotag_download.ipynb | isaac-chung/insight | 4459c72ea5f54a5da3709c28b647f5de2a732766 | [
"MIT"
] | 8 | 2020-07-16T11:54:53.000Z | 2020-09-28T14:06:53.000Z | notebooks/1-flickr_api_images_geotag_download.ipynb | isaac-chung/insight | 4459c72ea5f54a5da3709c28b647f5de2a732766 | [
"MIT"
] | 2 | 2020-06-23T17:23:45.000Z | 2020-07-03T01:38:49.000Z | 34.628215 | 215 | 0.477646 | [
[
[
"from time import time\nimport secrets\nimport flickrapi\nimport requests\nimport os\nimport pandas as pd\nimport pickle\nimport logging\n\n\ndef get_photos(image_tag):\n\n # setup dataframe for data\n raw_photos = pd.DataFrame(columns=['latitude', 'longitude','farm','server','id','secret'])\n \n # initialize api\n flickr = flickrapi.FlickrAPI(secrets.api_key, secrets.api_secret, format='parsed-json')\n\n errors = ''\n try:\n # search photos based on settings\n photos = flickr.photos.search(tags=image_tag,\n sort='relevance',\n content_type=1, #photos only\n extras='description,geo,url_c',\n has_geo=1,\n geo_context=2, #outdoors\n per_page=100,\n page=1\n )\n\n # append photo details: description and getags\n raw_photos = raw_photos.append(pd.DataFrame(photos['photos']['photo'])\n [['latitude', 'longitude','farm','server','id','secret']],\n ignore_index=True)\n\n # construct url from pieces\n raw_photos['url'] = 'https://farm'+ raw_photos.farm.astype(str) + '.staticflickr.com/' + raw_photos.server.astype(str) + '/'+ raw_photos.id.astype(str) + '_' + raw_photos.secret.astype(str) + '.jpg'\n \n # need a try/except here for images less than 'per page'\n print('..downloading photos')\n download_images(raw_photos, image_tag)\n \n # save data\n print('..saving metadata')\n with open('data/%s/%s.pkl' %(image_tag, image_tag), 'wb') as f:\n pickle.dump(raw_photos, f)\n f.close()\n \n del raw_photos\n \n except:\n print('Could not get info for: %s. '%image_tag)\n errors = image_tag\n\n return errors\n\n\ndef create_folder(path):\n if not os.path.isdir(path):\n os.makedirs(path)\n\n\ndef download_images(df, keyword):\n path = ''.join(['data/',keyword])\n create_folder(path)\n\n print('...df length: %d' %len(df.index))\n print('...going through each row of dataframe')\n for idx, row in df.iterrows():\n try:\n image_path = ''.join([path,'/',row.id,'.jpg'])\n response = requests.get(row.url)#, stream=True)\n\n with open(image_path, 'wb') as outfile:\n outfile.write(response.content)\n outfile.close()\n \n except:\n print('...Error occured at idx: %d'%idx)\n\n print('...download completed.')",
"_____no_output_____"
],
[
"places = pd.read_csv('IndoorOutdoor_places205.csv', names=['key','label'])",
"_____no_output_____"
],
[
"places.head()",
"_____no_output_____"
],
[
"# retrieve all outdoor scene categories. We clean up the 'key' column, remove duplicates, and re-index the dataframe.\nplaces['key'] = places['key'].str[3:].str.split('/',1,expand=True)\nplaces = places[places.label == 2]\nplaces = places.drop_duplicates(ignore_index=True)\nplaces['key'] = places['key'].str.strip('\\'')\nplaces['key'] = places['key'].replace(to_replace='_',value=' ',regex=True)\nplaces.head(-20)",
"_____no_output_____"
],
[
"places.count() #should have 132",
"_____no_output_____"
],
[
"errors = []\nfor idx, row in places.iterrows():\n\n # change this idx when it crashes. It will give an error for a few indices. It probably means Flickr does not have \n # geotagged images for these keywords. We skip over those. Should have a total of 130 keywords at the end.\n if idx < 0:\n pass\n else:\n start = time()\n error = get_photos(row.key)\n end = time()\n print('%20s in %.2e seconds.' %(row.key, end-start)) # should vary between 3-8 seconds depending on the keyword.\n \n if error != '':\n errors.append(error)",
"..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n abbey in 1.23e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n alley in 1.18e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n amphitheater in 1.27e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n amusement park in 1.28e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n aqueduct in 1.29e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n arch in 1.47e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n apartment building in 1.33e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n badlands in 1.21e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n bamboo forest in 1.22e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n baseball field in 1.23e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...Error occured at idx: 22\n...download completed.\n..saving metadata\n basilica in 2.28e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n bayou in 1.21e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...Error occured at idx: 75\n...download completed.\n..saving metadata\n boardwalk in 1.53e+01 seconds.\n..downloading photos\n...df length: 56\n...going through each row of dataframe\n...download completed.\n..saving metadata\n boat deck in 7.77e+00 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n botanical garden in 1.16e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n bridge in 1.18e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n building facade in 1.32e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n butte in 1.40e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n campsite in 1.74e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n canyon in 1.48e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n castle in 1.64e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n cemetery in 1.52e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n chalet in 1.61e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...Error occured at idx: 13\n...download completed.\n..saving metadata\n coast in 2.25e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n construction site in 1.61e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n corn field in 1.60e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n cottage garden in 1.63e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n courthouse in 1.59e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...Error occured at idx: 55\n...download completed.\n..saving metadata\n courtyard in 1.53e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n creek in 1.62e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n crevasse in 1.57e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n crosswalk in 1.70e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n cathedral in 1.56e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n church in 1.61e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n dam in 1.55e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n dock in 1.47e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n driveway in 1.46e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n desert in 1.46e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n doorway in 1.52e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...Error occured at idx: 43\n...Error occured at idx: 44\n...Error occured at idx: 82\n...download completed.\n..saving metadata\n excavation in 2.66e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n fairway in 1.36e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n fire escape in 1.51e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n fire station in 1.98e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n forest path in 2.50e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n forest road in 1.58e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n formal garden in 1.45e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n fountain in 1.42e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n field in 1.52e+01 seconds.\n..downloading photos\n...df length: 100\n...going through each row of dataframe\n...download completed.\n..saving metadata\n garbage dump in 1.59e+01 seconds.\n..downloading photos\n...df length: 99\n...going through each row of dataframe\n...download completed.\n..saving metadata\n gas station in 1.55e+01 seconds.\n"
],
[
"# we test loading the pickle file.\nkeyword = 'basilica'\nwith open('data/%s/%s.pkl' %(keyword,keyword), 'rb') as f:\n test = pickle.load(f)\n f.close()",
"_____no_output_____"
],
[
"test.head()",
"_____no_output_____"
],
[
"# we test loading the image.\nfrom PIL import Image\n\nimage = Image.open('data/%s/%s.jpg'%(keyword,test.id[0]))\nimage.show()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d087418a36320b75558da7c1e8b3ef0a87e07d0d | 37,799 | ipynb | Jupyter Notebook | examples/network-analysis/resting-state-graph-theory-nodal-dev.ipynb | tsalo/IDConn | b3f5a673067efd8c77d56cd7c0a70693d281b39f | [
"MIT"
] | 3 | 2020-01-17T18:20:17.000Z | 2021-06-24T19:37:49.000Z | examples/network-analysis/resting-state-graph-theory-nodal-dev.ipynb | 62442katieb/idconn-retrieval | 8d4322f9106194a5b80fb19c192d911ff6d222ec | [
"MIT"
] | 6 | 2020-10-01T18:41:04.000Z | 2022-03-01T17:48:05.000Z | notebooks/network-analysis/resting-state-graph-theory-nodal-dev.ipynb | NBCLab/IDConn | 0677e372c02fe35be28f70567e71e040e1d2a023 | [
"MIT"
] | 3 | 2020-10-01T17:56:34.000Z | 2021-06-17T21:16:31.000Z | 44.106184 | 1,965 | 0.488267 | [
[
[
"import numpy as np\nimport pandas as pd\nfrom os import makedirs\nfrom os.path import join, exists\n#from nilearn.input_data import NiftiLabelsMasker\nfrom nilearn.connectome import ConnectivityMeasure\nfrom nilearn.plotting import plot_anat, plot_roi\nimport bct\n#from nipype.interfaces.fsl import InvWarp, ApplyWarp\nimport datetime\n\nsubjects = ['101', '102', '103', '104', '106', '107', '108', '110', '212', '213',\n '214', '215', '216', '217', '218', '219', '320', '321', '322', '323',\n '324', '325', '327', '328', '329', '330', '331', '332', '333', '334',\n '335', '336', '337', '338', '339', '340', '341', '342', '343', '344',\n '345', '346', '347', '348', '349', '350', '451', '452', '453', '455',\n '456', '457', '458', '459', '460', '462', '463', '464', '465', '467',\n '468', '469', '470', '502', '503', '571', '572', '573', '574', '575',\n '577', '578', '579', '580', '581', '582', '584', '585', '586', '587',\n '588', '589', '590', '591', '592', '593', '594', '595', '596', '597',\n '598', '604', '605', '606', '607', '608', '609', '610', '611', '612',\n '613', '614', '615', '616', '617', '618', '619', '620', '621', '622',\n '623', '624', '625', '626', '627', '628', '629', '630', '631', '633',\n '634']\n#subjects = ['101', '102']\n\n\nsink_dir = '/Users/katherine/Dropbox/Projects/physics-retrieval/data/output'\n\nshen = '/home/kbott006/physics-retrieval/shen2015_2mm_268_parcellation.nii.gz'\ncraddock = '/home/kbott006/physics-retrieval/craddock2012_tcorr05_2level_270_2mm.nii.gz'\nmasks = {'shen2015': shen, 'craddock2012': craddock}\n\nsessions = [0,1]\nsesh = ['pre', 'post']\ntasks = ['rest']\n\nkappa_upper = 0.21\nkappa_lower = 0.31\n\nlab_notebook_dir = sink_dir\nindex = pd.MultiIndex.from_product([subjects, sessions], names=['subject', 'session'])\nlab_notebook = pd.DataFrame(index=index, columns=['start', 'end', 'errors'])\n\ncorrelation_measure = ConnectivityMeasure(kind='correlation')\n\n\nindex = pd.MultiIndex.from_product([subjects, sessions, tasks, masks.keys()], names=['subject', 'session', 'task', 'mask'])\ndf = pd.DataFrame(columns=['lEff1', 'clustCoeff1'], index=index, dtype=np.float64)\n\n",
"_____no_output_____"
],
[
"for subject in subjects:\n for session in sessions:\n lab_notebook.at[(subject, session),'start'] = str(datetime.datetime.now())\n for task in tasks:\n for mask in masks.keys():\n try:\n #shen_masker = NiftiLabelsMasker(xfmd_masks['shen2015'], background_label=0, standardize=True, detrend=True,t_r=3.)\n #craddock_masker = NiftiLabelsMasker(xfmd_masks['craddock2012'], background_label=0, standardize=True, detrend=True,t_r=3.)\n\n #confounds = '/home/data/nbc/physics-learning/anxiety-physics/output/{1}/{0}/{0}_confounds.txt'.format(subject, sesh[session])\n #epi_data = join(data_dir, subject, 'session-{0}'.format(session), 'resting-state/resting-state-0/endor1.feat', 'filtered_func_data.nii.gz')\n\n #shen_ts = shen_masker.fit_transform(epi_data, confounds)\n #shen_corrmat = correlation_measure.fit_transform([shen_ts])[0]\n #np.savetxt(join(sink_dir, sesh[session], subject, '{0}-session-{1}-rest_network_corrmat_shen2015.csv'.format(subject, session)), shen_corrmat, delimiter=\",\")\n corrmat = np.genfromtxt(join(sink_dir, '{0}-session-{1}-{2}_network_corrmat_{3}.csv'.format(subject, session, task, mask)), delimiter=\",\")\n print(corrmat.shape)\n #craddock_ts = craddock_masker.fit_transform(epi_data, confounds)\n #craddock_corrmat = correlation_measure.fit_transform([craddock_ts])[0]\n #np.savetxt(join(sink_dir, sesh[session], subject, '{0}-session-{1}-rest_network_corrmat_craddock2012.csv'.format(subject, session)), craddock_corrmat, delimiter=\",\")\n\n ge_s = []\n ge_c = []\n \n md_s = []\n md_c = []\n for p in np.arange(kappa_upper, kappa_lower, 0.02):\n thresh = bct.threshold_proportional(corrmat, p, copy=True)\n\n #network measures of interest here\n #global efficiency\n ge = bct.efficiency_wei(thresh, local=True)\n ge_s.append(ge)\n\n #modularity\n md = bct.clustering_coef_wu(thresh)\n md_s.append(md)\n\n ge_s = np.asarray(ge_s)\n md_s = np.asarray(md_s)\n leff = np.trapz(ge_s, dx=0.01, axis=0)\n print('local efficiency:', leff[0])\n ccoef = np.trapz(md_s, dx=0.01, axis=0)\n for j in np.arange(1, 270):\n df.at[(subject, session, task, mask), 'lEff{0}'.format(j)] = leff[j-1]\n df.at[(subject, session, task, mask), 'clustCoeff{0}'.format(j)] = ccoef[j-1]\n #df.to_csv(join(sink_dir, 'resting-state_graphtheory_shen+craddock.csv'), sep=',')\n lab_notebook.at[(subject, session),'end'] = str(datetime.datetime.now())\n except Exception as e:\n print(e, subject, session)\n lab_notebook.at[(subject,session),'errors'] = [e, str(datetime.datetime.now())]\n df.to_csv(join(sink_dir, 'resting-state_nodal-graphtheory_shen+craddock.csv'), sep=',')\n\ndf.to_csv(join(sink_dir, 'resting-state_nodal-graphtheory_shen+craddock_{0}.csv'.format(str(datetime.datetime.today()))), sep=',')\nlab_notebook.to_csv(join(lab_notebook_dir, 'LOG_resting-state-graph-theory_{0}.csv'.format(str(datetime.datetime.now()))))\n",
"(268, 268)\n"
],
[
"df",
"_____no_output_____"
],
[
"for j in np.arange(1, 269):\n print(ccoef[j-1])",
"0.007454690022501217\n0.007900775309480372\n0.0066918149977028265\n0.0069473206797935234\n0.0061713957389498125\n0.007169088245703788\n0.007405656577953213\n0.007198982729770417\n0.006381464545964363\n0.008434875432099458\n0.007322785219902214\n0.00597890874127783\n0.010217386515798819\n0.01151190372943397\n0.006403568750196864\n0.007708082284526093\n0.010005592207775492\n0.005331474734393296\n0.010359476804879897\n0.007097862545142596\n0.00544149690951881\n0.007480607090201493\n0.009793320202272731\n0.008607460556293738\n0.006706738646748877\n0.007154021121443947\n0.009111038566471434\n0.009318180823282118\n0.007436189354768462\n0.007428551266208059\n0.006844995047909842\n0.00619398333767998\n0.008455564433308433\n0.006803008123760667\n0.0074765325509648355\n0.009170952036467514\n0.007564604806441582\n0.009067524648310916\n0.007418657765489023\n0.009978812081225908\n0.006468073406582229\n0.010578806984904522\n0.009024685493513965\n0.011516528515934982\n0.009027238817043464\n0.009759536357200373\n0.006844007866736957\n0.005627610118184987\n0.007399809178692203\n0.007658773371390281\n0.006978452939680824\n0.007428725801579782\n0.012493287445854201\n0.00987697501581777\n0.005905732421435669\n0.008850098129033285\n0.007479402334059943\n0.006130758105047002\n0.009894063533633876\n0.0072001389000191625\n0.010802777088219584\n0.01036026970350266\n0.014179956743524343\n0.007982369123724593\n0.008632484056717977\n0.008117545489235517\n0.006482268011477272\n0.004995244557554969\n0.008371178590653535\n0.010739041009687401\n0.005340619448368994\n0.007529704921689973\n0.007110184953230469\n0.007753081595121785\n0.0070779451593416245\n0.0063211550657684065\n0.005359841252025639\n0.008515900292386244\n0.007182989791556011\n0.00971495830533502\n0.007323359512446779\n0.009379657400623777\n0.006406921964178703\n0.009783274954527639\n0.006491285739205132\n0.01021215928721404\n0.009131707167423009\n0.009257047923788831\n0.006885754003028502\n0.0061945876267550685\n0.008789931701780225\n0.0059411774718214775\n0.006334106627727209\n0.012664126343104657\n0.006732716291243277\n0.007216176147241363\n0.006412067433756979\n0.00691180112800548\n0.006537676016771487\n0.00699313247634324\n0.005867702527438163\n0.009479938326661998\n0.0070062206489902076\n0.00849042766494522\n0.007171557830421295\n0.006305363326821156\n0.009265943897591784\n0.008096824767548014\n0.005606762272534143\n0.007663845786166117\n0.007939177433871758\n0.008898344658449738\n0.011965967891872296\n0.008100793617987085\n0.0061563720377973245\n0.008302331077502811\n0.008500052793668682\n0.007390520814550478\n0.013334466070761594\n0.0067423312145490945\n0.008721091437969212\n0.007590942511475046\n0.006463186494907702\n0.008662256996360498\n0.013493137896641555\n0.00978846602765069\n0.005982950935885339\n0.008972019833125617\n0.00636486980449611\n0.0062091321295313796\n0.004992972521212461\n0.011919119851944372\n0.006339669831388179\n0.007378581980799708\n0.007332768650855528\n0.007115910829182025\n0.009688199457046638\n0.012358477722873532\n0.006425366286030285\n0.006494408388943105\n0.007655417705639864\n0.010462063372521023\n0.00955062449792673\n0.007050133061045565\n0.009493618348909454\n0.0066654906225125975\n0.007096619064021988\n0.007980831809707538\n0.007510577566175675\n0.008700678973997194\n0.012067994277006007\n0.009092451428942532\n0.006695051941081671\n0.009134755133439237\n0.006849165494179275\n0.009053307565058934\n0.00849102798780018\n0.0051159333452482705\n0.007110192698058497\n0.013447579168319313\n0.011248284249215031\n0.00850281981980662\n0.00836160052287269\n0.007282025419299353\n0.006313300138663247\n0.007144040668116743\n0.011580130927456387\n0.010491748993854607\n0.0070561092164905075\n0.007376248305195933\n0.007157630940914202\n0.008025218889713059\n0.008371539540681405\n0.008631654291197022\n0.006960357504359898\n0.00714277234291987\n0.006694660549041042\n0.007985228276052754\n0.006618412918250177\n0.007714452495884776\n0.010713268308774145\n0.0048873689627006445\n0.006939242795162175\n0.0074449725331089504\n0.008728382783062661\n0.006706727015052753\n0.006779480450696063\n0.006733372738731279\n0.007522493040019421\n0.0076356454135822985\n0.009281674454565519\n0.005822898788868961\n0.006600840231165335\n0.008831199864525538\n0.00593403036607229\n0.006693511149134872\n0.008218555857136541\n0.007073013748701679\n0.008881904981191489\n0.010721599517571354\n0.00813047700327866\n0.006553137262720536\n0.012656347721008952\n0.005630397494446256\n0.008101817779173633\n0.007917126343472806\n0.006373240509452029\n0.006529931517194996\n0.007356465141330117\n0.005936560150373943\n0.008183237929328032\n0.010044610261126765\n0.007369228194804916\n0.005424217598738534\n0.0067440388797445445\n0.0072750488586147835\n0.01033185852839098\n0.00755743787104767\n0.008482836655785152\n0.007531534219560368\n0.007076840360665373\n0.0074430272040579785\n0.007904525469316864\n0.011656621817938018\n0.007239618228365027\n0.010011945800586777\n0.007290695898234303\n0.009073961615673747\n0.00844416082458456\n0.00544639860771959\n0.010870311148675718\n0.007958931210237002\n0.008323400799451063\n0.012375058689491463\n0.013007573590442259\n0.008236057303498068\n0.006492772904954503\n0.005944586019102326\n0.010941650413672416\n0.007177486459489015\n0.0071652291781964545\n0.0077019197816761194\n0.01216516403894956\n0.007174148094543908\n0.009529739723168607\n0.008176634940071537\n0.007140994048343346\n0.004704254326358062\n0.007983416190091299\n0.008002646867894245\n0.007620287879237748\n0.007651042680808209\n0.009859686804454088\n0.008186876866106568\n0.006591683065592398\n0.007869002329506934\n0.012177219372273083\n0.0058558889936394165\n0.007666298161626041\n0.009397298813640764\n0.008276544064133997\n0.006784501120803278\n0.0070460235596249525\n0.007506240601026315\n0.007256635114751549\n0.009159139565998991\n0.006851797932013248\n0.006258589355094694\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code"
]
] |
d08745c39af97f52a62255cf23773f4b6e0359b2 | 873,471 | ipynb | Jupyter Notebook | jupyter_notebooks/notebooks/NB2_CIV-gradient_descent.ipynb | jbRothschild/mlreview_notebooks | 355c6c1380e66fcffab889369f47858df798a271 | [
"MIT"
] | null | null | null | jupyter_notebooks/notebooks/NB2_CIV-gradient_descent.ipynb | jbRothschild/mlreview_notebooks | 355c6c1380e66fcffab889369f47858df798a271 | [
"MIT"
] | null | null | null | jupyter_notebooks/notebooks/NB2_CIV-gradient_descent.ipynb | jbRothschild/mlreview_notebooks | 355c6c1380e66fcffab889369f47858df798a271 | [
"MIT"
] | null | null | null | 1,397.5536 | 301,676 | 0.954866 | [
[
[
"# Notebook 2: Gradient Descent",
"_____no_output_____"
],
[
"## Learning Goal\n\nThe goal of this notebook is to gain intuition for various gradient descent methods by visualizing and applying these methods to some simple two-dimensional surfaces. Methods studied include ordinary gradient descent, gradient descent with momentum, NAG, ADAM, and RMSProp.\n\n\n## Overview\n\nIn this notebook, we will visualize what different gradient descent methods are doing using some simple surfaces. From the onset, we emphasize that doing gradient descent on the surfaces is different from performing gradient descent on a loss function in Machine Learning (ML). The reason is that in ML not only do we want to find good minima, we want to find good minima that generalize well to new data. Despite this crucial difference, we can still build intuition about gradient descent methods by applying them to simple surfaces (see related blog posts [here](http://ruder.io/optimizing-gradient-descent/) and [here](http://tiao.io/notes/visualizing-and-animating-optimization-algorithms-with-matplotlib/)).\n\n## Surfaces\n\nWe will consider three simple surfaces: a quadratic minimum of the form $$z=ax^2+by^2,$$ a saddle-point of the form $$z=ax^2-by^2,$$ and [Beale's Function](https://en.wikipedia.org/wiki/Test_functions_for_optimization), a convex function often used to test optimization problems of the form:\n$$z(x,y) = (1.5-x+xy)^2+(2.25-x+xy^2)^2+(2.625-x+xy^3)^2$$\n\nThese surfaces can be plotted using the cells below. \n",
"_____no_output_____"
]
],
[
[
"#This cell sets up basic plotting functions awe\n#we will use to visualize the gradient descent routines.\n\n#Make plots interactive\n#%matplotlib notebook\n\n#Make plots static\n%matplotlib inline\n\n#Make 3D plots\nfrom mpl_toolkits.mplot3d import Axes3D\nimport matplotlib.pyplot as plt\nfrom matplotlib import cm\n#from matplotlib import animation\nfrom IPython.display import HTML\nfrom matplotlib.colors import LogNorm\n#from itertools import zip_longest\n\n#Import Numpy\nimport numpy as np\n\n#Define function for plotting \n\ndef plot_surface(x, y, z, azim=-60, elev=40, dist=10, cmap=\"RdYlBu_r\"):\n\n fig = plt.figure()\n ax = fig.add_subplot(111, projection='3d')\n plot_args = {'rstride': 1, 'cstride': 1, 'cmap':cmap,\n 'linewidth': 20, 'antialiased': True,\n 'vmin': -2, 'vmax': 2}\n ax.plot_surface(x, y, z, **plot_args)\n ax.view_init(azim=azim, elev=elev)\n ax.dist=dist\n ax.set_xlim(-1, 1)\n ax.set_ylim(-1, 1)\n ax.set_zlim(-2, 2)\n \n plt.xticks([-1, -0.5, 0, 0.5, 1], [\"-1\", \"-1/2\", \"0\", \"1/2\", \"1\"])\n plt.yticks([-1, -0.5, 0, 0.5, 1], [\"-1\", \"-1/2\", \"0\", \"1/2\", \"1\"])\n ax.set_zticks([-2, -1, 0, 1, 2])\n ax.set_zticklabels([\"-2\", \"-1\", \"0\", \"1\", \"2\"])\n \n ax.set_xlabel(\"x\", fontsize=18)\n ax.set_ylabel(\"y\", fontsize=18)\n ax.set_zlabel(\"z\", fontsize=18)\n return fig, ax;\n\ndef overlay_trajectory_quiver(ax,obj_func,trajectory, color='k'):\n xs=trajectory[:,0]\n ys=trajectory[:,1]\n zs=obj_func(xs,ys)\n ax.quiver(xs[:-1], ys[:-1], zs[:-1], xs[1:]-xs[:-1], ys[1:]-ys[:-1],zs[1:]-zs[:-1],color=color,arrow_length_ratio=0.3)\n \n return ax;\n\ndef overlay_trajectory(ax,obj_func,trajectory,label,color='k'):\n xs=trajectory[:,0]\n ys=trajectory[:,1]\n zs=obj_func(xs,ys)\n ax.plot(xs,ys,zs, color, label=label)\n \n return ax;\n\n \ndef overlay_trajectory_contour_M(ax,trajectory, label,color='k',lw=2):\n xs=trajectory[:,0]\n ys=trajectory[:,1]\n ax.plot(xs,ys, color, label=label,lw=lw)\n ax.plot(xs[-1],ys[-1],color+'>', markersize=14)\n return ax;\n\ndef overlay_trajectory_contour(ax,trajectory, label,color='k',lw=2):\n xs=trajectory[:,0]\n ys=trajectory[:,1]\n ax.plot(xs,ys, color, label=label,lw=lw)\n return ax;",
"_____no_output_____"
],
[
"#DEFINE SURFACES WE WILL WORK WITH\n\n#Define monkey saddle and gradient\ndef monkey_saddle(x,y):\n return x**3 - 3*x*y**2\n\ndef grad_monkey_saddle(params):\n x=params[0]\n y=params[1]\n grad_x= 3*x**2-3*y**2\n grad_y= -6*x*y\n return [grad_x,grad_y]\n\n#Define saddle surface\n\ndef saddle_surface(x,y,a=1,b=1):\n return a*x**2-b*y**2\n\ndef grad_saddle_surface(params,a=1,b=1):\n x=params[0]\n y=params[1]\n grad_x= a*x\n grad_y= -1*b*y\n return [grad_x,grad_y]\n\n\n# Define minima_surface\n\ndef minima_surface(x,y,a=1,b=1):\n return a*x**2+b*y**2-1\n\ndef grad_minima_surface(params,a=1,b=1):\n x=params[0]\n y=params[1]\n grad_x= 2*a*x\n grad_y= 2*b*y\n return [grad_x,grad_y]\n\n\ndef beales_function(x,y):\n return np.square(1.5-x+x*y)+np.square(2.25-x+x*y*y)+np.square(2.625-x+x*y**3)\n return f\n\ndef grad_beales_function(params):\n x=params[0]\n y=params[1]\n grad_x=2*(1.5-x+x*y)*(-1+y)+2*(2.25-x+x*y**2)*(-1+y**2)+2*(2.625-x+x*y**3)*(-1+y**3)\n grad_y=2*(1.5-x+x*y)*x+4*(2.25-x+x*y**2)*x*y+6*(2.625-x+x*y**3)*x*y**2\n return [grad_x,grad_y]\n\ndef contour_beales_function():\n #plot beales function\n x, y = np.meshgrid(np.arange(-4.5, 4.5, 0.2), np.arange(-4.5, 4.5, 0.2))\n fig, ax = plt.subplots(figsize=(10, 6))\n z=beales_function(x,y)\n cax = ax.contour(x, y, z, levels=np.logspace(0, 5, 35), norm=LogNorm(), cmap=\"RdYlBu_r\")\n ax.plot(3,0.5, 'r*', markersize=18)\n\n ax.set_xlabel('$x$')\n ax.set_ylabel('$y$')\n\n ax.set_xlim((-4.5, 4.5))\n ax.set_ylim((-4.5, 4.5))\n \n return fig,ax\n \n#Make plots of surfaces\nplt.close() # closes previous plots\nx, y = np.mgrid[-1:1:31j, -1:1:31j]\nfig1,ax1=plot_surface(x,y,monkey_saddle(x,y))\nfig2,ax2=plot_surface(x,y,saddle_surface(x,y))\nfig3,ax3=plot_surface(x,y,minima_surface(x,y,5),0)\n\n#Contour plot of Beale's Function\n\nfig4,ax4 =contour_beales_function()\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"## Gradient descent with and without momentum\n\nIn this notebook, we will visualize various gradient descent algorithms used in machine learning. We will be especially interested in trying to understand how various hyperparameters -- especially the learning rate -- affect our performance. Here, we confine ourselves primarily to looking at the performance in the absence of noise. However, we encourage the reader to experiment with playing with the noise strength below and seeing what differences introducing stochasticity makes. \n\nThroughout, we denote the parameters by $\\theta$ and the energy function we are trying to minimize by $E(\\theta)$.\n\n<b>Gradient Descent</b>\n\nWe start by considering a simple gradient descent method. In this method,\nwe will take steps in the direction of the local gradient. Given some parameters $\\theta$, we adjust the parameters at each iteration so that\n\n$$\\theta_{t+1}= \\theta_t - \\eta_t \\nabla_\\theta E(\\theta),$$\n\nwhere we have introduced the learning rate $\\eta_t$ that controls how large a step we take. In general, the algorithm is extremely sensitive to the choice of $\\eta_t$. If $\\eta_t$ is too large, then one can wildly oscillate around minima and miss important structure at small scales. This problem is amplified if our gradient computations are noisy and inexact (as is often the case in machine learning applications). If $\\eta_t$ is too small, then the learning/minimization procedure becomes extremely slow. This raises the natural question: <i> What sets the natural scale for the learning rate and how can we adaptively choose it?</i> We discuss this extensively in Section IV of the review.\n\n<b>Gradient Descent with Momentum</b>\nOne problem with gradient descent is that it has no memory of where the \"ball rolling down the hill\" comes from. This can be an issue when there are many shallow minima in our landscape. If we make an analogy with a ball rolling down a hill, the lack of memory is equivalent to having no inertia or momentum (i.e. completely overdamped dynamics). Without momentum, the ball has no kinetic energy and cannot climb out of shallow minima. \n\nMomentum becomes especially important when we start thinking about stochastic gradient descent with noisy, stochastic estimates of the gradient. In this case, we should remember where we were coming from and not react drastically to each new update.\n\n\n\nInspired by this, we can add a memory or momentum term to the stochastic gradient descent term above:\n\n$$\nv_{t}=\\gamma v_{t-1}+\\eta_{t}\\nabla_\\theta E(\\theta_t),\\\\\n\\theta_{t+1}= \\theta_t -v_{t},\n$$\n\nwith $0\\le \\gamma < 1$ called the momentum parameter. When $\\gamma=0$, this reduces to ordinary gradient descent, and increasing $\\gamma$ increases the inertial contribution to the gradient. From the equations above, we can see that typical memory lifetimes of the gradient is given by $(1-\\gamma)^{-1}$. For $\\gamma=0$ as in gradient descent, the lifetime is just one step. For $\\gamma=0.9$, we typically remember a gradient for ten steps. We will call this gradient descent with classical momentum or CM for short.\n\nA final widely used variant of gradient descent with momentum is called the Nesterov accelerated gradient (NAG). In NAG, rather than calculating the gradient at the current position, one calculates the gradient at the position momentum will carry us to at time $t+1$, namely, $\\theta_t -\\gamma v_{t-1}$. Thus, the update becomes\n$$\nv_{t}=\\gamma v_{t-1}+\\eta_{t}\\nabla_\\theta E(\\theta_t-\\gamma v_{t-1})\\\\\n\\theta_{t+1}= \\theta_t -v_{t}\n$$",
"_____no_output_____"
]
],
[
[
"#This writes a simple gradient descent, gradient descent+ momentum,\n#nesterov. \n\n#Mean-gradient based methods\ndef gd(grad, init, n_epochs=1000, eta=10**-4, noise_strength=0):\n #This is a simple optimizer\n params=np.array(init)\n param_traj=np.zeros([n_epochs+1,2])\n param_traj[0,]=init\n v=0;\n for j in range(n_epochs):\n noise=noise_strength*np.random.randn(params.size)\n v=eta*(np.array(grad(params))+noise)\n params=params-v\n param_traj[j+1,]=params\n return param_traj\n\n\ndef gd_with_mom(grad, init, n_epochs=5000, eta=10**-4, gamma=0.9,noise_strength=0):\n params=np.array(init)\n param_traj=np.zeros([n_epochs+1,2])\n param_traj[0,]=init\n v=0\n for j in range(n_epochs):\n noise=noise_strength*np.random.randn(params.size)\n v=gamma*v+eta*(np.array(grad(params))+noise)\n params=params-v\n param_traj[j+1,]=params\n return param_traj\n\ndef NAG(grad, init, n_epochs=5000, eta=10**-4, gamma=0.9,noise_strength=0):\n params=np.array(init)\n param_traj=np.zeros([n_epochs+1,2])\n param_traj[0,]=init\n v=0\n for j in range(n_epochs):\n noise=noise_strength*np.random.randn(params.size)\n params_nesterov=params-gamma*v\n v=gamma*v+eta*(np.array(grad(params_nesterov))+noise)\n params=params-v\n param_traj[j+1,]=params\n return param_traj",
"_____no_output_____"
]
],
[
[
"## Experiments with GD, CM, and NAG\n\nBefore introducing more complicated situations, let us experiment with these methods to gain some intuition.\n\nLet us look at the dependence of GD on learning rate in a simple quadratic minima of the form $z=ax^2+by^2-1$. Make plots below for $\\eta=0.1,0.5,1,1.01$ and $a=1$ and $b=1$. (to do this, you would have to add additional arguments to the function `gd` above in order to pass the new values of `a` and `b`; otherwise the default values `a=1` and `b=1` will be used by the gradient)\n<ul>\n<li>\nWhat are the qualitatively different behaviors that arise as $\\eta$ is increased?\n<li> What does this tell us about the importance of choosing learning parameters? How do these change if we change $a$ and $b$ above? In particular how does anisotropy change the learning behavior?\n<li> Make similar plots for CM and NAG? How do the learning rates for these procedures compare with those for GD?\n</ul>\n",
"_____no_output_____"
]
],
[
[
"# Investigate effect of learning rate in GD\nplt.close()\na,b = 1.0,1.0\nx, y = np.meshgrid(np.arange(-4.5, 4.5, 0.2), np.arange(-4.5, 4.5, 0.2))\nfig, ax = plt.subplots(figsize=(10, 6))\nz=np.abs(minima_surface(x,y,a,b))\nax.contour(x, y, z, levels=np.logspace(0.0, 5, 35), norm=LogNorm(), cmap=\"RdYlBu_r\")\nax.plot(0,0, 'r*', markersize=18)\n\n#initial point\ninit1=[-2,4]\ninit2=[-1.7,4]\ninit3=[-1.5,4]\ninit4=[-3,4.5]\neta1=0.1\neta2=0.5\neta3=1\neta4=1.01\ngd_1=gd(grad_minima_surface,init1, n_epochs=100, eta=eta1)\ngd_2=gd(grad_minima_surface,init2, n_epochs=100, eta=eta2)\ngd_3=gd(grad_minima_surface,init3, n_epochs=100, eta=eta3)\ngd_4=gd(grad_minima_surface,init4, n_epochs=10, eta=eta4)\n#print(gd_1)\noverlay_trajectory_contour(ax,gd_1,'$\\eta=$%s'% eta1,'g--*', lw=0.5)\noverlay_trajectory_contour(ax,gd_2,'$\\eta=$%s'% eta2,'b-<', lw=0.5)\noverlay_trajectory_contour(ax,gd_3,'$\\eta=$%s'% eta3,'->', lw=0.5)\noverlay_trajectory_contour(ax,gd_4,'$\\eta=$%s'% eta4,'c-o', lw=0.5)\nplt.legend(loc=2)\nplt.show()\nfig.savefig(\"GD3regimes.pdf\", bbox_inches='tight')\n",
"_____no_output_____"
]
],
[
[
"\n## Gradient Descents that utilize the second moment\n\nIn stochastic gradient descent, with and without momentum, we still have to specify a schedule for tuning the learning rates $\\eta_t$ as a function of time. As discussed in Sec. IV in the context of Newton's method, this presents a number of dilemmas. The learning rate is limited by the steepest direction which can change depending on where in the landscape we are. To circumvent this problem, ideally our algorithm would take large steps in shallow, flat directions and small steps in steep, narrow directions. Second-order methods accomplish this by calculating or approximating the Hessian and normalizing the learning rate by the curvature. However, this is very computationally expensive for extremely large models. Ideally, we would like to be able to adaptively change our step size to match the landscape without paying the steep computational price of calculating or approximating Hessians.\n\nRecently, a number of methods have been introduced that accomplish this by tracking not only the gradient but also the second moment of the gradient. These methods include AdaGrad, AdaDelta, RMS-Prop, and ADAM. Here, we discuss the latter of these two as representatives of this class of algorithms.\n\n\nIn RMS prop (Root-Mean-Square propagation), in addition to keeping a running average of the first moment of the gradient, we also keep track of the second moment through a moving average. The update rule for RMS prop is given by\n$$\n\\mathbf{g}_t = \\nabla_\\theta E(\\boldsymbol{\\theta}) \\\\\n\\mathbf{s}_t =\\beta \\mathbf{s}_{t-1} +(1-\\beta)\\mathbf{g}_t^2 \\nonumber \\\\\n\\boldsymbol{\\theta}_{t+1}=\\boldsymbol{\\theta}_t + \\eta_t { \\mathbf{g}_t \\over \\sqrt{\\mathbf{s}_t +\\epsilon}}, \\nonumber \\\\\n$$\nwhere $\\beta$ controls the averaging time of the second moment and is typically taken to be about $\\beta=0.9$, $\\eta_t$ is a learning rate typically chosen to be $10^{-3}$, and $\\epsilon\\sim 10^{-8}$ is a small regularization constant to prevent divergences. It is clear from this formula that the learning rate is reduced in directions where the norm of the gradient is consistently large. This greatly speeds up the convergence by allowing us to use a larger learning rate for flat directions.\n\nA related algorithm is the ADAM optimizer. In ADAM, we keep a running average of both the first and second moment of the gradient and use this information to adaptively change the learning rate for different parameters. In addition to keeping a running average of the first and second moments of the gradient, ADAM performs an additional bias correction to account for the fact that we are estimating the first two moments of the gradient using a running average (denoted by the hats in the update rule below). The update rule for ADAM is given by (where multiplication and division are understood to be element wise operations)\n$$\n\\mathbf{g}_t = \\nabla_\\theta E(\\boldsymbol{\\theta}) \\\\\n\\mathbf{m}_t = \\beta_1 \\mathbf{m}_{t-1} + (1-\\beta_1) \\mathbf{g}_t \\nonumber \\\\\n\\mathbf{s}_t =\\beta_2 \\mathbf{s}_{t-1} +(1-\\beta_2)\\mathbf{g}_t^2 \\nonumber \\\\\n\\hat{\\mathbf{m}}_t={\\mathbf{m}_t \\over 1-\\beta_1} \\nonumber \\\\\n\\hat{\\mathbf{s}}_t ={\\mathbf{s}_t \\over1-\\beta_2} \\nonumber \\\\\n\\boldsymbol{\\theta}_{t+1}=\\boldsymbol{\\theta}_t + \\eta_t { \\hat{\\mathbf{m}}_t \\over \\sqrt{\\hat{\\mathbf{s}}_t +\\epsilon}}, \\nonumber \n$$\nwhere $\\beta_1$ and $\\beta_2$ set the memory lifetime of the first and second moment and are typically take to be $0.9$ and $0.99$ respectively, and $\\eta$ and $\\epsilon$ are identical\nto RMSprop.\n\n\n",
"_____no_output_____"
]
],
[
[
"################################################################################\n# Methods that exploit first and second moments of gradient: RMS-PROP and ADAMS\n################################################################################\n\ndef rms_prop(grad, init, n_epochs=5000, eta=10**-3, beta=0.9,epsilon=10**-8,noise_strength=0):\n params=np.array(init)\n param_traj=np.zeros([n_epochs+1,2])\n param_traj[0,]=init#Import relevant packages\n grad_sq=0;\n for j in range(n_epochs):\n noise=noise_strength*np.random.randn(params.size)\n g=np.array(grad(params))+noise\n grad_sq=beta*grad_sq+(1-beta)*g*g\n v=eta*np.divide(g,np.sqrt(grad_sq+epsilon))\n params= params-v\n param_traj[j+1,]=params\n return param_traj\n \n \ndef adams(grad, init, n_epochs=5000, eta=10**-4, gamma=0.9, beta=0.99,epsilon=10**-8,noise_strength=0):\n params=np.array(init)\n param_traj=np.zeros([n_epochs+1,2])\n param_traj[0,]=init\n v=0;\n grad_sq=0;\n for j in range(n_epochs):\n noise=noise_strength*np.random.randn(params.size)\n g=np.array(grad(params))+noise\n v=gamma*v+(1-gamma)*g\n grad_sq=beta*grad_sq+(1-beta)*g*g\n v_hat=v/(1-gamma)\n grad_sq_hat=grad_sq/(1-beta)\n params=params-eta*np.divide(v_hat,np.sqrt(grad_sq_hat+epsilon))\n param_traj[j+1,]=params\n return param_traj",
"_____no_output_____"
]
],
[
[
"## Experiments with ADAM and RMSprop\n\nIn this section, we will experiment with ADAM and RMSprop. To do so, we will use a function commonly used in optimization protocols:\n\n$$\nf(x,y)=(1.5-x+xy)^2+(2.25-x+xy^2)^2+(2.625-x+xy^3)^2.\n$$\n\nThis function has a global minimum at $(x,y)=(3,0.5)$. We will use GD, GD with classical momentum, NAG, RMSprop, and ADAM to find minima starting at different initial conditions.\n\nOne of the things you should experiment with is the learning rate and the number of steps, $N_{\\mathrm{steps}}$ we take. Initially, we have set $N_{\\mathrm{steps}}=10^4$ and the learning rate for ADAM/RMSprop to $\\eta=10^{-3}$ and the learning rate for the remaining methods to $10^{-6}$.\n<ul>\n<li> Examine the plot for these default values. What do you see?\n<li> Make a plot when the learning rate of all methods is $\\eta=10^{-6}$? How does your plot change?\n<li> Now set the learning rate for all algorithms to $\\eta=10^{-3}$? What goes wrong? Why?\n</ul>",
"_____no_output_____"
]
],
[
[
"plt.close()\n#Make static plot of the results\nNsteps=10**4\nlr_l=10**-3\nlr_s=10**-6\n\ninit1=np.array([4,3])\nfig1, ax1=contour_beales_function()\n\ngd_trajectory1=gd(grad_beales_function,init1,Nsteps, eta=lr_s, noise_strength=0)\ngdm_trajectory1=gd_with_mom(grad_beales_function,init1,Nsteps,eta=lr_s, gamma=0.9,noise_strength=0)\nNAG_trajectory1=NAG(grad_beales_function,init1,Nsteps,eta=lr_s, gamma=0.9,noise_strength=0)\nrms_prop_trajectory1=rms_prop(grad_beales_function,init1,Nsteps,eta=lr_l, beta=0.9,epsilon=10**-8,noise_strength=0)\nadam_trajectory1=adams(grad_beales_function,init1,Nsteps,eta=lr_l, gamma=0.9, beta=0.99,epsilon=10**-8,noise_strength=0)\n\noverlay_trajectory_contour_M(ax1,gd_trajectory1, 'GD','k')\noverlay_trajectory_contour_M(ax1,gd_trajectory1, 'GDM','m')\noverlay_trajectory_contour_M(ax1,NAG_trajectory1, 'NAG','c--')\noverlay_trajectory_contour_M(ax1,rms_prop_trajectory1,'RMS', 'b-.')\noverlay_trajectory_contour_M(ax1,adam_trajectory1,'ADAMS', 'r')\n\nplt.legend(loc=2)\n\n#init2=np.array([1.5,1.5])\n#gd_trajectory2=gd(grad_beales_function,init2,Nsteps, eta=10**-6, noise_strength=0)\n#gdm_trajectory2=gd_with_mom(grad_beales_function,init2,Nsteps,eta=10**-6, gamma=0.9,noise_strength=0)\n#NAG_trajectory2=NAG(grad_beales_function,init2,Nsteps,eta=10**-6, gamma=0.9,noise_strength=0)\n#rms_prop_trajectory2=rms_prop(grad_beales_function,init2,Nsteps,eta=10**-3, beta=0.9,epsilon=10**-8,noise_strength=0)\n#adam_trajectory2=adams(grad_beales_function,init2,Nsteps,eta=10**-3, gamma=0.9, beta=0.99,epsilon=10**-8,noise_strength=0)\n#overlay_trajectory_contour_M(ax1,gdm_trajectory2, 'GDM','m')\n#overlay_trajectory_contour_M(ax1,NAG_trajectory2, 'NAG','c--')\n#overlay_trajectory_contour_M(ax1,rms_prop_trajectory2,'RMS', 'b-.')\n#overlay_trajectory_contour_M(ax1,adam_trajectory2,'ADAMS', 'r')\n\ninit3=np.array([-1,4])\n\ngd_trajectory3=gd(grad_beales_function,init3,10**5, eta=lr_s, noise_strength=0)\ngdm_trajectory3=gd_with_mom(grad_beales_function,init3,10**5,eta=lr_s, gamma=0.9,noise_strength=0)\nNAG_trajectory3=NAG(grad_beales_function,init3,Nsteps,eta=lr_s, gamma=0.9,noise_strength=0)\nrms_prop_trajectory3=rms_prop(grad_beales_function,init3,Nsteps,eta=lr_l, beta=0.9,epsilon=10**-8,noise_strength=0)\nadam_trajectory3=adams(grad_beales_function,init3,Nsteps,eta=lr_l, gamma=0.9, beta=0.99,epsilon=10**-8,noise_strength=0)\n\noverlay_trajectory_contour_M(ax1,gd_trajectory3, 'GD','k')\noverlay_trajectory_contour_M(ax1,gdm_trajectory3, 'GDM','m')\noverlay_trajectory_contour_M(ax1,NAG_trajectory3, 'NAG','c--')\noverlay_trajectory_contour_M(ax1,rms_prop_trajectory3,'RMS', 'b-.')\noverlay_trajectory_contour_M(ax1,adam_trajectory3,'ADAMS', 'r')\n\ninit4=np.array([-2,-4])\n\ngd_trajectory4=gd(grad_beales_function,init4,Nsteps, eta=lr_s, noise_strength=0)\ngdm_trajectory4=gd_with_mom(grad_beales_function,init4,Nsteps,eta=lr_s, gamma=0.9,noise_strength=0)\nNAG_trajectory4=NAG(grad_beales_function,init4,Nsteps,eta=lr_s, gamma=0.9,noise_strength=0)\nrms_prop_trajectory4=rms_prop(grad_beales_function,init4,Nsteps,eta=lr_l, beta=0.9,epsilon=10**-8,noise_strength=0)\nadam_trajectory4=adams(grad_beales_function,init4,Nsteps,eta=lr_l, gamma=0.9, beta=0.99,epsilon=10**-8,noise_strength=0)\n\noverlay_trajectory_contour_M(ax1,gd_trajectory4, 'GD','k')\noverlay_trajectory_contour_M(ax1,gdm_trajectory4, 'GDM','m')\noverlay_trajectory_contour_M(ax1,NAG_trajectory4, 'NAG','c--')\noverlay_trajectory_contour_M(ax1,rms_prop_trajectory4,'RMS', 'b-.')\noverlay_trajectory_contour_M(ax1,adam_trajectory4,'ADAMS', 'r')\n\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0875842ae11558760c53a77c47df6d0127083db | 570,463 | ipynb | Jupyter Notebook | Data Science Course/1. Programming/3. Python/Module 8 - Linear Regression/Practice Problem/01-Linear Regression Project.ipynb | tensorbored/career-now-program | 3c942fb1c4cd3d7f3bd4a30436b2735577d45dcd | [
"MIT"
] | null | null | null | Data Science Course/1. Programming/3. Python/Module 8 - Linear Regression/Practice Problem/01-Linear Regression Project.ipynb | tensorbored/career-now-program | 3c942fb1c4cd3d7f3bd4a30436b2735577d45dcd | [
"MIT"
] | null | null | null | Data Science Course/1. Programming/3. Python/Module 8 - Linear Regression/Practice Problem/01-Linear Regression Project.ipynb | tensorbored/career-now-program | 3c942fb1c4cd3d7f3bd4a30436b2735577d45dcd | [
"MIT"
] | null | null | null | 567.624876 | 352,496 | 0.944207 | [
[
[
"____\n\n<center> <h1 style=\"background-color:#975be5; color:white\"><br>01-Linear Regression Project<br></h1></center>\n\n____",
"_____no_output_____"
],
[
"<div align=\"right\">\n <b><a href=\"https://keytodatascience.com/\">KeytoDataScience.com </a></b>\n</div>",
"_____no_output_____"
],
[
"Congratulations !!\n\nKeytoDataScience just got some contract work with an Ecommerce company based in New York City that sells clothing online but they also have in-store style and clothing advice sessions. \n\nCustomers come in to the store, have sessions/meetings with a personal stylist, then they can go home and order either on a mobile app or website for the clothes they want.\n\n__The company is trying to decide whether to focus their efforts on their mobile app experience or their website. They've hired you on contract on behalf of KeytoDataScience to help them figure it out!__\n\nLet's get started!",
"_____no_output_____"
],
[
"Just follow the steps below to analyze the customer data (Emails and Addresses in data set are fake).",
"_____no_output_____"
],
[
"## 1 Imports\n\n**Import pandas, numpy, matplotlib, and seaborn. (You'll import sklearn as you need it.)**",
"_____no_output_____"
],
[
"## 2 Get the Data\n\nWe'll work with the Ecommerce Customers csv file from the company. It has Customer info, suchas Email, Address, and their color Avatar. Then it also has numerical value columns:\n\n* Avg. Session Length: Average session of in-store style advice sessions.\n* Time on App: Average time spent on App in minutes\n* Time on Website: Average time spent on Website in minutes\n* Length of Membership: How many years the customer has been a member. \n\n**Read in the Ecommerce Customers csv file as a DataFrame called customers.**",
"_____no_output_____"
],
[
"**Check the head of customers, and check out its info() and describe() methods.**",
"_____no_output_____"
],
[
"## 3 Exploratory Data Analysis\n\n**Let's explore the data!**\n\nFor the rest of the exercise we'll only be using the numerical data of the csv file.\n\n**Use seaborn to create a jointplot to compare the Time on Website and Yearly Amount Spent columns. Does the correlation make sense?**",
"_____no_output_____"
],
[
"**Do the same but with the Time on App column instead.**",
"_____no_output_____"
],
[
"**Use jointplot to create a 2D hex bin plot comparing Time on App and Length of Membership.**",
"_____no_output_____"
],
[
"**Let's explore these types of relationships across the entire data set. Use [pairplot](https://stanford.edu/~mwaskom/software/seaborn/tutorial/axis_grids.html#plotting-pairwise-relationships-with-pairgrid-and-pairplot) to recreate the plot below.(Don't worry about the the colors)**",
"_____no_output_____"
],
[
"**Based off this plot what looks to be the most correlated feature with Yearly Amount Spent?**",
"_____no_output_____"
]
],
[
[
"# Length of Membership ",
"_____no_output_____"
]
],
[
[
"**Create a linear model plot (using seaborn's lmplot) of Yearly Amount Spent vs. Length of Membership.**",
"_____no_output_____"
],
[
"## 4 Training and Testing Data\n\nNow that we've explored the data a bit, let's go ahead and split the data into training and testing sets.\n** Set a variable X equal to the numerical features of the customers and a variable y equal to the \"Yearly Amount Spent\" column. **",
"_____no_output_____"
],
[
"**Use model_selection.train_test_split from sklearn to split the data into training and testing sets. Set test_size=0.3 and random_state=101**",
"_____no_output_____"
],
[
"## 5 Training the Model\n\nNow its time to train our model on our training data!\n\n**Import LinearRegression from sklearn.linear_model**",
"_____no_output_____"
],
[
"**Create an instance of a LinearRegression() model named lm.**",
"_____no_output_____"
],
[
"**Train/fit lm on the training data.**",
"_____no_output_____"
],
[
"**Print out the coefficients of the model**",
"_____no_output_____"
],
[
"## 6 Predicting Test Data\nNow that we have fit our model, let's evaluate its performance by predicting off the test values!\n\n**Use lm.predict() to predict off the X_test set of the data.**",
"_____no_output_____"
],
[
"** Create a scatterplot of the real test values versus the predicted values. **",
"_____no_output_____"
],
[
"## 7 Evaluating the Model\n\n__Let's evaluate our model performance by calculating:__\n- R-squared (R2) or Explained variance score\n- Mean Absolute Error\n- Mean Squared Error\n- Root Mean Squared Error.",
"_____no_output_____"
],
[
"## 8 Residuals\n\nYou should have gotten a very good model with a good fit. Let's quickly explore the residuals to make sure everything was okay with our data. \n\n**Plot a histogram of the residuals and make sure it looks normally distributed. Use either seaborn distplot, or just plt.hist().**",
"_____no_output_____"
],
[
"## 9 Conclusion\nWe still want to figure out the answer to the original question, do we focus our efforst on mobile app or website development? Or maybe that doesn't even really matter, and Membership Time is what is really important. Let's see if we can interpret the coefficients at all to get an idea.\n\n**Recreate the dataframe below.**",
"_____no_output_____"
],
[
"**How can you interpret these coefficients?**",
"_____no_output_____"
],
[
"Interpreting the coefficients:\n\n- Holding all other features fixed, a 1 unit increase in **Avg. Session Length** is associated with an **increase of 25.98 total dollars spent**.\n- Holding all other features fixed, a 1 unit increase in **Time on App** is associated with an **increase of 38.59 total dollars spent**.\n- Holding all other features fixed, a 1 unit increase in **Time on Website** is associated with an **increase of 0.19 total dollars spent**.\n- Holding all other features fixed, a 1 unit increase in **Length of Membership** is associated with an **increase of 61.27 total dollars spent**.",
"_____no_output_____"
],
[
"**Do you think the company should focus more on their mobile app or on their website?**",
"_____no_output_____"
],
[
"\nThis is tricky, there are two ways to think about this: Develop the Website to catch up to the performance of the mobile app, or develop the app more since that is what is working better. This sort of answer really depends on the other factors going on at the company, you would probably want to explore the relationship between Length of Membership and the App or the Website before coming to a conclusion!\n",
"_____no_output_____"
],
[
"____\n\n<center> <h1 style=\"background-color:#975be5; color:white\"><br>Great Job!<br></h1><br></center>\n\n____",
"_____no_output_____"
],
[
"Congrats on your contract work! The company loved the insights!",
"_____no_output_____"
],
[
"<div align=\"right\">\n <b><a href=\"https://keytodatascience.com/\">KeytoDataScience.com </a></b>\n</div>",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d087646bec44e08f35467722db456a242a956d1b | 8,751 | ipynb | Jupyter Notebook | Final Dataset/Untitled.ipynb | sriharivishnudas/student-data-classifier | 12c207f4da5fd58d8ba49977da7f617d48d5c071 | [
"MIT"
] | null | null | null | Final Dataset/Untitled.ipynb | sriharivishnudas/student-data-classifier | 12c207f4da5fd58d8ba49977da7f617d48d5c071 | [
"MIT"
] | 1 | 2019-07-23T06:03:50.000Z | 2019-07-23T06:03:50.000Z | Final Dataset/Untitled.ipynb | SushilEze/Student-Data-Classifier | 546ad2b66149eb50008da3cd10245c9c41eae9a8 | [
"MIT"
] | null | null | null | 104.178571 | 1,421 | 0.697406 | [
[
[
"import pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"X = pd.read_csv('final-text.csv')\nX.head()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
d08767ebc8ca77b3f06d7384bfa9d25b62e49806 | 19,145 | ipynb | Jupyter Notebook | emc_512/lab/Python/03-lab-ex.ipynb | antelk/teaching | 4482456b8f51ff20e9560cbb6409f93c8a6d0d1d | [
"MIT"
] | null | null | null | emc_512/lab/Python/03-lab-ex.ipynb | antelk/teaching | 4482456b8f51ff20e9560cbb6409f93c8a6d0d1d | [
"MIT"
] | null | null | null | emc_512/lab/Python/03-lab-ex.ipynb | antelk/teaching | 4482456b8f51ff20e9560cbb6409f93c8a6d0d1d | [
"MIT"
] | null | null | null | 32.284992 | 478 | 0.516636 | [
[
[
"# 3. laboratorijska vježba",
"_____no_output_____"
]
],
[
[
"# učitavanje potrebnih biblioteka\n\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport scipy.signal as ss",
"_____no_output_____"
],
[
"#@title pomoćna funkcija\n# izvršite ovu ćeliju ali se ne opterećujte detaljima implementacije\n\ndef plot_frequency_response(f, Hm, fc=None, ylim_min=None):\n \"\"\"Grafički prikaz prijenosne funkcije filtra.\n \n Args\n f (numpy.ndarray) : frekvencije\n Hm (numpy.ndarray) : apsolutne vrijednosti prijenosne funkcije\n fc (number) : cutoff frekvencija\n ylim_min (number): minimalna vrijednost na y-osi za dB skalu\n\n Returns\n (matplotlib.figure.Figure, matplotlib.axes._subplots.AxesSubplot)\n \"\"\"\n Hc = 1 / np.sqrt(2)\n if fc is None:\n fc_idx = np.where(np.isclose(Hm, Hc, rtol=1e-03))[0][0]\n fc = f[fc_idx]\n H_db = 20 * np.log10(Hm)\n \n fig, ax = plt.subplots(nrows=2, ncols=2, figsize=(12, 7.5))\n\n ax[0, 0].plot(f, Hm, label='$H(f)$')\n ax[0, 0].plot(fc, Hc, 'o', label='$H(f_c)$')\n ax[0, 0].vlines(fc, Hm.min(), Hc, linestyle='--')\n ax[0, 0].annotate(f'$f_c = {fc:.3f}$ Hz\\n$H(f_c)={Hc:.3f}$', (fc * 1.4, Hc))\n ax[0, 0].set_xscale('log')\n ax[0, 0].set_ylabel('$|V_{out}$ / $V_{in}$|')\n ax[0, 0].set_title('log scale')\n ax[0, 0].legend(loc='lower left')\n ax[0, 0].grid()\n \n ax[0, 1].plot(f, Hm, label='$H(f)$')\n ax[0, 1].plot(fc, Hc, 'o', label='$H(f_c)$')\n ax[0, 1].annotate(f'$f_c = {fc:.3f}$ Hz\\n$H(f_c)={Hc:.3f}$', (fc * 1.4, Hc))\n ax[0, 1].set_title('linear scale')\n ax[0, 1].legend()\n ax[0, 1].grid()\n\n ax[1, 0].plot(f, H_db, label='$H_{dB}(f)$')\n ax[1, 0].plot(fc, H_db.max() - 3, 'o', label='$H_{dB}(f_c)$')\n ax[1, 0].vlines(fc, H_db.min(), H_db.max() - 3, linestyle='--')\n ax[1, 0].annotate(f'$f_c = {fc:.3f}$ Hz\\n$H(f_c)={H_db.max() - 3:.3f} dB$',\n (fc * 1.4, H_db.max() - 3))\n ax[1, 0].set_xscale('log')\n ax[1, 0].set_xlabel('$f$ [Hz]')\n ax[1, 0].set_ylabel('$20 \\\\cdot \\\\log$ |$V_{out}$ / $V_{in}$|')\n if ylim_min:\n ax[1, 0].set_ylim((ylim_min, 10))\n ax[1, 0].legend(loc='lower left')\n ax[1, 0].grid()\n\n ax[1, 1].plot(f, H_db, label='$H_{dB}(f)$')\n ax[1, 1].plot(fc, H_db.max() - 3, 'o', label='$H_{dB}(f_c)$')\n ax[1, 1].annotate(f'$f_c = {fc:.3f}$ Hz\\n$H(f_c)={H_db.max() - 3:.3f} dB$',\n (fc * 1.4, H_db.max() - 3))\n ax[1, 1].set_xlabel('$f$ [Hz]')\n if ylim_min:\n ax[1, 1].set_ylim((ylim_min, 10))\n ax[1, 1].legend()\n ax[1, 1].grid()\n\n fig.tight_layout\n return fig, ax",
"_____no_output_____"
]
],
[
[
"### Pasivni visoko-propusni filtri\n\nRealizacija visoko-propusnog filtra u ovom slučaju se ostvaruje korištenjem otpornika i zavojnice povezanih u seriju, pri čemu se izlaz promatra kao napon na zavojnici, $V_{out}$ Uz pretpostavku da je signal na ulazu, $V_{in}$, sinusoidalni naponski izvor, analizu možemo prebaciti u frekvencijsku domenu koristeći impedancijski model. Na ovaj način izbjegavamo potrebu za korištenjem diferencijalnog računa i čitav proračun se svodi na jednostavni algebarski problem. \n\n<center>\n<img src=\"https://upload.wikimedia.org/wikipedia/commons/thumb/b/bb/Series-RL.svg/768px-Series-RL.svg.png\" alt=\"simple-rl-highpass\" width=\"400\"/>\n</center>\n\nIzraz za funkciju prijenosnog odziva dobijamo kao omjer izlaznog i ulaznog napona. Izlazni napon - napon na zavojnici, $V_{out}$, definiramo kroz podjelu ulaznog napona na sljedeći način:\n\n$$\n\\begin{align}\n V_{out} &= \\frac{Z_l}{Z_l + Z_r} \\cdot V_{in} \\\\\n H(\\omega) = \\frac{V_{out}}{V_{in}} &= \\frac{Z_l}{Z_l + Z_r} = \\frac{j\\omega L}{j\\omega L + R} = \\frac{1}{1+R/(j\\omega L)}\n\\end{align}\n$$\n\nKako je $H$ funkcija frekvencije, imamo dva ruba slučaja:\n* za iznimno niske frekvencije kada je $\\omega \\sim 0$ slijedi da je $H(\\omega) \\rightarrow 0$;\n* za iznimno visoke frekvencije kada $\\omega \\rightarrow \\infty$ slijedi da je $H(\\omega) = 0$.\n\nPotrebno je dodatno definirati već spomenutu *cut-off* frekvenciju, $f_c$, za koju amplituda funkcije frekvencijskog odziva, $H$, pada za $\\sqrt 2$ puta, odnosno za $3$ dB:\n$$\n\\begin{align}\n f_c &= \\frac{R}{2 \\pi L}\n\\end{align}\n$$\n\nLink za interaktivni rad sa pasivnim visoko-propusnim filtrom: http://sim.okawa-denshi.jp/en/LRtool.php\n\n#### Zadatak 1\n\nPrvi zadatak je implementirati funkciju `cutoff_frequency` koja na ulazu prima iznose otpora, `R`, i zavojnice, `L`, a na izlazu daje *cutoff* frekvenciju visoko-propusnog filtra.",
"_____no_output_____"
]
],
[
[
"def cutoff_frequency(R, L):\n \"\"\"Cutoff frekvencija visoko-propusnog RL filtra.\n \n Args:\n R (number) : vrijednost otpora otpornika\n L (number) : induktivitet zavojnice\n \n Returns:\n number\n \"\"\"\n #######################################################\n ## TO-DO: implementiraj proračun cutoff frekvencije ##\n # Nakon toga zakomentiraj sljedeću liniju.\n raise NotImplementedError('Implementiraj proračun cutoff frekvencije.')\n #######################################################\n\n # definiraj cutoff frekvenciju\n fc = ...\n return fc",
"_____no_output_____"
]
],
[
[
"Kolika je *cutoff* frekvencija za otpor od $200 \\Omega$ i induktivitet zavojnice od $100 mH$?",
"_____no_output_____"
]
],
[
[
"R = ... # otpor\nL = ... # induktivitet\n\nfc = cutoff_frequency(...) # cutoff frekvencija\n\nprint(f'R = {R/1000} kΩ')\nprint(f'L = {L*1000} mH')\nprint(f'cutoff frekvencija iznosi {fc:.2f} Hz, '\n 'očekivana vrijednost je 318.31 Hz')",
"_____no_output_____"
]
],
[
[
"#### Zadatak 2\n\nDrugi zadatak je implementirati funkciju `rl_highpass` koja na ulazu prima iznose otpora, `R`, induktiviteta, `L`, i frekvenciju, `f`, a na izlazu daje prijenosni odziv pasivnog visoko-propusnog RL filtra.",
"_____no_output_____"
]
],
[
[
"def rl_highpass(R, L, f):\n \"\"\"Funkcija prijenosnog odziva RL visoko-propusnog filtra.\n \n Args:\n R (number) : vrijednost otpora otpornika\n L (number) : induktivitet\n f (number or numpy.ndarray) : frekvencija/e\n \n Returns:\n float or numpy.ndarray\n \"\"\"\n ######################################################\n ## TO-DO: implementiraj funkciju prijenosnog odziva ##\n # Nakon toga zakomentiraj sljedeću liniju.\n raise NotImplementedError('Implementiraj funckiju prijenosnog odziva.')\n ######################################################\n\n # definiraj funkciju prijenosnog pazeći da `f` može biti ili broj (int,\n # float) ili 1-D niz (`numpy.ndarray`)\n H = ...\n return H",
"_____no_output_____"
]
],
[
[
"Kolika je vrijednost prijenosne funkcije pri *cutoff* frekvencija za otpor od $200 \\Omega$ i induktivitet zavojnice od $100 mH$?",
"_____no_output_____"
]
],
[
[
"R = ... # otpor\nL = ... # induktivitet\n\nHc = rl_highpass(...) # prijenosna funkcija pri cutoff frekvenciji\n\nprint(f'R = {R:.2f} Ω')\nprint(f'C = {L * 1000:.2f} mH')\nprint(f'pojačanje pri cutoff frekvenciji iznosi {abs(Hc):.4f}, '\n 'očekivana vrijednost je 1/√2\\n\\n'\n 'provjerite ispravnost dobivenog rezutltata')",
"_____no_output_____"
],
[
"# ćelija za provjeru rezultata\n\n",
"_____no_output_____"
]
],
[
[
"Pretvorite vrijednost prijenosne funkcije pri *cutoff* frekvenciju u decibele i uvjerite se u tvrdnju da amplituda funkcije frekvencijskog odziva, $H$, pada za $3$ dB pri *cutoff* frekvenciji.",
"_____no_output_____"
]
],
[
[
"Hc_dB = ... # pretvorba prijenosne funkcije pri cutoff frekvenciji u dB skalu\nprint(Hc_dB)",
"_____no_output_____"
]
],
[
[
"Za raspon od $10000$ vrijednosti frekvencija do $10 kHz$ te za otpor od $200 \\Omega$ i induktivitet zavojnice od $100 mH$, izračunajte vrijednosti prijenosne funkcije.",
"_____no_output_____"
]
],
[
[
"f = np.linspace(..., num=10000)\nH = rl_highpass(...) # prijenosna funkcija",
"_____no_output_____"
]
],
[
[
"S obzirom da su vrijednosti prijenosne funkcije kompleksne veličine, razmilite što je potrebno napraviti s njima prije nego ih grafički prikažemo?",
"_____no_output_____"
]
],
[
[
"Hm = ... # konverzija u apsolutne vrijednosti",
"_____no_output_____"
]
],
[
[
"Vizualizirajte ovisnost prijenosne funkcije o frekvenciji koristeći `matplotlib` i funkciju `matplotlib.pyplot.plot`.",
"_____no_output_____"
]
],
[
[
"plt.plot(...)\nplt.xlabel('f [Hz]')\nplt.ylabel('H(f)')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Vizualizirajte sada rezultate koristeći već implementiranu funkciju `plot_frequency_response`.\n\nNapomena: za provjeru načina korištenja prethodne funkcije koristite sljedeću naredbu:\n\n```python\nhelp(plot_frequency_response)\n```\n\nili jednostavno\n\n```python\nplot_frequency_response?\n```",
"_____no_output_____"
]
],
[
[
"# provjerite način korištenja funkcije\n\n",
"_____no_output_____"
],
[
"fig, ax = plot_frequency_response(...) # grafički prikaz dobivenih rezultata",
"_____no_output_____"
]
],
[
[
"### Strujno-naponska karakteristika RL visoko-propusnog filtra",
"_____no_output_____"
]
],
[
[
"def time_constant(L, R):\n \"\"\"Vremenska konstanta RL visoko-propusnog filtra.\n \n Args:\n R (number) : vrijednost otpora otpornika\n L (number) : induktivitet\n \n Returns:\n float or numpy.ndarray\n \"\"\"\n ##################################################################\n ## TO-DO: implementiraj fnkciju koja racuna vremensku konstantu ##\n # Nakon toga zakomentiraj sljedeću liniju.\n raise NotImplementedError('Implementiraj vremensku konstantu.')\n ##################################################################\n \n # definiraj vremensku konstantu\n tau = ...\n return tau",
"_____no_output_____"
],
[
"tau = time_constant(L, R) # vremenska konstanta",
"_____no_output_____"
]
],
[
[
"Koja fizikalna veličina je pridružena vremenskoj konstanti? Objasni.",
"_____no_output_____"
]
],
[
[
"def rl_current(t, t_switch, V, R, L):\n \"\"\"Struja kroz RL visoko-propusni filtar.\n \n Args:\n t (number or numpy.ndarray) : trenutak/ci u kojima računamo\n vrijednost struje\n t_switch (number) : treneutak promjene predznaka struje\n V (number) : napon na ulazu\n R (number) : vrijednost otpora otpornika\n L (number) : induktivitet\n \n Returns:\n float or numpy.ndarray\n \"\"\"\n I0 = V / R\n i = np.where(t < t_switch,\n I0 * (1 - np.exp((-R / L) * t)),\n I0 * np.exp((-R / L) * (t - t_switch)))\n return i",
"_____no_output_____"
],
[
"V = 5 # napon na ulazu\ntau = time_constant(L, R) # vremenska konstanta filtra\nt_switch = tau * 4.4 # vrijeme promjene predznaka struje\nT = 2 * t_switch # period\nt = np.linspace(0, T) # vremenska serija trenutaka u kojima evaluiramo vrijednost struje\ni_rl = rl_current(t, t_switch, V, R, L) # RL struja\ni = V / R * np.sin(2 * np.pi * t / T) # sinusna struja",
"_____no_output_____"
],
[
"# vizualizacija RL struje\n\nplt.figure()\nplt.plot(t, i_rl, label='struja')\nplt.plot(t, i, label='on-off ciklus')\nplt.plot([t.min(), t_switch, t.max()], [0, 0, 0], 'rx')\nplt.hlines(0, t.min(), t.max(), 'k')\nplt.vlines(t_switch, i.min(), i.max(), 'k')\nplt.xlabel('t [s]')\nplt.ylabel('i(t) [A]')\nplt.legend()\nplt.grid()\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Pojasno propusni filtri\nSljedeći kod koristi više različitih tipova pojasno-propusnih filtara (Hamming, Kaiser, Remez) i uspoređuje ih s idealnom prijenosnom funkcijom.",
"_____no_output_____"
]
],
[
[
"def bandpass_firwin(ntaps, lowcut, highcut, fs, window='hamming'):\n taps = ss.firwin(ntaps, [lowcut, highcut], nyq=0.5 * fs, pass_zero=False,\n window=window, scale=False)\n return taps\n\n\ndef bandpass_kaiser(ntaps, lowcut, highcut, fs, width):\n atten = ss.kaiser_atten(ntaps, width / (0.5 * fs))\n beta = ss.kaiser_beta(atten)\n taps = ss.firwin(ntaps, [lowcut, highcut], nyq=0.5 * fs, pass_zero=False,\n window=('kaiser', beta), scale=False)\n return taps\n\n\ndef bandpass_remez(ntaps, lowcut, highcut, fs, width):\n delta = 0.5 * width\n edges = [0,\n lowcut - delta,\n lowcut + delta,\n highcut - delta,\n highcut + delta,\n 0.5 * fs,\n ]\n taps = ss.remez(ntaps, edges, [0, 1, 0], Hz=fs)\n return taps",
"_____no_output_____"
],
[
"fs = 63.0\nlowcut = 0.7\nhighcut = 4.0\nntaps = 128\n\ntaps_hamming = bandpass_firwin(ntaps, lowcut, highcut, fs)\ntaps_kaiser16 = bandpass_kaiser(ntaps, lowcut, highcut, fs, width=1.6)\ntaps_kaiser10 = bandpass_kaiser(ntaps, lowcut, highcut, fs, width=1.0)\ntaps_remez = bandpass_remez(ntaps, lowcut, highcut, fs=fs, width=1.0)",
"_____no_output_____"
],
[
"plt.figure()\nw, h = ss.freqz(taps_hamming, 1, worN=2000)\nplt.plot(fs * 0.5 / np.pi * w, abs(h), label='Hammingov prozor')\nw, h = ss.freqz(taps_kaiser16, 1, worN=2000)\nplt.plot(fs * 0.5 / np.pi * w, abs(h), label='Kaiser, širina = 1.6')\nw, h = ss.freqz(taps_kaiser10, 1, worN=2000)\nplt.plot(fs * 0.5/ np.pi * w, abs(h), label='Kaiser, širina = 1.0')\nw, h = ss.freqz(taps_remez, 1, worN=2000)\nplt.plot(fs * 0.5 / np.pi * w, abs(h), label=f'Remez, širina = 1.0')\nh = np.where((fs * 0.5 / np.pi * w < lowcut) | (fs * 0.5 / np.pi * w > highcut), 0, 1)\nplt.plot(fs * 0.5 / np.pi * w, h, 'k-', label='idealna karakteristika')\nplt.fill_between(fs * 0.5 / np.pi * w, h, color='gray', alpha=0.1)\nplt.xlim(0, 8.0)\nplt.grid()\nplt.legend(loc='upper right')\nplt.xlabel('f (Hz)')\nplt.ylabel('H(f)')\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d0876ccf7c280d5f5ecad402a570e0a9d5b065ae | 27,199 | ipynb | Jupyter Notebook | analysis/.ipynb_checkpoints/milestone2-checkpoint.ipynb | data301-2020-winter1/course-project-solo_311 | 4e7a36ca377a87852e5cba7583d0d203d190d3c2 | [
"MIT"
] | null | null | null | analysis/.ipynb_checkpoints/milestone2-checkpoint.ipynb | data301-2020-winter1/course-project-solo_311 | 4e7a36ca377a87852e5cba7583d0d203d190d3c2 | [
"MIT"
] | null | null | null | analysis/.ipynb_checkpoints/milestone2-checkpoint.ipynb | data301-2020-winter1/course-project-solo_311 | 4e7a36ca377a87852e5cba7583d0d203d190d3c2 | [
"MIT"
] | null | null | null | 33.829602 | 150 | 0.337071 | [
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"df = (\n df.assign(year=pd.to_datetime(df['year'], errors = 'coerce').dt.year)\n .dropna()\n .reset_index()\n .drop(columns=['index'])\n)\ndf['year']=df['year'].astype(int)\n",
"_____no_output_____"
],
[
"def load_and_process(url_or_path_to_csv_file):\n # Method Chain 1 (Load data and deal with missing data)\n\n df1 = (\n pd.read_excel(url_or_path_to_csv_file)\n .dropna()\n .reset_index()\n )\n\n # Method Chain 2 (Create new columns, drop others, and do processing)\n\n df2 = (\n df1\n .assign(year=pd.to_datetime(df['year'], errors = 'coerce').dt.year)\n .dropna()\n .reset_index()\n .drop(columns=['index'])\n )\n df2['year']=df2['year'].astype(int) #For some reason this did not work when it was in the method chain\n\n # Make sure to return the latest dataframe\n\n return df2 ",
"_____no_output_____"
],
[
"df = load_and_process(r\"C:\\Users\\Nolan\\Desktop\\DATA301\\course-project-solo_311\\data\\raw\\Meteorite_Landings.xlsx\")",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"from scripts import project_functions\ndf = project_functions.load_and_process(r\"C:\\Users\\Nolan\\Desktop\\DATA301\\course-project-solo_311\\data\\raw\\Meteorite_Landings.xlsx\")\ndf",
"_____no_output_____"
],
[
"df3 = (\n df.assign(year = df['year'].astype(int))\n .query('year < 2021')\n\n)\n",
"_____no_output_____"
],
[
"df3['year'] = df3['year']<2021",
"_____no_output_____"
],
[
"df3",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0877b90bf97891e283033353e812f062be91ad6 | 2,200 | ipynb | Jupyter Notebook | 19Octubre.ipynb | Erik-Silver/daa_2021_1 | 030421851f3fd436dc5c880703b72987ef0a40dd | [
"MIT"
] | null | null | null | 19Octubre.ipynb | Erik-Silver/daa_2021_1 | 030421851f3fd436dc5c880703b72987ef0a40dd | [
"MIT"
] | null | null | null | 19Octubre.ipynb | Erik-Silver/daa_2021_1 | 030421851f3fd436dc5c880703b72987ef0a40dd | [
"MIT"
] | null | null | null | 24.444444 | 230 | 0.409091 | [
[
[
"<a href=\"https://colab.research.google.com/github/Erik-Silver/daa_2021_1/blob/master/19Octubre.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"a2d = [[3,2,1],[6,4,8],[7,4,2]]\nn=3\nprint(a2d)\n\ntotal = 0 #1\nprint(\"Nivel 1\")\nfor ren in range(n):\n sumaRenglon =0\n print(\"Nivel 2\")\n for col in range(n):\n sumaRenglon += a2d[ren][col]\n total += a2d[ren][col]\n print(\"Nivel 3\")\nprint(total)",
"[[3, 2, 1], [6, 4, 8], [7, 4, 2]]\nNivel 1\nNivel 2\nNivel 3\nNivel 3\nNivel 3\nNivel 2\nNivel 3\nNivel 3\nNivel 3\nNivel 2\nNivel 3\nNivel 3\nNivel 3\n37\n"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
]
] |
d0877ee9eb22b6c136dea76994e9796de81ba483 | 88,680 | ipynb | Jupyter Notebook | Lab Experiments/Experiment-1 060720/ML_Experiment_1_060720.ipynb | rohitsmittal7/J045-ML-Sem-V | 782b18904e7ea7fd83c86a4e98f1872b4e97e596 | [
"Apache-2.0"
] | null | null | null | Lab Experiments/Experiment-1 060720/ML_Experiment_1_060720.ipynb | rohitsmittal7/J045-ML-Sem-V | 782b18904e7ea7fd83c86a4e98f1872b4e97e596 | [
"Apache-2.0"
] | null | null | null | Lab Experiments/Experiment-1 060720/ML_Experiment_1_060720.ipynb | rohitsmittal7/J045-ML-Sem-V | 782b18904e7ea7fd83c86a4e98f1872b4e97e596 | [
"Apache-2.0"
] | null | null | null | 55.080745 | 9,934 | 0.613036 | [
[
[
"# **EXPERIMENT 1**",
"_____no_output_____"
],
[
"Aim: Exploring variable in a dataset\n\nObjectives:\n\nExploring Variables in a Dataset\n\nLearn how to open and examine a dataset.\n\nPractice classifying variables by their type: quantitative or categorical.\n\nLearn how to handle categorical variables whose values are numerically coded.\n\nLink to experiment: https://upscfever.com/upsc-fever/en/data/en-exercises-1.html\n",
"_____no_output_____"
]
],
[
[
"import pandas as pd \nimport numpy as np \nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"depression = pd.read_csv('https://raw.githubusercontent.com/kopalsharma19/J045-ML-Sem-V/master/Lab%20Experiments/Experiment-1%20060720/depression.csv')\nfriends = pd.read_csv('https://raw.githubusercontent.com/kopalsharma19/J045-ML-Sem-V/master/Lab%20Experiments/Experiment-1%20060720/friends.csv')\nactor_age = pd.read_csv('https://raw.githubusercontent.com/kopalsharma19/J045-ML-Sem-V/master/Lab%20Experiments/Experiment-1%20060720/actor_age.csv')\ngrad_data = pd.read_csv('https://raw.githubusercontent.com/kopalsharma19/J045-ML-Sem-V/master/Lab%20Experiments/Experiment-1%20060720/grad_data.csv')\nratings = pd.read_csv('https://raw.githubusercontent.com/kopalsharma19/J045-ML-Sem-V/master/Lab%20Experiments/Experiment-1%20060720/ratings.csv')\n",
"_____no_output_____"
]
],
[
[
"## **Question 1**\nWhat are the categorical variables in depression dataset?\n\n",
"_____no_output_____"
]
],
[
[
"depression.head(10), depression.dtypes",
"_____no_output_____"
]
],
[
[
"The categorical Variables in depression dataset are-\n1. Hospt\n2. Treat\n3. Outcome\n4. Gender",
"_____no_output_____"
],
[
"## **QUESTION 2**\nWhat are the quantitative variables in depression dataset?\n\n",
"_____no_output_____"
]
],
[
[
"depression.head(10), depression.dtypes\n",
"_____no_output_____"
]
],
[
[
"Quantitative variables in depression dataset are-\n1. Time\n2. AcuteT\n3. Age",
"_____no_output_____"
],
[
"# **QUESTION 3**\nDescribe the distribution of the variable \"friends\" in dataset - Survey that asked 1,200 U.S. college students about their body perception\n",
"_____no_output_____"
]
],
[
[
"print(\"Datatype\\n\", friends.dtypes)\nprint(\"\\n\")\nprint(\"Shape of Dataset - \", friends.shape)\n",
"Datatype\n Unnamed: 0 int64\nFriends object\ndtype: object\n\n\nShape of Dataset - (1200, 2)\n"
],
[
"friends.Friends.value_counts()\n",
"_____no_output_____"
],
[
"friends.Friends.value_counts().plot(kind='pie')\n",
"_____no_output_____"
]
],
[
[
"## **QUESTION 4**\nDescribe the distribution of the ages of the Best Actor Oscar winners. Be sure to address shape, center, spread and outliers (Dataset - Best Actor Oscar winners (1970-2013))\n",
"_____no_output_____"
]
],
[
[
"actor_age.describe()\n",
"_____no_output_____"
],
[
"np.median(actor_age['Age'])\n",
"_____no_output_____"
],
[
"actor_age.boxplot(column='Age')\n",
"_____no_output_____"
],
[
"actor_age.shape\n",
"_____no_output_____"
],
[
"actor_age.hist(column='Age')\n",
"_____no_output_____"
]
],
[
[
"Shape: Skewed to the right, 44 rows and 1 column\n\nCenter (Median): 43.5\n\nSpread: The standard deviation is 9.749153\n\nOutlier: 76, there are no lower outliers\n\n",
"_____no_output_____"
],
[
"## **QUESTION 5**\nGetting information from the output: \n\na. How many observations are in this data set? \n\nb. What is the mean age of the actors who won the Oscar? \n\nc. What is the five-number summary of the distribution? (Dataset - Best Actor Oscar winners (1970-2013))\n",
"_____no_output_____"
]
],
[
[
"actor_age.describe()",
"_____no_output_____"
]
],
[
[
"a) No. of Observations (count)- 44\n\nb) Mean age of actors (mean)- 44.977273\n\nc) Five-number summary of distribution is\n\nmin- 29\n\nFirst Quartile (25%)- 38\n\nSecond Quartile (Median) (50%)- 43.5\n\nThird Quartile (75%)- 50\n\nmax- 76\n",
"_____no_output_____"
],
[
"## **QUESTION 6**\nGet information from the five-number summary:\n\na. Half of the actors won the Oscar before what age? \n\nb. What is the range covered by all the actors' ages? \n\nc. What is the range covered by the middle 50% of the ages? (Dataset - Best Actor Oscar winners (1970-2013))\n",
"_____no_output_____"
]
],
[
[
"actor_age.describe()",
"_____no_output_____"
]
],
[
[
"a) Half of the actors won oscar before the age of 43.5\n\nb) Range of age for all actors in 29-76\n\nc) Range covered by middle 50% of the ages- 38-50.25\n",
"_____no_output_____"
],
[
"## **QUESTION 7**\nWhat are the standard deviations of the three rating distributions? \n\nWas your intuition correct? \n\n(Dataset - 27 students in the class were asked to rate the instructor on a number scale of 1 to 9)\n",
"_____no_output_____"
]
],
[
[
"ratings.head(10)",
"_____no_output_____"
],
[
"ratings.describe()",
"_____no_output_____"
]
],
[
[
"Standard Deviation for Class. I- 1.568929\n\nStandard Deviation for Class. II- 4.0\n\nStandard Deviation for Class. III- 2.631174\n\n\nNo my intuition wasn't correct. ",
"_____no_output_____"
],
[
"## **QUESTION 8**\nAssume that the average rating in each of the three classes is 5 (which should be visually reasonably clear from the histograms), and recall the interpretation of the SD as a \"typical\" or \"average\" distance between the data points and their mean. \n\nJudging from the table and the histograms, which class would have the largest standard deviation, and which one would have the smallest standard deviation? Explain your reasoning \n\n(Dataset - 27 students in the class were asked to rate the instructor on a number scale of 1 to 9)",
"_____no_output_____"
]
],
[
[
"ratings.head()",
"_____no_output_____"
],
[
"ratings.describe()",
"_____no_output_____"
],
[
"ratings.hist(column='Class.I')\n",
"_____no_output_____"
],
[
"ratings.hist(column='Class.II')\n",
"_____no_output_____"
],
[
"ratings.hist(column='Class.III')\n",
"_____no_output_____"
]
],
[
[
"Seeing the tables and histograms\n\nClass 1 has the least standard deviation as maximum values lie in the center. \n\nClass 2 has the most standard deviation as maximum values lie at different ends of the histogram and very few in the center. \n\n\n",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d087ae4b6d36b7d83405486b37e53bb230b5f141 | 678,122 | ipynb | Jupyter Notebook | Python/maps/plot_map.ipynb | Duseong/CAM-chem | 4e1ae05dde5730f693abcc106b8c8b966c7ed275 | [
"Apache-2.0"
] | 5 | 2018-04-13T16:49:54.000Z | 2022-02-24T23:22:04.000Z | Python/maps/plot_map.ipynb | Duseong/CAM-chem | 4e1ae05dde5730f693abcc106b8c8b966c7ed275 | [
"Apache-2.0"
] | 2 | 2018-07-10T20:39:13.000Z | 2018-07-10T21:07:16.000Z | Python/maps/plot_map.ipynb | Duseong/CAM-chem | 4e1ae05dde5730f693abcc106b8c8b966c7ed275 | [
"Apache-2.0"
] | 4 | 2020-07-22T21:03:46.000Z | 2021-12-05T09:36:09.000Z | 774.111872 | 323,668 | 0.910897 | [
[
[
"# Example Map Plotting",
"_____no_output_____"
],
[
"### At the start of a Jupyter notebook you need to import all modules that you will use",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport xarray as xr\nimport matplotlib.pyplot as plt\nimport numpy as np\nfrom scipy.interpolate import griddata\nimport cartopy\nimport cartopy.crs as ccrs # For plotting maps\nimport cartopy.feature as cfeature # For plotting maps\nfrom cartopy.util import add_cyclic_point # For plotting maps\nimport datetime",
"_____no_output_____"
]
],
[
[
"### Define the directories and file of interest for your results. This can be shortened to less lines as well.",
"_____no_output_____"
]
],
[
[
"#result_dir = \"/home/buchholz/Documents/code_database/untracked/my-notebook/Janyl_plotting/\"\nresult_dir = \"../../data/\"\nfile = \"CAM_chem_merra2_FCSD_1deg_QFED_monthly_2019.nc\"\n#the netcdf file is now held in an xarray dataset named 'nc' and can be referenced later in the notebook\nnc_load = xr.open_dataset(result_dir+file)\n#to see what the netCDF file contains, just call the variable you read it into\nnc_load",
"_____no_output_____"
]
],
[
[
"### Extract the variable of choice at the time and level of choice",
"_____no_output_____"
]
],
[
[
"#extract grid variables\nlat = nc_load['lat']\nlon = nc_load['lon']\n\n#extract variable\nvar_sel = nc_load['PM25']\nprint(var_sel)\n#print(var_sel[0][0][0][0])\n\n#select the surface level at a specific time and convert to ppbv from vmr\n#var_srf = var_sel.isel(time=0, lev=55)\n#select the surface level for an average over three times and convert to ppbv from vmr\nvar_srf = var_sel.isel(time=[2,3,4], lev=55) # MAM chosen\nvar_srf = var_srf.mean('time')\nvar_srf = var_srf*1e09 # 10-9 to ppb\nprint(var_srf.shape)",
"<xarray.DataArray 'PM25' (time: 6, lev: 56, lat: 192, lon: 288)>\n[18579456 values with dtype=float32]\nCoordinates:\n * lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 87.17 88.12 89.06 90.0\n * lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8\n * lev (lev) float64 1.868 2.353 2.948 3.677 ... 947.5 962.5 977.5 992.5\n * time (time) datetime64[ns] 2019-02-01 2019-03-01 ... 2019-07-01\nAttributes:\n mdims: 1\n units: kg/m3\n long_name: PM2.5 concentration\n cell_methods: time: mean\n(192, 288)\n"
],
[
"# Add cyclic point to avoid white line over Africa\nvar_srf_cyc, lon_cyc = add_cyclic_point(var_srf, coord=lon) ",
"_____no_output_____"
]
],
[
[
"### Plot the value over a specific region",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(20,8))\n\n#Define projection\nax = plt.axes(projection=ccrs.PlateCarree())\n\n#define contour levels\nclev = np.arange(0, 100, 1)\n\n#plot the data\nplt.contourf(lon_cyc,lat,var_srf_cyc,clev,cmap='Spectral_r',extend='both')\n\n# add coastlines\n#ax.coastlines() \nax.add_feature(cfeature.COASTLINE)\n\n#add lat lon grids\nax.gridlines(draw_labels=True, color='grey', alpha=0.5, linestyle='--')\n\n#longitude limits in degrees\nax.set_xlim(20,120)\n#latitude limits in degrees\nax.set_ylim(5,60)\n\n# Title\nplt.title(\"CAM-chem 2019 O$_{3}$\")\n\n#axes\n# y-axis\nax.text(-0.09, 0.55, 'Latitude', va='bottom', ha='center',\n rotation='vertical', rotation_mode='anchor',\n transform=ax.transAxes)\n# x-axis\nax.text(0.5, -0.10, 'Longitude', va='bottom', ha='center',\n rotation='horizontal', rotation_mode='anchor',\n transform=ax.transAxes)\n# legend\nax.text(1.18, 0.5, 'O$_{3}$ (ppb)', va='bottom', ha='center',\n rotation='vertical', rotation_mode='anchor',\n transform=ax.transAxes)\n\nplt.colorbar()\nplt.show() ",
"_____no_output_____"
]
],
[
[
"### Add location markers",
"_____no_output_____"
]
],
[
[
"##Now lets look at the sufrace plot again, but this time add markers for observations at several points.\n#first we need to define our observational data into an array\n#this can also be imported from text files using various routines\n# Kyzylorda, Urzhar, Almaty, Balkhash\nobs_lat = np.array([44.8488,47.0870,43.2220,46.2161])\nobs_lon = np.array([65.4823,81.6315,76.8512,74.3775])\nobs_names = [\"Kyzylorda\", \"Urzhar\", \"Almaty\", \"Balkhash\"]\nnum_obs = obs_lat.shape[0]",
"_____no_output_____"
],
[
"plt.figure(figsize=(20,8))\n\n#Define projection\nax = plt.axes(projection=ccrs.PlateCarree())\n\n#define contour levels\nclev = np.arange(0, 100, 1)\n\n#plot the data\nplt.contourf(lon_cyc,lat,var_srf_cyc,clev,cmap='Spectral_r')\n\n# add coastlines\nax.add_feature(cfeature.COASTLINE)\nax.add_feature(cfeature.BORDERS)\n\n#add lat lon grids\nax.gridlines(draw_labels=True, color='grey', alpha=0.5, linestyle='--')\n\n#longitude limits in degrees\nax.set_xlim(20,120)\n#latitude limits in degrees\nax.set_ylim(5,60)\n\n# Title\nplt.title(\"CAM-chem 2019 O$_{3}$\")\n\n#axes\n# y-axisCOUNTRY\nax.text(-0.09, 0.55, 'Latitude', va='bottom', ha='center',\n rotation='vertical', rotation_mode='anchor',\n transform=ax.transAxes)\n# x-axis\nax.text(0.5, -0.10, 'Longitude', va='bottom', ha='center',\n rotation='horizontal', rotation_mode='anchor',\n transform=ax.transAxes)\n# legend\nax.text(1.18, 0.5, 'O$_{3}$ (ppb)', va='bottom', ha='center',\n rotation='vertical', rotation_mode='anchor',\n transform=ax.transAxes)\n\n#convert your observation lat/lon to Lambert-Conformal grid points\n#xpt,ypt = m(obs_lon,obs_lat)\n\n#to specify the color of each point it is easiest plot individual points in a loop\nfor i in range(num_obs):\n plt.plot(obs_lon[i], obs_lat[i], linestyle='none', marker=\"o\", markersize=8, alpha=0.8, c=\"black\", markeredgecolor=\"black\", markeredgewidth=1, transform=ccrs.PlateCarree())\n plt.text(obs_lon[i] - 0.8, obs_lat[i] - 0.5, obs_names[i], fontsize=20, horizontalalignment='right', transform=ccrs.PlateCarree())\n\n \nplt.colorbar()\nplt.show() ",
"_____no_output_____"
],
[
"cartopy.config['data_dir']",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d087c8b135ae86dfb22adc7b6250b597f8f8df83 | 25,840 | ipynb | Jupyter Notebook | examples/_working_Convert Single Image to Seven Tilechain-MULTI-THREADS.ipynb | netmanchris/pylifxtiles | f9a77fe0beaabff4c792032d7778a8ad2815e2bd | [
"Apache-2.0"
] | 6 | 2020-04-27T00:55:47.000Z | 2020-10-11T19:16:38.000Z | examples/_working_Convert Single Image to Seven Tilechain-MULTI-THREADS.ipynb | netmanchris/pylifxtiles | f9a77fe0beaabff4c792032d7778a8ad2815e2bd | [
"Apache-2.0"
] | null | null | null | examples/_working_Convert Single Image to Seven Tilechain-MULTI-THREADS.ipynb | netmanchris/pylifxtiles | f9a77fe0beaabff4c792032d7778a8ad2815e2bd | [
"Apache-2.0"
] | null | null | null | 66.770026 | 1,355 | 0.671246 | [
[
[
"# Description\n\nThis notebook documents allows the following on a group seven LIFX Tilechain with 5 Tiles\nlaid out horizontaly as following\n\n\nT1 [0] [1] [2] [3] [4]\n\nT2 [0] [1] [2] [3] [4]\n\nT3 [0] [1] [2] [3] [4]\n\nT4 [0] [1] [2] [3] [4]\n\nT5 [0] [1] [2] [3] [4]\n\nT6 [0] [1] [2] [3] [4]\n\nT7 [0] [1] [2] [3] [4]\n\n\nCare should be taken to ensure that the LIFX Tiles are all facing up to ensure that the 0,0 position is in the expected place. \n\nProgram will perform the following\n\n- take a jpg or png located in the same folder as the notebook and create a image to display across all 4 tilechains or 20 tiles. Image will be reduced from original size to a 32x40 matrix so resolution will not be great. You've been warned.\n\n",
"_____no_output_____"
]
],
[
[
"!pip install pylifxtiles\n!pip install thread",
"Requirement already satisfied: pylifxtiles in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (0.1.0)\nRequirement already satisfied: requests in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from pylifxtiles) (2.23.0)\nRequirement already satisfied: lifxlan in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from pylifxtiles) (1.2.5)\nRequirement already satisfied: jupyter in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from pylifxtiles) (1.0.0)\nRequirement already satisfied: nose in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from pylifxtiles) (1.3.7)\nRequirement already satisfied: matplotlib in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from pylifxtiles) (3.2.1)\nRequirement already satisfied: Pillow in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from pylifxtiles) (7.1.1)\nRequirement already satisfied: certifi>=2017.4.17 in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from requests->pylifxtiles) (2020.4.5.1)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from requests->pylifxtiles) (1.25.9)\nRequirement already satisfied: idna<3,>=2.5 in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from requests->pylifxtiles) (2.9)\nRequirement already satisfied: chardet<4,>=3.0.2 in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from requests->pylifxtiles) (3.0.4)\nRequirement already satisfied: bitstring in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from lifxlan->pylifxtiles) (3.1.6)\nRequirement already satisfied: netifaces in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from lifxlan->pylifxtiles) (0.10.9)\nRequirement already satisfied: qtconsole in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from jupyter->pylifxtiles) (4.7.3)\nRequirement already satisfied: ipywidgets in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from jupyter->pylifxtiles) (7.5.1)\nRequirement already satisfied: notebook in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from jupyter->pylifxtiles) (6.0.3)\nRequirement already satisfied: ipykernel in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from jupyter->pylifxtiles) (5.2.1)\nRequirement already satisfied: jupyter-console in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from jupyter->pylifxtiles) (6.1.0)\nRequirement already satisfied: nbconvert in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from jupyter->pylifxtiles) (5.6.1)\nRequirement already satisfied: kiwisolver>=1.0.1 in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from matplotlib->pylifxtiles) (1.2.0)\nRequirement already satisfied: numpy>=1.11 in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from matplotlib->pylifxtiles) (1.18.3)\nRequirement already satisfied: cycler>=0.10 in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from matplotlib->pylifxtiles) (0.10.0)\nRequirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from matplotlib->pylifxtiles) (2.4.7)\nRequirement already satisfied: python-dateutil>=2.1 in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from matplotlib->pylifxtiles) (2.8.1)\nRequirement already satisfied: jupyter-core in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from qtconsole->jupyter->pylifxtiles) (4.6.3)\nRequirement already satisfied: traitlets in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from qtconsole->jupyter->pylifxtiles) (4.3.3)\nRequirement already satisfied: pygments in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from qtconsole->jupyter->pylifxtiles) (2.6.1)\nRequirement already satisfied: jupyter-client>=4.1 in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from qtconsole->jupyter->pylifxtiles) (6.1.3)\nRequirement already satisfied: qtpy in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from qtconsole->jupyter->pylifxtiles) (1.9.0)\nRequirement already satisfied: pyzmq>=17.1 in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from qtconsole->jupyter->pylifxtiles) (19.0.0)\nRequirement already satisfied: ipython-genutils in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from qtconsole->jupyter->pylifxtiles) (0.2.0)\nRequirement already satisfied: widgetsnbextension~=3.5.0 in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from ipywidgets->jupyter->pylifxtiles) (3.5.1)\nRequirement already satisfied: nbformat>=4.2.0 in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from ipywidgets->jupyter->pylifxtiles) (5.0.6)\nRequirement already satisfied: ipython>=4.0.0; python_version >= \"3.3\" in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from ipywidgets->jupyter->pylifxtiles) (7.13.0)\nRequirement already satisfied: prometheus-client in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from notebook->jupyter->pylifxtiles) (0.7.1)\nRequirement already satisfied: tornado>=5.0 in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from notebook->jupyter->pylifxtiles) (6.0.4)\nRequirement already satisfied: Send2Trash in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from notebook->jupyter->pylifxtiles) (1.5.0)\nRequirement already satisfied: jinja2 in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from notebook->jupyter->pylifxtiles) (2.11.2)\nRequirement already satisfied: terminado>=0.8.1 in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from notebook->jupyter->pylifxtiles) (0.8.3)\nRequirement already satisfied: appnope; platform_system == \"Darwin\" in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from ipykernel->jupyter->pylifxtiles) (0.1.0)\nRequirement already satisfied: prompt-toolkit!=3.0.0,!=3.0.1,<3.1.0,>=2.0.0 in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from jupyter-console->jupyter->pylifxtiles) (3.0.5)\nRequirement already satisfied: defusedxml in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from nbconvert->jupyter->pylifxtiles) (0.6.0)\nRequirement already satisfied: pandocfilters>=1.4.1 in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from nbconvert->jupyter->pylifxtiles) (1.4.2)\nRequirement already satisfied: mistune<2,>=0.8.1 in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from nbconvert->jupyter->pylifxtiles) (0.8.4)\nRequirement already satisfied: entrypoints>=0.2.2 in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from nbconvert->jupyter->pylifxtiles) (0.3)\nRequirement already satisfied: bleach in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from nbconvert->jupyter->pylifxtiles) (3.1.4)\nRequirement already satisfied: testpath in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from nbconvert->jupyter->pylifxtiles) (0.4.4)\nRequirement already satisfied: six in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from cycler>=0.10->matplotlib->pylifxtiles) (1.14.0)\nRequirement already satisfied: decorator in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from traitlets->qtconsole->jupyter->pylifxtiles) (4.4.2)\nRequirement already satisfied: jsonschema!=2.5.0,>=2.4 in /Users/christopheryoung/PycharmProjects/netmanchrisLIFXTilesPrivate/venv/lib/python3.7/site-packages (from nbformat>=4.2.0->ipywidgets->jupyter->pylifxtiles) (3.2.0)\n"
],
[
"#Main Program for Convert Single Image to Tiles\n\n# Full running function with all dependencies\n#imports RGB to HSBK conversion function from LIFX LAN library\nimport _thread as thread\nfrom lifxlan import LifxLAN\nfrom lifxlan.utils import RGBtoHSBK\nfrom pylifxtiles import tiles\nfrom pylifxtiles import actions\nfrom matplotlib import image\nfrom PIL import Image\n\n# modify this variable to the name of the specific LIFX Tilechain as shown in the LIFX app\n\nsource_image = './images/meghan.jpg'\n\n\ndef main():\n lan = LifxLAN()\n tilechain_lights = lan.get_tilechain_lights()\n print(len(tilechain_lights))\n if len(tilechain_lights) != 0:\n for tile in tilechain_lights:\n if tile.get_label() == 'T1':\n print(tile.get_label())\n T1 = tile\n if tile.get_label() =='T2':\n print(tile.get_label())\n T2 = tile\n if tile.get_label() == 'T3':\n print(tile.get_label())\n T3 = tile\n if tile.get_label() == 'T4':\n print(tile.get_label())\n T4 = tile\n if tile.get_label() == 'T5':\n print(tile.get_label())\n T5 = tile\n if tile.get_label() == 'T6':\n print(tile.get_label())\n T6 = tile\n if tile.get_label() == 'T7':\n print(tile.get_label())\n T7 = tile\n tc_list = [ T1, T2, T3, T4, T5, T6, T7]\n try:\n thread.start_new_thread(display_image,(source_image,(40,56), tc_list))\n except KeyboardInterrupt:\n print(\"Done.\")\n\n\n\n\n#combined function\n\n# resize image and force a new shape and save to disk\ndef display_image(image_to_display,image_size, tilechain_list):\n # load the image\n my_image = Image.open(image_to_display)\n # report the size of the image\n #print(my_image.size)\n # resize image and ignore original aspect ratio\n img_resized = my_image.resize(image_size)\n #changing the file extension from jpg to png changes output brightness. You might need to play with this. \n img_resized.save('./images/resized_image.jpg')\n data = image.imread('./images/resized_image.jpg')\n target_tcs = []\n for row in data:\n temp_row = []\n for pixel in row:\n temp_row.append(RGBtoHSBK(pixel))\n target_tcs.append(temp_row)\n #print (\"length of target_tcs is \" + str(len(target_tcs)))\n tcsplit = tiles.split_tilechains(target_tcs)\n #print (\"legnth of tcssplit is \" + str(len(tcsplit)))\n #print (\"length tilelist is \" + str(len(tilechain_list)))\n for tile in range(len(tilechain_list)):\n print (tile)\n tilechain_list[tile].set_tilechain_colors(tiles.split_combined_matrix(tcsplit[tile]),rapid=True)\n \n \nif __name__ == \"__main__\":\n main()",
"23\nT4\nT5\nT3\nT6\nT1\nT7\nT2\n0\n1\n2\n3\n4\n5\n6\n"
]
],
[
[
"# test write to three tiles",
"_____no_output_____"
]
],
[
[
"#Main Program for Convert Single Image to Tiles\n\n# Full running function with all dependencies\n#imports RGB to HSBK conversion function from LIFX LAN library\nfrom lifxlan import LifxLAN\nfrom lifxlan.utils import RGBtoHSBK\nfrom pylifxtiles import tiles\nfrom pylifxtiles import actions\nfrom matplotlib import image\nfrom PIL import Image\n\n# modify this variable to the name of the specific LIFX Tilechain as shown in the LIFX app\n\nsource_image = './images/Youtubelogo.jpg'\n\n\ndef main():\n lan = LifxLAN()\n tilechain_lights = lan.get_tilechain_lights()\n print(len(tilechain_lights))\n if len(tilechain_lights) != 0:\n for tile in tilechain_lights:\n if tile.get_label() == 'T1':\n print(tile.get_label())\n T1 = tile\n if tile.get_label() =='T2':\n print(tile.get_label())\n T2 = tile\n if tile.get_label() == 'T3':\n print(tile.get_label())\n T3 = tile\n if tile.get_label() == 'T4':\n print(tile.get_label())\n T4 = tile\n tc_list = [T2, T3, T4]\n try:\n display_image(source_image,(40,24), tc_list)\n except KeyboardInterrupt:\n print(\"Done.\")\n\n\n\n\n#combined function\n\n# resize image and force a new shape and save to disk\ndef display_image(image_to_display,image_size, tilechain_list):\n # load the image\n my_image = Image.open(image_to_display)\n # report the size of the image\n #print(my_image.size)\n # resize image and ignore original aspect ratio\n img_resized = my_image.resize(image_size)\n #changing the file extension from jpg to png changes output brightness. You might need to play with this. \n img_resized.save('./images/resized_image.jpg')\n data = image.imread('./images/resized_image.jpg')\n target_tcs = []\n for row in data:\n temp_row = []\n for pixel in row:\n temp_row.append(RGBtoHSBK(pixel))\n target_tcs.append(temp_row)\n print (\"length of target_tcs is \" + str(len(target_tcs)))\n tcsplit = tiles.split_tilechains(target_tcs)\n print (\"legnth of tcssplit is \" + str(len(tcsplit)))\n print (\"length tilelist is \" + str(len(tilechain_list)))\n for tile in range(len(tilechain_list)):\n print (tile)\n tilechain_list[tile].set_tilechain_colors(tiles.split_combined_matrix(tcsplit[tile]),rapid=True)\n \nif __name__ == \"__main__\":\n main()",
"0\n"
],
[
"import threading",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d087d819d36daef82bcc6e0d8a10b49da3f1ba51 | 98,321 | ipynb | Jupyter Notebook | model_tempetes/notebooks/vulnerability_explo.ipynb | allezalex/batch8_worldbank | 33046c0127f5bdb5df9e670e13e9450585b45d4d | [
"MIT"
] | 7 | 2020-11-11T19:49:27.000Z | 2021-03-26T13:51:20.000Z | model_tempetes/notebooks/vulnerability_explo.ipynb | allezalex/batch8_worldbank | 33046c0127f5bdb5df9e670e13e9450585b45d4d | [
"MIT"
] | 7 | 2020-10-14T22:53:21.000Z | 2020-12-16T11:03:57.000Z | model_tempetes/notebooks/vulnerability_explo.ipynb | allezalex/batch8_worldbank | 33046c0127f5bdb5df9e670e13e9450585b45d4d | [
"MIT"
] | 16 | 2020-09-30T19:28:03.000Z | 2020-12-22T11:43:46.000Z | 61.876023 | 16,340 | 0.664639 | [
[
[
"import pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport statsmodels.api as sm",
"_____no_output_____"
]
],
[
[
"# Import Risk INFORM index",
"_____no_output_____"
]
],
[
[
"path = \"C:\\\\batch8_worldbank\\\\datasets\\\\tempetes\\\\INFORM_Risk_2021.xlsx\"",
"_____no_output_____"
],
[
"xl = pd.ExcelFile(path)\nxl.sheet_names",
"_____no_output_____"
],
[
"inform_df = xl.parse(xl.sheet_names[2])\ninform_df.columns = inform_df.iloc[0]\ninform_df = inform_df[2:]\ninform_df.head()",
"_____no_output_____"
]
],
[
[
"# Import emdat",
"_____no_output_____"
]
],
[
[
"path = \"C:\\\\batch8_worldbank\\\\datasets\\\\tempetes\\\\wb_disasters_bdd.xlsx\"",
"_____no_output_____"
],
[
"disasters_df = pd.read_excel(path)\ndisasters_df.head()",
"_____no_output_____"
],
[
"disasters_df['ISO']",
"_____no_output_____"
],
[
"max(disasters_df['Year'])",
"_____no_output_____"
]
],
[
[
"# Filter on storms",
"_____no_output_____"
]
],
[
[
"storms_df = disasters_df[disasters_df[\"Disaster Type\"]==\"Storm\"]",
"_____no_output_____"
]
],
[
[
"# Number of storms, nb people affected and total damages by country by decade",
"_____no_output_____"
]
],
[
[
"nb_storms_by_year_by_country = storms_df.groupby([\"Start Year\", \"ISO\"]).aggregate({\"Disaster Type\":\"count\", \"No Affected\": \"sum\", \"Total Damages ('000 US$)\":\"sum\"})\nnb_storms_by_year_by_country = nb_storms_by_year_by_country.reset_index()\nnb_storms_by_year_by_country = nb_storms_by_year_by_country.rename(columns={\"Start Year\": \"year\", \"Disaster Type\": \"storms_count\", \"No Affected\": \"total_nb_affected\", \"Total Damages ('000 US$)\": \"total_damages\"})",
"_____no_output_____"
],
[
"nb_storms_by_year_by_country[\"decade\"] = nb_storms_by_year_by_country[\"year\"].apply(lambda row: (row//10)*10)\nnb_storms_by_decade_by_country = nb_storms_by_year_by_country.groupby([\"decade\", \"ISO\"]).aggregate({\"storms_count\":\"sum\", \"total_nb_affected\":\"sum\", \"total_damages\":\"sum\"})\nnb_storms_by_decade_by_country = nb_storms_by_decade_by_country.reset_index()",
"_____no_output_____"
],
[
"nb_storms_by_decade_by_country.head()",
"_____no_output_____"
],
[
"max(nb_storms_by_decade_by_country[\"decade\"])",
"_____no_output_____"
]
],
[
[
"# Keep observations on decades 2000, 2010 and 2020 to increase nb of datapoints",
"_____no_output_____"
]
],
[
[
"nb_storms_by_decade_by_country_2020 = nb_storms_by_decade_by_country[nb_storms_by_decade_by_country[\"decade\"]>=2000]",
"_____no_output_____"
],
[
"nb_storms_by_decade_by_country_2020.head()",
"_____no_output_____"
],
[
"nb_storms_by_decade_by_country_2020.shape",
"_____no_output_____"
],
[
"nb_storms_by_decade_by_country_2020.columns",
"_____no_output_____"
],
[
"inform_df.columns",
"_____no_output_____"
],
[
"# Merge on ISO",
"_____no_output_____"
],
[
"nb_storms_by_decade_by_country_2020_with_inform = pd.merge(nb_storms_by_decade_by_country_2020, inform_df, how=\"left\", left_on=\"ISO\", right_on=\"ISO3\")",
"_____no_output_____"
],
[
"nb_storms_by_decade_by_country_2020_with_inform.head()",
"_____no_output_____"
],
[
"nb_storms_by_decade_by_country_2020_with_inform.shape",
"_____no_output_____"
],
[
"nb_storms_by_decade_by_country_2020_with_inform_filt_col = nb_storms_by_decade_by_country_2020_with_inform[[\"decade\", \"ISO\", \"storms_count\", \"total_nb_affected\", \"total_damages\",\"INFORM RISK\"]]",
"_____no_output_____"
],
[
"nb_storms_by_decade_by_country_2020_with_inform_filt_col.dtypes",
"_____no_output_____"
],
[
"nb_storms_by_decade_by_country_2020_with_inform_filt_col[\"INFORM RISK\"] = nb_storms_by_decade_by_country_2020_with_inform_filt_col[\"INFORM RISK\"].astype(\"float\")",
"C:\\Users\\clotilde.pety\\AppData\\Local\\Continuum\\anaconda3\\lib\\site-packages\\ipykernel_launcher.py:1: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n \"\"\"Entry point for launching an IPython kernel.\n"
],
[
"nb_storms_by_decade_by_country_2020_with_inform_filt_col.head()",
"_____no_output_____"
],
[
"nb_storms_inform_by_country_cor = nb_storms_by_decade_by_country_2020_with_inform_filt_col[[\"ISO\", \"storms_count\", \"total_nb_affected\", \"total_damages\",\"INFORM RISK\"]]\ncorr = nb_storms_inform_by_country_cor.corr()\nsm.graphics.plot_corr(corr, xnames=list(corr.columns))\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Keep observations on decades 2010 and 2020",
"_____no_output_____"
]
],
[
[
"nb_storms_inform_by_country_2010_2020 = nb_storms_by_decade_by_country_2020_with_inform_filt_col[nb_storms_by_decade_by_country_2020_with_inform_filt_col[\"decade\"]>=2010]",
"_____no_output_____"
],
[
"nb_storms_inform_by_country_2010_2020_cor = nb_storms_inform_by_country_2010_2020[[\"ISO\", \"storms_count\", \"total_nb_affected\", \"total_damages\",\"INFORM RISK\"]]\ncorr = nb_storms_inform_by_country_2010_2020_cor.corr()\nsm.graphics.plot_corr(corr, xnames=list(corr.columns))\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Keep observations on decade 2020 (decade of INFORM index)",
"_____no_output_____"
]
],
[
[
"nb_storms_inform_by_country_2020_only = nb_storms_by_decade_by_country_2020_with_inform_filt_col[nb_storms_by_decade_by_country_2020_with_inform_filt_col[\"decade\"]==2020]",
"_____no_output_____"
],
[
"nb_storms_inform_by_country_2020_only.head()",
"_____no_output_____"
],
[
"nb_storms_inform_by_country_2020_only_cor = nb_storms_inform_by_country_2020_only[[\"ISO\", \"storms_count\", \"total_nb_affected\", \"total_damages\",\"INFORM RISK\"]]\ncorr = nb_storms_inform_by_country_2020_only_cor.corr()\nsm.graphics.plot_corr(corr, xnames=list(corr.columns))\nplt.show()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d087e0b52565b6d6eca8b5b0ffaf467bd710adfa | 41,413 | ipynb | Jupyter Notebook | Db2 Jupyter Extensions Tutorial.ipynb | DB2-Samples/db2jupyter | 25929617ebf87483f0b452991ecb1e30f615c718 | [
"Apache-2.0"
] | 30 | 2017-10-25T04:10:02.000Z | 2022-03-09T03:17:35.000Z | Db2 Jupyter Extensions Tutorial.ipynb | IMUK23/db2jupyter | 2f5d9eb2de739280e5940134dffbc76dbef4da5d | [
"Apache-2.0"
] | 4 | 2017-09-30T10:54:35.000Z | 2021-01-08T09:37:04.000Z | Db2 Jupyter Extensions Tutorial.ipynb | IMUK23/db2jupyter | 2f5d9eb2de739280e5940134dffbc76dbef4da5d | [
"Apache-2.0"
] | 50 | 2017-10-25T13:31:08.000Z | 2021-12-03T00:51:16.000Z | 32.253115 | 578 | 0.602275 | [
[
[
"# Db2 Jupyter Notebook Extensions Tutorial\n\nThe SQL code tutorials for Db2 rely on a Jupyter notebook extension, commonly refer to as a \"magic\" command. The beginning of all of the notebooks begin with the following command which will load the extension and allow the remainder of the notebook to use the %sql magic command.\n<pre>\n%run db2.ipynb\n</pre>\nThe cell below will load the Db2 extension. Note that it will take a few seconds for the extension to load, so you should generally wait until the \"Db2 Extensions Loaded\" message is displayed in your notebook. ",
"_____no_output_____"
]
],
[
[
"%run db2.ipynb",
"_____no_output_____"
]
],
[
[
"## Options\nThere are two options that can be set with the **`%sql`** command. These options are:\n- **`MAXROWS n`** - The maximum number of rows that you want to display as part of a SQL statement. Setting MAXROWS to -1 will return all output, while maxrows of 0 will suppress all output.\n- **`RUNTIME n`** - When using the timer option on a SQL statement, the statement will execute for **`n`** number of seconds. The result that is returned is the number of times the SQL statement executed rather than the execution time of the statement. The default value for runtime is one second, so if the SQL is very complex you will need to increase the run time.\n\nTo set an option use the following syntax:\n```\n%sql option option_name value option_name value ....\n```\nThe following example sets all three options:\n```\n%sql option maxrows 100 runtime 2\n```\nThe values will be saved between Jupyter notebooks sessions.",
"_____no_output_____"
],
[
"## Connections to Db2\n\nBefore any SQL commands can be issued, a connection needs to be made to the Db2 database that you will be using. The connection can be done manually (through the use of the CONNECT command), or automatically when the first `%sql` command is issued.\n\nThe Db2 magic command tracks whether or not a connection has occured in the past and saves this information between notebooks and sessions. When you start up a notebook and issue a command, the program will reconnect to the database using your credentials from the last session. In the event that you have not connected before, the system will prompt you for all the information it needs to connect. This information includes:\n\n- Database name (SAMPLE) \n- Hostname - localhost (enter an IP address if you need to connect to a remote server) \n- PORT - 50000 (this is the default but it could be different) \n- Userid - DB2INST1 \n- Password - No password is provided so you have to enter a value \n- Maximum Rows - 10 lines of output are displayed when a result set is returned \n\nThere will be default values presented in the panels that you can accept, or enter your own values. All of the information will be stored in the directory that the notebooks are stored on. Once you have entered the information, the system will attempt to connect to the database for you and then you can run all of the SQL scripts. More details on the CONNECT syntax will be found in a section below.\n\nIf you have credentials available from Db2 on Cloud or DSX, place the contents of the credentials into a variable and then use the `CONNECT CREDENTIALS <var>` syntax to connect to the database.\n```Python\ndb2blu = { \"uid\" : \"xyz123456\", ...}\n%sql CONNECT CREDENTIALS db2blu\n```\n\nIf the connection is successful using the credentials, the variable will be saved to disk so that you can connected from within another notebook using the same syntax.\n\nThe next statement will force a CONNECT to occur with the default values. If you have not connected before, it will prompt you for the information.",
"_____no_output_____"
]
],
[
[
"%sql CONNECT",
"_____no_output_____"
]
],
[
[
"## Line versus Cell Command\nThe Db2 extension is made up of one magic command that works either at the LINE level (`%sql`) or at the CELL level (`%%sql`). If you only want to execute a SQL command on one line in your script, use the `%sql` form of the command. If you want to run a larger block of SQL, then use the `%%sql` form. Note that when you use the `%%sql` form of the command, the entire contents of the cell is considered part of the command, so you cannot mix other commands in the cell.\n\nThe following is an example of a line command:",
"_____no_output_____"
]
],
[
[
"%sql VALUES 'HELLO THERE'",
"_____no_output_____"
]
],
[
[
"If you have SQL that requires multiple lines, of if you need to execute many lines of SQL, then you should \nbe using the CELL version of the `%sql` command. To start a block of SQL, start the cell with `%%sql` and do not place any SQL following the command. Subsequent lines can contain SQL code, with each SQL statement delimited with the semicolon (`;`). You can change the delimiter if required for procedures, etc... More details on this later.",
"_____no_output_____"
]
],
[
[
"%%sql\nVALUES\n 1,\n 2,\n 3",
"_____no_output_____"
]
],
[
[
"If you are using a single statement then there is no need to use a delimiter. However, if you are combining a number of commands then you must use the semicolon.",
"_____no_output_____"
]
],
[
[
"%%sql\nDROP TABLE STUFF;\nCREATE TABLE STUFF (A INT);\nINSERT INTO STUFF VALUES\n 1,2,3;\nSELECT * FROM STUFF;",
"_____no_output_____"
]
],
[
[
"The script will generate messages and output as it executes. Each SQL statement that generates results will have a table displayed with the result set. If a command is executed, the results of the execution get listed as well. The script you just ran probably generated an error on the DROP table command.",
"_____no_output_____"
],
[
"## Options\nBoth forms of the `%sql` command have options that can be used to change the behavior of the code. For both forms of the command (`%sql`, `%%sql`), the options must be on the same line as the command:\n<pre>\n%sql -t ...\n%%sql -t\n</pre>\n\nThe only difference is that the `%sql` command can have SQL following the parameters, while the `%%sql` requires the SQL to be placed on subsequent lines.\n\nThere are a number of parameters that you can specify as part of the `%sql` statement. \n\n* `-d` - Use alternative delimiter\n* `-t` - Time the statement execution\n* `-q` - Suppress messages \n* `-j` - JSON formatting of a column\n* `-a` - Show all output\n* `-pb` - Bar chart of results\n* `-pp` - Pie chart of results \n* `-pl` - Line chart of results\n* `-i` - Interactive mode with Pixiedust\n* `-sampledata` Load the database with the sample EMPLOYEE and DEPARTMENT tables\n* `-r` - Return the results into a variable (list of rows)\n* `-e` - Echo macro substitution\n\nMultiple parameters are allowed on a command line. Each option should be separated by a space:\n<pre>\n%sql -a -j ...\n</pre>\n\nA `SELECT` statement will return the results as a dataframe and display the results as a table in the notebook. If you use the assignment statement, the dataframe will be placed into the variable and the results will not be displayed:\n<pre>\nr = %sql SELECT * FROM EMPLOYEE\n</pre>\n\nThe sections below will explain the options in more detail.",
"_____no_output_____"
],
[
"## Delimiters\nThe default delimiter for all SQL statements is the semicolon. However, this becomes a problem when you try to create a trigger, function, or procedure that uses SQLPL (or PL/SQL). Use the `-d` option to turn the SQL delimiter into the at (`@`) sign and `-q` to suppress error messages. The semi-colon is then ignored as a delimiter.\n\nFor example, the following SQL will use the `@` sign as the delimiter.",
"_____no_output_____"
]
],
[
[
"%%sql -d -q\nDROP TABLE STUFF\n@\nCREATE TABLE STUFF (A INT)\n@\nINSERT INTO STUFF VALUES\n 1,2,3\n@\nSELECT * FROM STUFF\n@",
"_____no_output_____"
]
],
[
[
"The delimiter change will only take place for the statements following the `%%sql` command. Subsequent cells\nin the notebook will still use the semicolon. You must use the `-d` option for every cell that needs to use the\nsemicolon in the script.",
"_____no_output_____"
],
[
"## Limiting Result Sets\nThe default number of rows displayed for any result set is 10. You have the option of changing this option when initially connecting to the database. If you want to override the number of rows display you can either update\nthe control variable, or use the -a option. The `-a` option will display all of the rows in the answer set. For instance, the following SQL will only show 10 rows even though we inserted 15 values:",
"_____no_output_____"
]
],
[
[
"%sql values 1,2,3,4,5,6,7,8,9,10,11,12,13,14,15",
"_____no_output_____"
]
],
[
[
"You will notice that the displayed result will split the visible rows to the first 5 rows and the last 5 rows.\nUsing the `-a` option will display all values in a scrollable table.",
"_____no_output_____"
]
],
[
[
"%sql -a values 1,2,3,4,5,6,7,8,9,10,11,12,13,14,15",
"_____no_output_____"
]
],
[
[
"To change the default value of rows displayed, you can use the `%sql option maxrow` command to set the value to something else. A value of 0 or -1 means unlimited output. ",
"_____no_output_____"
]
],
[
[
"%sql option maxrows 5\n%sql values 1,2,3,4,5,6,7,8,9,10,11,12,13,14,15",
"_____no_output_____"
]
],
[
[
"A special note regarding the output from a `SELECT` statement. If the SQL statement is the last line of a block, the results will be displayed by default (unless you assigned the results to a variable). If the SQL is in the middle of a block of statements, the results will not be displayed. To explicitly display the results you must use the display function (or pDisplay if you have imported another library like pixiedust which overrides the pandas display function). ",
"_____no_output_____"
]
],
[
[
"# Set the maximum back\n%sql option maxrows 10\n%sql values 1,2,3,4,5,6,7,8,9,10,11,12,13,14,15",
"_____no_output_____"
]
],
[
[
"## Quiet Mode\nEvery SQL statement will result in some output. You will either get an answer set (`SELECT`), or an indication if\nthe command worked. For instance, the following set of SQL will generate some error messages since the tables \nwill probably not exist:",
"_____no_output_____"
]
],
[
[
"%%sql\nDROP TABLE TABLE_NOT_FOUND;\nDROP TABLE TABLE_SPELLED_WRONG;",
"_____no_output_____"
]
],
[
[
"If you know that these errors may occur you can silence them with the -q option.",
"_____no_output_____"
]
],
[
[
"%%sql -q\nDROP TABLE TABLE_NOT_FOUND;\nDROP TABLE TABLE_SPELLED_WRONG;",
"_____no_output_____"
]
],
[
[
"SQL output will not be suppressed, so the following command will still show the results.",
"_____no_output_____"
]
],
[
[
"%%sql -q\nDROP TABLE TABLE_NOT_FOUND;\nDROP TABLE TABLE_SPELLED_WRONG;\nVALUES 1,2,3;",
"_____no_output_____"
]
],
[
[
"## Variables in %sql Blocks",
"_____no_output_____"
],
[
"Python variables can be passed to a `%sql` line command, and to a `%%sql` block. For both forms of the `%sql` command you can pass variables by placing a colon in front of the variable name.\n```python\n%sql SELECT * FROM EMPLOYEE WHERE EMPNO = :empno\n```\n\nThe following example illustrates the use of a variable in the SQL.",
"_____no_output_____"
]
],
[
[
"empno = '000010'\n%sql SELECT * FROM EMPLOYEE WHERE EMPNO = :empno",
"_____no_output_____"
]
],
[
[
"You can doublecheck that the substitution took place by using the `-e` option which echos the SQL command after substitution.",
"_____no_output_____"
]
],
[
[
"%sql -e SELECT * FROM EMPLOYEE WHERE EMPNO = :empno",
"_____no_output_____"
]
],
[
[
"Note that the variable `:empno` did not have quotes around it, although it is a string value. The `%sql` call will examine the contents of the variable and add quotes around strings so you do not have to supply them in the SQL command.\n\nVariables can also be array types. Arrays are expanded into multiple values, each separated by commas. This is useful when building SQL `IN` lists. The following example searches for 3 employees based on their employee number.",
"_____no_output_____"
]
],
[
[
"empnos = ['000010','000020','000030']\n%sql SELECT * FROM EMPLOYEE WHERE EMPNO IN (:empnos)",
"_____no_output_____"
]
],
[
[
"You can reference individual array items using this technique as well. If you wanted to search for only the first value in the `empnos` array, use `:empnos[0]` instead.",
"_____no_output_____"
]
],
[
[
"%sql SELECT * FROM EMPLOYEE WHERE EMPNO IN (:empnos[0])",
"_____no_output_____"
]
],
[
[
"One final type of variable substitution that is allowed is for dictionaries. Python dictionaries resemble JSON objects and can be used to insert JSON values into Db2. For instance, the following variable contains company information in a JSON structure.",
"_____no_output_____"
]
],
[
[
"customer = {\n \"name\" : \"Aced Hardware Stores\",\n \"city\" : \"Rockwood\",\n \"employees\" : 14\n}",
"_____no_output_____"
]
],
[
[
"Db2 has builtin functions for dealing with JSON objects. There is another Jupyter notebook which goes through this in detail. Rather than using those functions, the following code will create a Db2 table with a string column that will contain the contents of this JSON record.",
"_____no_output_____"
]
],
[
[
"%%sql\nDROP TABLE SHOWJSON;\nCREATE TABLE SHOWJSON (INJSON VARCHAR(256));",
"_____no_output_____"
]
],
[
[
"To insert the Dictionary (JSON Record) into this Db2 table, you only need to use the variable name as one of the fields being inserted.",
"_____no_output_____"
]
],
[
[
"%sql INSERT INTO SHOWJSON VALUES :customer",
"_____no_output_____"
]
],
[
[
"Selecting from this table will show that the data has been inserted as a string. ",
"_____no_output_____"
]
],
[
[
"%sql select * from showjson",
"_____no_output_____"
]
],
[
[
"If you want to retrieve the data from a column that contains JSON records, you must use the `-j` flag to insert the contents back into a variable.",
"_____no_output_____"
]
],
[
[
"v = %sql -j SELECT * FROM SHOWJSON",
"_____no_output_____"
]
],
[
[
"The variable `v` now contains the original JSON record for you to use.",
"_____no_output_____"
]
],
[
[
"v",
"_____no_output_____"
]
],
[
[
"## SQL Character Strings\n\nCharacter strings require special handling when dealing with Db2. The single quote character `'` is reserved for delimiting string constants, while the double quote `\"` is used for naming columns that require special characters. You cannot use the double quote character to delimit strings that happen to contain the single quote character. What Db2 requires you do is placed two quotes in a row to have them interpreted as a single quote character. For instance, the next statement will select one employee from the table who has a quote in their last name: `O'CONNELL`.",
"_____no_output_____"
]
],
[
[
"%sql SELECT * FROM EMPLOYEE WHERE LASTNAME = 'O''CONNELL'",
"_____no_output_____"
]
],
[
[
"Python handles quotes differently! You can assign a string to a Python variable using single or double quotes. The following assignment statements are not identical!",
"_____no_output_____"
]
],
[
[
"lastname = \"O'CONNELL\"\nprint(lastname)\nlastname = 'O''CONNELL'\nprint(lastname)",
"_____no_output_____"
]
],
[
[
"If you use the same syntax as Db2, Python will remove the quote in the string! It interprets this as two strings (O and CONNELL) being concatentated together. That probably isn't what you want! So the safest approach is to use double quotes around your string when you assign it to a variable. Then you can use the variable in the SQL statement as shown in the following example.",
"_____no_output_____"
]
],
[
[
"lastname = \"O'CONNELL\"\n%sql -e SELECT * FROM EMPLOYEE WHERE LASTNAME = :lastname",
"_____no_output_____"
]
],
[
[
"Notice how the string constant was updated to contain two quotes when inserted into the SQL statement. This is done automatically by the `%sql` magic command, so there is no need to use the two single quotes when assigning a string to a variable. However, you must use the two single quotes when using constants in a SQL statement.",
"_____no_output_____"
],
[
"## Builtin Variables\nThere are 5 predefined variables defined in the program:\n\n- database - The name of the database you are connected to\n- uid - The userid that you connected with\n- hostname = The IP address of the host system\n- port - The port number of the host system\n- max - The maximum number of rows to return in an answer set\n\nTheses variables are all part of a structure called _settings. To retrieve a value, use the syntax:\n```python\ndb = _settings['database']\n```\n\nThere are also 3 variables that contain information from the last SQL statement that was executed.\n\n- sqlcode - SQLCODE from the last statement executed\n- sqlstate - SQLSTATE from the last statement executed\n- sqlerror - Full error message returned on last statement executed\n\nYou can access these variables directly in your code. The following code segment illustrates the use of the SQLCODE variable.",
"_____no_output_____"
]
],
[
[
"empnos = ['000010','999999']\nfor empno in empnos:\n ans1 = %sql -r SELECT SALARY FROM EMPLOYEE WHERE EMPNO = :empno\n if (sqlcode != 0):\n print(\"Employee \"+ empno + \" left the company!\")\n else:\n print(\"Employee \"+ empno + \" salary is \" + str(ans1[1][0]))",
"_____no_output_____"
]
],
[
[
"## Timing SQL Statements\nSometimes you want to see how the execution of a statement changes with the addition of indexes or other\noptimization changes. The `-t` option will run the statement on the LINE or one SQL statement in the CELL for \nexactly one second. The results will be displayed and optionally placed into a variable. The syntax of the\ncommand is:\n<pre>\nsql_time = %sql -t SELECT * FROM EMPLOYEE\n</pre>\nFor instance, the following SQL will time the VALUES clause.",
"_____no_output_____"
]
],
[
[
"%sql -t VALUES 1,2,3,4,5,6,7,8,9",
"_____no_output_____"
]
],
[
[
"When timing a statement, no output will be displayed. If your SQL statement takes longer than one second you\nwill need to modify the runtime options. You can use the `%sql option runtime` command to change the duration the statement runs.",
"_____no_output_____"
]
],
[
[
"%sql option runtime 5\n%sql -t VALUES 1,2,3,4,5,6,7,8,9\n%sql option runtime 1",
"_____no_output_____"
]
],
[
[
"## JSON Formatting\nDb2 supports querying JSON that is stored in a column within a table. Standard output would just display the \nJSON as a string. For instance, the following statement would just return a large string of output.",
"_____no_output_____"
]
],
[
[
"%%sql \nVALUES \n '{\n \"empno\":\"000010\",\n \"firstnme\":\"CHRISTINE\",\n \"midinit\":\"I\",\n \"lastname\":\"HAAS\",\n \"workdept\":\"A00\",\n \"phoneno\":[3978],\n \"hiredate\":\"01/01/1995\",\n \"job\":\"PRES\",\n \"edlevel\":18,\n \"sex\":\"F\",\n \"birthdate\":\"08/24/1963\",\n \"pay\" : {\n \"salary\":152750.00,\n \"bonus\":1000.00,\n \"comm\":4220.00}\n }'",
"_____no_output_____"
]
],
[
[
"Adding the -j option to the `%sql` (or `%%sql`) command will format the first column of a return set to better\ndisplay the structure of the document. Note that if your answer set has additional columns associated with it, they will not be displayed in this format.",
"_____no_output_____"
]
],
[
[
"%%sql -j \nVALUES \n '{\n \"empno\":\"000010\",\n \"firstnme\":\"CHRISTINE\",\n \"midinit\":\"I\",\n \"lastname\":\"HAAS\",\n \"workdept\":\"A00\",\n \"phoneno\":[3978],\n \"hiredate\":\"01/01/1995\",\n \"job\":\"PRES\",\n \"edlevel\":18,\n \"sex\":\"F\",\n \"birthdate\":\"08/24/1963\",\n \"pay\" : {\n \"salary\":152750.00,\n \"bonus\":1000.00,\n \"comm\":4220.00}\n }'",
"_____no_output_____"
]
],
[
[
"JSON fields can be inserted into Db2 columns using Python dictionaries. This makes the input and output of JSON fields much simpler. For instance, the following code will create a Python dictionary which is similar to a JSON record.",
"_____no_output_____"
]
],
[
[
"employee = {\n \"firstname\" : \"John\",\n \"lastname\" : \"Williams\",\n \"age\" : 45\n}",
"_____no_output_____"
]
],
[
[
"The field can be inserted into a character column (or BSON if you use the JSON functions) by doing a direct variable insert.",
"_____no_output_____"
]
],
[
[
"%%sql -q\nDROP TABLE SHOWJSON;\nCREATE TABLE SHOWJSON(JSONIN VARCHAR(128));",
"_____no_output_____"
]
],
[
[
"An insert would use a variable parameter (colon in front of the variable) instead of a character string.",
"_____no_output_____"
]
],
[
[
"%sql INSERT INTO SHOWJSON VALUES (:employee)\n%sql SELECT * FROM SHOWJSON",
"_____no_output_____"
]
],
[
[
"An assignment statement to a variable will result in an equivalent Python dictionary type being created. Note that we must use the raw `-j` flag to make sure we only get the data and not a data frame.",
"_____no_output_____"
]
],
[
[
"x = %sql -j SELECT * FROM SHOWJSON\nprint(\"First Name is \" + x[0][\"firstname\"] + \" and the last name is \" + x[0]['lastname'])",
"_____no_output_____"
]
],
[
[
"## Plotting\nSometimes it would be useful to display a result set as either a bar, pie, or line chart. The first one or two\ncolumns of a result set need to contain the values need to plot the information.\n\nThe three possible plot options are:\n \n* `-pb` - bar chart (x,y)\n* `-pp` - pie chart (y)\n* `-pl` - line chart (x,y)\n\nThe following data will be used to demonstrate the different charting options.",
"_____no_output_____"
]
],
[
[
"%sql values 1,2,3,4,5",
"_____no_output_____"
]
],
[
[
"Since the results only have one column, the pie, line, and bar charts will not have any labels associated with\nthem. The first example is a bar chart.",
"_____no_output_____"
]
],
[
[
"%sql -pb values 1,2,3,4,5",
"_____no_output_____"
]
],
[
[
"The same data as a pie chart.",
"_____no_output_____"
]
],
[
[
"%sql -pp values 1,2,3,4,5",
"_____no_output_____"
]
],
[
[
"And finally a line chart.",
"_____no_output_____"
]
],
[
[
"%sql -pl values 1,2,3,4,5",
"_____no_output_____"
]
],
[
[
"If you retrieve two columns of information, the first column is used for the labels (X axis or pie slices) and \nthe second column contains the data. ",
"_____no_output_____"
]
],
[
[
"%sql -pb values ('A',1),('B',2),('C',3),('D',4),('E',5)",
"_____no_output_____"
]
],
[
[
"For a pie chart, the first column is used to label the slices, while the data comes from the second column.",
"_____no_output_____"
]
],
[
[
"%sql -pp values ('A',1),('B',2),('C',3),('D',4),('E',5)",
"_____no_output_____"
]
],
[
[
"Finally, for a line chart, the x contains the labels and the y values are used.",
"_____no_output_____"
]
],
[
[
"%sql -pl values ('A',1),('B',2),('C',3),('D',4),('E',5)",
"_____no_output_____"
]
],
[
[
"The following SQL will plot the number of employees per department.",
"_____no_output_____"
]
],
[
[
"%%sql -pb\nSELECT WORKDEPT, COUNT(*) \n FROM EMPLOYEE\nGROUP BY WORKDEPT",
"_____no_output_____"
]
],
[
[
"The final option for plotting data is to use interactive mode `-i`. This will display the data using an open-source project called Pixiedust. You can view the results in a table and then interactively create a plot by dragging and dropping column names into the appropriate slot. The next command will place you into interactive mode.",
"_____no_output_____"
]
],
[
[
"%sql -i select * from employee",
"_____no_output_____"
]
],
[
[
"## Sample Data\nMany of the Db2 notebooks depend on two of the tables that are found in the `SAMPLE` database. Rather than\nhaving to create the entire `SAMPLE` database, this option will create and populate the `EMPLOYEE` and \n`DEPARTMENT` tables in your database. Note that if you already have these tables defined, they will not be dropped.",
"_____no_output_____"
]
],
[
[
"%sql -sampledata",
"_____no_output_____"
]
],
[
[
"## Result Sets \nBy default, any `%sql` block will return the contents of a result set as a table that is displayed in the notebook. The results are displayed using a feature of pandas dataframes. The following select statement demonstrates a simple result set.",
"_____no_output_____"
]
],
[
[
"%sql select * from employee fetch first 3 rows only",
"_____no_output_____"
]
],
[
[
"You can assign the result set directly to a variable.",
"_____no_output_____"
]
],
[
[
"x = %sql select * from employee fetch first 3 rows only",
"_____no_output_____"
]
],
[
[
"The variable x contains the dataframe that was produced by the `%sql` statement so you access the result set by using this variable or display the contents by just referring to it in a command line.",
"_____no_output_____"
]
],
[
[
"x",
"_____no_output_____"
]
],
[
[
"There is an additional way of capturing the data through the use of the `-r` flag.\n<pre>\nvar = %sql -r select * from employee\n</pre>\nRather than returning a dataframe result set, this option will produce a list of rows. Each row is a list itself. The column names are found in row zero (0) and the data rows start at 1. To access the first column of the first row, you would use var[1][0] to access it.",
"_____no_output_____"
]
],
[
[
"rows = %sql -r select * from employee fetch first 3 rows only\nprint(rows[1][0])",
"_____no_output_____"
]
],
[
[
"The number of rows in the result set can be determined by using the length function and subtracting one for the header row.",
"_____no_output_____"
]
],
[
[
"print(len(rows)-1)",
"_____no_output_____"
]
],
[
[
"If you want to iterate over all of the rows and columns, you could use the following Python syntax instead of\ncreating a for loop that goes from 0 to 41.",
"_____no_output_____"
]
],
[
[
"for row in rows:\n line = \"\"\n for col in row:\n line = line + str(col) + \",\"\n print(line)",
"_____no_output_____"
]
],
[
[
"If you don't want the header row, modify the first line to start at the first row instead of row zero.",
"_____no_output_____"
]
],
[
[
"for row in rows[1:]:\n line = \"\"\n for col in row:\n line = line + str(col) + \",\"\n print(line)",
"_____no_output_____"
]
],
[
[
"Since the data may be returned in different formats (like integers), you should use the str() function\nto convert the values to strings. Otherwise, the concatenation function used in the above example will fail. For\ninstance, the 6th field is a birthdate field. If you retrieve it as an individual value and try and concatenate a string to it, you get the following error.",
"_____no_output_____"
]
],
[
[
"try:\n print(\"Birth Date=\"+rows[1][6])\nexcept Exception as err:\n print(\"Oops... Something went wrong!\")\n print(err)",
"_____no_output_____"
]
],
[
[
"You can fix this problem by adding the str function to convert the date.",
"_____no_output_____"
]
],
[
[
"print(\"Birth Date=\"+str(rows[1][6]))",
"_____no_output_____"
]
],
[
[
"## Development SQL\nThe previous set of `%sql` and `%%sql` commands deals with SQL statements and commands that are run in an interactive manner. There is a class of SQL commands that are more suited to a development environment where code is iterated or requires changing input. The commands that are associated with this form of SQL are:\n- AUTOCOMMIT\n- COMMIT/ROLLBACK\n- PREPARE \n- EXECUTE\n\nIn addition, the `sqlcode`, `sqlstate` and `sqlerror` fields are populated after every statement so you can use these variables to test for errors.\n\nAutocommit is the default manner in which SQL statements are executed. At the end of the successful completion of a statement, the results are commited to the database. There is no concept of a transaction where multiple DML/DDL statements are considered one transaction. The `AUTOCOMMIT` command allows you to turn autocommit `OFF` or `ON`. This means that the set of SQL commands run after the `AUTOCOMMIT OFF` command are executed are not commited to the database until a `COMMIT` or `ROLLBACK` command is issued.\n\n`COMMIT (WORK)` will finalize all of the transactions (`COMMIT`) to the database and `ROLLBACK` will undo all of the changes. If you issue a `SELECT` statement during the execution of your block, the results will reflect all of your changes. If you `ROLLBACK` the transaction, the changes will be lost.\n\n`PREPARE` is typically used in a situation where you want to repeatidly execute a SQL statement with different variables without incurring the SQL compilation overhead. For instance:\n```\nx = %sql PREPARE SELECT LASTNAME FROM EMPLOYEE WHERE EMPNO=?\nfor y in ['000010','000020','000030']:\n %sql execute :x using :y\n```\n`EXECUTE` is used to execute a previously compiled statement. ",
"_____no_output_____"
],
[
"## Db2 CONNECT Statement\nAs mentioned at the beginning of this notebook, connecting to Db2 is automatically done when you issue your first\n`%sql` statement. Usually the program will prompt you with what options you want when connecting to a database. The other option is to use the CONNECT statement directly. The CONNECT statement is similar to the native Db2\nCONNECT command, but includes some options that allow you to connect to databases that has not been\ncatalogued locally.\n\nThe CONNECT command has the following format:\n<pre>\n%sql CONNECT TO <database> USER <userid> USING <password | ?> HOST <ip address> PORT <port number>\n</pre>\nIf you use a \"?\" for the password field, the system will prompt you for a password. This avoids typing the \npassword as clear text on the screen. If a connection is not successful, the system will print the error\nmessage associated with the connect request.\n\nIf the connection is successful, the parameters are saved on your system and will be used the next time you\nrun a SQL statement, or when you issue the %sql CONNECT command with no parameters.\n\nIf you want to force the program to connect to a different database (with prompting), use the CONNECT RESET command. The next time you run a SQL statement, the program will prompt you for the the connection\nand will force the program to reconnect the next time a SQL statement is executed.",
"_____no_output_____"
],
[
"#### Credits: IBM 2018, George Baklarz [[email protected]]",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
d087e61df27e48769b814128c818ce2af4e58d35 | 62,123 | ipynb | Jupyter Notebook | NLP/NLP Restaurant Reviews.ipynb | InternityFoundation/Machine-Learning-Shaurya | 1135bfdcf12c17e4ba6f14609107d81328fb05f2 | [
"Apache-2.0"
] | 3 | 2020-02-07T03:33:13.000Z | 2021-06-05T09:48:52.000Z | NLP/NLP Restaurant Reviews.ipynb | InternityFoundation/Machine-Learning-Shaurya | 1135bfdcf12c17e4ba6f14609107d81328fb05f2 | [
"Apache-2.0"
] | null | null | null | NLP/NLP Restaurant Reviews.ipynb | InternityFoundation/Machine-Learning-Shaurya | 1135bfdcf12c17e4ba6f14609107d81328fb05f2 | [
"Apache-2.0"
] | 1 | 2018-08-07T15:51:17.000Z | 2018-08-07T15:51:17.000Z | 43.68706 | 147 | 0.543003 | [
[
[
"import os\nprint(os.listdir())",
"['.ipynb_checkpoints', 'Natural Language Processing Restaurant review.py', 'Natural_Language_Processing', 'NLP Restaurant Reviews.ipynb']\n"
],
[
"#Import Library\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"#Import dataset\ndataset = pd.read_csv(\"Natural_Language_Processing/Restaurant_Reviews.tsv\",encoding='latin-1',delimiter='\\t',quoting=3)\n#We do not use a csv file as delimiter in csv is a comma and in tsv delimiter is a tab space.\n#quoting parameter will ignore the double quotes in dataset, quoting=3",
"_____no_output_____"
],
[
"dataset.head(8)",
"_____no_output_____"
],
[
"dataset.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1000 entries, 0 to 999\nData columns (total 2 columns):\nReview 1000 non-null object\nLiked 1000 non-null int64\ndtypes: int64(1), object(1)\nmemory usage: 15.7+ KB\n"
],
[
"#Cleaning the text\nimport re \nimport nltk\n#Download stopword list\nnltk.download('stopwords')\n#Import stopwords\nfrom nltk.corpus import stopwords",
"[nltk_data] Downloading package stopwords to\n[nltk_data] C:\\Users\\shaur\\AppData\\Roaming\\nltk_data...\n[nltk_data] Package stopwords is already up-to-date!\n"
],
[
"corpus = []\nfor i in range(0,1000):\n #Step1\n review = re.sub('[^a-zA-Z]',' ', dataset['Review'][i])\n #print review\n #Step2\n review = review.lower()\n #Step3 : remove non-significant words example the,at,in,a,this\n #split reviews\n review = review.split()\n review = [word for word in review if not word in set(stopwords.words('english'))]\n #Stemming\n from nltk.stem.porter import PorterStemmer\n ps = PorterStemmer()\n review = [ps.stem(word) for word in review if not word in set(stopwords.words('english'))]\n review = \" \".join(review)\n corpus.append(review)",
"_____no_output_____"
],
[
"corpus",
"_____no_output_____"
],
[
"#Creating the Bag of Words Model\nfrom sklearn.feature_extraction.text import CountVectorizer\ncv = CountVectorizer(max_features=1500)\nX = cv.fit_transform(corpus).toarray()",
"_____no_output_____"
],
[
"X",
"_____no_output_____"
],
[
"y = dataset.iloc[:, 1].values",
"_____no_output_____"
],
[
"y",
"_____no_output_____"
],
[
"#Most common models used for NLP are Naive Bayes and Decision Tree\n\nfrom sklearn.model_selection import train_test_split\nX_train,X_test,y_train,y_test = train_test_split(X,y,test_size=0.20,random_state=0)\n\n\n#fit data to NB\nfrom sklearn.naive_bayes import GaussianNB\n\nclassifier = GaussianNB()\nclassifier.fit(X_train,y_train)\n\n#detect things using Confussion matrix\nfrom sklearn.metrics import confusion_matrix,accuracy_score\ncm = confusion_matrix(y_test,classifier.predict(X_test))\nprint(cm)",
"[[55 42]\n [12 91]]\n"
],
[
"accuracy_score(y_test,classifier.predict(X_test))",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d087f03e4eec7c06970dbc425f5b009ae79a1421 | 201,946 | ipynb | Jupyter Notebook | Session9/Day4/Matched_filter_tutorial.ipynb | jlmciver/LSSTC-DSFP-Sessions | f0ee7a5ce4badbd4e163a4ea180b7f2afcd29532 | [
"MIT"
] | null | null | null | Session9/Day4/Matched_filter_tutorial.ipynb | jlmciver/LSSTC-DSFP-Sessions | f0ee7a5ce4badbd4e163a4ea180b7f2afcd29532 | [
"MIT"
] | null | null | null | Session9/Day4/Matched_filter_tutorial.ipynb | jlmciver/LSSTC-DSFP-Sessions | f0ee7a5ce4badbd4e163a4ea180b7f2afcd29532 | [
"MIT"
] | null | null | null | 251.802993 | 44,332 | 0.920246 | [
[
[
"# Welcome to the matched filtering tutorial! \n\n### Installation \n\nMake sure you have PyCBC and some basic lalsuite tools installed. You can do this in a terminal with pip:",
"_____no_output_____"
]
],
[
[
"! pip install lalsuite pycbc",
"_____no_output_____"
]
],
[
[
"<span style=\"color:gray\">Jess notes: this notebook was made with a PyCBC 1.8.0 kernel. </span>\n\n### Learning goals \n\nWith this tutorial, you learn how to:\n\n* Generate source waveforms detectable by LIGO, Virgo, KAGRA\n* Use PyCBC to run a matched filter search on gravitational wave detector data \n* Estimate the significance of a trigger given a background distribution\n* **Challenge**: Code up a trigger coincidence algorithm \n\nThis tutorial borrows heavily from tutorials made for the [LIGO-Virgo Open Data Workshop](https://www.gw-openscience.org/static/workshop1/course.html) by Alex Nitz. You can find PyCBC documentation and additional examples [here](http://pycbc.org/pycbc/latest/html/py-modindex.html). \n\nLet's get started!\n\n___",
"_____no_output_____"
],
[
"## Generate a gravitational wave signal waveform\n\nWe'll use a popular waveform approximant ([SOEBNRv4](https://arxiv.org/pdf/1611.03703.pdf)) to generate waveforms that would be detectable by LIGO, Virgo, or KAGRA. \n\nFirst we import the packages we'll need. ",
"_____no_output_____"
]
],
[
[
"from pycbc.waveform import get_td_waveform\nimport pylab",
"_____no_output_____"
]
],
[
[
"Let's see what these waveforms look like for different component masses. We'll assume the two compact object have masses equal to each other, and we'll set a lower frequency bound of 30 Hz (determined by the sensitivity of our detectors).\n\nWe can also set a time sample rate with `get_td_waveform`. Let's try a rate of 4096 Hz. \n\nLet's make a plot of the plus polarization (`hp`) to get a feel for what the waveforms look like.",
"_____no_output_____"
]
],
[
[
"for m in [5, 10, 30, 100]:\n hp, hc = get_td_waveform(approximant=\"SEOBNRv4_opt\",\n mass1=m,\n mass2=m,\n delta_t=1.0/4096,\n f_lower=30)\n pylab.plot(hp.sample_times, hp, label='$M_{\\odot 1,2}=%s$' % m)\npylab.legend(loc='upper left')\npylab.ylabel('GW strain (plus polarization)')\npylab.grid()\npylab.xlabel('Time (s)')\npylab.show()",
"_____no_output_____"
]
],
[
[
" Now let's see what happens if we decrease the lower frequency bound from 30 Hz to 15 Hz. ",
"_____no_output_____"
]
],
[
[
"for m in [5, 10, 30, 100]:\n hp, hc = get_td_waveform(approximant=\"SEOBNRv4_opt\",\n mass1=m,\n mass2=m,\n delta_t=1.0/4096,\n f_lower=15)\n \n pylab.plot(hp.sample_times, hp, label='$M_{\\odot 1,2}=%s$' % m)\npylab.legend(loc='upper left')\npylab.ylabel('GW strain (plus polarization)')\npylab.grid()\npylab.xlabel('Time (s)')\npylab.show()",
"_____no_output_____"
]
],
[
[
"---\n\n### Exercise 1\n\nWhat happens to the waveform when the total mass (let's say 20 M<sub>sol</sub>) stays the same, but the mass ratio between the component masses changes? \n\nCompare the waveforms for a m<sub>1</sub> = m<sub>2</sub> = 10 M<sub>sol</sub> system, and a m<sub>1</sub> = 2 M<sub>sol</sub>, m<sub>2</sub> = 18 M<sub>sol</sub> system. What do you notice? \n\n",
"_____no_output_____"
]
],
[
[
"# complete ",
"_____no_output_____"
]
],
[
[
"### Exercise 2 \n\nHow much longer (in signal duration) would LIGO and Virgo (and KAGRA) be able to detect a 1.4-1.4 M<sub>sol</sub> binary neutron star system if our detectors were sensitive down to 10 Hz instead of 30 Hz? ** Note you'll need to use a different waveform approximant here. Try TaylorF2.** \n\n<span style=\"color:gray\">Jess notes: this would be a major benefit of next-generation (\"3G\") ground-based gravitational wave detectors.</span>",
"_____no_output_____"
]
],
[
[
"# complete ",
"_____no_output_____"
]
],
[
[
"---\n\n### Distance vs. signal amplitude\n\nLet's see what happens when we scale the distance (in units of Megaparsecs) for a system with a total mass of 20 M<sub>sol</sub>. \n\n<span style=\"color:gray\">Note: redshift effects are not included here.</span>",
"_____no_output_____"
]
],
[
[
"for d in [100, 500, 1000]:\n hp, hc = get_td_waveform(approximant=\"SEOBNRv4_opt\",\n mass1=10,\n mass2=10,\n delta_t=1.0/4096,\n f_lower=30,\n distance=d)\n \n pylab.plot(hp.sample_times, hp, label='Distance=%s Mpc' % d)\npylab.grid()\npylab.xlabel('Time (s)')\npylab.ylabel('GW strain (plus polarization)')\npylab.legend(loc='upper left')\npylab.show()",
"_____no_output_____"
]
],
[
[
"---\n\n## Run a matched filter search on gravitational wave detector data\n\nPyCBC also maintains a catalog of open data as PyCBC time series objects, easy to manipulate with PyCBC tools. Let's try using that and importing the data around the first detection, GW150914. \n",
"_____no_output_____"
]
],
[
[
"import pylab\nfrom pycbc.catalog import Merger\nfrom pycbc.filter import resample_to_delta_t, highpass\n\nmerger = Merger(\"GW150914\")\n\n# Get the data from the Hanford detector\nstrain = merger.strain('H1')",
"_____no_output_____"
]
],
[
[
"### Data pre-conditioning \n\nOnce we've imported the open data from this alternate source, the first thing we'll need to do is **pre-condition** the data. This serves a few purposes: \n* 1) reduces the dynamic range of the data\n* 2) supresses high amplitudes at low frequencies, which can introduce numerical artifacts\n* 3) if we don't need high frequency information, downsampling allows us to compute our matched filter result faster\n\nLet's try highpassing above 15 Hz and downsampling to 2048 Hz, and we'll make a plot to see what the result looks like:",
"_____no_output_____"
]
],
[
[
"# Remove the low frequency content and downsample the data to 2048Hz\nstrain = resample_to_delta_t(highpass(strain, 15.0), 1.0/2048)\n\npylab.plot(strain.sample_times, strain)\npylab.xlabel('Time (s)')",
"_____no_output_____"
]
],
[
[
"Notice the large amplitude excursions in the data at the start and end of our data segment. This is **spectral leakage** caused by filters we applied to the boundaries ringing off the discontinuities where the data suddenly starts and ends (for a time up to the length of the filter).\n\nTo avoid this we should trim the ends of the data in all steps of our filtering. Let's try cropping a couple seconds off of either side. ",
"_____no_output_____"
]
],
[
[
"# Remove 2 seconds of data from both the beginning and end\nconditioned = strain.crop(2, 2)\n\npylab.plot(conditioned.sample_times, conditioned)\npylab.xlabel('Time (s)')",
"_____no_output_____"
]
],
[
[
"That's better. \n\n### Calculating the spectral density of the data\n\nOptimal matched filtering requires *whitening*; weighting the frequency components of the potential signal and data by the estimated noise amplitude.\n\nLet's compute the power spectral density (PSD) of our conditioned data. \n",
"_____no_output_____"
]
],
[
[
"from pycbc.psd import interpolate, inverse_spectrum_truncation\n# Estimate the power spectral density\n\n# We use 4 second samles of our time series in Welch method.\npsd = conditioned.psd(4)\n\n# Now that we have the psd we need to interpolate it to match our data\n# and then limit the filter length of 1 / PSD. After this, we can\n# directly use this PSD to filter the data in a controlled manner\n\npsd = interpolate(psd, conditioned.delta_f)\n\n# 1/PSD will now act as a filter with an effective length of 4 seconds\n# Since the data has been highpassed above 15 Hz, and will have low values\n# below this we need to informat the function to not include frequencies\n# below this frequency. \npsd = inverse_spectrum_truncation(psd, 4 * conditioned.sample_rate,\n low_frequency_cutoff=15)",
"_____no_output_____"
]
],
[
[
"\n----\n\n### Define a signal model \n\nRecall that matched filtering is essentially integrating the inner product between your data and your signal model in frequency or time (after weighting frequencies correctly) as you slide your signal model over your data in time. \n\nIf there is a signal in the data that matches your 'template', we will see a large value of this inner product (the SNR, or 'signal to noise ratio') at that time.\n\nIn a full search, we would grid over the parameters and calculate the SNR time series for each template in our template bank\n\nHere we'll define just one template. Let's assume equal masses (which is within the posterior probability of GW150914). Because we want to match our signal model with each time sample in our data, let's also rescale our signal model vector to match the same number of time samples as our data vector (**<- very important!**). \n\nLet's also plot the output to see what it looks like. ",
"_____no_output_____"
]
],
[
[
"m = 36 # Solar masses\nhp, hc = get_td_waveform(approximant=\"SEOBNRv4_opt\",\n mass1=m,\n mass2=m,\n delta_t=conditioned.delta_t,\n f_lower=20)\n\n# We should resize the vector of our template to match our data\nhp.resize(len(conditioned))\npylab.plot(hp)\npylab.xlabel('Time samples')",
"_____no_output_____"
]
],
[
[
"Note that the waveform template currently begins at the start of the vector. However, we want our SNR time series (the inner product between our data and our template) to track with the approximate merger time. To do this, we need to shift our template so that the merger is approximately at the first bin of the data.\n\nFor this reason, waveforms returned from `get_td_waveform` have their merger stamped with time zero, so we can easily shift the merger into the right position to compute our SNR time series. \n\nLet's try shifting our template time and plot the output. ",
"_____no_output_____"
]
],
[
[
"template = hp.cyclic_time_shift(hp.start_time)\npylab.plot(template)\npylab.xlabel('Time samples')",
"_____no_output_____"
]
],
[
[
"---\n\n### Calculate an SNR time series\n\nNow that we've pre-conditioned our data and defined a signal model, we can compute the output of our matched filter search. ",
"_____no_output_____"
]
],
[
[
"from pycbc.filter import matched_filter\nimport numpy\n\nsnr = matched_filter(template, conditioned,\n psd=psd, low_frequency_cutoff=20)\n\npylab.figure(figsize=[10, 4])\npylab.plot(snr.sample_times, abs(snr))\npylab.xlabel('Time (s)')\npylab.ylabel('SNR')",
"_____no_output_____"
]
],
[
[
"Note that as we expect, there is some corruption at the start and end of our SNR time series by the template filter and the PSD filter. \n\nTo account for this, we can smoothly zero out 4 seconds (the length of the PSD filter) at the beginning and end for the PSD filtering. \n\nWe should remove an 4 additional seconds at the beginning to account for the template length, although this is somewhat generous for so short a template. A longer signal such as from a BNS, would require much more padding at the beginning of the vector.",
"_____no_output_____"
]
],
[
[
"snr = snr.crop(4 + 4, 4)\n\npylab.figure(figsize=[10, 4])\npylab.plot(snr.sample_times, abs(snr))\npylab.ylabel('Signal-to-noise')\npylab.xlabel('Time (s)')\npylab.show()",
"_____no_output_____"
]
],
[
[
"Finally, now that the output is properly cropped, we can find the peak of our SNR time series and estimate the merger time and associated SNR of any event candidate within the data. ",
"_____no_output_____"
]
],
[
[
"peak = abs(snr).numpy().argmax()\nsnrp = snr[peak]\ntime = snr.sample_times[peak]\n\nprint(\"We found a signal at {}s with SNR {}\".format(time, \n abs(snrp)))",
"We found a signal at 1126259462.42s with SNR 19.6770890131\n"
]
],
[
[
"You found the first gravitational wave detection in LIGO Hanford data! Nice work. \n\n---\n\n### Exercise 3 \n\nHow does the SNR change if you re-compute the matched filter result using a signal model with compenent masses that are closer to the current estimates for GW150914, say m<sub>1</sub> = 36 M<sub>sol</sub> and m<sub>2</sub> = 31 M<sub>sol</sub>? \n",
"_____no_output_____"
]
],
[
[
"# complete",
"_____no_output_____"
]
],
[
[
"### Exercise 4 \n\n**Network SNR** is the quadrature sum of the single-detector SNR from each contributing detector. GW150914 was detected by H1 and L1. Try calculating the network SNR (you'll need to estimate the SNR in L1 first), and compare your answer to the network PyCBC SNR as reported in the [GWTC-1 catalog](https://arxiv.org/abs/1811.12907).",
"_____no_output_____"
]
],
[
[
"# complete ",
"_____no_output_____"
]
],
[
[
"---\n\n## Estimate the single-detector significance of an event candidate\n\nGreat, we found a large spike in SNR! What are the chances this is a real astrophysical signal? How often would detector noise produce this by chance?\n\nLet's plot a histogram of SNR values output by our matched filtering analysis for this time and see how much this trigger stands out. \n",
"_____no_output_____"
]
],
[
[
"# import what we need\nfrom scipy.stats import norm\nfrom math import pi \nfrom math import exp\n\n# make a histogram of SNR values \nbackground = (abs(snr))\n\n# plot the histogram to check out any other outliers\npylab.hist(background, bins=50)\npylab.xlabel('SNR')\npylab.semilogy()\n\n# use norm.fit to fit a normal (Gaussian) distribution\n(mu, sigma) = norm.fit(background)\n\n# print out the mean and standard deviation of the fit\nprint('The fit mean = %f and the fit std dev = %f' )%(mu, sigma)\n\n",
"The fit mean = 1.295883 and the fit std dev = 0.739471\n"
]
],
[
[
"### Exercise 5 \n\nAt what single-detector SNR is the significance of a trigger > 5 sigma? \n\nRemember that sigma is constant for a normal distribution (read: this should be simple multiplication now that we have estimated what 1 sigma is). ",
"_____no_output_____"
]
],
[
[
"# complete ",
"_____no_output_____"
]
],
[
[
"---\n\n## Challenge\n\nOur match filter analysis assumes the noise is *stationary* and *Gaussian*, which is not a good assumption, and this short data set isn't representative of all the various things that can go bump in the detector (remember the phone?). \n\n**The simple significance estimate above won't work as soon as we encounter a glitch!** We need a better noise background estimate, and we can leverage our detector network to help make our signals stand out from our background. \n\nObserving a gravitational wave signal between detectors is an important cross-check to minimize the impact of transient detector noise. Our strategy:\n\n* We look for loud triggers within a time window to identify foreground events that occur within the gravitational wave travel time (v=c) between detectors, but could come from any sky position. \n* We use time slides to estimate the noise background for a network of detectors. \n\nIf you still have time, try coding up an algorithm that checks for time coincidence between triggers in different detectors. Remember that the maximum gravitational wave travel time between LIGO detectors is ~10 ms. Check your code with the GPS times for the H1 and L1 triggers you identified for GW150914. ",
"_____no_output_____"
]
],
[
[
"# complete if time ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d087f4932e77ca3f851de5a0d63a1f781074571a | 3,000 | ipynb | Jupyter Notebook | notebook/numpy_image_alpha_blend.ipynb | puyopop/python-snippets | 9d70aa3b2a867dd22f5a5e6178a5c0c5081add73 | [
"MIT"
] | 174 | 2018-05-30T21:14:50.000Z | 2022-03-25T07:59:37.000Z | notebook/numpy_image_alpha_blend.ipynb | puyopop/python-snippets | 9d70aa3b2a867dd22f5a5e6178a5c0c5081add73 | [
"MIT"
] | 5 | 2019-08-10T03:22:02.000Z | 2021-07-12T20:31:17.000Z | notebook/numpy_image_alpha_blend.ipynb | puyopop/python-snippets | 9d70aa3b2a867dd22f5a5e6178a5c0c5081add73 | [
"MIT"
] | 53 | 2018-04-27T05:26:35.000Z | 2022-03-25T07:59:37.000Z | 17.34104 | 98 | 0.483 | [
[
[
"import numpy as np\nfrom PIL import Image",
"_____no_output_____"
],
[
"src1 = np.array(Image.open('data/src/lena.jpg'))\nsrc2 = np.array(Image.open('data/src/rocket.jpg').resize(src1.shape[1::-1], Image.BILINEAR))",
"_____no_output_____"
],
[
"print(src1.dtype)",
"uint8\n"
],
[
"dst = src1 * 0.5 + src2 * 0.5",
"_____no_output_____"
],
[
"print(dst.dtype)",
"float64\n"
],
[
"Image.fromarray(dst.astype(np.uint8)).save('data/dst/numpy_image_alpha_blend.jpg')",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"dst = src1 * 0.5 + src2 * 0.2 + (96, 128, 160)",
"_____no_output_____"
],
[
"print(dst.max())",
"311.1\n"
],
[
"dst = dst.clip(0, 255)",
"_____no_output_____"
],
[
"print(dst.max())",
"255.0\n"
],
[
"Image.fromarray(dst.astype(np.uint8)).save('data/dst/numpy_image_alpha_blend_gamma.jpg')",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
d0880703ec0ffaa2fd86dce7349aeb4ed1113275 | 3,017 | ipynb | Jupyter Notebook | InOrderGraph.ipynb | davedecoder/coding_challenges | aaa572d304ce77fddf94566de28e02b6a27de1ba | [
"MIT"
] | null | null | null | InOrderGraph.ipynb | davedecoder/coding_challenges | aaa572d304ce77fddf94566de28e02b6a27de1ba | [
"MIT"
] | null | null | null | InOrderGraph.ipynb | davedecoder/coding_challenges | aaa572d304ce77fddf94566de28e02b6a27de1ba | [
"MIT"
] | null | null | null | 21.55 | 61 | 0.485582 | [
[
[
"from typing import List",
"_____no_output_____"
],
[
"# Definition for a binary tree node.\nclass TreeNode:\n def __init__(self, x):\n self.val = x\n self.left = None\n self.right = None",
"_____no_output_____"
],
[
"class Solution:\n def listInOrder(self, node:TreeNode) -> List:\n pre_list = list()\n if node.left is not None:\n pre_list += self.listInOrder(node.left)\n pre_list += [node.val]\n if node.right is not None:\n pre_list += self.listInOrder(node.right)\n return pre_list\n \n ",
"_____no_output_____"
],
[
"node = TreeNode(1)\nnode_b = TreeNode(2)\nnode_c = TreeNode(3)\n\n\nnode.left = node_b\nnode.right = node_c\ns = Solution()\nresult = s.listInOrder(node)\nassert result == [2,1,3]",
"_____no_output_____"
],
[
"node = TreeNode(1)\nnode_b = TreeNode(2)\nnode_c = TreeNode(3)\nnode_d = TreeNode(4)\nnode_e = TreeNode(5)\n\n\nnode.left = node_b\nnode.right = node_c\nnode_b.left = node_d\nnode_b.right = node_e\ns = Solution()\nresult = s.listInOrder(node)\nassert result == [4,2,5,1,3]",
"_____no_output_____"
],
[
"node = TreeNode(1)\nnode_b = TreeNode(3)\nnode_c = TreeNode(6)\nnode_d = TreeNode(7)\nnode_e = TreeNode(8)\nnode_f = TreeNode(9)\n\n\nnode.right = node_b\nnode_b.left = node_c\nnode_b.right = node_d\nnode_c.left = node_e\nnode_c.right = node_f\n\ns = Solution()\nresult = s.listInOrder(node)\nassert result == [1,8,6,9,3,7]",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d08808e27563aaec4c2d498dd1dc76b92f6b19db | 34,484 | ipynb | Jupyter Notebook | IBM_AI/5_TensorFlow/ML0120EN-4.1-Review-RBMMNIST.ipynb | merula89/cousera_notebooks | caa529a7abd3763d26f3f2add7c3ab508fbb9bd2 | [
"MIT"
] | null | null | null | IBM_AI/5_TensorFlow/ML0120EN-4.1-Review-RBMMNIST.ipynb | merula89/cousera_notebooks | caa529a7abd3763d26f3f2add7c3ab508fbb9bd2 | [
"MIT"
] | null | null | null | IBM_AI/5_TensorFlow/ML0120EN-4.1-Review-RBMMNIST.ipynb | merula89/cousera_notebooks | caa529a7abd3763d26f3f2add7c3ab508fbb9bd2 | [
"MIT"
] | null | null | null | 38.486607 | 798 | 0.609993 | [
[
[
"<a href=\"https://www.bigdatauniversity.com\"><img src=\"https://ibm.box.com/shared/static/qo20b88v1hbjztubt06609ovs85q8fau.png\" width=\"400px\" align=\"center\"></a>\n\n<h1 align=\"center\"><font size=\"5\">RESTRICTED BOLTZMANN MACHINES</font></h1>",
"_____no_output_____"
],
[
"<h3>Introduction</h3>\n<b>Restricted Boltzmann Machine (RBM):</b> RBMs are shallow neural nets that learn to reconstruct data by themselves in an unsupervised fashion. \n\n\n<h4>Why are RBMs important?</h4>\nIt can automatically extract <b>meaningful</b> features from a given input.\n\n\n<h4>How does it work?</h4>\nRBM is a 2 layer neural network. Simply, RBM takes the inputs and translates those into a set of binary values that represents them in the hidden layer. Then, these numbers can be translated back to reconstruct the inputs. Through several forward and backward passes, the RBM will be trained, and a trained RBM can reveal which features are the most important ones when detecting patterns. \n\n\n<h4>What are the applications of RBM?</h4>\nRBM is useful for <a href='http://www.cs.utoronto.ca/~hinton/absps/netflixICML.pdf'> Collaborative Filtering</a>, dimensionality reduction, classification, regression, feature learning, topic modeling and even <b>Deep Belief Networks</b>.\n\n\n\n<h4>Is RBM a generative or Discriminative model?</h4>\nRBM is a generative model. Let me explain it by first, see what is different between discriminative and generative models: \n\n<b>Discriminative:</b> Consider a classification problem in which we want to learn to distinguish between Sedan cars (y = 1) and SUV cars (y = 0), based on some features of cars. Given a training set, an algorithm like logistic regression tries to find a straight line—that is, a decision boundary—that separates the suv and sedan. \n<b>Generative:</b> looking at cars, we can build a model of what Sedan cars look like. Then, looking at SUVs, we can build a separate model of what SUV cars look like. Finally, to classify a new car, we can match the new car against the Sedan model, and match it against the SUV model, to see whether the new car looks more like the SUV or Sedan. \n\nGenerative Models specify a probability distribution over a dataset of input vectors. We can do both supervise and unsupervised tasks with generative models:\n<ul>\n <li>In an unsupervised task, we try to form a model for P(x), where P is the probability given x as an input vector.</li>\n <li>In the supervised task, we first form a model for P(x|y), where P is the probability of x given y(the label for x). For example, if y = 0 indicates whether a car is a SUV or y = 1 indicates indicate a car is a Sedan, then p(x|y = 0) models the distribution of SUVs’ features, and p(x|y = 1) models the distribution of Sedans’ features. If we manage to find P(x|y) and P(y), then we can use <code>Bayes rule</code> to estimate P(y|x), because: $$p(y|x) = \\frac{p(x|y)p(y)}{p(x)}$$</li>\n</ul>\nNow the question is, can we build a generative model, and then use it to create synthetic data by directly sampling from the modeled probability distributions? Lets see. ",
"_____no_output_____"
],
[
"<h2>Table of Contents</h2>\n<ol>\n <li><a href=\"#ref1\">Initialization</a></li>\n <li><a href=\"#ref2\">RBM layers</a></li>\n <li><a href=\"#ref3\">What RBM can do after training?</a></li>\n <li><a href=\"#ref4\">How to train the model?</a></li>\n <li><a href=\"#ref5\">Learned features</a></li>\n</ol>\n<p></p>\n</div>\n<br>\n\n<hr>",
"_____no_output_____"
],
[
"<a id=\"ref1\"></a>\n<h3>Initialization</h3>\n\nFirst we have to load the utility file which contains different utility functions that are not connected\nin any way to the networks presented in the tutorials, but rather help in\nprocessing the outputs into a more understandable way.",
"_____no_output_____"
]
],
[
[
"import urllib.request\nwith urllib.request.urlopen(\"http://deeplearning.net/tutorial/code/utils.py\") as url:\n response = url.read()\ntarget = open('utils.py', 'w')\ntarget.write(response.decode('utf-8'))\ntarget.close()",
"_____no_output_____"
]
],
[
[
"Now, we load in all the packages that we use to create the net including the TensorFlow package:",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nimport numpy as np\nfrom tensorflow.examples.tutorials.mnist import input_data\n#!pip install pillow\nfrom PIL import Image\nfrom utils import tile_raster_images\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"<hr>",
"_____no_output_____"
],
[
"<a id=\"ref2\"></a>\n<h3>RBM layers</h3>\n\nAn RBM has two layers. The first layer of the RBM is called the <b>visible</b> (or input layer). Imagine that our toy example, has only vectors with 7 values, so the visible layer must have j=7 input nodes. \nThe second layer is the <b>hidden</b> layer, which possesses i neurons in our case. Each hidden node can have either 0 or 1 values (i.e., si = 1 or si = 0) with a probability that is a logistic function of the inputs it receives from the other j visible units, called for example, p(si = 1). For our toy sample, we'll use 2 nodes in the hidden layer, so i = 2.\n\n<center><img src=\"https://ibm.box.com/shared/static/eu26opvcefgls6vnwuo29uwp0nudmokh.png\" alt=\"RBM Model\" style=\"width: 400px;\"></center>",
"_____no_output_____"
],
[
" \n\nEach node in the first layer also has a <b>bias</b>. We will denote the bias as “v_bias” for the visible units. The <b>v_bias</b> is shared among all visible units.\n\nHere we define the <b>bias</b> of second layer as well. We will denote the bias as “h_bias” for the hidden units. The <b>h_bias</b> is shared among all hidden units",
"_____no_output_____"
]
],
[
[
"v_bias = tf.placeholder(\"float\", [7])\nh_bias = tf.placeholder(\"float\", [2])",
"_____no_output_____"
]
],
[
[
"We have to define weights among the input layer and hidden layer nodes. In the weight matrix, the number of rows are equal to the input nodes, and the number of columns are equal to the output nodes. Let <b>W</b> be the Tensor of 7x2 (7 - number of visible neurons, 2 - number of hidden neurons) that represents weights between neurons. ",
"_____no_output_____"
]
],
[
[
"W = tf.constant(np.random.normal(loc=0.0, scale=1.0, size=(7, 2)).astype(np.float32))",
"_____no_output_____"
]
],
[
[
"<hr>",
"_____no_output_____"
],
[
"<a id=\"ref3\"></a>\n<h3>What RBM can do after training?</h3>\nThink RBM as a model that has been trained based on images of a dataset of many SUV and Sedan cars. Also, imagine that the RBM network has only two hidden nodes, one for the weight and, and one for the size of cars, which in a sense, their different configurations represent different cars, one represent SUV cars and one for Sedan. In a training process, through many forward and backward passes, RBM adjust its weights to send a stronger signal to either the SUV node (0, 1) or the Sedan node (1, 0) in the hidden layer, given the pixels of images. Now, given a SUV in hidden layer, which distribution of pixels should we expect? RBM can give you 2 things. First, it encodes your images in hidden layer. Second, it gives you the probability of observing a case, given some hidden values.\n\n\n<h3>How to inference?</h3>\n\nRBM has two phases:\n<ul>\n <li>Forward Pass</li> \n <li>Backward Pass or Reconstruction</li>\n</ul>\n\n<b>Phase 1) Forward pass:</b> Input one training sample (one image) <b>X</b> through all visible nodes, and pass it to all hidden nodes. Processing happens in each node in the hidden layer. This computation begins by making stochastic decisions about whether to transmit that input or not (i.e. to determine the state of each hidden layer). At the hidden layer's nodes, <b>X</b> is multiplied by a <b>$W_{ij}$</b> and added to <b>h_bias</b>. The result of those two operations is fed into the sigmoid function, which produces the node’s output, $p({h_j})$, where j is the unit number. \n\n\n$p({h_j})= \\sigma(\\sum_i w_{ij} x_i)$, where $\\sigma()$ is the logistic function.\n\n\nNow lets see what $p({h_j})$ represents. In fact, it is the probabilities of the hidden units. And, all values together are called <b>probability distribution</b>. That is, RBM uses inputs x to make predictions about hidden node activations. For example, imagine that the values of $h_p$ for the first training item is [0.51 0.84]. It tells you what is the conditional probability for each hidden neuron to be at Phase 1): \n<ul>\n <li>p($h_{1}$ = 1|V) = 0.51</li>\n <li>($h_{2}$ = 1|V) = 0.84</li> \n</ul>\n\nAs a result, for each row in the training set, <b>a vector/tensor</b> is generated, which in our case it is of size [1x2], and totally n vectors ($p({h})$=[nx2]). \n\nWe then turn unit $h_j$ on with probability $p(h_{j}|V)$, and turn it off with probability $1 - p(h_{j}|V)$.\n\nTherefore, the conditional probability of a configuration of h given v (for a training sample) is:\n\n$$p(\\mathbf{h} \\mid \\mathbf{v}) = \\prod_{j=0}^H p(h_j \\mid \\mathbf{v})$$",
"_____no_output_____"
],
[
"Now, sample a hidden activation vector <b>h</b> from this probability distribution $p({h_j})$. That is, we sample the activation vector from the probability distribution of hidden layer values. ",
"_____no_output_____"
],
[
"Before we go further, let's look at a toy example for one case out of all input. Assume that we have a trained RBM, and a very simple input vector such as [1.0, 0.0, 0.0, 1.0, 0.0, 0.0, 0.0], lets see what would be the output of forward pass:",
"_____no_output_____"
]
],
[
[
"sess = tf.Session()\nX = tf.constant([[1.0, 0.0, 0.0, 1.0, 0.0, 0.0, 0.0]])\nv_state = X\nprint (\"Input: \", sess.run(v_state))\n\nh_bias = tf.constant([0.1, 0.1])\nprint (\"hb: \", sess.run(h_bias))\nprint (\"w: \", sess.run(W))\n\n# Calculate the probabilities of turning the hidden units on:\nh_prob = tf.nn.sigmoid(tf.matmul(v_state, W) + h_bias) #probabilities of the hidden units\nprint (\"p(h|v): \", sess.run(h_prob))\n\n# Draw samples from the distribution:\nh_state = tf.nn.relu(tf.sign(h_prob - tf.random_uniform(tf.shape(h_prob)))) #states\nprint (\"h0 states:\", sess.run(h_state))",
"_____no_output_____"
]
],
[
[
"<b>Phase 2) Backward Pass (Reconstruction):</b>\nThe RBM reconstructs data by making several forward and backward passes between the visible and hidden layers.\n\nSo, in the second phase (i.e. reconstruction phase), the samples from the hidden layer (i.e. h) play the role of input. That is, <b>h</b> becomes the input in the backward pass. The same weight matrix and visible layer biases are used to go through the sigmoid function. The produced output is a reconstruction which is an approximation of the original input.",
"_____no_output_____"
]
],
[
[
"vb = tf.constant([0.1, 0.2, 0.1, 0.1, 0.1, 0.2, 0.1])\nprint (\"b: \", sess.run(vb))\nv_prob = sess.run(tf.nn.sigmoid(tf.matmul(h_state, tf.transpose(W)) + vb))\nprint (\"p(vi∣h): \", v_prob)\nv_state = tf.nn.relu(tf.sign(v_prob - tf.random_uniform(tf.shape(v_prob))))\nprint (\"v probability states: \", sess.run(v_state))",
"_____no_output_____"
]
],
[
[
"RBM learns a probability distribution over the input, and then, after being trained, the RBM can generate new samples from the learned probability distribution. As you know, <b>probability distribution</b>, is a mathematical function that provides the probabilities of occurrence of different possible outcomes in an experiment.\n\nThe (conditional) probability distribution over the visible units v is given by\n\n$p(\\mathbf{v} \\mid \\mathbf{h}) = \\prod_{i=0}^V p(v_i \\mid \\mathbf{h}),$\n\n\nwhere,\n\n$p(v_i \\mid \\mathbf{h}) = \\sigma\\left( a_i + \\sum_{j=0}^H w_{ji} h_j \\right)$\n\nso, given current state of hidden units and weights, what is the probability of generating [1. 0. 0. 1. 0. 0. 0.] in reconstruction phase, based on the above <b>probability distribution</b> function?",
"_____no_output_____"
]
],
[
[
"inp = sess.run(X)\nprint(inp)\nprint(v_prob[0])\nv_probability = 1\nfor elm, p in zip(inp[0],v_prob[0]) :\n if elm ==1:\n v_probability *= p\n else:\n v_probability *= (1-p)\nv_probability",
"_____no_output_____"
]
],
[
[
"How similar X and V vectors are? Of course, the reconstructed values most likely will not look anything like the input vector because our network has not trained yet. Our objective is to train the model in such a way that the input vector and reconstructed vector to be same. Therefore, based on how different the input values look to the ones that we just reconstructed, the weights are adjusted. ",
"_____no_output_____"
],
[
"<hr>",
"_____no_output_____"
],
[
"\n<h2>MNIST</h2>\n",
"_____no_output_____"
],
[
"We will be using the MNIST dataset to practice the usage of RBMs. The following cell loads the MNIST dataset.",
"_____no_output_____"
]
],
[
[
"mnist = input_data.read_data_sets(\"MNIST_data/\", one_hot=True)\ntrX, trY, teX, teY = mnist.train.images, mnist.train.labels, mnist.test.images, mnist.test.labels",
"_____no_output_____"
]
],
[
[
"Lets look at the dimension of the images.",
"_____no_output_____"
]
],
[
[
"trX[1].shape",
"_____no_output_____"
]
],
[
[
"MNIST images have 784 pixels, so the visible layer must have 784 input nodes. For our case, we'll use 50 nodes in the hidden layer, so i = 50.",
"_____no_output_____"
]
],
[
[
"vb = tf.placeholder(\"float\", [784])\nhb = tf.placeholder(\"float\", [50])",
"_____no_output_____"
]
],
[
[
"Let <b>W</b> be the Tensor of 784x50 (784 - number of visible neurons, 50 - number of hidden neurons) that represents weights between the neurons. ",
"_____no_output_____"
]
],
[
[
"W = tf.placeholder(\"float\", [784, 50])",
"_____no_output_____"
]
],
[
[
"Lets define the visible layer:",
"_____no_output_____"
]
],
[
[
"v0_state = tf.placeholder(\"float\", [None, 784])",
"_____no_output_____"
]
],
[
[
"Now, we can define hidden layer:",
"_____no_output_____"
]
],
[
[
"h0_prob = tf.nn.sigmoid(tf.matmul(v0_state, W) + hb) #probabilities of the hidden units\nh0_state = tf.nn.relu(tf.sign(h0_prob - tf.random_uniform(tf.shape(h0_prob)))) #sample_h_given_X",
"_____no_output_____"
]
],
[
[
"Now, we define reconstruction part:",
"_____no_output_____"
]
],
[
[
"v1_prob = tf.nn.sigmoid(tf.matmul(h0_state, tf.transpose(W)) + vb) \nv1_state = tf.nn.relu(tf.sign(v1_prob - tf.random_uniform(tf.shape(v1_prob)))) #sample_v_given_h",
"_____no_output_____"
]
],
[
[
"<h3>What is objective function?</h3>\n\n<b>Goal</b>: Maximize the likelihood of our data being drawn from that distribution\n\n<b>Calculate error:</b> \nIn each epoch, we compute the \"error\" as a sum of the squared difference between step 1 and step n,\ne.g the error shows the difference between the data and its reconstruction.\n\n<b>Note:</b> tf.reduce_mean computes the mean of elements across dimensions of a tensor.",
"_____no_output_____"
]
],
[
[
"err = tf.reduce_mean(tf.square(v0_state - v1_state))",
"_____no_output_____"
]
],
[
[
"<a id=\"ref4\"></a>\n<h3>How to train the model?</h3>\n<b>Warning!!</b> The following part discuss how to train the model which needs some algebra background. Still, you can skip this part and run the next cells.\n\nAs mentioned, we want to give a high probability to the input data we train on. So, in order to train an RBM, we have to maximize the product of probabilities assigned to all rows v (images) in the training set V (a matrix, where each row of it is treated as a visible vector v):\n\n<img src=\"https://wikimedia.org/api/rest_v1/media/math/render/svg/d42e9f5aad5e1a62b11b119c9315236383c1864a\">\n\n\nWhich is equivalent, maximizing the expected log probability of V:\n\n\n<img src=\"https://wikimedia.org/api/rest_v1/media/math/render/svg/ba0ceed99dca5ff1d21e5ace23f5f2223f19efc0\">\n\n\nSo, we have to update the weights wij to increase p(v) for all v in our training data during training. So we have to calculate the derivative:\n\n\n$$\\frac{\\partial \\log p(\\mathbf v)}{\\partial w_{ij}}$$\n\nThis cannot be easily done by typical <b>gradient descent (SGD)</b>, so we can use another approach, which has 2 steps:\n<ol>\n <li>Gibbs Sampling</li>\n <li>Contrastive Divergence</li>\n</ol> \n \n<h3>Gibbs Sampling</h3> \nFirst, given an input vector v we are using p(h|v) for prediction of the hidden values h. \n<ul>\n <li>$p(h|v) = sigmoid(X \\otimes W + hb)$</li>\n <li>h0 = sampleProb(h0)</li>\n</ul>\n \nThen, knowing the hidden values, we use p(v|h) for reconstructing of new input values v. \n<ul>\n <li>$p(v|h) = sigmoid(h0 \\otimes transpose(W) + vb)$</li>\n <li>$v1 = sampleProb(v1)$ (Sample v given h)</li>\n</ul>\n \nThis process is repeated k times. After k iterations we obtain an other input vector vk which was recreated from original input values v0 or X.\n\nReconstruction steps:\n<ul>\n <li> Get one data point from data set, like <i>x</i>, and pass it through the net</li>\n <li>Pass 0: (x) $\\Rightarrow$ (h0) $\\Rightarrow$ (v1) (v1 is reconstruction of the first pass)</li>\n <li>Pass 1: (v1) $\\Rightarrow$ (h1) $\\Rightarrow$ (v2) (v2 is reconstruction of the second pass)</li>\n <li>Pass 2: (v2) $\\Rightarrow$ (h2) $\\Rightarrow$ (v3) (v3 is reconstruction of the third pass)</li>\n <li>Pass n: (vk) $\\Rightarrow$ (hk+1) $\\Rightarrow$ (vk+1)(vk is reconstruction of the nth pass)</li>\n</ul>\n \n<h4>What is sampling here (sampleProb)?</h4>\n\nIn forward pass: We randomly set the values of each hi to be 1 with probability $sigmoid(v \\otimes W + hb)$. \n- To sample h given v means to sample from the conditional probability distribution P(h|v). It means that you are asking what are the probabilities of getting a specific set of values for the hidden neurons, given the values v for the visible neurons, and sampling from this probability distribution. \nIn reconstruction: We randomly set the values of each vi to be 1 with probability $ sigmoid(h \\otimes transpose(W) + vb)$.\n\n<h3>contrastive divergence (CD-k)</h3>\nThe update of the weight matrix is done during the Contrastive Divergence step. \n\nVectors v0 and vk are used to calculate the activation probabilities for hidden values h0 and hk. The difference between the outer products of those probabilities with input vectors v0 and vk results in the update matrix:\n\n\n$\\Delta W =v0 \\otimes h0 - vk \\otimes hk$ \n\nContrastive Divergence is actually matrix of values that is computed and used to adjust values of the W matrix. Changing W incrementally leads to training of W values. Then on each step (epoch), W is updated to a new value W' through the equation below:\n\n$W' = W + alpha * \\Delta W$ \n\n \n<b>What is Alpha?</b> \nHere, alpha is some small step rate and is also known as the \"learning rate\".\n\n\n",
"_____no_output_____"
],
[
"Ok, lets assume that k=1, that is we just get one more step:",
"_____no_output_____"
]
],
[
[
"h1_prob = tf.nn.sigmoid(tf.matmul(v1_state, W) + hb)\nh1_state = tf.nn.relu(tf.sign(h1_prob - tf.random_uniform(tf.shape(h1_prob)))) #sample_h_given_X",
"_____no_output_____"
],
[
"alpha = 0.01\nW_Delta = tf.matmul(tf.transpose(v0_state), h0_prob) - tf.matmul(tf.transpose(v1_state), h1_prob)\nupdate_w = W + alpha * W_Delta\nupdate_vb = vb + alpha * tf.reduce_mean(v0_state - v1_state, 0)\nupdate_hb = hb + alpha * tf.reduce_mean(h0_state - h1_state, 0)",
"_____no_output_____"
]
],
[
[
"Let's start a session and initialize the variables:",
"_____no_output_____"
]
],
[
[
"cur_w = np.zeros([784, 50], np.float32)\ncur_vb = np.zeros([784], np.float32)\ncur_hb = np.zeros([50], np.float32)\nprv_w = np.zeros([784, 50], np.float32)\nprv_vb = np.zeros([784], np.float32)\nprv_hb = np.zeros([50], np.float32)\nsess = tf.Session()\ninit = tf.global_variables_initializer()\nsess.run(init)",
"_____no_output_____"
]
],
[
[
"Lets look at the error of the first run:",
"_____no_output_____"
]
],
[
[
"sess.run(err, feed_dict={v0_state: trX, W: prv_w, vb: prv_vb, hb: prv_hb})",
"_____no_output_____"
],
[
"#Parameters\nepochs = 5\nbatchsize = 100\nweights = []\nerrors = []\n\nfor epoch in range(epochs):\n for start, end in zip( range(0, len(trX), batchsize), range(batchsize, len(trX), batchsize)):\n batch = trX[start:end]\n cur_w = sess.run(update_w, feed_dict={ v0_state: batch, W: prv_w, vb: prv_vb, hb: prv_hb})\n cur_vb = sess.run(update_vb, feed_dict={v0_state: batch, W: prv_w, vb: prv_vb, hb: prv_hb})\n cur_hb = sess.run(update_hb, feed_dict={ v0_state: batch, W: prv_w, vb: prv_vb, hb: prv_hb})\n prv_w = cur_w\n prv_vb = cur_vb\n prv_hb = cur_hb\n if start % 10000 == 0:\n errors.append(sess.run(err, feed_dict={v0_state: trX, W: cur_w, vb: cur_vb, hb: cur_hb}))\n weights.append(cur_w)\n print ('Epoch: %d' % epoch,'reconstruction error: %f' % errors[-1])\nplt.plot(errors)\nplt.xlabel(\"Batch Number\")\nplt.ylabel(\"Error\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"What is the final weight after training?",
"_____no_output_____"
]
],
[
[
"uw = weights[-1].T\nprint (uw) # a weight matrix of shape (50,784)",
"_____no_output_____"
]
],
[
[
"<a id=\"ref5\"></a>\n<h3>Learned features</h3> ",
"_____no_output_____"
],
[
"We can take each hidden unit and visualize the connections between that hidden unit and each element in the input vector. In our case, we have 50 hidden units. Lets visualize those.",
"_____no_output_____"
],
[
"Let's plot the current weights:\n<b>tile_raster_images</b> helps in generating an easy to grasp image from a set of samples or weights. It transform the <b>uw</b> (with one flattened image per row of size 784), into an array (of size $25\\times20$) in which images are reshaped and laid out like tiles on a floor.",
"_____no_output_____"
]
],
[
[
"tile_raster_images(X=cur_w.T, img_shape=(28, 28), tile_shape=(5, 10), tile_spacing=(1, 1))\nimport matplotlib.pyplot as plt\nfrom PIL import Image\n%matplotlib inline\nimage = Image.fromarray(tile_raster_images(X=cur_w.T, img_shape=(28, 28) ,tile_shape=(5, 10), tile_spacing=(1, 1)))\n### Plot image\nplt.rcParams['figure.figsize'] = (18.0, 18.0)\nimgplot = plt.imshow(image)\nimgplot.set_cmap('gray') ",
"_____no_output_____"
]
],
[
[
"Each tile in the above visualization corresponds to a vector of connections between a hidden unit and visible layer's units. ",
"_____no_output_____"
],
[
"Let's look at one of the learned weights corresponding to one of hidden units for example. In this particular square, the gray color represents weight = 0, and the whiter it is, the more positive the weights are (closer to 1). Conversely, the darker pixels are, the more negative the weights. The positive pixels will increase the probability of activation in hidden units (after multiplying by input/visible pixels), and negative pixels will decrease the probability of a unit hidden to be 1 (activated). So, why is this important? So we can see that this specific square (hidden unit) can detect a feature (e.g. a \"/\" shape) and if it exists in the input.",
"_____no_output_____"
]
],
[
[
"from PIL import Image\nimage = Image.fromarray(tile_raster_images(X =cur_w.T[10:11], img_shape=(28, 28),tile_shape=(1, 1), tile_spacing=(1, 1)))\n### Plot image\nplt.rcParams['figure.figsize'] = (4.0, 4.0)\nimgplot = plt.imshow(image)\nimgplot.set_cmap('gray') ",
"_____no_output_____"
]
],
[
[
"Let's look at the reconstruction of an image now. Imagine that we have a destructed image of figure 3. Lets see if our trained network can fix it:\n\nFirst we plot the image:",
"_____no_output_____"
]
],
[
[
"!wget -O destructed3.jpg https://ibm.box.com/shared/static/vvm1b63uvuxq88vbw9znpwu5ol380mco.jpg\nimg = Image.open('destructed3.jpg')\nimg",
"_____no_output_____"
]
],
[
[
"Now let's pass this image through the net:",
"_____no_output_____"
]
],
[
[
"# convert the image to a 1d numpy array\nsample_case = np.array(img.convert('I').resize((28,28))).ravel().reshape((1, -1))/255.0",
"_____no_output_____"
]
],
[
[
"Feed the sample case into the network and reconstruct the output:",
"_____no_output_____"
]
],
[
[
"hh0_p = tf.nn.sigmoid(tf.matmul(v0_state, W) + hb)\n#hh0_s = tf.nn.relu(tf.sign(hh0_p - tf.random_uniform(tf.shape(hh0_p)))) \nhh0_s = tf.round(hh0_p)\nhh0_p_val,hh0_s_val = sess.run((hh0_p, hh0_s), feed_dict={ v0_state: sample_case, W: prv_w, hb: prv_hb})\nprint(\"Probability nodes in hidden layer:\" ,hh0_p_val)\nprint(\"activated nodes in hidden layer:\" ,hh0_s_val)\n\n# reconstruct\nvv1_p = tf.nn.sigmoid(tf.matmul(hh0_s_val, tf.transpose(W)) + vb)\nrec_prob = sess.run(vv1_p, feed_dict={ hh0_s: hh0_s_val, W: prv_w, vb: prv_vb})",
"_____no_output_____"
]
],
[
[
"Here we plot the reconstructed image:",
"_____no_output_____"
]
],
[
[
"img = Image.fromarray(tile_raster_images(X=rec_prob, img_shape=(28, 28),tile_shape=(1, 1), tile_spacing=(1, 1)))\nplt.rcParams['figure.figsize'] = (4.0, 4.0)\nimgplot = plt.imshow(img)\nimgplot.set_cmap('gray') ",
"_____no_output_____"
]
],
[
[
"<hr>\n\n## Want to learn more?\n\nRunning deep learning programs usually needs a high performance platform. __PowerAI__ speeds up deep learning and AI. Built on IBM’s Power Systems, __PowerAI__ is a scalable software platform that accelerates deep learning and AI with blazing performance for individual users or enterprises. The __PowerAI__ platform supports popular machine learning libraries and dependencies including TensorFlow, Caffe, Torch, and Theano. You can use [PowerAI on IMB Cloud](https://cocl.us/ML0120EN_PAI).\n\nAlso, you can use __Watson Studio__ to run these notebooks faster with bigger datasets.__Watson Studio__ is IBM’s leading cloud solution for data scientists, built by data scientists. With Jupyter notebooks, RStudio, Apache Spark and popular libraries pre-packaged in the cloud, __Watson Studio__ enables data scientists to collaborate on their projects without having to install anything. Join the fast-growing community of __Watson Studio__ users today with a free account at [Watson Studio](https://cocl.us/ML0120EN_DSX).This is the end of this lesson. Thank you for reading this notebook, and good luck on your studies.",
"_____no_output_____"
],
[
"### Thanks for completing this lesson!\n\nNotebook created by: <a href = \"https://ca.linkedin.com/in/saeedaghabozorgi\">Saeed Aghabozorgi</a>",
"_____no_output_____"
],
[
"### References:\nhttps://en.wikipedia.org/wiki/Restricted_Boltzmann_machine \nhttp://deeplearning.net/tutorial/rbm.html \nhttp://www.cs.utoronto.ca/~hinton/absps/netflixICML.pdf<br>\nhttp://imonad.com/rbm/restricted-boltzmann-machine/ \n",
"_____no_output_____"
],
[
"<hr>\n\nCopyright © 2018 [Cognitive Class](https://cocl.us/DX0108EN_CC). This notebook and its source code are released under the terms of the [MIT License](https://bigdatauniversity.com/mit-license/).",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d0880acda4e8f300372a2bde2ddcf06557cd3559 | 73,711 | ipynb | Jupyter Notebook | module4-classification-metrics/Lesley_Rich_224_assignment.ipynb | terrainthesky-hub/DS-Unit-2-Kaggle-Challenge | e3cf2e5afe7e859dd8dbbc2808e0e0f4bef7ea23 | [
"MIT"
] | null | null | null | module4-classification-metrics/Lesley_Rich_224_assignment.ipynb | terrainthesky-hub/DS-Unit-2-Kaggle-Challenge | e3cf2e5afe7e859dd8dbbc2808e0e0f4bef7ea23 | [
"MIT"
] | null | null | null | module4-classification-metrics/Lesley_Rich_224_assignment.ipynb | terrainthesky-hub/DS-Unit-2-Kaggle-Challenge | e3cf2e5afe7e859dd8dbbc2808e0e0f4bef7ea23 | [
"MIT"
] | null | null | null | 97.115942 | 20,666 | 0.773982 | [
[
[
"<a href=\"https://colab.research.google.com/github/terrainthesky-hub/DS-Unit-2-Kaggle-Challenge/blob/master/module4-classification-metrics/Lesley_Rich_224_assignment.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"\n\n#Confusion matrix is at the bottom!! **************\n\nimport pandas as pd\nimport os\n\nfrom sklearn.model_selection import train_test_split\n\ntrain = pd.merge(pd.read_csv(r'C:\\Users\\Lesley\\Downloads\\train_features.csv'), pd.read_csv(r'C:\\Users\\Lesley\\Downloads\\train_labels.csv'))\n\ntest = pd.read_csv(r'C:\\Users\\Lesley\\Downloads\\test_features.csv')\n\ntrain, val = train_test_split(train, train_size=0.80, test_size=0.20,\n stratify=train['status_group'], random_state=42)\n\ntrain.shape, val.shape, test.shape\n",
"_____no_output_____"
],
[
"import numpy as np\n\ndef wrangle(X):\n \"\"\"Wrangle train, validate, and test sets in the same way\"\"\"\n \n # Prevent SettingWithCopyWarning\n X = X.copy()\n \n # About 3% of the time, latitude has small values near zero,\n # outside Tanzania, so we'll treat these values like zero.\n X['latitude'] = X['latitude'].replace(-2e-08, 0)\n \n # When columns have zeros and shouldn't, they are like null values.\n # So we will replace the zeros with nulls, and impute missing values later.\n # Also create a \"missing indicator\" column, because the fact that\n # values are missing may be a predictive signal.\n cols_with_zeros = ['longitude', 'latitude', 'construction_year', \n 'gps_height', 'population']\n for col in cols_with_zeros:\n X[col] = X[col].replace(0, np.nan)\n X[col+'_MISSING'] = X[col].isnull()\n \n # Drop duplicate columns\n duplicates = ['quantity_group', 'payment_type']\n X = X.drop(columns=duplicates)\n \n # Drop recorded_by (never varies) and id (always varies, random)\n unusable_variance = ['recorded_by']\n X = X.drop(columns=unusable_variance)\n \n # Convert date_recorded to datetime\n X['date_recorded'] = pd.to_datetime(X['date_recorded'], infer_datetime_format=True)\n \n # Extract components from date_recorded, then drop the original column\n X['year_recorded'] = X['date_recorded'].dt.year\n X['month_recorded'] = X['date_recorded'].dt.month\n X['day_recorded'] = X['date_recorded'].dt.day\n X = X.drop(columns='date_recorded')\n \n # Engineer feature: how many years from construction_year to date_recorded\n X['years'] = X['year_recorded'] - X['construction_year']\n X['years_MISSING'] = X['years'].isnull()\n \n # return the wrangled dataframe\n return X\n\ntrain = wrangle(train)\nval = wrangle(val)\ntest = wrangle(test)",
"_____no_output_____"
],
[
"target = 'status_group'\n\ntrain_features = train.drop(columns=[target, 'id'])\n\nnumeric_features = train_features.select_dtypes(include='number').columns.tolist()\n\ncardinality = train_features.select_dtypes(exclude='number').nunique()\n\ncategorical_features = cardinality[cardinality <= 50].index.tolist()\n\nfeatures = numeric_features + categorical_features",
"_____no_output_____"
],
[
"\nX_train = train[features]\ny_train = train[target]\nX_val = val[features]\ny_val = val[target]\nX_test = test[features]",
"_____no_output_____"
],
[
"import category_encoders as ce\nfrom sklearn.impute import SimpleImputer\nfrom sklearn.model_selection import cross_val_score\nfrom sklearn.pipeline import make_pipeline\nfrom sklearn.feature_selection import f_regression, SelectKBest\nfrom sklearn.ensemble import RandomForestClassifier\n",
"_____no_output_____"
],
[
"pipeline = make_pipeline(\n ce.OrdinalEncoder(),\n SimpleImputer(),\n RandomForestClassifier()\n)\n\nk = 3\nscore = cross_val_score(pipeline, X_train, y_train, cv=k,\n scoring='accuracy')\nprint(f'Accuracy for {k} folds', score)",
"Accuracy for 3 folds [0.79261364 0.79955808 0.79236111]\n"
],
[
"from scipy.stats import randint, uniform\nfrom sklearn.model_selection import GridSearchCV, RandomizedSearchCV\n\npipeline = make_pipeline(\n ce.OrdinalEncoder(),\n SimpleImputer(),\n RandomForestClassifier()\n)\n\nparam_distributions = {\n 'simpleimputer__strategy': ['mean', 'median'],\n 'randomforestclassifier__n_estimators': [23 ,24, 25, 26, 27, 28, 29, 30],\n 'randomforestclassifier__max_depth': [5, 10, 15, 20, 25, None],\n 'randomforestclassifier__max_features': uniform(0, 1),\n 'randomforestclassifier__min_samples_leaf': [1, 2, 3, 4, 5, 6, 7, 8, 9, 10],\n 'randomforestclassifier__min_samples_split': [5,6, 7, 8, 9, 10, 11, 12, 13, 14, 15]\n}\n\nsearch = RandomizedSearchCV(\n pipeline,\n param_distributions=param_distributions,\n n_iter=10,\n cv=3,\n scoring='accuracy',\n verbose=10,\n return_train_score=True,\n n_jobs=-1)\n\nsearch.fit(X_train, y_train);",
"Fitting 3 folds for each of 10 candidates, totalling 30 fits\n"
],
[
"pipeline.named_steps['randomforestclassifier']",
"_____no_output_____"
],
[
"pipeline = search.best_estimator_",
"_____no_output_____"
],
[
"pipeline",
"_____no_output_____"
],
[
"print('Best hyperparameters', search.best_params_)",
"Best hyperparameters {'randomforestclassifier__max_depth': 20, 'randomforestclassifier__max_features': 0.4105325914269071, 'randomforestclassifier__min_samples_leaf': 2, 'randomforestclassifier__min_samples_split': 11, 'randomforestclassifier__n_estimators': 29, 'simpleimputer__strategy': 'mean'}\n"
],
[
"\nsklearn.__version__",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"!pip install --user --upgrade scikit-learn",
"Collecting scikit-learn\n Downloading https://files.pythonhosted.org/packages/1f/e3/e400f94e368a7b0d2432a88ab671a7f27c9159f177bbed68f7cce83b5848/scikit_learn-0.22.2.post1-cp37-cp37m-win_amd64.whl (6.5MB)\nRequirement already satisfied, skipping upgrade: numpy>=1.11.0 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from scikit-learn) (1.16.5)\nRequirement already satisfied, skipping upgrade: joblib>=0.11 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from scikit-learn) (0.14.1)\nRequirement already satisfied, skipping upgrade: scipy>=0.17.0 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from scikit-learn) (1.4.1)\nInstalling collected packages: scikit-learn\nSuccessfully installed scikit-learn-0.22.2.post1\n"
],
[
"import sklearn\n\nsklearn.__version__",
"_____no_output_____"
],
[
"y_pred = pipeline.predict(X_test)",
"_____no_output_____"
],
[
"path=r'C:\\Users\\Lesley\\Desktop\\Lambda\\Lesley_Rich'\nsubmission = test[['id']].copy()\nsubmission['status_group'] = y_pred\n# submission['status_group']\nsubmission.to_csv(path+'DecisionTreeWaterPumpSub3.csv', index=False)",
"_____no_output_____"
],
[
"from sklearn.metrics import plot_confusion_matrix\nimport matplotlib.pyplot as plt\n\nplot_confusion_matrix(pipeline, X_val, y_val, values_format='.0f', xticks_rotation='vertical')\nplt.show()",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"Requirement already up-to-date: tensorflow in c:\\users\\lesley\\anaconda3\\lib\\site-packages (2.1.0)\nRequirement already satisfied, skipping upgrade: protobuf>=3.8.0 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from tensorflow) (3.11.3)\nRequirement already satisfied, skipping upgrade: wrapt>=1.11.1 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from tensorflow) (1.11.2)\nRequirement already satisfied, skipping upgrade: six>=1.12.0 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from tensorflow) (1.12.0)\nRequirement already satisfied, skipping upgrade: absl-py>=0.7.0 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from tensorflow) (0.9.0)\nRequirement already satisfied, skipping upgrade: keras-applications>=1.0.8 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from tensorflow) (1.0.8)\nRequirement already satisfied, skipping upgrade: numpy<2.0,>=1.16.0 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from tensorflow) (1.16.5)\nRequirement already satisfied, skipping upgrade: opt-einsum>=2.3.2 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from tensorflow) (3.2.0)\nRequirement already satisfied, skipping upgrade: grpcio>=1.8.6 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from tensorflow) (1.27.2)\nRequirement already satisfied, skipping upgrade: wheel>=0.26; python_version >= \"3\" in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from tensorflow) (0.33.6)\nRequirement already satisfied, skipping upgrade: tensorflow-estimator<2.2.0,>=2.1.0rc0 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from tensorflow) (2.1.0)\nRequirement already satisfied, skipping upgrade: termcolor>=1.1.0 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from tensorflow) (1.1.0)\nRequirement already satisfied, skipping upgrade: keras-preprocessing>=1.1.0 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from tensorflow) (1.1.0)\nRequirement already satisfied, skipping upgrade: scipy==1.4.1; python_version >= \"3\" in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from tensorflow) (1.4.1)\nRequirement already satisfied, skipping upgrade: gast==0.2.2 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from tensorflow) (0.2.2)\nRequirement already satisfied, skipping upgrade: google-pasta>=0.1.6 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from tensorflow) (0.2.0)\nRequirement already satisfied, skipping upgrade: tensorboard<2.2.0,>=2.1.0 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from tensorflow) (2.1.1)\nRequirement already satisfied, skipping upgrade: astor>=0.6.0 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from tensorflow) (0.8.1)\nRequirement already satisfied, skipping upgrade: setuptools in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from protobuf>=3.8.0->tensorflow) (41.4.0)\nRequirement already satisfied, skipping upgrade: h5py in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from keras-applications>=1.0.8->tensorflow) (2.9.0)\nRequirement already satisfied, skipping upgrade: markdown>=2.6.8 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from tensorboard<2.2.0,>=2.1.0->tensorflow) (3.2.1)\nRequirement already satisfied, skipping upgrade: google-auth-oauthlib<0.5,>=0.4.1 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from tensorboard<2.2.0,>=2.1.0->tensorflow) (0.4.1)\nRequirement already satisfied, skipping upgrade: google-auth<2,>=1.6.3 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from tensorboard<2.2.0,>=2.1.0->tensorflow) (1.11.3)\nRequirement already satisfied, skipping upgrade: werkzeug>=0.11.15 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from tensorboard<2.2.0,>=2.1.0->tensorflow) (0.16.0)\nRequirement already satisfied, skipping upgrade: requests<3,>=2.21.0 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from tensorboard<2.2.0,>=2.1.0->tensorflow) (2.22.0)\nRequirement already satisfied, skipping upgrade: requests-oauthlib>=0.7.0 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from google-auth-oauthlib<0.5,>=0.4.1->tensorboard<2.2.0,>=2.1.0->tensorflow) (1.3.0)\nRequirement already satisfied, skipping upgrade: cachetools<5.0,>=2.0.0 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from google-auth<2,>=1.6.3->tensorboard<2.2.0,>=2.1.0->tensorflow) (4.0.0)\nRequirement already satisfied, skipping upgrade: rsa<4.1,>=3.1.4 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from google-auth<2,>=1.6.3->tensorboard<2.2.0,>=2.1.0->tensorflow) (4.0)\nRequirement already satisfied, skipping upgrade: pyasn1-modules>=0.2.1 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from google-auth<2,>=1.6.3->tensorboard<2.2.0,>=2.1.0->tensorflow) (0.2.8)\nRequirement already satisfied, skipping upgrade: chardet<3.1.0,>=3.0.2 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from requests<3,>=2.21.0->tensorboard<2.2.0,>=2.1.0->tensorflow) (3.0.4)\nRequirement already satisfied, skipping upgrade: idna<2.9,>=2.5 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from requests<3,>=2.21.0->tensorboard<2.2.0,>=2.1.0->tensorflow) (2.8)\nRequirement already satisfied, skipping upgrade: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from requests<3,>=2.21.0->tensorboard<2.2.0,>=2.1.0->tensorflow) (1.24.2)\nRequirement already satisfied, skipping upgrade: certifi>=2017.4.17 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from requests<3,>=2.21.0->tensorboard<2.2.0,>=2.1.0->tensorflow) (2019.9.11)\nRequirement already satisfied, skipping upgrade: oauthlib>=3.0.0 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from requests-oauthlib>=0.7.0->google-auth-oauthlib<0.5,>=0.4.1->tensorboard<2.2.0,>=2.1.0->tensorflow) (3.1.0)\nRequirement already satisfied, skipping upgrade: pyasn1>=0.1.3 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from rsa<4.1,>=3.1.4->google-auth<2,>=1.6.3->tensorboard<2.2.0,>=2.1.0->tensorflow) (0.4.8)\n"
],
[
"",
"Collecting Keras\n Downloading https://files.pythonhosted.org/packages/ad/fd/6bfe87920d7f4fd475acd28500a42482b6b84479832bdc0fe9e589a60ceb/Keras-2.3.1-py2.py3-none-any.whl (377kB)\nRequirement already satisfied: numpy>=1.9.1 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from Keras) (1.16.5)\nRequirement already satisfied: six>=1.9.0 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from Keras) (1.12.0)\nRequirement already satisfied: pyyaml in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from Keras) (5.1.2)\nRequirement already satisfied: keras-applications>=1.0.6 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from Keras) (1.0.8)\nRequirement already satisfied: scipy>=0.14 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from Keras) (1.4.1)\nRequirement already satisfied: h5py in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from Keras) (2.9.0)\nRequirement already satisfied: keras-preprocessing>=1.0.5 in c:\\users\\lesley\\anaconda3\\lib\\site-packages (from Keras) (1.1.0)\nInstalling collected packages: Keras\nSuccessfully installed Keras-2.3.1\n"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0882d23e3cff795d343968aef96b402e085e34a | 10,902 | ipynb | Jupyter Notebook | Python Basic Programming/Programming Assignment_23.ipynb | Sayan97/Python | 13b28a9187bc8ad95948f89081bea8603197c791 | [
"MIT"
] | null | null | null | Python Basic Programming/Programming Assignment_23.ipynb | Sayan97/Python | 13b28a9187bc8ad95948f89081bea8603197c791 | [
"MIT"
] | null | null | null | Python Basic Programming/Programming Assignment_23.ipynb | Sayan97/Python | 13b28a9187bc8ad95948f89081bea8603197c791 | [
"MIT"
] | null | null | null | 18.699828 | 111 | 0.444322 | [
[
[
"### Question 1\n#### Create a function that takes a number as an argument and returns True or False depending\n#### on whether the number is symmetrical or not. A number is symmetrical when it is the same as\n#### its reverse.\n#### Examples\n#### is_symmetrical(7227) ➞ True\n#### is_symmetrical(12567) ➞ False\n#### is_symmetrical(44444444) ➞ True\n#### is_symmetrical(9939) ➞ False\n#### is_symmetrical(1112111) ➞ True",
"_____no_output_____"
]
],
[
[
"def is_symmetrical(n):\n rev = str(n)[::-1]\n #print(rev)\n if rev == str(n):\n return True\n return False\n ",
"_____no_output_____"
],
[
"is_symmetrical(7227)",
"_____no_output_____"
],
[
"is_symmetrical(12567)",
"_____no_output_____"
],
[
"is_symmetrical(44444444)",
"_____no_output_____"
],
[
"is_symmetrical(9939)",
"_____no_output_____"
],
[
"is_symmetrical(1112111)",
"_____no_output_____"
]
],
[
[
"### Question 2\n#### Given a string of numbers separated by a comma and space, return the product of the\n#### numbers.\n#### Examples\n#### multiply_nums("2, 3") ➞ 6\n#### multiply_nums("1, 2, 3, 4") ➞ 24\n#### multiply_nums("54, 75, 453, 0") ➞ 0\n#### multiply_nums("10, -2") ➞ -20",
"_____no_output_____"
]
],
[
[
"def multiply_nums(s):\n s = s.replace(' ', \"\")\n s = s.split(',')\n sum = 1\n for i in s:\n sum = sum * int(i)\n return sum",
"_____no_output_____"
],
[
"multiply_nums(\"2, 3\")",
"_____no_output_____"
],
[
"multiply_nums(\"1, 2, 3, 4\")",
"_____no_output_____"
],
[
"multiply_nums(\"54, 75, 453, 0\")",
"_____no_output_____"
],
[
"multiply_nums(\"10, -2\")",
"_____no_output_____"
]
],
[
[
"### Question 3\n#### Create a function that squares every digit of a number.\n#### Examples\n#### square_digits(9119) ➞ 811181\n#### square_digits(2483) ➞ 416649\n#### square_digits(3212) ➞ 9414\n#### Notes\n#### The function receives an integer and must return an integer.",
"_____no_output_____"
]
],
[
[
"def square_digits(n):\n sq = ''.join(str(int(i)**2) for i in str(n))\n return int(sq)\n \n ",
"_____no_output_____"
],
[
"square_digits(9119)",
"_____no_output_____"
],
[
"square_digits(2483)",
"_____no_output_____"
],
[
"square_digits(3212)",
"_____no_output_____"
]
],
[
[
"### Question 4\n#### Create a function that sorts a list and removes all duplicate items from it.\n#### Examples\n#### setify([1, 3, 3, 5, 5]) ➞ [1, 3, 5]\n#### setify([4, 4, 4, 4]) ➞ [4]\n#### setify([5, 7, 8, 9, 10, 15]) ➞ [5, 7, 8, 9, 10, 15]\n#### setify([3, 3, 3, 2, 1]) ➞ [1, 2, 3]",
"_____no_output_____"
]
],
[
[
"def setify(l):\n m = []\n l.sort()\n l = set(l)\n for i in l:\n m.append(i)\n return m\n ",
"_____no_output_____"
],
[
"setify([1, 3, 3, 5, 5])",
"_____no_output_____"
],
[
"setify([4, 4, 4, 4])",
"_____no_output_____"
],
[
"setify([5, 7, 8, 9, 10, 15])",
"_____no_output_____"
],
[
"setify([3, 3, 3, 2, 1])",
"_____no_output_____"
]
],
[
[
"### Question 5\n#### Create a function that returns the mean of all digits.\n#### Examples\n#### mean(42) ➞ 3\n#### mean(12345) ➞ 3\n#### mean(666) ➞ 6\n#### Notes\n#### The mean of all digits is the sum of digits / how many digits there are (e.g. mean of digits in\n#### 512 is (5+1+2)/3(number of digits) = 8/3=2).\n#### The mean will always be an integer.",
"_____no_output_____"
]
],
[
[
"def mean(n):\n sum = 0\n lenth = len(str(n))\n for i in str(n):\n sum = sum+ int(i)\n return int(sum/lenth)\n \n ",
"_____no_output_____"
],
[
"mean(42)",
"_____no_output_____"
],
[
"mean(12345)",
"_____no_output_____"
],
[
"mean(666)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d0883cefa79336375414a72bd6eb5bad65ab9d30 | 508,491 | ipynb | Jupyter Notebook | Breast_Cancer_Wisconsin(Progonostic).ipynb | DJLee68/Bioinformatics_AI | 4e9ed7fa29e598f4b5be433d53ae15742847f595 | [
"MIT"
] | 1 | 2019-11-29T07:20:20.000Z | 2019-11-29T07:20:20.000Z | Breast_Cancer_Wisconsin(Progonostic).ipynb | leedongjae0611/Bioinformatics_DL | 4e9ed7fa29e598f4b5be433d53ae15742847f595 | [
"MIT"
] | null | null | null | Breast_Cancer_Wisconsin(Progonostic).ipynb | leedongjae0611/Bioinformatics_DL | 4e9ed7fa29e598f4b5be433d53ae15742847f595 | [
"MIT"
] | 1 | 2021-10-04T14:17:24.000Z | 2021-10-04T14:17:24.000Z | 358.59732 | 449,960 | 0.90779 | [
[
[
"import numpy as np\nimport pandas as pd\nfrom sklearn.preprocessing import MinMaxScaler\nfrom sklearn.preprocessing import StandardScaler",
"_____no_output_____"
],
[
"df_train = pd.read_excel('wpbc.train.xlsx')\ndf_test = pd.read_excel('wpbc.test.xlsx')",
"_____no_output_____"
],
[
"train = df_train\ntest = df_test",
"_____no_output_____"
],
[
"train.shape",
"_____no_output_____"
],
[
"test.shape",
"_____no_output_____"
],
[
"train.describe()",
"_____no_output_____"
],
[
"import seaborn\nimport matplotlib.pyplot as plt\n\ndef plot_df(df, name):\n corr = df[df.columns].corr()\n mask = np.zeros_like(corr, dtype=np.bool)\n mask[np.triu_indices_from(mask)] = True\n plt.figure(figsize=(20, 15))\n seaborn.set(font_scale=1.2)\n seaborn.heatmap(corr, mask=mask, center=0, annot=True,\n square=True, linewidths=3, alpha=0.7)\n plt.title(name)",
"_____no_output_____"
],
[
"plot_df(train, 'Train')",
"_____no_output_____"
],
[
"print(train.columns)",
"Index(['class', 'f1', 'f2', 'f3', 'f4', 'f5', 'f6', 'f7', 'f8', 'f9', 'f10',\n 'f11', 'f12', 'f13', 'f14', 'f15', 'f16', 'f17', 'f18', 'f19', 'f20',\n 'f21', 'f22', 'f23', 'f24', 'f25', 'f26', 'f27', 'f28', 'f29', 'f30',\n 'f31', 'f32', 'f33'],\n dtype='object')\n"
],
[
"class_name = input(\"Chooese the class: \")",
"Chooese the class: class\n"
],
[
"minmax_scaler = MinMaxScaler()\nstandard_scaler = StandardScaler()",
"_____no_output_____"
],
[
"temp_tr_ans = train[class_name]\ntemp_ts_ans = test[class_name]\nclass_count = len(temp_tr_ans.unique())\nprint(class_count)",
"2\n"
],
[
"tr_data = train.drop([class_name], axis=1)\nts_data = test.drop([class_name], axis=1)",
"_____no_output_____"
],
[
"# #결측치 채우기 if 결측치가 0일 경우\n\n# from sklearn.impute import SimpleImputer\n\n# rep_0 = SimpleImputer(missing_values=0, strategy=\"mean\")\n\n# tr_data = rep_0.fit_transform(tr_data)\n# ts_data = rep_0.fit_transform(ts_data)\n\n#결측치 채우기 if 결측치가 ?일 경우 - 먼저 ?를 특정한수(ex.333)으로 변경\n\nfrom sklearn.impute import SimpleImputer\n\nrep_0 = SimpleImputer(missing_values=333, strategy=\"mean\")\n\ntr_data = rep_0.fit_transform(tr_data)\nts_data = rep_0.fit_transform(ts_data)",
"_____no_output_____"
],
[
"mm_tr_data = minmax_scaler.fit_transform(tr_data)\nmm_ts_data = minmax_scaler.transform(ts_data)\nstd_tr_data = standard_scaler.fit_transform(tr_data)\nstd_ts_data = standard_scaler.transform(ts_data)\ntr_ans, _ = pd.factorize(temp_tr_ans, sort=True)\nts_ans, _ = pd.factorize(temp_ts_ans, sort=True)",
"_____no_output_____"
],
[
"tr_ans",
"_____no_output_____"
],
[
"import tensorflow as tf\nfrom sklearn.model_selection import GridSearchCV\nfrom sklearn.model_selection import RandomizedSearchCV\nfrom sklearn.model_selection import ParameterGrid\nfrom sklearn.metrics import precision_score, recall_score, f1_score, accuracy_score\nfrom tensorflow.keras.wrappers.scikit_learn import KerasClassifier\nfrom tensorflow.keras.models import Sequential\nfrom tensorflow.keras.layers import Dense\nfrom tensorflow.keras.layers import Activation\nfrom tensorflow.keras.layers import BatchNormalization\nfrom tensorflow.keras.layers import Dropout\nfrom sklearn import metrics\nfrom tensorflow.keras.regularizers import l2\n",
"_____no_output_____"
],
[
"from sklearn.metrics import precision_recall_fscore_support\nfrom sklearn.metrics import confusion_matrix",
"_____no_output_____"
],
[
"# real Version\n\n\ndef create_model(hidden_layers = 1, neurons =1, init_mode = 'uniform', \n activation = 'elu', kernel_regularizer=l2(0.001)):\n model = Sequential()\n model.add(Dense(neurons, input_dim=len(mm_tr_data.T), kernel_initializer=init_mode, activation=activation))\n for i in range(hidden_layers):\n \n model.add(Dense(neurons, kernel_initializer=init_mode, kernel_regularizer=kernel_regularizer))\n model.add(BatchNormalization())\n model.add(Activation(activation))\n model.add(Dropout(0.2))\n \n if class_count == 2: \n model.add(Dense(1,activation='sigmoid'))\n model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])\n elif class_count != 2:\n model.add(Dense(class_count, activation='softmax'))\n model.compile(loss='sparse_categorical_crossentropy', optimizer='adam', metrics=['accuracy'])\n return model",
"_____no_output_____"
],
[
"keras_model = KerasClassifier(build_fn=create_model, epochs=64, batch_size=16)",
"_____no_output_____"
],
[
"leaky_relu = tf.nn.leaky_relu\nhidden_layers = [4,8,12]\nneurons = [32, 64, 128]\nactivation = ['elu', leaky_relu]\ninit_mode = ['glorot_uniform', 'he_normal']",
"_____no_output_____"
],
[
"param_grid = dict(hidden_layers = hidden_layers, neurons = neurons, init_mode = init_mode, activation = activation)\nminmax_grid = GridSearchCV(estimator=keras_model, param_grid=param_grid, n_jobs= -1, cv=3)\nstd_grid = GridSearchCV(estimator=keras_model, param_grid=param_grid, n_jobs= -1, cv=3)",
"_____no_output_____"
],
[
"import warnings\nwarnings.filterwarnings(\"ignore\")",
"_____no_output_____"
],
[
"minmax_grid_result = minmax_grid.fit(mm_tr_data, tr_ans)",
"WARNING: Logging before flag parsing goes to stderr.\nW0709 21:04:36.602367 7620 deprecation.py:506] From C:\\Users\\User\\Anaconda3\\lib\\site-packages\\tensorflow\\python\\ops\\init_ops.py:1251: calling VarianceScaling.__init__ (from tensorflow.python.ops.init_ops) with dtype is deprecated and will be removed in a future version.\nInstructions for updating:\nCall initializer instance with the dtype argument instead of passing it to the constructor\nW0709 21:04:37.682612 7620 deprecation.py:323] From C:\\Users\\User\\Anaconda3\\lib\\site-packages\\tensorflow\\python\\ops\\nn_impl.py:180: add_dispatch_support.<locals>.wrapper (from tensorflow.python.ops.array_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse tf.where in 2.0, which has the same broadcast rule as np.where\n"
],
[
"std_grid_result = std_grid.fit(std_tr_data, tr_ans)",
"Epoch 1/64\n119/119 [==============================] - 1s 4ms/sample - loss: 1.1203 - acc: 0.5294\nEpoch 2/64\n119/119 [==============================] - 0s 210us/sample - loss: 1.0099 - acc: 0.5210\nEpoch 3/64\n119/119 [==============================] - 0s 218us/sample - loss: 0.9820 - acc: 0.5882\nEpoch 4/64\n119/119 [==============================] - 0s 210us/sample - loss: 0.8849 - acc: 0.6639\nEpoch 5/64\n119/119 [==============================] - 0s 185us/sample - loss: 0.9679 - acc: 0.6050\nEpoch 6/64\n119/119 [==============================] - 0s 168us/sample - loss: 0.8611 - acc: 0.7227\nEpoch 7/64\n119/119 [==============================] - 0s 160us/sample - loss: 0.8248 - acc: 0.6891\nEpoch 8/64\n119/119 [==============================] - 0s 160us/sample - loss: 0.8453 - acc: 0.6471\nEpoch 9/64\n119/119 [==============================] - 0s 168us/sample - loss: 0.8563 - acc: 0.6975\nEpoch 10/64\n119/119 [==============================] - 0s 160us/sample - loss: 0.7789 - acc: 0.7899\nEpoch 11/64\n119/119 [==============================] - 0s 150us/sample - loss: 0.7985 - acc: 0.7731\nEpoch 12/64\n119/119 [==============================] - 0s 151us/sample - loss: 0.8003 - acc: 0.7311\nEpoch 13/64\n119/119 [==============================] - 0s 160us/sample - loss: 0.7646 - acc: 0.7479\nEpoch 14/64\n119/119 [==============================] - 0s 151us/sample - loss: 0.7984 - acc: 0.7479\nEpoch 15/64\n119/119 [==============================] - 0s 151us/sample - loss: 0.7570 - acc: 0.7731\nEpoch 16/64\n119/119 [==============================] - 0s 143us/sample - loss: 0.7384 - acc: 0.7899\nEpoch 17/64\n119/119 [==============================] - 0s 143us/sample - loss: 0.7701 - acc: 0.7983\nEpoch 18/64\n119/119 [==============================] - 0s 134us/sample - loss: 0.7619 - acc: 0.7731\nEpoch 19/64\n119/119 [==============================] - 0s 151us/sample - loss: 0.7112 - acc: 0.7731\nEpoch 20/64\n119/119 [==============================] - 0s 143us/sample - loss: 0.7655 - acc: 0.7563\nEpoch 21/64\n119/119 [==============================] - 0s 151us/sample - loss: 0.7517 - acc: 0.8235\nEpoch 22/64\n119/119 [==============================] - 0s 134us/sample - loss: 0.6950 - acc: 0.7815\nEpoch 23/64\n119/119 [==============================] - 0s 143us/sample - loss: 0.7366 - acc: 0.7479\nEpoch 24/64\n119/119 [==============================] - 0s 134us/sample - loss: 0.7286 - acc: 0.7899\nEpoch 25/64\n119/119 [==============================] - 0s 143us/sample - loss: 0.7633 - acc: 0.7815\nEpoch 26/64\n119/119 [==============================] - 0s 151us/sample - loss: 0.6882 - acc: 0.8151\nEpoch 27/64\n119/119 [==============================] - 0s 143us/sample - loss: 0.7259 - acc: 0.7395\nEpoch 28/64\n119/119 [==============================] - 0s 143us/sample - loss: 0.7039 - acc: 0.7563\nEpoch 29/64\n119/119 [==============================] - ETA: 0s - loss: 0.5918 - acc: 0.812 - 0s 143us/sample - loss: 0.7030 - acc: 0.7647\nEpoch 30/64\n119/119 [==============================] - 0s 143us/sample - loss: 0.6753 - acc: 0.7815\nEpoch 31/64\n119/119 [==============================] - 0s 143us/sample - loss: 0.6808 - acc: 0.7899\nEpoch 32/64\n119/119 [==============================] - 0s 143us/sample - loss: 0.6645 - acc: 0.8319\nEpoch 33/64\n119/119 [==============================] - 0s 151us/sample - loss: 0.6539 - acc: 0.8067\nEpoch 34/64\n119/119 [==============================] - 0s 143us/sample - loss: 0.6981 - acc: 0.8067\nEpoch 35/64\n119/119 [==============================] - 0s 143us/sample - loss: 0.6853 - acc: 0.7899\nEpoch 36/64\n119/119 [==============================] - 0s 143us/sample - loss: 0.6669 - acc: 0.8235\nEpoch 37/64\n119/119 [==============================] - 0s 151us/sample - loss: 0.7513 - acc: 0.7899\nEpoch 38/64\n119/119 [==============================] - 0s 151us/sample - loss: 0.7016 - acc: 0.8235\nEpoch 39/64\n119/119 [==============================] - 0s 143us/sample - loss: 0.6555 - acc: 0.8067\nEpoch 40/64\n119/119 [==============================] - 0s 143us/sample - loss: 0.6496 - acc: 0.8319\nEpoch 41/64\n119/119 [==============================] - 0s 134us/sample - loss: 0.6182 - acc: 0.8403\nEpoch 42/64\n119/119 [==============================] - 0s 143us/sample - loss: 0.6536 - acc: 0.8487\nEpoch 43/64\n119/119 [==============================] - 0s 143us/sample - loss: 0.6480 - acc: 0.8151\nEpoch 44/64\n119/119 [==============================] - 0s 151us/sample - loss: 0.6096 - acc: 0.8487\nEpoch 45/64\n119/119 [==============================] - 0s 134us/sample - loss: 0.6137 - acc: 0.7983\nEpoch 46/64\n119/119 [==============================] - 0s 134us/sample - loss: 0.6003 - acc: 0.8571\nEpoch 47/64\n119/119 [==============================] - 0s 143us/sample - loss: 0.6299 - acc: 0.8235\nEpoch 48/64\n119/119 [==============================] - 0s 134us/sample - loss: 0.6316 - acc: 0.8235\nEpoch 49/64\n119/119 [==============================] - 0s 134us/sample - loss: 0.6445 - acc: 0.8067\nEpoch 50/64\n119/119 [==============================] - 0s 134us/sample - loss: 0.5773 - acc: 0.8571\nEpoch 51/64\n119/119 [==============================] - 0s 143us/sample - loss: 0.7007 - acc: 0.7647\nEpoch 52/64\n119/119 [==============================] - 0s 143us/sample - loss: 0.6185 - acc: 0.8235\nEpoch 53/64\n119/119 [==============================] - 0s 143us/sample - loss: 0.5499 - acc: 0.8908\nEpoch 54/64\n119/119 [==============================] - 0s 143us/sample - loss: 0.5785 - acc: 0.8403\nEpoch 55/64\n119/119 [==============================] - 0s 143us/sample - loss: 0.5841 - acc: 0.8235\nEpoch 56/64\n119/119 [==============================] - 0s 151us/sample - loss: 0.5546 - acc: 0.8824\nEpoch 57/64\n119/119 [==============================] - 0s 149us/sample - loss: 0.6050 - acc: 0.8487\nEpoch 58/64\n119/119 [==============================] - 0s 143us/sample - loss: 0.6338 - acc: 0.8067\nEpoch 59/64\n119/119 [==============================] - 0s 151us/sample - loss: 0.5868 - acc: 0.8235\nEpoch 60/64\n119/119 [==============================] - 0s 134us/sample - loss: 0.5432 - acc: 0.8655\nEpoch 61/64\n119/119 [==============================] - 0s 143us/sample - loss: 0.5517 - acc: 0.8487\nEpoch 62/64\n119/119 [==============================] - 0s 136us/sample - loss: 0.5655 - acc: 0.8571\nEpoch 63/64\n119/119 [==============================] - 0s 143us/sample - loss: 0.6095 - acc: 0.8403\nEpoch 64/64\n119/119 [==============================] - 0s 137us/sample - loss: 0.5886 - acc: 0.8319\n"
],
[
"print(\"Scaler = minmax\")\nprint(\"Best: %f using %s\" % (minmax_grid_result.best_score_, minmax_grid_result.best_params_))\nmeans = minmax_grid_result.cv_results_['mean_test_score']\nstds = minmax_grid_result.cv_results_['std_test_score']\nparams = minmax_grid_result.cv_results_['params']\n\nfor mean, stdev, param in zip(means, stds, params):\n print(\"%f (%f) with: %r\" % (mean, stdev, param))",
"Scaler = minmax\nBest: 0.823529 using {'activation': 'elu', 'hidden_layers': 12, 'init_mode': 'glorot_uniform', 'neurons': 32}\n0.739496 (0.030880) with: {'activation': 'elu', 'hidden_layers': 4, 'init_mode': 'glorot_uniform', 'neurons': 32}\n0.731092 (0.027080) with: {'activation': 'elu', 'hidden_layers': 4, 'init_mode': 'glorot_uniform', 'neurons': 64}\n0.647059 (0.183241) with: {'activation': 'elu', 'hidden_layers': 4, 'init_mode': 'glorot_uniform', 'neurons': 128}\n0.739496 (0.043669) with: {'activation': 'elu', 'hidden_layers': 4, 'init_mode': 'he_normal', 'neurons': 32}\n0.714286 (0.051309) with: {'activation': 'elu', 'hidden_layers': 4, 'init_mode': 'he_normal', 'neurons': 64}\n0.663866 (0.117253) with: {'activation': 'elu', 'hidden_layers': 4, 'init_mode': 'he_normal', 'neurons': 128}\n0.680672 (0.121162) with: {'activation': 'elu', 'hidden_layers': 8, 'init_mode': 'glorot_uniform', 'neurons': 32}\n0.714286 (0.086957) with: {'activation': 'elu', 'hidden_layers': 8, 'init_mode': 'glorot_uniform', 'neurons': 64}\n0.655462 (0.135400) with: {'activation': 'elu', 'hidden_layers': 8, 'init_mode': 'glorot_uniform', 'neurons': 128}\n0.756303 (0.074817) with: {'activation': 'elu', 'hidden_layers': 8, 'init_mode': 'he_normal', 'neurons': 32}\n0.722689 (0.041129) with: {'activation': 'elu', 'hidden_layers': 8, 'init_mode': 'he_normal', 'neurons': 64}\n0.680672 (0.153345) with: {'activation': 'elu', 'hidden_layers': 8, 'init_mode': 'he_normal', 'neurons': 128}\n0.823529 (0.053624) with: {'activation': 'elu', 'hidden_layers': 12, 'init_mode': 'glorot_uniform', 'neurons': 32}\n0.663866 (0.199239) with: {'activation': 'elu', 'hidden_layers': 12, 'init_mode': 'glorot_uniform', 'neurons': 64}\n0.638655 (0.144869) with: {'activation': 'elu', 'hidden_layers': 12, 'init_mode': 'glorot_uniform', 'neurons': 128}\n0.815126 (0.060439) with: {'activation': 'elu', 'hidden_layers': 12, 'init_mode': 'he_normal', 'neurons': 32}\n0.739496 (0.096480) with: {'activation': 'elu', 'hidden_layers': 12, 'init_mode': 'he_normal', 'neurons': 64}\n0.672269 (0.024102) with: {'activation': 'elu', 'hidden_layers': 12, 'init_mode': 'he_normal', 'neurons': 128}\n0.747899 (0.003009) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 4, 'init_mode': 'glorot_uniform', 'neurons': 32}\n0.764706 (0.010725) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 4, 'init_mode': 'glorot_uniform', 'neurons': 64}\n0.714286 (0.034792) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 4, 'init_mode': 'glorot_uniform', 'neurons': 128}\n0.798319 (0.072511) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 4, 'init_mode': 'he_normal', 'neurons': 32}\n0.655462 (0.161641) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 4, 'init_mode': 'he_normal', 'neurons': 64}\n0.697479 (0.039416) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 4, 'init_mode': 'he_normal', 'neurons': 128}\n0.731092 (0.114790) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 8, 'init_mode': 'glorot_uniform', 'neurons': 32}\n0.722689 (0.056663) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 8, 'init_mode': 'glorot_uniform', 'neurons': 64}\n0.756303 (0.013558) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 8, 'init_mode': 'glorot_uniform', 'neurons': 128}\n0.747899 (0.057636) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 8, 'init_mode': 'he_normal', 'neurons': 32}\n0.680672 (0.103698) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 8, 'init_mode': 'he_normal', 'neurons': 64}\n0.714286 (0.043774) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 8, 'init_mode': 'he_normal', 'neurons': 128}\n0.764706 (0.025250) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 12, 'init_mode': 'glorot_uniform', 'neurons': 32}\n0.714286 (0.075590) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 12, 'init_mode': 'glorot_uniform', 'neurons': 64}\n0.680672 (0.101387) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 12, 'init_mode': 'glorot_uniform', 'neurons': 128}\n0.764706 (0.025250) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 12, 'init_mode': 'he_normal', 'neurons': 32}\n0.773109 (0.020676) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 12, 'init_mode': 'he_normal', 'neurons': 64}\n0.697479 (0.037526) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 12, 'init_mode': 'he_normal', 'neurons': 128}\n"
],
[
"print(\"Scaler = standard\")\nprint(\"Best: %f using %s\" % (std_grid_result.best_score_, std_grid_result.best_params_))\nmeans = std_grid_result.cv_results_['mean_test_score']\nstds = std_grid_result.cv_results_['std_test_score']\nparams = std_grid_result.cv_results_['params']\n\nfor mean, stdev, param in zip(means, stds, params):\n print(\"%f (%f) with: %r\" % (mean, stdev, param))",
"Scaler = standard\nBest: 0.798319 using {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 4, 'init_mode': 'he_normal', 'neurons': 32}\n0.731092 (0.068187) with: {'activation': 'elu', 'hidden_layers': 4, 'init_mode': 'glorot_uniform', 'neurons': 32}\n0.697479 (0.075222) with: {'activation': 'elu', 'hidden_layers': 4, 'init_mode': 'glorot_uniform', 'neurons': 64}\n0.714286 (0.061086) with: {'activation': 'elu', 'hidden_layers': 4, 'init_mode': 'glorot_uniform', 'neurons': 128}\n0.739496 (0.025426) with: {'activation': 'elu', 'hidden_layers': 4, 'init_mode': 'he_normal', 'neurons': 32}\n0.739496 (0.069513) with: {'activation': 'elu', 'hidden_layers': 4, 'init_mode': 'he_normal', 'neurons': 64}\n0.680672 (0.034445) with: {'activation': 'elu', 'hidden_layers': 4, 'init_mode': 'he_normal', 'neurons': 128}\n0.739496 (0.065318) with: {'activation': 'elu', 'hidden_layers': 8, 'init_mode': 'glorot_uniform', 'neurons': 32}\n0.680672 (0.049467) with: {'activation': 'elu', 'hidden_layers': 8, 'init_mode': 'glorot_uniform', 'neurons': 64}\n0.697479 (0.044428) with: {'activation': 'elu', 'hidden_layers': 8, 'init_mode': 'glorot_uniform', 'neurons': 128}\n0.697479 (0.058228) with: {'activation': 'elu', 'hidden_layers': 8, 'init_mode': 'he_normal', 'neurons': 32}\n0.714286 (0.099577) with: {'activation': 'elu', 'hidden_layers': 8, 'init_mode': 'he_normal', 'neurons': 64}\n0.680672 (0.082033) with: {'activation': 'elu', 'hidden_layers': 8, 'init_mode': 'he_normal', 'neurons': 128}\n0.655462 (0.158290) with: {'activation': 'elu', 'hidden_layers': 12, 'init_mode': 'glorot_uniform', 'neurons': 32}\n0.655462 (0.087286) with: {'activation': 'elu', 'hidden_layers': 12, 'init_mode': 'glorot_uniform', 'neurons': 64}\n0.655462 (0.082332) with: {'activation': 'elu', 'hidden_layers': 12, 'init_mode': 'glorot_uniform', 'neurons': 128}\n0.731092 (0.052060) with: {'activation': 'elu', 'hidden_layers': 12, 'init_mode': 'he_normal', 'neurons': 32}\n0.680672 (0.101387) with: {'activation': 'elu', 'hidden_layers': 12, 'init_mode': 'he_normal', 'neurons': 64}\n0.680672 (0.082033) with: {'activation': 'elu', 'hidden_layers': 12, 'init_mode': 'he_normal', 'neurons': 128}\n0.747899 (0.018080) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 4, 'init_mode': 'glorot_uniform', 'neurons': 32}\n0.647059 (0.037874) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 4, 'init_mode': 'glorot_uniform', 'neurons': 64}\n0.722689 (0.057932) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 4, 'init_mode': 'glorot_uniform', 'neurons': 128}\n0.798319 (0.036849) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 4, 'init_mode': 'he_normal', 'neurons': 32}\n0.697479 (0.075222) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 4, 'init_mode': 'he_normal', 'neurons': 64}\n0.722689 (0.023559) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 4, 'init_mode': 'he_normal', 'neurons': 128}\n0.781513 (0.076279) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 8, 'init_mode': 'glorot_uniform', 'neurons': 32}\n0.663866 (0.076140) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 8, 'init_mode': 'glorot_uniform', 'neurons': 64}\n0.705882 (0.027381) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 8, 'init_mode': 'glorot_uniform', 'neurons': 128}\n0.781513 (0.033485) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 8, 'init_mode': 'he_normal', 'neurons': 32}\n0.739496 (0.054826) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 8, 'init_mode': 'he_normal', 'neurons': 64}\n0.714286 (0.015345) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 8, 'init_mode': 'he_normal', 'neurons': 128}\n0.764706 (0.025250) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 12, 'init_mode': 'glorot_uniform', 'neurons': 32}\n0.773109 (0.034297) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 12, 'init_mode': 'glorot_uniform', 'neurons': 64}\n0.697479 (0.039416) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 12, 'init_mode': 'glorot_uniform', 'neurons': 128}\n0.764706 (0.025250) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 12, 'init_mode': 'he_normal', 'neurons': 32}\n0.747899 (0.041106) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 12, 'init_mode': 'he_normal', 'neurons': 64}\n0.697479 (0.058228) with: {'activation': <function leaky_relu at 0x000001F54D2D2598>, 'hidden_layers': 12, 'init_mode': 'he_normal', 'neurons': 128}\n"
],
[
"pred = minmax_grid.predict(mm_ts_data)\n\naccuracy = accuracy_score(pred, ts_ans)\nts_ans = ts_ans.astype(float)\nprecision, recall, fbeta_score, support = precision_recall_fscore_support(ts_ans, pred)\nconf_mat = confusion_matrix(ts_ans, pred)\nprint(\"Accuracy = \", accuracy)\nprint(\"Confusion Matrix\")\nprint(\"{0}\".format(metrics.confusion_matrix(ts_ans, pred)))\nprint(\"\")\nprint(\"Classification Report\")\nprint(metrics.classification_report(ts_ans, pred))",
"Accuracy = 0.7848101265822784\nConfusion Matrix\n[[55 5]\n [12 7]]\n\nClassification Report\n precision recall f1-score support\n\n 0.0 0.82 0.92 0.87 60\n 1.0 0.58 0.37 0.45 19\n\n micro avg 0.78 0.78 0.78 79\n macro avg 0.70 0.64 0.66 79\nweighted avg 0.76 0.78 0.77 79\n\n"
],
[
"pred = std_grid.predict(std_ts_data)\n\naccuracy = accuracy_score(pred, ts_ans)\nts_ans = ts_ans.astype(float)\nprecision, recall, fbeta_score, support = precision_recall_fscore_support(ts_ans, pred)\nconf_mat = confusion_matrix(ts_ans, pred)\nprint(\"Accuracy = \", accuracy)\nprint(\"Confusion Matrix\")\nprint(\"{0}\".format(metrics.confusion_matrix(ts_ans, pred)))\nprint(\"\")\nprint(\"Classification Report\")\nprint(metrics.classification_report(ts_ans, pred))",
"Accuracy = 0.7468354430379747\nConfusion Matrix\n[[54 6]\n [14 5]]\n\nClassification Report\n precision recall f1-score support\n\n 0.0 0.79 0.90 0.84 60\n 1.0 0.45 0.26 0.33 19\n\n micro avg 0.75 0.75 0.75 79\n macro avg 0.62 0.58 0.59 79\nweighted avg 0.71 0.75 0.72 79\n\n"
],
[
"# # testbed Version\n\n\n# def create_model(hidden_layers = 1, neurons =1, init_mode = 'uniform', activation = 'elu'):\n# model = Sequential()\n# model.add(Dense(neurons, input_dim=len(tr_data.T), kernel_initializer=init_mode, activation=activation))\n\n# for i in range(hidden_layers):\n \n# model.add(Dense(neurons, kernel_initializer=init_mode))\n# model.add(BatchNormalization())\n# model.add(Activation(activation))\n# model.add(Dropout(0.2))\n \n# if class_count == 2: \n# model.add(Dense(1,activation='sigmoid'))\n# model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])\n# elif class_count != 2:\n# model.add(Dense(class_count-1, activation='softmax'))\n# model.compile(loss='sparse_categorical_crossentropy', optimizer='adam', metrics=['accuracy'])\n\n# return model\n\n\n# hidden_layers = [5, 10]\n# neurons = [32, 64]\n# activation = ['elu']\n# init_mode = ['he_uniform']\n\n# keras_model = KerasClassifier(build_fn=create_model, epochs=4, batch_size=4)\n\n# param_grid = dict(hidden_layers = hidden_layers, neurons = neurons, init_mode = init_mode, activation = activation)\n# grid = GridSearchCV(estimator=keras_model, param_grid=param_grid, n_jobs= -1, cv=2)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0885205fdc339e70510024973d48fab9f6a6944 | 635,460 | ipynb | Jupyter Notebook | Differential_Imaging.ipynb | Max-FM/IAA-Social-Distancing | 4490cd0c163b8e045c75aea96087882655785542 | [
"MIT"
] | null | null | null | Differential_Imaging.ipynb | Max-FM/IAA-Social-Distancing | 4490cd0c163b8e045c75aea96087882655785542 | [
"MIT"
] | null | null | null | Differential_Imaging.ipynb | Max-FM/IAA-Social-Distancing | 4490cd0c163b8e045c75aea96087882655785542 | [
"MIT"
] | null | null | null | 2,104.172185 | 473,621 | 0.960725 | [
[
[
"<a href=\"https://colab.research.google.com/github/Max-FM/IAA-Social-Distancing/blob/master/Differential_Imaging.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"#Differential Imaging\n\n**Warning:** This notebook will likely cause Google Colab to crash. It is advised to run the notebook locally, either by downloading and running through Jupyter or by connecting to a local runtime.\n\n**Disclaimer:** Satellite images are not publicly available in the GitHub repository in order to avoid potential legal issues. The images used are available internally to other researchers at the University of Portsmouth [here](https://drive.google.com/drive/folders/1GGK6HksIM7jISqC71g0KpzSJnPjFkWO2?usp=sharing). Access is restricted to external persons and all external access requests will be denied. Should the user wish to acquire the images themselves, the corresponding shapefiles are publicly available in the repository.",
"_____no_output_____"
],
[
"###Import Files",
"_____no_output_____"
]
],
[
[
"import rasterio as rio\nimport rioxarray as riox\n\nimport numpy as np\nimport xarray as xr\n\nimport matplotlib.pyplot as plt\n\nfrom glob import glob",
"_____no_output_____"
]
],
[
[
"###Define Filepaths",
"_____no_output_____"
]
],
[
[
"fdir = '/home/foxleym/Downloads'\n\nfilepaths = glob(f'{fdir}/Southsea2020_PSScene4Band_Explorer/files/*_SR_clip.tif')",
"_____no_output_____"
]
],
[
[
"###Create 4-Band Median Raster",
"_____no_output_____"
]
],
[
[
"blueList = []\ngreenList = []\nredList = []\nnirList = []\n\nfor i, file in enumerate(filepaths):\n blueList.append(riox.open_rasterio(file)[0,:,:])\n greenList.append(riox.open_rasterio(file)[1,:,:])\n redList.append(riox.open_rasterio(file)[2,:,:])\n nirList.append(riox.open_rasterio(file)[3,:,:])\n \nblue_median = xr.concat(blueList, \"t\").median(dim=\"t\")\ngreen_median = xr.concat(greenList, \"t\").median(dim=\"t\")\nred_median = xr.concat(redList, \"t\").median(dim=\"t\")\nnir_median = xr.concat(nirList, \"t\").median(dim=\"t\")\n\nmedian_raster = xr.concat([blue_median, green_median, red_median, nir_median], dim='band')\n\ndel(blueList, greenList, redList, nirList, blue_median, green_median, red_median, nir_median)\n\nmedian_raster.rio.to_raster(f'{fdir}/Southsea2020_PSScene4Band_Explorer/Southsea2020Median.tif')",
"_____no_output_____"
]
],
[
[
"###Obtain Median RBG Raster and Plot",
"_____no_output_____"
]
],
[
[
"def normalize(array):\n \"\"\"Normalizes numpy arrays into scale 0.0 - 1.0\"\"\"\n array_min, array_max = array.min(), array.max()\n return ((array - array_min)/(array_max - array_min))\n\ndef make_composite(band_1, band_2, band_3):\n \"\"\"Converts three raster bands into a composite image\"\"\"\n return normalize(np.dstack((band_1, band_2, band_3)))\n\nb, g, r, nir = median_raster\n\nrgb = make_composite(r, g, b)\n\nplt.figure(figsize=(15,15))\nplt.imshow(rgb)\nplt.xticks([])\nplt.yticks([])",
"_____no_output_____"
]
],
[
[
"###Perform Image Subtractions",
"_____no_output_____"
]
],
[
[
"subtractions = []\n\nfor f in filepaths:\n fname = f.split('/')[-1].split('.')[0] \n \n raster = riox.open_rasterio(f)\n \n subtraction = raster - median_raster\n\n subtractions.append(subtraction)\n \n subtraction.rio.to_raster(f'{fdir}/Southsea2020_PSScene4Band_Explorer/files/{fname}_MEDDIFF.tif')",
"_____no_output_____"
]
],
[
[
"###Convert to RBG and Plot",
"_____no_output_____"
]
],
[
[
"b_0, g_0, r_0, nir_0 = raster \nb_med, g_med, r_med, nir_med = median_raster\nb_sub, g_sub, r_sub, nir_sub = subtractions[0]\n\nrgb_0 = make_composite(r_0, g_0, b_0)\nrgb_med = make_composite(r_med, g_med, b_med)\nrgb_sub = make_composite(r_sub, g_sub, b_sub)\n\nrgb_list = [rgb_0, rgb_med, rgb_sub]\n\nfig, ax = plt.subplots(nrows = 3, figsize=(15,15))\n\n\nfor i, rgb in enumerate(rgb_list):\n ax[i].imshow(rgb)\n ax[i].set_xticks([])\n ax[i].set_yticks([])\n \nplt.tight_layout()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d088580e483750d9336eb13567227f277dcdb1ce | 37,169 | ipynb | Jupyter Notebook | Experiment 1/Thurber.ipynb | NishenPather/Final-Year-Project-Automatic-Differentiation-for-Large-Scale-Optimisation- | 58d9bc721c1d0c308206820a257eb2073540b0d2 | [
"MIT"
] | null | null | null | Experiment 1/Thurber.ipynb | NishenPather/Final-Year-Project-Automatic-Differentiation-for-Large-Scale-Optimisation- | 58d9bc721c1d0c308206820a257eb2073540b0d2 | [
"MIT"
] | null | null | null | Experiment 1/Thurber.ipynb | NishenPather/Final-Year-Project-Automatic-Differentiation-for-Large-Scale-Optimisation- | 58d9bc721c1d0c308206820a257eb2073540b0d2 | [
"MIT"
] | null | null | null | 52.498588 | 12,324 | 0.621163 | [
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport torch\nimport pandas as pd\nfrom scipy.misc import derivative\nimport time",
"_____no_output_____"
],
[
"data= pd.read_csv(\"Thurber_Data.txt\",names=['y','x'], sep=\" \")\n\ndata",
"_____no_output_____"
],
[
"y = torch.from_numpy(data['y'].to_numpy(np.float64))\nx = torch.from_numpy(data['x'].to_numpy(np.float64))\n\n# b = torch.tensor([1000,1000,400,40,0.7,0.3,0.03],requires_grad=True)\nb = torch.tensor([1300,1500,500,75,1,0.4,0.05],requires_grad=True)",
"_____no_output_____"
],
[
"plt.plot(x.numpy(),y.numpy())",
"_____no_output_____"
],
[
"## Numerical Differentiation\n\n# b = np.array([1000,1000,400,40,0.7,0.3,0.03]).reshape(-1,1)\nb = np.array([1300,1500,500,75,1,0.4,0.05]).reshape(-1,1)\n\nx=x.detach().numpy()\n\nu=0.1 #beta multiply identity matrix\nbeta = 10\nV_prev = 0\n\n \n\ndef f0(b0):\n return (b0 + b[1]*x + b[2]*np.square(x) + b[3]*np.power(x,3)) / (1 + b[4]*x + b[5]*np.square(x) + b[6]*np.power(x,3))-y.detach().numpy() \ndef f1(b1):\n return(b[0] + b1*x + b[2]*np.square(x) + b[3]*np.power(x,3)) / (1 + b[4]*x + b[5]*np.square(x) + b[6]*np.power(x,3))-y.detach().numpy() \ndef f2(b2):\n return (b[0] + b[1]*x + b2*np.square(x) + b[3]*np.power(x,3)) / (1 + b[4]*x + b[5]*np.square(x) + b[6]*np.power(x,3))-y.detach().numpy() \ndef f3(b3):\n return (b[0] + b[1]*x + b[2]*np.square(x) + b3*np.power(x,3)) / (1 + b[4]*x + b[5]*np.square(x) + b[6]*np.power(x,3))-y.detach().numpy() \ndef f4(b4):\n return (b[0] + b[1]*x + b[2]*np.square(x) + b[3]*np.power(x,3)) / (1 + b4*x + b[5]*np.square(x) + b[6]*np.power(x,3))-y.detach().numpy() \ndef f5(b5):\n return (b[0] + b[1]*x + b[2]*np.square(x) + b[3]*np.power(x,3)) / (1 + b[4]*x + b5*np.square(x) + b[6]*np.power(x,3))-y.detach().numpy() \ndef f6(b6):\n return (b[0] + b[1]*x + b[2]*np.square(x) + b[3]*np.power(x,3)) / (1 + b[4]*x + b[5]*np.square(x) + b6*np.power(x,3))-y.detach().numpy() \n\n\nstart_time = time.time()\nfor c in range(500):\n \n y_pred = (b[0] + b[1]*x + b[2]*np.square(x) + b[3]*np.power(x,3)) / (1 + b[4]*x + b[5]*np.square(x) + b[6]*np.power(x,3)) \n error = (y_pred - y.detach().numpy()).reshape(-1,1)\n \n d_b0 = derivative(f0,b[0] , dx=1e-6)\n d_b1 = derivative(f1,b[1] , dx=1e-6)\n d_b2 = derivative(f2,b[2] , dx=1e-6)\n d_b3 = derivative(f3,b[3] , dx=1e-6)\n d_b4 = derivative(f4,b[4] , dx=1e-6)\n d_b5 = derivative(f5,b[5] , dx=1e-6)\n d_b6 = derivative(f6,b[6] , dx=1e-6)\n \n jacobian = np.transpose(np.array([d_b0,d_b1,d_b2,d_b3,d_b4,d_b5,d_b6]))\n dParam = np.matmul(np.matmul(np.linalg.inv((np.matmul(np.transpose(jacobian),jacobian)+u*np.identity(len(b)))),np.transpose(jacobian)),error)\n b -= dParam\n V = np.sum(np.square(error))\n if(V > V_prev):\n u *= beta\n else:\n u /= beta\n V_prev = V\n print(\"c: \",c,\" error: \",V,\" B:\", b)\n \n if V < 5.6427082397E+03:\n break\nprint(\"time taken to execute: \",time.time()-start_time) ",
"c: 0 error: 85873749.82313623 B: [[1.28876115e+03]\n [1.48774765e+03]\n [5.64667557e+02]\n [7.26321563e+01]\n [9.64730544e-01]\n [3.83839365e-01]\n [4.75818827e-02]]\nc: 1 error: 809864.7781325612 B: [[1.28873351e+03]\n [1.48053008e+03]\n [5.73153864e+02]\n [7.38695466e+01]\n [9.57666228e-01]\n [3.91839558e-01]\n [4.88232240e-02]]\nc: 2 error: 29138.167807215676 B: [[1.28828556e+03]\n [1.48372881e+03]\n [5.77964762e+02]\n [7.44602219e+01]\n [9.60421346e-01]\n [3.95372769e-01]\n [4.88327849e-02]]\nc: 3 error: 5729.369919273743 B: [[1.28815047e+03]\n [1.49273988e+03]\n [5.84340084e+02]\n [7.56311000e+01]\n [9.67216583e-01]\n [3.98388309e-01]\n [5.01698589e-02]]\nc: 4 error: 5645.0781185217365 B: [[1.28815104e+03]\n [1.48962170e+03]\n [5.82218436e+02]\n [7.52173149e+01]\n [9.65299874e-01]\n [3.97521813e-01]\n [4.94404511e-02]]\nc: 5 error: 5643.34191181903 B: [[1.28813380e+03]\n [1.49208993e+03]\n [5.83944636e+02]\n [7.55547174e+01]\n [9.66978592e-01]\n [3.98284784e-01]\n [4.99289617e-02]]\nc: 6 error: 5643.031742030124 B: [[1.28814286e+03]\n [1.49043349e+03]\n [5.82788445e+02]\n [7.53286708e+01]\n [9.65862103e-01]\n [3.97775513e-01]\n [4.95962764e-02]]\nc: 7 error: 5642.842314029481 B: [[1.28813703e+03]\n [1.49153155e+03]\n [5.83554311e+02]\n [7.54783915e+01]\n [9.66600963e-01]\n [3.98112306e-01]\n [4.98175526e-02]]\nc: 8 error: 5642.772567207946 B: [[1.28814122e+03]\n [1.49078281e+03]\n [5.83031651e+02]\n [7.53762292e+01]\n [9.66095745e-01]\n [3.97881997e-01]\n [4.96674611e-02]]\nc: 9 error: 5642.736292344363 B: [[1.28813853e+03]\n [1.49128262e+03]\n [5.83380355e+02]\n [7.54443970e+01]\n [9.66432333e-01]\n [3.98035450e-01]\n [4.97680205e-02]]\nc: 10 error: 5642.721299097057 B: [[1.28814040e+03]\n [1.49094421e+03]\n [5.83144165e+02]\n [7.53982276e+01]\n [9.66204139e-01]\n [3.97931420e-01]\n [4.97000959e-02]]\nc: 11 error: 5642.714047304371 B: [[1.28813917e+03]\n [1.49117097e+03]\n [5.83302383e+02]\n [7.54291573e+01]\n [9.66356888e-01]\n [3.98001057e-01]\n [4.97456969e-02]]\nc: 12 error: 5642.7109027479355 B: [[1.28814001e+03]\n [1.49101795e+03]\n [5.83195595e+02]\n [7.54082823e+01]\n [9.66253741e-01]\n [3.97954036e-01]\n [4.97149624e-02]]\nc: 13 error: 5642.709434876036 B: [[1.28813945e+03]\n [1.49112085e+03]\n [5.83267402e+02]\n [7.54223194e+01]\n [9.66323087e-01]\n [3.97985651e-01]\n [4.97356400e-02]]\nc: 14 error: 5642.708786600741 B: [[1.28813983e+03]\n [1.49105113e+03]\n [5.83218738e+02]\n [7.54128072e+01]\n [9.66276057e-01]\n [3.97964207e-01]\n [4.97216571e-02]]\nc: 15 error: 5642.708489322984 B: [[1.28813958e+03]\n [1.49109814e+03]\n [5.83251547e+02]\n [7.54192204e+01]\n [9.66307754e-01]\n [3.97978659e-01]\n [4.97310936e-02]]\nc: 16 error: 5642.708352971062 B: [[1.28813975e+03]\n [1.49106660e+03]\n [5.83229540e+02]\n [7.54149183e+01]\n [9.66286508e-01]\n [3.97968973e-01]\n [4.97247509e-02]]\nc: 17 error: 5642.708290633586 B: [[1.28813963e+03]\n [1.49108791e+03]\n [5.83244416e+02]\n [7.54178261e+01]\n [9.66300878e-01]\n [3.97975526e-01]\n [4.97290298e-02]]\nc: 18 error: 5642.708263209245 B: [[1.28813971e+03]\n [1.49107371e+03]\n [5.83234511e+02]\n [7.54158896e+01]\n [9.66291324e-01]\n [3.97971170e-01]\n [4.97261677e-02]]\nc: 19 error: 5642.708249757279 B: [[1.28813966e+03]\n [1.49108307e+03]\n [5.83241033e+02]\n [7.54171650e+01]\n [9.66297605e-01]\n [3.97974032e-01]\n [4.97280617e-02]]\nc: 20 error: 5642.7082442536675 B: [[1.28813969e+03]\n [1.49107692e+03]\n [5.83236749e+02]\n [7.54163273e+01]\n [9.66293482e-01]\n [3.97972155e-01]\n [4.97268150e-02]]\nc: 21 error: 5642.708241521348 B: [[1.28813967e+03]\n [1.49108105e+03]\n [5.83239626e+02]\n [7.54168896e+01]\n [9.66296257e-01]\n [3.97973419e-01]\n [4.97276471e-02]]\nc: 22 error: 5642.7082406169775 B: [[1.28813969e+03]\n [1.49107787e+03]\n [5.83237397e+02]\n [7.54164548e+01]\n [9.66294076e-01]\n [3.97972421e-01]\n [4.97270311e-02]]\nc: 23 error: 5642.708240212967 B: [[1.28813968e+03]\n [1.49107994e+03]\n [5.83238841e+02]\n [7.54167371e+01]\n [9.66295469e-01]\n [3.97973057e-01]\n [4.97274477e-02]]\nc: 24 error: 5642.708239852662 B: [[1.28813968e+03]\n [1.49107856e+03]\n [5.83237882e+02]\n [7.54165497e+01]\n [9.66294544e-01]\n [3.97972636e-01]\n [4.97271700e-02]]\nc: 25 error: 5642.708239791194 B: [[1.28813968e+03]\n [1.49107963e+03]\n [5.83238634e+02]\n [7.54166963e+01]\n [9.66295281e-01]\n [3.97972973e-01]\n [4.97273768e-02]]\nc: 26 error: 5642.70823971716 B: [[1.28813968e+03]\n [1.49107927e+03]\n [5.83238388e+02]\n [7.54166476e+01]\n [9.66295068e-01]\n [3.97972879e-01]\n [4.97272840e-02]]\nc: 27 error: 5642.708239672714 B: [[1.28813968e+03]\n [1.49107922e+03]\n [5.83238345e+02]\n [7.54166397e+01]\n [9.66295002e-01]\n [3.97972843e-01]\n [4.97272953e-02]]\ntime taken to execute: 0.07010626792907715\n"
],
[
"def Jacobian(loss,params,numParams):\n \n jacobian = torch.empty(len(loss), numParams) \n \n for i in range(len(loss)):\n \n loss[i].backward(retain_graph=True)\n \n for n in range(numParams):\n jacobian[i][n] = params.grad[n]\n params.grad.zero_() \n return jacobian ",
"_____no_output_____"
],
[
"## Automatic Differentiation\nnum_param = len(b)\nu=0.1 #beta multiply identity matrix\nbeta = 10\nerror_prev = 0\nstart_time = time.time()\nfor c in range(200):\n \n y_pred = (b[0] + b[1]*x + b[2]*torch.square(x) + b[3]*torch.pow(x,3)) / (1 + b[4]*x + b[5]*torch.square(x) + b[6]*torch.pow(x,3))\n loss = y_pred-y\n error = torch.sum(torch.square(loss)) #residual sum of squares\n print(\"\",c,\" error is: \",error.detach().numpy(),\" b is \", b.detach().numpy())\n jacobian = Jacobian(loss,b,len(b))\n\n dParam = torch.matmul(torch.matmul(torch.inverse(torch.matmul(torch.transpose(jacobian,-1,0),jacobian)+u*torch.eye(num_param, num_param)),torch.transpose(jacobian,-1,0)),loss.float())\n with torch.no_grad():\n b -=dParam\n \n if(error > error_prev):\n u *= beta\n else:\n u /= beta\n error_prev = error\n if error< 5.642708245E+03: #3.9050739624 given residual sum of squares\n break\nprint(\"time taken to execute: \",time.time()-start_time) ",
"_____no_output_____"
],
[
"plt.plot(y_pred.detach(),'g.', y,'r')",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d08858342a4c6fb9f4303a0ac81bb9c35a5713a7 | 12,195 | ipynb | Jupyter Notebook | DirectProgramming/DPC++/Jupyter/oneapi-essentials-training/07_DPCPP_Library/oneDPL_gamma_correction.ipynb | praveenkk123/oneAPI-samples | cde7bd306b608f5e42ac1df46f182dfa396ad302 | [
"MIT"
] | 310 | 2020-07-09T01:00:11.000Z | 2022-03-31T17:52:14.000Z | DirectProgramming/DPC++/Jupyter/oneapi-essentials-training/07_DPCPP_Library/oneDPL_gamma_correction.ipynb | praveenkk123/oneAPI-samples | cde7bd306b608f5e42ac1df46f182dfa396ad302 | [
"MIT"
] | 438 | 2020-06-30T23:25:19.000Z | 2022-03-31T00:37:13.000Z | DirectProgramming/DPC++/Jupyter/oneapi-essentials-training/07_DPCPP_Library/oneDPL_gamma_correction.ipynb | aaronkintel/oneAPI-samples | 5634b1077e5327076c749064369fc3d033bb45db | [
"MIT"
] | 375 | 2020-06-04T22:58:24.000Z | 2022-03-30T11:04:18.000Z | 35.762463 | 317 | 0.591472 | [
[
[
"# oneDPL- Gamma Correction example\n\n\n\n",
"_____no_output_____"
],
[
"#### Sections\n- [Gamma Correction](#Gamma-Correction)\n- [Why use buffer iterators?](#Why-use-buffer-iterators?)\n- _Lab Exercise:_ [Gamma Correction](#Lab-Exercise:-Gamma-Correction)\n- [Image outputs](#Image-outputs)",
"_____no_output_____"
],
[
"## Learning Objectives\n\n* Build a sample __DPC++ application__ to perform Image processing (gamma correction) using oneDPL.",
"_____no_output_____"
],
[
"## Gamma Correction\n\nGamma correction is an image processing algorithm where we enhance the image brightness and contrast levels to have a better view of the image.\n\nBelow example creates a bitmap image, and applies the gamma to the image using the DPC++ library offloading to a device. Once we run the program we can view the original image and the gamma corrected image in the corresponding cells below \n\nIn the below program we write a data parallel algorithm using the DPC++ library to leverage the computational power in __heterogenous computers__. The DPC++ platform model includes a host computer and a device. The host offloads computation to the device, which could be a __GPU, FPGA, or a multi-core CPU__.\n\n We create a buffer, being responsible for moving data around and counting dependencies. DPC++ Library provides `oneapi::dpl::begin()` and `oneapi::dpl::end()` interfaces for getting buffer iterators and we implemented as below.\n \n \n \n### Why use buffer iterators?\n\nUsing buffer iterators will ensure that memory is not copied back and forth in between each algorithm execution on device. The code example below shows how the same example above is implemented using buffer iterators which make sure the memory stays on device until the buffer is destructed.\n \nPass the policy object to the `std::for_each` Parallel STL algorithm, which is defined in the oneapi::dpl::execution namespace and pass the __'begin'__ and __'end'__ buffer iterators as the second and third arguments. \n\nThe `oneapi::dpl::execution::dpcpp_default` object is a predefined object of the device_policy class, created with a default kernel name and a default queue. Use it to create customized policy objects, or to pass directly when invoking an algorithm.\nThe Parallel STL API handles the data transfer and compute.\n\n### Lab Exercise: Gamma Correction\n* In this example the student will learn how to use oneDPL library to perform the gamma correction.\n* Follow the __Steps 1 to 3__ in the below code to create a SYCL buffer, create buffer iterators, and then call the std::for each function with DPC++ support. \n\n1. Select the code cell below, __follow the STEPS 1 to 3__ in the code comments, click run ▶ to save the code to file.\n2. Next run ▶ the cell in the __Build and Run__ section below the code to compile and execute the code.",
"_____no_output_____"
]
],
[
[
"%%writefile gamma-correction/src/main.cpp\n//==============================================================\n// Copyright © 2019 Intel Corporation\n//\n// SPDX-License-Identifier: MIT\n// =============================================================\n\n#include <oneapi/dpl/algorithm>\n#include <oneapi/dpl/execution>\n#include <oneapi/dpl/iterator>\n#include <iomanip>\n#include <iostream>\n#include <CL/sycl.hpp>\n\n#include \"utils.hpp\"\n\nusing namespace sycl;\nusing namespace std;\n\nint main() {\n // Image size is width x height\n int width = 1440;\n int height = 960;\n\n Img<ImgFormat::BMP> image{width, height};\n ImgFractal fractal{width, height};\n\n // Lambda to process image with gamma = 2\n auto gamma_f = [](ImgPixel &pixel) {\n auto v = (0.3f * pixel.r + 0.59f * pixel.g + 0.11f * pixel.b) / 255.0f;\n\n auto gamma_pixel = static_cast<uint8_t>(255 * v * v);\n if (gamma_pixel > 255) gamma_pixel = 255;\n pixel.set(gamma_pixel, gamma_pixel, gamma_pixel, gamma_pixel);\n };\n\n // fill image with created fractal\n int index = 0;\n image.fill([&index, width, &fractal](ImgPixel &pixel) {\n int x = index % width;\n int y = index / width;\n\n auto fractal_pixel = fractal(x, y);\n if (fractal_pixel < 0) fractal_pixel = 0;\n if (fractal_pixel > 255) fractal_pixel = 255;\n pixel.set(fractal_pixel, fractal_pixel, fractal_pixel, fractal_pixel);\n\n ++index;\n });\n\n string original_image = \"fractal_original.png\";\n string processed_image = \"fractal_gamma.png\";\n Img<ImgFormat::BMP> image2 = image;\n image.write(original_image);\n\n // call standard serial function for correctness check\n image.fill(gamma_f);\n\n // use default policy for algorithms execution\n auto policy = oneapi::dpl::execution::dpcpp_default;\n // We need to have the scope to have data in image2 after buffer's destruction\n {\n // ****Step 1: Uncomment the below line to create a buffer, being responsible for moving data around and counting dependencies \n //buffer<ImgPixel> b(image2.data(), image2.width() * image2.height());\n\n // create iterator to pass buffer to the algorithm\n // **********Step 2: Uncomment the below lines to create buffer iterators. These are passed to the algorithm\n //auto b_begin = oneapi::dpl::begin(b);\n //auto b_end = oneapi::dpl::end(b);\n\n //*****Step 3: Uncomment the below line to call std::for_each with DPC++ support \n //std::for_each(policy, b_begin, b_end, gamma_f);\n }\n\n image2.write(processed_image);\n // check correctness\n if (check(image.begin(), image.end(), image2.begin())) {\n cout << \"success\\n\";\n } else {\n cout << \"fail\\n\";\n return 1;\n }\n cout << \"Run on \"\n << policy.queue().get_device().template get_info<info::device::name>()\n << \"\\n\";\n cout << \"Original image is in \" << original_image << \"\\n\";\n cout << \"Image after applying gamma correction on the device is in \"\n << processed_image << \"\\n\";\n\n return 0;\n}",
"Overwriting gamma-correction/src/main.cpp\n"
]
],
[
[
"#### Build and Run\nSelect the cell below and click run ▶ to compile and execute the code:",
"_____no_output_____"
]
],
[
[
"! chmod 755 q; chmod 755 run_gamma_correction.sh; if [ -x \"$(command -v qsub)\" ]; then ./q run_gamma_correction.sh; else ./run_gamma_correction.sh; fi",
"_____no_output_____"
]
],
[
[
"_If the Jupyter cells are not responsive or if they error out when you compile the code samples, please restart the Jupyter Kernel: \n\"Kernel->Restart Kernel and Clear All Outputs\" and compile the code samples again_",
"_____no_output_____"
],
[
"### Image outputs\nonce you run the program sucessfuly it creates gamma corrected image and the original image. You can see the difference by running the two cells below and visually compare it. ",
"_____no_output_____"
],
[
"##### View the gamma corrected Image\nSelect the cell below and click run ▶ to view the generated image using gamma correction:",
"_____no_output_____"
]
],
[
[
"from IPython.display import display, Image\ndisplay(Image(filename='gamma-correction/build/src/fractal_gamma.png'))",
"_____no_output_____"
]
],
[
[
"##### View the original Image\nSelect the cell below and click run ▶ to view the generated image using gamma correction:",
"_____no_output_____"
]
],
[
[
"from IPython.display import display, Image\ndisplay(Image(filename='gamma-correction/build/src/fractal_original.png'))",
"_____no_output_____"
]
],
[
[
"# Summary\nIn this module you will have learned how to apply gamma correction to Images using Data Parallel C++ Library",
"_____no_output_____"
],
[
"<html><body><span style=\"color:Red\"><h1>Reset Notebook</h1></span></body></html>\n\n##### Should you be experiencing any issues with your notebook or just want to start fresh run the below cell.\n\n",
"_____no_output_____"
]
],
[
[
"from IPython.display import display, Markdown, clear_output\nimport ipywidgets as widgets\nbutton = widgets.Button(\n description='Reset Notebook',\n disabled=False,\n button_style='', # 'success', 'info', 'warning', 'danger' or ''\n tooltip='This will update this notebook, overwriting any changes.',\n icon='check' # (FontAwesome names without the `fa-` prefix)\n)\nout = widgets.Output()\ndef on_button_clicked(_):\n # \"linking function with output\"\n with out:\n # what happens when we press the button\n clear_output()\n !rsync -a --size-only /data/oneapi_workshop/oneAPI_Essentials/07_DPCPP_Library/ ~/oneAPI_Essentials/07_DPCPP_Library\n print('Notebook reset -- now click reload on browser.')\n# linking button and function together using a button's method\nbutton.on_click(on_button_clicked)\n# displaying button and its output together\nwidgets.VBox([button,out])",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
d0885e53dc0345019e5f2eda0bca524081482e74 | 65,778 | ipynb | Jupyter Notebook | tutorials/tutorial3_adversarial_attack_Gradient.ipynb | airbus/decomon | f3668fbd8edd0def4e23aa0634eebfec58349c35 | [
"MIT"
] | 11 | 2021-11-03T12:09:50.000Z | 2022-02-20T21:42:13.000Z | tutorials/tutorial3_adversarial_attack_Gradient.ipynb | airbus/decomon | f3668fbd8edd0def4e23aa0634eebfec58349c35 | [
"MIT"
] | 1 | 2022-02-18T13:40:46.000Z | 2022-02-18T13:40:46.000Z | tutorials/tutorial3_adversarial_attack_Gradient.ipynb | airbus/decomon | f3668fbd8edd0def4e23aa0634eebfec58349c35 | [
"MIT"
] | null | null | null | 65.062315 | 8,844 | 0.678373 | [
[
[
"# DECOMON tutorial #3 \n## Local Robustness to Adversarial Attacks for classification tasks",
"_____no_output_____"
],
[
"## Introduction\n\nAfter training a model, we want to make sure that the model will give the same output for any images \"close\" to the initial one, showing some robustness to perturbation. \n\nIn this notebook, we start from a classifier built on MNIST dataset that given a hand-written digit as input will predict the digit. This will be the first part of the notebook.\n\n<img src=\"./data/Plot-of-a-Subset-of-Images-from-the-MNIST-Dataset.png\" alt=\"examples of hand-written digit\" width=\"600\"/>\n\nIn the second part of the notebook, we will investigate the robustness of this model to unstructured modification of the input space: adversarial attacks. For this kind of attacks, **we vary the magnitude of the perturbation of the initial image** and want to assess that despite this noise, the classifier's prediction remain unchanged.\n\n<img src=\"./data/illustration_adv_attacks.jpeg\" alt=\"examples of hand-written digit\" width=\"600\"/>\n\nWhat we will show is the use of decomon module to assess the robustness of the prediction towards noise.",
"_____no_output_____"
],
[
"## The notebook\n\n### imports",
"_____no_output_____"
]
],
[
[
"import os\nimport tensorflow.keras as keras\nimport matplotlib.pyplot as plt\nimport matplotlib.patches as patches\n%matplotlib inline\nimport numpy as np\nimport tensorflow.keras.backend as K\nfrom tensorflow.keras.models import Sequential, Model, load_model\nfrom tensorflow.keras.layers import Dense\nfrom tensorflow.keras.datasets import mnist\nfrom ipywidgets import interact, interactive, fixed, interact_manual\nfrom ipykernel.pylab.backend_inline import flush_figures\nimport ipywidgets as widgets\nimport time\nimport sys\nsys.path.append('..')\nimport os.path\nimport os\nimport pickle as pkl\nfrom contextlib import closing\nimport time",
"_____no_output_____"
],
[
"import tensorflow as tf",
"_____no_output_____"
],
[
"import decomon",
"_____no_output_____"
],
[
"from decomon.wrapper import refine_boxes",
"_____no_output_____"
],
[
"x_min = np.ones((3, 4, 5))\nx_max = 2*x_min",
"_____no_output_____"
],
[
"refine_boxes(x_min, x_max, 10)",
"> \u001b[0;32m/Users/ducoffe/Documents/Code/open_sourcing/Airbus/decomon/decomon/wrapper.py\u001b[0m(1065)\u001b[0;36msplit\u001b[0;34m()\u001b[0m\n\u001b[0;32m 1063 \u001b[0;31m \u001b[0msplit_value\u001b[0m \u001b[0;34m=\u001b[0m \u001b[0mnp\u001b[0m\u001b[0;34m.\u001b[0m\u001b[0marray\u001b[0m\u001b[0;34m(\u001b[0m\u001b[0;34m[\u001b[0m\u001b[0mmid_x\u001b[0m\u001b[0;34m[\u001b[0m\u001b[0mi\u001b[0m\u001b[0;34m,\u001b[0m \u001b[0mindex_k\u001b[0m\u001b[0;34m,\u001b[0m \u001b[0mj\u001b[0m\u001b[0;34m[\u001b[0m\u001b[0mi\u001b[0m\u001b[0;34m]\u001b[0m\u001b[0;34m]\u001b[0m \u001b[0;32mfor\u001b[0m \u001b[0mi\u001b[0m \u001b[0;32min\u001b[0m \u001b[0mindex_0\u001b[0m\u001b[0;34m]\u001b[0m\u001b[0;34m)\u001b[0m\u001b[0;34m\u001b[0m\u001b[0;34m\u001b[0m\u001b[0m\n\u001b[0m\u001b[0;32m 1064 \u001b[0;31m \u001b[0;32mimport\u001b[0m \u001b[0mpdb\u001b[0m\u001b[0;34m;\u001b[0m \u001b[0mpdb\u001b[0m\u001b[0;34m.\u001b[0m\u001b[0mset_trace\u001b[0m\u001b[0;34m(\u001b[0m\u001b[0;34m)\u001b[0m\u001b[0;34m\u001b[0m\u001b[0;34m\u001b[0m\u001b[0m\n\u001b[0m\u001b[0;32m-> 1065 \u001b[0;31m \u001b[0;32mfor\u001b[0m \u001b[0mi\u001b[0m \u001b[0;32min\u001b[0m \u001b[0mindex_0\u001b[0m\u001b[0;34m:\u001b[0m\u001b[0;34m\u001b[0m\u001b[0;34m\u001b[0m\u001b[0m\n\u001b[0m\u001b[0;32m 1066 \u001b[0;31m \u001b[0mX_min_\u001b[0m\u001b[0;34m[\u001b[0m\u001b[0mi\u001b[0m\u001b[0;34m,\u001b[0m \u001b[0;36m1\u001b[0m\u001b[0;34m,\u001b[0m \u001b[0mindex_k\u001b[0m\u001b[0;34m,\u001b[0m \u001b[0mj\u001b[0m\u001b[0;34m[\u001b[0m\u001b[0mi\u001b[0m\u001b[0;34m]\u001b[0m\u001b[0;34m]\u001b[0m \u001b[0;34m=\u001b[0m \u001b[0msplit_value\u001b[0m\u001b[0;34m[\u001b[0m\u001b[0mi\u001b[0m\u001b[0;34m]\u001b[0m\u001b[0;34m\u001b[0m\u001b[0;34m\u001b[0m\u001b[0m\n\u001b[0m\u001b[0;32m 1067 \u001b[0;31m \u001b[0mX_max_\u001b[0m\u001b[0;34m[\u001b[0m\u001b[0mi\u001b[0m\u001b[0;34m,\u001b[0m \u001b[0;36m0\u001b[0m\u001b[0;34m,\u001b[0m \u001b[0mindex_k\u001b[0m\u001b[0;34m,\u001b[0m \u001b[0mj\u001b[0m\u001b[0;34m[\u001b[0m\u001b[0mi\u001b[0m\u001b[0;34m]\u001b[0m\u001b[0;34m]\u001b[0m \u001b[0;34m=\u001b[0m \u001b[0msplit_value\u001b[0m\u001b[0;34m[\u001b[0m\u001b[0mi\u001b[0m\u001b[0;34m]\u001b[0m\u001b[0;34m\u001b[0m\u001b[0;34m\u001b[0m\u001b[0m\n\u001b[0m\nipdb> p split_value.shape\n(3, 1)\nipdb> c\n> \u001b[0;32m/Users/ducoffe/Documents/Code/open_sourcing/Airbus/decomon/decomon/wrapper.py\u001b[0m(1065)\u001b[0;36msplit\u001b[0;34m()\u001b[0m\n\u001b[0;32m 1063 \u001b[0;31m \u001b[0msplit_value\u001b[0m \u001b[0;34m=\u001b[0m \u001b[0mnp\u001b[0m\u001b[0;34m.\u001b[0m\u001b[0marray\u001b[0m\u001b[0;34m(\u001b[0m\u001b[0;34m[\u001b[0m\u001b[0mmid_x\u001b[0m\u001b[0;34m[\u001b[0m\u001b[0mi\u001b[0m\u001b[0;34m,\u001b[0m \u001b[0mindex_k\u001b[0m\u001b[0;34m,\u001b[0m \u001b[0mj\u001b[0m\u001b[0;34m[\u001b[0m\u001b[0mi\u001b[0m\u001b[0;34m]\u001b[0m\u001b[0;34m]\u001b[0m \u001b[0;32mfor\u001b[0m \u001b[0mi\u001b[0m \u001b[0;32min\u001b[0m \u001b[0mindex_0\u001b[0m\u001b[0;34m]\u001b[0m\u001b[0;34m)\u001b[0m\u001b[0;34m\u001b[0m\u001b[0;34m\u001b[0m\u001b[0m\n\u001b[0m\u001b[0;32m 1064 \u001b[0;31m \u001b[0;32mimport\u001b[0m \u001b[0mpdb\u001b[0m\u001b[0;34m;\u001b[0m \u001b[0mpdb\u001b[0m\u001b[0;34m.\u001b[0m\u001b[0mset_trace\u001b[0m\u001b[0;34m(\u001b[0m\u001b[0;34m)\u001b[0m\u001b[0;34m\u001b[0m\u001b[0;34m\u001b[0m\u001b[0m\n\u001b[0m\u001b[0;32m-> 1065 \u001b[0;31m \u001b[0;32mfor\u001b[0m \u001b[0mi\u001b[0m \u001b[0;32min\u001b[0m \u001b[0mindex_0\u001b[0m\u001b[0;34m:\u001b[0m\u001b[0;34m\u001b[0m\u001b[0;34m\u001b[0m\u001b[0m\n\u001b[0m\u001b[0;32m 1066 \u001b[0;31m \u001b[0mX_min_\u001b[0m\u001b[0;34m[\u001b[0m\u001b[0mi\u001b[0m\u001b[0;34m,\u001b[0m \u001b[0;36m1\u001b[0m\u001b[0;34m,\u001b[0m \u001b[0mindex_k\u001b[0m\u001b[0;34m,\u001b[0m \u001b[0mj\u001b[0m\u001b[0;34m[\u001b[0m\u001b[0mi\u001b[0m\u001b[0;34m]\u001b[0m\u001b[0;34m]\u001b[0m \u001b[0;34m=\u001b[0m \u001b[0msplit_value\u001b[0m\u001b[0;34m[\u001b[0m\u001b[0mi\u001b[0m\u001b[0;34m]\u001b[0m\u001b[0;34m\u001b[0m\u001b[0;34m\u001b[0m\u001b[0m\n\u001b[0m\u001b[0;32m 1067 \u001b[0;31m \u001b[0mX_max_\u001b[0m\u001b[0;34m[\u001b[0m\u001b[0mi\u001b[0m\u001b[0;34m,\u001b[0m \u001b[0;36m0\u001b[0m\u001b[0;34m,\u001b[0m \u001b[0mindex_k\u001b[0m\u001b[0;34m,\u001b[0m \u001b[0mj\u001b[0m\u001b[0;34m[\u001b[0m\u001b[0mi\u001b[0m\u001b[0;34m]\u001b[0m\u001b[0;34m]\u001b[0m \u001b[0;34m=\u001b[0m \u001b[0msplit_value\u001b[0m\u001b[0;34m[\u001b[0m\u001b[0mi\u001b[0m\u001b[0;34m]\u001b[0m\u001b[0;34m\u001b[0m\u001b[0;34m\u001b[0m\u001b[0m\n\u001b[0m\nipdb> p split_value.shape\n(3, 2)\nipdb> c\n> \u001b[0;32m/Users/ducoffe/Documents/Code/open_sourcing/Airbus/decomon/decomon/wrapper.py\u001b[0m(1065)\u001b[0;36msplit\u001b[0;34m()\u001b[0m\n\u001b[0;32m 1063 \u001b[0;31m \u001b[0msplit_value\u001b[0m \u001b[0;34m=\u001b[0m \u001b[0mnp\u001b[0m\u001b[0;34m.\u001b[0m\u001b[0marray\u001b[0m\u001b[0;34m(\u001b[0m\u001b[0;34m[\u001b[0m\u001b[0mmid_x\u001b[0m\u001b[0;34m[\u001b[0m\u001b[0mi\u001b[0m\u001b[0;34m,\u001b[0m \u001b[0mindex_k\u001b[0m\u001b[0;34m,\u001b[0m \u001b[0mj\u001b[0m\u001b[0;34m[\u001b[0m\u001b[0mi\u001b[0m\u001b[0;34m]\u001b[0m\u001b[0;34m]\u001b[0m \u001b[0;32mfor\u001b[0m \u001b[0mi\u001b[0m \u001b[0;32min\u001b[0m \u001b[0mindex_0\u001b[0m\u001b[0;34m]\u001b[0m\u001b[0;34m)\u001b[0m\u001b[0;34m\u001b[0m\u001b[0;34m\u001b[0m\u001b[0m\n\u001b[0m\u001b[0;32m 1064 \u001b[0;31m \u001b[0;32mimport\u001b[0m \u001b[0mpdb\u001b[0m\u001b[0;34m;\u001b[0m \u001b[0mpdb\u001b[0m\u001b[0;34m.\u001b[0m\u001b[0mset_trace\u001b[0m\u001b[0;34m(\u001b[0m\u001b[0;34m)\u001b[0m\u001b[0;34m\u001b[0m\u001b[0;34m\u001b[0m\u001b[0m\n\u001b[0m\u001b[0;32m-> 1065 \u001b[0;31m \u001b[0;32mfor\u001b[0m \u001b[0mi\u001b[0m \u001b[0;32min\u001b[0m \u001b[0mindex_0\u001b[0m\u001b[0;34m:\u001b[0m\u001b[0;34m\u001b[0m\u001b[0;34m\u001b[0m\u001b[0m\n\u001b[0m\u001b[0;32m 1066 \u001b[0;31m \u001b[0mX_min_\u001b[0m\u001b[0;34m[\u001b[0m\u001b[0mi\u001b[0m\u001b[0;34m,\u001b[0m \u001b[0;36m1\u001b[0m\u001b[0;34m,\u001b[0m \u001b[0mindex_k\u001b[0m\u001b[0;34m,\u001b[0m \u001b[0mj\u001b[0m\u001b[0;34m[\u001b[0m\u001b[0mi\u001b[0m\u001b[0;34m]\u001b[0m\u001b[0;34m]\u001b[0m \u001b[0;34m=\u001b[0m \u001b[0msplit_value\u001b[0m\u001b[0;34m[\u001b[0m\u001b[0mi\u001b[0m\u001b[0;34m]\u001b[0m\u001b[0;34m\u001b[0m\u001b[0;34m\u001b[0m\u001b[0m\n\u001b[0m\u001b[0;32m 1067 \u001b[0;31m \u001b[0mX_max_\u001b[0m\u001b[0;34m[\u001b[0m\u001b[0mi\u001b[0m\u001b[0;34m,\u001b[0m \u001b[0;36m0\u001b[0m\u001b[0;34m,\u001b[0m \u001b[0mindex_k\u001b[0m\u001b[0;34m,\u001b[0m \u001b[0mj\u001b[0m\u001b[0;34m[\u001b[0m\u001b[0mi\u001b[0m\u001b[0;34m]\u001b[0m\u001b[0;34m]\u001b[0m \u001b[0;34m=\u001b[0m \u001b[0msplit_value\u001b[0m\u001b[0;34m[\u001b[0m\u001b[0mi\u001b[0m\u001b[0;34m]\u001b[0m\u001b[0;34m\u001b[0m\u001b[0;34m\u001b[0m\u001b[0m\n\u001b[0m\nipdb> p split_value.shape\n(3, 4)\nipdb> c\n"
]
],
[
[
"### load images\n\nWe load MNIST data from keras datasets. \n",
"_____no_output_____"
]
],
[
[
"ara\nimg_rows, img_cols = 28, 28\n(x_train, y_train_), (x_test, y_test_) = mnist.load_data()\nx_train = x_train.reshape((-1, 784))\nx_test = x_test.reshape((-1, 784))\nx_train = x_train.astype('float32')\nx_test = x_test.astype('float32')\nx_train /= 255\nx_test /= 255\ny_train = keras.utils.to_categorical(y_train_)\ny_test = keras.utils.to_categorical(y_test_)",
"_____no_output_____"
]
],
[
[
"\n\n### learn the model (classifier for MNIST images)\n\nFor the model, we use a small fully connected network. It is made of 6 layers with 100 units each and ReLU activation functions. **Decomon** is compatible with a large set of Keras layers, so do not hesitate to modify the architecture.\n",
"_____no_output_____"
]
],
[
[
"model = Sequential()\nmodel.add(Dense(100, activation='relu', input_dim=784))\nmodel.add(Dense(100, activation='relu'))\nmodel.add(Dense(10, activation='softmax'))",
"_____no_output_____"
],
[
"model.compile('adam', 'categorical_crossentropy', metrics='acc')\n\nmodel.fit(x_train, y_train, batch_size=32, shuffle=True, validation_split=0.2, epochs=5)",
"Epoch 1/5\n1500/1500 [==============================] - 2s 1ms/step - loss: 0.4626 - acc: 0.8668 - val_loss: 0.1406 - val_acc: 0.9591\nEpoch 2/5\n1500/1500 [==============================] - 1s 916us/step - loss: 0.1183 - acc: 0.9643 - val_loss: 0.1176 - val_acc: 0.9661\nEpoch 3/5\n1500/1500 [==============================] - 1s 909us/step - loss: 0.0794 - acc: 0.9741 - val_loss: 0.1068 - val_acc: 0.9688\nEpoch 4/5\n1500/1500 [==============================] - 1s 896us/step - loss: 0.0586 - acc: 0.9810 - val_loss: 0.1010 - val_acc: 0.9709\nEpoch 5/5\n1500/1500 [==============================] - 1s 912us/step - loss: 0.0426 - acc: 0.9862 - val_loss: 0.1226 - val_acc: 0.9669\n"
],
[
"model.evaluate(x_test, y_test, batch_size=32)",
"313/313 [==============================] - 0s 920us/step - loss: 0.1193 - acc: 0.9666\n"
]
],
[
[
"After training, we see that the assessment of performance of the model on data that was not seen during training shows pretty good results: around 0.97 (maximum value is 1). It means that out of 100 images, the model was able to guess the correct digit for 97 images. But how can we guarantee that we will get this performance for images different from the ones in the test dataset? \n\n- If we perturbate a \"little\" an image that was well predicted, will the model stay correct? \n- Up to which perturbation? \n- Can we guarantee that the model will output the same digit for a given perturbation? \n\nThis is where decomon comes in. \n\n<img src=\"./data/decomon.jpg\" alt=\"Decomon!\" width=\"400\"/>\n\n",
"_____no_output_____"
],
[
"### Applying Decomon for Local Robustness to misclassification\n\nIn this section, we detail how to prove local robustness to misclassification. Misclassification can be studied with the global optimisation of a function f:\n\n$$ f(x; \\Omega) = \\max_{z\\in \\Omega} \\text{NN}_{j\\not= i}(z) - \\text{NN}_i(z)\\;\\; \\text{s.t}\\;\\; i = argmax\\;\\text{NN}(x)$$\n\nIf the maximum of f is **negative**, this means that whathever the input sample from the domain, the value outputs by the neural network NN for class i will always be greater than the value output for another class. Hence, there will be no misclassification possible. This is **adversarial robustness**.\n\n<img src=\"./data/tuto_3_formal_robustness.png\" alt=\"Decomon!\" width=\"400\"/>\n\nIn that order, we will use the [decomon](https://gheprivate.intra.corp/CRT-DataScience/decomon/tree/master/decomon) library. Decomon combines several optimization trick, including linear relaxation\nto get state-of-the-art outer approximation.\n\nTo use **decomon** for **adversarial robustness** we first need the following imports:\n+ *from decomon.models import convert*: to convert our current Keras model into another neural network nn_model. nn_model will output the same prediction that our model and adds extra information that will be used to derive our formal bounds. For a sake of clarity, how to get such bounds is hidden to the user\n\n+ *from decomon import get_adv_box*: a genereric method to get an upper bound of the funtion f described previously. If the returned value is negative, then we formally assess the robustness to misclassification.\n\n+ *from decomon import check_adv_box*: a generic method that computes the maximum of a lower bound of f. Eventually if this value is positive, it demonstrates that the function f takes positive value. It results that a positive value formally proves the existence of misclassification.\n",
"_____no_output_____"
]
],
[
[
"import decomon\nfrom decomon.models import convert\nfrom decomon import get_adv_box, get_upper_box, get_lower_box, check_adv_box, get_upper_box",
"_____no_output_____"
]
],
[
[
"For computational efficiency, we convert the model into its decomon version once and for all.\nNote that the decomon method will work on the non-converted model. To obtain more refined guarantees, we activate an option denoted **forward**. You can speed up the method by removing this option in the convert method.",
"_____no_output_____"
]
],
[
[
"decomon_model = convert(model)",
"_____no_output_____"
],
[
"from decomon import build_formal_adv_model",
"_____no_output_____"
],
[
"adv_model = build_formal_adv_model(decomon_model)",
"_____no_output_____"
],
[
"x_=x_train[:1]\neps=1e-2\nz = np.concatenate([x_[:, None]-eps, x_[:, None]+eps], 1)\n",
"_____no_output_____"
],
[
"get_adv_box(decomon_model, x_,x_, source_labels=y_train[0].argmax())",
"_____no_output_____"
],
[
"adv_model.predict([x_, z, y_train[:1]])",
"_____no_output_____"
],
[
"# compute gradient\nimport tensorflow as tf",
"_____no_output_____"
],
[
"x_tensor = tf.convert_to_tensor(x_, dtype=tf.float32)",
"_____no_output_____"
],
[
"from tensorflow.keras.layers import Concatenate",
"_____no_output_____"
],
[
"with tf.GradientTape() as t:\n t.watch(x_tensor)\n z_tensor = Concatenate(1)([x_tensor[:,None]-eps,\\\n x_tensor[:, None]+eps])\n output = adv_model([x_, z_tensor, y_train[:1]])",
"_____no_output_____"
],
[
"result = output\ngradients = t.gradient(output, x_tensor)\nmask = gradients.numpy()\n# scale between 0 and 1.\nmask = (mask-mask.min())",
"_____no_output_____"
],
[
"plt.imshow(gradients.numpy().reshape((28,28)))",
"_____no_output_____"
],
[
"img_mask = np.zeros((784,))\nimg_mask[np.argsort(mask[0])[::-1][:100]]=1",
"_____no_output_____"
],
[
"plt.imshow(img_mask.reshape((28,28)))\n",
"_____no_output_____"
],
[
"plt.imshow(mask.reshape((28,28)))",
"_____no_output_____"
],
[
"plt.imshow(x_.reshape((28,28)))",
"_____no_output_____"
]
],
[
[
"We offer an interactive visualisation of the basic adversarial robustness method from decomon **get_adv_upper**. We randomly choose 10 test images use **get_adv_upper** to assess their robustness to misclassification pixel perturbations. The magnitude of the noise on each pixel is independent and bounded by the value of the variable epsilon. The user can reset the examples and vary the noise amplitude.\n\nNote one of the main advantage of decomon: **we can assess robustness on batches of data!**\n\nCircled in <span style=\"color:green\">green</span> are examples that are formally assessed to be robust, <span style=\"color:orange\">orange</span> examples that could be robust and <span style=\"color:red\">red</span> examples that are formally non robust",
"_____no_output_____"
]
],
[
[
"def frame(epsilon, reset=0, filename='./data/.hidden_index.pkl'):\n n_cols = 5\n n_rows = 2\n n_samples = n_cols*n_rows\n if reset:\n index = np.random.permutation(len(x_test))[:n_samples]\n \n with closing(open(filename, 'wb')) as f:\n pkl.dump(index, f)\n # save data\n else:\n # check that file exists\n \n if os.path.isfile(filename):\n with closing(open(filename, 'rb')) as f:\n index = pkl.load(f)\n else: \n index = np.arange(n_samples)\n with closing(open(filename, 'wb')) as f:\n pkl.dump(index, f)\n #x = np.concatenate([x_test[0:1]]*10, 0)\n x = x_test[index]\n\n x_min = np.maximum(x - epsilon, 0)\n x_max = np.minimum(x + epsilon, 1)\n\n n_cols = 5\n n_rows = 2\n fig, axs = plt.subplots(n_rows, n_cols)\n \n fig.set_figheight(n_rows*fig.get_figheight())\n fig.set_figwidth(n_cols*fig.get_figwidth())\n plt.subplots_adjust(hspace=0.2) # increase vertical separation\n axs_seq = axs.ravel()\n\n source_label = np.argmax(model.predict(x), 1)\n start_time = time.process_time()\n upper = get_adv_box(decomon_model, x_min, x_max, source_labels=source_label)\n lower = check_adv_box(decomon_model, x_min, x_max, source_labels=source_label)\n end_time = time.process_time()\n count = 0\n time.sleep(1)\n r_time = \"{:.2f}\".format(end_time - start_time)\n fig.suptitle('Formal Robustness to Adversarial Examples with eps={} running in {} seconds'.format(epsilon, r_time), fontsize=16)\n for i in range(n_cols):\n for j in range(n_rows):\n\n ax= axs[j, i]\n ax.imshow(x[count].reshape((28,28)), cmap='Greys')\n robust='ROBUST'\n if lower[count]>=0:\n color='red'\n robust='NON ROBUST'\n elif upper[count]<0:\n color='green'\n else:\n color='orange'\n robust='MAYBE ROBUST'\n \n ax.get_xaxis().set_visible(False)\n ax.get_yaxis().set_visible(False)\n # Create a Rectangle patch\n rect = patches.Rectangle((0,0),27,27,linewidth=3,edgecolor=color,facecolor='none')\n ax.add_patch(rect)\n ax.set_title(robust)\n count+=1\n \ninteract(frame, epsilon = widgets.FloatSlider(value=0.,\n min=0.,\n max=5./255.,\n step=0.0001, continuous_update=False, readout_format='.4f',),\n reset = widgets.IntSlider(value=0.,\n min=0,\n max=1,\n step=1, continuous_update=False),\n fast = widgets.IntSlider(value=1.,\n min=0,\n max=1,\n step=1, continuous_update=False)\n )",
"_____no_output_____"
]
],
[
[
"As explained previously, the method **get_adv_upper** output a constant upper bound that is valid on the whole domain.\nSometimes, this bound can be too lose and needs to be refined by splitting the input domain into sub domains.\nSeveral heuristics are possible and you are free to develop your own or take an existing one of the shelf.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d088680e4976789ebc021bd7a1f13a2caf7f1a5c | 7,286 | ipynb | Jupyter Notebook | examples/cifar10/Build the model.ipynb | juliagusak/neural-ode-metasolver | a5ca6ae0c00d2a8da3a5f4b77ee20fb151674d22 | [
"BSD-3-Clause"
] | 25 | 2021-03-16T13:40:45.000Z | 2021-08-12T04:54:39.000Z | examples/cifar10/Build the model.ipynb | MetaSolver/icml2021 | 619774abe4a834ae371434af8b23379e9524e7da | [
"BSD-3-Clause"
] | null | null | null | examples/cifar10/Build the model.ipynb | MetaSolver/icml2021 | 619774abe4a834ae371434af8b23379e9524e7da | [
"BSD-3-Clause"
] | 1 | 2021-03-31T02:58:03.000Z | 2021-03-31T02:58:03.000Z | 38.962567 | 128 | 0.501235 | [
[
[
"import argparse\nimport copy\nimport sys\n\nsys.path.append('../../')\nimport sopa.src.models.odenet_cifar10.layers as cifar10_models\nfrom sopa.src.models.odenet_cifar10.utils import *",
"_____no_output_____"
],
[
"parser = argparse.ArgumentParser()\n# Architecture params\nparser.add_argument('--is_odenet', type=eval, default=True, choices=[True, False])\nparser.add_argument('--network', type=str, choices=['metanode34', 'metanode18', 'metanode10', 'metanode6', 'metanode4',\n 'premetanode34', 'premetanode18', 'premetanode10', 'premetanode6',\n 'premetanode4'],\n default='premetanode10')\nparser.add_argument('--in_planes', type=int, default=64)\n\n# Type of layer's output normalization\nparser.add_argument('--normalization_resblock', type=str, default='NF',\n choices=['BN', 'GN', 'LN', 'IN', 'NF'])\nparser.add_argument('--normalization_odeblock', type=str, default='NF',\n choices=['BN', 'GN', 'LN', 'IN', 'NF'])\nparser.add_argument('--normalization_bn1', type=str, default='NF',\n choices=['BN', 'GN', 'LN', 'IN', 'NF'])\nparser.add_argument('--num_gn_groups', type=int, default=32, help='Number of groups for GN normalization')\n\n# Type of layer's weights normalization\nparser.add_argument('--param_normalization_resblock', type=str, default='PNF',\n choices=['WN', 'SN', 'PNF'])\nparser.add_argument('--param_normalization_odeblock', type=str, default='PNF',\n choices=['WN', 'SN', 'PNF'])\nparser.add_argument('--param_normalization_bn1', type=str, default='PNF',\n choices=['WN', 'SN', 'PNF'])\n# Type of activation\nparser.add_argument('--activation_resblock', type=str, default='ReLU',\n choices=['ReLU', 'GeLU', 'Softsign', 'Tanh', 'AF'])\nparser.add_argument('--activation_odeblock', type=str, default='ReLU',\n choices=['ReLU', 'GeLU', 'Softsign', 'Tanh', 'AF'])\nparser.add_argument('--activation_bn1', type=str, default='ReLU',\n choices=['ReLU', 'GeLU', 'Softsign', 'Tanh', 'AF'])\n\nargs, unknown_args = parser.parse_known_args()",
"_____no_output_____"
],
[
"# Initialize Neural ODE model\nconfig = copy.deepcopy(args)\n\nnorm_layers = (get_normalization(config.normalization_resblock),\n get_normalization(config.normalization_odeblock),\n get_normalization(config.normalization_bn1))\nparam_norm_layers = (get_param_normalization(config.param_normalization_resblock),\n get_param_normalization(config.param_normalization_odeblock),\n get_param_normalization(config.param_normalization_bn1))\nact_layers = (get_activation(config.activation_resblock),\n get_activation(config.activation_odeblock),\n get_activation(config.activation_bn1))\n\nmodel = getattr(cifar10_models, config.network)(norm_layers, param_norm_layers, act_layers,\n config.in_planes, is_odenet=config.is_odenet)",
"_____no_output_____"
],
[
"model",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code"
]
] |
d08887154591d4e17dc8427ea72b6c79676badf6 | 2,047 | ipynb | Jupyter Notebook | 003-largest-prime-factor.ipynb | arkeros/projecteuler | c95db97583034af8fc61d5786692d82eabe50c12 | [
"MIT"
] | 2 | 2017-02-19T12:37:13.000Z | 2021-01-19T04:58:09.000Z | 003-largest-prime-factor.ipynb | arkeros/projecteuler | c95db97583034af8fc61d5786692d82eabe50c12 | [
"MIT"
] | null | null | null | 003-largest-prime-factor.ipynb | arkeros/projecteuler | c95db97583034af8fc61d5786692d82eabe50c12 | [
"MIT"
] | 4 | 2018-01-05T14:29:09.000Z | 2020-01-27T13:37:40.000Z | 18.441441 | 77 | 0.469468 | [
[
[
"# Largest prime factor\n## Problem 3 \n\n<div class=\"problem_content\" role=\"problem\">\n<p>The prime factors of 13195 are 5, 7, 13 and 29.</p>\n<p>What is the largest prime factor of the number 600851475143 ?</p>\n</div>",
"_____no_output_____"
]
],
[
[
"from math import sqrt\n\ndef factorize(n, start=2):\n p = start\n while p <= sqrt(n):\n q, r = divmod(n, p)\n if r == 0:\n return [p] + factorize(q, p)\n p += 1\n return [n]",
"_____no_output_____"
],
[
"assert factorize(13195) == [5, 7, 13, 29]",
"_____no_output_____"
],
[
"def solve():\n return max(factorize(600851475143))",
"_____no_output_____"
]
],
[
[
"## Solution",
"_____no_output_____"
]
],
[
[
"solve()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.