code
stringlengths 501
5.19M
| package
stringlengths 2
81
| path
stringlengths 9
304
| filename
stringlengths 4
145
|
|---|---|---|---|
import boto3
from AWSIoTPythonSDK.MQTTLib import AWSIoTMQTTClient
import logging
import time
import argparse
# Custom MQTT message callback
def customCallback(client, userdata, message):
print("Received a new message: ")
print(message.payload)
print("from topic: ")
print(message.topic)
print("--------------\n\n")
# Read in command-line parameters
parser = argparse.ArgumentParser()
parser.add_argument("-e", "--endpoint", action="store", required=True, dest="host", help="Your AWS IoT custom endpoint")
parser.add_argument("-r", "--rootCA", action="store", required=True, dest="rootCAPath", help="Root CA file path")
parser.add_argument("-C", "--CognitoIdentityPoolID", action="store", required=True, dest="cognitoIdentityPoolID",
help="Your AWS Cognito Identity Pool ID")
parser.add_argument("-id", "--clientId", action="store", dest="clientId", default="basicPubSub_CognitoSTS",
help="Targeted client id")
parser.add_argument("-t", "--topic", action="store", dest="topic", default="sdk/test/Python", help="Targeted topic")
args = parser.parse_args()
host = args.host
rootCAPath = args.rootCAPath
clientId = args.clientId
cognitoIdentityPoolID = args.cognitoIdentityPoolID
topic = args.topic
# Configure logging
logger = logging.getLogger("AWSIoTPythonSDK.core")
logger.setLevel(logging.DEBUG)
streamHandler = logging.StreamHandler()
formatter = logging.Formatter('%(asctime)s - %(name)s - %(levelname)s - %(message)s')
streamHandler.setFormatter(formatter)
logger.addHandler(streamHandler)
# Cognito auth
identityPoolID = cognitoIdentityPoolID
region = host.split('.')[2]
cognitoIdentityClient = boto3.client('cognito-identity', region_name=region)
# identityPoolInfo = cognitoIdentityClient.describe_identity_pool(IdentityPoolId=identityPoolID)
# print identityPoolInfo
temporaryIdentityId = cognitoIdentityClient.get_id(IdentityPoolId=identityPoolID)
identityID = temporaryIdentityId["IdentityId"]
temporaryCredentials = cognitoIdentityClient.get_credentials_for_identity(IdentityId=identityID)
AccessKeyId = temporaryCredentials["Credentials"]["AccessKeyId"]
SecretKey = temporaryCredentials["Credentials"]["SecretKey"]
SessionToken = temporaryCredentials["Credentials"]["SessionToken"]
# Init AWSIoTMQTTClient
myAWSIoTMQTTClient = AWSIoTMQTTClient(clientId, useWebsocket=True)
# AWSIoTMQTTClient configuration
myAWSIoTMQTTClient.configureEndpoint(host, 443)
myAWSIoTMQTTClient.configureCredentials(rootCAPath)
myAWSIoTMQTTClient.configureIAMCredentials(AccessKeyId, SecretKey, SessionToken)
myAWSIoTMQTTClient.configureAutoReconnectBackoffTime(1, 32, 20)
myAWSIoTMQTTClient.configureOfflinePublishQueueing(-1) # Infinite offline Publish queueing
myAWSIoTMQTTClient.configureDrainingFrequency(2) # Draining: 2 Hz
myAWSIoTMQTTClient.configureConnectDisconnectTimeout(10) # 10 sec
myAWSIoTMQTTClient.configureMQTTOperationTimeout(5) # 5 sec
# Connect and subscribe to AWS IoT
myAWSIoTMQTTClient.connect()
myAWSIoTMQTTClient.subscribe(topic, 1, customCallback)
time.sleep(2)
# Publish to the same topic in a loop forever
loopCount = 0
while True:
myAWSIoTMQTTClient.publish(topic, "New Message " + str(loopCount), 1)
loopCount += 1
time.sleep(1) | AWSIoTPythonSDK | /AWSIoTPythonSDK-1.5.2.tar.gz/AWSIoTPythonSDK-1.5.2/samples/basicPubSub/basicPubSub_CognitoSTS.py | basicPubSub_CognitoSTS.py |
from AWSIoTPythonSDK.MQTTLib import AWSIoTMQTTClient
import logging
import time
import argparse
# General message notification callback
def customOnMessage(message):
print("Received a new message: ")
print(message.payload)
print("from topic: ")
print(message.topic)
print("--------------\n\n")
# Suback callback
def customSubackCallback(mid, data):
print("Received SUBACK packet id: ")
print(mid)
print("Granted QoS: ")
print(data)
print("++++++++++++++\n\n")
# Puback callback
def customPubackCallback(mid):
print("Received PUBACK packet id: ")
print(mid)
print("++++++++++++++\n\n")
# Read in command-line parameters
parser = argparse.ArgumentParser()
parser.add_argument("-e", "--endpoint", action="store", required=True, dest="host", help="Your AWS IoT custom endpoint")
parser.add_argument("-r", "--rootCA", action="store", required=True, dest="rootCAPath", help="Root CA file path")
parser.add_argument("-c", "--cert", action="store", dest="certificatePath", help="Certificate file path")
parser.add_argument("-k", "--key", action="store", dest="privateKeyPath", help="Private key file path")
parser.add_argument("-p", "--port", action="store", dest="port", type=int, help="Port number override")
parser.add_argument("-w", "--websocket", action="store_true", dest="useWebsocket", default=False,
help="Use MQTT over WebSocket")
parser.add_argument("-id", "--clientId", action="store", dest="clientId", default="basicPubSub",
help="Targeted client id")
parser.add_argument("-t", "--topic", action="store", dest="topic", default="sdk/test/Python", help="Targeted topic")
args = parser.parse_args()
host = args.host
rootCAPath = args.rootCAPath
certificatePath = args.certificatePath
privateKeyPath = args.privateKeyPath
port = args.port
useWebsocket = args.useWebsocket
clientId = args.clientId
topic = args.topic
if args.useWebsocket and args.certificatePath and args.privateKeyPath:
parser.error("X.509 cert authentication and WebSocket are mutual exclusive. Please pick one.")
exit(2)
if not args.useWebsocket and (not args.certificatePath or not args.privateKeyPath):
parser.error("Missing credentials for authentication.")
exit(2)
# Port defaults
if args.useWebsocket and not args.port: # When no port override for WebSocket, default to 443
port = 443
if not args.useWebsocket and not args.port: # When no port override for non-WebSocket, default to 8883
port = 8883
# Configure logging
logger = logging.getLogger("AWSIoTPythonSDK.core")
logger.setLevel(logging.DEBUG)
streamHandler = logging.StreamHandler()
formatter = logging.Formatter('%(asctime)s - %(name)s - %(levelname)s - %(message)s')
streamHandler.setFormatter(formatter)
logger.addHandler(streamHandler)
# Init AWSIoTMQTTClient
myAWSIoTMQTTClient = None
if useWebsocket:
myAWSIoTMQTTClient = AWSIoTMQTTClient(clientId, useWebsocket=True)
myAWSIoTMQTTClient.configureEndpoint(host, port)
myAWSIoTMQTTClient.configureCredentials(rootCAPath)
else:
myAWSIoTMQTTClient = AWSIoTMQTTClient(clientId)
myAWSIoTMQTTClient.configureEndpoint(host, port)
myAWSIoTMQTTClient.configureCredentials(rootCAPath, privateKeyPath, certificatePath)
# AWSIoTMQTTClient connection configuration
myAWSIoTMQTTClient.configureAutoReconnectBackoffTime(1, 32, 20)
myAWSIoTMQTTClient.configureOfflinePublishQueueing(-1) # Infinite offline Publish queueing
myAWSIoTMQTTClient.configureDrainingFrequency(2) # Draining: 2 Hz
myAWSIoTMQTTClient.configureConnectDisconnectTimeout(10) # 10 sec
myAWSIoTMQTTClient.configureMQTTOperationTimeout(5) # 5 sec
myAWSIoTMQTTClient.onMessage = customOnMessage
# Connect and subscribe to AWS IoT
myAWSIoTMQTTClient.connect()
# Note that we are not putting a message callback here. We are using the general message notification callback.
myAWSIoTMQTTClient.subscribeAsync(topic, 1, ackCallback=customSubackCallback)
time.sleep(2)
# Publish to the same topic in a loop forever
loopCount = 0
while True:
myAWSIoTMQTTClient.publishAsync(topic, "New Message " + str(loopCount), 1, ackCallback=customPubackCallback)
loopCount += 1
time.sleep(1) | AWSIoTPythonSDK | /AWSIoTPythonSDK-1.5.2.tar.gz/AWSIoTPythonSDK-1.5.2/samples/basicPubSub/basicPubSubAsync.py | basicPubSubAsync.py |
from AWSIoTPythonSDK.MQTTLib import AWSIoTMQTTClient
import logging
import time
import argparse
class CallbackContainer(object):
def __init__(self, client):
self._client = client
def messagePrint(self, client, userdata, message):
print("Received a new message: ")
print(message.payload)
print("from topic: ")
print(message.topic)
print("--------------\n\n")
def messageForward(self, client, userdata, message):
topicRepublish = message.topic + "/republish"
print("Forwarding message from: %s to %s" % (message.topic, topicRepublish))
print("--------------\n\n")
self._client.publishAsync(topicRepublish, str(message.payload), 1, self.pubackCallback)
def pubackCallback(self, mid):
print("Received PUBACK packet id: ")
print(mid)
print("++++++++++++++\n\n")
def subackCallback(self, mid, data):
print("Received SUBACK packet id: ")
print(mid)
print("Granted QoS: ")
print(data)
print("++++++++++++++\n\n")
# Read in command-line parameters
parser = argparse.ArgumentParser()
parser.add_argument("-e", "--endpoint", action="store", required=True, dest="host", help="Your AWS IoT custom endpoint")
parser.add_argument("-r", "--rootCA", action="store", required=True, dest="rootCAPath", help="Root CA file path")
parser.add_argument("-c", "--cert", action="store", dest="certificatePath", help="Certificate file path")
parser.add_argument("-k", "--key", action="store", dest="privateKeyPath", help="Private key file path")
parser.add_argument("-p", "--port", action="store", dest="port", type=int, help="Port number override")
parser.add_argument("-w", "--websocket", action="store_true", dest="useWebsocket", default=False,
help="Use MQTT over WebSocket")
parser.add_argument("-id", "--clientId", action="store", dest="clientId", default="basicPubSub",
help="Targeted client id")
parser.add_argument("-t", "--topic", action="store", dest="topic", default="sdk/test/Python", help="Targeted topic")
args = parser.parse_args()
host = args.host
rootCAPath = args.rootCAPath
certificatePath = args.certificatePath
privateKeyPath = args.privateKeyPath
port = args.port
useWebsocket = args.useWebsocket
clientId = args.clientId
topic = args.topic
if args.useWebsocket and args.certificatePath and args.privateKeyPath:
parser.error("X.509 cert authentication and WebSocket are mutual exclusive. Please pick one.")
exit(2)
if not args.useWebsocket and (not args.certificatePath or not args.privateKeyPath):
parser.error("Missing credentials for authentication.")
exit(2)
# Port defaults
if args.useWebsocket and not args.port: # When no port override for WebSocket, default to 443
port = 443
if not args.useWebsocket and not args.port: # When no port override for non-WebSocket, default to 8883
port = 8883
# Configure logging
logger = logging.getLogger("AWSIoTPythonSDK.core")
logger.setLevel(logging.DEBUG)
streamHandler = logging.StreamHandler()
formatter = logging.Formatter('%(asctime)s - %(name)s - %(levelname)s - %(message)s')
streamHandler.setFormatter(formatter)
logger.addHandler(streamHandler)
# Init AWSIoTMQTTClient
myAWSIoTMQTTClient = None
if useWebsocket:
myAWSIoTMQTTClient = AWSIoTMQTTClient(clientId, useWebsocket=True)
myAWSIoTMQTTClient.configureEndpoint(host, port)
myAWSIoTMQTTClient.configureCredentials(rootCAPath)
else:
myAWSIoTMQTTClient = AWSIoTMQTTClient(clientId)
myAWSIoTMQTTClient.configureEndpoint(host, port)
myAWSIoTMQTTClient.configureCredentials(rootCAPath, privateKeyPath, certificatePath)
# AWSIoTMQTTClient connection configuration
myAWSIoTMQTTClient.configureAutoReconnectBackoffTime(1, 32, 20)
myAWSIoTMQTTClient.configureOfflinePublishQueueing(-1) # Infinite offline Publish queueing
myAWSIoTMQTTClient.configureDrainingFrequency(2) # Draining: 2 Hz
myAWSIoTMQTTClient.configureConnectDisconnectTimeout(10) # 10 sec
myAWSIoTMQTTClient.configureMQTTOperationTimeout(5) # 5 sec
myCallbackContainer = CallbackContainer(myAWSIoTMQTTClient)
# Connect and subscribe to AWS IoT
myAWSIoTMQTTClient.connect()
# Perform synchronous subscribes
myAWSIoTMQTTClient.subscribe(topic, 1, myCallbackContainer.messageForward)
myAWSIoTMQTTClient.subscribe(topic + "/republish", 1, myCallbackContainer.messagePrint)
time.sleep(2)
# Publish to the same topic in a loop forever
loopCount = 0
while True:
myAWSIoTMQTTClient.publishAsync(topic, "New Message " + str(loopCount), 1, ackCallback=myCallbackContainer.pubackCallback)
loopCount += 1
time.sleep(1) | AWSIoTPythonSDK | /AWSIoTPythonSDK-1.5.2.tar.gz/AWSIoTPythonSDK-1.5.2/samples/basicPubSub/basicPubSub_APICallInCallback.py | basicPubSub_APICallInCallback.py |
from AWSIoTPythonSDK.MQTTLib import AWSIoTMQTTShadowClient
import logging
import time
import json
import argparse
# Shadow JSON schema:
#
# Name: Bot
# {
# "state": {
# "desired":{
# "property":<INT VALUE>
# }
# }
# }
# Custom Shadow callback
def customShadowCallback_Delta(payload, responseStatus, token):
# payload is a JSON string ready to be parsed using json.loads(...)
# in both Py2.x and Py3.x
print(responseStatus)
payloadDict = json.loads(payload)
print("++++++++DELTA++++++++++")
print("property: " + str(payloadDict["state"]["property"]))
print("version: " + str(payloadDict["version"]))
print("+++++++++++++++++++++++\n\n")
# Read in command-line parameters
parser = argparse.ArgumentParser()
parser.add_argument("-e", "--endpoint", action="store", required=True, dest="host", help="Your AWS IoT custom endpoint")
parser.add_argument("-r", "--rootCA", action="store", required=True, dest="rootCAPath", help="Root CA file path")
parser.add_argument("-c", "--cert", action="store", dest="certificatePath", help="Certificate file path")
parser.add_argument("-k", "--key", action="store", dest="privateKeyPath", help="Private key file path")
parser.add_argument("-p", "--port", action="store", dest="port", type=int, help="Port number override")
parser.add_argument("-w", "--websocket", action="store_true", dest="useWebsocket", default=False,
help="Use MQTT over WebSocket")
parser.add_argument("-n", "--thingName", action="store", dest="thingName", default="Bot", help="Targeted thing name")
parser.add_argument("-id", "--clientId", action="store", dest="clientId", default="basicShadowDeltaListener",
help="Targeted client id")
args = parser.parse_args()
host = args.host
rootCAPath = args.rootCAPath
certificatePath = args.certificatePath
privateKeyPath = args.privateKeyPath
port = args.port
useWebsocket = args.useWebsocket
thingName = args.thingName
clientId = args.clientId
if args.useWebsocket and args.certificatePath and args.privateKeyPath:
parser.error("X.509 cert authentication and WebSocket are mutual exclusive. Please pick one.")
exit(2)
if not args.useWebsocket and (not args.certificatePath or not args.privateKeyPath):
parser.error("Missing credentials for authentication.")
exit(2)
# Port defaults
if args.useWebsocket and not args.port: # When no port override for WebSocket, default to 443
port = 443
if not args.useWebsocket and not args.port: # When no port override for non-WebSocket, default to 8883
port = 8883
# Configure logging
logger = logging.getLogger("AWSIoTPythonSDK.core")
logger.setLevel(logging.DEBUG)
streamHandler = logging.StreamHandler()
formatter = logging.Formatter('%(asctime)s - %(name)s - %(levelname)s - %(message)s')
streamHandler.setFormatter(formatter)
logger.addHandler(streamHandler)
# Init AWSIoTMQTTShadowClient
myAWSIoTMQTTShadowClient = None
if useWebsocket:
myAWSIoTMQTTShadowClient = AWSIoTMQTTShadowClient(clientId, useWebsocket=True)
myAWSIoTMQTTShadowClient.configureEndpoint(host, port)
myAWSIoTMQTTShadowClient.configureCredentials(rootCAPath)
else:
myAWSIoTMQTTShadowClient = AWSIoTMQTTShadowClient(clientId)
myAWSIoTMQTTShadowClient.configureEndpoint(host, port)
myAWSIoTMQTTShadowClient.configureCredentials(rootCAPath, privateKeyPath, certificatePath)
# AWSIoTMQTTShadowClient configuration
myAWSIoTMQTTShadowClient.configureAutoReconnectBackoffTime(1, 32, 20)
myAWSIoTMQTTShadowClient.configureConnectDisconnectTimeout(10) # 10 sec
myAWSIoTMQTTShadowClient.configureMQTTOperationTimeout(5) # 5 sec
# Connect to AWS IoT
myAWSIoTMQTTShadowClient.connect()
# Create a deviceShadow with persistent subscription
deviceShadowHandler = myAWSIoTMQTTShadowClient.createShadowHandlerWithName(thingName, True)
# Listen on deltas
deviceShadowHandler.shadowRegisterDeltaCallback(customShadowCallback_Delta)
# Loop forever
while True:
time.sleep(1) | AWSIoTPythonSDK | /AWSIoTPythonSDK-1.5.2.tar.gz/AWSIoTPythonSDK-1.5.2/samples/basicShadow/basicShadowDeltaListener.py | basicShadowDeltaListener.py |
from AWSIoTPythonSDK.MQTTLib import AWSIoTMQTTShadowClient
import logging
import time
import json
import argparse
# Shadow JSON schema:
#
# Name: Bot
# {
# "state": {
# "desired":{
# "property":<INT VALUE>
# }
# }
# }
# Custom Shadow callback
def customShadowCallback_Update(payload, responseStatus, token):
# payload is a JSON string ready to be parsed using json.loads(...)
# in both Py2.x and Py3.x
if responseStatus == "timeout":
print("Update request " + token + " time out!")
if responseStatus == "accepted":
payloadDict = json.loads(payload)
print("~~~~~~~~~~~~~~~~~~~~~~~")
print("Update request with token: " + token + " accepted!")
print("property: " + str(payloadDict["state"]["desired"]["property"]))
print("~~~~~~~~~~~~~~~~~~~~~~~\n\n")
if responseStatus == "rejected":
print("Update request " + token + " rejected!")
def customShadowCallback_Delete(payload, responseStatus, token):
if responseStatus == "timeout":
print("Delete request " + token + " time out!")
if responseStatus == "accepted":
print("~~~~~~~~~~~~~~~~~~~~~~~")
print("Delete request with token: " + token + " accepted!")
print("~~~~~~~~~~~~~~~~~~~~~~~\n\n")
if responseStatus == "rejected":
print("Delete request " + token + " rejected!")
# Read in command-line parameters
parser = argparse.ArgumentParser()
parser.add_argument("-e", "--endpoint", action="store", required=True, dest="host", help="Your AWS IoT custom endpoint")
parser.add_argument("-r", "--rootCA", action="store", required=True, dest="rootCAPath", help="Root CA file path")
parser.add_argument("-c", "--cert", action="store", dest="certificatePath", help="Certificate file path")
parser.add_argument("-k", "--key", action="store", dest="privateKeyPath", help="Private key file path")
parser.add_argument("-p", "--port", action="store", dest="port", type=int, help="Port number override")
parser.add_argument("-w", "--websocket", action="store_true", dest="useWebsocket", default=False,
help="Use MQTT over WebSocket")
parser.add_argument("-n", "--thingName", action="store", dest="thingName", default="Bot", help="Targeted thing name")
parser.add_argument("-id", "--clientId", action="store", dest="clientId", default="basicShadowUpdater", help="Targeted client id")
args = parser.parse_args()
host = args.host
rootCAPath = args.rootCAPath
certificatePath = args.certificatePath
privateKeyPath = args.privateKeyPath
port = args.port
useWebsocket = args.useWebsocket
thingName = args.thingName
clientId = args.clientId
if args.useWebsocket and args.certificatePath and args.privateKeyPath:
parser.error("X.509 cert authentication and WebSocket are mutual exclusive. Please pick one.")
exit(2)
if not args.useWebsocket and (not args.certificatePath or not args.privateKeyPath):
parser.error("Missing credentials for authentication.")
exit(2)
# Port defaults
if args.useWebsocket and not args.port: # When no port override for WebSocket, default to 443
port = 443
if not args.useWebsocket and not args.port: # When no port override for non-WebSocket, default to 8883
port = 8883
# Configure logging
logger = logging.getLogger("AWSIoTPythonSDK.core")
logger.setLevel(logging.DEBUG)
streamHandler = logging.StreamHandler()
formatter = logging.Formatter('%(asctime)s - %(name)s - %(levelname)s - %(message)s')
streamHandler.setFormatter(formatter)
logger.addHandler(streamHandler)
# Init AWSIoTMQTTShadowClient
myAWSIoTMQTTShadowClient = None
if useWebsocket:
myAWSIoTMQTTShadowClient = AWSIoTMQTTShadowClient(clientId, useWebsocket=True)
myAWSIoTMQTTShadowClient.configureEndpoint(host, port)
myAWSIoTMQTTShadowClient.configureCredentials(rootCAPath)
else:
myAWSIoTMQTTShadowClient = AWSIoTMQTTShadowClient(clientId)
myAWSIoTMQTTShadowClient.configureEndpoint(host, port)
myAWSIoTMQTTShadowClient.configureCredentials(rootCAPath, privateKeyPath, certificatePath)
# AWSIoTMQTTShadowClient configuration
myAWSIoTMQTTShadowClient.configureAutoReconnectBackoffTime(1, 32, 20)
myAWSIoTMQTTShadowClient.configureConnectDisconnectTimeout(10) # 10 sec
myAWSIoTMQTTShadowClient.configureMQTTOperationTimeout(5) # 5 sec
# Connect to AWS IoT
myAWSIoTMQTTShadowClient.connect()
# Create a deviceShadow with persistent subscription
deviceShadowHandler = myAWSIoTMQTTShadowClient.createShadowHandlerWithName(thingName, True)
# Delete shadow JSON doc
deviceShadowHandler.shadowDelete(customShadowCallback_Delete, 5)
# Update shadow in a loop
loopCount = 0
while True:
JSONPayload = '{"state":{"desired":{"property":' + str(loopCount) + '}}}'
deviceShadowHandler.shadowUpdate(JSONPayload, customShadowCallback_Update, 5)
loopCount += 1
time.sleep(1) | AWSIoTPythonSDK | /AWSIoTPythonSDK-1.5.2.tar.gz/AWSIoTPythonSDK-1.5.2/samples/basicShadow/basicShadowUpdater.py | basicShadowUpdater.py |

## 1. OVERVIEW
`AWSOM` stands for: **A**mazing **W**ays to **S**earch and **O**rganise
**M**edia. `AWSOM` is a media automation toolkit, originally intended to
automate the Adobe Creative Cloud video production workflow at
[South West London TV](https://www.southwestlondon.tv).
This Python package contains a mixture of OS-independent scripts for managing
common (desktop/LAN) automation tasks such as renaming media files and making backups, as well
as some exciting automations built on top of the superb `Pymiere` library by
[Quentin Masingarbe](https://github.com/qmasingarbe/pymiere). Unfortunately
`Pymiere` currently only works on **Windows 10**, so the corresponding `AWSOM`
automations are limited to Windows users as well (for now).
If you're interested in exploring the rest of the **AWSOM** toolkit, which is
primarily aimed at serious YouTubers, video editors, and power-users of online
video such as journalists, researchers, and teachers, please follow us on Twitter
and start a conversation:

https://twitter.com/AppAwsom
Both this package and a snazzy new web version of `AWSOM` which we hope to launch soon provide a
tonne of powerful features completely free, but certain 'Pro' features like
Automatic Speech Recognition/Subtitling use third party services and may require
a registered account with us or them to access on a pay-per-use basis.
## 2. INSTALLATION
Very lightweight installation with only three dependencies: `pymiere`, `cleverdict` and `pysimplegui`.
pip install AWSOM
or to cover all bases...
python -m pip install AWSOM --upgrade --user
## 3. BASIC USE
`AWSOM` currently supports Sony's commonly used XDCAM format. More formats will be added soon, or feel free to get involved and contribute one!
1. Connect and power on your camera/storage device, then open up your Python interpreter/IDE:
```
import AWSOM
AWSOM.ingest(from_device=True)
```
2. Follow the (beautiful `PySimpleGui`) prompts to give your project a name and category/prefix, and point it to a template `.prproj` file to copy from.
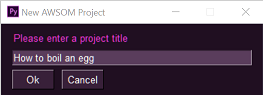
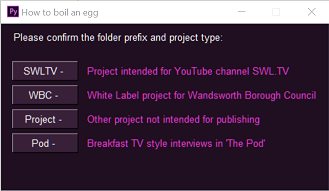
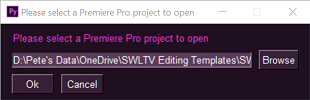
3. Go and have a cup of coffee, knowing that when you come back all the fiddly, non-creative, importy-draggy stuff will be done and you can get on with *actual* editing!
* New project Folder created on your main work hard drive.
* All media and metadata copied to subfolder `XDROOT`.
* All clips, thumbnails and XML files renamed to include the project name.
* `MEDIAPRO.XML` updated with new names.
* Your selected template .prproj file opened in Premiere Pro.
* Template file save with new project name to new folder.
* Rushes bin created if not already in the template.
* All clips imported into the Rushes bin.
* Rushes sequence created if not already in the template.
* All clips inserted in the Rushes sequence, all ready and waiting for you!
## 4. UNDER THE BONNET
None of `AWSOM`'s automations for `Adobe Premiere Pro` would be possible without `Pymiere`.
`AWSOM`'s library of choice for user interaction is `PySimpleGui` which creates beautiful looking popups, input windows and output displays with very few lines of code. We think their documentation is really fun and accessible too which makes the learning-curve for newcomers really painless.
Internally, `AWSOM` makes extensive use of `CleverDict`, a handy custom data type which allows
developers to flexibly switch between Python dictionary `{key: value}` notation
and `object.attribute` notation. For more information about `CleverDict` see:
https://pypi.org/project/cleverdict/
`AWSOM` uses `pathlib` in preference to `os` wherever possible for handling files, directories, and drives.
The primary class used in `AWSOM` is `Project` which encapsulates all the control data used by the rest of the main program.
Functions and methods are generally as 'atomic' as possible, i.e. one function generally does just one thing and is kept as short as reasonably possible. The exception to this are *workflow functions* like `ingest()` which by their nature chain together potentially long sequences of individual functions, passing a `Project` object between them.
## 5. CONTRIBUTING
Please join our virtual team if you have an interest in video editing, production, workflow automation or simply have an idea for improving this package. We're particularly keen to connect with anyone who can help us make `Pymiere` work on other Operating Systems and folk already active in the Adobe/Premiere/ExtendScript space or working on tools for Speech Recognition, Subtitles, Media Content Management, and online video generally (especially but not only YouTube). We're also on the lookout for professional help with UX/UI design and all things HTML/CSS to take our web app version of `AWSOM` to the next level.
Our preferred process for onboarding new contributors is as follows:
1. Say hello to us on [Twitter](https://twitter.com/AppAwsom) initially so we can "put a face to the name".
2. Fork this repository. Also **STAR** this repository for bonus karma!
3. Create new branches with the following standardised names as required:
* `cosmetic`: for reformatting and changes to comments, README, or user input/output e.g. print(), input() and GUI.
* `enhancements`: for new features and extensions to old features
* `refactoring`: for better ways to code existing features
* `tests`: for new or better test cases
* `bugfix`: for solutions to existing issues
* `miscellaneous`: for anything else
4. We're a naively zealous fan of *Test Driven Development*, so please start by creating a separate `test_xyz.py` script for any coding changes, and document your tests (and any new code) clearly enough that they'll tell us everything we need to know about your rationale and implementation approach.
5. When you're ready and any new code passes all your/our tests, create a *Pull Request* from one of your branches (above) back to the `main` branch of this repository.
If you'd be kind enough to follow that approach it will speed things on their way and cause less brain-ache for us, thanks!
| AWSOM | /AWSOM-0.14.tar.gz/AWSOM-0.14/README.md | README.md |
##########
AWS Scout2
##########
.. image:: https://travis-ci.org/nccgroup/Scout2.svg?branch=master
:target: https://travis-ci.org/nccgroup/Scout2
.. image:: https://coveralls.io/repos/github/nccgroup/Scout2/badge.svg?branch=master
:target: https://coveralls.io/github/nccgroup/Scout2
.. image:: https://badge.fury.io/py/AWSScout2.svg
:target: https://badge.fury.io/py/AWSScout2
:align: right
***********
Description
***********
Scout2 is a security tool that lets AWS administrators assess their
environment's security posture. Using the AWS API, Scout2 gathers configuration
data for manual inspection and highlights high-risk areas automatically. Rather
than pouring through dozens of pages on the web, Scout2 supplies a clear view of
the attack surface automatically.
**Note:** Scout2 is stable and actively maintained, but a number of features and
internals may change. As such, please bear with us as we find time to work on,
and improve, the tool. Feel free to report a bug with details (*e.g.* console
output using the "--debug" argument), request a new feature, or send a pull
request.
************
Installation
************
Install via `pip`_:
::
$ pip install awsscout2
Install from source:
::
$ git clone https://github.com/nccgroup/Scout2
$ cd Scout2
$ pip install -r requirements.txt
$ python setup.py install
************
Requirements
************
Computing resources
-------------------
Scout2 is a multi-threaded tool that fetches and stores your AWS account's configuration settings in memory during
runtime. It is expected that the tool will run with no issues on any modern laptop or equivalent VM.
**Running Scout2 in a VM with limited computing resources such as a t2.micro instance is not intended and will likely
result in the process being killed.**
Python
------
Scout2 is written in Python and supports the following versions:
* 2.7
* 3.3
* 3.4
* 3.5
* 3.6
AWS Credentials
---------------
To run Scout2, you will need valid AWS credentials (*e.g* Access Key ID and Secret Access Key).
The role, or user account, associated with these credentials requires read-only access for all resources in a number of
services, including but not limited to CloudTrail, EC2, IAM, RDS, Redshift, and S3.
The following AWS Managed Policies can be attached to the principal in order to grant necessary permissions:
* ReadOnlyAccess
* SecurityAudit
Compliance with AWS' Acceptable Use Policy
------------------------------------------
Use of Scout2 does not require AWS users to complete and submit the AWS
Vulnerability / Penetration Testing Request Form. Scout2 only performs AWS API
calls to fetch configuration data and identify security gaps, which is not
considered security scanning as it does not impact AWS' network and
applications.
Usage
-----
After performing a number of AWS API calls, Scout2 will create a local HTML report and open it in the default browser.
Using a computer already configured to use the AWS CLI, boto3, or another AWS SDK, you may use Scout2 using the
following command:
::
$ Scout2
**Note:** EC2 instances with an IAM role fit in this category.
If multiple profiles are configured in your .aws/credentials and .aws/config files, you may specify which credentials
to use with the following command:
::
$ Scout2 --profile <PROFILE_NAME>
If you have a CSV file containing the API access key ID and secret, you may run Scout2 with the following command:
::
$ Scout2 --csv-credentials <CREDENTIALS.CSV>
**********************
Advanced documentation
**********************
The following command will provide the list of available command line options:
::
$ Scout2 --help
For further details, checkout our Wiki pages at https://github.com/nccgroup/Scout2/wiki.
*******
License
*******
GPLv2: See LICENSE.
.. _pip: https://pip.pypa.io/en/stable/index.html
| AWSScout2 | /AWSScout2-3.2.1.tar.gz/AWSScout2-3.2.1/README.rst | README.rst |
# AWS External Account Scanner
> Xenos, is Greek for stranger.
AWSXenos will list all the trust relationships in all the IAM roles, and S3 buckets, in an AWS account and give you a breakdown of all the accounts that have trust relationships to your account. It will also highlight whether the trusts have an [external ID](https://docs.aws.amazon.com/IAM/latest/UserGuide/id_roles_create_for-user_externalid.html) or not.
This tool reports against the [Trusted Relationship Technique](https://attack.mitre.org/techniques/T1199/) of the ATT&CK Framework.
* For the "known" accounts list AWSXenos uses a modified version of [known AWS Account IDs](https://github.com/rupertbg/aws-public-account-ids).
* For the Org accounts list AWSXenos query AWS Organizations.
* AWS Services are classified separately.
* Everything else falls under unknown account
## Example

## Why
Access Analyzer falls short because:
1. You need to enable it in every region.
2. Identified external entities might be known entities. E.g. a trusted third party vendor or a vendor you no longer trust. An Account number is seldom useful.
3. Zone of trust is a fixed set of the AWS organisation. You won’t know if a trust between sandbox->prod has been established.
4. Does not identify AWS Service principals. This is mainly important because of [Wiz's AWSConfig, et al vulnverabilities](http://i.blackhat.com/USA21/Wednesday-Handouts/us-21-Breaking-The-Isolation-Cross-Account-AWS-Vulnerabilities.pdf)
## How to run
### Cli
```sh
pip install AWSXenos
awsxenos --reporttype HTML -w report.html
awsxenos --reporttype JSON -w report.json
```
You will get an HTML and JSON report.
See [example report](example/example.html)
### Library
```python
from awsxenos.scan import Scan
from awsxenos.report import Report
s = Scan()
r = Report(s.findings, s.known_accounts_data)
json_summary = r.JSON_report()
html_summary = r.HTML_report()
```
### IAM Permissions
Permissions required.
```json
{
"Version": "2012-10-17",
"Statement": [
{
"Action": [
"iam:ListRoles"
"organizations:ListAccounts",
"s3:ListAllMyBuckets",
"s3:GetBucketPolicy",
"s3:GetBucketAcl"
],
"Effect": "Allow",
"Resource": "*"
}
]
}
```
## Development
```sh
python3 -m env venv
source /env/bin/activate
pip install -r requirements.txt
```
## I want to add more known accounts
Create a PR or raise an issue. Contributions are welcome.
## Features
- [x] IAM Roles
- [x] S3 Bucket Policies and ACLs
- [x] Use as library
- [x] HTML and JSON output
- [x] Supports AWS Services
## TODO
- [ ] Add support for more resource policies services, e.g. SecretsManager, KSM, SNS, SQS, Lambda
- [ ] Add support for Cognito, RAM
- [ ] Add support for VPCE
| AWSXenos | /AWSXenos-0.0.2.tar.gz/AWSXenos-0.0.2/README.md | README.md |
from collections import defaultdict
from typing import List, Dict, DefaultDict
import json
from jinja2 import Environment, FileSystemLoader # type: ignore
from policyuniverse.arn import ARN # type: ignore
from awsxenos.finding import AccountType, Finding
from awsxenos import package_path
class Report:
def __init__(self, findings: DefaultDict[str, AccountType], account_info: DefaultDict[str, Dict]) -> None:
self.summary = self._summarise(findings, account_info)
def _summarise(
self, findings: DefaultDict[str, AccountType], account_info: DefaultDict[str, Dict]
) -> DefaultDict[str, List]:
summary = defaultdict(list)
for resource, accounttype in findings.items():
# Refactor
# for account_type, principal in finding
if accounttype.known_accounts:
for finding in accounttype.known_accounts:
role_arn = ARN(finding.principal)
summary["known_accounts"].append(
{
"ARN": resource,
"principal": accounttype.known_accounts,
"external_info": account_info[role_arn.account_number],
"external_id": finding.external_id,
}
)
if accounttype.org_accounts:
for finding in accounttype.org_accounts:
role_arn = ARN(finding.principal)
summary["org_accounts"].append(
{
"ARN": resource,
"principal": accounttype.org_accounts,
"external_info": account_info[role_arn.account_number],
}
)
if accounttype.aws_services:
for finding in accounttype.aws_services:
role_arn = ARN(finding.principal)
summary["aws_services"].append(
{
"ARN": resource,
"principal": accounttype.aws_services,
"external_info": account_info[role_arn.tech],
}
)
if accounttype.unknown_accounts:
for finding in accounttype.unknown_accounts:
role_arn = ARN(finding.principal)
summary["unknown_accounts"].append(
{
"ARN": resource,
"principal": accounttype.unknown_accounts,
"external_info": account_info[role_arn.account_number],
"external_id": finding.external_id,
}
)
return summary
def JSON_report(self) -> str:
"""Return the Findings in JSON format
Returns:
str: Return the Findings in JSON format
"""
return json.dumps(self.summary, indent=4, default=str)
def HTML_report(self) -> str:
"""Generate an HTML report based on the template.html
Returns:
str: return HTML
"""
jinja_env = Environment(loader=FileSystemLoader(package_path.resolve().parent))
template = jinja_env.get_template("template.html")
return template.render(summary=self.summary) | AWSXenos | /AWSXenos-0.0.2.tar.gz/AWSXenos-0.0.2/awsxenos/report.py | report.py |
import argparse
from collections import defaultdict
from re import I
from typing import Any, Optional, Dict, List, DefaultDict, Set
import json
import sys
import boto3 # type: ignore
from botocore.exceptions import ClientError # type: ignore
from policyuniverse.arn import ARN # type: ignore
from policyuniverse.policy import Policy # type: ignore
from policyuniverse.statement import Statement, ConditionTuple # type: ignore
from awsxenos.finding import AccountType, Finding
from awsxenos.report import Report
from awsxenos import package_path
class Scan:
def __init__(self, exclude_service: Optional[bool] = True, exclude_aws: Optional[bool] = True) -> None:
self.known_accounts_data = defaultdict(dict) # type: DefaultDict[str, Dict[Any, Any]]
self.findings = defaultdict(AccountType) # type: DefaultDict[str, AccountType]
self._buckets = self.list_account_buckets()
self.roles = self.get_roles(exclude_service, exclude_aws)
self.accounts = self.get_all_accounts()
self.bucket_policies = self.get_bucket_policies()
self.bucket_acls = self.get_bucket_acls()
for resource in ["roles", "bucket_policies", "bucket_acls"]:
if resource != "bucket_acls":
self.findings.update(self.collate_findings(self.accounts, getattr(self, resource)))
else:
self.findings.update(self.collate_acl_findings(self.accounts, getattr(self, resource)))
def get_org_accounts(self) -> DefaultDict[str, Dict]:
"""Get Account Ids from the AWS Organization
Returns:
DefaultDict: Key of Account Ids. Value of other Information
"""
accounts = defaultdict(dict) # type: DefaultDict[str, Dict]
orgs = boto3.client("organizations")
paginator = orgs.get_paginator("list_accounts")
try:
account_iterator = paginator.paginate()
for account_resp in account_iterator:
for account in account_resp["Accounts"]:
accounts[account["Id"]] = account
return accounts
except Exception as e:
print("[!] - Failed to get organization accounts")
print(e)
return accounts
def get_bucket_acls(self) -> DefaultDict[str, List[Dict[Any, Any]]]:
bucket_acls = defaultdict(str)
buckets = self._buckets
s3 = boto3.client("s3")
for bucket in buckets["Buckets"]:
bucket_arn = f'arn:aws:s3:::{bucket["Name"]}'
try:
bucket_acls[bucket_arn] = s3.get_bucket_acl(Bucket=bucket["Name"])["Grants"]
except ClientError as e:
if e.response["Error"]["Code"] == "AccessDenied":
bucket_acls[bucket_arn] = [
{
"Grantee": {"DisplayName": "AccessDenied", "ID": "AccessDenied", "Type": "CanonicalUser"},
"Permission": "FULL_CONTROL",
}
]
else:
print(e)
continue
return bucket_acls
def get_bucket_policies(self) -> DefaultDict[str, Dict[Any, Any]]:
"""Get a dictionary of buckets and their policies from the AWS Account
Returns:
DefaultDict[str, str]: Key of BucketARN, Value of PolicyDocument
"""
bucket_policies = defaultdict(str)
buckets = self._buckets
s3 = boto3.client("s3")
for bucket in buckets["Buckets"]:
bucket_arn = f'arn:aws:s3:::{bucket["Name"]}'
try:
bucket_policies[bucket_arn] = json.loads(s3.get_bucket_policy(Bucket=bucket["Name"])["Policy"])
except ClientError as e:
if e.response["Error"]["Code"] == "AccessDenied":
bucket_policies[bucket_arn] = {
"Version": "2012-10-17",
"Statement": [
{
"Sid": "AccessDeniedOnResource",
"Effect": "Allow",
"Principal": {"AWS": ["arn:aws:iam::111122223333:root"]},
"Action": ["s3:*"],
"Resource": f"{bucket_arn}",
}
],
}
continue
elif e.response["Error"]["Code"] == "NoSuchBucketPolicy":
continue
else:
print(e)
continue
return bucket_policies
def get_roles(
self, exclude_service: Optional[bool] = True, exclude_aws: Optional[bool] = True
) -> DefaultDict[str, Dict[Any, Any]]:
"""Get a dictionary of roles and their policies from the AWS Account
Args:
exclude_service (Optional[bool], optional): exclude roles starting with /service-role/. Defaults to True.
exclude_aws (Optional[bool], optional): exclude roles starting with /aws-service-role/. Defaults to True.
Returns:
DefaultDict[str, str]: Key of RoleNames, Value of AssumeRolePolicyDocument
"""
roles = defaultdict(str)
iam = boto3.client("iam")
paginator = iam.get_paginator("list_roles")
role_iterator = paginator.paginate()
for role_resp in role_iterator:
for role in role_resp["Roles"]:
if role["Path"] == "/service-role/" and exclude_service:
continue
elif role["Path"].startswith("/aws-service-role/") and exclude_aws:
continue
else:
roles[role["Arn"]] = role["AssumeRolePolicyDocument"]
return roles
def list_account_buckets(self) -> Dict[str, Dict[Any, Any]]:
s3 = boto3.client("s3")
return s3.list_buckets()
def get_all_accounts(self) -> DefaultDict[str, Set]:
"""Get all known accounts and from the AWS Organization
Returns:
DefaultDict[str, Set]: Key of account type. Value account ids
"""
accounts = defaultdict(set) # type: DefaultDict[str, Set]
with open(f"{package_path.resolve().parent}/accounts.json", "r") as f:
accounts_file = json.load(f)
for account in accounts_file:
self.known_accounts_data[account["id"]] = account
accounts["known_accounts"] = set(self.known_accounts_data.keys())
# Populate Org accounts
org_accounts = self.get_org_accounts()
aws_canonical_user = self._buckets["Owner"]
# Add to the set of org_accounts
accounts["org_accounts"] = set(org_accounts.keys())
accounts["org_accounts"].add(aws_canonical_user["ID"])
# Combine the metadata
self.known_accounts_data[aws_canonical_user["ID"]] = {"owner": aws_canonical_user["DisplayName"]}
self.known_accounts_data = self.known_accounts_data | org_accounts # type: ignore
return accounts
def collate_acl_findings(
self, accounts: DefaultDict[str, Set], resources: DefaultDict[str, List[Dict[Any, Any]]]
) -> DefaultDict[str, AccountType]:
"""Combine all accounts with all the acls to classify findings
Args:
accounts (DefaultDict[str, Set]): [description]
resources (DefaultDict[str, List[Dict[Any, Any]]]): [description]
Returns:
DefaultDict[str, AccountType]: [description]
"""
findings = defaultdict(AccountType) # type: DefaultDict[str, AccountType]
for resource, grants in resources.items():
for grant in grants:
if grant["Grantee"]["ID"] == self._buckets["Owner"]["ID"]:
continue # Don't add if the ACL is of the same account
elif grant["Grantee"]["ID"] in accounts["known_accounts"]:
findings[resource].known_accounts.append(Finding(principal=grant["Grantee"]["ID"], external_id=True))
elif grant["Grantee"]["ID"] in accounts["org_accounts"]:
findings[resource].org_accounts.append(Finding(principal=grant["Grantee"]["ID"], external_id=True))
else:
findings[resource].unknown_accounts.append(
Finding(principal=grant["Grantee"]["ID"], external_id=True)
)
return findings
def collate_findings(
self, accounts: DefaultDict[str, Set], resources: DefaultDict[str, Dict[Any, Any]]
) -> DefaultDict[str, AccountType]:
"""Combine all accounts with all the resources to classify findings
Args:
accounts (DefaultDict[str, Set]): Key of account type. Value account ids
resources (DefaultDict[str, Dict[Any, Any]]): Key ResourceIdentifier. Value Dict PolicyDocument
Returns:
DefaultDict[str, AccountType]: Key of ARN, Value of AccountType
"""
findings = defaultdict(AccountType) # type: DefaultDict[str, AccountType]
for resource, policy_document in resources.items():
try:
policy = Policy(policy_document)
except:
print(policy_document)
continue
for unparsed_principal in policy.whos_allowed():
try:
principal = ARN(unparsed_principal.value) # type: Any
except Exception as e:
print(e)
findings[resource].known_accounts.append(Finding(principal=unparsed_principal, external_id=True))
continue
# Check if Principal is an AWS Service
if principal.service:
findings[resource].aws_services.append(Finding(principal=principal.arn, external_id=True))
# Check against org_accounts
elif principal.account_number in accounts["org_accounts"]:
findings[resource].org_accounts.append(Finding(principal=principal.arn, external_id=True))
# Check against known external accounts
elif (
principal.account_number in accounts["known_accounts"]
or ConditionTuple(category="saml-endpoint", value="https://signin.aws.amazon.com/saml")
in policy.whos_allowed()
):
sts_set = False
for pstate in policy.statements:
if "sts" in pstate.action_summary():
try:
conditions = [
k.lower() for k in list(pstate.statement["Condition"]["StringEquals"].keys())
]
if "sts:externalid" in conditions:
findings[resource].known_accounts.append(
Finding(principal=principal.arn, external_id=True)
)
except:
findings[resource].known_accounts.append(
Finding(principal=principal.arn, external_id=False)
)
finally:
sts_set = True
break
if not sts_set:
findings[resource].known_accounts.append(Finding(principal=principal.arn, external_id=False))
# Unknown Account
else:
sts_set = False
for pstate in policy.statements:
if "sts" in pstate.action_summary():
try:
conditions = [
k.lower() for k in list(pstate.statement["Condition"]["StringEquals"].keys())
]
if "sts:externalid" in conditions:
findings[resource].unknown_accounts.append(
Finding(principal=principal.arn, external_id=True)
)
except:
findings[resource].unknown_accounts.append(
Finding(principal=principal.arn, external_id=False)
)
finally:
break
if not sts_set:
findings[resource].unknown_accounts.append(Finding(principal=principal.arn, external_id=False))
return findings
def cli():
parser = argparse.ArgumentParser(description="Scan an AWS Account for external trusts")
parser.add_argument(
"--reporttype",
dest="reporttype",
action="store",
default="all",
help="Type of report to generate. JSON or HTML",
)
parser.add_argument(
"--include_service_roles",
dest="service_roles",
action="store_false",
default=False,
help="Include service roles in the report",
)
parser.add_argument(
"--include_aws_service_roles",
dest="aws_service_roles",
action="store_false",
default=False,
help="Include AWS roles in the report",
)
parser.add_argument(
"-w",
"--write-output",
dest="write_output",
action="store",
default=False,
help="Path to write output",
)
args = parser.parse_args()
reporttype = args.reporttype
service_roles = args.service_roles
aws_service_roles = args.aws_service_roles
write_output = args.write_output
s = Scan(service_roles, aws_service_roles)
r = Report(s.findings, s.known_accounts_data)
if reporttype.lower() == "json":
summary = r.JSON_report()
elif reporttype.lower() == "html":
summary = r.HTML_report()
else:
summary = r.JSON_report()
if write_output:
with open(f"{write_output}", "w") as f:
f.write(summary)
sys.stdout.write(summary)
if __name__ == "__main__":
cli() | AWSXenos | /AWSXenos-0.0.2.tar.gz/AWSXenos-0.0.2/awsxenos/scan.py | scan.py |
import inspect
import types
__all__ = ["aliases", "expose", "make_callable", "AWSpiderPlugin"]
EXPOSED_FUNCTIONS = {}
CALLABLE_FUNCTIONS = {}
MEMOIZED_FUNCTIONS = {}
FUNCTION_ALIASES = {}
def aliases(*args):
def decorator(f):
FUNCTION_ALIASES[id(f)] = args
return f
return decorator
def expose(func=None, interval=0, name=None, memoize=False):
if func is not None:
EXPOSED_FUNCTIONS[id(func)] = {"interval":interval, "name":name}
return func
def decorator(f):
EXPOSED_FUNCTIONS[id(f)] = {"interval":interval, "name":name}
return f
return decorator
def make_callable(func=None, interval=0, name=None, memoize=False):
if func is not None:
CALLABLE_FUNCTIONS[id(func)] = {"interval":interval, "name":name}
return func
def decorator(f):
CALLABLE_FUNCTIONS[id(f)] = {"interval":interval, "name":name}
return f
return decorator
class AWSpiderPlugin(object):
def __init__(self, spider):
self.spider = spider
check_method = lambda x:isinstance(x[1], types.MethodType)
instance_methods = filter(check_method, inspect.getmembers(self))
for instance_method in instance_methods:
instance_id = id(instance_method[1].__func__)
if instance_id in EXPOSED_FUNCTIONS:
self.spider.expose(
instance_method[1],
interval=EXPOSED_FUNCTIONS[instance_id]["interval"],
name=EXPOSED_FUNCTIONS[instance_id]["name"])
if instance_id in FUNCTION_ALIASES:
for name in FUNCTION_ALIASES[instance_id]:
self.spider.expose(
instance_method[1],
interval=EXPOSED_FUNCTIONS[instance_id]["interval"],
name=name)
if instance_id in CALLABLE_FUNCTIONS:
self.spider.expose(
instance_method[1],
interval=CALLABLE_FUNCTIONS[instance_id]["interval"],
name=CALLABLE_FUNCTIONS[instance_id]["name"])
if instance_id in FUNCTION_ALIASES:
for name in CALLABLE_FUNCTIONS[instance_id]:
self.spider.expose(
instance_method[1],
interval=CALLABLE_FUNCTIONS[instance_id]["interval"],
name=name)
def setReservationFastCache(self, uuid, data):
return self.spider.setReservationFastCache(uuid, data)
def setReservationCache(self, uuid, data):
return self.spider.setReservationCache(uuid, data)
def getPage(self, *args, **kwargs):
return self.spider.getPage(*args, **kwargs) | AWSpider | /AWSpider-0.3.2.12.tar.gz/AWSpider-0.3.2.12/awspider/plugin.py | plugin.py |
from twisted.internet.defer import DeferredList
from twisted.web.client import HTTPClientFactory, _parse
from twisted.internet import reactor
import xml.etree.cElementTree as ET
import logging
logger = logging.getLogger("main")
import random
import socket
def getPage( url, method='GET', postdata=None, headers=None, agent="AWSpider", timeout=60, cookies=None, followRedirect=1 ):
scheme, host, port, path = _parse(url)
factory = HTTPClientFactory(
url,
method=method,
postdata=postdata,
headers=headers,
agent=agent,
timeout=timeout,
cookies=cookies,
followRedirect=followRedirect
)
if scheme == 'https':
from twisted.internet import ssl
contextFactory = ssl.ClientContextFactory()
reactor.connectSSL(host, port, factory, contextFactory, timeout=timeout)
else:
reactor.connectTCP(host, port, factory, timeout=timeout)
return factory.deferred
class NetworkAddressGetter():
local_ip = None
public_ip = None
def __init__( self ):
self.ip_functions = [self.getDomaintools, self.getIPPages]
random.shuffle(self.ip_functions)
def __call__( self ):
d = self.getAmazonIPs()
d.addCallback( self._getAmazonIPsCallback )
d.addErrback( self.getPublicIP )
return d
def getAmazonIPs( self ):
logger.debug( "Getting local IP from Amazon." )
a = getPage( "http://169.254.169.254/2009-04-04/meta-data/local-ipv4", timeout=5 )
logger.debug( "Getting public IP from Amazon." )
b = getPage( "http://169.254.169.254/2009-04-04/meta-data/public-ipv4", timeout=5 )
d = DeferredList([a,b], consumeErrors=True)
return d
def _getAmazonIPsCallback( self, data ):
if data[0][0] == True:
self.local_ip = data[0][1]
logger.debug( "Got local IP %s from Amazon." % self.local_ip )
else:
logger.debug( "Could not get local IP from Amazon." )
if data[1][0] == True:
public_ip = data[1][1]
logger.debug( "Got public IP %s from Amazon." % public_ip )
response = {}
if self.local_ip is not None:
response["local_ip"] = self.local_ip
response["public_ip"] = public_ip
return response
else:
logger.debug( "Could not get public IP from Amazon." )
raise Exception( "Could not get public IP from Amazon." )
def getPublicIP( self, error=None ):
if len(self.ip_functions) > 0:
func = self.ip_functions.pop()
d = func()
d.addCallback( self._getPublicIPCallback )
d.addErrback( self.getPublicIP )
return d
else:
logger.error( "Unable to get public IP address. Check your network connection" )
response = {}
if self.local_ip is not None:
response["local_ip"] = self.local_ip
else:
response["local_ip"] = socket.gethostbyname(socket.gethostname())
return response
def _getPublicIPCallback( self, public_ip ):
response = {}
response["public_ip"] = public_ip
if self.local_ip is not None:
response["local_ip"] = self.local_ip
else:
response["local_ip"] = socket.gethostbyname(socket.gethostname())
return response
def getIPPages(self):
logger.debug( "Getting public IP from ippages.com." )
d = getPage( "http://www.ippages.com/xml/", timeout=5 )
d.addCallback( self._getIPPagesCallback )
return d
def _getIPPagesCallback(self, data ):
domaintools_xml = ET.XML( data )
public_ip = domaintools_xml.find("ip").text
logger.debug( "Got public IP %s from ippages.com." % public_ip )
return public_ip
def getDomaintools(self):
logger.debug( "Getting public IP from domaintools.com." )
d = getPage( "http://ip-address.domaintools.com/myip.xml", timeout=5 )
d.addCallback( self._getDomaintoolsCallback )
return d
def _getDomaintoolsCallback(self, data):
domaintools_xml = ET.XML( data )
public_ip = domaintools_xml.find("ip_address").text
logger.debug( "Got public IP %s from domaintools.com." % public_ip )
return public_ip
def getNetworkAddress():
n = NetworkAddressGetter()
d = n()
d.addCallback( _getNetworkAddressCallback )
return d
def _getNetworkAddressCallback( data ):
return data
if __name__ == "__main__":
import logging.handlers
handler = logging.StreamHandler()
formatter = logging.Formatter("%(levelname)s: %(message)s %(pathname)s:%(lineno)d")
handler.setFormatter(formatter)
logger.addHandler(handler)
logger.setLevel(logging.DEBUG)
reactor.callWhenRunning( getNetworkAddress )
reactor.run() | AWSpider | /AWSpider-0.3.2.12.tar.gz/AWSpider-0.3.2.12/awspider/networkaddress.py | networkaddress.py |
import cPickle
import twisted.python.failure
import datetime
import dateutil.parser
import hashlib
import logging
import time
import copy
from twisted.internet.defer import maybeDeferred
from .requestqueuer import RequestQueuer
from .unicodeconverter import convertToUTF8, convertToUnicode
from .exceptions import StaleContentException
class ReportedFailure(twisted.python.failure.Failure):
pass
# A UTC class.
class CoordinatedUniversalTime(datetime.tzinfo):
ZERO = datetime.timedelta(0)
def utcoffset(self, dt):
return self.ZERO
def tzname(self, dt):
return "UTC"
def dst(self, dt):
return self.ZERO
UTC = CoordinatedUniversalTime()
LOGGER = logging.getLogger("main")
class PageGetter:
def __init__(self,
s3,
aws_s3_http_cache_bucket,
time_offset=0,
rq=None):
"""
Create an S3 based HTTP cache.
**Arguments:**
* *s3* -- S3 client object.
* *aws_s3_http_cache_bucket* -- S3 bucket to use for the HTTP cache.
**Keyword arguments:**
* *rq* -- Request Queuer object. (Default ``None``)
"""
self.s3 = s3
self.aws_s3_http_cache_bucket = aws_s3_http_cache_bucket
self.time_offset = time_offset
if rq is None:
self.rq = RequestQueuer()
else:
self.rq = rq
def clearCache(self):
"""
Clear the S3 bucket containing the S3 cache.
"""
d = self.s3.emptyBucket(self.aws_s3_http_cache_bucket)
return d
def getPage(self,
url,
method='GET',
postdata=None,
headers=None,
agent="AWSpider",
timeout=60,
cookies=None,
follow_redirect=1,
prioritize=False,
hash_url=None,
cache=0,
content_sha1=None,
confirm_cache_write=False):
"""
Make a cached HTTP Request.
**Arguments:**
* *url* -- URL for the request.
**Keyword arguments:**
* *method* -- HTTP request method. (Default ``'GET'``)
* *postdata* -- Dictionary of strings to post with the request.
(Default ``None``)
* *headers* -- Dictionary of strings to send as request headers.
(Default ``None``)
* *agent* -- User agent to send with request. (Default
``'AWSpider'``)
* *timeout* -- Request timeout, in seconds. (Default ``60``)
* *cookies* -- Dictionary of strings to send as request cookies.
(Default ``None``).
* *follow_redirect* -- Boolean switch to follow HTTP redirects.
(Default ``True``)
* *prioritize* -- Move this request to the front of the request
queue. (Default ``False``)
* *hash_url* -- URL string used to indicate a common resource.
Example: "http://digg.com" and "http://www.digg.com" could both
use hash_url, "http://digg.com" (Default ``None``)
* *cache* -- Cache mode. ``1``, immediately return contents of
cache if available. ``0``, check resource, return cache if not
stale. ``-1``, ignore cache. (Default ``0``)
* *content_sha1* -- SHA-1 hash of content. If this matches the
hash of data returned by the resource, raises a
StaleContentException.
* *confirm_cache_write* -- Wait to confirm cache write before returning.
"""
request_kwargs = {
"method":method.upper(),
"postdata":postdata,
"headers":headers,
"agent":agent,
"timeout":timeout,
"cookies":cookies,
"follow_redirect":follow_redirect,
"prioritize":prioritize}
cache = int(cache)
if cache not in [-1,0,1]:
raise Exception("Unknown caching mode.")
if not isinstance(url, str):
url = convertToUTF8(url)
if hash_url is not None and not isinstance(hash_url, str):
hash_url = convertToUTF8(hash_url)
# Create request_hash to serve as a cache key from
# either the URL or user-provided hash_url.
if hash_url is None:
request_hash = hashlib.sha1(cPickle.dumps([
url,
headers,
agent,
cookies])).hexdigest()
else:
request_hash = hashlib.sha1(cPickle.dumps([
hash_url,
headers,
agent,
cookies])).hexdigest()
if request_kwargs["method"] != "GET":
d = self.rq.getPage(url, **request_kwargs)
d.addCallback(self._checkForStaleContent, content_sha1, request_hash)
return d
if cache == -1:
# Cache mode -1. Bypass cache entirely.
LOGGER.debug("Getting request %s for URL %s." % (request_hash, url))
d = self.rq.getPage(url, **request_kwargs)
d.addCallback(self._returnFreshData,
request_hash,
url,
confirm_cache_write)
d.addErrback(self._requestWithNoCacheHeadersErrback,
request_hash,
url,
confirm_cache_write,
request_kwargs)
d.addCallback(self._checkForStaleContent, content_sha1, request_hash)
return d
elif cache == 0:
# Cache mode 0. Check cache, send cached headers, possibly use cached data.
LOGGER.debug("Checking S3 Head object request %s for URL %s." % (request_hash, url))
# Check if there is a cache entry, return headers.
d = self.s3.headObject(self.aws_s3_http_cache_bucket, request_hash)
d.addCallback(self._checkCacheHeaders,
request_hash,
url,
request_kwargs,
confirm_cache_write,
content_sha1)
d.addErrback(self._requestWithNoCacheHeaders,
request_hash,
url,
request_kwargs,
confirm_cache_write)
d.addCallback(self._checkForStaleContent, content_sha1, request_hash)
return d
elif cache == 1:
# Cache mode 1. Use cache immediately, if possible.
LOGGER.debug("Getting S3 object request %s for URL %s." % (request_hash, url))
d = self.s3.getObject(self.aws_s3_http_cache_bucket, request_hash)
d.addCallback(self._returnCachedData, request_hash)
d.addErrback(self._requestWithNoCacheHeaders,
request_hash,
url,
request_kwargs,
confirm_cache_write)
d.addCallback(self._checkForStaleContent, content_sha1, request_hash)
return d
def _checkCacheHeaders(self,
data,
request_hash,
url,
request_kwargs,
confirm_cache_write,
content_sha1):
LOGGER.debug("Got S3 Head object request %s for URL %s." % (request_hash, url))
http_history = {}
#if "content-length" in data["headers"] and int(data["headers"]["content-length"][0]) == 0:
# raise Exception("Zero Content length, do not use as cache.")
if "content-sha1" in data["headers"]:
http_history["content-sha1"] = data["headers"]["content-sha1"][0]
# Filter?
if "request-failures" in data["headers"]:
http_history["request-failures"] = data["headers"]["request-failures"][0].split(",")
if "content-changes" in data["headers"]:
http_history["content-changes"] = data["headers"]["content-changes"][0].split(",")
# If cached data is not stale, return it.
if "cache-expires" in data["headers"]:
expires = dateutil.parser.parse(data["headers"]["cache-expires"][0])
now = datetime.datetime.now(UTC)
if expires > now:
if "content-sha1" in http_history and http_history["content-sha1"] == content_sha1:
LOGGER.debug("Raising StaleContentException (1) on %s" % request_hash)
raise StaleContentException()
LOGGER.debug("Cached data %s for URL %s is not stale. Getting from S3." % (request_hash, url))
d = self.s3.getObject(self.aws_s3_http_cache_bucket, request_hash)
d.addCallback(self._returnCachedData, request_hash)
d.addErrback(
self._requestWithNoCacheHeaders,
request_hash,
url,
request_kwargs,
confirm_cache_write,
http_history=http_history)
return d
modified_request_kwargs = copy.deepcopy(request_kwargs)
# At this point, cached data may or may not be stale.
# If cached data has an etag header, include it in the request.
if "cache-etag" in data["headers"]:
modified_request_kwargs["etag"] = data["headers"]["cache-etag"][0]
# If cached data has a last-modified header, include it in the request.
if "cache-last-modified" in data["headers"]:
modified_request_kwargs["last_modified"] = data["headers"]["cache-last-modified"][0]
LOGGER.debug("Requesting %s for URL %s with etag and last-modified headers." % (request_hash, url))
# Make the request. A callback means a 20x response. An errback
# could be a 30x response, indicating the cache is not stale.
d = self.rq.getPage(url, **modified_request_kwargs)
d.addCallback(
self._returnFreshData,
request_hash,
url,
confirm_cache_write,
http_history=http_history)
d.addErrback(
self._handleRequestWithCacheHeadersError,
request_hash,
url,
request_kwargs,
confirm_cache_write,
data,
http_history,
content_sha1)
return d
def _returnFreshData(self,
data,
request_hash,
url,
confirm_cache_write,
http_history=None):
LOGGER.debug("Got request %s for URL %s." % (request_hash, url))
data["pagegetter-cache-hit"] = False
data["content-sha1"] = hashlib.sha1(data["response"]).hexdigest()
if http_history is not None and "content-sha1" in http_history:
if http_history["content-sha1"] == data["content-sha1"]:
return data
d = maybeDeferred(self._storeData,
data,
request_hash,
confirm_cache_write,
http_history=http_history)
d.addErrback(self._storeDataErrback, data, request_hash)
return d
def _requestWithNoCacheHeaders(self,
error,
request_hash,
url,
request_kwargs,
confirm_cache_write,
http_history=None):
try:
error.raiseException()
except StaleContentException, e:
LOGGER.debug("Raising StaleContentException (2) on %s" % request_hash)
raise StaleContentException()
except Exception, e:
pass
# No header stored in the cache. Make the request.
LOGGER.debug("Unable to find header for request %s on S3, fetching from %s." % (request_hash, url))
d = self.rq.getPage(url, **request_kwargs)
d.addCallback(
self._returnFreshData,
request_hash,
url,
confirm_cache_write,
http_history=http_history)
d.addErrback(
self._requestWithNoCacheHeadersErrback,
request_hash,
url,
confirm_cache_write,
request_kwargs,
http_history=http_history)
return d
def _requestWithNoCacheHeadersErrback(self,
error,
request_hash,
url,
confirm_cache_write,
request_kwargs,
http_history=None):
LOGGER.error(error.value.__dict__)
LOGGER.error("Unable to get request %s for URL %s.\n%s" % (
request_hash,
url,
error))
if http_history is None:
http_history = {}
if "request-failures" not in http_history:
http_history["request-failures"] = [str(int(self.time_offset + time.time()))]
else:
http_history["request-failures"].append(str(int(self.time_offset + time.time())))
http_history["request-failures"] = http_history["request-failures"][-3:]
LOGGER.debug("Writing data for failed request %s to S3." % request_hash)
headers = {}
headers["request-failures"] = ",".join(http_history["request-failures"])
d = self.s3.putObject(
self.aws_s3_http_cache_bucket,
request_hash,
"",
content_type="text/plain",
headers=headers)
if confirm_cache_write:
d.addCallback(self._requestWithNoCacheHeadersErrbackCallback, error)
return d
return error
def _requestWithNoCacheHeadersErrbackCallback(self, data, error):
return error
def _handleRequestWithCacheHeadersError(self,
error,
request_hash,
url,
request_kwargs,
confirm_cache_write,
data,
http_history,
content_sha1):
if error.value.status == "304":
if "content-sha1" in http_history and http_history["content-sha1"] == content_sha1:
LOGGER.debug("Raising StaleContentException (3) on %s" % request_hash)
raise StaleContentException()
LOGGER.debug("Request %s for URL %s hasn't been modified since it was last downloaded. Getting data from S3." % (request_hash, url))
d = self.s3.getObject(self.aws_s3_http_cache_bucket, request_hash)
d.addCallback(self._returnCachedData, request_hash)
d.addErrback(
self._requestWithNoCacheHeaders,
request_hash,
url,
request_kwargs,
confirm_cache_write,
http_history=http_history)
return d
else:
if http_history is None:
http_history = {}
if "request-failures" not in http_history:
http_history["request-failures"] = [str(int(self.time_offset + time.time()))]
else:
http_history["request-failures"].append(str(int(self.time_offset + time.time())))
http_history["request-failures"] = http_history["request-failures"][-3:]
LOGGER.debug("Writing data for failed request %s to S3. %s" % (request_hash, error))
headers = {}
for key in data["headers"]:
headers[key] = data["headers"][key][0]
headers["request-failures"] = ",".join(http_history["request-failures"])
d = self.s3.putObject(
self.aws_s3_http_cache_bucket,
request_hash,
data["response"],
content_type=data["headers"]["content-type"][0],
headers=headers)
if confirm_cache_write:
d.addCallback(self._handleRequestWithCacheHeadersErrorCallback, error)
return d
return ReportedFailure(error)
def _handleRequestWithCacheHeadersErrorCallback(self, data, error):
return ReportedFailure(error)
def _returnCachedData(self, data, request_hash):
LOGGER.debug("Got request %s from S3." % (request_hash))
data["pagegetter-cache-hit"] = True
data["status"] = 304
data["message"] = "Not Modified"
if "content-sha1" in data["headers"]:
data["content-sha1"] = data["headers"]["content-sha1"][0]
del data["headers"]["content-sha1"]
else:
data["content-sha1"] = hashlib.sha1(data["response"]).hexdigest()
if "cache-expires" in data["headers"]:
data["headers"]["expires"] = data["headers"]["cache-expires"]
del data["headers"]["cache-expires"]
if "cache-etag" in data["headers"]:
data["headers"]["etag"] = data["headers"]["cache-etag"]
del data["headers"]["cache-etag"]
if "cache-last-modified" in data["headers"]:
data["headers"]["last-modified"] = data["headers"]["cache-last-modified"]
del data["headers"]["cache-last-modified"]
return data
def _storeData(self,
data,
request_hash,
confirm_cache_write,
http_history=None):
if len(data["response"]) == 0:
return self._storeDataErrback(Failure(exc_value=Exception("Response data is of length 0")), response_data, request_hash)
#data["content-sha1"] = hashlib.sha1(data["response"]).hexdigest()
if http_history is None:
http_history = {}
if "content-sha1" not in http_history:
http_history["content-sha1"] = data["content-sha1"]
if "content-changes" not in http_history:
http_history["content-changes"] = []
if data["content-sha1"] != http_history["content-sha1"]:
http_history["content-changes"].append(str(int(self.time_offset + time.time())))
http_history["content-changes"] = http_history["content-changes"][-10:]
LOGGER.debug("Writing data for request %s to S3." % request_hash)
headers = {}
http_history["content-changes"] = filter(lambda x:len(x) > 0, http_history["content-changes"])
headers["content-changes"] = ",".join(http_history["content-changes"])
headers["content-sha1"] = data["content-sha1"]
if "cache-control" in data["headers"]:
if "no-cache" in data["headers"]["cache-control"][0]:
return data
if "expires" in data["headers"]:
headers["cache-expires"] = data["headers"]["expires"][0]
if "etag" in data["headers"]:
headers["cache-etag"] = data["headers"]["etag"][0]
if "last-modified" in data["headers"]:
headers["cache-last-modified"] = data["headers"]["last-modified"][0]
if "content-type" in data["headers"]:
content_type = data["headers"]["content-type"][0]
d = self.s3.putObject(
self.aws_s3_http_cache_bucket,
request_hash,
data["response"],
content_type=content_type,
headers=headers)
if confirm_cache_write:
d.addCallback(self._storeDataCallback, data)
d.addErrback(self._storeDataErrback, data, request_hash)
return d
return data
def _storeDataCallback(self, data, response_data):
return response_data
def _storeDataErrback(self, error, response_data, request_hash):
LOGGER.error("Error storing data for %s" % (request_hash))
return response_data
def _checkForStaleContent(self, data, content_sha1, request_hash):
if "content-sha1" not in data:
data["content-sha1"] = hashlib.sha1(data["response"]).hexdigest()
if content_sha1 == data["content-sha1"]:
LOGGER.debug("Raising StaleContentException (4) on %s" % request_hash)
raise StaleContentException(content_sha1)
else:
return data | AWSpider | /AWSpider-0.3.2.12.tar.gz/AWSpider-0.3.2.12/awspider/pagegetter.py | pagegetter.py |
import codecs
#######################################################
#
# Based on Beautiful Soup's Unicode, Dammit
# http://www.crummy.com/software/BeautifulSoup/
#
#######################################################
# Autodetects character encodings.
# Download from http://chardet.feedparser.org/
try:
import chardet
# import chardet.constants
# chardet.constants._debug = 1
except ImportError:
chardet = None
# cjkcodecs and iconv_codec make Python know about more character encodings.
# Both are available from http://cjkpython.i18n.org/
# They're built in if you use Python 2.4.
try:
import cjkcodecs.aliases
except ImportError:
pass
try:
import iconv_codec
except ImportError:
pass
def convertToUnicode( s ):
s = UnicodeConverter( s ).unicode
if isinstance( s, unicode ):
return s
else:
return None
def convertToUTF8( s ):
s = UnicodeConverter( s ).unicode
if isinstance( s, unicode ):
return s.encode("utf-8")
else:
return None
class UnicodeConverter:
"""A class for detecting the encoding of a *ML document and
converting it to a Unicode string. If the source encoding is
windows-1252, can replace MS smart quotes with their HTML or XML
equivalents."""
# This dictionary maps commonly seen values for "charset" in HTML
# meta tags to the corresponding Python codec names. It only covers
# values that aren't in Python's aliases and can't be determined
# by the heuristics in find_codec.
CHARSET_ALIASES = { "macintosh" : "mac-roman",
"x-sjis" : "shift-jis" }
def __init__(self, markup):
self.markup = markup
documentEncoding = None
sniffedEncoding = None
self.triedEncodings = []
if markup == '' or isinstance(markup, unicode):
self.originalEncoding = None
self.unicode = unicode(markup)
return
u = None
# If no luck and we have auto-detection library, try that:
if not u and chardet and not isinstance(self.markup, unicode):
u = self._convertFrom(chardet.detect(self.markup)['encoding'])
# As a last resort, try utf-8 and windows-1252:
if not u:
for proposed_encoding in ("utf-8", "windows-1252"):
u = self._convertFrom(proposed_encoding)
if u: break
self.unicode = u
if not u: self.originalEncoding = None
def _subMSChar(self, orig):
"""Changes a MS smart quote character to an XML or HTML
entity."""
sub = self.MS_CHARS.get(orig)
if type(sub) == types.TupleType:
if self.smartQuotesTo == 'xml':
sub = '&#x%s;' % sub[1]
else:
sub = '&%s;' % sub[0]
return sub
def _convertFrom(self, proposed):
proposed = self.find_codec(proposed)
if not proposed or proposed in self.triedEncodings:
return None
self.triedEncodings.append(proposed)
markup = self.markup
try:
# print "Trying to convert document to %s" % proposed
u = self._toUnicode(markup, proposed)
self.markup = u
self.originalEncoding = proposed
except Exception, e:
# print "That didn't work!"
# print e
return None
#print "Correct encoding: %s" % proposed
return self.markup
def _toUnicode(self, data, encoding):
'''Given a string and its encoding, decodes the string into Unicode.
encoding is a string recognized by encodings.aliases'''
# strip Byte Order Mark (if present)
if (len(data) >= 4) and (data[:2] == '\xfe\xff') \
and (data[2:4] != '\x00\x00'):
encoding = 'utf-16be'
data = data[2:]
elif (len(data) >= 4) and (data[:2] == '\xff\xfe') \
and (data[2:4] != '\x00\x00'):
encoding = 'utf-16le'
data = data[2:]
elif data[:3] == '\xef\xbb\xbf':
encoding = 'utf-8'
data = data[3:]
elif data[:4] == '\x00\x00\xfe\xff':
encoding = 'utf-32be'
data = data[4:]
elif data[:4] == '\xff\xfe\x00\x00':
encoding = 'utf-32le'
data = data[4:]
newdata = unicode(data, encoding)
return newdata
def find_codec(self, charset):
return self._codec(self.CHARSET_ALIASES.get(charset, charset)) \
or (charset and self._codec(charset.replace("-", ""))) \
or (charset and self._codec(charset.replace("-", "_"))) \
or charset
def _codec(self, charset):
if not charset: return charset
codec = None
try:
codecs.lookup(charset)
codec = charset
except (LookupError, ValueError):
pass
return codec
EBCDIC_TO_ASCII_MAP = None
def _ebcdic_to_ascii(self, s):
c = self.__class__
if not c.EBCDIC_TO_ASCII_MAP:
emap = (0,1,2,3,156,9,134,127,151,141,142,11,12,13,14,15,
16,17,18,19,157,133,8,135,24,25,146,143,28,29,30,31,
128,129,130,131,132,10,23,27,136,137,138,139,140,5,6,7,
144,145,22,147,148,149,150,4,152,153,154,155,20,21,158,26,
32,160,161,162,163,164,165,166,167,168,91,46,60,40,43,33,
38,169,170,171,172,173,174,175,176,177,93,36,42,41,59,94,
45,47,178,179,180,181,182,183,184,185,124,44,37,95,62,63,
186,187,188,189,190,191,192,193,194,96,58,35,64,39,61,34,
195,97,98,99,100,101,102,103,104,105,196,197,198,199,200,
201,202,106,107,108,109,110,111,112,113,114,203,204,205,
206,207,208,209,126,115,116,117,118,119,120,121,122,210,
211,212,213,214,215,216,217,218,219,220,221,222,223,224,
225,226,227,228,229,230,231,123,65,66,67,68,69,70,71,72,
73,232,233,234,235,236,237,125,74,75,76,77,78,79,80,81,
82,238,239,240,241,242,243,92,159,83,84,85,86,87,88,89,
90,244,245,246,247,248,249,48,49,50,51,52,53,54,55,56,57,
250,251,252,253,254,255)
import string
c.EBCDIC_TO_ASCII_MAP = string.maketrans( \
''.join(map(chr, range(256))), ''.join(map(chr, emap)))
return s.translate(c.EBCDIC_TO_ASCII_MAP)
MS_CHARS = { '\x80' : ('euro', '20AC'),
'\x81' : ' ',
'\x82' : ('sbquo', '201A'),
'\x83' : ('fnof', '192'),
'\x84' : ('bdquo', '201E'),
'\x85' : ('hellip', '2026'),
'\x86' : ('dagger', '2020'),
'\x87' : ('Dagger', '2021'),
'\x88' : ('circ', '2C6'),
'\x89' : ('permil', '2030'),
'\x8A' : ('Scaron', '160'),
'\x8B' : ('lsaquo', '2039'),
'\x8C' : ('OElig', '152'),
'\x8D' : '?',
'\x8E' : ('#x17D', '17D'),
'\x8F' : '?',
'\x90' : '?',
'\x91' : ('lsquo', '2018'),
'\x92' : ('rsquo', '2019'),
'\x93' : ('ldquo', '201C'),
'\x94' : ('rdquo', '201D'),
'\x95' : ('bull', '2022'),
'\x96' : ('ndash', '2013'),
'\x97' : ('mdash', '2014'),
'\x98' : ('tilde', '2DC'),
'\x99' : ('trade', '2122'),
'\x9a' : ('scaron', '161'),
'\x9b' : ('rsaquo', '203A'),
'\x9c' : ('oelig', '153'),
'\x9d' : '?',
'\x9e' : ('#x17E', '17E'),
'\x9f' : ('Yuml', ''),} | AWSpider | /AWSpider-0.3.2.12.tar.gz/AWSpider-0.3.2.12/awspider/unicodeconverter.py | unicodeconverter.py |
from twisted.internet import reactor
from twisted.internet.protocol import ClientCreator
from twisted.conch.telnet import Telnet
from twisted.internet.defer import Deferred
import datetime
import time
import calendar
import random
import logging
logger = logging.getLogger("main")
timeservers=["time.nist.gov", "time-a.nist.gov", "time-b.nist.gov", "time-nw.nist.gov", "nist1-ny.WiTime.net", "nist1-dc.WiTime.net", "nist1.aol-va.symmetricom.com", "nist1.columbiacountyga.gov", "nist.expertsmi.com", "nist.netservicesgroup.com", "time-a.timefreq.bldrdoc.gov", "time-c.timefreq.bldrdoc.gov", "utcnist.colorado.edu", "utcnist2.colorado.edu", "ntp-nist.ldsbc.edu", "nist1.aol-ca.symmetricom.com", "nist1.symmetricom.com", "nist1-sj.WiTime.net", "nist1-la.WiTime.net"]
class SimpleTelnet(Telnet):
def __init__( self, *args, **kwargs ):
self.deferred = Deferred()
self.data = []
Telnet.__init__(self, *args, **kwargs )
def dataReceived(self, data):
self.data.append( data )
def connectionLost( self, reason ):
self.deferred.callback( "".join(self.data) )
def getTimeOffset():
client = ClientCreator(reactor, SimpleTelnet)
server = timeservers.pop(0)
logger.debug( "Requesting time from %s." % server )
d = client.connectTCP(server, 13, timeout=5)
d.addCallback( _getTimeOffsetCallback, server )
d.addErrback( _getTimeOffsetErrback, 0 )
return d
def _getTimeOffsetErrback( error, count ):
if count < 5:
client = ClientCreator(reactor, SimpleTelnet)
server = timeservers.pop()
logger.debug( "Attempt %s failed, requesting time from %s." % (count + 1, server) )
d = client.connectTCP(server, 13, timeout=5)
d.addCallback( _getTimeOffsetCallback, server )
d.addErrback( _getTimeOffsetErrback, count + 1 )
return d
else:
logger.debug( "Could not fetch time after %s attempts." % count )
return error
def _getTimeOffsetCallback( simple_telnet, server ):
logger.debug( "Connected to time server %s." % server )
simple_telnet.deferred.addCallback( _getTimeOffsetCallback2, server )
return simple_telnet.deferred
def _getTimeOffsetCallback2( data, server ):
logger.debug( "Got time from %s." % server )
t = datetime.datetime(
2000 + int(data[7:9]),
int(data[10:12]),
int(data[13:15]),
int(data[16:18]),
int(data[19:21]),
int(data[22:24]) )
offset = calendar.timegm( t.timetuple() ) - time.time()
return offset | AWSpider | /AWSpider-0.3.2.12.tar.gz/AWSpider-0.3.2.12/awspider/timeoffset.py | timeoffset.py |
import urllib
import time
from twisted.internet.defer import Deferred
from twisted.internet import reactor, ssl
from twisted.web.client import HTTPClientFactory, _parse
import dateutil.parser
from .unicodeconverter import convertToUTF8
from OpenSSL import SSL
import logging
LOGGER = logging.getLogger("main")
class AllCipherSSLClientContextFactory(ssl.ClientContextFactory):
"""A context factory for SSL clients that uses all ciphers."""
def getContext(self):
context = SSL.Context(self.method)
context.set_cipher_list("ALL")
return context
class RequestQueuer(object):
"""
HTTP Request Queuer
"""
# Dictionary of lists of pending requests, by host
pending_reqs = {}
# Dictonary of timestamps - via time() - of last requests, by host
last_req = {}
# Dictonary of integer counts of active requests, by host
active_reqs = {}
# Dictionary of user specified minimum request intervals, by host
min_req_interval_per_hosts = {}
max_reqs_per_hosts_per_sec = {}
# Dictionary of user specified maximum simultaneous requests, by host
max_simul_reqs_per_hosts = {}
def __init__(self, max_simultaneous_requests=50,
max_requests_per_host_per_second=1,
max_simultaneous_requests_per_host=5):
"""
Set the maximum number of simultaneous requests for a particular host.
**Keyword arguments:**
* *max_simultaneous_requests* -- Maximum number of simultaneous
requests RequestQueuer should make across all hosts. (Default 50)
* *max_requests_per_host_per_second* -- Maximum number of requests
per host per second. If set to 1, RequestQueuer will not make more
than 1 request per second to a host. If set to 0, RequestQueuer
will not limit the request rate. Can be overridden for an
individual host using ``setHostMaxRequestsPerSecond()`` (Default 1)
* *max_simultaneous_requests_per_host* -- Maximum number of
simultaneous requests per host. If set to 1, RequestQueuer will
not make more than 1 simultaneous request per host. If set to 0,
RequestQueuer will not limit the number of simultaneous requests.
Can be overridden for an individual host using
``setHostMaxSimultaneousRequests()`` (Default 5)
"""
if max_simultaneous_requests == 0:
self.max_simul_reqs = 100000
else:
self.max_simul_reqs = int(max_simultaneous_requests)
# self.min_req_interval_per_host is the global minimum request
# interval. Can be overridden by self.min_req_interval_per_hosts[].
self.max_reqs_per_host_per_sec = max_requests_per_host_per_second
if max_requests_per_host_per_second == 0:
self.min_req_interval_per_host = 0
else:
max_req_per_host_per_sec = float(max_requests_per_host_per_second)
self.min_req_interval_per_host = 1.0 / max_req_per_host_per_sec
# self.max_simul_reqs_per_host is the global maximum simultaneous
# request count. Can be overridden by self.max_simul_reqs_per_hosts[].
if max_simultaneous_requests_per_host == 0:
self.max_simul_reqs_per_host = self.max_simul_reqs
else:
self.max_simul_reqs_per_host = int(max_simultaneous_requests_per_host)
def getPending(self):
"""
Return the number of pending requests.
"""
return sum([len(x) for x in self.pending_reqs.values()])
def getActive(self):
"""
Return the number of active requests.
"""
return sum(self.active_reqs.values())
def getActiveRequestsByHost(self):
"""
Return a dictionary of the number of active requests by host.
"""
return self.active_reqs
def getPendingRequestsByHost(self):
"""
Return a dictionary of the number of pending requests by host.
"""
reqs = [(x[0], len(x[1])) for x in self.pending_reqs.items()]
return dict(reqs)
def setHostMaxRequestsPerSecond(self, host, max_requests_per_second):
"""
Set the maximum number of requests per second for a particular host.
**Arguments:**
* *host* -- Hostname. (Example, ``"google.com"``)
* *max_requests_per_second* -- Maximum number of requests to the
host per second. If set to 1, RequestQueuer will not make more
than 1 request per second to the host. If set to 0, RequestQueuer
will not limit the request rate to the host.
"""
self.max_reqs_per_hosts_per_sec[host] = max_requests_per_second
if max_requests_per_second == 0:
self.min_req_interval_per_hosts[host] = 0
else:
min_req_interval = 1.0 / float(max_requests_per_second)
self.min_req_interval_per_hosts[host] = min_req_interval
def getHostMaxRequestsPerSecond(self, host):
"""
Get the maximum number of requests per second for a particular host.
**Arguments:**
* *host* -- Hostname. (Example, ``"google.com"``)
"""
if host in self.max_reqs_per_hosts_per_sec:
return self.max_reqs_per_hosts_per_sec[host]
else:
return self.max_reqs_per_host_per_sec
def setHostMaxSimultaneousRequests(self, host, max_simultaneous_requests):
"""
Set the maximum number of simultaneous requests for a particular host.
**Arguments:**
* *host* -- Hostname. (Example, ``"google.com"``)
* *max_simultaneous_requests* -- Maximum number of simultaneous
requests to the host. If set to 1, RequestQueuer will not make
more than 1 simultaneous request to the host. If set to 0,
RequestQueuer will not limit the number of simultaneous requests.
"""
if max_simultaneous_requests == 0:
self.max_simul_reqs_per_hosts[host] = self.max_simul_reqs
else:
self.max_simul_reqs_per_hosts[host] = int(max_simultaneous_requests)
def getHostMaxSimultaneousRequests(self, host):
"""
Get the maximum number of simultaneous requests for a particular host.
**Arguments:**
* *host* -- Hostname. (Example, ``"google.com"``)
"""
if host in self.max_simul_reqs_per_hosts:
return self.max_simul_reqs_per_hosts[host]
else:
return self.max_simul_reqs_per_host
def getPage(self,
url,
last_modified=None,
etag=None,
method='GET',
postdata=None,
headers=None,
agent="RequestQueuer",
timeout=60,
cookies=None,
follow_redirect=True,
prioritize=False
):
"""
Make an HTTP Request.
**Arguments:**
* *url* -- URL for the request.
**Keyword arguments:**
* *last_modified* -- Last modified date string to send as a request
header. (Default ``None``)
* *etag* -- Etag string to send as a request header. (Default
``None``)
* *method* -- HTTP request method. (Default ``'GET'``)
* *postdata* -- Dictionary of strings to post with the request.
(Default ``None``)
* *headers* -- Dictionary of strings to send as request headers.
(Default ``None``)
* *agent* -- User agent to send with request. (Default
``'RequestQueuer'``)
* *timeout* -- Request timeout, in seconds. (Default ``60``)
* *cookies* -- Dictionary of strings to send as request cookies.
(Default ``None``).
* *follow_redirect* -- Boolean switch to follow HTTP redirects.
(Default ``True``)
* *prioritize* -- Move this request to the front of the request
queue. (Default ``False``)
"""
if headers is None:
headers={}
if postdata is not None:
if isinstance(postdata, dict):
for key in postdata:
postdata[key] = convertToUTF8(postdata[key])
postdata = urllib.urlencode(postdata)
else:
convertToUTF8(postdata)
if method.lower() == "post":
headers["content-type"] = "application/x-www-form-urlencoded"
if last_modified is not None:
time_tuple = dateutil.parser.parse(last_modified).timetuple()
time_string = time.strftime("%a, %d %b %Y %T %z", time_tuple)
headers['If-Modified-Since'] = time_string
if etag is not None:
headers["If-None-Match"] = etag
req = {
"url":convertToUTF8(url),
"method":method,
"postdata":postdata,
"headers":headers,
"agent":agent,
"timeout":timeout,
"cookies":cookies,
"follow_redirect":follow_redirect,
"deferred":Deferred()
}
host = _parse(req["url"])[1]
if host not in self.pending_reqs:
self.pending_reqs[host] = []
if prioritize:
self.pending_reqs[host].insert(0, req)
else:
self.pending_reqs[host].append(req)
self._checkActive()
return req["deferred"]
def _hostRequestCheck(self, host):
if host not in self.pending_reqs:
return False
if host in self.last_req:
if host in self.min_req_interval_per_hosts:
if time.time() - self.last_req[host] < \
self.min_req_interval_per_hosts[host]:
return False
else:
if time.time() - self.last_req[host] < \
self.min_req_interval_per_host:
return False
if host in self.active_reqs:
if host in self.max_simul_reqs_per_hosts:
if self.active_reqs[host] > self.max_simul_reqs_per_hosts[host]:
return False
else:
if self.active_reqs[host] > self.max_simul_reqs_per_host:
return False
return True
def _checkActive(self):
while self.getActive() < self.max_simul_reqs and self.getPending() > 0:
hosts = self.pending_reqs.keys()
dispatched_requests = False
for host in hosts:
if len(self.pending_reqs[host]) == 0:
del self.pending_reqs[host]
elif self._hostRequestCheck(host):
dispatched_requests = True
req = self.pending_reqs[host].pop(0)
d = self._getPage(req)
d.addCallback(self._requestComplete, req["deferred"], host)
d.addErrback(self._requestError, req["deferred"], host)
self.last_req[host] = time.time()
self.active_reqs[host] = self.active_reqs.get(host, 0) + 1
if not dispatched_requests:
break
if self.getPending() > 0:
reactor.callLater(.1, self._checkActive)
def _requestComplete(self, response, deferred, host):
self.active_reqs[host] -= 1
self._checkActive()
deferred.callback(response)
return None
def _requestError(self, error, deferred, host):
self.active_reqs[host] -= 1
self._checkActive()
deferred.errback(error)
return None
def _getPage(self, req):
scheme, host, port = _parse(req['url'])[0:3]
factory = HTTPClientFactory(
req['url'],
method=req['method'],
postdata=req['postdata'],
headers=req['headers'],
agent=req['agent'],
timeout=req['timeout'],
cookies=req['cookies'],
followRedirect=req['follow_redirect']
)
if scheme == 'https':
reactor.connectSSL(
host,
port,
factory,
AllCipherSSLClientContextFactory(),
timeout=req['timeout']
)
else:
reactor.connectTCP(host, port, factory, timeout=req['timeout'])
factory.deferred.addCallback(self._getPageComplete, factory)
factory.deferred.addErrback(self._getPageError, factory)
return factory.deferred
def _getPageComplete(self, response, factory):
return {
"response":response,
"headers":factory.response_headers,
"status":int(factory.status),
"message":factory.message
}
def _getPageError(self, error, factory):
if hasattr(factory, "response_headers") \
and factory.response_headers is not None:
error.value.headers = factory.response_headers
return error | AWSpider | /AWSpider-0.3.2.12.tar.gz/AWSpider-0.3.2.12/awspider/requestqueuer.py | requestqueuer.py |
from twisted.web import server
import simplejson
from twisted.internet import reactor
from ..evaluateboolean import evaluateBoolean
import urlparse
from .base import BaseResource
class ExecutionResource(BaseResource):
def __init__(self, executionserver):
self.executionserver = executionserver
BaseResource.__init__(self)
def render(self, request):
request.setHeader('Content-type', 'text/javascript; charset=UTF-8')
if len(request.postpath) > 0:
if request.postpath[0] == "coordinate":
reactor.callLater(5, self.executionserver.coordinate)
return simplejson.dumps(True)
elif request.postpath[0] == "server":
return simplejson.dumps(self.executionserver.getServerData())
elif request.postpath[0] == "getpage" and "url" in request.args:
#print "Recieving peering request for %s" % request.args["url"][0]
kwargs = {}
if "method" in request.args:
kwargs["method"] = request.args["method"][0]
if "postdata" in request.args:
kwargs["postdata"] = urlparse.parse_qs(request.args["postdata"][0])
if "headers" in request.args:
kwargs["headers"] = urlparse.parse_qs(request.args["headers"][0])
if "cookies" in request.args:
kwargs["cookies"] = urlparse.parse_qs(request.args["cookies"][0])
if "agent" in request.args:
kwargs["agent"] = request.args["agent"][0]
if "timeout" in request.args:
kwargs["timeout"] = int(request.args["timeout"][0])
if "followRedirect" in request.args:
kwargs["followRedirect"] = evaluateBoolean(request.args["followRedirect"][0])
if "url_hash" in request.args:
kwargs["url_hash"] = request.args["url_hash"][0]
if "cache" in request.args:
kwargs["cache"] = int(request.args["cache"][0])
if "prioritize" in request.args:
kwargs["prioritize"] = evaluateBoolean(request.args["prioritize"][0])
d = self.executionserver.pg.getPage(request.args["url"][0], **kwargs)
d.addCallback(self._getpageCallback, request)
d.addErrback(self._errorResponse)
d.addCallback(self._immediateResponse, request)
return server.NOT_DONE_YET
message = "No such resource."
request.setResponseCode(404, message)
self._immediateResponse(simplejson.dumps({"error":message}), request)
def _getpageCallback(self, data, request):
request.setResponseCode(data["status"], data["message"])
for header in data["headers"]:
request.setHeader(header, data["headers"][header][0])
return self._immediateResponse(data["response"], request) | AWSpider | /AWSpider-0.3.2.12.tar.gz/AWSpider-0.3.2.12/awspider/resources/execution.py | execution.py |
from twisted.web import server
from .base import BaseResource
from twisted.python.failure import Failure
from twisted.internet.defer import DeferredList
class InterfaceResource(BaseResource):
isLeaf = True
def __init__(self, interfaceserver):
self.interfaceserver = interfaceserver
BaseResource.__init__(self)
def render(self, request):
request.setHeader('Content-type', 'text/javascript; charset=UTF-8')
if len(request.postpath) > 0:
if request.postpath[0] == "show_reservation":
if "uuid" in request.args:
deferreds = []
for uuid in request.args["uuid"]:
deferreds.append(self.interfaceserver.showReservation(uuid))
d = DeferredList(deferreds, consumeErrors=True)
d.addCallback(self._showReservationCallback, request.args["uuid"])
d.addCallback(self._successResponse)
d.addErrback(self._errorResponse)
d.addCallback(self._immediateResponse, request)
return server.NOT_DONE_YET
else:
return self._errorResponse(Failure(exc_value=Exception("Parameter UUID is required.")))
elif request.postpath[0] == "execute_reservation":
if "uuid" in request.args:
d = self.interfaceserver.executeReservation(request.args["uuid"][0])
d.addCallback(self._successResponse)
d.addErrback(self._errorResponse)
d.addCallback(self._immediateResponse, request)
return server.NOT_DONE_YET
else:
return self._errorResponse(Failure(exc_value=Exception("Parameter UUID is required.")))
elif request.postpath[0] == "delete_reservation":
if "uuid" in request.args:
d = self.interfaceserver.deleteReservation(request.args["uuid"][0])
d.addCallback(self._successResponse)
d.addErrback(self._errorResponse)
d.addCallback(self._immediateResponse, request)
return server.NOT_DONE_YET
else:
return self._errorResponse(Failure(exc_value=Exception("Parameter UUID is required.")))
def _showReservationCallback(self, data, uuids):
print data[0][1]
response = {}
for i in range(0, len(uuids)):
if data[i][0] == True:
response[uuids[i]] = data[i][1]
else:
response[uuids[i]] = {"error":str(data[i][1].value)}
return response | AWSpider | /AWSpider-0.3.2.12.tar.gz/AWSpider-0.3.2.12/awspider/resources/interface.py | interface.py |
from twisted.internet.defer import Deferred, DeferredList
from twisted.web import server
from twisted.internet import reactor
from .base import BaseServer, LOGGER
from ..resources import DataResource
class DataServer(BaseServer):
def __init__(self,
aws_access_key_id,
aws_secret_access_key,
aws_s3_storage_bucket,
aws_sdb_reservation_domain,
port=5002,
log_file='dataserver.log',
log_directory=None,
log_level="debug",
name=None,
max_simultaneous_requests=50):
if name == None:
name = "AWSpider Data Server UUID: %s" % self.uuid
resource = DataResource(self)
self.site_port = reactor.listenTCP(port, server.Site(resource))
BaseServer.__init__(
self,
aws_access_key_id,
aws_secret_access_key,
aws_s3_storage_bucket=aws_s3_storage_bucket,
aws_sdb_reservation_domain=aws_sdb_reservation_domain,
log_file=log_file,
log_directory=log_directory,
log_level=log_level,
name=name,
max_simultaneous_requests=max_simultaneous_requests,
port=port)
def clearStorage(self):
return self.s3.emptyBucket(self.aws_s3_storage_bucket)
def getData(self, uuid):
LOGGER.debug("Getting %s from S3." % uuid)
d = self.s3.getObject(self.aws_s3_storage_bucket, uuid)
d.addCallback(self._getCallback, uuid)
d.addErrback(self._getErrback, uuid)
return d
def _getCallback(self, data, uuid):
LOGGER.debug("Got %s from S3." % (uuid))
return cPickle.loads(data["response"])
def _getErrback(self, error, uuid):
LOGGER.error("Could not get %s from S3.\n%s" % (uuid, error))
return error
def shutdown(self):
deferreds = []
LOGGER.debug("%s stopping on main HTTP interface." % self.name)
d = self.site_port.stopListening()
if isinstance(d, Deferred):
deferreds.append(d)
if len(deferreds) > 0:
d = DeferredList(deferreds)
d.addCallback(self._shutdownCallback)
return d
else:
return self._shutdownCallback(None)
def _shutdownCallback(self, data):
return BaseServer.shutdown(self) | AWSpider | /AWSpider-0.3.2.12.tar.gz/AWSpider-0.3.2.12/awspider/servers/data.py | data.py |
from twisted.internet.defer import Deferred, DeferredList, maybeDeferred
from twisted.web.resource import Resource
from twisted.internet import reactor
from twisted.internet import task
from twisted.web import server
from .base import BaseServer, LOGGER, PRETTYPRINTER
from ..resources import AdminResource
from ..aws import sdb_now_add
class AdminServer(BaseServer):
peercheckloop = None
exposed_functions = []
exposed_function_resources = {}
def __init__(self,
aws_access_key_id,
aws_secret_access_key,
aws_sdb_reservation_domain,
aws_s3_http_cache_bucket=None,
aws_s3_storage_bucket=None,
aws_sdb_coordination_domain=None,
port=5003,
log_file='adminserver.log',
log_directory=None,
log_level="debug",
name=None,
time_offset=None,
peer_check_interval=60):
if name == None:
name = "AWSpider Admin Server UUID: %s" % self.uuid
resource = AdminResource(self)
self.site_port = reactor.listenTCP(port, server.Site(resource))
self.peer_check_interval = int(peer_check_interval)
BaseServer.__init__(
self,
aws_access_key_id,
aws_secret_access_key,
aws_s3_http_cache_bucket=aws_s3_http_cache_bucket,
aws_sdb_reservation_domain=aws_sdb_reservation_domain,
aws_s3_storage_bucket=aws_s3_storage_bucket,
aws_sdb_coordination_domain=aws_sdb_coordination_domain,
log_file=log_file,
log_directory=log_directory,
log_level=log_level,
name=name,
port=port,
time_offset=time_offset)
def start(self):
reactor.callWhenRunning(self._start)
return self.start_deferred
def _start(self):
deferreds = []
if self.time_offset is None:
deferreds.append(self.getTimeOffset())
d = DeferredList(deferreds, consumeErrors=True)
d.addCallback(self._startCallback)
def _startCallback(self, data):
for row in data:
if row[0] == False:
d = self.shutdown()
d.addCallback(self._startHandleError, row[1])
return d
d = BaseServer.start(self)
d.addCallback(self._startCallback2)
def _startCallback2(self, data):
LOGGER.debug("At callback.")
if self.shutdown_trigger_id is not None:
if self.aws_sdb_coordination_domain is not None:
self.peercheckloop = task.LoopingCall(self.peerCheck)
self.peercheckloop.start(self.peer_check_interval)
def shutdown(self):
deferreds = []
LOGGER.debug("%s stopping on main HTTP interface." % self.name)
d = self.site_port.stopListening()
if isinstance(d, Deferred):
deferreds.append(d)
if len(deferreds) > 0:
d = DeferredList(deferreds)
d.addCallback(self._shutdownCallback)
return d
else:
return self._shutdownCallback(None)
def _shutdownCallback(self, data):
return BaseServer.shutdown(self)
def peerCheck(self):
sql = "SELECT * FROM `%s` WHERE created > '%s'" % (
self.aws_sdb_coordination_domain,
sdb_now_add(self.peer_check_interval * -2,
offset=self.time_offset))
LOGGER.debug("Querying SimpleDB, \"%s\"" % sql)
d = self.sdb.select(sql)
d.addCallback(self._peerCheckCallback)
d.addErrback(self._peerCheckErrback)
return d
def _peerCheckCallback(self, data):
LOGGER.info("Got server data:\n%s" % PRETTYPRINTER.pformat(data.values()))
ip_addresses = [x['public_ip'][0] for x in data.values()]
LOGGER.info("%s machines responding. %s" % (len(data), ip_addresses))
def _peerCheckErrback(self, error):
LOGGER.error("Could not query SimpleDB for peers: %s" % str(error))
def clearHTTPCache(self):
return self.s3.emptyBucket(self.aws_s3_http_cache_bucket) | AWSpider | /AWSpider-0.3.2.12.tar.gz/AWSpider-0.3.2.12/awspider/servers/admin.py | admin.py |
import cPickle
import hashlib
import inspect
import logging
import logging.handlers
import os
import time
from decimal import Decimal
from uuid import uuid4
from twisted.internet import reactor
from twisted.internet.threads import deferToThread
from twisted.internet.defer import Deferred, DeferredList, maybeDeferred
from ..aws import AmazonS3, AmazonSDB
from ..aws import sdb_now
from ..exceptions import DeleteReservationException
from ..pagegetter import PageGetter
from ..requestqueuer import RequestQueuer
from ..timeoffset import getTimeOffset
import pprint
PRETTYPRINTER = pprint.PrettyPrinter(indent=4)
LOGGER = logging.getLogger("main")
class ReservationCachingException(Exception):
pass
class BaseServer(object):
logging_handler = None
shutdown_trigger_id = None
uuid = uuid4().hex
start_time = time.time()
active_jobs = {}
reserved_arguments = [
"reservation_function_name",
"reservation_created",
"reservation_next_request",
"reservation_error",
"reservation_cache"]
functions = {}
reservation_fast_caches = {}
def __init__(self,
aws_access_key_id,
aws_secret_access_key,
aws_s3_http_cache_bucket=None,
aws_sdb_reservation_domain=None,
aws_s3_storage_bucket=None,
aws_s3_reservation_cache_bucket=None,
aws_sdb_coordination_domain=None,
max_simultaneous_requests=100,
max_requests_per_host_per_second=0,
max_simultaneous_requests_per_host=0,
log_file=None,
log_directory=None,
log_level="debug",
name=None,
time_offset=None,
port=8080):
if name == None:
name = "AWSpider Server UUID: %s" % self.uuid
self.port = port
self.time_offset = time_offset
self.name = name
self.start_deferred = Deferred()
self.rq = RequestQueuer(
max_simultaneous_requests=int(max_simultaneous_requests),
max_requests_per_host_per_second=int(max_requests_per_host_per_second),
max_simultaneous_requests_per_host=int(max_simultaneous_requests_per_host))
self.rq.setHostMaxRequestsPerSecond("127.0.0.1", 0)
self.rq.setHostMaxSimultaneousRequests("127.0.0.1", 0)
self.aws_s3_reservation_cache_bucket = aws_s3_reservation_cache_bucket
self.aws_access_key_id = aws_access_key_id
self.aws_secret_access_key = aws_secret_access_key
self.aws_s3_http_cache_bucket = aws_s3_http_cache_bucket
self.aws_s3_storage_bucket = aws_s3_storage_bucket
self.aws_sdb_reservation_domain = aws_sdb_reservation_domain
self.aws_sdb_coordination_domain = aws_sdb_coordination_domain
self.s3 = AmazonS3(
self.aws_access_key_id,
self.aws_secret_access_key,
rq=self.rq)
self.sdb = AmazonSDB(
self.aws_access_key_id,
self.aws_secret_access_key,
rq=self.rq)
self.pg = PageGetter(
self.s3,
self.aws_s3_http_cache_bucket,
rq=self.rq)
self._setupLogging(log_file, log_directory, log_level)
if self.name is not None:
LOGGER.info("Successfully loaded %s configuration." % self.name)
def _setupLogging(self, log_file, log_directory, log_level):
if log_directory is None:
self.logging_handler = logging.StreamHandler()
else:
self.logging_handler = logging.handlers.TimedRotatingFileHandler(
os.path.join(log_directory, log_file),
when='D',
interval=1)
log_format = "%(levelname)s: %(message)s %(pathname)s:%(lineno)d"
self.logging_handler.setFormatter(logging.Formatter(log_format))
LOGGER.addHandler(self.logging_handler)
log_level = log_level.lower()
log_levels = {
"debug":logging.DEBUG,
"info":logging.INFO,
"warning":logging.WARNING,
"error":logging.ERROR,
"critical":logging.CRITICAL
}
if log_level in log_levels:
LOGGER.setLevel(log_levels[log_level])
else:
LOGGER.setLevel(logging.DEBUG)
def start(self):
reactor.callWhenRunning(self._start)
return self.start_deferred
def start(self):
reactor.callWhenRunning(self._baseStart)
return self.start_deferred
def _baseStart(self):
LOGGER.critical("Checking S3 and SDB setup.")
deferreds = []
if self.aws_s3_reservation_cache_bucket is not None:
deferreds.append(
self.s3.checkAndCreateBucket(self.aws_s3_reservation_cache_bucket))
if self.aws_s3_http_cache_bucket is not None:
deferreds.append(
self.s3.checkAndCreateBucket(self.aws_s3_http_cache_bucket))
if self.aws_sdb_reservation_domain is not None:
deferreds.append(
self.sdb.checkAndCreateDomain(self.aws_sdb_reservation_domain))
if self.aws_s3_storage_bucket is not None:
deferreds.append(
self.s3.checkAndCreateBucket(self.aws_s3_storage_bucket))
if self.aws_sdb_coordination_domain is not None:
deferreds.append(
self.sdb.checkAndCreateDomain(self.aws_sdb_coordination_domain))
d = DeferredList(deferreds, consumeErrors=True)
d.addCallback(self._baseStartCallback)
def _baseStartCallback(self, data):
for row in data:
if row[0] == False:
d = self.shutdown()
d.addCallback(self._startHandleError, row[1])
return d
self.shutdown_trigger_id = reactor.addSystemEventTrigger(
'before',
'shutdown',
self.shutdown)
LOGGER.critical("Starting %s" % self.name)
self._baseStartCallback2(None)
def _baseStartCallback2(self, data):
self.start_deferred.callback(True)
def _startHandleError(self, data, error):
self.start_deferred.errback(error)
def shutdown(self):
LOGGER.debug("%s waiting for shutdown." % self.name)
d = Deferred()
reactor.callLater(0, self._waitForShutdown, d)
return d
def _waitForShutdown(self, shutdown_deferred):
if self.rq.getPending() > 0 or self.rq.getActive() > 0:
LOGGER.debug("%s waiting for shutdown." % self.name)
reactor.callLater(1, self._waitForShutdown, shutdown_deferred)
return
self.shutdown_trigger_id = None
LOGGER.debug("%s shut down." % self.name)
LOGGER.removeHandler(self.logging_handler)
shutdown_deferred.callback(True)
def getTimeOffset(self):
d = getTimeOffset()
d.addCallback(self._getTimeOffsetCallback)
d.addErrback(self._getTimeOffsetErrback)
return d
def _getTimeOffsetCallback(self, time_offset):
self.time_offset = time_offset
LOGGER.info("Got time offset for sync: %s" % self.time_offset)
def _getTimeOffsetErrback(self, error):
if self.time_offset is None:
message = "Could not get time offset for sync."
LOGGER.critical(message)
raise Exception(message)
def callExposedFunction(self, func, kwargs, function_name, reservation_fast_cache=None, uuid=None):
if uuid is not None:
self.active_jobs[uuid] = True
if self.functions[function_name]["get_reservation_uuid"]:
kwargs["reservation_uuid"] = uuid
if self.functions[function_name]["check_reservation_fast_cache"] and \
reservation_fast_cache is not None:
kwargs["reservation_fast_cache"] = reservation_fast_cache
elif self.functions[function_name]["check_reservation_fast_cache"]:
kwargs["reservation_fast_cache"] = None
if self.functions[function_name]["check_reservation_cache"] and \
self.aws_s3_reservation_cache_bucket is not None:
d = self.getReservationCache(uuid)
d.addCallback(self._reservationCacheCallback,
func,
kwargs,
function_name,
uuid)
d.addErrback(self._reservationCacheErrback,
func,
kwargs,
function_name,
uuid)
return d
elif self.functions[function_name]["check_reservation_cache"]:
kwargs["reservation_cache"] = None
d = maybeDeferred(func, **kwargs)
d.addCallback(self._callExposedFunctionCallback, function_name, uuid)
d.addErrback(self._callExposedFunctionErrback, function_name, uuid)
return d
def _reservationCacheCallback(self, data, func, kwargs, function_name, uuid):
LOGGER.debug("Got reservation cache for %s" % uuid)
kwargs["reservation_cache"] = data
d = maybeDeferred(func, **kwargs)
d.addCallback(self._callExposedFunctionCallback, function_name, uuid)
d.addErrback(self._callExposedFunctionErrback, function_name, uuid)
return d
def _reservationCacheErrback(self, error, func, kwargs, function_name, uuid):
LOGGER.debug("Could not get reservation cache for %s" % uuid)
kwargs["reservation_cache"] = None
d = maybeDeferred(func, **kwargs)
d.addCallback(self._callExposedFunctionCallback, function_name, uuid)
d.addErrback(self._callExposedFunctionErrback, function_name, uuid)
return d
def _callExposedFunctionErrback(self, error, function_name, uuid):
if uuid is not None:
del self.active_jobs[uuid]
try:
error.raiseException()
except DeleteReservationException, e:
if uuid is not None:
self.deleteReservation(uuid)
message = """Error with %s, %s.\n%s
Reservation deleted at request of the function.""" % (
function_name,
uuid,
error)
LOGGER.error(message)
return
except:
pass
if uuid is None:
LOGGER.error("Error with %s.\n%s" % (function_name, error))
else:
LOGGER.error("Error with %s.\nUUID:%s\n%s" % (
function_name,
uuid,
error))
return error
def _callExposedFunctionCallback(self, data, function_name, uuid):
LOGGER.debug("Function %s returned successfully." % (function_name))
# If the UUID is None, this is a one-off type of thing.
if uuid is None:
return data
# If the data is None, there's nothing to store.
if data is None:
del self.active_jobs[uuid]
return None
# If we have an place to store the response on S3, do it.
if self.aws_s3_storage_bucket is not None:
LOGGER.debug("Putting result for %s, %s on S3." % (function_name, uuid))
pickled_data = cPickle.dumps(data)
d = self.s3.putObject(
self.aws_s3_storage_bucket,
uuid,
pickled_data,
content_type="text/plain",
gzip=True)
d.addCallback(self._exposedFunctionCallback2, data, uuid)
d.addErrback(self._exposedFunctionErrback2, data, function_name, uuid)
return d
return data
def _exposedFunctionErrback2(self, error, data, function_name, uuid):
del self.active_jobs[uuid]
LOGGER.error("Could not put results of %s, %s on S3.\n%s" % (function_name, uuid, error))
return data
def _exposedFunctionCallback2(self, s3_callback_data, data, uuid):
del self.active_jobs[uuid]
return data
def expose(self, *args, **kwargs):
return self.makeCallable(expose=True, *args, **kwargs)
def makeCallable(self, func, interval=0, name=None, expose=False):
argspec = inspect.getargspec(func)
# Get required / optional arguments
arguments = argspec[0]
if len(arguments) > 0 and arguments[0:1][0] == 'self':
arguments.pop(0)
kwarg_defaults = argspec[3]
if kwarg_defaults is None:
kwarg_defaults = []
required_arguments = arguments[0:len(arguments) - len(kwarg_defaults)]
optional_arguments = arguments[len(arguments) - len(kwarg_defaults):]
# Reservation cache is stored on S3
if "reservation_cache" in required_arguments:
del required_arguments[required_arguments.index("reservation_cache")]
check_reservation_cache = True
elif "reservation_cache" in optional_arguments:
del optional_arguments[optional_arguments.index("reservation_cache")]
check_reservation_cache = True
else:
check_reservation_cache = False
# Reservation fast cache is stored on SDB with the reservation
if "reservation_fast_cache" in required_arguments:
del required_arguments[required_arguments.index("reservation_fast_cache")]
check_reservation_fast_cache = True
elif "reservation_fast_cache" in optional_arguments:
del optional_arguments[optional_arguments.index("reservation_fast_cache")]
check_reservation_fast_cache = True
else:
check_reservation_fast_cache = False
# Indicates whether to send the reservation's UUID to the function
if "reservation_uuid" in required_arguments:
del required_arguments[required_arguments.index("reservation_uuid")]
get_reservation_uuid = True
elif "reservation_uuid" in optional_arguments:
del optional_arguments[optional_arguments.index("reservation_uuid")]
get_reservation_uuid = True
else:
get_reservation_uuid = False
# Get function name, usually class/method
if name is not None:
function_name = name
elif hasattr(func, "im_class"):
function_name = "%s/%s" % (func.im_class.__name__, func.__name__)
else:
function_name = func.__name__
function_name = function_name.lower()
# Make sure the function isn't using any reserved arguments.
for key in required_arguments:
if key in self.reserved_arguments:
message = "Required argument name '%s' used in function %s is reserved." % (key, function_name)
LOGGER.error(message)
raise Exception(message)
for key in optional_arguments:
if key in self.reserved_arguments:
message = "Optional argument name '%s' used in function %s is reserved." % (key, function_name)
LOGGER.error(message)
raise Exception(message)
# Make sure we don't already have a function with the same name.
if function_name in self.functions:
raise Exception("A method or function with the name %s is already callable." % function_name)
# Add it to our list of callable functions.
self.functions[function_name] = {
"function":func,
"interval":interval,
"required_arguments":required_arguments,
"optional_arguments":optional_arguments,
"check_reservation_cache":check_reservation_cache,
"check_reservation_fast_cache":check_reservation_fast_cache,
"get_reservation_uuid":get_reservation_uuid
}
LOGGER.info("Function %s is now callable." % function_name)
return function_name
def getPage(self, *args, **kwargs):
return self.pg.getPage(*args, **kwargs)
def setHostMaxRequestsPerSecond(self, *args, **kwargs):
return self.rq.setHostMaxRequestsPerSecond(*args, **kwargs)
def setHostMaxSimultaneousRequests(self, *args, **kwargs):
return self.rq.setHostMaxSimultaneousRequests(*args, **kwargs)
def deleteReservation(self, uuid, function_name="Unknown"):
LOGGER.info("Deleting reservation %s, %s." % (function_name, uuid))
deferreds = []
deferreds.append(self.sdb.delete(self.aws_sdb_reservation_domain, uuid))
deferreds.append(self.s3.deleteObject(self.aws_s3_storage_bucket, uuid))
d = DeferredList(deferreds)
d.addCallback(self._deleteReservationCallback, function_name, uuid)
d.addErrback(self._deleteReservationErrback, function_name, uuid)
return d
def _deleteReservationCallback(self, data, function_name, uuid):
LOGGER.info("Reservation %s, %s successfully deleted." % (function_name, uuid))
return True
def _deleteReservationErrback(self, error, function_name, uuid ):
LOGGER.error("Error deleting reservation %s, %s.\n%s" % (function_name, uuid, error))
return False
def deleteHTTPCache(self):
deferreds = []
if self.aws_s3_http_cache_bucket is not None:
deferreds.append(
self.s3.emptyBucket(self.aws_s3_http_cache_bucket))
if len(deferreds) > 0:
d = DeferredList(deferreds, consumeErrors=True)
d.addCallback(self._deleteHTTPCacheCallback)
return d
else:
return True
def _deleteHTTPCacheCallback(self, data):
deferreds = []
if self.aws_s3_http_cache_bucket is not None:
deferreds.append(
self.s3.deleteBucket(self.aws_s3_http_cache_bucket))
if len(deferreds) > 0:
d = DeferredList(deferreds, consumeErrors=True)
d.addCallback(self._deleteHTTPCacheCallback2)
return d
else:
return True
def _deleteHTTPCacheCallback2(self, data):
return True
def getServerData(self):
running_time = time.time() - self.start_time
cost = (self.sdb.box_usage * .14) * (60*60*24*30.4) / (running_time)
active_requests_by_host = self.rq.getActiveRequestsByHost()
pending_requests_by_host = self.rq.getPendingRequestsByHost()
data = {
"load_avg":[str(Decimal(str(x), 2)) for x in os.getloadavg()],
"running_time":running_time,
"cost":cost,
"active_requests_by_host":active_requests_by_host,
"pending_requests_by_host":pending_requests_by_host,
"active_requests":self.rq.getActive(),
"pending_requests":self.rq.getPending(),
"current_timestamp":sdb_now(offset=self.time_offset)
}
LOGGER.debug("Got server data:\n%s" % PRETTYPRINTER.pformat(data))
return data
def getReservationCache(self, uuid):
if self.aws_s3_reservation_cache_bucket is None:
raise ReservationCachingException("No reservation cache bucket is specified.")
d = self.s3.getObject(
self.aws_s3_reservation_cache_bucket,
uuid)
d.addCallback(self._getReservationCacheCallback)
return d
def _getReservationCacheCallback(self, data):
return cPickle.loads(data["response"])
def setReservationFastCache(self, uuid, data):
if not isinstance(data, str):
raise Exception("ReservationFastCache must be a string.")
if uuid is None:
return None
self.reservation_fast_caches[uuid] = data
def setReservationCache(self, uuid, data):
if uuid is None:
return None
if self.aws_s3_reservation_cache_bucket is None:
raise ReservationCachingException("No reservation cache bucket is specified.")
d = self.s3.putObject(
self.aws_s3_reservation_cache_bucket,
uuid,
cPickle.dumps(data))
return d | AWSpider | /AWSpider-0.3.2.12.tar.gz/AWSpider-0.3.2.12/awspider/servers/base.py | base.py |
import cPickle
import time
import pprint
import re
from twisted.web.client import _parse
from uuid import uuid5, NAMESPACE_DNS
from twisted.internet.defer import Deferred, DeferredList, maybeDeferred
from twisted.internet import task
from twisted.internet import reactor
from twisted.web import server
from .base import BaseServer, LOGGER
from ..aws import sdb_now, sdb_now_add
from ..resources import ExecutionResource
from ..networkaddress import getNetworkAddress
PRETTYPRINTER = pprint.PrettyPrinter(indent=4)
class ExecutionServer(BaseServer):
peers = {}
peer_uuids = []
reportjobspeedloop = None
jobsloop = None
queryloop = None
coordinateloop = None
uuid_limits = {'start':None, 'end':None}
public_ip = None
local_ip = None
network_information = {}
queued_jobs = {}
job_queue = []
job_count = 0
query_start_time = None
simultaneous_jobs = 25
querying_for_jobs = False
reservation_update_queue = []
current_sql = ""
last_job_query_count = 0
def __init__(self,
aws_access_key_id,
aws_secret_access_key,
aws_sdb_reservation_domain,
aws_s3_http_cache_bucket=None,
aws_s3_storage_bucket=None,
aws_sdb_coordination_domain=None,
aws_s3_reservation_cache_bucket=None,
max_simultaneous_requests=50,
max_requests_per_host_per_second=0,
max_simultaneous_requests_per_host=0,
port=5001,
log_file='executionserver.log',
log_directory=None,
log_level="debug",
name=None,
time_offset=None,
peer_check_interval=60,
reservation_check_interval=60,
hammer_prevention=False):
if name == None:
name = "AWSpider Execution Server UUID: %s" % self.uuid
self.network_information["port"] = port
self.hammer_prevention = hammer_prevention
self.peer_check_interval = int(peer_check_interval)
self.reservation_check_interval = int(reservation_check_interval)
resource = ExecutionResource(self)
self.site_port = reactor.listenTCP(port, server.Site(resource))
BaseServer.__init__(
self,
aws_access_key_id,
aws_secret_access_key,
aws_s3_reservation_cache_bucket=aws_s3_reservation_cache_bucket,
aws_s3_http_cache_bucket=aws_s3_http_cache_bucket,
aws_sdb_reservation_domain=aws_sdb_reservation_domain,
aws_s3_storage_bucket=aws_s3_storage_bucket,
aws_sdb_coordination_domain=aws_sdb_coordination_domain,
max_simultaneous_requests=max_simultaneous_requests,
max_requests_per_host_per_second=max_requests_per_host_per_second,
max_simultaneous_requests_per_host=max_simultaneous_requests_per_host,
log_file=log_file,
log_directory=log_directory,
log_level=log_level,
name=name,
time_offset=time_offset,
port=port)
def start(self):
reactor.callWhenRunning(self._start)
return self.start_deferred
def _start(self):
deferreds = []
deferreds.append(self.getNetworkAddress())
if self.time_offset is None:
deferreds.append(self.getTimeOffset())
d = DeferredList(deferreds, consumeErrors=True)
d.addCallback(self._startCallback)
def _startCallback(self, data):
for row in data:
if row[0] == False:
d = self.shutdown()
d.addCallback(self._startHandleError, row[1])
return d
d = BaseServer.start(self)
d.addCallback(self._startCallback2)
def _startCallback2(self, data):
if self.shutdown_trigger_id is not None:
self.reportjobspeedloop = task.LoopingCall(self.reportJobSpeed)
self.reportjobspeedloop.start(60)
self.jobsloop = task.LoopingCall(self.executeJobs)
self.jobsloop.start(1)
if self.aws_sdb_coordination_domain is not None:
self.peerCheckRequest()
d = maybeDeferred(self.coordinate)
d.addCallback(self._startCallback3)
else:
self.queryloop = task.LoopingCall(self.query)
self.queryloop.start(self.reservation_check_interval)
def _startCallback3(self, data):
self.coordinateloop = task.LoopingCall(self.coordinate)
self.coordinateloop.start(self.peer_check_interval)
self.queryloop = task.LoopingCall(self.query)
self.queryloop.start(self.reservation_check_interval)
def shutdown(self):
LOGGER.critical("Shutting down.")
self.job_queue = []
self.queued_jobs = {}
deferreds = []
LOGGER.debug("%s stopping on main HTTP interface." % self.name)
d = self.site_port.stopListening()
if isinstance(d, Deferred):
deferreds.append(d)
if self.reportjobspeedloop is not None:
LOGGER.debug("Stopping report job speed loop.")
d = self.reportjobspeedloop.stop()
if isinstance(d, Deferred):
deferreds.append(d)
if self.jobsloop is not None:
LOGGER.debug("Stopping jobs loop.")
d = self.jobsloop.stop()
if isinstance(d, Deferred):
deferreds.append(d)
if self.queryloop is not None:
LOGGER.debug("Stopping query loop.")
d = self.queryloop.stop()
if isinstance(d, Deferred):
deferreds.append(d)
if self.coordinateloop is not None:
LOGGER.debug("Stopping coordinating loop.")
d = self.coordinateloop.stop()
if isinstance(d, Deferred):
deferreds.append(d)
LOGGER.debug("Removing data from SDB coordination domain.")
d = self.sdb.delete(self.aws_sdb_coordination_domain, self.uuid)
d.addCallback(self.peerCheckRequest)
deferreds.append(d)
if len(deferreds) > 0:
d = DeferredList(deferreds)
d.addCallback(self._shutdownCallback)
return d
else:
return self._shutdownCallback(None)
def _shutdownCallback(self, data):
return BaseServer.shutdown(self)
def peerCheckRequest(self, data=None):
LOGGER.debug("Signaling peers.")
deferreds = []
for uuid in self.peers:
if uuid != self.uuid and self.peers[uuid]["active"]:
LOGGER.debug("Signaling %s to check peers." % self.peers[uuid]["uri"])
d = self.rq.getPage(
self.peers[uuid]["uri"] + "/coordinate",
prioritize=True)
d.addCallback(
self._peerCheckRequestCallback,
self.peers[uuid]["uri"])
d.addErrback(
self._peerCheckRequestErrback,
self.peers[uuid]["uri"])
deferreds.append(d)
if len(deferreds) > 0:
LOGGER.debug("Combinining shutdown signal deferreds.")
return DeferredList(deferreds, consumeErrors=True)
return True
def _peerCheckRequestErrback(self, error, uri):
LOGGER.debug("Could not get %s/coordinate: %s" % (uri, str(error)))
def _peerCheckRequestCallback(self, data, uri):
LOGGER.debug("Got %s/coordinate." % uri)
def getNetworkAddress(self):
d = getNetworkAddress()
d.addCallback(self._getNetworkAddressCallback)
d.addErrback(self._getNetworkAddressErrback)
return d
def _getNetworkAddressCallback(self, data):
if "public_ip" in data:
self.public_ip = data["public_ip"]
self.network_information["public_ip"] = self.public_ip
if "local_ip" in data:
self.local_ip = data["local_ip"]
self.network_information["local_ip"] = self.local_ip
def _getNetworkAddressErrback(self, error):
message = "Could not get network address."
LOGGER.error(message)
raise Exception(message)
def coordinate(self):
server_data = self.getServerData()
attributes = {
"created":sdb_now(offset=self.time_offset),
"load_avg":server_data["load_avg"],
"running_time":server_data["running_time"],
"cost":server_data["cost"],
"active_requests":server_data["active_requests"],
"pending_requests":server_data["pending_requests"],
"current_timestamp":server_data["current_timestamp"],
"job_queue":len(self.job_queue),
"active_jobs":len(self.active_jobs),
"queued_jobs":len(self.queued_jobs),
"current_sql":self.current_sql.replace("\n", ""),
"last_job_query_count":self.last_job_query_count}
if self.uuid_limits["start"] is None and self.uuid_limits["end"] is not None:
attributes["range"] = "Start - %s" % self.uuid_limits["end"]
elif self.uuid_limits["start"] is not None and self.uuid_limits["end"] is None:
attributes["range"] = "%s - End" % self.uuid_limits["start"]
elif self.uuid_limits["start"] is None and self.uuid_limits["end"] is None:
attributes["range"] = "Full range"
else:
attributes["range"] = "%s - %s" % (self.uuid_limits["start"], self.uuid_limits["end"])
attributes.update(self.network_information)
d = self.sdb.putAttributes(
self.aws_sdb_coordination_domain,
self.uuid,
attributes,
replace=attributes.keys())
d.addCallback(self._coordinateCallback)
d.addErrback(self._coordinateErrback)
return d
def _coordinateCallback(self, data):
sql = "SELECT public_ip, local_ip, port FROM `%s` WHERE created > '%s'" % (
self.aws_sdb_coordination_domain,
sdb_now_add(self.peer_check_interval * -2,
offset=self.time_offset))
LOGGER.debug("Querying SimpleDB, \"%s\"" % sql)
d = self.sdb.select(sql)
d.addCallback(self._coordinateCallback2)
d.addErrback(self._coordinateErrback)
return d
def _coordinateCallback2(self, discovered):
existing_peers = set(self.peers.keys())
discovered_peers = set(discovered.keys())
new_peers = discovered_peers - existing_peers
old_peers = existing_peers - discovered_peers
for uuid in old_peers:
LOGGER.debug("Removing peer %s" % uuid)
if uuid in self.peers:
del self.peers[uuid]
deferreds = []
for uuid in new_peers:
if uuid == self.uuid:
self.peers[uuid] = {
"uri":"http://127.0.0.1:%s" % self.port,
"local_ip":"127.0.0.1",
"port":self.port,
"active":True
}
else:
deferreds.append(self.verifyPeer(uuid, discovered[uuid]))
if len(new_peers) > 0:
if len(deferreds) > 0:
d = DeferredList(deferreds, consumeErrors=True)
d.addCallback(self._coordinateCallback3)
return d
else:
self._coordinateCallback3(None) #Just found ourself.
elif len(old_peers) > 0:
self._coordinateCallback3(None)
else:
pass # No old, no new.
def _coordinateCallback3(self, data):
LOGGER.debug("Re-organizing peers.")
for uuid in self.peers:
if "local_ip" in self.peers[uuid]:
self.peers[uuid]["uri"] = "http://%s:%s" % (
self.peers[uuid]["local_ip"],
self.peers[uuid]["port"])
self.peers[uuid]["active"] = True
self.rq.setHostMaxRequestsPerSecond(
self.peers[uuid]["local_ip"],
0)
self.rq.setHostMaxSimultaneousRequests(
self.peers[uuid]["local_ip"],
0)
elif "public_ip" in self.peers[uuid]:
self.peers[uuid]["uri"] = "http://%s:%s" % (
self.peers[uuid]["public_ip"],
self.peers[uuid]["port"])
self.peers[uuid]["active"] = True
self.rq.setHostMaxRequestsPerSecond(
self.peers[uuid]["public_ip"],
0)
self.rq.setHostMaxSimultaneousRequests(
self.peers[uuid]["public_ip"],
0)
else:
LOGGER.error("Peer %s has no local or public IP. This should not happen." % uuid)
self.peer_uuids = self.peers.keys()
self.peer_uuids.sort()
LOGGER.debug("Peers updated to: %s" % self.peers)
# Set UUID peer limits by splitting up lexicographical namespace using hex values.
peer_count = len(self.peers)
splits = [hex(4096/peer_count * x)[2:] for x in range(1, peer_count)]
splits = zip([None] + splits, splits + [None])
splits = [{"start":x[0], "end":x[1]} for x in splits]
if self.uuid in self.peer_uuids:
self.uuid_limits = splits[self.peer_uuids.index(self.uuid)]
else:
self.uuid_limits = {"start":None, "end":None}
job_queue_length = len(self.job_queue)
if self.uuid_limits["start"] is None and self.uuid_limits["end"] is not None:
self.job_queue = filter(self.testJobByEnd, self.job_queue)
elif self.uuid_limits["start"] is not None and self.uuid_limits["end"] is None:
self.job_queue = filter(self.testJobByStart, self.job_queue)
elif self.uuid_limits["start"] is not None and self.uuid_limits["end"] is not None:
self.job_queue = filter(self.testJobByStartAndEnd, self.job_queue)
self.queued_jobs = dict((x["uuid"], True) for x in self.job_queue)
LOGGER.info("Abandoned %s jobs that were out of range." % (job_queue_length - len(self.job_queue)))
LOGGER.debug("Updated UUID limits to: %s" % self.uuid_limits)
def _coordinateErrback(self, error):
LOGGER.error("Could not query SimpleDB for peers: %s" % str(error))
def testJobByEnd(self, job):
return job["uuid"] < self.uuid_limits["end"]
def testJobByStart(self, job):
return job["uuid"] > self.uuid_limits["start"]
def testJobByStartAndEnd(self, job):
return (job["uuid"] < self.uuid_limits["end"] and job["uuid"] > self.uuid_limits["start"])
def verifyPeer(self, uuid, peer):
LOGGER.debug("Verifying peer %s" % uuid)
deferreds = []
if "port" in peer:
port = int(peer["port"][0])
else:
port = self.port
if uuid not in self.peers:
self.peers[uuid] = {}
self.peers[uuid]["active"] = False
self.peers[uuid]["port"] = port
if "local_ip" in peer:
local_ip = peer["local_ip"][0]
local_url = "http://%s:%s/server" % (local_ip, port)
d = self.rq.getPage(local_url, timeout=5, prioritize=True)
d.addCallback(self._verifyPeerLocalIPCallback, uuid, local_ip, port)
deferreds.append(d)
if "public_ip" in peer:
public_ip = peer["public_ip"][0]
public_url = "http://%s:%s/server" % (public_ip, port)
d = self.rq.getPage(public_url, timeout=5, prioritize=True)
d.addCallback(self._verifyPeerPublicIPCallback, uuid, public_ip, port)
deferreds.append(d)
if len(deferreds) > 0:
d = DeferredList(deferreds, consumeErrors=True)
return d
else:
return None
def _verifyPeerLocalIPCallback(self, data, uuid, local_ip, port):
LOGGER.debug("Verified local IP for %s" % uuid)
self.peers[uuid]["local_ip"] = local_ip
def _verifyPeerPublicIPCallback(self, data, uuid, public_ip, port):
LOGGER.debug("Verified public IP for %s" % uuid)
self.peers[uuid]["public_ip"] = public_ip
def getPage(self, *args, **kwargs):
if not self.hammer_prevention or len(self.peer_uuids) == 0:
return self.pg.getPage(*args, **kwargs)
else:
scheme, host, port, path = _parse(args[0])
peer_key = int(uuid5(NAMESPACE_DNS, host).int % len(self.peer_uuids))
peer_uuid = self.peer_uuids[peer_key]
if peer_uuid == self.uuid or self.peers[peer_uuid]["active"] == False:
return self.pg.getPage(*args, **kwargs)
else:
parameters = {}
parameters["url"] = args[0]
if "method" in kwargs:
parameters["method"] = kwargs["method"]
if "postdata" in kwargs:
parameters["postdata"] = urllib.urlencode(kwargs["postdata"])
if "headers" in kwargs:
parameters["headers"] = urllib.urlencode(kwargs["headers"])
if "cookies" in kwargs:
parameters["cookies"] = urllib.urlencode(kwargs["cookies"])
if "agent" in kwargs:
parameters["agent"] = kwargs["agent"]
if "timeout" in kwargs:
parameters["timeout"] = kwargs["timeout"]
if "followRedirect" in kwargs:
parameters["followRedirect"] = kwargs["followRedirect"]
if "url_hash" in kwargs:
parameters["url_hash"] = kwargs["url_hash"]
if "cache" in kwargs:
parameters["cache"] = kwargs["cache"]
if "prioritize" in kwargs:
parameters["prioritize"] = kwargs["prioritize"]
url = "%s/getpage?%s" % (
self.peers[peer_uuid]["uri"],
urllib.urlencode(parameters))
LOGGER.debug("Rerouting request for %s to %s" % (args[0], url))
d = self.rq.getPage(url, prioritize=True)
d.addErrback(self._getPageErrback, args, kwargs)
return d
def _getPageErrback(self, error, args, kwargs):
LOGGER.error(args[0] + ":" + str(error))
return self.pg.getPage(*args, **kwargs)
def queryByUUID(self, uuid):
sql = "SELECT * FROM `%s` WHERE itemName() = '%s'" % (
self.aws_sdb_reservation_domain,
uuid)
LOGGER.debug("Querying SimpleDB, \"%s\"" % sql)
d = self.sdb.select(sql)
d.addCallback(self._queryCallback2)
d.addErrback(self._queryErrback)
return d
def query(self, data=None):
if len(self.job_queue) > 1000:
LOGGER.debug("Skipping query. %s jobs already active." % len(self.job_queue))
return
if self.querying_for_jobs:
LOGGER.debug("Skipping query. Already querying for jobs.")
return
self.querying_for_jobs = True
if self.uuid_limits["start"] is None and self.uuid_limits["end"] is not None:
uuid_limit_clause = "AND itemName() < '%s'" % self.uuid_limits["end"]
elif self.uuid_limits["start"] is not None and self.uuid_limits["end"] is None:
uuid_limit_clause = "AND itemName() > '%s'" % self.uuid_limits["start"]
elif self.uuid_limits["start"] is None and self.uuid_limits["end"] is None:
uuid_limit_clause = ""
else:
uuid_limit_clause = "AND itemName() BETWEEN '%s' AND '%s'" % (
self.uuid_limits["start"],
self.uuid_limits["end"])
sql = """SELECT *
FROM `%s`
WHERE
reservation_next_request < '%s' %s
LIMIT 2500""" % (
self.aws_sdb_reservation_domain,
sdb_now(offset=self.time_offset),
uuid_limit_clause)
sql = re.sub(r"\s\s*", " ", sql);
self.current_sql = sql
LOGGER.debug("Querying SimpleDB, \"%s\"" % sql)
d = self.sdb.select(sql, max_results=5000)
d.addCallback(self._queryCallback)
d.addErrback(self._queryErrback)
def _queryErrback(self, error):
self.querying_for_jobs = False
LOGGER.error("Unable to query SimpleDB.\n%s" % error)
def _queryCallback(self, data):
LOGGER.info("Fetched %s jobs." % len(data))
self.querying_for_jobs = False
# Iterate through the reservation data returned from SimpleDB
self.last_job_query_count = len(data)
for uuid in data:
if uuid in self.active_jobs or uuid in self.queued_jobs:
continue
kwargs_raw = {}
reserved_arguments = {}
# Load attributes into dicts for use by the system or custom functions.
for key in data[uuid]:
if key in self.reserved_arguments:
reserved_arguments[key] = data[uuid][key][0]
else:
kwargs_raw[key] = data[uuid][key][0]
# Check for the presence of all required system attributes.
if "reservation_function_name" not in reserved_arguments:
LOGGER.error("Reservation %s does not have a function name." % uuid)
self.deleteReservation(uuid)
continue
function_name = reserved_arguments["reservation_function_name"]
if function_name not in self.functions:
LOGGER.error("Unable to process function %s for UUID: %s" % (function_name, uuid))
continue
if "reservation_created" not in reserved_arguments:
LOGGER.error("Reservation %s, %s does not have a created time." % (function_name, uuid))
self.deleteReservation(uuid, function_name=function_name)
continue
if "reservation_next_request" not in reserved_arguments:
LOGGER.error("Reservation %s, %s does not have a next request time." % (function_name, uuid))
self.deleteReservation(uuid, function_name=function_name)
continue
if "reservation_error" not in reserved_arguments:
LOGGER.error("Reservation %s, %s does not have an error flag." % (function_name, uuid))
self.deleteReservation(uuid, function_name=function_name)
continue
# Load custom function.
if function_name in self.functions:
exposed_function = self.functions[function_name]
else:
LOGGER.error("Could not find function %s." % function_name)
continue
# Check for required / optional arguments.
kwargs = {}
for key in kwargs_raw:
if key in exposed_function["required_arguments"]:
kwargs[key] = kwargs_raw[key]
if key in exposed_function["optional_arguments"]:
kwargs[key] = kwargs_raw[key]
has_required_arguments = True
for key in exposed_function["required_arguments"]:
if key not in kwargs:
has_required_arguments = False
LOGGER.error("%s, %s does not have required argument %s." % (function_name, uuid, key))
if not has_required_arguments:
continue
self.queued_jobs[uuid] = True
job = {"exposed_function":exposed_function,
"kwargs":kwargs,
"function_name":function_name,
"uuid":uuid}
if "reservation_cache" in reserved_arguments:
LOGGER.debug("Using reservation fast cache for %s, %s on on SimpleDB." % (function_name, uuid))
job["reservation_cache"] = reserved_arguments["reservation_cache"]
else:
job["reservation_cache"] = None
self.job_queue.append(job)
self.job_count = 0
self.query_start_time = time.time()
self.executeJobs()
def reportJobSpeed(self):
if self.query_start_time is not None and self.job_count > 0:
seconds_per_job = (time.time() - self.query_start_time) / self.job_count
LOGGER.info("Average execution time: %s, %s active." % (seconds_per_job, len(self.active_jobs)))
else:
LOGGER.info("No average speed to report yet.")
def executeJobs(self, data=None):
while len(self.job_queue) > 0 and len(self.active_jobs) < self.simultaneous_jobs:
job = self.job_queue.pop(0)
exposed_function = job["exposed_function"]
kwargs = job["kwargs"]
function_name = job["function_name"]
uuid = job["uuid"]
del self.queued_jobs[uuid]
LOGGER.debug("Calling %s with args %s" % (function_name, kwargs))
d = self.callExposedFunction(
exposed_function["function"],
kwargs,
function_name,
uuid=uuid,
reservation_fast_cache=job["reservation_cache"])
d.addCallback(self._jobCountCallback)
d.addErrback(self._jobCountErrback)
d.addCallback(self._setNextRequest, uuid, exposed_function["interval"], function_name)
def _jobCountCallback(self, data=None):
self.job_count += 1
def _jobCountErrback(self, error):
self.job_count += 1
def _setNextRequest(self, data, uuid, exposed_function_interval, function_name):
reservation_next_request_parameters = {
"reservation_next_request":sdb_now_add(
exposed_function_interval,
offset=self.time_offset)}
if uuid in self.reservation_fast_caches:
LOGGER.debug("Set reservation fast cache for %s, %s on on SimpleDB." % (function_name, uuid))
reservation_next_request_parameters["reservation_cache"] = self.reservation_fast_caches[uuid]
del self.reservation_fast_caches[uuid]
self.reservation_update_queue.append((
uuid,
reservation_next_request_parameters))
if len(self.reservation_update_queue) > 25:
self._sendReservationUpdateQueue()
def _sendReservationUpdateQueue(self, data=None):
if len(self.reservation_update_queue) < 25 and (len(self.active_jobs) > 0 or len(self.job_queue) > 0):
return
if len(self.reservation_update_queue) == 0:
return
LOGGER.debug("Sending reservation queue. Current length is %s" % (
len(self.reservation_update_queue)))
reservation_updates = self.reservation_update_queue[0:25]
replace=["reservation_next_request", "reservation_cache"]
reservation_replaces = [(x[0], replace) for x in reservation_updates]
reservation_updates = dict(reservation_updates)
reservation_replaces = dict(reservation_replaces)
self.reservation_update_queue = self.reservation_update_queue[25:]
d = self.sdb.batchPutAttributes(
self.aws_sdb_reservation_domain,
reservation_updates,
replace_by_item_name=reservation_replaces)
d.addCallback(
self._sendReservationUpdateQueueCallback,
reservation_updates.keys())
d.addErrback(
self._sendReservationUpdateQueueErrback,
reservation_updates.keys(),
reservation_updates)
if len(self.reservation_update_queue) > 0:
reactor.callLater(0, self._sendReservationUpdateQueue)
def _sendReservationUpdateQueueCallback(self, data, uuids):
LOGGER.debug("Set next request for %s on on SimpleDB." % uuids)
def _sendReservationUpdateQueueErrback(self, error, uuids, reservation_updates):
LOGGER.error("Unable to set next request for %s on SimpleDB. Adding back to update queue.\n%s" % (uuids, error.value))
self.reservation_update_queue.extend(reservation_updates.items()) | AWSpider | /AWSpider-0.3.2.12.tar.gz/AWSpider-0.3.2.12/awspider/servers/execution.py | execution.py |
import pprint
from uuid import uuid4
from twisted.internet.defer import Deferred, DeferredList, maybeDeferred
from twisted.web.resource import Resource
from twisted.internet import reactor
from twisted.web import server
from .base import BaseServer, LOGGER
from ..resources import InterfaceResource, ExposedResource
from ..aws import sdb_now
from ..evaluateboolean import evaluateBoolean
PRETTYPRINTER = pprint.PrettyPrinter(indent=4)
class InterfaceServer(BaseServer):
exposed_functions = []
exposed_function_resources = {}
def __init__(self,
aws_access_key_id,
aws_secret_access_key,
aws_sdb_reservation_domain,
aws_s3_reservation_cache_bucket=None,
aws_s3_http_cache_bucket=None,
aws_s3_storage_bucket=None,
aws_sdb_coordination_domain=None,
max_simultaneous_requests=50,
max_requests_per_host_per_second=1,
max_simultaneous_requests_per_host=5,
port=5000,
log_file='interfaceserver.log',
log_directory=None,
log_level="debug",
name=None,
time_offset=None):
if name == None:
name = "AWSpider Interface Server UUID: %s" % self.uuid
resource = Resource()
interface_resource = InterfaceResource(self)
resource.putChild("interface", interface_resource)
self.function_resource = Resource()
resource.putChild("function", self.function_resource)
self.site_port = reactor.listenTCP(port, server.Site(resource))
BaseServer.__init__(
self,
aws_access_key_id,
aws_secret_access_key,
aws_s3_reservation_cache_bucket=aws_s3_reservation_cache_bucket,
aws_s3_http_cache_bucket=aws_s3_http_cache_bucket,
aws_sdb_reservation_domain=aws_sdb_reservation_domain,
aws_s3_storage_bucket=aws_s3_storage_bucket,
aws_sdb_coordination_domain=aws_sdb_coordination_domain,
max_simultaneous_requests=max_simultaneous_requests,
max_requests_per_host_per_second=max_requests_per_host_per_second,
max_simultaneous_requests_per_host=max_simultaneous_requests_per_host,
log_file=log_file,
log_directory=log_directory,
log_level=log_level,
name=name,
time_offset=time_offset,
port=port)
def start(self):
reactor.callWhenRunning(self._start)
return self.start_deferred
def _start(self):
deferreds = []
if self.time_offset is None:
deferreds.append(self.getTimeOffset())
d = DeferredList(deferreds, consumeErrors=True)
d.addCallback(self._startCallback)
def _startCallback(self, data):
for row in data:
if row[0] == False:
d = self.shutdown()
d.addCallback(self._startHandleError, row[1])
return d
d = BaseServer.start(self)
def shutdown(self):
deferreds = []
LOGGER.debug("%s stopping on main HTTP interface." % self.name)
d = self.site_port.stopListening()
if isinstance(d, Deferred):
deferreds.append(d)
if len(deferreds) > 0:
d = DeferredList(deferreds)
d.addCallback(self._shutdownCallback)
return d
else:
return self._shutdownCallback(None)
def _shutdownCallback(self, data):
return BaseServer.shutdown(self)
def makeCallable(self, func, interval=0, name=None, expose=False):
function_name = BaseServer.makeCallable(
self,
func,
interval=interval,
name=name,
expose=expose)
if expose:
self.exposed_functions.append(function_name)
er = ExposedResource(self, function_name)
function_name_parts = function_name.split("/")
if len(function_name_parts) > 1:
if function_name_parts[0] in self.exposed_function_resources:
r = self.exposed_function_resources[function_name_parts[0]]
else:
r = Resource()
self.exposed_function_resources[function_name_parts[0]] = r
self.function_resource.putChild(function_name_parts[0], r)
r.putChild(function_name_parts[1], er)
else:
self.function_resource.putChild(function_name_parts[0], er)
LOGGER.info("Function %s is now available via the HTTP interface." % function_name)
def createReservation(self, function_name, **kwargs):
if not isinstance(function_name, str):
for key in self.functions:
if self.functions[key]["function"] == function_name:
function_name = key
break
if function_name not in self.functions:
raise Exception("Function %s does not exist." % function_name)
function = self.functions[function_name]
filtered_kwargs = {}
for key in function["required_arguments"]:
if key in kwargs:
#filtered_kwargs[key] = convertToUTF8(kwargs[key])
filtered_kwargs[key] = kwargs[key]
else:
raise Exception("Required parameter '%s' not found. Required parameters are %s. Optional parameters are %s." % (key, function["required_arguments"], function["optional_arguments"]))
for key in function["optional_arguments"]:
if key in kwargs:
#filtered_kwargs[key] = convertToUTF8(kwargs[key])
filtered_kwargs[key] = kwargs[key]
if function["interval"] > 0:
reserved_arguments = {}
reserved_arguments["reservation_function_name"] = function_name
reserved_arguments["reservation_created"] = sdb_now(offset=self.time_offset)
reserved_arguments["reservation_next_request"] = reserved_arguments["reservation_created"]
reserved_arguments["reservation_error"] = "0"
arguments = {}
arguments.update(reserved_arguments)
arguments.update(filtered_kwargs)
uuid = uuid4().hex
LOGGER.debug("Creating reservation on SimpleDB for %s, %s." % (function_name, uuid))
a = self.sdb.putAttributes(self.aws_sdb_reservation_domain, uuid, arguments)
a.addCallback(self._createReservationCallback, function_name, uuid)
a.addErrback(self._createReservationErrback, function_name, uuid)
if "call_immediately" in kwargs and not evaluateBoolean(kwargs["call_immediately"]):
d = DeferredList([a], consumeErrors=True)
else:
LOGGER.debug("Calling %s immediately with arguments:\n%s" % (function_name, PRETTYPRINTER.pformat(filtered_kwargs)))
self.active_jobs[uuid] = True
b = self.callExposedFunction(function["function"], filtered_kwargs, function_name, uuid=uuid)
d = DeferredList([a,b], consumeErrors=True)
d.addCallback(self._createReservationCallback2, function_name, uuid)
d.addErrback(self._createReservationErrback2, function_name, uuid)
return d
else:
LOGGER.debug("Calling %s with arguments:\n%s" % (function_name, PRETTYPRINTER.pformat(filtered_kwargs)))
d = self.callExposedFunction(function["function"], filtered_kwargs, function_name)
return d
def _createReservationCallback(self, data, function_name, uuid):
LOGGER.error(data)
LOGGER.debug("Created reservation on SimpleDB for %s, %s." % (function_name, uuid))
return uuid
def _createReservationErrback(self, error, function_name, uuid):
LOGGER.error("Unable to create reservation on SimpleDB for %s:%s, %s.\n" % (function_name, uuid, error))
return error
def _createReservationCallback2(self, data, function_name, uuid):
for row in data:
if row[0] == False:
raise row[1]
if len(data) == 1:
return {data[0][1]:{}}
else:
return {data[0][1]:data[1][1]}
def _createReservationErrback2(self, error, function_name, uuid):
LOGGER.error("Unable to create reservation for %s:%s, %s.\n" % (function_name, uuid, error))
return error
def showReservation(self, uuid):
d = self.sdb.getAttributes(self.aws_sdb_reservation_domain, uuid)
return d
def executeReservation(self, uuid):
sql = "SELECT * FROM `%s` WHERE itemName() = '%s'" % (self.aws_sdb_reservation_domain, uuid)
LOGGER.debug("Querying SimpleDB, \"%s\"" % sql)
d = self.sdb.select(sql)
d.addCallback(self._executeReservationCallback)
d.addErrback(self._executeReservationErrback)
return d
def _executeReservationCallback(self, data):
if len(data) == 0:
raise Exception("Could not find reservation.")
uuid = data.keys()[0]
kwargs_raw = {}
reserved_arguments = {}
# Load attributes into dicts for use by the system or custom functions.
for key in data[uuid]:
if key in self.reserved_arguments:
reserved_arguments[key] = data[uuid][key][0]
else:
kwargs_raw[key] = data[uuid][key][0]
# Check to make sure the custom function is present.
function_name = reserved_arguments["reservation_function_name"]
if function_name not in self.functions:
raise Exception("Unable to process function %s for UUID: %s" % (function_name, uuid))
return
# Check for the presence of all required system attributes.
if "reservation_function_name" not in reserved_arguments:
self.deleteReservation(uuid)
raise Exception("Reservation %s does not have a function name." % uuid)
if "reservation_created" not in reserved_arguments:
self.deleteReservation(uuid, function_name=function_name)
raise Exception("Reservation %s, %s does not have a created time." % (function_name, uuid))
if "reservation_next_request" not in reserved_arguments:
self.deleteReservation(uuid, function_name=function_name)
raise Exception("Reservation %s, %s does not have a next request time." % (function_name, uuid))
if "reservation_error" not in reserved_arguments:
self.deleteReservation(uuid, function_name=function_name)
raise Exception("Reservation %s, %s does not have an error flag." % (function_name, uuid))
# Load custom function.
if function_name in self.functions:
exposed_function = self.functions[function_name]
else:
raise Exception("Could not find function %s." % function_name)
return
# Check for required / optional arguments.
kwargs = {}
for key in kwargs_raw:
if key in exposed_function["required_arguments"]:
kwargs[key] = kwargs_raw[key]
if key in exposed_function["optional_arguments"]:
kwargs[key] = kwargs_raw[key]
has_reqiured_arguments = True
for key in exposed_function["required_arguments"]:
if key not in kwargs:
has_reqiured_arguments = False
raise Exception("%s, %s does not have required argument %s." % (function_name, uuid, key))
LOGGER.debug("Executing function.\n%s" % function_name)
return self.callExposedFunction(exposed_function["function"], kwargs, function_name, uuid=uuid)
def _executeReservationErrback(self, error):
LOGGER.error("Unable to query SimpleDB.\n%s" % error) | AWSpider | /AWSpider-0.3.2.12.tar.gz/AWSpider-0.3.2.12/awspider/servers/interface.py | interface.py |
from uuid import UUID, uuid4
import time
import random
import logging
import logging.handlers
from heapq import heappush, heappop
from twisted.internet import reactor, task
from twisted.web import server
from twisted.enterprise import adbapi
from MySQLdb.cursors import DictCursor
from twisted.internet.defer import Deferred, inlineCallbacks, DeferredList
from twisted.internet import task
from twisted.internet.threads import deferToThread
from txamqp.content import Content
from .base import BaseServer, LOGGER
from ..resources2 import SchedulerResource
from ..amqp import amqp as AMQP
from ..resources import ExposedResource
from twisted.web.resource import Resource
class SchedulerServer(BaseServer):
exposed_functions = []
exposed_function_resources = {}
name = "AWSpider Schedule Server UUID: %s" % str(uuid4())
heap = []
unscheduled_items = []
enqueueCallLater = None
statusloop = None
amqp_queue_size = 0
def __init__(self,
mysql_username,
mysql_password,
mysql_host,
mysql_database,
amqp_host,
amqp_username,
amqp_password,
amqp_vhost,
amqp_queue,
amqp_exchange,
amqp_port=5672,
mysql_port=3306,
port=5004,
service_mapping=None,
log_file='schedulerserver.log',
log_directory=None,
log_level="debug"):
self.function_resource = Resource()
# Create MySQL connection.
self.mysql = adbapi.ConnectionPool(
"MySQLdb",
db=mysql_database,
port=mysql_port,
user=mysql_username,
passwd=mysql_password,
host=mysql_host,
cp_reconnect=True,
cursorclass=DictCursor)
# Resource Mappings
self.service_mapping = service_mapping
# AMQP connection parameters
self.amqp_host = amqp_host
self.amqp_vhost = amqp_vhost
self.amqp_port = amqp_port
self.amqp_username = amqp_username
self.amqp_password = amqp_password
self.amqp_queue = amqp_queue
self.amqp_exchange = amqp_exchange
# HTTP interface
resource = SchedulerResource(self)
self.function_resource = Resource()
resource.putChild("function", self.function_resource)
self.site_port = reactor.listenTCP(port, server.Site(resource))
# Logging, etc
BaseServer.__init__(
self,
log_file=log_file,
log_directory=log_directory,
log_level=log_level)
function_name = BaseServer.makeCallable(
self,
self.remoteAddToHeap,
interval=0,
name=None,
expose=True)
self.exposed_functions.append(function_name)
er = ExposedResource(self, function_name)
function_name_parts = function_name.split("/")
if len(function_name_parts) > 1:
if function_name_parts[0] in self.exposed_function_resources:
r = self.exposed_function_resources[function_name_parts[0]]
else:
r = Resource()
self.exposed_function_resources[function_name_parts[0]] = r
self.function_resource.putChild(function_name_parts[0], r)
r.putChild(function_name_parts[1], er)
else:
self.function_resource.putChild(function_name_parts[0], er)
LOGGER.info("Function %s is now available via the HTTP interface."
% function_name)
function_name = BaseServer.makeCallable(
self,
self.remoteRemoveFromHeap,
interval=0,
name=None,
expose=True)
self.exposed_functions.append(function_name)
er = ExposedResource(self, function_name)
function_name_parts = function_name.split("/")
if len(function_name_parts) > 1:
if function_name_parts[0] in self.exposed_function_resources:
r = self.exposed_function_resources[function_name_parts[0]]
else:
r = Resource()
self.exposed_function_resources[function_name_parts[0]] = r
self.function_resource.putChild(function_name_parts[0], r)
r.putChild(function_name_parts[1], er)
else:
self.function_resource.putChild(function_name_parts[0], er)
LOGGER.info("Function %s is now available via the HTTP interface."
% function_name)
def start(self):
reactor.callWhenRunning(self._start)
return self.start_deferred
@inlineCallbacks
def _start(self):
# Load in names of functions supported by plugins
self.function_names = self.functions.keys()
LOGGER.info('Connecting to broker.')
self.conn = yield AMQP.createClient(
self.amqp_host,
self.amqp_vhost,
self.amqp_port)
yield self.conn.authenticate(self.amqp_username, self.amqp_password)
self.chan = yield self.conn.channel(1)
yield self.chan.channel_open()
# Create Queue
yield self.chan.queue_declare(
queue=self.amqp_queue,
durable=False,
exclusive=False,
auto_delete=False)
# Create Exchange
yield self.chan.exchange_declare(
exchange=self.amqp_exchange,
type="fanout",
durable=False,
auto_delete=False)
yield self.chan.queue_bind(
queue=self.amqp_queue,
exchange=self.amqp_exchange)
# Build heap from data in MySQL
yield self._loadFromMySQL()
self.statusloop = task.LoopingCall(self.queueStatusCheck)
self.statusloop.start(60)
def _loadFromMySQL(self, start=0):
# Select the entire spider_service DB, 10k rows at at time.
sql = """SELECT uuid, type
FROM spider_service
ORDER BY id LIMIT %s, 10000
""" % start
LOGGER.debug(sql)
d = self.mysql.runQuery(sql)
d.addCallback(self._loadFromMySQLCallback, start)
d.addErrback(self._loadFromMySQLErrback)
return d
def _loadFromMySQLCallback(self, data, start):
# Add rows to heap. The second argument is interval, would be
# based on the plugin's interval setting, random for now.
for row in data:
self.addToHeap(row["uuid"], row["type"])
# Load next chunk.
if len(data) >= 10000:
return self._loadFromMySQL(start=start + 10000)
# Done loading, start queuing
self.enqueue()
d = BaseServer.start(self)
return d
def _loadFromMySQLErrback(self, error):
return error
@inlineCallbacks
def shutdown(self):
LOGGER.debug("Closting connection")
try:
self.enqueueCallLater.cancel()
except:
pass
# Shut things down
LOGGER.info('Closing broker connection')
yield self.chan.channel_close()
chan0 = yield self.conn.channel(0)
yield chan0.connection_close()
LOGGER.info('Closing MYSQL Connnection Pool')
yield self.mysql.close()
# def enqueue(self):
# # Defer this to a thread so we don't block on the web interface.
# deferToThread(self._enqueue)
@inlineCallbacks
def queueStatusCheck(self):
yield self.chan.queue_bind(
queue=self.amqp_queue,
exchange=self.amqp_exchange)
queue_status = yield self.chan.queue_declare(
queue=self.amqp_queue,
passive=True)
self.amqp_queue_size = queue_status.fields[1]
LOGGER.debug('AMQP queue size: %d' % self.amqp_queue_size)
def enqueue(self):
now = int(time.time())
# Compare the heap min timestamp with now().
# If it's time for the item to be queued, pop it, update the
# timestamp and add it back to the heap for the next go round.
queue_items = []
if self.amqp_queue_size < 100000:
queue_items_a = queue_items.append
LOGGER.debug("%s:%s" % (self.heap[0][0], now))
while self.heap[0][0] < now and len(queue_items) < 1000:
job = heappop(self.heap)
uuid = UUID(bytes=job[1][0])
if not uuid.hex in self.unscheduled_items:
queue_items_a(job[1][0])
new_job = (now + job[1][1], job[1])
heappush(self.heap, new_job)
else:
self.unscheduled_items.remove(uuid.hex)
else:
LOGGER.critical('AMQP queue is at or beyond max limit (%d/100000)'
% self.amqp_queue_size)
# add items to amqp
if queue_items:
LOGGER.info('Found %d new uuids, adding them to the queue'
% len(queue_items))
msgs = [Content(uuid) for uuid in queue_items]
deferreds = [self.chan.basic_publish(
exchange=self.amqp_exchange, content=msg) for msg in msgs]
d = DeferredList(deferreds, consumeErrors=True)
d.addCallbacks(self._addToQueueComplete, self._addToQueueErr)
else:
self.enqueueCallLater = reactor.callLater(1, self.enqueue)
def _addToQueueComplete(self, data):
LOGGER.info('Completed adding items into the queue...')
self.enqueueCallLater = reactor.callLater(2, self.enqueue)
def _addToQueueErr(self, error):
LOGGER.error(error.printBriefTraceback)
raise
def remoteAddToHeap(self, uuid, type):
LOGGER.debug('remoteAddToHeap: uuid=%s, type=%s' % (uuid, type))
pass
def remoteRemoveFromHeap(self, uuid):
LOGGER.debug('remoteRemoveFromHeap: uuid=%s' % uuid)
def createReservation(self, function_name, **kwargs):
LOGGER.debug('%s Called' % function_name)
if function_name == 'schedulerserver/remoteaddtoheap':
LOGGER.debug('remoteaddtoheap has been called')
LOGGER.debug('kwargs: %s' % repr(kwargs))
if set(('uuid', 'type')).issubset(set(kwargs)):
LOGGER.debug('\tUUID: %s\n\tType: %s'
% (kwargs['uuid'], kwargs['type']))
if kwargs['uuid']:
self.addToHeap(kwargs['uuid'], kwargs['type'])
return {}
else:
return {'error':
'Required parameters are uuid and type'}
elif function_name == 'schedulerserver/remoteremovefromheap':
LOGGER.debug('remoteremovefromheap has been called')
LOGGER.debug('kwargs: %s' % repr(kwargs))
if 'uuid' in kwargs:
LOGGER.debug('UUID: %s' % kwargs['uuid'])
if kwargs['uuid']:
self.removeFromHeap(kwargs['uuid'])
return {}
else:
return {'error':
'Required parameters are uuid'}
return
def addToHeap(self, uuid, type):
# lookup if type is in the service_mapping, if it is
# then rewrite type to the proper resource
if not uuid in self.unscheduled_items:
if self.service_mapping and type in self.service_mapping:
LOGGER.info('Remapping resource %s to %s'
% (type, self.service_mapping[type]))
type = self.service_mapping[type]
try:
# Make sure the uuid is in bytes
uuid_bytes = UUID(uuid).bytes
except ValueError:
LOGGER.error('Cound not turn UUID into byes using string %s'
% uuid)
return
if type in self.functions and 'interval' in self.functions[type]:
interval = int(self.functions[type]['interval'])
else:
LOGGER.error('Could not find interval for type %s' % type)
return
enqueue_time = int(time.time() + interval)
# Add a UUID to the heap.
LOGGER.debug('Adding %s to heap with time %s and interval of %s'
% (uuid, enqueue_time, interval))
heappush(self.heap, (enqueue_time, (uuid_bytes, interval)))
else:
LOGGER.info('Unscheduling %s' % uuid)
self.unscheduled_items.remove(uuid)
def removeFromHeap(self, uuid):
LOGGER.info('Removing %s from heap' % uuid)
self.unscheduled_items.append(uuid) | AWSpider | /AWSpider-0.3.2.12.tar.gz/AWSpider-0.3.2.12/awspider/servers2/scheduler.py | scheduler.py |
import cPickle
import urllib
import inspect
import logging
import logging.handlers
import os
import time
from decimal import Decimal
from uuid import uuid4
from twisted.internet import reactor
from twisted.internet.threads import deferToThread
from twisted.internet.defer import Deferred, DeferredList, maybeDeferred
from ..aws import AmazonS3
from ..exceptions import DeleteReservationException
from ..pagegetter import PageGetter
from ..requestqueuer import RequestQueuer
import pprint
from boto.ec2.connection import EC2Connection
PRETTYPRINTER = pprint.PrettyPrinter(indent=4)
LOGGER = logging.getLogger("main")
class ReservationCachingException(Exception):
pass
class BaseServer(object):
scheduler_server = None
logging_handler = None
shutdown_trigger_id = None
uuid = uuid4().hex
start_time = time.time()
active_jobs = {}
reserved_arguments = [
"reservation_function_name",
"reservation_created",
"reservation_next_request",
"reservation_error"]
functions = {}
reservation_fast_caches = {}
def __init__(self,
aws_access_key_id=None,
aws_secret_access_key=None,
aws_s3_http_cache_bucket=None,
aws_s3_storage_bucket=None,
scheduler_server_group=None,
max_simultaneous_requests=100,
max_requests_per_host_per_second=0,
max_simultaneous_requests_per_host=0,
log_file=None,
log_directory=None,
log_level="debug",
port=8080):
self.start_deferred = Deferred()
self.rq = RequestQueuer(
max_simultaneous_requests=int(max_simultaneous_requests),
max_requests_per_host_per_second=int(max_requests_per_host_per_second),
max_simultaneous_requests_per_host=int(max_simultaneous_requests_per_host))
self.rq.setHostMaxRequestsPerSecond("127.0.0.1", 0)
self.rq.setHostMaxSimultaneousRequests("127.0.0.1", 0)
self.aws_access_key_id = aws_access_key_id
self.aws_secret_access_key = aws_secret_access_key
self.aws_s3_http_cache_bucket = aws_s3_http_cache_bucket
self.aws_s3_storage_bucket = aws_s3_storage_bucket
self.s3 = AmazonS3(
self.aws_access_key_id,
self.aws_secret_access_key,
rq=self.rq)
self.scheduler_server_group=scheduler_server_group
self.pg = PageGetter(
self.s3,
self.aws_s3_http_cache_bucket,
rq=self.rq)
self._setupLogging(log_file, log_directory, log_level)
def _setupLogging(self, log_file, log_directory, log_level):
if log_directory is None:
self.logging_handler = logging.StreamHandler()
else:
self.logging_handler = logging.handlers.TimedRotatingFileHandler(
os.path.join(log_directory, log_file),
when='D',
interval=1)
log_format = "%(levelname)s: %(message)s %(pathname)s:%(lineno)d"
self.logging_handler.setFormatter(logging.Formatter(log_format))
LOGGER.addHandler(self.logging_handler)
log_level = log_level.lower()
log_levels = {
"debug":logging.DEBUG,
"info":logging.INFO,
"warning":logging.WARNING,
"error":logging.ERROR,
"critical":logging.CRITICAL
}
if log_level in log_levels:
LOGGER.setLevel(log_levels[log_level])
else:
LOGGER.setLevel(logging.DEBUG)
def start(self):
reactor.callWhenRunning(self._start)
return self.start_deferred
def start(self):
reactor.callWhenRunning(self._baseStart)
return self.start_deferred
def _testAWSCredentials(self):
if self.aws_access_key_id is None:
raise Exception("AWS Access Key ID is required.")
if self.aws_secret_access_key is None:
raise Exception("AWS Secret Access Key ID is required.")
def _baseStart(self):
LOGGER.critical("Checking S3 setup.")
deferreds = []
if self.scheduler_server is None:
deferreds.append(deferToThread(self.setSchedulerServer))
if self.aws_s3_http_cache_bucket is not None:
self._testAWSCredentials()
deferreds.append(
self.s3.checkAndCreateBucket(self.aws_s3_http_cache_bucket))
if self.aws_s3_storage_bucket is not None:
self._testAWSCredentials()
deferreds.append(
self.s3.checkAndCreateBucket(self.aws_s3_storage_bucket))
d = DeferredList(deferreds, consumeErrors=True)
d.addCallback(self._baseStartCallback)
def _baseStartCallback(self, data):
for row in data:
if row[0] == False:
d = self.shutdown()
d.addCallback(self._startHandleError, row[1])
return d
self.shutdown_trigger_id = reactor.addSystemEventTrigger(
'before',
'shutdown',
self.shutdown)
LOGGER.critical("Starting.")
self._baseStartCallback2(None)
def _baseStartCallback2(self, data):
self.start_deferred.callback(True)
def _startHandleError(self, data, error):
self.start_deferred.errback(error)
def shutdown(self):
LOGGER.debug("Waiting for shutdown.")
d = Deferred()
reactor.callLater(0, self._waitForShutdown, d)
return d
def _waitForShutdown(self, shutdown_deferred):
if self.rq.getPending() > 0 or self.rq.getActive() > 0:
LOGGER.debug("Waiting for shutdown.")
reactor.callLater(1, self._waitForShutdown, shutdown_deferred)
return
self.shutdown_trigger_id = None
LOGGER.debug("Shut down.")
LOGGER.removeHandler(self.logging_handler)
shutdown_deferred.callback(True)
def callExposedFunction(self, func, kwargs, function_name, reservation_fast_cache=None, uuid=None):
if uuid is not None:
self.active_jobs[uuid] = True
if self.functions[function_name]["get_reservation_uuid"]:
kwargs["reservation_uuid"] = uuid
if self.functions[function_name]["check_reservation_fast_cache"] and \
reservation_fast_cache is not None:
kwargs["reservation_fast_cache"] = reservation_fast_cache
elif self.functions[function_name]["check_reservation_fast_cache"]:
kwargs["reservation_fast_cache"] = None
d = maybeDeferred(func, **kwargs)
d.addCallback(self._callExposedFunctionCallback, function_name, uuid)
d.addErrback(self._callExposedFunctionErrback, function_name, uuid)
return d
def _callExposedFunctionErrback(self, error, function_name, uuid):
if uuid is not None and uuid in self.active_jobs:
del self.active_jobs[uuid]
try:
error.raiseException()
except DeleteReservationException:
if uuid is not None:
self.deleteReservation(uuid)
message = """Error with %s, %s.\n%s
Reservation deleted at request of the function.""" % (
function_name,
uuid,
error)
LOGGER.debug(message)
return
except:
pass
if uuid is None:
LOGGER.error("Error with %s.\n%s" % (function_name, error))
else:
LOGGER.error("Error with %s.\nUUID:%s\n%s" % (
function_name,
uuid,
error))
return error
def setSchedulerServer(self):
try:
LOGGER.info('Locating scheduler server for security group: %s' % self.scheduler_server_group)
if self.scheduler_server_group and self.aws_access_key_id and self.aws_secret_access_key:
conn = EC2Connection(self.aws_access_key_id, self.aws_secret_access_key)
scheduler_hostnames = []
scheduler_hostnames_a = scheduler_hostnames.append
for reservation in conn.get_all_instances():
for reservation_group in reservation.groups:
if reservation_group.id == self.scheduler_server_group:
for instance in reservation.instances:
if instance.state == "running":
scheduler_hostnames_a(instance.private_dns_name)
if scheduler_hostnames:
self.scheduler_server = scheduler_hostnames[0]
if not self.scheduler_server:
LOGGER.error('No scheduler server found, using 0.0.0.0')
self.scheduler_server = "0.0.0.0"
LOGGER.debug('Scheduler Server found at %s' % self.scheduler_server)
except Exception, e:
LOGGER.error('setSchedulerServer Error: %s' % e)
def _callExposedFunctionCallback(self, data, function_name, uuid):
LOGGER.debug("Function %s returned successfully." % (function_name))
# If the UUID is None, this is a one-off type of thing.
if uuid is None:
return data
# If the data is None, there's nothing to store.
if data is None:
del self.active_jobs[uuid]
return None
# If we have an place to store the response on S3, do it.
if self.aws_s3_storage_bucket is not None:
LOGGER.debug("Putting result for %s, %s on S3." % (function_name, uuid))
pickled_data = cPickle.dumps(data)
d = self.s3.putObject(
self.aws_s3_storage_bucket,
uuid,
pickled_data,
content_type="text/plain",
gzip=True)
d.addCallback(self._exposedFunctionCallback2, data, uuid)
d.addErrback(self._exposedFunctionErrback2, data, function_name, uuid)
return d
return data
def _exposedFunctionErrback2(self, error, data, function_name, uuid):
if uuid in self.active_jobs:
del self.active_jobs[uuid]
LOGGER.error("Could not put results of %s, %s on S3.\n%s" % (function_name, uuid, error))
return data
def _exposedFunctionCallback2(self, s3_callback_data, data, uuid):
if uuid in self.active_jobs:
del self.active_jobs[uuid]
return data
def expose(self, *args, **kwargs):
return self.makeCallable(expose=True, *args, **kwargs)
def makeCallable(self, func, interval=0, name=None, expose=False):
argspec = inspect.getargspec(func)
# Get required / optional arguments
arguments = argspec[0]
if len(arguments) > 0 and arguments[0:1][0] == 'self':
arguments.pop(0)
kwarg_defaults = argspec[3]
if kwarg_defaults is None:
kwarg_defaults = []
required_arguments = arguments[0:len(arguments) - len(kwarg_defaults)]
optional_arguments = arguments[len(arguments) - len(kwarg_defaults):]
# Reservation fast cache is stored on with the reservation
if "reservation_fast_cache" in required_arguments:
del required_arguments[required_arguments.index("reservation_fast_cache")]
check_reservation_fast_cache = True
elif "reservation_fast_cache" in optional_arguments:
del optional_arguments[optional_arguments.index("reservation_fast_cache")]
check_reservation_fast_cache = True
else:
check_reservation_fast_cache = False
# Indicates whether to send the reservation's UUID to the function
if "reservation_uuid" in required_arguments:
del required_arguments[required_arguments.index("reservation_uuid")]
get_reservation_uuid = True
elif "reservation_uuid" in optional_arguments:
del optional_arguments[optional_arguments.index("reservation_uuid")]
get_reservation_uuid = True
else:
get_reservation_uuid = False
# Get function name, usually class/method
if name is not None:
function_name = name
elif hasattr(func, "im_class"):
function_name = "%s/%s" % (func.im_class.__name__, func.__name__)
else:
function_name = func.__name__
function_name = function_name.lower()
# Make sure the function isn't using any reserved arguments.
for key in required_arguments:
if key in self.reserved_arguments:
message = "Required argument name '%s' used in function %s is reserved." % (key, function_name)
LOGGER.error(message)
raise Exception(message)
for key in optional_arguments:
if key in self.reserved_arguments:
message = "Optional argument name '%s' used in function %s is reserved." % (key, function_name)
LOGGER.error(message)
raise Exception(message)
# Make sure we don't already have a function with the same name.
if function_name in self.functions:
raise Exception("A method or function with the name %s is already callable." % function_name)
# Add it to our list of callable functions.
self.functions[function_name] = {
"function":func,
"interval":interval,
"required_arguments":required_arguments,
"optional_arguments":optional_arguments,
"check_reservation_fast_cache":check_reservation_fast_cache,
"get_reservation_uuid":get_reservation_uuid
}
LOGGER.info("Function %s is now callable." % function_name)
return function_name
def getPage(self, *args, **kwargs):
return self.pg.getPage(*args, **kwargs)
def setHostMaxRequestsPerSecond(self, *args, **kwargs):
return self.rq.setHostMaxRequestsPerSecond(*args, **kwargs)
def setHostMaxSimultaneousRequests(self, *args, **kwargs):
return self.rq.setHostMaxSimultaneousRequests(*args, **kwargs)
def deleteReservation(self, uuid, function_name="Unknown"):
LOGGER.info("Deleting reservation %s, %s." % (function_name, uuid))
parameters = {'uuid': uuid}
query_string = urllib.urlencode(parameters)
url = 'http://%s:%s/function/schedulerserver/remoteremovefromheap?%s' % (self.scheduler_server, self.schedulerserver_port, query_string)
deferreds = []
# deferreds.append(self.getPage(url=url))
deferreds.append(self.s3.deleteObject(self.aws_s3_storage_bucket, uuid))
d = DeferredList(deferreds)
d.addCallback(self._deleteReservationCallback, function_name, uuid)
d.addErrback(self._deleteReservationErrback, function_name, uuid)
return d
def _deleteReservationCallback(self, data, function_name, uuid):
LOGGER.info("Reservation %s, %s successfully deleted." % (function_name, uuid))
return True
def _deleteReservationErrback(self, error, function_name, uuid ):
LOGGER.error("Error deleting reservation %s, %s.\n%s" % (function_name, uuid, error))
return False
def deleteHTTPCache(self):
deferreds = []
if self.aws_s3_http_cache_bucket is not None:
deferreds.append(
self.s3.emptyBucket(self.aws_s3_http_cache_bucket))
if len(deferreds) > 0:
d = DeferredList(deferreds, consumeErrors=True)
d.addCallback(self._deleteHTTPCacheCallback)
return d
else:
return True
def _deleteHTTPCacheCallback(self, data):
deferreds = []
if self.aws_s3_http_cache_bucket is not None:
deferreds.append(
self.s3.deleteBucket(self.aws_s3_http_cache_bucket))
if len(deferreds) > 0:
d = DeferredList(deferreds, consumeErrors=True)
d.addCallback(self._deleteHTTPCacheCallback2)
return d
else:
return True
def _deleteHTTPCacheCallback2(self, data):
return True
def getServerData(self):
running_time = time.time() - self.start_time
active_requests_by_host = self.rq.getActiveRequestsByHost()
pending_requests_by_host = self.rq.getPendingRequestsByHost()
data = {
"load_avg":[str(Decimal(str(x), 2)) for x in os.getloadavg()],
"running_time":running_time,
"active_requests_by_host":active_requests_by_host,
"pending_requests_by_host":pending_requests_by_host,
"active_requests":self.rq.getActive(),
"pending_requests":self.rq.getPending()
}
LOGGER.debug("Got server data:\n%s" % PRETTYPRINTER.pformat(data))
return data
def setReservationFastCache(self, uuid, data):
if not isinstance(data, str):
raise Exception("ReservationFastCache must be a string.")
if uuid is None:
return None
self.reservation_fast_caches[uuid] = data | AWSpider | /AWSpider-0.3.2.12.tar.gz/AWSpider-0.3.2.12/awspider/servers2/base.py | base.py |
import pprint
import urllib
from uuid import uuid4
from twisted.internet.defer import Deferred, DeferredList, maybeDeferred
from twisted.internet.threads import deferToThread
from twisted.web.resource import Resource
from twisted.internet import reactor
from twisted.web import server
from .base import BaseServer, LOGGER
from ..resources import InterfaceResource, ExposedResource
from ..aws import sdb_now
from ..evaluateboolean import evaluateBoolean
PRETTYPRINTER = pprint.PrettyPrinter(indent=4)
class InterfaceServer(BaseServer):
exposed_functions = []
exposed_function_resources = {}
name = "AWSpider Interface Server UUID: %s" % str(uuid4())
def __init__(self,
aws_access_key_id,
aws_secret_access_key,
aws_s3_http_cache_bucket=None,
aws_s3_storage_bucket=None,
remote_scheduler=True,
scheduler_server_group='flavors_spider_production',
schedulerserver_port=5004,
max_simultaneous_requests=50,
max_requests_per_host_per_second=1,
max_simultaneous_requests_per_host=5,
port=5000,
log_file='interfaceserver.log',
log_directory=None,
log_level="debug"):
self.aws_access_key_id=aws_access_key_id
self.aws_secret_access_key=aws_secret_access_key
self.scheduler_server_group=scheduler_server_group
self.schedulerserver_port=schedulerserver_port
self.remote_scheduler=remote_scheduler
resource = Resource()
interface_resource = InterfaceResource(self)
resource.putChild("interface", interface_resource)
self.function_resource = Resource()
resource.putChild("function", self.function_resource)
self.site_port = reactor.listenTCP(port, server.Site(resource))
BaseServer.__init__(
self,
aws_access_key_id,
aws_secret_access_key,
aws_s3_http_cache_bucket=aws_s3_http_cache_bucket,
aws_s3_storage_bucket=aws_s3_storage_bucket,
scheduler_server_group=scheduler_server_group,
max_simultaneous_requests=max_simultaneous_requests,
max_requests_per_host_per_second=max_requests_per_host_per_second,
max_simultaneous_requests_per_host=max_simultaneous_requests_per_host,
log_file=log_file,
log_directory=log_directory,
log_level=log_level,
port=port)
def start(self):
reactor.callWhenRunning(self._start)
return self.start_deferred
def _start(self):
deferreds = []
d = DeferredList(deferreds, consumeErrors=True)
d.addCallback(self._startCallback)
def _startCallback(self, data):
for row in data:
if row[0] == False:
d = self.shutdown()
d.addCallback(self._startHandleError, row[1])
return d
d = BaseServer.start(self)
def shutdown(self):
deferreds = []
LOGGER.debug("%s stopping on main HTTP interface." % self.name)
d = self.site_port.stopListening()
if isinstance(d, Deferred):
deferreds.append(d)
if len(deferreds) > 0:
d = DeferredList(deferreds)
d.addCallback(self._shutdownCallback)
return d
else:
return self._shutdownCallback(None)
def _shutdownCallback(self, data):
return BaseServer.shutdown(self)
def makeCallable(self, func, interval=0, name=None, expose=False):
function_name = BaseServer.makeCallable(
self,
func,
interval=interval,
name=name,
expose=expose)
if expose:
self.exposed_functions.append(function_name)
er = ExposedResource(self, function_name)
function_name_parts = function_name.split("/")
if len(function_name_parts) > 1:
if function_name_parts[0] in self.exposed_function_resources:
r = self.exposed_function_resources[function_name_parts[0]]
else:
r = Resource()
self.exposed_function_resources[function_name_parts[0]] = r
self.function_resource.putChild(function_name_parts[0], r)
r.putChild(function_name_parts[1], er)
else:
self.function_resource.putChild(function_name_parts[0], er)
LOGGER.info("Function %s is now available via the HTTP interface." % function_name)
def createReservation(self, function_name, **kwargs):
uuid = None
if not isinstance(function_name, str):
for key in self.functions:
if self.functions[key]["function"] == function_name:
function_name = key
break
if function_name not in self.functions:
raise Exception("Function %s does not exist." % function_name)
function = self.functions[function_name]
if function["interval"] > 0:
uuid = uuid4().hex
d = self.callExposedFunction(
self.functions[function_name]["function"],
kwargs,
function_name,
uuid=uuid)
d.addCallback(self._createReservationCallback, function_name, uuid)
d.addErrback(self._createReservationErrback, function_name, uuid)
return d
def _createReservationCallback(self, data, function_name, uuid):
if self.remote_scheduler:
parameters = {
'uuid': uuid,
'type': function_name
}
query_string = urllib.urlencode(parameters)
url = 'http://%s:%s/function/schedulerserver/remoteaddtoheap?%s' % (self.scheduler_server, self.schedulerserver_port, query_string)
LOGGER.info('Sending UUID to scheduler: %s' % url)
d = self.getPage(url=url)
d.addCallback(self._createReservationCallback2, function_name, uuid, data)
d.addErrback(self._createReservationErrback, function_name, uuid)
return d
else:
self._createReservationCallback2(data, function_name, uuid, data)
def _createReservationCallback2(self, data, function_name, uuid, reservation_data):
LOGGER.debug("Function %s returned successfully." % (function_name))
if not uuid:
return reservation_data
else:
return {uuid: reservation_data}
def _createReservationErrback(self, error, function_name, uuid):
LOGGER.error("Unable to create reservation for %s:%s, %s.\n" % (function_name, uuid, error))
return error | AWSpider | /AWSpider-0.3.2.12.tar.gz/AWSpider-0.3.2.12/awspider/servers2/interface.py | interface.py |
from .base import BaseServer, LOGGER
from ..resources2 import WorkerResource
from ..networkaddress import getNetworkAddress
from ..amqp import amqp as AMQP
from ..resources import InterfaceResource, ExposedResource
from MySQLdb.cursors import DictCursor
from twisted.internet import reactor, protocol, task
from twisted.enterprise import adbapi
from twisted.web import server
from twisted.protocols.memcache import MemCacheProtocol, DEFAULT_PORT
from twisted.internet.defer import Deferred, DeferredList, maybeDeferred, inlineCallbacks
from twisted.internet.threads import deferToThread
from uuid import UUID, uuid4
import pprint
import simplejson
PRETTYPRINTER = pprint.PrettyPrinter(indent=4)
class WorkerServer(BaseServer):
public_ip = None
local_ip = None
network_information = {}
simultaneous_jobs = 75
jobs_complete = 0
job_queue = []
job_queue_a = job_queue.append
jobsloop = None
pending_dequeue = False
def __init__(self,
aws_access_key_id,
aws_secret_access_key,
aws_s3_http_cache_bucket=None,
aws_s3_storage_bucket=None,
mysql_username=None,
mysql_password=None,
mysql_host=None,
mysql_database=None,
amqp_host=None,
amqp_username=None,
amqp_password=None,
amqp_vhost=None,
amqp_queue=None,
amqp_exchange=None,
memcached_host=None,
scheduler_server_group='flavors_spider_production',
schedulerserver_port=5004,
service_mapping=None,
service_args_mapping=None,
amqp_port=5672,
amqp_prefetch_count=200,
mysql_port=3306,
memcached_port=11211,
max_simultaneous_requests=100,
max_requests_per_host_per_second=0,
max_simultaneous_requests_per_host=0,
port=5005,
log_file='workerserver.log',
log_directory=None,
log_level="debug"):
self.network_information["port"] = port
# Create MySQL connection.
self.mysql = adbapi.ConnectionPool(
"MySQLdb",
db=mysql_database,
port=mysql_port,
user=mysql_username,
passwd=mysql_password,
host=mysql_host,
cp_reconnect=True,
cursorclass=DictCursor)
# Create Memcached client
self.memcached_host = memcached_host
self.memcached_port = memcached_port
self.memc_ClientCreator = protocol.ClientCreator(
reactor, MemCacheProtocol)
#Schedule Server
self.scheduler_server_group=scheduler_server_group
self.schedulerserver_port=schedulerserver_port
# Resource Mappings
self.service_mapping = service_mapping
self.service_args_mapping = service_args_mapping
# HTTP interface
resource = WorkerResource(self)
self.site_port = reactor.listenTCP(port, server.Site(resource))
# Create AMQP Connection
# AMQP connection parameters
self.amqp_host = amqp_host
self.amqp_vhost = amqp_vhost
self.amqp_port = amqp_port
self.amqp_username = amqp_username
self.amqp_password = amqp_password
self.amqp_queue = amqp_queue
self.amqp_exchange = amqp_exchange
self.amqp_prefetch_count = amqp_prefetch_count
BaseServer.__init__(
self,
aws_access_key_id=aws_access_key_id,
aws_secret_access_key=aws_secret_access_key,
aws_s3_http_cache_bucket=aws_s3_http_cache_bucket,
aws_s3_storage_bucket=aws_s3_storage_bucket,
scheduler_server_group=scheduler_server_group,
max_simultaneous_requests=max_simultaneous_requests,
max_requests_per_host_per_second=max_requests_per_host_per_second,
max_simultaneous_requests_per_host=max_simultaneous_requests_per_host,
log_file=log_file,
log_directory=log_directory,
log_level=log_level)
def start(self):
reactor.callWhenRunning(self._start)
return self.start_deferred
@inlineCallbacks
def _start(self):
yield self.getNetworkAddress()
# Create memcached client
self.memc = yield self.memc_ClientCreator.connectTCP(self.memcached_host, self.memcached_port)
LOGGER.info('Connecting to broker.')
self.conn = yield AMQP.createClient(
self.amqp_host,
self.amqp_vhost,
self.amqp_port)
self.auth = yield self.conn.authenticate(
self.amqp_username,
self.amqp_password)
self.chan = yield self.conn.channel(2)
yield self.chan.channel_open()
yield self.chan.basic_qos(prefetch_count=self.amqp_prefetch_count)
# Create Queue
yield self.chan.queue_declare(
queue=self.amqp_queue,
durable=False,
exclusive=False,
auto_delete=False)
# Create Exchange
yield self.chan.exchange_declare(
exchange=self.amqp_exchange,
type="fanout",
durable=False,
auto_delete=False)
yield self.chan.queue_bind(
queue=self.amqp_queue,
exchange=self.amqp_exchange)
yield self.chan.basic_consume(queue=self.amqp_queue,
no_ack=False,
consumer_tag="awspider_consumer")
self.queue = yield self.conn.queue("awspider_consumer")
yield BaseServer.start(self)
self.jobsloop = task.LoopingCall(self.executeJobs)
self.jobsloop.start(0.2)
LOGGER.info('Starting dequeueing thread...')
self.dequeue()
@inlineCallbacks
def shutdown(self):
LOGGER.debug("Closing connection")
try:
self.jobsloop.cancel()
except:
pass
# Shut things down
LOGGER.info('Closing broker connection')
yield self.chan.channel_close()
chan0 = yield self.conn.channel(0)
yield chan0.connection_close()
LOGGER.info('Closing MYSQL Connnection Pool')
yield self.mysql.close()
@inlineCallbacks
def reconnectMemcache(self):
self.memc = yield self.memc_ClientCreator.connectTCP(self.memcached_host, self.memcached_port)
def dequeue(self):
LOGGER.debug('Pending Deuque: %s / Completed Jobs: %d / Queued Jobs: %d / Active Jobs: %d' % (self.pending_dequeue, self.jobs_complete, len(self.job_queue), len(self.active_jobs)))
if len(self.job_queue) <= self.amqp_prefetch_count and not self.pending_dequeue:
self.pending_dequeue = True
LOGGER.debug('Fetching from queue')
d = self.queue.get()
d.addCallback(self._dequeueCallback)
d.addErrback(self._dequeueErrback)
else:
reactor.callLater(1, self.dequeue)
def _dequeueErrback(self, error):
LOGGER.error('Dequeue Error: %s' % error)
self.pending_dequeue = False
reactor.callLater(0, self.dequeue)
return error
def _dequeueCallback(self, msg):
if msg.delivery_tag:
LOGGER.debug('basic_ack for delivery_tag: %s' % msg.delivery_tag)
d = self.chan.basic_ack(msg.delivery_tag)
d.addCallback(self._dequeueCallback2, msg)
d.addErrback(self.basicAckErrback)
return d
else:
self._dequeueCallback2(data=True, msg=msg)
def _dequeueCallback2(self, data, msg):
LOGGER.debug('fetched msg from queue: %s' % repr(msg))
# Get the hex version of the UUID from byte string we were sent
uuid = UUID(bytes=msg.content.body).hex
d = self.getJob(uuid, msg.delivery_tag)
d.addCallback(self._dequeueCallback3, msg)
d.addErrback(self._dequeueErrback)
def _dequeueCallback3(self, job, msg):
# Load custom function.
if job is not None:
if job['function_name'] in self.functions:
LOGGER.debug('Successfully pulled job off of AMQP queue')
job['exposed_function'] = self.functions[job['function_name']]
if not job.has_key('kwargs'):
job['kwargs'] = self.mapKwargs(job)
if not job.has_key('delivery_tag'):
job['delivery_tag'] = msg.delivery_tag
# If function asked for fast_cache, try to fetch it from memcache
# while it's queued. Go ahead and add it to the queue in the meantime
# to speed things up.
job["reservation_fast_cache"] = None
if self.functions[job['function_name']]["check_reservation_fast_cache"]:
d = self.getReservationFastCache(job['uuid'])
d.addCallback(self._dequeueCallback4, job)
self.job_queue_a(job)
else:
LOGGER.error("Could not find function %s." % job['function_name'])
self.pending_dequeue = False
reactor.callLater(0, self.dequeue)
def _dequeueCallback4(self, data, job):
job["reservation_fast_cache"] = data
def executeJobs(self):
while len(self.job_queue) > 0 and len(self.active_jobs) < self.simultaneous_jobs:
job = self.job_queue.pop(0)
exposed_function = job["exposed_function"]
kwargs = job["kwargs"]
function_name = job["function_name"]
if job.has_key('uuid'):
uuid = job["uuid"]
else:
# assign a temp uuid
uuid = UUID(bytes=msg.content.body).hex
d = self.callExposedFunction(
exposed_function["function"],
kwargs,
function_name,
reservation_fast_cache=job["reservation_fast_cache"],
uuid=uuid)
d.addCallback(self._executeJobCallback, job)
d.addErrback(self.workerErrback, 'Execute Jobs', job['delivery_tag'])
def _executeJobCallback(self, data, job):
self.jobs_complete += 1
LOGGER.debug('Completed Jobs: %d / Queued Jobs: %d / Active Jobs: %d' % (self.jobs_complete, len(self.job_queue), len(self.active_jobs)))
# Save account info in memcached for up to 7 days
if job.has_key('exposed_function'):
del(job['exposed_function'])
d = self.memc.set(job['uuid'], simplejson.dumps(job), 60*60*24*7)
d.addCallback(self._executeJobCallback2)
d.addErrback(self.workerErrback, 'Execute Jobs', job['delivery_tag'])
return d
def _executeJobCallback2(self, data):
return
def workerErrback(self, error, function_name='Worker', delivery_tag=None):
LOGGER.error('%s Error: %s' % (function_name, str(error)))
LOGGER.debug('Queued Jobs: %d / Active Jobs: %d' % (len(self.job_queue), len(self.active_jobs)))
LOGGER.debug('Active Jobs List: %s' % repr(self.active_jobs))
self.pending_dequeue = False
if 'not connected' in str(error):
LOGGER.info('Attempting to reconnect to memcached...')
d = self.reconnectMemcache()
d.addCallback(self._reconnectMemcacheCallback)
return d
else:
return error
def _reconnectMemcacheCallback(self, data):
return
def _basicAckCallback(self, data):
return
def basicAckErrback(self, error):
LOGGER.error('basic_ack Error: %s' % (error))
return
def getJob(self, uuid, delivery_tag):
d = self.memc.get(uuid)
d.addCallback(self._getJobCallback, uuid, delivery_tag)
d.addErrback(self.workerErrback, 'Get Job', delivery_tag)
return d
def _getJobCallback(self, account, uuid, delivery_tag):
job = account[1]
if not job:
LOGGER.debug('Could not find uuid in memcached: %s' % uuid)
sql = "SELECT account_id, type FROM spider_service WHERE uuid = '%s'" % uuid
d = self.mysql.runQuery(sql)
d.addCallback(self.getAccountMySQL, uuid, delivery_tag)
d.addErrback(self.workerErrback, 'Get Job Callback', delivery_tag)
return d
else:
LOGGER.debug('Found uuid in memcached: %s' % uuid)
return simplejson.loads(job)
def getAccountMySQL(self, spider_info, uuid, delivery_tag):
if spider_info:
account_type = spider_info[0]['type'].split('/')[0]
sql = "SELECT * FROM content_%saccount WHERE account_id = %d" % (account_type.lower(), spider_info[0]['account_id'])
d = self.mysql.runQuery(sql)
d.addCallback(self.createJob, spider_info, uuid, delivery_tag)
d.addErrback(self.workerErrback, 'Get MySQL Account', delivery_tag)
return d
LOGGER.debug('No spider_info given for uuid %s' % uuid)
return None
def createJob(self, account_info, spider_info, uuid, delivery_tag):
job = {}
account = account_info[0]
function_name = spider_info[0]['type']
job['type'] = function_name.split('/')[1]
if self.service_mapping and self.service_mapping.has_key(function_name):
LOGGER.debug('Remapping resource %s to %s' % (function_name, self.service_mapping[function_name]))
function_name = self.service_mapping[function_name]
job['function_name'] = function_name
job['uuid'] = uuid
job['account'] = account
job['delivery_tag'] = delivery_tag
return job
def mapKwargs(self, job):
kwargs = {}
service_name = job['function_name'].split('/')[0]
# remap some fields that differ from the plugin and the database
if service_name in self.service_args_mapping:
for key in self.service_args_mapping[service_name]:
if key in job['account']:
job['account'][self.service_args_mapping[service_name][key]] = job['account'][key]
# apply job fields to req and optional kwargs
for arg in job['exposed_function']['required_arguments']:
if arg in job:
kwargs[arg] = job[arg]
elif arg in job['account']:
kwargs[arg] = job['account'][arg]
for arg in job['exposed_function']['optional_arguments']:
if arg in job['account']:
kwargs[arg] = job['account'][arg]
LOGGER.debug('Function: %s\nKWARGS: %s' %(job['function_name'], repr(kwargs)))
return kwargs
def getNetworkAddress(self):
d = getNetworkAddress()
d.addCallback(self._getNetworkAddressCallback)
d.addErrback(self._getNetworkAddressErrback)
return d
def _getNetworkAddressCallback(self, data):
if "public_ip" in data:
self.public_ip = data["public_ip"]
self.network_information["public_ip"] = self.public_ip
if "local_ip" in data:
self.local_ip = data["local_ip"]
self.network_information["local_ip"] = self.local_ip
def _getNetworkAddressErrback(self, error):
message = "Could not get network address."
LOGGER.error(message)
raise Exception(message)
def getReservationFastCache(self, uuid):
d = self.memc.get("%s_fc" % uuid)
d.addCallback(self._getReservationFastCacheCallback, uuid)
d.addErrback(self._getReservationFastCacheErrback, uuid)
return d
def _getReservationFastCacheCallback(self, data, uuid):
flags, value = data
if value:
LOGGER.debug("Successfully got Fast Cache for %s" % uuid)
return value
else:
LOGGER.debug("Could not get Fast Cache (2) for %s" % uuid)
return None
def _getReservationFastCacheErrback(self, error, uuid):
LOGGER.debug("Could not get Fast Cache (1) for %s" % uuid)
return None
def setReservationFastCache(self, uuid, data):
if not isinstance(data, str):
raise Exception("ReservationFastCache must be a string.")
if uuid is None:
return None
d = self.memc.set("%s_fc" % uuid, data, 60*60*24*7)
d.addCallback(self._setReservationFastCacheCallback, uuid)
d.addErrback(self._setReservationFastCacheErrback)
def _setReservationFastCacheCallback(self, data, uuid):
LOGGER.debug("Successfully set Fast Cache for %s" % uuid)
def _setReservationFastCacheErrback(self, error):
LOGGER.error(str(error)) | AWSpider | /AWSpider-0.3.2.12.tar.gz/AWSpider-0.3.2.12/awspider/servers2/worker.py | worker.py |
v0_8 = '''<?xml version="1.0"?>
<!--
Copyright Notice
================
(c) Copyright JPMorgan Chase Bank, Cisco Systems, Inc., Envoy Technologies Inc.,
iMatix Corporation, ION Technologies, Red Hat, Inc.,
TWIST Process Innovations, and 29West Inc. 2006. All rights reserved.
License
=======
JPMorgan Chase Bank, Cisco Systems, Inc., Envoy Technologies Inc., iMatix
Corporation, ION Technologies, Red Hat, Inc., TWIST Process Innovations, and
29West Inc. (collectively, the "Authors") each hereby grants to you a worldwide,
perpetual, royalty-free, nontransferable, nonexclusive license to
(i) copy, display, and implement the Advanced Messaging Queue Protocol
("AMQP") Specification and (ii) the Licensed Claims that are held by
the Authors, all for the purpose of implementing the Advanced Messaging
Queue Protocol Specification. Your license and any rights under this
Agreement will terminate immediately without notice from
any Author if you bring any claim, suit, demand, or action related to
the Advanced Messaging Queue Protocol Specification against any Author.
Upon termination, you shall destroy all copies of the Advanced Messaging
Queue Protocol Specification in your possession or control.
As used hereunder, "Licensed Claims" means those claims of a patent or
patent application, throughout the world, excluding design patents and
design registrations, owned or controlled, or that can be sublicensed
without fee and in compliance with the requirements of this
Agreement, by an Author or its affiliates now or at any
future time and which would necessarily be infringed by implementation
of the Advanced Messaging Queue Protocol Specification. A claim is
necessarily infringed hereunder only when it is not possible to avoid
infringing it because there is no plausible non-infringing alternative
for implementing the required portions of the Advanced Messaging Queue
Protocol Specification. Notwithstanding the foregoing, Licensed Claims
shall not include any claims other than as set forth above even if
contained in the same patent as Licensed Claims; or that read solely
on any implementations of any portion of the Advanced Messaging Queue
Protocol Specification that are not required by the Advanced Messaging
Queue Protocol Specification, or that, if licensed, would require a
payment of royalties by the licensor to unaffiliated third parties.
Moreover, Licensed Claims shall not include (i) any enabling technologies
that may be necessary to make or use any Licensed Product but are not
themselves expressly set forth in the Advanced Messaging Queue Protocol
Specification (e.g., semiconductor manufacturing technology, compiler
technology, object oriented technology, networking technology, operating
system technology, and the like); or (ii) the implementation of other
published standards developed elsewhere and merely referred to in the
body of the Advanced Messaging Queue Protocol Specification, or
(iii) any Licensed Product and any combinations thereof the purpose or
function of which is not required for compliance with the Advanced
Messaging Queue Protocol Specification. For purposes of this definition,
the Advanced Messaging Queue Protocol Specification shall be deemed to
include both architectural and interconnection requirements essential
for interoperability and may also include supporting source code artifacts
where such architectural, interconnection requirements and source code
artifacts are expressly identified as being required or documentation to
achieve compliance with the Advanced Messaging Queue Protocol Specification.
As used hereunder, "Licensed Products" means only those specific portions
of products (hardware, software or combinations thereof) that implement
and are compliant with all relevant portions of the Advanced Messaging
Queue Protocol Specification.
The following disclaimers, which you hereby also acknowledge as to any
use you may make of the Advanced Messaging Queue Protocol Specification:
THE ADVANCED MESSAGING QUEUE PROTOCOL SPECIFICATION IS PROVIDED "AS IS,"
AND THE AUTHORS MAKE NO REPRESENTATIONS OR WARRANTIES, EXPRESS OR
IMPLIED, INCLUDING, BUT NOT LIMITED TO, WARRANTIES OF MERCHANTABILITY,
FITNESS FOR A PARTICULAR PURPOSE, NON-INFRINGEMENT, OR TITLE; THAT THE
CONTENTS OF THE ADVANCED MESSAGING QUEUE PROTOCOL SPECIFICATION ARE
SUITABLE FOR ANY PURPOSE; NOR THAT THE IMPLEMENTATION OF THE ADVANCED
MESSAGING QUEUE PROTOCOL SPECIFICATION WILL NOT INFRINGE ANY THIRD PARTY
PATENTS, COPYRIGHTS, TRADEMARKS OR OTHER RIGHTS.
THE AUTHORS WILL NOT BE LIABLE FOR ANY DIRECT, INDIRECT, SPECIAL,
INCIDENTAL OR CONSEQUENTIAL DAMAGES ARISING OUT OF OR RELATING TO ANY
USE, IMPLEMENTATION OR DISTRIBUTION OF THE ADVANCED MESSAGING QUEUE
PROTOCOL SPECIFICATION.
The name and trademarks of the Authors may NOT be used in any manner,
including advertising or publicity pertaining to the Advanced Messaging
Queue Protocol Specification or its contents without specific, written
prior permission. Title to copyright in the Advanced Messaging Queue
Protocol Specification will at all times remain with the Authors.
No other rights are granted by implication, estoppel or otherwise.
Upon termination of your license or rights under this Agreement, you
shall destroy all copies of the Advanced Messaging Queue Protocol
Specification in your possession or control.
Trademarks
==========
"JPMorgan", "JPMorgan Chase", "Chase", the JPMorgan Chase logo and the
Octagon Symbol are trademarks of JPMorgan Chase & Co.
IMATIX and the iMatix logo are trademarks of iMatix Corporation sprl.
IONA, IONA Technologies, and the IONA logos are trademarks of IONA
Technologies PLC and/or its subsidiaries.
LINUX is a trademark of Linus Torvalds. RED HAT and JBOSS are registered
trademarks of Red Hat, Inc. in the US and other countries.
Java, all Java-based trademarks and OpenOffice.org are trademarks of
Sun Microsystems, Inc. in the United States, other countries, or both.
Other company, product, or service names may be trademarks or service
marks of others.
Links to full AMQP specification:
=================================
http://www.envoytech.org/spec/amq/
http://www.iona.com/opensource/amqp/
http://www.redhat.com/solutions/specifications/amqp/
http://www.twiststandards.org/tiki-index.php?page=AMQ
http://www.imatix.com/amqp
-->
<amqp major="8" minor="0" port="5672" comment="AMQ protocol 0.80">
AMQ Protocol 0.80
<!--
======================================================
== CONSTANTS
======================================================
-->
<constant name="frame method" value="1"/>
<constant name="frame header" value="2"/>
<constant name="frame body" value="3"/>
<constant name="frame oob method" value="4"/>
<constant name="frame oob header" value="5"/>
<constant name="frame oob body" value="6"/>
<constant name="frame trace" value="7"/>
<constant name="frame heartbeat" value="8"/>
<constant name="frame min size" value="4096"/>
<constant name="frame end" value="206"/>
<constant name="reply success" value="200">
Indicates that the method completed successfully. This reply code is
reserved for future use - the current protocol design does not use
positive confirmation and reply codes are sent only in case of an
error.
</constant>
<constant name="not delivered" value="310" class="soft error">
The client asked for a specific message that is no longer available.
The message was delivered to another client, or was purged from the
queue for some other reason.
</constant>
<constant name="content too large" value="311" class="soft error">
The client attempted to transfer content larger than the server
could accept at the present time. The client may retry at a later
time.
</constant>
<constant name="connection forced" value="320" class="hard error">
An operator intervened to close the connection for some reason.
The client may retry at some later date.
</constant>
<constant name="invalid path" value="402" class="hard error">
The client tried to work with an unknown virtual host or cluster.
</constant>
<constant name="access refused" value="403" class="soft error">
The client attempted to work with a server entity to which it has
no due to security settings.
</constant>
<constant name="not found" value="404" class="soft error">
The client attempted to work with a server entity that does not exist.
</constant>
<constant name="resource locked" value="405" class="soft error">
The client attempted to work with a server entity to which it has
no access because another client is working with it.
</constant>
<constant name="frame error" value="501" class="hard error">
The client sent a malformed frame that the server could not decode.
This strongly implies a programming error in the client.
</constant>
<constant name="syntax error" value="502" class="hard error">
The client sent a frame that contained illegal values for one or more
fields. This strongly implies a programming error in the client.
</constant>
<constant name="command invalid" value="503" class="hard error">
The client sent an invalid sequence of frames, attempting to perform
an operation that was considered invalid by the server. This usually
implies a programming error in the client.
</constant>
<constant name="channel error" value="504" class="hard error">
The client attempted to work with a channel that had not been
correctly opened. This most likely indicates a fault in the client
layer.
</constant>
<constant name="resource error" value="506" class="hard error">
The server could not complete the method because it lacked sufficient
resources. This may be due to the client creating too many of some
type of entity.
</constant>
<constant name="not allowed" value="530" class="hard error">
The client tried to work with some entity in a manner that is
prohibited by the server, due to security settings or by some other
criteria.
</constant>
<constant name="not implemented" value="540" class="hard error">
The client tried to use functionality that is not implemented in the
server.
</constant>
<constant name="internal error" value="541" class="hard error">
The server could not complete the method because of an internal error.
The server may require intervention by an operator in order to resume
normal operations.
</constant>
<!--
======================================================
== DOMAIN TYPES
======================================================
-->
<domain name="access ticket" type="short">
access ticket granted by server
<doc>
An access ticket granted by the server for a certain set of access
rights within a specific realm. Access tickets are valid within the
channel where they were created, and expire when the channel closes.
</doc>
<assert check="ne" value="0"/>
</domain>
<domain name="class id" type="short"/>
<domain name="consumer tag" type="shortstr">
consumer tag
<doc>
Identifier for the consumer, valid within the current connection.
</doc>
<rule implement="MUST">
The consumer tag is valid only within the channel from which the
consumer was created. I.e. a client MUST NOT create a consumer in
one channel and then use it in another.
</rule>
</domain>
<domain name="delivery tag" type="longlong">
server-assigned delivery tag
<doc>
The server-assigned and channel-specific delivery tag
</doc>
<rule implement="MUST">
The delivery tag is valid only within the channel from which the
message was received. I.e. a client MUST NOT receive a message on
one channel and then acknowledge it on another.
</rule>
<rule implement="MUST">
The server MUST NOT use a zero value for delivery tags. Zero is
reserved for client use, meaning "all messages so far received".
</rule>
</domain>
<domain name="exchange name" type="shortstr">
exchange name
<doc>
The exchange name is a client-selected string that identifies
the exchange for publish methods. Exchange names may consist
of any mixture of digits, letters, and underscores. Exchange
names are scoped by the virtual host.
</doc>
<assert check="length" value="127"/>
</domain>
<domain name="known hosts" type="shortstr">
list of known hosts
<doc>
Specifies the list of equivalent or alternative hosts that the server
knows about, which will normally include the current server itself.
Clients can cache this information and use it when reconnecting to a
server after a failure.
</doc>
<rule implement="MAY">
The server MAY leave this field empty if it knows of no other
hosts than itself.
</rule>
</domain>
<domain name="method id" type="short"/>
<domain name="no ack" type="bit">
no acknowledgement needed
<doc>
If this field is set the server does not expect acknowledgments
for messages. That is, when a message is delivered to the client
the server automatically and silently acknowledges it on behalf
of the client. This functionality increases performance but at
the cost of reliability. Messages can get lost if a client dies
before it can deliver them to the application.
</doc>
</domain>
<domain name="no local" type="bit">
do not deliver own messages
<doc>
If the no-local field is set the server will not send messages to
the client that published them.
</doc>
</domain>
<domain name="path" type="shortstr">
<doc>
Must start with a slash "/" and continue with path names
separated by slashes. A path name consists of any combination
of at least one of [A-Za-z0-9] plus zero or more of [.-_+!=:].
</doc>
<assert check="notnull"/>
<assert check="syntax" rule="path"/>
<assert check="length" value="127"/>
</domain>
<domain name="peer properties" type="table">
<doc>
This string provides a set of peer properties, used for
identification, debugging, and general information.
</doc>
<rule implement="SHOULD">
The properties SHOULD contain these fields:
"product", giving the name of the peer product, "version", giving
the name of the peer version, "platform", giving the name of the
operating system, "copyright", if appropriate, and "information",
giving other general information.
</rule>
</domain>
<domain name="queue name" type="shortstr">
queue name
<doc>
The queue name identifies the queue within the vhost. Queue
names may consist of any mixture of digits, letters, and
underscores.
</doc>
<assert check="length" value="127"/>
</domain>
<domain name="redelivered" type="bit">
message is being redelivered
<doc>
This indicates that the message has been previously delivered to
this or another client.
</doc>
<rule implement="SHOULD">
The server SHOULD try to signal redelivered messages when it can.
When redelivering a message that was not successfully acknowledged,
the server SHOULD deliver it to the original client if possible.
</rule>
<rule implement="MUST">
The client MUST NOT rely on the redelivered field but MUST take it
as a hint that the message may already have been processed. A
fully robust client must be able to track duplicate received messages
on non-transacted, and locally-transacted channels.
</rule>
</domain>
<domain name="reply code" type="short">
reply code from server
<doc>
The reply code. The AMQ reply codes are defined in AMQ RFC 011.
</doc>
<assert check="notnull"/>
</domain>
<domain name="reply text" type="shortstr">
localised reply text
<doc>
The localised reply text. This text can be logged as an aid to
resolving issues.
</doc>
<assert check="notnull"/>
</domain>
<class name="connection" handler="connection" index="10">
<!--
======================================================
== CONNECTION
======================================================
-->
work with socket connections
<doc>
The connection class provides methods for a client to establish a
network connection to a server, and for both peers to operate the
connection thereafter.
</doc>
<doc name="grammar">
connection = open-connection *use-connection close-connection
open-connection = C:protocol-header
S:START C:START-OK
*challenge
S:TUNE C:TUNE-OK
C:OPEN S:OPEN-OK | S:REDIRECT
challenge = S:SECURE C:SECURE-OK
use-connection = *channel
close-connection = C:CLOSE S:CLOSE-OK
/ S:CLOSE C:CLOSE-OK
</doc>
<chassis name="server" implement="MUST"/>
<chassis name="client" implement="MUST"/>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name="start" synchronous="1" index="10">
start connection negotiation
<doc>
This method starts the connection negotiation process by telling
the client the protocol version that the server proposes, along
with a list of security mechanisms which the client can use for
authentication.
</doc>
<rule implement="MUST">
If the client cannot handle the protocol version suggested by the
server it MUST close the socket connection.
</rule>
<rule implement="MUST">
The server MUST provide a protocol version that is lower than or
equal to that requested by the client in the protocol header. If
the server cannot support the specified protocol it MUST NOT send
this method, but MUST close the socket connection.
</rule>
<chassis name="client" implement="MUST"/>
<response name="start-ok"/>
<field name="version major" type="octet">
protocol major version
<doc>
The protocol major version that the server agrees to use, which
cannot be higher than the client's major version.
</doc>
</field>
<field name="version minor" type="octet">
protocol major version
<doc>
The protocol minor version that the server agrees to use, which
cannot be higher than the client's minor version.
</doc>
</field>
<field name="server properties" domain="peer properties">
server properties
</field>
<field name="mechanisms" type="longstr">
available security mechanisms
<doc>
A list of the security mechanisms that the server supports, delimited
by spaces. Currently ASL supports these mechanisms: PLAIN.
</doc>
<see name="security mechanisms"/>
<assert check="notnull"/>
</field>
<field name="locales" type="longstr">
available message locales
<doc>
A list of the message locales that the server supports, delimited
by spaces. The locale defines the language in which the server
will send reply texts.
</doc>
<rule implement="MUST">
All servers MUST support at least the en_US locale.
</rule>
<assert check="notnull"/>
</field>
</method>
<method name="start-ok" synchronous="1" index="11">
select security mechanism and locale
<doc>
This method selects a SASL security mechanism. ASL uses SASL
(RFC2222) to negotiate authentication and encryption.
</doc>
<chassis name="server" implement="MUST"/>
<field name="client properties" domain="peer properties">
client properties
</field>
<field name="mechanism" type="shortstr">
selected security mechanism
<doc>
A single security mechanisms selected by the client, which must be
one of those specified by the server.
</doc>
<rule implement="SHOULD">
The client SHOULD authenticate using the highest-level security
profile it can handle from the list provided by the server.
</rule>
<rule implement="MUST">
The mechanism field MUST contain one of the security mechanisms
proposed by the server in the Start method. If it doesn't, the
server MUST close the socket.
</rule>
<assert check="notnull"/>
</field>
<field name="response" type="longstr">
security response data
<doc>
A block of opaque data passed to the security mechanism. The contents
of this data are defined by the SASL security mechanism. For the
PLAIN security mechanism this is defined as a field table holding
two fields, LOGIN and PASSWORD.
</doc>
<assert check="notnull"/>
</field>
<field name="locale" type="shortstr">
selected message locale
<doc>
A single message local selected by the client, which must be one
of those specified by the server.
</doc>
<assert check="notnull"/>
</field>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name="secure" synchronous="1" index="20">
security mechanism challenge
<doc>
The SASL protocol works by exchanging challenges and responses until
both peers have received sufficient information to authenticate each
other. This method challenges the client to provide more information.
</doc>
<chassis name="client" implement="MUST"/>
<response name="secure-ok"/>
<field name="challenge" type="longstr">
security challenge data
<doc>
Challenge information, a block of opaque binary data passed to
the security mechanism.
</doc>
<see name="security mechanisms"/>
</field>
</method>
<method name="secure-ok" synchronous="1" index="21">
security mechanism response
<doc>
This method attempts to authenticate, passing a block of SASL data
for the security mechanism at the server side.
</doc>
<chassis name="server" implement="MUST"/>
<field name="response" type="longstr">
security response data
<doc>
A block of opaque data passed to the security mechanism. The contents
of this data are defined by the SASL security mechanism.
</doc>
<assert check="notnull"/>
</field>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name="tune" synchronous="1" index="30">
propose connection tuning parameters
<doc>
This method proposes a set of connection configuration values
to the client. The client can accept and/or adjust these.
</doc>
<chassis name="client" implement="MUST"/>
<response name="tune-ok"/>
<field name="channel max" type="short">
proposed maximum channels
<doc>
The maximum total number of channels that the server allows
per connection. Zero means that the server does not impose a
fixed limit, but the number of allowed channels may be limited
by available server resources.
</doc>
</field>
<field name="frame max" type="long">
proposed maximum frame size
<doc>
The largest frame size that the server proposes for the
connection. The client can negotiate a lower value. Zero means
that the server does not impose any specific limit but may reject
very large frames if it cannot allocate resources for them.
</doc>
<rule implement="MUST">
Until the frame-max has been negotiated, both peers MUST accept
frames of up to 4096 octets large. The minimum non-zero value for
the frame-max field is 4096.
</rule>
</field>
<field name="heartbeat" type="short">
desired heartbeat delay
<doc>
The delay, in seconds, of the connection heartbeat that the server
wants. Zero means the server does not want a heartbeat.
</doc>
</field>
</method>
<method name="tune-ok" synchronous="1" index="31">
negotiate connection tuning parameters
<doc>
This method sends the client's connection tuning parameters to the
server. Certain fields are negotiated, others provide capability
information.
</doc>
<chassis name="server" implement="MUST"/>
<field name="channel max" type="short">
negotiated maximum channels
<doc>
The maximum total number of channels that the client will use
per connection. May not be higher than the value specified by
the server.
</doc>
<rule implement="MAY">
The server MAY ignore the channel-max value or MAY use it for
tuning its resource allocation.
</rule>
<assert check="notnull"/>
<assert check="le" method="tune" field="channel max"/>
</field>
<field name="frame max" type="long">
negotiated maximum frame size
<doc>
The largest frame size that the client and server will use for
the connection. Zero means that the client does not impose any
specific limit but may reject very large frames if it cannot
allocate resources for them. Note that the frame-max limit
applies principally to content frames, where large contents
can be broken into frames of arbitrary size.
</doc>
<rule implement="MUST">
Until the frame-max has been negotiated, both peers must accept
frames of up to 4096 octets large. The minimum non-zero value for
the frame-max field is 4096.
</rule>
</field>
<field name="heartbeat" type="short">
desired heartbeat delay
<doc>
The delay, in seconds, of the connection heartbeat that the client
wants. Zero means the client does not want a heartbeat.
</doc>
</field>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name="open" synchronous="1" index="40">
open connection to virtual host
<doc>
This method opens a connection to a virtual host, which is a
collection of resources, and acts to separate multiple application
domains within a server.
</doc>
<rule implement="MUST">
The client MUST open the context before doing any work on the
connection.
</rule>
<chassis name="server" implement="MUST"/>
<response name="open-ok"/>
<response name="redirect"/>
<field name="virtual host" domain="path">
virtual host name
<assert check="regexp" value="^[a-zA-Z0-9/-_]+$"/>
<doc>
The name of the virtual host to work with.
</doc>
<rule implement="MUST">
If the server supports multiple virtual hosts, it MUST enforce a
full separation of exchanges, queues, and all associated entities
per virtual host. An application, connected to a specific virtual
host, MUST NOT be able to access resources of another virtual host.
</rule>
<rule implement="SHOULD">
The server SHOULD verify that the client has permission to access
the specified virtual host.
</rule>
<rule implement="MAY">
The server MAY configure arbitrary limits per virtual host, such
as the number of each type of entity that may be used, per
connection and/or in total.
</rule>
</field>
<field name="capabilities" type="shortstr">
required capabilities
<doc>
The client may specify a number of capability names, delimited by
spaces. The server can use this string to how to process the
client's connection request.
</doc>
</field>
<field name="insist" type="bit">
insist on connecting to server
<doc>
In a configuration with multiple load-sharing servers, the server
may respond to a Connection.Open method with a Connection.Redirect.
The insist option tells the server that the client is insisting on
a connection to the specified server.
</doc>
<rule implement="SHOULD">
When the client uses the insist option, the server SHOULD accept
the client connection unless it is technically unable to do so.
</rule>
</field>
</method>
<method name="open-ok" synchronous="1" index="41">
signal that the connection is ready
<doc>
This method signals to the client that the connection is ready for
use.
</doc>
<chassis name="client" implement="MUST"/>
<field name="known hosts" domain="known hosts"/>
</method>
<method name="redirect" synchronous="1" index="50">
asks the client to use a different server
<doc>
This method redirects the client to another server, based on the
requested virtual host and/or capabilities.
</doc>
<rule implement="SHOULD">
When getting the Connection.Redirect method, the client SHOULD
reconnect to the host specified, and if that host is not present,
to any of the hosts specified in the known-hosts list.
</rule>
<chassis name="client" implement="MAY"/>
<field name="host" type="shortstr">
server to connect to
<doc>
Specifies the server to connect to. This is an IP address or a
DNS name, optionally followed by a colon and a port number. If
no port number is specified, the client should use the default
port number for the protocol.
</doc>
<assert check="notnull"/>
</field>
<field name="known hosts" domain="known hosts"/>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name="close" synchronous="1" index="60">
request a connection close
<doc>
This method indicates that the sender wants to close the connection.
This may be due to internal conditions (e.g. a forced shut-down) or
due to an error handling a specific method, i.e. an exception. When
a close is due to an exception, the sender provides the class and
method id of the method which caused the exception.
</doc>
<rule implement="MUST">
After sending this method any received method except the Close-OK
method MUST be discarded.
</rule>
<rule implement="MAY">
The peer sending this method MAY use a counter or timeout to
detect failure of the other peer to respond correctly with
the Close-OK method.
</rule>
<rule implement="MUST">
When a server receives the Close method from a client it MUST
delete all server-side resources associated with the client's
context. A client CANNOT reconnect to a context after sending
or receiving a Close method.
</rule>
<chassis name="client" implement="MUST"/>
<chassis name="server" implement="MUST"/>
<response name="close-ok"/>
<field name="reply code" domain="reply code"/>
<field name="reply text" domain="reply text"/>
<field name="class id" domain="class id">
failing method class
<doc>
When the close is provoked by a method exception, this is the
class of the method.
</doc>
</field>
<field name="method id" domain="class id">
failing method ID
<doc>
When the close is provoked by a method exception, this is the
ID of the method.
</doc>
</field>
</method>
<method name="close-ok" synchronous="1" index="61">
confirm a connection close
<doc>
This method confirms a Connection.Close method and tells the
recipient that it is safe to release resources for the connection
and close the socket.
</doc>
<rule implement="SHOULD">
A peer that detects a socket closure without having received a
Close-Ok handshake method SHOULD log the error.
</rule>
<chassis name="client" implement="MUST"/>
<chassis name="server" implement="MUST"/>
</method>
</class>
<class name="channel" handler="channel" index="20">
<!--
======================================================
== CHANNEL
======================================================
-->
work with channels
<doc>
The channel class provides methods for a client to establish a virtual
connection - a channel - to a server and for both peers to operate the
virtual connection thereafter.
</doc>
<doc name="grammar">
channel = open-channel *use-channel close-channel
open-channel = C:OPEN S:OPEN-OK
use-channel = C:FLOW S:FLOW-OK
/ S:FLOW C:FLOW-OK
/ S:ALERT
/ functional-class
close-channel = C:CLOSE S:CLOSE-OK
/ S:CLOSE C:CLOSE-OK
</doc>
<chassis name="server" implement="MUST"/>
<chassis name="client" implement="MUST"/>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name="open" synchronous="1" index="10">
open a channel for use
<doc>
This method opens a virtual connection (a channel).
</doc>
<rule implement="MUST">
This method MUST NOT be called when the channel is already open.
</rule>
<chassis name="server" implement="MUST"/>
<response name="open-ok"/>
<field name="out of band" type="shortstr">
out-of-band settings
<doc>
Configures out-of-band transfers on this channel. The syntax and
meaning of this field will be formally defined at a later date.
</doc>
<assert check="null"/>
</field>
</method>
<method name="open-ok" synchronous="1" index="11">
signal that the channel is ready
<doc>
This method signals to the client that the channel is ready for use.
</doc>
<chassis name="client" implement="MUST"/>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name="flow" synchronous="1" index="20">
enable/disable flow from peer
<doc>
This method asks the peer to pause or restart the flow of content
data. This is a simple flow-control mechanism that a peer can use
to avoid oveflowing its queues or otherwise finding itself receiving
more messages than it can process. Note that this method is not
intended for window control. The peer that receives a request to
stop sending content should finish sending the current content, if
any, and then wait until it receives a Flow restart method.
</doc>
<rule implement="MAY">
When a new channel is opened, it is active. Some applications
assume that channels are inactive until started. To emulate this
behaviour a client MAY open the channel, then pause it.
</rule>
<rule implement="SHOULD">
When sending content data in multiple frames, a peer SHOULD monitor
the channel for incoming methods and respond to a Channel.Flow as
rapidly as possible.
</rule>
<rule implement="MAY">
A peer MAY use the Channel.Flow method to throttle incoming content
data for internal reasons, for example, when exchangeing data over a
slower connection.
</rule>
<rule implement="MAY">
The peer that requests a Channel.Flow method MAY disconnect and/or
ban a peer that does not respect the request.
</rule>
<chassis name="server" implement="MUST"/>
<chassis name="client" implement="MUST"/>
<response name="flow-ok"/>
<field name="active" type="bit">
start/stop content frames
<doc>
If 1, the peer starts sending content frames. If 0, the peer
stops sending content frames.
</doc>
</field>
</method>
<method name="flow-ok" index="21">
confirm a flow method
<doc>
Confirms to the peer that a flow command was received and processed.
</doc>
<chassis name="server" implement="MUST"/>
<chassis name="client" implement="MUST"/>
<field name="active" type="bit">
current flow setting
<doc>
Confirms the setting of the processed flow method: 1 means the
peer will start sending or continue to send content frames; 0
means it will not.
</doc>
</field>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name="alert" index="30">
send a non-fatal warning message
<doc>
This method allows the server to send a non-fatal warning to the
client. This is used for methods that are normally asynchronous
and thus do not have confirmations, and for which the server may
detect errors that need to be reported. Fatal errors are handled
as channel or connection exceptions; non-fatal errors are sent
through this method.
</doc>
<chassis name="client" implement="MUST"/>
<field name="reply code" domain="reply code"/>
<field name="reply text" domain="reply text"/>
<field name="details" type="table">
detailed information for warning
<doc>
A set of fields that provide more information about the
problem. The meaning of these fields are defined on a
per-reply-code basis (TO BE DEFINED).
</doc>
</field>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name="close" synchronous="1" index="40">
request a channel close
<doc>
This method indicates that the sender wants to close the channel.
This may be due to internal conditions (e.g. a forced shut-down) or
due to an error handling a specific method, i.e. an exception. When
a close is due to an exception, the sender provides the class and
method id of the method which caused the exception.
</doc>
<rule implement="MUST">
After sending this method any received method except
Channel.Close-OK MUST be discarded.
</rule>
<rule implement="MAY">
The peer sending this method MAY use a counter or timeout to detect
failure of the other peer to respond correctly with Channel.Close-OK..
</rule>
<chassis name="client" implement="MUST"/>
<chassis name="server" implement="MUST"/>
<response name="close-ok"/>
<field name="reply code" domain="reply code"/>
<field name="reply text" domain="reply text"/>
<field name="class id" domain="class id">
failing method class
<doc>
When the close is provoked by a method exception, this is the
class of the method.
</doc>
</field>
<field name="method id" domain="method id">
failing method ID
<doc>
When the close is provoked by a method exception, this is the
ID of the method.
</doc>
</field>
</method>
<method name="close-ok" synchronous="1" index="41">
confirm a channel close
<doc>
This method confirms a Channel.Close method and tells the recipient
that it is safe to release resources for the channel and close the
socket.
</doc>
<rule implement="SHOULD">
A peer that detects a socket closure without having received a
Channel.Close-Ok handshake method SHOULD log the error.
</rule>
<chassis name="client" implement="MUST"/>
<chassis name="server" implement="MUST"/>
</method>
</class>
<class name="access" handler="connection" index="30">
<!--
======================================================
== ACCESS CONTROL
======================================================
-->
work with access tickets
<doc>
The protocol control access to server resources using access tickets.
A client must explicitly request access tickets before doing work.
An access ticket grants a client the right to use a specific set of
resources - called a "realm" - in specific ways.
</doc>
<doc name="grammar">
access = C:REQUEST S:REQUEST-OK
</doc>
<chassis name="server" implement="MUST"/>
<chassis name="client" implement="MUST"/>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name="request" synchronous="1" index="10">
request an access ticket
<doc>
This method requests an access ticket for an access realm.
The server responds by granting the access ticket. If the
client does not have access rights to the requested realm
this causes a connection exception. Access tickets are a
per-channel resource.
</doc>
<rule implement="MUST">
The realm name MUST start with either "/data" (for application
resources) or "/admin" (for server administration resources).
If the realm starts with any other path, the server MUST raise
a connection exception with reply code 403 (access refused).
</rule>
<rule implement="MUST">
The server MUST implement the /data realm and MAY implement the
/admin realm. The mapping of resources to realms is not
defined in the protocol - this is a server-side configuration
issue.
</rule>
<chassis name="server" implement="MUST"/>
<response name="request-ok"/>
<field name="realm" domain="path">
name of requested realm
<rule implement="MUST">
If the specified realm is not known to the server, the server
must raise a channel exception with reply code 402 (invalid
path).
</rule>
</field>
<field name="exclusive" type="bit">
request exclusive access
<doc>
Request exclusive access to the realm. If the server cannot grant
this - because there are other active tickets for the realm - it
raises a channel exception.
</doc>
</field>
<field name="passive" type="bit">
request passive access
<doc>
Request message passive access to the specified access realm.
Passive access lets a client get information about resources in
the realm but not to make any changes to them.
</doc>
</field>
<field name="active" type="bit">
request active access
<doc>
Request message active access to the specified access realm.
Acvtive access lets a client get create and delete resources in
the realm.
</doc>
</field>
<field name="write" type="bit">
request write access
<doc>
Request write access to the specified access realm. Write access
lets a client publish messages to all exchanges in the realm.
</doc>
</field>
<field name="read" type="bit">
request read access
<doc>
Request read access to the specified access realm. Read access
lets a client consume messages from queues in the realm.
</doc>
</field>
</method>
<method name="request-ok" synchronous="1" index="11">
grant access to server resources
<doc>
This method provides the client with an access ticket. The access
ticket is valid within the current channel and for the lifespan of
the channel.
</doc>
<rule implement="MUST">
The client MUST NOT use access tickets except within the same
channel as originally granted.
</rule>
<rule implement="MUST">
The server MUST isolate access tickets per channel and treat an
attempt by a client to mix these as a connection exception.
</rule>
<chassis name="client" implement="MUST"/>
<field name="ticket" domain="access ticket"/>
</method>
</class>
<class name="exchange" handler="channel" index="40">
<!--
======================================================
== EXCHANGES (or "routers", if you prefer)
== (Or matchers, plugins, extensions, agents,... Routing is just one of
== the many fun things an exchange can do.)
======================================================
-->
work with exchanges
<doc>
Exchanges match and distribute messages across queues. Exchanges can be
configured in the server or created at runtime.
</doc>
<doc name="grammar">
exchange = C:DECLARE S:DECLARE-OK
/ C:DELETE S:DELETE-OK
</doc>
<chassis name="server" implement="MUST"/>
<chassis name="client" implement="MUST"/>
<rule implement="MUST">
<test>amq_exchange_19</test>
The server MUST implement the direct and fanout exchange types, and
predeclare the corresponding exchanges named amq.direct and amq.fanout
in each virtual host. The server MUST also predeclare a direct
exchange to act as the default exchange for content Publish methods
and for default queue bindings.
</rule>
<rule implement="SHOULD">
<test>amq_exchange_20</test>
The server SHOULD implement the topic exchange type, and predeclare
the corresponding exchange named amq.topic in each virtual host.
</rule>
<rule implement="MAY">
<test>amq_exchange_21</test>
The server MAY implement the system exchange type, and predeclare the
corresponding exchanges named amq.system in each virtual host. If the
client attempts to bind a queue to the system exchange, the server
MUST raise a connection exception with reply code 507 (not allowed).
</rule>
<rule implement="MUST">
<test>amq_exchange_22</test>
The default exchange MUST be defined as internal, and be inaccessible
to the client except by specifying an empty exchange name in a content
Publish method. That is, the server MUST NOT let clients make explicit
bindings to this exchange.
</rule>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name="declare" synchronous="1" index="10">
declare exchange, create if needed
<doc>
This method creates an exchange if it does not already exist, and if the
exchange exists, verifies that it is of the correct and expected class.
</doc>
<rule implement="SHOULD">
<test>amq_exchange_23</test>
The server SHOULD support a minimum of 16 exchanges per virtual host
and ideally, impose no limit except as defined by available resources.
</rule>
<chassis name="server" implement="MUST"/>
<response name="declare-ok"/>
<field name="ticket" domain="access ticket">
<doc>
When a client defines a new exchange, this belongs to the access realm
of the ticket used. All further work done with that exchange must be
done with an access ticket for the same realm.
</doc>
<rule implement="MUST">
The client MUST provide a valid access ticket giving "active" access
to the realm in which the exchange exists or will be created, or
"passive" access if the if-exists flag is set.
</rule>
</field>
<field name="exchange" domain="exchange name">
<rule implement="MUST">
<test>amq_exchange_15</test>
Exchange names starting with "amq." are reserved for predeclared
and standardised exchanges. If the client attempts to create an
exchange starting with "amq.", the server MUST raise a channel
exception with reply code 403 (access refused).
</rule>
<assert check="regexp" value="^[a-zA-Z0-9-_.:]+$"/>
</field>
<field name="type" type="shortstr">
exchange type
<doc>
Each exchange belongs to one of a set of exchange types implemented
by the server. The exchange types define the functionality of the
exchange - i.e. how messages are routed through it. It is not valid
or meaningful to attempt to change the type of an existing exchange.
</doc>
<rule implement="MUST">
<test>amq_exchange_16</test>
If the exchange already exists with a different type, the server
MUST raise a connection exception with a reply code 507 (not allowed).
</rule>
<rule implement="MUST">
<test>amq_exchange_18</test>
If the server does not support the requested exchange type it MUST
raise a connection exception with a reply code 503 (command invalid).
</rule>
<assert check="regexp" value="^[a-zA-Z0-9-_.:]+$"/>
</field>
<field name="passive" type="bit">
do not create exchange
<doc>
If set, the server will not create the exchange. The client can use
this to check whether an exchange exists without modifying the server
state.
</doc>
<rule implement="MUST">
<test>amq_exchange_05</test>
If set, and the exchange does not already exist, the server MUST
raise a channel exception with reply code 404 (not found).
</rule>
</field>
<field name="durable" type="bit">
request a durable exchange
<doc>
If set when creating a new exchange, the exchange will be marked as
durable. Durable exchanges remain active when a server restarts.
Non-durable exchanges (transient exchanges) are purged if/when a
server restarts.
</doc>
<rule implement="MUST">
<test>amq_exchange_24</test>
The server MUST support both durable and transient exchanges.
</rule>
<rule implement="MUST">
The server MUST ignore the durable field if the exchange already
exists.
</rule>
</field>
<field name="auto delete" type="bit">
auto-delete when unused
<doc>
If set, the exchange is deleted when all queues have finished
using it.
</doc>
<rule implement="SHOULD">
<test>amq_exchange_02</test>
The server SHOULD allow for a reasonable delay between the point
when it determines that an exchange is not being used (or no longer
used), and the point when it deletes the exchange. At the least it
must allow a client to create an exchange and then bind a queue to
it, with a small but non-zero delay between these two actions.
</rule>
<rule implement="MUST">
<test>amq_exchange_25</test>
The server MUST ignore the auto-delete field if the exchange already
exists.
</rule>
</field>
<field name="internal" type="bit">
create internal exchange
<doc>
If set, the exchange may not be used directly by publishers, but
only when bound to other exchanges. Internal exchanges are used to
construct wiring that is not visible to applications.
</doc>
</field>
<field name = "nowait" type = "bit">
do not send a reply method
<doc>
If set, the server will not respond to the method. The client should
not wait for a reply method. If the server could not complete the
method it will raise a channel or connection exception.
</doc>
</field>
<field name="arguments" type="table">
arguments for declaration
<doc>
A set of arguments for the declaration. The syntax and semantics
of these arguments depends on the server implementation. This
field is ignored if passive is 1.
</doc>
</field>
</method>
<method name="declare-ok" synchronous="1" index="11">
confirms an exchange declaration
<doc>
This method confirms a Declare method and confirms the name of the
exchange, essential for automatically-named exchanges.
</doc>
<chassis name="client" implement="MUST"/>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name="delete" synchronous="1" index="20">
delete an exchange
<doc>
This method deletes an exchange. When an exchange is deleted all queue
bindings on the exchange are cancelled.
</doc>
<chassis name="server" implement="MUST"/>
<response name="delete-ok"/>
<field name="ticket" domain="access ticket">
<rule implement="MUST">
The client MUST provide a valid access ticket giving "active"
access rights to the exchange's access realm.
</rule>
</field>
<field name="exchange" domain="exchange name">
<rule implement="MUST">
<test>amq_exchange_11</test>
The exchange MUST exist. Attempting to delete a non-existing exchange
causes a channel exception.
</rule>
<assert check="notnull"/>
</field>
<field name="if unused" type="bit">
delete only if unused
<doc>
If set, the server will only delete the exchange if it has no queue
bindings. If the exchange has queue bindings the server does not
delete it but raises a channel exception instead.
</doc>
<rule implement="SHOULD">
<test>amq_exchange_12</test>
If set, the server SHOULD delete the exchange but only if it has
no queue bindings.
</rule>
<rule implement="SHOULD">
<test>amq_exchange_13</test>
If set, the server SHOULD raise a channel exception if the exchange is in
use.
</rule>
</field>
<field name = "nowait" type = "bit">
do not send a reply method
<doc>
If set, the server will not respond to the method. The client should
not wait for a reply method. If the server could not complete the
method it will raise a channel or connection exception.
</doc>
</field>
</method>
<method name="delete-ok" synchronous="1" index="21">
confirm deletion of an exchange
<doc>
This method confirms the deletion of an exchange.
</doc>
<chassis name="client" implement="MUST"/>
</method>
</class>
<class name="queue" handler="channel" index="50">
<!--
======================================================
== QUEUES
======================================================
-->
work with queues
<doc>
Queues store and forward messages. Queues can be configured in the server
or created at runtime. Queues must be attached to at least one exchange
in order to receive messages from publishers.
</doc>
<doc name="grammar">
queue = C:DECLARE S:DECLARE-OK
/ C:BIND S:BIND-OK
/ C:PURGE S:PURGE-OK
/ C:DELETE S:DELETE-OK
</doc>
<chassis name="server" implement="MUST"/>
<chassis name="client" implement="MUST"/>
<rule implement="MUST">
<test>amq_queue_33</test>
A server MUST allow any content class to be sent to any queue, in any
mix, and queue and delivery these content classes independently. Note
that all methods that fetch content off queues are specific to a given
content class.
</rule>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name="declare" synchronous="1" index="10">
declare queue, create if needed
<doc>
This method creates or checks a queue. When creating a new queue
the client can specify various properties that control the durability
of the queue and its contents, and the level of sharing for the queue.
</doc>
<rule implement="MUST">
<test>amq_queue_34</test>
The server MUST create a default binding for a newly-created queue
to the default exchange, which is an exchange of type 'direct'.
</rule>
<rule implement="SHOULD">
<test>amq_queue_35</test>
The server SHOULD support a minimum of 256 queues per virtual host
and ideally, impose no limit except as defined by available resources.
</rule>
<chassis name="server" implement="MUST"/>
<response name="declare-ok"/>
<field name="ticket" domain="access ticket">
<doc>
When a client defines a new queue, this belongs to the access realm
of the ticket used. All further work done with that queue must be
done with an access ticket for the same realm.
</doc>
<doc>
The client provides a valid access ticket giving "active" access
to the realm in which the queue exists or will be created, or
"passive" access if the if-exists flag is set.
</doc>
</field>
<field name="queue" domain="queue name">
<rule implement="MAY">
<test>amq_queue_10</test>
The queue name MAY be empty, in which case the server MUST create
a new queue with a unique generated name and return this to the
client in the Declare-Ok method.
</rule>
<rule implement="MUST">
<test>amq_queue_32</test>
Queue names starting with "amq." are reserved for predeclared and
standardised server queues. If the queue name starts with "amq."
and the passive option is zero, the server MUST raise a connection
exception with reply code 403 (access refused).
</rule>
<assert check="regexp" value="^[a-zA-Z0-9-_.:]*$"/>
</field>
<field name="passive" type="bit">
do not create queue
<doc>
If set, the server will not create the queue. The client can use
this to check whether a queue exists without modifying the server
state.
</doc>
<rule implement="MUST">
<test>amq_queue_05</test>
If set, and the queue does not already exist, the server MUST
respond with a reply code 404 (not found) and raise a channel
exception.
</rule>
</field>
<field name="durable" type="bit">
request a durable queue
<doc>
If set when creating a new queue, the queue will be marked as
durable. Durable queues remain active when a server restarts.
Non-durable queues (transient queues) are purged if/when a
server restarts. Note that durable queues do not necessarily
hold persistent messages, although it does not make sense to
send persistent messages to a transient queue.
</doc>
<rule implement="MUST">
<test>amq_queue_03</test>
The server MUST recreate the durable queue after a restart.
</rule>
<rule implement="MUST">
<test>amq_queue_36</test>
The server MUST support both durable and transient queues.
</rule>
<rule implement="MUST">
<test>amq_queue_37</test>
The server MUST ignore the durable field if the queue already
exists.
</rule>
</field>
<field name="exclusive" type="bit">
request an exclusive queue
<doc>
Exclusive queues may only be consumed from by the current connection.
Setting the 'exclusive' flag always implies 'auto-delete'.
</doc>
<rule implement="MUST">
<test>amq_queue_38</test>
The server MUST support both exclusive (private) and non-exclusive
(shared) queues.
</rule>
<rule implement="MUST">
<test>amq_queue_04</test>
The server MUST raise a channel exception if 'exclusive' is specified
and the queue already exists and is owned by a different connection.
</rule>
</field>
<field name="auto delete" type="bit">
auto-delete queue when unused
<doc>
If set, the queue is deleted when all consumers have finished
using it. Last consumer can be cancelled either explicitly or because
its channel is closed. If there was no consumer ever on the queue, it
won't be deleted.
</doc>
<rule implement="SHOULD">
<test>amq_queue_02</test>
The server SHOULD allow for a reasonable delay between the point
when it determines that a queue is not being used (or no longer
used), and the point when it deletes the queue. At the least it
must allow a client to create a queue and then create a consumer
to read from it, with a small but non-zero delay between these
two actions. The server should equally allow for clients that may
be disconnected prematurely, and wish to re-consume from the same
queue without losing messages. We would recommend a configurable
timeout, with a suitable default value being one minute.
</rule>
<rule implement="MUST">
<test>amq_queue_31</test>
The server MUST ignore the auto-delete field if the queue already
exists.
</rule>
</field>
<field name = "nowait" type = "bit">
do not send a reply method
<doc>
If set, the server will not respond to the method. The client should
not wait for a reply method. If the server could not complete the
method it will raise a channel or connection exception.
</doc>
</field>
<field name="arguments" type="table">
arguments for declaration
<doc>
A set of arguments for the declaration. The syntax and semantics
of these arguments depends on the server implementation. This
field is ignored if passive is 1.
</doc>
</field>
</method>
<method name="declare-ok" synchronous="1" index="11">
confirms a queue definition
<doc>
This method confirms a Declare method and confirms the name of the
queue, essential for automatically-named queues.
</doc>
<chassis name="client" implement="MUST"/>
<field name="queue" domain="queue name">
<doc>
Reports the name of the queue. If the server generated a queue
name, this field contains that name.
</doc>
<assert check="notnull"/>
</field>
<field name="message count" type="long">
number of messages in queue
<doc>
Reports the number of messages in the queue, which will be zero
for newly-created queues.
</doc>
</field>
<field name="consumer count" type="long">
number of consumers
<doc>
Reports the number of active consumers for the queue. Note that
consumers can suspend activity (Channel.Flow) in which case they
do not appear in this count.
</doc>
</field>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name="bind" synchronous="1" index="20">
bind queue to an exchange
<doc>
This method binds a queue to an exchange. Until a queue is
bound it will not receive any messages. In a classic messaging
model, store-and-forward queues are bound to a dest exchange
and subscription queues are bound to a dest_wild exchange.
</doc>
<rule implement="MUST">
<test>amq_queue_25</test>
A server MUST allow ignore duplicate bindings - that is, two or
more bind methods for a specific queue, with identical arguments
- without treating these as an error.
</rule>
<rule implement="MUST">
<test>amq_queue_39</test>
If a bind fails, the server MUST raise a connection exception.
</rule>
<rule implement="MUST">
<test>amq_queue_12</test>
The server MUST NOT allow a durable queue to bind to a transient
exchange. If the client attempts this the server MUST raise a
channel exception.
</rule>
<rule implement="SHOULD">
<test>amq_queue_13</test>
Bindings for durable queues are automatically durable and the
server SHOULD restore such bindings after a server restart.
</rule>
<rule implement="MUST">
<test>amq_queue_17</test>
If the client attempts to an exchange that was declared as internal,
the server MUST raise a connection exception with reply code 530
(not allowed).
</rule>
<rule implement="SHOULD">
<test>amq_queue_40</test>
The server SHOULD support at least 4 bindings per queue, and
ideally, impose no limit except as defined by available resources.
</rule>
<chassis name="server" implement="MUST"/>
<response name="bind-ok"/>
<field name="ticket" domain="access ticket">
<doc>
The client provides a valid access ticket giving "active"
access rights to the queue's access realm.
</doc>
</field>
<field name = "queue" domain = "queue name">
<doc>
Specifies the name of the queue to bind. If the queue name is
empty, refers to the current queue for the channel, which is
the last declared queue.
</doc>
<doc name = "rule">
If the client did not previously declare a queue, and the queue
name in this method is empty, the server MUST raise a connection
exception with reply code 530 (not allowed).
</doc>
<doc name = "rule" test = "amq_queue_26">
If the queue does not exist the server MUST raise a channel exception
with reply code 404 (not found).
</doc>
</field>
<field name="exchange" domain="exchange name">
The name of the exchange to bind to.
<rule implement="MUST">
<test>amq_queue_14</test>
If the exchange does not exist the server MUST raise a channel
exception with reply code 404 (not found).
</rule>
</field>
<field name="routing key" type="shortstr">
message routing key
<doc>
Specifies the routing key for the binding. The routing key is
used for routing messages depending on the exchange configuration.
Not all exchanges use a routing key - refer to the specific
exchange documentation. If the routing key is empty and the queue
name is empty, the routing key will be the current queue for the
channel, which is the last declared queue.
</doc>
</field>
<field name = "nowait" type = "bit">
do not send a reply method
<doc>
If set, the server will not respond to the method. The client should
not wait for a reply method. If the server could not complete the
method it will raise a channel or connection exception.
</doc>
</field>
<field name="arguments" type="table">
arguments for binding
<doc>
A set of arguments for the binding. The syntax and semantics of
these arguments depends on the exchange class.
</doc>
</field>
</method>
<method name="bind-ok" synchronous="1" index="21">
confirm bind successful
<doc>
This method confirms that the bind was successful.
</doc>
<chassis name="client" implement="MUST"/>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name="purge" synchronous="1" index="30">
purge a queue
<doc>
This method removes all messages from a queue. It does not cancel
consumers. Purged messages are deleted without any formal "undo"
mechanism.
</doc>
<rule implement="MUST">
<test>amq_queue_15</test>
A call to purge MUST result in an empty queue.
</rule>
<rule implement="MUST">
<test>amq_queue_41</test>
On transacted channels the server MUST not purge messages that have
already been sent to a client but not yet acknowledged.
</rule>
<rule implement="MAY">
<test>amq_queue_42</test>
The server MAY implement a purge queue or log that allows system
administrators to recover accidentally-purged messages. The server
SHOULD NOT keep purged messages in the same storage spaces as the
live messages since the volumes of purged messages may get very
large.
</rule>
<chassis name="server" implement="MUST"/>
<response name="purge-ok"/>
<field name="ticket" domain="access ticket">
<doc>
The access ticket must be for the access realm that holds the
queue.
</doc>
<rule implement="MUST">
The client MUST provide a valid access ticket giving "read" access
rights to the queue's access realm. Note that purging a queue is
equivalent to reading all messages and discarding them.
</rule>
</field>
<field name = "queue" domain = "queue name">
<doc>
Specifies the name of the queue to purge. If the queue name is
empty, refers to the current queue for the channel, which is
the last declared queue.
</doc>
<doc name = "rule">
If the client did not previously declare a queue, and the queue
name in this method is empty, the server MUST raise a connection
exception with reply code 530 (not allowed).
</doc>
<doc name = "rule" test = "amq_queue_16">
The queue must exist. Attempting to purge a non-existing queue
causes a channel exception.
</doc>
</field>
<field name = "nowait" type = "bit">
do not send a reply method
<doc>
If set, the server will not respond to the method. The client should
not wait for a reply method. If the server could not complete the
method it will raise a channel or connection exception.
</doc>
</field>
</method>
<method name="purge-ok" synchronous="1" index="31">
confirms a queue purge
<doc>
This method confirms the purge of a queue.
</doc>
<chassis name="client" implement="MUST"/>
<field name="message count" type="long">
number of messages purged
<doc>
Reports the number of messages purged.
</doc>
</field>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name="delete" synchronous="1" index="40">
delete a queue
<doc>
This method deletes a queue. When a queue is deleted any pending
messages are sent to a dead-letter queue if this is defined in the
server configuration, and all consumers on the queue are cancelled.
</doc>
<rule implement="SHOULD">
<test>amq_queue_43</test>
The server SHOULD use a dead-letter queue to hold messages that
were pending on a deleted queue, and MAY provide facilities for
a system administrator to move these messages back to an active
queue.
</rule>
<chassis name="server" implement="MUST"/>
<response name="delete-ok"/>
<field name="ticket" domain="access ticket">
<doc>
The client provides a valid access ticket giving "active"
access rights to the queue's access realm.
</doc>
</field>
<field name = "queue" domain = "queue name">
<doc>
Specifies the name of the queue to delete. If the queue name is
empty, refers to the current queue for the channel, which is the
last declared queue.
</doc>
<doc name = "rule">
If the client did not previously declare a queue, and the queue
name in this method is empty, the server MUST raise a connection
exception with reply code 530 (not allowed).
</doc>
<doc name = "rule" test = "amq_queue_21">
The queue must exist. Attempting to delete a non-existing queue
causes a channel exception.
</doc>
</field>
<field name="if unused" type="bit">
delete only if unused
<doc>
If set, the server will only delete the queue if it has no
consumers. If the queue has consumers the server does does not
delete it but raises a channel exception instead.
</doc>
<rule implement="MUST">
<test>amq_queue_29</test>
<test>amq_queue_30</test>
The server MUST respect the if-unused flag when deleting a queue.
</rule>
</field>
<field name="if empty" type="bit">
delete only if empty
<test>amq_queue_27</test>
<doc>
If set, the server will only delete the queue if it has no
messages. If the queue is not empty the server raises a channel
exception.
</doc>
</field>
<field name = "nowait" type = "bit">
do not send a reply method
<doc>
If set, the server will not respond to the method. The client should
not wait for a reply method. If the server could not complete the
method it will raise a channel or connection exception.
</doc>
</field>
</method>
<method name="delete-ok" synchronous="1" index="41">
confirm deletion of a queue
<doc>
This method confirms the deletion of a queue.
</doc>
<chassis name="client" implement="MUST"/>
<field name="message count" type="long">
number of messages purged
<doc>
Reports the number of messages purged.
</doc>
</field>
</method>
</class>
<class name="basic" handler="channel" index="60">
<!--
======================================================
== BASIC MIDDLEWARE
======================================================
-->
work with basic content
<doc>
The Basic class provides methods that support an industry-standard
messaging model.
</doc>
<doc name = "grammar">
basic = C:QOS S:QOS-OK
/ C:CONSUME S:CONSUME-OK
/ C:CANCEL S:CANCEL-OK
/ C:PUBLISH content
/ S:RETURN content
/ S:DELIVER content
/ C:GET ( S:GET-OK content / S:GET-EMPTY )
/ C:ACK
/ C:REJECT
</doc>
<chassis name = "server" implement = "MUST" />
<chassis name = "client" implement = "MAY" />
<doc name = "rule" test = "amq_basic_08">
The server SHOULD respect the persistent property of basic messages
and SHOULD make a best-effort to hold persistent basic messages on a
reliable storage mechanism.
</doc>
<doc name = "rule" test = "amq_basic_09">
The server MUST NOT discard a persistent basic message in case of a
queue overflow. The server MAY use the Channel.Flow method to slow
or stop a basic message publisher when necessary.
</doc>
<doc name = "rule" test = "amq_basic_10">
The server MAY overflow non-persistent basic messages to persistent
storage and MAY discard or dead-letter non-persistent basic messages
on a priority basis if the queue size exceeds some configured limit.
</doc>
<doc name = "rule" test = "amq_basic_11">
The server MUST implement at least 2 priority levels for basic
messages, where priorities 0-4 and 5-9 are treated as two distinct
levels. The server MAY implement up to 10 priority levels.
</doc>
<doc name = "rule" test = "amq_basic_12">
The server MUST deliver messages of the same priority in order
irrespective of their individual persistence.
</doc>
<doc name = "rule" test = "amq_basic_13">
The server MUST support both automatic and explicit acknowledgements
on Basic content.
</doc>
<!-- These are the properties for a Basic content -->
<field name = "content type" type = "shortstr">
MIME content type
</field>
<field name = "content encoding" type = "shortstr">
MIME content encoding
</field>
<field name = "headers" type = "table">
Message header field table
</field>
<field name = "delivery mode" type = "octet">
Non-persistent (1) or persistent (2)
</field>
<field name = "priority" type = "octet">
The message priority, 0 to 9
</field>
<field name = "correlation id" type = "shortstr">
The application correlation identifier
</field>
<field name = "reply to" type = "shortstr">
The destination to reply to
</field>
<field name = "expiration" type = "shortstr">
Message expiration specification
</field>
<field name = "message id" type = "shortstr">
The application message identifier
</field>
<field name = "timestamp" type = "timestamp">
The message timestamp
</field>
<field name = "type" type = "shortstr">
The message type name
</field>
<field name = "user id" type = "shortstr">
The creating user id
</field>
<field name = "app id" type = "shortstr">
The creating application id
</field>
<field name = "cluster id" type = "shortstr">
Intra-cluster routing identifier
</field>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name = "qos" synchronous = "1" index = "10">
specify quality of service
<doc>
This method requests a specific quality of service. The QoS can
be specified for the current channel or for all channels on the
connection. The particular properties and semantics of a qos method
always depend on the content class semantics. Though the qos method
could in principle apply to both peers, it is currently meaningful
only for the server.
</doc>
<chassis name = "server" implement = "MUST" />
<response name = "qos-ok" />
<field name = "prefetch size" type = "long">
prefetch window in octets
<doc>
The client can request that messages be sent in advance so that
when the client finishes processing a message, the following
message is already held locally, rather than needing to be sent
down the channel. Prefetching gives a performance improvement.
This field specifies the prefetch window size in octets. The
server will send a message in advance if it is equal to or
smaller in size than the available prefetch size (and also falls
into other prefetch limits). May be set to zero, meaning "no
specific limit", although other prefetch limits may still apply.
The prefetch-size is ignored if the no-ack option is set.
</doc>
<doc name = "rule" test = "amq_basic_17">
The server MUST ignore this setting when the client is not
processing any messages - i.e. the prefetch size does not limit
the transfer of single messages to a client, only the sending in
advance of more messages while the client still has one or more
unacknowledged messages.
</doc>
</field>
<field name = "prefetch count" type = "short">
prefetch window in messages
<doc>
Specifies a prefetch window in terms of whole messages. This
field may be used in combination with the prefetch-size field;
a message will only be sent in advance if both prefetch windows
(and those at the channel and connection level) allow it.
The prefetch-count is ignored if the no-ack option is set.
</doc>
<doc name = "rule" test = "amq_basic_18">
The server MAY send less data in advance than allowed by the
client's specified prefetch windows but it MUST NOT send more.
</doc>
</field>
<field name = "global" type = "bit">
apply to entire connection
<doc>
By default the QoS settings apply to the current channel only. If
this field is set, they are applied to the entire connection.
</doc>
</field>
</method>
<method name = "qos-ok" synchronous = "1" index = "11">
confirm the requested qos
<doc>
This method tells the client that the requested QoS levels could
be handled by the server. The requested QoS applies to all active
consumers until a new QoS is defined.
</doc>
<chassis name = "client" implement = "MUST" />
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name = "consume" synchronous = "1" index = "20">
start a queue consumer
<doc>
This method asks the server to start a "consumer", which is a
transient request for messages from a specific queue. Consumers
last as long as the channel they were created on, or until the
client cancels them.
</doc>
<doc name = "rule" test = "amq_basic_01">
The server SHOULD support at least 16 consumers per queue, unless
the queue was declared as private, and ideally, impose no limit
except as defined by available resources.
</doc>
<chassis name = "server" implement = "MUST" />
<response name = "consume-ok" />
<field name = "ticket" domain = "access ticket">
<doc name = "rule">
The client MUST provide a valid access ticket giving "read" access
rights to the realm for the queue.
</doc>
</field>
<field name = "queue" domain = "queue name">
<doc>
Specifies the name of the queue to consume from. If the queue name
is null, refers to the current queue for the channel, which is the
last declared queue.
</doc>
<doc name = "rule">
If the client did not previously declare a queue, and the queue name
in this method is empty, the server MUST raise a connection exception
with reply code 530 (not allowed).
</doc>
</field>
<field name = "consumer tag" domain = "consumer tag">
<doc>
Specifies the identifier for the consumer. The consumer tag is
local to a connection, so two clients can use the same consumer
tags. If this field is empty the server will generate a unique
tag.
</doc>
<doc name = "rule" test = "todo">
The tag MUST NOT refer to an existing consumer. If the client
attempts to create two consumers with the same non-empty tag
the server MUST raise a connection exception with reply code
530 (not allowed).
</doc>
</field>
<field name = "no local" domain = "no local" />
<field name = "no ack" domain = "no ack" />
<field name = "exclusive" type = "bit">
request exclusive access
<doc>
Request exclusive consumer access, meaning only this consumer can
access the queue.
</doc>
<doc name = "rule" test = "amq_basic_02">
If the server cannot grant exclusive access to the queue when asked,
- because there are other consumers active - it MUST raise a channel
exception with return code 403 (access refused).
</doc>
</field>
<field name = "nowait" type = "bit">
do not send a reply method
<doc>
If set, the server will not respond to the method. The client should
not wait for a reply method. If the server could not complete the
method it will raise a channel or connection exception.
</doc>
</field>
</method>
<method name = "consume-ok" synchronous = "1" index = "21">
confirm a new consumer
<doc>
The server provides the client with a consumer tag, which is used
by the client for methods called on the consumer at a later stage.
</doc>
<chassis name = "client" implement = "MUST" />
<field name = "consumer tag" domain = "consumer tag">
<doc>
Holds the consumer tag specified by the client or provided by
the server.
</doc>
</field>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name = "cancel" synchronous = "1" index = "30">
end a queue consumer
<doc test = "amq_basic_04">
This method cancels a consumer. This does not affect already
delivered messages, but it does mean the server will not send any
more messages for that consumer. The client may receive an
abitrary number of messages in between sending the cancel method
and receiving the cancel-ok reply.
</doc>
<doc name = "rule" test = "todo">
If the queue no longer exists when the client sends a cancel command,
or the consumer has been cancelled for other reasons, this command
has no effect.
</doc>
<chassis name = "server" implement = "MUST" />
<response name = "cancel-ok" />
<field name = "consumer tag" domain = "consumer tag" />
<field name = "nowait" type = "bit">
do not send a reply method
<doc>
If set, the server will not respond to the method. The client should
not wait for a reply method. If the server could not complete the
method it will raise a channel or connection exception.
</doc>
</field>
</method>
<method name = "cancel-ok" synchronous = "1" index = "31">
confirm a cancelled consumer
<doc>
This method confirms that the cancellation was completed.
</doc>
<chassis name = "client" implement = "MUST" />
<field name = "consumer tag" domain = "consumer tag" />
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name = "publish" content = "1" index = "40">
publish a message
<doc>
This method publishes a message to a specific exchange. The message
will be routed to queues as defined by the exchange configuration
and distributed to any active consumers when the transaction, if any,
is committed.
</doc>
<chassis name = "server" implement = "MUST" />
<field name = "ticket" domain = "access ticket">
<doc name = "rule">
The client MUST provide a valid access ticket giving "write"
access rights to the access realm for the exchange.
</doc>
</field>
<field name = "exchange" domain = "exchange name">
<doc>
Specifies the name of the exchange to publish to. The exchange
name can be empty, meaning the default exchange. If the exchange
name is specified, and that exchange does not exist, the server
will raise a channel exception.
</doc>
<doc name = "rule" test = "amq_basic_06">
The server MUST accept a blank exchange name to mean the default
exchange.
</doc>
<doc name = "rule" test = "amq_basic_14">
If the exchange was declared as an internal exchange, the server
MUST raise a channel exception with a reply code 403 (access
refused).
</doc>
<doc name = "rule" test = "amq_basic_15">
The exchange MAY refuse basic content in which case it MUST raise
a channel exception with reply code 540 (not implemented).
</doc>
</field>
<field name = "routing key" type = "shortstr">
Message routing key
<doc>
Specifies the routing key for the message. The routing key is
used for routing messages depending on the exchange configuration.
</doc>
</field>
<field name = "mandatory" type = "bit">
indicate mandatory routing
<doc>
This flag tells the server how to react if the message cannot be
routed to a queue. If this flag is set, the server will return an
unroutable message with a Return method. If this flag is zero, the
server silently drops the message.
</doc>
<doc name = "rule" test = "amq_basic_07">
The server SHOULD implement the mandatory flag.
</doc>
</field>
<field name = "immediate" type = "bit">
request immediate delivery
<doc>
This flag tells the server how to react if the message cannot be
routed to a queue consumer immediately. If this flag is set, the
server will return an undeliverable message with a Return method.
If this flag is zero, the server will queue the message, but with
no guarantee that it will ever be consumed.
</doc>
<doc name = "rule" test = "amq_basic_16">
The server SHOULD implement the immediate flag.
</doc>
</field>
</method>
<method name = "return" content = "1" index = "50">
return a failed message
<doc>
This method returns an undeliverable message that was published
with the "immediate" flag set, or an unroutable message published
with the "mandatory" flag set. The reply code and text provide
information about the reason that the message was undeliverable.
</doc>
<chassis name = "client" implement = "MUST" />
<field name = "reply code" domain = "reply code" />
<field name = "reply text" domain = "reply text" />
<field name = "exchange" domain = "exchange name">
<doc>
Specifies the name of the exchange that the message was
originally published to.
</doc>
</field>
<field name = "routing key" type = "shortstr">
Message routing key
<doc>
Specifies the routing key name specified when the message was
published.
</doc>
</field>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name = "deliver" content = "1" index = "60">
notify the client of a consumer message
<doc>
This method delivers a message to the client, via a consumer. In
the asynchronous message delivery model, the client starts a
consumer using the Consume method, then the server responds with
Deliver methods as and when messages arrive for that consumer.
</doc>
<doc name = "rule" test = "amq_basic_19">
The server SHOULD track the number of times a message has been
delivered to clients and when a message is redelivered a certain
number of times - e.g. 5 times - without being acknowledged, the
server SHOULD consider the message to be unprocessable (possibly
causing client applications to abort), and move the message to a
dead letter queue.
</doc>
<chassis name = "client" implement = "MUST" />
<field name = "consumer tag" domain = "consumer tag" />
<field name = "delivery tag" domain = "delivery tag" />
<field name = "redelivered" domain = "redelivered" />
<field name = "exchange" domain = "exchange name">
<doc>
Specifies the name of the exchange that the message was
originally published to.
</doc>
</field>
<field name = "routing key" type = "shortstr">
Message routing key
<doc>
Specifies the routing key name specified when the message was
published.
</doc>
</field>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name = "get" synchronous = "1" index = "70">
direct access to a queue
<doc>
This method provides a direct access to the messages in a queue
using a synchronous dialogue that is designed for specific types of
application where synchronous functionality is more important than
performance.
</doc>
<response name = "get-ok" />
<response name = "get-empty" />
<chassis name = "server" implement = "MUST" />
<field name = "ticket" domain = "access ticket">
<doc name = "rule">
The client MUST provide a valid access ticket giving "read"
access rights to the realm for the queue.
</doc>
</field>
<field name = "queue" domain = "queue name">
<doc>
Specifies the name of the queue to consume from. If the queue name
is null, refers to the current queue for the channel, which is the
last declared queue.
</doc>
<doc name = "rule">
If the client did not previously declare a queue, and the queue name
in this method is empty, the server MUST raise a connection exception
with reply code 530 (not allowed).
</doc>
</field>
<field name = "no ack" domain = "no ack" />
</method>
<method name = "get-ok" synchronous = "1" content = "1" index = "71">
provide client with a message
<doc>
This method delivers a message to the client following a get
method. A message delivered by 'get-ok' must be acknowledged
unless the no-ack option was set in the get method.
</doc>
<chassis name = "client" implement = "MAY" />
<field name = "delivery tag" domain = "delivery tag" />
<field name = "redelivered" domain = "redelivered" />
<field name = "exchange" domain = "exchange name">
<doc>
Specifies the name of the exchange that the message was originally
published to. If empty, the message was published to the default
exchange.
</doc>
</field>
<field name = "routing key" type = "shortstr">
Message routing key
<doc>
Specifies the routing key name specified when the message was
published.
</doc>
</field>
<field name = "message count" type = "long" >
number of messages pending
<doc>
This field reports the number of messages pending on the queue,
excluding the message being delivered. Note that this figure is
indicative, not reliable, and can change arbitrarily as messages
are added to the queue and removed by other clients.
</doc>
</field>
</method>
<method name = "get-empty" synchronous = "1" index = "72">
indicate no messages available
<doc>
This method tells the client that the queue has no messages
available for the client.
</doc>
<chassis name = "client" implement = "MAY" />
<field name = "cluster id" type = "shortstr">
Cluster id
<doc>
For use by cluster applications, should not be used by
client applications.
</doc>
</field>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name = "ack" index = "80">
acknowledge one or more messages
<doc>
This method acknowledges one or more messages delivered via the
Deliver or Get-Ok methods. The client can ask to confirm a
single message or a set of messages up to and including a specific
message.
</doc>
<chassis name = "server" implement = "MUST" />
<field name = "delivery tag" domain = "delivery tag" />
<field name = "multiple" type = "bit">
acknowledge multiple messages
<doc>
If set to 1, the delivery tag is treated as "up to and including",
so that the client can acknowledge multiple messages with a single
method. If set to zero, the delivery tag refers to a single
message. If the multiple field is 1, and the delivery tag is zero,
tells the server to acknowledge all outstanding mesages.
</doc>
<doc name = "rule" test = "amq_basic_20">
The server MUST validate that a non-zero delivery-tag refers to an
delivered message, and raise a channel exception if this is not the
case.
</doc>
</field>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name = "reject" index = "90">
reject an incoming message
<doc>
This method allows a client to reject a message. It can be used to
interrupt and cancel large incoming messages, or return untreatable
messages to their original queue.
</doc>
<doc name = "rule" test = "amq_basic_21">
The server SHOULD be capable of accepting and process the Reject
method while sending message content with a Deliver or Get-Ok
method. I.e. the server should read and process incoming methods
while sending output frames. To cancel a partially-send content,
the server sends a content body frame of size 1 (i.e. with no data
except the frame-end octet).
</doc>
<doc name = "rule" test = "amq_basic_22">
The server SHOULD interpret this method as meaning that the client
is unable to process the message at this time.
</doc>
<doc name = "rule">
A client MUST NOT use this method as a means of selecting messages
to process. A rejected message MAY be discarded or dead-lettered,
not necessarily passed to another client.
</doc>
<chassis name = "server" implement = "MUST" />
<field name = "delivery tag" domain = "delivery tag" />
<field name = "requeue" type = "bit">
requeue the message
<doc>
If this field is zero, the message will be discarded. If this bit
is 1, the server will attempt to requeue the message.
</doc>
<doc name = "rule" test = "amq_basic_23">
The server MUST NOT deliver the message to the same client within
the context of the current channel. The recommended strategy is
to attempt to deliver the message to an alternative consumer, and
if that is not possible, to move the message to a dead-letter
queue. The server MAY use more sophisticated tracking to hold
the message on the queue and redeliver it to the same client at
a later stage.
</doc>
</field>
</method>
<method name = "recover" index = "100">
redeliver unacknowledged messages. This method is only allowed on non-transacted channels.
<doc>
This method asks the broker to redeliver all unacknowledged messages on a
specifieid channel. Zero or more messages may be redelivered.
</doc>
<chassis name = "server" implement = "MUST" />
<field name = "requeue" type = "bit">
requeue the message
<doc>
If this field is zero, the message will be redelivered to the original recipient. If this bit
is 1, the server will attempt to requeue the message, potentially then delivering it to an
alternative subscriber.
</doc>
</field>
<doc name="rule">
The server MUST set the redelivered flag on all messages that are resent.
</doc>
<doc name="rule">
The server MUST raise a channel exception if this is called on a transacted channel.
</doc>
</method>
</class>
<class name="file" handler="channel" index="70">
<!--
======================================================
== FILE TRANSFER
======================================================
-->
work with file content
<doc>
The file class provides methods that support reliable file transfer.
File messages have a specific set of properties that are required for
interoperability with file transfer applications. File messages and
acknowledgements are subject to channel transactions. Note that the
file class does not provide message browsing methods; these are not
compatible with the staging model. Applications that need browsable
file transfer should use Basic content and the Basic class.
</doc>
<doc name = "grammar">
file = C:QOS S:QOS-OK
/ C:CONSUME S:CONSUME-OK
/ C:CANCEL S:CANCEL-OK
/ C:OPEN S:OPEN-OK C:STAGE content
/ S:OPEN C:OPEN-OK S:STAGE content
/ C:PUBLISH
/ S:DELIVER
/ S:RETURN
/ C:ACK
/ C:REJECT
</doc>
<chassis name = "server" implement = "MAY" />
<chassis name = "client" implement = "MAY" />
<doc name = "rule">
The server MUST make a best-effort to hold file messages on a
reliable storage mechanism.
</doc>
<doc name = "rule">
The server MUST NOT discard a file message in case of a queue
overflow. The server MUST use the Channel.Flow method to slow or stop
a file message publisher when necessary.
</doc>
<doc name = "rule">
The server MUST implement at least 2 priority levels for file
messages, where priorities 0-4 and 5-9 are treated as two distinct
levels. The server MAY implement up to 10 priority levels.
</doc>
<doc name = "rule">
The server MUST support both automatic and explicit acknowledgements
on file content.
</doc>
<!-- These are the properties for a File content -->
<field name = "content type" type = "shortstr">
MIME content type
</field>
<field name = "content encoding" type = "shortstr">
MIME content encoding
</field>
<field name = "headers" type = "table">
Message header field table
</field>
<field name = "priority" type = "octet">
The message priority, 0 to 9
</field>
<field name = "reply to" type = "shortstr">
The destination to reply to
</field>
<field name = "message id" type = "shortstr">
The application message identifier
</field>
<field name = "filename" type = "shortstr">
The message filename
</field>
<field name = "timestamp" type = "timestamp">
The message timestamp
</field>
<field name = "cluster id" type = "shortstr">
Intra-cluster routing identifier
</field>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name = "qos" synchronous = "1" index = "10">
specify quality of service
<doc>
This method requests a specific quality of service. The QoS can
be specified for the current channel or for all channels on the
connection. The particular properties and semantics of a qos method
always depend on the content class semantics. Though the qos method
could in principle apply to both peers, it is currently meaningful
only for the server.
</doc>
<chassis name = "server" implement = "MUST" />
<response name = "qos-ok" />
<field name = "prefetch size" type = "long">
prefetch window in octets
<doc>
The client can request that messages be sent in advance so that
when the client finishes processing a message, the following
message is already held locally, rather than needing to be sent
down the channel. Prefetching gives a performance improvement.
This field specifies the prefetch window size in octets. May be
set to zero, meaning "no specific limit". Note that other
prefetch limits may still apply. The prefetch-size is ignored
if the no-ack option is set.
</doc>
</field>
<field name = "prefetch count" type = "short">
prefetch window in messages
<doc>
Specifies a prefetch window in terms of whole messages. This
is compatible with some file API implementations. This field
may be used in combination with the prefetch-size field; a
message will only be sent in advance if both prefetch windows
(and those at the channel and connection level) allow it.
The prefetch-count is ignored if the no-ack option is set.
</doc>
<doc name = "rule">
The server MAY send less data in advance than allowed by the
client's specified prefetch windows but it MUST NOT send more.
</doc>
</field>
<field name = "global" type = "bit">
apply to entire connection
<doc>
By default the QoS settings apply to the current channel only. If
this field is set, they are applied to the entire connection.
</doc>
</field>
</method>
<method name = "qos-ok" synchronous = "1" index = "11">
confirm the requested qos
<doc>
This method tells the client that the requested QoS levels could
be handled by the server. The requested QoS applies to all active
consumers until a new QoS is defined.
</doc>
<chassis name = "client" implement = "MUST" />
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name = "consume" synchronous = "1" index = "20">
start a queue consumer
<doc>
This method asks the server to start a "consumer", which is a
transient request for messages from a specific queue. Consumers
last as long as the channel they were created on, or until the
client cancels them.
</doc>
<doc name = "rule">
The server SHOULD support at least 16 consumers per queue, unless
the queue was declared as private, and ideally, impose no limit
except as defined by available resources.
</doc>
<chassis name = "server" implement = "MUST" />
<response name = "consume-ok" />
<field name = "ticket" domain = "access ticket">
<doc name = "rule">
The client MUST provide a valid access ticket giving "read" access
rights to the realm for the queue.
</doc>
</field>
<field name = "queue" domain = "queue name">
<doc>
Specifies the name of the queue to consume from. If the queue name
is null, refers to the current queue for the channel, which is the
last declared queue.
</doc>
<doc name = "rule">
If the client did not previously declare a queue, and the queue name
in this method is empty, the server MUST raise a connection exception
with reply code 530 (not allowed).
</doc>
</field>
<field name = "consumer tag" domain = "consumer tag">
<doc>
Specifies the identifier for the consumer. The consumer tag is
local to a connection, so two clients can use the same consumer
tags. If this field is empty the server will generate a unique
tag.
</doc>
<doc name = "rule" test = "todo">
The tag MUST NOT refer to an existing consumer. If the client
attempts to create two consumers with the same non-empty tag
the server MUST raise a connection exception with reply code
530 (not allowed).
</doc>
</field>
<field name = "no local" domain = "no local" />
<field name = "no ack" domain = "no ack" />
<field name = "exclusive" type = "bit">
request exclusive access
<doc>
Request exclusive consumer access, meaning only this consumer can
access the queue.
</doc>
<doc name = "rule" test = "amq_file_00">
If the server cannot grant exclusive access to the queue when asked,
- because there are other consumers active - it MUST raise a channel
exception with return code 405 (resource locked).
</doc>
</field>
<field name = "nowait" type = "bit">
do not send a reply method
<doc>
If set, the server will not respond to the method. The client should
not wait for a reply method. If the server could not complete the
method it will raise a channel or connection exception.
</doc>
</field>
</method>
<method name = "consume-ok" synchronous = "1" index = "21">
confirm a new consumer
<doc>
This method provides the client with a consumer tag which it MUST
use in methods that work with the consumer.
</doc>
<chassis name = "client" implement = "MUST" />
<field name = "consumer tag" domain = "consumer tag">
<doc>
Holds the consumer tag specified by the client or provided by
the server.
</doc>
</field>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name = "cancel" synchronous = "1" index = "30">
end a queue consumer
<doc>
This method cancels a consumer. This does not affect already
delivered messages, but it does mean the server will not send any
more messages for that consumer.
</doc>
<chassis name = "server" implement = "MUST" />
<response name = "cancel-ok" />
<field name = "consumer tag" domain = "consumer tag" />
<field name = "nowait" type = "bit">
do not send a reply method
<doc>
If set, the server will not respond to the method. The client should
not wait for a reply method. If the server could not complete the
method it will raise a channel or connection exception.
</doc>
</field>
</method>
<method name = "cancel-ok" synchronous = "1" index = "31">
confirm a cancelled consumer
<doc>
This method confirms that the cancellation was completed.
</doc>
<chassis name = "client" implement = "MUST" />
<field name = "consumer tag" domain = "consumer tag" />
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name = "open" synchronous = "1" index = "40">
request to start staging
<doc>
This method requests permission to start staging a message. Staging
means sending the message into a temporary area at the recipient end
and then delivering the message by referring to this temporary area.
Staging is how the protocol handles partial file transfers - if a
message is partially staged and the connection breaks, the next time
the sender starts to stage it, it can restart from where it left off.
</doc>
<response name = "open-ok" />
<chassis name = "server" implement = "MUST" />
<chassis name = "client" implement = "MUST" />
<field name = "identifier" type = "shortstr">
staging identifier
<doc>
This is the staging identifier. This is an arbitrary string chosen
by the sender. For staging to work correctly the sender must use
the same staging identifier when staging the same message a second
time after recovery from a failure. A good choice for the staging
identifier would be the SHA1 hash of the message properties data
(including the original filename, revised time, etc.).
</doc>
</field>
<field name = "content size" type = "longlong">
message content size
<doc>
The size of the content in octets. The recipient may use this
information to allocate or check available space in advance, to
avoid "disk full" errors during staging of very large messages.
</doc>
<doc name = "rule">
The sender MUST accurately fill the content-size field.
Zero-length content is permitted.
</doc>
</field>
</method>
<method name = "open-ok" synchronous = "1" index = "41">
confirm staging ready
<doc>
This method confirms that the recipient is ready to accept staged
data. If the message was already partially-staged at a previous
time the recipient will report the number of octets already staged.
</doc>
<response name = "stage" />
<chassis name = "server" implement = "MUST" />
<chassis name = "client" implement = "MUST" />
<field name = "staged size" type = "longlong">
already staged amount
<doc>
The amount of previously-staged content in octets. For a new
message this will be zero.
</doc>
<doc name = "rule">
The sender MUST start sending data from this octet offset in the
message, counting from zero.
</doc>
<doc name = "rule">
The recipient MAY decide how long to hold partially-staged content
and MAY implement staging by always discarding partially-staged
content. However if it uses the file content type it MUST support
the staging methods.
</doc>
</field>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name = "stage" content = "1" index = "50">
stage message content
<doc>
This method stages the message, sending the message content to the
recipient from the octet offset specified in the Open-Ok method.
</doc>
<chassis name = "server" implement = "MUST" />
<chassis name = "client" implement = "MUST" />
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name = "publish" index = "60">
publish a message
<doc>
This method publishes a staged file message to a specific exchange.
The file message will be routed to queues as defined by the exchange
configuration and distributed to any active consumers when the
transaction, if any, is committed.
</doc>
<chassis name = "server" implement = "MUST" />
<field name = "ticket" domain = "access ticket">
<doc name = "rule">
The client MUST provide a valid access ticket giving "write"
access rights to the access realm for the exchange.
</doc>
</field>
<field name = "exchange" domain = "exchange name">
<doc>
Specifies the name of the exchange to publish to. The exchange
name can be empty, meaning the default exchange. If the exchange
name is specified, and that exchange does not exist, the server
will raise a channel exception.
</doc>
<doc name = "rule">
The server MUST accept a blank exchange name to mean the default
exchange.
</doc>
<doc name = "rule">
If the exchange was declared as an internal exchange, the server
MUST respond with a reply code 403 (access refused) and raise a
channel exception.
</doc>
<doc name = "rule">
The exchange MAY refuse file content in which case it MUST respond
with a reply code 540 (not implemented) and raise a channel
exception.
</doc>
</field>
<field name = "routing key" type = "shortstr">
Message routing key
<doc>
Specifies the routing key for the message. The routing key is
used for routing messages depending on the exchange configuration.
</doc>
</field>
<field name = "mandatory" type = "bit">
indicate mandatory routing
<doc>
This flag tells the server how to react if the message cannot be
routed to a queue. If this flag is set, the server will return an
unroutable message with a Return method. If this flag is zero, the
server silently drops the message.
</doc>
<doc name = "rule" test = "amq_file_00">
The server SHOULD implement the mandatory flag.
</doc>
</field>
<field name = "immediate" type = "bit">
request immediate delivery
<doc>
This flag tells the server how to react if the message cannot be
routed to a queue consumer immediately. If this flag is set, the
server will return an undeliverable message with a Return method.
If this flag is zero, the server will queue the message, but with
no guarantee that it will ever be consumed.
</doc>
<doc name = "rule" test = "amq_file_00">
The server SHOULD implement the immediate flag.
</doc>
</field>
<field name = "identifier" type = "shortstr">
staging identifier
<doc>
This is the staging identifier of the message to publish. The
message must have been staged. Note that a client can send the
Publish method asynchronously without waiting for staging to
finish.
</doc>
</field>
</method>
<method name = "return" content = "1" index = "70">
return a failed message
<doc>
This method returns an undeliverable message that was published
with the "immediate" flag set, or an unroutable message published
with the "mandatory" flag set. The reply code and text provide
information about the reason that the message was undeliverable.
</doc>
<chassis name = "client" implement = "MUST" />
<field name = "reply code" domain = "reply code" />
<field name = "reply text" domain = "reply text" />
<field name = "exchange" domain = "exchange name">
<doc>
Specifies the name of the exchange that the message was
originally published to.
</doc>
</field>
<field name = "routing key" type = "shortstr">
Message routing key
<doc>
Specifies the routing key name specified when the message was
published.
</doc>
</field>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name = "deliver" index = "80">
notify the client of a consumer message
<doc>
This method delivers a staged file message to the client, via a
consumer. In the asynchronous message delivery model, the client
starts a consumer using the Consume method, then the server
responds with Deliver methods as and when messages arrive for
that consumer.
</doc>
<doc name = "rule">
The server SHOULD track the number of times a message has been
delivered to clients and when a message is redelivered a certain
number of times - e.g. 5 times - without being acknowledged, the
server SHOULD consider the message to be unprocessable (possibly
causing client applications to abort), and move the message to a
dead letter queue.
</doc>
<chassis name = "client" implement = "MUST" />
<field name = "consumer tag" domain = "consumer tag" />
<field name = "delivery tag" domain = "delivery tag" />
<field name = "redelivered" domain = "redelivered" />
<field name = "exchange" domain = "exchange name">
<doc>
Specifies the name of the exchange that the message was originally
published to.
</doc>
</field>
<field name = "routing key" type = "shortstr">
Message routing key
<doc>
Specifies the routing key name specified when the message was
published.
</doc>
</field>
<field name = "identifier" type = "shortstr">
staging identifier
<doc>
This is the staging identifier of the message to deliver. The
message must have been staged. Note that a server can send the
Deliver method asynchronously without waiting for staging to
finish.
</doc>
</field>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name = "ack" index = "90">
acknowledge one or more messages
<doc>
This method acknowledges one or more messages delivered via the
Deliver method. The client can ask to confirm a single message or
a set of messages up to and including a specific message.
</doc>
<chassis name = "server" implement = "MUST" />
<field name = "delivery tag" domain = "delivery tag" />
<field name = "multiple" type = "bit">
acknowledge multiple messages
<doc>
If set to 1, the delivery tag is treated as "up to and including",
so that the client can acknowledge multiple messages with a single
method. If set to zero, the delivery tag refers to a single
message. If the multiple field is 1, and the delivery tag is zero,
tells the server to acknowledge all outstanding mesages.
</doc>
<doc name = "rule">
The server MUST validate that a non-zero delivery-tag refers to an
delivered message, and raise a channel exception if this is not the
case.
</doc>
</field>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name = "reject" index = "100">
reject an incoming message
<doc>
This method allows a client to reject a message. It can be used to
return untreatable messages to their original queue. Note that file
content is staged before delivery, so the client will not use this
method to interrupt delivery of a large message.
</doc>
<doc name = "rule">
The server SHOULD interpret this method as meaning that the client
is unable to process the message at this time.
</doc>
<doc name = "rule">
A client MUST NOT use this method as a means of selecting messages
to process. A rejected message MAY be discarded or dead-lettered,
not necessarily passed to another client.
</doc>
<chassis name = "server" implement = "MUST" />
<field name = "delivery tag" domain = "delivery tag" />
<field name = "requeue" type = "bit">
requeue the message
<doc>
If this field is zero, the message will be discarded. If this bit
is 1, the server will attempt to requeue the message.
</doc>
<doc name = "rule">
The server MUST NOT deliver the message to the same client within
the context of the current channel. The recommended strategy is
to attempt to deliver the message to an alternative consumer, and
if that is not possible, to move the message to a dead-letter
queue. The server MAY use more sophisticated tracking to hold
the message on the queue and redeliver it to the same client at
a later stage.
</doc>
</field>
</method>
</class>
<class name="stream" handler="channel" index="80">
<!--
======================================================
== STREAMING
======================================================
-->
work with streaming content
<doc>
The stream class provides methods that support multimedia streaming.
The stream class uses the following semantics: one message is one
packet of data; delivery is unacknowleged and unreliable; the consumer
can specify quality of service parameters that the server can try to
adhere to; lower-priority messages may be discarded in favour of high
priority messages.
</doc>
<doc name = "grammar">
stream = C:QOS S:QOS-OK
/ C:CONSUME S:CONSUME-OK
/ C:CANCEL S:CANCEL-OK
/ C:PUBLISH content
/ S:RETURN
/ S:DELIVER content
</doc>
<chassis name = "server" implement = "MAY" />
<chassis name = "client" implement = "MAY" />
<doc name = "rule">
The server SHOULD discard stream messages on a priority basis if
the queue size exceeds some configured limit.
</doc>
<doc name = "rule">
The server MUST implement at least 2 priority levels for stream
messages, where priorities 0-4 and 5-9 are treated as two distinct
levels. The server MAY implement up to 10 priority levels.
</doc>
<doc name = "rule">
The server MUST implement automatic acknowledgements on stream
content. That is, as soon as a message is delivered to a client
via a Deliver method, the server must remove it from the queue.
</doc>
<!-- These are the properties for a Stream content -->
<field name = "content type" type = "shortstr">
MIME content type
</field>
<field name = "content encoding" type = "shortstr">
MIME content encoding
</field>
<field name = "headers" type = "table">
Message header field table
</field>
<field name = "priority" type = "octet">
The message priority, 0 to 9
</field>
<field name = "timestamp" type = "timestamp">
The message timestamp
</field>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name = "qos" synchronous = "1" index = "10">
specify quality of service
<doc>
This method requests a specific quality of service. The QoS can
be specified for the current channel or for all channels on the
connection. The particular properties and semantics of a qos method
always depend on the content class semantics. Though the qos method
could in principle apply to both peers, it is currently meaningful
only for the server.
</doc>
<chassis name = "server" implement = "MUST" />
<response name = "qos-ok" />
<field name = "prefetch size" type = "long">
prefetch window in octets
<doc>
The client can request that messages be sent in advance so that
when the client finishes processing a message, the following
message is already held locally, rather than needing to be sent
down the channel. Prefetching gives a performance improvement.
This field specifies the prefetch window size in octets. May be
set to zero, meaning "no specific limit". Note that other
prefetch limits may still apply.
</doc>
</field>
<field name = "prefetch count" type = "short">
prefetch window in messages
<doc>
Specifies a prefetch window in terms of whole messages. This
field may be used in combination with the prefetch-size field;
a message will only be sent in advance if both prefetch windows
(and those at the channel and connection level) allow it.
</doc>
</field>
<field name = "consume rate" type = "long">
transfer rate in octets/second
<doc>
Specifies a desired transfer rate in octets per second. This is
usually determined by the application that uses the streaming
data. A value of zero means "no limit", i.e. as rapidly as
possible.
</doc>
<doc name = "rule">
The server MAY ignore the prefetch values and consume rates,
depending on the type of stream and the ability of the server
to queue and/or reply it. The server MAY drop low-priority
messages in favour of high-priority messages.
</doc>
</field>
<field name = "global" type = "bit">
apply to entire connection
<doc>
By default the QoS settings apply to the current channel only. If
this field is set, they are applied to the entire connection.
</doc>
</field>
</method>
<method name = "qos-ok" synchronous = "1" index = "11">
confirm the requested qos
<doc>
This method tells the client that the requested QoS levels could
be handled by the server. The requested QoS applies to all active
consumers until a new QoS is defined.
</doc>
<chassis name = "client" implement = "MUST" />
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name = "consume" synchronous = "1" index = "20">
start a queue consumer
<doc>
This method asks the server to start a "consumer", which is a
transient request for messages from a specific queue. Consumers
last as long as the channel they were created on, or until the
client cancels them.
</doc>
<doc name = "rule">
The server SHOULD support at least 16 consumers per queue, unless
the queue was declared as private, and ideally, impose no limit
except as defined by available resources.
</doc>
<doc name = "rule">
Streaming applications SHOULD use different channels to select
different streaming resolutions. AMQP makes no provision for
filtering and/or transforming streams except on the basis of
priority-based selective delivery of individual messages.
</doc>
<chassis name = "server" implement = "MUST" />
<response name = "consume-ok" />
<field name = "ticket" domain = "access ticket">
<doc name = "rule">
The client MUST provide a valid access ticket giving "read" access
rights to the realm for the queue.
</doc>
</field>
<field name = "queue" domain = "queue name">
<doc>
Specifies the name of the queue to consume from. If the queue name
is null, refers to the current queue for the channel, which is the
last declared queue.
</doc>
<doc name = "rule">
If the client did not previously declare a queue, and the queue name
in this method is empty, the server MUST raise a connection exception
with reply code 530 (not allowed).
</doc>
</field>
<field name = "consumer tag" domain = "consumer tag">
<doc>
Specifies the identifier for the consumer. The consumer tag is
local to a connection, so two clients can use the same consumer
tags. If this field is empty the server will generate a unique
tag.
</doc>
<doc name = "rule" test = "todo">
The tag MUST NOT refer to an existing consumer. If the client
attempts to create two consumers with the same non-empty tag
the server MUST raise a connection exception with reply code
530 (not allowed).
</doc>
</field>
<field name = "no local" domain = "no local" />
<field name = "exclusive" type = "bit">
request exclusive access
<doc>
Request exclusive consumer access, meaning only this consumer can
access the queue.
</doc>
<doc name = "rule" test = "amq_file_00">
If the server cannot grant exclusive access to the queue when asked,
- because there are other consumers active - it MUST raise a channel
exception with return code 405 (resource locked).
</doc>
</field>
<field name = "nowait" type = "bit">
do not send a reply method
<doc>
If set, the server will not respond to the method. The client should
not wait for a reply method. If the server could not complete the
method it will raise a channel or connection exception.
</doc>
</field>
</method>
<method name = "consume-ok" synchronous = "1" index = "21">
confirm a new consumer
<doc>
This method provides the client with a consumer tag which it may
use in methods that work with the consumer.
</doc>
<chassis name = "client" implement = "MUST" />
<field name = "consumer tag" domain = "consumer tag">
<doc>
Holds the consumer tag specified by the client or provided by
the server.
</doc>
</field>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name = "cancel" synchronous = "1" index = "30">
end a queue consumer
<doc>
This method cancels a consumer. Since message delivery is
asynchronous the client may continue to receive messages for
a short while after canceling a consumer. It may process or
discard these as appropriate.
</doc>
<chassis name = "server" implement = "MUST" />
<response name = "cancel-ok" />
<field name = "consumer tag" domain = "consumer tag" />
<field name = "nowait" type = "bit">
do not send a reply method
<doc>
If set, the server will not respond to the method. The client should
not wait for a reply method. If the server could not complete the
method it will raise a channel or connection exception.
</doc>
</field>
</method>
<method name = "cancel-ok" synchronous = "1" index = "31">
confirm a cancelled consumer
<doc>
This method confirms that the cancellation was completed.
</doc>
<chassis name = "client" implement = "MUST" />
<field name = "consumer tag" domain = "consumer tag" />
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name = "publish" content = "1" index = "40">
publish a message
<doc>
This method publishes a message to a specific exchange. The message
will be routed to queues as defined by the exchange configuration
and distributed to any active consumers as appropriate.
</doc>
<chassis name = "server" implement = "MUST" />
<field name = "ticket" domain = "access ticket">
<doc name = "rule">
The client MUST provide a valid access ticket giving "write"
access rights to the access realm for the exchange.
</doc>
</field>
<field name = "exchange" domain = "exchange name">
<doc>
Specifies the name of the exchange to publish to. The exchange
name can be empty, meaning the default exchange. If the exchange
name is specified, and that exchange does not exist, the server
will raise a channel exception.
</doc>
<doc name = "rule">
The server MUST accept a blank exchange name to mean the default
exchange.
</doc>
<doc name = "rule">
If the exchange was declared as an internal exchange, the server
MUST respond with a reply code 403 (access refused) and raise a
channel exception.
</doc>
<doc name = "rule">
The exchange MAY refuse stream content in which case it MUST
respond with a reply code 540 (not implemented) and raise a
channel exception.
</doc>
</field>
<field name = "routing key" type = "shortstr">
Message routing key
<doc>
Specifies the routing key for the message. The routing key is
used for routing messages depending on the exchange configuration.
</doc>
</field>
<field name = "mandatory" type = "bit">
indicate mandatory routing
<doc>
This flag tells the server how to react if the message cannot be
routed to a queue. If this flag is set, the server will return an
unroutable message with a Return method. If this flag is zero, the
server silently drops the message.
</doc>
<doc name = "rule" test = "amq_stream_00">
The server SHOULD implement the mandatory flag.
</doc>
</field>
<field name = "immediate" type = "bit">
request immediate delivery
<doc>
This flag tells the server how to react if the message cannot be
routed to a queue consumer immediately. If this flag is set, the
server will return an undeliverable message with a Return method.
If this flag is zero, the server will queue the message, but with
no guarantee that it will ever be consumed.
</doc>
<doc name = "rule" test = "amq_stream_00">
The server SHOULD implement the immediate flag.
</doc>
</field>
</method>
<method name = "return" content = "1" index = "50">
return a failed message
<doc>
This method returns an undeliverable message that was published
with the "immediate" flag set, or an unroutable message published
with the "mandatory" flag set. The reply code and text provide
information about the reason that the message was undeliverable.
</doc>
<chassis name = "client" implement = "MUST" />
<field name = "reply code" domain = "reply code" />
<field name = "reply text" domain = "reply text" />
<field name = "exchange" domain = "exchange name">
<doc>
Specifies the name of the exchange that the message was
originally published to.
</doc>
</field>
<field name = "routing key" type = "shortstr">
Message routing key
<doc>
Specifies the routing key name specified when the message was
published.
</doc>
</field>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name = "deliver" content = "1" index = "60">
notify the client of a consumer message
<doc>
This method delivers a message to the client, via a consumer. In
the asynchronous message delivery model, the client starts a
consumer using the Consume method, then the server responds with
Deliver methods as and when messages arrive for that consumer.
</doc>
<chassis name = "client" implement = "MUST" />
<field name = "consumer tag" domain = "consumer tag" />
<field name = "delivery tag" domain = "delivery tag" />
<field name = "exchange" domain = "exchange name">
<doc>
Specifies the name of the exchange that the message was originally
published to.
</doc>
</field>
<field name = "queue" domain = "queue name">
<doc>
Specifies the name of the queue that the message came from. Note
that a single channel can start many consumers on different
queues.
</doc>
<assert check = "notnull" />
</field>
</method>
</class>
<class name="tx" handler="channel" index="90">
<!--
======================================================
== TRANSACTIONS
======================================================
-->
work with standard transactions
<doc>
Standard transactions provide so-called "1.5 phase commit". We can
ensure that work is never lost, but there is a chance of confirmations
being lost, so that messages may be resent. Applications that use
standard transactions must be able to detect and ignore duplicate
messages.
</doc>
<rule implement="SHOULD">
An client using standard transactions SHOULD be able to track all
messages received within a reasonable period, and thus detect and
reject duplicates of the same message. It SHOULD NOT pass these to
the application layer.
</rule>
<doc name="grammar">
tx = C:SELECT S:SELECT-OK
/ C:COMMIT S:COMMIT-OK
/ C:ROLLBACK S:ROLLBACK-OK
</doc>
<chassis name="server" implement="SHOULD"/>
<chassis name="client" implement="MAY"/>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name="select" synchronous="1" index="10">
select standard transaction mode
<doc>
This method sets the channel to use standard transactions. The
client must use this method at least once on a channel before
using the Commit or Rollback methods.
</doc>
<chassis name="server" implement="MUST"/>
<response name="select-ok"/>
</method>
<method name="select-ok" synchronous="1" index="11">
confirm transaction mode
<doc>
This method confirms to the client that the channel was successfully
set to use standard transactions.
</doc>
<chassis name="client" implement="MUST"/>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name="commit" synchronous="1" index="20">
commit the current transaction
<doc>
This method commits all messages published and acknowledged in
the current transaction. A new transaction starts immediately
after a commit.
</doc>
<chassis name="server" implement="MUST"/>
<response name="commit-ok"/>
</method>
<method name="commit-ok" synchronous="1" index="21">
confirm a successful commit
<doc>
This method confirms to the client that the commit succeeded.
Note that if a commit fails, the server raises a channel exception.
</doc>
<chassis name="client" implement="MUST"/>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name="rollback" synchronous="1" index="30">
abandon the current transaction
<doc>
This method abandons all messages published and acknowledged in
the current transaction. A new transaction starts immediately
after a rollback.
</doc>
<chassis name="server" implement="MUST"/>
<response name="rollback-ok"/>
</method>
<method name="rollback-ok" synchronous="1" index="31">
confirm a successful rollback
<doc>
This method confirms to the client that the rollback succeeded.
Note that if an rollback fails, the server raises a channel exception.
</doc>
<chassis name="client" implement="MUST"/>
</method>
</class>
<class name="dtx" handler="channel" index="100">
<!--
======================================================
== DISTRIBUTED TRANSACTIONS
======================================================
-->
work with distributed transactions
<doc>
Distributed transactions provide so-called "2-phase commit". The
AMQP distributed transaction model supports the X-Open XA
architecture and other distributed transaction implementations.
The Dtx class assumes that the server has a private communications
channel (not AMQP) to a distributed transaction coordinator.
</doc>
<doc name="grammar">
dtx = C:SELECT S:SELECT-OK
C:START S:START-OK
</doc>
<chassis name="server" implement="MAY"/>
<chassis name="client" implement="MAY"/>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name="select" synchronous="1" index="10">
select standard transaction mode
<doc>
This method sets the channel to use distributed transactions. The
client must use this method at least once on a channel before
using the Start method.
</doc>
<chassis name="server" implement="MUST"/>
<response name="select-ok"/>
</method>
<method name="select-ok" synchronous="1" index="11">
confirm transaction mode
<doc>
This method confirms to the client that the channel was successfully
set to use distributed transactions.
</doc>
<chassis name="client" implement="MUST"/>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name="start" synchronous="1" index="20">
start a new distributed transaction
<doc>
This method starts a new distributed transaction. This must be
the first method on a new channel that uses the distributed
transaction mode, before any methods that publish or consume
messages.
</doc>
<chassis name="server" implement="MAY"/>
<response name="start-ok"/>
<field name="dtx identifier" type="shortstr">
transaction identifier
<doc>
The distributed transaction key. This identifies the transaction
so that the AMQP server can coordinate with the distributed
transaction coordinator.
</doc>
<assert check="notnull"/>
</field>
</method>
<method name="start-ok" synchronous="1" index="21">
confirm the start of a new distributed transaction
<doc>
This method confirms to the client that the transaction started.
Note that if a start fails, the server raises a channel exception.
</doc>
<chassis name="client" implement="MUST"/>
</method>
</class>
<class name="tunnel" handler="tunnel" index="110">
<!--
======================================================
== TUNNEL
======================================================
-->
methods for protocol tunneling.
<doc>
The tunnel methods are used to send blocks of binary data - which
can be serialised AMQP methods or other protocol frames - between
AMQP peers.
</doc>
<doc name="grammar">
tunnel = C:REQUEST
/ S:REQUEST
</doc>
<chassis name="server" implement="MAY"/>
<chassis name="client" implement="MAY"/>
<field name="headers" type="table">
Message header field table
</field>
<field name="proxy name" type="shortstr">
The identity of the tunnelling proxy
</field>
<field name="data name" type="shortstr">
The name or type of the message being tunnelled
</field>
<field name="durable" type="octet">
The message durability indicator
</field>
<field name="broadcast" type="octet">
The message broadcast mode
</field>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name="request" content="1" index="10">
sends a tunnelled method
<doc>
This method tunnels a block of binary data, which can be an
encoded AMQP method or other data. The binary data is sent
as the content for the Tunnel.Request method.
</doc>
<chassis name="server" implement="MUST"/>
<field name="meta data" type="table">
meta data for the tunnelled block
<doc>
This field table holds arbitrary meta-data that the sender needs
to pass to the recipient.
</doc>
</field>
</method>
</class>
<class name="test" handler="channel" index="120">
<!--
======================================================
== TEST - CHECK FUNCTIONAL CAPABILITIES OF AN IMPLEMENTATION
======================================================
-->
test functional primitives of the implementation
<doc>
The test class provides methods for a peer to test the basic
operational correctness of another peer. The test methods are
intended to ensure that all peers respect at least the basic
elements of the protocol, such as frame and content organisation
and field types. We assume that a specially-designed peer, a
"monitor client" would perform such tests.
</doc>
<doc name="grammar">
test = C:INTEGER S:INTEGER-OK
/ S:INTEGER C:INTEGER-OK
/ C:STRING S:STRING-OK
/ S:STRING C:STRING-OK
/ C:TABLE S:TABLE-OK
/ S:TABLE C:TABLE-OK
/ C:CONTENT S:CONTENT-OK
/ S:CONTENT C:CONTENT-OK
</doc>
<chassis name="server" implement="MUST"/>
<chassis name="client" implement="SHOULD"/>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name="integer" synchronous="1" index="10">
test integer handling
<doc>
This method tests the peer's capability to correctly marshal integer
data.
</doc>
<chassis name="client" implement="MUST"/>
<chassis name="server" implement="MUST"/>
<response name="integer-ok"/>
<field name="integer 1" type="octet">
octet test value
<doc>
An octet integer test value.
</doc>
</field>
<field name="integer 2" type="short">
short test value
<doc>
A short integer test value.
</doc>
</field>
<field name="integer 3" type="long">
long test value
<doc>
A long integer test value.
</doc>
</field>
<field name="integer 4" type="longlong">
long-long test value
<doc>
A long long integer test value.
</doc>
</field>
<field name="operation" type="octet">
operation to test
<doc>
The client must execute this operation on the provided integer
test fields and return the result.
</doc>
<assert check="enum">
<value name="add">return sum of test values</value>
<value name="min">return lowest of test values</value>
<value name="max">return highest of test values</value>
</assert>
</field>
</method>
<method name="integer-ok" synchronous="1" index="11">
report integer test result
<doc>
This method reports the result of an Integer method.
</doc>
<chassis name="client" implement="MUST"/>
<chassis name="server" implement="MUST"/>
<field name="result" type="longlong">
result value
<doc>
The result of the tested operation.
</doc>
</field>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name="string" synchronous="1" index="20">
test string handling
<doc>
This method tests the peer's capability to correctly marshal string
data.
</doc>
<chassis name="client" implement="MUST"/>
<chassis name="server" implement="MUST"/>
<response name="string-ok"/>
<field name="string 1" type="shortstr">
short string test value
<doc>
An short string test value.
</doc>
</field>
<field name="string 2" type="longstr">
long string test value
<doc>
A long string test value.
</doc>
</field>
<field name="operation" type="octet">
operation to test
<doc>
The client must execute this operation on the provided string
test fields and return the result.
</doc>
<assert check="enum">
<value name="add">return concatentation of test strings</value>
<value name="min">return shortest of test strings</value>
<value name="max">return longest of test strings</value>
</assert>
</field>
</method>
<method name="string-ok" synchronous="1" index="21">
report string test result
<doc>
This method reports the result of a String method.
</doc>
<chassis name="client" implement="MUST"/>
<chassis name="server" implement="MUST"/>
<field name="result" type="longstr">
result value
<doc>
The result of the tested operation.
</doc>
</field>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name="table" synchronous="1" index="30">
test field table handling
<doc>
This method tests the peer's capability to correctly marshal field
table data.
</doc>
<chassis name="client" implement="MUST"/>
<chassis name="server" implement="MUST"/>
<response name="table-ok"/>
<field name="table" type="table">
field table of test values
<doc>
A field table of test values.
</doc>
</field>
<field name="integer op" type="octet">
operation to test on integers
<doc>
The client must execute this operation on the provided field
table integer values and return the result.
</doc>
<assert check="enum">
<value name="add">return sum of numeric field values</value>
<value name="min">return min of numeric field values</value>
<value name="max">return max of numeric field values</value>
</assert>
</field>
<field name="string op" type="octet">
operation to test on strings
<doc>
The client must execute this operation on the provided field
table string values and return the result.
</doc>
<assert check="enum">
<value name="add">return concatenation of string field values</value>
<value name="min">return shortest of string field values</value>
<value name="max">return longest of string field values</value>
</assert>
</field>
</method>
<method name="table-ok" synchronous="1" index="31">
report table test result
<doc>
This method reports the result of a Table method.
</doc>
<chassis name="client" implement="MUST"/>
<chassis name="server" implement="MUST"/>
<field name="integer result" type="longlong">
integer result value
<doc>
The result of the tested integer operation.
</doc>
</field>
<field name="string result" type="longstr">
string result value
<doc>
The result of the tested string operation.
</doc>
</field>
</method>
<!-- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -->
<method name="content" synchronous="1" content="1" index="40">
test content handling
<doc>
This method tests the peer's capability to correctly marshal content.
</doc>
<chassis name="client" implement="MUST"/>
<chassis name="server" implement="MUST"/>
<response name="content-ok"/>
</method>
<method name="content-ok" synchronous="1" content="1" index="41">
report content test result
<doc>
This method reports the result of a Content method. It contains the
content checksum and echoes the original content as provided.
</doc>
<chassis name="client" implement="MUST"/>
<chassis name="server" implement="MUST"/>
<field name="content checksum" type="long">
content hash
<doc>
The 32-bit checksum of the content, calculated by adding the
content into a 32-bit accumulator.
</doc>
</field>
</method>
</class>
</amqp>''' | AWSpider | /AWSpider-0.3.2.12.tar.gz/AWSpider-0.3.2.12/awspider/amqp/specs/__init__.py | __init__.py |
import base64
import hmac
import hashlib
import urllib
from datetime import datetime
import xml.etree.cElementTree as ET
from .lib import safe_quote_tuple, etree_to_dict
from ..requestqueuer import RequestQueuer
PA_NAMESPACE = "{http://webservices.amazon.com/AWSECommerceService/2009-10-01}"
class AmazonProductAdvertising:
"""
Amazon Product Advertising API.
"""
host = "ecs.amazonaws.com"
def __init__(self, aws_access_key_id, aws_secret_access_key, rq=None):
"""
**Arguments:**
* *aws_access_key_id* -- Amazon AWS access key ID
* *aws_secret_access_key* -- Amazon AWS secret access key
**Keyword arguments:**
* *rq* -- Optional RequestQueuer object.
"""
if rq is None:
self.rq = RequestQueuer()
else:
self.rq = rq
self.rq.setHostMaxRequestsPerSecond(self.host, 0)
self.rq.setHostMaxSimultaneousRequests(self.host, 0)
self.aws_access_key_id = aws_access_key_id
self.aws_secret_access_key = aws_secret_access_key
def itemSearch(self, **kwargs):
if "MerchantId" not in kwargs:
kwargs["MerchantId"] = "All"
if "SearchIndex" not in kwargs:
kwargs["SearchIndex"] = "All"
parameters = kwargs
parameters["Operation"] = "ItemSearch"
d = self._request(parameters)
d.addCallback(self._itemSearchCallback)
return d
def _itemSearchCallback(self, data):
xml = ET.fromstring(data["response"])
errors = xml.findall(".//%sError" % PA_NAMESPACE)
if len(errors) > 0:
message = errors[0].find("./%sMessage" % PA_NAMESPACE).text
raise Exception(message)
items = xml.findall(".//%sItem" % PA_NAMESPACE)
results = []
for item in items:
results.append(self._processItem(item))
return results
def _processItem(self, item_etree):
item = {}
item["asin"] = item_etree.find("./%sASIN" % PA_NAMESPACE).text
item["detailpageurl"] = item_etree.find("./%sDetailPageURL" % PA_NAMESPACE).text
# Attributes
attributes = item_etree.find("./%sItemAttributes" % PA_NAMESPACE).getchildren()
for element in attributes:
tag = element.tag.replace(PA_NAMESPACE, "")
item[tag.lower()] = element.text
# Links
links = item_etree.findall(".//%sItemLink" % PA_NAMESPACE)
if len(links) > 0:
link_list = []
for link in links:
link_list.append({
"description":link.find("./%sDescription" % PA_NAMESPACE).text,
"url":link.find("./%sURL" % PA_NAMESPACE).text})
item["links"] = link_list
# Sales Rank
sales_rank = item_etree.find(".//%sSalesRank" % PA_NAMESPACE)
if sales_rank is not None:
item["sales_rank"] = sales_rank.text
# Images
item.update(self._processImageSet(item_etree))
image_sets = item_etree.findall(".//%sImageSet" % PA_NAMESPACE)
if len(image_sets) > 0:
item["image_sets"] = {}
for image_set in image_sets:
item["image_sets"][image_set.attrib["Category"]] = self._processImageSet(image_set)
# Subjects
subjects = item_etree.findall(".//%sSubject" % PA_NAMESPACE)
if len(subjects) > 0:
item["subjects"] = []
for subject in subjects:
item["subjects"].append(subject.text)
# Offer summary
offer_summary = item_etree.find(".//%sOfferSummary" % PA_NAMESPACE)
item["offer_summary"] = etree_to_dict(
offer_summary,
namespace=PA_NAMESPACE,
tag_list=False,
convert_camelcase=True)
# Offers
total_offers = item_etree.find(".//%sTotalOffers" % PA_NAMESPACE)
if total_offers is not None:
item["total_offers"] = total_offers.text
total_offer_pages = item_etree.find(".//%sTotalOfferPages" % PA_NAMESPACE)
if total_offer_pages is not None:
item["total_offer_pages"] = total_offer_pages.text
offers = item_etree.findall(".//%sOffer" % PA_NAMESPACE)
if len(offers) > 0:
item["offers"] = []
for offer in offers:
item["offers"].append(etree_to_dict(
offer,
namespace=PA_NAMESPACE,
tag_list=False,
convert_camelcase=True))
# Reviews
average_rating = item_etree.find(".//%sAverageRating" % PA_NAMESPACE)
if average_rating is not None:
item["average_rating"] = average_rating.text
total_reviews = item_etree.find(".//%sTotalReviews" % PA_NAMESPACE)
if total_reviews is not None:
item["total_reviews"] = total_reviews.text
total_review_pages = item_etree.find(".//%sTotalReviewPages" % PA_NAMESPACE)
if total_review_pages is not None:
item["total_review_pages"] = total_review_pages.text
reviews = item_etree.findall(".//%sReview" % PA_NAMESPACE)
if len(reviews) > 0:
item["reviews"] = []
for review in reviews:
item["reviews"].append(etree_to_dict(
review,
namespace=PA_NAMESPACE,
tag_list=False,
convert_camelcase=True))
# Editorial reviews
editorial_reviews = item_etree.findall(".//%sEditorialReview" % PA_NAMESPACE)
if len(editorial_reviews) > 0:
item["editorial_reviews"] = []
for review in editorial_reviews:
item["editorial_reviews"].append(etree_to_dict(
review,
namespace=PA_NAMESPACE,
tag_list=False,
convert_camelcase=True))
# Similar products
similar_products = item_etree.findall(".//%sSimilarProduct" % PA_NAMESPACE)
if len(similar_products) > 0:
item["similar_products"] = []
for product in similar_products:
item["similar_products"].append(etree_to_dict(
product,
namespace=PA_NAMESPACE,
tag_list=False,
convert_camelcase=True))
# Tags
if item_etree.find(".//%sTags" % PA_NAMESPACE) is not None:
item["tag_information"] = {}
distinct_tags = item_etree.find(".//%sDistinctTags" % PA_NAMESPACE)
if distinct_tags is not None:
item["tag_information"]["distinct_tags"] = distinct_tags.text
distinct_items = item_etree.find(".//%sDistinctItems" % PA_NAMESPACE)
if distinct_items is not None:
item["tag_information"]["distinct_items"] = distinct_items.text
distinct_users = item_etree.find(".//%sDistinctUsers" % PA_NAMESPACE)
if distinct_users is not None:
item["tag_information"]["distinct_users"] = distinct_users.text
total_usages = item_etree.find(".//%sTotalUsages" % PA_NAMESPACE)
if total_usages is not None:
item["tag_information"]["total_usages"] = total_usages.text
first_tagging = item_etree.find(".//%sFirstTagging" % PA_NAMESPACE)
if first_tagging is not None:
item["tag_information"]["first_tagging"] = etree_to_dict(
first_tagging,
namespace=PA_NAMESPACE,
tag_list=False,
convert_camelcase=True)
last_tagging = item_etree.find(".//%sLastTagging" % PA_NAMESPACE)
if last_tagging is not None:
item["tag_information"]["last_tagging"] = etree_to_dict(
last_tagging,
namespace=PA_NAMESPACE,
tag_list=False,
convert_camelcase=True)
tags = item_etree.findall(".//%sTag" % PA_NAMESPACE)
if len(tags) > 0:
item["tags"] = []
for tag in tags:
item["tags"].append(etree_to_dict(
tag,
namespace=PA_NAMESPACE,
tag_list=False,
convert_camelcase=True))
# BrowseNodes
browse_nodes = item_etree.find(".//%sBrowseNodes" % PA_NAMESPACE)
if browse_nodes is not None:
item["browse_nodes"] = []
for browse_node in browse_nodes.getchildren():
item["browse_nodes"].append(etree_to_dict(
browse_node,
namespace=PA_NAMESPACE,
tag_list=False,
convert_camelcase=True))
# Lists
listmania_lists = item_etree.findall(".//%sListmaniaList" % PA_NAMESPACE)
if len(listmania_lists) > 0:
item["listmania_lists"] = []
for listmania_list in listmania_lists:
item["listmania_lists"].append(etree_to_dict(
listmania_list,
namespace=PA_NAMESPACE,
tag_list=False,
convert_camelcase=True))
return item
def _processImageSet(self, item_etree):
item = {}
swatch_image = item_etree.find("./%sSwatchImage" % PA_NAMESPACE)
if swatch_image is not None:
item["swatch_image"] = swatch_image.find("./%sURL" % PA_NAMESPACE).text
small_image = item_etree.find("./%sSmallImage" % PA_NAMESPACE)
if small_image is not None:
item["small_image"] = small_image.find("./%sURL" % PA_NAMESPACE).text
thumbnail_image = item_etree.find("./%sThumbnailImage" % PA_NAMESPACE)
if thumbnail_image is not None:
item["thumbnail_image"] = thumbnail_image.find("./%sURL" % PA_NAMESPACE).text
tiny_image = item_etree.find("./%sTinyImage" % PA_NAMESPACE)
if tiny_image is not None:
item["tiny_image"] = tiny_image.find("./%sURL" % PA_NAMESPACE).text
medium_image = item_etree.find("./%sMediumImage" % PA_NAMESPACE)
if medium_image is not None:
item["medium_image"] = medium_image.find("./%sURL" % PA_NAMESPACE).text
large_image = item_etree.find("./%sLargeImage" % PA_NAMESPACE)
if large_image is not None:
item["large_image"] = large_image.find("./%sURL" % PA_NAMESPACE).text
return item
def _request(self, parameters):
"""
Add authentication parameters and make request to Amazon.
**Arguments:**
* *parameters* -- Key value pairs of parameters
"""
parameters["Service"] = "AWSECommerceService"
parameters = self._getAuthorization("GET", parameters)
query_string = urllib.urlencode(parameters)
url = "https://%s/onca/xml?%s" % (self.host, query_string)
d = self.rq.getPage(url, method="GET")
return d
def _canonicalize(self, parameters):
"""
Canonicalize parameters for use with AWS Authorization.
**Arguments:**
* *parameters* -- Key value pairs of parameters
**Returns:**
* A safe-quoted string representation of the parameters.
"""
parameters = parameters.items()
parameters.sort(lambda x, y:cmp(x[0], y[0]))
return "&".join([safe_quote_tuple(x) for x in parameters])
def _getAuthorization(self, method, parameters):
"""
Create authentication parameters.
**Arguments:**
* *method* -- HTTP method of the request
* *parameters* -- Key value pairs of parameters
**Returns:**
* A dictionary of authorization parameters
"""
signature_parameters = {
"AWSAccessKeyId":self.aws_access_key_id,
'Timestamp':datetime.utcnow().isoformat()[0:19]+"+00:00",
"AWSAccessKeyId":self.aws_access_key_id,
"Version":"2009-10-01"
}
signature_parameters.update(parameters)
query_string = self._canonicalize(signature_parameters)
string_to_sign = "%(method)s\n%(host)s\n%(resource)s\n%(qs)s" % {
"method":method,
"host":self.host.lower(),
"resource":"/onca/xml",
"qs":query_string,
}
args = [self.aws_secret_access_key, string_to_sign, hashlib.sha256]
signature = base64.encodestring(hmac.new(*args).digest()).strip()
signature_parameters.update({'Signature': signature})
return signature_parameters | AWSpider | /AWSpider-0.3.2.12.tar.gz/AWSpider-0.3.2.12/awspider/aws/productadvertising.py | productadvertising.py |
import base64
import hmac
import hashlib
import urllib
import xml.etree.cElementTree as ET
from datetime import datetime, timedelta
import simplejson
from ..unicodeconverter import convertToUTF8
from ..requestqueuer import RequestQueuer
from .lib import return_true, safe_quote_tuple
SQS_NAMESPACE = "{http://queue.amazonaws.com/doc/2009-02-01/}"
class AmazonSQS:
"""
Amazon Simple Queue Service API.
"""
host = "queue.amazonaws.com"
def __init__(self, aws_access_key_id, aws_secret_access_key, rq=None):
"""
**Arguments:**
* *aws_access_key_id* -- Amazon AWS access key ID string
* *aws_secret_access_key* -- Amazon AWS secret access key string
**Keyword arguments:**
* *rq* -- Optional RequestQueuer object.
"""
if rq is None:
self.rq = RequestQueuer()
else:
self.rq = rq
self.rq.setHostMaxRequestsPerSecond(self.host, 0)
self.rq.setHostMaxSimultaneousRequests(self.host, 0)
self.aws_access_key_id = aws_access_key_id
self.aws_secret_access_key = aws_secret_access_key
def listQueues(self, name_prefix=None):
"""
Return a list of your queues. The maximum number of queues
that can be returned is 1000. If you specify a value for
the optional name_prefix parameter, only queues with
a name beginning with the specified value are returned.
See: http://docs.amazonwebservices.com/AWSSimpleQueueService/2009-02-01/SQSDeveloperGuide/index.html?Query_QueryListQueues.html
**Keyword arguments:**
* *name_prefix* -- String to use for filtering the list results.
Only those queues whose name begins with the specified
string are returned. (Default None)
****Returns:****
* Deferred of a list of queue paths that can be used as the
'resource' argument for other class methods.
"""
parameters = {
"Action":"ListQueues"}
if name_prefix is not None:
parameters["QueueNamePrefix"] = name_prefix
d = self._request(parameters)
d.addCallback(self._listQueuesCallback)
return d
def _listQueuesCallback(self, data):
xml = ET.fromstring(data["response"])
queue_urls = xml.findall(".//%sQueueUrl" % SQS_NAMESPACE)
host_string = "https://%s" % self.host
queue_paths = [x.text.replace(host_string, "") for x in queue_urls]
return queue_paths
def createQueue(self, name, visibility_timeout=None):
"""
Create a new queue.
See: http://docs.amazonwebservices.com/AWSSimpleQueueService/2009-02-01/SQSDeveloperGuide/index.html?Query_QueryCreateQueue.html
**Arguments:**
* *name* -- The name to use for the queue created.
**Keyword arguments:**
* *visibility_timeout* -- The visibility timeout (in seconds) to use
for this queue. (Default None)
**Returns:**
* Deferred of a queue path string that can be used as the 'resource' argument
for other class methods.
"""
name = convertToUTF8(name)
parameters = {
"Action":"CreateQueue",
"QueueName":name}
if visibility_timeout is not None:
parameters["DefaultVisibilityTimeout"] = visibility_timeout
d = self._request(parameters)
d.addCallback(self._createQueueCallback)
d.addErrback(self._genericErrback)
return d
def _createQueueCallback(self, data):
xml = ET.fromstring(data["response"])
queue_url = xml.find(".//%sQueueUrl" % SQS_NAMESPACE).text
queue_path = queue_url.replace("https://%s" % self.host, "")
return queue_path
def deleteQueue(self, resource):
"""
Delete the queue specified by resource, regardless of whether the
queue is empty. If the specified queue does not exist, returns
a successful response.
See: http://docs.amazonwebservices.com/AWSSimpleQueueService/2009-02-01/SQSDeveloperGuide/index.html?Query_QueryDeleteQueue.html
**Arguments:**
* *resource* -- The path of the queue.
**Returns:**
* Deferred of boolean True.
"""
parameters = {"Action":"DeleteQueue"}
d = self._request(parameters, resource=resource, method="DELETE")
d.addCallback(return_true)
d.addErrback(self._genericErrback)
return d
def setQueueAttributes(self, resource, visibility_timeout=None,
policy=None):
"""
Set attributes of a queue.
See: http://docs.amazonwebservices.com/AWSSimpleQueueService/2009-02-01/SQSDeveloperGuide/index.html?Query_QuerySetQueueAttributes.html
**Keyword arguments:**
* *visibility_timeout* -- The visibility timeout (in seconds) to use
for this queue. (Default None)
* *policy* -- Python object representing a valid policy. (Default None)
See: http://docs.amazonwebservices.com/AWSSimpleQueueService/2009-02-01/SQSDeveloperGuide/index.html?AccessPolicyLanguage_Concepts.html
**Arguments:**
* *resource* -- The path of the queue.
**Returns:**
* Deferred of boolean True.
"""
parameters = {"Action":"SetQueueAttributes"}
attributes = {}
if policy is not None:
attributes["Policy"] = simplejson.dumps(policy)
if visibility_timeout is not None:
attributes["VisibilityTimeout"] = visibility_timeout
attr_count = 1
for name in attributes:
parameters["Attribute.%s.Name" % attr_count] = name
parameters["Attribute.%s.Value" % attr_count] = attributes[name]
attr_count += 1
d = self._request(parameters, resource=resource)
d.addCallback(return_true)
d.addErrback(self._genericErrback)
return d
def getQueueAttributes(self, resource, name=None):
"""
Get attributes of a queue.
See: http://docs.amazonwebservices.com/AWSSimpleQueueService/2009-02-01/SQSDeveloperGuide/index.html?Query_QueryGetQueueAttributes.html
**Keyword arguments:**
* *name* -- The attribute you want to get. (Default All.) Valid
values are ``(All | ApproximateNumberOfMessages | VisibilityTimeout
| CreatedTimestamp | LastModifiedTimestamp | Policy)``
**Arguments:**
* *resource* -- The path of the queue.
**Returns:**
* Deferred of a dictionary of the requested attributes.
"""
attributes = [
"All",
"ApproximateNumberOfMessages",
"VisibilityTimeout",
"CreatedTimestamp",
"LastModifiedTimestamp",
"Policy"]
if name is not None:
if name not in attributes:
raise Exception("Unknown attribute name '%s'." % name)
else:
name = "All"
parameters = {
"Action":"GetQueueAttributes",
"AttributeName":name}
d = self._request(parameters, resource=resource, method="DELETE")
d.addCallback(self._getQueueAttributesCallback)
d.addErrback(self._genericErrback)
return d
def _getQueueAttributesCallback(self, data):
attributes = {}
xml = ET.fromstring(data["response"])
xml_attributes = xml.findall(".//%sAttribute" % SQS_NAMESPACE)
for attribute in xml_attributes:
name = attribute.find(".//%sName" % SQS_NAMESPACE).text
value = attribute.find(".//%sValue" % SQS_NAMESPACE).text
attributes[name] = value
if "Policy" in attributes:
attributes["Policy"] = simplejson.loads(attributes["Policy"])
integer_attribute_names = [
'CreatedTimestamp',
'ApproximateNumberOfMessages',
'LastModifiedTimestamp',
'VisibilityTimeout']
for name in integer_attribute_names:
if name in attributes:
attributes[name] = int(attributes[name])
return attributes
def addPermission(self, resource, label, aws_account_ids, actions=None):
"""
Add a permission to a queue for a specific principal. This allows
for sharing access to the queue.
See: http://docs.amazonwebservices.com/AWSSimpleQueueService/2009-02-01/SQSDeveloperGuide/index.html?Query_QueryAddPermission.html
**Arguments:**
* *resource* -- The path of the queue.
* *label* -- The unique identification string of the permission you're
setting.
* *aws_account_ids* -- A string or list of strings of (a) valid
12-digit AWS account number(s), with or without hyphens.
**Keyword arguments:**
* *actions* -- A string or list of strings of actions you want to
allow for the specified principal. Default '*'. Valid values are (* |
SendMessage | ReceiveMessage | DeleteMessage | ChangeMessageVisibility
| GetQueueAttributes)
**Returns:**
* Deferred of boolean True.
"""
if actions is None:
actions = ["*"]
if isinstance(actions, str) or isinstance(actions, unicode):
actions = [str(actions)]
if not isinstance(actions, list):
raise Exception("Actions must be a string or list of strings.")
if isinstance(aws_account_ids, str) or \
isinstance(aws_account_ids, unicode):
aws_account_ids = [str(aws_account_ids)]
if not isinstance(aws_account_ids, list):
message = "aws_account_ids must be a string or list of strings."
raise Exception(message)
aws_account_ids = [x.replace("-", "") for x in aws_account_ids]
action_options = [
"*",
"SendMessage",
"ReceiveMessage",
"DeleteMessage",
"ChangeMessageVisibility",
"GetQueueAttributes"]
for action in actions:
if action not in action_options:
raise Exception("Unknown action name '%s'." % action)
if len(actions) == 0:
actions.append("*")
parameters = {
"Action":"AddPermission",
"Label":label}
action_count = 1
for name in actions:
parameters["ActionName.%s" % action_count] = name
action_count += 1
aws_account_id_count = 1
for name in aws_account_ids:
parameters["AWSAccountId.%s" % aws_account_id_count] = name
aws_account_id_count += 1
d = self._request(parameters, resource=resource)
d.addCallback(return_true)
d.addErrback(self._genericErrback)
return d
def removePermission(self, resource, label):
"""
Revokes any permissions in the queue policy that matches the
label parameter.
See: http://docs.amazonwebservices.com/AWSSimpleQueueService/2009-02-01/SQSDeveloperGuide/index.html?Query_QueryRemovePermission.html
**Arguments:**
* *resource -- The path of the queue.
* *label -- The identfication of the permission you want to remove.
**Returns:**
* Deferred of boolean True.
"""
parameters = {
"Action":"RemovePermission",
"Label":label}
d = self._request(parameters, resource=resource)
d.addCallback(return_true)
d.addErrback(self._genericErrback)
return d
def sendMessage(self, resource, message):
"""
Deliver a message to the specified queue. The maximum allowed message
size is 8 KB.
See: http://docs.amazonwebservices.com/AWSSimpleQueueService/2009-02-01/SQSDeveloperGuide/index.html?Query_QuerySendMessage.html
**Arguments:**
* *resource* -- The path of the queue.
* *message* -- The message to send. Characters must be in: ( #x9 | #xA |
#xD | [#x20 to #xD7FF] | [#xE000 to #xFFFD] | [#x10000 to #x10FFFF] )
See: http://www.w3.org/TR/REC-xml/#charsets
**Returns:**
* Deferred of a string with a Message ID.
"""
parameters = {
"Action":"SendMessage",
"MessageBody":message}
d = self._request(parameters, resource=resource)
d.addCallback(self._sendMessageCallback)
d.addErrback(self._genericErrback)
return d
def _sendMessageCallback(self, data):
xml = ET.fromstring(data["response"])
return xml.find(".//%sMessageId" % SQS_NAMESPACE).text
def receiveMessage(self, resource, visibility_timeout=None,
max_number_of_messages=1, get_sender_id=False,
get_sent_timestamp=False):
"""
Retrieve one or more messages from the specified queue.
See: http://docs.amazonwebservices.com/AWSSimpleQueueService/2009-02-01/SQSDeveloperGuide/index.html?Query_QueryReceiveMessage.html
**Arguments:**
* *resource* -- The path of the queue.
**Keyword arguments:**
* *visibility_timeout* -- The duration (in seconds) that the received
messages are hidden from subsequent retrieve requests after being
retrieved by a ReceiveMessage request. (Default None)
* *max_number_of_messages* -- Maximum number of messages to return. 1-10.
(Default 1)
* *get_sender_id* -- Retrieve the sender id attribute. (Default False)
* *get_sent_timestamp* -- Retrieve the timestamp attribute. (Default False)
**Returns:**
* Deferred of dictionary with attributes 'md5', 'id', 'receipt_handle',
'body'. Optionally 'sender_id' and 'sent_timestamp'.
"""
parameters = {
"Action":"ReceiveMessage"}
if visibility_timeout is not None:
parameters["VisibilityTimeout"] = int(visibility_timeout)
if int(max_number_of_messages) > 1:
parameters["MaxNumberOfMessages"] = int(max_number_of_messages)
attr_count = 1
if get_sender_id == True:
parameters["AttributeName.%s" % attr_count] = "SenderId"
attr_count += 1
if get_sent_timestamp == True:
parameters["AttributeName.%s" % attr_count] = "SentTimestamp"
attr_count += 1
d = self._request(parameters, resource=resource)
d.addCallback(self._receiveMessageCallback)
d.addErrback(self._genericErrback)
return d
def _receiveMessageCallback(self, data):
messages = []
xml = ET.fromstring(data["response"])
message_elements = xml.findall(".//%sMessage" % SQS_NAMESPACE)
for element in message_elements:
message = {}
attributes = element.findall(".//%sAttributes" % SQS_NAMESPACE)
for attribute in attributes:
name = attribute.find(".//%sName" % SQS_NAMESPACE).text
value = attribute.find(".//%sValue" % SQS_NAMESPACE).text
if name == "SenderId":
message['sender_id'] = value
elif name == "SentTimestamp":
message['sent_timestamp'] = value
message["md5"] = element.find(".//%sMD5OfBody" % SQS_NAMESPACE).text
message["id"] = element.find(".//%sMessageId" % SQS_NAMESPACE).text
message["receipt_handle"] = element.find(".//%sReceiptHandle" % SQS_NAMESPACE).text
message["body"] = element.find(".//%sBody" % SQS_NAMESPACE).text
messages.append(message)
return messages
def deleteMessage(self, resource, receipt_handle):
"""
Delete the specified message from the specified queue.
See: http://docs.amazonwebservices.com/AWSSimpleQueueService/2009-02-01/SQSDeveloperGuide/index.html?Query_QueryDeleteMessage.html
**Arguments:**
* *resource* -- The path of the queue.
* *receipt_handle* -- The receipt handle associated with the message
you want to delete.
**Returns:**
* Deferred of boolean True.
"""
parameters = {
"Action":"DeleteMessage",
"ReceiptHandle":receipt_handle}
d = self._request(parameters, resource=resource)
d.addCallback(return_true)
d.addErrback(self._genericErrback)
return d
def changeMessageVisibility(self, resource, receipt_handle,
visibility_timeout):
"""
Change the visibility timeout of a specified message in a queue to
a new value.
See: http://docs.amazonwebservices.com/AWSSimpleQueueService/2009-02-01/SQSDeveloperGuide/index.html?Query_QueryChangeMessageVisibility.html
**Arguments:**
* *resource* -- The path of the queue.
* *receipt_handle* -- The receipt handle associated with the message
you want to delete.
* *visibility_timeout* -- The new value for the message's visibility timeout (in seconds).
**Returns:**
* Deferred of boolean True.
"""
parameters = {
"Action":"ChangeMessageVisibility",
"ReceiptHandle":receipt_handle,
"VisibilityTimeout":int(visibility_timeout)}
d = self._request(parameters, resource=resource)
d.addCallback(return_true)
d.addErrback(self._genericErrback)
return d
def _genericErrback(self, error):
"""
Use server specified error when possible.
**Arguments:**
* *error* -- Twisted error
**Returns:**
* Twisted error
"""
if hasattr(error, "value"):
if hasattr(error.value, "response"):
xml = ET.fromstring(error.value.response)
message = xml.find(".//%sMessage" % SQS_NAMESPACE).text
raise Exception(message)
return error
def _canonicalize(self, parameters):
"""
Canonicalize parameters for use with AWS Authorization.
**Arguments:**
* *parameters* -- Key value pairs of parameters
**Returns:**
* A safe-quoted string representation of the parameters.
"""
parameters = parameters.items()
# Alphebetize key-value pairs.
parameters.sort(lambda x, y:cmp(x[0], y[0]))
# Safe-quote and combine parameters into a string
return "&".join([safe_quote_tuple(x) for x in parameters])
def _request(self, parameters, method="GET", resource="/"):
"""
Add authentication parameters and make request to Amazon.
**Arguments:**
* *parameters* -- Key value pairs of parameters
**Keyword arguments:**
* *method* -- HTTP request method
* *resource* -- Requested server resource
**Returns:**
* Deferred of server response.
"""
parameters = self._getAuthorization(method, parameters,
resource=resource)
query_string = urllib.urlencode(parameters)
url = "https://%s%s?%s" % (self.host, resource, query_string)
#print url
d = self.rq.getPage(url, method=method)
return d
def _getAuthorization(self, method, parameters, resource="/"):
"""
Create authentication parameters.
**Arguments:**
* *method* -- HTTP method of the request
* *parameters* -- Key value pairs of parameters
**Returns:**
* A dictionary of authorization parameters
"""
expires = datetime.utcnow() + timedelta(30)
signature_parameters = {
"AWSAccessKeyId":self.aws_access_key_id,
"SignatureVersion":"2",
"SignatureMethod":"HmacSHA256",
'Expires':"%s+00:00" % expires.isoformat()[0:19],
"AWSAccessKeyId":self.aws_access_key_id,
"Version":"2009-02-01"}
signature_parameters.update(parameters)
query_string = self._canonicalize(signature_parameters)
string_to_sign = "%(method)s\n%(host)s\n%(resource)s\n%(qs)s" % {
"method":method,
"host":self.host.lower(),
"resource":resource,
"qs":query_string}
args = [self.aws_secret_access_key, string_to_sign, hashlib.sha256]
signature = base64.encodestring(hmac.new(*args).digest()).strip()
signature_parameters.update({'Signature':signature})
return signature_parameters | AWSpider | /AWSpider-0.3.2.12.tar.gz/AWSpider-0.3.2.12/awspider/aws/sqs.py | sqs.py |
import cStringIO
import gzip as gzip_package
import base64
import hmac
import hashlib
import logging
import time
import urllib
import xml.etree.cElementTree as ET
from twisted.internet import reactor
from twisted.internet.defer import Deferred, DeferredList, deferredGenerator, waitForDeferred
from ..unicodeconverter import convertToUTF8
from ..requestqueuer import RequestQueuer
from .lib import return_true, etree_to_dict
S3_NAMESPACE = "{http://s3.amazonaws.com/doc/2006-03-01/}"
LOGGER = logging.getLogger("main")
class AmazonS3:
"""
Amazon Simple Storage Service API.
"""
ACCEPTABLE_ERROR_CODES = [400, 403, 404, 409]
host = "s3.amazonaws.com"
reserved_headers = ["x-amz-id-2", "x-amz-request-id", "date", "last-modified", "etag", "content-type", "content-length", "server"]
def __init__(self, aws_access_key_id, aws_secret_access_key, rq=None):
"""
**Arguments:**
* *aws_access_key_id* -- Amazon AWS access key ID
* *aws_secret_access_key* -- Amazon AWS secret access key
**Keyword arguments:**
* *rq* -- Optional RequestQueuer object.
"""
if rq is None:
self.rq = RequestQueuer()
else:
self.rq = rq
self.rq.setHostMaxRequestsPerSecond(self.host, 0)
self.rq.setHostMaxSimultaneousRequests(self.host, 0)
self.aws_access_key_id = aws_access_key_id
self.aws_secret_access_key = aws_secret_access_key
def checkAndCreateBucket(self, bucket):
"""
Check for a bucket's existence. If it does not exist,
create it.
**Arguments:**
* *bucket* -- Bucket name
"""
d = self.getBucket(bucket)
d.addErrback(self._checkAndCreateBucketErrback, bucket)
return d
def _checkAndCreateBucketErrback(self, error, bucket):
if int(error.value.status) == 404:
d = self.putBucket(bucket)
d.addErrback(self._checkAndCreateBucketErrback2, bucket)
return d
raise Exception("Could not find or create bucket '%s'." % bucket_name)
def _checkAndCreateBucketErrback2( self, error, bucket_name):
raise Exception("Could not create bucket '%s'" % bucket_name)
def emptyBucket(self, bucket):
"""
Delete all items in a bucket.
**Arguments:**
* *bucket* -- Bucket name
"""
d = self.getBucket(bucket)
d.addCallback(self._emptyBucketCallback, bucket)
return d
def _emptyBucketCallback(self, result, bucket):
xml = ET.XML(result["response"])
key_nodes = xml.findall(".//%sKey" % S3_NAMESPACE)
delete_deferreds = []
for node in key_nodes:
delete_deferreds.append(self.deleteObject(bucket, node.text))
if len(delete_deferreds) == 0:
return True
d = DeferredList(delete_deferreds)
if xml.find('.//%sIsTruncated' % S3_NAMESPACE).text == "false":
d.addCallback(return_true)
else:
d.addCallback(self._emptyBucketCallbackRepeat, bucket)
return d
def _emptyBucketCallbackRepeat(self, data, bucket):
return self.emptyBucket(bucket)
# def listObjects(self, bucket, marker=None):
# """
# List information about the objects in the bucket.
#
# **Arguments:**
# * *bucket* -- Bucket name
# """
# bucket = convertToUTF8(bucket)
# path = bucket
# if marker is not None:
# path = urllib.urlencode({"marker":marker})
# headers = self._getAuthorization("GET", "", "", {}, "/" + path)
# url = "http://%s/%s" % (self.host, path)
# d = self.rq.getPage(url, method="GET", headers=headers)
# d.addCallback(self._listObjectsCallback, bucket)
# d.addErrback(self._genericErrback, url, method="GET", headers=headers)
# return d
#
# @deferredGenerator
# def _listObjectsCallback(self, result, bucket):
# xml = ET.XML(result["response"])
# data = etree_to_dict(xml, namespace='{http://s3.amazonaws.com/doc/2006-03-01/}')
# for obj in data["Contents"]:
# for key in obj:
# obj[key] = obj[key][0]
# for key in obj["Owner"]:
# obj["Owner"][key] = obj["Owner"][key][0]
# yield obj
#
# if data["IsTruncated"][0] == "true":
# d = self.listObjects(bucket, marker=obj["Key"])
# wfd = waitForDeferred(d)
# yield wfd
def getBucket(self, bucket):
"""
List information about the objects in the bucket.
**Arguments:**
* *bucket* -- Bucket name
"""
bucket = convertToUTF8(bucket)
headers = self._getAuthorization("GET", "", "", {}, "/" + bucket)
url = "http://%s/%s" % (self.host, bucket)
d = self.rq.getPage(url, method="GET", headers=headers)
d.addErrback(self._genericErrback, url, method="GET", headers=headers)
return d
def putBucket(self, bucket):
"""
Creates a new bucket.
**Arguments:**
* *bucket* -- Bucket name
"""
bucket = convertToUTF8(bucket)
headers = {
"Content-Length": 0
}
auth = self._getAuthorization("PUT", "", "", headers, "/" + bucket)
headers.update(auth)
url = "http://%s/%s" % (self.host, bucket)
d = self.rq.getPage(url, method="PUT", headers=headers)
d.addErrback(self._genericErrback, url, method="PUT", headers=headers)
return d
def deleteBucket(self, bucket):
"""
Delete the bucket.
**Arguments:**
* *bucket* -- Bucket name
"""
bucket = convertToUTF8(bucket)
headers = {
"Content-Length": 0
}
auth = self._getAuthorization("DELETE", "", "", headers, "/" + bucket)
headers.update(auth)
url = "http://%s/%s" % (self.host, bucket)
d = self.rq.getPage(url, method="DELETE", headers=headers)
d.addErrback(self._genericErrback, url, method="DELETE",
headers=headers)
return d
def headObject(self, bucket, key):
"""
Retrieve information about a specific object or object size, without
actually fetching the object itself.
**Arguments:**
* *bucket* -- Bucket name
* *key* -- Key name
"""
bucket = convertToUTF8(bucket)
key = convertToUTF8(key)
path = "/" + bucket + "/" + key
headers = self._getAuthorization("HEAD", "", "", {}, path)
url = "http://%s/%s/%s" % (self.host, bucket, key)
d = self.rq.getPage(url, method="HEAD", headers=headers)
d.addCallback(self._getObjectCallback)
d.addErrback(self._genericErrback, url, method="HEAD", headers=headers)
return d
def _decodeAmazonHeaders(self, headers):
"""
Remove custom header prefix from header dictionary keys.
"""
for key in headers.keys():
if ("x-amz-meta-%s" % key.lower()) in self.reserved_headers:
message = "Header %s is reserved for use by Amazon S3." % key
LOGGER.critical(message)
raise Exception(message)
keys = headers.keys()
values = headers.values()
meta = "x-amz-meta-"
return dict(zip([x.replace(meta,"") for x in keys], values))
def getObject(self, bucket, key):
"""
Returns object directly from Amazon S3 using a client/server
delivery mechanism.
**Arguments:**
* *bucket* -- Bucket name
* *key* -- Key name
"""
bucket = convertToUTF8(bucket)
key = convertToUTF8(key)
path = "/" + bucket + "/" + key
headers = self._getAuthorization("GET", "", "", {}, path)
url = "http://%s/%s/%s" % (self.host, bucket, key)
d = self.rq.getPage(url, method="GET", headers=headers)
d.addCallback(self._getObjectCallback)
d.addErrback(self._genericErrback, url, method="GET", headers=headers)
return d
def _getObjectCallback(self, data):
if "content-encoding" in data["headers"]:
if "gzip" in data["headers"]["content-encoding"]:
compressedstream = cStringIO.StringIO(data["response"])
zfile = gzip_package.GzipFile(fileobj=compressedstream)
data["response"] = zfile.read()
data["headers"] = self._decodeAmazonHeaders(data["headers"])
return data
def _encodeAmazonHeaders(self, headers):
"""
Prepend custom header prefix to header dictionary keys.
"""
headers = dict([(x[0].lower(), x[1]) for x in headers.items()])
for header in self.reserved_headers:
if header in headers:
del headers[header]
keys = headers.keys()
values = headers.values()
#for key in keys:
# if key.lower() in self.reserved_headers:
# message = "Header %s is reserved for use by Amazon S3." % key
# LOGGER.error(message)
# raise Exception(message)
meta = "x-amz-meta-"
return dict(zip(["%s%s" % (meta, x) for x in keys], values))
def putObject(self, bucket, key, data, content_type="text/html",
public=True, headers=None, gzip=False):
"""
Add an object to a bucket.
**Arguments:**
* *bucket* -- Bucket name
* *key* -- Key name
* *data* -- Data string
**Keyword arguments:**
* *content_type* -- Content type header (Default 'text/html')
* *public* -- Boolean flag representing access (Default True)
* *headers* -- Custom header dictionary (Default empty dictionary)
* *gzip* -- Boolean flag to gzip data (Default False)
"""
bucket = convertToUTF8(bucket)
key = convertToUTF8(key)
if not isinstance(data, str):
data = convertToUTF8(data)
if headers is None:
headers = {}
# Wrap user-defined headers in with the Amazon custom header prefix.
headers = self._encodeAmazonHeaders(headers)
if gzip:
# Gzip that bastard!
headers["content-encoding"] = "gzip"
zbuf = cStringIO.StringIO()
zfile = gzip_package.GzipFile(None, 'wb', 9, zbuf)
zfile.write(data)
zfile.close()
data = zbuf.getvalue()
content_md5 = base64.encodestring(hashlib.md5(data).digest()).strip()
if public:
headers['x-amz-acl'] = 'public-read'
else:
headers['x-amz-acl'] = 'private'
headers.update({
'Content-Length':len(data),
'Content-Type':content_type,
'Content-MD5':content_md5
})
path = "/" + bucket + "/" + key
auth = self._getAuthorization("PUT", content_md5, content_type,
headers, path)
headers.update(auth)
url = "http://%s/%s/%s" % (self.host, bucket, key)
d = self.rq.getPage(url, method="PUT", headers=headers, postdata=data)
d.addErrback(self._genericErrback, url, method="PUT", headers=headers,
postdata=data)
return d
def deleteObject(self, bucket, key):
"""
Remove the specified object from Amazon S3.
**Arguments:**
* *bucket* -- Bucket name
* *key* -- Key name
"""
bucket = convertToUTF8(bucket)
key = convertToUTF8(key)
path = "/" + bucket + "/" + key
headers = self._getAuthorization("DELETE", "", "", {}, path)
url = "http://%s/%s/%s" % (self.host, bucket, key)
d = self.rq.getPage(url, method="DELETE", headers=headers)
d.addErrback(self._genericErrback, url, method="DELETE",
headers=headers)
return d
def _genericErrback(self, error, url, method="GET", headers=None,
postdata=None, count=0):
if headers is None:
headers = {}
if "status" in error.value.__dict__:
# 204, empty response but otherwise OK, as in the case of a delete.
# Move on, nothing to see here.
if int(error.value.status) == 204:
return {
"response":error.value.response,
"status":int(error.value.status),
"headers":error.value.response,
"message":error.value.headers
}
# Something other than a 40x error. Something is wrong, but let's
# try that again a few times.
elif int(error.value.status) not in self.ACCEPTABLE_ERROR_CODES \
and count < 3:
d = self.rq.getPage(url,
method=method,
headers=headers,
postdata=postdata
)
d.addErrback(self._genericErrback,
url,
method=method,
headers=headers,
postdata=postdata,
count=count + 1
)
return d
# 404 or other normal error, pass it along.
else:
return error
else:
return error
def _canonicalize(self, headers):
"""
Canonicalize headers for use with AWS Authorization.
**Arguments:**
* *headers* -- Key value pairs of headers
**Returns:**
* A string representation of the parameters.
"""
keys = [k for k in headers.keys() if k.startswith("x-amz-")]
keys.sort(key = str.lower)
return "\n".join(["%s:%s" % (x, headers[x]) for x in keys])
def _getAuthorization(self, method, content_hash, content_type,
headers, resource):
"""
Create authentication headers.
**Arguments:**
* *method* -- HTTP method of the request
* *parameters* -- Key value pairs of parameters
**Returns:**
* A dictionary of authorization parameters
"""
date = time.strftime("%a, %d %b %Y %H:%M:%S +0000", time.gmtime())
amazon_headers = self._canonicalize(headers)
if len(amazon_headers):
data = "%s\n%s\n%s\n%s\n%s\n%s" % (
method,
content_hash,
content_type,
date,
amazon_headers,
resource
)
else:
data = "%s\n%s\n%s\n%s\n%s%s" % (
method,
content_hash,
content_type,
date,
amazon_headers,
resource
)
args = [self.aws_secret_access_key, data, hashlib.sha1]
signature = base64.encodestring(hmac.new(*args).digest()).strip()
authorization = "AWS %s:%s" % (self.aws_access_key_id, signature)
return {'Authorization': authorization, 'Date':date, "Host":self.host} | AWSpider | /AWSpider-0.3.2.12.tar.gz/AWSpider-0.3.2.12/awspider/aws/s3.py | s3.py |
import base64
import hmac
import hashlib
import urllib
import xml.etree.cElementTree as ET
from datetime import datetime
import time
import dateutil.parser
import logging
from twisted.internet.defer import DeferredList
from ..requestqueuer import RequestQueuer
from .lib import etree_to_dict, safe_quote_tuple
LOGGER = logging.getLogger("main")
SDB_NAMESPACE = "{http://sdb.amazonaws.com/doc/2009-04-15/}"
def base10toN(num,n):
"""Change a to a base-n number.
Up to base-36 is supported without special notation."""
num_rep={10:'a',
11:'b',
12:'c',
13:'d',
14:'e',
15:'f',
16:'g',
17:'h',
18:'i',
19:'j',
20:'k',
21:'l',
22:'m',
23:'n',
24:'o',
25:'p',
26:'q',
27:'r',
28:'s',
29:'t',
30:'u',
31:'v',
32:'w',
33:'x',
34:'y',
35:'z'}
new_num_string=''
current=num
while current!=0:
remainder=current%n
if 36>remainder>9:
remainder_string=num_rep[remainder]
elif remainder>=36:
remainder_string='('+str(remainder)+')'
else:
remainder_string=str(remainder)
new_num_string=remainder_string+new_num_string
current=current/n
return new_num_string
def base10to36(i):
return base10toN(i, 36)
def base36to10(s):
return int(s, 36)
def sdb_now(offset=0):
"""Return an 11 character, zero padded string with the current Unixtime.
**Keyword arguments:**
* *offset* -- Offset in seconds. (Default 0)
"""
return str(int(offset + time.time())).zfill(11)
def sdb_now_add(seconds, offset=0):
"""Return an 11 character, zero padded string with the current Unixtime
plus an integer.
**Arguments:**
* *seconds* -- Seconds to add to the current time.
**Keyword arguments:**
* *offset* -- Offset in seconds. (Default 0)
"""
return str(int(offset + time.time() + seconds)).zfill(11)
def sdb_parse_time(date_string, offset=0):
"""Parse a date string, then return an 11 character, zero padded
string with the current Unixtime plus an integer.
**Arguments:**
* *date_string* -- Date string
**Keyword arguments:**
* *offset* -- Offset in seconds. (Default 0)
"""
parsed_time = time.mktime(dateutil.parser.parse(date_string).timetuple())
return str(int(offset + parsed_time)).zfill(11)
def sdb_latitude(latitude):
"""Return an 8 character, zero padded string version of the
latitude parameter.
**Arguments:**
* *latitude* -- Latitude.
"""
adjusted = (90 + float(latitude)) * 100000
return str(int(adjusted)).zfill(8)
def sdb_longitude(longitude):
"""Return an 8 character, zero padded string version of the
longitude parameter.
**Arguments:**
* *longitude* -- Longitude.
"""
adjusted = (180 + float(longitude)) * 100000
return str(int(adjusted)).zfill(8)
class AmazonSDB:
"""
Amazon Simple Database API.
"""
host = "sdb.amazonaws.com"
box_usage = 0.0
def __init__(self, aws_access_key_id, aws_secret_access_key, rq=None):
"""
**Arguments:**
* *aws_access_key_id* -- Amazon AWS access key ID
* *aws_secret_access_key* -- Amazon AWS secret access key
**Keyword arguments:**
* *rq* -- Optional RequestQueuer object.
"""
if rq is None:
self.rq = RequestQueuer()
else:
self.rq = rq
self.aws_access_key_id = aws_access_key_id
self.aws_secret_access_key = aws_secret_access_key
self.rq.setHostMaxRequestsPerSecond(self.host, 0)
self.rq.setHostMaxSimultaneousRequests(self.host, 0)
def copyDomain(self, source_domain, destination_domain):
"""
Copy all elements of a source domain to a destination domain.
**Arguments:**
* *source_domain* -- Source domain name
* *destination_domain* -- Destination domain name
"""
d = self.checkAndCreateDomain(destination_domain)
d.addCallback(self._copyDomainCallback, source_domain,
destination_domain)
return d
def _copyDomainCallback(self, data, source_domain, destination_domain):
return self._copyDomainCallback2(source_domain, destination_domain)
def _copyDomainCallback2(self, source_domain, destination_domain,
next_token=None, total_box_usage=0):
parameters = {}
parameters["Action"] = "Select"
parameters["SelectExpression"] = "SELECT * FROM `%s`" % source_domain
if next_token is not None:
parameters["NextToken"] = next_token
d = self._request(parameters)
d.addCallback(self._copyDomainCallback3,
source_domain=source_domain,
destination_domain=destination_domain,
total_box_usage=total_box_usage)
d.addErrback(self._genericErrback)
return d
def _copyDomainCallback3(self, data, source_domain, destination_domain,
total_box_usage=0):
xml = ET.fromstring(data["response"])
box_usage = float(xml.find(".//%sBoxUsage" % SDB_NAMESPACE).text)
self.box_usage += box_usage
total_box_usage += box_usage
next_token_element = xml.find(".//%sNextToken" % SDB_NAMESPACE)
if next_token_element is not None:
next_token = next_token_element.text
else:
next_token = None
items = xml.findall(".//%sItem" % SDB_NAMESPACE)
results = {}
for item in items:
key = item.find("./%sName" % SDB_NAMESPACE).text
attributes = item.findall("%sAttribute" % SDB_NAMESPACE)
attribute_dict = {}
for attribute in attributes:
attr_name = attribute.find("./%sName" % SDB_NAMESPACE).text
attr_value = attribute.find("./%sValue" % SDB_NAMESPACE).text
if attr_name in attribute_dict:
attribute_dict[attr_name].append(attr_value)
else:
attribute_dict[attr_name] = [attr_value]
results[key] = attribute_dict
deferreds = []
for key in results:
d = self.putAttributes(destination_domain, key, results[key])
d.addErrback(self._copyPutAttributesErrback, destination_domain, key, results[key])
deferreds.append(d)
d = DeferredList(deferreds, consumeErrors=True)
d.addCallback(self._copyDomainCallback4, source_domain,
destination_domain, next_token=next_token, total_box_usage=total_box_usage)
return d
def _copyDomainCallback4(self, data, source_domain, destination_domain,
next_token=None, total_box_usage=0):
for row in data:
if row[0] == False:
raise row[1]
if next_token is not None:
return self._copyDomainCallback2(
source_domain=source_domain,
destination_domain=destination_domain,
next_token=next_token,
total_box_usage=total_box_usage)
LOGGER.debug("""CopyDomain:\n%s -> %s\nBox usage: %s""" % (
source_domain,
destination_domain,
total_box_usage))
return True
def _copyPutAttributesErrback(self, error, destination_domain, key, attributes, count=0):
if count < 3:
d = self.putAttributes(destination_domain, key, attributes)
d.addErrback(self._copyPutAttributesErrback, destination_domain, key, attributes, count=count + 1)
return d
return error
def checkAndCreateDomain(self, domain):
"""
Check for a SimpleDB domain's existence. If it does not exist,
create it.
**Arguments:**
* *domain* -- Domain name
"""
d = self.domainMetadata(domain)
d.addErrback(self._checkAndCreateDomainErrback, domain)
return d
def _checkAndCreateDomainErrback(self, error, domain):
if hasattr(error, "value") and hasattr(error.value, "status"):
if int(error.value.status) == 400:
d = self.createDomain(domain)
d.addErrback(self._checkAndCreateDomainErrback2, domain)
return d
message = "Could not find or create domain '%s'." % domain
raise Exception(message)
def _checkAndCreateDomainErrback2(self, error, domain):
message = "Could not create domain '%s'" % domain
raise Exception(message)
def createDomain(self, domain):
"""
Create a SimpleDB domain.
**Arguments:**
* *domain* -- Domain name
"""
parameters = {
"Action":"CreateDomain",
"DomainName":domain
}
d = self._request(parameters)
d.addCallback(self._createDomainCallback, domain)
d.addErrback(self._genericErrback)
return d
def _createDomainCallback(self, data, domain):
xml = ET.fromstring(data["response"])
box_usage = float(xml.find(".//%sBoxUsage" % SDB_NAMESPACE).text)
self.box_usage += box_usage
LOGGER.debug("Created SimpleDB domain '%s'. Box usage: %s" % (domain,
box_usage))
return True
def deleteDomain(self, domain):
"""
Delete a SimpleDB domain.
**Arguments:**
* *domain* -- Domain name
"""
parameters = {}
parameters["Action"] = "DeleteDomain"
parameters["DomainName"] = domain
d = self._request(parameters)
d.addCallback(self._deleteDomainCallback, domain)
d.addErrback(self._genericErrback)
return d
def _deleteDomainCallback(self, data, domain):
xml = ET.fromstring(data["response"])
box_usage = float(xml.find(".//%sBoxUsage" % SDB_NAMESPACE).text)
self.box_usage += box_usage
LOGGER.debug("Deleted SimpleDB domain '%s'. Box usage: %s" % (domain,
box_usage))
return True
def listDomains(self):
"""
List SimpleDB domains associated with an account.
"""
return self._listDomains()
def _listDomains(self,
next_token=None,
previous_results=None,
total_box_usage=0):
parameters = {}
parameters["Action"] = "ListDomains"
if next_token is not None:
parameters["NextToken"] = next_token
d = self._request(parameters)
d.addCallback(self._listDomainsCallback,
previous_results=previous_results,
total_box_usage=total_box_usage)
d.addErrback(self._genericErrback)
return d
def _listDomainsCallback(self,
data,
previous_results=None,
total_box_usage=0):
xml = ET.fromstring(data["response"])
box_usage = float(xml.find(".//%sBoxUsage" % SDB_NAMESPACE).text)
self.box_usage += box_usage
total_box_usage += box_usage
xml_response = etree_to_dict(xml, namespace=SDB_NAMESPACE)
if "DomainName" in xml_response["ListDomainsResult"][0]:
results = xml_response["ListDomainsResult"][0]["DomainName"]
else:
results = []
if previous_results is not None:
results.extend(previous_results)
if "NextToken" in xml_response["ListDomainsResult"]:
next_token = xml_response["ListDomainsResult"][0]["NextToken"][0]
return self._listDomains(next_token=next_token,
previous_results=results,
total_box_usage=total_box_usage)
LOGGER.debug("Listed domains. Box usage: %s" % total_box_usage)
return results
def domainMetadata(self, domain):
"""
Return meta-information about a domain.
**Arguments:**
* *domain* -- Domain name
"""
parameters = {}
parameters["Action"] = "DomainMetadata"
parameters["DomainName"] = domain
d = self._request(parameters)
d.addCallback(self._domainMetadataCallback, domain)
d.addErrback(self._genericErrback)
return d
def _domainMetadataCallback(self, data, domain):
xml = ET.fromstring(data["response"])
box_usage = float(xml.find(".//%sBoxUsage" % SDB_NAMESPACE).text)
self.box_usage += box_usage
LOGGER.debug("Got SimpleDB domain '%s' metadata. Box usage: %s" % (
domain,
box_usage))
xml_response = etree_to_dict(xml, namespace=SDB_NAMESPACE)
return xml_response["DomainMetadataResult"][0]
def batchPutAttributes(self, domain, attributes_by_item_name,
replace_by_item_name=None):
"""
Batch put attributes into domain.
**Arguments:**
* *domain* -- Domain name
* *attributes_by_item_name* -- Dictionary of dictionaries. First
level keys are the item name, value is dictionary of key/value
pairs. Example: ``{"item_name":{"attribute_name":"value"}}``
**Keyword arguments:**
* *replace_by_item_name* -- Dictionary of lists. First level keys
are the item names, value is a list of of attributes that should
be overwritten. ``{"item_name":["attribute_name"]}`` (Default
empty dictionary)
"""
if replace_by_item_name is None:
replace_by_item_name = {}
if len(attributes_by_item_name) > 25:
raise Exception("Too many items in batchPutAttributes. Up to 25 items per call allowed.")
for item_name in replace_by_item_name:
if not isinstance(replace_by_item_name[item_name], list):
raise Exception("Replace argument '%s' must be a list." % item_name)
for item_name in attributes_by_item_name:
if not isinstance(attributes_by_item_name[item_name], dict):
raise Exception("Attributes argument '%s' must be a dictionary." % item_name)
parameters = {}
parameters["Action"] = "BatchPutAttributes"
parameters["DomainName"] = domain
i = 0
for item_name in attributes_by_item_name:
parameters["Item.%s.ItemName" % i] = item_name
attributes_list = []
for attribute in attributes_by_item_name[item_name].items():
# If the attribute is a list, split into multiple attributes.
if isinstance(attribute[1], list):
for value in attribute[1]:
attributes_list.append((attribute[0], value))
else:
attributes_list.append(attribute)
j = 0
for attribute in attributes_list:
parameters["Item.%s.Attribute.%s.Name" % (i,j)] = attribute[0]
parameters["Item.%s.Attribute.%s.Value" % (i,j)] = attribute[1]
if item_name in replace_by_item_name:
if attribute[0] in replace_by_item_name[item_name]:
parameters["Item.%s.Attribute.%s.Replace" % (i,j)] = "true"
j += 1
i += 1
d = self._request(parameters)
d.addCallback(
self._batchPutAttributesCallback,
domain,
attributes_by_item_name)
d.addErrback(self._genericErrback)
return d
def _batchPutAttributesCallback(self,
data,
domain,
attributes_by_item_name):
xml = ET.fromstring(data["response"])
box_usage = float(xml.find(".//%sBoxUsage" % SDB_NAMESPACE).text)
self.box_usage += box_usage
LOGGER.debug("""Batch put attributes %s in SimpleDB domain '%s'. Box usage: %s""" % (
attributes_by_item_name,
domain,
box_usage))
return True
def putAttributes(self, domain, item_name, attributes, replace=None):
"""
Put attributes into domain at item_name.
**Arguments:**
* *domain* -- Domain name
* *item_name* -- Item name
* *attributes* -- Dictionary of attributes
**Keyword arguments:**
* *replace* -- List of attributes that should be overwritten
(Default empty list)
"""
if replace is None:
replace = []
if not isinstance(replace, list):
raise Exception("Replace argument must be a list.")
if not isinstance(attributes, dict):
raise Exception("Attributes argument must be a dictionary.")
parameters = {}
parameters["Action"] = "PutAttributes"
parameters["DomainName"] = domain
parameters["ItemName"] = item_name
attributes_list = []
for attribute in attributes.items():
# If the attribute is a list, split into multiple attributes.
if isinstance(attribute[1], list):
for value in attribute[1]:
attributes_list.append((attribute[0], value))
else:
attributes_list.append(attribute)
i = 0
for attribute in attributes_list:
parameters["Attribute.%s.Name" % i] = attribute[0]
parameters["Attribute.%s.Value" % i] = attribute[1]
if attribute[0] in replace:
parameters["Attribute.%s.Replace" % i] = "true"
i += 1
d = self._request(parameters)
d.addCallback(self._putAttributesCallback, domain, item_name, attributes)
d.addErrback(self._genericErrback)
return d
def _putAttributesCallback(self, data, domain, item_name, attributes):
xml = ET.fromstring(data["response"])
box_usage = float(xml.find(".//%sBoxUsage" % SDB_NAMESPACE).text)
self.box_usage += box_usage
LOGGER.debug("""Put attributes %s on '%s' in SimpleDB domain '%s'. Box usage: %s""" % (
attributes,
item_name,
domain,
box_usage))
return True
def getAttributes(self, domain, item_name, attribute_name=None):
"""
Get one or all attributes from domain at item_name.
**Arguments:**
* *domain* -- Domain name
* *item_name* -- Item name
**Keyword arguments:**
* *attribute_name* -- Name of specific attribute to get (Default None)
"""
parameters = {}
parameters["Action"] = "GetAttributes"
parameters["DomainName"] = domain
parameters["ItemName"] = item_name
if attribute_name is not None:
parameters["AttributeName"] = attribute_name
d = self._request(parameters)
d.addCallback(self._getAttributesCallback, domain, item_name)
d.addErrback(self._genericErrback)
return d
def _getAttributesCallback(self, data, domain, item_name):
xml = ET.fromstring(data["response"])
box_usage = float(xml.find(".//%sBoxUsage" % SDB_NAMESPACE).text)
self.box_usage += box_usage
LOGGER.debug("""Got attributes from '%s' in SimpleDB domain '%s'. Box usage: %s""" % (
item_name,
domain,
box_usage))
xml_response = etree_to_dict(xml, namespace=SDB_NAMESPACE)
attributes = {}
if xml_response["GetAttributesResult"][0] is None:
raise Exception("Item does not exist.")
for attribute in xml_response["GetAttributesResult"][0]['Attribute']:
if attribute["Name"][0] not in attributes:
attributes[attribute["Name"][0]] = []
attributes[attribute["Name"][0]].extend(attribute["Value"])
return attributes
def delete(self, domain, item_name):
"""
Delete all attributes from domain at item_name.
**Arguments:**
* *domain* -- Domain name
* *item_name* -- Item name
"""
return self.deleteAttributes(domain, item_name)
def deleteAttributes(self, domain, item_name, attributes=None):
"""
Delete one or all attributes from domain at item_name.
**Arguments:**
* *domain* -- Domain name
* *item_name* -- Item name
**Keyword arguments:**
* *attributes* -- List of attribute names, or dictionary of
attribute name / value pairs. (Default empty dict)
"""
if attributes is None:
attributes = {}
if not isinstance(attributes, dict) and \
not isinstance(attributes, list):
message = "Attributes parameter must be a dictionary or a list."
raise Exception(message)
parameters = {}
parameters["Action"] = "DeleteAttributes"
parameters["DomainName"] = domain
parameters["ItemName"] = item_name
if isinstance(attributes, dict):
attr_count = 1
for key in attributes:
parameters["Attribute.%s.Name" % attr_count] = key
parameters["Attribute.%s.Value" % attr_count] = attributes[key]
attr_count += 1
if isinstance(attributes, list):
attr_count = 0
for key in attributes:
parameters["Attribute.%s.Name" % attr_count] = key
attr_count += 1
d = self._request(parameters)
d.addCallback(self._deleteAttributesCallback, domain, item_name)
d.addErrback(self._genericErrback)
return d
def _deleteAttributesCallback(self, data, domain, item_name):
xml = ET.fromstring(data["response"])
box_usage = float(xml.find(".//%sBoxUsage" % SDB_NAMESPACE).text)
self.box_usage += box_usage
LOGGER.debug("""Deleted attributes from '%s' in SimpleDB domain '%s'. Box usage: %s""" % (
item_name,
domain,
box_usage))
return True
def select(self, select_expression, max_results=0):
"""
Run a select query
**Arguments:**
* *select_expression* -- Select expression
"""
if "count(" in select_expression.lower():
return self._selectCount(select_expression)
return self._select(select_expression, max_results=max_results)
def _selectCount(self, select_expression, next_token=None,
previous_count=0,
total_box_usage=0):
parameters = {}
parameters["Action"] = "Select"
parameters["SelectExpression"] = select_expression
if next_token is not None:
parameters["NextToken"] = next_token
d = self._request(parameters)
d.addCallback(self._selectCountCallback,
select_expression=select_expression,
previous_count=previous_count,
total_box_usage=total_box_usage)
d.addErrback(self._genericErrback)
return d
def _selectCountCallback(self, data, select_expression=None,
previous_count=0,
total_box_usage=0):
xml = ET.fromstring(data["response"])
box_usage = float(xml.find(".//%sBoxUsage" % SDB_NAMESPACE).text)
self.box_usage += box_usage
total_box_usage += box_usage
next_token_element = xml.find(".//%sNextToken" % SDB_NAMESPACE)
if next_token_element is not None:
next_token = next_token_element.text
else:
next_token = None
count = previous_count + int(xml.find(".//%sValue" % SDB_NAMESPACE).text)
if next_token is not None:
return self._selectCount(select_expression, next_token=next_token,
previous_count=count,
total_box_usage=total_box_usage)
LOGGER.debug("""Select:\n'%s'\nBox usage: %s""" % (
select_expression,
total_box_usage))
return count
def _select(self, select_expression, next_token=None,
previous_results=None,
total_box_usage=0,
max_results=0):
parameters = {}
parameters["Action"] = "Select"
parameters["SelectExpression"] = select_expression
if next_token is not None:
parameters["NextToken"] = next_token
d = self._request(parameters)
d.addCallback(self._selectCallback,
select_expression=select_expression,
previous_results=previous_results,
total_box_usage=total_box_usage,
max_results=max_results)
d.addErrback(self._genericErrback)
return d
def _selectCallback(self, data, select_expression=None,
previous_results=None,
total_box_usage=0,
max_results=0):
if previous_results is not None:
results = previous_results
else:
results = {}
xml = ET.fromstring(data["response"])
box_usage = float(xml.find(".//%sBoxUsage" % SDB_NAMESPACE).text)
self.box_usage += box_usage
total_box_usage += box_usage
next_token_element = xml.find(".//%sNextToken" % SDB_NAMESPACE)
if next_token_element is not None:
next_token = next_token_element.text
else:
next_token = None
items = xml.findall(".//%sItem" % SDB_NAMESPACE)
for item in items:
key = item.find("./%sName" % SDB_NAMESPACE).text
attributes = item.findall("%sAttribute" % SDB_NAMESPACE)
attribute_dict = {}
for attribute in attributes:
attr_name = attribute.find("./%sName" % SDB_NAMESPACE).text
attr_value = attribute.find("./%sValue" % SDB_NAMESPACE).text
if attr_name in attribute_dict:
attribute_dict[attr_name].append(attr_value)
else:
attribute_dict[attr_name] = [attr_value]
results[key] = attribute_dict
if next_token is not None:
if max_results == 0 or len(results) < max_results:
return self._select(select_expression, next_token=next_token,
previous_results=results,
total_box_usage=total_box_usage,
max_results=max_results)
LOGGER.debug("""Select:\n'%s'\nBox usage: %s""" % (
select_expression,
total_box_usage))
return results
def _request(self, parameters):
"""
Add authentication parameters and make request to Amazon.
**Arguments:**
* *parameters* -- Key value pairs of parameters
"""
parameters = self._getAuthorization("GET", parameters)
query_string = urllib.urlencode(parameters)
url = "https://%s/?%s" % (self.host, query_string)
if len(url) > 4096:
del parameters['Signature']
parameters = self._getAuthorization("POST", parameters)
query_string = urllib.urlencode(parameters)
url = "https://%s" % (self.host)
d = self.rq.getPage(url, method="POST", postdata=query_string)
return d
else:
d = self.rq.getPage(url, method="GET")
return d
def _canonicalize(self, parameters):
"""
Canonicalize parameters for use with AWS Authorization.
**Arguments:**
* *parameters* -- Key value pairs of parameters
**Returns:**
* A safe-quoted string representation of the parameters.
"""
parameters = parameters.items()
parameters.sort(lambda x, y:cmp(x[0], y[0]))
return "&".join([safe_quote_tuple(x) for x in parameters])
def _getAuthorization(self, method, parameters):
"""
Create authentication parameters.
**Arguments:**
* *method* -- HTTP method of the request
* *parameters* -- Key value pairs of parameters
**Returns:**
* A dictionary of authorization parameters
"""
signature_parameters = {
"AWSAccessKeyId":self.aws_access_key_id,
"SignatureVersion":"2",
"SignatureMethod":"HmacSHA256",
'Timestamp':datetime.utcnow().isoformat()[0:19]+"+00:00",
"AWSAccessKeyId":self.aws_access_key_id,
"Version":"2009-04-15"
}
signature_parameters.update(parameters)
query_string = self._canonicalize(signature_parameters)
string_to_sign = "%(method)s\n%(host)s\n%(resource)s\n%(qs)s" % {
"method":method,
"host":self.host.lower(),
"resource":"/",
"qs":query_string,
}
args = [self.aws_secret_access_key, string_to_sign, hashlib.sha256]
signature = base64.encodestring(hmac.new(*args).digest()).strip()
signature_parameters.update({'Signature': signature})
return signature_parameters
def _genericErrback(self, error):
if hasattr(error, "value"):
if hasattr(error.value, "response"):
xml = ET.XML(error.value.response)
try:
LOGGER.debug(xml.find(".//Message").text)
except Exception, e:
pass
return error | AWSpider | /AWSpider-0.3.2.12.tar.gz/AWSpider-0.3.2.12/awspider/aws/sdb.py | sdb.py |
<img src="https://github.com/rob-dalton/rob-dalton.github.io/blob/master/images/awspot/awspot_logo.png" alt="awspot-logo" width="420">
<hr>
Command line utility to easily manage Amazon AWS spot resources.
*NOTE: The only available resource type is `ec2`. Spot fleet and EMR management are in progress.*
## Setup
1. [Install and configure awscli.](https://aws.amazon.com/cli/)
2. Run `pip install awspot`
## Usage
Please refer to [the project docs](https://rob-dalton.github.io/awspot) to learn more.
| AWSpot | /AWSpot-0.0.5.tar.gz/AWSpot-0.0.5/README.md | README.md |
import argparse
import base64
import json
import logging
import os
import time
import typing
from typing import List
from .base import Ec2BaseAction
class Launch(Ec2BaseAction):
""" Action class for launching ec2 spot instances. """
def _parse_args(self, parser, args):
parser.add_argument('-n', '--name', type=str, required=True,
help='name for instance')
parser.add_argument('-s', '--specification', type=str, required=True,
help='path to specification JSON file')
parser.add_argument('-p', '--price', type=str, required=True,
help='max bid price for instance')
parser.add_argument('-u', '--userdata', type=str, default=None,
help='optional path to userdata shell script')
return parser.parse_args(args)
def execute(self):
""" Launch ec2 spot instance, store information by name. """
# load launch specifications and base64 encoded userdata
with open(self.args.specification) as f:
launch_spec = json.loads(f.read())
if self.args.userdata is not None:
with open(self.args.userdata) as f:
userdata_str = f.read()
userdata_bytes = base64.b64encode(bytes(userdata_str, 'utf-8'))
userdata = userdata_bytes.decode('utf-8')
launch_spec['UserData'] = userdata
# request instance
request = self.client.request_spot_instances(
LaunchSpecification=launch_spec,
SpotPrice=self.args.price
)
request_id = request['SpotInstanceRequests'][0].get('SpotInstanceRequestId')
if request_id is None:
print('Request submission failed.')
return False
else:
print(f"\nSpot instance request submitted.\nSpotInstanceRequestId: {request_id}\n")
# check request state every 1.5 seconds, get instance id when fulfilled
holding = ['pending-evaluation','not-scheduled-yet',
'pending-fulfillment']
request_status = 'pending-evaluation'
while request_status in holding:
request_state = self.client.describe_spot_instance_requests(
SpotInstanceRequestIds=[request_id]
)['SpotInstanceRequests'][0]
request_status = request_state['Status'].get('Code')
time.sleep(1.5)
# handle successful fulfillment
if request_status == 'fulfilled':
instance_id = request_state.get('InstanceId')
print(f"Spot instance request fulfilled.\nInstanceId: {instance_id}\n")
# Add name tag to resource
self.client.create_tags(
Resources=[instance_id],
Tags=[{'Key': 'Name',
'Value': self.args.name}]
)
return True
# handle failed fulfillment
else:
self.client.cancel_spot_instance_requests(
SpotInstanceRequestIds=[request_id]
)
print(f"Spot instance request cancelled.\nReason: {request_status}\n")
return False | AWSpot | /AWSpot-0.0.5.tar.gz/AWSpot-0.0.5/awspot/actions/ec2/launch.py | launch.py |
import base64
import json
import logging
import os
import subprocess
import time
import typing
from typing import List
from .base import Ec2BaseAction
class Ssh(Ec2BaseAction):
""" Class for managing ec2 spot instances. """
def _parse_args(self, parser, args):
parser.add_argument('-n', '--name', type=str, required=True,
help='name for instance')
parser.add_argument('-i', '--identity_file', type=str, required=True,
help='path to .pem file')
parser.add_argument('-u', '--user', type=str, required=True,
help='user to login as')
parser.add_argument('--forward_agent', action='store_true',
help='forward SSH agent from local machine')
parser.add_argument('--create_ssh_profile', action='store_true',
help='create profile for awspot ssh config file')
parser.add_argument('--remove_ssh_profile', action='store_true',
help='remove profile for awspot ssh config file')
return parser.parse_args()
def create_ssh_profile(self, public_dns: str):
""" Add profile to ssh config file. """
config_path = os.path.expanduser("~") + "/.awspot/ssh_config"
profile = f"\nHost {self.args.name}\n HostName {public_dns}"
profile += f"\n User {self.args.user}\n IdentityFile {self.args.identity_file}\n"
if self.args.forward_agent:
profile += f"\n ForwardAgent Yes\n"
with open(config_path, 'a') as f:
f.write(profile)
print(f'\nProfile for {self.args.name} successfully created.\n')
def remove_ssh_profile(self):
config_path = os.path.expanduser("~") + "/.awspot/ssh_config"
# get lines
with open(config_path, 'r') as f:
lines = f.readlines()
# iterate over lines
with open(config_path, 'w') as f:
ignore = False
for line in lines:
# ignore lines from target Host til next host or EOF
if f"Host {self.args.name}" in line:
ignore = True
continue
if ignore:
if line[:5]=="Host ":
ignore = False
else:
continue
f.write(line)
def execute(self):
# TODO: Automatically detect if profile exists for instance by name
# Use if so. Removes requirement for -i and -u args
instance = self._find_instance_by_name(self.args.name)
public_dns = instance['PublicDnsName']
if self.args.create_ssh_profile:
self.create_ssh_profile(public_dns)
elif self.args.remove_ssh_profile:
self.remove_ssh_profile()
else:
subprocess.run(["ssh", "-i", self.args.identity_file,
f"{self.args.user}@{public_dns}"]) | AWSpot | /AWSpot-0.0.5.tar.gz/AWSpot-0.0.5/awspot/actions/ec2/ssh.py | ssh.py |
import copy
import pickle
from django.conf import settings
from django.core.exceptions import ValidationError
from django.core.mail import EmailMessage, EmailMultiAlternatives, get_connection
from premailer import transform
from .settings import EMAIL_BACKEND, EMAIL_BACKUP_LIST
def _serialize_email_message(email_message):
message_dict = {
'subject': email_message.subject,
'body': email_message.body,
'from_email': email_message.from_email,
'to': email_message.to,
'bcc': email_message.bcc,
'attachments': [],
'headers': email_message.extra_headers,
'cc': email_message.cc,
'reply_to': email_message.reply_to
}
if hasattr(email_message, 'alternatives'):
message_dict['alternatives'] = email_message.alternatives
if email_message.content_subtype != EmailMessage.content_subtype:
message_dict["content_subtype"] = email_message.content_subtype
if email_message.mixed_subtype != EmailMessage.mixed_subtype:
message_dict["mixed_subtype"] = email_message.mixed_subtype
attachments = email_message.attachments
for attachment in attachments:
attach = pickle.dumps(attachment)
message_dict['attachments'].append(attach)
return message_dict
def _deserialize_email_message(serialized_email_message):
message_kwargs = copy.deepcopy(serialized_email_message) # prevents missing items on retry
# remove items from message_kwargs until only valid EmailMessage/EmailMultiAlternatives
# kwargs are left and save the removed items to be used as EmailMessage/EmailMultiAlternatives
# attributes later
message_attributes = ['content_subtype', 'mixed_subtype']
attributes_to_copy = {}
for attr in message_attributes:
if attr in message_kwargs:
attributes_to_copy[attr] = message_kwargs.pop(attr)
# remove attachments from message_kwargs then reinsert after base64 decoding
attachments = message_kwargs.pop('attachments')
message_kwargs['attachments'] = []
for attachment in attachments:
attach = pickle.loads(attachment)
message_kwargs['attachments'].append(attach)
if 'alternatives' in message_kwargs:
message = EmailMultiAlternatives(
connection=get_connection(backend=EMAIL_BACKEND),
**message_kwargs,
)
else:
message = EmailMessage(
connection=get_connection(backend=EMAIL_BACKEND),
**message_kwargs,
)
# set attributes on message with items removed from message_kwargs earlier
for attr, val in attributes_to_copy.items():
setattr(message, attr, val)
return message
def send_email(
subject,
body,
mail_to,
reply_to=None,
bcc=None,
from_email=settings.DEFAULT_FROM_EMAIL,
attachments=None,
alternative=None
):
bcc_list: list = bcc + EMAIL_BACKUP_LIST if bcc else EMAIL_BACKUP_LIST
if alternative is None:
email_message = _email_message_simple(
subject=_email_subject_format(subject),
body=body,
mail_to=mail_to,
reply_to=reply_to,
bcc=bcc_list,
from_email=from_email,
)
else:
email_message = _email_message_alternatives(
subject=_email_subject_format(subject),
body=body,
mail_to=mail_to,
alternative=alternative,
reply_to=reply_to,
bcc=bcc_list,
from_email=from_email,
)
if attachments is not None:
for attachment in attachments:
email_message.attach(*attachment)
email_message.send()
def _email_message_simple(subject, body, mail_to, reply_to, bcc, from_email):
email_message = EmailMessage(
subject=subject,
body=transform(body),
from_email=from_email,
to=mail_to,
reply_to=reply_to,
bcc=bcc,
)
email_message.content_subtype = 'html'
return email_message
def _email_message_alternatives(subject, body, mail_to, alternative, reply_to, bcc, from_email):
email_message = EmailMultiAlternatives(
subject=subject,
body=body,
from_email=from_email,
to=mail_to,
reply_to=reply_to,
bcc=bcc,
)
if 'content' in alternative and 'mimetype' in alternative:
content = alternative['content']
mimetype = alternative['mimetype']
if 'html' in mimetype:
content = transform(content)
email_message.attach_alternative(content, mimetype)
else:
raise ValidationError('invalid alternative: Unable to add alternative to email')
email_message.mixed_subtype = 'related'
return email_message
def _email_subject_format(subject):
if hasattr(settings, 'EMAIL_SUBJECT'):
return settings.EMAIL_SUBJECT.format(subject)
return subject | AX3-Email | /ax3_email-1.0.13-py3-none-any.whl/ax3_email/utils.py | utils.py |
from django.apps import apps
from django.core.cache import cache
from django.utils import timezone
from django.utils.module_loading import import_string
from . import data, settings
from .api import AX3Client
from .cache_keys import CACHE_KEY_BANK_LIST, CACHE_KEY_IDENTIFICATION_TYPE_LIST
from .exceptions import MercadopagoError
from .models import MercadopagoAccessToken
def refresh_bank_list_cache():
mercado_pago = AX3Client()
response = mercado_pago.payment_methods.list()
for item in response.data:
if item['id'] != 'pse':
continue
bank_list = [(x['id'], x['description']) for x in item.get('financial_institutions', [])]
if bank_list:
cache.set(CACHE_KEY_BANK_LIST, bank_list, timeout=None)
def refresh_document_types_cache():
mercado_pago = AX3Client()
response = mercado_pago.identification_types.list()
bank_list = [(x['id'], x['name']) for x in response.data]
cache.set(CACHE_KEY_IDENTIFICATION_TYPE_LIST, bank_list, timeout=None)
def create_mercadopago_user(user_dict: dict, retries: int = 3) -> str:
"""user_dict must have following keys: first_name, last_name, email"""
mercadopago = AX3Client()
response = mercadopago.customers.search(email=user_dict['email'])
if response.total > 0:
return response.results[0]['id']
response = mercadopago.customers.create(**user_dict)
return response.data['id']
def update_payment(mercadopago_payment_id: int):
mercado_pago = AX3Client()
response = mercado_pago.payments.get(mercadopago_payment_id)
payment = apps.get_model(settings.PAYMENT_MODEL).objects.filter(
id=response.data['external_reference'].replace(settings.REFERENCE_PREFIX, '')
).first()
if payment and response.status_code == 200 and 'status' in response.data:
old_status = payment.payment_status
new_status = data.MERCADOPAGO_STATUS_MAP[response.data['status']]
payment.payment_response = response.data
payment.payment_status = new_status
payment.save(update_fields=['payment_response', 'payment_status'])
if old_status != new_status:
try:
if payment.payment_status == data.APPROVED_CHOICE:
usecase = import_string(settings.PAID_USECASE)(payment=payment)
usecase.execute()
elif payment.payment_status in [
data.CANCELLED_CHOICE,
data.REJECTED_CHOICE,
data.REFUNDED_CHOICE
]:
usecase = import_string(settings.REJECTED_USECASE)(payment=payment)
usecase.execute()
except ImportError:
pass
def create_seller_token(code):
mercado_pago = AX3Client()
response = mercado_pago.marketplace_tokens.create(code=code)
MercadopagoAccessToken.objects.create(
user_id=response.data['user_id'],
access_token=response.data['access_token'],
public_key=response.data['public_key'],
refresh_token=response.data['refresh_token'],
token_type=response.data['token_type'],
expires_in=timezone.localtime() + timezone.timedelta(seconds=response.data['expires_in']),
response_json=response.data,
)
def refresh_seller_token():
token = MercadopagoAccessToken.objects.first()
if not token:
raise MercadopagoError('Ensure create the first token using create_seller_token function')
mercado_pago = AX3Client()
response = mercado_pago.marketplace_tokens.refresh(refresh_token=token.refresh_token)
MercadopagoAccessToken.objects.create(
user_id=response.data['user_id'],
access_token=response.data['access_token'],
public_key=response.data['public_key'],
refresh_token=response.data['refresh_token'],
token_type=response.data['token_type'],
expires_in=timezone.localtime() + timezone.timedelta(seconds=response.data['expires_in']),
response_json=response.data,
) | AX3-Mercadopago | /AX3_Mercadopago-0.3.7-py3-none-any.whl/ax3_mercadopago/utils.py | utils.py |
from urllib.parse import urlencode
import requests
from mercadopago import api
from mercadopago.client import BaseClient
from . import exceptions, settings
from .models import MercadopagoAccessToken
class CardTokenAPI(api.CardTokenAPI):
_base_path = '/v1/card_tokens'
params = {'public_key': settings.PUBLIC_KEY}
def create(self, **data):
return self._client.post('/', params=self.params, json=data)
def get(self, token_id):
return self._client.get('/{id}', {'id': token_id}, params=self.params)
def update(self, token_id, public_key, **data):
return self._client.put('/{id}', {'id': token_id}, params=self.params, json=data)
class MarketplaceOAuthTokenAPI(api.API):
_base_path = '/oauth/token'
_redirect_uri = settings.MARKETPLACE_REDIRECT_URI
def create(self, code):
params = {
'client_secret': settings.ACCESS_TOKEN,
'grant_type': 'authorization_code',
'code': code,
'redirect_uri': self._redirect_uri,
}
return self._client.post('/', params=params)
def refresh(self, refresh_token):
params = {
'client_secret': settings.ACCESS_TOKEN,
'grant_type': 'refresh_token',
'refresh_token': refresh_token,
}
return self._client.post('/', params=params)
def get_auth_uri(self):
return 'https://auth.mercadopago.com.co/authorization?{}'.format(
urlencode({
'client_id': settings.MARKETPLACE_APP_ID,
'redirect_uri': self._redirect_uri,
'response_type': 'code',
'platform_id': 'mp',
})
)
class AX3Client(BaseClient):
base_url = 'https://api.mercadopago.com'
def __init__(self, access_token=None):
self._session = requests.Session()
if settings.PLATFORM_ID:
self._session.headers['x-platform-id'] = settings.PLATFORM_ID
if not access_token:
access_token = settings.ACCESS_TOKEN
self._access_token = access_token
def _handle_request_error(self, error):
if isinstance(error, requests.HTTPError):
status = error.response.status_code
if status == 400:
raise exceptions.BadRequestError(error)
if status == 401:
raise exceptions.AuthenticationError(error)
if status == 404:
raise exceptions.NotFoundError(error)
raise exceptions.MercadopagoError(error)
def request(self, method, path, path_args=None, **kwargs):
if path_args is None:
path_args = {}
if 'params' not in kwargs:
kwargs['params'] = {}
if MarketplaceOAuthTokenAPI._base_path not in path:
kwargs['params']['access_token'] = self.access_token
if settings.MARKETPLACE_SELLER and api.PaymentAPI._base_path in path:
seller_token = MercadopagoAccessToken.objects.first()
if not seller_token:
raise exceptions.AuthenticationError(
'Ensure create the first token using create_seller_token function, maybe you '
'need generate code to create token, use marketplace_tokens.get_auth_uri '
'to get auth url and paste it on the browser'
)
kwargs['params']['access_token'] = seller_token.access_token
url = self.base_url + path.format(**path_args)
return self._request(method, url, **kwargs)
@property
def access_token(self):
return self._access_token
@property
def marketplace_tokens(self):
return MarketplaceOAuthTokenAPI(self)
@property
def card_tokens(self):
return CardTokenAPI(self)
@property
def customers(self):
return api.CustomerAPI(self)
@property
def identification_types(self):
return api.IdentificationTypeAPI(self)
@property
def invoices(self):
return api.InvoiceAPI(self)
@property
def merchant_orders(self):
return api.MerchantOrderAPI(self)
@property
def payment_methods(self):
return api.PaymentMethodAPI(self)
@property
def payments(self):
return api.PaymentAPI(self)
@property
def advanced_payments(self):
return api.AdvancedPaymentAPI(self)
@property
def chargebacks(self):
return api.ChargebackAPI(self)
@property
def plans(self):
return api.PlanAPI(self)
@property
def preapprovals(self):
return api.PreapprovalAPI(self)
@property
def preferences(self):
return api.PreferenceAPI(self)
@property
def money_requests(self):
return api.MoneyRequestAPI(self)
@property
def shipping_options(self):
return api.ShippingOptionAPI(self)
@property
def pos(self):
return api.PosAPI(self)
@property
def account(self):
return api.AccountAPI(self)
@property
def users(self):
return api.UsersAPI(self)
@property
def sites(self):
return api.SiteAPI(self) | AX3-Mercadopago | /AX3_Mercadopago-0.3.7-py3-none-any.whl/ax3_mercadopago/api.py | api.py |
# AX3 OTP Auth
AX3 OTP Auth is a very simple Django library for generating and verifying one-time passwords using HTOP guidelines.
## Installation
Axes is easy to install from the PyPI package:
$ pip install django-axes
After installing the package, the project settings need to be configured.
**1.** Add ``ax3_OTP`` to your ``INSTALLED_APPS``::
INSTALLED_APPS = [
'django.contrib.admin',
'django.contrib.auth',
'django.contrib.contenttypes',
'django.contrib.sessions',
'django.contrib.messages',
'django.contrib.staticfiles',
# Axes app can be in any position in the INSTALLED_APPS list.
'ax3_OTP_Auth',
]
**2.** Add ``ax3_OTP_Auth.backends.OTPAuthBackend`` to the top of ``AUTHENTICATION_BACKENDS``:
AUTHENTICATION_BACKENDS = [
'ax3_OTP_Auth.backends.OTPAuthBackend',
# Django ModelBackend is the default authentication backend.
'django.contrib.auth.backends.ModelBackend',
]
**3.** Add the following to your urls.py:
urlpatterns = [
path('OTP-Auth/', include('ax3_OTP_Auth.urls', namespace='otp_auth')),
]
**4.** Create html button to your template:
<button class="js-otp-auth" type="button" otp-login="{% url 'otp_auth:start' %}" otp-redirect="{% url 'login' %}">
Login
</button>
**5.** Create Javascript for open OTP window:
$(() => {
$('.js-otp-auth').on('click', function () {
let redirect = $(this).attr('otp-redirect');
let OTPLoginUrl = $(this).attr('otp-login');
let width = 420;
let height = 470;
let top = (screen.height / 2) - (height / 2);
let left = (screen.width / 2) - (width / 2);
window.open(`${window.origin}${OTPLoginUrl}?redirect=${redirect}`, '_blank', `location=yes, scrollbars=yes, status=yes, width=${width}, height=${height}, top=${top}, left=${left}`);
});
});
## Configuration
If your need pass any param for whole pipeline you can use `OTP_AUTH_PARAMS`:
`OTP_AUTH_PARAMS = ['param']`
If your need change life time cache value you can use `OTP_AUTH_TTL`:
`OTP_AUTH_TTL = 60 * 60 * 5 # 5 minutes`
If your need change sms message:
`OTP_AUTH_MESSAGE = 'Utiliza {} como código de inicio de sesión.`
Configure countries allowed list:
COLOMBIA = 57
ARGENTINA = 54
BOLIVIA = 591
CHILE = 56
COSTA_RICA = 506
CUBA = 53
DOMINICAN_REPUBLIC = 809
ECUADOR = 593
GUATEMALA = 502
MEXICO = 52
PERU = 51
OTP_AUTH_COUNTRIES_CODES = [57, 54]
Change color, brand name and logo using this variables:
OTP_PRIMARY_COLOR = '#eb6806'
OTP_BACKGROUND_BTN = '#eb6806'
OTP_BACKGROUND_BTN_HOVER = '#000'
OTP_COLOR_TEXT_BTN = '#fff'
OTP_COLOR_TEXT_BTN_HOVER = '#fff'
OTP_BRAND_NAME = 'Axiacore'
OTP_BRAND_IMG = 'user-relative-path'
## NSN Configuration
AX3 OTP use NSN AWS service for sending messages, please create a group and AIM user with the following policy:
{
"Version": "2012-10-17",
"Statement": [
{
"Effect": "Allow",
"Action": [
"sns:Publish",
"sns:SetSMSAttributes",
"sns:GetSMSAttributes"
],
"Resource": "*"
}
]
}
Set AIM user credentials to your settings:
OTP_AUTH_AWS_ACCESS_KEY_ID = ''
OTP_AUTH_AWS_SECRET_ACCESS_KEY = ''
OTP_AUTH_AWS_DEFAULT_REGION = 'us-west-2'
## Authentication and Authorization
Authenticated user requires an OTP, this OTP was sent by AWS SNS service, once the code is valid, the system returns a token that must then be used to obtain the phone number which was requested. for this purpose you can use 'get_phone_number':
hotp = HOTP(session_key=request.session.session_key)
phone_number = htop.get_phone_number(code='123')
## Custom SMS Gateway
Set ``OTP_CUSTOM_SMS_GATEWAY`` to your settings with the path of your function and the function must be receive ``country_code``, ``phone_number`` and ``message``
OTP_CUSTOM_SMS_GATEWAY = 'app.utils.send_sms'
## Style SASS
For development porpuse is necessary to compile the SASS files before you commit any change.
Install node from this link:
https://nodejs.org/en/
Then install sass
$ sudo npm install -g sass
It ask you for a password, write de password of the user of the computer.
## Compile SASS
To change the styles of the web page you need to do edit the Sass's files and
then run this command on the root folder of the project to compile it to CSS:
$ sass --watch ax3_OTP_Auth/static/otp_auth/sass/styles.sass ax3_OTP_Auth/static/otp_auth/css/styles.css
| AX3-OTP-Auth | /AX3%20OTP%20Auth-1.0.5.tar.gz/AX3 OTP Auth-1.0.5/README.md | README.md |
from secrets import token_urlsafe
from django.core.cache import cache
from django.utils.module_loading import import_string
import boto3
import pyotp
from . import settings
class HOTP:
def __init__(self, unique_id: str, digits: int = 6):
self._unique_id = unique_id
self._digits = digits
self._ttl = settings.OTP_AUTH_TTL
def _create_secret(self, secret: str) -> str:
cache.set('{}.secret'.format(self._unique_id), secret, timeout=self._ttl)
return secret
def _create_counter(self) -> str:
try:
cache.incr('{}.counter'.format(self._unique_id))
except ValueError:
cache.set('{}.counter'.format(self._unique_id), 1, timeout=self._ttl)
return cache.get('{}.counter'.format(self._unique_id))
def _create_token(self, phone_number: int) -> str:
token = token_urlsafe()
cache.set(token, phone_number, timeout=self._ttl)
return token
def _get_secret(self):
return cache.get('{}.secret'.format(self._unique_id))
def _get_counter(self):
return cache.get('{}.counter'.format(self._unique_id))
def _send_sms(self, sms_code: int, country_code: str, phone_number: int):
message = settings.OTP_AUTH_MESSAGE.format(sms_code)
if settings.OTP_CUSTOM_SMS_GATEWAY:
gateway = import_string(settings.OTP_CUSTOM_SMS_GATEWAY)
gateway(country_code=country_code, phone_number=phone_number, message=message)
else:
sns = boto3.client(
'sns',
aws_access_key_id=settings.AWS_ACCESS_KEY_ID,
aws_secret_access_key=settings.AWS_SECRET_ACCESS_KEY,
region_name=settings.AWS_DEFAULT_REGION
)
sns.publish(
PhoneNumber=f'+{country_code}{phone_number}',
Message=message,
MessageAttributes={
'AWS.SNS.SMS.SMSType': {
'DataType': 'String',
'StringValue': 'Transactional'
}
}
)
def create(self, country_code: str, phone_number: int):
secret = self._create_secret(secret=pyotp.random_base32(length=32))
counter = self._create_counter()
hotp = pyotp.HOTP(secret, digits=self._digits)
self._send_sms(
sms_code=hotp.at(counter),
country_code=country_code,
phone_number=phone_number
)
def verify(self, sms_code: int, phone_number: int) -> str:
secret = self._get_secret()
count = self._get_counter()
if count and secret:
hotp = pyotp.HOTP(secret, digits=self._digits)
if hotp.verify(sms_code, count):
return self._create_token(phone_number=phone_number)
return None
def get_phone_number(self, token: str) -> int:
phone_number = cache.get(token)
cache.delete(token)
cache.delete_pattern('{}.*'.format(self._unique_id))
return phone_number | AX3-OTP-Auth | /AX3%20OTP%20Auth-1.0.5.tar.gz/AX3 OTP Auth-1.0.5/ax3_OTP_Auth/hotp.py | hotp.py |
from urllib import parse
from secrets import token_urlsafe
from django.shortcuts import redirect
from django.urls import reverse
from django.views.generic import FormView, TemplateView
from .forms import StartForm, VerifyForm
from .hotp import HOTP
from .settings import OTP_AUTH_PARAMS, LOGIN_URL
class StartView(FormView):
template_name = 'otp_auth/start.html'
form_class = StartForm
def render_to_response(self, context, **response_kwargs):
response = super().render_to_response(context, **response_kwargs)
if not self.request.COOKIES.get('otp_unique_id', None):
response.set_cookie('otp_unique_id', token_urlsafe())
return response
def form_valid(self, form):
unique_id = self.request.COOKIES.get('otp_unique_id', None)
if not unique_id:
return redirect('otp_auth:start')
hotp = HOTP(unique_id=unique_id)
hotp.create(
country_code=form.cleaned_data['country_code'],
phone_number=form.cleaned_data['phone_number']
)
params = {
'country_code': form.cleaned_data['country_code'],
'phone_number': form.cleaned_data['phone_number'],
'redirect': self.request.GET.get('redirect', reverse(LOGIN_URL)),
}
if OTP_AUTH_PARAMS:
for param in OTP_AUTH_PARAMS:
params[param] = self.request.GET.get(param)
return redirect('{}?{}'.format(
reverse('otp_auth:verify'),
parse.urlencode(params, safe='/'))
)
class VerifyView(FormView):
template_name = 'otp_auth/verify.html'
form_class = VerifyForm
def get_initial(self):
initial = super().get_initial()
initial.update({
'country_code': self.request.GET.get('country_code'),
'phone_number': self.request.GET.get('phone_number'),
})
return initial
def get_context_data(self, **kwargs):
context = super().get_context_data(**kwargs)
params = {'redirect': self.request.GET.get('redirect', reverse(LOGIN_URL))}
if OTP_AUTH_PARAMS:
for param in OTP_AUTH_PARAMS:
params[param] = self.request.GET.get(param)
context['sms_url'] = '{}?{}'.format(
reverse('otp_auth:start'),
parse.urlencode(params, safe='/')
)
return context
def form_valid(self, form):
cleaned_data = form.cleaned_data
hotp = HOTP(unique_id=self.request.COOKIES.get('otp_unique_id', ''))
token = hotp.verify(cleaned_data.get('code', ''), phone_number=cleaned_data['phone_number'])
if not token:
form.add_error('code', 'Código no es valido')
return self.form_invalid(form)
params = {'token': token, 'redirect': self.request.GET.get('redirect', reverse(LOGIN_URL))}
if OTP_AUTH_PARAMS:
for param in OTP_AUTH_PARAMS:
params[param] = self.request.GET.get(param)
return redirect('{}?{}'.format(reverse('otp_auth:done'), parse.urlencode(params, safe='/')))
class DoneView(TemplateView):
template_name = 'otp_auth/done.html'
def get_context_data(self, **kwargs):
context = super().get_context_data(**kwargs)
params = {'token': self.request.GET['token']}
if OTP_AUTH_PARAMS:
for param in OTP_AUTH_PARAMS:
params[param] = self.request.GET.get(param)
context['redirect'] = '{}?{}'.format(
self.request.GET.get('redirect', reverse(LOGIN_URL)),
parse.urlencode(params, safe='/')
)
return context | AX3-OTP-Auth | /AX3%20OTP%20Auth-1.0.5.tar.gz/AX3 OTP Auth-1.0.5/ax3_OTP_Auth/views.py | views.py |
from io import BytesIO
from django.core.exceptions import ValidationError
from django.db.models import ImageField
from django.utils.translation import gettext_lazy as _
from PIL import Image
from resizeimage import resizeimage
from resizeimage.imageexceptions import ImageSizeError
class OptimizedImageField(ImageField):
def __init__(self, *args, **kwargs):
"""
`optimized_image_output_size` must be a tuple and `optimized_image_resize_method` can be
'crop', 'cover', 'contain', 'width', 'height' or 'thumbnail'.
"""
self.optimized_image_output_size = kwargs.pop('optimized_image_output_size', None)
self.optimized_image_resize_method = kwargs.pop('optimized_image_resize_method', 'cover')
self.optimized_file_formats = kwargs.pop('optimized_file_formats', ['JPEG', 'PNG', 'GIF'])
self.optimized_image_quality = kwargs.pop('optimized_image_quality', 75)
super().__init__(*args, **kwargs)
def deconstruct(self):
name, path, args, kwargs = super().deconstruct()
# Remove the arguments from the kwargs dict.
kwargs.pop('optimized_image_output_size', None)
kwargs.pop('optimized_image_resize_method', None)
kwargs.pop('optimized_file_formats', None)
kwargs.pop('optimized_image_quality', None)
return name, path, args, kwargs
def save_form_data(self, instance, data):
updating_image = data and getattr(instance, self.name) != data
if updating_image:
if self.optimized_image_quality < 0 or self.optimized_image_quality > 100:
raise ValidationError({self.name: [_('The allowed quality is from 0 to 100')]})
try:
data = self.optimize_image(
image_data=data,
output_size=self.optimized_image_output_size,
resize_method=self.optimized_image_resize_method,
quality=self.optimized_image_quality,
)
except ImageSizeError:
raise ValidationError({self.name: [_('Image too small to be scaled')]})
except OSError:
raise ValidationError({self.name: [_('The image is invalid or corrupted')]})
super().save_form_data(instance, data)
def optimize_image(self, image_data, output_size, resize_method, quality):
"""Optimize an image that has not been saved to a file."""
img = Image.open(image_data)
if img.format not in self.optimized_file_formats:
raise ValidationError({self.name: [_('Image format unsupported')]})
# GIF files needs strict size validation
if img.format == 'GIF' and output_size and output_size != (img.width, img.height):
raise ValidationError({self.name: [_('GIF image size unsupported')]})
# Check if is a supported format for optimization
if img.format not in ['JPEG', 'PNG']:
return image_data
# If output_size content 0.
if output_size and output_size[0] == 0:
output_size = (img.width, output_size[1])
elif output_size and output_size[1] == 0:
output_size = (output_size[0], img.height)
# If output_size is set, resize the image with the selected resize_method.
if output_size and output_size != (img.width, img.height):
output_image = resizeimage.resize(resize_method, img, output_size)
else:
output_image = img
# If the file extension is JPEG, convert the output_image to RGB
if img.format == 'JPEG':
output_image = output_image.convert('RGB')
bytes_io = BytesIO()
output_image.save(bytes_io, format=img.format, optimize=True, quality=quality)
image_data.seek(0)
image_data.file.write(bytes_io.getvalue())
image_data.file.truncate()
return image_data | AX3-model-extras | /AX3_model_extras-2.0.0-py3-none-any.whl/ax3_model_extras/fields.py | fields.py |
====
AXUI
====
.. image:: https://readthedocs.org/projects/axui/badge/?version=latest
:target: https://readthedocs.org/projects/axui/?badge=latest
:alt: Documentation Status
.. image:: https://landscape.io/github/xcgspring/AXUI/master/landscape.svg?style=flat
:target: https://landscape.io/github/xcgspring/AXUI/master
:alt: Code Health
.. image:: https://img.shields.io/pypi/v/AXUI.svg
:target: https://pypi.python.org/pypi/AXUI/
:alt: Latest AXUI version
.. image:: https://img.shields.io/pypi/dm/AXUI.svg
:target: https://pypi.python.org/pypi/AXUI/
:alt: Number of PyPI downloads
AXUI is short for "Auto eXecute UI", is an UI automation framework, target to minimize the gap between tools and testers.
AXUI provides testers a powerful, unified, easy to use interface for common met platforms, like windows desktop, web, Android, IOS...
AXUI features
==============
1. AXUI provide a plug-in mechanism for automation guy to extend support for different UI
2. AXUI provide built-in drivers for:
- `windows native UIAutomation Client API <https://msdn.microsoft.com/en-us/library/windows/desktop/ee684021(v=vs.85).aspx>`_ for windows desktop UI
- `selenium project <https://github.com/SeleniumHQ/selenium>`_ for web UI
- `appium project <https://github.com/appium/appium>`_ for Android and IOS UI
3. AXUI provide an unified, easy to use python interface for use in test scripts
4. AXUI separate UI logic from test scripts, make test scripts more readable and easier to maintain
5. AXUI provide mechanism to handle auto met UI automation issues, like UI response time
An overview of AXUI structure
=============================
.. image:: http://axui.readthedocs.org/en/latest/_images/AXUI_structure.PNG
:target: http://axui.readthedocs.org/en/latest/_images/AXUI_structure.PNG
:alt: AXUI structure
code demonstrations
==============================
This code is in ``example/selenium``, it's a simple example to demonstrate how AXUI separate UI logic from test script.
Though this example give us a impression that AXUI add extra complexities but doesn't improve code readability.
Image that an app contains a lot of UI Elements, and the search identifiers split into multiple test scripts, then AXUI can gather all UI identifiers into one appmap, and make your scripts clean to read and maintain.
*Original*::
import selenium.webdriver as webdriver
browser = webdriver.Chrome(executable_path = r"chromedriver.exe")
browser.get(r"http://www.bing.com")
searchEdit = browser.find_element_by_id("sb_form_q")
goButton = browser.find_element_by_id("sb_form_go")
searchEdit.send_keys("AXUI")
goButton.click()
*AXUI AppMap*::
<AXUI:app_map xmlns:AXUI="AXUI" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" xsi:schemaLocation="AXUI AXUI_app_map.xsd">
<AXUI:funcs>
<AXUI:func name="go_to_bing" description="">
<AXUI:step type="GUI" cmd='browser.BrowserPattern.get "http://www.bing.com"'/>
</AXUI:func>
</AXUI:funcs>
<AXUI:UI_elements>
<AXUI:Root_element name="browser" >
<AXUI:UI_element name="searchEdit" identifier="id='sb_form_q'" start_func="go_to_bing"/>
<AXUI:UI_element name="goButton" identifier="id='sb_form_go'" start_func="go_to_bing"/>
</AXUI:Root_element>
</AXUI:UI_elements>
</AXUI:app_map>
*AXUI Code*::
import AXUI
config_file = "selenium.cfg"
app_map = "www.bing.com.xml"
AXUI.Config(config_file)
appmap = AXUI.AppMap(app_map)
appmap.browser.start(browser_name="CHROME", executable_path = r"chromedriver.exe")
appmap.browser.searchEdit.Keyboard.input("AXUI")
appmap.browser.goButton.Mouse.left_click()
More details, please check `AXUI documents <http://axui.readthedocs.org/en/latest/index.html>`_
To have quick experience about AXUI, please check `AXUI samples <http://axui.readthedocs.org/en/latest/Appendices.html#samples>`_
| AXUI | /AXUI-0.2.4.zip/AXUI-0.2.4/README.rst | README.rst |
import datetime
import logging
import logging.config
import os
from logging.config import dictConfig
BASE_LOG_PATH = "/data/var/log/{% project_name %}"
if not os.path.exists(BASE_LOG_PATH):
os.makedirs(BASE_LOG_PATH)
class LogLevelFilter(logging.Filter):
def __init__(self, pass_level):
self.pass_level = pass_level
def filter(self, record):
print(record.levelno)
if self.pass_level == record.levelno:
return True
return False
def config_log():
dictConfig({
'version': 1,
'disable_existing_loggers': False,
'loggers': {
"gunicorn": {
"level": "INFO",
"handlers": ["info", "error", "warning"],
"propagate": 1,
"qualname": "gunicorn"
},
"console": {
"level": "DEBUG",
"handlers": ["console"],
"propagate": 0,
"qualname": "console"
}
},
'filters': {
'info_filter': {
'()': "config.log_config.LogLevelFilter",
"pass_level": logging.INFO
},
'error_filter': {
'()': "config.log_config.LogLevelFilter",
"pass_level": logging.ERROR
},
'warning_filter': {
'()': "config.log_config.LogLevelFilter",
"pass_level": logging.WARNING
}
},
'handlers': {
"info": {
"class": 'concurrent_log.ConcurrentTimedRotatingFileHandler',
'backupCount': 14,
'when': 'midnight',
'delay': False,
'level': 'INFO',
"formatter": "generic",
"filters": ['info_filter'],
"filename": os.path.join(BASE_LOG_PATH, 'info.log')
},
"error": {
"class": 'concurrent_log.ConcurrentTimedRotatingFileHandler',
'backupCount': 14,
'when': 'midnight',
'delay': False,
'level': 'ERROR',
"formatter": "generic",
"filters": ['error_filter'],
"filename": os.path.join(BASE_LOG_PATH, 'error.log')
},
"warning": {
"class": 'concurrent_log.ConcurrentTimedRotatingFileHandler',
'backupCount': 14,
'when': 'midnight',
'delay': False,
'level': 'WARNING',
"formatter": "generic",
"filters": ['warning_filter'],
"filename": os.path.join(BASE_LOG_PATH, 'warning.log')
},
"console": {
"class": "logging.StreamHandler",
"level": "DEBUG",
"formatter": "console"
}
},
'formatters': {
"generic": {
"format": "[process=%(process)d] "
"[tx_id=] [level=%(levelname)s] "
"[timestamp=%(asctime)s] "
"[bu_id=JT_AILab] [app_id={% project_name %}] %(message)s",
"datefmt": "%Y-%m-%d %H:%M:%S",
"class": "logging.Formatter"
},
"console": {
"format": "'[%(levelname)s][%(asctime)s] %(message)s'",
"class": "logging.Formatter"
}
}
}) | AXX-AIAPI | /AXX_AIAPI-1.3.0-py3-none-any.whl/axx_aiapp/templates/project_template/project_name/config/log_config.py | log_config.py |
import time
import logging
import logging.config
import os
from logging.handlers import TimedRotatingFileHandler
import portalocker.constants as porta_lock_const
from portalocker.utils import Lock as PortaLock
class ConcurrentLogFileLock(PortaLock):
def __init__(self, filename, *args, **kwargs):
PortaLock.__init__(self, self.get_lock_filename(filename), *args, **kwargs)
def get_lock_filename(self, log_file_name):
"""
定义日志文件锁名称,类似于 `.__file.lock`,其中file与日志文件baseFilename一致
:return: 锁文件名称
"""
if log_file_name.endswith(".log"):
lock_file = log_file_name[:-4]
else:
lock_file = log_file_name
lock_file += ".lock"
lock_path, lock_name = os.path.split(lock_file)
# hide the file on Unix and generally from file completion
lock_name = ".__" + lock_name
return os.path.join(lock_path, lock_name)
class ConcurrentTimedRotatingFileHandler(TimedRotatingFileHandler):
# 上一次翻转时间
before_rollover_at = -1
def __init__(self, filename, *args, **kwargs):
TimedRotatingFileHandler.__init__(self, filename, *args, **kwargs)
file_path = os.path.split(filename)[0]
if not os.path.exists(file_path):
os.makedirs(file_path)
self.concurrent_lock = ConcurrentLogFileLock(filename, flags=porta_lock_const.LOCK_EX)
def emit(self, record) -> None:
"""
本方法继承Python标准库,修改的部分已在下方使用注释标记出
本次改动主要是对日志文件进行加锁,并且保证在多进程环境下日志内容切割正确
"""
# 此行为新增代码,尝试获取非重入进程锁,阻塞,直到成功获取
with self.concurrent_lock:
try:
if self.shouldRollover(record):
self.doRollover()
"""
如果日志内容创建时间小于上一次翻转时间,不能记录在baseFilename文件中,否则正常记录
处理日志写入哪个日志文件,修改开始
"""
if record.created <= ConcurrentTimedRotatingFileHandler.before_rollover_at:
currentTime = int(record.created)
# v 引用Python3.7标准库logging.TimedRotatingFileHandler.doRollover(110:124)中翻转目标文件名生成代码 v
dstNow = time.localtime(currentTime)[-1]
t = self.computeRollover(currentTime) - self.interval
if self.utc:
timeTuple = time.gmtime(t)
else:
timeTuple = time.localtime(t)
dstThen = timeTuple[-1]
if dstNow != dstThen:
if dstNow:
addend = 3600
else:
addend = -3600
timeTuple = time.localtime(t + addend)
dfn = self.rotation_filename(self.baseFilename + "." +
time.strftime(self.suffix, timeTuple))
# ^ 引用标准库TimedRotatingFileHandler中翻转目标文件名生成规则代码 ^
# 如果back_count值设置的过低,会出现日志文件实际数量大于设置值
# 因为当日志写入负载过高时,之前的某个时刻产生的日志会延迟到现在才进行写入,在写入时又找不到与时间对应的日志文件,
# 则会再创建一个与日志创建时刻对应的日志文件进行写入。
# 对应的日志文件是指达到翻转条件后创建的翻转文件,文件命名规则与标准库一致。
self._do_write_record(dfn, record)
else:
logging.FileHandler.emit(self, record)
"""
处理日志写入哪个日志文件,修改结束
"""
except Exception:
self.handleError(record)
def doRollover(self):
"""
本方法继承Python标准库,修改的部分已在下方使用注释标记出
"""
if self.stream:
self.stream.close()
self.stream = None
# get the time that this sequence started at and make it a TimeTuple
currentTime = int(time.time())
dstNow = time.localtime(currentTime)[-1]
t = self.rolloverAt - self.interval
if self.utc:
timeTuple = time.gmtime(t)
else:
timeTuple = time.localtime(t)
dstThen = timeTuple[-1]
if dstNow != dstThen:
if dstNow:
addend = 3600
else:
addend = -3600
timeTuple = time.localtime(t + addend)
dfn = self.rotation_filename(self.baseFilename + "." +
time.strftime(self.suffix, timeTuple))
"""
如果翻转文件已经生成,则说明其他进程已经处理过翻转
处理日志文件已经翻转当前进程中未写入文件的日志副本,修改开始
"""
# 直接修改静态变量,因为代码执行到此处已经获取到非重入进程锁,保证同一时间只有一个线程对变量进行修改
# 由于Python GIL,同一时间同一进程内只有一个线程运行,线程切换后缓存自动失效,即其他线程可以看见修改后的最新值
# 记录每一次触发翻转动作的时间,不管反转是否真的执行
ConcurrentTimedRotatingFileHandler.before_rollover_at = self.rolloverAt
if os.path.exists(dfn):
# 因为进程变量不会在内存同步,所以存在其他进程已经翻转过日志文件当时当前进程中还标识为未翻转
# 日志内容创建时间如果小于等于下一个处理翻转时刻,则将日志写入反转后的日志文件,而不是当前的baseFilename
# 当前磁盘上的baseFilename对于当前进程中的标识副本来说已经是翻转后要写入的文件
# 所以当文件存在时,本次不再进行翻转动作
pass
else:
self.rotate(self.baseFilename, dfn)
"""
处理日志文件已经翻转当前进程中未写入文件的日志副本,修改结束
"""
if self.backupCount > 0:
for s in self.getFilesToDelete():
os.remove(s)
if not self.delay:
self.stream = self._open()
newRolloverAt = self.computeRollover(currentTime)
while newRolloverAt <= currentTime:
newRolloverAt = newRolloverAt + self.interval
# If DST changes and midnight or weekly rollover, adjust for this.
if (self.when == 'MIDNIGHT' or self.when.startswith('W')) and not self.utc:
dstAtRollover = time.localtime(newRolloverAt)[-1]
if dstNow != dstAtRollover:
if not dstNow: # DST kicks in before next rollover, so we need to deduct an hour
addend = -3600
else: # DST bows out before next rollover, so we need to add an hour
addend = 3600
newRolloverAt += addend
# 此刻,当前进程中的标识副本已经同步为最新
self.rolloverAt = newRolloverAt
def _do_write_record(self, dfn, record):
"""
将日志内容写入指定文件
:param dfn: 指定日志文件
:param record: 日志内容
"""
with open(dfn, mode="a", encoding=self.encoding) as file:
file.write(self.format(record) + self.terminator)
# 添加当前类到 "logging.handlers" 模块中,使logging.config.fileConfig()进行配置时可以使用
import logging.handlers
logging.handlers.ConcurrentTimedRotatingFileHandler = ConcurrentTimedRotatingFileHandler | AXX-AIAPI | /AXX_AIAPI-1.3.0-py3-none-any.whl/axx_aiapp/templates/project_template/project_name/concurrent_log/__init__.py | __init__.py |
import os
import pymysql
from config import config
class MySQLTemplate:
"""
简单封装了几个方法,其余方法可根据实际情况自行封装
"""
@staticmethod
def open(cursor):
"""
创建-打开链接方法
:param cursor:
:return:
"""
env = os.environ.get('AI_ENV', 'default')
pool = config.get(env).POOL
conn = pool.connection()
cursor = conn.cursor(cursor=cursor)
return conn, cursor
@staticmethod
def close(conn, cursor):
"""
关闭链接方法
:param conn:
:param cursor:
:return:
"""
# conn.commit()
cursor.close()
conn.close()
@classmethod
def fetch_one(cls, sql, args=(), cursor=pymysql.cursors.DictCursor):
"""
查询单个数据
:param sql: SQL语句
:param args: 对应参数
:param cursor:
:return:成功返回数据,失败返回false
"""
conn, cursor = cls.open(cursor)
try:
cursor.execute(sql, args)
obj = cursor.fetchone()
return obj
except Exception as e:
return False
finally:
cls.close(conn, cursor)
@classmethod
def fetch_all(cls, sql, args=(), cursor=pymysql.cursors.DictCursor):
"""
查询所有复合条件的
:param sql: SQL语句
:param args: 参数
:param cursor:
:return: 成功返回数据,失败返回false
"""
conn, cursor = cls.open(cursor)
try:
cursor.execute(sql, args)
obj = cursor.fetchall()
return obj
except Exception as e:
return False
finally:
cls.close(conn, cursor)
@classmethod
def insert(cls, sql, args=(), cursor=pymysql.cursors.DictCursor):
"""
插入数据
:param sql:
:param args:
:param cursor:
:return: 成功返回ID 失败返回false
"""
conn, cursor = cls.open(cursor)
try:
conn.begin()
cursor.execute(sql, args)
_id = cursor.lastrowid
conn.commit()
return _id
except Exception as e:
conn.rollback()
return False
finally:
cls.close(conn, cursor)
@classmethod
def delete(cls, sql, args=(), cursor=pymysql.cursors.DictCursor):
"""
删除数据 , 建议采用逻辑删除,即数据库字段标志位,而非物理闪出去
:param sql:
:param args:
:param cursor:
:return: 成功返回true 失败返回false
"""
conn, cursor = cls.open(cursor)
try:
conn.begin()
cursor.execute(sql, args)
conn.commit()
return True
except Exception as e:
conn.rollback()
return False
finally:
cls.close(conn, cursor)
@classmethod
def update(cls, sql, args=(), cursor=pymysql.cursors.DictCursor):
"""
更新数据
:param sql:
:param args:
:param cursor:
:return: 成功返回true 失败返回false
"""
conn, cursor = cls.open(cursor)
try:
conn.begin()
cursor.execute(sql, args)
conn.commit()
return True
except Exception as e:
conn.rollback()
return False
finally:
cls.close(conn, cursor) | AXX-AIAPI | /AXX_AIAPI-1.3.0-py3-none-any.whl/axx_aiapp/templates/project_template/project_name/db/__init__.py | __init__.py |
import json
from flask import Blueprint, jsonify, current_app
from werkzeug.exceptions import HTTPException
geh = Blueprint('common', __name__)
class APIResponse:
"""
接口响应通用格式
含 code(响应状态码)msg(响应消息)data(响应数据三部分)
"""
__default_succeed = {
'code': 200,
'msg': 'Success',
'data': None
}
__default_failed = {
'code': 500,
'msg': 'Server Failed',
'data': None
}
@classmethod
def success(cls, data=None):
"""
返回成功响应
:param data:
:return:
"""
rsp = dict(cls.__default_succeed)
if data is not None:
rsp['data'] = data
return rsp
@classmethod
def failed(cls, msg=None, code=None):
"""
返回失败响应
:param msg:
:param code:
:return:
"""
rsp = dict(cls.__default_failed)
if code is not None:
rsp['code'] = code
if msg is not None:
rsp['msg'] = msg
return rsp
class AIException(Exception):
"""
自定义异常类,抛出此异常可被全局异常处理器捕捉并包装成通用响应体返回
"""
def __init__(self, message, code=None):
Exception.__init__(self)
self.message = message
self.code = code
def get_response(self):
return APIResponse.failed(self.message, self.code)
@geh.app_errorhandler(AIException)
def handle_invalid_usage(error):
"""
拦截所有AIException类型异常并进行包装返回
:param error: AIException类型异常
:return:
"""
response = None
if isinstance(error, AIException):
apiRes = error.get_response()
response = jsonify(apiRes)
current_app.logger.info("[code=%d][msg=%s][data=%s]" % (apiRes["code"], apiRes["msg"], apiRes["data"]))
current_app.logger.exception("[code=%d][msg=%s][data=%s]" % (apiRes["code"], apiRes["msg"], apiRes["data"]))
return response
# @geh.app_errorhandler(HTTPException)
# def handle_invalid_usage(error):
# """
# 拦截所有HTTPException异常并进行包装返回,包含框架HTTP协议自身产生的,以及代码中通过abort抛出的
# :param error:
# :return:
# """
# response = None
# if issubclass(type(error), HTTPException):
# apiRes = APIResponse.failed(error.name, error.code)
# response = jsonify(apiRes)
# current_app.logger.info("[code=%d][msg=%s][data=%s]" % (apiRes["code"], apiRes["msg"], apiRes["data"]))
# return response
@geh.app_errorhandler(Exception)
def handle_exception(error):
"""
拦截出上述异常外的所有Exception异常并进行包装返回
:param error:
:return:
"""
if issubclass(type(error), HTTPException):
return error
apiRes = APIResponse.failed("internal error", 500)
response = jsonify(apiRes)
current_app.logger.exception("[code=%d][msg=%s][data=%s]" % (apiRes["code"], apiRes["msg"], apiRes["data"]))
return response | AXX-AIAPI | /AXX_AIAPI-1.3.0-py3-none-any.whl/axx_aiapp/templates/project_template/project_name/common/__init__.py | __init__.py |
import datetime
import logging
import logging.config
import os
from logging.config import dictConfig
BASE_LOG_PATH = "/data/var/log/{% project_name %}"
if not os.path.exists(BASE_LOG_PATH):
os.makedirs(BASE_LOG_PATH)
class LogLevelFilter(logging.Filter):
def __init__(self, pass_level):
self.pass_level = pass_level
def filter(self, record):
print(record.levelno)
if self.pass_level == record.levelno:
return True
return False
def config_log():
dictConfig({
'version': 1,
'disable_existing_loggers': False,
'loggers': {
"gunicorn": {
"level": "INFO",
"handlers": ["info", "error", "warning"],
"propagate": 1,
"qualname": "gunicorn"
},
"console": {
"level": "DEBUG",
"handlers": ["console"],
"propagate": 0,
"qualname": "console"
}
},
'filters': {
'info_filter': {
'()': "config.log_config.LogLevelFilter",
"pass_level": logging.INFO
},
'error_filter': {
'()': "config.log_config.LogLevelFilter",
"pass_level": logging.ERROR
},
'warning_filter': {
'()': "config.log_config.LogLevelFilter",
"pass_level": logging.WARNING
}
},
'handlers': {
"info": {
"class": 'concurrent_log.ConcurrentTimedRotatingFileHandler',
'backupCount': 14,
'when': 'midnight',
'delay': False,
'level': 'INFO',
"formatter": "generic",
"filters": ['info_filter'],
"filename": os.path.join(BASE_LOG_PATH, 'info.log')
},
"error": {
"class": 'concurrent_log.ConcurrentTimedRotatingFileHandler',
'backupCount': 14,
'when': 'midnight',
'delay': False,
'level': 'ERROR',
"formatter": "generic",
"filters": ['error_filter'],
"filename": os.path.join(BASE_LOG_PATH, 'error.log')
},
"warning": {
"class": 'concurrent_log.ConcurrentTimedRotatingFileHandler',
'backupCount': 14,
'when': 'midnight',
'delay': False,
'level': 'WARNING',
"formatter": "generic",
"filters": ['warning_filter'],
"filename": os.path.join(BASE_LOG_PATH, 'warning.log')
},
"console": {
"class": "logging.StreamHandler",
"level": "DEBUG",
"formatter": "console"
}
},
'formatters': {
"generic": {
"format": "[process=%(process)d] "
"[tx_id=] [level=%(levelname)s] "
"[timestamp=%(asctime)s] "
"[bu_id=JT_AILab] [app_id={% project_name %}] %(message)s",
"datefmt": "%Y-%m-%d %H:%M:%S",
"class": "logging.Formatter"
},
"console": {
"format": "'[%(levelname)s][%(asctime)s] %(message)s'",
"class": "logging.Formatter"
}
}
}) | AXX-AIAPI | /AXX_AIAPI-1.3.0-py3-none-any.whl/AXX_AIAPI-1.3.0.data/data/log_config.py | log_config.py |
AYABInterface
=============
.. image:: https://travis-ci.org/fossasia/AYABInterface.svg
:target: https://travis-ci.org/fossasia/AYABInterface
:alt: Build Status
.. image:: https://ci.appveyor.com/api/projects/status/a6yhbt0rqvb212s7?svg=true
:target: https://ci.appveyor.com/project/AllYarnsAreBeautiful/AYABInterface
:alt: AppVeyor CI build status (Windows)
.. image:: https://codeclimate.com/github/fossasia/AYABInterface/badges/gpa.svg
:target: https://codeclimate.com/github/fossasia/AYABInterface
:alt: Code Climate
.. image:: https://codeclimate.com/github/fossasia/AYABInterface/badges/coverage.svg
:target: https://codeclimate.com/github/fossasia/AYABInterface/coverage
:alt: Test Coverage
.. image:: https://codeclimate.com/github/fossasia/AYABInterface/badges/issue_count.svg
:target: https://codeclimate.com/github/fossasia/AYABInterface
:alt: Issue Count
.. image:: https://badge.fury.io/py/AYABInterface.svg
:target: https://pypi.python.org/pypi/AYABInterface
:alt: Python Package Version on Pypi
.. image:: https://img.shields.io/pypi/dm/AYABInterface.svg
:target: https://pypi.python.org/pypi/AYABInterface#downloads
:alt: Downloads from Pypi
.. image:: https://readthedocs.org/projects/ayabinterface/badge/?version=latest
:target: http://ayabinterface.readthedocs.io/en/latest/?badge=latest
:alt: Documentation Status
.. image:: https://landscape.io/github/fossasia/AYABInterface/master/landscape.svg?style=flat
:target: https://landscape.io/github/fossasia/AYABInterface/master
:alt: Code Health
.. image:: https://badge.waffle.io/fossasia/AYABInterface.svg?label=ready&title=issues%20ready
:target: https://waffle.io/fossasia/AYABInterface
:alt: Issues ready to work on
A Python library with the interface to the AYAB shield.
For installation instructions and more, `see the documentation
<http://AYABInterface.readthedocs.io/>`__.
| AYABInterface | /AYABInterface-0.0.9-py3-none-any.whl/AYABInterface-0.0.9.dist-info/DESCRIPTION.rst | DESCRIPTION.rst |
import requests
# AZ Studio版权所有
# 该项目仅供娱乐 请勿用于违法事业 违者与AZ Studio无关
def getmusic(keyword):
#获得指定关键词的歌曲 关键词可为 歌手/歌曲名/专辑名
#keyword:关键词
#返回值说明:正常返回列表 "Error 0":没有结果 "Error 1":用户未输入
headers = {
"Accept": "application/json, text/plain, */*",
"Accept-Encoding": "gzip, deflate, br",
"Accept-Language": "zh-CN,zh;q=0.9",
"Connection": "keep-alive",
"Cookie": "Hm_lvt_cdb524f42f0ce19b169a8071123a4797=1690028471; _ga=GA1.2.291025627.1690028471; _gid=GA1.2.1906759595.1690028471; SL_G_WPT_TO=zh; SL_GWPT_Show_Hide_tmp=1; SL_wptGlobTipTmp=1; Hm_lpvt_cdb524f42f0ce19b169a8071123a4797=1690028564; _gat=1; Hm_Iuvt_cdb524f42f0ce19b169b8072123a4727=WjT5ktibAJwfEFyQFeJAEFcTxpwYHCeK; _ga_ETPBRPM9ML=GS1.2.1690028471.1.1.1690028577.47.0.0",
"Host": "www.kuwo.cn",
"Referer": "https://www.kuwo.cn/search/list?key=%E7%AC%BC",
"Sec-Fetch-Dest": "empty",
"Sec-Fetch-Mode": "cors",
"Sec-Fetch-Site": "same-origin",
"Secret": "09362e5991f0846bff719a26e1a0e0ac7bd0b3c9f8ab5fdf000c7b88ecfa727000a0ba6e",
"User-Agent": "Mozilla/5.0 (Windows NT 10.0; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/114.0.0.0 Safari/537.36",
"sec-ch-ua": "\"Not.A/Brand\";v=\"8\", \"Chromium\";v=\"114\", \"Google Chrome\";v=\"114\"",
"sec-ch-ua-mobile": "?0",
"sec-ch-ua-platform": "\"Windows\""
}
search_url = 'https://www.kuwo.cn/api/www/search/searchMusicBykeyWord?'
search_data = {
'key': keyword,
'pn': '1',
'rn': '20',
'httpsStatus': '1',
'reqId': '7d9e2c60-288a-11ee-9cdf-3f476f1ba25e',
"plat":"web_www"
}
if len(keyword)>0:
response_data = requests.get(search_url, params=search_data, headers=headers, timeout=20).json()
songs_data = response_data['data']['list']
if int(response_data['data']['total']) <= 0:
return "Error 0"
else:
data=[]
for i in range(len(songs_data)):
try:
data.append({"songname":songs_data[i]['name'],"singer":songs_data[i]['artist'],"album":songs_data[i]['album'],"pic":songs_data[i]['albumpic'],"rid":songs_data[i]['rid'],"reqId":response_data['reqId']})
except:
pass
return data
else:
return "Error 1"
def geturl(rid,reqId):
#获取音乐的音频地址
#rid:你要解析的歌曲在getmusic返回的rid reqId:你要解析的歌曲在getmusic返回的reqId
#返回值:正常返回音频链接 "Error 3":歌曲需要单曲付费或rid/reqId不正确
headers = {
"Accept": "application/json, text/plain, */*",
"Accept-Encoding": "gzip, deflate, br",
"Accept-Language": "zh-CN,zh;q=0.9",
"Connection": "keep-alive",
"Cookie": "Hm_lvt_cdb524f42f0ce19b169a8071123a4797=1690028471; _ga=GA1.2.291025627.1690028471; _gid=GA1.2.1906759595.1690028471; SL_G_WPT_TO=zh; SL_GWPT_Show_Hide_tmp=1; SL_wptGlobTipTmp=1; Hm_lpvt_cdb524f42f0ce19b169a8071123a4797=1690028564; _gat=1; Hm_Iuvt_cdb524f42f0ce19b169b8072123a4727=WjT5ktibAJwfEFyQFeJAEFcTxpwYHCeK; _ga_ETPBRPM9ML=GS1.2.1690028471.1.1.1690028577.47.0.0",
"Host": "www.kuwo.cn",
"Referer": "https://www.kuwo.cn/search/list?key=%E7%AC%BC",
"Sec-Fetch-Dest": "empty",
"Sec-Fetch-Mode": "cors",
"Sec-Fetch-Site": "same-origin",
"Secret": "09362e5991f0846bff719a26e1a0e0ac7bd0b3c9f8ab5fdf000c7b88ecfa727000a0ba6e",
"User-Agent": "Mozilla/5.0 (Windows NT 10.0; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/114.0.0.0 Safari/537.36",
"sec-ch-ua": "\"Not.A/Brand\";v=\"8\", \"Chromium\";v=\"114\", \"Google Chrome\";v=\"114\"",
"sec-ch-ua-mobile": "?0",
"sec-ch-ua-platform": "\"Windows\""
}
music_url = 'https://www.kuwo.cn/api/v1/www/music/playUrl?mid={}&type=music&httpsStatus=1&reqId={}&plat=web_www&from='.format(rid, reqId)
response_data = requests.get(music_url,headers=headers).json()
try:
song_url = response_data['data'].get('url')
except:
return "Error 3"
return song_url | AZMusicAPI | /AZMusicAPI-1.2.8-py3-none-any.whl/MusicAPI/AZMusicAPI.py | AZMusicAPI.py |
import requests
# AZ Studio版权所有
# 该项目仅供娱乐 请勿用于违法事业 违者与AZ Studio无关
def getmusic(keyword):
#获得指定关键词的歌曲 关键词可为 歌手/歌曲名/专辑名
#keyword:关键词
#返回值说明:正常返回列表 "Error 0":没有结果 "Error 1":用户未输入
headers = {
"Accept": "application/json, text/plain, */*",
"Accept-Encoding": "gzip, deflate, br",
"Accept-Language": "zh-CN,zh;q=0.9",
"Connection": "keep-alive",
"Cookie": "Hm_lvt_cdb524f42f0ce19b169a8071123a4797=1690028471; _ga=GA1.2.291025627.1690028471; _gid=GA1.2.1906759595.1690028471; SL_G_WPT_TO=zh; SL_GWPT_Show_Hide_tmp=1; SL_wptGlobTipTmp=1; Hm_lpvt_cdb524f42f0ce19b169a8071123a4797=1690028564; _gat=1; Hm_Iuvt_cdb524f42f0ce19b169b8072123a4727=WjT5ktibAJwfEFyQFeJAEFcTxpwYHCeK; _ga_ETPBRPM9ML=GS1.2.1690028471.1.1.1690028577.47.0.0",
"Host": "www.kuwo.cn",
"Referer": "https://www.kuwo.cn/search/list?key=%E7%AC%BC",
"Sec-Fetch-Dest": "empty",
"Sec-Fetch-Mode": "cors",
"Sec-Fetch-Site": "same-origin",
"Secret": "09362e5991f0846bff719a26e1a0e0ac7bd0b3c9f8ab5fdf000c7b88ecfa727000a0ba6e",
"User-Agent": "Mozilla/5.0 (Windows NT 10.0; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/114.0.0.0 Safari/537.36",
"sec-ch-ua": "\"Not.A/Brand\";v=\"8\", \"Chromium\";v=\"114\", \"Google Chrome\";v=\"114\"",
"sec-ch-ua-mobile": "?0",
"sec-ch-ua-platform": "\"Windows\""
}
search_url = 'https://www.kuwo.cn/api/www/search/searchMusicBykeyWord?'
search_data = {
'key': keyword,
'pn': '1',
'rn': '20',
'httpsStatus': '1',
'reqId': '7d9e2c60-288a-11ee-9cdf-3f476f1ba25e',
"plat":"web_www"
}
if len(keyword)>0:
response_data = requests.get(search_url, params=search_data, headers=headers, timeout=20).json()
songs_data = response_data['data']['list']
if int(response_data['data']['total']) <= 0:
return "Error 0"
else:
data=[]
for i in range(len(songs_data)):
try:
data.append({"songname":songs_data[i]['name'],"singer":songs_data[i]['artist'],"album":songs_data[i]['album'],"pic":songs_data[i]['albumpic'],"rid":songs_data[i]['rid'],"reqId":response_data['reqId']})
except:
pass
return data
else:
return "Error 1"
def geturl(rid,reqId):
#获取音乐的音频地址
#rid:你要解析的歌曲在getmusic返回的rid reqId:你要解析的歌曲在getmusic返回的reqId
#返回值:正常返回音频链接 "Error 3":歌曲需要单曲付费或rid/reqId不正确
headers = {
"Accept": "application/json, text/plain, */*",
"Accept-Encoding": "gzip, deflate, br",
"Accept-Language": "zh-CN,zh;q=0.9",
"Connection": "keep-alive",
"Cookie": "Hm_lvt_cdb524f42f0ce19b169a8071123a4797=1690028471; _ga=GA1.2.291025627.1690028471; _gid=GA1.2.1906759595.1690028471; SL_G_WPT_TO=zh; SL_GWPT_Show_Hide_tmp=1; SL_wptGlobTipTmp=1; Hm_lpvt_cdb524f42f0ce19b169a8071123a4797=1690028564; _gat=1; Hm_Iuvt_cdb524f42f0ce19b169b8072123a4727=WjT5ktibAJwfEFyQFeJAEFcTxpwYHCeK; _ga_ETPBRPM9ML=GS1.2.1690028471.1.1.1690028577.47.0.0",
"Host": "www.kuwo.cn",
"Referer": "https://www.kuwo.cn/search/list?key=%E7%AC%BC",
"Sec-Fetch-Dest": "empty",
"Sec-Fetch-Mode": "cors",
"Sec-Fetch-Site": "same-origin",
"Secret": "09362e5991f0846bff719a26e1a0e0ac7bd0b3c9f8ab5fdf000c7b88ecfa727000a0ba6e",
"User-Agent": "Mozilla/5.0 (Windows NT 10.0; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/114.0.0.0 Safari/537.36",
"sec-ch-ua": "\"Not.A/Brand\";v=\"8\", \"Chromium\";v=\"114\", \"Google Chrome\";v=\"114\"",
"sec-ch-ua-mobile": "?0",
"sec-ch-ua-platform": "\"Windows\""
}
music_url = 'https://www.kuwo.cn/api/v1/www/music/playUrl?mid={}&type=music&httpsStatus=1&reqId={}&plat=web_www&from='.format(rid, reqId)
response_data = requests.get(music_url,headers=headers).json()
try:
song_url = response_data['data'].get('url')
except:
return "Error 3"
return song_url | AZMusicAPI | /AZMusicAPI-1.2.8-py3-none-any.whl/MusicAPI/MusicAPI.py | MusicAPI.py |
import requests
# AZ Studio版权所有
# 该项目仅供娱乐 请勿用于违法事业 违者与AZ Studio无关
def getmusic(keyword):
#获得指定关键词的歌曲 关键词可为 歌手/歌曲名/专辑名
#keyword:关键词
#返回值说明:正常返回列表 "Error 0":没有结果 "Error 1":用户未输入
headers = {
"Accept": "application/json, text/plain, */*",
"Accept-Encoding": "gzip, deflate, br",
"Accept-Language": "zh-CN,zh;q=0.9",
"Connection": "keep-alive",
"Cookie": "Hm_lvt_cdb524f42f0ce19b169a8071123a4797=1690028471; _ga=GA1.2.291025627.1690028471; _gid=GA1.2.1906759595.1690028471; SL_G_WPT_TO=zh; SL_GWPT_Show_Hide_tmp=1; SL_wptGlobTipTmp=1; Hm_lpvt_cdb524f42f0ce19b169a8071123a4797=1690028564; _gat=1; Hm_Iuvt_cdb524f42f0ce19b169b8072123a4727=WjT5ktibAJwfEFyQFeJAEFcTxpwYHCeK; _ga_ETPBRPM9ML=GS1.2.1690028471.1.1.1690028577.47.0.0",
"Host": "www.kuwo.cn",
"Referer": "https://www.kuwo.cn/search/list?key=%E7%AC%BC",
"Sec-Fetch-Dest": "empty",
"Sec-Fetch-Mode": "cors",
"Sec-Fetch-Site": "same-origin",
"Secret": "09362e5991f0846bff719a26e1a0e0ac7bd0b3c9f8ab5fdf000c7b88ecfa727000a0ba6e",
"User-Agent": "Mozilla/5.0 (Windows NT 10.0; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/114.0.0.0 Safari/537.36",
"sec-ch-ua": "\"Not.A/Brand\";v=\"8\", \"Chromium\";v=\"114\", \"Google Chrome\";v=\"114\"",
"sec-ch-ua-mobile": "?0",
"sec-ch-ua-platform": "\"Windows\""
}
search_url = 'https://www.kuwo.cn/api/www/search/searchMusicBykeyWord?'
search_data = {
'key': keyword,
'pn': '1',
'rn': '20',
'httpsStatus': '1',
'reqId': '7d9e2c60-288a-11ee-9cdf-3f476f1ba25e',
"plat":"web_www"
}
if len(keyword)>0:
response_data = requests.get(search_url, params=search_data, headers=headers, timeout=20).json()
songs_data = response_data['data']['list']
if int(response_data['data']['total']) <= 0:
return "Error 0"
else:
data=[]
for i in range(len(songs_data)):
try:
data.append({"songname":songs_data[i]['name'],"singer":songs_data[i]['artist'],"album":songs_data[i]['album'],"pic":songs_data[i]['albumpic'],"rid":songs_data[i]['rid'],"reqId":response_data['reqId']})
except:
pass
return data
else:
return "Error 1"
def geturl(rid,reqId):
#获取音乐的音频地址
#rid:你要解析的歌曲在getmusic返回的rid reqId:你要解析的歌曲在getmusic返回的reqId
#返回值:正常返回音频链接 "Error 3":歌曲需要单曲付费或rid/reqId不正确
headers = {
"Accept": "application/json, text/plain, */*",
"Accept-Encoding": "gzip, deflate, br",
"Accept-Language": "zh-CN,zh;q=0.9",
"Connection": "keep-alive",
"Cookie": "Hm_lvt_cdb524f42f0ce19b169a8071123a4797=1690028471; _ga=GA1.2.291025627.1690028471; _gid=GA1.2.1906759595.1690028471; SL_G_WPT_TO=zh; SL_GWPT_Show_Hide_tmp=1; SL_wptGlobTipTmp=1; Hm_lpvt_cdb524f42f0ce19b169a8071123a4797=1690028564; _gat=1; Hm_Iuvt_cdb524f42f0ce19b169b8072123a4727=WjT5ktibAJwfEFyQFeJAEFcTxpwYHCeK; _ga_ETPBRPM9ML=GS1.2.1690028471.1.1.1690028577.47.0.0",
"Host": "www.kuwo.cn",
"Referer": "https://www.kuwo.cn/search/list?key=%E7%AC%BC",
"Sec-Fetch-Dest": "empty",
"Sec-Fetch-Mode": "cors",
"Sec-Fetch-Site": "same-origin",
"Secret": "09362e5991f0846bff719a26e1a0e0ac7bd0b3c9f8ab5fdf000c7b88ecfa727000a0ba6e",
"User-Agent": "Mozilla/5.0 (Windows NT 10.0; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/114.0.0.0 Safari/537.36",
"sec-ch-ua": "\"Not.A/Brand\";v=\"8\", \"Chromium\";v=\"114\", \"Google Chrome\";v=\"114\"",
"sec-ch-ua-mobile": "?0",
"sec-ch-ua-platform": "\"Windows\""
}
music_url = 'https://www.kuwo.cn/api/v1/www/music/playUrl?mid={}&type=music&httpsStatus=1&reqId={}&plat=web_www&from='.format(rid, reqId)
response_data = requests.get(music_url,headers=headers).json()
try:
song_url = response_data['data'].get('url')
except:
return "Error 3"
return song_url | AZMusicAPI | /AZMusicAPI-1.2.8-py3-none-any.whl/MusicAPI/__init__.py | __init__.py |
class AadhaarExtractor:
# assume inputs are file name and not cv2 images #give underscore for private member functions
# and pil images
def __init__(self, data1=None): # and pil images
def invalidate():
if type(data1) == str:
import re
if re.match(r'.*\.(jpg|png|jpeg)',data1,re.M|re.I)!= None:
return False
return True
### check if the input string seems like a image file ###
if type(data1)!=str :
raise ValueError("Invalid input: Give file paths only")
elif invalidate():
raise ValueError("Only image files possible")
self.data1 = data1
# FIXED: check for invalid inputs
self.mainlist = None
self.process()
def load(self, data1): # can load and jsonify be merged?
self.data1 = data1
self.extract_details()
def extract(self):
import json
# try:
# f = open(jsonpath, 'r+')
# except FileNotFoundError: # for the first time
# f = open(jsonpath, 'w+')
# try:
# maindict = json.load(f)
# except ValueError: # for the first time
# print("value error")
# maindict = {}
# if self.maindict['aadhaar_no'] in maindict.keys():
# choice = input(
# "This aadhar number is already present in the database:\n Do you want to update the the data for this aadhaar number (y\n)?")
# if choice.lower() == 'n':
# f.close()
# return self.maindict
# maindict[self.maindict['aadhaar_no']] = self.maindict
# f.seek(0)
# json.dump(maindict, f, indent=2)
# f.close()
return self.mainlist
def file_type(self, file):
# import re
# if re.match(".*\.pdf$", filePath, re.M | re.I):
if file.content_type == r'application/pdf':
return 'pdf'
# if re.match(".*\.(png|jpg|jpeg|bmp|svg)$", filePath, re.M | re.I): # changed and made more flexible
if file.content_type == r'image/jpeg':
return 'img'
return 0
def process(self):
# if self.file_type(data) == 'pdf':
# dict = self.extract_from_pdf(data)
# elif self.file_type(data) == 'img':
self.mainlist = self.extract_from_images()
# else:
# pass
# def extract_details(self):
# if self.data1 != None:
# mainlist = self.give_details_back()
def extract_from_images(self):
import numpy as np
from ultralytics import YOLO
import cv2
import pytesseract
### specify that pytesseract needs to be explicitly installed ###
import logging
import os
logging.basicConfig(level=logging.NOTSET)
# Get the absolute path of the current file
current_dir = os.path.dirname(os.path.abspath(__file__))
# Construct the absolute path to the resource file
MODEL_PATH = os.path.join(current_dir, 'best.pt')
# MODEL_PATH = r"best.pt"
def filter_tuples(lst):
##### filters the list so that only one instance of each class is present ########
d = {}
for tup in lst:
key = tup[1]
value = tup[2]
if key not in d:
d[key] = (tup, value)
else:
if value > d[key][1]:
d[key] = (tup, value)
return [tup for key, (tup, value) in d.items()]
def clean_words(name):
name = name.replace("8", "B")
name = name.replace("0", "D")
name = name.replace("6", "G")
name = name.replace("1", "I")
name = name.replace('5', 'S')
return name
def clean_dob(dob):
dob = dob.strip()
dob = dob.replace('l', '/')
dob = dob.replace('L', '/')
dob = dob.replace('I', '/')
dob = dob.replace('i', '/')
dob = dob.replace('|', '/')
dob = dob.replace('\"', '/1')
# dob = dob.replace(":","")
dob = dob.replace(" ", "")
return dob
def validate_aadhaar_numbers(candidate):
if candidate == None :
return True
candidate = candidate.replace(' ', '')
# The multiplication table
d = [
[0, 1, 2, 3, 4, 5, 6, 7, 8, 9],
[1, 2, 3, 4, 0, 6, 7, 8, 9, 5],
[2, 3, 4, 0, 1, 7, 8, 9, 5, 6],
[3, 4, 0, 1, 2, 8, 9, 5, 6, 7],
[4, 0, 1, 2, 3, 9, 5, 6, 7, 8],
[5, 9, 8, 7, 6, 0, 4, 3, 2, 1],
[6, 5, 9, 8, 7, 1, 0, 4, 3, 2],
[7, 6, 5, 9, 8, 2, 1, 0, 4, 3],
[8, 7, 6, 5, 9, 3, 2, 1, 0, 4],
[9, 8, 7, 6, 5, 4, 3, 2, 1, 0]
]
# permutation table p
p = [
[0, 1, 2, 3, 4, 5, 6, 7, 8, 9],
[1, 5, 7, 6, 2, 8, 3, 0, 9, 4],
[5, 8, 0, 3, 7, 9, 6, 1, 4, 2],
[8, 9, 1, 6, 0, 4, 3, 5, 2, 7],
[9, 4, 5, 3, 1, 2, 6, 8, 7, 0],
[4, 2, 8, 6, 5, 7, 3, 9, 0, 1],
[2, 7, 9, 3, 8, 0, 6, 4, 1, 5],
[7, 0, 4, 6, 9, 1, 3, 2, 5, 8]
]
# inverse table inv
inv = [0, 4, 3, 2, 1, 5, 6, 7, 8, 9]
# print("sonddffsddsdd")
# print(len(candidate))
lastDigit = candidate[-1]
c = 0
array = [int(i) for i in candidate if i != ' ']
array.pop()
array.reverse()
for i in range(len(array)):
c = d[c][p[((i + 1) % 8)][array[i]]] # use verheoffs algorithm to validate
if inv[c] == int(lastDigit):
return True
return False
# file.seek(0)
# img_bytes = file.read()
# img_array = np.frombuffer(img_bytes, dtype=np.uint8)
img = cv2.imread(self.data1)
og_height,og_width,_ = img.shape
height_scaling = og_height/640
width_scaling = og_height/640
# TODO for all the self.data types it should support
# img = cv2.imdecode(img_array, cv2.IMREAD_COLOR)
img = cv2.resize(img, (640, 640))
# cv2.imshow("sone", img)
# cv2.waitKey(0)
# cv2.destroyAllWindows()
model = YOLO(MODEL_PATH)
results = model(img)
# rois = []
roidata = []
##### this will create a roidata which will be list of tuples with roi image, cls and confidence #####
for result in results:
# cls = result.boxes.cls
# boxes = result.boxes.xyxy
boxes = result.boxes
for box in boxes:
# x1,y1,x2,y2 = box
l = box.boxes.flatten().tolist()
roicoords = list(map(int, l[0:4]))
# roicoords: x1,y1,x2,y2
confidence, cls = l[4:]
cls = int(cls)
# l = list(box)
# x1,y1,x2,y2 = list(map(int,l))
# print(x1, x2, y1, y2)
# img = cv2.rectangle(img, (x1, y1), (x2, y2), (0, 255, 0), 2)
# cv2.putText(img, str(cls), (x1, y1), cv2.FONT_HERSHEY_SIMPLEX, 2, 255)
# roi = img[y1:y2, x1:x2]
# rois.append(roi)
templist = [roicoords, cls,confidence]
roidata.append(templist)
# cv2.imshow("s", img)
# cv2.waitKey(0)
# cv2.destroyAllWindows()
# print(roidata)
index = {0: "aadhaar_no",
1: "dob",
2: "gender",
3: "name",
4: "address"}
# logging.info('BEFORE FILTERING :')
# logging.info(len(roidata))
# TODO there is no flag to filter roidata , introduce later
# roidata = filter_tuples(roidata)
# maindict = {}
# maindict['aadhaar_no'] = maindict['dob'] = maindict['gender'] = maindict['address'] = maindict['name'] = maindict['phonenumber'] = maindict['vid'] = maindict['enrollment_number'] = None
# logging.info('AFTER FILTERING :')
# logging.info(len(roidata))
img = cv2.cvtColor(img,cv2.COLOR_BGR2GRAY)
for data in roidata:
cls = data[1]
x1,y1,x2,y2 = data[0]
# data = cv2.cvtColor(data, cv2.COLOR_BGR2GRAY)
cropped = img[y1:y2, x1:x2]
info = pytesseract.image_to_string(cropped).strip()
# logging.info(str(cls)+'-'+info)
data[0][0] = x1*width_scaling
data[0][1] = y1*height_scaling
data[0][2] = x2*width_scaling
data[0][3] = y2*height_scaling
#### the scaling is reconverted to the original size dimensions ####
data[1] = index[cls]
# change from indexes to name if the class
if info != None :
if cls == 3:
info = clean_words(info)
elif cls == 1:
info = clean_dob(info)
elif cls == 0:
info = info.replace(' ','')
if len(info) == 12:
try:
if not validate_aadhaar_numbers(info):
info = "INVALID AADHAAR NUMBER"
except ValueError:
info = None
else:
info = None
data.append(info)
# extracted text cleaned up :FIXED
# TODO extract these fields too
# maindict['phonenumber'] = None
# maindict['vid'] = None
# maindict['enrollment_no'] = None
# FIXED: the coords are for 640:640 , fix it for original coords
return roidata
def extract_from_pdf(self, file):
def extract_pymupdf(file):
# Usinf pymupdf
import fitz # this is pymupdf
# extract text page by page
with fitz.open(stream=file.stream.read(), filetype='pdf') as doc:
pymupdf_text = ""
if (doc.is_encrypted):
passw = input("Enter the password")
# TODO display this message where?
doc.authenticate(password=passw)
for page in doc:
pymupdf_text += page.get_text("Text")
return pymupdf_text
def get_details(txt):
import re
pattern = re.compile(
r'Enrolment No\.: (?P<enrolment_no>[^\n]*)\nTo\n[^\n]*\n(?P<name>[^\n]*)\n(?P<relation>[S,W,D])\/O: (?P<fathers_name>[^\n]*)\n(?P<address>.*)(?P<phonenumber>\d{10})\n(?P<aadhaar_number>^\d{4} \d{4} \d{4}\n).*(?P<vid>\d{4} \d{4} \d{4} \d{4})\n.*DOB: (?P<dob>[^\n]*)\n.*(?P<gender>MALE|FEMALE|Female|Male)',
re.M | re.A | re.S)
# gets all info in one match(enrolment to V) which can then be parsed by the groups
return pattern.search(txt)
def get_enrolment_no(txt):
return get_details(txt).group('enrolment_no')
def get_name(txt):
return get_details(txt).group('name')
def get_fathers_name(txt):
matchobj = get_details(txt)
relation = matchobj.group('fathers_name')
if matchobj.group('relation').lower() == 'w':
return None
return relation
def get_husbands_name(txt):
matchobj = get_details(txt)
return matchobj.group('fathers_name')
def get_address(txt):
return get_details(txt).group('address')
def get_phonenumber(txt):
return get_details(txt).group('phonenumber')
def get_aadhaarnumber(txt):
return get_details(txt).group('aadhaar_number').strip()
def get_vid(txt):
return get_details(txt).group('vid')
def get_gender(txt):
return get_details(txt).group('gender')
def get_dob(txt):
return get_details(txt).group('dob')
def get_details_pdf(file):
import re
txt = extract_pymupdf(file)
dict = {'vid': get_vid(txt),
'enrollment_no': get_enrolment_no(txt), # Fathers name':get_fathers_name(txt),
'name': get_name(txt), # if dict['Fathers name'] == None :
'address': get_address(txt),
'phonenumber': get_phonenumber(txt), # dict['Husbands name']=get_husbands_name(txt)
'aadhaar_no': get_aadhaarnumber(txt),
'sex': get_gender(txt),
'dob': get_dob(txt)}
# ,'ID Type':'Aadhaar'}
return dict
return get_details_pdf(file)
if __name__ == '__main__':
obj = AadhaarExtractor(r"C:\Users\91886\Desktop\cardF.jpg")
print(obj.extract())
# file = open(r"C:\Users\91886\Desktop\cardF.jpg",'r')
# obj.load(file)
# print(obj.to_json())
# | Aadhaar-extractor | /Aadhaar_extractor-0.0.5-py3-none-any.whl/AADHAAR_EXTRACTOR/Extractor.py | Extractor.py |
import os
import os.path
import json
import sys
import pytesseract
import re
import csv
from PIL import Image
import spacy
def adharOcr(image_path):
img = Image.open(image_path)
text = pytesseract.image_to_string(img)
name = None
gender = None
ayear = None
uid = None
yearline = []
genline = []
nameline = []
text1 = []
text2 = []
genderStr = '(Female|Male|emale|male|ale|FEMALE|MALE|EMALE)$'
lines = text
#print(text)
#Searching for Year of Birth
dob_regex = r'\d{2}/\d{2}/\d{4}'
dob_match = re.search(dob_regex, text)
if dob_match:
dob = dob_match.group()
#print("DOB found:", dob)
# else:
# print("DOB not found")
#Searching for Gender
gender_regex = r'[M|F]'
gender_match = re.search(gender_regex, text)
if gender_match:
gender = gender_match.group()
# if gender == 'M':
# print("Gender: Male")
# elif gender == 'F':
# print("Gender: Female")
# else:
# print("Gender not found")
# Searching for Aadhar number
clean_text = text.replace("_", " ")
uid_regex = r"\b\d{4}[\s-]?\d{4}[\s-]?\d{4}\b"
uid_match = re.search(uid_regex, clean_text)
if uid_match:
uid = uid_match.group()
# print("UID: ", uid)
# else:
# print("UID not found")
#Searching for Name
name_regex = r'[A-Z][a-z]+(?: [A-Z][a-z]+)*'
match = re.search(name_regex, text)
if match:
name = match.group()
# print(f"Name found: {name}")
# else:
# print("Name not found")
# Making tuples of data
data = {
'gender': gender,
'dob': dob,
'aadhar': uid,
'name':name
}
with open('aadhar_data.json', 'w') as outfile:
json.dump(data, outfile)
with open('aadhar_data.json', 'r') as file:
data = json.load(file)
print(data['name'])
print("-------------------------------")
print(data['aadhar'])
print("-------------------------------")
print(data['dob'])
print("-------------------------------")
print(data['gender'])
print("-------------------------------") | Aadhar-OCR | /Aadhar%20OCR-0.0.1.tar.gz/Aadhar OCR-0.0.1/__init__.py | __init__.py |
# Installation
Install `AaditsHangman` with `pip install AaditsHangman`.
This will install the module, and will also install other dependencies that this module needs.
# Opening up the game
Play the game by opening up some editor (or ig typing `python` in terminal also works)
Then type this following code:
```py
from AaditsHangman import Hangman
hangman = Hangman()
hangman.new_game()
```
A tkinter window should open on your screen.
Have Fun playing!
# Tips on the game and how to play
1. The computer will choose the word, and you have to guess it
2. There is a timer on the top right corner of the window. When you get the word OR you run out of tries, that timer will end. See how fast you can get it!
3. Instead of pressing submit on your window, you can just press `Enter` on your keyboard!
4. The hangman is hung when you use up nine tries.
5. The `Submit` button automatically becomes `New Game` when the game is over. You can press `Enter` for that also.
| AaditsHangman | /AaditsHangman-0.0.4.tar.gz/AaditsHangman-0.0.4/README.md | README.md |
<h1><img src="https://aaio.io/assets/landing/img/logo-m.svg" width=30 height=30> AAIO</h1>
A Library for easy work with [Aaio API](https://wiki.aaio.io/), in the Python programming language.
Библиотека для легкой работы с [Aaio API](https://wiki.aaio.io/), на языке программирования Python.
## What is available in this library? - Что имеется в данной библиотеке?
- Creating a bill for payment - Создание счета для оплаты
- Quick check of payment status - Быстрая проверка статуса оплаты
- Get balance - Получение баланса
- The largest number of payment methods - Наибольшее количество способов оплаты
## Installation - Установка
Required version [Python](https://www.python.org/): not lower than 3.7
Требуемая версия [Python](https://www.python.org/): не ниже 3.7
```cmd
pip install AaioAPI
```
## Using - Использование
To get started, you need to register and get all the necessary store data [via this link on the official AAIO website](https://aaio.io/cabinet/merchants/)
Чтобы начать работу, вам необходимо зарегистрироваться и получить все необходимые данные магазина [по этой ссылке на оф.сайте AAIO](https://aaio.io/cabinet/merchants/)
### Get balance - Получение баланса
Чтобы получить доступ к балансу, скопируйте ваш [API Ключ](https://aaio.io/cabinet/api/)
``` python
import AaioAPI
client = 'your_api_key'
balance = AaioAPI.get_balance(client)
balance = balance['balance']
# balance = {
# "type": "success",
# "balance": 50.43, // Текущий доступный баланс
# "referral": 0, // Текущий реферальный баланс
# "hold": 1.57 // Текущий замороженный баланс
# }
print(balance)
```
### Example of creating an invoice and receiving a payment link - Пример создания счета и получения ссылки на оплату
Здесь вам понадобятся данные вашего магазина
``` python
from AaioAPI import Aaio
import AaioAPI, time
payment = Aaio()
merchant_id = 'your_shop_id' # ID магазина
amount = 25 # Сумма к оплате
currency = 'RUB' # Валюта заказа
secret = 'your_secret_key' # Секретный ключ №1 из настроек магазина
desc = 'Test payment.' # Описание заказа
url_aaio = AaioAPI.pay(merchant_id, amount, currency, secret, desc)
print(url_aaio) # Ссылка на оплату
```
### Example of a status check - Пример проверки статуса
Проверяем статус платежа каждые 5 секунд с помощью цикла
```python
while True:
AaioAPI.check_payment(url_aaio, payment)
if payment.is_expired(): # Если счет просрочен
print("Invoice was expired")
break
elif payment.is_success(): # Если оплата прошла успешно
print("Payment was succesful")
break
else: # Или если счет ожидает оплаты
print("Invoice wasn't paid. Please pay the bill")
time.sleep(5)
```
### Full Code - Полный код
```python
from AaioAPI import Aaio
import AaioAPI, time
payment = Aaio()
merchant_id = 'your_shop_id' # ID магазина
amount = 25 # Сумма к оплате
currency = 'RUB' # Валюта заказа
secret = 'your_secret_key' # Секретный ключ №1 из настроек магазина
desc = 'Test payment.' # Описание заказа
url_aaio = AaioAPI.pay(merchant_id, amount, currency, secret, desc)
print(url_aaio) # Ссылка на оплату
while True:
AaioAPI.check_payment(url_aaio, payment)
if payment.is_expired(): # Если счет просрочен
print("Invoice was expired")
break
elif payment.is_success(): # Если оплата прошла успешно
print("Payment was succesful")
break
else: # Или если счет ожидает оплаты
print("Invoice wasn't paid. Please pay the bill")
time.sleep(5)
```
## License
MIT | AaioAPI | /AaioAPI-1.0.2.tar.gz/AaioAPI-1.0.2/README.md | README.md |
# AaioAsync
> Fully async python wrapper for Aaio.io API
## Installing
pip install AaioAsync
## Code example
```python
import asyncio
from AaioAsync import AaioAsync
aaio = AaioAsync("API Key", "Shop ID", "Secretkey №1")
async def main():
balance = await aaio.getbalance()
print('Доступно >>> ', balance.balance)
print('В холде >>> ', balance.hold)
print('Реферальный >>> ', balance.referral)
info = await aaio.getorderinfo('HelloAaio')
print('ID >>> ', info.order_id)
print('Сумма >>> ', info.amount)
print('Дата >>> ', info.date)
print('Валюта >>>', info.currency)
asyncio.run(main())
```
## Output
```Python
Доступно >>> 122783.43
В холде >>> 10267.3
Реферальный >>> 3245.92
ID >>> HelloAaio
Сумма >>> 7510.0
Дата >>> 2023-07-29 23:21:20
Валюта >>> RUB
```
## Docs
> Go to https://wiki.aaio.io/ for more information about working with acquiring
| AaioAsync | /AaioAsync-0.1.7.tar.gz/AaioAsync-0.1.7/README.md | README.md |
v =========== * =========== * =========== * =========== * =========== * =========== v
Project : Aalmond!
Contents: This file contains three (3) Function Definitions that provides Primary EDA (Exploratory Data Analysis) functionality:
Function: Dataframe Vital Stats, Outliers Detection, Data View from various sections of a DataFrame.
1. vitalStats() : Displays Pandas DataFrame Vital Stats, an exended output of seeded describe()
2. showdfQ() : Displays data rows of a Pandas DataFrame selectively from Mid, Mid Q1, Mid Q3 section of a Dataframe
3. showOutL() : Displays and/or Imputes Outlier values, based on IQR Method, of Pandas DataFrame Numeric Column(s)
Usage, Parameter & General Notes documented inside each function as Docstring.
^ =========== * =========== * =========== * =========== * =========== * =========== ^
| Aalmond | /Aalmond-0.1.tar.gz/Aalmond-0.1/README.txt | README.txt |
v =========== * =========== * =========== * =========== * =========== * =========== v
Project : Aalmond!
Contents: This file contains three (3) Function Definitions that provides Primary EDA (Exploratory Data Analysis) functionality:
Function: Dataframe Vital Stats, Outliers Detection, Data View from various sections of a DataFrame.
1. vitalStats() : Displays Pandas DataFrame Vital Stats, an exended output of seeded describe()
2. showdfQ() : Displays data rows of a Pandas DataFrame selectively from Mid, Mid Q1, Mid Q3 section of a Dataframe
3. showOutL() : Displays and/or Imputes Outlier values, based on IQR Method, of Pandas DataFrame Numeric Column(s)
Usage, Parameter & General Notes documented inside each function as Docstring.
^ =========== * =========== * =========== * =========== * =========== * =========== ^
| Aalmond | /Aalmond-0.1.tar.gz/Aalmond-0.1/.ipynb_checkpoints/README-checkpoint.txt | README-checkpoint.txt |
# LordOfTheRings SDK
## Installation
```bash
pip install AaronBlaser-SDK==1.0.1
```
## Usage
1. Import the LordOfTheRings class
```python
from lordoftherings import LordOfTheRings
```
2. Initialize the class with your API Token
```python
api = LordOfTheRings('YOUR API TOKEN')
```
3. Make calls to the API
- /movie
```python
all_movies = api.movies().get_all().fetch()
```
- /movie/{id}
```python
movie = api.movies("5cd95395de30eff6ebccde56").get()
```
- /movie/{id}/quote
```python
movie_quotes = api.movies("5cd95395de30eff6ebccde5b").quotes.get_all().fetch()
```
- /quote
```python
quotes = api.quotes().get_all().fetch()
```
- /quote/{id}
```python
single_quote = api.quotes("5cd96e05de30eff6ebccebc9").get()
```
4. Make calls using sorting, filtering, and pagination
- Filtering
```python
match = api.movies().get_all().filter_by(name='The Lord of the Rings Series').fetch()
negate_match = api.movies().get_all().filter_by(name=('not', 'The Lord of the Rings Series')).fetch()
include = api.movies().get_all().filter_by(name=['The Lord of the Rings Series', 'The Desolation of Smaug']).fetch()
exclude = api.movies().get_all().filter_by(name=[('not', 'The Lord of the Rings Series'), ('not', 'The Desolation of Smaug')]).fetch()
less_than = api.movies().get_all().filter_by(budgetInMillions=('<', 100)).fetch()
greater_than = api.movies().get_all().filter_by(runtimeInMinutes=('>', 160)).fetch()
```
- Sorting
```python
sorted_movies = api.movies().get_all().sort_by("name:asc").fetch()
```
- Pagination
```python
paged_movies_1 = api.movies().get_all().limit(2).fetch()
paged_movies_2 = api.movies().get_all().limit(3).page(2).fetch()
paged_movies_3 = api.movies().get_all().limit(1).page(1).offset(1).fetch()
```
- Combine all Three
```python
filter_sorted_paged = api.movies().get_all().limit(5).filter_by(name=('not', 'The Lord of the Rings Series')).sort_by('name:asc').fetch()
```
## Testing
1. Unit Tests
There is a file called test_lordoftherings.py in the tests folder that has some unit tests in it. Run this file to run the tests
- Replace with your token
```python
self.auth_token = 'YOUR API TOKEN'
```
2. Example Usage File
There is a file called test_usage.py in the tests that has many examples of how you can use this SDK. Add your auth token and uncomment
the examples you want to run, then run the file.
- Replace with your token
```python
api = LordOfTheRings('YOUR API TOKEN')
```
| AaronBlaser-SDK | /AaronBlaser-SDK-1.0.1.tar.gz/AaronBlaser-SDK-1.0.1/README.md | README.md |
import requests
import time
class APIResource:
"""Class that is used to represent something that will be returned from the api
Attributes
----------
api : LordOfTheRings
The instance of the LordOfTheRings class that this resource will use to make requests
endpoint : str
The endpoint associated with this API resource
id : str, optional
The id of the resource you wish to get from the api.
"""
def __init__(self, api, endpoint, id=None):
"""Initialize an APIResource instance.
Parameters
----------
api : LordOfTheRings
The instance of the LordOfTheRings class that this resource will use to make requests.
endpoint : str
The endpoint associated with this API resource.
id : str, optional
The unique identifier of this resource (default is None).
"""
self.api = api
self.endpoint = endpoint
self.id = id
def get_all(self):
"""Fetch all resources of this type
Returns
-------
ChainableMethods
An instance of ChainableMethods to allow for option chaining like filtering, sorting, or pagination.
"""
return ChainableMethods(self)
def get(self):
"""Fetch a single resource by the id of the resource
Returns
-------
dict
The JSON response with the data from the resource that was queried
Raises
------
Exception
If the ID of the resource has not been set.
"""
if self.id:
return self.api.request('GET', f'{self.endpoint}/{self.id}')
else:
raise Exception('Resource ID not provided')
class ChainableMethods:
"""Class to allow chainable methods on a resource.
Attributes
----------
resource : APIResource
The resource that will have method chaining.
params : dict
A dictionary of parameters for the API request.
"""
def __init__(self, resource):
"""Initialize a ChainableMethods instance.
Parameters
----------
resource : APIResource
The resource on which the methods should be chained.
"""
self.resource = resource
self.params = {}
def limit(self, limit):
"""Set the limit parameter for the request, limitting the amount of records returned by the api
Parameters
----------
limit : int
The maximum number of results to return.
Returns
-------
ChainableMethods
The current instance to allow method chaining.
"""
self.params['limit'] = limit
return self
def page(self, page):
"""Set the page parameter for the request, specifying which page of results should be returned
Parameters
----------
page : int
The page of results to return.
Returns
-------
ChainableMethods
The current instance to allow method chaining.
"""
self.params['page'] = page
return self
def offset(self, offset):
"""Set the offset parameter for the request.
Parameters
----------
offset : int
The number of results to skip before starting to fetch.
Returns
-------
ChainableMethods
The current instance to allow method chaining.
"""
self.params['offset'] = offset
return self
def filter_by(self, **filters):
"""Set filter parameters for the request.
Parameters
----------
**filters : dict
The filters to apply to the request.
Returns
-------
ChainableMethods
The current instance to allow method chaining.
"""
for key, value in filters.items():
if isinstance(value, str):
# match
self.params[key] = value
elif isinstance(value, tuple):
if value[0] == 'not':
# negate match
self.params[f"{key}!"] = value[1]
elif value[0] in ('<', '>'):
# less than, greater than or equal to
self.params[f"{key}{value[0]}"] = value[1]
elif isinstance(value, list):
if all(isinstance(v, str) for v in value):
# include
self.params[key] = ','.join(value)
elif all(isinstance(v, tuple) and v[0] == 'not' for v in value):
# exclude
self.params[f"{key}!"] = ','.join(v[1] for v in value)
return self
def sort_by(self, sort_by):
"""Set the sort parameter for the request.
Parameters
----------
sort_by : str
The field by which to sort the results. For example, name:asc
Returns
-------
ChainableMethods
The current instance to allow method chaining.
"""
self.params['sort'] = sort_by
return self
def fetch(self):
"""Make the API request and return the response.
Returns
-------
dict
The JSON response from the server.
"""
return self.resource.api.request('GET', self.resource.endpoint, params=self.params)
class Movie(APIResource):
"""A class representing a movie resource.
Attributes
----------
quotes : Quotable
A Quotable instance composed with the movie, allowing the user to get quotes for a particular movie
"""
def __init__(self, api, id=None):
"""Initialize a Movie instance.
Parameters
----------
api : LordOfTheRings
The main API instance.
id : str, optional
The ID of the movie.
"""
endpoint = 'movie'
super().__init__(api, endpoint, id)
self.quotes = Quotable(api, endpoint, self.id)
class Quote(APIResource):
"""A class representing a quote resource."""
def __init__(self, api, id=None):
"""Initialize a Quote instance.
Parameters
----------
api : LordOfTheRings
The main API instance.
id : str, optional
The ID of the quote.
"""
endpoint = 'quote'
super().__init__(api, endpoint, id)
class Quotable(APIResource):
"""A class representing a quotable resource, that can have quotes in the form of {resource}/{id}/quote"""
def __init__(self, api, parent_endpoint, parent_id):
"""Initialize a Quotable instance.
Parameters
----------
api : LordOfTheRings
The main API instance.
parent_endpoint : str
The endpoint of the parent resource.
parent_id : str
The ID of the parent resource.
"""
super().__init__(api, f'{parent_endpoint}/{parent_id}/quote')
class LordOfTheRings:
"""The client for interfacing with the Lord of The Rings API.
Attributes
----------
base_url : str
The base URL for the API.
auth_token : str
The authentication token for the API.
session : requests.Session
The session for making HTTP requests.
max_retries : int
The maximum number of retries when a request fails.
failure_delay : int
The delay (in seconds) between retries.
"""
def __init__(self, auth_token, max_retries=3, failure_delay=1):
"""Initialize a LordOfTheRings client instance.
Parameters
----------
auth_token : str
The authentication token for the API.
max_retries : int, optional
The maximum number of retries when a request fails.
failure_delay : int, optional
The delay (in seconds) between retries.
"""
self.base_url = 'https://the-one-api.dev/v2/'
self.auth_token = auth_token
self.session = requests.Session()
self.max_retries = max_retries
self.failure_delay = failure_delay
if auth_token:
self.session.headers.update({'Authorization': f'Bearer {auth_token}'})
def request(self, method, endpoint, **kwargs):
"""Send a request to the API.
Parameters
----------
method : str
The HTTP method ('GET', 'POST', etc.).
endpoint : str
The API endpoint (excluding the base URL).
**kwargs
Any other parameters to pass to requests.Session.request.
Returns
-------
dict
The JSON response from the API.
Raises
------
Exception
If the request fails after max_retries.
"""
url = f'{self.base_url.rstrip("/")}/{endpoint.lstrip("/")}'
for _ in range(self.max_retries):
response = self.session.request(method, url, **kwargs)
if response.status_code == 429: # Rate limit error
print("Rate limit reached. Retrying...")
time.sleep(self.failure_delay)
continue
elif response.status_code >= 500: # Server error
print("Server error. Retrying...")
time.sleep(self.failure_delay)
continue
elif response.status_code >= 400: # Client error
raise Exception(f'Request failed with status {response.status_code}')
else:
return response.json()
raise Exception('Max retries exceeded')
def movies(self, id=None):
"""Get a movie resource.
Parameters
----------
id : str, optional
The ID of the movie.
Returns
-------
Movie
A Movie instance.
"""
return Movie(self, id)
def quotes(self, id=None):
"""Get a quote resource.
Parameters
----------
id : str, optional
The ID of the quote.
Returns
-------
Quote
A Quote instance.
"""
return Quote(self, id) | AaronBlaser-SDK | /AaronBlaser-SDK-1.0.1.tar.gz/AaronBlaser-SDK-1.0.1/lordoftherings/lordoftherings.py | lordoftherings.py |
from messaging_client.messaging import Messaging
from time import sleep
from threading import Thread
import sys, readline, os
NAME = input("Username> ")
ADMIN = input("Admin? t/f> ")
if ADMIN.lower() == "t":
ADMIN = True
elif ADMIN.lower() == "f":
ADMIN = False
else:
raise ValueError("Use t or f")
M = Messaging(NAME, ADMIN)
#Number of messages to grab at start
START_MESSAGES = 10
def print_with_input(text):
"""
Prints around an input prompt
Thank you jwmullally!
https://stackoverflow.com/questions/2082387/reading-input-from-raw-input-without-having-the-prompt-overwritten-by-other-th
"""
sys.stdout.write('\r'+' '*(len(readline.get_line_buffer())+2)+'\r')
print(text)
sys.stdout.write('> ' + readline.get_line_buffer())
sys.stdout.flush()
def print_message(content):
print_with_input("{}: {}".format(content["n"], content["c"]))
def recieve_messages():
delay = 1
while True:
message = M.poll()
if message["new"]:
print_message(message["content"])
sleep(delay)
def send_messages():
while True:
message = input("> ")
#Forgot where I found this \/ solution, remove the prompt after input
sys.stdout.write("\033[A \033[A \n")
sys.stdout.flush()
M.send_message(message)
def start():
try:
os.system('clear')
i = START_MESSAGES
while i != 0:
#Grab last x messages
message = M.get_message(M.last_message_id-i)
print_message(message)
i -= 1
#Start thread to poll for messages
recieve_thread = Thread(target=recieve_messages)
recieve_thread.daemon = True
recieve_thread.start()
#Start waiting for input
send_messages()
except KeyboardInterrupt:
print("Good Bye!")
#Fixs buggy behavior when exiting
os.system("reset")
sys.exit()
except:
e = sys.exc_info()[0]
print("Unexpected error {}".format(e))
#Fixs buggy behavior when exiting
os.system("reset")
sys.exit()
if __name__ == "__main__":
start() | AaronOS_Messaging | /AaronOS_Messaging-0.1.1.tar.gz/AaronOS_Messaging-0.1.1/messaging_client/main.py | main.py |
import re
import numpy as np
from AaronTools import addlogger
from AaronTools.const import UNIT, PHYSICAL
from AaronTools.utils.utils import float_num
class Signal:
"""
parent class for each signal in a spectrum
"""
# attribute for the x position of this signal
x_attr = None
required_attrs = ()
nested = None
def __init__(self, x_var, **kwargs):
for attr in self.required_attrs:
setattr(self, attr, None)
for arg in kwargs:
setattr(self, arg, kwargs[arg])
setattr(self, self.x_attr, x_var)
@addlogger
class Signals:
"""
parent class for storing data for different signals in the
spectrum and plotting a simulated spectrum
"""
# label for x axis - should be set by child classes
x_label = None
LOG = None
def __init__(self, data, style="gaussian", *args, **kwargs):
self.data = []
if isinstance(data[0], Signal):
self.data = data
return
lines = False
if isinstance(data, str):
lines = data.splitlines()
if lines and style == "gaussian":
self.parse_gaussian_lines(lines, *args, **kwargs)
elif lines and style == "orca":
self.parse_orca_lines(lines, *args, **kwargs)
elif lines and style == "psi4":
self.parse_psi4_lines(lines, *args, **kwargs)
elif lines and style == "qchem":
self.parse_qchem_lines(lines, *args, **kwargs)
else:
raise NotImplementedError("cannot parse data for %s" % style)
def parse_gaussian_lines(self, lines, *args, **kwargs):
"""parse data from Gaussian output files related to this spectrum"""
raise NotImplementedError(
"parse_gaussian_lines not implemented by %s" %
self.__class__.__name__
)
def parse_orca_lines(self, lines, *args, **kwargs):
"""parse data from ORCA output files related to this spectrum"""
raise NotImplementedError(
"parse_orca_lines not implemented by %s" %
self.__class__.__name__
)
def parse_psi4_lines(self, lines, *args, **kwargs):
"""parse data from Psi4 output files related to this spectrum"""
raise NotImplementedError(
"parse_psi4_lines not implemented by %s" %
self.__class__.__name__
)
def parse_qchem_lines(self, lines, *args, **kwargs):
"""parse data from Q-Chem output files related to this spectrum"""
raise NotImplementedError(
"parse_qchem_lines not implemented by %s" %
self.__class__.__name__
)
def filter_data(self, signal):
"""
used to filter out some data from the spectrum (e.g.
imaginary modes from an IR spec)
return False if signal should not be in the spectrum
"""
return True
def get_spectrum_functions(
self,
fwhm=15.0,
peak_type="pseudo-voigt",
voigt_mixing=0.5,
scalar_scale=0.0,
linear_scale=0.0,
quadratic_scale=0.0,
intensity_attr="intensity",
data_attr="data",
):
"""
returns a list of functions that can be evaluated to
produce a spectrum
fwhm - full width at half max of each peak
peak_type - gaussian, lorentzian, pseudo-voigt, or delta
voigt_mixing - ratio of pseudo-voigt that is gaussian
scalar_scale - shift x data
linear_scale - scale x data
quadratic_scale - scale x data
x' = (1 - linear_scale * x - quadratic_scale * x^2 - scalar_scale)
intensity_attr - attribute of Signal used for the intensity
of that signal
data_attr - attribute of self for the list of Signal()
"""
data = getattr(self, data_attr)
x_attr = data[0].x_attr
# scale x positions
if not data[0].nested:
x_positions = np.array(
[getattr(d, x_attr) for d in data if self.filter_data(d)]
)
intensities = [
getattr(d, intensity_attr) for d in data if self.filter_data(d)
]
else:
x_positions = []
intensities = []
x_positions.extend(
[getattr(d, x_attr) for d in data if self.filter_data(d)]
)
intensities.extend(
[getattr(d, intensity_attr) for d in data if self.filter_data(d)]
)
for nest in data[0].nested:
for d in data:
nest_attr = getattr(d, nest)
if isinstance(nest_attr, dict):
for value in nest_attr.values():
if hasattr(value, "__iter__"):
for item in value:
x_positions.append(getattr(item, x_attr))
intensities.append(getattr(item, intensity_attr))
else:
x_positions.append(getattr(value, x_attr))
intensities.append(getattr(value, intensity_attr))
elif hasattr(nest_attr, "__iter__"):
for item in nest_attr:
x_positions.append(getattr(item, x_attr))
intensities.append(getattr(item, intensity_attr))
else:
x_positions.append(getattr(nest_attr, x_attr))
intensities.append(getattr(nest_attr, intensity_attr))
x_positions = np.array(x_positions)
x_positions -= (
linear_scale * x_positions + quadratic_scale * x_positions ** 2
)
x_positions += scalar_scale
e_factor = -4 * np.log(2) / fwhm ** 2
sigma = fwhm / (2 * np.sqrt(2 * np.log(2)))
functions = []
for x_pos, intensity in zip(x_positions, intensities):
if intensity is not None:
if peak_type.lower() == "gaussian":
functions.append(
lambda x, x0=x_pos, inten=intensity: inten
* np.exp(e_factor * (x - x0) ** 2)
* fwhm / (2 * np.sqrt(2 * np.log(2)))
)
elif peak_type.lower() == "lorentzian":
functions.append(
lambda x, x0=x_pos, inten=intensity: inten
* (
0.5 * fwhm
/ (np.pi * ((x - x0) ** 2 + (0.5 * fwhm) ** 2))
)
)
elif peak_type.lower() == "pseudo-voigt":
functions.append(
lambda x, x0=x_pos, inten=intensity: inten
* (
(1 - voigt_mixing)
* (
(0.5 * fwhm) ** 2
/ (((x - x0) ** 2 + (0.5 * fwhm) ** 2))
)
+ voigt_mixing
* np.exp(-(x - x0) ** 2 / (2 * sigma ** 2))
)
)
elif peak_type.lower() == "delta":
functions.append(
lambda x, x0=x_pos, inten=intensity: inten
* int(x == x0)
)
return functions, x_positions, intensities
@staticmethod
def get_plot_data(
functions,
signal_centers,
point_spacing=None,
transmittance=False,
peak_type="pseudo-voigt",
normalize=True,
fwhm=15.0,
change_x_unit_func=None,
show_functions=None,
):
"""
returns arrays of x_values, y_values for a spectrum
point_spacing - spacing between points, default is higher resolution around
each peak (i.e. not uniform)
this is pointless if peak_type == delta
fwhm - full width at half max
transmittance - if true, take 10^(2 - y_values) before returning
to get transmittance as a %
peak_type - pseudo-voigt, gaussian, lorentzian, or delta
voigt_mixing - fraction of pseudo-voigt that is gaussian
linear_scale - subtract linear_scale * frequency off each mode
quadratic_scale - subtract quadratic_scale * frequency^2 off each mode
"""
other_y_list = []
if peak_type.lower() != "delta":
if point_spacing is not None and peak_type.lower():
x_values = []
x = -point_spacing
stop = max(signal_centers)
if peak_type.lower() != "delta":
stop += 5 * fwhm
while x < stop:
x += point_spacing
x_values.append(x)
x_values = np.array(x_values)
else:
x_values = np.linspace(
0,
max(signal_centers) - 10 * fwhm,
num=100
).tolist()
for freq in signal_centers:
x_values.extend(
np.linspace(
max(freq - (7.5 * fwhm), 0),
freq + (7.5 * fwhm),
num=75,
).tolist()
)
x_values.append(freq)
if not point_spacing:
x_values = np.array(list(set(x_values)))
x_values.sort()
y_values = np.sum([f(x_values) for f in functions], axis=0)
if show_functions:
for (ndx1, ndx2) in show_functions:
other_y_list.append(
np.sum(
[f(x_values) for f in functions[ndx1: ndx2]],
axis=0,
)
)
else:
x_values = []
y_values = []
for freq, func in zip(signal_centers, functions):
y_values.append(func(freq))
x_values.append(freq)
y_values = np.array(y_values)
if len(y_values) == 0:
Signals.LOG.warning("nothing to plot")
return None
if normalize or transmittance:
max_val = abs(max(y_values.max(), y_values.min(), key=abs))
y_values /= max_val
for y_vals in other_y_list:
y_vals /= max_val
if transmittance:
y_values = np.array([10 ** (2 - y) for y in y_values])
for i in range(0, len(other_y_list)):
other_y_list[i] = np.array(
[10 ** (2 - y) for y in other_y_list[i]]
)
if change_x_unit_func:
x_values, ndx = change_x_unit_func(x_values)
y_values = y_values[ndx]
for i in range(0, len(other_y_list)):
other_y_list[i] = other_y_list[i][ndx]
return x_values, y_values, other_y_list
@classmethod
def plot_spectrum(
cls,
figure,
x_values,
y_values,
other_y_values=None,
other_y_style=None,
centers=None,
widths=None,
exp_data=None,
reverse_x=None,
y_label=None,
plot_type="transmittance",
x_label=r"wavenumber (cm$^{-1}$)",
peak_type="pseudo-voigt",
rotate_x_ticks=False,
):
"""
plot the x_data and y_data on figure (matplotlib figure)
this is intended for IR spectra
centers - array-like of float, plot is split into sections centered
on the frequency specified by centers
default is to not split into sections
widths - array-like of float, defines the width of each section
exp_data - other data to plot
should be a list of (x_data, y_data, color)
reverse_x - if True, 0 cm^-1 will be on the right
"""
if not centers:
# if no centers were specified, pretend they were so we
# can do everything the same way
axes = [figure.subplots(nrows=1, ncols=1)]
y_nonzero = np.nonzero(y_values)[0]
x_values = np.array(x_values)
widths = [max(x_values[y_nonzero])]
centers = [max(x_values[y_nonzero]) / 2]
else:
n_sections = len(centers)
figure.subplots_adjust(wspace=0.05)
# sort the sections so we don't jump around
widths = [
x
for _, x in sorted(
zip(centers, widths),
key=lambda p: p[0],
reverse=reverse_x,
)
]
centers = sorted(centers, reverse=reverse_x)
axes = figure.subplots(
nrows=1,
ncols=n_sections,
sharey=True,
gridspec_kw={"width_ratios": widths},
)
if not hasattr(axes, "__iter__"):
# only one section was specified (e.g. zooming in on a peak)
# make sure axes is iterable
axes = [axes]
for i, ax in enumerate(axes):
if i == 0:
ax.set_ylabel(y_label)
# need to split plot into sections
# put a / on the border at the top and bottom borders
# of the plot
if len(axes) > 1:
ax.spines["right"].set_visible(False)
ax.tick_params(labelright=False, right=False)
ax.plot(
[1, 1],
[0, 1],
marker=((-1, -1), (1, 1)),
markersize=5,
linestyle="none",
color="k",
mec="k",
mew=1,
clip_on=False,
transform=ax.transAxes,
)
elif i == len(axes) - 1 and len(axes) > 1:
# last section needs a set of / too, but on the left side
ax.spines["left"].set_visible(False)
ax.tick_params(labelleft=False, left=False)
ax.plot(
[0, 0],
[0, 1],
marker=((-1, -1), (1, 1)),
markersize=5,
linestyle="none",
color="k",
mec="k",
mew=1,
clip_on=False,
transform=ax.transAxes,
)
elif len(axes) > 1:
# middle sections need two sets of /
ax.spines["right"].set_visible(False)
ax.spines["left"].set_visible(False)
ax.tick_params(
labelleft=False, labelright=False, left=False, right=False
)
ax.plot(
[0, 0],
[0, 1],
marker=((-1, -1), (1, 1)),
markersize=5,
linestyle="none",
label="Silence Between Two Subplots",
color="k",
mec="k",
mew=1,
clip_on=False,
transform=ax.transAxes,
)
ax.plot(
[1, 1],
[0, 1],
marker=((-1, -1), (1, 1)),
markersize=5,
label="Silence Between Two Subplots",
linestyle="none",
color="k",
mec="k",
mew=1,
clip_on=False,
transform=ax.transAxes,
)
if peak_type.lower() != "delta":
ax.plot(
x_values,
y_values,
color="k",
linewidth=1,
label="computed",
)
if other_y_values:
for y_vals, style in zip(other_y_values, other_y_style):
ax.plot(
x_values,
y_vals,
color=style[0],
linestyle=style[1],
linewidth=1,
label=style[2],
zorder=-1,
)
else:
if plot_type.lower() == "transmittance":
ax.vlines(
x_values,
y_values,
[100 for y in y_values],
linewidth=1,
colors=["k" for x in x_values],
label="computed",
)
ax.hlines(
100,
0,
max(4000, *x_values),
linewidth=1,
colors=["k" for y in y_values],
label="computed",
)
if other_y_values:
for y_vals, style in zip(other_y_values, other_y_style):
ax.vlines(
x_values,
y_vals,
[100 for y in y_vals],
colors=[style[0] for x in x_values],
linestyles=style[1],
linewidth=1,
label=style[2],
zorder=-1,
)
else:
ax.vlines(
x_values,
[0 for y in y_values],
y_values,
linewidth=1,
colors=["k" for x in x_values],
label="computed",
)
ax.hlines(
0,
0,
max(4000, *x_values),
linewidth=1,
colors=["k" for y in y_values],
label="computed",
)
if other_y_values:
for y_vals, style in zip(other_y_values, other_y_style):
ax.vlines(
x_values,
[0 for y in y_vals],
y_vals,
colors=[style[0] for x in x_values],
linestyles=style[1],
linewidth=1,
label=style[2],
zorder=-1,
)
if exp_data:
for x, y, color in exp_data:
ax.plot(
x,
y,
color=color,
zorder=1,
linewidth=1,
label="observed",
)
center = centers[i]
width = widths[i]
high = center + width / 2
low = center - width / 2
if reverse_x:
ax.set_xlim(high, low)
else:
ax.set_xlim(low, high)
# b/c we're doing things in sections, we can't add an x-axis label
# well we could, but which section would be put it one?
# it wouldn't be centered
# so instead the x-axis label is this
figure.text(
0.5, 0.0, x_label, ha="center", va="bottom"
)
if rotate_x_ticks:
figure.autofmt_xdate(rotation=-45, ha="center")
@classmethod
def get_mixed_signals(
cls,
signal_groups,
weights,
fractions=None,
data_attr="data",
**kwargs,
):
"""
get signals for a mixture of components or conformers
signal_groups - list of Signals() instances or list of lists of Signals()
a list of Signals() is a group of conformers
a list of lists of Signals() are the different components
weights - weights for each conformer, organized according to signal_groups
fractions - fraction of each component in the mixture
default: all components have equal fractions
data_attr - attribute of Signals() for data
**kwargs - passed to cls.__init__, along with a new list of data
"""
if not hasattr(signal_groups[0], "__iter__"):
signal_groups = [signal_groups]
if not hasattr(weights[0], "__iter__"):
weights = [weights]
if fractions is None:
fractions = np.ones(len(signal_groups))
new_data = []
for group, weighting, fraction in zip(signal_groups, weights, fractions):
for signals, weight in zip(group, weighting):
data = getattr(signals, data_attr)
for d in data:
x_val = getattr(d, d.x_attr)
vals = d.__dict__
data_cls = d.__class__
new_vals = dict()
for key, item in vals.items():
if isinstance(item, float):
new_vals[key] = fraction * weight * item
else:
new_vals[key] = item
if d.nested:
if not isinstance(d.nested, str):
for attr in d.nested:
nest = getattr(d, attr)
nest_vals = dict()
if isinstance(nest, dict):
for k, items in nest.items():
for i, item in enumerate(items):
nest_x_val = getattr(item, item.x_attr)
vals = item.__dict__
nest_cls = item.__class__
for k2, j in vals.items():
if isinstance(item, float):
nest_vals[k2] = fraction * weight * j
else:
nest_vals[k2] = j
new_vals[attr][k][i] = nest_cls(
nest_x_val, **nest_vals
)
elif hasattr(nest, "__iter__"):
for i, item in enumerate(nest):
nest_x_val = getattr(item, item.x_attr)
vals = item.__dict__
nest_cls = item.__class__
for k, j in vals.items():
if isinstance(item, float):
nest_vals[k] = fraction * weight * j
else:
nest_vals[k] = j
new_vals[attr][i] = nest_cls(
nest_x_val, **nest_vals
)
else:
nest_x_val = getattr(nest, nest.x_attr)
vals = nest.__dict__
nest_cls = nest.__class__
for k, j in vals.items():
if isinstance(nest, float):
nest_vals[k] = fraction * weight * j
else:
nest_vals[k] = j
new_vals[attr][k] = nest_cls(
nest_x_val, **nest_vals
)
new_data.append(data_cls(x_val, **new_vals))
return cls(new_data, **kwargs)
class HarmonicVibration(Signal):
x_attr = "frequency"
required_attrs = (
"intensity", "vector", "symmetry", "rotation", "raman_activity", "forcek",
)
class AnharmonicVibration(Signal):
x_attr = "frequency"
required_attrs = (
"intensity", "harmonic", "overtones", "combinations",
"rotation", "raman_activity",
)
nested = ("overtones", "combinations")
@property
def harmonic_frequency(self):
return self.harmonic.frequency
@property
def delta_anh(self):
return self.frequency - self.harmonic.frequency
class Frequency(Signals):
"""for spectra in the IR/NIR region based on vibrational modes"""
def __init__(self, *args, harmonic=True, hpmodes=None, **kwargs):
super().__init__(*args, harmonic=harmonic, hpmodes=hpmodes, **kwargs)
self.anharm_data = None
self.imaginary_frequencies = None
self.real_frequencies = None
self.lowest_frequency = None
self.by_frequency = {}
self.is_TS = None
self.sort_frequencies()
def parse_gaussian_lines(
self, lines, *args, hpmodes=None, harmonic=True, **kwargs
):
if harmonic:
return self._parse_harmonic_gaussian(lines, hpmodes=hpmodes)
return self._parse_anharmonic_gaussian(lines)
def _parse_harmonic_gaussian(self, lines, hpmodes):
if hpmodes is None:
raise TypeError(
"hpmodes argument required when data is a string"
)
num_head = 0
for line in lines:
if "Harmonic frequencies" in line:
num_head += 1
if hpmodes and num_head != 2:
raise RuntimeError("Log file damaged, cannot get frequencies")
num_head = 0
idx = -1
modes = []
for k, line in enumerate(lines):
if "Harmonic frequencies" in line:
num_head += 1
if hpmodes and num_head == 2:
# if hpmodes, want just the first set of freqs
break
continue
if "Frequencies" in line and (
(hpmodes and "---" in line) or ("--" in line and not hpmodes)
):
for i, symm in zip(
float_num.findall(line), lines[k - 1].split()
):
self.data += [HarmonicVibration(float(i), symmetry=symm)]
modes += [[]]
idx += 1
continue
if ("Force constants" in line and "---" in line and hpmodes) or (
"Frc consts" in line and "--" in line and not hpmodes
):
force_constants = float_num.findall(line)
for i in range(-len(force_constants), 0, 1):
self.data[i].forcek = float(force_constants[i])
continue
if ("Reduced masses" in line and "---" in line and hpmodes) or (
"Red. masses" in line and "--" in line and not hpmodes
):
red_masses = float_num.findall(line)
for i in range(-len(red_masses), 0, 1):
self.data[i].red_mass = float(red_masses[i])
continue
if ("Rot. strength" in line and "---" in line and hpmodes) or (
"Rot. str." in line and "--" in line and not hpmodes
):
roational_strength = float_num.findall(line)
for i in range(-len(roational_strength), 0, 1):
self.data[i].rotation = float(roational_strength[i])
continue
if ("Raman Activities" in line and "---" in line and hpmodes) or (
"Raman Activ" in line and "--" in line and not hpmodes
):
roational_strength = float_num.findall(line)
for i in range(-len(roational_strength), 0, 1):
self.data[i].raman_activity = float(roational_strength[i])
continue
if "IR Inten" in line and (
(hpmodes and "---" in line) or (not hpmodes and "--" in line)
):
intensities = float_num.findall(line)
for i in range(-len(force_constants), 0, 1):
self.data[i].intensity = float(intensities[i])
continue
if hpmodes:
match = re.search(
r"^\s+\d+\s+\d+\s+\d+(\s+[+-]?\d+\.\d+)+$", line
)
if match is None:
continue
values = float_num.findall(line)
coord = int(values[0]) - 1
atom = int(values[1]) - 1
moves = values[3:]
for i, m in enumerate(moves):
tmp = len(moves) - i
mode = modes[-tmp]
try:
vector = mode[atom]
except IndexError:
vector = [0, 0, 0]
modes[-tmp] += [[]]
vector[coord] = m
modes[-tmp][atom] = vector
else:
match = re.search(r"^\s+\d+\s+\d+(\s+[+-]?\d+\.\d+)+$", line)
if match is None:
continue
values = float_num.findall(line)
atom = int(values[0]) - 1
moves = np.array(values[2:], dtype=np.float)
n_moves = len(moves) // 3
for i in range(-n_moves, 0):
modes[i].append(
moves[3 * n_moves + 3 * i : 4 * n_moves + 3 * i]
)
for mode, data in zip(modes, self.data):
data.vector = np.array(mode, dtype=np.float64)
def _parse_anharmonic_gaussian(self, lines):
reading_combinations = False
reading_overtones = False
reading_fundamentals = False
combinations = []
overtones = []
fundamentals = []
mode_re = re.compile(r"(\d+)\((\d+)\)")
for line in lines:
if "---" in line or "Mode" in line or not line.strip():
continue
if "Fundamental Bands" in line:
reading_fundamentals = True
continue
if "Overtones" in line:
reading_overtones = True
continue
if "Combination Bands" in line:
reading_combinations = True
continue
if reading_combinations:
info = line.split()
mode1 = mode_re.search(info[0])
mode2 = mode_re.search(info[1])
ndx_1 = int(mode1.group(1))
exp_1 = int(mode1.group(2))
ndx_2 = int(mode2.group(1))
exp_2 = int(mode2.group(2))
harm_freq = float(info[2])
anharm_freq = float(info[3])
anharm_inten = float(info[4])
harm_inten = 0
combinations.append(
(
ndx_1,
ndx_2,
exp_1,
exp_2,
anharm_freq,
anharm_inten,
harm_freq,
harm_inten,
)
)
elif reading_overtones:
info = line.split()
mode = mode_re.search(info[0])
ndx = int(mode.group(1))
exp = int(mode.group(2))
harm_freq = float(info[1])
anharm_freq = float(info[2])
anharm_inten = float(info[3])
harm_inten = 0
overtones.append(
(
ndx,
exp,
anharm_freq,
anharm_inten,
harm_freq,
harm_inten,
)
)
elif reading_fundamentals:
info = line.split()
harm_freq = float(info[1])
anharm_freq = float(info[2])
anharm_inten = float(info[4])
harm_inten = float(info[3])
fundamentals.append(
(anharm_freq, anharm_inten, harm_freq, harm_inten)
)
self.anharm_data = []
for i, mode in enumerate(
sorted(fundamentals, key=lambda pair: pair[2])
):
self.anharm_data.append(
AnharmonicVibration(mode[0], intensity=mode[1], harmonic=self.data[i])
)
self.anharm_data[-1].overtones = []
self.anharm_data[-1].combinations = dict()
for overtone in overtones:
ndx = len(fundamentals) - overtone[0]
data = self.anharm_data[ndx]
harm_data = HarmonicVibration(overtone[4], intensity=overtone[5])
data.overtones.append(
AnharmonicVibration(
overtone[2], intensity=overtone[3], harmonic=harm_data
)
)
for combo in combinations:
ndx1 = len(fundamentals) - combo[0]
ndx2 = len(fundamentals) - combo[1]
data = self.anharm_data[ndx1]
harm_data = HarmonicVibration(combo[6], intensity=combo[7])
data.combinations[ndx2] = [
AnharmonicVibration(combo[4], intensity=combo[5], harmonic=harm_data)
]
def parse_qchem_lines(self, lines, *args, **kwargs):
num_head = 0
modes = []
for k, line in enumerate(lines):
if "Frequency:" in line:
ndx = 0
for i in float_num.findall(line):
self.data += [HarmonicVibration(float(i))]
modes += [[]]
continue
if "Force Cnst:" in line:
force_constants = float_num.findall(line)
for i in range(-len(force_constants), 0, 1):
self.data[i].forcek = float(force_constants[i])
continue
if "Red. Mass:" in line:
red_masses = float_num.findall(line)
for i in range(-len(red_masses), 0, 1):
self.data[i].red_mass = float(red_masses[i])
continue
if "IR Intens:" in line:
intensities = float_num.findall(line)
for i in range(-len(force_constants), 0, 1):
self.data[i].intensity = float(intensities[i])
continue
if "Raman Intens:" in line:
intensities = float_num.findall(line)
for i in range(-len(force_constants), 0, 1):
self.data[i].raman_activity = float(intensities[i])
continue
match = re.search(r"^\s?[A-Z][a-z]?\s+(\s+[+-]?\d+\.\d+)+$", line)
if match is None:
continue
ndx += 1
values = float_num.findall(line)
moves = np.array(values, dtype=np.float)
n_moves = len(moves) // 3
for i in range(-n_moves, 0):
modes[i].append(
moves[3 * n_moves + 3 * i : 4 * n_moves + 3 * i]
)
for mode, data in zip(modes, self.data):
data.vector = np.array(mode, dtype=np.float64)
def parse_orca_lines(self, lines, *args, **kwargs):
"""parse lines of orca output related to frequency
hpmodes is not currently used"""
# vibrational frequencies appear as a list, one per line
# block column 0 is the index of the mode
# block column 1 is the frequency in 1/cm
# skip line one b/c its just "VIBRATIONAL FREQUENCIES" with the way we got the lines
for n, line in enumerate(lines[1:]):
if line == "NORMAL MODES":
break
freq = line.split()[1]
self.data += [HarmonicVibration(float(freq))]
# all 3N modes are printed with six modes in each block
# each column corresponds to one mode
# the rows of the columns are x_1, y_1, z_1, x_2, y_2, z_2, ...
displacements = np.zeros((len(self.data), len(self.data)))
carryover = 0
start = 0
stop = 6
for i, line in enumerate(lines[n + 2 :]):
if "IR SPECTRUM" in line:
break
if i % (len(self.data) + 1) == 0:
carryover = i // (len(self.data) + 1)
start = 6 * carryover
stop = start + 6
continue
ndx = (i % (len(self.data) + 1)) - 1
mode_info = line.split()[1:]
displacements[ndx][start:stop] = [float(x) for x in mode_info]
# reshape columns into Nx3 arrays
for k, data in enumerate(self.data):
data.vector = np.reshape(
displacements[:, k], (len(self.data) // 3, 3)
)
# purge rotational and translational modes
n_data = len(self.data)
k = 0
while k < n_data:
if self.data[k].frequency == 0:
del self.data[k]
n_data -= 1
else:
k += 1
for k, line in enumerate(lines):
if line.strip() == "IR SPECTRUM":
order = lines[k + 1].split()
if "Int" in order:
ndx = order.index("Int")
else:
ndx = order.index("T**2") - 1
intensity_start = k + 2
# IR intensities are only printed for vibrational
# the first column is the index of the mode
# the second column is the frequency
# the third is the intensity, which we read next
t = 0
for line in lines[intensity_start:]:
if not re.match(r"\s*\d+:", line):
continue
ir_info = line.split()
inten = float(ir_info[ndx])
self.data[t].intensity = inten
t += 1
if t >= len(self.data):
break
for k, line in enumerate(lines):
if line.strip() == "RAMAN SPECTRUM":
t = 0
for line in lines[k + 1:]:
if not re.match(r"\s*\d+:", line):
continue
ir_info = line.split()
inten = float(ir_info[2])
self.data[t].raman_activity = inten
t += 1
if t >= len(self.data):
break
def parse_psi4_lines(self, lines, *args, **kwargs):
"""parse lines of psi4 output related to frequencies
hpmodes is not used"""
# normal mode info appears in blocks, with up to 3 modes per block
# at the top is the index of the normal mode
# next is the frequency in wavenumbers (cm^-1)
# after a line of '-----' are the normal displacements
read_displacement = False
modes = []
for n, line in enumerate(lines):
if len(line.strip()) == 0:
read_displacement = False
for i, data in enumerate(self.data[-nmodes:]):
data.vector = np.array(modes[i])
elif read_displacement:
info = [float(x) for x in line.split()[2:]]
for i, mode in enumerate(modes):
mode.append(info[3 * i : 3 * (i + 1)])
elif line.strip().startswith("Vibration"):
nmodes = len(line.split()) - 1
elif line.strip().startswith("Freq"):
freqs = [-1 * float(x.strip("i")) if x.endswith("i") else float(x) for x in line.split()[2:]]
for freq in freqs:
self.data.append(HarmonicVibration(float(freq)))
elif line.strip().startswith("Force const"):
force_consts = [float(x) for x in line.split()[3:]]
for i, data in enumerate(self.data[-nmodes:]):
data.forcek = force_consts[i]
elif line.strip().startswith("Irrep"):
# sometimes psi4 doesn't identify the irrep of a mode, so we can't
# use line.split()
symm = [
x.strip() if x.strip() else None
for x in [line[31:40], line[51:60], line[71:80]]
]
for i, data in enumerate(self.data[-nmodes:]):
data.symmetry = symm[i]
elif line.strip().startswith("----"):
read_displacement = True
modes = [[] for i in range(0, nmodes)]
def sort_frequencies(self):
self.imaginary_frequencies = []
self.real_frequencies = []
for data in self.data:
freq = data.frequency
if freq < 0:
self.imaginary_frequencies += [freq]
elif freq > 0:
self.real_frequencies += [freq]
self.by_frequency[freq] = {
"intensity": data.intensity,
}
if hasattr(data, "vector"):
# anharmonic data might not have a vector
self.by_frequency[freq]["vector"] = data.vector
if len(self.data) > 0:
self.lowest_frequency = self.data[0].frequency
else:
self.lowest_frequency = None
self.is_TS = True if len(self.imaginary_frequencies) == 1 else False
def filter_data(self, signal):
return signal.frequency > 0
def plot_ir(
self,
figure,
centers=None,
widths=None,
exp_data=None,
plot_type="transmittance",
peak_type="pseudo-voigt",
reverse_x=True,
y_label=None,
point_spacing=None,
normalize=True,
fwhm=15.0,
anharmonic=False,
rotate_x_ticks=False,
show_functions=None,
**kwargs,
):
"""
plot IR data on figure
figure - matplotlib figure
centers - array-like of float, plot is split into sections centered
on the frequency specified by centers
default is to not split into sections
widths - array-like of float, defines the width of each section
exp_data - other data to plot
should be a list of (x_data, y_data, color)
reverse_x - if True, 0 cm^-1 will be on the right
plot_type - see Frequency.get_plot_data
peak_type - any value allowed by Frequency.get_plot_data
kwargs - keywords for Frequency.get_spectrum_functions
"""
if "intensity_attr" not in kwargs:
intensity_attr = "intensity"
if plot_type.lower() == "vcd":
intensity_attr = "rotation"
elif plot_type.lower() == "raman":
intensity_attr = "raman_activity"
elif plot_type.lower() == "absorbance":
intensity_attr = "intensity"
elif plot_type.lower() == "transmittance":
intensity_attr = "intensity"
else:
self.LOG.warning("unrecognized plot type: %s\nDefaulting to absorbance" % plot_type)
kwargs["intensity_attr"] = intensity_attr
data_attr = "data"
if anharmonic:
data_attr = "anharm_data"
functions, frequencies, intensities = self.get_spectrum_functions(
peak_type=peak_type,
fwhm=fwhm,
data_attr=data_attr,
**kwargs,
)
other_y_style = None
ndx_list = None
if show_functions is not None:
ndx_list = [info[0] for info in show_functions]
other_y_style = list(info[1:] for info in show_functions)
data = self.get_plot_data(
functions,
frequencies,
fwhm=fwhm,
transmittance=plot_type.lower().startswith("transmittance"),
peak_type=peak_type,
point_spacing=point_spacing,
normalize=normalize,
show_functions=ndx_list,
)
if data is None:
return
x_values, y_values, other_y_values = data
if y_label is None and plot_type.lower().startswith("transmittance"):
y_label = "Transmittance (%)"
elif y_label is None and plot_type.lower() == "absorbance":
y_label = "Absorbance (arb.)"
elif y_label is None and plot_type.lower() == "vcd":
y_label = "ΔAbsorbance (arb.)"
elif y_label is None and plot_type.lower() == "raman":
y_label = "Activity (arb.)"
self.plot_spectrum(
figure,
x_values,
y_values,
other_y_values=other_y_values,
other_y_style=other_y_style,
centers=centers,
widths=widths,
exp_data=exp_data,
reverse_x=reverse_x,
peak_type=peak_type,
plot_type=plot_type,
y_label=y_label,
rotate_x_ticks=rotate_x_ticks,
)
class ValenceExcitation(Signal):
x_attr = "excitation_energy"
required_attrs = (
"rotatory_str_len", "rotatory_str_vel", "oscillator_str",
"oscillator_str_vel", "symmetry", "multiplicity",
)
@property
def dipole_str_len(self):
return self.oscillator_str / self.excitation_energy
@property
def dipole_str_vel(self):
return self.oscillator_str_vel / self.excitation_energy
class TransientExcitation(ValenceExcitation):
x_attr = "excitation_energy"
required_attrs = (
"rotatory_str_len", "rotatory_str_vel", "oscillator_str",
"oscillator_str_vel", "symmetry", "multiplicity",
)
def __init__(self, *args, **kwargs):
super().__init__(*args, **kwargs)
class ValenceExcitations(Signals):
def __init__(self, *args, **kwargs):
self.transient_data = None
super().__init__(*args, **kwargs)
def parse_gaussian_lines(self, lines, *args, **kwargs):
i = 0
nrgs = []
rotatory_str_len = []
rotatory_str_vel = []
oscillator_str = []
oscillator_vel = []
symmetry = []
multiplicity = []
while i < len(lines):
if "Ground to excited state transition electric" in lines[i]:
i += 2
line = lines[i]
while line and line.split()[0].isdigit():
oscillator_str.append(float(line.split()[-1]))
i += 1
line = lines[i]
elif "Ground to excited state transition velocity" in lines[i]:
i += 2
line = lines[i]
while line and line.split()[0].isdigit():
oscillator_vel.append(float(line.split()[-1]))
i += 1
line = lines[i]
elif "R(length)" in lines[i]:
i += 1
line = lines[i]
while line and line.split()[0].isdigit():
rotatory_str_len.append(float(line.split()[-1]))
i += 1
line = lines[i]
elif "R(velocity)" in lines[i]:
i += 1
line = lines[i]
while line and line.split()[0].isdigit():
rotatory_str_vel.append(float(line.split()[-2]))
i += 1
line = lines[i]
elif re.search(r"Excited State\s*\d+:", lines[i]):
excitation_data = re.search(
r"Excited State\s*\d+:\s*([\D]+)-([\S]+)\s+(\d+\.\d+)",
lines[i],
)
multiplicity.append(excitation_data.group(1))
symmetry.append(excitation_data.group(2))
nrgs.append(float(excitation_data.group(3)))
i += 1
else:
i += 1
for nrg, rot_len, rot_vel, osc_len, osc_vel, sym, mult in zip(
nrgs, rotatory_str_len, rotatory_str_vel, oscillator_str,
oscillator_vel, symmetry, multiplicity,
):
self.data.append(
ValenceExcitation(
nrg, rotatory_str_len=rot_len,
rotatory_str_vel=rot_vel, oscillator_str=osc_len,
oscillator_str_vel=osc_vel, symmetry=sym,
multiplicity=mult,
)
)
def parse_orca_lines(self, lines, *args, **kwargs):
i = 0
nrgs = []
corr = []
rotatory_str_len = []
rotatory_str_vel = []
oscillator_str = []
oscillator_vel = []
multiplicity = []
mult = "Singlet"
transient_oscillator_str = []
transient_oscillator_vel = []
transient_rot_str = []
transient_nrg = []
while i < len(lines):
line = lines[i]
if "SINGLETS" in line:
mult = "Singlet"
i += 1
elif "TRIPLETS" in line:
mult = "Triplet"
i += 1
elif re.search("IROOT=.+?(\d+\.\d+)\seV", line):
info = re.search("IROOT=.+?(\d+\.\d+)\seV", line)
nrgs.append(float(info.group(1)))
i += 1
elif line.startswith("STATE"):
info = re.search("STATE\s*\d+:\s*E=\s*\S+\s*au\s*(\d+\.\d+)", line)
nrgs.append(float(info.group(1)))
multiplicity.append(mult)
i += 1
elif "ABSORPTION SPECTRUM VIA TRANSITION ELECTRIC DIPOLE MOMENTS" in line and "TRANSIENT" not in line:
i += 5
line = lines[i]
while line.strip():
info = line.split()
oscillator_str.append(float(info[3]))
i += 1
line = lines[i]
elif "ABSORPTION SPECTRUM VIA TRANSITION VELOCITY DIPOLE MOMENTS" in line and "TRANSIENT" not in line:
i += 5
line = lines[i]
while line.strip():
info = line.split()
oscillator_vel.append(float(info[3]))
i += 1
line = lines[i]
elif line.endswith("CD SPECTRUM") and "TRANSIENT" not in line:
i += 5
line = lines[i]
while line.strip():
info = line.split()
rotatory_str_len.append(float(info[3]))
i += 1
line = lines[i]
elif "CD SPECTRUM VIA TRANSITION VELOCITY DIPOLE MOMENTS" in line and "TRANSIENT" not in line:
i += 5
line = lines[i]
while line.strip():
info = line.split()
rotatory_str_vel.append(float(info[3]))
i += 1
line = lines[i]
elif "TRANSIENT ABSORPTION SPECTRUM VIA TRANSITION ELECTRIC DIPOLE MOMENTS" in line:
i += 5
line = lines[i]
while line.strip():
info = line.split()
transient_oscillator_str.append(float(info[3]))
transient_nrg.append(self.nm_to_ev(float(info[2])))
i += 1
line = lines[i]
elif "TRANSIENT ABSORPTION SPECTRUM VIA TRANSITION VELOCITY DIPOLE MOMENTS" in line:
i += 5
line = lines[i]
while line.strip():
info = line.split()
transient_oscillator_vel.append(float(info[3]))
i += 1
line = lines[i]
elif "TRANSIENT CD SPECTRUM" in line:
i += 5
line = lines[i]
while line.strip():
info = line.split()
transient_rot_str.append(float(info[3]))
i += 1
line = lines[i]
elif "CALCULATED SOLVENT SHIFTS" in line:
i += 8
line = lines[i]
while line.strip():
info = line.split()
corr.append(float(info[-1]))
i += 1
line = lines[i]
else:
i += 1
if corr:
for i in range(0, len(nrgs)):
nrgs[i] = corr[i]
if not multiplicity:
multiplicity = [None for x in nrgs]
if not rotatory_str_vel:
rotatory_str_vel = [None for x in rotatory_str_len]
if not oscillator_vel:
oscillator_vel = [None for x in oscillator_str]
for nrg, rot_len, rot_vel, osc_len, osc_vel, mult in zip(
nrgs, rotatory_str_len, rotatory_str_vel, oscillator_str,
oscillator_vel, multiplicity,
):
self.data.append(
ValenceExcitation(
nrg, rotatory_str_len=rot_len,
rotatory_str_vel=rot_vel, oscillator_str=osc_len,
oscillator_str_vel=osc_vel, multiplicity=mult,
)
)
for nrg, rot_len, osc_len, osc_vel in zip(
transient_nrg,
transient_rot_str,
transient_oscillator_str,
transient_oscillator_vel,
):
if not hasattr(self, "transient_data") or not self.transient_data:
self.transient_data = []
self.transient_data.append(
TransientExcitation(
nrg,
rotatory_str_len=rot_len,
oscillator_str=osc_len,
oscillator_str_vel=osc_vel,
)
)
def parse_psi4_lines(self, lines, *args, **kwargs):
symmetry = []
energy = []
oscillator_str = []
oscillator_str_vel = []
rotation_str = []
rotation_str_vel = []
for line in lines:
if "->" not in line and line.split()[0].isdigit():
info = line.split()
symmetry.append(info[1])
energy.append(float(info[2]))
oscillator_str.append(float(info[6]))
rotation_str.append(float(info[7]))
rotation_str_vel.append(float(info[7]))
oscillator_str_vel.append(None)
elif re.search("\| State\s*\d+", line):
info = re.search("(\d+\.\d+)\s*eV", line)
energy.append(float(info.group(1)))
elif re.search("Oscillator strength \(length", line):
oscillator_str.append(float(line.split()[-1]))
elif re.search("Oscillator strength \(velocity", line):
oscillator_str_vel.append(float(line.split()[-1]))
elif re.search("Rotational strength \(length", line):
rotation_str.append(float(line.split()[-1]))
elif re.search("Rotational strength \(velocity", line):
rotation_str_vel.append(float(line.split()[-1]))
elif line.split()[0].isdigit():
info = line[46:].split()
symmetry.append(line.split("(")[1].split(")")[0])
energy.append(float(info[0]))
oscillator_str_vel.append(float(info[2]))
oscillator_str.append(float(info[3]))
rotation_str.append(float(info[4]))
rotation_str_vel.append(float(info[5]))
lists = [
symmetry, energy, oscillator_str_vel, oscillator_str,
rotation_str, rotation_str_vel,
]
max_list = max(lists, key=len)
for l in lists:
while len(l) < len(max_list):
l.append(None)
for nrg, sym, osc_v, osc, r_l, r_v in zip(
energy, symmetry, oscillator_str_vel, oscillator_str,
rotation_str, rotation_str_vel,
):
self.data.append(
ValenceExcitation(
nrg, symmetry=sym, oscillator_str=osc, rotatory_str_len=r_l,
rotatory_str_vel=r_v, oscillator_str_vel=osc_v,
)
)
def parse_qchem_lines(self, lines, *args, **kwargs):
multiplicity = []
energy = []
oscillator_str = []
symmetry = []
rotation_str = []
rotation_str_vel = []
for line in lines:
if re.search("Excited state\s+\d+: excitation energy", line):
energy.append(float(line.split()[-1]))
if re.search("Multiplicity:", line):
multiplicity.append(line.split()[-1])
if re.search("Strength", line):
oscillator_str.append(float(line.split()[-1]))
if re.search("Excited state\s+\d+\s*\(", line):
info = re.search("\((\S+), (\S+)\)", line)
multiplicity.append(info.group(1).capitalize())
symmetry.append(info.group(2))
if re.search("Excitation energy:", line):
if len(energy) > len(oscillator_str):
oscillator_str.append(0)
energy.append(float(line.split()[-2]))
if re.search("Osc. strength:", line):
oscillator_str.append(float(line.split()[-1]))
if re.search("State B:", line):
symmetry.append(line.split("/")[-1])
if re.search("Oscillator strength", line):
oscillator_str.append(float(line.split()[-1]))
if re.search("Energy GAP", line):
energy.append(float(line.split()[-2]))
if re.search("Rotatory strength, length gauge", line):
rotation_str.append(float(line.split()[-1]))
if re.search("Rotatory strength, velocity gauge", line):
rotation_str_vel.append(float(line.split()[-1]))
lists = [
symmetry, energy, oscillator_str, multiplicity,
rotation_str, rotation_str_vel,
]
max_list = max(lists, key=len)
for l in lists:
while len(l) < len(max_list):
l.append(None)
for nrg, mult, osc, symm, rot, rot_vel in zip(
energy, multiplicity, oscillator_str, symmetry,
rotation_str, rotation_str_vel,
):
self.data.append(
ValenceExcitation(
nrg, multiplicity=mult, oscillator_str=osc, symmetry=symm,
rotatory_str_len=rot, rotatory_str_vel=rot_vel,
)
)
@staticmethod
def nm_to_ev(x):
"""convert x nm to eV"""
if isinstance(x, float):
return PHYSICAL.SPEED_OF_LIGHT * 1e7 * PHYSICAL.PLANCK * UNIT.JOULE_TO_EV / x
x = np.array(x)
ndx = np.where(x > 0)
return PHYSICAL.SPEED_OF_LIGHT * 1e7 * PHYSICAL.PLANCK * UNIT.JOULE_TO_EV / x[ndx], ndx
@staticmethod
def ev_to_nm(x):
"""convert x eV to nm"""
if isinstance(x, float):
return PHYSICAL.SPEED_OF_LIGHT * 1e7 * PHYSICAL.PLANCK * UNIT.JOULE_TO_EV / x
x = np.array(x)
ndx = np.where(x > 0)
return PHYSICAL.SPEED_OF_LIGHT * 1e7 * PHYSICAL.PLANCK * UNIT.JOULE_TO_EV / x[ndx], ndx
def plot_uv_vis(
self,
figure,
centers=None,
widths=None,
exp_data=None,
plot_type="uv-vis-veloctiy",
peak_type="gaussian",
reverse_x=False,
y_label=None,
point_spacing=None,
normalize=True,
fwhm=15.0,
units="nm",
rotate_x_ticks=False,
show_functions=None,
transient=False,
**kwargs,
):
"""
plot IR data on figure
figure - matplotlib figure
centers - array-like of float, plot is split into sections centered
on the frequency specified by centers
default is to not split into sections
widths - array-like of float, defines the width of each section
exp_data - other data to plot
should be a list of (x_data, y_data, color)
reverse_x - if True, 0 cm^-1 will be on the right
plot_type - see Frequency.get_plot_data
peak_type - any value allowed by Frequency.get_plot_data
kwargs - keywords for Frequency.get_spectrum_functions
"""
data_attr = "data"
if transient:
data_attr = "transient_data"
if "intensity_attr" not in kwargs:
intensity_attr = "oscillator_str"
if plot_type.lower() == "uv-vis-velocity":
intensity_attr = "oscillator_str_vel"
elif plot_type.lower() == "transmittance-velocity":
intensity_attr = "oscillator_str_vel"
elif plot_type.lower() == "transmittance":
intensity_attr = "oscillator_str"
elif plot_type.lower() == "uv-vis":
intensity_attr = "oscillator_str"
elif plot_type.lower() == "ecd":
intensity_attr = "rotatory_str_len"
elif plot_type.lower() == "ecd-velocity":
intensity_attr = "rotatory_str_vel"
else:
self.LOG.warning("unrecognized plot type: %s\nDefaulting to uv-vis" % plot_type)
kwargs["intensity_attr"] = intensity_attr
if getattr(self.data[0], kwargs["intensity_attr"]) is None:
raise RuntimeError("no data was parsed for %s" % kwargs["intensity_attr"])
if not centers and units == "nm":
data_list = getattr(self, data_attr)
data_min = None
data_max = None
for data in data_list:
wavelength = self.ev_to_nm(data.excitation_energy)
if data_min is None or wavelength < data_min:
data_min = wavelength
if data_max is None or wavelength > data_max:
data_max = wavelength
centers = [(data_min + data_max) / 2]
widths = [1.5 * (data_max - data_min)]
change_x_unit_func = None
x_label = "wavelength (nm)"
change_x_unit_func = self.ev_to_nm
if units == "eV":
change_x_unit_func = None
x_label = r"$h\nu$ (eV)"
functions, energies, intensities = self.get_spectrum_functions(
peak_type=peak_type,
fwhm=fwhm,
data_attr=data_attr,
**kwargs,
)
other_y_style = None
ndx_list = None
if show_functions is not None:
ndx_list = [info[0] for info in show_functions]
other_y_style = list(info[1:] for info in show_functions)
data = self.get_plot_data(
functions,
energies,
fwhm=fwhm,
transmittance=plot_type.lower().startswith("transmittance"),
peak_type=peak_type,
point_spacing=point_spacing,
change_x_unit_func=change_x_unit_func,
normalize=normalize,
show_functions=ndx_list,
)
if data is None:
return
x_values, y_values, other_y_values = data
if y_label is None and plot_type.lower().startswith("transmittance"):
y_label = "Transmittance (%)"
elif y_label is None and "uv-vis" in plot_type.lower():
y_label = "Absorbance (arb.)"
elif y_label is None and "ecd" in plot_type.lower():
y_label = "ΔAbsorbance (arb.)"
self.plot_spectrum(
figure,
x_values,
y_values,
other_y_values=other_y_values,
other_y_style=other_y_style,
centers=centers,
widths=widths,
exp_data=exp_data,
reverse_x=reverse_x,
peak_type=peak_type,
plot_type=plot_type,
x_label=x_label,
y_label=y_label,
rotate_x_ticks=rotate_x_ticks,
) | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/spectra.py | spectra.py |
import os
import re
import sys
from copy import deepcopy
from io import IOBase, StringIO
from math import ceil
import numpy as np
from AaronTools import addlogger
from AaronTools.atoms import Atom
from AaronTools.const import ELEMENTS, PHYSICAL, UNIT
from AaronTools.orbitals import Orbitals
from AaronTools.spectra import Frequency, ValenceExcitations
from AaronTools.theory import *
from AaronTools.utils.utils import (
is_alpha,
is_int,
is_num,
float_num,
)
read_types = [
"xyz",
"log",
"com",
"gjf",
"sd",
"sdf",
"mol",
"mol2",
"out",
"dat",
"fchk",
"crest",
"xtb",
"sqmout",
"47",
"31",
"qout",
]
write_types = ["xyz", "com", "inp", "inq", "in", "sqmin", "cube"]
file_type_err = "File type not yet implemented: {}"
NORM_FINISH = "Normal termination"
ORCA_NORM_FINISH = "****ORCA TERMINATED NORMALLY****"
PSI4_NORM_FINISH = "*** Psi4 exiting successfully. Buy a developer a beer!"
ERROR = {
"Convergence failure -- run terminated.": "SCF_CONV",
"Inaccurate quadrature in CalDSu": "CONV_CDS",
"Error termination request processed by link 9999": "CONV_LINK",
"FormBX had a problem": "FBX",
"NtrErr Called from FileIO": "CHK",
"Wrong number of Negative eigenvalues": "EIGEN",
"Erroneous write": "QUOTA",
"Atoms too close": "CLASH",
"The combination of multiplicity": "CHARGEMULT",
"Bend failed for angle": "REDUND",
"Linear angle in Bend": "REDUND",
"Error in internal coordinate system": "COORD",
"galloc: could not allocate memory": "GALLOC",
"Error imposing constraints": "CONSTR",
"End of file reading basis center.": "BASIS_READ",
"Atomic number out of range for .* basis set.": "BASIS",
"Unrecognized atomic symbol": "ATOM",
"malloc failed.": "MEM",
"A syntax error was detected in the input line": "SYNTAX",
"Unknown message": "UNKNOWN",
}
ERROR_ORCA = {
"SCF NOT CONVERGED AFTER": "SCF_CONV",
# ORCA doesn't actually exit if the SCF doesn't converge...
# "CONV_CDS": "",
"The optimization did not converge but reached the maximum number": "OPT_CONV",
# ORCA still prints the normal finish line if opt doesn't converge...
# "FBX": "",
# "CHK": "",
# "EIGEN": "", <- ORCA doesn't seem to have this
# "QUOTA": "",
"Zero distance between atoms": "CLASH", # <- only get an error if atoms are literally on top of each other
"Error : multiplicity": "CHARGEMULT",
# "REDUND": "",
# "REDUND": "",
# "GALLOC": "",
# "CONSTR": "",
"The basis set was either not assigned or not available for this element": "BASIS",
"Element name/number, dummy atom or point charge expected": "ATOM",
"Error (ORCA_SCF): Not enough memory available!": "MEM",
"WARNING: Analytical MP2 frequency calculations": "NUMFREQ",
"WARNING: Analytical Hessians are not yet implemented for meta-GGA functionals": "NUMFREQ",
"ORCA finished with error return": "UNKNOWN",
"UNRECOGNIZED OR DUPLICATED KEYWORD(S) IN SIMPLE INPUT LINE": "TYPO",
}
# some exceptions are listed in https://psicode.org/psi4manual/master/_modules/psi4/driver/p4util/exceptions.html
ERROR_PSI4 = {
"PsiException: Could not converge SCF iterations": "SCF_CONV",
"psi4.driver.p4util.exceptions.SCFConvergenceError: Could not converge SCF iterations": "SCF_CONV",
"OptimizationConvergenceError": "OPT_CONV",
"TDSCFConvergenceError": "TDCF_CONV",
"The INTCO_EXCEPTion handler": "INT_COORD",
# ^ this is basically psi4's FBX
# "CONV_CDS": "",
# "CONV_LINK": "",
# "FBX": "",
# "CHK": "",
# "EIGEN": "", <- psi4 doesn't seem to have this
# "QUOTA": "",
# "ValidationError:": "INPUT", <- generic input error, CHARGEMULT and CLASH would also get caught by this
"qcelemental.exceptions.ValidationError: Following atoms are too close:": "CLASH",
"qcelemental.exceptions.ValidationError: Inconsistent or unspecified chg/mult": "CHARGEMULT",
"MissingMethodError": "INVALID_METHOD",
# "REDUND": "",
# "REDUND": "",
# "GALLOC": "",
# "CONSTR": "",
"psi4.driver.qcdb.exceptions.BasisSetNotFound: BasisSet::construct: Unable to find a basis set for": "BASIS",
"qcelemental.exceptions.NotAnElementError": "ATOM",
"psi4.driver.p4util.exceptions.ValidationError: set_memory()": "MEM",
# ERROR_PSI4[""] = "UNKNOWN",
"Could not converge backtransformation.": "ICOORDS",
}
def step2str(step):
if int(step) == step:
return str(int(step))
else:
return str(step).replace(".", "-")
def str2step(step_str):
if "-" in step_str:
return float(step_str.replace("-", "."))
else:
return float(step_str)
def expected_inp_ext(exec_type):
"""
extension expected for an input file for exec_type
Gaussian - .com (.gjf on windows)
ORCA - .inp
Psi4 - .in
SQM - .mdin
qchem - .inp
"""
if exec_type.lower() == "gaussian":
if sys.platform.startswith("win"):
return ".gjf"
return ".com"
if exec_type.lower() == "orca":
return ".inp"
if exec_type.lower() == "psi4":
return ".in"
if exec_type.lower() == "sqm":
return ".mdin"
if exec_type.lower() == "qchem":
return ".inp"
def expected_out_ext(exec_type):
"""
extension expected for an input file for exec_type
Gaussian - .log
ORCA - .out
Psi4 - .out
SQM - .mdout
qchem - .out
"""
if exec_type.lower() == "gaussian":
return ".log"
if exec_type.lower() == "orca":
return ".out"
if exec_type.lower() == "psi4":
return ".out"
if exec_type.lower() == "sqm":
return ".mdout"
if exec_type.lower() == "qchem":
return ".out"
class FileWriter:
@classmethod
def write_file(
cls, geom, style=None, append=False, outfile=None, *args, **kwargs
):
"""
Writes file from geometry in the specified style
:geom: the Geometry to use
:style: the file type style to generate
Currently supported options: xyz (default), com, inp, in
if outfile has one of these extensions, default is that style
:append: for *.xyz, append geometry to the same file
:outfile: output destination - default is
[geometry name] + [extension] or [geometry name] + [step] + [extension]
if outfile is False, no output file will be written, but the contents will be returned
:theory: for com, inp, and in files, an object with a get_header and get_footer method
"""
if isinstance(outfile, str) and style is None:
name, ext = os.path.splitext(outfile)
style = ext.strip(".")
elif style is None:
style = "xyz"
if style.lower() not in write_types:
if style.lower() == "gaussian":
style = "com"
elif style.lower() == "orca":
style = "inp"
elif style.lower() == "psi4":
style = "in"
elif style.lower() == "sqm":
style = "sqmin"
elif style.lower() == "qchem":
style = "inq"
else:
raise NotImplementedError(file_type_err.format(style))
if (
outfile is None
and os.path.dirname(geom.name)
and not os.access(os.path.dirname(geom.name), os.W_OK)
):
os.makedirs(os.path.dirname(geom.name))
if style.lower() == "xyz":
out = cls.write_xyz(geom, append, outfile)
elif style.lower() == "com":
if "theory" in kwargs:
theory = kwargs["theory"]
del kwargs["theory"]
out = cls.write_com(geom, theory, outfile, **kwargs)
else:
raise TypeError(
"when writing 'com/gjf' files, **kwargs must include: theory=Aaron.Theory() (or AaronTools.Theory())"
)
elif style.lower() == "inp":
if "theory" in kwargs:
theory = kwargs["theory"]
del kwargs["theory"]
out = cls.write_inp(geom, theory, outfile, **kwargs)
else:
raise TypeError(
"when writing 'inp' files, **kwargs must include: theory=Aaron.Theory() (or AaronTools.Theory())"
)
elif style.lower() == "in":
if "theory" in kwargs:
theory = kwargs["theory"]
del kwargs["theory"]
out = cls.write_in(geom, theory, outfile, **kwargs)
else:
raise TypeError(
"when writing 'in' files, **kwargs must include: theory=Aaron.Theory() (or AaronTools.Theory())"
)
elif style.lower() == "sqmin":
if "theory" in kwargs:
theory = kwargs["theory"]
del kwargs["theory"]
out = cls.write_sqm(geom, theory, outfile, **kwargs)
else:
raise TypeError(
"when writing 'sqmin' files, **kwargs must include: theory=Aaron.Theory() (or AaronTools.Theory())"
)
elif style.lower() == "inq":
if "theory" in kwargs:
theory = kwargs["theory"]
del kwargs["theory"]
out = cls.write_inq(geom, theory, outfile, **kwargs)
else:
raise TypeError(
"when writing 'inq' files, **kwargs must include: theory=Aaron.Theory() (or AaronTools.Theory())"
)
elif style.lower() == "cube":
out = cls.write_cube(geom, outfile=outfile, **kwargs)
return out
@classmethod
def write_xyz(cls, geom, append, outfile=None):
mode = "a" if append else "w"
fmt = "{:3s} {: 10.5f} {: 10.5f} {: 10.5f}\n"
s = "%i\n" % len(geom.atoms)
s += "%s\n" % geom.comment
for atom in geom.atoms:
s += fmt.format(atom.element, *atom.coords)
if outfile is None:
# if no output file is specified, use the name of the geometry
with open(geom.name + ".xyz", mode) as f:
f.write(s)
elif outfile is False:
# if no output file is desired, just return the file contents
return s.strip()
else:
# write output to the requested destination
with open(outfile, mode) as f:
f.write(s)
return
@classmethod
def write_com(
cls, geom, theory, outfile=None, return_warnings=False, **kwargs
):
"""
write Gaussian input file for given Theory() and Geometry()
geom - Geometry()
theory - Theory()
outfile - None, False, or str
None - geom.name + ".com" is used as output destination
False - return contents of the input file as a str
str - output destination
return_warnings - True to return a list of warnings (e.g. basis
set might be misspelled
kwargs - passed to Theory methods (make_header, make_molecule, etc.)
"""
# get file content string
header, header_warnings = theory.make_header(
geom, return_warnings=True, **kwargs
)
mol, mol_warnings = theory.make_molecule(
geom, return_warnings=True, **kwargs
)
footer, footer_warnings = theory.make_footer(
geom, return_warnings=True, **kwargs
)
s = header + mol + footer
warnings = header_warnings + mol_warnings + footer_warnings
if outfile is None:
# if outfile is not specified, name file in Aaron format
if "step" in kwargs:
outfile = "{}.{}.com".format(geom.name, step2str(kwargs["step"]))
else:
outfile = "{}.com".format(geom.name)
if outfile is False:
if return_warnings:
return s, warnings
return s
else:
fname = os.path.basename(outfile)
name, ext = os.path.splitext(fname)
# could use jinja, but it's one thing...
s = s.replace("{{ name }}", name)
with open(outfile, "w") as f:
f.write(s)
if return_warnings:
return warnings
return
@classmethod
def write_inp(
cls, geom, theory, outfile=None, return_warnings=False, **kwargs
):
"""
write ORCA input file for the given Theory() and Geometry()
geom - Geometry()
theory - Theory()
outfile - None, False, or str
None - geom.name + ".inp" is used as output destination
False - return contents of the input file as a str
str - output destination
return_warnings - True to return a list of warnings (e.g. basis
set might be misspelled
kwargs - passed to Theory methods (make_header, make_molecule, etc.)
"""
fmt = "{:<3s} {: 9.5f} {: 9.5f} {: 9.5f}\n"
header, warnings = theory.make_header(
geom, style="orca", return_warnings=True, **kwargs
)
footer = theory.make_footer(
geom, style="orca", return_warnings=False, **kwargs
)
s = header
for atom in geom.atoms:
s += fmt.format(atom.element, *atom.coords)
s += "*\n"
s += footer
if outfile is None:
# if outfile is not specified, name file in Aaron format
if "step" in kwargs:
outfile = "{}.{}.inp".format(geom.name, step2str(kwargs["step"]))
else:
outfile = "{}.inp".format(geom.name)
if outfile is False:
if return_warnings:
return s, warnings
return s
else:
fname = os.path.basename(outfile)
name, ext = os.path.splitext(fname)
# could use jinja, but it's one thing...
s = s.replace("{{ name }}", name)
with open(outfile, "w") as f:
f.write(s)
if return_warnings:
return warnings
@classmethod
def write_inq(
cls, geom, theory, outfile=None, return_warnings=False, **kwargs
):
"""
write QChem input file for the given Theory() and Geometry()
geom - Geometry()
theory - Theory()
outfile - None, False, or str
None - geom.name + ".inq" is used as output destination
False - return contents of the input file as a str
str - output destination
return_warnings - True to return a list of warnings (e.g. basis
set might be misspelled
kwargs - passed to Theory methods (make_header, make_molecule, etc.)
"""
fmt = "{:<3s} {: 9.5f} {: 9.5f} {: 9.5f}\n"
header, header_warnings = theory.make_header(
geom, style="qchem", return_warnings=True, **kwargs
)
mol, mol_warnings = theory.make_molecule(
geom, style="qchem", return_warnings=True, **kwargs
)
out = header + mol
warnings = header_warnings + mol_warnings
if outfile is None:
# if outfile is not specified, name file in Aaron format
if "step" in kwargs:
outfile = "{}.{}.inq".format(geom.name, step2str(kwargs["step"]))
else:
outfile = "{}.inq".format(geom.name)
if outfile is False:
if return_warnings:
return out, warnings
return out
else:
fname = os.path.basename(outfile)
name, ext = os.path.splitext(fname)
# could use jinja, but it's one thing...
out = out.replace("{{ name }}", name)
with open(outfile, "w") as f:
f.write(out)
if return_warnings:
return warnings
@classmethod
def write_in(
cls, geom, theory, outfile=None, return_warnings=False, **kwargs
):
"""
write Psi4 input file for the given Theory() and Geometry()
geom - Geometry()
theory - Theory()
outfile - None, False, or str
None - geom.name + ".com" is used as output destination
False - return contents of the input file as a str
str - output destination
return_warnings - True to return a list of warnings (e.g. basis
set might be misspelled
kwargs - passed to Theory methods (make_header, make_molecule, etc.)
"""
header, header_warnings = theory.make_header(
geom, style="psi4", return_warnings=True, **kwargs
)
mol, mol_warnings = theory.make_molecule(
geom, style="psi4", return_warnings=True, **kwargs
)
footer, footer_warnings = theory.make_footer(
geom, style="psi4", return_warnings=True, **kwargs
)
s = header + mol + footer
warnings = header_warnings + mol_warnings + footer_warnings
if outfile is None:
# if outfile is not specified, name file in Aaron format
if "step" in kwargs:
outfile = "{}.{}.in".format(geom.name, step2str(kwargs["step"]))
else:
outfile = "{}.in".format(geom.name)
if outfile is False:
if return_warnings:
return s, warnings
return s
else:
fname = os.path.basename(outfile)
name, ext = os.path.splitext(fname)
# could use jinja, but it's one thing...
s = s.replace("{{ name }}", name)
with open(outfile, "w") as f:
f.write(s)
if return_warnings:
return warnings
@classmethod
def write_sqm(
cls, geom, theory, outfile=None, return_warnings=False, **kwargs
):
"""
write SQM input file for the given Theory() and Geometry()
geom - Geometry()
theory - Theory()
outfile - None, False, or str
None - geom.name + ".com" is used as output destination
False - return contents of the input file as a str
str - output destination
return_warnings - True to return a list of warnings (e.g. basis
set might be misspelled
kwargs - passed to Theory methods (make_header, make_molecule, etc.)
"""
header, header_warnings = theory.make_header(
geom, style="sqm", return_warnings=True, **kwargs
)
mol, mol_warnings = theory.make_molecule(
geom, style="sqm", return_warnings=True, **kwargs
)
s = header + mol
warnings = header_warnings + mol_warnings
if outfile is None:
# if outfile is not specified, name file in Aaron format
if "step" in kwargs:
outfile = "{}.{}.com".format(
geom.name, step2str(kwargs["step"])
)
else:
outfile = "{}.com".format(geom.name)
if outfile is False:
if return_warnings:
return s, warnings
return s
else:
fname = os.path.basename(outfile)
name, ext = os.path.splitext(fname)
# could use jinja, but it's one thing...
s = s.replace("{{ name }}", name)
with open(outfile, "w") as f:
f.write(s)
if return_warnings:
return warnings
@classmethod
def write_cube(
cls,
geom,
orbitals=None,
outfile=None,
kind="homo",
padding=4.0,
spacing=0.2,
alpha=True,
xyz=False,
n_jobs=1,
delta=0.1,
**kwargs,
):
"""
write a cube file for a molecular orbital
geom - geometry
orbitals - Orbitals()
outfile - output destination
mo - index of molecular orbital or "homo" for ground state
highest occupied molecular orbital or "lumo" for first
ground state unoccupied MO
can also be an array of MO coefficients
ao - index of atomic orbital to print
padding - padding around geom's coordinates
spacing - targeted spacing between points
n_jobs - number of parallel threads to use
this is on top of NumPy's multithreading, so
if NumPy uses 8 threads and n_jobs=2, you can
expect to see 16 threads in use
delta - see Orbitals.fukui_donor_value or fukui_acceptor_value
"""
if orbitals is None:
raise RuntimeError(
"no Orbitals() instance given to FileWriter.write_cube"
)
n_pts1, n_pts2, n_pts3, v1, v2, v3, com, u = orbitals.get_cube_array(
geom,
standard_axes=xyz,
spacing=spacing,
padding=padding,
)
mo = None
if kind.lower() == "homo":
mo = max(orbitals.n_alpha, orbitals.n_beta) - 1
elif kind.lower() == "lumo":
mo = max(orbitals.n_alpha, orbitals.n_beta)
elif kind.lower().startswith("mo"):
mo = int(kind.split()[-1])
elif kind.lower().startswith("ao"):
mo = np.zeros(orbitals.n_mos)
mo[int(kind.split()[-1])] = 1
s = ""
s += " %s\n" % geom.comment
s += " %s\n" % kind
# the '-' in front of the number of atoms indicates that this is
# MO info so there's an extra data entry between the molecule
# and the function values
bohr_com = com / UNIT.A0_TO_BOHR
if isinstance(mo, int):
s += " -"
else:
s += " "
s += "%i %13.5f %13.5f %13.5f 1\n" % (
len(geom.atoms), *bohr_com,
)
# the basis vectors of cube files are ordered based on the
# spacing between points along that axis
# or maybe it's the number of points?
# we use the first one
for n, v in sorted(
zip([n_pts1, n_pts2, n_pts3], [v1, v2, v3]),
key=lambda p: np.linalg.norm(p[1]),
):
bohr_v = v / UNIT.A0_TO_BOHR
s += " %5i %13.5f %13.5f %13.5f\n" % (
n, *bohr_v
)
# contruct an array of points for the grid
coords, n_list = orbitals.get_cube_points(
n_pts1, n_pts2, n_pts3, v1, v2, v3, com
)
# write the structure in bohr
for atom in geom.atoms:
s += " %5i %13.5f %13.5f %13.5f %13.5f\n" % (
ELEMENTS.index(atom.element),
ELEMENTS.index(atom.element),
atom.coords[0] / UNIT.A0_TO_BOHR,
atom.coords[1] / UNIT.A0_TO_BOHR,
atom.coords[2] / UNIT.A0_TO_BOHR,
)
# extra section - only for MO data
if isinstance(mo, int):
s += " %5i %5i\n" % (1, mo + 1)
# get values for this MO
if kind.lower() == "density":
val = orbitals.density_value(coords, n_jobs=n_jobs)
elif kind.lower() == "fukui donor":
val = orbitals.fukui_donor_value(
coords, n_jobs=n_jobs, delta=delta
)
elif kind.lower() == "fukui acceptor":
val = orbitals.fukui_acceptor_value(
coords, n_jobs=n_jobs, delta=delta
)
elif kind.lower() == "fukui dual":
val = orbitals.fukui_dual_value(
coords, n_jobs=n_jobs, delta=delta
)
else:
val = orbitals.mo_value(mo, coords, n_jobs=n_jobs)
# write to a file
for n1 in range(0, n_list[0]):
for n2 in range(0, n_list[1]):
val_ndx = n1 * n_list[2] * n_list[1] + n2 * n_list[2]
val_subset = val[val_ndx : val_ndx + n_list[2]]
for i, v in enumerate(val_subset):
if abs(v) < 1e-30:
v = 0
s += "%13.5e" % v
if (i + 1) % 6 == 0:
s += "\n"
if (i + 1) % 6 != 0:
s += "\n"
if outfile is None:
# if no output file is specified, use the name of the geometry
with open(geom.name + ".cube", "w") as f:
f.write(s)
elif outfile is False:
# if no output file is desired, just return the file contents
return s
else:
# write output to the requested destination
with open(outfile, "w") as f:
f.write(s)
return
@addlogger
class FileReader:
"""
Attributes:
name ''
file_type ''
comment ''
atoms [Atom]
other {}
"""
LOG = None
LOGLEVEL = "DEBUG"
def __init__(
self,
fname,
get_all=False,
just_geom=True,
freq_name=None,
conf_name=None,
nbo_name=None,
max_length=10000000,
):
"""
:fname: either a string specifying the file name of the file to read
or a tuple of (str(name), str(file_type), str(content))
:get_all: if true, optimization steps are also saved in
self.all_geom; otherwise only saves last geometry
:just_geom: if true, does not store other information, such as
frequencies, only what is needed to construct a Geometry() obj
:freq_name: Name of the file containing the frequency output. Only use
if this information is in a different file than `fname` (eg: xtb runs
using the --hess runtype option)
:nbo_name: Name of the file containing the NBO orbital coefficients
in the AO basis. Only used when reading *.47 files.
:max_length: maximum array size to store from FCHK files
any array that would be larger than this will be the
size the array would be
"""
# Initialization
self.name = ""
self.file_type = ""
self.comment = ""
self.atoms = []
self.other = {}
self.content = None
self.all_geom = None
# get file name and extention
if isinstance(fname, str):
self.name, self.file_type = os.path.splitext(fname)
self.file_type = self.file_type.lower()[1:]
elif isinstance(fname, (tuple, list)):
self.name = fname[0]
self.file_type = fname[1]
self.content = fname[2]
if self.file_type not in read_types:
raise NotImplementedError(file_type_err.format(self.file_type))
# Fill in attributes with geometry information
if self.content is None:
self.read_file(
get_all, just_geom,
freq_name=freq_name,
conf_name=conf_name,
nbo_name=nbo_name,
max_length=max_length,
)
elif isinstance(self.content, str):
f = StringIO(self.content)
elif isinstance(self.content, IOBase):
f = self.content
if self.content is not None:
if self.file_type == "log":
self.read_log(f, get_all, just_geom)
elif any(self.file_type == ext for ext in ["sd", "sdf", "mol"]):
self.read_sd(f)
elif self.file_type == "xyz":
self.read_xyz(f, get_all)
elif self.file_type == "mol2":
self.read_mol2(f, get_all)
elif any(self.file_type == ext for ext in ["com", "gjf"]):
self.read_com(f)
elif self.file_type == "out":
self.read_orca_out(f, get_all, just_geom)
elif self.file_type == "dat":
self.read_psi4_out(f, get_all, just_geom)
elif self.file_type == "fchk":
self.read_fchk(f, just_geom, max_length=max_length)
elif self.file_type == "crest":
self.read_crest(f, conf_name=conf_name)
elif self.file_type == "xtb":
self.read_xtb(f, freq_name=freq_name)
elif self.file_type == "sqmout":
self.read_sqm(f)
elif self.file_type == "47":
self.read_nbo_47(f, nbo_name=nbo_name)
elif self.file_type == "31":
self.read_nbo_31(f, nbo_name=nbo_name)
elif self.file_type == "qout":
self.read_qchem_out(f, get_all, just_geom)
def read_file(
self, get_all=False, just_geom=True,
freq_name=None, conf_name=None, nbo_name=None,
max_length=10000000,
):
"""
Reads geometry information from fname.
Parameters:
get_all If false (default), only keep the last geom
If true, self is last geom, but return list
of all others encountered
nbo_name nbo output file containing coefficients to
map AO's to orbitals
max_length max. array size for arrays to store in FCHK
files - anything larger will be the size
the array would be
"""
if os.path.isfile(self.name):
f = open(self.name)
else:
fname = ".".join([self.name, self.file_type])
fname = os.path.expanduser(fname)
if os.path.isfile(fname):
f = open(fname)
else:
raise FileNotFoundError(
"Error while looking for %s: could not find %s or %s in %s"
% (self.name, fname, self.name, os.getcwd())
)
if self.file_type == "xyz":
self.read_xyz(f, get_all)
elif self.file_type == "log":
self.read_log(f, get_all, just_geom)
elif any(self.file_type == ext for ext in ["com", "gjf"]):
self.read_com(f)
elif any(self.file_type == ext for ext in ["sd", "sdf", "mol"]):
self.read_sd(f)
elif self.file_type == "mol2":
self.read_mol2(f)
elif self.file_type == "out":
self.read_orca_out(f, get_all, just_geom)
elif self.file_type == "dat":
self.read_psi4_out(f, get_all, just_geom)
elif self.file_type == "fchk":
self.read_fchk(f, just_geom, max_length=max_length)
elif self.file_type == "crest":
self.read_crest(f, conf_name=conf_name)
elif self.file_type == "xtb":
self.read_xtb(f, freq_name=freq_name)
elif self.file_type == "sqmout":
self.read_sqm(f)
elif self.file_type == "47":
self.read_nbo_47(f, nbo_name=nbo_name)
elif self.file_type == "31":
self.read_nbo_31(f, nbo_name=nbo_name)
elif self.file_type == "qout":
self.read_qchem_out(f, get_all, just_geom)
f.close()
return
def skip_lines(self, f, n):
for i in range(n):
f.readline()
return
def read_xyz(self, f, get_all=False):
self.all_geom = []
# number of atoms
f.readline()
# comment
self.comment = f.readline().strip()
# atom info
atom_count = 0
for line in f:
line = line.strip()
if line == "":
continue
try:
int(line)
if get_all:
self.all_geom += [
(deepcopy(self.comment), deepcopy(self.atoms))
]
self.comment = f.readline().strip()
self.atoms = []
atom_count = 0
except ValueError:
line = line.split()
atom_count += 1
self.atoms += [Atom(element=line[0], coords=line[1:4], name=str(atom_count))]
# if get_all:
# self.all_geom += [(deepcopy(self.comment), deepcopy(self.atoms))]
def read_sd(self, f, get_all=False):
self.all_geom = []
lines = f.readlines()
progress = 0
for i, line in enumerate(lines):
progress += 1
if "$$$$" in line:
progress = 0
if get_all:
self.all_geom.append(
[deepcopy(self.comment), deepcopy(self.atoms)]
)
continue
if progress == 3:
self.comment = line.strip()
if progress == 4:
counts = line.split()
natoms = int(counts[0])
nbonds = int(counts[1])
if progress == 5:
self.atoms = []
for line in lines[i : i + natoms]:
atom_info = line.split()
self.atoms += [
Atom(element=atom_info[3], coords=atom_info[0:3])
]
for line in lines[i + natoms : i + natoms + nbonds]:
a1, a2 = [int(x) - 1 for x in line.split()[0:2]]
self.atoms[a1].connected.add(self.atoms[a2])
self.atoms[a2].connected.add(self.atoms[a1])
for j, a in enumerate(self.atoms):
a.name = str(j + 1)
self.other["charge"] = 0
for line in lines[i + natoms + nbonds:]:
if "CHG" in line:
self.other["charge"] += int(line.split()[-1])
if "$$$$" in line:
break
def read_mol2(self, f, get_all=False):
"""
read TRIPOS mol2
"""
atoms = []
lines = f.readlines()
i = 0
while i < len(lines):
if lines[i].startswith("@<TRIPOS>MOLECULE"):
self.comment = lines[i + 1]
info = lines[i + 2].split()
n_atoms = int(info[0])
n_bonds = int(info[1])
i += 3
elif lines[i].startswith("@<TRIPOS>ATOM"):
for j in range(0, n_atoms):
i += 1
info = lines[i].split()
# name = info[1]
coords = np.array([float(x) for x in info[2:5]])
element = re.match("([A-Za-z]+)", info[5]).group(1)
atoms.append(
Atom(element=element, coords=coords, name=str(j + 1))
)
self.atoms = atoms
elif lines[i].startswith("@<TRIPOS>BOND"):
for j in range(0, n_bonds):
i += 1
info = lines[i].split()
a1, a2 = [int(ndx) - 1 for ndx in info[1:3]]
self.atoms[a1].connected.add(self.atoms[a2])
self.atoms[a2].connected.add(self.atoms[a1])
i += 1
def read_psi4_out(self, f, get_all=False, just_geom=True):
uv_vis = ""
def get_atoms(f, n):
rv = []
self.skip_lines(f, 1)
n += 2
line = f.readline()
i = 0
mass = 0
while line.strip():
i += 1
line = line.strip()
atom_info = line.split()
element = atom_info[0]
# might be a ghost atom - like for sapt
if "Gh" in element:
element = element.strip("Gh(").strip(")")
coords = np.array([float(x) for x in atom_info[1:-1]])
rv += [Atom(element=element, coords=coords, name=str(i))]
mass += float(atom_info[-1])
line = f.readline()
n += 1
return rv, mass, n
line = f.readline()
n = 1
read_geom = False
while line != "":
if "* O R C A *" in line:
self.file_type = "out"
return self.read_orca_out(
f, get_all=get_all, just_geom=just_geom
)
if "A Quantum Leap Into The Future Of Chemistry" in line:
self.file_type = "qout"
return self.read_qchem_out(
f, get_all=get_all, just_geom=just_geom
)
if line.startswith(" Geometry (in Angstrom), charge"):
if not just_geom:
self.other["charge"] = int(line.split()[5].strip(","))
self.other["multiplicity"] = int(
line.split()[8].strip(":")
)
elif line.strip() == "SCF":
read_geom = True
elif line.strip().startswith("Center") and read_geom:
read_geom = False
if get_all and len(self.atoms) > 0:
if self.all_geom is None:
self.all_geom = []
self.all_geom += [
(deepcopy(self.atoms), deepcopy(self.other))
]
self.atoms, mass, n = get_atoms(f, n)
if not just_geom:
self.other["mass"] = mass
self.other["mass"] *= UNIT.AMU_TO_KG
if just_geom:
line = f.readline()
n += 1
continue
else:
if line.strip().startswith("Total Energy ="):
self.other["energy"] = float(line.split()[-1])
elif line.strip().startswith("Total E0"):
self.other["energy"] = float(line.split()[-2])
elif line.strip().startswith("Correction ZPE"):
self.other["ZPVE"] = float(line.split()[-4])
elif line.strip().startswith("Total ZPE"):
self.other["E_ZPVE"] = float(line.split()[-2])
elif line.strip().startswith("Total H, Enthalpy"):
self.other["enthalpy"] = float(line.split()[-2])
elif line.strip().startswith("Total G, Free"):
self.other["free_energy"] = float(line.split()[-2])
self.other["temperature"] = float(line.split()[-4])
elif "symmetry no. =" in line:
self.other["rotational_symmetry_number"] = int(
line.split()[-1].strip(")")
)
elif (
line.strip().startswith("Rotational constants:")
and line.strip().endswith("[cm^-1]")
and "rotational_temperature" not in self.other
):
self.other["rotational_temperature"] = [
float(x) if is_num(x) else 0
for x in line.split()[-8:-1:3]
]
self.other["rotational_temperature"] = [
x
* PHYSICAL.SPEED_OF_LIGHT
* PHYSICAL.PLANCK
/ PHYSICAL.KB
for x in self.other["rotational_temperature"]
]
elif line.startswith(" Vibration "):
freq_str = ""
while not line.strip().startswith("=="):
freq_str += line
line = f.readline()
n += 1
self.other["frequency"] = Frequency(
freq_str, hpmodes=False, style="psi4"
)
elif PSI4_NORM_FINISH in line:
self.other["finished"] = True
elif line.startswith(" Convergence Criteria"):
# for tolerances:
# psi4 puts '*' next to converged values and 'o' in place of things that aren't monitored
grad = {}
dE_tol = line[24:38]
if "o" in dE_tol:
dE_tol = None
else:
dE_tol = dE_tol.split()[0]
max_f_tol = line[38:52]
if "o" in max_f_tol:
max_f_tol = None
else:
max_f_tol = max_f_tol.split()[0]
rms_f_tol = line[52:66]
if "o" in rms_f_tol:
rms_f_tol = None
else:
rms_f_tol = rms_f_tol.split()[0]
max_d_tol = line[66:80]
if "o" in max_d_tol:
max_d_tol = None
else:
max_d_tol = max_d_tol.split()[0]
rms_d_tol = line[80:94]
if "o" in rms_d_tol:
rms_d_tol = None
else:
rms_d_tol = rms_d_tol.split()[0]
line = f.readline()
line = f.readline()
n += 2
# for convergence:
# psi4 puts '*' next to converged values and 'o' next to things that aren't monitored
if dE_tol is not None:
dE_conv = line[24:38]
dE = float(dE_conv.split()[0])
grad["Delta E"] = {}
grad["Delta E"]["value"] = dE
grad["Delta E"]["converged"] = "*" in dE_conv
if max_f_tol is not None:
max_f_conv = line[38:52]
max_f = float(max_f_conv.split()[0])
grad["Max Force"] = {}
grad["Max Force"]["value"] = max_f
grad["Max Force"]["converged"] = "*" in max_f_conv
if rms_f_tol is not None:
rms_f_conv = line[52:66]
rms_f = float(rms_f_conv.split()[0])
grad["RMS Force"] = {}
grad["RMS Force"]["value"] = rms_f
grad["RMS Force"]["converged"] = "*" in rms_f_conv
if max_d_tol is not None:
max_d_conv = line[66:80]
max_d = float(max_d_conv.split()[0])
grad["Max Disp"] = {}
grad["Max Disp"]["value"] = max_d
grad["Max Disp"]["converged"] = "*" in max_d_conv
if rms_d_tol is not None:
rms_d_conv = line[80:94]
rms_d = float(rms_d_conv.split()[0])
grad["RMS Disp"] = {}
grad["RMS Disp"]["value"] = rms_d
grad["RMS Disp"]["converged"] = "*" in max_d_conv
self.other["gradient"] = grad
elif "Total Gradient" in line:
gradient = np.zeros((len(self.atoms), 3))
self.skip_lines(f, 2)
n += 2
for i in range(0, len(self.atoms)):
n += 1
line = f.readline()
info = line.split()
gradient[i] = np.array([float(x) for x in info[1:]])
self.other["forces"] = -gradient
elif "SAPT Results" in line:
self.skip_lines(f, 1)
n += 1
while "Total sSAPT" not in line:
n += 1
line = f.readline()
if "---" in line:
break
if len(line.strip()) > 0:
if "Special recipe" in line:
continue
item = line[:26].strip()
val = 1e-3 * float(line[34:47])
self.other[item] = val
elif "SCF energy" in line:
self.other["SCF energy"] = float(line.split()[-1])
elif "correlation energy" in line and "=" in line:
item = line.split("=")[0].strip()
self.other[item] = float(line.split()[-1])
elif "Full point group" in line:
self.other["full_point_group"] = line.split()[-1]
elif "Molecular point group" in line:
self.other["molecular_point_group"] = line.split()[-1]
elif (
"total energy" in line
and "=" in line
or re.search("\(.\) energy", line)
):
item = line.split("=")[0].strip().strip("*").strip()
self.other[item] = float(line.split()[-1])
# hopefully the highest level energy gets printed last
self.other["energy"] = self.other[item]
elif "Total Energy" in line and "=" in line:
item = line.split("=")[0].strip().strip("*").strip()
self.other[item] = float(line.split()[-2])
# hopefully the highest level energy gets printed last
self.other["energy"] = self.other[item]
elif "Correlation Energy" in line and "=" in line:
item = line.split("=")[0].strip().strip("*").strip()
if "DFT Exchange-Correlation" in item:
self.other[item] = float(line.split()[-1])
else:
self.other[item] = float(line.split()[-2])
elif "Ground State -> Excited State Transitions" in line:
self.skip_lines(f, 3)
n += 3
line = f.readline()
s = ""
while line.strip():
s += line
n += 1
line = f.readline()
self.other["uv_vis"] = ValenceExcitations(s, style="psi4")
elif "Excitation Energy" in line and "Rotatory" in line:
self.skip_lines(f, 2)
n += 2
line = f.readline()
s = ""
while line.strip():
s += line
n += 1
line = f.readline()
self.other["uv_vis"] = ValenceExcitations(s, style="psi4")
elif re.search("\| State\s*\d+", line):
# read energies from property calculation
uv_vis += line
elif "Excited state properties:" in line:
# read osc str or rotation from property calculation
while line.strip():
uv_vis += line
n += 1
line = f.readline()
if "Oscillator" in uv_vis or "Rotation" in uv_vis:
self.other["uv_vis"] = ValenceExcitations(uv_vis, style="psi4")
if "error" not in self.other:
for err in ERROR_PSI4:
if err in line:
self.other["error"] = ERROR_PSI4[err]
self.other["error_msg"] = line.strip()
line = f.readline()
n += 1
def read_orca_out(self, f, get_all=False, just_geom=True):
"""read orca output file"""
nrg_regex = re.compile("(?:[A-Za-z]+\s+)?E\((.*)\)\s*\.\.\.\s*(.*)$")
def add_grad(grad, name, line):
grad[name] = {}
grad[name]["value"] = line.split()[-3]
grad[name]["converged"] = line.split()[-1] == "YES"
def get_atoms(f, n):
"""parse atom info"""
rv = []
self.skip_lines(f, 1)
n += 2
line = f.readline()
i = 0
while line.strip():
i += 1
line = line.strip()
atom_info = line.split()
element = atom_info[0]
coords = np.array([float(x) for x in atom_info[1:]])
rv += [Atom(element=element, coords=coords, name=str(i))]
line = f.readline()
n += 1
return rv, n
line = f.readline()
n = 1
while line != "":
if (
"Psi4: An Open-Source Ab Initio Electronic Structure Package"
in line
):
self.file_type = "dat"
return self.read_psi4_out(
f, get_all=get_all, just_geom=just_geom
)
if (
"A Quantum Leap Into The Future Of Chemistry"
in line
):
self.file_type = "qout"
return self.read_qchem_out(
f, get_all=get_all, just_geom=just_geom
)
if line.startswith("CARTESIAN COORDINATES (ANGSTROEM)"):
if get_all and len(self.atoms) > 0:
if self.all_geom is None:
self.all_geom = []
self.all_geom += [
(deepcopy(self.atoms), deepcopy(self.other))
]
self.atoms, n = get_atoms(f, n)
if just_geom:
line = f.readline()
n += 1
continue
else:
nrg = nrg_regex.match(line)
if nrg is not None:
nrg_type = nrg.group(1)
# for some reason, ORCA prints MP2 correlation energy
# as E(MP2) for CC jobs
if nrg_type == "MP2":
nrg_type = "MP2 CORR"
self.other["E(%s)" % nrg_type] = float(nrg.group(2))
if line.startswith("FINAL SINGLE POINT ENERGY"):
# if the wavefunction doesn't converge, ORCA prints a message next
# to the energy so we can't use line.split()[-1]
self.other["energy"] = float(line.split()[4])
if line.startswith("TOTAL SCF ENERGY"):
self.skip_lines(f, 2)
line = f.readline()
n += 3
self.other["SCF energy"] = float(line.split()[3])
elif "TOTAL ENERGY:" in line:
item = line.split()[-5] + " energy"
self.other[item] = float(line.split()[-2])
elif "CORRELATION ENERGY" in line and "Eh" in line:
item = line.split()[-6] + " correlation energy"
self.other[item] = float(line.split()[-2])
elif re.match("E\(\S+\)\s+...\s+-?\d+\.\d+$", line):
nrg = re.match("(E\(\S+\))\s+...\s+(-?\d+\.\d+)$", line)
self.other["energy"] = float(nrg.group(2))
self.other[nrg.group(1)] = float(nrg.group(2))
elif line.startswith("CARTESIAN GRADIENT"):
gradient = np.zeros((len(self.atoms), 3))
self.skip_lines(f, 2)
n += 2
for i in range(0, len(self.atoms)):
n += 1
line = f.readline()
# orca prints a warning before gradient if some
# coordinates are constrained
if line.startswith("WARNING:"):
continue
info = line.split()
gradient[i] = np.array([float(x) for x in info[3:]])
self.other["forces"] = -gradient
elif line.startswith("VIBRATIONAL FREQUENCIES"):
stage = "frequencies"
freq_str = "VIBRATIONAL FREQUENCIES\n"
self.skip_lines(f, 4)
n += 5
line = f.readline()
while not (stage == "THERMO" and line == "\n") and line:
if "--" not in line and line != "\n":
freq_str += line
if "NORMAL MODES" in line:
stage = "modes"
self.skip_lines(f, 6)
n += 6
if "RAMAN SPECTRUM" in line:
stage = "RAMAN"
self.skip_lines(f, 2)
n += 2
if "IR SPECTRUM" in line:
stage = "IR"
self.skip_lines(f, 2)
n += 2
if "THERMOCHEMISTRY" in line:
stage = "THERMO"
n += 1
line = f.readline()
self.other["frequency"] = Frequency(
freq_str, hpmodes=False, style="orca"
)
elif line.startswith("Temperature"):
self.other["temperature"] = float(line.split()[2])
elif line.startswith("Total Mass"):
# this may only get printed for freq jobs
self.other["mass"] = float(line.split()[3])
self.other["mass"] *= UNIT.AMU_TO_KG
elif line.startswith(" Total Charge"):
self.other["charge"] = int(line.split()[-1])
elif line.startswith(" Multiplicity"):
self.other["multiplicity"] = int(line.split()[-1])
elif "rotational symmetry number" in line:
# TODO: make this cleaner
self.other["rotational_symmetry_number"] = int(
line.split()[-2]
)
elif "Symmetry Number:" in line:
self.other["rotational_symmetry_number"] = int(
line.split()[-1]
)
elif line.startswith("Zero point energy"):
self.other["ZPVE"] = float(line.split()[4])
elif line.startswith("Total Enthalpy"):
self.other["enthalpy"] = float(line.split()[3])
elif line.startswith("Final Gibbs"):
# NOTE - Orca seems to only print Grimme's Quasi-RRHO free energy
# RRHO can be computed in AaronTool's CompOutput by setting the w0 to 0
self.other["free_energy"] = float(line.split()[5])
elif line.startswith("Rotational constants in cm-1:"):
# orca doesn't seem to print rotational constants in older versions
self.other["rotational_temperature"] = [
float(x) for x in line.split()[-3:]
]
self.other["rotational_temperature"] = [
x
* PHYSICAL.SPEED_OF_LIGHT
* PHYSICAL.PLANCK
/ PHYSICAL.KB
for x in self.other["rotational_temperature"]
]
elif "Point Group:" in line:
self.other["full_point_group"] = line.split()[2][:-1]
elif "Symmetry Number" in line:
self.other["rotational_symmetry_number"] = int(
line.split()[-1]
)
elif "sn is the rotational symmetry number" in line:
# older versions of orca print this differently
self.other["rotational_symmetry_number"] = int(
line.split()[-2]
)
elif "Geometry convergence" in line:
grad = {}
self.skip_lines(f, 2)
n += 3
line = f.readline()
while line and re.search("\w", line):
if re.search("Energy\schange", line):
add_grad(grad, "Delta E", line)
elif re.search("RMS\sgradient", line):
add_grad(grad, "RMS Force", line)
elif re.search("MAX\sgradient", line):
add_grad(grad, "Max Force", line)
elif re.search("RMS\sstep", line):
add_grad(grad, "RMS Disp", line)
elif re.search("MAX\sstep", line):
add_grad(grad, "Max Disp", line)
line = f.readline()
n += 1
self.other["gradient"] = grad
elif "MAYER POPULATION ANALYSIS" in line:
self.skip_lines(f, 2)
n += 2
line = f.readline()
data = dict()
headers = []
while line.strip():
info = line.split()
header = info[0]
name = " ".join(info[2:])
headers.append(header)
data[header] = (name, [])
line = f.readline()
self.skip_lines(f, 1)
n += 1
for i in range(0, len(self.atoms)):
line = f.readline()
info = line.split()[2:]
for header, val in zip(headers, info):
data[header][1].append(float(val))
for header in headers:
self.other[data[header][0]] = np.array(data[header][1])
elif line.startswith("LOEWDIN ATOMIC CHARGES"):
self.skip_lines(f, 1)
n += 1
charges = np.zeros(len(self.atoms))
for i in range(0, len(self.atoms)):
line = f.readline()
n += 1
charges[i] = float(line.split()[-1])
self.other["Löwdin Charges"] = charges
elif line.startswith("BASIS SET IN INPUT FORMAT"):
# read basis set primitive info
self.skip_lines(f, 3)
n += 3
line = f.readline()
n += 1
self.other["basis_set_by_ele"] = dict()
while "--" not in line and line != "":
new_gto = re.search("NewGTO\s+(\S+)", line)
if new_gto:
ele = new_gto.group(1)
line = f.readline()
n += 1
primitives = []
while "end" not in line and line != "":
shell_type, n_prim = line.split()
n_prim = int(n_prim)
exponents = []
con_coeffs = []
for i in range(0, n_prim):
line = f.readline()
n += 1
info = line.split()
exponent = float(info[1])
con_coeff = [float(x) for x in info[2:]]
exponents.append(exponent)
con_coeffs.extend(con_coeff)
primitives.append(
(
shell_type,
n_prim,
exponents,
con_coeffs,
)
)
line = f.readline()
n += 1
self.other["basis_set_by_ele"][ele] = primitives
line = f.readline()
n += 1
elif "EXCITED STATES" in line or re.search("STEOM.* RESULTS", line) or line.startswith("APPROXIMATE EOM LHS"):
s = ""
done = False
while not done:
s += line
n += 1
line = f.readline()
if (
"ORCA-CIS/TD-DFT FINISHED WITHOUT ERROR" in line or
re.search("TDM done", line) or
"TIMINGS" in line or
line == ""
):
done = True
self.other["uv_vis"] = ValenceExcitations(s, style="orca")
elif line.startswith("MOLECULAR ORBITALS"):
# read molecular orbitals
self.skip_lines(f, 1)
n += 1
line = f.readline()
self.other["alpha_coefficients"] = []
self.other["beta_coefficients"] = []
self.other["alpha_nrgs"] = []
self.other["beta_nrgs"] = []
self.other["alpha_occupancies"] = []
self.other["beta_occupancies"] = []
at_info = re.compile(
"\s*(\d+)\S+\s+\d+(?:s|p[xyz]|d(?:z2|xz|yz|x2y2|xy)|[fghi][\+\-]?\d+)"
)
if self.other["multiplicity"] != 1:
args = [
("alpha_coefficients", "beta_coefficients"),
("alpha_nrgs", "beta_nrgs"),
("alpha_occupancies", "beta_occupancies"),
]
else:
args = [
("alpha_coefficients",),
("alpha_nrgs",),
("alpha_occupancies",),
]
for coeff_name, nrg_name, occ_name in zip(*args):
self.other["shell_to_atom"] = []
mo_coefficients = []
orbit_nrgs = []
occupancy = []
while line.strip() != "":
at_match = at_info.match(line)
if at_match:
ndx = int(at_match.group(1))
self.other["shell_to_atom"].append(ndx)
coeffs = []
# there might not always be a space between the coefficients
# so we can't just split(), but they are formatted(-ish)
for coeff in re.findall("-?\d+\.\d\d\d\d\d\d", line[16:]):
coeffs.append(float(coeff))
for coeff, mo in zip(coeffs, mo_coefficients):
mo.append(coeff)
elif "--" not in line:
orbit_nrgs = occupancy
occupancy = [float(x) for x in line.split()]
elif "--" in line:
self.other[nrg_name].extend(orbit_nrgs)
self.other[occ_name].extend(occupancy)
if mo_coefficients:
self.other[coeff_name].extend(
mo_coefficients
)
if not all(
len(coeff) == len(mo_coefficients[0])
for coeff in mo_coefficients
):
self.LOG.warning(
"orbital coefficients may not "
"have been parsed correctly"
)
mo_coefficients = [[] for x in orbit_nrgs]
orbit_nrgs = []
line = f.readline()
n += 1
self.other[coeff_name].extend(mo_coefficients)
line = f.readline()
elif line.startswith("N(Alpha) "):
self.other["n_alpha"] = int(
np.rint(float(line.split()[2]))
)
elif line.startswith("N(Beta) "):
self.other["n_beta"] = int(np.rint(float(line.split()[2])))
elif ORCA_NORM_FINISH in line:
self.other["finished"] = True
# TODO E_ZPVE
if "error" not in self.other:
for err in ERROR_ORCA:
if err in line:
self.other["error"] = ERROR_ORCA[err]
self.other["error_msg"] = line.strip()
break
line = f.readline()
n += 1
if not just_geom:
if "finished" not in self.other:
self.other["finished"] = False
if (
"alpha_coefficients" in self.other
and "basis_set_by_ele" in self.other
):
self.other["orbitals"] = Orbitals(self)
def read_qchem_out(self, f, get_all=False, just_geom=True):
"""read qchem output file"""
def get_atoms(f, n):
"""parse atom info"""
rv = []
self.skip_lines(f, 2)
n += 1
line = f.readline()
i = 0
while "--" not in line:
i += 1
line = line.strip()
atom_info = line.split()
element = atom_info[1]
coords = np.array([float(x) for x in atom_info[2:5]])
rv += [Atom(element=element, coords=coords, name=str(i))]
line = f.readline()
n += 1
return rv, n
def add_grad(grad, name, line):
grad[name] = {}
grad[name]["value"] = line.split()[-3]
grad[name]["converged"] = line.split()[-1] == "YES"
line = f.readline()
n = 1
while line != "":
if (
"Psi4: An Open-Source Ab Initio Electronic Structure Package"
in line
):
self.file_type = "dat"
return self.read_psi4_out(
f, get_all=get_all, just_geom=just_geom
)
if "* O R C A *" in line:
self.file_type = "out"
return self.read_orca_out(
f, get_all=get_all, just_geom=just_geom
)
if (
"A Quantum Leap Into The Future Of Chemistry"
in line
):
self.file_type = "qout"
return self.read_qchem_out(
f, get_all=get_all, just_geom=just_geom
)
if "Standard Nuclear Orientation (Angstroms)" in line:
if get_all and len(self.atoms) > 0:
if self.all_geom is None:
self.all_geom = []
self.all_geom += [
(deepcopy(self.atoms), deepcopy(self.other))
]
self.atoms, n = get_atoms(f, n)
if just_geom:
line = f.readline()
n += 1
continue
else:
if "energy in the final basis set" in line:
self.other["energy"] = float(line.split()[-1])
if "SCF" in line:
self.other["scf_energy"] = self.other["energy"]
if re.search(r"energy\s+=\s+-?\d+\.\d+", line):
info = re.search(r"\s*([\S\s]+)\s+energy\s+=\s+(-?\d+\.\d+)", line)
kind = info.group(1)
if len(kind.split()) <= 2:
val = float(info.group(2))
if "correlation" not in kind and len(kind.split()) <= 2:
self.other["E(%s)" % kind.split()[0]] = val
self.other["energy"] = val
else:
self.other["E(corr)(%s)" % kind.split()[0]] = val
if "Total energy:" in line:
self.other["energy"] = float(line.split()[-2])
#MPn energy is printed as EMPn(SDQ)
if re.search("EMP\d(?:[A-Z]+)?\s+=\s*-?\d+.\d+$", line):
self.other["energy"] = float(line.split()[-1])
self.other["E(%s)" % line.split()[0][1:]] = self.other["energy"]
if "Molecular Point Group" in line:
self.other["full_point_group"] = line.split()[3]
if "Largest Abelian Subgroup" in line:
self.other["abelian_subgroup"] = line.split()[3]
if "Ground-State Mulliken Net Atomic Charges" in line:
charges = []
self.skip_lines(f, 3)
n += 2
line = f.readline()
while "--" not in line:
charge = float(line.split()[-1])
charges.append(charge)
line = f.readline()
n += 1
self.other["Mulliken Charges"] = charges
if "Cnvgd?" in line:
grad = {}
line = f.readline()
while line and re.search("\w", line):
if re.search("Energy\schange", line):
add_grad(grad, "Delta E", line)
elif re.search("Displacement", line):
add_grad(grad, "Disp", line)
elif re.search("Gradient", line):
add_grad(grad, "Max Disp", line)
line = f.readline()
n += 1
self.other["gradient"] = grad
if "VIBRATIONAL ANALYSIS" in line:
freq_str = ""
self.skip_lines(f, 10)
n += 9
line = f.readline()
while "STANDARD THERMODYNAMIC QUANTITIES" not in line:
n += 1
freq_str += line
line = f.readline()
self.other["frequency"] = Frequency(
freq_str, style="qchem",
)
self.other["temperature"] = float(line.split()[4])
if "Rotational Symmetry Number is" in line:
self.other["rotational_symmetry_number"] = int(line.split()[-1])
if "Molecular Mass:" in line:
self.other["mass"] = float(line.split()[-2]) * UNIT.AMU_TO_KG
if "$molecule" in line.lower():
line = f.readline()
while "$end" not in line.lower() and line:
if re.search("\d+\s+\d+", line):
match = re.search("^\s*(\d+)\s+(\d+)\s*$", line)
self.other["charge"] = int(match.group(1))
self.other["multiplicity"] = int(match.group(2))
break
line = f.readline()
if "Principal axes and moments of inertia" in line:
self.skip_lines(f, 1)
line = f.readline()
rot_consts = np.array([
float(x) for x in line.split()[2:]
])
rot_consts *= UNIT.AMU_TO_KG
rot_consts *= UNIT.A0_TO_BOHR ** 2
rot_consts *= 1e-20
rot_consts = PHYSICAL.PLANCK ** 2 / (8 * np.pi ** 2 * rot_consts * PHYSICAL.KB)
self.other["rotational_temperature"] = rot_consts
if line.startswith("Mult"):
self.other["multiplicity"] = int(line.split()[1])
# TD-DFT excitations
if re.search("TDDFT.* Excitation Energies", line):
excite_s = ""
self.skip_lines(f, 2)
line = f.readline()
n += 3
while "---" not in line and line:
excite_s += line
line = f.readline()
n += 1
self.other["uv_vis"] = ValenceExcitations(
excite_s, style="qchem",
)
# ADC excitations
if re.search("Excited State Summary", line):
excite_s = ""
self.skip_lines(f, 2)
line = f.readline()
n += 3
while "===" not in line and line:
excite_s += line
line = f.readline()
n += 1
self.other["uv_vis"] = ValenceExcitations(
excite_s, style="qchem",
)
# EOM excitations
if re.search("Start computing the transition properties", line):
excite_s = ""
line = f.readline()
n += 1
while "All requested transition properties have been computed" not in line and line:
excite_s += line
line = f.readline()
n += 1
self.other["uv_vis"] = ValenceExcitations(
excite_s, style="qchem",
)
if "Thank you very much for using Q-Chem" in line:
self.other["finished"] = True
line = f.readline()
n += 1
if not just_geom and "finished" not in self.other:
self.other["finished"] = False
def read_log(self, f, get_all=False, just_geom=True):
def get_atoms(f, n):
rv = self.atoms
self.skip_lines(f, 4)
line = f.readline()
n += 5
atnum = 0
while "--" not in line:
line = line.strip()
line = line.split()
for l in line:
try:
float(l)
except ValueError:
msg = "Error detected with log file on line {}"
raise IOError(msg.format(n))
try:
rv[atnum].coords = np.array(line[3:], dtype=float)
except IndexError:
pass
#print(atnum)
atnum += 1
line = f.readline()
n += 1
return rv, n
def get_input(f, n):
rv = []
line = f.readline()
n += 1
match = re.search(
"Charge\s*=\s*(-?\d+)\s*Multiplicity\s*=\s*(\d+)", line
)
if match is not None:
self.other["charge"] = int(match.group(1))
self.other["multiplicity"] = int(match.group(2))
line = f.readline()
n += 1
a = 0
while len(line.split()) > 1:
line = line.split()
if len(line) == 5:
flag = not(bool(line[1]))
a += 1
rv += [Atom(element=line[0], flag=flag, coords=line[2:], name=str(a))]
elif len(line) == 4:
a += 1
rv += [Atom(element=line[0], coords=line[1:], name=str(a))]
line = f.readline()
n += 1
return rv, n
def get_params(f, n):
rv = []
self.skip_lines(f, 2)
n += 3
line = f.readline()
if "Definition" in line:
definition = True
else:
definition = False
self.skip_lines(f, 1)
n += 2
line = f.readline()
while "--" not in line:
line = line.split()
param = line[1]
if definition:
val = float(line[3])
else:
val = float(line[2])
rv.append((param, val))
line = f.readline()
n += 1
return rv, n
def get_modredundant(f, n):
"""read constraints for modredundant section"""
rv = {}
line = f.readline()
n += 1
while line.strip():
atom_match = re.search("X\s+(\d+)\s+F", line)
bond_match = re.search("B\s+(\d+)\s+(\d+)\s+F", line)
angle_match = re.search("A\s+(\d+)\s+(\d+)\s+(\d+)\s+F", line)
torsion_match = re.search(
"D\s+(\d+)\s+(\d+)\s+(\d+)\s+(\d+)\s+F", line
)
if atom_match:
if "atoms" not in rv:
rv["atoms"] = ""
else:
rv["atoms"] += ","
rv["atoms"] += atom_match.group(1)
elif bond_match:
if "bonds" not in rv:
rv["bonds"] = []
rv["bonds"].append(
",".join([bond_match.group(1), bond_match.group(2)])
)
elif angle_match:
if "angles" not in rv:
rv["angles"] = []
rv["angles"].append(
",".join(
[
angle_match.group(1),
angle_match.group(2),
angle_match.group(3),
]
)
)
elif torsion_match:
if "torsions" not in rv:
rv["torsions"] = []
rv["torsions"].append(
",".join(
[
torsion_match.group(1),
torsion_match.group(2),
torsion_match.group(3),
torsion_match.group(4),
]
)
)
line = f.readline()
n += 1
return rv, n
self.all_geom = []
line = f.readline()
self.other["archive"] = ""
constraints = {}
self.other["opt_steps"] = 0
found_archive = False
n = 1
route = None
while line != "":
# route
# we need to grab the route b/c sometimes 'hpmodes' can get split onto multiple lines:
# B3LYP/genecp EmpiricalDispersion=GD3 int=(grid=superfinegrid) freq=(h
# pmodes,noraman,temperature=313.15)
if line.strip().startswith("#") and route is None:
route = ""
while "------" not in line:
route += line.strip()
n += 1
line = f.readline()
# archive entry
if line.strip().startswith("1\\1\\"):
found_archive = True
line = "@" + line.strip()[4:]
if found_archive and line.strip().endswith("@"):
self.other["archive"] = self.other["archive"][:-2] + "\\\\"
found_archive = False
elif found_archive:
self.other["archive"] += line.strip()
# input atom specs and charge/mult
if "Symbolic Z-matrix:" in line:
self.atoms, n = get_input(f, n)
#Pseudopotential info
if "Pseudopotential Parameters" in line:
self.other["ECP"] = []
self.skip_lines(f, 4)
n += 5
line = f.readline()
while "=====" not in line:
line = line.split()
if line[0].isdigit() and line[1].isdigit():
ele = line[1]
n += 1
line = f.readline().split()
if line[0] != "No":
self.other["ECP"].append(ELEMENTS[int(ele)])
n += 1
line = f.readline()
# geometry
if re.search("(Standard|Input) orientation:", line):
if get_all and len(self.atoms) > 0:
self.all_geom += [
(deepcopy(self.atoms), deepcopy(self.other))
]
self.atoms, n = get_atoms(f, n)
self.other["opt_steps"] += 1
if re.search(
"The following ModRedundant input section has been read:", line
):
constraints, n = get_modredundant(f, n)
if just_geom:
line = f.readline()
n += 1
continue
# z-matrix parameters
if re.search("Optimized Parameters", line):
self.other["params"], n = get_params(f, n)
# status
if NORM_FINISH in line:
self.other["finished"] = True
# read energies from different methods
if "SCF Done" in line:
tmp = [word.strip() for word in line.split()]
idx = tmp.index("=")
self.other["energy"] = float(tmp[idx + 1])
self.other["scf_energy"] = float(tmp[idx + 1])
else:
nrg_match = re.search("\s+(E\(\S+\))\s*=\s*(\S+)", line)
# ^ matches many methods
# will also match the SCF line (hence the else here)
# the match in the SCF line could be confusing b/c
# the SCF line could be
# SCF Done: E(RB2PLYPD3) = -76.2887108570 A.U. after 10 cycles
# and later on, there will be a line...
# E2(B2PLYPD3) = -0.6465105880D-01 E(B2PLYPD3) = -0.76353361915801D+02
# this will give:
# * E(RB2PLYPD3) = -76.2887108570
# * E(B2PLYPD3) = -76.353361915801
# very similar names for very different energies...
if nrg_match:
self.other["energy"] = float(nrg_match.group(2).replace("D", "E"))
self.other[nrg_match.group(1)] = self.other["energy"]
# CC energy
if line.startswith(" CCSD(T)= "):
self.other["energy"] = float(line.split()[-1].replace("D", "E"))
self.other["E(CCSD(T))"] = self.other["energy"]
# MP energies
mp_match = re.search("([RU]MP\d+(?:\(\S+\))?)\s*=\s*(\S+)", line)
if mp_match:
self.other["energy"] = float(mp_match.group(2).replace("D", "E"))
self.other["E(%s)" % mp_match.group(1)] = self.other["energy"]
if "Molecular mass:" in line:
self.other["mass"] = float(float_num.search(line).group(0))
self.other["mass"] *= UNIT.AMU_TO_KG
# Frequencies
if route is not None and "hpmodes" in route.lower():
self.other["hpmodes"] = True
if "Harmonic frequencies" in line:
freq_str = line
line = f.readline()
while line != "\n":
n += 1
freq_str += line
line = f.readline()
if "hpmodes" not in self.other:
self.other["hpmodes"] = False
self.other["frequency"] = Frequency(
freq_str, hpmodes=self.other["hpmodes"]
)
if "Anharmonic Infrared Spectroscopy" in line:
self.skip_lines(f, 5)
n += 5
anharm_str = ""
combinations_read = False
combinations = False
line = f.readline()
while not combinations_read:
n += 1
anharm_str += line
if "Combination Bands" in line:
combinations = True
line = f.readline()
if combinations and line == "\n":
combinations_read = True
self.other["frequency"].parse_gaussian_lines(
anharm_str.splitlines(), harmonic=False,
)
# X matrix for anharmonic
if "Total Anharmonic X Matrix" in line:
self.skip_lines(f, 1)
n += 1
n_freq = len(self.other["frequency"].data)
n_sections = int(np.ceil(n_freq / 5))
x_matrix = np.zeros((n_freq, n_freq))
for section in range(0, n_sections):
header = f.readline()
n += 1
for j in range(5 * section, n_freq):
line = f.readline()
n += 1
ll = 5 * section
ul = 5 * section + min(j - ll + 1, 5)
x_matrix[j, ll:ul] = [
float(x.replace("D", "e"))
for x in line.split()[1:]
]
x_matrix += np.tril(x_matrix, k=-1).T
self.other["X_matrix"] = x_matrix
if "Total X0" in line:
self.other["X0"] = float(line.split()[5])
# TD-DFT output
if line.strip().startswith("Ground to excited state"):
uv_vis = ""
highest_state = 0
done = False
read_states = False
while not done:
n += 1
uv_vis += line
if not read_states and line.strip() and line.split()[0].isdigit():
state = int(line.split()[0])
if state > highest_state:
highest_state = state
if line.strip().startswith("Ground to excited state transition velocity"):
read_states = True
if re.search("Excited State\s*%i:" % highest_state, line):
done = True
if line.strip().startswith("Total Energy, E"):
nrg = re.search(
r"Total Energy, E\((\S+)\)\s*=\s*(-?\d+\.\d+)", line
)
self.other["E(%s)" % nrg.group(1)] = float(nrg.group(2))
self.other["energy"] = float(nrg.group(2))
line = f.readline()
self.other["uv_vis"] = ValenceExcitations(
uv_vis, style="gaussian"
)
# Thermo
if re.search("Temperature\s*\d+\.\d+", line):
self.other["temperature"] = float(
float_num.search(line).group(0)
)
if "Rotational constants (GHZ):" in line:
rot = float_num.findall(line)
rot = [
float(r) * PHYSICAL.PLANCK * (10 ** 9) / PHYSICAL.KB
for r in rot
]
self.other["rotational_temperature"] = rot
# rotational constants from anharmonic frequency jobs
if "Rotational Constants (in MHz)" in line:
self.skip_lines(f, 2)
n += 2
equilibrium_rotational_temperature = np.zeros(3)
ground_rotational_temperature = np.zeros(3)
centr_rotational_temperature = np.zeros(3)
for i in range(0, 3):
line = f.readline()
n += 1
info = line.split()
Be = float(info[1])
B00 = float(info[3])
B0 = float(info[5])
equilibrium_rotational_temperature[i] = Be
ground_rotational_temperature[i] = B00
centr_rotational_temperature[i] = B0
equilibrium_rotational_temperature *= (
PHYSICAL.PLANCK * 1e6 / PHYSICAL.KB
)
ground_rotational_temperature *= (
PHYSICAL.PLANCK * 1e6 / PHYSICAL.KB
)
centr_rotational_temperature *= (
PHYSICAL.PLANCK * 1e6 / PHYSICAL.KB
)
self.other[
"equilibrium_rotational_temperature"
] = equilibrium_rotational_temperature
self.other[
"ground_rotational_temperature"
] = ground_rotational_temperature
self.other[
"centr_rotational_temperature"
] = centr_rotational_temperature
if "Sum of electronic and zero-point Energies=" in line:
self.other["E_ZPVE"] = float(float_num.search(line).group(0))
if "Sum of electronic and thermal Enthalpies=" in line:
self.other["enthalpy"] = float(float_num.search(line).group(0))
if "Sum of electronic and thermal Free Energies=" in line:
self.other["free_energy"] = float(
float_num.search(line).group(0)
)
if "Zero-point correction=" in line:
self.other["ZPVE"] = float(float_num.search(line).group(0))
if "Rotational symmetry number" in line:
self.other["rotational_symmetry_number"] = int(
re.search("\d+", line).group(0)
)
# Gradient
if re.search("Threshold\s+Converged", line) is not None:
line = f.readline()
n += 1
grad = {}
def add_grad(line, name, grad):
line = line.split()
grad[name] = {
"value": line[2],
"threshold": line[3],
"converged": True if line[4] == "YES" else False,
}
return grad
while line != "":
if "Predicted change in Energy" in line:
break
if re.search("Maximum\s+Force", line) is not None:
grad = add_grad(line, "Max Force", grad)
if re.search("RMS\s+Force", line) is not None:
grad = add_grad(line, "RMS Force", grad)
if re.search("Maximum\s+Displacement", line) is not None:
grad = add_grad(line, "Max Disp", grad)
if re.search("RMS\s+Displacement", line) is not None:
grad = add_grad(line, "RMS Disp", grad)
line = f.readline()
n += 1
self.other["gradient"] = grad
# electronic properties
if "Electrostatic Properties (Atomic Units)" in line:
self.skip_lines(f, 5)
n += 5
self.other["electric_potential"] = []
self.other["electric_field"] = []
line = f.readline()
while "--" not in line:
info = line.split()
self.other["electric_potential"].append(float(info[2]))
self.other["electric_field"].append([float(x) for x in info[3:]])
line = f.readline()
n += 1
self.other["electric_potential"] = np.array(self.other["electric_potential"])
self.other["electric_field"] = np.array(self.other["electric_field"])
# optical features
if "[Alpha]" in line:
alpha_match = re.search("\[Alpha\].*\(\s*(.*\s?.*)\)\s*=\s*(-?\d+\.\d+)", line)
self.other["optical_rotation_(%s)" % alpha_match.group(1)] = \
float(alpha_match.group(2))
# symmetry
if "Full point group" in line:
self.other["full_point_group"] = line.split()[-3]
if "Largest Abelian subgroup" in line:
self.other["abelian_subgroup"] = line.split()[-3]
if "Largest concise Abelian subgroup" in line:
self.other["concise_abelian_subgroup"] = line.split()[-3]
# forces
if "Forces (Hartrees/Bohr)" in line:
gradient = np.zeros((len(self.atoms), 3))
self.skip_lines(f, 2)
n += 2
for i in range(0, len(self.atoms)):
n += 1
line = f.readline()
info = line.split()
gradient[i] = np.array([float(x) for x in info[2:]])
self.other["forces"] = gradient
# nbo stuff
if "N A T U R A L A T O M I C O R B I T A L A N D" in line:
self.read_nbo(f)
# atomic charges
charge_match = re.search("(\S+) charges:\s*$", line)
if charge_match:
self.skip_lines(f, 1)
n += 1
charges = []
for i in range(0, len(self.atoms)):
line = f.readline()
n += 1
charges.append(float(line.split()[2]))
self.atoms[i].charge = float(line.split()[2])
self.other[charge_match.group(1) + " Charges"] = charges
# capture errors
# only keep first error, want to fix one at a time
if "error" not in self.other:
for err in ERROR:
if re.search(err, line):
self.other["error"] = ERROR[err]
self.other["error_msg"] = line.strip()
break
line = f.readline()
n += 1
if not just_geom:
if route is not None:
other_kwargs = {GAUSSIAN_ROUTE: {}}
route_spec = re.compile("(\w+)=?\((.*)\)")
method_and_basis = re.search(
"#(?:[NnPpTt]\s+?)(\S+)|#\s*?(\S+)", route
)
if method_and_basis is not None:
if method_and_basis.group(2):
method_info = method_and_basis.group(2).split("/")
else:
method_info = method_and_basis.group(1).split("/")
method = method_info[0]
if len(method_info) > 1:
basis = method_info[1]
else:
basis = None
route_options = route.split()
job_type = []
grid = None
solvent = None
for option in route_options:
if option.startswith("#"):
continue
elif option.startswith(method):
continue
option_lower = option.lower()
if option_lower.startswith("opt"):
ts = False
match = route_spec.search(option)
if match:
options = match.group(2).split(",")
elif option_lower.startswith("opt="):
options = ["".join(option.split("=")[1:])]
else:
if not constraints:
# if we didn't read constraints, try using flagged atoms instead
from AaronTools.finders import FlaggedAtoms
constraints = {"atoms": FlaggedAtoms}
if not any(atom.flag for atom in self.atoms):
constraints = None
job_type.append(
OptimizationJob(constraints=constraints)
)
continue
other_kwargs[GAUSSIAN_ROUTE]["opt"] = []
for opt in options:
if opt.lower() == "ts":
ts = True
else:
other_kwargs[GAUSSIAN_ROUTE]["opt"].append(
opt
)
job_type.append(
OptimizationJob(
transition_state=ts,
constraints=constraints,
)
)
elif option_lower.startswith("freq"):
temp = 298.15
match = route_spec.search(option)
if match:
options = match.group(2).split(",")
elif option_lower.startswith("freq="):
options = "".join(option.split("=")[1:])
else:
job_type.append(FrequencyJob())
continue
other_kwargs[GAUSSIAN_ROUTE]["freq"] = []
for opt in options:
if opt.lower().startswith("temp"):
temp = float(opt.split("=")[1])
else:
other_kwargs[GAUSSIAN_ROUTE][
"freq"
].append(opt)
job_type.append(FrequencyJob(temperature=temp))
elif option_lower == "sp":
job_type.append(SinglePointJob())
elif option_lower.startswith("int"):
match = route_spec.search(option)
if match:
options = match.group(2).split(",")
elif option_lower.startswith("freq="):
options = "".join(option.split("=")[1:])
else:
job_type.append(FrequencyJob())
continue
for opt in options:
if opt.lower().startswith("grid"):
grid_name = opt.split("=")[1]
grid = IntegrationGrid(grid_name)
else:
if (
"Integral"
not in other_kwargs[GAUSSIAN_ROUTE]
):
other_kwargs[GAUSSIAN_ROUTE][
"Integral"
] = []
other_kwargs[GAUSSIAN_ROUTE][
"Integral"
].append(opt)
else:
# TODO: parse solvent
match = route_spec.search(option)
if match:
keyword = match.group(1)
options = match.group(2).split(",")
other_kwargs[GAUSSIAN_ROUTE][keyword] = options
elif "=" in option:
keyword = option.split("=")[0]
options = "".join(option.split("=")[1:])
other_kwargs[GAUSSIAN_ROUTE][keyword] = [
options
]
else:
other_kwargs[GAUSSIAN_ROUTE][option] = []
continue
self.other["other_kwargs"] = other_kwargs
try:
theory = Theory(
charge=self.other["charge"],
multiplicity=self.other["multiplicity"],
job_type=job_type,
basis=basis,
method=method,
grid=grid,
solvent=solvent,
)
theory.kwargs = self.other["other_kwargs"]
self.other["theory"] = theory
except KeyError:
# if there is a serious error, too little info may be available
# to properly create the theory object
pass
for i, a in enumerate(self.atoms):
a.name = str(i + 1)
if "finished" not in self.other:
self.other["finished"] = False
if "error" not in self.other:
self.other["error"] = None
return
def read_com(self, f):
found_atoms = False
found_constraint = False
atoms = []
other = {}
for line in f:
# header
if line.startswith("%"):
continue
if line.startswith("#"):
method = re.search("^#([NnPpTt]\s+?)(\S+)|^#\s*?(\S+)", line)
# route can be #n functional/basis ...
# or #functional/basis ...
# or # functional/basis ...
if method.group(3):
other["method"] = method.group(3)
else:
other["method"] = method.group(2)
if "temperature=" in line:
other["temperature"] = re.search(
"temperature=(\d+\.?\d*)", line
).group(1)
if "solvent=" in line:
other["solvent"] = re.search(
"solvent=(\S+)\)", line
).group(1)
if "scrf=" in line:
# solvent model should be non-greedy b/c solvent name can have commas
other["solvent_model"] = re.search(
"scrf=\((\S+?),", line
).group(1)
if "EmpiricalDispersion=" in line:
other["emp_dispersion"] = re.search(
"EmpiricalDispersion=(\S+)", line
).group(1)
if "int=(grid" in line or "integral=(grid" in line.lower():
other["grid"] = re.search(
"(?:int||Integral)=\(grid[(=](\S+?)\)", line
).group(1)
# comments can be multiple lines long
# but there should be a blank line between the route and the comment
# and another between the comment and the charge+mult
blank_lines = 0
while blank_lines < 2:
line = f.readline().strip()
if len(line) == 0:
blank_lines += 1
else:
if "comment" not in other:
other["comment"] = ""
other["comment"] += "%s\n" % line
other["comment"] = (
other["comment"].strip() if "comment" in other else ""
)
line = f.readline()
if len(line.split()) > 1:
line = line.split()
else:
line = line.split(",")
other["charge"] = int(line[0])
other["multiplicity"] = int(line[1])
found_atoms = True
continue
# constraints
if found_atoms and line.startswith("B") and line.endswith("F"):
found_constraint = True
if "constraint" not in other:
other["constraint"] = []
other["constraint"] += [float_num.findall(line)]
continue
# footer
if found_constraint:
if "footer" not in other:
other["footer"] = ""
other["footer"] += line
continue
# atom coords
nums = float_num.findall(line)
line = line.split()
if len(line) == 5 and is_alpha(line[0]) and len(nums) == 4:
if not is_int(line[1]):
continue
a = Atom(element=line[0], coords=nums[1:], flag=nums[0])
atoms += [a]
elif len(line) == 4 and is_alpha(line[0]) and len(nums) == 3:
a = Atom(element=line[0], coords=nums)
atoms += [a]
else:
continue
for i, a in enumerate(atoms):
a.name = str(i + 1)
self.atoms = atoms
self.other = other
return
def read_fchk(self, f, just_geom=True, max_length=10000000):
def parse_to_list(
i, lines, length, data_type, debug=False, max_length=max_length,
):
"""takes a block in an fchk file and turns it into an array
block headers all end with N= <int>
the length of the array will be <int>
the data type is specified by data_type"""
i += 1
line = f.readline()
# print("first line", line)
items_per_line = len(line.split())
# print("items per line", items_per_line)
total_items = items_per_line
num_lines = ceil(length / items_per_line)
# print("lines in block", num_lines)
block = [line]
for k in range(0, num_lines - 1):
line = f.readline()
if max_length < length:
continue
block.append(line)
if max_length < length:
return length, i + num_lines
block = " ".join(block)
if debug:
print("full block")
print(block)
return (
np.fromstring(block, count=length, dtype=data_type, sep=" "),
i + num_lines,
)
self.atoms = []
atom_numbers = []
atom_coords = []
other = {}
int_info = re.compile("([\S\s]+?)\s*I\s*(N=)?\s*(-?\d+)")
real_info = re.compile(
"([\S\s]+?)\s*R\s*(N=)\s*(-?\d+\.?\d*[Ee]?[+-]?\d*)"
)
char_info = re.compile(
"([\S\s]+?)\s*C\s*(N=)?\s*(-?\d+\.?\d*[Ee]?[+-]?\d*)"
)
theory = Theory()
line = f.readline()
i = 0
while line != "":
if i == 0:
other["comment"] = line.strip()
elif i == 1:
i += 1
line = f.readline()
job_info = line.split()
if job_info[0] == "SP":
theory.job_type = [SinglePointJob()]
elif job_info[0] == "FOPT":
theory.job_type[OptimizationJob()]
elif job_info[0] == "FTS":
theory.job_type = [OptimizationJob(transition_state=True)]
elif job_info[0] == "FORCE":
theory.job_type = [ForceJob()]
elif job_info[0] == "FREQ":
theory.job_type = [FrequencyJob()]
theory.method = job_info[1]
if len(job_info) > 2:
theory.basis = job_info[2]
i += 1
line = f.readline()
continue
int_match = int_info.match(line)
real_match = real_info.match(line)
char_match = char_info.match(line)
if int_match is not None:
data = int_match.group(1)
# print("int", data)
value = int_match.group(3)
if data == "Charge" and not just_geom:
theory.charge = int(value)
elif data == "Multiplicity" and not just_geom:
theory.multiplicity = int(value)
elif data == "Atomic numbers":
atom_numbers, i = parse_to_list(i, f, int(value), int)
elif not just_geom:
if int_match.group(2):
other[data], i = parse_to_list(
i, f, int(value), int
)
else:
other[data] = int(value)
elif real_match is not None:
data = real_match.group(1)
# print("real", data)
value = real_match.group(3)
if data == "Current cartesian coordinates":
atom_coords, i = parse_to_list(i, f, int(value), float)
elif data == "Total Energy":
other["energy"] = float(value)
elif not just_geom:
if real_match.group(2):
other[data], i = parse_to_list(
i, f, int(value), float
)
else:
other[data] = float(value)
# elif char_match is not None:
# data = char_match.group(1)
# value = char_match.group(3)
# if not just_geom:
# other[data] = lines[i + 1]
# i += 1
line = f.readline()
i += 1
self.other = other
self.other["theory"] = theory
if isinstance(atom_coords, int):
raise RuntimeError(
"max. array size is insufficient to parse atom data\n"
"must be at least %i" % atom_coords
)
coords = np.reshape(atom_coords, (len(atom_numbers), 3))
for n, (atnum, coord) in enumerate(zip(atom_numbers, coords)):
atom = Atom(
element=ELEMENTS[atnum],
coords=UNIT.A0_TO_BOHR * coord,
name=str(n + 1),
)
self.atoms.append(atom)
try:
self.other["orbitals"] = Orbitals(self)
except (NotImplementedError, KeyError):
pass
except (TypeError, ValueError) as err:
self.LOG.warning(
"could not create Orbitals, try increasing the max.\n"
"array size to read from FCHK files\n\n"
"%s" % err
)
for key in [
"Alpha MO coefficients", "Beta MO coefficients",
"Shell types", "Shell to atom map", "Contraction coefficients",
"Primitive exponents", "Number of primitives per shell",
"Coordinates of each shell",
]:
if key in self.other and isinstance(self.other[key], int):
self.LOG.warning(
"size of %s is > %i: %i" % (key, max_length, self.other[key])
)
def read_nbo(self, f):
"""
read nbo data
"""
line = f.readline()
while line:
if "natural bond orbitals (summary):" in line.lower():
break
if "NATURAL POPULATIONS:" in line:
self.skip_lines(f, 3)
ao_types = []
ao_atom_ndx = []
nao_types = []
occ = []
nrg = []
blank_lines = 0
while blank_lines <= 1:
match = re.search(
"\d+\s+[A-Z][a-z]?\s+(\d+)\s+(\S+)\s+([\S\s]+?)(-?\d+\.\d+)\s+(-?\d+\.\d+)",
line
)
if match:
ao_atom_ndx.append(int(match.group(1)) - 1)
ao_types.append(match.group(2))
nao_types.append(match.group(3))
occ.append(float(match.group(4)))
nrg.append(float(match.group(5)))
blank_lines = 0
else:
blank_lines += 1
line = f.readline()
self.other["ao_types"] = ao_types
self.other["ao_atom_ndx"] = ao_atom_ndx
self.other["nao_type"] = nao_types
self.other["ao_occ"] = occ
self.other["ao_nrg"] = nrg
if "Summary of Natural Population Analysis:" in line:
self.skip_lines(f, 5)
core_occ = []
val_occ = []
rydberg_occ = []
nat_q = []
line = f.readline()
while "==" not in line:
info = line.split()
core_occ.append(float(info[3]))
val_occ.append(float(info[4]))
rydberg_occ.append(float(info[5]))
nat_q.append(float(info[2]))
line = f.readline()
self.other["Natural Charges"] = nat_q
self.other["core_occ"] = core_occ
self.other["valence_occ"] = val_occ
self.other["rydberg_occ"] = rydberg_occ
if "Wiberg bond index matrix in the NAO basis" in line:
dim = len(self.other["Natural Charges"])
bond_orders = np.zeros((dim, dim))
done = False
j = 0
for block in range(0, ceil(dim / 9)):
offset = 9 * j
self.skip_lines(f, 3)
for i in range(0, dim):
line = f.readline()
for k, bo in enumerate(line.split()[2:]):
bo = float(bo)
bond_orders[i][offset + k] = bo
j += 1
self.other["wiberg_nao"] = bond_orders
line = f.readline()
def read_crest(self, f, conf_name=None):
"""
conf_name = False to skip conformer loading (doesn't get written until crest job is done)
"""
if conf_name is None:
conf_name = os.path.join(
os.path.dirname(self.name), "crest_conformers.xyz"
)
line = True
self.other["finished"] = False
self.other["error"] = None
while line:
line = f.readline()
if "terminated normally" in line:
self.other["finished"] = True
elif "population of lowest" in line:
self.other["best_pop"] = float(float_num.findall(line)[0])
elif "ensemble free energy" in line:
self.other["free_energy"] = (
float(float_num.findall(line)[0]) / UNIT.HART_TO_KCAL
)
elif "ensemble entropy" in line:
self.other["entropy"] = (
float(float_num.findall(line)[1]) / UNIT.HART_TO_KCAL
)
elif "ensemble average energy" in line:
self.other["avg_energy"] = (
float(float_num.findall(line)[0]) / UNIT.HART_TO_KCAL
)
elif "E lowest" in line:
self.other["energy"] = float(float_num.findall(line)[0])
elif "T /K" in line:
self.other["temperature"] = float(float_num.findall(line)[0])
elif (
line.strip()
.lower()
.startswith(("forrtl", "warning", "*warning"))
):
self.other["error"] = "UNKNOWN"
if "error_msg" not in self.other:
self.other["error_msg"] = ""
self.other["error_msg"] += line
elif "-chrg" in line:
self.other["charge"] = int(float_num.findall(line)[0])
elif "-uhf" in line:
self.other["multiplicity"] = (
int(float_num.findall(line)[0]) + 1
)
if self.other["finished"] and conf_name:
self.other["conformers"] = FileReader(
conf_name,
get_all=True,
).all_geom
self.comment, self.atoms = self.other["conformers"][0]
self.other["conformers"] = self.other["conformers"][1:]
def read_xtb(self, f, freq_name=None):
line = True
self.other["finished"] = False
self.other["error"] = None
self.atoms = []
self.comment = ""
while line:
line = f.readline()
if "Optimized Geometry" in line:
line = f.readline()
n_atoms = int(line.strip())
line = f.readline()
self.comment = " ".join(line.strip().split()[2:])
for i in range(n_atoms):
line = f.readline()
elem, x, y, z = line.split()
self.atoms.append(Atom(element=elem, coords=[x, y, z]))
if "normal termination" in line:
self.other["finished"] = True
if "abnormal termination" in line:
self.other["error"] = "UNKNOWN"
if line.strip().startswith("#ERROR"):
if "error_msg" not in self.other:
self.other["error_msg"] = ""
self.other["error_msg"] += line
if "charge" in line and ":" in line:
self.other["charge"] = int(float_num.findall(line)[0])
if "spin" in line and ":" in line:
self.other["multiplicity"] = (
2 * float(float_num.findall(line)[0]) + 1
)
if "total energy" in line:
self.other["energy"] = (
float(float_num.findall(line)[0]) * UNIT.HART_TO_KCAL
)
if "zero point energy" in line:
self.other["ZPVE"] = (
float(float_num.findall(line)[0]) * UNIT.HART_TO_KCAL
)
if "total free energy" in line:
self.other["free_energy"] = (
float(float_num.findall(line)[0]) * UNIT.HART_TO_KCAL
)
if "electronic temp." in line:
self.other["temperature"] = float(float_num.findall(line)[0])
if freq_name is not None:
with open(freq_name) as f_freq:
self.other["frequency"] = Frequency(f_freq.read())
def read_sqm(self, f):
lines = f.readlines()
self.other["finished"] = False
self.atoms = []
i = 0
while i < len(lines):
line = lines[i]
if "Atomic Charges for Step" in line:
elements = []
for info in lines[i + 2 :]:
if not info.strip() or not info.split()[0].isdigit():
break
ele = info.split()[1]
elements.append(ele)
i += len(elements) + 2
if "Final Structure" in line:
k = 0
for info in lines[i + 4 :]:
data = info.split()
coords = np.array([x for x in data[4:7]])
self.atoms.append(
Atom(
name=str(k + 1),
coords=coords,
element=elements[k],
)
)
k += 1
if k == len(elements):
break
i += k + 4
if "Calculation Completed" in line:
self.other["finished"] = True
if "Total SCF energy" in line:
self.other["energy"] = (
float(line.split()[4]) / UNIT.HART_TO_KCAL
)
i += 1
if not self.atoms:
# there's no atoms if there's an error
# error is probably on the last line
self.other["error"] = "UNKNOWN"
self.other["error_msg"] = line
def read_nbo_47(self, f, nbo_name=None):
lines = f.readlines()
bohr = False
i = 0
while i < len(lines):
line = lines[i]
if line.startswith(" $"):
section = line.split()[0]
if section.startswith("$COORD"):
i += 1
self.atoms = []
line = lines[i]
while not line.startswith(" $END"):
if re.search("\d+\s+\d+(?:\s+-?\d+\.\d+\s){3}", line):
info = line.split()
ndx = int(info[0])
coords = [float(x) for x in info[2:5]]
self.atoms.append(
Atom(
element=ELEMENTS[ndx],
name=str(len(self.atoms) + 1),
coords=np.array(coords),
)
)
i += 1
line = lines[i]
elif section.startswith("$BASIS"):
reading_centers = False
reading_labels = False
i += 1
line = lines[i]
while not line.startswith(" $END"):
if "CENTER" in line.upper():
self.other["shell_to_atom"] = [
int(x) for x in line.split()[2:]
]
reading_centers = True
reading_labels = False
elif "LABEL" in line.upper():
self.other["momentum_label"] = [
int(x) for x in line.split()[2:]
]
reading_labels = True
reading_centers = False
elif reading_centers:
self.other["shell_to_atom"].extend(
[int(x) for x in line.split()]
)
elif reading_labels:
self.other["momentum_label"].extend(
[int(x) for x in line.split()]
)
i += 1
line = lines[i]
elif section.startswith("$CONTRACT"):
int_sections = {
"NCOMP": "funcs_per_shell",
"NPRIM": "n_prim_per_shell",
"NPTR": "start_ndx",
}
float_sections = {
"EXP": "exponents",
"CS": "s_coeff",
"CP": "p_coeff",
"CD": "d_coeff",
"CF": "f_coeff",
}
i += 1
line = lines[i]
while not line.startswith(" $END"):
if any(line.strip().startswith(section) for section in int_sections):
section = line.split()[0]
self.other[int_sections[section]] = [
int(x) for x in line.split()[2:]
]
i += 1
line = lines[i]
while "=" not in line and "$" not in line:
self.other[int_sections[section]].extend([
int(x) for x in line.split()
])
i += 1
line = lines[i]
elif any(line.strip().startswith(section) for section in float_sections):
section = line.split()[0]
self.other[float_sections[section]] = [
float(x) for x in line.split()[2:]
]
i += 1
line = lines[i]
while "=" not in line and "$" not in line:
self.other[float_sections[section]].extend([
float(x) for x in line.split()
])
i += 1
line = lines[i]
else:
i += 1
line = lines[i]
elif section.startswith("$GENNBO"):
if "BOHR" in section.upper():
bohr = True
nbas = re.search("NBAS=(\d+)", line)
n_funcs = int(nbas.group(1))
if "CUBICF" in section.upper():
self.LOG.warning("cubic F shell will not be handled correctly")
i += 1
if nbo_name is not None:
self._read_nbo_coeffs(nbo_name)
def _read_nbo_coeffs(self, nbo_name):
"""
read coefficients in AO basis for NBO's/NLHO's/NAO's/etc.
called by methods that read NBO input (.47) or output files (.31)
"""
with open(nbo_name, "r") as f2:
lines = f2.readlines()
kind = re.search("P?(\S+)s", lines[1]).group(1)
desc_file = os.path.splitext(nbo_name)[0] + ".46"
if os.path.exists(desc_file):
with open(desc_file, "r") as f3:
desc_lines = f3.readlines()
for k, line in enumerate(desc_lines):
if kind in line:
self.other["orbit_kinds"] = []
n_orbits = int(line.split()[1])
k += 1
while len(self.other["orbit_kinds"]) < n_orbits:
self.other["orbit_kinds"].extend([
desc_lines[k][i: i + 10]
for i in range(1, len(desc_lines[k]) - 1, 10)
])
k += 1
else:
self.LOG.warning(
"no .46 file found - orbital descriptions will be unavialable"
)
j = 3
self.other["alpha_coefficients"] = []
while len(self.other["alpha_coefficients"]) < sum(self.other["funcs_per_shell"]):
mo_coeff = []
while len(mo_coeff) < sum(self.other["funcs_per_shell"]):
mo_coeff.extend([float(x) for x in lines[j].split()])
j += 1
self.other["alpha_coefficients"].append(mo_coeff)
self.other["orbitals"] = Orbitals(self)
def read_nbo_31(self, f, nbo_name=None):
lines = f.readlines()
comment = lines[0].strip()
info = lines[3].split()
n_atoms = int(info[0])
self.atoms = []
for i in range(5, 5 + n_atoms):
atom_info = lines[i].split()
ele = ELEMENTS[int(atom_info[0])]
coords = np.array([float(x) for x in atom_info[1:4]])
self.atoms.append(
Atom(
element=ele,
coords=coords,
name=str(i-4),
)
)
i = n_atoms + 6
line = lines[i]
self.other["shell_to_atom"] = []
self.other["momentum_label"] = []
self.other["funcs_per_shell"] = []
self.other["start_ndx"] = []
self.other["n_prim_per_shell"] = []
while "---" not in line:
info = line.split()
ndx = int(info[0])
funcs = int(info[1])
start_ndx = int(info[2])
n_prim = int(info[3])
self.other["shell_to_atom"].extend([ndx for j in range(0, funcs)])
self.other["funcs_per_shell"].append(funcs)
self.other["start_ndx"].append(start_ndx)
self.other["n_prim_per_shell"].append(n_prim)
i += 1
line = lines[i]
momentum_labels = [int(x) for x in line.split()]
self.other["momentum_label"].extend(momentum_labels)
i += 1
line = lines[i]
i += 1
self.other["exponents"] = []
line = lines[i]
while line.strip() != "":
exponents = [float(x) for x in line.split()]
self.other["exponents"].extend(exponents)
i += 1
line = lines[i]
i += 1
self.other["s_coeff"] = []
line = lines[i]
while line.strip() != "":
coeff = [float(x) for x in line.split()]
self.other["s_coeff"].extend(coeff)
i += 1
line = lines[i]
i += 1
self.other["p_coeff"] = []
line = lines[i]
while line.strip() != "":
coeff = [float(x) for x in line.split()]
self.other["p_coeff"].extend(coeff)
i += 1
line = lines[i]
i += 1
self.other["d_coeff"] = []
line = lines[i]
while line.strip() != "":
coeff = [float(x) for x in line.split()]
self.other["d_coeff"].extend(coeff)
i += 1
line = lines[i]
i += 1
self.other["f_coeff"] = []
line = lines[i]
while line.strip() != "":
coeff = [float(x) for x in line.split()]
self.other["f_coeff"].extend(coeff)
i += 1
line = lines[i]
i += 1
self.other["g_coeff"] = []
line = lines[i]
while line.strip() != "":
coeff = [float(x) for x in line.split()]
self.other["g_coeff"].extend(coeff)
i += 1
line = lines[i]
if nbo_name is not None:
self._read_nbo_coeffs(nbo_name) | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/fileIO.py | fileIO.py |
import json
import os
import re
import numpy as np
from AaronTools import addlogger
from AaronTools.const import (
CONNECTIVITY,
EIJ,
ELEMENTS,
MASS,
RADII,
RIJ,
SATURATION,
TMETAL,
VDW_RADII,
)
warn_LJ = set([])
class BondOrder:
bonds = {}
warn_atoms = set([])
warn_str = (
"\n"
+ " Could not get bond order for: "
+ " {} {}"
+ " using bond order of 1"
)
# warn(s.format(a1.element, a2.element))
def __init__(self):
if BondOrder.bonds:
return
with open(
os.path.join(
os.path.dirname(__file__), "calculated_bond_lengths.json"
)
) as f:
BondOrder.bonds = json.load(f)
@classmethod
def key(cls, a1, a2):
if isinstance(a1, Atom):
a1 = a1.element
if isinstance(a2, Atom):
a2 = a2.element
return " ".join(sorted([a1, a2]))
@classmethod
def get(cls, a1, a2):
"""determines bond order between two atoms based on bond length"""
try:
bonds = cls.bonds[cls.key(a1, a2)]
except KeyError:
if a1.element == "H" or a2.element == "H":
return 1
else:
BondOrder.warn_atoms.add((a1.element, a2.element))
return 1
dist = a1.dist(a2)
closest = 0, None # (bond order, length diff)
for order, length in bonds.items():
diff = abs(length - dist)
if closest[1] is None or diff < closest[1]:
closest = order, diff
return float(closest[0])
@addlogger
class Atom:
"""
Attributes:
element str
coords np.array(float)
flag bool true if frozen, false if relaxed
name str form of \d+(\.\d+)*
tags set
charge float
connected set(Atom)
constraint set(Atom) for determining constrained bonds
_rank
_radii float for calculating if bonded
_connectivity int max connections without hypervalence
_saturation int max connections without hypervalence or charges
"""
LOG = None
_bo = BondOrder()
def __init__(
self, element="", coords=None, flag=False, name="", tags=None, charge=None
):
super().__setattr__("_hashed", False)
if coords is None:
coords = []
if tags is None:
tags = []
# for BqO to have a ghost atom with oxygen basis functions
ele = str(element).strip()
element = ele.capitalize()
if "-" in ele:
element = "-".join(e.capitalize() for e in ele.split("-"))
if element == "":
self.element = element
self._radii = None
self._connectivity = None
elif element not in ELEMENTS and not element.endswith("Bq"):
raise ValueError("Unknown element detected: %s" % element)
else:
self.element = element
self.reset()
self.coords = np.array(coords, dtype=float)
self.flag = bool(flag)
self.name = str(name).strip()
if hasattr(tags, "__iter__") and not isinstance(tags, str):
self.tags = set(tags)
else:
self.tags = set([tags])
self.charge = charge
if charge:
self.charge = float(charge)
self.connected = set([])
self.constraint = set([])
self._rank = None
# utilities
def __float__(self):
"""
converts self.name from a string to a floating point number
"""
rv = self.name.split(".")
if len(rv) == 0:
return float(0)
if len(rv) == 1:
return float(rv[0])
rv = "{}.{}".format(rv[0], rv[1])
return float(rv)
def __lt__(self, other):
"""
sorts by canonical smiles rank, then by neighbor ID, then by name
more connections first
then, more non-H bonds first
then, higher atomic number first
then, higher number of attached hydrogens first
then, lower sorting name first
"""
if (
self._rank is not None
and other._rank is not None
and self._rank != other._rank
):
return self._rank > other._rank
if self._rank is None or other._rank is None:
# if the ranks are the same, we have little reason to
# believe the invariants will differ
# print("getting invariants during <", self._rank, other._rank)
a = self.get_invariant()
b = other.get_invariant()
if a != b:
return a > b
# print("using names")
a = self.name.split(".")
b = other.name.split(".")
while len(a) < len(b):
a += ["0"]
while len(b) < len(a):
b += ["0"]
for i, j in zip(a, b):
try:
if int(i) != int(j):
return int(i) < int(j)
except ValueError:
pass
return True
def __str__(self):
s = ""
s += "{:>4s} ".format(self.name)
s += "{:<3s} ".format(self.element)
for c in self.coords:
s += " {: 10.6f}".format(c)
return s
def __repr__(self):
s = ""
s += "{:>3s} ".format(self.element)
for c in self.coords:
s += " {: 13.8f}".format(c)
s += " {: 2d}".format(-1 if self.flag else 0)
s += " {:>4s}".format(self.name)
s += " ({:d})".format(self._rank) if self._rank is not None else ""
return s
def _set_radii(self):
"""Sets atomic radii"""
try:
self._radii = float(RADII[self.element])
except KeyError:
self.LOG.warning("Radii not found for element: %s" % self.element)
return
def __setattr__(self, attr, val):
if (
not self._hashed
or (attr == "_hashed" and val)
or (attr != "element" and attr != "coords")
):
super().__setattr__(attr, val)
else:
raise RuntimeError(
"Atom %s's Geometry has been hashed and can no longer be changed\n"
% self.name
+ "setattr was called to set %s to %s" % (attr, val)
)
def _set_vdw(self):
"""Sets atomic radii"""
try:
self._vdw = float(VDW_RADII[self.element])
except KeyError:
self.LOG.warning(
"VDW Radii not found for element: %s" % self.element
)
self._vdw = 0
return
def _set_connectivity(self):
"""Sets theoretical maximum connectivity.
If # connections > self._connectivity, then atom is hyper-valent
"""
try:
self._connectivity = int(CONNECTIVITY[self.element])
except KeyError:
pass
# self.LOG.warning(
# "Connectivity not found for element: " + self.element
# )
return
def _set_saturation(self):
"""Sets theoretical maximum connectivity without the atom having a formal charge.
If # connections > self._saturation, then atom is hyper-valent or has a non-zero formal charge
"""
try:
self._saturation = int(SATURATION[self.element])
except KeyError:
if self.element not in TMETAL:
self.LOG.warning(
"Saturation not found for element: " + self.element
)
return
@property
def is_dummy(self):
return re.match("(X$|[A-Z][a-z]?-Bq|Bq)", self.element)
def reset(self):
if self.is_dummy:
self._vdw = 0
self._connectivity = 1000
self._saturation = 0
self._radii = 0
else:
self._set_radii()
self._set_vdw()
self._set_connectivity()
self._set_saturation()
def add_tag(self, *args):
for a in args:
if hasattr(a, "__iter__") and not isinstance(a, str):
self.tags = self.tags.union(set(a))
else:
self.tags.add(a)
return
def get_invariant(self):
"""
gets initial invariant
(1) number of non-hydrogen connections (\d{1}): nconn
(2) sum of bond order of non-hydrogen bonds * 10 (\d{2}): nB
(3) atomic number (\d{3}): z
#(4) sign of charge (\d{1})
#(5) absolute charge (\d{1})
(6) number of attached hydrogens (\d{1}): nH
"""
heavy = set([x for x in self.connected if x.element != "H"])
# number of non-hydrogen connections:
nconn = len(heavy)
# number of bonds with heavy atoms
nB = 0
for h in heavy:
nB += BondOrder.get(h, self)
# number of connected hydrogens
nH = len(self.connected - heavy)
# atomic number
z = ELEMENTS.index(self.element)
return "{:01d}{:03d}{:03d}{:01d}".format(
int(nconn), int(nB * 10), int(z), int(nH)
)
def get_neighbor_id(self):
"""
gets initial invariant based on self's element and the element of
the atoms connected to self
"""
# atomic number
z = ELEMENTS.index(self.element)
heavy = [
ELEMENTS.index(x.element)
for x in self.connected
if x.element != "H"
]
# number of non-hydrogen connections
# number of bonds with heavy atoms and their element
t = []
for h in sorted(set(heavy)):
t.extend([h, heavy.count(h)])
# number of connected hydrogens
nH = len(self.connected) - len(heavy)
fmt = "%03i%02i" + (len(set(heavy)) * "%03i%02i") + "%02i"
s = fmt % (z, len(heavy), *t, nH)
return s
def copy(self):
rv = Atom()
for key, val in self.__dict__.items():
if key == "connected":
continue
if key == "constraint":
continue
if key == "_hashed":
continue
try:
rv.__dict__[key] = val.copy()
except AttributeError:
rv.__dict__[key] = val
# ignore chimerax objects so seqcrow doesn't print a
# warning when a geometry is copied
if "chimerax" in val.__class__.__module__:
continue
if val.__class__.__module__ != "builtins":
self.LOG.warning(
"No copy method for {}: in-place changes may occur".format(
type(val)
)
)
return rv
# measurement
def is_connected(self, other, tolerance=None):
"""determines if distance between atoms is small enough to be bonded"""
return self.dist_is_connected(other, self.dist(other), tolerance)
def dist_is_connected(self, other, dist_to_other, tolerance):
"""
determines if distance between atoms is small enough to be bonded
used to optimize connected checks when distances can be quickly precalculated
like with scipy.spatial.distance_matrix
"""
if tolerance is None:
tolerance = 0.3
if self._radii is None:
self._set_radii()
if other._radii is None:
other._set_radii()
cutoff = self._radii + other._radii + tolerance
return dist_to_other < cutoff
def add_bond_to(self, other):
"""add self and other to eachother's connected attribute"""
self.connected.add(other)
other.connected.add(self)
def bond(self, other):
"""returns the vector self-->other"""
return np.array(other.coords) - np.array(self.coords)
def dist(self, other):
"""returns the distance between self and other"""
return np.linalg.norm(self.bond(other))
def angle(self, a1, a3):
"""returns the a1-self-a3 angle"""
v1 = self.bond(a1)
v2 = self.bond(a3)
dot = np.dot(v1, v2)
# numpy is still unhappy with this sometimes
# every now and again, the changeElement cls test will "fail" b/c
# numpy throws a warning here
if abs(dot / (self.dist(a1) * self.dist(a3))) >= 1:
return 0
else:
return np.arccos(dot / (self.dist(a1) * self.dist(a3)))
def mass(self):
"""returns atomic mass"""
if self.element in MASS:
return MASS[self.element]
elif not self.is_dummy:
self.LOG.warning("no mass for %s" % self.element)
return 0
def rij(self, other):
try:
rv = RIJ[self.element + other.element]
except KeyError:
try:
rv = RIJ[other.element + self.element]
except KeyError:
warn_LJ.add("".join(sorted([self.element, other.element])))
return 0
return rv
def eij(self, other):
try:
rv = EIJ[self.element + other.element]
except KeyError:
try:
rv = EIJ[other.element + self.element]
except KeyError:
warn_LJ.add("".join(sorted([self.element, other.element])))
return 0
return rv
def bond_order(self, other):
return BondOrder.get(self, other)
@classmethod
def get_shape(cls, shape_name):
"""returns dummy atoms in an idealized vsepr geometry
shape_name can be:
point
linear 1
linear 2
bent 2 tetrahedral
bent 2 planar
trigonal planar
bent 3 tetrahedral
t shaped
tetrahedral
sawhorse
seesaw
square planar
trigonal pyramidal
trigonal bipyramidal
square pyramidal
pentagonal
hexagonal
trigonal prismatic
pentagonal pyramidal
octahedral
capped octahedral
hexagonal pyramidal
pentagonal bipyramidal
capped trigonal prismatic
heptagonal
hexagonal bipyramidal
heptagonal pyramidal
octagonal
square antiprismatic
trigonal dodecahedral
capped cube
biaugmented trigonal prismatic
cubic
elongated trigonal bipyramidal
capped square antiprismatic
enneagonal
heptagonal bipyramidal
hula-hoop
triangular cupola
tridiminished icosahedral
muffin
octagonal pyramidal
tricapped trigonal prismatic
"""
if shape_name == "point":
return cls.linear_shape()[0:1]
elif shape_name == "linear 1":
return cls.linear_shape()[0:2]
elif shape_name == "linear 2":
return cls.linear_shape()
elif shape_name == "bent 2 tetrahedral":
return cls.tetrahedral_shape()[0:3]
elif shape_name == "bent 2 planar":
return cls.trigonal_planar_shape()[0:3]
elif shape_name == "trigonal planar":
return cls.trigonal_planar_shape()
elif shape_name == "bent 3 tetrahedral":
return cls.tetrahedral_shape()[0:4]
elif shape_name == "t shaped":
return cls.octahedral_shape()[0:4]
elif shape_name == "tetrahedral":
return cls.tetrahedral_shape()
elif shape_name == "sawhorse":
return (
cls.trigonal_bipyramidal_shape()[0:3]
+ cls.trigonal_bipyramidal_shape()[-2:]
)
elif shape_name == "seesaw":
return cls.octahedral_shape()[0:3] + cls.octahedral_shape()[-2:]
elif shape_name == "square planar":
return cls.octahedral_shape()[0:5]
elif shape_name == "trigonal pyramidal":
return cls.trigonal_bipyramidal_shape()[0:5]
elif shape_name == "trigonal bipyramidal":
return cls.trigonal_bipyramidal_shape()
elif shape_name == "square pyramidal":
return cls.octahedral_shape()[0:6]
elif shape_name == "pentagonal":
return cls.pentagonal_bipyraminal_shape()[0:6]
elif shape_name == "hexagonal":
return cls.hexagonal_bipyramidal_shape()[0:7]
elif shape_name == "trigonal prismatic":
return cls.trigonal_prismatic_shape()
elif shape_name == "pentagonal pyramidal":
return cls.pentagonal_bipyraminal_shape()[0:7]
elif shape_name == "octahedral":
return cls.octahedral_shape()
elif shape_name == "capped octahedral":
return cls.capped_octahedral_shape()
elif shape_name == "hexagonal pyramidal":
return cls.hexagonal_bipyramidal_shape()[0:8]
elif shape_name == "pentagonal bipyramidal":
return cls.pentagonal_bipyraminal_shape()
elif shape_name == "capped trigonal prismatic":
return cls.capped_trigonal_prismatic_shape()
elif shape_name == "heptagonal":
return cls.heptagonal_bipyramidal_shape()[0:8]
elif shape_name == "hexagonal bipyramidal":
return cls.hexagonal_bipyramidal_shape()
elif shape_name == "heptagonal pyramidal":
return cls.heptagonal_bipyramidal_shape()[0:9]
elif shape_name == "octagonal":
return cls.octagonal_pyramidal_shape()[0:9]
elif shape_name == "square antiprismatic":
return cls.square_antiprismatic_shape()
elif shape_name == "trigonal dodecahedral":
return cls.trigonal_dodecahedral_shape()
elif shape_name == "capped cube":
return cls.capped_cube_shape()
elif shape_name == "biaugmented trigonal prismatic":
return cls.biaugmented_trigonal_prismatic_shape()
elif shape_name == "cubic":
return cls.cubic_shape()
elif shape_name == "elongated trigonal bipyramidal":
return cls.elongated_trigonal_bipyramidal_shape()
elif shape_name == "capped square antiprismatic":
return cls.capped_square_antiprismatic_shape()
elif shape_name == "enneagonal":
return cls.enneagonal_shape()
elif shape_name == "heptagonal bipyramidal":
return cls.heptagonal_bipyramidal_shape()
elif shape_name == "hula-hoop":
return cls.hula_hoop_shape()
elif shape_name == "triangular cupola":
return cls.triangular_cupola_shape()
elif shape_name == "tridiminished icosahedral":
return cls.tridiminished_icosahedral_shape()
elif shape_name == "muffin":
return cls.muffin_shape()
elif shape_name == "octagonal pyramidal":
return cls.octagonal_pyramidal_shape()
elif shape_name == "tricapped trigonal prismatic":
return cls.tricapped_trigonal_prismatic_shape()
else:
raise RuntimeError(
"no shape method is defined for %s" % shape_name
)
@classmethod
def linear_shape(cls):
"""returns a list of 3 dummy atoms in a linear shape"""
center = Atom("X", np.zeros(3), name="0")
pos1 = Atom("X", np.array([1.0, 0.0, 0.0]), name="1")
pos2 = Atom("X", np.array([-1.0, 0.0, 0.0]), name="2")
return [center, pos1, pos2]
@classmethod
def trigonal_planar_shape(cls):
"""returns a list of 4 dummy atoms in a trigonal planar shape"""
positions = cls.trigonal_bipyramidal_shape()
return positions[:-2]
@classmethod
def tetrahedral_shape(cls):
"""returns a list of 5 dummy atoms in a tetrahedral shape"""
center = Atom("X", np.zeros(3), name="0")
angle = np.deg2rad(109.471 / 2)
pos1 = Atom(
"X", np.array([np.cos(angle), -np.sin(angle), 0.0]), name="1"
)
pos2 = Atom(
"X", np.array([np.cos(angle), np.sin(angle), 0.0]), name="2"
)
pos3 = Atom(
"X", np.array([-np.cos(angle), 0.0, np.sin(angle)]), name="3"
)
pos4 = Atom(
"X", np.array([-np.cos(angle), 0.0, -np.sin(angle)]), name="4"
)
return [center, pos1, pos2, pos3, pos4]
@classmethod
def trigonal_bipyramidal_shape(cls):
"""returns a list of 6 dummy atoms in a trigonal bipyramidal shape"""
center = Atom("X", np.zeros(3), name="0")
angle = np.deg2rad(120)
pos1 = Atom("X", np.array([1.0, 0.0, 0.0]), name="1")
pos2 = Atom(
"X", np.array([np.cos(angle), np.sin(angle), 0.0]), name="2"
)
pos3 = Atom(
"X", np.array([np.cos(angle), -np.sin(angle), 0.0]), name="3"
)
pos4 = Atom("X", np.array([0.0, 0.0, 1.0]), name="4")
pos5 = Atom("X", np.array([0.0, 0.0, -1.0]), name="5")
return [center, pos1, pos2, pos3, pos4, pos5]
@classmethod
def octahedral_shape(cls):
"""returns a list of 7 dummy atoms in an octahedral shape"""
center = Atom("X", np.zeros(3), name="0")
pos1 = Atom("X", np.array([1.0, 0.0, 0.0]), name="1")
pos2 = Atom("X", np.array([0.0, 1.0, 0.0]), name="2")
pos3 = Atom("X", np.array([-1.0, 0.0, 0.0]), name="3")
pos4 = Atom("X", np.array([0.0, -1.0, 0.0]), name="4")
pos5 = Atom("X", np.array([0.0, 0.0, 1.0]), name="5")
pos6 = Atom("X", np.array([0.0, 0.0, -1.0]), name="6")
return [center, pos1, pos2, pos3, pos4, pos5, pos6]
@classmethod
def trigonal_prismatic_shape(cls):
center = Atom("X", np.zeros(3), name="0")
pos1 = Atom("X", np.array([-0.6547, -0.3780, 0.6547]), name="1")
pos2 = Atom("X", np.array([-0.6547, -0.3780, -0.6547]), name="2")
pos3 = Atom("X", np.array([0.6547, -0.3780, 0.6547]), name="3")
pos4 = Atom("X", np.array([0.6547, -0.3780, -0.6547]), name="4")
pos5 = Atom("X", np.array([0.0, 0.7559, 0.6547]), name="5")
pos6 = Atom("X", np.array([0.0, 0.7559, -0.6547]), name="6")
return [center, pos1, pos2, pos3, pos4, pos5, pos6]
@classmethod
def capped_octahedral_shape(cls):
center = Atom("X", np.zeros(3), name="0")
pos1 = Atom("X", np.array([0.0, 0.0, 1.0]), name="1")
pos2 = Atom("X", np.array([0.9777, 0.0, 0.2101]), name="2")
pos3 = Atom("X", np.array([0.1698, 0.9628, 0.2101]), name="3")
pos4 = Atom("X", np.array([-0.9187, 0.3344, 0.2102]), name="4")
pos5 = Atom("X", np.array([-0.4888, -0.8467, 0.2102]), name="5")
pos6 = Atom("X", np.array([0.3628, -0.6284, -0.6881]), name="6")
pos7 = Atom("X", np.array([-0.2601, 0.4505, -0.8540]), name="7")
return [center, pos1, pos2, pos3, pos4, pos5, pos6, pos7]
@classmethod
def capped_trigonal_prismatic_shape(cls):
center = Atom("X", np.zeros(3), name="0")
pos1 = Atom("X", np.array([0.0, 0.0, 1.0]), name="1")
pos2 = Atom("X", np.array([0.6869, 0.6869, 0.2374]), name="2")
pos3 = Atom("X", np.array([-0.6869, 0.6869, 0.2374]), name="3")
pos4 = Atom("X", np.array([0.6869, -0.6869, 0.2374]), name="4")
pos5 = Atom("X", np.array([-0.6869, -0.6869, 0.2374]), name="5")
pos6 = Atom("X", np.array([0.6175, 0.0, -0.7866]), name="6")
pos7 = Atom("X", np.array([-0.6175, 0.0, -0.7866]), name="7")
return [center, pos1, pos2, pos3, pos4, pos5, pos6, pos7]
@classmethod
def pentagonal_bipyraminal_shape(cls):
center = Atom("X", np.zeros(3), name="0")
pos1 = Atom("X", np.array([1.0, 0.0, 0.0]), name="1")
pos2 = Atom("X", np.array([0.3090, 0.9511, 0.0]), name="2")
pos3 = Atom("X", np.array([-0.8090, 0.5878, 0.0]), name="3")
pos4 = Atom("X", np.array([-0.8090, -0.5878, 0.0]), name="4")
pos5 = Atom("X", np.array([0.3090, -0.9511, 0.0]), name="5")
pos6 = Atom("X", np.array([0.0, 0.0, 1.0]), name="6")
pos7 = Atom("X", np.array([0.0, 0.0, -1.0]), name="7")
return [center, pos1, pos2, pos3, pos4, pos5, pos6, pos7]
@classmethod
def biaugmented_trigonal_prismatic_shape(cls):
center = Atom("X", np.zeros(3), name="0")
pos1 = Atom("X", np.array([-0.6547, -0.3780, 0.6547]), name="1")
pos2 = Atom("X", np.array([-0.6547, -0.3780, -0.6547]), name="2")
pos3 = Atom("X", np.array([0.6547, -0.3780, 0.6547]), name="3")
pos4 = Atom("X", np.array([0.6547, -0.3780, -0.6547]), name="4")
pos5 = Atom("X", np.array([0.0, 0.7559, 0.6547]), name="5")
pos6 = Atom("X", np.array([0.0, 0.7559, -0.6547]), name="6")
pos7 = Atom("X", np.array([0.0, -1.0, 0.0]), name="7")
pos8 = Atom("X", np.array([-0.8660, 0.5, 0.0]), name="8")
return [center, pos1, pos2, pos3, pos4, pos5, pos6, pos7, pos8]
@classmethod
def cubic_shape(cls):
center = Atom("X", np.zeros(3), name="0")
pos1 = Atom("X", np.array([0.5775, 0.5774, 0.5774]), name="1")
pos2 = Atom("X", np.array([0.5775, 0.5774, -0.5774]), name="2")
pos3 = Atom("X", np.array([0.5775, -0.5774, 0.5774]), name="3")
pos4 = Atom("X", np.array([-0.5775, 0.5774, 0.5774]), name="4")
pos5 = Atom("X", np.array([0.5775, -0.5774, -0.5774]), name="5")
pos6 = Atom("X", np.array([-0.5775, 0.5774, -0.5774]), name="6")
pos7 = Atom("X", np.array([-0.5775, -0.5774, 0.5774]), name="7")
pos8 = Atom("X", np.array([-0.5775, -0.5774, -0.5774]), name="8")
return [center, pos1, pos2, pos3, pos4, pos5, pos6, pos7, pos8]
@classmethod
def elongated_trigonal_bipyramidal_shape(cls):
center = Atom("X", np.zeros(3), name="0")
pos1 = Atom("X", np.array([0.6547, 0.0, 0.7559]), name="1")
pos2 = Atom("X", np.array([-0.6547, 0.0, 0.7559]), name="2")
pos3 = Atom("X", np.array([0.6547, 0.6547, -0.3780]), name="3")
pos4 = Atom("X", np.array([-0.6547, 0.6547, -0.3780]), name="4")
pos5 = Atom("X", np.array([0.6547, -0.6547, -0.3780]), name="5")
pos6 = Atom("X", np.array([-0.6547, -0.6547, -0.3780]), name="6")
pos7 = Atom("X", np.array([1.0, 0.0, 0.0]), name="7")
pos8 = Atom("X", np.array([-1.0, 0.0, 0.0]), name="8")
return [center, pos1, pos2, pos3, pos4, pos5, pos6, pos7, pos8]
@classmethod
def hexagonal_bipyramidal_shape(cls):
center = Atom("X", np.zeros(3), name="0")
pos1 = Atom("X", np.array([0.0, -1.0, 0.0]), name="1")
pos2 = Atom("X", np.array([0.8660, -0.5, 0.0]), name="2")
pos3 = Atom("X", np.array([0.8660, 0.5, 0.0]), name="3")
pos4 = Atom("X", np.array([0.0, 1.0, 0.0]), name="4")
pos5 = Atom("X", np.array([-0.8660, 0.5, 0.0]), name="5")
pos6 = Atom("X", np.array([-0.8660, -0.5, 0.0]), name="6")
pos7 = Atom("X", np.array([0.0, 0.0, 1.0]), name="7")
pos8 = Atom("X", np.array([0.0, 0.0, -1.0]), name="8")
return [center, pos1, pos2, pos3, pos4, pos5, pos6, pos7, pos8]
@classmethod
def square_antiprismatic_shape(cls):
center = Atom("X", np.zeros(3), name="0")
pos1 = Atom("X", np.array([0.0, 0.0, 1.0]), name="1")
pos2 = Atom("X", np.array([0.9653, 0.0, 0.2612]), name="2")
pos3 = Atom("X", np.array([-0.5655, 0.7823, 0.2612]), name="3")
pos4 = Atom("X", np.array([-0.8825, -0.3912, 0.2612]), name="4")
pos5 = Atom("X", np.array([0.1999, -0.9444, 0.2612]), name="5")
pos6 = Atom("X", np.array([0.3998, 0.7827, -0.4776]), name="6")
pos7 = Atom("X", np.array([-0.5998, 0.1620, -0.7836]), name="7")
pos8 = Atom("X", np.array([0.4826, -0.3912, -0.7836]), name="8")
return [center, pos1, pos2, pos3, pos4, pos5, pos6, pos7, pos8]
@classmethod
def trigonal_dodecahedral_shape(cls):
center = Atom("X", np.zeros(3), name="0")
pos1 = Atom("X", np.array([-0.5997, 0.0, 0.8002]), name="1")
pos2 = Atom("X", np.array([0.0, -0.9364, 0.3509]), name="2")
pos3 = Atom("X", np.array([0.5998, 0.0, 0.8002]), name="3")
pos4 = Atom("X", np.array([0.0, 0.9364, 0.3509]), name="4")
pos5 = Atom("X", np.array([-0.9364, 0.0, -0.3509]), name="5")
pos6 = Atom("X", np.array([0.0, -0.5997, -0.8002]), name="6")
pos7 = Atom("X", np.array([0.9365, 0.0, -0.3509]), name="7")
pos8 = Atom("X", np.array([0.0, 0.5997, -0.8002]), name="8")
return [center, pos1, pos2, pos3, pos4, pos5, pos6, pos7, pos8]
@classmethod
def heptagonal_bipyramidal_shape(cls):
center = Atom("X", np.zeros(3), name="0")
pos1 = Atom("X", np.array([1.0, 0.0, 0.0]), name="1")
pos2 = Atom("X", np.array([0.6235, 0.7818, 0.0]), name="2")
pos3 = Atom("X", np.array([-0.2225, 0.9749, 0.0]), name="3")
pos4 = Atom("X", np.array([-0.9010, 0.4339, 0.0]), name="4")
pos5 = Atom("X", np.array([-0.9010, -0.4339, 0.0]), name="5")
pos6 = Atom("X", np.array([-0.2225, -0.9749, 0.0]), name="6")
pos7 = Atom("X", np.array([0.6235, -0.7818, 0.0]), name="7")
pos8 = Atom("X", np.array([0.0, 0.0, 1.0]), name="8")
pos9 = Atom("X", np.array([0.0, 0.0, -1.0]), name="9")
return [center, pos1, pos2, pos3, pos4, pos5, pos6, pos7, pos8, pos9]
@classmethod
def capped_cube_shape(cls):
center = Atom("X", np.zeros(3), name="0")
pos1 = Atom("X", np.array([0.6418, 0.6418, 0.4196]), name="1")
pos2 = Atom("X", np.array([0.6418, -0.6418, 0.4196]), name="2")
pos3 = Atom("X", np.array([-0.6418, 0.6418, 0.4196]), name="3")
pos4 = Atom("X", np.array([-0.6418, -0.6418, 0.4196]), name="4")
pos5 = Atom("X", np.array([0.5387, 0.5387, -0.6478]), name="5")
pos6 = Atom("X", np.array([0.5387, -0.5387, -0.6478]), name="6")
pos7 = Atom("X", np.array([-0.5387, 0.5387, -0.6478]), name="7")
pos8 = Atom("X", np.array([-0.5387, -0.5387, -0.6478]), name="8")
pos9 = Atom("X", np.array([0.0, 0.0, 1.0]), name="9")
return [center, pos1, pos2, pos3, pos4, pos5, pos6, pos7, pos8, pos9]
@classmethod
def capped_square_antiprismatic_shape(cls):
center = Atom("X", np.zeros(3), name="0")
pos1 = Atom("X", np.array([0.9322, 0.0, 0.3619]), name="1")
pos2 = Atom("X", np.array([-0.9322, 0.0, 0.3619]), name="2")
pos3 = Atom("X", np.array([0.0, 0.9322, 0.3619]), name="3")
pos4 = Atom("X", np.array([0.0, -0.9322, 0.3619]), name="4")
pos5 = Atom("X", np.array([0.5606, 0.5606, -0.6095]), name="5")
pos6 = Atom("X", np.array([-0.5606, 0.5606, -0.6095]), name="6")
pos7 = Atom("X", np.array([-0.5606, -0.5606, -0.6095]), name="7")
pos8 = Atom("X", np.array([0.5606, -0.5606, -0.6095]), name="8")
pos9 = Atom("X", np.array([0.0, 0.0, 1.0]), name="9")
return [center, pos1, pos2, pos3, pos4, pos5, pos6, pos7, pos8, pos9]
@classmethod
def enneagonal_shape(cls):
center = Atom("X", np.zeros(3), name="0")
pos1 = Atom("X", np.array([1.0, 0.0, 0.0]), name="1")
pos2 = Atom("X", np.array([0.7660, 0.6428, 0.0]), name="2")
pos3 = Atom("X", np.array([0.1736, 0.9848, 0.0]), name="3")
pos4 = Atom("X", np.array([-0.5, 0.8660, 0.0]), name="4")
pos5 = Atom("X", np.array([-0.9397, 0.3420, 0.0]), name="5")
pos6 = Atom("X", np.array([-0.9397, -0.3420, 0.0]), name="6")
pos7 = Atom("X", np.array([-0.5, -0.8660, 0.0]), name="7")
pos8 = Atom("X", np.array([0.1736, -0.9848, 0.0]), name="8")
pos9 = Atom("X", np.array([0.7660, -0.6428, 0.0]), name="9")
return [center, pos1, pos2, pos3, pos4, pos5, pos6, pos7, pos8, pos9]
@classmethod
def hula_hoop_shape(cls):
center = Atom("X", np.zeros(3), name="0")
pos1 = Atom("X", np.array([1.0, 0.0, 0.0]), name="1")
pos2 = Atom("X", np.array([0.5, 0.8660, 0.0]), name="2")
pos3 = Atom("X", np.array([-0.5, 0.8660, 0.0]), name="3")
pos4 = Atom("X", np.array([-1.0, 0.0, 0.0]), name="4")
pos5 = Atom("X", np.array([-0.5, -0.8660, 0.0]), name="5")
pos6 = Atom("X", np.array([0.5, -0.8660, 0.0]), name="6")
pos7 = Atom("X", np.array([0.0, 0.0, 1.0]), name="7")
pos8 = Atom("X", np.array([0.5, 0.0, -0.8660]), name="8")
pos9 = Atom("X", np.array([-0.5, 0.0, -0.8660]), name="9")
return [center, pos1, pos2, pos3, pos4, pos5, pos6, pos7, pos8, pos9]
@classmethod
def triangular_cupola_shape(cls):
center = Atom("X", np.zeros(3), name="0")
pos1 = Atom("X", np.array([1.0, 0.0, 0.0]), name="1")
pos2 = Atom("X", np.array([0.5, 0.8660, 0.0]), name="2")
pos3 = Atom("X", np.array([-0.5, 0.8660, 0.0]), name="3")
pos4 = Atom("X", np.array([-1.0, 0.0, 0.0]), name="4")
pos5 = Atom("X", np.array([0.5, -0.8660, 0.0]), name="5")
pos6 = Atom("X", np.array([-0.5, -0.8660, 0.0]), name="6")
pos7 = Atom("X", np.array([0.5, 0.2887, -0.8165]), name="7")
pos8 = Atom("X", np.array([-0.5, 0.2887, -0.8165]), name="8")
pos9 = Atom("X", np.array([0.0, -0.5774, -0.8165]), name="9")
return [center, pos1, pos2, pos3, pos4, pos5, pos6, pos7, pos8, pos9]
@classmethod
def tridiminished_icosahedral_shape(cls):
center = Atom("X", np.zeros(3), name="0")
pos1 = Atom("X", np.array([-0.2764, 0.8507, -0.4472]), name="1")
pos2 = Atom("X", np.array([-0.8944, 0.0, -0.4472]), name="2")
pos3 = Atom("X", np.array([-0.2764, -0.8507, -0.4472]), name="3")
pos4 = Atom("X", np.array([0.7236, -0.5257, -0.4472]), name="4")
pos5 = Atom("X", np.array([0.8944, 0.0, 0.4472]), name="5")
pos6 = Atom("X", np.array([0.2764, 0.8507, 0.4472]), name="6")
pos7 = Atom("X", np.array([-0.7236, -0.5257, 0.4472]), name="7")
pos8 = Atom("X", np.array([0.0, 0.0, 1.0]), name="8")
pos9 = Atom("X", np.array([0.0, 0.0, -1.0]), name="9")
return [center, pos1, pos2, pos3, pos4, pos5, pos6, pos7, pos8, pos9]
@classmethod
def muffin_shape(cls):
center = Atom("X", np.zeros(3), name="0")
pos1 = Atom("X", np.array([0.0, 0.9875, 0.1579]), name="1")
pos2 = Atom("X", np.array([0.9391, 0.3051, 0.1579]), name="2")
pos3 = Atom("X", np.array([0.5804, -0.7988, 0.1579]), name="3")
pos4 = Atom("X", np.array([-0.5804, -0.7988, 0.1579]), name="4")
pos5 = Atom("X", np.array([-0.9391, 0.3055, 0.1579]), name="5")
pos6 = Atom("X", np.array([-0.5799, -0.3356, -0.7423]), name="6")
pos7 = Atom("X", np.array([0.5799, -0.3356, -0.7423]), name="7")
pos8 = Atom("X", np.array([0.0, 0.6694, -0.7429]), name="8")
pos9 = Atom("X", np.array([0.0, 0.0, 1.0]), name="9")
return [center, pos1, pos2, pos3, pos4, pos5, pos6, pos7, pos8, pos9]
@classmethod
def octagonal_pyramidal_shape(cls):
center = Atom("X", np.zeros(3), name="0")
pos1 = Atom("X", np.array([0.7071, -0.7071, 0.0]), name="1")
pos2 = Atom("X", np.array([1.0, 0.0, 0.0]), name="2")
pos3 = Atom("X", np.array([0.7071, 0.7071, 0.0]), name="3")
pos4 = Atom("X", np.array([0.0, 1.0, 0.0]), name="4")
pos5 = Atom("X", np.array([-0.7071, 0.7071, 0.0]), name="5")
pos6 = Atom("X", np.array([-1.0, 0.0, 0.0]), name="6")
pos7 = Atom("X", np.array([-0.7071, -0.7071, 0.0]), name="7")
pos8 = Atom("X", np.array([0.0, -1.0, 0.0]), name="8")
pos9 = Atom("X", np.array([0.0, 0.0, -1.0]), name="9")
return [center, pos1, pos2, pos3, pos4, pos5, pos6, pos7, pos8, pos9]
@classmethod
def tricapped_trigonal_prismatic_shape(cls):
center = Atom("X", np.zeros(3), name="0")
pos1 = Atom("X", np.array([0.0, 0.0, 1.0]), name="1")
pos2 = Atom("X", np.array([-0.2357, 0.9129, 0.3333]), name="2")
pos3 = Atom("X", np.array([-0.9428, 0.0, 0.3333]), name="3")
pos4 = Atom("X", np.array([0.2357, -0.9129, 0.3333]), name="4")
pos5 = Atom("X", np.array([0.9428, 0.0, 0.3333]), name="5")
pos6 = Atom("X", np.array([0.5303, 0.6847, -0.5]), name="6")
pos7 = Atom("X", np.array([-0.5303, -0.6847, -0.5]), name="7")
pos8 = Atom("X", np.array([-0.5893, 0.4564, -0.6667]), name="8")
pos9 = Atom("X", np.array([0.5893, -0.4564, -0.6667]), name="9")
return [center, pos1, pos2, pos3, pos4, pos5, pos6, pos7, pos8, pos9]
@staticmethod
def new_shape(old_shape, new_connectivity, bond_change):
"""returns the name of the expected vsepr geometry when the number of bonds
changes by +/- 1
old_shape - :str: vsepr geometry name
new_connectivity - :int: connectivity (see Atom._connectivity)
bond_change - :int: +1 or -1, indicating that the number of bonds is changing by 1"""
if old_shape == "point":
if bond_change == 1:
return "linear 1"
else:
return None
elif old_shape == "linear 1":
if bond_change == 1:
return "linear 2"
elif bond_change == -1:
return None
elif old_shape == "linear 2":
if bond_change == 1:
if new_connectivity is not None and new_connectivity > 4:
return "t shaped"
else:
return "trigonal planar"
elif bond_change == -1:
return "linear 1"
elif old_shape == "bent 2 tetrahedral":
if bond_change == 1:
return "bent 3 tetrahedral"
elif bond_change == -1:
return "linear 1"
elif old_shape == "bent 2 planar":
if bond_change == 1:
return "trigonal planar"
elif bond_change == -1:
return "linear 1"
elif old_shape == "trigonal planar":
if bond_change == 1:
return "tetrahedral"
elif bond_change == -1:
if new_connectivity == 4:
return "bent 2 tetrahedral"
else:
return "bent 2 planar"
elif old_shape == "bent 3 tetrahedral":
if bond_change == 1:
return "tetrahedral"
elif bond_change == -1:
return "bent 2 tetrahedral"
elif old_shape == "t shaped":
if bond_change == 1:
if new_connectivity == 6:
return "square planar"
else:
return "sawhorse"
elif bond_change == -1:
return "linear 2"
elif old_shape == "tetrahedral":
if bond_change == 1:
return "trigonal bipyramidal"
elif bond_change == -1:
return "bent 3 tetrahedral"
elif old_shape == "square planar":
if bond_change == 1:
return "trigonal bipyramidal"
elif bond_change == -1:
return "t shaped"
elif old_shape == "trigonal bipyramidal":
if bond_change == 1:
return "octahedral"
elif bond_change == -1:
return "sawhorse"
elif old_shape == "octahedral":
if bond_change == -1:
return "trigonal bipyramid"
else:
raise RuntimeError("no shape method is defined for %s" % old_shape)
def get_vsepr(self):
"""determine vsepr geometry around an atom
returns shape as a string and the score assigned to that shape
returns None if self has > 6 bonds
scores > 0.5 are generally questionable
see atom.get_shape for a list of shapes
"""
# shapes with a code in the commend next to them are from Simas et. al. Inorg. Chem. 2018, 57, 17, 10557–10567
# determine what geometries to try based on the number of bonded atoms
try_shapes = {}
if len(self.connected) == 0:
try_shapes["point"] = Atom.get_shape("point")
elif len(self.connected) == 1:
try_shapes["linear 1"] = Atom.get_shape("linear 1")
elif len(self.connected) == 2:
try_shapes["linear 2"] = Atom.get_shape("linear 2")
try_shapes["bent 2 planar"] = Atom.get_shape("bent 2 planar")
try_shapes["bent 2 tetrahedral"] = Atom.get_shape(
"bent 2 tetrahedral"
)
elif len(self.connected) == 3:
try_shapes["trigonal planar"] = Atom.get_shape("trigonal planar")
try_shapes["bent 3 tetrahedral"] = Atom.get_shape(
"bent 3 tetrahedral"
)
try_shapes["t shaped"] = Atom.get_shape("t shaped")
elif len(self.connected) == 4:
try_shapes["tetrahedral"] = Atom.get_shape("tetrahedral")
try_shapes["sawhorse"] = Atom.get_shape("sawhorse")
try_shapes["seesaw"] = Atom.get_shape("seesaw")
try_shapes["square planar"] = Atom.get_shape("square planar")
try_shapes["trigonal pyramidal"] = Atom.get_shape(
"trigonal pyramidal"
)
elif len(self.connected) == 5:
try_shapes["trigonal bipyramidal"] = Atom.get_shape(
"trigonal bipyramidal"
)
try_shapes["square pyramidal"] = Atom.get_shape("square pyramidal")
try_shapes["pentagonal"] = Atom.get_shape("pentagonal") # PP-5
elif len(self.connected) == 6:
try_shapes["octahedral"] = Atom.get_shape("octahedral")
try_shapes["hexagonal"] = Atom.get_shape("hexagonal") # HP-6
try_shapes["trigonal prismatic"] = Atom.get_shape(
"trigonal prismatic"
) # TPR-6
try_shapes["pentagonal pyramidal"] = Atom.get_shape(
"pentagonal pyramidal"
) # PPY-6
elif len(self.connected) == 7:
try_shapes["capped octahedral"] = Atom.get_shape(
"capped octahedral"
) # COC-7
try_shapes["capped trigonal prismatic"] = Atom.get_shape(
"capped trigonal prismatic"
) # CTPR-7
try_shapes["heptagonal"] = Atom.get_shape("heptagonal") # HP-7
try_shapes["hexagonal pyramidal"] = Atom.get_shape(
"hexagonal pyramidal"
) # HPY-7
try_shapes["pentagonal bipyramidal"] = Atom.get_shape(
"pentagonal bipyramidal"
) # PBPY-7
elif len(self.connected) == 8:
try_shapes["biaugmented trigonal prismatic"] = Atom.get_shape(
"biaugmented trigonal prismatic"
) # BTPR-8
try_shapes["cubic"] = Atom.get_shape("cubic") # CU-8
try_shapes["elongated trigonal bipyramidal"] = Atom.get_shape(
"elongated trigonal bipyramidal"
) # ETBPY-8
try_shapes["hexagonal bipyramidal"] = Atom.get_shape(
"hexagonal bipyramidal"
) # HBPY-8
try_shapes["heptagonal pyramidal"] = Atom.get_shape(
"heptagonal pyramidal"
) # HPY-8
try_shapes["octagonal"] = Atom.get_shape("octagonal") # OP-8
try_shapes["square antiprismatic"] = Atom.get_shape(
"square antiprismatic"
) # SAPR-8
try_shapes["trigonal dodecahedral"] = Atom.get_shape(
"trigonal dodecahedral"
) # TDD-8
elif len(self.connected) == 9:
try_shapes["capped cube"] = Atom.get_shape("capped cube") # CCU-9
try_shapes["capped square antiprismatic"] = Atom.get_shape(
"capped square antiprismatic"
) # CSAPR-9
try_shapes["enneagonal"] = Atom.get_shape("enneagonal") # EP-9
try_shapes["heptagonal bipyramidal"] = Atom.get_shape(
"heptagonal bipyramidal"
) # HBPY-9
try_shapes["hula-hoop"] = Atom.get_shape("hula-hoop") # HH-9
try_shapes["triangular cupola"] = Atom.get_shape(
"triangular cupola"
) # JTC-9
try_shapes["tridiminished icosahedral"] = Atom.get_shape(
"tridiminished icosahedral"
) # JTDIC-9
try_shapes["muffin"] = Atom.get_shape("muffin") # MFF-9
try_shapes["octagonal pyramidal"] = Atom.get_shape(
"octagonal pyramidal"
) # OPY-9
try_shapes["tricapped trigonal prismatic"] = Atom.get_shape(
"tricapped trigonal prismatic"
) # TCTPR-9
else:
return None, None
# make a copy of the atom and the atoms bonded to it
# set each bond length to 1 to more easily compare to the
# idealized shapes from Atom
adjusted_shape = [atom.copy() for atom in [self, *self.connected]]
for atom in adjusted_shape:
atom.coords -= self.coords
for atom in adjusted_shape[1:]:
atom.coords /= atom.dist(adjusted_shape[0])
Y = np.array([position.coords for position in adjusted_shape])
r1 = np.matmul(np.transpose(Y), Y)
u, s1, vh = np.linalg.svd(r1)
best_score = None
best_shape = None
for shape in try_shapes:
X = np.array([position.coords for position in try_shapes[shape]])
r2 = np.matmul(np.transpose(X), X)
u, s2, vh = np.linalg.svd(r2)
score = sum([abs(x1 - x2) for x1, x2 in zip(s1, s2)])
if best_score is None or score < best_score:
best_score = score
best_shape = shape
return best_shape, best_score | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/atoms.py | atoms.py |
import os
HOME = os.path.expanduser("~")
if "AARONLIB" in os.environ:
AARONLIB = os.path.abspath(os.environ["AARONLIB"])
else:
AARONLIB = os.path.join(HOME, "Aaron_libs")
AARONTOOLS = os.path.dirname(os.path.abspath(__file__))
CONNECTIVITY_THRESHOLD = 0.5
D_CUTOFF = 0.35
RMSD_CUTOFF = 0.15
ELEMENTS = [
"Bq",
"H",
"He",
"Li",
"Be",
"B",
"C",
"N",
"O",
"F",
"Ne",
"Na",
"Mg",
"Al",
"Si",
"P",
"S",
"Cl",
"Ar",
"K",
"Ca",
"Sc",
"Ti",
"V",
"Cr",
"Mn",
"Fe",
"Co",
"Ni",
"Cu",
"Zn",
"Ga",
"Ge",
"As",
"Se",
"Br",
"Kr",
"Rb",
"Sr",
"Y",
"Zr",
"Nb",
"Mo",
"Tc",
"Ru",
"Rh",
"Pd",
"Ag",
"Cd",
"In",
"Sn",
"Sb",
"Te",
"I",
"Xe",
"Cs",
"Ba",
"La",
"Ce",
"Pr",
"Nd",
"Pm",
"Sm",
"Eu",
"Gd",
"Tb",
"Dy",
"Ho",
"Er",
"Tm",
"Yb",
"Lu",
"Hf",
"Ta",
"W",
"Re",
"Os",
"Ir",
"Pt",
"Au",
"Hg",
"Tl",
"Pb",
"Bi",
"Po",
"At",
"Rn",
"Fr",
"Ra",
"Ac",
"Th",
"Pa",
"U",
"Np",
"Pu",
"Am",
"Cm",
"Bk",
"Cf",
"Es",
"Fm",
"Md",
"No",
"Lr",
"Rf",
"Db",
"Sg",
"Bh",
"Hs",
"Mt",
"Ds",
"Rg",
"Cn",
"Nh",
"Fl",
"Mc",
"Lv",
"Ts",
"Og",
"X",
]
TMETAL = {
"Sc": 1.44,
"Ti": 1.32,
"V": 1.22,
"Cr": 1.18,
"Mn": 1.17,
"Fe": 1.17,
"Co": 1.16,
"Ni": 1.15,
"Cu": 1.17,
"Zn": 1.25,
"Y": 1.62,
"Zr": 1.45,
"Nb": 1.34,
"Mo": 1.30,
"Tc": 1.27,
"Ru": 1.25,
"Rh": 1.25,
"Pd": 1.28,
"Ag": 1.34,
"Cd": 1.48,
"La": 1.69,
"Lu": 1.60,
"Hf": 1.44,
"Ta": 1.34,
"W": 1.30,
"Re": 1.28,
"Os": 1.26,
"Ir": 1.27,
"Pt": 1.30,
"Au": 1.34,
"Hg": 1.49,
}
METAL = [
"Li",
"Be",
"Na",
"Mg",
"K",
"Ca",
"Rb",
"Sr",
"Cs",
"Ba",
"Fr",
"Ra",
"Al",
"Ga",
"In",
"Sn",
"Tl",
"Pb",
"Bi",
"Nh",
"Fl",
"Mc",
"Lv",
"La",
"Ce",
"Pr",
"Nd",
"Pm",
"Sm",
"Eu",
"Gd",
"Tb",
"Dy",
"Ho",
"Er",
"Tm",
"Yb",
"Lu",
"Ac",
"Th",
"Pa",
"U",
"Np",
"Pu",
"Am",
"Cm",
"Bk",
"Cf",
"Es",
"Fm",
"Md",
"No",
"Lr",
] + list(TMETAL.keys())
RADII = {
"H": 0.32,
"He": 0.93,
"Li": 1.23,
"Be": 0.90,
"B": 0.82,
"C": 0.77,
"N": 0.75,
"O": 0.73,
"F": 0.72,
"Ne": 0.71,
"Na": 1.54,
"Mg": 1.36,
"Al": 1.18,
"Si": 1.11,
"P": 1.06,
"S": 1.02,
"Cl": 0.99,
"Ar": 0.98,
"K": 2.03,
"Ca": 1.74,
"Sc": 1.44,
"Ti": 1.32,
"V": 1.22,
"Cr": 1.18,
"Mn": 1.17,
"Fe": 1.17,
"Co": 1.16,
"Ni": 1.15,
"Cu": 1.17,
"Zn": 1.25,
"Ga": 1.26,
"Ge": 1.22,
"As": 1.20,
"Se": 1.16,
"Br": 1.14,
"Kr": 1.12,
"Rb": 2.16,
"Sr": 1.91,
"Y": 1.62,
"Zr": 1.45,
"Nb": 1.34,
"Mo": 1.30,
"Tc": 1.27,
"Ru": 1.25,
"Rh": 1.25,
"Pd": 1.28,
"Ag": 1.34,
"Cd": 1.48,
"In": 1.44,
"Sn": 1.41,
"Sb": 1.40,
"Te": 1.36,
"I": 1.33,
"Xe": 1.31,
"Cs": 2.35,
"Ba": 1.98,
"La": 1.69,
"Lu": 1.60,
"Hf": 1.44,
"Ta": 1.34,
"W": 1.30,
"Re": 1.28,
"Os": 1.26,
"Ir": 1.27,
"Pt": 1.30,
"Au": 1.34,
"Hg": 1.49,
"Tl": 1.48,
"Pb": 1.47,
"Bi": 1.46,
"X": 0,
}
CONNECTIVITY = {
"H": 1,
"B": 4,
"C": 4,
"N": 4,
"O": 4,
"F": 4,
"Si": 6,
"Rh": 6,
"Fe": 6,
"Ni": 6,
"Cu": 6,
"Ru": 6,
"Pd": 6,
"Ir": 6,
"P": 4,
"S": 4,
"Cl": 4,
"I": 6,
"Br": 6,
"X": 1000,
"Pt": 6,
"Au": 6,
}
SATURATION = {
"H": 1,
"B": 3,
"C": 4,
"N": 3,
"O": 2,
"F": 1,
"Si": 6,
"P": 4,
"S": 4,
"Cl": 1,
"I": 1,
"Br": 1,
"X": 1000,
}
ELECTRONEGATIVITY = {
"H": 2.20,
"He": None,
"Li": 0.98,
"Be": 1.57,
"B": 2.04,
"C": 2.55,
"N": 3.04,
"O": 3.44,
"F": 3.98,
"Ne": None,
"Na": 0.93,
"Mg": 1.31,
"Al": 1.61,
"Si": 1.90,
"P": 2.19,
"S": 2.58,
"Cl": 3.16,
"Ar": None,
"K": 0.82,
"Ca": 1.00,
"Sc": 1.36,
"Ti": 1.54,
"V": 1.63,
"Cr": 1.66,
"Mn": 1.55,
"Fe": 1.83,
"Co": 1.88,
"Ni": 1.91,
"Cu": 1.90,
"Zn": 1.65,
"Ga": 1.81,
"Ge": 2.01,
"As": 2.18,
"Se": 2.55,
"Br": 2.96,
"Kr": 3.00,
"Rb": 0.82,
"Sr": 0.95,
"Y": 1.22,
"Zr": 1.33,
"Nb": 1.6,
"Mo": 2.16,
"Tc": 1.9,
"Ru": 2.2,
"Rh": 2.28,
"Pd": 2.20,
"Ag": 1.93,
"Cd": 1.69,
"In": 1.78,
"Sn": 1.96,
"Sb": 2.05,
"Te": 2.1,
"I": 2.66,
"Xe": 2.6,
"Cs": 0.79,
"Ba": 0.89,
"La": 1.10,
"Ce": 1.12,
"Pr": 1.13,
"Nd": 1.14,
"Pm": None,
"Sm": 1.17,
"Eu": None,
"Gd": 1.20,
"Tb": None,
"Dy": 1.22,
"Ho": 1.23,
"Er": 1.24,
"Tm": 1.25,
"Yb": None,
"Lu": 1.27,
"Hf": 1.3,
"Ta": 1.5,
"W": 2.36,
"Re": 1.9,
"Os": 2.2,
"Ir": 2.20,
"Pt": 2.28,
"Au": 2.54,
"Hg": 2.00,
"Tl": 1.62,
"Pb": 2.33,
"Bi": 2.02,
"Po": 2.0,
"At": 2.2,
"Rn": None,
"Fr": 0.7,
"Ra": 0.9,
"Ac": 1.1,
"Th": 1.3,
"Pa": 1.5,
"U": 1.38,
"Np": 1.36,
"Pu": 1.28,
"Am": 1.3,
"Cm": 1.3,
"Bk": 1.3,
"Cf": 1.3,
"Es": 1.3,
"Fm": 1.3,
"Md": 1.3,
"No": 1.3,
}
MASS = {
"X": 0.0,
"H": 1.00782503207,
"He": 4.00260325415,
"Li": 7.016004548,
"Be": 9.012182201,
"B": 11.009305406,
"C": 12.0,
"N": 14.00307400478,
"O": 15.99491461956,
"F": 18.998403224,
"Ne": 19.99244017542,
"Na": 22.98976928087,
"Mg": 23.985041699,
"Al": 26.981538627,
"Si": 27.97692653246,
"P": 30.973761629,
"S": 31.972070999,
"Cl": 34.968852682,
"Ar": 39.96238312251,
"K": 38.963706679,
"Ca": 39.962590983,
"Sc": 44.955911909,
"Ti": 47.947946281,
"V": 50.943959507,
"Cr": 51.940507472,
"Mn": 54.938045141,
"Fe": 55.934937475,
"Co": 58.933195048,
"Ni": 57.935342907,
"Cu": 62.929597474,
"Zn": 63.929142222,
"Ga": 68.925573587,
"Ge": 73.921177767,
"As": 74.921596478,
"Se": 79.916521271,
"Br": 78.918337087,
"Kr": 85.910610729,
"Rb": 84.911789737,
"Sr": 87.905612124,
"Y": 88.905848295,
"Zr": 89.904704416,
"Nb": 92.906378058,
"Mo": 97.905408169,
"Tc": 98.906254747,
"Ru": 101.904349312,
"Rh": 102.905504292,
"Pd": 105.903485715,
"Ag": 106.90509682,
"Cd": 113.90335854,
"In": 114.903878484,
"Sn": 119.902194676,
"Sb": 120.903815686,
"Te": 129.906224399,
"I": 126.904472681,
"Xe": 131.904153457,
"Cs": 132.905451932,
"Ba": 137.905247237,
"La": 138.906353267,
"Lu": 174.940771819,
"Hf": 179.946549953,
"Ta": 180.947995763,
"W": 183.950931188,
"Re": 186.955753109,
"Os": 191.96148069,
"Ir": 192.96292643,
"Pt": 194.964791134,
"Au": 196.966568662,
"Hg": 201.970643011,
"Tl": 204.974427541,
"Pb": 207.976652071,
"Bi": 208.980398734,
}
EIJ = {
"CC": 0.1500,
"CN": 0.1549,
"NC": 0.1549,
"CO": 0.1732,
"OC": 0.1732,
"CP": 0.1732,
"PC": 0.1732,
"CS": 0.1732,
"SC": 0.1732,
"CH": 0.0548,
"HC": 0.0548,
"CFe": 0.0387,
"FeC": 0.0387,
"CF": 0.1095,
"FC": 0.1095,
"CCl": 0.2035,
"ClC": 0.2035,
"CBr": 0.2416,
"BrC": 0.2416,
"CI": 0.2877,
"IC": 0.2877,
"CMg": 0.3623,
"MgC": 0.3623,
"CZn": 0.2872,
"ZnC": 0.2872,
"CCa": 0.2872,
"CaC": 0.2872,
"NC": 0.1549,
"CN": 0.1549,
"NN": 0.1600,
"NO": 0.1789,
"ON": 0.1789,
"NP": 0.1789,
"PN": 0.1789,
"NS": 0.1789,
"SN": 0.1789,
"NH": 0.0566,
"HN": 0.0566,
"NFe": 0.0400,
"FeN": 0.0400,
"NF": 0.1131,
"FN": 0.1131,
"NCl": 0.2101,
"ClN": 0.2101,
"NBr": 0.2495,
"BrN": 0.2495,
"NI": 0.2972,
"IN": 0.2972,
"NMg": 0.3742,
"MgN": 0.3742,
"NZn": 0.2966,
"ZnN": 0.2966,
"NCa": 0.2966,
"CaN": 0.2966,
"OC": 0.1732,
"CO": 0.1732,
"ON": 0.1789,
"NO": 0.1789,
"OO": 0.2000,
"OP": 0.2000,
"PO": 0.2000,
"OS": 0.2000,
"SO": 0.2000,
"OH": 0.0632,
"HO": 0.0632,
"OFe": 0.0447,
"FeO": 0.0447,
"OF": 0.1265,
"FO": 0.1265,
"OCl": 0.2349,
"ClO": 0.2349,
"OBr": 0.2789,
"BrO": 0.2789,
"OI": 0.3323,
"IO": 0.3323,
"OMg": 0.4183,
"MgO": 0.4183,
"OZn": 0.3317,
"ZnO": 0.3317,
"OCa": 0.3317,
"CaO": 0.3317,
"PC": 0.1732,
"CP": 0.1732,
"PN": 0.1789,
"NP": 0.1789,
"PO": 0.2000,
"OP": 0.2000,
"PP": 0.2000,
"PS": 0.2000,
"SP": 0.2000,
"PH": 0.0632,
"HP": 0.0632,
"PFe": 0.0447,
"FeP": 0.0447,
"PF": 0.1265,
"FP": 0.1265,
"PCl": 0.2349,
"ClP": 0.2349,
"PBr": 0.2789,
"BrP": 0.2789,
"PI": 0.3323,
"IP": 0.3323,
"PMg": 0.4183,
"MgP": 0.4183,
"PZn": 0.3317,
"ZnP": 0.3317,
"PCa": 0.3317,
"CaP": 0.3317,
"SC": 0.1732,
"CS": 0.1732,
"SN": 0.1789,
"NS": 0.1789,
"SO": 0.2000,
"OS": 0.2000,
"SP": 0.2000,
"PS": 0.2000,
"SS": 0.2000,
"SH": 0.0632,
"HS": 0.0632,
"SFe": 0.0447,
"FeS": 0.0447,
"SF": 0.1265,
"FS": 0.1265,
"SCl": 0.2349,
"ClS": 0.2349,
"SBr": 0.2789,
"BrS": 0.2789,
"SI": 0.3323,
"IS": 0.3323,
"SMg": 0.4183,
"MgS": 0.4183,
"SZn": 0.3317,
"ZnS": 0.3317,
"SCa": 0.3317,
"CaS": 0.3317,
"HC": 0.0548,
"CH": 0.0548,
"HN": 0.0566,
"NH": 0.0566,
"HO": 0.0632,
"OH": 0.0632,
"HP": 0.0632,
"PH": 0.0632,
"HS": 0.0632,
"SH": 0.0632,
"HH": 0.0200,
"HFe": 0.0141,
"FeH": 0.0141,
"HF": 0.0400,
"FH": 0.0400,
"HCl": 0.0743,
"ClH": 0.0743,
"HBr": 0.0882,
"BrH": 0.0882,
"HI": 0.1051,
"IH": 0.1051,
"HMg": 0.1323,
"MgH": 0.1323,
"HZn": 0.1049,
"ZnH": 0.1049,
"HCa": 0.1049,
"CaH": 0.1049,
"FeC": 0.0387,
"CFe": 0.0387,
"FeN": 0.0400,
"NFe": 0.0400,
"FeO": 0.0447,
"OFe": 0.0447,
"FeP": 0.0447,
"PFe": 0.0447,
"FeS": 0.0447,
"SFe": 0.0447,
"FeH": 0.0141,
"HFe": 0.0141,
"FeFe": 0.0100,
"FeF": 0.0283,
"FFe": 0.0283,
"FeCl": 0.0525,
"ClFe": 0.0525,
"FeBr": 0.0624,
"BrFe": 0.0624,
"FeI": 0.0743,
"IFe": 0.0743,
"FeMg": 0.0935,
"MgFe": 0.0935,
"FeZn": 0.0742,
"ZnFe": 0.0742,
"FeCa": 0.0742,
"CaFe": 0.0742,
"FC": 0.1095,
"CF": 0.1095,
"FN": 0.1131,
"NF": 0.1131,
"FO": 0.1265,
"OF": 0.1265,
"FP": 0.1265,
"PF": 0.1265,
"FS": 0.1265,
"SF": 0.1265,
"FH": 0.0400,
"HF": 0.0400,
"FFe": 0.0283,
"FeF": 0.0283,
"FF": 0.0800,
"FCl": 0.1486,
"ClF": 0.1486,
"FBr": 0.1764,
"BrF": 0.1764,
"FI": 0.2101,
"IF": 0.2101,
"FMg": 0.2646,
"MgF": 0.2646,
"FZn": 0.2098,
"ZnF": 0.2098,
"FCa": 0.2098,
"CaF": 0.2098,
"ClC": 0.2035,
"CCl": 0.2035,
"ClN": 0.2101,
"NCl": 0.2101,
"ClO": 0.2349,
"OCl": 0.2349,
"ClP": 0.2349,
"PCl": 0.2349,
"ClS": 0.2349,
"SCl": 0.2349,
"ClH": 0.0743,
"HCl": 0.0743,
"ClFe": 0.0525,
"FeCl": 0.0525,
"ClF": 0.1486,
"FCl": 0.1486,
"ClCl": 0.2760,
"ClBr": 0.3277,
"BrCl": 0.3277,
"ClI": 0.3903,
"ICl": 0.3903,
"ClMg": 0.4914,
"MgCl": 0.4914,
"ClZn": 0.3896,
"ZnCl": 0.3896,
"ClCa": 0.3896,
"CaCl": 0.3896,
"BrC": 0.2416,
"CBr": 0.2416,
"BrN": 0.2495,
"NBr": 0.2495,
"BrO": 0.2789,
"OBr": 0.2789,
"BrP": 0.2789,
"PBr": 0.2789,
"BrS": 0.2789,
"SBr": 0.2789,
"BrH": 0.0882,
"HBr": 0.0882,
"BrFe": 0.0624,
"FeBr": 0.0624,
"BrF": 0.1764,
"FBr": 0.1764,
"BrCl": 0.3277,
"ClBr": 0.3277,
"BrBr": 0.3890,
"BrI": 0.4634,
"IBr": 0.4634,
"BrMg": 0.5834,
"MgBr": 0.5834,
"BrZn": 0.4625,
"ZnBr": 0.4625,
"BrCa": 0.4625,
"CaBr": 0.4625,
"IC": 0.2877,
"CI": 0.2877,
"IN": 0.2972,
"NI": 0.2972,
"IO": 0.3323,
"OI": 0.3323,
"IP": 0.3323,
"PI": 0.3323,
"IS": 0.3323,
"SI": 0.3323,
"IH": 0.1051,
"HI": 0.1051,
"IFe": 0.0743,
"FeI": 0.0743,
"IF": 0.2101,
"FI": 0.2101,
"ICl": 0.3903,
"ClI": 0.3903,
"IBr": 0.4634,
"BrI": 0.4634,
"II": 0.5520,
"IMg": 0.6950,
"MgI": 0.6950,
"IZn": 0.5510,
"ZnI": 0.5510,
"ICa": 0.5510,
"CaI": 0.5510,
"MgC": 0.3623,
"CMg": 0.3623,
"MgN": 0.3742,
"NMg": 0.3742,
"MgO": 0.4183,
"OMg": 0.4183,
"MgP": 0.4183,
"PMg": 0.4183,
"MgS": 0.4183,
"SMg": 0.4183,
"MgH": 0.1323,
"HMg": 0.1323,
"MgFe": 0.0935,
"FeMg": 0.0935,
"MgF": 0.2646,
"FMg": 0.2646,
"MgCl": 0.4914,
"ClMg": 0.4914,
"MgBr": 0.5834,
"BrMg": 0.5834,
"MgI": 0.6950,
"IMg": 0.6950,
"MgMg": 0.8750,
"MgZn": 0.6937,
"ZnMg": 0.6937,
"MgCa": 0.6937,
"CaMg": 0.6937,
"ZnC": 0.2872,
"CZn": 0.2872,
"ZnN": 0.2966,
"NZn": 0.2966,
"ZnO": 0.3317,
"OZn": 0.3317,
"ZnP": 0.3317,
"PZn": 0.3317,
"ZnS": 0.3317,
"SZn": 0.3317,
"ZnH": 0.1049,
"HZn": 0.1049,
"ZnFe": 0.0742,
"FeZn": 0.0742,
"ZnF": 0.2098,
"FZn": 0.2098,
"ZnCl": 0.3896,
"ClZn": 0.3896,
"ZnBr": 0.4625,
"BrZn": 0.4625,
"ZnI": 0.5510,
"IZn": 0.5510,
"ZnMg": 0.6937,
"MgZn": 0.6937,
"ZnZn": 0.5500,
"ZnCa": 0.5500,
"CaZn": 0.5500,
"CaC": 0.2872,
"CCa": 0.2872,
"CaN": 0.2966,
"NCa": 0.2966,
"CaO": 0.3317,
"OCa": 0.3317,
"CaP": 0.3317,
"PCa": 0.3317,
"CaS": 0.3317,
"SCa": 0.3317,
"CaH": 0.1049,
"HCa": 0.1049,
"CaFe": 0.0742,
"FeCa": 0.0742,
"CaF": 0.2098,
"FCa": 0.2098,
"CaCl": 0.3896,
"ClCa": 0.3896,
"CaBr": 0.4625,
"BrCa": 0.4625,
"CaI": 0.5510,
"ICa": 0.5510,
"CaMg": 0.6937,
"MgCa": 0.6937,
"CaZn": 0.5500,
"ZnCa": 0.5500,
"CaCa": 0.5500,
"X": 0,
}
RIJ = {
"CC": 4.00,
"CN": 3.75,
"NC": 3.75,
"CO": 3.60,
"OC": 3.60,
"CP": 4.10,
"PC": 4.10,
"CS": 4.00,
"SC": 4.00,
"CH": 3.00,
"HC": 3.00,
"CFe": 2.65,
"FeC": 2.65,
"CF": 3.54,
"FC": 3.54,
"CCl": 4.04,
"ClC": 4.04,
"CBr": 4.17,
"BrC": 4.17,
"CI": 4.36,
"IC": 4.36,
"CMg": 2.65,
"MgC": 2.65,
"CZn": 2.74,
"ZnC": 2.74,
"CCa": 2.99,
"CaC": 2.99,
"NC": 3.75,
"CN": 3.75,
"NN": 3.50,
"NO": 3.35,
"ON": 3.35,
"NP": 3.85,
"PN": 3.85,
"NS": 3.75,
"SN": 3.75,
"NH": 2.75,
"HN": 2.75,
"NFe": 2.40,
"FeN": 2.40,
"NF": 3.29,
"FN": 3.29,
"NCl": 3.79,
"ClN": 3.79,
"NBr": 3.92,
"BrN": 3.92,
"NI": 4.11,
"IN": 4.11,
"NMg": 2.40,
"MgN": 2.40,
"NZn": 2.49,
"ZnN": 2.49,
"NCa": 2.74,
"CaN": 2.74,
"OC": 3.60,
"CO": 3.60,
"ON": 3.35,
"NO": 3.35,
"OO": 3.20,
"OP": 3.70,
"PO": 3.70,
"OS": 3.60,
"SO": 3.60,
"OH": 2.60,
"HO": 2.60,
"OFe": 2.25,
"FeO": 2.25,
"OF": 3.15,
"FO": 3.15,
"OCl": 3.65,
"ClO": 3.65,
"OBr": 3.77,
"BrO": 3.77,
"OI": 3.96,
"IO": 3.96,
"OMg": 2.25,
"MgO": 2.25,
"OZn": 2.34,
"ZnO": 2.34,
"OCa": 2.59,
"CaO": 2.59,
"PC": 4.10,
"CP": 4.10,
"PN": 3.85,
"NP": 3.85,
"PO": 3.70,
"OP": 3.70,
"PP": 4.20,
"PS": 4.10,
"SP": 4.10,
"PH": 3.10,
"HP": 3.10,
"PFe": 2.75,
"FeP": 2.75,
"PF": 3.65,
"FP": 3.65,
"PCl": 4.14,
"ClP": 4.14,
"PBr": 4.27,
"BrP": 4.27,
"PI": 4.46,
"IP": 4.46,
"PMg": 2.75,
"MgP": 2.75,
"PZn": 2.84,
"ZnP": 2.84,
"PCa": 3.09,
"CaP": 3.09,
"SC": 4.00,
"CS": 4.00,
"SN": 3.75,
"NS": 3.75,
"SO": 3.60,
"OS": 3.60,
"SP": 4.10,
"PS": 4.10,
"SS": 4.00,
"SH": 3.00,
"HS": 3.00,
"SFe": 2.65,
"FeS": 2.65,
"SF": 3.54,
"FS": 3.54,
"SCl": 4.04,
"ClS": 4.04,
"SBr": 4.17,
"BrS": 4.17,
"SI": 4.36,
"IS": 4.36,
"SMg": 2.65,
"MgS": 2.65,
"SZn": 2.74,
"ZnS": 2.74,
"SCa": 2.99,
"CaS": 2.99,
"HC": 3.00,
"CH": 3.00,
"HN": 2.75,
"NH": 2.75,
"HO": 2.60,
"OH": 2.60,
"HP": 3.10,
"PH": 3.10,
"HS": 3.00,
"SH": 3.00,
"HH": 2.00,
"HFe": 1.65,
"FeH": 1.65,
"HF": 2.54,
"FH": 2.54,
"HCl": 3.04,
"ClH": 3.04,
"HBr": 3.17,
"BrH": 3.17,
"HI": 3.36,
"IH": 3.36,
"HMg": 1.65,
"MgH": 1.65,
"HZn": 1.74,
"ZnH": 1.74,
"HCa": 1.99,
"CaH": 1.99,
"FeC": 2.65,
"CFe": 2.65,
"FeN": 2.40,
"NFe": 2.40,
"FeO": 2.25,
"OFe": 2.25,
"FeP": 2.75,
"PFe": 2.75,
"FeS": 2.65,
"SFe": 2.65,
"FeH": 1.65,
"HFe": 1.65,
"FeFe": 1.30,
"FeF": 2.19,
"FFe": 2.19,
"FeCl": 2.69,
"ClFe": 2.69,
"FeBr": 2.81,
"BrFe": 2.81,
"FeI": 3.01,
"IFe": 3.01,
"FeMg": 1.30,
"MgFe": 1.30,
"FeZn": 1.39,
"ZnFe": 1.39,
"FeCa": 1.64,
"CaFe": 1.64,
"FC": 3.54,
"CF": 3.54,
"FN": 3.29,
"NF": 3.29,
"FO": 3.15,
"OF": 3.15,
"FP": 3.65,
"PF": 3.65,
"FS": 3.54,
"SF": 3.54,
"FH": 2.54,
"HF": 2.54,
"FFe": 2.19,
"FeF": 2.19,
"FF": 3.09,
"FCl": 3.59,
"ClF": 3.59,
"FBr": 3.71,
"BrF": 3.71,
"FI": 3.90,
"IF": 3.90,
"FMg": 2.19,
"MgF": 2.19,
"FZn": 2.29,
"ZnF": 2.29,
"FCa": 2.54,
"CaF": 2.54,
"ClC": 4.04,
"CCl": 4.04,
"ClN": 3.79,
"NCl": 3.79,
"ClO": 3.65,
"OCl": 3.65,
"ClP": 4.14,
"PCl": 4.14,
"ClS": 4.04,
"SCl": 4.04,
"ClH": 3.04,
"HCl": 3.04,
"ClFe": 2.69,
"FeCl": 2.69,
"ClF": 3.59,
"FCl": 3.59,
"ClCl": 4.09,
"ClBr": 4.21,
"BrCl": 4.21,
"ClI": 4.40,
"ICl": 4.40,
"ClMg": 2.69,
"MgCl": 2.69,
"ClZn": 2.79,
"ZnCl": 2.79,
"ClCa": 3.04,
"CaCl": 3.04,
"BrC": 4.17,
"CBr": 4.17,
"BrN": 3.92,
"NBr": 3.92,
"BrO": 3.77,
"OBr": 3.77,
"BrP": 4.27,
"PBr": 4.27,
"BrS": 4.17,
"SBr": 4.17,
"BrH": 3.17,
"HBr": 3.17,
"BrFe": 2.81,
"FeBr": 2.81,
"BrF": 3.71,
"FBr": 3.71,
"BrCl": 4.21,
"ClBr": 4.21,
"BrBr": 4.33,
"BrI": 4.53,
"IBr": 4.53,
"BrMg": 2.81,
"MgBr": 2.81,
"BrZn": 2.91,
"ZnBr": 2.91,
"BrCa": 3.16,
"CaBr": 3.16,
"IC": 4.36,
"CI": 4.36,
"IN": 4.11,
"NI": 4.11,
"IO": 3.96,
"OI": 3.96,
"IP": 4.46,
"PI": 4.46,
"IS": 4.36,
"SI": 4.36,
"IH": 3.36,
"HI": 3.36,
"IFe": 3.01,
"FeI": 3.01,
"IF": 3.90,
"FI": 3.90,
"ICl": 4.40,
"ClI": 4.40,
"IBr": 4.53,
"BrI": 4.53,
"II": 4.72,
"IMg": 3.01,
"MgI": 3.01,
"IZn": 3.10,
"ZnI": 3.10,
"ICa": 3.35,
"CaI": 3.35,
"MgC": 2.65,
"CMg": 2.65,
"MgN": 2.40,
"NMg": 2.40,
"MgO": 2.25,
"OMg": 2.25,
"MgP": 2.75,
"PMg": 2.75,
"MgS": 2.65,
"SMg": 2.65,
"MgH": 1.65,
"HMg": 1.65,
"MgFe": 1.30,
"FeMg": 1.30,
"MgF": 2.19,
"FMg": 2.19,
"MgCl": 2.69,
"ClMg": 2.69,
"MgBr": 2.81,
"BrMg": 2.81,
"MgI": 3.01,
"IMg": 3.01,
"MgMg": 1.30,
"MgZn": 1.39,
"ZnMg": 1.39,
"MgCa": 1.64,
"CaMg": 1.64,
"ZnC": 2.74,
"CZn": 2.74,
"ZnN": 2.49,
"NZn": 2.49,
"ZnO": 2.34,
"OZn": 2.34,
"ZnP": 2.84,
"PZn": 2.84,
"ZnS": 2.74,
"SZn": 2.74,
"ZnH": 1.74,
"HZn": 1.74,
"ZnFe": 1.39,
"FeZn": 1.39,
"ZnF": 2.29,
"FZn": 2.29,
"ZnCl": 2.79,
"ClZn": 2.79,
"ZnBr": 2.91,
"BrZn": 2.91,
"ZnI": 3.10,
"IZn": 3.10,
"ZnMg": 1.39,
"MgZn": 1.39,
"ZnZn": 1.48,
"ZnCa": 1.73,
"CaZn": 1.73,
"CaC": 2.99,
"CCa": 2.99,
"CaN": 2.74,
"NCa": 2.74,
"CaO": 2.59,
"OCa": 2.59,
"CaP": 3.09,
"PCa": 3.09,
"CaS": 2.99,
"SCa": 2.99,
"CaH": 1.99,
"HCa": 1.99,
"CaFe": 1.64,
"FeCa": 1.64,
"CaF": 2.54,
"FCa": 2.54,
"CaCl": 3.04,
"ClCa": 3.04,
"CaBr": 3.16,
"BrCa": 3.16,
"CaI": 3.35,
"ICa": 3.35,
"CaMg": 1.64,
"MgCa": 1.64,
"CaZn": 1.73,
"ZnCa": 1.73,
"CaCa": 1.98,
"X": 0,
}
ATOM_TYPES = [
"c",
"c1",
"c2",
"c3",
"ca",
"n",
"n1",
"n2",
"n3",
"n4",
"na",
"nh",
"no",
"f",
"cl",
"br",
"i",
"o",
"oh",
"os",
"s2",
"sh",
"ss",
"s4",
"s6",
"hc",
"ha",
"hn",
"ho",
"hs",
"hp",
"p2",
"p3",
"p4",
"p5",
"h1",
"h2",
"h3",
"h4",
"h5",
"nb",
"nc",
"sx",
"sy",
"cc",
"ce",
"cp",
"cu",
"cv",
"cx",
"cy",
"pb",
"pc",
"pd",
"pe",
"px",
"py",
"H",
"HC",
"HO",
"HS",
"HW",
"H2",
"H3",
"C",
"CA",
"CB",
"CC",
"CK",
"CM",
"CN",
"CQ",
"CR",
"CT",
"CV",
"CW",
"C*",
"CD",
"CE",
"CF",
"CG",
"CH",
"CI",
"CJ",
"CP",
"C2",
"C3",
"N",
"NA",
"NB",
"NC",
"NT",
"N2",
"N3",
"N*",
"O",
"OH",
"OS",
"OW",
"O2",
"S",
"SH",
"P",
"CU",
"CO",
"I",
"IM",
"MG",
"QC",
"QK",
"QL",
"QN",
"QR",
"LP",
"AH",
"BH",
"HT",
"HY",
"AC",
"BC",
"CS",
"OA",
"OB",
"OE",
"OT",
]
# main group vdw radii from J. Phys. Chem. A 2009, 113, 19, 5806–5812
# (DOI: 10.1021/jp8111556)
# transition metals are crystal radii from Batsanov, S.S. Van der Waals
# Radii of Elements. Inorganic Materials 37, 871–885 (2001).
# (DOI: 10.1023/A:1011625728803)
# transition metals are indented a bit more than the rest
VDW_RADII = {
"H": 1.10,
"He": 1.40,
"Li": 1.81,
"Be": 1.53,
"B": 1.92,
"C": 1.70,
"N": 1.55,
"O": 1.52,
"F": 1.47,
"Ne": 1.54,
"Na": 2.27,
"Mg": 1.73,
"Al": 1.84,
"Si": 2.10,
"P": 1.80,
"S": 1.80,
"Cl": 1.75,
"Ar": 1.88,
"K": 2.75,
"Ca": 2.31,
"Sc": 2.3,
"Ti": 2.15,
"V": 2.05,
"Cr": 2.05,
"Mn": 2.05,
"Fe": 2.05,
"Co": 2.0,
"Ni": 2.0,
"Cu": 2.0,
"Zn": 2.1,
"Ga": 1.87,
"Ge": 2.11,
"As": 1.85,
"Se": 1.90,
"Br": 1.83,
"Kr": 2.02,
"Rb": 3.03,
"Sr": 2.49,
"Y": 2.4,
"Zr": 2.3,
"Nb": 2.15,
"Mo": 2.1,
"Tc": 2.05,
"Ru": 2.05,
"Rh": 2.0,
"Pd": 2.05,
"Ag": 2.1,
"Cd": 2.2,
"In": 1.93,
"Sn": 2.17,
"Sb": 2.06,
"Te": 2.06,
"I": 1.98,
"Xe": 2.16,
"Cs": 3.43,
"Ba": 2.68,
"La": 2.5,
"Hf": 2.25,
"Ta": 2.2,
"W": 2.1,
"Re": 2.05,
"Os": 2.0,
"Ir": 2.0,
"Pt": 2.05,
"Au": 2.1,
"Hg": 2.05,
"Tl": 1.96,
"Pb": 2.02,
"Bi": 2.07,
"Po": 1.97,
"At": 2.02,
"Rn": 2.20,
"Fr": 3.48,
"Ra": 2.83,
"X": 0,
}
# J. Phys. Chem. 1964, 68, 3, 441–451 (DOI: 10.1021/j100785a001)
BONDI_RADII = {
"H": 1.20,
"He": 1.40,
"C": 1.70,
"N": 1.55,
"O": 1.52,
"F": 1.47,
"Ne": 1.54,
"Si": 2.10,
"P": 1.80,
"S": 1.80,
"Cl": 1.75,
"Ar": 1.88,
"As": 1.85,
"Se": 1.90,
"Br": 1.85,
"Kr": 2.02,
"Te": 2.06,
"I": 1.98,
"Xe": 2.16,
}
FREQUENCY_SCALE_LIBS = {
"NIST CCCBDB": (
"https://cccbdb.nist.gov/vibscalejust.asp",
{
"HF": {
"STO-3G": 0.817,
"3-21G": 0.906,
"3-21G(d)": 0.903,
"6-31G": 0.903,
"6-31G(d)": 0.899,
"6-31G(d,p)": 0.903,
"6-31+G(d,p)": 0.904,
"6-311G(d)": 0.904,
"6-311G(d,p)": 0.909,
"6-31G(2df,p)": 0.906,
"6-311+G(3df,2p)": 0.909,
"6-311+G(3df,2pd)": 0.906,
"TZVP": 0.909,
"cc-pVDZ": 0.908,
"cc-pVTZ": 0.910,
"cc-pVQZ": 0.908,
"aug-cc-pVDZ": 0.911,
"aug-cc-pVTZ": 0.910,
"aug-cc-pVQZ": 0.909,
"cc-pV(T+d)Z": 0.910,
"cc-pCVDZ": 0.916,
"cc-pCVTZ": 0.913,
"daug-cc-pVDZ": 0.912,
"daug-cc-pVTZ": 0.905,
"Sadlej_pVTZ": 0.913,
},
"ROHF": {
"3-21G": 0.907,
"3-21G(d)": 0.909,
"6-31G": 0.895,
"6-31G(d)": 0.890,
"6-31G(d,p)": 0.855,
"6-31+G(d,p)": 0.856,
"6-311G(d)": 0.856,
"6-311G(d,p)": 0.913,
"6-311+G(3df,2p)": 0.909,
"cc-pVDZ": 0.861,
"cc-pVTZ": 0.901,
},
"LSDA": {
"STO-3G": 0.896,
"3-21G": 0.984,
"3-21G(d)": 0.982,
"6-31G": 0.980,
"6-31G(d)": 0.981,
"6-31G(d,p)": 0.981,
"6-31+G(d,p)": 0.985,
"6-311G(d)": 0.984,
"6-311G(d,p)": 0.988,
"6-31G(2df,p)": 0.984,
"TZVP": 0.988,
"cc-pVDZ": 0.989,
"cc-pVTZ": 0.989,
"aug-cc-pVDZ": 0.989,
"aug-cc-pVTZ": 0.991,
"cc-pV(T+d)Z": 0.990,
},
"BLYP": {
"STO-3G": 0.925,
"3-21G": 0.995,
"3-21G(d)": 0.994,
"6-31G": 0.992,
"6-31G(d)": 0.992,
"6-31G(d,p)": 0.992,
"6-31+G(d,p)": 0.995,
"6-311G(d)": 0.998,
"6-311G(d,p)": 0.996,
"6-31G(2df,p)": 0.995,
"6-311+G(3df,2p)": 0.995,
"TZVP": 0.998,
"cc-pVDZ": 1.002,
"cc-pVTZ": 0.997,
"aug-cc-pVDZ": 0.998,
"aug-cc-pVTZ": 0.997,
"cc-pV(T+d)Z": 0.996,
},
"B1B95": {
"STO-3G": 0.883,
"3-21G": 0.957,
"3-21G(d)": 0.955,
"6-31G": 0.954,
"6-31G(d)": 0.949,
"6-31G(d,p)": 0.955,
"6-31+G(d,p)": 0.957,
"6-311G(d)": 0.959,
"6-311G(d,p)": 0.960,
"6-31G(2df,p)": 0.958,
"TZVP": 0.957,
"cc-pVDZ": 0.961,
"cc-pVTZ": 0.957,
"aug-cc-pVDZ": 0.958,
"aug-cc-pVTZ": 0.959,
"cc-pV(T+d)Z": 0.957,
},
"B3LYP": {
"STO-3G": 0.892,
"3-21G": 0.965,
"3-21G(d)": 0.962,
"6-31G": 0.962,
"6-31G(d)": 0.960,
"6-31G(d,p)": 0.961,
"6-31+G(d,p)": 0.964,
"6-311G(d)": 0.966,
"6-311G(d,p)": 0.967,
"6-31G(2df,p)": 0.965,
"6-311+G(3df,2p)": 0.967,
"6-311+G(3df,2pd)": 0.964,
"TZVP": 0.965,
"cc-pVDZ": 0.970,
"cc-pVTZ": 0.967,
"cc-pVQZ": 0.969,
"aug-cc-pVDZ": 0.970,
"aug-cc-pVTZ": 0.968,
"aug-cc-pVQZ": 0.969,
"cc-pV(T+d)Z": 0.965,
"Sadlej_pVTZ": 0.972,
},
"B3LYP (ultrafine grid)": {
"STO-3G": 0.892,
"3-21G": 0.965,
"3-21G(d)": 0.962,
"6-31G": 0.962,
"6-31G(d)": 0.958,
"6-31G(d,p)": 0.961,
"6-31+G(d,p)": 0.963,
"6-311G(d)": 0.966,
"6-311G(d,p)": 0.967,
"6-31G(2df,p)": 0.965,
"6-311+G(3df,2pd)": 0.970,
"TZVP": 0.963,
"cc-pVDZ": 0.970,
"cc-pVTZ": 0.967,
"aug-cc-pVDZ": 0.970,
"aug-cc-pVTZ": 0.968,
},
"B3PW91": {
"STO-3G": 0.885,
"3-21G": 0.961,
"3-21G(d)": 0.959,
"6-31G": 0.958,
"6-31G(d)": 0.957,
"6-31G(d,p)": 0.958,
"6-31+G(d,p)": 0.960,
"6-311G(d)": 0.963,
"6-311G(d,p)": 0.963,
"6-31G(2df,p)": 0.961,
"6-311+G(3df,2p)": 0.957,
"TZVP": 0.964,
"cc-pVDZ": 0.965,
"cc-pVTZ": 0.962,
"aug-cc-pVDZ": 0.965,
"aug-cc-pVTZ": 0.965,
"cc-pV(T+d)Z": 0.964,
},
"mPW1PW91": {
"STO-3G": 0.879,
"3-21G": 0.955,
"3-21G(d)": 0.950,
"6-31G": 0.947,
"6-31G(d)": 0.948,
"6-31G(d,p)": 0.952,
"6-31+G(d,p)": 0.952,
"6-311G(d)": 0.954,
"6-311G(d,p)": 0.957,
"6-31G(2df,p)": 0.955,
"TZVP": 0.954,
"cc-pVDZ": 0.958,
"cc-pVTZ": 0.959,
"aug-cc-pVDZ": 0.958,
"aug-cc-pVTZ": 0.958,
"cc-pV(T+d)Z": 0.958,
},
"M06-2X": {
"3-21G": 0.959,
"3-21G(d)": 0.947,
"6-31G(d)": 0.947,
"6-31G(d,p)": 0.950,
"6-31+G(d,p)": 0.952,
"6-31G(2df,p)": 0.952,
"TZVP": 0.946,
"cc-pVTZ": 0.955,
"aug-cc-pVTZ": 0.956,
},
"PBEPBE": {
"STO-3G": 0.914,
"3-21G": 0.991,
"3-21G(d)": 0.954,
"6-31G": 0.986,
"6-31G(d)": 0.986,
"6-31G(d,p)": 0.986,
"6-31+G(d,p)": 0.989,
"6-311G(d)": 0.990,
"6-311G(d,p)": 0.991,
"6-31G(2df,p)": 0.990,
"6-311+G(3df,2p)": 0.992,
"6-311+G(3df,2pd)": 0.990,
"TZVP": 0.989,
"cc-pVDZ": 0.994,
"cc-pVTZ": 0.993,
"aug-cc-pVDZ": 0.994,
"aug-cc-pVTZ": 0.994,
"cc-pV(T+d)Z": 0.993,
"Sadlej_pVTZ": 0.995,
},
"PBEPBE (ultrafine grid)": {
"STO-3G": 0.914,
"3-21G": 0.991,
"3-21G(d)": 0.954,
"6-31G": 0.986,
"6-31G(d)": 0.984,
"6-31G(d,p)": 0.986,
"6-31+G(d,p)": 0.989,
"6-311G(d)": 0.990,
"6-311G(d,p)": 0.991,
"6-31G(2df,p)": 0.990,
"6-311+G(3df,2pd)": 0.990,
"TZVP": 0.989,
"cc-pVDZ": 0.994,
"cc-pVTZ": 0.993,
"aug-cc-pVDZ": 0.994,
"aug-cc-pVTZ": 0.989,
},
"PBE0": {
"STO-3G": 0.882,
"3-21G": 0.960,
"3-21G(d)": 0.960,
"6-31G": 0.956,
"6-31G(d)": 0.950,
"6-31G(d,p)": 0.953,
"6-31+G(d,p)": 0.955,
"6-311G(d)": 0.959,
"6-311G(d,p)": 0.959,
"6-31G(2df,p)": 0.957,
"TZVP": 0.960,
"cc-pVDZ": 0.962,
"cc-pVTZ": 0.961,
"aug-cc-pVDZ": 0.962,
"aug-cc-pVTZ": 0.962,
},
"HSEh1PBE": {
"STO-3G": 0.883,
"3-21G": 0.963,
"3-21G(d)": 0.960,
"6-31G": 0.957,
"6-31G(d)": 0.951,
"6-31G(d,p)": 0.954,
"6-31+G(d,p)": 0.955,
"6-311G(d)": 0.960,
"6-311G(d,p)": 0.960,
"6-31G(2df,p)": 0.958,
"TZVP": 0.960,
"cc-pVDZ": 0.962,
"cc-pVTZ": 0.961,
"aug-cc-pVDZ": 0.962,
"aug-cc-pVTZ": 0.962,
},
"TPSSh": {
"3-21G": 0.969,
"3-21G(d)": 0.966,
"6-31G": 0.962,
"6-31G(d)": 0.959,
"6-31G(d,p)": 0.959,
"6-31+G(d,p)": 0.963,
"6-311G(d)": 0.963,
"6-31G(2df,p)": 0.965,
"TZVP": 0.964,
"cc-pVDZ": 0.972,
"cc-pVTZ": 0.968,
"aug-cc-pVDZ": 0.967,
"aug-cc-pVTZ": 0.965,
},
"ωB97X-D": {
"3-21G(d)": 0.948,
"6-31G(d)": 0.949,
"6-31+G(d,p)": 0.952,
"6-311G(d,p)": 0.957,
"TZVP": 0.955,
"cc-pVDZ": 0.953,
"cc-pVTZ": 0.956,
"aug-cc-pVDZ": 0.957,
"aug-cc-pVTZ": 0.957,
},
"B97-D3": {
"3-21G": 0.983,
"6-31G(d)": 0.980,
"6-31+G(d,p)": 0.983,
"6-311G(d,p)": 0.986,
"6-311+G(3df,2p)": 0.987,
"6-311+G(3df,2pd)": 0.986,
"TZVP": 0.986,
"cc-pVDZ": 0.992,
"cc-pVTZ": 0.986,
"aug-cc-pVTZ": 0.985,
},
"MP2": {
"STO-3G": 0.872,
"3-21G": 0.955,
"3-21G(d)": 0.951,
"6-31G": 0.957,
"6-31G(d)": 0.943,
"6-31G(d,p)": 0.937,
"6-31+G(d,p)": 0.941,
"6-311G(d)": 0.950,
"6-311G(d,p)": 0.950,
"6-31G(2df,p)": 0.945,
"6-311+G(3df,2p)": 0.943,
"6-311+G(3df,2pd)": 0.950,
"TZVP": 0.948,
"cc-pVDZ": 0.953,
"cc-pVTZ": 0.950,
"cc-pVQZ": 0.948,
"aug-cc-pVDZ": 0.959,
"aug-cc-pVTZ": 0.953,
"aug-cc-pVQZ": 0.950,
"cc-pV(T+d)Z": 0.953,
"cc-pCVDZ": 0.956,
"cc-pCVTZ": 0.953,
"Sadlej_pVTZ": 0.962,
},
"MP2=Full": {
"STO-3G": 0.889,
"3-21G": 0.955,
"3-21G(d)": 0.948,
"6-31G": 0.950,
"6-31G(d)": 0.942,
"6-31G(d,p)": 0.934,
"6-31+G(d,p)": 0.939,
"6-311G(d)": 0.947,
"6-311G(d,p)": 0.949,
"6-31G(2df,p)": 0.940,
"6-311+G(3df,2p)": 0.943,
"TZVP": 0.953,
"cc-pVDZ": 0.950,
"cc-pVTZ": 0.949,
"cc-pVQZ": 0.957,
"aug-cc-pVDZ": 0.969,
"aug-cc-pVTZ": 0.951,
"aug-cc-pVQZ": 0.956,
"cc-pV(T+d)Z": 0.948,
"cc-pCVDZ": 0.955,
"cc-pCVTZ": 0.951,
},
"MP3": {
"STO-3G": 0.894,
"3-21G": 0.968,
"3-21G(d)": 0.965,
"6-31G": 0.966,
"6-31G(d)": 0.939,
"6-31G(d,p)": 0.935,
"6-31+G(d,p)": 0.931,
"TZVP": 0.935,
"cc-pVDZ": 0.948,
"cc-pVTZ": 0.945,
},
"MP3=Full": {
"6-31G(d)": 0.938,
"6-31+G(d,p)": 0.932,
"6-311G(d)": 0.904,
"TZVP": 0.934,
"cc-pVDZ": 0.940,
"cc-pVTZ": 0.933,
},
"MP4": {
"3-21G": 0.970,
"3-21G(d)": 0.944,
"6-31G": 0.944,
"6-31G(d)": 0.955,
"6-31G(d,p)": 0.944,
"6-31+G(d,p)": 0.944,
"6-311G(d)": 0.959,
"6-311G(d,p)": 0.970,
"6-311+G(3df,2p)": 0.944,
"TZVP": 0.963,
"cc-pVDZ": 0.967,
"cc-pVTZ": 0.969,
"aug-cc-pVDZ": 0.977,
"aug-cc-pVTZ": 0.973,
},
"MP4=Full": {
"3-21G": 0.979,
"6-31G(d)": 0.962,
"6-311G(d,p)": 0.962,
"TZVP": 0.966,
"cc-pVDZ": 0.965,
"cc-pVTZ": 0.963,
"aug-cc-pVDZ": 0.975,
"aug-cc-pVTZ": 0.969,
},
"B2PLYP": {
"6-31G(d)": 0.949,
"6-31+G(d,p)": 0.952,
"6-31G(2df,p)": 0.955,
"TZVP": 0.954,
"cc-pVDZ": 0.958,
"cc-pVTZ": 0.959,
"cc-pVQZ": 0.957,
"aug-cc-pVTZ": 0.961,
},
"B2PLYP=Full": {
"3-21G": 0.952,
"6-31G(d)": 0.948,
"6-31+G(d,p)": 0.951,
"6-311G(d)": 0.904,
"TZVP": 0.954,
"cc-pVDZ": 0.959,
"cc-pVTZ": 0.956,
"aug-cc-pVDZ": 0.962,
"aug-cc-pVTZ": 0.959,
},
"B2PLYP=Full (ultrafine grid)": {
"6-31G(d)": 0.949,
"cc-pVDZ": 0.958,
"cc-pVTZ": 0.955,
"aug-cc-pVDZ": 0.962,
"aug-cc-pVTZ": 0.959,
},
"CID": {
"3-21G": 0.932,
"3-21G(d)": 0.931,
"6-31G": 0.935,
"6-31G(d)": 0.924,
"6-31G(d,p)": 0.924,
"6-31+G(d,p)": 0.924,
"6-311G(d)": 0.929,
"6-311+G(3df,2p)": 0.924,
"cc-pVDZ": 0.924,
"cc-pVTZ": 0.927,
},
"CISD": {
"3-21G": 0.941,
"3-21G(d)": 0.934,
"6-31G": 0.938,
"6-31G(d)": 0.926,
"6-31G(d,p)": 0.918,
"6-31+G(d,p)": 0.922,
"6-311G(d)": 0.925,
"6-311+G(3df,2p)": 0.922,
"cc-pVDZ": 0.922,
"cc-pVTZ": 0.930,
},
"QCISD": {
"3-21G": 0.969,
"3-21G(d)": 0.961,
"6-31G": 0.964,
"6-31G(d)": 0.952,
"6-31G(d,p)": 0.941,
"6-31+G(d,p)": 0.945,
"6-311G(d)": 0.957,
"6-311G(d,p)": 0.954,
"6-31G(2df,p)": 0.947,
"6-311+G(3df,2p)": 0.954,
"TZVP": 0.955,
"cc-pVDZ": 0.959,
"cc-pVTZ": 0.956,
"aug-cc-pVDZ": 0.969,
"aug-cc-pVTZ": 0.962,
"cc-pV(T+d)Z": 0.955,
},
"QCISD(T)": {
"3-21G": 0.954,
"3-21G(d)": 0.954,
"6-31G": 0.954,
"6-31G(d)": 0.959,
"6-31G(d,p)": 0.937,
"6-31+G(d,p)": 0.939,
"6-311G(d)": 0.963,
"6-311+G(3df,2p)": 0.954,
"TZVP": 0.963,
"cc-pVDZ": 0.953,
"cc-pVTZ": 0.949,
"aug-cc-pVDZ": 0.978,
"aug-cc-pVTZ": 0.967,
},
"QCISD(T)=Full": {
"cc-pVDZ": 0.959,
"cc-pVTZ": 0.957,
"aug-cc-pVDZ": 0.970,
},
"CCD": {
"3-21G": 0.972,
"3-21G(d)": 0.957,
"6-31G": 0.960,
"6-31G(d)": 0.947,
"6-31G(d,p)": 0.938,
"6-31+G(d,p)": 0.942,
"6-311G(d)": 0.955,
"6-311G(d,p)": 0.955,
"6-31G(2df,p)": 0.947,
"6-311+G(3df,2p)": 0.943,
"TZVP": 0.948,
"cc-pVDZ": 0.957,
"cc-pVTZ": 0.934,
"aug-cc-pVDZ": 0.965,
"aug-cc-pVTZ": 0.957,
"cc-pV(T+d)Z": 0.952,
},
"CCSD": {
"3-21G": 0.943,
"3-21G(d)": 0.943,
"6-31G": 0.943,
"6-31G(d)": 0.944,
"6-31G(d,p)": 0.933,
"6-31+G(d,p)": 0.934,
"6-311G(d)": 0.954,
"6-31G(2df,p)": 0.946,
"6-311+G(3df,2p)": 0.943,
"TZVP": 0.954,
"cc-pVDZ": 0.947,
"cc-pVTZ": 0.941,
"cc-pVQZ": 0.951,
"aug-cc-pVDZ": 0.963,
"aug-cc-pVTZ": 0.956,
"aug-cc-pVQZ": 0.953,
},
"CCSD=Full": {
"6-31G(d)": 0.950,
"6-31G(2df,p)": 0.942,
"TZVP": 0.948,
"cc-pVTZ": 0.948,
"aug-cc-pVTZ": 0.951,
},
"CCSD(T)": {
"3-21G": 0.991,
"3-21G(d)": 0.943,
"6-31G": 0.943,
"6-31G(d)": 0.962,
"6-31G(d,p)": 0.949,
"6-31+G(d,p)": 0.960,
"6-311G(d)": 0.963,
"6-311G(d,p)": 0.965,
"6-311+G(3df,2p)": 0.987,
"TZVP": 0.963,
"cc-pVDZ": 0.979,
"cc-pVTZ": 0.975,
"cc-pVQZ": 0.970,
"aug-cc-pVDZ": 0.963,
"aug-cc-pVTZ": 0.970,
"aug-cc-pVQZ": 0.961,
"cc-pV(T+d)Z": 0.965,
"cc-pCVDZ": 0.971,
"cc-pCVTZ": 0.966,
},
"CCSD(T)=Full": {
"6-31G(d)": 0.971,
"TZVP": 0.956,
"cc-pVDZ": 0.963,
"cc-pVTZ": 0.958,
"cc-pVQZ": 0.966,
"aug-cc-pVDZ": 0.971,
"aug-cc-pVTZ": 0.964,
"aug-cc-pVQZ": 0.958,
"cc-pV(T+d)Z": 0.959,
"cc-pCVDZ": 0.969,
"cc-pCVTZ": 0.966,
},
"AM1": 0.954,
"PM3": 0.974,
"PM6": 1.062,
"AMBER": 1.000,
"DREIDING": 0.936,
}
),
"UMN CTC (v5)": (
"https://comp.chem.umn.edu/freqscale/index.html",
{
"B1B95": {
"6-31+G(d,p)": 0.946,
"MG3S": 0.948,
},
"B1LYP": {
"MG3S": 0.955,
},
"B3LYP": {
"6-31G(d)": 0.952,
"6-31G(2df,2p)": 0.955,
"MG3S": 0.960,
"aug-cc-pVTZ": 0.959,
"def2-TZVP": 0.960,
"ma-TZVP": 0.960,
"6-311++G(d,p)": 0.959,
},
"B3P86": {
"6-31G(d)": 0.946,
},
"B3PW91": {
"6-31G(d)": 0.947,
},
"B97-3": {
"def2-TZVP": 0.949,
"ma-TZVP": 0.950,
"MG3S": 0.947,
},
"B98": {
"def2-TZVP": 0.958,
"ma-TZVP": 0.959,
"MG3S": 0.956,
},
"BB1K": {
"MG3S": 0.932,
"6-31+G(d,p)": 0.929,
},
"BB95": {
"6-31+G(d,p)": 0.985,
"MG3S": 0.986,
},
"BLYP": {
"6-311G(df,p)": 0.987,
"6-31G(d)": 0.983,
"MG3S": 0.987,
},
"BMK": {
"ma-TZVP": 0.947,
"MG3S": 0.945,
},
"BP86": {
"6-31G(d)": 0.981,
"ma-TZVP": 0.988,
},
"BPW60": {
"6-311+G(d,p)": 0.947,
},
"BPW63": {
"MG3S": 0.936,
},
"CAM-B3LYP": {
"ma-TZVP": 0.951,
},
"CASPT2(11,9)": {
"aug-cc-pVTZ": 0.932,
},
"CCSD(T)": {
"jul-cc-pVTZ": 0.958,
"aug-cc-pVTZ": 0.961,
},
"CCSD(T)-F12": {
"jul-cc-pVTZ": 0.955,
},
"CCSD(T)-F12a": {
"cc-pVDZ-F12": 0.957,
"cc-pVTZ-F12": 0.958,
"jun-cc-pVTZ": 0.958,
},
"CCSD": {
"jul-cc-pVTZ": 0.948,
},
"CCSD-F12": {
"jul-cc-pVTZ": 0.946,
},
"DCSD-F12a": {
"cc-pVDZ-F12": 0.942,
},
"DF-CCSD(T)-F12b": {
"cc-pVDZ-F12": 0.957,
"jun-cc-pVDZ-F12": 0.955,
},
"G96LYP80": {
"6-311+G(d,p)": 0.924,
},
"G96LYP82": {
"MG3S": 0.920,
},
"GAM": {
"def2-TZVP": 0.955,
"ma-TZVP": 0.956,
},
"HF": {
"3-21G": 0.895,
"6-31+G(d)": 0.887,
"6-31+G(d,p)": 0.891,
"6-311G(d,p)": 0.896,
"6-311G(df,p)": 0.896,
"6-31G(d)": 0.885,
"6-31G(d,p)": 0.889,
"MG3S": 0.895,
},
"HFLYP": {
"MG3S": 0.876,
},
"HSEh1PBE": {
"ma-TZVP": 0.954,
},
"M05": {
"aug-cc-pVTZ": 0.953,
"def2-TZVP": 0.952,
"ma-TZVP": 0.954,
"maug-cc-pVTZ": 0.953,
"MG3S": 0.951,
},
"M05-2X": {
"6-31+G(d,p)": 0.936,
"aug-cc-pVTZ": 0.939,
"def2-TZVPP": 0.938,
"ma-TZVP": 0.940,
"maug-cc-pVTZ": 0.939,
"MG3S": 0.937,
},
"M06": {
"6-31+G(d,p)": 0.955,
"6-311+G(d,p)": 0.957,
"aug-cc-pVTZ": 0.958,
"def2-SVP": 0.957,
"def2-TZVP": 0.956,
"def2-TZVPP": 0.963,
"ma-TZVP": 0.956,
"maug-cc-pVTZ": 0.956,
"MG3S": 0.955,
},
"M06-2X": {
"6-31G(d,p)": 0.940,
"6-31+G(d,p)": 0.942,
"6-311G(d,p)": 0.943,
"6-311+G(d,p)": 0.944,
"6-311++G(d,p)": 0.944,
"aug-cc-pVDZ": 0.954,
"aug-cc-pVTZ": 0.946,
"def2-TZVP": 0.946,
"def2-QZVP": 0.945,
"def2-TZVPP": 0.945,
"jul-cc-pVDZ": 0.952,
"jul-cc-pVTZ": 0.946,
"jun-cc-pVDZ": 0.951,
"jun-cc-pVTZ": 0.946,
"may-cc-pVTZ": 0.946,
"ma-TZVP": 0.947,
"maug-cc-pV(T+d)Z": 0.945,
"MG3S": 0.944,
},
"M06CR": {
"MG3S": 0.955,
},
"M06-HF": {
"6-31+G(d,p)": 0.931,
"aug-cc-pVTZ": 0.936,
"def2-TZVPP": 0.932,
"ma-TZVP": 0.932,
"maug-cc-pVTZ": 0.934,
"MG3S": 0.930,
},
"M06-L": {
"6-31G(d)": 0.951,
"6-31G(d,p)": 0.952,
"6-31+G(d,p)": 0.953,
"aug-cc-pVTZ": 0.955,
"aug-cc-pV(T+d)Z": 0.955,
"aug-cc-pVTZ-pp": 0.955,
"def2-TZVP": 0.951,
"def2-TZVPP": 0.951,
"ma-TZVP": 0.956,
"maug-cc-pVTZ": 0.952,
"MG3S": 0.952,
},
"M06-L(DKH2)": {
"aug-cc-pwcVTZ-DK": 0.959,
},
"M08-HX": {
"6-31+G(d,p)": 0.947,
"aug-cc-pVTZ": 0.950,
"cc-pVTZ+": 0.949,
"def2-TZVPP": 0.948,
"jun-cc-pVTZ": 0.949,
"may-cc-pVTZ": 0.949,
"ma-TZVP": 0.951,
"maug-cc-pVTZ": 0.951,
"MG3S": 0.948,
},
"M08-SO": {
"6-31+G(d,p)": 0.954,
"aug-cc-pVTZ": 0.959,
"cc-pVTZ+": 0.956,
"def2-TZVPP": 0.954,
"ma-TZVP": 0.958,
"jun-cc-pVTZ": 0.958,
"maug-cc-pVTZ": 0.957,
"MG3": 0.959,
"MG3S": 0.956,
"MG3SXP": 0.957,
},
"M11-L": {
"maug-cc-pVTZ": 0.962,
},
"MN11-L": {
"MG3S": 0.959,
},
"MN12-L": {
"jul-cc-pVDZ": 0.950,
"MG3S": 0.959,
},
"MN12-SX": {
"6-311++G(d,p)": 0.950,
"jul-cc-pVDZ": 0.954,
},
"MN15": {
"aug-cc-pVTZ": 0.950,
"def2-SVP": 0.947,
"ma-TZVP": 0.950,
},
"MN15-L": {
"def2-TZVP": 0.955,
"MG3S": 0.947,
"maug-cc-pVTZ": 0.954,
},
"MOHLYP": {
"ma-TZVP": 1.000,
"MG3S": 0.995,
},
"MP2 (frozen core)": {
"6-31+G(d,p)": 0.943,
"6-311G(d,p)": 0.945,
"6-31G(d)": 0.939,
"6-31G(d,p)": 0.933,
"cc-pVDZ": 0.952,
"cc-pVTZ": 0.950,
},
"MP2 (full)": {
"6-31G(d)": 0.938,
},
"MP4 (SDQ)": {
"jul-cc-pVTZ": 0.948,
},
"MPW1B95": {
"6-31+G(d,p)": 0.945,
"MG3": 0.945,
"MG3S": 0.947,
},
"MPW1K": {
"6-31+G(d,p)": 0.924,
"aug-cc-pVDTZ": 0.934,
"aug-cc-pVTZ": 0.930,
"jul-cc-pVDZ": 0.932,
"jul-cc-pVTZ": 0.929,
"jun-cc-pVDZ": 0.930,
"jun-cc-pVTZ": 0.929,
"ma-TZVP": 0.931,
"MG3": 0.928,
"MG3S": 0.931,
"MIDI!": 0.928,
"MIDIY": 0.922,
},
"MPW3LYP": {
"6-31+G(d,p)": 0.955,
"6-311+G(2d,p)": 0.960,
"6-31G(d)": 0.951,
"ma-TZVP": 0.960,
"MG3S": 0.956,
},
"MPW74": {
"6-311+G(d,p)": 0.925,
},
"MPW76": {
"MG3S": 0.956,
},
"MPWB1K": {
"6-31+G(d,p)": 0.926,
"MG3S": 0.929
},
"MPWLYP1M": {
"ma-TZVP": 0.983,
},
"OreLYP": {
"ma-TZVP": 0.984,
"def2-TZVP": 0.982,
},
"PBE": {
"def2-TZVP": 0.985,
"MG3S": 0.985,
"ma-TZVP": 0.987,
},
"PBE0": {
"6-31G(d,p)": 0.980,
"aug-cc-pVTZ": 0.986,
"def2-TZVP": 0.985,
"ma-TZVP": 0.987,
"MG3S": 0.950,
},
"PBE1KCIS": {
"MG3": 0.955,
"MG3S": 0.955,
},
"PW6B95": {
"def2-TZVP": 0.949,
},
"PWB6K": {
"cc-pVDZ": 0.928,
},
"QCISD": {
"cc-pVTZ": 0.950,
"MG3S": 0.953,
},
"QCISD(FC)": {
"6-31G(d)": 0.948,
},
"QCISD(T)": {
"aug-cc-pVQZ": 0.963,
},
"revM06": {
"ma-TZVP": 0.945,
"M3GS": 0.943,
},
"revM06-L": {
"def2-TZVP": 0.947,
},
"revTPSS": {
"def2-TZVP": 0.972,
"ma-TZVP": 0.973,
},
"SOGGA": {
"ma-TZVP": 0.991,
},
"τHCTHhyb": {
"ma-TZVP": 0.963,
},
"TPSS1KCIS": {
"def2-TZVP": 0.956,
"ma-TZVP": 0.957,
},
"TPSSh": {
"MG3S": 0.963,
},
"VSXC": {
"MG3S": 0.962,
},
"ωB97": {
"def2-TZVP": 0.944,
"ma-TZVP": 0.945,
},
"ωB97X-D": {
"6-31G(d,p)": 0.943,
"6-31+G(d,p)": 0.946,
"def2-TZVP": 0.945,
"ma-TZVP": 0.946,
"maug-cc-pVTZ": 0.949,
},
"X1B95": {
"6-31+G(d,p)": 0.943,
"MG3S": 0.946,
},
"XB1K": {
"6-31+G(d,p)": 0.927,
"MG3S": 0.930,
},
"AM1": 0.923,
"PM3": 0.916,
"PM6": 1.050,
"PM7": 1.050,
}
),
}
class PHYSICAL:
# Physical constants
BOLTZMANN = 0.001987204 # kcal/mol
R = 0.001987204 # kcal/mol
KB = 1.380662e-23 # J/K
ROOM_TEMPERATURE = 298.15 # K
PLANCK = 6.62606957e-34
SPEED_OF_LIGHT = 29979245800 # cm/s
GAS_CONSTANT = 1.987204 # cal/mol
STANDARD_PRESSURE = 101317 # Pa
class UNIT:
# Unit conversion
AMU_TO_KG = 1.66053886e-27
HART_TO_KCAL = 627.5095
HART_TO_JOULE = 4.3597441775e-18
A0_TO_METER = 5.291772109217e-11
A0_TO_BOHR = 0.52917720859
HARTREE_TO_WAVENUMBER = 219474.6
HART_TO_EV = 27.211386245988
JOULE_TO_EV = 6.241509074e18
ATM_TO_PASCAL = 101325 | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/const.py | const.py |
import configparser
from difflib import SequenceMatcher as seqmatch
import itertools as it
import os
import re
from getpass import getuser
import AaronTools
from AaronTools import addlogger
from AaronTools.const import AARONLIB, AARONTOOLS
from AaronTools.theory import (
GAUSSIAN_COMMENT,
GAUSSIAN_CONSTRAINTS,
GAUSSIAN_COORDINATES,
GAUSSIAN_GEN_BASIS,
GAUSSIAN_GEN_ECP,
GAUSSIAN_POST,
GAUSSIAN_PRE_ROUTE,
GAUSSIAN_ROUTE,
ORCA_BLOCKS,
ORCA_COMMENT,
ORCA_COORDINATES,
ORCA_ROUTE,
PSI4_AFTER_JOB,
PSI4_BEFORE_GEOM,
PSI4_BEFORE_JOB,
PSI4_COMMENT,
PSI4_JOB,
PSI4_MOLECULE,
PSI4_OPTKING,
PSI4_SETTINGS,
PSI4_SOLVENT,
SQM_COMMENT,
SQM_QMMM,
QCHEM_MOLECULE,
QCHEM_REM,
QCHEM_COMMENT,
QCHEM_SETTINGS,
Theory,
)
from AaronTools.theory.implicit_solvent import ImplicitSolvent
from AaronTools.theory.job_types import job_from_string
THEORY_OPTIONS = [
"GAUSSIAN_COMMENT",
"GAUSSIAN_CONSTRAINTS",
"GAUSSIAN_COORDINATES",
"GAUSSIAN_GEN_BASIS",
"GAUSSIAN_GEN_ECP",
"GAUSSIAN_POST",
"GAUSSIAN_PRE_ROUTE",
"GAUSSIAN_ROUTE",
"ORCA_BLOCKS",
"ORCA_COMMENT",
"ORCA_COORDINATES",
"ORCA_ROUTE",
"PSI4_AFTER_JOB",
"PSI4_BEFORE_GEOM",
"PSI4_BEFORE_JOB",
"PSI4_COMMENT",
"PSI4_MOLECULE",
"PSI4_JOB",
"PSI4_OPTKING",
"PSI4_SETTINGS",
"SQM_COMMENT",
"SQM_QMMM",
"QCHEM_MOLECULE",
"QCHEM_REM",
"QCHEM_COMMENT",
"QCHEM_SETTINGS",
]
@addlogger
class Config(configparser.ConfigParser):
"""
Reads configuration information from INI files found at:
$QCHASM/AaronTools/config.ini
$AARONLIB/config.ini
./config.ini or /path/to/file supplied during initialization
Access to configuration information available using dictionary notation.
eg: self[`section_name`][`option_name`] returns `option_value`
See help(configparser.ConfigParser) for more information
"""
LOG = None
SPEC_ATTRS = [
"_changes",
"_changed_list",
"_args",
"_kwargs",
"infile",
"metadata",
]
@classmethod
def _process_content(cls, filename, quiet=True):
"""
process file content to handle optional default section header
"""
contents = filename
if os.path.isfile(filename):
try:
with open(filename) as f:
contents = f.read()
except Exception as e:
if not quiet:
cls.LOG.INFO("failed to read %s: %s", filename, e)
return ""
try:
configparser.ConfigParser().read_string(contents)
except configparser.MissingSectionHeaderError:
contents = "[DEFAULT]\n" + contents
return contents
def __init__(
self, infile=None, quiet=False, skip_user_default=False, **kwargs
):
"""
infile: the configuration file to read
quiet: prints helpful status information
skip_user_default: change to True to skip importing user's default config files
**kwargs: passed to initialization of parent class
"""
configparser.ConfigParser.__init__(
self, interpolation=None, comment_prefixes=("#"), **kwargs
)
self.infile = infile
if not quiet:
print("Reading configuration...")
self._read_config(infile, quiet, skip_user_default)
# enforce case-sensitivity in certain sections
for section in self:
if section in [
"Substitution",
"Mapping",
"Configs",
"Results",
]:
continue
for option, value in list(self[section].items()):
if section == "Geometry" and option.lower().startswith(
"structure"
):
del self[section][option]
option = option.split(".")
option[0] = option[0].lower()
option = ".".join(option)
self[section][option] = value
continue
if section == "Geometry" and "structure" in value.lower():
re.sub("structure", "structure", value, flags=re.I)
self[section][option] = value
if option.lower() != option:
self[section][option.lower()] = value
del self[section][option]
# handle included sections
self._parse_includes()
if infile is not None:
self.read(infile)
# set additional default values
if infile:
if "top_dir" not in self["DEFAULT"]:
self["DEFAULT"]["top_dir"] = os.path.dirname(
os.path.abspath(infile)
)
if "name" not in self["DEFAULT"]:
self["DEFAULT"]["name"] = ".".join(
os.path.relpath(
infile, start=self["DEFAULT"]["top_dir"]
).split(".")[:-1]
)
else:
if "top_dir" not in self["DEFAULT"]:
self["DEFAULT"]["top_dir"] = os.path.abspath(os.path.curdir)
# handle substitutions/mapping
self._changes = {}
self._changed_list = []
self._parse_changes()
# for passing to Theory(*args, **kwargs)
self._args = []
self._kwargs = {}
# metadata is username and project name
self.metadata = {
"user": self.get(
"DEFAULT",
"user",
fallback=self.get("HPC", "user", fallback=getuser()),
),
"project": self.get("DEFAULT", "project", fallback=""),
}
def get(self, section, option, *, junk="_ ", max_junk=1, **kwargs):
"""
see ConfigParser.get for details
junk are characters that are not important in the option name
max_junk - number of allowed junk characters
e.g. junk="_" with max_junk=1 when looking for
'empirical dispersion' will match 'empirical_dispersion'
"""
# dummy class to allow user to specify whatever they
# want for fallback - even False and None
class NoFallback:
pass
fallback = NoFallback
if "fallback" in kwargs:
fallback = kwargs.pop("fallback")
# see if option is present as given
out = super().get(section, option, fallback=NoFallback, **kwargs)
# otherwise, look through the rest of the options to see if one
# is basically the same but with some extra junk characters
# e.g. 'empirical_dispersion' instead of 'empirical dispersion'
if out is NoFallback and junk and self.has_section(section):
for test_opt in self.options(section):
# calculate similarity
similarity = seqmatch(lambda x: x in junk, option, test_opt).ratio()
# must have no more than max_junk junk characters
if similarity >= 1 - max_junk / len(option) and any(
x in test_opt for x in junk
):
out = super().get(section, test_opt, fallback=NoFallback, **kwargs)
break
if out is NoFallback and fallback is NoFallback:
raise configparser.NoOptionError(section, option)
if out is NoFallback:
out = fallback
return out
def optionxform(self, option):
return str(option)
def __str__(self):
rv = ""
for section in self:
if "." in section:
continue
rv += "[{}]\n".format(section)
for option, value in self[section].items():
rv += "{} = {}\n".format(option, value)
rv += "\n"
return rv
def copy(self):
config = Config(infile=None, quiet=True)
for section in config.sections():
config.remove_section(section)
for option in list(config["DEFAULT"].keys()):
config.remove_option("DEFAULT", option)
for section in ["DEFAULT"] + self.sections():
try:
config.add_section(section)
except (configparser.DuplicateSectionError, ValueError):
pass
for key, val in self[section].items():
config[section][key] = val
for section in self.SPEC_ATTRS:
setattr(config, section, getattr(self, section))
return config
def for_change(self, change, structure=None):
this_config = self.copy()
if structure is not None:
this_config["Job"]["name"] = structure.name
if change:
this_config["Job"]["name"] = os.path.join(
change, this_config["Job"]["name"]
)
this_config._changes = {change: self._changes[change]}
return this_config
def _parse_changes(self):
for section in ["Substitution", "Mapping"]:
if section not in self:
continue
if self[section].getboolean("reopt", fallback=True):
self._changes[""] = ({}, None)
for key, val in self[section].items():
if key in self["DEFAULT"]:
continue
del self[section][key]
key = "\n".join(["".join(k.split()) for k in key.split("\n")])
val = "\n".join(["".join(v.split()) for v in val.split("\n")])
self[section][key] = val
for key, val in self[section].items():
if key in self["DEFAULT"] or key == "reopt":
continue
if "=" not in val:
val = [v.strip() for v in val.split(",")]
else:
tmp = [v.strip() for v in val.split(";")]
val = []
for t in tmp:
t = t.strip()
if not t:
continue
elif "\n" in t:
val += t.split("\n")
else:
val += [t]
tmp = {}
for i, v in enumerate(val):
if i == 0 and len(v.split("=")) == 1:
v = "{}={}".format(key, v)
val[i] = v
del self[section][key]
key = ""
self[section]["~PLACEHOLDER~"] = ";".join(val)
v = v.split("=")
if (
not key.startswith("&combinations")
and "(" not in v[0]
):
v[0] = v[0].split(",")
else:
v[0] = [v[0]]
for k in v[0]:
tmp[k] = v[1]
val = tmp
# handle request for combinations
if key.startswith("&combination"):
atoms = []
subs = []
# val <= { "2, 4": "H, CH3", "7, 9": "OH, NH2", .. }
for k, v in val.items():
if "(" not in k:
# regular substituents
atoms.append(k.split(","))
else:
# ring substitutions
atoms.append(re.findall("\(.*?\)", k))
subs.append([None] + [i for i in v.strip().split(",")])
# atoms <= [ [2, 4], [7, 9], .. ]
# subs <= [ [None, H, CH3], [None, OH, NH2], .. ]
for combo in it.product(*[range(len(s)) for s in subs]):
# combos <= (0, 0,..), (0,.., 1),..(1,.. 0),..(1,.., 1),..
if not any(combo):
# skip if no substitutions
# (already included if reopt=True)
continue
name = []
tmp = {}
for i, p in enumerate(combo):
# don't add subsitution if sub == None
if subs[i][p] is None:
continue
name.append(subs[i][p])
for a in atoms[i]:
tmp[a] = subs[i][p]
name = "_".join(name)
self._changes[name] = (
tmp,
section,
)
else:
if isinstance(val, list):
name = "_".join(val)
val = {key: ",".join(val)}
elif not key:
name = "_".join(
[
"_".join([v] * len(k.split(",")))
for k, v in val.items()
]
)
self[section][name] = self[section]["~PLACEHOLDER~"]
del self[section]["~PLACEHOLDER~"]
else:
name = key
self._changes[name] = (val, section)
def parse_functions(self):
"""
Evaluates functions supplied in configuration file
Functions indicated by "%{...}"
Pulls in values of options indicated by $option_name
Eg:
ppn = 4
memory = %{ $ppn * 2 }GB --> memory = 8GB
"""
func_patt = re.compile("(%{(.*?)})")
attr_patt = re.compile("\$([a-zA-Z0-9_:]+)")
for section in ["DEFAULT"] + self.sections():
# evaluate functions
for key, val in self[section].items():
match_list = func_patt.findall(val)
while match_list:
match = match_list.pop()
eval_match = match[1]
for attr in attr_patt.findall(match[1]):
if ":" in attr:
from_section, option = attr.split(":")
else:
option, from_section = attr, section
option = self[from_section][option]
eval_match = eval_match.replace("$" + attr, option, 1)
try:
eval_match = eval(eval_match, {})
except TypeError as e:
raise TypeError(
"{} for\n\t[{}]\n\t{} = {}\nin config file. Could not evaluate {}".format(
e.args[0], section, key, val, eval_match
)
)
except (NameError, SyntaxError):
if attr_patt.findall(eval_match):
eval_match = "%{" + eval_match.strip() + "}"
else:
eval_match = eval_match.strip()
val = val.replace(match[0], str(eval_match))
self[section][key] = val
match_list = func_patt.findall(val)
def getlist(self, section, option, *args, delim=",", **kwargs):
"""returns a list of option values by splitting on the delimiter specified by delim"""
raw = self.get(section, option, *args, **kwargs)
out = [x.strip() for x in raw.split(delim) if len(x.strip()) > 0]
return out
def read(self, filename, quiet=True):
try:
self.read_string(self._process_content(filename, quiet=quiet))
except configparser.ParsingError:
pass
def _read_config(self, infile, quiet, skip_user_default):
"""
Reads configuration information from `infile` after pulling defaults
"""
filenames = [
os.path.join(AARONTOOLS, "config.ini"),
]
if not skip_user_default:
filenames += [os.path.join(AARONLIB, "config.ini")]
if infile:
filenames += [infile]
local_only = False
job_include = None
for i, filename in enumerate(filenames):
if not quiet:
if os.path.isfile(filename):
print(" ✓", end=" ")
else:
print(" ✗", end=" ")
print(filename)
content = self._process_content(filename)
self.read(content, quiet=quiet)
job_include = self.get("Job", "include", fallback=job_include)
if filename != infile:
self.remove_option("Job", "include")
# local_only can only be overridden at the user level if "False" in the system config file
if i == 0:
local_only = self["DEFAULT"].getboolean("local_only")
elif local_only:
self["DEFAULT"]["local_only"] = str(local_only)
if "Job" in self:
type_spec = [
re.search("(?<!_)type", option) for option in self["Job"]
]
else:
type_spec = []
if not any(type_spec):
self.set("Job", "include", job_include)
def get_other_kwargs(self, section="Theory"):
"""
Returns dict() that can be unpacked and passed to Geometry.write along with a theory
Example:
[Theory]
route = pop NBORead
opt MaxCycle=1000, NoEigenTest
end_of_file = $nbo RESONANCE NBOSUM E2PERT=0.0 NLMO BNDIDX $end
this adds opt(MaxCycle=1000,NoEigenTest) pop=NBORead to the route with any other
pop or opt options being added by the job type
'two-layer' options can also be specified as a python dictionary
the following is equivalent to the above example:
[Theory]
route = {"pop":["NBORead"], "opt":["MaxCycle=1000", NoEigenTest"]}
end_of_file = $nbo RESONANCE NBOSUM E2PERT=0.0 NLMO BNDIDX $end
"""
# these need to be dicts
two_layer = [
GAUSSIAN_ROUTE,
GAUSSIAN_PRE_ROUTE,
ORCA_BLOCKS,
PSI4_JOB,
QCHEM_REM,
QCHEM_SETTINGS,
PSI4_SOLVENT,
]
# these need to be dicts, but can only have one value
two_layer_single_value = [
PSI4_OPTKING,
PSI4_SETTINGS,
PSI4_MOLECULE,
]
# these need to be lists
one_layer = [
GAUSSIAN_COMMENT,
GAUSSIAN_CONSTRAINTS,
GAUSSIAN_POST,
ORCA_COMMENT,
ORCA_ROUTE,
PSI4_AFTER_JOB,
PSI4_BEFORE_GEOM,
PSI4_BEFORE_JOB,
PSI4_COMMENT,
QCHEM_MOLECULE,
QCHEM_COMMENT,
]
theory_kwargs = [
"method",
"charge",
"multiplicity",
"type",
"basis",
"ecp",
"grid",
"empirical_dispersion",
]
# two layer options are separated by newline
# individual options are split on white space, with the first defining the primary layer
out = {}
for option in two_layer:
value = self[section].get(option, fallback=False)
value = self._kwargs.get(option, value)
if value:
if isinstance(value, dict):
out[option] = value
elif "{" in value:
# if it's got brackets, it's probably a python-looking dictionary
# eval it instead of parsing
out[option] = eval(value, {})
else:
out[option] = {}
for v in value.splitlines():
key = v.split()[0]
if len(v.split()) > 1:
info = v.split()[1].split(",")
else:
info = []
out[option][key] = [x.strip() for x in info]
for option in two_layer_single_value:
value = self.get(section, option, fallback=False)
value = self._kwargs.get(option, value)
if value:
if "{" in value:
out[option] = eval(value, {})
else:
out[option] = {}
for v in value.splitlines():
key = v.split()[0]
if len(v.split()) > 1:
info = [v.split()[1]]
else:
info = []
out[option][key] = [x.strip() for x in info]
for option in one_layer:
value = self[section].get(option, fallback=False)
value = self._kwargs.get(option, value)
if value:
out[option] = value.splitlines()
for option in theory_kwargs:
value = self[section].get(option, fallback=False)
if value:
out[option] = value
return out
def get_constraints(self, geometry):
constraints = {}
try:
con_list = re.findall("\(.*?\)", self["Geometry"]["constraints"])
except KeyError:
try:
geometry.parse_comment()
con_list = geometry.other["constraint"]
except KeyError:
raise RuntimeError(
"Constraints for forming/breaking bonds must be specified for TS search"
)
for con in con_list:
tmp = []
try:
for c in eval(con, {}):
tmp += geometry.find(str(c))
except TypeError:
for c in con:
tmp += geometry.find(str(c))
con = [a.name for a in tmp]
if len(con) == 1:
constraints.setdefault("atoms", [])
constraints["atoms"] += [con]
elif len(con) == 2:
constraints.setdefault("bonds", [])
constraints["bonds"] += [con]
elif len(con) == 3:
constraints.setdefault("angles", [])
constraints["angles"] += [con]
elif len(con) == 4:
constraints.setdefault("torsions", [])
constraints["torsions"] += [con]
return constraints
def get_theory(self, geometry, section="Theory"):
"""
Get the theory object according to configuration information
"""
if not self.has_section(section):
self.LOG.warning(
'config has no "%s" section, switching to "Theory"' % section
)
section = "Theory"
kwargs = self.get_other_kwargs(section=section)
theory = Theory(*self._args, geometry=geometry, **kwargs)
# build ImplicitSolvent object
if self[section].get("solvent", fallback="gas") == "gas":
theory.solvent = None
elif self[section]["solvent"]:
solvent_model = self.get(section, "solvent_model", fallback=False)
theory.solvent = ImplicitSolvent(
solvent_model,
self[section]["solvent"],
)
# build JobType list
job_type = self["Job"].get("type", fallback=False)
if job_type:
theory.job_type = []
numerical = self[section].get("numerical", fallback=False)
temperature = self[section].get("temperature", fallback=298.15)
try:
constraints = self.get_constraints(theory.geometry)
except RuntimeError:
constraints = None
theory.geometry.freeze()
theory.geometry.relax(self._changed_list)
info = {
"numerical": numerical,
"temperature": temperature,
"constraints": constraints,
"geometry": theory.geometry,
}
try:
theory.job_type += [job_from_string(job_type, **info)]
except ValueError:
raise ValueError("cannot parse job type: %s" % ".".join(job_type))
else:
# default to opt+freq
theory.job_type = [
OptimizationJob(geometry=geometry),
FrequencyJob(
numerical=self[section].get("numerical", fallback=False),
temperature=self[section].get(
"temperature", fallback=None
),
),
]
# return updated theory object
return theory
def get_template(self):
# captures name placeholder and iterator from for-loop initilaizer
for_patt = re.compile("&for\s+(.+)\s+in\s+(.+)")
# captures structure_dict-style structure/suffix -> (structure['suffix'], suffix)
parsed_struct_patt = re.compile("(structure\['(\S+?)'\])\.?")
# captures config-style structure/suffix -> (structure.suffix, suffix)
structure_patt = re.compile("(structure\.(\S+?))\.?")
def get_multiple(filenames, path=None, suffix=None):
rv = []
for name in filenames:
kind = "Minimum"
if name.startswith("TS"):
kind = "TS"
if path is not None:
name = os.path.join(path, name)
if not os.path.isfile(name):
continue
geom = AaronTools.geometry.Geometry(name)
if suffix is not None:
geom.name += ".{}".format(suffix)
rv += [(geom, kind)]
return rv
def structure_assignment(line):
# assignments must be done outside of eval()
# left -> structure.suffix -> structure_dict["suffix"]
# right -> eval(right)
# left = right -> structure_dict[suffix] = eval(right)
left = line.split("=")[0].strip()
right = line.split("=")[1].strip()
suffix_match = parsed_struct_patt.search(left)
if suffix_match is None:
raise RuntimeError(
"Can only assign to Geometry objects with names of the form `structure.suffix`"
)
suffix = suffix_match.group(2)
structure_dict[suffix] = eval(right, eval_dict)
structure_dict[suffix].name = ".".join(
structure_dict[suffix].name.split(".")[:-1] + [suffix]
)
if structure_dict[suffix].name.startswith("TS"):
kind_dict[suffix] = "TS"
def structure_suffix_parse(line, for_loop=None):
if for_loop is not None:
for_match, it_val = for_loop
for structure_match in structure_patt.findall(line):
# if our suffix is not the iterator, keep it's value for the dict key
if for_loop is None or structure_match[1] != for_match.group(
1
):
suffix = structure_match[1]
else:
suffix = str(it_val)
# change to dict-style syntax (structure.suffix -> structure["suffix"])
line = line.replace(
structure_match[0],
"structure['{}']".format(suffix),
)
if suffix not in structure_dict:
structure_dict[suffix] = AaronTools.geometry.Geometry()
kind_dict[suffix] = None
return line
structure_dict = {}
kind_dict = {}
structure_list = []
# load templates from AARONLIB
if "Reaction" in self:
path = None
if "template" in self["Reaction"]:
path = os.path.join(
AARONLIB,
"template_geoms",
self["Reaction"]["reaction"],
self["Reaction"]["template"],
)
for dirpath, dirnames, filenames in os.walk(path):
structure_list += get_multiple(filenames, path=dirpath)
else:
path = os.path.join(
AARONLIB,
"template_geoms",
self["Reaction"]["reaction"],
)
for dirpath, dirnames, filenames in os.walk(path):
structure_list += get_multiple(filenames, path=dirpath)
for structure, kind in structure_list:
structure.name = os.path.relpath(structure.name, path)
if not self.has_section("Geometry"):
return structure_list
# load templates from config[Geometry]
# store in structure_dict, keyed by structure option suffix
# `structure.suffix = geom.xyz` store as {suffix: geom.xyz}
# `structure = geom.xyz` (no suffix), store as {"": geom.xyz}
if "structure" in self["Geometry"]:
structure_dict[""] = self["Geometry"]["structure"]
else:
for key in self["Geometry"]:
if key.startswith("structure."):
suffix = ".".join(key.split(".")[1:])
structure_dict[suffix] = self["Geometry"][key]
# create Geometry objects
pop_sd = set([])
for suffix, structure in structure_dict.items():
if structure is not None and os.path.isdir(structure):
# if structure is a directory
for dirpath, dirnames, filenames in os.walk(structure):
structure_list += get_multiple(
filenames, path=dirpath, suffix=suffix
)
elif structure is not None:
try:
# if structure is a filename
structure = AaronTools.geometry.Geometry(structure)
except FileNotFoundError:
# if structure is a filename
structure = AaronTools.geometry.Geometry(
os.path.join(self["DEFAULT"]["top_dir"], structure)
)
except (IndexError, NotImplementedError):
if "coordination_complex" in structure.lower():
shape = None
center = None
ligands = None
for line in structure.splitlines():
line = line.strip()
if "coordination_complex" in line.lower():
shape = re.split("[:=]", line)[1].strip()
if "center" in line.lower():
center = re.split("[:=]", line)[1].strip()
if "ligands" in line.lower():
ligands = (
re.split("[:=]", line)[1].strip().split()
)
for (
geom
) in AaronTools.geometry.Geometry.get_coordination_complexes(
center=center, ligands=ligands, shape=shape
)[
0
]:
if suffix:
geom.name += "." + suffix
structure_list += [(geom, None)]
structure = None
pop_sd.add(suffix)
else:
# if structure is a smiles string
structure = AaronTools.geometry.Geometry.from_string(
structure
)
# adjust structure attributes
if structure is not None:
if "name" in self["Job"]:
structure.name = self["Job"]["name"]
if "Geometry" in self and "comment" in self["Geometry"]:
structure.comment = self["Geometry"]["comment"]
structure.parse_comment()
structure_dict[suffix] = structure
kind_dict[suffix] = None
for s in pop_sd:
del structure_dict[s]
# for loop for structure modification/creation
# structure.suffix = geom.xyz
# &for name in <iterator>:
# structure.name = structure.suffix.copy()
# structure.name.method_call(*args, **kwargs)
if "Geometry" in self:
for key in self["Geometry"]:
if not key.startswith("&for"):
continue
for_match = for_patt.search(key)
if for_match is None:
raise SyntaxError(
"Malformed &for loop specification in config"
)
lines = self["Geometry"][key].split("\n")
for it_val in eval(for_match.group(2), {}):
eval_dict = {
"Geometry": AaronTools.geometry.Geometry,
"structure": structure_dict,
for_match.group(1): it_val,
}
for line in lines:
line = line.strip()
if not line:
continue
line = structure_suffix_parse(
line,
for_loop=(for_match, it_val),
)
if "=" in line:
structure_assignment(line)
else:
eval(line, eval_dict)
# add structure_dict to structure list
try:
padding = max(
[
len(suffix)
for suffix in structure_dict.keys()
if suffix.isnumeric()
]
)
except ValueError:
padding = 0
for suffix in structure_dict:
geom = structure_dict[suffix]
if suffix:
geom.name = "{}.{}".format(
self["Job"]["name"], suffix.zfill(padding)
)
structure_list += [(geom, kind_dict[suffix])]
# apply functions found in [Geometry] section
if "Geometry" in self and "&call" in self["Geometry"]:
eval_dict = {
"Geometry": AaronTools.geometry.Geometry,
"structure": structure_dict,
}
lines = self["Geometry"]["&call"]
for line in lines.split("\n"):
line = structure_suffix_parse(line)
if parsed_struct_patt.search(line.strip()):
try:
eval(line.strip(), eval_dict)
except SyntaxError:
structure_assignment(line)
for suffix in structure_dict:
val = (structure_dict[suffix], kind_dict[suffix])
if val not in structure_list:
structure_list += [val]
elif line.strip():
for structure, kind in structure_list:
eval_dict["structure"] = structure
eval(line.strip(), eval_dict)
return structure_list
def make_changes(self, structure):
if not self._changes:
return structure
changed = []
for name, (changes, kind) in self._changes.items():
for key, val in changes.items():
if kind == "Substitution" and "(" not in key:
# regular substitutions
for k in key.split(","):
k = k.strip()
if val.lower() == "none":
structure -= structure.get_fragment(k)
else:
sub = structure.substitute(val, k)
for atom in sub:
changed += [atom.name]
elif kind == "Substitution":
# fused ring substitutions
target_patt = re.compile("\((.*?)\)")
for k in target_patt.findall(key):
k = [i.strip() for i in k.split(",")]
if val.lower() == "none":
structure -= structure.get_fragment(*k)
else:
sub = structure.ring_substitute(k, val)
for atom in sub:
changed += [atom.name]
elif kind == "Mapping":
key = [k.strip() for k in key.split(",")]
new_ligands = structure.map_ligand(val, key)
for ligand in new_ligands:
for atom in ligand:
changed += [atom.name]
try:
con_list = list(
eval(
"[{}]".format(self["Geometry"].get("constraints", "")), {}
)
)
except KeyError:
structure.parse_comment()
try:
con_list = structure.other["constraint"]
except KeyError:
con_list = []
for con in con_list:
for c in con:
try:
changed.remove(str(c))
except ValueError:
pass
self._changed_list = changed
return structure
def _parse_includes(self, section=None):
"""
Moves option values from subsections into parent section
Eg:
[HPC]
include = Wheeler
ppn = 12
[HPC.Wheeler]
nodes = 1
queue = wheeler_q
^^^evaluates to:
[Job]
nodes = 1
ppn = 12
queue = wheeler_q
"""
if section is None:
section_list = self.sections()
elif isinstance(section, str):
section_list = [section]
else:
section_list = section
for section in section_list:
# add requested subsections to parent section
if self.has_option(section, "include"):
if section == "Job" and self[section]["include"] == "detect":
continue
include_section = self[section]["include"].split(".")
if include_section[0] in self.sections():
# include specifies full section name, eg:
# include = Job.Minimum --> [Job.Minimum]
include_section = ".".join(include_section)
else:
# short-form of include, eg:
# [Job]
# include = Minimum
# --> [Job.Minimum]
include_section = [section] + include_section
include_section = ".".join(include_section)
for key, val in self[include_section].items():
self[section][key] = val
# handle non-default capitalization of default section
if section.lower() == "default":
for key, val in self[section].items():
self["DEFAULT"][key] = val
def as_dict(self, spec=None, skip=None):
"""
Forms a metadata spec dictionary from configuration info
:spec: (dict) if given, append key/vals to that dict
:skip: (list) skip storing stuff according to (section, option) or attrs
section, option, and attrs are strings that can be regex (full match only)
eg: skip=[("Job", ".*"), "conformer"] will skip everything in the Job
section and the Config.conformer attribute
"""
if spec is None:
spec = {}
if skip is None:
skip = []
skip_attrs = []
skip_sections = []
skip_options = []
for s in skip:
if isinstance(s, tuple):
skip_sections.append(s[0])
skip_options.append(s[1])
else:
skip_attrs.append(s)
for attr in self.SPEC_ATTRS:
for s in skip_attrs:
if re.fullmatch(s, attr):
break
else:
spec[attr] = getattr(self, attr)
for section in ["DEFAULT"] + self.sections():
if "." in section:
# these are include sections that should already be pulled into
# the main body of the config file
continue
for option in self[section]:
for i, s in enumerate(skip_sections):
o = skip_options[i]
if re.fullmatch(s, section) and re.fullmatch(o, option):
break
else:
# only include default options once, unless they are overridden
if (
section != "DEFAULT"
and option in self["DEFAULT"]
and self["DEFAULT"][option] == self[section][option]
):
continue
spec[
"{}/{}".format(section, option.replace(".", "_"))
] = self[section][option]
return spec
def read_spec(self, spec):
"""
Loads configuration metadata from spec dictionaries
"""
for attr in spec:
if attr in self.SPEC_ATTRS:
setattr(self, attr, spec[attr])
if "/" in attr:
section, key = attr.split("/")
if section not in self:
self.add_section(section)
self[section][key] = spec[attr]
def for_step(self, step=None, parse_functions=True):
"""
Generates a config copy with only options for the given step
"""
config = self.copy()
# find step-specific options
for section in ["DEFAULT"] + config.sections():
for key, val in config[section].items():
remove_key = key
key = key.strip().split()
if len(key) == 1:
continue
try:
key_step = float(key[0])
except ValueError:
continue
key = " ".join(key[1:])
# screen based on step
if key_step == float(step):
config[section][key] = val
# clean up metadata
del config[section][remove_key]
# other job-specific additions
if config.has_section("HPC") and "host" in config["HPC"]:
try:
config["HPC"]["work_dir"] = config["HPC"].get("remote_dir")
except TypeError as e:
raise RuntimeError(
"Must specify remote working directory for HPC (remote_dir = /path/to/HPC/work/dir)"
) from e
else:
if not config.has_section("HPC"):
config.add_section("HPC")
config["HPC"]["work_dir"] = config["DEFAULT"].get("top_dir")
# parse user-supplied functions in config file
if parse_functions:
config.parse_functions()
return config | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/config.py | config.py |
import os
import re
import numpy as np
from AaronTools.const import (
AARONLIB,
AARONTOOLS,
BONDI_RADII,
ELEMENTS,
VDW_RADII,
)
from AaronTools.fileIO import read_types
from AaronTools.finders import BondedTo, CloserTo, NotAny
from AaronTools.geometry import Geometry
from AaronTools.substituent import Substituent
from AaronTools.utils.utils import perp_vector
class Component(Geometry):
"""
Attributes:
:name: str
:comment: str
:atoms: list(Atom)
:other: dict()
:substituents: list(Substituent) substituents detected
:backbone: list(Atom) the backbone atoms
:key_atoms: list(Atom) the atoms used for mapping
"""
AARON_LIBS = os.path.join(AARONLIB, "Ligands")
BUILTIN = os.path.join(AARONTOOLS, "Ligands")
FROM_SUBSTITUENTS = set([])
def __init__(
self,
structure,
name="",
comment=None,
tag=None,
to_center=None,
key_atoms=None,
detect_backbone=True,
):
"""
comp is either a file, a geometry, or an atom list
"""
super().__init__()
self.name = name
self.comment = comment
self.other = {}
self.substituents = []
self.backbone = None
self.key_atoms = []
if isinstance(structure, str) and not os.access(structure, os.R_OK):
for ext in read_types:
if structure.endswith(".%s" % ext):
structure = structure[: -(1 + len(ext))]
for lib in [Component.AARON_LIBS, Component.BUILTIN]:
if not os.path.exists(lib):
continue
flig = None
for f in os.listdir(lib):
name, ext = os.path.splitext(f)
if not any(".%s" % x == ext for x in read_types):
continue
match = structure == name
if match:
flig = os.path.join(lib, f)
break
if flig:
break
else:
try:
structure = Substituent(structure)
Component.FROM_SUBSTITUENTS.add(structure.name)
self.__init__(structure, comment="K:1")
return
except Exception:
raise FileNotFoundError(
"Cannot find ligand in library:", structure
)
structure = flig
super().__init__(structure, name, comment)
if tag is not None:
for a in self.atoms:
a.add_tag(tag)
self.other = self.parse_comment()
try:
self.key_atoms = self.find("key")
except LookupError:
if "key_atoms" in self.other:
self.key_atoms = [
self.atoms[i] for i in self.other["key_atoms"]
]
if key_atoms is not None:
self.key_atoms = self.find(key_atoms)
for a in self.key_atoms:
a.tags.add("key")
if detect_backbone:
self.detect_backbone(to_center)
self.rebuild()
def __lt__(self, other):
if len(self) != len(other):
return len(self) < len(other)
for a, b in zip(sorted(self.atoms), sorted(other.atoms)):
if a < b:
return True
return False
@classmethod
def list(
cls,
name_regex=None,
coordinating_elements=None,
denticity=None,
include_ext=False,
):
names = []
for lib in [cls.AARON_LIBS, cls.BUILTIN]:
if not os.path.exists(lib):
continue
for f in os.listdir(lib):
name, ext = os.path.splitext(f)
if not any(".%s" % x == ext for x in read_types):
continue
if name in names:
continue
name_ok = True
elements_ok = True
denticity_ok = True
if (
name_regex is not None
and re.search(name_regex, name, re.IGNORECASE) is None
):
name_ok = False
if coordinating_elements is not None:
geom = Geometry(
os.path.join(lib, name + ext),
refresh_connected=False,
refresh_ranks=False,
)
# geom = cls(name)
elements = [
geom.atoms[i].element for i in geom.other["key_atoms"]
]
if not all(
elements.count(x) == coordinating_elements.count(x)
for x in coordinating_elements
) or not all(
coordinating_elements.count(x) == elements.count(x)
for x in elements
):
elements_ok = False
if denticity is not None:
geom = cls(name)
if len(geom.find("key")) != denticity:
denticity_ok = False
if name_ok and elements_ok and denticity_ok:
if include_ext:
names.append(name + ext)
else:
names.append(name)
return names + sorted(cls.FROM_SUBSTITUENTS)
def c2_symmetric(self, to_center=None, tolerance=0.1):
"""determine if center-key atom axis is a C2 axis"""
# determine ranks
ranks = self.canonical_rank(
update=False,
break_ties=False,
invariant=False,
)
# remove the rank of atoms that are along the c2 axis
ranks_off_c2_axis = []
if to_center is None:
center = np.zeros(3)
else:
center = self.COM(to_center)
v = self.COM(self.key_atoms) - center
v /= np.linalg.norm(v)
for atom, rank in zip(self.atoms, ranks):
dist_along_v = np.dot(atom.coords - center, v)
if (
abs(np.linalg.norm(atom.coords - center) - dist_along_v)
< tolerance
):
continue
ranks_off_c2_axis.append(rank)
return all([ranks.count(x) % 2 == 0 for x in set(ranks_off_c2_axis)])
def sterimol(self, to_center=None, bisect_L=False, **kwargs):
"""
calculate ligand sterimol parameters for the ligand
to_center - atom the ligand is coordinated to
bisect_L - L axis will bisect (or analogous for higher denticity
ligands) the L-M-L angle
Default - center to centroid of key atoms
**kwargs - arguments passed to Geometry.sterimol
"""
if to_center is not None:
center = self.find(to_center)
else:
center = self.find(
[BondedTo(atom) for atom in self.key_atoms], NotAny(self.atoms)
)
if len(center) != 1:
raise TypeError(
"wrong number of center atoms specified;\n"
"expected 1, got %i" % len(center)
)
center = center[0]
if bisect_L:
L_axis = np.zeros(3)
for atom in self.key_atoms:
v = center.bond(atom)
v /= np.linalg.norm(v)
v /= len(self.key_atoms)
L_axis += v
else:
L_axis = self.COM(self.key_atoms) - center.coords
L_axis /= np.linalg.norm(L_axis)
return super().sterimol(L_axis, center, self.atoms, **kwargs)
def copy(self, atoms=None, name=None, comment=None):
rv = super().copy()
return Component(rv)
def rebuild(self):
sub_atoms = []
for sub in sorted(self.substituents):
tmp = [sub.atoms[0]]
tmp += sorted(sub.atoms[1:])
for t in tmp:
if t in sub_atoms:
continue
if self.backbone and t in self.backbone:
continue
sub_atoms += [t]
if self.backbone is None:
self.backbone = [a for a in self.atoms if a not in sub_atoms]
self.backbone = sorted(self.backbone)
self.atoms = self.backbone + sub_atoms
def get_frag_list(self, targets=None, max_order=None):
"""
find fragments connected by only one bond
(both fragments contain no overlapping atoms)
"""
if targets:
atoms = self.find(targets)
else:
atoms = self.atoms
frag_list = []
for i, a in enumerate(atoms[:-1]):
for b in atoms[i + 1 :]:
if b not in a.connected:
continue
frag_a = self.get_fragment(a, b)
frag_b = self.get_fragment(b, a)
if len(frag_a) == len(frag_b) and sorted(
frag_a, key=lambda x: ELEMENTS.index(x.element)
) == sorted(frag_b, key=lambda x: ELEMENTS.index(x.element)):
continue
if len(frag_a) == 1 and frag_a[0].element == "H":
continue
if len(frag_b) == 1 and frag_b[0].element == "H":
continue
if max_order is not None and a.bond_order(b) > max_order:
continue
if (frag_a, a, b) not in frag_list:
frag_list += [(frag_a, a, b)]
if (frag_b, b, a) not in frag_list:
frag_list += [(frag_b, b, a)]
return frag_list
def detect_backbone(self, to_center=None):
"""
Detects backbone and substituents attached to backbone
Will tag atoms as 'backbone' or by substituent name
:to_center: the atoms connected to the metal/active center
"""
# we must remove any tags already made
for a in self.atoms:
a.tags.discard("backbone")
self.backbone = []
if self.substituents is not None:
for sub in self.substituents:
for a in sub.atoms:
a.tags.discard(sub.name)
self.substituents = []
# get all possible fragments connected by one bond
frag_list = self.get_frag_list()
# get atoms connected to center
if to_center is not None:
to_center = self.find(to_center)
else:
try:
to_center = self.find("key")
except LookupError:
to_center = []
try:
center = self.find("center")
to_center += list(c.connected for c in center)
except LookupError:
center = []
new_tags = {} # hold atom tag options until assignment determined
subs_found = {} # for testing which sub assignment is best
sub_atoms = set([]) # holds atoms assigned to substituents
for frag_tup in sorted(frag_list, key=lambda x: len(x[0])):
frag, start, end = frag_tup
if frag[0] != start:
frag = self.reorder(start=start, targets=frag)[0]
# if frag contains atoms from to_center, it's part of backbone
is_backbone = False
for a in frag:
if to_center and a in to_center:
is_backbone = True
break
# skip substituent assignment if part of backbone
if is_backbone:
continue
# try to find fragment in substituent library
try:
sub = Substituent(frag, end=end)
except LookupError:
continue
if not to_center and len(frag) > len(self.atoms) - len(sub_atoms):
break
# save atoms and tags if found
sub_atoms = sub_atoms.union(set(frag))
subs_found[sub.name] = len(sub.atoms)
for a in sub.atoms:
if a in new_tags:
new_tags[a] += [sub.name]
else:
new_tags[a] = [sub.name]
# save substituent
self.substituents += [sub]
# tag substituents
for a in new_tags:
tags = new_tags[a]
if len(tags) > 1:
# if multiple substituent assignments possible,
# want to keep the largest one (eg: tBu instead of Me)
sub_length = []
for t in tags:
sub_length += [subs_found[t]]
max_length = max(sub_length)
if max_length < 0:
max_length = min(sub_length)
keep = sub_length.index(max_length)
a.add_tag(tags[keep])
else:
a.add_tag(tags[0])
# tag backbone
for a in set(self.atoms) - set(sub_atoms):
a.add_tag("backbone")
self.backbone += [a]
if not self.backbone:
self.backbone = None
return
def capped_backbone(self, to_center=None, as_copy=True):
if as_copy:
comp = self.copy()
else:
comp = self
if comp.backbone is None:
comp.detect_backbone()
subs = []
for sub in comp.substituents:
subs += [comp.remove_fragment(sub.atoms, sub.end, ret_frag=True)]
if as_copy:
comp.substituents = None
return comp, subs
else:
return subs
def minimize_sub_torsion(self, geom=None, **kwargs):
"""
rotates substituents to minimize LJ potential
geom: calculate LJ potential between self and another geometry-like
object, instead of just within self
"""
if geom is None:
geom = self
if self.substituents is None:
self.detect_backbone()
return super().minimize_sub_torsion(geom, **kwargs)
def sub_rotate(self, start, angle=None):
start = self.find_exact(start)[0]
for sub in self.substituents:
if sub.atoms[0] == start:
break
end = sub.end
if angle is None:
angle = sub.conf_angle
if not angle:
return
self.change_dihedral(
start, end, angle, fix=4, adjust=True, as_group=True
)
def cone_angle(
self, center=None, method="exact", return_cones=False, radii="umn"
):
"""
returns cone angle in degrees
center - Atom() that this component is coordinating
used as the apex of the cone
method (str) can be:
'Tolman' - Tolman cone angle for unsymmetric ligands
See J. Am. Chem. Soc. 1974, 96, 1, 53–60 (DOI: 10.1021/ja00808a009)
NOTE: this does not make assumptions about the geometry
NOTE: only works with monodentate and bidentate ligands
'exact' - cone angle from Allen et. al.
See Bilbrey, J.A., Kazez, A.H., Locklin, J. and Allen, W.D.
(2013), Exact ligand cone angles. J. Comput. Chem., 34:
1189-1197. (DOI: 10.1002/jcc.23217)
return_cones - return cone apex, center of base, and base radius list
the sides of the cones will be 5 angstroms long
for Tolman cone angles, multiple cones will be returned, one for
each substituent coming off the coordinating atom
radii: 'bondi' - Bondi vdW radii
'umn' - vdW radii from Mantina, Chamberlin, Valero, Cramer, and Truhlar
dict() with elements as keys and radii as values
"""
if method.lower() == "tolman":
CITATION = "doi:10.1021/ja00808a009"
elif method.lower() == "exact":
CITATION = "doi:10.1002/jcc.23217"
self.LOG.citation(CITATION)
key = self.find("key")
center = self.find_exact(center)[0]
L_axis = self.COM(key) - center.coords
L_axis /= np.linalg.norm(L_axis)
if isinstance(radii, dict):
radii_dict = radii
elif radii.lower() == "bondi":
radii_dict = BONDI_RADII
elif radii.lower() == "umn":
radii_dict = VDW_RADII
# list of cone data for printing bild file or w/e
cones = []
if method.lower() == "tolman":
total_angle = 0
if len(key) > 2:
raise NotImplementedError(
"Tolman cone angle not implemented for tridentate or more ligands"
)
elif len(key) == 2:
key1, key2 = key
try:
bridge_path = self.shortest_path(key1, key2)
except LookupError:
bridge_path = False
for key_atom in key:
L_axis = key_atom.coords - center.coords
L_axis /= np.linalg.norm(L_axis)
bonded_atoms = self.find(BondedTo(key_atom))
for bonded_atom in bonded_atoms:
frag = self.get_fragment(bonded_atom, key_atom)
use_bridge = False
if any(k in frag for k in key):
# fragment on bidentate ligands that connects to
# the other coordinating atom
k = self.find(frag, key)[0]
# the bridge might be part of a ring (e.g. BPY)
# to avoid double counting the bridge, check if the
# first atom in the fragment is the first atom on the
# path from one key atom to the other
if frag[0] in bridge_path:
use_bridge = True
if use_bridge:
# angle between one L-M bond and L-M-L bisecting vector
tolman_angle = center.angle(k, key_atom) / 2
else:
tolman_angle = None
# for bidentate ligands with multiple bridges across, only use atoms that
# are closer to the key atom we are looking at right now
if len(key) == 2:
if bridge_path:
if key_atom is key1:
other_key = key2
else:
other_key = key1
frag = self.find(
frag,
CloserTo(
key_atom, other_key, include_ties=True
),
)
# some ligands like DuPhos have rings on the phosphorous atom
# we only want ones that are closer to the the substituent end
frag = self.find(frag, CloserTo(bonded_atom, key_atom))
# Geometry(frag).write(outfile="frag%s.xyz" % bonded_atom.name)
for atom in frag:
beta = np.arcsin(
radii_dict[atom.element] / atom.dist(center)
)
v = center.bond(atom) / center.dist(atom)
c = np.linalg.norm(v - L_axis)
test_angle = beta + np.arccos((c ** 2 - 2) / -2)
if (
tolman_angle is None
or test_angle > tolman_angle
):
tolman_angle = test_angle
scale = 5 * np.cos(tolman_angle)
cones.append(
(
center.coords + scale * L_axis,
center.coords,
scale * abs(np.tan(tolman_angle)),
)
)
total_angle += 2 * tolman_angle / len(bonded_atoms)
if return_cones:
return np.rad2deg(total_angle), cones
return np.rad2deg(total_angle)
elif method.lower() == "exact":
beta = np.zeros(len(self.atoms), dtype=float)
test_one_atom_axis = None
max_beta = None
for i, atom in enumerate(self.atoms):
beta[i] = np.arcsin(
radii_dict[atom.element] / atom.dist(center)
)
if max_beta is None or beta[i] > max_beta:
max_beta = beta[i]
test_one_atom_axis = center.bond(atom)
# check to see if all other atoms are in the shadow of one atom
# e.g. cyano, carbonyl
overshadowed_list = []
for i, atom in enumerate(self.atoms):
rhs = beta[i]
if (
np.dot(center.bond(atom), test_one_atom_axis)
/ (center.dist(atom) * np.linalg.norm(test_one_atom_axis))
<= 1
):
rhs += np.arccos(
np.dot(center.bond(atom), test_one_atom_axis)
/ (
center.dist(atom)
* np.linalg.norm(test_one_atom_axis)
)
)
lhs = max_beta
if lhs >= rhs:
# print(atom, "is overshadowed")
overshadowed_list.append(atom)
break
# all atoms are in the cone - we're done
if len(overshadowed_list) == len(self.atoms):
scale = 5 * np.cos(max_beta)
cones.append(
(
center.coords + scale * test_one_atom_axis,
center.coords,
scale
* abs(
np.linalg.norm(test_one_atom_axis)
* np.tan(max_beta)
),
)
)
if return_cones:
return np.rad2deg(2 * max_beta), cones
return np.rad2deg(2 * max_beta)
overshadowed_list = []
for i, atom1 in enumerate(self.atoms):
for j, atom2 in enumerate(self.atoms[:i]):
rhs = beta[i]
if (
np.dot(center.bond(atom1), center.bond(atom2))
/ (center.dist(atom1) * center.dist(atom2))
<= 1
):
rhs += np.arccos(
np.dot(center.bond(atom1), center.bond(atom2))
/ (center.dist(atom1) * center.dist(atom2))
)
lhs = beta[j]
if lhs >= rhs:
overshadowed_list.append(atom1)
break
# winow list to ones that aren't in the shadow of another
atom_list = [
atom for atom in self.atoms if atom not in overshadowed_list
]
# check pairs of atoms
max_a = None
aij = None
bij = None
cij = None
for i, atom1 in enumerate(atom_list):
ndx_i = self.atoms.index(atom1)
for j, atom2 in enumerate(atom_list[:i]):
ndx_j = self.atoms.index(atom2)
beta_ij = np.arccos(
np.dot(center.bond(atom1), center.bond(atom2))
/ (atom1.dist(center) * atom2.dist(center))
)
test_alpha = (beta[ndx_i] + beta[ndx_j] + beta_ij) / 2
if max_a is None or test_alpha > max_a:
max_a = test_alpha
mi = center.bond(atom1)
mi /= np.linalg.norm(mi)
mj = center.bond(atom2)
mj /= np.linalg.norm(mj)
aij = np.sin(
0.5 * (beta_ij + beta[ndx_i] - beta[ndx_j])
) / np.sin(beta_ij)
bij = np.sin(
0.5 * (beta_ij - beta[ndx_i] + beta[ndx_j])
) / np.sin(beta_ij)
cij = 0
norm = (
aij * mi
+ bij * mj
+ cij * np.cross(mi, mj) / np.sin(bij)
)
# r = 0.2 * np.tan(max_a)
# print(
# ".cone %.3f %.3f %.3f 0.0 0.0 0.0 %.3f open" % (
# 0.2 * norm[0], 0.2 * norm[1], 0.2 * norm[2], r
# )
# )
overshadowed_list = []
rhs = max_a
for atom in atom_list:
ndx_i = self.atoms.index(atom)
lhs = beta[ndx_i] + np.arccos(
np.dot(center.bond(atom), norm) / center.dist(atom)
)
# this should be >=, but there can be numerical issues
if rhs > lhs or np.isclose(rhs, lhs):
overshadowed_list.append(atom)
# the cone fits all atoms, we're done
if len(overshadowed_list) == len(atom_list):
scale = 5 * np.cos(max_a)
cones.append(
(
center.coords + (scale * norm),
center.coords,
scale * abs(np.tan(max_a)),
)
)
if return_cones:
return np.rad2deg(2 * max_a), cones
return np.rad2deg(2 * max_a)
centroid = self.COM()
c_vec = centroid - center.coords
c_vec /= np.linalg.norm(c_vec)
min_alpha = None
c = 0
for i, atom1 in enumerate(atom_list):
for j, atom2 in enumerate(atom_list[:i]):
for k, atom3 in enumerate(atom_list[i + 1 :]):
c += 1
ndx_i = self.atoms.index(atom1)
ndx_j = self.atoms.index(atom2)
ndx_k = self.atoms.index(atom3)
# print(atom1.name, atom2.name, atom3.name)
mi = center.bond(atom1)
mi /= np.linalg.norm(center.dist(atom1))
mj = center.bond(atom2)
mj /= np.linalg.norm(center.dist(atom2))
mk = center.bond(atom3)
mk /= np.linalg.norm(center.dist(atom3))
gamma_ijk = np.dot(mi, np.cross(mj, mk))
# M = np.column_stack((mi, mj, mk))
# N = gamma_ijk * np.linalg.inv(M)
N = np.column_stack(
(
np.cross(mj, mk),
np.cross(mk, mi),
np.cross(mi, mj),
)
)
u = np.array(
[
np.cos(beta[ndx_i]),
np.cos(beta[ndx_j]),
np.cos(beta[ndx_k]),
]
)
v = np.array(
[
np.sin(beta[ndx_i]),
np.sin(beta[ndx_j]),
np.sin(beta[ndx_k]),
]
)
P = np.dot(N.T, N)
A = np.dot(u.T, np.dot(P, u))
B = np.dot(v.T, np.dot(P, v))
C = np.dot(u.T, np.dot(P, v))
D = gamma_ijk ** 2
# beta_ij = np.dot(center.bond(atom1), center.bond(atom2))
# beta_ij /= atom1.dist(center) * atom2.dist(center)
# beta_ij = np.arccos(beta_ij)
# beta_jk = np.dot(center.bond(atom2), center.bond(atom3))
# beta_jk /= atom2.dist(center) * atom3.dist(center)
# beta_jk = np.arccos(beta_jk)
# beta_ik = np.dot(center.bond(atom1), center.bond(atom3))
# beta_ik /= atom1.dist(center) * atom3.dist(center)
# beta_ik = np.arccos(beta_ik)
#
# D = 1 - np.cos(beta_ij) ** 2 - np.cos(beta_jk) ** 2 - np.cos(beta_ik) ** 2
# D += 2 * np.cos(beta_ik) * np.cos(beta_jk) * np.cos(beta_ij)
# this should be equal to the other D
t1 = (A - B) ** 2 + 4 * C ** 2
t2 = 2 * (A - B) * (A + B - 2 * D)
t3 = (A + B - 2 * D) ** 2 - 4 * C ** 2
w_lt = (-t2 - np.sqrt(t2 ** 2 - 4 * t1 * t3)) / (
2 * t1
)
w_gt = (-t2 + np.sqrt(t2 ** 2 - 4 * t1 * t3)) / (
2 * t1
)
alpha1 = np.arccos(w_lt) / 2
alpha2 = (2 * np.pi - np.arccos(w_lt)) / 2
alpha3 = np.arccos(w_gt) / 2
alpha4 = (2 * np.pi - np.arccos(w_gt)) / 2
for alpha in [alpha1, alpha2, alpha3, alpha4]:
if alpha < max_a:
continue
if min_alpha is not None and alpha >= min_alpha:
continue
lhs = (
A * np.cos(alpha) ** 2 + B * np.sin(alpha) ** 2
)
lhs += 2 * C * np.sin(alpha) * np.cos(alpha)
if not np.isclose(lhs, D):
continue
# print(lhs, D)
p = np.dot(
N, u * np.cos(alpha) + v * np.sin(alpha)
)
norm = p / gamma_ijk
for atom in atom_list:
ndx = self.atoms.index(atom)
rhs = beta[ndx]
d = np.dot(
center.bond(atom), norm
) / center.dist(atom)
if abs(d) < 1:
rhs += np.arccos(d)
if not alpha >= rhs:
break
else:
if min_alpha is None or alpha < min_alpha:
# print("min_alpha set", alpha)
min_alpha = alpha
min_norm = norm
# r = 2 * np.tan(min_alpha)
# print(
# ".cone %.3f %.3f %.3f 0.0 0.0 0.0 %.3f open" % (
# 2 * norm[0], 2 * norm[1], 2 * norm[2], r
# )
# )
scale = 5 * np.cos(min_alpha)
cones.append(
(
center.coords + scale * min_norm,
center.coords,
scale * abs(np.tan(min_alpha)),
)
)
if return_cones:
return np.rad2deg(2 * min_alpha), cones
return np.rad2deg(2 * min_alpha)
else:
raise NotImplementedError(
"cone angle type is not implemented: %s" % method
) | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/component.py | component.py |
<a href="https://badge.fury.io/py/AaronTools"><img src="https://badge.fury.io/py/AaronTools.svg" alt="PyPI version" height="18"></a>
# AaronTools.py
AaronTools provides a collection of tools for automating routine tasks encountered when running quantum chemistry computations.
These tools can be used either directly within a Python script using AaronTools objects, or via a series of command-line scripts.
See the <a href="https://github.com/QChASM/AaronTools.py/wiki">Wiki</a> for installation and usage.
AaronTools is described in
"QChASM: Quantum Chemistry Automation and Structure Manipulation" <a href="http://dx.doi.org/10.1002/wcms.1510" target="_blank"><i>WIREs Comp. Mol. Sci.</i> <b>11</b>, e1510 (2021)</a>.
A Perl implementation of AaronTools is also <a href="https://github.com/QChASM/AaronTools">available here.</a>
However, users are <em>strongly urged</em> to use the Python version since it has far more powerful features and, unlike the Perl version, will continue to be developed and supported.
## Citation
If you use the Python AaronTools, please cite:
V. M. Ingman, A. J. Schaefer, L. R. Andreola, and S. E. Wheeler "QChASM: Quantum Chemistry Automation and Structure Manipulation" <a href="http://dx.doi.org/10.1002/wcms.1510" target="_blank"><i>WIREs Comp. Mol. Sci.</i> <b>11</b>, e1510 (2021)</a>
## Contact
If you have any questions or would like to discuss bugs or additional needed features, feel free to contact us at [email protected]
| AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/README.md | README.md |
import json
from inspect import signature
import numpy as np
from AaronTools.atoms import Atom
from AaronTools.comp_output import CompOutput
from AaronTools.component import Component
from AaronTools.spectra import Frequency, HarmonicVibration
from AaronTools.finders import (
Finder,
AnyNonTransitionMetal,
AnyTransitionMetal,
NotAny,
get_class,
)
from AaronTools.geometry import Geometry
from AaronTools.substituent import Substituent
from AaronTools.theory import (
Theory,
ImplicitSolvent,
OptimizationJob,
FrequencyJob,
ForceJob,
SinglePointJob,
BasisSet,
Basis,
ECP,
)
class ATEncoder(json.JSONEncoder):
def default(self, obj):
"""
Calls appropriate encoding method for supported AaronTools types.
If type not supported, calls the default `default` method
"""
if isinstance(obj, Atom):
return self._encode_atom(obj)
elif isinstance(obj, Geometry):
return self._encode_geometry(obj)
elif isinstance(obj, CompOutput):
return self._encode_comp_output(obj)
elif isinstance(obj, Frequency):
return self._encode_frequency(obj)
elif isinstance(obj, Theory):
return self._encode_theory(obj)
elif isinstance(obj, Finder):
return self._encode_finder(obj)
else:
super().default(obj)
def _encode_atom(self, obj):
"""
Encodes the data necessary to re-inialize an equivalent atom.
Note: constraint information is lost and must be re-initialized by the
parent geometry through the use of Geometry.parse_comment()
"""
rv = {"_type": obj.__class__.__name__}
rv["element"] = obj.element
rv["coords"] = list(obj.coords)
rv["flag"] = obj.flag
rv["name"] = obj.name
rv["tags"] = list(sorted(obj.tags))
rv["charge"] = obj.charge
rv["_rank"] = obj._rank
return rv
def _encode_geometry(self, obj):
"""
Encodes data necessary to re-initialize a geometry object.
"""
rv = {"_type": obj.__class__.__name__}
# for Geometry and all child classes
rv["name"] = obj.name
rv["atoms"] = obj.atoms
rv["connectivity"] = []
for a in obj.atoms:
rv["connectivity"] += [[obj.atoms.index(b) for b in a.connected]]
# for Geometry and all child classes but Substituent
if hasattr(obj, "comment"):
rv["comment"] = obj.comment
# for Catalyst child classes
if isinstance(obj, Geometry) and obj.components:
# comment
obj.fix_comment()
rv["comment"] = obj.comment
# for Substituent child class
if hasattr(obj, "conf_num"):
rv["conf_num"] = obj.conf_num
if hasattr(obj, "conf_angle"):
rv["conf_angle"] = obj.conf_angle
if hasattr(obj, "end"):
rv["end"] = obj.end
# for Component child class
if hasattr(obj, "key_atoms"):
rv["key_atoms"] = obj.key_atoms
return rv
def _encode_comp_output(self, obj):
rv = {"_type": obj.__class__.__name__}
rv["geometry"] = obj.geometry
rv["opts"] = obj.opts
rv["frequency"] = obj.frequency
rv["archive"] = obj.archive
rv["E_ZPVE"] = obj.E_ZPVE
rv["ZPVE"] = obj.ZPVE
rv["energy"] = obj.energy
rv["enthalpy"] = obj.enthalpy
rv["free_energy"] = obj.free_energy
rv["grimme_g"] = obj.grimme_g
rv["mass"] = obj.mass
rv["charge"] = obj.charge
rv["multiplicity"] = obj.multiplicity
rv["temperature"] = obj.temperature
rv["gradient"] = obj.gradient
rv["rotational_symmetry_number"] = obj.rotational_symmetry_number
rv["rotational_temperature"] = obj.rotational_temperature
rv["error"] = obj.error
rv["error_msg"] = obj.error_msg
rv["finished"] = obj.finished
return rv
def _encode_frequency(self, obj):
rv = {"_type": obj.__class__.__name__}
data = []
for d in obj.data:
entry = {}
for k, v in d.__dict__.items():
if isinstance(v, np.ndarray):
v = v.tolist()
entry[k] = v
data += [entry.copy()]
rv["data"] = data
return rv
def _encode_theory(self, obj):
rv = {"_type": obj.__class__.__name__}
if obj.method:
rv["method"] = obj.method.name
rv["semi-empirical"] = obj.method.is_semiempirical
if obj.grid:
rv["grid"] = obj.grid.name
if obj.empirical_dispersion:
rv["disp"] = obj.empirical_dispersion.name
if obj.solvent:
rv["solvent model"] = obj.solvent.solvent_model
rv["solvent"] = obj.solvent.solvent
if obj.processors:
rv["nproc"] = obj.processors
if obj.memory:
rv["mem"] = obj.memory
if obj.kwargs:
rv["other"] = obj.kwargs
if obj.job_type:
rv["jobs"] = {}
for job in obj.job_type:
job_type = job.__class__.__name__
rv["jobs"][job_type] = {}
for arg in signature(job.__init__).parameters:
if arg == "self" or arg == "geometry" or "*" in arg:
continue
try:
rv["jobs"][job_type][arg] = getattr(job, arg)
except AttributeError:
pass
if obj.basis:
rv["basis"] = {"name": [], "elements":[], "file":[], "auxiliary":[]}
if obj.basis.basis:
for basis in obj.basis.basis:
rv["basis"]["name"].append(basis.name)
rv["basis"]["elements"].append([])
for ele in basis.ele_selection:
if isinstance(ele, str):
rv["basis"]["elements"][-1].append(ele)
elif isinstance(ele, AnyTransitionMetal):
rv["basis"]["elements"][-1].append("tm")
elif isinstance(ele, AnyNonTransitionMetal):
rv["basis"]["elements"][-1].append("!tm")
if basis.not_anys:
for ele in basis.not_anys:
if isinstance(ele, str):
rv["basis"]["elements"][-1].append("!%s" % ele)
elif isinstance(ele, AnyTransitionMetal):
rv["basis"]["elements"][-1].append("!tm")
elif isinstance(ele, AnyNonTransitionMetal):
rv["basis"]["elements"][-1].append("!!tm")
rv["basis"]["file"].append(basis.user_defined)
rv["basis"]["auxiliary"].append(basis.aux_type)
if obj.basis.ecp:
rv["ecp"] = {"name": [], "elements":[], "file":[]}
for basis in obj.basis.ecp:
rv["ecp"]["name"].append(basis.name)
rv["ecp"]["elements"].append([])
for ele in basis.ele_selection:
if isinstance(ele, str):
rv["ecp"]["elements"][-1].append(ele)
elif isinstance(ele, AnyTransitionMetal):
rv["ecp"]["elements"][-1].append("tm")
elif isinstance(ele, AnyNonTransitionMetal):
rv["ecp"]["elements"][-1].append("!tm")
if basis.not_anys:
for ele in basis.not_anys:
if isinstance(ele, str):
rv["ecp"]["elements"][-1].append("!%s" % ele)
elif isinstance(ele, AnyTransitionMetal):
rv["ecp"]["elements"][-1].append("!tm")
elif isinstance(ele, AnyNonTransitionMetal):
rv["ecp"]["elements"][-1].append("!!tm")
rv["ecp"]["file"].append(basis.user_defined)
if obj.kwargs:
rv["other"] = obj.kwargs
return rv
def _encode_finder(self, obj):
rv = {"_type": "Finder"}
rv["_spec_type"] = obj.__class__.__name__
rv["kwargs"] = obj.__dict__
for kw in rv["kwargs"]:
if isinstance(rv["kwargs"][kw], np.ndarray):
rv["kwargs"][kw] = rv["kwargs"][kw].tolist()
return rv
class ATDecoder(json.JSONDecoder):
with_progress = False
def __init__(self, *args, **kwargs):
json.JSONDecoder.__init__(
self, object_hook=self.object_hook, *args, **kwargs
)
def object_hook(self, obj):
if "_type" not in obj:
return obj
if obj["_type"] == "Atom":
return self._decode_atom(obj)
if obj["_type"] == "Substituent":
return self._decode_substituent(obj)
if obj["_type"] in ["Geometry", "Component"]:
return self._decode_geometry(obj)
if obj["_type"] == "Frequency":
return self._decode_frequency(obj)
if obj["_type"] == "CompOutput":
return self._decode_comp_output(obj)
if obj["_type"] == "Theory":
return self._decode_theory(obj)
if obj["_type"] == "Finder":
return self._decode_finder(obj)
def _decode_atom(self, obj):
kwargs = {}
for key in ["element", "coords", "flag", "name", "tags", "charge"]:
if key not in obj:
continue
kwargs[key] = obj[key]
rv = Atom(**kwargs)
rv._rank = obj["_rank"]
return rv
def _decode_geometry(self, obj):
if ATDecoder.with_progress:
print("Loading structure", obj["name"], " " * 50, end="\r")
kwargs = {"structure": obj["atoms"]}
for key in ["name", "comment"]:
kwargs[key] = obj[key]
geom = Geometry(**kwargs, refresh_connected=False, refresh_ranks=False)
for i, connected in enumerate(obj["connectivity"]):
for c in connected:
geom.atoms[i].connected.add(geom.atoms[c])
if obj["_type"] == "Component":
key_atom_names = [a.name for a in obj["key_atoms"]]
return Component(geom, key_atoms=key_atom_names)
else:
return geom
def _decode_substituent(self, obj):
kwargs = {}
for key in ["name", "end", "conf_num", "conf_angle"]:
kwargs[key] = obj[key]
ranks = [a._rank for a in obj["atoms"]]
obj = self._decode_geometry(obj)
for a, r in zip(obj.atoms, ranks):
a._rank = r
return Substituent(obj, **kwargs)
def _decode_frequency(self, obj):
data = []
for d in obj["data"]:
kw = {k:v for k, v in d.items()}
freq = kw.pop("frequency")
data += [
HarmonicVibration(freq, **kw)
]
return Frequency(data)
def _decode_comp_output(self, obj):
keys = [
"geometry",
"opts",
"frequency",
"archive",
"energy",
"enthalpy",
"free_energy",
"grimme_g",
"gradient",
"frequency",
"E_ZPVE",
"ZPVE",
"mass",
"temperature",
"multiplicity",
"charge",
"rotational_temperature",
"rotational_symmetry_number",
"error",
"error_msg",
"finished",
]
rv = CompOutput()
for key in keys:
rv.__dict__[key] = obj[key]
return rv
def _decode_theory(self, obj):
rv = Theory()
if "method" in obj:
rv.method = obj["method"]
if "semi-empirical" in obj:
rv.method.is_semiempirical = obj["semi-empirical"]
if "grid" in obj:
rv.grid = obj["grid"]
if "solvent model" in obj and "solvent" in obj:
rv.solvent = ImplicitSolvent(obj["solvent model"], obj["solvent"])
if "disp" in obj:
rv.empirical_dispersion = obj["disp"]
if "nproc" in obj:
rv.processors = obj["nproc"]
if "mem" in obj:
rv.memory = obj["mem"]
if "jobs" in obj:
jobs = []
for job in obj["jobs"]:
if job == "OptimizationJob":
jobs.append(OptimizationJob(**obj["jobs"][job]))
elif job == "FrequencyJob":
jobs.append(FrequencyJob(**obj["jobs"][job]))
elif job == "SinglePointJob":
jobs.append(SinglePointJob(**obj["jobs"][job]))
elif job == "ForceJob":
jobs.append(ForceJob(**obj["jobs"][job]))
rv.job_type = jobs
if "basis" in obj or "ecp" in obj:
rv.basis = BasisSet([], [])
if "basis" in obj:
for name, aux_type, file, elements in zip(
obj["basis"]["name"],
obj["basis"]["auxiliary"],
obj["basis"]["file"],
obj["basis"]["elements"],
):
rv.basis.basis.append(
Basis(
name,
elements=elements,
aux_type=aux_type,
user_defined=file,
)
)
if "ecp" in obj:
for name, file, elements in zip(
obj["ecp"]["name"],
obj["ecp"]["file"],
obj["ecp"]["elements"],
):
rv.basis.ecp.append(
ECP(
name,
elements=elements,
user_defined=file,
)
)
if "other" in obj:
rv.kwargs = obj["other"]
return rv
def _decode_finder(self, obj):
specific_type = obj["_spec_type"]
kwargs = obj["kwargs"]
cls = get_class(specific_type)
args = []
sig = signature(cls.__init__)
for param in sig.parameters.values():
if param.name in kwargs and (
param.kind == param.POSITIONAL_ONLY or
param.kind == param.POSITIONAL_OR_KEYWORD
):
args.append(kwargs.pop(param.name))
return cls(*args, **kwargs) | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/json_extension.py | json_extension.py |
import os
import re
import subprocess
from time import sleep
from jinja2 import Environment, FileSystemLoader, Template
from AaronTools import addlogger
from AaronTools.const import AARONLIB
USER = os.getenv("USER")
QUEUE_TYPE = os.getenv("QUEUE_TYPE", "None").upper()
class JobControl:
pass
@addlogger
class SubmitProcess:
"""class for submitting jobs to the queue
attributes:
name: name of job and input file minus the extension
exe: type of input file (com, in, inp)
directory: directory the input file is in
walltime: allocated walltime in hours
processors: allocated processors
memory: allocated memory in GB
template: template job file"""
LOG = None
def __init__(self, fname, walltime, processors, memory, template=None):
"""fname: str - path to input file (e.g. /home/CoolUser/CoolStuff/neat.com
walltime: int/str - walltime in hours
processors: int/str - allocated processors
memory: int/str - allocated memory in GB
template: str - path to template file; if template is None, will look for
psi4.job, orca.job, or gaussian.job (depending on
extension on fname)"""
directory, filename = os.path.split(fname)
self.name, exe = os.path.splitext(filename)
self.exe = exe[1:]
self.directory = os.path.abspath(directory)
self.walltime = walltime
self.processors = processors
self.memory = memory
self.template = template
if not isinstance(template, Template):
self.set_template(template)
@staticmethod
def unfinished_jobs_in_dir(directory, retry=True):
"""returns list(jobids (str)) of jobids in directory
retry: bool - if there's an error while checking the queue, sleep 300s and try again"""
if QUEUE_TYPE == "LSF":
args = ["bjobs", "-l", "2"]
proc = subprocess.Popen(
args, stdout=subprocess.PIPE, stderr=subprocess.PIPE
)
out, err = proc.communicate()
if len(err) != 0 and retry:
SubmitProcess.LOG.warning(
"error checking queue: %s\nsleeping 300s before trying again"
% err.decode("utf-8")
)
sleep(300)
return SubmitProcess.unfinished_jobs_in_dir(directory, retry)
else:
out = out.decode("utf-8")
out = out.replace("\s", "")
out = out.replace("\r", "")
out = out.replace("\n", "")
jobs = re.findall("(Job<\d+>.*RUNLIMIT)", out)
job_ids = []
for job in jobs:
test = re.match("Job<(\d+)>\S+CWD<.+%s>" % directory, job)
if test:
job_ids.append(test.group(1))
return job_ids
elif QUEUE_TYPE == "PBS":
args = ["qstat", "-fx"]
proc = subprocess.Popen(
args, stdout=subprocess.PIPE, stderr=subprocess.PIPE
)
out, err = proc.communicate()
if len(err) != 0 and retry:
SubmitProcess.LOG.warning(
"error checking queue: %s\nsleeping 300s before trying again"
% err.decode("utf-8")
)
sleep(300)
return SubmitProcess.unfinished_jobs_in_dir(directory, retry)
else:
out = out.decode("utf-8")
out = out.replace("\n", "")
out = out.replace("\r", "")
jobs = re.findall("<Job>(.+?)<\/Job>", out)
job_ids = []
for job in jobs:
# Q - queued
# R - running
# S - suspended
test = re.match(
"<Job_Id>(\d+).+<job_state>[QRS].+PBS_O_WORKDIR=[^,<>]*%s"
% directory,
job,
)
if test:
job_ids.append(test.group(1))
return job_ids
elif QUEUE_TYPE == "SLURM":
args = ["squeue", "-o", "%i#%Z#%t", "-u", USER]
proc = subprocess.Popen(
args, stdout=subprocess.PIPE, stderr=subprocess.PIPE
)
out, err = proc.communicate()
if len(err) != 0 and retry:
SubmitProcess.LOG.warning(
"error checking queue: %s\nsleeping 300s before trying again"
% err.decode("utf-8")
)
sleep(300)
return SubmitProcess.unfinished_jobs_in_dir(directory, retry)
else:
out = out.decode("utf-8")
job_ids = []
for job in out.splitlines():
jobid, job_path, job_status = job.split("#")
if directory.endswith(job_path) and job_status in [
"R",
"PD",
]:
job_ids.append(jobid)
return job_ids
elif QUEUE_TYPE == "SGE":
# for SGE, we first grab the job ids for the jobs the user is running
# we then call qstat again to get the directory those jobs are in
args = ["qstat", "-s", "pr", "-u", USER]
proc = subprocess.Popen(
args, stdout=subprocess.PIPE, stderr=subprocess.PIPE
)
out, err = proc.communicate()
if len(err) != 0 and retry:
SubmitProcess.LOG.warning(
"error checking queue: %s\nsleeping 300s before trying again"
% err.decode("utf-8")
)
sleep(300)
return SubmitProcess.unfinished_jobs_in_dir(directory, retry)
else:
out = out.decode("utf-8")
jobs = re.findall("^\s*?(\w+)", out)
# first line is a header
jobs.pop(0)
jlist = ",".join(jobs)
args = ["qstat", "-j", jlist]
proc = subprocess.Popen(
args, stdout=subprocess.PIPE, stderr=subprocess.PIPE
)
out, err = proc.communicate()
if len(err) != 0 and retry:
SubmitProcess.LOG.warning(
"error checking queue: %s\nsleeping 300s before trying again"
% err.decode("utf-8")
)
sleep(300)
return SubmitProcess.unfinished_jobs_in_dir(
directory, retry
)
else:
out = out.decode("utf-8")
job_ids = []
for line in out.splitlines():
job_number = re.search("job_number:\s+(\d+)", line)
workdir = re.search(
"sge_o_workdir:\s+[\S]+%s$" % directory, line
)
if job_number:
job = job_number.group(1)
if workdir:
job_ids.append(job)
return job_ids
def submit(self, wait=False, quiet=True, **opts):
"""submit job to the queue
wait: bool/int - do not leave the function until any job in the directory
finishes (polled every 5 minutes or 'wait' seconds)
opts: dict() used to render template; keys are template variables (e.g. exec_memory)
and values are the corresponding values
"""
job_file = os.path.join(self.directory, self.name + ".job")
opts["name"] = self.name
opts["walltime"] = self.walltime
opts["processors"] = self.processors
opts["memory"] = self.memory
tm = self.template.render(**opts)
if not os.path.isdir(self.directory):
os.mkdirs(self.directory)
with open(job_file, "w") as f:
f.write(tm)
stdin = None
if QUEUE_TYPE == "LSF":
args = ["bsub"]
stdin = open(job_file, "r")
elif QUEUE_TYPE == "SLURM":
args = ["sbatch", job_file]
elif QUEUE_TYPE == "PBS":
args = ["qsub", job_file]
elif QUEUE_TYPE == "SGE":
args = ["qsub", job_file]
else:
raise NotImplementedError(
"%s queues not supported, only LSF, SLURM, PBS, and SGE"
% QUEUE_TYPE
)
proc = subprocess.Popen(
args,
cwd=self.directory,
stdin=stdin,
stdout=subprocess.PIPE,
stderr=subprocess.PIPE,
)
self.submit_out, self.submit_err = proc.communicate()
if len(self.submit_err) != 0:
raise RuntimeError(
"error with submitting job %s: %s"
% (self.name, self.submit_err.decode("utf-8"))
)
if not quiet:
print(self.submit_out.decode("utf-8").strip())
if wait is not False:
if wait is True:
wait_time = 30
else:
wait_time = abs(wait)
sleep(wait_time)
while len(self.unfinished_jobs_in_dir(self.directory)) != 0:
# print(self.unfinished_jobs_in_dir(self.directory))
sleep(wait_time)
return
def set_template(self, filename):
"""
sets job template to filename
AARONLIB directories are searched
"""
environment = Environment(loader=FileSystemLoader(AARONLIB))
if filename is None:
if self.exe == "com" or self.exe == "gjf":
filename = "Gaussian_template.txt"
elif self.exe == "inp":
filename = "ORCA_template.txt"
elif self.exe == "in":
filename = "Psi4_template.txt"
self.template = environment.get_template(filename) | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/job_control.py | job_control.py |
import itertools
import os
import re
from glob import glob
from AaronTools.const import AARONLIB, AARONTOOLS
from AaronTools.fileIO import FileReader, read_types
from AaronTools.geometry import Geometry
class Ring(Geometry):
"""
Attributes:
name
atoms
end
"""
AARON_LIBS = os.path.join(AARONLIB, "Rings")
BUILTIN = os.path.join(AARONTOOLS, "Rings")
def __init__(self, frag, name=None, end=None):
"""
frag is either a file sub, a geometry, or an atom list
name is a name
end is a list of atoms that defines which part of the ring is not part of the fragment
"""
super().__init__()
self.end = end
if isinstance(frag, (Geometry, list)):
# we can create ring object from a geometry
if isinstance(frag, Ring):
self.name = name if name else frag.name
self.end = end if end else frag.end
elif isinstance(frag, Geometry):
self.name = name if name else frag.name
self.end = end if end else None
else:
self.name = name
try:
self.atoms = frag.atoms
except AttributeError:
self.atoms = frag
else: # or we can create from file
# find ring xyz file
fring = None
for lib in [Ring.AARON_LIBS, Ring.BUILTIN]:
if not os.path.exists(lib):
continue
for f in os.listdir(lib):
name, ext = os.path.splitext(f)
if not any(".%s" % x == ext for x in read_types):
continue
match = frag == name
if match:
fring = os.path.join(lib, f)
break
if fring:
break
# or assume we were given a file name instead
if not fring and ".xyz" in frag:
fring = frag
frag = os.path.basename(frag).rstrip(".xyz")
if fring is None:
raise RuntimeError("ring name not recognized: %s" % frag)
# load in atom info
from_file = FileReader(fring)
self.name = frag
self.comment = from_file.comment
self.atoms = from_file.atoms
self.refresh_connected()
end_info = re.search("E:(\d+)", self.comment)
if end_info is not None:
self.end = [
self.find(end)[0]
for end in re.findall("\d+", self.comment)
]
else:
self.end = None
@classmethod
def from_string(cls, name, end_length, end_atom=None, form="smiles"):
"""create ring fragment from string
name str identifier for ring
end_length int number of atoms in ring end
end_atom identifiers identifier for ring end
form str type of identifier (smiles, iupac)
"""
ring = Geometry.from_string(name, form)
if end_atom is not None and end_length is not None:
ring = cls(ring)
end_atom = ring.find(end_atom)[0]
ring.find_end(end_length, end_atom)
return ring
elif end_length is not None:
ring = cls(ring)
ring.find_end(end_length)
return ring
else:
return cls(ring, name=name)
@classmethod
def list(cls, include_ext=False):
names = []
for lib in [cls.AARON_LIBS, cls.BUILTIN]:
if not os.path.exists(lib):
continue
for f in os.listdir(lib):
name, ext = os.path.splitext(os.path.basename(f))
if not any(".%s" % x == ext for x in read_types):
continue
if name in names:
continue
if include_ext:
names.append(name + ext)
else:
names.append(name)
return names
def copy(self):
dup = super().copy()
dup.end = dup.find([atom.name for atom in self.end])
return Ring(dup, end=dup.end)
def find_end(self, path_length, start=[]):
"""finds a path around self that is path_length long and starts with start"""
def linearly_connected(atom_list):
"""returns true if every atom in atom_list is connected to another atom in
the list without backtracking"""
# start shouldn't be end
if atom_list[0] == atom_list[-1]:
return False
# first and second atoms should be bonded
elif atom_list[0] not in atom_list[1].connected:
return False
# last and second to last atoms should be bonded
elif atom_list[-1] not in atom_list[-2].connected:
return False
# all other atoms should be conneced to exactly 2 atoms
elif any(
[
sum([atom1 in atom2.connected for atom2 in atom_list]) != 2
for atom1 in atom_list[1:-1]
]
):
return False
# first two atoms should only be connected to one atom unless they are connected to each other
elif (
sum(
[
sum([atom_list[0] in atom.connected])
+ sum([atom_list[-1] in atom.connected])
for atom in atom_list
]
)
> 2
and atom_list[0] not in atom_list[-1].connected
):
return False
else:
return True
self.end = None
if start:
start_atoms = self.find(start)
else:
start_atoms = []
usable_atoms = []
for atom in self.atoms:
if atom not in start_atoms:
if hasattr(atom, "_connectivity"):
if atom._connectivity > 1:
usable_atoms.append(atom)
else:
usable_atoms.append(atom)
for path in itertools.permutations(
usable_atoms, path_length - len(start_atoms)
):
full_path = start_atoms + list(path)
if linearly_connected(full_path) or path_length == 1:
self.end = list(full_path)
break
if self.end is None:
raise LookupError(
"unable to find %i long path starting with %s around %s"
% (path_length, start, self.name)
)
else:
self.comment = "E:" + ",".join(
[str(self.atoms.index(a) + 1) for a in self.end]
) | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/ring.py | ring.py |
from collections.abc import MutableSequence
import numpy as np
from AaronTools import addlogger, getlogger
from AaronTools.atoms import Atom
from AaronTools.const import PHYSICAL, UNIT
from AaronTools.fileIO import FileReader
from AaronTools.geometry import Geometry
from AaronTools.utils.utils import float_vec, uptri2sym
def obj_to_dict(obj, skip_attrs=None):
# log = getlogger(level="debug")
if skip_attrs is None:
skip_attrs = []
if isinstance(obj, Geometry):
return obj.comment, [str(a) for a in obj]
rv = {}
if hasattr(obj, "__dict__"):
for attr in obj.__dict__:
if attr in skip_attrs:
continue
val = getattr(obj, attr)
if isinstance(val, MutableSequence):
val = [obj_to_dict(v) for v in val]
else:
val = obj_to_dict(val)
rv[str(attr)] = val
return rv
return obj
@addlogger
class CompOutput:
"""
Attributes:
geometry the last Geometry
opts list of Geometry for each optimization steps
frequency Frequency object
archive a string containing the archive entry
energy, enthalpy, free_energy, grimme_g,
mass, temperature, rotational_temperature,
multiplicity, charge, rotational_symmetry_number
error, error_msg, finished,
gradient, E_ZPVE, ZPVE
"""
ELECTRONIC_ENERGY = "NRG"
ZEROPOINT_ENERGY = "ZPE"
RRHO_ENTHALPY = "ENTHALPY"
QUASI_HARMONIC = "QHARM"
QUASI_RRHO = "QRRHO"
RRHO = "RRHO"
LOG = None
LOGLEVEL = "debug"
def __init__(
self,
fname="",
get_all=True,
freq_name=None,
conf_name=None,
):
self.geometry = None
self.opts = None
self.opt_steps = None
self.frequency = None
self.archive = None
self.other = None
self.conformers = None
self.gradient, self.E_ZPVE, self.ZPVE = ({}, None, None)
self.energy, self.enthalpy = (None, None)
self.free_energy, self.grimme_g = (None, None)
self.mass, self.temperature = (None, None)
self.multiplicity, self.charge = (None, None)
self.rotational_temperature = None
self.rotational_symmetry_number = None
self.error, self.error_msg, self.finished = (None, None, None)
# these will be pulled out of FileReader.other dict
keys = [
"opt_steps",
"energy",
"error",
"error_msg",
"gradient",
"finished",
"frequency",
"mass",
"temperature",
"rotational_temperature",
"free_energy",
"multiplicity",
"charge",
"E_ZPVE",
"ZPVE",
"rotational_symmetry_number",
"enthalpy",
"archive",
]
if isinstance(fname, (str, tuple)) and len(fname) > 0:
from_file = FileReader(
fname,
get_all,
just_geom=False,
freq_name=freq_name,
conf_name=conf_name,
)
elif isinstance(fname, FileReader):
from_file = fname
else:
return
if from_file.atoms:
self.geometry = Geometry(
from_file.atoms, comment=from_file.comment, name=from_file.name,
)
if from_file.all_geom:
self.opts = []
for g in from_file.all_geom:
self.opts += [Geometry(g[0])]
if "conformers" in from_file.other:
self.conformers = []
for comment, atoms in from_file.other["conformers"]:
self.conformers.append(Geometry(atoms, comment=comment))
del from_file.other["conformers"]
for k in keys:
if k in from_file.other:
setattr(self, k, from_file.other[k])
else:
setattr(self, k, None)
self.other = {k:v for k, v in from_file.other.items() if k not in keys}
if self.rotational_temperature is None and self.geometry:
self.compute_rot_temps()
if self.frequency:
self.grimme_g = self.calc_Grimme_G()
# recalculate ZPVE b/c our constants and the ones in various programs
# might be slightly different
self.ZPVE = self.calc_zpe()
self.E_ZPVE = self.energy + self.ZPVE
@staticmethod
def boltzmann_weights(
thermo_cos,
nrg_cos=None,
weighting="RRHO",
temperature=298.15,
v0=100,
):
"""
returns boltzmann weights
thermo_cos - list of CompOutput instances for thermochem corrections
nrg_cos - list of CompOutput to take the electronic energy from
order should correspond to thermo_cos
if not given, the energies from thermo_cos are used
weighting - type of energy to use for weighting
can be:
"NRG"
"ZPE"
"ENTHALPY"
"QHARM"
"QRRHO"
"RRHO"
temperature - temperature in K
v0 - parameter for quasi free energy corrections
"""
if not nrg_cos:
nrg_cos = thermo_cos
energies = np.array([co.energy for co in nrg_cos])
corr = None
if weighting == CompOutput.ZEROPOINT_ENERGY:
corr = np.array([co.ZPVE for co in thermo_cos])
elif weighting == CompOutput.RRHO_ENTHALPY:
corr = np.array([
co.therm_corr(temperature=temperature, v0=v0)[1] for
co in thermo_cos
])
elif weighting == CompOutput.QUASI_HARMONIC:
corr = np.array([
co.calc_G_corr(temperature=temperature, v0=v0, method=weighting) for
co in thermo_cos
])
elif weighting == CompOutput.QUASI_RRHO:
corr = np.array([
co.calc_G_corr(temperature=temperature, v0=v0, method=weighting) for
co in thermo_cos
])
elif weighting == CompOutput.RRHO:
corr = np.array([
co.calc_G_corr(temperature=temperature, v0=v0, method=weighting) for
co in thermo_cos
])
if corr is not None:
try:
energies += corr
except ValueError:
raise RuntimeError(
"number of single point energies (%i) "
"does not match number of thermochemical "
"corrections (%i)" % (len(energies), len(corr))
)
relative = energies - min(energies)
w = np.exp(
-relative * UNIT.HART_TO_KCAL / (PHYSICAL.R * temperature)
)
return w / sum(w)
def to_dict(self, skip_attrs=None):
return obj_to_dict(self, skip_attrs=skip_attrs)
def get_progress(self):
rv = ""
grad = self.gradient
if not grad:
rv += "Progress not found"
return rv
for name in grad:
rv += "{:>9}:{}/{:<3} ".format(
name,
grad[name]["value"],
"YES" if grad[name]["converged"] else "NO",
)
return rv.rstrip()
def calc_zpe(self, anharmonic=False):
"""returns ZPVE correction"""
hc = PHYSICAL.PLANCK * PHYSICAL.SPEED_OF_LIGHT / UNIT.HART_TO_JOULE
if anharmonic:
vib = sum(self.frequency.real_frequencies)
x = np.tril(self.other["X_matrix"]).sum()
x0 = self.other["X0"]
zpve = hc * (0.5 * vib + 0.25 * x + x0)
else:
vib = sum(self.frequency.real_frequencies)
zpve = 0.5 * hc * vib
return zpve
def therm_corr(self, temperature=None, v0=100, method="RRHO", pressure=1):
"""
returns thermal correction to energy, enthalpy correction to energy, and entropy
for the specified cutoff frequency and temperature
in that order (Hartrees for corrections, Eh/K for entropy)
temperature: float, temperature in K- None will use self.temperature
pressure: float, pressure in atm
v0: float, cutoff/damping parameter for quasi G corrections
method: str - type of quasi treatment:
RRHO - no quasi treatment
QRRHO - Grimme's quasi-RRHO
see Grimme, S. (2012), Supramolecular Binding Thermodynamics by
Dispersion‐Corrected Density Functional Theory. Chem. Eur. J.,
18: 9955-9964. (DOI: 10.1002/chem.201200497) for details
QHARM - Truhlar's quasi-harmonic
see J. Phys. Chem. B 2011, 115, 49, 14556–14562
(DOI: 10.1021/jp205508z) for details
"""
if self.frequency is None:
msg = "Vibrational frequencies not found, "
msg += "cannot calculate vibrational entropy."
raise AttributeError(msg)
rot = [temp for temp in self.rotational_temperature if temp != 0]
T = temperature if temperature is not None else self.temperature
if T == 0:
return 0, 0, 0
if pressure is None:
pressure = PHYSICAL.STANDARD_PRESSURE
else:
pressure *= UNIT.ATM_TO_PASCAL
mass = self.mass
sigmar = self.rotational_symmetry_number
if sigmar is None and len(self.geometry.atoms) == 1:
sigmar = 3
mult = self.multiplicity
freqs = np.array(self.frequency.real_frequencies)
vib_unit_convert = (
PHYSICAL.SPEED_OF_LIGHT * PHYSICAL.PLANCK / PHYSICAL.KB
)
vibtemps = np.array(
[f_i * vib_unit_convert for f_i in freqs if f_i > 0]
)
if method == "QHARM":
harm_vibtemps = np.array(
[
f_i * vib_unit_convert
if f_i > v0
else v0 * vib_unit_convert
for f_i in freqs
if f_i > 0
]
)
else:
harm_vibtemps = vibtemps
Bav = PHYSICAL.PLANCK ** 2 / (24 * np.pi ** 2 * PHYSICAL.KB)
Bav *= sum([1 / r for r in rot])
# Translational
qt = 2 * np.pi * mass * PHYSICAL.KB * T / (PHYSICAL.PLANCK ** 2)
qt = qt ** (3 / 2)
qt *= PHYSICAL.KB * T / pressure
St = PHYSICAL.GAS_CONSTANT * (np.log(qt) + (5 / 2))
Et = 3 * PHYSICAL.GAS_CONSTANT * T / 2
# Electronic
Se = PHYSICAL.GAS_CONSTANT * (np.log(mult))
# Rotational
if all(r == np.inf for r in rot):
# atomic
qr = 1
Sr = 0
elif len(rot) == 3:
# non linear molecules
qr = np.sqrt(np.pi) / sigmar
qr *= T ** (3 / 2) / np.sqrt(rot[0] * rot[1] * rot[2])
Sr = PHYSICAL.GAS_CONSTANT * (np.log(qr) + 3 / 2)
elif len(rot) == 2:
# linear molecules
qr = (1 / sigmar) * (T / np.sqrt(rot[0] * rot[1]))
Sr = PHYSICAL.GAS_CONSTANT * (np.log(qr) + 1)
else:
# atoms
qr = 1
Sr = 0
if all(r == np.inf for r in rot):
Er = 0
else:
Er = len(rot) * PHYSICAL.GAS_CONSTANT * T / 2
# Vibrational
if method == self.QUASI_HARMONIC:
Sv = np.sum(
harm_vibtemps / (T * (np.exp(harm_vibtemps / T) - 1))
- np.log(1 - np.exp(-harm_vibtemps / T))
)
elif method == self.RRHO:
Sv = np.sum(
vibtemps / (T * (np.exp(vibtemps / T) - 1))
- np.log(1 - np.exp(-vibtemps / T))
)
elif method == self.QUASI_RRHO:
mu = PHYSICAL.PLANCK
mu /= 8 * np.pi ** 2 * freqs * PHYSICAL.SPEED_OF_LIGHT
mu = mu * Bav / (mu + Bav)
Sr_eff = 1 / 2 + np.log(
np.sqrt(
8
* np.pi ** 3
* mu
* PHYSICAL.KB
* T
/ PHYSICAL.PLANCK ** 2
)
)
weights = weight = 1 / (1 + (v0 / freqs) ** 4)
Sv = np.sum(
weights
* (
harm_vibtemps / (T * (np.exp(harm_vibtemps / T) - 1))
- np.log(1 - np.exp(-harm_vibtemps / T))
)
+ (1 - weights) * Sr_eff
)
Ev = np.sum(vibtemps * (1.0 / 2 + 1 / (np.exp(vibtemps / T) - 1)))
Ev *= PHYSICAL.GAS_CONSTANT
Sv *= PHYSICAL.GAS_CONSTANT
Ecorr = (Et + Er + Ev) / (UNIT.HART_TO_KCAL * 1000)
Hcorr = Ecorr + (
PHYSICAL.GAS_CONSTANT * T / (UNIT.HART_TO_KCAL * 1000)
)
Stot = (St + Sr + Sv + Se) / (UNIT.HART_TO_KCAL * 1000)
return Ecorr, Hcorr, Stot
def calc_G_corr(self, temperature=None, v0=0, method="RRHO", **kwargs):
"""
returns quasi rrho free energy correction (Eh)
temperature: float, temperature; default is self.temperature
v0: float, parameter for quasi-rrho or quasi-harmonic entropy
method: str (RRHO, QRRHO, QHARM) method for treating entropy
see CompOutput.therm_corr for references
"""
Ecorr, Hcorr, Stot = self.therm_corr(temperature, v0, method, **kwargs)
T = temperature if temperature is not None else self.temperature
Gcorr_qRRHO = Hcorr - T * Stot
return Gcorr_qRRHO
def calc_Grimme_G(self, temperature=None, v0=100, **kwargs):
"""
returns quasi rrho free energy (Eh)
see Grimme, S. (2012), Supramolecular Binding Thermodynamics by
Dispersion‐Corrected Density Functional Theory. Chem. Eur. J.,
18: 9955-9964. (DOI: 10.1002/chem.201200497) for details
"""
Gcorr_qRRHO = self.calc_G_corr(
temperature=temperature, v0=v0, method=self.QUASI_RRHO, **kwargs
)
return Gcorr_qRRHO + self.energy
def bond_change(self, atom1, atom2, threshold=0.25):
""""""
ref = self.opts[0]
d_ref = ref.atoms[atom1].dist(ref.atoms[atom2])
n = len(self.opts) - 1
for i, step in enumerate(self.opts[::-1]):
d = step.atoms[atom1].dist(step.atoms[atom2])
if abs(d_ref - d) < threshold:
n = len(self.opts) - 1 - i
break
return n
def parse_archive(self):
"""
Reads info from archive string
Returns: a dictionary with the parsed information
"""
def grab_coords(line):
rv = {}
for i, word in enumerate(line.split("\\")):
word = word.split(",")
if i == 0:
rv["charge"] = int(word[0])
rv["multiplicity"] = int(word[1])
rv["atoms"] = []
continue
rv["atoms"] += [
Atom(element=word[0], coords=word[1:4], name=str(i))
]
return rv
rv = {}
lines = iter(self.archive.split("\\\\"))
for line in lines:
line = line.strip()
if not line:
continue
if line.startswith("@"):
line = line[1:]
for word in line.split("\\"):
if "summary" not in rv:
rv["summary"] = [word]
elif word not in rv["summary"]:
rv["summary"] += [word]
continue
if line.startswith("#"):
if "route" not in rv:
rv["route"] = line
elif isinstance(rv["route"], list):
# for compound jobs, like opt freq
rv["route"] += [line]
else:
# for compound jobs, like opt freq
rv["route"] = [rv["route"]] + [line]
line = next(lines).strip()
if "comment" not in line:
rv["comment"] = line
line = next(lines).strip()
for key, val in grab_coords(line).items():
rv[key] = val
continue
words = iter(line.split("\\"))
for word in words:
if not word:
# get rid of pesky empty elements
continue
if "=" in word:
key, val = word.split("=")
rv[key.lower()] = float_vec(val)
else:
if "hessian" not in rv:
rv["hessian"] = uptri2sym(
float_vec(word),
3 * len(rv["atoms"]),
col_based=True,
)
else:
rv["gradient"] = float_vec(word)
return rv
def follow(self, reverse=False, step=0.1):
"""
Follow imaginary mode
"""
# get geometry and frequency objects
geom = self.geometry.copy()
freq = self.frequency
# make sure geom is a TS and has computed frequencies available
if freq is None:
raise AttributeError("Frequencies for this geometry not found.")
if not freq.is_TS:
raise RuntimeError("Geometry not a transition state")
# get displacement vectors for imaginary frequency
img_mode = freq.imaginary_frequencies[0]
vector = freq.by_frequency[img_mode]["vector"]
# apply transformation to geometry and return it
for i, a in enumerate(geom.atoms):
if reverse:
a.coords -= vector[i] * step
else:
a.coords += vector[i] * step
return geom
def compute_rot_temps(self):
"""
sets self's 'rotational_temperature' attribute by using self.geometry
not recommended b/c atoms should be specific isotopes, but this uses
average atomic weights
exists because older versions of ORCA don't print rotational temperatures
"""
COM = self.geometry.COM(mass_weight=True)
self.geometry.coord_shift(-COM)
inertia_mat = np.zeros((3, 3))
for atom in self.geometry.atoms:
for i in range(0, 3):
for j in range(0, 3):
if i == j:
inertia_mat[i][j] += sum(
[
atom.mass() * atom.coords[k] ** 2
for k in range(0, 3)
if k != i
]
)
else:
inertia_mat[i][j] -= (
atom.mass() * atom.coords[i] * atom.coords[j]
)
principal_inertia, vecs = np.linalg.eigh(inertia_mat)
principal_inertia *= UNIT.AMU_TO_KG * 1e-20
# rotational constants in Hz
rot_consts = [
PHYSICAL.PLANCK / (8 * np.pi ** 2 * moment)
for moment in principal_inertia
if moment > 0
]
self.rotational_temperature = [
PHYSICAL.PLANCK * const / PHYSICAL.KB for const in rot_consts
]
# shift geometry back
self.geometry.coord_shift(COM) | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/comp_output.py | comp_output.py |
import numpy as np
from scipy.integrate import quad as integrate
class Pathway:
"""
interpolating between multiple Geometries
Attributes:
geometry - structure for which coordinates are interpolated
num_geom - number of geometries
n_cart - number of Cartesian coordinates
basis - matrix representation of basis used to interpolate between geometries
basis_inverse - inverse of basis
region_length - length of each spline subregion
"""
def __init__(
self,
geometry,
coordinate_array,
other_vars=None,
basis=None,
mass_weighted=False,
):
"""
geometry - Geometry()
coordinate_array - np.array(float, shape=(N, n_atoms, 3))
coordinates for the geometry at N different points
other_vars - dict('variable name':[float])
dictionary of other variables (e.g. energy)
basis - list(np.array(float, shape=(n_atoms,3)))
coordinate displacement matrices (shape n_atoms x 3)
mass_weighted - bool, True if supplied modes are mass-weighted
"""
self.geometry = geometry
self.coordsets = coordinate_array
if other_vars:
self.other_vars = other_vars
else:
self.other_vars = {}
if basis is None:
# if no modes are given, Cartesian coordinates are used
self.gen_basis([], mass_weighted)
else:
self.gen_basis(basis, mass_weighted)
# set up coordinate and energy functions
self.gen_func()
@property
def num_geom(self):
"""returns number of input coordinate sets"""
return self.coordsets.shape[0]
def gen_basis(self, modes, remove_mass_weight=False):
"""
modes - list of coordinate displacement matrices
remove_mass_weight bool, True to remove mass-weighting from displacement coordinates
"""
basis = []
n_cart = 3 * len(self.geometry.atoms)
# go through each of the supplied coordinate displacement matrices
# remove mass-weighting if needed, and reshape
for mode in modes:
no_mass_mode = []
for i, atom in enumerate(self.geometry.atoms):
if remove_mass_weight:
no_mass_mode.append(mode[i] * atom.mass())
else:
no_mass_mode.append(mode[i])
basis.append(np.array(np.reshape(no_mass_mode, n_cart)))
# we need n_cart basis vectors
if len(basis) < n_cart:
if basis:
raise RuntimeError(
"number of basis vectors (%i) is less than 3N (%i)"
% (len(basis), n_cart)
)
# if we don't have any, this is equivalent to using each atom's
# x, y, and z coordinates as our basis
basis = np.identity(n_cart)
basis = np.transpose(basis)
self.basis = basis
self.basis_inverse = np.linalg.inv(basis)
def gen_func(self):
"""
generate Cartesian-displacement-representation coordinate and
miscellaneous variable interpolation functions
sets self.coord_func, self.dcoord_func_dt, self.E_func, and self.dE_func_dt
"""
basis_rep = []
n_cart = 3 * len(self.geometry.atoms)
for xyz in self.coordsets:
d_xyz = xyz - self.coordsets[0]
a = Pathway.dxyz_to_q(d_xyz, self.basis_inverse)
basis_rep.append(np.reshape(a, (n_cart, 1)))
basis_rep = np.reshape(np.array(basis_rep), (n_cart * self.num_geom, 1))
# get cubic spline coefficients for the subregions
# solved by mat * coord = basis -> coord = mat^-1 * basis
mat = Pathway.get_splines_mat(self.num_geom)
# basis is a matrix with basis rep. coefficients, or zeros for derivative rows in M
basis = np.zeros((4 * (self.num_geom - 1), n_cart))
for i in range(0, self.num_geom - 1):
for j in range(0, n_cart):
basis[2 * i][j] = basis_rep[i * n_cart + j][0]
basis[2 * i + 1][j] = basis_rep[(i + 1) * n_cart + j][0]
mat_i = np.linalg.inv(mat)
coord = np.dot(mat_i, basis)
# get arc length for each region
arc_length = Pathway.get_arc_length(coord)
# region_length = [simpson(arc_length, m, m+1) for m in range(0, self.num_geom-1)]
region_length = [
integrate(arc_length, m, m + 1)[0] for m in range(0, self.num_geom - 1)
]
self.region_length = region_length
# set self's coordinate function
# coordinates are coefficients for Cartesian displacement representation
self.coord_func, self.dcoord_func_dt = self.get_coord_func(
coord, region_length
)
self.var_func = {}
self.dvar_func_dt = {}
for var in self.other_vars:
c_var = np.dot(mat_i, Pathway.get_splines_vector(self.other_vars[var]))
self.var_func[var], self.dvar_func_dt[var] = Pathway.get_var_func(
c_var, region_length
)
def geom_func(self, t):
"""returns a Geometry from the interpolated pathway at point t
t float point on pathway {t|0 <= t <= 1}"""
geom = self.geometry.copy()
geom.update_geometry(self.coords_func(t))
return geom
def coords_func(self, t):
"""returns Cartesian coordinates for the geometry at point t"""
Q = self.coord_func(t)
return self.q_to_xyz(Q)
def dcoords_dt_func(self, t):
"""returns derivative of Cartesian coordinates for the geometry at point t"""
Q = self.dcoord_func_dt(t)
return self.q_to_xyz(Q)
def get_coord_func(self, coord, region_length):
"""
returns function for Cartesian displacement coordinate as a function of t (t [0, 1])
and a derivative of this function
coord - array-like(float, shape = (4*n_subregions, n_cart))
matrix of cubic polynomial coefficients
region_length - array-like(float)
arc length of each subregion
"""
n_cart = 3 * len(self.geometry.atoms)
def coord_fun(t):
# map input t to s and region number
s, r = Pathway.t_to_s(t, region_length)
# evaluate polynomial
q = np.array(
[
coord[4 * r][i] * (s - r) ** 3
+ coord[4 * r + 1][i] * (s - r) ** 2
+ coord[4 * r + 2][i] * (s - r)
+ coord[4 * r + 3][i]
for i in range(0, n_cart)
]
)
return q
def dcoord_dt(t):
s, r = Pathway.t_to_s(t, region_length)
q = np.array(
[
3 * coord[4 * r][i] * (s - r) ** 2
+ 2 * coord[4 * r + 1][i] * (s - r)
+ coord[4 * r + 2][i]
for i in range(0, n_cart)
]
)
return q
return coord_fun, dcoord_dt
@staticmethod
def get_var_func(c_var, region_length):
"""just like get_coord_func, but for other variables"""
def var_func(t):
s, r = Pathway.t_to_s(t, region_length)
var = (
c_var[4 * r] * (s - r) ** 3
+ c_var[4 * r + 1] * (s - r) ** 2
+ c_var[4 * r + 2] * (s - r)
+ c_var[4 * r + 3]
)
return var
def dvardt_func(t):
s, r = Pathway.t_to_s(t, region_length)
dvar = (
3 * c_var[4 * r] * (s - r) ** 2
+ 2 * c_var[4 * r + 1] * (s - r)
+ c_var[4 * r + 2]
)
return dvar
return var_func, dvardt_func
@staticmethod
def dxyz_to_q(dxyz, basis_inverse):
"""
converts Cartesian changes (dxyz) to whatever basis set basis_inverse
is (normal mode displacements/Cartesian)
returns a vector containing the coefficients of each basis matrix
"""
q = np.reshape(dxyz, 3 * len(dxyz))
a = np.dot(basis_inverse, q)
return a
def q_to_xyz(self, current_q):
"""converts coordinates for self.basis to Cartesian"""
coords = self.coordsets[0].copy()
for i, mode in enumerate(np.transpose(self.basis)):
coords += current_q[i] * np.reshape(
mode, (len(self.geometry.atoms), 3)
)
return coords
@staticmethod
def get_splines_mat(n_nodes):
"""generate matrix for fitting cubic splines to data
matrix is 4*n_regions x 4*n_regions (n_regions = n_nodes-1)
additional contraints (that might not be valid) as that
the first derivatives are 0 at both ends of the interpolation
region (e.g. f'(0) = 0 and f'(1) = 0)"""
mat = np.zeros((4 * (n_nodes - 1), 4 * (n_nodes - 1)))
# function values are equal where regions meet
for i in range(0, n_nodes - 1):
mat[2 * i][4 * (i + 1) - 1] = 1
for k in range(4 * i, 4 * (i + 1)):
mat[2 * i + 1][k] = 1
# 1st derivatives are equal where regions meet
for i in range(0, n_nodes - 2):
j = 2 * (n_nodes - 1) + i
mat[j][4 * i] = 3
mat[j][4 * i + 1] = 2
mat[j][4 * i + 2] = 1
mat[j][4 * i + 6] = -1
# 2nd derivatives are equal where regions meet
for i in range(0, n_nodes - 2):
j = 3 * (n_nodes - 1) - 1 + i
mat[j][4 * i] = 6
mat[j][4 * i + 1] = 2
mat[j][4 * i + 5] = -2
# 1st derivatives are 0 at the ends
mat[-2][2] = 1
mat[-1][-2] = 1
mat[-1][-3] = 2
mat[-1][-4] = 3
return mat
@staticmethod
def get_splines_vector(data):
"""organize data into a vector that can be used with cubic splines matrix"""
n_regions = len(data) - 1
v = np.zeros(4 * (n_regions))
for i in range(0, n_regions):
v[2 * i] = data[i]
v[2 * i + 1] = data[i + 1]
return v
@staticmethod
def t_to_s(t, region_length):
"""
maps t ([0, 1]) to s (changes linearly with displacement coordinates
need to map b/c cubic splines polynomials generated for interpolation subregions
should be given an input between 0 and 1, no matter where they are on the
whole interpolation
s float point on interpolation arc
region_length list(float) arc length of each region
returns s, r
r is the region number
s is the point in that region
"""
n_regions = len(region_length)
path_length = sum(region_length)
region_start = [
sum(region_length[:i]) for i in range(0, len(region_length))
]
u = t * path_length
r = 0
for l in range(0, n_regions):
if u > region_start[l]:
r = l
s = r + (u - region_start[r]) / region_length[r]
return s, r
@staticmethod
def s_to_t(s, region_length):
"""
map s (changes linearly with displacement coordinate) to t (ranges from 0 to 1)
s float point on interpolation arc
region_length list(float) arc length of each region
returns t float
"""
n_regions = len(region_length)
path_length = sum(region_length)
region_start = [
sum(region_length[:i]) for i in range(0, len(region_length))
]
r = int(s)
while r >= (n_regions):
r -= 1
u = (s - r) * region_length[r] + region_start[r]
t = u / path_length
return t
@staticmethod
def get_arc_length(coord):
"""
returns a function that can be integrated to determine the arc length
of interpolation splines before normalization
coord - array-like(float, shape = (4*n_subregions, n_cart))
matrix of cubic polynomial coefficients
returns function(s)
"""
n_cart = len(coord[0])
def unnormalized_func(s):
r = int(s)
if r == s:
r = int(s - 1)
f = 0
for i in range(0, n_cart):
f += (
3 * coord[4 * r][i] * (s - r) ** 2
+ 2 * coord[4 * r + 1][i] * (s - r)
+ coord[4 * r + 2][i]
) ** 2
return np.sqrt(f)
return unnormalized_func | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/pathway.py | pathway.py |
import sys
import inspect
from collections import deque
import numpy as np
from AaronTools import addlogger
def get_class(name):
"""returns the finder class with the given name"""
for obj_name, obj in inspect.getmembers(sys.modules[__name__]):
if obj_name == name and inspect.isclass(obj):
return obj
raise ValueError("no finder named %s in AaronTools.finders" % name)
class Finder:
def get_matching_atoms(self, atoms, geometry=None):
"""overwrite with function that returns list(Atom) of the atoms that
match your Finder's criteria
geometry is an optional argument that could be used to e.g. find
atoms a certain number of bonds"""
pass
class BondsFrom(Finder):
"""exact number of bonds from specified atom
avoid: bonding path cannot pass through these atoms"""
def __init__(self, central_atom, number_of_bonds, avoid=None):
super().__init__()
self.central_atom = central_atom
self.number_of_bonds = number_of_bonds
self.avoid = avoid
def __repr__(self):
return "atoms %i bonds from %s" % (self.number_of_bonds, self.central_atom)
def get_matching_atoms(self, atoms, geometry):
"""returns List(Atom) that are a certain number of bonds away from the given atom"""
matching_atoms = []
for atom in atoms:
try:
path = geometry.shortest_path(atom, self.central_atom, avoid=self.avoid)
except LookupError:
continue
if len(path) - 1 == self.number_of_bonds:
matching_atoms.append(atom)
return matching_atoms
class WithinBondsOf(BondsFrom):
"""within a specified number of bonds from the atom"""
def __init__(self, central_atom, number_of_bonds, **kwargs):
super().__init__(central_atom, number_of_bonds)
def __repr__(self):
return "atoms within %i bonds of %s" % (self.number_of_bonds, self.central_atom)
def get_matching_atoms(self, atoms, geometry):
"""returns List(Atom) that are a certain number of bonds away from the given atom"""
matching_atoms = []
stack = deque([self.central_atom])
next_stack = deque([])
frag = [self.central_atom]
n_bonds = 1
while stack and self.number_of_bonds:
next_connected = stack.popleft()
connected = next_connected.connected - set(frag)
if n_bonds < self.number_of_bonds:
next_stack.extend(connected)
matching_atoms.extend(connected)
frag += connected
if not stack:
n_bonds += 1
stack = next_stack
next_stack = deque([])
return matching_atoms
class BondedTo(Finder):
"""returns all atoms that are bonded to the specified atom"""
def __init__(self, atom):
super().__init__()
self.atom = atom
def __repr__(self):
return "atoms bonded to %s" % self.atom
def get_matching_atoms(self, atoms, geometry=None):
"""returns list(Atom) that are within a radius of a point"""
return [atom for atom in atoms if atom in self.atom.connected]
class WithinRadiusFromPoint(Finder):
"""within a specified radius of a point"""
def __init__(self, point, radius):
super().__init__()
self.point = np.array(point)
self.radius = radius
def __repr__(self):
return "atoms within %.2f angstroms of (%.2f, %.2f, %.2f)" % (self.radius, *self.point)
def get_matching_atoms(self, atoms, geometry=None):
"""returns list(Atom) that are within a radius of a point"""
keep = np.arange(0, len(atoms), dtype=int)
coords = np.array([atom.coords for atom in atoms])
coords -= self.point
mask = np.where(coords[:, 0] < self.radius)
coords = coords[mask]
keep = keep[mask]
mask = np.where(coords[:, 1] < self.radius)
coords = coords[mask]
keep = keep[mask]
mask = np.where(coords[:, 2] < self.radius)
coords = coords[mask]
keep = keep[mask]
dist = np.linalg.norm(coords, axis=1)
mask = np.where(dist < self.radius)
keep = keep[mask]
matching_atoms = [atoms[k] for k in keep]
return matching_atoms
class WithinRadiusFromAtom(Finder):
"""within a specified radius of a point"""
def __init__(self, atom, radius):
super().__init__()
self.atom = atom
self.radius = radius
def __repr__(self):
return "atoms within %.2f angstroms of %s" % (self.radius, self.atom)
def get_matching_atoms(self, atoms, geometry=None):
"""returns list(Atom) that are within a radius of a point"""
keep = np.arange(0, len(atoms), dtype=int)
coords = np.array([atom.coords for atom in atoms])
coords -= self.atom.coords
mask = np.where(coords[:, 0] < self.radius)
coords = coords[mask]
keep = keep[mask]
mask = np.where(coords[:, 1] < self.radius)
coords = coords[mask]
keep = keep[mask]
mask = np.where(coords[:, 2] < self.radius)
coords = coords[mask]
keep = keep[mask]
dist = np.linalg.norm(coords, axis=1)
mask = np.where(dist < self.radius)
keep = keep[mask]
matching_atoms = [atoms[k] for k in keep]
return matching_atoms
class NotAny(Finder):
"""atoms not matching specifiers/Finders"""
def __init__(self, *critera, **kwargs):
"""critera can be any number of Finders and/or other atom specifiers (tags, elements, etc.)"""
super().__init__()
if not critera and "critera" in kwargs:
critera = kwargs["critera"]
if len(critera) == 1:
if isinstance(critera[0], tuple):
critera = critera[0]
self.critera = critera
def __repr__(self):
return "not any of: %s" % ", ".join([str(x) for x in self.critera])
def get_matching_atoms(self, atoms, geometry):
"""returns List(Atom) that do not match any of the critera"""
unmatching_atoms = []
for criterion in self.critera:
try:
unmatch = geometry.find(criterion)
unmatching_atoms.extend(unmatch)
except LookupError:
pass
return [atom for atom in atoms if atom not in set(unmatching_atoms)]
class AnyTransitionMetal(Finder):
"""any atoms that are transition metals"""
def __init__(self):
super().__init__()
def __repr__(self):
return "any transition metal"
def get_matching_atoms(self, atoms, geometry=None):
"""returns List(Atom) of atoms that are metals"""
from AaronTools.const import TMETAL
return [atom for atom in atoms if atom.element in TMETAL]
class AnyNonTransitionMetal(NotAny):
"""any atoms that are not transition metals"""
def __init__(self, *a, **kw):
super().__init__(AnyTransitionMetal())
def __repr__(self):
return "any non-transition metal"
class HasAttribute(Finder):
"""all atoms with the specified attribute"""
def __init__(self, attribute_name):
super().__init__()
self.attribute_name = attribute_name
def __repr__(self):
return "atoms with the '%s' attribute" % self.attribute_name
def get_matching_atoms(self, atoms, geometry=None):
"""returns List(Atom) of atoms that have the attribute"""
return [atom for atom in atoms if hasattr(atom, self.attribute_name)]
@addlogger
class VSEPR(Finder):
"""atoms with the specified VSEPR geometry
see Atom.get_shape for a list of valid vsepr strings"""
LOG = None
def __init__(self, vsepr, cutoff=0.5):
super().__init__()
self.vsepr = vsepr
if any(vsepr == x for x in ["triangular cupola", "heptagonal bipyramidal"]):
self.LOG.warning(
"triangular cupola and heptagonal bipyramidal cannot be distinguished"
)
self.cutoff = cutoff
def __repr__(self):
return "atoms with %s shape" % self.vsepr
def get_matching_atoms(self, atoms, geometry=None):
matching_atoms = []
for atom in atoms:
out = atom.get_vsepr()
if out is not None:
shape, score = atom.get_vsepr()
if shape == self.vsepr and score < self.cutoff:
matching_atoms.append(atom)
return matching_atoms
class BondedElements(Finder):
"""atoms bonded to the specified neighboring elements
if match_exact=True (default), elements must match exactly
e.g. BondedElements('C') will find
atoms bonded to only one carbon and nothing else"""
def __init__(self, *args, match_exact=True, **kwargs):
super().__init__()
if not args and "elements" in kwargs:
args = kwargs["elements"]
self.elements = list(args)
self.match_exact = match_exact
def __repr__(self):
if len(self.elements) == 0:
return "atoms bonded to nothing"
elif len(self.elements) == 1:
return "atoms bonded to %s" % self.elements[0]
else:
return "atoms bonded to %s and %s" % (", ".join(self.elements[:-1]), self.elements[-1])
def get_matching_atoms(self, atoms, geometry=None):
matching_atoms = []
if self.match_exact:
ref = "".join(sorted(self.elements))
else:
ref = self.elements
for atom in atoms:
if self.match_exact:
ele_list = [a.element for a in [ele for ele in atom.connected]]
test = "".join(sorted(ele_list))
if ref == test:
matching_atoms.append(atom)
else:
bonded_eles = [bonded_atom.element for bonded_atom in atom.connected]
if all([ele in bonded_eles for ele in self.elements]):
matching_atoms.append(atom)
return matching_atoms
class NumberOfBonds(Finder):
"""atoms with the specified number of bonds"""
def __init__(self, num_bonds):
super().__init__()
self.num_bonds = num_bonds
def __repr__(self):
return "atoms with %i bonds" % self.num_bonds
def get_matching_atoms(self, atoms, geometry=None):
return [atom for atom in atoms if len(atom.connected) == self.num_bonds]
class ChiralCentres(Finder):
"""chiral centers
atoms with a non-planar VSEPR geometry with all bonded groups
being distinct
for rings, looks for a set of unique canonical ranks for atoms that
are all the same number of bonds away from one atom"""
#IUPAC spelling
def __init__(self, RS_only=False):
"""RS_only: bool - if True, do not identify chiral centers that are chiral because they
are connected to multiple chiral fragments with the same chirality
this corresponds to R/S centers
False will include r/s centers as well
"""
super().__init__()
self.RS_only = RS_only
def __repr__(self):
return "chiral centers"
def get_matching_atoms(self, atoms, geometry):
from AaronTools.geometry import Geometry
from AaronTools.symmetry import PointGroup
# from time import perf_counter
#
# start = perf_counter()
matching_atoms = []
# b/c they are connected to chiral fragments
geometry.refresh_ranks()
chiral_atoms_changed = True
ranks = geometry.canonical_rank(break_ties=False, update=False, invariant=True)
frags = []
properly_shaped_atoms = []
for atom in geometry.atoms:
if len(atom.connected) < 3:
continue
vsepr, _ = atom.get_vsepr()
if vsepr in ['trigonal planar', 't shaped', 'sqaure planar']:
continue
properly_shaped_atoms.append(atom)
frags.append([])
single_atoms = dict()
for bonded_atom in atom.connected:
frag = geometry.get_fragment(bonded_atom, atom, as_object=False)
frags[-1].append(frag)
# keep track of single atom fragments to more quickly
# eliminate atoms that aren't chiral
if len(frag) == 1:
if frag[0].element in single_atoms:
single_atoms[frag[0].element] += 1
if single_atoms[frag[0].element] >= len(atom.connected) / 2:
frags.pop(-1)
properly_shaped_atoms.pop(-1)
break
else:
single_atoms[frag[0].element] = 1
# print(properly_shaped_atoms)
# need to do multiple passes b/c sometimes atoms are chiral
# because of other chiral centers
k = 0
while chiral_atoms_changed:
chiral_atoms_changed = False
k += 1
#skip atoms we've already found
for ndx, atom in enumerate(properly_shaped_atoms):
if atom in matching_atoms:
continue
neighbor_ranks = [
ranks[geometry.atoms.index(bonded_atom)]
for bonded_atom in atom.connected
]
# first iteration should only look for centers that are chiral
# because the fragments are different
if k == 1 and len(atom.connected) <= 4 and all(
neighbor_ranks.count(rank) == 1 for rank in neighbor_ranks
):
matching_atoms.append(atom)
chiral_atoms_changed = True
elif k == 1 and len(atom.connected) > 4:
test_geom = Geometry(
[atom, *atom.connected], refresh_ranks=False, refresh_connected=False
)
groups = [ranks[geometry.atoms.index(a)] for a in test_geom.atoms]
pg = PointGroup(test_geom, groups=groups, center=atom.coords)
print(pg.name)
if pg.name == "C1":
matching_atoms.append(atom)
chiral_atoms_changed = True
# more iterations should only look for centers that are
# chiral because of the presence of other chiral centers
elif k > 1 and all(
neighbor_ranks.count(rank) <= len(atom.connected) / 2 for rank in neighbor_ranks
):
chiral = True
for i, frag1 in enumerate(frags[ndx]):
#get the ranks of the atoms in this fragment
ranks_1 = [ranks[geometry.atoms.index(atom)] for atom in frag1]
for frag2 in frags[ndx][:i]:
same = True
ranks_2 = [ranks[geometry.atoms.index(atom)] for atom in frag2]
if len(frag1) != len(frag2):
same = False
continue
for a, b in zip(sorted(ranks_1), sorted(ranks_2)):
# want correct elements
if a != b:
same = False
break
for a, b in zip(sorted(frag1), sorted(frag2)):
# and other chiral atoms
# correct connected elements
for o, l in zip(
sorted([aa.element for aa in a.connected]),
sorted([bb.element for bb in b.connected]),
):
if o != l:
same = False
break
if a is b:
break
if not self.RS_only and a in matching_atoms and b in matching_atoms:
#use RMSD to see if they have the same handedness
a_connected = sorted(a.connected)
b_connected = sorted(b.connected)
a_targets = [a] + list(a_connected)
b_targets = [b] + list(b_connected)
if geometry.RMSD(
geometry,
targets=a_targets,
ref_targets=b_targets,
sort=False,
align=False,
) < 0.1:
same = False
break
# I'm not sure why this code was here...
# ring_atoms = [
# bonded_atom for bonded_atom in atom.connected
# if bonded_atom in frag1 and bonded_atom in frag2
# ]
# if len(ring_atoms) > 0:
# #this is a ring
# #look at the rank of all atoms that are n bonds away from this atom
# #if the ranks are ever all different, this is a chiral center
# n_bonds = 1
# acceptable_nbonds = True
# while acceptable_nbonds:
# try:
# atoms_within_nbonds = geometry.find(BondsFrom(atom, n_bonds))
# nbonds_ranks = [
# ranks[geometry.atoms.index(a)] for a in atoms_within_nbonds
# ]
# if all(nbonds_ranks.count(r) == 1 for r in nbonds_ranks):
# same = False
# acceptable_nbonds = False
# elif not self.RS_only:
# # need to find things in the ring that are chiral
# # b/c of other chiral centers
# for n, atom1 in enumerate(atoms_within_nbonds):
# for m, atom2 in enumerate(atoms_within_nbonds[n+1:]):
# p = m + n + 1
# if nbonds_ranks[n] == nbonds_ranks[p]:
# a_connected = sorted(atom1.connected)
# b_connected = sorted(atom2.connected)
# a_targets = [atom1] + list(a_connected)
# b_targets = [atom2] + list(b_connected)
# if geometry.RMSD(
# geometry,
# targets=a_targets,
# ref_targets=b_targets,
# sort=False,
# align=False,
# ) < 0.1:
# same = False
# break
# if not same:
# break
#
# n_bonds += 1
# except LookupError:
# acceptable_nbonds = False
#
# if not same:
# break
if same:
chiral = False
break
if chiral:
chiral_atoms_changed = True
matching_atoms.append(atom)
if self.RS_only:
break
# stop = perf_counter()
# print("took %.3fs" % (stop - start))
return matching_atoms
#alternative spelling
ChiralCenters = ChiralCentres
class FlaggedAtoms(Finder):
"""
atoms with a non-zero flag
"""
# useful for finding constrained atoms
def __repr__(self):
return "flagged atoms"
def get_matching_atoms(self, atoms, geometry):
return [atom for atom in atoms if atom.flag]
class CloserTo(Finder):
"""
atoms closer to atom1 than atom2 (based on bonds, not actual distance)
"""
def __init__(self, atom1, atom2, include_ties=False):
super().__init__()
self.atom1 = atom1
self.atom2 = atom2
self.include_ties = include_ties
def __repr__(self):
return "atoms closer to %s than %s" % (self.atom1, self.atom2)
def get_matching_atoms(self, atoms, geometry):
matching_atoms = []
for atom in atoms:
if atom is self.atom1 and atom is not self.atom2:
matching_atoms.append(atom)
continue
try:
d1 = len(geometry.shortest_path(self.atom1, atom))
except LookupError:
d1 = False
try:
d2 = len(geometry.shortest_path(self.atom2, atom))
except LookupError:
d2 = False
if d1 is not False and d2 is not False and d1 <= d2:
if self.include_ties:
matching_atoms.append(atom)
elif d1 < d2:
matching_atoms.append(atom)
elif d1 is not False and d2 is False:
matching_atoms.append(atom)
return matching_atoms
class AmideCarbon(Finder):
"""
amide carbons
trigonal planar carbons bonded to a linear oxygen and a
nitrogen with 3 bonds
"""
def __repr__(self):
return "amide carbons"
def get_matching_atoms(self, atoms, geometry):
matching_atoms = []
carbons = geometry.find("C", VSEPR("trigonal planar"))
oxygens = geometry.find("O", VSEPR("linear 1"))
nitrogens = geometry.find("N", NumberOfBonds(3))
for carbon in carbons:
if (
any(atom in oxygens for atom in carbon.connected)
and any(atom in nitrogens for atom in carbon.connected)
):
matching_atoms.append(carbon)
return matching_atoms
class Bridgehead(Finder):
"""
bridgehead atoms
can specify ring sizes that the atoms bridge
"""
def __init__(self, ring_sizes=None, match_exact=False):
"""
ring_sizes - list of int, size of rings (e.g. [6, 6] for atoms that bridge
two 6-membered rings)
not specifying yields bridgehead atoms for any ring size
match_exact - bool, if True, return atoms only bridging the specified rings
if False, the ring_sizes is taken as a minimum (e.g.
ring_size=[6, 6], match_exact=False would also yield atoms
bridging three 6-membered rings or two six-membered rings and
a five-membered ring)
"""
self.ring_sizes = ring_sizes
self.match_exact = match_exact
def __repr__(self):
if self.ring_sizes:
return "bridgeheads of %s-member rings" % " or ".join([str(x) for x in self.ring_sizes])
return "bridgehead atoms"
def get_matching_atoms(self, atoms, geometry):
matching_atoms = []
for atom1 in atoms:
matching = True
if self.ring_sizes:
unfound_rings = list(self.ring_sizes)
n_rings = 0
for i, atom2 in enumerate(atom1.connected):
for atom3 in list(atom1.connected)[:i]:
try:
path = geometry.shortest_path(atom2, atom3, avoid=atom1)
n_rings += 1
if self.ring_sizes:
ring_size = len(path) + 1
if ring_size in unfound_rings:
unfound_rings.remove(ring_size)
elif self.match_exact:
matching = False
break
except LookupError:
pass
if not matching:
break
if self.ring_sizes and not unfound_rings and matching:
matching_atoms.append(atom1)
elif n_rings > 1 and not self.ring_sizes:
matching_atoms.append(atom1)
return matching_atoms
class SpiroCenters(Finder):
"""
atom in two different rings with no other common atoms
"""
def __init__(self, ring_sizes=None, match_exact=False):
"""
ring_sizes - list of int, size of rings (e.g. [6, 6] for atoms that bridge
two 6-membered rings)
not specifying yields bridgehead atoms for any ring size
match_exact - bool, if True, return atoms only bridging the specified rings
if False, the ring_sizes is taken as a minimum (e.g.
ring_size=[6, 6], match_exact=False would also yield atoms
bridging three 6-membered rings or two six-membered rings and
a five-membered ring)
"""
self.ring_sizes = ring_sizes
self.match_exact = match_exact
def __repr__(self):
if self.ring_sizes:
return "atoms in different %s-member rings" % " or ".join(
[str(x) for x in self.ring_sizes]
)
return "spiro atoms"
def get_matching_atoms(self, atoms, geometry):
matching_atoms = []
for atom1 in atoms:
matching = True
if self.ring_sizes:
unfound_rings = list(self.ring_sizes)
n_rings = 0
rings = []
for i, atom2 in enumerate(atom1.connected):
for atom3 in list(atom1.connected)[:i]:
try:
path = geometry.shortest_path(atom2, atom3, avoid=atom1)
for ring in rings:
if any(atom in path for atom in ring):
continue
rings.append(path)
except LookupError:
pass
for i, ring in enumerate(rings):
bad_ring = False
for ring2 in rings[:i]:
if any(atom in ring for atom in ring2):
bad_ring = True
break
if bad_ring:
continue
n_rings += 1
if self.ring_sizes:
ring_size = len(path) + 1
if ring_size in unfound_rings:
unfound_rings.remove(ring_size)
elif self.match_exact:
matching = False
break
if not matching:
break
if self.ring_sizes and not unfound_rings and matching:
matching_atoms.append(atom1)
elif n_rings > 1 and not self.ring_sizes:
matching_atoms.append(atom1)
return matching_atoms | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/finders.py | finders.py |
"""for fragments attached to a structure by one bond"""
import json
import os
import re
import sys
from copy import deepcopy
import numpy as np
from AaronTools import addlogger
from AaronTools.const import AARONLIB, AARONTOOLS, BONDI_RADII, VDW_RADII
from AaronTools.fileIO import FileReader, read_types
from AaronTools.geometry import Geometry
from AaronTools.utils.utils import boltzmann_average
@addlogger
class Substituent(Geometry):
"""
Attributes:
name
atoms
end the atom substituent is connected to
conf_num number of conformers
conf_angle angle to rotate by to make next conformer
"""
LOG = None
AARON_LIBS = os.path.join(AARONLIB, "Subs")
BUILTIN = os.path.join(AARONTOOLS, "Substituents")
CACHE_FILE = os.path.join(AARONLIB, "cache", "substituents.json")
try:
with open(CACHE_FILE) as f:
cache = json.load(f)
except (FileNotFoundError, json.decoder.JSONDecodeError):
cache = {}
cache["lengths"] = {} # for storing number of atoms in each sub
def __init__(
self,
sub=None,
name=None,
targets=None,
end=None,
conf_num=None,
conf_angle=None,
detect=True,
):
"""
sub is either a file sub, a geometry, or an atom list
"""
super().__init__()
self.name = name
self.atoms = []
self.end = end
self.conf_angle = conf_angle
self.conf_num = conf_num
self.comment = None
if sub is None:
return
if isinstance(sub, (Geometry, list)):
# we can create substituent object from fragment
if isinstance(sub, Substituent):
self.name = name if name else sub.name
self.conf_num = conf_num if conf_num else sub.conf_num
self.conf_angle = conf_angle if conf_angle else sub.conf_angle
self.comment = sub.comment
elif isinstance(sub, Geometry):
self.name = name if name else sub.name
self.conf_num = conf_num
self.conf_angle = conf_angle
self.comment = sub.comment
else:
self.name = name
self.conf_num = conf_num
self.conf_angle = conf_angle
# save atom info
if targets is None:
try:
self.atoms = sub.atoms
except AttributeError:
self.atoms = sub
else:
self.atoms = sub.find(targets)
# detect sub and conformer info
if detect and (not conf_num or not conf_angle):
if not self.detect_sub():
LookupError(
"Substituent not found in library: " + str(self.name)
)
else:
# or we can create from file
# find substituent xyz file
fsub = None
for lib in [Substituent.AARON_LIBS, Substituent.BUILTIN]:
if not os.path.exists(lib):
continue
for f in os.listdir(lib):
name, ext = os.path.splitext(f)
if not any(".%s" % x == ext for x in read_types):
continue
match = sub == name
if match:
fsub = os.path.join(lib, f)
break
if fsub:
break
# or assume we were given a file name instead
if not fsub and ".xyz" in sub:
fsub = sub
sub = os.path.basename(sub).rstrip(".xyz")
if fsub is None:
match = re.search(r"^{X(.*)}$", sub)
if match:
fsub = Geometry.from_string("Cl" + match.group(1))
fsub.coord_shift(-fsub.atoms[0].coords)
bond = fsub.bond(fsub.atoms[0], fsub.atoms[1])
x_axis = np.array([1.0, 0.0, 0.0])
rot_axis = np.cross(bond, x_axis)
if np.linalg.norm(rot_axis):
bond /= np.linalg.norm(bond)
rot_axis /= np.linalg.norm(rot_axis)
angle = np.arccos(np.dot(bond, x_axis))
fsub.rotate(rot_axis, angle)
self.atoms = fsub.atoms[1:]
self.refresh_connected()
self.name = match.group(1)
self.LOG.warning("Conformer info not loaded for" + sub)
return
else:
raise RuntimeError(
"substituent name not recognized: %s" % sub
)
# load in atom info
from_file = FileReader(fsub)
self.name = sub
self.comment = from_file.comment
self.atoms = from_file.atoms
self.refresh_connected()
if targets is not None:
self.atoms = self.find(targets)
# set conformer info
conf_info = re.search(r"CF:(\d+),(\d+)", self.comment)
if conf_info is not None:
self.conf_num = int(conf_info.group(1))
self.conf_angle = np.deg2rad(float(conf_info.group(2)))
else:
self.LOG.warning("Conformer info not loaded for" + f)
if not self.name:
self.name = "sub"
if self.name == "sub" and end is not None:
self.name += "-{}".format(end.name)
def __lt__(self, other):
if self.end < other.end and not other.end < self.end:
return True
if len(self.atoms) != len(other.atoms):
return len(self.atoms) < len(other.atoms)
for a, b in zip(
self.reorder(start=self.atoms[0])[0],
other.reorder(start=other.atoms[0])[0],
):
if a < b and not b < a:
return True
return False
@staticmethod
def weighted_sterimol(substituents, energies, temperature, *args, **kwargs):
"""
returns Boltzmann-averaged sterimol parameters for the substituents
substituents - list of Substituent instances
energies - numpy array, energy in kcal/mol; ith energy corresponds to ith substituent
temperature - temperature in K
*args, **kwargs - passed to Substituent.sterimol()
"""
CITATION = "doi:10.1021/acscatal.8b04043"
Substituent.LOG.citation(CITATION)
values = {
"B1": [],
"B2": [],
"B3": [],
"B4": [],
"B5": [],
"L": [],
}
rv = dict()
for sub in substituents:
data = sub.sterimol(*args, **kwargs)
for key in data.keys():
values[key].append(data[key])
for key in values.keys():
values[key] = np.array(values[key])
rv[key] = boltzmann_average(energies, values[key], temperature)
return rv
@classmethod
def from_string(
cls,
name,
conf_num=None,
conf_angle=None,
form="smiles",
debug=False,
strict_use_rdkit=False,
):
"""
creates a substituent from a string
name str identifier for substituent
conf_num int number of conformers expected for hierarchical conformer generation
conf_angle int angle between conformers
form str type of identifier (smiles, iupac)
"""
# convert whatever format we"re given to smiles
# then grab the structure from cactus site
from AaronTools.finders import BondedTo
accepted_forms = ["iupac", "smiles"]
if form not in accepted_forms:
raise NotImplementedError(
"cannot create substituent given %s; use one of %s" % form,
str(accepted_forms),
)
rad = re.compile(r"\[\S+?\]")
elements = re.compile(r"[A-Z][a-z]?")
if form == "smiles":
smiles = name
elif form == "iupac":
smiles = cls.iupac2smiles(name)
if debug:
print("radical smiles:", smiles, file=sys.stderr)
# radical atom is the first atom in []
# charged atoms are also in []
my_rad = None
radicals = rad.findall(smiles)
if radicals:
for rad in radicals:
if "." in rad:
my_rad = rad
break
elif "+" not in rad and "-" not in rad:
my_rad = rad
break
if my_rad is None:
if radicals:
cls.LOG.warning(
"radical atom may be ambiguous, be sure to check output: %s"
% smiles
)
my_rad = radicals[0]
else:
raise RuntimeError(
"could not determine radical site on %s; radical site is expected to be in []"
% smiles
)
# construct a modified smiles string with (Cl) right after the radical center
# keep track of the position of this added Cl
# (use Cl instead of H b/c explicit H"s don"t always play nice with RDKit)
pos1 = smiles.index(my_rad)
pos2 = smiles.index(my_rad) + len(my_rad)
previous_atoms = elements.findall(smiles[:pos1])
rad_pos = len(previous_atoms)
if "+" not in my_rad and "-" not in my_rad:
mod_smiles = (
smiles[:pos1]
+ re.sub(r"H\d+", "", my_rad[1:-1])
+ "(Cl)"
+ smiles[pos2:]
)
else:
mod_smiles = (
smiles[:pos1]
+ my_rad[:-1].rstrip("H")
+ "]"
+ "(Cl)"
+ smiles[pos2:]
)
mod_smiles = mod_smiles.replace(".", "")
if debug:
print("modified smiles:", mod_smiles, file=sys.stderr)
print("radical position:", rad_pos, file=sys.stderr)
# grab structure from cactus/RDKit
geom = Geometry.from_string(
mod_smiles, form="smiles", strict_use_rdkit=strict_use_rdkit
)
# the Cl we added is in the same position in the structure as in the smiles string
rad = geom.atoms[rad_pos]
added_Cl = [atom for atom in rad.connected if atom.element == "Cl"][0]
# move the added H to the origin
geom.coord_shift(-added_Cl.coords)
# get the atom bonded to this H
# also move the atom on H to the front of the atoms list to have the expected connectivity
bonded_atom = geom.find(BondedTo(added_Cl))[0]
geom.atoms = [bonded_atom] + [
atom for atom in geom.atoms if atom != bonded_atom
]
bonded_atom.connected.discard(added_Cl)
# align the H-atom bond with the x-axis to have the expected orientation
bond = deepcopy(bonded_atom.coords)
bond /= np.linalg.norm(bond)
x_axis = np.array([1.0, 0.0, 0.0])
rot_axis = np.cross(x_axis, bond)
if abs(np.linalg.norm(rot_axis)) > np.finfo(float).eps:
rot_axis /= np.linalg.norm(rot_axis)
angle = np.arccos(np.dot(bond, x_axis))
geom.rotate(rot_axis, -angle)
else:
try:
import rdkit
except ImportError:
# if the bonded_atom is already on the x axis, we will instead
# rotate about the y axis by 180 degrees
angle = np.pi
geom.rotate(np.array([0.0, 1.0, 0.0]), -angle)
out = cls(
[atom for atom in geom.atoms if atom is not added_Cl],
conf_num=conf_num,
conf_angle=conf_angle,
detect=False,
)
out.refresh_connected()
out.refresh_ranks()
return out
def copy(self, end=None):
"""
creates a new copy of the geometry
parameters:
atoms (list): defaults to all atoms
name (str): defaults to NAME_copy
"""
rv = super().copy(copy_atoms=False)
rv = Substituent(
rv,
end=end,
conf_angle=self.conf_angle,
conf_num=self.conf_num,
detect=False,
)
if end is not None:
rv.atoms[0].connected.add(rv.end)
return rv
@classmethod
def list(cls, include_ext=False):
"""list substituents available from AaronTools or the user's library"""
names = []
for lib in [cls.AARON_LIBS, cls.BUILTIN]:
if not os.path.exists(lib):
continue
for f in os.listdir(lib):
name, ext = os.path.splitext(os.path.basename(f))
if not any(".%s" % x == ext for x in read_types):
continue
if name in names:
continue
if include_ext:
names.append(name + ext)
else:
names.append(name)
return names
def detect_sub(self):
"""
detects conformer information for a substituent by searching the
substituent library
"""
sub_lengths = Substituent.cache["lengths"]
found = False
cache_changed = False
# temporarily detach end from sub so the connectivity is same as
# for the library substituent by itself
atoms_bonded_to_end = [
atom for atom in self.atoms if self.end in atom.connected
]
ranked = False
for atom in atoms_bonded_to_end:
atom.connected.remove(self.end)
for lib in [Substituent.AARON_LIBS, Substituent.BUILTIN]:
if not os.path.exists(lib):
continue
for filename in os.listdir(lib):
name, ext = os.path.splitext(filename)
if not any(".%s" % x == ext for x in read_types):
continue
# test number of atoms against cache
if (
name in sub_lengths
and len(self.atoms) != sub_lengths[name]
):
continue
# use Geometry until we've done all the checks we can do without
# determining connectivity
# (for performance reasons)
init_ref = Geometry(
os.path.join(lib, name + ext),
refresh_connected=False,
refresh_ranks=False,
)
# add to cache
sub_lengths[name] = len(init_ref.atoms)
cache_changed = True
# want same number of atoms
if len(self.atoms) != len(init_ref.atoms):
continue
# same number of each element
ref_eles = [atom.element for atom in init_ref.atoms]
test_eles = [atom.element for atom in self.atoms]
ref_counts = {
ele: ref_eles.count(ele) for ele in set(ref_eles)
}
test_counts = {
ele: test_eles.count(ele) for ele in set(test_eles)
}
if ref_counts != test_counts:
continue
if not ranked:
self.refresh_ranks()
ranked = True
ref_sub = Substituent(init_ref, detect=False)
ref_sub.name = name
ref_sub.refresh_connected()
ref_sub.refresh_ranks()
for a, b in zip(sorted(self.atoms), sorted(ref_sub.atoms)):
# want correct elements
if a.element != b.element:
break
# and correct connections
if len(a.connected) != len(b.connected):
break
# and correct connected elements
failed = False
for i, j in zip(
sorted([aa.element for aa in a.connected]),
sorted([bb.element for bb in b.connected]),
):
if i != j:
failed = True
break
if failed:
break
else:
# if found, save name and conf info
self.name = ref_sub.name
self.comment = ref_sub.comment
conf_info = re.search(r"CF:(\d+),(\d+)", ref_sub.comment)
if conf_info is not None:
self.conf_num = int(conf_info.group(1))
self.conf_angle = np.deg2rad(float(conf_info.group(2)))
found = True
break
for atom in atoms_bonded_to_end:
atom.connected.add(self.end)
# update cache
if cache_changed:
Substituent.cache["lengths"] = sub_lengths
if not os.path.exists(os.path.dirname(Substituent.CACHE_FILE)):
os.makedirs(os.path.dirname(Substituent.CACHE_FILE))
with open(Substituent.CACHE_FILE, "w") as f:
json.dump(Substituent.cache, f)
return found
def sterimol(self, return_vector=False, radii="bondi", old_L=False, **kwargs):
"""
returns sterimol parameter values in a dictionary
keys are B1, B2, B3, B4, B5, and L
see Verloop, A. and Tipker, J. (1976), Use of linear free energy
related and other parameters in the study of fungicidal
selectivity. Pestic. Sci., 7: 379-390.
(DOI: 10.1002/ps.2780070410)
return_vector: bool/returns dict of tuple(vector start, vector end) instead
radii: "bondi" - Bondi vdW radii
"umn" - vdW radii from Mantina, Chamberlin, Valero, Cramer, and Truhlar
old_L: bool - True: use original L (ideal bond length between first substituent
atom and hydrogen + 0.40 angstrom
False: use AaronTools definition
AaronTools' definition of the L parameter is different than the original
STERIMOL program. In STERIMOL, the van der Waals radii of the substituent is
projected onto a plane parallel to the bond between the molecule and the substituent.
The L parameter is 0.40 Å plus the distance from the first substituent atom to the
outer van der Waals surface of the projection along the bond vector. This 0.40 Å is
a correction for STERIMOL using a hydrogen to represent the molecule, when a carbon
would be more likely. In AaronTools the substituent is projected the same, but L is
calculated starting from the van der Waals radius of the first substituent atom
instead. This means AaronTools will give the same L value even if the substituent
is capped with something besides a hydrogen. When comparing AaronTools' L values
with STERIMOL (using the same set of radii for the atoms), the values usually
differ by < 0.1 Å.
"""
from AaronTools.finders import BondedTo
CITATION = "doi:10.1002/ps.2780070410"
self.LOG.citation(CITATION)
if self.end is None:
raise RuntimeError(
"cannot calculate sterimol values for substituents without end"
)
atom1 = self.find(BondedTo(self.end))[0]
atom2 = self.end
if isinstance(radii, dict):
radii_dict = radii
elif radii.lower() == "bondi":
radii_dict = BONDI_RADII
elif radii.lower() == "umn":
radii_dict = VDW_RADII
if old_L:
from AaronTools.atoms import Atom, BondOrder
bo = BondOrder
key = bo.key(atom1, Atom(element="H"))
dx = bo.bonds[key]["1.0"] + 0.4
def L_func(atom, start, radius, L_axis, dx=dx, atom1=atom1):
test_v = start.bond(atom)
test_L = (
np.dot(test_v, L_axis)
- start.dist(atom1)
+ dx
+ radius
)
start_x = atom1.coords - dx * L_axis
L_vec = (start_x, start_x + test_L * L_axis)
return test_L, L_vec
else:
r1 = radii_dict[atom1.element]
def L_func(atom, start, radius, L_axis, atom1=atom1, r1=r1):
test_v = start.bond(atom)
test_L = (
np.dot(test_v, L_axis)
- start.dist(atom1)
+ r1
+ radius
)
start_x = atom1.coords - r1 * L_axis
L_vec = (start_x, start_x + test_L * L_axis)
return test_L, L_vec
L_axis = atom2.bond(atom1)
L_axis /= np.linalg.norm(L_axis)
return super().sterimol(
L_axis,
atom2,
self.atoms,
L_func=L_func,
return_vector=return_vector,
radii=radii,
**kwargs,
)
def align_to_bond(self, bond):
"""
align substituent to a bond vector
"""
bond /= np.linalg.norm(bond)
x_axis = np.array([1.0, 0.0, 0.0])
rot_axis = np.cross(x_axis, bond)
if np.linalg.norm(rot_axis) > 1e-4:
rot_axis /= np.linalg.norm(rot_axis)
else:
rot_axis = np.array([0.0, 1.0, 0.0])
angle = np.arccos(np.dot(bond, x_axis))
self.rotate(rot_axis, angle)
def align_to_x_axis(self):
bond = self.bond(self.end, self.atoms[0])
x_axis = np.array([1.0, 0.0, 0.0])
rot_axis = np.cross(bond, x_axis)
if np.linalg.norm(rot_axis) > 1e-4:
bond /= np.linalg.norm(bond)
rot_axis /= np.linalg.norm(rot_axis)
angle = np.arccos(np.dot(bond, x_axis))
self.rotate(rot_axis, angle, center=self.end.coords)
def sub_rotate(self, angle=None, reverse=False):
"""
rotates substituent about bond w/ rest of geometry
:angle: in radians
"""
if angle is None:
angle = self.conf_angle
if reverse:
angle *= -1
axis = self.atoms[0].bond(self.end)
self.rotate(axis, angle, center=self.end)
def rebuild(self):
start = self.atoms.pop(0)
super().rebuild()
self.atoms = [start] + self.atoms | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/substituent.py | substituent.py |
import concurrent.futures
import numpy as np
from scipy.spatial import distance_matrix
from scipy.special import factorial2
from AaronTools import addlogger
from AaronTools.const import ELEMENTS, UNIT, VDW_RADII, BONDI_RADII
from AaronTools.utils.utils import lebedev_sphere, gauss_legendre_grid
@addlogger
class Orbitals:
"""
stores functions for the shells in a basis set
for evaluation at arbitrary points
attributes:
basis_functions - list(len=n_shell) of lists(len=n_prim_per_shell)
of functions
function takes the arguments:
r2 - float array like, squared distance from the
shell's center to each point being evaluated
x - float or array like, distance from the shell's
center to the point(s) being evaluated along
the x axis
y and z - same as x for the corresponding axis
mo_coeffs - list(len=funcs_per_shell), MO coefficients
for the functions in this shell (e.g. 3
coefficients for the p shell); order
might depend on input file format
for example, FCHK files will be px, py, pz
ORCA files will be pz, px, py
funcs_per_shell - list(len=n_shell), number of basis functions for
each shell
alpha_coefficients - array(shape=(n_mos, n_mos)), coefficients of
molecular orbitals for alpha electrons
beta_coefficients - same as alpha_coefficients for beta electrons
shell_coords - array(shape=(n_shells, 3)), coordinates of each shell
in Angstroms
shell_types - list(str, len=n_shell), type of each shell (e.g. s,
p, sp, 5d, 6d...)
n_shell - number of shells
n_prim_per_shell - list(len=n_shell), number of primitives per shell
n_mos - number of molecular orbitals
exponents - array, exponents for primitives in Eh
each shell
alpha_nrgs - array(len=n_mos), energy of alpha MO's
beta_nrgs - array(len=n_mos), energy of beta MO's
contraction_coeff - array, contraction coefficients for each primitive
in each shell
n_alpha - int, number of alpha electrons
n_beta - int, number of beta electrons
"""
LOG = None
def __init__(self, filereader):
if filereader.file_type == "fchk":
self._load_fchk_data(filereader)
elif filereader.file_type == "out":
self._load_orca_out_data(filereader)
elif filereader.file_type == "47" or filereader.file_type == "31":
self._load_nbo_data(filereader)
else:
raise NotImplementedError(
"cannot load orbital info from %s files" % filereader.file_type
)
def _load_fchk_data(self, filereader):
from scipy.special import factorial2
if "Coordinates of each shell" in filereader.other:
self.shell_coords = np.reshape(
filereader.other["Coordinates of each shell"],
(len(filereader.other["Shell types"]), 3),
)
else:
center_coords = []
for ndx in filereader.other["Shell to atom map"]:
center_coords.append(filereader.atoms[ndx - 1].coords)
self.center_coords = np.array(center_coords)
self.shell_coords *= UNIT.A0_TO_BOHR
self.contraction_coeff = filereader.other["Contraction coefficients"]
self.exponents = filereader.other["Primitive exponents"]
self.n_prim_per_shell = filereader.other["Number of primitives per shell"]
self.alpha_nrgs = filereader.other["Alpha Orbital Energies"]
self.scf_density = filereader.other["Total SCF Density"]
self.beta_nrgs = None
if "Beta Orbital Energies" in filereader.other:
self.beta_nrgs = filereader.other["Beta Orbital Energies"]
self.funcs_per_shell = []
def gau_norm(a, l):
"""
normalization for gaussian primitives that depends on
the exponential (a) and the total angular momentum (l)
"""
t1 = np.sqrt((2 * a) ** (l + 3 / 2)) / (np.pi ** (3.0 / 4))
t2 = np.sqrt(2 ** l / factorial2(2 * l - 1))
return t1 * t2
# get functions for norm of s, p, 5d, and 7f
s_norm = lambda a, l=0: gau_norm(a, l)
p_norm = lambda a, l=1: gau_norm(a, l)
d_norm = lambda a, l=2: gau_norm(a, l)
f_norm = lambda a, l=3: gau_norm(a, l)
g_norm = lambda a, l=4: gau_norm(a, l)
h_norm = lambda a, l=5: gau_norm(a, l)
i_norm = lambda a, l=6: gau_norm(a, l)
self.basis_functions = list()
self.n_mos = 0
self.shell_types = []
shell_i = 0
for n_prim, shell in zip(
self.n_prim_per_shell,
filereader.other["Shell types"],
):
exponents = self.exponents[shell_i : shell_i + n_prim]
con_coeff = self.contraction_coeff[shell_i : shell_i + n_prim]
if shell == 0:
# s functions
self.shell_types.append("s")
self.n_mos += 1
self.funcs_per_shell.append(1)
norms = s_norm(exponents)
if n_prim > 1:
def s_shell(
r2, x, y, z, mo_coeffs,
alpha=exponents, con_coeff=con_coeff, norms=norms
):
e_r2 = np.exp(np.outer(-alpha, r2))
return mo_coeffs[0] * np.dot(con_coeff * norms, e_r2)
else:
def s_shell(
r2, x, y, z, mo_coeff,
alpha=exponents, con_coeff=con_coeff, norms=norms
):
e_r2 = np.exp(-alpha * r2)
return mo_coeff * con_coeff * norms * e_r2
self.basis_functions.append(s_shell)
elif shell == 1:
# p functions
self.shell_types.append("p")
self.n_mos += 3
self.funcs_per_shell.append(3)
norms = p_norm(exponents)
if n_prim > 1:
def p_shell(
r2, x, y, z, mo_coeffs,
alpha=exponents, con_coeff=con_coeff, norms=norms
):
e_r2 = np.exp(np.outer(-alpha, r2))
s_val = np.dot(con_coeff * norms, e_r2)
res = np.zeros(len(r2))
if mo_coeffs[0] != 0:
res += mo_coeffs[0] * x
if mo_coeffs[1] != 0:
res += mo_coeffs[1] * y
if mo_coeffs[2] != 0:
res += mo_coeffs[2] * z
return res * s_val
else:
def p_shell(
r2, x, y, z, mo_coeffs,
alpha=exponents, con_coeff=con_coeff, norms=norms
):
e_r2 = np.exp(-alpha * r2)
s_val = con_coeff * norms * e_r2
res = np.zeros(len(r2))
if mo_coeffs[0] != 0:
res += mo_coeffs[0] * x
if mo_coeffs[1] != 0:
res += mo_coeffs[1] * y
if mo_coeffs[2] != 0:
res += mo_coeffs[2] * z
return res * s_val
self.basis_functions.append(p_shell)
elif shell == -1:
# s=p functions
self.shell_types.append("sp")
self.n_mos += 4
self.funcs_per_shell.append(4)
norm_s = s_norm(exponents)
norm_p = p_norm(exponents)
sp_coeff = filereader.other["P(S=P) Contraction coefficients"][shell_i: shell_i + n_prim]
if n_prim > 1:
def sp_shell(
r2, x, y, z, mo_coeffs,
alpha=exponents,
s_coeff=con_coeff,
p_coeff=sp_coeff,
s_norms=norm_s,
p_norms=norm_p,
):
e_r2 = np.exp(np.outer(-alpha, r2))
sp_val_s = np.dot(s_coeff * s_norms, e_r2)
sp_val_p = np.dot(p_coeff * p_norms, e_r2)
s_res = np.zeros(len(r2))
p_res = np.zeros(len(r2))
if mo_coeffs[0] != 0:
s_res += mo_coeffs[0]
if mo_coeffs[1] != 0:
p_res += mo_coeffs[1] * x
if mo_coeffs[2] != 0:
p_res += mo_coeffs[2] * y
if mo_coeffs[3] != 0:
p_res += mo_coeffs[3] * z
return s_res * sp_val_s + p_res * sp_val_p
else:
def sp_shell(
r2, x, y, z, mo_coeffs,
alpha=exponents,
s_coeff=con_coeff,
p_coeff=sp_coeff,
s_norms=norm_s,
p_norms=norm_p,
):
e_r2 = np.exp(-alpha * r2)
sp_val_s = s_coeff * s_norms * e_r2
sp_val_p = p_coeff * p_norms * e_r2
s_res = np.zeros(len(r2))
p_res = np.zeros(len(r2))
if mo_coeffs[0] != 0:
s_res += mo_coeffs[0]
if mo_coeffs[1] != 0:
p_res += mo_coeffs[1] * x
if mo_coeffs[2] != 0:
p_res += mo_coeffs[2] * y
if mo_coeffs[3] != 0:
p_res += mo_coeffs[3] * z
return s_res * sp_val_s + p_res * sp_val_p
self.basis_functions.append(sp_shell)
elif shell == 2:
# cartesian d functions
self.shell_types.append("6d")
self.n_mos += 6
self.funcs_per_shell.append(6)
norms = d_norm(exponents)
if n_prim > 1:
def d_shell(
r2, x, y, z, mo_coeffs,
alpha=exponents, con_coeff=con_coeff, norms=norms
):
e_r2 = np.exp(np.outer(-alpha, r2))
s_val = np.dot(con_coeff * norms, e_r2)
res = np.zeros(len(r2))
if mo_coeffs[0] != 0:
xx = x ** 2
res += mo_coeffs[0] * xx
if mo_coeffs[1] != 0:
yy = y ** 2
res += mo_coeffs[1] * yy
if mo_coeffs[2] != 0:
zz = z ** 2
res += mo_coeffs[2] * zz
if mo_coeffs[3] != 0:
xy = np.sqrt(3) * x * y
res += mo_coeffs[3] * xy
if mo_coeffs[4] != 0:
xz = np.sqrt(3) * x * z
res += mo_coeffs[4] * xz
if mo_coeffs[5] != 0:
yz = np.sqrt(3) * y * z
res += mo_coeffs[5] * yz
return res * s_val
else:
def d_shell(
r2, x, y, z, mo_coeffs,
alpha=exponents, con_coeff=con_coeff, norms=norms
):
e_r2 = np.exp(-alpha * r2)
s_val = con_coeff * norms * e_r2
res = np.zeros(len(r2))
if mo_coeffs[0] != 0:
xx = x ** 2
res += mo_coeffs[0] * xx
if mo_coeffs[1] != 0:
yy = y ** 2
res += mo_coeffs[1] * yy
if mo_coeffs[2] != 0:
zz = z ** 2
res += mo_coeffs[2] * zz
if mo_coeffs[3] != 0:
xy = np.sqrt(3) * x * y
res += mo_coeffs[3] * xy
if mo_coeffs[4] != 0:
xz = np.sqrt(3) * x * z
res += mo_coeffs[4] * xz
if mo_coeffs[5] != 0:
yz = np.sqrt(3) * y * z
res += mo_coeffs[5] * yz
return res * s_val
self.basis_functions.append(d_shell)
elif shell == -2:
# pure d functions
self.shell_types.append("5d")
self.n_mos += 5
self.funcs_per_shell.append(5)
norms = d_norm(exponents)
if n_prim > 1:
def d_shell(
r2, x, y, z, mo_coeffs,
alpha=exponents, con_coeff=con_coeff, norms=norms
):
e_r2 = np.exp(np.outer(-alpha, r2))
s_val = np.dot(con_coeff * norms, e_r2)
res = np.zeros(len(r2))
if mo_coeffs[0] != 0:
z2r2 = 0.5 * (3 * z ** 2 - r2)
res += mo_coeffs[0] * z2r2
if mo_coeffs[1] != 0:
xz = np.sqrt(3) * x * z
res += mo_coeffs[1] * xz
if mo_coeffs[2] != 0:
yz = np.sqrt(3) * y * z
res += mo_coeffs[2] * yz
if mo_coeffs[3] != 0:
x2y2 = np.sqrt(3) * (x ** 2 - y ** 2) / 2
res += mo_coeffs[3] * x2y2
if mo_coeffs[4] != 0:
xy = np.sqrt(3) * x * y
res += mo_coeffs[4] * xy
return res * s_val
else:
def d_shell(
r2, x, y, z, mo_coeffs,
alpha=exponents, con_coeff=con_coeff, norms=norms
):
e_r2 = np.exp(-alpha * r2)
s_val = con_coeff * norms * e_r2
res = np.zeros(len(r2))
if mo_coeffs[0] != 0:
z2r2 = 0.5 * (3 * z ** 2 - r2)
res += mo_coeffs[0] * z2r2
if mo_coeffs[1] != 0:
xz = np.sqrt(3) * x * z
res += mo_coeffs[1] * xz
if mo_coeffs[2] != 0:
yz = np.sqrt(3) * y * z
res += mo_coeffs[2] * yz
if mo_coeffs[3] != 0:
x2y2 = np.sqrt(3) * (x ** 2 - y ** 2) / 2
res += mo_coeffs[3] * x2y2
if mo_coeffs[4] != 0:
xy = np.sqrt(3) * x * y
res += mo_coeffs[4] * xy
return res * s_val
self.basis_functions.append(d_shell)
elif shell == 3:
# 10f functions
self.shell_types.append("10f")
self.n_mos += 10
self.funcs_per_shell.append(10)
norms = f_norm(exponents)
def f_shell(
r2,
x,
y,
z,
mo_coeffs,
alpha=exponents,
con_coeff=con_coeff,
norms=norms,
):
e_r2 = np.exp(np.outer(-alpha, r2))
s_val = np.dot(con_coeff * norms, e_r2)
res = np.zeros(len(r2))
# ** 3 takes ~6x longer than x * x * x or x ** 2 * x
if mo_coeffs[0] != 0:
xxx = x * x * x
res += mo_coeffs[0] * xxx
if mo_coeffs[1] != 0:
yyy = y * y * y
res += mo_coeffs[1] * yyy
if mo_coeffs[2] != 0:
zzz = z * z * z
res += mo_coeffs[2] * zzz
if mo_coeffs[3] != 0:
xyy = np.sqrt(5) * x * y ** 2
res += mo_coeffs[3] * xyy
if mo_coeffs[4] != 0:
xxy = np.sqrt(5) * x ** 2 * y
res += mo_coeffs[4] * xxy
if mo_coeffs[5] != 0:
xxz = np.sqrt(5) * x ** 2 * z
res += mo_coeffs[5] * xxz
if mo_coeffs[6] != 0:
xzz = np.sqrt(5) * x * z ** 2
res += mo_coeffs[6] * xzz
if mo_coeffs[7] != 0:
yzz = np.sqrt(5) * y * z ** 2
res += mo_coeffs[7] * yzz
if mo_coeffs[8] != 0:
yyz = np.sqrt(5) * y ** 2 * z
res += mo_coeffs[8] * yyz
if mo_coeffs[9] != 0:
xyz = np.sqrt(15) * x * y * z
res += mo_coeffs[9] * xyz
return res * s_val
self.basis_functions.append(f_shell)
elif shell == -3:
# pure f functions
self.shell_types.append("7f")
self.n_mos += 7
self.funcs_per_shell.append(7)
norms = f_norm(exponents)
def f_shell(
r2, x, y, z, mo_coeffs,
alpha=exponents, con_coeff=con_coeff, norms=norms
):
e_r2 = np.exp(np.outer(-alpha, r2))
s_val = np.dot(con_coeff * norms, e_r2)
res = np.zeros(len(r2))
if mo_coeffs[0] != 0:
z3zr2 = z * (5 * z ** 2 - 3 * r2) / 2
res += mo_coeffs[0] * z3zr2
if mo_coeffs[1] != 0:
xz2xr2 = np.sqrt(3) * x * (5 * z ** 2 - r2) / (2 * np.sqrt(2))
res += mo_coeffs[1] * xz2xr2
if mo_coeffs[2] != 0:
yz2yr2 = np.sqrt(3) * y * (5 * z ** 2 - r2) / (2 * np.sqrt(2))
res += mo_coeffs[2] * yz2yr2
if mo_coeffs[3] != 0:
x2zr2z = np.sqrt(15) * z * (x ** 2 - y ** 2) / 2
res += mo_coeffs[3] * x2zr2z
if mo_coeffs[4] != 0:
xyz = np.sqrt(15) * x * y * z
res += mo_coeffs[4] * xyz
if mo_coeffs[5] != 0:
x3r2x = np.sqrt(5) * x * (x ** 2 - 3 * y ** 2) / (2 * np.sqrt(2))
res += mo_coeffs[5] * x3r2x
if mo_coeffs[6] != 0:
x2yy3 = np.sqrt(5) * y * (3 * x ** 2 - y ** 2) / (2 * np.sqrt(2))
res += mo_coeffs[6] * x2yy3
return res * s_val
self.basis_functions.append(f_shell)
# elif shell == 4:
elif False:
# TODO: validate these - order might be wrong
self.shell_types.append("15g")
self.n_mos += 15
self.funcs_per_shell.append(15)
norms = g_norm(exponents)
def g_shell(
r2, x, y, z, mo_coeffs,
alpha=exponents, con_coeffs=con_coeff, norms=norms
):
e_r2 = np.exp(np.outer(-alpha, r2))
s_val = np.dot(con_coeff * norms, e_r2)
res = np.zeros(len(r2))
if mo_coeffs[0] != 0:
x4 = (x ** 2) ** 2
res += mo_coeffs[0] * x4
if mo_coeffs[1] != 0:
x3y = (x ** 3) * x * y
res += mo_coeffs[1] * x3y
if mo_coeffs[2] != 0:
x3z = (x ** 3) * x * z
res += mo_coeffs[2] * x3z
if mo_coeffs[3] != 0:
x2y2 = (x ** 2) * (y ** 2)
res += mo_coeffs[3] * x2y2
if mo_coeffs[4] != 0:
x2yz = (x ** 2) * y * z
res += mo_coeffs[4] * x2yz
if mo_coeffs[5] != 0:
x2z2 = (x ** 2) * (z ** 2)
res += mo_coeffs[5] * x2z2
if mo_coeffs[6] != 0:
xy3 = x * y * y ** 2
res += mo_coeffs[6] * xy3
if mo_coeffs[7] != 0:
xy2z = x * z * y ** 2
res += mo_coeffs[7] * xy2z
if mo_coeffs[8] != 0:
xyz2 = x * y * z ** 2
res += mo_coeffs[8] * xyz2
if mo_coeffs[9] != 0:
xz3 = x * z * z ** 2
res += mo_coeffs[9] * xz3
if mo_coeffs[10] != 0:
y4 = (y ** 2) ** 2
res += mo_coeffs[10] * y4
if mo_coeffs[11] != 0:
y3z = (y ** 2) * y * z
res += mo_coeffs[11] * y3z
if mo_coeffs[12] != 0:
y2z2 = (y * z) ** 2
res += mo_coeffs[12] * y2z2
if mo_coeffs[13] != 0:
yz3 = y * z * z ** 2
res += mo_coeffs[13] * yz3
if mo_coeffs[14] != 0:
z4 = (z ** 2) ** 2
res += mo_coeffs[14] * z4
return res * s_val
self.basis_functions.append(g_shell)
elif shell == -4:
self.shell_types.append("9g")
self.n_mos += 9
self.funcs_per_shell.append(9)
norms = g_norm(exponents)
def g_shell(
r2, x, y, z, mo_coeffs,
alpha=exponents, con_coeffs=con_coeff, norms=norms
):
e_r2 = np.exp(np.outer(-alpha, r2))
s_val = np.dot(con_coeff * norms, e_r2)
res = np.zeros(len(r2))
if mo_coeffs[0] != 0:
z4 = (35 * (z ** 4) - 30 * (r2 * z ** 2) + 3 * r2 ** 2) / 8
res += mo_coeffs[0] * z4
if mo_coeffs[1] != 0:
z3x = np.sqrt(10) * (x * z * (7 * z ** 2 - 3 * r2)) / 4
res += mo_coeffs[1] * z3x
if mo_coeffs[2] != 0:
z3y = np.sqrt(10) * (y * z * (7 * z ** 2 - 3 * r2)) / 4
res += mo_coeffs[2] * z3y
if mo_coeffs[3] != 0:
z2x2y2 = np.sqrt(5) * (x ** 2 - y ** 2) * (7 * z ** 2 - r2) / 4
res += mo_coeffs[3] * z2x2y2
if mo_coeffs[4] != 0:
z2xy = np.sqrt(5) * x * y * (7 * z ** 2 - r2) / 2
res += mo_coeffs[4] * z2xy
if mo_coeffs[5] != 0:
zx3 = np.sqrt(70) * x * z * (x ** 2 - 3 * y ** 2) / 4
res += mo_coeffs[5] * zx3
if mo_coeffs[6] != 0:
zy3 = np.sqrt(70) * z * y * (3 * x ** 2 - y ** 2) / 4
res += mo_coeffs[6] * zy3
if mo_coeffs[7] != 0:
x2 = x ** 2
y2 = y ** 2
x4y4 = np.sqrt(35) * (x2 * (x2 - 3 * y2) - y2 * (3 * x2 - y2)) / 8
res += mo_coeffs[7] * x4y4
if mo_coeffs[8] != 0:
xyx2y2 = np.sqrt(35) * x * y * (x ** 2 - y ** 2) / 2
res += mo_coeffs[8] * xyx2y2
return res * s_val
self.basis_functions.append(g_shell)
elif shell == -5:
self.shell_types.append("11h")
self.n_mos += 11
self.funcs_per_shell.append(11)
norms = h_norm(exponents)
def h_shell(
r2, x, y, z, mo_coeffs,
alpha=exponents, con_coeffs=con_coeff, norms=norms
):
e_r2 = np.exp(np.outer(-alpha, r2))
s_val = np.dot(con_coeff * norms, e_r2)
res = np.zeros(len(r2))
z2 = z ** 2
if mo_coeffs[0] != 0:
z5z3r2zr4 = z * (63 * z2 ** 2 - 70 * z2 * r2 + 15 * r2 ** 2) / 8
res += mo_coeffs[0] * z5z3r2zr4
if mo_coeffs[1] != 0:
xz4xz2r2xr4 = np.sqrt(15) * x * (21 * z2 ** 2 - 14 * z2 * r2 + r2 ** 2) / 8
res += mo_coeffs[1] * xz4xz2r2xr4
if mo_coeffs[2] != 0:
yz4yz2r2yr4 = np.sqrt(15) * y * (21 * z2 ** 2 - 14 * z2 * r2 + r2 ** 2) / 8
res += mo_coeffs[2] * yz4yz2r2yr4
if mo_coeffs[3] != 0:
x2y3z3zr2 = np.sqrt(105) * (x ** 2 - y ** 2) * (3 * z2 - r2) * z / 4
res += mo_coeffs[3] * x2y3z3zr2
if mo_coeffs[4] != 0:
xyz3zr2 = np.sqrt(105) * x * y * z * (3 * z2 - r2) / 2
res += mo_coeffs[4] * xyz3zr2
if mo_coeffs[5] != 0:
xx2y2z2r2 = 35 * x * (x ** 2 - 3 * y ** 2) * (9 * z2 - r2) / (8 * np.sqrt(70))
res += mo_coeffs[5] * xx2y2z2r2
if mo_coeffs[6] != 0:
yx2y2z2r2 = 35 * y * (3 * x ** 2 - y ** 2) * (9 * z2 - r2) / (8 * np.sqrt(70))
res += mo_coeffs[6] * yx2y2z2r2
if mo_coeffs[7] != 0:
zx4x2y2y4 = 105 * z * ((x ** 2) ** 2 - 6 * (x * y) ** 2 + (y ** 2) ** 2) / (8 * np.sqrt(35))
res += mo_coeffs[7] * zx4x2y2y4
if mo_coeffs[8] != 0:
zx3yxy3 = 105 * x * y * z * (4 * x ** 2 - 4 * y ** 2) / (8 * np.sqrt(35))
res += mo_coeffs[8] * zx3yxy3
if mo_coeffs[9] != 0:
xx4y2x2y4 = 21 * x * ((x ** 2) ** 2 - 10 * (x * y) ** 2 + 5 * (y ** 2) ** 2) / (8 * np.sqrt(14))
res += mo_coeffs[9] * xx4y2x2y4
if mo_coeffs[10] != 0:
yx4y2x2y4 = 21 * y * (5 * (x ** 2) ** 2 - 10 * (x * y) ** 2 + (y ** 2) ** 2) / (8 * np.sqrt(14))
res += mo_coeffs[10] * yx4y2x2y4
return res * s_val
self.basis_functions.append(h_shell)
elif shell == -6:
self.shell_types.append("13i")
self.n_mos += 13
self.funcs_per_shell.append(13)
norms = i_norm(exponents)
def i_shell(
r2, x, y, z, mo_coeffs,
alpha=exponents, con_coeffs=con_coeff, norms=norms
):
e_r2 = np.exp(np.outer(-alpha, r2))
s_val = np.dot(con_coeff * norms, e_r2)
res = np.zeros(len(r2))
z2 = z ** 2
if mo_coeffs[0] != 0:
z6z4r2z2r4r6 = (231 * z2 * z2 ** 2 - 315 * z2 ** 2 * r2 + 105 * z2 * r2 ** 2 - 5 * r2 * r2 ** 2) / 16
res += mo_coeffs[0] * z6z4r2z2r4r6
if mo_coeffs[1] != 0:
xz5z3r2zr4 = np.sqrt(21) * x * z * (33 * z2 ** 2 - 30 * z2 * r2 + 5 * r2 ** 2) / 8
res += mo_coeffs[1] * xz5z3r2zr4
if mo_coeffs[2] != 0:
yz5z3r2zr4 = np.sqrt(21) * y * z * (33 * z2 ** 2 - 30 * z2 * r2 + 5 * r2 ** 2) / 8
res += mo_coeffs[2] * yz5z3r2zr4
if mo_coeffs[3] != 0:
x2y2z4z2r2r3 = 105 * (x ** 2 - y ** 2) * (33 * z2 ** 2 - 18 * z2 * r2 + r2 ** 2) / (16 * np.sqrt(210))
res += mo_coeffs[3] * x2y2z4z2r2r3
if mo_coeffs[4] != 0:
xyz4z2r2r4 = 105 * x * y * (33 * z2 ** 2 - 18 * z2 * r2 + r2 ** 2) / (8 * np.sqrt(210))
res += mo_coeffs[4] * xyz4z2r2r4
if mo_coeffs[5] != 0:
xx2y2z3zr2 = 105 * x * z * (x ** 2 - 3 * y ** 2) * (11 * z2 - 3 * r2) / (8 * np.sqrt(210))
res += mo_coeffs[5] * xx2y2z3zr2
if mo_coeffs[6] != 0:
yx2y2z3zr2 = 105 * y * z * (3 * x ** 2 - y ** 2) * (11 * z2 - 3 * r2) / (8 * np.sqrt(210))
res += mo_coeffs[6] * yx2y2z3zr2
if mo_coeffs[7] != 0:
x4x2y2y4z2r2 = np.sqrt(63) * ((x ** 2) ** 2 - 6 * (x * y) ** 2 + (y ** 2) ** 2) * (11 * z2 - r2) / 16
res += mo_coeffs[7] * x4x2y2y4z2r2
if mo_coeffs[8] != 0:
xyx2y2z2r2 = np.sqrt(63) * x * y * (x ** 2 - y ** 2) * (11 * z2 - r2) / 4
res += mo_coeffs[8] * xyx2y2z2r2
if mo_coeffs[9] != 0:
xzx4x2y2y4 = 231 * x * z * ((x ** 2) ** 2 - 10 * (x * y) ** 2 + 5 * (y ** 2) ** 2) / (8 * np.sqrt(154))
res += mo_coeffs[9] * xzx4x2y2y4
if mo_coeffs[10] != 0:
yzx4x2y2y4 = 231 * y * z * (5 * (x ** 2) ** 2 - 10 * (x * y) ** 2 + (y ** 2) ** 2) / (8 * np.sqrt(154))
res += mo_coeffs[10] * yzx4x2y2y4
if mo_coeffs[11] != 0:
x6x4y2x2y4y6 = 231 * ((x * x ** 2) ** 2 - 15 * (x ** 2 * y) ** 2 + 15 * (x * y ** 2) ** 2 - (y * y ** 2) ** 2) / (16 * np.sqrt(462))
res += mo_coeffs[11] * x6x4y2x2y4y6
if mo_coeffs[12] != 0:
yx5x3y3xy5 = 231 * x * y * (6 * (x ** 2) ** 2 - 20 * (x * y) ** 2 + 6 * (y ** 2) ** 2) / (16 * np.sqrt(462))
res += mo_coeffs[12] * yx5x3y3xy5
return res * s_val
self.basis_functions.append(i_shell)
else:
self.LOG.warning("cannot parse shell with type %i" % shell)
shell_i += n_prim
self.alpha_coefficients = np.reshape(
filereader.other["Alpha MO coefficients"],
(self.n_mos, self.n_mos),
)
if "Beta MO coefficients" in filereader.other:
self.beta_coefficients = np.reshape(
filereader.other["Beta MO coefficients"],
(self.n_mos, self.n_mos),
)
else:
self.beta_coefficients = None
self.n_alpha = filereader.other["Number of alpha electrons"]
if "Number of beta electrons" in filereader.other:
self.n_beta = filereader.other["Number of beta electrons"]
def _load_nbo_data(self, filereader):
self.basis_functions = []
self.exponents = np.array(filereader.other["exponents"])
self.alpha_coefficients = np.array(filereader.other["alpha_coefficients"])
self.beta_coefficients = None
self.shell_coords = []
self.funcs_per_shell = []
self.shell_types = []
self.n_shell = len(filereader.other["n_prim_per_shell"])
self.alpha_nrgs = [0 for x in self.alpha_coefficients]
self.n_mos = len(self.alpha_coefficients)
self.n_alpha = 0
self.n_beta = 0
self.beta_nrgs = None
label_i = 0
# NBO includes normalization constant with the contraction coefficient
# so we don't have a gau_norm function like gaussian or orca
for n_prim, n_funcs, shell_i in zip(
filereader.other["n_prim_per_shell"],
filereader.other["funcs_per_shell"],
filereader.other["start_ndx"],
):
shell_i -= 1
exponents = self.exponents[shell_i: shell_i + n_prim]
shell_funcs = []
con_coeffs = []
shell_type = []
self.funcs_per_shell.append(n_funcs)
self.shell_coords.append(
filereader.atoms[filereader.other["shell_to_atom"][label_i] - 1].coords
)
for i in range(0, n_funcs):
shell = filereader.other["momentum_label"][label_i]
label_i += 1
# XXX: each function is treated as a different
# shell because NBO allows them to be in any order
# I think that technically means the functions in
# the d shell for example don't need to be next
# to each other
if shell < 100:
shell_type.append("s")
# s - shell can be 1 or 51
con_coeff = filereader.other["s_coeff"][shell_i: shell_i + n_prim]
con_coeffs.append(con_coeff)
def s_shell(
r2, x, y, z, s_val
):
return s_val
shell_funcs.append(s_shell)
elif shell < 200:
# p - shell can be 101, 102, 103, 151, 152, 153
con_coeff = filereader.other["p_coeff"][shell_i: shell_i + n_prim]
con_coeffs.append(con_coeff)
if shell == 101 or shell == 151:
shell_type.append("px")
def px_shell(
r2, x, y, z, s_val
):
return s_val * x
shell_funcs.append(px_shell)
elif shell == 102 or shell == 152:
shell_type.append("py")
def py_shell(
r2, x, y, z, s_val
):
return s_val * y
shell_funcs.append(py_shell)
elif shell == 103 or shell == 153:
shell_type.append("pz")
def pz_shell(
r2, x, y, z, s_val
):
return s_val * z
shell_funcs.append(pz_shell)
elif shell < 300:
con_coeff = filereader.other["d_coeff"][shell_i: shell_i + n_prim]
con_coeffs.append(con_coeff)
if shell == 201:
shell_type.append("dxx")
def dxx_shell(
r2, x, y, z, s_val
):
return s_val * x * x
shell_funcs.append(dxx_shell)
elif shell == 202:
shell_type.append("dxy")
def dxy_shell(
r2, x, y, z, s_val
):
return s_val * x * y
shell_funcs.append(dxy_shell)
elif shell == 203:
shell_type.append("dxz")
def dxz_shell(
r2, x, y, z, s_val
):
return s_val * x * z
shell_funcs.append(dxz_shell)
elif shell == 204:
shell_type.append("dyy")
def dyy_shell(
r2, x, y, z, s_val
):
return s_val * y * y
shell_funcs.append(dyy_shell)
elif shell == 205:
shell_type.append("dyz")
def dyz_shell(
r2, x, y, z, s_val
):
return s_val * y * z
shell_funcs.append(dyz_shell)
elif shell == 206:
shell_type.append("dzz")
def dzz_shell(
r2, x, y, z, s_val
):
return s_val * z * z
shell_funcs.append(dzz_shell)
elif shell == 251:
shell_type.append("5dxy")
def dxy_shell(
r2, x, y, z, s_val
):
return np.sqrt(3) * s_val * x * y
shell_funcs.append(dxy_shell)
elif shell == 252:
shell_type.append("5dxz")
def dxz_shell(
r2, x, y, z, s_val
):
return np.sqrt(3) * s_val * x * z
shell_funcs.append(dxz_shell)
elif shell == 253:
shell_type.append("5dyz")
def dyz_shell(
r2, x, y, z, s_val
):
return np.sqrt(3) * s_val * y * z
shell_funcs.append(dyz_shell)
elif shell == 254:
shell_type.append("5dx2-y2")
def dx2y2_shell(
r2, x, y, z, s_val
):
return np.sqrt(3) * s_val * (x ** 2 - y ** 2) / 2
shell_funcs.append(dx2y2_shell)
elif shell == 255:
shell_type.append("5dz2")
def dz2_shell(
r2, x, y, z, s_val
):
return (3 * z ** 2 - r2) * s_val / 2
shell_funcs.append(dz2_shell)
elif shell < 400:
con_coeff = filereader.other["f_coeff"][shell_i: shell_i + n_prim]
con_coeffs.append(con_coeff)
if shell == 301:
shell_type.append("fxxx")
def fxxx_shell(
r2, x, y, z, s_val
):
return s_val * x * x * x
shell_funcs.append(fxxx_shell)
if shell == 302:
shell_type.append("fxxy")
def fxxy_shell(
r2, x, y, z, s_val
):
return s_val * x * x * y
shell_funcs.append(fxxy_shell)
if shell == 303:
shell_type.append("fxxz")
def fxxz_shell(
r2, x, y, z, s_val
):
return s_val * x * x * z
shell_funcs.append(fxxz_shell)
if shell == 304:
shell_type.append("fxyy")
def fxyy_shell(
r2, x, y, z, s_val
):
return s_val * x * y * y
shell_funcs.append(fxyy_shell)
if shell == 305:
shell_type.append("fxyz")
def fxyz_shell(
r2, x, y, z, s_val
):
return s_val * x * y * z
shell_funcs.append(fxyz_shell)
if shell == 306:
shell_type.append("fxzz")
def fxzz_shell(
r2, x, y, z, s_val
):
return s_val * x * z * z
shell_funcs.append(fxzz_shell)
if shell == 307:
shell_type.append("fyyy")
def fyyy_shell(
r2, x, y, z, s_val
):
return s_val * y * y * y
shell_funcs.append(fyyy_shell)
if shell == 308:
shell_type.append("fyyz")
def fyyz_shell(
r2, x, y, z, s_val
):
return s_val * y * y * z
shell_funcs.append(fyyz_shell)
if shell == 309:
shell_type.append("fyzz")
def fyzz_shell(
r2, x, y, z, s_val
):
return s_val * y * z * z
shell_funcs.append(fyzz_shell)
if shell == 310:
shell_type.append("fzzz")
def fzzz_shell(
r2, x, y, z, s_val
):
return s_val * z * z * z
shell_funcs.append(fzzz_shell)
if shell == 351:
shell_type.append("7fz3-zr2")
def fz3zr2_shell(
r2, x, y, z, s_val
):
return s_val * z * (5 * z ** 2 - 3 * r2) / 2
shell_funcs.append(fz3zr2_shell)
if shell == 352:
shell_type.append("7fxz2-xr2")
def fxz2xr2_shell(
r2, x, y, z, s_val
):
return np.sqrt(3) * s_val * x * (5 * z ** 2 - r2) / (2 * np.sqrt(2))
shell_funcs.append(fxz2xr2_shell)
if shell == 353:
shell_type.append("7fyz2-yr2")
def fyz2yr2_shell(
r2, x, y, z, s_val
):
return s_val * y * (5 * z ** 2 - r2) / (2 * np.sqrt(2))
shell_funcs.append(fyz2yr2_shell)
if shell == 354:
shell_type.append("7fzx2-zy2")
def fzx2zy2_shell(
r2, x, y, z, s_val
):
return np.sqrt(15) * s_val * z * (x ** 2 - y ** 2) / 2
shell_funcs.append(fzx2zy2_shell)
if shell == 355:
shell_type.append("7fxyz")
def fxyz_shell(
r2, x, y, z, s_val
):
return np.sqrt(15) * s_val * x * y * z
shell_funcs.append(fxyz_shell)
if shell == 356:
shell_type.append("7fx3-xy2")
def fx3xy2_shell(
r2, x, y, z, s_val
):
return np.sqrt(5) * s_val * x * (x ** 2 - 3 * y ** 2) / (2 * np.sqrt(2))
shell_funcs.append(fx3xy2_shell)
if shell == 357:
shell_type.append("7fyx2-y3")
def fyx2y3_shell(
r2, x, y, z, s_val
):
return np.sqrt(5) * s_val * y * (3 * x ** 2 - y ** 2) / (2 * np.sqrt(2))
shell_funcs.append(fyx2y3_shell)
elif shell < 500:
# I can't tell what NBO does with g orbitals
# I don't have any reference to compare to
self.LOG.warning(
"g shell results have not been verified for NBO\n"
"any LCAO's may be invalid"
)
con_coeff = filereader.other["g_coeff"][shell_i: shell_i + n_prim]
con_coeffs.append(con_coeff)
if shell == 401:
shell_type.append("gxxxx")
def gxxxx_shell(
r2, x, y, z, s_val
):
return s_val * x * x * x * x
shell_funcs.append(gxxxx_shell)
if shell == 402:
shell_type.append("gxxxy")
def gxxxy_shell(
r2, x, y, z, s_val
):
return s_val * x * x * x * y
shell_funcs.append(gxxxy_shell)
if shell == 403:
shell_type.append("gxxxz")
def gxxxz_shell(
r2, x, y, z, s_val
):
return s_val * x * x * x * z
shell_funcs.append(gxxxz_shell)
if shell == 404:
shell_type.append("gxxyy")
def gxxyy_shell(
r2, x, y, z, s_val
):
return s_val * x * x * y * y
shell_funcs.append(gxxyy_shell)
if shell == 405:
shell_type.append("gxxyz")
def gxxyz_shell(
r2, x, y, z, s_val
):
return s_val * x * x * y * z
shell_funcs.append(gxxyz_shell)
if shell == 406:
shell_type.append("gxxzz")
def gxxzz_shell(
r2, x, y, z, s_val
):
return s_val * x * x * z * z
shell_funcs.append(gxxzz_shell)
if shell == 407:
shell_type.append("gxyyy")
def gxyyy_shell(
r2, x, y, z, s_val
):
return s_val * x * y * y * y
shell_funcs.append(gxyyy_shell)
if shell == 408:
shell_type.append("gxyyz")
def gxyyz_shell(
r2, x, y, z, s_val
):
return s_val * x * y * y * z
shell_funcs.append(gxyyz_shell)
if shell == 409:
shell_type.append("gxyzz")
def gxyzz_shell(
r2, x, y, z, s_val
):
return s_val * x * y * z * z
shell_funcs.append(gxyzz_shell)
if shell == 410:
shell_type.append("gxzzz")
def gxzzz_shell(
r2, x, y, z, s_val
):
return s_val * x * z * z * z
shell_funcs.append(gxzzz_shell)
if shell == 411:
shell_type.append("gyyyy")
def gyyyy_shell(
r2, x, y, z, s_val
):
return s_val * y * y * y * y
shell_funcs.append(gyyyy_shell)
if shell == 412:
shell_type.append("gyyyz")
def gyyyz_shell(
r2, x, y, z, s_val
):
return s_val * y * y * y * z
shell_funcs.append(gyyyz_shell)
if shell == 413:
shell_type.append("gyyzz")
def gyyzz_shell(
r2, x, y, z, s_val
):
return s_val * y * y * z * z
shell_funcs.append(gyyzz_shell)
if shell == 414:
shell_type.append("gyzzz")
def gyzzz_shell(
r2, x, y, z, s_val
):
return s_val * y * z * z * z
shell_funcs.append(gyzzz_shell)
if shell == 415:
shell_type.append("gzzzz")
def gzzzz_shell(
r2, x, y, z, s_val
):
return s_val * z * z * z * z
shell_funcs.append(gzzzz_shell)
if shell == 451:
shell_type.append("9gz4")
def gz4_shell(
r2, x, y, z, s_val
):
return s_val * (35 * (z ** 2) ** 2 - 30 * z ** 2 * r2 + 3 * r2 ** 2) / 8
shell_funcs.append(gz4_shell)
if shell == 452:
shell_type.append("9gz3x")
def gz3x_shell(
r2, x, y, z, s_val
):
return s_val * np.sqrt(10) * (x * z * (7 * z ** 2 - 3 * r2)) / 4
shell_funcs.append(gz3x_shell)
if shell == 453:
shell_type.append("9gz3y")
def gz3y_shell(
r2, x, y, z, s_val
):
return s_val * np.sqrt(10) * (y * z * (7 * z ** 2 - 3 * r2)) / 4
shell_funcs.append(gz3y_shell)
if shell == 454:
shell_type.append("9gz2x2-z2y2")
def gz2x2y2_shell(
r2, x, y, z, s_val
):
return s_val * np.sqrt(5) * (x ** 2 - y ** 2) * (7 * z ** 2 - r2) / 4
shell_funcs.append(gz2x2y2_shell)
if shell == 455:
shell_type.append("9gz2xy")
def gz2xy_shell(
r2, x, y, z, s_val
):
return s_val * np.sqrt(5) * x * y * (7 * z ** 2 - r2) / 2
shell_funcs.append(gz2xy_shell)
if shell == 456:
shell_type.append("9gzx3")
def gzx3_shell(
r2, x, y, z, s_val
):
return s_val * np.sqrt(70) * x * z * (x ** 2 - 3 * y ** 2) / 4
shell_funcs.append(gzx3_shell)
if shell == 457:
shell_type.append("9gzy3")
def gzy3_shell(
r2, x, y, z, s_val
):
return s_val * np.sqrt(70) * z * y * (3 * x ** 2 - y ** 2) / 4
shell_funcs.append(gzy3_shell)
if shell == 458:
shell_type.append("9gx4y4")
def gx4y4_shell(
r2, x, y, z, s_val
):
x2 = x ** 2
y2 = y ** 2
return s_val * np.sqrt(35) * (x2 * (x2 - 3 * y2) - y2 * (3 * x2 - y2)) / 8
shell_funcs.append(gx4y4_shell)
if shell == 459:
shell_type.append("9gxyx2y2")
def gxyx2y2_shell(
r2, x, y, z, s_val
):
return s_val * np.sqrt(35) * x * y * (x ** 2 - y ** 2) / 2
shell_funcs.append(gxyx2y2_shell)
else:
self.LOG.warning("cannot handle shells with momentum label %i" % shell)
def eval_shells(
r2, x, y, z, mo_coeffs,
alpha=exponents,
con_coeffs=con_coeffs,
shell_funcs=shell_funcs
):
e_r2 = np.exp(np.outer(-alpha, r2))
res = np.zeros(len(r2))
last_con_coeff = None
for mo_coeff, con_coeff, func in zip(mo_coeffs, con_coeffs, shell_funcs):
if mo_coeff == 0:
continue
if last_con_coeff is None or any(
x - y != 0 for x, y in zip(last_con_coeff, con_coeff)
):
s_val = np.dot(con_coeff, e_r2)
last_con_coeff = con_coeff
res += mo_coeff * func(r2, x, y, z, s_val)
return res
self.basis_functions.append(eval_shells)
self.shell_types.append(", ".join(shell_type))
self.shell_coords = np.array(self.shell_coords)
def _load_orca_out_data(self, filereader):
self.shell_coords = []
self.basis_functions = []
self.alpha_nrgs = np.array(filereader.other["alpha_nrgs"])
self.alpha_coefficients = np.array(filereader.other["alpha_coefficients"])
if not filereader.other["beta_nrgs"]:
self.beta_nrgs = None
self.beta_coefficients = None
else:
self.beta_nrgs = np.array(filereader.other["beta_nrgs"])
self.beta_coefficients = np.array(filereader.other["beta_coefficients"])
self.shell_types = []
self.funcs_per_shell = []
self.n_aos = 0
self.n_mos = 0
def gau_norm(a, l):
"""
normalization for gaussian primitives that depends on
the exponential (a) and the total angular momentum (l)
"""
t1 = np.sqrt((2 * a) ** (l + 3 / 2)) / (np.pi ** (3.0 / 4))
t2 = np.sqrt(2 ** l / factorial2(2 * l - 1))
return t1 * t2
# get functions for norm of s, p, 5d, and 7f
s_norm = lambda a, l=0: gau_norm(a, l)
p_norm = lambda a, l=1: gau_norm(a, l)
d_norm = lambda a, l=2: gau_norm(a, l)
f_norm = lambda a, l=3: gau_norm(a, l)
g_norm = lambda a, l=4: gau_norm(a, l)
h_norm = lambda a, l=5: gau_norm(a, l)
i_norm = lambda a, l=6: gau_norm(a, l)
# ORCA order differs from FCHK in a few places:
# pz, px, py instead of ox, py, pz
# f(3xy^2 - x^3) instead of f(x^3 - 3xy^2)
# f(y^3 - 3x^2y) instead of f(3x^2y - y^3)
# ORCA doesn't seem to print the coordinates of each
# shell, but they should be the same as the atom coordinates
for atom in filereader.atoms:
ele = atom.element
for shell_type, n_prim, exponents, con_coeff in filereader.other[
"basis_set_by_ele"
][ele]:
self.shell_coords.append(atom.coords)
exponents = np.array(exponents)
con_coeff = np.array(con_coeff)
if shell_type.lower() == "s":
self.shell_types.append("s")
self.funcs_per_shell.append(1)
self.n_aos += 1
norms = s_norm(exponents)
if n_prim > 1:
def s_shell(
r2, x, y, z, mo_coeff,
alpha=exponents,
con_coeff=con_coeff,
norms=norms
):
e_r2 = np.exp(np.outer(-alpha, r2))
return mo_coeff[0] * np.dot(con_coeff * norms, e_r2)
else:
def s_shell(
r2, x, y, z, mo_coeff,
alpha=exponents,
con_coeff=con_coeff,
norms=norms
):
e_r2 = np.exp(-alpha * r2)
return mo_coeff * con_coeff * norms * e_r2
self.basis_functions.append(s_shell)
elif shell_type.lower() == "p":
self.shell_types.append("p")
self.funcs_per_shell.append(3)
self.n_aos += 3
norms = p_norm(exponents)
def p_shell(
r2,
x,
y,
z,
mo_coeffs,
alpha=exponents,
con_coeff=con_coeff,
norms=norms,
):
e_r2 = np.exp(np.outer(-alpha, r2))
s_val = np.dot(con_coeff * norms, e_r2)
if isinstance(r2, float):
res = 0
else:
res = np.zeros(len(r2))
if mo_coeffs[0] != 0:
res += mo_coeffs[0] * z
if mo_coeffs[1] != 0:
res += mo_coeffs[1] * x
if mo_coeffs[2] != 0:
res += mo_coeffs[2] * y
return res * s_val
self.basis_functions.append(p_shell)
elif shell_type.lower() == "d":
self.shell_types.append("5d")
self.funcs_per_shell.append(5)
self.n_aos += 5
norms = d_norm(exponents)
def d_shell(
r2,
x,
y,
z,
mo_coeffs,
alpha=exponents,
con_coeff=con_coeff,
norms=norms,
):
e_r2 = np.exp(np.outer(-alpha, r2))
s_val = np.dot(con_coeff * norms, e_r2)
if isinstance(r2, float):
res = 0
else:
res = np.zeros(len(r2))
if mo_coeffs[0] != 0:
z2r2 = 0.5 * (3 * z * z - r2)
res += mo_coeffs[0] * z2r2
if mo_coeffs[1] != 0:
xz = np.sqrt(3) * x * z
res += mo_coeffs[1] * xz
if mo_coeffs[2] != 0:
yz = np.sqrt(3) * y * z
res += mo_coeffs[2] * yz
if mo_coeffs[3] != 0:
x2y2 = np.sqrt(3) * (x ** 2 - y ** 2) / 2
res += mo_coeffs[3] * x2y2
if mo_coeffs[4] != 0:
xy = np.sqrt(3) * x * y
res += mo_coeffs[4] * xy
return res * s_val
self.basis_functions.append(d_shell)
elif shell_type.lower() == "f":
self.shell_types.append("7f")
self.funcs_per_shell.append(7)
self.n_aos += 7
norms = f_norm(exponents)
def f_shell(
r2,
x,
y,
z,
mo_coeffs,
alpha=exponents,
con_coeff=con_coeff,
norms=norms,
):
e_r2 = np.exp(np.outer(-alpha, r2))
s_val = np.dot(con_coeff * norms, e_r2)
if isinstance(r2, float):
res = 0
else:
res = np.zeros(len(r2))
if mo_coeffs[0] != 0:
z3zr2 = z * (5 * z ** 2 - 3 * r2) / 2
res += mo_coeffs[0] * z3zr2
if mo_coeffs[1] != 0:
xz2xr2 = (
np.sqrt(3)
* x
* (5 * z ** 2 - r2)
/ (2 * np.sqrt(2))
)
res += mo_coeffs[1] * xz2xr2
if mo_coeffs[2] != 0:
yz2yr2 = (
np.sqrt(3)
* y
* (5 * z ** 2 - r2)
/ (2 * np.sqrt(2))
)
res += mo_coeffs[2] * yz2yr2
if mo_coeffs[3] != 0:
x2zr2z = np.sqrt(15) * z * (x ** 2 - y ** 2) / 2
res += mo_coeffs[3] * x2zr2z
if mo_coeffs[4] != 0:
xyz = np.sqrt(15) * x * y * z
res += mo_coeffs[4] * xyz
if mo_coeffs[5] != 0:
x3r2x = (
np.sqrt(5)
* x
* (3 * y ** 2 - x ** 2)
/ (2 * np.sqrt(2))
)
res += mo_coeffs[5] * x3r2x
if mo_coeffs[6] != 0:
x2yy3 = (
np.sqrt(5)
* y
* (y ** 2 - 3 * x ** 2)
/ (2 * np.sqrt(2))
)
res += mo_coeffs[6] * x2yy3
return res * s_val
self.basis_functions.append(f_shell)
elif shell_type.lower() == "g":
self.shell_types.append("9g")
self.funcs_per_shell.append(9)
self.n_aos += 9
norms = g_norm(exponents)
def g_shell(
r2, x, y, z, mo_coeffs,
alpha=exponents, con_coeffs=con_coeff, norms=norms
):
e_r2 = np.exp(np.outer(-alpha, r2))
s_val = np.dot(con_coeff * norms, e_r2)
res = np.zeros(len(r2))
if mo_coeffs[0] != 0:
z4 = (35 * (z ** 4) - 30 * (r2 * z ** 2) + 3 * r2 ** 2) / 8
res += mo_coeffs[0] * z4
if mo_coeffs[1] != 0:
z3x = np.sqrt(10) * (x * z * (7 * z ** 2 - 3 * r2)) / 4
res += mo_coeffs[1] * z3x
if mo_coeffs[2] != 0:
z3y = np.sqrt(10) * (y * z * (7 * z ** 2 - 3 * r2)) / 4
res += mo_coeffs[2] * z3y
if mo_coeffs[3] != 0:
z2x2y2 = np.sqrt(5) * (x ** 2 - y ** 2) * (7 * z ** 2 - r2) / 4
res += mo_coeffs[3] * z2x2y2
if mo_coeffs[4] != 0:
z2xy = np.sqrt(5) * x * y * (7 * z ** 2 - r2) / 2
res += mo_coeffs[4] * z2xy
if mo_coeffs[5] != 0:
zx3 = -np.sqrt(70) * x * z * (x ** 2 - 3 * y ** 2) / 4
res += mo_coeffs[5] * zx3
if mo_coeffs[6] != 0:
zy3 = -np.sqrt(70) * z * y * (3 * x ** 2 - y ** 2) / 4
res += mo_coeffs[6] * zy3
if mo_coeffs[7] != 0:
x2 = x ** 2
y2 = y ** 2
x4y4 = -np.sqrt(35) * (x2 * (x2 - 3 * y2) - y2 * (3 * x2 - y2)) / 8
res += mo_coeffs[7] * x4y4
if mo_coeffs[8] != 0:
xyx2y2 = -np.sqrt(35) * x * y * (x ** 2 - y ** 2) / 2
res += mo_coeffs[8] * xyx2y2
return res * s_val
self.basis_functions.append(g_shell)
elif shell_type.lower() == "h":
self.shell_types.append("11h")
self.funcs_per_shell.append(11)
self.n_aos += 11
norms = h_norm(exponents)
def h_shell(
r2, x, y, z, mo_coeffs,
alpha=exponents, con_coeffs=con_coeff, norms=norms
):
e_r2 = np.exp(np.outer(-alpha, r2))
s_val = np.dot(con_coeff * norms, e_r2)
res = np.zeros(len(r2))
z2 = z ** 2
if mo_coeffs[0] != 0:
z5z3r2zr4 = z * (63 * z2 ** 2 - 70 * z2 * r2 + 15 * r2 ** 2) / 8
res += mo_coeffs[0] * z5z3r2zr4
if mo_coeffs[1] != 0:
xz4xz2r2xr4 = np.sqrt(15) * x * (21 * z2 ** 2 - 14 * z2 * r2 + r2 ** 2) / 8
res += mo_coeffs[1] * xz4xz2r2xr4
if mo_coeffs[2] != 0:
yz4yz2r2yr4 = np.sqrt(15) * y * (21 * z2 ** 2 - 14 * z2 * r2 + r2 ** 2) / 8
res += mo_coeffs[2] * yz4yz2r2yr4
if mo_coeffs[3] != 0:
x2y3z3zr2 = np.sqrt(105) * (x ** 2 - y ** 2) * (3 * z2 - r2) * z / 4
res += mo_coeffs[3] * x2y3z3zr2
if mo_coeffs[4] != 0:
xyz3zr2 = np.sqrt(105) * x * y * z * (3 * z2 - r2) / 2
res += mo_coeffs[4] * xyz3zr2
if mo_coeffs[5] != 0:
xx2y2z2r2 = -35 * x * (x ** 2 - 3 * y ** 2) * (9 * z2 - r2) / (8 * np.sqrt(70))
res += mo_coeffs[5] * xx2y2z2r2
if mo_coeffs[6] != 0:
yx2y2z2r2 = -35 * y * (3 * x ** 2 - y ** 2) * (9 * z2 - r2) / (8 * np.sqrt(70))
res += mo_coeffs[6] * yx2y2z2r2
if mo_coeffs[7] != 0:
zx4x2y2y4 = -105 * z * ((x ** 2) ** 2 - 6 * (x * y) ** 2 + (y ** 2) ** 2) / (8 * np.sqrt(35))
res += mo_coeffs[7] * zx4x2y2y4
if mo_coeffs[8] != 0:
zx3yxy3 = -105 * x * y * z * (4 * x ** 2 - 4 * y ** 2) / (8 * np.sqrt(35))
res += mo_coeffs[8] * zx3yxy3
if mo_coeffs[9] != 0:
xx4y2x2y4 = 21 * x * ((x ** 2) ** 2 - 10 * (x * y) ** 2 + 5 * (y ** 2) ** 2) / (8 * np.sqrt(14))
res += mo_coeffs[9] * xx4y2x2y4
if mo_coeffs[10] != 0:
yx4y2x2y4 = 21 * y * (5 * (x ** 2) ** 2 - 10 * (x * y) ** 2 + (y ** 2) ** 2) / (8 * np.sqrt(14))
res += mo_coeffs[10] * yx4y2x2y4
return res * s_val
self.basis_functions.append(h_shell)
elif shell_type.lower() == "i":
self.shell_types.append("13i")
self.funcs_per_shell.append(13)
self.n_aos += 13
norms = i_norm(exponents)
def i_shell(
r2, x, y, z, mo_coeffs,
alpha=exponents, con_coeffs=con_coeff, norms=norms
):
e_r2 = np.exp(np.outer(-alpha, r2))
s_val = np.dot(con_coeff * norms, e_r2)
res = np.zeros(len(r2))
z2 = z ** 2
if mo_coeffs[0] != 0:
z6z4r2z2r4r6 = (231 * z2 * z2 ** 2 - 315 * z2 ** 2 * r2 + 105 * z2 * r2 ** 2 - 5 * r2 * r2 ** 2) / 16
res += mo_coeffs[0] * z6z4r2z2r4r6
if mo_coeffs[1] != 0:
xz5z3r2zr4 = np.sqrt(21) * x * z * (33 * z2 ** 2 - 30 * z2 * r2 + 5 * r2 ** 2) / 8
res += mo_coeffs[1] * xz5z3r2zr4
if mo_coeffs[2] != 0:
yz5z3r2zr4 = np.sqrt(21) * y * z * (33 * z2 ** 2 - 30 * z2 * r2 + 5 * r2 ** 2) / 8
res += mo_coeffs[2] * yz5z3r2zr4
if mo_coeffs[3] != 0:
x2y2z4z2r2r3 = 105 * (x ** 2 - y ** 2) * (33 * z2 ** 2 - 18 * z2 * r2 + r2 ** 2) / (16 * np.sqrt(210))
res += mo_coeffs[3] * x2y2z4z2r2r3
if mo_coeffs[4] != 0:
xyz4z2r2r4 = 105 * x * y * (33 * z2 ** 2 - 18 * z2 * r2 + r2 ** 2) / (8 * np.sqrt(210))
res += mo_coeffs[4] * xyz4z2r2r4
if mo_coeffs[5] != 0:
xx2y2z3zr2 = -105 * x * z * (x ** 2 - 3 * y ** 2) * (11 * z2 - 3 * r2) / (8 * np.sqrt(210))
res += mo_coeffs[5] * xx2y2z3zr2
if mo_coeffs[6] != 0:
yx2y2z3zr2 = -105 * y * z * (3 * x ** 2 - y ** 2) * (11 * z2 - 3 * r2) / (8 * np.sqrt(210))
res += mo_coeffs[6] * yx2y2z3zr2
if mo_coeffs[7] != 0:
x4x2y2y4z2r2 = -np.sqrt(63) * ((x ** 2) ** 2 - 6 * (x * y) ** 2 + (y ** 2) ** 2) * (11 * z2 - r2) / 16
res += mo_coeffs[7] * x4x2y2y4z2r2
if mo_coeffs[8] != 0:
xyx2y2z2r2 = -np.sqrt(63) * x * y * (x ** 2 - y ** 2) * (11 * z2 - r2) / 4
res += mo_coeffs[8] * xyx2y2z2r2
if mo_coeffs[9] != 0:
xzx4x2y2y4 = 231 * x * z * ((x ** 2) ** 2 - 10 * (x * y) ** 2 + 5 * (y ** 2) ** 2) / (8 * np.sqrt(154))
res += mo_coeffs[9] * xzx4x2y2y4
if mo_coeffs[10] != 0:
yzx4x2y2y4 = 231 * y * z * (5 * (x ** 2) ** 2 - 10 * (x * y) ** 2 + (y ** 2) ** 2) / (8 * np.sqrt(154))
res += mo_coeffs[10] * yzx4x2y2y4
if mo_coeffs[11] != 0:
x6x4y2x2y4y6 = 231 * ((x * x ** 2) ** 2 - 15 * (x ** 2 * y) ** 2 + 15 * (x * y ** 2) ** 2 - (y * y ** 2) ** 2) / (16 * np.sqrt(462))
res += mo_coeffs[11] * x6x4y2x2y4y6
if mo_coeffs[12] != 0:
yx5x3y3xy5 = 231 * x * y * (6 * (x ** 2) ** 2 - 20 * (x * y) ** 2 + 6 * (y ** 2) ** 2) / (16 * np.sqrt(462))
res += mo_coeffs[12] * yx5x3y3xy5
return res * s_val
self.basis_functions.append(i_shell)
else:
self.LOG.warning(
"cannot handle shell of type %s" % shell_type
)
self.n_mos = len(self.alpha_coefficients)
if "n_alpha" not in filereader.other:
tot_electrons = sum(
ELEMENTS.index(atom.element) for atom in filereader.atoms
)
self.n_beta = tot_electrons // 2
self.n_alpha = tot_electrons - self.n_beta
else:
self.n_alpha = filereader.other["n_alpha"]
self.n_beta = filereader.other["n_beta"]
def _get_value(self, coords, arr):
"""returns value for the MO coefficients in arr"""
ao = 0
prev_center = None
if coords.ndim == 1:
val = 0
else:
val = np.zeros(len(coords))
for coord, shell, n_func, shell_type in zip(
self.shell_coords,
self.basis_functions,
self.funcs_per_shell,
self.shell_types,
):
# don't calculate distances until we find an AO
# in this shell that has a non-zero MO coefficient
if not np.count_nonzero(arr[ao : ao + n_func]):
ao += n_func
continue
# print(shell_type, arr[ao : ao + n_func])
# don't recalculate distances unless this shell's coordinates
# differ from the previous
if (
prev_center is None
or np.linalg.norm(coord - prev_center) > 1e-13
):
prev_center = coord
d_coord = (coords - coord) / UNIT.A0_TO_BOHR
if coords.ndim == 1:
r2 = np.dot(d_coord, d_coord)
else:
r2 = np.sum(d_coord * d_coord, axis=1)
if coords.ndim == 1:
res = shell(
r2,
d_coord[0],
d_coord[1],
d_coord[2],
arr[ao : ao + n_func],
)
else:
res = shell(
r2,
d_coord[:, 0],
d_coord[:, 1],
d_coord[:, 2],
arr[ao : ao + n_func],
)
val += res
ao += n_func
return val
def mo_value(self, mo, coords, alpha=True, n_jobs=1):
"""
get the MO evaluated at the specified coords
m - index of molecular orbital or an array of MO coefficients
coords - numpy array of points (N,3) or (3,)
alpha - use alpha coefficients (default)
n_jobs - number of parallel threads to use
this is on top of NumPy's multithreading, so
if NumPy uses 8 threads and n_jobs=2, you can
expect to see 16 threads in use
"""
# val is the running sum of MO values
if alpha:
coeff = self.alpha_coefficients
else:
coeff = self.beta_coefficients
if isinstance(mo, int):
coeff = coeff[mo]
else:
coeff = mo
# calculate AO values for each shell at each point
# multiply by the MO coefficient and add to val
if n_jobs > 1:
# get all shells grouped by coordinates
# this reduces the number of times we will need to
# calculate the distance from all the coords to
# a shell's center
prev_coords = []
arrays = []
ndx = 0
add_to = 0
for i, coord in enumerate(self.shell_coords):
for j, prev_coord in enumerate(prev_coords):
if np.linalg.norm(coord - prev_coord) < 1e-13:
add_to = j
break
else:
prev_coords.append(coord)
add_to = len(arrays)
arrays.append(np.zeros(self.n_mos))
arrays[add_to][ndx : ndx + self.funcs_per_shell[i]] = coeff[
ndx : ndx + self.funcs_per_shell[i]
]
ndx += self.funcs_per_shell[i]
with concurrent.futures.ThreadPoolExecutor(
max_workers=n_jobs
) as executor:
out = [executor.submit(self._get_value, coords, arr) for arr in arrays]
return sum([shells.result() for shells in out])
val = self._get_value(coords, coeff)
return val
def density_value(
self,
coords,
n_jobs=1,
alpha_occ=None,
beta_occ=None,
low_mem=False,
):
"""
returns the eletron density
coords - coordinates to calculate e density at
n_jobs - number of concurrent threads to use in calculation
alpha_occ - array of alpha occupancies
if not specified, defaults to lowest self.n_alpha
orbitals
beta_occ - same at alpha_occ, but for beta electrons
"""
if low_mem:
return self._low_mem_density_value(
coords,
n_jobs=n_jobs,
alpha_occ=alpha_occ,
beta_occ=beta_occ,
)
# set default occupancy
if alpha_occ is None:
if not self.n_alpha:
self.LOG.warning("number of alpha electrons was not read")
alpha_occ = np.zeros(self.n_mos, dtype=int)
alpha_occ[0:self.n_alpha] = 1
if beta_occ is None:
beta_occ = np.zeros(self.n_mos, dtype=int)
beta_occ[0:self.n_beta] = 1
# val is output data
# func_vals is the value of each basis function
# at all coordinates
if coords.ndim == 1:
val = 0
func_vals = np.zeros(
len(self.basis_functions), dtype="float32"
)
else:
val = np.zeros(len(coords))
func_vals = np.zeros(
(self.n_mos, *coords.shape,)
)
# get values of basis functions at all points
arrays = np.eye(self.n_mos)
if n_jobs > 1:
# get all shells grouped by coordinates
# this reduces the number of times we will need to
# calculate the distance from all the coords to
# a shell's center
with concurrent.futures.ThreadPoolExecutor(
max_workers=n_jobs
) as executor:
out = [
executor.submit(self._get_value, coords, arr) for arr in arrays
]
data = np.array([shells.result() for shells in out])
else:
data = np.array([
self._get_value(coords, arr) for arr in arrays
])
# multiply values by orbital coefficients and square
for i, occ in enumerate(alpha_occ):
if occ == 0:
continue
val += occ * np.dot(data.T, self.alpha_coefficients[i]) ** 2
if self.beta_coefficients is not None:
for i, occ in enumerate(beta_occ):
if occ == 0:
continue
val += occ * np.dot(data.T, self.beta_coefficients[i]) ** 2
else:
val *= 2
return val
def _low_mem_density_value(
self,
coords,
n_jobs=1,
alpha_occ=None,
beta_occ=None
):
"""
returns the eletron density
same at self.density_value, but uses less memory at
the cost of performance
"""
# set initial occupancies
if alpha_occ is None:
if not self.n_alpha:
self.LOG.warning("number of alpha electrons was not read")
alpha_occ = np.zeros(self.n_mos, dtype=int)
alpha_occ[0:self.n_alpha] = 1
if beta_occ is None:
beta_occ = np.zeros(self.n_mos, dtype=int)
beta_occ[0:self.n_beta] = 1
# val is output array
if coords.ndim == 1:
val = 0
else:
val = np.zeros(len(coords), dtype="float32")
# calculate each occupied orbital
# square it can add to output
if n_jobs > 1:
with concurrent.futures.ThreadPoolExecutor(
max_workers=n_jobs
) as executor:
out = [
executor.submit(self.mo_value, i, coords, n_jobs=1)
for i, occ in enumerate(alpha_occ) if occ != 0
]
val += sum([occ * orbit.result() ** 2 for orbit, occ in zip(out, alpha_occ)])
if self.beta_coefficients is not None:
with concurrent.futures.ThreadPoolExecutor(
max_workers=n_jobs
) as executor:
out = [
executor.submit(self.mo_value, i, coords, alpha=False, n_jobs=1)
for i, occ in enumerate(beta_occ) if occ != 0
]
val += sum([occ * orbit.result() ** 2 for orbit, occ in zip(out, beta_occ)])
else:
val *= 2
else:
for i in range(0, self.n_alpha):
val += self.mo_value(i, coords) ** 2
if self.beta_coefficients is not None:
for i in range(0, self.n_beta):
val += self.mo_value(i, coords, alpha=False) ** 2
else:
val *= 2
return val
def fukui_donor_value(self, coords, delta=0.1, **kwargs):
"""
orbital-weighted fukui donor function
electron density change for removing an electron
orbital weighting from DOI 10.1002/jcc.24699 accounts
for nearly degenerate orbitals
coords - coordinate to evaluate function at
delta - parameter for weighting
kwargs - passed to density_value
"""
CITATION = "doi:10.1002/jcc.24699"
self.LOG.citation(CITATION)
if self.beta_coefficients is None:
homo_nrg = self.alpha_nrgs[self.n_alpha - 1]
lumo_nrg = self.alpha_nrgs[self.n_alpha]
chem_pot = 0.5 * (lumo_nrg + homo_nrg)
minus_e = np.zeros(self.n_mos)
for i in range(0, self.n_alpha):
minus_e[i] = np.exp(
-((chem_pot - self.alpha_nrgs[i]) / delta) ** 2
)
minus_e /= sum(minus_e)
minus_density = self.density_value(
coords, alpha_occ=minus_e, beta_occ=None, **kwargs
)
else:
homo_nrg = self.alpha_nrgs[self.n_alpha - 1]
if self.n_beta > self.n_alpha:
homo_nrg = self.beta_nrgs[self.n_beta - 1]
lumo_nrg = self.alpha_nrgs[self.n_alpha]
if self.n_beta > self.n_alpha:
lumo_nrg = self.beta_nrgs[self.n_beta]
chem_pot = 0.5 * (lumo_nrg + homo_nrg)
alpha_occ = beta_occ = np.zeros(self.n_mos)
for i in range(0, self.n_alpha):
alpha_occ[i] = np.exp(
-((chem_pot - self.alpha_nrgs[i]) / delta) ** 2
)
for i in range(0, self.n_beta):
beta_occ[i] = np.exp(
-((chem_pot - self.beta_nrgs[i]) / delta) ** 2
)
alpha_occ /= sum(alpha_occ)
beta_occ /= sum(beta_occ)
minus_density = self.density_value(
coords, alpha_occ=alpha_occ, beta_occ=beta_occ, **kwargs
)
return minus_density
def fukui_acceptor_value(self, coords, delta=0.1, **kwargs):
"""
orbital-weighted fukui acceptor function
electron density change for removing an electron
orbital weighting from DOI 10.1021/acs.jpca.9b07516 accounts
for nearly degenerate orbitals
coords - coordinate to evaluate function at
delta - parameter for weighting
kwargs - passed to density_value
"""
CITATION = "doi:10.1021/acs.jpca.9b07516"
self.LOG.citation(CITATION)
alpha_occ = np.zeros(self.n_mos)
alpha_occ[self.n_alpha - 1] = 1
beta_occ = None
if self.beta_coefficients is not None:
beta_occ = np.zeros(self.n_mos)
beta_occ[self.n_beta - 1] = 1
if self.beta_coefficients is None:
homo_nrg = self.alpha_nrgs[self.n_alpha - 1]
lumo_nrg = self.alpha_nrgs[self.n_alpha]
chem_pot = 0.5 * (lumo_nrg + homo_nrg)
plus_e = np.zeros(self.n_mos)
for i in range(self.n_alpha, self.n_mos):
plus_e[i] = np.exp(
-((chem_pot - self.alpha_nrgs[i]) / delta) ** 2
)
plus_e /= sum(plus_e)
plus_density = self.density_value(
coords, alpha_occ=plus_e, beta_occ=beta_occ, **kwargs
)
else:
homo_nrg = self.alpha_nrgs[self.n_alpha - 1]
if self.n_beta > self.n_alpha:
homo_nrg = self.beta_nrgs[self.n_beta - 1]
lumo_nrg = self.alpha_nrgs[self.n_alpha]
if self.n_beta > self.n_alpha:
lumo_nrg = self.beta_nrgs[self.n_beta]
chem_pot = 0.5 * (lumo_nrg + homo_nrg)
alpha_occ = np.zeros(self.n_mos)
beta_occ = np.zeros(self.n_mos)
for i in range(self.n_alpha, self.n_mos):
alpha_occ[i] = np.exp(
-((chem_pot - self.alpha_nrgs[i]) / delta) ** 2
)
for i in range(self.n_beta, self.n_mos):
beta_occ[i] = np.exp(
-((chem_pot - self.beta_nrgs[i]) / delta) ** 2
)
alpha_occ /= sum(alpha_occ)
beta_occ /= sum(beta_occ)
plus_density = self.density_value(
coords, alpha_occ=alpha_occ, beta_occ=beta_occ, **kwargs
)
return plus_density
def fukui_dual_value(self, coords, delta=0.1, **kwargs):
CITATION = "doi:10.1021/acs.jpca.9b07516"
self.LOG.citation(CITATION)
alpha_occ = np.zeros(self.n_mos)
alpha_occ[self.n_alpha - 1] = 1
beta_occ = None
if self.beta_coefficients is not None:
beta_occ = np.zeros(self.n_mos)
beta_occ[self.n_beta - 1] = 1
if self.beta_coefficients is None:
homo_nrg = self.alpha_nrgs[self.n_alpha - 1]
lumo_nrg = self.alpha_nrgs[self.n_alpha]
chem_pot = 0.5 * (lumo_nrg + homo_nrg)
plus_e = np.zeros(self.n_mos)
minus_e = np.zeros(self.n_mos)
for i in range(0, self.n_alpha):
minus_e[i] = np.exp(
-((chem_pot - self.alpha_nrgs[i]) / delta) ** 2
)
for i in range(self.n_alpha, self.n_mos):
plus_e[i] = np.exp(
-((chem_pot - self.alpha_nrgs[i]) / delta) ** 2
)
minus_e /= sum(minus_e)
plus_e /= sum(plus_e)
dual_density = self.density_value(
coords, alpha_occ=plus_e - minus_e, beta_occ=beta_occ, **kwargs
)
else:
homo_nrg = self.alpha_nrgs[self.n_alpha - 1]
if self.n_beta > self.n_alpha:
homo_nrg = self.beta_nrgs[self.n_beta - 1]
lumo_nrg = self.alpha_nrgs[self.n_alpha]
if self.n_beta > self.n_alpha:
lumo_nrg = self.beta_nrgs[self.n_beta]
chem_pot = 0.5 * (lumo_nrg + homo_nrg)
alpha_occ = np.zeros(self.n_mos)
beta_occ = np.zeros(self.n_mos)
for i in range(0, self.n_alpha):
alpha_occ[i] = -np.exp(
-((chem_pot - self.alpha_nrgs[i]) / delta) ** 2
)
for i in range(0, self.n_beta):
beta_occ[i] = -np.exp(
-((chem_pot - self.beta_nrgs[i]) / delta) ** 2
)
for i in range(self.n_alpha, self.n_mos):
alpha_occ[i] = np.exp(
-((chem_pot - self.alpha_nrgs[i]) / delta) ** 2
)
for i in range(self.n_beta, self.n_mos):
beta_occ[i] = np.exp(
-((chem_pot - self.beta_nrgs[i]) / delta) ** 2
)
alpha_occ[self.n_alpha:] /= abs(sum(alpha_occ[self.n_alpha:]))
beta_occ[self.n_beta:] /= abs(sum(beta_occ[self.n_beta:]))
alpha_occ[:self.n_alpha] /= sum(alpha_occ[:self.n_alpha])
beta_occ[:self.n_beta] /= sum(beta_occ[:self.n_beta])
dual_density = self.density_value(
coords, alpha_occ=alpha_occ, beta_occ=beta_occ, **kwargs
)
return dual_density
@staticmethod
def get_cube_array(
geom,
padding=4,
spacing=0.2,
standard_axes=False,
):
"""returns n_pts1, n_pts2, n_pts3, v1, v2, v3, com, u
n_pts1 is the number of points along the first axis
n_pts2 ... second axis
n_pts3 ... third axis
v1 is the vector for the first axis, norm should be close to spacing
v2 ... second axis
v3 ... third axis
com is the center of the cube
u is a rotation matrix for the v1, v2, v3 axes relative to xyz
geom - Geometry() used to define the cube
padding - extra space around atoms in angstrom
spacing - distance between adjacent points in angstrom
standard_axes - True to use x, y, and z axes
by default, the cube will be oriented to fit
the geom and have the smallest volume possible
"""
def get_standard_axis():
"""returns info to set up a grid along the x, y, and z axes"""
geom_coords = geom.coords
# get range of geom's coordinates
x_min = np.min(geom_coords[:, 0])
x_max = np.max(geom_coords[:, 0])
y_min = np.min(geom_coords[:, 1])
y_max = np.max(geom_coords[:, 1])
z_min = np.min(geom_coords[:, 2])
z_max = np.max(geom_coords[:, 2])
# add padding, figure out vectors
r1 = 2 * padding + x_max - x_min
n_pts1 = int(r1 // spacing) + 1
d1 = r1 / (n_pts1 - 1)
v1 = np.array((d1, 0., 0.))
r2 = 2 * padding + y_max - y_min
n_pts2 = int(r2 // spacing) + 1
d2 = r2 / (n_pts2 - 1)
v2 = np.array((0., d2, 0.))
r3 = 2 * padding + z_max - z_min
n_pts3 = int(r3 // spacing) + 1
d3 = r3 / (n_pts3 - 1)
v3 = np.array((0., 0., d3))
com = np.array([x_min, y_min, z_min]) - padding
u = np.eye(3)
return n_pts1, n_pts2, n_pts3, v1, v2, v3, com, u
if standard_axes:
n_pts1, n_pts2, n_pts3, v1, v2, v3, com, u = get_standard_axis()
else:
test_coords = geom.coords - geom.COM()
covar = np.dot(test_coords.T, test_coords)
try:
# use SVD on the coordinate covariance matrix
# this decreases the volume of the box we're making
# that means less work for higher resolution
# for many structures, this only decreases the volume
# by like 5%
u, s, vh = np.linalg.svd(covar)
v1 = u[:, 0]
v2 = u[:, 1]
v3 = u[:, 2]
# change basis of coordinates to the singular vectors
# this is how we determine the range + padding
new_coords = np.dot(test_coords, u)
xr_max = np.max(new_coords[:, 0])
xr_min = np.min(new_coords[:, 0])
yr_max = np.max(new_coords[:, 1])
yr_min = np.min(new_coords[:, 1])
zr_max = np.max(new_coords[:, 2])
zr_min = np.min(new_coords[:, 2])
com = np.array([xr_min, yr_min, zr_min]) - padding
# move the COM back to the xyz space of the original molecule
com = np.dot(u, com)
com += geom.COM()
r1 = 2 * padding + np.linalg.norm(xr_max - xr_min)
r2 = 2 * padding + np.linalg.norm(yr_max - yr_min)
r3 = 2 * padding + np.linalg.norm(zr_max - zr_min)
n_pts1 = int(r1 // spacing) + 1
n_pts2 = int(r2 // spacing) + 1
n_pts3 = int(r3 // spacing) + 1
v1 = v1 * r1 / (n_pts1 - 1)
v2 = v2 * r2 / (n_pts2 - 1)
v3 = v3 * r3 / (n_pts3 - 1)
except np.linalg.LinAlgError:
n_pts1, n_pts2, n_pts3, v1, v2, v3, com, u = get_standard_axis()
return n_pts1, n_pts2, n_pts3, v1, v2, v3, com, u
@staticmethod
def get_cube_points(
n_pts1, n_pts2, n_pts3, v1, v2, v3, com, sort=True
):
"""
returns coords, n_list
coords is an array of points in the cube
n_list specifies where each point is along the axes
e.g. 5th point along v1, 4th point along v2, 0th point along v3
"""
v_list = [v1, v2, v3]
n_list = [n_pts1, n_pts2, n_pts3]
if sort:
v_list = []
n_list = []
for n, v in sorted(
zip([n_pts1, n_pts2, n_pts3], [v1, v2, v3]),
key=lambda p: np.linalg.norm(p[1]),
):
v_list.append(v)
n_list.append(n)
ndx = (
np.vstack(
np.mgrid[
0 : n_list[0],
0 : n_list[1],
0 : n_list[2],
]
)
.reshape(3, np.prod(n_list))
.T
)
coords = np.matmul(ndx, v_list)
del ndx
coords += com
return coords, n_list
def memory_estimate(
self,
func_name,
n_points=None,
low_mem=False,
n_jobs=1,
apoints=None,
rpoints=None,
n_atoms=None,
):
"""
returns the estimated memory use (in GB) for calling the
specified function on the specified number of points
if func_name is a condensed fukui function, apoints,
and rpoints must be given
otherwise, n_points must be given
"""
test_array = np.ones(1)
if test_array.dtype == np.float64:
# bytes - 8 bits per byte
num_size = 8
else:
# hopefully float32
num_size = 4
size = n_points
if any(func_name == x for x in [
"density_value",
"fukui_acceptor_value",
"fukui_donor_value",
"fukui_dual_value",
]
):
size *= num_size * 4 * max(n_jobs, n_atoms)
if not low_mem:
size *= self.n_mos / (2 * max(n_jobs, n_atoms))
elif func_name == "mo_value":
size *= num_size * (4 * n_jobs + max(n_atoms - n_jobs, 0))
elif any(func_name == x for x in [
"condensed_fukui_acceptor_values",
"condensed_fukui_donor_values",
"condensed_fukui_dual_values",
]
):
density_size = self.memory_estimate(
"density_value",
n_points=apoints * rpoints,
n_jobs=n_jobs,
n_atoms=n_atoms,
low_mem=low_mem,
)
mat_size = num_size * n_atoms * rpoints * apoints
size = max(density_size, mat_size)
return size * 1e-9
def voronoi_integral(
self,
target,
geom,
*args,
rpoints=32,
apoints=1454,
func=None,
rmax=None,
**kwargs,
):
"""
integrates func in the Voronoi cell of the specified target
geom - Geometry() target belongs to
args - passed to func
rpoints - radial points used for Gauss-Legendre integral
apoints - angular points for Lebedev integral
func - function to evaluate
kwargs - passed to func
"""
atom = geom.find(target)[0]
if rmax is None:
rmax = 10 * atom._vdw
rgrid, rweights = gauss_legendre_grid(
start=0, stop=rmax, num=rpoints
)
# grab Lebedev grid for unit sphere at origin
agrid, aweights = lebedev_sphere(
radius=1, center=np.zeros(3), num=apoints
)
# TODO: switch to np.zeros((n_ang * n_rad, 3))
# this eliminates appending
# build a list of points and weights around the atom
all_points = np.empty((0, 3))
weights = np.empty(0)
for rvalue, rweight in zip(rgrid, rweights):
agrid_r = agrid * rvalue
agrid_r += atom.coords
all_points = np.append(all_points, agrid_r, axis=0)
weights = np.append(weights, rweight * aweights)
# find points that are closest to this atom
# than any other
dist_mat = distance_matrix(geom.coords, all_points)
atom_ndx = geom.atoms.index(atom)
mask = np.argmin(dist_mat, axis=0) == atom_ndx
voronoi_points = all_points[mask]
voronoi_weights = weights[mask]
# evaluate function
vals = func(voronoi_points, *args, **kwargs)
# multiply values by weights, add them up, and return the sum
return np.dot(vals, voronoi_weights)
def power_integral(
self,
target,
geom,
*args,
radii="umn",
rpoints=32,
apoints=1454,
func=None,
rmax=None,
**kwargs,
):
"""
integrates func in the power cell of the specified target
power diagrams are a form of weighted Voronoi diagrams
that form cells based on the smallest d^2 - r^2
see wikipedia article: https://en.wikipedia.org/wiki/Power_diagram
radii - "bondi" - Bondi vdW radii
"umn" - vdW radii from Mantina, Chamberlin, Valero, Cramer, and Truhlar
dict() - radii are values and elements are keys
list() - list of radii corresponding to targets
geom - Geometry() target belongs to
args - passed to func
rpoints - radial points used for Gauss-Legendre integral
apoints - angular points for Lebedev integral
func - function to evaluate
kwargs - passed to func
"""
if func is None:
func = self.density_value
target = geom.find(target)[0]
target_ndx = geom.atoms.index(target)
radius_list = []
radii_dict = None
if isinstance(radii, dict):
radii_dict = radii
elif isinstance(radii, list):
radius_list = radii
elif radii.lower() == "bondi":
radii_dict = BONDI_RADII
elif radii.lower() == "umn":
radii_dict = VDW_RADII
else:
raise TypeError(
"radii must be list, dict, \"UMN\", or \"BONDI\": %s" % radii
)
if not radius_list:
for atom in geom.atoms:
radius_list.append(radii_dict[atom.element])
radius_list = np.array(radius_list)
if rmax is None:
rmax = 5 * radius_list[target_ndx]
radius_list = radius_list ** 2
rgrid, rweights = gauss_legendre_grid(
start=0, stop=rmax, num=rpoints
)
# grab Lebedev grid for unit sphere at origin
agrid, aweights = lebedev_sphere(
radius=1, center=np.zeros(3), num=apoints
)
# TODO: switch to np.zeros((n_ang * n_rad, 3))
# this eliminates appending
# build a list of points and weights around the atom
power_points = np.empty((0, 3))
power_weights = np.empty(0)
atom_ndx = geom.atoms.index(target)
found_pts = False
for rvalue, rweight in zip(rgrid, rweights):
agrid_r = agrid * rvalue
agrid_r += target.coords
dist_mat = distance_matrix(geom.coords, agrid_r) ** 2
dist_mat = np.transpose(dist_mat.T - radius_list)
mask = np.argmin(dist_mat, axis=0) == atom_ndx
# find points that are closest to this atom's vdw sphere
# than any other
if any(mask):
power_points = np.append(power_points, agrid_r[mask], axis=0)
power_weights = np.append(power_weights, rweight * aweights[mask])
found_pts = True
elif found_pts:
break
# with open("test_%s.bild" % target.name, "w") as f:
# s = ""
# for p in power_points:
# s += ".sphere %.4f %.4f %.4f 0.05\n" % tuple(p)
# f.write(s)
# evaluate function
vals = func(power_points, *args, **kwargs)
# multiply values by weights, add them up, and return the sum
return np.dot(vals, power_weights)
def condensed_fukui_donor_values(
self,
geom,
*args,
**kwargs,
):
"""
uses power_integral to integrate the fukui_donor_value
for all atoms in geom
values are normalized so they sum to 1
geom - Geometry()
args and kwargs are passed to power_integral
returns array for each atom's condensed Fukui donor values
"""
out = np.zeros(len(geom.atoms))
for i, atom in enumerate(geom.atoms):
out[i] = self.power_integral(
atom, geom, *args, func=self.fukui_donor_value, **kwargs,
)
out /= sum(out)
return out
def condensed_fukui_acceptor_values(
self,
geom,
*args,
**kwargs,
):
"""
uses power_integral to integrate the fukui_acceptor_value
for all atoms in geom
values are normalized so they sum to 1
geom - Geometry()
args and kwargs are passed to power_integral
returns array for each atom's condensed Fukui acceptor values
"""
out = np.zeros(len(geom.atoms))
for i, atom in enumerate(geom.atoms):
out[i] = self.power_integral(
atom, geom, *args, func=self.fukui_acceptor_value, **kwargs,
)
out /= sum(out)
return out
def condensed_fukui_dual_values(
self,
geom,
*args,
**kwargs,
):
"""
returns the difference between condensed_fukui_acceptor_values
and condensed_fukui_donor_values
"""
out = self.condensed_fukui_acceptor_values(
geom, *args, **kwargs,
) - self.condensed_fukui_donor_values(
geom, *args, **kwargs,
)
return out | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/orbitals.py | orbitals.py |
import numpy as np
from scipy.spatial import distance_matrix
from AaronTools import addlogger
from AaronTools.utils.prime_numbers import Primes
from AaronTools.utils.utils import (
rotation_matrix, mirror_matrix, proj, angle_between_vectors, perp_vector
)
class SymmetryElement:
def __init__(self, order, center):
self.order = order
self.operation = np.identity(3)
self.translation = center
def perp_dist(self, coords):
"""distance from each coordinate perpendicular to this symmetry element"""
return np.zeros(len(coords))
def apply_operation(self, coords):
"""returns coords with the symmetry operation applied"""
coords = coords - self.translation
coords = np.matmul(coords, self.operation)
coords += self.translation
return coords
def apply_operation_without_translation(self, coords):
"""
returns coords with the symmetry operation applied but without
translating the coordinates to or from self's center
"""
coords = np.matmul(coords, self.operation)
return coords
def error(self, geom=None, tolerance=None, groups=None, coords=None):
"""
error in this symmetry element for the given geometry
either geom or coords and groups must be given
if groups is not given and geom is, atoms will be grouped by element
"""
if coords is None:
coords = geom.coords
full_coords2 = self.apply_operation(coords)
error = 0
# compute distances between the initial coords and
# the coords after applying the symmetry operation
# but only calculate distances for atoms that might
# be symmetry-equivalent (i.e. in the same inital
# group, which typically is based on what the atom's
# neighbors are)
if groups is not None:
group_names = groups
else:
group_names = geom.elements
for group in set(group_names):
ndx = (group_names == group).nonzero()[0]
coords1 = np.take(coords, ndx, axis=0)
coords2 = np.take(full_coords2, ndx, axis=0)
dist_mat = distance_matrix(coords1, coords2)
perp_dist = self.perp_dist(coords1)
# treat values less than 1 as 1 to avoid numerical nonsense
perp_dist = np.maximum(perp_dist, np.ones(len(ndx)))
error_mat = dist_mat / perp_dist
min_d = max(np.min(error_mat, axis=1))
if min_d > error:
error = min_d
return error
def equivalent_positions(self, coords, groups):
"""
return an array with the indices that are equivalent after
applying this operation
for example:
ndx = element.equivalent_positions(geom.coords, groups)
coords[ndx] should be equal to element.apply_operation(geom.coords)
"""
eq_ndx = np.zeros(len(coords), dtype=int)
init_partitions = dict()
init_ndx = dict()
for i, (coord, group) in enumerate(zip(coords, groups)):
init_partitions.setdefault(group, [])
init_partitions[group].append(coord)
init_ndx.setdefault(group, [])
init_ndx[group].append(i)
for group in init_partitions:
coords = init_partitions[group]
new_coords = self.apply_operation(coords)
dist = distance_matrix(coords, new_coords)
closest_ndx = np.argmin(dist, axis=1)
for i, (atom, ndx) in enumerate(zip(init_partitions[group], closest_ndx)):
j = init_ndx[group][i]
k = init_ndx[group][ndx]
eq_ndx[j] = k
return eq_ndx
@property
def trace(self):
"""trace of this symmetry element's matrix"""
return np.trace(self.operation)
class Identity(SymmetryElement):
def __init__(self):
self.translation = np.zeros(3)
self.operation = np.eye(3)
def __repr__(self):
return "E"
def __lt__(self, other):
return False
class ProperRotation(SymmetryElement):
"""proper rotation"""
def __init__(self, center, axis, n, exp=1):
self.order = n
self.operation = rotation_matrix(
2 * np.pi * exp / n,
axis,
renormalize=False,
)
self.translation = center
self.axis = axis
self.n = n
self.exp = exp
def __repr__(self):
if self.exp > 1:
return "C%i^%i (%5.2f %5.2f %5.2f)" % (
self.n,
self.exp,
*self.axis,
)
return "C%i (%5.2f %5.2f %5.2f)" % (
self.n,
*self.axis,
)
def __lt__(self, other):
if isinstance(other, Identity) or isinstance(other, InversionCenter):
return False
if isinstance(other, ProperRotation):
if self.n == other.n:
return self.exp > other.exp
return self.n < other.n
return False
def perp_dist(self, coords):
v = coords - self.translation
n = np.dot(v, self.axis)
p = np.outer(n, self.axis)
return np.linalg.norm(v - p, axis=1)
class MirrorPlane(SymmetryElement):
"""mirror plane"""
def __init__(self, center, axis, label=None):
self.order = 2
self.translation = center
self.axis = axis
self.operation = mirror_matrix(axis)
self.label = label
def __repr__(self):
if self.label:
return "sigma_%s (%5.2f %5.2f %5.2f)" % (self.label, *self.axis)
return "sigma (%5.2f %5.2f %5.2f)" % tuple(self.axis)
def __lt__(self, other):
if not isinstance(other, MirrorPlane):
return True
if self.label and other.label:
return self.label < other.label
return True
def perp_dist(self, coords):
v = coords - self.translation
return np.dot(v, self.axis[:, None]).flatten()
class InversionCenter(SymmetryElement):
"""inversion center"""
def __init__(self, center):
self.order = 2
self.operation = -np.identity(3)
self.translation = center
def __lt__(self, other):
if isinstance(other, Identity):
return True
return False
def __repr__(self):
return "i (%.2f %.2f %.2f)" % (
*self.translation,
)
def perp_dist(self, coords):
v = coords - self.translation
return np.linalg.norm(v, axis=1)
class ImproperRotation(SymmetryElement):
"""improper rotation"""
def __init__(self, center, axis, n, exp=1):
self.order = n
self.operation = np.matmul(
rotation_matrix(
2 * np.pi * exp / n,
axis,
renormalize=False,
),
mirror_matrix(axis)
)
self.axis = axis
self.translation = center
self.n = n
self.exp = exp
def __repr__(self):
if self.exp > 1:
return "S%i^%i (%5.2f %5.2f %5.2f)" % (
self.n,
self.exp,
*self.axis,
)
return "S%i (%5.2f %5.2f %5.2f)" % (
self.n,
*self.axis,
)
def __lt__(self, other):
if (
isinstance(other, Identity) or
isinstance(other, ProperRotation) or
isinstance(other, InversionCenter)
):
return True
if isinstance(other, ImproperRotation):
if self.n == other.n:
return self.exp > other.exp
return self.n < other.n
return False
def perp_dist(self, coords):
v = coords - self.translation
n = np.dot(v, self.axis)
p = np.outer(n, self.axis)
ax_dist = np.linalg.norm(v - p, axis=1)
sig_dist = np.dot(v, self.axis[:, None]).flatten()
return np.minimum(ax_dist, sig_dist)
@addlogger
class PointGroup:
LOG = None
def __init__(
self,
geom,
tolerance=0.1,
max_rotation=6,
rotation_tolerance=0.01,
groups=None,
center=None
):
self.geom = geom
self.center = center
if self.center is None:
self.center = geom.COM()
self.elements = self.get_symmetry_elements(
geom,
tolerance=tolerance,
max_rotation=max_rotation,
groups=groups,
rotation_tolerance=rotation_tolerance,
)
self.name = self.determine_point_group(
rotation_tolerance=rotation_tolerance
)
def get_symmetry_elements(
self,
geom,
tolerance=0.1,
max_rotation=6,
rotation_tolerance=0.01,
groups=None,
):
"""
determine what symmetry elements are valid for geom
geom - Geometry()
tolerance - maximum error for an element to be valid
max_rotation - maximum n for Cn (Sn can be 2x this)
rotation_tolerance - tolerance in radians for angle between
axes to be for them to be considered parallel/antiparallel/orthogonal
returns list(SymmetryElement)
"""
CITATION = "doi:10.1002/jcc.22995"
self.LOG.citation(CITATION)
# atoms are grouped based on what they are bonded to
# if there's not many atoms, don't bother splitting them up
# based on ranks
if groups is not None:
atom_ids = np.array(groups)
self.initial_groups = atom_ids
else:
atom_ids = np.array(
geom.canonical_rank(
update=False,
break_ties=False,
invariant=False,
)
)
self.initial_groups = atom_ids
coords = geom.coords
moments, axes = geom.get_principle_axes()
axes = axes.T
valid = [Identity()]
degeneracy = np.ones(3, dtype=int)
for i, m1 in enumerate(moments):
for j, m2 in enumerate(moments):
if i == j:
continue
if np.isclose(m1, m2, rtol=tolerance, atol=tolerance):
degeneracy[i] += 1
com = self.center
inver = InversionCenter(com)
error = inver.error(geom, tolerance, groups=atom_ids)
if error <= tolerance:
valid.append(inver)
if any(np.isclose(m, 0) for m in moments):
return valid
ortho_to = []
for vec, degen in zip(axes, degeneracy):
if any(d > 1 for d in degeneracy) and degen == 1:
ortho_to.append(vec)
elif all(d == 1 for d in degeneracy):
ortho_to.append(vec)
# find vectors from COM to each atom
# these might be proper rotation axes
atom_axes = geom.coords - com
# find vectors normal to each pair of atoms
# these might be normal to a miror plane
atom_pair_norms = []
for i, v in enumerate(atom_axes):
dv = atom_axes - v
c2 = np.linalg.norm(dv, axis=1) ** 2
angles = np.arccos(-0.5 * (c2 - 2))
mask2 = angles > rotation_tolerance
mask3 = angles < np.pi - rotation_tolerance
mask = np.logical_and(mask2, mask3)
pair_n = np.cross(v, atom_axes[mask])
norms = np.linalg.norm(pair_n, axis=1)
pair_n = np.take(pair_n, np.nonzero(norms), axis=0)[0]
norms = np.take(norms, np.nonzero(norms), axis=0)
pair_n /= norms.T
atom_pair_norms.extend(pair_n.tolist())
atom_pair_norms = np.array(atom_pair_norms)
# find vectors to the midpoints between each
# pair of like atoms
# these might be proper rotations
atom_pairs = []
for atom_id in set(atom_ids):
ndx = (atom_ids == atom_id).nonzero()[0]
subset_axes = np.take(atom_axes, ndx, axis=0)
for i, v in enumerate(subset_axes):
mask = np.ones(len(subset_axes), dtype=bool)
mask[i] = False
pair_v = subset_axes[mask] + v
norms = np.linalg.norm(pair_v, axis=1)
pair_v = np.take(pair_v, np.nonzero(norms), axis=0)[0]
norms = np.take(norms, np.nonzero(norms), axis=0)
pair_v /= norms.T
atom_pairs.extend(pair_v.tolist())
atom_pairs = np.array(atom_pairs)
norms = np.linalg.norm(atom_axes, axis=1)
# don't want axis for an atom that is at the COM (0-vector)
atom_axes = np.take(atom_axes, np.nonzero(norms), axis=0)[0]
# normalize
norms = np.take(norms, np.nonzero(norms))
atom_axes /= norms.T
# s = ""
# for v in atom_axes:
# s += ".arrow %f %f %f " % tuple(com)
# end = com + 2 * v
# s += "%f %f %f\n" % tuple(end)
# with open("test2.bild", "w") as f:
# f.write(s)
# remove parallel/antiparallel axes for single atoms
# print(atom_axes)
mask = np.ones(len(atom_axes), dtype=bool)
for i, v in enumerate(atom_axes):
if not mask[i]:
continue
dv = atom_axes - v
c2 = np.linalg.norm(dv, axis=1) ** 2
angles = np.arccos(-0.5 * (c2 - 2))
# print(", ".join(["%.2f" % a for a in angles]))
mask2 = angles > rotation_tolerance
mask3 = angles < np.pi - rotation_tolerance
mask[:i] *= np.logical_and(mask2, mask3)[:i]
# print(mask)
atom_axes = atom_axes[mask]
# s = ""
# for v in atom_axes:
# s += ".arrow %f %f %f " % tuple(com)
# end = com + 2 * v
# s += "%f %f %f\n" % tuple(end)
# with open("test2.bild", "w") as f:
# f.write(s)
# remove parallel/antiparallel axes for pairs of atoms
mask = np.ones(len(atom_pairs), dtype=bool)
for i, v in enumerate(atom_pairs):
if not mask[i]:
continue
dv = np.delete(atom_pairs, i, axis=0) - v
c2 = np.linalg.norm(dv, axis=1) ** 2
angles = np.arccos(-0.5 * (c2 - 2))
mask2 = angles > rotation_tolerance
mask3 = angles < np.pi - rotation_tolerance
mask4 = np.logical_and(mask2, mask3)
mask[:i] *= mask4[:i]
mask[i + 1:] *= mask4[i:]
atom_pairs = atom_pairs[mask]
# remove parallel/antiparallel norms for pairs of atoms
mask = np.ones(len(atom_pair_norms), dtype=bool)
for i, v in enumerate(atom_pair_norms):
if not mask[i]:
continue
dv = np.delete(atom_pair_norms, i, axis=0) - v
c2 = np.linalg.norm(dv, axis=1) ** 2
angles = np.arccos(-0.5 * (c2 - 2))
mask2 = angles > rotation_tolerance
mask3 = angles < np.pi - rotation_tolerance
mask4 = np.logical_and(mask2, mask3)
mask[:i] *= mask4[:i]
mask[i + 1:] *= mask4[i:]
atom_pair_norms = atom_pair_norms[mask]
# s = ""
# for v in atom_pair_norms:
# s += ".arrow %f %f %f " % tuple(com)
# end = com + 2 * v
# s += "%f %f %f\n" % tuple(end)
# with open("test2.bild", "w") as f:
# f.write(s)
if len(atom_pairs):
# remove axes for pairs of atoms that are parallel/antiparallel
# to axes for single atoms
mask = np.ones(len(atom_pairs), dtype=bool)
for i, v in enumerate(atom_axes):
dv = atom_pairs - v
c2 = np.linalg.norm(dv, axis=1) ** 2
angles = np.arccos(-0.5 * (c2 - 2))
mask2 = angles > rotation_tolerance
mask3 = angles < np.pi - rotation_tolerance
mask *= np.logical_and(mask2, mask3)
atom_pairs = atom_pairs[mask]
if len(atom_pair_norms):
# remove norms for pairs of atoms that are parallel/antiparallel
# to axes for single atoms
mask = np.ones(len(atom_pair_norms), dtype=bool)
for i, v in enumerate(atom_axes):
dv = atom_pair_norms - v
c2 = np.linalg.norm(dv, axis=1) ** 2
angles = np.arccos(-0.5 * (c2 - 2))
mask2 = angles > rotation_tolerance
mask3 = angles < np.pi - rotation_tolerance
mask *= np.logical_and(mask2, mask3)
atom_pair_norms = atom_pair_norms[mask]
# s = ""
# for v in atom_pair_norms:
# s += ".arrow %f %f %f " % tuple(com)
# end = com + 2 * v
# s += "%f %f %f\n" % tuple(end)
# with open("test2.bild", "w") as f:
# f.write(s)
# remove axes for single atoms that are parallel/antiparallel
# to moment of inertia axes
mask = np.ones(len(atom_axes), dtype=bool)
for i, v in enumerate(axes):
dv = atom_axes - v
c2 = np.linalg.norm(dv, axis=1) ** 2
angles = np.arccos(-0.5 * (c2 - 2))
mask2 = angles > rotation_tolerance
mask3 = angles < np.pi - rotation_tolerance
mask *= np.logical_and(mask2, mask3)
atom_axes = atom_axes[mask]
# remove axes for pairs of atoms that are parallel/antiparallel
# to moment of inertia axes
if len(atom_pairs):
mask = np.ones(len(atom_pairs), dtype=bool)
for i, v in enumerate(axes):
dv = atom_pairs - v
c2 = np.linalg.norm(dv, axis=1) ** 2
angles = np.arccos(-0.5 * (c2 - 2))
mask2 = angles > rotation_tolerance
mask3 = angles < np.pi - rotation_tolerance
mask *= np.logical_and(mask2, mask3)
atom_pairs = atom_pairs[mask]
# remove norms for pairs of atoms that are parallel/antiparallel
# to moment of inertia axes
if len(atom_pair_norms):
mask = np.ones(len(atom_pair_norms), dtype=bool)
for i, v in enumerate(axes):
dv = atom_pair_norms - v
c2 = np.linalg.norm(dv, axis=1) ** 2
angles = np.arccos(-0.5 * (c2 - 2))
mask2 = angles > rotation_tolerance
mask3 = angles < np.pi - rotation_tolerance
mask *= np.logical_and(mask2, mask3)
atom_pair_norms = atom_pair_norms[mask]
# s = ""
# for v in atom_pair_norms:
# s += ".arrow %f %f %f " % tuple(com)
# end = com + 2 * v
# s += "%f %f %f\n" % tuple(end)
# with open("test2.bild", "w") as f:
# f.write(s)
# remove axes that are not orthogonal to moments of inertia axes
if ortho_to:
mask = np.ones(len(atom_axes), dtype=bool)
pair_mask = np.ones(len(atom_pairs), dtype=bool)
pair_mask_norms = np.ones(len(atom_pair_norms), dtype=bool)
for v in ortho_to:
dv = atom_axes - v
c2 = np.linalg.norm(dv, axis=1) ** 2
angles = np.arccos(-0.5 * (c2 - 2))
mask1 = abs(angles - np.pi / 2) < rotation_tolerance
mask *= mask1
if len(atom_pairs):
dv = atom_pairs - v
c2 = np.linalg.norm(dv, axis=1) ** 2
angles = np.arccos(-0.5 * (c2 - 2))
pair_mask = abs(angles - np.pi / 2) < rotation_tolerance
atom_pairs = atom_pairs[pair_mask]
if len(atom_pair_norms):
dv = atom_pair_norms - v
c2 = np.linalg.norm(dv, axis=1) ** 2
angles = np.arccos(-0.5 * (c2 - 2))
pair_mask_norms = abs(angles - np.pi / 2) < rotation_tolerance
atom_pair_norms = atom_pair_norms[pair_mask_norms]
atom_axes = atom_axes[mask]
for v in axes:
mask = np.ones(len(atom_axes), dtype=bool)
pair_mask = np.ones(len(atom_pairs), dtype=bool)
pair_mask_norms = np.ones(len(atom_pair_norms), dtype=bool)
dv = atom_axes - v
c2 = np.linalg.norm(dv, axis=1) ** 2
angles = np.arccos(-0.5 * (c2 - 2))
mask1 = angles > rotation_tolerance
mask2 = angles < np.pi - rotation_tolerance
mask *= np.logical_and(mask1, mask2)
if len(atom_pairs):
dv = atom_pairs - v
c2 = np.linalg.norm(dv, axis=1) ** 2
angles = np.arccos(-0.5 * (c2 - 2))
pair_mask1 = angles > rotation_tolerance
pair_mask2 = angles < np.pi - rotation_tolerance
pair_mask *= np.logical_and(pair_mask1, pair_mask2)
atom_pairs = atom_pairs[pair_mask]
if len(atom_pair_norms):
dv = atom_pair_norms - v
c2 = np.linalg.norm(dv, axis=1) ** 2
angles = np.arccos(-0.5 * (c2 - 2))
atom_pair_norms1 = angles > rotation_tolerance
atom_pair_norms2 = angles < np.pi - rotation_tolerance
pair_mask_norms *= np.logical_and(atom_pair_norms1, atom_pair_norms2)
atom_pair_norms = atom_pair_norms[pair_mask_norms]
if len(atom_pairs) and len(atom_axes):
for v in atom_pairs:
mask = np.ones(len(atom_axes), dtype=bool)
dv = atom_axes - v
c2 = np.linalg.norm(dv, axis=1) ** 2
angles = np.arccos(-0.5 * (c2 - 2))
mask1 = angles > rotation_tolerance
mask2 = angles < np.pi - rotation_tolerance
mask *= np.logical_and(mask1, mask2)
atom_axes = atom_axes[mask]
# s = ""
# for v in ortho_to:
# s += ".arrow %f %f %f " % tuple(com)
# end = com + 2 * v
# s += "%f %f %f\n" % tuple(end)
# with open("test2.bild", "w") as f:
# f.write(s)
checked_axes = 0
# find proper rotations along the axes we've found:
# * moments of inertia axes
# * COM -> atom vectors
# * COM -> midpoint of atom paris
# also grab axes for checking mirror planes
check_axes = []
primes = dict()
args = tuple([arg for arg in [axes, atom_axes, atom_pairs] if len(arg)])
principal_axis = None
for ax in np.concatenate(args):
max_n = None
found_n = []
for n in range(2, max_rotation + 1):
if n not in primes:
primes[n] = Primes.primes_below(n // 2)
# print(n, primes[n])
skip = False
for prime in primes[n]:
if n % prime == 0 and prime not in found_n:
# print("skipping", n)
skip = True
break
# if max_n and max_n % n != 0:
# # the highest order proper rotation axis must be
# # divisible by all other coincident axes
# continue
# look for C5^2 stuff
# for exp in range(1, 1 + n // 2):
for exp in range(1, 2):
if exp > 1 and n % exp == 0:
# skip things like C4^2 b/c that's just C2
continue
# see if the error associated with the element is reasonable
rot = ProperRotation(com, ax, n, exp)
error = rot.error(tolerance, groups=atom_ids, coords=coords)
checked_axes += 1
if error <= tolerance:
# print(geom.atoms[i])
# s = ".arrow %f %f %f " % tuple(com)
# end = com + 2 * ax
# s += "%f %f %f\n" % tuple(end)
# with open("test.bild", "a") as f:
# f.write(s)
valid.append(rot)
if principal_axis is None or rot.n > principal_axis[0].n:
principal_axis = [rot]
elif principal_axis is not None and rot.n == principal_axis[0].n:
principal_axis.append(rot)
found_n.append(n)
if n > 2:
# for Cn n != 2, add an element that is the same
# except the axis of rotation is antiparallel
rot2 = ProperRotation(com, -ax, n, exp)
valid.append(rot2)
if not max_n:
max_n = n
check_axes.append(ax)
elif exp == 1:
# can't have Cn^y if you don't have Cn
break
if degeneracy[0] == 3:
# spherical top molecules need more checks related to C2 axes
c2_axes = list(
filter(
lambda ele: isinstance(ele, ProperRotation) and ele.n == 2,
valid,
)
)
# TODO: replace with array operations like before
for i, c2_1 in enumerate(c2_axes):
for c2_2 in c2_axes[:i]:
test_axes = []
if len(c2_axes) == 3:
# T groups - check midpoint
for c2_3 in c2_axes[i:]:
axis = c2_1.axis + c2_2.axis + c2_3.axis
test_axes.append(axis)
axis = c2_1.axis + c2_2.axis - c2_3.axis
test_axes.append(axis)
axis = c2_1.axis - c2_2.axis + c2_3.axis
test_axes.append(axis)
axis = c2_1.axis - c2_2.axis - c2_3.axis
test_axes.append(axis)
else:
# O, I groups - check cross product
test_axes.append(np.cross(c2_1.axis, c2_2.axis))
for axis in test_axes:
norm = np.linalg.norm(axis)
if norm < 1e-5:
continue
axis /= norm
dup = False
for element in valid:
if isinstance(element, ProperRotation):
if 1 - abs(np.dot(element.axis, axis)) < rotation_tolerance:
dup = True
break
if dup:
continue
max_n = None
for n in range(max_rotation, 1, -1):
if max_n and max_n % n != 0:
continue
# for exp in range(1, 1 + n // 2):
for exp in range(1, 2):
if exp > 1 and n % exp == 0:
continue
rot = ProperRotation(com, axis, n, exp)
checked_axes += 1
error = rot.error(tolerance, groups=atom_ids, coords=coords)
if error <= tolerance:
if principal_axis is None or rot.n > principal_axis[0].n:
principal_axis = [rot]
elif principal_axis is not None and rot.n == principal_axis[0].n:
principal_axis.append(rot)
valid.append(rot)
if not max_n:
max_n = n
check_axes.append(ax)
if n > 2:
rot2 = ProperRotation(com, -axis, n, exp)
valid.append(rot2)
elif exp == 1:
break
# improper rotations
# coincident with proper rotations and can be 1x or 2x
# the order of the proper rotation
for element in valid:
if not isinstance(element, ProperRotation):
continue
if element.exp != 1:
continue
for x in [1, 2]:
if x * element.n == 2:
# S2 is inversion - we already checked i
continue
# for exp in range(1, 1 + (x * element.n) // 2):
for exp in range(1, 2):
if exp > 1 and (x * element.n) % exp == 0:
continue
for element2 in valid:
if isinstance(element2, ImproperRotation):
angle = angle_between_vectors(element2.axis, element.axis)
if (
element2.exp == exp and
(
angle < rotation_tolerance or
angle > (np.pi - rotation_tolerance)
) and
element2.n == x * element.n
):
break
else:
imp_rot = ImproperRotation(
element.translation,
element.axis,
x * element.n,
exp,
)
error = imp_rot.error(tolerance, groups=atom_ids, coords=coords)
if error <= tolerance:
valid.append(imp_rot)
rot2 = ImproperRotation(
element.translation,
-element.axis,
x * element.n,
exp
)
valid.append(rot2)
elif exp == 1:
break
c2_axes = list(
filter(
lambda ele: isinstance(ele, ProperRotation) and ele.n == 2 and ele.exp == 1,
valid,
)
)
c2_vectors = np.array([c2.axis for c2 in c2_axes])
sigma_norms = []
if bool(principal_axis) and len(c2_vectors) and principal_axis[0].n != 2:
for ax in principal_axis:
perp = np.cross(ax.axis, c2_vectors)
norms = np.linalg.norm(perp, axis=1)
mask = np.nonzero(norms)
perp = perp[mask]
norms = norms[mask]
perp /= norms[:, None]
sigma_norms.extend(perp)
sigma_norms = np.array(sigma_norms)
mask = np.ones(len(sigma_norms), dtype=bool)
for i, v in enumerate(sigma_norms):
if not mask[i]:
continue
dv = np.delete(sigma_norms, i, axis=0) - v
c2 = np.linalg.norm(dv, axis=1) ** 2
angles = np.arccos(-0.5 * (c2 - 2))
mask2 = angles > rotation_tolerance
mask3 = angles < np.pi - rotation_tolerance
mask4 = np.logical_and(mask2, mask3)
mask[:i] *= mask4[:i]
mask[i + 1:] *= mask4[i:]
sigma_norms = sigma_norms[mask]
c2_vectors = np.append(c2_vectors, [-c2.axis for c2 in c2_axes], axis=0)
# mirror axes
# for I, O - only check c2 axes
if (
degeneracy[0] != 3 or
not c2_axes or
(degeneracy[0] == 3 and len(c2_axes) == 3)
):
if len(atom_pair_norms):
mask = np.ones(len(atom_pair_norms), dtype=bool)
for i, v in enumerate(axes):
dv = atom_pair_norms - v
c2 = np.linalg.norm(dv, axis=1) ** 2
angles = np.arccos(-0.5 * (c2 - 2))
mask2 = angles > rotation_tolerance
mask3 = angles < np.pi - rotation_tolerance
mask *= np.logical_and(mask2, mask3)
atom_pair_norms = atom_pair_norms[mask]
mask = np.ones(len(atom_pair_norms), dtype=bool)
for i, v in enumerate(check_axes):
dv = atom_pair_norms - v
c2 = np.linalg.norm(dv, axis=1) ** 2
angles = np.arccos(-0.5 * (c2 - 2))
mask2 = angles > rotation_tolerance
mask3 = angles < np.pi - rotation_tolerance
mask *= np.logical_and(mask2, mask3)
atom_pair_norms = atom_pair_norms[mask]
if check_axes:
check_axes = np.array(check_axes)
mask = np.ones(len(check_axes), dtype=bool)
for i, v in enumerate(axes):
dv = check_axes - v
c2 = np.linalg.norm(dv, axis=1) ** 2
angles = np.arccos(-0.5 * (c2 - 2))
mask2 = angles > rotation_tolerance
mask3 = angles < np.pi - rotation_tolerance
mask *= np.logical_and(mask2, mask3)
check_axes = check_axes[mask]
mask = np.ones(len(check_axes), dtype=bool)
for i, v in enumerate(atom_axes):
dv = check_axes - v
c2 = np.linalg.norm(dv, axis=1) ** 2
angles = np.arccos(-0.5 * (c2 - 2))
mask2 = angles > rotation_tolerance
mask3 = angles < np.pi - rotation_tolerance
mask *= np.logical_and(mask2, mask3)
check_axes = check_axes[mask]
mask = np.ones(len(check_axes), dtype=bool)
for i, v in enumerate(atom_pair_norms):
dv = check_axes - v
c2 = np.linalg.norm(dv, axis=1) ** 2
angles = np.arccos(-0.5 * (c2 - 2))
mask2 = angles > rotation_tolerance
mask3 = angles < np.pi - rotation_tolerance
mask *= np.logical_and(mask2, mask3)
check_axes = check_axes[mask]
if len(sigma_norms):
sigma_norms = np.array(sigma_norms)
mask = np.ones(len(sigma_norms), dtype=bool)
for i, v in enumerate(axes):
dv = sigma_norms - v
c2 = np.linalg.norm(dv, axis=1) ** 2
angles = np.arccos(-0.5 * (c2 - 2))
angles = np.nan_to_num(angles)
mask2 = angles > rotation_tolerance
mask3 = angles < np.pi - rotation_tolerance
mask *= np.logical_and(mask2, mask3)
sigma_norms = sigma_norms[mask]
mask = np.ones(len(sigma_norms), dtype=bool)
for i, v in enumerate(atom_axes):
dv = sigma_norms - v
c2 = np.linalg.norm(dv, axis=1) ** 2
angles = np.arccos(-0.5 * (c2 - 2))
mask2 = angles > rotation_tolerance
mask3 = angles < np.pi - rotation_tolerance
mask *= np.logical_and(mask2, mask3)
sigma_norms = sigma_norms[mask]
mask = np.ones(len(sigma_norms), dtype=bool)
for i, v in enumerate(atom_pair_norms):
dv = sigma_norms - v
c2 = np.linalg.norm(dv, axis=1) ** 2
angles = np.arccos(-0.5 * (c2 - 2))
mask2 = angles > rotation_tolerance
mask3 = angles < np.pi - rotation_tolerance
mask *= np.logical_and(mask2, mask3)
sigma_norms = sigma_norms[mask]
mask = np.ones(len(sigma_norms), dtype=bool)
for i, v in enumerate(check_axes):
dv = sigma_norms - v
c2 = np.linalg.norm(dv, axis=1) ** 2
angles = np.arccos(-0.5 * (c2 - 2))
mask2 = angles > rotation_tolerance
mask3 = angles < np.pi - rotation_tolerance
mask *= np.logical_and(mask2, mask3)
sigma_norms = sigma_norms[mask]
# print("axes")
# for ax in axes:
# print(ax)
#
# print("atom_axes")
# for ax in atom_axes:
# print(ax)
#
# print("atom_pair_norms")
# for ax in atom_pair_norms:
# print(ax)
#
# print("check_axes")
# for ax in check_axes:
# print(ax)
#
# print("sigma_norms")
# for ax in sigma_norms:
# print(ax)
args = tuple(
[arg for arg in [
axes, atom_axes, atom_pair_norms, check_axes, sigma_norms
] if len(arg)]
)
for ax in np.concatenate(args):
mirror = MirrorPlane(com, ax)
error = mirror.error(tolerance, groups=atom_ids, coords=coords)
if error <= tolerance:
valid.append(mirror)
else:
for i, ax in enumerate(c2_axes):
mirror = MirrorPlane(com, ax.axis)
error = mirror.error(tolerance, groups=atom_ids, coords=coords)
if error <= tolerance:
valid.append(mirror)
min_atoms = None
if principal_axis:
for ele in valid:
if not isinstance(ele, MirrorPlane):
continue
for ax in principal_axis:
c2 = np.linalg.norm(ax.axis - ele.axis) ** 2
angle = np.arccos(-0.5 * (c2 - 2))
if angle < rotation_tolerance or angle > np.pi - rotation_tolerance:
# mirror plane normal is parallel to principal_axis
ele.label = "h"
break
elif abs(angle - np.pi / 2) < rotation_tolerance:
# mirror plane normal is perpendicular to principal_axis
ele.label = "v"
if ele.label == "v":
# determine number of atoms in sigma_v in case we need to
# differentiate from sigma_d later
perp_dist = ele.perp_dist(geom.coords)
atoms_contained = sum([1 if abs(d) < tolerance else 0 for d in perp_dist])
if min_atoms is None or atoms_contained < min_atoms:
min_atoms = atoms_contained
if c2_axes:
# check for sigma_d
# should be a sigma_v that bisects two C2 axes
# if a molecule has different sigma_v planes that
# bisect C2 axes and contain different numbers of
# atoms, it is convention to label just the sigma_v
# planes that contain the fewest atoms as sigma_d
for ele in valid:
if not isinstance(ele, MirrorPlane):
continue
if principal_axis and ele.label != "v":
continue
perp_dist = ele.perp_dist(geom.coords)
atoms_contained = sum([1 if abs(d) < tolerance else 0 for d in perp_dist])
if min_atoms is not None and atoms_contained != min_atoms:
continue
perp_v = perp_vector(ele.axis)
c2 = np.linalg.norm(c2_vectors - perp_v, axis=1) ** 2
angles = np.arccos(-0.5 * (c2 - 2))
for i, angle1 in enumerate(angles):
for j, angle2 in enumerate(angles[:i]):
if abs(np.dot(c2_vectors[i], c2_vectors[j])) == 1:
continue
if abs(angle1 - angle2) < rotation_tolerance:
ele.label = "d"
break
if ele.label == "d":
break
# s = ""
# colors = ["black", "red", "blue", "green", "purple", "yellow", "cyan"]
# for element in valid:
# # if isinstance(element, ProperRotation):
# # s += ".color %s\n" % colors[element.n - 2]
# # s += ".note C%i\n" % element.n
# # s += ".arrow %f %f %f " % tuple(com)
# # end = com + 2 * np.sqrt(element.n) * element.axis
# # s += "%f %f %f\n" % tuple(end)
# if isinstance(element, MirrorPlane):
# s += ".arrow %f %f %f " % tuple(com)
# end = com + 2 * element.axis
# s += "%f %f %f\n" % tuple(end)
#
# with open("test.bild", "w") as f:
# f.write(s)
return valid
def determine_point_group(self, rotation_tolerance=0.01):
"""
determines point group of self by looing at self.elements
rotation_tolerance - tolerance in radians for axes to be
considered parallel/antiparallel/orthogonal
returns str for point group name
"""
moments, axes = self.geom.get_principle_axes()
linear = False
if any(np.isclose(m, 0) for m in moments):
linear = True
if linear:
if any(isinstance(ele, InversionCenter) for ele in self.elements):
return "D_inf_h"
else:
return "C_inf_v"
Cn = dict()
has_inversion = False
has_mirror = False
has_sig_h = False
has_sig_d = False
has_sig_v = False
for ele in self.elements:
if isinstance(ele, ProperRotation):
if ele.n not in Cn and ele.exp == 1:
Cn[ele.n] = []
Cn[ele.n].append(ele)
if isinstance(ele, InversionCenter):
has_inversion = True
if isinstance(ele, MirrorPlane):
has_mirror = True
if ele.label == "d":
has_sig_d = True
elif ele.label == "h":
has_sig_h = True
elif ele.label == "v":
has_sig_v = True
if Cn:
max_n = max(Cn.keys())
if max_n == 5 and len(Cn[5]) >= 12:
if has_mirror:
return "Ih"
return "I"
if max_n == 4 and len(Cn[4]) >= 6:
if has_mirror:
return "Oh"
return "O"
if max_n == 3 and len(Cn[3]) >= 8:
for ele in self.elements:
if has_sig_d:
return "Td"
if has_mirror:
return "Th"
return "T"
n_sig_v = len([
ele for ele in self.elements
if isinstance(ele, MirrorPlane) and (ele.label == "v" or ele.label == "d")
])
if n_sig_v > max_n:
self.LOG.warning(
"%i-fold rotation found, but %i sigma_v planes found\n" % (
max_n,
n_sig_v,
) +
"you may need to increase the maximum order of proper rotational axes that are checked"
)
prin_ax = Cn[max_n][0]
n_perp = 0
if 2 in Cn:
for c2 in Cn[2]:
angle = angle_between_vectors(
c2.axis,
prin_ax.axis,
renormalize=False
)
if abs(angle - np.pi / 2) < rotation_tolerance:
n_perp += 1
if n_perp >= max_n:
if has_sig_h:
return "D%ih" % max_n
if n_sig_v >= max_n:
return "D%id" % max_n
return "D%i" % max_n
if has_sig_h:
return "C%ih" % max_n
if n_sig_v >= max_n:
return "C%iv" % max_n
for ele in self.elements:
if isinstance(ele, ImproperRotation) and ele.n == 2 * max_n:
return "S%i" % (2 * max_n)
return "C%i" % max_n
if has_mirror:
return "Cs"
if has_inversion:
return "Ci"
return "C1"
@property
def symmetry_number(self):
"""external symmetry number"""
n = 1
for ele in self.elements:
if isinstance(ele, ProperRotation):
if ele.n > n:
n = ele.n
elif isinstance(ele, ImproperRotation):
if ele.n / 2 > n:
n = ele.n
if self.name.startswith("D"):
n *= 2
if self.name.startswith("T"):
n *= 4
if self.name.startswith("O"):
n *= 6
if self.name.startswith("I"):
n *= 12
return n
def equivalent_positions(self):
"""returns a list of lists of atoms that are symmetry-equivalent"""
equivs = [set([atom]) for atom in self.geom.atoms]
init_coords = self.geom.coords
init_partitions = dict()
for atom, group in zip(self.geom.atoms, self.initial_groups):
init_partitions.setdefault(group, [])
init_partitions[group].append(atom)
init_partition_coords = {
group: self.geom.coordinates(atoms) for group, atoms in init_partitions.items()
}
for ele in self.elements:
if isinstance(ele, Identity):
continue
for group in init_partitions:
coords = init_partition_coords[group]
new_coords = ele.apply_operation(coords)
dist = distance_matrix(coords, new_coords)
closest_ndx = np.argmin(dist, axis=1)
for i, (atom, ndx) in enumerate(zip(init_partitions[group], closest_ndx)):
if i == ndx:
continue
j = self.geom.atoms.index(atom)
k = self.geom.atoms.index(init_partitions[group][ndx])
equivs[j] = {*equivs[j], init_partitions[group][ndx]}
equivs[k] = {*equivs[k], atom}
# for e in equivs:
# for atom in sorted(e, key=lambda a: int(a.name)):
# print(atom)
# print("----------------")
out = []
for eq in equivs:
for group in out:
if any(atom in group for atom in eq):
group.extend(eq)
break
else:
out.append(list(eq))
return out
@property
def optically_chiral(self):
"""is this point group optically_chiral?"""
found_mirror = False
found_inversion = False
found_improper = False
for ele in self.elements:
if isinstance(ele, MirrorPlane):
# S1
found_mirror = True
elif isinstance(ele, InversionCenter):
# S2
found_inversion = True
elif isinstance(ele, ImproperRotation):
# Sn
found_improper = True
return not (found_mirror or found_inversion or found_improper)
def idealize_geometry(self):
"""
adjust coordinates of self.geom to better match this point group
also re-determines point group and symmetry elements
"""
com = self.geom.COM()
coords = self.geom.coords
centered_coords = coords - com
out = np.zeros((len(self.geom.atoms), 3))
n_ele = len(self.elements)
principal_axis = None
# try moving axes of elements to more ideal positions relative to
# a principal axis
# for example, some point groups have elements with axes that
# are 30, 45, 60, and 90 degrees from a principal axis
# if we find an element close to one of these angles, we move it
# to that angle
# many symmetry elements will also be regularly spaced around
# the principal_axis
# e.g. D6h has 6 C2 axes perpendicular to the C6, with each
# being 30 degrees apart
# so this looks for C2 axes (and others) that are perpendicular
# to the principal axis and looks for C2 axes that are
# 2 * x * pi / N (order of principal axis) apart from one of
# the C2 axes
# it turns out none of that really helps, so if False
if False:
# if principal_axis:
principal_axis.sort(
key=lambda ele: ele.error(
geom=self.geom,
groups=self.initial_groups,
)
)
for ax in principal_axis:
non_ideal_axes = dict()
for ele in sorted(self.elements):
if not hasattr(ele, "axis"):
continue
if ax is ele:
continue
dv = ax.axis - ele.axis
c2 = np.linalg.norm(dv) ** 2
angle = np.arccos(-0.5 * (c2 - 2))
if np.isclose(angle, 0):
continue
for n in range(1, ax.n):
test_angle = np.arccos(1 / np.sqrt(n))
if np.isclose(angle, np.pi / 2, atol=5e-3):
non_ideal_axes.setdefault(np.pi / 2, [])
perp_axis = np.cross(ax.axis, ele.axis)
perp_axis = np.cross(perp_axis, ax.axis)
perp_axis /= np.linalg.norm(perp_axis)
if np.dot(perp_axis, ele.axis) < 0:
perp_axis *= -1
ele.axis = perp_axis
non_ideal_axes[np.pi / 2].append(ele)
continue
if np.isclose(angle, test_angle, atol=5e-3):
non_ideal_axes.setdefault(test_angle, [])
non_ideal_axes[test_angle].append(ele)
if n == 1:
continue
test_angle = np.arccos(1. / n)
if any(np.isclose(a, test_angle) for a in non_ideal_axes.keys()):
continue
if np.isclose(angle, test_angle, atol=5e-3):
non_ideal_axes.setdefault(test_angle, [])
non_ideal_axes[test_angle].append(ele)
for angle in non_ideal_axes:
# print(angle)
prop_rots = dict()
improp_rots = dict()
mirror = []
for ele in non_ideal_axes[angle]:
# print("\t", ele)
if isinstance(ele, MirrorPlane):
mirror.append(ele)
elif isinstance(ele, ProperRotation):
prop_rots.setdefault(ele.n, [])
prop_rots[ele.n].append(ele)
elif isinstance(ele, ImproperRotation):
improp_rots.setdefault(ele.n, [])
improp_rots[ele.n].append(ele)
for n in prop_rots:
prop_rots[n].sort(
key=lambda ele: ele.error(
geom=self.geom,
groups=self.initial_groups,
)
)
for n in improp_rots:
improp_rots[n].sort(
key=lambda ele: ele.error(
geom=self.geom,
groups=self.initial_groups,
)
)
mirror.sort(
key=lambda ele: ele.error(
geom=self.geom,
groups=self.initial_groups,
)
)
for i in range(1, 2 * ax.n):
test_angle = i * np.pi / (2 * ax.n)
mat = rotation_matrix(test_angle, ax.axis)
for n in improp_rots:
new_v = np.matmul(mat, improp_rots[n][0].axis)
for ele in improp_rots[n][1:]:
if np.isclose(np.dot(new_v, ele.axis), 1):
ele.axis = new_v
break
else:
print("no C%i axis at angle" % n, test_angle)
for n in improp_rots:
new_v = np.matmul(mat, improp_rots[n][0].axis)
for ele in improp_rots[n][1:]:
if np.isclose(np.dot(new_v, ele.axis), 1):
ele.axis = new_v
for mir in mirror:
new_v = np.matmul(mat, mirror[0].axis)
for ele in mirror[1:]:
if np.isclose(np.dot(new_v, ele.axis), 1):
ele.axis = new_v
# apply each operation and average the coordinates of the
# equivalent positions
max_n = 0
for ele in self.elements:
equiv = ele.equivalent_positions(coords, self.initial_groups)
out += ele.apply_operation_without_translation(
centered_coords[equiv]
) / n_ele
if isinstance(ele, ProperRotation):
if ele.n > max_n:
max_n = ele.n
self.geom.coords = out + com
self.elements = self.get_symmetry_elements(
self.geom,
max_rotation=max_n,
groups=self.initial_groups,
)
self.determine_point_group()
def total_error(self, return_max=False):
tot_error = 0
max_error = 0
max_ele = None
for ele in self.elements:
error = ele.error(geom=self.geom, groups=self.initial_groups)
tot_error += error
if error > max_error:
max_error = error
max_ele = ele
if return_max:
return tot_error, max_error, max_ele
return tot_error | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/symmetry.py | symmetry.py |
import configparser
import inspect
import logging
import os
import re
from AaronTools.const import AARONLIB, AARONTOOLS
config = configparser.ConfigParser(interpolation=None, comment_prefixes=("#"))
for filename in [
os.path.join(AARONTOOLS, "config.ini"),
os.path.join(AARONLIB, "config.ini"),
]:
try:
config.read(filename)
except FileNotFoundError:
continue
except configparser.MissingSectionHeaderError:
# add global options to default section
with open(filename) as f:
contents = "[DEFAULT]\n" + f.read()
config.read_string(contents)
if "log_level" in config["DEFAULT"]:
LOGLEVEL = config["DEFAULT"]["log_level"].upper()
else:
LOGLEVEL = "WARNING"
if "print_citations" in config["DEFAULT"]:
PRINT_CITATIONS = config["DEFAULT"].getboolean("print_citations")
else:
PRINT_CITATIONS = False
try:
SAVE_CITATIONS = config["DEFAULT"].getboolean("save_citations")
except ValueError:
SAVE_CITATIONS = config["DEFAULT"].get("save_citations")
if SAVE_CITATIONS is False:
SAVE_CITATIONS = None
logging.logThreads = 0
logging.logProcesses = 0
logging.captureWarnings(True)
class CustomFilter(logging.Filter):
def __init__(self, name="", level=None, override=None, cite=False):
super().__init__(name=name)
self.level = logging.WARNING
if isinstance(level, str):
level = getattr(logging, level.upper())
if level is not None:
self.level = level
self.override = {}
if override is not None:
self.override = override
self.cite = cite
def filter(self, record):
if record.funcName == "citation":
found = False
for frame in reversed(inspect.stack()):
if found:
record.funcName = frame.function
break
if frame.function == "_callTestMethod":
found = True
else:
record.funcName = inspect.stack()[-2].function
record.levelname = "CITATION"
if not self.cite:
return False
self.parse_message(record)
return True
for level, func_list in self.override.items():
if isinstance(level, str):
level = getattr(logging, level.upper())
if record.funcName not in func_list:
continue
if record.levelno < level:
return False
self.parse_message(record)
return True
if record.levelno < self.level:
return False
self.parse_message(record)
return True
def parse_message(self, record):
"""
Formats message to print prettily to console
"""
if isinstance(record.msg, str):
record.msg = re.sub(
"\n(\S)", lambda x: "\n %s" % x.group(1), record.msg
)
msg = ["\n "]
for word in re.findall("\S+\s*", record.getMessage()):
if len("".join(msg).split("\n")[-1]) + len(word) < 80:
msg.append(word)
else:
msg.append("\n {}".format(word))
record.getMessage = lambda: "".join(msg)
class CitationHandler(logging.FileHandler):
def __init__(self, filename, **kwargs):
filename = os.path.expandvars(filename)
if not os.path.exists(os.path.dirname(filename)):
# might be trying to put citations in $AARONLIB, but user
# didn't bother to set the environment variable and just
# uses the default
from AaronTools.const import AARONLIB
if "$AARONLIB" in filename:
filename = filename.replace("$AARONLIB", AARONLIB)
elif "${AARONLIB}" in filename:
filename = filename.replace("${AARONLIB}", AARONLIB)
elif "%AARONLIB%" in filename:
filename = filename.replace("%AARONLIB%", AARONLIB)
super().__init__(filename, **kwargs)
def emit(self, record):
"""
Adds a record to the citation file if it's not already present
"""
if record.levelname != "CITATION":
return
msg = record.msg.replace("\n ", " ")
record.getMessage = lambda: "".join(msg)
# check for duplicates
dupe = False
with open(self.baseFilename) as f:
for line in f.readlines():
if line.strip() == self.format(record):
dupe = True
break
if not dupe:
super().emit(record)
class ATLogger(logging.Logger):
def __init__(
self, name, level=None, override=None, fmt=None, add_hdlrs=None
):
"""
:level: the log level to use
:override: dict(level=funcName) to override loglevel for certain funcitons
:fmt: formatting string (optional)
:add_hdlrs: list(str(handlerName)) or list(Handler())
"""
super().__init__(name, level=1)
if level is None:
level = LOGLEVEL
if isinstance(level, str):
level = getattr(logging, level.upper())
self.level = level
if fmt is None:
fmt = "%(levelname)s %(name)s.%(funcName)s %(message)s"
formatter = logging.Formatter(fmt=fmt)
handlers = [(logging.StreamHandler(), PRINT_CITATIONS)]
if SAVE_CITATIONS is not None and os.access(SAVE_CITATIONS, os.W_OK):
handlers += [(CitationHandler(SAVE_CITATIONS), True)]
if add_hdlrs is not None:
for hdlr in add_hdlrs:
if isinstance(hdlr, str):
hdlr = getattr(logging, hdlr)
handlers.append((hdlr(), PRINT_CITATIONS))
else:
handlers.append(hdlr, PRINT_CITATIONS)
for hdlr, cite in handlers:
hdlr.setFormatter(formatter)
hdlr.addFilter(
CustomFilter(
name=name, level=self.level, override=override, cite=cite
)
)
self.addHandler(hdlr)
def citation(self, msg, *args, **kwargs):
self.info(msg, *args, **kwargs)
def getlogger(name=None, level=None, override=None, fmt=None):
"""
Get the logger without using the class decorator
:level: the log level to apply, defaults to WARNING
:override: a dictionary of the form {new_level: function_name_list} will apply the
`new_level` to log records produced from functions with names in
`function_name_list`, eg:
override={"DEBUG": ["some_function"]}
will set the log level to DEBUG for any messages produced during the run of
some_function()
"""
if name is None:
package = None
for frame in reversed(inspect.stack()):
res = inspect.getargvalues(frame.frame)
if "__name__" in res.locals and name is None:
name = res.locals["__name__"]
if "__package__" in res.locals and package is None:
package = res.locals["__package__"]
if name is not None and package is not None:
break
name = "{}{}{}".format(
name if name is not None else "",
"." if package is not None else "",
package if package is not None else "",
)
log = ATLogger(name, level=level, override=override, fmt=fmt)
return log
def addlogger(cls):
"""
Import this function and use it as a class decorator.
Log messages using the created LOG class attribute.
Useful class attributes to set that will be picked up by this decorator:
:LOG: Will be set to the logger instance during class initialization
:LOGLEVEL: Set this to use a different log level than what is in your config. Only
do this for testing purposes, and do not include it when pushing commits to the
master AaronTools branch.
:LOGLEVEL_OVERRIDE: Use this dict to override the log level set in the config file
for records originating in particular functions. Keys are log levels, values
are lists of strings corresponding to function names (default: {})
Example:
```
from AaronTools import addlogger
@addlogger
class Someclass:
LOG = None
LOGLEVEL = "WARNING"
LOGLEVEL_OVERRIDE = {"DEBUG": ["some_function"]}
# this won't be printed b/c "INFO" < LOGLEVEL
LOG.info("loading class")
def some_function(self):
# this message will be printed thanks to LOGLEVEL_OVERRIDE
self.LOG.debug("function called")
```
"""
name = "{}.{}".format(cls.__module__, cls.__name__)
level = None
if hasattr(cls, "LOGLEVEL") and cls.LOGLEVEL is not None:
level = cls.LOGLEVEL
override = None
if hasattr(cls, "LOGLEVEL_OVERRIDE") and cls.LOGLEVEL_OVERRIDE is not None:
override = cls.LOGLEVEL_OVERRIDE
cls.LOG = ATLogger(name, level=level, override=override)
return cls | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/__init__.py | __init__.py |
import re
import cclib
from AaronTools.atoms import Atom
from AaronTools.const import ELEMENTS, PHYSICAL, UNIT
from AaronTools.utils.utils import is_alpha, is_int
float_num = re.compile("[-+]?\d+\.?\d*")
READTYPES = ["XYZ", "Gaussian"]
WRITETYPES = ["XYZ", "Gaussian"]
NORM_FINISH = "Normal termination"
ERRORS = {
"NtrErr Called from FileIO": "CHK", # delete
"Wrong number of Negative eigenvalues": "EIGEN", # opt=noeigen
"Convergence failure -- run terminated.": "CONV", # scf=xqc
# check quota and alert user; REMOVE error from end of file!
"Erroneous write": "QUOTA",
"Atoms too close": "CLASH", # flag as CLASH
# die and alert user to check catalyst structure or fix input file
"The combination of multiplicity": "CHARGEMULT",
"Bend failed for angle": "REDUND", # Using opt=cartesian
"Unknown message": "UNKNOWN",
}
class FileReader:
"""
:name: file name without extension
:file_ext: file extension
:file_type:
:comment:
:atoms: list of Atom()
:all_geom: list of [Atom()] if optimization steps requested
:other: dict() containing additional info
"""
def __init__(self, filename, get_all=False, just_geom=True):
"""
:filename: a file name or a tuple(file_name, file_extension, IOstream)
:get_all: if true, optimization steps are also saved in
self.other['all_geom']; otherwise only saves last geometry
:just_geom: if true, does not store other information, such as
frequencies, only what is needed to construct a Geometry() obj
"""
if isinstance(filename, str):
self.name, self.file_ext = filename.rsplit(".", 1)
else:
self.name, self.file_ext = filename[:2]
filename = filename[2]
self.file_type = ""
self.comment = ""
self.atoms = []
self.all_geom = None
self.other = {}
try:
parser = cclib.io.ccopen(filename)
data = parser.parse()
self.file_type = str(parser).split()[0].split(".")[-1]
self.other = data.__dict__
except AttributeError:
if self.file_ext == "com":
self.file_type = "Gaussian"
self.read_com(filename)
return
for key in self.other:
print(key)
print(self.other[key])
for i, (n, c) in enumerate(zip(data.atomnos, data.atomcoords[-1])):
self.atoms += [Atom(element=ELEMENTS[n], coords=c, name=i + 1)]
if len(data.atomcoords) == 1:
# if > 1, there are more geometries to handle
del self.other["atomnos"]
del self.other["atomcoords"]
elif get_all:
# only handle them if get_all is true
self.all_geom = []
for i, coords in enumerate(data.atomcoords[:-1]):
atoms = []
for j, (n, c) in enumerate(zip(data.atomnos, coords)):
self.atoms += [
Atom(element=ELEMENTS[n], coords=c, name=j + 1)
]
self.all_geom += [atoms]
# cclib doesn't store XYZ file comments
if self.file_type == "XYZ":
self.read_xyz(filename)
# Grab things cclib doesn't from log files
if self.file_type == "Gaussian" and self.file_ext == "log":
self.read_log(filename)
# fix naming conventions
self.fix_names()
return
def read_log(self, filename):
# Grab things cclib doesn't from log files
if not self.other["metadata"]["success"]:
if isinstance(filename, str):
f = open(filename)
else:
f = filename
for line in f:
if "Molecular mass" in line:
self.other["mass"] = float(float_num.search(line).group(0))
self.other["mass"] *= UNIT.AMU_TO_KG
if "Rotational constants (GHZ):" in line:
rot = float_num.findall(line)
rot = [
float(r) * PHYSICAL.PLANCK * (10 ** 9) / PHYSICAL.KB
for r in rot
]
self.other["rotational_temperature"] = rot
def read_xyz(self, filename):
if isinstance(filename, str):
f = open(filename)
else:
f = filename
f.readline()
self.comment = f.readline().strip()
f.close()
def read_com(self, filename):
if isinstance(filename, str):
f = open(filename)
else:
f = filename
atoms = []
other = {}
found_atoms = False
found_constraint = False
for line in f:
# header
if line.startswith("%"):
# checkfile spec
other["checkfile"] = line.strip().split("=")[1]
continue
if line.startswith("#"):
match = re.search("^#(\S+)", line).group(1)
other["method"] = match.split("/")[0]
other["basis"] = match.split("/")[1]
if "temperature=" in line:
other["temperature"] = re.search(
"temperature=(\d+\.?\d*)", line
).group(1)
if "solvent=" in line:
other["solvent"] = re.search(
"solvent=(\S+)\)", line
).group(1)
if "scrf=" in line:
other["solvent_model"] = re.search(
"scrf=\((\S+),", line
).group(1)
if "EmpiricalDispersion=" in line:
other["emp_dispersion"] = re.search(
"EmpiricalDispersion=(\s+)", line
).group(1)
if "int=(grid(" in line:
other["grid"] = re.search("int=\(grid(\S+)", line).group(1)
for _ in range(4):
line = f.readline()
line = line.split()
other["charge"] = line[0]
other["mult"] = line[1]
found_atoms = True
continue
# constraints
if found_atoms and line.startswith("B") and line.endswith("F"):
found_constraint = True
if "constraint" not in other:
other["constraint"] = []
other["constraint"] += [float_num.findall(line)]
continue
# footer
if found_constraint:
if "footer" not in other:
other["footer"] = ""
other["footer"] += line
continue
# atom coords
nums = float_num.findall(line)
line = line.split()
if len(line) == 5 and is_alpha(line[0]) and len(nums) == 4:
if not is_int(line[1]):
continue
a = Atom(element=line[0], coords=nums[1:], flag=nums[0])
atoms += [a]
elif len(line) == 4 and is_alpha(line[0]) and len(nums) == 3:
a = Atom(element=line[0], coords=nums)
atoms += [a]
else:
continue
self.atoms = atoms
self.other = other
return
def fix_names(self):
if "metadata" in self.other:
if "success" in self.other["metadata"]:
self.other["finished"] = self.other["metadata"]["success"] | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/new_fileIO.py | new_fileIO.py |
import itertools
import re
import ssl
from collections import deque
from copy import deepcopy
import concurrent.futures
import numpy as np
from scipy.spatial import distance_matrix
import AaronTools
import AaronTools.utils.utils as utils
from AaronTools import addlogger
from AaronTools.atoms import Atom
from AaronTools.config import Config
from AaronTools.const import BONDI_RADII, D_CUTOFF, ELEMENTS, TMETAL, VDW_RADII
from AaronTools.fileIO import FileReader, FileWriter
from AaronTools.finders import Finder
from AaronTools.utils.prime_numbers import Primes
COORD_THRESHOLD = 0.2
CACTUS_HOST = "https://cactus.nci.nih.gov"
OPSIN_HOST = "https://opsin.ch.cam.ac.uk"
DEFAULT_CONFIG = Config(quiet=True)
if not DEFAULT_CONFIG["DEFAULT"].getboolean("local_only"):
import urllib.parse
from urllib.error import HTTPError
from urllib.request import urlopen
@addlogger
class Geometry:
"""
Attributes:
name
comment
atoms
other
_iter_idx
"""
# AaronTools.addlogger decorator will add logger to this class attribute
LOG = None
# decorator uses this to set log level (defaults to WARNING if None)
# LOGLEVEL = "INFO"
# add to this dict to override log level for specific functions
# keys are log level, values are lists of function names
# LOGLEVEL_OVERRIDE = {"DEBUG": "find"}
Primes()
def __init__(
self,
structure="",
name="",
comment="",
components=None,
refresh_connected=True,
refresh_ranks=True,
):
"""
:structure: can be a Geometry(), a FileReader(), a file name, or a
list of atoms
:name: str
:comment: str
:components: list of AaronTools.component.Component() or None
:refresh_connected: usually True - determine connectivity
:refresh_ranks: usually True - rank atoms, only False when loading from database
"""
super().__setattr__("_hashed", False)
self.name = name
self.comment = comment
self.atoms = []
self.center = None
self.components = components
self.other = {}
self._iter_idx = None
self._sigmat = None
self._epsmat = None
if isinstance(structure, Geometry):
# new from geometry
self.atoms = structure.atoms
if not name:
self.name = structure.name
if not comment:
self.comment = structure.comment
return
elif isinstance(structure, FileReader):
# get info from FileReader object
from_file = structure
elif isinstance(structure, str) and structure:
# parse file
from_file = FileReader(structure)
elif hasattr(structure, "__iter__") and structure:
for a in structure:
if not isinstance(a, Atom):
raise TypeError
else:
# list of atoms supplied
self.atoms = structure
if refresh_connected:
# SEQCROW sometimes uses refresh_connected=False to keep
# the connectivity the same as what's on screen
self.refresh_connected()
if refresh_ranks:
self.refresh_ranks()
return
else:
return
# only get here if we were given a file reader object or a file name
self.name = from_file.name
self.comment = from_file.comment
self.atoms = from_file.atoms
self.other = self.parse_comment()
if refresh_connected:
# some file types contain connectivity info (e.g. sd) - might not want
# to overwrite that
self.refresh_connected()
if refresh_ranks:
self.refresh_ranks()
return
# class methods
@classmethod
def iupac2smiles(cls, name):
if DEFAULT_CONFIG["DEFAULT"].getboolean("local_only"):
raise PermissionError(
"Converting IUPAC to SMILES failed. External network lookup disallowed."
)
# opsin seems to be better at iupac names with radicals
url_smi = "{}/opsin/{}.smi".format(
OPSIN_HOST, urllib.parse.quote(name)
)
try:
smiles = (
urlopen(url_smi, context=ssl.SSLContext())
.read()
.decode("utf8")
)
except HTTPError:
raise RuntimeError(
"%s is not a valid IUPAC name or https://opsin.ch.cam.ac.uk is down"
% name
)
return smiles
@classmethod
def from_string(cls, name, form="smiles", strict_use_rdkit=False):
"""get Geometry from string
form=iupac -> iupac to smiles from opsin API
--> form=smiles
form=smiles -> structure from cactvs API/RDKit
"""
# CC and HOH are special-cased because they are used in
# the automated testing and we don't want that to fail
# b/c cactus is down and the user doesn't have rdkit
# these structures are from NIST
if name == "CC":
return cls([
Atom("C", coords=[0.0, 0.0, 0.7680], name="1"),
Atom("C", coords=[0.0, 0.0, -0.7680], name="2"),
Atom("H", coords=[-1.0192, 0.0, 1.1573], name="3"),
Atom("H", coords=[0.5096, 0.8826, 1.1573], name="4"),
Atom("H", coords=[0.5096, -0.8826, 1.1573], name="5"),
Atom("H", coords=[1.0192, 0.0, -1.1573], name="6"),
Atom("H", coords=[-0.5096, -0.8826, -1.1573], name="7"),
Atom("H", coords=[-0.5096, 0.8826, -1.1573], name="8"),
])
elif name == "HOH":
return cls([
Atom("H", coords=[0.0, 0.7572, -0.4692], name="1"),
Atom("O", coords=[0.0, 0.0, 0.0], name="2"),
Atom("H", coords=[0.0, -0.7572, -0.4692], name="3"),
])
def get_cactus_sd(smiles):
if DEFAULT_CONFIG["DEFAULT"].getboolean("local_only"):
raise PermissionError(
"Cannot retrieve structure from {}. External network lookup disallowed.".format(
CACTUS_HOST
)
)
url_sd = "{}/cgi-bin/translate.tcl?smiles={}&format=sdf&astyle=kekule&dim=3D&file=".format(
CACTUS_HOST, urllib.parse.quote(smiles)
)
s_sd_get = urlopen(url_sd, context=ssl.SSLContext())
msg, status = s_sd_get.msg, s_sd_get.status
if msg != "OK":
cls.LOG.error(
"Issue contacting %s for SMILES lookup (status: %s)",
CACTUS_HOST,
status,
)
raise IOError
s_sd_get = s_sd_get.read().decode("utf8")
try:
tmp_url = re.search(
'User-defined exchange format file: <a href="(.*)"',
s_sd_get,
).group(1)
except AttributeError as err:
if re.search("You entered an invalid SMILES", s_sd_get):
cls.LOG.error(
"Invalid SMILES encountered: %s (consult %s for syntax help)",
smiles,
"https://cactus.nci.nih.gov/translate/smiles.html",
)
exit(1)
raise IOError(err)
new_url = "{}{}".format(CACTUS_HOST, tmp_url)
s_sd = (
urlopen(new_url, context=ssl.SSLContext())
.read()
.decode("utf8")
)
return s_sd
if DEFAULT_CONFIG["DEFAULT"].getboolean("local_only"):
strict_use_rdkit = True
accepted_forms = ["iupac", "smiles"]
if form not in accepted_forms:
raise NotImplementedError(
"cannot create substituent given %s; use one of %s" % form,
str(accepted_forms),
)
if form == "smiles":
smiles = name
elif form == "iupac":
smiles = cls.iupac2smiles(name)
try:
import rdkit.Chem.AllChem as rdk
m = rdk.MolFromSmiles(smiles)
if m is None and not strict_use_rdkit:
s_sd = get_cactus_sd(smiles)
elif m:
mh = rdk.AddHs(m)
rdk.EmbedMolecule(mh, randomSeed=0x421C52)
s_sd = rdk.MolToMolBlock(mh)
else:
raise RuntimeError(
"Could not load {} with RDKit".format(smiles)
)
except ImportError:
s_sd = get_cactus_sd(smiles)
try:
f = FileReader((name, "sd", s_sd))
is_sdf = True
except ValueError:
# for some reason, CACTUS is giving xyz files instead of sdf...
is_sdf = False
try:
f = FileReader((name, "xyz", s_sd))
except ValueError:
cls.LOG.error("Error loading geometry:\n %s", s_sd)
raise
return cls(f, refresh_connected=not is_sdf)
@classmethod
def get_coordination_complexes(
cls,
center,
ligands,
shape,
c2_symmetric=None,
minimize=False,
session=None,
):
"""
get all unique coordination complexes
uses templates from Inorg. Chem. 2018, 57, 17, 10557–10567
center - str, element of center atom
ligands - list of ligand names in the ligand library
shape - str, e.g. octahedral - see Atom.get_shape
c2_symmetric - list of bools, specifies which of the bidentate ligands are C2-symmetric
if this list is as long as the ligands list, the nth item corresponds
to the nth ligand
otherwise, the nth item indicate the symmetry of the nth bidentate ligand
minimize - bool, use minimize=True when mapping ligands (see Geometry.map_ligand)
returns a list of cls containing all unique coordination complexes and the
general formula of the complexes
"""
import os.path
from AaronTools.atoms import BondOrder
from AaronTools.component import Component
from AaronTools.const import AARONTOOLS
if c2_symmetric is None:
c2_symmetric = []
for lig in ligands:
comp = Component(lig)
if not len(comp.key_atoms) == 2:
c2_symmetric.append(False)
continue
c2_symmetric.append(comp.c2_symmetric())
bo = BondOrder()
# create a geometry with the specified shape
# change the elements from dummy atoms to something else
start_atoms = Atom.get_shape(shape)
n_coord = len(start_atoms) - 1
start_atoms[0].element = center
start_atoms[0].reset()
for atom in start_atoms[1:]:
start_atoms[0].connected.add(atom)
atom.connected.add(start_atoms[0])
atom.element = "B"
atom.reset()
geom = cls(start_atoms, refresh_connected=False, refresh_ranks=False)
# we'll need to determine the formula of the requested complex
# monodentate ligands are a, b, etc
# symmetric bidentate are AA, BB, etc
# asymmetric bidentate are AB, CD, etc
# ligands are sorted monodentate, followed by symmetric bidentate, followed by
# asymmetric bidentate, then by decreasing count
# e.g., Ca(CO)2(ACN)4 is Ma4b2
alphabet = "abcdefghi"
symmbet = ["AA", "BB", "CC", "DD"]
asymmbet = ["AB", "CD", "EF", "GH"]
monodentate_names = []
symm_bidentate_names = []
asymm_bidentate_names = []
n_bidentate = 0
# determine types of ligands
for i, lig in enumerate(ligands):
comp = Component(lig)
if len(comp.key_atoms) == 1:
monodentate_names.append(lig)
elif len(comp.key_atoms) == 2:
if len(ligands) == len(c2_symmetric):
c2 = c2_symmetric[i]
else:
c2 = c2_symmetric[n_bidentate]
n_bidentate += 1
if c2:
symm_bidentate_names.append(lig)
else:
asymm_bidentate_names.append(lig)
else:
# tridentate or something
raise NotImplementedError(
"can only attach mono- and bidentate ligands: %s (%i)"
% (lig, len(comp.key_atoms))
)
coord_num = len(monodentate_names) + 2 * (
len(symm_bidentate_names) + len(asymm_bidentate_names)
)
if coord_num != n_coord:
raise RuntimeError(
"coordination number (%i) does not match sum of ligand denticity (%i)"
% (n_coord, coord_num)
)
# start putting formula together
cc_type = "M"
this_name = center
# sorted by name count is insufficient when there's multiple monodentate ligands
# with the same count (e.g. Ma3b3)
# add the index in the library to offset this
monodentate_names = sorted(
monodentate_names,
key=lambda x: 10000 * monodentate_names.count(x)
+ Component.list().index(x),
reverse=True,
)
for i, mono_lig in enumerate(
sorted(
set(monodentate_names),
key=lambda x: 10000 * monodentate_names.count(x)
+ Component.list().index(x),
reverse=True,
)
):
cc_type += alphabet[i]
this_name += "(%s)" % mono_lig
if monodentate_names.count(mono_lig) > 1:
cc_type += "%i" % monodentate_names.count(mono_lig)
this_name += "%i" % monodentate_names.count(mono_lig)
symm_bidentate_names = sorted(
symm_bidentate_names,
key=lambda x: 10000 * symm_bidentate_names.count(x)
+ Component.list().index(x),
reverse=True,
)
for i, symbi_lig in enumerate(
sorted(
set(symm_bidentate_names),
key=lambda x: 10000 * symm_bidentate_names.count(x)
+ Component.list().index(x),
reverse=True,
)
):
cc_type += "(%s)" % symmbet[i]
this_name += "(%s)" % symbi_lig
if symm_bidentate_names.count(symbi_lig) > 1:
cc_type += "%i" % symm_bidentate_names.count(symbi_lig)
this_name += "%i" % symm_bidentate_names.count(symbi_lig)
asymm_bidentate_names = sorted(
asymm_bidentate_names,
key=lambda x: 10000 * asymm_bidentate_names.count(x)
+ Component.list().index(x),
reverse=True,
)
for i, asymbi_lig in enumerate(
sorted(
set(asymm_bidentate_names),
key=lambda x: 10000 * asymm_bidentate_names.count(x)
+ Component.list().index(x),
reverse=True,
)
):
cc_type += "(%s)" % asymmbet[i]
this_name += "(%s)" % asymbi_lig
if asymm_bidentate_names.count(asymbi_lig) > 1:
cc_type += "%i" % asymm_bidentate_names.count(asymbi_lig)
this_name += "%i" % asymm_bidentate_names.count(asymbi_lig)
# load the key atoms for ligand mapping from the template file
libdir = os.path.join(
AARONTOOLS, "coordination_complex", shape, cc_type
)
if not os.path.exists(libdir):
raise RuntimeError("no templates for %s %s" % (cc_type, shape))
geoms = []
for f in os.listdir(libdir):
mappings = np.loadtxt(
os.path.join(libdir, f), dtype=str, delimiter=",", ndmin=2
)
point_group, subset = f.rstrip(".csv").split("_")
# for each possible structure, create a copy of the original template shape
# attach ligands in the order they would appear in the formula
for i, mapping in enumerate(mappings):
geom_copy = geom.copy()
geom_copy.center = [geom_copy.atoms[0]]
geom_copy.components = [
Component([atom]) for atom in geom_copy.atoms[1:]
]
start = 0
for lig in monodentate_names:
key = mapping[start]
start += 1
comp = Component(lig)
d = 2.5
# adjust distance to key atoms to what they should be for the new ligand
try:
d = bo.bonds[bo.key(center, comp.key_atoms[0])]["1.0"]
except KeyError:
pass
geom_copy.change_distance(
geom_copy.atoms[0], key, dist=d, fix=1
)
# attach ligand
geom_copy.map_ligand(comp, key, minimize=minimize)
for key in comp.key_atoms:
geom_copy.atoms[0].connected.add(key)
key.connected.add(geom_copy.atoms[0])
for lig in symm_bidentate_names:
keys = mapping[start : start + 2]
start += 2
comp = Component(lig)
for old_key, new_key in zip(keys, comp.key_atoms):
d = 2.5
try:
d = bo.bonds[bo.key(center, new_key)]["1.0"]
except KeyError:
pass
geom_copy.change_distance(
geom_copy.atoms[0],
old_key,
dist=d,
fix=1,
as_group=False,
)
geom_copy.map_ligand(comp, keys, minimize=minimize)
for key in comp.key_atoms:
geom_copy.atoms[0].connected.add(key)
key.connected.add(geom_copy.atoms[0])
for lig in asymm_bidentate_names:
keys = mapping[start : start + 2]
start += 2
comp = Component(lig)
for old_key, new_key in zip(keys, comp.key_atoms):
d = 2.5
try:
d = bo.bonds[bo.key(center, new_key)]["1.0"]
except KeyError:
pass
geom_copy.change_distance(
geom_copy.atoms[0],
old_key,
dist=d,
fix=1,
as_group=False,
)
geom_copy.map_ligand(comp, keys, minimize=minimize)
for key in comp.key_atoms:
geom_copy.atoms[0].connected.add(key)
key.connected.add(geom_copy.atoms[0])
geom_copy.name = "%s-%i_%s_%s" % (
this_name,
i + 1,
point_group,
subset,
)
geoms.append(geom_copy)
return geoms, cc_type
@classmethod
def get_diastereomers(cls, geometry, minimize=True):
"""returns a list of all diastereomers for detected chiral centers"""
from AaronTools.finders import ChiralCenters, Bridgehead, NotAny, SpiroCenters
from AaronTools.ring import Ring
from AaronTools.substituent import Substituent
if not isinstance(geometry, Geometry):
geometry = Geometry(geometry)
updating_diastereomer = geometry.copy()
if not getattr(updating_diastereomer, "substituents", False):
updating_diastereomer.substituents = []
# we can invert any chiral center that isn't part of a
# fused ring unless it's a spiro center
chiral_centers = updating_diastereomer.find(ChiralCenters())
spiro_chiral = updating_diastereomer.find(SpiroCenters(), chiral_centers)
ring_centers = updating_diastereomer.find(
chiral_centers, Bridgehead(), NotAny(spiro_chiral)
)
chiral_centers = [c for c in chiral_centers if c not in ring_centers]
diastereomer_count = [2 for c in chiral_centers]
mod_array = []
for i in range(0, len(diastereomer_count)):
mod_array.append(1)
for j in range(i + 1, len(diastereomer_count)):
mod_array[i] *= diastereomer_count[j]
diastereomers = [updating_diastereomer.copy()]
previous_diastereomer = 0
for d in range(1, int(np.prod(diastereomer_count))):
for i, center in enumerate(chiral_centers):
flip = int(d / mod_array[i]) % diastereomer_count[i]
flip -= int(previous_diastereomer / mod_array[i]) % diastereomer_count[i]
if flip == 0:
continue
updating_diastereomer.change_chirality(center)
diastereomers.append(updating_diastereomer.copy())
previous_diastereomer = d
if minimize:
for diastereomer in diastereomers:
diastereomer.minimize_sub_torsion(increment=15)
return diastereomers
@staticmethod
def weighted_percent_buried_volume(
geometries, energies, temperature, *args, **kwargs
):
"""
Boltzmann-averaged percent buried volume
geometries - list of Geometry instances
energies - numpy array, energy in kcal/mol; ith energy corresponds to ith substituent
temperature - temperature in K
*args, **kwargs - passed to Geometry.percent_buried_volume()
"""
values = []
for geom in geometries:
values.append(geom.percent_buried_volume(*args, **kwargs))
rv = utils.boltzmann_average(
energies,
np.array(values),
temperature,
)
return rv
# attribute access
def _stack_coords(self, atoms=None):
"""
Generates a N x 3 coordinate matrix for atoms
Note: the matrix rows are copies of, not references to, the
Atom.coords objects. Run Geometry.update_geometry(matrix) after
using this method to save changes.
"""
if atoms is None:
atoms = self.atoms
else:
atoms = self.find(atoms)
rv = np.zeros((len(atoms), 3), dtype=float)
for i, a in enumerate(atoms):
rv[i] = a.coords[:]
return rv
@property
def elements(self):
"""returns list of elements composing the atoms in the geometry"""
return np.array([a.element for a in self.atoms])
@property
def coords(self):
"""
array of coordinates (read only)
"""
return self.coordinates()
@coords.setter
def coords(self, value):
"""
set coordinates
"""
for a, c in zip(self.atoms, value):
a.coords = np.array(c, dtype=float)
def coordinates(self, atoms=None):
"""
returns N x 3 coordinate matrix for requested atoms
(defaults to all atoms)
"""
if atoms is None:
return self._stack_coords()
return self._stack_coords(atoms)
# utilities
def __str__(self):
s = ""
for a in self:
s += a.__str__() + "\n"
return s
def __repr__(self):
"""string representation"""
s = ""
for a in self:
s += a.__repr__() + "\n"
return s
def __eq__(self, other):
"""
two geometries equal if:
same number of atoms
same numbers of elements
coordinates of atoms similar
"""
if id(self) == id(other):
return True
if len(self.atoms) != len(other.atoms):
return False
self_eles = [atom.element for atom in self.atoms]
other_eles = [atom.element for atom in other.atoms]
self_counts = {ele: self_eles.count(ele) for ele in set(self_eles)}
other_counts = {ele: other_eles.count(ele) for ele in set(other_eles)}
if self_counts != other_counts:
return False
rmsd = self.RMSD(other, sort=False)
return rmsd < COORD_THRESHOLD
def __add__(self, other):
if isinstance(other, Atom):
other = [other]
elif not isinstance(other, list):
other = other.atoms
self.atoms += other
return self
def __sub__(self, other):
if isinstance(other, Atom):
other = [other]
elif not isinstance(other, list):
other = other.atoms
for o in other:
self.atoms.remove(o)
for a in self.atoms:
if a.connected & set(other):
a.connected = a.connected - set(other)
return self
def __iter__(self):
self._iter_idx = -1
return self
def __next__(self):
if self._iter_idx + 1 < len(self.atoms):
self._iter_idx += 1
return self.atoms[self._iter_idx]
raise StopIteration
def __len__(self):
return len(self.atoms)
def __setattr__(self, attr, val):
if attr == "_hashed" and not val:
raise RuntimeError("can only set %s to True" % attr)
if not self._hashed or (self._hashed and attr != "atoms"):
super().__setattr__(attr, val)
else:
raise RuntimeError(
"cannot change atoms attribute of HashableGeometry"
)
def __hash__(self):
# hash depends on atom elements, connectivity, order, and coordinates
# reorient using principle axes
coords = self.coords
coords -= self.COM()
mat = np.matmul(coords.T, coords)
vals = np.linalg.svd(mat, compute_uv=False)
t = [int(v * 3) for v in vals]
for atom, coord in zip(self.atoms, coords):
# only use the first 3 decimal places of coordinates b/c numerical issues
t.append(int(atom.get_neighbor_id()))
if not atom._hashed:
atom.connected = frozenset(atom.connected)
atom.coords.setflags(write=False)
atom._hashed = True
# make sure atoms don't move
# if atoms move, te hash value could change making it impossible to access
# items in a dictionary with this instance as the key
if not self._hashed:
self.LOG.warning(
"Geometry `%s` has been hashed and will no longer be editable.\n"
"Use Geometry.copy to get an editable duplicate of this instance",
self.name,
)
self.atoms = tuple(self.atoms)
self._hashed = True
return hash(tuple(t))
def tag(self, tag, targets=None):
if targets is None:
targets = self.atoms
else:
targets = self.find(targets)
for atom in targets:
atom.tags.add(tag)
def write(self, name=None, *args, **kwargs):
"""
Write geometry to a file
:name: defaults to self.name
:style: defaults to xyz
*args and **kwargs for styles:
xyz
:append: True/False
com
:step:
:theory: the Theory specification object
:**kwargs: Additional route arguments in key=val format
"""
tmp = self.name
if name is not None:
self.name = name
out = FileWriter.write_file(self, *args, **kwargs)
self.name = tmp
if out is not None:
return out
def copy(self, atoms=None, name=None, comment=None, copy_atoms=True):
"""
creates a new copy of the geometry
parameters:
atoms (list): defaults to all atoms
name (str): defaults to NAME_copy
"""
if name is None:
name = self.name
if comment is None:
comment = self.comment
atoms = self._fix_connectivity(atoms, copy=copy_atoms)
if hasattr(self, "components") and self.components is not None and comment is None:
self.fix_comment()
return Geometry(atoms, name, comment=comment)
def parse_comment(self):
"""
Saves auxillary data found in comment line
"""
if not self.comment:
return {}
rv = {}
# constraints
match = re.search("F:([0-9;-]+)", self.comment)
if match is not None:
rv["constraint"] = []
for a in self.atoms:
a.constraint = set([])
match = match.group(1).split(";")
for m in match:
if m == "":
continue
m = m.split("-")
m = [int(i) for i in m]
if len(m) == 2:
for i, j in zip(m[:-1], m[1:]):
a = self.find(str(i))[0]
b = self.find(str(j))[0]
a.constraint.add((b, a.dist(b)))
b.constraint.add((a, b.dist(a)))
rv["constraint"] += [m]
# active centers
match = re.search("C:([0-9,]+)", self.comment)
if match is not None:
rv["center"] = []
match = match.group(1).split(",")
for m in match:
if m == "":
continue
a = self.atoms[int(m) - 1]
a.add_tag("center")
rv["center"] += [a]
# ligand
match = re.search("L:([0-9;,-]+)", self.comment)
if match is not None:
rv["ligand"] = []
match = match.group(1).split(";")
for submatch in match:
tmp = []
for m in submatch.split(","):
if m == "":
continue
if "-" not in m:
a = self.atoms[int(m) - 1]
tmp += [a]
continue
m = m.split("-")
for i in range(int(m[0]) - 1, int(m[1])):
try:
a = self.atoms[i]
except IndexError:
continue
tmp += [a]
rv["ligand"] += [tmp]
# key atoms
match = re.search("K:([0-9,;]+)", self.comment)
if match is not None:
rv["key_atoms"] = []
match = match.group(1).split(";")
for m in match:
if m == "":
continue
m = m.split(",")
for i in m:
if i == "":
continue
rv["key_atoms"] += [int(i) - 1]
self.other = rv
return rv
def fix_comment(self):
if not hasattr(self, "components"):
return
elif self.components is None:
self.detect_components()
new_comment = ""
# center
if self.center:
new_comment += "C:"
for c in self.center:
new_comment += "{},".format(self.atoms.index(c) + 1)
else:
new_comment = new_comment[:-1]
# key atoms
new_comment += " K:"
for frag in sorted(self.components):
tmp = ""
for key in sorted(frag.key_atoms, key=self.atoms.index):
tmp += "{},".format(self.atoms.index(key) + 1)
if tmp:
new_comment += tmp[:-1] + ";"
if new_comment[-3:] == " K:":
new_comment = new_comment[:-3]
else:
new_comment = new_comment[:-1]
# constrained bonds
constrained = self.get_constraints()
if constrained:
new_comment += " F:"
for cons in constrained:
ids = [cons[0] + 1]
ids += [cons[1] + 1]
new_comment += "{}-{};".format(*sorted(ids))
else:
new_comment = new_comment[:-1]
# components
if self.components:
new_comment += " L:"
for lig in sorted(self.components):
ids = sorted([1 + self.atoms.index(a) for a in lig])
tmp = []
for i in ids:
if i == ids[0]:
tmp = [i]
continue
if i == tmp[-1] + 1:
tmp += [i]
elif len(tmp) == 1:
new_comment += "{},".format(tmp[0])
tmp = [i]
else:
new_comment += "{}-{},".format(tmp[0], tmp[-1])
tmp = [i]
if len(tmp) == 1:
new_comment += "{},".format(tmp[0])
else:
new_comment += "{}-{},".format(tmp[0], tmp[-1])
new_comment = new_comment[:-1] + ";"
new_comment = new_comment[:-1]
# save new comment (original comment still in self.other)
self.comment = new_comment
def _flag(self, flag, targets=None):
"""
freezes targets if <flag> is True,
relaxes targets if <flag> is False
"""
if isinstance(targets, Config):
if targets._changed_list is not None:
targets = targets._changed_list
else:
raise RuntimeError(
"Substitutions/Mappings requested, but not performed"
)
if targets is not None:
try:
targets = self.find(targets)
except LookupError:
targets = self.atoms
else:
targets = self.atoms
for a in targets:
a.flag = flag
return
def freeze(self, targets=None):
"""
freezes atoms in the geometry
"""
self._flag(True, targets)
def relax(self, targets=None):
"""
relaxes atoms in the geometry
"""
self._flag(False, targets)
def get_constraints(self, as_index=True):
rv = {}
for i, a in enumerate(self.atoms[:-1]):
if not a.constraint:
continue
for j, b in enumerate(self.atoms[i:]):
for atom, dist in a.constraint:
if b == atom:
if as_index:
rv[(i, i + j)] = dist
else:
rv[(a, b)] = dist
break
return rv
def get_connectivity(self):
rv = []
for atom in self.atoms:
rv += [atom.connected]
return rv
def get_frag_list(self, targets=None, max_order=None):
"""
find fragments connected by only one bond
(both fragments contain no overlapping atoms)
"""
if targets:
atoms = self.find(targets)
else:
atoms = self.atoms
frag_list = []
for i, a in enumerate(atoms[:-1]):
for b in atoms[i + 1 :]:
if b not in a.connected:
continue
frag_a = self.get_fragment(a, b)
frag_b = self.get_fragment(b, a)
if sorted(frag_a) == sorted(frag_b):
continue
if len(frag_a) == 1 and frag_a[0].element == "H":
continue
if len(frag_b) == 1 and frag_b[0].element == "H":
continue
if max_order is not None and a.bond_order(b) > max_order:
continue
if (frag_a, a, b) not in frag_list:
frag_list += [(frag_a, a, b)]
if (frag_b, b, a) not in frag_list:
frag_list += [(frag_b, b, a)]
return frag_list
def detect_substituents(self):
"""sets self.substituents to a list of substituents"""
from AaronTools.substituent import Substituent
# TODO: allow detection of specific substituents only
# -check fragment length and elements against
# that of the specified substituent
# copy-pasted from Component.detect_backbone, but
# removed stuff that refers to the center/backbone
if not hasattr(self, "substituents") or self.substituents is None:
self.substituents = []
frag_list = self.get_frag_list()
new_tags = {} # hold atom tag options until assignment determined
subs_found = {} # for testing which sub assignment is best
sub_atoms = set([]) # holds atoms assigned to substituents
for frag_tup in sorted(frag_list, key=lambda x: len(x[0])):
frag, start, end = frag_tup
if frag[0] != start:
frag = self.reorder(start=start, targets=frag)[0]
# try to find fragment in substituent library
try:
sub = Substituent(frag, end=end)
except LookupError:
continue
# substituents with more than half of self's atoms are ignored
if len(frag) > len(self.atoms) - len(frag):
continue
# save atoms and tags if found
sub_atoms = sub_atoms.union(set(frag))
subs_found[sub.name] = len(sub.atoms)
for a in sub.atoms:
if a in new_tags:
new_tags[a] += [sub.name]
else:
new_tags[a] = [sub.name]
# save substituent
self.substituents += [sub]
# tag substituents
for a in new_tags:
tags = new_tags[a]
if len(tags) > 1:
# if multiple substituent assignments possible,
# want to keep the largest one (eg: tBu instead of Me)
sub_length = []
for t in tags:
sub_length += [subs_found[t]]
max_length = max(sub_length)
if max_length < 0:
max_length = min(sub_length)
keep = sub_length.index(max_length)
a.add_tag(tags[keep])
else:
a.add_tag(tags[0])
def find(self, *args, debug=False):
"""
finds atom in geometry
Parameters:
*args are tags, names, elements, or a Finder subclass
args=(['this', 'that'], 'other') will find atoms for which
('this' || 'that') && 'other' == True
Returns:
[Atom()] or []
Raises:
LookupError when the tags/names provided do not exist
However, it will return empty list if valid tag/names were provided
but were screened out using the && argument form
"""
def _find(arg):
"""find a single atom"""
# print(arg)
if isinstance(arg, Atom):
# print("atom")
return [arg]
rv = []
if isinstance(arg, Finder):
# print("finder")
rv += arg.get_matching_atoms(self.atoms, self)
name_str = re.compile("^(\*|\d)+(\.?\*|\.\d+)*$")
if isinstance(arg, str) and name_str.match(arg) is not None:
# print("name")
test_name = arg.replace(".", "\.")
test_name = test_name.replace("*", "(\.?\d+\.?)*")
test_name = re.compile("^" + test_name + "$")
# this is a name
for a in self.atoms:
if test_name.search(a.name) is not None:
rv += [a]
elif arg == "all":
rv += [a for a in self.atoms]
elif isinstance(arg, str) and len(arg.split(",")) > 1:
# print("comma list")
list_style = arg.split(",")
if len(list_style) > 1:
for i in list_style:
if len(i.split("-")) > 1:
rv += _find_between(i)
else:
rv += _find(i)
elif (
isinstance(arg, str)
and len(arg.split("-")) > 1
and not re.search("[A-Za-z]", arg)
):
# print("range list")
rv += _find_between(arg)
elif isinstance(arg, str) and arg in ELEMENTS:
# print("element")
# this is an element
for a in self.atoms:
if a.element == arg:
rv += [a]
else:
# print("tag")
# this is a tag
for a in self.atoms:
if arg in a.tags:
rv += [a]
return rv
def _find_between(arg):
"""find sequence of atoms"""
def _name2ints(name):
name = name.split(".")
return [int(i) for i in name]
a1, a2 = tuple(arg.split("-"))
a1 = _find(a1)[0]
a2 = _find(a2)[0]
rv = []
for a in self.atoms:
# keep if a.name is between a1.name and a2.name
test_name = _name2ints(a.name)
a1_name = _name2ints(a1.name)
a2_name = _name2ints(a2.name)
for tn, a1n, a2n in zip(test_name, a1_name, a2_name):
if tn < a1n:
# don't want if test atom sorts before a1
break
if tn > a2n:
# don't want if test atom sorts after a2
break
else:
rv += _find(a)
return rv
if len(args) == 1:
if isinstance(args[0], tuple):
args = args[0]
rv = []
for a in args:
if hasattr(a, "__iter__") and not isinstance(a, str):
# argument is a list of sub-arguments
# OR condition
tmp = []
for i in a:
tmp += _find(i)
rv += [tmp]
else:
rv += [_find(a)]
# error if no atoms found (no error if AND filters out all found atoms)
if len(rv) == 1:
if len(rv[0]) == 0:
raise LookupError(
"Could not find atom: %s on\n%s\n%s"
% ("; ".join([str(x) for x in args]), self.name, str(self))
)
return rv[0]
# exclude atoms not fulfilling AND requirement
tmp = []
for i in rv[0]:
good = True
for j in rv[1:]:
if i not in j:
good = False
if good:
tmp += [i]
return tmp
def find_exact(self, *args):
"""
finds exactly the same number of atoms as arguments used.
Raises LookupError if wrong number of atoms found
"""
rv = []
err = "Wrong number of atoms found: "
is_err = False
for arg in args:
try:
a = self.find(arg)
except LookupError:
a = []
if len(a) != 1:
is_err = True
err += "{} (found {}), ".format(arg, len(a))
else:
rv += a
if is_err:
err = err[:-2]
raise LookupError(err)
return tuple(rv)
def _fix_connectivity(self, atoms=None, copy=True):
"""
for fixing the connectivity for a set of atoms when grabbing
a fragment or copying atoms, ensures atom references are sane
:atoms: the atoms to fix connectivity for; connections to atoms
outside of this list are severed in the resulting list
:copy: perform a deepcopy of the atom list
"""
if atoms is None:
atoms = self.atoms
else:
atoms = self.find(atoms)
connectivity = []
for a in atoms:
connectivity += [
[atoms.index(i) for i in a.connected if i in atoms]
]
if copy:
atoms = [a.copy() for a in atoms]
for a, con in zip(atoms, connectivity):
a.connected = set([])
for c in con:
a.connected.add(atoms[c])
return atoms
def refresh_connected(self, targets=None, threshold=None):
"""
reset connected atoms
atoms are connected if their distance from each other is less than
the sum of their covalent radii plus a threshold
"""
# clear current connectivity
if targets is None:
targets = self.atoms
else:
targets = self.find(targets)
# reset the connectivity
for a in targets:
if targets is not self.atoms:
for b in a.connected:
b.connected.discard(a)
a.connected = set([])
D = distance_matrix(
self.coordinates(targets), self.coordinates(targets)
)
# determine connectivity
for i, a in enumerate(targets):
for j, b in enumerate(targets[:i]):
if a.dist_is_connected(b, D[i, j], threshold):
a.add_bond_to(b)
def refresh_ranks(self, invariant=True):
rank = self.canonical_rank(invariant=invariant)
for a, r in zip(self.atoms, rank):
a._rank = r
return
def get_invariants(self, heavy_only=False):
"""
returns a list of invariants for the specified targets
see Atom.get_invariant for some more details
"""
targets = self.atoms
if heavy_only:
targets = [a for a in targets if a.element != "H"]
indices = {a: i for i, a in enumerate(targets)}
target_set = set(targets)
coords = self.coordinates(targets)
def get_bo(atom1, atom2, dist):
"""
atom1, atom2 - Atom()
dist - float, distance between atom1 and atom2
returns a bond order (float) or 1 if we don't have
bond info for these atoms' elements
"""
try:
bonds = atom1._bo.bonds[atom1._bo.key(atom1, atom2)]
closest = 0, None
for order, length in bonds.items():
diff = abs(length - dist)
if closest[1] is None or diff < closest[1]:
closest = order, diff
return float(closest[0])
except KeyError:
return 1
atom_numbers = [ELEMENTS.index(a.element) for a in targets]
hydrogen_bonds = np.zeros(len(targets))
bo_sums = np.zeros(len(targets))
heavy_bonds = np.zeros(len(targets))
for i, atom1 in enumerate(targets):
if not atom1.connected:
continue
dists = np.linalg.norm(np.array([atom1.coords - atom2.coords for atom2 in atom1.connected]), axis=1)
for k, atom2 in enumerate(atom1.connected):
if atom2 in target_set:
j = indices[atom2]
else:
j = None
if atom2.element == "H":
hydrogen_bonds[i] += 1
else:
heavy_bonds[i] += 1
if atom1.element == "H":
hydrogen_bonds[j] += 1
elif j is not None:
heavy_bonds[j] += 1
if atom1.element != "H" or atom2.element != "H":
bond_order = get_bo(atom1, atom2, dists[k])
if atom2.element != "H":
bo_sums[i] += bond_order
if j is not None and atom1.element != "H":
bo_sums[j] += bond_order
elif j is None and atom2.element == "H":
hydrogen_bonds[i] += 1
elif j is None:
heavy_bonds[i] += 1
bo_sums[i] += bond_order
invariants = []
for nconn, nB, z, nH in zip(
heavy_bonds, bo_sums, atom_numbers, hydrogen_bonds
):
invariants.append(
"{:01d}{:03d}{:03d}{:01d}".format(
int(nconn), int(nB * 10), int(z), int(nH)
)
)
return invariants
def canonical_rank(
self, heavy_only=False, break_ties=True, update=True, invariant=True
):
"""
determine canonical ranking for atoms
invariant: bool - if True, use invariant described in J. Chem. Inf. Comput. Sci. 1989, 29, 2, 97–101
(DOI: 10.1021/ci00062a008)
if False, use neighbor IDs
algorithm described in J. Chem. Inf. Model. 2015, 55, 10, 2111–2120
(DOI: 10.1021/acs.jcim.5b00543)
"""
CITATION = "doi:10.1021/ci00062a008"
self.LOG.citation(CITATION)
CITATION = "doi:10.1021/acs.jcim.5b00543"
self.LOG.citation(CITATION)
primes = Primes.list(len(self.atoms))
# list of atoms we are ranking
atoms = []
# list of ranks corresponding to each atom
ranks = []
# index of each atom (faster than using atoms.index,
# particularly for larger structures
indices = dict()
# set of atoms for performance reasons
atoms_set = set()
# using the catalyst's center can make it difficult
# to compare C2 symmetric ligands
# center = list(filter(lambda x: "center" in x.tags, self))
# if center:
# center = self.COM(targets=center)
# else:
center = self.COM()
def neighbors_rank(ranks):
# partitions key is product of rank and neighbors' rank
# use prime numbers for product so products are distinct
# eg: primes[2]*primes[2] != primes[1]*primes[4]
# some high-symmetry molecules can get a rank greater than
# the number of atoms
# I've had this issue with adamantane (Td)
# this is a lazy fix that reduces the rank of some atoms by 1
while max(ranks) >= len(ranks):
for i in range(1, max(ranks) + 1):
if ranks.count(i - 1) == 0:
for j in range(1, max(ranks)):
if ranks[j] >= i:
ranks[j] -= 1
partitions = {}
for i, a in enumerate(atoms):
key = primes[ranks[i]]
for b in a.connected.intersection(atoms_set):
# print(indices[b], ranks[indices[b]])
key *= primes[ranks[indices[b]]]
partitions.setdefault(ranks[i], {})
partitions[ranks[i]].setdefault(key, [])
partitions[ranks[i]][key] += [i]
return update_ranks(ranks, partitions)
def update_ranks(ranks, partitions):
new_ranks = ranks.copy()
for rank, key_dict in partitions.items():
if len(key_dict) == 1:
continue
for key in sorted(key_dict.keys()):
for idx in key_dict[key]:
new_ranks[idx] = rank
rank += len(key_dict[key])
return new_ranks
def tie_break(ranks):
"""
Uses atom angles around COM -> shared_atom axis to break ties[
Does not break symmetry (eg: pentane carbons still [0, 2, 4, 2, 0]
because C2 and C4 are ~180 deg apart relative to COM-C5 axis)
"""
def get_angle(vi, vj, norm):
dot = np.dot(vi, vj)
cross = np.cross(vi, vj)
det = np.dot(norm, cross)
rv = np.arctan2(det, dot)
return round(rv, 1)
def get_start(connected, center, norm):
# if we can, use the COM of tied atoms as reference 0-deg
start = self.COM(targets=[atoms[c] for c in connected])
start -= center
if np.linalg.norm(np.cross(start, norm)) > 1e-2:
return start
# if there's one atom that is closest/farthest to center,
# use that as start
start_min = None, None
start_max = None, None
for c in connected:
dist = np.linalg.norm(atoms[c].coords - center)
if start_min[0] is None or dist < start_min[1]:
start_min = [c], dist
elif dist == start_min[1]:
start_min = start_min[0] + [c], dist
if start_max[0] is None or dist < start_max[1]:
start_max = [c], dist
elif dist == start_max[1]:
start_max = start_max[0] + [c], dist
if len(start_min[0]) == 1:
start = atoms[start_min[0][0]].coords - center
return start
if len(start_max[0]) == 1:
start = atoms[start_max[0][0]].coords - center
return start
# otherwise, try to use COM of equally close/far atoms
if len(start_min[0]) < len(connected):
start = self.COM(targets=[atoms[c] for c in start_min[0]])
start -= center
if np.linalg.norm(np.cross(start, norm)) > 1e-2:
return start
if len(start_max[0]) < len(connected):
start = self.COM(targets=[atoms[c] for c in start_max[0]])
start -= center
if np.linalg.norm(np.cross(start, norm)) > 1e-2:
return start
# if all else fails, just use the first atom I guess...
return atoms[connected[0]].coords - center
partitions = {}
for i, rank in enumerate(ranks):
partitions.setdefault(rank, {})
partitions[rank].setdefault(rank, [])
partitions[rank][rank] += [i]
new_partitions = partitions.copy()
# norm = self.get_principle_axes()
# norm = norm[1][:, 0] - center
for rank, rank_dict in partitions.items():
idx_list = rank_dict[rank]
if len(idx_list) == 1:
continue
# split ties into groups connected to same atom
groups = {}
for i in idx_list[:-1]:
a = atoms[i]
for j in idx_list[1:]:
b = atoms[j]
connected = a.connected & b.connected
if len(connected) == 1:
k = connected.pop()
if k in atoms:
k = indices[k]
else:
continue
groups.setdefault(k, set([i]))
groups[k] |= set([j])
# atoms in each group sorted in counter clockwise order
# around axis centered at shared atom and orthogonal to COM
for shared_idx, connected in groups.items():
connected = sorted(connected)
start = atoms[shared_idx].coords - center
norm = np.cross(start, center)
angles = {}
for c in connected:
this = atoms[c].coords - center
angle = get_angle(start, this, norm)
angles.setdefault(angle, [])
angles[angle] += [c]
if len(angles) == 1 and atoms[shared_idx].connected - set(
[atoms[c] for c in connected]
):
tmp_center = self.COM(
atoms[shared_idx].connected
- set([atoms[c] for c in connected])
)
start = atoms[shared_idx].coords - tmp_center
norm = np.cross(start, tmp_center)
angles = {}
for c in connected:
this = atoms[c].coords - tmp_center
angle = get_angle(start, this, norm)
angles.setdefault(angle, [])
angles[angle] += [c]
for i, angle in enumerate(sorted(angles.keys())):
new_partitions[rank].setdefault(rank + i, [])
new_partitions[rank][rank + i] += angles[angle]
for idx in angles[angle]:
if idx in new_partitions[rank][rank]:
new_partitions[rank][rank].remove(idx)
return update_ranks(ranks, new_partitions)
# rank all atoms the same initially
c = 0
for a in self.atoms:
if heavy_only and a.element == "H":
continue
atoms += [a]
ranks += [0]
indices[a] = c
c += 1
atoms_set = set(atoms)
# partition and re-rank using invariants
partitions = {}
if invariant:
invariants = self.get_invariants(heavy_only=heavy_only)
else:
invariants = [a.get_neighbor_id() for a in atoms]
for i, (a, id) in enumerate(zip(atoms, invariants)):
partitions.setdefault(id, [])
partitions[id] += [i]
new_rank = 0
for key in sorted(partitions.keys()):
idx_list = partitions[key]
for idx in idx_list:
ranks[idx] = new_rank
new_rank += len(idx_list)
# re-rank using neighbors until no change
for i in range(0, min(500, len(ranks))):
new_ranks = neighbors_rank(ranks)
if ranks == new_ranks:
break
ranks = new_ranks
else:
self.LOG.warning(
"Max cycles reached in canonical sorting (neighbor-ranks)"
)
# break ties using spatial positions
# AND update neighbors until no change
if break_ties:
for i in range(0, min(500, len(ranks))):
new_ranks = tie_break(ranks)
new_ranks = neighbors_rank(new_ranks)
if ranks == new_ranks:
break
ranks = new_ranks
else:
self.LOG.warning(
"Max cycles reached in canonical sorting (tie-breaking)"
)
return ranks
def element_counts(self):
eles = dict()
for ele in self.elements:
if ele not in eles:
eles[ele] = 0
eles[ele] += 1
return eles
def reorder(
self,
start=None,
targets=None,
heavy_only=False,
):
"""
Returns:
list(ordered_targets), list(non_targets)
Depth-first reorder of atoms based on canonical ranking
"""
if not targets:
targets = self.atoms
else:
targets = self.find(targets)
if heavy_only:
targets = [t for t in targets if t.element != "H"]
non_targets = [a for a in self.atoms if a not in targets]
# get starting atom
if not start:
order = [sorted(targets)[0]]
else:
order = sorted(self.find(start))
start = sorted(order)
stack = []
for s in start:
stack += sorted(s.connected)
atoms_left = set(targets) - set(order) - set(stack)
while len(stack) > 0:
this = stack.pop()
if heavy_only and this.element == "H":
continue
if this in order:
continue
order += [this]
connected = set(this.connected & atoms_left)
atoms_left -= connected
stack += sorted(connected)
if not stack and atoms_left:
stack += [sorted(atoms_left)[0]]
atoms_left -= set(stack)
return order, non_targets
def rebuild(self):
atoms = []
if self.components:
if self.center:
atoms += self.center
for comp in sorted(self.components):
comp.rebuild()
atoms += comp.atoms
self.atoms = atoms
self.fix_comment()
self.refresh_ranks()
def detect_components(self, center=None):
from AaronTools.component import Component
self.components = []
if center is None:
self.center = []
else:
self.center = self.find(center)
# get center
if not self.center:
for a in self.atoms:
if a.element in TMETAL.keys():
# detect transition metal center
if a not in self.center:
self.center += [a]
a.add_tag("center")
if "center" in a.tags:
# center provided by comment line in xyz file
if a not in self.center:
self.center += [a]
# label key atoms:
for i, a in enumerate(self.atoms):
if "key_atoms" not in self.other:
break
if i in self.other["key_atoms"]:
a.add_tag("key")
else:
del self.other["key_atoms"]
# get components
self.components = self.detect_fragments(self.atoms)
# rename
for i, frag in enumerate(self.components):
name = self.name + ".{:g}".format(
min(
[
float(a.name)
if utils.is_num(a.name)
else frag.index(a)
for a in frag
]
)
)
self.components[i] = Component(frag, name)
self.rebuild()
return
def detect_fragments(self, targets, avoid=None):
"""
Returns a list of Geometries in which the connection to other
atoms in the larger geometry must go through the center atoms
eg: L1--C--L2 will give two fragments, L1 and L2
( /
L1/
"""
def add_tags(frag):
for f in frag:
found.add(f)
for c in self.center:
if f in c.connected:
f.add_tag("key")
if avoid is None and self.center:
avoid = self.center
found = set([])
rv = []
if "ligand" in self.other:
for ligand in self.other["ligand"]:
frag = set(self.find(ligand))
frag -= found
add_tags(frag)
rv += [sorted(frag)]
found.union(frag)
for a in targets:
if a in found:
continue
if avoid:
if a in avoid:
continue
frag = set(self.get_fragment(a, avoid))
frag -= found
add_tags(frag)
rv += [sorted(frag)]
found.union(frag)
else:
frag = set([a])
queue = a.connected.copy()
while queue:
this = queue.pop()
if this in frag:
continue
frag.add(this)
queue = queue.union(this.connected)
frag -= found
add_tags(frag)
rv += [sorted(frag)]
found = found.union(frag)
return rv
def shortest_path(self, atom1, atom2, avoid=None):
"""
Uses Dijkstra's algorithm to find shortest path between atom1 and atom2
avoid: atoms to avoid on the path
"""
a1 = self.find(atom1)[0]
a2 = self.find(atom2)[0]
if avoid is None:
path = utils.shortest_path(self, a1, a2)
else:
avoid = self.find(avoid)
graph = [
[
self.atoms.index(j)
for j in i.connected
if j in self.atoms and j not in avoid
]
if i not in avoid
else []
for i in self.atoms
]
path = utils.shortest_path(
graph, self.atoms.index(a1), self.atoms.index(a2)
)
if not path:
raise LookupError(
"could not determine best path between {} and {}".format(
atom1, atom2
)
)
return [self.atoms[i] for i in path]
# geometry measurement
def bond(self, a1, a2):
"""takes two atoms and returns the bond vector"""
a1, a2 = self.find_exact(a1, a2)
return a1.bond(a2)
def angle(self, a1, a2, a3=None):
"""returns a1-a2-a3 angle"""
a1, a2, a3 = self.find_exact(a1, a2, a3)
return a2.angle(a1, a3)
def dihedral(self, a1, a2, a3, a4):
"""measures dihedral angle of a1 and a4 with respect to a2-a3 bond"""
a1, a2, a3, a4 = self.find_exact(a1, a2, a3, a4)
b12 = a1.bond(a2)
b23 = a2.bond(a3)
b34 = a3.bond(a4)
dihedral = np.cross(np.cross(b12, b23), np.cross(b23, b34))
dihedral = np.dot(dihedral, b23) / np.linalg.norm(b23)
dihedral = np.arctan2(
dihedral, np.dot(np.cross(b12, b23), np.cross(b23, b34))
)
return dihedral
def COM(self, targets=None, heavy_only=False, mass_weight=True):
"""
calculates center of mass of the target atoms
returns a vector from the origin to the center of mass
parameters:
targets (list) - the atoms to use in calculation (defaults to all)
heavy_only (bool) - exclude hydrogens (defaults to False)
"""
# get targets
if targets:
targets = self.find(targets)
else:
targets = list(self.atoms)
# screen hydrogens if necessary
if heavy_only:
targets = [a for a in targets if a.element != "H"]
coords = self.coordinates(targets)
if mass_weight:
total_mass = 0
for i in range(0, len(coords)):
coords[i] *= targets[i].mass()
total_mass += targets[i].mass()
# COM = (1/M) * sum(m * r) = sum(m*r) / sum(m)
center = np.mean(coords, axis=0)
if mass_weight and total_mass:
return center * len(targets) / total_mass
return center
def RMSD(
self,
ref,
align=False,
heavy_only=False,
sort=True,
targets=None,
ref_targets=None,
debug=False,
weights=None,
ref_weights=None,
):
"""
calculates the RMSD between two geometries
Returns: rmsd (float)
:ref: (Geometry) the geometry to compare to
:align: (bool) if True (default), align self to other;
if False, just calculate the RMSD
:heavy_only: (bool) only use heavy atoms (default False)
:targets: (list) the atoms in `self` to use in calculation
:ref_targets: (list) the atoms in the reference geometry to use
:sort: (bool) canonical sorting of atoms before comparing
:debug: returns RMSD and Geometry([ref_targets]), Geometry([targets])
:weights: (list(float)) weights to apply to targets
:ref_weights: (list(float)) weights to apply to ref_targets
"""
def _RMSD(ref, other):
"""
ref and other are lists of atoms
returns rmsd, angle, vector
rmsd (float)
angle (float) angle to rotate by
vector (np.array(float)) the rotation axis
"""
matrix = np.zeros((4, 4), dtype=np.float64)
for i, a in enumerate(ref):
pt1 = a.coords
try:
pt2 = other[i].coords
except IndexError:
break
matrix += utils.quat_matrix(pt2, pt1)
eigenval, eigenvec = np.linalg.eigh(matrix)
val = eigenval[0]
vec = eigenvec.T[0]
if val > 0:
# val is the SD
rmsd = np.sqrt(val / len(ref))
else:
# negative numbers that are basically zero, like -1e-16
rmsd = 0
# sometimes it freaks out if the coordinates are right on
# top of each other and gives overly large rmsd/rotation
# I think this is a numpy precision problem, may want to
# try scipy.linalg to see if that helps?
tmp = sum(
[
np.linalg.norm(a.coords - b.coords) ** 2
for a, b in zip(ref, other)
]
)
tmp = np.sqrt(tmp / len(ref))
if tmp < rmsd:
rmsd = tmp
vec = np.array([0, 0, 0])
return rmsd, vec
# get target atoms
tmp = targets
if targets is not None:
targets = self.find(targets)
else:
targets = self.atoms
if ref_targets is not None:
ref_targets = ref.find(ref_targets)
elif tmp is not None:
ref_targets = ref.find(tmp)
else:
ref_targets = ref.atoms
# screen out hydrogens if heavy_only requested
if heavy_only:
targets = [a for a in targets if a.element != "H"]
ref_targets = [a for a in ref_targets if a.element != "H"]
this = Geometry([t.copy() for t in targets])
ref = Geometry([r.copy() for r in ref_targets])
if weights is not None:
for w, a in zip(weights, this.atoms):
a.coords *= w
if ref_weights is not None:
for w, a in zip(ref_weights, ref.atoms):
a.coords *= w
# align center of mass to origin
com = this.COM()
ref_com = ref.COM()
this.coord_shift(-com)
ref.coord_shift(-ref_com)
# try current ordering
min_rmsd = _RMSD(ref.atoms, this.atoms)
# try canonical ordering
if sort:
this.atoms = this.reorder()[0]
ref.atoms = ref.reorder()[0]
this_ranks = [a._rank for a in this.atoms]
ref_ranks = [a._rank for a in ref.atoms]
if any(this_ranks.count(r) > 1 for r in this_ranks) or any(
ref_ranks.count(r) > 1 for r in ref_ranks
):
# if there are atoms with the same rank, align both ref and this
# to their principle axes and use the distance between the atoms
# in the structures to determine the order
this_atoms = []
ref_atoms = [a for a in ref.atoms]
_, this_axes = this.get_principle_axes()
_, ref_axes = ref.get_principle_axes()
this_coords = this.coords - this.COM()
ref_coords = ref.coords - ref.COM()
# align this to ref using the matrix that aligns this's principle axes
# to ref's principle axes
H = np.dot(ref_coords.T, this_coords)
u, s, vh = np.linalg.svd(H, compute_uv=True)
d = 1.0
if np.linalg.det(np.matmul(vh.T, u.T)) < 0:
d = 1.0
m = np.diag([1.0, 1.0, d])
R = np.matmul(vh.T, m)
R = np.matmul(R, u.T)
this_coords = np.dot(this_coords, R)
# find the closest atom in this to the closest atom in ref
dist = distance_matrix(ref_coords, this_coords)
for i, r_a in enumerate(ref.atoms):
min_dist = None
match = None
for j, t_a in enumerate(this.atoms):
if t_a.element != r_a.element:
continue
if t_a in this_atoms:
continue
if min_dist is None or dist[i, j] < min_dist:
min_dist = dist[i, j]
match = t_a
if match is not None:
this_atoms.append(match)
else:
ref_atoms.remove(r_a)
# if we didn't find any matches or not all atoms are matched,
# use the original order
# otherwise, use the order determined by distances
if len(ref_atoms) == len(this_atoms) and ref_atoms:
res = _RMSD(ref_atoms, this_atoms)
else:
res = _RMSD(ref.atoms, this.atoms)
else:
res = _RMSD(ref.atoms, this.atoms)
if res[0] < min_rmsd[0]:
min_rmsd = res
rmsd, vec = min_rmsd
# return rmsd
if not align:
if debug:
return this, ref, rmsd, vec
else:
return rmsd
# or update geometry and return rmsd
self.coord_shift(-com)
if np.linalg.norm(vec) > 0:
self.rotate(vec)
self.coord_shift(ref_com)
if debug:
this.rotate(vec)
return this, ref, rmsd, vec
else:
return rmsd
def get_near(self, ref, dist, by_bond=False, include_ref=False):
"""
Returns: list of atoms within a distance or number of bonds of a
reference point, line, plane, atom, or list of atoms
:ref: the point (eg: [0, 0, 0]), line (eg: ['*', 0, 0]), plane
(eg: ['*', '*', 0]), atom, or list of atoms
:dist: the distance threshold or number of bonds away threshold, is an
inclusive upper bound (uses `this <= dist`)
:by_bond: if true, `dist` is interpreted as the number of bonds away
instead of distance in angstroms
NOTE: by_bond=True means that ref must be an atom or list of atoms
:include_ref: if Atom or list(Atom) given as ref, include these in the
returned list, (default=False, do not include ref in returned list)
"""
if dist < 0:
raise ValueError(
"Distance or number of bonds threshold must be positive"
)
if not hasattr(ref, "iter") and isinstance(ref, Atom):
ref = [ref]
rv = []
# find atoms within number of bonds away
if by_bond:
dist_err = "by_bond=True only applicable for integer bonds away"
ref_err = (
"by_bond=True only applicable for ref of type Atom() or "
"list(Atom())"
)
if int(dist) != dist:
raise ValueError(dist_err)
for r in ref:
if not isinstance(r, Atom):
raise TypeError(ref_err)
stack = set(ref)
rv = set([])
while dist > 0:
dist -= 1
new_stack = set([])
for s in stack:
rv = rv.union(s.connected)
new_stack = new_stack.union(s.connected)
stack = new_stack
if not include_ref:
rv = rv - set(ref)
return sorted(rv)
# find atoms within distance
if isinstance(ref, Atom):
ref = [ref.coords]
elif isinstance(ref, list):
new_ref = []
just_nums = []
for r in ref:
if isinstance(r, Atom):
new_ref += [r.coords]
elif isinstance(r, list):
new_ref += [r]
else:
just_nums += [r]
if len(just_nums) % 3 != 0:
raise ValueError(
"coordinates (or wildcards) must be passed in sets of "
"three: [x, y, z]"
)
else:
while len(just_nums) > 0:
new_ref += [just_nums[-3:]]
just_nums = just_nums[:-3]
mask = [False, False, False]
for r in new_ref:
for i, x in enumerate(r):
if x == "*":
mask[i] = True
r[i] = 0
for a in self.atoms:
coords = a.coords.copy()
for i, x in enumerate(mask):
if x:
coords[i] = 0
if np.linalg.norm(np.array(r, dtype=float) - coords) <= dist:
rv += [a]
if not include_ref:
for r in ref:
if isinstance(r, Atom) and r in rv:
rv.remove(r)
return rv
def get_principle_axes(self, mass_weight=True, center=None):
"""
Return: [principal moments], [principle axes]
"""
if center is None:
COM = self.COM(mass_weight=mass_weight)
else:
COM = center
I_CM = np.zeros((3, 3))
for a in self:
if mass_weight:
mass = a.mass()
else:
mass = 1
coords = a.coords - COM
I_CM[0, 0] += mass * (coords[1] ** 2 + coords[2] ** 2)
I_CM[1, 1] += mass * (coords[0] ** 2 + coords[2] ** 2)
I_CM[2, 2] += mass * (coords[0] ** 2 + coords[1] ** 2)
I_CM[0, 1] -= mass * (coords[0] * coords[1])
I_CM[0, 2] -= mass * (coords[0] * coords[2])
I_CM[1, 2] -= mass * (coords[1] * coords[2])
I_CM[1, 0] = I_CM[0, 1]
I_CM[2, 0] = I_CM[0, 2]
I_CM[2, 1] = I_CM[1, 2]
return np.linalg.eigh(I_CM)
def LJ_energy(self, other=None, use_prev_params=False):
"""
computes LJ energy using autodock parameters
use_prev_params - use same sigma/epsilon as the last time LJ_energy was called
useful for methods that make repetitive LJ_energy calls, like
minimize_torsion
"""
if (
use_prev_params
and self._sigmat is not None
and self._sigmat.shape != (len(self), len(other))
):
sigmat = self._sigmat
epsmat = self._epsmat
else:
sigmat = np.array(
[[a.rij(b) for a in self.atoms] for b in self.atoms]
)
epsmat = np.array(
[[a.eij(b) for a in self.atoms] for b in self.atoms]
)
if other is None or other is self:
D = distance_matrix(self.coords, self.coords)
np.fill_diagonal(D, 1)
else:
if hasattr(other, "coords"):
D = distance_matrix(self.coords, other.coords)
other = other.atoms
sigmat = np.array(
[[a.rij(b) for a in other] for b in self.atoms]
)
epsmat = np.array(
[[a.eij(b) for a in other] for b in self.atoms]
)
else:
D = distance_matrix(
self.coords, np.array([a.coords for a in other])
)
self._sigmat = sigmat
self._epsmat = epsmat
repmat = sigmat / D
repmat = repmat ** 2
repmat = repmat ** 3
attmat = repmat ** 2
if other is None or other is self:
nrgmat = np.tril(epsmat * (attmat - repmat), -1)
else:
nrgmat = epsmat * (attmat - repmat)
return np.sum(nrgmat)
def examine_constraints(self, thresh=None):
"""
Determines if constrained atoms are too close/ too far apart
Returns: (atom1, atom2, flag) where flag is 1 if atoms too close,
-1 if atoms to far apart (so one can multiply a distance to change
by the flag and it will adjust in the correct direction)
"""
rv = []
if thresh is None:
thresh = D_CUTOFF
constraints = self.get_constraints()
# con of form (atom_name_1, atom_name_2, original_distance)
for con in constraints:
if len(con) != 2:
continue
dist = self.atoms[con[0]].dist(self.atoms[con[1]])
if dist - constraints[con] > thresh:
# current > constraint: atoms too far apart
# want to move closer together
rv += [(con[0], con[1], -1)]
elif constraints[con] - dist > thresh:
# constraint > current: atoms too close together
# want to move farther apart
rv += [(con[0], con[1], 1)]
return rv
def compare_connectivity(self, ref, thresh=None, return_idx=False):
"""
Compares connectivity of self relative to ref
Returns: broken, formed
:broken: set of atom name pairs for which a bond broke
:formed: set of atom name pairs for which a bond formed
:ref: the structure to compare to (str(path), FileReader, or Geometry)
ref.atoms should be in the same order as self.atoms
:thresh: allow for connectivity changes as long as the difference
between bond distances is below a threshold
:by_name: if True (default) lookup atoms by name, otherwise compare
based on index in atom list
"""
broken = set([])
formed = set([])
if not isinstance(ref, Geometry):
ref = Geometry(ref)
not_found = set(self.atoms)
for i, r in enumerate(ref.atoms):
s = self.find(r.name)[0]
not_found.remove(s)
conn = set(self.find(i.name)[0] for i in r.connected)
if not conn ^ s.connected:
continue
for c in conn - s.connected:
if thresh is not None:
dist = r.dist(ref.find(c.name)[0]) - s.dist(c)
if abs(dist) <= thresh:
continue
if return_idx:
broken.add(tuple(sorted([i, self.atoms.index(c)])))
else:
broken.add(tuple(sorted([s.name, c.name])))
for c in s.connected - conn:
if thresh is not None:
dist = r.dist(ref.find(c.name)[0]) - s.dist(c)
if abs(dist) <= thresh:
continue
if return_idx:
broken.add(tuple(sorted([i, self.atoms.index(c)])))
else:
formed.add(tuple(sorted([s.name, c.name])))
return broken, formed
def percent_buried_volume(
self,
center=None,
targets=None,
radius=3.5,
radii="umn",
scale=1.17,
exclude=None,
method="lebedev",
rpoints=20,
apoints=1454,
min_iter=25,
basis=None,
n_threads=1,
):
"""
calculates % buried volume (%V_bur) using Monte-Carlo or Gauss-Legendre/Lebedev integration
see Organometallics 2008, 27, 12, 2679–2681 (DOI: 10.1021/om8001119) for details
center - center atom(s) or np.array of coordinates
if more than one atom is specified, the sphere will be centered on
the centroid between the atoms
targets - atoms to use in calculation, defaults to all non-center if there
is only one center, otherwise all atoms
radius - sphere radius around center atom
radii - "umn" or "bondi", VDW radii to use
can also be a dict() with atom symbols as the keys and
their respective radii as the values
scale - scale VDW radii by this
method - integration method (MC or lebedev)
rpoints - number of radial shells for Lebedev integration
apoints - number of angular points for Lebedev integration
min_iter - minimum number of iterations for MC integration
each iteration is a batch of 3000 points
iterations will continue beyond min_iter if the volume has not converged
basis - change of basis matrix
will cause %Vbur to be returned as a tuple for different quadrants (I, II, III, IV)
n_threads - number of threads to use for MC integration
using multiple threads doesn't benefit performance very much
"""
# NOTE - it would be nice to multiprocess the MC integration (or
# split up the shells for the Lebedev integration, but...
# python's multiprocessing doesn't let you spawn processes
# outside of the __name__ == '__main__' context
# determine center if none was specified
if center is None:
if self.center is None:
self.detect_components()
center = self.center
center_coords = self.COM(center)
else:
try:
center = self.find(center)
center_coords = self.COM(center)
except LookupError:
# assume an array was given
center_coords = center
# determine atoms if none were specified
if targets is None:
if center is None:
targets = self.atoms
else:
if len(center) == 1:
targets = [
atom for atom in self.atoms if atom not in center
]
else:
targets = [atom for atom in self.atoms]
else:
targets = self.find(targets)
# VDW radii to use
if isinstance(radii, dict):
radii_dict = radii
elif radii.lower() == "umn":
radii_dict = VDW_RADII
elif radii.lower() == "bondi":
radii_dict = BONDI_RADII
else:
raise RuntimeError(
"received %s for radii, must be umn or bondi" % radii
)
# list of scaled VDW radii for each atom that's close enough to
# the center of the sphere
radius_list = []
atoms_within_radius = []
# determine which atom's radii extend within the sphere
# reduces the number of distances we need to calculate
# also determine innermost and outermost atom edges (minr and maxr)
# so we can skip integration shells that don't contain atoms
minr = radius
maxr = 0.0
for atom in targets:
if exclude is not None and atom in exclude:
continue
d = np.linalg.norm(center_coords - atom.coords)
inner_edge = d - scale * radii_dict[atom.element]
outer_edge = inner_edge + 2 * scale * radii_dict[atom.element]
if inner_edge < radius:
atoms_within_radius.append(atom)
if inner_edge < minr:
minr = inner_edge
if outer_edge > maxr:
maxr = outer_edge
maxr = min(maxr, radius)
if minr < 0:
minr = 0
# sort atoms based on their distance to the center
# this makes is so we usually break out of looping over the atoms faster
atoms_within_radius.sort(
key=lambda a, c=center_coords: np.linalg.norm(a.coords - c)
)
for atom in atoms_within_radius:
radius_list.append(scale * radii_dict[atom.element])
radius_list = np.array(radius_list)
coords = self.coordinates(atoms_within_radius)
# Monte-Carlo integration
if method.lower() == "mc":
n_samples = 3000
def get_iter_vol(n_samples=n_samples):
"""get the buried points and total points for one MC batch"""
if basis is None:
buried_points = 0
tot_points = 0
else:
buried_points = np.zeros(8)
tot_points = np.zeros(8)
# get a random point uniformly distributed inside the sphere
# only sample points between minr and maxr because maybe that makes
# things converge faster
r = (maxr - minr) * np.random.uniform(0, 1, n_samples) ** (
1 / 3
)
r += minr
z = np.random.uniform(-1, 1, n_samples)
theta = np.arcsin(z) + np.pi / 2
phi = np.random.uniform(0, 2 * np.pi, n_samples)
x = r * np.sin(theta) * np.cos(phi)
y = r * np.sin(theta) * np.sin(phi)
z *= r
xyz = np.array([x, y, z]).T
if basis is not None:
# determine what quadrant this point is in, add it to the appropriate bin
map_xyz = np.dot(xyz, basis)
signs = np.sign(map_xyz)
oct_0 = np.where(np.dot(signs, [1, 1, 1]) > 2, 1, 0)
tot_points[0] += sum(oct_0)
oct_1 = np.where(np.dot(signs, [-1, 1, 1]) >= 2, 1, 0)
tot_points[1] += sum(oct_1)
oct_2 = np.where(np.dot(signs, [-1, -1, 1]) > 2, 1, 0)
tot_points[2] += sum(oct_2)
oct_3 = np.where(np.dot(signs, [1, -1, 1]) >= 2, 1, 0)
tot_points[3] += sum(oct_3)
oct_4 = np.where(np.dot(signs, [1, -1, -1]) > 2, 1, 0)
tot_points[4] += sum(oct_4)
oct_5 = np.where(np.dot(signs, [-1, -1, -1]) >= 2, 1, 0)
tot_points[5] += sum(oct_5)
oct_6 = np.where(np.dot(signs, [-1, 1, -1]) > 2, 1, 0)
tot_points[6] += sum(oct_6)
oct_7 = np.where(np.dot(signs, [1, 1, -1]) >= 2, 1, 0)
tot_points[7] += sum(oct_7)
xyz += center_coords
# see if the point is inside of any atom's
# scaled VDW radius
D = distance_matrix(xyz, coords)
diff_mat = D - radius_list
if basis is None:
buried_points += np.sum(np.any(diff_mat <= 0, axis=1))
else:
mask = np.any(diff_mat <= 0, axis=1)
buried_coords = map_xyz[mask]
signs = np.sign(buried_coords)
oct_0 = np.where(np.dot(signs, [1, 1, 1]) > 2, 1, 0)
buried_points[0] += sum(oct_0)
oct_1 = np.where(np.dot(signs, [-1, 1, 1]) > 2, 1, 0)
buried_points[1] += sum(oct_1)
oct_2 = np.where(np.dot(signs, [-1, -1, 1]) > 2, 1, 0)
buried_points[2] += sum(oct_2)
oct_3 = np.where(np.dot(signs, [1, -1, 1]) > 2, 1, 0)
buried_points[3] += sum(oct_3)
oct_4 = np.where(np.dot(signs, [1, -1, -1]) > 2, 1, 0)
buried_points[4] += sum(oct_4)
oct_5 = np.where(np.dot(signs, [-1, -1, -1]) > 2, 1, 0)
buried_points[5] += sum(oct_5)
oct_6 = np.where(np.dot(signs, [-1, 1, -1]) > 2, 1, 0)
buried_points[6] += sum(oct_6)
oct_7 = np.where(np.dot(signs, [1, 1, -1]) > 2, 1, 0)
buried_points[7] += sum(oct_7)
return buried_points, tot_points
dV = []
i = 0
if basis is None:
prev_vol = cur_vol = 0
buried_points = 0
tot_points = 0
else:
prev_vol = np.zeros(8)
cur_vol = np.zeros(8)
buried_points = np.zeros(8)
tot_points = np.zeros(8)
# determine %V_bur
# do at least 75000 total points, but keep going until
# the last 5 changes are all less than 1e-4
while i < min_iter or not (
all(dv < 2e-4 for dv in dV[-5:]) and np.mean(dV[-5:]) < 1e-4
):
if n_threads == 1:
iter_buried, iter_tot = get_iter_vol()
buried_points += iter_buried
tot_points += iter_tot
if basis is None:
cur_vol = float(buried_points) / (float((i + 1) * n_samples))
dV.append(abs(cur_vol - prev_vol))
prev_vol = cur_vol
else:
cur_vol = np.divide(buried_points, tot_points) / 8
dV.append(abs(sum(cur_vol) - sum(prev_vol)))
prev_vol = cur_vol
else:
with concurrent.futures.ThreadPoolExecutor(
max_workers=n_threads
) as executor:
out = [executor.submit(get_iter_vol) for k in range(0, n_threads)]
results = [data.result() for data in out]
for k in range(0, n_threads):
buried_points += results[k][0]
if basis is None:
cur_vol = float(buried_points) / (float((i + k + 1) * n_samples))
dV.append(abs(cur_vol - prev_vol))
prev_vol = cur_vol
else:
tot_points += results[k][1]
cur_vol = np.divide(buried_points, tot_points) / 8
dV.append(abs(sum(cur_vol) - sum(prev_vol)))
prev_vol = cur_vol
i += n_threads
between_v = cur_vol * (maxr ** 3 - minr ** 3)
tot_v = radius ** 3
return 100 * between_v / tot_v
# default to Gauss-Legendre integration over Lebedev spheres
else:
# grab radial grid points and weights for range (minr, maxr)
rgrid, rweights = utils.gauss_legendre_grid(
start=minr, stop=maxr, num=rpoints
)
# grab Lebedev grid for unit sphere at origin
agrid, aweights = utils.lebedev_sphere(
radius=1, center=np.zeros(3), num=apoints
)
# value of integral (without 4 pi r^2) for each shell
if basis is not None:
shell_values = np.zeros((8, rpoints))
else:
shell_values = np.zeros(rpoints)
# loop over radial shells
for i, rvalue in enumerate(rgrid):
# collect non-zero weights in inside_weights, then sum after looping over shell
# scale grid point to radius and shift to center
agrid_r = agrid * rvalue
if basis is not None:
map_agrid_r = np.dot(agrid_r, basis)
agrid_r += center_coords
D = distance_matrix(agrid_r, coords)
diff_mat = D - radius_list
mask = np.any(diff_mat <= 0, axis=1)
if basis is None:
shell_values[i] = sum(aweights[mask])
else:
mask = np.any(diff_mat <= 0, axis=1)
buried_coords = map_agrid_r[mask]
buried_weights = aweights[mask]
signs = np.sign(buried_coords)
# dot product should be 3, but > 2 allows for
# numerical error
oct_0 = np.where(np.dot(signs, [1, 1, 1]) > 2, 1, 0)
shell_values[0][i] += np.dot(oct_0, buried_weights)
oct_1 = np.where(np.dot(signs, [-1, 1, 1]) >= 2, 1, 0)
shell_values[1][i] += np.dot(oct_1, buried_weights)
oct_2 = np.where(np.dot(signs, [-1, -1, 1]) > 2, 1, 0)
shell_values[2][i] += np.dot(oct_2, buried_weights)
oct_3 = np.where(np.dot(signs, [1, -1, 1]) >= 2, 1, 0)
shell_values[3][i] += np.dot(oct_3, buried_weights)
oct_4 = np.where(np.dot(signs, [1, -1, -1]) > 2, 1, 0)
shell_values[4][i] += np.dot(oct_4, buried_weights)
oct_5 = np.where(np.dot(signs, [-1, -1, -1]) >= 2, 1, 0)
shell_values[5][i] += np.dot(oct_5, buried_weights)
oct_6 = np.where(np.dot(signs, [-1, 1, -1]) > 2, 1, 0)
shell_values[6][i] += np.dot(oct_6, buried_weights)
oct_7 = np.where(np.dot(signs, [1, 1, -1]) >= 2, 1, 0)
shell_values[7][i] += np.dot(oct_7, buried_weights)
if basis is not None:
# return a list of buried volume in each quadrant
return [
300
* np.dot(shell_values[k] * rgrid ** 2, rweights)
/ (radius ** 3)
for k in range(0, 8)
]
else:
# return buried volume
return (
300
* np.dot(shell_values * rgrid ** 2, rweights)
/ (radius ** 3)
)
def steric_map(
self,
center=None,
key_atoms=None,
radii="umn",
radius=3.5,
oop_vector=None,
ip_vector=None,
return_basis=False,
num_pts=100,
shape="circle",
):
"""
returns x, y, z, min_alt, max_alt or x, y, z, min_alt, max_alt, basis, atoms if return_basis is True
x - x coordinates for grid
y - y coordinates for grid
z - altitude levels; points where no atoms are will be -1000
min_alt - minimum altitude (above -1000)
max_alt - maximum altitute
basis - basis to e.g. reorient structure with np.dot(self.coords, basis)
atoms - list of atoms that are in the steric map
a contour plot can be created with this data - see stericMap.py command line script
parameters:
center - atom, list of atoms, or array specifiying the origin
key_atoms - list of ligand key atoms. Atoms on these ligands will be in the steric map.
radii - "umn", "bondi", or dict() specifying the VDW radii to use
oop_vector - None or array specifying the direction out of the plane of the steric map
if None, oop_vector is determined using the average vector from the key
atoms to the center atom
ip_vector - None or array specifying a vector in the plane of the steric map
if None, ip_vector is determined as the plane of best fit through the
key_atoms and the center
return_basis - whether or not to return a change of basis matrix
num_pts - number of points along x and y axis to use
shape - "circle" or "square"
"""
# determine center if none was specified
if center is None:
if self.center is None:
self.detect_components()
center = self.center
center_coords = self.COM(center)
else:
try:
center = self.find(center)
center_coords = self.COM(center)
except LookupError:
# assume an array was given
center_coords = center
# VDW radii to use
if isinstance(radii, dict):
radii_dict = radii
elif radii.lower() == "umn":
radii_dict = VDW_RADII
elif radii.lower() == "bondi":
radii_dict = BONDI_RADII
else:
raise RuntimeError(
"received %s for radii, must be umn or bondi" % radii
)
if key_atoms is None:
key_atoms = []
if self.components is None:
self.detect_components()
for comp in self.components:
key_atoms.extend(comp.key_atoms)
else:
key_atoms = self.find(key_atoms)
targets = []
for key in key_atoms:
if key not in targets:
if isinstance(center, Atom) or (
hasattr(center, "__iter__")
and all(isinstance(a, Atom) for a in center)
):
targets.extend(self.get_fragment(key, center))
else:
targets.extend(self.get_all_connected(key))
if oop_vector is None:
oop_vector = np.zeros(3)
for atom in key_atoms:
oop_vector += center_coords - atom.coords
oop_vector /= np.linalg.norm(oop_vector)
if ip_vector is None:
if len(key_atoms) == 1:
ip_vector = utils.perp_vector(oop_vector)
x_vec = np.cross(ip_vector, oop_vector)
else:
coords = [atom.coords for atom in key_atoms]
coords.append(center_coords)
coords = np.array(coords)
ip_vector = utils.perp_vector(coords)
x_vec = np.cross(ip_vector, oop_vector)
x_vec /= np.linalg.norm(x_vec)
ip_vector = -np.cross(x_vec, oop_vector)
else:
x_vec = np.cross(ip_vector, oop_vector)
basis = np.array([x_vec, ip_vector, oop_vector]).T
coords = self.coordinates(targets) - center_coords
new_coords = np.dot(coords, basis)
dist_ip = distance_matrix(new_coords[:, 0:2], [np.zeros(2)])[:, 0]
atoms_within_radius = []
radius_list = []
for i, atom in enumerate(targets):
if (
shape == "circle"
and dist_ip[i] - radii_dict[atom.element] < radius
):
atoms_within_radius.append(atom)
radius_list.append(radii_dict[atom.element])
elif (
shape == "square"
and dist_ip[i] - radii_dict[atom.element] < np.sqrt(2) * radius
):
atoms_within_radius.append(atom)
radius_list.append(radii_dict[atom.element])
atom_coords = np.dot(
self.coordinates(atoms_within_radius) - center_coords, basis
)
x = np.linspace(-radius, radius, num=num_pts)
y = np.linspace(-radius, radius, num=num_pts)
z = -1000 * np.ones((num_pts, num_pts))
max_alt = None
min_alt = None
for i in range(0, num_pts):
for j in range(0, num_pts):
if shape == "circle" and x[i] ** 2 + y[j] ** 2 > radius ** 2:
continue
for k in range(0, len(atoms_within_radius)):
w = np.sqrt(
(x[i] - atom_coords[k][0]) ** 2
+ (y[j] - atom_coords[k][1]) ** 2
)
if w < radius_list[k]:
h = np.sqrt(radius_list[k] ** 2 - w ** 2)
alt = atom_coords[k, 2] + h
# matplotlib mirrors z?
if alt > z[j, i]:
z[j, i] = alt
if max_alt is None or alt > max_alt:
max_alt = alt
if min_alt is None or alt < min_alt:
min_alt = alt
if return_basis:
return x, y, z, min_alt, max_alt, basis, atoms_within_radius
return x, y, z, min_alt, max_alt
def sterimol(
self,
L_axis,
start_atom,
targets,
L_func=None,
return_vector=False,
radii="bondi",
at_L=None,
max_error=None,
):
"""
returns sterimol parameter values in a dictionary
keys are B1, B2, B3, B4, B5, and L
B1 is determined numerically; B2-B4 depend on B1
B5 and L are analytical (unless L_func is not analytical)
see Verloop, A. and Tipker, J. (1976), Use of linear free energy
related and other parameters in the study of fungicidal
selectivity. Pestic. Sci., 7: 379-390.
(DOI: 10.1002/ps.2780070410)
return_vector: bool/returns dict of tuple(vector start, vector end) instead
radii: "bondi" - Bondi vdW radii
"umn" - vdW radii from Mantina, Chamberlin, Valero, Cramer, and Truhlar
dict() - radii are values and elements are keys
list() - list of radii corresponding to targets
L_axis: vector defining L-axis
targets: atoms to include in the parameter calculation
L_func: function to evaluate for getting the L value and vector
for each atom
takes positional arguments:
atom: Atom() - atom being checked
start: Atom() - start_atom
radius: vdw radius of atom
L_axis: unit vector for L-axis
if L_func is not given, the default is the distance from
start_atom to the furthest vdw radius projected onto the
L-axis
return_vector - returned dictionary will have tuples of start, end
for vectors to represent the parameters in 3D space
at_L - L value to calculate sterimol parameters at
Used for Sterimol2Vec
max_error - max. error in angstroms for B1
higher error can sometimes make the calculation
go slightly faster
max_error=None will have an error for B1 of at most
(sum of radii tangent to B1 face) * (1 - cos(0.5 degrees))
"""
from scipy.spatial import ConvexHull
CITATION = "doi:10.1002/ps.2780070410"
if at_L:
CITATION += "; doi:10.5281/zenodo.4702098"
self.LOG.citation(CITATION)
targets = self.find(targets)
start = self.find(start_atom)
if len(start) != 1:
raise TypeError(
"start must be exactly 1 atom, %i found for %s"
% (
len(start),
repr(start_atom),
)
)
start = start[0]
L_axis /= np.linalg.norm(L_axis)
if not L_func:
def L_func(atom, start, radius, L_axis):
test_v = start.bond(atom)
test_L = np.dot(test_v, L_axis) + radius
vec = (start.coords, start.coords + test_L * L_axis)
return test_L, vec
radius_list = []
radii_dict = None
if isinstance(radii, dict):
radii_dict = radii
elif isinstance(radii, list):
radius_list = radii
elif radii.lower() == "bondi":
radii_dict = BONDI_RADII
elif radii.lower() == "umn":
radii_dict = VDW_RADII
B1 = None
B2 = None
B3 = None
B4 = None
B5 = None
L = None
vector = {
"B1": None,
"B2": None,
"B3": None,
"B4": None,
"B5": None,
"L": None,
}
# for B1, we're going to use ConvexHull to find the minimum distance
# from one face of a bounding box
# to do this, we're going to project the substituent in a plane
# perpendicular to the L-axis and get a set of points along the
# vdw radii of the atoms
# ConvexHull will take these points and figure out which ones
# are on the outside (vertices)
# we then just need to find the bounding box with the minimum distance
# from L-axis to one side of the box
points = np.empty((0,2))
ndx = np.empty(0, dtype=int)
# just grab a random vector perpendicular to the L-axis
# it doesn't matter really
ip_vector = utils.perp_vector(L_axis)
x_vec = np.cross(ip_vector, L_axis)
x_vec /= np.linalg.norm(x_vec)
basis = np.array([x_vec, ip_vector, L_axis]).T
if not radius_list:
radius_list = []
coords = self.coordinates(targets)
L_vals = []
for i, atom in enumerate(targets):
test_v = start.bond(atom)
if radii_dict is not None:
radius_list.append(radii_dict[atom.element])
# L
test_L, L_vec = L_func(atom, start, radius_list[i], L_axis)
L_vals.append(test_L)
if L is None or test_L > L:
L = test_L
vector["L"] = L_vec
num_pts = 360
if max_error is not None:
# max error estimate is:
# (sum of atom radii that are tangent to B1 face) *
# (1 - cos(360 degrees / (2 * number of points)))
# we don't know B1 until after we pick num_pts, so
# we don't know which atoms determine the B1 face here
# but it is either one or two atoms and we guess
# it's the two largest atoms
num_pts = int(
2 / np.arccos(
1 - max_error / sum(
np.argsort(radius_list)[:-2][::-1]
)
)
)
v = np.linspace(0, 2 * np.pi, num=num_pts)
b1_points = np.stack(
(np.cos(v), np.sin(v)), axis=1
)
std_ndx = np.ones(num_pts, dtype=int)
# if a specific L value was requested, only check atoms
# with a radius that intersects the plane at that L
# value
# do this by setting the radii of atoms that don't intersect
# that plane to -1 so they are skipped later
# adjust the radii of atoms that do intersect to be the
# radius of the circle formed by the interection of the
# plane with the VDW sphere and adjust the coordinates
# so it loos like the atom is in that plane
if at_L is not None:
if not any(L >= at_L for L in L_vals):
at_L = max(L_vals)
if all(L < at_L for L in L_vals):
at_L = 0
L_vec = vector["L"][1] - vector["L"][0]
L_vec *= at_L / np.linalg.norm(L_vec)
vector["L"] = (vector["L"][0], vector["L"][0] + L_vec)
L = at_L
for i in range(0, len(coords)):
if L_vals[i] - 2 * radius_list[i] > at_L:
radius_list[i] = -1
continue
if L_vals[i] < at_L:
radius_list[i] = -1
continue
diff = L_vals[i] - radius_list[i] - at_L
radius_list[i] = np.sqrt(radius_list[i] ** 2 - diff ** 2)
coords[i] -= diff * L_axis
for i, (rad, coord) in enumerate(zip(radius_list, coords)):
if rad < 0:
continue
# B1-4 stuff - we come back to this later
test_v = coord - start.coords
new_coords = np.dot(test_v, basis)
# in plane coordinates - z-axis is L-axis, which
# we don't care about for B1
ip_coords = new_coords[0:2]
ndx = np.append(ndx, (i * std_ndx))
grid = rad * b1_points
grid += ip_coords
points = np.append(points, grid, axis=0)
# B5
# find distance along L-axis, then subtract this from vector from
# vector from molecule to this atom to get the B5 vector
# add the atom's radius to get the full B5
b = np.dot(test_v, L_axis)
test_B5_v = test_v - (b * L_axis)
test_B5 = np.linalg.norm(test_B5_v) + rad
if B5 is None or test_B5 > B5:
B5 = test_B5
start_x = coord - test_B5_v
if np.linalg.norm(test_B5_v) > 3 * np.finfo(float).eps:
perp_vec = test_B5_v
else:
# this atom might be along the L-axis, in which case use
# any vector orthogonal to L-axis
v_n = test_v / np.linalg.norm(test_v)
perp_vec = utils.perp_vector(L_axis)
perp_vec -= np.dot(v_n, perp_vec) * v_n
end = start_x + test_B5 * (perp_vec / np.linalg.norm(perp_vec))
vector["B5"] = (start_x, end)
hull = ConvexHull(points)
# import matplotlib.pyplot as plt
# for i, pt in enumerate(points):
# color = "blue"
# if self.atoms[ndx[i]].element == "H":
# color = "white"
# if self.atoms[ndx[i]].element == "C":
# color = "#5c5c5c"
# if self.atoms[ndx[i]].element == "F":
# color = "#90e050"
# if self.atoms[ndx[i]].element == "O":
# color = "#ff0000"
# plt.plot(*pt, 'o', markersize=1, color=color)
# # plt.plot(points[:, 0], points[:, 1], 'o', markersize=0.1)
# plt.plot(0, 0, 'kx')
# plt.plot(
# [*points[hull.vertices, 0], points[hull.vertices[0], 0]],
# [*points[hull.vertices, 1], points[hull.vertices[0], 1]],
# 'ro-',
# markersize=3,
# )
#
# ax = plt.gca()
# ax.set_aspect('equal')
# ax.set_facecolor("#dddddd")
# go through each edge, find a vector perpendicular to the one
# defined by the edge that passes through the origin
# the length of the shortest of these vectors is B1
tangents = points[hull.vertices[1:]] - points[hull.vertices[:-1]]
tangents = np.append(
tangents,
[points[hull.vertices[-1]] - points[hull.vertices[0]]],
axis=0,
)
tangents = tangents / np.linalg.norm(tangents, axis=1)[:, None]
paras = np.sum(
tangents * points[hull.vertices], axis=1
)
norms = points[hull.vertices] - paras[:, None] * tangents
norm_mags = np.linalg.norm(norms, axis=1)
B1_ndx = np.argmin(norm_mags)
B1 = norm_mags[B1_ndx]
b1_atom_coords = coords[ndx[hull.vertices[B1_ndx]]]
test_v = b1_atom_coords - start.coords
test_B1_v = test_v - (np.dot(test_v, L_axis) * L_axis)
start_x = b1_atom_coords - test_B1_v
end = x_vec * norms[B1_ndx][0] + ip_vector * norms[B1_ndx][1]
end += start_x
vector["B1"] = (start_x, end)
# figure out B2-4
# these need to be sorted in increasing order
# for now, they will just be Bpar for the one opposite B1
# and Bperp1 and Bperp2 for the ones perpendicular to B1
b1_norm = end - start_x
b1_norm /= np.linalg.norm(b1_norm)
b1_perp = np.cross(L_axis, b1_norm)
b1_perp /= np.linalg.norm(b1_perp)
Bpar = None
Bperp1 = None
Bperp2 = None
perp_vec1 = None
perp_vec2 = None
for rad, coord in zip(radius_list, coords):
if rad < 0:
continue
test_v = coord - start.coords
b = np.dot(test_v, L_axis)
test_B_v = test_v - (b * L_axis)
test_par_vec = np.dot(test_B_v, b1_norm) * b1_norm
test_par_vec -= rad * b1_norm
start_x = coord - test_B_v
end = start_x + test_par_vec
test_Bpar = np.linalg.norm(end - start_x)
if Bpar is None or test_Bpar > Bpar:
Bpar = test_Bpar
par_vec = (start_x, end)
perp_vec = np.dot(test_B_v, b1_perp) * b1_perp
if (
np.dot(test_B_v, b1_perp) > 0
or abs(np.dot(b1_perp, test_B_v)) < 1e-3
):
test_perp_vec1 = perp_vec + rad * b1_perp
end = start_x + test_perp_vec1
test_Bperp1 = np.linalg.norm(end - start_x)
if Bperp1 is None or test_Bperp1 > Bperp1:
Bperp1 = test_Bperp1
perp_vec1 = (start_x, end)
if (
np.dot(test_B_v, b1_perp) < 0
or abs(np.dot(b1_perp, test_B_v)) < 1e-3
):
test_perp_vec2 = perp_vec - rad * b1_perp
end = start_x + test_perp_vec2
test_Bperp2 = np.linalg.norm(end - start_x)
if Bperp2 is None or test_Bperp2 > Bperp2:
Bperp2 = test_Bperp2
perp_vec2 = (start_x, end)
if perp_vec1 is None:
perp_vec1 = perp_vec2[0], -perp_vec2[1]
Bperp1 = Bperp2
if perp_vec2 is None:
perp_vec2 = perp_vec1[0], -perp_vec1[1]
Bperp2 = Bperp1
# put B2-4 in order
i = 0
Bs = [Bpar, Bperp1, Bperp2]
Bvecs = [par_vec, perp_vec1, perp_vec2]
while Bs:
max_b = max(Bs)
n = Bs.index(max_b)
max_v = Bvecs.pop(n)
Bs.pop(n)
if i == 0:
B4 = max_b
vector["B4"] = max_v
elif i == 1:
B3 = max_b
vector["B3"] = max_v
elif i == 2:
B2 = max_b
vector["B2"] = max_v
i += 1
params = {
"B1": B1,
"B2": B2,
"B3": B3,
"B4": B4,
"B5": B5,
"L": L,
}
# plt.plot(
# [0, norms[B1_ndx,0]],
# [0, norms[B1_ndx,1]],
# 'g-', markersize=10,
# )
# plt.show()
if return_vector:
return vector
return params
# geometry manipulation
def append_structure(self, structure):
from AaronTools.component import Component
if not isinstance(structure, Geometry):
structure = Component(structure)
if not self.components:
self.detect_components()
self.components += [structure]
self.rebuild()
def update_geometry(self, structure):
"""
Replace current coords with those from :structure:
:structure: a file name, atom list, Geometry or np.array() of shape Nx3
"""
if isinstance(structure, np.ndarray):
coords = structure
elements = None
else:
atoms = Geometry(structure).atoms
elements = [a.element for a in atoms]
coords = [a.coords for a in atoms]
if len(coords) != len(self.atoms):
raise RuntimeError(
"Updated geometry has different number of atoms"
)
for i, row in enumerate(coords):
if elements is not None and elements[i] != self.atoms[i].element:
raise RuntimeError(
"Updated coords atom order doesn't seem to match original "
"atom order. Stopping..."
)
self.atoms[i].coords = row
self.refresh_connected()
return
def get_all_connected(self, target):
"""returns a list of all elements on the target atom's monomer"""
def _get_all_connected(geom, target, avoid):
atoms = [target]
for atom in target.connected:
if atom not in avoid:
new_avoid = avoid + [target]
atoms.extend(
[
x
for x in _get_all_connected(geom, atom, new_avoid)
if x not in atoms
]
)
return atoms
target = self.find(target)[0]
atoms = _get_all_connected(self, target, [])
return atoms
def get_fragment(
self, start, stop=None, as_object=False, copy=False, biggest=False
):
"""
Returns:
[Atoms()] if as_object == False
Geometry() if as_object == True
:start: the atoms to start on
:stop: the atom(s) to avoid
stop=None will try all possibilities and return smallest fragment
:as_object: return as list (default) or Geometry object
:copy: whether or not to copy the atoms before returning the list;
copy will automatically fix connectivity information
:biggest: if stop=None, will return biggest possible fragment instead of smallest
"""
start = self.find(start)
if stop is None:
best = None
for stop in itertools.chain(*[s.connected for s in start]):
frag = self.get_fragment(start, stop, as_object, copy)
if (
best is None
or (len(frag) < len(best) and not biggest)
or (len(frag) > len(best) and biggest)
):
best = frag
return best
stop = self.find(stop)
stack = deque(start)
frag = start
while len(stack) > 0:
connected = stack.popleft()
connected = connected.connected - set(stop) - set(frag)
stack.extend(connected)
frag += connected
if as_object:
return self.copy(atoms=frag, comment="")
if copy:
return self._fix_connectivity(frag, copy=True)
return frag
def remove_fragment(self, start, avoid=None, add_H=True):
"""
Removes a fragment of the geometry
Returns:
(list) :start: + the removed fragment
:start: the atom of the fragment to be removed that attaches to the
rest of the geometry
:avoid: the atoms :start: is attached to that should be avoided
:add_H: default is to change :start: to H and update bond lengths, but
add_H=False overrides this behaviour
"""
start = self.find(start)
if avoid is not None:
avoid = self.find(avoid)
frag = self.get_fragment(start, avoid)[len(start) :]
self -= frag
rv = start + frag
# replace start with H
if add_H:
for a in start:
a.element = "H"
a._set_radii()
self.change_distance(a, a.connected - set(frag), fix=2)
return rv
def coord_shift(self, vector, targets=None):
"""
shifts the coordinates of the target atoms by a vector
parameters:
vector (np.array) - the shift vector
targets (list) - the target atoms to shift (default to all)
"""
if targets is None:
targets = self.atoms
else:
targets = self.find(targets)
if not isinstance(vector, np.ndarray):
vector = np.array(vector, dtype=np.float64)
for t in targets:
t.coords += vector
return
def change_distance(
self, a1, a2, dist=None, adjust=False, fix=0, as_group=True
):
"""For setting/adjusting bond length between atoms
Parameters:
a1_arg - the first atom
a2_arg - the second atom
dist - the distance to change by/to. Default is to set the bond
lenght to that determined by RADII
adjust - default is to set the bond length to `dist`, adjust=True
indicates the current bond length should be adjusted by
`dist`
fix - default is to move both a1 and a2 by half of `dist`, fix=1
will move only a2 and fix=2 will move only a1
as_group - default is to move the fragments connected to a1 and a2
as well, as_group=False will only move the requested
atom(s)
"""
a1, a2 = self.find_exact(a1, a2)
# determine new bond length
if isinstance(dist, str):
dist = float(dist)
if dist is None:
if hasattr(a1, "_radii") and hasattr(a2, "_radii"):
new_dist = a1._radii + a2._radii
elif not hasattr(a1, "_radii"):
self.LOG.warning("no radii for %s", a1)
return
elif not hasattr(a2, "_radii"):
self.LOG.warning("no radii for %s", a2)
return
elif adjust:
new_dist = a1.dist(a2) + dist
else:
new_dist = dist
dist = a1.dist(a2)
# adjustment vector for each atom
adj_a1 = (new_dist - dist) * a2.bond(a1) / a2.dist(a1)
adj_a2 = (new_dist - dist) * a1.bond(a2) / a1.dist(a2)
if fix == 0:
adj_a1 /= 2
adj_a2 /= 2
elif fix == 1:
adj_a1 = None
elif fix == 2:
adj_a2 = None
else:
raise ValueError(
"Bad parameter `fix` (should be 0, 1, or 2):", fix
)
# get atoms to adjust
if as_group:
a1 = self.get_fragment(a1, a2)
a2 = self.get_fragment(a2, a1)
else:
a1 = [a1]
a2 = [a2]
# translate atom(s)
for i in a1:
if adj_a1 is None:
break
i.coords += adj_a1
for i in a2:
if adj_a2 is None:
break
i.coords += adj_a2
return
def rotate_fragment(self, start, avoid, angle):
"""rotates the all atoms on the 'start' side of the
start-avoid bond about the bond vector by angle"""
start = self.find(start)[0]
avoid = self.find(avoid)[0]
shift = start.coords
self.coord_shift(-shift)
self.rotate(
start.bond(avoid),
angle=angle * 180 / np.pi,
targets=self.get_fragment(start, avoid),
)
self.coord_shift(shift)
def rotate(self, w, angle=None, targets=None, center=None):
"""
rotates target atoms by an angle about an axis
:w: (np.array) - the axis of rotation (doesnt need to be unit vector)
or a quaternion (angle not required then)
:angle: (float) - the angle by which to rotate (in radians)
:targets: (list) - the atoms to rotate (defaults to all)
:center: (Atom or list) - if provided, the atom (or COM of a list)
will be centered at the origin before rotation, then shifted
back after rotation
"""
if targets is None:
targets = self.atoms
else:
targets = self.find(targets)
# shift geometry to place center atom at origin
if center is not None:
if not (
hasattr(center, "__len__")
and all(isinstance(x, float) for x in center)
):
tmp = self.find(center)
if len(tmp) > 1:
center = deepcopy(self.COM(tmp))
else:
center = deepcopy(tmp[0].coords)
else:
center = deepcopy(center)
self.coord_shift(-1 * center)
if not isinstance(w, np.ndarray):
w = np.array(w, dtype=np.double)
if angle is not None and len(w) == 3:
w = w / np.linalg.norm(w)
q = np.hstack(([np.cos(angle / 2)], w * np.sin(angle / 2)))
elif len(w) != 4:
raise TypeError(
"""Vector `w` must be either a rotation vector (len 3)
or a quaternion (len 4). Angle parameter required if `w` is a
rotation vector"""
)
else:
q = w
q /= np.linalg.norm(q)
qs = q[0]
qv = q[1:]
xyz = self.coordinates(targets)
xprod = np.cross(qv, xyz)
qs_xprod = 2 * qs * xprod
qv_xprod = 2 * np.cross(qv, xprod)
xyz += qs_xprod + qv_xprod
for t, coord in zip(targets, xyz):
t.coords = coord
if center is not None:
self.coord_shift(center)
def mirror(self, plane="xy"):
"""
mirror self across a plane
plane can be xy, xz, yz or an array for a vector orthogonal to a plane
"""
eye = np.identity(3)
if isinstance(plane, str):
if plane.lower() == "xy":
eye[0, 0] *= -1
if plane.lower() == "xz":
eye[1, 1] *= -1
if plane.lower() == "yz":
eye[2, 2] *= -1
else:
eye = utils.mirror_matrix(plane)
self.update_geometry(np.dot(self.coords, eye))
def invert(self, plane="xy"):
"""
invert self's coordinates
"""
op = -np.identity(3)
self.update_geometry(np.dot(self.coords, op))
def change_angle(
self,
a1,
a2,
a3,
angle,
radians=True,
adjust=False,
fix=0,
as_group=True,
):
"""For setting/adjusting angle between atoms
Parameters:
a1 - the first atom
a2 - the second atom (vertex)
a3 - the third atom
angle - the angle to change by/to
radians - default units are radians, radians=False uses degrees
adjust - default is to set the angle to `angle`, adjust=True
indicates the current angle should be adjusted by `angle`
fix - default is to move both a1 and a3 by half of `angle`, fix=1
will move only a3 and fix=3 will move only a1
as_group - default is to move the fragments connected to a1 and a3
as well, as_group=False will only move the requested atom(s)
"""
try:
a1, a2, a3 = self.find([a1, a2, a3])
except ValueError:
raise LookupError(
"Bad atom request: {}, {}, {}".format(a1, a2, a3)
)
# get rotation vector
v1 = a2.bond(a1)
v2 = a2.bond(a3)
w = np.cross(v1, v2)
w = w / np.linalg.norm(w)
# determine rotation angle
if not radians:
angle = np.deg2rad(angle)
if not adjust:
angle -= a2.angle(a1, a3)
# get target fragments
a1_frag = self.get_fragment(a1, a2)
a3_frag = self.get_fragment(a3, a2)
# shift a2 to origin
self.coord_shift(-a2.coords, a1_frag)
self.coord_shift(-a2.coords, a3_frag)
# perform rotation
if fix == 0:
angle /= 2
self.rotate(w, -angle, a1_frag)
self.rotate(w, angle, a3_frag)
elif fix == 1:
self.rotate(w, angle, a3_frag)
elif fix == 3:
self.rotate(w, -angle, a1_frag)
else:
raise ValueError("fix must be 0, 1, 3 (supplied: {})".format(fix))
# shift a2 back to original location
self.coord_shift(a2.coords, a1_frag)
self.coord_shift(a2.coords, a3_frag)
def change_dihedral(self, *args, **kwargs):
"""
For setting/adjusting dihedrals
*args
:a1: the first atom
:a2: the second atom
:a3: the third atom (optional for adjust=True if as_group=True)
:a4: the fourth atom (optional for adjust=True if as_group=True)
:dihedral: the dihedral to change by/to
**kwargs
:fix: default is to move both a1 and a4 by half of `dihedral`,
fix=1 will move only a4 and fix=4 will move only a1
:adjust: default is to set the dihedral to `dihedral`, adjust=True
indicates the current dihedral should be adjusted by `dihedral`
:as_group: default is to move the fragments connected to a1 and a3
as well, as_group=False will only move the requested atom(s)
:radians: default units are degrees, radians=True to use radians
"""
fix = kwargs.get("fix", 0)
adjust = kwargs.get("adjust", False)
as_group = kwargs.get("as_group", True)
radians = kwargs.get("radians", False)
left_over = set(kwargs.keys()) - set(
["fix", "adjust", "as_group", "radians"]
)
if left_over:
raise SyntaxError(
"Unused **kwarg(s) provided: {}".format(left_over)
)
# get atoms
count = len(args)
if count == 3 and adjust:
# we can just define the bond to rotate about, as long as we are
# adjusting, not setting, the whole fragments on either side
as_group = True
a2, a3 = self.find_exact(*args[:2])
dihedral = args[2]
try:
a1 = next(iter(a2.connected - set([a2, a3])))
except StopIteration:
a1 = next(iter(set(self.atoms) - set([a2, a3])))
try:
a4 = next(iter(a3.connected - set([a1, a2, a3])))
except StopIteration:
a4 = next(iter(set(self.atoms) - set([a1, a2, a3])))
elif count != 5:
raise TypeError(
"Number of atom arguments provided insufficient to define "
+ "dihedral"
)
else:
a1, a2, a3, a4 = self.find_exact(*args[:4])
dihedral = args[4]
# get fragments
if as_group:
a2_frag = self.get_fragment(a2, a3)[1:]
a3_frag = self.get_fragment(a3, a2)[1:]
if any(atom in a2_frag for atom in a3_frag):
self.LOG.warning(
"changing dihedral that is part of a ring: %s %s", a2, a3
)
else:
a2_frag = [a1]
a3_frag = [a4]
# fix units
if not radians:
dihedral = np.deg2rad(dihedral)
# get adjustment
if not adjust:
dihedral -= self.dihedral(a1, a2, a3, a4)
# rotate fragments
if not a2_frag and not a3_frag:
raise RuntimeError(
"Cannot change dihedral, no fragments to target for rotation"
)
if not a2_frag and fix == 0:
fix = 1
if not a3_frag and fix == 0:
fix = 4
if fix == 0:
dihedral /= 2
self.rotate(a2.bond(a3), -dihedral, a2_frag, center=a2)
self.rotate(a2.bond(a3), dihedral, a3_frag, center=a3)
elif fix == 1:
self.rotate(a2.bond(a3), dihedral, a3_frag, center=a3)
elif fix == 4:
self.rotate(a2.bond(a3), -dihedral, a2_frag, center=a2)
else:
raise ValueError(
"`fix` must be 0, 1, or 4 (supplied: {})".format(fix)
)
def minimize_sub_torsion(
self, geom=None, all_frags=False, increment=30, allow_planar=False
):
"""rotate substituents to try to minimize LJ potential
geom: calculate LJ potential between self and another geometry-like
object, instead of just within self
all_frags: minimize rotatable bonds on substituents
allow_planar: allow substituents that start and end with atoms
with planar VSEPR geometries that are nearly
planar to be rotated
"""
# minimize torsion for each substituent
if not hasattr(self, "substituents") or self.substituents is None:
self.detect_substituents()
# we don't want to rotate any substituents that
# shouldn't be rotate-able
# filter out any substituents that start on a planar atom
# and end on a planar atom
if not allow_planar:
vsepr = [atom.get_vsepr()[0] for atom in self.atoms]
for i, sub in enumerate(sorted(self.substituents, reverse=True)):
if len(sub.atoms) < 2:
continue
if not allow_planar:
# don't rotate substituents that might be E/Z
vsepr_1 = vsepr[self.atoms.index(sub.end)]
vsepr_2 = vsepr[self.atoms.index(sub.atoms[0])]
if (
vsepr_1 and vsepr_2 and
"planar" in vsepr_1 and "planar" in vsepr_2
):
a1 = [a for a in sub.end.connected if a is not sub.atoms[0]][0]
a2 = [a for a in sub.atoms[0].connected if a is not sub.end][0]
angle = self.dihedral(a1, sub.end, sub.atoms[0], a2)
# ~5 degree tolerance for being planar
if any(np.isclose(angle, ref, atol=0.09) for ref in [np.pi, 0, -np.pi]):
continue
axis = sub.atoms[0].bond(sub.end)
center = sub.end
self.minimize_torsion(
sub.atoms, axis, center, geom, increment=increment
)
if all_frags:
for frag, a, b in self.get_frag_list(
targets=sub.atoms, max_order=1
):
axis = a.bond(b)
center = b.coords
self.minimize_torsion(frag, axis, center, geom)
def minimize_torsion(self, targets, axis, center, geom=None, increment=5):
"""
Rotate :targets: to minimize the LJ potential
:targets: the target atoms to rotate
:axis: the axis by which to rotate
:center: where to center before rotation
:geom: calculate LJ potential between self and another geometry-like
object, instead of just within self
"""
targets = Geometry(
self.find(targets),
refresh_connected=False,
refresh_ranks=False,
)
if geom is None or geom is self:
from AaronTools.finders import NotAny
try:
geom = Geometry(
self.find(NotAny(targets)),
refresh_connected=False,
refresh_ranks=False,
)
except LookupError:
return
E_min = None
angle_min = None
# copied an reorganized some stuff from Geometry.rotate for
# performance reasons
if hasattr(center, "__iter__") and all(
isinstance(x, float) for x in center
):
center_coords = center
else:
center_coords = self.COM(center)
axis = axis / np.linalg.norm(axis)
q = np.hstack(
(
[np.cos(np.deg2rad(increment) / 2)],
axis * np.sin(np.deg2rad(increment) / 2),
)
)
q /= np.linalg.norm(q)
qs = q[0]
qv = q[1:]
# rotate targets by increment and save lowest energy
angle = 0
xyz = targets.coords
for inc in range(0, 360, increment):
angle += increment
xyz -= center_coords
xprod = np.cross(qv, xyz)
qs_xprod = 2 * qs * xprod
qv_xprod = 2 * np.cross(qv, xprod)
xyz += qs_xprod + qv_xprod
xyz += center_coords
for t, coord in zip(targets.atoms, xyz):
t.coords = coord
energy = targets.LJ_energy(other=geom, use_prev_params=True)
if E_min is None or energy < E_min:
E_min = energy
angle_min = angle
# rotate to min angle
self.rotate(
axis,
np.deg2rad(angle_min - angle),
targets=targets,
center=center_coords,
)
return
def substitute(self, sub, target, attached_to=None, minimize=False):
"""
substitutes fragment containing `target` with substituent `sub`
if attached_to is provided, this is the atom where the substituent is attached
if attached_to==None, replace the smallest fragment containing `target`
minimize - bool, rotate sub to lower LJ potential
"""
from AaronTools.component import Component
# set up substituent
if not isinstance(sub, AaronTools.substituent.Substituent):
sub = AaronTools.substituent.Substituent(sub)
sub.refresh_connected()
# determine target and atoms defining connection bond
target = self.find(target)
# if we have components, do the substitution to the component
# otherwise, just do it on self
geom = self
if hasattr(self, "components") and self.components is not None:
for comp in self.components:
if target in comp:
geom = comp
break
# attached_to is provided or is the atom giving the
# smallest target fragment
if attached_to is not None:
attached_to = geom.find_exact(attached_to)
else:
smallest_frag = None
smallest_attached_to = None
# get all possible connection points
attached_to = set()
for t in target:
attached_to = attached_to | (t.connected - set(target))
# find smallest fragment
for e in attached_to:
frag = geom.get_fragment(target, e)
if smallest_frag is None or len(frag) < len(smallest_frag):
smallest_frag = frag
smallest_attached_to = e
attached_to = [smallest_attached_to]
if len(attached_to) != 1:
raise NotImplementedError(
"Can only replace substituents with one point of attachment"
)
attached_to = attached_to[0]
sub.end = attached_to
# determine which atom of target fragment is connected to attached_to
sub_attach = attached_to.connected & set(target)
if len(sub_attach) > 1:
raise NotImplementedError(
"Can only replace substituents with one point of attachment"
)
if len(sub_attach) < 1:
raise LookupError("attached_to atom not connected to targets")
sub_attach = sub_attach.pop()
# manipulate substituent geometry; want sub.atoms[0] -> sub_attach
# attached_to == sub.end
# sub_attach will eventually be sub.atoms[0]
# move attached_to to the origin
shift = np.array([x for x in attached_to.coords])
geom.coord_shift(-1 * shift)
# align substituent to current bond
bond = geom.bond(attached_to, sub_attach)
sub.align_to_bond(bond)
# shift geometry back and shift substituent to appropriate place
geom.coord_shift(shift)
sub.coord_shift(shift)
# tag and update name for sub atoms
for i, s in enumerate(sub.atoms):
s.add_tag(sub.name)
if i > 0:
s.name = sub_attach.name + "." + s.name
else:
s.name = sub_attach.name
# add first atoms of new substituent where the target atoms were
# add the rest of the new substituent at the end
old = geom.get_fragment(target, attached_to)
for i, a in enumerate(old):
if i == len(sub.atoms):
break
geom.atoms.insert(geom.atoms.index(old[i]), sub.atoms[i])
sub.atoms[i].name = old[i].name
else:
if len(sub.atoms) > len(old):
geom += sub.atoms[i + 1 :]
# remove old substituent
geom -= old
attached_to.connected.discard(sub_attach)
# fix connections (in lieu of geom.refresh_connected(), since clashing may occur)
attached_to.connected.add(sub.atoms[0])
sub.atoms[0].connected.add(attached_to)
# fix bond distance
geom.change_distance(attached_to, sub.atoms[0], as_group=True, fix=1)
# clean up changes
if isinstance(geom, Component):
self.substituents += [sub]
self.detect_backbone(to_center=self.backbone)
self.rebuild()
self.refresh_ranks()
if minimize:
self.minimize_torsion(sub.atoms, bond, shift)
return sub
def find_substituent(self, start, for_confs=False):
"""
Finds a substituent based on a given atom (matches start==sub.atoms[0])
:start: the first atom of the subsituent, where it connects to sub.end
:for_confs: if true(default), only consider substituents that need to
be rotated to generate conformers
"""
start = self.find(start)[0]
for sub in self.get_substituents(for_confs):
if sub.atoms[0] == start:
return sub
else:
if for_confs:
for sub in self.get_substituents(for_confs=not for_confs):
if sub.atoms[0] == start:
return None
msg = "Could not find substituent starting at atom {}."
raise LookupError(msg.format(start.name))
if not hasattr(self, "substituents") or self.substituents is None:
self.substituents = []
self.substituents.append(sub)
def get_substituents(self, for_confs=True):
"""
Returns list of all substituents found on all components
:for_confs: if true (default), returns only substituents that need to
be rotated to generate conformers
"""
rv = []
if self.components is None:
self.detect_components()
for comp in self.components:
if comp.substituents is None:
comp.detect_backbone()
for sub in comp.substituents:
if for_confs and (sub.conf_num is None or sub.conf_num <= 1):
continue
rv += [sub]
return rv
def ring_substitute(
self, targets, ring_fragment, minimize=False, flip_walk=False
):
"""
take ring, reorient it, put it on self and replace targets with atoms
on the ring fragment
ring_fragment - Ring instance or name of ring in the library
minimize - try other rings with the same name (appended with a number)
in the library to see if they fit better
flip_walk - also flip the rings when minimizing to see if that fits better
"""
def attach_short(geom, walk, ring_fragment):
"""for when walk < end, rmsd and remove end[1:-1]"""
# align ring's end to geom's walk
ring_fragment.RMSD(
geom,
align=True,
targets=ring_fragment.end,
ref_targets=walk,
sort=False,
)
ring_waddle(geom, targets, [walk[1], walk[-2]], ring_fragment)
for atom in ring_fragment.end[1:-1]:
for t in atom.connected:
if t not in ring_fragment.end:
ring_fragment.remove_fragment(t, atom, add_H=False)
ring_fragment -= t
ring_fragment -= atom
geom.remove_fragment([walk[0], walk[-1]], walk[1:-1], add_H=False)
geom -= [walk[0], walk[-1]]
walk[1].connected.add(ring_fragment.end[0])
walk[-2].connected.add(ring_fragment.end[-1])
ring_fragment.end[-1].connected.add(walk[-2])
ring_fragment.end[0].connected.add(walk[1])
ring_fragment.end = walk[1:-1]
geom.atoms.extend(ring_fragment.atoms)
geom.refresh_ranks()
def ring_waddle(geom, targets, walk_end, ring):
"""adjusted the new bond lengths by moving the ring in a 'waddling' motion
pivot on one end atom to adjust the bond lenth of the other, then do
the same with the other end atom"""
if hasattr(ring.end[0], "_radii") and hasattr(
walk_end[0], "_radii"
):
d1 = ring.end[0]._radii + walk_end[0]._radii
else:
d1 = ring.end[0].dist(walk_end[0])
v1 = ring.end[-1].bond(walk_end[0])
v2 = ring.end[-1].bond(ring.end[0])
v1_n = np.linalg.norm(v1)
v2_n = np.linalg.norm(v2)
target_angle = np.arccos(
(d1 ** 2 - v1_n ** 2 - v2_n ** 2) / (-2.0 * v1_n * v2_n)
)
current_angle = ring.end[-1].angle(ring.end[0], walk_end[0])
ra = target_angle - current_angle
rv = np.cross(v1, v2)
ring.rotate(rv, ra, center=ring.end[-1])
if hasattr(ring.end[-1], "_radii") and hasattr(
walk_end[-1], "_radii"
):
d1 = ring.end[-1]._radii + walk_end[-1]._radii
else:
d1 = ring.end[-1].dist(walk_end[-1])
v1 = ring.end[0].bond(walk_end[-1])
v2 = ring.end[0].bond(ring.end[-1])
v1_n = np.linalg.norm(v1)
v2_n = np.linalg.norm(v2)
target_angle = np.arccos(
(d1 ** 2 - v1_n ** 2 - v2_n ** 2) / (-2.0 * v1_n * v2_n)
)
current_angle = ring.end[0].angle(ring.end[-1], walk_end[-1])
ra = target_angle - current_angle
rv = np.cross(v1, v2)
ring.rotate(rv, ra, center=ring.end[0])
def clashing(geom, ring):
from AaronTools.finders import NotAny
geom_coords = geom.coordinates(NotAny(ring.atoms))
dist_mat = distance_matrix(geom_coords, ring.coords)
if np.any(dist_mat < 0.75):
return True
return False
from AaronTools.ring import Ring
if not isinstance(ring_fragment, Ring):
ring_fragment = Ring(ring_fragment)
targets = self.find(targets)
# we want to keep atom naming conventions consistent with regular substitutions
for atom in ring_fragment.atoms:
atom.name = "{}.{}".format(targets[0].name, atom.name)
# find a path between the targets
walk = self.shortest_path(*targets)
if len(ring_fragment.end) != len(walk):
ring_fragment.find_end(len(walk), start=ring_fragment.end)
if len(walk) == len(ring_fragment.end) and len(walk) != 2:
if not minimize:
attach_short(self, walk, ring_fragment)
else:
# to minimize, check VSEPR on self's atoms attached to targets
# lower deviation is better
# do this for the original ring and also try flipping the ring
# ring is flipped by reversing walk
# check for other rings in the library with ring.\d+
# e.g. cyclohexane.2
vsepr1, _ = walk[1].get_vsepr()
vsepr2, _ = walk[-2].get_vsepr()
geom = self.copy()
test_walk = [
geom.atoms[i]
for i in [self.atoms.index(atom) for atom in walk]
]
frag = ring_fragment.copy()
attach_short(geom, test_walk, frag)
new_vsepr1, score1 = test_walk[1].get_vsepr()
new_vsepr2, score2 = test_walk[-2].get_vsepr()
new_vsepr3, score3 = frag.end[0].get_vsepr()
new_vsepr4, score4 = frag.end[-1].get_vsepr()
score = score1 + score2 + score3 + score4
min_diff = score
min_ring = 0
# print("%s score: %.3f" % (ring_fragment.name, score))
if flip_walk:
geom = self.copy()
test_walk = [
geom.atoms[i]
for i in [self.atoms.index(atom) for atom in walk]
][::-1]
frag = ring_fragment.copy()
attach_short(geom, test_walk, frag)
new_vsepr1, score1 = test_walk[1].get_vsepr()
new_vsepr2, score2 = test_walk[-2].get_vsepr()
new_vsepr3, score3 = frag.end[0].get_vsepr()
new_vsepr4, score4 = frag.end[-1].get_vsepr()
score = score1 + score2 + score3 + score4
if score < min_diff and not clashing(geom, frag):
min_ring = 1
min_diff = score
# print("flipped %s score: %.3f" % (ring_fragment.name, score))
# check other rings in library
# for these, flip the ring end instead of walk
for ring_name in Ring.list():
if re.search("%s\.\d+" % ring_fragment.name, ring_name):
test_ring_0 = Ring(ring_name)
geom = self.copy()
test_walk = [
geom.atoms[i]
for i in [self.atoms.index(atom) for atom in walk]
]
if len(test_ring_0.end) != len(walk):
test_ring_0.find_end(
len(walk), start=test_ring_0.end
)
frag = test_ring_0.copy()
attach_short(geom, test_walk, frag)
new_vsepr1, score1 = test_walk[1].get_vsepr()
new_vsepr2, score2 = test_walk[-2].get_vsepr()
new_vsepr3, score3 = frag.end[0].get_vsepr()
new_vsepr4, score4 = frag.end[-1].get_vsepr()
score = score1 + score2 + score3 + score4
if score < min_diff and not clashing(geom, frag):
min_ring = test_ring_0
min_diff = score
# print("%s score: %.3f" % (ring_name, score))
if flip_walk:
test_ring_1 = Ring(ring_name)
test_ring_1.end.reverse()
geom = self.copy()
test_walk = [
geom.atoms[i]
for i in [
self.atoms.index(atom) for atom in walk
]
]
if len(test_ring_0.end) != len(walk):
test_ring_0.find_end(
len(walk), start=test_ring_0.end
)
frag = test_ring_1.copy()
attach_short(geom, test_walk, frag)
new_vsepr1, score1 = test_walk[1].get_vsepr()
new_vsepr2, score2 = test_walk[-2].get_vsepr()
new_vsepr3, score3 = frag.end[0].get_vsepr()
new_vsepr4, score4 = frag.end[-1].get_vsepr()
score = score1 + score2 + score3 + score4
# print("flipped %s score: %.3f" % (ring_name, score))
if score < min_diff and not clashing(geom, frag):
min_ring = test_ring_1
min_diff = score
if not isinstance(min_ring, Ring) and min_ring == 0:
walk = self.shortest_path(*targets)
attach_short(self, walk, ring_fragment)
elif not isinstance(min_ring, Ring) and min_ring == 1:
walk = self.shortest_path(*targets)[::-1]
attach_short(self, walk, ring_fragment)
else:
walk = self.shortest_path(*targets)
attach_short(self, walk, min_ring)
elif not walk[1:-1]:
raise ValueError(
"insufficient information to close ring - selected atoms are bonded to each other: %s"
% (" ".join(str(a) for a in targets))
)
else:
raise ValueError(
"this ring is not appropriate to connect\n%s\nand\n%s:\n%s\nspacing is %i; expected %i"
% (
targets[0],
targets[1],
ring_fragment.name,
len(ring_fragment.end),
len(walk),
)
)
# AaronJr needs to know this when relaxing changes
return ring_fragment.atoms
def change_element(
self,
target,
new_element,
adjust_bonds=False,
adjust_hydrogens=False,
hold_steady=None,
):
"""change the element of an atom on self
target - target atom
new_element - str: element of new atom
adjust_bonds - bool: adjust distance to bonded atoms
adjust_hydrogens - bool: try to add or remove hydrogens and guess how many
hydrogens to add or remove
tuple(int, str): remove specified number of hydrogens and
set the geometry to the specified shape
(see Atom.get_shape for a list of shapes)
hold_steady - atom: atom bonded to target that will be held steady when
adjusting bonds; Default - longest fragment
"""
def get_corresponding_shape(target, shape_object, frags):
"""
returns shape object, but where shape_object.atoms are lined up with
target.connected as much as possible
"""
shape_object.coord_shift(
target.coords - shape_object.atoms[0].coords
)
if len(frags) == 0:
return shape_object
# to minimize changes to the structure, things are aligned to the largest fragment
max_frag = sorted(frags, key=len, reverse=True)[0]
angle = target.angle(shape_object.atoms[1], max_frag[0])
v1 = target.bond(max_frag[0])
v2 = shape_object.atoms[0].bond(shape_object.atoms[1])
v1 /= np.linalg.norm(v1)
v2 /= np.linalg.norm(v2)
rv = np.cross(v1, v2)
# could have numerical issues
# avoid those, but check to see if the angle is 180 degrees
if abs(np.linalg.norm(rv)) < 10 ** -3 or abs(angle) < 10 ** -3:
if np.dot(v1, v2) == -1:
rv = np.array([v1[2], v1[0], v1[1]])
shape_object.rotate(rv, np.pi, center=target)
angle = 0
if abs(np.linalg.norm(rv)) > 10 ** -3 and abs(angle) > 10 ** -3:
shape_object.rotate(rv, -angle, center=target)
# rotate about the vector from the center (target atom) to the first
# atom in shape (which will be the largest fragment) to get the shape object
# as lined up with the rest of the connected atoms as possible
rv = target.bond(shape_object.atoms[1])
min_dev = None
min_angle = 0
inc = 5
angle = 0
while angle < 360:
angle += inc
shape_object.rotate(rv, np.deg2rad(inc), center=target)
previous_positions = [0]
dev = 0
for j, frag in enumerate(sorted(frags, key=len, reverse=True)):
if j == 0:
continue
v1 = target.bond(frag[0])
max_overlap = None
corresponding_position = None
for i, position in enumerate(shape_object.atoms[1:]):
if i in previous_positions:
continue
v2 = shape_object.atoms[0].bond(position)
d = np.dot(v1, v2)
if max_overlap is None or d > max_overlap:
max_overlap = d
corresponding_position = i
if corresponding_position is None:
continue
previous_positions.append(corresponding_position)
dev += (
max_overlap
- (
np.linalg.norm(frag[0].coords)
* np.linalg.norm(
shape_object.atoms[
corresponding_position + 1
].coords
)
)
) ** 2
if min_dev is None or dev < min_dev:
min_dev = dev
min_angle = angle
shape_object.rotate(rv, np.deg2rad(min_angle), center=target)
return shape_object
target = self.find(target)
if len(target) > 1:
raise RuntimeError(
"only one atom's element can be changed at a time (%i attempted)"
% len(target)
)
else:
target = target[0]
# new_atom is only used to determine how many H's to add
new_atom = Atom(
element=new_element, name=target.name, coords=target.coords
)
if adjust_hydrogens is True:
# try to determine how many hydrogens to add based on how many hydrogens are currently
# bonded and what the saturation of the atoms is
# e.g. C(H3) -> N(H?)
# C's saturation is 4, it's missing one
# N's saturation is 3, it should be missing one: N(H2)
if hasattr(target, "_saturation") and hasattr(
new_atom, "_saturation"
):
change_hydrogens = new_atom._saturation - target._saturation
new_shape = None
else:
raise RuntimeError(
"H adjust requested, but saturation is not known for %s"
% ", ".join(
[
atom.element
for atom in [target, new_atom]
if not hasattr(atom, "_saturation")
]
)
)
elif isinstance(adjust_hydrogens, tuple):
# tuple of (change in hydrogens, vsepr shape) was given
change_hydrogens, new_shape = adjust_hydrogens
if callable(change_hydrogens):
change_hydrogens = change_hydrogens(target)
else:
# no change was requested, only the element will change
# and maybe bond lengths
change_hydrogens = 0
new_shape = None
if change_hydrogens != 0 or new_shape is not None:
# if we're removing hydrogens, check if we have enough to remove
if change_hydrogens < 0:
n_hygrogens = sum(
[1 for atom in target.connected if atom.element == "H"]
)
if n_hygrogens + change_hydrogens < 0:
raise RuntimeError(
"cannot remove %i hydrogens from an atom with %i hydrogens"
% (abs(change_hydrogens), n_hygrogens)
)
# get vsepr geometry
old_shape, score = target.get_vsepr()
if new_shape is None:
shape = old_shape
if hasattr(new_atom, "_connectivity"):
new_connectivity = new_atom._connectivity
else:
new_connectivity = None
# if we're changing the number of hydrogens, but no shape was specified,
# we will remove hydrogens one by one and see what shape we end up with
# shape changes are based on rules like adding a bond to a trigonal planar
# atom will cause it to be tetrahedral (e.g. carbocation gaining a hydride)
for i in range(0, abs(change_hydrogens)):
shape = Atom.new_shape(
shape, new_connectivity, np.sign(change_hydrogens)
)
if shape is None:
raise RuntimeError(
"shape changed from %s to None" % old_shape
)
new_shape = shape
shape_object = Geometry(Atom.get_shape(new_shape))
if (
len(shape_object.atoms[1:]) - len(target.connected)
!= change_hydrogens
):
# insufficient to change the shape
# we cannot delete random fragments
raise RuntimeError(
"number of positions changed by %i, but a change of %i hydrogens was attempted"
% (
len(shape_object.atoms[1:]) - len(target.connected),
change_hydrogens,
)
)
# get each branch off of the target atom
frags = [
self.get_fragment(atom, target) for atom in target.connected
]
if new_shape != old_shape or change_hydrogens == 0:
if change_hydrogens < 0:
# remove extra hydrogens
shape_object = get_corresponding_shape(
target, shape_object, frags
)
removed_Hs = 1
while removed_Hs <= abs(change_hydrogens):
H_atom = [
atom
for atom in target.connected
if atom.element == "H"
][0]
self -= H_atom
removed_Hs += 1
# get fragments after hydrogens have been removed
frags = [
self.get_fragment(atom, target)
for atom in target.connected
]
# align the shape object
# the shape object's first atom will be on top of target
# the second will be lined up with the largest fragment on target
# the rest are rotated to minimize deviation from the remaining groups
shape_object = get_corresponding_shape(
target, shape_object, frags
)
# ring detection - remove ring fragments because those are more difficult to adjust
remove_frags = []
for i, frag1 in enumerate(frags):
for frag2 in frags[i + 1 :]:
dups = [atom for atom in frag2 if atom in frag1]
if len(dups) != 0:
remove_frags.append(frag1)
remove_frags.append(frag2)
# add Hs if needed
if change_hydrogens > 0:
# determine which connected atom is occupying which position on the shape
shape_object = get_corresponding_shape(
target, shape_object, frags
)
positions = []
for j, frag in enumerate(
sorted(frags, key=len, reverse=True)
):
v2 = target.bond(frag[0])
max_overlap = None
position = None
for i, pos in enumerate(shape_object.atoms[1:]):
v1 = shape_object.atoms[0].bond(pos)
if i in positions:
continue
d = np.dot(v1, v2)
if max_overlap is None or d > max_overlap:
max_overlap = d
position = i
positions.append(position)
# add hydrogens to positions that are not occupied
for open_position in [
i + 1
for i in range(0, len(shape_object.atoms[1:]))
if i not in positions
]:
# add one because the 0th "position" of the shape is the central atom
H_atom = Atom(
element="H",
coords=shape_object.atoms[open_position].coords,
name=str(len(self.atoms) + 1),
)
self.change_distance(target, H_atom, fix=1)
self += H_atom
target.connected.add(H_atom)
H_atom.connected.add(target)
frags.append([H_atom])
# for each position on the new idealized geometry, find the fragment
# that corresponds to it the best
# reorient that fragment to match the idealized geometry
previous_positions = []
frag_atoms = []
first_frag = None
if hold_steady:
hold_steady = self.find(hold_steady)
for j, frag in enumerate(sorted(frags, key=len, reverse=True)):
# print(j, frag)
if j == 0 or (hold_steady and frag[0] in hold_steady):
# skip the first fragment
# that's already aligned with one of the atoms on shape_object
first_frag = frag
continue
frag_atoms.extend(frag)
v1 = target.bond(frag[0])
max_overlap = None
corresponding_position = None
for i, position in enumerate(shape_object.atoms[2:]):
if i in previous_positions:
continue
v2 = shape_object.atoms[0].bond(position)
d = np.dot(v1, v2)
# determine by max. bond overlap
if max_overlap is None or d > max_overlap:
max_overlap = d
corresponding_position = i
previous_positions.append(corresponding_position)
corresponding_position += 2
v1 = target.bond(frag[0])
v1 /= np.linalg.norm(v1)
v2 = shape_object.atoms[0].bond(
shape_object.atoms[corresponding_position]
)
v2 /= np.linalg.norm(v2)
rv = np.cross(v1, v2)
if np.linalg.norm(rv) < 10 ** -3:
continue
c = np.linalg.norm(v1 - v2)
if abs((c ** 2 - 2.0) / -2.0) >= 1:
continue
angle = np.arccos((c ** 2 - 2.0) / -2.0)
self.rotate(rv, angle, targets=frag, center=target)
# rotate the normal of this atom to be parallel to the largest group
# this makes changing to trigonal planar look cleaner
# don't do this if it isn't planar
if "planar" in new_shape:
for frag in frags:
if first_frag and frag is first_frag:
stop = frag[0]
other_vsepr = stop.get_vsepr()[0]
if (
isinstance(other_vsepr, str)
and "planar" in other_vsepr
):
min_torsion = None
for atom in target.connected:
if atom is stop:
continue
for a4 in stop.connected:
if a4 is target:
continue
torsion = self.dihedral(
atom, target, stop, a4
)
# print("checking", atom, a4, torsion, min_torsion)
if min_torsion is None or abs(
torsion
) < abs(min_torsion):
min_torsion = torsion
if (
min_torsion is not None
and abs(min_torsion) > 1e-2
):
angle = min_torsion
targs = []
self.write(outfile="test_ele.xyz")
for f in frags:
if f is frag:
continue
targs.extend(f)
self.rotate(
target.bond(stop),
angle,
targets=targs,
center=target,
)
for frag in frags:
if first_frag and frag is not first_frag:
stop = frag[0]
if len(stop.connected) > 1:
other_vsepr = stop.get_vsepr()[0]
if (
isinstance(other_vsepr, str)
and "planar" in other_vsepr
):
min_torsion = None
for atom in target.connected:
if atom is stop:
continue
for atom2 in stop.connected:
if atom2 is target:
continue
torsion = self.dihedral(
atom, target, stop, atom2
)
if min_torsion is None or abs(
torsion
) < abs(min_torsion):
min_torsion = torsion
if (
min_torsion is not None
and abs(min_torsion) > 1e-2
):
angle = -1 * min_torsion
targs = frag
self.rotate(
target.bond(stop),
angle,
targets=targs,
center=target,
)
self.refresh_ranks()
target.element = new_element
# these methods are normally called when an atom is instantiated
target.reset()
# fix bond lengths if requested
# try to guess the bond order based on saturation
if adjust_bonds:
from AaronTools.atoms import BondOrder
bo = BondOrder()
if hold_steady:
hold_steady = self.find(hold_steady)
frags = [
self.get_fragment(atom, target) for atom in target.connected
]
target_bo = 1
if hasattr(target, "_saturation"):
target_bo = max(
1 + target._saturation - len(target.connected), 1
)
for i, frag in enumerate(sorted(frags, key=len, reverse=True)):
frag_bo = 1
if hasattr(frag[0], "_saturation"):
frag_bo = max(
1 + frag[0]._saturation - len(frag[0].connected), 1
)
expected_bo = "%.1f" % float(min(frag_bo, target_bo))
# print(expected_bo)
key = bo.key(frag[0], target)
try:
expected_dist = bo.bonds[key][expected_bo]
except KeyError:
expected_dist = None
if hold_steady:
self.change_distance(
target,
frag[0],
as_group=True,
dist=expected_dist,
fix=2 if frag[0] in hold_steady else 1,
)
else:
self.change_distance(
target,
frag[0],
as_group=True,
dist=expected_dist,
fix=2 if i == 0 else 1,
)
def map_ligand(self, ligands, old_keys, minimize=True):
"""
Maps new ligand according to key_map
Parameters:
:ligand: the name of a ligand in the ligand library
:old_keys: the key atoms of the old ligand to map to
"""
def get_rotation(old_axis, new_axis):
w = np.cross(old_axis, new_axis)
# if old and new axes are colinear, use perp_vector
if np.linalg.norm(w) <= 1e-4:
w = utils.perp_vector(old_axis)
angle = np.dot(old_axis, new_axis)
angle /= np.linalg.norm(old_axis)
angle /= np.linalg.norm(new_axis)
# occasionally there will be some round-off errors,
# so let's fix those before we take arccos
if angle > 1 + 10 ** -12 or angle < -1 - 10 ** -12:
# and check to make sure we aren't covering something
# more senister up...
raise ValueError("Bad angle value for arccos():", angle)
elif angle > 1:
angle = 1.0
elif angle < -1:
angle = -1.0
angle = np.arccos(angle)
return w, -1 * angle
def map_1_key(self, ligand, old_key, new_key):
# align new key to old key
shift = new_key.bond(old_key)
ligand.coord_shift(shift)
# rotate ligand
targets = [
atom for atom in self.center if atom.is_connected(old_key)
]
if len(targets) > 0:
new_axis = shift - new_key.coords
else:
targets = old_key.connected - set(self.center)
new_axis = (
ligand.COM(targets=new_key.connected) - new_key.coords
)
if not targets:
old_axis = old_key.coords - self.COM(self.center)
else:
old_axis = self.COM(targets=targets) - old_key.coords
w, angle = get_rotation(old_axis, new_axis)
if np.linalg.norm(w) > 1e-4:
ligand.rotate(w, angle, center=new_key)
return ligand
def map_2_key(old_ligand, ligand, old_keys, new_keys, rev_ang=False):
# align COM of key atoms
center = old_ligand.COM(targets=old_keys)
shift = center - ligand.COM(targets=new_keys)
ligand.coord_shift(shift)
remove_centers = []
old_walk = False
# bend around key axis
try:
old_walk = old_ligand.shortest_path(*old_keys)
except (LookupError, ValueError):
# for some ferrocene ligands, AaronTools misidentifies the Fe
# as another metal center
# we'll remove any centers that are on the path between the key atoms
# also, sometimes the ligand atoms don't have the center in their connected
# attribute, even though the center has the ligand atoms in its
# connected attribute
for center in self.center:
for key in old_keys:
if center.is_connected(key):
center.add_bond_to(key)
# print("old keys:", old_keys)
# print("old ligand:\n", old_ligand)
stop = [
atom
for atom in old_keys[0].connected
if atom not in old_ligand.atoms
]
if stop:
frag = old_ligand.get_fragment(
old_keys[0],
stop=stop,
)
if all(atom in frag for atom in old_keys):
old_walk = self.shortest_path(
*old_keys,
avoid=[
a
for a in self.center
if any(k.is_connected(a) for k in old_keys)
]
)
remove_centers = [
c for c in self.center if c in old_walk
]
else:
old_walk = [a for a in old_keys]
if old_walk and len(old_walk) == 2:
old_con = set([])
for k in old_keys:
for c in k.connected:
old_con.add(c)
if old_con:
old_vec = old_ligand.COM(targets=old_con) - center
else:
old_vec = center
elif old_walk:
old_vec = old_ligand.COM(targets=old_walk[1:-1]) - center
else:
old_vec = np.zeros(3)
for atom in old_keys:
if any(
bonded_atom not in old_ligand.atoms
for bonded_atom in atom.connected
):
v = atom.coords - self.COM(
targets=[
bonded_atom
for bonded_atom in atom.connected
if bonded_atom not in old_ligand.atoms
]
)
v /= np.linalg.norm(v)
old_vec += v
# print(atom)
# print("vec:", old_vec)
old_vec /= np.linalg.norm(old_vec)
new_walk = ligand.shortest_path(*new_keys)
if len(new_walk) == 2:
new_con = set([])
for k in new_keys:
for c in k.connected:
new_con.add(c)
new_vec = ligand.COM(targets=new_con) - center
else:
new_vec = ligand.COM(targets=new_walk[1:-1]) - center
w, angle = get_rotation(old_vec, new_vec)
if rev_ang:
angle = -angle
ligand.rotate(w, angle, center=center)
# rotate for best overlap
old_axis = old_keys[0].bond(old_keys[1])
new_axis = new_keys[0].bond(new_keys[1])
w, angle = get_rotation(old_axis, new_axis)
ligand.rotate(w, angle, center=center)
return remove_centers
def map_rot_frag(frag, a, b, ligand, old_key, new_key):
old_vec = old_key.coords - b.coords
new_vec = new_key.coords - b.coords
axis, angle = get_rotation(old_vec, new_vec)
ligand.rotate(b.bond(a), -1 * angle, targets=frag, center=b.coords)
for c in new_key.connected:
con_frag = ligand.get_fragment(new_key, c)
if len(con_frag) > len(frag):
continue
old_vec = self.COM(targets=old_key.connected)
old_vec -= old_key.coords
new_vec = ligand.COM(targets=new_key.connected)
new_vec -= new_key.coords
axis, angle = get_rotation(old_vec, new_vec)
ligand.rotate(
c.bond(new_key),
-1 * angle,
targets=con_frag,
center=new_key.coords,
)
def map_more_key(self, old_ligand, ligand, old_keys, new_keys):
# backbone fragments separated by rotatable bonds
frag_list = ligand.get_frag_list(max_order=1)
# ligand.write("ligand")
remove_centers = []
# get key atoms on each side of rotatable bonds
key_count = {}
for frag, a, b in frag_list:
n_keys = []
for i in frag:
if i not in ligand.key_atoms:
continue
n_keys += [i]
if len(n_keys) < 1 or len(n_keys) > 2:
continue
if a in ligand.key_atoms or b in ligand.key_atoms:
continue
if utils.same_cycle(ligand, a, b):
continue
if len(n_keys) not in key_count:
key_count[len(n_keys)] = [(frag, a, b)]
else:
key_count[len(n_keys)] += [(frag, a, b)]
partial_map = False
mapped_frags = []
for k in sorted(key_count.keys(), reverse=True):
if k == 2 and not partial_map:
frag, a, b = key_count[k][0]
ok = []
nk = []
for i, n in enumerate(new_keys):
if n not in frag:
continue
ok += [old_keys[i]]
nk += [n]
remove_centers.extend(
map_2_key(old_ligand, ligand, ok, nk)
)
partial_map = True
mapped_frags += [frag]
continue
if k == 1 and not partial_map:
frag, a, b = key_count[k][0]
for i, n in enumerate(new_keys):
if n not in frag:
continue
map_1_key(self, ligand, n, old_keys[i])
partial_map = True
mapped_frags += [frag]
break
continue
if k == 1 and partial_map:
for frag, a, b in key_count[k]:
for i, n in enumerate(new_keys):
if n not in frag:
continue
map_rot_frag(frag, a, b, ligand, old_keys[i], n)
mapped_frags += [frag]
break
return remove_centers
if not self.components:
self.detect_components()
# find old and new keys
old_keys = self.find(old_keys)
if isinstance(ligands, (str, Geometry)):
ligands = [ligands]
new_keys = []
for i, ligand in enumerate(ligands):
if not isinstance(ligand, AaronTools.component.Component):
ligand = AaronTools.component.Component(ligand)
ligands[i] = ligand
ligand.refresh_connected()
new_keys += ligand.key_atoms
if len(old_keys) != len(new_keys):
raise ValueError(
"Cannot map ligand. "
+ "Differing number of key atoms. "
+ "Old keys: "
+ ",".join([i.name for i in old_keys])
+ "; "
+ "New keys: "
+ ",".join([i.name for i in new_keys])
)
old_ligand = []
remove_components = []
for k in old_keys:
for i, c in enumerate(self.components):
if k in c.atoms and k not in old_ligand:
old_ligand.extend(c.atoms)
remove_components.append(i)
for i in remove_components:
self.components.pop(i)
for j in range(0, len(remove_components)):
if remove_components[j] > i:
remove_components[j] -= 1
old_ligand = Geometry(old_ligand)
start = 0
end = None
remove_centers = []
for i, ligand in enumerate(ligands):
end = start + len(ligand.key_atoms)
if len(ligand.key_atoms) == 1:
map_1_key(self, ligand, old_keys[start], new_keys[start])
elif len(ligand.key_atoms) == 2:
remove_centers.extend(
map_2_key(
old_ligand,
ligand,
old_keys[start:end],
new_keys[start:end],
)
)
else:
remove_centers.extend(
map_more_key(
self,
old_ligand,
ligand,
old_keys[start:end],
new_keys[start:end],
)
)
for a in ligand.atoms:
a.name = old_keys[start].name + "." + a.name
a.add_tag("ligand")
start = end
for atom in self.atoms:
if atom.connected & set(old_ligand.atoms):
atom.connected = atom.connected - set(old_ligand.atoms)
# remove extraneous centers, i.e. from ferrocene ligands
for rc in remove_centers:
self.center.remove(rc)
# add new
for ligand in ligands:
self.components += [ligand]
rv = ligands
self.rebuild()
# rotate monodentate to relieve clashing
for ligand in self.components:
if len(ligand.key_atoms) == 1:
targets = ligand.atoms
key = ligand.key_atoms[0]
if self.center:
start = self.COM(self.center)
end = key.coords
else:
start = key.coords
end = self.COM(key.connected)
axis = end - start
self.minimize_torsion(targets, axis, center=key, increment=10)
self.remove_clash()
if minimize:
self.minimize()
self.refresh_ranks()
return rv
def remove_clash(self, sub_list=None):
def get_clash(sub, scale):
"""
Returns: np.array(bend_axis) if clash found, False otherwise
"""
clashing = []
D = distance_matrix(self.coords, sub.coords)
for i, atom in enumerate(self.atoms):
if atom in sub.atoms or atom == sub.end:
continue
threshold = atom._radii
for j, sub_atom in enumerate(sub.atoms):
threshold += sub_atom._radii
threshold *= scale
dist = D[i, j]
if dist < threshold or dist < 0.8:
clashing += [(atom, threshold - dist)]
if not clashing:
return False
rot_axis = sub.atoms[0].bond(sub.end)
vector = np.array([0, 0, 0], dtype=float)
for a, w in clashing:
vector += a.bond(sub.end) * w
bend_axis = np.cross(rot_axis, vector)
return bend_axis
bad_subs = [] # substituents for which releif not found
# bend_angles = [8, -16, 32, -48, 68, -88]
# bend_back = np.deg2rad(20)
bend_angles = [8, 8, 8, 5, 5, 5]
bend_back = []
rot_angles = [8, -16, 32, -48]
rot_back = np.deg2rad(16)
scale = 0.75 # for scaling distance threshold
if sub_list is None:
sub_list = sorted(self.get_substituents())
try_twice = True
else:
scale = 0.65
sub_list = sorted(sub_list, reverse=True)
try_twice = False
for i, b in enumerate(bend_angles):
bend_angles[i] = -np.deg2rad(b)
for i, r in enumerate(rot_angles):
rot_angles[i] = np.deg2rad(r)
for sub in sub_list:
b, r = 0, 0 # bend_angle, rot_angle index counters
bend_axis = get_clash(sub, scale)
if bend_axis is False:
continue
else:
# try just rotating first
while r < len(rot_angles):
# try rotating
if r < len(rot_angles):
sub.sub_rotate(rot_angles[r])
r += 1
if get_clash(sub, scale) is False:
break
else:
sub.sub_rotate(rot_back)
r = 0
bend_axis = get_clash(sub, scale)
while b < len(bend_angles) and bend_axis is not False:
bend_back += [bend_axis]
# try bending
if b < len(bend_angles):
sub.rotate(bend_axis, bend_angles[b], center=sub.end)
b += 1
bend_axis = get_clash(sub, scale)
if bend_axis is False:
break
while r < len(rot_angles):
# try rotating
if r < len(rot_angles):
sub.sub_rotate(rot_angles[r])
r += 1
if get_clash(sub, scale) is False:
break
else:
sub.sub_rotate(rot_back)
r = 0
else:
# bend back to original if cannot automatically remove
# the clash, add to bad_sub list
bend_axis = get_clash(sub, scale)
if bend_axis is False:
break
for bend_axis in bend_back:
sub.rotate(bend_axis, -bend_angles[0], center=sub.end)
bad_subs += [sub]
# try a second time just in case other subs moved out of the way enough
# for the first subs encountered to work now
if try_twice and len(bad_subs) > 0:
bad_subs = self.remove_clash(bad_subs)
return bad_subs
def minimize(self):
"""
Rotates substituents in each component to minimize LJ_energy.
Different from Component.minimize_sub_torsion() in that it minimizes
with respect to the entire catalyst instead of just the component
"""
targets = {}
for sub in self.get_substituents(for_confs=True):
if len(sub.atoms):
continue
try:
targets[len(sub.atoms)] += [sub]
except KeyError:
targets[len(sub.atoms)] = [sub]
# minimize torsion for each substituent
# smallest to largest
for k in sorted(targets.keys()):
for sub in targets[k]:
axis = sub.atoms[0].bond(sub.end)
center = sub.end
self.minimize_torsion(sub.atoms, axis, center)
def next_conformer(self, conf_spec, skip_spec={}):
"""
Generates the next possible conformer
:conf_spec: {sub_start_number: conf_number}
:skip_spec: {sub_start_number: [skip_numbers]}
Returns:
conf_spec if there are still more conformers
{} if there are no more conformers to generate
"""
for start, conf_num in sorted(conf_spec.items()):
sub = self.find_substituent(start)
# skip conformer if signalled it's a repeat
skip = skip_spec.get(start, [])
if skip == "all" or conf_num == 0 or conf_num in skip:
if conf_num == sub.conf_num:
conf_spec[start] = 1
else:
conf_spec[start] += 1
continue
# reset conf if we hit max conf #
if conf_num == sub.conf_num:
sub.sub_rotate()
conf_spec[start] = 1
continue
# perform rotation
sub.sub_rotate()
conf_spec[start] += 1
self.remove_clash()
# continue if the same as cf1
angle = int(np.rad2deg((conf_spec[start] - 1) * sub.conf_angle))
if angle != 360 and angle != 0:
return conf_spec
else:
continue
else:
# we are done now
return {}
def make_conformer(self, conf_spec):
"""
Returns:
conf_spec, True if conformer generated (allowed by conf_spec),
conf_spec, False if not allowed or invalid
:conf_spec: dictionary of the form
{sub_start_number: (conf_number, [skip_numbers])}
"""
original = self.copy()
for start, conf_num in conf_spec.items():
current, skip = conf_spec[start]
# skip if flagged a repeat
if conf_num in skip or skip == "all":
self = original
return conf_spec, False
sub = self.find_substituent(start)
# validate conf_spec
if conf_num > sub.conf_num:
self = original
self.LOG.warning(
"Bad conformer number given: {} {} > {}".format(
sub.name, conf_num, sub.conf_num
)
)
return conf_spec, False
if conf_num > current:
n_rot = conf_num - current - 1
for _ in range(n_rot):
conf_spec[start][0] += 1
sub.rotate()
elif conf_num < current:
n_rot = current - conf_num - 1
for _ in range(n_rot):
conf_spec[start][0] -= 1
sub.rotate(reverse=True)
return conf_spec, True
def change_chirality(self, target):
"""
change chirality of the target atom
target should be a chiral center that is not a bridgehead
of a fused ring, though spiro centers are allowed
"""
# find two fragments
# rotate those about the vector that bisects the angle between them
# this effectively changes the chirality
target = self.find_exact(target)[0]
fragments = []
for a in target.connected:
frag = self.get_fragment(
a, target,
)
if sum(int(target in frag_atom.connected) for frag_atom in frag) == 1:
fragments.append([atom.copy() for atom in frag])
if len(fragments) == 2:
break
# if there are not two fragments not in a ring,
# this is a spiro center
# find a spiro ring and rotate that
a2 = None
if len(fragments) < 2:
for a1 in target.connected:
targets = self.get_fragment(
a1, stop=target,
)
a2 = [a for a in targets if a in target.connected and a is not a1]
if a2:
a2 = a2[0]
break
if not a2:
raise RuntimeError(
"could not find suitable groups to swap on %s" % target
)
v1 = target.bond(a1)
v1 /= np.linalg.norm(v1)
v2 = target.bond(a2)
v2 /= np.linalg.norm(v2)
rv = v1 + v2
self.rotate(
rv, angle=np.pi, center=target,
targets=targets,
)
else:
v1 = target.bond(fragments[0][0])
v1 /= np.linalg.norm(v1)
v2 = target.bond(fragments[1][0])
v2 /= np.linalg.norm(v2)
rv = v1 + v2
targets = [atom.name for atom in fragments[0]]
targets.extend([atom.name for atom in fragments[1]])
self.rotate(
rv, angle=np.pi, center=target,
targets=targets,
)
return targets | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/geometry.py | geometry.py |
from AaronTools import addlogger
from AaronTools.theory import GAUSSIAN_ROUTE, ORCA_ROUTE, QCHEM_REM
@addlogger
class EmpiricalDispersion:
"""try to keep emerpical dispersion keywords and settings consistent across file types"""
LOG = None
def __init__(self, name):
"""name can be (availability may vary):
Grimme D2 (or D2, -D2, GD2)
Zero-damped Grimme D3 (or D3, -D3, GD3)
Becke-Johnson damped Grimme D3 (or D3BJ, -D3BJ, GD3BJ)
Becke-Johnson damped modified Grimme D3 (or B3MBJ, -D3MBJ)
Petersson-Frisch (or PFD)
Grimme D4 (or D4, -D4, GD4)
Chai & Head-Gordon (or CHG, -CHG)
Nonlocal Approximation (or NL, NLA, -NL)
Pernal, Podeszwa, Patkowski, & Szalewicz (or DAS2009, -DAS2009)
Podeszwa, Katarzyna, Patkowski, & Szalewicz (or DAS2010, -DAS2010)
Coupled-Cluster Doubles (or CCD)
Řezác, Greenwell, & Beran (or DMP2)
Coupled-Cluster Doubles + Řezác, Greenwell, & Beran (or (CCD)DMP2)
or simply the keyword for the input file type you are using"""
self.name = name
def __eq__(self, other):
if self.__class__ is not other.__class__:
return False
if self.name == other.name:
return True
for d in [
["grimme d2", "d2", "-d2", "gd2"],
["grimme d3", "d3", "-d3", "gd3", "d3zero", "zero-damped grimme d3"],
["becke-johnson damped grimme d3", "d3bj", "-d3bj", "gd3bj"],
["becke-johnson damped modified grimme d3", "d3mbj", "-d3mbj"],
["petersson-frisch", "pfd"],
["grimme d4", "d4", "-d4", "gd4"],
["nonlocal approximation", "nl", "nla", "-nl"],
["coupled-cluster doubles", "ccd"],
]:
if self.name.lower() in d and other.name.lower() in d:
return True
return False
def copy(self):
new_dict = dict()
for key, value in self.__dict__.items():
try:
new_dict[key] = value.copy()
except AttributeError:
new_dict[key] = value
# ignore chimerax objects so seqcrow doesn't print a
# warning when a geometry is copied
if "chimerax" in value.__class__.__module__:
continue
if value.__class__.__module__ != "builtins":
self.LOG.warning(
"No copy method for {}: in-place changes may occur".format(
type(value)
)
)
return self.__class__(**new_dict)
def get_gaussian(self):
"""Acceptable dispersion methods for Gaussian are:
Grimme D2
Grimme D3
Becke-Johnson damped Grimme D3
Petersson-Frisch
Dispersion methods available in other software that will be modified are:
Grimme D4
undampened Grimme D3"""
if any(
self.name.upper() == name for name in [
"GRIMME D2", "GD2", "D2", "-D2"
]
):
return ({GAUSSIAN_ROUTE:{"EmpiricalDispersion":["GD2"]}}, None)
elif any(
self.name.upper() == name for name in [
"ZERO-DAMPED GRIMME D3", "GRIMME D3", "GD3", "D3", "-D3", "D3ZERO"
]
):
return ({GAUSSIAN_ROUTE:{"EmpiricalDispersion":["GD3"]}}, None)
elif any(
self.name.upper() == name for name in [
"BECKE-JOHNSON DAMPED GRIMME D3", "GD3BJ", "D3BJ", "-D3BJ"
]
):
return ({GAUSSIAN_ROUTE:{"EmpiricalDispersion":["GD3BJ"]}}, None)
elif any(
self.name.upper() == name for name in [
"PETERSSON-FRISCH", "PFD"
]
):
return ({GAUSSIAN_ROUTE:{"EmpiricalDispersion":["PFD"]}}, None)
#dispersions in ORCA but not Gaussian
elif self.name == "Grimme D4":
out_dict = {
GAUSSIAN_ROUTE: {
"EmpiricalDispersion":["GD3BJ"]
}
}
return (out_dict, "Grimme's D4 has no keyword in Gaussian, switching to GD3BJ")
#unrecognized
return (self.name, "unrecognized emperical dispersion: %s" % self.name)
def get_orca(self):
"""Acceptable keywords for ORCA are:
Grimme D2
Zero-damped Grimme D3
Becke-Johnson damped Grimme D3
Grimme D4"""
if any(
self.name.upper() == name for name in [
"GRIMME D2", "GD2", "D2", "-D2"
]
):
return ({ORCA_ROUTE:["D2"]}, None)
elif any(
self.name.upper() == name for name in [
"ZERO-DAMPED GRIMME D3", "GRIMME D3", "GD3", "D3", "-D3", "D3ZERO"
]
):
return ({ORCA_ROUTE:["D3ZERO"]}, None)
elif any(
self.name.upper() == name for name in [
"BECKE-JOHNSON DAMPED GRIMME D3", "GD3BJ", "D3BJ", "-D3BJ"
]
):
return ({ORCA_ROUTE:["D3BJ"]}, None)
elif any(
self.name.upper() == name for name in [
"GRIMME D4", "GD4", "D4", "-D4"
]
):
return ({ORCA_ROUTE:["D4"]}, None)
out_dict = {
ORCA_ROUTE: [self.name]
}
return(out_dict, "unrecognized emperical dispersion: %s" % self.name)
def get_psi4(self):
"""Acceptable keywords for Psi4 are:
Grimme D1
Grimme D2
Zero-damped Grimme D3
Becke-Johnson damped Grimme D3
Chai & Head-Gordon
Nonlocal Approximation
Pernal, Podeszwa, Patkowski, & Szalewicz
Podeszwa, Katarzyna, Patkowski, & Szalewicz
Řezác, Greenwell, & Beran"""
if any(
self.name.upper() == name for name in [
"GRIMME D1", "GD1", "D1", "-D1"
]
):
return ("-d1", None)
elif any(
self.name.upper() == name for name in [
"GRIMME D2", "GD2", "D2", "-D2"
]
):
return ("-d2", None)
elif any(
self.name.upper() == name for name in [
"ZERO-DAMPED GRIMME D3", "GRIMME D3", "GD3", "D3", "-D3", "D3ZERO"
]
):
return ("-d3", None)
elif any(
self.name.upper() == name for name in [
"BECKE-JOHNSON DAMPED GRIMME D3", "GD3BJ", "D3BJ", "-D3BJ"
]
):
return ("-d3bj", None)
elif any(
self.name.upper() == name for name in [
"BECKE-JOHNSON DAMPED MODIFIED GRIMME D3", "GD3MBJ", "D3MBJ", "-D3MBJ"
]
):
return ("-d3mbj", None)
elif any(
self.name.upper() == name for name in [
"CHAI & HEAD-GORDON", "CHG", "-CHG"
]
):
return ("-chg", None)
elif any(
self.name.upper() == name for name in [
"NONLOCAL APPROXIMATION", "NL", "NLA", "-NL"
]
):
return ("-nl", None)
elif any(
self.name.upper() == name for name in [
"PERNAL, PODESZWA, PATKOWSKI, & SZALEWICZ", "DAS2009", "-DAS2009"
]
):
return ("-das2009", None)
elif any(
self.name.upper() == name for name in [
"PODESZWA, KATARZYNA, PATKOWSKI, & SZALEWICZ", "DAS2010", "-DAS2010"
]
):
return ("-das2010", None)
elif any(
self.name.upper() == name for name in [
"COUPLED-CLUSTER DOUBLES", "CCD"
]
):
return ("(ccd)", None)
elif any(
self.name.upper() == name for name in [
"ŘEZÁC, GREENWELL, & BERAN", "DMP2"
]
):
return ("dmp2", None)
elif any(
self.name.upper() == name for name in [
"COUPLED-CLUSTER DOUBLES + ŘEZÁC, GREENWELL, & BERAN", "(CCD)DMP2"
]
):
return ("(ccd)dmp2", None)
else:
return (self.name, "unrecognized emperical dispersion: %s" % self.name)
def get_qchem(self):
"""Acceptable keywords for QChem are:
Grimme D2
Modified Zero-damped Grimme D3
Zero-damped Grimme D3
Becke-Johnson damped Grimme D3
Becke-Johnson damped modified Grimme D3
Chai & Head-Gordon
"""
if any(
self.name.upper() == name for name in [
"GRIMME D2", "GD2", "D2", "-D2", "EMPIRICAL_GRIMME",
]
):
return ({QCHEM_REM: {"DFT_D": "EMPIRICAL_GRIMME"}}, None)
elif any(
self.name.upper() == name for name in [
"ZERO-DAMPED GRIMME D3", "GRIMME D3", "GD3", "D3",
"-D3", "D3ZERO", "D3_ZERO",
]
):
return ({QCHEM_REM: {"DFT_D": "D3_ZERO"}}, None)
elif any(
self.name.upper() == name for name in [
"MODIFIED ZERO-DAMPED GRIMME D3", "D3_ZEROM",
]
):
# Smith et. al. modified D3
return ({QCHEM_REM: {"DFT_D": "D3_ZEROM"}}, None)
elif any(
self.name.upper() == name for name in [
"C6 ONLY GRIMME D3", "C<SUB>6</SUB> ONLY GRIMME D3", "D3_CSO",
]
):
return ({QCHEM_REM: {"DFT_D": "D3_CSO"}}, None)
elif any(
self.name.upper() == name for name in [
"OPTIMIZED POWER GRIMME D3", "D3_OP",
]
):
return ({QCHEM_REM: {"DFT_D": "D3_OP"}}, None)
elif any(
self.name.upper() == name for name in [
"BECKE-JOHNSON DAMPED GRIMME D3", "GD3BJ", "D3BJ",
"-D3BJ", "B3BJ",
]
):
return ({QCHEM_REM: {"DFT_D": "D3_BJ"}}, None)
elif any(
self.name.upper() == name for name in [
"BECKE-JOHNSON DAMPED MODIFIED GRIMME D3", "GD3MBJ",
"D3MBJ", "-D3MBJ", "D3_BJM",
]
):
return ({QCHEM_REM: {"DFT_D": "D3_BJM"}}, None)
elif any(
self.name.upper() == name for name in [
"CHAI & HEAD-GORDON", "CHG", "-CHG", "EMPIRICAL_CHG"
]
):
return ({QCHEM_REM: {"DFT_D": "EMPIRICAL_CHG"}}, None)
elif any(
self.name.upper() == name for name in [
"CALDEWEYHER ET. AL. D4", "D4",
]
):
return ({QCHEM_REM: {"DFT_D": "D4"}}, None)
else:
return (self.name, "unrecognized emperical dispersion: %s" % self.name) | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/theory/emp_dispersion.py | emp_dispersion.py |
import re
from AaronTools import addlogger
from AaronTools.const import ELEMENTS, UNIT
from AaronTools.theory import (
GAUSSIAN_COMMENT,
GAUSSIAN_CONSTRAINTS,
GAUSSIAN_COORDINATES,
GAUSSIAN_GEN_BASIS,
GAUSSIAN_GEN_ECP,
GAUSSIAN_POST,
GAUSSIAN_PRE_ROUTE,
GAUSSIAN_ROUTE,
ORCA_BLOCKS,
ORCA_COMMENT,
ORCA_COORDINATES,
ORCA_ROUTE,
PSI4_AFTER_JOB,
PSI4_BEFORE_GEOM,
PSI4_BEFORE_JOB,
PSI4_COMMENT,
PSI4_COORDINATES,
PSI4_JOB,
PSI4_MOLECULE,
PSI4_OPTKING,
PSI4_SETTINGS,
PSI4_SOLVENT,
SQM_COMMENT,
SQM_QMMM,
QCHEM_MOLECULE,
QCHEM_REM,
QCHEM_COMMENT,
QCHEM_SETTINGS,
FrequencyJob,
job_from_string,
)
from AaronTools.utils.utils import combine_dicts
from AaronTools.theory.basis import ECP, BasisSet
from AaronTools.theory.emp_dispersion import EmpiricalDispersion
from AaronTools.theory.grid import IntegrationGrid
from AaronTools.theory.job_types import JobType, SinglePointJob
from AaronTools.theory.method import KNOWN_SEMI_EMPIRICAL, Method, SAPTMethod
@addlogger
class Theory:
"""
A Theory object can be used to create an input file for different QM software.
The creation of a Theory object does not depend on the specific QM software
that is determined when the file is written
attribute names are the same as initialization keywords (with the exception of ecp, which
is added to the basis attribute)
valid initialization keywords are:
geometry - AaronTools Geometry
charge - total charge
multiplicity - electronic multiplicity
job_type - JobType or list(JobType)
method - Method object (or str - Method instance will be created)
basis - BasisSet object (or str - will be set to BasisSet(Basis(keyword)))
ecp - str parsable by BasisSet.parse_basis_str
empirical_dispersion - EmpiricalDispersion object (or str)
grid - IntegrationGrid object (or str)
solvent - ImplicitSolvent object
memory - int - allocated memory (GB)
processors - int - allocated cores
"""
ACCEPTED_INIT_KW = [
"geometry",
"memory",
"processors",
"job_type",
"solvent",
"grid",
"empirical_dispersion",
"basis",
"method",
]
# if there's a setting that should be an array and Psi4 errors out
# if it isn't an array, put it in this list (lower case)
# generally happens if the setting value isn't a string
# don't add settings that need > 1 value in the array
FORCED_PSI4_ARRAY = [
"cubeprop_orbitals",
"docc",
"frac_occ",
]
# commonly used settings that do not take array values
FORCED_PSI4_SINGLE = [
"reference",
"scf_type",
"freeze_core",
"diag_method",
"ex_level",
"fci",
"maxiter",
"t",
"p",
"opt_type",
"dft_radial_points",
"dft_spherical_points",
]
FORCED_PSI4_SOLVENT_SINGLE = [
"units",
"codata",
"type",
"npzfile",
"area",
"scaling",
"raddiset",
"minradius",
"mode",
"nonequilibrium",
"solvent",
"solvertype",
"matrixsymm",
"correction",
"diagonalintegrator",
"diagonalscaling",
"proberadius",
]
# some blocks need to go after the molecule
# because they refer to atom indices, so orca needs
# to read the structure first
ORCA_BLOCKS_AFTER_MOL = ["eprnmr"]
LOG = None
def __init__(
self,
charge=0,
multiplicity=1,
method=None,
basis=None,
ecp=None,
empirical_dispersion=None,
grid=None,
**kwargs,
):
if not isinstance(charge, list):
self.charge = int(charge)
else:
self.charge = charge
if not isinstance(multiplicity, list):
self.multiplicity = int(multiplicity)
else:
self.multiplicity = multiplicity
self.method = None
self.basis = None
self.empirical_dispersion = None
self.grid = None
self.solvent = None
self.job_type = None
self.processors = None
self.memory = None
self.geometry = None
self.kwargs = {}
for key in self.ACCEPTED_INIT_KW:
if key in kwargs:
self.__setattr__(key, kwargs[key])
del kwargs[key]
else:
self.__setattr__(key, None)
self.kwargs = kwargs
if isinstance(self.processors, str):
processors = re.search(r"(\d+)", self.processors)
if processors:
self.processors = processors.group(1)
if isinstance(self.memory, str):
memory = re.search(r"(\d+)", self.memory)
if memory:
self.memory = memory.group(1)
# if method, basis, etc aren't the expected classes, make them so
if method is not None:
if not isinstance(method, Method):
self.method = Method(
method, method.upper() in KNOWN_SEMI_EMPIRICAL
)
else:
self.method = method
if grid is not None:
if not isinstance(grid, IntegrationGrid):
self.grid = IntegrationGrid(grid)
else:
self.grid = grid
if basis is not None:
self.basis = BasisSet(basis=basis)
if ecp is not None:
if self.basis is None:
self.basis = BasisSet(ecp=ecp)
else:
self.basis.ecp = BasisSet(ecp=ecp).ecp
if empirical_dispersion is not None:
if not isinstance(empirical_dispersion, EmpiricalDispersion):
self.empirical_dispersion = EmpiricalDispersion(
empirical_dispersion
)
else:
self.empirical_dispersion = empirical_dispersion
if self.job_type is not None:
if isinstance(self.job_type, str):
self.job_type = job_from_string(self.job_type)
if isinstance(self.job_type, JobType):
self.job_type = [self.job_type]
# for i, job1 in enumerate(self.job_type):
# for job2 in self.job_type[i + 1 :]:
# if type(job1) is type(job2):
# self.LOG.warning(
# "multiple jobs of the same type: %s, %s"
# % (str(job1), str(job2))
# )
def __setattr__(self, attr, val):
if isinstance(val, str):
if attr == "method":
super().__setattr__(attr, Method(val))
elif attr == "basis":
super().__setattr__(attr, BasisSet(val))
elif attr == "empirical_dispersion":
super().__setattr__(attr, EmpiricalDispersion(val))
elif attr == "grid":
super().__setattr__(attr, IntegrationGrid(val))
elif attr == "job_type" and isinstance(val, str):
super().__setattr__(attr, [job_from_string(val)])
else:
super().__setattr__(attr, val)
elif attr == "job_type" and isinstance(val, JobType):
super().__setattr__(attr, [val])
else:
super().__setattr__(attr, val)
def __eq__(self, other):
if self.method != other.method:
# print("method")
return False
if self.charge != other.charge:
# print("charge")
return False
if self.multiplicity != other.multiplicity:
# print("multiplicity")
return False
if self.basis != other.basis:
# print("basis")
return False
if self.empirical_dispersion != other.empirical_dispersion:
# print("disp")
print(self.empirical_dispersion, other.empirical_dispersion)
return False
if self.grid != other.grid:
# print("grid")
return False
if self.solvent != other.solvent:
# print("solvent")
return False
if self.processors != other.processors:
# print("procs")
return False
if self.memory != other.memory:
# print("mem")
return False
if self.job_type and other.job_type:
if len(self.job_type) != len(other.job_type):
return False
for job1, job2 in zip(self.job_type, other.job_type):
if job1 != job2:
return False
return True
def copy(self):
new_dict = dict()
new_kwargs = dict()
for key, value in self.__dict__.items():
try:
if key == "kwargs":
new_kwargs = value.copy()
else:
new_dict[key] = value.copy()
except AttributeError:
new_dict[key] = value
# ignore chimerax objects so seqcrow doesn't print a
# warning when a geometry is copied
if "chimerax" in value.__class__.__module__:
continue
if value.__class__.__module__ != "builtins":
self.LOG.warning(
"No copy method for {}: in-place changes may occur".format(
type(value)
)
)
return self.__class__(**new_dict, **new_kwargs)
def make_header(
self,
geom=None,
style="gaussian",
conditional_kwargs=None,
sanity_check_method=False,
**kwargs,
):
"""
geom: Geometry
style: str, gaussian, orca, psi4, or sqm
conditional_kwargs: dict - keys are ORCA_*, PSI4_*, or GAUSSIAN_*
items in conditional_kwargs will only be added
to the input if they would otherwise be preset
e.g. if self.job_type is FrequencyJob and a Gaussian
input file is being written,
conditional_kwargs = {GAUSSIAN_ROUTE:{'opt':['noeigentest']}}
will not add opt=noeigentest to the route
but if it's an OptimizationJob, it will add opt=noeigentest
sanity_check_method: bool, check if method is available in recent version
of the target software package (Psi4 checks when its
footer is created)
kwargs: keywords are GAUSSIAN_*, ORCA_*, PSI4_*, or QCHEM_*
"""
if geom is None:
geom = self.geometry
if conditional_kwargs is None:
conditional_kwargs = {}
if geom is not None:
self.geometry = geom
if self.basis is not None:
self.basis.refresh_elements(self.geometry)
kwargs = combine_dicts(self.kwargs, kwargs)
other_kw_dict = {}
for keyword in kwargs:
if (
keyword.startswith("PSI4_")
or keyword.startswith("ORCA_")
or keyword.startswith("GAUSSIAN_")
or keyword.startswith("QCHEM_")
):
new_kw = eval(keyword)
other_kw_dict[new_kw] = kwargs[keyword]
elif hasattr(self, keyword):
if keyword == "method":
self.method = Method(kwargs[keyword])
elif keyword == "basis":
self.basis = BasisSet(kwargs[keyword])
elif keyword == "grid":
self.grid = IntegrationGrid(kwargs[keyword])
elif keyword == "empirical_dispersion":
self.grid = EmpiricalDispersion(kwargs[keyword])
elif keyword == "job_type":
if isinstance(kwargs[keyword], JobType):
self.job_type = [kwargs[keyword]]
else:
self.job_type = kwargs[keyword]
elif keyword in self.ACCEPTED_INIT_KW:
setattr(self, keyword, kwargs[keyword])
else:
other_kw_dict[keyword] = kwargs[keyword]
if style == "gaussian":
return self.get_gaussian_header(
conditional_kwargs=conditional_kwargs, **other_kw_dict
)
elif style == "orca":
return self.get_orca_header(
conditional_kwargs=conditional_kwargs, **other_kw_dict
)
elif style == "psi4":
return self.get_psi4_header(
conditional_kwargs=conditional_kwargs, **other_kw_dict
)
elif style == "sqm":
return self.get_sqm_header(
conditional_kwargs=conditional_kwargs, **other_kw_dict
)
elif style == "qchem":
return self.get_qchem_header(
conditional_kwargs=conditional_kwargs, **other_kw_dict
)
raise NotImplementedError("no get_header method for style: %s" % style)
def make_molecule(
self,
geom=None,
style="gaussian",
conditional_kwargs=None,
**kwargs,
):
"""
geom: Geometry()
style: gaussian, psi4, or sqm
conditional_kwargs: dict() of keyword: value pairs
kwargs: keywords are GAUSSIAN_*, ORCA_*, PSI4_*, or QCHEM_*
"""
if geom is None:
geom = self.geometry
if conditional_kwargs is None:
conditional_kwargs = {}
if self.basis is not None:
self.basis.refresh_elements(geom)
kwargs = combine_dicts(self.kwargs, kwargs)
other_kw_dict = {}
for keyword in kwargs:
if (
keyword.startswith("PSI4_")
or keyword.startswith("ORCA_")
or keyword.startswith("GAUSSIAN_")
or keyword.startswith("QCHEM_")
):
new_kw = eval(keyword)
other_kw_dict[new_kw] = kwargs[keyword]
elif hasattr(self, keyword):
if keyword == "method":
self.method = Method(kwargs[keyword])
elif keyword == "basis":
self.basis = BasisSet(kwargs[keyword])
elif keyword == "grid":
self.grid = IntegrationGrid(kwargs[keyword])
elif keyword == "empirical_dispersion":
self.grid = EmpiricalDispersion(kwargs[keyword])
elif keyword == "job_type":
if isinstance(kwargs[keyword], JobType):
self.job_type = [kwargs[keyword]]
else:
self.job_type = kwargs[keyword]
elif keyword in self.ACCEPTED_INIT_KW:
setattr(self, keyword, kwargs[keyword])
else:
other_kw_dict[keyword] = kwargs[keyword]
if style == "gaussian":
return self.get_gaussian_molecule(
conditional_kwargs=conditional_kwargs, **other_kw_dict
)
elif style == "psi4":
return self.get_psi4_molecule(
conditional_kwargs=conditional_kwargs, **other_kw_dict
)
elif style == "sqm":
return self.get_sqm_molecule(
conditional_kwargs=conditional_kwargs, **other_kw_dict
)
elif style == "qchem":
return self.get_qchem_molecule(
conditional_kwargs=conditional_kwargs, **other_kw_dict
)
NotImplementedError("no get_molecule method for style: %s" % style)
def make_footer(
self,
geom=None,
style="gaussian",
conditional_kwargs=None,
sanity_check_method=False,
**kwargs,
):
"""
geom: Geometry
style: str, gaussian or psi4
conditional_kwargs: dict, see make_header
sanity_check_method: bool, check if method is available in recent version
of the target software package (Psi4 only)
kwargs: keywords are GAUSSIAN_*, ORCA_*, or PSI4_*
"""
if geom is None:
geom = self.geometry
if conditional_kwargs is None:
conditional_kwargs = {}
if self.basis is not None:
self.basis.refresh_elements(geom)
kwargs = combine_dicts(self.kwargs, kwargs)
other_kw_dict = {}
for keyword in kwargs:
if (
keyword.startswith("PSI4_")
or keyword.startswith("ORCA_")
or keyword.startswith("GAUSSIAN_")
):
new_kw = eval(keyword)
other_kw_dict[new_kw] = kwargs[keyword]
elif hasattr(self, keyword):
if keyword == "method":
self.method = Method(kwargs[keyword])
elif keyword == "basis":
self.basis = BasisSet(kwargs[keyword])
elif keyword == "grid":
self.grid = IntegrationGrid(kwargs[keyword])
elif keyword == "empirical_dispersion":
self.grid = EmpiricalDispersion(kwargs[keyword])
elif keyword == "job_type":
if isinstance(kwargs[keyword], JobType):
self.job_type = [kwargs[keyword]]
else:
self.job_type = kwargs[keyword]
elif keyword in self.ACCEPTED_INIT_KW:
setattr(self, keyword, kwargs[keyword])
else:
other_kw_dict[keyword] = kwargs[keyword]
if style == "gaussian":
return self.get_gaussian_footer(
conditional_kwargs=conditional_kwargs, **other_kw_dict
)
elif style == "psi4":
return self.get_psi4_footer(
conditional_kwargs=conditional_kwargs, **other_kw_dict
)
elif style == "orca":
return self.get_orca_footer(
conditional_kwargs=conditional_kwargs, **other_kw_dict
)
NotImplementedError("no get_footer method for style: %s" % style)
def get_gaussian_header(
self,
return_warnings=False,
conditional_kwargs=None,
**other_kw_dict,
):
"""
write Gaussian09/16 input file header (up to charge mult)
other_kw_dict is a dictionary with file positions (using GAUSSIAN_*)
corresponding to options/keywords
returns warnings if a certain feature is not available in Gaussian
"""
if conditional_kwargs is None:
conditional_kwargs = {}
warnings = []
if self.job_type is not None:
for job in self.job_type[::-1]:
if hasattr(job, "geometry"):
job.geometry = self.geometry
job_dict = job.get_gaussian()
other_kw_dict = combine_dicts(job_dict, other_kw_dict)
if (
GAUSSIAN_COMMENT not in other_kw_dict
or not other_kw_dict[GAUSSIAN_COMMENT]
):
if self.geometry.comment:
other_kw_dict[GAUSSIAN_COMMENT] = [self.geometry.comment]
else:
other_kw_dict[GAUSSIAN_COMMENT] = [self.geometry.name]
# add EmpiricalDispersion info
if self.empirical_dispersion is not None:
disp, warning = self.empirical_dispersion.get_gaussian()
other_kw_dict = combine_dicts(other_kw_dict, disp)
if warning is not None:
warnings.append(warning)
# add Integral(grid=X)
if self.grid is not None:
grid, warning = self.grid.get_gaussian()
other_kw_dict = combine_dicts(other_kw_dict, grid)
if warning is not None:
warnings.append(warning)
# add implicit solvent
if self.solvent is not None:
solvent_info, warning = self.solvent.get_gaussian()
if warning is not None:
warnings.extend(warning)
other_kw_dict = combine_dicts(other_kw_dict, solvent_info)
other_kw_dict = combine_dicts(
other_kw_dict, conditional_kwargs, dict2_conditional=True
)
out_str = ""
# processors, memory, and other link 0 stuff
if self.processors is not None:
out_str += "%%NProcShared=%i\n" % self.processors
if self.memory is not None:
out_str += "%%Mem=%iGB\n" % self.memory
if GAUSSIAN_PRE_ROUTE in other_kw_dict:
for key in other_kw_dict[GAUSSIAN_PRE_ROUTE]:
out_str += "%%%s" % key
if other_kw_dict[GAUSSIAN_PRE_ROUTE][key]:
out_str += "=%s" % ",".join(
other_kw_dict[GAUSSIAN_PRE_ROUTE][key]
)
if not out_str.endswith("\n"):
out_str += "\n"
# start route line with method
out_str += "#n "
if self.method is not None:
func, warning = self.method.get_gaussian()
if warning is not None:
warnings.append(warning)
warning = self.method.sanity_check_method(func, "gaussian")
if warning:
warnings.append(warning)
out_str += "%s" % func
if not self.method.is_semiempirical and self.basis is not None:
(
basis_info,
basis_warnings,
) = self.basis.get_gaussian_basis_info()
warnings.extend(basis_warnings)
# check basis elements to make sure no element is
# in two basis sets or left out of any
if self.geometry is not None:
basis_warning = self.basis.check_for_elements(
self.geometry
)
if basis_warning is not None:
warnings.append(basis_warning)
if GAUSSIAN_ROUTE in basis_info:
out_str += "%s" % basis_info[GAUSSIAN_ROUTE]
out_str += " "
# add other route options
# only one option can be specfied
# e.g. for {'Integral':['grid=X', 'grid=Y']}, only grid=X will be used
if GAUSSIAN_ROUTE in other_kw_dict.keys():
for option in other_kw_dict[GAUSSIAN_ROUTE].keys():
known_opts = []
out_str += option
if option.lower() == "opt":
# need to specified CalcFC for gaussian ts optimization
if any(
x.lower() == "ts"
for x in other_kw_dict[GAUSSIAN_ROUTE][option]
) and not any(
x.lower() == y
for y in [
"calcfc",
"readfc",
"rcfc",
"readcartesianfc",
"calcall",
"calchffc",
]
for x in other_kw_dict[GAUSSIAN_ROUTE][option]
):
other_kw_dict[GAUSSIAN_ROUTE][option].append("CalcFC")
if other_kw_dict[GAUSSIAN_ROUTE][option] or (
other_kw_dict[GAUSSIAN_ROUTE][option]
and len(other_kw_dict[GAUSSIAN_ROUTE][option]) == 1
and (
"=" in other_kw_dict[GAUSSIAN_ROUTE][option][0]
or "(" in other_kw_dict[GAUSSIAN_ROUTE][option][0]
)
):
out_str += "=("
for x in other_kw_dict[GAUSSIAN_ROUTE][option]:
opt = x.split("=")[0]
if opt not in known_opts:
if known_opts:
out_str += ","
known_opts.append(opt)
out_str += x
out_str += ")"
elif (
other_kw_dict[GAUSSIAN_ROUTE][option]
and len(other_kw_dict[GAUSSIAN_ROUTE][option]) == 1
):
out_str += "=%s" % other_kw_dict[GAUSSIAN_ROUTE][option][0]
out_str += " "
out_str += "\n\n"
# add comment, removing any trailing newlines
if GAUSSIAN_COMMENT in other_kw_dict:
if other_kw_dict[GAUSSIAN_COMMENT]:
out_str += "\n".join(
[x.rstrip() for x in other_kw_dict[GAUSSIAN_COMMENT]]
)
else:
out_str += "comment"
if not out_str.endswith("\n"):
out_str += "\n"
out_str += "\n"
# charge mult
out_str += "%i %i\n" % (int(self.charge), int(self.multiplicity))
if return_warnings:
return out_str, warnings
return out_str
def get_gaussian_molecule(
self,
return_warnings=False,
conditional_kwargs=None,
**other_kw_dict,
):
"""
get molecule specification for gaussian input files
"""
if conditional_kwargs is None:
conditional_kwargs = {}
warnings = []
if self.job_type is not None:
for job in self.job_type[::-1]:
if hasattr(job, "geometry"):
job.geometry = self.geometry
job_dict = job.get_gaussian()
other_kw_dict = combine_dicts(job_dict, other_kw_dict)
other_kw_dict = combine_dicts(
other_kw_dict, conditional_kwargs, dict2_conditional=True
)
if GAUSSIAN_COORDINATES not in other_kw_dict:
other_kw_dict[GAUSSIAN_COORDINATES] = {}
if "coords" not in other_kw_dict[GAUSSIAN_COORDINATES]:
other_kw_dict[GAUSSIAN_COORDINATES][
"coords"
] = self.geometry.coords
s = ""
# atom specs need flag column before coords if any atoms frozen
has_frozen = False
for atom in self.geometry.atoms:
if atom.flag:
has_frozen = True
break
for atom, coord in zip(
self.geometry.atoms, other_kw_dict[GAUSSIAN_COORDINATES]["coords"]
):
s += "%-2s" % atom.element
if has_frozen:
s += " % 2d" % (-1 if atom.flag else 0)
for val in coord:
s += " "
if isinstance(val, float):
s += " %9.5f" % val
elif isinstance(val, str):
s += " %5s" % val
elif isinstance(val, int):
s += " %3i" % val
else:
warnings.append("unknown coordinate type: %s" % type(val))
s += "\n"
if (
"variables" in other_kw_dict[GAUSSIAN_COORDINATES]
and other_kw_dict[GAUSSIAN_COORDINATES]["variables"]
):
s += "Variable:\n"
for var in other_kw_dict[GAUSSIAN_COORDINATES]["variables"]:
s += "%4s = %9.5f\n" % tuple(var)
if (
"constants" in other_kw_dict[GAUSSIAN_COORDINATES]
and other_kw_dict[GAUSSIAN_COORDINATES]["constants"]
):
s += "Constant:\n"
for var in other_kw_dict[GAUSSIAN_COORDINATES]["constants"]:
s += "%4s = %9.5f\n" % tuple(var)
if return_warnings:
return s, warnings
return s
def get_gaussian_footer(
self,
return_warnings=False,
conditional_kwargs=None,
**other_kw_dict,
):
"""write footer of gaussian input file"""
if conditional_kwargs is None:
conditional_kwargs = {}
if self.job_type is not None:
for job in self.job_type[::-1]:
if hasattr(job, "geometry"):
job.geometry = self.geometry
job_dict = job.get_gaussian()
other_kw_dict = combine_dicts(job_dict, other_kw_dict)
other_kw_dict = combine_dicts(
other_kw_dict, conditional_kwargs, dict2_conditional=True
)
out_str = ""
warnings = []
# if method is not semi emperical, basis set might be gen or genecp
# get basis info (will be written after constraints)
if (
self.method is not None
and not self.method.is_semiempirical
and self.basis is not None
):
basis_info, warnings = self.basis.get_gaussian_basis_info()
elif (
self.method is not None
and not self.method.is_semiempirical
and self.basis is None
):
basis_info = {}
warnings.append("no basis specfied")
out_str += "\n"
# bond, angle, and torsion constraints
if GAUSSIAN_CONSTRAINTS in other_kw_dict:
for constraint in other_kw_dict[GAUSSIAN_CONSTRAINTS]:
out_str += constraint
out_str += "\n"
out_str += "\n"
# write gen info
if self.method is not None and not self.method.is_semiempirical:
if GAUSSIAN_GEN_BASIS in basis_info:
out_str += basis_info[GAUSSIAN_GEN_BASIS]
out_str += "\n"
if GAUSSIAN_GEN_ECP in basis_info:
out_str += basis_info[GAUSSIAN_GEN_ECP]
out_str += "\n"
# post info e.g. for NBOREAD
if GAUSSIAN_POST in other_kw_dict:
for item in other_kw_dict[GAUSSIAN_POST]:
out_str += item
out_str += " "
out_str += "\n"
out_str = out_str.rstrip()
# new lines
out_str += "\n\n\n"
if return_warnings:
return out_str, warnings
return out_str
def get_orca_header(
self,
return_warnings=False,
conditional_kwargs=None,
**other_kw_dict,
):
"""
get ORCA input file header
other_kw_dict is a dictionary with file positions (using ORCA_*)
corresponding to options/keywords
returns file content and warnings e.g. if a certain feature is not available in ORCA
returns str of header content
if return_warnings, returns str, list(warning)
"""
if conditional_kwargs is None:
conditional_kwargs = {}
if self.job_type is not None:
for job in self.job_type[::-1]:
if hasattr(job, "geometry"):
job.geometry = self.geometry
job_dict = job.get_orca()
other_kw_dict = combine_dicts(other_kw_dict, job_dict)
warnings = []
# if method isn't semi-empirical, get basis info to write later
if not self.method.is_semiempirical and self.basis is not None:
basis_info, basis_warnings = self.basis.get_orca_basis_info()
warnings.extend(basis_warnings)
if self.geometry is not None:
warning = self.basis.check_for_elements(self.geometry)
if warning is not None:
warnings.append(warning)
else:
basis_info = {}
other_kw_dict = combine_dicts(other_kw_dict, basis_info)
# get grid info
if self.grid is not None:
grid_info, warning = self.grid.get_orca()
if warning is not None:
warnings.append(warning)
if any(
"finalgrid" in x.lower() for x in other_kw_dict[ORCA_ROUTE]
):
grid_info[ORCA_ROUTE].pop(1)
other_kw_dict = combine_dicts(other_kw_dict, grid_info)
# add implicit solvent
if self.solvent is not None:
solvent_info, warning = self.solvent.get_orca()
warnings.extend(warning)
other_kw_dict = combine_dicts(other_kw_dict, solvent_info)
# dispersion
if self.empirical_dispersion is not None:
dispersion, warning = self.empirical_dispersion.get_orca()
if warning is not None:
warnings.append(warning)
other_kw_dict = combine_dicts(other_kw_dict, dispersion)
other_kw_dict = combine_dicts(
other_kw_dict, conditional_kwargs, dict2_conditional=True
)
# start building input file header
out_str = ""
# comment
if ORCA_COMMENT not in other_kw_dict:
if self.geometry.comment:
other_kw_dict[ORCA_COMMENT] = [self.geometry.comment]
else:
other_kw_dict[ORCA_COMMENT] = [self.geometry.name]
for comment in other_kw_dict[ORCA_COMMENT]:
for line in comment.split("\n"):
out_str += "#%s\n" % line
out_str += "! "
# method
if self.method is not None:
func, warning = self.method.get_orca()
if warning is not None:
warnings.append(warning)
warning = self.method.sanity_check_method(func, "orca")
if warning:
warnings.append(warning)
out_str += "%s" % func
# add other route options
if ORCA_ROUTE in other_kw_dict:
used_keywords = []
for kw in other_kw_dict[ORCA_ROUTE]:
if any(kw.lower() == used_kw for used_kw in used_keywords):
continue
used_keywords.append(kw.lower())
out_str += " %s" % kw
out_str += "\n"
# procs
if self.processors is not None:
out_str += "%%pal\n nprocs %i\nend\n" % self.processors
# orca memory is per core, so only specify it if processors are specified
if self.memory is not None:
out_str += "%%MaxCore %i\n" % (
int(1000 * self.memory / self.processors)
)
# add other blocks
if ORCA_BLOCKS in other_kw_dict:
for keyword in other_kw_dict[ORCA_BLOCKS]:
if any(keyword.lower() == name for name in self.ORCA_BLOCKS_AFTER_MOL):
continue
if any(other_kw_dict[ORCA_BLOCKS][keyword]):
used_settings = []
if keyword == "base":
out_str += "%%%s " % keyword
if isinstance(
other_kw_dict[ORCA_BLOCKS][keyword], str
):
out_str += (
'"%s"\n' % other_kw_dict[ORCA_BLOCKS][keyword]
)
else:
out_str += (
'"%s"\n'
% other_kw_dict[ORCA_BLOCKS][keyword][0]
)
else:
out_str += "%%%s\n" % keyword
for opt in other_kw_dict[ORCA_BLOCKS][keyword]:
if any(
keyword.lower() == block_name for block_name in [
"freq", "geom",
]
) and any(
opt.split()[0].lower() == prev_opt for prev_opt in used_settings
):
continue
used_settings.append(opt.split()[0].lower())
out_str += " %s\n" % opt
out_str += "end\n"
out_str += "\n"
# start of coordinate section - end of header
out_str += "*xyz %i %i\n" % (self.charge, self.multiplicity)
if return_warnings:
return out_str, warnings
return out_str
def get_orca_footer(
self,
return_warnings=False,
conditional_kwargs=None,
**other_kw_dict,
):
"""
get ORCA input file header
other_kw_dict is a dictionary with file positions (using ORCA_*)
corresponding to options/keywords
returns file content and warnings e.g. if a certain feature is not available in ORCA
returns str of header content
if return_warnings, returns str, list(warning)
"""
if conditional_kwargs is None:
conditional_kwargs = {}
if self.job_type is not None:
for job in self.job_type[::-1]:
if hasattr(job, "geometry"):
job.geometry = self.geometry
job_dict = job.get_orca()
other_kw_dict = combine_dicts(job_dict, other_kw_dict)
warnings = []
# if method isn't semi-empirical, get basis info to write later
if not self.method.is_semiempirical and self.basis is not None:
basis_info, basis_warnings = self.basis.get_orca_basis_info()
warnings.extend(basis_warnings)
if self.geometry is not None:
warning = self.basis.check_for_elements(self.geometry)
if warning is not None:
warnings.append(warning)
else:
basis_info = {}
other_kw_dict = combine_dicts(basis_info, other_kw_dict)
# get grid info
if self.grid is not None:
grid_info, warning = self.grid.get_orca()
if warning is not None:
warnings.append(warning)
if any(
"finalgrid" in x.lower() for x in other_kw_dict[ORCA_ROUTE]
):
grid_info[ORCA_ROUTE].pop(1)
other_kw_dict = combine_dicts(grid_info, other_kw_dict)
# add implicit solvent
if self.solvent is not None:
solvent_info, warning = self.solvent.get_orca()
warnings.extend(warning)
other_kw_dict = combine_dicts(solvent_info, other_kw_dict)
# dispersion
if self.empirical_dispersion is not None:
dispersion, warning = self.empirical_dispersion.get_orca()
if warning is not None:
warnings.append(warning)
other_kw_dict = combine_dicts(dispersion, other_kw_dict)
other_kw_dict = combine_dicts(
other_kw_dict, conditional_kwargs, dict2_conditional=True
)
# start building input file header
out_str = "\n"
# add other blocks
if ORCA_BLOCKS in other_kw_dict:
for keyword in other_kw_dict[ORCA_BLOCKS]:
if not any(keyword.lower() == name for name in self.ORCA_BLOCKS_AFTER_MOL):
continue
if any(other_kw_dict[ORCA_BLOCKS][keyword]):
if keyword == "base":
out_str += "%%%s " % keyword
if isinstance(
other_kw_dict[ORCA_BLOCKS][keyword], str
):
out_str += (
'"%s"\n' % other_kw_dict[ORCA_BLOCKS][keyword]
)
else:
out_str += (
'"%s"\n'
% other_kw_dict[ORCA_BLOCKS][keyword][0]
)
else:
out_str += "%%%s\n" % keyword
for opt in other_kw_dict[ORCA_BLOCKS][keyword]:
out_str += " %s\n" % opt
out_str += "end\n"
out_str += "\n"
if return_warnings:
return out_str, warnings
return out_str
def get_psi4_header(
self,
return_warnings=False,
conditional_kwargs=None,
**other_kw_dict,
):
"""
write Psi4 input file
other_kw_dict is a dictionary with file positions (using PSI4_*)
corresponding to options/keywords
returns file content and warnings e.g. if a certain feature is not available in Psi4
"""
if conditional_kwargs is None:
conditional_kwargs = {}
if self.job_type is not None:
for job in self.job_type[::-1]:
if hasattr(job, "geometry"):
job.geometry = self.geometry
job_dict = job.get_psi4()
other_kw_dict = combine_dicts(other_kw_dict, job_dict)
warnings = []
# add implicit solvent
if self.solvent is not None:
solvent_info, warning = self.solvent.get_psi4()
warnings.extend(warning)
other_kw_dict = combine_dicts(other_kw_dict, solvent_info)
# get basis info if method is not semi empirical
if not self.method.is_semiempirical and self.basis is not None:
basis_info, basis_warnings = self.basis.get_psi4_basis_info(
isinstance(self.method, SAPTMethod)
)
warnings.extend(basis_warnings)
if self.geometry is not None:
warning = self.basis.check_for_elements(self.geometry)
if warning is not None:
warnings.append(warning)
# aux basis sets might have a '%s' b/c the keyword to apply them depends on
# the method - replace %s with the appropriate thing for the method
for key in basis_info:
for i in range(0, len(basis_info[key])):
if "%s" in basis_info[key][i]:
if "cc" in self.method.name.lower():
basis_info[key][i] = basis_info[key][i].replace(
"%s", "cc"
)
elif "dct" in self.method.name.lower():
basis_info[key][i] = basis_info[key][i].replace(
"%s", "dct"
)
elif "mp2" in self.method.name.lower():
basis_info[key][i] = basis_info[key][i].replace(
"%s", "mp2"
)
elif isinstance(self.method, SAPTMethod):
basis_info[key][i] = basis_info[key][i].replace(
"%s", "sapt"
)
elif "scf" in self.method.name.lower():
basis_info[key][i] = basis_info[key][i].replace(
"%s", "scf"
)
elif "ci" in self.method.name.lower():
basis_info[key][i] = basis_info[key][i].replace(
"%s", "mcscf"
)
else:
basis_info = {}
combined_dict = combine_dicts(other_kw_dict, basis_info)
other_kw_dict = combine_dicts(
other_kw_dict, conditional_kwargs, dict2_conditional=True
)
# start building input file header
out_str = ""
# comment
if PSI4_COMMENT not in combined_dict:
if self.geometry.comment:
combined_dict[PSI4_COMMENT] = [self.geometry.comment]
else:
combined_dict[PSI4_COMMENT] = [self.geometry.name]
for comment in combined_dict[PSI4_COMMENT]:
for line in comment.split("\n"):
out_str += "#%s\n" % line
# procs
if self.processors is not None:
out_str += "set_num_threads(%i)\n" % self.processors
# mem
if self.memory is not None:
out_str += "memory %i GB\n" % self.memory
# before geometry options e.g. basis {} or building a dft method
if PSI4_BEFORE_GEOM in combined_dict:
if combined_dict[PSI4_BEFORE_GEOM]:
for opt in combined_dict[PSI4_BEFORE_GEOM]:
out_str += opt
out_str += "\n"
out_str += "\n"
if return_warnings:
return out_str, warnings
return out_str
def get_psi4_molecule(
self,
return_warnings=False,
conditional_kwargs=None,
**other_kw_dict,
):
"""
get molecule specification for psi4 input files
"""
if conditional_kwargs is None:
conditional_kwargs = {}
warnings = []
use_bohr = False
pubchem = False
use_molecule_array = False
if self.job_type is not None:
for job in self.job_type[::-1]:
if hasattr(job, "geometry"):
job.geometry = self.geometry
job_dict = job.get_psi4()
other_kw_dict = combine_dicts(job_dict, other_kw_dict)
other_kw_dict = combine_dicts(
other_kw_dict, conditional_kwargs, dict2_conditional=True
)
if PSI4_COORDINATES not in other_kw_dict:
other_kw_dict[PSI4_COORDINATES] = {}
if "coords" not in other_kw_dict[PSI4_COORDINATES]:
other_kw_dict[PSI4_COORDINATES]["coords"] = self.geometry.coords
s = ""
if isinstance(self.method, SAPTMethod) and not hasattr(
self.charge, "__iter__"
):
warnings.append(
"for a SAPTMethod, charge and multiplicity should both be lists\n"
"with the first item being the overall charge/multiplicity and\n"
"subsequent items being the charge/multiplicity of the\n"
"corresponding monomer"
)
return s, warnings
if (
isinstance(self.method, SAPTMethod)
and sum(self.multiplicity[1:]) - len(self.multiplicity[1:]) + 1
> self.multiplicity[0]
):
use_molecule_array = True
s += "mol = psi4.core.Molecule.from_arrays(\n"
s += " molecular_multiplicity=%i,\n" % self.multiplicity[0]
s += " molecular_charge=%i,\n" % self.charge[0]
if PSI4_MOLECULE in other_kw_dict:
for keyword in other_kw_dict[PSI4_MOLECULE]:
if other_kw_dict[keyword]:
s += " %s=%s,\n" % (
keyword,
repr(other_kw_dict[keyword][0]),
)
else:
s += "molecule {\n"
if isinstance(self.method, SAPTMethod):
if not hasattr(self.charge, "__iter__"):
warnings.append(
"for a SAPTMethod, charge and multiplicity should both be lists\n"
"with the first item being the overall charge/multiplicity and\n"
"subsequent items being the charge/multiplicity of the\n"
"corresponding monomer"
)
s += " %2i %i\n" % (self.charge[0], self.multiplicity[0])
if len(self.charge) > 1 and self.charge[0] != sum(
self.charge[1:]
):
warnings.append(
"total charge is not equal to sum of monomer charges"
)
else:
s += " %2i %i\n" % (self.charge, self.multiplicity)
if PSI4_MOLECULE in other_kw_dict:
for keyword in other_kw_dict[PSI4_MOLECULE]:
if other_kw_dict[PSI4_MOLECULE][keyword]:
opt = other_kw_dict[PSI4_MOLECULE][keyword][0]
if (
"pubchem" in keyword.lower()
and not keyword.strip().endswith(":")
):
keyword = keyword.strip() + ":"
pubchem = True
s += " %s %s\n" % (keyword.strip(), opt)
if keyword == "units":
if opt.lower() in ["bohr", "au", "a.u."]:
use_bohr = True
else:
s += " %s\n" % keyword
if use_molecule_array:
# psi4 input is VERY different for sapt jobs with the low-spin
# combination of fragments
monomers = [comp.atoms for comp in self.geometry.components]
atoms_in_monomer = []
seps = []
for i, m1 in enumerate(self.geometry.components[:-1]):
seps.append(0)
for m2 in monomers[: i + 1]:
seps[-1] += len(m2)
s += " fragment_separators=%s,\n" % repr(seps)
s += " elez=%s,\n" % repr(
[
ELEMENTS.index(atom.element)
for monomer in monomers
for atom in monomer
]
)
s += " fragment_multiplicities=%s,\n" % repr(
self.multiplicity[1:]
)
s += " fragment_charges=%s,\n" % repr(self.charge[1:])
s += " geom=["
i = 0
for monomer in monomers:
s += "\n"
for atom in monomer:
if atom not in atoms_in_monomer:
atoms_in_monomer.append(atom)
else:
warnings.append("atom in two monomers: %s" % atom.name)
ndx = self.geometry.atoms.index(atom)
coord = other_kw_dict[PSI4_COORDINATES]["coords"][ndx]
for val in coord:
s += " "
if isinstance(val, float):
if use_bohr:
s += "%9.5f," % (val / UNIT.A0_TO_BOHR)
else:
s += "%9.5f," % val
else:
warnings.append(
"unknown coordinate type: %s" % type(val)
)
s += "\n"
s += " ],\n"
s += ")\n\n"
s += "activate(mol)\n"
if len(atoms_in_monomer) != len(self.geometry.atoms):
from AaronTools.finders import NotAny
warnings.append(
"there are atoms not in any monomers: %s"
% (
", ".join(
[
atom.name
for atom in self.geometry.find(
NotAny(atoms_in_monomer)
)
]
)
)
)
elif isinstance(self.method, SAPTMethod):
monomers = [comp.atoms for comp in self.geometry.components]
atoms_in_monomer = []
for monomer, mult, charge in zip(
monomers, self.multiplicity[1:], self.charge[1:]
):
s += " --\n"
s += " %2i %i\n" % (charge, mult)
for atom in monomer:
s += " %2s" % atom.element
if atom not in atoms_in_monomer:
atoms_in_monomer.append(atom)
else:
warnings.append("atom in two monomers: %s" % atom.name)
ndx = self.geometry.atoms.index(atom)
coord = other_kw_dict[PSI4_COORDINATES]["coords"][ndx]
for val in coord:
s += " "
if isinstance(val, float):
if use_bohr:
s += " %9.5f" % (val / UNIT.A0_TO_BOHR)
else:
s += " %9.5f" % val
elif isinstance(val, str):
s += " %9s" % val
else:
warnings.append(
"unknown coordinate type: %s" % type(val)
)
s += "\n"
if "variables" in other_kw_dict[PSI4_COORDINATES]:
for (name, val, angstrom) in other_kw_dict[PSI4_COORDINATES][
"variables"
]:
if use_bohr and angstrom:
val /= UNIT.A0_TO_BOHR
s += " %3s = %9.5f\n" % (name, val)
s += "}\n"
if len(atoms_in_monomer) != len(self.geometry.atoms):
from AaronTools.finders import NotAny
warnings.append(
"there are atoms not in any monomers: %s"
% (
", ".join(
[
atom.name
for atom in self.geometry.find(
NotAny(atoms_in_monomer)
)
]
)
)
)
elif not pubchem:
for atom, coord in zip(
self.geometry.atoms, other_kw_dict[PSI4_COORDINATES]["coords"]
):
s += " %2s" % atom.element
for val in coord:
s += " "
if isinstance(val, float):
s += " %9.5f" % val
elif isinstance(val, str):
s += " %9s" % val
else:
warnings.append(
"unknown coordinate type: %s" % type(val)
)
s += "\n"
if "variables" in other_kw_dict[PSI4_COORDINATES]:
for (name, val, angstrom) in other_kw_dict[PSI4_COORDINATES][
"variables"
]:
if use_bohr and angstrom:
val /= UNIT.A0_TO_BOHR
s += " %3s = %9.5f\n" % (name, val)
s += "}\n"
else:
s += "}\n"
if return_warnings:
return s, warnings
return s
def get_psi4_footer(
self,
return_warnings=False,
conditional_kwargs=None,
**other_kw_dict,
):
"""
get psi4 footer
"""
if conditional_kwargs is None:
conditional_kwargs = {}
warnings = []
if self.job_type is not None:
for job in self.job_type[::-1]:
if hasattr(job, "geometry"):
job.geometry = self.geometry
job_dict = job.get_psi4()
other_kw_dict = combine_dicts(job_dict, other_kw_dict)
# add implicit solvent
if self.solvent is not None:
solvent_info, warning = self.solvent.get_psi4()
other_kw_dict = combine_dicts(other_kw_dict, solvent_info)
# grid
if self.grid is not None:
grid_info, warning = self.grid.get_psi4()
if warning is not None:
warnings.append(warning)
other_kw_dict = combine_dicts(other_kw_dict, grid_info)
other_kw_dict = combine_dicts(
other_kw_dict, conditional_kwargs, dict2_conditional=True
)
out_str = "\n"
# settings
# a setting will only get added if its list has at least
# one item, but only the first item will be used
if PSI4_SETTINGS in other_kw_dict and any(
other_kw_dict[PSI4_SETTINGS][setting]
for setting in other_kw_dict[PSI4_SETTINGS]
):
out_str += "set {\n"
for setting in other_kw_dict[PSI4_SETTINGS]:
if other_kw_dict[PSI4_SETTINGS][setting]:
if isinstance(other_kw_dict[PSI4_SETTINGS][setting], str):
val = other_kw_dict[PSI4_SETTINGS][setting]
else:
if len(
other_kw_dict[PSI4_SETTINGS][setting]
) == 1 and (
not any(
array_setting == setting.strip().lower()
for array_setting in self.FORCED_PSI4_ARRAY
)
or any(
single_setting == setting.strip().lower()
for single_setting in self.FORCED_PSI4_SINGLE
)
):
val = other_kw_dict[PSI4_SETTINGS][setting][0]
else:
# array of values
val = "["
val += ",".join(
[
"%s" % v
for v in other_kw_dict[PSI4_SETTINGS][
setting
]
]
)
val += "]"
out_str += " %-20s %s\n" % (setting, val)
out_str += "}\n\n"
if PSI4_SOLVENT in other_kw_dict:
out_str += "pcm = {\n"
for setting in other_kw_dict[PSI4_SOLVENT]:
if other_kw_dict[PSI4_SOLVENT][setting]:
if isinstance(other_kw_dict[PSI4_SOLVENT][setting], str):
val = other_kw_dict[PSI4_SOLVENT][setting]
out_str += " %s = %s\n" % (setting, val)
else:
if any(
single_setting == setting.strip().lower()
for single_setting in self.FORCED_PSI4_SOLVENT_SINGLE
):
val = other_kw_dict[PSI4_SOLVENT][setting][0]
out_str += " %s = %s\n" % (setting, val)
else:
# array of values
if not out_str.endswith("\n\n") and not out_str.endswith("{\n"):
out_str += "\n"
out_str += " %s {\n" % setting
for val in other_kw_dict[PSI4_SOLVENT][setting]:
out_str += " %s\n" % val
out_str += " }\n\n"
out_str += "}\n\n"
if PSI4_OPTKING in other_kw_dict and any(
other_kw_dict[PSI4_OPTKING][setting]
for setting in other_kw_dict[PSI4_OPTKING]
):
out_str += "set optking {\n"
for setting in other_kw_dict[PSI4_OPTKING]:
if other_kw_dict[PSI4_OPTKING][setting]:
out_str += " %-20s %s\n" % (
setting,
other_kw_dict[PSI4_OPTKING][setting][0],
)
out_str += "}\n\n"
# method is method name + dispersion if there is dispersion
method = self.method.get_psi4()[0]
if self.empirical_dispersion is not None:
disp = self.empirical_dispersion.get_psi4()[0]
if "%s" in method:
method = method % disp
else:
method += disp
elif "%s" in method:
method = method.replace("%s", "")
warning = self.method.sanity_check_method(method, "psi4")
if warning:
warnings.append(warning)
# after job stuff - replace METHOD with method
if PSI4_BEFORE_JOB in other_kw_dict:
for opt in other_kw_dict[PSI4_BEFORE_JOB]:
if "$METHOD" in opt:
opt = opt.replace("$METHOD", "'%s'" % method)
out_str += opt
out_str += "\n"
# for each job, start with nrg = f('method'
# unless return_wfn=True, then do nrg, wfn = f('method'
if PSI4_JOB in other_kw_dict:
for func in other_kw_dict[PSI4_JOB].keys():
if any(
[
"return_wfn" in kwarg
and ("True" in kwarg or "on" in kwarg)
for kwarg in other_kw_dict[PSI4_JOB][func]
]
):
out_str += "nrg, wfn = %s('%s'" % (func, method)
else:
out_str += "nrg = %s('%s'" % (func, method)
known_kw = []
for keyword in other_kw_dict[PSI4_JOB][func]:
key = keyword.split("=")[0].strip()
if key not in known_kw:
known_kw.append(key)
out_str += ", "
out_str += keyword.replace("$METHOD", "'%s'" % method)
out_str += ")\n"
# after job stuff - replace METHOD with method
if PSI4_AFTER_JOB in other_kw_dict:
for opt in other_kw_dict[PSI4_AFTER_JOB]:
if "$METHOD" in opt:
opt = opt.replace("$METHOD", "'%s'" % method)
out_str += opt
out_str += "\n"
if return_warnings:
return out_str, warnings
return out_str
def get_xtb_cmdline(self, config):
"""
Uses the config and job type to set command line options for xtb and crest jobs
Returns a dictionary of option=val pairs; val is None when option doesn't take
an argument. This dict should be parsed by the caller into the command line
string.
"""
if len(self.job_type) > 1:
raise NotImplementedError(
"Multiple job types not supported for crest/xtb"
)
cmdline = {}
job_type = self.job_type[0]
style = config["Job"]["exec_type"]
if style not in ["xtb", "crest"]:
raise NotImplementedError(
"Wrong executable type: %s (can only get command line options "
"for `xtb` or `crest`)" % style
)
# pull in stuff set by resolve_error
if config._args:
for arg in config._args:
cmdline[arg] = None
if config._kwargs:
for key, val in config._kwargs.items():
cmdline[key] = val
# run types
if "gfn" in config["Theory"]:
cmdline["--gfn"] = config["Job"]["gfn"]
if (
style == "xtb"
and hasattr(job_type, "transition_state")
and job_type.transition_state
):
cmdline["--optts"] = None
elif style == "xtb":
cmdline["--opt"] = None
# charge/mult/temp
if self.charge != 0:
cmdline["--chrg"] = self.charge
if self.multiplicity != 1:
cmdline["--uhf"] = self.multiplicity - 1
if style == "crest":
cmdline["--temp"] = config["Theory"].get(
"temperature", fallback="298"
)
else:
cmdline["--etemp"] = config["Theory"].get(
"temperature", fallback="298"
)
# screening parameters
if (
style == "crest"
and "energy_cutoff" in config["Job"]
and config["Job"].getfloat("energy_cutoff") > 6
):
cmdline["--ewin"] = config["Job"]["energy_cutoff"]
if (
style == "crest"
and "rmsd_cutoff" in config["Job"]
and config["Job"].getfloat("rmsd_cutoff") < 0.125
):
cmdline["--rthr"] = config["Job"]["rmsd_cutoff"]
# solvent stuff
if (
"solvent_model" in config["Theory"]
and config["Theory"]["solvent_model"] == "alpb"
):
solvent = config["Theory"]["solvent"].split()
if len(solvent) > 1:
solvent, ref = solvent
elif len(solvent) == 1:
solvent, ref = solvent[0], None
else:
raise ValueError
if solvent.lower() not in [
"acetone",
"acetonitrile",
"aniline",
"benzaldehyde",
"benzene",
"ch2cl2",
"chcl3",
"cs2",
"dioxane",
"dmf",
"dmso",
"ether",
"ethylacetate",
"furane",
"hexandecane",
"hexane",
"methanol",
"nitromethane",
"octanol",
"woctanol",
"phenol",
"toluene",
"thf",
"water",
]:
raise ValueError("%s is not a supported solvent" % solvent)
if ref is not None and ref.lower() not in ["reference", "bar1m"]:
raise ValueError(
"%s Gsolv reference state not supported" % ref
)
if style.lower() == "crest" or ref is None:
cmdline["--alpb"] = "{}".format(solvent)
else:
cmdline["--alpb"] = "{} {}".format(solvent, ref)
elif (
"solvent_model" in config["Theory"]
and config["Theory"]["solvent_model"] == "gbsa"
):
solvent = config["Theory"]["solvent"].split()
if len(solvent) > 1:
solvent, ref = solvent
else:
solvent, ref = solvent[0], None
if solvent.lower() not in [
"acetone",
"acetonitrile",
"benzene",
"ch2cl2",
"chcl3",
"cs2",
"dmf",
"dmso",
"ether",
"h2o",
"methanol",
"n-hexane",
"thf",
"toluene",
]:
gfn = config["Theory"].get("gfn", fallback="2")
if gfn != "1" and solvent.lower() in ["benzene"]:
raise ValueError("%s is not a supported solvent" % solvent)
elif gfn != "2" and solvent.lower() in ["DMF", "n-hexane"]:
raise ValueError("%s is not a supported solvent" % solvent)
else:
raise ValueError("%s is not a supported solvent" % solvent)
if ref is not None and ref.lower() not in ["reference", "bar1m"]:
raise ValueError(
"%s Gsolv reference state not supported" % ref
)
if style.lower() == "crest" or ref is None:
cmdline["--gbsa"] = "{}".format(solvent)
else:
cmdline["--gbsa"] = "{} {}".format(solvent, ref)
other = config["Job"].get("cmdline", fallback="").split()
i = 0
while i < len(other):
if other[i].startswith("-"):
key = other[i]
cmdline[key] = None
else:
cmdline[key] = other[i]
i += 1
return cmdline
def get_xcontrol(self, config, ref=None):
if len(self.job_type) > 1:
raise NotImplementedError(
"Multiple job types not supported for crest/xtb"
)
job_type = self.job_type[0]
return job_type.get_xcontrol(config, ref=ref)
def get_sqm_header(
self,
return_warnings=False,
conditional_kwargs=None,
**other_kw_dict,
):
"""retruns header, warnings_list for sqm job"""
warnings = []
if conditional_kwargs is None:
conditional_kwargs = {}
# job stuff
if self.job_type is not None:
for job in self.job_type[::-1]:
if hasattr(job, "geometry"):
job.geometry = self.geometry
job_dict, job_warnings = job.get_sqm()
warnings.extend(job_warnings)
other_kw_dict = combine_dicts(job_dict, other_kw_dict)
s = ""
# charge and mult
other_kw_dict = combine_dicts(
{
SQM_QMMM: {
"qmcharge": [str(self.charge)],
"spin": [str(self.multiplicity)],
}
},
other_kw_dict,
)
# comment
if SQM_COMMENT not in other_kw_dict:
if self.geometry.comment:
other_kw_dict[SQM_COMMENT] = [self.geometry.comment]
else:
other_kw_dict[SQM_COMMENT] = [self.geometry.name]
for comment in other_kw_dict[SQM_COMMENT]:
for line in comment.split("\n"):
s += "%s" % line
s += "\n"
# method
if self.method:
method = self.method.get_sqm()
warning = self.method.sanity_check_method(method, "sqm")
if warning:
warnings.append(warning)
other_kw_dict = combine_dicts(
other_kw_dict,
{SQM_QMMM: {"qm_theory": [method]}}
)
other_kw_dict = combine_dicts(
other_kw_dict, conditional_kwargs, dict2_conditional=True
)
# options
s += " &qmmm\n"
for key in other_kw_dict[SQM_QMMM]:
if not other_kw_dict[SQM_QMMM][key]:
continue
s += " %s=" % key
option = other_kw_dict[SQM_QMMM][key][0]
if re.match("-?\d+", option):
s += "%s,\n" % option
elif any(option.lower() == b for b in [".true.", ".false."]):
s += "%s,\n" % option
else:
s += "'%s',\n" % option
s += " /\n"
return s, warnings
def get_sqm_molecule(
self,
**kwargs,
):
"""returns molecule specification for sqm input"""
warnings = []
s = ""
for atom in self.geometry.atoms:
s += " %2i %2s %9.5f %9.5f %9.5f\n" % (
ELEMENTS.index(atom.element),
atom.element,
atom.coords[0],
atom.coords[1],
atom.coords[2],
)
return s.rstrip(), warnings
def get_qchem_header(
self,
return_warnings=False,
conditional_kwargs=None,
**other_kw_dict,
):
"""
write QChem input file header (up to charge mult)
other_kw_dict is a dictionary with file positions (using QCHEM_*)
corresponding to options/keywords
returns warnings if a certain feature is not available in QChem
"""
if conditional_kwargs is None:
conditional_kwargs = {}
warnings = []
job_type_count = 0
if self.job_type is not None:
for i, job in enumerate(self.job_type[::-1]):
if hasattr(job, "geometry"):
job.geometry = self.geometry
job_dict = job.get_qchem()
if isinstance(job, FrequencyJob) and job.temperature != 298.15:
warnings.append(
"thermochemistry data in the output file will be for 298.15 K\n"
"in spite of the user setting %.2f K\n" % (job.temperature) + \
"free energy corrections can be calculated at different\n"
"temperatures using AaronTools grabThermo.py script or\n"
"SEQCROW's Procress QM Thermochemistry tool"
)
if QCHEM_REM in job_dict and any(key.lower() == "job_type" for key in job_dict[QCHEM_REM]):
job_type_count += 1
if job_type_count > 1:
raise NotImplementedError("cannot put multiple JOB_TYPE entries in one Q-Chem header")
other_kw_dict = combine_dicts(job_dict, other_kw_dict)
if (
QCHEM_COMMENT not in other_kw_dict
or not other_kw_dict[QCHEM_COMMENT]
):
if self.geometry.comment:
other_kw_dict[QCHEM_COMMENT] = [self.geometry.comment]
else:
other_kw_dict[QCHEM_COMMENT] = [self.geometry.name]
if QCHEM_SETTINGS in other_kw_dict:
other_kw_dict = combine_dicts(
other_kw_dict,
{QCHEM_SETTINGS: {QCHEM_COMMENT: other_kw_dict[QCHEM_COMMENT]}},
)
else:
other_kw_dict[QCHEM_SETTINGS] = {QCHEM_COMMENT: other_kw_dict[QCHEM_COMMENT]}
# add memory info
if self.memory:
other_kw_dict = combine_dicts(
other_kw_dict,
{QCHEM_REM: {"MEM_TOTAL": str(1000 * self.memory)}}
)
# add EmpiricalDispersion info
if self.empirical_dispersion is not None:
disp, warning = self.empirical_dispersion.get_qchem()
other_kw_dict = combine_dicts(other_kw_dict, disp)
if warning is not None:
warnings.append(warning)
# add Integral(grid=X)
if self.grid is not None:
grid, warning = self.grid.get_qchem()
other_kw_dict = combine_dicts(other_kw_dict, grid)
if warning is not None:
warnings.append(warning)
# add implicit solvent
if self.solvent is not None:
solvent_info, warning = self.solvent.get_qchem()
if warning is not None:
warnings.extend(warning)
other_kw_dict = combine_dicts(other_kw_dict, solvent_info)
if self.method is not None:
func, warning = self.method.get_qchem()
if warning is not None:
warnings.append(warning)
warning = self.method.sanity_check_method(func, "qchem")
if warning:
warnings.append(warning)
# Q-Chem seems to still require a basis set for HF-3c
# if not self.method.is_semiempirical and self.basis is not None:
if self.basis is not None:
(
basis_info,
basis_warnings,
) = self.basis.get_qchem_basis_info(self.geometry)
warnings.extend(basis_warnings)
# check basis elements to make sure no element is
# in two basis sets or left out of any
other_kw_dict = combine_dicts(
other_kw_dict,
basis_info,
)
if self.geometry is not None:
basis_warning = self.basis.check_for_elements(
self.geometry,
)
if basis_warning is not None:
warnings.append(basis_warning)
other_kw_dict = combine_dicts(
other_kw_dict, {QCHEM_REM: {"METHOD": func}},
)
other_kw_dict = combine_dicts(
other_kw_dict, conditional_kwargs, dict2_conditional=True
)
out_str = ""
if QCHEM_REM in other_kw_dict and QCHEM_SETTINGS in other_kw_dict:
other_kw_dict[QCHEM_SETTINGS] = combine_dicts(
{"rem": other_kw_dict[QCHEM_REM]}, other_kw_dict[QCHEM_SETTINGS],
)
elif QCHEM_REM in other_kw_dict:
other_kw_dict[QCHEM_SETTINGS] = {"rem": other_kw_dict[QCHEM_REM]}
else:
warnings.append("no REM section")
if self.memory:
other_kw_dict = combine_dicts(
other_kw_dict,
{"rem": {"MEM_TOTAL": "%i" % (1000 * self.memory)}}
)
if QCHEM_SETTINGS in other_kw_dict:
for section in other_kw_dict[QCHEM_SETTINGS]:
settings = other_kw_dict[QCHEM_SETTINGS][section]
out_str += "$%s\n" % section
for setting in settings:
if not setting:
continue
if isinstance(settings, dict):
opt = settings[setting]
if not opt:
continue
if isinstance(opt, str):
val = opt
out_str += " %-20s = %s\n" % (setting, val)
elif isinstance(opt, dict):
for s, v in opt.items():
out_str += " %-20s = %s\n" % (s, v)
else:
if len(opt) == 1:
val = opt[0]
out_str += " %-20s = %s\n" % (setting, val)
elif not opt:
out_str += " %-20s\n" % setting
else:
if section.lower() == "rem" and setting.lower() == "job_type":
raise NotImplementedError(
"cannot put multiple JOB_TYPE entries in one Q-Chem header"
)
out_str += " %-20s = %s\n" % (setting, ", ".join(opt))
elif hasattr(setting, "__iter__") and not isinstance(setting, str):
for val in setting:
out_str += " %s\n" % val
else:
out_str += " %s\n" % setting
out_str += "$end\n\n"
return out_str, warnings
def get_qchem_molecule(
self,
return_warnings=False,
conditional_kwargs=None,
**other_kw_dict,
):
if conditional_kwargs is None:
conditional_kwargs = {}
warnings = []
if self.job_type is not None:
for job in self.job_type[::-1]:
if hasattr(job, "geometry"):
job.geometry = self.geometry
job_dict = job.get_qchem()
other_kw_dict = combine_dicts(job_dict, other_kw_dict)
if (
GAUSSIAN_COMMENT not in other_kw_dict
or not other_kw_dict[GAUSSIAN_COMMENT]
):
if self.geometry.comment:
other_kw_dict[GAUSSIAN_COMMENT] = [self.geometry.comment]
else:
other_kw_dict[GAUSSIAN_COMMENT] = [self.geometry.name]
# add EmpiricalDispersion info
if self.empirical_dispersion is not None:
disp, warning = self.empirical_dispersion.get_qchem()
other_kw_dict = combine_dicts(other_kw_dict, disp)
if warning is not None:
warnings.append(warning)
# add Integral(grid=X)
if self.grid is not None:
grid, warning = self.grid.get_qchem()
other_kw_dict = combine_dicts(other_kw_dict, grid)
if warning is not None:
warnings.append(warning)
# add implicit solvent
if self.solvent is not None:
solvent_info, warning = self.solvent.get_qchem()
if warning is not None:
warnings.extend(warning)
other_kw_dict = combine_dicts(other_kw_dict, solvent_info)
other_kw_dict = combine_dicts(
other_kw_dict, conditional_kwargs, dict2_conditional=True
)
out_str = "$molecule\n %i %i\n" % (
self.charge, self.multiplicity
)
if QCHEM_MOLECULE in other_kw_dict:
for line in other_kw_dict[QCHEM_MOLECULE]:
out_str += " %-20s\n" % line
elif not self.geometry:
warnings.append("no molecule")
for atom in self.geometry.atoms:
out_str += " %-2s" % atom.element
out_str += " %9.5f %9.5f %9.5f\n" % tuple(atom.coords)
out_str += "$end\n"
return out_str, warnings | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/theory/theory.py | theory.py |
from AaronTools import addlogger
from AaronTools.theory import (
GAUSSIAN_ROUTE, ORCA_BLOCKS, ORCA_ROUTE, PSI4_SETTINGS, PSI4_SOLVENT
)
class ImplicitSolvent:
"""implicit solvent info"""
KNOWN_GAUSSIAN_SOLVENTS = [
"Water",
"Acetonitrile",
"Methanol",
"Ethanol",
"IsoQuinoline",
"Quinoline",
"Chloroform",
"DiethylEther",
"DichloroMethane",
"DiChloroEthane",
"CarbonTetraChloride",
"Benzene",
"Toluene",
"ChloroBenzene",
"NitroMethane",
"Heptane",
"CycloHexane",
"Aniline",
"Acetone",
"TetraHydroFuran",
"DiMethylSulfoxide",
"Argon",
"Krypton",
"Xenon",
"n-Octanol",
"1,1,1-TriChloroEthane",
"1,1,2-TriChloroEthane",
"1,2,4-TriMethylBenzene",
"1,2-DiBromoEthane",
"1,2-EthaneDiol",
"1,4-Dioxane",
"1-Bromo-2-MethylPropane",
"1-BromoOctane",
"1-BromoPentane",
"1-BromoPropane",
"1-Butanol",
"1-ChloroHexane",
"1-ChloroPentane",
"1-ChloroPropane",
"1-Decanol",
"1-FluoroOctane",
"1-Heptanol",
"1-Hexanol",
"1-Hexene",
"1-Hexyne",
"1-IodoButane",
"1-IodoHexaDecane",
"1-IodoPentane",
"1-IodoPropane",
"1-NitroPropane",
"1-Nonanol",
"1-Pentanol",
"1-Pentene",
"1-Propanol",
"2,2,2-TriFluoroEthanol",
"2,2,4-TriMethylPentane",
"2,4-DiMethylPentane",
"2,4-DiMethylPyridine",
"2,6-DiMethylPyridine",
"2-BromoPropane",
"2-Butanol",
"2-ChloroButane",
"2-Heptanone",
"2-Hexanone",
"2-MethoxyEthanol",
"2-Methyl-1-Propanol",
"2-Methyl-2-Propanol",
"2-MethylPentane",
"2-MethylPyridine",
"2-NitroPropane",
"2-Octanone",
"2-Pentanone",
"2-Propanol",
"2-Propen-1-ol",
"3-MethylPyridine",
"3-Pentanone",
"4-Heptanone",
"4-Methyl-2-Pentanone",
"4-MethylPyridine",
"5-Nonanone",
"AceticAcid",
"AcetoPhenone",
"a-ChloroToluene",
"Anisole",
"Benzaldehyde",
"BenzoNitrile",
"BenzylAlcohol",
"BromoBenzene",
"BromoEthane",
"Bromoform",
"Butanal",
"ButanoicAcid",
"Butanone",
"ButanoNitrile",
"ButylAmine",
"ButylEthanoate",
"CarbonDiSulfide",
"Cis-1,2-DiMethylCycloHexane",
"Cis-Decalin",
"CycloHexanone",
"CycloPentane",
"CycloPentanol",
"CycloPentanone",
"Decalin-mixture",
"DiBromoMethane",
"DiButylEther",
"DiEthylAmine",
"DiEthylSulfide",
"DiIodoMethane",
"DiIsoPropylEther",
"DiMethylDiSulfide",
"DiPhenylEther",
"DiPropylAmine",
"E-1,2-DiChloroEthene",
"E-2-Pentene",
"EthaneThiol",
"EthylBenzene",
"EthylEthanoate",
"EthylMethanoate",
"EthylPhenylEther",
"FluoroBenzene",
"Formamide",
"FormicAcid",
"HexanoicAcid",
"IodoBenzene",
"IodoEthane",
"IodoMethane",
"IsoPropylBenzene",
"m-Cresol",
"Mesitylene",
"MethylBenzoate",
"MethylButanoate",
"MethylCycloHexane",
"MethylEthanoate",
"MethylMethanoate",
"MethylPropanoate",
"m-Xylene",
"n-ButylBenzene",
"n-Decane",
"n-Dodecane",
"n-Hexadecane",
"n-Hexane",
"NitroBenzene",
"NitroEthane",
"n-MethylAniline",
"n-MethylFormamide-mixture",
"n,n-DiMethylAcetamide",
"n,n-DiMethylFormamide",
"n-Nonane",
"n-Octane",
"n-Pentadecane",
"n-Pentane",
"n-Undecane",
"o-ChloroToluene",
"o-Cresol",
"o-DiChloroBenzene",
"o-NitroToluene",
"o-Xylene",
"Pentanal",
"PentanoicAcid",
"PentylAmine",
"PentylEthanoate",
"PerFluoroBenzene",
"p-IsoPropylToluene",
"Propanal",
"PropanoicAcid",
"PropanoNitrile",
"PropylAmine",
"PropylEthanoate",
"p-Xylene",
"Pyridine",
"sec-ButylBenzene",
"tert-ButylBenzene",
"TetraChloroEthene",
"TetraHydroThiophene-s,s-dioxide",
"Tetralin",
"Thiophene",
"Thiophenol",
"trans-Decalin",
"TriButylPhosphate",
"TriChloroEthene",
"TriEthylAmine",
"Xylene-mixture",
"Z-1,2-DiChloroEthene",
]
KNOWN_GAUSSIAN_MODELS = [
"SMD",
"CPCM",
"PCM",
"DIPOLE",
"IPCM",
"ISODENSITY",
"IEFPCM",
"SCIPCM",
]
KNOWN_ORCA_CPCM_SOLVENTS = [
"Water",
"Acetonitrile",
"Acetone",
"Ammonia",
"Ethanol",
"Methanol",
"CH2Cl2",
"CCl4",
"DMF",
"DMSO",
"Pyridine",
"THF",
"Chloroform",
"Hexane",
"Benzene",
"CycloHexane",
"Octanol",
"Toluene",
]
KNOWN_ORCA_SMD_SOLVENTS = [
"1,1,1-TRICHLOROETHANE",
"CYCLOPENTANE",
"1,1,2-TRICHLOROETHANE",
"CYCLOPENTANOL",
"1,2,4-TRIMETHYLBENZENE",
"CYCLOPENTANONE",
"1,2-DIBROMOETHANE",
"DECALIN (CIS/TRANS MIXTURE)",
"1,2-DICHLOROETHANE",
"CIS-DECALIN",
"1,2-ETHANEDIOL",
"N-DECANE",
"1,4-DIOXANE",
"DIBROMOMETHANE",
"1-BROMO-2-METHYLPROPANE",
"DIBUTYLETHER",
"1-BROMOOCTANE",
"O-DICHLOROBENZENE",
"1-BROMOPENTANE",
"E-1,2-DICHLOROETHENE",
"1-BROMOPROPANE",
"Z-1,2-DICHLOROETHENE",
"1-BUTANOL",
"DICHLOROMETHANE",
"1-CHLOROHEXANE",
"DIETHYL ETHER",
"1-CHLOROPENTANE",
"DIETHYL SULFIDE",
"1-CHLOROPROPANE",
"DIETHYLAMINE",
"1-DECANOL",
"DIIODOMETHANE",
"1-FLUOROOCTANE",
"DIISOPROPYL ETHER",
"1-HEPTANOL",
"CIS-1,2-DIMETHYLCYCLOHEXANE",
"1-HEXANOL",
"DIMETHYL DISULFIDE",
"1-HEXENE",
"N,N-DIMETHYLACETAMIDE",
"1-HEXYNE",
"N,N-DIMETHYLFORMAMIDE",
"DMF",
"1-IODOBUTANE",
"DIMETHYLSULFOXIDE",
"DMSO",
"1-IODOHEXADECANE",
"DIPHENYLETHER",
"1-IODOPENTANE",
"DIPROPYLAMINE",
"1-IODOPROPANE",
"N-DODECANE",
"1-NITROPROPANE",
"ETHANETHIOL",
"1-NONANOL",
"ETHANOL",
"1-OCTANOL",
"ETHYL ETHANOATE",
"1-PENTANOL",
"ETHYL METHANOATE",
"1-PENTENE",
"ETHYL PHENYL ETHER",
"1-PROPANOL",
"ETHYLBENZENE",
"2,2,2-TRIFLUOROETHANOL",
"FLUOROBENZENE",
"2,2,4-TRIMETHYLPENTANE",
"FORMAMIDE",
"2,4-DIMETHYLPENTANE",
"FORMIC ACID",
"2,4-DIMETHYLPYRIDINE",
"N-HEPTANE",
"2,6-DIMETHYLPYRIDINE",
"N-HEXADECANE",
"2-BROMOPROPANE",
"N-HEXANE",
"2-BUTANOL",
"HEXANOIC ACID",
"2-CHLOROBUTANE",
"IODOBENZENE",
"2-HEPTANONE",
"IODOETHANE",
"2-HEXANONE",
"IODOMETHANE",
"2-METHOXYETHANOL",
"ISOPROPYLBENZENE",
"2-METHYL-1-PROPANOL",
"P-ISOPROPYLTOLUENE",
"2-METHYL-2-PROPANOL",
"MESITYLENE",
"2-METHYLPENTANE",
"METHANOL",
"2-METHYLPYRIDINE",
"METHYL BENZOATE",
"2-NITROPROPANE",
"METHYL BUTANOATE",
"2-OCTANONE",
"METHYL ETHANOATE",
"2-PENTANONE",
"METHYL METHANOATE",
"2-PROPANOL",
"METHYL PROPANOATE",
"2-PROPEN-1-OL",
"N-METHYLANILINE",
"E-2-PENTENE",
"METHYLCYCLOHEXANE",
"3-METHYLPYRIDINE",
"N-METHYLFORMAMIDE (E/Z MIXTURE)",
"3-PENTANONE",
"NITROBENZENE",
"PhNO2",
"4-HEPTANONE",
"NITROETHANE",
"4-METHYL-2-PENTANONE",
"NITROMETHANE",
"MeNO2",
"4-METHYLPYRIDINE",
"O-NITROTOLUENE",
"5-NONANONE",
"N-NONANE",
"ACETIC ACID",
"N-OCTANE",
"ACETONE",
"N-PENTADECANE",
"ACETONITRILE",
"MeCN",
"PENTANAL",
"ACETOPHENONE",
"N-PENTANE",
"ANILINE",
"PENTANOIC ACID",
"ANISOLE",
"PENTYL ETHANOATE",
"BENZALDEHYDE",
"PENTYLAMINE",
"BENZENE",
"PERFLUOROBENZENE",
"BENZONITRILE",
"PROPANAL",
"BENZYL ALCOHOL",
"PROPANOIC ACID",
"BROMOBENZENE",
"PROPANONITRILE",
"BROMOETHANE",
"PROPYL ETHANOATE",
"BROMOFORM",
"PROPYLAMINE",
"BUTANAL",
"PYRIDINE",
"BUTANOIC ACID",
"TETRACHLOROETHENE",
"BUTANONE",
"TETRAHYDROFURAN",
"THF",
"BUTANONITRILE",
"TETRAHYDROTHIOPHENE-S,S-DIOXIDE",
"BUTYL ETHANOATE",
"TETRALIN",
"BUTYLAMINE",
"THIOPHENE",
"N-BUTYLBENZENE",
"THIOPHENOL",
"SEC-BUTYLBENZENE",
"TOLUENE",
"TERT-BUTYLBENZENE",
"TRANS-DECALIN",
"CARBON DISULFIDE",
"TRIBUTYLPHOSPHATE",
"CARBON TETRACHLORIDE",
"CCl4",
"TRICHLOROETHENE",
"CHLOROBENZENE",
"TRIETHYLAMINE",
"CHLOROFORM",
"N-UNDECANE",
"A-CHLOROTOLUENE",
"WATER",
"O-CHLOROTOLUENE",
"XYLENE (MIXTURE)",
"M-CRESOL",
"M-XYLENE",
"O-CRESOL",
"O-XYLENE",
"CYCLOHEXANE",
"P-XYLENE",
"CYCLOHEXANONE",
]
KNOWN_ORCA_MODELS = ["SMD", "CPCM", "C-PCM", "PCM"]
KNOWN_PSI4_SOLVENTS = [
"water",
"propylene carbonate",
"dimethylsolfoxide",
"nitromethane",
"aceotonitrile",
"methanol",
"ethanol",
"acetone",
"1,2-dichloroethane",
"methylenechloride",
"CH2Cl2",
"tetrahydrofuran",
"aniline",
"chlorobenzene",
"chloroform",
"toluene",
"1,4-dioxane",
"benzene",
"carbon tetrachloride",
"cyclohexane",
"n-heptane",
]
KNOWN_PSI4_MODELS = ["CPCM", "IEFPCM"]
LOG = None
def __init__(self, solvent_model, solvent):
self.solvent_model = solvent_model
self.solvent = solvent
def __repr__(self):
return "%s(%s)" % (self.solvent_model.upper(), self.solvent.lower())
def __eq__(self, other):
return repr(self) == repr(other)
def copy(self):
new_dict = dict()
for key, value in self.__dict__.items():
try:
new_dict[key] = value.copy()
except AttributeError:
new_dict[key] = value
# ignore chimerax objects so seqcrow doesn't print a
# warning when a geometry is copied
if "chimerax" in value.__class__.__module__:
continue
if value.__class__.__module__ != "builtins":
self.LOG.warning(
"No copy method for {}: in-place changes may occur".format(
type(value)
)
)
return self.__class__(**new_dict)
def get_gaussian(self):
"""returns dict() with solvent information for gaussian input files"""
# need to check if solvent model is available
warnings = []
if self.solvent.lower() == "gas":
# all gas, no solvent
return (dict(), warnings)
if not any(
self.solvent_model.upper() == model for model in self.KNOWN_GAUSSIAN_MODELS
):
warnings.append(
"solvent model is not available in Gaussian: %s\nuse one of: %s"
% (
self.solvent_model,
" ".join(
[
"SMD",
"CPCM",
"PCM",
"DIPOLE",
"IPCM",
"ISODENSITY",
"IEFPCM",
"SCIPCM",
]
),
)
)
# check some orca solvent keywords and switch to gaussian ones
solvent = self.solvent
if solvent.lower() == "chcl2":
solvent = "DiChloroMethane"
elif solvent.lower() == "ccl4":
solvent = "CarbonTetraChloride"
elif solvent.lower() == "THF":
solvent = "TetraHydroFuran"
else:
if not any(
solvent.lower() == gaussian_sol.lower()
for gaussian_sol in self.KNOWN_GAUSSIAN_SOLVENTS
):
warnings.append(
"solvent is unknown to Gaussian: %s\n" % solvent +
"see AaronTools.theory.implicit_solvent.KNOWN_GAUSSIAN_SOLVENTS"
)
# route option: scrf(model,solvent=solvent solvent_model)
return (
{GAUSSIAN_ROUTE: {"scrf": [self.solvent_model, "solvent=%s" % solvent]}},
warnings,
)
def get_orca(self):
"""returns dict() with solvent information for orca input files"""
warnings = []
if self.solvent.lower() == "gas":
return (dict(), warnings)
if not any(
self.solvent_model.upper() == model for model in self.KNOWN_ORCA_MODELS
):
warnings.append(
"solvent model is not available in ORCA: %s\nuse CPCM or SMD"
% self.solvent_model
)
out = {}
cpcm = True
# route option: CPCM(solvent name)
# if using smd, add block %cpcm smd true end
if self.solvent_model.upper() == "SMD":
cpcm = False
out[ORCA_BLOCKS] = {"cpcm": ["smd true"]}
solvent = self.solvent
# orca has different solvents for cpcm and smd...
# check both lists, might be able to switch a gaussian keyword to the correct orca one
if cpcm:
if solvent.lower() == "dichloromethane":
solvent = "CH2Cl2"
elif solvent.lower() == "carbontetrachloride":
solvent = "CCl4"
elif solvent.lower() == "tetrahydrofuran":
solvent = "THF"
else:
if not any(
solvent.lower() == orca_sol.lower()
for orca_sol in self.KNOWN_ORCA_CPCM_SOLVENTS
):
warnings.append(
"solvent is unknown to ORCA: %s\n" % solvent +
"see AaronTools.theory.implicit_solvent.KNOWN_ORCA_CPCM_SOLVENTS"
)
else:
# TODO: look for gaussian/orca pcm solvent names that need to change
if not any(
solvent.lower() == orca_sol.lower()
for orca_sol in self.KNOWN_ORCA_SMD_SOLVENTS
):
warnings.append(
"solvent is unknown to ORCA: %s\n" % solvent +
"see AaronTools.theory.implicit_solvent.KNOWN_ORCA_SMD_SOLVENTS"
)
out[ORCA_ROUTE] = ["CPCM(%s)" % solvent]
return (out, warnings)
def get_psi4(self):
"""returns dict() with solvent information for psi4 input files"""
warnings = []
if self.solvent.lower() == "gas":
return (dict(), warnings)
if not any(
self.solvent_model.upper() == model for model in self.KNOWN_ORCA_MODELS
):
warnings.append(
"solvent model is not available in ORCA: %s\nuse CPCM or SMD"
% self.solvent_model
)
out = {}
# route option: CPCM(solvent name)
# if using smd, add block %cpcm smd true end
out[PSI4_SETTINGS] = {"pcm": "true"}
solvent = self.solvent
# orca has different solvents for cpcm and smd...
# check both lists, might be able to switch a gaussian keyword to the correct orca one
if solvent.lower() == "dichloromethane":
solvent = "CH2Cl2"
elif solvent.lower() == "carbontetrachloride":
solvent = "carbon tetrachloride"
elif solvent.lower() == "thf":
solvent = "tetrahydrofuran"
else:
if not any(
solvent.lower() == orca_sol.lower()
for orca_sol in self.KNOWN_ORCA_CPCM_SOLVENTS
):
warnings.append(
"solvent may be unknown to Psi4: %s\n" % solvent +
"see AaronTools.theory.implicit_solvent.KNOWN_PSI4_SOLVENTS"
)
out[PSI4_SOLVENT] = {
"Units": "Angstrom",
"Medium":
[
"SolverType = %s" % self.solvent_model,
"Solvent = \"%s\"" % solvent,
],
"Mode": "Implicit"
}
return (out, warnings)
def get_qchem(self):
"""returns dict() with solvent information for qchem input files"""
warnings = []
if self.solvent.lower() == "gas":
return (dict(), warnings)
raise NotImplementedError(
"cannot create solvent info for Q-Chem input files"
) | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/theory/implicit_solvent.py | implicit_solvent.py |
import re
from AaronTools import addlogger
from AaronTools.theory import (
GAUSSIAN_ROUTE, PSI4_SETTINGS, ORCA_ROUTE, ORCA_BLOCKS, QCHEM_REM
)
class IntegrationGrid:
"""
used to try to keep integration grid settings more
easily when writing different input files
"""
LOG = None
def __init__(self, name):
"""
name: str, Gaussian keyword (e.g. SuperFineGrid),
ORCA keyword (e.g. Grid7),
or "(radial, angular)"
ORCA can only use ORCA grid keywords
Gaussian can use its keywords and will try to use ORCA keywords, and
can use "(radial, angular)" or radialangular
Psi4 will use "(radial, angular)" and will try to use ORCA or Gaussian keywords
"""
self.name = name
def __eq__(self, other):
if self.__class__ is not other.__class__:
return False
if self.name == other.name:
return True
return self.get_gaussian()[0] == other.get_gaussian()[0]
def copy(self):
new_dict = dict()
for key, value in self.__dict__.items():
try:
new_dict[key] = value.copy()
except AttributeError:
new_dict[key] = value
# ignore chimerax objects so seqcrow doesn't print a
# warning when a geometry is copied
if "chimerax" in value.__class__.__module__:
continue
if value.__class__.__module__ != "builtins":
self.LOG.warning(
"No copy method for {}: in-place changes may occur".format(
type(value)
)
)
return self.__class__(**new_dict)
def get_gaussian(self):
"""
gets gaussian integration grid info and a warning as tuple(dict, str or None)
dict is of the form {GAUSSIAN_ROUTE:[x]}
"""
if self.name.lower() == "ultrafine":
return ({GAUSSIAN_ROUTE:{"Integral":["grid=UltraFine"]}}, None)
elif self.name.lower() == "finegrid":
return ({GAUSSIAN_ROUTE:{"Integral":["grid=FineGrid"]}}, None)
elif self.name.lower() == "superfinegrid":
return ({GAUSSIAN_ROUTE:{"Integral":["grid=SuperFineGrid"]}}, None)
# Grids available in ORCA but not Gaussian
# uses n_rad from K-Kr as specified in ORCA 4.2.1 manual (section 9.3)
elif self.name.lower() == "grid2":
out_dict = {
GAUSSIAN_ROUTE: {
"Integral":["grid=45110"]
}
}
return (out_dict, "Approximating ORCA Grid 2")
elif self.name.lower() == "grid3":
out_dict = {
GAUSSIAN_ROUTE: {
"Integral":["grid=45194"]
}
}
return (out_dict, "Approximating ORCA Grid 3")
elif self.name.lower() == "grid4":
out_dict = {
GAUSSIAN_ROUTE: {
"Integral":["grid=45302"]
}
}
return (out_dict, "Approximating ORCA Grid 4")
elif self.name.lower() == "grid5":
out_dict = {
GAUSSIAN_ROUTE: {
"Integral":["grid=50434"]
}
}
return (out_dict, "Approximating ORCA Grid 5")
elif self.name.lower() == "grid6":
out_dict = {
GAUSSIAN_ROUTE: {
"Integral":["grid=55590"]
}
}
return (out_dict, "Approximating ORCA Grid 6")
elif self.name.lower() == "grid7":
out_dict = {
GAUSSIAN_ROUTE: {
"Integral":["grid=60770"]
}
}
return (out_dict, "Approximating ORCA Grid 7")
# grid format may be (int, int)
# or just int
match = re.match(r"\(\s*?(\d+)\s*?,\s*?(\d+)?\s*\)", self.name)
match_digit = re.match(r"-?\d+?", self.name)
if match:
r_pts = int(match.group(1))
a_pts = int(match.group(2))
out_dict = {
GAUSSIAN_ROUTE: {
"Integral":["grid=%i%03i" % (r_pts, a_pts)]
}
}
return (out_dict, None)
elif match_digit:
out_dict = {
GAUSSIAN_ROUTE: {
"Integral":["grid=%s" % self.name]
}
}
return (out_dict, None)
out_dict = {
GAUSSIAN_ROUTE: {
"Integral":["grid=%s" % self.name]
}
}
return (out_dict, "grid may not be available in Gaussian")
def get_orca(self):
"""
translates grid to something ORCA accepts
returns tuple(dict(ORCA_ROUTE:[self.name]), None) or
tuple(dict(ORCA_BLOCKS:{"method":[str]}), None)
"""
if self.name.lower() == "ultrafine":
out_dict = {
ORCA_BLOCKS: {
"method": [
"AngularGrid Lebedev590",
"IntAcc 4.0",
]
}
}
return (out_dict, "approximating UltraFineGrid")
elif self.name.lower() == "finegrid":
out_dict = {
ORCA_BLOCKS: {
"method": [
"AngularGrid Lebedev302",
"IntAcc 4.0",
]
}
}
return (out_dict, "approximating FineGrid")
elif self.name.lower() == "superfinegrid":
#radial is 175 for 1st row, 250 for later rows
out_dict = {
ORCA_ROUTE: [
"Grid7",
"FinalGrid7",
],
}
return (
out_dict,
"could not set SuperFineGrid equivalent - using largest ORCA grid keyword",
)
elif re.match("defgrid\d", self.name.lower()):
warnings = None
grid_n = re.match("defgrid(\d)", self.name.lower()).group(1)
if int(grid_n) < 1 or int(grid_n) > 3:
warnings = "grid may not be available"
out_dict = {
ORCA_ROUTE: [self.name]
}
return (out_dict, warnings)
elif self.name.lower().startswith("grid"):
# orca grid keyword
out_dict = {
ORCA_ROUTE: [
self.name,
"Final%s" % self.name,
]
}
return (out_dict, None)
# grid format may be (int, int)
match = re.match(r"\(\s*?(\d+)\s*?,\s*?(\d+)?\s*\)", self.name)
if match:
r_pts = int(match.group(1))
a_pts = int(match.group(2))
int_acc = -((r_pts / -5) + 2 - 8) / 3
out_dict = {
ORCA_BLOCKS: {
"method": [
"AngularGrid Lebedev%i" % a_pts,
"IntAcc %.1f" % int_acc,
]
}
}
return (out_dict, None)
raise RuntimeError(
"could not determine acceptable ORCA grid settings for %s" % self.name
)
def get_psi4(self):
"""
returns ({PSI4_SETTINGS:{"dft_radial_points":["n"], "dft_spherical_points":["m"]}}, warning)
"""
if self.name.lower() == "ultrafine":
out_dict = {
PSI4_SETTINGS: {
"dft_radial_points": ["99"],
"dft_spherical_points": ["590"],
}
}
return (out_dict, None)
elif self.name.lower() == "finegrid":
out_dict = {
PSI4_SETTINGS: {
"dft_radial_points": ["75"],
"dft_spherical_points": ["302"],
}
}
return (out_dict, None)
elif self.name.lower() == "superfinegrid":
# radial is 175 for 1st row, 250 for later rows
out_dict = {
PSI4_SETTINGS: {
"dft_radial_points": ["250"],
"dft_spherical_points": ["974"],
}
}
return (out_dict, "Approximating Gaussian SuperFineGrid")
# uses radial from K-Kr as specified in ORCA 4.2.1 manual (section 9.3)
elif self.name.lower() == "grid2":
out_dict = {
PSI4_SETTINGS: {
"dft_radial_points":["45"],
"dft_spherical_points": ["110"],
}
}
return (out_dict, "Approximating ORCA Grid 2")
elif self.name.lower() == "grid3":
out_dict = {
PSI4_SETTINGS: {
"dft_radial_points": ["45"],
"dft_spherical_points":["194"],
}
}
return (out_dict, "Approximating ORCA Grid 3")
elif self.name.lower() == "grid4":
out_dict = {
PSI4_SETTINGS: {
"dft_radial_points":["45"],
"dft_spherical_points": ["302"],
}
}
return (out_dict, "Approximating ORCA Grid 4")
elif self.name.lower() == "grid5":
out_dict = {
PSI4_SETTINGS: {
"dft_radial_points":["50"],
"dft_spherical_points": ["434"],
}
}
return (out_dict, "Approximating ORCA Grid 5")
elif self.name.lower() == "grid6":
out_dict = {
PSI4_SETTINGS: {
"dft_radial_points": ["55"],
"dft_spherical_points": ["590"],
}
}
return (out_dict, "Approximating ORCA Grid 6")
elif self.name.lower() == "grid7":
out_dict = {
PSI4_SETTINGS: {
"dft_radial_points": ["60"],
"dft_spherical_points": ["770"],
}
}
return (out_dict, "Approximating ORCA Grid 7")
# grid format may be (int, int)
match = re.match(r"\(\s*?(\d+)\s*?,\s*?(\d+)?\s*\)", self.name)
if match:
r_pts = int(match.group(1))
a_pts = int(match.group(2))
out_dict = {
PSI4_SETTINGS: {
"dft_radial_points": [r_pts],
"dft_spherical_points": [a_pts],
}
}
return (out_dict, None)
raise RuntimeError(
"could not determine acceptable Psi4 grid settings for %s" % self.name
)
def get_qchem(self):
if self.name.lower() == "sg-1":
out_dict = {
QCHEM_REM: {
"XC_GRID": "1",
}
}
return (out_dict, None)
elif self.name.lower() == "sg-2":
out_dict = {
QCHEM_REM: {
"XC_GRID": "2",
}
}
return (out_dict, None)
elif self.name.lower() == "sg-3":
out_dict = {
QCHEM_REM: {
"XC_GRID": "3",
}
}
return (out_dict, None)
elif self.name.lower() == "ultrafine":
out_dict = {
QCHEM_REM: {
"XC_GRID": "%2i%06i" % (99, 590),
}
}
return (out_dict, None)
elif self.name.lower() == "finegrid":
out_dict = {
QCHEM_REM: {
"XC_GRID": "%2i%06i" % (75, 302),
}
}
return (out_dict, None)
elif self.name.lower() == "superfinegrid":
# radial is 175 for 1st row, 250 for later rows
out_dict = {
QCHEM_REM: {
"XC_GRID": "%2i%06i" % (250, 974),
}
}
return (out_dict, "Approximating Gaussian SuperFineGrid")
# grid format may be "(int, int)"
match = re.match(r"\(\s*?(\d+)\s*?,\s*?(\d+)?\s*\)", self.name)
if match:
r_pts = int(match.group(1))
a_pts = int(match.group(2))
out_dict = {
QCHEM_REM: {
"XC_GRID": "%2i%06i" % (r_pts, a_pts),
}
}
return (out_dict, None)
raise RuntimeError(
"could not determine acceptable QChem grid settings for %s" % self.name
) | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/theory/grid.py | grid.py |
import numpy as np
from AaronTools import addlogger
from AaronTools.finders import FlaggedAtoms
from AaronTools.theory import (
GAUSSIAN_CONSTRAINTS,
GAUSSIAN_COORDINATES,
GAUSSIAN_ROUTE,
ORCA_BLOCKS,
ORCA_ROUTE,
PSI4_BEFORE_GEOM,
PSI4_COORDINATES,
PSI4_JOB,
PSI4_OPTKING,
PSI4_SETTINGS,
PSI4_BEFORE_JOB,
SQM_QMMM,
QCHEM_REM,
QCHEM_SETTINGS,
)
from AaronTools.utils.utils import range_list, combine_dicts
def job_from_string(name, **kwargs):
"""
return a job type for the given string
valid names are:
"opt" or "conf" with ".ts", ".transition_state", ".change", and ".con" extensions
* .ts and .transition_state indicate a transition state optimization
* .con indicates a constrained optimization - "constraints" should
be in kwargs and the value should be a dictionary conformable with
the keyword of OptimizationJob
"freq" with ".num" extensions
* .num indicates a numerical frequnecy, as does kwargs["numerical"] = True
kwargs can also have a "temperature" key
"sp" or "energy" or "single-point"
"force" or "gradient" with a ".num" extension
* .num indicates a numerical frequnecy, as does kwargs["numerical"] = True
"""
ext = None
if "." in name:
ext = name.split(".")[-1].lower()
if any(name.lower().startswith(x) for x in ["opt", "conf"]):
geom = kwargs.get("geometry", None)
constraints = kwargs.get("constraints", None)
if ext and (ext.startswith("ts") or ext.startswith("transition")):
return OptimizationJob(transition_state=True, geometry=geom)
if ext and ext.startswith("con") and constraints:
return OptimizationJob(constraints=constraints)
if ext and ext.startswith("change"):
return OptimizationJob(
constraints={"atoms": FlaggedAtoms()}, geometry=geom
)
return OptimizationJob(geometry=geom)
if name.lower().startswith("freq"):
numerical = kwargs.get("numerical", False)
numerical = numerical or (ext and ext.startswith("num"))
temperature = kwargs.get("temperature", 298.15)
return FrequencyJob(numerical=numerical, temperature=temperature)
if any(name.lower().startswith(x) for x in ["sp", "energy", "single-point"]):
return SinglePointJob()
if any(name.lower().startswith(x) for x in ["force", "gradient"]):
numerical = kwargs.get("numerical", False)
numerical = numerical or (ext and ext.startswith("num"))
return ForceJob(numerical=numerical)
raise ValueError("cannot determine job type from string: %s" % name)
class JobType:
"""
parent class of all job types
initialization keywords should be the same as attribute names
"""
def __init__(self):
pass
def __eq__(self, other):
if self.__class__ is not other.__class__:
return False
return self.__dict__ == other.__dict__
def get_gaussian(self):
"""overwrite to return dict with GAUSSIAN_* keys"""
pass
def get_orca(self):
"""overwrite to return dict with ORCA_* keys"""
pass
def get_psi4(self):
"""overwrite to return dict with PSI4_* keys"""
pass
def get_sqm(self):
"""overwrite to return a dict with SQM_* keys"""
pass
def get_qchem(self):
"""overwrite to return a dict with QCHEM_* keys"""
pass
@staticmethod
def resolve_error(error, theory, exec_type, geometry=None):
"""returns a copy of theory or modifies theory to attempt
to resolve an error
theory will be modified if it is not possible for the current theory
to work for any job
if the error is specific to the molecule and settings, theory will
be copied, modified, and returned
raises NotImplementedError if this job type has no fix for
the error code
error: error code (e.g. SCF_CONV; see fileIO ERROR)
theory: Theory() instance
exec_type: software program (i.e. gaussian, orca, etc.)
optional kwargs:
geometry: Geometry(), structure might be adjusted slightly if
there are close contacts
"""
if error.upper() == "CLASH":
# if there is a clash, rotate substituents to mitigate clashing
if geometry:
geom_copy = geometry.copy()
bad_subs = geom_copy.remove_clash()
if not bad_subs:
geometry.update_structure(geom_copy.coords)
return None
if error.upper() == "SCF_CONV":
if exec_type.lower() == "gaussian":
# SCF convergence issue, try different SCF algorithm
out_theory = theory.copy()
out_theory.kwargs = combine_dicts(
{
GAUSSIAN_ROUTE: {"scf": ["xqc"]}
},
out_theory.kwargs
)
return out_theory
if exec_type.lower() == "orca":
# SCF convergence issue, orca recommends ! SlowConv
# and increasing SCF iterations
out_theory = theory.copy()
out_theory.kwargs = combine_dicts(
{
ORCA_ROUTE: ["SlowConv"],
ORCA_BLOCKS: {"scf": ["MaxIter 500"]}
},
out_theory.kwargs
)
return out_theory
if exec_type.lower() == "psi4":
out_theory = theory.copy()
# if theory.charge < 0:
# # if the charge is negative, try adding two electrons
# # to get a guess that might work
# # as well as HF with a small basis set
# out_theory.kwargs = combine_dicts(
# {
# PSI4_BEFORE_JOB: {
# [
# "mol = get_active_molecule()",
# "mol.set_molecular_charge(%i)" % (theory.charge + 2),
# "nrg = energy($METHOD)",
# "mol.set_molecular_charge(%i)" % theory.charge,
# ]
# }
# },
# out_theory.kwargs,
# )
# ^ this doesn't seem to help in general
# do 500 iterations and dampen
out_theory.kwargs = combine_dicts(
{
PSI4_SETTINGS: {
"damping_percentage": "15",
"maxiter": "500",
},
},
out_theory.kwargs,
)
raise NotImplementedError(
"cannot fix %s errors for %s; check your input" % (error, exec_type)
)
@addlogger
class OptimizationJob(JobType):
"""optimization job"""
LOG = None
def __init__(
self,
transition_state=False,
constraints=None,
geometry=None,
):
"""use transition_state=True to do a TS optimization
constraints - dict with keys:
**** available for ORCA, Gaussian, and Psi4 ****
'atoms': atom identifiers/finders - atoms to constrain
'bonds': list(atom idenifiers/finders) - distances to constrain
each atom identifier in the list should result in exactly 2 atoms
'angles': list(atom idenifiers/finders) - 1-3 angles to constrain
each atom identifier should result in exactly 3 atoms
'torsions': list(atom identifiers/finders) - constrained dihedral angles
each atom identifier should result in exactly 4 atoms
**** available for Gaussian and Psi4 ****
'x': list(atom identifiers/finders) - constrain the x coordinate of
these atoms
similarly, 'y' and 'z' are also accepted
'xgroup': list(tuple(list(atom idenifiers), x_val, hold)) -
constrain the x coordinate of these atoms to be the same
x_val - set x-coordinate to this value
hold - hold this value constant during the optimization
if 'hold' is omitted, the value will not be held
constant during the optimization
e.g. 'xgroup':[("1-6", 0, False), ("13-24", 3.25, False)]
this will keep atoms 1-6 and 13-24 in parallel planes, while also
allowing those planes to move
'ygroup' and 'zgroup' are also available, with analagous options
*** NOTE ***
for Gaussian, 'bonds', 'angles', and 'torsions' constraints cannot be mixed
with 'x', 'y', 'z', 'xgroup', 'ygroup', or 'zgroup' constraints
geometry - Geoemtry, will be set when using an AaronTools FileWriter"""
super().__init__()
self.transition_state = transition_state
self.constraints = constraints
self.geometry = geometry
def get_gaussian(self):
"""returns a dict with keys: GAUSSIAN_ROUTE, GAUSSIAN_CONSTRAINTS"""
if self.transition_state:
out = {GAUSSIAN_ROUTE: {"Opt": ["ts"]}}
else:
out = {GAUSSIAN_ROUTE: {"Opt": []}}
coords = self.geometry.coords.tolist()
vars = []
consts = []
use_zmat = False
group_count = 1
if self.constraints is not None and any(
self.constraints[key] for key in self.constraints.keys()
):
for key in self.constraints:
if key not in [
"x",
"y",
"z",
"xgroup",
"ygroup",
"zgroup",
"atoms",
"bonds",
"angles",
"torsions",
]:
raise NotImplementedError(
"%s constraints cannot be generated for Gaussian" % key
)
out[GAUSSIAN_CONSTRAINTS] = []
if "x" in self.constraints and self.constraints["x"]:
x_atoms = self.geometry.find(self.constraints["x"])
for i, atom in enumerate(self.geometry.atoms):
if atom in x_atoms:
var_name = "x%i" % (i + 1)
consts.append((var_name, atom.coords[0]))
coords[i] = [var_name, coords[i][1], coords[i][2]]
if not use_zmat:
use_zmat = True
out[GAUSSIAN_ROUTE]["Opt"].append("Z-Matrix")
if "y" in self.constraints and self.constraints["y"]:
y_atoms = self.geometry.find(self.constraints["y"])
for i, atom in enumerate(self.geometry.atoms):
if atom in y_atoms:
var_name = "y%i" % (i + 1)
consts.append((var_name, atom.coords[1]))
coords[i] = [coords[i][0], var_name, coords[i][2]]
if not use_zmat:
use_zmat = True
out[GAUSSIAN_ROUTE]["Opt"].append("Z-Matrix")
if "z" in self.constraints and self.constraints["z"]:
z_atoms = self.geometry.find(self.constraints["z"])
for i, atom in enumerate(self.geometry.atoms):
if atom in z_atoms:
var_name = "z%i" % (i + 1)
consts.append((var_name, atom.coords[2]))
coords[i] = [coords[i][0], coords[i][1], var_name]
if not use_zmat:
use_zmat = True
out[GAUSSIAN_ROUTE]["Opt"].append("Z-Matrix")
if "xgroup" in self.constraints:
for constraint in self.constraints["xgroup"]:
if len(constraint) == 3:
finders, val, hold = constraint
else:
finders, val = constraint
hold = False
x_atoms = self.geometry.find(finders)
var_name = "gx%i" % group_count
group_count += 1
if hold:
consts.append([var_name, val])
else:
vars.append([var_name, val])
for i, atom in enumerate(self.geometry.atoms):
if atom in x_atoms:
coords[i] = [var_name, coords[i][1], coords[i][2]]
if not use_zmat:
use_zmat = True
out[GAUSSIAN_ROUTE]["Opt"].append("Z-Matrix")
if "ygroup" in self.constraints:
for constraint in self.constraints["ygroup"]:
if len(constraint) == 3:
finders, val, hold = constraint
else:
finders, val = constraint
hold = False
y_atoms = self.geometry.find(finders)
var_name = "gy%i" % group_count
group_count += 1
if hold:
consts.append([var_name, val])
else:
vars.append([var_name, val])
for i, atom in enumerate(self.geometry.atoms):
if atom in y_atoms:
coords[i] = [coords[i][0], var_name, coords[i][2]]
if not use_zmat:
use_zmat = True
out[GAUSSIAN_ROUTE]["Opt"].append("Z-Matrix")
if "zgroup" in self.constraints:
for constraint in self.constraints["zgroup"]:
if len(constraint) == 3:
finders, val, hold = constraint
else:
finders, val = constraint
hold = False
z_atoms = self.geometry.find(finders)
var_name = "gz%i" % group_count
group_count += 1
if hold:
consts.append([var_name, val])
else:
vars.append([var_name, val])
for i, atom in enumerate(self.geometry.atoms):
if atom in z_atoms:
coords[i] = [coords[i][0], coords[i][1], var_name]
if not use_zmat:
use_zmat = True
out[GAUSSIAN_ROUTE]["Opt"].append("Z-Matrix")
if "atoms" in self.constraints and self.constraints["atoms"]:
try:
atoms = self.geometry.find(self.constraints["atoms"])
except LookupError as e:
self.LOG.warning(e)
atoms = []
for atom in atoms:
ndx = self.geometry.atoms.index(atom) + 1
if not use_zmat:
out[GAUSSIAN_CONSTRAINTS].append("%2i F" % ndx)
else:
for j, coord in enumerate(coords[ndx - 1]):
if isinstance(coord, str):
var_name = coord
for k, var in enumerate(vars):
if var[0] == coord and not var[
0
].startswith("g"):
vars.pop(k)
break
else:
var_name = "%s%i" % (
["x", "y", "z"][j],
ndx,
)
coords[ndx - 1][j] = var_name
else:
var_name = "%s%i" % (["x", "y", "z"][j], ndx)
coords[ndx - 1][j] = var_name
if not any(
const[0] == var_name for const in consts
):
consts.append([var_name, atom.coords[j]])
if not use_zmat:
if "ModRedundant" not in out[GAUSSIAN_ROUTE]["Opt"]:
out[GAUSSIAN_ROUTE]["Opt"].append("ModRedundant")
if "bonds" in self.constraints:
for constraint in self.constraints["bonds"]:
atom1, atom2 = self.geometry.find(constraint)
ndx1 = self.geometry.atoms.index(atom1) + 1
ndx2 = self.geometry.atoms.index(atom2) + 1
if not use_zmat:
out[GAUSSIAN_CONSTRAINTS].append(
"B %2i %2i F" % (ndx1, ndx2)
)
else:
raise NotImplementedError(
"cannot apply bond constraints when using Cartesian Z-Matrix, which"
+ " is necessitated by x, y, or z constraints"
)
if "ModRedundant" not in out[GAUSSIAN_ROUTE]["Opt"]:
out[GAUSSIAN_ROUTE]["Opt"].append("ModRedundant")
if "angles" in self.constraints:
for constraint in self.constraints["angles"]:
atom1, atom2, atom3 = self.geometry.find(constraint)
ndx1 = self.geometry.atoms.index(atom1) + 1
ndx2 = self.geometry.atoms.index(atom2) + 1
ndx3 = self.geometry.atoms.index(atom3) + 1
if not use_zmat:
out[GAUSSIAN_CONSTRAINTS].append(
"A %2i %2i %2i F" % (ndx1, ndx2, ndx3)
)
else:
raise NotImplementedError(
"cannot apply angle constraints when using Cartesian Z-Matrix, which"
+ " is necessitated by x, y, or z constraints"
)
if "ModRedundant" not in out[GAUSSIAN_ROUTE]["Opt"]:
out[GAUSSIAN_ROUTE]["Opt"].append("ModRedundant")
if "torsions" in self.constraints:
for constraint in self.constraints["torsions"]:
atom1, atom2, atom3, atom4 = self.geometry.find(constraint)
ndx1 = self.geometry.atoms.index(atom1) + 1
ndx2 = self.geometry.atoms.index(atom2) + 1
ndx3 = self.geometry.atoms.index(atom3) + 1
ndx4 = self.geometry.atoms.index(atom4) + 1
if not use_zmat:
out[GAUSSIAN_CONSTRAINTS].append(
"D %2i %2i %2i %2i F" % (ndx1, ndx2, ndx3, ndx4)
)
else:
raise NotImplementedError(
"cannot apply torsion constraints when using Cartesian Z-Matrix,"
+ "which is necessitated by x, y, or z constraints"
)
if "ModRedundant" not in out[GAUSSIAN_ROUTE]["Opt"]:
out[GAUSSIAN_ROUTE]["Opt"].append("ModRedundant")
if consts or vars:
for i, coord in enumerate(coords):
for j, ax in enumerate(["x", "y", "z"]):
if isinstance(coord[j], float):
var_name = "%s%i" % (ax, i + 1)
vars.append((var_name, coord[j]))
coord[j] = var_name
if consts or vars:
for coord in coords:
coord.insert(0, 0)
out[GAUSSIAN_COORDINATES] = {
"coords": coords,
"variables": vars,
"constants": consts,
}
return out
def get_orca(self):
"""returns a dict with keys: ORCA_ROUTE, ORCA_BLOCKS"""
if self.transition_state:
out = {ORCA_ROUTE: ["OptTS"]}
else:
out = {ORCA_ROUTE: ["Opt"]}
if self.constraints is not None and any(
self.constraints[key] for key in self.constraints.keys()
):
for key in self.constraints:
if key not in [
"atoms",
"bonds",
"angles",
"torsions",
]:
raise NotImplementedError(
"%s constraints cannot be generated for ORCA" % key
)
out[ORCA_BLOCKS] = {"geom": ["Constraints"]}
if "atoms" in self.constraints:
for constraint in self.constraints["atoms"]:
atom1 = self.geometry.find(constraint)[0]
ndx1 = self.geometry.atoms.index(atom1)
out_str = " {C %2i C}" % (ndx1)
out[ORCA_BLOCKS]["geom"].append(out_str)
if "bonds" in self.constraints:
for constraint in self.constraints["bonds"]:
atom1, atom2 = self.geometry.find(constraint)
ndx1 = self.geometry.atoms.index(atom1)
ndx2 = self.geometry.atoms.index(atom2)
out_str = " {B %2i %2i C}" % (ndx1, ndx2)
out[ORCA_BLOCKS]["geom"].append(out_str)
if "angles" in self.constraints:
for constraint in self.constraints["angles"]:
atom1, atom2, atom3 = self.geometry.find(constraint)
ndx1 = self.geometry.atoms.index(atom1)
ndx2 = self.geometry.atoms.index(atom2)
ndx3 = self.geometry.atoms.index(atom3)
out_str = " {A %2i %2i %2i C}" % (ndx1, ndx2, ndx3)
out[ORCA_BLOCKS]["geom"].append(out_str)
if "torsions" in self.constraints:
for constraint in self.constraints["torsions"]:
atom1, atom2, atom3, atom4 = self.geometry.find(constraint)
ndx1 = self.geometry.atoms.index(atom1)
ndx2 = self.geometry.atoms.index(atom2)
ndx3 = self.geometry.atoms.index(atom3)
ndx4 = self.geometry.atoms.index(atom4)
out_str = " {D %2i %2i %2i %2i C}" % (
ndx1,
ndx2,
ndx3,
ndx4,
)
out[ORCA_BLOCKS]["geom"].append(out_str)
out[ORCA_BLOCKS]["geom"].append("end")
return out
def get_psi4(self):
"""returns a dict with keys: PSI4_JOB, PSI4_OPTKING, PSI4_BEFORE_GEOM"""
if self.transition_state:
out = {
PSI4_JOB: {"optimize": []},
PSI4_SETTINGS: {"opt_type": ["ts"]},
}
else:
out = {PSI4_JOB: {"optimize": []}}
coords = self.geometry.coords.tolist()
vars = []
group_count = 1
freeze_str = ""
freeze_str += 'freeze_list = """\n'
add_freeze_list = False
# constraints
if self.constraints is not None and any(
[self.constraints[key] for key in self.constraints.keys()]
):
for key in self.constraints:
if key not in [
"x",
"y",
"z",
"xgroup",
"ygroup",
"zgroup",
"atoms",
"bonds",
"angles",
"torsions",
]:
raise NotImplementedError(
"%s constraints cannot be generated for Psi4" % key
)
out[PSI4_OPTKING] = {}
if (
"x" in self.constraints
and self.constraints["x"]
and self.geometry is not None
):
add_freeze_list = True
atoms = self.geometry.find(self.constraints["x"])
for atom in atoms:
freeze_str += " %2i x\n" % (
self.geometry.atoms.index(atom) + 1
)
if (
"y" in self.constraints
and self.constraints["y"]
and self.geometry is not None
):
add_freeze_list = True
atoms = self.geometry.find(self.constraints["y"])
for atom in atoms:
freeze_str += " %2i y\n" % (
self.geometry.atoms.index(atom) + 1
)
if (
"z" in self.constraints
and self.constraints["z"]
and self.geometry is not None
):
add_freeze_list = True
atoms = self.geometry.find(self.constraints["z"])
for atom in atoms:
freeze_str += " %2i z\n" % (
self.geometry.atoms.index(atom) + 1
)
if (
"atoms" in self.constraints
and self.constraints["atoms"]
and self.geometry is not None
):
add_freeze_list = True
atoms = self.geometry.find(self.constraints["atoms"])
for atom in atoms:
freeze_str += " %2i xyz\n" % (
self.geometry.atoms.index(atom) + 1
)
if "xgroup" in self.constraints:
for constraint in self.constraints["xgroup"]:
if len(constraint) == 3:
finders, val, hold = constraint
else:
finders, val = constraint
hold = False
x_atoms = self.geometry.find(finders)
var_name = "gx%i" % group_count
group_count += 1
if hold:
add_freeze_list = True
for i, atom in enumerate(x_atoms):
freeze_str += " %2i x\n" % (
self.geometry.atoms.index(atom) + 1
)
coords[i][0] = val
else:
vars.append([var_name, val, True])
for i, atom in enumerate(self.geometry.atoms):
if atom in x_atoms:
coords[i] = [
coords[i][0],
coords[i][1],
var_name,
]
if "ygroup" in self.constraints:
for constraint in self.constraints["ygroup"]:
if len(constraint) == 3:
finders, val, hold = constraint
else:
finders, val = constraint
hold = False
y_atoms = self.geometry.find(finders)
var_name = "gy%i" % group_count
group_count += 1
if hold:
add_freeze_list = True
for i, atom in enumerate(y_atoms):
freeze_str += " %2i y\n" % (
self.geometry.atoms.index(atom) + 1
)
coords[i][1] = val
else:
vars.append([var_name, val, True])
for i, atom in enumerate(self.geometry.atoms):
if atom in y_atoms:
coords[i] = [
coords[i][0],
coords[i][1],
var_name,
]
if "zgroup" in self.constraints:
for constraint in self.constraints["zgroup"]:
if len(constraint) == 3:
finders, val, hold = constraint
else:
finders, val = constraint
hold = False
z_atoms = self.geometry.find(finders)
var_name = "gz%i" % group_count
group_count += 1
if hold:
add_freeze_list = True
for i, atom in enumerate(z_atoms):
freeze_str += " %2i z\n" % (
self.geometry.atoms.index(atom) + 1
)
coords[i][2] = val
else:
vars.append([var_name, val, True])
for i, atom in enumerate(self.geometry.atoms):
if atom in z_atoms:
coords[i] = [
coords[i][0],
coords[i][1],
var_name,
]
if add_freeze_list:
freeze_str += '"""\n'
freeze_str += " \n"
out[PSI4_BEFORE_GEOM] = [freeze_str]
out[PSI4_OPTKING]["frozen_cartesian"] = ["$freeze_list"]
if "bonds" in self.constraints:
if self.constraints["bonds"] and self.geometry is not None:
out_str = '("\n'
for bond in self.constraints["bonds"]:
atom1, atom2 = self.geometry.find(bond)
out_str += " %2i %2i\n" % (
self.geometry.atoms.index(atom1) + 1,
self.geometry.atoms.index(atom2) + 1,
)
out_str += ' ")\n'
out[PSI4_OPTKING]["frozen_distance"] = [out_str]
if "angles" in self.constraints:
if self.constraints["angles"] and self.geometry is not None:
out_str = '("\n'
for angle in self.constraints["angles"]:
atom1, atom2, atom3 = self.geometry.find(angle)
out_str += " %2i %2i %2i\n" % (
self.geometry.atoms.index(atom1) + 1,
self.geometry.atoms.index(atom2) + 1,
self.geometry.atoms.index(atom3) + 1,
)
out_str += ' ")\n'
out[PSI4_OPTKING]["frozen_bend"] = [out_str]
if "torsions" in self.constraints:
if self.constraints["torsions"] and self.geometry is not None:
out_str += '("\n'
for torsion in self.constraints["torsions"]:
atom1, atom2, atom3, atom4 = self.geometry.find(
torsion
)
out_str += " %2i %2i %2i %2i\n" % (
self.geometry.atoms.index(atom1) + 1,
self.geometry.atoms.index(atom2) + 1,
self.geometry.atoms.index(atom3) + 1,
self.geometry.atoms.index(atom4) + 1,
)
out_str += ' ")\n'
out[PSI4_OPTKING]["frozen_dihedral"] = [out_str]
if vars:
out[PSI4_COORDINATES] = {
"coords": coords,
"variables": vars,
}
return out
def get_xcontrol(self, config, ref=None):
"""
Generates xcontrol file constraints
Returns: dict(xcontrol)
"""
if ref is None:
ref = "ref.xyz"
xcontrol = ""
# only put constraints in xcontrol file so this works with Crest also
frozen = [i + 1 for i, a in enumerate(self.geometry) if bool(a.flag)]
if frozen:
frozen = range_list(frozen)
xcontrol += "$fix\n"
xcontrol += " atoms: {}\n".format(frozen)
xcontrol += " freeze: {}\n".format(frozen)
elif self.constraints:
xcontrol += "$constrain\n"
xcontrol += " force constant={}\n".format(
config["Job"].get("constrain_force", fallback="0.5")
)
xcontrol += " reference={}\n".format(
config["Job"].get("constrain_ref", fallback=ref)
)
constrained = set([])
for bond in self.constraints.get("bonds", []):
bond = self.geometry.find(bond)
constrained.update(bond)
xcontrol += " distance: {},{},auto\n".format(
*(self.geometry.atoms.index(c) + 1 for c in bond)
)
for angle in self.constraints.get("angles", []):
angle = self.geometry.find(angle)
constrained.update(angle)
xcontrol += " angle: {},{},{},auto\n".format(
*(self.geometry.atoms.index(c) + 1 for c in angle)
)
for dihedral in self.constraints.get("torsions", []):
dihedral = self.geometry.find(dihedral)
constrained.update(dihedral)
xcontrol += " dihedral: {},{},{},{},auto\n".format(
*(self.geometry.atoms.index(c) + 1 for c in dihedral)
)
relaxed = {
i + 1
for i, a in enumerate(self.geometry.atoms)
if a not in constrained
}
relaxed = range_list(relaxed)
xcontrol += "$metadyn\n"
xcontrol += " atoms: {}\n".format(relaxed)
xcontrol += "$end\n"
return xcontrol
def get_sqm(self):
"""returns a dict(), warnings for optimization jobs"""
warnings = []
if self.transition_state:
warnings.append("cannot do TS optimization with sqm")
if self.constraints:
warnings.append("cannot constrain sqm optimization")
return dict(), warnings
def get_qchem(self):
if self.transition_state:
out = {QCHEM_REM: {"JOB_TYPE": "TS"}}
else:
out = {QCHEM_REM: {"JOB_TYPE": "OPT"}}
# constraints
if self.constraints is not None and any(
[self.constraints[key] for key in self.constraints.keys()]
):
out[QCHEM_SETTINGS] = {"opt": []}
constraints = None
fixed = None
x_atoms = []
y_atoms = []
z_atoms = []
xyz_atoms = []
if "x" in self.constraints:
x_atoms = self.geometry.find(self.constraints["x"])
if "y" in self.constraints:
y_atoms = self.geometry.find(self.constraints["y"])
if "z" in self.constraints:
z_atoms = self.geometry.find(self.constraints["z"])
if "atoms" in self.constraints:
xyz_atoms = self.geometry.find(self.constraints["atoms"])
if any([x_atoms, y_atoms, z_atoms, xyz_atoms]):
fixed = "FIXED"
for atom in x_atoms:
fixed += "\n %i X" % (self.geometry.atoms.index(atom) + 1)
if atom in xyz_atoms:
fixed += "YZ"
continue
if atom in y_atoms:
fixed += "Y"
if atom in z_atoms:
fixed += "Z"
for atom in y_atoms:
if atom in x_atoms:
continue
fixed += "\n %i " % (self.geometry.atoms.index(atom) + 1)
if atom in xyz_atoms:
fixed += "XYZ"
continue
fixed += "Y"
if atom in z_atoms:
fixed += "Z"
for atom in y_atoms:
if atom in x_atoms or atom in y_atoms:
continue
fixed += "\n %i " % (self.geometry.atoms.index(atom) + 1)
if atom in xyz_atoms:
fixed += "XYZ"
continue
fixed += "Z"
for atom in xyz_atoms:
if any(atom in l for l in [x_atoms, y_atoms, z_atoms]):
continue
fixed += "\n %i XYZ" % (self.geometry.atoms.index(atom) + 1)
if "bonds" in self.constraints:
if constraints is None:
constraints = "CONSTRAINT\n"
for bond in self.constraints["bonds"]:
atom1, atom2 = self.geometry.find(bond)
constraints += " STRE %2i %2i %9.5f\n" % (
self.geometry.atoms.index(atom1) + 1,
self.geometry.atoms.index(atom2) + 1,
atom1.dist(atom2),
)
if "angles" in self.constraints:
if constraints is None:
constraints = "CONSTRAINT\n"
for angle in self.constraints["angles"]:
atom1, atom2, atom3 = self.geometry.find(angle)
constraints += " BEND %2i %2i %2i %9.5f\n" % (
self.geometry.atoms.index(atom1) + 1,
self.geometry.atoms.index(atom2) + 1,
self.geometry.atoms.index(atom3) + 1,
np.rad2deg(atom2.angle(atom1, atom3)),
)
if "torsions" in self.constraints:
if constraints is None:
constraints = "CONSTRAINT\n"
for angle in self.constraints["torsions"]:
atom1, atom2, atom3, atom4 = self.geometry.find(angle)
constraints += " TORS %2i %2i %2i %2i %9.5f\n" % (
self.geometry.atoms.index(atom1) + 1,
self.geometry.atoms.index(atom2) + 1,
self.geometry.atoms.index(atom3) + 1,
self.geometry.atoms.index(atom4) + 1,
np.rad2deg(
self.geometry.dihedral(
atom1, atom2, atom3, atom4,
)
),
)
if fixed:
fixed += "\n ENDFIXED"
out[QCHEM_SETTINGS]["opt"].append(fixed)
if constraints:
constraints += " ENDCONSTRAINT"
out[QCHEM_SETTINGS]["opt"].append(constraints)
return out
@staticmethod
def resolve_error(error, theory, exec_type, geometry=None):
"""
resolves optimization-specific errors
errors resolved by JobType take priority
optional kwargs:
geometry: Geometry(), structure might be adjusted slightly if
the software had an issue with generating internal coordinates
"""
try:
return super(OptimizationJob, OptimizationJob).resolve_error(
error, theory, exec_type, geometry=geometry
)
except NotImplementedError:
pass
if exec_type.lower() == "gaussian":
if error.upper() == "CONV_LINK":
# optimization out of steps, add more steps
out_theory = theory.copy()
out_theory.kwargs = combine_dicts(
{GAUSSIAN_ROUTE: {"opt": ["MaxCycles=300"]}}, out_theory.kwargs,
)
return out_theory
if error.upper() == "FBX":
# FormBX error, just restart the job
# adjusting the geometry slightly can help
if geometry:
coords = geometry.coords
scale = 1e-3
coords += scale * np.random.random_sample - scale / 2
geometry.update_structure(coords)
return None
if error.upper() == "REDUND":
# internal coordinate error, just restart the job
if geometry:
coords = geometry.coords
scale = 1e-3
coords += scale * np.random.random_sample - scale / 2
geometry.update_structure(coords)
return None
if exec_type.lower() == "orca":
if error.upper() == "OPT_CONV":
# optimization out of steps, add more steps
out_theory = theory.copy()
out_theory.kwargs = combine_dicts(
{ORCA_BLOCKS: {"geom": ["MaxIter 300"]}}, out_theory.kwargs,
)
return out_theory
if exec_type.lower() == "psi4":
if error.upper() == "ICOORD":
out_theory = theory.copy()
out_theory.kwargs = combine_dicts(
{PSI4_OPTKING: {"opt_coordinates": "cartesian"}}, out_theory.kwargs,
)
return out_theory
raise NotImplementedError(
"cannot fix %s errors for %s; check your input" % (error, exec_type)
)
class FrequencyJob(JobType):
"""frequnecy job"""
def __init__(self, numerical=False, temperature=None):
"""
temperature in K for thermochem info, defaults to 298.15 K
"""
super().__init__()
if temperature is None:
temperature = 298.15
self.numerical = numerical
self.temperature = temperature
def get_gaussian(self):
"""returns a dict with keys: GAUSSIAN_ROUTE"""
out = {
GAUSSIAN_ROUTE: {
"Freq": ["temperature=%.2f" % float(self.temperature)]
}
}
if self.numerical:
out[GAUSSIAN_ROUTE]["Freq"].append("Numerical")
return out
def get_orca(self):
"""returns a dict with keys: ORCA_ROUTE"""
out = {ORCA_BLOCKS: {"freq": ["Temp %.2f" % self.temperature]}}
if self.numerical:
out[ORCA_ROUTE] = ["NumFreq"]
else:
out[ORCA_ROUTE] = ["Freq"]
return out
def get_psi4(self):
"""returns a dict with keys: PSI4_JOB"""
out = {
PSI4_JOB: {"frequencies": []},
PSI4_SETTINGS: {"T": ["%.2f" % self.temperature]},
}
if self.numerical:
out[PSI4_JOB]["frequencies"].append('dertype="gradient"')
return out
def get_sqm(self):
raise NotImplementedError("cannot build frequnecy job input for sqm")
def get_qchem(self):
out = {QCHEM_REM: {"JOB_TYPE": "Freq"}}
if self.numerical:
out[QCHEM_REM]["FD_DERIVATIVE_TYPE"] = "1"
return out
@staticmethod
def resolve_error(error, theory, exec_type, geometry=None):
"""
resolves frequnecy-specific errors
errors resolved by JobType take priority
"""
try:
return super(FrequencyJob, FrequencyJob).resolve_error(
error, theory, exec_type, geometry=geometry
)
except NotImplementedError:
pass
if exec_type.lower() == "orca":
if error.upper() == "NUMFREQ":
# analytical derivatives are not available
for job in theory.job_type:
if isinstance(job, FrequencyJob):
job.numerical = True
return None
raise NotImplementedError(
"cannot fix %s errors for %s; check your input" % (error, exec_type)
)
class SinglePointJob(JobType):
"""single point energy"""
def get_gaussian(self):
"""returns a dict with keys: GAUSSIAN_ROUTE"""
return {GAUSSIAN_ROUTE: {"SP": []}}
def get_orca(self):
"""returns a dict with keys: ORCA_ROUTE"""
return {ORCA_ROUTE: ["SP"]}
def get_psi4(self):
"""returns a dict with keys: PSI4_JOB"""
return {PSI4_JOB: {"energy": []}}
def get_sqm(self):
"""returns a dict with keys: SQM_QMMM"""
return {SQM_QMMM: {"maxcyc": ["0"]}}
def get_qchem(self):
out = {QCHEM_REM: {"JOB_TYPE": "SP"}}
return out
class ForceJob(JobType):
"""force/gradient job"""
def __init__(self, numerical=False):
super().__init__()
self.numerical = numerical
def get_gaussian(self):
"""returns a dict with keys: GAUSSIAN_ROUTE"""
out = {GAUSSIAN_ROUTE: {"force": []}}
if self.numerical:
out[GAUSSIAN_ROUTE]["force"].append("EnGrad")
return out
def get_orca(self):
"""returns a dict with keys: ORCA_ROUTE"""
return {ORCA_ROUTE: ["NumGrad" if self.numerical else "EnGrad"]}
def get_psi4(self):
"""returns a dict with keys: PSI4_JOB"""
out = {PSI4_JOB: {"gradient": []}}
if self.numerical:
out[PSI4_JOB]["gradient"].append("dertype='energy'")
return out
def get_qchem(self):
out = {QCHEM_REM: {"JOB_TYPE": "Force"}}
return out | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/theory/job_types.py | job_types.py |
import os
import re
from warnings import warn
from AaronTools import addlogger
from AaronTools.const import ELEMENTS
from AaronTools.finders import (
AnyNonTransitionMetal,
AnyTransitionMetal,
NotAny,
)
from AaronTools.theory import (
GAUSSIAN_GEN_BASIS,
GAUSSIAN_GEN_ECP,
GAUSSIAN_ROUTE,
ORCA_BLOCKS,
ORCA_ROUTE,
PSI4_BEFORE_GEOM,
QCHEM_REM,
QCHEM_SETTINGS,
)
@addlogger
class Basis:
"""
has attributes:
name - same as initialization keyword
elements - same as initialization keyword
aux_type - same as initialization keyword
elements - list of element symbols for elements this basis applies to
updated with Basis.refresh_elements
Basis.refresh_elements is called when writing an input file
ele_selection - list of finders used to determine which elements this basis applies to
not_anys - list of finders used to determine which elements this basis does not apply to
"""
LOG = None
default_elements = [AnyTransitionMetal(), AnyNonTransitionMetal()]
def __init__(self, name, elements=None, aux_type=None, user_defined=False):
"""
name - basis set base name (e.g. 6-31G)
elements - list of element symbols or finders to determine the basis set applies to
elements may also be 'tm' or 'all' to indicate any transition metal and
all elements, respectively
elements may start with '!' to exclude that element from the basis
for example, elements='!H' will apply to default elements, minus H
aux_type - str - ORCA: one of BasisSet.ORCA_AUX; Psi4: one of BasisSet.PSI4_AUX
user_defined - path to file containing basis info from basissetexchange.org or similar
False for builtin basis sets
"""
self.name = name
if elements is None:
self.elements = []
self.ele_selection = self.default_elements
self.not_anys = []
else:
# a list of elements or other identifiers was given
# if it's an element with a ! in front, add that element to not_anys
# otherwise, add the appropriate thing to ele_selection
if not hasattr(elements, "__iter__") or isinstance(elements, str):
elements = [elements]
self.elements = elements
ele_selection = []
not_anys = []
for ele in elements:
not_any = False
if isinstance(ele, str) and ele.startswith("!"):
ele = ele.lstrip("!")
not_any = True
if ele.lower() == "all":
if not_any:
not_anys.append(AnyTransitionMetal())
not_anys.append(AnyNonTransitionMetal())
else:
ele_selection.append(AnyTransitionMetal())
ele_selection.append(AnyNonTransitionMetal())
elif ele.lower() == "tm" and ele != "Tm":
if not_any:
ele_selection.append(AnyNonTransitionMetal())
else:
ele_selection.append(AnyTransitionMetal())
elif ele.lower() == "!tm" and ele != "!Tm":
if not_any:
ele_selection.append(AnyNonTransitionMetal())
else:
ele_selection.append(AnyNonTransitionMetal())
elif isinstance(ele, str) and ele in ELEMENTS:
if not_any:
not_anys.append(ele)
else:
ele_selection.append(ele)
else:
warn("element not known: %s" % repr(ele))
if not ele_selection and not_anys:
# if only not_anys were given, fall back to the default elements
ele_selection = self.default_elements
self.ele_selection = ele_selection
self.not_anys = not_anys
self.aux_type = aux_type
self.user_defined = user_defined
def __repr__(self):
return "%s(%s)%s" % (
self.name,
" ".join(self.elements),
"" if not self.aux_type else "/%s" % self.aux_type
)
def __lt__(self, other):
if self.name < other.name:
return True
elif self.name == other.name and self.elements and other.elements:
return self.elements[0] < other.elements[0]
return False
def __eq__(self, other):
if self.__class__ is not other.__class__:
return False
if (
self.get_gaussian(self.name).lower()
!= other.get_gaussian(other.name).lower()
):
return False
if self.aux_type != other.aux_type:
return False
for obj, obj2 in zip([self, other], [other, self]):
for finder in obj.ele_selection:
if isinstance(finder, str):
if finder not in obj2.ele_selection:
return False
else:
for finder2 in obj2.ele_selection:
if repr(finder) == repr(finder2):
break
else:
return False
return True
def copy(self):
new_dict = dict()
for key, value in self.__dict__.items():
try:
new_dict[key] = value.copy()
except AttributeError:
new_dict[key] = value
# ignore chimerax objects so seqcrow doesn't print a
# warning when a geometry is copied
if "chimerax" in value.__class__.__module__:
continue
if value.__class__.__module__ != "builtins":
self.LOG.warning(
"No copy method for {}: in-place changes may occur".format(
type(value)
)
)
return self.__class__(**new_dict)
def refresh_elements(self, geometry):
"""sets self's elements for the geometry"""
atoms = geometry.find(self.ele_selection, NotAny(*self.not_anys))
elements = set([atom.element for atom in atoms])
self.elements = sorted(elements)
@staticmethod
def sanity_check_basis(name, program):
import os.path
from difflib import SequenceMatcher as seqmatch
from re import IGNORECASE, match
from AaronTools.const import AARONTOOLS
from numpy import argsort, loadtxt
warning = None
if program.lower() == "gaussian":
valid = loadtxt(
os.path.join(
AARONTOOLS, "theory", "valid_basis_sets", "gaussian.txt"
),
dtype=str,
)
elif program.lower() == "orca":
valid = loadtxt(
os.path.join(
AARONTOOLS, "theory", "valid_basis_sets", "orca.txt"
),
dtype=str,
)
elif program.lower() == "psi4":
valid = loadtxt(
os.path.join(
AARONTOOLS, "theory", "valid_basis_sets", "psi4.txt"
),
dtype=str,
)
elif program.lower() == "qchem":
valid = loadtxt(
os.path.join(
AARONTOOLS, "theory", "valid_basis_sets", "qchem.txt"
),
dtype=str,
)
else:
raise NotImplementedError(
"cannot validate basis names for %s" % program
)
if not any(
# need to escape () b/c they aren't capturing groups, it's ccsd(t) or something
match(
"%s$"
% (basis.replace("(", "\(").replace(")", "\)"))
.replace("*", "\*")
.replace("+", "\+"),
name,
flags=IGNORECASE,
)
for basis in valid
):
warning = (
"basis '%s' may not be available in %s\n" % (name, program)
+ "if this is incorrect, please submit a bug report at https://github.com/QChASM/AaronTools.py/issues"
)
# try to suggest alternatives that have similar names
simm = [
seqmatch(
lambda x: x in "-_()/", name.upper(), test_basis.upper()
).ratio()
for test_basis in valid
]
ndx = argsort(simm)[-5:][::-1]
warning += "\npossible misspelling of:\n"
warning += "\n".join([valid[i] for i in ndx])
return warning
@staticmethod
def get_gaussian(name):
"""
returns the Gaussian09/16 name of the basis set
currently just removes the hyphen from the Karlsruhe def2 ones
"""
if name.startswith("def2-"):
return name.replace("def2-", "def2", 1)
return name
@staticmethod
def get_orca(name):
"""
returns the ORCA name of the basis set
currently just adds hyphen to Karlsruhe basis if it isn't there
"""
if name.startswith("def2") and not re.match("def2(?:-|$)", name):
return name.replace("def2", "def2-", 1)
return name
@staticmethod
def get_psi4(name):
"""
returns the Psi4 name of the basis set
currently just adds hyphen to Karlsruhe basis if it isn't there
"""
if name.startswith("def2") and not name.startswith("def2-"):
return name.replace("def2", "def2-", 1)
# pople basis sets don't have commas
# e.g. 6-31G(d,p) -> 6-31G(d_p)
if name.startswith("6-31") or name.startswith("3-21"):
name = name.replace(",", "_")
return name
@staticmethod
def get_qchem(name):
"""
returns the Psi4 name of the basis set
currently just adds hyphen to Karlsruhe basis if it isn't there
"""
if name.startswith("def2") and not name.startswith("def2-"):
return name.replace("def2", "def2-", 1)
return name
class ECP(Basis):
"""ECP - aux info will be ignored"""
default_elements = (AnyTransitionMetal(), )
def __init__(self, *args, **kw):
super().__init__(*args, **kw)
def __eq__(self, other):
if not isinstance(other, ECP):
return False
return super().__eq__(other)
@staticmethod
def sanity_check_basis(name, program):
import os.path
from difflib import SequenceMatcher as seqmatch
from re import IGNORECASE, match
from AaronTools.const import AARONTOOLS
from numpy import argsort, loadtxt
warning = None
if program.lower() == "gaussian":
valid = loadtxt(
os.path.join(
AARONTOOLS,
"theory",
"valid_basis_sets",
"gaussian_ecp.txt",
),
dtype=str,
)
elif program.lower() == "orca":
valid = loadtxt(
os.path.join(
AARONTOOLS, "theory", "valid_basis_sets", "orca_ecp.txt"
),
dtype=str,
)
elif program.lower() == "qchem":
valid = loadtxt(
os.path.join(
AARONTOOLS, "theory", "valid_basis_sets", "qchem_ecp.txt"
),
dtype=str,
)
else:
raise NotImplementedError(
"cannot validate ECP names for %s" % program
)
if not any(
# need to escape () b/c they aren't capturing groups, it's ccsd(t) or something
match(
"%s$"
% (basis.replace("(", "\(").replace(")", "\)"))
.replace("*", "\*")
.replace("+", "\+"),
name,
flags=IGNORECASE,
)
for basis in valid
):
warning = (
"ECP '%s' may not be available in %s\n" % (name, program)
+ "if this is incorrect, please submit a bug report at https://github.com/QChASM/AaronTools.py/issues"
)
# try to suggest alternatives that have similar names
simm = [
seqmatch(
lambda x: x in "-_()/", name.upper(), test_basis.upper()
).ratio()
for test_basis in valid
]
ndx = argsort(simm)[-5:][::-1]
warning += "\npossible misspelling of:\n"
warning += "\n".join([valid[i] for i in ndx])
return warning
@addlogger
class BasisSet:
"""used to more easily get basis set info for writing input files"""
LOG = None
ORCA_AUX = ["C", "J", "JK", "CABS", "OptRI CABS"]
PSI4_AUX = [
"JK", "RI", "DF SCF", "DF SAPT", "DF GUESS", "DF SAD", "DF MP2",
"DF CC", "DF DCT", "DF MCSCF", "DF ELST"
]
QCHEM_AUX = ["RI", "J", "K", "corr"]
def __init__(self, basis=None, ecp=None):
"""
basis: list(Basis), Basis, str, or None
ecp: list(ECP) or None
"""
if isinstance(basis, str):
basis = self.parse_basis_str(basis, cls=Basis)
elif isinstance(basis, Basis):
basis = [basis]
elif isinstance(basis, BasisSet):
if ecp is None:
ecp = basis.ecp
basis = basis.basis
if isinstance(ecp, str):
if ecp.split():
ecp = self.parse_basis_str(ecp, cls=ECP)
else:
ecp = [ECP(ecp)]
elif isinstance(ecp, ECP):
ecp = [ecp]
elif isinstance(ecp, BasisSet):
ecp = ecp.ecp
self.basis = basis
self.ecp = ecp
@property
def elements_in_basis(self):
"""returns a list of elements in self's basis"""
elements = []
if self.basis is not None:
for basis in self.basis:
elements.extend(basis.elements)
return elements
@staticmethod
def parse_basis_str(basis_str, cls=Basis):
"""
parse basis set specification string and returns list(cls)
cls should be Basis or ECP (or subclasses of these)
basis info should have:
- a list of elements before basis set name (e.g. C H N O)
- other element keywords are 'tm' (transition metals) and 'all' (all elements)
- can also put "!" before an element to exclude it from the basis set
- auxilliary type before basis name (e.g. auxilliary C)
- basis set name
- path to basis set file right after basis set name if the basis is not builtin
- path cannot contain spaces
Example:
"!H !tm def2-SVPD /home/CoolUser/basis_sets/def2svpd.gbs H def2-SVP Ir SDD
"""
info = basis_str.split()
i = 0
basis_sets = []
elements = []
aux_type = None
user_defined = False
while i < len(info):
if info[i].lstrip("!") in ELEMENTS or any(
info[i].lower().lower().lstrip("!") == x for x in ["all", "tm"]
):
elements.append(info[i])
elif info[i].lower().startswith("aux"):
try:
aux_type = info[i + 1]
i += 1
if any(aux_type.lower() == x for x in ["df", "optri"]):
aux_type += " %s" % info[i + 1]
i += 1
except:
raise RuntimeError(
'error while parsing basis set string: %s\nfound "aux"'
+ ", but no auxilliary type followed" % basis_str
)
else:
basis_name = info[i]
try:
# TODO: allow spaces in paths
if (
# os thinks I have a file named "aux" somewhere on my computer
# I don't see it, but basis file names cannot start with 'aux'
os.path.exists(info[i + 1])
and not info[i + 1].lower().startswith("aux")
) or os.sep in info[i + 1]:
user_defined = info[i + 1]
i += 1
except IndexError:
pass
if not elements:
elements = None
basis_sets.append(
cls(
basis_name,
elements=elements,
aux_type=aux_type,
user_defined=user_defined,
)
)
elements = []
aux_type = None
user_defined = False
i += 1
return basis_sets
def __eq__(self, other):
if self.__class__ is not other.__class__:
return False
if self.basis and other.basis:
if len(self.basis) != len(other.basis):
return False
for b1, b2 in zip(sorted(self.basis), sorted(other.basis)):
if b1 != b2:
return False
else:
if self.basis != other.basis:
return False
if self.ecp and other.ecp:
if len(self.ecp) != len(other.ecp):
return False
for b1, b2 in zip(sorted(self.ecp), sorted(other.ecp)):
if b1 != b2:
return False
else:
if bool(self.ecp) != bool(other.ecp):
return False
return True
def copy(self):
new_dict = dict()
for key, value in self.__dict__.items():
try:
new_dict[key] = value.copy()
except AttributeError:
new_dict[key] = value
# ignore chimerax objects so seqcrow doesn't print a
# warning when a geometry is copied
if "chimerax" in value.__class__.__module__:
continue
if value.__class__.__module__ != "builtins":
self.LOG.warning(
"No copy method for {}: in-place changes may occur".format(
type(value)
)
)
return self.__class__(**new_dict)
def add_ecp(self, ecp):
"""add ecp to this BasisSet
ecp - ECP"""
if self.ecp is None:
self.ecp = []
self.ecp.append(ecp)
def add_basis(self, basis):
"""add basis to this BasisSet
basis - Basis"""
if self.basis is None:
self.basis = []
self.basis.append(basis)
def refresh_elements(self, geometry):
"""evaluate element specifications for each basis and ecp to
make them compatible with the supplied geometry"""
if self.basis is not None:
for basis in self.basis:
basis.refresh_elements(geometry)
if self.ecp is not None:
for ecp in self.ecp:
ecp.refresh_elements(geometry)
def get_gaussian_basis_info(self):
"""returns dict used by get_gaussian_header/footer with basis info"""
info = {}
warnings = []
if self.basis is not None:
# check if we need to use gen or genecp:
# -a basis set is user-defined (stored in an external file e.g. from the BSE)
# -multiple basis sets
# -an ecp
if (
all([basis == self.basis[0] for basis in self.basis])
and not self.basis[0].user_defined
and self.ecp is None
):
basis_name = Basis.get_gaussian(self.basis[0].name)
warning = self.basis[0].sanity_check_basis(
basis_name, "gaussian"
)
if warning:
warnings.append(warning)
info[GAUSSIAN_ROUTE] = "/%s" % basis_name
else:
if self.ecp is None or all(
not ecp.elements for ecp in self.ecp
):
info[GAUSSIAN_ROUTE] = "/gen"
else:
info[GAUSSIAN_ROUTE] = "/genecp"
out_str = ""
# gaussian flips out if you specify basis info for an element that
# isn't on the molecule, so make sure the basis set has an element
for basis in self.basis:
if basis.elements and not basis.user_defined:
out_str += " ".join([ele for ele in basis.elements])
out_str += " 0\n"
basis_name = Basis.get_gaussian(basis.name)
warning = basis.sanity_check_basis(
basis_name, "gaussian"
)
if warning:
warnings.append(warning)
out_str += basis_name
out_str += "\n****\n"
for basis in self.basis:
if basis.elements:
if basis.user_defined:
if os.path.exists(basis.user_defined):
with open(basis.user_defined, "r") as f:
lines = f.readlines()
i = 0
while i < len(lines):
test = lines[i].strip()
if not test or test.startswith("!"):
i += 1
continue
ele = test.split()[0]
while i < len(lines):
if ele in basis.elements:
out_str += lines[i]
if lines[i].startswith("****"):
break
i += 1
i += 1
# if the file does not exists, just insert the path as an @ file
else:
out_str += "@%s\n" % basis.user_defined
info[GAUSSIAN_GEN_BASIS] = out_str
if self.ecp is not None:
out_str = ""
for basis in self.ecp:
if basis.elements and not basis.user_defined:
out_str += " ".join([ele for ele in basis.elements])
out_str += " 0\n"
basis_name = Basis.get_gaussian(basis.name)
warning = basis.sanity_check_basis(basis_name, "gaussian")
if warning:
warnings.append(warning)
out_str += basis_name
out_str += "\n"
for basis in self.ecp:
if basis.elements:
if basis.user_defined:
if os.path.exists(basis.user_defined):
with open(basis.user_defined, "r") as f:
lines = f.readlines()
i = 0
while i < len(lines):
test = lines[i].strip()
if not test or test.startswith("!"):
i += 1
continue
ele = test.split()[0]
while i < len(lines):
if ele in basis.elements:
out_str += lines[i]
if lines[i].startswith("****"):
break
i += 1
i += 1
else:
out_str += "@%s\n" % basis.user_defined
info[GAUSSIAN_GEN_ECP] = out_str
if self.basis is None:
info[GAUSSIAN_ROUTE] = " Pseudo=Read"
return info, warnings
def get_orca_basis_info(self):
"""return dict for get_orca_header"""
# TODO: warn if basis should be f12
info = {ORCA_BLOCKS: {"basis": []}, ORCA_ROUTE: []}
warnings = []
first_basis = []
if self.basis is not None:
for basis in self.basis:
if basis.elements:
if basis.aux_type is None:
if basis.aux_type not in first_basis:
if not basis.user_defined:
basis_name = Basis.get_orca(basis.name)
warning = Basis.sanity_check_basis(
basis_name, "orca"
)
if warning:
warnings.append(warning)
out_str = basis_name
info[ORCA_ROUTE].append(out_str)
first_basis.append(basis.aux_type)
else:
out_str = 'GTOName "%s"' % basis.user_defined
info[ORCA_BLOCKS]["basis"].append(out_str)
first_basis.append(basis.aux_type)
else:
for ele in basis.elements:
out_str = "newGTO %-2s " % ele
if not basis.user_defined:
basis_name = Basis.get_orca(basis.name)
warning = Basis.sanity_check_basis(
basis_name, "orca"
)
if warning:
warnings.append(warning)
out_str += '"%s" end' % basis_name
else:
out_str += '"%s" end' % basis.user_defined
info[ORCA_BLOCKS]["basis"].append(out_str)
elif basis.aux_type.upper() == "C":
if basis.aux_type not in first_basis:
if not basis.user_defined:
basis_name = Basis.get_orca(basis.name) + "/C"
warning = Basis.sanity_check_basis(
basis_name, "orca"
)
if warning:
warnings.append(warning)
out_str = "%s" % basis_name
info[ORCA_ROUTE].append(out_str)
first_basis.append(basis.aux_type)
else:
out_str = (
'AuxCGTOName "%s"' % basis.user_defined
)
info[ORCA_BLOCKS]["basis"].append(out_str)
first_basis.append(basis.aux_type)
else:
for ele in basis.elements:
out_str = "newAuxCGTO %-2s " % ele
if not basis.user_defined:
basis_name = (
Basis.get_orca(basis.name) + "/C"
)
warning = Basis.sanity_check_basis(
basis_name, "orca"
)
if warning:
warnings.append(warning)
out_str += '"%s" end' % basis_name
else:
out_str += '"%s" end' % basis.user_defined
info[ORCA_BLOCKS]["basis"].append(out_str)
elif basis.aux_type.upper() == "J":
if basis.aux_type not in first_basis:
if not basis.user_defined:
basis_name = Basis.get_orca(basis.name) + "/J"
warning = Basis.sanity_check_basis(
basis_name, "orca"
)
if warning:
warnings.append(warning)
out_str = "%s" % basis_name
info[ORCA_ROUTE].append(out_str)
first_basis.append(basis.aux_type)
else:
out_str = (
'AuxJGTOName "%s"' % basis.user_defined
)
info[ORCA_BLOCKS]["basis"].append(out_str)
first_basis.append(basis.aux_type)
else:
for ele in basis.elements:
out_str = "newAuxJGTO %-2s " % ele
if not basis.user_defined:
basis_name = (
Basis.get_orca(basis.name) + "/J"
)
warning = Basis.sanity_check_basis(
basis_name, "orca"
)
if warning:
warnings.append(warning)
out_str += '"%s" end' % basis_name
else:
out_str += '"%s" end' % basis.user_defined
info[ORCA_BLOCKS]["basis"].append(out_str)
elif basis.aux_type.upper() == "JK":
if basis.aux_type not in first_basis:
if not basis.user_defined:
basis_name = Basis.get_orca(basis.name) + "/JK"
warning = Basis.sanity_check_basis(
basis_name, "orca"
)
if warning:
warnings.append(warning)
out_str = "%s" % basis_name
info[ORCA_ROUTE].append(out_str)
first_basis.append(basis.aux_type)
else:
out_str = (
'AuxJKGTOName "%s"' % basis.user_defined
)
info[ORCA_BLOCKS]["basis"].append(out_str)
first_basis.append(basis.aux_type)
else:
for ele in basis.elements:
out_str = "newAuxJKGTO %-2s " % ele
if not basis.user_defined:
basis_name = (
Basis.get_orca(basis.name) + "/JK"
)
warning = Basis.sanity_check_basis(
basis_name, "orca"
)
if warning:
warnings.append(warning)
out_str += '"%s" end' % basis_name
else:
out_str += '"%s" end' % basis.user_defined
info[ORCA_BLOCKS]["basis"].append(out_str)
elif basis.aux_type.upper() == "CABS":
if basis.aux_type not in first_basis:
if not basis.user_defined:
basis_name = (
Basis.get_orca(basis.name) + "-CABS"
)
warning = Basis.sanity_check_basis(
basis_name, "orca"
)
if warning:
warnings.append(warning)
out_str = "%s" % basis_name
info[ORCA_ROUTE].append(out_str)
first_basis.append(basis.aux_type)
else:
out_str = (
'CABSGTOName "%s"' % basis.user_defined
)
info[ORCA_BLOCKS]["basis"].append(out_str)
first_basis.append(basis.aux_type)
else:
for ele in basis.elements:
out_str = "newCABSGTO %-2s " % ele
if not basis.user_defined:
basis_name = (
Basis.get_orca(basis.name) + "-CABS"
)
warning = Basis.sanity_check_basis(
basis_name, "orca"
)
if warning:
warnings.append(warning)
out_str += '"%s" end' % basis_name
else:
out_str += '"%s" end' % basis.user_defined
info[ORCA_BLOCKS]["basis"].append(out_str)
elif basis.aux_type.upper() == "OPTRI CABS":
if basis.aux_type not in first_basis:
if not basis.user_defined:
basis_name = (
Basis.get_orca(basis.name) + "-OptRI"
)
warning = Basis.sanity_check_basis(
basis_name, "orca"
)
if warning:
warnings.append(warning)
out_str = "%s" % basis_name
info[ORCA_ROUTE].append(out_str)
first_basis.append(basis.aux_type)
else:
out_str = (
'CABSGTOName "%s"' % basis.user_defined
)
info[ORCA_BLOCKS]["basis"].append(out_str)
first_basis.append(basis.aux_type)
else:
for ele in basis.elements:
out_str = "newCABSGTO %-2s " % ele
if not basis.user_defined:
basis_name = (
Basis.get_orca(basis.name) + "-OptRI"
)
warning = Basis.sanity_check_basis(
basis_name, "orca"
)
if warning:
warnings.append(warning)
out_str += '"%s" end' % basis_name
else:
out_str += '"%s" end' % basis.user_defined
info[ORCA_BLOCKS]["basis"].append(out_str)
if self.ecp is not None:
for basis in self.ecp:
if basis.elements and not basis.user_defined:
for ele in basis.elements:
out_str = "newECP %-2s " % ele
basis_name = Basis.get_orca(basis.name)
warning = basis.sanity_check_basis(basis_name, "orca")
if warning:
warnings.append(warning)
out_str += '"%s" end' % basis_name
info[ORCA_BLOCKS]["basis"].append(out_str)
elif basis.elements and basis.user_defined:
# TODO: check if this works
out_str = 'GTOName "%s"' % basis.user_defined
info[ORCA_BLOCKS]["basis"].append(out_str)
return info, warnings
def get_psi4_basis_info(self, sapt=False):
"""
sapt: bool, use df_basis_sapt instead of df_basis_scf for jk basis
return dict for get_psi4_header
"""
out_str = dict()
warnings = []
if self.basis is not None:
for basis in self.basis:
if basis.user_defined:
continue
aux_type = basis.aux_type
basis_name = basis.get_psi4(basis.name)
# JK and RI will try to guess what basis set is being requested
# specifying "DF X" as the aux type will give more control
# but the user will have to request -ri or -jkfit explicitly
if isinstance(aux_type, str):
aux_type = aux_type.upper()
else:
aux_type = "basis"
if aux_type == "JK":
aux_type = "df_basis_scf"
basis_name += "-jkfit"
elif aux_type == "DF SCF":
aux_type = "df_basis_scf"
elif aux_type == "DF SAPT":
aux_type = "df_basis_sapt"
elif aux_type == "DF GUESS":
aux_type = "df_basis_guess"
elif aux_type == "DF SAD":
aux_type = "df_basis_sad"
elif aux_type == "DF MP2":
aux_type = "df_basis_mp2"
elif aux_type == "DF DCT":
aux_type = "df_basis_dct"
elif aux_type == "DF MCSCF":
aux_type = "df_basis_mcscf"
elif aux_type == "DF CC":
aux_type = "df_basis_cc"
elif aux_type == "DF ELST":
aux_type = "df_basis_elst"
elif aux_type == "RI":
aux_type = "df_basis_%s"
if sapt:
aux_type = "df_basis_sapt"
if basis_name.lower() in ["sto-3g", "3-21g"]:
basis_name += "-rifit"
else:
basis_name += "-ri"
warning = basis.sanity_check_basis(basis_name, "psi4")
if warning:
warnings.append(warning)
if aux_type not in out_str:
out_str[aux_type] = "%s {\n" % aux_type
out_str[aux_type] += " assign %s\n" % basis_name
else:
for ele in basis.elements:
out_str[aux_type] += " assign %-2s %s\n" % (
ele, basis_name
)
if any(basis.user_defined for basis in self.basis):
for basis in self.basis:
if basis.user_defined:
aux_type = basis.aux_type
if not aux_type:
aux_type = ""
aux_type = aux_type.upper()
if os.path.exists(basis.user_defined):
if aux_type not in out_str:
if not aux_type:
out_str[aux_type] = "basis {\n"
elif aux_type == "JK" and sapt:
out_str[aux_type] = "df_basis_sapt {\n"
elif aux_type == "JK":
out_str[aux_type] = "df_basis_scf {\n"
elif aux_type == "RI":
out_str[aux_type] = "df_basis_%s {\n"
out_str[aux_type] += "\n[%s]\n" % basis.name
with open(basis.user_defined, "r") as f:
lines = [
line.rstrip()
for line in f.readlines()
if line.strip()
and not line.startswith("!")
]
out_str[aux_type] += "\n".join(lines)
out_str[aux_type] += "\n\n"
s = "}\n\n".join(out_str.values())
s += "}"
info = {PSI4_BEFORE_GEOM: [s]}
return info, warnings
def check_for_elements(self, geometry, count_ecps=False):
"""checks to make sure each element is in a basis set"""
warning = ""
# assume all elements aren't in a basis set, remove from the list if they have a basis
# need to check each type of aux basis
elements = list(set([str(atom.element) for atom in geometry.atoms]))
if self.basis is not None:
elements_without_basis = {None: elements.copy()}
for basis in self.basis:
if basis.aux_type not in elements_without_basis:
elements_without_basis[basis.aux_type] = [
str(e) for e in elements
]
for element in basis.elements:
if element in elements_without_basis[basis.aux_type]:
elements_without_basis[basis.aux_type].remove(element)
if count_ecps and self.ecp:
for basis in self.basis:
if basis.aux_type != None and basis.aux_type != "no":
continue
for ecp in self.ecp:
for element in ecp.elements:
print("removing", element)
if element in elements_without_basis[basis.aux_type]:
elements_without_basis[basis.aux_type].remove(element)
if any(
elements_without_basis[aux]
for aux in elements_without_basis.keys()
):
for aux in elements_without_basis.keys():
if elements_without_basis[aux]:
if aux is not None and aux != "no":
warning += "%s ha%s no auxiliary %s basis; " % (
", ".join(elements_without_basis[aux]),
"s"
if len(elements_without_basis[aux]) == 1
else "ve",
aux,
)
else:
warning += "%s ha%s no basis; " % (
", ".join(elements_without_basis[aux]),
"s"
if len(elements_without_basis[aux]) == 1
else "ve",
)
return warning.strip("; ")
return None
def get_qchem_basis_info(self, geom):
"""returns dict used by get_qchem_header with basis info"""
info = {QCHEM_REM: dict(), QCHEM_SETTINGS: dict()}
warnings = []
if self.basis:
no_aux_basis = [basis for basis in self.basis if not basis.aux_type]
other_basis = [basis for basis in self.basis if basis not in no_aux_basis]
aux_j_basis = [basis for basis in other_basis if basis.aux_type.lower() == "j"]
aux_k_basis = [basis for basis in other_basis if basis.aux_type.lower() == "k"]
aux_corr_basis = [basis for basis in other_basis if basis.aux_type.lower() == "corr"]
aux_ri_basis = [basis for basis in other_basis if basis.aux_type.lower() == "ri"]
else:
no_aux_basis = []
aux_j_basis = []
aux_k_basis = []
aux_corr_basis = []
aux_ri_basis = []
for basis_list, label in zip(
[no_aux_basis, aux_j_basis, aux_k_basis, aux_corr_basis, aux_ri_basis],
["BASIS", "AUX_BASIS_J", "AUX_BASIS_K", "AUX_BASIS_CORR", "AUX_BASIS"],
):
if basis_list:
# check if we need to use gen or mixed:
# -a basis set is user-defined (stored in an external file e.g. from the BSE)
# -multiple basis sets
if (
all([basis == basis_list[0] for basis in basis_list])
and not basis_list[0].user_defined
):
basis_name = Basis.get_qchem(basis_list[0].name)
warning = basis_list[0].sanity_check_basis(
basis_name, "qchem"
)
if warning:
warnings.append(warning)
info[QCHEM_REM][label] = "%s" % basis_name
elif not any(basis.user_defined for basis in basis_list):
info[QCHEM_REM][label] = "General"
else:
info[QCHEM_REM][label] = "MIXED"
if any(x == info[QCHEM_REM][label] for x in ["MIXED", "General"]):
out_str = ""
for basis in basis_list:
if basis.elements and not basis.user_defined:
if info[QCHEM_REM][label] == "General":
for ele in basis.elements:
out_str += "%-2s 0\n " % ele
basis_name = Basis.get_qchem(basis.name)
warning = basis.sanity_check_basis(
basis_name, "qchem"
)
if warning:
warnings.append(warning)
out_str += basis_name
out_str += "\n ****\n "
else:
atoms = geom.find(ele)
for atom in atoms:
out_str += "%s %i\n " % (atom.element, geom.atoms.index(atom) + 1)
basis_name = Basis.get_qchem(basis.name)
warning = basis.sanity_check_basis(
basis_name, "qchem"
)
if warning:
warnings.append(warning)
out_str += basis_name
out_str += "\n ****\n "
for basis in basis_list:
if basis.elements and basis.user_defined:
if os.path.exists(basis.user_defined):
with open(basis.user_defined, "r") as f:
lines = f.readlines()
for element in basis.elements:
atoms = geom.find(element)
for atom in atoms:
i = 0
while i < len(lines):
test = lines[i].strip()
if not test or test.startswith("!") or test.startswith("$"):
i += 1
continue
ele = test.split()[0]
if ele == atom.element:
out_str += "%s %i\n" % (ele, geom.atoms.index(atom))
i += 1
while i < len(lines):
if ele == atom.element:
out_str += lines[i]
if lines[i].startswith("****"):
break
i += 1
i += 1
# if the file does not exists, just insert the path as an @ file
else:
warnings.append("file not found: %s" % basis.user_defined)
info[QCHEM_SETTINGS][label.lower()] = [out_str.strip()]
if self.ecp is not None and any(ecp.elements for ecp in self.ecp):
# check if we need to use gen:
# -a basis set is user-defined (stored in an external file e.g. from the BSE)
if (
all([ecp == self.ecp[0] for ecp in self.ecp])
and not self.ecp[0].user_defined
and not self.basis
):
basis_name = ECP.get_qchem(self.ecp[0].name)
warning = self.ecp[0].sanity_check_basis(
basis_name, "qchem"
)
if warning:
warnings.append(warning)
if QCHEM_REM not in info:
info[QCHEM_REM] = {"ECP": "%s" % basis_name}
else:
info[QCHEM_REM]["ECP"] = "%s" % basis_name
elif not any(basis.user_defined for basis in self.basis):
if QCHEM_REM not in info:
info[QCHEM_REM] = {"ECP": "General"}
else:
info[QCHEM_REM]["ECP"] = "General"
else:
if QCHEM_REM not in info:
info[QCHEM_REM] = {"ECP": "MIXED"}
else:
info[QCHEM_REM]["ECP"] = "MIXED"
if any(x == info[QCHEM_REM]["ECP"] for x in ["MIXED", "General"]):
out_str = ""
for basis in self.ecp:
if basis.elements and not basis.user_defined:
if info[QCHEM_REM]["ECP"] == "General":
for ele in basis.elements:
out_str += "%-2s 0\n " % ele
basis_name = ECP.get_qchem(basis.name)
warning = basis.sanity_check_basis(
basis_name, "qchem"
)
if warning:
warnings.append(warning)
out_str += basis_name
out_str += "\n ****\n "
else:
atoms = geom.find(element)
for atom in atoms:
out_str += "%s %i\n " % (atom.element, geom.atoms.index(atom) + 1)
basis_name = ECP.get_qchem(basis.name)
warning = basis.sanity_check_basis(
basis_name, "qchem"
)
if warning:
warnings.append(warning)
out_str += basis_name
out_str += "\n ****\n "
for ecp in self.ecp:
if ecp.elements:
if ecp.user_defined:
if os.path.exists(ecp.user_defined):
with open(ecp.user_defined, "r") as f:
lines = f.readlines()
for element in ecp.elements:
atoms = geom.find(element)
for atom in atoms:
i = 0
while i < len(lines):
test = lines[i].strip()
if not test or test.startswith("!") or test.startswith("$"):
i += 1
continue
ele = test.split()[0]
if ele == atom.element:
out_str += "%s %i\n" % (ele, geom.atoms.index(atom))
i += 1
while i < len(lines):
if ele == atom.element:
out_str += lines[i]
if lines[i].startswith("****"):
break
i += 1
i += 1
# if the file does not exists, just insert the path as an @ file
else:
warnings.append("file not found: %s" % ecp.user_defined)
if QCHEM_SETTINGS not in info:
info[QCHEM_SETTINGS] = {"ecp": [out_str.strip()]}
else:
info[QCHEM_SETTINGS]["ecp"] = [out_str.strip()]
return info, warnings | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/theory/basis.py | basis.py |
import re
from AaronTools import addlogger
KNOWN_SEMI_EMPIRICAL = [
"AM1",
"PM3",
"PM6",
"PM7",
"HF-3C",
"PM3MM",
"PDDG",
"RM1",
"MNDO",
"PM3-PDDG",
"MNDO-PDDG",
"PM3-CARB1",
"MNDO/d",
"AM1/d",
"DFTB2",
"DFTB3",
"AM1-D*",
"PM6-D",
"AM1-DH+",
"PM6-DH+",
]
class Method:
"""functional object
used to ensure the proper keyword is used
e.g.
using Functional('PBE0') will use PBE1PBE in a gaussian input file"""
LOG = None
def __init__(self, name, is_semiempirical=False):
"""
name: str, functional name
is_semiempirical: bool, basis set is not required
"""
self.name = name
self.is_semiempirical = is_semiempirical
def __eq__(self, other):
if self.__class__ is not other.__class__:
return False
return (
self.get_gaussian()[0].lower() == other.get_gaussian()[0].lower() and
self.is_semiempirical == other.is_semiempirical
)
@staticmethod
def sanity_check_method(name, program):
"""
check to see if method is available in the specified program
name - str, name of method
program, str, gaussian, orca, psi4, or qchem
"""
import os.path
from difflib import SequenceMatcher as seqmatch
from numpy import argsort, loadtxt
from AaronTools.const import AARONTOOLS
warning = None
prefix = ""
if program.lower() == "gaussian":
valid = loadtxt(os.path.join(AARONTOOLS, "theory", "valid_methods", "gaussian.txt"), dtype=str)
prefix = "(?:RO|R|U)?"
elif program.lower() == "orca":
valid = loadtxt(os.path.join(AARONTOOLS, "theory", "valid_methods", "orca.txt"), dtype=str)
elif program.lower() == "psi4":
valid = loadtxt(os.path.join(AARONTOOLS, "theory", "valid_methods", "psi4.txt"), dtype=str)
elif program.lower() == "sqm":
valid = loadtxt(os.path.join(AARONTOOLS, "theory", "valid_methods", "sqm.txt"), dtype=str)
elif program.lower() == "qchem":
valid = loadtxt(os.path.join(AARONTOOLS, "theory", "valid_methods", "qchem.txt"), dtype=str)
else:
raise NotImplementedError("cannot validate method names for %s" % program)
if not any(
# need to escape () b/c they aren't capturing groups, it's ccsd(t) or something
re.match(
"%s%s$" % (prefix, method.replace("(", "\(").replace(")", "\)").replace("+", "\+")), name, flags=re.IGNORECASE
) for method in valid
):
warning = "method '%s' may not be available in %s\n" % (name, program) + \
"if this is incorrect, please submit a bug report at https://github.com/QChASM/AaronTools.py/issues"
# try to suggest alternatives that have similar names
simm = [
seqmatch(
lambda x: x in "-_()/", name.upper(), test_method.upper()
).ratio() for test_method in valid
]
ndx = argsort(simm)[-5:][::-1]
warning += "\npossible misspelling of:\n"
warning += "\n".join([valid[i] for i in ndx])
return warning
def copy(self):
new_dict = dict()
for key, value in self.__dict__.items():
try:
new_dict[key] = value.copy()
except AttributeError:
new_dict[key] = value
# ignore chimerax objects so seqcrow doesn't print a
# warning when a geometry is copied
if "chimerax" in value.__class__.__module__:
continue
if value.__class__.__module__ != "builtins":
self.LOG.warning(
"No copy method for {}: in-place changes may occur".format(
type(value)
)
)
return self.__class__(**new_dict)
def get_gaussian(self):
"""maps proper functional name to one Gaussian accepts"""
warning = None
if self.name.lower() == "ωb97x-d" or self.name.lower() == "wb97x-d":
return ("wB97XD", None)
elif self.name == "Gaussian's B3LYP":
return ("B3LYP", None)
elif self.name.lower() == "b97-d":
return ("B97D", None)
elif self.name.lower().startswith("m06-"):
return (self.name.upper().replace("M06-", "M06", 1), None)
elif self.name.upper() == "PBE0":
return ("PBE1PBE", None)
#methods available in ORCA but not Gaussian
elif self.name.lower() == "ωb97x-d3":
return ("wB97XD", "ωB97X-D3 is not available in Gaussian, switching to ωB97X-D2")
elif self.name.lower() == "b3lyp":
return ("B3LYP", None)
name = self.name.replace('ω', 'w')
return name, warning
def get_orca(self):
"""maps proper functional name to one ORCA accepts"""
warning = None
if (
self.name == "ωB97X-D" or
any(
test == self.name.lower() for test in ["wb97xd", "wb97x-d"]
)
):
return ("wB97X-D3", "ωB97X-D may refer to ωB97X-D2 or ωB97X-D3 - using the latter")
elif self.name == "ωB97X-D3":
return ("wB97X-D3", None)
elif any(self.name.upper() == name for name in ["B97-D", "B97D"]):
return ("B97-D", None)
elif self.name == "Gaussian's B3LYP":
return ("B3LYP/G", None)
elif self.name.upper() == "M06-L":
return ("M06L", None)
elif self.name.upper() == "M06-2X":
return ("M062X", None)
elif self.name.upper() == "PBE1PBE":
return ("PBE0", None)
name = self.name.replace('ω', 'w')
return name, warning
def get_psi4(self):
"""maps proper functional name to one Psi4 accepts"""
if self.name.lower() == 'wb97xd':
return "wB97X-D", None
elif self.name.upper() == 'B97D':
return ("B97-D", None)
elif self.name.upper() == "PBE1PBE":
return ("PBE0", None)
elif self.name.upper() == "M062X":
return ("M06-2X", None)
elif self.name.upper() == "M06L":
return ("M06-L", None)
# the functionals havent been combined with dispersion yet, so
# we aren't checking if the method is available
return self.name.replace('ω', 'w'), None
def get_sqm(self):
"""get method name that is appropriate for sqm"""
return self.name
def get_qchem(self):
"""maps proper functional name to one Psi4 accepts"""
if re.match("[wω]b97[xm]?[^-xm]", self.name.lower()) and self.name.lower() != "wb97m(2)":
name = re.match("([wω]?)b97([xm]?)([\S]+)", self.name.lower())
return ("%sB97%s%s" % (
name.group(1) if name.group(1) else "",
name.group(2).upper() if name.group(2) else "",
"-%s" % name.group(3).upper() if name.group(3) else "",
),
None
)
elif self.name.upper() == 'B97D':
return ("B97-D", None)
elif self.name.upper() == "M062X":
return ("M06-2X", None)
elif self.name.upper() == "M06L":
return ("M06-L", None)
# the functionals havent been combined with dispersion yet, so
# we aren't checking if the method is available
return self.name.replace('ω', 'w'), None
class SAPTMethod(Method):
"""
method used to differentiate between regular methods and sapt
methods because the molecule will need to be split into monomers
if using a sapt method, the geometry given to Theory or Geometry.write
should have a 'components' attribute with each monomer being a coordinate
the charge and multiplicity given to Theory should be a list, with the first
item in each list being the overall charge/multiplicity and the subsequent items
being the charge/multiplicity of the monomers (components)
"""
def __init__(self, *args, **kwargs):
super().__init__(*args, **kwargs) | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/theory/method.py | method.py |
import time
import unittest
from os.path import dirname
import numpy as np
from AaronTools.geometry import Geometry
prefix = dirname(__file__)
def rmsd_tol(geom, superTight=False, superLoose=False):
"""
Automatically determine a reasonable rmsd tolerance for the input
geometry based on its size and number of atoms
"""
tolerance = len(geom.atoms) ** (
2 - int(superTight) + int(superLoose)
) * np.sqrt(np.finfo(float).eps)
com = geom.COM()
max_d = None
for atom in geom.atoms:
d = np.linalg.norm(atom.coords - com)
if max_d is None or d > max_d:
max_d = d
tolerance *= max_d * (2 - int(superTight) + int(superLoose))
tolerance = tolerance ** (2 / (4 - int(superTight) + int(superLoose)))
return tolerance
def check_atom_list(ref, comp):
rv = True
for i, j in zip(ref, comp):
rv &= i.__repr__() == j.__repr__()
return rv
def validate(test, ref, thresh=None, heavy_only=False, sort=True, debug=False):
"""
Validates `test` geometry against `ref` geometry
Returns: True if validation passed, False if failed
:test: the geometry to validate
:ref: the reference geometry
:thresh: the RMSD threshold
if thresh is a number: use that as threshold
if thresh is None: use rmsd_tol() to determine
if thresh is "tight": use rmsd_tol(superTight=True)
if thresh is "loose": use rmsd_tol(superLoose=True)
:sort: allow canonical sorting of atoms
:debug: print info useful for debugging
"""
if debug:
print("before alignment")
print(ref.write("ref", outfile=False))
print(test.write("test", outfile=False))
if thresh is None:
thresh = rmsd_tol(ref)
try:
thresh = float(thresh)
except ValueError:
if thresh.lower() == "tight":
thresh = rmsd_tol(ref, superTight=True)
elif thresh.lower() == "loose":
thresh = rmsd_tol(ref, superLoose=True)
else:
raise ValueError("Bad threshold provided")
# elements should all be the same
t_el = sorted([t.element for t in test.atoms])
r_el = sorted([r.element for r in ref.atoms])
if len(t_el) != len(r_el):
if debug:
print(
"wrong number of atoms: {} (test) vs. {} (ref)".format(
len(t_el), len(r_el)
)
)
return False
for t, r in zip(t_el, r_el):
if t != r:
if debug:
print("elements don't match")
return False
# and RMSD should be below a threshold
rmsd = test.RMSD(
ref, align=debug, heavy_only=heavy_only, sort=sort, debug=debug
)
if debug:
print("after alignment")
print(ref.write("ref", outfile=False))
print(test.write("test", outfile=False))
if debug:
print("RMSD:", rmsd[2], "\tTHRESH:", thresh)
rmsd[0].write("ref")
rmsd[1].write("test")
rmsd = rmsd[2]
return rmsd < thresh
class TestWithTimer(unittest.TestCase):
test_count = 0
total_time = 0
this_class = None
last_class = None
last_result = None
errors = 0
fails = 0
last_errors = 0
last_fails = 0
@classmethod
def setUpClass(cls):
TestWithTimer.total_time = time.time()
@classmethod
def tearDownClass(cls):
TestWithTimer.total_time = time.time() - TestWithTimer.total_time
print(TestWithTimer.get_status())
if TestWithTimer.errors - TestWithTimer.last_errors:
status = "ERROR"
elif TestWithTimer.fails - TestWithTimer.last_fails:
status = "FAIL"
else:
status = "ok"
TestWithTimer.last_errors = TestWithTimer.errors
TestWithTimer.last_fails = TestWithTimer.fails
print(
"Ran %d test in %.4fs %s"
% (
TestWithTimer.last_result.testsRun - TestWithTimer.test_count,
TestWithTimer.total_time,
status,
)
)
TestWithTimer.test_count = TestWithTimer.last_result.testsRun
print(unittest.TextTestResult.separator2)
@classmethod
def get_status(cls):
if TestWithTimer.errors != len(TestWithTimer.last_result.errors):
status = "ERROR"
elif TestWithTimer.fails != len(TestWithTimer.last_result.failures):
status = "FAIL"
else:
status = "ok"
TestWithTimer.errors = len(TestWithTimer.last_result.errors)
TestWithTimer.fails = len(TestWithTimer.last_result.failures)
return status
def setUp(self):
self.start_time = time.time()
def tearDown(self):
t = time.time() - self.start_time
TestWithTimer.last_result = self._outcome.result
TestWithTimer.this_class, self.test_name = self.id().split(".")[-2:]
status = TestWithTimer.get_status()
if TestWithTimer.this_class != TestWithTimer.last_class:
TestWithTimer.last_class = TestWithTimer.this_class
print(TestWithTimer.this_class)
else:
print(status)
print(
"\b %2d. %-30s %.4fs "
% (
TestWithTimer.last_result.testsRun - TestWithTimer.test_count,
self.test_name,
t,
),
end="",
) | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/test/__init__.py | __init__.py |
import sys
import argparse
import numpy as np
from AaronTools.fileIO import FileReader, read_types
from AaronTools.geometry import Geometry
from AaronTools.symmetry import (
PointGroup,
InversionCenter,
ProperRotation,
ImproperRotation,
MirrorPlane,
)
from AaronTools.utils.utils import perp_vector, glob_files
pg_parser = argparse.ArgumentParser(
description="print point group",
formatter_class=argparse.RawTextHelpFormatter
)
pg_parser.add_argument(
"infile", metavar="input file",
type=str,
nargs="*",
default=[sys.stdin],
help="a coordinate file"
)
pg_parser.add_argument(
"-o", "--output",
type=str,
default=False,
required=False,
dest="outfile",
help="output destination \nDefault: stdout"
)
pg_parser.add_argument(
"-if", "--input-format",
type=str,
default=None,
dest="input_format",
choices=read_types,
help="file format of input - xyz is assumed if input is stdin"
)
pg_parser.add_argument(
"-t", "--tolerance",
default=0.1,
type=float,
dest="tolerance",
help="tolerance for determining if a symmetry element is valid\n"
"for the input structure(s)\nDefault: 0.1"
)
pg_parser.add_argument(
"-a", "--axis-tolerance",
default=0.01,
type=float,
dest="rotation_tolerance",
help="tolerance for determining if two axes are coincident or orthogonal"
"\nDefault: 0.01"
)
pg_parser.add_argument(
"-n", "--max-n",
default=6,
type=int,
dest="max_n",
help="max. order for proper rotation axes (improper rotations can be 2x this)"
"\nDefault: 6"
)
pg_parser.add_argument(
"-v", "--verbose",
action="store_true",
default=False,
dest="print_eles",
help="print all symmetry elements",
)
pg_parser.add_argument(
"-b", "--bild",
action="store_true",
default=False,
dest="print_bild",
help="print Chimera(X) bild file to display various symmetry elements",
)
args = pg_parser.parse_args()
s = ""
for f in glob_files(args.infile, parser=pg_parser):
if isinstance(f, str):
if args.input_format is not None:
infile = FileReader((f, args.input_format, None), just_geom=True)
else:
infile = FileReader(f, just_geom=True)
else:
if args.input_format is not None:
infile = FileReader(("from stdin", args.input_format[0], f), just_geom=True)
else:
if len(sys.argv) >= 1:
infile = FileReader(("from stdin", "xyz", f), just_geom=True)
geom = Geometry(infile)
pg = PointGroup(
geom,
tolerance=args.tolerance,
rotation_tolerance=args.rotation_tolerance,
max_rotation=args.max_n,
)
if args.print_bild:
inv = ""
mirror = ""
prots = ""
irots = ""
for ele in sorted(pg.elements, reverse=True):
if isinstance(ele, InversionCenter):
inv += ".note %s\n" % repr(ele)
inv += ".color plum\n"
inv += ".sphere %.5f %.5f %.5f 0.3\n" % tuple(pg.center)
elif isinstance(ele, ProperRotation):
prots += ".note %s\n" % repr(ele)
prots += ".color red\n"
prots += ".arrow %.5f %.5f %.5f " % tuple(pg.center)
end = pg.center + ele.n * np.sqrt(ele.exp) * ele.axis
prots += "%.5f %.5f %.5f 0.05\n" % tuple(end)
elif isinstance(ele, ImproperRotation):
irots += ".note %s\n" % repr(ele)
irots += ".color blue\n"
irots += ".arrow %.5f %.5f %.5f " % tuple(pg.center)
end = pg.center + np.sqrt(ele.n) * np.sqrt(ele.exp) * ele.axis
irots += "%.5f %.5f %.5f 0.05\n" % tuple(end)
irots += ".transparency 25\n"
z = ele.axis
x = perp_vector(z)
y = np.cross(x, z)
for angle in np.linspace(0, 2 * np.pi, num=25):
pt2 = ele.n ** 0.9 * x * np.cos(angle)
pt2 += ele.n ** 0.9 * y * np.sin(angle)
pt2 += pg.center
if angle > 0:
irots += ".polygon %6.3f %6.3f %6.3f" % tuple(pt1)
irots += " %6.3f %6.3f %6.3f" % tuple(pg.center)
irots += " %6.3f %6.3f %6.3f" % tuple(pt2)
irots += "\n"
pt1 = pt2
elif isinstance(ele, MirrorPlane):
mirror += ".note %s\n" % repr(ele)
if ele.label is None:
mirror += ".color purple\n"
elif ele.label == "h":
mirror += ".color black\n"
elif ele.label == "v":
mirror += ".color chocolate\n"
elif ele.label == "d":
mirror += ".color teal\n"
mirror += ".transparency 40\n"
z = ele.axis
x = perp_vector(z)
y = np.cross(x, z)
for angle in np.linspace(0, 2 * np.pi, num=25):
pt2 = 5 * x * np.cos(angle)
pt2 += 5 * y * np.sin(angle)
pt2 += pg.center
if angle > 0:
mirror += ".polygon %6.3f %6.3f %6.3f" % tuple(pt1)
mirror += " %6.3f %6.3f %6.3f" % tuple(pg.center)
mirror += " %6.3f %6.3f %6.3f" % tuple(pt2)
mirror += "\n"
pt1 = pt2
if args.outfile:
with open(args.outfile, "w") as f:
f.write("\n".join([inv, prots, irots, mirror]))
else:
if inv:
print(inv)
if prots:
print(prots)
if irots:
print(irots)
if mirror:
print(mirror)
else:
s += "%s: %s\n" % (f, pg.name)
if args.print_eles:
for ele in sorted(pg.elements, reverse=True):
s += "\t%s\n" % repr(ele)
if not args.print_bild:
if args.outfile:
with open(args.outfile, "w") as f:
f.write(s)
else:
print(s) | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/bin/pointGroup.py | pointGroup.py |
import sys
import argparse
from AaronTools.geometry import Geometry
from AaronTools.fileIO import FileReader, read_types
from AaronTools.utils.utils import get_filename, glob_files
cube_parser = argparse.ArgumentParser(
description="print a cube file for a molecular orbital",
formatter_class=argparse.RawTextHelpFormatter
)
cube_parser.add_argument(
"infile", metavar="input file",
type=str,
nargs="*",
default=[sys.stdin],
help="an FCHK file, ORCA output with MO's, or NBO files"
)
cube_parser.add_argument(
"-o", "--output",
type=str,
default=False,
dest="outfile",
help="output destination\n" +
"$INFILE will be replaced with the name of the input file\n" +
"Default: stdout"
)
info = cube_parser.add_mutually_exclusive_group(required=False)
info.add_argument(
"-mo", "--molecular-orbital",
dest="mo_ndx",
default="homo",
help="index of molecular orbital to print (0-indexed)\n"
"can also give 'homo' or 'lumo' for highest occupied or\n"
"lowest unoccupied molecular orbital\n"
"Default: highest occupied MO in the ground state"
)
info.add_argument(
"-ao", "--atomic-orbital",
dest="ao_ndx",
default=None,
help="index of atomic orbital to print (0-indexed)"
)
info.add_argument(
"-ed", "--electron-density",
dest="density",
default=False,
action="store_true",
help="print electron density"
)
info.add_argument(
"-fd", "--fukui-donor",
dest="fukui_donor",
default=False,
action="store_true",
help="print Fukui donor values\n"
"see DOI 10.1002/jcc.24699 for weighting method"
)
info.add_argument(
"-fa", "--fukui-acceptor",
dest="fukui_acceptor",
default=False,
action="store_true",
help="print Fukui acceptor values\n"
"see DOI 10.1021/acs.jpca.9b07516 for weighting method"
)
info.add_argument(
"-f2", "--fukui-dual",
dest="fukui_dual",
default=False,
action="store_true",
help="print Fukui dual values\n"
"see DOI 10.1021/acs.jpca.9b07516 for weighting method"
)
cube_parser.add_argument(
"-d", "--delta",
type=float,
dest="delta",
default=0.1,
help="delta parameter for weighting orbitals in Fukui functions\n"
"Default: 0.1 Hartree",
)
cube_parser.add_argument(
"-s", "--spacing",
type=float,
dest="spacing",
default=0.2,
help="spacing between points in the cube file\n"
"Default: 0.2",
)
cube_parser.add_argument(
"-p", "--padding",
type=float,
dest="padding",
default=4,
help="extra space around the molecule\n"
"Default: 4"
)
cube_parser.add_argument(
"-xyz", "--standard-axes",
action="store_true",
dest="xyz",
default=False,
help="use x, y, and z axes to define the directions\n"
"Default: determine directions using SVD"
)
cube_parser.add_argument(
"-nt", "--number-of-threads",
type=int,
default=1,
dest="n_jobs",
help="number of threads to use when evaluating basis functions"
"this is on top of NumPy's multithreading,\n"
"so if NumPy uses 8 threads and n_jobs=2, you can\n"
"expect to see 16 threads in use\n"
"Default: 1"
)
cube_parser.add_argument(
"-nbo", "--nbo-file",
type=str,
default=None,
dest="nbo_name",
help="file containing coefficients for NBO's (e.g. *.37 file)"
"ignored unless input file is a *.47 file"
)
cube_parser.add_argument(
"-m", "--max-array",
type=int,
default=10000000,
dest="max_length",
help="max. array size to read from FCHK files\n"
"a reasonable size for setting parsing orbital data\n"
"can improve performance when reading large FCHK files\n"
"too small of a value will prevent orbital data from\n"
"being parsed\n"
"Default: 10000000",
)
args = cube_parser.parse_args()
kind = args.mo_ndx
if args.density:
kind = "density"
elif args.fukui_donor:
kind = "fukui donor"
elif args.fukui_acceptor:
kind = "fukui acceptor"
elif args.fukui_dual:
kind = "fukui dual"
elif args.ao_ndx:
kind = "AO %s" % args.ao_ndx
elif args.mo_ndx.isdigit():
kind = "MO %s" % args.mo_ndx
for f in glob_files(args.infile, parser=cube_parser):
if isinstance(f, str):
infile = FileReader(
f, just_geom=False, nbo_name=args.nbo_name, max_length=args.max_length
)
elif len(sys.argv) >= 1:
infile = FileReader(
("from stdin", "fchk", f),
just_geom=False,
nbo_name=args.nbo_name,
max_length=args.max_length,
)
geom = Geometry(infile, refresh_connected=False, refresh_ranks=False)
out = geom.write(
outfile=False,
orbitals=infile.other["orbitals"],
padding=args.padding,
kind=kind,
spacing=args.spacing,
style="cube",
xyz=args.xyz,
delta=args.delta,
n_jobs=args.n_jobs,
)
if not args.outfile:
print(out)
else:
outfile = args.outfile
if "$INFILE" in outfile:
outfile = outfile.replace("$INFILE", get_filename(f))
with open(outfile, "w") as f:
f.write(out) | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/bin/printCube.py | printCube.py |
import argparse
from sys import stdin
from AaronTools.fileIO import FileReader, read_types
from AaronTools.geometry import Geometry
from AaronTools.ring import Ring
from AaronTools.utils.utils import get_filename, glob_files
ring_parser = argparse.ArgumentParser(
description="close rings on a geometry",
formatter_class=argparse.RawTextHelpFormatter
)
ring_parser.add_argument(
"infile",
metavar="input file",
type=str,
nargs="*",
default=[stdin],
help="a coordinate file"
)
ring_parser.add_argument(
"-o", "--output",
type=str,
default=False,
required=False,
metavar="output destination",
dest="outfile",
help="output destination\n" +
"$INFILE will be replaced with the name of the input file\n" +
"Default: stdout"
)
ring_parser.add_argument(
"-ls", "--list",
action="store_const",
const=True,
default=False,
required=False,
dest="list_avail",
help="list available rings"
)
ring_parser.add_argument(
"-if", "--input-format",
type=str,
nargs=1,
default=None,
choices=read_types,
dest="input_format",
help="file format of input - xyz is assumed if input is stdin"
)
ring_parser.add_argument(
"-r", "--ring",
metavar=("atom1", "atom2", "ring"),
type=str,
nargs=3,
action="append",
default=None,
required=False,
dest="substitutions",
help="substitution instructions \n" +
"atom1 and atom2 specify the position to add the new ring"
)
ring_parser.add_argument(
"-m", "--minimize",
action="store_const",
const=True,
default=False,
required=False,
dest="minimize",
help="try to minimize structure difference"
)
ring_parser.add_argument(
"-f", "--flip-rings",
action="store_const",
const=True,
default=False,
required=False,
dest="flip",
help="also try swapping target order when minimizing"
)
args = ring_parser.parse_args()
if args.list_avail:
s = ""
for i, name in enumerate(sorted(Ring.list())):
s += "%-20s" % name
# if (i + 1) % 3 == 0:
if (i + 1) % 1 == 0:
s += "\n"
print(s.strip())
exit(0)
for infile in glob_files(args.infile, parser=ring_parser):
if isinstance(infile, str):
if args.input_format is not None:
f = FileReader((infile, args.input_format, infile))
else:
f = FileReader(infile)
else:
if args.input_format is not None:
f = FileReader(("from stdin", args.input_format, stdin))
else:
f = FileReader(("from stdin", "xyz", stdin))
geom = Geometry(f)
targets = {}
for sub_info in args.substitutions:
ring = sub_info[2]
ring_geom = Ring(ring)
key = ",".join(sub_info[:2])
if key in targets:
targets[key].append(ring_geom)
else:
targets[key] = [ring_geom]
for key in targets:
for ring_geom in targets[key]:
geom.ring_substitute(
key,
ring_geom,
minimize=args.minimize,
flip_walk=args.flip,
)
if args.outfile:
outfile = args.outfile
if "$INFILE" in outfile:
outfile = outfile.replace("$INFILE", get_filename(infile))
geom.write(append=True, outfile=outfile)
else:
print(geom.write(outfile=False)) | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/bin/closeRing.py | closeRing.py |
import sys
import argparse
import copy
import numpy as np
import matplotlib.pyplot as plt
from AaronTools.geometry import Geometry
from AaronTools.fileIO import FileReader, read_types
from AaronTools.utils.utils import rotation_matrix, get_filename, glob_files
steric_parser = argparse.ArgumentParser(
description="create a steric map for a ligand",
formatter_class=argparse.RawTextHelpFormatter
)
steric_parser.add_argument(
"infile",
metavar="input file",
type=str,
nargs="*",
default=[sys.stdin],
help="a coordinate file",
)
steric_parser.add_argument(
"-o",
"--output",
type=str,
default=False,
required=False,
dest="outfile",
help="output destination\n" +
"$INFILE will be replaced with the name of the input file\n" +
"Default: show plot",
)
steric_parser.add_argument(
"-if",
"--input-format",
type=str,
default=None,
dest="input_format",
choices=read_types,
help="file format of input - xyz is assumed if input is stdin"
)
steric_parser.add_argument(
"-k",
"--key-atoms",
default=None,
required=False,
dest="key",
help="atoms coordinated to the center\n" +
"these atoms\" ligands will be shown on the map",
)
steric_parser.add_argument(
"-c",
"--center",
action="append",
default=None,
required=False,
dest="center",
help="atom the sphere is centered on\n" +
"Default: detect metal center (centroid of all metals if multiple are present)",
)
steric_parser.add_argument(
"-v",
"--vdw-radii",
default="umn",
choices=["umn", "bondi"],
dest="radii",
help="VDW radii to use in calculation\n" +
"umn: main group vdw radii from J. Phys. Chem. A 2009, 113, 19, 5806–5812\n" +
" (DOI: 10.1021/jp8111556)\n" +
" transition metals are crystal radii from Batsanov, S.S. Van der Waals\n" +
" Radii of Elements. Inorganic Materials 37, 871–885 (2001).\n" +
" (DOI: 10.1023/A:1011625728803)\n" +
"bondi: radii from J. Phys. Chem. 1964, 68, 3, 441–451 (DOI: 10.1021/j100785a001)\n" +
"Default: umn",
)
steric_parser.add_argument(
"-r",
"--radius",
default=3.5,
type=float,
dest="radius",
help="radius around center\nDefault: 3.5 Ångström"
)
steric_parser.add_argument(
"-oop",
"--out-of-plane",
default=None,
nargs=3,
type=float,
metavar=("x_oop", "y_oop", "z_oop"),
dest="oop_vector",
help="list of three numbers defining a vector perpendicular to\n" +
"the desired steric map",
)
steric_parser.add_argument(
"-ip",
"--in-plane",
default=None,
nargs=3,
type=float,
metavar=("x_ip", "y_ip", "z_ip"),
dest="ip_vector",
help="list of three numbers defining the \"y axis\" of the steric map",
)
steric_parser.add_argument(
"-n", "--number-of-points",
default=100,
type=int,
dest="num_pts",
help="number of points along x and y axes\nDefault: 100",
)
steric_parser.add_argument(
"-amin", "--altitude-minimum",
default=None,
type=float,
dest="min",
help="manually set the lower cutoff of the altitude map",
)
steric_parser.add_argument(
"-amax",
"--altitude-maximum",
default=None,
type=float,
dest="max",
help="manually set the upper cutoff of the altitude map",
)
steric_parser.add_argument(
"-p", "--projection-shape",
choices=("circle", "square"),
default="circle",
dest="shape",
help="shape of steric map\n" +
"note that buried volume values with the square shape are still based\n" +
"on a sphere around the center\n" +
"Default: circle",
)
vbur_options = steric_parser.add_argument_group("Buried volume options")
vbur_options.add_argument(
"-vbur", "--buried-volume",
nargs="?",
default=False,
choices=("Lebedev", "MC"),
# this allows these choices to be case-insensitive, but we can still
# show normal upper- and lowercase in the help page
type=lambda x: x.capitalize() if x.lower() == "lebedev" else x.upper(),
dest="vbur",
help="show buried volume in each quadrant using the specified method\n" +
"Default: do not include %%Vbur",
)
vbur_options.add_argument(
"-rp", "--radial-points",
type=int,
default=20,
choices=[20, 32, 64, 75, 99, 127],
dest="rpoints",
help="number of radial shells for Lebedev integration\n" +
"lower values are faster, but at the cost of accuracy\n" +
"Default: 20"
)
vbur_options.add_argument(
"-ap", "--angular-points",
type=int,
default=1454,
choices=[110, 194, 302, 590, 974, 1454, 2030, 2702, 5810],
dest="apoints",
help="number of angular points for Lebedev integration\n" +
"lower values are faster, but at the cost of accuracy\n" +
"Default: 1454"
)
vbur_options.add_argument(
"-i",
"--minimum-iterations",
type=int,
default=25,
metavar="ITERATIONS",
dest="min_iter",
help="minimum iterations - each is a batch of 3000 points\n" +
"MC will continue after this until convergence criteria are met\n" +
"Default: 25",
)
vbur_options.add_argument(
"-s",
"--scale",
type=float,
dest="scale",
default=1.17,
help="scale VDW radii by this amount\nDefault: 1.17"
)
args = steric_parser.parse_args()
oop_vector = args.oop_vector
if args.oop_vector is not None:
oop_vector = np.array(args.oop_vector)
ip_vector = args.ip_vector
if args.ip_vector is not None:
ip_vector = np.array(args.ip_vector)
if args.vbur is None:
args.vbur = "Lebedev"
for f in glob_files(args.infile, parser=steric_parser):
if isinstance(f, str):
if args.input_format is not None:
infile = FileReader((f, args.input_format[0], None))
else:
infile = FileReader(f, just_geom=False)
else:
if args.input_format is not None:
infile = FileReader(("from stdin", args.input_format[0], f))
else:
if len(sys.argv) >= 1:
infile = FileReader(("from stdin", "xyz", f))
geom = Geometry(infile)
x, y, z, min_alt, max_alt, basis, targets = geom.steric_map(
center=args.center,
key_atoms=args.key,
radii=args.radii,
return_basis=True,
num_pts=args.num_pts,
oop_vector=oop_vector,
ip_vector=ip_vector,
shape=args.shape,
)
if args.ip_vector is None or args.oop_vector is None:
print(f)
if args.oop_vector is None:
z_vec = np.squeeze(basis[:, 2])
print("out-of-plane vector: %s" % " ".join(["%6.3f" % yi for yi in z_vec]))
if args.ip_vector is None:
z_vec = np.squeeze(basis[:, 2])
y_vec = np.squeeze(basis[:, 1])
r15 = rotation_matrix(np.deg2rad(15), z_vec)
yr = y_vec
for i in range(1, 24):
yr = np.dot(r15, yr)
print("in-plane vector rotated by %5.1f degrees: %s" % (
(15 * i), " ".join(["%6.3f" % yi for yi in yr])
))
if args.min is not None:
min_alt = args.min
if args.max is not None:
max_alt = args.max
fig, ax = plt.subplots()
steric_map = ax.contourf(
x,
y,
z,
extend="min",
cmap=copy.copy(plt.cm.get_cmap("jet")),
levels=np.linspace(min_alt, max_alt, num=20),
)
steric_map.cmap.set_under("w")
steric_lines = ax.contour(
x,
y,
z,
extend="min",
colors="k",
levels=np.linspace(min_alt, max_alt, num=20),
)
bar = fig.colorbar(steric_map, format="%.1f")
bar.set_label("altitude (Å)")
ax.set_aspect("equal")
if args.vbur:
vbur = geom.percent_buried_volume(
center=args.center,
targets=targets,
radius=args.radius,
radii=args.radii,
scale=args.scale,
method=args.vbur,
rpoints=args.rpoints,
apoints=args.apoints,
basis=basis,
min_iter=args.min_iter,
)
ax.hlines(0, -args.radius, args.radius, color="k")
ax.vlines(0, -args.radius, args.radius, color="k")
vbur_1 = vbur[0] + vbur[7]
vbur_2 = vbur[1] + vbur[6]
vbur_3 = vbur[2] + vbur[5]
vbur_4 = vbur[3] + vbur[4]
ax.text(+0.7 * args.radius, +0.9 * args.radius, "%.1f%%" % vbur_1)
ax.text(-0.9 * args.radius, +0.9 * args.radius, "%.1f%%" % vbur_2)
ax.text(-0.9 * args.radius, -0.9 * args.radius, "%.1f%%" % vbur_3)
ax.text(+0.7 * args.radius, -0.9 * args.radius, "%.1f%%" % vbur_4)
if not args.outfile:
plt.show()
else:
outfile = args.outfile
if "$INFILE" in outfile:
outfile = outfile.replace("$INFILE", get_filename(f))
plt.savefig(outfile, dpi=500) | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/bin/stericMap.py | stericMap.py |
import argparse
import sys
from AaronTools.fileIO import FileReader
from AaronTools.utils.utils import get_filename, glob_files
from matplotlib import rcParams
import matplotlib.pyplot as plt
import numpy as np
rcParams["savefig.dpi"] = 300
peak_types = ["pseudo-voigt", "gaussian", "lorentzian", "delta"]
plot_types = ["transmittance", "transmittance-velocity", "uv-vis", "uv-vis-velocity", "ecd", "ecd-velocity"]
def peak_type(x):
out = [y for y in peak_types if y.startswith(x)]
if out:
return out[0]
raise TypeError(
"peak type must be one of: %s" % ", ".join(
peak_types
)
)
def plot_type(x):
out = [y for y in plot_types if y.startswith(x)]
if out:
return out[0]
raise TypeError(
"plot type must be one of: %s" % ", ".join(
plot_types
)
)
uvvis_parser = argparse.ArgumentParser(
description="plot UV/vis spectrum",
formatter_class=argparse.RawTextHelpFormatter
)
uvvis_parser.add_argument(
"infiles", metavar="files",
type=str,
nargs="+",
help="TD-DFT or EOM job output file(s)"
)
uvvis_parser.add_argument(
"-o", "--output",
type=str,
default=None,
dest="outfile",
help="output destination\nDefault: show plot",
)
uvvis_parser.add_argument(
"-t", "--plot-type",
type=plot_type,
choices=plot_types,
default="uv-vis-velocity",
dest="plot_type",
help="type of plot\nDefault: uv-vis-velocity (absorbance)",
)
uvvis_parser.add_argument(
"-u", "--transient",
action="store_true",
dest="transient",
help="use transient excitation data",
)
uvvis_parser.add_argument(
"-ev", "--electron-volt",
action="store_true",
default=False,
dest="ev_unit",
help="use eV on x axis instead of nm",
)
peak_options = uvvis_parser.add_argument_group("peak options")
peak_options.add_argument(
"-p", "--peak-type",
type=peak_type,
choices=peak_types,
default="gaussian",
dest="peak_type",
help="function for peaks\nDefault: gaussian",
)
peak_options.add_argument(
"-m", "--voigt-mixing",
type=float,
default=0.5,
dest="voigt_mixing",
help="fraction of pseudo-Voigt that is Gaussian\nDefault: 0.5",
)
peak_options.add_argument(
"-fwhm", "--full-width-half-max",
type=float,
default=0.5,
dest="fwhm",
help="full width at half max. of peaks\nDefault: 0.5 eV",
)
uvvis_parser.add_argument(
"-s", "--point-spacing",
default=None,
type=float,
dest="point_spacing",
help="spacing between each x value\n"
"Default: a non-uniform spacing that is more dense near peaks",
)
scale_options = uvvis_parser.add_argument_group("scale energies (in eV)")
scale_options.add_argument(
"-ss", "--scalar-shift",
type=float,
default=0.0,
dest="scalar_scale",
help="subtract scalar shift from each excitation\n"
"Default: 0 (no shift)",
)
scale_options.add_argument(
"-l", "--linear-scale",
type=float,
default=0.0,
dest="linear_scale",
help="subtract linear_scale * energy from each excitation\n"
"Default: 0 (no scaling)",
)
scale_options.add_argument(
"-q", "--quadratic-scale",
type=float,
default=0.0,
dest="quadratic_scale",
help="subtract quadratic_scale * energy^2 from each excitation\n"
"Default: 0 (no scaling)",
)
center_centric = uvvis_parser.add_argument_group("x-centered interruptions")
center_centric.add_argument(
"-sc", "--section-centers",
type=lambda x: [float(v) for v in x.split(",")],
dest="centers",
default=None,
help="split plot into sections with a section centered on each of the specified values\n"
"values should be separated by commas"
)
center_centric.add_argument(
"-sw", "--section-widths",
type=lambda x: [float(v) for v in x.split(",")],
dest="widths",
default=None,
help="width of each section specified by -c/--centers\n"
"should be separated by commas, with one for each section"
)
minmax_centric = uvvis_parser.add_argument_group("x-range interruptions")
minmax_centric.add_argument(
"-r", "--ranges",
type=lambda x: [[float(v) for v in r.split("-")] for r in x.split(",")],
dest="ranges",
default=None,
help="split plot into sections (e.g. 200-350,400-650)"
)
uvvis_parser.add_argument(
"-fw", "--figure-width",
type=float,
dest="fig_width",
help="width of figure in inches"
)
uvvis_parser.add_argument(
"-fh", "--figure-height",
type=float,
dest="fig_height",
help="height of figure in inches"
)
uvvis_parser.add_argument(
"-csv", "--experimental-csv",
type=str,
nargs="+",
dest="exp_data",
help="CSV file containing observed spectrum data, which will be plotted on top\n"
"frequency job files should not come directly after this flag"
)
uvvis_parser.add_argument(
"-rx", "--rotate-x-ticks",
action="store_true",
dest="rotate_x_ticks",
default=False,
help="rotate x-axis tick labels by 45 degrees"
)
args = uvvis_parser.parse_args()
if bool(args.centers) != bool(args.widths):
sys.stderr.write(
"both -sw/--section-widths and -sc/--section-centers must be specified"
)
sys.exit(2)
if args.ranges and bool(args.ranges) == bool(args.widths):
sys.stderr.write(
"cannot use -r/--ranges with -sw/--section-widths"
)
sys.exit(2)
centers = args.centers
widths = args.widths
if args.ranges:
centers = []
widths = []
for (xmin, xmax) in args.ranges:
centers.append((xmin + xmax) / 2)
widths.append(abs(xmax - xmin))
units = "nm"
if args.ev_unit:
units = "eV"
exp_data = None
if args.exp_data:
exp_data = []
for f in args.exp_data:
data = np.loadtxt(f, delimiter=",")
for i in range(1, data.shape[1]):
exp_data.append((data[:,0], data[:,i], None))
for f in glob_files(args.infiles, parser=uvvis_parser):
fr = FileReader(f, just_geom=False)
uv_vis = fr.other["uv_vis"]
fig = plt.gcf()
fig.clear()
uv_vis.plot_uv_vis(
fig,
centers=centers,
widths=widths,
plot_type=args.plot_type,
peak_type=args.peak_type,
fwhm=args.fwhm,
point_spacing=args.point_spacing,
voigt_mixing=args.voigt_mixing,
scalar_scale=args.scalar_scale,
linear_scale=args.linear_scale,
quadratic_scale=args.quadratic_scale,
exp_data=exp_data,
units=units,
rotate_x_ticks=args.rotate_x_ticks,
transient=args.transient,
)
if args.fig_width:
fig.set_figwidth(args.fig_width)
if args.fig_height:
fig.set_figheight(args.fig_height)
if args.outfile:
outfile_name = args.outfile.replace("$INFILE", get_filename(f))
plt.savefig(outfile_name, dpi=300)
else:
plt.show() | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/bin/plotUVVis.py | plotUVVis.py |
import sys
import argparse
from AaronTools.geometry import Geometry
from AaronTools.fileIO import FileReader, read_types
from AaronTools.utils.utils import get_filename, glob_files
xyz_parser = argparse.ArgumentParser(
description="print structure in xyz format",
formatter_class=argparse.RawTextHelpFormatter
)
xyz_parser.add_argument(
"infile", metavar="input file",
type=str,
nargs="*",
default=[sys.stdin],
help="a coordinate file"
)
xyz_parser.add_argument(
"-o", "--output",
type=str,
default=False,
required=False,
dest="outfile",
help="output destination\n" +
"$INFILE will be replaced with the name of the input file\n" +
"Default: stdout"
)
xyz_parser.add_argument(
"-if", "--input-format",
type=str,
default=None,
dest="input_format",
choices=read_types,
help="file format of input - xyz is assumed if input is stdin"
)
xyz_parser.add_argument(
"-c", "--comment",
type=str,
default=None,
required=False,
dest="comment",
help="comment line"
)
xyz_parser.add_argument(
"-a", "--append",
action="store_true",
default=False,
required=False,
dest="append",
help="append structures to output file if it already exists\nDefault: false"
)
args = xyz_parser.parse_args()
for f in glob_files(args.infile, parser=xyz_parser):
if isinstance(f, str):
if args.input_format is not None:
infile = FileReader((f, args.input_format, None))
else:
infile = FileReader(f)
else:
if args.input_format is not None:
infile = FileReader(("from stdin", args.input_format, f))
else:
if len(sys.argv) >= 1:
infile = FileReader(("from stdin", "xyz", f))
geom = Geometry(infile, refresh_connected=False, refresh_ranks=False)
if args.comment:
geom.comment = args.comment
else:
geom.comment = f
if not args.outfile:
print(geom.write(outfile=False))
else:
outfile = args.outfile
if "$INFILE" in outfile:
outfile = outfile.replace("$INFILE", get_filename(f))
geom.write(append=args.append, outfile=outfile) | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/bin/printXYZ.py | printXYZ.py |
import argparse
import os
import sys
import numpy as np
from AaronTools.geometry import Geometry
from AaronTools.fileIO import read_types, FileReader
from AaronTools.utils.utils import glob_files, get_filename
remove_frag_parser = argparse.ArgumentParser(
description="remove a fragment from a molecule",
formatter_class=argparse.RawTextHelpFormatter,
)
remove_frag_parser.add_argument(
"infile", metavar="input file",
type=str,
nargs="*",
default=[sys.stdin],
help="a coordinate file"
)
remove_frag_parser.add_argument(
"-if", "--input-format",
type=str,
default=None,
dest="input_format",
choices=read_types,
help="file format of input - xyz is assumed if input is stdin",
)
remove_frag_parser.add_argument(
"-o",
"--output",
type=str,
default=False,
required=False,
metavar="output destination",
dest="outfile",
help="output destination\n" +
"$INFILE will be replaced with the name of the input file\n" +
"Default: stdout",
)
remove_frag_parser.add_argument(
"-t",
"--targets",
type=str,
required=True,
dest="target",
help="fragment atom connected to the rest of the molecule (1-indexed)",
)
remove_frag_parser.add_argument(
"-k",
"--keep-group",
type=str,
required=False,
default=None,
dest="avoid",
help="atom on the molecule that is connected to the fragment being removed\nDefault: longest fragment",
)
remove_frag_parser.add_argument(
"-a",
"--add-hydrogen",
action="store_true",
required=False,
default=False,
dest="add_H",
help="add hydrogen to cap where the fragment was removed",
)
args = remove_frag_parser.parse_args()
for infile in glob_files(args.infile, parser=remove_frag_parser):
if isinstance(infile, str):
if args.input_format is not None:
f = FileReader((infile, args.input_format[0], infile))
else:
f = FileReader(infile)
else:
if args.input_format is not None:
f = FileReader(("from stdin", args.input_format[0], infile))
else:
f = FileReader(("from stdin", "xyz", infile))
geom = Geometry(f)
for atom in geom.find(args.target):
if atom in geom.atoms:
geom.remove_fragment(atom, avoid=args.avoid, add_H=args.add_H)
if not args.add_H:
geom -= atom
if args.outfile:
outfile = args.outfile
if "$INFILE" in outfile:
outfile = outfile.replace("$INFILE", get_filename(infile))
geom.write(append=False, outfile=outfile)
else:
s = geom.write(outfile=False)
print(s) | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/bin/removeFragment.py | removeFragment.py |
import argparse
import sys
from AaronTools.fileIO import FileReader
from AaronTools.utils.utils import get_filename, glob_files
from matplotlib import rcParams
import matplotlib.pyplot as plt
import numpy as np
rcParams["savefig.dpi"] = 300
peak_types = ["pseudo-voigt", "gaussian", "lorentzian", "delta"]
plot_types = ["transmittance", "absorbance", "vcd", "raman"]
def peak_type(x):
out = [y for y in peak_types if y.startswith(x)]
if out:
return out[0]
raise TypeError(
"peak type must be one of: %s" % ", ".join(
peak_types
)
)
def plot_type(x):
out = [y for y in plot_types if y.startswith(x)]
if out:
return out[0]
raise TypeError(
"plot type must be one of: %s" % ", ".join(
plot_types
)
)
ir_parser = argparse.ArgumentParser(
description="plot IR spectrum",
formatter_class=argparse.RawTextHelpFormatter
)
ir_parser.add_argument(
"infiles", metavar="files",
type=str,
nargs="+",
help="frequency job output file(s)"
)
ir_parser.add_argument(
"-o", "--output",
type=str,
default=None,
dest="outfile",
help="output destination\nDefault: show plot",
)
ir_parser.add_argument(
"-t", "--plot-type",
type=plot_type,
choices=plot_types,
default="transmittance",
dest="plot_type",
help="type of plot\nDefault: transmittance",
)
# TODO: figure out more anharmonic options
# anharmonic_options = ir_parser.add_argument_group("anharmonic options")
ir_parser.add_argument(
"-na", "--harmonic",
action="store_false",
default=True,
dest="anharmonic",
help="force to use harmonic frequencies when anharmonic data is in the file",
)
peak_options = ir_parser.add_argument_group("peak options")
peak_options.add_argument(
"-p", "--peak-type",
type=peak_type,
choices=peak_types,
default="pseudo-voigt",
dest="peak_type",
help="function for peaks\nDefault: pseudo-voigt",
)
peak_options.add_argument(
"-m", "--voigt-mixing",
type=float,
default=0.5,
dest="voigt_mixing",
help="fraction of pseudo-Voigt that is Gaussian\nDefault: 0.5",
)
peak_options.add_argument(
"-fwhm", "--full-width-half-max",
type=float,
default=15.0,
dest="fwhm",
help="full width at half max. of peaks\nDefault: 15 cm^-1",
)
ir_parser.add_argument(
"-s", "--point-spacing",
default=None,
type=float,
dest="point_spacing",
help="spacing between each x value\n"
"Default: a non-uniform spacing that is more dense near peaks",
)
scale_options = ir_parser.add_argument_group("scale frequencies")
scale_options.add_argument(
"-l", "--linear-scale",
type=float,
default=0.0,
dest="linear_scale",
help="subtract linear_scale * frequency from each mode (i.e. this is 1 - λ)\n"
"Default: 0 (no scaling)",
)
scale_options.add_argument(
"-q", "--quadratic-scale",
type=float,
default=0.0,
dest="quadratic_scale",
help="subtract quadratic_scale * frequency^2 from each mode\n"
"Default: 0 (no scaling)",
)
ir_parser.add_argument(
"-nr", "--no-reverse",
action="store_false",
default=True,
dest="reverse_x",
help="do not reverse x-axis",
)
center_centric = ir_parser.add_argument_group("x-centered interruptions")
center_centric.add_argument(
"-sc", "--section-centers",
type=lambda x: [float(v) for v in x.split(",")],
dest="centers",
default=None,
help="split plot into sections with a section centered on each of the specified values\n"
"values should be separated by commas"
)
center_centric.add_argument(
"-sw", "--section-widths",
type=lambda x: [float(v) for v in x.split(",")],
dest="widths",
default=None,
help="width of each section specified by -c/--centers\n"
"should be separated by commas, with one for each section"
)
minmax_centric = ir_parser.add_argument_group("x-range interruptions")
minmax_centric.add_argument(
"-r", "--ranges",
type=lambda x: [[float(v) for v in r.split("-")] for r in x.split(",")],
dest="ranges",
default=None,
help="split plot into sections (e.g. 0-1900,2900-3300)"
)
ir_parser.add_argument(
"-fw", "--figure-width",
type=float,
dest="fig_width",
help="width of figure in inches"
)
ir_parser.add_argument(
"-fh", "--figure-height",
type=float,
dest="fig_height",
help="height of figure in inches"
)
ir_parser.add_argument(
"-csv", "--experimental-csv",
type=str,
nargs="+",
dest="exp_data",
help="CSV file containing observed spectrum data, which will be plotted on top\n"
"frequency job files should not come directly after this flag"
)
ir_parser.add_argument(
"-rx", "--rotate-x-ticks",
action="store_true",
dest="rotate_x_ticks",
default=False,
help="rotate x-axis tick labels by 45 degrees"
)
args = ir_parser.parse_args()
if bool(args.centers) != bool(args.widths):
sys.stderr.write(
"both -sw/--section-widths and -sc/--section-centers must be specified"
)
sys.exit(2)
if args.ranges and bool(args.ranges) == bool(args.widths):
sys.stderr.write(
"cannot use -r/--ranges with -sw/--section-widths"
)
sys.exit(2)
centers = args.centers
widths = args.widths
if args.ranges:
centers = []
widths = []
for (xmin, xmax) in args.ranges:
centers.append((xmin + xmax) / 2)
widths.append(abs(xmax - xmin))
exp_data = None
if args.exp_data:
exp_data = []
for f in args.exp_data:
data = np.loadtxt(f, delimiter=",")
for i in range(1, data.shape[1]):
exp_data.append((data[:,0], data[:,i], None))
for f in glob_files(args.infiles, parser=ir_parser):
fr = FileReader(f, just_geom=False)
freq = fr.other["frequency"]
fig = plt.gcf()
fig.clear()
freq.plot_ir(
fig,
centers=centers,
widths=widths,
plot_type=args.plot_type,
peak_type=args.peak_type,
reverse_x=args.reverse_x,
fwhm=args.fwhm,
point_spacing=args.point_spacing,
voigt_mixing=args.voigt_mixing,
linear_scale=args.linear_scale,
quadratic_scale=args.quadratic_scale,
exp_data=exp_data,
anharmonic=freq.anharm_data and args.anharmonic,
rotate_x_ticks=args.rotate_x_ticks,
)
if args.fig_width:
fig.set_figwidth(args.fig_width)
if args.fig_height:
fig.set_figheight(args.fig_height)
if args.outfile:
outfile_name = args.outfile.replace("$INFILE", get_filename(f))
plt.savefig(outfile_name, dpi=300)
else:
plt.show() | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/bin/plotIR.py | plotIR.py |
import argparse
import sys
from warnings import warn
from AaronTools.comp_output import CompOutput
from AaronTools.fileIO import FileReader
from AaronTools.geometry import Geometry
from AaronTools.spectra import ValenceExcitations
from AaronTools.utils.utils import get_filename, glob_files
from matplotlib import rcParams
import matplotlib.pyplot as plt
import numpy as np
rcParams["savefig.dpi"] = 300
peak_types = ["pseudo-voigt", "gaussian", "lorentzian", "delta"]
plot_types = [
"transmittance", "transmittance-velocity",
"uv-vis", "uv-vis-velocity",
"ecd", "ecd-velocity"
]
weight_types = ["electronic", "zero-point", "enthalpy", "free", "quasi-rrho", "quasi-harmonic"]
def peak_type(x):
out = [y for y in peak_types if y.startswith(x)]
if out:
return out[0]
raise TypeError(
"peak type must be one of: %s" % ", ".join(
peak_types
)
)
def plot_type(x):
out = [y for y in plot_types if y.startswith(x)]
if out:
return out[0]
raise TypeError(
"plot type must be one of: %s" % ", ".join(
plot_types
)
)
def weight_type(x):
out = [y for y in weight_types if y.startswith(x)]
if len(out) == 1:
return out[0]
raise TypeError(
"weight type must be one of: %s" % ", ".join(
weight_types
)
)
uvvis_parser = argparse.ArgumentParser(
description="plot Boltzmann-averaged UV/vis spectrum",
formatter_class=argparse.RawTextHelpFormatter
)
uvvis_parser.add_argument(
"infiles", metavar="files",
type=str,
nargs="+",
help="TD-DFT or EOM job output files"
)
uvvis_parser.add_argument(
"-o", "--output",
type=str,
default=None,
dest="outfile",
help="output destination\n"
"if the file extension is .csv, a CSV file will be written\n"
"Default: show plot"
)
uvvis_parser.add_argument(
"-t", "--plot-type",
type=plot_type,
choices=plot_types,
default="uv-vis-velocity",
dest="plot_type",
help="type of plot\nDefault: uv-vis-velocity",
)
uvvis_parser.add_argument(
"-u", "--transient",
action="store_true",
dest="transient",
help="use transient excitation data",
)
uvvis_parser.add_argument(
"-ev", "--electron-volt",
action="store_true",
default=False,
dest="ev_unit",
help="use eV on x axis instead of nm",
)
peak_options = uvvis_parser.add_argument_group("peak options")
peak_options.add_argument(
"-p", "--peak-type",
type=peak_type,
choices=peak_types,
default="gaussian",
dest="peak_type",
help="function for peaks\nDefault: gaussian",
)
peak_options.add_argument(
"-m", "--voigt-mixing",
type=float,
default=0.5,
dest="voigt_mixing",
help="fraction of pseudo-Voigt that is Gaussian\nDefault: 0.5",
)
peak_options.add_argument(
"-fwhm", "--full-width-half-max",
type=float,
default=0.5,
dest="fwhm",
help="full width at half max. of peaks\nDefault: 0.5 eV",
)
uvvis_parser.add_argument(
"-s", "--point-spacing",
default=None,
type=float,
dest="point_spacing",
help="spacing between each x value\n"
"Default: a non-uniform spacing that is more dense near peaks",
)
scale_options = uvvis_parser.add_argument_group("scale energies (in eV)")
scale_options.add_argument(
"-ss", "--scalar-shift",
type=float,
default=0.0,
dest="scalar_scale",
help="subtract scalar shift from each excitation\n"
"Default: 0 (no shift)",
)
scale_options.add_argument(
"-l", "--linear-scale",
type=float,
default=0.0,
dest="linear_scale",
help="subtract linear_scale * energy from each excitation\n"
"Default: 0 (no scaling)",
)
scale_options.add_argument(
"-q", "--quadratic-scale",
type=float,
default=0.0,
dest="quadratic_scale",
help="subtract quadratic_scale * energy^2 from each excitation\n"
"Default: 0 (no scaling)",
)
center_centric = uvvis_parser.add_argument_group("x-centered interruptions")
center_centric.add_argument(
"-sc", "--section-centers",
type=lambda x: [float(v) for v in x.split(",")],
dest="centers",
default=None,
help="split plot into sections with a section centered on each of the specified values\n"
"values should be separated by commas"
)
center_centric.add_argument(
"-sw", "--section-widths",
type=lambda x: [float(v) for v in x.split(",")],
dest="widths",
default=None,
help="width of each section specified by -sc/--section-centers\n"
"should be separated by commas, with one for each section"
)
minmax_centric = uvvis_parser.add_argument_group("x-range interruptions")
minmax_centric.add_argument(
"-r", "--ranges",
type=lambda x: [[float(v) for v in r.split("-")] for r in x.split(",")],
dest="ranges",
default=None,
help="split plot into sections (e.g. 200-350,400-650)"
)
uvvis_parser.add_argument(
"-fw", "--figure-width",
type=float,
dest="fig_width",
help="width of figure in inches"
)
uvvis_parser.add_argument(
"-fh", "--figure-height",
type=float,
dest="fig_height",
help="height of figure in inches"
)
uvvis_parser.add_argument(
"-csv", "--experimental-csv",
type=str,
nargs="+",
dest="exp_data",
help="CSV file containing observed spectrum data, which will be plotted on top\n"
"frequency job files should not come directly after this flag"
)
energy_options = uvvis_parser.add_argument_group("energy weighting")
energy_options.add_argument(
"-w", "--weighting-energy",
type=weight_type,
dest="weighting",
default="quasi-rrho",
choices=weight_types,
help="type of energy to use for Boltzmann weighting\n"
"Default: quasi-rrho",
)
energy_options.add_argument(
"-freq", "--frequency-files",
type=str,
nargs="+",
default=None,
dest="freq_files",
help="frequency jobs to use for thermochem"
)
energy_options.add_argument(
"-sp", "--single-point-files",
type=str,
nargs="+",
default=None,
required=False,
dest="sp_files",
help="single point energies to use for thermochem\n"
"Default: TD-DFT energies from INFILES"
)
energy_options.add_argument(
"-temp", "--temperature",
type=float,
dest="temperature",
default=298.15,
help="temperature (K) to use for weighting\n"
"Default: 298.15",
)
energy_options.add_argument(
"-w0", "--frequency-cutoff",
type=float,
dest="w0",
default=100,
help="cutoff frequency for quasi free energy corrections (1/cm)\n" +
"Default: 100 cm^-1",
)
uvvis_parser.add_argument(
"-rx", "--rotate-x-ticks",
action="store_true",
dest="rotate_x_ticks",
default=False,
help="rotate x-axis tick labels by 45 degrees"
)
args = uvvis_parser.parse_args()
if bool(args.centers) != bool(args.widths):
sys.stderr.write(
"both -sw/--section-widths and -sc/--section-centers must be specified"
)
sys.exit(2)
if args.ranges and bool(args.ranges) == bool(args.widths):
sys.stderr.write(
"cannot use -r/--ranges with -sw/--section-widths"
)
sys.exit(2)
centers = args.centers
widths = args.widths
if args.ranges:
centers = []
widths = []
for (xmin, xmax) in args.ranges:
centers.append((xmin + xmax) / 2)
widths.append(abs(xmax - xmin))
units = "nm"
if args.ev_unit:
units = "eV"
exp_data = None
if args.exp_data:
exp_data = []
for f in args.exp_data:
data = np.loadtxt(f, delimiter=",")
for i in range(1, data.shape[1]):
exp_data.append((data[:,0], data[:,i], None))
filereaders = []
for f in glob_files(args.infiles, parser=uvvis_parser):
fr = FileReader(f, just_geom=False)
filereaders.append(fr)
sp_cos = []
if args.sp_files is None:
sp_cos = [CompOutput(f) for f in filereaders]
else:
for f in glob_files(args.sp_files, parser=uvvis_parser):
co = CompOutput(f)
sp_cos.append(co)
compouts = []
if args.freq_files:
for f in glob_files(args.freq_files, parser=uvvis_parser):
co = CompOutput(f)
compouts.append(co)
if (args.weighting == "electronic" or "frequency" in fr.other) and not compouts:
compouts = [CompOutput(fr) for fr in filereaders]
for i, (fr, sp, freq) in enumerate(zip(filereaders, sp_cos, compouts)):
geom = Geometry(fr)
rmsd = geom.RMSD(sp.geometry, sort=True)
if rmsd > 1e-2:
print(
"TD-DFT structure might not match SP energy file:\n"
"%s %s RMSD = %.2f" % (fr.name, sp.geometry.name, rmsd)
)
rmsd = geom.RMSD(freq.geometry, sort=True)
if rmsd > 1e-2:
print(
"TD-DFT structure might not match frequency file:\n"
"%s %s RMSD = %.2f" % (fr.name, freq.geometry.name, rmsd)
)
for freq2 in compouts[:i]:
rmsd = freq.geometry.RMSD(freq2.geometry, sort=True)
if rmsd < 1e-2:
print(
"two frequency files appear to be identical:\n"
"%s %s RMSD = %.2f" % (freq2.geometry.name, freq.geometry.name, rmsd)
)
if args.weighting == "electronic":
weighting = CompOutput.ELECTRONIC_ENERGY
elif args.weighting == "zero-point":
weighting = CompOutput.ZEROPOINT_ENERGY
elif args.weighting == "enthalpy":
weighting = CompOutput.RRHO_ENTHALPY
elif args.weighting == "free":
weighting = CompOutput.RRHO
elif args.weighting == "quasi-rrho":
weighting = CompOutput.QUASI_RRHO
elif args.weighting == "quasi-harmonic":
weighting = CompOutput.QUASI_HARMONIC
weights = CompOutput.boltzmann_weights(
compouts,
nrg_cos=sp_cos,
temperature=args.temperature,
weighting=weighting,
v0=args.w0,
)
data_attr = "data"
if all(fr.other["uv_vis"].transient_data for fr in filereaders) and args.transient:
data_attr = "transient_data"
mixed_uvvis = ValenceExcitations.get_mixed_signals(
[fr.other["uv_vis"] for fr in filereaders],
weights=weights,
data_attr=data_attr,
)
if not args.outfile or not args.outfile.lower().endswith("csv"):
fig = plt.gcf()
fig.clear()
mixed_uvvis.plot_uv_vis(
fig,
centers=centers,
widths=widths,
plot_type=args.plot_type,
peak_type=args.peak_type,
fwhm=args.fwhm,
point_spacing=args.point_spacing,
voigt_mixing=args.voigt_mixing,
scalar_scale=args.scalar_scale,
linear_scale=args.linear_scale,
quadratic_scale=args.quadratic_scale,
exp_data=exp_data,
units=units,
rotate_x_ticks=args.rotate_x_ticks,
)
if args.fig_width:
fig.set_figwidth(args.fig_width)
if args.fig_height:
fig.set_figheight(args.fig_height)
if args.outfile:
plt.savefig(args.outfile, dpi=300)
else:
plt.show()
else:
intensity_attr = "dipole_str"
if args.plot_type.lower() == "uv-vis-veloctiy":
intensity_attr = "dipole_vel"
if args.plot_type.lower() == "ecd":
intensity_attr = "rotatory_str_len"
if args.plot_type.lower() == "ecd-velocity":
intensity_attr = "rotatory_str_vel"
change_x_unit_func = ValenceExcitations.ev_to_nm
x_label = "wavelength (nm)"
if units == "eV":
change_x_unit_func = None
x_label = r"$h\nu$ (eV)"
funcs, x_positions, intensities = mixed_uvvis.get_spectrum_functions(
fwhm=args.fwhm,
peak_type=args.peak_type,
voigt_mixing=args.voigt_mixing,
scalar_scale=args.scalar_scale,
linear_scale=args.linear_scale,
quadratic_scale=args.quadratic_scale,
intensity_attr=intensity_attr,
)
x_values, y_values, _ = mixed_uvvis.get_plot_data(
funcs,
x_positions,
point_spacing=args.point_spacing,
transmittance=args.plot_type == "transmittance",
peak_type=args.peak_type,
change_x_unit_func=change_x_unit_func,
fwhm=args.fwhm,
)
if "transmittance" in args.plot_type.lower():
y_label = "Transmittance (%)"
elif args.plot_type.lower() == "uv-vis":
y_label = "Absorbance (arb.)"
elif args.plot_type.lower() == "uv-vis-velocity":
y_label = "Absorbance (arb.)"
elif args.plot_type.lower() == "ecd":
y_label = "delta_Absorbance (arb.)"
elif args.plot_type.lower() == "ecd-velocity":
y_label = "delta_Absorbance (arb.)"
else:
y_label = "Absorbance (arb.)"
with open(args.outfile, "w") as f:
s = ",".join([x_label, y_label])
s += "\n"
for x, y in zip(x_values, y_values):
s += ",".join(["%.4f" % z for z in [x, y]])
s += "\n"
f.write(s) | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/bin/plotAverageUVVis.py | plotAverageUVVis.py |
import sys
import argparse
from AaronTools.comp_output import CompOutput
from AaronTools.fileIO import FileReader
from AaronTools.utils.utils import glob_files
stat_parser = argparse.ArgumentParser(
description="prints status of optimization job",
formatter_class=argparse.RawTextHelpFormatter
)
stat_parser.add_argument(
"infile",
metavar="input file",
type=str,
nargs="*",
default=[sys.stdin],
help="input optimization file (i.e. Gaussian output where \"opt\" was specified)"
)
stat_parser.add_argument(
"-if", "--input-format",
type=str,
nargs=1,
default=None,
dest="input_format",
choices=["log", "out", "dat"],
help="file format of input - required if input is stdin"
)
stat_parser.add_argument(
"-o", "--output",
type=str,
nargs="+",
default=None,
required=False,
dest="outfile",
help="output destination\nDefault: stdout"
)
args = stat_parser.parse_args()
s = ""
header_vals = [None]
for f in glob_files(args.infile, parser=stat_parser):
if isinstance(f, str):
if args.input_format is not None:
infile = FileReader((f, args.input_format[0], None), just_geom=False)
else:
infile = FileReader(f, just_geom=False)
else:
if args.input_format is not None:
infile = FileReader(("from stdin", args.input_format[0], f), just_geom=False)
else:
stat_parser.print_help()
raise RuntimeError(
"when no input file is given, stdin is read and a format must be specified"
)
co = CompOutput(infile)
if co.gradient.keys() and (
not all(
x in header_vals for x in co.gradient.keys()
) or not all(
x in co.gradient for x in header_vals
)
):
header_vals = [x for x in sorted(co.gradient.keys())]
header = " Filename Step " + " ".join(
["%14s" % crit for crit in header_vals]
)
header += "\n"
s += header
s += "%30s" % f
s += "%8s" % co.opt_steps
if co.gradient.keys():
for crit in header_vals:
col = "%.2e/%s" % (
float(co.gradient[crit]["value"]), "YES" if co.gradient[crit]["converged"] else "NO"
)
s += " %14s" % col
if (
"error" in infile.other and
infile.other["error"] is not None and
infile.other["error"] != "UNKNOWN"
):
s += " %s" % infile.other["error_msg"]
elif not co.gradient.keys():
s += " no progress found"
elif co.finished:
s += " finished"
else:
s += " not finished"
s += "\n"
if not args.outfile:
print(s.rstrip())
else:
with open(
args.outfile,
"a"
) as f:
f.write(s.rstrip()) | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/bin/grabStatus.py | grabStatus.py |
import sys
from os.path import splitext
import argparse
from AaronTools.geometry import Geometry
from AaronTools.fileIO import FileReader, read_types
from AaronTools.theory import *
from AaronTools.utils.utils import combine_dicts, get_filename, glob_files
theory_parser = argparse.ArgumentParser(
description="print Gaussian, ORCA, Psi4, SQM, or QChem input file",
formatter_class=argparse.RawTextHelpFormatter
)
theory_parser.add_argument(
"infile", metavar="input file",
type=str,
nargs="*",
default=[sys.stdin],
help="a coordinate file",
)
theory_parser.add_argument(
"-o", "--output",
type=str,
default=False,
required=False,
dest="outfile",
help="output destination\n" +
"$INFILE will be replaced with the name of the input file\n" +
"Default: stdout"
)
theory_parser.add_argument(
"-if", "--input-format",
type=str,
default=None,
dest="input_format",
choices=read_types,
help="file format of input - xyz is assumed if input is stdin",
)
theory_parser.add_argument(
"-of", "--output-format",
type=str,
default=None,
dest="out_format",
choices=["gaussian", "orca", "psi4", "sqm", "qchem"],
help="file format of output",
)
theory_parser.add_argument(
"-c", "--comment",
action="append",
nargs="+",
default=[],
dest="comments",
help="comment to put in the output file\ninput file(s) should not be right after comments",
)
theory_parser.add_argument(
"-q", "--charge",
type=int,
dest="charge",
default=None,
help="net charge\nDefault: 0 or what is found in the input file",
)
theory_parser.add_argument(
"-mult", "--multiplicity",
type=int,
dest="multiplicity",
default=None,
help="electronic multiplicity\nDefault: 1 or what is found in the input file",
)
theory_parser.add_argument(
"-p", "--cores",
type=int,
dest="processors",
default=None,
required=False,
help="number of cpu cores to use",
)
theory_parser.add_argument(
"-mem", "--memory",
type=int,
dest="memory",
default=None,
required=False,
help="total memory in GB\n" +
"Note: ORCA and Gaussian only use this to limit the storage-intensive\n" +
" portions of the calculation (e.g. integrals, wavefunction info)",
)
theory_parser.add_argument(
"-up", "--use-previous",
action="store_true",
default=False,
required=False,
dest="use_prev",
help="use settings that can be parsed from the input file",
)
theory_options = theory_parser.add_argument_group("Theory options")
theory_options.add_argument(
"-m", "--method",
type=str,
dest="method",
required=False,
help="method (e.g. B3LYP or MP2)",
)
theory_options.add_argument(
"-b", "--basis",
nargs="+",
type=str,
action="append",
required=False,
default=None,
dest="basis",
help="basis set or list of elements and basis set (e.g. C O N aug-cc-pvtz)\n" +
"elements can be prefixed with ! to exclude them from the basis\n" +
"tm is a synonym for d-block elements\n" +
"auxilliary basis sets can be specified by putting aux X before the basis\n" +
"set name, where X is the auxilliary type (e.g. aux JK cc-pVDZ for cc-pVDZ/JK)\n" +
"a path to a file containing a basis set definition (like one\n" +
"downloaded from basissetexchange.org) can be placed after the\n" +
"basis set name\n" +
"the file's contents should be appropriate for the software package you are using"
)
theory_options.add_argument(
"-ecp", "--pseudopotential",
nargs="+",
type=str,
action="append",
required=False,
default=None,
dest="ecp",
help="ECP or list of elements and ECP (e.g. Pt LANL2DZ)\n" +
"elements can be prefixed with ! to exclude them from the ECP\n" +
"tm is a synonym for d-block elements\n" +
"a path to a file containing a basis set definition (like one\n" +
"downloaded from basissetexchange.org) can be placed after the\n" +
"basis set name\n" +
"the file's contents should be appropriate for the software package you are using"
)
theory_options.add_argument(
"-ed", "--empirical-dispersion",
required=False,
default=None,
dest="empirical_dispersion",
help="empirical dispersion keyword",
)
theory_options.add_argument(
"-sv", "--solvent",
required=False,
default=None,
dest="solvent",
help="solvent",
)
theory_options.add_argument(
"-sm", "--solvent-model",
required=False,
default=None,
dest="solvent_model",
help="implicit solvent model",
)
theory_options.add_argument(
"-g", "--grid",
required=False,
default=None,
dest="grid",
help="integration grid",
)
job_options = theory_parser.add_argument_group("Job options")
job_options.add_argument(
"-opt", "--optimize",
action="store_true",
required=False,
dest="optimize",
help="request geometry optimization job",
)
job_options.add_argument(
"-freq", "--frequencies",
action="store_true",
required=False,
default=False,
dest="freq",
help="request vibrational frequencies job",
)
job_options.add_argument(
"-e", "--energy",
action="store_true",
required=False,
default=False,
dest="energy",
help="request single point energy job",
)
opt_type = theory_parser.add_argument_group("Optimization options")
opt_type.add_argument(
"-ts", "--transition-state",
action="store_true",
default=False,
dest="ts",
help="request transition state optimization",
)
opt_type.add_argument(
"-ca", "--constrained-atoms",
nargs=1,
type=str,
action="append",
default=None,
dest="atoms",
help="comma- or hyphen-separated list of atoms (1-indexed) to constrain during optimization",
)
opt_type.add_argument(
"-cb", "--constrain-bond",
nargs=1,
action="append",
default=None,
dest="bonds",
help="list of comma-separated atom pairs\n" +
"the distance between the atoms in each pair will be constrained during optimization",
)
opt_type.add_argument(
"-cang", "--constrain-angle",
type=str,
nargs=1,
action="append",
default=None,
dest="angles",
help="list of comma-separated atom trios\n" +
"the angle defined by each trio will be constrained during optimization",
)
opt_type.add_argument(
"-ct", "--constrain-torsion",
type=str,
nargs=1,
action="append",
default=None,
dest="torsions",
help="list of comma-separated atom quartets\n" +
"the torsional angle defined by each quartet will be constrained during optimization",
)
opt_type.add_argument(
"-cx", "--constrained-x",
nargs=1,
type=str,
action="append",
default=None,
dest="x",
help="comma- or hyphen-separated list of atoms (1-indexed) to constrain the x coordinate of\n" +
"available for Gaussian, Psi4, and Q-Chem",
)
opt_type.add_argument(
"-cy", "--constrained-y",
nargs=1,
type=str,
action="append",
default=None,
dest="y",
help="comma- or hyphen-separated list of atoms (1-indexed) to constrain the y coordinate of\n" +
"available for Gaussian, Psi4, and Q-Chem",
)
opt_type.add_argument(
"-cz", "--constrained-z",
nargs=1,
type=str,
action="append",
default=None,
dest="z",
help="comma- or hyphen-separated list of atoms (1-indexed) to constrain the z coordinate of\n" +
"available for Gaussian, Psi4, and Q-Chem",
)
opt_type.add_argument(
"-gx", "--grouped-x",
nargs=2,
type=str,
action="append",
default=None,
dest="xgroup",
metavar=("atoms", "value"),
help="comma- or hyphen-separated list of atoms (1-indexed) to keep in the same yz plane\n" +
"available for Gaussian and Psi4",
)
opt_type.add_argument(
"-gy", "--grouped-y",
nargs=2,
type=str,
action="append",
default=None,
dest="ygroup",
metavar=("atoms", "value"),
help="comma- or hyphen-separated list of atoms (1-indexed) to keep in the same xz plane\n" +
"available for Gaussian, Psi4",
)
opt_type.add_argument(
"-gz", "--grouped-z",
nargs=2,
type=str,
action="append",
default=None,
dest="zgroup",
metavar=("atoms", "value"),
help="comma- or hyphen-separated list of atoms (1-indexed) to keep in the same xy plane\n" +
"available for Gaussian and Psi4",
)
freq_type = theory_parser.add_argument_group("Frequency options")
freq_type.add_argument(
"-n", "--numerical",
action="store_true",
default=False,
dest="numerical",
help="request numerical frequencies",
)
freq_type.add_argument(
"-t", "--temperature",
type=float,
default=298.15,
dest="temperature",
help="temperature for calculated thermochemical corrections\nDefault: 298.15",
)
qchem_options = theory_parser.add_argument_group("Q-Chem-specific options")
qchem_options.add_argument(
"--rem",
action="append",
nargs="+",
default=[],
dest=QCHEM_REM,
metavar=("KEYWORD", "OPTION"),
help="REM options\nexample: --rem MAX_SCF_CYCLES 300\n" +
"input file(s) should not be right after --rem",
)
qchem_options.add_argument(
"--section",
nargs="+",
action="append",
default=[],
dest=QCHEM_SETTINGS,
metavar=("SECTION_NAME", "TEXT"),
help="add text to a section"
"input file(s) should not be right after --section",
)
orca_options = theory_parser.add_argument_group("ORCA-specific options")
orca_options.add_argument(
"--simple",
action="append",
default=[],
dest=ORCA_ROUTE,
help="keywords for simple input",
)
orca_options.add_argument(
"--block",
nargs=3,
action="append",
default=[],
dest=ORCA_BLOCKS,
metavar=("BLOCK", "OPTION", "VALUE"),
help="blocks and block options\nexample: --block scf maxiter 500",
)
psi4_options = theory_parser.add_argument_group("Psi4-specific options")
psi4_options.add_argument(
"--before-molecule",
action="append",
default=[],
dest=PSI4_BEFORE_GEOM,
metavar="BEFORE MOL",
help="line to add before the molecule specification",
)
psi4_options.add_argument(
"--before-job",
action="append",
nargs="+",
default=[],
dest=PSI4_BEFORE_JOB,
metavar="BEFORE JOB",
help="line to add before the job\ninput file(s) should not be right after --before-job",
)
psi4_options.add_argument(
"--after-job",
action="append",
nargs="+",
default=[],
dest=PSI4_AFTER_JOB,
metavar="AFTER JOB",
help="line to add after the job\ninput file(s) should not be right after --after-job",
)
psi4_options.add_argument(
"--job",
action="append",
nargs="+",
default=[],
dest=PSI4_JOB,
metavar="JOB",
help="other jobs to add\nexample: --job hessian\ninput file(s) should not be right after --job",
)
psi4_options.add_argument(
"--setting",
action="append",
nargs=2,
default=[],
dest=PSI4_SETTINGS,
metavar=("SETTING", "VALUE"),
help="settings\nexample: --setting reference uhf",
)
psi4_options.add_argument(
"--pcm-solver",
action="append",
nargs="+",
default=[],
dest=PSI4_SOLVENT,
metavar=("SETTING", "VALUE"),
help="settings\nexample: --pcm-solver Cavity 'RadiiSet = UFF' 'Area = 0.3'" +
"\ninput file(s) should not be right after --pcm-solver",
)
psi4_options.add_argument(
"--optking",
action="append",
nargs=2,
default=[],
dest=PSI4_OPTKING,
metavar=("SETTING", "VALUE"),
help="optking settings",
)
psi4_options.add_argument(
"--molecule",
action="append",
nargs="+",
default=[],
dest=PSI4_MOLECULE,
metavar=("SETTING", "VALUE"),
help="options to add to the molecule section\n" +
"example: --molecule units bohr\ninput file(s) should not be right after --molecule",
)
gaussian_options = theory_parser.add_argument_group("Gaussian-specific options")
gaussian_options.add_argument(
"--route",
action="append",
nargs="+",
default=[],
dest=GAUSSIAN_ROUTE,
metavar=("KEYWORD", "OPTION"),
help="route options\nexample: --route freq hpmodes\n" +
"input file(s) should not be right after --route",
)
gaussian_options.add_argument(
"--link0",
action="append",
nargs="+",
default=[],
dest=GAUSSIAN_PRE_ROUTE,
metavar=("COMMAND", "VALUE"),
help="Link 0 commands (without %%)\n" +
"example: --link0 chk asdf.chk\ninput file(s) should not be right after --link0",
)
gaussian_options.add_argument(
"--end-of-file",
action="append",
default=[],
dest=GAUSSIAN_POST,
metavar="input",
help="line to add to the end of the file (e.g. for NBORead)",
)
args = theory_parser.parse_args()
if not args.method and not args.use_prev:
sys.stderr.write("no method specified; -m/--method or -u/--use-previous is required")
theory_parser.print_help()
sys.exit(1)
kwargs = {}
blocks = getattr(args, ORCA_BLOCKS)
if blocks:
kwargs[ORCA_BLOCKS] = {}
for block in blocks:
block_name = block[0]
if block_name not in kwargs[ORCA_BLOCKS]:
kwargs[ORCA_BLOCKS][block_name] = []
kwargs[ORCA_BLOCKS][block_name].append("\t".join(block[1:]))
for pos in [
PSI4_SETTINGS, PSI4_MOLECULE, PSI4_JOB, PSI4_OPTKING, PSI4_SOLVENT,
GAUSSIAN_ROUTE, GAUSSIAN_PRE_ROUTE, QCHEM_REM, QCHEM_SETTINGS,
]:
opts = getattr(args, pos)
if opts:
if pos not in kwargs:
kwargs[pos] = {}
for opt in opts:
setting = opt.pop(0)
if setting.lower() == "rem" and pos == QCHEM_SETTINGS:
s = " ".join(["'%s'" % val if " " in val else val for val in opt])
raise TypeError("use --rem %s instead of --section rem %s" % (s, s))
if setting not in kwargs[pos]:
kwargs[pos][setting] = []
kwargs[pos][setting].extend(opt)
for pos in [ORCA_ROUTE]:
opt = getattr(args, pos)
if opt:
if pos not in kwargs:
kwargs[pos] = []
kwargs[pos].extend(opt)
for pos in [PSI4_BEFORE_GEOM, PSI4_AFTER_JOB, PSI4_BEFORE_JOB, GAUSSIAN_POST]:
opt = getattr(args, pos)
if opt:
if pos not in kwargs:
kwargs[pos] = []
kwargs[pos].extend([" ".join(word) for word in opt])
if args.comments:
kwargs["comments"] = [" ".join(comment) for comment in args.comments]
# Theory() is made for each file because we might be using things from the input file
for f in glob_files(args.infile, parser=theory_parser):
if isinstance(f, str):
if args.input_format is not None:
infile = FileReader((f, args.input_format[0], None), just_geom=False, get_all=True)
else:
infile = FileReader(f, just_geom=False, get_all=True)
else:
if args.input_format is not None:
infile = FileReader(
("from stdin", args.input_format[0], f), just_geom=False, get_all=True
)
else:
if len(sys.argv) >= 1:
infile = FileReader(("from stdin", "xyz", f), just_geom=False, get_all=True)
geom = Geometry(infile)
if args.method is None and args.use_prev:
if "method" in infile.other:
method = infile.other["method"].split("/")[0]
elif "theory" in infile.other:
method = infile.other["theory"].method
elif args.method is not None:
method = args.method
else:
raise RuntimeError(
"method was not determined from %s and was not specified with --method" % f
)
if args.basis is not None:
basis_sets = []
for basis in args.basis:
basis_sets.append(
BasisSet.parse_basis_str(" ".join(basis))[0]
)
elif args.use_prev:
if "method" in infile.other:
basis_sets = method.split("/")[-1]
elif "theory" in infile.other:
basis_sets = infile.other["theory"].basis.basis
else:
basis_sets = None
else:
basis_sets = None
if args.ecp is not None:
ecps = []
for ecp in args.ecp:
ecps.append(
BasisSet.parse_basis_str(" ".join(ecp), cls=ECP)[0]
)
elif args.use_prev:
if "theory" in infile.other:
ecps = infile.other["theory"].basis.ecp
else:
ecps = None
else:
ecps = None
if ecps is None and basis_sets is None:
basis_set = None
else:
basis_set = BasisSet(basis_sets, ecps)
if args.solvent is not None or args.solvent_model is not None:
if args.solvent_model is None or args.solvent is None:
raise RuntimeError("--solvent and --solvent-model must both be specified")
solvent = ImplicitSolvent(args.solvent_model, args.solvent)
else:
solvent = None
job_types = []
if not args.use_prev or (args.optimize or args.freq or args.energy):
if args.optimize:
constraints = {}
if args.atoms is not None:
constraints["atoms"] = []
for constraint in args.atoms:
constraints["atoms"].extend(geom.find(constraint))
if args.bonds is not None:
constraints["bonds"] = []
for bond in args.bonds:
bonded_atoms = geom.find(bond)
if len(bonded_atoms) != 2:
raise RuntimeError(
"not exactly 2 atoms specified in a bond constraint\n" +
"use the format --constrain-bond 1,2"
)
constraints["bonds"].append(bonded_atoms)
if args.angles is not None:
constraints["angles"] = []
for angle in args.angle:
angle_atoms = geom.find(angle)
if len(angle_atoms) != 3:
raise RuntimeError(
"not exactly 3 atoms specified in a angle constraint\n" +
"use the format --constrain-angle 1,2,3"
)
constraints["angles"].append(angle_atoms)
if args.torsions is not None:
constraints["torsions"] = []
for torsion in args.torsions:
torsion_atoms = geom.find(torsion)
if len(torsion_atoms) != 4:
raise RuntimeError(
"not exactly 4 atoms specified in a torsion constraint\n" +
"use the format --constrain-torsion 1,2,3,4"
)
constraints["torsions"].append(torsion_atoms)
if args.x is not None:
constraints["x"] = []
for constraint in args.x:
constraints["x"].extend(geom.find(constraint))
if args.y is not None:
constraints["y"] = []
for constraint in args.y:
constraints["y"].extend(geom.find(constraint))
if args.z is not None:
constraints["z"] = []
for constraint in args.z:
constraints["z"].extend(geom.find(constraint))
if args.xgroup is not None:
constraints["xgroup"] = []
for constraint, val in args.xgroup:
constraints["xgroup"].append((geom.find(constraint), float(val)))
if args.ygroup is not None:
constraints["ygroup"] = []
for constraint, val in args.ygroup:
constraints["ygroup"].append((geom.find(constraint), float(val)))
if args.zgroup is not None:
constraints["zgroup"] = []
for constraint, val in args.zgroup:
constraints["zgroup"].append((geom.find(constraint), float(val)))
if not constraints.keys():
constraints = None
job_types.append(OptimizationJob(transition_state=args.ts, constraints=constraints))
if args.freq:
job_types.append(FrequencyJob(numerical=args.numerical, temperature=args.temperature))
if args.energy:
job_types.append(SinglePointJob())
elif args.use_prev and "theory" in infile.other:
job_types = infile.other["theory"].job_type
grid = args.grid
if args.use_prev and "theory" in infile.other and not grid:
grid = infile.other["theory"].grid
if args.charge is None:
if "charge" in infile.other:
charge = infile.other["charge"]
else:
charge = 0
else:
charge = args.charge
if args.multiplicity is None:
if "multiplicity" in infile.other:
multiplicity = infile.other["multiplicity"]
else:
multiplicity = 1
else:
multiplicity = args.multiplicity
other_kwargs = {}
theory = Theory(
method=method,
basis=basis_set,
grid=grid,
solvent=solvent,
job_type=job_types,
empirical_dispersion=args.empirical_dispersion,
charge=charge,
multiplicity=multiplicity,
processors=args.processors,
memory=args.memory,
)
if args.out_format:
style = args.out_format
else:
if args.outfile:
style = splitext(args.outfile)[-1].lstrip(".")
else:
raise RuntimeError("file format must be specified if no output file is specified")
if args.use_prev and "other_kwargs" in infile.other:
other_kwargs = combine_dicts(other_kwargs, infile.other["other_kwargs"])
other_kwargs = combine_dicts(kwargs, other_kwargs)
if args.outfile:
outfile = args.outfile
if "$INFILE" in outfile:
outfile = outfile.replace("$INFILE", get_filename(f))
warnings = geom.write(
append=True,
outfile=outfile,
style=style,
theory=theory,
return_warnings=True,
**other_kwargs
)
else:
out, warnings = geom.write(
append=True,
outfile=False,
style=style,
theory=theory,
return_warnings=True,
**other_kwargs
)
print(out)
if warnings:
for warning in warnings:
Geometry.LOG.warning(warning) | AaronTools | /AaronTools-1.0b14.tar.gz/AaronTools-1.0b14/bin/makeInput.py | makeInput.py |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.