code
stringlengths 501
5.19M
| package
stringlengths 2
81
| path
stringlengths 9
304
| filename
stringlengths 4
145
|
|---|---|---|---|
import numpy as np
import h5py
import glob
import six
import os
from .groupcat import gcPath, offsetPath
from .util import partTypeNum
def treePath(basePath, treeName, chunkNum=0):
""" Return absolute path to a SubLink HDF5 file (modify as needed). """
# tree_path = '/trees/' + treeName + '/' + 'tree_extended.' + str(chunkNum) + '.hdf5'
tree_path = os.path.join('trees', treeName, 'tree_extended.' + str(chunkNum) + '.hdf5')
_path = os.path.join(basePath, tree_path)
if len(glob.glob(_path)):
return _path
# new path scheme
_path = os.path.join(basePath, os.path.pardir, 'postprocessing', tree_path)
if len(glob.glob(_path)):
return _path
# try one or more alternative path schemes before failing
_path = os.path.join(basePath, 'postprocessing', tree_path)
if len(glob.glob(_path)):
return _path
raise ValueError("Could not construct treePath from basePath = '{}'".format(basePath))
def treeOffsets(basePath, snapNum, id, treeName):
""" Handle offset loading for a SubLink merger tree cutout. """
# old or new format
if 'fof_subhalo' in gcPath(basePath, snapNum) or treeName == "SubLink_gal":
# load groupcat chunk offsets from separate 'offsets_nnn.hdf5' files
with h5py.File(offsetPath(basePath, snapNum), 'r') as f:
groupFileOffsets = f['FileOffsets/Subhalo'][()]
offsetFile = offsetPath(basePath, snapNum)
prefix = 'Subhalo/' + treeName + '/'
groupOffset = id
else:
# load groupcat chunk offsets from header of first file
with h5py.File(gcPath(basePath, snapNum), 'r') as f:
groupFileOffsets = f['Header'].attrs['FileOffsets_Subhalo']
# calculate target groups file chunk which contains this id
groupFileOffsets = int(id) - groupFileOffsets
fileNum = np.max(np.where(groupFileOffsets >= 0))
groupOffset = groupFileOffsets[fileNum]
offsetFile = gcPath(basePath, snapNum, fileNum)
prefix = 'Offsets/Subhalo_Sublink'
with h5py.File(offsetFile, 'r') as f:
# load the merger tree offsets of this subgroup
RowNum = f[prefix+'RowNum'][groupOffset]
LastProgID = f[prefix+'LastProgenitorID'][groupOffset]
SubhaloID = f[prefix+'SubhaloID'][groupOffset]
return RowNum, LastProgID, SubhaloID
offsetCache = dict()
def subLinkOffsets(basePath, treeName, cache=True):
# create quick offset table for rows in the SubLink files
if cache is True:
cache = offsetCache
if type(cache) is dict:
path = os.path.join(basePath, treeName)
try:
return cache[path]
except KeyError:
pass
search_path = treePath(basePath, treeName, '*')
numTreeFiles = len(glob.glob(search_path))
if numTreeFiles == 0:
raise ValueError("No tree files found! for path '{}'".format(search_path))
offsets = np.zeros(numTreeFiles, dtype='int64')
for i in range(numTreeFiles-1):
with h5py.File(treePath(basePath, treeName, i), 'r') as f:
offsets[i+1] = offsets[i] + f['SubhaloID'].shape[0]
if type(cache) is dict:
cache[path] = offsets
return offsets
def loadTree(basePath, snapNum, id, fields=None, onlyMPB=False, onlyMDB=False, treeName="SubLink", cache=True):
""" Load portion of Sublink tree, for a given subhalo, in its existing flat format.
(optionally restricted to a subset fields)."""
# the tree is all subhalos between SubhaloID and LastProgenitorID
RowNum, LastProgID, SubhaloID = treeOffsets(basePath, snapNum, id, treeName)
if RowNum == -1:
print('Warning, empty return. Subhalo [%d] at snapNum [%d] not in tree.' % (id, snapNum))
return None
rowStart = RowNum
rowEnd = RowNum + (LastProgID - SubhaloID)
# make sure fields is not a single element
if isinstance(fields, six.string_types):
fields = [fields]
offsets = subLinkOffsets(basePath, treeName, cache)
# find the tree file chunk containing this row
rowOffsets = rowStart - offsets
try:
fileNum = np.max(np.where(rowOffsets >= 0))
except ValueError as err:
print("ERROR: ", err)
print("rowStart = {}, offsets = {}, rowOffsets = {}".format(rowStart, offsets, rowOffsets))
print(np.where(rowOffsets >= 0))
raise
fileOff = rowOffsets[fileNum]
# load only main progenitor branch? in this case, get MainLeafProgenitorID now
if onlyMPB:
with h5py.File(treePath(basePath, treeName, fileNum), 'r') as f:
MainLeafProgenitorID = f['MainLeafProgenitorID'][fileOff]
# re-calculate rowEnd
rowEnd = RowNum + (MainLeafProgenitorID - SubhaloID)
# load only main descendant branch (e.g. from z=0 descendant to current subhalo)
if onlyMDB:
with h5py.File(treePath(basePath, treeName, fileNum),'r') as f:
RootDescendantID = f['RootDescendantID'][fileOff]
# re-calculate tree subset (rowStart), either single branch to root descendant, or
# subset of tree ending at this subhalo if this subhalo is not on the MPB of that
# root descendant
rowStart = RowNum - (SubhaloID - RootDescendantID) + 1
rowEnd = RowNum + 1
fileOff -= (rowEnd - rowStart)
# calculate number of rows to load
nRows = rowEnd - rowStart + 1
# read
result = {'count': nRows}
with h5py.File(treePath(basePath, treeName, fileNum), 'r') as f:
# if no fields requested, return all fields
if not fields:
fields = list(f.keys())
if fileOff + nRows > f['SubfindID'].shape[0]:
raise Exception('Should not occur. Each tree is contained within a single file.')
# loop over each requested field
for field in fields:
if field not in f.keys():
raise Exception("SubLink tree does not have field ["+field+"]")
# read
result[field] = f[field][fileOff:fileOff+nRows]
# only a single field? then return the array instead of a single item dict
if len(fields) == 1:
return result[fields[0]]
return result
def maxPastMass(tree, index, partType='stars'):
""" Get maximum past mass (of the given partType) along the main branch of a subhalo
specified by index within this tree. """
ptNum = partTypeNum(partType)
branchSize = tree['MainLeafProgenitorID'][index] - tree['SubhaloID'][index] + 1
masses = tree['SubhaloMassType'][index: index + branchSize, ptNum]
return np.max(masses)
def numMergers(tree, minMassRatio=1e-10, massPartType='stars', index=0):
""" Calculate the number of mergers in this sub-tree (optionally above some mass ratio threshold). """
# verify the input sub-tree has the required fields
reqFields = ['SubhaloID', 'NextProgenitorID', 'MainLeafProgenitorID',
'FirstProgenitorID', 'SubhaloMassType']
if not set(reqFields).issubset(tree.keys()):
raise Exception('Error: Input tree needs to have loaded fields: '+', '.join(reqFields))
numMergers = 0
invMassRatio = 1.0 / minMassRatio
# walk back main progenitor branch
rootID = tree['SubhaloID'][index]
fpID = tree['FirstProgenitorID'][index]
while fpID != -1:
fpIndex = index + (fpID - rootID)
fpMass = maxPastMass(tree, fpIndex, massPartType)
# explore breadth
npID = tree['NextProgenitorID'][fpIndex]
while npID != -1:
npIndex = index + (npID - rootID)
npMass = maxPastMass(tree, npIndex, massPartType)
# count if both masses are non-zero, and ratio exceeds threshold
if fpMass > 0.0 and npMass > 0.0:
ratio = npMass / fpMass
if ratio >= minMassRatio and ratio <= invMassRatio:
numMergers += 1
npID = tree['NextProgenitorID'][npIndex]
fpID = tree['FirstProgenitorID'][fpIndex]
return numMergers | ALMASim | /ALMASim-1.4.tar.gz/ALMASim-1.4/submodules/illustris_python/sublink.py | sublink.py |
groupcat.py: File I/O related to the FoF and Subfind group catalogs. """
from __future__ import print_function
import six
from os.path import isfile,expanduser
import numpy as np
import h5py
def gcPath(basePath, snapNum, chunkNum=0):
""" Return absolute path to a group catalog HDF5 file (modify as needed). """
gcPath = basePath + '/groups_%03d/' % snapNum
filePath1 = gcPath + 'groups_%03d.%d.hdf5' % (snapNum, chunkNum)
filePath2 = gcPath + 'fof_subhalo_tab_%03d.%d.hdf5' % (snapNum, chunkNum)
if isfile(expanduser(filePath1)):
return filePath1
return filePath2
def offsetPath(basePath, snapNum):
""" Return absolute path to a separate offset file (modify as needed). """
offsetPath = basePath + '/../postprocessing/offsets/offsets_%03d.hdf5' % snapNum
return offsetPath
def loadObjects(basePath, snapNum, gName, nName, fields):
""" Load either halo or subhalo information from the group catalog. """
result = {}
# make sure fields is not a single element
if isinstance(fields, six.string_types):
fields = [fields]
# load header from first chunk
with h5py.File(gcPath(basePath, snapNum), 'r') as f:
header = dict(f['Header'].attrs.items())
if 'N'+nName+'_Total' not in header and nName == 'subgroups':
nName = 'subhalos' # alternate convention
result['count'] = f['Header'].attrs['N' + nName + '_Total']
if not result['count']:
print('warning: zero groups, empty return (snap=' + str(snapNum) + ').')
return result
# if fields not specified, load everything
if not fields:
fields = list(f[gName].keys())
for field in fields:
# verify existence
if field not in f[gName].keys():
raise Exception("Group catalog does not have requested field [" + field + "]!")
# replace local length with global
shape = list(f[gName][field].shape)
shape[0] = result['count']
# allocate within return dict
result[field] = np.zeros(shape, dtype=f[gName][field].dtype)
# loop over chunks
wOffset = 0
for i in range(header['NumFiles']):
f = h5py.File(gcPath(basePath, snapNum, i), 'r')
if not f['Header'].attrs['N'+nName+'_ThisFile']:
continue # empty file chunk
# loop over each requested field
for field in fields:
if field not in f[gName].keys():
raise Exception("Group catalog does not have requested field [" + field + "]!")
# shape and type
shape = f[gName][field].shape
# read data local to the current file
if len(shape) == 1:
result[field][wOffset:wOffset+shape[0]] = f[gName][field][0:shape[0]]
else:
result[field][wOffset:wOffset+shape[0], :] = f[gName][field][0:shape[0], :]
wOffset += shape[0]
f.close()
# only a single field? then return the array instead of a single item dict
if len(fields) == 1:
return result[fields[0]]
return result
def loadSubhalos(basePath, snapNum, fields=None):
""" Load all subhalo information from the entire group catalog for one snapshot
(optionally restrict to a subset given by fields). """
return loadObjects(basePath, snapNum, "Subhalo", "subgroups", fields)
def loadHalos(basePath, snapNum, fields=None):
""" Load all halo information from the entire group catalog for one snapshot
(optionally restrict to a subset given by fields). """
return loadObjects(basePath, snapNum, "Group", "groups", fields)
def loadHeader(basePath, snapNum):
""" Load the group catalog header. """
with h5py.File(gcPath(basePath, snapNum), 'r') as f:
header = dict(f['Header'].attrs.items())
return header
def load(basePath, snapNum):
""" Load complete group catalog all at once. """
r = {}
r['subhalos'] = loadSubhalos(basePath, snapNum)
r['halos'] = loadHalos(basePath, snapNum)
r['header'] = loadHeader(basePath, snapNum)
return r
def loadSingle(basePath, snapNum, haloID=-1, subhaloID=-1):
""" Return complete group catalog information for one halo or subhalo. """
if (haloID < 0 and subhaloID < 0) or (haloID >= 0 and subhaloID >= 0):
raise Exception("Must specify either haloID or subhaloID (and not both).")
gName = "Subhalo" if subhaloID >= 0 else "Group"
searchID = subhaloID if subhaloID >= 0 else haloID
# old or new format
if 'fof_subhalo' in gcPath(basePath, snapNum):
# use separate 'offsets_nnn.hdf5' files
with h5py.File(offsetPath(basePath, snapNum), 'r') as f:
offsets = f['FileOffsets/'+gName][()]
else:
# use header of group catalog
with h5py.File(gcPath(basePath, snapNum), 'r') as f:
offsets = f['Header'].attrs['FileOffsets_'+gName]
offsets = searchID - offsets
fileNum = np.max(np.where(offsets >= 0))
groupOffset = offsets[fileNum]
# load halo/subhalo fields into a dict
result = {}
with h5py.File(gcPath(basePath, snapNum, fileNum), 'r') as f:
for haloProp in f[gName].keys():
result[haloProp] = f[gName][haloProp][groupOffset]
return result | ALMASim | /ALMASim-1.4.tar.gz/ALMASim-1.4/submodules/illustris_python/groupcat.py | groupcat.py |
import numpy as np
import h5py
import six
from .groupcat import gcPath, offsetPath
from os.path import isfile
def treePath(basePath, chunkNum=0):
""" Return absolute path to a LHaloTree HDF5 file (modify as needed). """
filePath_list = [ basePath + '/trees/treedata/' + 'trees_sf1_135.' + str(chunkNum) + '.hdf5',
basePath + '/../postprocessing/trees/LHaloTree/trees_sf1_099.' + str(chunkNum) + '.hdf5', #new path scheme for TNG
basePath + '/../postprocessing/trees/LHaloTree/trees_sf1_080.' + str(chunkNum) + '.hdf5', #Thesan
]
for filePath in filePath_list:
if isfile(filePath):
return filePath
raise ValueError("No tree file found!")
def treeOffsets(basePath, snapNum, id):
""" Handle offset loading for a LHaloTree merger tree cutout. """
# load groupcat chunk offsets from header of first file (old or new format)
if 'fof_subhalo' in gcPath(basePath, snapNum):
# load groupcat chunk offsets from separate 'offsets_nnn.hdf5' files
with h5py.File(offsetPath(basePath, snapNum), 'r') as f:
groupFileOffsets = f['FileOffsets/Subhalo'][()]
offsetFile = offsetPath(basePath, snapNum)
prefix = 'Subhalo/LHaloTree/'
groupOffset = id
else:
# load groupcat chunk offsets from header of first file
with h5py.File(gcPath(basePath, snapNum), 'r') as f:
groupFileOffsets = f['Header'].attrs['FileOffsets_Subhalo']
# calculate target groups file chunk which contains this id
groupFileOffsets = int(id) - groupFileOffsets
fileNum = np.max(np.where(groupFileOffsets >= 0))
groupOffset = groupFileOffsets[fileNum]
offsetFile = gcPath(basePath, snapNum, fileNum)
prefix = 'Offsets/Subhalo_LHaloTree'
with h5py.File(offsetFile, 'r') as f:
# load the merger tree offsets of this subgroup
TreeFile = f[prefix+'File'][groupOffset]
TreeIndex = f[prefix+'Index'][groupOffset]
TreeNum = f[prefix+'Num'][groupOffset]
return TreeFile, TreeIndex, TreeNum
def singleNodeFlat(conn, index, data_in, data_out, count, onlyMPB):
""" Recursive helper function: Add a single tree node. """
data_out[count] = data_in[index]
count += 1
count = recProgenitorFlat(conn, index, data_in, data_out, count, onlyMPB)
return count
def recProgenitorFlat(conn, start_index, data_in, data_out, count, onlyMPB):
""" Recursive helper function: Flatten out the unordered LHaloTree, one data field at a time. """
firstProg = conn["FirstProgenitor"][start_index]
if firstProg < 0:
return count
# depth-ordered traversal (down mpb)
count = singleNodeFlat(conn, firstProg, data_in, data_out, count, onlyMPB)
# explore breadth
if not onlyMPB:
nextProg = conn["NextProgenitor"][firstProg]
while nextProg >= 0:
count = singleNodeFlat(conn, nextProg, data_in, data_out, count, onlyMPB)
nextProg = conn["NextProgenitor"][nextProg]
firstProg = conn["FirstProgenitor"][firstProg]
return count
def loadTree(basePath, snapNum, id, fields=None, onlyMPB=False):
""" Load portion of LHaloTree, for a given subhalo, re-arranging into a flat format. """
TreeFile, TreeIndex, TreeNum = treeOffsets(basePath, snapNum, id)
if TreeNum == -1:
print('Warning, empty return. Subhalo [%d] at snapNum [%d] not in tree.' % (id, snapNum))
return None
# config
gName = 'Tree' + str(TreeNum) # group name containing this subhalo
nRows = None # we do not know in advance the size of the tree
# make sure fields is not a single element
if isinstance(fields, six.string_types):
fields = [fields]
fTree = h5py.File(treePath(basePath, TreeFile), 'r')
# if no fields requested, return everything
if not fields:
fields = list(fTree[gName].keys())
# verify existence of requested fields
for field in fields:
if field not in fTree[gName].keys():
raise Exception('Error: Requested field '+field+' not in tree.')
# load connectivity for this entire TreeX group
connFields = ['FirstProgenitor', 'NextProgenitor']
conn = {}
for field in connFields:
conn[field] = fTree[gName][field][:]
# determine sub-tree size with dummy walk
dummy = np.zeros(conn['FirstProgenitor'].shape, dtype='int32')
nRows = singleNodeFlat(conn, TreeIndex, dummy, dummy, 0, onlyMPB)
result = {}
result['count'] = nRows
# walk through connectivity, one data field at a time
for field in fields:
# load field for entire tree? doing so is much faster than randomly accessing the disk
# during walk, assuming that the sub-tree is a large fraction of the full tree, and that
# the sub-tree is large in the absolute sense. the decision is heuristic, and can be
# modified (if you have the tree on a fast SSD, could disable the full load).
if nRows < 1000: # and float(nRows)/len(result['FirstProgenitor']) > 0.1
# do not load, walk with single disk reads
full_data = fTree[gName][field]
else:
# pre-load all, walk in-memory
full_data = fTree[gName][field][:]
# allocate the data array in the sub-tree
dtype = fTree[gName][field].dtype
shape = list(fTree[gName][field].shape)
shape[0] = nRows
data = np.zeros(shape, dtype=dtype)
# walk the tree, depth-first
count = singleNodeFlat(conn, TreeIndex, full_data, data, 0, onlyMPB)
# save field
result[field] = data
fTree.close()
# only a single field? then return the array instead of a single item dict
if len(fields) == 1:
return result[fields[0]]
return result | ALMASim | /ALMASim-1.4.tar.gz/ALMASim-1.4/submodules/illustris_python/lhalotree.py | lhalotree.py |
snapshot.py: File I/O related to the snapshot files. """
from __future__ import print_function
import numpy as np
import h5py
import six
from os.path import isfile
from .util import partTypeNum
from .groupcat import gcPath, offsetPath
def snapPath(basePath, snapNum, chunkNum=0):
""" Return absolute path to a snapshot HDF5 file (modify as needed). """
snapPath = basePath + '/snapdir_' + str(snapNum).zfill(3) + '/'
filePath1 = snapPath + 'snap_' + str(snapNum).zfill(3) + '.' + str(chunkNum) + '.hdf5'
filePath2 = filePath1.replace('/snap_', '/snapshot_')
if isfile(filePath1):
return filePath1
return filePath2
def getNumPart(header):
""" Calculate number of particles of all types given a snapshot header. """
if 'NumPart_Total_HighWord' not in header:
return header['NumPart_Total'] # new uint64 convention
nTypes = 6
nPart = np.zeros(nTypes, dtype=np.int64)
for j in range(nTypes):
nPart[j] = header['NumPart_Total'][j] | (header['NumPart_Total_HighWord'][j] << 32)
return nPart
def loadSubset(basePath, snapNum, partType, fields=None, subset=None, mdi=None, sq=True, float32=False):
""" Load a subset of fields for all particles/cells of a given partType.
If offset and length specified, load only that subset of the partType.
If mdi is specified, must be a list of integers of the same length as fields,
giving for each field the multi-dimensional index (on the second dimension) to load.
For example, fields=['Coordinates', 'Masses'] and mdi=[1, None] returns a 1D array
of y-Coordinates only, together with Masses.
If sq is True, return a numpy array instead of a dict if len(fields)==1.
If float32 is True, load any float64 datatype arrays directly as float32 (save memory). """
result = {}
ptNum = partTypeNum(partType)
gName = "PartType" + str(ptNum)
# make sure fields is not a single element
if isinstance(fields, six.string_types):
fields = [fields]
# load header from first chunk
with h5py.File(snapPath(basePath, snapNum), 'r') as f:
header = dict(f['Header'].attrs.items())
nPart = getNumPart(header)
# decide global read size, starting file chunk, and starting file chunk offset
if subset:
offsetsThisType = subset['offsetType'][ptNum] - subset['snapOffsets'][ptNum, :]
fileNum = np.max(np.where(offsetsThisType >= 0))
fileOff = offsetsThisType[fileNum]
numToRead = subset['lenType'][ptNum]
else:
fileNum = 0
fileOff = 0
numToRead = nPart[ptNum]
result['count'] = numToRead
if not numToRead:
# print('warning: no particles of requested type, empty return.')
return result
# find a chunk with this particle type
i = 1
while gName not in f:
f = h5py.File(snapPath(basePath, snapNum, i), 'r')
i += 1
# if fields not specified, load everything
if not fields:
fields = list(f[gName].keys())
for i, field in enumerate(fields):
# verify existence
if field not in f[gName].keys():
raise Exception("Particle type ["+str(ptNum)+"] does not have field ["+field+"]")
# replace local length with global
shape = list(f[gName][field].shape)
shape[0] = numToRead
# multi-dimensional index slice load
if mdi is not None and mdi[i] is not None:
if len(shape) != 2:
raise Exception("Read error: mdi requested on non-2D field ["+field+"]")
shape = [shape[0]]
# allocate within return dict
dtype = f[gName][field].dtype
if dtype == np.float64 and float32: dtype = np.float32
result[field] = np.zeros(shape, dtype=dtype)
# loop over chunks
wOffset = 0
origNumToRead = numToRead
while numToRead:
f = h5py.File(snapPath(basePath, snapNum, fileNum), 'r')
# no particles of requested type in this file chunk?
if gName not in f:
f.close()
fileNum += 1
fileOff = 0
continue
# set local read length for this file chunk, truncate to be within the local size
numTypeLocal = f['Header'].attrs['NumPart_ThisFile'][ptNum]
numToReadLocal = numToRead
if fileOff + numToReadLocal > numTypeLocal:
numToReadLocal = numTypeLocal - fileOff
#print('['+str(fileNum).rjust(3)+'] off='+str(fileOff)+' read ['+str(numToReadLocal)+\
# '] of ['+str(numTypeLocal)+'] remaining = '+str(numToRead-numToReadLocal))
# loop over each requested field for this particle type
for i, field in enumerate(fields):
# read data local to the current file
if mdi is None or mdi[i] is None:
result[field][wOffset:wOffset+numToReadLocal] = f[gName][field][fileOff:fileOff+numToReadLocal]
else:
result[field][wOffset:wOffset+numToReadLocal] = f[gName][field][fileOff:fileOff+numToReadLocal, mdi[i]]
wOffset += numToReadLocal
numToRead -= numToReadLocal
fileNum += 1
fileOff = 0 # start at beginning of all file chunks other than the first
f.close()
# verify we read the correct number
if origNumToRead != wOffset:
raise Exception("Read ["+str(wOffset)+"] particles, but was expecting ["+str(origNumToRead)+"]")
# only a single field? then return the array instead of a single item dict
if sq and len(fields) == 1:
return result[fields[0]]
return result
def getSnapOffsets(basePath, snapNum, id, type):
""" Compute offsets within snapshot for a particular group/subgroup. """
r = {}
# old or new format
if 'fof_subhalo' in gcPath(basePath, snapNum):
# use separate 'offsets_nnn.hdf5' files
with h5py.File(offsetPath(basePath, snapNum), 'r') as f:
groupFileOffsets = f['FileOffsets/'+type][()]
r['snapOffsets'] = np.transpose(f['FileOffsets/SnapByType'][()]) # consistency
else:
# load groupcat chunk offsets from header of first file
with h5py.File(gcPath(basePath, snapNum), 'r') as f:
groupFileOffsets = f['Header'].attrs['FileOffsets_'+type]
r['snapOffsets'] = f['Header'].attrs['FileOffsets_Snap']
# calculate target groups file chunk which contains this id
groupFileOffsets = int(id) - groupFileOffsets
fileNum = np.max(np.where(groupFileOffsets >= 0))
groupOffset = groupFileOffsets[fileNum]
# load the length (by type) of this group/subgroup from the group catalog
with h5py.File(gcPath(basePath, snapNum, fileNum), 'r') as f:
r['lenType'] = f[type][type+'LenType'][groupOffset, :]
# old or new format: load the offset (by type) of this group/subgroup within the snapshot
if 'fof_subhalo' in gcPath(basePath, snapNum):
with h5py.File(offsetPath(basePath, snapNum), 'r') as f:
r['offsetType'] = f[type+'/SnapByType'][id, :]
else:
with h5py.File(gcPath(basePath, snapNum, fileNum), 'r') as f:
r['offsetType'] = f['Offsets'][type+'_SnapByType'][groupOffset, :]
return r
def loadSubhalo(basePath, snapNum, id, partType, fields=None):
""" Load all particles/cells of one type for a specific subhalo
(optionally restricted to a subset fields). """
# load subhalo length, compute offset, call loadSubset
subset = getSnapOffsets(basePath, snapNum, id, "Subhalo")
return loadSubset(basePath, snapNum, partType, fields, subset=subset)
def loadHalo(basePath, snapNum, id, partType, fields=None):
""" Load all particles/cells of one type for a specific halo
(optionally restricted to a subset fields). """
# load halo length, compute offset, call loadSubset
subset = getSnapOffsets(basePath, snapNum, id, "Group")
return loadSubset(basePath, snapNum, partType, fields, subset=subset) | ALMASim | /ALMASim-1.4.tar.gz/ALMASim-1.4/submodules/build/lib/illustris_python/snapshot.py | snapshot.py |
import numpy as np
import h5py
import glob
import six
import os
from .groupcat import gcPath, offsetPath
from .util import partTypeNum
def treePath(basePath, treeName, chunkNum=0):
""" Return absolute path to a SubLink HDF5 file (modify as needed). """
# tree_path = '/trees/' + treeName + '/' + 'tree_extended.' + str(chunkNum) + '.hdf5'
tree_path = os.path.join('trees', treeName, 'tree_extended.' + str(chunkNum) + '.hdf5')
_path = os.path.join(basePath, tree_path)
if len(glob.glob(_path)):
return _path
# new path scheme
_path = os.path.join(basePath, os.path.pardir, 'postprocessing', tree_path)
if len(glob.glob(_path)):
return _path
# try one or more alternative path schemes before failing
_path = os.path.join(basePath, 'postprocessing', tree_path)
if len(glob.glob(_path)):
return _path
raise ValueError("Could not construct treePath from basePath = '{}'".format(basePath))
def treeOffsets(basePath, snapNum, id, treeName):
""" Handle offset loading for a SubLink merger tree cutout. """
# old or new format
if 'fof_subhalo' in gcPath(basePath, snapNum) or treeName == "SubLink_gal":
# load groupcat chunk offsets from separate 'offsets_nnn.hdf5' files
with h5py.File(offsetPath(basePath, snapNum), 'r') as f:
groupFileOffsets = f['FileOffsets/Subhalo'][()]
offsetFile = offsetPath(basePath, snapNum)
prefix = 'Subhalo/' + treeName + '/'
groupOffset = id
else:
# load groupcat chunk offsets from header of first file
with h5py.File(gcPath(basePath, snapNum), 'r') as f:
groupFileOffsets = f['Header'].attrs['FileOffsets_Subhalo']
# calculate target groups file chunk which contains this id
groupFileOffsets = int(id) - groupFileOffsets
fileNum = np.max(np.where(groupFileOffsets >= 0))
groupOffset = groupFileOffsets[fileNum]
offsetFile = gcPath(basePath, snapNum, fileNum)
prefix = 'Offsets/Subhalo_Sublink'
with h5py.File(offsetFile, 'r') as f:
# load the merger tree offsets of this subgroup
RowNum = f[prefix+'RowNum'][groupOffset]
LastProgID = f[prefix+'LastProgenitorID'][groupOffset]
SubhaloID = f[prefix+'SubhaloID'][groupOffset]
return RowNum, LastProgID, SubhaloID
offsetCache = dict()
def subLinkOffsets(basePath, treeName, cache=True):
# create quick offset table for rows in the SubLink files
if cache is True:
cache = offsetCache
if type(cache) is dict:
path = os.path.join(basePath, treeName)
try:
return cache[path]
except KeyError:
pass
search_path = treePath(basePath, treeName, '*')
numTreeFiles = len(glob.glob(search_path))
if numTreeFiles == 0:
raise ValueError("No tree files found! for path '{}'".format(search_path))
offsets = np.zeros(numTreeFiles, dtype='int64')
for i in range(numTreeFiles-1):
with h5py.File(treePath(basePath, treeName, i), 'r') as f:
offsets[i+1] = offsets[i] + f['SubhaloID'].shape[0]
if type(cache) is dict:
cache[path] = offsets
return offsets
def loadTree(basePath, snapNum, id, fields=None, onlyMPB=False, onlyMDB=False, treeName="SubLink", cache=True):
""" Load portion of Sublink tree, for a given subhalo, in its existing flat format.
(optionally restricted to a subset fields)."""
# the tree is all subhalos between SubhaloID and LastProgenitorID
RowNum, LastProgID, SubhaloID = treeOffsets(basePath, snapNum, id, treeName)
if RowNum == -1:
print('Warning, empty return. Subhalo [%d] at snapNum [%d] not in tree.' % (id, snapNum))
return None
rowStart = RowNum
rowEnd = RowNum + (LastProgID - SubhaloID)
# make sure fields is not a single element
if isinstance(fields, six.string_types):
fields = [fields]
offsets = subLinkOffsets(basePath, treeName, cache)
# find the tree file chunk containing this row
rowOffsets = rowStart - offsets
try:
fileNum = np.max(np.where(rowOffsets >= 0))
except ValueError as err:
print("ERROR: ", err)
print("rowStart = {}, offsets = {}, rowOffsets = {}".format(rowStart, offsets, rowOffsets))
print(np.where(rowOffsets >= 0))
raise
fileOff = rowOffsets[fileNum]
# load only main progenitor branch? in this case, get MainLeafProgenitorID now
if onlyMPB:
with h5py.File(treePath(basePath, treeName, fileNum), 'r') as f:
MainLeafProgenitorID = f['MainLeafProgenitorID'][fileOff]
# re-calculate rowEnd
rowEnd = RowNum + (MainLeafProgenitorID - SubhaloID)
# load only main descendant branch (e.g. from z=0 descendant to current subhalo)
if onlyMDB:
with h5py.File(treePath(basePath, treeName, fileNum),'r') as f:
RootDescendantID = f['RootDescendantID'][fileOff]
# re-calculate tree subset (rowStart), either single branch to root descendant, or
# subset of tree ending at this subhalo if this subhalo is not on the MPB of that
# root descendant
rowStart = RowNum - (SubhaloID - RootDescendantID) + 1
rowEnd = RowNum + 1
fileOff -= (rowEnd - rowStart)
# calculate number of rows to load
nRows = rowEnd - rowStart + 1
# read
result = {'count': nRows}
with h5py.File(treePath(basePath, treeName, fileNum), 'r') as f:
# if no fields requested, return all fields
if not fields:
fields = list(f.keys())
if fileOff + nRows > f['SubfindID'].shape[0]:
raise Exception('Should not occur. Each tree is contained within a single file.')
# loop over each requested field
for field in fields:
if field not in f.keys():
raise Exception("SubLink tree does not have field ["+field+"]")
# read
result[field] = f[field][fileOff:fileOff+nRows]
# only a single field? then return the array instead of a single item dict
if len(fields) == 1:
return result[fields[0]]
return result
def maxPastMass(tree, index, partType='stars'):
""" Get maximum past mass (of the given partType) along the main branch of a subhalo
specified by index within this tree. """
ptNum = partTypeNum(partType)
branchSize = tree['MainLeafProgenitorID'][index] - tree['SubhaloID'][index] + 1
masses = tree['SubhaloMassType'][index: index + branchSize, ptNum]
return np.max(masses)
def numMergers(tree, minMassRatio=1e-10, massPartType='stars', index=0):
""" Calculate the number of mergers in this sub-tree (optionally above some mass ratio threshold). """
# verify the input sub-tree has the required fields
reqFields = ['SubhaloID', 'NextProgenitorID', 'MainLeafProgenitorID',
'FirstProgenitorID', 'SubhaloMassType']
if not set(reqFields).issubset(tree.keys()):
raise Exception('Error: Input tree needs to have loaded fields: '+', '.join(reqFields))
numMergers = 0
invMassRatio = 1.0 / minMassRatio
# walk back main progenitor branch
rootID = tree['SubhaloID'][index]
fpID = tree['FirstProgenitorID'][index]
while fpID != -1:
fpIndex = index + (fpID - rootID)
fpMass = maxPastMass(tree, fpIndex, massPartType)
# explore breadth
npID = tree['NextProgenitorID'][fpIndex]
while npID != -1:
npIndex = index + (npID - rootID)
npMass = maxPastMass(tree, npIndex, massPartType)
# count if both masses are non-zero, and ratio exceeds threshold
if fpMass > 0.0 and npMass > 0.0:
ratio = npMass / fpMass
if ratio >= minMassRatio and ratio <= invMassRatio:
numMergers += 1
npID = tree['NextProgenitorID'][npIndex]
fpID = tree['FirstProgenitorID'][fpIndex]
return numMergers | ALMASim | /ALMASim-1.4.tar.gz/ALMASim-1.4/submodules/build/lib/illustris_python/sublink.py | sublink.py |
groupcat.py: File I/O related to the FoF and Subfind group catalogs. """
from __future__ import print_function
import six
from os.path import isfile,expanduser
import numpy as np
import h5py
def gcPath(basePath, snapNum, chunkNum=0):
""" Return absolute path to a group catalog HDF5 file (modify as needed). """
gcPath = basePath + '/groups_%03d/' % snapNum
filePath1 = gcPath + 'groups_%03d.%d.hdf5' % (snapNum, chunkNum)
filePath2 = gcPath + 'fof_subhalo_tab_%03d.%d.hdf5' % (snapNum, chunkNum)
if isfile(expanduser(filePath1)):
return filePath1
return filePath2
def offsetPath(basePath, snapNum):
""" Return absolute path to a separate offset file (modify as needed). """
offsetPath = basePath + '/../postprocessing/offsets/offsets_%03d.hdf5' % snapNum
return offsetPath
def loadObjects(basePath, snapNum, gName, nName, fields):
""" Load either halo or subhalo information from the group catalog. """
result = {}
# make sure fields is not a single element
if isinstance(fields, six.string_types):
fields = [fields]
# load header from first chunk
with h5py.File(gcPath(basePath, snapNum), 'r') as f:
header = dict(f['Header'].attrs.items())
if 'N'+nName+'_Total' not in header and nName == 'subgroups':
nName = 'subhalos' # alternate convention
result['count'] = f['Header'].attrs['N' + nName + '_Total']
if not result['count']:
print('warning: zero groups, empty return (snap=' + str(snapNum) + ').')
return result
# if fields not specified, load everything
if not fields:
fields = list(f[gName].keys())
for field in fields:
# verify existence
if field not in f[gName].keys():
raise Exception("Group catalog does not have requested field [" + field + "]!")
# replace local length with global
shape = list(f[gName][field].shape)
shape[0] = result['count']
# allocate within return dict
result[field] = np.zeros(shape, dtype=f[gName][field].dtype)
# loop over chunks
wOffset = 0
for i in range(header['NumFiles']):
f = h5py.File(gcPath(basePath, snapNum, i), 'r')
if not f['Header'].attrs['N'+nName+'_ThisFile']:
continue # empty file chunk
# loop over each requested field
for field in fields:
if field not in f[gName].keys():
raise Exception("Group catalog does not have requested field [" + field + "]!")
# shape and type
shape = f[gName][field].shape
# read data local to the current file
if len(shape) == 1:
result[field][wOffset:wOffset+shape[0]] = f[gName][field][0:shape[0]]
else:
result[field][wOffset:wOffset+shape[0], :] = f[gName][field][0:shape[0], :]
wOffset += shape[0]
f.close()
# only a single field? then return the array instead of a single item dict
if len(fields) == 1:
return result[fields[0]]
return result
def loadSubhalos(basePath, snapNum, fields=None):
""" Load all subhalo information from the entire group catalog for one snapshot
(optionally restrict to a subset given by fields). """
return loadObjects(basePath, snapNum, "Subhalo", "subgroups", fields)
def loadHalos(basePath, snapNum, fields=None):
""" Load all halo information from the entire group catalog for one snapshot
(optionally restrict to a subset given by fields). """
return loadObjects(basePath, snapNum, "Group", "groups", fields)
def loadHeader(basePath, snapNum):
""" Load the group catalog header. """
with h5py.File(gcPath(basePath, snapNum), 'r') as f:
header = dict(f['Header'].attrs.items())
return header
def load(basePath, snapNum):
""" Load complete group catalog all at once. """
r = {}
r['subhalos'] = loadSubhalos(basePath, snapNum)
r['halos'] = loadHalos(basePath, snapNum)
r['header'] = loadHeader(basePath, snapNum)
return r
def loadSingle(basePath, snapNum, haloID=-1, subhaloID=-1):
""" Return complete group catalog information for one halo or subhalo. """
if (haloID < 0 and subhaloID < 0) or (haloID >= 0 and subhaloID >= 0):
raise Exception("Must specify either haloID or subhaloID (and not both).")
gName = "Subhalo" if subhaloID >= 0 else "Group"
searchID = subhaloID if subhaloID >= 0 else haloID
# old or new format
if 'fof_subhalo' in gcPath(basePath, snapNum):
# use separate 'offsets_nnn.hdf5' files
with h5py.File(offsetPath(basePath, snapNum), 'r') as f:
offsets = f['FileOffsets/'+gName][()]
else:
# use header of group catalog
with h5py.File(gcPath(basePath, snapNum), 'r') as f:
offsets = f['Header'].attrs['FileOffsets_'+gName]
offsets = searchID - offsets
fileNum = np.max(np.where(offsets >= 0))
groupOffset = offsets[fileNum]
# load halo/subhalo fields into a dict
result = {}
with h5py.File(gcPath(basePath, snapNum, fileNum), 'r') as f:
for haloProp in f[gName].keys():
result[haloProp] = f[gName][haloProp][groupOffset]
return result | ALMASim | /ALMASim-1.4.tar.gz/ALMASim-1.4/submodules/build/lib/illustris_python/groupcat.py | groupcat.py |
import numpy as np
import h5py
import six
from .groupcat import gcPath, offsetPath
from os.path import isfile
def treePath(basePath, chunkNum=0):
""" Return absolute path to a LHaloTree HDF5 file (modify as needed). """
filePath_list = [ basePath + '/trees/treedata/' + 'trees_sf1_135.' + str(chunkNum) + '.hdf5',
basePath + '/../postprocessing/trees/LHaloTree/trees_sf1_099.' + str(chunkNum) + '.hdf5', #new path scheme for TNG
basePath + '/../postprocessing/trees/LHaloTree/trees_sf1_080.' + str(chunkNum) + '.hdf5', #Thesan
]
for filePath in filePath_list:
if isfile(filePath):
return filePath
raise ValueError("No tree file found!")
def treeOffsets(basePath, snapNum, id):
""" Handle offset loading for a LHaloTree merger tree cutout. """
# load groupcat chunk offsets from header of first file (old or new format)
if 'fof_subhalo' in gcPath(basePath, snapNum):
# load groupcat chunk offsets from separate 'offsets_nnn.hdf5' files
with h5py.File(offsetPath(basePath, snapNum), 'r') as f:
groupFileOffsets = f['FileOffsets/Subhalo'][()]
offsetFile = offsetPath(basePath, snapNum)
prefix = 'Subhalo/LHaloTree/'
groupOffset = id
else:
# load groupcat chunk offsets from header of first file
with h5py.File(gcPath(basePath, snapNum), 'r') as f:
groupFileOffsets = f['Header'].attrs['FileOffsets_Subhalo']
# calculate target groups file chunk which contains this id
groupFileOffsets = int(id) - groupFileOffsets
fileNum = np.max(np.where(groupFileOffsets >= 0))
groupOffset = groupFileOffsets[fileNum]
offsetFile = gcPath(basePath, snapNum, fileNum)
prefix = 'Offsets/Subhalo_LHaloTree'
with h5py.File(offsetFile, 'r') as f:
# load the merger tree offsets of this subgroup
TreeFile = f[prefix+'File'][groupOffset]
TreeIndex = f[prefix+'Index'][groupOffset]
TreeNum = f[prefix+'Num'][groupOffset]
return TreeFile, TreeIndex, TreeNum
def singleNodeFlat(conn, index, data_in, data_out, count, onlyMPB):
""" Recursive helper function: Add a single tree node. """
data_out[count] = data_in[index]
count += 1
count = recProgenitorFlat(conn, index, data_in, data_out, count, onlyMPB)
return count
def recProgenitorFlat(conn, start_index, data_in, data_out, count, onlyMPB):
""" Recursive helper function: Flatten out the unordered LHaloTree, one data field at a time. """
firstProg = conn["FirstProgenitor"][start_index]
if firstProg < 0:
return count
# depth-ordered traversal (down mpb)
count = singleNodeFlat(conn, firstProg, data_in, data_out, count, onlyMPB)
# explore breadth
if not onlyMPB:
nextProg = conn["NextProgenitor"][firstProg]
while nextProg >= 0:
count = singleNodeFlat(conn, nextProg, data_in, data_out, count, onlyMPB)
nextProg = conn["NextProgenitor"][nextProg]
firstProg = conn["FirstProgenitor"][firstProg]
return count
def loadTree(basePath, snapNum, id, fields=None, onlyMPB=False):
""" Load portion of LHaloTree, for a given subhalo, re-arranging into a flat format. """
TreeFile, TreeIndex, TreeNum = treeOffsets(basePath, snapNum, id)
if TreeNum == -1:
print('Warning, empty return. Subhalo [%d] at snapNum [%d] not in tree.' % (id, snapNum))
return None
# config
gName = 'Tree' + str(TreeNum) # group name containing this subhalo
nRows = None # we do not know in advance the size of the tree
# make sure fields is not a single element
if isinstance(fields, six.string_types):
fields = [fields]
fTree = h5py.File(treePath(basePath, TreeFile), 'r')
# if no fields requested, return everything
if not fields:
fields = list(fTree[gName].keys())
# verify existence of requested fields
for field in fields:
if field not in fTree[gName].keys():
raise Exception('Error: Requested field '+field+' not in tree.')
# load connectivity for this entire TreeX group
connFields = ['FirstProgenitor', 'NextProgenitor']
conn = {}
for field in connFields:
conn[field] = fTree[gName][field][:]
# determine sub-tree size with dummy walk
dummy = np.zeros(conn['FirstProgenitor'].shape, dtype='int32')
nRows = singleNodeFlat(conn, TreeIndex, dummy, dummy, 0, onlyMPB)
result = {}
result['count'] = nRows
# walk through connectivity, one data field at a time
for field in fields:
# load field for entire tree? doing so is much faster than randomly accessing the disk
# during walk, assuming that the sub-tree is a large fraction of the full tree, and that
# the sub-tree is large in the absolute sense. the decision is heuristic, and can be
# modified (if you have the tree on a fast SSD, could disable the full load).
if nRows < 1000: # and float(nRows)/len(result['FirstProgenitor']) > 0.1
# do not load, walk with single disk reads
full_data = fTree[gName][field]
else:
# pre-load all, walk in-memory
full_data = fTree[gName][field][:]
# allocate the data array in the sub-tree
dtype = fTree[gName][field].dtype
shape = list(fTree[gName][field].shape)
shape[0] = nRows
data = np.zeros(shape, dtype=dtype)
# walk the tree, depth-first
count = singleNodeFlat(conn, TreeIndex, full_data, data, 0, onlyMPB)
# save field
result[field] = data
fTree.close()
# only a single field? then return the array instead of a single item dict
if len(fields) == 1:
return result[fields[0]]
return result | ALMASim | /ALMASim-1.4.tar.gz/ALMASim-1.4/submodules/build/lib/illustris_python/lhalotree.py | lhalotree.py |
# ALMa - An Active Leanring (data) Manager
ALMa elimiates the need for bookkeeping when using Active Learning. Read the blog post
on [Active Learning with ALMa](https://www.lighttag.io/blog/active-learning-manager/)
Made with heart by LightTag - The Text Annotation Tool For Teams.
We use ALMa to facilitate multi annotator active learning. Originally developed as a contribution for [Modal](https://github.com/modAL-python/modAL)
but moved to it's own library
## Install
```
pip install ALMa
```
## Use
Check out the full [example for text classification]('./examples/text_text_classification_with_modAL')
```python
from ALMa import ActiveLearningManager
manager = ActiveLearningManager(my_featurized_data, sources=optional_original_data)
learner = #...some active learning learner
for index in range(N_QUERIES):
index_to_label, query_instance = learner.query(manager.unlabeld)
original_ix = manager.get_original_index_from_unlabeled_index(index_to_label)
y = original_labels_train[original_ix]
label = (index_to_label, y)
manager.add_labels(labels)
learner.teach(X=manager.labeled, y=manager.labels)
```
| ALMa | /ALMa-0.0.3.tar.gz/ALMa-0.0.3/README.md | README.md |
# ALP4lib
ALP4lib is a Python module to control Vialux DMDs based on ALP4.X API.
This is not an independant open source module, it uses the .ddl files provided by [Vialux](http://www.vialux.de/en/).
This software is experimental, use it at your own risk.
## What is it?
This module wraps the basic function of the Vialux dlls to control a digitial micro-mirror device with a Vialux board.
Vialux provides dlls and also modules for Matlab and Labview but not for Python.
This code is tested with a device using the 4.3 version of the ALP API, other versions may have issues.
LED control related functions are not implemented.
Please read the ALP API description provided with the [Vialux](http://www.vialux.de/en/) ALP installation.
## Requirements
* Windows 32 or 64,
* Vialux drivers and the ALP4.X dll files available for download on [Vialux website](http://www.vialux.de/en/),
* Compatible Python 2.7 and 3.X.
## Citing the code
If the code was helpful to your work, please consider citing it:
[](https://zenodo.org/badge/latestdoi/70229567)
## Installation
### Manual installation
Just copy the ALP4.py file in the working directory.
### Automatic installation
To automatically download and copy the module in the python directory (so it can be available from anywhere), run the command:
```shell
pip install ALP4lib
```
or
```shell
easy_install ALP4lib
```
### Installation from source (Github)
To install the latest version from Github, clone the repository, and install the package with the the following command.
```shell script
python setup.py install
```
Instead of the normal installation, if you want to install ALP4lib in [development mode](https://setuptools.readthedocs.io/en/latest/userguide/development_mode.html), use:
```shell script
python setup.py develop
```
## Copy the .dll
The win32 ALPX.dll files should be directly in the working directory and the win64 dll with the same name in a /x64 subfolder.
Alternatively, a different dll directory can be set at the initialization of the DMD handler object.
The dlls have the following names respectively for the 4.1, 4.2 and 4.3 versions of the ALP API: 'alp41.dll', 'alp42.dll' and 'alp4395.dll'.
## A simple example
```python
import numpy as np
from ALP4 import *
import time
# Load the Vialux .dll
DMD = ALP4(version = '4.3')
# Initialize the device
DMD.Initialize()
# Binary amplitude image (0 or 1)
bitDepth = 1
imgBlack = np.zeros([DMD.nSizeY,DMD.nSizeX])
imgWhite = np.ones([DMD.nSizeY,DMD.nSizeX])*(2**8-1)
imgSeq = np.concatenate([imgBlack.ravel(),imgWhite.ravel()])
# Allocate the onboard memory for the image sequence
DMD.SeqAlloc(nbImg = 2, bitDepth = bitDepth)
# Send the image sequence as a 1D list/array/numpy array
DMD.SeqPut(imgData = imgSeq)
# Set image rate to 50 Hz
DMD.SetTiming(pictureTime = 20000)
# Run the sequence in an infinite loop
DMD.Run()
time.sleep(10)
# Stop the sequence display
DMD.Halt()
# Free the sequence from the onboard memory
DMD.FreeSeq()
# De-allocate the device
DMD.Free()
```
| ALP4lib | /ALP4lib-1.0.1.tar.gz/ALP4lib-1.0.1/README.md | README.md |
import ctypes as ct
import platform
import numpy as np
import six
if six.PY3:
import winreg as _winreg
else:
import _winreg
# Standard parameter
ALP_DEFAULT = 0
ALP_ENABLE = 1
## Return codes
ALP_OK = 0x00000000 # Successfull execution
ALP_NOT_ONLINE = 1001 # The specified ALP has not been found or is not ready.
## parameters ##
## AlpDevInquire
ALP_DEVICE_NUMBER = 2000 # Serial number of the ALP device
ALP_VERSION = 2001 # Version number of the ALP device
ALP_DEV_STATE = 2002 # current ALP status, see above
ALP_AVAIL_MEMORY = 2003 # ALP on-board sequence memory available for further sequence
# allocation (AlpSeqAlloc); number of binary pictures
# Temperatures. Data format: signed long with 1 LSB=1/256 degrees C
ALP_DDC_FPGA_TEMPERATURE = 2050 # V4100 Rev B: LM95231. External channel: DDC FPGAs Temperature Diode
ALP_APPS_FPGA_TEMPERATURE = 2051 # V4100 Rev B: LM95231. External channel: Application FPGAs Temperature Diode
ALP_PCB_TEMPERATURE = 2052 # V4100 Rev B: LM95231. Internal channel. "Board temperature"
# AlpDevControl - ControlTypes & ControlValues
ALP_SYNCH_POLARITY = 2004 # Select frame synch output signal polarity
ALP_TRIGGER_EDGE = 2005 # Select active input trigger edge (slave mode)
ALP_LEVEL_HIGH = 2006 # Active high synch output
ALP_LEVEL_LOW = 2007 # Active low synch output
ALP_EDGE_FALLING = 2008 # High to low signal transition
ALP_EDGE_RISING = 2009 # Low to high signal transition
ALP_TRIGGER_TIME_OUT = 2014 # trigger time-out (slave mode)
ALP_TIME_OUT_ENABLE = 0 # Time-out enabled (default)
ALP_TIME_OUT_DISABLE = 1 # Time-out disabled */
ALP_USB_CONNECTION = 2016 # Re-connect after a USB interruption
ALP_DEV_DMDTYPE = 2021 # Select DMD type; only allowed for a new allocated ALP-3 device
ALP_DMDTYPE_XGA = 1 # 1024*768 mirror pixels (0.7" Type A, D3000)
ALP_DMDTYPE_SXGA_PLUS = 2 # 1400*1050 mirror pixels (0.95" Type A, D3000)
ALP_DMDTYPE_1080P_095A = 3 # 1920*1080 mirror pixels (0.95" Type A, D4x00)
ALP_DMDTYPE_XGA_07A = 4 # 1024*768 mirror pixels (0.7" Type A, D4x00)
ALP_DMDTYPE_XGA_055A = 5 # 1024*768 mirror pixels (0.55" Type A, D4x00)
ALP_DMDTYPE_XGA_055X = 6 # 1024*768 mirror pixels (0.55" Type X, D4x00)
ALP_DMDTYPE_WUXGA_096A = 7 # 1920*1200 mirror pixels (0.96" Type A, D4100)
ALP_DMDTYPE_WQXGA_400MHZ_090A = 8 # 2560*1600 mirror pixels (0.90" Type A, DLPC910) at standard clock rate (400 MHz)
ALP_DMDTYPE_WQXGA_480MHZ_090A = 9 # WQXGA at extended clock rate (480 MHz); WARNING: This mode requires temperature control of DMD
ALP_DMDTYPE_WXGA_S450 = 12 # # 1280x800 (DLP650LNIR WXGA S450)
ALP_DMDTYPE_DISCONNECT = 255 # behaves like 1080p (D4100)
ALP_DEV_DISPLAY_HEIGHT = 2057 # number of mirror rows on the DMD
ALP_DEV_DISPLAY_WIDTH = 2058 # number of mirror columns on the DMD
ALP_DEV_DMD_MODE = 2064 # query/set DMD PWR_FLOAT mode, valid options: ALP_DMD_RESUME (normal operation: "wake up DMD"), ALP_DMD_POWER_FLOAT
ALP_DMD_RESUME = 0 # default mode, Wake up DMD; Auto-Shutdown on loss of supply voltage safely switches off DMD
ALP_DMD_POWER_FLOAT = 1 # power down, release micro mirrors from deflected state
ALP_PWM_LEVEL = 2063 # PWM pin duty-cycle as percentage: 0..100%; after AlpDevAlloc: 0%
# AlpDevControlEx
ALP_DEV_DYN_SYNCH_OUT1_GATE = 2023
ALP_DEV_DYN_SYNCH_OUT2_GATE = 2024
ALP_DEV_DYN_SYNCH_OUT3_GATE = 2025
class tAlpDynSynchOutGate(ct.Structure):
# For ControlType ALP_DEV_DYN_TRIG_OUT[1..3]_GATE of function AlpDevControlEx
# Configure compiler to not insert padding bytes! (e.g. #pragma pack)
_fields_ = [("Period", ct.c_ubyte), # Period=1..16 enables output; 0: tri-state
("Polarity", ct.c_ubyte), # 0: active pulse is low, 1: high
("Gate", ct.c_ubyte * 16)] # #Period number of bytes; each one is 0 or 1
# Period bytes are used!
# AlpSeqControl - ControlTypes
ALP_SEQ_REPEAT = 2100 # Non-continuous display of a sequence (AlpProjStart) allows
# for configuring the number of sequence iterations.
ALP_SEQ_REPETE = ALP_SEQ_REPEAT # According to the typo made in primary documentation (ALP API description)
ALP_FIRSTFRAME = 2101 # First image of this sequence to be displayed.
ALP_LASTFRAME = 2102 # Last image of this sequence to be displayed.
ALP_BITNUM = 2103 # A sequence can be displayed with reduced bit depth for faster speed.
ALP_BIN_MODE = 2104 # Binary mode: select from ALP_BIN_NORMAL and ALP_BIN_UNINTERRUPTED (AlpSeqControl)
ALP_BIN_NORMAL = 2105 # Normal operation with progammable dark phase
ALP_BIN_UNINTERRUPTED = 2106 # Operation without dark phase
ALP_PWM_MODE = 2107 # ALP_DEFAULT, ALP_FLEX_PWM
ALP_FLEX_PWM = 3 # ALP_PWM_MODE: all bit planes of the sequence are displayed as
# fast as possible in binary uninterrupted mode;
# use ALP_SLAVE mode to achieve a custom pulse-width modulation timing for generating gray-scale
ALP_DATA_FORMAT = 2110 # Data format and alignment
ALP_DATA_MSB_ALIGN = 0 # Data is MSB aligned (default)
ALP_DATA_LSB_ALIGN = 1 # Data is LSB aligned
ALP_DATA_BINARY_TOPDOWN = 2 # Data is packed binary, top row first; bit7 of a byte = leftmost of 8 pixels
ALP_DATA_BINARY_BOTTOMUP = 3 # Data is packed binary, bottom row first
# XGA: one pixel row occupies 128 byte of binary data.
# Byte0.Bit7 = top left pixel (TOPDOWN format)
# 1080p and WUXGA: one pixel row occupies 256 byte of binary data.
# Byte0.Bit7 = top left pixel (TOPDOWN format)
# SXGA+: one pixel row occupies 176 byte of binary data. First byte ignored.
# Byte1.Bit7 = top left pixel (TOPDOWN format)
ALP_SEQ_PUT_LOCK = 2119 # ALP_DEFAULT: Lock Sequence Memory in AlpSeqPut;
# Not ALP_DEFAULT: do not lock, instead allow writing sequence image data even currently displayed
ALP_FIRSTLINE = 2111 # Start line position at the first image
ALP_LASTLINE = 2112 # Stop line position at the last image
ALP_LINE_INC = 2113 # Line shift value for the next frame
ALP_SCROLL_FROM_ROW = 2123 # combined value from ALP_FIRSTFRAME and ALP_FIRSTLINE
ALP_SCROLL_TO_ROW = 2124 # combined value from ALP_LASTFRAME and ALP_LASTLINE
# Frame Look Up Table (FLUT): sequence settings select how to use the FLUT.
# The look-up table itself is shared across all sequences.
# (use ALP_FLUT_SET_MEMORY controls for accessing it)
ALP_FLUT_MODE = 2118 # Select Frame LookUp Table usage mode:
ALP_FLUT_NONE = 0 # linear addressing, do not use FLUT (default)
ALP_FLUT_9BIT = 1 # Use FLUT for frame addressing: 9-bit entries
ALP_FLUT_18BIT = 2 # Use FLUT for frame addressing: 18-bit entries
ALP_FLUT_ENTRIES9 = 2120 # Determine number of FLUT entries; default=1
# Entries: supports all values from 1 to ALP_FLUT_MAX_ENTRIES9
ALP_FLUT_OFFSET9 = 2122 # Determine offset of FLUT index; default=0
# Offset supports multiples of 256;
# For ALP_FLUT_18BIT, the effective index is half of the 9-bit index.
# --> "ALP_FLUT_ENTRIES18" and "ALP_FLUT_FRAME_OFFSET18" are 9-bit settings divided by 2
# The API does not reject overflow! (FRAME_OFFSET+ENTRIES > MAX_ENTRIES).
# The user is responsible for correct settings.
ALP_SEQ_DMD_LINES = 2125 # Area of Interest: Value = MAKELONG(StartRow, RowCount)
ALP_X_SHEAR_SELECT = 2132 # ALP_DEFAULT or ALP_ENABLE
ALP_DMD_MASK_SELECT = 2134 # ALP_DEFAULT or ALP_ENABLE, ALP_DMD_MASK_16X8
ALP_DMD_MASK_16X16 = ALP_ENABLE
ALP_DMD_MASK_16X8 = 2 # XGA only
def MAKELONG(StartRow, RowCount):
return StartRow + RowCount << 16
# AlpSeqInquire
ALP_BITPLANES = 2200 # Bit depth of the pictures in the sequence
ALP_PICNUM = 2201 # Number of pictures in the sequence
ALP_PICTURE_TIME = 2203 # Time between the start of consecutive pictures in the sequence in microseconds,
# the corresponding in frames per second is
# picture rate [fps] = 1 000 000 / ALP_PICTURE_TIME [µs]
ALP_ILLUMINATE_TIME = 2204 # Duration of the display of one picture in microseconds
ALP_SYNCH_DELAY = 2205 # Delay of the start of picture display with respect
# to the frame synch output (master mode) in microseconds
ALP_SYNCH_PULSEWIDTH = 2206 # Duration of the active frame synch output pulse in microseconds
ALP_TRIGGER_IN_DELAY = 2207 # Delay of the start of picture display with respect to the
# active trigger input edge in microseconds
ALP_MAX_SYNCH_DELAY = 2209 # Maximal duration of frame synch output to projection delay in microseconds
ALP_MAX_TRIGGER_IN_DELAY = 2210 # Maximal duration of trigger input to projection delay in microseconds
ALP_MIN_PICTURE_TIME = 2211 # Minimum time between the start of consecutive pictures in microseconds
ALP_MIN_ILLUMINATE_TIME = 2212 # Minimum duration of the display of one picture in microseconds
# depends on ALP_BITNUM and ALP_BIN_MODE
ALP_MAX_PICTURE_TIME = 2213 # Maximum value of ALP_PICTURE_TIME
# ALP_PICTURE_TIME = ALP_ON_TIME + ALP_OFF_TIME
# ALP_ON_TIME may be smaller than ALP_ILLUMINATE_TIME
ALP_ON_TIME = 2214 # Total active projection time
ALP_OFF_TIME = 2215 # Total inactive projection time
# AlpProjInquire & AlpProjControl & ...Ex - InquireTypes, ControlTypes & Values */
ALP_PROJ_MODE = 2300 # Select from ALP_MASTER and ALP_SLAVE mode */
ALP_MASTER = 2301 # The ALP operation is controlled by internal */
# timing, a synch signal is sent out for any */
# picture displayed */
ALP_SLAVE = 2302 # The ALP operation is controlled by external */
# trigger, the next picture in a sequence is */
# displayed after the detection of an external */
# input trigger signal. */
ALP_PROJ_STEP = 2329 # ALP operation should run in ALP_MASTER mode,
# but each frame is repeatedly displayed
# until a trigger event is received.
# Values (conditions): ALP_LEVEL_HIGH |
# LOW, ALP_EDGE_RISING | FALLING.
# ALP_DEFAULT disables the trigger and
# makes the sequence progress "as usual".
# If an event is "stored" in edge mode due
# to a past edge, then it will be
# discarded during
# AlpProjControl(ALP_PROJ_STEP). */
ALP_PROJ_SYNC = 2303 # Select from ALP_SYNCHRONOUS and ALP_ASYNCHRONOUS mode */
ALP_SYNCHRONOUS = 2304 # The calling program gets control back after completion */
# of sequence display. */
ALP_ASYNCHRONOUS = 2305 # The calling program gets control back immediatelly. */
ALP_PROJ_INVERSION = 2306 # Reverse dark into bright */
ALP_PROJ_UPSIDE_DOWN = 2307 # Turn the pictures upside down */
ALP_PROJ_STATE = 2400 # Inquire only */
ALP_FLUT_MAX_ENTRIES9 = 2324 # Inquire FLUT size */
# Transfer FLUT memory to ALP. Use AlpProjControlEx and pUserStructPtr of type tFlutWrite. */
ALP_FLUT_WRITE_9BIT = 2325 # 9-bit look-up table entries */
ALP_FLUT_WRITE_18BIT = 2326 # 18-bit look-up table entries */
# for ALP_FLUT_WRITE_9BIT, ALP_FLUT_WRITE_18BIT (both versions share the same data type),
# to be used with AlpProjControlEx */
class tFlutWrite(ct.Structure):
_fields_ = [("nOffset", ct.c_long), # first LUT entry to transfer (write FrameNumbers[0] to LUT[nOffset]):
# number of 9-bit or 18-bit entries to transfer;
# For nSize=ALP_DEFAULT(0) the API sets nSize to its maximum value. This
# requires nOffset=0
("nSize", ct.c_long),
# nOffset+nSize must not exceed ALP_FLUT_MAX_ENTRIES9 (ALP_FLUT_WRITE_9BIT)
# or ALP_FLUT_MAX_ENTRIES9/2 (ALP_FLUT_WRITE_18BIT).
# The ALP API reads only the first nSize entries from this array. It
# extracts 9 or 18 least significant bits from each entry.
("FrameNumbers", ct.c_ulong * 4096)]
ALP_X_SHEAR = 2337 # use AlpProjControlEx and UserStructPtr of type tAlpShearTable
class tAlpShearTable(ct.Structure): # AlpProjControlEx, ControlType ALP_X_SHEAR
_fields_ = [("nOffset", ct.c_long),
("nSize", ct.c_long),
("nShiftDistance", ct.c_long * 2048)] # values range from 0 to 511
ALP_DMD_MASK_WRITE = 2339 # use AlpProjControlEx and UserStructPtr of type tAlpDmdMask
class tAlpDmdMask(ct.Structure): # AlpProjControlEx, ControlType ALP_DMD_MASK_WRITE
_fields_ = [("nRowOffset", ct.c_long), # Bitmap position in a 16x16 mask, ALP_DEFAULT=0
("nRowCount", ct.c_long), # rows to be written or ALP_DEFAULT (full DMD 16x16 mask)
("Bitmap", ct.c_ubyte * 2048)] # each bit controls a block of DMD pixels
## Sequence Queue API Extension:
ALP_PROJ_QUEUE_MODE = 2314
ALP_PROJ_LEGACY = 0 # ALP_DEFAULT: emulate legacy mode: 1 waiting position. AlpProjStart replaces enqueued and still waiting sequences */
ALP_PROJ_SEQUENCE_QUEUE = 1 # manage active sequences in a queue */
ALP_PROJ_QUEUE_ID = 2315 # provide the QueueID (ALP_ID) of the most recently enqueued sequence (or ALP_INVALID_ID) */
ALP_PROJ_QUEUE_MAX_AVAIL = 2316 # total number of waiting positions in the sequence queue */
ALP_PROJ_QUEUE_AVAIL = 2317 # number of available waiting positions in the queue */
# bear in mind that when a sequence runs, it is already dequeued and does not consume a waiting position any more */
ALP_PROJ_PROGRESS = 2318 # (AlpProjInquireEx) inquire detailled progress of the running sequence and the queue */
ALP_PROJ_RESET_QUEUE = 2319 # Remove all enqueued sequences from the queue. The currently running sequence is not affected. ControlValue must be ALP_DEFAULT */
ALP_PROJ_ABORT_SEQUENCE = 2320 # abort the current sequence (ControlValue=ALP_DEFAULT) or a specific sequence (ControlValue=QueueID); abort after last frame of current iteration */
ALP_PROJ_ABORT_FRAME = 2321 # similar, but abort after next frame */
# Only one abort request can be active at a time. If it is requested to
# abort another sequence before the old request is completed, then
# AlpProjControl returns ALP_NOT_IDLE. (Please note, that AlpProjHalt
# and AlpDevHalt work anyway.) If the QueueID points to a sequence
# behind an indefinitely started one (AlpProjStartCont) then it returns
# ALP_PARM_INVALID in order to prevent dead-locks. */
ALP_PROJ_WAIT_UNTIL = 2323 # When does AlpProjWait complete regarding the last frame? or after picture time of last frame */
ALP_PROJ_WAIT_PIC_TIME = 0 # ALP_DEFAULT: AlpProjWait returns after picture time */
ALP_PROJ_WAIT_ILLU_TIME = 1 # AlpProjWait returns after illuminate time (except binary uninterrupted sequences, because an "illuminate time" is not applicable there) */
# for AlpProjInquireEx(ALP_PROJ_PROGRESS):
class tAlpProjProgress(ct.Structure):
_fields_ = [("CurrentQueueId", ct.c_ulong),
("SequenceId", ct.c_ulong), # Consider that a sequence can be enqueued multiple times!
("nWaitingSequences", ct.c_ulong), # number of sequences waiting in the queue
# track iterations and frames: device-internal counters are incompletely
# reported; The API description contains more details on that.
("nSequenceCounter", ct.c_ulong), # Number of iterations to be done
("nSequenceCounterUnderflow", ct.c_ulong), # nSequenceCounter can
# underflow (for indefinitely long Sequences: AlpProjStartCont);
# nSequenceCounterUnderflow is 0 before, and non-null afterwards
("nFrameCounter", ct.c_ulong), # Frames left inside current iteration
("nPictureTime", ct.c_ulong), # micro seconds of each frame; this is
# reported, because the picture time of the original sequence could
# already have changed in between
("nFramesPerSubSequence", ct.c_ulong), # Each sequence iteration
# displays this number of frames. It is reported to the user just for
# convenience, because it depends on different parameters. */
("nFlagse", ct.c_ulong)]
# may be a combination of ALP_FLAG_SEQUENCE_ABORTING | SEQUENCE_INDEFINITE | QUEUE_IDLE | FRAME_FINISHED
ALP_FLAG_QUEUE_IDLE = ct.c_ulong(1)
ALP_FLAG_SEQUENCE_ABORTING = ct.c_ulong(2)
ALP_FLAG_SEQUENCE_INDEFINITE = ct.c_ulong(4) # AlpProjStartCont: this sequence runs indefinitely long, until aborted
ALP_FLAG_FRAME_FINISHED = ct.c_ulong(8) # illumination of last frame finished, picture time still progressing
ALP_FLAG_RSVD0 = ct.c_ulong(16) # reserved
# for AlpSeqPutEx():
class tAlpLinePut(ct.Structure):
_fields_ = [("TransferMode", ct.c_long), # common first member of AlpSeqPutEx' UserStructPtr argument
("PicOffset", ct.c_long),
("PicLoad", ct.c_long),
("LineOffset", ct.c_long),
("LineLoad", ct.c_long)]
ALP_PUT_LINES = ct.c_long(1) # not ulong; need to be long in the tAlpLinePut struct
ALP_ERRORS = {1001: 'The specified ALP device has not been found or is not ready.',
1002: 'The ALP device is not in idle state.',
1003: 'The specified ALP device identifier is not valid.',
1004: 'The specified ALP device is already allocated.',
1005: 'One of the parameters is invalid.',
1006: 'Error accessing user data.',
1007: 'The requested memory is not available (full?).',
1008: 'The sequence specified is currently in use.',
1009: 'The ALP device has been stopped while image data transfer was active.',
1010: 'Initialization error.',
1011: 'Communication error.',
1012: 'The specified ALP has been removed.',
1013: 'The onboard FPGA is unconfigured.',
1014: 'The function is not supported by this version of the driver file VlxUsbLd.sys.',
1018: 'Waking up the DMD from PWR_FLOAT did not work (ALP_DMD_POWER_FLOAT)',
1019: 'Support in ALP drivers missing. Update drivers and power-cycle device.',
1020: 'SDRAM Initialization failed.'}
class ALPError(Exception):
def __init__(self, error_code):
super(ALPError, self).__init__(ALP_ERRORS[error_code])
def afficheur(bitPlane):
nSizeX = 2560
nSizeY = 1600
display = np.zeros((nSizeY, nSizeX))
for jj in range(nSizeY):
for ii in range(nSizeX // 8):
Q = bitPlane[jj * nSizeX // 8 + ii]
R = [0, 0, 0, 0, 0, 0, 0, 0]
k = 7
while Q != 0:
R[k] = (Q % 2)
Q = Q // 2
k -= 1
for ll in range(8):
display[jj, ii * 8 + ll] = R[ll]
return display
def img_to_bitplane(imgArray, bitShift):
"""
Convert a binary image into a bitplane.
"""
bitPlane = np.packbits(imgArray).tolist()
return bitPlane
class ALP4(object):
"""
This class controls a Vialux DMD board based on the Vialux ALP 4.X API.
"""
def __init__(self, version='4.3', libDir=None):
os_type = platform.system()
if libDir is None:
try:
reg = _winreg.ConnectRegistry(None, _winreg.HKEY_LOCAL_MACHINE)
key = _winreg.OpenKey(reg, r"SOFTWARE\ViALUX\ALP-" + version)
libDir = (_winreg.QueryValueEx(key, "Path"))[0] + "/ALP-{0} API/".format(version)
except EnvironmentError:
raise ValueError("Cannot auto detect libDir! Please specify it manually.")
if libDir.endswith('/'):
libPath = libDir
else:
libPath = libDir + '/'
## Load the ALP dll
if (os_type == 'Windows'):
if (ct.sizeof(ct.c_voidp) == 8): ## 64bit
libPath += 'x64/'
elif not (ct.sizeof(ct.c_voidp) == 4): ## 32bit
raise OSError('System not supported.')
else:
raise OSError('System not supported.')
if (version == '4.1'):
libPath += 'alpD41.dll'
elif (version == '4.2'):
libPath += 'alpD41.dll'
elif (version == '4.3'):
libPath += 'alp4395.dll'
print('Loading library: ' + libPath)
self._ALPLib = ct.CDLL(libPath)
## Class parameters
# ID of the current ALP device
self.ALP_ID = ct.c_ulong(0)
# Type of DMD found
self.DMDType = ct.c_long(0)
# Pointer to the last stored image sequence
self._lastDDRseq = None
# List of all Sequences
self.Seqs = []
def _checkError(self, returnValue, errorString, warning=False):
if not (returnValue == ALP_OK):
if not warning:
raise ALPError(returnValue)
else:
print(errorString + '\n' + ALP_ERRORS[returnValue])
def Initialize(self, DeviceNum=None):
"""
Initialize the communication with the DMD.
Usage:
Initialize(DeviceNum = None)
PARAMETERS
----------
DeviceNum : int
Serial number of the DMD to initialize, useful for multiple DMD control.
If not specify, open the first available DMD.
"""
if DeviceNum is None:
DeviceNum = ct.c_long(ALP_DEFAULT)
self._checkError(self._ALPLib.AlpDevAlloc(DeviceNum, ALP_DEFAULT, ct.byref(self.ALP_ID)), 'Cannot open DMD.')
self._checkError(self._ALPLib.AlpDevInquire(self.ALP_ID, ALP_DEV_DMDTYPE, ct.byref(self.DMDType)),
'Inquery fails.')
nSizeX = ct.c_long(0)
self._checkError(self._ALPLib.AlpDevInquire(self.ALP_ID, ALP_DEV_DISPLAY_WIDTH, ct.byref(nSizeX)),
'Inquery ALP_DEV_DISPLAY_WIDTH fails.')
self.nSizeX = nSizeX.value
nSizeY = ct.c_long(0)
self._checkError(self._ALPLib.AlpDevInquire(self.ALP_ID, ALP_DEV_DISPLAY_HEIGHT, ct.byref(nSizeY)),
'Inquery ALP_DEV_DISPLAY_HEIGHT fails.')
self.nSizeY = nSizeY.value
print('DMD found, resolution = ' + str(self.nSizeX) + ' x ' + str(self.nSizeY) + '.')
def SeqAlloc(self, nbImg=1, bitDepth=1):
"""
This function provides ALP memory for a sequence of pictures. All pictures of a sequence have the
same bit depth. The function allocates memory from the ALP board RAM. The user has no direct
read/write access. ALP functions provide data transfer using the sequence memory identifier
(SequenceId) of type ALP_ID.
Pictures can be loaded into the ALP RAM using the SeqPut function.
The availability of ALP memory can be tested using the DevInquire function.
When a sequence is no longer required, release it using SeqFree.
Usage:
SeqAlloc(nbImg = 1, bitDepth = 1)
PARAMETERS
----------
nbImg : int
Number of images in the sequence.
bitDepth : int
Quantization of the image between 1 (on/off) and 8 (256 pwm grayscale levels).
See ALPLib.AlpSeqAlloc in the ALP API description for more information.
RETURNS
-------
SequenceID : ctypes c_ulong
Id of the created sequence.
This id is stored internally as the last created sequence and
erase the previous one. When a sequence relasted function is used without
specifying a SequenceId, it will use the stored SequenceId.
"""
SequenceId = ct.c_long(0)
self.Seqs.append(SequenceId) # Put SequenceId in list of all Sequences to keep track of them
# Allocate memory on the DDR RAM for the sequence of image.
self._checkError(
self._ALPLib.AlpSeqAlloc(self.ALP_ID, ct.c_long(bitDepth), ct.c_long(nbImg), ct.byref(SequenceId)),
'Cannot allocate image sequence.')
self._lastDDRseq = SequenceId
return SequenceId
def SeqPutEx(self, imgData, LineOffset, LineLoad, SequenceId=None, PicOffset=0, PicLoad=0, dataFormat='Python'):
"""
Image data transfer using AlpSeqPut is based on whole DMD frames. Applications that only
update small regions inside a frame suffer from overhead of this default behavior. An extended
ALP API function is available to reduce this overhead.
The AlpSeqPutEx function offers the same functionality as the standard function (AlpSeqPut),
but in addition, it is possible to select a section within a sequence frame using the
LineOffset and LineLoad parameters of the tAlpLinePut data-structure (see below) and update
only this section of the SDRAM-memory associated with the sequence for a range of
sequence-pictures (selected via the PicOffset and PicLoad parameters of tAlpLinePut in
similarity to AlpSeqPut).
This results in accelerated transfer-time of small image data updates (due to the fact that the
amount of transferred data is reduced).
Therefore, the user only passes the lines of the pictures he wants to update via the UserArrayPtr
(that would be PicLoad*LineLoad lines in total).
PARAMETERS
----------
imgData : list, 1D array or 1D ndarray
Data stream corresponding to a sequence of nSizeX by nSizeX images.
Values has to be between 0 and 255.
LineOffset : int
Defines the offset of the frame-section. The frame-data of this section is transferred
for each of the frames selected with PicOffset and PicLoad. The value of this
parameter must be greater or equal to zero, otherwise ALP_PARM_INVALID is returned.
LineLoad : int
Defines the size of the frame-section. If the value of the parameter is
less than zero or if LineOffset+LineLoad exceeds the number of lines
per sequence-frame, ALP_PARM_INVALID is returned. If LineLoad is
zero, this value is adjusted to include all lines of the frame, starting at
line LineOffset
SequenceId : ctypes c_long
Sequence identifier. If not specified, set the last sequence allocated in the DMD board memory
PicOffset : int, optional
Picture number in the sequence (starting at 0) where the data upload is
started; the meaning depends upon ALP_DATA_FORMAT.
By default, PifOffset = 0.
PicLoad : int, optional
number of pictures that are to be loaded into the sequence memory.
Depends on ALP_DATA_FORMAT.
PicLoad = 0 correspond to a complete sequence.
By default, PicLoad = 0.
dataFormat : string, optional
Specify the type of data sent as image.
Should be ' Python' or 'C'.
If the data is of Python format, it is converted into a C array before sending to the DMD via the dll.
By default dataFormat = 'Python'
"""
if not SequenceId:
SequenceId = self._lastDDRseq
LinePutParam = tAlpLinePut(ALP_PUT_LINES,
ct.c_long(PicOffset),
ct.c_long(PicLoad),
ct.c_long(LineOffset),
ct.c_long(LineLoad))
if dataFormat not in ['Python', 'C']:
raise ValueError('dataFormat must be one of "Python" or "C"')
if dataFormat == 'Python':
pImageData = imgData.astype(np.uint8).ctypes.data_as(ct.c_void_p)
elif dataFormat == 'C':
pImageData = ct.cast(imgData, ct.c_void_p)
self._checkError(self._ALPLib.AlpSeqPutEx(self.ALP_ID, SequenceId, LinePutParam, pImageData),
'Cannot send image sequence to device.')
def SeqPut(self, imgData, SequenceId=None, PicOffset=0, PicLoad=0, dataFormat='Python'):
"""
This function allows loading user supplied data via the USB connection into the ALP memory of a
previously allocated sequence (AlpSeqAlloc) or a part of such a sequence. The loading operation can
run concurrently to the display of other sequences. Data cannot be loaded into sequences that are
currently started for display. Note: This protection can be disabled by ALP_SEQ_PUT_LOCK.
The function loads PicNum pictures into the ALP memory reserved for the specified sequence starting
at picture PicOffset. The calling program is suspended until the loading operation is completed.
The ALP API compresses image data before sending it over USB. This results in a virtual
improvement of data transfer speed. Compression ratio is expected to vary depending on image data.
Incompressible data do not cause overhead delays.
Usage:
SeqPut(imgData, nbImg = 1, bitDepth = 1)
PARAMETERS
----------
imgData : list, 1D array or 1D ndarray
Data stream corresponding to a sequence of nSizeX by nSizeX images.
Values has to be between 0 and 255.
SequenceId : ctypes c_long
Sequence identifier. If not specified, set the last sequence allocated in the DMD board memory
PicOffset : int, optional
Picture number in the sequence (starting at 0) where the data upload is
started; the meaning depends upon ALP_DATA_FORMAT.
By default, PifOffset = 0.
PicLoad : int, optional
number of pictures that are to be loaded into the sequence memory.
Depends on ALP_DATA_FORMAT.
PicLoad = 0 correspond to a complete sequence.
By default, PicLoad = 0.
dataFormat : string, optional
Specify the type of data sent as image.
Should be ' Python' or 'C'.
If the data is of Python format, it is converted into a C array before sending to the DMD via the dll.
By default dataFormat = 'Python'
SEE ALSO
--------
See ALPLib.AlpSeqPut in the ALP API description for more information.
"""
if not SequenceId:
SequenceId = self._lastDDRseq
if dataFormat == 'Python':
pImageData = imgData.astype(np.uint8).ctypes.data_as(ct.c_void_p)
elif dataFormat == 'C':
pImageData = ct.cast(imgData, ct.c_void_p)
else:
raise ValueError('dataFormat must be one of "Python" or "C"')
self._checkError(
self._ALPLib.AlpSeqPut(self.ALP_ID, SequenceId, ct.c_long(PicOffset), ct.c_long(PicLoad), pImageData),
'Cannot send image sequence to device.')
def ImgToBitPlane(self, imgArray, bitShift=0):
"""
Create a bit plane from the imgArray.
The bit plane is an (nSizeX x nSizeY / 8) array containing only the bit values
corresponding to the bit number bitShift.
For a bit depth = 8, 8 bit planes can be extracted from the imgArray by iterating ImgToBitPlane.
WARNING: It is recommended to directly generate images as bitplanes for better performances.
Usage:
ImgToBitPlane(imgArray,bitShift = 0)
PARAMETERS
----------
imgArray: 1D array or list
An image of the same resolution as the DMD (nSizeX by nSizeY).
bitShift: int, optional
Bit plane to extract form the imgArray (0 to 8),
Has to be <= bit depth.
RETURNS
-------
bitPlane: list
Array (nSizeX x nSizeY)/8
"""
return img_to_bitplane(imgArray, bitShift)
def SetTiming(self, SequenceId=None, illuminationTime=None, pictureTime=None, synchDelay=None,
synchPulseWidth=None, triggerInDelay=None):
"""
Set the timing properties of the sequence to display.
Usage:
SetTiming( SequenceId = None, illuminationTime = None, pictureTime = None, synchDelay = None, \
synchPulseWidth = None, triggerInDelay = None)
PARAMETERS
----------
SequenceId : c_ulong, optional
Identified of the sequence. If not specified, set the last sequence allocated in the DMD board memory
illuminationTime: c_ulong, optional
Display time of a single image of the sequence in microseconds.
If not specified, use the highest possible value compatible with pictureTime.
pictureTime : int, optional
Time between the start of two consecutive picture, up to 10^7 microseconds = 10 seconds.
With illuminationTime, it sets the display rate.
If not specified, the value is set to minimize the dark time according illuminationTime.
If illuminationTime is also not specified, set to a frame rate of 30Hz.
synchDelay : Specifies the time delay between the start of the output sync pulse and the start of the display (master mode).
Value between 0 and 130,000 microseconds. Set to 0 if not specified.
synchPulseWidth : Duration of the sync output pulse.
By default equals synchDelay + illuminationTime in normal mode.
By default equals ALP_ILLUMINATION_TIME in binary uninterrupted mode.
triggerInDelay : Length of the trigger signal in microseconds, set to 0 by default.
SEE ALSO
--------
See ALPLib.AlpSeqAlloc in the ALP API description for more information.
"""
if (SequenceId is None) and (self._lastDDRseq):
SequenceId = self._lastDDRseq
if (SequenceId is None):
raise ValueError('No sequence to display.')
if (synchDelay is None):
synchDelay = ALP_DEFAULT
if (synchPulseWidth is None):
synchPulseWidth = ALP_DEFAULT
if (triggerInDelay is None):
triggerInDelay = ALP_DEFAULT
if (illuminationTime is None):
illuminationTime = ALP_DEFAULT
if (pictureTime is None):
pictureTime = ALP_DEFAULT
self._checkError(
self._ALPLib.AlpSeqTiming(self.ALP_ID, SequenceId, ct.c_long(illuminationTime), ct.c_long(pictureTime),
ct.c_long(synchDelay), ct.c_long(synchPulseWidth), ct.c_long(triggerInDelay)),
'Cannot set timing.')
def DevInquire(self, inquireType):
"""
Ask the controller board the value of a specified parameter about the ALP device.
Usage: Inquire(request)
PARAMETERS
----------
inquireType : ctypes c_ulong
Sepcifies the type of value to return.
RETURNS
-------
value : c_double
Value of the requested parameter.
SEE ALSO
--------
See AlpDevInquire in the ALP API description for request types.
"""
ret = ct.c_long(0)
self._checkError(self._ALPLib.AlpDevInquire(self.ALP_ID, inquireType, ct.byref(ret)), 'Error sending request.')
return ret.value
def SeqInquire(self, inquireType, SequenceId=None):
"""
Ask the controller board the value of a specified parameter about an image sequence.
Usage: Inquire(self, inquireType, SequenceId = None)
PARAMETERS
----------
inquireType : ctypes c_ulong
Sepcifies the type of value to return.
SequenceId : ctyles c_long, optional
Identified of the sequence. If not specified, set the last sequence allocated in the DMD board memory
RETURNS
-------
value : int
Value of the requested parameter.
SEE ALSO
--------
See AlpSeqInquire in the ALP API description for request types.
"""
ret = ct.c_long(0)
if (SequenceId is None) and (self._lastDDRseq):
SequenceId = self._lastDDRseq
self._checkError(self._ALPLib.AlpSeqInquire(self.ALP_ID, SequenceId, inquireType, ct.byref(ret)),
'Error sending request.')
return ret.value
def ProjInquire(self, inquireType, SequenceId=None):
"""
Usage: ProjInquire(self, inquireType, SequenceId = None)
PARAMETERS
----------
request : ctypes c_ulong
Sepcifies the type of value to return.
SequenceId : ctyles c_long, optional
Identified of the sequence. If not specified, set the last sequence allocated in the DMD board memory
RETURNS
-------
value : int
Value of the requested parameter.
SEE ALSO
--------
See AlpProjInquire in the ALP API description for request types.
"""
ret = ct.c_long(0)
if (SequenceId is None) and (self._lastDDRseq):
SequenceId = self._lastDDRseq
self._checkError(self._ALPLib.AlpProjInquire(self.ALP_ID, SequenceId, inquireType, ct.byref(ret)),
'Error sending request.')
return ret.value
def ProjInquireEx(self, inquireType, SequenceId=None):
"""
Data objects that do not fit into a simple 32-bit number can be inquired using this function.
Meaning and layout of the data depend on the InquireType.
Usage: ProjInquireEx(self, inquireType, UserStructPtr, SequenceId = None)
PARAMETERS
----------
inquireType : ctypes c_ulong
Sepcifies the type of value to return.
SequenceId : ctypes c_long, optional
Identified of the sequence. If not specified, set the last sequence allocated in the DMD board memory
RETURNS
-------
UserStructPtr : ctypes POINTER
Pointer to a data structure which shall be filled out by AlpSeqInquireEx.
SEE ALSO
--------
See AlpProjInquireEx in the ALP API description for request types.
"""
UserStructPtr = ct.c_double(0)
if (SequenceId is None) and (self._lastDDRseq):
SequenceId = self._lastDDRseq
self._checkError(self._ALPLib.AlpProjInquire(self.ALP_ID, SequenceId, inquireType, ct.byref(UserStructPtr)),
'Error sending request.')
return UserStructPtr
def DevControl(self, controlType, value):
"""
This function is used to change the display properties of the ALP.
The default values are assigned during device allocation by AllocateSequence.
Usage: Control(self, controlType, value)
PARAMETERS
----------
controlType: ctypes c_ulong
Specifies the type of value to set.
SEE ALSO
--------
See AlpDevControl in the ALP API description for control types.
"""
self._checkError(self._ALPLib.AlpDevControl(self.ALP_ID, controlType, ct.c_long(value)),
'Error sending request.')
def DevControlEx(self, controlType, userStruct):
"""
Data objects that do not fit into a simple 32-bit number can be written using this function. Meaning and
layout of the data depend on the ControlType.
Usage: Control(self, controlType, value)
PARAMETERS
----------
controlType : ctypes c_ulong
Specifies the type of value to set.
userStruct : tAlpDynSynchOutGate structure
It contains synch parameters.
SEE ALSO
--------
See AlpDevControlEx in the ALP API description for control types.
"""
self._checkError(self._ALPLib.AlpDevControlEx(self.ALP_ID, controlType, userStruct.byref()),
'Error sending request.')
def ProjControl(self, controlType, value):
"""
This function controls the system parameters that are in effect for all sequences. These parameters
are maintained until they are modified again or until the ALP is freed. Default values are in effect after
ALP allocation. All parameters can be read out using the AlpProjInquire function.
This function is only allowed if the ALP is in idle wait state (ALP_PROJ_IDLE), which can be enforced
by the AlpProjHalt function.
Usage: Control(self, controlType, value)
PARAMETERS
----------
controlType : attribute flag (ctypes c_ulong)
Specify the paramter to set.
value : c_double
Value of the parameter to set.
SEE ALSO
--------
See AlpProjControl in the ALP API description for control types.
"""
self._checkError(self._ALPLib.AlpProjControl(self.ALP_ID, controlType, ct.c_long(value)),
'Error sending request.')
def ProjControlEx(self, controlType, pointerToStruct):
"""
Data objects that do not fit into a simple 32-bit number can be written using this function. These
objects are unique to the ALP device, so they may affect display of all sequences.
Meaning and layout of the data depend on the ControlType.
Usage: Control(self, controlType, value)
PARAMETERS
----------
controlType : attribute flag (ctypes c_ulong)
Specify the paramter to set.
pointerToStruct : ctypes POINTER
Pointer to a tFlutWrite structure. Create a tFlutWrite object and pass it to the function using ctypes.byref
(Requires importing ctypes)
SEE ALSO
--------
See AlpProjControlEx in the ALP API description for control types.
"""
self._checkError(self._ALPLib.AlpProjContro(self.ALP_ID, controlType, pointerToStruct),
'Error sending request.')
def SeqControl(self, controlType, value, SequenceId=None):
"""
This function is used to change the display properties of a sequence.
The default values are assigned during sequence allocation by AlpSeqAlloc.
It is allowed to change settings of sequences that are currently in use.
However the new settings become effective after restart using AlpProjStart or AlpProjStartCont.
Usage: SeqControl(self, controlType, value, SequenceId = None)
PARAMETERS
----------
controlType : attribute flag (ctypes c_ulong)
Specify the paramter to set.
value : ctypes c_double
Value of the parameter to set.
SequenceId : ctypes c_long, optional
Identified of the sequence. If not specified, set the last sequence allocated in the DMD board memory
SEE ALSO
--------
See AlpSeqControl in the ALP API description for control types.
"""
if (SequenceId is None) and (self._lastDDRseq):
SequenceId = self._lastDDRseq
self._checkError(self._ALPLib.AlpSeqControl(self.ALP_ID, SequenceId, controlType, ct.c_long(value)),
'Error sending request.')
def FreeSeq(self, SequenceId=None):
"""
Frees a previously allocated sequence. The ALP memory reserved for the specified sequence in the device DeviceId is released.
Usage: FreeSeq(SequenceId = None)
PARAMETERS
----------
SequenceId : ctypes c_long, optional
Identified of the sequence. If not specified, free the last sequence allocated in the DMD board memory
"""
if (SequenceId is None) and (self._lastDDRseq):
SequenceId = self._lastDDRseq
self.Seqs.remove(SequenceId) # Removes the last SequenceId from sequence list
self._checkError(self._ALPLib.AlpSeqFree(self.ALP_ID, SequenceId), 'Unable to free the image sequence.',
warning=True)
def Run(self, SequenceId=None, loop=True):
"""
Display a sequence loaded into the DDR memory.
Usage: Run( SequenceId = None, loop = True)
PARAMETERS
----------
SequenceId : ctypes c_ulong
Id of the sequence to run.
If no sequence pointer is given, display the last sequence stored.
loop : bool
If True, display the sequence continuously using ALPLib.AlpProjStartCont.
If False, display it once using ALPLib.AlpProjStart. Set to True by default.
SEE ALSO
--------
See ALPLib.AlpProjStart and ALPLib.AlpProjStartCont in the ALP API description for more information.
"""
if (SequenceId is None) and (self._lastDDRseq):
SequenceId = self._lastDDRseq
if (SequenceId is None):
self._raiseError('No sequence to display.')
if loop:
self._checkError(self._ALPLib.AlpProjStartCont(self.ALP_ID, SequenceId), 'Cannot launch sequence.')
else:
self._checkError(self._ALPLib.AlpProjStart(self.ALP_ID, SequenceId), 'Cannot launch sequence.')
def Wait(self):
"""
This function is used to wait for the completion of the running sequence display.
Usage: Wait()
"""
self._checkError(self._ALPLib.AlpProjWait(self.ALP_ID), 'Cannot go in wait mode.')
def Halt(self):
"""
This function puts the ALP in an idle wait state. Current sequence display is canceled
(ALP_PROJ_IDLE) and the loading of sequences is aborted (AlpSeqPut).
Usage: Halt()
"""
self._checkError(self._ALPLib.AlpDevHalt(self.ALP_ID), 'Cannot stop device.')
def Free(self):
"""
This function de-allocates a previously allocated ALP device. The memory reserved by calling
AlpSeqAlloc is also released.
The ALP has to be in idle wait state, see also AlpDevHalt.
Usage: Free()
"""
self._checkError(self._ALPLib.AlpDevFree(self.ALP_ID), 'Cannot free device.')
del self._ALPLib | ALP4lib | /ALP4lib-1.0.1.tar.gz/ALP4lib-1.0.1/src/ALP4.py | ALP4.py |
ALPAFA
======
ALPAFA (/ˌælˈpæfə/, Algorithm for Lexicocentric Parameter Acquisition by Feature Assignment) is a
Python implementation of the algorithm described in chapter 2 of my 2015 PhD thesis, `Algorithmic
Acquisition of Focus Parameters <http://ling.auf.net/lingbuzz/003006>`_ (AAFP), which grew out of
an attempt to formalise certain proposals of the `Rethinking Comparative Syntax
<http://recos-dtal.mml.cam.ac.uk/>`_ (ReCoS) project. The algorithm takes a set of heads, each of
which is specified for a number of discoverable properties, and uses a "prominence" order of
properties to construct a minimal categorial system. This is achieved by attempting to assign each
new feature to all heads of the "largest" existing category, and, if this fails, making the
"smallest" categorial division possible. This algorithm is a putative component of domain-general
categorisation processes that is designed to capture the structured typological and historical
syntactic variation seen crosslinguistically through third factor principles. The motivation for
and behaviour of this algorithm is described in detail in chapters 1 and 2 of AAFP, including
extensions for unvalued features, movement triggers, and c-selection. Chapter 10 presents and
compares worked examples of inputs to the algorithm for toy fragment grammars of 6 varieties.
Links
=====
* AAFP: http://ling.auf.net/lingbuzz/003006
* ReCoS project: http://recos-dtal.mml.cam.ac.uk/
* Github: https://www.github.com/timothybazalgette/alpafa
* PyPI: https://pypi.python.org/pypi/alpafa
Installation
============
Install with pip:
``$ pip install alpafa`` or ``$ pip3 install alpafa``
Input file format
=================
Input files are closely based on the set notation used for input specifications in AAFP, but are
somewhat simplified for ease of creation and reading. They should be plain text files with UTF-8
compatible encodings. Place each head name on a separate line, followed by a colon and a
comma-separated list of properties. The prominence order should be placed on another line, starting
``prominence=``, followed by a comma-separated list of property names. Properties that are sets or
ordered pairs remain the same as in AAFP, though all sets must be given in full (i.e. no set-builder
notation). All spaces and blank lines will be ignored. An example specification for a toy fragment
of English is as follows::
Cmat: comp, {T}}
Cwh: comp, int, <whq, m>, {T}
Crel: n, comp, nom, {T}
Csub: comp, arg, {T}
T: <phi, m>, {V, Copadj}
V: v, cat, {Csub, D, Dwh}
Copadj: v, {A}
D: n, arg, {Crel, φ}
Dwh: n, arg, wh, whq, {Crel, φ}
only: invis, excl, {D}
OpCR: invis, {φ, N}
φ: n, nom, phi, low, {noun}, {N}
ind: n
N: n, cat, noun, low
A: cat
Focfeat: invis, foc, feat, {Cmat, Cwh, Crel, Csub, T, V, Copadj, D, Dwh, only, OpCR, φ, ind, N, A, Focfeat}
prominence = n, v, cat, noun, comp, arg, wh, whq, nom, phi, int, invis, excl, feat, foc, low
Included with the source code are example input files for the 6 toy fragment grammars in AAFP
chapter 10.
Ouput
=====
ALPAFA defaults to outputting a list of the heads with their categorial and dependent features,
along with a brief description of the algorithm's operation. Feature bundles are separated by tabs
for easy formatting when pasted into word processors - I may incorporate prettier printing in later
versions. There are a number of options for more detailed output of the algorithm's operation and
the categories created by it, discussed in the following section. ::
Cmat [-N,-V,-CAT,+COMP,-ARG,-INT] (-N,-V,-CAT,-COMP,-INVIS)
Cwh [-N,-V,-CAT,+COMP,-ARG,+INT] (-N,-V,-CAT,-COMP,-INVIS) [uwhq^]
Crel [+N,-CAT,+COMP,-ARG,+NOM,-LOW] (-N,-V,-CAT,-COMP,-INVIS)
Csub [-N,-V,-CAT,+COMP,+ARG] (-N,-V,-CAT,-COMP,-INVIS)
T [-N,-V,-CAT,-COMP,-ARG,-INVIS] (-N,+V) [uphi^]
V [-N,+V,+CAT] (-CAT,+ARG)
Copadj [-N,+V,-CAT,-COMP,-ARG] (-N,-V,+CAT)
D [+N,-CAT,-COMP,+ARG,-WH,-LOW] (+N,-CAT,-ARG,+NOM)
Dwh [+N,-CAT,-COMP,+ARG,+WH,-LOW] (+N,-CAT,-ARG,+NOM) [vwhq]
only [-N,-V,-CAT,-COMP,-ARG,+INVIS,+EXCL] (+N,-CAT,-COMP,+ARG,-WH)
OpCR [-N,-V,-CAT,-COMP,-ARG,+INVIS,-EXCL,-FEAT] (+N,+LOW)
φ [+N,-CAT,-COMP,-ARG,+NOM,+LOW] (+N,+CAT) [vphi,unoun]
ind [+N,-CAT,-COMP,-ARG,-NOM,-LOW]
N [+N,+CAT,+LOW] [vnoun]
A [-N,-V,+CAT]
Focfeat [-N,-V,-CAT,-COMP,-ARG,+INVIS,-EXCL,+FEAT] () [vfoc]
Over 82 loops, 28 of which were non-vacuous, ALPAFA created 67 categories using 12 pairs of categorial features, and assigned 16 non-categorial features.
Usage
=====
ALPAFA is implemented as a command line utility. Use the following syntax to read from an input file
and write the output of ALPAFA to a file (note that this will overwrite existing files of the same
name as the output):
``$ alpafa input_file output_file``
More complex options can be seen with ``$ alpafa -h`` or ``$ alpafa --help``::
usage: alpafa [-h] [--no_uf] [--no_cselect] [--log] [--categories]
[--dependents]
input_file output_file
Applies the algorithm from AAFP to a correctly formatted input file.
positional arguments:
input_file correctly formatted UTF-8 input file
output_file name of file to output
optional arguments:
-h, --help show this help message and exit
--no_uf do not implement unvalued features
--no_cselect do not implement c-selection
--log include a log of algorithm operations
--categories list all categories before heads
--dependents list all dependent features below their relevant categories
(implies --categories)
| ALPAFA | /ALPAFA-0.2.tar.gz/ALPAFA-0.2/README.rst | README.rst |
class FeatureBearer():
'''Defines some display methods common to Head and Category'''
def __init__(self):
self.d = []
self.feats = []
self.c_feat = None
self.category_properties = [] # properties that are used to define categories
def dstring(self, c_select=False):
'''Returns a string of self's categorial feature bundle.
:param c_select: c-selectional feature bundle.
'''
def catfeats():
'''Converts d to a readable +/- feature string.'''
for i, digit in enumerate(self.d):
if digit == 2:
yield '+' + self.category_properties[i]
elif digit == 1:
yield '-' + self.category_properties[i]
if c_select:
return '(' + ','.join(catfeats()) + ')'
return '[' + ','.join(catfeats()) + ']'
def c_select(self):
'''Returns a str of self's c-selectional feature.'''
if self.c_feat is None:
return ''
return self.c_feat.dstring(c_select=True)
def featstring(self):
'''Returns a str of self's non-categorial feature bundle'''
if self.feats:
return '[' + ','.join(self.feats) + ']'
return ''
class Head(FeatureBearer):
'''Instances of this class correspond to linguistic heads, and recieve their name and properties
from the input. During the course of the algorithm they are divided into categories, and
assigned categorial, non-categorial, and c-selectional features.
'''
def __init__(self, name, properties):
FeatureBearer.__init__(self)
self.name = name
self.properties = properties
def __repr__(self):
return self.name
def __str__(self):
return self.name
def spec(self, c_select_choice=False):
'''Returns a full (tabbed) specifcation str for self.
:param c_select_choice: include c-selectional features
'''
if c_select_choice is True:
spec = (self.name, self.dstring(), self.c_select(), self.featstring())
else:
spec = (self.name, self.dstring(), self.featstring())
return '\t'.join(spec).rstrip('\t')
class Category(FeatureBearer):
'''Instances of this class correspond to categories of linguistic heads, defined by a bundle of
categorial features (d). Initially the only category is the category of all heads, but in during
the course of the algorithm this is divided into smaller categories (by Category.divide), each
of which can be assigned non-categorial and c-selectional features (by Category.assign).
'''
def __init__(self, container, d, contents):
FeatureBearer.__init__(self)
self.container = container # lexicon containing the category
self.log = container.log # log of operations carried out by the algorithm
self.category_properties = container.category_properties # overriding base class
self.d = d # overriding base class
self.contents = contents
def __repr__(self):
return self.dstring()
def __str__(self):
return self.dstring()
def __iter__(self):
return iter(self.contents)
def __len__(self):
'''len(X) returns the number of categorial features that X bears.'''
return sum(1 for i in self.d if i != 0)
def __ge__(self, other):
'''X >= Y iff Y is a subcategory of X: i.e. Y has a superset of X's categorial features.'''
for i, j in zip(self.d, other.d):
if i != 0 and i != j:
return False
return True
def lhd(self):
'''Returns a display string of the values of l, h, and d.'''
l = len(self)
h = len(self.contents)
d = ''.join(str(i) for i in self.d)
return 'l = {}, h = {}, d = {}'.format(l, h, d)
def spec(self, cats_dep_choice=False):
'''Returns a full (tabbed) specifcation str for self, including non-categorial and
c-selectional features if specified by the user.
:param cats_dep_choice: show dependent features beneath categories
'''
spec = (self.dstring(), self.lhd(), ','.join(str(head) for head in self.contents))
if cats_dep_choice and (self.c_feat or self.feats):
return '\t'.join(spec) + '\n' + self.c_select() + self.featstring()
return '\t'.join(spec)
def assign(self, feat):
'''Assigns a non-categorial or c-selectional feature to self and all the heads it contains.
'''
if isinstance(feat, Category):
self.c_feat = feat
for head in self:
head.c_feat = feat
self.log.append('Assign {} to {}'.format(self.c_select(), self.dstring()))
else:
self.feats.append(feat)
for head in self:
head.feats.append(feat)
self.log.append('Assign [{}] to {}'.format(feat, self.dstring()))
def divide(self, headswithp):
'''Divide any subcategories of self that have at least one head in headswithp and at least
one not in headswithp new [+P] and [-P] variants. Returns a list of newly created
categories.
'''
new_cats = []
minus_p = {head for head in self.contents if head not in headswithp}
# Extend head features.
for head in self.container.heads:
if head in headswithp:
head.d = head.d[:-1] + [2]
elif head in minus_p:
head.d = head.d[:-1] + [1]
for cat in list(self.container):
if (self >= cat and
headswithp.intersection(set(cat.contents)) != set() and
set(cat.contents) - headswithp != set()):
# i.e. cat is a subcat of self, and contains all heads with p and at least one head
# without p
cat_ps = [head for head in cat if head in headswithp]
cat_others = [head for head in cat if head in minus_p]
cat_plus_p = Category(self.container, cat.d[:-1] + [2], cat_ps)
cat_minus_p = Category(self.container, cat.d[:-1] + [1], cat_others)
self.container.categories.extend([cat_plus_p, cat_minus_p])
new_cats.extend(self.container.categories[-2:])
self.log.append('Divide {} into [{}]'
.format(self.dstring(), self.category_properties[-1]))
return new_cats
class Lexicon():
'''Defines a "container" for all the Heads, which are then divided up into categories by the
assignment of features to these heads via a specified prominence order. Lexicon._learn starts
the main algorithm, which also uses Lexicon._add_dependent_feature and
Lexicon._divide_categories, with the help of Lexicon.headswith. The remaining methods are
largely for display purposes.
'''
def __init__(self, prominence, heads, uf_choice=True, c_select_choice=True):
''':param uf_choice: implement unvalued features
:param c_select_choice: implement c-selection
'''
# parameters
self.uf_choice = uf_choice
self.c_select_choice = c_select_choice
# core setup
self.prominence = prominence
self.heads = heads
self.initial_prominence = list(prominence)
self._invis_index = None # position of the special "invis" cateorial feature
self.category_properties = [] # properties that are used to define categories
self.log = [] # log of operations carried out by the algorithm
self.categories = [Category(self, [], self.heads)]
for head in self.heads:
head.container = self
head.category_properties = self.category_properties
if self.c_select_choice:
self.prominence.append(self.categories[0])
self._learn()
def __iter__(self):
return iter(self.categories)
def __repr__(self):
return '\n'.join(cat.spec() for cat in self)
def headswith(self, p):
'''Helper function for the main algorithm. Returns a set of the heads bearing the current
property p, and a bool stating whether or not that property triggers movement.
'''
move = False
if isinstance(p, Category):
headswithp = {head for head in self.heads if
set([h.name for h in p.contents]) in head.properties}
return headswithp, move
else:
headswithp = {head for head in self.heads if p in head.properties}
if headswithp == set():
# no singleton sets, so must be a movement feature
headswithp = {head for head in self.heads if (list(p)[0], 'm') in head.properties}
move = True
return headswithp, move
#---------------------------------------------------------------------------------------------------
# Main body of the algorithm, annotated with the step numbers from (103) of AAFP chapter 1:
#---------------------------------------------------------------------------------------------------
def _learn(self):
'''Loops over the prominence order, first attempting to assign p as a dependent feature of
the "largest" possible category, and if this fails, dividing the "smallest" possible
category (and its relevant subcategories) into +P and -P variants.
'''
# (i, xv) identify next undescribed property p
for p in self.prominence:
non_cat = False
headswithp, move = self.headswith(p)
# (ii) search for "largest" category coextensive with p
for cat in self:
if set(cat.contents) == headswithp:
non_cat = True
# go to (iii)
self._add_dependent_feature(p, cat, move)
break
if non_cat is False and isinstance(p, str) and headswithp != set():
# (x) is p a bare property?
# go to (xi)
self._divide_categories(p, headswithp)
self.acquired = True
def _add_dependent_feature(self, prop, category, move):
'''Adds the appropriate dependent feature, depending on the nature of prop. Equivalent to
the schema in (102) of AAFP chapter 2. Note that move is a bool.
'''
if isinstance(prop, str):
# (iii) is p a bare property?
if self.uf_choice:
# (iv) assign [vp], add {p} to prominence
category.assign('v' + prop)
self.prominence.append({prop})
else:
# (iv) assign [vp]
category.assign(prop)
elif isinstance(prop, Category):
# (v) is p a category?
# (vi) assign corresponding c-selectional feature
if category.c_feat is None:
category.assign(prop)
elif move:
# (vii) does p trigger movement?
# (viii) assign [up^]
category.assign('u' + list(prop)[0] + '^') # (viii)
else:
# (ix) assign [up]
category.assign('u' + list(prop)[0]) # (ix)
def _divide_categories(self, prop, headswithprop):
'''Divides the chosen category and all relevant subcategories into +PROP and -PROP variants.
'''
if prop == 'invis':
self._invis_index = len(self.category_properties)
# add new categorial feature to lexicon
self.category_properties.append(prop.upper())
for cat in self:
cat.d = cat.d + [0]
for head in self.heads:
head.d = head.d + [0]
# (xi) search for "smallest" category
for category in reversed(self.categories):
if headswithprop < set(category.contents):
# (xii, xiii) assign categorial features to the appropriate heads and categories
new_cats = category.divide(headswithprop)
break
# (xiv) reorder categories and append new visible categories to prominence
self.categories.sort(key=lambda k: (-len(k), len(k.contents), k.d), reverse=True)
if self.c_select_choice:
if self._invis_index is not None:
new_cats = [cat for cat in new_cats if cat.d[self._invis_index] != 2]
self.prominence.extend([cat for cat in self if cat in new_cats])
#---------------------------------------------------------------------------------------------------
def display(self, divlog_choice=True, cats_choice=True, cats_dep_choice=False):
'''Takes a number of optional parameters, and returns a tabbed specification of the lexicon.
:param divlog_choice: display self.log
:param cats_choice: display categories
:param cats_dep_choice: display dependent features below categories
'''
output = ''
if divlog_choice:
output += '\n'.join(self.log) + '\n\n'
if cats_choice:
output += '\n'.join(cat.spec(cats_dep_choice=cats_dep_choice) for cat in self) + '\n\n'
output += '\n'.join(head.spec(self.c_select_choice) for head in self.heads)
output += '\n\n' + self.stats()
return output
def stats(self):
'''Returns a string containing some information on the algorithm's behaviour.'''
def agree(number, y=False):
'''Returns a tuple of number and its agreement inflection.
:param y: for irregular "y/ies" plurals
'''
agree = [number]
if y:
if number == 1:
agree += ['y']
else:
agree += ['ies']
else:
if number == 1:
agree += ['']
else:
agree += ['s']
return tuple(agree)
loops = agree(len(self.prominence))
nonvacs = len(self.log)
cats = agree(len(self.categories), y=True)
catfeats = agree(len(self.category_properties))
noncatfeats = agree(len(self.log) - len(self.category_properties))
stats = 'Over {} loop{}, '.format(*loops)
if loops == 1:
stats += 'which was non-vacuous, '
elif loops == nonvacs:
stats += 'all of which were non-vacuous, '
elif loops != 0:
stats += '{} of which were non-vacuous, '.format(nonvacs)
stats += 'ALPAFA created {} categor{} '.format(*cats)
stats += 'using {} pair{} of categorial features, '.format(*catfeats)
stats += 'and assigned {} non-categorial feature{}.'.format(*noncatfeats)
return stats | ALPAFA | /ALPAFA-0.2.tar.gz/ALPAFA-0.2/alpafa/alpafa.py | alpafa.py |
from .alpafa import Head
class ParserError(Exception):
'''For parser-specific errors.'''
pass
class ParseHead():
'''Parser for input file lines which represent a head. Designed to be created once per line,
with self.parse immediately being called, which returns a parsed frozenset of properties.'''
def __init__(self, linenum, line):
self._linenum = linenum # for more informative errors
self._name, separator, properties = line.partition(':')
if not (self._name and separator):
raise ParserError("Invalid syntax on line {} of input".format(self._linenum))
self._properties = properties.split(',') # split but unparsed properties list
# set during self.parse
self._index = None # current position in parsing this list
self._item = None # current object in list
self._setstart = None
self._tupstart = None
# always have the same initial values
self._prevobjend = 0 # position of last set or tuple
self._final_properties = []
def _open_bracket(self, tup=False):
'''Called when the parser comes across an open bracket - checks for illicit embeddings,
and updates instance attributes appropriately.
:param tup: < rather than {
'''
if self._tupstart is not None or self._setstart is not None:
raise ParserError("Set inside set on line {} of input".format(self._linenum))
# we're done with the chunk of properties up to here
self._final_properties += self._properties[self._prevobjend:self._index]
if tup:
self._tupstart = int(self._index)
else:
self._setstart = int(self._index)
def _close_bracket(self, tup=False):
'''Called when the parser comes across a closed bracket - checks for invalid closings (and
makes sure ordered pairs are well-formed), then adds the new subobject to
self.final_properties and updates instance attributes appropriately.
:param tup: > rather than }
'''
if tup and (self._tupstart is None or self._setstart is not None or
self._index - self._tupstart != 1 or self._item != 'm>'):
raise ParserError("Invalid ordered set on line {} of input".format(self._linenum))
if not tup and (self._setstart is None or self._tupstart is not None):
raise ParserError("Invalid set on line {} of input".format(self._linenum))
if tup:
newtup = self._properties[self._tupstart:self._index+1]
newtup[0] = newtup[0].lstrip('<')
newtup[-1] = newtup[-1].rstrip('>')
self._final_properties.append(tuple(newtup))
self._tupstart = None
else:
newset = self._properties[self._setstart:self._index+1]
newset[0] = newset[0].lstrip('{')
newset[-1] = newset[-1].rstrip('}')
self._final_properties.append(frozenset(newset))
self._setstart = None
self._prevobjend = self._index+1
def parse(self):
'''Parses self._properties into frozenset of properties, which is combined with self.Name to
return a Head object.
'''
for i, item in enumerate(self._properties):
self._index = i
self._item = item
if item.startswith('{'):
self._open_bracket()
elif item.startswith('<'):
self._open_bracket(tup=True)
if item.endswith('}'):
self._close_bracket()
elif item.endswith('>'):
self._close_bracket(tup=True)
if self._setstart is not None or self._tupstart is not None:
raise ParserError("Unclosed bracket on line {} of input".format(linenum))
self._final_properties += self._properties[self._prevobjend:]
return Head(self._name, frozenset(self._final_properties))
def parse_file(input_file):
'''Takes a correctly formatted input file and returns a parsed prominence order and list of
Head objects.
'''
prominence = []
heads = []
with open(input_file, encoding='utf-8') as f:
for i, line in enumerate(f):
line = line.replace(' ', '').strip()
if line.startswith("prominence="):
prominence = line[11:].split(',')
elif line != '':
heads.append(ParseHead(i+1, line).parse())
if not heads:
raise ParserError("No heads found")
if not prominence:
raise ParserError("No prominence order found")
if '' in prominence:
raise ParserError("Zero length feature in prominence order")
return prominence, heads | ALPAFA | /ALPAFA-0.2.tar.gz/ALPAFA-0.2/alpafa/parse.py | parse.py |
import argparse
from .alpafa import Lexicon
from .parse import parse_file, ParserError
def set_args():
'''Sets command line parameters, and runs ALPAFA.'''
parser = argparse.ArgumentParser(prog='alpafa',
description='Applies the algorithm from AAFP to a correctly \
formatted input file.')
parser.add_argument('input_file', help='correctly formatted UTF-8 input file')
parser.add_argument('output_file', help='name of file to output')
parser.add_argument('--no_uf', dest='uf', action='store_false',
help='do not implement unvalued features')
parser.add_argument('--no_cselect', dest='cselect', action='store_false',
help='do not implement c-selection')
parser.add_argument('--log', dest='log', action='store_true',
help='include a log of algorithm operations')
parser.add_argument('--categories', dest='cats', action='store_true',
help='list all categories before heads')
parser.add_argument('--dependents', dest='dependents', action='store_true',
help='list all dependent features below their relevant categories (implies \
--categories)')
args = parser.parse_args()
if args.dependents:
args.cats = True
return(args.input_file, args.output_file, args.uf, args.cselect, args.log, args.cats,
args.dependents)
def run_alpafa(input_file, output_file, uf, cselect, log, cats, dependents):
'''Parse an input file, and apply ALPAFA to its contents, printing the output to a specified
file.
'''
try:
prominence, heads = parse_file(input_file)
except FileNotFoundError as e:
print('alpafa: input failure: ' + str(e)[10:])
return
except ParserError as e:
print('alpafa: parsing failure: {}'.format(e))
return
lex = Lexicon(prominence, heads, uf, cselect)
with open(output_file, 'w', encoding='utf-8') as f:
f.write(lex.display(log, cats, dependents))
print(lex.stats())
def main():
run_alpafa(*set_args()) | ALPAFA | /ALPAFA-0.2.tar.gz/ALPAFA-0.2/alpafa/cli.py | cli.py |
# AdventureLand Python Client
This is meant to be a python client for the game [Adventure Land - The Code MMORPG](https://adventure.land). It's heavily inspired by and based on earthiverse's typescript-based [ALClient](https://github.com/earthiverse/ALClient).
Currently a major work in progress; but the current state is available through [pip](https://pypi.org/project/pip/) for simple installation and use.
## Requirements
This package currently requires `aiohttp`, `aiosignal`, `async-timeout`, `attrs`, `bidict`, `charset-normalizer`, `frozenlist`, `idna`, `igraph`, `multidict`, `python-engineio`, `setuptools`, `texttables`, `ujson`, and `yarl`. (Technially only some of these are dependencies of ALPC; others are dependencies of various dependencies).
[b]All[/b] of these requirements are installable through pip, and their individual installations are also taken care of through the install of ALPC.
## Installation
The PyPI page can be found [here]. In order to install, simply install the package using pip like so:
<details><summary>Unix/Linux</summary>
```
python3 -m pip install --upgrade ALPC
```
</details>
<details><summary>Windows</summary>
```
py -m pip install --upgrade ALPC
```
</details>
* <small>*Note: This package was developed with Python 3.10.4; therefore, I cannot guarantee that it will work with anything below that. In fact, due to current bugs, I cannot even guarantee that it will work perfectly **with** that.*</small>
## Usage
* First: be sure to install the package from PyPI using pip.
* Second: create a `credentials.json` file like so:
```json
{
'email': '[email protected]',
'password': 'yourpassword'
}
```
* Third: create a python file like so:
```python
import aiohttp
import asyncio
import logging
import sys
import ALPC as AL
logging.root.setLevel(logging.INFO)
async def main():
async with aiohttp.ClientSession() as session:
print('Logging in...')
await AL.Game.loginJSONFile(session, '..\credentials.json')
print('Successfully logged in!')
print('Getting G Data...')
await AL.Game.getGData(session, True, True)
print('Obtained G Data!')
print('Preparing pathfinder...')
await AL.Pathfinder.prepare(AL.Game.G)
print('Pathfinder prepared!')
print('Starting character...')
char = await AL.Game.startCharacter(session, 'WarriorSurge', 'US', 'I')
print('Moving to main...')
await char.smartMove('main')
print('Moving to halloween...')
await char.smartMove('halloween')
print('Moving to desertland...')
await char.smartMove('desertland')
print('Returning to main...')
await char.smartMove('main')
print('Disconnecting...')
await char.disconnect()
# this part is technically only required if you're running on windows due to hinkyness involving windows OS and asyncio
if sys.platform == 'win32':
asyncio.set_event_loop_policy(asyncio.WindowsSelectorEventLoopPolicy())
asyncio.run(main())
```
* Fourth: run your python file; you should get this as a result:
```
Logging in...
Successfully logged in!
Getting G Data...
Obtained G Data!
Preparing pathfinder...
Pathfinder prepared!
Starting character...
Moving to main...
Moving to halloween...
Moving to desertland...
Returning to main...
Disconnecting...
```
## Final Notes
* AS STATED, THIS PACKAGE IS STILL A WORK IN PROGRESS. If you have ANY issues at all or any suggestions or come accross any bugs, feel free to either submit them to the issues tab or submit your info to the existing issue if your bug is already there.
<<<<<<< HEAD
* Currently, there is no full support for the individual classes within the game; there is only support for basic attacks, movement, and item usage. My current focus is somewhat split between fixing the existing issues and completing the missing pieces (along with school and the fact that I work 40+ hours a week...so please have patience).
=======
* Currently, there is no full support for the individual classes within the game; there is only support for basic attacks, movement, and item usage. My current focus is somewhat split between fixing the existing issues and completing the missing pieces (along with school and the fact that I work 40+ hours a week...so please have patience).
>>>>>>> f2c5275bd8cb993590afe68dedaf42c714de8493
| ALPC | /ALPC-0.1.0.tar.gz/ALPC-0.1.0/README.md | README.md |
# ALS.Liam (version 0.10)
Overview
---
_**Liam**_ is a Python module used to visualize CCD data that was collected
on the **Scattering Chamber** located at **Beamline 4.0.2**
(_a.k.a. **BL402**_) of the **Advanced Light Source** (Berkeley, CA USA).
This module utilizes the _**ALS.Milo**_ package for processing the data.
It is distributed under the _namespace package_, _**ALS**_.
Installation
---
### Install from PyPI
**_ALS.Liam_** can be installed from PyPI using `pip`.
The following example shows how.
```bash
>> sudo python -m pip install ALS.Liam -vv
```
### Install from local repository (download)
**_ALS.Liam_** can be installed from a local copy of the project repository
using `pip`. The following example shows how.
```bash
>> cd ALS.Liam-0.10.0/ # Local directory of project repository
>> sudo python -m pip install . -vv
```
Background information
---
Data from the BL402 Scattering Chamber is stored in two types of files:
* **FITS files**: Each image captured by the CCD (_a.k.a._ the camera) is
stored in a separate file using the _FITS_ format. _More details below._
https://fits.gsfc.nasa.gov/fits_documentation.html
* **Scan summary files**: When a scan sequence is run to collect data, a text
file is created to summarize the parameters of the scan and the data collected.
These files typically end with the extension "*-AI.txt".
Every _scan summary file_ contains a header that describes the scan and the
types of data recorded, followed by data rows -- one row per data point. An
_Instrument Scan_ provides an _image filename_ in each data row that can be
used to access the CCD images recorded during the scan.
Using the FITS Viewer to visualize your data
---
To start the viewer, run the following command in your terminal or prompt:
```bash
python fitsViewer.py
```
### Load data files
* Click `Load data file` button to open a file selection dialog.
* Select file for display. This can be a `.FITS` file, a `*-AI.txt` file, or
many image file types (`.png`, `.jpg`, etc.)
* If the file type you are looking for is not displayer or not selectable, it
might be necessary to change the file filter to the appropriate file extension.
* Mac: For OS X, the file filter might be hidden. If so, click the
`Options` button in the file selection dialog.
* Selecting a `*-AI.txt` file will allow you to browse all FITS files that
were collected as part of this data scan
### Viewing the data
Image data is displayed in the central region. File name and location are
displayed near the top of the window. To the right of the image is a color
scale bar and histogram of the data intensity values. The color scale of the
image can be adjusted with these controls (see _Adjusting the color scale_).
To the left and also to the bottom of the plot are 1D representations of the
image intensity, collapsed along the orthogonal dimension. The rows and columns
can be restricted for these 1D plots using control bars in the image plot
(see _Setting ROI horizontal and vertical limits_). The cursor displays the
row, column, and intensity value for the pixel under the cross-hair.
### Zoom or pan 2D data
The image magnification (zoom) can be changed by using the middle mouse button
(or scroll wheel). Clicking-and-dragging the mouse on the image will recenter
(pan) the image by the amount that you drag it.
### Adjusting the color scale
Drag the yellow bars (between the intensity labels and the color bar) to adjust
the upper or lower limit of color scaling. There are options for autoscaling
the intensity (covers full range) or toggling between linear and log intensity
scale; these are activated by the corresponding checkboxes. The colored arrows
to the right of the color bar allow the color scale to be manipulated. These
arrows can be relocated, added (by double-clicking in an empty space), removed
(right-click for context menu), or be changed to other colors (click on arrow).
### Setting ROI horizontal and vertical limits
A Region of Interest (ROI) can be selected from the image by setting horizontal
and/or vertical limits. This is done by dragging the yellow or blue bars that
are initially located at the left and bottom edges of the image. The
highlighted blue (yellow) regions are averaged to generate the blue (yellow)
1D plots. Initially the 1D plots average data across the entire image before
the selection bars are first moved. Returning the selection bars to their
initial position recovers this initial state. Intensity values for the 1D
plots are displayed in the average counts per pixel.
### Navigate FITS files within a scan set
Click `Prev` or `Next` buttons to display the previous or next image in the
scan data set. `First` and `Last` buttons will display the first or last image
in the scan data set. The image number can be typed directly into the entry
field to the left of these buttons. If an out-of-range value is entered, it
will automatically select the nearest available image.
Navigation is only possible for `*-AI.txt` files. The image number persists
between data sets; i.e., it does not change when you load a new data set
(unless it is out-of-bounds for the new data set).
### Reloading (incomplete) data sets
Click `Reload` to reload the data file with the most recent information. This
is most useful for datasets (`*-AI.txt` files) that were incomplete when
initially loaded (because data was still being captured or transfered).
### Export plots
Right-click on image or plot, then select `Export`. Select region, format, and
other options. Data can be exported to file or copied to clipboard.
Notes for Developers
---
### Additional test data
Additional CCD data files and scan sets can be downloaded from these links.
Unzip and place the contents into the `test_data` folder.
* CCD Scan 8032: [https://zenodo.org/record/3923169#.Xvs-hi2ZPxg
](https://zenodo.org/record/3923169#.Xvs-hi2ZPxg)
* CCD Scan 8044: [https://zenodo.org/record/3923175#.XvqZcS2ZPxg
](https://zenodo.org/record/3923175#.XvqZcS2ZPxg)
Copyright Notice
---
ALS.Liam: BL402 CCD image viewer for RSXD data, Copyright (c) 2017-2019, 2021,
The Regents of the University of California, through Lawrence Berkeley
National Laboratory (subject to receipt of any required approvals from the
U.S. Dept. of Energy). All rights reserved.
If you have questions about your rights to use or distribute this software,
please contact Berkeley Lab's Intellectual Property Office at [email protected].
NOTICE. This Software was developed under funding from the U.S. Department of
Energy and the U.S. Government consequently retains certain rights. As such,
the U.S. Government has been granted for itself and others acting on its
behalf a paid-up, nonexclusive, irrevocable, worldwide license in the
Software to reproduce, distribute copies to the public, prepare derivative
works, and perform publicly and display publicly, and to permit other to do
so. | ALS.Liam | /ALS.Liam-0.10.0.tar.gz/ALS.Liam-0.10.0/README.md | README.md |
from __future__ import print_function
try:
import configparser
except ImportError:
import ConfigParser as configparser
import errno
import json
import os
import re
import subprocess
import sys
class VersioneerConfig:
"""Container for Versioneer configuration parameters."""
def get_root():
"""Get the project root directory.
We require that all commands are run from the project root, i.e. the
directory that contains setup.py, setup.cfg, and versioneer.py .
"""
root = os.path.realpath(os.path.abspath(os.getcwd()))
setup_py = os.path.join(root, "setup.py")
versioneer_py = os.path.join(root, "versioneer.py")
if not (os.path.exists(setup_py) or os.path.exists(versioneer_py)):
# allow 'python path/to/setup.py COMMAND'
root = os.path.dirname(os.path.realpath(os.path.abspath(sys.argv[0])))
setup_py = os.path.join(root, "setup.py")
versioneer_py = os.path.join(root, "versioneer.py")
if not (os.path.exists(setup_py) or os.path.exists(versioneer_py)):
err = ("Versioneer was unable to run the project root directory. "
"Versioneer requires setup.py to be executed from "
"its immediate directory (like 'python setup.py COMMAND'), "
"or in a way that lets it use sys.argv[0] to find the root "
"(like 'python path/to/setup.py COMMAND').")
raise VersioneerBadRootError(err)
try:
# Certain runtime workflows (setup.py install/develop in a setuptools
# tree) execute all dependencies in a single python process, so
# "versioneer" may be imported multiple times, and python's shared
# module-import table will cache the first one. So we can't use
# os.path.dirname(__file__), as that will find whichever
# versioneer.py was first imported, even in later projects.
me = os.path.realpath(os.path.abspath(__file__))
me_dir = os.path.normcase(os.path.splitext(me)[0])
vsr_dir = os.path.normcase(os.path.splitext(versioneer_py)[0])
if me_dir != vsr_dir:
print("Warning: build in %s is using versioneer.py from %s"
% (os.path.dirname(me), versioneer_py))
except NameError:
pass
return root
def get_config_from_root(root):
"""Read the project setup.cfg file to determine Versioneer config."""
# This might raise EnvironmentError (if setup.cfg is missing), or
# configparser.NoSectionError (if it lacks a [versioneer] section), or
# configparser.NoOptionError (if it lacks "VCS="). See the docstring at
# the top of versioneer.py for instructions on writing your setup.cfg .
setup_cfg = os.path.join(root, "setup.cfg")
parser = configparser.SafeConfigParser()
with open(setup_cfg, "r") as f:
parser.readfp(f)
VCS = parser.get("versioneer", "VCS") # mandatory
def get(parser, name):
if parser.has_option("versioneer", name):
return parser.get("versioneer", name)
return None
cfg = VersioneerConfig()
cfg.VCS = VCS
cfg.style = get(parser, "style") or "pep440-auto"
cfg.versionfile_source = get(parser, "versionfile_source")
cfg.versionfile_build = get(parser, "versionfile_build")
cfg.tag_prefix = get(parser, "tag_prefix")
if cfg.tag_prefix in ("''", '""'):
cfg.tag_prefix = ""
cfg.parentdir_prefix = get(parser, "parentdir_prefix")
cfg.verbose = get(parser, "verbose")
return cfg
class NotThisMethod(Exception):
"""Exception raised if a method is not valid for the current scenario."""
# these dictionaries contain VCS-specific tools
LONG_VERSION_PY = {}
HANDLERS = {}
def register_vcs_handler(vcs, method): # decorator
"""Decorator to mark a method as the handler for a particular VCS."""
def decorate(f):
"""Store f in HANDLERS[vcs][method]."""
if vcs not in HANDLERS:
HANDLERS[vcs] = {}
HANDLERS[vcs][method] = f
return f
return decorate
def run_command(commands, args, cwd=None, verbose=False, hide_stderr=False,
env=None):
"""Call the given command(s)."""
assert isinstance(commands, list)
p = None
for c in commands:
try:
dispcmd = str([c] + args)
# remember shell=False, so use git.cmd on windows, not just git
p = subprocess.Popen([c] + args, cwd=cwd, env=env,
stdout=subprocess.PIPE,
stderr=(subprocess.PIPE if hide_stderr
else None))
break
except EnvironmentError:
e = sys.exc_info()[1]
if e.errno == errno.ENOENT:
continue
if verbose:
print("unable to run %s" % dispcmd)
print(e)
return None, None
else:
if verbose:
print("unable to find command, tried %s" % (commands,))
return None, None
stdout = p.communicate()[0].strip()
if sys.version_info[0] >= 3:
stdout = stdout.decode()
if p.returncode != 0:
if verbose:
print("unable to run %s (error)" % dispcmd)
print("stdout was %s" % stdout)
return None, p.returncode
return stdout, p.returncode
def unique(seq):
"""Find unique elements in a list, without sorting
Implementation does not depend on `numpy`
* borrowed from https://stackoverflow.com/a/480227
* credit to Markus Jarderot
+ https://www.peterbe.com/plog/uniqifiers-benchmark
"""
seen = set()
seen_add = seen.add
return [x for x in seq if not (x in seen or seen_add(x))]
LONG_VERSION_PY['git'] = '''
# This file helps to compute a version number in source trees obtained from
# git-archive tarball (such as those provided by githubs download-from-tag
# feature). Distribution tarballs (built by setup.py sdist) and build
# directories (produced by setup.py build) will contain a much shorter file
# that just contains the computed version number.
# This file is released into the public domain. Generated by
# versioneer-0.18 (https://github.com/warner/python-versioneer)
"""Git implementation of _version.py."""
import errno
import os
import re
import subprocess
import sys
def get_keywords():
"""Get the keywords needed to look up the version information."""
# these strings will be replaced by git during git-archive.
# setup.py/versioneer.py will grep for the variable names, so they must
# each be defined on a line of their own. _version.py will just call
# get_keywords().
git_refnames = "%(DOLLAR)sFormat:%%d%(DOLLAR)s"
git_full = "%(DOLLAR)sFormat:%%H%(DOLLAR)s"
git_date = "%(DOLLAR)sFormat:%%ci%(DOLLAR)s"
keywords = {"refnames": git_refnames, "full": git_full, "date": git_date}
return keywords
class VersioneerConfig:
"""Container for Versioneer configuration parameters."""
def get_config():
"""Create, populate and return the VersioneerConfig() object."""
# these strings are filled in when 'setup.py versioneer' creates
# _version.py
cfg = VersioneerConfig()
cfg.VCS = "git"
cfg.style = "%(STYLE)s"
cfg.tag_prefix = "%(TAG_PREFIX)s"
cfg.parentdir_prefix = "%(PARENTDIR_PREFIX)s"
cfg.versionfile_source = "%(VERSIONFILE_SOURCE)s"
cfg.verbose = False
return cfg
class NotThisMethod(Exception):
"""Exception raised if a method is not valid for the current scenario."""
LONG_VERSION_PY = {}
HANDLERS = {}
def register_vcs_handler(vcs, method): # decorator
"""Decorator to mark a method as the handler for a particular VCS."""
def decorate(f):
"""Store f in HANDLERS[vcs][method]."""
if vcs not in HANDLERS:
HANDLERS[vcs] = {}
HANDLERS[vcs][method] = f
return f
return decorate
def run_command(commands, args, cwd=None, verbose=False, hide_stderr=False,
env=None):
"""Call the given command(s)."""
assert isinstance(commands, list)
p = None
for c in commands:
try:
dispcmd = str([c] + args)
# remember shell=False, so use git.cmd on windows, not just git
p = subprocess.Popen([c] + args, cwd=cwd, env=env,
stdout=subprocess.PIPE,
stderr=(subprocess.PIPE if hide_stderr
else None))
break
except EnvironmentError:
e = sys.exc_info()[1]
if e.errno == errno.ENOENT:
continue
if verbose:
print("unable to run %%s" %% dispcmd)
print(e)
return None, None
else:
if verbose:
print("unable to find command, tried %%s" %% (commands,))
return None, None
stdout = p.communicate()[0].strip()
if sys.version_info[0] >= 3:
stdout = stdout.decode()
if p.returncode != 0:
if verbose:
print("unable to run %%s (error)" %% dispcmd)
print("stdout was %%s" %% stdout)
return None, p.returncode
return stdout, p.returncode
def versions_from_parentdir(parentdir_prefix, root, verbose):
"""Try to determine the version from the parent directory name.
Source tarballs conventionally unpack into a directory that includes both
the project name and a version string. We will also support searching up
two directory levels for an appropriately named parent directory
"""
rootdirs = []
for i in range(3):
dirname = os.path.basename(root)
if dirname.startswith(parentdir_prefix):
return {"version": dirname[len(parentdir_prefix):],
"full-revisionid": None,
"dirty": False, "error": None, "date": None}
else:
rootdirs.append(root)
root = os.path.dirname(root) # up a level
if verbose:
print("Tried directories %%s but none started with prefix %%s" %%
(str(rootdirs), parentdir_prefix))
raise NotThisMethod("rootdir doesn't start with parentdir_prefix")
@register_vcs_handler("git", "get_keywords")
def git_get_keywords(versionfile_abs):
"""Extract version information from the given file."""
# the code embedded in _version.py can just fetch the value of these
# keywords. When used from setup.py, we don't want to import _version.py,
# so we do it with a regexp instead. This function is not used from
# _version.py.
keywords = {}
try:
f = open(versionfile_abs, "r")
for line in f.readlines():
if line.strip().startswith("git_refnames ="):
mo = re.search(r'=\s*"(.*)"', line)
if mo:
keywords["refnames"] = mo.group(1)
if line.strip().startswith("git_full ="):
mo = re.search(r'=\s*"(.*)"', line)
if mo:
keywords["full"] = mo.group(1)
if line.strip().startswith("git_date ="):
mo = re.search(r'=\s*"(.*)"', line)
if mo:
keywords["date"] = mo.group(1)
f.close()
except EnvironmentError:
pass
return keywords
@register_vcs_handler("git", "keywords")
def git_versions_from_keywords(keywords, tag_prefix, verbose):
"""Get version information from git keywords."""
if not keywords:
raise NotThisMethod("no keywords at all, weird")
date = keywords.get("date")
if date is not None:
# git-2.2.0 added "%%cI", which expands to an ISO-8601 -compliant
# datestamp. However we prefer "%%ci" (which expands to an "ISO-8601
# -like" string, which we must then edit to make compliant), because
# it's been around since git-1.5.3, and it's too difficult to
# discover which version we're using, or to work around using an
# older one.
date = date.strip().replace(" ", "T", 1).replace(" ", "", 1)
refnames = keywords["refnames"].strip()
if refnames.startswith("$Format"):
if verbose:
print("keywords are unexpanded, not using")
raise NotThisMethod("unexpanded keywords, not a git-archive tarball")
refs = set([r.strip() for r in refnames.strip("()").split(",")])
# starting in git-1.8.3, tags are listed as "tag: foo-1.0" instead of
# just "foo-1.0". If we see a "tag: " prefix, prefer those.
TAG = "tag: "
tags = set([r[len(TAG):] for r in refs if r.startswith(TAG)])
if not tags:
# Either we're using git < 1.8.3, or there really are no tags. We use
# a heuristic: assume all version tags have a digit. The old git %%d
# expansion behaves like git log --decorate=short and strips out the
# refs/heads/ and refs/tags/ prefixes that would let us distinguish
# between branches and tags. By ignoring refnames without digits, we
# filter out many common branch names like "release" and
# "stabilization", as well as "HEAD" and "master".
tags = set([r for r in refs if re.search(r'\d', r)])
if verbose:
print("discarding '%%s', no digits" %% ",".join(refs - tags))
if verbose:
print("likely tags: %%s" %% ",".join(sorted(tags)))
for ref in sorted(tags):
# sorting will prefer e.g. "2.0" over "2.0rc1"
if ref.startswith(tag_prefix):
r = ref[len(tag_prefix):]
if verbose:
print("picking %%s" %% r)
return {"version": r,
"full-revisionid": keywords["full"].strip(),
"dirty": False, "error": None,
"date": date}
# no suitable tags, so version is "0+unknown", but full hex is still there
if verbose:
print("no suitable tags, using unknown + full revision id")
return {"version": "0+unknown",
"full-revisionid": keywords["full"].strip(),
"dirty": False, "error": "no suitable tags", "date": None}
@register_vcs_handler("git", "pieces_from_vcs")
def git_pieces_from_vcs(tag_prefix, root, verbose, run_command=run_command):
"""Get version from 'git describe' in the root of the source tree.
This only gets called if the git-archive 'subst' keywords were *not*
expanded, and _version.py hasn't already been rewritten with a short
version string, meaning we're inside a checked out source tree.
"""
GITS = ["git"]
if sys.platform == "win32":
GITS = ["git.cmd", "git.exe"]
out, rc = run_command(GITS, ["rev-parse", "--git-dir"], cwd=root,
hide_stderr=True)
if rc != 0:
if verbose:
print("Directory %%s not under git control" %% root)
raise NotThisMethod("'git rev-parse --git-dir' returned error")
# if there is a tag matching tag_prefix, this yields TAG-NUM-gHEX[-dirty]
# if there isn't one, this yields HEX[-dirty] (no NUM)
describe_out, rc = run_command(GITS, ["describe", "--tags", "--dirty",
"--always", "--long",
"--match", "%%s*" %% tag_prefix],
cwd=root)
# --long was added in git-1.5.5
if describe_out is None:
raise NotThisMethod("'git describe' failed")
describe_out = describe_out.strip()
full_out, rc = run_command(GITS, ["rev-parse", "HEAD"], cwd=root)
if full_out is None:
raise NotThisMethod("'git rev-parse' failed")
full_out = full_out.strip()
pieces = {}
pieces["long"] = full_out
pieces["short"] = full_out[:7] # maybe improved later
pieces["error"] = None
# parse describe_out. It will be like TAG-NUM-gHEX[-dirty] or HEX[-dirty]
# TAG might have hyphens.
git_describe = describe_out
# look for -dirty suffix
dirty = git_describe.endswith("-dirty")
pieces["dirty"] = dirty
if dirty:
git_describe = git_describe[:git_describe.rindex("-dirty")]
# now we have TAG-NUM-gHEX or HEX
# Determine which branch is active (ie, checked-out)
branch_name, rc = run_command(
GITS, ["rev-parse", "--abbrev-ref", "HEAD"], cwd=root)
pieces["branch"] = branch_name
if "-" in git_describe:
# TAG-NUM-gHEX
mo = re.search(r'^(.+)-(\d+)-g([0-9a-f]+)$', git_describe)
if not mo:
# unparseable. Maybe git-describe is misbehaving?
pieces["error"] = ("unable to parse git-describe output: '%%s'"
%% describe_out)
return pieces
# tag
full_tag = mo.group(1)
if not full_tag.startswith(tag_prefix):
if verbose:
fmt = "tag '%%s' doesn't start with prefix '%%s'"
print(fmt %% (full_tag, tag_prefix))
pieces["error"] = ("tag '%%s' doesn't start with prefix '%%s'"
%% (full_tag, tag_prefix))
return pieces
pieces["closest-tag"] = full_tag[len(tag_prefix):]
# distance: number of commits since tag
pieces["distance"] = int(mo.group(2))
# Added to deal with merged branches
history_search_args = ["--ancestry-path"]
if (branch_name == "master"):
# Count distance only along `master`; ignore merge paths
history_search_args = ["--ancestry-path", "--first-parent"]
distance_to_tag, rc = run_command(
GITS, ["rev-list", "--count",] + history_search_args +
["%%s..HEAD" % pieces["closest-tag"] ], cwd=root)
pieces["distance"] = int(distance_to_tag)
# commit: short hex revision ID
pieces["short"] = mo.group(3)
else:
# HEX: no tags
pieces["closest-tag"] = None
count_out, rc = run_command(GITS, ["rev-list", "HEAD", "--count"],
cwd=root)
pieces["distance"] = int(count_out) # total number of commits
##################################################
# Added for `develop` branch from `master`
# `HEAD` might have branched from untagged commit on `master`
merge_base_master, rc = run_command(
GITS, ["merge-base", "master", "HEAD", "-a"], cwd=root)
distance_to_master, rc = run_command(
GITS, ["rev-list", "--count", "--ancestry-path",
"%%s..HEAD" %% merge_base_master], cwd=root)
pieces["distance-to-master"] = int(distance_to_master)
##################################################
# Added for branches other than `master`
if pieces["closest-tag"]:
# TAG exists
master_to_tag, rc = run_command(
GITS, ["rev-list", "--count", "--ancestry-path", "--first-parent",
"%%s..%%s" % (pieces["closest-tag"], merge_base_master) ],
cwd=root)
pieces["master-to-tag"] = int(master_to_tag)
else:
pieces["master-to-tag"] = pieces["distance"]
##################################################
# Added for "feature" branch from `develop`
pieces["develop"] = None
pieces["distance-to-develop"] = None
branch_develop, rc = run_command(
GITS, ["rev-parse", "--verify", "refs/heads/develop"], cwd=root)
branch_develop_exists = not bool(rc)
if branch_develop_exists:
merge_base_develop, rc = run_command(
GITS, ["merge-base", "develop", "HEAD", "-a"], cwd=root)
distance_to_develop, rc = run_command(
GITS, ["rev-list", "--count", "--ancestry-path",
"%%s..HEAD" %% merge_base_develop], cwd=root)
pieces["develop"] = branch_develop
pieces["distance-to-develop"] = int(distance_to_develop)
# commit date: see ISO-8601 comment in git_versions_from_keywords()
date = run_command(GITS, ["show", "-s", "--format=%%ci", "HEAD"],
cwd=root)[0].strip()
pieces["date"] = date.strip().replace(" ", "T", 1).replace(" ", "", 1)
return pieces
def plus_or_dot(pieces):
"""Return a + if we don't already have one, else return a ."""
if "+" in pieces.get("closest-tag", ""):
return "."
return "+"
def render_pep440(pieces):
"""Build up version string, with post-release "local version identifier".
Our goal: TAG[+DISTANCE.gHEX[.dirty]] . Note that if you
get a tagged build and then dirty it, you'll get TAG+0.gHEX.dirty
Exceptions:
1: no tags. git_describe was just HEX. 0+untagged.DISTANCE.gHEX[.dirty]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"] or pieces["dirty"]:
rendered += plus_or_dot(pieces)
rendered += "%%d.g%%s" %% (pieces["distance"], pieces["short"])
if pieces["dirty"]:
rendered += ".dirty"
else:
# exception #1
rendered = "0+untagged.%%d.g%%s" %% (pieces["distance"],
pieces["short"])
if pieces["dirty"]:
rendered += ".dirty"
return rendered
def render_pep440_pre(pieces):
"""TAG[.post.devDISTANCE] -- No -dirty.
Exceptions:
1: no tags. 0.post.devDISTANCE
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"]:
rendered += ".post.dev%%d" %% pieces["distance"]
else:
# exception #1
rendered = "0.post.dev%%d" %% pieces["distance"]
return rendered
def render_pep440_post(pieces):
"""TAG[.postDISTANCE[.dev0]+gHEX] .
The ".dev0" means dirty. Note that .dev0 sorts backwards
(a dirty tree will appear "older" than the corresponding clean one),
but you shouldn't be releasing software with -dirty anyways.
Exceptions:
1: no tags. 0.postDISTANCE[.dev0]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"] or pieces["dirty"]:
rendered += ".post%%d" %% pieces["distance"]
if pieces["dirty"]:
rendered += ".dev0"
rendered += plus_or_dot(pieces)
rendered += "g%%s" %% pieces["short"]
else:
# exception #1
rendered = "0.post%%d" %% pieces["distance"]
if pieces["dirty"]:
rendered += ".dev0"
rendered += "+g%%s" %% pieces["short"]
return rendered
def render_pep440_old(pieces):
"""TAG[.postDISTANCE[.dev0]] .
The ".dev0" means dirty.
Eexceptions:
1: no tags. 0.postDISTANCE[.dev0]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"] or pieces["dirty"]:
rendered += ".post%%d" %% pieces["distance"]
if pieces["dirty"]:
rendered += ".dev0"
else:
# exception #1
rendered = "0.post%%d" %% pieces["distance"]
if pieces["dirty"]:
rendered += ".dev0"
return rendered
def render_pep440_micro(pieces):
"""Build up version string, with post-release "local version identifier".
Our goal: TAG.DISTANCE[+gHEX.dirty]
Exceptions:
1: no tags. git_describe was just HEX. 0+untagged.DISTANCE.gHEX[.dirty]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
rendered += ".%%d" %% pieces["distance"]
if pieces["dirty"]:
rendered += plus_or_dot(pieces)
rendered += "g%%s" %% pieces["short"]
rendered += ".dirty"
else:
# exception #1
rendered = "0+untagged.%%d.g%%s" %% (pieces["distance"],
pieces["short"])
if pieces["dirty"]:
rendered += ".dirty"
return rendered
def render_pep440_develop(pieces):
"""Build up version string, used within development branch of repository.
Our goal: MERGE-POINT.post.devN[+gHEX.dirty]
+) MERGE-POINT = Most recent common ancestor for `develop` and `master`
*) Does not yet handle branch from `release-*`
+) N = DISTANCE from the MERGE-POINT
Exceptions:
1: no tags. 0.post.devDISTANCE[+gHEX.dirty]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
distance_to_merge = pieces["distance-to-master"]
# distance_merge_to_tag = (pieces["distance"] - distance_to_merge)
distance_merge_to_tag = pieces["master-to-tag"]
if (distance_merge_to_tag > 0):
rendered += ".%%d" %% distance_merge_to_tag
rendered += ".post.dev%%d" %% distance_to_merge
else:
# exception #1
rendered = "0.post.dev%%d" %% (pieces["distance"] - 1)
if pieces["dirty"]:
rendered += plus_or_dot(pieces)
rendered += "g%%s" %% pieces["short"]
rendered += ".dirty"
return rendered
def render_pep440_feature(pieces):
"""Build up version string, used within "feature" branch of repository.
Our goal: MERGE-POINT.post.devN+gHEX.BRANCH-NAME.M[.dirty]
+) MERGE-POINT = Most recent common ancestor for `develop` and `master`
*) Does not yet handle branch from `release-*`
+) N = DISTANCE from the MERGE-POINT of `develop` and `master`
+) M = DISTANCE from the MERGE-POINT of "feature" and `develop`
Exceptions:
1: no tags. 0.post.devDISTANCE+gHEX[.dirty]
"""
if pieces["closest-tag"] and pieces["develop"]:
rendered = pieces["closest-tag"]
distance_to_develop = pieces["distance-to-develop"]
distance_to_merge = pieces["distance-to-master"]
# distance_merge_to_tag = (pieces["distance"] - distance_to_merge)
distance_merge_to_tag = pieces["master-to-tag"]
distance_dev_to_merge = (distance_to_merge - distance_to_develop)
if (distance_merge_to_tag > 0):
rendered += ".%%d" %% distance_merge_to_tag
rendered += ".post.dev%%d" %% distance_dev_to_merge
rendered += plus_or_dot(pieces)
rendered += "g%%s" %% pieces["short"]
rendered += ".%%s" %% pieces["branch"]
rendered += ".%%d" %% distance_to_develop
else:
# exception #1
rendered = "0.post.dev%%d" %% (pieces["distance"] - 1)
rendered += plus_or_dot(pieces)
rendered += "g%%s" %% pieces["short"]
if pieces["dirty"]:
rendered += ".dirty"
return rendered
def render_pep440_auto(pieces):
"""Build up version string, based on branch of repository.
Our goal: If on development branch, use render_pep440_develop()
Otherwise use render_pep440_micro()
Exceptions:
+) NONE
"""
if pieces["branch"] == "master":
# logging.debug("Rendering: render_pep440_micro()")
rendered = render_pep440_micro(pieces)
elif pieces["branch"] == "develop":
# logging.debug("Rendering: render_pep440_develop()")
rendered = render_pep440_develop(pieces)
elif pieces["branch"].startswith("release"):
# render_pep440_release() not yet implemented
rendered = render_git_describe(pieces)
else:
# logging.debug("Rendering: render_pep440_feature()")
rendered = render_pep440_feature(pieces)
return rendered
def render_git_describe(pieces):
"""TAG[-DISTANCE-gHEX][-dirty].
Like 'git describe --tags --dirty --always'.
Exceptions:
1: no tags. HEX[-dirty] (note: no 'g' prefix)
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"]:
rendered += "-%%d-g%%s" %% (pieces["distance"], pieces["short"])
else:
# exception #1
rendered = pieces["short"]
if pieces["dirty"]:
rendered += "-dirty"
return rendered
def render_git_describe_long(pieces):
"""TAG-DISTANCE-gHEX[-dirty].
Like 'git describe --tags --dirty --always -long'.
The distance/hash is unconditional.
Exceptions:
1: no tags. HEX[-dirty] (note: no 'g' prefix)
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
rendered += "-%%d-g%%s" %% (pieces["distance"], pieces["short"])
else:
# exception #1
rendered = pieces["short"]
if pieces["dirty"]:
rendered += "-dirty"
return rendered
def render(pieces, style):
"""Render the given version pieces into the requested style."""
if pieces["error"]:
return {"version": "unknown",
"full-revisionid": pieces.get("long"),
"dirty": None,
"error": pieces["error"],
"date": None}
if not style or style == "default":
style = "pep440" # the default
if style == "pep440":
rendered = render_pep440(pieces)
elif style == "pep440-pre":
rendered = render_pep440_pre(pieces)
elif style == "pep440-post":
rendered = render_pep440_post(pieces)
elif style == "pep440-old":
rendered = render_pep440_old(pieces)
elif style == "pep440-micro":
rendered = render_pep440_micro(pieces)
elif style == "pep440-develop":
rendered = render_pep440_develop(pieces)
elif style == "pep440-auto":
rendered = render_pep440_auto(pieces)
elif style == "git-describe":
rendered = render_git_describe(pieces)
elif style == "git-describe-long":
rendered = render_git_describe_long(pieces)
else:
raise ValueError("unknown style '%%s'" %% style)
return {"version": rendered, "full-revisionid": pieces["long"],
"dirty": pieces["dirty"], "error": None,
"date": pieces.get("date")}
def get_versions():
"""Get version information or return default if unable to do so."""
# I am in _version.py, which lives at ROOT/VERSIONFILE_SOURCE. If we have
# __file__, we can work backwards from there to the root. Some
# py2exe/bbfreeze/non-CPython implementations don't do __file__, in which
# case we can only use expanded keywords.
cfg = get_config()
verbose = cfg.verbose
try:
return git_versions_from_keywords(get_keywords(), cfg.tag_prefix,
verbose)
except NotThisMethod:
pass
try:
root = os.path.realpath(__file__)
# versionfile_source is the relative path from the top of the source
# tree (where the .git directory might live) to this file. Invert
# this to find the root from __file__.
for i in cfg.versionfile_source.split('/'):
root = os.path.dirname(root)
except NameError:
return {"version": "0+unknown", "full-revisionid": None,
"dirty": None,
"error": "unable to find root of source tree",
"date": None}
try:
pieces = git_pieces_from_vcs(cfg.tag_prefix, root, verbose)
return render(pieces, cfg.style)
except NotThisMethod:
pass
try:
if cfg.parentdir_prefix:
return versions_from_parentdir(cfg.parentdir_prefix, root, verbose)
except NotThisMethod:
pass
return {"version": "0+unknown", "full-revisionid": None,
"dirty": None,
"error": "unable to compute version", "date": None}
'''
@register_vcs_handler("git", "get_keywords")
def git_get_keywords(versionfile_abs):
"""Extract version information from the given file."""
# the code embedded in _version.py can just fetch the value of these
# keywords. When used from setup.py, we don't want to import _version.py,
# so we do it with a regexp instead. This function is not used from
# _version.py.
keywords = {}
try:
f = open(versionfile_abs, "r")
for line in f.readlines():
if line.strip().startswith("git_refnames ="):
mo = re.search(r'=\s*"(.*)"', line)
if mo:
keywords["refnames"] = mo.group(1)
if line.strip().startswith("git_full ="):
mo = re.search(r'=\s*"(.*)"', line)
if mo:
keywords["full"] = mo.group(1)
if line.strip().startswith("git_date ="):
mo = re.search(r'=\s*"(.*)"', line)
if mo:
keywords["date"] = mo.group(1)
f.close()
except EnvironmentError:
pass
return keywords
@register_vcs_handler("git", "keywords")
def git_versions_from_keywords(keywords, tag_prefix, verbose):
"""Get version information from git keywords."""
if not keywords:
raise NotThisMethod("no keywords at all, weird")
date = keywords.get("date")
if date is not None:
# git-2.2.0 added "%cI", which expands to an ISO-8601 -compliant
# datestamp. However we prefer "%ci" (which expands to an "ISO-8601
# -like" string, which we must then edit to make compliant), because
# it's been around since git-1.5.3, and it's too difficult to
# discover which version we're using, or to work around using an
# older one.
date = date.strip().replace(" ", "T", 1).replace(" ", "", 1)
refnames = keywords["refnames"].strip()
if refnames.startswith("$Format"):
if verbose:
print("keywords are unexpanded, not using")
raise NotThisMethod("unexpanded keywords, not a git-archive tarball")
refs = set([r.strip() for r in refnames.strip("()").split(",")])
# starting in git-1.8.3, tags are listed as "tag: foo-1.0" instead of
# just "foo-1.0". If we see a "tag: " prefix, prefer those.
TAG = "tag: "
tags = set([r[len(TAG):] for r in refs if r.startswith(TAG)])
if not tags:
# Either we're using git < 1.8.3, or there really are no tags. We use
# a heuristic: assume all version tags have a digit. The old git %d
# expansion behaves like git log --decorate=short and strips out the
# refs/heads/ and refs/tags/ prefixes that would let us distinguish
# between branches and tags. By ignoring refnames without digits, we
# filter out many common branch names like "release" and
# "stabilization", as well as "HEAD" and "master".
tags = set([r for r in refs if re.search(r'\d', r)])
if verbose:
print("discarding '%s', no digits" % ",".join(refs - tags))
if verbose:
print("likely tags: %s" % ",".join(sorted(tags)))
for ref in sorted(tags):
# sorting will prefer e.g. "2.0" over "2.0rc1"
if ref.startswith(tag_prefix):
r = ref[len(tag_prefix):]
if verbose:
print("picking %s" % r)
return {"version": r,
"full-revisionid": keywords["full"].strip(),
"dirty": False, "error": None,
"date": date}
# no suitable tags, so version is "0+unknown", but full hex is still there
if verbose:
print("no suitable tags, using unknown + full revision id")
return {"version": "0+unknown",
"full-revisionid": keywords["full"].strip(),
"dirty": False, "error": "no suitable tags", "date": None}
@register_vcs_handler("git", "pieces_from_vcs")
def git_pieces_from_vcs(tag_prefix, root, verbose, run_command=run_command):
"""Get version from 'git describe' in the root of the source tree.
This only gets called if the git-archive 'subst' keywords were *not*
expanded, and _version.py hasn't already been rewritten with a short
version string, meaning we're inside a checked out source tree.
"""
GITS = ["git"]
if sys.platform == "win32":
GITS = ["git.cmd", "git.exe"]
out, rc = run_command(GITS, ["rev-parse", "--git-dir"], cwd=root,
hide_stderr=True)
if rc != 0:
if verbose:
print("Directory %s not under git control" % root)
raise NotThisMethod("'git rev-parse --git-dir' returned error")
# if there is a tag matching tag_prefix, this yields TAG-NUM-gHEX[-dirty]
# if there isn't one, this yields HEX[-dirty] (no NUM)
describe_out, rc = run_command(GITS, ["describe", "--tags", "--dirty",
"--always", "--long",
"--match", "%s*" % tag_prefix],
cwd=root)
# --long was added in git-1.5.5
if describe_out is None:
raise NotThisMethod("'git describe' failed")
describe_out = describe_out.strip()
full_out, rc = run_command(GITS, ["rev-parse", "HEAD"], cwd=root)
if full_out is None:
raise NotThisMethod("'git rev-parse' failed")
full_out = full_out.strip()
pieces = {}
pieces["long"] = full_out
pieces["short"] = full_out[:7] # maybe improved later
pieces["error"] = None
# parse describe_out. It will be like TAG-NUM-gHEX[-dirty] or HEX[-dirty]
# TAG might have hyphens.
git_describe = describe_out
# look for -dirty suffix
dirty = git_describe.endswith("-dirty")
pieces["dirty"] = dirty
if dirty:
git_describe = git_describe[:git_describe.rindex("-dirty")]
# now we have TAG-NUM-gHEX or HEX
# Determine which branch is active (ie, checked-out)
branch_name, rc = run_command(
GITS, ["rev-parse", "--abbrev-ref", "HEAD"], cwd=root)
pieces["branch"] = branch_name
if "-" in git_describe:
# TAG-NUM-gHEX
mo = re.search(r'^(.+)-(\d+)-g([0-9a-f]+)$', git_describe)
if not mo:
# unparseable. Maybe git-describe is misbehaving?
pieces["error"] = ("unable to parse git-describe output: '%s'"
% describe_out)
return pieces
# tag
full_tag = mo.group(1)
if not full_tag.startswith(tag_prefix):
if verbose:
fmt = "tag '%s' doesn't start with prefix '%s'"
print(fmt % (full_tag, tag_prefix))
pieces["error"] = ("tag '%s' doesn't start with prefix '%s'"
% (full_tag, tag_prefix))
return pieces
pieces["closest-tag"] = full_tag[len(tag_prefix):]
# distance: number of commits since tag
pieces["distance"] = int(mo.group(2))
# Added to deal with merged branches
history_search_args = ["--ancestry-path"]
if (branch_name == "master"):
# Count distance only along `master`; ignore merge paths
history_search_args = ["--ancestry-path", "--first-parent"]
distance_to_tag, rc = run_command(
GITS, ["rev-list", "--count",] + history_search_args +
["%s..HEAD" % pieces["closest-tag"] ], cwd=root)
pieces["distance"] = int(distance_to_tag)
# commit: short hex revision ID
pieces["short"] = mo.group(3)
else:
# HEX: no tags
pieces["closest-tag"] = None
count_out, rc = run_command(GITS, ["rev-list", "HEAD", "--count"],
cwd=root)
pieces["distance"] = int(count_out) # total number of commits
##################################################
# Added for `develop` branch from `master`
# `HEAD` might have branched from untagged commit on `master`
merge_base_master, rc = run_command(
GITS, ["merge-base", "master", "HEAD", "-a"], cwd=root)
distance_to_master, rc = run_command(
GITS, ["rev-list", "--count", "--ancestry-path",
"%s..HEAD" % merge_base_master], cwd=root)
pieces["distance-to-master"] = int(distance_to_master)
##################################################
# Added for branches other than `master`
if pieces["closest-tag"]:
# TAG exists
master_to_tag, rc = run_command(
GITS, ["rev-list", "--count", "--ancestry-path", "--first-parent",
"%s..%s" % (pieces["closest-tag"], merge_base_master) ],
cwd=root)
pieces["master-to-tag"] = int(master_to_tag)
else:
pieces["master-to-tag"] = pieces["distance"]
##################################################
# Added for "feature" branch from `develop`
pieces["develop"] = None
pieces["distance-to-develop"] = None
branch_develop, rc = run_command(
GITS, ["rev-parse", "--verify", "refs/heads/develop"], cwd=root)
branch_develop_exists = not bool(rc)
if branch_develop_exists:
merge_base_develop, rc = run_command(
GITS, ["merge-base", "develop", "HEAD", "-a"], cwd=root)
distance_to_develop, rc = run_command(
GITS, ["rev-list", "--count", "--ancestry-path",
"%s..HEAD" % merge_base_develop], cwd=root)
pieces["develop"] = branch_develop
pieces["distance-to-develop"] = int(distance_to_develop)
# commit date: see ISO-8601 comment in git_versions_from_keywords()
date = run_command(GITS, ["show", "-s", "--format=%ci", "HEAD"],
cwd=root)[0].strip()
pieces["date"] = date.strip().replace(" ", "T", 1).replace(" ", "", 1)
##################################################
# Added to retrieve list of authors
(authors_raw, rc) = run_command(GITS, ["log", "--pretty=%an"], cwd=root)
authors = [author.strip() for author in authors_raw.split('\n')]
authors_unique = unique(authors)
pieces["authors"] = list(reversed(authors_unique))
return pieces
def do_vcs_install(manifest_in, versionfile_source, ipy):
"""Git-specific installation logic for Versioneer.
For Git, this means creating/changing .gitattributes to mark _version.py
for export-subst keyword substitution.
"""
GITS = ["git"]
if sys.platform == "win32":
GITS = ["git.cmd", "git.exe"]
files = [manifest_in, versionfile_source]
if ipy:
files.append(ipy)
try:
me = __file__
if me.endswith(".pyc") or me.endswith(".pyo"):
me = os.path.splitext(me)[0] + ".py"
versioneer_file = os.path.relpath(me)
except NameError:
versioneer_file = "versioneer.py"
files.append(versioneer_file)
present = False
try:
f = open(".gitattributes", "r")
for line in f.readlines():
if line.strip().startswith(versionfile_source):
if "export-subst" in line.strip().split()[1:]:
present = True
f.close()
except EnvironmentError:
pass
if not present:
f = open(".gitattributes", "a+")
f.write("%s export-subst\n" % versionfile_source)
f.close()
files.append(".gitattributes")
run_command(GITS, ["add", "--"] + files)
def versions_from_parentdir(parentdir_prefix, root, verbose):
"""Try to determine the version from the parent directory name.
Source tarballs conventionally unpack into a directory that includes both
the project name and a version string. We will also support searching up
two directory levels for an appropriately named parent directory
"""
rootdirs = []
for i in range(3):
dirname = os.path.basename(root)
if dirname.startswith(parentdir_prefix):
return {"version": dirname[len(parentdir_prefix):],
"full-revisionid": None,
"dirty": False, "error": None, "date": None}
else:
rootdirs.append(root)
root = os.path.dirname(root) # up a level
if verbose:
print("Tried directories %s but none started with prefix %s" %
(str(rootdirs), parentdir_prefix))
raise NotThisMethod("rootdir doesn't start with parentdir_prefix")
SHORT_VERSION_PY = """
# This file was generated by 'versioneer.py' (0.18) from
# revision-control system data, or from the parent directory name of an
# unpacked source archive. Distribution tarballs contain a pre-generated copy
# of this file.
import json
version_json = '''
%s
''' # END VERSION_JSON
def get_versions():
return json.loads(version_json)
"""
def versions_from_file(filename):
"""Try to determine the version from _version.py if present."""
try:
with open(filename) as f:
contents = f.read()
except EnvironmentError:
raise NotThisMethod("unable to read _version.py")
mo = re.search(r"version_json = '''\n(.*)''' # END VERSION_JSON",
contents, re.M | re.S)
if not mo:
mo = re.search(r"version_json = '''\r\n(.*)''' # END VERSION_JSON",
contents, re.M | re.S)
if not mo:
raise NotThisMethod("no version_json in _version.py")
return json.loads(mo.group(1))
def write_to_version_file(filename, versions):
"""Write the given version number to the given _version.py file."""
os.unlink(filename)
contents = json.dumps(versions, sort_keys=True,
indent=1, separators=(",", ": "))
with open(filename, "w") as f:
f.write(SHORT_VERSION_PY % contents)
print("set %s to '%s'" % (filename, versions["version"]))
def plus_or_dot(pieces):
"""Return a + if we don't already have one, else return a ."""
if "+" in pieces.get("closest-tag", ""):
return "."
return "+"
def render_pep440(pieces):
"""Build up version string, with post-release "local version identifier".
Our goal: TAG[+DISTANCE.gHEX[.dirty]] . Note that if you
get a tagged build and then dirty it, you'll get TAG+0.gHEX.dirty
Exceptions:
1: no tags. git_describe was just HEX. 0+untagged.DISTANCE.gHEX[.dirty]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"] or pieces["dirty"]:
rendered += plus_or_dot(pieces)
rendered += "%d.g%s" % (pieces["distance"], pieces["short"])
if pieces["dirty"]:
rendered += ".dirty"
else:
# exception #1
rendered = "0+untagged.%d.g%s" % (pieces["distance"],
pieces["short"])
if pieces["dirty"]:
rendered += ".dirty"
return rendered
def render_pep440_pre(pieces):
"""TAG[.post.devDISTANCE] -- No -dirty.
Exceptions:
1: no tags. 0.post.devDISTANCE
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"]:
rendered += ".post.dev%d" % pieces["distance"]
else:
# exception #1
rendered = "0.post.dev%d" % pieces["distance"]
return rendered
def render_pep440_post(pieces):
"""TAG[.postDISTANCE[.dev0]+gHEX] .
The ".dev0" means dirty. Note that .dev0 sorts backwards
(a dirty tree will appear "older" than the corresponding clean one),
but you shouldn't be releasing software with -dirty anyways.
Exceptions:
1: no tags. 0.postDISTANCE[.dev0]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"] or pieces["dirty"]:
rendered += ".post%d" % pieces["distance"]
if pieces["dirty"]:
rendered += ".dev0"
rendered += plus_or_dot(pieces)
rendered += "g%s" % pieces["short"]
else:
# exception #1
rendered = "0.post%d" % pieces["distance"]
if pieces["dirty"]:
rendered += ".dev0"
rendered += "+g%s" % pieces["short"]
return rendered
def render_pep440_old(pieces):
"""TAG[.postDISTANCE[.dev0]] .
The ".dev0" means dirty.
Eexceptions:
1: no tags. 0.postDISTANCE[.dev0]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"] or pieces["dirty"]:
rendered += ".post%d" % pieces["distance"]
if pieces["dirty"]:
rendered += ".dev0"
else:
# exception #1
rendered = "0.post%d" % pieces["distance"]
if pieces["dirty"]:
rendered += ".dev0"
return rendered
def render_pep440_micro(pieces):
"""Build up version string, with post-release "local version identifier".
Our goal: TAG.DISTANCE[+gHEX.dirty]
Exceptions:
1: no tags. git_describe was just HEX. 0+untagged.DISTANCE.gHEX[.dirty]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
rendered += ".%d" % pieces["distance"]
if pieces["dirty"]:
rendered += plus_or_dot(pieces)
rendered += "g%s" % pieces["short"]
rendered += ".dirty"
else:
# exception #1
rendered = "0+untagged.%d.g%s" % (pieces["distance"],
pieces["short"])
if pieces["dirty"]:
rendered += ".dirty"
return rendered
def render_pep440_develop(pieces):
"""Build up version string, used within development branch of repository.
Our goal: MERGE-POINT.post.devN[+gHEX.dirty]
+) MERGE-POINT = Most recent common ancestor for `develop` and `master`
*) Does not yet handle branch from `release-*`
+) N = DISTANCE from the MERGE-POINT
Exceptions:
1: no tags. 0.post.devDISTANCE[+gHEX.dirty]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
distance_to_merge = pieces["distance-to-master"]
# distance_merge_to_tag = (pieces["distance"] - distance_to_merge)
distance_merge_to_tag = pieces["master-to-tag"]
if (distance_merge_to_tag > 0):
rendered += ".%d" % distance_merge_to_tag
rendered += ".post.dev%d" % distance_to_merge
else:
# exception #1
rendered = "0.post.dev%d" % (pieces["distance"] - 1)
if pieces["dirty"]:
rendered += plus_or_dot(pieces)
rendered += "g%s" % pieces["short"]
rendered += ".dirty"
return rendered
def render_pep440_feature(pieces):
"""Build up version string, used within "feature" branch of repository.
Our goal: MERGE-POINT.post.devN+gHEX.BRANCH-NAME.M[.dirty]
+) MERGE-POINT = Most recent common ancestor for `develop` and `master`
*) Does not yet handle branch from `release-*`
+) N = DISTANCE from the MERGE-POINT of `develop` and `master`
+) M = DISTANCE from the MERGE-POINT of "feature" and `develop`
Exceptions:
1: no tags. 0.post.devDISTANCE+gHEX[.dirty]
"""
if pieces["closest-tag"] and pieces["develop"]:
rendered = pieces["closest-tag"]
distance_to_develop = pieces["distance-to-develop"]
distance_to_merge = pieces["distance-to-master"]
# distance_merge_to_tag = (pieces["distance"] - distance_to_merge)
distance_merge_to_tag = pieces["master-to-tag"]
distance_dev_to_merge = (distance_to_merge - distance_to_develop)
if (distance_merge_to_tag > 0):
rendered += ".%d" % distance_merge_to_tag
rendered += ".post.dev%d" % distance_dev_to_merge
rendered += plus_or_dot(pieces)
rendered += "g%s" % pieces["short"]
rendered += ".%s" % pieces["branch"]
rendered += ".%d" % distance_to_develop
else:
# exception #1
rendered = "0.post.dev%d" % (pieces["distance"] - 1)
rendered += plus_or_dot(pieces)
rendered += "g%s" % pieces["short"]
if pieces["dirty"]:
rendered += ".dirty"
return rendered
def render_pep440_auto(pieces):
"""Build up version string, based on branch of repository.
Our goal: If on development branch, use render_pep440_develop()
Otherwise use render_pep440_micro()
Exceptions:
+) NONE
"""
if pieces["branch"] == "master":
# logging.debug("Rendering: render_pep440_micro()")
rendered = render_pep440_micro(pieces)
elif pieces["branch"] == "develop":
# logging.debug("Rendering: render_pep440_develop()")
rendered = render_pep440_develop(pieces)
elif pieces["branch"].startswith("release"):
# render_pep440_release() not yet implemented
rendered = render_git_describe(pieces)
else:
# logging.debug("Rendering: render_pep440_feature()")
rendered = render_pep440_feature(pieces)
return rendered
def render_git_describe(pieces):
"""TAG[-DISTANCE-gHEX][-dirty].
Like 'git describe --tags --dirty --always'.
Exceptions:
1: no tags. HEX[-dirty] (note: no 'g' prefix)
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"]:
rendered += "-%d-g%s" % (pieces["distance"], pieces["short"])
else:
# exception #1
rendered = pieces["short"]
if pieces["dirty"]:
rendered += "-dirty"
return rendered
def render_git_describe_long(pieces):
"""TAG-DISTANCE-gHEX[-dirty].
Like 'git describe --tags --dirty --always -long'.
The distance/hash is unconditional.
Exceptions:
1: no tags. HEX[-dirty] (note: no 'g' prefix)
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
rendered += "-%d-g%s" % (pieces["distance"], pieces["short"])
else:
# exception #1
rendered = pieces["short"]
if pieces["dirty"]:
rendered += "-dirty"
return rendered
def render(pieces, style):
"""Render the given version pieces into the requested style."""
if pieces["error"]:
return {"version": "unknown",
"full-revisionid": pieces.get("long"),
"dirty": None,
"error": pieces["error"],
"date": None,
"authors": None,
}
if not style or style == "default":
style = "pep440-auto" # the default
if style == "pep440":
rendered = render_pep440(pieces)
elif style == "pep440-pre":
rendered = render_pep440_pre(pieces)
elif style == "pep440-post":
rendered = render_pep440_post(pieces)
elif style == "pep440-old":
rendered = render_pep440_old(pieces)
elif style == "pep440-micro":
rendered = render_pep440_micro(pieces)
elif style == "pep440-develop":
rendered = render_pep440_develop(pieces)
elif style == "pep440-auto":
rendered = render_pep440_auto(pieces)
elif style == "git-describe":
rendered = render_git_describe(pieces)
elif style == "git-describe-long":
rendered = render_git_describe_long(pieces)
else:
raise ValueError("unknown style '%s'" % style)
return {"version": rendered, "full-revisionid": pieces["long"],
"dirty": pieces["dirty"], "error": None,
"date": pieces.get("date"), "authors": pieces["authors"]}
class VersioneerBadRootError(Exception):
"""The project root directory is unknown or missing key files."""
def get_versions(verbose=False):
"""Get the project version from whatever source is available.
Returns dict with two keys: 'version' and 'full'.
"""
if "versioneer" in sys.modules:
# see the discussion in cmdclass.py:get_cmdclass()
del sys.modules["versioneer"]
root = get_root()
cfg = get_config_from_root(root)
assert cfg.VCS is not None, "please set [versioneer]VCS= in setup.cfg"
handlers = HANDLERS.get(cfg.VCS)
assert handlers, "unrecognized VCS '%s'" % cfg.VCS
verbose = verbose or cfg.verbose
assert cfg.versionfile_source is not None, \
"please set versioneer.versionfile_source"
assert cfg.tag_prefix is not None, "please set versioneer.tag_prefix"
versionfile_abs = os.path.join(root, cfg.versionfile_source)
# extract version from first of: _version.py, VCS command (e.g. 'git
# describe'), parentdir. This is meant to work for developers using a
# source checkout, for users of a tarball created by 'setup.py sdist',
# and for users of a tarball/zipball created by 'git archive' or github's
# download-from-tag feature or the equivalent in other VCSes.
get_keywords_f = handlers.get("get_keywords")
from_keywords_f = handlers.get("keywords")
if get_keywords_f and from_keywords_f:
try:
keywords = get_keywords_f(versionfile_abs)
ver = from_keywords_f(keywords, cfg.tag_prefix, verbose)
if verbose:
print("got version from expanded keyword %s" % ver)
return ver
except NotThisMethod:
pass
try:
ver = versions_from_file(versionfile_abs)
if verbose:
print("got version from file %s %s" % (versionfile_abs, ver))
return ver
except NotThisMethod:
pass
from_vcs_f = handlers.get("pieces_from_vcs")
if from_vcs_f:
try:
pieces = from_vcs_f(cfg.tag_prefix, root, verbose)
ver = render(pieces, cfg.style)
if verbose:
print("got version from VCS %s" % ver)
return ver
except NotThisMethod:
pass
try:
if cfg.parentdir_prefix:
ver = versions_from_parentdir(cfg.parentdir_prefix, root, verbose)
if verbose:
print("got version from parentdir %s" % ver)
return ver
except NotThisMethod:
pass
if verbose:
print("unable to compute version")
return {"version": "0+unknown", "full-revisionid": None,
"dirty": None, "error": "unable to compute version",
"date": None,
"authors": [],
}
def get_version():
"""Get the short version string for this project."""
return get_versions()["version"]
def get_cmdclass():
"""Get the custom setuptools/distutils subclasses used by Versioneer."""
if "versioneer" in sys.modules:
del sys.modules["versioneer"]
# this fixes the "python setup.py develop" case (also 'install' and
# 'easy_install .'), in which subdependencies of the main project are
# built (using setup.py bdist_egg) in the same python process. Assume
# a main project A and a dependency B, which use different versions
# of Versioneer. A's setup.py imports A's Versioneer, leaving it in
# sys.modules by the time B's setup.py is executed, causing B to run
# with the wrong versioneer. Setuptools wraps the sub-dep builds in a
# sandbox that restores sys.modules to it's pre-build state, so the
# parent is protected against the child's "import versioneer". By
# removing ourselves from sys.modules here, before the child build
# happens, we protect the child from the parent's versioneer too.
# Also see https://github.com/warner/python-versioneer/issues/52
cmds = {}
# we add "version" to both distutils and setuptools
from distutils.core import Command
class cmd_version(Command):
description = "report generated version string"
user_options = []
boolean_options = []
def initialize_options(self):
pass
def finalize_options(self):
pass
def run(self):
vers = get_versions(verbose=True)
print("Version: %s" % vers["version"])
print(" full-revisionid: %s" % vers.get("full-revisionid"))
print(" dirty: %s" % vers.get("dirty"))
print(" date: %s" % vers.get("date"))
if vers["error"]:
print(" error: %s" % vers["error"])
cmds["version"] = cmd_version
# we override "build_py" in both distutils and setuptools
#
# most invocation pathways end up running build_py:
# distutils/build -> build_py
# distutils/install -> distutils/build ->..
# setuptools/bdist_wheel -> distutils/install ->..
# setuptools/bdist_egg -> distutils/install_lib -> build_py
# setuptools/install -> bdist_egg ->..
# setuptools/develop -> ?
# pip install:
# copies source tree to a tempdir before running egg_info/etc
# if .git isn't copied too, 'git describe' will fail
# then does setup.py bdist_wheel, or sometimes setup.py install
# setup.py egg_info -> ?
# we override different "build_py" commands for both environments
if "setuptools" in sys.modules:
from setuptools.command.build_py import build_py as _build_py
else:
from distutils.command.build_py import build_py as _build_py
class cmd_build_py(_build_py):
def run(self):
root = get_root()
cfg = get_config_from_root(root)
versions = get_versions()
_build_py.run(self)
# now locate _version.py in the new build/ directory and replace
# it with an updated value
if cfg.versionfile_build:
target_versionfile = os.path.join(self.build_lib,
cfg.versionfile_build)
print("UPDATING %s" % target_versionfile)
write_to_version_file(target_versionfile, versions)
cmds["build_py"] = cmd_build_py
if "cx_Freeze" in sys.modules: # cx_freeze enabled?
from cx_Freeze.dist import build_exe as _build_exe
# nczeczulin reports that py2exe won't like the pep440-style string
# as FILEVERSION, but it can be used for PRODUCTVERSION, e.g.
# setup(console=[{
# "version": versioneer.get_version().split("+", 1)[0], # FILEVERSION
# "product_version": versioneer.get_version(),
# ...
class cmd_build_exe(_build_exe):
def run(self):
root = get_root()
cfg = get_config_from_root(root)
versions = get_versions()
target_versionfile = cfg.versionfile_source
print("UPDATING %s" % target_versionfile)
write_to_version_file(target_versionfile, versions)
_build_exe.run(self)
os.unlink(target_versionfile)
with open(cfg.versionfile_source, "w") as f:
LONG = LONG_VERSION_PY[cfg.VCS]
f.write(LONG %
{"DOLLAR": "$",
"STYLE": cfg.style,
"TAG_PREFIX": cfg.tag_prefix,
"PARENTDIR_PREFIX": cfg.parentdir_prefix,
"VERSIONFILE_SOURCE": cfg.versionfile_source,
})
cmds["build_exe"] = cmd_build_exe
del cmds["build_py"]
if 'py2exe' in sys.modules: # py2exe enabled?
try:
from py2exe.distutils_buildexe import py2exe as _py2exe # py3
except ImportError:
from py2exe.build_exe import py2exe as _py2exe # py2
class cmd_py2exe(_py2exe):
def run(self):
root = get_root()
cfg = get_config_from_root(root)
versions = get_versions()
target_versionfile = cfg.versionfile_source
print("UPDATING %s" % target_versionfile)
write_to_version_file(target_versionfile, versions)
_py2exe.run(self)
os.unlink(target_versionfile)
with open(cfg.versionfile_source, "w") as f:
LONG = LONG_VERSION_PY[cfg.VCS]
f.write(LONG %
{"DOLLAR": "$",
"STYLE": cfg.style,
"TAG_PREFIX": cfg.tag_prefix,
"PARENTDIR_PREFIX": cfg.parentdir_prefix,
"VERSIONFILE_SOURCE": cfg.versionfile_source,
})
cmds["py2exe"] = cmd_py2exe
# we override different "sdist" commands for both environments
if "setuptools" in sys.modules:
from setuptools.command.sdist import sdist as _sdist
else:
from distutils.command.sdist import sdist as _sdist
class cmd_sdist(_sdist):
def run(self):
versions = get_versions()
self._versioneer_generated_versions = versions
# unless we update this, the command will keep using the old
# version
self.distribution.metadata.version = versions["version"]
return _sdist.run(self)
def make_release_tree(self, base_dir, files):
root = get_root()
cfg = get_config_from_root(root)
_sdist.make_release_tree(self, base_dir, files)
# now locate _version.py in the new base_dir directory
# (remembering that it may be a hardlink) and replace it with an
# updated value
target_versionfile = os.path.join(base_dir, cfg.versionfile_source)
print("UPDATING %s" % target_versionfile)
write_to_version_file(target_versionfile,
self._versioneer_generated_versions)
cmds["sdist"] = cmd_sdist
return cmds
CONFIG_ERROR = """
setup.cfg is missing the necessary Versioneer configuration. You need
a section like:
[versioneer]
VCS = git
style = pep440
versionfile_source = src/myproject/_version.py
versionfile_build = myproject/_version.py
tag_prefix =
parentdir_prefix = myproject-
You will also need to edit your setup.py to use the results:
import versioneer
setup(version=versioneer.get_version(),
cmdclass=versioneer.get_cmdclass(), ...)
Please read the docstring in ./versioneer.py for configuration instructions,
edit setup.cfg, and re-run the installer or 'python versioneer.py setup'.
"""
SAMPLE_CONFIG = """
# See the docstring in versioneer.py for instructions. Note that you must
# re-run 'versioneer.py setup' after changing this section, and commit the
# resulting files.
[versioneer]
#VCS = git
#style = pep440
#versionfile_source =
#versionfile_build =
#tag_prefix =
#parentdir_prefix =
"""
INIT_PY_SNIPPET = """
from ._version import get_versions
__version__ = get_versions()['version']
del get_versions
"""
def do_setup():
"""Main VCS-independent setup function for installing Versioneer."""
root = get_root()
try:
cfg = get_config_from_root(root)
except (EnvironmentError, configparser.NoSectionError,
configparser.NoOptionError) as e:
if isinstance(e, (EnvironmentError, configparser.NoSectionError)):
print("Adding sample versioneer config to setup.cfg",
file=sys.stderr)
with open(os.path.join(root, "setup.cfg"), "a") as f:
f.write(SAMPLE_CONFIG)
print(CONFIG_ERROR, file=sys.stderr)
return 1
print(" creating %s" % cfg.versionfile_source)
with open(cfg.versionfile_source, "w") as f:
LONG = LONG_VERSION_PY[cfg.VCS]
f.write(LONG % {"DOLLAR": "$",
"STYLE": cfg.style,
"TAG_PREFIX": cfg.tag_prefix,
"PARENTDIR_PREFIX": cfg.parentdir_prefix,
"VERSIONFILE_SOURCE": cfg.versionfile_source,
})
ipy = os.path.join(os.path.dirname(cfg.versionfile_source),
"__init__.py")
if os.path.exists(ipy):
try:
with open(ipy, "r") as f:
old = f.read()
except EnvironmentError:
old = ""
if INIT_PY_SNIPPET not in old:
print(" appending to %s" % ipy)
with open(ipy, "a") as f:
f.write(INIT_PY_SNIPPET)
else:
print(" %s unmodified" % ipy)
else:
print(" %s doesn't exist, ok" % ipy)
ipy = None
# Make sure both the top-level "versioneer.py" and versionfile_source
# (PKG/_version.py, used by runtime code) are in MANIFEST.in, so
# they'll be copied into source distributions. Pip won't be able to
# install the package without this.
manifest_in = os.path.join(root, "MANIFEST.in")
simple_includes = set()
try:
with open(manifest_in, "r") as f:
for line in f:
if line.startswith("include "):
for include in line.split()[1:]:
simple_includes.add(include)
except EnvironmentError:
pass
# That doesn't cover everything MANIFEST.in can do
# (http://docs.python.org/2/distutils/sourcedist.html#commands), so
# it might give some false negatives. Appending redundant 'include'
# lines is safe, though.
if "versioneer.py" not in simple_includes:
print(" appending 'versioneer.py' to MANIFEST.in")
with open(manifest_in, "a") as f:
f.write("include versioneer.py\n")
else:
print(" 'versioneer.py' already in MANIFEST.in")
if cfg.versionfile_source not in simple_includes:
print(" appending versionfile_source ('%s') to MANIFEST.in" %
cfg.versionfile_source)
with open(manifest_in, "a") as f:
f.write("include %s\n" % cfg.versionfile_source)
else:
print(" versionfile_source already in MANIFEST.in")
# Make VCS-specific changes. For git, this means creating/changing
# .gitattributes to mark _version.py for export-subst keyword
# substitution.
do_vcs_install(manifest_in, cfg.versionfile_source, ipy)
return 0
def scan_setup_py():
"""Validate the contents of setup.py against Versioneer's expectations."""
found = set()
setters = False
errors = 0
with open("setup.py", "r") as f:
for line in f.readlines():
if "import versioneer" in line:
found.add("import")
if "versioneer.get_cmdclass()" in line:
found.add("cmdclass")
if "versioneer.get_version()" in line:
found.add("get_version")
if "versioneer.VCS" in line:
setters = True
if "versioneer.versionfile_source" in line:
setters = True
if len(found) != 3:
print("")
print("Your setup.py appears to be missing some important items")
print("(but I might be wrong). Please make sure it has something")
print("roughly like the following:")
print("")
print(" import versioneer")
print(" setup( version=versioneer.get_version(),")
print(" cmdclass=versioneer.get_cmdclass(), ...)")
print("")
errors += 1
if setters:
print("You should remove lines like 'versioneer.VCS = ' and")
print("'versioneer.versionfile_source = ' . This configuration")
print("now lives in setup.cfg, and should be removed from setup.py")
print("")
errors += 1
return errors
if __name__ == "__main__":
cmd = sys.argv[1]
if cmd == "setup":
errors = do_setup()
errors += scan_setup_py()
if errors:
sys.exit(1) | ALS.Liam | /ALS.Liam-0.10.0.tar.gz/ALS.Liam-0.10.0/versioneer.py | versioneer.py |
import sip
sip.setapi('QVariant', 2) # Call this before referencing QtCore
sip.setapi('QString', 2) # Call this before referencing QtCore
# from PyQt4 import QtGui, QtCore
# from PyQt4.QtCore import Qt, QString, QVariant, QRect, QRectF, QSize, QPoint
from PyQt4.QtCore import Qt, QRect, QRectF, QSize, QPoint
from PyQt4.QtCore import QObject, SIGNAL, SLOT, QCoreApplication
from PyQt4.QtCore import QAbstractTableModel, QModelIndex
from PyQt4.QtGui import QApplication, QMainWindow, QTextEdit, QLabel, QStyle
from PyQt4.QtGui import QStyledItemDelegate, QStyleOptionViewItemV4
from PyQt4.QtGui import QTextDocument, QPushButton, QStyleOptionButton
from PyQt4.QtGui import QStyleFactory, QGraphicsProxyWidget, QCheckBox
from PyQt4.QtGui import QSizePolicy, QFileDialog, QPalette, QColor
from PyQt4.QtGui import QHeaderView
# from PyQt4.QtGui import QCommonStyle, QCleanlooksStyle, QPlastiqueStyle
# from PyQt4.QtGui import QMacStyle, QWindowsStyle, QWindowsXPStyle
import pyqtgraph as pg
import als.liam.fitsViewerGui as fitsViewerGui
from als.liam.fitsParams import fitsParamsModel
from als.liam.fitsParams import qtFloatDelegate, qtHtmlDelegate
from als.liam.fitsParams import qtDateDelegate, qtTimeDelegate
from als.liam.fitsParams import fitsParamsLeftColModel, fitsParamsRightColModel
from PIL import Image
from numpy import nan, float32, sqrt, round, floor, array, log10
from numpy.random import random_integers, randint
from astropy.io import fits
import pandas as pd
import os
import sys
from als.milo.qimage import Diffractometer402, CcdImageFromFITS
try:
#_fromUtf8 = QString.fromUtf8
pass
except AttributeError:
def _fromUtf8(s):
return s
try:
_encoding = QApplication.UnicodeUTF8
def _translate(context, text, disambig):
return QApplication.translate(context, text, disambig, _encoding)
except AttributeError:
def _translate(context, text, disambig):
return QApplication.translate(context, text, disambig)
pg.setConfigOptions(imageAxisOrder="row-major")
def show_motor_params(primary_hdu):
# print 'Beamline Energy : ' + str(primary_hdu['Beamline Energy']) + ' eV'
print 'Mono Energy : ' + str(primary_hdu['Mono Energy']) + ' eV'
print 'Bottom Rotary Seal: ' + str(primary_hdu['Bottom Rotary Seal']) + ' deg'
print 'Top Rotary Seal : ' + str(primary_hdu['Top Rotary Seal']) + ' deg'
print 'Flip : ' + str(primary_hdu['Flip']) + ' deg'
print 'Twice Top Offset : ' + str(primary_hdu['Twice Top Offset']) + ' deg'
# energy = primary_hdu['Beamline Energy']
energy = primary_hdu['Mono Energy']
bottom = primary_hdu['Bottom Rotary Seal']
top = primary_hdu['Top Rotary Seal']
flip = primary_hdu['Flip']
offset_top = primary_hdu['Twice Top Offset'] / 2
offset_flip = 0
offset_ccd = -18.68
if ('Top Offset' in primary_hdu):
offset_top = primary_hdu['Top Offset']
print 'Top Offset : ' + str(offset_top) + ' deg'
if ('Flip Offset' in primary_hdu):
offset_flip = primary_hdu['Flip Offset']
print 'Flip Offset : ' + str(offset_flip) + ' deg'
if ('CCD Offset' in primary_hdu):
offset_ccd = primary_hdu['CCD Offset']
print 'CCD Offset : ' + str(offset_ccd) + ' deg'
wavelength = 1239.842 / energy
twotheta = bottom - offset_ccd
truetop = top - offset_top
incidence = bottom - truetop
chi = flip - offset_flip
print ''
print 'Wavelength : ' + str(wavelength) + ' nm'
print 'Detector : ' + str(twotheta) + ' deg'
print 'Incidence : ' + str(incidence) + ' deg'
print 'Chi : ' + str(chi) + ' deg'
def load_image(filename):
hdulist = fits.open(filename)
# print "We opened an image. It's filename is ", filename
# print "here's some info"
# print hdulist.info()
# show_motor_params(hdulist[0].header)
# let's assume, the last entry is the image.
return (hdulist[0].header, hdulist[-1])
class FitsViewerApp(QMainWindow, fitsViewerGui.Ui_MainWindow):
def __init__(self, parent=None):
super(FitsViewerApp, self).__init__(parent)
self.setupUi(self)
self.setWindowTitle(
_translate("MainWindow", "FITS data viewer", None))
glImage = self.glFitsImage
glBoxImageTitle = (0, 0, 1, 3)
glBoxImageSubtitle = (1, 0, 1, 3)
glBoxPlotRows = (2, 0)
glBoxImageView = (2, 1)
glBoxLutHistog = (2, 2)
glBoxPlotCols = (3, 1)
glBoxColorScaling = (3, 2)
self.txtImageTitle = pg.LabelItem(
"Click 'Load data file' button to begin...", size='24pt')
self.txtImageSubtitle = pg.LabelItem("Default data displayed")
glImage.addItem(self.txtImageTitle, *glBoxImageTitle)
glImage.addItem(self.txtImageSubtitle, *glBoxImageSubtitle)
self.txtImageTitle.setSizePolicy(
QSizePolicy(QSizePolicy.Minimum, QSizePolicy.Minimum))
self.txtImageSubtitle.setSizePolicy(
QSizePolicy(QSizePolicy.Minimum, QSizePolicy.Minimum))
self.image_array = randint(512, 2**16, size=(512, 512))
image_array = self.image_array
(imgRows, imgCols) = image_array.shape
self.imv = glImage.addViewBox(*glBoxImageView)
imv = self.imv
# imv.invertY(True) # Need to invert if image is not FITS
img = pg.ImageItem(image_array, border=0.5)
self.image = img
self.image.setCursor(Qt.CrossCursor)
imv.addItem(img)
imv.setAspectLocked()
# imv.setLimits(
# maxXRange=imgCols, maxYRange=imgRows,
# xMin=0, xMax=imgCols, yMin=0, yMax=imgRows,
# )
self.linRegRows = pg.LinearRegionItem(
values=[0, 0],
orientation=pg.LinearRegionItem.Horizontal,
brush=(0, 0, 255, 64),
# pen='b',
bounds=(0, imgRows),
)
self.linRegCols = pg.LinearRegionItem(
values=[0, 0],
orientation=pg.LinearRegionItem.Vertical,
brush=(255, 255, 0, 64),
# pen = 'y',
bounds=(0, imgCols),
)
imv.addItem(self.linRegRows)
imv.addItem(self.linRegCols)
# imv.setPredefinedGradient("flame")
# imv.setLookupTable(xxx)
# vbLutHistog = glImage.addViewBox(0, 1)
self.lutHistog = pg.HistogramLUTItem(img)
lutHistog = self.lutHistog
# lutHistog.setImageItem(img)
lutHistog.plot.rotate(-90)
lutHistog.plot.setLogMode(False, True)
lutHistog.plot.rotate(90)
# lutHistog.sigLevelsChanged.connect(self.onImageLevelsChanged)
lutHistog.sigLevelChangeFinished.connect(self.onImageLevelsChanged)
lutHistog.gradient.loadPreset("flame")
glImage.addItem(lutHistog, *glBoxLutHistog)
# viewLutHistog = glImage.addViewBox(0, 2)
# viewLutHistog.addItem(lutHistog)
self.plotFitsRows = glImage.addPlot(*glBoxPlotRows)
self.plotFitsCols = glImage.addPlot(*glBoxPlotCols)
self.plotFitsRowsData = self.plotFitsRows.plot(
image_array.mean(axis=1),
fillLevel=image_array.min(),
# -image_array.mean(axis=1),
# fillLevel = -(image_array.min()),
# fillLevel = (-image_array.astype(int)).max(),
fillBrush='y')
self.plotFitsRowsData.scale(-1, 1)
self.plotFitsRowsData.rotate(90)
self.plotFitsRows.setYLink(imv)
self.plotFitsRows.invertX(True)
# self.plotFitsRows.invertY(True)
self.plotFitsColsData = self.plotFitsCols.plot(
image_array.mean(axis=0),
fillLevel=image_array.min(),
fillBrush='b')
self.plotFitsCols.setXLink(imv)
# self.plotFitsRows.rotate(90)
glImage.ci.layout.setColumnMaximumWidth(glBoxPlotRows[1], 100)
glImage.ci.layout.setRowMaximumHeight(glBoxPlotCols[0], 100)
glImageScale = pg.GraphicsLayout()
proxy = QGraphicsProxyWidget()
self.chkAutoScale = QCheckBox('Auto scale')
self.chkAutoScale.setChecked(True)
self.chkAutoScale.stateChanged.connect(self.onAutoScaleChanged)
# self.chkAutoScale.setSizePolicy(
# QSizePolicy(QSizePolicy.Minimum, QSizePolicy.Minimum))
proxy.setWidget(self.chkAutoScale)
glImageScale.addItem(proxy, 0, 0)
proxy = QGraphicsProxyWidget()
# self.chkLogScale = QCheckBox('Log scale', self)
self.chkLogScale = QCheckBox('Log scale')
self.chkLogScale.setChecked(False)
self.chkLogScale.stateChanged.connect(self.onLogScaleChanged)
# self.chkLogScale.setSizePolicy(
# QSizePolicy(QSizePolicy.Minimum, QSizePolicy.Minimum))
proxy.setWidget(self.chkLogScale)
glImageScale.addItem(proxy, 1, 0)
glImageScale.setSizePolicy(
QSizePolicy(QSizePolicy.Minimum, QSizePolicy.Minimum))
glImage.addItem(glImageScale, *glBoxColorScaling)
# SLOT('onLinRegColsChanged(QObject)')
# SLOT('onLinRegRowsChanged(QObject)')
self.linRegCols.sigRegionChanged.connect(self.onLinRegColsChanged)
self.linRegRows.sigRegionChanged.connect(self.onLinRegRowsChanged)
# QObject.connect(
# # self.linRegCols, SIGNAL('sigRangeChanged(QObject)'),
# self.linRegCols, SIGNAL('sigRangeChanged'),
# self.onLinRegColsChanged)
# QObject.connect(
# # self.linRegRows, SIGNAL('sigRangeChanged(QObject)'),
# self.linRegRows, SIGNAL('sigRangeChanged'),
# self.onLinRegRowsChanged)
imv.autoRange()
self.spinDataFrameNum.setMinimum(0)
self.spinDataFrameNum.setMaximum(0)
self.image_cursor_label = None
img.scene().sigMouseMoved.connect(self.onMouseMoved)
self.btnSelectDataFile.clicked.connect(self.selectDataFile)
self.btnReloadDataFile.clicked.connect(self.loadDataFile)
self.spinDataFrameNum.valueChanged.connect(self.updateDataFrame)
self.btnFirstDataFrame.clicked.connect(self.gotoFirstDataFrame)
self.btnPrevDataFrame.clicked.connect(self.gotoPrevDataFrame)
self.btnNextDataFrame.clicked.connect(self.gotoNextDataFrame)
self.btnLastDataFrame.clicked.connect(self.gotoLastDataFrame)
# Project directory is parent of "als/liam/"
self.project_directory = os.path.dirname(
os.path.dirname(
os.path.dirname(
os.path.abspath(__file__)
)
)
)
self.data_directory = "{}/test_data/".format(self.project_directory)
self.working_directory = str(self.data_directory)
self.data_pathnames = []
self.data_frame_num = 0
# self.selectDataFile()
# palSelectButton = QPalette()
# palSelectButton.setColor(QPalette.Button, QColor(Qt.yellow))
# self.btnSelectDataFile.setAutoFillBackground(True)
# self.btnSelectDataFile.setPalette(palSelectButton)
self.cbPauseParamUpdates.setChecked(False)
self.cbPauseParamUpdates.stateChanged.connect(
self.onPauseFitsParamsUpdatesChanged)
self.model_fits_params_left = fitsParamsLeftColModel()
self.viewFitsParamsLeft.setModel(self.model_fits_params_left)
self.viewFitsParamsLeft.setItemDelegateForColumn(
0,
qtHtmlDelegate(self) )
self.viewFitsParamsLeft.setItemDelegateForColumn(
1,
qtFloatDelegate(self, decimals=3) )
self.viewFitsParamsLeft.setItemDelegateForColumn(
2,
qtHtmlDelegate(self) )
self.viewFitsParamsLeft.resizeColumnsToContents()
self.viewFitsParamsLeft.horizontalHeader().setResizeMode(
1, QHeaderView.Stretch)
self.viewFitsParamsLeft.horizontalHeader().setDefaultAlignment(
Qt.AlignRight)
self.viewFitsParamsLeft.horizontalHeader().hide()
self.viewFitsParamsLeft.verticalHeader().hide()
# self.viewFitsParamsLeft.setHorizontalScrollBarPolicy(
# Qt.ScrollBarAlwaysOff)
# self.viewFitsParamsLeft.setVerticalScrollBarPolicy(
# Qt.ScrollBarAlwaysOff)
# self.viewFitsParamsLeft.adjustSize()
fitsParamsRightColModel
self.model_fits_params_right = fitsParamsRightColModel()
self.viewFitsParamsRight.setModel(self.model_fits_params_right)
self.viewFitsParamsRight.setItemDelegateForColumn(
0,
qtHtmlDelegate(self) )
self.viewFitsParamsRight.setItemDelegateForColumn(
1,
qtFloatDelegate(self, decimals=3) )
self.viewFitsParamsRight.setItemDelegateForColumn(
2,
qtHtmlDelegate(self) )
self.viewFitsParamsRight.resizeColumnsToContents()
self.viewFitsParamsRight.horizontalHeader().setResizeMode(
1, QHeaderView.Stretch)
self.viewFitsParamsRight.horizontalHeader().setDefaultAlignment(
Qt.AlignRight)
self.viewFitsParamsRight.horizontalHeader().hide()
self.viewFitsParamsRight.verticalHeader().hide()
self.viewFitsParamsRight.setSpan(0, 0, 1, 3)
self.viewFitsParamsRight.setSpan(1, 0, 1, 3)
self.viewFitsParamsRight.setItemDelegateForRow(
0,
qtDateDelegate(self) )
self.viewFitsParamsRight.setItemDelegateForRow(
1,
qtTimeDelegate(self) )
# self.setStyleSheet(".sub { vertical-align: sub }")
# self.setStyleSheet(".sup { vertical-align: super }")
def onLinRegColsChanged(self, linReg):
# self.plotFitsRows.setYRange(linReg.getRegion())
image_array = self.image_array
(minCol, maxCol) = linReg.getRegion()
(minCol, maxCol) = (int(round(minCol)), int(round(maxCol)))
if (minCol == maxCol):
minCol = 0
maxCol = image_array.shape[1]
# print "onLinRegRowsChanged:", linReg.getRegion()
self.plotFitsRowsData.setData(image_array[:, minCol:maxCol].mean(axis=1))
# self.plotFitsRowsData.setData(-image_array[:, minCol:maxCol].mean(axis=1))
def onLinRegRowsChanged(self, linReg):
# self.plotFitsCols.setXRange(linReg.getRegion())
image_array = self.image_array
(minRow, maxRow) = linReg.getRegion()
(minRow, maxRow) = (int(round(minRow)), int(round(maxRow)))
if (minRow == maxRow):
minRow = 0
maxRow = image_array.shape[0]
# print "onLinRegColsChanged:", linReg.getRegion()
self.plotFitsColsData.setData(image_array[minRow:maxRow].mean(axis=0))
def onLogScaleChanged(self, state):
log_mode = (state & Qt.Checked)
lutHistog_levels = self.lutHistog.getLevels()
# if self.lutHistog.axis.logMode:
# lutHistog_levels = 10 ** array(lutHistog_levels)
image_array = self.image_array
if log_mode:
image_array = log10(self.image_array)
lutHistog_levels = log10(lutHistog_levels)
else:
lutHistog_levels = 10 ** array(lutHistog_levels)
fill_level = image_array.min()
self.plotFitsRows.getAxis("bottom").setLogMode(log_mode)
self.plotFitsCols.getAxis("left").setLogMode(log_mode)
self.plotFitsRowsData.rotate(-90)
self.plotFitsRowsData.setLogMode(False, log_mode)
self.plotFitsRowsData.setFillLevel(fill_level)
self.plotFitsRowsData.rotate(90)
self.plotFitsColsData.setLogMode(False, log_mode)
self.plotFitsColsData.setFillLevel(fill_level)
lutHistog = self.lutHistog
lutHistog.sigLevelChangeFinished.disconnect(self.onImageLevelsChanged)
self.image.setImage(image_array)
self.lutHistog.setLevels(*lutHistog_levels)
self.lutHistog.axis.setLogMode(log_mode)
lutHistog.sigLevelChangeFinished.connect(self.onImageLevelsChanged)
def onAutoScaleChanged(self, state):
auto_scale_mode = (state & Qt.Checked)
log_mode = self.chkLogScale.isChecked()
image_array = self.image_array
if log_mode:
image_array = log10(self.image_array)
if auto_scale_mode:
self.lutHistog.sigLevelChangeFinished.disconnect(
self.onImageLevelsChanged)
self.lutHistog.autoHistogramRange()
self.lutHistog.setLevels(image_array.min(), image_array.max())
self.lutHistog.sigLevelChangeFinished.connect(
self.onImageLevelsChanged)
def onImageLevelsChanged(self, lutHistog):
self.chkAutoScale.setChecked(False)
# log_mode = self.chkLogScale.isChecked()
#
# lutHistog_levels = lutHistog.getLevels()
# print "lutHistog_levels:", lutHistog_levels
# # if lutHistog.axis.logMode:
# if log_mode:
# lutHistog_levels = 10 ** array(lutHistog_levels)
# print "[LOG] lutHistog_levels:", lutHistog_levels
#
# # self.image
# print "getLevels():", lutHistog.imageItem().getLevels()
# lutHistog.imageItem().setLevels(lutHistog_levels)
# print "[AFTER] getLevels():", lutHistog.imageItem().getLevels()
# # lutHistog.update()
def onPauseFitsParamsUpdatesChanged(self, state):
if self.data_pathnames:
self.loadDataFile()
def keyPressEvent(self, keyEvent):
if ((
(keyEvent.key() == Qt.Key_Q) and
(keyEvent.modifiers() == Qt.ControlModifier)
) or
(
(keyEvent.key() == Qt.Key_W) and
(keyEvent.modifiers() == Qt.ControlModifier)
)):
print "QUIT"
QCoreApplication.instance().quit()
def onMouseMoved(self, event):
# pos = event.pos()
pos = [event.x(), event.y()]
if self.image.sceneBoundingRect().contains(event):
# self.image.setCursor(Qt.CrossCursor)
view_pos = self.imv.mapToView(event)
# image_pos = self.imv.mapFromViewToItem(
# self.image, view_pos)
image_pos = self.image.mapFromScene(event)
# print "\nEvent Pos:", event
# print "\tView Pos:", view_pos
# print "\tImage Pos:", image_pos
# pos = [event.x(), event.y()]
# pos = [view_pos.x(), view_pos.y()]
pos = [image_pos.x(), image_pos.y()]
# coords = tuple([int(round(coord)) for coord in pos])
coords = tuple([int(floor(coord)) for coord in pos])
# coord_text = "({0:d}, {1:d})".format(*coords)
coord_text = "(r:{1:d}, c:{0:d})".format(*coords)
coord_text += "\nIntemsity:{0:0,.0f}".format(
self.image_array[coords[1], coords[0]],
)
self.image.setToolTip(coord_text)
# if self.image_cursor_label is None:
# label = pg.TextItem(coord_text, anchor=(1,0), fill=(0,0,0,80))
# label.setParentItem(self.image)
# # label.setPos(*coords)
# # label.setPos(event)
# # label.setPos(view_pos)
# label.setPos(image_pos)
# self.image_cursor_label = label
# else:
# label = self.image_cursor_label
# label.setText(coord_text)
# # label.setPos(*coords)
# # label.setPos(event)
# # label.setPos(view_pos)
# label.setPos(image_pos)
# anchor_point = [1, 0]
# if not label.viewRect().contains(
# label.boundingRect().topLeft() ):
# anchor_point[0] = 0
# if not label.viewRect().contains(
# label.boundingRect().bottomRight() ):
# anchor_point[1] = 1
# label.setAnchor(anchor_point)
# else:
# if self.image_cursor_label is not None:
# self.image_cursor_label.scene().removeItem(
# self.image_cursor_label)
# self.image_cursor_label = None
def selectDataFile(self):
# data_pathname = "{}Log_NiFe_00d_1340-060.fits".format(self.data_directory)
# data_pathname = "{}NiFe_00d_1340-060.fits".format(self.data_directory)
# self.data_pathnames = [data_pathname]
# self.data_frame_num = 1
# png_filename = "{}NiFe_00d_1340-060.png".format(self.data_directory)
# self.image_array = array(Image.open(png_filename))
# # self.image_array = array(Image.open(png_filename).convert("L"))
# # print "self.image_array:", self.image_array
# # print "self.image_array.shape:", self.image_array.shape
# # exit()
# QFileDialog.setFileMode(QFileDialog.ExistingFile)
selected_pathname = str(QFileDialog.getOpenFileName(
self,
"Select data file",
self.working_directory,
# "FITS (*.fits);; Images (*.png *.jpg *.jpeg *.gif *.tiff);; AI files (*-AI.txt)")
"FITS, PNG, AI files (*.fits *.png *-AI.txt)")
)
# print "selected_pathname:", selected_pathname
self.raise_()
self.activateWindow()
data_frame_num = self.data_frame_num
selected_pathname_parts = selected_pathname.split('/')
data_path = "/".join(selected_pathname_parts[:-1])
self.working_directory = data_path
if selected_pathname.endswith("-AI.txt"):
# data_file = open(selected_pathname, 'r')
# file_line = data_file.readline().rstrip('\r\n')
# imgscan_header_linenum = 0
# while not file_line.startswith("Frame"):
# file_line = data_file.readline().rstrip('\r\n')
# imgscan_header_linenum++
# data_file.close()
with open(selected_pathname, 'r') as data_file:
for (header_linenum, file_line) in enumerate(data_file):
# print (header_linenum, file_line)
if file_line.startswith("Frame"):
break
if file_line.startswith("Time"):
break
if file_line[0].isdigit():
header_linenum -= 1
break
# check for missing header line
# print "header_linenum:", header_linenum
# header_linenum = 9
imgscan_data = pd.read_table(
selected_pathname,
delimiter='\t',
header=header_linenum,
skip_blank_lines=False,
)
# imgscan_num_rows = len(imgscan_data)
column_map = dict({"PNG Image filename": "Filename"})
imgscan_data.rename(columns=column_map, inplace=True)
for detector_path in ["WinView CCD", "LightField PI-MTE"]:
if os.path.exists("{:s}/{:s}/".format(
data_path,
detector_path,
)):
break # Detector image directory was found
else:
detector_path = ""
self.data_pathnames = ["{:s}/{:s}/{:s}.fits".format(
data_path,
detector_path,
filename,
) for filename in imgscan_data["Filename"] ]
if (self.data_frame_num > len(self.data_pathnames) ):
# self.data_frame_num = len(self.data_pathnames)
data_frame_num = len(self.data_pathnames)
if (len(self.data_pathnames) > 0):
# self.data_frame_num = 1
data_frame_num = 1
else:
self.data_pathnames = [selected_pathname]
# self.data_frame_num = 1
data_frame_num = 1
self.editDataFilename.setText(selected_pathname)
self.spinDataFrameNum.setMinimum(1)
self.spinDataFrameNum.setMaximum(len(self.data_pathnames))
# self.spinDataFrameNum.setValue(self.data_frame_num)
self.spinDataFrameNum.setValue(data_frame_num)
self.loadDataFile(fullView=True)
def loadDataFile(self, fullView=False):
data_pathname = self.data_pathnames[self.data_frame_num - 1]
# print "data_pathname:", data_pathname
data_pathname_parts = data_pathname.split('/')
data_path = " / ".join(data_pathname_parts[:-1])
data_filename = data_pathname_parts[-1]
self.txtImageTitle.setText(data_filename)
self.txtImageSubtitle.setText(data_path)
if data_filename.lower().endswith(".fits"):
# hdu_pair = load_image(data_pathname)
# hdu_header = hdu_pair[0]
# hdu_image = hdu_pair[-1]
hdulist = fits.open(data_pathname)
ccd_image = CcdImageFromFITS(
hdulist,
# offset_diffractometer = offset_angles
)
# self.image_array = hdu_image.data
self.image_array = ccd_image.data
self.imv.invertY(False) # Need to invert if image is not FITS
if self.cbPauseParamUpdates.isChecked():
self.updateFitsParameters(None)
else:
self.updateFitsParameters(ccd_image)
else:
self.image_array = array(Image.open(data_pathname))
self.imv.invertY(True) # Need to invert if image is not FITS
self.updateFitsParameters(None)
image_array = self.image_array
log_mode = self.chkLogScale.isChecked()
if log_mode:
image_array = log10(self.image_array)
(imgRows, imgCols) = image_array.shape
self.image.setImage(image_array)
self.image.setLevels(self.lutHistog.getLevels())
self.linRegRows.setBounds((0, imgRows))
self.linRegCols.setBounds((0, imgCols))
fill_level = image_array.min()
self.plotFitsRowsData.setFillLevel(fill_level)
self.plotFitsColsData.setFillLevel(fill_level)
if fullView:
self.imv.autoRange()
self.onLinRegRowsChanged(self.linRegRows)
self.onLinRegColsChanged(self.linRegCols)
def updateDataFrame(self, value):
self.data_frame_num = int(value)
self.loadDataFile()
def gotoFirstDataFrame(self):
self.spinDataFrameNum.setValue(self.spinDataFrameNum.minimum())
def gotoPrevDataFrame(self):
self.spinDataFrameNum.stepDown()
def gotoNextDataFrame(self):
self.spinDataFrameNum.stepUp()
def gotoLastDataFrame(self):
self.spinDataFrameNum.setValue(self.spinDataFrameNum.maximum())
def updateFitsParameters(self, ccdImage=None):
self.model_fits_params_left.loadParams(ccdImage)
self.model_fits_params_right.loadParams(ccdImage)
def main():
app = QApplication(sys.argv)
# print "QStyleFactory.keys():", [str(s) for s in QStyleFactory.keys()]
# print "app.style():", app.style(), type(app.style())
# print QApplication.style().metaObject().className(), "\n"
# for key in QStyleFactory.keys():
# st = QStyleFactory.create(key)
# print key, st.metaObject().className(), type(app.style())
# app.setStyle("Windows")
mainWin = FitsViewerApp()
mainWin.show()
mainWin.raise_()
# mainWin.activateWindow()
app.exec_()
if __name__ == '__main__':
main() | ALS.Liam | /ALS.Liam-0.10.0.tar.gz/ALS.Liam-0.10.0/als/liam/fitsViewer.py | fitsViewer.py |
__version__ = None # This will be assigned later; see below
__date__ = None # This will be assigned later; see below
__credits__ = None # This will be assigned later; see below
try:
from als.liam._version import git_pieces_from_vcs as _git_pieces_from_vcs
from als.liam._version import run_command, register_vcs_handler
from als.liam._version import render as _render
from als.liam._version import render_pep440_auto
from als.liam._version import render_pep440_micro, render_pep440_develop
from als.liam._version import get_versions as _get_versions
from als.liam._version import get_config, get_keywords
from als.liam._version import git_versions_from_keywords
from als.liam._version import versions_from_parentdir
from als.liam._version import NotThisMethod
except ImportError:
# Assumption is that _version.py was generated by 'versioneer.py'
# for tarball distribution, which contains only static JSON version data
from als.liam._version import get_versions
# from als.liam._version import get_versions as _get_versions
#
# def get_versions():
# """Get version information or return default if unable to do so.
#
# Extension to ._version.get_versions()
#
# Additional functionality:
# Returns list of authors found in `git`
# """
# default_keys_values = {
# "version": "0+unknown",
# "full-revisionid": None,
# "dirty": None,
# "error": "unable to compute version",
# "date": None,
# "authors": [],
# }
#
# return_key_values = _get_versions()
# return_key_values = dict(
# default_keys_values.items() + return_key_values.items()
# )
# return return_key_values
else:
import os
import sys
import numpy as np
@register_vcs_handler("git", "pieces_from_vcs")
def git_pieces_from_vcs(
tag_prefix, root, verbose, run_command=run_command):
"""Get version information from 'git' in the root of the source tree.
Extension to ._version.git_pieces_from_vcs()
Additional functionality:
Extracts all commit authors, sorts unique authors chronologically,
then adds them to `pieces["authors"]`, where `pieces` is the object
that was returned by ._version.git_pieces_from_vcs()
"""
pieces = _git_pieces_from_vcs(
tag_prefix, root, verbose, run_command=run_command)
GITS = ["git"]
if sys.platform == "win32":
GITS = ["git.cmd", "git.exe"]
##################################################
# Added to retrieve list of authors
(authors_raw, rc) = run_command(
GITS, ["log", "--pretty=%an"], cwd=root)
authors = [author.strip() for author in authors_raw.split('\n')]
(authors_unique, authors_indices) = np.unique(
authors, return_index=True)
pieces["authors"] = list(reversed(np.array(authors)[authors_indices]))
return pieces
def render(pieces, style):
"""Render the given version pieces into the requested style."""
if pieces["error"]:
return {"version": "unknown",
"full-revisionid": pieces.get("long"),
"dirty": None,
"error": pieces["error"],
"date": None,
"authors": None,
}
if not style or style == "default":
style = "pep440-auto" # the default
if style == "pep440-micro":
rendered = render_pep440_micro(pieces)
elif style == "pep440-develop":
rendered = render_pep440_develop(pieces)
elif style == "pep440-auto":
rendered = render_pep440_auto(pieces)
else:
return_key_values = _render(pieces, style)
return_key_values["authors"] = pieces["authors"]
return return_key_values
return {"version": rendered, "full-revisionid": pieces["long"],
"dirty": pieces["dirty"], "error": None,
"date": pieces.get("date"), "authors": pieces["authors"]}
def get_versions():
"""Get version information or return default if unable to do so.
Extension to ._version.get_versions()
Additional functionality:
Returns list of authors found in `git`
"""
# I am in _version.py, which lives at ROOT/VERSIONFILE_SOURCE.
# If we have __file__, we can work backwards from there to the root.
# Some py2exe/bbfreeze/non-CPython implementations don't do __file__,
# in which case we can only use expanded keywords.
cfg = get_config()
verbose = cfg.verbose
default_keys_values = {
"version": "0+unknown",
"full-revisionid": None,
"dirty": None,
"error": "unable to compute version",
"date": None,
"authors": [],
}
try:
return git_versions_from_keywords(get_keywords(), cfg.tag_prefix,
verbose)
except NotThisMethod:
pass
try:
root = os.path.realpath(__file__)
# versionfile_source is the relative path from the top of the
# source tree (where the .git directory might live) to this file.
# Invert this to find the root from __file__.
for i in cfg.versionfile_source.split('/'):
root = os.path.dirname(root)
except NameError:
return default_keys_values
try:
pieces = git_pieces_from_vcs(cfg.tag_prefix, root, verbose)
return render(pieces, cfg.style)
except NotThisMethod:
pass
try:
if cfg.parentdir_prefix:
return versions_from_parentdir(
cfg.parentdir_prefix, root, verbose)
except NotThisMethod:
pass
return_key_values = _get_versions()
return_key_values = dict(
default_keys_values.items() + return_key_values.items()
)
return return_key_values
__version__ = get_versions()["version"]
__date__ = get_versions()["date"]
__credits__ = get_versions()["authors"]
del get_versions | ALS.Liam | /ALS.Liam-0.10.0.tar.gz/ALS.Liam-0.10.0/als/liam/version.py | version.py |
import sip
sip.setapi('QVariant', 2) # Call this before referencing QtCore
sip.setapi('QString', 2) # Call this before referencing QtCore
# from PyQt4 import QtGui, QtCore
# from PyQt4.QtCore import Qt, QString, QVariant, QRect, QRectF, QSize, QPoint
from PyQt4.QtCore import Qt, QRect, QRectF, QSize, QPoint
from PyQt4.QtCore import QAbstractTableModel, QModelIndex
from PyQt4.QtGui import QApplication, QMainWindow, QTextEdit, QLabel, QStyle
from PyQt4.QtGui import QStyledItemDelegate, QStyleOptionViewItemV4
from PyQt4.QtGui import QTextDocument, QPushButton, QStyleOptionButton
from PyQt4.QtGui import QStyleFactory
# from PyQt4.QtGui import QCommonStyle, QCleanlooksStyle, QPlastiqueStyle
# from PyQt4.QtGui import QMacStyle, QWindowsStyle, QWindowsXPStyle
from datetime import datetime, date
from numpy import nan, float32, sqrt, array, isnan
import pandas as pd
import itertools as it
import sys
from als.milo.qimage import Diffractometer402, CcdImageFromFITS, Polarization
try:
# _fromUtf8 = QString.fromUtf8
pass
except AttributeError:
def _fromUtf8(s):
return s
try:
_encoding = QApplication.UnicodeUTF8
def _translate(context, text, disambig):
return QApplication.translate(context, text, disambig, _encoding)
except AttributeError:
def _translate(context, text, disambig):
return QApplication.translate(context, text, disambig)
# ----------------------------------------------------------------
# ----------------------------------------------------------------
class fitsParamsModel(QAbstractTableModel):
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def __init__(self, parent=None, ccdImage=None):
super(fitsParamsModel, self).__init__(parent)
self.numRows = 0
self.numCols = 3
labels = []
values = []
units = []
(
self.labels_col,
self.values_col,
self.units_col,
) = range(self.numCols)
self.df = pd.DataFrame(
{
"label": labels,
"value": values,
"unit": units,
},
)
self.df = self.df.reindex_axis([
"label",
"value",
"unit",
], axis=1)
self.loadParams(ccdImage)
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def loadParams(self, ccdImage=None):
self.ccdImage = ccdImage
if ccdImage is None:
self.df.loc[:, "value"] = nan
self.dataChanged.emit(
self.index(0, self.values_col),
self.index(len(self.df), self.values_col) )
return()
self.loadParamsFromCcdFits(ccdImage)
self.dataChanged.emit(
self.index(0, self.values_col),
self.index(len(self.df), self.values_col) )
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def clearParams(self):
self.loadParams(ccdImage=None)
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def loadParamsFromCcdFits(self, ccdImage):
return()
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def rowCount(self, parent=None):
if (parent is None) or (parent == QModelIndex()):
return(self.numRows)
return(0)
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def columnCount(self, parent=None):
if (parent is None) or (parent == QModelIndex()):
return(self.numCols)
return(0)
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def data(self, index, role = Qt.DisplayRole):
if (role == Qt.DisplayRole):
return self.df.iloc[index.row(), index.column()]
# if (role == Qt.TextAlignmentRole):
# if (index.column() == self.units_col):
# return Qt.AlignLeft
# else:
# return Qt.AlignRight
return None
# ----------------------------------------------------------------
# ----------------------------------------------------------------
class fitsParamsLeftColModel(fitsParamsModel):
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def __init__(self, parent=None, ccdImage=None):
super(fitsParamsLeftColModel, self).__init__(parent, ccdImage)
table_contents = array([
["Exposure time", nan, "sec."],
[" ", nan, " "],
[" ", nan, " "],
["Energy", nan, "eV"],
[" ", nan, " "],
["Detector, <i>2θ</i>", nan, "deg."],
["(2D) Incidence, <i>α</i>", nan, "deg."],
["(2D) Exit, <i>β</i>", nan, "deg."],
["Transverse, <i>χ</i>", nan, "deg."],
[" ", nan, " "],
["X", nan, "mm"],
["Y", nan, "mm"],
["Z", nan, "mm"],
[" ", nan, " "],
# ["q<span class='sub'>X</span>", nan, "nm<span class='sup'>-1</span>"],
["q<sub>X</sub>", nan, "nm<sup>-1</sup>"],
["q<sub>Y</sub>", nan, "nm<sup>-1</sup>"],
["q<sub>Z</sub>", nan, "nm<sup>-1</sup>"],
])
(self.numRows, self.numCols) = table_contents.shape
# (
# labels_col,
# values_col,
# units_col,
# ) = range(self.numCols)
labels = table_contents[:, self.labels_col]
values = table_contents[:, self.values_col]
units = table_contents[:, self.units_col]
blankline = it.count(0)
indices = [
"expTime",
"blank_{0:d}".format(blankline.next()),
"blank_{0:d}".format(blankline.next()),
"energy",
"blank_{0:d}".format(blankline.next()),
"twotheta",
"incidence2D",
"exit2D",
"transverse",
"blank_{0:d}".format(blankline.next()),
"x",
"y",
"z",
"blank_{0:d}".format(blankline.next()),
"qx",
"qy",
"qz",
]
self.df = pd.DataFrame(
{
"label": labels,
"value": values,
"unit": units,
},
index = pd.Index(indices),
)
self.df = self.df.reindex_axis([
"label",
"value",
"unit",
], axis=1)
# print self.df["values"]
# self.df["values"] = self.df["values"].astype(float32)
# print self.df["values"]
# print self.df
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def loadParamsFromCcdFits(self, ccdImage):
diffractometer = ccdImage._diffractometer
primary_hdu = ccdImage.hdulist[0].header
qimage_df = ccdImage.qvalues_df()
(num_rows, num_cols) = ccdImage.data.shape
center_pixel = (
# (qimage_df["row"] == int(num_rows*(460./512.))) &
(qimage_df["row"] == int(num_rows - num_rows*(460./512.))) &
(qimage_df["col"] == int(num_cols/2.)) )
self.df.loc["expTime", "value"] = ccdImage.exposure_time
self.df.loc["energy", "value"] = diffractometer.energy
self.df.loc["twotheta", "value"] = diffractometer.twotheta
self.df.loc["incidence2D", "value"] = diffractometer.incidence
self.df.loc["exit2D", "value"] = diffractometer.exit_angle
self.df.loc["transverse", "value"] = diffractometer.transverse
self.df.loc["x", "value"] = primary_hdu['X Position']
self.df.loc["y", "value"] = primary_hdu['Y Position']
self.df.loc["z", "value"] = primary_hdu['Z Position']
self.df.loc["qx", "value"] = qimage_df.loc[center_pixel, "Qx"].values
self.df.loc["qy", "value"] = qimage_df.loc[center_pixel, "Qy"].values
self.df.loc["qz", "value"] = qimage_df.loc[center_pixel, "Qz"].values
# print self.df["values"]
# self.df["values"] = self.df["values"].astype(float32)
# print self.df["values"]
# print self.df
# ----------------------------------------------------------------
# ----------------------------------------------------------------
class fitsParamsRightColModel(fitsParamsModel):
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def __init__(self, parent=None, ccdImage=None):
super(fitsParamsRightColModel, self).__init__(parent, ccdImage)
table_contents = array([
["Date", nan, " "],
["Timestamp", nan, " "],
[" ", nan, " "],
["Polarization", nan, " "],
[" ", nan, " "],
["Bottom Seal", nan, "deg."],
["Top Seal", nan, "deg."],
["Top Seal <i>offset</i>", nan, "deg."],
["Flip", nan, "deg."],
["Flip <i>offset</i>", nan, "deg."],
[" ", nan, " "],
["Temperature A", nan, "K"],
["Temperature B", nan, "K"],
[" ", nan, " "],
["|q|", nan, "nm<sup>-1</sup>"],
["q<sub>In-Plane</sub>", nan, "nm<sup>-1</sup>"],
["q<sub>Out-of-Plane</sub>", nan, "nm<sup>-1</sup>"],
])
(self.numRows, self.numCols) = table_contents.shape
# (
# labels_col,
# values_col,
# units_col,
# ) = range(self.numCols)
labels = table_contents[:, self.labels_col]
values = table_contents[:, self.values_col]
units = table_contents[:, self.units_col]
blankline = it.count(0)
indices = [
"dateStamp",
"timeStamp".format(blankline.next()),
"blank_{0:d}".format(blankline.next()),
"polarization".format(blankline.next()),
"blank_{0:d}".format(blankline.next()),
"bottom",
"top",
"topOffset",
"flip",
"flipOffset",
"blank_{0:d}".format(blankline.next()),
"tempA",
"tempB",
"blank_{0:d}".format(blankline.next()),
"qmag",
"qip",
"qop",
]
self.df = pd.DataFrame(
{
"label": labels,
"value": values,
"unit": units,
},
index = pd.Index(indices),
)
self.df = self.df.reindex_axis([
"label",
"value",
"unit",
], axis=1)
# print self.df["values"]
# self.df["values"] = self.df["values"].astype(float32)
# print self.df["values"]
# print self.df
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def loadParamsFromCcdFits(self, ccdImage):
diffractometer = ccdImage._diffractometer
primary_hdu = ccdImage.hdulist[0].header
qimage_df = ccdImage.qvalues_df()
(num_rows, num_cols) = ccdImage.data.shape
center_pixel = (
(qimage_df["row"] == int(num_rows*(460./512.))) &
(qimage_df["col"] == int(num_cols/2.)) )
value = primary_hdu.get('DATE', None)
if value is not None:
if 'T' in value:
image_date = datetime.strptime(value,
"%Y-%m-%dT%H:%M:%S").date()
image_time = datetime.strptime(value,
"%Y-%m-%dT%H:%M:%S").time()
else:
image_date = datetime.strptime(value, "%Y-%m-%d").date()
image_time = datetime.strptime(value, "%H:%M:%S").time()
else:
value = primary_hdu.get('DATETIME', None)
if value is not None:
image_date = datetime.strptime(value,
"%Y/%m/%d %H:%M:%S").date()
image_time = datetime.strptime(value,
"%Y/%m/%d %H:%M:%S").time()
else:
image_date = datetime.strptime("1970-01-01", "%Y-%m-%d").date()
image_time = datetime.strptime("1970-01-01", "%H:%M:%S").time()
pol = diffractometer.polarization
pol_state_text = dict({
Polarization.UNDEFINED: "N/A",
Polarization.CIRCULAR: "Circ.",
Polarization.LINEAR: "Lin.",
})
pol_value_text = dict({
Polarization.UNDEFINED: nan,
Polarization.CIRCULAR: pol.circular_degree,
Polarization.LINEAR: pol.linear_angle,
})
for key_entry in ('TempCtrlrA', 'Temperature A',
'Lakeshore Temp Controller A', ):
value = primary_hdu.get(key_entry, None)
if value is not None:
temperatureA = value
break
else:
temperatureA = 298 # Kelvin
for key_entry in ('TempCtrlrB', 'Temperature B',
'Lakeshore Temp Controller B', ):
value = primary_hdu.get(key_entry, None)
if value is not None:
temperatureB = value
break
else:
temperatureB = 298 # Kelvin
qxy = sqrt(qimage_df.loc[center_pixel, "Qx"].values**2 +
qimage_df.loc[center_pixel, "Qy"].values**2)
qmag = sqrt(qimage_df.loc[center_pixel, "Qz"].values**2 + qxy**2)
self.df.loc["polarization", "unit"] = pol_state_text[pol.state]
self.df.loc["dateStamp", "value"] = image_date
self.df.loc["timeStamp", "value"] = image_time
self.df.loc["polarization", "value"] = pol_value_text[pol.state]
self.df.loc["bottom", "value"] = diffractometer.bottom_angle
self.df.loc["top", "value"] = diffractometer.top_angle
self.df.loc["topOffset", "value"] = diffractometer.offset_top
self.df.loc["flip", "value"] = diffractometer.flip_angle
self.df.loc["flipOffset", "value"] = diffractometer.offset_flip
self.df.loc["tempA", "value"] = temperatureA
self.df.loc["tempB", "value"] = temperatureB
self.df.loc["qmag", "value"] = qmag
self.df.loc["qip", "value"] = qxy
self.df.loc["qop", "value"] = qimage_df.loc[center_pixel, "Qz"].values
# print self.df["values"]
# self.df["values"] = self.df["values"].astype(float32)
# print self.df["values"]
# print self.df
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# def data(self, index, role = Qt.DisplayRole):
# if (role == Qt.DisplayRole):
# value = super(fitsParamsRightColModel, self).data(index, role)
# if ( (self.df.index[index.row()] == "dateStamp")
# and (index.column() == self.values_col)
# ):
# print "dateStamp"
# # print value.strftime("%A, %B %d, %Y")
# print value.strftime("%c")
# return value.strftime("%c")
# return value.strftime("%A, %B %d, %Y")
# if ( (self.df.index[index.row()] == "timeStamp")
# and (index.column() == self.values_col)
# ):
# print "timeStamp"
# print value.strftime("%H : %M : %S . %f")
# return value.strftime("%H : %M : %S . %f")
# return value
# return None
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def data(self, index, role = Qt.DisplayRole):
if (role == Qt.DisplayRole):
value = super(fitsParamsRightColModel, self).data(index, role)
if (self.df.index[index.row()] == "dateStamp"):
return self.df.iloc[index.row(), self.values_col]
if (self.df.index[index.row()] == "timeStamp"):
return self.df.iloc[index.row(), self.values_col]
return value
return None
# ----------------------------------------------------------------
# ----------------------------------------------------------------
class qtLabelDelegate(QStyledItemDelegate):
def __init__(self, parent=None):
super(QStyledItemDelegate, self).__init__(parent)
def paint (self, painter, option, index):
return
# # item = QLabel(QString(index.data()), self)
# # item = QLabel(str(index.data()), self)
# item = QLabel(index.data().toString(), self.parent())
# item.setDisabled(True)
# item.setAutoFillBackground(True)
# # item.render(painter)
# item.drawContents(painter)
# update sizeHint() ?
# ----------------------------------------------------------------
# ----------------------------------------------------------------
class qtHtmlDelegate(QStyledItemDelegate):
def __init__(self, parent=None):
super(QStyledItemDelegate, self).__init__(parent)
def paint(self, painter, option, index):
options = QStyleOptionViewItemV4(option)
self.initStyleOption(options, index)
# print "option.displayAlignment:"
# print "\tLeft :", (option.displayAlignment & Qt.AlignLeft)
# print "\tRight:", (option.displayAlignment & Qt.AlignRight)
# if (option.displayAlignment & Qt.AlignLeft): print "Left"
# if (option.displayAlignment & Qt.AlignRight): print "Right"
painter.save()
doc = QTextDocument()
doc.setHtml(options.text)
options.text = ""
options.widget.style().drawControl(
QStyle.CE_ItemViewItem, options, painter)
# Assuming .alignment() & Qt.AlignVCenter
doc_size = self.sizeHint(option, index)
height_offset = (options.rect.height() - doc_size.height()) / 2.
# print "\n", index.row(), ",", index.column(), ":"
# print "options.rect.height():", options.rect.height()
# print "doc_size.height():", doc_size.height()
# print "height_offset:", height_offset
# print "doc_size.width():", doc_size.width()
height_offset = max(0, height_offset)
painter.translate(
options.rect.left(),
options.rect.top() + height_offset)
clip = QRectF(0, 0, options.rect.width(), options.rect.height())
doc.drawContents(painter, clip)
painter.restore()
def sizeHint(self, option, index):
options = QStyleOptionViewItemV4(option)
self.initStyleOption(options, index)
doc = QTextDocument()
doc.setHtml(options.text)
# doc.setTextWidth(options.rect.width()) # For multiline text
return QSize(doc.idealWidth(), doc.size().height())
# ----------------------------------------------------------------
# ----------------------------------------------------------------
class qtFloatDelegate(QStyledItemDelegate):
def __init__(self, parent=None, decimals=6):
super(QStyledItemDelegate, self).__init__(parent)
self.decimals = decimals
# def paint(self, painter, option, index):
# painter.save()
#
# # value = float(index.data().toPyObject())
# value = float(index.data())
# # painter.drawText(
# # option.rect,
# # Qt.AlignRight & Qt.AlignVCenter,
# # "{0:0.{1:d}f}".format(value, self.decimals))
#
# # Assuming .alignment() & Qt.AlignRight & Qt.AlignVCenter
# doc_size = self.sizeHint(option, index)
# height_offset = (option.rect.height() - doc_size.height()) / 2.
# print "\n", index.row(), ",", index.column(), ":"
# print "option.rect.height():", option.rect.height()
# print "doc_size.height():", doc_size.height()
# print "height_offset:", height_offset
# print "doc_size.width():", doc_size.width()
# height_offset = max(0, height_offset)
# height_offset = 0
# painter.translate(
# # option.rect.right() - doc_size.width(),
# option.rect.left(),
# # option.rect.left() + doc_size.width()/2.,
# option.rect.top() + height_offset)
# painter.drawText(
# option.rect,
# # Qt.AlignRight & Qt.AlignVCenter,
# Qt.AlignLeft & Qt.AlignTop,
# "{0:0.{1:d}f}".format(value, self.decimals))
#
# painter.restore()
def paint(self, painter, option, index):
options = QStyleOptionViewItemV4(option)
self.initStyleOption(options, index)
painter.save()
doc = QTextDocument()
# value = float(index.data().toPyObject())
value = float(index.data())
doc.setPlainText("{0:0.{1:d}f}".format(value, self.decimals))
if isnan(value):
doc.setPlainText("")
options.text = ""
options.widget.style().drawControl(
QStyle.CE_ItemViewItem, options, painter)
# Assuming .alignment() & Qt.AlignRight & Qt.AlignVCenter
doc_size = self.sizeHint(option, index)
height_offset = (options.rect.height() - doc_size.height()) / 2.
# print "\n", index.row(), ",", index.column(), ":"
# print "options.rect.height():", options.rect.height()
# print "doc_size.height():", doc_size.height()
# print "height_offset:", height_offset
# print "doc_size.width():", doc_size.width()
height_offset = max(0, height_offset)
painter.translate(
options.rect.right() - doc_size.width(),
options.rect.top() + height_offset)
clip = QRectF(0, 0, options.rect.width(), options.rect.height())
doc.drawContents(painter, clip)
painter.restore()
def sizeHint(self, option, index):
options = QStyleOptionViewItemV4(option)
self.initStyleOption(options, index)
# print "index ({0:d}, {1:d}): data =".format(index.row(), index.column()), index.data().toPyObject()
# print "index ({0:d}, {1:d}): data =".format(index.row(), index.column()), index.data()
doc = QTextDocument()
# value = float(index.data().toPyObject())
value = float(index.data())
doc.setPlainText("{0:0.{1:d}f}".format(value, self.decimals))
# doc.setTextWidth(options.rect.width()) # For multiline text
return QSize(doc.idealWidth(), doc.size().height())
# ----------------------------------------------------------------
# ----------------------------------------------------------------
class qtButtonDelegate(QStyledItemDelegate):
def __init__(self, parent=None, decimals=6):
super(QStyledItemDelegate, self).__init__(parent)
def paint(self, painter, option, index):
# options = QStyleOptionViewItemV4(option)
# self.initStyleOption(options, index)
painter.save()
doc = QPushButton(index.data(), self.parent() )
# options.text = ""
# options.widget.style().drawControl(
# QStyle.CE_PushButton, options, painter)
btn_option = QStyleOptionButton()
# btn_option.initFrom(option)
# print "btn_option.state:", type(btn_option.state), btn_option.state
# print "QStyle.State_Enabled:", type(QStyle.State_Enabled), QStyle.State_Enabled
# print "QStyle.State_Raised:", type(QStyle.State_Raised), QStyle.State_Raised
print "btn_option.features:", btn_option.features
# btn_option.features = QStyleOptionButton.None
# btn_option.features = QStyleOptionButton.Flat
btn_option.state = QStyle.State_Enabled or QStyle.State_Raised
btn_option.state = QStyle.State_Raised
btn_option.rect = option.rect
btn_option.text = index.data()
doc.resize(doc.minimumSizeHint())
# Assuming .alignment() & Qt.AlignCenter
doc_size = self.sizeHint(option, index)
btn_option.rect = QRect(
QPoint(option.rect.left(), option.rect.top()), doc_size)
btn_option.rect = QRect(
QPoint(option.rect.left()+5, option.rect.top()+5),
QSize(doc_size.width()-10, doc_size.height()-10) )
btn_option.rect = option.rect.adjusted(1, 1, -1, -1)
btn_option.rect = option.rect.adjusted(2, 2, -2, -2)
btn_option.rect = option.rect.adjusted(15, 5, -15, -5)
height_offset = (option.rect.height() - doc_size.height()) / 2.
width_offset = (option.rect.width() - doc_size.width()) / 2.
print "\n", index.row(), ",", index.column(), ":"
print "option.rect.height():", option.rect.height()
print "doc_size.height():", doc_size.height()
print "height_offset:", height_offset
print "doc_size.width():", doc_size.width()
height_offset = max(0, height_offset)
width_offset = max(0, width_offset)
# painter.translate(
# option.rect.left() + width_offset,
# option.rect.top() + height_offset)
# clip = QRectF(0, 0, option.rect.width(), option.rect.height())
# doc.drawContents(painter, clip)
# doc.style().drawControl(
QApplication.style().drawControl(
QStyle.CE_PushButton, btn_option, painter)
painter.restore()
def sizeHint(self, option, index):
# options = QStyleOptionViewItemV4(option)
# self.initStyleOption(options, index)
# print "index ({0:d}, {1:d}): data =".format(index.row(), index.column()), index.data().toPyObject()
print "index ({0:d}, {1:d}): data =".format(index.row(), index.column()), index.data()
doc = QPushButton(index.data())
print "doc.sizeHint():", doc.sizeHint()
print "doc.minimumSizeHint():", doc.minimumSizeHint()
doc.resize(doc.minimumSizeHint())
print "doc.size():", doc.size()
print "doc.rect():", doc.rect().left(), doc.rect().top(), doc.rect().right(), doc.rect().bottom()
print "doc.contentsRect():", doc.contentsRect().left(), doc.contentsRect().top(), doc.contentsRect().right(), doc.contentsRect().bottom()
print "doc.contentsMargins():", doc.contentsMargins().left(), doc.contentsMargins().top(), doc.contentsMargins().right(), doc.contentsMargins().bottom()
# doc.setTextWidth(options.rect.width()) # For multiline text
# return QSize(doc.idealWidth(), doc.size().height())
return doc.size()
# ----------------------------------------------------------------
# ----------------------------------------------------------------
class qtDateDelegate(QStyledItemDelegate):
def __init__(self, parent=None):
super(QStyledItemDelegate, self).__init__(parent)
def paint(self, painter, option, index):
options = QStyleOptionViewItemV4(option)
self.initStyleOption(options, index)
painter.save()
doc = QTextDocument()
value = index.data()
try:
doc.setPlainText(value.strftime("%A, %B %d, %Y"))
except:
doc.setPlainText("")
options.text = ""
options.widget.style().drawControl(
QStyle.CE_ItemViewItem, options, painter)
# Assuming .alignment() & Qt.AlignLeft & Qt.AlignVCenter
doc_size = self.sizeHint(option, index)
height_offset = (options.rect.height() - doc_size.height()) / 2.
# print "\n", index.row(), ",", index.column(), ":"
# print "options.rect.height():", options.rect.height()
# print "doc_size.height():", doc_size.height()
# print "height_offset:", height_offset
# print "doc_size.width():", doc_size.width()
height_offset = max(0, height_offset)
painter.translate(
# options.rect.right() - doc_size.width(),
options.rect.left(),
options.rect.top() + height_offset)
clip = QRectF(0, 0, options.rect.width(), options.rect.height())
doc.drawContents(painter, clip)
painter.restore()
def sizeHint(self, option, index):
options = QStyleOptionViewItemV4(option)
self.initStyleOption(options, index)
# print "index ({0:d}, {1:d}): data =".format(index.row(), index.column()), index.data().toPyObject()
# print "index ({0:d}, {1:d}): data =".format(index.row(), index.column()), index.data()
doc = QTextDocument()
value = index.data()
try:
doc.setPlainText(value.strftime("%A, %B %d, %Y"))
except:
doc.setPlainText("")
# doc.setTextWidth(options.rect.width()) # For multiline text
return QSize(doc.idealWidth(), doc.size().height())
# ----------------------------------------------------------------
# ----------------------------------------------------------------
class qtTimeDelegate(QStyledItemDelegate):
def __init__(self, parent=None):
super(QStyledItemDelegate, self).__init__(parent)
def paint(self, painter, option, index):
options = QStyleOptionViewItemV4(option)
self.initStyleOption(options, index)
painter.save()
doc = QTextDocument()
value = index.data()
try:
doc.setPlainText("Time stamp: " + value.strftime("%H:%M:%S"))
except:
doc.setPlainText("")
options.text = ""
options.widget.style().drawControl(
QStyle.CE_ItemViewItem, options, painter)
# Assuming .alignment() & Qt.AlignLeft & Qt.AlignVCenter
doc_size = self.sizeHint(option, index)
height_offset = (options.rect.height() - doc_size.height()) / 2.
# print "\n", index.row(), ",", index.column(), ":"
# print "options.rect.height():", options.rect.height()
# print "doc_size.height():", doc_size.height()
# print "height_offset:", height_offset
# print "doc_size.width():", doc_size.width()
height_offset = max(0, height_offset)
painter.translate(
# options.rect.right() - doc_size.width(),
options.rect.left(),
options.rect.top() + height_offset)
clip = QRectF(0, 0, options.rect.width(), options.rect.height())
doc.drawContents(painter, clip)
painter.restore()
def sizeHint(self, option, index):
options = QStyleOptionViewItemV4(option)
self.initStyleOption(options, index)
# print "index ({0:d}, {1:d}): data =".format(index.row(), index.column()), index.data().toPyObject()
# print "index ({0:d}, {1:d}): data =".format(index.row(), index.column()), index.data()
doc = QTextDocument()
value = index.data()
try:
doc.setPlainText("Time stamp: " + value.strftime("%H:%M:%S"))
except:
doc.setPlainText("")
# doc.setTextWidth(options.rect.width()) # For multiline text
return QSize(doc.idealWidth(), doc.size().height())
# ----------------------------------------------------------------
# ---------------------------------------------------------------- | ALS.Liam | /ALS.Liam-0.10.0.tar.gz/ALS.Liam-0.10.0/als/liam/fitsParams.py | fitsParams.py |
# ALS.Milo (version 0.18)
Overview
---
_**Milo**_ is a suite of Python modules used to read, process, and analyze
data that was collected on the **Scattering Chamber** located at
**Beamline 4.0.2** (_a.k.a. **BL402**_) of the **Advanced Light Source**
(Berkeley, CA USA).
It is distributed under the _namespace package_, _**ALS**_.
How to cite this software
---
If you use this software in your research, please include the following
citation in your publication:
* Shafer, Padraic. (2014, August 3). ALS.Milo (Version unspecified).
http://doi.org/10.5281/zenodo.3779551
Installation
---
### Install from PyPI
**_ALS.Milo_** can be installed from PyPI using `pip`.
The following example shows how.
```bash
>> sudo python -m pip install ALS.Milo -vv
```
### Install from Anaconda
**_ALS.Milo_** can be installed from Anaconda Cloud.
The following example shows how.
```bash
>> conda config --add channels padraicshafer # Required for ALS.Milo
>> conda config --add channels pshafer # Required for dependency
>> conda install ALS.Milo
```
### Install from local repository (download)
**_ALS.Milo_** can be installed from a local copy of the project repository
using `pip`. The following example shows how.
```bash
>> cd ALS.Milo-0.18.1/ # Local directory of project repository
>> sudo pip install .
```
Background information
---
Data from the BL402 Scattering Chamber is stored in two types of files:
* **FITS files**: Each image captured by the CCD (_a.k.a._ the camera) is
stored in a separate file using the _FITS_ format. _More details below._
https://fits.gsfc.nasa.gov/fits_documentation.html
* **Scan summary files**: When a scan sequence is run to collect data, a text
file is created to summarize the parameters of the scan and the data collected.
Every _scan summary file_ contains a header that describes the scan and the
types of data recorded, followed by data rows -- one row per data point. An
_Instrument Scan_ provides an _image filename_ in each data row that can be
used to access the CCD images recorded during the scan.
Usage
---
`class CcdImageFromFITS` encapsulates the operations of loading CCD data
from a FITS file (including header information), accessing that data, and
converting CCD screen coordinates (pixels) into reciprocal space coordinates.
The following command can be used
to access the `CcdImageFromFITS` class in the `qimage` module.
```python
from als.milo.qimage import CcdImageFromFITS
```
To read a FITS file use the constructor:
`CcdImageFromFITS(`*`filename`*`)` ,
where _filename_ is a reachable (relative or absolute) file path to the FITS
file.
The two-dimensional array of data can be accessed through the instance member,
`data`.
```python
ccd_image = CcdImageFromFITS("NiFe_8044-00024.fits")
ccd_image.data # 2D array with shape (num_rows, num_columns)
```
Reciprocal space coordinates are calculated for each pixel using the member
function, `qvalues_df()`. The return value is a PANDAS dataframe (_df_) where
each row corresponds to a single pixel. The columns `["Qx", "Qy", "Qz"]` are
the diffractometer coordinates of the reciprocal space vector in units of
nm<sup>-1</sup> (_Q_ = 2π / _d_). Intensity of each pixel is in column,
`"Counts"`.
```python
ccd_image = CcdImageFromFITS("NiFe_8044-00024.fits")
q_ccd_df = ccd_image.qvalues_df()
q_ccd_df["Qx", "Qy", "Qz", "Counts"] # CCD data in reciprocal space
```
Copyright Notice
---
ALS.Milo: BL402 RSXD Data Analysis, Copyright (c) 2014-2021, The Regents of
the University of California, through Lawrence Berkeley National Laboratory
(subject to receipt of any required approvals from the U.S. Dept. of Energy).
All rights reserved.
If you have questions about your rights to use or distribute this software,
please contact Berkeley Lab's Intellectual Property Office at [email protected].
NOTICE. This Software was developed under funding from the U.S. Department of
Energy and the U.S. Government consequently retains certain rights. As such,
the U.S. Government has been granted for itself and others acting on its
behalf a paid-up, nonexclusive, irrevocable, worldwide license in the
Software to reproduce, distribute copies to the public, prepare derivative
works, and perform publicly and display publicly, and to permit other to do
so. | ALS.Milo | /ALS.Milo-0.18.1.tar.gz/ALS.Milo-0.18.1/README.md | README.md |
from __future__ import print_function
from future import standard_library
standard_library.install_aliases()
from builtins import str
from builtins import range
from builtins import object
try:
import configparser
except ImportError:
import ConfigParser as configparser
import errno
import json
import os
import re
import subprocess
import sys
class VersioneerConfig(object):
"""Container for Versioneer configuration parameters."""
def get_root():
"""Get the project root directory.
We require that all commands are run from the project root, i.e. the
directory that contains setup.py, setup.cfg, and versioneer.py .
"""
root = os.path.realpath(os.path.abspath(os.getcwd()))
setup_py = os.path.join(root, "setup.py")
versioneer_py = os.path.join(root, "versioneer.py")
if not (os.path.exists(setup_py) or os.path.exists(versioneer_py)):
# allow 'python path/to/setup.py COMMAND'
root = os.path.dirname(os.path.realpath(os.path.abspath(sys.argv[0])))
setup_py = os.path.join(root, "setup.py")
versioneer_py = os.path.join(root, "versioneer.py")
if not (os.path.exists(setup_py) or os.path.exists(versioneer_py)):
err = ("Versioneer was unable to run the project root directory. "
"Versioneer requires setup.py to be executed from "
"its immediate directory (like 'python setup.py COMMAND'), "
"or in a way that lets it use sys.argv[0] to find the root "
"(like 'python path/to/setup.py COMMAND').")
raise VersioneerBadRootError(err)
try:
# Certain runtime workflows (setup.py install/develop in a setuptools
# tree) execute all dependencies in a single python process, so
# "versioneer" may be imported multiple times, and python's shared
# module-import table will cache the first one. So we can't use
# os.path.dirname(__file__), as that will find whichever
# versioneer.py was first imported, even in later projects.
me = os.path.realpath(os.path.abspath(__file__))
me_dir = os.path.normcase(os.path.splitext(me)[0])
vsr_dir = os.path.normcase(os.path.splitext(versioneer_py)[0])
if me_dir != vsr_dir:
print("Warning: build in %s is using versioneer.py from %s"
% (os.path.dirname(me), versioneer_py))
except NameError:
pass
return root
def get_config_from_root(root):
"""Read the project setup.cfg file to determine Versioneer config."""
# This might raise EnvironmentError (if setup.cfg is missing), or
# configparser.NoSectionError (if it lacks a [versioneer] section), or
# configparser.NoOptionError (if it lacks "VCS="). See the docstring at
# the top of versioneer.py for instructions on writing your setup.cfg .
setup_cfg = os.path.join(root, "setup.cfg")
parser = configparser.SafeConfigParser()
with open(setup_cfg, "r") as f:
parser.readfp(f)
VCS = parser.get("versioneer", "VCS") # mandatory
def get(parser, name):
if parser.has_option("versioneer", name):
return parser.get("versioneer", name)
return None
cfg = VersioneerConfig()
cfg.VCS = VCS
cfg.style = get(parser, "style") or "pep440-auto"
cfg.versionfile_source = get(parser, "versionfile_source")
cfg.versionfile_build = get(parser, "versionfile_build")
cfg.tag_prefix = get(parser, "tag_prefix")
if cfg.tag_prefix in ("''", '""'):
cfg.tag_prefix = ""
cfg.parentdir_prefix = get(parser, "parentdir_prefix")
cfg.verbose = get(parser, "verbose")
return cfg
class NotThisMethod(Exception):
"""Exception raised if a method is not valid for the current scenario."""
# these dictionaries contain VCS-specific tools
LONG_VERSION_PY = {}
HANDLERS = {}
def register_vcs_handler(vcs, method): # decorator
"""Decorator to mark a method as the handler for a particular VCS."""
def decorate(f):
"""Store f in HANDLERS[vcs][method]."""
if vcs not in HANDLERS:
HANDLERS[vcs] = {}
HANDLERS[vcs][method] = f
return f
return decorate
def run_command(commands, args, cwd=None, verbose=False, hide_stderr=False,
env=None):
"""Call the given command(s)."""
assert isinstance(commands, list)
p = None
for c in commands:
try:
dispcmd = str([c] + args)
# remember shell=False, so use git.cmd on windows, not just git
p = subprocess.Popen([c] + args, cwd=cwd, env=env,
stdout=subprocess.PIPE,
stderr=(subprocess.PIPE if hide_stderr
else None))
break
except EnvironmentError:
e = sys.exc_info()[1]
if e.errno == errno.ENOENT:
continue
if verbose:
print("unable to run %s" % dispcmd)
print(e)
return None, None
else:
if verbose:
print("unable to find command, tried %s" % (commands,))
return None, None
stdout = p.communicate()[0].strip()
if sys.version_info[0] >= 3:
stdout = stdout.decode()
if p.returncode != 0:
if verbose:
print("unable to run %s (error)" % dispcmd)
print("stdout was %s" % stdout)
return None, p.returncode
return stdout, p.returncode
def unique(seq):
"""Find unique elements in a list, without sorting
Implementation does not depend on `numpy`
* borrowed from https://stackoverflow.com/a/480227
* credit to Markus Jarderot
+ https://www.peterbe.com/plog/uniqifiers-benchmark
"""
seen = set()
seen_add = seen.add
return [x for x in seq if not (x in seen or seen_add(x))]
LONG_VERSION_PY['git'] = '''
# This file helps to compute a version number in source trees obtained from
# git-archive tarball (such as those provided by githubs download-from-tag
# feature). Distribution tarballs (built by setup.py sdist) and build
# directories (produced by setup.py build) will contain a much shorter file
# that just contains the computed version number.
# This file is released into the public domain. Generated by
# versioneer-0.18 (https://github.com/warner/python-versioneer)
"""Git implementation of _version.py."""
from __future__ import print_function
from builtins import str
from builtins import range
from builtins import object
import errno
import os
import re
import subprocess
import sys
def get_keywords():
"""Get the keywords needed to look up the version information."""
# these strings will be replaced by git during git-archive.
# setup.py/versioneer.py will grep for the variable names, so they must
# each be defined on a line of their own. _version.py will just call
# get_keywords().
git_refnames = "%(DOLLAR)sFormat:%%d%(DOLLAR)s"
git_full = "%(DOLLAR)sFormat:%%H%(DOLLAR)s"
git_date = "%(DOLLAR)sFormat:%%ci%(DOLLAR)s"
keywords = {"refnames": git_refnames, "full": git_full, "date": git_date}
return keywords
class VersioneerConfig(object):
"""Container for Versioneer configuration parameters."""
def get_config():
"""Create, populate and return the VersioneerConfig() object."""
# these strings are filled in when 'setup.py versioneer' creates
# _version.py
cfg = VersioneerConfig()
cfg.VCS = "git"
cfg.style = "%(STYLE)s"
cfg.tag_prefix = "%(TAG_PREFIX)s"
cfg.parentdir_prefix = "%(PARENTDIR_PREFIX)s"
cfg.versionfile_source = "%(VERSIONFILE_SOURCE)s"
cfg.verbose = False
return cfg
class NotThisMethod(Exception):
"""Exception raised if a method is not valid for the current scenario."""
LONG_VERSION_PY = {}
HANDLERS = {}
def register_vcs_handler(vcs, method): # decorator
"""Decorator to mark a method as the handler for a particular VCS."""
def decorate(f):
"""Store f in HANDLERS[vcs][method]."""
if vcs not in HANDLERS:
HANDLERS[vcs] = {}
HANDLERS[vcs][method] = f
return f
return decorate
def run_command(commands, args, cwd=None, verbose=False, hide_stderr=False,
env=None):
"""Call the given command(s)."""
assert isinstance(commands, list)
p = None
for c in commands:
try:
dispcmd = str([c] + args)
# remember shell=False, so use git.cmd on windows, not just git
p = subprocess.Popen([c] + args, cwd=cwd, env=env,
stdout=subprocess.PIPE,
stderr=(subprocess.PIPE if hide_stderr
else None))
break
except EnvironmentError:
e = sys.exc_info()[1]
if e.errno == errno.ENOENT:
continue
if verbose:
print("unable to run %%s" %% dispcmd)
print(e)
return None, None
else:
if verbose:
print("unable to find command, tried %%s" %% (commands,))
return None, None
stdout = p.communicate()[0].strip()
if sys.version_info[0] >= 3:
stdout = stdout.decode()
if p.returncode != 0:
if verbose:
print("unable to run %%s (error)" %% dispcmd)
print("stdout was %%s" %% stdout)
return None, p.returncode
return stdout, p.returncode
def versions_from_parentdir(parentdir_prefix, root, verbose):
"""Try to determine the version from the parent directory name.
Source tarballs conventionally unpack into a directory that includes both
the project name and a version string. We will also support searching up
two directory levels for an appropriately named parent directory
"""
rootdirs = []
for i in range(3):
dirname = os.path.basename(root)
if dirname.startswith(parentdir_prefix):
return {"version": dirname[len(parentdir_prefix):],
"full-revisionid": None,
"dirty": False, "error": None, "date": None}
else:
rootdirs.append(root)
root = os.path.dirname(root) # up a level
if verbose:
print("Tried directories %%s but none started with prefix %%s" %%
(str(rootdirs), parentdir_prefix))
raise NotThisMethod("rootdir doesn't start with parentdir_prefix")
@register_vcs_handler("git", "get_keywords")
def git_get_keywords(versionfile_abs):
"""Extract version information from the given file."""
# the code embedded in _version.py can just fetch the value of these
# keywords. When used from setup.py, we don't want to import _version.py,
# so we do it with a regexp instead. This function is not used from
# _version.py.
keywords = {}
try:
f = open(versionfile_abs, "r")
for line in f.readlines():
if line.strip().startswith("git_refnames ="):
mo = re.search(r'=\s*"(.*)"', line)
if mo:
keywords["refnames"] = mo.group(1)
if line.strip().startswith("git_full ="):
mo = re.search(r'=\s*"(.*)"', line)
if mo:
keywords["full"] = mo.group(1)
if line.strip().startswith("git_date ="):
mo = re.search(r'=\s*"(.*)"', line)
if mo:
keywords["date"] = mo.group(1)
f.close()
except EnvironmentError:
pass
return keywords
@register_vcs_handler("git", "keywords")
def git_versions_from_keywords(keywords, tag_prefix, verbose):
"""Get version information from git keywords."""
if not keywords:
raise NotThisMethod("no keywords at all, weird")
date = keywords.get("date")
if date is not None:
# git-2.2.0 added "%%cI", which expands to an ISO-8601 -compliant
# datestamp. However we prefer "%%ci" (which expands to an "ISO-8601
# -like" string, which we must then edit to make compliant), because
# it's been around since git-1.5.3, and it's too difficult to
# discover which version we're using, or to work around using an
# older one.
date = date.strip().replace(" ", "T", 1).replace(" ", "", 1)
refnames = keywords["refnames"].strip()
if refnames.startswith("$Format"):
if verbose:
print("keywords are unexpanded, not using")
raise NotThisMethod("unexpanded keywords, not a git-archive tarball")
refs = set([r.strip() for r in refnames.strip("()").split(",")])
# starting in git-1.8.3, tags are listed as "tag: foo-1.0" instead of
# just "foo-1.0". If we see a "tag: " prefix, prefer those.
TAG = "tag: "
tags = set([r[len(TAG):] for r in refs if r.startswith(TAG)])
if not tags:
# Either we're using git < 1.8.3, or there really are no tags. We use
# a heuristic: assume all version tags have a digit. The old git %%d
# expansion behaves like git log --decorate=short and strips out the
# refs/heads/ and refs/tags/ prefixes that would let us distinguish
# between branches and tags. By ignoring refnames without digits, we
# filter out many common branch names like "release" and
# "stabilization", as well as "HEAD" and "master".
tags = set([r for r in refs if re.search(r'\d', r)])
if verbose:
print("discarding '%%s', no digits" %% ",".join(refs - tags))
if verbose:
print("likely tags: %%s" %% ",".join(sorted(tags)))
for ref in sorted(tags):
# sorting will prefer e.g. "2.0" over "2.0rc1"
if ref.startswith(tag_prefix):
r = ref[len(tag_prefix):]
if verbose:
print("picking %%s" %% r)
return {"version": r,
"full-revisionid": keywords["full"].strip(),
"dirty": False, "error": None,
"date": date}
# no suitable tags, so version is "0+unknown", but full hex is still there
if verbose:
print("no suitable tags, using unknown + full revision id")
return {"version": "0+unknown",
"full-revisionid": keywords["full"].strip(),
"dirty": False, "error": "no suitable tags", "date": None}
@register_vcs_handler("git", "pieces_from_vcs")
def git_pieces_from_vcs(tag_prefix, root, verbose, run_command=run_command):
"""Get version from 'git describe' in the root of the source tree.
This only gets called if the git-archive 'subst' keywords were *not*
expanded, and _version.py hasn't already been rewritten with a short
version string, meaning we're inside a checked out source tree.
"""
GITS = ["git"]
if sys.platform == "win32":
GITS = ["git.cmd", "git.exe"]
out, rc = run_command(GITS, ["rev-parse", "--git-dir"], cwd=root,
hide_stderr=True)
if rc != 0:
if verbose:
print("Directory %%s not under git control" %% root)
raise NotThisMethod("'git rev-parse --git-dir' returned error")
# if there is a tag matching tag_prefix, this yields TAG-NUM-gHEX[-dirty]
# if there isn't one, this yields HEX[-dirty] (no NUM)
describe_out, rc = run_command(GITS, ["describe", "--tags", "--dirty",
"--always", "--long",
"--match", "%%s*" %% tag_prefix],
cwd=root)
# --long was added in git-1.5.5
if describe_out is None:
raise NotThisMethod("'git describe' failed")
describe_out = describe_out.strip()
full_out, rc = run_command(GITS, ["rev-parse", "HEAD"], cwd=root)
if full_out is None:
raise NotThisMethod("'git rev-parse' failed")
full_out = full_out.strip()
pieces = {}
pieces["long"] = full_out
pieces["short"] = full_out[:7] # maybe improved later
pieces["error"] = None
# parse describe_out. It will be like TAG-NUM-gHEX[-dirty] or HEX[-dirty]
# TAG might have hyphens.
git_describe = describe_out
# look for -dirty suffix
dirty = git_describe.endswith("-dirty")
pieces["dirty"] = dirty
if dirty:
git_describe = git_describe[:git_describe.rindex("-dirty")]
# now we have TAG-NUM-gHEX or HEX
# Determine which branch is active (ie, checked-out)
branch_name, rc = run_command(
GITS, ["rev-parse", "--abbrev-ref", "HEAD"], cwd=root)
pieces["branch"] = branch_name
if "-" in git_describe:
# TAG-NUM-gHEX
mo = re.search(r'^(.+)-(\d+)-g([0-9a-f]+)$', git_describe)
if not mo:
# unparseable. Maybe git-describe is misbehaving?
pieces["error"] = ("unable to parse git-describe output: '%%s'"
%% describe_out)
return pieces
# tag
full_tag = mo.group(1)
if not full_tag.startswith(tag_prefix):
if verbose:
fmt = "tag '%%s' doesn't start with prefix '%%s'"
print(fmt %% (full_tag, tag_prefix))
pieces["error"] = ("tag '%%s' doesn't start with prefix '%%s'"
%% (full_tag, tag_prefix))
return pieces
pieces["closest-tag"] = full_tag[len(tag_prefix):]
# distance: number of commits since tag
pieces["distance"] = int(mo.group(2))
# Added to deal with merged branches
history_search_args = ["--ancestry-path"]
if (branch_name == "master"):
# Count distance only along `master`; ignore merge paths
history_search_args = ["--ancestry-path", "--first-parent"]
distance_to_tag, rc = run_command(
GITS, ["rev-list", "--count",] + history_search_args +
["%%s..HEAD" % pieces["closest-tag"] ], cwd=root)
pieces["distance"] = int(distance_to_tag)
# commit: short hex revision ID
pieces["short"] = mo.group(3)
else:
# HEX: no tags
pieces["closest-tag"] = None
count_out, rc = run_command(GITS, ["rev-list", "HEAD", "--count"],
cwd=root)
pieces["distance"] = int(count_out) # total number of commits
##################################################
# Added for `develop` branch from `master`
# `HEAD` might have branched from untagged commit on `master`
merge_base_master, rc = run_command(
GITS, ["merge-base", "master", "HEAD", "-a"], cwd=root)
distance_to_master, rc = run_command(
GITS, ["rev-list", "--count", "--ancestry-path",
"%%s..HEAD" %% merge_base_master], cwd=root)
pieces["distance-to-master"] = int(distance_to_master)
##################################################
# Added for branches other than `master`
if pieces["closest-tag"]:
# TAG exists
master_to_tag, rc = run_command(
GITS, ["rev-list", "--count", "--ancestry-path", "--first-parent",
"%%s..%%s" % (pieces["closest-tag"], merge_base_master) ],
cwd=root)
pieces["master-to-tag"] = int(master_to_tag)
else:
pieces["master-to-tag"] = pieces["distance"]
##################################################
# Added for "feature" branch from `develop`
pieces["develop"] = None
pieces["distance-to-develop"] = None
branch_develop, rc = run_command(
GITS, ["rev-parse", "--verify", "refs/heads/develop"], cwd=root)
branch_develop_exists = not bool(rc)
if branch_develop_exists:
merge_base_develop, rc = run_command(
GITS, ["merge-base", "develop", "HEAD", "-a"], cwd=root)
distance_to_develop, rc = run_command(
GITS, ["rev-list", "--count", "--ancestry-path",
"%%s..HEAD" %% merge_base_develop], cwd=root)
pieces["develop"] = branch_develop
pieces["distance-to-develop"] = int(distance_to_develop)
# commit date: see ISO-8601 comment in git_versions_from_keywords()
date = run_command(GITS, ["show", "-s", "--format=%%ci", "HEAD"],
cwd=root)[0].strip()
pieces["date"] = date.strip().replace(" ", "T", 1).replace(" ", "", 1)
return pieces
def plus_or_dot(pieces):
"""Return a + if we don't already have one, else return a ."""
if "+" in pieces.get("closest-tag", ""):
return "."
return "+"
def render_pep440(pieces):
"""Build up version string, with post-release "local version identifier".
Our goal: TAG[+DISTANCE.gHEX[.dirty]] . Note that if you
get a tagged build and then dirty it, you'll get TAG+0.gHEX.dirty
Exceptions:
1: no tags. git_describe was just HEX. 0+untagged.DISTANCE.gHEX[.dirty]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"] or pieces["dirty"]:
rendered += plus_or_dot(pieces)
rendered += "%%d.g%%s" %% (pieces["distance"], pieces["short"])
if pieces["dirty"]:
rendered += ".dirty"
else:
# exception #1
rendered = "0+untagged.%%d.g%%s" %% (pieces["distance"],
pieces["short"])
if pieces["dirty"]:
rendered += ".dirty"
return rendered
def render_pep440_pre(pieces):
"""TAG[.post.devDISTANCE] -- No -dirty.
Exceptions:
1: no tags. 0.post.devDISTANCE
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"]:
rendered += ".post.dev%%d" %% pieces["distance"]
else:
# exception #1
rendered = "0.post.dev%%d" %% pieces["distance"]
return rendered
def render_pep440_post(pieces):
"""TAG[.postDISTANCE[.dev0]+gHEX] .
The ".dev0" means dirty. Note that .dev0 sorts backwards
(a dirty tree will appear "older" than the corresponding clean one),
but you shouldn't be releasing software with -dirty anyways.
Exceptions:
1: no tags. 0.postDISTANCE[.dev0]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"] or pieces["dirty"]:
rendered += ".post%%d" %% pieces["distance"]
if pieces["dirty"]:
rendered += ".dev0"
rendered += plus_or_dot(pieces)
rendered += "g%%s" %% pieces["short"]
else:
# exception #1
rendered = "0.post%%d" %% pieces["distance"]
if pieces["dirty"]:
rendered += ".dev0"
rendered += "+g%%s" %% pieces["short"]
return rendered
def render_pep440_old(pieces):
"""TAG[.postDISTANCE[.dev0]] .
The ".dev0" means dirty.
Eexceptions:
1: no tags. 0.postDISTANCE[.dev0]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"] or pieces["dirty"]:
rendered += ".post%%d" %% pieces["distance"]
if pieces["dirty"]:
rendered += ".dev0"
else:
# exception #1
rendered = "0.post%%d" %% pieces["distance"]
if pieces["dirty"]:
rendered += ".dev0"
return rendered
def render_pep440_micro(pieces):
"""Build up version string, with post-release "local version identifier".
Our goal: TAG.DISTANCE[+gHEX.dirty]
Exceptions:
1: no tags. git_describe was just HEX. 0+untagged.DISTANCE.gHEX[.dirty]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
rendered += ".%%d" %% pieces["distance"]
if pieces["dirty"]:
rendered += plus_or_dot(pieces)
rendered += "g%%s" %% pieces["short"]
rendered += ".dirty"
else:
# exception #1
rendered = "0+untagged.%%d.g%%s" %% (pieces["distance"],
pieces["short"])
if pieces["dirty"]:
rendered += ".dirty"
return rendered
def render_pep440_develop(pieces):
"""Build up version string, used within development branch of repository.
Our goal: MERGE-POINT.post.devN[+gHEX.dirty]
+) MERGE-POINT = Most recent common ancestor for `develop` and `master`
*) Does not yet handle branch from `release-*`
+) N = DISTANCE from the MERGE-POINT
Exceptions:
1: no tags. 0.post.devDISTANCE[+gHEX.dirty]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
distance_to_merge = pieces["distance-to-master"]
# distance_merge_to_tag = (pieces["distance"] - distance_to_merge)
distance_merge_to_tag = pieces["master-to-tag"]
if (distance_merge_to_tag > 0):
rendered += ".%%d" %% distance_merge_to_tag
rendered += ".post.dev%%d" %% distance_to_merge
else:
# exception #1
rendered = "0.post.dev%%d" %% (pieces["distance"] - 1)
if pieces["dirty"]:
rendered += plus_or_dot(pieces)
rendered += "g%%s" %% pieces["short"]
rendered += ".dirty"
return rendered
def render_pep440_feature(pieces):
"""Build up version string, used within "feature" branch of repository.
Our goal: MERGE-POINT.post.devN+gHEX.BRANCH-NAME.M[.dirty]
+) MERGE-POINT = Most recent common ancestor for `develop` and `master`
*) Does not yet handle branch from `release-*`
+) N = DISTANCE from the MERGE-POINT of `develop` and `master`
+) M = DISTANCE from the MERGE-POINT of "feature" and `develop`
Exceptions:
1: no tags. 0.post.devDISTANCE+gHEX[.dirty]
"""
if pieces["closest-tag"] and pieces["develop"]:
rendered = pieces["closest-tag"]
distance_to_develop = pieces["distance-to-develop"]
distance_to_merge = pieces["distance-to-master"]
# distance_merge_to_tag = (pieces["distance"] - distance_to_merge)
distance_merge_to_tag = pieces["master-to-tag"]
distance_dev_to_merge = (distance_to_merge - distance_to_develop)
if (distance_merge_to_tag > 0):
rendered += ".%%d" %% distance_merge_to_tag
rendered += ".post.dev%%d" %% distance_dev_to_merge
rendered += plus_or_dot(pieces)
rendered += "g%%s" %% pieces["short"]
rendered += ".%%s" %% pieces["branch"]
rendered += ".%%d" %% distance_to_develop
else:
# exception #1
rendered = "0.post.dev%%d" %% (pieces["distance"] - 1)
rendered += plus_or_dot(pieces)
rendered += "g%%s" %% pieces["short"]
if pieces["dirty"]:
rendered += ".dirty"
return rendered
def render_pep440_auto(pieces):
"""Build up version string, based on branch of repository.
Our goal: If on development branch, use render_pep440_develop()
Otherwise use render_pep440_micro()
Exceptions:
+) NONE
"""
if pieces["branch"] == "master":
# logging.debug("Rendering: render_pep440_micro()")
rendered = render_pep440_micro(pieces)
elif pieces["branch"] == "develop":
# logging.debug("Rendering: render_pep440_develop()")
rendered = render_pep440_develop(pieces)
elif pieces["branch"].startswith("release"):
# render_pep440_release() not yet implemented
rendered = render_git_describe(pieces)
else:
# logging.debug("Rendering: render_pep440_feature()")
rendered = render_pep440_feature(pieces)
return rendered
def render_git_describe(pieces):
"""TAG[-DISTANCE-gHEX][-dirty].
Like 'git describe --tags --dirty --always'.
Exceptions:
1: no tags. HEX[-dirty] (note: no 'g' prefix)
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"]:
rendered += "-%%d-g%%s" %% (pieces["distance"], pieces["short"])
else:
# exception #1
rendered = pieces["short"]
if pieces["dirty"]:
rendered += "-dirty"
return rendered
def render_git_describe_long(pieces):
"""TAG-DISTANCE-gHEX[-dirty].
Like 'git describe --tags --dirty --always -long'.
The distance/hash is unconditional.
Exceptions:
1: no tags. HEX[-dirty] (note: no 'g' prefix)
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
rendered += "-%%d-g%%s" %% (pieces["distance"], pieces["short"])
else:
# exception #1
rendered = pieces["short"]
if pieces["dirty"]:
rendered += "-dirty"
return rendered
def render(pieces, style):
"""Render the given version pieces into the requested style."""
if pieces["error"]:
return {"version": "unknown",
"full-revisionid": pieces.get("long"),
"dirty": None,
"error": pieces["error"],
"date": None}
if not style or style == "default":
style = "pep440" # the default
if style == "pep440":
rendered = render_pep440(pieces)
elif style == "pep440-pre":
rendered = render_pep440_pre(pieces)
elif style == "pep440-post":
rendered = render_pep440_post(pieces)
elif style == "pep440-old":
rendered = render_pep440_old(pieces)
elif style == "pep440-micro":
rendered = render_pep440_micro(pieces)
elif style == "pep440-develop":
rendered = render_pep440_develop(pieces)
elif style == "pep440-auto":
rendered = render_pep440_auto(pieces)
elif style == "git-describe":
rendered = render_git_describe(pieces)
elif style == "git-describe-long":
rendered = render_git_describe_long(pieces)
else:
raise ValueError("unknown style '%%s'" %% style)
return {"version": rendered, "full-revisionid": pieces["long"],
"dirty": pieces["dirty"], "error": None,
"date": pieces.get("date")}
def get_versions():
"""Get version information or return default if unable to do so."""
# I am in _version.py, which lives at ROOT/VERSIONFILE_SOURCE. If we have
# __file__, we can work backwards from there to the root. Some
# py2exe/bbfreeze/non-CPython implementations don't do __file__, in which
# case we can only use expanded keywords.
cfg = get_config()
verbose = cfg.verbose
try:
return git_versions_from_keywords(get_keywords(), cfg.tag_prefix,
verbose)
except NotThisMethod:
pass
try:
root = os.path.realpath(__file__)
# versionfile_source is the relative path from the top of the source
# tree (where the .git directory might live) to this file. Invert
# this to find the root from __file__.
for i in cfg.versionfile_source.split('/'):
root = os.path.dirname(root)
except NameError:
return {"version": "0+unknown", "full-revisionid": None,
"dirty": None,
"error": "unable to find root of source tree",
"date": None}
try:
pieces = git_pieces_from_vcs(cfg.tag_prefix, root, verbose)
return render(pieces, cfg.style)
except NotThisMethod:
pass
try:
if cfg.parentdir_prefix:
return versions_from_parentdir(cfg.parentdir_prefix, root, verbose)
except NotThisMethod:
pass
return {"version": "0+unknown", "full-revisionid": None,
"dirty": None,
"error": "unable to compute version", "date": None}
'''
@register_vcs_handler("git", "get_keywords")
def git_get_keywords(versionfile_abs):
"""Extract version information from the given file."""
# the code embedded in _version.py can just fetch the value of these
# keywords. When used from setup.py, we don't want to import _version.py,
# so we do it with a regexp instead. This function is not used from
# _version.py.
keywords = {}
try:
f = open(versionfile_abs, "r")
for line in f.readlines():
if line.strip().startswith("git_refnames ="):
mo = re.search(r'=\s*"(.*)"', line)
if mo:
keywords["refnames"] = mo.group(1)
if line.strip().startswith("git_full ="):
mo = re.search(r'=\s*"(.*)"', line)
if mo:
keywords["full"] = mo.group(1)
if line.strip().startswith("git_date ="):
mo = re.search(r'=\s*"(.*)"', line)
if mo:
keywords["date"] = mo.group(1)
f.close()
except EnvironmentError:
pass
return keywords
@register_vcs_handler("git", "keywords")
def git_versions_from_keywords(keywords, tag_prefix, verbose):
"""Get version information from git keywords."""
if not keywords:
raise NotThisMethod("no keywords at all, weird")
date = keywords.get("date")
if date is not None:
# git-2.2.0 added "%cI", which expands to an ISO-8601 -compliant
# datestamp. However we prefer "%ci" (which expands to an "ISO-8601
# -like" string, which we must then edit to make compliant), because
# it's been around since git-1.5.3, and it's too difficult to
# discover which version we're using, or to work around using an
# older one.
date = date.strip().replace(" ", "T", 1).replace(" ", "", 1)
refnames = keywords["refnames"].strip()
if refnames.startswith("$Format"):
if verbose:
print("keywords are unexpanded, not using")
raise NotThisMethod("unexpanded keywords, not a git-archive tarball")
refs = set([r.strip() for r in refnames.strip("()").split(",")])
# starting in git-1.8.3, tags are listed as "tag: foo-1.0" instead of
# just "foo-1.0". If we see a "tag: " prefix, prefer those.
TAG = "tag: "
tags = set([r[len(TAG):] for r in refs if r.startswith(TAG)])
if not tags:
# Either we're using git < 1.8.3, or there really are no tags. We use
# a heuristic: assume all version tags have a digit. The old git %d
# expansion behaves like git log --decorate=short and strips out the
# refs/heads/ and refs/tags/ prefixes that would let us distinguish
# between branches and tags. By ignoring refnames without digits, we
# filter out many common branch names like "release" and
# "stabilization", as well as "HEAD" and "master".
tags = set([r for r in refs if re.search(r'\d', r)])
if verbose:
print("discarding '%s', no digits" % ",".join(refs - tags))
if verbose:
print("likely tags: %s" % ",".join(sorted(tags)))
for ref in sorted(tags):
# sorting will prefer e.g. "2.0" over "2.0rc1"
if ref.startswith(tag_prefix):
r = ref[len(tag_prefix):]
if verbose:
print("picking %s" % r)
return {"version": r,
"full-revisionid": keywords["full"].strip(),
"dirty": False, "error": None,
"date": date}
# no suitable tags, so version is "0+unknown", but full hex is still there
if verbose:
print("no suitable tags, using unknown + full revision id")
return {"version": "0+unknown",
"full-revisionid": keywords["full"].strip(),
"dirty": False, "error": "no suitable tags", "date": None}
@register_vcs_handler("git", "pieces_from_vcs")
def git_pieces_from_vcs(tag_prefix, root, verbose, run_command=run_command):
"""Get version from 'git describe' in the root of the source tree.
This only gets called if the git-archive 'subst' keywords were *not*
expanded, and _version.py hasn't already been rewritten with a short
version string, meaning we're inside a checked out source tree.
"""
GITS = ["git"]
if sys.platform == "win32":
GITS = ["git.cmd", "git.exe"]
out, rc = run_command(GITS, ["rev-parse", "--git-dir"], cwd=root,
hide_stderr=True)
if rc != 0:
if verbose:
print("Directory %s not under git control" % root)
raise NotThisMethod("'git rev-parse --git-dir' returned error")
# if there is a tag matching tag_prefix, this yields TAG-NUM-gHEX[-dirty]
# if there isn't one, this yields HEX[-dirty] (no NUM)
describe_out, rc = run_command(GITS, ["describe", "--tags", "--dirty",
"--always", "--long",
"--match", "%s*" % tag_prefix],
cwd=root)
# --long was added in git-1.5.5
if describe_out is None:
raise NotThisMethod("'git describe' failed")
describe_out = describe_out.strip()
full_out, rc = run_command(GITS, ["rev-parse", "HEAD"], cwd=root)
if full_out is None:
raise NotThisMethod("'git rev-parse' failed")
full_out = full_out.strip()
pieces = {}
pieces["long"] = full_out
pieces["short"] = full_out[:7] # maybe improved later
pieces["error"] = None
# parse describe_out. It will be like TAG-NUM-gHEX[-dirty] or HEX[-dirty]
# TAG might have hyphens.
git_describe = describe_out
# look for -dirty suffix
dirty = git_describe.endswith("-dirty")
pieces["dirty"] = dirty
if dirty:
git_describe = git_describe[:git_describe.rindex("-dirty")]
# now we have TAG-NUM-gHEX or HEX
# Determine which branch is active (ie, checked-out)
branch_name, rc = run_command(
GITS, ["rev-parse", "--abbrev-ref", "HEAD"], cwd=root)
pieces["branch"] = branch_name
if "-" in git_describe:
# TAG-NUM-gHEX
mo = re.search(r'^(.+)-(\d+)-g([0-9a-f]+)$', git_describe)
if not mo:
# unparseable. Maybe git-describe is misbehaving?
pieces["error"] = ("unable to parse git-describe output: '%s'"
% describe_out)
return pieces
# tag
full_tag = mo.group(1)
if not full_tag.startswith(tag_prefix):
if verbose:
fmt = "tag '%s' doesn't start with prefix '%s'"
print(fmt % (full_tag, tag_prefix))
pieces["error"] = ("tag '%s' doesn't start with prefix '%s'"
% (full_tag, tag_prefix))
return pieces
pieces["closest-tag"] = full_tag[len(tag_prefix):]
# distance: number of commits since tag
pieces["distance"] = int(mo.group(2))
# Added to deal with merged branches
history_search_args = ["--ancestry-path"]
if (branch_name == "master"):
# Count distance only along `master`; ignore merge paths
history_search_args = ["--ancestry-path", "--first-parent"]
distance_to_tag, rc = run_command(
GITS, ["rev-list", "--count",] + history_search_args +
["%s..HEAD" % pieces["closest-tag"] ], cwd=root)
pieces["distance"] = int(distance_to_tag)
# commit: short hex revision ID
pieces["short"] = mo.group(3)
else:
# HEX: no tags
pieces["closest-tag"] = None
count_out, rc = run_command(GITS, ["rev-list", "HEAD", "--count"],
cwd=root)
pieces["distance"] = int(count_out) # total number of commits
##################################################
# Added for `develop` branch from `master`
# `HEAD` might have branched from untagged commit on `master`
merge_base_master, rc = run_command(
GITS, ["merge-base", "master", "HEAD", "-a"], cwd=root)
distance_to_master, rc = run_command(
GITS, ["rev-list", "--count", "--ancestry-path",
"%s..HEAD" % merge_base_master], cwd=root)
pieces["distance-to-master"] = int(distance_to_master)
##################################################
# Added for branches other than `master`
if pieces["closest-tag"]:
# TAG exists
master_to_tag, rc = run_command(
GITS, ["rev-list", "--count", "--ancestry-path", "--first-parent",
"%s..%s" % (pieces["closest-tag"], merge_base_master) ],
cwd=root)
pieces["master-to-tag"] = int(master_to_tag)
else:
pieces["master-to-tag"] = pieces["distance"]
##################################################
# Added for "feature" branch from `develop`
pieces["develop"] = None
pieces["distance-to-develop"] = None
branch_develop, rc = run_command(
GITS, ["rev-parse", "--verify", "refs/heads/develop"], cwd=root)
branch_develop_exists = not bool(rc)
if branch_develop_exists:
merge_base_develop, rc = run_command(
GITS, ["merge-base", "develop", "HEAD", "-a"], cwd=root)
distance_to_develop, rc = run_command(
GITS, ["rev-list", "--count", "--ancestry-path",
"%s..HEAD" % merge_base_develop], cwd=root)
pieces["develop"] = branch_develop
pieces["distance-to-develop"] = int(distance_to_develop)
# commit date: see ISO-8601 comment in git_versions_from_keywords()
date = run_command(GITS, ["show", "-s", "--format=%ci", "HEAD"],
cwd=root)[0].strip()
pieces["date"] = date.strip().replace(" ", "T", 1).replace(" ", "", 1)
##################################################
# Added to retrieve list of authors
(authors_raw, rc) = run_command(GITS, ["log", "--pretty=%an"], cwd=root)
authors = [author.strip() for author in authors_raw.split('\n')]
authors_unique = unique(authors)
pieces["authors"] = list(reversed(authors_unique))
return pieces
def do_vcs_install(manifest_in, versionfile_source, ipy):
"""Git-specific installation logic for Versioneer.
For Git, this means creating/changing .gitattributes to mark _version.py
for export-subst keyword substitution.
"""
GITS = ["git"]
if sys.platform == "win32":
GITS = ["git.cmd", "git.exe"]
files = [manifest_in, versionfile_source]
if ipy:
files.append(ipy)
try:
me = __file__
if me.endswith(".pyc") or me.endswith(".pyo"):
me = os.path.splitext(me)[0] + ".py"
versioneer_file = os.path.relpath(me)
except NameError:
versioneer_file = "versioneer.py"
files.append(versioneer_file)
present = False
try:
f = open(".gitattributes", "r")
for line in f.readlines():
if line.strip().startswith(versionfile_source):
if "export-subst" in line.strip().split()[1:]:
present = True
f.close()
except EnvironmentError:
pass
if not present:
f = open(".gitattributes", "a+")
f.write("%s export-subst\n" % versionfile_source)
f.close()
files.append(".gitattributes")
run_command(GITS, ["add", "--"] + files)
def versions_from_parentdir(parentdir_prefix, root, verbose):
"""Try to determine the version from the parent directory name.
Source tarballs conventionally unpack into a directory that includes both
the project name and a version string. We will also support searching up
two directory levels for an appropriately named parent directory
"""
rootdirs = []
for i in range(3):
dirname = os.path.basename(root)
if dirname.startswith(parentdir_prefix):
return {"version": dirname[len(parentdir_prefix):],
"full-revisionid": None,
"dirty": False, "error": None, "date": None}
else:
rootdirs.append(root)
root = os.path.dirname(root) # up a level
if verbose:
print("Tried directories %s but none started with prefix %s" %
(str(rootdirs), parentdir_prefix))
raise NotThisMethod("rootdir doesn't start with parentdir_prefix")
SHORT_VERSION_PY = """
# This file was generated by 'versioneer.py' (0.18) from
# revision-control system data, or from the parent directory name of an
# unpacked source archive. Distribution tarballs contain a pre-generated copy
# of this file.
import json
version_json = '''
%s
''' # END VERSION_JSON
def get_versions():
return json.loads(version_json)
"""
def versions_from_file(filename):
"""Try to determine the version from _version.py if present."""
try:
with open(filename) as f:
contents = f.read()
except EnvironmentError:
raise NotThisMethod("unable to read _version.py")
mo = re.search(r"version_json = '''\n(.*)''' # END VERSION_JSON",
contents, re.M | re.S)
if not mo:
mo = re.search(r"version_json = '''\r\n(.*)''' # END VERSION_JSON",
contents, re.M | re.S)
if not mo:
raise NotThisMethod("no version_json in _version.py")
return json.loads(mo.group(1))
def write_to_version_file(filename, versions):
"""Write the given version number to the given _version.py file."""
os.unlink(filename)
contents = json.dumps(versions, sort_keys=True,
indent=1, separators=(",", ": "))
with open(filename, "w") as f:
f.write(SHORT_VERSION_PY % contents)
print("set %s to '%s'" % (filename, versions["version"]))
def plus_or_dot(pieces):
"""Return a + if we don't already have one, else return a ."""
if "+" in pieces.get("closest-tag", ""):
return "."
return "+"
def render_pep440(pieces):
"""Build up version string, with post-release "local version identifier".
Our goal: TAG[+DISTANCE.gHEX[.dirty]] . Note that if you
get a tagged build and then dirty it, you'll get TAG+0.gHEX.dirty
Exceptions:
1: no tags. git_describe was just HEX. 0+untagged.DISTANCE.gHEX[.dirty]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"] or pieces["dirty"]:
rendered += plus_or_dot(pieces)
rendered += "%d.g%s" % (pieces["distance"], pieces["short"])
if pieces["dirty"]:
rendered += ".dirty"
else:
# exception #1
rendered = "0+untagged.%d.g%s" % (pieces["distance"],
pieces["short"])
if pieces["dirty"]:
rendered += ".dirty"
return rendered
def render_pep440_pre(pieces):
"""TAG[.post.devDISTANCE] -- No -dirty.
Exceptions:
1: no tags. 0.post.devDISTANCE
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"]:
rendered += ".post.dev%d" % pieces["distance"]
else:
# exception #1
rendered = "0.post.dev%d" % pieces["distance"]
return rendered
def render_pep440_post(pieces):
"""TAG[.postDISTANCE[.dev0]+gHEX] .
The ".dev0" means dirty. Note that .dev0 sorts backwards
(a dirty tree will appear "older" than the corresponding clean one),
but you shouldn't be releasing software with -dirty anyways.
Exceptions:
1: no tags. 0.postDISTANCE[.dev0]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"] or pieces["dirty"]:
rendered += ".post%d" % pieces["distance"]
if pieces["dirty"]:
rendered += ".dev0"
rendered += plus_or_dot(pieces)
rendered += "g%s" % pieces["short"]
else:
# exception #1
rendered = "0.post%d" % pieces["distance"]
if pieces["dirty"]:
rendered += ".dev0"
rendered += "+g%s" % pieces["short"]
return rendered
def render_pep440_old(pieces):
"""TAG[.postDISTANCE[.dev0]] .
The ".dev0" means dirty.
Eexceptions:
1: no tags. 0.postDISTANCE[.dev0]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"] or pieces["dirty"]:
rendered += ".post%d" % pieces["distance"]
if pieces["dirty"]:
rendered += ".dev0"
else:
# exception #1
rendered = "0.post%d" % pieces["distance"]
if pieces["dirty"]:
rendered += ".dev0"
return rendered
def render_pep440_micro(pieces):
"""Build up version string, with post-release "local version identifier".
Our goal: TAG.DISTANCE[+gHEX.dirty]
Exceptions:
1: no tags. git_describe was just HEX. 0+untagged.DISTANCE.gHEX[.dirty]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
rendered += ".%d" % pieces["distance"]
if pieces["dirty"]:
rendered += plus_or_dot(pieces)
rendered += "g%s" % pieces["short"]
rendered += ".dirty"
else:
# exception #1
rendered = "0+untagged.%d.g%s" % (pieces["distance"],
pieces["short"])
if pieces["dirty"]:
rendered += ".dirty"
return rendered
def render_pep440_develop(pieces):
"""Build up version string, used within development branch of repository.
Our goal: MERGE-POINT.post.devN[+gHEX.dirty]
+) MERGE-POINT = Most recent common ancestor for `develop` and `master`
*) Does not yet handle branch from `release-*`
+) N = DISTANCE from the MERGE-POINT
Exceptions:
1: no tags. 0.post.devDISTANCE[+gHEX.dirty]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
distance_to_merge = pieces["distance-to-master"]
# distance_merge_to_tag = (pieces["distance"] - distance_to_merge)
distance_merge_to_tag = pieces["master-to-tag"]
if (distance_merge_to_tag > 0):
rendered += ".%d" % distance_merge_to_tag
rendered += ".post.dev%d" % distance_to_merge
else:
# exception #1
rendered = "0.post.dev%d" % (pieces["distance"] - 1)
if pieces["dirty"]:
rendered += plus_or_dot(pieces)
rendered += "g%s" % pieces["short"]
rendered += ".dirty"
return rendered
def render_pep440_feature(pieces):
"""Build up version string, used within "feature" branch of repository.
Our goal: MERGE-POINT.post.devN+gHEX.BRANCH-NAME.M[.dirty]
+) MERGE-POINT = Most recent common ancestor for `develop` and `master`
*) Does not yet handle branch from `release-*`
+) N = DISTANCE from the MERGE-POINT of `develop` and `master`
+) M = DISTANCE from the MERGE-POINT of "feature" and `develop`
Exceptions:
1: no tags. 0.post.devDISTANCE+gHEX[.dirty]
"""
if pieces["closest-tag"] and pieces["develop"]:
rendered = pieces["closest-tag"]
distance_to_develop = pieces["distance-to-develop"]
distance_to_merge = pieces["distance-to-master"]
# distance_merge_to_tag = (pieces["distance"] - distance_to_merge)
distance_merge_to_tag = pieces["master-to-tag"]
distance_dev_to_merge = (distance_to_merge - distance_to_develop)
if (distance_merge_to_tag > 0):
rendered += ".%d" % distance_merge_to_tag
rendered += ".post.dev%d" % distance_dev_to_merge
rendered += plus_or_dot(pieces)
rendered += "g%s" % pieces["short"]
rendered += ".%s" % pieces["branch"]
rendered += ".%d" % distance_to_develop
else:
# exception #1
rendered = "0.post.dev%d" % (pieces["distance"] - 1)
rendered += plus_or_dot(pieces)
rendered += "g%s" % pieces["short"]
if pieces["dirty"]:
rendered += ".dirty"
return rendered
def render_pep440_auto(pieces):
"""Build up version string, based on branch of repository.
Our goal: If on development branch, use render_pep440_develop()
Otherwise use render_pep440_micro()
Exceptions:
+) NONE
"""
if pieces["branch"] == "master":
# logging.debug("Rendering: render_pep440_micro()")
rendered = render_pep440_micro(pieces)
elif pieces["branch"] == "develop":
# logging.debug("Rendering: render_pep440_develop()")
rendered = render_pep440_develop(pieces)
elif pieces["branch"].startswith("release"):
# render_pep440_release() not yet implemented
rendered = render_git_describe(pieces)
else:
# logging.debug("Rendering: render_pep440_feature()")
rendered = render_pep440_feature(pieces)
return rendered
def render_git_describe(pieces):
"""TAG[-DISTANCE-gHEX][-dirty].
Like 'git describe --tags --dirty --always'.
Exceptions:
1: no tags. HEX[-dirty] (note: no 'g' prefix)
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"]:
rendered += "-%d-g%s" % (pieces["distance"], pieces["short"])
else:
# exception #1
rendered = pieces["short"]
if pieces["dirty"]:
rendered += "-dirty"
return rendered
def render_git_describe_long(pieces):
"""TAG-DISTANCE-gHEX[-dirty].
Like 'git describe --tags --dirty --always -long'.
The distance/hash is unconditional.
Exceptions:
1: no tags. HEX[-dirty] (note: no 'g' prefix)
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
rendered += "-%d-g%s" % (pieces["distance"], pieces["short"])
else:
# exception #1
rendered = pieces["short"]
if pieces["dirty"]:
rendered += "-dirty"
return rendered
def render(pieces, style):
"""Render the given version pieces into the requested style."""
if pieces["error"]:
return {"version": "unknown",
"full-revisionid": pieces.get("long"),
"dirty": None,
"error": pieces["error"],
"date": None,
"authors": None,
}
if not style or style == "default":
style = "pep440-auto" # the default
if style == "pep440":
rendered = render_pep440(pieces)
elif style == "pep440-pre":
rendered = render_pep440_pre(pieces)
elif style == "pep440-post":
rendered = render_pep440_post(pieces)
elif style == "pep440-old":
rendered = render_pep440_old(pieces)
elif style == "pep440-micro":
rendered = render_pep440_micro(pieces)
elif style == "pep440-develop":
rendered = render_pep440_develop(pieces)
elif style == "pep440-auto":
rendered = render_pep440_auto(pieces)
elif style == "git-describe":
rendered = render_git_describe(pieces)
elif style == "git-describe-long":
rendered = render_git_describe_long(pieces)
else:
raise ValueError("unknown style '%s'" % style)
return {"version": rendered, "full-revisionid": pieces["long"],
"dirty": pieces["dirty"], "error": None,
"date": pieces.get("date"), "authors": pieces["authors"]}
class VersioneerBadRootError(Exception):
"""The project root directory is unknown or missing key files."""
def get_versions(verbose=False):
"""Get the project version from whatever source is available.
Returns dict with two keys: 'version' and 'full'.
"""
if "versioneer" in sys.modules:
# see the discussion in cmdclass.py:get_cmdclass()
del sys.modules["versioneer"]
root = get_root()
cfg = get_config_from_root(root)
assert cfg.VCS is not None, "please set [versioneer]VCS= in setup.cfg"
handlers = HANDLERS.get(cfg.VCS)
assert handlers, "unrecognized VCS '%s'" % cfg.VCS
verbose = verbose or cfg.verbose
assert cfg.versionfile_source is not None, \
"please set versioneer.versionfile_source"
assert cfg.tag_prefix is not None, "please set versioneer.tag_prefix"
versionfile_abs = os.path.join(root, cfg.versionfile_source)
# extract version from first of: _version.py, VCS command (e.g. 'git
# describe'), parentdir. This is meant to work for developers using a
# source checkout, for users of a tarball created by 'setup.py sdist',
# and for users of a tarball/zipball created by 'git archive' or github's
# download-from-tag feature or the equivalent in other VCSes.
get_keywords_f = handlers.get("get_keywords")
from_keywords_f = handlers.get("keywords")
if get_keywords_f and from_keywords_f:
try:
keywords = get_keywords_f(versionfile_abs)
ver = from_keywords_f(keywords, cfg.tag_prefix, verbose)
if verbose:
print("got version from expanded keyword %s" % ver)
return ver
except NotThisMethod:
pass
try:
ver = versions_from_file(versionfile_abs)
if verbose:
print("got version from file %s %s" % (versionfile_abs, ver))
return ver
except NotThisMethod:
pass
from_vcs_f = handlers.get("pieces_from_vcs")
if from_vcs_f:
try:
pieces = from_vcs_f(cfg.tag_prefix, root, verbose)
ver = render(pieces, cfg.style)
if verbose:
print("got version from VCS %s" % ver)
return ver
except NotThisMethod:
pass
try:
if cfg.parentdir_prefix:
ver = versions_from_parentdir(cfg.parentdir_prefix, root, verbose)
if verbose:
print("got version from parentdir %s" % ver)
return ver
except NotThisMethod:
pass
if verbose:
print("unable to compute version")
return {"version": "0+unknown", "full-revisionid": None,
"dirty": None, "error": "unable to compute version",
"date": None,
"authors": [],
}
def get_version():
"""Get the short version string for this project."""
return get_versions()["version"]
def get_cmdclass():
"""Get the custom setuptools/distutils subclasses used by Versioneer."""
if "versioneer" in sys.modules:
del sys.modules["versioneer"]
# this fixes the "python setup.py develop" case (also 'install' and
# 'easy_install .'), in which subdependencies of the main project are
# built (using setup.py bdist_egg) in the same python process. Assume
# a main project A and a dependency B, which use different versions
# of Versioneer. A's setup.py imports A's Versioneer, leaving it in
# sys.modules by the time B's setup.py is executed, causing B to run
# with the wrong versioneer. Setuptools wraps the sub-dep builds in a
# sandbox that restores sys.modules to it's pre-build state, so the
# parent is protected against the child's "import versioneer". By
# removing ourselves from sys.modules here, before the child build
# happens, we protect the child from the parent's versioneer too.
# Also see https://github.com/warner/python-versioneer/issues/52
cmds = {}
# we add "version" to both distutils and setuptools
from distutils.core import Command
class cmd_version(Command):
description = "report generated version string"
user_options = []
boolean_options = []
def initialize_options(self):
pass
def finalize_options(self):
pass
def run(self):
vers = get_versions(verbose=True)
print("Version: %s" % vers["version"])
print(" full-revisionid: %s" % vers.get("full-revisionid"))
print(" dirty: %s" % vers.get("dirty"))
print(" date: %s" % vers.get("date"))
if vers["error"]:
print(" error: %s" % vers["error"])
cmds["version"] = cmd_version
# we override "build_py" in both distutils and setuptools
#
# most invocation pathways end up running build_py:
# distutils/build -> build_py
# distutils/install -> distutils/build ->..
# setuptools/bdist_wheel -> distutils/install ->..
# setuptools/bdist_egg -> distutils/install_lib -> build_py
# setuptools/install -> bdist_egg ->..
# setuptools/develop -> ?
# pip install:
# copies source tree to a tempdir before running egg_info/etc
# if .git isn't copied too, 'git describe' will fail
# then does setup.py bdist_wheel, or sometimes setup.py install
# setup.py egg_info -> ?
# we override different "build_py" commands for both environments
if "setuptools" in sys.modules:
from setuptools.command.build_py import build_py as _build_py
else:
from distutils.command.build_py import build_py as _build_py
class cmd_build_py(_build_py):
def run(self):
root = get_root()
cfg = get_config_from_root(root)
versions = get_versions()
_build_py.run(self)
# now locate _version.py in the new build/ directory and replace
# it with an updated value
if cfg.versionfile_build:
target_versionfile = os.path.join(self.build_lib,
cfg.versionfile_build)
print("UPDATING %s" % target_versionfile)
write_to_version_file(target_versionfile, versions)
cmds["build_py"] = cmd_build_py
if "cx_Freeze" in sys.modules: # cx_freeze enabled?
from cx_Freeze.dist import build_exe as _build_exe
# nczeczulin reports that py2exe won't like the pep440-style string
# as FILEVERSION, but it can be used for PRODUCTVERSION, e.g.
# setup(console=[{
# "version": versioneer.get_version().split("+", 1)[0], # FILEVERSION
# "product_version": versioneer.get_version(),
# ...
class cmd_build_exe(_build_exe):
def run(self):
root = get_root()
cfg = get_config_from_root(root)
versions = get_versions()
target_versionfile = cfg.versionfile_source
print("UPDATING %s" % target_versionfile)
write_to_version_file(target_versionfile, versions)
_build_exe.run(self)
os.unlink(target_versionfile)
with open(cfg.versionfile_source, "w") as f:
LONG = LONG_VERSION_PY[cfg.VCS]
f.write(LONG %
{"DOLLAR": "$",
"STYLE": cfg.style,
"TAG_PREFIX": cfg.tag_prefix,
"PARENTDIR_PREFIX": cfg.parentdir_prefix,
"VERSIONFILE_SOURCE": cfg.versionfile_source,
})
cmds["build_exe"] = cmd_build_exe
del cmds["build_py"]
if 'py2exe' in sys.modules: # py2exe enabled?
try:
from py2exe.distutils_buildexe import py2exe as _py2exe # py3
except ImportError:
from py2exe.build_exe import py2exe as _py2exe # py2
class cmd_py2exe(_py2exe):
def run(self):
root = get_root()
cfg = get_config_from_root(root)
versions = get_versions()
target_versionfile = cfg.versionfile_source
print("UPDATING %s" % target_versionfile)
write_to_version_file(target_versionfile, versions)
_py2exe.run(self)
os.unlink(target_versionfile)
with open(cfg.versionfile_source, "w") as f:
LONG = LONG_VERSION_PY[cfg.VCS]
f.write(LONG %
{"DOLLAR": "$",
"STYLE": cfg.style,
"TAG_PREFIX": cfg.tag_prefix,
"PARENTDIR_PREFIX": cfg.parentdir_prefix,
"VERSIONFILE_SOURCE": cfg.versionfile_source,
})
cmds["py2exe"] = cmd_py2exe
# we override different "sdist" commands for both environments
if "setuptools" in sys.modules:
from setuptools.command.sdist import sdist as _sdist
else:
from distutils.command.sdist import sdist as _sdist
class cmd_sdist(_sdist):
def run(self):
versions = get_versions()
self._versioneer_generated_versions = versions
# unless we update this, the command will keep using the old
# version
self.distribution.metadata.version = versions["version"]
return _sdist.run(self)
def make_release_tree(self, base_dir, files):
root = get_root()
cfg = get_config_from_root(root)
_sdist.make_release_tree(self, base_dir, files)
# now locate _version.py in the new base_dir directory
# (remembering that it may be a hardlink) and replace it with an
# updated value
target_versionfile = os.path.join(base_dir, cfg.versionfile_source)
print("UPDATING %s" % target_versionfile)
write_to_version_file(target_versionfile,
self._versioneer_generated_versions)
cmds["sdist"] = cmd_sdist
return cmds
CONFIG_ERROR = """
setup.cfg is missing the necessary Versioneer configuration. You need
a section like:
[versioneer]
VCS = git
style = pep440
versionfile_source = src/myproject/_version.py
versionfile_build = myproject/_version.py
tag_prefix =
parentdir_prefix = myproject-
You will also need to edit your setup.py to use the results:
import versioneer
setup(version=versioneer.get_version(),
cmdclass=versioneer.get_cmdclass(), ...)
Please read the docstring in ./versioneer.py for configuration instructions,
edit setup.cfg, and re-run the installer or 'python versioneer.py setup'.
"""
SAMPLE_CONFIG = """
# See the docstring in versioneer.py for instructions. Note that you must
# re-run 'versioneer.py setup' after changing this section, and commit the
# resulting files.
[versioneer]
#VCS = git
#style = pep440
#versionfile_source =
#versionfile_build =
#tag_prefix =
#parentdir_prefix =
"""
INIT_PY_SNIPPET = """
from ._version import get_versions
__version__ = get_versions()['version']
del get_versions
"""
def do_setup():
"""Main VCS-independent setup function for installing Versioneer."""
root = get_root()
try:
cfg = get_config_from_root(root)
except (EnvironmentError, configparser.NoSectionError,
configparser.NoOptionError) as e:
if isinstance(e, (EnvironmentError, configparser.NoSectionError)):
print("Adding sample versioneer config to setup.cfg",
file=sys.stderr)
with open(os.path.join(root, "setup.cfg"), "a") as f:
f.write(SAMPLE_CONFIG)
print(CONFIG_ERROR, file=sys.stderr)
return 1
print(" creating %s" % cfg.versionfile_source)
with open(cfg.versionfile_source, "w") as f:
LONG = LONG_VERSION_PY[cfg.VCS]
f.write(LONG % {"DOLLAR": "$",
"STYLE": cfg.style,
"TAG_PREFIX": cfg.tag_prefix,
"PARENTDIR_PREFIX": cfg.parentdir_prefix,
"VERSIONFILE_SOURCE": cfg.versionfile_source,
})
ipy = os.path.join(os.path.dirname(cfg.versionfile_source),
"__init__.py")
if os.path.exists(ipy):
try:
with open(ipy, "r") as f:
old = f.read()
except EnvironmentError:
old = ""
if INIT_PY_SNIPPET not in old:
print(" appending to %s" % ipy)
with open(ipy, "a") as f:
f.write(INIT_PY_SNIPPET)
else:
print(" %s unmodified" % ipy)
else:
print(" %s doesn't exist, ok" % ipy)
ipy = None
# Make sure both the top-level "versioneer.py" and versionfile_source
# (PKG/_version.py, used by runtime code) are in MANIFEST.in, so
# they'll be copied into source distributions. Pip won't be able to
# install the package without this.
manifest_in = os.path.join(root, "MANIFEST.in")
simple_includes = set()
try:
with open(manifest_in, "r") as f:
for line in f:
if line.startswith("include "):
for include in line.split()[1:]:
simple_includes.add(include)
except EnvironmentError:
pass
# That doesn't cover everything MANIFEST.in can do
# (http://docs.python.org/2/distutils/sourcedist.html#commands), so
# it might give some false negatives. Appending redundant 'include'
# lines is safe, though.
if "versioneer.py" not in simple_includes:
print(" appending 'versioneer.py' to MANIFEST.in")
with open(manifest_in, "a") as f:
f.write("include versioneer.py\n")
else:
print(" 'versioneer.py' already in MANIFEST.in")
if cfg.versionfile_source not in simple_includes:
print(" appending versionfile_source ('%s') to MANIFEST.in" %
cfg.versionfile_source)
with open(manifest_in, "a") as f:
f.write("include %s\n" % cfg.versionfile_source)
else:
print(" versionfile_source already in MANIFEST.in")
# Make VCS-specific changes. For git, this means creating/changing
# .gitattributes to mark _version.py for export-subst keyword
# substitution.
do_vcs_install(manifest_in, cfg.versionfile_source, ipy)
return 0
def scan_setup_py():
"""Validate the contents of setup.py against Versioneer's expectations."""
found = set()
setters = False
errors = 0
with open("setup.py", "r") as f:
for line in f.readlines():
if "import versioneer" in line:
found.add("import")
if "versioneer.get_cmdclass()" in line:
found.add("cmdclass")
if "versioneer.get_version()" in line:
found.add("get_version")
if "versioneer.VCS" in line:
setters = True
if "versioneer.versionfile_source" in line:
setters = True
if len(found) != 3:
print("")
print("Your setup.py appears to be missing some important items")
print("(but I might be wrong). Please make sure it has something")
print("roughly like the following:")
print("")
print(" import versioneer")
print(" setup( version=versioneer.get_version(),")
print(" cmdclass=versioneer.get_cmdclass(), ...)")
print("")
errors += 1
if setters:
print("You should remove lines like 'versioneer.VCS = ' and")
print("'versioneer.versionfile_source = ' . This configuration")
print("now lives in setup.cfg, and should be removed from setup.py")
print("")
errors += 1
return errors
if __name__ == "__main__":
cmd = sys.argv[1]
if cmd == "setup":
errors = do_setup()
errors += scan_setup_py()
if errors:
sys.exit(1) | ALS.Milo | /ALS.Milo-0.18.1.tar.gz/ALS.Milo-0.18.1/versioneer.py | versioneer.py |
from __future__ import print_function
from __future__ import division
from builtins import zip
from builtins import str
from builtins import range
from builtins import object
__author__ = "Padraic Shafer"
__copyright__ = "Copyright (c) 2014-2021, Padraic Shafer"
__credits__ = [__author__, ]
__license__ = ""
__maintainer__ = "Padraic Shafer"
__email__ = "[email protected]"
__status__ = "Development"
# Allow import of package siblings when module is run as script
import pkgscript
if (__name__ == "__main__") and (__package__ is None):
pkgscript.import_parent_packages("als.milo", globals())
from als.milo import __version__, __date__
from numbers import Real
from datetime import datetime, date
from astropy.io import fits
from numpy import pi, cos, sin, deg2rad, rad2deg, arctan2, arcsin
from numpy import array, empty, full, newaxis
from numpy import linspace, arange, tile, hstack, vstack
from numpy import matrix, dot, cross, outer, sum, product, zeros, roots
from numpy import square, sqrt, around, isfinite, nan, isnan
from numpy.linalg import norm, solve, lstsq, tensorsolve
from scipy.optimize import brentq
import argparse
import pandas as pd
class Photon( object ):
"""Photon: set or access (energy / wavelength / q) of the photon
energy (eV) / wavelength (nm) / q (1 / nm) are always synchronized
according to relationship:
q == 2*pi / wavelength;
wavelength == 1239.842 eV*nm / energy
"""
energy_at_1nm = 1239.842 # eV
def __init__(self, *arguments, **keywords):
"""Initial value can be supplied in one of several formats
Due to the multi-faceted nature of this class,
keyword arguments are preferred
***Highest priority***
energy = value, hv = value
wavelength = value, wl = value, lambda_ = value
q = value, momentum = value
***Lowest priority***
If no keywords are given, then value of first arbitrary argument
is used as the value for energy
(as if input were energy = value)
"""
if keywords:
value = keywords.get('energy', None)
if value is not None:
self.energy = value
return
value = keywords.get('hv', None)
if value is not None:
self.energy = value
return
value = keywords.get('wavelength', None)
if value is not None:
self.wavelength = value
return
value = keywords.get('wl', None)
if value is not None:
self.wavelength = value
return
value = keywords.get('lambda_', None)
if value is not None:
self.wavelength = value
return
value = keywords.get('q', None)
if value is not None:
self.q = value
return
value = keywords.get('momentum', None)
if value is not None:
self.q = value
return
elif arguments:
value = arguments[0]
if isinstance(value, Real):
self.energy = value
return
# handle undefined input
pass
@property
def energy(self):
"""Access the energy (in eV) of the Photon"""
return self._energy
@energy.setter
def energy(self, value):
# Does not check for valid value
self._energy = value
return value
@property
def wavelength(self):
"""Access the wavelength (in nm) of the Photon"""
return Photon.energy_at_1nm / self._energy
@wavelength.setter
def wavelength(self, value):
# Does not check for valid value
self._energy = Photon.energy_at_1nm / value
return value
@property
def q(self):
"""Access the momentum (in 1/nm) of the Photon"""
return 2*pi * self._energy / Photon.energy_at_1nm
@q.setter
def q(self, value):
# Does not check for valid value
self._energy = value * Photon.energy_at_1nm / (2*pi)
return value
class Polarization( object ):
"""Polarization: set or access polarization of the x-ray beamline
State can be circular, linear, or undefined
Degree of circular polarization is given as ratio, -1 <= ratio <= 1
Angle of linear polarization axis is given in degrees from horizontal
"""
# Allowed polarization states
UNDEFINED = -1
CIRCULAR = 0
LINEAR = 1
# Offsets for internal calculations of state / value
_OFFSET_CIRCULAR = 0
_OFFSET_LINEAR = 100
def __init__(self, init_value=0., *arguments, **keywords):
"""init_value should be supplied in the following range(s):
0 -- Horizontal linear polarization
-1 <= init_value <= 1 -- Degree of circular polarization
2 -- Vertical linear polarization
100 <= init_value <= 190 -- 100 + angle of linear polarization
State is set to circular, linear, or undefined based on init_value
"""
self.value = float(init_value)
@property
def value(self):
"""Access the raw value of the x-ray polarization"""
return self._value
@value.setter
def value(self, new_value):
self._value = new_value
# might need to adjust comparisons for floating point rounding
if (0 < abs(new_value) <= 1):
self._state = Polarization.CIRCULAR
self._offset_circular = Polarization._OFFSET_CIRCULAR
self._offset_linear = float('NaN')
elif (100 <= new_value <= 190):
self._state = Polarization.LINEAR
self._offset_circular = float('NaN')
self._offset_linear = Polarization._OFFSET_LINEAR
elif (0 <= new_value <= 0):
self._value = 100
self._state = Polarization.LINEAR
self._offset_circular = float('NaN')
self._offset_linear = Polarization._OFFSET_LINEAR
elif (2 <= new_value <= 2):
self._value = 190
self._state = Polarization.LINEAR
self._offset_circular = float('NaN')
self._offset_linear = Polarization._OFFSET_LINEAR
else:
self._state = Polarization.UNDEFINED
self._offset_circular = float('NaN')
self._offset_linear = float('NaN')
return new_value
@property
def circular_degree(self):
"""Access the degree of circular polarization for the x-ray
Given as ratio, -1 <= degree_of_polarization <= 1
"""
return (self._value - self._offset_circular)
@circular_degree.setter
def circular_degree(self, degree_of_polarization):
# Error checking performed by @value.setter
self.value = degree_of_polarization
return self.value
@property
def linear_angle(self):
"""Access the angle of linear polarization for the x-ray
Angle is in degrees from horizontal
"""
return (self._value - self._offset_linear)
@linear_angle.setter
def linear_angle(self, angle):
# Error checking performed by @value.setter
self.value = angle + Polarization._OFFSET_LINEAR
return self.value
@property
def state(self):
"""Access the polarization state of the x-ray"""
return self._state
class SampleLattice( object ):
"""SampleLattice: set or access lattice parameters of sample
x_unit (x = a,b,c) = unit vector of fundamental lattice translation
x (x = a,b,c) = distance of fundamental lattice translation
x_vect (x = a,b,c) = x * x_unit
reset_axes: Sets a_unit // x, b_unit // y, c_unit // z
permute_axes: Cycles lattice fundamental directions so that (x,y,z) //
(a,b,c)_unit -> (a,b,c)_unit -> (a,b,c)_unit // (x,y,z)
rotate_by: Rotate sample by XX degrees about given axis
normal: lattice vector normal to sample stage (z)
longitudinal: lattice vector longitudinal to sample stage (y)
transverse: lattice vector transverse to sample stage (x)
"""
# set parameter modes
CCD, DIODE_WIDE, DIODE_HIRES = list( range(3) )
energy_at_1nm = 1239.842 # eV
def __init__(self, *arguments, **keywords):
"""Initial value can be supplied in one of several formats
Due to the multi-faceted nature of this class,
keyword arguments are preferred
***Highest priority***
energy = value, hv = value
wavelength = value, wl = value, lambda_ = value
q = value, momentum = value
***Lowest priority***
If no keywords are given, then value of first arbitrary argument
is used as the value for energy
(as if input were energy = value)
"""
if keywords:
value = keywords.get('energy', None)
if value is not None:
self.energy = value
return
value = keywords.get('hv', None)
if value is not None:
self.energy = value
return
value = keywords.get('wavelength', None)
if value is not None:
self.wavelength = value
return
value = keywords.get('wl', None)
if value is not None:
self.wavelength = value
return
value = keywords.get('lambda_', None)
if value is not None:
self.wavelength = value
return
value = keywords.get('q', None)
if value is not None:
self.q = value
return
value = keywords.get('momentum', None)
if value is not None:
self.q = value
return
elif arguments:
value = arguments[0]
if isinstance(value, Real):
self.energy = value
return
# handle undefined input
pass
@property
def energy(self):
"""Access the energy (in eV) of the Photon"""
return self._energy
@energy.setter
def energy(self, value):
# Does not check for valid value
self._energy = value
return value
@property
def wavelength(self):
"""Access the wavelength (in nm) of the Photon"""
return Photon.energy_at_1nm / self._energy
@wavelength.setter
def wavelength(self, value):
# Does not check for valid value
self._energy = Photon.energy_at_1nm / value
return value
@property
def q(self):
"""Access the momentum (in 1/nm) of the Photon"""
return 2*pi * self._energy / Photon.energy_at_1nm
@q.setter
def q(self, value):
# Does not check for valid value
self._energy = value * Photon.energy_at_1nm / (2*pi)
return value
class Diffractometer( object ):
"""Diffractometer: diffractometer settings and readings"""
def __init__(self, twotheta=0., incidence=0., transverse=0., azimuth=0.,
energy=700., polarization=0., intensity0=1.,
temperature=298, distance=1.,
*arguments, **keywords):
self.twotheta = twotheta
self.incidence = incidence
self.transverse = transverse
self.azimuth = azimuth
self.photon = Photon(energy)
self.polarization = polarization
self.intensity0 = intensity0
self.temperature = temperature
@property
def twotheta(self):
"""Access the detector position, 2-Theta (in degrees)"""
return self._twotheta
@twotheta.setter
def twotheta(self, value):
# Does not check for valid value
self._twotheta = value
return value
@property
def incidence(self):
"""Access the 2D incidence angle (in degrees) within Th-2Th plane"""
return self._incidence
@incidence.setter
def incidence(self, value):
# Does not check for valid value
self._incidence = value
return value
@property
def exit_angle(self):
"""Access the 2D exit angle (in degrees) within Th-2Th plane"""
return (self._twotheta - self._incidence)
@exit_angle.setter
def exit_angle(self, value):
# Does not check for valid value
# self._twotheta = self._incidence + value
self._incidence = self._twotheta - value
return value
@property
def transverse(self):
"""Access the chi angle (in degrees), transverse to Th-2Th plane"""
return self._transverse
@transverse.setter
def transverse(self, value):
# Does not check for valid value
self._transverse = value
return value
@property
def azimuth(self):
"""Access the sample stage's azimuth angle (in degrees), phi"""
return self._azimuth
@azimuth.setter
def azimuth(self, value):
# Does not check for valid value
self._azimuth = value
return value
@property
def energy(self):
"""Access the energy (in eV) of the Photon"""
return self.photon.energy
@energy.setter
def energy(self, value):
# Does not check for valid value
self.photon.energy = value
return value
@property
def wavelength(self):
"""Access the wavelength (in nm) of the Photon"""
return self.photon.wavelength
@wavelength.setter
def wavelength(self, value):
# Does not check for valid value
self.photon.wavelength = value
return value
@property
def q_xray(self):
"""Access the momentum (in 1/nm) of the Photon"""
return self.photon.q
@q_xray.setter
def q_xray(self, value):
# Does not check for valid value
self.photon.q = value
return value
@property
def polarization(self):
"""Access the Polarization object for the x-ray polarization"""
return self._polarization
@polarization.setter
def polarization(self, new_polarization):
# Does not check for valid value
if not isinstance(new_polarization, Polarization):
new_polarization = Polarization(new_polarization)
self._polarization = new_polarization
return new_polarization
@property
def intensity0(self):
"""Access the incoming flux, I0, (in arb. units)"""
return self._intensity0
@intensity0.setter
def intensity0(self, value):
# Does not check for valid value
self._intensity0 = value
return value
@property
def temperature(self):
"""Access the temperature of the sample stage (in Kelvin)"""
return self._temperature
@temperature.setter
def temperature(self, value):
# Does not check for valid value
self._temperature = value
return value
@property
def exposure_time(self):
"""Access the exposure (acquisition) time (in seconds)"""
return self._exposure_time
@exposure_time.setter
def exposure_time(self, value):
# Does not check for valid value
self._exposure_time = value
return value
@property
def distance(self):
"""Access the distance between sample and detector (in mm)"""
return self._distance
@distance.setter
def distance(self, value):
# Does not check for valid value
self._distance = value
return value
class Diffractometer402( Diffractometer ):
"""Diffractometer402:
Settings and readings and scattering chamber at ALS beamline 4.0.2
"""
# Detector modes
CCD, DIODE_WIDE, DIODE_HIRES = list( range(3) )
DIODE = DIODE_WIDE
# Defaults
_DISTANCE_SAMPLE_TO_CCD = 1.125 * 142.265 # mm
_DISTANCE_SAMPLE_TO_DIODE = 7.75 * 25.4 # mm
default_chamber_params = dict({
'Beamline Energy': 700.,
'EPU Polarization': 0.,
'Bottom Rotary Seal': 15.,
'Top Rotary Seal': 137.5,
'Flip': -2.,
'Flip Offset': -1.,
'Top Offset': 2.,
# 'Twice Top Offset': 4.,
# 'CCD Offset': -18.68,
# 'DATE': "2015-02-12",
'DATE': date.today().strftime("%Y-%m-%d"),
# 'Counter 2': 340*1000.,
'I0 BL': 340*1000.,
'TempCtrlrA': 296.,
'TempCtrlrB': 295.,
})
def __init__(self, param_dict=None, *arguments, **keywords):
"""Initialize the Diffractometer using dictionary value pairs,
or provide default values as needed
"""
# Do NOT call __init__() for base class
# Diffractometer.__init__(self, **keywords)
if param_dict is None:
# Need to provide some default values
param_dict = self.default_chamber_params
# return
value = param_dict.get('DATE', None)
if value is not None:
if 'T' in value:
image_date = datetime.strptime(value,
"%Y-%m-%dT%H:%M:%S").date()
else:
image_date = datetime.strptime(value, "%Y-%m-%d").date()
else:
value = param_dict.get('DATETIME', None)
if value is not None:
image_date = datetime.strptime(value,
"%Y/%m/%d %H:%M:%S").date()
else:
image_date = datetime.strptime("1970-01-01", "%Y-%m-%d").date()
# print image_date
value = param_dict.get('EXPOSURE', None)
if value is not None:
self.exposure_time = value
value = param_dict.get('Detector Mode', None)
if value is not None:
self.detector = value
else:
value = keywords.get('detector', None)
if value is not None:
self.detector = value
else:
self.detector = Diffractometer402.CCD
value = param_dict.get('Beamline Energy', None)
if value is not None:
energy = value
value = param_dict.get('EPU Polarization', None)
if value is not None:
polarization = value
value = param_dict.get('Bottom Rotary Seal', None)
if value is not None:
# self.bottom_angle = value
bottom_angle = value
value = param_dict.get('Top Rotary Seal', None)
if value is not None:
# self.top_angle = value
top_angle = value
value = param_dict.get('Flip', None)
if value is not None:
# self.flip_angle = value
flip_angle = value
else:
value = param_dict.get('Flip Position', None)
if value is not None:
# self.flip_angle = value
flip_angle = value
value = param_dict.get('Flip Offset', None)
# if value is not None:
# self.offset_flip = value
# else:
# self.offset_flip = 0.0
# # Potentially a useful default value
# # self.offset_flip = -2.0
if value is not None:
self._offset_flip = value
else:
self._offset_flip = 0.0
# Potentially a useful default value
# self._offset_flip = -2.0
value = param_dict.get('Top Offset', None)
# if value is not None:
# self.offset_top = value
# else:
# value = keywords.get('Twice Top Offset', None)
# if value is not None:
# self.offset_top = (value / 2)
# else:
# self.offset_top = 0.0
# # Potentially a useful default value
# # self.offset_top = 2.0
if value is not None:
self._offset_top = value
else:
value = keywords.get('Twice Top Offset', None)
if value is not None:
self._offset_top = (value / 2.0)
else:
self._offset_top = 0.0
# Potentially a useful default value
# self._offset_top = 2.0
value = param_dict.get('CCD Offset', None)
if value is not None:
self.offset_CCD = value
else:
# # value = keywords.get('DATE', None)
# value = param_dict.get('DATE', None)
# image_date = datetime.strptime(value, "%Y-%m-%d").date()
if image_date > date(2014, 6, 1):
self.offset_CCD = -18.68 # degrees
else:
self.offset_CCD = -18.63 # degrees
value = param_dict.get('Hi-Res Offset', None)
if value is not None:
self.offset_diode_hires = value
else:
# # value = keywords.get('DATE', None)
# value = param_dict.get('DATE', None)
# image_date = datetime.strptime(value, "%Y-%m-%d").date()
if image_date > date(2014, 6, 1):
self.offset_diode_hires = 5.976 # degrees
else:
self.offset_diode_hires = 0.00 # degrees
value = param_dict.get('Azimuth', None)
if value is not None:
self.azimuth = value
else:
value = keywords.get('azimuth', None)
if value is not None:
self.azimuth = value
else:
self.azimuth = 0.0
value = param_dict.get('I0 BL', None)
# print "CCD I0 BL:", value
if value is not None:
self.intensity0 = value
else:
value = param_dict.get('Counter 2', None)
# print "CCD Counter 2:", value
if value is not None:
self.intensity0 = value
# print "self.intensity0:", self.intensity0
for key_entry in ('TempCtrlrA', 'Temperature A',
'Lakeshore Temp Controller A',
'TempCtrlrB', 'Temperature B',
'Lakeshore Temp Controller B', ):
value = param_dict.get(key_entry, None)
if value is not None:
self.temperature = value
break
else:
self.temperature = 298 # Kelvin
# TO DO: Gracefully handle inconsistent derived motor values
# value = param_dict.get('CCD 2-Theta (Th-2Th)', None)
# if value is not None:
# self.twotheta = value
self.offset_diode_wide = 0.00 # degrees
self._distance_CCD = Diffractometer402._DISTANCE_SAMPLE_TO_CCD
self._distance_diode = Diffractometer402._DISTANCE_SAMPLE_TO_DIODE
# Allow for keyword to change the distance
if (self.detector == self.CCD):
self.offset_detector = self.offset_CCD
distance = self._distance_CCD
elif (self.detector == self.DIODE_WIDE):
self.offset_detector = self.offset_diode_wide
distance = self._distance_diode
elif (self.detector == self.DIODE_HIRES):
self.offset_detector = self.offset_diode_hires
distance = self._distance_diode
# twotheta = self.bottom_angle - self.offset_detector
# top_true = self.top_angle - self.offset_top
# incidence = self.bottom_angle - top_true
# transverse = self.flip_angle - self.offset_flip
twotheta = bottom_angle - self.offset_detector
top_true = top_angle - self.offset_top
incidence = bottom_angle - top_true
transverse = flip_angle - self.offset_flip
self.twotheta = twotheta
self.incidence = incidence
self.transverse = transverse
# self.azimuth = azimuth
self.photon = Photon(energy)
self.polarization = polarization
# self.intensity0 = intensity0
# self.temperature = temperature
self.distance = distance
# This appears to be resolved as of 2015-02-12 edit
# -------------------------------------------------
# NOTE: default values will always override param_dict values
# Need to either provide defaults or overrides
# NOTE: Base class initialization duplicated with overrides
# and also with param_dict values
# REQUIRES SIMPLIFICATION
@property
def twotheta(self):
"""Access the detector position, 2-Theta (in degrees)"""
return self._twotheta
@twotheta.setter
def twotheta(self, value):
# Does not check for valid value
self._twotheta = value
return value
@property
def incidence(self):
"""Access the 2D incidence angle (in degrees) within Th-2Th plane"""
return self._incidence
@incidence.setter
def incidence(self, value):
# Does not check for valid value
self._incidence = value
return value
@property
def exit_angle(self):
"""Access the 2D exit angle (in degrees) within Th-2Th plane"""
return (self._twotheta - self._incidence)
@exit_angle.setter
def exit_angle(self, value):
# Does not check for valid value
# self._twotheta = self._incidence + value
self._incidence = self._twotheta - value
return value
@property
def transverse(self):
"""Access the chi angle (in degrees), transverse to Th-2Th plane"""
return self._transverse
@transverse.setter
def transverse(self, value):
# Does not check for valid value
self._transverse = value
return value
@property
def azimuth(self):
"""Access the sample stage's azimuth angle (in degrees), phi"""
return self._azimuth
@azimuth.setter
def azimuth(self, value):
# Does not check for valid value
self._azimuth = value
return value
@property
def bottom_angle(self):
"""Access the stored value of the Bottom Rotary Seal MOTOR"""
return (self.twotheta + self.offset_detector)
@property
def top_angle(self):
"""Access the stored value of the Top Rotary Seal MOTOR"""
return ( (self.bottom_angle - self.incidence) + self.offset_top)
@property
def flip_angle(self):
"""Access the stored value of the Flip MOTOR"""
return (self.transverse + self.offset_flip)
@property
def offset_top(self):
"""Access the stored offset of the Top Rotary Seal MOTOR"""
return self._offset_top
@property
def offset_flip(self):
"""Access the stored offset of the Flip MOTOR"""
return self._offset_flip
def offset(self, motor, value = 0, motor_unchanged = True):
"""Update the stored offset of a MOTOR
motor: name (or short name) of motor to which offset is applied
value: new value of the motor's offset
motor_unchanged: True = motor value unchanged, update ideal angle
False = ideal angle unchanged, update motor value
"""
twotheta = self.twotheta
incidence = self.incidence
transverse = self.transverse
bottom_angle = self.bottom_angle
top_angle = self.top_angle
flip_angle = self.flip_angle
motor = motor.lower()
if (
(motor == "top") or
(motor == "top motor") or
(motor == "top seal") or
(motor == "top rotary seal")
):
self._offset_top = value
if (motor_unchanged == False):
# top_true = self._bottom_angle - self.incidence
# self._top_angle = top_true + self.offset_top
pass
else:
# top_true = self._top_angle - self.offset_top
# self.incidence = self._bottom_angle - top_true
top_true = top_angle - self.offset_top
self.incidence = bottom_angle - top_true
elif (
(motor == "flip") or
(motor == "flip motor")
):
self._offset_flip = value
if (motor_unchanged == False):
# self._flip_angle = self.transverse + self.offset_flip
pass
else:
# self.transverse = self._flip_angle - self.offset_flip
self.transverse = flip_angle - self.offset_flip
else:
# print "Bad motor name supplied to offset() method"
pass
def offset_step(self, motor, value_step = 0, motor_unchanged = True):
"""Update the stored offset of a MOTOR, relative to existing offset
motor: name (or short name) of motor to which offset is applied
value_step: add to existing value of the motor's offset
motor_unchanged: True = motor value unchanged, update ideal angle
False = ideal angle unchanged, update motor value
"""
twotheta = self.twotheta
incidence = self.incidence
transverse = self.transverse
bottom_angle = self.bottom_angle
top_angle = self.top_angle
flip_angle = self.flip_angle
offset_top = self.offset_top
offset_flip = self.offset_flip
motor = motor.lower()
if (
(motor == "top") or
(motor == "top motor") or
(motor == "top seal") or
(motor == "top rotary seal")
):
self._offset_top += value_step
if (motor_unchanged == False):
# top_true = self._bottom_angle - self.incidence
# self._top_angle = top_true + self.offset_top
pass
else:
# top_true = self._top_angle - self.offset_top
# self.incidence = self._bottom_angle - top_true
top_true = top_angle - self.offset_top
self.incidence = bottom_angle - top_true
elif (
(motor == "flip") or
(motor == "flip motor")
):
self._offset_flip += value_step
if (motor_unchanged == False):
# self._flip_angle = self.transverse + self.offset_flip
pass
else:
# self.transverse = self._flip_angle - self.offset_flip
self.transverse = flip_angle - self.offset_flip
# Hack for allowing ideal angles (not just MOTORs)
elif (
(motor == "incidence") # Ideal angle, not MOTOR
):
self._offset_top -= value_step # opposite rotation sense
if (motor_unchanged == False):
# top_true = self._bottom_angle - self.incidence
# self._top_angle = top_true + self.offset_top
pass
else:
# top_true = self._top_angle - self.offset_top
# self.incidence = self._bottom_angle - top_true
top_true = top_angle - self.offset_top
self.incidence = bottom_angle - top_true
elif (
(motor == "transverse") # Ideal angle, not MOTOR
):
self._offset_flip += value_step # same rotation sense
if (motor_unchanged == False):
# self._flip_angle = self.transverse + self.offset_flip
pass
else:
# self.transverse = self._flip_angle - self.offset_flip
self.transverse = flip_angle - self.offset_flip
else:
# print "Bad motor name supplied to offset() method"
pass
def valid_motors(self):
"""Checks whether motor values are valid
returns: True if motor values are valid; False otherwise
"""
if ( (self.detector == self.CCD) and (self.bottom_angle < 6.0) ):
return False
if (self.bottom_angle > 144.0):
return False
if (self.flip_angle < -5.0) or (self.flip_angle > 95.0):
return False
return True
class CcdImage(object):
"""CcdImage: wrapper for 2D data + header info from diffractometer image"""
def __init__(self):
pass
class CcdImageFromFITS( CcdImage ):
"""CCDImageFromFITS: CCDImage created from FITS file"""
def __init__(self, filename_or_hdulist, offset_diffractometer=None):
"""Extract relevant data from FITS file"""
if isinstance(filename_or_hdulist, fits.HDUList):
self.hdulist = filename_or_hdulist
else:
# check for errors during load
self.hdulist = fits.open(filename_or_hdulist)
self._build_diffractometer(offset_diffractometer)
def _build_diffractometer(self, offset_diffractometer=None):
"""Extract relevant diffractometer data"""
self._repair_fits_header()
self._diffractometer = Diffractometer402(
param_dict = self.hdulist[0].header)
if offset_diffractometer:
self._diffractometer.azimuth += offset_diffractometer.azimuth
self._diffractometer.transverse += offset_diffractometer.transverse
self._diffractometer.incidence += offset_diffractometer.incidence
# Assumes last image has intensity map
self.data = self.hdulist[-1].data
def _repair_fits_header(self):
"""Repair AI channels in diffractometer data"""
primary_hdu = self.hdulist[0].header
# z_motor = primary_hdu['Z']
z_ai = primary_hdu['Z Position']
z_motor = primary_hdu.get('Z', z_ai)
ai_ratio = int(z_ai / z_motor)
# for key in primary_hdu.keys(): # list() needed for Python3 ?
for key in list(primary_hdu.keys()):
if primary_hdu.comments[key] == "Analog Input":
# print key, ":", primary_hdu[key]
primary_hdu[key] /= ai_ratio
# print key, ":", primary_hdu[key]
@property
def intensity0(self):
"""Access the incoming flux, I0, (in arb. units)"""
return self._diffractometer.intensity0
@property
def exposure_time(self):
"""Access the exposure (acquisition) time (in seconds)"""
return self._diffractometer.exposure_time
def qimage(self, output_filename=None):
"""Convert pixel map of intensities into reciprocal space map"""
diffr = self._diffractometer
image_array = self.data
# wavelength = diffr.photon.wavelength
twotheta = diffr.twotheta
incidence = diffr.incidence
chi = diffr.transverse
phi = diffr.azimuth
distance_ccd = diffr.distance
ccd_center_room = [cos(deg2rad(twotheta)), 0 , sin(deg2rad(twotheta))]
ccd_center_room = array(ccd_center_room)
ccd_center_room *= distance_ccd
ccd_tilt_axisY = 0
ccd_unit_normal = [cos(deg2rad(twotheta)) - ccd_tilt_axisY,
0 ,
sin(deg2rad(twotheta)) - ccd_tilt_axisY]
ccd_unit_Y = [0, 1, 0]
ccd_unit_X = cross(ccd_unit_normal, ccd_unit_Y)
raw_pixel_size = 0.0135 # size in mm
raw_pixel_num = 2048 # pixels / side
num_cols, num_rows = image_array.shape
ccd_pixel_size_X = raw_pixel_size * raw_pixel_num / num_cols
ccd_pixel_size_Y = raw_pixel_size * raw_pixel_num / num_rows
ccd_center_pixel_X = float(num_cols - 1) / 2 # Assumes 0...numCols-1
ccd_center_pixel_Y = num_rows * (float(1024 - 460) / 1024) - 1; # Assumes 0...numRows-1
ccd_pixel_Y = linspace(
(0 - ccd_center_pixel_Y) * ccd_pixel_size_Y,
(num_rows - ccd_center_pixel_Y) * ccd_pixel_size_Y,
num=num_rows,
endpoint=False
)
ccd_pixel_X = linspace(
(0 - ccd_center_pixel_X) * ccd_pixel_size_X,
(num_cols - ccd_center_pixel_X) * ccd_pixel_size_X,
num=num_cols,
endpoint=False
)
# print ccd_pixel_Y, ccd_pixel_X
ccd_pixel_room_rows = outer(ccd_pixel_Y, ccd_unit_Y)
ccd_pixel_room_cols = outer(ccd_pixel_X, ccd_unit_X)
ccd_pixel_room = (ccd_pixel_room_rows[:, None] + ccd_pixel_room_cols +
ccd_center_room)
# print "ccd_pixel_room.shape", ccd_pixel_room.shape
ccd_pixel_room = ccd_pixel_room.reshape(num_rows * num_cols, 3)
# print "ccd_pixel_room.shape (flattened)", ccd_pixel_room.shape
ccd_pixel_intensities = array(image_array)
ccd_pixel_intensities = ccd_pixel_intensities.reshape(
num_rows * num_cols)
ccd_pixel_room_norms = norm(ccd_pixel_room, axis=-1)
ccd_pixel_room_norm = ccd_pixel_room / ccd_pixel_room_norms[:, newaxis]
# print "ccd_pixel_room_norm.shape", ccd_pixel_room_norm.shape
q_xray = diffr.photon.q # 2*pi / d [1 / nm]
qxyz_sphere = ccd_pixel_room_norm * q_xray + array([-q_xray, 0, 0])
incidence_rad = deg2rad(incidence)
chi_rad = deg2rad(chi)
phi_rad = deg2rad(phi)
incidence_rotation = array([
[ cos(incidence_rad), 0, -sin(incidence_rad) ],
[ 0, 1, 0 ],
[ sin(incidence_rad), 0, cos(incidence_rad) ] ])
chi_rotation = array([
[ 1, 0, 0 ],
[ 0, cos(chi_rad), -sin(chi_rad) ],
[ 0, sin(chi_rad), cos(chi_rad) ] ])
# need to verify sign of sin() components
# this version (mathematica, not IGOR) appears correct
phi_rotation = array([
[ cos(phi_rad), -sin(phi_rad), 0 ],
[ sin(phi_rad), cos(phi_rad), 0 ],
[ 0, 0, 1 ] ])
qxyz_normal = array([
[1, 0, 0],
[0, 1, 0],
[0, 0, 1] ])
# lattice_a = 42 #nm
# lattice_spacing = array([lattice_a, lattice_a, 1])
# q_lattice_spacing = 2 * pi / lattice_spacing
q_lattice_spacing = array([1, 1, 1])
qxyz_rotated = incidence_rotation.dot(
chi_rotation).dot(phi_rotation).dot(qxyz_normal)
# print "qxyz_rotated.shape", qxyz_rotated.shape
# Rotation leaves new unit vectors as columns
# Divide 1st COLUMN by 2*pi/a
# Divide 2nd COLUMN by 2*pi/b
# Divide 3rd COLUMN by 2*pi/c
hkl_rotated = qxyz_rotated / q_lattice_spacing[newaxis, :]
# print "hkl_rotated.shape", hkl_rotated.shape
hkl_map = qxyz_sphere.dot(hkl_rotated)
# print "hkl_map.shape", hkl_map.shape
ccd_int_qxqy = array([
hkl_map[:, 0],
hkl_map[:,1],
ccd_pixel_intensities])
# print "ccd_int_qxqy.shape", ccd_int_qxqy.shape
ccd_int_qxqy = ccd_int_qxqy.transpose()
# print "ccd_int_qxqy.shape (transposed)", ccd_int_qxqy.shape
return( ccd_int_qxqy )
def qvalues_df(self, output_filename=None):
"""Convert pixel map of intensities into reciprocal space map
*) output_filename: optional storage location of calculated data
*) Returns PANDAS dataframe containing:
row, col, Qx, Qy, Qz, intensity (in "Counts")
"""
diffr = self._diffractometer
image_array = self.data
# wavelength = diffr.photon.wavelength
twotheta = diffr.twotheta
incidence = diffr.incidence
chi = diffr.transverse
phi = diffr.azimuth
distance_ccd = diffr.distance
ccd_center_room = [cos(deg2rad(twotheta)), 0 , sin(deg2rad(twotheta))]
ccd_center_room = array(ccd_center_room)
ccd_center_room *= distance_ccd
ccd_tilt_axisY = 0
ccd_unit_normal = [cos(deg2rad(twotheta)) - ccd_tilt_axisY,
0 ,
sin(deg2rad(twotheta)) - ccd_tilt_axisY]
ccd_unit_Y = [0, 1, 0]
ccd_unit_X = cross(ccd_unit_normal, ccd_unit_Y)
raw_pixel_size = 0.0135 # size in mm
raw_pixel_num = 2048 # pixels / side
num_cols, num_rows = image_array.shape
ccd_pixel_size_X = raw_pixel_size * raw_pixel_num / num_cols
ccd_pixel_size_Y = raw_pixel_size * raw_pixel_num / num_rows
ccd_center_pixel_X = float(num_cols - 1) / 2 # Assumes 0...numCols-1
ccd_center_pixel_Y = num_rows * (float(1024 - 460) / 1024) - 1; # Assumes 0...numRows-1
ccd_pixel_Y = linspace(
(0 - ccd_center_pixel_Y) * ccd_pixel_size_Y,
(num_rows - ccd_center_pixel_Y) * ccd_pixel_size_Y,
num=num_rows,
endpoint=False
)
ccd_pixel_X = linspace(
(0 - ccd_center_pixel_X) * ccd_pixel_size_X,
(num_cols - ccd_center_pixel_X) * ccd_pixel_size_X,
num=num_cols,
endpoint=False
)
# print ccd_pixel_Y, ccd_pixel_X
ccd_pixel_room_rows = outer(ccd_pixel_Y, ccd_unit_Y)
ccd_pixel_room_cols = outer(ccd_pixel_X, ccd_unit_X)
ccd_pixel_room = (ccd_pixel_room_rows[:, None] + ccd_pixel_room_cols +
ccd_center_room)
# print "ccd_pixel_room.shape", ccd_pixel_room.shape
ccd_pixel_room = ccd_pixel_room.reshape(num_rows * num_cols, 3)
# print "ccd_pixel_room.shape (flattened)", ccd_pixel_room.shape
ccd_pixel_intensities = array(image_array)
ccd_pixel_intensities = ccd_pixel_intensities.reshape(
num_rows * num_cols)
ccd_pixel_room_norms = norm(ccd_pixel_room, axis=-1)
ccd_pixel_room_norm = ccd_pixel_room / ccd_pixel_room_norms[:, newaxis]
# print "ccd_pixel_room_norm.shape", ccd_pixel_room_norm.shape
q_xray = diffr.photon.q # 2*pi / d [1 / nm]
qxyz_sphere = ccd_pixel_room_norm * q_xray + array([-q_xray, 0, 0])
incidence_rad = deg2rad(incidence)
chi_rad = deg2rad(chi)
phi_rad = deg2rad(phi)
incidence_rotation = array([
[ cos(incidence_rad), 0, -sin(incidence_rad) ],
[ 0, 1, 0 ],
[ sin(incidence_rad), 0, cos(incidence_rad) ] ])
chi_rotation = array([
[ 1, 0, 0 ],
[ 0, cos(chi_rad), -sin(chi_rad) ],
[ 0, sin(chi_rad), cos(chi_rad) ] ])
# need to verify sign of sin() components
# this version (mathematica, not IGOR) appears correct
phi_rotation = array([
[ cos(phi_rad), -sin(phi_rad), 0 ],
[ sin(phi_rad), cos(phi_rad), 0 ],
[ 0, 0, 1 ] ])
qxyz_normal = array([
[1, 0, 0],
[0, 1, 0],
[0, 0, 1] ])
# lattice_a = 42 #nm
# lattice_spacing = array([lattice_a, lattice_a, 1])
# q_lattice_spacing = 2 * pi / lattice_spacing
q_lattice_spacing = array([1, 1, 1])
qxyz_rotated = incidence_rotation.dot(
chi_rotation).dot(phi_rotation).dot(qxyz_normal)
# print "qxyz_rotated.shape", qxyz_rotated.shape
# Rotation leaves new unit vectors as columns
# Divide 1st COLUMN by 2*pi/a
# Divide 2nd COLUMN by 2*pi/b
# Divide 3rd COLUMN by 2*pi/c
hkl_rotated = qxyz_rotated / q_lattice_spacing[newaxis, :]
# print "hkl_rotated.shape", hkl_rotated.shape
hkl_map = qxyz_sphere.dot(hkl_rotated)
# print "hkl_map.shape", hkl_map.shape
ccd_int_qxqy = array([
hkl_map[:, 0],
hkl_map[:,1],
ccd_pixel_intensities])
# print "ccd_int_qxqy.shape", ccd_int_qxqy.shape
ccd_int_qxqy = ccd_int_qxqy.transpose()
# print "ccd_int_qxqy.shape (transposed)", ccd_int_qxqy.shape
row_values = tile(arange(num_rows), (num_cols, 1)).transpose()
row_values = row_values.reshape(num_rows * num_cols)
col_values = tile(arange(num_cols), (num_rows, 1))
col_values = col_values.reshape(num_rows * num_cols)
df_qxyz = pd.DataFrame({
"row": row_values,
"col": col_values,
"Qx": hkl_map[:, 0],
"Qy": hkl_map[:, 1],
"Qz": hkl_map[:, 2],
"Counts": ccd_pixel_intensities,
})
return( df_qxyz )
class QSpacePath(object):
"""QSpacePath: Build a path through reciprocal (Q) space
*) Export scan files for use at ALS BL 4.0.2 Diffractometer
*) Qx is along relative azimuth = 0 deg (uses offset)
*) Qy is along relative azimuth = 90 deg (uses offset)
*) Qz is along azimuthal axis
*) Q1 is along absolute azimuth = 0 deg (offset = 0)
*) Q2 is along absolute azimuth = 90 deg (offset = 0)
*) Q3 is along azimuthal axis
"""
def __init__(self,
diffractometer=None,
offset_diffractometer=None):
"""Attach a diffractometer for generating QSpacePath angles
diffractometer: diffractometer object to generate angles, motions
offset_diffractometer: Extract relevant angle offsets
for calibrating exported scans
"""
self._build_diffractometer(diffractometer, offset_diffractometer)
self.angles = None
def _build_diffractometer(self,
diffractometer=None,
offset_diffractometer=None):
"""Attach a diffractometer for generating QSpacePath angles
diffractometer: diffractometer object to generate angles, motions
offset_diffractometer: Extract relevant angle offsets
for calibrating exported scans
"""
if diffractometer:
self._diffractometer = diffractometer
else:
self._diffractometer = Diffractometer402()
if offset_diffractometer:
self._diffractometer.azimuth += offset_diffractometer.azimuth
# Move sample, rather than move model...opposite offsets
self._diffractometer.offset_step(
"transverse", -offset_diffractometer.transverse)
self._diffractometer.offset_step(
"incidence", -offset_diffractometer.incidence)
def lin_path(self,
q_start = array([0, 0, 0]),
q_stop = array([0, 0, 1]),
num_samples = 31 ):
"""Define a linear path through reciprocal (Q) space
q_start: starting point, array([Qx, Qy, Qz]) in 1/nm
q_stop: ending point, array([Qx, Qy, Qz]) in 1/nm
num_samples: number of points (M) to generate
set(self.angles):
M x 3 array([2theta, incidence, transverse (chi)])
"""
qx_values = linspace(q_start[0], q_stop[0], num_samples, endpoint=True)
qy_values = linspace(q_start[1], q_stop[1], num_samples, endpoint=True)
qz_values = linspace(q_start[2], q_stop[2], num_samples, endpoint=True)
q_values = array([qx_values, qy_values, qz_values]).transpose()
self.angles = self.q2angles(q_values)
def q2angles(self, q_points = None):
"""Convert an array of reciprocal (Q) space points into ideal angles
*** Assumes that ideal angles are calculated at fixed azimuth
q_points: M x 3 array([Qx, Qy, Qz]) in 1/nm
returns: M x 3 array([2theta, incidence, transverse (chi)])
"""
if q_points is None:
q_points = array([
[0, 0, 0],
])
diffr = self._diffractometer
# incidence_rad = deg2rad(incidence)
# transverse_rad = deg2rad(transverse)
# azimuth_rad = deg2rad(azimuth)
azimuth_rad = deg2rad(diffr.azimuth)
# incidence_rotation = array([
# [ cos(incidence_rad), 0, -sin(incidence_rad) ],
# [ 0, 1, 0 ],
# [ sin(incidence_rad), 0, cos(incidence_rad) ] ])
#
# transverse_rotation = array([
# [ 1, 0, 0 ],
# [ 0, cos(transverse_rad), -sin(transverse_rad) ],
# [ 0, sin(transverse_rad), cos(transverse_rad) ] ])
azimuth_rotation = array([
[ cos(azimuth_rad), -sin(azimuth_rad), 0 ],
[ sin(azimuth_rad), cos(azimuth_rad), 0 ],
[ 0, 0, 1 ] ])
# Convert q_points (relative to sample azimuth = 0)
# into q123_points (has no azimuthal motion)
q123_points = [azimuth_rotation.dot(q_point) for q_point in q_points]
# transverse_solutions = [
# brentq(f, -pi, pi, args=(q123_point)
# ) for q123_point in q123_points]
# transverse, chi = arctan(Q2 / Q3)
transverse_angles = rad2deg([(
arctan2(q123_point[1], q123_point[2])
) for q123_point in q123_points])
# incidence_offset = arctan(Q1 / norm(Q2,Q3) )
incidence_angles = rad2deg([(
arctan2(q123_point[0], norm(q123_point[1:]) )
) for q123_point in q123_points])
# theta = arcsin(Q / 2*Q_xray)
theta_angles = rad2deg([(
arcsin(norm(q123_point) / (2 * diffr.photon.q) )
) for q123_point in q123_points])
twoTheta_angles = 2 * theta_angles
incidence_angles += theta_angles
angles = array([
twoTheta_angles,
incidence_angles,
transverse_angles,
]).transpose()
return angles
def is_accessible(self, twotheta, incidence, transverse, tolerance=2.0):
"""True if reciprocal space is accessible at angles requested
tolerance = tolerable incidence (in deg.) beyond accessible limits
"""
if ( isnan(twotheta) or isnan(incidence) or isnan(transverse) ):
return False
if (-tolerance < incidence) and (incidence < (twotheta + tolerance) ):
return True
else:
return False
def angles2motors(self, twotheta, incidence, transverse,
validate_motors = True):
"""Convert ideal angles into ALS BL 4.0.2 motor values"""
if ( isnan(twotheta) or isnan(incidence) or isnan(transverse) ):
return (nan, nan, nan)
diffr = self._diffractometer
diffr.twotheta = twotheta
diffr.incidence = incidence
diffr.transverse = transverse
if (validate_motors == True) and not diffr.valid_motors():
return (nan, nan, nan)
return (diffr.bottom_angle, diffr.top_angle, diffr.flip_angle)
def export_scanfile(self,
output_filename="test.scn",
export_energy = True,
export_polarization = True):
"""Convert reciprocal space path into a scan file (ALS BL 4.0.2)"""
diffr = self._diffractometer
accessible_angles = self.angles[
array([
array(self.is_accessible(*tuple(angle_values) )
) for angle_values in self.angles])
]
motors = array([
array(self.angles2motors(*tuple(angle_values) )
) for angle_values in accessible_angles])
# Remove 'nan' rows
motors = motors[isfinite(motors).all(axis=1)]
col_names = []
col_values = []
if (export_energy == True):
energies = full(len(motors), diffr.energy)
col_names += ["Mono Energy", "EPU Energy"]
col_values += [energies, energies]
if (export_polarization == True):
polarizations = full(len(motors), diffr.polarization.value)
col_names += ["EPU Polarization"]
col_values += [polarizations]
col_names += ["Bottom Rotary Seal", "Top Rotary Seal", "Flip"]
if len(col_values) > 0:
motor_values = hstack((
array(col_values).transpose(),
around(motors, 3),
))
else:
motor_values = around(motors, 3)
scan_motors = pd.DataFrame(motor_values, columns=col_names)
scan_motors.to_csv(
output_filename,
sep = '\t',
index = False,
line_terminator = "\r\n",
)
class ResonanceProfile(object):
"""ResonanceProfile: Photon energy changes at constant reciprocal point, Q
*) Export scan files for use at ALS BL 4.0.2 Diffractometer
*) Qx is along relative azimuth = 0 deg (uses offset)
*) Qy is along relative azimuth = 90 deg (uses offset)
*) Qz is along azimuthal axis
*) Q1 is along absolute azimuth = 0 deg (offset = 0)
*) Q2 is along absolute azimuth = 90 deg (offset = 0)
*) Q3 is along azimuthal axis
"""
def __init__(self,
q_value,
diffractometer=None,
offset_diffractometer=None):
"""Constant reciprocal point, Q = (Qx, Qy, Qz)
Attach a diffractometer for generating ResonanceProfile angles
q_value: reciprocal point, array([Qx, Qy, Qz]) in 1/nm
diffractometer: diffractometer object to generate angles, motions
offset_diffractometer: Extract relevant angle offsets
for calibrating exported scans
"""
if len(q_value) == 3:
self.q = array([ q_value ])
else:
self.q = array([ [0, 0, 1] ]) # Better default available?
self._build_diffractometer(diffractometer, offset_diffractometer)
self.angles = None
def _build_diffractometer(self,
diffractometer=None,
offset_diffractometer=None):
"""Attach a diffractometer for generating QSpacePath angles
diffractometer: diffractometer object to generate angles, motions
offset_diffractometer: Extract relevant angle offsets
for calibrating exported scans
"""
if diffractometer:
self._diffractometer = diffractometer
else:
self._diffractometer = Diffractometer402()
if offset_diffractometer:
self._diffractometer.azimuth += offset_diffractometer.azimuth
# Move sample, rather than move model...opposite offsets
self._diffractometer.offset_step(
"transverse", -offset_diffractometer.transverse)
self._diffractometer.offset_step(
"incidence", -offset_diffractometer.incidence)
def new_spectrum(self,
energies):
"""Define a resonance profile for the energies
energies: array() of energy values defining the resonance profile
set(self.angles):
M x 3 array([2theta, incidence, transverse (chi)])
Adjusted for each energy to maintain constant Q
"""
self.energies = energies
diffr = self._diffractometer
q_value = self.q
angles = []
for energy in energies:
diffr.energy = energy
angles.append( self.q2angles(q_value)[0] )
self.angles = array(angles)
def q2angles(self, q_points = None):
"""Convert an array of reciprocal (Q) space points into ideal angles
*** Assumes that ideal angles are calculated at fixed azimuth
q_points: M x 3 array([Qx, Qy, Qz]) in 1/nm
returns: M x 3 array([2theta, incidence, transverse (chi)])
"""
if q_points is None:
q_points = array([
[0, 0, 0],
])
diffr = self._diffractometer
# incidence_rad = deg2rad(incidence)
# transverse_rad = deg2rad(transverse)
# azimuth_rad = deg2rad(azimuth)
azimuth_rad = deg2rad(diffr.azimuth)
# incidence_rotation = array([
# [ cos(incidence_rad), 0, -sin(incidence_rad) ],
# [ 0, 1, 0 ],
# [ sin(incidence_rad), 0, cos(incidence_rad) ] ])
#
# transverse_rotation = array([
# [ 1, 0, 0 ],
# [ 0, cos(transverse_rad), -sin(transverse_rad) ],
# [ 0, sin(transverse_rad), cos(transverse_rad) ] ])
azimuth_rotation = array([
[ cos(azimuth_rad), -sin(azimuth_rad), 0 ],
[ sin(azimuth_rad), cos(azimuth_rad), 0 ],
[ 0, 0, 1 ] ])
# Convert q_points (relative to sample azimuth = 0)
# into q123_points (has no azimuthal motion)
q123_points = [azimuth_rotation.dot(q_point) for q_point in q_points]
# transverse_solutions = [
# brentq(f, -pi, pi, args=(q123_point)
# ) for q123_point in q123_points]
# transverse, chi = arctan(Q2 / Q3)
transverse_angles = rad2deg([(
arctan2(q123_point[1], q123_point[2])
) for q123_point in q123_points])
# incidence_offset = arctan(Q1 / norm(Q2,Q3) )
incidence_angles = rad2deg([(
arctan2(q123_point[0], norm(q123_point[1:]) )
) for q123_point in q123_points])
# theta = arcsin(Q / 2*Q_xray)
theta_angles = rad2deg([(
arcsin(norm(q123_point) / (2 * diffr.photon.q) )
) for q123_point in q123_points])
twoTheta_angles = 2 * theta_angles
incidence_angles += theta_angles
angles = array([
twoTheta_angles,
incidence_angles,
transverse_angles,
]).transpose()
return angles
def is_accessible(self, twotheta, incidence, transverse, tolerance=2.0):
"""True if reciprocal space is accessible at angles requested
tolerance = tolerable incidence (in deg.) beyond accessible limits
"""
if ( isnan(twotheta) or isnan(incidence) or isnan(transverse) ):
return False
if (-tolerance < incidence) and (incidence < (twotheta + tolerance) ):
return True
else:
return False
def angles2motors(self, twotheta, incidence, transverse,
validate_motors = True):
"""Convert ideal angles into ALS BL 4.0.2 motor values"""
if ( isnan(twotheta) or isnan(incidence) or isnan(transverse) ):
return (nan, nan, nan)
diffr = self._diffractometer
diffr.twotheta = twotheta
diffr.incidence = incidence
diffr.transverse = transverse
if (validate_motors == True) and not diffr.valid_motors():
return (nan, nan, nan)
return (diffr.bottom_angle, diffr.top_angle, diffr.flip_angle)
def export_scanfile(self,
output_filename="test.scn",
polarizations = None,
alternate_polarization = False,
export_polarization = True):
"""Convert resonance profile into a scan file (ALS BL 4.0.2)"""
diffr = self._diffractometer
accessible_angles = self.angles[
array([
array(self.is_accessible(*tuple(angle_values) )
) for angle_values in self.angles])
]
motors = array([
array(self.angles2motors(*tuple(angle_values) )
) for angle_values in accessible_angles])
# Remove 'nan' rows
motors = motors[isfinite(motors).all(axis=1)]
polarization_values = None
energy_list = []
pol_list = []
motor_list = []
if (export_polarization == True) and (polarizations is None):
polarization_values = full(len(motors), diffr.polarization.value)
if (polarizations is not None) and (alternate_polarization == False):
for polarization in polarizations:
energy_list.append(self.energies)
pol_list.append( full(len(motors), polarization) )
motor_list.append(motors)
energy_values = hstack(( tuple(energy_list) ))
polarization_values = hstack(( tuple(pol_list) ))
# motor_values = hstack(( tuple(motor_list) ))
motor_values = vstack(( tuple(motor_list) ))
if (polarizations is not None) and (alternate_polarization == True):
for (energy, motor_positions) in zip(self.energies, motors):
energy_list.append( full(len(polarizations), energy) )
pol_list.append(polarizations)
motor_list.append(
full(
(len(polarizations), len(motor_positions)),
motor_positions)
)
energy_values = hstack(( tuple(energy_list) ))
polarization_values = hstack(( tuple(pol_list) ))
# motor_values = hstack(( tuple(motor_list) ))
motor_values = vstack(( tuple(motor_list) ))
col_names = ["Mono Energy", "EPU Energy"]
col_values = [energy_values, energy_values]
if (polarization_values is not None):
col_names += ["EPU Polarization"]
col_values += [polarization_values]
col_names += ["Bottom Rotary Seal", "Top Rotary Seal", "Flip"]
motor_entries = hstack((
array(col_values).transpose(),
around(motor_values, 3),
))
scan_motors = pd.DataFrame(motor_entries, columns=col_names)
scan_motors.to_csv(
output_filename,
sep = '\t',
index = False,
line_terminator = "\r\n",
)
class DetectorPath(object):
"""DetectorPath: Build a path through detector angle space
*) Export scan files for use at ALS BL 4.0.2 Diffractometer
*) Qx is along relative azimuth = 0 deg (uses offset)
*) Qy is along relative azimuth = 90 deg (uses offset)
*) Qz is along azimuthal axis
*) Q1 is along absolute azimuth = 0 deg (offset = 0)
*) Q2 is along absolute azimuth = 90 deg (offset = 0)
*) Q3 is along azimuthal axis
"""
def __init__(self,
diffractometer=None,
offset_diffractometer=None):
"""Attach a diffractometer for generating DetectorPath angles
diffractometer: diffractometer object to generate angles, motions
offset_diffractometer: Extract relevant angle offsets
for calibrating exported scans
"""
self._build_diffractometer(diffractometer, offset_diffractometer)
self._incidence = self._diffractometer.incidence
self._transverse = self._diffractometer._transverse
self._azimuth = self._diffractometer._azimuth
self.angles = None
def _build_diffractometer(self,
diffractometer=None,
offset_diffractometer=None):
"""Attach a diffractometer for generating DetectorPath angles
diffractometer: diffractometer object to generate angles, motions
offset_diffractometer: Extract relevant angle offsets
for calibrating exported scans
"""
if diffractometer:
self._diffractometer = diffractometer
else:
self._diffractometer = Diffractometer402()
if offset_diffractometer:
self._diffractometer.azimuth += offset_diffractometer.azimuth
# Move sample, rather than move model...opposite offsets
self._diffractometer.offset_step(
"transverse", -offset_diffractometer.transverse)
self._diffractometer.offset_step(
"incidence", -offset_diffractometer.incidence)
@property
def incidence(self):
"""Access the 2D incidence angle (in degrees) within Th-2Th plane"""
return self._incidence
@incidence.setter
def incidence(self, value):
# Does not check for valid value
self._incidence = value
return value
@property
def transverse(self):
"""Access the chi angle (in degrees), transverse to Th-2Th plane"""
return self._transverse
@transverse.setter
def transverse(self, value):
# Does not check for valid value
self._transverse = value
return value
@property
def azimuth(self):
"""Access the sample stage's azimuth angle (in degrees), phi"""
return self._azimuth
@azimuth.setter
def azimuth(self, value):
# Does not check for valid value
self._azimuth = value
return value
def lin_path(self,
start = 0., # degrees
stop = None, # degrees
num_samples=1,
incidence = None, # degrees
transverse = None, # degrees
**kwargs):
"""Define a linear path through detector angle space
start: starting point, two_theta in degrees
stop: ending point, two_theta in degrees
num_samples: number of points (M) to generate
set(self.angles):
M x 3 array([2theta, incidence, transverse (chi)])
"""
stop = stop or start
if incidence is None:
incidence = self._diffractometer.incidence
self.incidence = incidence
if transverse is None:
transverse = self._diffractometer.transverse
self.transverse = transverse
twoTheta_angles = linspace(start, stop, num_samples, endpoint=True)
incidence_angle = self.incidence
transverse_angle = self.transverse
self.angles = array([[
twoTheta_angle,
incidence_angle,
transverse_angle,
] for twoTheta_angle in twoTheta_angles])
def is_accessible(self, twotheta, incidence, transverse, tolerance=2.0):
"""True if reciprocal space is accessible at angles requested
tolerance = tolerable incidence (in deg.) beyond accessible limits
"""
if ( isnan(twotheta) or isnan(incidence) or isnan(transverse) ):
return False
if (-tolerance < incidence) and (incidence < (twotheta + tolerance) ):
return True
else:
return False
def angles2motors(self, twotheta, incidence, transverse,
validate_motors = True):
"""Convert ideal angles into ALS BL 4.0.2 motor values"""
if ( isnan(twotheta) or isnan(incidence) or isnan(transverse) ):
return (nan, nan, nan)
diffr = self._diffractometer
diffr.twotheta = twotheta
diffr.incidence = incidence
diffr.transverse = transverse
if (validate_motors == True) and not diffr.valid_motors():
return (nan, nan, nan)
return (diffr.bottom_angle, diffr.top_angle, diffr.flip_angle)
def export_scanfile(self,
output_filename="test.scn",
export_energy = True,
export_polarization = True):
"""Convert reciprocal space path into a scan file (ALS BL 4.0.2)"""
diffr = self._diffractometer
accessible_angles = self.angles[
array([
array(self.is_accessible(*tuple(angle_values) )
) for angle_values in self.angles])
]
motors = array([
array(self.angles2motors(*tuple(angle_values) )
) for angle_values in accessible_angles])
# Remove 'nan' rows
motors = motors[isfinite(motors).all(axis=1)]
col_names = []
col_values = []
if (export_energy == True):
energies = full(len(motors), diffr.energy)
col_names += ["Mono Energy", "EPU Energy"]
col_values += [energies, energies]
if (export_polarization == True):
polarizations = full(len(motors), diffr.polarization.value)
col_names += ["EPU Polarization"]
col_values += [polarizations]
col_names += ["Bottom Rotary Seal", "Top Rotary Seal", "Flip"]
if len(col_values) > 0:
motor_values = hstack((
array(col_values).transpose(),
around(motors, 3),
))
else:
motor_values = around(motors, 3)
scan_motors = pd.DataFrame(motor_values, columns=col_names)
scan_motors.to_csv(
output_filename,
sep = '\t',
index = False,
line_terminator = "\r\n",
)
#~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# MAIN: Testing of module
#~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def print_photon(photon):
"""Print properties of the photon"""
print("****************************")
print(" energy (eV) = " + str(photon.energy) )
print(" lambda (nm) = " + str(photon.wavelength) )
print(" q (1/nm) = " + str(photon.q) )
def test_photon():
"""Basic tests of the class<Photon>"""
print("\nTesting class<Photon>")
xray = Photon(energy=700)
print_photon(xray)
xray.energy = 642
print_photon(xray)
xray.wavelength = 2
print_photon(xray)
xray.q = 2
print_photon(xray)
def print_polarization(pol):
"""Print properties of the polarization"""
state_text = dict({
-1: "UNDEFINED",
0: "CIRCULAR",
1: "LINEAR" })
print("****************************")
print(" value = " + str(pol.value) )
print(" state = " + (state_text[pol.state] +
" (" + str(pol.state) + ")")
)
print(" deg. circ. = " + str(pol.circular_degree) )
print(" lin. angle. (deg) = " + str(pol.linear_angle) )
def test_polarization():
"""Basic tests of the class<Polarization>"""
print("\nTesting class<Polarization>")
pol = Polarization(0)
print_polarization(pol)
pol.value = 100
print_polarization(pol)
pol.value = 190
print_polarization(pol)
pol.value = 200
print_polarization(pol)
pol.value = 0.9
print_polarization(pol)
pol.value = 1
print_polarization(pol)
pol.value = -1
print_polarization(pol)
pol.value = 1.
print_polarization(pol)
pol.value = -1.
print_polarization(pol)
def print_diffractometer(diffr):
"""Print properties of the diffractometer"""
print("****************************")
print(" twotheta (deg) = " + str(diffr.twotheta) )
print(" incidence (deg) = " + str(diffr.incidence) )
print(" transverse (deg) = " + str(diffr.transverse) )
print(" azimuth (deg) = " + str(diffr.azimuth) )
print(" photon...")
print_photon(diffr.photon)
print(" polarization...")
print_polarization(diffr.polarization)
print(" intensity0 (counts) = " + str(diffr.intensity0) )
print(" temperature (K) = " + str(diffr.temperature) )
print(" distance (mm) = " + str(diffr.distance) )
def test_diff402():
"""Basic tests of the class<Diffractometer402>"""
print("\nTesting class<Diffractometer402>")
chamber_params = dict({
'Beamline Energy': 700,
'EPU Polarization': 0,
'Bottom Rotary Seal': 15,
'Top Rotary Seal': 137,
'Flip': -2,
'Flip Offset': -1,
'Top Offset': 2,
'Twice Top Offset': 4,
'CCD Offset': -18.68,
'DATE': "2015-02-12",
'Counter 2': 340*1000,
'TempCtrlrA': 296,
'TempCtrlrB': 295 })
chamber = Diffractometer402(param_dict=chamber_params)
print_diffractometer(chamber)
class SmartFormatter(argparse.HelpFormatter):
"""Copied from 'Anthon' (https://stackoverflow.com/a/22157136)"""
def _split_lines(self, text, width):
"""Allows '\n' in help strings that start with 'R|'"""
if text.startswith('R|'):
return text[2:].splitlines()
# this is the RawTextHelpFormatter._split_lines
return argparse.HelpFormatter._split_lines(self, text, width)
def main():
parser = argparse.ArgumentParser(
usage=__doc__,
description="Command line usage: Basic tests of module functionality",
formatter_class=SmartFormatter,
)
# parser.add_argument(
# "--version",
# action="store_true",
# help="Display PEP440 version identifier",
# )
parser.add_argument(
"--version",
action="version",
version="{} {}".format(__package__, __version__),
help="Display PEP440 version identifier",
)
parser.add_argument(
'--test',
action='append',
default=[],
help="R|Name of test to run; "
"Multiple tests allowed by repeating this option."
"\ne.g., --test diffractometer --test photon --test pol"
"{}{}{}".format(
"\nd[iff[r[actometer]]]: {}".format(test_diff402.__doc__),
"\nph[oton]: {}".format(test_photon.__doc__),
"\npol[arization]: {}".format(test_polarization.__doc__),
),
)
args = parser.parse_args()
# Tests of module functionality
for module_test in args.test:
if module_test in ["diffractometer", "diffract", "diffr", "diff", "d"]:
test_diff402()
elif module_test in ["photon", "ph"]:
test_photon()
elif module_test in ["polarization", "pol"]:
test_polarization()
else:
pass
if __name__ == "__main__":
main() | ALS.Milo | /ALS.Milo-0.18.1.tar.gz/ALS.Milo-0.18.1/als/milo/qimage.py | qimage.py |
from __future__ import print_function
from __future__ import absolute_import
from __future__ import division
from builtins import str
from builtins import zip
__author__ = "Padraic Shafer"
__copyright__ = "Copyright (c) 2018-2021, Padraic Shafer"
__credits__ = [__author__, ]
__license__ = ""
__maintainer__ = "Padraic Shafer"
__email__ = "[email protected]"
__status__ = "Development"
from als.milo import __version__, __date__
import logging
import sys
import os
from collections import namedtuple
import numpy as np
import pandas as pd
from numpy import pi, cos, sin, deg2rad, cross, array, empty, matrix, dot
from numpy import newaxis, isfinite
from numpy import linspace, outer, sum, product, zeros, roots, square, sqrt
from numpy.linalg import norm, solve, lstsq, tensorsolve
from astropy.io import fits
from sys import exit
import matplotlib.pyplot as plt
import matplotlib.cm as cmx
from .qimage import Diffractometer402, Polarization, print_diffractometer
from .qimage import QSpacePath, ResonanceProfile
from .qscan import AngleOffsets, OrthoLatticeABC, HKL
from .qscan import OrthoReciprocalLatticeQabc, OrthoReciprocalVector
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# DATA STRUCTURES ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# class Defaults:
# *) Default values used internally by functions in this script
Defaults = namedtuple("Defaults", ["output_dir"])
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# GLOBALS ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# __defaults__ = Defaults(
# output_dir=os.getcwd(),
# )
__default_output_dir = os.getcwd()
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def get_default_chamber_params():
"""Get dict() of suitable default_chamber_params that can be customized.
RETURNS: Copy of Diffractometer402.default_chamber_params
"""
return(Diffractometer402.default_chamber_params.copy())
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def set_output_dir(format, **kwargs):
"""Sets default output directory using supplied format and keyword params.
format: Format string for output directory using keywords;
*) {base}/... is implied
kwargs: (keyword = value) pairs for keywords used in 'format'
RETURNS: Generated output directory as string
!!! Side Effects !!!
*) generated directory is created if it does not already exist
*) generated directory is set as global __defaults__.output_dir
"""
global __default_output_dir
if not format.startswith("{base}"):
format = "{base}/" + format
output_dir = format.format(**kwargs)
if not os.path.exists(output_dir):
os.makedirs(output_dir)
__default_output_dir = output_dir
return(output_dir)
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def get_output_dir():
"""Gets default output path.
RETURNS: global __defaults__.output_dir
"""
global __default_output_dir
return(__default_output_dir)
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def show_motor_params(primary_hdu):
"""Dumps motor values from CCD .fits header to the console standard output.
primary_hdu: Metadata from primary Header Data Unit of FITS file
RETURNS: None
!!! Side Effects !!!
*) Prints selected metadata from HDU to console standard output.
"""
('Beamline Energy : ' + str(primary_hdu['Beamline Energy']) + ' eV')
logging.info('Bottom Rotary Seal: ' + str(primary_hdu['Bottom Rotary Seal']) + ' deg')
logging.info('Top Rotary Seal : ' + str(primary_hdu['Top Rotary Seal']) + ' deg')
logging.info('Flip : ' + str(primary_hdu['Flip']) + ' deg')
logging.info('Twice Top Offset : ' + str(primary_hdu['Twice Top Offset']) + ' deg')
energy = primary_hdu['Beamline Energy']
bottom = primary_hdu['Bottom Rotary Seal']
top = primary_hdu['Top Rotary Seal']
flip = primary_hdu['Flip']
offset_top = primary_hdu['Twice Top Offset'] / 2
offset_flip = 0
offset_ccd = -18.68
if ('Top Offset' in primary_hdu):
offset_top = primary_hdu['Top Offset']
logging.info('Top Offset : ' + str(offset_top) + ' deg')
if ('Flip Offset' in primary_hdu):
offset_flip = primary_hdu['Flip Offset']
logging.info('Flip Offset : ' + str(offset_flip) + ' deg')
if ('CCD Offset' in primary_hdu):
offset_ccd = primary_hdu['CCD Offset']
logging.info('CCD Offset : ' + str(offset_ccd) + ' deg')
wavelength = 1239.842 / energy
twotheta = bottom - offset_ccd
truetop = top - offset_top
incidence = bottom - truetop
chi = flip - offset_flip
logging.info('')
logging.info('Wavelength : ' + str(wavelength) + ' nm')
logging.info('Detector : ' + str(twotheta) + ' deg')
logging.info('Incidence : ' + str(incidence) + ' deg')
logging.info('Chi : ' + str(chi) + ' deg')
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def load_image(fits_file_path):
"""Reads BL402 CCD header data from FITS file (.fits).
fits_file_path: Fully qualified file name for FITS file.
RETURNS: (CCD_image_metadata, CCD_image_HDU)
*) CCD_image_metadata: Metadata from primary Header Data Unit
*) CCD_image_HDU: Header Data Unit for CCD image data
!!! Side Effects !!!
*) Prints selected metadata from HDU to console standard output.
"""
hdulist = fits.open(fits_file_path)
logging.info("Opening FITS data file: {}".format(fits_file_path))
logging.info("...data structure: {}".format(hdulist.info()))
show_motor_params(hdulist[0].header)
# let's assume, the last entry is the image.
return (hdulist[0].header, hdulist[-1])
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def plot_image_model(image_array, ccd_pairs=None, max_I=None, min_I=None):
"""Display the CCD image data, overlaid with supplied RLU (h,k,L) points.
image_array: 2D array of intensity data to plot.
*) Row order is automatically reversed to accommodate FITS format.
ccd_pairs: 2D coordinates, in units of image pixels.
*) Pixels at these coordinates are indicated by colored markers.
*) Supplied values typically correspond to (h,k,L) points in model.
max_I: Upper limit of color scale is mapped to this intensity value.
min_I: Lower limit of color scale is mapped to this intensity value.
RETURNS: None
!!! Side Effects !!!
*) Displays plotted data on screen.
*) Left panel shows color scale mapped linearly to intensity
*) Right panel shows color scale mapped to log(intensity)
"""
if ccd_pairs is None:
ccd_pairs = array([
[ [0, 0] ]
])
logging.info(
"plot_image_model (ccd_pairs) [{}]: {}".format(
ccd_pairs.shape,
ccd_pairs,
)
)
ccd_pairs_shape = array(ccd_pairs.shape)
ccd_pairs_flat_shape = ccd_pairs_shape[-2:].copy()
ccd_pairs_flat_shape[0] *= product(ccd_pairs_shape[:-2])
ccd_pairs = ccd_pairs.reshape(*ccd_pairs_flat_shape)
logging.debug(
"plot_image_model (reshaped ccd_pairs) [{}]: {}".format(
ccd_pairs.shape,
ccd_pairs,
)
)
img_display_array = image_array[::-1]
if max_I is None:
max_I = img_display_array.max()
if min_I is None:
min_I = img_display_array.min()
ax1 = plt.subplot(1,2,1)
im1 = ax1.imshow(img_display_array)
im1.set_clim(min_I, max_I)
plt.colorbar(im1)
ax2 = plt.subplot(1,2,2)
im2 = ax2.imshow(np.log10(abs(img_display_array)))
plt.colorbar(im2)
ax1.scatter(x=ccd_pairs[:,0], y=(1024-ccd_pairs[:,-1]), s=40, facecolors='none', edgecolors='m')
ax1.set_xlim([0,1024])
ax1.set_ylim([1024,0])
ax2.scatter(x=ccd_pairs[:,0], y=(1024-ccd_pairs[:,-1]), s=40, facecolors='none', edgecolors='m')
ax2.set_xlim([0,1024])
ax2.set_ylim([1024,0])
# plt.show()
plt.show(block=True)
return
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def ccd_rlu_points(primary_hdu, image_array, hk_pairs=None, lattice=None):
"""Return an array of CCD coordinates, corresponding to RLU (h,k) points.
primary_hdu: Metadata from primary Header Data Unit of FITS file.
*) Used to construct reciprocal space model for this CCD image.
image_array: 2D array of intensity data, used for coordinate mapping.
*) Array shape defines CCD image pixel coordinates for hk_pairs.
hk_pairs: Array of 2D coordinates, (h, k), in reciprocal lattice units.
*) Array can have any shape, with innermost indices being 2D.
*) e.g., [M x N x 2]
*) DEFAULT value: [[ [0, 0] ]]; shape = [1 x 1 x 2]
lattice: OrthoLatticeABC object.
*) DEFAULT: a=b=c=(2*pi) ; all angles = 0. (qa,qb,qc)==(h,k,L)
RETURNS: Array of 2D coordinates, in units of image pixels.
*) Array has same shape as input array, hk_pairs
!!! Side Effects !!!
*) None
"""
if hk_pairs is None:
hk_pairs = array([
[ [0, 0] ]
])
hk_pairs_shape = array(hk_pairs.shape)
# logging.debug("%s: %s\n%s",
# "hk_pairs_shape (#1)",
# hk_pairs_shape.shape,
# hk_pairs_shape)
# .copy() necessary to not overwrite original
hk_pairs_flat_shape = hk_pairs_shape[-2:].copy()
# logging.debug("%s: %s\n%s",
# "hk_pairs_shape (#2)",
# hk_pairs_shape.shape,
# hk_pairs_shape)
hk_pairs_flat_shape[0] *= product(hk_pairs_shape[:-2])
# logging.debug("%s: %s\n%s",
# "hk_pairs_shape (#3)",
# hk_pairs_shape.shape,
# hk_pairs_shape)
hk_pairs = hk_pairs.reshape(*hk_pairs_flat_shape)
# logging.debug("%s: %s\n%s",
# "hk_pairs_shape",
# hk_pairs_shape.shape,
# hk_pairs_shape)
# logging.debug("%s: %s\n%s",
# "hk_pairs_flat_shape",
# hk_pairs_flat_shape.shape,
# hk_pairs_flat_shape)
# logging.debug("%s: %s\n%s",
# "hk_pairs",
# hk_pairs.shape,
# hk_pairs)
# exit()
if lattice is None:
lattice = qscan.OrthoLatticeABC(
a=2*pi, # nm
b=2*pi, # nm
c=2*pi, # nm
offsets=qscan.AngleOffsets(
incidence = 0, # degrees
transverse = 0, # degrees
azimuth = 0, # degrees
)
)
# TODO: Replace header extraction with CcdImageFromFITS object
energy = primary_hdu['Beamline Energy']
bottom = primary_hdu['Bottom Rotary Seal']
top = primary_hdu['Top Rotary Seal']
flip = primary_hdu['Flip']
offset_top = primary_hdu['Twice Top Offset'] / 2
offset_flip = 90
offset_ccd = -18.68 # For data captured after June 2014
# offset_ccd = -18.63
if ('Top Offset' in primary_hdu):
offset_top = primary_hdu['Top Offset']
logging.info('Top Offset : ' + str(offset_top) + ' deg')
if ('Flip Offset' in primary_hdu):
offset_flip = primary_hdu['Flip Offset']
logging.info('Flip Offset : ' + str(offset_flip) + ' deg')
if ('CCD Offset' in primary_hdu):
offset_ccd = primary_hdu['CCD Offset']
logging.info('CCD Offset : ' + str(offset_ccd) + ' deg')
wavelength = 1239.842 / energy
twotheta = bottom - offset_ccd
truetop = top - offset_top
incidence = bottom - truetop
chi = flip - offset_flip # transverse
phi = 0 # azimuth
# Adjustments to alignment
incidence += lattice.offsets.incidence
chi += lattice.offsets.transverse
phi += lattice.offsets.azimuth
q_xray = 2 * pi * energy / 1239.842 # 2*pi / d [1 / nm]
incidence_rad = deg2rad(incidence)
chi_rad = deg2rad(chi)
phi_rad = deg2rad(phi)
incidence_rotation = array([
[ cos(incidence_rad), 0, -sin(incidence_rad) ],
[ 0, 1, 0 ],
[ sin(incidence_rad), 0, cos(incidence_rad) ] ])
chi_rotation = array([
[ 1, 0, 0 ],
[ 0, cos(chi_rad), -sin(chi_rad) ],
[ 0, sin(chi_rad), cos(chi_rad) ] ])
# need to verify sign of sin() components
# this version (mathematica, not IGOR) appears correct
phi_rotation = array([
[ cos(phi_rad), -sin(phi_rad), 0 ],
[ sin(phi_rad), cos(phi_rad), 0 ],
[ 0, 0, 1 ] ])
qxyz_unit = array([
[1, 0, 0],
[0, 1, 0],
[0, 0, 1] ])
# logging.debug(dot(array([[1,2],[3,4]]), array([[1,3],[5,7]])))
# logging.debug(array([[1,2],[3,4]]).dot(array([[1,3],[5,7]])))
# logging.debug(array([[1,3],[5,7]]).dot(array([[1,2],[3,4]])))
# a.dot(b) == dot(a, b) == a.b
# logging.debug(array([[1,2],[3,4]]).dot(array([[1,3],[5,7]])).dot(array([[10,20],[30,40]])))
# logging.debug(dot(array([[1,2],[3,4]]), dot(array([[1,3],[5,7]]), array([[10,20],[30,40]]))))
# a.dot(b).dot(c) == dot(a, dot(b, c)) == a.b.c
lattice_a = lattice.a # nm
lattice_b = lattice.b # nm
lattice_spacing = array([lattice_a, lattice_b, 1])
q_lattice_spacing = 2 * pi / lattice_spacing
qxyz_rotated = incidence_rotation.dot(chi_rotation).dot(phi_rotation).dot(qxyz_unit)
# Rotation leaves new unit vectors as columns
# qxyz_rotated = incidence_rotation.dot(qxyz_unit)
# logging.debug("qxyz_rotated (incid): %s --> %s\n%s",
# incidence,
# qxyz_rotated.shape,
# qxyz_rotated)
# qxyz_rotated = chi_rotation.dot(qxyz_unit)
# logging.debug("qxyz_rotated (chi): %s --> %s\n%s",
# chi,
# qxyz_rotated.shape,
# qxyz_rotated)
# qxyz_rotated = phi_rotation.dot(qxyz_unit)
# logging.debug("qxyz_rotated (phi): %s --> %s\n%s",
# phi,
# qxyz_rotated.shape,
# qxyz_rotated)
# Divide 1st COLUMN by 2*pi/a
# Divide 2nd COLUMN by 2*pi/b
# Divide 3rd COLUMN by 2*pi/c
hkl_rotated = qxyz_rotated / q_lattice_spacing[newaxis, :]
# logging.debug("%s: %s\n%s",
# "qxyz_rotated",
# qxyz_rotated.shape,
# qxyz_rotated)
# logging.debug("%s: %s\n%s",
# "q_lattice_spacing",
# q_lattice_spacing.shape,
# q_lattice_spacing)
# logging.debug("%s: %s\n%s",
# "hkl_rotated",
# hkl_rotated.shape,
# hkl_rotated)
h_unit = hkl_rotated[:, 0]
k_unit = hkl_rotated[:, 1]
L_unit = hkl_rotated[:, 2]
h_unit /= norm(h_unit)
k_unit /= norm(k_unit)
L_unit /= norm(L_unit)
# logging.debug("%s: %s\n%s",
# "h_unit",
# h_unit.shape,
# h_unit)
# logging.debug("%s: %s\n%s",
# "k_unit",
# k_unit.shape,
# k_unit)
# logging.debug("%s: %s\n%s",
# "L_unit",
# L_unit.shape,
# L_unit)
sphere_center_q = array([-q_xray, 0, 0])
sphere_center_q_proj_hk = array([
sphere_center_q.dot(h_unit),
sphere_center_q.dot(k_unit) ])
# sphere_center_q_proj_h = sphere_center_q.dot(h_unit)
# sphere_center_q_proj_k = sphere_center_q.dot(k_unit)
# logging.debug("%s: %s\n%s",
# "hk_pairs",
# hk_pairs.shape,
# hk_pairs)
# logging.debug("%s: %s\n%s",
# "q_lattice_spacing[newaxis,:-1]",
# q_lattice_spacing[newaxis,:-1].shape,
# q_lattice_spacing[newaxis,:-1])
# logging.debug("%s: %s\n%s",
# "(hk_pairs * q_lattice_spacing[newaxis,:-1])",
# (hk_pairs * q_lattice_spacing[newaxis,:-1]).shape,
# (hk_pairs * q_lattice_spacing[newaxis,:-1]))
# exit()
# Next command has same effect with(out) using 'newaxis'
# hk_offset_from_sphere_center_q = hk_pairs * q_lattice_spacing[newaxis,:-1]
hk_offset_from_sphere_center_q = hk_pairs * q_lattice_spacing[:-1]
# logging.debug("%s: %s\n%s",
# "hk_offset_from_sphere_center_q (#1)",
# hk_offset_from_sphere_center_q.shape,
# hk_offset_from_sphere_center_q)
hk_offset_from_sphere_center_q -= sphere_center_q_proj_hk
logging.debug("hk_offset_from_sphere_center_q (#2)", end=' ')
logging.debug(hk_offset_from_sphere_center_q.shape, end=' ')
logging.debug(hk_offset_from_sphere_center_q)
hk_circle_center_q = \
hk_offset_from_sphere_center_q[:, :, newaxis] \
* array([h_unit, k_unit])[newaxis, :, :]
# logging.debug("%s: %s\n%s",
# "hk_circle_center_q (#1)",
# hk_circle_center_q.shape,
# hk_circle_center_q)
hk_circle_center_q += array([sphere_center_q, sphere_center_q])
logging.debug("hk_circle_center_q (#2)", end=' ')
logging.debug(hk_circle_center_q.shape, end=' ')
logging.debug(hk_circle_center_q)
# logging.debug("%s: %s\n%s",
# "array([h_unit, k_unit]) * hk_circle_center_q",
# (array([h_unit, k_unit]) * hk_circle_center_q).shape,
# array([h_unit, k_unit]) * hk_circle_center_q)
# logging.debug("%s: %s\n%s",
# "array([h_unit, k_unit])[newaxis, :, :] * hk_circle_center_q",
# (array([h_unit, k_unit])[newaxis, :, :] * hk_circle_center_q).shape,
# array([h_unit, k_unit])[newaxis, :, :] * hk_circle_center_q)
# Next command has same effect with(out) using 'newaxis'
# hk_linsolve_vector = array([h_unit, k_unit])[newaxis, :, :] * hk_circle_center_q
hk_linsolve_vector = array([h_unit, k_unit]) * hk_circle_center_q
hk_linsolve_vector = sum(hk_linsolve_vector, axis=-1)
# logging.debug("%s: %s\n%s",
# "hk_linsolve_vector",
# hk_linsolve_vector.shape,
# hk_linsolve_vector)
linsolve_vector = \
zeros( (hk_linsolve_vector.shape[0], hk_linsolve_vector.shape[1] + 1) )
linsolve_vector[:, :-1] = hk_linsolve_vector
logging.debug("linsolve_vector", end=' ')
logging.debug(linsolve_vector.shape, end=' ')
logging.debug(linsolve_vector)
# exit()
linsolve_matrix = hkl_rotated.transpose()
hk_plane_intersection_vector = cross(h_unit, k_unit)
# logging.debug("%s: %s --> %s\n%s",
# "hk_plane_intersection_vector",
# hk_plane_intersection_vector.shape,
# norm(hk_plane_intersection_vector),
# hk_plane_intersection_vector)
hk_plane_intersection_vector /= norm(hk_plane_intersection_vector)
logging.debug("hk_plane_intersection_vector", end=' ')
logging.debug(hk_plane_intersection_vector.shape, end=' ')
logging.debug(hk_plane_intersection_vector)
# logging.debug("%s: %s\n%s",
# "hk_plane_intersection_vector vs. L_unit",
# (hk_plane_intersection_vector - L_unit).shape,
# (hk_plane_intersection_vector - L_unit))
# logging.debug("%s: %s\n%s",
# "hk_plane_intersection_vector vs. qxyz_rotated[:,2]",
# (hk_plane_intersection_vector - qxyz_rotated[:,2]).shape,
# (hk_plane_intersection_vector - qxyz_rotated[:,2]))
# exit()
# Only works if h, k, L are orthogonal
# if L_unit[0] != 0 : # L has component along room-X
# linsolve_matrix[-1] = array([1, 0, 0])
# solve(linsolve_matrix, linsolve_vector)
# elif L_unit[1] != 0 : # L has component along room-Y
# Pass
# elif L_unit[2] != 0 : # L has component along room-Z
# Pass
# else:
# return() # Return array of complex-valued pairs [INVALID]
if hk_plane_intersection_vector[0] != 0 : # vector has component along room-X
linsolve_matrix[-1] = array([1, 0, 0])
hk_plane_intersection_origin = solve(
linsolve_matrix[newaxis, :, :], linsolve_vector)
logging.debug("hk_plane_intersection_origin", end=' ')
logging.debug(hk_plane_intersection_origin.shape, end=' ')
logging.debug(hk_plane_intersection_origin)
# exit()
elif hk_plane_intersection_vector[1] != 0 : # vector has component along room-Y
linsolve_matrix[-1] = array([0, 1, 0])
hk_plane_intersection_origin = solve(
linsolve_matrix[newaxis, :, :], linsolve_vector)
logging.debug("hk_plane_intersection_origin", end=' ')
logging.debug(hk_plane_intersection_origin.shape, end=' ')
logging.debug(hk_plane_intersection_origin)
# exit()
elif hk_plane_intersection_vector[2] != 0 : # vector has component along room-Z
linsolve_matrix[-1] = array([0, 0, 1])
hk_plane_intersection_origin = solve(
linsolve_matrix[newaxis, :, :], linsolve_vector)
logging.debug("hk_plane_intersection_origin", end=' ')
logging.debug(hk_plane_intersection_origin.shape, end=' ')
logging.debug(hk_plane_intersection_origin)
# exit()
else:
# Return array of INVALID pairs
# ccd_hk_pairs_pixels = array([float('NaN'), float('NaN'), float('NaN')])
ccd_coords_pairs_shape = empty(hk_pairs_shape.shape[0] + 1)
ccd_coords_pairs_shape[:-2] = hk_pairs_shape[:-1]
ccd_coords_pairs_shape[-2] = 2
ccd_coords_pairs_shape[-1] = 2
# ccd_hk_pairs_pixels = ccd_hk_pairs_pixels.reshape(*ccd_coords_pairs_shape)
ccd_hk_pairs_pixels = empty( tuple(ccd_coords_pairs_shape) )
ccd_hk_pairs_pixels.fill( float('NaN') )
logging.debug("ccd_hk_pairs_pixels", end=' ')
logging.debug(ccd_hk_pairs_pixels.shape, end=' ')
logging.debug(ccd_hk_pairs_pixels)
return (ccd_hk_pairs_pixels)
# a = hk_plane_intersection_vector[newaxis, :]
a = hk_plane_intersection_vector
b = hk_plane_intersection_origin - sphere_center_q
logging.debug("a", end=' ')
logging.debug(a.shape, end=' ')
logging.debug(a)
logging.debug("b", end=' ')
logging.debug(b.shape, end=' ')
logging.debug(b)
logging.debug("a.dot(a)", end=' ')
logging.debug((a.dot(a)).shape, end=' ')
logging.debug(a.dot(a))
# logging.debug("%s: %s\n%s",
# "a.dot(b)",
# (a.dot(b)).shape,
# a.dot(b))
logging.debug("a.dot(b)", end=' ')
logging.debug(sum(a * b, axis=-1).shape, end=' ')
logging.debug(sum(a * b, axis=-1))
# logging.debug("%s: %s\n%s",
# "b.dot(b)",
# (b.dot(b)).shape,
# b.dot(b))
logging.debug("b.dot(b)", end=' ')
logging.debug(sum(b * b, axis=-1).shape, end=' ')
logging.debug(sum(b * b, axis=-1))
logging.debug("-B / 2A", end=' ')
logging.debug((-2 * sum(a * b, axis=-1) / (2 * a.dot(a))).shape, end=' ')
logging.debug(-2 * sum(a * b, axis=-1) / (2 * a.dot(a)))
logging.debug("SQRT(B^2 - 4AC) / 2A", end=' ')
logging.debug((sqrt(4 * square(sum(a * b, axis=-1)) \
- 4 * a.dot(a) * (sum(b * b, axis=-1) - q_xray * q_xray)) \
/ (2 * a.dot(a))).shape, end=' ')
logging.debug(sqrt(4 * square(sum(a * b, axis=-1)) \
- 4 * a.dot(a) * (sum(b * b, axis=-1) - q_xray * q_xray)) \
/ (2 * a.dot(a))
)
t0 = -2 * sum(a * b, axis=-1) / (2 * a.dot(a))
t_delta = sqrt(4 * square(sum(a * b, axis=-1)) \
- 4 * a.dot(a) * (sum(b * b, axis=-1) - q_xray * q_xray)) \
/ (2 * a.dot(a))
t_12 = empty((t0.shape[0], 2))
t_12[:, 0] = t0 - t_delta
t_12[:, 1] = t0 + t_delta
logging.debug("t_12", end=' ')
logging.debug(t_12.shape, end=' ')
logging.debug(t_12)
# logging.debug(hk_plane_intersection_vector[newaxis, :, newaxis] * t_12[:, newaxis, :])
# intersections = hk_plane_intersection_origin[:, :, newaxis] \
# + hk_plane_intersection_vector[newaxis, :, newaxis] * t_12[:, newaxis, :]
intersections = hk_plane_intersection_origin[:, newaxis, :] \
+ hk_plane_intersection_vector[newaxis, newaxis, :] * t_12[:, :, newaxis]
logging.debug("intersections", end=' ')
logging.debug(intersections.shape, end=' ')
logging.debug(intersections)
exit_vectors = intersections - sphere_center_q[newaxis, newaxis, :]
logging.debug("exit_vectors", end=' ')
logging.debug(exit_vectors.shape, end=' ')
logging.debug(exit_vectors)
# circle_intersections = roots([a.dot(a), 2*a.dot(b), b.dot(b) - q_xray*q_xray])
# logging.debug("%s: %s\n%s",
# "circle_intersections",
# circle_intersections.shape,
# circle_intersections)
# exit()
distance_ccd = 1.120 * 142.265
twotheta_rad = deg2rad(twotheta)
ccd_center_room = [cos(twotheta_rad), 0 , sin(twotheta_rad)]
ccd_center_room = array(ccd_center_room)
# logging.debug("%s: %s deg. == %s rad.",
# "Two-Theta: ",
# twotheta,
# deg2rad(twotheta))
# logging.debug("%s: %s",
# "ccd_center_room unit: ",
# ccd_center_room)
# logging.debug("%s: %s",
# "ccd_center_room length: ",
# distance_ccd)
ccd_center_room *= distance_ccd
ccd_tilt_axisY = 0
ccd_unit_normal = array(
[cos(twotheta_rad) - ccd_tilt_axisY,
0 ,
sin(twotheta_rad) - ccd_tilt_axisY] )
ccd_unit_Y = array( [0, 1, 0] )
ccd_unit_X = cross(ccd_unit_normal, ccd_unit_Y)
raw_pixel_size = 0.0135 # size in mm
raw_pixel_num = 2048 # pixels / side
num_cols, num_rows = image_array.shape
ccd_pixel_size_X = raw_pixel_size * raw_pixel_num / num_cols
ccd_pixel_size_Y = raw_pixel_size * raw_pixel_num / num_rows
ccd_center_pixel_X = float(num_cols - 1) / 2 # Assumes 0...numCols-1
ccd_center_pixel_Y = num_rows * (float(1024 - 460) / 1024) - 1; # Assumes 0...numRows-1
# logging.debug("%s: %s, %s",
# "ccd_center_pixel: ",
# ccd_center_pixel_X,
# ccd_center_pixel_Y)
# logging.debug("%s: %s, %s",
# "ccd_pixel_size: ",
# ccd_pixel_size_X,
# ccd_pixel_size_Y)
# logging.debug("%s: %s",
# "ccd_center_room: ",
# ccd_center_room)
# Solve for intersections of exit_vectors (lines) and CCD (plane)
# Build linear equations matrices, using permutation of exit vector components
# Matrices intentionally over-determined
linsolve_matrix_row1 = array([ [ 0, 0, 0],
[ 0, 0, 1],
[ 0, -1 , 0] ])
linsolve_matrix_row2 = array([ [ 0, 0, -1],
[ 0, 0, 0],
[ 1, 0 , 0] ])
linsolve_matrix_row3 = array([ [ 0, 1, 0],
[-1, 0, 0],
[ 0, 0 , 0] ])
linsolve_matrix_rows = array([ linsolve_matrix_row1,
linsolve_matrix_row2,
linsolve_matrix_row3 ])
logging.debug("linsolve_matrix_rows", end=' ')
logging.debug(linsolve_matrix_rows.shape, end=' ')
logging.debug(linsolve_matrix_rows)
linsolve_matrix_3x3 = linsolve_matrix_rows[newaxis, newaxis, :, :, :] \
* exit_vectors[:, :, newaxis, newaxis, :]
logging.debug("linsolve_matrix_3x3 (#1)", end=' ')
logging.debug(linsolve_matrix_3x3.shape, end=' ')
logging.debug(linsolve_matrix_3x3)
linsolve_matrix_3x3 = sum(linsolve_matrix_3x3, axis=-1)
logging.debug("linsolve_matrix_3x3 (#2)", end=' ')
logging.debug(linsolve_matrix_3x3.shape, end=' ')
logging.debug(linsolve_matrix_3x3)
linsolve_matrix = empty(( linsolve_matrix_3x3.shape[0],
linsolve_matrix_3x3.shape[1],
linsolve_matrix_3x3.shape[2] + 1,
linsolve_matrix_3x3.shape[3] ))
linsolve_matrix[:, :, 1:, :] = linsolve_matrix_3x3
linsolve_matrix[:, :, 0, :] = ccd_unit_normal
logging.debug("linsolve_matrix (#3)", end=' ')
logging.debug(linsolve_matrix.shape, end=' ')
logging.debug(linsolve_matrix)
# exit()
linsolve_vector = zeros(4)
linsolve_vector[0] = ccd_unit_normal.dot(ccd_center_room)
logging.debug("linsolve_vector", end=' ')
logging.debug(linsolve_vector.shape, end=' ')
logging.debug(linsolve_vector)
# ccd_hk_pairs_room = tensorsolve(
# linsolve_matrix, linsolve_vector[newaxis, newaxis, :])
# Must use lstsq() for over-determined system
# ...solution is contained in first index of returned array
ccd_hk_pairs_room = array([
[
lstsq(exit_matrix, linsolve_vector)[0]
if isfinite(exit_matrix).all() else
# inaccessible (h,k) pair, return INVALID coordinate
array([float('NaN'), float('NaN'), float('NaN')])
for exit_matrix in hk_pair ]
for hk_pair in linsolve_matrix
])
logging.debug("ccd_hk_pairs_room", end=' ')
logging.debug(ccd_hk_pairs_room.shape, end=' ')
logging.debug(ccd_hk_pairs_room)
exit_vectors_proj_ccd = sum(ccd_hk_pairs_room * exit_vectors, axis=-1)
# ccd_hk_pairs_room = array([
# [
# if exit_proj < 0:
# array([1j, 1j]) # INVALID pair; projected AWAY from CCD
# else:
#
# for exit_proj in hk_pair ]
# for hk_pair in exit_vectors_proj_ccd
# ])
# ccd_hk_pairs_room = array([
# [
# if exit_proj < 0:
# array([1j, 1j]) # INVALID pair; projected AWAY from CCD
# else:
# hk_proj_room
# for exit_proj, hk_proj_room in zip(exit_pair, hk_pair) ]
# for exit_pair, hk_pair in zip(exit_vectors_proj_ccd, ccd_hk_pairs_room)
# ])
ccd_hk_pairs_room = array([
[
# array([1j, 1j, 1j]) # INVALID pair; projected AWAY from CCD
# INVALID pair; projected AWAY from CCD
array([float('NaN'), float('NaN'), float('NaN')])
if exit_proj < 0 else
hk_proj_room
for exit_proj, hk_proj_room in zip(exit_pair, hk_pair) ]
for exit_pair, hk_pair in zip(exit_vectors_proj_ccd, ccd_hk_pairs_room)
])
logging.debug("ccd_hk_pairs_room (#2)", end=' ')
logging.debug(ccd_hk_pairs_room.shape, end=' ')
logging.debug(ccd_hk_pairs_room)
# exit()
ccd_hk_pairs_room -= ccd_center_room
logging.debug("ccd_hk_pairs_room (#3)", end=' ')
logging.debug(ccd_hk_pairs_room.shape, end=' ')
logging.debug(ccd_hk_pairs_room)
ccd_hk_pairs_pixels = ccd_hk_pairs_room[:, :, :, newaxis] \
* array([ccd_unit_X, ccd_unit_Y]).transpose()[newaxis, newaxis, :, :]
ccd_hk_pairs_pixels = sum(ccd_hk_pairs_pixels, axis=-2)
logging.debug("ccd_hk_pairs_pixels (#1)", end=' ')
logging.debug(ccd_hk_pairs_pixels.shape, end=' ')
logging.debug(ccd_hk_pairs_pixels)
ccd_hk_pairs_pixels /= \
array([ccd_pixel_size_X, ccd_pixel_size_Y])[newaxis, newaxis, :]
logging.debug("ccd_hk_pairs_pixels (#2)", end=' ')
logging.debug(ccd_hk_pairs_pixels.shape, end=' ')
logging.debug(ccd_hk_pairs_pixels)
ccd_hk_pairs_pixels += \
array([ccd_center_pixel_X, ccd_center_pixel_Y])[newaxis, newaxis, :]
logging.debug("ccd_hk_pairs_pixels (#3)", end=' ')
logging.debug(ccd_hk_pairs_pixels.shape, end=' ')
logging.debug(ccd_hk_pairs_pixels)
# exit()
ccd_coords_pairs_shape = empty(hk_pairs_shape.shape[0] + 1, dtype=int)
ccd_coords_pairs_shape[:-2] = hk_pairs_shape[:-1]
ccd_coords_pairs_shape[-2] = 2
ccd_coords_pairs_shape[-1] = 2
ccd_hk_pairs_pixels = ccd_hk_pairs_pixels.reshape(*ccd_coords_pairs_shape)
logging.debug("ccd_hk_pairs_pixels (#4)", end=' ')
logging.debug(ccd_hk_pairs_pixels.shape, end=' ')
logging.debug(ccd_hk_pairs_pixels)
return (ccd_hk_pairs_pixels)
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def main(args=None):
"""The main routine."""
if args is None:
args = sys.argv[1:]
for arg in args:
# if arg.lower() == "--version":
# print __version__
pass
print("Usage details")
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
if __name__ == '__main__':
main() | ALS.Milo | /ALS.Milo-0.18.1.tar.gz/ALS.Milo-0.18.1/als/milo/qalign.py | qalign.py |
from __future__ import division
from future.utils import iteritems
__author__ = "Padraic Shafer"
__copyright__ = "Copyright (c) 2014-2021, Padraic Shafer"
__credits__ = [__author__, ]
__license__ = ""
__maintainer__ = "Padraic Shafer"
__email__ = "[email protected]"
__status__ = "Development"
from als.milo import __version__, __date__
import scipy
from numpy import pi, cos, sin, deg2rad, nan
from numpy import array, empty, matrix, newaxis, zeros, arange
from numpy import cross, dot, linspace, outer, sum, product, mean
from numpy import roots, square, sqrt, isfinite
from numpy import logical_and, logical_or, any, all, unique
from numpy import append
from numpy.linalg import norm, solve, lstsq, tensorsolve
from astropy.io import fits
from sys import exit
import os
import matplotlib.pyplot as plt
import matplotlib.cm as cmx
import matplotlib as mpl
from matplotlib.dates import DayLocator, HourLocator, DateFormatter, num2date
from matplotlib.ticker import FuncFormatter
from datetime import datetime, date
import pandas as pd
#---------------------------------------------------------------------------
# FUNCTIONS
#---------------------------------------------------------------------------
def std_datetime_formatter(d, pos=None):
"""Formats time axis of pyplot using actual date+time
*) Returns label to be displayed on time axis
"""
dt = num2date(d)
if (pos == 0):
fmt = "%b. %d %H:%M"
elif (dt.strftime("%H") == "00"):
fmt = "%b. %d %H:%M"
elif (int(dt.strftime("%H")) < 4):
fmt = "%b. %d %H:%M"
else:
fmt = "%H:%M"
label = dt.strftime(fmt)
return label
def anon_datetime_formatter(d, pos=None, start=datetime(1970, 1, 1) ):
"""Formats time axis of pyplot using relative date+time
*) Returns label to be displayed on time axis
"""
dt = num2date(d)
dt = dt.replace(tzinfo=None)
dt_diff = dt - start
days = 1 + dt_diff.days
hours = dt_diff.seconds // 3600
mins = (dt_diff.seconds % 3600) // 60
secs = (dt_diff.seconds % 60) + dt_diff.microseconds
if (pos == 0):
label = "[ Day {0:0d} ] {1:2d} hrs".format(days, hours)
elif (hours == 0):
label = "[ Day {0:0d} ] {1:2d} hrs".format(days, hours)
elif (hours < 4):
label = "[ Day {0:0d} ] {1:2d} hrs".format(days, hours)
else:
label = "{1:2d} hrs".format(days, hours)
return label
def anondate_datetime_formatter(d, pos=None, start=datetime(1970, 1, 1) ):
"""Formats time axis of pyplot using relative date + actual time
*) Returns label to be displayed on time axis
"""
dt = num2date(d)
# dt = dt.replace(tzinfo=None)
days = 1 + dt.toordinal() - start.toordinal()
if (pos == 0):
label = "[ Day {0:0d} ] ".format(days)
elif (dt.strftime("%H") == "00"):
label = "[ Day {0:0d} ] ".format(days)
elif (int(dt.strftime("%H")) < 4):
label = "[ Day {0:0d} ] ".format(days)
else:
label = ""
label += dt.strftime("%H:%M")
return label
def relative_mins_formatter(d, pos=None, start=datetime(1970, 1, 1) ):
"""Formats time axis of pyplot using relative date+time in minutes
*) Returns label to be displayed on time axis
"""
dt = num2date(d)
dt = dt.replace(tzinfo=None)
dt_diff = dt - start
days = 1 + dt_diff.days
hours = dt_diff.seconds // 3600
mins = (dt_diff.seconds % 3600) // 60
secs = (dt_diff.seconds % 60) + dt_diff.microseconds
if (pos == 0):
label = "[ Day {0:0d} ] {1:2d} min".format(days, mins)
elif (hours == 0):
label = "[ Day {0:0d} ] {1:2d} min".format(days, mins)
elif (hours < 4):
label = "[ Day {0:0d} ] {1:2d} min".format(days, mins)
else:
label = "{1:2d} min".format(days, hours)
return label
#---------------------------------------------------------------------------
def make_plot_curve(**kwargs):
"""Generate a dict of curve params for the plot_XXXXX() functions
*) User-supplied keywords override default paramaters
*) Returns a dict of curve params for the plot_XXXXX() functions
"""
dict_plot_curve = dict({
"df": None, # PANDAS dataframe
"x_col": "", # str name of column for abcissa (X, horizontal axis)
"y_col": "", # str name of column for ordinate (Y, vertical axis)
"z_col": "", # str name of column for intensity (Z, "color" axis)
"label": "",
})
for (key, value) in iteritems(kwargs):
dict_plot_curve[key] = value
return(dict_plot_curve)
#---------------------------------------------------------------------------
def make_plot_params(**kwargs):
"""Generate a dict of options for the plot_XXXXX() functions
*) User-supplied keywords override default paramaters
*) Returns a dict of options for the plot_XXXXX() functions
"""
dict_plot_params = dict({
"colors": ['b','g','k'],
"legend_loc": 0,
"title": "",
"xlabel": "",
"ylabel": "",
"xlim": None,
"ylim": None,
"clim": None,
"hlines": array([]),
"vlines": array([]),
"fig_size": (12, 8),
"filename": None,
"hide": False, # Set to True to not display the plot
"clear": True, # Set to False to leave previous figure in plot
"vs_time": False, # True: plot vs. date+time; False: plot vs. index
})
for (key, value) in iteritems(kwargs):
dict_plot_params[key] = value
return(dict_plot_params)
#---------------------------------------------------------------------------
def plot_scatter(
curve_array = array([]),
plot_options = make_plot_params()
):
"""Generates a scatter plot of supplied curves
*) curve_array: array of curve 'dict's; see make_plot_curve()
*) plot_options: dict of plotting options; see make_plot_params()
*) Returns current (figure, axis)
"""
mpl.rcParams.update({'font.size': 22})
if plot_options["clear"]:
plt.clf()
[plt.plot(
curve["df"][curve["x_col"] ],
curve["df"][curve["y_col"] ],
# plot_options["colors"][i % len(plot_options["colors"])],
color=plot_options["colors"][i % len(plot_options["colors"])],
linewidth=2,
label=curve["label"],
) for (i, curve) in enumerate(curve_array)]
plt.legend(frameon=False, loc=plot_options["legend_loc"])
# plt.gcf().autofmt_xdate()
# date_plot_format = DateFormatter("%m/%d %H:%M")
# date_plot_format = FuncFormatter(anondate_datetime_formatter)
# date_plot_format = FuncFormatter(std_datetime_formatter)
fig = plt.gcf()
ax = fig.gca()
# ax.xaxis.set_major_formatter(date_plot_format)
# ax.xaxis.set_major_locator(HourLocator(interval=4))
# ax.xaxis.set_minor_locator(HourLocator())
plt.title(plot_options["title"])
plt.xlabel(plot_options["xlabel"])
plt.ylabel(plot_options["ylabel"])
if plot_options["xlim"]:
plt.xlim(plot_options["xlim"])
if plot_options["ylim"]:
plt.ylim(plot_options["ylim"])
[plt.axhline(
value, color='k', ls='--') for value in plot_options["hlines"] ]
[plt.axvline(
value, color='k', ls='--') for value in plot_options["vlines"] ]
#ax.autoscale_view()
# plt.plot(time_endpoints, [50, 50], 'g--')
# plt.plot(time_endpoints, [30, 30], 'c--')
# plt.plot(time_endpoints, [10, 10], 'k--')
# plt.plot(time_endpoints, [0, 0], 'k--')
fig.set_size_inches(plot_options["fig_size"])
if plot_options["filename"]:
plt.savefig(plot_options["filename"])
if not plot_options["hide"]:
plt.show()
return(fig, ax)
#---------------------------------------------------------------------------
def plot_history(
curve_array = array([]),
plot_options = make_plot_params()
):
"""Generates a "time" plot of supplied curves
*) curve_array: array of curve 'dict's; see make_plot_curve()
*) plot_options: dict of plotting options; see make_plot_params()
*) Returns current (figure, axis)
"""
mpl.rcParams.update({'font.size': 22})
if plot_options["clear"]:
plt.clf()
if plot_options["vs_time"]:
[plt.plot(
curve["df"][curve["x_col"] ].tolist(),
curve["df"][curve["y_col"] ],
plot_options["colors"][i % len(plot_options["colors"])],
linewidth=2,
label=curve["label"],
) for (i, curve) in enumerate(curve_array)]
else:
[plt.plot(
curve["df"].index.tolist(),
curve["df"][curve["y_col"] ],
plot_options["colors"][i % len(plot_options["colors"])],
linewidth=2,
label=curve["label"],
) for (i, curve) in enumerate(curve_array)]
plt.gcf().autofmt_xdate()
plt.legend(frameon=False, loc=plot_options["legend_loc"])
fig = plt.gcf()
ax = fig.gca()
if plot_options["vs_time"]:
def elapsed_mins_formatter(d, pos=None):
return relative_mins_formatter(d, pos,
start=curve["df"].loc[0, curve["x_col"] ])
fig.autofmt_xdate()
# date_plot_format = FuncFormatter("%m/%d %H:%M")
# date_plot_format = FuncFormatter(anondate_datetime_formatter)
# date_plot_format = FuncFormatter(std_datetime_formatter)
date_plot_format = FuncFormatter(elapsed_mins_formatter)
ax.xaxis.set_major_formatter(date_plot_format)
ax.xaxis.set_major_locator(HourLocator(interval=4))
ax.xaxis.set_minor_locator(HourLocator())
plt.title(plot_options["title"])
plt.xlabel(plot_options["xlabel"])
plt.ylabel(plot_options["ylabel"])
if plot_options["xlim"]:
plt.xlim(plot_options["xlim"])
if plot_options["ylim"]:
plt.ylim(plot_options["ylim"])
[plt.axhline(
value, color='k', ls='--') for value in plot_options["hlines"] ]
[plt.axvline(
value, color='k', ls='--') for value in plot_options["vlines"] ]
#ax.autoscale_view()
# plt.plot(time_endpoints, [50, 50], 'g--')
# plt.plot(time_endpoints, [30, 30], 'c--')
# plt.plot(time_endpoints, [10, 10], 'k--')
# plt.plot(time_endpoints, [0, 0], 'k--')
fig.set_size_inches(plot_options["fig_size"])
if plot_options["filename"]:
plt.savefig(plot_options["filename"])
if not plot_options["hide"]:
plt.show()
return(fig, ax)
#---------------------------------------------------------------------------
def plot_points2D(
curve_array = array([]),
plot_options = make_plot_params()
):
"""Generates a scatter plot of supplied curves
*) curve_array: array of curve 'dict's; see make_plot_curve()
*) plot_options: dict of plotting options; see make_plot_params()
*) Returns current (figure, axis)
"""
mpl.rcParams.update({'font.size': 22})
if plot_options["clear"]:
plt.clf()
cm = plt.get_cmap()
scalarMap = cmx.ScalarMappable(cmap=cm)
def get_intensity_cmap(
intensity_values = array([])
):
scalarMap.set_array(intensity_values)
scalarMap.autoscale()
cm = scalarMap.get_cmap()
# print scalarMap.get_clim()
cm_array = [get_intensity_cmap(curve["df"][curve["z_col"] ])
for curve in curve_array]
[plt.scatter(
x = curve["df"][curve["x_col"] ],
y = curve["df"][curve["y_col"] ],
c = curve["df"][curve["z_col"] ],
s=4,
cmap = cm_array[i],
edgecolors='none',
# label=curve["label"],
) for (i, curve) in enumerate(curve_array)]
# plt.legend(frameon=False, loc=plot_options["legend_loc"])
# plt.gcf().autofmt_xdate()
# date_plot_format = DateFormatter("%m/%d %H:%M")
# date_plot_format = FuncFormatter(anondate_datetime_formatter)
# date_plot_format = FuncFormatter(std_datetime_formatter)
fig = plt.gcf()
ax = fig.gca()
# ax.xaxis.set_major_formatter(date_plot_format)
# ax.xaxis.set_major_locator(HourLocator(interval=4))
# ax.xaxis.set_minor_locator(HourLocator())
plt.title(plot_options["title"])
plt.xlabel(plot_options["xlabel"])
plt.ylabel(plot_options["ylabel"])
if plot_options["xlim"]:
plt.xlim(plot_options["xlim"])
if plot_options["ylim"]:
plt.ylim(plot_options["ylim"])
if plot_options["clim"]:
plt.clim(plot_options["clim"])
[plt.axhline(
value, color='k', ls='--') for value in plot_options["hlines"] ]
[plt.axvline(
value, color='k', ls='--') for value in plot_options["vlines"] ]
#ax.autoscale_view()
# plt.plot(time_endpoints, [50, 50], 'g--')
# plt.plot(time_endpoints, [30, 30], 'c--')
# plt.plot(time_endpoints, [10, 10], 'k--')
# plt.plot(time_endpoints, [0, 0], 'k--')
fig.set_size_inches(plot_options["fig_size"])
if plot_options["filename"]:
plt.savefig(plot_options["filename"])
if not plot_options["hide"]:
plt.show()
return(fig, ax)
#~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# MAIN body
#~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
if __name__ == "__main__":
plot_scatter(
array([
make_plot_curve(**{
"df": data_by_avgLoop.xs(z_value, level=z_col),
"x_col": field_col,
# "y_col": diff_tey_norm0,
"y_col": tey_norm0_asym,
"label": "Z = {0:3.1f} mm".format(z_value),
}),
]),
make_plot_params(**{
"title": "CD vs. Field\n",
"xlabel": "Magnetic Field [ T ]",
"ylabel": "Circular Difference [ a.u. ]\n",
"fig_size": (12, 24),
"filename": "{0:s}field-loops_{1:5d}{2:s}_avg.pdf".format(
output_path,
traj_base_num,
input_postfix,
),
"clear": False,
"hide": True,
})
)
exit() | ALS.Milo | /ALS.Milo-0.18.1.tar.gz/ALS.Milo-0.18.1/als/milo/miloplot.py | miloplot.py |
from __future__ import print_function
from __future__ import division
from builtins import object
__author__ = "Padraic Shafer"
__copyright__ = "Copyright (c) 2018-2021, Padraic Shafer"
__credits__ = [__author__, ]
__license__ = ""
__maintainer__ = "Padraic Shafer"
__email__ = "[email protected]"
__status__ = "Development"
import logging
import sys
import os
from collections import namedtuple
import numpy as np
import pandas as pd
# from .qimage import Diffractometer402, Polarization
# from .qimage import QSpacePath, ResonanceProfile
from als.milo.qimage import Diffractometer402, Polarization
from als.milo.qimage import QSpacePath, ResonanceProfile
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# DATA STRUCTURES ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# class OrthoLatticeABC:
# *) Assumes orthorhombic lattice; c // z;
# *) (a, b) have azimuthal rotation from (x, y), defined by 'offsets.azimuth'
# *) Fine tuning of lattice relative to diffractometer achieved by 'offsets'
OrthoLatticeABC = namedtuple("OrthoLatticeABC", ['a', 'b', 'c', "offsets"])
# class AngleOffsets:
# *) Corrections to alignment angle offsets,
# determined by manual alignment of model to data
AngleOffsets = namedtuple(
"AngleOffsets", ["incidence", "transverse", "azimuth"])
# class HKL:
# *) Relative coordinates of reciprocal space vector
HKL = namedtuple("HKL", ['h', 'k', 'L'])
# class Defaults:
# *) Default values used internally by functions in this script
Defaults = namedtuple("Defaults", ["output_dir"])
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
class OrthoReciprocalLatticeQabc(object):
"""Reciprocal lattice vectors (qa, qb, qc) for lattice vectors (a, b, c)
*) Assumes orthorhombic lattice; c // z;
*) (a, b) have azimuthal rotation from (x, y) defined in 'offsets'
*) qa = 2*pi/a (parallel to a)
*) qb = 2*pi/b (parallel to b)
*) qc = 2*pi/c (parallel to c)
"""
def __init__(self, lattice, **keywords):
"""Initialize reciprocal lattice from OrthoLatticeABC
"""
# The following assignments can be consolidated using self.lattice?
# self.a = lattice.a
# self.b = lattice.b
# self.c = lattice.c
# self.offsets = lattice.offsets
self.lattice = lattice
# The following assignments are redundant?
# self.qa = 2*np.pi / self.a
# self.qb = 2*np.pi / self.b
# self.qc = 2*np.pi / self.c
@property
def lattice(self):
"""Returns a OrthoLatticeABC object"""
return OrthoLatticeABC(
a = self._a,
b = self._b,
c = self._c,
offsets = self._offsets,
)
@lattice.setter
def lattice(self, lattice_obj):
# Does not check for valid value
self.a = lattice_obj.a
self.b = lattice_obj.b
self.c = lattice_obj.c
self.offsets = lattice_obj.offsets
return lattice_obj
@property
def offsets(self):
"""Access the lattice angle offsets (in degrees)"""
return self._offsets
@offsets.setter
def offsets(self, value):
# Does not check for valid value
self._offsets = value
return value
@property
def a(self):
"""Access the lattice parameter, a (in nm)"""
return self._a
@a.setter
def a(self, value):
# Does not check for valid value
self._a = value
self._qa = 2 * np.pi / self._a
return value
@property
def b(self):
"""Access the lattice parameter, b (in nm)"""
return self._b
@b.setter
def b(self, value):
# Does not check for valid value
self._b = value
self._qb = 2 * np.pi / self._b
return value
@property
def c(self):
"""Access the lattice parameter, c (in nm)"""
return self._c
@c.setter
def c(self, value):
# Does not check for valid value
self._c = value
self._qc = 2 * np.pi / self._c
return value
@property
def qa(self):
"""Access the lattice parameter, qa (in nm)"""
return self._qa
@qa.setter
def qa(self, value):
# Does not check for valid value
self._qa = value
self._a = 2 * np.pi / self._qa
return value
@property
def qb(self):
"""Access the lattice parameter, qb (in nm)"""
return self._qb
@qb.setter
def qb(self, value):
# Does not check for valid value
self._qb = value
self._b = 2 * np.pi / self._qb
return value
@property
def qc(self):
"""Access the lattice parameter, qc (in nm)"""
return self._qc
@qc.setter
def qc(self, value):
# Does not check for valid value
self._qc = value
self._c = 2 * np.pi / self._qc
return value
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
class OrthoReciprocalVector(object):
"""Reciprocal space vector (qa, qb, qc) for lattice vectors (a, b, c)
*) Assumes orthorhombic lattice; c // z;
*) (a, b) have azimuthal rotation from (x, y) defined in 'offsets'
*) qa = h * rlattice.qa; rlattice.qa = 2*pi/a (parallel to a)
*) qb = k * rlattice.qb; rlattice.qb = 2*pi/b (parallel to b)
*) qc = L * rlattice.qc; rlattice.qc = 2*pi/c (parallel to c)
"""
def __init__(self, lattice, qa=0, qb=0, qc=0, **keywords):
"""Initialize reciprocal space vector from OrthoLatticeABC
"""
self._rlattice = OrthoReciprocalLatticeQabc(lattice)
self.qa = qa
self.qb = qb
self.qc = qc
@classmethod
def fromQabc(cls, lattice, qa=0, qb=0, qc=0, **keywords):
"""Initialize reciprocal space vector using (qa, qb, qc)
"""
return cls(lattice, qa, qb, qc, **keywords)
@classmethod
def fromHKL(cls, lattice, h=0, k=0, L=0, **keywords):
"""Initialize reciprocal space vector using (h, k, L)
"""
rlattice = OrthoReciprocalLatticeQabc(lattice)
qa = h * rlattice.qa
qb = k * rlattice.qb
qc = L * rlattice.qc
return cls(lattice, qa, qb, qc, **keywords)
@property
def rlattice(self):
"""Access OrthoReciprocalLatticeQabc object"""
return self._rlattice
@rlattice.setter
def rlattice(self, rlattice_obj):
# Does not check for valid value
self._rlattice = rlattice_obj
self._h = self._qa / rlattice_obj.qa
self._k = self._qb / rlattice_obj.qb
self._L = self._qc / rlattice_obj.qc
return value
@property
def h(self):
"""Access the relative reciprocal vector, h (in rlu)"""
return self._h
@h.setter
def h(self, value):
# Does not check for valid value
self._h = value
self._qa = self._h * rlattice_obj.qa
return value
@property
def k(self):
"""Access the relative reciprocal vector, k (in rlu)"""
return self._k
@k.setter
def k(self, value):
# Does not check for valid value
self._k = value
self._qb = self._k * rlattice_obj.qb
return value
@property
def L(self):
"""Access the relative reciprocal vector, L (in rlu)"""
return self._L
@L.setter
def L(self, value):
# Does not check for valid value
self._L = value
self._qc = self._L * rlattice_obj.qc
return value
@property
def qa(self):
"""Access the absolute reciprocal vector, qa (in nm)"""
return self._qa
@qa.setter
def qa(self, value):
# Does not check for valid value
self._qa = value
self._h = self._qa / self._rlattice.qa
return value
@property
def qb(self):
"""Access the absolute reciprocal vector, qb (in nm)"""
return self._qb
@qb.setter
def qb(self, value):
# Does not check for valid value
self._qb = value
self._k = self._qb / self._rlattice.qb
return value
@property
def qc(self):
"""Access the absolute reciprocal vector, qc (in nm)"""
return self._qc
@qc.setter
def qc(self, value):
# Does not check for valid value
self._qc = value
self._L = self._qc / self._rlattice.qc
return value
def hkL(self):
return(self.h, self.k, self.L)
def qABC(self):
return(self.qa, self.qb, self.qc)
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# GLOBALS ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# __defaults__ = Defaults(
# output_dir=os.getcwd(),
# )
__default_output_dir = os.getcwd()
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def get_default_chamber_params():
"""Get dict() of suitable default_chamber_params that can be customized.
RETURNS: Copy of Diffractometer402.default_chamber_params
"""
return(Diffractometer402.default_chamber_params.copy())
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def set_output_dir(format, **kwargs):
"""Sets default output directory using supplied format and keyword params.
format: Format string for output directory using keywords;
*) {base}/... is implied
kwargs: (keyword = value) pairs for keywords used in 'format'
RETURNS: Generated output directory as string
!!! Side Effects !!!
*) generated directory is created if it does not already exist
*) generated directory is set as global __defaults__.output_dir
"""
global __default_output_dir
if not format.startswith("{base}"):
format = "{base}/" + format
output_dir = format.format(**kwargs)
if not os.path.exists(output_dir):
os.makedirs(output_dir)
__default_output_dir = output_dir
return(output_dir)
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def get_output_dir():
"""Gets default output path.
RETURNS: global __defaults__.output_dir
"""
global __default_output_dir
return(__default_output_dir)
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def get_scan_header_line_number(scan_file_path):
"""Extract line number of motor headers from scan file.
scan_file_path: Fully qualified path (dir + file) of scan file.
RETURNS: Zero-based line number of motor header row.
-1 = Not found
"""
with open(scan_file_path, 'r') as scan_file:
for (header_linenum, file_line) in enumerate(scan_file):
logging.debug(header_linenum, file_line)
if file_line[0].isdigit() or file_line.lower().startswith("file"):
header_linenum -= 1
return(header_linenum)
return(-1)
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def import_scan_file(scan_file_path):
"""Import motor positions from scan file into PANDAS DataFrame.
scan_file_path: Fully qualified path (dir + file) of scan file.
RETURNS: PANDAS DataFrame of imported motor positions
"""
header_linenum = get_scan_header_line_number(scan_file_path)
df = pd.read_table(
scan_file_path,
delimiter='\t',
header=header_linenum,
skip_blank_lines=False,
)
return(df)
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def calculate_qABC(df, diffractometer=None, inplace=False):
"""Calculate (qa, qb, qc) for PANDAS DataFrame.
df: PANDAS DataFrame containing motor positions.
diffractometer: Diffractometer402 object for interpreting motor values.
inplace: False = returns copy of df; True = returns original df.
RETURNS: PANDAS DataFrame with additional (qa, qb, qc) columns
"""
if inplace:
df_new = df
else:
df_new = df.copy()
df_new["qa"] = np.nan
df_new["qb"] = np.nan
df_new["qc"] = np.nan
if diffractometer is None:
diffractometer = Diffractometer402(
param_dict=get_default_chamber_params(),
)
if "Beamline Energy" in df_new.columns:
energy_col = "Beamline Energy"
elif "Mono Energy" in df_new.columns:
energy_col = "Mono Energy"
else:
logging.error("Energy column not found in DataFrame")
if "EPU Polarization" in df_new.columns:
pol_col = "EPU Polarization"
else:
pol_col = None
if "Flip Position" in df_new.columns:
flip_col = "Flip Position"
elif "Flip" in df_new.columns:
flip_col = "Flip"
else:
logging.error("Flip column not found in DataFrame")
if "Top Offset" in df_new.columns:
offset_top_col = "Top Offset"
else:
offset_top_col = None
if "Flip Offset" in df_new.columns:
offset_flip_col = "Flip Offset"
else:
offset_flip_col = None
if "I0 BL" in df_new.columns:
i0_col = "I0 BL"
elif "Counter 2" in df_new.columns:
i0_col = "Counter 2"
else:
i0_col = None
detector = diffractometer.detector
azimuth = diffractometer.azimuth
for (i, row) in df_new.iterrows():
if pol_col is None:
pol_value = diffractometer.polarization.value
else:
pol_value = row[pol_col]
if offset_top_col is None:
offset_top = diffractometer.offset_top
else:
offset_top = row[offset_top_col]
if offset_flip_col is None:
offset_flip = diffractometer.offset_flip
else:
offset_flip = row[offset_flip_col]
if i0_col is None:
i0_value = 1.
else:
i0_value = row[i0_col]
diffractometer_params = get_default_chamber_params()
diffractometer_params_new = dict({
"Beamline Energy": row[energy_col],
"EPU Polarization": pol_value,
"Bottom Rotary Seal": row["Bottom Rotary Seal"],
"Top Rotary Seal": row["Top Rotary Seal"],
"Flip": row[flip_col],
"Azimuth": azimuth,
"Top Offset": offset_top,
"Flip Offset": offset_flip,
"I0 BL": i0_value,
"Detector Mode": detector,
})
diffractometer_params = dict(
list( diffractometer_params.items() )
+ list( diffractometer_params_new.items() )
)
diffractometer = Diffractometer402(diffractometer_params)
if pol_col is not None:
diffractometer.polarization.value = row[pol_col]
# q_magnitude = 2 * q_photon * sin(twotheta / 2)
# (!) q_magnitude can be < 0 (if twotheta < 0)
q_magnitude = 2 * diffractometer.photon.q * np.sin(
np.deg2rad(diffractometer.twotheta / 2.)
)
# if (q_c > 0) and (q_a > 0) --> (incidence > (twotheta / 2))
# if (q_c < 0) and (q_a < 0) --> (incidence > (twotheta / 2))
angle_qa_from_qbc = (
diffractometer.incidence - (diffractometer.twotheta / 2.)
)
angle_qa_from_qbc_radians = np.deg2rad(angle_qa_from_qbc)
q_a = q_magnitude * np.sin(angle_qa_from_qbc_radians)
# (!) q_bc can be < 0 (if twotheta < 0)
q_bc = q_magnitude * np.cos(angle_qa_from_qbc_radians)
transverse_radians = np.deg2rad(diffractometer.transverse)
# if (q_c > 0) and (q_b > 0) --> (transverse > 0)
# if (q_c < 0) and (q_b < 0) --> (transverse > 0)
q_b = q_bc * np.sin(transverse_radians)
# sign(q_c) == sign(twotheta)
q_c = q_bc * np.cos(transverse_radians)
df_new.loc[i, "qa"] = q_a
df_new.loc[i, "qb"] = q_b
df_new.loc[i, "qc"] = q_c
logging.info("Index: {0:d}".format(i))
logging.info("\ttwotheta: {0:0.3f}".format(diffractometer.twotheta))
logging.info("\tincidence: {0:0.3f}".format(diffractometer.incidence))
logging.info("\ttransverse: {0:0.3f}".format(diffractometer.transverse))
logging.info("\tq_a: {0:0.3f}".format(q_a))
logging.info("\tq_b: {0:0.3f}".format(q_b))
logging.info("\tq_c: {0:0.3f}".format(q_c))
logging.info("\tq_bc: {0:0.3f}".format(q_bc))
logging.info("\tangle_qx_from_qyz: {0:0.3f}".format(angle_qa_from_qbc))
return(df_new)
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def calculate_hkl(df, lattice, inplace=False):
"""Calculate (h, k, L) for PANDAS DataFrame.
df: PANDAS DataFrame containing (qa, qb, qc) positions.
lattice: OrthoLatticeABC object.
inplace: False = returns copy of df; True = returns original df.
RETURNS: PANDAS DataFrame with additional (h, k, L) columns
"""
if inplace:
df_new = df
else:
df_new = df.copy()
if ( ("qa" not in df_new.columns)
or ("qb" not in df_new.columns)
or ("qc" not in df_new.columns)
):
logging.error("DataFrame must contain (qa, qb, qc) columns")
df_new["h"] = np.nan
df_new["k"] = np.nan
df_new["L"] = np.nan
for (i, row) in df_new.iterrows():
rlattice = OrthoReciprocalVector.fromQabc(
lattice, row["qa"], row["qb"], row["qc"])
df_new.loc[i, "h"] = rlattice.h
df_new.loc[i, "k"] = rlattice.k
df_new.loc[i, "L"] = rlattice.L
return(df_new)
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def make_hkl_scan(
lattice, hkl_start, hkl_stop=None, nsteps=None, diffract_params=None,
output_dir=None, output_file_base=None, output_file_ext=".scn",
export_energy=True, export_polarization=False,
energy_format="{:+0.1f}",
angle_format="{:+0.1f}",
rlu_format="{:+0.2f}",
**kwargs
):
"""Generate a motor scan file for a linear path through reciprocal space.
lattice: OrthoLatticeABC object
hkl_start: Starting (h,k,L) of linear path through reciprocal space
hkl_stop: Final (h,k,L) of linear path through reciprocal space
*) DEFAULT: hkl_start
nsteps: Number of steps along linear path through reciprocal space
*) DEFAULT: either 1 (hkl_stop == hkl_start), or else 2 steps
diffract_params: Motor settings for Diffractometer402 object
*) DEFAULT: Diffractometer402.default_chamber_params
output_dir: Directory path for motor scan file
*) DEFAULT: global __defaults__.output_path
output_file_base: File base name for motor scan file
*) DEFAULT: verbose informative file name
output_file_ext: File extension for motor scan file
export_energy: If True, scan file contains x-ray energy
export_polarization: If True, scan file contains x-ray polarization
RETURNS: PANDAS DataFrame containing scan meta information
!!! Side Effects !!!
*) output_dir is created if it does not already exist
*) motor scan file is created at specified path
"""
if hkl_stop is None:
hkl_stop = hkl_start
if nsteps is None:
if hkl_stop == hkl_start:
nsteps = 1
else:
nsteps = 2
q_start = OrthoReciprocalVector.fromHKL(lattice, **hkl_start._asdict())
q_stop = OrthoReciprocalVector.fromHKL(lattice, **hkl_stop._asdict())
if diffract_params is None:
diffract_params = get_default_chamber_params()
diffractometer = Diffractometer402(
param_dict=diffract_params,
)
energy = diffractometer.energy
polarization = diffractometer.polarization
if (polarization.state == Polarization.LINEAR):
pol_format = "{:0.0f}"
else:
pol_format = "{:+0.2f}"
offset_angles = Diffractometer402(
param_dict=get_default_chamber_params(),
)
offset_angles.incidence = lattice.offsets.incidence
offset_angles.transverse = lattice.offsets.transverse
offset_angles.azimuth = lattice.offsets.azimuth
qpath = QSpacePath(
diffractometer=diffractometer,
offset_diffractometer=offset_angles)
qpath.lin_path(
np.array(q_start.qABC()),
np.array(q_stop.qABC()),
nsteps
)
if output_dir is None:
output_dir = get_output_dir()
if output_file_base is None:
output_file_base = "{name}_phi{azim}_{temp}K"
if export_energy:
output_file_base += "_{hv}eV"
if export_polarization:
output_file_base += "_pol{pol}"
output_file_base += "__{h0}_{k0}_{L0}__{hN}_{kN}_{LN}"
output_file_path = "{dir}{file_base}{ext}".format(
dir = output_dir,
file_base = output_file_base,
ext = output_file_ext,
).format(
azim = angle_format.format(lattice.offsets.azimuth),
hv = energy_format.format(energy),
pol = pol_format.format(polarization.value),
h0 = "H" + rlu_format.format(q_start.h),
k0 = "K" + rlu_format.format(q_start.k),
L0 = "L" + rlu_format.format(q_start.L),
hkL0 = "H{0}_K{0}_L{0}".format(rlu_format).format(*q_start.hkL()),
hN = "H" + rlu_format.format(q_stop.h),
kN = "K" + rlu_format.format(q_stop.k),
LN = "L" + rlu_format.format(q_stop.L),
hkLN = "H{0}_K{0}_L{0}".format(rlu_format).format(*q_stop.hkL()),
**kwargs
)
qpath.export_scanfile(
output_file_path,
export_energy=export_energy,
export_polarization=export_polarization)
df = import_scan_file(output_file_path)
if ( ("Beamline Energy" not in df.columns)
and ("Mono Energy" not in df.columns)
and ("EPU Energy" not in df.columns)
):
df["Mono Energy"] = energy
df["EPU Energy"] = energy
calculate_qABC(df, diffractometer=diffractometer, inplace=True)
logging.info("Q values calculated from scan file {}:\n{}".format(
output_file_path,
df,
)
)
calculate_hkl(df, lattice=lattice, inplace=True)
logging.info("RLU values calculated from scan file {}:\n{}".format(
output_file_path,
df,
)
)
df.to_csv(
"{path_base}_HKL{ext}".format(
path_base = output_file_path.split(output_file_ext)[0],
ext = output_file_ext,
),
sep = '\t',
index = False,
# line_terminator = "\r\n",
line_terminator = "\n",
)
return(df)
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def make_constQ_energy_scan(
lattice, hkl_point, energy_values, diffract_params=None,
output_dir=None, output_file_base=None, output_file_ext=".scn",
polarizations = None, alternate_polarization = False,
export_polarization=True,
**kwargs
):
"""Generate a motor scan file for a linear path through reciprocal space.
lattice: OrthoLatticeABC object
hkl_point: Constant (h,k,L) indices of reciprocal space vector
energy_values: Ordered list of x-ray energy values to use for scan
diffract_params: Motor settings for Diffractometer402 object
*) DEFAULT: Diffractometer402.default_chamber_params
output_dir: Directory path for motor scan file
*) DEFAULT: global __defaults__.output_path
output_file_base: File base name for motor scan file
*) DEFAULT: verbose informative file name
output_file_ext: File extension for motor scan file
polarizations: Ordered list of polarization values to use for scan
alternate_polarization: If True, x-ray polarization alternates
... between supplied values for each x-ray energy;
If False, polarization changes for each subsequent energy sub-scan
export_polarization: If True, scan file contains x-ray polarization
RETURNS: PANDAS DataFrame containing scan meta information
!!! Side Effects !!!
*) output_dir is created if it does not already exist
*) motor scan file is created at specified path
"""
q_point = OrthoReciprocalVector.fromHKL(lattice, **hkl_point._asdict())
if diffract_params is None:
diffract_params = get_default_chamber_params()
diffractometer = Diffractometer402(
param_dict=diffract_params,
)
offset_angles = Diffractometer402(
param_dict=get_default_chamber_params(),
)
offset_angles.incidence = lattice.offsets.incidence
offset_angles.transverse = lattice.offsets.transverse
offset_angles.azimuth = lattice.offsets.azimuth
res_profile = ResonanceProfile(
np.array(q_point.qABC()),
diffractometer=diffractometer,
offset_diffractometer=offset_angles)
res_profile.new_spectrum(energy_values)
if output_dir is None:
output_dir = get_output_dir()
if output_file_base is None:
output_file_base = "{name}_phi{azim}_{temp}K"
output_file_base += "_{h0}_{k0}_{L0}"
output_file_path = "{dir}{file_base}{ext}".format(
dir = output_dir,
file_base = output_file_base,
ext = output_file_ext,
).format(
azim = "{:+0.1f}".format(lattice.offsets.azimuth),
h0 = "H{:+0.2f}".format(q_point.h),
k0 = "K{:+0.2f}".format(q_point.k),
L0 = "L{:+0.2f}".format(q_point.L),
hkL0 = "H{:+0.2f}_K{:+0.2f}_L{:+0.2f}".format(*q_point.hkL()),
**kwargs
)
res_profile.export_scanfile(
output_file_path,
polarizations = polarizations,
alternate_polarization = alternate_polarization)
return
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
def main(args=None):
"""The main routine."""
if args is None:
args = sys.argv[1:]
for arg in args:
# if arg.lower() == "--version":
# print __version__
pass
print("Usage details")
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
if __name__ == '__main__':
main() | ALS.Milo | /ALS.Milo-0.18.1.tar.gz/ALS.Milo-0.18.1/als/milo/qscan.py | qscan.py |
__version__ = None # This will be assigned later; see below
__date__ = None # This will be assigned later; see below
__credits__ = None # This will be assigned later; see below
try:
from als.milo._version import git_pieces_from_vcs as _git_pieces_from_vcs
from als.milo._version import run_command, register_vcs_handler
from als.milo._version import render as _render
from als.milo._version import render_pep440_auto
from als.milo._version import render_pep440_micro, render_pep440_develop
from als.milo._version import get_versions as _get_versions
from als.milo._version import get_config, get_keywords
from als.milo._version import git_versions_from_keywords
from als.milo._version import versions_from_parentdir
from als.milo._version import NotThisMethod
except ImportError:
# Assumption is that _version.py was generated by 'versioneer.py'
# for tarball distribution, which contains only static JSON version data
from als.milo._version import get_versions
# from als.milo._version import get_versions as _get_versions
#
# def get_versions():
# """Get version information or return default if unable to do so.
#
# Extension to ._version.get_versions()
#
# Additional functionality:
# Returns list of authors found in `git`
# """
# default_keys_values = {
# "version": "0+unknown",
# "full-revisionid": None,
# "dirty": None,
# "error": "unable to compute version",
# "date": None,
# "authors": [],
# }
#
# return_key_values = _get_versions()
# return_key_values = dict(
# default_keys_values.items() + return_key_values.items()
# )
# return return_key_values
else:
import os
import sys
import numpy as np
@register_vcs_handler("git", "pieces_from_vcs")
def git_pieces_from_vcs(
tag_prefix, root, verbose, run_command=run_command):
"""Get version information from 'git' in the root of the source tree.
Extension to ._version.git_pieces_from_vcs()
Additional functionality:
Extracts all commit authors, sorts unique authors chronologically,
then adds them to `pieces["authors"]`, where `pieces` is the object
that was returned by ._version.git_pieces_from_vcs()
"""
pieces = _git_pieces_from_vcs(
tag_prefix, root, verbose, run_command=run_command)
GITS = ["git"]
if sys.platform == "win32":
GITS = ["git.cmd", "git.exe"]
##################################################
# Added to retrieve list of authors
(authors_raw, rc) = run_command(
GITS, ["log", "--pretty=%an"], cwd=root)
authors = [author.strip() for author in authors_raw.split('\n')]
(authors_unique, authors_indices) = np.unique(
authors, return_index=True)
pieces["authors"] = list(reversed(np.array(authors)[authors_indices]))
return pieces
def render(pieces, style):
"""Render the given version pieces into the requested style."""
if pieces["error"]:
return {"version": "unknown",
"full-revisionid": pieces.get("long"),
"dirty": None,
"error": pieces["error"],
"date": None,
"authors": None,
}
if not style or style == "default":
style = "pep440-auto" # the default
if style == "pep440-micro":
rendered = render_pep440_micro(pieces)
elif style == "pep440-develop":
rendered = render_pep440_develop(pieces)
elif style == "pep440-auto":
rendered = render_pep440_auto(pieces)
else:
return_key_values = _render(pieces, style)
return_key_values["authors"] = pieces["authors"]
return return_key_values
return {"version": rendered, "full-revisionid": pieces["long"],
"dirty": pieces["dirty"], "error": None,
"date": pieces.get("date"), "authors": pieces["authors"]}
def get_versions():
"""Get version information or return default if unable to do so.
Extension to ._version.get_versions()
Additional functionality:
Returns list of authors found in `git`
"""
# I am in _version.py, which lives at ROOT/VERSIONFILE_SOURCE.
# If we have __file__, we can work backwards from there to the root.
# Some py2exe/bbfreeze/non-CPython implementations don't do __file__,
# in which case we can only use expanded keywords.
cfg = get_config()
verbose = cfg.verbose
default_keys_values = {
"version": "0+unknown",
"full-revisionid": None,
"dirty": None,
"error": "unable to compute version",
"date": None,
"authors": [],
}
try:
return git_versions_from_keywords(get_keywords(), cfg.tag_prefix,
verbose)
except NotThisMethod:
pass
try:
root = os.path.realpath(__file__)
# versionfile_source is the relative path from the top of the
# source tree (where the .git directory might live) to this file.
# Invert this to find the root from __file__.
for i in cfg.versionfile_source.split('/'):
root = os.path.dirname(root)
except NameError:
return default_keys_values
try:
pieces = git_pieces_from_vcs(cfg.tag_prefix, root, verbose)
return render(pieces, cfg.style)
except NotThisMethod:
pass
try:
if cfg.parentdir_prefix:
return versions_from_parentdir(
cfg.parentdir_prefix, root, verbose)
except NotThisMethod:
pass
return_key_values = _get_versions()
return_key_values = dict(
list( default_keys_values.items() )
+ list( return_key_values.items() )
)
return return_key_values
__version__ = get_versions()["version"]
__date__ = get_versions()["date"]
__credits__ = get_versions()["authors"]
del get_versions | ALS.Milo | /ALS.Milo-0.18.1.tar.gz/ALS.Milo-0.18.1/als/milo/version.py | version.py |
<h1 align="center">ALSHBG</h1>
<p align="center">It is a project that includes all social networking sites, and the most important features of the project is a private library whose work is to verify the availability of the user name if it is available or not available on more than one social site</p>
## Installation :
```
pip install ALSHBG
```
## ***The method of work***
* Open a file in a text editor and name the file eg
ALSHBG.py
For example, we will check the username on Tiktok
```python
python3 ALSHBG.py
from ALSHBG import check
ck = check()
BG = ck.Tiktok("username")
print(BG)
```
* If the result is 200, this indicates that your Tiktok username is available☑️
* If the result is 404, this indicates that the Tiktok username is not available ❎
## ***The sites available in the project are***
* Instagram
* Tiktok
* Snapchat
* Telegramg
* Tellonym
* Twitch
* Xbox
* github
* Reddit
* Gmail
* Hotmail
* Yahoo
* Steam
* Sony
* Like
## Follow us on social media
[](https://t.me/ALSH_3K)
[](https://youtube.com/channel/UCUNbzQRjfAXGCKI1LY72DTA)
[](https://Instagram.com/alsh_bg)
[](https://t.me/XXCBB)
| ALSHBG | /ALSHBG-0.0.5.tar.gz/ALSHBG-0.0.5/README.md | README.md |
<h1 align="center">ALU-PYQUEST🐍💻</h1>
<img src="https://github.com/Elhameed/ALU-PYQUEST/blob/main/img/pythonquiz.jpg" />
🐍 ALU PYQUEST 🎮 is a game designed to 🧐 test your knowledge of Python programming language. It consists of a series of 🤔 questions related to the Python curriculum, presented one at a time ⏰, starting from beginner's level to advanced level. You will have a limited time ⏳ to answer each question, and for each ✅ correct answer, you will earn 🔟 points, while for each ❌ incorrect answer or if you run out of time ⌛, 5 points will be deducted from your score.
## Table of Contents 📑
- [Project Overview](#project-overview)
- [Prerequisites](#prerequisites)
- [Installation](#installation)
- [Updating ALU-PyQuest](#updating-alu-pyquest)
- [How To Play](#how-to-play)
- [Screenshots](#screenshots)
- [Demo Video](#demo-video)
- [License](#license)
- [Contributors](#contributors)
- [Acknowlegdement](#acknowledgement)
<h2 id="project-overview">Project Overview 📋</h2>
🎮 "ALU PyQuest" 🐍 is a Python-based scavenger hunt-style game designed for Software Engineering students to reinforce their understanding of the Python curriculum. 🕵️♀️ The game presents a series of questions from beginner's to advanced level, with a limited time ⏰ to answer each question. The program utilizes Python functions such as Input/Output, loops, conditional statements, data structures, exception handling, scoring system 📈, and a high score feature 🏆 to check your scores and provide an interactive and fun learning experience for students 🎓.
<h2 id="prerequisites">Prerequisites 📚</h2>
Before installing ALU-PYQUEST, you will need to have pip installed. To check if you have pip installed, open your terminal window and run the following command:
```sh
pip --version
```
If you see a version number in the output, it means that pip is installed. If not, you can install pip by running the following commands in your terminal:
```sh
sudo apt-get update
sudo apt-get install python3-pip
```
Verify that pip has been installed correctly by running:
```sh
pip --version
```
You should see something like:
```sh
user@User:~$ pip --version
pip 20.0.2 from /usr/lib/python3/dist-packages/pip (python 3.8)
user@User:~$
```
<h2 id="installation">Installation 💻</h2>
To install ALU-PYQUEST, please follow the steps below:
1. Make sure you have Python installed on your computer. If you do not have Python installed, download and install the latest version of Python from the official website: [https://www.python.org/downloads/](https://www.python.org/downloads/)
2. Open the command prompt or terminal on your computer 💻.
3. Install ALU-PyQuest using the following command:
```sh
pip install ALU-PyQuest
```
4. Once the installation is complete, you can launch the game by using the following command:
```sh
PyQuest
```
5. The game 🎮 will start, and you will be presented with a series of questions related to the Python programming language 🐍 from beginner to advanced level 📈.
That's it! You're all set to play ALU-PyQuest 🎉.
<h2 id="updating-alu-pyquest">Updating ALU-PyQuest 🔄</h2>
To update ALU-PYQUEST to the latest version, you can run the following command in your terminal or command prompt:
```sh
pip install --upgrade ALU-PYQUEST
```
This will update your installation of ALU-PYQUEST to the latest version available on PyPI.
<h2 id="how-to-play">How To Play 🎮</h2>
1. Open the command prompt or terminal on your computer.
2. Run the following command to start the game:
```sh
user@User:~/$ PyQuest
```
4. Read the question and choose the correct answer from the given options.
5. You have `25` seconds to answer each question.
6. For each correct answer, you will earn `10` points.
7. For each incorrect answer or if you run out of time, `5` points will be deducted from your score.
8. At the end of the game, you will see your total score.
9. You will also be provided with the option to view your high scores.
<h2 id="screenshots">Screenshots 📷</h2>
<div>
<img src="https://github.com/Elhameed/ALU-PYQUEST/blob/main/img/screenshot1.PNG" />
<img src="https://github.com/Elhameed/ALU-PYQUEST/blob/main/img/screenshot2.PNG" />
<img src="https://github.com/Elhameed/ALU-PYQUEST/blob/main/img/screenshot3.PNG" />
</div>
<h2 id="demo-video">Demo Video 🎥</h2>
You can find a demo of the game at the following link: <a href="https://youtu.be/ym5cIpgxvMQ" target="_blank">https://youtu.be/ym5cIpgxvMQ</a>
<h2 id="license">License 📄</h2>
ALU-PYQUEST is licensed under the MIT License. See the [LICENSE](./LICENSE) file for more information.
<h2 id="contributors">Contributors 👥</h2>
- [Sadick Achuli](https://github.com/Sadickachuli)
- [Abdulhameed Teniola Ajani](https://github.com/Elhameed)
- [Noella Uwayo](https://github.com/n-uwayo)
- [Sabir Walid](https://github.com/SabirWalid)
- [Mohammed Yasin](https://github.com/MohamedAYasin)
- [Iranzi Prince](https://github.com/iranziprince01)
- [Innocent Manzi](https://github.com/innocentmanzi)
- [Iraduhaye Bukuru Paterne](https://github.com/IraduhayeBukuruPaterne1)
<h2 id="acknowledgement">Acknowledgement 🙏</h2>
- 👏🏼 Special thanks to our instructor, [Mr. Hervé Musangwa](https://www.linkedin.com/in/hervé-musangwa-67478a112/) for reviewing our project and providing feedback that helped improve its quality.
- 👨🏫 We also want to acknowledge the BSE Faculty for challenging us to apply the technologies we learned during the term to develop a project that showcases our skills and knowledge in the field.
----------
- If you found ALU-PYQUEST useful, please consider giving this repo a star ⭐️!
| ALU-PyQuest | /ALU-PyQuest-0.1.9.tar.gz/ALU-PyQuest-0.1.9/README.md | README.md |
PROBLEM_SETS = [
{
"question": "What is the difference between an instance attribute and a class attribute in Python?",
"options": {
"a": "Instance attributes are defined within a method, while class attributes are defined outside of a method.",
"b": "Instance attributes are accessible only within the class, while class attributes are accessible outside of the class.",
"c": "Instance attributes are specific to an instance of a class, while class attributes are shared among all instances of a class.",
"d": "There is no difference between the two types of attributes."
},
"answer": "c"
},
{
"question": "What is the difference between an import statement and a from-import statement in Python?",
"options": {
"a": "An import statement imports an entire module, while a from-import statement imports a specific function or class from a module.",
"b": "An import statement is used for built-in Python modules, while a from-import statement is used for user-defined modules.",
"c": "An import statement is used for user-defined modules, while a from-import statement is used for built-in Python modules.",
"d": "There is no difference between the two types of statements."
},
"answer": "a"
},
{
"question": "What is the output of the following code?\n import math\n print(math.ceil(4.2))",
"options": {
"a": "4",
"b": "5",
"c": "6",
"d": "Error"
},
"answer": "b"
},
{
"question": "What is the output of the following code?\n try:\n x = 1 / 0\nexcept ZeroDivisionError:\n print('Division by zero.')",
"options": {
"a": "Division by zero.",
"b": "ZeroDivisionError.",
"c": "None",
"d": "Error"
},
"answer": "a"
},
{
"question": "What is a method in Python?",
"options": {
"a": "A function that is defined inside a class.",
"b": "A function that is defined outside a class.",
"c": "A variable that is defined inside a class.",
"d": "A variable that is defined outside a class."
},
"answer": "a"
},
{
"question": "What is the output of the following code?\nclass Person:\n def init(self, name, age):\n self.name = name\n self.age = age\n\nperson = Person('John', 25)\nprint(person.name)",
"options": {
"a": "John",
"b": "25",
"c": "Person",
"d": "Error"
},
"answer": "a"
},
{
"question": "What is the output of the following code?\nclass A:\n def init(self):\n self.x = 1\n\nclass B(A):\n def init(self):\n super().init()\n self.y = 2\n\nb = B()\nprint(b.x + b.y)",
"options": {
"a": "1",
"b": "2",
"c": "3",
"d": "Error"
},
"answer": "c"
},
{
"question": "What is the difference between a set and a list in Python?",
"options": {
"a": "A set is immutable and a list is mutable.",
"b": "A set is mutable and a list is immutable.",
"c": "Both sets and lists are immutable.",
"d": "Both sets and lists are mutable."
},
"answer": "b"
},
{
"question": "What is the output of the following code?\n\n class Animal:\n\t def init(self, name):\n\t\t self.name = name\n\n class Dog(Animal):\n\t\t def bark(self):\n\t\t\t person1 = Person(\"John\", 25)\n\t\t\t print(person1.name)\n",
"options": {
"a": "age 25",
"b": "john 25",
"c": "John",
"d": "None of the above"
},
"answer": "d"
},
{
"question": "What is the output of the following code?\n\n class Animal:\n\t def __init__(self, name):\n\t self.name = name\n\n class Dog(Animal):\n\tdef bark(self):\n\tprint('Woof!')\n\nmy_dog = Dog('Fido')\nprint(my_dog.name)\nmy_dog.bark()\n",
"options": {
"a": "Woof",
"b": "error",
"c": "Fido",
"d": "Woof! Fido"
},
"answer": "d"
},
{
"question": "What's the output of the following Python code?\n\ndef add_numbers(x, y):\n\t return x + y\n\nresult = add_numbers(5, 7)\nprint(result)",
"options": {
"a": "10",
"b": "11",
"c": "12",
"d": "13"
},
"answer": "c"
},
{
"question": "How do I use functions in Python?",
"options": {
"a": "By using the def keyword to define a function and calling the function by passing arguments",
"b": "By using the for loop to define a function and calling the function by passing arguments",
"c": "By using the if-else statement to define a function and calling the function by passing arguments",
"d": "None of the above"
},
"answer": "a"
},
{
"question": "How can I deal with modules in Python?",
"options": {
"a": "Using import statement",
"b": "Using from...import statement",
"c": "Using as statement",
"d": "All of the above"
},
"answer": "d"
},
{
"question": "What is the output of the following code?\n\ndef greet(name):\n\tprint('Hello, ' + name + '!')\n\nimport mymodule\n\nmymodule.greet('Alice')\n",
"options": {
"a": "Hello, mymodule!",
"b": "Hello, Alice!",
"c": "Hello, World!",
"d": "An error occurs"
},
"answer": "b"
},
{
"question": "What is the output of the following code?\n\nx = 5\ny = 10\nx, y = y, x\nprint(x, y)",
"options": {
"a": "5 10",
"b": "10 5",
"c": "Error",
"d": "None of the above"
},
"answer": "b"
},
{
"question": "What is the output of the following code?\n\nmy_list = [1, 2, 3, 4, 5]\nprint(my_list[::2])",
"options": {
"a": "[1, 3, 5]",
"b": "[2, 4]",
"c": "[1, 2, 3, 4, 5]",
"d": "[1, 4]"
},
"answer": "a"
},
{
"question": "What does the map() function do in Python?",
"options": {
"a": " It applies a function to each element of an iterable and returns an iterator.",
"b": "It combines two or more iterables into a single iterator.",
"c": "It sorts an iterable in ascending or descending order.",
"d": "It creates a dictionary from an iterable."
},
"answer": "a"
},
{
"question": "What is the purpose of the pass keyword in Python?",
"options": {
"a": "It is used to continue to the next iteration of a loop",
"b": "It is used to break out of a loop",
"c": "It is used to define a function without any content.",
"d": "It is used to raise an exception."
},
"answer": "c"
},
{
"question": "What does the join() method do in Python?",
"options": {
"a": "Concatenates two strings together.",
"b": "Removes all whitespace from a string.",
"c": "Splits a string into a list of substrings.",
"d": "Concatenates a list of strings into a single string with a delimiter between each element."
},
"answer": "d"
},
{
"question": "What is the difference between a local variable and a global variable in Python?",
"options": {
"a": "A local variable is declared inside a function and can be accessed anywhere in the program, while a global variable is declared outside a function and can only be accessed within the function where it is declared.",
"b": "A local variable is declared inside a function and can only be accessed within that function, while a global variable is declared outside a function and can be accessed anywhere in the program.",
"c": "A local variable can only be assigned a value once, while a global variable can be assigned multiple values.",
"d": "A local variable is declared using the local keyword, while a global variable is declared using the global keyword."
},
"answer": "b"
},
{
"question": "Which of the following is a valid way to open a file in Python for writing?",
"options": {
"a": "file = open(\"myfile.txt\", \"rb\")",
"b": "file = open(\"myfile.txt\", \"w\")",
"c": "file = open(\"myfile.txt\", \"read\")",
"d": "file = open(\"myfile.txt\", \"a+\")"
},
"answer": "b"
},
{
"question": "Which of the following is NOT a valid numeric type in Python?",
"options": {
"a": "int",
"b": "float",
"c": "double",
"d": "complex"
},
"answer": "c"
},
{
"question": "What is the output of the following code?\n\nfor i in range(2, 10, 2):\n print(i)",
"options": {
"a": "2 4 6 8",
"b": "2 4 8",
"c": "2 6",
"d": "4 8"
},
"answer": "a"
},
{
"question": "What is the value of x after the following code executes?\n\nx = 'Python'\nx += ' is great!'\n",
"options": {
"a": "'Python is great!'",
"b": "'Pythonis great!'",
"c": "'Python isgreat!'",
"d": "'Python is great'"
},
"answer": "a"
},
{
"question 6": "What is the difference between a list and a tuple in Python?",
"options": {
"a": "Lists are immutable while tuples are mutable",
"b": "Tuples are immutable while lists are mutable",
"c": "Lists and tuples are both immutable",
"d": "Lists and tuples are both mutable"
},
"answer": "b"
},
{
"question": "What does the map() function do in Python?",
"options": {
"a": " It applies a function to each element of an iterable and returns an iterator.",
"b": "It combines two or more iterables into a single iterator.",
"c": "It sorts an iterable in ascending or descending order.",
"d": "It creates a dictionary from an iterable."
},
"answer": "a"
},
{
"question": "What is the purpose of the pass keyword in Python?",
"options": {
"a": "It is used to continue to the next iteration of a loop",
"b": "It is used to break out of a loop",
"c": "It is used to define a function without any content.",
"d": "It is used to raise an exception."
},
"answer": "c"
},
{
"question": "What does the join() method do in Python?",
"options": {
"a": "Concatenates two strings together.",
"b": "Removes all whitespace from a string.",
"c": "Splits a string into a list of substrings.",
"d": "Concatenates a list of strings into a single string with a delimiter between each element."
},
"answer": "d"
},
{
"question": "What is the difference between a local variable and a global variable in Python?",
"options": {
"a": "A local variable is declared inside a function and can be accessed anywhere in the program, while a global variable is declared outside a function and can only be accessed within the function where it is declared.",
"b": "A local variable is declared inside a function and can only be accessed within that function, while a global variable is declared outside a function and can be accessed anywhere in the program.",
"c": "A local variable can only be assigned a value once, while a global variable can be assigned multiple values.",
"d": "A local variable is declared using the local keyword, while a global variable is declared using the global keyword."
},
"answer": "b"
},
{
"question": "Which of the following is a valid way to open a file in Python for writing?",
"options": {
"a": "file = open(\"myfile.txt\", \"rb\")",
"b": "file = open(\"myfile.txt\", \"w\")",
"c": "file = open(\"myfile.txt\", \"read\")",
"d": "file = open(\"myfile.txt\", \"a+\")"
},
"answer": "b"
},
{
"question": "Which of the following is NOT a valid way to create an empty list in Python?",
"options": {
"a": "my_list = []",
"b": "my_list = list()",
"c": "my_list = list([])",
"d": "my_list = ()"
},
"answer": "d"
},
{
"question": "What is the output of the following code?\n\nimport math\nprint(math.pi)",
"options": {
"a": "3.14",
"b": "3.14159265359",
"c": "22/7",
"d": "Undefined"
},
"answer": "b"
},
{
"question": "Which keyword is used in Python to handle exceptions?",
"options": {
"a": "throw",
"b": "except",
"c": "catch",
"d": "try"
},
"answer": "d"
},
{
"question": "Which of the following is an example of inheritance in Python?",
"options": {
"a": "class Car:\n def init(self):\n self.color = 'red'\n\nclass SportsCar(Car):\n def init(self):\n super().init()\n self.top_speed = 200",
"b": "class Animal:\n def init(self):\n self.name = 'Animal'\n def make_sound(self):\n print('generic animal sound')\n\nclass Dog(Animal):\n def make_sound(self):\n print('bark')",
"c": "class Rectangle:\n def init(self, width, height):\n self.width = width\n self.height = height\n\nclass Square(Rectangle):\n def init(self, side_length):\n super().init(side_length, side_length)",
"d": "class Person:\n def init(self, name):\n self.name = name\n\nperson1 = Person('Alice')\nperson2 = Person('Bob')"
},
"answer": "a"
},
{
"question": "What is the output of the following code?\n\nclass MyClass:\n def init(self):\n self.my_var = 42\n\nmy_obj = MyClass()\nprint(my_obj.my_var)",
"options": {
"a": "MyClass",
"b": "my_obj",
"c": "42",
"d": "Undefined"
},
"answer": "c"
},
{
"question": "What is the purpose of the __init__ method in a Python class?",
"options": {
"a": "To create an instance of the class.",
"b": "To define the class hierarchy.",
"c": "To define class methods.",
"d": "To initialize the attributes of an object created from the class."
},
"answer": "d"
},
{
"question": "What is the output of the following code?\n\nmy_list = [1, 2, 3, 4]\nprint(my_list[2:])",
"options": {
"a": "[1, 2]",
"b": "[3, 4]",
"c": "[2, 3, 4]",
"d": "[1, 2, 3]"
},
"answer": "b"
},
{
"question": "What is the purpose of the super() function in Python?",
"options": {
"a": "To call the parent class method from within a subclass method.",
"b": "To create a new instance of a class.",
"c": "To access the attributes of a superclass.",
"d": "To define a new class."
},
"answer": "a"
},
{
"question": "Which of the following is NOT a valid way to import a module in Python?",
"options": {
"a": "import my_module",
"b": "from my_module import my_function",
"c": "import my_module.my_function",
"d": "from my_module import *"
},
"answer": "c"
},
{
"question": "What do these lines print?\n\nclass User:\n\tid = 1:\n\n\n User.id = 98\n\nu = User\nu.id = 89\nprint(u.id)",
"options": {
"a": "89",
"b": "1",
"c": "none",
"d": "100"
},
"answer": "a"
},
{
"question": "Which one of the following data structures in python is immutable?",
"options": {
"a": "Tuple",
"b": "Set",
"c": "Dictionary",
"d": "List"
},
"answer": "a"
},
{
"question": "Which of these definitions correctly describes a module?",
"options": {
"a": "Any program that reuses code",
"b": "Defines the specification of how it is to be used",
"c": "Denoted by triple quotes for providing the specification of certain program elements",
"d": "Design and implementation of specific functionality to be incorporated into a program"
},
"answer": "d"
},
{
"question": "To include the use of functions which are present in the random library, we must use the option:",
"options": {
"a": "random.h",
"b": "import.random",
"c": "import random",
"d": "random.random"
},
"answer": "c"
},
{
"question": "The output of the following Python code is either 1 or 2.\nimport random\nrandom.randint(1,2)",
"options": {
"a": "True",
"b": "False"
},
"answer": "a"
},
{
"question": "What is the output of the following code set([1, 2, 3, 2, 1])?",
"options": {
"a": "set([1, 2, 3])",
"b": "set[1, 2, 3, 2, 1]",
"c": "{1, 2, 3, 2, 1}",
"d": "{1, 2, 3}"
},
"answer": "d"
},
{
"question": "What do these lines print?\na = [5, 7, 9, 11]\na[-1]",
"options": {
"a": "-1",
"b": "[11, 9, 7, 5]",
"c": "11",
"d": "7"
},
"answer": "c"
},
{
"question": "What do these lines print?\nfor i in range(5, 8):\n print(i, end=\" \")",
"options": {
"a": "5 6 7 8",
"b": "5 6 7",
"c": "0 5 6 7",
"d": "5,6,7"
},
"answer": "b"
},
{
"question": "What does this python program output?\ndef dem(name, age):\n print(name, age)\ndem(\"Ben\", 25)",
"options": {
"a": "25",
"b": "Ben",
"c": "Ben 25",
"d": "error"
},
"answer": "c"
},
{
"question": "Which of the following is correct?",
"options": {
"a": "defunct(a, b)",
"b": "def add(a, b):",
"c": "define add(a + b):",
"d": "define add(a + b)"
},
"answer": "b"
},
{
"question": "What do these lines print?\na = { 'id': 89, 'name': \"John\" }\na.get('age', 0)",
"options": {
"a": "'age'",
"b": "89",
"c": "0",
"d": "Nothing"
},
"answer": "c"
},
{
"question": "What do these lines print?\nfor i in [1, 3, 4, 2]:\n\tprint(i, end=\"-\")",
"options": {
"a": "1 2 3 4",
"b": "1, 3, 4, 2, 0",
"c": "1--3--4--2",
"d": "1-3-4-2"
},
"answer": "d"
},
{
"question": "What do these lines print?\na = { 'id': 89, 'name': \"John\" }\na.get('age')",
"options": {
"a": "'age'",
"b": "89",
"c": "12",
"d": "Nothing"
},
"answer": "d"
},
{
"question": "Which of the following data structure in python does not allow duplicates?",
"options": {
"a": "Tuple",
"b": "Set",
"c": "Dictionary",
"d": "List"
},
"answer": "c"
},
{
"question": "What is setattr() used for?",
"options": {
"a": "To access the attribute of the object",
"b": "To set an attribute",
"c": "To check if an attribute exists or not",
"d": "To delete an attribute"
},
"answer": "b"
},
{
"question": "Which of the following is true about dictionaries in python?",
"options": {
"a": "Dictionaries are ordered",
"b": "Dictionaries can have duplicate keys",
"c": "Dictionaries can have mutable values",
"d": "Dictionaries can be accessed using any index"
},
"answer": "a"
},
{
"question": "What will be the output of the following Python program?\n\ni = 0\nwhile i < 5:\n\tprint(i)\ni += 1\nif i == 3:\n\tbreak\nelse:\n\tprint(0)",
"options": {
"a": "error",
"b": "0 1 2 0",
"c": "0 1 2",
"d": "none of the mentioned"
},
"answer": "c"
},
{
"question": "What is the output of the following code dict(zip(['a', 'b', 'c'], [1, 2, 3]))?",
"options": {
"a": "{'a': 1, 'b': 2, 'c': 3}",
"b": "{1: 'a', 2: 'b', 3: 'c'}",
"c": "[('a', 1), ('b', 2), ('c', 3)]",
"d": "{(1, 'a'), (2, 'b'), (3, 'c')}"
},
"answer": "a"
},
{
"question": "Which of the following method can be used to add an item to a set?",
"options": {
"a": "add()",
"b": "append()",
"c": "extend()",
"d": "insert()"
},
"answer": "a"
},
{
"question": "Which of the following methods can be used to remove an item from a set?",
"options": {
"a": "remove()",
"b": "pop()",
"c": "delete()",
"d": "discard()"
},
"answer": "b"
},
{
"question": "What will be the output of the following Python code?\n\nclass change:\n\tdef __init__(self, x, y, z):\n\tself.a = x + y + z\n\tx = change(1,2,3)\n\ty = getattr(x, 'a')\n\tsetattr(x, 'a', y+1)\n\tprint(x.a)",
"options": {
"a": "6",
"b": "7",
"c": "0",
"d": "Error"
},
"answer": "b"
},
{
"question": "Which of these definitions correctly describes a module?",
"options": {
"a": "Any program that reuses code",
"b": "Defines the specification of how it is to be used",
"c": "Denoted by triple quotes for providing the specification of certain program elements",
"d": "Design and implementation of specific functionality to be incorporated into a program"
},
"answer": "d"
},
{
"question": "To include the use of functions which are present in the random library, we must use the option:",
"options": {
"a": "random.h",
"b": "import.random",
"c": "import random",
"d": "random.random"
},
"answer": "c"
},
{
"question": "The output of the following Python code is either 1 or 2.\nimport random\nrandom.randint(1,2)",
"options": {
"a": "True",
"b": "False"
},
"answer": "a"
},
{
"question": "What do these lines print?\na = [5, 7, 9, 11]\na[-1]",
"options": {
"a": "-1",
"b": "[11, 9, 7, 5]",
"c": "11",
"d": "7"
},
"answer": "c"
},
{
"question": "What do these lines print?\nfor i in range(5, 8):\n print(i, end=\" \")",
"options": {
"a": "5 6 7 8",
"b": "5 6 7",
"c": "0 5 6 7",
"d": "5,6,7"
},
"answer": "b"
},
{
"question": "What does this python program output?\ndef dem(name, age):\n print(name, age)\ndem(\"Ben\", 25)",
"options": {
"a": "25",
"b": "Ben",
"c": "Ben 25",
"d": "error"
},
"answer": "c"
},
{
"question": "Which of the following is correct?",
"options": {
"a": "defunct(a, b)",
"b": "def add(a, b):",
"c": "define add(a + b):",
"d": "define add(a + b)"
},
"answer": "b"
},
{
"question": "What do these lines print?\na = { 'id': 89, 'name': \"John\" }\na.get('age', 0)",
"options": {
"a": "'age'",
"b": "89",
"c": "0",
"d": "Nothing"
},
"answer": "c"
},
{
"question": "What do these lines print?\nfor i in [1, 3, 4, 2]:\n\tprint(i, end=\"-\")",
"options": {
"a": "1 2 3 4",
"b": "1, 3, 4, 2, 0",
"c": "1--3--4--2",
"d": "1-3-4-2"
},
"answer": "d"
},
{
"question": "What do these lines print?\na = { 'id': 89, 'name': \"John\" }\na.get('age')",
"options": {
"a": "'age'",
"b": "89",
"c": "12",
"d": "Nothing"
},
"answer": "d"
},
{
"question": "What will be the output of the following Python code?\ni = 1\nwhile True:\n\tif i%3 == 0:\n\tbreak\nprint(i)\n\ni + = 1\n",
"options": {
"a": "1 2 3",
"b": "error",
"c": "1 2",
"d": "none of the mentioned"
},
"answer": "b"
},
{
"question": "The following python program can work with ____ parameters.\ndef f(x):\n\tdef f1(*args, **kwargs):\n\tprint(\"Sanfoundry\")\nreturn x(*args, **kwargs)\nreturn f1\n",
"options": {
"a": "any number of",
"b": "0",
"c": "1",
"d": "2"
},
"answer": "a"
},
{
"question": " What will be the output of the following Python function?\nmin(max(False,-3,-4), 2,7)\n",
"options": {
"a": "-4",
"b": "-2",
"c": "-3",
"d": "7"
},
"answer": "c"
},
{
"question": "What will be the output of the following Python code?\nx = 'abcd'\nfor i in x:\n\tprint(i.upper())\n",
"options": {
"a": "A b C d",
"b": "a b c d",
"c": "error",
"d": "A B C D"
},
"answer": "d"
},
{
"question": " What will be the output of the following Python code?\nx = 'abcd'\nfor i in range(len(x)):\n\tprint(i)\n",
"options": {
"a": "-4",
"b": "-2",
"c": "-3",
"d": "False"
},
"answer": "d"
},
{
"question": "What is a generator in Python?",
"options": {
"a": "The function that generates random numbers of specified count",
"b": "The function that generates an error when a warning occurs",
"c": "The function that gives a statement as output rather than a return",
"d": "The function that returns a generic object"
},
"answer": "c"
},
{
"question": "Which of the following is true about the stack?",
"options": {
"a": "Stack following LIFO order",
"b": "The time complexity to push and pop an element is O(1)",
"c": "It is a linear data Structure",
"d": "All of the above"
},
"answer": "d"
},
{
"question": "Which of these definitions correctly describes a module?",
"options": {
"a": "Any program that reuses code",
"b": "Defines the specification of how it is to be used",
"c": "Denoted by triple quotes for providing the specification of certain program elements",
"d": "Design and implementation of specific functionality to be incorporated into a program"
},
"answer": "d"
},
{
"question": "To include the use of functions which are present in the random library, we must use the option:",
"options": {
"a": "random.h",
"b": "import.random",
"c": "import random",
"d": "random.random"
},
"answer": "c"
},
{
"question": "The output of the following Python code is either 1 or 2.\nimport random\nrandom.randint(1,2)",
"options": {
"a": "True",
"b": "False"
},
"answer": "a"
},
{
"question": "Which of the following is true about tuple in python?",
"options": {
"a": "Tuple can be modified after creation",
"b": "Tuple can be used as key in a dictionary",
"c": "Tuple can be accessed using an index",
"d": "Tuple can have duplicate"
},
"answer": "c"
},
{
"question": "What do these lines print?\na = [5, 7, 9, 11]\na[-1]",
"options": {
"a": "-1",
"b": "[11, 9, 7, 5]",
"c": "11",
"d": "7"
},
"answer": "c"
},
{
"question": "What do these lines print?\nfor i in range(5, 8):\n print(i, end=\" \")",
"options": {
"a": "5 6 7 8",
"b": "5 6 7",
"c": "0 5 6 7",
"d": "5,6,7"
},
"answer": "b"
},
{
"question": "Which of the following methods can be used to open a file in python for writing?",
"options": {
"a": "open('filename.txt', 'r')",
"b": "open('filename.txt', 'w')",
"c": "open('filename.txt', 'a')",
"d": "open('filename.txt', 'x')"
},
"answer": "b"
},
{
"question": "What does this python program output?\ndef dem(name, age):\n print(name, age)\ndem(\"Ben\", 25)",
"options": {
"a": "25",
"b": "Ben",
"c": "Ben 25",
"d": "error"
},
"answer": "c"
},
{
"question": "Which of the following is correct?",
"options": {
"a": "defunct(a, b)",
"b": "def add(a, b):",
"c": "define add(a + b):",
"d": "define add(a + b)"
},
"answer": "b"
},
{
"question": "What do these lines print?\na = { 'id': 89, 'name': \"John\" }\na.get('age', 0)",
"options": {
"a": "'age'",
"b": "89",
"c": "0",
"d": "Nothing"
},
"answer": "c"
},
{
"question": "What do these lines print?\nfor i in [1, 3, 4, 2]:\n\tprint(i, end=\"-\")",
"options": {
"a": "1 2 3 4",
"b": "1, 3, 4, 2, 0",
"c": "1--3--4--2",
"d": "1-3-4-2"
},
"answer": "d"
},
{
"question": "What do these lines print?\na = { 'id': 89, 'name': \"John\" }\na.get('age')",
"options": {
"a": "'age'",
"b": "89",
"c": "12",
"d": "Nothing"
},
"answer": "d"
},
{
"question": "What does the join() method do in Python?",
"options": {
"a": "Concatenates two strings together.",
"b": "Removes all whitespace from a string.",
"c": "Splits a string into a list of substrings.",
"d": "Concatenates a list of strings into a single string with a delimiter between each element."
},
"answer": "d"
},
{
"question": "What is the output of the following code?\n\nfor i in range(2, 10, 2):\n print(i)",
"options": {
"a": "2 4 6 8",
"b": "2 4 8",
"c": "2 6",
"d": "4 8"
},
"answer": "a"
},
{
"question": "Which of these definitions correctly describes a module?",
"options": {
"a": "Any program that reuses code",
"b": "Defines the specification of how it is to be used",
"c": "Denoted by triple quotes for providing the specification of certain program elements",
"d": "Design and implementation of specific functionality to be incorporated into a program"
},
"answer": "d"
},
{
"question": "Which of the following methods can be used to open a file in python for writing?",
"options": {
"a": "open('filename.txt', 'r')",
"b": "open('filename.txt', 'w')",
"c": "open('filename.txt', 'a')",
"d": "open('filename.txt', 'x')"
},
"answer": "b"
},
{
"question": "What do these lines print?\nfor i in [1, 3, 4, 2]:\n\tprint(i, end=\"-\")",
"options": {
"a": "1 2 3 4",
"b": "1, 3, 4, 2, 0",
"c": "1--3--4--2",
"d": "1-3-4-2"
},
"answer": "d"
},
{
"question": "What do these lines print?\na = { 'id': 89, 'name': \"John\" }\na.get('age', 0)",
"options": {
"a": "'age'",
"b": "89",
"c": "0",
"d": "Nothing"
},
"answer": "c"
},
{
"question": "Which of the following is correct?",
"options": {
"a": "defunct(a, b)",
"b": "def add(a, b):",
"c": "define add(a + b):",
"d": "define add(a + b)"
},
"answer": "b"
},
{
"question": "To include the use of functions which are present in the random library, we must use the option:",
"options": {
"a": "random.h",
"b": "import.random",
"c": "import random",
"d": "random.random"
},
"answer": "c"
},
{
"question": "What do these lines print?\nfor i in [1, 3, 4, 2]:\n\tprint(i, end=\"-\")",
"options": {
"a": "1 2 3 4",
"b": "1, 3, 4, 2, 0",
"c": "1--3--4--2",
"d": "1-3-4-2"
},
"answer": "d"
},
{
"question": "What do these lines print?\na = { 'id': 89, 'name': \"John\" }\na.get('age')",
"options": {
"a": "'age'",
"b": "89",
"c": "12",
"d": "Nothing"
},
"answer": "d"
}
] | ALU-PyQuest | /ALU-PyQuest-0.1.9.tar.gz/ALU-PyQuest-0.1.9/src/medium_questions.py | medium_questions.py |
# advanced_questions.py
# Store advanced level questions as a list of dictionaries
PROBLEM_SETS = [
{
"question": "What is the output of the following code?\n\nmy_list = [1, 2, 3, 4, 5]\nnew_list = [x for x in my_list if x % 2 == 0]\nprint(new_list)",
"options": {
"a": "[2, 4]",
"b": "[1, 3, 5]",
"c": "[2, 4, 6]",
"d": "[1, 2, 3, 4, 5]"
},
"answer": "a"
},
{
"question": "What is the output of the following code?\n\ndef my_func(a, b, c):\n print(a, b, c)\n\nargs = (1, 2, 3)\nmy_func(*args)",
"options": {
"a": "1 2 3",
"b": "(1, 2, 3)",
"c": "TypeError: my_func() takes 3 positional arguments but 4 were given",
"d": "SyntaxError: invalid syntax"
},
"answer": "a"
},
{
"question": "What is the output of the following code?\n\ndef my_func(a, b, c=0):\n print(a, b, c)\n\nmy_func(1, 2)",
"options": {
"a": "1 2 0",
"b": "1 2",
"c": "TypeError: my_func() missing 1 required positional argument: 'b'",
"d": "SyntaxError: invalid syntax"
},
"answer": "a"
},
{
"question": "What is a metaclass in Python?",
"options": {
"a": "A class used to define the behavior of other classes.",
"b": "A class used to define the behavior of other metaclasses.",
"c": "A class used to define the behavior of class instances.",
"d": "A class used to define the behavior of function calls."
},
"answer": "a"
},
{
"question": "What is the purpose of the __slots__ attribute in Python classes?",
"options": {
"a": "It is used to define a set of allowed attributes for a class instance",
"b": "It is used to define a set of forbidden attributes for a class instance",
"c": "It is used to define a set of class-level attributes",
"d": "It is used to define a set of static attributes."
},
"answer": "a"
},
{
"question": "Which of the following is not a Python built-in module for working with files?",
"options": {
"a": "os",
"b": "io",
"c": "pathlib",
"d": "file"
},
"answer": "d"
},
{
"question": "Which of the following is a Python decorator that can be used for caching expensive function calls?",
"options": {
"a": "@staticmethod",
"b": "@classmethod",
"c": "@property",
"d": "@lru_cache"
},
"answer": "d"
},
{
"question": "What is the difference between a shallow copy and a deep copy in Python?",
"options": {
"a": "A shallow copy creates a copy of the object's reference while a deep copy creates a copy of the object's data.",
"b": "A shallow copy creates a new object while a deep copy does not.",
"c": "A shallow copy only copies the first level of the object while a deep copy copies all levels.",
"d": "A shallow copy is slower than a deep copy."
},
"answer": "a"
},
{
"question": "What is the output of the following code?\n\nimport re\nmy_string = 'The quick brown fox jumps over the lazy dog'\nresult = re.findall('[aeiou]', my_string)\nprint(result)",
"options": {
"a": "['a', 'e', 'i', 'o', 'u']",
"b": "['The', 'quick', 'brown', 'fox', 'jumps', 'over', 'the', 'lazy', 'dog']",
"c": "['aeiou']",
"d": "SyntaxError: invalid syntax"
},
"answer": "a"
},
{
"question": "What is the output of the following code?\n\nmy_dict = {1: 'one', 2: 'two', 3: 'three'}\nfor key, value in my_dict.items():\n print(key, value)",
"options": {
"a": "1 'one', 2 'two', 3 'three'",
"b": "1 one, 2 two, 3 three",
"c": "['one', 'two', 'three']",
"d": "SyntaxError: invalid syntax"
},
"answer": "b"
},
{
"question": "What is the difference between the print() function and the return statement in Python?",
"options": {
"a": "print() displays output to the console, while return statement returns a value from a function",
"b": "print() is used for debugging purposes, while return statement is used to terminate a loop",
"c": "print() and return statement are interchangeable and can be used interchangeably",
"d": "print() and return statement are not valid functions in Python"
},
"answer": "a"
},
{
"question": "What is the purpose of the urllib module in Python?",
"options": {
"a": "To perform HTTP requests and handle responses",
"b": "To generate random numbers",
"c": "To manipulate strings",
"d": "To perform mathematical operations"
},
"answer": "a"
},
{
"question": "What is the purpose of test-driven development (TDD) in software development?",
"options": {
"a": "To ensure that code is thoroughly tested before it is released to production",
"b": "To write tests after the code has been written to ensure it works properly",
"c": "To write code without testing and fix any bugs that arise later",
"d": "To write code quickly without worrying about testing"
},
"answer": "a"
},
{
"question": "What is object-relational mapping (ORM) in Python?",
"options": {
"a": "A technique for mapping objects in Python to relational database tables",
"b": "A technique for mapping Python functions to RESTful APIs",
"c": "A technique for mapping Python modules to object-oriented programming (OOP) concepts",
"d": "A technique for mapping Python scripts to network protocols"
},
"answer": "a"
},
{
"question": "What is the difference between the input() function and the raw_input() function in Python 2.x?",
"options": {
"a": "The input() function returns a string, while the raw_input() function returns the input as it is typed",
"b": "The raw_input() function is not a valid function in Python 2.x",
"c": "The input() function is not a valid function in Python 2.x",
"d": "The input() and raw_input() functions are interchangeable and can be used interchangeably in Python 2.x"
},
"answer": "a"
},
{
"question": "Which of the following is a valid way to open a file named \"example.txt\" in read mode?",
"options": {
"a": "file = open(\"example.txt\", mode=\"w\")",
"b": "file = open(\"example.txt\", mode=\"r+\")"
},
"answer": "b"
},
{
"question": "What is the output of the following code?\n\ndef greet(name):\n return f\"Hello, {name}!\"\n\nprint(greet(\"Alice\"))",
"options": {
"a": "Hello, Alice!",
"b": "Hello, !",
"c": "TypeError: greet() missing 1 required positional argument: 'name'",
"d": "SyntaxError: invalid syntax"
},
"answer": "a"
},
{
"question": "What is the output of the following code?\n lst = [1, 2, 3, 4, 5]\n new_lst = [num * 2 for num in lst if num % 2 == 0]\n print(new_lst)",
"options": {
"a": "[2, 4, 6, 8, 10]",
"b": "[4, 8]",
"c": "[2, 6, 10]",
"d": "[1, 2, 3, 4, 5]"
},
"answer": "b"
},
{
"question": "Which of the following is not a built-in function in Python?",
"options": {
"a": "sum()",
"b": "len()",
"c": "sort()",
"d": "range()"
},
"answer": "c"
},
{
"question": "What is the output of the following code?\n a = [1, 2, 3]\n b = a\n b[0] = 0\n print(a)",
"options": {
"a": "[0, 2, 3]",
"b": "[1, 2, 3]",
"c": "[0, 1, 2, 3]",
"d": "[1, 0, 2, 3]"
},
"answer": "a"
},
{
"question": "What is the output of the following code?\n class MyClass:\n\t def init(self, num):\n\t\t self.num = num\n a = MyClass(5)\n b = MyClass(10)\n print(a.num + b.num)",
"options": {
"a": "15",
"b": "'5 10'",
"c": "TypeError",
"d": "AttributeError"
},
"answer": "a"
},
{
"question": "What is the output of the following code?\n def my_func(a, b, c, d):\n\t print(a, b, c, d)\n lst = [1, 2, 3, 4]\n my_func(*lst)",
"options": {
"a": "TypeError",
"b": "1 2 3 4",
"c": "4 3 2 1",
"d": "1 4 2 3"
},
"answer": "b"
},
{
"question": "What is the output of the following code?\n def my_func(a, b=2, c=3):\n\t print(a, b, c)\n my_func(1, c=4)",
"options": {
"a": "1 2 3",
"b": "1 3 4",
"c": "1 2 4",
"d": "SyntaxError"
},
"answer": "c"
},
{
"question": "Which of the following is true about Python's Global Interpreter Lock (GIL)?",
"options": {
"a": "It allows multiple threads to execute Python code simultaneously.",
"b": "It prevents deadlocks from occurring in Python programs.",
"c": "It ensures that only one thread executes Python bytecode at a time.",
"d": "It prevents race conditions from occurring in Python programs."
},
"answer": "c"
},
{
"question": "What is the output of the following code?\n def my_gen(n):\n\t for i in range(n):\n\t\t yield i\n gen = my_gen(3)\n print(next(gen), next(gen), next(gen), next(gen))",
"options": {
"a": "0 1 2 StopIteration",
"b": "0 1 2 None",
"c": "0 1 2 TypeError",
"d": "SyntaxError"
},
"answer": "a"
},
{
"question": "What is the output of the following code?\n x = 10\n y = 20\n x, y = y, x\n print(x, y)",
"options": {
"a": "10 20",
"b": "20 10",
"c": "SyntaxError",
"d": "TypeError"
},
"answer": "b"
},
{
"question": "What is the output of the following code?\n def my_func(x):\n\t if x % 2 == 0:\n\t\t return True\n\t else:\n\t\t return False\n lst = [1, 2, 3, 4, 5]\n new_lst = filter(my_func, lst)\n print(list(new_lst))",
"options": {
"a": "[1, 3, 5]",
"b": "[2, 4]",
"c": "[False, True, False, True, False]",
"d": "TypeError"
},
"answer": "b"
},
{
"question": "Which of the following statements is true about Python's asyncio library?",
"options": {
"a": "It allows multiple threads to execute in parallel",
"b": "It is used for parallel processing of CPU-bound tasks",
"c": "It is based on the concept of coroutines",
"d": "It is not compatible with Python 3"
},
"answer": "c"
},
{
"question": "What will this code print \nnum1 = 4\nnum2 = 2\nres = num1 * num2\n\nprint(\"Multiplication is\", res)",
"options": {
"a": "4",
"b": "42",
"c": "8",
"d": "6"
},
"answer": "c"
},
{
"question": "What does this function do?\ndef write_file(filename="", text=""):\n\t\twith open(filename, \"w\", encoding=\"utf-8\") as f:\n\t\treturn f.write(text)",
"options": {
"a": "a function that writes a string to a text file (UTF8) and returns the number of characters written",
"b": "a function that appends a string at the end of a text file (UTF8) and returns the number of characters added",
"c": "afunction that returns an object (Python data structure) represented by a JSON string",
"d": "a function that writes an Object to a text file, using a JSON representation"
},
"answer": "a"
},
{
"question": "What does the function below do?\nimport json\n\ndef load_from_json_file(filename):\n\t\twith open(filename) as f:\n\t\treturn json.load(f)",
"options": {
"a": "writes an Object to a text file, using a JSON representation",
"b": "returns an object (Python data structure) represented by a JSON string",
"c": "creates an Object from a JSON file",
"d": "returns the JSON representation of an object (string)"
},
"answer": "c"
},
{
"question": "What function can be used to read input from the user in Python3?",
"options": {
"a": "raw_input()",
"b": "input()",
"c": "read_input()",
"d": "read_input()"
},
"answer": "b"
},
{
"question": "Which Python module is used to create network sockets?",
"options": {
"a": "Urllib",
"b": "network",
"c": "request",
"d": "socket"
},
"answer": "d"
},
{
"question": "What is the first step in test-driven development?",
"options": {
"a": "Write the code",
"b": "Debug the code",
"c": "Deploy the code",
"d": "Write the tests"
},
"answer": "d"
},
{
"question": "In object-relational mapping (ORM), what is an entity?",
"options": {
"a": "A database table",
"b": "A database column",
"c": "A Python class",
"d": "An SQL query"
},
"answer": "c"
},
{
"question": "What is a database connection pool?",
"options": {
"a": "A distributed database",
"b": "A cache of database connections that can be reused",
"c": "A backup copy of a database",
"d": "A group of users who share access to a database"
},
"answer": "b"
},
{
"question": "When reading a file in Python, which method can be used to read the entire file as a single string?",
"options": {
"a": "read()",
"b": "readline()",
"c": "readlines()",
"d": "file()"
},
"answer": "a"
},
{
"question": "Which of the following is not a benefit of using test-driven development (TDD)?",
"options": {
"a": "Faster development time",
"b": "Reduced maintenance costs",
"c": "Increased risk of bugs",
"d": "Improved code quality"
},
"answer": "c"
},
{
"question": "What is the output of the below code?\n\nmyList=[1,2,3,5,3,4,6,9]\nmyList[-6:6]",
"options": {
"a": "[]",
"b": "[3, 5, 3, 4]",
"c": "[4, 3, 5, 3]",
"d": "Index Error"
},
"answer": "b"
},
{
"question": "What is the run time of the below code?\n\nfor i in range(n):\n\tj=1\nwhile(j<n):\n\nprint(i,j)\nj*=2",
"options": {
"a": "O(n)",
"b": "O(n^2)",
"c": "O(log(n))",
"d": "O(n*log(n))"
},
"answer": "d"
},
{
"question": "What is the method that is bound to class but not the instance?",
"options": {
"a": "Static method",
"b": "Class method",
"c": "Main method",
"d": "None of the above"
},
"answer": "b"
},
{
"question": "Which Python module is used to create network sockets?",
"options": {
"a": "Urllib",
"b": "network",
"c": "request",
"d": "socket"
},
"answer": "d"
},
{
"question": "In object-relational mapping (ORM), what is an entity?",
"options": {
"a": "A database table",
"b": "A database column",
"c": "A Python class",
"d": "An SQL query"
},
"answer": "c"
},
{
"question": "When reading a file in Python, which method can be used to read the entire file as a single string?",
"options": {
"a": "read()",
"b": "readline()",
"c": "readlines()",
"d": "file()"
},
"answer": "a"
},
{
"question": "Which of the following is the correct statement about the statement below?\n\n z = {\"x\":0, \"y\":1}",
"options": {
"a": "x dictionary z is created",
"b": "x and y are the keys of dictionary z",
"c": "0 and 1 are the values of dictionary z",
"d": "All of the above"
},
"answer": "d"
},
{
"question": "What will be the output of the following Python code?\n i = 5\n while True:\n if i%0O11 == 0:\n break\n print(i)\n i += 1",
"options": {
"a": "5 6 7 8 9 10",
"b": "5 6 7 8",
"c": "5 6",
"d": "error"
},
"answer": "b"
},
{
"question": "What will be the output of the following Python code?\n i = 1\n while True:\n if i%3 == 0:\n break\n print(i)\n i + = 1",
"options": {
"a": "1 2",
"b": "1 2 3",
"c": "error",
"d": " none of the mentioned"
},
"answer": "c"
},
{
"question": "What will be the output of the following Python code?\n lamb = lambda x: x ** 3\n print(lamb(5))",
"options": {
"a": "15.",
"b": "555",
"c": "125",
"d": "none of the mentioned"
},
"answer": "c"
},
{
"question": "What will be the output of the following Python code?\n i = 1\n while True:\n if i%3 == 0:\n break\n print(i)\n i + = 1",
"options": {
"a": "1 2",
"b": "1 2 3",
"c": "error",
"d": " none of the mentioned"
},
"answer": "c"
},
{
"question": "What is the run time of the below code?\n\nfor i in range(n):\n\tj=1\nwhile(j<n):\n\nprint(i,j)\nj*=2",
"options": {
"a": "O(n)",
"b": "O(n^2)",
"c": "O(log(n))",
"d": "O(n*log(n))"
},
"answer": "d"
},
{
"question": "What will be the output of the following Python code?\n lamb = lambda x: x ** 3\n print(lamb(5))",
"options": {
"a": "15.",
"b": "555",
"c": "125",
"d": "none of the mentioned"
},
"answer": "c"
},
{
"question": "What is the output of the following code snippet?\n\n def foo():\n print(\"Start\")\n yield 1\n print(\"Middle\")\n yield 2\n print(\"End\")\n\n for i in foo():\n print(i)",
"options": {
"a": "Start Middle End",
"b": "Start 1 Middle 2 End",
"c": "Start 1 2 Middle End",
"d": "Start Middle 1 2 End"
},
"answer": "b"
},
{
"question": "What is the output of the following code?",
"options": {
"a": "42",
"b": "None",
"c": "coroutine object at 0x...",
"d": "Raises a TypeError exception."
},
"answer": "a"
},
{
"question": "Which of the following is a valid way to redirect standard output to a file in Python?",
"options": {
"a": "sys.stdout.write(\"Hello world\")",
"b": "sys.stderr.write(\"Hello world\")",
"c": "with open(\"output.txt\", \"w\") as f: f.write(\"Hello world\")",
"d": "print(\"Hello world\", file=open(\"output.txt\", \"w\"))"
},
"answer": "d"
},
{
"question": "What is the output of the following code?\nimport asyncio\n\nasync def coro():\n await asyncio.sleep(1)\n return 42\n\nasync def main():\n result = await coro()\n print(result)\n\nloop = asyncio.get_event_loop()\nloop.run_until_complete(main())",
"options": {
"a": "42",
"b": "\"None\"",
"c": "\"coroutine object at 0x...\"",
"d": "Raises a TypeError exception."
},
"answer": "a"
},
{
"question": "What is the output of the following code?",
"options": {
"a": "\"Something went wrong\"",
"b": "\"CustomException: Something went wrong\"",
"c": "\"Exception: Something went wrong\"",
"d": "Raises a TypeError exception."
},
"answer": "a"
},
{
"question": "What is the output of the following code?\nimport subprocess\n\ntry:\n output = subprocess.check_output([\"echo\", \"Hello world\"])\n print(output)\nexcept subprocess.CalledProcessError:\n print(\"Command failed\")\n",
"options": {
"a": "\"Hello world\n\"",
"b": "\"Hello world\"",
"c": "\"Command failed\"",
"d": "Raises a TypeError exception."
},
"answer": "a"
},
{
"question": "What is the output of the following code?\n\nimport asyncio\n\nasync def coro():\n print('coro started')\n await asyncio.sleep(1)\n print('coro ended')\n\nloop = asyncio.get_event_loop()\nloop.run_until_complete(coro())",
"options": {
"a": "'coro started', followed by 'coro ended' after 1 second.",
"b": "'coro started', followed by 'coro ended' immediately.",
"c": "'coro ended', followed by 'coro started' after 1 second.",
"d": "'coro ended', followed by 'coro started' immediately."
},
"answer": "a"
},
{
"question": "Which of the following is a valid way to write test cases for a REST API using Python's unittest module?",
"options": {
"a": "Use the requests module to send HTTP requests and check the responses.",
"b": "Use the unittest.mock module to mock the API responses and test the behavior of the code.",
"c": "Use the unittest.TestCase.assert*() methods to check the results of API calls.",
"d": "Use the unittest.skip() decorator to skip tests that require a running server."
},
"answer": "a"
},
{
"question": "Which of the following is a correct statement about Python's garbage collection system?",
"options": {
"a": "Python uses reference counting to keep track of object lifetimes.",
"b": "Python's garbage collector runs in a separate thread to avoid blocking the main thread.",
"c": "Python's garbage collector uses a generational algorithm to optimize memory usage.",
"d": "Python's garbage collector only runs when the system is low on memory."
},
"answer": "c"
},
{
"question": "Which of the following is a valid way to serialize and deserialize complex data structures using Python's pickle module?",
"options": {
"a": "pickle.dumps(data, protocol=pickle.HIGHEST_PROTOCOL)",
"b": "pickle.dump(data, file)",
"c": "pickle.loads(data)",
"d": "pickle.load(file)"
},
"answer": "b"
},
{
"question": "Which of the following is a difference between multiprocessing and threading in Python?",
"options": {
"a": "multiprocessing can run multiple processes on different CPUs, while threading can only run on a single CPU.",
"b": "multiprocessing is more memory-efficient than threading.",
"c": "multiprocessing uses more memory than threading.",
"d": "threading is more CPU-efficient than multiprocessing."
},
"answer": "c"
},
{
"question": "Which of the following is a valid way to execute a shell command asynchronously in Python?",
"options": {
"a": "os.system(\"ls -l\")",
"b": "subprocess.call(\"ls -l\", shell=True)",
"c": "subprocess.Popen(\"ls -l\", shell=True)",
"d": "os.execvp(\"ls\", [\"-l\"])"
},
"answer": "c"
},
{
"question": "Which of the following is a valid way to create a custom exception in Python?",
"options": {
"a": "class CustomException(BaseException): pass",
"b": "class CustomException(Exception): pass",
"c": "class CustomException(RuntimeError): pass",
"d": "class CustomException(ValueError): pass"
},
"answer": "b"
},
{
"question":"Which of the following is not a complex number?",
"options": {
"a": "k = 2 + 3j",
"b": "k = complex(2, 3)",
"c": "k = 2 + 3l",
"d": "k = 2 + 3J"
},
"answer": "c"
},
{
"question": "Study the following program:\n z = \"xyz\"\n j = \"j\"\n while j in z:\n pass\nWhat will be the output of this statement?",
"options": {
"a": "xyz",
"b": "No output",
"c": "x y z",
"d": "j j j j j j j.."
},
"answer": "b"
},
{
"question": "What error will occur when you execute the following code?\nMANGO = APPLE",
"options": {
"a": "NameError",
"b": "SyntaxError",
"c": "TypeError",
"d": "ValueError"
},
"answer": "a"
},
{
"question": "What will be the output of the following Python code snippet?\n a = [0, 1, 2, 3]\n i = -2\n for i not in a:\n print(i)\n i += 1",
"options": {
"a": "-2 -1",
"b": "0",
"c": " error",
"d": "none of the mentioned"
},
"answer": "c"
},
{
"question": "What will be the output of the following Python code?\n x = (i for i in range(3))\n for i in x:\n print(i)",
"options": {
"a": " 0 1 2",
"b": "error",
"c": "0 1 2 0 1 2",
"d": "none of the mentioned"
},
"answer": "a"
}
] | ALU-PyQuest | /ALU-PyQuest-0.1.9.tar.gz/ALU-PyQuest-0.1.9/src/advanced_questions.py | advanced_questions.py |
# Store beginner level questions as a list of dictionaries
PROBLEM_SETS = [
{
"question": "Which of these is an example of a simple program in Python?",
"options": {
"a": "print('Hello, World!')\n >>> Hello, World!",
"b": "def hello():\n\t print('Hello, Boss!')\n >>> Hello, World!",
"c": "print('Hello, Python!') >>> Hello, World!",
"d": "def world():\n\t print('Hello, Word!') >>> Hello, World!"
},
"answer": "a"
},
{
"question": "What's the correct way to declare variables in Python?",
"options": {
"a": "set x = 5, y = 10, z = x + y, print(z)\n >>> Output: 15",
"b": "x = 5\ny = 10\nz = x + y\nprint(z)\n >>> Output: 15",
"c": "x = 5, y = 10, z = x + y\nprint(z)\n >>> Output: 15",
"d": "set x = 5\nset y = 10\nset z = x + y\nprint(z)\n >>> Output: 15"
},
"answer": "b"
},
{
"question": "How do I add a single item to a list in Python?",
"options": {
"a": "my_list = [1, 2, 3, 4, 5]\nmy_list.append(6)\nprint(my_list)\n >> [1, 2, 3, 4, 5, 6]",
"b": "my_list = [1, 2, 3, 4, 5]\nmy_list.add(6)\nprint(my_list) >>\n [1, 2, 3, 4, 5, 6]",
"c": "my_list = [1, 2, 3, 4, 5]\nmy_list.extend(6)\nprint(my_list) >>\n [1, 2, 3, 4, 5, 6]",
"d": "my_list = [1, 2, 3, 4, 5]\nmy_list.insert(6)\nprint(my_list) >>\n [1, 2, 3, 4, 5, 6]"
},
"answer": "a"
},
{
"question": "What is the output of the following code?,\n\n list = ['apple', 'banana', 'cherry']\nlist.append('orange')\nprint(list)",
"options": {
"a": "['apple', 'banana', 'cherry']",
"b": "['apple', 'banana', 'cherry', 'orange']",
"c": "['apple', 'banana', 'cherry', 'orange', 'grape', 'kiwi']",
"d": "Error"
},
"answer": "b"
},
{
"question": "What's the output of this code?\n\n for i in range(5):\n\t print(i)\n",
"options": {
"a": "0 1 2 3 4",
"b": "0, 1, 2, 3, 4",
"c": "01234",
"d": "54321"
},
"answer": "a"
},
{
"question": "Which of the following is a correct way to declare a variable in Python?\n",
"options": {
"a": "1var = 'Hello World'",
"b": "var-1 = 'Hello World'",
"c": "var_1 = 'Hello World'",
"d": "Var$1 = 'Hello World'"
},
"answer": "c"
},
{
"question": "What is the output of the following code?\n\n x = 5 \n y = 2 \n print(x+y)\n print(x-y)\n print(x*y)\n print(x/y)\n",
"options": {
"a": "7, 3, 10, 2.5",
"b": "3, 7, 10, 2.5",
"c": "7, 3, 10, 2",
"d": "3, 7, 10, 2"
},
"answer": "a"
},
{
"question": "What is the output of the following code?\n\n x = 'Hello'\n y = 'World'\n print(x+y)\n",
"options": {
"a": "Hello World",
"b": "HelloWorld",
"c": "Hello+World",
"d": "None of the above"
},
"answer": "a"
},
{
"question": "Which of the following is the correct syntax to declare a list in Python?\n",
"options": {
"a": "list = [1, 2, 3]",
"b": "list = {1, 2, 3}",
"c": "list = (1, 2, 3)",
"d": "list = '1, 2, 3'"
},
"answer": "a"
},
{
"question": "What is the output of the following code?\n\n x = 5 \n y = 10\n if x>y:\n\tprint('x is greater than y')\nelse:\n\tprint('y is greater than x')\n",
"options": {
"a": "x is greater than y",
"b": "y is greater than x",
"c": "x and y are equal",
"d": "Syntax error"
},
"answer": "b"
},
{
"question": "What is the output of the following code?\n\n x = 0\n while x < 5:\n\tprint(x)\n\tx+=1\n",
"options": {
"a": "0 1 2 3 4",
"b": "5",
"c": "Infinite loop",
"d": "None of the above"
},
"answer": "a"
},
{
"question": "What is the output of the following code?\n\n for x in range(1,6):\n\tprint(x)",
"options": {
"a": "1 2 3 4 5",
"b": "0 1 2 3 4",
"c": "6 7 8 9 10",
"d": "None of the above"
},
"answer": "a"
},
{
"question": "What does the 'print()' function do?",
"options": {
"a": "It prompts the user to input a value",
"b": "It adds two numbers together",
"c": "It displays a message or value on the screen",
"d": "It converts a value to a string"
},
"answer": "c"
},
{
"question": "What is the result of the expression '5 + 7'?",
"options": {
"a": "12",
"b": "13",
"c": "14",
"d": "15"
},
"answer": "a"
},
{
"question": "What is the correct way to declare a variable in Python?",
"options": {
"a": "var x = 5",
"b": "int x = 5",
"c": "x = 5",
"d": "x := 5"
},
"answer": "c"
},
{
"question": "What is the output of the code 'print(len('hello'))'?",
"options": {
"a": "4",
"b": "5",
"c": "6",
"d": "7"
},
"answer": "b"
},
{
"question": "What is the correct way to define a function in Python?",
"options": {
"a": "function myFunction():",
"b": "def myFunction():",
"c": "def myFunction:",
"d": "myFunction():"
},
"answer": "b"
},
{
"question": "What do these lines print?\n\n class User:\n\tid = 1\n\nprint(User.id)\n",
"options": {
"a": "89",
"b": "None",
"c": "1",
"d": "98"
},
"answer": "c"
},
{
"question": "what will this line print?\n hub = 1\n if hub < 3:\n\t print('Less than 3')\n else:\n\t print('Greater that or equal to 3')",
"options": {
"a": "Less than 3",
"b": "Greater than or equal to 3",
"c": "syntax error",
"d": "The code will not print anything"
},
"answer": "a"
},
{
"question": "what will this line print?\n name = \"Adam\"\n print(\"Hello, \"+name)\n",
"options": {
"a": "Hello Adam",
"b": "Hello",
"c": "syntax error",
"d": "Adam Hello"
},
"answer": "a"
},
{
"question": "what will this line print?\n q = True\n p = False\n result = q and p\n print(result)",
"options": {
"a": "True",
"b": "False",
"c": "Error",
"d": "I don't know"
},
"answer": "b"
},
{
"question": "what will this line print?\n w = 9\n z = 6\n result = w < z\n print(result)",
"options": {
"a": "True",
"b": "False",
"c": "Error",
"d": "I don't know"
},
"answer": "b"
},
{
"question": "what will this line print?\n age = 50\n print(\"My age is \"+ age)",
"options": {
"a": "My age is 50",
"b": "My age",
"c": "syntax error",
"d": "My age 50"
},
"answer": "c"
},
{
"question": "what will this line print?\n for r in range(7):\n print(r)",
"options": {
"a": "0 1 2 3 4 5 6",
"b": "0 1 2 3 4 5 6 7",
"c": "0 1 2 3 4 5",
"d": "None of the above"
},
"answer": "a"
},
{
"question": "what will this line print?\n my_list = [1, 2, 3, 4, 5]\n for my in my_list:\n\t print(my)",
"options": {
"a": "1 2 3 4 5",
"b": "[1, 2, 3, 4, 5]",
"c": "1, 2, 3",
"d": "syntax error"
},
"answer": "a"
},
{
"question": "what will this line print?\n x = 15\n y = 5\n if x > y:\n\t print(\"x is number 24\")\n else:\n\t print(\"y is number 25\")",
"options": {
"a": "x is number 24",
"b": "y is number 25",
"c": "x is number 25",
"d": "syntax error"
},
"answer": "a"
},
{
"question": "what will this line print?\n first_list = [1 2 3 4 5]\n print(len(first_list))",
"options": {
"a": "1",
"b": "5",
"c": "4",
"d": "syntax error"
},
"answer": "b"
},
{
"question": "what will this line print?\n x = 8\n y = 5\n result = x - y\n print(result)",
"options": {
"a": "3",
"b": "5",
"c": "8",
"d": "syntax error"
},
"answer": "a"
},
{
"question": "Why are local variable names beginning with an underscore discouraged?",
"options": {
"a": "they are used to indicate a private variables of a class",
"b": "they confuse the interpreter",
"c": "they are used to indicate global variables",
"d": "they slow down execution"
},
"answer": "a"
},
{
"question": "What will be the output of the following code?\nx = 10\nif x > 5:\n print('x is greater than 5')\nelse:\n print('x is less than or equal to 5')",
"options": {
"a": "x is greater than 5",
"b": "x is less than or equal to 5",
"c": "Syntax error",
"d": "None of the above"
},
"answer": "a"
},
{
"question": "What will be the output of the following code?\nage = 20\nif age >= 18 and age <= 25:\n print('Age is between 18 and 25')\nelse:\n print('Age is not between 18 and 25')",
"options": {
"a": "Age is between 18 and 26",
"b": "Age is between 18 and 25",
"c": "Syntax error",
"d": "None of the above"
},
"answer": "b"
},
{
"question": "What keyword would you use to add an alternative condition to an if statement?",
"options": {
"a": "else if",
"b": "elseif",
"c": "elif",
"d": "None of the above"
},
"answer": "c"
},
{
"question": "What is the output of the following code:\n\n x = 6\n y = 2\n print(x ** y)\n print(x // y)",
"options": {
"a": "66\n\t 0",
"b": "36\n\t 0",
"c": "66\n\t 3",
"d": "36\n\t 3"
},
"answer": "d"
},
{
"question": "Find the output of the given Python program?\n\n a = 25\n if a < 15:\n\t print(\"Hi\")\n elif a <= 30:\n\t print(\"Hello\")\n else:\n\t print(\"Know Program\")",
"options": {
"a": "Hi",
"b": "Hello",
"c": "Know Program",
"d": "Compiled successfully, No output"
},
"answer": "b"
},
{
"question": "Select the correct output of the following String operations\n\nstr1 = 'Welcome'\nprint (str1[:6] + ' John')",
"options": {
"a": "Welcome John",
"b": "WelcomJohn",
"c": "Welcom John",
"d": "None of the above"
},
"answer": "c"
},
{
"question": "Guess the correct output of the following String operations\n\nstr1 = 'Welcome'\nprint(str1*2)",
"options": {
"a": "WelcomeWelcome",
"b": "TypeError unsupported operand type(s)",
"c": "Welcome",
"d": "Welcome2"
},
"answer": "a"
},
{
"question": "What is the output of the following\n\nl = [None] * 10\nprint(len(l))",
"options": {
"a": "10",
"b": "0",
"c": "Syntax Error",
"d": "I don't know"
},
"answer": "a"
},
{
"question": "What does pip stand for python?",
"options": {
"a": "Pip Installs Python",
"b": "Pip Installs Packages",
"c": "Preferred Installer Program",
"d": "All of the mentioned"
},
"answer": "c"
},
{
"question": "Which of the following would give an error?",
"options": {
"a": "list1 = []",
"b": "list1 = [] * 3",
"c": "list1 = [2, 8, 7]",
"d": "None of the above"
},
"answer": "b"
},
{
"question": "What will be the output of the following Python code?\n\n>>> str1 = 'hello'\n>>> str2 = ','\n>>> str3 = 'world'\n>>> str1[-1:]",
"options": {
"a": "olleh",
"b": "hello",
"c": "h",
"d": "o"
},
"answer": "d"
},
{
"question": "Suppose list1 is [1, 3, 2], What is list1 * 2?",
"options": {
"a": "[2, 6, 4]",
"b": "[1, 3, 2, 1, 3]",
"c": "[1, 3, 2, 1, 3, 2]",
"d": "[1, 3, 2, 3, 2, 1]"
},
"answer": "c"
},
{
"question": "Which one of the following is a valid Python if statement.",
"options": {
"a": "if a >= 2:",
"b": "if (a >= 2)",
"c": "if (a => 22)",
"d": "if a >= 22"
},
"answer": "a"
},
{
"question": "What is the output of the following assignment operator?\n y = 10\n x = y += 2\n print(x)",
"options": {
"a": "12",
"b": "10",
"c": "8",
"d": "Syntax error"
},
"answer": "d"
},
{
"question": "Suppose listExample is [‘h’,’e’,’l’,’l’,’o’], what is len(listExample)?",
"options": {
"a": "5",
"b": "4",
"c": "None",
"d": "Error"
},
"answer": "a"
},
{
"question": "Which of the following is used to define a block of code in Python language?",
"options": {
"a": "Indentation",
"b": "Key",
"c": "Brackets",
"d": "All of the mentioned"
},
"answer": "a"
},
{
"question": "Which keyword is used for function in Python language?",
"options": {
"a": "Function",
"b": "def",
"c": "Fun",
"d": "Define"
},
"answer": "b"
},
{
"question": "Which of the following functions can help us to find the version of python that we are currently working on?",
"options": {
"a": "sys.version(1)",
"b": "sys.version(0)",
"c": "sys.version()",
"d": "sys.version"
},
"answer": "d"
},
{
"question": "What is the maximum possible length of an identifier?",
"options": {
"a": "16",
"b": "34",
"c": "64",
"d": "none of the mentioned"
},
"answer": "d"
},
{
"question": "Which of the following is used to define a block of code in Python language?",
"options": {
"a": "Indentation",
"b": "Key",
"c": "Brackets",
"d": "All of the mentioned"
},
"answer": "a"
},
{
"question": "Which of the following functions can help us to find the version of python that we are currently working on?",
"options": {
"a": "sys.version(1)",
"b": "sys.version(0)",
"c": "sys.version()",
"d": "sys.version"
},
"answer": "d"
},
{
"question": "What will be the output of the following Python code?\n\n>>> str1 = 'hello'\n>>> str2 = ','\n>>> str3 = 'world'\n>>> str1[-1:]",
"options": {
"a": "olleh",
"b": "hello",
"c": "h",
"d": "o"
},
"answer": "d"
},
{
"question": "Why are local variable names beginning with an underscore discouraged?",
"options": {
"a": "they are used to indicate a private variables of a class",
"b": "they confuse the interpreter",
"c": "they are used to indicate global variables",
"d": "they slow down execution"
},
"answer": "a"
},
{
"question": "Select the correct output of the following String operations\n\nstr1 = 'Welcome'\nprint (str1[:6] + ' John')",
"options": {
"a": "Welcome John",
"b": "WelcomJohn",
"c": "Welcom John",
"d": "None of the above"
},
"answer": "c"
},
{
"question": "Which of the following declarations is incorrect?",
"options": {
"a": "_x = 2",
"b": "__x = 3",
"c": "__xyz__ = 5",
"d": "None of these"
},
"answer": "d"
},
{
"question": "what will this line print?\n first_list = [1 2 3 4 5]\n print(len(first_list))",
"options": {
"a": "1",
"b": "5",
"c": "4",
"d": "syntax error"
},
"answer": "b"
},
{
"question": "What will be the output of the following code?\nage = 20\nif age >= 18 and age <= 25:\n print('Age is between 18 and 25')\nelse:\n print('Age is not between 18 and 25')",
"options": {
"a": "Age is between 18 and 26",
"b": "Age is between 18 and 25",
"c": "Syntax error",
"d": "None of the above"
},
"answer": "b"
},
{
"question": "Which of the following functions can help us to find the version of python that we are currently working on?",
"options": {
"a": "sys.version(1)",
"b": "sys.version(0)",
"c": "sys.version()",
"d": "sys.version"
},
"answer": "d"
},
{
"question": "What is the output of print(2 + 2 * 3)?",
"options": {
"a": "6",
"b": "8",
"c": "12",
"d": "None of the above"
},
"answer": "b"
},
{
"question": "What is the output of print([1, 2, 3] + [4, 5, 6])?",
"options": {
"a": "[1, 2, 3, 4, 5, 6]",
"b": "[1, 4, 2, 5, 3, 6]",
"c": "[[1, 2, 3], [4, 5, 6]]",
"d": "None of the above"
},
"answer": "a"
},
{
"question": "What is the output of print(1 == \"1\")?",
"options": {
"a": "True",
"b": "False",
"c": "Error",
"d": "None of the above"
},
"answer": "b"
},
{
"question": "What is the output of print(\"Hello, World!\"[1:5])?",
"options": {
"a": "ello",
"b": "Hell",
"c": "World",
"d": "None of the above"
},
"answer": "a"
},
{
"question": "What is the output of print(not True)?",
"options": {
"a": "True",
"b": "False",
"c": "Error",
"d": "None of the above"
},
"answer": "b"
},
{
"question": "What is the syntax to print \"Hello, World!\" in Python?",
"options":{
"a": "print('Hello, World!')",
"b": "print('Hello World!')",
"c": "print(Hello, World!)",
"d": "print('Hello', 'World!')"
},
"answer": "a"
},
{
"question":"What is the result of 7 % 3 in Python?",
"options": {
"a": "1",
"b": "2",
"c": "3",
"d": "4"
},
"answer": "b"
},
{
"question":"How do you concatenate two strings in Python?",
"options": {
"a": "Using the '+' operator",
"b": "Using the '-' operator",
"c": "Using the '*' operator",
"d": "Using the '/' operator"
},
"answer": "a"
},
{
"question":"How do you access the first element of a list in Python?",
"options": {
"a": "list[0]",
"b": "list[1]",
"c": "list[-1]",
"d": "list[-2]"
},
"answer": "a"
},
{
"question":"What is the syntax to write an if statement in Python?",
"options": {
"a": "if condition:",
"b": "if (condition)",
"c": "if {condition}",
"d": "if [condition]"
},
"answer": "a"
},
{
"question":"What is the syntax to write a for loop in Python?",
"options": {
"a": "for i in range(n):",
"b": "for i in n:",
"c": "for i in list:",
"d": "for i in dictionary:"
},
"answer": "a"
},
{
"question":"How do you exit a loop prematurely in Python?",
"options": {
"a": "break",
"b": "continue",
"c": "pass",
"d": "return"
},
"answer": "a"
},
{
"question":"How do you find the length of a string in Python?",
"options": {
"a": "len(string)",
"b": "string.length()",
"c": "string.len()",
"d": "length(string)"
},
"answer": "a"
},
{
"question":"How do you add an element to the end of a list in Python?",
"options": {
"a": "list.append(element)",
"b": "list.add(element)",
"c": "list.insert(element)",
"d": "list.extend(element)"
},
"answer": "a"
},
{
"question":"What is the difference between '==' and '!=' operators in Python?",
"options": {
"a": "'==' checks for equality, '!=' checks for inequality",
"b": "'==' checks for inequality, '!=' checks for equality",
"c": "'==' checks for identity, '!=' checks for equality",
"d": "'==' checks for equality, '!=' checks for identity"
},
"answer": "a"
},
{
"question":"Write a Python program to print the even numbers from 0 to 10.",
"options": {
"a": "for i in range(0, 11, 2):\n print(i)",
"b": "for i in range(11):\n if i % 2 == 0:\n print(i)",
"c": "for i in range(11):\n if i % 2 != 0:\n print(i)",
"d": "for i in range(0, 10):\n if i % 2 == 0:\n print(i)"
},
"answer": "a"
},
{
"question":"Write a Python program to find the maximum element in a list.",
"options": {
"a": "my_list = [1, 2, 3, 4, 5]\nprint(max(my_list))",
"b": "my_list = [1, 2, 3, 4, 5]\nmax_element = my_list[0]\nfor i in my_list:\n if i > max_element:\n max_element = i\nprint(max_element)",
"c": "my_list = [1, 2, 3, 4, 5]\nmy_list.sort()\nprint(my_list[-1])",
"d": "my_list = [1, 2, 3, 4, 5]\nmy_list.reverse()\nprint(my_list[0])"
},
"answer": "a"
},
{
"question":"What's the output of the following code?\n x = 5\n y = 3\n print(x % y)",
"options": {
"a": "1",
"b": "2",
"c": "3",
"d": "4"
},
"answer": "b"
},
{
"question": "Study the following program:\n a = 1\n while True:\n if a % 7 == 0:\n break\n print(a)\n a += 1\nWhich of the following is correct output of this program?",
"options": {
"a": "1 2 3 4 5",
"b": "1 2 3 4 5 6",
"c": "1 2 3 4 5 6 7",
"d": "Invalid syntax"
},
"answer": "b"
},
{
"question": "Study the following program:\n i = 0\n while i < 5:\n print(i)\n i += 1\n if i == 3:\n break\nelse:\n print(0)\nWhat will be the output of this statement?",
"options": {
"a": "1 2 3",
"b": "0 1 2 3",
"c": "0 1 2",
"d": "3 2 1"
},
"answer": "c"
},
{
"question": "Study the following program:\n i = 0\n while i < 3:\n print(i)\n i += 1\nelse:\n print(0)\nWhat will be the output of this statement?",
"options": {
"a": "0 1",
"b": "0 1 2",
"c": "0 1 2 0",
"d": "0 1 2 3"
},
"answer": "d"
},
{
"question": "What will be the output of the following Python code?\nfor i in range(10):\n if i == 5:\n break\nelse:\n print(i)\nprint(i)\nprint(\"Here\")",
"options": {
"a": "0 1 2 3 4 Here",
"b": "0 1 2 3 4 5 Here",
"c": "0 1 2 3 4",
"d": "1 2 3 4 5"
},
"answer": "c"
},
{
"question": "What will be the output of the following Python code?\n string = \"my name is x\"\n for i in string:\n print(i, end=\", \")",
"options": {
"a": "m, y, , n, a, m, e, , i, s, , x,",
"b": " m, y, , n, a, m, e, , i, s, , x",
"c": "my, name, is, x,",
"d": "error"
},
"answer": "a"
}
] | ALU-PyQuest | /ALU-PyQuest-0.1.9.tar.gz/ALU-PyQuest-0.1.9/src/beginner_questions.py | beginner_questions.py |
import time
import random
import src.beginner_questions
import src.medium_questions
import src.advanced_questions
# Define the game rules and guidelines as a string variable:
RULES = """
Welcome to ALU PyQuest!
The goal of this game is to test your knowledge of Python programming language.
You will be presented with a series of questions related to the Python curriculum. The questions will be presented one at a time, starting from beginner's level to advanced level, and you will have a limited time to answer each question.
For each correct answer, you will earn 10 points, and for each incorrect answer or if you run out of time, 5 points will be deducted from your score. You will have 25 seconds for each question.
In total, you will be asked 12 questions, 4 questions for each level of difficulty.
At the end of the game, your total score will be displayed. Good luck!
"""
#function to load and save high scores to a file
HIGH_SCORES_FILE = 'high_scores.txt'
def load_high_scores():
high_scores = []
try:
with open(HIGH_SCORES_FILE, 'r') as f:
for line in f:
name, score = line.strip().split(',')
high_scores.append((name, int(score)))
except FileNotFoundError:
pass # If the file doesn't exist, return an empty list
return high_scores
def save_high_scores(high_scores):
with open(HIGH_SCORES_FILE, 'w') as f:
for name, score in high_scores:
f.write(f"{name},{score}\n")
#function to generate random questions from beginner to advanced
BEGINNER_PROBLEMS = src.beginner_questions.PROBLEM_SETS
MEDIUM_PROBLEMS = src.medium_questions.PROBLEM_SETS
ADVANCED_PROBLEMS = src.advanced_questions.PROBLEM_SETS
def generate_problem_sets(difficulty_level):
if difficulty_level == 'beginner':
return random.sample(BEGINNER_PROBLEMS, k=4)
elif difficulty_level == 'medium':
return random.sample(MEDIUM_PROBLEMS, k=4)
elif difficulty_level == 'advanced':
return random.sample(ADVANCED_PROBLEMS, k=4)
# Define a function to play game
def playgame():
#print initial message
print("Loading the game...")
time.sleep(2)
print(RULES)
#load and display high scores
high_scores = load_high_scores()
if high_scores:
print("========== High Scores ==========")
for i, (name, score) in enumerate(high_scores[:5]):
print(f"{i+1}. {name}: {score}")
#start the game
ready = input("Are you ready to start the game? Enter(yes/no): ")
if ready.lower() == "yes":
print("\nGreat! Let's start the game. \n")
print("Loading the game...")
time.sleep(1.5)
print("Please wait...")
time.sleep(1.5)
print("Ready!")
time.sleep(1.5)
difficulty_levels = ['beginner', 'medium', 'advanced']
score = 0
for level in difficulty_levels:
problem_set = generate_problem_sets(level)
print(f"\nLevel: {level.capitalize()}")
counter = 1
for problem in problem_set:
print(f"\n{counter}. {problem['question']}")
for option in problem['options']:
print(f"{option}: {problem['options'][option]}")
start_time = time.time()
answer = input("Enter your answer: ")
elapsed_time = time.time() - start_time
if answer.strip().lower() == problem['answer'].lower() and elapsed_time <= 25:
score += 10
print("Correct!")
elif elapsed_time > 25:
score -= 5
print(f"The correct answer is: {problem['answer']}")
print(f"You took too long to answer the question. Time elapsed: {elapsed_time:.2f} seconds")
else:
score -= 5
print(f"Incorrect! The correct answer is: {problem['answer']}")
print(f"Your current score is: {score}")
counter += 1
time.sleep(1)
print(f"\nYour final score is: {score}")
#Stores user score
name = input("Enter your name: ")
high_scores.append((name, score))
high_scores.sort(key=lambda x: x[1], reverse=True)
if len(high_scores) > 5:
high_scores = high_scores[:5]
save_high_scores(high_scores)
#asks user to view high score
view_high_scores = input("\nDo you want to view high scores? Enter(yes/no): ")
if view_high_scores.lower() == "yes":
print("\n========== High Scores ==========")
for i, (name, score) in enumerate(high_scores[:5]):
print(f"{i+1}. {name}: {score}")
play_again = input("\nDo you want to play again? Enter(yes/no): ")
while play_again.lower() == 'yes':
print("Loading the game...")
time.sleep(1.5)
print("Please wait...")
time.sleep(1.5)
print("Ready!")
time.sleep(1.5)
difficulty_levels = ['beginner', 'medium', 'advanced']
score = 0
for level in difficulty_levels:
problem_set = generate_problem_sets(level)
print(f"\nLevel: {level.capitalize()}")
counter = 1
for problem in problem_set:
print(f"\n{counter}. {problem['question']}")
for option in problem['options']:
print(f"{option}: {problem['options'][option]}")
start_time = time.time()
answer = input("Enter your answer: ")
elapsed_time = time.time() - start_time
if answer.strip().lower() == problem['answer'].lower() and elapsed_time <= 25:
score += 10
print("Correct!")
elif elapsed_time > 25:
score -= 5
print(f"The correct answer is: {problem['answer']}")
print(f"You took too long to answer the question. Time elapsed: {elapsed_time:.2f} seconds")
else:
score -= 5
print(f"Incorrect! The correct answer is: {problem['answer']}")
print(f"Your current score is: {score}")
counter += 1
time.sleep(1)
print(f"\nYour final score is: {score}")
#store users score
name = input("Enter your name: ")
high_scores.append((name, score))
high_scores.sort(key=lambda x: x[1], reverse=True)
if len(high_scores) > 5:
high_scores = high_scores[:5]
save_high_scores(high_scores)
#asks user to view high core
view_high_scores = input("\nDo you want to view high scores? Enter(yes/no): ")
if view_high_scores.lower() == "yes":
print("\n========== High Scores ==========")
for i, (name, score) in enumerate(high_scores[:5]):
print(f"{i+1}. {name}: {score}")
play_again = input("\nDo you want to play again? Enter(yes/no): ")
else:
print("\nThank you for playing! Goodbye.")
else:
print("\nNo problem. See you again!\n")
playgame() | ALU-PyQuest | /ALU-PyQuest-0.1.9.tar.gz/ALU-PyQuest-0.1.9/src/quiz.py | quiz.py |
# ALazyQiwi - Async Library for qiwi famrs
1. Install package
- Windows -> `$ pip install LazyQiwi`
- Mac OS & Linux -> `$ pip3 install LazyQiwi`
## Usage
### Import in your project
`from ALazyQiwi import LazyQiwi`
### Initialize
`api = LazyQiwi(token="")`
### Get account balance
```
async def bal():
token = "<your token here>"
api = LazyQiwi(token)
return await api.account_balance()
asyncio.run(bal())
>>> [0.0]
```
### Calculate transfer commission
```
async def main():
token = ""
api = LazyQiwi(token)
ww_amount = 1000
reciver = '7910XXXXXXX'
coms = await api.get_commission(ww_amount, reciver)
return coms
asyncio.run(main())
>>> 10
```
### Transfer money
```
async def transfer():
token = "<your token here>"
api = LazyQiwi(token)
await api.pay(account='7910XXXXXXX', amount=10.35, comment="Test transfer")
asyncio.run(transfer())
>>> None
```
### Create account information dump
```
# In one method
async def main():
token = "<your token here>"
api = LazyQiwi(token)
data = await api.create_account_dump()
print(data)
asyncio.run(main())
```
#### Get current account infromation
```
async def main():
token = "<your token here>"
api = LazyQiwi(token)
basic_data = await api.get_basic_info() # returns dict
account_number = basic_data['basic']['number']['number']
language = basic_data['basic']['language']
personal_data = await api.get_personal_info(account_number) # returns dict
limits = await api.get_accounts_limits(account_number, language) # returns dict
restrictions = await api.get_account_restriction(account_number) # returns list
print(
basic_data, personal_data, limits, restrictions
)
asyncio.run(main())
```
### Errors handlers
`InvalidTokenError` - token is invalid
`InRequestError` - trying to send http request and got error
`APIError` - if got error after server return result | ALazyQiwi | /ALazyQiwi-1.2.tar.gz/ALazyQiwi-1.2/README.md | README.md |
# ALife Data Standards - Python Development Utilities
[](https://github.com/alife-data-standards/alife-std-dev-python/actions/workflows/CI.yaml)
[](https://www.codacy.com/gh/alife-data-standards/alife-std-dev-python/dashboard?utm_source=github.com&utm_medium=referral&utm_content=alife-data-standards/alife-std-dev-python&utm_campaign=Badge_Grade)
[](https://codecov.io/gh/alife-data-standards/alife-std-dev-python)

This is the repository for the ALifeStdDev Python package, which contains Python
development utilities for working with [standardized](https://github.com/alife-data-standards/alife-data-standards)
ALife data.
# Installation Instructions
ALifeStdDev can be installed using pip:
```
pip install ALifeStdDev
```
# Usage Instructions
To load a single submodule,
```python3
from ALifeStdDev import phylogeny as asd_phylo
asd_phylo.load_phylogeny_to_pandas_df("myfile.csv")
```
To load the library as a flat namespace,
```python3
from ALifeStdDev import ALifeStdDev as asd
asd.load_phylogeny_to_pandas_df("myfile.csv")
```
| ALifeStdDev | /ALifeStdDev-0.2.4.tar.gz/ALifeStdDev-0.2.4/README.md | README.md |
MIT License
Copyright (c) 2018-2021 Alexis Huvier
Permission is hereby granted, free of charge, to any person obtaining a copy
of this software and associated documentation files (the "Software"), to deal
in the Software without restriction, including without limitation the rights
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
copies of the Software, and to permit persons to whom the Software is
furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all
copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
SOFTWARE. | AList | /AList-2.0.0-py3-none-any.whl/AList-2.0.0.dist-info/LICENSE.md | LICENSE.md |
from tkinter import ttk
from tkinter import *
from alist.utils import Config
from alist.pages import *
from alist.data import *
class Main(Tk):
def __init__(self):
super(Main, self).__init__()
self.mal = MALProvider()
self.imager = ImageProvider()
self.translator = TranslationProvider(self)
self.myanime = MyAnimeListProvider()
self.mymanga = MyMangaListProvider()
self.mal_export = MALExporter(self)
self.mal_import = MALImporter(self)
self.config = Config()
self.tk.call('source', 'alist/themes/' + self.config.get("theme", "azure-dark") + '.tcl')
ttk.Style().theme_use(self.config.get("theme", "azure-dark"))
self.title("AList")
self.geometry("1280x800")
self.resizable(width=False, height=False)
self.menu = Menu(self)
self.page = Frame(self)
self.page.pack(side=RIGHT)
self.current_page = ""
self.show_page("accueil")
self.mainloop()
def show_page(self, page):
if self.current_page != page:
self.page.destroy()
if page == "accueil" or (page == "reload" and self.current_page == "accueil"):
self.page = Accueil(self)
elif page == "list_anime" or (page == "reload" and self.current_page == "list_anime"):
self.page = ListAnime(self)
elif page == "list_manga" or (page == "reload" and self.current_page == "list_manga"):
self.page = ListManga(self)
elif page == "my_anime" or (page == "reload" and self.current_page == "my_anime"):
self.page = MyAnime(self)
elif page == "my_manga" or (page == "reload" and self.current_page == "my_manga"):
self.page = MyManga(self)
elif page == "parameters" or (page == "reload" and self.current_page == "parameters"):
self.page = Parameters(self)
elif page.startswith("anime ") or (page == "reload" and self.current_page.startswith("anime ")):
self.page = Anime(self, int(page.split(" ")[1]))
elif page.startswith("manga ") or (page == "reload" and self.current_page.startswith("manga ")):
self.page = Manga(self, int(page.split(" ")[1]))
elif page.startswith("modifanime ") or (page == "reload" and self.current_page.startswith("modifanime ")):
self.page = ModifAnime(self, int(page.split(" ")[1]))
elif page.startswith("modifmanga ") or (page == "reload" and self.current_page.startswith("modifmanga ")):
self.page = ModifManga(self, int(page.split(" ")[1]))
else:
print("ERROR : Unknown Page ("+page+")")
if page != "reload":
self.current_page = page
def launch():
Main()
if __name__ == "__main__":
Main() | AList | /AList-2.0.0-py3-none-any.whl/alist/AList.py | AList.py |
from lxml import etree
from tkinter import simpledialog
STATUS_AL_MAL = {
"A voir": "Plan to Watch",
"MA voir": "Plan to Read",
"En visionnement": "Watching",
"MEn visionnement": "Reading",
"Fini": "Completed",
"Abandonné": "Dropped"
}
class MALExporter:
def __init__(self, main):
self.main = main
def export(self, type_, file):
if type_ == "anime":
mal = etree.Element("myanimelist")
myinfo = etree.SubElement(mal, "myinfo")
username = etree.SubElement(myinfo, "user_name")
username.text = simpledialog.askstring("AList - Export MAL", "Pseudo MyAnimeList :")
export_type = etree.SubElement(myinfo, "user_export_type")
export_type.text = "1"
for i in self.main.myanime.get_all():
anime = etree.SubElement(mal, "anime")
infos = {
"series_animedb_id": str(i["id"]),
"series_title": str(i["name"]),
"series_episodes": str(i["max_ep"]),
"series_type": str(i["type"]),
"my_watched_episodes": str(i["ep"]),
"my_status": STATUS_AL_MAL[i["status"]],
"update_on_import": str(1)
}
for k, v in infos.items():
temp = etree.SubElement(anime, k)
temp.text = v
with open(file, "w") as f:
f.write(etree.tostring(mal, pretty_print=True).decode("utf-8"))
else:
mal = etree.Element("myanimelist")
myinfo = etree.SubElement(mal, "myinfo")
username = etree.SubElement(myinfo, "user_name")
username.text = simpledialog.askstring("AList - Export MAL", "Pseudo MyAnimeList :")
export_type = etree.SubElement(myinfo, "user_export_type")
export_type.text = "2"
for i in self.main.mymanga.get_all():
manga = etree.SubElement(mal, "manga")
if i["status"] == "En visionnement":
status = "MEn visionnement"
elif i["status"] == "A voir":
status = "MA voir"
else:
status = i["status"]
infos = {
"manga_mangadb_id": str(i["id"]),
"manga_title": str(i["name"]),
"manga_volumes": str(i["max_vol"]),
"manga_chaptes": str(i["max_chap"]),
"my_read_volumes": str(i["vol"]),
"my_read_chapters": str(i["chap"]),
"my_status": STATUS_AL_MAL[status],
"update_on_import": str(1)
}
for k, v in infos.items():
temp = etree.SubElement(manga, k)
temp.text = v
with open(file, "w") as f:
f.write(etree.tostring(mal, pretty_print=True).decode("utf-8")) | AList | /AList-2.0.0-py3-none-any.whl/alist/data/mal_export.py | mal_export.py |
from tkinter import ttk, Canvas
class ScrollFrame(ttk.Frame):
def __init__(self, parent, *args, **kwargs):
super().__init__(parent, *args, **kwargs)
self.canvas = Canvas(self, borderwidth=0)
self.viewport = ttk.Frame(self.canvas, width=kwargs["width"])
self.vsb = ttk.Scrollbar(self, orient="vertical", command=self.canvas.yview)
self.canvas.configure(yscrollcommand=self.vsb.set)
self.vsb.pack(side="right", fill="y")
self.canvas.pack(side="left", fill="both", expand=True)
self.canvas_window = self.canvas.create_window((0, 0), window=self.viewport, anchor="nw", tags="self.viewport")
self.viewport.bind("<Configure>", self.on_frame_configure)
self.canvas.bind("<Configure>", self.on_canvas_configure)
self.bind("<Enter>", self.bind_mousewheel)
self.bind("<Leave>", self.unbind_mousewheel)
self.on_frame_configure(None)
def bind_mousewheel(self, evt):
# Windows
self.canvas.bind_all("<MouseWheel>", self.on_canvas_mousewheel)
# Linux
self.canvas.bind_all("<Button-4>", self.on_canvas_mousewheel)
self.canvas.bind_all("<Button-5>", self.on_canvas_mousewheel)
def unbind_mousewheel(self, evt):
# Windows
self.canvas.unbind_all("<MouseWheel>")
# Linux
self.canvas.unbind_all("<Button-4>")
self.canvas.unbind_all("<Button-5>")
def on_frame_configure(self, event):
self.canvas.configure(scrollregion=self.canvas.bbox("all"))
def on_canvas_configure(self, event):
canvas_width = event.width
self.canvas.itemconfig(self.canvas_window, width=canvas_width)
def on_canvas_mousewheel(self, event):
if self.canvas.winfo_exists():
if event.num == 5 or event.delta == -120:
self.canvas.yview_scroll(1, "units")
if event.num == 4 or event.delta == 120:
self.canvas.yview_scroll(-1, "units")
else:
self.unbind_mousewheel(None) | AList | /AList-2.0.0-py3-none-any.whl/alist/utils/scroll_frame.py | scroll_frame.py |
from tkinter import ttk, RIGHT, BOTH, LEFT, StringVar, SUNKEN, filedialog
from alist.pages.right_page import RightPage
from alist.utils import ScrollFrame
class MyAnime(RightPage):
def __init__(self, main):
super(MyAnime, self).__init__(main)
title = ttk.Label(self, text="Mes Animes", font="-size 22 -weight bold")
title.pack(pady=15)
top_frame = ttk.Frame(self)
self.search = StringVar(self)
search_entry = ttk.Entry(top_frame, textvariable=self.search, width=30)
search_entry.pack(side=LEFT, padx=10)
valid_search = ttk.Button(top_frame, text="Rechercher", width=20, command=self.validate_search)
valid_search.pack(side=LEFT, padx=(10, 30))
self.etat = StringVar(self)
self.etat.set("Tous")
etat_select = ttk.OptionMenu(top_frame, self.etat, "Tous", "Tous", "A voir", "En visionnement", "Fini",
"Abandonné")
etat_select["width"] = 30
etat_select.pack(side=LEFT, padx=(30, 10))
valid_etat = ttk.Button(top_frame, text="Afficher", width=20, command=self.validate_state)
valid_etat.pack(side=LEFT, padx=10)
top_frame.pack(pady=10)
bottom_frame = ttk.Frame(self)
self.exp_imp = StringVar(self)
self.exp_imp.set("MyAnimeList")
exp_imp = ttk.OptionMenu(bottom_frame, self.exp_imp, "MyAnimeList")
exp_imp["width"] = 30
exp_imp.pack(side=LEFT, padx=20)
import_ = ttk.Button(bottom_frame, text="Import", width=20, command=self.import_)
import_.pack(side=LEFT, padx=10)
export = ttk.Button(bottom_frame, text="Export", width=20, command=self.export_)
export.pack(side=LEFT, padx=10)
bottom_frame.pack(pady=10)
self.result_frame = ScrollFrame(self, width=1080, height=800)
self.result_frame.pack_propagate(False)
self.result_frame.grid_propagate(False)
self.result_frame.pack(pady=(10, 0))
self.validate_state()
self.pack(side=RIGHT, fill=BOTH)
def import_(self):
if self.exp_imp.get() == "MyAnimeList":
file = filedialog.askopenfilename(parent=self, title="AList - Import MAL",
filetypes=(("Fichier MAL", ".xml"),), multiple=False)
if file:
self.main.mal_import.import_("anime", file)
self.main.show_page("reload")
def export_(self):
if self.exp_imp.get() == "MyAnimeList":
file = filedialog.asksaveasfilename(parent=self, title="AList - Export MAL",
filetypes=(("Fichier MAL", ".xml"),))
if file:
self.main.mal_export.export("anime", file)
def validate_search(self):
self.reload_results(self.main.myanime.search(self.search.get()))
def validate_state(self):
if self.etat.get() == "Tous":
self.reload_results(self.main.myanime.get_all())
else:
self.reload_results(self.main.myanime.get_all_state(self.etat.get()))
def reload_results(self, results):
self.result_frame.destroy()
self.result_frame = ScrollFrame(self, width=1080, height=800)
self.result_frame.pack_propagate(False)
self.result_frame.grid_propagate(False)
for i, anime in enumerate(results):
temp = ttk.Frame(self.result_frame.viewport, relief=SUNKEN)
temp.pack_propagate(False)
temp.config(width=400, height=220)
title = ttk.Label(temp, text=anime["name"], font="-size 13")
title.pack(pady=15)
status = ttk.Label(temp, text="Statut : "+anime["status"])
status.pack(pady=5)
ep = ttk.Label(temp, text="Episodes : "+str(anime["ep"])+"/"+str(anime["max_ep"]))
ep.pack(pady=10)
buttons = ttk.Frame(temp)
delete = ttk.Button(buttons, text="Supprimer", width=10,
command=lambda a=anime: self.delete_anime(a["id"]))
delete.pack(side=LEFT, padx=10)
more_info = ttk.Button(buttons, text="Plus d'info", width=10,
command=lambda a=anime: self.main.show_page("anime "+str(a["id"])))
more_info.pack(side=RIGHT, padx=10)
modify = ttk.Button(buttons, text="Modifier", width=10,
command=lambda a=anime: self.main.show_page("modifanime "+str(a["id"])))
modify.pack(padx=10)
buttons.pack(pady=(15, 3))
buttons2 = ttk.Frame(temp)
down = ttk.Button(buttons2, text="-1", width=3, command=lambda a=anime: self.modify_ep(a, a["ep"] - 1))
down.pack(side=LEFT, padx=10)
up = ttk.Button(buttons2, text="+1", width=3, command=lambda a=anime: self.modify_ep(a, a["ep"] + 1))
up.pack(side=RIGHT, padx=10)
buttons2.pack(pady=(3, 15))
temp.grid(row=i // 2, column=i % 2, pady=20)
self.result_frame.viewport.columnconfigure(0, weight=1)
self.result_frame.viewport.columnconfigure(1, weight=1)
self.result_frame.pack(pady=(10, 0))
def modify_ep(self, anime, nb):
if nb > anime["max_ep"]:
self.main.myanime.modify(anime["id"], ep=anime["max_ep"])
elif nb < 0:
self.main.myanime.modify(anime["id"], ep=0)
else:
self.main.myanime.modify(anime["id"], ep=nb)
self.main.show_page("reload")
def delete_anime(self, mal_id):
self.main.myanime.delete(mal_id)
self.main.show_page("reload") | AList | /AList-2.0.0-py3-none-any.whl/alist/pages/my_anime.py | my_anime.py |
from tkinter import ttk, BOTH, StringVar, RIGHT, LEFT
from alist.pages.right_page import RightPage
from alist.utils import ScrollFrame
class ListManga(RightPage):
def __init__(self, main):
super(ListManga, self).__init__(main)
self.current_page = 1
self.current_display = ""
title = ttk.Label(self, text="Liste Manga", font="-size 22 -weight bold")
title.pack(pady=15)
top_frame = ttk.Frame(self)
self.search = StringVar(self)
search_entry = ttk.Entry(top_frame, textvariable=self.search, width=30)
search_entry.pack(side=LEFT, padx=10)
valid_entry = ttk.Button(top_frame, text="Rechercher", command=self.validate_search, width=20)
valid_entry.pack(side=LEFT, padx=(10, 30))
self.top = StringVar(self)
self.top.set("Top Global")
top_select = ttk.OptionMenu(top_frame, self.top, "Top Global", "Top Global", "Top Manga", "Top Novels",
"Top Oneshots", "Top Doujin", "Top Manhwa", "Top Manhua", "Top Populaire",
"Top Favoris")
top_select["width"] = 30
top_select.pack(side=LEFT, padx=(30, 10))
valid_top = ttk.Button(top_frame, text="Afficher le top", command=self.validate_top, width=20)
valid_top.pack(side=LEFT, padx=10)
top_frame.pack(pady=20)
bottom_frame = ttk.Frame(self)
left_page = ttk.Button(bottom_frame, text="<-", width=20, command=self.previous_page)
left_page.pack(side=LEFT, padx=20)
self.num_page = ttk.Label(bottom_frame, text="Page 1")
self.num_page.pack(side=LEFT, padx=20)
right_page = ttk.Button(bottom_frame, text="->", width=20, command=self.next_page)
right_page.pack(side=LEFT, padx=20)
bottom_frame.pack(pady=10)
self.result_frame = ScrollFrame(self, width=1080, height=600)
self.result_frame.pack_propagate(False)
self.result_frame.grid_propagate(False)
self.result_frame.pack(pady=(20, 0))
self.pack(side=RIGHT, fill=BOTH)
def next_page(self):
if self.current_display.startswith("search "):
self.current_page += 1
self.reload_results(
self.main.mal.search("manga", " ".join(self.current_display.split(" ")[:1]), self.current_page)
)
else:
temp = {
"Top Global": None,
"Top Manga": "manga",
"Top Novels": "novels",
"Top Oneshots": "oneshots",
"Top Doujin": "doujin",
"Top Manhwa": "manhwa",
"Top Manhua": "manhua",
"Top Populaire": "bypopularity",
"Top Favoris": "favorite"
}
self.current_page += 1
self.reload_results(self.main.mal.top("manga", self.current_page, temp[self.current_display]))
self.num_page["text"] = "Page "+str(self.current_page)
def previous_page(self):
if self.current_page != 1:
if self.current_display.startswith("search "):
self.current_page -= 1
self.reload_results(
self.main.mal.search("manga", " ".join(self.current_display.split(" ")[:1]), self.current_page)
)
else:
temp = {
"Top Global": None,
"Top Manga": "manga",
"Top Novels": "novels",
"Top Oneshots": "oneshots",
"Top Doujin": "doujin",
"Top Manhwa": "manhwa",
"Top Manhua": "manhua",
"Top Populaire": "bypopularity",
"Top Favoris": "favorite"
}
self.current_page -= 1
self.reload_results(self.main.mal.top("manga", self.current_page, temp[self.current_display]))
self.num_page["text"] = "Page " + str(self.current_page)
def validate_search(self):
self.current_display = "search "+self.search.get()
self.current_page = 1
self.num_page["text"] = "Page "+str(self.current_page)
self.reload_results(self.main.mal.search("manga", self.search.get()))
def validate_top(self):
temp = {
"Top Global": None,
"Top Manga": "manga",
"Top Novels": "novels",
"Top Oneshots": "oneshots",
"Top Doujin": "doujin",
"Top Manhwa": "manhwa",
"Top Manhua": "manhua",
"Top Populaire": "bypopularity",
"Top Favoris": "favorite"
}
self.current_display = self.top.get()
self.current_page = 1
self.num_page["text"] = "Page "+str(self.current_page)
self.reload_results(self.main.mal.top("manga", 1, temp[self.top.get()]))
def reload_results(self, results):
self.result_frame.destroy()
self.result_frame = ScrollFrame(self, width=1080, height=600)
self.result_frame.pack_propagate(False)
self.result_frame.grid_propagate(False)
for i, manga in enumerate(results):
temp = ttk.Frame(self.result_frame.viewport)
image = ttk.Label(temp)
self.main.imager.apply_image_on_label("manga_" + str(manga["mal_id"]) + ".jpg", manga["image_url"], image)
image.pack()
title = ttk.Button(temp, text=manga["title"],
command=lambda m=manga: self.main.show_page("manga "+str(m["mal_id"])))
title.pack(pady=10)
temp.grid(row=i // 2, column=i % 2, pady=20)
self.result_frame.viewport.columnconfigure(0, weight=1)
self.result_frame.viewport.columnconfigure(1, weight=1)
self.result_frame.pack(pady=(20, 0)) | AList | /AList-2.0.0-py3-none-any.whl/alist/pages/list_manga.py | list_manga.py |
from tkinter import ttk, RIGHT, BOTH, LEFT
from alist.pages.right_page import RightPage
from alist.utils import utils
class Manga(RightPage):
def __init__(self, main, mal_id):
super(Manga, self).__init__(main)
self.mal_id = mal_id
self.manga = self.main.mal.manga(mal_id)
title = ttk.Label(self, text=self.manga["title"], font="-size 20 -weight bold")
title.pack(pady=15)
manga = ttk.Frame(self)
image = ttk.Label(manga)
self.main.imager.apply_image_on_label("manga_" + str(self.manga["mal_id"]) + ".jpg",
self.manga["image_url"],
image)
image.pack(side=RIGHT, padx=50)
type_ = ttk.Label(manga, text="Type : Manga")
type_.pack(pady=5)
if self.manga["title_english"]:
en_title = ttk.Label(manga, text="Titre Anglais : "+self.manga["title_english"])
else:
en_title = ttk.Label(manga, text="Titre Anglais : Aucun")
en_title.pack(pady=5)
if len(self.manga["authors"][0]["name"].split(", ")) == 2:
author = ttk.Label(manga, text="Auteur principal : "+self.manga["authors"][0]["name"].split(", ")[1]+" " +
self.manga["authors"][0]["name"].split(", ")[0])
else:
author = ttk.Label(manga, text="Auteur pricipal : "+self.manga["authors"][0]["name"])
author.pack(pady=5)
genres = ["Genres : "]
for k, v in enumerate(self.manga["genres"]):
if k != 0:
if k % 7 == 0: # Retour à ligne tous les 5 genres
genres.append(", \n")
else:
genres.append(", ")
genres.append(self.main.translator.manuel_translate(v["name"]))
genre = ttk.Label(manga, text="".join(genres), justify="center")
genre.pack(pady=5)
if self.manga["status"].startswith("Finished"):
status = ttk.Label(manga, text="Statut : Fini")
elif self.manga["status"] == "Publishing":
status = ttk.Label(manga, text="Statut : En cours en publication")
elif self.manga["status"] == "Not yet aired":
status = ttk.Label(manga, text="Statut : A Venir")
else:
status = ttk.Label(manga, text="Statut : Inconnu")
status.pack(pady=5)
if self.manga["volumes"]:
if self.manga["chapters"]:
vol_chap = ttk.Label(manga, text="Nombre de Volumes : " + str(self.manga["volumes"]) +
" Nombre de Chapitres : "+str(self.manga["chapters"]))
else:
vol_chap = ttk.Label(manga, text="Nombre de Volumes : " + str(self.manga["volumes"]) +
" Nombre de Chapitres : 0")
else:
if self.manga["chapters"]:
vol_chap = ttk.Label(manga, text="Nombre de Volumes : 0 Nombre de Chapitres : " +
str(self.manga["chapters"]))
else:
vol_chap = ttk.Label(manga, text="Nombre de Volumes : 0 Nombre de Chapitres : 0")
vol_chap.pack(pady=5)
stats_list = []
if self.manga["rank"]:
stats_list.append("Top : "+str(self.manga["rank"]))
else:
stats_list.append("Top : Inconnu")
if self.manga["score"]:
stats_list.append(" Score : "+str(self.manga["score"]))
else:
stats_list.append(" Score : Inconnu")
if self.manga["popularity"]:
stats_list.append(" Popularité : "+str(self.manga["popularity"]))
else:
stats_list.append(" Popularité : Inconnue")
stats = ttk.Label(manga, text="".join(stats_list))
stats.pack(pady=5)
synopsis_list = ["Synopsis :\n"]
if self.manga["synopsis"]:
mots = self.main.translator.translate(self.manga["synopsis"]).split(" ")
nb = 80
lignes = 0
for i in mots:
if nb - len(i) <= 0:
nb = 80 - len(i)
lignes += 1
if lignes == 20:
synopsis_list.append("...")
break
else:
synopsis_list.extend(("\n", i, " "))
else:
synopsis_list.extend((i, " "))
nb -= len(i)
else:
synopsis_list.append("Aucun")
synopsis = ttk.Label(manga, text="".join(synopsis_list), justify="center")
synopsis.pack(pady=20)
manga.pack()
buttons = ttk.Frame(self)
lien = ttk.Button(buttons, text="Lien MAL", width=20,
command=lambda: utils.open_url(self.manga["url"]))
lien.pack(side=LEFT, padx=20)
add_list = ttk.Button(buttons, text="Ajouter à ma liste", width=20, command=self.add_to_list)
add_list.pack(side=RIGHT, padx=20)
buttons.pack(pady=10)
self.pack(side=RIGHT, fill=BOTH)
def add_to_list(self):
if self.manga["chapters"]:
if self.manga["volumes"]:
self.main.mymanga.add(self.manga["mal_id"], self.manga["title"], self.manga["volumes"],
self.manga["chapters"], self.manga["type"])
else:
self.main.mymanga.add(self.manga["mal_id"], self.manga["title"], 0, self.manga["chapters"],
self.manga["type"])
else:
if self.manga["volumes"]:
self.main.mymanga.add(self.manga["mal_id"], self.manga["title"], self.manga["volumes"], 0,
self.manga["type"])
else:
self.main.mymanga.add(self.manga["mal_id"], self.manga["title"], 0, 0, self.manga["type"]) | AList | /AList-2.0.0-py3-none-any.whl/alist/pages/manga.py | manga.py |
from tkinter import ttk, RIGHT, BOTH, LEFT
from alist.pages.right_page import RightPage
from alist.utils import utils
class Anime(RightPage):
def __init__(self, main, mal_id):
super(Anime, self).__init__(main)
self.mal_id = mal_id
self.anime = self.main.mal.anime(mal_id)
title = ttk.Label(self, text=self.anime["title"], font="-size 20 -weight bold")
title.pack(pady=15)
anime = ttk.Frame(self)
image = ttk.Label(anime)
self.main.imager.apply_image_on_label("anime_" + str(self.anime["mal_id"]) + ".jpg",
self.anime["image_url"],
image)
image.pack(side=RIGHT, padx=50)
type_ = ttk.Label(anime, text="Type : Anime")
type_.pack(pady=5)
if self.anime["title_english"]:
en_title = ttk.Label(anime, text="Titre Anglais : "+self.anime["title_english"])
else:
en_title = ttk.Label(anime, text="Titre Anglais : Aucun")
en_title.pack(pady=5)
author = ttk.Label(anime, text="Studio Principal : "+self.anime["studios"][0]["name"])
author.pack(pady=5)
genres = ["Genres : "]
for k, v in enumerate(self.anime["genres"]):
if k != 0:
if k % 7 == 0: # Retour à ligne tous les 5 genres
genres.append(", \n")
else:
genres.append(", ")
genres.append(self.main.translator.manuel_translate(v["name"]))
genre = ttk.Label(anime, text="".join(genres), justify="center")
genre.pack(pady=5)
if self.anime["status"].startswith("Finished"):
status = ttk.Label(anime, text="Statut : Fini")
elif self.anime["status"] == "Publishing":
status = ttk.Label(anime, text="Statut : En cours en publication")
elif self.anime["status"] == "Not yet aired":
status = ttk.Label(anime, text="Statut : A Venir")
else:
status = ttk.Label(anime, text="Statut : Inconnu")
status.pack(pady=5)
if self.anime["episodes"]:
ep = ttk.Label(anime, text="Nombre d'Episodes : "+str(self.anime["episodes"]))
else:
ep = ttk.Label(anime, text="Nombre d'Episodes : 0")
ep.pack(pady=5)
stats_list = []
if self.anime["rank"]:
stats_list.append("Top : "+str(self.anime["rank"]))
else:
stats_list.append("Top : Inconnu")
if self.anime["score"]:
stats_list.append(" Score : "+str(self.anime["score"]))
else:
stats_list.append(" Score : Inconnu")
if self.anime["popularity"]:
stats_list.append(" Popularité : "+str(self.anime["popularity"]))
else:
stats_list.append(" Popularité : Inconnue")
stats = ttk.Label(anime, text="".join(stats_list))
stats.pack(pady=5)
synopsis_list = ["Synopsis :\n"]
if self.anime["synopsis"]:
mots = self.main.translator.translate(self.anime["synopsis"]).split(" ")
nb = 80
lignes = 0
for i in mots:
if nb - len(i) <= 0:
nb = 80 - len(i)
lignes += 1
if lignes == 20:
synopsis_list.append("...")
break
else:
synopsis_list.extend(("\n", i, " "))
else:
synopsis_list.extend((i, " "))
nb -= len(i)
else:
synopsis_list.append("Aucun")
synopsis = ttk.Label(anime, text="".join(synopsis_list), justify="center")
synopsis.pack(pady=20)
anime.pack()
buttons = ttk.Frame(self)
lien = ttk.Button(buttons, text="Lien MAL", width=20,
command=lambda: utils.open_url(self.anime["url"]))
lien.pack(side=LEFT, padx=20)
trailer = ttk.Button(buttons, text="Trailer", width=20,
command=lambda: utils.open_url(self.anime["trailer_url"]))
trailer.pack(side=RIGHT, padx=20)
add_list = ttk.Button(buttons, text="Ajouter à ma liste", width=20, command=self.add_to_list)
add_list.pack(padx=20)
buttons.pack(pady=10)
self.pack(side=RIGHT, fill=BOTH)
def add_to_list(self):
if self.anime["episodes"]:
self.main.myanime.add(self.anime["mal_id"], self.anime["title"], self.anime["episodes"], self.anime["type"])
else:
self.main.myanime.add(self.anime["mal_id"], self.anime["title"], 0, self.anime["type"]) | AList | /AList-2.0.0-py3-none-any.whl/alist/pages/anime.py | anime.py |
from tkinter import ttk, RIGHT, BOTH, StringVar, LEFT, SUNKEN, filedialog
from alist.pages.right_page import RightPage
from alist.utils import ScrollFrame
class MyManga(RightPage):
def __init__(self, main):
super(MyManga, self).__init__(main)
title = ttk.Label(self, text="Mes Mangas", font="-size 22 -weight bold")
title.pack(pady=15)
top_frame = ttk.Frame(self)
self.search = StringVar(self)
search_entry = ttk.Entry(top_frame, textvariable=self.search, width=30)
search_entry.pack(side=LEFT, padx=10)
valid_search = ttk.Button(top_frame, text="Rechercher", width=20, command=self.validate_search)
valid_search.pack(side=LEFT, padx=(10, 30))
self.etat = StringVar(self)
self.etat.set("Tous")
etat_select = ttk.OptionMenu(top_frame, self.etat, "Tous", "Tous", "A voir", "En visionnement", "Fini",
"Abandonné")
etat_select["width"] = 30
etat_select.pack(side=LEFT, padx=(30, 10))
valid_etat = ttk.Button(top_frame, text="Afficher", width=20, command=self.validate_state)
valid_etat.pack(side=LEFT, padx=10)
top_frame.pack(pady=10)
bottom_frame = ttk.Frame(self)
self.exp_imp = StringVar(self)
self.exp_imp.set("MyAnimeList")
exp_imp = ttk.OptionMenu(bottom_frame, self.exp_imp, "MyAnimeList")
exp_imp["width"] = 30
exp_imp.pack(side=LEFT, padx=20)
import_ = ttk.Button(bottom_frame, text="Import", width=20, command=self.import_)
import_.pack(side=LEFT, padx=10)
export = ttk.Button(bottom_frame, text="Export", width=20, command=self.export_)
export.pack(side=LEFT, padx=10)
bottom_frame.pack(pady=10)
self.result_frame = ScrollFrame(self, width=1080, height=800)
self.result_frame.pack_propagate(False)
self.result_frame.grid_propagate(False)
self.result_frame.pack(pady=(10, 0))
self.validate_state()
self.pack(side=RIGHT, fill=BOTH)
def import_(self):
if self.exp_imp.get() == "MyAnimeList":
file = filedialog.askopenfilename(parent=self, title="AList - Import MAL",
filetypes=(("Fichier MAL", ".xml"),), multiple=False)
if file:
self.main.mal_import.import_("manga", file)
self.main.show_page("reload")
def export_(self):
if self.exp_imp.get() == "MyAnimeList":
file = filedialog.asksaveasfilename(parent=self, title="AList - Export MAL",
filetypes=(("Fichier MAL", ".xml"),))
if file:
self.main.mal_export.export("manga", file)
def validate_search(self):
self.reload_results(self.main.mymanga.search(self.search.get()))
def validate_state(self):
if self.etat.get() == "Tous":
self.reload_results(self.main.mymanga.get_all())
else:
self.reload_results(self.main.mymanga.get_all_state(self.etat.get()))
def reload_results(self, results):
self.result_frame.destroy()
self.result_frame = ScrollFrame(self, width=1080, height=800)
self.result_frame.pack_propagate(False)
self.result_frame.grid_propagate(False)
for i, manga in enumerate(results):
temp = ttk.Frame(self.result_frame.viewport, relief=SUNKEN)
temp.pack_propagate(False)
temp.config(width=400, height=220)
title = ttk.Label(temp, text=manga["name"], font="-size 13")
title.pack(pady=10)
status = ttk.Label(temp, text="Statut : "+manga["status"])
status.pack(pady=5)
ep = ttk.Label(temp, text="Volumes : "+str(manga["vol"])+"/"+str(manga["max_vol"]))
ep.pack(pady=5)
ep = ttk.Label(temp, text="Chapitres : "+str(manga["chap"])+"/"+str(manga["max_chap"]))
ep.pack(pady=5)
buttons = ttk.Frame(temp)
delete = ttk.Button(buttons, text="Supprimer", width=10,
command=lambda a=manga: self.delete_manga(a["id"]))
delete.pack(side=LEFT, padx=10)
more_info = ttk.Button(buttons, text="Plus d'info",
command=lambda a=manga: self.main.show_page("manga "+str(a["id"])))
more_info.pack(side=RIGHT, padx=10)
modify = ttk.Button(buttons, text="Modifier",
command=lambda a=manga: self.main.show_page("modifmanga "+str(a["id"])))
modify.pack(padx=10)
buttons.pack(pady=(15, 3))
buttons2 = ttk.Frame(temp)
down_vol = ttk.Button(buttons2, text="-1 Vol", width=8,
command=lambda a=manga: self.modify_vol_chap(a, "vol", a["vol"] - 1))
down_vol.pack(side=LEFT, padx=10)
down_chap = ttk.Button(buttons2, text="-1 Chap", width=8,
command=lambda a=manga: self.modify_vol_chap(a, "chap", a["chap"] - 1))
down_chap.pack(side=LEFT, padx=10)
up_vol = ttk.Button(buttons2, text="+1 Vol", width=8,
command=lambda a=manga: self.modify_vol_chap(a, "vol", a["vol"] + 1))
up_vol.pack(side=RIGHT, padx=10)
up_chap = ttk.Button(buttons2, text="+1 Chap", width=8,
command=lambda a=manga: self.modify_vol_chap(a, "chap", a["chap"] + 1))
up_chap.pack(side=RIGHT, padx=10)
buttons2.pack(pady=(3, 15))
temp.grid(row=i // 2, column=i % 2, pady=20)
self.result_frame.viewport.columnconfigure(0, weight=1)
self.result_frame.viewport.columnconfigure(1, weight=1)
self.result_frame.pack(pady=(10, 0))
def modify_vol_chap(self, manga, type_, nb):
if nb > manga["max_"+type_]:
eval("self.main.mymanga.modify(manga[\"id\"], "+type_+"=manga[\"max_\""+type_+"])")
elif nb < 0:
eval("self.main.mymanga.modify(manga[\"id\"], "+type_+"=0)")
else:
eval("self.main.mymanga.modify(manga[\"id\"], "+type_+"=nb)")
self.main.show_page("reload")
def delete_manga(self, mal_id):
self.main.mymanga.delete(mal_id)
self.main.show_page("reload") | AList | /AList-2.0.0-py3-none-any.whl/alist/pages/my_manga.py | my_manga.py |
from tkinter import ttk, BOTH, StringVar, RIGHT, LEFT
from alist.pages.right_page import RightPage
from alist.utils import ScrollFrame
class ListAnime(RightPage):
def __init__(self, main):
super(ListAnime, self).__init__(main)
self.current_page = 1
self.current_display = ""
title = ttk.Label(self, text="Liste Animes", font="-size 22 -weight bold")
title.pack(pady=15)
top_frame = ttk.Frame(self)
self.search = StringVar(self)
search_entry = ttk.Entry(top_frame, textvariable=self.search, width=30)
search_entry.pack(side=LEFT, padx=10)
valid_search = ttk.Button(top_frame, text="Rechercher", command=self.validate_search, width=20)
valid_search.pack(side=LEFT, padx=(10, 30))
self.top = StringVar(self)
self.top.set("Top Global")
top_select = ttk.OptionMenu(top_frame, self.top, "Top Global", "Top Global", "Top Sortie", "Top A Venir",
"Top TV", "Top Film", "Top OVA", "Top Spécial", "Top Populaire", "Top Favoris")
top_select["width"] = 30
top_select.pack(side=LEFT, padx=(30, 10))
valid_top = ttk.Button(top_frame, text="Afficher le Top", command=self.validate_top, width=20)
valid_top.pack(side=LEFT, padx=10)
top_frame.pack(pady=20)
bottom_frame = ttk.Frame(self)
left_page = ttk.Button(bottom_frame, text="<-", width=20, command=self.previous_page)
left_page.pack(side=LEFT, padx=20)
self.num_page = ttk.Label(bottom_frame, text="Page 1")
self.num_page.pack(side=LEFT, padx=20)
right_page = ttk.Button(bottom_frame, text="->", width=20, command=self.next_page)
right_page.pack(side=LEFT, padx=20)
bottom_frame.pack(pady=10)
self.result_frame = ScrollFrame(self, width=1080, height=600)
self.result_frame.pack_propagate(False)
self.result_frame.grid_propagate(False)
self.result_frame.pack(pady=(20, 0))
self.pack(side=RIGHT, fill=BOTH)
def next_page(self):
if self.current_display.startswith("search "):
self.current_page += 1
self.reload_results(
self.main.mal.search("anime", " ".join(self.current_display.split(" ")[:1]), self.current_page)
)
else:
temp = {
"Top Global": None,
"Top Sortie": "airing",
"Top A Venir": "upcoming",
"Top TV": "tv",
"Top Film": "movie",
"Top OVA": "ova",
"Top Spécial": "special",
"Top Populaire": "bypopularity",
"Top Favoris": "favorite"
}
self.current_page += 1
self.reload_results(self.main.mal.top("anime", self.current_page, temp[self.current_display]))
self.num_page["text"] = "Page "+str(self.current_page)
def previous_page(self):
if self.current_page != 1:
if self.current_display.startswith("search "):
self.current_page -= 1
self.reload_results(
self.main.mal.search("anime", " ".join(self.current_display.split(" ")[:1]), self.current_page)
)
else:
temp = {
"Top Global": None,
"Top Sortie": "airing",
"Top A Venir": "upcoming",
"Top TV": "tv",
"Top Film": "movie",
"Top OVA": "ova",
"Top Spécial": "special",
"Top Populaire": "bypopularity",
"Top Favoris": "favorite"
}
self.current_page -= 1
self.reload_results(self.main.mal.top("anime", self.current_page, temp[self.current_display]))
self.num_page["text"] = "Page "+str(self.current_page)
def validate_search(self):
self.current_display = "search "+self.search.get()
self.current_page = 1
self.num_page["text"] = "Page "+str(self.current_page)
self.reload_results(self.main.mal.search('anime', self.search.get()))
def validate_top(self):
temp = {
"Top Global": None,
"Top Sortie": "airing",
"Top A Venir": "upcoming",
"Top TV": "tv",
"Top Film": "movie",
"Top OVA": "ova",
"Top Spécial": "special",
"Top Populaire": "bypopularity",
"Top Favoris": "favorite"
}
self.current_display = self.top.get()
self.current_page = 1
self.num_page["text"] = "Page "+str(self.current_page)
self.reload_results(self.main.mal.top("anime", 1, temp[self.top.get()]))
def reload_results(self, results):
self.result_frame.destroy()
self.result_frame = ScrollFrame(self, width=1080, height=600)
self.result_frame.pack_propagate(False)
self.result_frame.grid_propagate(False)
for i, anime in enumerate(results):
temp = ttk.Frame(self.result_frame.viewport)
image = ttk.Label(temp)
self.main.imager.apply_image_on_label("anime_" + str(anime["mal_id"]) + ".jpg", anime["image_url"], image)
image.pack()
title = ttk.Button(temp, text=anime["title"],
command=lambda a=anime: self.main.show_page("anime "+str(a["mal_id"])))
title.pack(pady=10)
temp.grid(row=i // 2, column=i % 2, pady=20)
self.result_frame.viewport.columnconfigure(0, weight=1)
self.result_frame.viewport.columnconfigure(1, weight=1)
self.result_frame.pack(pady=(20, 0)) | AList | /AList-2.0.0-py3-none-any.whl/alist/pages/list_anime.py | list_anime.py |
import math
import matplotlib.pyplot as plt
from .Generaldistribution import Distribution
class Gaussian(Distribution):
""" Gaussian distribution class for calculating and
visualizing a Gaussian distribution.
Attributes:
mean (float) representing the mean value of the distribution
stdev (float) representing the standard deviation of the distribution
data_list (list of floats) a list of floats extracted from the data file
"""
def __init__(self, mu=0, sigma=1):
Distribution.__init__(self, mu, sigma)
def calculate_mean(self):
"""Function to calculate the mean of the data set.
Args:
None
Returns:
float: mean of the data set
"""
avg = 1.0 * sum(self.data) / len(self.data)
self.mean = avg
return self.mean
def calculate_stdev(self, sample=True):
"""Function to calculate the standard deviation of the data set.
Args:
sample (bool): whether the data represents a sample or population
Returns:
float: standard deviation of the data set
"""
if sample:
n = len(self.data) - 1
else:
n = len(self.data)
mean = self.calculate_mean()
sigma = 0
for d in self.data:
sigma += (d - mean) ** 2
sigma = math.sqrt(sigma / n)
self.stdev = sigma
return self.stdev
def plot_histogram(self):
"""Function to output a histogram of the instance variable data using
matplotlib pyplot library.
Args:
None
Returns:
None
"""
plt.hist(self.data)
plt.title('Histogram of Data')
plt.xlabel('data')
plt.ylabel('count')
def pdf(self, x):
"""Probability density function calculator for the gaussian distribution.
Args:
x (float): point for calculating the probability density function
Returns:
float: probability density function output
"""
return (1.0 / (self.stdev * math.sqrt(2*math.pi))) * math.exp(-0.5*((x - self.mean) / self.stdev) ** 2)
def plot_histogram_pdf(self, n_spaces = 50):
"""Function to plot the normalized histogram of the data and a plot of the
probability density function along the same range
Args:
n_spaces (int): number of data points
Returns:
list: x values for the pdf plot
list: y values for the pdf plot
"""
mu = self.mean
sigma = self.stdev
min_range = min(self.data)
max_range = max(self.data)
# calculates the interval between x values
interval = 1.0 * (max_range - min_range) / n_spaces
x = []
y = []
# calculate the x values to visualize
for i in range(n_spaces):
tmp = min_range + interval*i
x.append(tmp)
y.append(self.pdf(tmp))
# make the plots
fig, axes = plt.subplots(2,sharex=True)
fig.subplots_adjust(hspace=.5)
axes[0].hist(self.data, density=True)
axes[0].set_title('Normed Histogram of Data')
axes[0].set_ylabel('Density')
axes[1].plot(x, y)
axes[1].set_title('Normal Distribution for \n Sample Mean and Sample Standard Deviation')
axes[0].set_ylabel('Density')
plt.show()
return x, y
def __add__(self, other):
"""Function to add together two Gaussian distributions
Args:
other (Gaussian): Gaussian instance
Returns:
Gaussian: Gaussian distribution
"""
result = Gaussian()
result.mean = self.mean + other.mean
result.stdev = math.sqrt(self.stdev ** 2 + other.stdev ** 2)
return result
def __repr__(self):
"""Function to output the characteristics of the Gaussian instance
Args:
None
Returns:
string: characteristics of the Gaussian
"""
return "mean {}, standard deviation {}".format(self.mean, self.stdev) | ALust-probability | /ALust_probability-0.1.tar.gz/ALust_probability-0.1/ALust_probability/Gaussiandistribution.py | Gaussiandistribution.py |
import math
import matplotlib.pyplot as plt
from .Generaldistribution import Distribution
class Binomial(Distribution):
""" Binomial distribution class for calculating and
visualizing a Binomial distribution.
Attributes:
mean (float) representing the mean value of the distribution
stdev (float) representing the standard deviation of the distribution
data_list (list of floats) a list of floats to be extracted from the data file
p (float) representing the probability of an event occurring
n (int) number of trials
TODO: Fill out all functions below
"""
def __init__(self, prob=.5, size=20):
self.n = size
self.p = prob
Distribution.__init__(self, self.calculate_mean(), self.calculate_stdev())
def calculate_mean(self):
"""Function to calculate the mean from p and n
Args:
None
Returns:
float: mean of the data set
"""
self.mean = self.p * self.n
return self.mean
def calculate_stdev(self):
"""Function to calculate the standard deviation from p and n.
Args:
None
Returns:
float: standard deviation of the data set
"""
self.stdev = math.sqrt(self.n * self.p * (1 - self.p))
return self.stdev
def replace_stats_with_data(self):
"""Function to calculate p and n from the data set
Args:
None
Returns:
float: the p value
float: the n value
"""
self.n = len(self.data)
self.p = 1.0 * sum(self.data) / len(self.data)
self.mean = self.calculate_mean()
self.stdev = self.calculate_stdev()
def plot_bar(self):
"""Function to output a histogram of the instance variable data using
matplotlib pyplot library.
Args:
None
Returns:
None
"""
plt.bar(x = ['0', '1'], height = [(1 - self.p) * self.n, self.p * self.n])
plt.title('Bar Chart of Data')
plt.xlabel('outcome')
plt.ylabel('count')
def pdf(self, k):
"""Probability density function calculator for the gaussian distribution.
Args:
x (float): point for calculating the probability density function
Returns:
float: probability density function output
"""
a = math.factorial(self.n) / (math.factorial(k) * (math.factorial(self.n - k)))
b = (self.p ** k) * (1 - self.p) ** (self.n - k)
return a * b
def plot_bar_pdf(self):
"""Function to plot the pdf of the binomial distribution
Args:
None
Returns:
list: x values for the pdf plot
list: y values for the pdf plot
"""
x = []
y = []
# calculate the x values to visualize
for i in range(self.n + 1):
x.append(i)
y.append(self.pdf(i))
# make the plots
plt.bar(x, y)
plt.title('Distribution of Outcomes')
plt.ylabel('Probability')
plt.xlabel('Outcome')
plt.show()
return x, y
def __add__(self, other):
"""Function to add together two Binomial distributions with equal p
Args:
other (Binomial): Binomial instance
Returns:
Binomial: Binomial distribution
"""
try:
assert self.p == other.p, 'p values are not equal'
except AssertionError as error:
raise
result = Binomial()
result.n = self.n + other.n
result.p = self.p
result.calculate_mean()
result.calculate_stdev()
return result
def __repr__(self):
"""Function to output the characteristics of the Binomial instance
Args:
None
Returns:
string: characteristics of the Gaussian
"""
return "mean {}, standard deviation {}, p {}, n {}".\
format(self.mean, self.stdev, self.p, self.n) | ALust-probability | /ALust_probability-0.1.tar.gz/ALust_probability-0.1/ALust_probability/Binomialdistribution.py | Binomialdistribution.py |
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from scipy import ndimage
import sys
from .grid_utils import get_filtered_frame
from scipy.spatial import ConvexHull, convex_hull_plot_2d
def get_object_center(obj_id, labeled_image):
""" Returns index of center pixel of the given object id from labeled
image. The center is calculated as the median pixel of the object extent;
it is not a true centroid. """
obj_index = np.argwhere(labeled_image == obj_id)
center = np.median(obj_index, axis=0).astype('i')
return center
def get_obj_extent(labeled_image, obj_label):
""" Takes in labeled image and finds the radius, area, and center of the
given object. """
obj_index = np.argwhere(labeled_image == obj_label)#vi tri lab
xlength = np.max(obj_index[:, 0]) - np.min(obj_index[:, 0]) + 1
ylength = np.max(obj_index[:, 1]) - np.min(obj_index[:, 1]) + 1
obj_radius = np.max((xlength, ylength))/2#ban kinh max
obj_center = np.round(np.median(obj_index, axis=0), 0)#vitri trung tam
obj_area = len(obj_index[:, 0])#pham vi o dong
obj_extent = {'obj_center': obj_center, 'obj_radius': obj_radius,
'obj_area': obj_area, 'obj_index': obj_index}
return obj_extent
def init_current_objects(first_frame, second_frame, pairs, counter):
""" Returns a dictionary for objects with unique ids and their
corresponding ids in frame1 and frame1. This function is called when
echoes are detected after a period of no echoes. """
nobj = np.max(first_frame)
id1 = np.arange(nobj) + 1
uid = counter.next_uid(count=nobj)
id2 = pairs
obs_num = np.zeros(nobj, dtype='i')
origin = np.array(['-1']*nobj)
current_objects = {'id1': id1, 'uid': uid, 'id2': id2,
'obs_num': obs_num, 'origin': origin}
current_objects = attach_last_heads(first_frame, second_frame,
current_objects)
return current_objects, counter
def update_current_objects(frame1, frame2, pairs, old_objects, counter):
""" Removes dead objects, updates living objects, and assigns new uids to
new-born objects. """
nobj = np.max(frame1)
id1 = np.arange(nobj) + 1
uid = np.array([], dtype='str')
obs_num = np.array([], dtype='i')
origin = np.array([], dtype='str')
for obj in np.arange(nobj) + 1:
if obj in old_objects['id2']:
obj_index = old_objects['id2'] == obj
uid = np.append(uid, old_objects['uid'][obj_index])
obs_num = np.append(obs_num, old_objects['obs_num'][obj_index] + 1)
origin = np.append(origin, old_objects['origin'][obj_index])
else:
# obj_orig = get_origin_uid(obj, frame1, old_objects)
obj_orig = '-1'
origin = np.append(origin, obj_orig)
if obj_orig != '-1':
uid = np.append(uid, counter.next_cid(obj_orig))
else:
uid = np.append(uid, counter.next_uid())
obs_num = np.append(obs_num, 0)
id2 = pairs
current_objects = {'id1': id1, 'uid': uid, 'id2': id2,
'obs_num': obs_num, 'origin': origin}
current_objects = attach_last_heads(frame1, frame2, current_objects)
return current_objects, counter
def attach_last_heads(frame1, frame2, current_objects):
""" Attaches last heading information to current_objects dictionary. """
nobj = len(current_objects['uid'])
heads = np.ma.empty((nobj, 2))
for obj in range(nobj):
if ((current_objects['id1'][obj] > 0) and
(current_objects['id2'][obj] > 0)):
center1 = get_object_center(current_objects['id1'][obj], frame1)
center2 = get_object_center(current_objects['id2'][obj], frame2)
heads[obj, :] = center2 - center1
else:
heads[obj, :] = np.ma.array([-999, -999], mask=[True, True])
current_objects['last_heads'] = heads
return current_objects
def check_isolation(raw, filtered, grid_size, params):
""" Returns list of booleans indicating object isolation. Isolated objects
are not connected to any other objects by pixels greater than ISO_THRESH,
and have at most one peak. """
nobj = np.max(filtered)
min_size = params['MIN_SIZE'] / np.prod(grid_size[1:]/1000)
iso_filtered = get_filtered_frame(raw,
min_size,
params['ISO_THRESH'])
nobj_iso = np.max(iso_filtered)
iso = np.empty(nobj, dtype='bool')
for iso_id in np.arange(nobj_iso) + 1:
obj_ind = np.where(iso_filtered == iso_id)
objects = np.unique(filtered[obj_ind])
objects = objects[objects != 0]
if len(objects) == 1 and single_max(obj_ind, raw, params):
iso[objects - 1] = True
else:
iso[objects - 1] = False
return iso
def single_max(obj_ind, raw, params):
""" Returns True if object has at most one peak. """
max_proj = np.max(raw, axis=0)
smooth = ndimage.filters.gaussian_filter(max_proj, params['ISO_SMOOTH'])
padded = np.pad(smooth, 1, mode='constant')
obj_ind = [axis + 1 for axis in obj_ind] # adjust for padding
maxima = 0
for pixel in range(len(obj_ind[0])):
ind_0 = obj_ind[0][pixel]
ind_1 = obj_ind[1][pixel]
neighborhood = padded[(ind_0-1):(ind_0+2), (ind_1-1):(ind_1+2)]
max_ind = np.unravel_index(neighborhood.argmax(), neighborhood.shape)
if max_ind == (1, 1):
maxima += 1
if maxima > 1:
return False
return True
def get_object_prop(image1, grid1, field, record, params):
""" Returns dictionary of object properties for all objects found in
image1. """
id1 = []
center = []
grid_x = []
grid_y = []
area = []
longitude = []
latitude = []
field_max = []
field_mean = []
nobj = np.max(image1)
#soodong=[]
unit_dim = record.grid_size
unit_alt = unit_dim[0]/1000
unit_area = 1 #(unit_dim[1]*unit_dim[2])/(1000**2)
unit_vol = (unit_dim[0]*unit_dim[1]*unit_dim[2])/(1000**3)
raw3D = grid1['data']
get_items = []
lonoj=[]
latoj=[]
for obj in np.arange(nobj) + 1:
try:
obj_index = np.argwhere(image1 == obj)
this_centroid = np.round(np.mean(obj_index, axis=0), 3)
rounded = np.round(this_centroid).astype('i')
c_x = grid1['x'][rounded[1]]
c_y = grid1['y'][rounded[0]]
longitude.append(np.round(grid1['x'][rounded[1]], 4))
latitude.append(np.round(grid1['y'][rounded[0]], 4))
id1.append(obj)
# 2D frame stats
center.append(np.median(obj_index, axis=0))
grid_x.append(this_centroid[1])
grid_y.append(this_centroid[0])
area.append(obj_index.shape[0] * unit_area)
lo_A=grid1['x'][obj_index[:,1]]
la_A=grid1['y'][obj_index[:,0]]
points = np.vstack((lo_A,la_A)).T
hull = ConvexHull(points)
lonoj.append(points[hull.simplices][:,:,0].ravel().astype('float32'))
latoj.append(points[hull.simplices][:,:,1].ravel().astype('float32'))
# raw 3D grid stats
obj_slices = [raw3D[:, ind[0], ind[1]] for ind in obj_index]
field_max.append(np.nanmax(obj_slices))
field_mean.append(np.nanmean(obj_slices))
filtered_slices = [obj_slice > params['FIELD_THRESH']
for obj_slice in obj_slices]
get_items.append(obj - 1)
except IndexError:
pass
# cell isolation
isolation = check_isolation(raw3D, image1, record.grid_size, params)
objprop = {'id1': id1,
'center': center,
'grid_x': grid_x,
'grid_y': grid_y,
'area': area,
'field_max': field_max,
'field_mean':field_mean,
'lon': longitude,
'lat': latitude,
'isolated': isolation,
'ok_items': get_items,
'lonoj': lonoj,
'latoj': latoj}
return objprop
def write_tracks(old_tracks, record, current_objects, obj_props):
""" Writes all cell information to tracks dataframe. """
sys.stdout.flush()
sys.stdout.write('\rWriting tracks for scan %s \n'%(record.scan))
sys.stdout.flush()
nobj = len(obj_props['id1'])
scan_num = [record.scan] * nobj
gi = obj_props['ok_items']
uid = current_objects['uid'][gi]
new_tracks = pd.DataFrame({
'scan': scan_num,
'uid': uid,
'time': record.time,
'grid_x': obj_props['grid_x'],
'grid_y': obj_props['grid_y'],
'lon': obj_props['lon'],
'lat': obj_props['lat'],
'area': obj_props['area'],
'max': obj_props['field_max'],
'mean': obj_props['field_mean'],
'isolated': obj_props['isolated'][gi],
'lonoj': obj_props['lonoj'],
'latoj': obj_props['latoj']
})
new_tracks.set_index(['scan', 'uid'], inplace=True)
tracks = old_tracks.append(new_tracks)
return tracks | AM-project1 | /AM_project1-0.0.2.tar.gz/AM_project1-0.0.2/AM_project1/objects.py | objects.py |
import datetime
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from scipy import ndimage
def parse_grid_datetime(grid_obj):
""" Obtains datetime object from the data dictionary. """
#dt_string = grid_obj.time['units'].split(' ')[-1]
#date = dt_string[:10]
##time = dt_string[11:19]
#dt = datetime.datetime.strptime(date + ' ' + time, '%Y-%m-%d %H:%M:%S')
return grid_obj['time']
def get_grid_size(grid_obj):
""" Calculates grid size per dimension given a grid object. """
return np.array(grid_obj['data'].shape)
def get_radar_info(radar):
info = {'radar_lon': radar[0],
'radar_lat': radar[0]}
return info
def get_grid_alt(grid_size, alt_meters=1500):
""" Returns z-index closest to alt_meters. """
return np.int(np.round(alt_meters/grid_size[0]))
def get_vert_projection(grid, thresh=40):
""" Returns boolean vertical projection from grid. """
return np.any(grid > thresh, axis=0)
def get_filtered_frame(grid, min_size, thresh):
""" Returns a labeled frame from gridded radar data. Smaller objects
are removed and the rest are labeled. """
echo_height = get_vert_projection(grid, thresh)
labeled_echo = ndimage.label(echo_height)[0]
frame = clear_small_echoes(labeled_echo, min_size)
return frame
def clear_small_echoes(label_image, min_size):
""" Takes in binary image and clears objects less than min_size. """
flat_image = pd.Series(label_image.flatten())
flat_image = flat_image[flat_image > 0]
size_table = flat_image.value_counts(sort=False)
small_objects = size_table.keys()[size_table < min_size]
for obj in small_objects:
label_image[label_image == obj] = 0
label_image = ndimage.label(label_image)
return label_image[0]
def extract_grid_data(grid_obj, field, grid_size, params):
""" Returns filtered grid frame and raw grid slice at global shift
altitude. """
try:
masked = grid_obj['data'].filled(0)
except AttributeError:
masked = grid_obj['data']
gs_alt = params['GS_ALT']
raw = masked[0 , :, :]#value
frame = get_filtered_frame(masked, params['MIN_SIZE'],
params['FIELD_THRESH'])
return raw, frame | AM-project1 | /AM_project1-0.0.2.tar.gz/AM_project1-0.0.2/AM_project1/grid_utils.py | grid_utils.py |
import string
import numpy as np
import pandas as pd
from datetime import datetime, timedelta
from netCDF4 import num2date, date2num
from .grid_utils import parse_grid_datetime, get_grid_size
class Counter(object):
"""
Counter objects generate and keep track of unique cell ids.
Currently only the uid attribute is used, but this framework can
accomodate further development of merge/split detection.
Attributes
----------
uid : int
Last uid assigned.
cid : dict
Record of cell genealogy.
"""
def __init__(self):
""" uid is an integer that tracks the number of independently formed
cells. The cid dictionary keeps track of 'children' --i.e., cells that
have split off from another cell. """
self.uid = -1
self.cid = {}
def next_uid(self, count=1):
""" Incremented for every new independently formed cell. """
new_uids = self.uid + np.arange(count) + 1
self.uid += count
return np.array([str(uid) for uid in new_uids])
def next_cid(self, pid):
""" Returns parent uid with appended letter to denote child. """
if pid in self.cid.keys():
self.cid[pid] += 1
else:
self.cid[pid] = 0
letter = string.ascii_lowercase[self.cid[pid]]
return pid + letter
class Record(object):
"""
Record objects keep track of information related to the shift correction
process.
Attributes
----------
scan : int
Index of the current scan.
time : datetime
Time corresponding to scan.
interval : timedelta
Temporal difference between the next scan and the current scan.
interval_ratio : float
Ratio of current interval to previous interval.
grid_size : array of floats
Length 3 array containing z, y, and x mesh size in meters.
shifts : dataframe
Records inputs of shift correction process. See matching.correct_shift.
new_shfits : dataframe
Row of new shifts to be added to shifts dataframe.
correction_tally : dict
Tallies correction cases for performance analysis.
Shift Correction Case Guide:
case0 - new object, local_shift and global_shift disagree, returns global
case1 - new object, returns local_shift
case2 - local disagrees with last head and global, returns last head
case3 - local disagrees with last head, returns local
case4 - local and last head agree, returns average of both
case5 - flow regions empty or at edge of frame, returns global_shift
"""
def __init__(self, grid_obj):
self.scan = -1
self.time = None
self.interval = None
self.interval_ratio = None
self.grid_size = get_grid_size(grid_obj)
self.shifts = pd.DataFrame()
self.new_shifts = pd.DataFrame()
self.correction_tally = {'case0': 0, 'case1': 0, 'case2': 0,
'case3': 0, 'case4': 0, 'case5': 0}
def count_case(self, case_num):
""" Updates correction_tally dictionary. This is used to monitor the
shift correction process. """
self.correction_tally['case' + str(case_num)] += 1
def record_shift(self, corr, gl_shift, l_heads, local_shift, case):
""" Records corrected shift, phase shift, global shift, and last
heads per object per timestep. This information can be used to
monitor and refine the shift correction algorithm in the
correct_shift function. """
if l_heads is None:
l_heads = np.ma.array([-999, -999], mask=[True, True])
new_shift_record = pd.DataFrame()
new_shift_record['scan'] = [self.scan]
new_shift_record['uid'] = ['uid']
new_shift_record['corrected'] = [corr]
new_shift_record['global'] = [gl_shift]
new_shift_record['last_heads'] = [l_heads]
new_shift_record['phase'] = [local_shift]
new_shift_record['case'] = [case]
self.new_shifts = self.new_shifts.append(new_shift_record)
def add_uids(self, current_objects):
""" Because of the chronology of the get_tracks process, object uids
cannot be added to the shift record at the time of correction, so they
must be added later in the process. """
if len(self.new_shifts) > 0:
self.new_shifts['uid'] = current_objects['uid']
self.new_shifts.set_index(['scan', 'uid'], inplace=True)
self.shifts = self.shifts.append(self.new_shifts)
self.new_shifts = pd.DataFrame()
def update_scan_and_time(self, grid_obj1, grid_obj2=None):
""" Updates the scan number and associated time. This information is
used for obtaining object properties as well as for the interval ratio
correction of last_heads vectors. """
self.scan += 1
self.time = parse_grid_datetime(grid_obj1)
if grid_obj2 is None:
# tracks for last scan are being written
return
time2 = parse_grid_datetime(grid_obj2)
old_diff = self.interval
self.interval = time2 - self.time
if old_diff is not None:
self.interval_ratio = self.interval.seconds/old_diff.seconds
def spl(present, time):
out = []
start = True
a = ''.join(list(present.astype(int).astype(str)))
b = list(present.astype(int).astype(str))
ii = 0
for k, g in groupby(a):
gg = list(g)
if len(gg) == 1:
b[ii] = str(1 - int(gg[0]))
ii += len(gg)
ii = 0
for k, g in groupby(''.join(b)):
G = list(g)
if k == '1':
kk = 0
for i in G:
d1 = time[kk+ii]
try:
d2 = time[kk+ii+1]
except IndexError:
break
if (d2 - d1) > 24*60**2:
break
kk += 1
if (ii+kk-1) - ii > 5:
out.append((ii, ii+kk-1))
ii += len(G)
return out
def get_grids(group, slices, lon, lat, varname='rain_rate'):
x = group.variables[varname].shape[1]
y = group.variables[varname].shape[2]
for s in range(slices[0], slices[-1]+1):
yield {'x': lon, 'y': lat,
'data': group.variables[varname][s].reshape(1, x, y),
'time': num2date(group.variables['time'][s],
group.variables['time'].units)}
def get_times(time, start=None, end=None, isfile=None):
'''Get the start and end index for a given period'''
if type(end) == type('a') and type(start) == type('a'):
end = datetime.strptime(end, '%Y-%m-%d %H:%M')
start = datetime.strptime(start, '%Y-%m-%d %H:%M')
start = date2num([start], time.units)
end = date2num([end], time.units)
e_idx = np.argmin(np.fabs(time[:] - end))+1
s_idx = np.argmin(np.fabs(time[:] - start))
return [(s_idx, e_idx)]
elif type(isfile) == type(None):
isifle = np.ones_like(time.shape[0])
return spl(isfile, time) | AM-project1 | /AM_project1-0.0.2.tar.gz/AM_project1-0.0.2/AM_project1/helpers.py | helpers.py |
import numpy as np
from scipy import optimize
from .phase_correlation import get_ambient_flow
from .objects import get_obj_extent
LARGE_NUM = 1000
def euclidean_dist(vec1, vec2):
""" Computes euclidean distance. """
vec1 = np.array(vec1)
vec2 = np.array(vec2)
dist = np.sqrt(sum((vec1-vec2)**2))
return dist
def get_sizeChange(size1, size2):
""" Returns change in size of an echo as the ratio of the larger size to
the smaller, minus 1. """
if (size1 < 5) and (size2 < 5):
return 0
elif size1 >= size2:
return size1/size2 - 1
else:
return size2/size1 - 1
def find_objects(search_box, image2):
""" Identifies objects found in the search region. """
if not search_box['valid']:
obj_found = np.array(-1)
else:
search_area = image2[search_box['x1']:search_box['x2'],
search_box['y1']:search_box['y2']]
obj_found = np.unique(search_area)
return obj_found
def shifts_disagree(shift1, shift2, record, params):
""" Returns True if shift disparity greater than MAX_SHIFT_DISP
parameter. """
shift1 = shift1*record.grid_size[1:]
shift2 = shift2*record.grid_size[1:]
shift_disparity = euclidean_dist(shift1, shift2)
return shift_disparity/record.interval.seconds > params['MAX_SHIFT_DISP']
def clip_shift(shift, record, params):
""" Clips shift according to MAX_FLOW_MAG paramter. """#so sanh vs toc do toi da m/s
shift_meters = shift * record.grid_size[1:]
shift_mag = np.linalg.norm(shift_meters)
velocity = shift_mag/record.interval.seconds
unit = shift_meters/shift_mag
#print(velocity,unit,record.grid_size[1:],record.interval.seconds)
if velocity > params['MAX_FLOW_MAG']:
clipped = unit * params['MAX_FLOW_MAG'] * record.interval.seconds
clipped_pix = clipped/record.grid_size[1:]
return clipped_pix
else:
return shift
def correct_shift(local_shift, current_objects, obj_id1, global_shift, record,
params):
""" Takes in flow vector based on local phase correlation (see
get_std_flow) and compares it to the last headings of the object and
the global_shift vector for that timestep. Corrects accordingly.
Note: At the time of this function call, current_objects has not yet been
updated for the current frame1 and frame2, so the id2s in current_objects
correspond to the objects in the current frame1. """
global_shift = clip_shift(global_shift, record, params)
#print('global_shift in correct_shift',global_shift)
if current_objects is None:
last_heads = None
else:
obj_index = current_objects['id2'] == obj_id1
last_heads = current_objects['last_heads'][obj_index].flatten()
last_heads = np.round(last_heads * record.interval_ratio, 2)
if len(last_heads) == 0:
last_heads = None
if last_heads is None:
if shifts_disagree(local_shift, global_shift, record, params):
case = 0
corrected_shift = global_shift
else:
case = 1
corrected_shift = (local_shift + global_shift)/2
elif shifts_disagree(local_shift, last_heads, record, params):
if shifts_disagree(local_shift, global_shift, record, params):
case = 2
corrected_shift = last_heads
else:
case = 3
corrected_shift = local_shift
else:
case = 4
corrected_shift = (local_shift + last_heads)/2
corrected_shift = np.round(corrected_shift, 2)
record.count_case(case)
record.record_shift(corrected_shift, global_shift,
last_heads, local_shift, case)
return corrected_shift#hieu chinh lai vectoc
def predict_search_extent(obj1_extent, shift, params, grid_size):
""" Predicts search extent/region for the object in image2 given
the image shift. """# tim vung anh huong cua o dong du doan
shifted_center = obj1_extent['obj_center'] + shift
search_radius_r = params['SEARCH_MARGIN'] / grid_size[1]
search_radius_c = params['SEARCH_MARGIN'] / grid_size[2]
x1 = shifted_center[0] - search_radius_r
x2 = shifted_center[0] + search_radius_r + 1
y1 = shifted_center[1] - search_radius_c
y2 = shifted_center[1] + search_radius_c + 1
x1 = np.int(x1)
x2 = np.int(x2)
y1 = np.int(y1)
y2 = np.int(y2)
return {'x1': x1, 'x2': x2, 'y1': y1, 'y2': y2,
'center_pred': shifted_center, 'valid': True}
def check_search_box(search_box, img_dims):
""" Checks if search_box is within the boundaries of the frame. Clips to
edges of frame if out of bounds. Marks as invalid if too small. """
if search_box['x1'] < 0:
search_box['x1'] = 0
if search_box['y1'] < 0:
search_box['y1'] = 0
if search_box['x2'] > img_dims[0]:
search_box['x2'] = img_dims[0]
if search_box['y2'] > img_dims[1]:
search_box['y2'] = img_dims[1]
if ((search_box['x2'] - search_box['x1'] < 5) or
(search_box['y2'] - search_box['y1'] < 5)):
search_box['valid'] = False
return search_box
def get_disparity(obj_found, image2, search_box, obj1_extent):
""" Computes disparities for objects in obj_found. """
dist_pred = np.empty(0)
change = np.empty(0)
for target_obj in obj_found:
target_extent = get_obj_extent(image2, target_obj)
euc_dist = euclidean_dist(target_extent['obj_center'],
search_box['center_pred'])
dist_pred = np.append(dist_pred, euc_dist)
size_changed = get_sizeChange(target_extent['obj_area'],
obj1_extent['obj_area'])
change = np.append(change, size_changed)
disparity = dist_pred + change
return disparity
def get_disparity_all(obj_found, image2, search_box, obj1_extent):
""" Returns disparities of all objects found within the search box. """
if np.max(obj_found) <= 0:
disparity = np.array([LARGE_NUM])
else:
obj_found = obj_found[obj_found > 0]
disparity = get_disparity(obj_found, image2,
search_box, obj1_extent)
return disparity
def save_obj_match(obj_id1, obj_found, disparity, obj_match, params):
""" Saves disparity values in obj_match matrix. If disparity is greater than
MAX_DISPARITY, saves a large number. """
disparity[disparity > params['MAX_DISPARITY']] = LARGE_NUM
if np.max(obj_found) > 0:
obj_found = obj_found[obj_found > 0]
obj_found = obj_found - 1
obj_id1 = obj_id1 - 1
obj_match[obj_id1, obj_found] = disparity
return obj_match
def locate_allObjects(image1, image2, global_shift, current_objects, record,
params):
""" Matches all the objects in image1 to objects in image2. This is the
main function called on a pair of images. """#ket noi id
nobj1 = np.max(image1)
nobj2 = np.max(image2)
if (nobj2 == 0) or (nobj1 == 0):
print('No echoes to track!')
return
obj_match = np.full((nobj1, np.max((nobj1, nobj2))),
LARGE_NUM, dtype='f')
index_obj={}
for obj_id1 in np.arange(nobj1) + 1:
obj1_extent = get_obj_extent(image1, obj_id1)#dua ra cac ttobj_extent = {'obj_center': obj_center, 'obj_radius': obj_radius,
#'obj_area': obj_area, 'obj_index': obj_index}
#print('obj1_extent',obj1_extent['obj_index'])
index_obj[str(obj_id1)]= obj1_extent['obj_index']#them 27042021
shift = get_ambient_flow(obj1_extent, image1,
image2, params, record.grid_size)#lay vectoc cua 1 o dong obj_id1
if shift is None:
record.count_case(5)
shift = global_shift
shift = correct_shift(shift, current_objects, obj_id1,
global_shift, record, params)
search_box = predict_search_extent(obj1_extent, shift,
params, record.grid_size)# du doan khung anh huong của o dong
search_box = check_search_box(search_box, image2.shape)# check pham vi co nam trong khung 2
objs_found = find_objects(search_box, image2)# tim tat ca cac o dong tren khung thuc te o thoi diem sau.
disparity = get_disparity_all(objs_found, image2,
search_box, obj1_extent)# tinh toan khoang cach cua odong k1 vs ca odong tim dc trong k2
#print('disparity',disparity)
obj_match = save_obj_match(obj_id1, objs_found, disparity, obj_match,
params)
return obj_match,index_obj
def match_pairs(obj_match, params):
""" Matches objects into pairs given a disparity matrix and removes
bad matches. Bad matches have a disparity greater than the maximum
threshold. """
pairs = optimize.linear_sum_assignment(obj_match)
for id1 in pairs[0]:
if obj_match[id1, pairs[1][id1]] > params['MAX_DISPARITY']:
pairs[1][id1] = -1 # -1 indicates the object has died
pairs = pairs[1] + 1 # ids in current_objects are 1-indexed
return pairs
def get_pairs(image1, image2, global_shift, current_objects, record, params):
""" Given two images, this function identifies the matching objects and
pairs them appropriately. See disparity function. """ #image1 -lab,global_shift - vectoc tong,
nobj1 = np.max(image1)
nobj2 = np.max(image2)
if nobj1 == 0:
print('No echoes found in the first scan.')
return
elif nobj2 == 0:
zero_pairs = np.zeros(nobj1)
return zero_pairs
obj_match,index_obj = locate_allObjects(image1,
image2,
global_shift,
current_objects,
record,
params)
pairs = match_pairs(obj_match, params)
return pairs | AM-project1 | /AM_project1-0.0.2.tar.gz/AM_project1-0.0.2/AM_project1/matching.py | matching.py |
import copy
import datetime
import sys
import numpy as np
import pandas as pd
from .grid_utils import get_grid_size, get_radar_info, extract_grid_data
from .helpers import Record, Counter
from .phase_correlation import get_global_shift
from .matching import get_pairs
from .objects import init_current_objects, update_current_objects
from .objects import get_object_prop, write_tracks
# Tracking Parameter Defaults
FIELD_THRESH = 32
ISO_THRESH = 4
ISO_SMOOTH = 4
MIN_SIZE = 8
SEARCH_MARGIN = 250
FLOW_MARGIN = 750
MAX_DISPARITY = 999
MAX_FLOW_MAG = 50
MAX_SHIFT_DISP = 15
GS_ALT = 1500
"""
Tracking Parameter Guide
------------------------
FIELD_THRESH : units of 'field' attribute
The threshold used for object detection. Detected objects are connnected
pixels above this threshold.
ISO_THRESH : units of 'field' attribute
Used in isolated cell classification. Isolated cells must not be connected
to any other cell by contiguous pixels above this threshold.
ISO_SMOOTH : pixels
Gaussian smoothing parameter in peak detection preprocessing. See
single_max in tint.objects.
MIN_SIZE : square kilometers
The minimum size threshold in pixels for an object to be detected.
SEARCH_MARGIN : meters
The radius of the search box around the predicted object center.
FLOW_MARGIN : meters
The margin size around the object extent on which to perform phase
correlation.
MAX_DISPARITY : float
Maximum allowable disparity value. Larger disparity values are sent to
LARGE_NUM.
MAX_FLOW_MAG : meters per second
Maximum allowable global shift magnitude. See get_global_shift in
tint.phase_correlation.
MAX_SHIFT_DISP : meters per second
Maximum magnitude of difference in meters per second for two shifts to be
considered in agreement. See correct_shift in tint.matching.
GS_ALT : meters
Altitude in meters at which to perform phase correlation for global shift
calculation. See correct_shift in tint.matching.
"""
class Cell_tracks(object):
def __init__(self, field='reflectivity'):
self.params = {'FIELD_THRESH': FIELD_THRESH,
'MIN_SIZE': MIN_SIZE,
'SEARCH_MARGIN': SEARCH_MARGIN,
'FLOW_MARGIN': FLOW_MARGIN,
'MAX_FLOW_MAG': MAX_FLOW_MAG,
'MAX_DISPARITY': MAX_DISPARITY,
'MAX_SHIFT_DISP': MAX_SHIFT_DISP,
'ISO_THRESH': ISO_THRESH,
'ISO_SMOOTH': ISO_SMOOTH,
'GS_ALT': GS_ALT}
self.field = field
self.grid_size = None
self.radar_info = None
self.last_grid = None
self.counter = None
self.record = None
self.current_objects = None
self.tracks = pd.DataFrame()
self.odong = None
self.__saved_record = None
self.__saved_counter = None
self.__saved_objects = None
def __save(self):
""" Saves deep copies of record, counter, and current_objects. """
self.__saved_record = copy.deepcopy(self.record)
self.__saved_counter = copy.deepcopy(self.counter)
self.__saved_objects = copy.deepcopy(self.current_objects)
def __load(self):
""" Loads saved copies of record, counter, and current_objects. If new
tracks are appended to existing tracks via the get_tracks method, the
most recent scan prior to the addition must be overwritten to link up
with the new scans. Because of this, record, counter and
current_objects must be reverted to their state in the penultimate
iteration of the loop in get_tracks. See get_tracks for details. """
self.record = self.__saved_record
self.counter = self.__saved_counter
self.current_objects = self.__saved_objects
def get_tracks(self, grids, c):
""" Obtains tracks given a list of data arrays. This is the
primary method of the tracks class. This method makes use of all of the
functions and helper classes defined above. """
start_time = datetime.datetime.now()
ncells = 0
if self.record is None:
# tracks object being initialized
grid_obj2 = next(grids)
self.grid_size = get_grid_size(grid_obj2)
self.radar_info = get_radar_info(c)
self.counter = Counter()
self.record = Record(grid_obj2)
else:
# tracks object being updated
grid_obj2 = self.last_grid
self.tracks.drop(self.record.scan + 1) # last scan is overwritten
if self.current_objects is None:
newRain = True
else:
newRain = False
raw2, frame2 = extract_grid_data(grid_obj2, self.field, self.grid_size,
self.params)
self.odong=[]
while grid_obj2 is not None:
grid_obj1 = grid_obj2
raw1 = raw2
frame1 = frame2
try:
grid_obj2 = next(grids)
except StopIteration:
grid_obj2 = None
if grid_obj2 is not None:
self.record.update_scan_and_time(grid_obj1, grid_obj2)
raw2, frame2 = extract_grid_data(grid_obj2,
self.field,
self.grid_size,
self.params)
else:
self.__save()
self.last_grid = grid_obj1
self.record.update_scan_and_time(grid_obj1)
raw2 = None
frame2 = np.zeros_like(frame1)
if np.nanmax(frame1) == 0:
newRain = True
sys.stdout.flush()
sys.stdout.write('\rNo cells found in scan %s '%(self.record.scan))
sys.stdout.flush()
self.current_objects = None
continue
ncells += 1
global_shift = get_global_shift(raw1, raw2, self.params)
pairs = get_pairs(frame1,
frame2,
global_shift,
self.current_objects,
self.record,
self.params)
if newRain:
# first nonempty scan after a period of empty scans
self.current_objects, self.counter = init_current_objects(
frame1,
frame2,
pairs,
self.counter
)
newRain = False
else:
self.current_objects, self.counter = update_current_objects(
frame1,
frame2,
pairs,
self.current_objects,
self.counter
)
#try:
obj_props = get_object_prop(frame1, grid_obj1, self.field,
self.record, self.params)
self.record.add_uids(self.current_objects)
self.tracks = write_tracks(self.tracks, self.record,
self.current_objects, obj_props)
#except IndexError:
#obj_props = None
self.odong.append(frame1)
del grid_obj1, raw1, frame1, global_shift, pairs, obj_props
# scan loop end
self.__load()
time_elapsed = datetime.datetime.now() - start_time
print('time elapsed', np.round(time_elapsed.seconds/60, 1), 'minutes')
return ncells | AM-project1 | /AM_project1-0.0.2.tar.gz/AM_project1-0.0.2/AM_project1/tracks.py | tracks.py |
import numpy as np
from scipy import ndimage
def get_ambient_flow(obj_extent, img1, img2, params, grid_size):
margin_r = params['FLOW_MARGIN'] / grid_size[1]
margin_c = params['FLOW_MARGIN'] / grid_size[2]
row_lb = obj_extent['obj_center'][0] - obj_extent['obj_radius'] - margin_r
row_ub = obj_extent['obj_center'][0] + obj_extent['obj_radius'] + margin_r
col_lb = obj_extent['obj_center'][1] - obj_extent['obj_radius'] - margin_c
col_ub = obj_extent['obj_center'][1] + obj_extent['obj_radius'] + margin_c
row_lb = np.int(row_lb)
row_ub = np.int(row_ub)
col_lb = np.int(col_lb)
col_ub = np.int(col_ub)
dims = img1.shape
row_lb = np.max([row_lb, 0])
row_ub = np.min([row_ub, dims[0]])
col_lb = np.max([col_lb, 0])
col_ub = np.max([col_ub, dims[1]])
flow_region1 = np.copy(img1[row_lb:row_ub+1, col_lb:col_ub+1])
flow_region2 = np.copy(img2[row_lb:row_ub+1, col_lb:col_ub+1])
flow_region1[flow_region1 != 0] = 1
flow_region2[flow_region2 != 0] = 1#mang 2 chieu
return fft_flowvectors(flow_region1, flow_region2)
def fft_flowvectors(im1, im2, global_shift=False):
""" Estimates flow vectors in two images using cross covariance. """
if not global_shift and (np.max(im1) == 0 or np.max(im2) == 0):
return None
crosscov = fft_crosscov(im1, im2)
sigma = (1/8) * min(crosscov.shape)
cov_smooth = ndimage.filters.gaussian_filter(crosscov, sigma)
dims = np.array(im1.shape)
pshift = np.argwhere(cov_smooth == np.max(cov_smooth))[0]
pshift = (pshift+1) - np.round(dims/2, 0)
return pshift
def fft_crosscov(im1, im2):
""" Computes cross correlation matrix using FFT method. """
fft1_conj = np.conj(np.fft.fft2(im1))
fft2 = np.fft.fft2(im2)
normalize = abs(fft2*fft1_conj)
normalize[normalize == 0] = 1
cross_power_spectrum = (fft2*fft1_conj)/normalize
crosscov = np.fft.ifft2(cross_power_spectrum)
crosscov = np.real(crosscov)
return fft_shift(crosscov)
def fft_shift(fft_mat):
""" Rearranges the cross correlation matrix so that 'zero' frequency or DC
component is in the middle of the matrix. Taken from stackoverflow Que.
30630632. """
if type(fft_mat) is np.ndarray:
rd2 = np.int(fft_mat.shape[0]/2)
cd2 = np.int(fft_mat.shape[1]/2)
quad1 = fft_mat[:rd2, :cd2]
quad2 = fft_mat[:rd2, cd2:]
quad3 = fft_mat[rd2:, cd2:]
quad4 = fft_mat[rd2:, :cd2]
centered_t = np.concatenate((quad4, quad1), axis=0)
centered_b = np.concatenate((quad3, quad2), axis=0)
centered = np.concatenate((centered_b, centered_t), axis=1)
return centered
else:
print('input to fft_shift() should be a matrix')
return
def get_global_shift(im1, im2, params):
""" Returns standardazied global shift vector. im1 and im2 are full frames
of raw DBZ values. """
if im2 is None:
return None
shift = fft_flowvectors(im1, im2, global_shift=True)
return shift | AM-project1 | /AM_project1-0.0.2.tar.gz/AM_project1-0.0.2/AM_project1/phase_correlation.py | phase_correlation.py |
# recommend_reactions.py
"""
Predicts annotations of reaction(s) using a local XML file
and the reaction ID.
Usage: python recommend_reaction.py files/BIOMD0000000190.xml --cutoff 0.6 --outfile res.csv
"""
import argparse
import os
from os.path import dirname, abspath
import pandas as pd
import sys
sys.path.insert(0, dirname(dirname(abspath(__file__))))
from AMAS import constants as cn
from AMAS import recommender
def main():
parser = argparse.ArgumentParser(description='Recommend reaction annotations of an SBML model and save results')
parser.add_argument('model', type=str, help='SBML model file (.xml)')
# One or more reaction IDs can be given
parser.add_argument('--reactions', type=str, help='ID(s) of reaction(s) to be recommended. ' +\
'If not provided, all reactions will be used', nargs='*')
parser.add_argument('--min_len', type=int, help='Minimum number of reaction components (reactants and products) ' +\
'to be used for prediction. ' +\
'Reactions with fewer components than this value ' +\
'will be ignored. Default is zero.', nargs='?', default=0)
parser.add_argument('--cutoff', type=float, help='Match score cutoff.', nargs='?', default=0.0)
parser.add_argument('--mssc', type=str,
help='Match score selection criteria (MSSC). ' +\
'Choose either "top" or "above". "top" recommends ' +\
'the best candidates that are above the cutoff, ' +\
'and "above" recommends all candidates that are above ' +\
'the cutoff. Default is "top"',
nargs='?',
default='top')
parser.add_argument('--outfile', type=str, help='File path to save recommendation.', nargs='?',
default=os.path.join(os.getcwd(), 'reaction_rec.csv'))
args = parser.parse_args()
recom = recommender.Recommender(libsbml_fpath=args.model)
one_fpath = args.model
reacts = args.reactions
min_len = args.min_len
cutoff = args.cutoff
mssc = args.mssc.lower()
outfile = args.outfile
#
recom = recommender.Recommender(libsbml_fpath=one_fpath)
# # if nothing is given, predict all IDs
if reacts is None:
reacts = recom.getReactionIDs()
print("...\nAnalyzing %d reaction(s)...\n" % len(reacts))
res_tab = recom.recommendReactions(ids=reacts,
mssc=mssc,
cutoff=cutoff,
min_len=min_len,
outtype='table')
recom.saveToCSV(res_tab, outfile)
if isinstance(res_tab, pd.DataFrame):
print("Recommendations saved as:\n%s\n" % os.path.abspath(outfile))
if __name__ == '__main__':
main() | AMAS-sb | /AMAS_sb-1.0.1-py3-none-any.whl/AMAS_sb-1.0.1.data/scripts/recommend_reactions.py | recommend_reactions.py |
# recommend_annotation.py
"""
Predicts annotations of species and reactions using a local XML file
and the reaction ID.
This is a combined version of recommend_species and recommend_reaction,
but is more convenient because user will just get the updated XML file or whole recommendations.
Usage: python recommend_reaction.py files/BIOMD0000000190.xml --cutoff 0.6 --save csv --outfile res.csv
"""
import argparse
import itertools
import libsbml
import numpy as np
import os
from os.path import dirname, abspath
import sys
sys.path.insert(0, dirname(dirname(abspath(__file__))))
from AMAS import constants as cn
from AMAS import iterator as it
from AMAS import species_annotation as sa
from AMAS import reaction_annotation as ra
from AMAS import recommender
def main():
parser = argparse.ArgumentParser(description='Recommend annotations of an SBML model ' +\
'(for both species and reactions) and save results.')
parser.add_argument('model', type=str, help='SBML model file (.xml).')
# One or more reaction IDs can be given
parser.add_argument('--cutoff', type=float, help='Match score cutoff.', nargs='?', default=0.0)
parser.add_argument('--optimize', type=str, help='Whether to optimize or not. ' +\
'If y or yes is given, predictions will be ' +\
'optimized. N or no will not optimize predictions.',
nargs='?',
default='no')
parser.add_argument('--mssc', type=str,
help='Match score selection criteria (MSSC). ' +\
'Choose either "top" or "above". "top" recommends ' +\
'the best candidates that are above the cutoff, ' +\
'and "above" recommends all candidates that are above ' +\
'the cutoff. Default is "top"',
nargs='?',
default='top')
parser.add_argument('--save', type=str,
help='Either "sbml" or "csv". ' +\
'If "sbml" is chosen, model will be automatically ' +\
'annotated with recommended candidates and saved. ' +\
'If "csv" is chosen, recommendations will be saved ' +\
'as a csv file. Default is "csv".',
nargs='?',
default='sbml')
parser.add_argument('--outfile', type=str, help='Path to save an output file.', nargs='?')
args = parser.parse_args()
recom = recommender.Recommender(libsbml_fpath=args.model)
one_fpath = args.model
cutoff = args.cutoff
optim_raw = args.optimize
if optim_raw.lower() in ['y', 'yes']:
optim = True
else:
optim = False
mssc = args.mssc.lower()
save = args.save
outfile = args.outfile
#
recom = recommender.Recommender(libsbml_fpath=one_fpath)
specs = recom.getSpeciesIDs()
print("...\nAnalyzing %d species...\n" % len(specs))
reacts = recom.getReactionIDs()
print("...\nAnalyzing %d reaction(s)...\n" % len(reacts))
res_tab = recom.recommendAnnotation(mssc=mssc,
cutoff=cutoff,
optimize=optim,
outtype='table')
if save == 'csv':
if outfile is None:
outfile = os.path.join(os.getcwd(), 'recommendations.csv')
recom.saveToCSV(res_tab, outfile)
else:
if outfile is None:
outfile = os.path.join(os.getcwd(), 'updated_model.xml')
res_sbml = recom.getSBMLDocument(sbml_document=recom.sbml_document,
chosen=res_tab,
auto_feedback=True)
libsbml.writeSBMLToFile(res_sbml, outfile)
print("Recommendations saved as:\n%s\n" % os.path.abspath(outfile))
if __name__ == '__main__':
main() | AMAS-sb | /AMAS_sb-1.0.1-py3-none-any.whl/AMAS_sb-1.0.1.data/scripts/recommend_annotation.py | recommend_annotation.py |
# recommend_species.py
"""
Predicts annotations of species using a local XML file
and the species ID.
Usage: python recommend_species.py files/BIOMD0000000190.xml --min_len 2 --cutoff 0.6 --outfile res.csv
"""
import argparse
import os
from os.path import dirname, abspath
import pandas as pd
import sys
sys.path.insert(0, dirname(dirname(abspath(__file__))))
from AMAS import constants as cn
from AMAS import recommender
def main():
parser = argparse.ArgumentParser(description='Recommend species annotations of an SBML model and save results')
parser.add_argument('model', type=str, help='SBML model file (.xml)')
# One or more species IDs can be given
parser.add_argument('--species', type=str, help='ID(s) of species to be recommended. ' +\
'If not provided, all species will be used', nargs='*')
parser.add_argument('--min_len', type=int, help='Minimum length of species names to be used for prediction. ' +\
'Species with names that are at least as long as this value ' +\
'will be analyzed. Default is zero', nargs='?', default=0)
parser.add_argument('--cutoff', type=float, help='Match score cutoff', nargs='?', default=0.0)
parser.add_argument('--mssc', type=str,
help='Match score selection criteria (MSSC). ' +\
'Choose either "top" or "above". "top" recommends ' +\
'the best candidates that are above the cutoff, ' +\
'and "above" recommends all candidates that are above ' +\
'the cutoff. Default is "top"',
nargs='?',
default='top')
parser.add_argument('--outfile', type=str, help='File path to save recommendation.', nargs='?',
default=os.path.join(os.getcwd(), 'species_rec.csv'))
args = parser.parse_args()
recom = recommender.Recommender(libsbml_fpath=args.model)
one_fpath = args.model
specs = args.species
min_len = args.min_len
cutoff = args.cutoff
mssc = args.mssc.lower()
outfile = args.outfile
#
recom = recommender.Recommender(libsbml_fpath=one_fpath)
# # if nothing is given, predict all IDs
if specs is None:
specs = recom.getSpeciesIDs()
print("...\nAnalyzing %d species...\n" % len(specs))
res_tab = recom.recommendSpecies(ids=specs,
mssc=mssc,
cutoff=cutoff,
min_len=min_len,
outtype='table')
recom.saveToCSV(res_tab, outfile)
if isinstance(res_tab, pd.DataFrame):
print("Recommendations saved as:\n%s\n" % os.path.abspath(outfile))
if __name__ == '__main__':
main() | AMAS-sb | /AMAS_sb-1.0.1-py3-none-any.whl/AMAS_sb-1.0.1.data/scripts/recommend_species.py | recommend_species.py |
# recommend_reaction.py
"""
Predicts annotations of reaction(s) using a local XML file
and the reaction ID.
Usage: python recommend_reaction.py files/BIOMD0000000190.xml --min_score 0.6 --outfile res.csv
"""
import argparse
import os
from os.path import dirname, abspath
import sys
sys.path.insert(0, dirname(dirname(abspath(__file__))))
from AMAS import constants as cn
from AMAS import recommender
def main():
parser = argparse.ArgumentParser(description='Recommend reaction annotations of an SBML model and save results')
parser.add_argument('model', type=str, help='SBML model file (.xml)')
# One or more reaction IDs can be given
parser.add_argument('--reaction', type=str, help='ID(s) of reaction(s) to be recommended. ' +\
'If not provided, all reactions will be used', nargs='*')
parser.add_argument('--reject', type=int, help='number of the components of each reaction to reject. ' +\
'Only reactions with components greater than this value ' +\
'will be used. Default is zero', nargs='?', default=0)
parser.add_argument('--cutoff', type=float, help='minimum match score cutoff', nargs='?', default=0.0)
parser.add_argument('--method', type=str,
help='Choose either "top" or "above". "top" recommends ' +\
'the best candidates that are above the cutoff, ' +\
'and "above" recommends all candidates that are above ' +\
'the cutoff. Default is "top"',
nargs='?',
default='top')
parser.add_argument('--outfile', type=str, help='file path to save recommendation', nargs='?',
default=os.path.join(os.getcwd(), 'reaction_rec.csv'))
args = parser.parse_args()
recom = recommender.Recommender(libsbml_fpath=args.model)
one_fpath = args.model
reacts = args.reaction
reject = args.reject
cutoff = args.cutoff
method = args.method
outfile = args.outfile
recom = recommender.Recommender(libsbml_fpath=one_fpath)
recom.current_type = 'reaction'
# if nothing is given, predict all IDs
if reacts is None:
reacts = recom.getReactionIDs()
print("...\nAnalyzing %d reaction(s)...\n" % len(reacts))
# removing ids with less components than 'reject'
filt_reacts = [val for val in reacts \
if len(recom.reactions.reaction_components[val]) > reject]
# stops if all elements were removed by filtering...
if len(filt_reacts) == 0:
print("No element found after the element filter.")
return None
res = recom.getReactionListRecommendation(pred_ids=filt_reacts, get_df=True)
for idx, one_df in enumerate(res):
filt_df = recom.autoSelectAnnotation(df=one_df,
min_score=cutoff,
method=method)
recom.updateSelection(filt_reacts[idx], filt_df)
# save file to csv
recom.saveToCSV(outfile)
print("Recommendations saved as:\n%s\n" % os.path.abspath(outfile))
if __name__ == '__main__':
main() | AMAS-sb | /AMAS_sb-1.0.1-py3-none-any.whl/AMAS_sb-1.0.1.data/scripts/recommend_reaction.py | recommend_reaction.py |
# update_annotation.py
"""
Set annotation of a model file
Usage: python update_annotation.py res.csv files/BIOMD0000000190.xml BIOMD0000000190_upd.xml
"""
import argparse
import itertools
import libsbml
import numpy as np
import os
from os.path import dirname, abspath
import pandas as pd
import sys
sys.path.insert(0, dirname(dirname(abspath(__file__))))
from AMAS import constants as cn
from AMAS import annotation_maker as am
from AMAS import tools
def main():
parser = argparse.ArgumentParser(description='Update annotations of a model using user\'s feedback file (.csv)')
parser.add_argument('infile', type=str, help='path of a model file (.xml) to update annotation')
parser.add_argument('feedback', type=str, help='path of the file (.csv) containing user\'s feedback')
parser.add_argument('outfile', type=str, help='file path to save model with updated annotations')
# csv file with user choice
args = parser.parse_args()
user_csv = pd.read_csv(args.feedback)
# Only takes cells with values 'add' or 'delete'
chosen = user_csv[(user_csv['UPDATE ANNOTATION']=='add') |\
(user_csv['UPDATE ANNOTATION']=='delete')]
outfile = args.outfile
reader = libsbml.SBMLReader()
document = reader.readSBML(args.infile)
model = document.getModel()
ELEMENT_FUNC = {'species': model.getSpecies,
'reaction': model.getReaction}
element_types = list(np.unique(chosen['type']))
for one_type in element_types:
maker = am.AnnotationMaker(one_type)
ACTION_FUNC = {'delete': maker.deleteAnnotation,
'add': maker.addAnnotation}
df_type = chosen[chosen['type']==one_type]
uids = list(np.unique(df_type['id']))
meta_ids = {val:list(df_type[df_type['id']==val]['meta id'])[0] for val in uids}
# going through one id at a time
for one_id in uids:
orig_str = ELEMENT_FUNC[one_type](one_id).getAnnotationString()
df_id = df_type[df_type['id']==one_id]
dels = list(df_id[df_id[cn.DF_UPDATE_ANNOTATION_COL]=='delete'].loc[:, 'annotation'])
adds_raw = list(df_id[df_id[cn.DF_UPDATE_ANNOTATION_COL]=='add'].loc[:, 'annotation'])
# existing annotations to be kept
keeps = list(df_id[df_id[cn.DF_UPDATE_ANNOTATION_COL]=='keep'].loc[:, 'annotation'])
adds = list(set(adds_raw + keeps))
# if type is 'reaction', need to map rhea terms back to ec/kegg terms to delete them.
if one_type == 'reaction':
rhea_del_terms = list(set(itertools.chain(*[tools.getAssociatedTermsToRhea(val) for val in dels])))
deled = maker.deleteAnnotation(rhea_del_terms, orig_str)
elif one_type == 'species':
deled = maker.deleteAnnotation(dels, orig_str)
added = maker.addAnnotation(adds, deled, meta_ids[one_id])
ELEMENT_FUNC[one_type](one_id).setAnnotation(added)
libsbml.writeSBMLToFile(document, outfile)
print("...\nUpdated model file saved as:\n%s\n" % os.path.abspath(outfile))
if __name__ == '__main__':
main() | AMAS-sb | /AMAS_sb-1.0.1-py3-none-any.whl/AMAS_sb-1.0.1.data/scripts/update_annotation.py | update_annotation.py |
import copy
import itertools
import numpy as np
import os
from AMAS import constants as cn
from AMAS import tools
# from AMAS import species_annotation as sa
# from AMAS import reaction_annotation as ra
# Keys when evaluating match results.
NEW_SCORE = 'new_score'
OLD_SCORE = 'old_score'
INCREASED = 'is_increased'
# Max limit for iteration
MAX_ITER = 3
class Iterator(object):
def __init__(self,
cur_spec_formula,
reaction_cl,
reactions_to_update=None):
"""
Ideally, arguments should be directly from
the relevant species.formula and reactions.candidates.
Returns may be dictionaries
for species.candidates and reactions.candidates;
(not 100% confirmed yet)
Parameters
----------
cur_spec_formula: dict
{species_id: [predicted-formulas]}
Current (most recent) annotation of species
reaction_cl: AMAS.reaction_annotation.ReactionAnnotation
reaction_annotation class instance with
loaded SBML model information (reaction_components, etc.)
reaction_to_update: list-str
List of reactions to update; if None, use all reactions
from self.reactions.candidates
"""
self.orig_spec_formula = cur_spec_formula
# Storing reaction candidates separately,
# as it may be different than self.reactions.candidates
self.reactions = reaction_cl
if reactions_to_update:
self.r2upd = reactions_to_update
else:
self.r2upd = list(reaction_cl.candidates.keys())
def getDictOfRheaComponentFormula(self, inp_rhea):
"""
Get a dictionary {chebi_id: formula}
from a given rhea term.
Rhea term -> CheBI IDs -> Formulas
Parameters
----------
str: inp_rhea
A Rhea identifier
Returns
-------
: dict
{chebi_id: formula-str}
"""
chebis = cn.REF_RHEA2CHEBI[inp_rhea]
return {val:cn.REF_CHEBI2FORMULA[val] for val in chebis \
if val in cn.REF_CHEBI2FORMULA.keys()}
def getDictMatchByItem(self,
chebi2ref_formula,
spec2pred_formula):
"""
Get match between two keys,
where there are exactly
one matching items.
If all items are matched by 1-1
(i.e., one species - one chebi),
return the fully matched dictionary.
(i.e., improve precision)
If neither, return None.
(i.e., nothing to update)
Parameters
----------
chebi2ref_formula: dict
{chebi_term: a_species_formula(string)}
spec2pred_formula: dict
{species_id: [predicted_formulas]}
Returns
-------
dict/None
{species_id: [chebi_term]}
"""
match_dict = {one_k:[spec_id for spec_id in spec2pred_formula.keys() \
if chebi2ref_formula[one_k] in spec2pred_formula[spec_id]
] \
for one_k in chebi2ref_formula.keys()}
unmatched_species = [val for val in spec2pred_formula.keys() \
if val not in list(itertools.chain(*match_dict.values()))]
unmatched_chebi = [val for val in match_dict.keys() if not match_dict[val]]
if len(unmatched_species) == 1 and len(unmatched_chebi) == 1:
return {unmatched_species[0]: unmatched_chebi}
# reverse match_dict into the proper return format.
elif all([len(val[1])==1 for val in list(match_dict.items())]):
return {match_dict[k][0]: [k] for k in match_dict.keys()}
else:
return None
def getDictsToUpdate(self, reaction_id):
"""
Using self.getDictMatchByItem(),
get dictionaries to update
Parameters
----------
str: reaction_id
Returns
-------
match_res: dict
{species_id: [ChEBI terms]}
match_res_formula: dict
{species_id: [formula-str]}
"""
one_rhea = self.reactions.candidates[reaction_id][0][0]
# match_res will look like {species_id: [CHEBI term]}
# filter to have only keys and items of one reaction
filt_spec_formula = {k:self.orig_spec_formula[k] \
for k in self.reactions.reaction_components[reaction_id]}
upd_spec_chebi = self.getDictMatchByItem(chebi2ref_formula=self.getDictOfRheaComponentFormula(one_rhea),
spec2pred_formula=filt_spec_formula)
if upd_spec_chebi:
upd_spec_formula = {k:[cn.REF_CHEBI2FORMULA[chebi] \
for chebi in upd_spec_chebi[k]] for k in upd_spec_chebi.keys()}
else:
upd_spec_formula = None
return upd_spec_chebi, upd_spec_formula
def getUpdatedMatchScore(self, cur_spec_formulas, inp_spec2formula_dict):
"""
Check whether it improves reaction measures;
if new value (sum of maximum match score per reaction)
increased, return True; otherwise return False.
Parameters
----------
cur_spec_formulas: dict
{'species_id': [formula-str]}
Dictionary to be updated
inp_spec_2formula_dict: dict
{'species_id': [formula-str]}
Dictionary to update
Returns
-------
: dict
"""
cur_spec_formulas.update(inp_spec2formula_dict)
new_pred_res = self.reactions.getRScores(spec_dict=cur_spec_formulas,
reacs=list(self.r2upd),
mssc='top',
cutoff=0.0)
old_pred_res = self.reactions.getRScores(spec_dict=self.orig_spec_formula,
reacs=list(self.r2upd),
mssc='top',
cutoff=0.0)
# since candidates are already sorted,
# just check the match score (index '1') of the very first candidate tuple (index '0')
new_pred_val = np.mean([new_pred_res[k][0][1] \
for k in new_pred_res.keys()])
old_pred_val = np.mean([old_pred_res[k][0][1] \
for k in old_pred_res.keys()])
return {NEW_SCORE: new_pred_val,
OLD_SCORE: old_pred_val,
INCREASED: new_pred_val>old_pred_val}
def match(self):
"""
Use self.runOneMatchCycle()
and determine the final products to return.
Will be used by the recommender or the user.
"""
all_upd_spec_chebi = dict()
for _ in range(MAX_ITER):
upd_spec_chebi = self.runOneMatchCycle()
if upd_spec_chebi:
all_upd_spec_chebi.update(upd_spec_chebi)
# Update the formula attribute for the next iteration
for one_k in upd_spec_chebi.keys():
self.orig_spec_formula[one_k] = [cn.REF_CHEBI2FORMULA[val] \
for val in upd_spec_chebi[one_k] \
if val in cn.REF_CHEBI2FORMULA.keys()]
else:
break
# Maybe run reaction once, and return final results :)
return all_upd_spec_chebi
def runOneMatchCycle(self):
"""
Using the methohds & information,
determine species to update.
(Reaction will be updated in the following steps).
This method will directly used by
the Recommender, or even the user.
Returns
-------
combine_upd_spec2chebi: dict
{species_id: [ChEBI terms]}
"""
combine_upd_spec2chebi = dict()
# Use reactions existing in self.r2upd
for one_reaction in self.r2upd:
one_rhea_tup = self.reactions.candidates[one_reaction]
one_rhea = one_rhea_tup[0][0]
pred_spec_formulas = self.orig_spec_formula
one_rhea2formula = self.getDictOfRheaComponentFormula(inp_rhea=one_rhea)
upd_spec2chebi, upd_spec2formula = self.getDictsToUpdate(reaction_id=one_reaction)
# Meaning, when examining match scores we only consider
# individual updates; not cumulated updtaes (so we don't use combine_spec2chhebi below)
if upd_spec2formula:
upd_val = self.getUpdatedMatchScore(cur_spec_formulas = copy.deepcopy(self.orig_spec_formula),
inp_spec2formula_dict = upd_spec2formula)
if upd_val[INCREASED]:
# update combine_upd_spec2chebi by combining the elements.
for k in upd_spec2chebi.keys():
if k in combine_upd_spec2chebi.keys():
combine_upd_spec2chebi[k] = list(set(combine_upd_spec2chebi[k] + upd_spec2chebi[k]))
else:
combine_upd_spec2chebi[k] = upd_spec2chebi[k]
return combine_upd_spec2chebi | AMAS-sb | /AMAS_sb-1.0.1-py3-none-any.whl/AMAS/iterator.py | iterator.py |
from AMAS import constants as cn
import itertools
import numpy as np
import re
def applyMSSC(pred,
mssc,
cutoff):
"""
Apply MSSC to a predicted results.
If cutoff is too high,
return an empty list.
Parameters
----------
pred: list-tuple
[(CHEBI:XXXXX, 1.0), etc.]
mssc: string
cutoff: float
Returns
-------
filt: list-tuple
[(CHEBI:XXXXX, 1.0), etc.]
"""
filt_pred = [val for val in pred if val[1]>=cutoff]
if not filt_pred:
return []
if mssc == 'top':
max_val = np.max([val[1] for val in filt_pred])
res_pred = [val for val in filt_pred if val[1]==max_val]
elif mssc == 'above':
res_pred = filt_pred
return res_pred
def extractExistingSpeciesAnnotation(inp_model, qualifier=cn.CHEBI):
"""
Get existing annotation of species
that contains ChEBI terms
Parameters
---------
qualifier: str
'chebi' or 'obo.chebi'?
"""
exist_raw = {val.getId():getQualifierFromString(val.getAnnotationString(), [cn.CHEBI, cn.OBO_CHEBI]) \
for val in inp_model.getListOfSpecies()}
exist_filt = {val:exist_raw[val] for val in exist_raw.keys() \
if exist_raw[val]}
return exist_filt
def extractExistingReactionAnnotation(inp_model):
"""
Get existing annotation of reactions in Rhea
in Bi-directional format (RHEA:10003, etc.)
This will extract annotations from three
knowledge resources:
1. Rhea (mapped to BI-format)
2. KEGG (kegg.reaction mapped to Rhea-BI)
3. EC-Number (or ec-code, mapped to list of Rhea-BIs)
Once they are mapped to a list of Rhea terms,
a list of unique Rhea-Bi terms will be filtered
and be assigned to exist_annotation of the
reaction_annotation class instance.
Parameters
----------
inp_model: libsbml.Model
Returns
-------
dict
"""
exist_raw = {val.getId():extractRheaFromAnnotationString(val.getAnnotationString()) \
for val in inp_model.getListOfReactions()}
exist_filt = {val:exist_raw[val] for val in exist_raw.keys() \
if exist_raw[val]}
return exist_filt
def extractRheaFromAnnotationString(inp_str):
"""
Extract Rhea from existing annotation string,
by directly extracting Rhea,
and converting from KEGGG and EC-Code.
Parameters
----------
inp_str: str
Returns
-------
list-str
"""
exist_rheas = [formatRhea(val) for val in getQualifierFromString(inp_str, cn.RHEA)]
map_rhea_bis = [cn.REF_RHEA2MASTER[val] for val in exist_rheas if val in cn.REF_RHEA2MASTER.keys()]
exist_keggs = [cn.KEGG_HEADER+val for val in getQualifierFromString(inp_str, cn.KEGG_REACTION)]
map_kegg2rhea = list(itertools.chain(*[cn.REF_KEGG2RHEA[val] \
for val in exist_keggs if val in cn.REF_KEGG2RHEA.keys()]))
exist_ecs = [cn.EC_HEADER+val for val in getQualifierFromString(inp_str, cn.EC)]
map_ec2rhea = list(itertools.chain(*[cn.REF_EC2RHEA[val] \
for val in exist_ecs if val in cn.REF_EC2RHEA.keys()]))
return list(set(map_rhea_bis + map_kegg2rhea + map_ec2rhea))
def formatRhea(one_rhea):
"""
Format rhea values;
if 'RHEA:' is not in the name,
add it; if not, ignore it
Parameters
----------
str: one_rhea
Returns
-------
:str
"""
if one_rhea[:4].lower() == 'rhea':
str_to_add = one_rhea[5:]
else:
str_to_add = one_rhea
return cn.RHEA_HEADER + str_to_add
def getOntologyFromString(string_annotation,
bqbiol_qualifiers=['is', 'isVersionOf']):
"""
Parse string and return string annotation,
marked as <bqbiol:is> or <bqbiol:isVersionOf>.
If neither exists, return None.
Parameters
----------
string_annotation: str
bqbiol_qualifiers: str-list
Use 'is' and 'isVersionOf' by default
Returns
-------
list-tuple (ontology type, ontology id)
Return [] if none is provided
"""
combined_str = ''
for one_qualifier in bqbiol_qualifiers:
one_match = '<bqbiol:' + one_qualifier + \
'[^a-zA-Z].*?<\/bqbiol:' + \
one_qualifier + '>'
one_matched = re.findall(one_match,
string_annotation,
flags=re.DOTALL)
if len(one_matched)>0:
matched_filt = [s.replace(" ", "") for s in one_matched]
one_str = '\n'.join(matched_filt)
else:
one_str = ''
combined_str = combined_str + one_str
identifiers_list = re.findall('identifiers\.org/.*/', combined_str)
result_identifiers = [(r.split('/')[1],r.split('/')[2].replace('\"', '')) \
for r in identifiers_list]
return result_identifiers
def getQualifierFromString(input_str, qualifier):
"""
Parses string and returns an identifier.
If not, return None.
Qualifier is allowed to be
either a string or a list of string.
Parameters
----------
str/list-str: (list of) string_annotation
Returns
-------
str (ontology Id)
Returns an empty list if none is provided
"""
ontologies = getOntologyFromString(input_str)
# To make sure it works, make it lower
if isinstance(qualifier, str):
qualifier_list = [val for val in ontologies if val[0].lower()==qualifier.lower()]
elif isinstance(qualifier, list):
lower_qualifiers = [q.lower() for q in qualifier]
qualifier_list = [val for val in ontologies \
if val[0].lower() in lower_qualifiers]
if qualifier_list:
return [val[1] for val in qualifier_list]
else:
return []
def getPrecision(ref, pred, mean=True):
"""
(A model element-agnostic
version of the method.)
A complementary term of 'recall',
precision is the fraction of correct
elements detected from all detected elements.
Parameters
----------
ref: dict
{id: [str-annotation, e,g., formula/Rhea]}
pred: dict
{id: [str-annotation, e,g., formula/Rhea]}
mean: bool
If True, get model-level average
If False, get value of each ID
Returns
-------
: float/dict{id: float}
Depending on the 'mean' argument
"""
ref_keys = set(ref.keys())
pred_keys = set(pred.keys())
precision = dict()
# select species that can be evaluated
species_to_test = ref_keys.intersection(pred_keys)
# go through each species
for one_k in species_to_test:
num_intersection = len(set(ref[one_k]).intersection(pred[one_k]))
num_predicted = len(set(pred[one_k]))
precision[one_k] = num_intersection / num_predicted
# return value is rounded up to the three decimal places
if mean:
return np.round(np.mean([precision[val] for val in precision.keys()]), cn.ROUND_DIGITS)
else:
return {val:np.round(precision[val],cn.ROUND_DIGITS) for val in precision.keys()}
def getRecall(ref, pred, mean=True):
"""
(A model element-agnostic
version of the method.)
A precise version of 'accuracy',
recall is the fraction of correct
elements detected.
Arguments are given as dictionaries.
Parameters
----------
ref: dict
{id: [str-annotation, e,g., formula/Rhea]}
Annotations from reference. Considered 'correct'
pred: dict
{id: [str-annotation, e,g., formula/Rhea]}
Annotations to be evaluated.
mean: bool
If True, get the average across the keys.
If False, get value of each key.
Returns
-------
float/dict{id: float}
Depending on the 'mean' argument
"""
ref_keys = set(ref.keys())
pred_keys = set(pred.keys())
# select species that can be evaluated
species_to_test = ref_keys.intersection(pred_keys)
recall = dict()
# go through each species
for one_k in species_to_test:
num_intersection = len(set(ref[one_k]).intersection(pred[one_k]))
recall[one_k] = num_intersection / len(set(ref[one_k]))
if mean:
return np.round(np.mean([recall[val] for val in recall.keys()]), cn.ROUND_DIGITS)
else:
return {val:np.round(recall[val],cn.ROUND_DIGITS) for val in recall.keys()}
def transformCHEBIToFormula(inp_list, ref_to_formula_dict):
"""
transform input list of CHEBI terms
to list of annotations.
Parameters
----------
inp_list: str-list
Returns
-------
res: str-list
"""
inp_formulas = [ref_to_formula_dict[val] for val in inp_list \
if val in ref_to_formula_dict.keys()]
res = list(set([val for val in inp_formulas if val is not None]))
return res
def updateDictKeyToList(inp_orig_dict, inp_new_dict):
"""
Update inp_orig_dict using inp_up_dict.
If key of inp_up_dict is already in inp_orig_dict,
simply append the item list,
otherwise create a new list with a single item.
Parameters
----------
inp_orig_dict: dict
{key: [items]}
inp_new_dict: dict
{key: [items]} / {key: item}
Returns
-------
res_dict: dict
{key: [list of items]}
"""
res_dict = inp_orig_dict.copy()
# If nothing to update; return original dictionary
if inp_new_dict is None:
return res_dict
for one_k in inp_new_dict.keys():
# make item to a list, it is already not
if isinstance(inp_new_dict[one_k], list):
itm2add = inp_new_dict[one_k]
else:
itm2add = [inp_new_dict[one_k]]
if one_k in res_dict.keys():
res_dict[one_k] = list(set(res_dict[one_k] + itm2add))
else:
res_dict[one_k] = itm2add
return res_dict
def getAssociatedTermsToRhea(inp_rhea):
"""
Get a list of associated terms
of a rhea term.
The resulting list will contain
the original rhea term,
associated EC & KEGG numbers.
Parameters
----------
inp_rhea: str
Returns
-------
: list-str
"""
if inp_rhea in cn.REF_RHEA2ECKEGG.keys():
return cn.REF_RHEA2ECKEGG[inp_rhea] + [inp_rhea]
else:
return [inp_rhea] | AMAS-sb | /AMAS_sb-1.0.1-py3-none-any.whl/AMAS/tools.py | tools.py |
import collections
import compress_pickle
import os
# Folder for reference data
CUR_DIR = os.path.dirname(os.path.realpath(__file__))
REF_DIR = os.path.join(CUR_DIR, 'files')
TEST_DIR = os.path.join(CUR_DIR, os.pardir, 'tests')
# Strings used in the modules
CANDIDATES = 'candidates'
CHEBI = 'chebi'
OBO_CHEBI = 'obo.chebi'
EC = 'ec-code'
EC_HEADER = 'EC:'
KEGG_HEADER = 'KEGG:'
GO = 'go'
RHEA = "rhea"
RHEA_HEADER = 'RHEA:'
KEGG_REACTION = "kegg.reaction"
MATCH_SCORE = "match_score"
NAME_USED = "name_used"
FORMULA = "formula"
QUERY_DF = 'query_df'
RECALL = 'recall'
PRECISION = 'precision'
# For resulting DataFrame
DF_MATCH_SCORE_COL = 'match score'
DF_UPDATE_ANNOTATION_COL = 'UPDATE ANNOTATION'
# Tolerance to determine identical numerical values
TOLERANCE = 0.00001
# Digits to be rounded up
ROUND_DIGITS = 3
# Default URLs for CHEBI/Rhea
CHEBI_DEFAULT_URL = 'https://www.ebi.ac.uk/chebi/searchId.do?chebiId=CHEBI%3A'
RHEA_DEFAULT_URL = 'https://www.rhea-db.org/rhea/'
# Output; namedtuple 'Recommendation'
Recommendation = collections.namedtuple('Recommendation',
['id', 'candidates', 'urls', 'labels'])
with open(os.path.join(REF_DIR, 'chebi_shortened_formula_comp.lzma'), 'rb') as f:
REF_CHEBI2FORMULA = compress_pickle.load(f)
with open(os.path.join(REF_DIR, 'chebi2label.lzma'), 'rb') as f:
REF_CHEBI2LABEL = compress_pickle.load(f)
with open(os.path.join(REF_DIR, 'ec2mrhea.lzma'), 'rb') as handle:
REF_EC2RHEA = compress_pickle.load(handle)
with open(os.path.join(REF_DIR, 'kegg2mrhea.lzma'), 'rb') as handle:
REF_KEGG2RHEA = compress_pickle.load(handle)
with open(os.path.join(REF_DIR, 'rhea_all2master.lzma'), 'rb') as f:
REF_RHEA2MASTER = compress_pickle.load(f)
with open(os.path.join(REF_DIR, 'mrhea2chebi_prime.lzma'), 'rb') as f:
REF_RHEA2CHEBI = compress_pickle.load(f)
with open(os.path.join(REF_DIR, 'rhea2label.lzma'), 'rb') as f:
REF_RHEA2LABEL = compress_pickle.load(f)
with open(os.path.join(REF_DIR, 'mrhea2eckegg.lzma'), 'rb') as f:
REF_RHEA2ECKEGG = compress_pickle.load(f) | AMAS-sb | /AMAS_sb-1.0.1-py3-none-any.whl/AMAS/constants.py | constants.py |
# recommend_reactions.py
"""
Predicts annotations of reaction(s) using a local XML file
and the reaction ID.
Usage: python recommend_reaction.py files/BIOMD0000000190.xml --cutoff 0.6 --outfile res.csv
"""
import argparse
import os
from os.path import dirname, abspath
import pandas as pd
import sys
sys.path.insert(0, dirname(dirname(abspath(__file__))))
from AMAS import constants as cn
from AMAS import recommender
def main():
parser = argparse.ArgumentParser(description='Recommend reaction annotations of an SBML model and save results')
parser.add_argument('model', type=str, help='SBML model file (.xml)')
# One or more reaction IDs can be given
parser.add_argument('--reactions', type=str, help='ID(s) of reaction(s) to be recommended. ' +\
'If not provided, all reactions will be used', nargs='*')
parser.add_argument('--min_len', type=int, help='Minimum number of reaction components (reactants and products) ' +\
'to be used for prediction. ' +\
'Reactions with fewer components than this value ' +\
'will be ignored. Default is zero.', nargs='?', default=0)
parser.add_argument('--cutoff', type=float, help='Match score cutoff.', nargs='?', default=0.0)
parser.add_argument('--mssc', type=str,
help='Match score selection criteria (MSSC). ' +\
'Choose either "top" or "above". "top" recommends ' +\
'the best candidates that are above the cutoff, ' +\
'and "above" recommends all candidates that are above ' +\
'the cutoff. Default is "top"',
nargs='?',
default='top')
parser.add_argument('--outfile', type=str, help='File path to save recommendation.', nargs='?',
default=os.path.join(os.getcwd(), 'reaction_rec.csv'))
args = parser.parse_args()
recom = recommender.Recommender(libsbml_fpath=args.model)
one_fpath = args.model
reacts = args.reactions
min_len = args.min_len
cutoff = args.cutoff
mssc = args.mssc.lower()
outfile = args.outfile
#
recom = recommender.Recommender(libsbml_fpath=one_fpath)
# # if nothing is given, predict all IDs
if reacts is None:
reacts = recom.getReactionIDs()
print("...\nAnalyzing %d reaction(s)...\n" % len(reacts))
res_tab = recom.recommendReactions(ids=reacts,
mssc=mssc,
cutoff=cutoff,
min_len=min_len,
outtype='table')
recom.saveToCSV(res_tab, outfile)
if isinstance(res_tab, pd.DataFrame):
print("Recommendations saved as:\n%s\n" % os.path.abspath(outfile))
if __name__ == '__main__':
main() | AMAS-sb | /AMAS_sb-1.0.1-py3-none-any.whl/AMAS/recommend_reactions.py | recommend_reactions.py |
# recommend_annotation.py
"""
Predicts annotations of species and reactions using a local XML file
and the reaction ID.
This is a combined version of recommend_species and recommend_reaction,
but is more convenient because user will just get the updated XML file or whole recommendations.
Usage: python recommend_reaction.py files/BIOMD0000000190.xml --cutoff 0.6 --save csv --outfile res.csv
"""
import argparse
import itertools
import libsbml
import numpy as np
import os
from os.path import dirname, abspath
import sys
sys.path.insert(0, dirname(dirname(abspath(__file__))))
from AMAS import constants as cn
from AMAS import iterator as it
from AMAS import species_annotation as sa
from AMAS import reaction_annotation as ra
from AMAS import recommender
def main():
parser = argparse.ArgumentParser(description='Recommend annotations of an SBML model ' +\
'(for both species and reactions) and save results.')
parser.add_argument('model', type=str, help='SBML model file (.xml).')
# One or more reaction IDs can be given
parser.add_argument('--cutoff', type=float, help='Match score cutoff.', nargs='?', default=0.0)
parser.add_argument('--optimize', type=str, help='Whether to optimize or not. ' +\
'If y or yes is given, predictions will be ' +\
'optimized. N or no will not optimize predictions.',
nargs='?',
default='no')
parser.add_argument('--mssc', type=str,
help='Match score selection criteria (MSSC). ' +\
'Choose either "top" or "above". "top" recommends ' +\
'the best candidates that are above the cutoff, ' +\
'and "above" recommends all candidates that are above ' +\
'the cutoff. Default is "top"',
nargs='?',
default='top')
parser.add_argument('--save', type=str,
help='Either "sbml" or "csv". ' +\
'If "sbml" is chosen, model will be automatically ' +\
'annotated with recommended candidates and saved. ' +\
'If "csv" is chosen, recommendations will be saved ' +\
'as a csv file. Default is "csv".',
nargs='?',
default='sbml')
parser.add_argument('--outfile', type=str, help='Path to save an output file.', nargs='?')
args = parser.parse_args()
recom = recommender.Recommender(libsbml_fpath=args.model)
one_fpath = args.model
cutoff = args.cutoff
optim_raw = args.optimize
if optim_raw.lower() in ['y', 'yes']:
optim = True
else:
optim = False
mssc = args.mssc.lower()
save = args.save
outfile = args.outfile
#
recom = recommender.Recommender(libsbml_fpath=one_fpath)
specs = recom.getSpeciesIDs()
print("...\nAnalyzing %d species...\n" % len(specs))
reacts = recom.getReactionIDs()
print("...\nAnalyzing %d reaction(s)...\n" % len(reacts))
res_tab = recom.recommendAnnotation(mssc=mssc,
cutoff=cutoff,
optimize=optim,
outtype='table')
if save == 'csv':
if outfile is None:
outfile = os.path.join(os.getcwd(), 'recommendations.csv')
recom.saveToCSV(res_tab, outfile)
else:
if outfile is None:
outfile = os.path.join(os.getcwd(), 'updated_model.xml')
res_sbml = recom.getSBMLDocument(sbml_document=recom.sbml_document,
chosen=res_tab,
auto_feedback=True)
libsbml.writeSBMLToFile(res_sbml, outfile)
print("Recommendations saved as:\n%s\n" % os.path.abspath(outfile))
if __name__ == '__main__':
main() | AMAS-sb | /AMAS_sb-1.0.1-py3-none-any.whl/AMAS/recommend_annotation.py | recommend_annotation.py |
from AMAS import constants as cn
from AMAS import tools
import collections
import compress_pickle
import editdistance
import itertools
import libsbml
import numpy as np
import operator
import os
import pandas as pd
import pickle
import re
import warnings
with open(os.path.join(cn.REF_DIR, 'chebi_low_synonyms_comp.lzma'), 'rb') as f:
CHEBI_LOW_SYNONYMS = compress_pickle.load(f)
CHARCOUNT_COMB_DF = compress_pickle.load(os.path.join(cn.REF_DIR, 'charcount_df_scaled.lzma'),
compression="lzma")
CHARCOUNT_DF = CHARCOUNT_COMB_DF.iloc[:, :-2]
CHEBI_DF = CHARCOUNT_COMB_DF.iloc[:, -2:]
class SpeciesAnnotation(object):
def __init__(self, libsbml_fpath=None,
inp_tuple=None):
"""
Parameters
----------
libsbml_fpath: str
File path of an SBMl (.xml) model
inp_tuple: tuple
Tuple of model information,
first element (index 0) is information on
species names,
second element (index 1) is existing
ChEBI information.
({species_id: species_display_name},
{species_id: ChEBI terms})
"""
# self.exist_annotation stores existing CHEBI annotations in the model
# If none exists, set None
if libsbml_fpath is not None:
reader = libsbml.SBMLReader()
document = reader.readSBML(libsbml_fpath)
self.model = document.getModel()
self.names = {val.getId():val.name for val in self.model.getListOfSpecies()}
self.exist_annotation = tools.extractExistingSpeciesAnnotation(self.model)
exist_annotation_formula_raw = {k:tools.transformCHEBIToFormula(self.exist_annotation[k], cn.REF_CHEBI2FORMULA) \
for k in self.exist_annotation.keys()}
self.exist_annotation_formula = {val:exist_annotation_formula_raw[val] for val in exist_annotation_formula_raw.keys() \
if exist_annotation_formula_raw[val]}
# inp_tuple: ({species_id:species_name}, {species_id: [CHEBI annotations]})
elif inp_tuple is not None:
self.model = None
self.names = inp_tuple[0]
self.exist_annotation = inp_tuple[1]
exist_annotation_formula_raw = {k:tools.transformCHEBIToFormula(inp_tuple[1][k], cn.REF_CHEBI2FORMULA) \
for k in inp_tuple[1].keys()}
self.exist_annotation_formula = {val:exist_annotation_formula_raw[val] for val in exist_annotation_formula_raw.keys() \
if exist_annotation_formula_raw[val]}
else:
self.model = None
self.names = None
self.exist_annotation = None
self.exist_annotation_formula = None
# Below are predicted annotations in dictionary format
# Once created, each will be {species_ID: float/str-list}
self.candidates = dict()
self.formula = dict()
def getCScores(self,
inp_strs,
mssc,
cutoff,
ref_df=CHARCOUNT_DF,
chebi_df=CHEBI_DF):
"""
Compute the eScores
of query strings with
all possible ChEBI terms.
A sorted list of tuples
(CHEBI:XXXXX, eScore)
will be returned.
Only unique strings
will be calculated to avoid
cases such as {'a': 'a',
'a': 'b'}.
Parameters
----------
inp_strs: list-str
List of strings
mssc: match score selection criteria
'top' will recommend candidates with
the highest match score above cutoff
'above' will recommend all candidates with
match scores above cutoff
cutoff: float
Cutoff value; only candidates with match score
at or above the cutoff will be recommended.
ref_df: DataFrame
Reference database
chebi_df: DataFrame
ChEBI information sharing the index with ref_df
Returns
-------
:dict
{one_str: [(CHEBI:XXXXX, 1.0), ...]}
"""
unq_strs = list(set(inp_strs))
one_query, name_used = self.prepareCounterQuery(specs=unq_strs,
ref_cols=ref_df.columns,
use_id=False)
multi_mat = ref_df.dot(one_query)
# updated code to avoid repeated prediction
cscores = dict()
multi_mat[cn.CHEBI] = chebi_df[cn.CHEBI]
for spec in inp_strs:
# Get max-value of each chebi term
g_res = multi_mat.loc[:,[cn.CHEBI, spec]].groupby([cn.CHEBI]).max()[spec]
spec_cscore = tools.applyMSSC(pred=zip(g_res.index, g_res),
mssc=mssc,
cutoff=cutoff)
cscores[spec] = spec_cscore
# cscores = dict()
# for spec in unq_strs:
# spec_cscore = tools.applyMSSC(pred=zip(chebi_df[cn.CHEBI], multi_mat[spec]),
# mssc=mssc,
# cutoff=cutoff)
# spec_cscore.sort(key=operator.itemgetter(1), reverse=True)
# cscores[spec] = spec_cscore
return cscores
def getOneEScore(self, one_s, two_s):
"""
Compute the eScore
of a pair of two strings using
the formula below:
1.0 - (editdistance(one_s, two_s) / max(len(one_s, two_s)))
Values should be between 0.0 and 1.0.
Parameters
----------
one_s: str
two_s: str
Returns
-------
: float (0.0-1.0)
"""
edist = editdistance.eval(one_s, two_s)/ max(len(one_s), len(two_s))
escore = 1.0 - edist
return escore
def getEScores(self,
inp_strs,
mssc,
cutoff):
"""
Compute the eScores
of a list of query strings with
all possible ChEBI terms.
A sorted list of tuples
(CHEBI:XXXXX, eScore)
will be returned.
Only unique strings
will be calculated.
Parameters
----------
inp_strs: str
List of strings
mssc: match score selection criteria
'top' will recommend candidates with
the highest match score above cutoff
'above' will recommend all candidates with
match scores above cutoff
cutoff: float
Cutoff value; only candidates with match score
at or above the cutoff will be recommended.
Returns
-------
:dict
{one_str: [(CHEBI:XXXXX, 1.0), ...]}
"""
unq_strs = list(set(inp_strs))
escores = dict()
for spec in unq_strs:
spec_escore = [(one_k, np.max([self.getOneEScore(spec.lower(), val) \
for val in CHEBI_LOW_SYNONYMS[one_k]])) \
for one_k in CHEBI_LOW_SYNONYMS.keys() \
if one_k in cn.REF_CHEBI2FORMULA.keys()]
mssc_escore = tools.applyMSSC(pred=spec_escore,
mssc=mssc,
cutoff=cutoff)
mssc_escore.sort(key=operator.itemgetter(1), reverse=True)
escores[spec] = mssc_escore
return escores
# Methods to use Cosine Similarity
def getCountOfIndividualCharacters(self, inp_str):
"""
Get a list of characters
between a-z and 0-9.
Parameters
----------
inp_str: str
Returns
-------
: list
"""
return collections.Counter(itertools.chain(*re.findall('[a-z0-9]+', inp_str.lower())))
def prepareCounterQuery(self,
specs,
ref_cols,
use_id=True):
"""
Prepare a query vector, which will be used
as a vector for predictor variables.
Input will be a list of
IDs using which names_used will be determined.
In addition, querys will also be scaled
by the length of each vector.
There is 'use_id' option, so
if False, directly use the string
instead of searching for used_name.
Parameters
----------
list-str: specs
IDs of species
ref_cols: list-str
Column names to use
use_id: bool
If False, directly use the string
If True, use getNameToUse
Returns
-------
: pandas.DataFrame
: dict
"""
name_used = dict()
query_mat = pd.DataFrame(0, index=ref_cols, columns=specs)
for one_spec in specs:
if use_id:
name2use = self.getNameToUse(one_spec)
# characters are lowered in getCountOfIndividualCharacters()
char_counts = self.getCountOfIndividualCharacters(name2use)
name_used[one_spec] = name2use
else:
name2use = one_spec
# characters are lowered in getCountOfIndividualCharacters()
char_counts = self.getCountOfIndividualCharacters(name2use)
name_used[one_spec] = name2use
for one_char in char_counts:
query_mat.loc[one_char, one_spec] = char_counts[one_char]
# Now, scale it using the vector distance
div_row = query_mat.apply(lambda col : np.sqrt(np.sum([val**2 for val in col])), axis = 0)
norm_query = query_mat.divide(div_row, axis=1)
return norm_query, name_used
def getNameToUse(self, inp_id):
"""
Get name to use;
If .name is not '', use it;
otherwise use ID
Parameters
----------
inp_id: ID of model element
Returns
-------
str
"""
species_name = self.names[inp_id]
if len(species_name) > 0:
res_name = species_name
else:
res_name = inp_id
return res_name
def updateSpeciesWithRecommendation(self, inp_recom):
"""
Update species_annotation class using
Recommendation namedtuple.
self.candidates is a sorted list of tuples,
(chebi_id: match_score)
self.formula is a unsorted list of unique formulas
Parameters
----------
inp_recom: cn.Recommendation
A namedtuple. Created by recom.getSpeciesAnnotation
Returns
-------
None
"""
self.candidates.update({inp_recom.id: inp_recom.candidates})
formulas2update = list(set([cn.REF_CHEBI2FORMULA[val[0]] \
for val in inp_recom.candidates \
if val[0] in cn.REF_CHEBI2FORMULA.keys()]))
self.formula.update({inp_recom.id: formulas2update})
return None
def updateSpeciesWithDict(self, inp_dict):
"""
A direct way of updating species annotations,
using ChEBI terms.
As match scores are given
when exact matches are found,
match scores were given as 1.0.
Parameters
----------
inp_dict: dict
{species_id: [chebi terms]}
Returns
-------
None
"""
info2upd_candidates = {k:[(val, 1.0) for val in inp_dict[k]] for k in inp_dict.keys()}
info2upd_formula = {k:[cn.REF_CHEBI2FORMULA[chebi] \
for chebi in inp_dict[k]] for k in inp_dict.keys()}
self.candidates.update(info2upd_candidates)
self.formula.update(info2upd_formula) | AMAS-sb | /AMAS_sb-1.0.1-py3-none-any.whl/AMAS/species_annotation.py | species_annotation.py |
from AMAS import constants as cn
from AMAS import tools
import compress_pickle
import itertools
import libsbml
import numpy as np
import operator
import os
import pandas as pd
with open(os.path.join(cn.REF_DIR, 'data2ref_mat.lzma'), 'rb') as handle:
REF_DAT = compress_pickle.load(handle)
# might need to be deleted after trying Jaccard Index
REF_NONZERO_COLS = compress_pickle.load(os.path.join(cn.REF_DIR, 'ref_nonzero_cols.lzma'),
compression="lzma",
set_default_extension=False)
# first of list is list of columns
cols = REF_DAT[0]
# second, list of indices
inds = REF_DAT[1]
# third, list of index (column, [non-zero rows])
ref_mat_pairs = REF_DAT[2]
REF_MAT = pd.DataFrame(0, index=inds, columns=cols)
for val in ref_mat_pairs:
REF_MAT.iloc[val[1], val[0]] = 1
class ReactionAnnotation(object):
def __init__(self, libsbml_fpath=None,
inp_tuple=None):
"""
Parameters
----------
libsbml_fpath: str
File path of an SBMl model (.xml)
inp_tuple: tuple
({reaction_id: [unique components (that is, species) of that reaction]},
{reaction_id: Rhea terms})
"""
# self.exist_annotation stores
# existing KEGG Reaction or Rhea annotations in the model.
# If none exists, set None.
if libsbml_fpath is not None:
reader = libsbml.SBMLReader()
document = reader.readSBML(libsbml_fpath)
self.model = document.getModel()
self.exist_annotation = tools.extractExistingReactionAnnotation(inp_model=self.model)
self.reaction_components = {val.getId():list(set([k.species for k in val.getListOfReactants()]+\
[k.species for k in val.getListOfProducts()])) \
for val in self.model.getListOfReactions()}
elif inp_tuple is not None:
self.model = None
self.reaction_components = inp_tuple[0]
self.exist_annotation = inp_tuple[1]
else:
self.model = None
self.reaction_components = None
self.exist_annotation = None
# Attributes after prediction
self.candidates = None
self.query_df = None
def getReactionComponents(self,
inp_reaction):
"""
Get component of reactions in species IDs
(both reactants and products)
of a reaction.
Parameters
----------
reaction_id: str/libsbml.Reaction
Returns
-------
None/str-list (list of species IDs)
"""
if isinstance(inp_reaction, libsbml.Reaction):
one_reaction = inp_reaction
elif isinstance(inp_reaction, str):
one_reaction = self.model.getReaction(inp_reaction)
else:
return None
reactants = [val.species for val in one_reaction.getListOfReactants()]
products = [val.species for val in one_reaction.getListOfProducts()]
r_components = list(set(reactants + products))
return r_components
def getRScores(self,
spec_dict,
reacs,
mssc,
cutoff,
ref_mat=REF_MAT):
"""
Get a sorted list of
Rhea-rScore tuples.
[(RHEA:XXXXX, 1.0), etc.]
Parameters
----------
dict: inp_spec_dict
Dictionoary, {species id: formula(str-list)}
reacs: str-list
IDs of reactions to predict annotatinos.
mssc: match score selection criteria
'top' will recommend candidates with
the highest match score above cutoff
'above' will recommend all candidates with
match scores above cutoff
cutoff: float
Cutoff value; only candidates with match score
at or above the cutoff will be recommended.
ref_mat: pd.DataFrame
Reference matrix
Returns
-------
:dict
{one_str: [(Rhea:XXXXX, 1.0), ...]}
"""
# def get_jaccard(inp1, inp2):
# """
# Get jaccard score of two lists;
# Parameters
# ----------
# inp1: list-str
# inp2: list-str
# Returns
# -------
# : float
# """
# inters = set(inp1).intersection(inp2)
# uni = set(inp1).union(inp2)
# return len(inters) / len(uni)
# r2comb_spec_formulas = dict()
# for one_rid in reacs:
# r2comb_spec_formulas[one_rid] = list(set(itertools.chain(*[spec_dict[spec] \
# for spec in self.reaction_components[one_rid]])))
# j_rscores = dict()
# for rid in r2comb_spec_formulas.keys():
# score = REF_NONZERO_COLS.apply(lambda x: get_jaccard(x, r2comb_spec_formulas[rid]))
# reac_rscore = tools.applyMSSC(pred=zip(score.index, score),
# mssc=mssc,
# cutoff=cutoff)
# reac_rscore.sort(key=operator.itemgetter(1), reverse=True)
# j_rscores[rid] = reac_rscore
# return j_rscores
# BELOW IS THE ORIGINAL MINI-MAX VERSION
# Get dictionary of reaction ID: species component
r2pred_spec_formulas = dict()
for one_rid in reacs:
r2pred_spec_formulas[one_rid] = {spec:spec_dict[spec] \
for spec in self.reaction_components[one_rid]}
# prepare query df for prediction
query_df = pd.DataFrame(0,
index=ref_mat.columns,
columns=reacs)
for one_rid in reacs:
one_set_species = r2pred_spec_formulas[one_rid]
# for each species element of the select reaction
for spec_key in one_set_species.keys():
one_spec = one_set_species[spec_key]
# For each one_rid, set the values 1.0
query_df.loc[[val for val in one_spec if val in query_df.index], one_rid] = 1
multi_mat = ref_mat.dot(query_df)
# new minimax of reference value
# max_multi_mat = np.max(multi_mat)
max_multi_mat = multi_mat.max()
query_colsum = pd.Series(0, index=max_multi_mat.index)
for idx in query_colsum.index:
query_colsum.at[idx] = np.min(np.sum(ref_mat.loc[multi_mat[multi_mat[idx]==max_multi_mat[idx]][idx].index,:],1))
# divided
div_mat = multi_mat.divide(query_colsum, axis=1)
rscores = dict()
for reac in reacs:
reac_rscore = tools.applyMSSC(pred=zip(div_mat.index, div_mat[reac]),
mssc=mssc,
cutoff=cutoff)
reac_rscore.sort(key=operator.itemgetter(1), reverse=True)
rscores[reac] = reac_rscore
return rscores
def getRheaElementNum(self,
inp_rhea,
inp_df=REF_MAT):
"""
Get Number of elements of
the given rhea term.
Parameters
----------
inp_rhea: str
Returns
-------
: int
"""
return len(inp_df.loc[inp_rhea, :].to_numpy().nonzero()[0]) | AMAS-sb | /AMAS_sb-1.0.1-py3-none-any.whl/AMAS/reaction_annotation.py | reaction_annotation.py |
# recommend_species.py
"""
Predicts annotations of species using a local XML file
and the species ID.
Usage: python recommend_species.py files/BIOMD0000000190.xml --min_len 2 --cutoff 0.6 --outfile res.csv
"""
import argparse
import os
from os.path import dirname, abspath
import pandas as pd
import sys
sys.path.insert(0, dirname(dirname(abspath(__file__))))
from AMAS import constants as cn
from AMAS import recommender
def main():
parser = argparse.ArgumentParser(description='Recommend species annotations of an SBML model and save results')
parser.add_argument('model', type=str, help='SBML model file (.xml)')
# One or more species IDs can be given
parser.add_argument('--species', type=str, help='ID(s) of species to be recommended. ' +\
'If not provided, all species will be used', nargs='*')
parser.add_argument('--min_len', type=int, help='Minimum length of species names to be used for prediction. ' +\
'Species with names that are at least as long as this value ' +\
'will be analyzed. Default is zero', nargs='?', default=0)
parser.add_argument('--cutoff', type=float, help='Match score cutoff', nargs='?', default=0.0)
parser.add_argument('--mssc', type=str,
help='Match score selection criteria (MSSC). ' +\
'Choose either "top" or "above". "top" recommends ' +\
'the best candidates that are above the cutoff, ' +\
'and "above" recommends all candidates that are above ' +\
'the cutoff. Default is "top"',
nargs='?',
default='top')
parser.add_argument('--outfile', type=str, help='File path to save recommendation.', nargs='?',
default=os.path.join(os.getcwd(), 'species_rec.csv'))
args = parser.parse_args()
recom = recommender.Recommender(libsbml_fpath=args.model)
one_fpath = args.model
specs = args.species
min_len = args.min_len
cutoff = args.cutoff
mssc = args.mssc.lower()
outfile = args.outfile
#
recom = recommender.Recommender(libsbml_fpath=one_fpath)
# # if nothing is given, predict all IDs
if specs is None:
specs = recom.getSpeciesIDs()
print("...\nAnalyzing %d species...\n" % len(specs))
res_tab = recom.recommendSpecies(ids=specs,
mssc=mssc,
cutoff=cutoff,
min_len=min_len,
outtype='table')
recom.saveToCSV(res_tab, outfile)
if isinstance(res_tab, pd.DataFrame):
print("Recommendations saved as:\n%s\n" % os.path.abspath(outfile))
if __name__ == '__main__':
main() | AMAS-sb | /AMAS_sb-1.0.1-py3-none-any.whl/AMAS/recommend_species.py | recommend_species.py |
# import collections
import compress_pickle
import copy
import fnmatch
import itertools
import libsbml
import numpy as np
import os
import pandas as pd
import re
from AMAS import annotation_maker as am
from AMAS import constants as cn
from AMAS import iterator as it
from AMAS import tools
from AMAS import species_annotation as sa
from AMAS import reaction_annotation as ra
ELEMENT_TYPES = ['species', 'reaction']
class Recommender(object):
def __init__(self,
libsbml_fpath=None,
libsbml_cl=None,
model_specs=None):
"""
Parameters
----------
libsbml_cl: libsbml.SBMLDocument
A libsbml document class instance
libsbml_fpath: str
File path of an SBML model
mdoel_specs: tuple/list
Iterable object of two tuples including model specifications
"""
# Document will be updated and saved if chosen.
self.sbml_document = None
# First of all, collect model information from libsbml model
# and send the informaton to create species/reaction annotations
fname = None
if libsbml_fpath:
spec_tuple, reac_tuple = self._parseSBML(libsbml_fpath)
# basically split fpath and use the last one
fname = libsbml_fpath.split('/')[-1]
elif libsbml_cl:
spec_tuple, reac_tuple = self._parseSBML(libsbml_cl)
elif model_specs:
spec_tuple = model_specs[0]
reac_tuple = model_specs[1]
else:
spec_tuple = None
reac_tuple = None
self.fname = fname
self.species = sa.SpeciesAnnotation(inp_tuple=spec_tuple)
self.reactions = ra.ReactionAnnotation(inp_tuple=reac_tuple)
# Below are elements to interact with user
self.current_type = None
self.just_displayed = None
self.selection = {val:dict() for val in ELEMENT_TYPES}
def getDataFrameFromRecommendation(self,
rec,
show_url=False):
"""
Get a pandas dataframe from
a single recommendation.
Parameters
----------
rec: cn.Recommendation
show_url: bool
If False, omit this column
Returns
-------
:str
"""
cands = [val[0] for val in rec.candidates]
match_scores = [val[1] for val in rec.candidates]
labels = rec.labels
# index starts from 1;
df = pd.DataFrame({'annotation':cands,
cn.DF_MATCH_SCORE_COL:match_scores,
'label':labels})
df.index.name = rec.id
if show_url:
urls = rec.urls
df['url'] = urls
return df
def getRecommendationFromDataFrame(self,
df):
"""
Convert dataframe back to
namedtuple Recommendation.
WARNING: it may not work with
empty dataframe, so be careful.
Parameters
----------
df: pandas.DataFrame
element_type: str
'species' or 'reaction'
Returns
-------
Recommendation (namedtuple)
"""
cands_tups = list(zip(df['annotation'], df['match score']))
one_annotation = cands_tups[0][0]
# indicating species
if one_annotation[:4] == 'CHEB':
default_url = cn.CHEBI_DEFAULT_URL
url_digit = 6
# indicating reaction
elif one_annotation[:4] == 'RHEA':
default_url = cn.RHEA_DEFAULT_URL
url_digit = 5
return cn.Recommendation(df.index.name,
list(zip(df['annotation'], df['match score'])),
[default_url + val[url_digit:] for val in df['annotation']],
list(df['label']))
def getMarkdownFromRecommendation(self,
rec,
show_url=False):
"""
Get a markdown using
a cn.Recommendation or pandas.DataFrame.
Parameters
----------
rec: cn.Recommendation/pandas.DataFrame
show_url: bool
If False, omit this column
Returns
-------
:str
"""
if isinstance(rec, pd.DataFrame):
# to deepcopy so original data doesn't get changed
# in line 156.
df = copy.deepcopy(rec)
idx_name = df.index.name.split(' ')
rec_id = idx_name[0]
else:
df = self.getDataFrameFromRecommendation(rec, show_url)
rec_id = rec.id
# In markdown, title is shown separately,
# so index name with element ID is removed;
df.index.name=None
df_str = df.to_markdown(tablefmt="grid", floatfmt=".03f", index=True)
# Centering and adding the title
len_first_line = len(df_str.split('\n')[0])
title_line = rec_id
title_line = title_line.center(len_first_line)
df_str = title_line + '\n' + df_str
return df_str
def getSpeciesRecommendation(self,
pred_str=None,
pred_id=None,
method='cdist',
mssc='top',
cutoff=0.0,
update=True,
get_df=False):
"""
Predict annotations of species using
the provided string or ID.
If pred_str is given, directly use the string;
if pred_id is given, determine the appropriate
name using the species ID.
Algorithmically, it is a special case of
self.getSpeciesListRecommendation.
Parameters
----------
pred_str: str
Species name to predict annotation with
pred_id: str
ID of species (search for name using it)
method: str
One of ['cdist', 'edist']
'cdist' represents Cosine Similarity
'edist' represents Edit Distance.
Default method id 'cdist'
mssc: match score selection criteria
'top' will recommend candidates with
the highest match score above cutoff
'above' will recommend all candidates with
match scores above cutoff
cutoff: float
Cutoff value; only candidates with match score
at or above the cutoff will be recommended.
update: bool
If true, update existing species annotations
(i.e., replace or create new entries)
in self.species.candidates and self.species.formula
get_df: bool
If true, return a pandas.DataFrame.
If False, return a cn.Recommendation
Returns
-------
cn.Recommendation (namedtuple) / str
"""
if pred_str:
result = self.getSpeciesListRecommendation(pred_strs=[pred_str],
method=method,
mssc=mssc,
cutoff=cutoff,
update=update,
get_df=get_df)
elif pred_id:
result = self.getSpeciesListRecommendation(pred_ids=[pred_id],
method=method,
mssc=mssc,
cutoff=cutoff,
update=update,
get_df=get_df)
return result[0]
def getSpeciesIDs(self, pattern=None, regex=False):
"""
Returns Species IDs that match the pattern.
The pattern is given as glob
If none is given, returns all available
species that exist in the model.
Parameters
---------
pattern: str/None
string pattern
reges: bool
if True, use regex
if False, use glob
Returns
-------
list-str/None
None returned if no match was found
"""
# list of species ids
specs = list(self.species.names.keys())
# returns a list of ids thta match pattern, if None, return all
if pattern is None:
return specs
else:
if regex:
re_pattern = pattern
else:
re_pattern = fnmatch.translate(pattern)
matched = [re.match(re_pattern, val) for val in specs]
filt_matched = [val.group(0) for val in matched if val]
if len(filt_matched)>0:
return filt_matched
else:
return None
def getSpeciesListRecommendation(self,
pred_strs=None,
pred_ids=None,
method='cdist',
mssc='top',
cutoff=0.0,
update=True,
get_df=False):
"""
Get annotation of multiple species,
given as a list (or an iterable object).
self.getSpeciesRecommendation is applied to
each element.
Parameters
----------
pred_strs: str-list
:Species names to predict annotations with
pred_ids: str-list
:Species IDs to predict annotations with
(model info should have been already loaded)
method: str
One of ['cdist', 'edist']
'cdist' represents Cosine Similarity
'edist' represents Edit Distance.
Default method id 'cdist'
mssc: match score selection criteria
'top' will recommend candidates with
the highest match score above cutoff
'above' will recommend all candidates with
match scores above cutoff
cutoff: float
Cutoff value; only candidates with match score
at or above the cutoff will be recommended.
update: bool
:If true, update the current annotations
(i.e., replace or create new entries)
in self.species.candidates and self.species.formula
get_df: bool
If True, return a list of pandas.DataFrame.
If False, return a list of cn.Recommendation
Returns
-------
list-Recommendation (list-namedtuple) / list-str
"""
scoring_methods = {'edist': self.species.getEScores,
'cdist': self.species.getCScores}
if pred_strs:
ids_dict = {k:k for k in pred_strs}
inp_strs = pred_strs
elif pred_ids:
ids_dict = {k:self.species.getNameToUse(inp_id=k) \
for k in pred_ids}
inp_strs = [ids_dict[k] for k in ids_dict.keys()]
pred_res = scoring_methods[method](inp_strs=inp_strs,
mssc=mssc,
cutoff=cutoff)
result = []
for spec in ids_dict.keys():
urls = [cn.CHEBI_DEFAULT_URL + val[0][6:] for val in pred_res[ids_dict[spec]]]
labels = [cn.REF_CHEBI2LABEL[val[0]] for val in pred_res[ids_dict[spec]]]
one_recom = cn.Recommendation(spec,
[(val[0], np.round(val[1], cn.ROUND_DIGITS)) \
for val in pred_res[ids_dict[spec]]],
urls,
labels)
result.append(one_recom)
if update:
_ = self.species.updateSpeciesWithRecommendation(one_recom)
if get_df:
return [self.getDataFrameFromRecommendation(rec=val) \
for val in result]
else:
return result
def getReactionRecommendation(self, pred_id,
use_exist_species_annotation=False,
spec_res=None,
spec_method='cdist',
mssc='top',
cutoff=0.0,
update=True,
get_df=False):
"""
Predict annotations of reactions using
the provided IDs (argument).
Can be either singular (string) or plural
Parameters
----------
pred_id: str
A single ID of reaction to annotate
use_exist_speices_annotation: bool
If True, use existing species annotation
spec_res: list-cn.Recommendation
If provided, species will not be predicted
for remaining species
spec_method: str
Method to use if to directly predict species annotation;
if 'cdist' Cosine Similarity
if 'edist' Edit distance
mssc: match score selection criteria
'top' will recommend candidates with
the highest match score above cutoff
'above' will recommend all candidates with
match scores above cutoff
cutoff: float
Cutoff value; only candidates with match score
at or above the cutoff will be recommended.
update: bool
If true, update existing species annotations
(i.e., replace or create new entries)
in self.reactions.candidates
get_df: bool
If True, return a pandas DataFrame.
If False, return a cn.Recommendation
Returns
-------
Recommendation (namedtuple) / str
"""
result = self.getReactionListRecommendation(pred_ids=[pred_id],
use_exist_species_annotation=use_exist_species_annotation,
spec_res=spec_res,
spec_method=spec_method,
mssc=mssc,
cutoff=cutoff,
update=update,
get_df=get_df)
return result[0]
def getReactionIDs(self, pattern=None, by_species=False, regex=False):
"""
Get IDs of reactions based on
the pattern.
If by_species is True, it retrieves
all reaction with the species that match
the pattern;
if False, it searches based on the ID of
reactions
Parameters
---------
pattern: str
Pattern
by_species: bool
If True, find species with pattern
If False, find reaction IDs
regex: bool
If True, use regex expression
If False, convert it to regex.
"""
reacts = list(self.reactions.reaction_components.keys())
if pattern is None:
return reacts
# returns a list of ids thta match pattern, if None, return all
if regex:
re_pattern = pattern
else:
re_pattern = fnmatch.translate(pattern)
if by_species:
specs2use = self.getSpeciesIDs(pattern=re_pattern, regex=True)
if any(specs2use):
comp_items = list(self.reactions.reaction_components.items())
result = [val[0] for val in comp_items \
if any(set(val[1]).intersection(specs2use))]
# if no species match was found
else:
return None
else:
matched = [re.match(re_pattern, val) for val in reacts]
result = [val.group(0) for val in matched if val]
return result
def getReactionListRecommendation(self, pred_ids,
use_exist_species_annotation=False,
spec_res=None,
spec_method='cdist',
mssc='top',
cutoff=0.0,
update=True,
get_df=False):
"""
Get annotation of multiple reactions.
Instead of applying getReactionRecommendation
for each reaction,
it'll predict all component species first
and proceed (this will reduce computational cost).
Parameters
----------
pred_ids: str-list
For now, it only accommodates calling by reaction IDs.
use_exist_species_annotation: tool
If True, search existing annotation for species
and predict the remaining species
spec_res: list-cn.Recommendation
If provided, species will not be predicted
for remaining species
spec_method: str
Method to use if to directly predict species annotation;
if 'cdist' Cosine Similarity
if 'edist' Edit distance
mssc: match score selection criteria
'top' will recommend candidates with
the highest match score above cutoff
'above' will recommend all candidates with
match scores above cutoff
cutoff: float
Cutoff value; only candidates with match score
at or above the cutoff will be recommended.
update: bool
If true, update existing species annotations
(i.e., replace or create new entries)
in self.reactions.candidates
get_df: bool
If True, return a list of pandas DataFrames.
If False, return a list of cn.Recommendation
Returns
-------
list-Reccommendation (list-namedtuple) / list-str
"""
# First, collect all species IDs to annotate
specs_to_annotate = list(set(itertools.chain(*[self.reactions.reaction_components[val] \
for val in pred_ids])))
if use_exist_species_annotation:
pred_formulas = {val:self.species.exist_annotation_formula[val] \
for val in specs_to_annotate \
if val in self.species.exist_annotation_formula.keys()}
else:
pred_formulas = {}
remaining_species = [val for val in specs_to_annotate if val not in pred_formulas.keys()]
# Get annotation of collected species
if len(remaining_species) > 0:
if spec_res:
spec_results = [val for val in spec_res if val.id in remaining_species]
else:
# No updates; use MSSC Top, cutoff 0.0.
spec_results = self.getSpeciesListRecommendation(pred_ids=remaining_species,
update=False,
method=spec_method)
for one_recom in spec_results:
chebis = [val[0] for val in one_recom.candidates]
forms = list(set([cn.REF_CHEBI2FORMULA[k] \
for k in chebis if k in cn.REF_CHEBI2FORMULA.keys()]))
pred_formulas[one_recom.id] = forms
# Predict reaction annotations.
pred_res = self.reactions.getRScores(spec_dict=pred_formulas,
reacs=pred_ids,
mssc=mssc,
cutoff=cutoff)
result = []
for reac in pred_res.keys():
urls = [cn.RHEA_DEFAULT_URL + val[0][5:] for val in pred_res[reac]]
labels = [cn.REF_RHEA2LABEL[val[0]] for val in pred_res[reac]]
one_recom = cn.Recommendation(reac,
[(val[0], np.round(val[1], cn.ROUND_DIGITS)) \
for val in pred_res[reac]],
urls,
labels)
result.append(one_recom)
if update:
self.reactions.candidates = pred_res
if get_df:
return [self.getDataFrameFromRecommendation(rec=val) \
for val in result]
else:
return result
def _parseSBML(self, sbml):
"""
Parse SBML file and return
two tuples, for species and reactions
respecitvely,
equivalent to model_specs.
Can use either libsbml.Document class
or a file path.
Parameters
----------
sbml: str(filepath)/libsbml.Document
Returns
-------
(tuple, tuple):
Two tuples to create species/reaction annotation classes
(species_tuple, reaction_tuple)
"""
if isinstance(sbml, str):
reader = libsbml.SBMLReader()
# Reading the model string file
with open(sbml, 'r') as f:
model_str = f.read()
self.sbml_document = reader.readSBMLFromString(model_str)
elif isinstance(sbml, libsbml.SBMLDocument):
self.sbml_document = sbml
model = self.sbml_document.getModel()
exist_spec_annotation = tools.extractExistingSpeciesAnnotation(model)
species_names = {val.getId():val.name for val in model.getListOfSpecies()}
species_tuple = (species_names, exist_spec_annotation)
#
reac_exist_annotation = tools.extractExistingReactionAnnotation(inp_model=model)
# Next, reaction components for each reaction
reac_components = {val.getId():list(set([k.species for k in val.getListOfReactants()]+\
[k.species for k in val.getListOfProducts()])) \
for val in model.getListOfReactions()}
reaction_tuple = (reac_components, reac_exist_annotation)
return species_tuple, reaction_tuple
def getSpeciesStatistics(self,
mssc='top',
cutoff=0.0,
model_mean=True):
"""
Get recall and precision
of species in a model, for both species and
reactions.
This method works only if there exists
annotation in model; otherwise
None will be returned.
In the result, values will be
returned after rounding to the two decimal places.
Parameters
----------
mssc: str
match score selection criteria
'top' will recommend candidates with
the highest match score above cutoff
'above' will recommend all candidates with
match scores above cutoff
cutoff: float
Cutoff value; only candidates with match score
at or above the cutoff will be recommended.
model_mean: bool
If True, get single float values for recall/precision.
If False, get a dictionary for recall/precision.
Returns
-------
None/dict
Return None if there is nothing to evaluate
(i.e., if there is no existing model annotation)
"""
# get dictionary of formulas if they exist
refs = {val:self.species.exist_annotation_formula[val] \
for val in self.species.exist_annotation_formula.keys() \
if self.species.exist_annotation_formula[val]}
specs2eval = list(refs.keys())
if len(specs2eval) == 0:
return None
preds_comb = self.getSpeciesListRecommendation(pred_ids=specs2eval,
mssc=mssc,
cutoff=cutoff)
chebi_preds = {val.id:[k[0] for k in val.candidates] \
for val in preds_comb}
preds = {k:[cn.REF_CHEBI2FORMULA[val] for val in chebi_preds[k] \
if val in cn.REF_CHEBI2FORMULA.keys()] \
for k in chebi_preds.keys()}
recall = tools.getRecall(ref=refs, pred=preds, mean=model_mean)
precision = tools.getPrecision(ref=refs, pred=preds, mean=model_mean)
return {cn.RECALL: recall, cn.PRECISION: precision}
def getReactionStatistics(self,
model_mean=True,
mssc='top',
cutoff=0.0):
"""
Get recall and precision
of reactions in a model, for both species and
reactions.
This method works only if there exists
annotation in model; otherwise
None will be returned.
In the result, values will be
returned after rounding to the two decimal places.
Parameters
----------
mssc: str
match score selection criteria
'top' will recommend candidates with
the highest match score above cutoff
'above' will recommend all candidates with
match scores above cutoff
cutoff: float
Cutoff value; only candidates with match score
at or above the cutoff will be recommended.
model_mean: bool
If True, get single float values for recall/precision.
If False, get a dictionary for recall/precision.
Returns
-------
None/dict
Return None if there is nothing to evaluate
(i.e., if there is no existing model annotation)
"""
# For reactions, component species should be
# predicted first.
refs = self.reactions.exist_annotation
if len(refs) == 0:
return None
specs2pred = list(set(itertools.chain(*([self.reactions.reaction_components[val] for val in refs.keys()]))))
## Use mssc top, cutoff 0.0.
preds_comb = self.getSpeciesListRecommendation(pred_ids=specs2pred,
mssc='top',
cutoff=0.0)
chebi_preds = {val.id:[k[0] for k in val.candidates] \
for val in preds_comb}
specs_predicted = {k:[cn.REF_CHEBI2FORMULA[val] for val in chebi_preds[k] \
if val in cn.REF_CHEBI2FORMULA.keys()] \
for k in chebi_preds.keys()}
reac_preds = self.reactions.getRScores(spec_dict=specs_predicted,
reacs=refs.keys(),
mssc='top',
cutoff=0.0)
preds = {k:[val[0] for val in reac_preds[k]] for k in reac_preds.keys()}
recall = tools.getRecall(ref=refs, pred=preds, mean=model_mean)
precision = tools.getPrecision(ref=refs, pred=preds, mean=model_mean)
return {cn.RECALL: recall, cn.PRECISION: precision}
def filterDataFrameByThreshold(self, df, min_score):
"""
Filter dataframe by min_score (threshold),
and returns the result;
Note that if no item meets the threshold,
it'll still return an empty dataframe.
Paramters
---------
df: pd.DataFrame
min_score: float (0.0-1.0)
Returns
-------
pd.DataFrame
"""
scores = df[cn.DF_MATCH_SCORE_COL]
filt_idx = scores[scores>=min_score].index
filt_df = df.loc[filt_idx, :]
return filt_df
def autoSelectAnnotation(self,
df,
cutoff=0.0,
mssc='top'):
"""
Choose annotations based on
the set threshold;
(1) if None meets the threshold, return an empty frame
(2) if multiple meet the threshold,
(a) if method is 'best':
(i) find the highest match score among them
(ii) return all above match score == highest_match_score
(b) if method is 'all':
(i) return all that is at or above min_score
Parameters
----------
df: pandas.DataFrame
cutoff: float (0.0-1.0)
Match score cutoff
mssc: str
Match score selection criteria;
either 'top' or 'above'.
Returns
-------
pandas.DataFrame
if nothing matched,
an empty dataframe is returned
"""
scores = df[cn.DF_MATCH_SCORE_COL]
# max_score: highest match score that exists
# min_score: cutoff
max_score = np.max(scores)
if max_score < cutoff:
# this will create an empty dataframe
filt_idx = scores[scores>=cutoff].index
elif mssc=='top':
filt_idx = scores[scores==max_score].index
else:
filt_idx = scores[scores>=cutoff].index
filt_df = df.loc[filt_idx, :]
return filt_df
def recommendAnnotation(self,
mssc='top',
cutoff=0.0,
optimize=False,
outtype='table'):
"""
Combine recommendSpecies and recommendReactions
methods; can optimize.
Parameters
----------
mssc: str
cutoff: float
optiimize: bool
outtype: str
If 'table', returns recommendation table
if 'sbml', returns an updated SBML model.
Returns
-------
pandas.DataFrame / str
"""
pred_spec = self.getSpeciesListRecommendation(pred_ids=self.getSpeciesIDs(),
mssc=mssc,
cutoff=cutoff,
get_df=True)
pred_reac = self.getReactionListRecommendation(pred_ids=self.getReactionIDs(),
mssc=mssc,
cutoff=cutoff,
get_df=True)
if optimize:
res_tab = self.optimizePrediction(pred_spec=pred_spec,
pred_reac=pred_reac)
else:
s_df = self.getRecomTable(element_type='species',
recommended=pred_spec)
r_df = self.getRecomTable(element_type='reaction',
recommended=pred_reac)
res_tab = pd.concat([s_df, r_df],
ignore_index=True)
if outtype == 'table':
return res_tab
elif outtype == 'sbml':
res_sbml = self.getSBMLDocument(sbml_document=self.sbml_document,
chosen=res_tab,
auto_feedback=True)
return libsbml.writeSBMLToString(res_sbml)
def recommendReactions(self,
ids=None,
min_len=0,
mssc='top',
cutoff=0.0,
outtype='table'):
"""
Recommend one or more ids of reactions
and returns a single dataframe or
a list of dataframes.
Parameters
----------
ids: str/list-str
If None, recommend all reactionos.
min_len: int
Minimum number of reaction components
to be returned for results
mssc: str
match score selection criteria.
'top' or 'above'
cutoff: float
MSSC cutoff
outtype: str
Either 'table' or 'sbml'.
'table' will return a pandas.DataFrame
'sbml' will return an sbml string
Returns
-------
None
"""
self.updateCurrentElementType('reaction')
if isinstance(ids, str):
reacs = [ids]
elif ids is None:
reacs = self.getReactionIDs()
else:
reacs = ids
filt_reacs = [val for val in reacs \
if len(self.reactions.reaction_components[val])>=min_len]
if len(filt_reacs) == 0:
print("No reaction after the element filter.\n")
return None
pred = self.getReactionListRecommendation(pred_ids=filt_reacs,
mssc=mssc,
cutoff=cutoff,
get_df=True)
res_table = self.getRecomTable(element_type='reaction',
recommended=pred)
if outtype == 'table':
return res_table
elif outtype == 'sbml':
res_sbml = self.getSBMLDocument(sbml_document=self.sbml_document,
chosen=res_table,
auto_feedback=True)
return libsbml.writeSBMLToString(res_sbml)
return None
def recommendSpecies(self,
ids=None,
min_len=0,
mssc='top',
cutoff=0.0,
outtype='table'):
"""
Recommend one or more ids of species
and returns a single dataframe or
a list of dataframes.
Parameters
----------
ids: str/list-str
If None, will predict all
min_len: int
Minimum length of species name
to be returned for results
mssc: str
Match score selection criteria.
cutoff: match score cutoff
If None given, returns all values.
outtype: str
Either 'table' or 'sbml'.
'table' will return a pandas.DataFrame
'sbml' will return an sbml string
Returns
-------
: pd.DataFrame/str/None
Either
"""
self.updateCurrentElementType('species')
if isinstance(ids, str):
specs = [ids]
elif ids is None:
specs = self.getSpeciesIDs()
else:
specs = ids
filt_specs = [val for val in specs \
if len(self.species.getNameToUse(val))>=min_len]
if len(filt_specs) == 0:
print("No species after the element filter.\n")
return None
pred = self.getSpeciesListRecommendation(pred_ids=filt_specs,
mssc=mssc,
cutoff=cutoff,
get_df=True)
res_table = self.getRecomTable(element_type='species',
recommended=pred)
if outtype == 'table':
return res_table
elif outtype == 'sbml':
res_sbml = self.getSBMLDocument(sbml_document=self.sbml_document,
chosen=res_table,
auto_feedback=True)
return libsbml.writeSBMLToString(res_sbml)
return None
def updateCurrentElementType(self, element_type):
"""
Updating self.current_type
indicator; updated when
recommendSpecies or recommendReactions
is called;
Parameters
----------
element_type: str
Either 'species' or 'reaction'
"""
self.current_type = element_type
def updateJustDisplayed(self, df_dict):
"""
Used it every time
result is shown to user.
called by
/recommendSpecies/recommendReactions/
/selectAnnotation/
For now, always in the format as
pandas.DataFrame.
Parameters
----------
df_dict: dict()
Dictionary of pandas.DataFrame
Returns
-------
None
"""
self.just_displayed = df_dict
def selectAnnotation(self, choice=None):
"""
Based on the previous recommendation,
determine the annotations to store.
If 'all' given in choice[1],
select all.
Parameters
----------
choice: tuple/list-tuple (str, int)
[(element ID, choice number)]
"""
# assumes self.just_displayced is {id: pd.dataframe}
sel_id = choice[0]
sel_idx = choice[1]
df = self.just_displayed[choice[0]]
if sel_idx == 'all':
result = df
else:
if isinstance(sel_idx, int):
chosen = [sel_idx]
else:
chosen = sel_idx
result = df.loc[chosen, :]
# Now, update the selected annotation
self.updateSelection(sel_id, result)
print("Selection updated.")
return None
def updateSelection(self, sel_id, sel_df):
"""
Direct result of selectAnnotation;
filtered or non-filtered
dictionary of dataframes.
By calling SaveFile,
All selected annotations will be
saved as an .xml file.
Parameters
----------
sel_id: str
sel_df: pandas.DataFrame
"""
self.selection[self.current_type].update({sel_id: sel_df})
def displaySelection(self):
"""
To assist user,
display all selected
annotations from
self.selection.
"""
for one_type in ELEMENT_TYPES:
type_selection = self.selection[one_type]
for k in type_selection.keys():
print(self.getMarkdownFromRecommendation(type_selection[k])+"\n")
def getRecomTable(self,
element_type,
recommended):
"""
Extend the dataframe using
results obtained by
self.get....ListRecommendation.
A new, extended dataframe will be
returned; to be either
saved as CSV or shown to the user.
Parameters
----------
element_type: str
either 'species' or 'reaction'
recommended: list-pandas.DataFrame
result of get....ListRecommendation method
Returns
-------
:pandas.DataFrame
a single dataframe
(not a list of dataframes)
"""
etype = element_type
model = self.sbml_document.getModel()
TYPE_EXISTING_ATTR = {'species': self.species.exist_annotation,
'reaction': self.reactions.exist_annotation}
ELEMENT_FUNC = {'species': model.getSpecies,
'reaction': model.getReaction}
TYPE_LABEL = {'species': cn.REF_CHEBI2LABEL,
'reaction': cn.REF_RHEA2LABEL}
pd.set_option('display.max_colwidth', 255)
edfs = []
for one_edf in recommended:
element_id = one_edf.index.name
if one_edf.shape[0] == 0:
continue
annotations = list(one_edf['annotation'])
match_scores = list(one_edf[cn.DF_MATCH_SCORE_COL])
labels = list(one_edf['label'])
# if there is existing annotation among predicted candidates;
if element_id in TYPE_EXISTING_ATTR[etype].keys():
existings = [1 if val in TYPE_EXISTING_ATTR[etype][element_id] else 0 \
for idx, val in enumerate(one_edf['annotation'])]
upd_annotation = ['keep' if val in TYPE_EXISTING_ATTR[etype][element_id] else 'ignore' \
for idx, val in enumerate(one_edf['annotation'])]
annotation2add_raw = [val for val in TYPE_EXISTING_ATTR[etype][element_id] \
if val not in list(one_edf['annotation'])]
# only use existing annotation that exists in the label dictionaries
annotation2add = [val for val in annotation2add_raw \
if val in TYPE_LABEL[etype].keys()]
# if there doesn't exist existing annotation among predicted candidates;
else:
existings = [0] * len(annotations)
upd_annotation = ['ignore'] * len(annotations)
annotation2add = []
# handling existing annotations that were not predicted
for new_anot in annotation2add:
annotations.append(new_anot)
if etype=='reaction':
match_scores.append(self.getMatchScoreOfRHEA(element_id, new_anot))
labels.append(cn.REF_RHEA2LABEL[new_anot])
elif etype=='species':
match_scores.append(self.getMatchScoreOfCHEBI(element_id, new_anot))
labels.append(cn.REF_CHEBI2LABEL[new_anot])
existings.append(1)
upd_annotation.append('keep')
new_edf = pd.DataFrame({'type': [etype]*len(annotations),
'id': [element_id]*len(annotations),
'display name': [ELEMENT_FUNC[etype](element_id).name]*len(annotations),
'meta id': [ELEMENT_FUNC[etype](element_id).meta_id]*len(annotations),
'annotation': annotations,
'annotation label': labels,
cn.DF_MATCH_SCORE_COL: match_scores,
'existing': existings,
cn.DF_UPDATE_ANNOTATION_COL: upd_annotation})
edfs.append(new_edf)
res = pd.concat(edfs, ignore_index=True)
res.insert(0, 'file', self.fname)
return res
def getSBMLDocument(self,
sbml_document,
chosen,
auto_feedback=False):
"""
Create an updated SBML document
based on the feedback.
If auto_feedback is True,
replace 'ignore' with 'add'
and subsequently update the file.
Parameters
----------
sbml_document: libsbml.SBMLDocument
chosen: pandas.DataFrame
Returns
-------
str
SBML document
"""
model = sbml_document.getModel()
if auto_feedback:
chosen.replace('ignore', 'add', inplace=True)
ELEMENT_FUNC = {'species': model.getSpecies,
'reaction': model.getReaction}
element_types = list(np.unique(chosen['type']))
for one_type in element_types:
maker = am.AnnotationMaker(one_type)
ACTION_FUNC = {'delete': maker.deleteAnnotation,
'add': maker.addAnnotation}
df_type = chosen[chosen['type']==one_type]
uids = list(np.unique(df_type['id']))
meta_ids = {val:list(df_type[df_type['id']==val]['meta id'])[0] for val in uids}
# going through one id at a time
for one_id in uids:
orig_str = ELEMENT_FUNC[one_type](one_id).getAnnotationString()
df_id = df_type[df_type['id']==one_id]
dels = list(df_id[df_id[cn.DF_UPDATE_ANNOTATION_COL]=='delete'].loc[:, 'annotation'])
adds_raw = list(df_id[df_id[cn.DF_UPDATE_ANNOTATION_COL]=='add'].loc[:, 'annotation'])
# existing annotations to be kept
keeps = list(df_id[df_id[cn.DF_UPDATE_ANNOTATION_COL]=='keep'].loc[:, 'annotation'])
# TODO: remove RHEA
adds_raw = list(set(adds_raw + keeps))
adds = []
for one_add in adds_raw:
# if it is rhea, only store number
if one_add[:4].lower()=='rhea':
adds.append(one_add[5:])
# if it is else, store as it is
else:
adds.append(one_add)
# if type is 'reaction', need to map rhea terms back to ec/kegg terms to delete them.
if one_type == 'reaction':
rhea_del_terms = list(set(itertools.chain(*[tools.getAssociatedTermsToRhea(val) for val in dels])))
deled = maker.deleteAnnotation(rhea_del_terms, orig_str)
elif one_type == 'species':
deled = maker.deleteAnnotation(dels, orig_str)
added = maker.addAnnotation(adds, deled, meta_ids[one_id])
ELEMENT_FUNC[one_type](one_id).setAnnotation(added)
return sbml_document
def optimizePrediction(self,
pred_spec,
pred_reac):
"""
Optimize prediction using iteration.
Parameters
----------
pred_spec: list-DataFrame
Result of getSpeciesListRecommendation
with get_df=True
pred_reac: list-DataFrame
Result of getReactionListRecommendation
with get_df=True
reactions_to_update: list
IDs of reactions
Returns
-------
fin_spec_recom: Recommendation (namedtuple)
fin_reac_recom: Recommendation (namedtuple)
"""
# filtering out reactions that can be updated
filt_reac = [val for val in pred_reac if val.shape[0]>0]
filt_reac_ids = [val.index.name for val in filt_reac]
spec_formulas = dict()
for one_rec in pred_spec:
formulas = list(set([cn.REF_CHEBI2FORMULA[k] \
for k in one_rec['annotation'] \
if k in cn.REF_CHEBI2FORMULA.keys()]))
spec_formulas[one_rec.index.name] = formulas
anot_iter = it.Iterator(cur_spec_formula=spec_formulas,
reaction_cl=self.reactions,
reactions_to_update=filt_reac_ids)
res_iter = anot_iter.match()
recoms_tobe_added = []
for one_spec in res_iter.keys():
pred_reac_ids = [val.index.name for val in pred_reac]
reacs_using_one_spec = [val for val in pred_reac_ids \
if one_spec in self.reactions.reaction_components[val]]
filt_pred_reac = [val for val in pred_reac if val.index.name in reacs_using_one_spec]
# match score of reactions using that species
# average of the [very first match score from each candidaets set]
adj_match_score = np.mean([val['match score'].iloc[0] for val in filt_pred_reac])
cands = res_iter[one_spec]
scores = [adj_match_score for val in cands]
labels = [cn.REF_CHEBI2LABEL[val] for val in cands]
adj_recom = pd.DataFrame({'annotation': cands,
'match score': scores,
'label': labels})
adj_recom.index.name = one_spec
recoms_tobe_added.append(adj_recom)
upd_spec_dfs = recoms_tobe_added + \
[val for val in pred_spec if val.index.name not in res_iter.keys()]
# need to be converted back to namedtuple DataFrame
upd_spec_recom = [self.getRecommendationFromDataFrame(val) for val in upd_spec_dfs]
upd_reac_dfs = self.getReactionListRecommendation(pred_ids=filt_reac_ids,
spec_res=upd_spec_recom,
get_df=True)
s_df = self.getRecomTable(element_type='species',
recommended=upd_spec_dfs)
r_df = self.getRecomTable(element_type='reaction',
recommended=upd_reac_dfs)
return pd.concat([s_df, r_df], ignore_index=True)
def saveToCSV(self, obj,
fpath="recommendation.csv"):
"""
Save a completed dataframe
to csv. Doesn't proceed if obj is None,
which indicates it didn't pass the element filter.
Parameters
----------
obj: pandas.DataFrame
Object that can be saved to csv.
fpath: str
Path of the csv file to be saved.
"""
if isinstance(obj, pd.DataFrame):
obj.to_csv(fpath, index=False)
# print a summary message
for one_type in ELEMENT_TYPES:
saved_elements = list(np.unique(obj[obj['type']==one_type]['id']))
self.printSummary(saved_elements, one_type)
# def saveToSBML(self,
# fpath='model_amas_annotations.xml',
# option='augment'):
# """
# Update and save model;
# How to distinguish species vs. reactions?
# by using self.current_element_type.
# If option is 'augment',
# it'll add candidate annotations to
# existing annotation string.
# If option is 'replace',
# create a new annotation string and
# replace whatevers exists.
# Default to 'augment'.
# Call annotation maker;
# Parameters
# ----------
# fpath: str
# Path to save file
# option: str
# Either 'augment' or 'replace'
# """
# model = self.sbml_document.getModel()
# ELEMENT_FUNC = {'species': model.getSpecies,
# 'reaction': model.getReaction}
# # dictionary with empty lists;
# saved_elements = {k:[] for k in ELEMENT_TYPES}
# for one_type in ELEMENT_TYPES:
# type_selection = self.selection[one_type]
# maker = am.AnnotationMaker(one_type)
# sel2save = type_selection
# for one_k in sel2save.keys():
# one_element = ELEMENT_FUNC[one_type](one_k)
# meta_id = one_element.meta_id
# df = sel2save[one_k]
# # cands2save = list(df['annotation'])
# cands2save = []
# for val2save in list(df['annotation']):
# if val2save[:4].lower() == 'rhea':
# cands2save.append(val2save[5:])
# else:
# cands2save.append(val2save)
# if cands2save:
# if option == 'augment':
# orig_annotation = one_element.getAnnotationString()
# annotation_str = maker.addAnnotation(cands2save,
# orig_annotation,
# meta_id)
# elif option == 'replace':
# annotation_str = maker.getAnnotationString(cands2save, meta_id)
# one_element.setAnnotation(annotation_str)
# saved_elements[one_type].append(one_k)
# else:
# continue
# # finally, write the sbml document
# libsbml.writeSBMLToFile(self.sbml_document, fpath)
# # Summary message
# for one_type in ELEMENT_TYPES:
# self.printSummary(saved_elements[one_type], one_type)
def printSummary(self, saved, element_type):
"""
Print out a summary of
saved element IDs and numbers.
Parameters
----------
saved: list-str
List of elements saved.
element_type: str
'species' or 'reaction'
"""
plural_str = {'species': '',
'reaction': '(s)'}
num_saved = len(saved)
if num_saved == 0:
return None
print("Annotation recommended for %d %s%s:\n[%s]\n" %\
(num_saved,
element_type,
plural_str[element_type],
', '.join(saved)))
def getMatchScoreOfCHEBI(self, inp_id, inp_chebi):
"""
Calculate match score of a species (by ID)
with a ChEBI term.
This is to inform user of how well it matches with
a specific ChEBI term.
If the ChEBI term somehow
doesn't exist in the dictionary,
0.0 will be returned.
Parameters
----------
inp_id: str
ID of a species
inp_chebi: str
A ChEBI term.
Returns
-------
res: float
"""
inp_str = self.species.getNameToUse(inp_id)
scores = self.species.getCScores(inp_strs=[inp_str],
mssc='above',
cutoff=0.0)[inp_str]
# searching for the match score
res = next((np.round(v[1], cn.ROUND_DIGITS) \
for v in scores if v[0] == inp_chebi), 0.0)
return res
def getMatchScoreOfRHEA(self, inp_id, inp_rhea):
"""
Calculate match score of a reaction (by ID)
with a Rhea term.
This is to inform user of how well it matches with
a specific Rhea term.
Parameters
----------
inp_id: str
ID of a species
inp_rhea: str
A Rhea term.
Returns
-------
res_match_score: float
"""
specs2predict = self.reactions.reaction_components[inp_id]
spec_results = self.getSpeciesListRecommendation(pred_ids=specs2predict,
update=False,
method='cdist',
mssc='top',
cutoff=0.0)
pred_formulas = dict()
for one_spec_res in spec_results:
chebis = [val[0] for val in one_spec_res.candidates]
forms = list(set([cn.REF_CHEBI2FORMULA[k] \
for k in chebis if k in cn.REF_CHEBI2FORMULA.keys()]))
pred_formulas[one_spec_res.id] = forms
scores = self.reactions.getRScores(spec_dict=pred_formulas,
reacs=[inp_id],
mssc='above',
cutoff=0.0)[inp_id]
# searching for the match score
res = next((np.round(v[1], cn.ROUND_DIGITS) \
for v in scores if v[0] == inp_rhea), 0.0)
return res | AMAS-sb | /AMAS_sb-1.0.1-py3-none-any.whl/AMAS/recommender.py | recommender.py |
# recommend_reaction.py
"""
Predicts annotations of reaction(s) using a local XML file
and the reaction ID.
Usage: python recommend_reaction.py files/BIOMD0000000190.xml --min_score 0.6 --outfile res.csv
"""
import argparse
import os
from os.path import dirname, abspath
import sys
sys.path.insert(0, dirname(dirname(abspath(__file__))))
from AMAS import constants as cn
from AMAS import recommender
def main():
parser = argparse.ArgumentParser(description='Recommend reaction annotations of an SBML model and save results')
parser.add_argument('model', type=str, help='SBML model file (.xml)')
# One or more reaction IDs can be given
parser.add_argument('--reaction', type=str, help='ID(s) of reaction(s) to be recommended. ' +\
'If not provided, all reactions will be used', nargs='*')
parser.add_argument('--reject', type=int, help='number of the components of each reaction to reject. ' +\
'Only reactions with components greater than this value ' +\
'will be used. Default is zero', nargs='?', default=0)
parser.add_argument('--cutoff', type=float, help='minimum match score cutoff', nargs='?', default=0.0)
parser.add_argument('--method', type=str,
help='Choose either "top" or "above". "top" recommends ' +\
'the best candidates that are above the cutoff, ' +\
'and "above" recommends all candidates that are above ' +\
'the cutoff. Default is "top"',
nargs='?',
default='top')
parser.add_argument('--outfile', type=str, help='file path to save recommendation', nargs='?',
default=os.path.join(os.getcwd(), 'reaction_rec.csv'))
args = parser.parse_args()
recom = recommender.Recommender(libsbml_fpath=args.model)
one_fpath = args.model
reacts = args.reaction
reject = args.reject
cutoff = args.cutoff
method = args.method
outfile = args.outfile
recom = recommender.Recommender(libsbml_fpath=one_fpath)
recom.current_type = 'reaction'
# if nothing is given, predict all IDs
if reacts is None:
reacts = recom.getReactionIDs()
print("...\nAnalyzing %d reaction(s)...\n" % len(reacts))
# removing ids with less components than 'reject'
filt_reacts = [val for val in reacts \
if len(recom.reactions.reaction_components[val]) > reject]
# stops if all elements were removed by filtering...
if len(filt_reacts) == 0:
print("No element found after the element filter.")
return None
res = recom.getReactionListRecommendation(pred_ids=filt_reacts, get_df=True)
for idx, one_df in enumerate(res):
filt_df = recom.autoSelectAnnotation(df=one_df,
min_score=cutoff,
method=method)
recom.updateSelection(filt_reacts[idx], filt_df)
# save file to csv
recom.saveToCSV(outfile)
print("Recommendations saved as:\n%s\n" % os.path.abspath(outfile))
if __name__ == '__main__':
main() | AMAS-sb | /AMAS_sb-1.0.1-py3-none-any.whl/AMAS/recommend_reaction.py | recommend_reaction.py |
# update_annotation.py
"""
Set annotation of a model file
Usage: python update_annotation.py res.csv files/BIOMD0000000190.xml BIOMD0000000190_upd.xml
"""
import argparse
import itertools
import libsbml
import numpy as np
import os
from os.path import dirname, abspath
import pandas as pd
import sys
sys.path.insert(0, dirname(dirname(abspath(__file__))))
from AMAS import constants as cn
from AMAS import annotation_maker as am
from AMAS import tools
def main():
parser = argparse.ArgumentParser(description='Update annotations of a model using user\'s feedback file (.csv)')
parser.add_argument('infile', type=str, help='path of a model file (.xml) to update annotation')
parser.add_argument('feedback', type=str, help='path of the file (.csv) containing user\'s feedback')
parser.add_argument('outfile', type=str, help='file path to save model with updated annotations')
# csv file with user choice
args = parser.parse_args()
user_csv = pd.read_csv(args.feedback)
# Only takes cells with values 'add' or 'delete'
chosen = user_csv[(user_csv['UPDATE ANNOTATION']=='add') |\
(user_csv['UPDATE ANNOTATION']=='delete')]
outfile = args.outfile
reader = libsbml.SBMLReader()
document = reader.readSBML(args.infile)
model = document.getModel()
ELEMENT_FUNC = {'species': model.getSpecies,
'reaction': model.getReaction}
element_types = list(np.unique(chosen['type']))
for one_type in element_types:
maker = am.AnnotationMaker(one_type)
ACTION_FUNC = {'delete': maker.deleteAnnotation,
'add': maker.addAnnotation}
df_type = chosen[chosen['type']==one_type]
uids = list(np.unique(df_type['id']))
meta_ids = {val:list(df_type[df_type['id']==val]['meta id'])[0] for val in uids}
# going through one id at a time
for one_id in uids:
orig_str = ELEMENT_FUNC[one_type](one_id).getAnnotationString()
df_id = df_type[df_type['id']==one_id]
dels = list(df_id[df_id[cn.DF_UPDATE_ANNOTATION_COL]=='delete'].loc[:, 'annotation'])
adds_raw = list(df_id[df_id[cn.DF_UPDATE_ANNOTATION_COL]=='add'].loc[:, 'annotation'])
# existing annotations to be kept
keeps = list(df_id[df_id[cn.DF_UPDATE_ANNOTATION_COL]=='keep'].loc[:, 'annotation'])
adds = list(set(adds_raw + keeps))
# if type is 'reaction', need to map rhea terms back to ec/kegg terms to delete them.
if one_type == 'reaction':
rhea_del_terms = list(set(itertools.chain(*[tools.getAssociatedTermsToRhea(val) for val in dels])))
deled = maker.deleteAnnotation(rhea_del_terms, orig_str)
elif one_type == 'species':
deled = maker.deleteAnnotation(dels, orig_str)
added = maker.addAnnotation(adds, deled, meta_ids[one_id])
ELEMENT_FUNC[one_type](one_id).setAnnotation(added)
libsbml.writeSBMLToFile(document, outfile)
print("...\nUpdated model file saved as:\n%s\n" % os.path.abspath(outfile))
if __name__ == '__main__':
main() | AMAS-sb | /AMAS_sb-1.0.1-py3-none-any.whl/AMAS/update_annotation.py | update_annotation.py |
import itertools
import re
RDF_TAG_ITEM = ['rdf:RDF',
'xmlns:rdf="http://www.w3.org/1999/02/22-rdf-syntax-ns#"',
'xmlns:dcterms="http://purl.org/dc/terms/"',
'xmlns:vcard4="http://www.w3.org/2006/vcard/ns#"',
'xmlns:bqbiol="http://biomodels.net/biology-qualifiers/"',
'xmlns:bqmodel="http://biomodels.net/model-qualifiers/"']
RDF_TAG = ' '.join(RDF_TAG_ITEM)
MATCH_SCORE_BY = {'species': 'by_name',
'reaction': 'by_component'}
KNOWLEDGE_RESOURCE = {'species': 'chebi',
'reaction': 'rhea'}
class AnnotationMaker(object):
def __init__(self,
element,
prefix='bqbiol:is'):
"""
Parameters
----------
element: str
Either 'species' or 'reaction'
This will determine
the type of match score
and the knowledge resource used.
"""
self.prefix = prefix
self.knowledge_resource = KNOWLEDGE_RESOURCE[element]
# Below is only used when annotation line is created;
self.version = 'v1'
self.element = element
self.score_by = MATCH_SCORE_BY[element]
def createAnnotationContainer(self, items):
"""
Create an empty annotation container
that will hold the annotation blocks
Parameters
----------
items: str-list
Returns
-------
list-str
"""
container =[]
for one_item in items:
one_t = self.createTag(one_item)
container = self.insertList(insert_from=one_t,
insert_to=container)
return container
def createAnnotationItem(self,
knowledge_resource,
identifier):
"""
Create a one-line annotation,
e.g., <rdf:li rdf:resource="http://identifiers.org/chebi/CHEBI:15414"/>
Parameters
----------
knowledge_resource: str
identifier: str
Returns
-------
str
"""
annotation_items = ['identifiers.org',
knowledge_resource,
identifier]
res = '<rdf:li rdf:resource="http://' + \
'/'.join(annotation_items) +\
'"/>'
return res
def createTag(self,
tag_str):
"""
Create a tag based on the given string
Parameters
---------
str: inp_str
Returns
-------
list-str
"""
head_str = tag_str
tail_str = tag_str.split(' ')[0]
res_tag = ['<'+head_str+'>', '</'+tail_str+'>']
return res_tag
def getAnnotationString(self,
candidates,
meta_id):
"""
Get a string of annotations,
using a list of strings.
(of candidates)
Can replace a whole annotation.
Parameters
----------
candidates: list-str
e.g., ['CHEBI:12345', 'CHEBI:98765']
meta_id: str
Meta ID of the element to be included in the annotation.
Returns
-------
str
"""
# First, construct an empty container
container_items = ['annotation',
RDF_TAG,
'rdf:Description rdf:about="#'+meta_id+'"',
self.prefix,
'rdf:Bag']
empty_container = self.createAnnotationContainer(container_items)
# Next, create annotation lines
items_from = []
for one_cand in candidates:
items_from.append(self.createAnnotationItem(KNOWLEDGE_RESOURCE[self.element],
one_cand))
#
result = self.insertList(insert_to=empty_container,
insert_from=items_from)
return ('\n').join(result)
def getIndent(self, num_indents=0):
"""
Parameters
----------
num_indents: int
Time of indentation
Returns
-------
:str
"""
return ' ' * (num_indents)
def insertEntry(self,
inp_str,
inp_list=[],
insert_loc=None):
"""
Insert a string into a list
Parameters
----------
inp_str: str
inp_list: list
New entry will be inserted in the middle.
If not specified, will create a new list
insert_loc: int
If None, choose based on the middle of inp_list
insert: bool
If None, just return the create tag
Returns
-------
: list-str
"""
if insert_loc:
idx_insert = insert_loc
else:
idx_insert = int(len(inp_list)/2)
val2insert = [self.getIndent(idx_insert) + inp_str]
return inp_list[:idx_insert] + val2insert + inp_list[idx_insert:]
def insertList(self,
insert_to,
insert_from,
start_loc=None):
"""
Insert a list to another list.
Parameters
----------
insert_to:list
List where new list is inserted to
inser_from: list
A list where items will be inserted from
start_loc: int
If not given, insert_from will be
added in the middle of insert_to
"""
if start_loc is None:
start_loc = int(len(insert_to)/2)
indents = self.getIndent(start_loc)
insert_from_indented = [indents+val for val in insert_from]
return insert_to[:start_loc] + \
insert_from_indented + \
insert_to[start_loc:]
def divideExistingAnnotation(self,
inp_str):
"""
Divide existing string annotation
into an empty container and
items;
Parameters
----------
inp_str: str
Returns
-------
:dict/None
Dictionary of container,
and items to be augmented
Return None if it cannot be divided
"""
template_container = []
items = []
# check if it can be divided
if '<rdf:Bag>' not in inp_str:
return None
exist_anot_list = inp_str.split('\n')
one_line = ''
while one_line.strip() != '<rdf:Bag>' and exist_anot_list:
one_line = exist_anot_list.pop(0)
template_container.append(one_line)
one_line = exist_anot_list.pop(0)
while one_line.strip() != '</rdf:Bag>' and exist_anot_list:
items.append(one_line.strip())
one_line = exist_anot_list.pop(0)
template_container.append(one_line)
while exist_anot_list:
one_line = exist_anot_list.pop(0)
template_container.append(one_line)
res = {'container': template_container,
'items': items}
return res
def addAnnotation(self,
terms,
annotation,
meta_id=None):
"""
Add terms to existing annotations
(meta id is supposed to be included
in the existing annotation)
Parameters
----------
terms: str-list
List of terms to be added
annotation: str
Existing element annotation
meta_id: str
Optional argument;
if not provided and is needed,
it'll extract appropriate one from annotation.
Returns
-------
:str
"""
annotation_dict = self.divideExistingAnnotation(annotation)
# TODO: if there is no existing annotations, create a new one
if annotation_dict is None:
if meta_id is None:
meta_id = tools.extractMetaID(annotation)
return self.getAnnotationString(terms, meta_id)
container = annotation_dict['container']
existing_items = annotation_dict['items']
existing_identifiers = []
for val in existing_items:
url = re.findall('"(.*?)"', val)[0]
existing_identifiers.append(url.split('/')[-1])
# duplicated terms will not be added
additional_identifiers = [val for val in terms \
if val not in existing_identifiers]
new_items = [self.createAnnotationItem(KNOWLEDGE_RESOURCE[self.element],one_cand) \
for one_cand in additional_identifiers]
items = existing_items + new_items
res = self.insertList(container, items)
return '\n'.join(res)
def deleteAnnotation(self,
terms,
annotation):
"""
Remove entire annotation by
returning a null string.
Parameters
----------
terms: str-list
List of terms to be removed
annotation: str
Existing element annotation
Returns
-------
:str
"""
annotation_dict = self.divideExistingAnnotation(annotation)
# if cannot parse annotation, return the original annotation
if annotation_dict is None:
return annotation
container = annotation_dict['container']
exist_items = annotation_dict['items']
# finding remaining items
rem_items = []
for val in exist_items:
if all([k not in val for k in terms]):
rem_items.append(val)
if rem_items:
res = self.insertList(container, rem_items)
return '\n'.join(res)
# if all items were deleted, return an empty string
else:
return ''
def extractMetaID(self,
inp_str):
"""
Extract meta id from
the given annotation string, by searching for
two strings: '#metaid_' and '">'.
If none found, return an emtpy string
Parameters
----------
inp_str: str
Annotation string
Returns
-------
:str
Extracted meta id
"""
metaid_re = re.search('rdf:about="#(.*)">', inp_str)
if metaid_re is None:
return ''
else:
return metaid_re.group(1) | AMAS-sb | /AMAS_sb-1.0.1-py3-none-any.whl/AMAS/annotation_maker.py | annotation_maker.py |
# AMAT
Aerocapture Mission Analysis Tool (AMAT) is designed to provide rapid mission
analysis capability for aerocapture and atmospheric Entry, Descent, and Landing (EDL)
mission concepts to the planetary science community. AMAT was developed at the
[Advanced Astrodynamics Concepts (AAC)](https://engineering.purdue.edu/AAC/)
research group at Purdue University.
See [AMAT documentation](https://amat.readthedocs.io) for more details.
[](https://amat.readthedocs.io/en/master/?badge=master) [](https://doi.org/10.21105/joss.03710) [](https://badge.fury.io/py/AMAT)
If you find AMAT useful in your work, please consider citing us:
Girija et al., (2021). AMAT: A Python package for rapid conceptual design of
aerocapture and atmospheric Entry, Descent, and Landing (EDL) missions in a
Jupyter environment. *Journal of Open Source Software*, 6(67), 3710,
[DOI: 10.21105/joss.03710](https://doi.org/10.21105/joss.03710)
## Capabilities
AMAT allows the user to simulate atmospheric entry trajectories, compute deceleration
and heating loads, compute aerocapture entry corridors and simulate aerocapture
trajectories. AMAT supports analysis for all atmosphere-bearing destinations
in the Solar System: Venus, Earth, Mars, Jupiter, Saturn, Titan, Uranus, and Neptune.
AMAT allows the calculation of launch performance for a set of launch vehicles.
AMAT allows the calculation of V-inf vector from a Lambert arc for an interplanetary
transfer. AMAT allows calculation of planetary approach trajectories for orbiters and entry
systems from a given V_inf vector, B-plane targeting, and deflection maneuvers.
AMAT allows the calculation of visibility of landers to Earth and relay orbiters
and compute telecom link budgets.
### Venus Aerocapture Trajectories
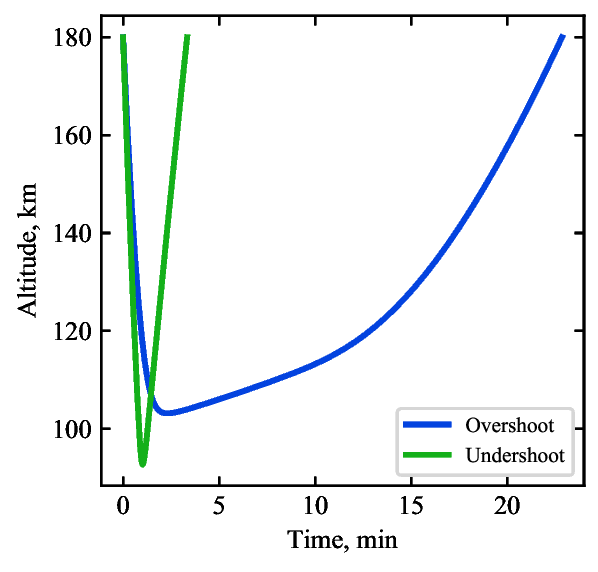
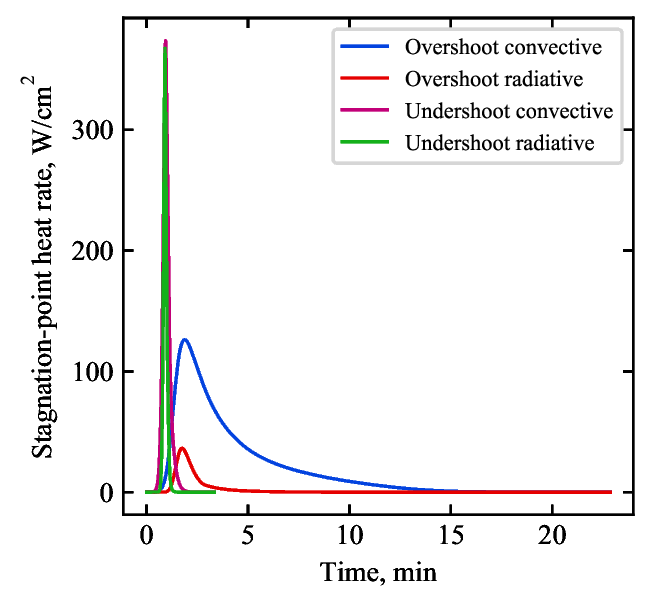
### Neptune Aerocapture Feasibility Chart

### Monte Carlo Simulations


### Examples
AMAT comes with a number of example
[Jupyter notebooks](https://amat.readthedocs.io/en/master/jupyter_link.html) to help users get started. The examples inculde:
1. Venus aerocapture assesssment study
2. Titan aerocapture assessment study
3. Neptune aerocapture using blunt-body aeroshells
4. Drag modulation aerocapture asssessment
5. Planetary probe entry at various Solar System destinations
## Features
* Easy installation,
* Many examples and recreation of results from published literature
* Extensive documented source code
* Can be used for probe and lander Entry, Descent, Landing (EDL) calculations
* Comes with a standard set of nominal atmospheric models
## Installation
Note: AMAT is currently supported on Python ``3.8``, ``3.9``, and ``3.10``.
There are two ways to install AMAT.
### Option 1: Install from pip (recommended for most users)
This is the recommended method if you do not plan or need to make changes
to the source code.
Note: Python Package Index limits the amount of additional data that can be
packaged in the distribution, hence all data cannot be included in the built version.
You will need to clone the GitHub repository to get the required
data files, examples, and start using AMAT.
Create a virtual environment and activate it. It is strongly recommended you install
AMAT inside a virtual environment to prevent it from affecting your global python
packages.
Change directory to where you want the virtual environment to be created.
* ```$ cd home/path```
#### On Linux/MacOS machines:
* ```$ python3 -m venv env1```
* ```$ source env1/bin/activate```
####On Windows machines (from Anaconda Prompt):
* ```$ conda create --name env1```
* ```$ conda activate env1```
* ```$ conda install pip```
#### Clone the repository and install AMAT:
* ```$ git clone https://github.com/athulpg007/AMAT.git```
* ```$ pip install AMAT```
Once AMAT is installed, run an example Jupyter notebook to check everything works correctly.
* ```$ cd AMAT/examples```
* ```$ jupyter-notebook```
Note that you will need jupyterlab and pandas (for some examples) to run
the example notebooks. Use ```pip install jupyterlab pandas``` to
install Jupyter and pandas if it is not already installed on your system.
This will display the full list of example Jupyter notebooks included with AMAT.
Open and run the ```example-01-hello-world``` notebook to get started with AMAT.
### Option 2: Install from setup.py (recommended for developers)
This is the recommended method if you need to make changes to the source code.
Create a virtual environment and activate it
following the steps at the beginning of Option 1.
Clone the GitHub repository and install AMAT using setup.py. The -e editable
flag allows changes you make to take effect when using AMAT.
* ```$ git clone https://github.com/athulpg007/AMAT.git```
* ```$ cd AMAT```
* ```$ python setup.py install -e```
* ```$ cd examples```
* ```$ jupyter-notebook```
Note that you will need jupyterlab and pandas (for some examples)
to run the example notebooks. Use ```pip install jupyterlab pandas```
to install Jupyter and pandas if it is not already installed on your system.
If you want to create a new distribution package:
* ```$ python3 setup.py sdist bdist_wheel```
To build docs locally if you made changes to the source code
(you must have the dependencies in ```docs/requirements.txt``` installed):
* ```$ cd AMAT/docs```
* ```$ make html```
## License
AMAT is an open source project licensed under the GNU General
Public License Version 3.
## Credits
Parts of the AMAT source code were originally developed in support of contracts between AAC and the Jet Propulsion Laboratory for various aerocapture mission studies between 2016 and 2020. Samples of atmospheric data from Global Reference Atmospheric Model (GRAM) software is used for illustration purpose only, and was developed by NASA Marshall Space Flight Center. The use of these GRAM models does not imply endorsement by NASA in any way whatsoever. A minimal working set of atmospheric profiles is included with AMAT to run the example notebooks. A minimal working interplanetary trajctory dataset is included with AMAT. The dataset was generated at Purdue University using the STOUR software package by Alec Mudek, and is also derived from trajectories published in the NASA Ice Giants Pre-Decadal Mission Study. The author plans to augment the interplanetary dataset with more publicly available information as it becomes available.
## Support and Contribution
If you wish to contribute or report an issue, feel free to [contact me](mailto:[email protected]) or to use the [issue tracker](https://github.com/athulpg007/AMAT/issues) and [pull requests](https://github.com/athulpg007/AMAT/pulls) from the [code repository](https://github.com/athulpg007/AMAT).
If you wish to make a contribution, you can do as follows:
* fork the GitHub repository
* create a feature branch from *master*
* add your feature and document it
* add tests to verify your feature is implemented correctly
* run all the tests to verify your change does not break something else
* open a pull request
## Extras
The AMAT repository includes representative atmospheric profiles for
Solar System bodies, an Excel sheet with a comprehensive literature review of
aerocapture, sample feasibility charts for aerocapture at all destinations,
reference journal articles (by the author).
## Reference Articles
Results from these articles are used as benchmark examples.
1. Craig, Scott, and James Evans Lyne. "Parametric Study of Aerocapture for Missions to Venus." Journal of Spacecraft and Rockets Vol. 42, No. 6, pp. 1035-1038. [DOI: 10.2514/1.2589](https://arc.aiaa.org/doi/10.2514/1.2589)
2. Putnam and Braun, "Drag-Modulation Flight-Control System Options for Planetary Aerocapture", Journal of Spacecraft and Rockets, Vol. 51, No. 1, 2014. [DOI:10.2514/1.A32589](https://arc.aiaa.org/doi/10.2514/1.A32589)
3. Lu, Ye, and Sarag J. Saikia. "Feasibility Assessment of Aerocapture for Future Titan Orbiter Missions." Journal of Spacecraft and Rockets Vol. 55, No. 5, pp. 1125-1135. [DOI: 10.2514/1.A34121](https://arc.aiaa.org/doi/10.2514/1.A34121)
4. Girija, A. P., Lu, Y., & Saikia, S. J. "Feasibility and Mass-Benefit Analysis of Aerocapture for Missions to Venus". Journal of Spacecraft and Rockets, Vol. 57, No. 1, pp. 58-73. [DOI: 10.2514/1.A34529](https://arc.aiaa.org/doi/10.2514/1.A34529)
5. Girija, A. P. et al. "Feasibility and Performance Analysis of Neptune
Aerocapture Using Heritage Blunt-Body Aeroshells", Journal of Spacecraft and Rockets, Vol. 57, No. 6, pp. 1186-1203. [DOI: 10.2514/1.A34719](https://arc.aiaa.org/doi/full/10.2514/1.A34719)
6. Girija A. P. et al. "Quantitative Assessment of Aerocapture and Applications to Future Missions", Journal of Spacecraft and Rockets, 2022.
[DOI: 10.2514/1.A35214](https://arc.aiaa.org/doi/full/10.2514/1.A35214) | AMAT | /AMAT-2.2.22.tar.gz/AMAT-2.2.22/README.md | README.md |
import math
import matplotlib.pyplot as plt
from .Generaldistribution import Distribution
class Gaussian(Distribution):
""" Gaussian distribution class for calculating and
visualizing a Gaussian distribution.
Attributes:
mean (float) representing the mean value of the distribution
stdev (float) representing the standard deviation of the distribution
data_list (list of floats) a list of floats extracted from the data file
"""
def __init__(self, mu=0, sigma=1):
Distribution.__init__(self, mu, sigma)
def calculate_mean(self):
"""Function to calculate the mean of the data set.
Args:
None
Returns:
float: mean of the data set
"""
avg = 1.0 * sum(self.data) / len(self.data)
self.mean = avg
return self.mean
def calculate_stdev(self, sample=True):
"""Function to calculate the standard deviation of the data set.
Args:
sample (bool): whether the data represents a sample or population
Returns:
float: standard deviation of the data set
"""
if sample:
n = len(self.data) - 1
else:
n = len(self.data)
mean = self.calculate_mean()
sigma = 0
for d in self.data:
sigma += (d - mean) ** 2
sigma = math.sqrt(sigma / n)
self.stdev = sigma
return self.stdev
def plot_histogram(self):
"""Function to output a histogram of the instance variable data using
matplotlib pyplot library.
Args:
None
Returns:
None
"""
plt.hist(self.data)
plt.title('Histogram of Data')
plt.xlabel('data')
plt.ylabel('count')
def pdf(self, x):
"""Probability density function calculator for the gaussian distribution.
Args:
x (float): point for calculating the probability density function
Returns:
float: probability density function output
"""
return (1.0 / (self.stdev * math.sqrt(2*math.pi))) * math.exp(-0.5*((x - self.mean) / self.stdev) ** 2)
def plot_histogram_pdf(self, n_spaces = 50):
"""Function to plot the normalized histogram of the data and a plot of the
probability density function along the same range
Args:
n_spaces (int): number of data points
Returns:
list: x values for the pdf plot
list: y values for the pdf plot
"""
mu = self.mean
sigma = self.stdev
min_range = min(self.data)
max_range = max(self.data)
# calculates the interval between x values
interval = 1.0 * (max_range - min_range) / n_spaces
x = []
y = []
# calculate the x values to visualize
for i in range(n_spaces):
tmp = min_range + interval*i
x.append(tmp)
y.append(self.pdf(tmp))
# make the plots
fig, axes = plt.subplots(2,sharex=True)
fig.subplots_adjust(hspace=.5)
axes[0].hist(self.data, density=True)
axes[0].set_title('Normed Histogram of Data')
axes[0].set_ylabel('Density')
axes[1].plot(x, y)
axes[1].set_title('Normal Distribution for \n Sample Mean and Sample Standard Deviation')
axes[0].set_ylabel('Density')
plt.show()
return x, y
def __add__(self, other):
"""Function to add together two Gaussian distributions
Args:
other (Gaussian): Gaussian instance
Returns:
Gaussian: Gaussian distribution
"""
result = Gaussian()
result.mean = self.mean + other.mean
result.stdev = math.sqrt(self.stdev ** 2 + other.stdev ** 2)
return result
def __repr__(self):
"""Function to output the characteristics of the Gaussian instance
Args:
None
Returns:
string: characteristics of the Gaussian
"""
return "mean {}, standard deviation {}".format(self.mean, self.stdev) | AMB-distribution | /AMB_distribution-0.1.tar.gz/AMB_distribution-0.1/AMB_distribution/Gaussiandistribution.py | Gaussiandistribution.py |
import math
import matplotlib.pyplot as plt
from .Generaldistribution import Distribution
class Binomial(Distribution):
""" Binomial distribution class for calculating and
visualizing a Binomial distribution.
Attributes:
mean (float) representing the mean value of the distribution
stdev (float) representing the standard deviation of the distribution
data_list (list of floats) a list of floats to be extracted from the data file
p (float) representing the probability of an event occurring
n (int) number of trials
TODO: Fill out all functions below
"""
def __init__(self, prob=.5, size=20):
self.n = size
self.p = prob
Distribution.__init__(self, self.calculate_mean(), self.calculate_stdev())
def calculate_mean(self):
"""Function to calculate the mean from p and n
Args:
None
Returns:
float: mean of the data set
"""
self.mean = self.p * self.n
return self.mean
def calculate_stdev(self):
"""Function to calculate the standard deviation from p and n.
Args:
None
Returns:
float: standard deviation of the data set
"""
self.stdev = math.sqrt(self.n * self.p * (1 - self.p))
return self.stdev
def replace_stats_with_data(self):
"""Function to calculate p and n from the data set
Args:
None
Returns:
float: the p value
float: the n value
"""
self.n = len(self.data)
self.p = 1.0 * sum(self.data) / len(self.data)
self.mean = self.calculate_mean()
self.stdev = self.calculate_stdev()
def plot_bar(self):
"""Function to output a histogram of the instance variable data using
matplotlib pyplot library.
Args:
None
Returns:
None
"""
plt.bar(x = ['0', '1'], height = [(1 - self.p) * self.n, self.p * self.n])
plt.title('Bar Chart of Data')
plt.xlabel('outcome')
plt.ylabel('count')
def pdf(self, k):
"""Probability density function calculator for the gaussian distribution.
Args:
x (float): point for calculating the probability density function
Returns:
float: probability density function output
"""
a = math.factorial(self.n) / (math.factorial(k) * (math.factorial(self.n - k)))
b = (self.p ** k) * (1 - self.p) ** (self.n - k)
return a * b
def plot_bar_pdf(self):
"""Function to plot the pdf of the binomial distribution
Args:
None
Returns:
list: x values for the pdf plot
list: y values for the pdf plot
"""
x = []
y = []
# calculate the x values to visualize
for i in range(self.n + 1):
x.append(i)
y.append(self.pdf(i))
# make the plots
plt.bar(x, y)
plt.title('Distribution of Outcomes')
plt.ylabel('Probability')
plt.xlabel('Outcome')
plt.show()
return x, y
def __add__(self, other):
"""Function to add together two Binomial distributions with equal p
Args:
other (Binomial): Binomial instance
Returns:
Binomial: Binomial distribution
"""
try:
assert self.p == other.p, 'p values are not equal'
except AssertionError as error:
raise
result = Binomial()
result.n = self.n + other.n
result.p = self.p
result.calculate_mean()
result.calculate_stdev()
return result
def __repr__(self):
"""Function to output the characteristics of the Binomial instance
Args:
None
Returns:
string: characteristics of the Gaussian
"""
return "mean {}, standard deviation {}, p {}, n {}".\
format(self.mean, self.stdev, self.p, self.n) | AMB-distribution | /AMB_distribution-0.1.tar.gz/AMB_distribution-0.1/AMB_distribution/Binomialdistribution.py | Binomialdistribution.py |
import tellurium as te
import matplotlib.pyplot as plt
def reactionAntimony(dict, currentK):
f = open("webapp/static/antimony1.txt", 'a')
reactantSpecies = dict['Reactants']
FixedReactants = dict['FixedReactants']
newStr = ''
for species in reactantSpecies:
for fixedSpecies in FixedReactants:
if species == fixedSpecies:
species = '$' + species
newStr = newStr + species + '+'
else:
newStr = newStr + species + '+'
newStr = newStr[:-1] + '->'
productSpecies = dict['Products']
FixedProducts = dict['FixedProducts']
for species in productSpecies:
for fixedSpecies in FixedProducts:
if species == fixedSpecies:
species = '$' + fixedSpecies
newStr = newStr + species + '+'
else:
newStr = newStr + species + '+'
rxnVals = dict['RxnConstant']
kVal = len(rxnVals)
kList = []
for i in range(kVal):
numStr = str(i + currentK)
kList.append('k' + numStr)
numConstants = i
currentK = currentK + kVal
newStr = newStr[:-1] + ' ; ' + kList[0] + '*'
for species in reactantSpecies:
newStr = newStr + species + '*'
newStr = newStr[:-1]
# print this line to the antimony file
f.write('\n')
f.write(newStr)
if dict['Reversibility'] == True:
secStr = ''
for species in productSpecies:
for fixedSpecies in FixedReactants:
if species == fixedSpecies:
species = '$' + fixedSpecies
secStr = secStr + species + '+'
else:
secStr = secStr + species + '+'
secStr = secStr[:-1] + '->'
for species in reactantSpecies:
for fixedSpecies in FixedReactants:
if species == fixedSpecies:
species = '$' + species
secStr = secStr + species + '+'
else:
secStr = secStr + species + '+'
secStr = secStr[:-1] + ' ; ' + kList[1] + '*'
for species in productSpecies:
secStr = secStr + species + '*'
secStr = secStr[:-1]
# print secStr to antimony file within the if statement
f.write('\n')
f.write(secStr)
f.close()
return currentK, kList
def conditionsAntimony(dict, kList):
f = open('webapp/static/antimony2.txt','a')
reactantIC = dict['ReactantIC']
reactantSpecies = dict['Reactants']
str1 = ''
count = 0
for species in reactantSpecies:
str1 += species + '=' + reactantIC[count] + ';'
count += 1
str1 = str1[:-1]
f.write('\n')
f.write(str1)
# print to IC file
productIC = dict['ProductIC']
productSpecies = dict['Products']
str2 = ''
count = 0
for species in productSpecies:
str2 += species + '=' + productIC[count] + ';'
count += 1
str2 = str2[:-1]
# print to IC file
f.write('\n')
f.write(str2)
rxnConstants = dict['RxnConstant']
str3 = ''
count = 0
for var in kList:
str3 += var + '=' + rxnConstants[count] + ';'
count += 1
str3 = str3[:-1]
f.write('\n')
f.write(str3)
f.close()
return str1, str2, str3
def resetFiles():
with open('webapp/static/antimony1.txt', 'r+') as file:
file.truncate(0)
file.close()
with open('webapp/static/antimony2.txt', 'r+') as file:
file.truncate(0)
file.close()
"""
Takes in given values as strings or booleans from the website program and saves
them to a dictionary
Inputs:
- reactants: (Str) comma separated string of a list of letters that represent reactants
- fixed_reactants: (Str) comma separated string of the reactants that are fixed values
- reactantIC: (Str) comma separated string of numbers that are the IC of the reactants
- products: (Str) comma separated string of a list of letters that represent products
- fixed_products: (Str) comma separated string of the products that are fixed values
- productIC: (Str) comma separated string of numbers that are the IC of the products
- reactionConstant: (Str) comma separates string of numbers that are the k values of reaction
- reversibility: (boolean) true if the reaction is reversible
Outputs:
- dict: a dictionary that contains the values as the inputs connected to a name
"""
def init(reactants, fixed_reactants, reactantIC, products, fixed_products, productIC, reactionConstant, reversibility):
dict = {}
dict['Reactants'] = reactants.split(',')
dict['FixedReactants'] = fixed_reactants.split(',')
dict['ReactantIC'] = reactantIC.split(',')
dict['Products'] = products.split(',')
dict['FixedProducts'] = fixed_products.split(',')
dict['ProductIC'] = productIC.split(',')
dict['RxnConstant'] = reactionConstant.split(',')
dict['Reversibility'] = reversibility
return dict
def runSim():
file1 = open('webapp/static/antimony1.txt','a')
file2 = open('webapp/static/antimony2.txt','r')
for line in file2:
file1.write(line)
print(line)
file1.close()
file2.close()
loadModel()
def loadModel():
r = te.loadAntimonyModel('webapp/static/antimony1.txt')
result = r.simulate()
r.plot()
plt.savefig("webapp/static/output.jpg")
def loadSBML(url):
r = te.loadSBMLModel(url)
result = r.simulate()
r.plot()
plt.savefig("webapp/static/output.jpg") | AMC-OWMV | /AMC_OWMV-1.0.1-py3-none-any.whl/webapp/antimonyTools.py | antimonyTools.py |
# Antimony Model Creator - Olivia Walsh, Matthew Van Ginneken
This package allows the user to visualize a reaction via a steady state graph. The user can create and submit their own reactions on the Flask Webpage, or they can submit an antimony txt file or a SBML file also through the webpage. The Webpage will run a simulation using the reactions with tellurium and show the steady state graph.
## <b>Installation </b>
To install this package the user should open up to their terminal and use pip install to install the package
> pip install AMC-OWMVG==1.1.0
Once the terminal has confirmed installation, the user must create a new folder in their desktop and move to that directory
> cd Desktop <br>
> cd newFolder (replace newFolder with the name of the folder)
Within this folder the user has to create a file and another folder called static. We recommend using an IDE or an editor to make these.
> static (a folder) <br>
> runApp.py <br>
Within the static folder the user should make two .txt files that are empty.
> antimony1.txt <br>
> antimony2.txt <br>
Going back to the file runApp.py the user should type out the following into the file
> import AMCowmvg <br>
> from AMCowmvg.testing import runApp <br>
> runApp()
The user should then run the runApp.py in their terminal
> python runApp.py
This will produce a few lines of response including a website link, the user should copy and paste this into their browser. The user now can create and visualize their reactions
##<b> Installation if you have access to the StarterAMC folder </b>
### <i>If the user has access to the StarterAMC folder they should download it to their desktop then run through the following commands </i>
To install this package the user should open up to their terminal and use pip install to install the package
> pip install AMC-OWMVG==1.1.0
Once the terminal has confirmed installation the user must navigate to the StarterAMC directory
> cd Desktop <br>
> cd StarterAMC
Once the user is in the correct directory they should run the runApp.py file from their terminal
> python runApp.py
This will produce a few lines of response including a website link, the user should copy and paste this into their browser. The user now can create and visualize their reactions
| AMCowmvg | /AMCowmvg-1.1.0.tar.gz/AMCowmvg-1.1.0/README.md | README.md |
# Arduino_Master_Delta(AMD) Version 2.2.0
## From Version 2.2 AMD will stand for Arduino-Matplotlib-DataScience since I've implemented various functions as in the new expansion in this single module.

___
# What's the difference between Alpha and Delta Versions ?
#### >>> Alpha version provides ` easier but approximate ` interface for data extraction and Visualization **_(Since X values are automatically generated and fit enables only approximation of the data plotting)_**.
#### >>> Delta version of Arduino_Master is designed to deliver `accurate visualization` and hence a bit more complex than Alpha versions.
#### >>> To ease using of functions, Delta version uses a specific type of input called `hybrid` to represent both the X and Y values of a given plot with just one variable.
#### >>> Default plotting style is 'dark_background' unlike 'ggplot' of Alpha.
#### >>> Link to Arduino_Master (Alpha Version) [Arduino_Master_Alpha](https://pypi.org/project/Arduino-Master/). Note that this module is officially not supported anymore. That is no updates would be added to it and also the availability of data science functions are less.
___
___
# What's New in 2.2.0 ?
#### >>> Bugs related to readSerial, writeSerial and dynamicSerial fixed.
#### >>> **`farFrom`** and **`minDeviation`** parameters added to **`filter`** function.
#### >>> Bugs in filter function that prevented the use of farFrom, closeTo, maxDeviation and minDeviation parameters together **_rectified_**.
#### >>> **`compress()`** function updated to work with hybrids too !! (Version-0.9.2)
#### >>> 'avg' can also be passed to **`closeTo`** parameter of the filter function !! (Version-1.2)
#### >>> **`below`** and **`above`** parameters added to filter function ! (Version-1.3)
#### >>> 9 new data science functions added !! (Version-1.4)
#### >>> hybridize function updated to check if both index and amplitude lists have equal number of elements ! (Version-1.5)
#### >>> **`equiAxis'** parameter added to Graph, compGraph and visualizeSmoothie functions !!! (Version-1.5)
#### >>> Font Bug in plotting functions **_rectified_** !! (Version-1.6)
#### >>> The debugging of the previuos bug which I cleared out in Version 1.6 gave some more bugs, I got it **_rectified_**. You guys know about this, You are programmers !! (Version-1.7)
#### >>> Change 1.6 as 1.7 in my previous point.Checked it, everything is fine to go now ! (Version-1.8)
#### >>> Included docstrings and compatibility checked with all operating systems and jupyter notebook ! (Version-2.0)
#### >>> Functions like densePop, scarcePop, remImp, detectImp, cleanImpulses, reduce, instAvg, smoothie, Graph, compGraph and visualizeSmoothie modified to work even if you pass a list at the place of a hybird. But for accurate Data visualizations I'd recommend using hybrids. However all those functions returns hybrids alone as usual. Maximum error tolerance from this version !!!!!!!!!!!!!!!!!! (Version-2.1)
#### >>> Data storage module compatibility added !! (Version-2.2.0)
#### >>> 6 New functions providing data storage and retrieval interface added !! (Version-2.2.0)
___
___
### AMD tutorial in this link -> [tutorial](https://github.com/SayadPervez/AMD--TUTORIAL/blob/master/README.md)
___
___
# Intro:
#### Embedded C is used to program microcontrollers like Arduino. However embedded C could never compete with Python's simplicity and functionality. Also Arduino being a microcontroller, we get a lot of garbage values which we require to filter before utilizing. This module eases the process of extracting and passing data to Arduino via Serial communication.
#### This Module also provides Easy and flexible Data Science functions for Data extraction, filtering, removing garbage values and Data Visualization !
___
___
# Installing via pip:
### Use the following command to install Arduino_Master using pip.
### **`pip install AMD`**
___
___
# Automatic Installation of other required Modules:
#### This module requires two more packages namely **`pyserial`** and **`matplotlib`**. Yet just by importing this module using the import statement or by using any function, unavailable modules will be installed and imported automatically. Make sure you have good internet connection and if still you get ModuleNotFound error, try installing these two modules manually via pip.
___
___
# Importing Functions:
#### **`from AMD import *`** statement is used to import all available functions from Arduino_Master. This version contains the following functions which we'll be discussing shortly. These functions can be grouped into 3 categories:
___
### [>> For Extracting and Writing data to Arduino:](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/Arduino_Data_Extraction_Functions.md)
#### [$ ardata](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/ardata().md)
#### [$ readSerial](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/readSerial.md)
#### [$ writeSerial](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/writeSerial.md)
#### [$ dynamicSerial](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/dynamicSerial.md)
___
### [>> Data Science enabled functions for filtering and visualizing Data:](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/DataScience/DataScience%20Main.md)
#### [$ hybridize](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/DataScience/hybrids.md)
#### [$ Graph](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/DataScience/Graph.md)
#### [$ compGraph](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/DataScience/compGraph.md)
#### [$ horizontal](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/DataScience/horizontal.md)
#### [$ vertical](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/DataScience/vertical.md)
#### [$ marker](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/DataScience/marker.md)
#### [$ most_frequent](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/DataScience/most_frequent.md)
#### [$ least_frequent](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/DataScience/least_frequent.md)
#### [$ compress](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/DataScience/compress.md)
#### [$ filter](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/DataScience/filter.md)
#### [$ densePop](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/DataScience/densePop.md)
#### [$ scarcePop](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/DataScience/scarcePop.md)
#### [$ remImp](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/DataScience/remImp.md)
#### [$ detectImp](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/DataScience/detectImp.md)
#### [$ cleanImpulses](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/DataScience/cleanImpulses.md)
#### [$ reduce](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/DataScience/reduce.md)
#### [$ instAvg](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/DataScience/instAbd.md)
#### [$ smoothie](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/DataScience/smoothie.md)
#### [$ visualizeSmoothie](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/DataScience/visualizeSmoothie.md)
___
### [ >> Data storage and extraction functions for AMD known as AMD CDB which stands for 'AMD Custom DataBase': ](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/AMD%20CDB.md)
#### [Intro to AMD CDB](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/DataBase/AMD%20Custom%20DataBase%20Methods.md)
#### [save](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/DataBase/save.md)
#### [appendSave](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/DataBase/appendSave.md)
#### [rewriteSave](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/DataBase/rewriteSave.md)
#### [extract](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/DataBase/extract.md)
#### [extractX](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/DataBase/extractX.md)
#### [extractY](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/DataBase/extractY.md)
___
___
### [Some projects that can be done with this module !!](https://github.com/SayadPervez/AMD-SEPERATE-DOCUMENTATION/blob/master/Examples.md)
___
___

### Sayad Pervez - the solo developer of AMD Module !
### E-Mail : [[email protected]]([email protected])
___
___
| AMD | /AMD-2.2.0.tar.gz/AMD-2.2.0/README.md | README.md |
[](https://doi.org/10.5281/zenodo.4003825) [](https://pypi.org/project/AMDirT) [](https://amdirt.readthedocs.io/en/dev/?badge=dev) [](https://github.com/SPAAM-community/AMDirT/actions/workflows/ci_test.yml)
# AMDirT
**AMDirT**: [**A**ncient**M**etagenome**Dir**](https://github.com/SPAAM-community/ancientmetagenomedir) **T**oolkit
AMDirT is a toolkit for interacting with the AncientMetagenomeDir metadata repository of ancient metagenomic samples and ancient microbial genomes. This tool provides ways to validate AncientMetagenomeDir submissions, explore and download sequencing data for ancient microbial and environmental (meta)genomes, and automatically prepare input samplesheets for a range of bioinformatic processing pipelines.
For documentation on using the tool, please see [How Tos](how_to/index), [Tutorials](/tutorials) and/or [Quick Reference](/reference).
## Install
### 1. With pip
```bash
pip install amdirt
```
### 2. With conda
```bash
conda install -c bioconda amdirt
```
### The latest development version, directly from GitHub
```bash
pip install --upgrade --force-reinstall git+https://github.com/SPAAM-community/AMDirT.git@dev
```
### The latest development version, with local changes
- Fork AMDirT on GitHub
- Clone your fork `git clone [your-AMDirT-fork]`
- Checkout the `dev` branch `git switch dev`
- Create the conda environment `conda env create -f environment.yml`
- Activate the environment `conda activate amdirt`
- Install amdirt in development mode `pip install -e .`
- In some cases you may need to force update streamlit with `pip install --upgrade steamlit`
To locally render documentation:
- `conda activate amdirt`
- Install additional requirements `cd docs && pip install -r requirements.txt`
- Build the HTML `make html`
- Open the `build/html/README.html` file in your browser
## More information
For more information, please see the AMDirT Documentation
- Stable: [amdirt.readthedocs.io/en/latest/](https://amdirt.readthedocs.io/en/latest/)
- Development version: [amdirt.readthedocs.io/en/dev/](https://amdirt.readthedocs.io/en/dev/)
| AMDirT | /AMDirT-1.4.3.tar.gz/AMDirT-1.4.3/README.md | README.md |
# AMDock: **A***ssisted* **M***olecular* **Dock***ing with Autodock4 and Autodock Vina*
AMDock (Assisted Molecular Docking) is a user-friendly graphical tool to assist in the docking of protein-ligand
complexes using Autodock-Vina or AutoDock4. This tool integrates several external programs for processing docking input
files, define the search space (box) and perform docking under user’s supervision.
**Version 1.6.x for Linux** (**Build 1.6.1-beta**)
**DOCUMENTATION**
Manual, tutorials and test files are located in **Doc** folder. (May be out of date. Please check the wiki)
**Cite us**
Valdes-Tresanco, M.S., Valdes-Tresanco, M.E., Valiente, P.A. and Moreno E. AMDock: a versatile graphical tool for
assisting molecular docking with Autodock Vina and Autodock4. Biol Direct 15, 12 (2020). https://doi.org/10.1186/s13062-020-00267-2
<a href="https://www.scimagojr.com/journalsearch.php?q=5800173376&tip=sid&exact=no" title="SCImago Journal & Country Rank"><img border="0" src="https://www.scimagojr.com/journal_img.php?id=5800173376" alt="SCImago Journal & Country Rank" /></a>
**INSTALL**
Installation can be carried out in two ways
1. (For Linux only) Using a conda environment.
Note: macOS users can use this procedure, however, PyMOL must be compiled using this environment's Python interpreter,
since there is no working version of PyMOL for mac in the anaconda repositories.
To do this, proceed as follows:
If you don't have conda installed, please visit the [Miniconda download page](https://docs.conda.io/en/latest/miniconda.html).
Those with an existing conda installation may wish to create a new conda "environment" to avoid conflicts with what
you already have installed. To do this:
conda create --name AMDock
conda activate AMDock
(Note that you would need to perform the "conda activate" step every time you wish to use AMDock in a new terminal;
it might be appropriate to add this to your start-up script. Creating a new environment should not be necessary if
you only use conda for AMDock.)
Once this is done, type:
conda install -c conda-forge pymol-open-source openbabel pdb2pqr
and finally:
python -m pip install git+https://github.com/Valdes-Tresanco-MS/AutoDockTools_py3
python -m pip install AMDock
2. Using the OS Python 3 environment. To do this, proceed as follows:
sudo apt install pymol openbabel
(Note that this version of AMDock works with openbabel 3.x)
python3 -m pip install pdb2pqr
python3 -m pip install git+https://github.com/Valdes-Tresanco-MS/AutoDockTools_py3
python3 -m pip install AMDock
Before using AMDock, you most install the PyMOL plugin (grid_amdock.py).
If it does not appear please follow the instructions:
- Download the grid_amdock.py file
- Open PyMOL > Plugins > Manager Plugins > Install New Plugin > Choose File and select the grid_amdock.py file
- Restart PyMOL
To open AMDock, type in the terminal:
AMDock
For macOS users,
To view the update history, please check Changes_History file
**TUTORIALS**
Please, check the wiki https://github.com/Valdes-Tresanco-MS/AMDock-win/wiki
[<img src="./AMDock/AMDock/images/jetbrains-variant-4.png" height="100" align="right" />](https://www.jetbrains.com/?from=https://github.com/Valdes-Tresanco-MS/AMDock)
## Support
This project is possible thanks to the Open Source license of the
[JetBrains](https://www.jetbrains.com/?from=https://github.com/Valdes-Tresanco-MS/AMDock
) programs.
| AMDock | /AMDock-1.6.2.tar.gz/AMDock-1.6.2/README.md | README.md |
# pylint:disable=invalid-name,import-outside-toplevel,missing-function-docstring
# pylint:disable=missing-class-docstring,too-many-branches,too-many-statements
# pylint:disable=raise-missing-from,too-many-lines,too-many-locals,import-error
# pylint:disable=too-few-public-methods,redefined-outer-name,consider-using-with
# pylint:disable=attribute-defined-outside-init,too-many-arguments
import configparser
import errno
import json
import os
import re
import subprocess
import sys
from typing import Callable, Dict
class VersioneerConfig:
"""Container for Versioneer configuration parameters."""
def get_root():
"""Get the project root directory.
We require that all commands are run from the project root, i.e. the
directory that contains setup.py, setup.cfg, and versioneer.py .
"""
root = os.path.realpath(os.path.abspath(os.getcwd()))
setup_py = os.path.join(root, "setup.py")
versioneer_py = os.path.join(root, "versioneer.py")
if not (os.path.exists(setup_py) or os.path.exists(versioneer_py)):
# allow 'python path/to/setup.py COMMAND'
root = os.path.dirname(os.path.realpath(os.path.abspath(sys.argv[0])))
setup_py = os.path.join(root, "setup.py")
versioneer_py = os.path.join(root, "versioneer.py")
if not (os.path.exists(setup_py) or os.path.exists(versioneer_py)):
err = ("Versioneer was unable to run the project root directory. "
"Versioneer requires setup.py to be executed from "
"its immediate directory (like 'python setup.py COMMAND'), "
"or in a way that lets it use sys.argv[0] to find the root "
"(like 'python path/to/setup.py COMMAND').")
raise VersioneerBadRootError(err)
try:
# Certain runtime workflows (setup.py install/develop in a setuptools
# tree) execute all dependencies in a single python process, so
# "versioneer" may be imported multiple times, and python's shared
# module-import table will cache the first one. So we can't use
# os.path.dirname(__file__), as that will find whichever
# versioneer.py was first imported, even in later projects.
my_path = os.path.realpath(os.path.abspath(__file__))
me_dir = os.path.normcase(os.path.splitext(my_path)[0])
vsr_dir = os.path.normcase(os.path.splitext(versioneer_py)[0])
if me_dir != vsr_dir:
print("Warning: build in %s is using versioneer.py from %s"
% (os.path.dirname(my_path), versioneer_py))
except NameError:
pass
return root
def get_config_from_root(root):
"""Read the project setup.cfg file to determine Versioneer config."""
# This might raise OSError (if setup.cfg is missing), or
# configparser.NoSectionError (if it lacks a [versioneer] section), or
# configparser.NoOptionError (if it lacks "VCS="). See the docstring at
# the top of versioneer.py for instructions on writing your setup.cfg .
setup_cfg = os.path.join(root, "setup.cfg")
parser = configparser.ConfigParser()
with open(setup_cfg, "r") as cfg_file:
parser.read_file(cfg_file)
VCS = parser.get("versioneer", "VCS") # mandatory
# Dict-like interface for non-mandatory entries
section = parser["versioneer"]
cfg = VersioneerConfig()
cfg.VCS = VCS
cfg.style = section.get("style", "")
cfg.versionfile_source = section.get("versionfile_source")
cfg.versionfile_build = section.get("versionfile_build")
cfg.tag_prefix = section.get("tag_prefix")
if cfg.tag_prefix in ("''", '""'):
cfg.tag_prefix = ""
cfg.parentdir_prefix = section.get("parentdir_prefix")
cfg.verbose = section.get("verbose")
return cfg
class NotThisMethod(Exception):
"""Exception raised if a method is not valid for the current scenario."""
# these dictionaries contain VCS-specific tools
LONG_VERSION_PY: Dict[str, str] = {}
HANDLERS: Dict[str, Dict[str, Callable]] = {}
def register_vcs_handler(vcs, method): # decorator
"""Create decorator to mark a method as the handler of a VCS."""
def decorate(f):
"""Store f in HANDLERS[vcs][method]."""
HANDLERS.setdefault(vcs, {})[method] = f
return f
return decorate
def run_command(commands, args, cwd=None, verbose=False, hide_stderr=False,
env=None):
"""Call the given command(s)."""
assert isinstance(commands, list)
process = None
for command in commands:
try:
dispcmd = str([command] + args)
# remember shell=False, so use git.cmd on windows, not just git
process = subprocess.Popen([command] + args, cwd=cwd, env=env,
stdout=subprocess.PIPE,
stderr=(subprocess.PIPE if hide_stderr
else None))
break
except OSError:
e = sys.exc_info()[1]
if e.errno == errno.ENOENT:
continue
if verbose:
print("unable to run %s" % dispcmd)
print(e)
return None, None
else:
if verbose:
print("unable to find command, tried %s" % (commands,))
return None, None
stdout = process.communicate()[0].strip().decode()
if process.returncode != 0:
if verbose:
print("unable to run %s (error)" % dispcmd)
print("stdout was %s" % stdout)
return None, process.returncode
return stdout, process.returncode
LONG_VERSION_PY['git'] = r'''
# This file helps to compute a version number in source trees obtained from
# git-archive tarball (such as those provided by githubs download-from-tag
# feature). Distribution tarballs (built by setup.py sdist) and build
# directories (produced by setup.py build) will contain a much shorter file
# that just contains the computed version number.
# This file is released into the public domain. Generated by
# versioneer-0.21 (https://github.com/python-versioneer/python-versioneer)
"""Git implementation of _version.py."""
import errno
import os
import re
import subprocess
import sys
from typing import Callable, Dict
def get_keywords():
"""Get the keywords needed to look up the version information."""
# these strings will be replaced by git during git-archive.
# setup.py/versioneer.py will grep for the variable names, so they must
# each be defined on a line of their own. _version.py will just call
# get_keywords().
git_refnames = "%(DOLLAR)sFormat:%%d%(DOLLAR)s"
git_full = "%(DOLLAR)sFormat:%%H%(DOLLAR)s"
git_date = "%(DOLLAR)sFormat:%%ci%(DOLLAR)s"
keywords = {"refnames": git_refnames, "full": git_full, "date": git_date}
return keywords
class VersioneerConfig:
"""Container for Versioneer configuration parameters."""
def get_config():
"""Create, populate and return the VersioneerConfig() object."""
# these strings are filled in when 'setup.py versioneer' creates
# _version.py
cfg = VersioneerConfig()
cfg.VCS = "git"
cfg.style = "%(STYLE)s"
cfg.tag_prefix = "%(TAG_PREFIX)s"
cfg.parentdir_prefix = "%(PARENTDIR_PREFIX)s"
cfg.versionfile_source = "%(VERSIONFILE_SOURCE)s"
cfg.verbose = False
return cfg
class NotThisMethod(Exception):
"""Exception raised if a method is not valid for the current scenario."""
LONG_VERSION_PY: Dict[str, str] = {}
HANDLERS: Dict[str, Dict[str, Callable]] = {}
def register_vcs_handler(vcs, method): # decorator
"""Create decorator to mark a method as the handler of a VCS."""
def decorate(f):
"""Store f in HANDLERS[vcs][method]."""
if vcs not in HANDLERS:
HANDLERS[vcs] = {}
HANDLERS[vcs][method] = f
return f
return decorate
def run_command(commands, args, cwd=None, verbose=False, hide_stderr=False,
env=None):
"""Call the given command(s)."""
assert isinstance(commands, list)
process = None
for command in commands:
try:
dispcmd = str([command] + args)
# remember shell=False, so use git.cmd on windows, not just git
process = subprocess.Popen([command] + args, cwd=cwd, env=env,
stdout=subprocess.PIPE,
stderr=(subprocess.PIPE if hide_stderr
else None))
break
except OSError:
e = sys.exc_info()[1]
if e.errno == errno.ENOENT:
continue
if verbose:
print("unable to run %%s" %% dispcmd)
print(e)
return None, None
else:
if verbose:
print("unable to find command, tried %%s" %% (commands,))
return None, None
stdout = process.communicate()[0].strip().decode()
if process.returncode != 0:
if verbose:
print("unable to run %%s (error)" %% dispcmd)
print("stdout was %%s" %% stdout)
return None, process.returncode
return stdout, process.returncode
def versions_from_parentdir(parentdir_prefix, root, verbose):
"""Try to determine the version from the parent directory name.
Source tarballs conventionally unpack into a directory that includes both
the project name and a version string. We will also support searching up
two directory levels for an appropriately named parent directory
"""
rootdirs = []
for _ in range(3):
dirname = os.path.basename(root)
if dirname.startswith(parentdir_prefix):
return {"version": dirname[len(parentdir_prefix):],
"full-revisionid": None,
"dirty": False, "error": None, "date": None}
rootdirs.append(root)
root = os.path.dirname(root) # up a level
if verbose:
print("Tried directories %%s but none started with prefix %%s" %%
(str(rootdirs), parentdir_prefix))
raise NotThisMethod("rootdir doesn't start with parentdir_prefix")
@register_vcs_handler("git", "get_keywords")
def git_get_keywords(versionfile_abs):
"""Extract version information from the given file."""
# the code embedded in _version.py can just fetch the value of these
# keywords. When used from setup.py, we don't want to import _version.py,
# so we do it with a regexp instead. This function is not used from
# _version.py.
keywords = {}
try:
with open(versionfile_abs, "r") as fobj:
for line in fobj:
if line.strip().startswith("git_refnames ="):
mo = re.search(r'=\s*"(.*)"', line)
if mo:
keywords["refnames"] = mo.group(1)
if line.strip().startswith("git_full ="):
mo = re.search(r'=\s*"(.*)"', line)
if mo:
keywords["full"] = mo.group(1)
if line.strip().startswith("git_date ="):
mo = re.search(r'=\s*"(.*)"', line)
if mo:
keywords["date"] = mo.group(1)
except OSError:
pass
return keywords
@register_vcs_handler("git", "keywords")
def git_versions_from_keywords(keywords, tag_prefix, verbose):
"""Get version information from git keywords."""
if "refnames" not in keywords:
raise NotThisMethod("Short version file found")
date = keywords.get("date")
if date is not None:
# Use only the last line. Previous lines may contain GPG signature
# information.
date = date.splitlines()[-1]
# git-2.2.0 added "%%cI", which expands to an ISO-8601 -compliant
# datestamp. However we prefer "%%ci" (which expands to an "ISO-8601
# -like" string, which we must then edit to make compliant), because
# it's been around since git-1.5.3, and it's too difficult to
# discover which version we're using, or to work around using an
# older one.
date = date.strip().replace(" ", "T", 1).replace(" ", "", 1)
refnames = keywords["refnames"].strip()
if refnames.startswith("$Format"):
if verbose:
print("keywords are unexpanded, not using")
raise NotThisMethod("unexpanded keywords, not a git-archive tarball")
refs = {r.strip() for r in refnames.strip("()").split(",")}
# starting in git-1.8.3, tags are listed as "tag: foo-1.0" instead of
# just "foo-1.0". If we see a "tag: " prefix, prefer those.
TAG = "tag: "
tags = {r[len(TAG):] for r in refs if r.startswith(TAG)}
if not tags:
# Either we're using git < 1.8.3, or there really are no tags. We use
# a heuristic: assume all version tags have a digit. The old git %%d
# expansion behaves like git log --decorate=short and strips out the
# refs/heads/ and refs/tags/ prefixes that would let us distinguish
# between branches and tags. By ignoring refnames without digits, we
# filter out many common branch names like "release" and
# "stabilization", as well as "HEAD" and "master".
tags = {r for r in refs if re.search(r'\d', r)}
if verbose:
print("discarding '%%s', no digits" %% ",".join(refs - tags))
if verbose:
print("likely tags: %%s" %% ",".join(sorted(tags)))
for ref in sorted(tags):
# sorting will prefer e.g. "2.0" over "2.0rc1"
if ref.startswith(tag_prefix):
r = ref[len(tag_prefix):]
# Filter out refs that exactly match prefix or that don't start
# with a number once the prefix is stripped (mostly a concern
# when prefix is '')
if not re.match(r'\d', r):
continue
if verbose:
print("picking %%s" %% r)
return {"version": r,
"full-revisionid": keywords["full"].strip(),
"dirty": False, "error": None,
"date": date}
# no suitable tags, so version is "0+unknown", but full hex is still there
if verbose:
print("no suitable tags, using unknown + full revision id")
return {"version": "0+unknown",
"full-revisionid": keywords["full"].strip(),
"dirty": False, "error": "no suitable tags", "date": None}
@register_vcs_handler("git", "pieces_from_vcs")
def git_pieces_from_vcs(tag_prefix, root, verbose, runner=run_command):
"""Get version from 'git describe' in the root of the source tree.
This only gets called if the git-archive 'subst' keywords were *not*
expanded, and _version.py hasn't already been rewritten with a short
version string, meaning we're inside a checked out source tree.
"""
GITS = ["git"]
TAG_PREFIX_REGEX = "*"
if sys.platform == "win32":
GITS = ["git.cmd", "git.exe"]
TAG_PREFIX_REGEX = r"\*"
_, rc = runner(GITS, ["rev-parse", "--git-dir"], cwd=root,
hide_stderr=True)
if rc != 0:
if verbose:
print("Directory %%s not under git control" %% root)
raise NotThisMethod("'git rev-parse --git-dir' returned error")
# if there is a tag matching tag_prefix, this yields TAG-NUM-gHEX[-dirty]
# if there isn't one, this yields HEX[-dirty] (no NUM)
describe_out, rc = runner(GITS, ["describe", "--tags", "--dirty",
"--always", "--long",
"--match",
"%%s%%s" %% (tag_prefix, TAG_PREFIX_REGEX)],
cwd=root)
# --long was added in git-1.5.5
if describe_out is None:
raise NotThisMethod("'git describe' failed")
describe_out = describe_out.strip()
full_out, rc = runner(GITS, ["rev-parse", "HEAD"], cwd=root)
if full_out is None:
raise NotThisMethod("'git rev-parse' failed")
full_out = full_out.strip()
pieces = {}
pieces["long"] = full_out
pieces["short"] = full_out[:7] # maybe improved later
pieces["error"] = None
branch_name, rc = runner(GITS, ["rev-parse", "--abbrev-ref", "HEAD"],
cwd=root)
# --abbrev-ref was added in git-1.6.3
if rc != 0 or branch_name is None:
raise NotThisMethod("'git rev-parse --abbrev-ref' returned error")
branch_name = branch_name.strip()
if branch_name == "HEAD":
# If we aren't exactly on a branch, pick a branch which represents
# the current commit. If all else fails, we are on a branchless
# commit.
branches, rc = runner(GITS, ["branch", "--contains"], cwd=root)
# --contains was added in git-1.5.4
if rc != 0 or branches is None:
raise NotThisMethod("'git branch --contains' returned error")
branches = branches.split("\n")
# Remove the first line if we're running detached
if "(" in branches[0]:
branches.pop(0)
# Strip off the leading "* " from the list of branches.
branches = [branch[2:] for branch in branches]
if "master" in branches:
branch_name = "master"
elif not branches:
branch_name = None
else:
# Pick the first branch that is returned. Good or bad.
branch_name = branches[0]
pieces["branch"] = branch_name
# parse describe_out. It will be like TAG-NUM-gHEX[-dirty] or HEX[-dirty]
# TAG might have hyphens.
git_describe = describe_out
# look for -dirty suffix
dirty = git_describe.endswith("-dirty")
pieces["dirty"] = dirty
if dirty:
git_describe = git_describe[:git_describe.rindex("-dirty")]
# now we have TAG-NUM-gHEX or HEX
if "-" in git_describe:
# TAG-NUM-gHEX
mo = re.search(r'^(.+)-(\d+)-g([0-9a-f]+)$', git_describe)
if not mo:
# unparsable. Maybe git-describe is misbehaving?
pieces["error"] = ("unable to parse git-describe output: '%%s'"
%% describe_out)
return pieces
# tag
full_tag = mo.group(1)
if not full_tag.startswith(tag_prefix):
if verbose:
fmt = "tag '%%s' doesn't start with prefix '%%s'"
print(fmt %% (full_tag, tag_prefix))
pieces["error"] = ("tag '%%s' doesn't start with prefix '%%s'"
%% (full_tag, tag_prefix))
return pieces
pieces["closest-tag"] = full_tag[len(tag_prefix):]
# distance: number of commits since tag
pieces["distance"] = int(mo.group(2))
# commit: short hex revision ID
pieces["short"] = mo.group(3)
else:
# HEX: no tags
pieces["closest-tag"] = None
count_out, rc = runner(GITS, ["rev-list", "HEAD", "--count"], cwd=root)
pieces["distance"] = int(count_out) # total number of commits
# commit date: see ISO-8601 comment in git_versions_from_keywords()
date = runner(GITS, ["show", "-s", "--format=%%ci", "HEAD"], cwd=root)[0].strip()
# Use only the last line. Previous lines may contain GPG signature
# information.
date = date.splitlines()[-1]
pieces["date"] = date.strip().replace(" ", "T", 1).replace(" ", "", 1)
return pieces
def plus_or_dot(pieces):
"""Return a + if we don't already have one, else return a ."""
if "+" in pieces.get("closest-tag", ""):
return "."
return "+"
def render_pep440(pieces):
"""Build up version string, with post-release "local version identifier".
Our goal: TAG[+DISTANCE.gHEX[.dirty]] . Note that if you
get a tagged build and then dirty it, you'll get TAG+0.gHEX.dirty
Exceptions:
1: no tags. git_describe was just HEX. 0+untagged.DISTANCE.gHEX[.dirty]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"] or pieces["dirty"]:
rendered += plus_or_dot(pieces)
rendered += "%%d.g%%s" %% (pieces["distance"], pieces["short"])
if pieces["dirty"]:
rendered += ".dirty"
else:
# exception #1
rendered = "0+untagged.%%d.g%%s" %% (pieces["distance"],
pieces["short"])
if pieces["dirty"]:
rendered += ".dirty"
return rendered
def render_pep440_branch(pieces):
"""TAG[[.dev0]+DISTANCE.gHEX[.dirty]] .
The ".dev0" means not master branch. Note that .dev0 sorts backwards
(a feature branch will appear "older" than the master branch).
Exceptions:
1: no tags. 0[.dev0]+untagged.DISTANCE.gHEX[.dirty]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"] or pieces["dirty"]:
if pieces["branch"] != "master":
rendered += ".dev0"
rendered += plus_or_dot(pieces)
rendered += "%%d.g%%s" %% (pieces["distance"], pieces["short"])
if pieces["dirty"]:
rendered += ".dirty"
else:
# exception #1
rendered = "0"
if pieces["branch"] != "master":
rendered += ".dev0"
rendered += "+untagged.%%d.g%%s" %% (pieces["distance"],
pieces["short"])
if pieces["dirty"]:
rendered += ".dirty"
return rendered
def pep440_split_post(ver):
"""Split pep440 version string at the post-release segment.
Returns the release segments before the post-release and the
post-release version number (or -1 if no post-release segment is present).
"""
vc = str.split(ver, ".post")
return vc[0], int(vc[1] or 0) if len(vc) == 2 else None
def render_pep440_pre(pieces):
"""TAG[.postN.devDISTANCE] -- No -dirty.
Exceptions:
1: no tags. 0.post0.devDISTANCE
"""
if pieces["closest-tag"]:
if pieces["distance"]:
# update the post release segment
tag_version, post_version = pep440_split_post(pieces["closest-tag"])
rendered = tag_version
if post_version is not None:
rendered += ".post%%d.dev%%d" %% (post_version+1, pieces["distance"])
else:
rendered += ".post0.dev%%d" %% (pieces["distance"])
else:
# no commits, use the tag as the version
rendered = pieces["closest-tag"]
else:
# exception #1
rendered = "0.post0.dev%%d" %% pieces["distance"]
return rendered
def render_pep440_post(pieces):
"""TAG[.postDISTANCE[.dev0]+gHEX] .
The ".dev0" means dirty. Note that .dev0 sorts backwards
(a dirty tree will appear "older" than the corresponding clean one),
but you shouldn't be releasing software with -dirty anyways.
Exceptions:
1: no tags. 0.postDISTANCE[.dev0]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"] or pieces["dirty"]:
rendered += ".post%%d" %% pieces["distance"]
if pieces["dirty"]:
rendered += ".dev0"
rendered += plus_or_dot(pieces)
rendered += "g%%s" %% pieces["short"]
else:
# exception #1
rendered = "0.post%%d" %% pieces["distance"]
if pieces["dirty"]:
rendered += ".dev0"
rendered += "+g%%s" %% pieces["short"]
return rendered
def render_pep440_post_branch(pieces):
"""TAG[.postDISTANCE[.dev0]+gHEX[.dirty]] .
The ".dev0" means not master branch.
Exceptions:
1: no tags. 0.postDISTANCE[.dev0]+gHEX[.dirty]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"] or pieces["dirty"]:
rendered += ".post%%d" %% pieces["distance"]
if pieces["branch"] != "master":
rendered += ".dev0"
rendered += plus_or_dot(pieces)
rendered += "g%%s" %% pieces["short"]
if pieces["dirty"]:
rendered += ".dirty"
else:
# exception #1
rendered = "0.post%%d" %% pieces["distance"]
if pieces["branch"] != "master":
rendered += ".dev0"
rendered += "+g%%s" %% pieces["short"]
if pieces["dirty"]:
rendered += ".dirty"
return rendered
def render_pep440_old(pieces):
"""TAG[.postDISTANCE[.dev0]] .
The ".dev0" means dirty.
Exceptions:
1: no tags. 0.postDISTANCE[.dev0]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"] or pieces["dirty"]:
rendered += ".post%%d" %% pieces["distance"]
if pieces["dirty"]:
rendered += ".dev0"
else:
# exception #1
rendered = "0.post%%d" %% pieces["distance"]
if pieces["dirty"]:
rendered += ".dev0"
return rendered
def render_git_describe(pieces):
"""TAG[-DISTANCE-gHEX][-dirty].
Like 'git describe --tags --dirty --always'.
Exceptions:
1: no tags. HEX[-dirty] (note: no 'g' prefix)
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"]:
rendered += "-%%d-g%%s" %% (pieces["distance"], pieces["short"])
else:
# exception #1
rendered = pieces["short"]
if pieces["dirty"]:
rendered += "-dirty"
return rendered
def render_git_describe_long(pieces):
"""TAG-DISTANCE-gHEX[-dirty].
Like 'git describe --tags --dirty --always -long'.
The distance/hash is unconditional.
Exceptions:
1: no tags. HEX[-dirty] (note: no 'g' prefix)
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
rendered += "-%%d-g%%s" %% (pieces["distance"], pieces["short"])
else:
# exception #1
rendered = pieces["short"]
if pieces["dirty"]:
rendered += "-dirty"
return rendered
def render(pieces, style):
"""Render the given version pieces into the requested style."""
if pieces["error"]:
return {"version": "unknown",
"full-revisionid": pieces.get("long"),
"dirty": None,
"error": pieces["error"],
"date": None}
if not style or style == "default":
style = "pep440" # the default
if style == "pep440":
rendered = render_pep440(pieces)
elif style == "pep440-branch":
rendered = render_pep440_branch(pieces)
elif style == "pep440-pre":
rendered = render_pep440_pre(pieces)
elif style == "pep440-post":
rendered = render_pep440_post(pieces)
elif style == "pep440-post-branch":
rendered = render_pep440_post_branch(pieces)
elif style == "pep440-old":
rendered = render_pep440_old(pieces)
elif style == "git-describe":
rendered = render_git_describe(pieces)
elif style == "git-describe-long":
rendered = render_git_describe_long(pieces)
else:
raise ValueError("unknown style '%%s'" %% style)
return {"version": rendered, "full-revisionid": pieces["long"],
"dirty": pieces["dirty"], "error": None,
"date": pieces.get("date")}
def get_versions():
"""Get version information or return default if unable to do so."""
# I am in _version.py, which lives at ROOT/VERSIONFILE_SOURCE. If we have
# __file__, we can work backwards from there to the root. Some
# py2exe/bbfreeze/non-CPython implementations don't do __file__, in which
# case we can only use expanded keywords.
cfg = get_config()
verbose = cfg.verbose
try:
return git_versions_from_keywords(get_keywords(), cfg.tag_prefix,
verbose)
except NotThisMethod:
pass
try:
root = os.path.realpath(__file__)
# versionfile_source is the relative path from the top of the source
# tree (where the .git directory might live) to this file. Invert
# this to find the root from __file__.
for _ in cfg.versionfile_source.split('/'):
root = os.path.dirname(root)
except NameError:
return {"version": "0+unknown", "full-revisionid": None,
"dirty": None,
"error": "unable to find root of source tree",
"date": None}
try:
pieces = git_pieces_from_vcs(cfg.tag_prefix, root, verbose)
return render(pieces, cfg.style)
except NotThisMethod:
pass
try:
if cfg.parentdir_prefix:
return versions_from_parentdir(cfg.parentdir_prefix, root, verbose)
except NotThisMethod:
pass
return {"version": "0+unknown", "full-revisionid": None,
"dirty": None,
"error": "unable to compute version", "date": None}
'''
@register_vcs_handler("git", "get_keywords")
def git_get_keywords(versionfile_abs):
"""Extract version information from the given file."""
# the code embedded in _version.py can just fetch the value of these
# keywords. When used from setup.py, we don't want to import _version.py,
# so we do it with a regexp instead. This function is not used from
# _version.py.
keywords = {}
try:
with open(versionfile_abs, "r") as fobj:
for line in fobj:
if line.strip().startswith("git_refnames ="):
mo = re.search(r'=\s*"(.*)"', line)
if mo:
keywords["refnames"] = mo.group(1)
if line.strip().startswith("git_full ="):
mo = re.search(r'=\s*"(.*)"', line)
if mo:
keywords["full"] = mo.group(1)
if line.strip().startswith("git_date ="):
mo = re.search(r'=\s*"(.*)"', line)
if mo:
keywords["date"] = mo.group(1)
except OSError:
pass
return keywords
@register_vcs_handler("git", "keywords")
def git_versions_from_keywords(keywords, tag_prefix, verbose):
"""Get version information from git keywords."""
if "refnames" not in keywords:
raise NotThisMethod("Short version file found")
date = keywords.get("date")
if date is not None:
# Use only the last line. Previous lines may contain GPG signature
# information.
date = date.splitlines()[-1]
# git-2.2.0 added "%cI", which expands to an ISO-8601 -compliant
# datestamp. However we prefer "%ci" (which expands to an "ISO-8601
# -like" string, which we must then edit to make compliant), because
# it's been around since git-1.5.3, and it's too difficult to
# discover which version we're using, or to work around using an
# older one.
date = date.strip().replace(" ", "T", 1).replace(" ", "", 1)
refnames = keywords["refnames"].strip()
if refnames.startswith("$Format"):
if verbose:
print("keywords are unexpanded, not using")
raise NotThisMethod("unexpanded keywords, not a git-archive tarball")
refs = {r.strip() for r in refnames.strip("()").split(",")}
# starting in git-1.8.3, tags are listed as "tag: foo-1.0" instead of
# just "foo-1.0". If we see a "tag: " prefix, prefer those.
TAG = "tag: "
tags = {r[len(TAG):] for r in refs if r.startswith(TAG)}
if not tags:
# Either we're using git < 1.8.3, or there really are no tags. We use
# a heuristic: assume all version tags have a digit. The old git %d
# expansion behaves like git log --decorate=short and strips out the
# refs/heads/ and refs/tags/ prefixes that would let us distinguish
# between branches and tags. By ignoring refnames without digits, we
# filter out many common branch names like "release" and
# "stabilization", as well as "HEAD" and "master".
tags = {r for r in refs if re.search(r'\d', r)}
if verbose:
print("discarding '%s', no digits" % ",".join(refs - tags))
if verbose:
print("likely tags: %s" % ",".join(sorted(tags)))
for ref in sorted(tags):
# sorting will prefer e.g. "2.0" over "2.0rc1"
if ref.startswith(tag_prefix):
r = ref[len(tag_prefix):]
# Filter out refs that exactly match prefix or that don't start
# with a number once the prefix is stripped (mostly a concern
# when prefix is '')
if not re.match(r'\d', r):
continue
if verbose:
print("picking %s" % r)
return {"version": r,
"full-revisionid": keywords["full"].strip(),
"dirty": False, "error": None,
"date": date}
# no suitable tags, so version is "0+unknown", but full hex is still there
if verbose:
print("no suitable tags, using unknown + full revision id")
return {"version": "0+unknown",
"full-revisionid": keywords["full"].strip(),
"dirty": False, "error": "no suitable tags", "date": None}
@register_vcs_handler("git", "pieces_from_vcs")
def git_pieces_from_vcs(tag_prefix, root, verbose, runner=run_command):
"""Get version from 'git describe' in the root of the source tree.
This only gets called if the git-archive 'subst' keywords were *not*
expanded, and _version.py hasn't already been rewritten with a short
version string, meaning we're inside a checked out source tree.
"""
GITS = ["git"]
TAG_PREFIX_REGEX = "*"
if sys.platform == "win32":
GITS = ["git.cmd", "git.exe"]
TAG_PREFIX_REGEX = r"\*"
_, rc = runner(GITS, ["rev-parse", "--git-dir"], cwd=root,
hide_stderr=True)
if rc != 0:
if verbose:
print("Directory %s not under git control" % root)
raise NotThisMethod("'git rev-parse --git-dir' returned error")
# if there is a tag matching tag_prefix, this yields TAG-NUM-gHEX[-dirty]
# if there isn't one, this yields HEX[-dirty] (no NUM)
describe_out, rc = runner(GITS, ["describe", "--tags", "--dirty",
"--always", "--long",
"--match",
"%s%s" % (tag_prefix, TAG_PREFIX_REGEX)],
cwd=root)
# --long was added in git-1.5.5
if describe_out is None:
raise NotThisMethod("'git describe' failed")
describe_out = describe_out.strip()
full_out, rc = runner(GITS, ["rev-parse", "HEAD"], cwd=root)
if full_out is None:
raise NotThisMethod("'git rev-parse' failed")
full_out = full_out.strip()
pieces = {}
pieces["long"] = full_out
pieces["short"] = full_out[:7] # maybe improved later
pieces["error"] = None
branch_name, rc = runner(GITS, ["rev-parse", "--abbrev-ref", "HEAD"],
cwd=root)
# --abbrev-ref was added in git-1.6.3
if rc != 0 or branch_name is None:
raise NotThisMethod("'git rev-parse --abbrev-ref' returned error")
branch_name = branch_name.strip()
if branch_name == "HEAD":
# If we aren't exactly on a branch, pick a branch which represents
# the current commit. If all else fails, we are on a branchless
# commit.
branches, rc = runner(GITS, ["branch", "--contains"], cwd=root)
# --contains was added in git-1.5.4
if rc != 0 or branches is None:
raise NotThisMethod("'git branch --contains' returned error")
branches = branches.split("\n")
# Remove the first line if we're running detached
if "(" in branches[0]:
branches.pop(0)
# Strip off the leading "* " from the list of branches.
branches = [branch[2:] for branch in branches]
if "master" in branches:
branch_name = "master"
elif not branches:
branch_name = None
else:
# Pick the first branch that is returned. Good or bad.
branch_name = branches[0]
pieces["branch"] = branch_name
# parse describe_out. It will be like TAG-NUM-gHEX[-dirty] or HEX[-dirty]
# TAG might have hyphens.
git_describe = describe_out
# look for -dirty suffix
dirty = git_describe.endswith("-dirty")
pieces["dirty"] = dirty
if dirty:
git_describe = git_describe[:git_describe.rindex("-dirty")]
# now we have TAG-NUM-gHEX or HEX
if "-" in git_describe:
# TAG-NUM-gHEX
mo = re.search(r'^(.+)-(\d+)-g([0-9a-f]+)$', git_describe)
if not mo:
# unparsable. Maybe git-describe is misbehaving?
pieces["error"] = ("unable to parse git-describe output: '%s'"
% describe_out)
return pieces
# tag
full_tag = mo.group(1)
if not full_tag.startswith(tag_prefix):
if verbose:
fmt = "tag '%s' doesn't start with prefix '%s'"
print(fmt % (full_tag, tag_prefix))
pieces["error"] = ("tag '%s' doesn't start with prefix '%s'"
% (full_tag, tag_prefix))
return pieces
pieces["closest-tag"] = full_tag[len(tag_prefix):]
# distance: number of commits since tag
pieces["distance"] = int(mo.group(2))
# commit: short hex revision ID
pieces["short"] = mo.group(3)
else:
# HEX: no tags
pieces["closest-tag"] = None
count_out, rc = runner(GITS, ["rev-list", "HEAD", "--count"], cwd=root)
pieces["distance"] = int(count_out) # total number of commits
# commit date: see ISO-8601 comment in git_versions_from_keywords()
date = runner(GITS, ["show", "-s", "--format=%ci", "HEAD"], cwd=root)[0].strip()
# Use only the last line. Previous lines may contain GPG signature
# information.
date = date.splitlines()[-1]
pieces["date"] = date.strip().replace(" ", "T", 1).replace(" ", "", 1)
return pieces
def do_vcs_install(manifest_in, versionfile_source, ipy):
"""Git-specific installation logic for Versioneer.
For Git, this means creating/changing .gitattributes to mark _version.py
for export-subst keyword substitution.
"""
GITS = ["git"]
if sys.platform == "win32":
GITS = ["git.cmd", "git.exe"]
files = [manifest_in, versionfile_source]
if ipy:
files.append(ipy)
try:
my_path = __file__
if my_path.endswith(".pyc") or my_path.endswith(".pyo"):
my_path = os.path.splitext(my_path)[0] + ".py"
versioneer_file = os.path.relpath(my_path)
except NameError:
versioneer_file = "versioneer.py"
files.append(versioneer_file)
present = False
try:
with open(".gitattributes", "r") as fobj:
for line in fobj:
if line.strip().startswith(versionfile_source):
if "export-subst" in line.strip().split()[1:]:
present = True
break
except OSError:
pass
if not present:
with open(".gitattributes", "a+") as fobj:
fobj.write(f"{versionfile_source} export-subst\n")
files.append(".gitattributes")
run_command(GITS, ["add", "--"] + files)
def versions_from_parentdir(parentdir_prefix, root, verbose):
"""Try to determine the version from the parent directory name.
Source tarballs conventionally unpack into a directory that includes both
the project name and a version string. We will also support searching up
two directory levels for an appropriately named parent directory
"""
rootdirs = []
for _ in range(3):
dirname = os.path.basename(root)
if dirname.startswith(parentdir_prefix):
return {"version": dirname[len(parentdir_prefix):],
"full-revisionid": None,
"dirty": False, "error": None, "date": None}
rootdirs.append(root)
root = os.path.dirname(root) # up a level
if verbose:
print("Tried directories %s but none started with prefix %s" %
(str(rootdirs), parentdir_prefix))
raise NotThisMethod("rootdir doesn't start with parentdir_prefix")
SHORT_VERSION_PY = """
# This file was generated by 'versioneer.py' (0.21) from
# revision-control system data, or from the parent directory name of an
# unpacked source archive. Distribution tarballs contain a pre-generated copy
# of this file.
import json
version_json = '''
%s
''' # END VERSION_JSON
def get_versions():
return json.loads(version_json)
"""
def versions_from_file(filename):
"""Try to determine the version from _version.py if present."""
try:
with open(filename) as f:
contents = f.read()
except OSError:
raise NotThisMethod("unable to read _version.py")
mo = re.search(r"version_json = '''\n(.*)''' # END VERSION_JSON",
contents, re.M | re.S)
if not mo:
mo = re.search(r"version_json = '''\r\n(.*)''' # END VERSION_JSON",
contents, re.M | re.S)
if not mo:
raise NotThisMethod("no version_json in _version.py")
return json.loads(mo.group(1))
def write_to_version_file(filename, versions):
"""Write the given version number to the given _version.py file."""
os.unlink(filename)
contents = json.dumps(versions, sort_keys=True,
indent=1, separators=(",", ": "))
with open(filename, "w") as f:
f.write(SHORT_VERSION_PY % contents)
print("set %s to '%s'" % (filename, versions["version"]))
def plus_or_dot(pieces):
"""Return a + if we don't already have one, else return a ."""
if "+" in pieces.get("closest-tag", ""):
return "."
return "+"
def render_pep440(pieces):
"""Build up version string, with post-release "local version identifier".
Our goal: TAG[+DISTANCE.gHEX[.dirty]] . Note that if you
get a tagged build and then dirty it, you'll get TAG+0.gHEX.dirty
Exceptions:
1: no tags. git_describe was just HEX. 0+untagged.DISTANCE.gHEX[.dirty]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"] or pieces["dirty"]:
rendered += plus_or_dot(pieces)
rendered += "%d.g%s" % (pieces["distance"], pieces["short"])
if pieces["dirty"]:
rendered += ".dirty"
else:
# exception #1
rendered = "0+untagged.%d.g%s" % (pieces["distance"],
pieces["short"])
if pieces["dirty"]:
rendered += ".dirty"
return rendered
def render_pep440_branch(pieces):
"""TAG[[.dev0]+DISTANCE.gHEX[.dirty]] .
The ".dev0" means not master branch. Note that .dev0 sorts backwards
(a feature branch will appear "older" than the master branch).
Exceptions:
1: no tags. 0[.dev0]+untagged.DISTANCE.gHEX[.dirty]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"] or pieces["dirty"]:
if pieces["branch"] != "master":
rendered += ".dev0"
rendered += plus_or_dot(pieces)
rendered += "%d.g%s" % (pieces["distance"], pieces["short"])
if pieces["dirty"]:
rendered += ".dirty"
else:
# exception #1
rendered = "0"
if pieces["branch"] != "master":
rendered += ".dev0"
rendered += "+untagged.%d.g%s" % (pieces["distance"],
pieces["short"])
if pieces["dirty"]:
rendered += ".dirty"
return rendered
def pep440_split_post(ver):
"""Split pep440 version string at the post-release segment.
Returns the release segments before the post-release and the
post-release version number (or -1 if no post-release segment is present).
"""
vc = str.split(ver, ".post")
return vc[0], int(vc[1] or 0) if len(vc) == 2 else None
def render_pep440_pre(pieces):
"""TAG[.postN.devDISTANCE] -- No -dirty.
Exceptions:
1: no tags. 0.post0.devDISTANCE
"""
if pieces["closest-tag"]:
if pieces["distance"]:
# update the post release segment
tag_version, post_version = pep440_split_post(pieces["closest-tag"])
rendered = tag_version
if post_version is not None:
rendered += ".post%d.dev%d" % (post_version+1, pieces["distance"])
else:
rendered += ".post0.dev%d" % (pieces["distance"])
else:
# no commits, use the tag as the version
rendered = pieces["closest-tag"]
else:
# exception #1
rendered = "0.post0.dev%d" % pieces["distance"]
return rendered
def render_pep440_post(pieces):
"""TAG[.postDISTANCE[.dev0]+gHEX] .
The ".dev0" means dirty. Note that .dev0 sorts backwards
(a dirty tree will appear "older" than the corresponding clean one),
but you shouldn't be releasing software with -dirty anyways.
Exceptions:
1: no tags. 0.postDISTANCE[.dev0]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"] or pieces["dirty"]:
rendered += ".post%d" % pieces["distance"]
if pieces["dirty"]:
rendered += ".dev0"
rendered += plus_or_dot(pieces)
rendered += "g%s" % pieces["short"]
else:
# exception #1
rendered = "0.post%d" % pieces["distance"]
if pieces["dirty"]:
rendered += ".dev0"
rendered += "+g%s" % pieces["short"]
return rendered
def render_pep440_post_branch(pieces):
"""TAG[.postDISTANCE[.dev0]+gHEX[.dirty]] .
The ".dev0" means not master branch.
Exceptions:
1: no tags. 0.postDISTANCE[.dev0]+gHEX[.dirty]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"] or pieces["dirty"]:
rendered += ".post%d" % pieces["distance"]
if pieces["branch"] != "master":
rendered += ".dev0"
rendered += plus_or_dot(pieces)
rendered += "g%s" % pieces["short"]
if pieces["dirty"]:
rendered += ".dirty"
else:
# exception #1
rendered = "0.post%d" % pieces["distance"]
if pieces["branch"] != "master":
rendered += ".dev0"
rendered += "+g%s" % pieces["short"]
if pieces["dirty"]:
rendered += ".dirty"
return rendered
def render_pep440_old(pieces):
"""TAG[.postDISTANCE[.dev0]] .
The ".dev0" means dirty.
Exceptions:
1: no tags. 0.postDISTANCE[.dev0]
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"] or pieces["dirty"]:
rendered += ".post%d" % pieces["distance"]
if pieces["dirty"]:
rendered += ".dev0"
else:
# exception #1
rendered = "0.post%d" % pieces["distance"]
if pieces["dirty"]:
rendered += ".dev0"
return rendered
def render_git_describe(pieces):
"""TAG[-DISTANCE-gHEX][-dirty].
Like 'git describe --tags --dirty --always'.
Exceptions:
1: no tags. HEX[-dirty] (note: no 'g' prefix)
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
if pieces["distance"]:
rendered += "-%d-g%s" % (pieces["distance"], pieces["short"])
else:
# exception #1
rendered = pieces["short"]
if pieces["dirty"]:
rendered += "-dirty"
return rendered
def render_git_describe_long(pieces):
"""TAG-DISTANCE-gHEX[-dirty].
Like 'git describe --tags --dirty --always -long'.
The distance/hash is unconditional.
Exceptions:
1: no tags. HEX[-dirty] (note: no 'g' prefix)
"""
if pieces["closest-tag"]:
rendered = pieces["closest-tag"]
rendered += "-%d-g%s" % (pieces["distance"], pieces["short"])
else:
# exception #1
rendered = pieces["short"]
if pieces["dirty"]:
rendered += "-dirty"
return rendered
def render(pieces, style):
"""Render the given version pieces into the requested style."""
if pieces["error"]:
return {"version": "unknown",
"full-revisionid": pieces.get("long"),
"dirty": None,
"error": pieces["error"],
"date": None}
if not style or style == "default":
style = "pep440" # the default
if style == "pep440":
rendered = render_pep440(pieces)
elif style == "pep440-branch":
rendered = render_pep440_branch(pieces)
elif style == "pep440-pre":
rendered = render_pep440_pre(pieces)
elif style == "pep440-post":
rendered = render_pep440_post(pieces)
elif style == "pep440-post-branch":
rendered = render_pep440_post_branch(pieces)
elif style == "pep440-old":
rendered = render_pep440_old(pieces)
elif style == "git-describe":
rendered = render_git_describe(pieces)
elif style == "git-describe-long":
rendered = render_git_describe_long(pieces)
else:
raise ValueError("unknown style '%s'" % style)
return {"version": rendered, "full-revisionid": pieces["long"],
"dirty": pieces["dirty"], "error": None,
"date": pieces.get("date")}
class VersioneerBadRootError(Exception):
"""The project root directory is unknown or missing key files."""
def get_versions(verbose=False):
"""Get the project version from whatever source is available.
Returns dict with two keys: 'version' and 'full'.
"""
if "versioneer" in sys.modules:
# see the discussion in cmdclass.py:get_cmdclass()
del sys.modules["versioneer"]
root = get_root()
cfg = get_config_from_root(root)
assert cfg.VCS is not None, "please set [versioneer]VCS= in setup.cfg"
handlers = HANDLERS.get(cfg.VCS)
assert handlers, "unrecognized VCS '%s'" % cfg.VCS
verbose = verbose or cfg.verbose
assert cfg.versionfile_source is not None, \
"please set versioneer.versionfile_source"
assert cfg.tag_prefix is not None, "please set versioneer.tag_prefix"
versionfile_abs = os.path.join(root, cfg.versionfile_source)
# extract version from first of: _version.py, VCS command (e.g. 'git
# describe'), parentdir. This is meant to work for developers using a
# source checkout, for users of a tarball created by 'setup.py sdist',
# and for users of a tarball/zipball created by 'git archive' or github's
# download-from-tag feature or the equivalent in other VCSes.
get_keywords_f = handlers.get("get_keywords")
from_keywords_f = handlers.get("keywords")
if get_keywords_f and from_keywords_f:
try:
keywords = get_keywords_f(versionfile_abs)
ver = from_keywords_f(keywords, cfg.tag_prefix, verbose)
if verbose:
print("got version from expanded keyword %s" % ver)
return ver
except NotThisMethod:
pass
try:
ver = versions_from_file(versionfile_abs)
if verbose:
print("got version from file %s %s" % (versionfile_abs, ver))
return ver
except NotThisMethod:
pass
from_vcs_f = handlers.get("pieces_from_vcs")
if from_vcs_f:
try:
pieces = from_vcs_f(cfg.tag_prefix, root, verbose)
ver = render(pieces, cfg.style)
if verbose:
print("got version from VCS %s" % ver)
return ver
except NotThisMethod:
pass
try:
if cfg.parentdir_prefix:
ver = versions_from_parentdir(cfg.parentdir_prefix, root, verbose)
if verbose:
print("got version from parentdir %s" % ver)
return ver
except NotThisMethod:
pass
if verbose:
print("unable to compute version")
return {"version": "0+unknown", "full-revisionid": None,
"dirty": None, "error": "unable to compute version",
"date": None}
def get_version():
"""Get the short version string for this project."""
return get_versions()["version"]
def get_cmdclass(cmdclass=None):
"""Get the custom setuptools/distutils subclasses used by Versioneer.
If the package uses a different cmdclass (e.g. one from numpy), it
should be provide as an argument.
"""
if "versioneer" in sys.modules:
del sys.modules["versioneer"]
# this fixes the "python setup.py develop" case (also 'install' and
# 'easy_install .'), in which subdependencies of the main project are
# built (using setup.py bdist_egg) in the same python process. Assume
# a main project A and a dependency B, which use different versions
# of Versioneer. A's setup.py imports A's Versioneer, leaving it in
# sys.modules by the time B's setup.py is executed, causing B to run
# with the wrong versioneer. Setuptools wraps the sub-dep builds in a
# sandbox that restores sys.modules to it's pre-build state, so the
# parent is protected against the child's "import versioneer". By
# removing ourselves from sys.modules here, before the child build
# happens, we protect the child from the parent's versioneer too.
# Also see https://github.com/python-versioneer/python-versioneer/issues/52
cmds = {} if cmdclass is None else cmdclass.copy()
# we add "version" to both distutils and setuptools
from distutils.core import Command
class cmd_version(Command):
description = "report generated version string"
user_options = []
boolean_options = []
def initialize_options(self):
pass
def finalize_options(self):
pass
def run(self):
vers = get_versions(verbose=True)
print("Version: %s" % vers["version"])
print(" full-revisionid: %s" % vers.get("full-revisionid"))
print(" dirty: %s" % vers.get("dirty"))
print(" date: %s" % vers.get("date"))
if vers["error"]:
print(" error: %s" % vers["error"])
cmds["version"] = cmd_version
# we override "build_py" in both distutils and setuptools
#
# most invocation pathways end up running build_py:
# distutils/build -> build_py
# distutils/install -> distutils/build ->..
# setuptools/bdist_wheel -> distutils/install ->..
# setuptools/bdist_egg -> distutils/install_lib -> build_py
# setuptools/install -> bdist_egg ->..
# setuptools/develop -> ?
# pip install:
# copies source tree to a tempdir before running egg_info/etc
# if .git isn't copied too, 'git describe' will fail
# then does setup.py bdist_wheel, or sometimes setup.py install
# setup.py egg_info -> ?
# we override different "build_py" commands for both environments
if 'build_py' in cmds:
_build_py = cmds['build_py']
elif "setuptools" in sys.modules:
from setuptools.command.build_py import build_py as _build_py
else:
from distutils.command.build_py import build_py as _build_py
class cmd_build_py(_build_py):
def run(self):
root = get_root()
cfg = get_config_from_root(root)
versions = get_versions()
_build_py.run(self)
# now locate _version.py in the new build/ directory and replace
# it with an updated value
if cfg.versionfile_build:
target_versionfile = os.path.join(self.build_lib,
cfg.versionfile_build)
print("UPDATING %s" % target_versionfile)
write_to_version_file(target_versionfile, versions)
cmds["build_py"] = cmd_build_py
if 'build_ext' in cmds:
_build_ext = cmds['build_ext']
elif "setuptools" in sys.modules:
from setuptools.command.build_ext import build_ext as _build_ext
else:
from distutils.command.build_ext import build_ext as _build_ext
class cmd_build_ext(_build_ext):
def run(self):
root = get_root()
cfg = get_config_from_root(root)
versions = get_versions()
_build_ext.run(self)
if self.inplace:
# build_ext --inplace will only build extensions in
# build/lib<..> dir with no _version.py to write to.
# As in place builds will already have a _version.py
# in the module dir, we do not need to write one.
return
# now locate _version.py in the new build/ directory and replace
# it with an updated value
target_versionfile = os.path.join(self.build_lib,
cfg.versionfile_build)
print("UPDATING %s" % target_versionfile)
write_to_version_file(target_versionfile, versions)
cmds["build_ext"] = cmd_build_ext
if "cx_Freeze" in sys.modules: # cx_freeze enabled?
from cx_Freeze.dist import build_exe as _build_exe
# nczeczulin reports that py2exe won't like the pep440-style string
# as FILEVERSION, but it can be used for PRODUCTVERSION, e.g.
# setup(console=[{
# "version": versioneer.get_version().split("+", 1)[0], # FILEVERSION
# "product_version": versioneer.get_version(),
# ...
class cmd_build_exe(_build_exe):
def run(self):
root = get_root()
cfg = get_config_from_root(root)
versions = get_versions()
target_versionfile = cfg.versionfile_source
print("UPDATING %s" % target_versionfile)
write_to_version_file(target_versionfile, versions)
_build_exe.run(self)
os.unlink(target_versionfile)
with open(cfg.versionfile_source, "w") as f:
LONG = LONG_VERSION_PY[cfg.VCS]
f.write(LONG %
{"DOLLAR": "$",
"STYLE": cfg.style,
"TAG_PREFIX": cfg.tag_prefix,
"PARENTDIR_PREFIX": cfg.parentdir_prefix,
"VERSIONFILE_SOURCE": cfg.versionfile_source,
})
cmds["build_exe"] = cmd_build_exe
del cmds["build_py"]
if 'py2exe' in sys.modules: # py2exe enabled?
from py2exe.distutils_buildexe import py2exe as _py2exe
class cmd_py2exe(_py2exe):
def run(self):
root = get_root()
cfg = get_config_from_root(root)
versions = get_versions()
target_versionfile = cfg.versionfile_source
print("UPDATING %s" % target_versionfile)
write_to_version_file(target_versionfile, versions)
_py2exe.run(self)
os.unlink(target_versionfile)
with open(cfg.versionfile_source, "w") as f:
LONG = LONG_VERSION_PY[cfg.VCS]
f.write(LONG %
{"DOLLAR": "$",
"STYLE": cfg.style,
"TAG_PREFIX": cfg.tag_prefix,
"PARENTDIR_PREFIX": cfg.parentdir_prefix,
"VERSIONFILE_SOURCE": cfg.versionfile_source,
})
cmds["py2exe"] = cmd_py2exe
# we override different "sdist" commands for both environments
if 'sdist' in cmds:
_sdist = cmds['sdist']
elif "setuptools" in sys.modules:
from setuptools.command.sdist import sdist as _sdist
else:
from distutils.command.sdist import sdist as _sdist
class cmd_sdist(_sdist):
def run(self):
versions = get_versions()
self._versioneer_generated_versions = versions
# unless we update this, the command will keep using the old
# version
self.distribution.metadata.version = versions["version"]
return _sdist.run(self)
def make_release_tree(self, base_dir, files):
root = get_root()
cfg = get_config_from_root(root)
_sdist.make_release_tree(self, base_dir, files)
# now locate _version.py in the new base_dir directory
# (remembering that it may be a hardlink) and replace it with an
# updated value
target_versionfile = os.path.join(base_dir, cfg.versionfile_source)
print("UPDATING %s" % target_versionfile)
write_to_version_file(target_versionfile,
self._versioneer_generated_versions)
cmds["sdist"] = cmd_sdist
return cmds
CONFIG_ERROR = """
setup.cfg is missing the necessary Versioneer configuration. You need
a section like:
[versioneer]
VCS = git
style = pep440
versionfile_source = src/myproject/_version.py
versionfile_build = myproject/_version.py
tag_prefix =
parentdir_prefix = myproject-
You will also need to edit your setup.py to use the results:
import versioneer
setup(version=versioneer.get_version(),
cmdclass=versioneer.get_cmdclass(), ...)
Please read the docstring in ./versioneer.py for configuration instructions,
edit setup.cfg, and re-run the installer or 'python versioneer.py setup'.
"""
SAMPLE_CONFIG = """
# See the docstring in versioneer.py for instructions. Note that you must
# re-run 'versioneer.py setup' after changing this section, and commit the
# resulting files.
[versioneer]
#VCS = git
#style = pep440
#versionfile_source =
#versionfile_build =
#tag_prefix =
#parentdir_prefix =
"""
OLD_SNIPPET = """
from ._version import get_versions
__version__ = get_versions()['version']
del get_versions
"""
INIT_PY_SNIPPET = """
from . import {0}
__version__ = {0}.get_versions()['version']
"""
def do_setup():
"""Do main VCS-independent setup function for installing Versioneer."""
root = get_root()
try:
cfg = get_config_from_root(root)
except (OSError, configparser.NoSectionError,
configparser.NoOptionError) as e:
if isinstance(e, (OSError, configparser.NoSectionError)):
print("Adding sample versioneer config to setup.cfg",
file=sys.stderr)
with open(os.path.join(root, "setup.cfg"), "a") as f:
f.write(SAMPLE_CONFIG)
print(CONFIG_ERROR, file=sys.stderr)
return 1
print(" creating %s" % cfg.versionfile_source)
with open(cfg.versionfile_source, "w") as f:
LONG = LONG_VERSION_PY[cfg.VCS]
f.write(LONG % {"DOLLAR": "$",
"STYLE": cfg.style,
"TAG_PREFIX": cfg.tag_prefix,
"PARENTDIR_PREFIX": cfg.parentdir_prefix,
"VERSIONFILE_SOURCE": cfg.versionfile_source,
})
ipy = os.path.join(os.path.dirname(cfg.versionfile_source),
"__init__.py")
if os.path.exists(ipy):
try:
with open(ipy, "r") as f:
old = f.read()
except OSError:
old = ""
module = os.path.splitext(os.path.basename(cfg.versionfile_source))[0]
snippet = INIT_PY_SNIPPET.format(module)
if OLD_SNIPPET in old:
print(" replacing boilerplate in %s" % ipy)
with open(ipy, "w") as f:
f.write(old.replace(OLD_SNIPPET, snippet))
elif snippet not in old:
print(" appending to %s" % ipy)
with open(ipy, "a") as f:
f.write(snippet)
else:
print(" %s unmodified" % ipy)
else:
print(" %s doesn't exist, ok" % ipy)
ipy = None
# Make sure both the top-level "versioneer.py" and versionfile_source
# (PKG/_version.py, used by runtime code) are in MANIFEST.in, so
# they'll be copied into source distributions. Pip won't be able to
# install the package without this.
manifest_in = os.path.join(root, "MANIFEST.in")
simple_includes = set()
try:
with open(manifest_in, "r") as f:
for line in f:
if line.startswith("include "):
for include in line.split()[1:]:
simple_includes.add(include)
except OSError:
pass
# That doesn't cover everything MANIFEST.in can do
# (http://docs.python.org/2/distutils/sourcedist.html#commands), so
# it might give some false negatives. Appending redundant 'include'
# lines is safe, though.
if "versioneer.py" not in simple_includes:
print(" appending 'versioneer.py' to MANIFEST.in")
with open(manifest_in, "a") as f:
f.write("include versioneer.py\n")
else:
print(" 'versioneer.py' already in MANIFEST.in")
if cfg.versionfile_source not in simple_includes:
print(" appending versionfile_source ('%s') to MANIFEST.in" %
cfg.versionfile_source)
with open(manifest_in, "a") as f:
f.write("include %s\n" % cfg.versionfile_source)
else:
print(" versionfile_source already in MANIFEST.in")
# Make VCS-specific changes. For git, this means creating/changing
# .gitattributes to mark _version.py for export-subst keyword
# substitution.
do_vcs_install(manifest_in, cfg.versionfile_source, ipy)
return 0
def scan_setup_py():
"""Validate the contents of setup.py against Versioneer's expectations."""
found = set()
setters = False
errors = 0
with open("setup.py", "r") as f:
for line in f.readlines():
if "import versioneer" in line:
found.add("import")
if "versioneer.get_cmdclass()" in line:
found.add("cmdclass")
if "versioneer.get_version()" in line:
found.add("get_version")
if "versioneer.VCS" in line:
setters = True
if "versioneer.versionfile_source" in line:
setters = True
if len(found) != 3:
print("")
print("Your setup.py appears to be missing some important items")
print("(but I might be wrong). Please make sure it has something")
print("roughly like the following:")
print("")
print(" import versioneer")
print(" setup( version=versioneer.get_version(),")
print(" cmdclass=versioneer.get_cmdclass(), ...)")
print("")
errors += 1
if setters:
print("You should remove lines like 'versioneer.VCS = ' and")
print("'versioneer.versionfile_source = ' . This configuration")
print("now lives in setup.cfg, and should be removed from setup.py")
print("")
errors += 1
return errors
if __name__ == "__main__":
cmd = sys.argv[1]
if cmd == "setup":
errors = do_setup()
errors += scan_setup_py()
if errors:
sys.exit(1) | AMDock | /AMDock-1.6.2.tar.gz/AMDock-1.6.2/versioneer.py | versioneer.py |
AMFM_decompy
=============
version 1.0.11
This python package provides the tools necessary for decomposing the voiced part
of a speech signal into its modulated components, aka AM-FM decomposition. This
designation is used due the fact that, in this method, the signal is modeled as
a sum of amplitude- and frequency-modulated components.
The goal is to overcome the drawbacks from Fourier-alike techniques, e.g. SFFT,
wavelets, etc, which are limited in the time-frequency analysis by the so-called
Heisenberg-Gabor inequality.
The algorithms here implemented are the QHM (Quasi-Harmonic Model), and its
upgrades, aQHM (adaptive Quasi-Harmonic Model) and eaQHM (extended adaptive
Quasi-Harmonic Model). Their formulation can be found at references [2-4].
Since that the tools mentioned above require a fundamental frequency reference,
the package also includes the pitch tracker YAAPT (Yet Another Algorithm for
Pitch Tracking) [1], which is extremely robust for both high quality and
telephone speech.
The study of AM-FM decomposition algorithms was the theme from my Master Thesis.
The original YAAPT program in MATLAB is provided for free by its authors, while
the QHM algorithms I implemented by myself also in MATLAB. I'm porting them now
to python because:
* the python language is easier to share, read and understand, making it a
better way to distribute the codes;
* is more resourceful than MATLAB (has different data structures, scripting
options, etc), which will be useful for me in future studies;
* the computational performance from its numeric and scientific packages (numpy
and scipy) is equivalent to MATLAB;
* python is free-to-use, while MATLAB is a proprietary software;
Evaluations and future expansions
=============
As for the algorithms computational performance, I optimized the YAAPT code, so
my pyhton version runs now about twice as fast as the original MATLAB one.
However, the QHM algorithms still run as fast as their counterparts in MATLAB.
That's because the main bottleneck of both versions are the matrix dot and
least-squares operations. Since numpy and MATLAB are already optimized to perform
these tasks using internal Fortran functions, as far as I investigated there's
no way to speed them up using Cython, for example. Nevertheless, numba and CUDA
seem to be promising tools to speed the code, so adding support to them is one
of my priorities for future releases.
In [1] the YAAPT is compared with well-known pitch trackers like the YIN and
the RAPT, and presents the best results. In fact, so far I've been using it,
the algorithm has been proved to be indeed very robust. It must be emphasized
that I merely translated the code, so I only have an average knowledge about
its theoretical formulation. For deep questions concerning it, I would advise
to contact the original authors.
The QHM-like algorithms present some stability problems concerning small
magnitude modulated components, which are already documented at [2,3]. In my
python code I implemented a workaround to this problem, but it is still a
sub-optimal solution.
Actually, I dedicated a chapter in my Master Thesis to a deeper study about
this problem and came up with a better solution. Unfortunately, due stupid
bureaucratic issues, I don't know if and when my work will be defended and
published (to be short, the deadline was expired because me and my advisor
needed more time to correct and improve the thesis text. Then we required a
prorrogation, but the lecturers board declined it. So, basically, I was expelled
from the post-gradute program with a finished and working thesis). Anyway, I'm
still trying to figure out do now with my work and as soon as find a solution,
I'll add my own contributions to this package.
IMPORTANT - Considerations about version 1.0.7 and later versions
=============
In the latest release of the original YAAPT MATLAB source code (YAAPT v4.0)
the default values from the following parameters have been altered:
* `frame_length` parameter changed from 25 ms to 35 ms;
* `nccf_thresh1` parameter changed from 0.25 to 0.3;
Moreover, a new parameter called `frame_lengtht` was added (please pay atention
to the extra "t" at the end), which name is quite similar to `frame_length`.
In order to avoid confusion between them, an alternative (and preferred) alias
for `frame_lengtht` called `tda_frame_length` was used in pYAAPT.py. Nevertheless,
both inputs (`frame_lengtht` and `tda_frame_length`) are accepted.
Due these modifications, if you were running AMFM_decompy 1.0.6 or earlier
versions with their default settings, you may obtain slightly different results
from the ones obtained by running AMFM_decompy 1.0.7 and later versions. with
the new default parameters.
Therefore, if you really need to obtain exactly the same results from previous
versions, you must provide the old parameter values to the yaapt function. For
example, a 1.0.6 or earlier code like
`pitch = pYAAPT.yaapt(signal)`
should be rewritten in the 1.0.7 and later versions as
`pitch = pYAAPT.yaapt(signal, **{'frame_length': 25.0, 'nccf_thresh1': 0.25, 'tda_frame_length': 25.0})`
Installation
=============
The pypi page https://pypi.python.org/pypi/AMFM_decompy/1.0.11 is recommended for
a quick installation. But you can also copy all directories here and then run
```python setup.py install```
in command line. After that, run the test script by typing
`AMFM_test.py`
to check if everything is ok (it can take couple of minutes to calculate the
results). This script is a example about how to use the package.
I've tested the installation script and the package itself in Linux and Windows
systems (but not in iOS) and everything went fine. So, if a problem comes up,
it must be probably something about python not finding the files paths.
How to use
=============
Check the AMFM_decompy pdf documentation included in the docs folder or the
online documentation at http://bjbschmitt.github.io/AMFM_decompy. The amfm_decompy
folder contains the sample.wav file that is used to ilustrate the package's code
examples.
Credits and Publications
=============
The original MATLAB YAAPT program was written by Hongbing Hu and Stephen
A.Zahorian from the Speech Communication Laboratory of the State University of
New York at Binghamton.
It is available at http://www.ws.binghamton.edu/zahorian as free software.
Further information about the program can be found at
[1] Stephen A. Zahorian, and Hongbing Hu, "A spectral/temporal method for robust
fundamental frequency tracking," J. Acoust. Soc. Am. 123(6), June 2008.
The QHM algorithm and its upgrades are formulated and presented in the following publications:
[2] Y. Pantazis, , PhD Thesis, University of Creta, 2010.
[3] Y. Pantazis, O. Rosec and Y. Stylianou, , IEEE Transactions on Audio, Speech and
Language Processing, vol. 19, n 2, 2011.
[4] G. P. Kafentzis, Y. Pantazis, O. Rosec and Y. Stylianou, , in IEEE International Conference on Acoustics,
Speech and Signal Processing (ICASSP), 2012.
Copyright and contact
=============
The AMFM_decompy is free to use, share and modify under the terms of the MIT
license.
Questions, comments, suggestions, and contributions are welcome. Please contact
me at
[email protected].
| AMFM-decompy | /AMFM_decompy-1.0.11.tar.gz/AMFM_decompy-1.0.11/README.txt | README.txt |
import numpy as np
from scipy.signal import lfilter
"""
Creates a signal object.
"""
class SignalObj(object):
def __init__(self, *args, **kwargs):
output_dtype = kwargs.get('output_dtype', 'f')
# Read the signal data from the path of a wav file.
if len(args) == 1 or 'name' in kwargs:
name = args[0] if len(args) == 1 else kwargs['name']
try:
from scipy.io import wavfile
except:
print("ERROR: Wav modules could not loaded!")
raise KeyboardInterrupt
self.fs, self.data = wavfile.read(name)
self.name = name
# Alternatively, read the signal from a Numpy array.
elif len(args) == 2 or all (k in kwargs.keys() for k in ('data','fs')):
data = args[0] if len(args) == 2 else kwargs['data']
fs = args[1] if len(args) == 2 else kwargs['fs']
self.data = data
self.fs = fs
# If the signal data is in the signed integer format (PCM), convert it
# to float.
if self.data.dtype.kind == 'i':
self.nbits = self.data.itemsize*8
self.data = pcm2float(self.data, output_dtype)
self.size = len(self.data)
self.fs = float(self.fs)
# Check if the wav file is stereo.
if self.size == self.data.size/2:
print("Warning: stereo wav file. Converting it to mono for the analysis.")
self.data = (self.data[:,0]+self.data[:,1])/2
"""
Filters the signal data by a bandpass filter object and decimate it.
"""
def filtered_version(self, bp_filter):
# Filter the signal.
tempData = lfilter(bp_filter.b, bp_filter.a, self.data)
# Decimate the filtered output.
self.filtered = tempData[0:self.size:bp_filter.dec_factor]
self.new_fs = self.fs/bp_filter.dec_factor
"""
Method that uses the pitch values to estimate the number of modulated
components in the signal.
"""
def set_nharm(self, pitch_track, n_harm_max):
n_harm = (self.fs/2)/np.amax(pitch_track) - 0.5
self.n_harm = int(np.floor(min(n_harm, n_harm_max)))
"""
Adds a zero-mean gaussian noise to the signal.
"""
def noiser(self, pitch_track, SNR):
self.clean = np.empty((self.size))
self.clean[:] = self.data
RMS = np.std(self.data[pitch_track > 0])
noise = np.random.normal(0, RMS/(10**(SNR/20)), self.size)
self.data += noise
"""
Transform a pcm raw signal into a float one, with values limited between -1 and
1.
"""
def pcm2float(sig, output_dtype=np.float64):
# Make sure it's a NumPy array.
sig = np.asarray(sig)
# Check if it is an array of signed integers.
assert sig.dtype.kind == 'i', "'sig' must be an array of signed integers!"
# Set the array output format. Accepts string as input argument for the
# desired output format (e.g. 'f').
out_dtype = np.dtype(output_dtype)
# Note that 'min' has a greater (by 1) absolute value than 'max'!
# Therefore, we use 'min' here to avoid clipping.
return sig.astype(out_dtype) / out_dtype.type(-np.iinfo(sig.dtype).min) | AMFM-decompy | /AMFM_decompy-1.0.11.tar.gz/AMFM_decompy-1.0.11/amfm_decompy/basic_tools.py | basic_tools.py |
import numpy as np
import numpy.lib.stride_tricks as stride_tricks
from scipy.signal import firwin, medfilt, lfilter
from scipy.signal.windows import hann, kaiser
import scipy.interpolate as scipy_interp
import amfm_decompy.basic_tools as basic
"""
--------------------------------------------
Classes.
--------------------------------------------
"""
"""
Auxiliary class to handle the class properties.
"""
class ClassProperty(object):
def __init__(self, initval=None):
self.val = initval
def __get__(self, obj, objtype):
return self.val
def __set__(self, obj, val):
self.val = val
"""
Creates a pitch object.
"""
class PitchObj(object):
PITCH_HALF = ClassProperty(0)
PITCH_HALF_SENS = ClassProperty(2.9)
PITCH_DOUBLE = ClassProperty(0)
PITCH_DOUBLE_SENS = ClassProperty(2.9)
SMOOTH_FACTOR = ClassProperty(5)
SMOOTH = ClassProperty(5)
PTCH_TYP = ClassProperty(100.0)
def __init__(self, frame_size, frame_jump, nfft=8192):
self.nfft = nfft
self.frame_size = frame_size
self.frame_jump = frame_jump
self.noverlap = self.frame_size-self.frame_jump
def set_energy(self, energy, threshold):
self.mean_energy = np.mean(energy)
self.energy = energy/self.mean_energy
self.vuv = (self.energy > threshold)
def set_frames_pos(self, frames_pos):
self.frames_pos = frames_pos
self.nframes = len(self.frames_pos)
def set_values(self, samp_values, file_size, interp_tech='pchip'):
self.samp_values = samp_values
self.fix()
self.values = self.upsample(self.samp_values, file_size, 0, 0,
interp_tech)
self.edges = self.edges_finder(self.values)
self.interpolate()
self.values_interp = self.upsample(self.samp_interp, file_size,
self.samp_interp[0],
self.samp_interp[-1], interp_tech)
"""
For the voiced/unvoiced version of the pitch data, finds the n samples where
the transitions between these two states occur.
"""
def edges_finder(self, values):
vec1 = (np.abs(values[1:]+values[:-1]) > 0)
vec2 = (np.abs(values[1:]*values[:-1]) == 0)
edges = np.logical_and(vec1, vec2)
# The previous logical operation detects where voiced/unvoiced transitions
# occur. Thus, a 'True' in the edges[n] sample indicates that the sample
# value[n+1] has a different state than value[n](i.e. if values[n] is
# voiced, then values[n+1] is unvoiced - and vice-versa). Consequently,
# the last sample from edges array will always be 'False' and is not
# calculated (because "there is no n+1 sample" for it. That's why
# len(edges) = len(values)-1). However, just for sake of comprehension
# (and also to avoid python warnings about array length mismatchs), I
# add a 'False' to edges the array. But in pratice, this 'False' is
# useless.
edges = np.append(edges,[False])
index = np.arange(len(values))
index = index[edges > 0]
return index.tolist()
"""
This method corresponds to the first half of the ptch_fix.m file. It tries
to fix half pitch and double pitch errors.
"""
def fix(self):
if self.PITCH_HALF > 0:
nz_pitch = self.samp_values[self.samp_values > 0]
idx = self.samp_values < (np.mean(nz_pitch)-self.PITCH_HALF_SENS *
np.std(nz_pitch))
if self.PITCH_HALF == 1:
self.samp_values[idx] = 0
elif self.PITCH_HALF == 2:
self.samp_values[idx] = 2*self.samp_values[idx]
if self.PITCH_DOUBLE > 0:
nz_pitch = self.samp_values[self.samp_values > 0]
idx = self.samp_values > (np.mean(nz_pitch)+self.PITCH_DOUBLE_SENS *
np.std(nz_pitch))
if self.PITCH_DOUBLE == 1:
self.samp_values[idx] = 0
elif self.PITCH_DOUBLE == 2:
self.samp_values[idx] = 0.5*self.samp_values[idx]
"""
Corresponds to the second half of the ptch_fix.m file. Creates the
interpolated pitch data.
"""
def interpolate(self):
pitch = np.zeros((self.nframes))
pitch[:] = self.samp_values
pitch2 = medfilt(self.samp_values, self.SMOOTH_FACTOR)
# This part in the original code is kind of confused and caused
# some problems with the extrapolated points before the first
# voiced frame and after the last voiced frame. So, I made some
# small modifications in order to make it work better.
edges = self.edges_finder(pitch)
first_sample = pitch[0]
last_sample = pitch[-1]
if len(np.nonzero(pitch2)[0]) < 2:
pitch[pitch == 0] = self.PTCH_TYP
else:
nz_pitch = pitch2[pitch2 > 0]
pitch2 = scipy_interp.pchip(np.nonzero(pitch2)[0],
nz_pitch)(range(self.nframes))
pitch[pitch == 0] = pitch2[pitch == 0]
if self.SMOOTH > 0:
pitch = medfilt(pitch, self.SMOOTH_FACTOR)
try:
if first_sample == 0:
pitch[:edges[0]-1] = pitch[edges[0]]
if last_sample == 0:
pitch[edges[-1]+1:] = pitch[edges[-1]]
except:
pass
self.samp_interp = pitch
"""
Upsample the pitch data so that it length becomes the same as the speech
value.
"""
def upsample(self, samp_values, file_size, first_samp=0, last_samp=0,
interp_tech='pchip'):
if interp_tech == 'step':
beg_pad = int((self.noverlap)/2)
up_version = np.zeros((file_size))
up_version[:beg_pad] = first_samp
up_version[beg_pad:beg_pad+self.frame_jump*self.nframes] = \
np.repeat(samp_values, self.frame_jump)
up_version[beg_pad+self.frame_jump*self.nframes:] = last_samp
elif interp_tech in ['pchip', 'spline']:
if np.amin(samp_values) > 0:
if interp_tech == 'pchip':
up_version = scipy_interp.pchip(self.frames_pos,
samp_values)(range(file_size))
elif interp_tech == 'spline':
tck, u_original = scipy_interp.splprep(
[self.frames_pos, samp_values],
u=self.frames_pos)
up_version = scipy_interp.splev(range(file_size), tck)[1]
else:
beg_pad = int((self.noverlap)/2)
up_version = np.zeros((file_size))
up_version[:beg_pad] = first_samp
voiced_frames = np.nonzero(samp_values)[0]
edges = np.nonzero((voiced_frames[1:]-voiced_frames[:-1]) > 1)[0]
edges = np.insert(edges, len(edges), len(voiced_frames)-1)
voiced_frames = np.split(voiced_frames, edges+1)[:-1]
for frame in voiced_frames:
up_interval = self.frames_pos[frame]
tot_interval = np.arange(int(up_interval[0]-(self.frame_jump/2)),
int(up_interval[-1]+(self.frame_jump/2)))
if interp_tech == 'pchip' and len(frame) > 2:
up_version[tot_interval] = scipy_interp.pchip(
up_interval,
samp_values[frame])(tot_interval)
elif interp_tech == 'spline' and len(frame) > 3:
tck, u_original = scipy_interp.splprep(
[up_interval, samp_values[frame]],
u=up_interval)
up_version[tot_interval] = scipy_interp.splev(tot_interval, tck)[1]
# MD: In case len(frame)==2, above methods fail.
#Use linear interpolation instead.
elif len(frame) > 1:
up_version[tot_interval] = scipy_interp.interp1d(
up_interval,
samp_values[frame],
fill_value='extrapolate')(tot_interval)
elif len(frame) == 1:
up_version[tot_interval] = samp_values[frame]
up_version[beg_pad+self.frame_jump*self.nframes:] = last_samp
return up_version
"""
Creates a bandpass filter object.
"""
class BandpassFilter(object):
def __init__(self, fs, parameters):
fs_min = 1000.0
if (fs > fs_min):
dec_factor = parameters['dec_factor']
else:
dec_factor = 1
filter_order = parameters['bp_forder']
f_hp = parameters['bp_low']
f_lp = parameters['bp_high']
f1 = f_hp/(fs/2)
f2 = f_lp/(fs/2)
self.b = firwin(filter_order+1, [f1, f2], pass_zero=False)
self.a = 1
self.dec_factor = dec_factor
"""
--------------------------------------------
Main function.
--------------------------------------------
"""
def yaapt(signal, **kwargs):
# Rename the YAAPT v4.0 parameter "frame_lengtht" to "tda_frame_length"
# (if provided).
if 'frame_lengtht' in kwargs:
if 'tda_frame_length' in kwargs:
warning_str = 'WARNING: Both "tda_frame_length" and "frame_lengtht" '
warning_str += 'refer to the same parameter. Therefore, the value '
warning_str += 'of "frame_lengtht" is going to be discarded.'
print(warning_str)
else:
kwargs['tda_frame_length'] = kwargs.pop('frame_lengtht')
#---------------------------------------------------------------
# Set the default values for the parameters.
#---------------------------------------------------------------
parameters = {}
parameters['frame_length'] = kwargs.get('frame_length', 35.0) #Length of each analysis frame (ms)
# WARNING: In the original MATLAB YAAPT 4.0 code the next parameter is called
# "frame_lengtht" which is quite similar to the previous one "frame_length".
# Therefore, I've decided to rename it to "tda_frame_length" in order to
# avoid confusion between them. Nevertheless, both inputs ("frame_lengtht"
# and "tda_frame_length") are accepted when the function is called.
parameters['tda_frame_length'] = \
kwargs.get('tda_frame_length', 35.0) #Frame length employed in the time domain analysis (ms)
parameters['frame_space'] = kwargs.get('frame_space', 10.0) #Spacing between analysis frames (ms)
parameters['f0_min'] = kwargs.get('f0_min', 60.0) #Minimum F0 searched (Hz)
parameters['f0_max'] = kwargs.get('f0_max', 400.0) #Maximum F0 searched (Hz)
parameters['fft_length'] = kwargs.get('fft_length', 8192) #FFT length
parameters['bp_forder'] = kwargs.get('bp_forder', 150) #Order of band-pass filter
parameters['bp_low'] = kwargs.get('bp_low', 50.0) #Low frequency of filter passband (Hz)
parameters['bp_high'] = kwargs.get('bp_high', 1500.0) #High frequency of filter passband (Hz)
parameters['nlfer_thresh1'] = kwargs.get('nlfer_thresh1', 0.75) #NLFER boundary for voiced/unvoiced decisions
parameters['nlfer_thresh2'] = kwargs.get('nlfer_thresh2', 0.1) #Threshold for NLFER definitely unvoiced
parameters['shc_numharms'] = kwargs.get('shc_numharms', 3) #Number of harmonics in SHC calculation
parameters['shc_window'] = kwargs.get('shc_window', 40.0) #SHC window length (Hz)
parameters['shc_maxpeaks'] = kwargs.get('shc_maxpeaks', 4) #Maximum number of SHC peaks to be found
parameters['shc_pwidth'] = kwargs.get('shc_pwidth', 50.0) #Window width in SHC peak picking (Hz)
parameters['shc_thresh1'] = kwargs.get('shc_thresh1', 5.0) #Threshold 1 for SHC peak picking
parameters['shc_thresh2'] = kwargs.get('shc_thresh2', 1.25) #Threshold 2 for SHC peak picking
parameters['f0_double'] = kwargs.get('f0_double', 150.0) #F0 doubling decision threshold (Hz)
parameters['f0_half'] = kwargs.get('f0_half', 150.0) #F0 halving decision threshold (Hz)
parameters['dp5_k1'] = kwargs.get('dp5_k1', 11.0) #Weight used in dynamic program
parameters['dec_factor'] = kwargs.get('dec_factor', 1) #Factor for signal resampling
parameters['nccf_thresh1'] = kwargs.get('nccf_thresh1', 0.3) #Threshold for considering a peak in NCCF
parameters['nccf_thresh2'] = kwargs.get('nccf_thresh2', 0.9) #Threshold for terminating serach in NCCF
parameters['nccf_maxcands'] = kwargs.get('nccf_maxcands', 3) #Maximum number of candidates found
parameters['nccf_pwidth'] = kwargs.get('nccf_pwidth', 5) #Window width in NCCF peak picking
parameters['merit_boost'] = kwargs.get('merit_boost', 0.20) #Boost merit
parameters['merit_pivot'] = kwargs.get('merit_pivot', 0.99) #Merit assigned to unvoiced candidates in
#defintely unvoiced frames
parameters['merit_extra'] = kwargs.get('merit_extra', 0.4) #Merit assigned to extra candidates
#in reducing F0 doubling/halving errors
parameters['median_value'] = kwargs.get('median_value', 7) #Order of medial filter
parameters['dp_w1'] = kwargs.get('dp_w1', 0.15) #DP weight factor for V-V transitions
parameters['dp_w2'] = kwargs.get('dp_w2', 0.5) #DP weight factor for V-UV or UV-V transitions
parameters['dp_w3'] = kwargs.get('dp_w3', 0.1) #DP weight factor of UV-UV transitions
parameters['dp_w4'] = kwargs.get('dp_w4', 0.9) #Weight factor for local costs
# Exclusive from pYAAPT.
parameters['spec_pitch_min_std'] = kwargs.get('spec_pitch_min_std', 0.05)
#Weight factor that sets a minimum
#spectral pitch standard deviation,
#which is calculated as
#min_std = pitch_avg*spec_pitch_min_std
#---------------------------------------------------------------
# Create the signal objects and filter them.
#---------------------------------------------------------------
fir_filter = BandpassFilter(signal.fs, parameters)
nonlinear_sign = basic.SignalObj(signal.data**2, signal.fs)
signal.filtered_version(fir_filter)
nonlinear_sign.filtered_version(fir_filter)
#---------------------------------------------------------------
# Create the pitch object.
#---------------------------------------------------------------
nfft = parameters['fft_length']
frame_size = int(np.fix(parameters['frame_length']*signal.fs/1000))
frame_jump = int(np.fix(parameters['frame_space']*signal.fs/1000))
pitch = PitchObj(frame_size, frame_jump, nfft)
assert pitch.frame_size > 15, 'Frame length value {} is too short.'.format(pitch.frame_size)
assert pitch.frame_size < 2048, 'Frame length value {} exceeds the limit.'.format(pitch.frame_size)
#---------------------------------------------------------------
# Calculate NLFER and determine voiced/unvoiced frames.
#---------------------------------------------------------------
nlfer(signal, pitch, parameters)
#---------------------------------------------------------------
# Calculate an approximate pitch track from the spectrum.
#---------------------------------------------------------------
spec_pitch, pitch_std = spec_track(nonlinear_sign, pitch, parameters)
#---------------------------------------------------------------
# Temporal pitch tracking based on NCCF.
#---------------------------------------------------------------
time_pitch1, time_merit1 = time_track(signal, spec_pitch, pitch_std, pitch,
parameters)
time_pitch2, time_merit2 = time_track(nonlinear_sign, spec_pitch, pitch_std,
pitch, parameters)
# Added in YAAPT 4.0
if time_pitch1.shape[1] < len(spec_pitch):
len_time = time_pitch1.shape[1]
len_spec = len(spec_pitch)
time_pitch1 = np.concatenate((time_pitch1, np.zeros((3,len_spec-len_time),
dtype=time_pitch1.dtype)),axis=1)
time_pitch2 = np.concatenate((time_pitch2, np.zeros((3,len_spec-len_time),
dtype=time_pitch2.dtype)),axis=1)
time_merit1 = np.concatenate((time_merit1, np.zeros((3,len_spec-len_time),
dtype=time_merit1.dtype)),axis=1)
time_merit2 = np.concatenate((time_merit2, np.zeros((3,len_spec-len_time),
dtype=time_merit2.dtype)),axis=1)
#---------------------------------------------------------------
# Refine pitch candidates.
#---------------------------------------------------------------
ref_pitch, ref_merit = refine(time_pitch1, time_merit1, time_pitch2,
time_merit2, spec_pitch, pitch, parameters)
#---------------------------------------------------------------
# Use dyanamic programming to determine the final pitch.
#---------------------------------------------------------------
final_pitch = dynamic(ref_pitch, ref_merit, pitch, parameters)
pitch.set_values(final_pitch, signal.size)
return pitch
"""
--------------------------------------------
Side functions.
--------------------------------------------
"""
"""
Normalized Low Frequency Energy Ratio function. Corresponds to the nlfer.m file,
but instead of returning the results to them function, encapsulates them in the
pitch object.
"""
def nlfer(signal, pitch, parameters):
#---------------------------------------------------------------
# Set parameters.
#---------------------------------------------------------------
N_f0_min = np.around((parameters['f0_min']*2/float(signal.new_fs))*pitch.nfft)
N_f0_max = np.around((parameters['f0_max']/float(signal.new_fs))*pitch.nfft)
window = hann(pitch.frame_size+2)[1:-1]
data = np.zeros((signal.size)) #Needs other array, otherwise stride and
data[:] = signal.filtered #windowing will modify signal.filtered
#---------------------------------------------------------------
# Main routine.
#---------------------------------------------------------------
samples = np.arange(int(np.fix(float(pitch.frame_size)/2)),
signal.size-int(np.fix(float(pitch.frame_size)/2)),
pitch.frame_jump)
data_matrix = np.empty((len(samples), pitch.frame_size))
data_matrix[:, :] = stride_matrix(data, len(samples),
pitch.frame_size, pitch.frame_jump)
data_matrix *= window
specData = np.fft.rfft(data_matrix, pitch.nfft)
frame_energy = np.abs(specData[:, int(N_f0_min-1):int(N_f0_max)]).sum(axis=1)
pitch.set_energy(frame_energy, parameters['nlfer_thresh1'])
pitch.set_frames_pos(samples)
"""
Spectral pitch tracking. Computes estimates of pitch using nonlinearly processed
speech (typically square or absolute value) and frequency domain processing.
Search for frequencies which have energy at multiplies of that frequency.
Corresponds to the spec_trk.m file.
"""
def spec_track(signal, pitch, parameters):
#---------------------------------------------------------------
# Set parameters.
#---------------------------------------------------------------
nframe_size = pitch.frame_size*2
maxpeaks = parameters['shc_maxpeaks']
delta = signal.new_fs/pitch.nfft
window_length = int(np.fix(parameters['shc_window']/delta))
half_window_length = int(np.fix(float(window_length)/2))
if not(window_length % 2):
window_length += 1
max_SHC = int(np.fix((parameters['f0_max']+parameters['shc_pwidth']*2)/delta))
min_SHC = int(np.ceil(parameters['f0_min']/delta))
num_harmonics = parameters['shc_numharms']
#---------------------------------------------------------------
# Main routine.
#---------------------------------------------------------------
cand_pitch = np.zeros((maxpeaks, pitch.nframes))
cand_merit = np.ones((maxpeaks, pitch.nframes))
data = np.append(signal.filtered,
np.zeros((1, nframe_size +
((pitch.nframes-1)*pitch.frame_jump-signal.size))))
#Compute SHC for voiced frame
window = kaiser(nframe_size, 0.5)
SHC = np.zeros((max_SHC))
row_mat_list = np.array([np.empty((max_SHC-min_SHC+1, window_length))
for x in range(num_harmonics+1)])
magnitude = np.zeros(int((half_window_length+(pitch.nfft/2)+1)))
for frame in np.where(pitch.vuv)[0].tolist():
fir_step = frame*pitch.frame_jump
data_slice = data[fir_step:fir_step+nframe_size]*window
data_slice -= np.mean(data_slice)
magnitude[half_window_length:] = np.abs(np.fft.rfft(data_slice,
pitch.nfft))
for idx,row_mat in enumerate(row_mat_list):
row_mat[:, :] = stride_matrix(magnitude[min_SHC*(idx+1):],
max_SHC-min_SHC+1,
window_length, idx+1)
SHC[min_SHC-1:max_SHC] = np.sum(np.prod(row_mat_list,axis=0),axis=1)
cand_pitch[:, frame], cand_merit[:, frame] = \
peaks(SHC, delta, maxpeaks, parameters)
#Extract the pitch candidates of voiced frames for the future pitch selection.
spec_pitch = cand_pitch[0, :]
voiced_cand_pitch = cand_pitch[:, cand_pitch[0, :] > 0]
voiced_cand_merit = cand_merit[:, cand_pitch[0, :] > 0]
num_voiced_cand = len(voiced_cand_pitch[0, :])
avg_voiced = np.mean(voiced_cand_pitch[0, :])
std_voiced = np.std(voiced_cand_pitch[0, :])
#Interpolation of the weigthed candidates.
delta1 = abs((voiced_cand_pitch - 0.8*avg_voiced))*(3-voiced_cand_merit)
index = delta1.argmin(0)
voiced_peak_minmrt = voiced_cand_pitch[index, range(num_voiced_cand)]
voiced_merit_minmrt = voiced_cand_merit[index, range(num_voiced_cand)]
voiced_peak_minmrt = medfilt(voiced_peak_minmrt,
max(1, parameters['median_value']-2))
#Replace the lowest merit candidates by the median smoothed ones
#computed from highest merit peaks above.
voiced_cand_pitch[index, range(num_voiced_cand)] = voiced_peak_minmrt
voiced_cand_merit[index, range(num_voiced_cand)] = voiced_merit_minmrt
#Use dynamic programming to find best overal path among pitch candidates.
#Dynamic weight for transition costs balance between local and
#transition costs.
weight_trans = parameters['dp5_k1']*std_voiced/avg_voiced
if num_voiced_cand > 2:
voiced_pitch = dynamic5(voiced_cand_pitch, voiced_cand_merit,
weight_trans, parameters['f0_min'])
voiced_pitch = medfilt(voiced_pitch, max(1, parameters['median_value']-2))
else:
if num_voiced_cand > 0:
voiced_pitch = (np.ones((num_voiced_cand)))*150.0
else:
voiced_pitch = np.array([150.0])
cand_pitch[0, 0] = 0
pitch_avg = np.mean(voiced_pitch)
pitch_std = np.maximum(np.std(voiced_pitch), pitch_avg*parameters['spec_pitch_min_std'])
spec_pitch[cand_pitch[0, :] > 0] = voiced_pitch[:]
if (spec_pitch[0] < pitch_avg/2):
spec_pitch[0] = pitch_avg
if (spec_pitch[-1] < pitch_avg/2):
spec_pitch[-1] = pitch_avg
spec_voiced = np.array(np.nonzero(spec_pitch)[0])
spec_pitch = scipy_interp.pchip(spec_voiced,
spec_pitch[spec_voiced])(range(pitch.nframes))
spec_pitch = lfilter(np.ones((3))/3, 1.0, spec_pitch)
spec_pitch[0] = spec_pitch[2]
spec_pitch[1] = spec_pitch[3]
return spec_pitch, pitch_std
"""
Temporal pitch tracking.
Corresponds to the tm_trk.m file.
"""
def time_track(signal, spec_pitch, pitch_std, pitch, parameters):
#---------------------------------------------------------------
# Set parameters.
#---------------------------------------------------------------
tda_frame_length = int(parameters['tda_frame_length']*signal.fs/1000)
tda_noverlap = tda_frame_length-pitch.frame_jump
tda_nframes = int((len(signal.data)-tda_noverlap)/pitch.frame_jump)
len_spectral = len(spec_pitch)
if tda_nframes < len_spectral:
spec_pitch = spec_pitch[:tda_nframes]
elif tda_nframes > len_spectral:
tda_nframes = len_spectral
merit_boost = parameters['merit_boost']
maxcands = parameters['nccf_maxcands']
freq_thresh = 5.0*pitch_std
spec_range = np.maximum(spec_pitch-2.0*pitch_std, parameters['f0_min'])
spec_range = np.vstack((spec_range,
np.minimum(spec_pitch+2.0*pitch_std, parameters['f0_max'])))
time_pitch = np.zeros((maxcands, tda_nframes))
time_merit = np.zeros((maxcands, tda_nframes))
#---------------------------------------------------------------
# Main routine.
#---------------------------------------------------------------
data = np.zeros((signal.size)) #Needs other array, otherwise stride and
data[:] = signal.filtered #windowing will modify signal.filtered
signal_frames = stride_matrix(data, tda_nframes,tda_frame_length,
pitch.frame_jump)
for frame in range(tda_nframes):
lag_min0 = (int(np.fix(signal.new_fs/spec_range[1, frame])) -
int(np.fix(parameters['nccf_pwidth']/2.0)))
lag_max0 = (int(np.fix(signal.new_fs/spec_range[0, frame])) +
int(np.fix(parameters['nccf_pwidth']/2.0)))
phi = crs_corr(signal_frames[frame, :], lag_min0, lag_max0)
time_pitch[:, frame], time_merit[:, frame] = \
cmp_rate(phi, signal.new_fs, maxcands, lag_min0, lag_max0, parameters)
diff = np.abs(time_pitch - spec_pitch)
match1 = (diff < freq_thresh)
match = ((1 - diff/freq_thresh) * match1)
time_merit = (((1+merit_boost)*time_merit) * match)
return time_pitch, time_merit
"""
Refines pitch candidates obtained from NCCF using spectral pitch track and
NLFER energy information.
Corresponds to the refine.m file.
"""
def refine(time_pitch1, time_merit1, time_pitch2, time_merit2, spec_pitch,
pitch, parameters):
#---------------------------------------------------------------
# Set parameters.
#---------------------------------------------------------------
nlfer_thresh2 = parameters['nlfer_thresh2']
merit_pivot = parameters['merit_pivot']
#---------------------------------------------------------------
# Main routine.
#---------------------------------------------------------------
time_pitch = np.append(time_pitch1, time_pitch2, 0)
time_merit = np.append(time_merit1, time_merit2, 0)
maxcands = time_pitch.shape[0]
idx = np.argsort(-time_merit, axis=0)
time_merit.sort(axis=0)
time_merit[:, :] = time_merit[::-1,:]
time_pitch = time_pitch[idx, range(pitch.nframes)]
best_pitch = medfilt(time_pitch[0, :], parameters['median_value'])*pitch.vuv
idx1 = pitch.energy <= nlfer_thresh2
idx2 = (pitch.energy > nlfer_thresh2) & (time_pitch[0, :] > 0)
idx3 = (pitch.energy > nlfer_thresh2) & (time_pitch[0, :] <= 0)
merit_mat = (time_pitch[1:maxcands-1, :] == 0) & idx2
merit_mat = np.insert(merit_mat, [0, maxcands-2],
np.zeros((1, pitch.nframes), dtype=bool), 0)
time_pitch[:, idx1] = 0
time_merit[:, idx1] = merit_pivot
time_pitch[maxcands-1, idx2] = 0.0
time_merit[maxcands-1, idx2] = 1.0-time_merit[0, idx2]
time_merit[merit_mat] = 0.0
time_pitch[0, idx3] = spec_pitch[idx3]
time_merit[0, idx3] = np.minimum(1, pitch.energy[idx3]/2.0)
time_pitch[1:maxcands, idx3] = 0.0
time_merit[1:maxcands, idx3] = 1.0-time_merit[0, idx3]
time_pitch[maxcands-2, :] = best_pitch
non_zero_frames = best_pitch > 0.0
time_merit[maxcands-2, non_zero_frames] = time_merit[0, non_zero_frames]
time_merit[maxcands-2, ~(non_zero_frames)] = 1.0-np.minimum(1,
pitch.energy[~(non_zero_frames)]/2.0)
time_pitch[maxcands-3, :] = spec_pitch
time_merit[maxcands-3, :] = pitch.energy/5.0
return time_pitch, time_merit
"""
Dynamic programming used to compute local and transition cost matrices,
enabling the lowest cost tracking of pitch candidates.
It uses NFLER from the spectrogram and the highly robust spectral F0 track,
plus the merits, for computation of the cost matrices.
Corresponds to the dynamic.m file.
"""
def dynamic(ref_pitch, ref_merit, pitch, parameters):
#---------------------------------------------------------------
# Set parameters.
#---------------------------------------------------------------
num_cands = ref_pitch.shape[0]
best_pitch = ref_pitch[num_cands-2, :]
mean_pitch = np.mean(best_pitch[best_pitch > 0])
dp_w1 = parameters['dp_w1']
dp_w2 = parameters['dp_w2']
dp_w3 = parameters['dp_w3']
dp_w4 = parameters['dp_w4']
#---------------------------------------------------------------
# Main routine.
#---------------------------------------------------------------
local_cost = 1 - ref_merit
trans_cmatrix = np.ones((num_cands, num_cands, pitch.nframes))
ref_mat1 = np.zeros((num_cands, num_cands, pitch.nframes))
ref_mat2 = np.zeros((num_cands, num_cands, pitch.nframes))
idx_mat1 = np.zeros((num_cands, num_cands, pitch.nframes), dtype=bool)
idx_mat2 = np.zeros((num_cands, num_cands, pitch.nframes), dtype=bool)
idx_mat3 = np.zeros((num_cands, num_cands, pitch.nframes), dtype=bool)
ref_mat1[:, :, 1:] = np.tile(ref_pitch[:, 1:].reshape(1, num_cands,
pitch.nframes-1), (num_cands, 1, 1))
ref_mat2[:, :, 1:] = np.tile(ref_pitch[:, :-1].reshape(num_cands, 1,
pitch.nframes-1), (1, num_cands, 1))
idx_mat1[:, :, 1:] = (ref_mat1[:, :, 1:] > 0) & (ref_mat2[:, :, 1:] > 0)
idx_mat2[:, :, 1:] = (((ref_mat1[:, :, 1:] == 0) & (ref_mat2[:, :, 1:] > 0)) |
((ref_mat1[:, :, 1:] > 0) & (ref_mat2[:, :, 1:] == 0)))
idx_mat3[:, :, 1:] = (ref_mat1[:, :, 1:] == 0) & (ref_mat2[:, :, 1:] == 0)
mat1_values = np.abs(ref_mat1-ref_mat2)/mean_pitch
benefit2 = np.insert(np.minimum(1, abs(pitch.energy[:-1]-pitch.energy[1:])),
0, 0)
benefit2 = np.tile(benefit2, (num_cands, num_cands, 1))
trans_cmatrix[idx_mat1] = dp_w1*mat1_values[idx_mat1]
trans_cmatrix[idx_mat2] = dp_w2*(1-benefit2[idx_mat2])
trans_cmatrix[idx_mat3] = dp_w3
trans_cmatrix = trans_cmatrix/dp_w4
path = path1(local_cost, trans_cmatrix, num_cands, pitch.nframes)
final_pitch = ref_pitch[path, range(pitch.nframes)]
return final_pitch
"""
--------------------------------------------
Auxiliary functions.
--------------------------------------------
"""
"""
Computes peaks in a frequency domain function associated with the peaks found
in each frame based on the correlation sequence.
Corresponds to the peaks.m file.
"""
def peaks(data, delta, maxpeaks, parameters):
#---------------------------------------------------------------
# Set parameters.
#---------------------------------------------------------------
PEAK_THRESH1 = parameters['shc_thresh1']
PEAK_THRESH2 = parameters['shc_thresh2']
epsilon = .00000000000001
width = int(np.fix(parameters['shc_pwidth']/delta))
if not(float(width) % 2):
width = width + 1
center = int(np.ceil(width/2))
min_lag = int(np.fix(parameters['f0_min']/delta - center))
max_lag = int(np.fix(parameters['f0_max']/delta + center))
if (min_lag < 1):
min_lag = 1
print('Min_lag is too low and adjusted ({}).'.format(min_lag))
if max_lag > (len(data) - width):
max_lag = len(data) - width
print('Max_lag is too high and adjusted ({}).'.format(max_lag))
pitch = np.zeros((maxpeaks))
merit = np.zeros((maxpeaks))
#---------------------------------------------------------------
# Main routine.
#---------------------------------------------------------------
max_data = max(data[min_lag:max_lag+1])
if (max_data > epsilon):
data = data/max_data
avg_data = np.mean(data[min_lag:max_lag+1])
if (avg_data > 1/PEAK_THRESH1):
pitch = np.zeros((maxpeaks))
merit = np.ones((maxpeaks))
return pitch, merit
#---------------------------------------------------------------
#Step1 (this step was implemented differently than in original version)
#---------------------------------------------------------------
numpeaks = 0
vec_back = (data[min_lag+center+1:max_lag-center+1] >
data[min_lag+center:max_lag-center])
vec_forw = (data[min_lag+center+1:max_lag-center+1] >
data[min_lag+center+2:max_lag-center+2])
above_thresh = (data[min_lag+center+1:max_lag-center+1] >
PEAK_THRESH2*avg_data)
peaks = np.logical_and(np.logical_and(vec_back, vec_forw), above_thresh)
for n in (peaks.ravel().nonzero()[0]+min_lag+center+1).tolist():
if np.argmax(data[n-center:n+center+1]) == center:
if numpeaks >= maxpeaks:
pitch = np.append(pitch, np.zeros((1)))
merit = np.append(merit, np.zeros((1)))
pitch[numpeaks] = float(n)*delta
merit[numpeaks] = data[n]
numpeaks += 1
#---------------------------------------------------------------
#Step2
#---------------------------------------------------------------
if (max(merit)/avg_data < PEAK_THRESH1):
pitch = np.zeros((maxpeaks))
merit = np.ones((maxpeaks))
return pitch, merit
#---------------------------------------------------------------
#Step3
#---------------------------------------------------------------
idx = (-merit).ravel().argsort().tolist()
merit = merit[idx]
pitch = pitch[idx]
numpeaks = min(numpeaks, maxpeaks)
pitch = np.append(pitch[:numpeaks], np.zeros((maxpeaks-numpeaks)))
merit = np.append(merit[:numpeaks], np.zeros((maxpeaks-numpeaks)))
#---------------------------------------------------------------
#Step4
#---------------------------------------------------------------
if (0 < numpeaks < maxpeaks):
pitch[numpeaks:maxpeaks] = pitch[0]
merit[numpeaks:maxpeaks] = merit[0]
else:
pitch = np.zeros((maxpeaks))
merit = np.ones((maxpeaks))
return np.transpose(pitch), np.transpose(merit)
"""
Dynamic programming used to compute local and transition cost matrices,
enabling the lowest cost tracking of pitch candidates.
It uses NFLER from the spectrogram and the highly robust spectral F0 track,
plus the merits, for computation of the cost matrices.
Corresponds to the dynamic5.m file.
"""
def dynamic5(pitch_array, merit_array, k1, f0_min):
num_cand = pitch_array.shape[0]
num_frames = pitch_array.shape[1]
local = 1-merit_array
trans = np.zeros((num_cand, num_cand, num_frames))
trans[:, :, 1:] = abs(pitch_array[:, 1:].reshape(1, num_cand, num_frames-1) -
pitch_array[:, :-1].reshape(num_cand, 1, num_frames-1))/f0_min
trans[:, :, 1:] = 0.05*trans[:, :, 1:] + trans[:, :, 1:]**2
trans = k1*trans
path = path1(local, trans, num_cand, num_frames)
final_pitch = pitch_array[path, range(num_frames)]
return final_pitch
"""
Finds the optimal path with the lowest cost if two matrice(Local cost matrix
and Transition cost) are given.
Corresponds to the path1.m file.
"""
def path1(local, trans, n_lin, n_col):
# Apparently the following lines are somehow kind of useless.
# Therefore, I removed them in the version 1.0.3.
# if n_lin >= 100:
# print 'Stop in Dynamic due to M>100'
# raise KeyboardInterrupt
#
# if n_col >= 1000:
# print 'Stop in Dynamic due to N>1000'
# raise KeyboardInterrupt
PRED = np.zeros((n_lin, n_col), dtype=int)
P = np.ones((n_col), dtype=int)
p_small = np.zeros((n_col), dtype=int)
PCOST = np.zeros((n_lin))
CCOST = np.zeros((n_lin))
PCOST = local[:, 0]
for I in range(1, n_col):
aux_matrix = PCOST+np.transpose(trans[:, :, I])
K = n_lin-np.argmin(aux_matrix[:, ::-1], axis=1)-1
PRED[:, I] = K
CCOST = PCOST[K]+trans[K, range(n_lin), I]
assert CCOST.any() < 1.0E+30, 'CCOST>1.0E+30, Stop in Dynamic'
CCOST = CCOST+local[:, I]
PCOST[:] = CCOST
J = n_lin - np.argmin(CCOST[::-1])-1
p_small[I] = J
P[-1] = p_small[-1]
for I in range(n_col-2, -1, -1):
P[I] = PRED[P[I+1], I+1]
return P
"""
Computes the NCCF (Normalized cross correlation Function) sequence based on
the RAPT algorithm discussed by DAVID TALKIN.
Corresponds to the crs_corr.m file.
"""
def crs_corr(data, lag_min, lag_max):
eps1 = 0.0
data_len = len(data)
N = data_len-lag_max
error_str = 'ERROR: Negative index in the cross correlation calculation of '
error_str += 'the pYAAPT time domain analysis. Please try to increase the '
error_str += 'value of the "tda_frame_length" parameter.'
assert N>0, error_str
phi = np.zeros((data_len))
data -= np.mean(data)
x_j = data[0:N]
x_jr = data[lag_min:lag_max+N]
p = np.dot(x_j, x_j)
x_jr_matrix = stride_matrix(x_jr, lag_max-lag_min, N, 1)
formula_nume = np.dot(x_jr_matrix, x_j)
formula_denom = np.sum(x_jr_matrix*x_jr_matrix, axis=1)*p + eps1
phi[lag_min:lag_max] = formula_nume/np.sqrt(formula_denom)
return phi
"""
Computes pitch estimates and the corresponding merit values associated with the
peaks found in each frame based on the correlation sequence.
Corresponds to the cmp_rate.m file.
"""
def cmp_rate(phi, fs, maxcands, lag_min, lag_max, parameters):
#---------------------------------------------------------------
# Set parameters.
#---------------------------------------------------------------
width = parameters['nccf_pwidth']
center = int(np.fix(width/2.0))
merit_thresh1 = parameters['nccf_thresh1']
merit_thresh2 = parameters['nccf_thresh2']
numpeaks = 0
pitch = np.zeros((maxcands))
merit = np.zeros((maxcands))
#---------------------------------------------------------------
# Main routine.
#(this step was implemented differently than in original version)
#---------------------------------------------------------------
vec_back = (phi[lag_min+center:lag_max-center+1] >
phi[lag_min+center-1:lag_max-center])
vec_forw = (phi[lag_min+center:lag_max-center+1] >
phi[lag_min+center+1:lag_max-center+2])
above_thresh = phi[lag_min+center:lag_max-center+1] > merit_thresh1
peaks = np.logical_and(np.logical_and(vec_back, vec_forw), above_thresh)
peaks = (peaks.ravel().nonzero()[0]+lag_min+center).tolist()
if np.amax(phi) > merit_thresh2 and len(peaks) > 0:
max_point = peaks[np.argmax(phi[peaks])]
pitch[numpeaks] = fs/float(max_point+1)
merit[numpeaks] = np.amax(phi[peaks])
numpeaks += 1
else:
for n in peaks:
if np.argmax(phi[n-center:n+center+1]) == center:
try:
pitch[numpeaks] = fs/float(n+1)
merit[numpeaks] = phi[n]
except:
pitch = np.hstack((pitch, fs/float(n+1)))
merit = np.hstack((merit, phi[n]))
numpeaks += 1
#---------------------------------------------------------------
# Sort the results.
#---------------------------------------------------------------
idx = (-merit).ravel().argsort().tolist()
merit = merit[idx[:maxcands]]
pitch = pitch[idx[:maxcands]]
if (np.amax(merit) > 1.0):
merit = merit/np.amax(merit)
return pitch, merit
"""
--------------------------------------------
Extra functions.
--------------------------------------------
"""
def stride_matrix(vector, n_lin, n_col, hop):
data_matrix = stride_tricks.as_strided(vector, shape=(n_lin, n_col),
strides=(vector.strides[0]*hop, vector.strides[0]))
return data_matrix | AMFM-decompy | /AMFM_decompy-1.0.11.tar.gz/AMFM_decompy-1.0.11/amfm_decompy/pYAAPT.py | pYAAPT.py |
import numpy as np
import scipy
"""
--------------------------------------------
Classes.
--------------------------------------------
"""
"""
Creates a single component object.
"""
class ComponentObj(object):
def __init__(self, H, harm):
self.mag = H[harm, 0, :]
self.phase = H[harm, 1, :]
self.freq = H[harm, 2, :]
"""
Synthsize the modulated component by using the extracted magnitude and
phase.
"""
def synthesize(self):
self.signal = 2*self.mag*np.cos(self.phase)
"""
Creates the output signal object (which, in its turn, is formed by n_harm
modulated components).
"""
class ModulatedSign(object):
def __init__(self, n_harm, file_size, fs, phase_tech='phase'):
self.n_harm = n_harm
self.size = file_size
self.fs = fs
self.H = np.zeros((self.n_harm, 3, self.size))
self.harmonics = [ComponentObj(self.H, i) for i in range(self.n_harm)]
self.error = np.zeros(self.size)
self.phase_tech = phase_tech
"""
Updates the 3-dimension array H, which stores the magnitude, phase and
frequency values from all components. Its first dimension refers to the
n_harm components, the second to the three composing parameters (where 0
stands for the magnitude, 1 for the phase and 2 for the frequency) and the
third dimension to the temporal axis.
"""
def update_values(self, a, freq, frame):
self.H[:, 0, frame] = np.abs(a)
self.H[:, 1, frame] = np.angle(a)
self.H[:, 2, frame] = freq
"""
Interpolate the parameters values when the extraction is not performed
sample-by-sample. While the interpolation from magnitude and frequency
is pretty straightforward, the phase one is not. Therefore, references
[1,2] present a solution for this problem.
"""
def interpolate_samp(self, samp_frames, pitch_track):
# Interpolation from magnitude and frequency.
for idx, func in [(0, 'linear'), (2, 'cubic')]:
f = scipy.interpolate.interp1d(samp_frames,
self.H[:, idx, samp_frames], kind=func)
self.H[:, idx, np.nonzero(pitch_track)[0]] = f(
np.nonzero(pitch_track)[0])
# Interpolation from phase.
step = samp_frames[1]-samp_frames[0]
sin_f = np.cumsum(np.sin(np.pi*np.arange(1, step)/step)).reshape(
1, step-1)
for idx, frame in np.ndenumerate(samp_frames[1:]):
if frame-samp_frames[idx] <= step:
cum_phase = np.cumsum(self.H[:, 2, samp_frames[idx]+1:frame+1],
axis=1)*2*np.pi
bad_phase = cum_phase[:, -1]+self.H[:, 1, samp_frames[idx]]
M = np.around(np.abs(self.H[:, 1, frame]-bad_phase)/(2*np.pi))
if frame-samp_frames[idx] < step:
end_step = frame-samp_frames[idx]
func = np.cumsum(np.sin(np.pi*np.arange(1, end_step) /
end_step)).reshape(1, end_step-1)
else:
func = sin_f
r_vec = (np.pi*(self.H[:, 1, frame]+2*np.pi*M-bad_phase) /
(2*(frame-samp_frames[idx]))).reshape(self.n_harm, 1)
new_phase = cum_phase[:, :-1]+r_vec*func + \
self.H[:, 1, samp_frames[idx]].reshape(self.n_harm, 1)
self.H[:, 1, samp_frames[idx]+1:frame] = ((new_phase + np.pi) %
(2*np.pi)-np.pi)
"""
Synthesize the final signal by initially creating each modulated component
and then summing all of them.
"""
def synthesize(self, N=None):
if N is None:
N = self.n_harm
[self.harmonics[i].synthesize()
for i in range(N)]
self.signal = sum([self.harmonics[i].signal
for i in range(self.n_harm)])
"""
Calculates the SRER (Signal-to-Reconstruction Error Ratio) for the
synthesized signal.
"""
def srer(self, orig_signal, pitch_track):
self.SRER = 20*np.log10(np.std(orig_signal[np.nonzero(pitch_track)[0]]) /
np.std(orig_signal[np.nonzero(pitch_track)[0]] -
self.signal[np.nonzero(pitch_track)[0]]))
"""
Extrapolates the phase at the border of the voiced frames by integrating
the edge frequency value. This procedure is necessary for posterior aQHM
calculations. Additionally, the method allows the replacement of the
extracted phase by the cumulative frequency. The objective is to provide
smoother bases for further aQHM and eaQHM calculations. Normally this is
not necessary, since that the interpolation process already smooths the
phase vector. But in a sample-by-sample extraction case, this substitution
is very helpful to avoid the degradation of aQHM and eaQHM performance
due the phase wild behaviour.
"""
def phase_edges(self, edges, window):
# Selects whether the phase itself or the cummulative frequency will be
# used.
if self.phase_tech is 'phase':
self.extrap_phase = np.unwrap(self.H[:, 1, :])
elif self.phase_tech is 'freq':
delta_phase = self.H[:, 1, edges[0]+1] - \
self.H[:, 2, edges[0]+1]*2*np.pi
self.extrap_phase = np.cumsum(self.H[:, 2, :], axis=1)*2*np.pi + \
delta_phase.reshape(self.n_harm, 1)
# Extrapolate the phase edges.
n_beg = -window.half_len_vec[::-1][:-1].reshape(1, window.N)
n_end = window.half_len_vec[1:].reshape(1, window.N)
for beg, end in zip(edges[::2], edges[1::2]):
old_phase = self.extrap_phase[:, beg+1].reshape(self.n_harm, 1)
freq = self.H[:, 2, beg+1].reshape(self.n_harm, 1)
self.extrap_phase[:, beg-window.N+1:beg+1] = \
2*np.pi*freq*n_beg+old_phase
old_phase = self.extrap_phase[:, end].reshape(self.n_harm, 1)
freq = self.H[:, 2, end].reshape(self.n_harm, 1)
self.extrap_phase[:, end+1:end+window.N+1] = \
2*np.pi*freq*n_end+old_phase
"""
Creates the sample window object.
"""
class SampleWindow(object):
def __init__(self, window_duration, fs):
self.dur = window_duration # in seconds
self.length = int(self.dur*fs+1)
if not self.length %2:
self.length -= 1
self.data = np.hamming(self.length)
self.data2 = self.data**2
self.N = int(self.dur*fs/2)
self.half_len_vec = np.arange(self.N+1)
self.len_vec = np.arange(-self.N, self.N+1)
self.a0 = 0.54**2 + (0.46**2)/2
self.a1 = 0.54*0.46
self.a2 = (0.46**2)/4
self.R0_diag = R_eq(0, g0, self)
self.R2_diag = sum(self.data2*(self.len_vec**2))
"""
--------------------------------------------
Main Functions.
--------------------------------------------
"""
"""
Main QHM function.
"""
def qhm(signal, pitch, window, samp_jump=None, N_iter=1, phase_tech='phase'):
return HM_run(qhm_iteration, signal, pitch, window, samp_jump, N_iter,
phase_tech)
"""
Main aQHM function.
"""
def aqhm(signal, previous_HM, pitch, window, samp_jump=None, N_iter=1,
N_runs=float('Inf'), phase_tech='phase', eaQHM_flag=False):
count = 1
outflag = False
while outflag is False:
func_options = [previous_HM, eaQHM_flag, 0]
HM = HM_run(aqhm_iteration, signal, pitch, window, samp_jump, N_iter,
phase_tech, func_options)
if count == 1:
previous_HM = HM
elif (count > 1 and HM.SRER > previous_HM.SRER):
previous_HM = HM
else:
outflag = True
count += 1
if count > N_runs:
outflag = True
return previous_HM
"""
Main eaQHM function (which in fact varies very few from the aQHM).
"""
def eaqhm(signal, previous_HM, pitch, window, samp_jump=None, N_iter=1,
N_runs=float('Inf'), phase_tech='phase'):
return aqhm(signal, previous_HM, pitch, window, samp_jump, N_iter, N_runs,
phase_tech, eaQHM_flag=True)
"""
Parser for the three algorithms.
"""
def HM_run(func, signal, pitch, window, samp_jump=None, N_iter=1,
phase_tech='phase', func_options=None):
# Creates the output signal object and the dummy frequency vector.
HM = ModulatedSign(signal.n_harm, signal.size, signal.fs, phase_tech)
freq = np.zeros(signal.n_harm)
# Selects whether the extration will be performed with temporal jumps or
# not.
if samp_jump is None:
voiced_frames = np.nonzero(pitch.values)[0]
else:
jump = int(np.fix(max(samp_jump*signal.fs, 1.0)))
voiced_frames = np.array([], dtype=int)
for beg, end in zip(pitch.edges[::2], pitch.edges[1::2]):
voiced_frames = np.append(voiced_frames, np.arange(
beg+1, end-1, jump))
voiced_frames = np.append(voiced_frames, end)
# Run the algorithm in the selected voiced frames.
for frame in voiced_frames:
# Uses the pitch value and the harmonic definition f_k = k*f0 to create
# a frequency reference vector, which is employed to keep each component
# within a frquency band and thus, avoiding least-squares instability.
f0_ref = pitch.values[frame]*np.arange(1, signal.n_harm+1)/signal.fs
# Set some algorithm options.
if func is qhm_iteration:
if frame-1 in pitch.edges[::2]:
freq[:] = f0_ref
func_options = freq
elif func is aqhm_iteration:
func_options[2] = frame
# Core algorithm function.
coef, freq, HM.error[frame] = func(
signal.data[frame-window.N:frame+window.N+1],
f0_ref, window, signal.fs, 20.0, func_options,
N_iter)
# Updates frame parameter values in the 3-dimension storage array H.
HM.update_values(coef[:signal.n_harm], freq, frame)
# If the extraction was performed with temporal jumps, interpolate the
# results.
if samp_jump is not None:
HM.interpolate_samp(voiced_frames, pitch.values)
HM.synthesize()
HM.srer(signal.data, pitch.values)
HM.phase_edges(pitch.edges, window)
return HM
"""
Core QHM function.
"""
def qhm_iteration(data, f0_ref, window, fs, max_step, freq, N_iter=1):
# Initialize and allocate variables.
K = len(freq)
coef = np.zeros((2*K))
E = np.ones((window.length, 2*K), dtype=complex)
E = exp_matrix(E, freq, window, K)
E_windowed = np.ones((window.length, 2*K), dtype=complex)
windowed_data = (window.data*data).reshape(window.length, 1)
# Run the QHM algorithm N_iter times.
for k in range(N_iter):
# Calculate the a and b coeficients via least-squares.
coef = least_squares(E, E_windowed, windowed_data, window, K)
# Set a magnitude reference, which is used to detect and supress
# erroneous magnitude spikes.
mag_ref = np.abs(coef[0])
# Updates the frequency values.
freq, ro = freq_correction(coef[:K], coef[K:], freq, f0_ref, mag_ref, K,
max_step, fs)
# Updates the complex exponentials matrix.
E = exp_matrix(E, freq, window, K)
# Compute the final coefficients values.
coef = least_squares(E, E_windowed, windowed_data, window, K)
# This part is a workaround not present in the original references [1-3].
# It was created to detect and supress erroneous magnitude spikes, which
# degradate the final synthsized signal and consequently, its SRER.
# Alternatively, the magnitude signals could be smoothed after extraction.
# For more details, check the README file.
cond = (np.abs(coef[:K]) < 5.5*np.abs(coef[0]))
if not cond.all():
freq[~cond] = f0_ref[~cond]
# Updates the complex exponentials matrix with the modified frequencies.
E = exp_matrix(E, freq, window, K)
# Recalculate the final coefficients.
coef = least_squares(E, E_windowed, windowed_data, window, K)
# Calculate the mean squared error between the original frame and the
# synthesized one.
err = error_calc(windowed_data, E, coef, window)
return coef, freq, err
"""
Core aQHM and eaQHM function.
"""
def aqhm_iteration(data, f0_ref, window, fs, max_step, func_options,
N_iter=1):
# Initialize and allocate variables.
previous_HM = func_options[0]
eaQHM_flag = func_options[1]
frame = func_options[2]
freq = previous_HM.H[:, 2, frame]
windowed_data = (window.data*data).reshape(window.length, 1)
# Set a magnitude reference, which is used to detect and supress
# erroneous magnitude spikes.
mag_ref = np.abs(previous_HM.H[0, 0, frame])
# Ajust the phase frame.
extrap_phase_center = previous_HM.extrap_phase[:, frame].reshape(
previous_HM.n_harm, 1)
phase_frame = previous_HM.extrap_phase[:, frame-window.N:frame+window.N+1] - \
extrap_phase_center
# Initialize the coefficients.
coef = np.vstack((previous_HM.H[:, 0, frame].reshape(previous_HM.n_harm, 1) *
np.exp(1j*extrap_phase_center), np.zeros((previous_HM.n_harm, 1))))[:, 0]
# Initialize the matrices.
E = np.ones((window.length, 2*previous_HM.n_harm), dtype=complex)
E_ro = np.ones((window.length, 2*previous_HM.n_harm), dtype=complex)
E_windowed = np.ones((window.length, 2*previous_HM.n_harm), dtype=complex)
E[:, :previous_HM.n_harm] = np.exp(1j*phase_frame.T)
# If the eaQHM algorithm was selected, ajust the exponential matrix with
# the normalized magnitude.
if eaQHM_flag:
mag_center = previous_HM.H[:, 0, frame].reshape(previous_HM.n_harm, 1)
mag_frame = previous_HM.H[:, 0, frame-window.N:frame+window.N+1] / \
mag_center
E[:, :previous_HM.n_harm] = mag_frame.T*E[:, :previous_HM.n_harm]
E[:, previous_HM.n_harm:] = E[:, :previous_HM.n_harm] * \
window.len_vec.reshape(window.length, 1)
# Run the aQHM/eaQHM algorithm N_iter times.
for k in range(N_iter):
# Calculate the a and b coeficients via least-squares.
coef = least_squares(E, E_windowed, windowed_data, window,
previous_HM.n_harm)
# Updates the frequency values.
freq, ro = freq_correction(coef[:previous_HM.n_harm],
coef[previous_HM.n_harm:], freq, f0_ref,
mag_ref, previous_HM.n_harm, max_step, fs)
# Updates the complex exponentials matrix.
E = E*exp_matrix(E_ro, ro/(2*np.pi), window, previous_HM.n_harm)
# Compute the final coefficients values.
coef = least_squares(E, E_windowed, windowed_data, window,
previous_HM.n_harm)
# This part is a workaround not present in the original references [1-3].
# It was created to detect and supress erroneous magnitude spikes, which
# degradate the final synthsized signal and consequently, its SRER.
# Alternatively, the magnitude signals could be smoothed after extraction.
# For more details, check the README file.
cond = (np.abs(coef[:previous_HM.n_harm]) < 5.5*mag_ref)
if not cond.all():
freq[~cond] = f0_ref[~cond]
# Since that the troubling aQHM/eaQHM exponentials are degradating the
# results, they are replaced by the QHM version, which is more stable.
E[:, ~np.append(cond, cond)] = exp_matrix(E_ro, freq, window,
previous_HM.n_harm)[:, ~np.append(cond, cond)]
# Recalculate the final coefficients.
coef = least_squares(E, E_windowed, windowed_data, window,
previous_HM.n_harm)
# Calculate the mean squared error between the original frame and the
# synthsized one.
err = error_calc(windowed_data, E, coef, window)
return coef, freq, err
"""
--------------------------------------------
Auxiliary Functions.
--------------------------------------------
"""
"""
Calculate the a and b coeficients via least-squares method.
"""
def least_squares(E, E_windowed, windowed_data, window, K):
R = np.zeros((2*K, 2*K), dtype=complex)
B = np.zeros((window.length, 1), dtype=complex)
E_windowed[:, :] = E*window.data.reshape(window.length, 1)
R = E_windowed.conj().T.dot(E_windowed)
B = E_windowed.conj().T.dot(windowed_data)
coef = np.linalg.solve(R, B)[:, 0]
return coef
"""
Calculates the frequency mismatch and updates the frequency values.
"""
def freq_correction(a, b, freq, f0_ref, mag_ref, n_harm, max_step, fs):
old_freq = np.zeros(n_harm)
old_freq[:] = freq[:]
ro = (a.real*b.imag-a.imag*b.real)/(np.abs(a)*np.abs(a))
# If the mismatch is too high (>20Hz), the frequency update is satured to
# this value. This avoids big fluctuations, which can spoil the algorithms
# convergence as whole.
over_ro = np.abs(ro) > max_step*2*np.pi/fs
ro[over_ro] = np.sign(ro[over_ro])*(max_step*2*np.pi/fs)
freq = freq+ro/(2*np.pi)
# Checks whether each component frequency lies within its spectral band and
# also checks whether there are magnitude spikes.
cond = ((np.round(freq/f0_ref[0]) != np.arange(n_harm)+1) |
(freq > 0.5) | (freq < 0) | (np.abs(a) > 5.5*mag_ref))
freq[cond] = f0_ref[cond]
return freq, (freq-old_freq)*(2*np.pi)
"""
Calculate the mean squared error between the original frame and the
synthsized one.
"""
def error_calc(windowed_data, E, coef, window):
h = E.dot(coef)
err = np.sum((windowed_data-2*h.real*window.data)**2)
return err
"""
Mounts the complex exponentials matrix.
"""
def exp_matrix(E, freq, window, K):
E[window.N+1:, :K] = np.exp(1j*np.pi*2*freq)
E[window.N+1:, :K] = np.cumprod(E[window.N+1:, :K], axis=0)
E[:window.N, :K] = np.conj(E[window.N+1:, :K][::-1, :])
E[:, K:] = E[:, :K]*window.len_vec.reshape(window.length, 1)
return E
"""
Some side functions found in reference [2].
"""
def g0(x, N):
if x != 0:
return np.sin((2*N+1)*x/2)/np.sin(x/2)
else:
return 2*N+1
def g1(x, N):
if x != 0:
return 1j*((np.sin(N*x)/(2*np.sin(x/2)**2)) -
N*(np.cos((2*N+1)*x/2)/np.sin(x/2)))
else:
return 0
def R_eq(delta_f, func, window):
return (window.a0*func(2*np.pi*delta_f, window.N) +
func(2*np.pi*(delta_f+1./(2*window.N)), window.N)*window.a1 +
func(2*np.pi*(delta_f-1./(2*window.N)), window.N)*window.a1 +
func(2*np.pi*(delta_f+1./window.N), window.N)*window.a2 +
func(2*np.pi*(delta_f-1./window.N), window.N)*window.a2) | AMFM-decompy | /AMFM_decompy-1.0.11.tar.gz/AMFM_decompy-1.0.11/amfm_decompy/pyQHM.py | pyQHM.py |
from typing import List
import numpy as np
import pandas as pd
class CallablePdSeries(pd.Series):
"""a pd.Series object with a dummy .__call__() method"""
def __call__(self, *args, **kwargs):
return pd.Series(self)
class CallablePdDataFrame(pd.DataFrame):
"""a pd.DataFrame object with a dummy .__call__() method"""
def __call__(self, *args, **kwargs):
return pd.DataFrame(self)
class CallableNpArray(np.ndarray):
"""a np.ndarray object with a dummy .__call__() method"""
def __new__(cls, arr):
obj = np.asarray(arr).view(cls)
return obj
def __array_finalize__(self, obj):
if obj is None: return
def __array_wrap__(self, out_arr, context=None):
return np.ndarray.__array_wrap__(self, out_arr, context).view(np.ndarray)
def __call__(self, *args, **kwargs):
return self.view(np.ndarray)
class CallableList(list):
"""a list object with a dummy .__call__() method"""
def __call__(self, col=None):
return list(self)
def make_callable(obj):
"""
returns the obj with a dummy __call__ method attached.
This means that calling `obj` and 'obj(()' and 'obj(arg1, arg2='foo') result in the same thing.
Used to make @property that in subclass may be callable.
"""
if isinstance(obj, np.ndarray):
return CallableNpArray(obj)
if isinstance(obj, pd.Series):
return CallablePdSeries(obj)
if isinstance(obj, pd.DataFrame):
return CallablePdDataFrame(obj)
if isinstance(obj, list):
return CallableList(obj)
class DummyCallable(type(obj)):
def __call__(self, *args, **kwargs):
return type(obj)(self)
return DummyCallable(obj)
class DefaultDfList(pd.DataFrame):
""""
You have the set source_list manually!
e.g.
dfl = DefaultDfList(df1)
dfl.source_list = [df1, df2]
"""
_internal_names = list(pd.DataFrame._internal_names) + ['source_list']
_internal_names_set = set(_internal_names)
def __call__(self, index=None):
if index is not None:
return self.source_list[index]
else:
return pd.DataFrame(self)
@property
def _constructor(self):
return DefaultDfList
class DefaultSeriesList(pd.Series):
_internal_names = list(pd.Series._internal_names) + ['source_list']
_internal_names_set = set(_internal_names)
def __call__(self, index=None):
if index is not None:
return self.source_list[index]
else:
return pd.Series(self)
@property
def _constructor(self):
return DefaultSeriesList
class DefaultNpArrayList(np.ndarray):
def __new__(cls, default_array, source_list):
obj = np.asarray(default_array).view(cls)
obj.source_list = source_list
return obj
def __array_finalize__(self, obj):
if obj is None: return
self.source_list = getattr(obj, 'source_list', None)
def __array_wrap__(self, out_arr, context=None):
return np.ndarray.__array_wrap__(self, out_arr, context).view(np.ndarray)
def __call__(self, index=None):
if index is not None:
return self.source_list[index]
return self.view(np.ndarray)
def default_list(source_list:List, default_index:int):
"""
Normally gives the default_index item in a list.
If used as a callable, you can specify a specific index.
Use to make @property that you can pass optional index parameter to
"""
if isinstance(source_list[default_index], pd.DataFrame):
df_list = DefaultDfList(source_list[default_index])
df_list.source_list = source_list
return df_list
if isinstance(source_list[default_index], pd.Series):
s_list = DefaultSeriesList(source_list[default_index])
s_list.source_list = source_list
return s_list
if isinstance(source_list[default_index], np.ndarray):
a_list = DefaultNpArrayList(source_list[default_index], source_list)
return a_list
class DefaultList(type(source_list[default_index])):
def __new__(cls, default_value, source_list):
obj = type(source_list[default_index]).__new__(cls, default_value)
return obj
def __init__(self, default_value, source_list):
super().__init__()
self.source_list = source_list
self.default_type = type(default_value)
def __call__(self, index=None):
if index is not None:
return self.source_list[index]
else:
return self.default_type(self)
return DefaultList(source_list[default_index], source_list)
class DefaultColumn2dNpArray(np.ndarray):
"""
a 2d numpy array that by default returns only a single default column array.
If used as a callable, you can specify a specific column to return.
Used to make @property that you can pass a column parameter to
"""
def __new__(cls, full_array, default_col):
obj = np.asarray(full_array[:, default_col]).view(cls)
obj.full_array = full_array
return obj
def __array_finalize__(self, obj):
if obj is None: return
self.full_array = getattr(obj, 'full_array', None)
def __array_wrap__(self, out_arr, context=None):
return np.ndarray.__array_wrap__(self, out_arr, context).view(np.ndarray)
def __call__(self, col=None):
if col is not None:
return self.full_array[:, col]
return self.view(np.ndarray)
def default_2darray(array_2d:np.ndarray, default_column:int):
"""
when used as property returns default_column col of array_2d
when used as callable __call__(col) returns column col of array_2d
Used to make a @property that you can pass an optional column parameter to
"""
return DefaultColumn2dNpArray(array_2d, default_column) | AMLBID | /Explainer/make_callables.py | make_callables.py |
__all__ = ['BaseExplainer',
'ClassifierExplainer',
'RegressionExplainer',
'RandomForestClassifierExplainer',
'RandomForestRegressionExplainer',
'XGBClassifierExplainer',
'XGBRegressionExplainer',
'ClassifierBunch', # deprecated
'RegressionBunch', # deprecated
'RandomForestClassifierBunch', # deprecated
'RandomForestRegressionBunch', # deprecated
]
from abc import ABC
import base64
from pathlib import Path
from typing import List, Union
import numpy as np
import pandas as pd
from pdpbox import pdp
import shap
from dtreeviz.trees import ShadowDecTree, dtreeviz
from sklearn.pipeline import Pipeline
from sklearn.metrics import roc_auc_score, accuracy_score, f1_score
from sklearn.metrics import precision_score, recall_score, log_loss
from sklearn.metrics import mean_squared_error, mean_absolute_error, r2_score
from sklearn.metrics import average_precision_score
from .explainer_methods import *
from .explainer_plots import *
from .make_callables import make_callable, default_list, default_2darray
import plotly.io as pio
pio.templates.default = "none"
class BaseExplainer(ABC):
""" """
def __init__(self, model,recommended_config, X, y=None, permutation_metric=r2_score,
shap="guess", X_background=None, model_output="raw",
cats=None, idxs=None, index_name=None, target=None,
descriptions=None,
n_jobs=None, permutation_cv=None, na_fill=-999):
"""Defines the basic functionality that is shared by both
ClassifierExplainer and RegressionExplainer.
Args:
model: a model with a scikit-learn compatible .fit and .predict methods
X (pd.DataFrame): a pd.DataFrame with your model features
y (pd.Series): Dependent variable of your model, defaults to None
permutation_metric (function or str): is a scikit-learn compatible
metric function (or string). Defaults to r2_score
shap (str): type of shap_explainer to fit: 'tree', 'linear', 'kernel'.
Defaults to 'guess'.
X_background (pd.DataFrame): background X to be used by shap
explainers that need a background dataset (e.g. shap.KernelExplainer
or shap.TreeExplainer with boosting models and
model_output='probability').
model_output (str): model_output of shap values, either 'raw',
'logodds' or 'probability'. Defaults to 'raw' for regression and
'probability' for classification.
cats ({dict, list}): dict of features that have been
onehotencoded. e.g. cats={'Sex':['Sex_male', 'Sex_female']}.
If all encoded columns are underscore-seperated (as above), can simply
pass a list of prefixes: cats=['Sex']. Allows to
group onehot encoded categorical variables together in
various plots. Defaults to None.
idxs (pd.Series): list of row identifiers. Can be names, id's, etc.
Defaults to X.index.
index_name (str): identifier for row indexes. e.g. index_name='Passenger'.
Defaults to X.index.name or idxs.name.
target: name of the predicted target, e.g. "Survival",
"Ticket price", etc. Defaults to y.name.
n_jobs (int): for jobs that can be parallelized using joblib,
how many processes to split the job in. For now only used
for calculating permutation importances. Defaults to None.
permutation_cv (int): If not None then permutation importances
will get calculated using cross validation across X.
This is for calculating permutation importances against
X_train. Defaults to None
na_fill (int): The filler used for missing values, defaults to -999.
"""
self._params_dict = dict(
shap=shap, model_output=model_output, cats=cats,
descriptions=descriptions, target=target, n_jobs=n_jobs,
permutation_cv=n_jobs, na_fill=na_fill, recommended_config=recommended_config)
if isinstance(model, Pipeline):
self.X, self.model = split_pipeline(model, X)
self.X_background, _ = split_pipeline(model, X_background, verbose=0)
else:
self.X, self.X_background = X, X_background
self.model = model
if y is not None:
self.y = pd.Series(y)
self.y_missing = False
else:
self.y = pd.Series(np.full(len(X), np.nan))
self.y_missing = True
if self.y.name is None: self.y.name = 'Target'
self.metric = permutation_metric
if shap == "guess":
shap_guess = guess_shap(self.model)
if shap_guess is not None:
model_str = str(type(self.model))\
.replace("'", "").replace("<", "").replace(">", "")\
.split(".")[-1]
#print(f"Note: shap=='guess' so guessing for {model_str}"
#f" shap='{shap_guess}'...")
self.shap = shap_guess
else:
raise ValueError(
"Parameter shap='gues'', but failed to to guess the type of "
"shap explainer to use. "
"Please explicitly pass a `shap` parameter to the explainer, "
"e.g. shap='tree', shap='linear', etc.")
else:
assert shap in ['tree', 'linear', 'deep', 'kernel'], \
"Only shap='guess', 'tree', 'linear', 'deep', or ' kernel' allowed."
self.shap = shap
self.model_output = model_output
self.cats, self.cats_dict = parse_cats(self.X, cats)
if idxs is not None:
assert len(idxs) == len(self.X) == len(self.y), \
("idxs should be same length as X but is not: "
f"len(idxs)={len(idxs)} but len(X)={len(self.X)}!")
self.idxs = pd.Index(idxs, dtype=str)
else:
self.idxs = X.index.astype(str)
self.X.index = self.idxs
self.y.index = self.idxs
if index_name is None:
if self.idxs.name is not None:
self.index_name = self.idxs.name.capitalize()
else:
self.index_name = "Index"
else:
self.index_name = index_name.capitalize()
self.descriptions = {} if descriptions is None else descriptions
self.target = target if target is not None else self.y.name
self.n_jobs = n_jobs
self.permutation_cv = permutation_cv
self.na_fill = na_fill
self.columns = self.X.columns.tolist()
self.pos_label = None
self.units = ""
self.is_classifier = False
self.is_regression = False
self.interactions_should_work = True
self.recommended_config=recommended_config
_ = self.shap_explainer
@classmethod
def from_file(cls, filepath):
"""Load an Explainer from file. Depending on the suffix of the filepath
will either load with pickle ('.pkl'), dill ('.dill') or joblib ('joblib').
If no suffix given, will try with joblib.
Args:
filepath {str, Path} the location of the stored Explainer
returns:
Explainer object
"""
filepath = Path(filepath)
if str(filepath).endswith(".pkl") or str(filepath).endswith(".pickle"):
import pickle
return pickle.load(open(filepath, "rb"))
elif str(filepath).endswith(".dill"):
import dill
return dill.load(open(filepath, "rb"))
else:
if not filepath.exists():
if (filepath.parent / (filepath.name + ".joblib")).exists():
filepath = filepath.parent / (filepath.name + ".joblib")
else:
raise ValueError(f"Cannot find file: {str(filepath)}")
import joblib
return joblib.load(filepath)
def dump(self, filepath):
"""
Dump the current Explainer to file. Depending on the suffix of the filepath
will either dump with pickle ('.pkl'), dill ('.dill') or joblib ('joblib').
If no suffix given, will dump with joblib and add '.joblib'
Args:
filepath (str, Path): filepath where to save the Explainer.
"""
filepath = Path(filepath)
if str(filepath).endswith(".pkl") or str(filepath).endswith(".pickle"):
import pickle
pickle.dump(self, open(str(filepath), "wb"))
elif str(filepath).endswith(".dill"):
import dill
dill.dump(self, open(str(filepath), "wb"))
elif str(filepath).endswith(".joblib"):
import joblib
joblib.dump(self, filepath)
else:
filepath = Path(filepath)
filepath = filepath.parent / (filepath.name + ".joblib")
import joblib
joblib.dump(self, filepath)
def to_yaml(self, filepath=None, return_dict=False,
modelfile="model.pkl",
datafile="data.csv",
index_col=None,
target_col=None,
explainerfile="explainer.joblib",
dashboard_yaml="dashboard.yaml"):
"""Returns a yaml configuration for the current Explainer
that can be used by the explainerdashboard CLI. Recommended filename
is `explainer.yaml`.
Args:
filepath ({str, Path}, optional): Filepath to dump yaml. If None
returns the yaml as a string. Defaults to None.
return_dict (bool, optional): instead of yaml return dict with config.
modelfile (str, optional): filename of model dump. Defaults to
`model.pkl`
datafile (str, optional): filename of datafile. Defaults to
`data.csv`.
index_col (str, optional): column to be used for idxs. Defaults to
self.idxs.name.
target_col (str, optional): column to be used for to split X and y
from datafile. Defaults to self.target.
explainerfile (str, optional): filename of explainer dump. Defaults
to `explainer.joblib`.
dashboard_yaml (str, optional): filename of the dashboard.yaml
configuration file. This will be used to determine which
properties to calculate before storing to disk.
Defaults to `dashboard.yaml`.
"""
import oyaml as yaml
yaml_config = dict(
explainer=dict(
modelfile=modelfile,
datafile=datafile,
explainerfile=explainerfile,
data_target=self.target,
data_index=self.idxs.name,
explainer_type="classifier" if self.is_classifier else "regression",
dashboard_yaml=dashboard_yaml,
params=self._params_dict))
if return_dict:
return yaml_config
if filepath is not None:
yaml.dump(yaml_config, open(filepath, "w"))
return
return yaml.dump(yaml_config)
def __len__(self):
return len(self.X)
def __contains__(self, index):
if self.get_int_idx(index) is not None:
return True
return False
def check_cats(self, col1, col2=None):
"""check whether should use cats=True based on col1 (and col2)
Args:
col1: First column
col2: Second column (Default value = None)
Returns:
Boolean whether cats should be True
"""
if col2 is None:
if col1 in self.columns:
return False
elif col1 in self.columns_cats:
return True
raise ValueError(f"Can't find {col1}.")
if col1 not in self.columns and col1 not in self.columns_cats:
raise ValueError(f"Can't find {col1}.")
if col2 not in self.columns and col2 not in self.columns_cats:
raise ValueError(f"Can't find {col2}.")
if col1 in self.columns and col2 in self.columns:
return False
if col1 in self.columns_cats and col2 in self.columns_cats:
return True
if col1 in self.columns_cats and not col2 in self.columns_cats:
raise ValueError(
f"{col1} is categorical but {col2} is not in columns_cats")
if col2 in self.columns_cats and not col1 in self.columns_cats:
raise ValueError(
f"{col2} is categorical but {col1} is not in columns_cats")
@property
def shap_explainer(self):
""" """
if not hasattr(self, '_shap_explainer'):
X_str = ", X_background" if self.X_background is not None else 'X'
NoX_str = ", X_background" if self.X_background is not None else ''
if self.shap == 'tree':
print("Generating self.shap_explainer = "
f"shap.TreeExplainer(model{NoX_str})")
self._shap_explainer = shap.TreeExplainer(self.model, self.X_background)
elif self.shap=='linear':
if self.X_background is None:
print(
"Warning: shap values for shap.LinearExplainer get "
"calculated against X_background, but paramater "
"X_background=None, so using X instead")
print(f"Generating self.shap_explainer = shap.LinearExplainer(model, {X_str})...")
self._shap_explainer = shap.LinearExplainer(self.model,
self.X_background if self.X_background is not None else self.X)
elif self.shap=='deep':
print(f"Generating self.shap_explainer = "
f"shap.DeepExplainer(model{NoX_str})")
self._shap_explainer = shap.DeepExplainer(self.model)
elif self.shap=='kernel':
if self.X_background is None:
print(
"Warning: shap values for shap.LinearExplainer get "
"calculated against X_background, but paramater "
"X_background=None, so using X instead")
print("Generating self.shap_explainer = "
f"shap.KernelExplainer(model, {X_str})...")
self._shap_explainer = shap.KernelExplainer(self.model,
self.X_background if self.X_background is not None else self.X)
return self._shap_explainer
def get_int_idx(self, index):
"""Turn str index into an int index
Args:
index(str or int):
Returns:
int index
"""
if isinstance(index, int):
if index >= 0 and index < len(self):
return index
elif isinstance(index, str):
if self.idxs is not None and index in self.idxs:
return self.idxs.get_loc(index)
return None
def get_index(self, index):
"""Turn int index into a str index
Args:
index(str or int):
Returns:
str index
"""
if isinstance(index, int) and index >= 0 and index < len(self):
return self.idxs[index]
elif isinstance(index, str) and index in self.idxs:
return index
return None
def random_index(self, y_min=None, y_max=None, pred_min=None, pred_max=None,
return_str=False, **kwargs):
"""random index following constraints
Args:
y_min: (Default value = None)
y_max: (Default value = None)
pred_min: (Default value = None)
pred_max: (Default value = None)
return_str: (Default value = False)
**kwargs:
Returns:
if y_values is given select an index for which y in y_values
if return_str return str index from self.idxs
"""
if pred_min is None:
pred_min = self.preds.min()
if pred_max is None:
pred_max = self.preds.max()
if not self.y_missing:
if y_min is None: y_min = self.y.min()
if y_max is None: y_max = self.y.max()
potential_idxs = self.y[(self.y>=y_min) &
(self.y <= y_max) &
(self.preds>=pred_min) &
(self.preds <= pred_max)].index
else:
potential_idxs = self.y[(self.preds>=pred_min) &
(self.preds <= pred_max)].index
if len(potential_idxs) > 0:
idx = np.random.choice(potential_idxs)
else:
return None
if return_str:
return idx
return idxs.get_loc(idx)
@property
def preds(self):
"""returns model model predictions"""
if not hasattr(self, '_preds'):
#print("Calculating predictions...", flush=True)
self._preds = self.model.predict(self.X).astype(np.float64)
return self._preds
@property
def pred_percentiles(self):
"""returns percentile rank of model predictions"""
if not hasattr(self, '_pred_percentiles'):
print("Calculating prediction percentiles...", flush=True)
self._pred_percentiles = (pd.Series(self.preds)
.rank(method='min')
.divide(len(self.preds))
.values)
return make_callable(self._pred_percentiles)
def columns_ranked_by_shap(self, cats=False, pos_label=None):
"""returns the columns of X, ranked by mean abs shap value
Args:
cats: Group categorical together (Default value = False)
pos_label: (Default value = None)
Returns:
list of columns
"""
if cats:
return self.mean_abs_shap_cats(pos_label).Feature.tolist()
else:
return self.mean_abs_shap(pos_label).Feature.tolist()
def n_features(self, cats=False):
"""number of features with cats=True or cats=False
Args:
cats: (Default value = False)
Returns:
int, number of features
"""
if cats:
return len(self.columns_cats)
else:
return len(self.columns)
def equivalent_col(self, col):
"""Find equivalent col in columns_cats or columns
if col in self.columns, return equivalent col in self.columns_cats,
e.g. equivalent_col('Gender_Male') -> 'Gender'
if col in self.columns_cats, return first one hot encoded col,
e.g. equivalent_col('Gender') -> 'Gender_Male'
(useful for switching between cats=True and cats=False, while
maintaining column selection)
Args:
col: col to get equivalent col for
Returns:
col
"""
if col in self.cats:
# first onehot-encoded columns
return self.cats_dict[col][0]
elif col in self.columns:
# the cat that the col belongs to
return [k for k, v in self.cats_dict.items() if col in v][0]
return None
def get_row_from_input(self, inputs:List, ranked_by_shap=False):
"""returns a single row pd.DataFrame from a given list of *inputs"""
if len(inputs)==1 and isinstance(inputs[0], list):
inputs = inputs[0]
elif len(inputs)==1 and isinstance(inputs[0], tuple):
inputs = list(inputs[0])
else:
inputs = list(inputs)
if len(inputs) == len(self.columns_cats):
cols = self.columns_ranked_by_shap(cats=True) if ranked_by_shap else self.columns_cats
df = pd.DataFrame(dict(zip(cols, inputs)), index=[0]).fillna(self.na_fill)
return df[self.columns_cats]
elif len(inputs) == len(self.columns):
cols = self.columns_ranked_by_shap() if ranked_by_shap else self.columns
df = pd.DataFrame(dict(zip(cols, inputs)), index=[0]).fillna(self.na_fill)
return df[self.columns]
else:
raise ValueError(f"len inputs {len(inputs)} should be the same length as either "
f"explainer.columns_cats ({len(self.columns_cats)}) or "
f"explainer.columns ({len(self.columns)})!")
def description(self, col):
"""returns the written out description of what feature col means
Args:
col(str): col to get description for
Returns:
str, description
"""
if col in self.descriptions.keys():
return self.descriptions[col]
elif self.equivalent_col(col) in self.descriptions.keys():
return self.descriptions[self.equivalent_col(col)]
return ""
def description_list(self, cols):
"""returns a list of descriptions of a list of cols
Args:
cols(list): cols to be converted to descriptions
Returns:
list of descriptions
"""
return [self.description(col) for col in cols]
def get_col(self, col):
"""return pd.Series with values of col
For categorical feature reverse engineers the onehotencoding.
Args:
col: column tof values to be returned
Returns:
pd.Series with values of col
"""
assert col in self.columns or col in self.cats, \
f"{col} not in columns!"
if col in self.X.columns:
return self.X[col]
elif col in self.cats:
return pd.Series(retrieve_onehot_value(
self.X, col, self.cats_dict[col]), name=col)
def get_col_value_plus_prediction(self, col, index=None, X_row=None, pos_label=None):
"""return value of col and prediction for either index or X_row
Args:
col: feature col
index (str or int, optional): index row
X_row (single row pd.DataFrame, optional): single row of features
Returns:
tupe(value of col, prediction for index)
"""
assert (col in self.X.columns) or (col in self.cats),\
f"{col} not in columns of dataset"
if index is not None:
assert index in self, f"index {index} not found"
idx = self.get_int_idx(index)
if col in self.X.columns:
col_value = self.X[col].iloc[idx]
elif col in self.cats:
col_value = retrieve_onehot_value(self.X, col, self.cats_dict[col])[idx]
if self.is_classifier:
if pos_label is None:
pos_label = self.pos_label
prediction = self.pred_probas(pos_label)[idx]
if self.model_output == 'probability':
prediction = 100*prediction
elif self.is_regression:
prediction = self.preds[idx]
return col_value, prediction
elif X_row is not None:
assert X_row.shape[0] == 1, "X_Row should be single row dataframe!"
if ((len(X_row.columns) == len(self.X_cats.columns)) and
(X_row.columns == self.X_cats.columns).all()):
X_row = X_cats_to_X(X_row, self.cats_dict, self.X.columns)
else:
assert (X_row.columns == self.X.columns).all(), \
"X_row should have the same columns as self.X or self.X_cats!"
if col in X_row.columns:
col_value = X_row[col].item()
elif col in self.cats:
col_value = retrieve_onehot_value(X_row, col, self.cats_dict[col]).item()
if self.is_classifier:
if pos_label is None:
pos_label = self.pos_label
prediction = self.model.predict_proba(X_row)[0][pos_label]
if self.model_output == 'probability':
prediction = 100*prediction
elif self.is_regression:
prediction = self.model.predict(X_row)[0]
return col_value, prediction
else:
raise ValueError("You need to pass either index or X_row!")
@property
def permutation_importances(self):
"""Permutation importances """
if not hasattr(self, '_perm_imps'):
print("Calculating importances...", flush=True)
self._perm_imps = cv_permutation_importances(
self.model, self.X, self.y, self.metric,
cv=self.permutation_cv,
n_jobs=self.n_jobs,
needs_proba=self.is_classifier)
return make_callable(self._perm_imps)
@property
def permutation_importances_cats(self):
"""permutation importances with categoricals grouped"""
if not hasattr(self, '_perm_imps_cats'):
self._perm_imps_cats = cv_permutation_importances(
self.model, self.X, self.y, self.metric,
cats_dict=self.cats_dict,
cv=self.permutation_cv,
n_jobs=self.n_jobs,
needs_proba=self.is_classifier)
return make_callable(self._perm_imps_cats)
@property
def X_cats(self):
"""X with categorical variables grouped together"""
if not hasattr(self, '_X_cats'):
self._X_cats = merge_categorical_columns(self.X, self.cats_dict)
return self._X_cats
@property
def columns_cats(self):
"""columns of X with categorical features grouped"""
if not hasattr(self, '_columns_cats'):
self._columns_cats = self.X_cats.columns.tolist()
return self._columns_cats
@property
def shap_base_value(self):
"""the intercept for the shap values.
(i.e. 'what would the prediction be if we knew none of the features?')
"""
if not hasattr(self, '_shap_base_value'):
# CatBoost needs shap values calculated before expected value
if not hasattr(self, "_shap_values"):
_ = self.shap_values
self._shap_base_value = self.shap_explainer.expected_value
if isinstance(self._shap_base_value, np.ndarray):
# shap library now returns an array instead of float
self._shap_base_value = self._shap_base_value.item()
return make_callable(self._shap_base_value)
@property
def shap_values(self):
"""SHAP values calculated using the shap library"""
if not hasattr(self, '_shap_values'):
print("Calculating shap values...", flush=True)
self._shap_values = self.shap_explainer.shap_values(self.X)
return make_callable(self._shap_values)
@property
def shap_values_cats(self):
"""SHAP values when categorical features have been grouped"""
if not hasattr(self, '_shap_values_cats'):
self._shap_values_cats = merge_categorical_shap_values(
self.X, self.shap_values, self.cats_dict)
return make_callable(self._shap_values_cats)
@property
def shap_interaction_values(self):
"""SHAP interaction values calculated using shap library"""
assert self.shap != 'linear', \
"Unfortunately shap.LinearExplainer does not provide " \
"shap interaction values! So no interactions tab!"
if not hasattr(self, '_shap_interaction_values'):
print("Calculating shap interaction values...", flush=True)
if self.shap == 'tree':
print("Reminder: TreeShap computational complexity is O(TLD^2), "
"where T is the number of trees, L is the maximum number of"
" leaves in any tree and D the maximal depth of any tree. So "
"reducing these will speed up the calculation.",
flush=True)
self._shap_interaction_values = \
self.shap_explainer.shap_interaction_values(self.X)
return make_callable(self._shap_interaction_values)
@property
def shap_interaction_values_cats(self):
"""SHAP interaction values with categorical features grouped"""
if not hasattr(self, '_shap_interaction_values_cats'):
self._shap_interaction_values_cats = \
merge_categorical_shap_interaction_values(
self.shap_interaction_values, self.X, self.X_cats, self.cats_dict)
return make_callable(self._shap_interaction_values_cats)
@property
def mean_abs_shap(self):
"""Mean absolute SHAP values per feature."""
if not hasattr(self, '_mean_abs_shap'):
self._mean_abs_shap = mean_absolute_shap_values(
self.columns, self.shap_values)
return make_callable(self._mean_abs_shap)
@property
def mean_abs_shap_cats(self):
"""Mean absolute SHAP values with categoricals grouped."""
if not hasattr(self, '_mean_abs_shap_cats'):
self._mean_abs_shap_cats = mean_absolute_shap_values(
self.columns, self.shap_values, self.cats_dict)
return make_callable(self._mean_abs_shap_cats)
def calculate_properties(self, include_interactions=True):
"""Explicitely calculates all lazily calculated properties.
Useful so that properties are not calculate multiple times in
parallel when starting a dashboard.
Args:
include_interactions(bool, optional, optional): shap interaction values can take a long
time to compute for larger datasets with more features. Therefore you
can choose not to calculate these, defaults to True
Returns:
"""
_ = (self.preds, self.pred_percentiles,
self.shap_base_value, self.shap_values,
self.mean_abs_shap)
if not self.y_missing:
_ = self.permutation_importances
if self.cats is not None:
_ = (self.mean_abs_shap_cats, self.X_cats,
self.shap_values_cats)
if self.interactions_should_work and include_interactions:
_ = self.shap_interaction_values
if self.cats is not None:
_ = self.shap_interaction_values_cats
def metrics(self, *args, **kwargs):
"""returns a dict of metrics.
Implemented by either ClassifierExplainer or RegressionExplainer
"""
return {}
def mean_abs_shap_df(self, topx=None, cutoff=None, cats=False, pos_label=None):
"""sorted dataframe with mean_abs_shap
returns a pd.DataFrame with the mean absolute shap values per features,
sorted rom highest to lowest.
Args:
topx(int, optional, optional): Only return topx most importance features, defaults to None
cutoff(float, optional, optional): Only return features with mean abs shap of at least cutoff, defaults to None
cats(bool, optional, optional): group categorical variables, defaults to False
pos_label: (Default value = None)
Returns:
pd.DataFrame: shap_df
"""
shap_df = self.mean_abs_shap_cats(pos_label) if cats \
else self.mean_abs_shap(pos_label)
if topx is None: topx = len(shap_df)
if cutoff is None: cutoff = shap_df['MEAN_ABS_SHAP'].min()
return (shap_df[shap_df['MEAN_ABS_SHAP'] >= cutoff]
.sort_values('MEAN_ABS_SHAP', ascending=False).head(topx))
def shap_top_interactions(self, col, topx=None, cats=False, pos_label=None):
"""returns the features that interact with feature col in descending order.
if shap interaction values have already been calculated, use those.
Otherwise use shap approximate_interactions or simply mean abs shap.
Args:
col(str): feature for which you want to get the interactions
topx(int, optional, optional): Only return topx features, defaults to None
cats(bool, optional, optional): Group categorical features, defaults to False
pos_label: (Default value = None)
Returns:
list: top_interactions
"""
if cats:
if hasattr(self, '_shap_interaction_values'):
col_idx = self.X_cats.columns.get_loc(col)
top_interactions = self.X_cats.columns[
np.argsort(
-np.abs(self.shap_interaction_values_cats(
pos_label)[:, col_idx, :]).mean(0))].tolist()
else:
top_interactions = self.mean_abs_shap_cats(pos_label)\
.Feature.values.tolist()
top_interactions.insert(0, top_interactions.pop(
top_interactions.index(col))) #put col first
if topx is None: topx = len(top_interactions)
return top_interactions[:topx]
else:
if hasattr(self, '_shap_interaction_values'):
col_idx = self.X.columns.get_loc(col)
top_interactions = self.X.columns[np.argsort(-np.abs(
self.shap_interaction_values(
pos_label)[:, col_idx, :]).mean(0))].tolist()
else:
if hasattr(shap, "utils"):
interaction_idxs = shap.utils.approximate_interactions(
col, self.shap_values(pos_label), self.X)
elif hasattr(shap, "common"):
# shap < 0.35 has approximate interactions in common
interaction_idxs = shap.common.approximate_interactions(
col, self.shap_values(pos_label), self.X)
top_interactions = self.X.columns[interaction_idxs].tolist()
#put col first
top_interactions.insert(0, top_interactions.pop(-1))
if topx is None: topx = len(top_interactions)
return top_interactions[:topx]
def shap_interaction_values_by_col(self, col, cats=False, pos_label=None):
"""returns the shap interaction values[np.array(N,N)] for feature col
Args:
col(str): features for which you'd like to get the interaction value
cats(bool, optional, optional): group categorical, defaults to False
pos_label: (Default value = None)
Returns:
np.array(N,N): shap_interaction_values
"""
if cats:
return self.shap_interaction_values_cats(pos_label)[:,
self.X_cats.columns.get_loc(col), :]
else:
return self.shap_interaction_values(pos_label)[:,
self.X.columns.get_loc(col), :]
def permutation_importances_df(self, topx=None, cutoff=None, cats=False,
pos_label=None):
"""dataframe with features ordered by permutation importance.
For more about permutation importances.
see https://explained.ai/rf-importance/index.html
Args:
topx(int, optional, optional): only return topx most important
features, defaults to None
cutoff(float, optional, optional): only return features with importance
of at least cutoff, defaults to None
cats(bool, optional, optional): Group categoricals, defaults to False
pos_label: (Default value = None)
Returns:
pd.DataFrame: importance_df
"""
if cats:
importance_df = self.permutation_importances_cats(pos_label)
else:
importance_df = self.permutation_importances(pos_label)
if topx is None: topx = len(importance_df)
if cutoff is None: cutoff = importance_df.Importance.min()
return importance_df[importance_df.Importance >= cutoff].head(topx)
def importances_df(self, kind="shap", topx=None, cutoff=None, cats=False,
pos_label=None):
"""wrapper function for mean_abs_shap_df() and permutation_importance_df()
Args:
kind(str): 'shap' or 'permutations' (Default value = "shap")
topx: only display topx highest features (Default value = None)
cutoff: only display features above cutoff (Default value = None)
cats: Group categoricals (Default value = False)
pos_label: Positive class (Default value = None)
Returns:
pd.DataFrame
"""
assert kind=='shap' or kind=='permutation', \
"kind should either be 'shap' or 'permutation'!"
if kind=='permutation':
return self.permutation_importances_df(topx, cutoff, cats, pos_label)
elif kind=='shap':
return self.mean_abs_shap_df(topx, cutoff, cats, pos_label)
def contrib_df(self, index=None, X_row=None, cats=True, topx=None, cutoff=None, sort='abs',
pos_label=None):
"""shap value contributions to the prediction for index.
Used as input for the plot_contributions() method.
Args:
index(int or str): index for which to calculate contributions
X_row (pd.DataFrame, single row): single row of feature for which
to calculate contrib_df. Can us this instead of index
cats(bool, optional, optional): Group categoricals, defaults to True
topx(int, optional, optional): Only return topx features, remainder
called REST, defaults to None
cutoff(float, optional, optional): only return features with at least
cutoff contributions, defaults to None
sort({'abs', 'high-to-low', 'low-to-high', 'importance'}, optional): sort by
absolute shap value, or from high to low, low to high, or
ordered by the global shap importances.
Defaults to 'abs'.
pos_label: (Default value = None)
Returns:
pd.DataFrame: contrib_df
"""
if pos_label is None:
pos_label = self.pos_label
if sort =='importance':
if cutoff is None:
cols = self.columns_ranked_by_shap(cats)
else:
cols = self.mean_abs_shap_df(cats=cats).query(f"MEAN_ABS_SHAP > {cutoff}").Feature.tolist()
if topx is not None:
cols = cols[:topx]
else:
cols = None
if X_row is not None:
if ((len(X_row.columns) == len(self.X_cats.columns)) and
(X_row.columns == self.X_cats.columns).all()):
if cats:
X_row_cats = X_row
X_row = X_cats_to_X(X_row, self.cats_dict, self.X.columns)
else:
assert (X_row.columns == self.X.columns).all(), \
"X_row should have the same columns as self.X or self.X_cats!"
X_row_cats = merge_categorical_columns(X_row, self.cats_dict)
shap_values = self.shap_explainer.shap_values(X_row)
if self.is_classifier:
if not isinstance(shap_values, list) and len(self.labels)==2:
shap_values = [-shap_values, shap_values]
shap_values = shap_values[self.get_pos_label_index(pos_label)]
if cats:
shap_values_cats = merge_categorical_shap_values(X_row, shap_values, self.cats_dict)
return get_contrib_df(self.shap_base_value(pos_label), shap_values_cats[0],
X_row_cats, topx, cutoff, sort, cols)
else:
return get_contrib_df(self.shap_base_value(pos_label), shap_values[0],
X_row, topx, cutoff, sort, cols)
elif index is not None:
idx = self.get_int_idx(index)
if cats:
return get_contrib_df(self.shap_base_value(pos_label),
self.shap_values_cats(pos_label)[idx],
self.X_cats.iloc[[idx]], topx, cutoff, sort, cols)
else:
return get_contrib_df(self.shap_base_value(pos_label),
self.shap_values(pos_label)[idx],
self.X.iloc[[idx]], topx, cutoff, sort, cols)
else:
raise ValueError("Either index or X_row should be passed!")
def contrib_summary_df(self, index=None, X_row=None, cats=True, topx=None, cutoff=None,
round=2, sort='abs', pos_label=None):
"""Takes a contrib_df, and formats it to a more human readable format
Args:
index: index to show contrib_summary_df for
X_row (pd.DataFrame, single row): single row of feature for which
to calculate contrib_df. Can us this instead of index
cats: Group categoricals (Default value = True)
topx: Only show topx highest features(Default value = None)
cutoff: Only show features above cutoff (Default value = None)
round: round figures (Default value = 2)
sort({'abs', 'high-to-low', 'low-to-high', 'importance'}, optional): sort by
absolute shap value, or from high to low, or low to high, or
ordered by the global shap importances.
Defaults to 'abs'.
pos_label: Positive class (Default value = None)
Returns:
pd.DataFrame
"""
idx = self.get_int_idx(index) # if passed str convert to int index
return get_contrib_summary_df(
self.contrib_df(idx, X_row, cats, topx, cutoff, sort, pos_label),
model_output=self.model_output, round=round, units=self.units, na_fill=self.na_fill)
def interactions_df(self, col, cats=False, topx=None, cutoff=None,
pos_label=None):
"""dataframe of mean absolute shap interaction values for col
Args:
col: Feature to get interactions_df for
cats: Group categoricals (Default value = False)
topx: Only display topx most important features (Default value = None)
cutoff: Only display features with mean abs shap of at least cutoff (Default value = None)
pos_label: Positive class (Default value = None)
Returns:
pd.DataFrame
"""
importance_df = mean_absolute_shap_values(
self.columns_cats if cats else self.columns,
self.shap_interaction_values_by_col(col, cats, pos_label))
if topx is None: topx = len(importance_df)
if cutoff is None: cutoff = importance_df.MEAN_ABS_SHAP.min()
return importance_df[importance_df.MEAN_ABS_SHAP >= cutoff].head(topx)
def formatted_contrib_df(self, index, round=None, lang='en', pos_label=None):
"""contrib_df formatted in a particular idiosyncratic way.
Additional language option for output in Dutch (lang='nl')
Args:
index(str or int): index to return contrib_df for
round(int, optional, optional): rounding of continuous features, defaults to 2
lang(str, optional, optional): language to name the columns, defaults to 'en'
pos_label: (Default value = None)
Returns:
pd.DataFrame: formatted_contrib_df
"""
cdf = self.contrib_df(index, cats=True, pos_label=pos_label).copy()
cdf.reset_index(inplace=True)
cdf.loc[cdf.col=='base_value', 'value'] = np.nan
cdf['row_id'] = self.get_int_idx(index)
cdf['name_id'] = index
cdf['cat_value'] = np.where(cdf.col.isin(self.cats), cdf.value, np.nan)
cdf['cont_value'] = np.where(cdf.col.isin(self.cats), np.nan, cdf.value)
if round is not None:
rounded_cont = np.round(cdf['cont_value'].values.astype(float), round)
cdf['value'] = np.where(cdf.col.isin(self.cats), cdf.cat_value, rounded_cont)
cdf['type'] = np.where(cdf.col.isin(self.cats), 'cat', 'cont')
cdf['abs_contribution'] = np.abs(cdf.contribution)
cdf = cdf[['row_id', 'name_id', 'contribution', 'abs_contribution',
'col', 'value', 'cat_value', 'cont_value', 'type', 'index']]
if lang == 'nl':
cdf.columns = ['row_id', 'name_id', 'SHAP', 'ABS_SHAP', 'Variabele', 'Waarde',
'Cat_Waarde', 'Cont_Waarde', 'Waarde_Type', 'Variabele_Volgorde']
return cdf
cdf.columns = ['row_id', 'name_id', 'SHAP', 'ABS_SHAP', 'Feature', 'Value',
'Cat_Value', 'Cont_Value', 'Value_Type', 'Feature_Order']
return cdf
def get_pdp_result(self, col, index=None, X_row=None, drop_na=True,
sample=500, num_grid_points=20, pos_label=None):
"""Uses the PDPBox to calculate partial dependences for feature col.
Args:
col(str): Feature to calculate partial dependences for
index(int or str, optional, optional): Index of row to put at iloc[0], defaults to None
X_row (single row pd.DataFrame): row of features to highlight in pdp
drop_na(bool, optional, optional): drop rows where col equals na_fill, defaults to True
sample(int, optional, optional): Number of rows to sample for plot, defaults to 500
num_grid_points(ints: int, optional, optional): Number of grid points to calculate, default 20
pos_label: (Default value = None)
Returns:
PDPBox.pdp_result: pdp_result
"""
assert col in self.X.columns or col in self.cats, \
f"{col} not in columns of dataset"
if col in self.columns and not col in self.columns_cats:
features = col
else:
features = self.cats_dict[col]
if index is not None:
index = self.get_index(index)
if len(features)==1 and drop_na: # regular col, not onehotencoded
sample_size=min(sample, len(self.X[(self.X[features[0]] != self.na_fill)])-1)
sampleX = pd.concat([
self.X[self.X.index==index],
self.X[(self.X.index != index) & (self.X[features[0]] != self.na_fill)]\
.sample(sample_size)],
ignore_index=True, axis=0)
else:
sample_size = min(sample, len(self.X)-1)
sampleX = pd.concat([
self.X[self.X.index==index],
self.X[(self.X.index!=index)].sample(sample_size)],
ignore_index=True, axis=0)
elif X_row is not None:
if ((len(X_row.columns) == len(self.X_cats.columns)) and
(X_row.columns == self.X_cats.columns).all()):
X_row = X_cats_to_X(X_row, self.cats_dict, self.X.columns)
else:
assert (X_row.columns == self.X.columns).all(), \
"X_row should have the same columns as self.X or self.X_cats!"
if len(features)==1 and drop_na: # regular col, not onehotencoded
sample_size=min(sample, len(self.X[(self.X[features[0]] != self.na_fill)])-1)
sampleX = pd.concat([
X_row,
self.X[(self.X[features[0]] != self.na_fill)]\
.sample(sample_size)],
ignore_index=True, axis=0)
else:
sample_size = min(sample, len(self.X)-1)
sampleX = pd.concat([
X_row,
self.X.sample(sample_size)],
ignore_index=True, axis=0)
else:
if len(features)==1 and drop_na: # regular col, not onehotencoded
sample_size=min(sample, len(self.X[(self.X[features[0]] != self.na_fill)])-1)
sampleX = self.X[(self.X[features[0]] != self.na_fill)]\
.sample(sample_size)
else:
sampleX = self.X.sample(min(sample, len(self.X)))
# if only a single value (i.e. not onehot encoded, take that value
# instead of list):
if len(features)==1: features=features[0]
pdp_result = pdp.pdp_isolate(
model=self.model, dataset=sampleX,
model_features=self.X.columns,
num_grid_points=num_grid_points, feature=features)
if isinstance(features, list):
# strip 'col_' from the grid points
if isinstance(pdp_result, list):
for i in range(len(pdp_result)):
pdp_result[i].feature_grids = \
np.array([f[len(col)+1:] if f.startswith(col+"_") else f
for f in pdp_result[i].feature_grids])
else:
pdp_result.feature_grids = \
np.array([f[len(col)+1:] if f.startswith(col+"_") else f
for f in pdp_result.feature_grids])
return pdp_result
def get_dfs(self, cats=True, round=None, lang='en', pos_label=None):
"""return three summary dataframes for storing main results
Returns three pd.DataFrames. The first with id, prediction, actual and
feature values, the second with only id and shap values. The third
is similar to contrib_df for every id.
These can then be used to build your own custom dashboard on these data,
for example using PowerBI.
Args:
cats(bool, optional, optional): group categorical variables, defaults to True
round(int, optional, optional): how to round shap values (Default value = None)
lang(str, optional, optional): language to format dfs in. Defaults to 'en', 'nl' also available
pos_label: (Default value = None)
Returns:
pd.DataFrame, pd.DataFrame, pd.DataFrame: cols_df, shap_df, contribs_df
"""
if cats:
cols_df = self.X_cats.copy()
shap_df = pd.DataFrame(self.shap_values_cats(pos_label), columns = self.X_cats.columns)
else:
cols_df = self.X.copy()
shap_df = pd.DataFrame(self.shap_values(pos_label), columns = self.X.columns)
actual_str = 'Uitkomst' if lang == 'nl' else 'Actual'
prediction_str = 'Voorspelling' if lang == 'nl' else 'Prediction'
cols_df.insert(0, actual_str, self.y )
if self.is_classifier:
cols_df.insert(0, prediction_str, self.pred_probas)
else:
cols_df.insert(0, prediction_str, self.preds)
cols_df.insert(0, 'name_id', self.idxs)
cols_df.insert(0, 'row_id', range(len(self)))
shap_df.insert(0, 'SHAP_base', np.repeat(self.shap_base_value, len(self)))
shap_df.insert(0, 'name_id', self.idxs)
shap_df.insert(0, 'row_id', range(len(self)))
contribs_df = None
for idx in range(len(self)):
fcdf = self.formatted_contrib_df(idx, round=round, lang=lang)
if contribs_df is None: contribs_df = fcdf
else: contribs_df = pd.concat([contribs_df, fcdf])
return cols_df, shap_df, contribs_df
def to_sql(self, conn, schema, name, if_exists='replace',
cats=True, round=None, lang='en', pos_label=None):
"""Writes three dataframes generated by .get_dfs() to a sql server.
Tables will be called name_COLS and name_SHAP and name_CONTRBIB
Args:
conn(sqlalchemy.engine.Engine or sqlite3.Connection):
database connecter acceptable for pd.to_sql
schema(str): schema to write to
name(str): name prefix of tables
cats(bool, optional, optional): group categorical variables, defaults to True
if_exists({'fail’, ‘replace’, ‘append’}, default ‘replace’, optional):
How to behave if the table already exists. (Default value = 'replace')
round(int, optional, optional): how to round shap values (Default value = None)
lang(str, optional, optional): language to format dfs in. Defaults to 'en', 'nl' also available
pos_label: (Default value = None)
Returns:
"""
cols_df, shap_df, contribs_df = self.get_dfs(cats, round, lang, pos_label)
cols_df.to_sql(con=conn, schema=schema, name=name+"_COLS",
if_exists=if_exists, index=False)
shap_df.to_sql(con=conn, schema=schema, name=name+"_SHAP",
if_exists=if_exists, index=False)
contribs_df.to_sql(con=conn, schema=schema, name=name+"_CONTRIB",
if_exists=if_exists, index=False)
def plot_importances(self, kind='shap', topx=None, cats=False, round=3, pos_label=None):
"""plot barchart of importances in descending order.
Args:
type(str, optional): shap' for mean absolute shap values, 'permutation' for
permutation importances, defaults to 'shap'
topx(int, optional, optional): Only return topx features, defaults to None
cats(bool, optional, optional): Group categoricals defaults to False
kind: (Default value = 'shap')
round: (Default value = 3)
pos_label: (Default value = None)
Returns:
plotly.fig: fig
"""
importances_df = self.importances_df(kind=kind, topx=topx, cats=cats, pos_label=pos_label)
if kind=='shap':
if self.target:
title = f"Average impact on predicted {self.target}<br>(mean absolute SHAP value)"
else:
title = 'Average impact on prediction<br>(mean absolute SHAP value)'
units = self.units
else:
title = f"Permutation Importances <br>(decrease in metric '{self.metric.__name__}'' with randomized feature)"
units = ""
if self.descriptions:
descriptions = self.description_list(importances_df.Feature)
return plotly_importances_plot(importances_df, descriptions, round=round, units=units, title=title)
else:
return plotly_importances_plot(importances_df, round=round, units=units, title=title)
def plot_interactions(self, col, cats=False, topx=None, pos_label=None):
"""plot mean absolute shap interaction value for col.
Args:
col: column for which to generate shap interaction value
cats(bool, optional, optional): Group categoricals defaults to False
topx(int, optional, optional): Only return topx features, defaults to None
pos_label: (Default value = None)
Returns:
plotly.fig: fig
"""
if col in self.cats:
cats = True
interactions_df = self.interactions_df(col, cats=cats, topx=topx, pos_label=pos_label)
title = f"Average interaction shap values for {col}"
return plotly_importances_plot(interactions_df, units=self.units, title=title)
def plot_shap_contributions(self, index=None, X_row=None, cats=True, topx=None, cutoff=None,
sort='abs', orientation='vertical', higher_is_better=True,
round=2, pos_label=None):
"""plot waterfall plot of shap value contributions to the model prediction for index.
Args:
index(int or str): index for which to display prediction
X_row (pd.DataFrame single row): a single row of a features to plot
shap contributions for. Can use this instead of index for
what-if scenarios.
cats(bool, optional, optional): Group categoricals, defaults to True
topx(int, optional, optional): Only display topx features,
defaults to None
cutoff(float, optional, optional): Only display features with at least
cutoff contribution, defaults to None
sort({'abs', 'high-to-low', 'low-to-high', 'importance'}, optional):
sort by absolute shap value, or from high to low,
or low to high, or by order of shap feature importance.
Defaults to 'abs'.
orientation({'vertical', 'horizontal'}): Horizontal or vertical bar chart.
Horizontal may be better if you have lots of features.
Defaults to 'vertical'.
higher_is_better (bool): if True, up=green, down=red. If false reversed.
Defaults to True.
round(int, optional, optional): round contributions to round precision,
defaults to 2
pos_label: (Default value = None)
Returns:
plotly.Fig: fig
"""
assert orientation in ['vertical', 'horizontal']
contrib_df = self.contrib_df(self.get_int_idx(index), X_row, cats, topx, cutoff, sort, pos_label)
return plotly_contribution_plot(contrib_df, model_output=self.model_output,
orientation=orientation, round=round, higher_is_better=higher_is_better,
target=self.target, units=self.units)
def plot_shap_summary(self, index=None, topx=None, cats=False, pos_label=None):
"""Plot barchart of mean absolute shap value.
Displays all individual shap value for each feature in a horizontal
scatter chart in descending order by mean absolute shap value.
Args:
index (str or int): index to highlight
topx(int, optional): Only display topx most important features, defaults to None
cats(bool, optional): Group categoricals , defaults to False
pos_label: positive class (Default value = None)
Returns:
plotly.Fig
"""
if self.is_classifier:
if pos_label is None:
pos_label = self.pos_label
pos_label_str = self.labels[self.get_pos_label_index(pos_label)]
if self.model_output == 'probability':
if self.target:
title = f"Impact of feature on predicted probability {self.target}={pos_label_str} <br> (SHAP values)"
else:
title = f"Impact of Feature on Prediction probability <br> (SHAP values)"
elif self.model_output == 'logodds':
title = f"Impact of Feature on predicted logodds <br> (SHAP values)"
elif self.is_regression:
if self.target:
title = f"Impact of Feature on Predicted {self.target} <br> (SHAP values)"
else:
title = f"Impact of Feature on Prediction<br> (SHAP values)"
if cats:
return plotly_shap_scatter_plot(
self.shap_values_cats(pos_label),
self.X_cats,
self.importances_df(kind='shap', topx=topx, cats=True, pos_label=pos_label)\
['Feature'].values.tolist(),
idxs=self.idxs.values,
highlight_index=index,
title=title,
na_fill=self.na_fill,
index_name=self.index_name)
else:
return plotly_shap_scatter_plot(
self.shap_values(pos_label),
self.X,
self.importances_df(kind='shap', topx=topx, cats=False, pos_label=pos_label)\
['Feature'].values.tolist(),
idxs=self.idxs.values,
highlight_index=index,
title=title,
na_fill=self.na_fill,
index_name=self.index_name)
def plot_shap_interaction_summary(self, col, index=None, topx=None, cats=False, pos_label=None):
"""Plot barchart of mean absolute shap interaction values
Displays all individual shap interaction values for each feature in a
horizontal scatter chart in descending order by mean absolute shap value.
Args:
col(type]): feature for which to show interactions summary
index (str or int): index to highlight
topx(int, optional): only show topx most important features, defaults to None
cats: group categorical features (Default value = False)
pos_label: positive class (Default value = None)
Returns:
fig
"""
if col in self.cats:
cats = True
interact_cols = self.shap_top_interactions(col, cats=cats, pos_label=pos_label)
if topx is None: topx = len(interact_cols)
title = f"Shap interaction values for {col}"
return plotly_shap_scatter_plot(
self.shap_interaction_values_by_col(col, cats=cats, pos_label=pos_label),
self.X_cats if cats else self.X, interact_cols[:topx], title=title,
idxs=self.idxs.values, highlight_index=index, na_fill=self.na_fill,
index_name=self.index_name)
def plot_shap_dependence(self, col, color_col=None, highlight_index=None, pos_label=None):
"""plot shap dependence
Plots a shap dependence plot:
- on the x axis the possible values of the feature `col`
- on the y axis the associated individual shap values
Args:
col(str): feature to be displayed
color_col(str): if color_col provided then shap values colored (blue-red)
according to feature color_col (Default value = None)
highlight_idx: individual observation to be highlighed in the plot.
(Default value = None)
pos_label: positive class (Default value = None)
Returns:
"""
cats = self.check_cats(col, color_col)
highlight_idx = self.get_int_idx(highlight_index)
if cats:
if col in self.cats:
return plotly_shap_violin_plot(
self.X_cats,
self.shap_values_cats(pos_label),
col,
color_col,
highlight_index=highlight_idx,
idxs=self.idxs.values,
index_name=self.index_name)
else:
return plotly_dependence_plot(
self.X_cats,
self.shap_values_cats(pos_label),
col,
color_col,
na_fill=self.na_fill,
units=self.units,
highlight_index=highlight_idx,
idxs=self.idxs.values,
index_name=self.index_name)
else:
return plotly_dependence_plot(
self.X,
self.shap_values(pos_label),
col,
color_col,
na_fill=self.na_fill,
units=self.units,
highlight_index=highlight_idx,
idxs=self.idxs.values,
index_name=self.index_name)
def plot_shap_interaction(self, col, interact_col, highlight_index=None,
pos_label=None):
"""plots a dependence plot for shap interaction effects
Args:
col(str): feature for which to find interaction values
interact_col(str): feature for which interaction value are displayed
highlight_idx(int, optional, optional): idx that will be highlighted, defaults to None
pos_label: (Default value = None)
Returns:
plotly.Fig: Plotly Fig
"""
cats = self.check_cats(col, interact_col)
highlight_idx = self.get_int_idx(highlight_index)
if cats and interact_col in self.cats:
return plotly_shap_violin_plot(
self.X_cats,
self.shap_interaction_values_by_col(col, cats, pos_label=pos_label),
interact_col, col, interaction=True, units=self.units,
highlight_index=highlight_idx, idxs=self.idxs.values,
index_name=self.index_name)
else:
return plotly_dependence_plot(self.X_cats if cats else self.X,
self.shap_interaction_values_by_col(col, cats, pos_label=pos_label),
interact_col, col, interaction=True, units=self.units,
highlight_index=highlight_idx, idxs=self.idxs.values,
index_name=self.index_name)
def plot_pdp(self, col, index=None, X_row=None, drop_na=True, sample=100,
gridlines=100, gridpoints=10, pos_label=None):
"""plot partial dependence plot (pdp)
returns plotly fig for a partial dependence plot showing ice lines
for num_grid_lines rows, average pdp based on sample of sample.
If index is given, display pdp for this specific index.
Args:
col(str): feature to display pdp graph for
index(int or str, optional, optional): index to highlight in pdp graph,
defaults to None
X_row (pd.Dataframe, single row, optional): a row of features to highlight
predictions for. Alternative to passing index.
drop_na(bool, optional, optional): if true drop samples with value
equal to na_fill, defaults to True
sample(int, optional, optional): sample size on which the average
pdp will be calculated, defaults to 100
gridlines(int, optional): number of ice lines to display,
defaults to 100
gridpoints(ints: int, optional): number of points on the x axis
to calculate the pdp for, defaults to 10
pos_label: (Default value = None)
Returns:
plotly.Fig: fig
"""
pdp_result = self.get_pdp_result(col, index, X_row,
drop_na=drop_na, sample=sample, num_grid_points=gridpoints, pos_label=pos_label)
units = "Predicted %" if self.model_output=='probability' else self.units
if index is not None:
col_value, pred = self.get_col_value_plus_prediction(col, index=index, pos_label=pos_label)
return plotly_pdp(pdp_result,
display_index=0, # the idx to be displayed is always set to the first row by self.get_pdp_result()
index_feature_value=col_value, index_prediction=pred,
feature_name=col,
num_grid_lines=min(gridlines, sample, len(self.X)),
target=self.target, units=units)
elif X_row is not None:
col_value, pred = self.get_col_value_plus_prediction(col, X_row=X_row, pos_label=pos_label)
return plotly_pdp(pdp_result,
display_index=0, # the idx to be displayed is always set to the first row by self.get_pdp_result()
index_feature_value=col_value, index_prediction=pred,
feature_name=col,
num_grid_lines=min(gridlines, sample, len(self.X)),
target=self.target, units=units)
else:
return plotly_pdp(pdp_result, feature_name=col,
num_grid_lines=min(gridlines, sample, len(self.X)),
target=self.target, units=units)
class ClassifierExplainer(BaseExplainer):
""" """
def __init__(self, model,recommended_config , X, y=None, permutation_metric=roc_auc_score,
shap='guess', X_background=None, model_output="probability",
cats=None, idxs=None, index_name=None, target=None,
descriptions=None, n_jobs=None, permutation_cv=None, na_fill=-999,
labels=None, pos_label=2):
"""
Explainer for classification models. Defines the shap values for
each possible class in the classification.
You assign the positive label class afterwards with e.g. explainer.pos_label=0
In addition defines a number of plots specific to classification problems
such as a precision plot, confusion matrix, roc auc curve and pr auc curve.
Compared to BaseExplainer defines two additional parameters
Args:
labels(list): list of str labels for the different classes,
defaults to e.g. ['0', '1'] for a binary classification
pos_label: class that should be used as the positive class,
defaults to 1
"""
super().__init__(model,recommended_config, X, y, permutation_metric,
shap, X_background, model_output,
cats, idxs, index_name, target, descriptions,
n_jobs, permutation_cv, na_fill)
assert hasattr(model, "predict_proba"), \
("for ClassifierExplainer, model should be a scikit-learn "
"compatible *classifier* model that has a predict_proba(...) "
f"method, so not a {type(model)}!")
self._params_dict = {**self._params_dict, **dict(
labels=labels, pos_label=pos_label)}
if labels is not None:
self.labels = labels
elif hasattr(self.model, 'classes_'):
self.labels = [str(cls) for cls in self.model.classes_]
else:
self.labels = [str(i) for i in range(self.y.nunique())]
self.pos_label = pos_label
self.is_classifier = True
if str(type(self.model)).endswith("RandomForestClassifier'>"):
self.__class__ = RandomForestClassifierExplainer
if str(type(self.model)).endswith("XGBClassifier'>"):
print(f"Detected XGBClassifier model: "
"Changing class type to XGBClassifierExplainer...",
flush=True)
self.__class__ = XGBClassifierExplainer
@property
def shap_explainer(self):
"""Initialize SHAP explainer.
Taking into account model type and model_output
"""
if not hasattr(self, '_shap_explainer'):
model_str = str(type(self.model)).replace("'", "").replace("<", "").replace(">", "").split(".")[-1]
if self.shap == 'tree':
if (str(type(self.model)).endswith("XGBClassifier'>") or
str(type(self.model)).endswith("LGBMClassifier'>") or
str(type(self.model)).endswith("CatBoostClassifier'>") or
str(type(self.model)).endswith("GradientBoostingClassifier'>") or
str(type(self.model)).endswith("HistGradientBoostingClassifier'>")
):
if self.model_output == "probability":
if self.X_background is None:
pass
self._shap_explainer = shap.TreeExplainer(
self.model,
self.X_background if self.X_background is not None else self.X,
model_output="probability",
feature_perturbation="interventional")
self.interactions_should_work = False
else:
self.model_output = "logodds"
print(f"Generating self.shap_explainer = shap.TreeExplainer(model{', X_background' if self.X_background is not None else ''})")
self._shap_explainer = shap.TreeExplainer(self.model, self.X_background)
else:
if self.model_output == "probability":
pass
self._shap_explainer = shap.TreeExplainer(self.model, self.X_background)
elif self.shap=='linear':
if self.model_output == "probability":
self.model_output = "logodds"
if self.X_background is None:
pass
self._shap_explainer = shap.LinearExplainer(self.model,
self.X_background if self.X_background is not None else self.X)
elif self.shap=='deep':
# print("Generating self.shap_explainer = shap.DeepExplainer(model{', X_background' if self.X_background is not None else ''})")
self._shap_explainer = shap.DeepExplainer(self.model, self.X_background)
elif self.shap=='kernel':
if self.X_background is None:
pass
if self.model_output != "probability":
self.model_output = 'probability'
self._shap_explainer = shap.KernelExplainer(self.model.predict_proba,
self.X_background if self.X_background is not None else self.X,
link="identity")
return self._shap_explainer
@property
def pos_label(self):
return self._pos_label
@pos_label.setter
def pos_label(self, label):
if label is None or isinstance(label, int) and label >=0 and label <len(self.labels):
self._pos_label = label
elif isinstance(label, str) and label in self.labels:
self._pos_label = self.labels.index(label)
else:
raise ValueError(f"'{label}' not in labels")
@property
def pos_label_str(self):
"""return str label of self.pos_label"""
return self.labels[self.pos_label]
def get_pos_label_index(self, pos_label):
"""return int index of pos_label_str"""
if isinstance(pos_label, int):
assert pos_label <= len(self.labels), \
f"pos_label {pos_label} is larger than number of labels!"
return pos_label
elif isinstance(pos_label, str):
assert pos_label in self.labels, \
f"Unknown pos_label. {pos_label} not in self.labels!"
return self.labels.index(pos_label)
raise ValueError("pos_label should either be int or str in self.labels!")
def get_prop_for_label(self, prop:str, label):
"""return property for a specific pos_label
Args:
prop: property to get for a certain pos_label
label: pos_label
Returns:
property
"""
tmp = self.pos_label
self.pos_label = label
ret = getattr(self, prop)
self.pos_label = tmp
return ret
@property
def y_binary(self):
"""for multiclass problems returns one-vs-rest array of [1,0] pos_label"""
if not hasattr(self, '_y_binaries'):
if not self.y_missing:
self._y_binaries = [np.where(self.y.values==i, 1, 0)
for i in range(self.y.nunique())]
else:
self._y_binaries = [self.y.values for i in range(len(self.labels))]
return default_list(self._y_binaries, self.pos_label)
@property
def pred_probas_raw(self):
"""returns pred_probas with probability for each class"""
if not hasattr(self, '_pred_probas'):
#print("Calculating prediction probabilities...", flush=True)
assert hasattr(self.model, 'predict_proba'), \
"model does not have a predict_proba method!"
self._pred_probas = self.model.predict_proba(self.X)
return self._pred_probas
@property
def pred_percentiles_raw(self):
""" """
if not hasattr(self, '_pred_percentiles_raw'):
#print("Calculating pred_percentiles...", flush=True)
self._pred_percentiles_raw = (pd.DataFrame(self.pred_probas_raw)
.rank(method='min')
.divide(len(self.pred_probas_raw))
.values)
return self._pred_percentiles_raw
@property
def pred_probas(self):
"""returns pred_proba for pos_label class"""
return default_2darray(self.pred_probas_raw, self.pos_label)
@property
def pred_percentiles(self):
"""returns ranks for pos_label class"""
return default_2darray(self.pred_percentiles_raw, self.pos_label)
@property
def permutation_importances(self):
"""Permutation importances"""
if not hasattr(self, '_perm_imps'):
#print("Calculating permutation importances (if slow, try setting n_jobs parameter)...", flush=True)
self._perm_imps = [cv_permutation_importances(
self.model, self.X, self.y, self.metric,
cv=self.permutation_cv,
needs_proba=self.is_classifier,
pos_label=label) for label in range(len(self.labels))]
return default_list(self._perm_imps, self.pos_label)
@property
def permutation_importances_cats(self):
"""permutation importances with categoricals grouped"""
if not hasattr(self, '_perm_imps_cats'):
#print("Calculating categorical permutation importances (if slow, try setting n_jobs parameter)...", flush=True)
self._perm_imps_cats = [cv_permutation_importances(
self.model, self.X, self.y, self.metric,
cats_dict=self.cats_dict,
cv=self.permutation_cv,
needs_proba=self.is_classifier,
pos_label=label) for label in range(len(self.labels))]
return default_list(self._perm_imps_cats, self.pos_label)
@property
def shap_base_value(self):
"""SHAP base value: average outcome of population"""
if not hasattr(self, '_shap_base_value'):
_ = self.shap_values() # CatBoost needs to have shap values calculated before expected value for some reason
self._shap_base_value = self.shap_explainer.expected_value
if isinstance(self._shap_base_value, np.ndarray) and len(self._shap_base_value) == 1:
self._shap_base_value = self._shap_base_value[0]
if isinstance(self._shap_base_value, np.ndarray):
self._shap_base_value = list(self._shap_base_value)
if len(self.labels)==2 and isinstance(self._shap_base_value, (np.floating, float)):
if self.model_output == 'probability':
self._shap_base_value = [1-self._shap_base_value, self._shap_base_value]
else: # assume logodds
self._shap_base_value = [-self._shap_base_value, self._shap_base_value]
assert len(self._shap_base_value)==len(self.labels),\
f"len(shap_explainer.expected_value)={len(self._shap_base_value)}"\
+ f"and len(labels)={len(self.labels)} do not match!"
if self.model_output == 'probability':
for shap_base_value in self._shap_base_value:
assert shap_base_value >= 0.0 and shap_base_value <= 1.0, \
(f"Shap base value does not look like a probability: {self._shap_base_value}. "
"Try setting model_output='logodds'.")
return default_list(self._shap_base_value, self.pos_label)
@property
def shap_values(self):
"""SHAP Values"""
if not hasattr(self, '_shap_values'):
#print("Calculating shap values...", flush=True)
self._shap_values = self.shap_explainer.shap_values(self.X)
if not isinstance(self._shap_values, list) and len(self.labels)==2:
self._shap_values = [-self._shap_values, self._shap_values]
assert len(self._shap_values)==len(self.labels),\
f"len(shap_values)={len(self._shap_values)}"\
+ f"and len(labels)={len(self.labels)} do not match!"
if self.model_output == 'probability':
for shap_values in self._shap_values:
assert np.all(shap_values >= -1.0) , \
(f"model_output=='probability but some shap values are < 1.0!"
"Try setting model_output='logodds'.")
for shap_values in self._shap_values:
assert np.all(shap_values <= 1.0) , \
(f"model_output=='probability but some shap values are > 1.0!"
"Try setting model_output='logodds'.")
return default_list(self._shap_values, self.pos_label)
@property
def shap_values_cats(self):
"""SHAP values with categoricals grouped together"""
if not hasattr(self, '_shap_values_cats'):
_ = self.shap_values
self._shap_values_cats = [
merge_categorical_shap_values(
self.X, sv, self.cats_dict) for sv in self._shap_values]
return default_list(self._shap_values_cats, self.pos_label)
@property
def shap_interaction_values(self):
"""SHAP interaction values"""
if not hasattr(self, '_shap_interaction_values'):
_ = self.shap_values #make sure shap values have been calculated
#print("Calculating shap interaction values...", flush=True)
if self.shap == 'tree':
print("Reminder: TreeShap computational complexity is O(TLD^2), "
"where T is the number of trees, L is the maximum number of"
" leaves in any tree and D the maximal depth of any tree. So "
"reducing these will speed up the calculation.",
flush=True)
self._shap_interaction_values = self.shap_explainer.shap_interaction_values(self.X)
if not isinstance(self._shap_interaction_values, list) and len(self.labels)==2:
if self.model_output == "probability":
self._shap_interaction_values = [1-self._shap_interaction_values,
self._shap_interaction_values]
else: # assume logodds so logodds of negative class is -logodds of positive class
self._shap_interaction_values = [-self._shap_interaction_values,
self._shap_interaction_values]
self._shap_interaction_values = [
normalize_shap_interaction_values(siv, self.shap_values)
for siv, sv in zip(self._shap_interaction_values, self._shap_values)]
return default_list(self._shap_interaction_values, self.pos_label)
@property
def shap_interaction_values_cats(self):
"""SHAP interaction values with categoricals grouped together"""
if not hasattr(self, '_shap_interaction_values_cats'):
_ = self.shap_interaction_values
self._shap_interaction_values_cats = [
merge_categorical_shap_interaction_values(
siv, self.X, self.X_cats, self.cats_dict)
for siv in self._shap_interaction_values]
return default_list(self._shap_interaction_values_cats, self.pos_label)
@property
def mean_abs_shap(self):
"""mean absolute SHAP values"""
if not hasattr(self, '_mean_abs_shap'):
_ = self.shap_values
self._mean_abs_shap = [mean_absolute_shap_values(
self.columns, sv) for sv in self._shap_values]
return default_list(self._mean_abs_shap, self.pos_label)
@property
def mean_abs_shap_cats(self):
"""mean absolute SHAP values with categoricals grouped together"""
if not hasattr(self, '_mean_abs_shap_cats'):
_ = self.shap_values
self._mean_abs_shap_cats = [
mean_absolute_shap_values(self.columns, sv, self.cats_dict)
for sv in self._shap_values]
return default_list(self._mean_abs_shap_cats, self.pos_label)
def cutoff_from_percentile(self, percentile, pos_label=None):
"""The cutoff equivalent to the percentile given
For example if you want the cutoff that splits the highest 20%
pred_proba from the lowest 80%, you would set percentile=0.8
and get the correct cutoff.
Args:
percentile(float): percentile to convert to cutoff
pos_label: positive class (Default value = None)
Returns:
cutoff
"""
if pos_label is None:
return pd.Series(self.pred_probas).nlargest(int((1-percentile)*len(self))).min()
else:
return pd.Series(self.pred_probas_raw[:, pos_label]).nlargest(int((1-percentile)*len(self))).min()
def percentile_from_cutoff(self, cutoff, pos_label=None):
"""The percentile equivalent to the cutoff given
For example if set the cutoff at 0.8, then what percentage
of pred_proba is above this cutoff?
Args:
cutoff (float): cutoff to convert to percentile
pos_label: positive class (Default value = None)
Returns:
percentile
"""
if cutoff is None:
return None
if pos_label is None:
return 1-(self.pred_probas < cutoff).mean()
else:
pos_label = self.get_pos_label_index(pos_label)
return 1-np.mean(self.pred_probas_raw[:, pos_label] < cutoff)
def metrics(self, cutoff=0.5, pos_label=None, **kwargs):
"""returns a dict with useful metrics for your classifier:
accuracy, precision, recall, f1, roc auc, pr auc, log loss
Args:
cutoff(float): cutoff used to calculate metrics (Default value = 0.5)
pos_label: positive class (Default value = None)
Returns:
dict
"""
if self.y_missing:
raise ValueError("No y was passed to explainer, so cannot calculate metrics!")
if pos_label is None: pos_label = self.pos_label
metrics_dict = {
'accuracy' : accuracy_score(self.y_binary(pos_label), np.where(self.pred_probas(pos_label) > cutoff, 1, 0)),
'precision' : precision_score(self.y_binary(pos_label), np.where(self.pred_probas(pos_label) > cutoff, 1, 0)),
'recall' : recall_score(self.y_binary(pos_label), np.where(self.pred_probas(pos_label) > cutoff, 1, 0)),
'f1' : f1_score(self.y_binary(pos_label), np.where(self.pred_probas(pos_label) > cutoff, 1, 0)),
'roc_auc_score' : roc_auc_score(self.y_binary(pos_label), self.pred_probas(pos_label)),
'pr_auc_score' : average_precision_score(self.y_binary(pos_label), self.pred_probas(pos_label)),
'log_loss' : log_loss(self.y_binary(pos_label), self.pred_probas(pos_label))
}
return metrics_dict
def metrics_descriptions(self, cutoff=0.5, pos_label=None):
metrics_dict = self.metrics(cutoff, pos_label)
metrics_descriptions_dict = {}
for k, v in metrics_dict.items():
if k == 'accuracy':
metrics_descriptions_dict[k] = f"{round(100*v, 2)}% of predicted labels was predicted correctly."
if k == 'precision':
metrics_descriptions_dict[k] = f"{round(100*v, 2)}% of predicted positive labels was predicted correctly."
if k == 'recall':
metrics_descriptions_dict[k] = f"{round(100*v, 2)}% of positive labels was predicted correctly."
if k == 'f1':
metrics_descriptions_dict[k] = f"The weighted average of precision and recall is {round(v, 2)}"
if k == 'roc_auc_score':
metrics_descriptions_dict[k] = f"The probability that a random positive label has a higher score than a random negative label is {round(100*v, 2)}%"
if k == 'pr_auc_score':
metrics_descriptions_dict[k] = f"The average precision score calculated for each recall threshold is {round(v, 2)}. This ignores true negatives."
if k == 'log_loss':
metrics_descriptions_dict[k] = f"A measure of how far the predicted label is from the true label on average in log space {round(v, 2)}"
return metrics_descriptions_dict
def get_pdp_result(self, col, index=None, X_row=None, drop_na=True,
sample=1000, num_grid_points=20, pos_label=None):
"""gets a the result out of the PDPBox library
Adjust for multiple labels.
Args:
col(str): Feature to display
index(str or int): index to add to plot (Default value = None)
X_row (pd.DataFrame, single row): single row of features to highlight
in pdp
drop_na(bool): drop value equal to self.fill_na (Default value = True)
sample(int): sample size to compute average pdp (Default value = 1000)
num_grid_points(int): number of horizontal breakpoints in pdp (Default value = 20)
pos_label: positive class (Default value = None)
Returns:
PDPBox pdp result
"""
if pos_label is None: pos_label = self.pos_label
pdp_result = super().get_pdp_result(
col, index, X_row, drop_na, sample, num_grid_points)
if len(self.labels)==2:
# for binary classifer PDPBox only gives pdp for the positive class.
# instead of a list of pdps for every class
# so we simply inverse when predicting the negative
if self.model_output == 'probability':
pdp_result.pdp = 100*pdp_result.pdp
pdp_result.ice_lines = pdp_result.ice_lines.multiply(100)
if pos_label==0:
if self.model_output == 'probability':
pdp_result.pdp = 100 - pdp_result.pdp
pdp_result.ice_lines = 100 - pdp_result.ice_lines
elif self.model_output == 'logodds':
pdp_result.pdp = -pdp_result.pdp
pdp_result.ice_lines = -pdp_result.ice_lines
return pdp_result
else:
pdp_result = pdp_result[pos_label]
if self.model_output == 'probability':
pdp_result.pdp = 100*pdp_result.pdp
pdp_result.ice_lines = pdp_result.ice_lines.multiply(100)
return pdp_result
def random_index(self, y_values=None, return_str=False,
pred_proba_min=None, pred_proba_max=None,
pred_percentile_min=None, pred_percentile_max=None, pos_label=None):
"""random index satisfying various constraint
Args:
y_values: list of labels to include (Default value = None)
return_str: return str from self.idxs (Default value = False)
pred_proba_min: minimum pred_proba (Default value = None)
pred_proba_max: maximum pred_proba (Default value = None)
pred_percentile_min: minimum pred_proba percentile (Default value = None)
pred_percentile_max: maximum pred_proba percentile (Default value = None)
pos_label: positive class (Default value = None)
Returns:
index
"""
# if pos_label is None: pos_label = self.pos_label
if (y_values is None
and pred_proba_min is None and pred_proba_max is None
and pred_percentile_min is None and pred_percentile_max is None):
potential_idxs = self.idxs.values
else:
if pred_proba_min is None: pred_proba_min = self.pred_probas(pos_label).min()
if pred_proba_max is None: pred_proba_max = self.pred_probas(pos_label).max()
if pred_percentile_min is None: pred_percentile_min = 0.0
if pred_percentile_max is None: pred_percentile_max = 1.0
if not self.y_missing:
if y_values is None: y_values = self.y.unique().tolist()
if not isinstance(y_values, list): y_values = [y_values]
y_values = [y if isinstance(y, int) else self.labels.index(y) for y in y_values]
potential_idxs = self.idxs[(self.y.isin(y_values)) &
(self.pred_probas(pos_label) >= pred_proba_min) &
(self.pred_probas(pos_label) <= pred_proba_max) &
(self.pred_percentiles(pos_label) > pred_percentile_min) &
(self.pred_percentiles(pos_label) <= pred_percentile_max)].values
else:
potential_idxs = self.idxs[
(self.pred_probas(pos_label) >= pred_proba_min) &
(self.pred_probas(pos_label) <= pred_proba_max) &
(self.pred_percentiles(pos_label) > pred_percentile_min) &
(self.pred_percentiles(pos_label) <= pred_percentile_max)].values
if len(potential_idxs) > 0:
idx = np.random.choice(potential_idxs)
else:
return None
if return_str:
return idx
return self.idxs.get_loc(idx)
def prediction_result_df(self, index=None, X_row=None, add_star=True, logodds=False, round=3):
"""returns a table with the predicted probability for each label for index
Args:
index ({int, str}): index
add_star(bool): add a star to the observed label
round (int): rounding to apply to pred_proba float
Returns:
pd.DataFrame
"""
if index is None and X_row is None:
raise ValueError("You need to either pass an index or X_row!")
if index is not None:
int_idx = self.get_int_idx(index)
pred_probas = self.pred_probas_raw[int_idx, :]
elif X_row is not None:
if X_row.columns.tolist()==self.X_cats.columns.tolist():
X_row = X_cats_to_X(X_row, self.cats_dict, self.X.columns)
pred_probas = self.model.predict_proba(X_row)[0, :]
preds_df = pd.DataFrame(dict(
label=self.labels,
probability=pred_probas))
if logodds:
try:
preds_df.loc[:, "logodds"] = \
preds_df.probability.apply(lambda p: np.log(p / (1-p)))
except ZeroDivisionError:
preds_df.loc[:, "logodds"] = preds_df.probability
if index is not None and not self.y_missing and not np.isnan(self.y[int_idx]):
preds_df.iloc[self.y[int_idx], 0] = f"{preds_df.iloc[self.y[int_idx], 0]}*"
return preds_df.round(round)
def precision_df(self, bin_size=None, quantiles=None, multiclass=False,
round=3, pos_label=None):
"""dataframe with predicted probabilities and precision
Args:
bin_size(float, optional, optional): group predictions in bins of size bin_size, defaults to 0.1
quantiles(int, optional, optional): group predictions in evenly sized quantiles of size quantiles, defaults to None
multiclass(bool, optional, optional): whether to calculate precision for every class (Default value = False)
round: (Default value = 3)
pos_label: (Default value = None)
Returns:
pd.DataFrame: precision_df
"""
if self.y_missing:
raise ValueError("No y was passed to explainer, so cannot calculate precision_df!")
assert self.pred_probas is not None
if pos_label is None: pos_label = self.pos_label
if bin_size is None and quantiles is None:
bin_size=0.1 # defaults to bin_size=0.1
if multiclass:
return get_precision_df(self.pred_probas_raw, self.y,
bin_size, quantiles,
round=round, pos_label=pos_label)
else:
return get_precision_df(self.pred_probas(pos_label), self.y_binary(pos_label),
bin_size, quantiles, round=round)
def lift_curve_df(self, pos_label=None):
"""returns a pd.DataFrame with data needed to build a lift curve
Args:
pos_label: (Default value = None)
Returns:
"""
if pos_label is None: pos_label = self.pos_label
return get_lift_curve_df(self.pred_probas(pos_label), self.y, pos_label)
def prediction_result_markdown(self, index, include_percentile=True, round=2, pos_label=None):
"""markdown of result of prediction for index
Args:
index(int or str): the index of the row for which to generate the prediction
include_percentile(bool, optional, optional): include the rank
percentile of the prediction, defaults to True
round(int, optional, optional): rounding to apply to results, defaults to 2
pos_label: (Default value = None)
**kwargs:
Returns:
str: markdown string
"""
int_idx = self.get_int_idx(index)
if pos_label is None: pos_label = self.pos_label
def display_probas(pred_probas_raw, labels, model_output='probability', round=2):
assert (len(pred_probas_raw.shape)==1 and len(pred_probas_raw) ==len(labels))
def log_odds(p, round=2):
return np.round(np.log(p / (1-p)), round)
for i in range(len(labels)):
proba_str = f"{np.round(100*pred_probas_raw[i], round)}%"
logodds_str = f"(logodds={log_odds(pred_probas_raw[i], round)})"
yield f"* {labels[i]}: {proba_str} {logodds_str if model_output=='logodds' else ''}\n"
model_prediction = "###### Prediction:\n\n"
if (isinstance(self.y[0], int) or
isinstance(self.y[0], np.int64)):
model_prediction += f"Observed {self.target}: {self.labels[self.y[int_idx]]}\n\n"
model_prediction += "Prediction probabilities per label:\n\n"
for pred in display_probas(
self.pred_probas_raw[int_idx],
self.labels, self.model_output, round):
model_prediction += pred
if include_percentile:
percentile = np.round(100*(1-self.pred_percentiles(pos_label)[int_idx]))
model_prediction += f'\nIn top {percentile}% percentile probability {self.labels[pos_label]}'
return model_prediction
def plot_precision(self, bin_size=None, quantiles=None, cutoff=None, multiclass=False, pos_label=None):
"""plot precision vs predicted probability
plots predicted probability on the x-axis and observed precision (fraction of actual positive
cases) on the y-axis.
Should pass either bin_size fraction of number of quantiles, but not both.
Args:
bin_size(float, optional): size of the bins on x-axis (e.g. 0.05 for 20 bins)
quantiles(int, optional): number of equal sized quantiles to split
the predictions by e.g. 20, optional)
cutoff: cutoff of model to include in the plot (Default value = None)
multiclass: whether to display all classes or only positive class,
defaults to False
pos_label: positive label to display, defaults to self.pos_label
Returns:
Plotly fig
"""
if pos_label is None: pos_label = self.pos_label
if bin_size is None and quantiles is None:
bin_size=0.1 # defaults to bin_size=0.1
precision_df = self.precision_df(
bin_size=bin_size, quantiles=quantiles, multiclass=multiclass, pos_label=pos_label)
return plotly_precision_plot(precision_df,
cutoff=cutoff, labels=self.labels, pos_label=pos_label)
def plot_cumulative_precision(self, percentile=None, pos_label=None):
"""plot cumulative precision
returns a cumulative precision plot, which is a slightly different
representation of a lift curve.
Args:
pos_label: positive label to display, defaults to self.pos_label
Returns:
plotly fig
"""
if pos_label is None: pos_label = self.pos_label
return plotly_cumulative_precision_plot(
self.lift_curve_df(pos_label=pos_label), labels=self.labels,
percentile=percentile, pos_label=pos_label)
def plot_confusion_matrix(self, cutoff=0.5, normalized=False, binary=False, pos_label=None):
"""plot of a confusion matrix.
Args:
cutoff(float, optional, optional): cutoff of positive class to
calculate confusion matrix for, defaults to 0.5
normalized(bool, optional, optional): display percentages instead
of counts , defaults to False
binary(bool, optional, optional): if multiclass display one-vs-rest
instead, defaults to False
pos_label: positive label to display, defaults to self.pos_label
Returns:
plotly fig
"""
if self.y_missing:
raise ValueError("No y was passed to explainer, so cannot plot confusion matrix!")
if pos_label is None: pos_label = self.pos_label
pos_label_str = self.labels[pos_label]
if binary:
if len(self.labels)==2:
def order_binary_labels(labels, pos_label):
pos_index = labels.index(pos_label)
return [labels[1-pos_index], labels[pos_index]]
labels = order_binary_labels(self.labels, pos_label_str)
else:
labels = ['Not ' + pos_label_str, pos_label_str]
return plotly_confusion_matrix(
self.y_binary(pos_label), np.where(self.pred_probas(pos_label) > cutoff, 1, 0),
percentage=normalized, labels=labels)
else:
return plotly_confusion_matrix(
self.y, self.pred_probas_raw.argmax(axis=1),
percentage=normalized, labels=self.labels)
def plot_lift_curve(self, cutoff=None, percentage=False, round=2, pos_label=None):
"""plot of a lift curve.
Args:
cutoff(float, optional): cutoff of positive class to calculate lift
(Default value = None)
percentage(bool, optional): display percentages instead of counts,
defaults to False
round: number of digits to round to (Default value = 2)
pos_label: positive label to display, defaults to self.pos_label
Returns:
plotly fig
"""
return plotly_lift_curve(self.lift_curve_df(pos_label), cutoff, percentage, round)
def plot_classification(self, cutoff=0.5, percentage=True, pos_label=None):
"""plot showing a barchart of the classification result for cutoff
Args:
cutoff(float, optional): cutoff of positive class to calculate lift
(Default value = 0.5)
percentage(bool, optional): display percentages instead of counts,
defaults to True
pos_label: positive label to display, defaults to self.pos_label
Returns:
plotly fig
"""
return plotly_classification_plot(self.pred_probas(pos_label), self.y, self.labels, cutoff, percentage=percentage)
def plot_roc_auc(self, cutoff=0.5, pos_label=None):
"""plots ROC_AUC curve.
The TPR and FPR of a particular cutoff is displayed in crosshairs.
Args:
cutoff: cutoff value to be included in plot (Default value = 0.5)
pos_label: (Default value = None)
Returns:
"""
if self.y_missing:
raise ValueError("No y was passed to explainer, so cannot plot roc auc!")
return plotly_roc_auc_curve(self.y_binary(pos_label), self.pred_probas(pos_label), cutoff=cutoff)
def plot_pr_auc(self, cutoff=0.5, pos_label=None):
"""plots PR_AUC curve.
the precision and recall of particular cutoff is displayed in crosshairs.
Args:
cutoff: cutoff value to be included in plot (Default value = 0.5)
pos_label: (Default value = None)
Returns:
"""
if self.y_missing:
raise ValueError("No y was passed to explainer, so cannot plot PR AUC!")
return plotly_pr_auc_curve(self.y_binary(pos_label), self.pred_probas(pos_label), cutoff=cutoff)
def plot_prediction_result(self, index=None, X_row=None, showlegend=True):
"""Returns a piechart with the predicted probabilities distribution
Args:
index ({int, str}): Index for which to display prediction
X_row (pd.DataFrame): single row of an input dataframe, e.g.
explainer.X.iloc[[0]]
showlegend (bool, optional): Display legend. Defaults to False.
Returns:
plotly.fig
"""
preds_df = self.prediction_result_df(index, X_row)
return plotly_prediction_piechart(preds_df, showlegend=showlegend)
def calculate_properties(self, include_interactions=True):
"""calculate all lazily calculated properties of explainer
Args:
include_interactions: (Default value = True)
Returns:
None
"""
_ = self.pred_probas
super().calculate_properties(include_interactions=include_interactions)
class RegressionExplainer(BaseExplainer):
""" """
def __init__(self, model, X, y=None, permutation_metric=r2_score,
shap="guess", X_background=None, model_output="raw",
cats=None, idxs=None, index_name=None, target=None,
descriptions=None, n_jobs=None, permutation_cv=None,
na_fill=-999, units=""):
"""Explainer for regression models.
In addition to BaseExplainer defines a number of plots specific to
regression problems such as a predicted vs actual and residual plots.
Combared to BaseExplainerBunch defines two additional parameters.
Args:
units(str): units to display for regression quantity
"""
super().__init__(model, X, y, permutation_metric,
shap, X_background, model_output,
cats, idxs, index_name, target, descriptions,
n_jobs, permutation_cv, na_fill)
self._params_dict = {**self._params_dict, **dict(units=units)}
self.units = units
self.is_regression = True
if str(type(self.model)).endswith("RandomForestRegressor'>"):
print(f"Changing class type to RandomForestRegressionExplainer...", flush=True)
self.__class__ = RandomForestRegressionExplainer
if str(type(self.model)).endswith("XGBRegressor'>"):
print(f"Changing class type to XGBRegressionExplainer...", flush=True)
self.__class__ = XGBRegressionExplainer
@property
def residuals(self):
"""residuals: y-preds"""
if not hasattr(self, '_residuals'):
print("Calculating residuals...")
self._residuals = self.y-self.preds
return self._residuals
@property
def abs_residuals(self):
"""absolute residuals"""
if not hasattr(self, '_abs_residuals'):
print("Calculating absolute residuals...")
self._abs_residuals = np.abs(self.residuals)
return self._abs_residuals
def random_index(self, y_min=None, y_max=None, pred_min=None, pred_max=None,
residuals_min=None, residuals_max=None,
abs_residuals_min=None, abs_residuals_max=None,
return_str=False, **kwargs):
"""random index following to various exclusion criteria
Args:
y_min: (Default value = None)
y_max: (Default value = None)
pred_min: (Default value = None)
pred_max: (Default value = None)
residuals_min: (Default value = None)
residuals_max: (Default value = None)
abs_residuals_min: (Default value = None)
abs_residuals_max: (Default value = None)
return_str: return the str index from self.idxs (Default value = False)
**kwargs:
Returns:
a random index that fits the exclusion criteria
"""
if self.y_missing:
if pred_min is None:
pred_min = self.preds.min()
if pred_max is None:
pred_max = self.preds.max()
potential_idxs = self.idxs[
(self.preds >= pred_min) &
(self.preds <= pred_max)].values
else:
if y_min is None:
y_min = self.y.min()
if y_max is None:
y_max = self.y.max()
if pred_min is None:
pred_min = self.preds.min()
if pred_max is None:
pred_max = self.preds.max()
if residuals_min is None:
residuals_min = self.residuals.min()
if residuals_max is None:
residuals_max = self.residuals.max()
if abs_residuals_min is None:
abs_residuals_min = self.abs_residuals.min()
if abs_residuals_max is None:
abs_residuals_max = self.abs_residuals.max()
potential_idxs = self.idxs[(self.y >= y_min) &
(self.y <= y_max) &
(self.preds >= pred_min) &
(self.preds <= pred_max) &
(self.residuals >= residuals_min) &
(self.residuals <= residuals_max) &
(self.abs_residuals >= abs_residuals_min) &
(self.abs_residuals <= abs_residuals_max)].values
if len(potential_idxs) > 0:
idx = np.random.choice(potential_idxs)
else:
return None
if return_str:
return idx
return self.idxs.get_loc(idx)
def prediction_result_markdown(self, index, include_percentile=True, round=2):
"""markdown of prediction result
Args:
index: row index to be predicted
include_percentile (bool): include line about prediciton percentile
round: (Default value = 2)
Returns:
str: markdown summary of prediction for index
"""
int_idx = self.get_int_idx(index)
model_prediction = "###### Prediction:\n"
model_prediction += f"Predicted {self.target}: {np.round(self.preds[int_idx], round)} {self.units}\n\n"
if not self.y_missing:
model_prediction += f"Observed {self.target}: {np.round(self.y[int_idx], round)} {self.units}\n\n"
model_prediction += f"Residual: {np.round(self.residuals[int_idx], round)} {self.units}\n\n"
if include_percentile:
percentile = np.round(100*(1-self.pred_percentiles[int_idx]))
model_prediction += f"\nIn top {percentile}% percentile predicted {self.target}"
return model_prediction
def prediction_result_df(self, index=None, X_row=None, round=3):
"""prediction result in dataframe format
Args:
index: row index to be predicted
round (int): rounding applied to floats (defaults to 3)
Returns:
pd.DataFrame
"""
if index is None and X_row is None:
raise ValueError("You need to either pass an index or X_row!")
if index is not None:
int_idx = self.get_int_idx(index)
preds_df = pd.DataFrame(columns = ["", self.target])
preds_df = preds_df.append(
pd.Series(("Predicted", str(np.round(self.preds[int_idx], round)) + f" {self.units}"),
index=preds_df.columns), ignore_index=True)
if not self.y_missing:
preds_df = preds_df.append(
pd.Series(("Observed", str(np.round(self.y[int_idx], round)) + f" {self.units}"),
index=preds_df.columns), ignore_index=True)
preds_df = preds_df.append(
pd.Series(("Residual", str(np.round(self.residuals[int_idx], round)) + f" {self.units}"),
index=preds_df.columns), ignore_index=True)
elif X_row is not None:
if X_row.columns.tolist()==self.X_cats.columns.tolist():
X_row = X_cats_to_X(X_row, self.cats_dict, self.X.columns)
assert np.all(X_row.columns==self.X.columns), \
("The column names of X_row should match X! Instead X_row.columns"
f"={X_row.columns.tolist()}...")
prediction = self.model.predict(X_row)[0]
preds_df = pd.DataFrame(columns = ["", self.target])
preds_df = preds_df.append(
pd.Series(("Predicted", str(np.round(prediction, round)) + f" {self.units}"),
index=preds_df.columns), ignore_index=True)
return preds_df
def metrics(self):
"""dict of performance metrics: rmse, mae and R^2"""
if self.y_missing:
raise ValueError("No y was passed to explainer, so cannot calculate metrics!")
metrics_dict = {
'root_mean_squared_error' : np.sqrt(mean_squared_error(self.y, self.preds)),
'mean_absolute_error' : mean_absolute_error(self.y, self.preds),
'R-squared' : r2_score(self.y, self.preds),
}
return metrics_dict
def metrics_descriptions(self):
metrics_dict = self.metrics()
metrics_descriptions_dict = {}
for k, v in metrics_dict.items():
if k == 'root_mean_squared_error':
metrics_descriptions_dict[k] = f"A measure of how close predicted value fits true values, where large deviations are punished more heavily. So the lower this number the better the model."
if k == 'mean_absolute_error':
metrics_descriptions_dict[k] = f"On average predictions deviate {round(v, 2)} {self.units} off the observed value of {self.target} (can be both above or below)"
if k == 'R-squared':
metrics_descriptions_dict[k] = f"{round(100*v, 2)}% of all variation in {self.target} was explained by the model."
return metrics_descriptions_dict
def plot_predicted_vs_actual(self, round=2, logs=False, log_x=False, log_y=False, **kwargs):
"""plot with predicted value on x-axis and actual value on y axis.
Args:
round(int, optional): rounding to apply to outcome, defaults to 2
logs (bool, optional): log both x and y axis, defaults to False
log_y (bool, optional): only log x axis. Defaults to False.
log_x (bool, optional): only log y axis. Defaults to False.
**kwargs:
Returns:
Plotly fig
"""
if self.y_missing:
raise ValueError("No y was passed to explainer, so cannot plot predicted vs actual!")
return plotly_predicted_vs_actual(self.y, self.preds,
target=self.target, units=self.units, idxs=self.idxs.values,
logs=logs, log_x=log_x, log_y=log_y, round=round,
index_name=self.index_name)
def plot_residuals(self, vs_actual=False, round=2, residuals='difference'):
"""plot of residuals. x-axis is the predicted outcome by default
Args:
vs_actual(bool, optional): use actual value for x-axis,
defaults to False
round(int, optional): rounding to perform on values, defaults to 2
residuals (str, {'difference', 'ratio', 'log-ratio'} optional):
How to calcualte residuals. Defaults to 'difference'.
Returns:
Plotly fig
"""
if self.y_missing:
raise ValueError("No y was passed to explainer, so cannot plot residuals!")
return plotly_plot_residuals(self.y, self.preds, idxs=self.idxs.values,
vs_actual=vs_actual, target=self.target,
units=self.units, residuals=residuals,
round=round, index_name=self.index_name)
def plot_residuals_vs_feature(self, col, residuals='difference', round=2,
dropna=True, points=True, winsor=0):
"""Plot residuals vs individual features
Args:
col(str): Plot against feature col
residuals (str, {'difference', 'ratio', 'log-ratio'} optional):
How to calcualte residuals. Defaults to 'difference'.
round(int, optional): rounding to perform on residuals, defaults to 2
dropna(bool, optional): drop missing values from plot, defaults to True.
points (bool, optional): display point cloud next to violin plot.
Defaults to True.
winsor (int, 0-50, optional): percentage of outliers to winsor out of
the y-axis. Defaults to 0.
Returns:
plotly fig
"""
if self.y_missing:
raise ValueError("No y was passed to explainer, so cannot plot residuals!")
assert col in self.columns or col in self.columns_cats, \
f'{col} not in columns or columns_cats!'
col_vals = self.X_cats[col] if self.check_cats(col) else self.X[col]
na_mask = col_vals != self.na_fill if dropna else np.array([True]*len(col_vals))
return plotly_residuals_vs_col(
self.y[na_mask], self.preds[na_mask], col_vals[na_mask],
residuals=residuals, idxs=self.idxs.values[na_mask], points=points,
round=round, winsor=winsor, index_name=self.index_name)
def plot_y_vs_feature(self, col, residuals='difference', round=2,
dropna=True, points=True, winsor=0):
"""Plot y vs individual features
Args:
col(str): Plot against feature col
round(int, optional): rounding to perform on residuals, defaults to 2
dropna(bool, optional): drop missing values from plot, defaults to True.
points (bool, optional): display point cloud next to violin plot.
Defaults to True.
winsor (int, 0-50, optional): percentage of outliers to winsor out of
the y-axis. Defaults to 0.
Returns:
plotly fig
"""
if self.y_missing:
raise ValueError("No y was passed to explainer, so cannot plot y vs feature!")
assert col in self.columns or col in self.columns_cats, \
f'{col} not in columns or columns_cats!'
col_vals = self.X_cats[col] if self.check_cats(col) else self.X[col]
na_mask = col_vals != self.na_fill if dropna else np.array([True]*len(col_vals))
return plotly_actual_vs_col(self.y[na_mask], self.preds[na_mask], col_vals[na_mask],
idxs=self.idxs.values[na_mask], points=points, round=round, winsor=winsor,
units=self.units, target=self.target, index_name=self.index_name)
def plot_preds_vs_feature(self, col, residuals='difference', round=2,
dropna=True, points=True, winsor=0):
"""Plot y vs individual features
Args:
col(str): Plot against feature col
round(int, optional): rounding to perform on residuals, defaults to 2
dropna(bool, optional): drop missing values from plot, defaults to True.
points (bool, optional): display point cloud next to violin plot.
Defaults to True.
winsor (int, 0-50, optional): percentage of outliers to winsor out of
the y-axis. Defaults to 0.
Returns:
plotly fig
"""
assert col in self.columns or col in self.columns_cats, \
f'{col} not in columns or columns_cats!'
col_vals = self.X_cats[col] if self.check_cats(col) else self.X[col]
na_mask = col_vals != self.na_fill if dropna else np.array([True]*len(col_vals))
return plotly_preds_vs_col(self.y[na_mask], self.preds[na_mask], col_vals[na_mask],
idxs=self.idxs.values[na_mask], points=points, round=round, winsor=winsor,
units=self.units, target=self.target, index_name=self.index_name)
class RandomForestExplainer(BaseExplainer):
"""RandomForestBunch allows for the analysis of individual DecisionTrees that
make up the RandomForest.
"""
@property
def is_tree_explainer(self):
"""this is either a RandomForestExplainer or XGBExplainer"""
return True
@property
def no_of_trees(self):
"""The number of trees in the RandomForest model"""
return len(self.model.estimators_)
@property
def graphviz_available(self):
""" """
if not hasattr(self, '_graphviz_available'):
try:
import graphviz.backend as be
cmd = ["dot", "-V"]
stdout, stderr = be.run(cmd, capture_output=True, check=True, quiet=True)
except:
print("""
WARNING: you don't seem to have graphviz in your path (cannot run 'dot -V'),
so no dtreeviz visualisation of decision trees will be shown on the shadow trees tab.
See https://github.com/parrt/dtreeviz for info on how to properly install graphviz
for dtreeviz.
""")
self._graphviz_available = False
else:
self._graphviz_available = True
return self._graphviz_available
@property
def decision_trees(self):
"""a list of ShadowDecTree objects"""
if not hasattr(self, '_decision_trees'):
#print("Calculating ShadowDecTree for each individual decision tree...", flush=True)
assert hasattr(self.model, 'estimators_'), \
"""self.model does not have an estimators_ attribute, so probably not
actually a sklearn RandomForest?"""
self._decision_trees = [
ShadowDecTree.get_shadow_tree(decision_tree,
self.X,
self.y,
feature_names=self.X.columns.tolist(),
target_name='target',
class_names = self.labels if self.is_classifier else None)
for decision_tree in self.model.estimators_]
return self._decision_trees
def decisiontree_df(self, tree_idx, index, pos_label=None):
"""dataframe with all decision nodes of a particular decision tree
Args:
tree_idx: the n'th tree in the random forest
index: row index
round: (Default value = 2)
pos_label: positive class (Default value = None)
Returns:
dataframe with summary of the decision tree path
"""
assert tree_idx >= 0 and tree_idx < len(self.decision_trees), \
f"tree index {tree_idx} outside 0 and number of trees ({len(self.decision_trees)}) range"
idx = self.get_int_idx(index)
assert idx >= 0 and idx < len(self.X), \
f"=index {idx} outside 0 and size of X ({len(self.X)}) range"
if self.is_classifier:
if pos_label is None: pos_label = self.pos_label
return get_decisiontree_df(self.decision_trees[tree_idx], self.X.iloc[idx],
pos_label=pos_label)
else:
return get_decisiontree_df(self.decision_trees[tree_idx], self.X.iloc[idx])
def decisiontree_summary_df(self, tree_idx, index, round=2, pos_label=None):
"""formats decisiontree_df in a slightly more human readable format.
Args:
tree_idx: the n'th tree in the random forest
index: row index
round: (Default value = 2)
pos_label: positive class (Default value = None)
Returns:
dataframe with summary of the decision tree path
"""
idx=self.get_int_idx(index)
return get_decisiontree_summary_df(self.decisiontree_df(tree_idx, idx, pos_label=pos_label),
classifier=self.is_classifier, round=round, units=self.units)
def decision_path_file(self, tree_idx, index, show_just_path=False):
"""get a dtreeviz visualization of a particular tree in the random forest.
Args:
tree_idx: the n'th tree in the random forest
index: row index
show_just_path (bool, optional): show only the path not rest of the
tree. Defaults to False.
Returns:
the path where the .svg file is stored.
"""
if not self.graphviz_available:
print("No graphviz 'dot' executable available!")
return None
idx = self.get_int_idx(index)
viz = dtreeviz(self.decision_trees[tree_idx],
X=self.X.iloc[idx, :],
fancy=False,
show_node_labels = False,
show_just_path=show_just_path)
return viz.save_svg()
def decision_path(self, tree_idx, index, show_just_path=False):
"""get a dtreeviz visualization of a particular tree in the random forest.
Args:
tree_idx: the n'th tree in the random forest
index: row index
show_just_path (bool, optional): show only the path not rest of the
tree. Defaults to False.
Returns:
a IPython display SVG object for e.g. jupyter notebook.
"""
if not self.graphviz_available:
print("No graphviz 'dot' executable available!")
return None
from IPython.display import SVG
svg_file = self.decision_path_file(tree_idx, index, show_just_path)
return SVG(open(svg_file,'rb').read())
def decision_path_encoded(self, tree_idx, index, show_just_path=False):
"""get a dtreeviz visualization of a particular tree in the random forest.
Args:
tree_idx: the n'th tree in the random forest
index: row index
show_just_path (bool, optional): show only the path not rest of the
tree. Defaults to False.
Returns:
a base64 encoded image, for inclusion in websites (e.g. dashboard)
"""
if not self.graphviz_available:
print("No graphviz 'dot' executable available!")
return None
svg_file = self.decision_path_file(tree_idx, index, show_just_path)
encoded = base64.b64encode(open(svg_file,'rb').read())
svg_encoded = 'data:image/svg+xml;base64,{}'.format(encoded.decode())
return svg_encoded
def plot_trees(self, index, highlight_tree=None, round=2,
higher_is_better=True, pos_label=None):
"""plot barchart predictions of each individual prediction tree
Args:
index: index to display predictions for
highlight_tree: tree to highlight in plot (Default value = None)
round: rounding of numbers in plot (Default value = 2)
higher_is_better (bool): flip red and green. Dummy bool for compatibility
with gbm plot_trees().
pos_label: positive class (Default value = None)
Returns:
"""
idx=self.get_int_idx(index)
assert idx is not None, 'invalid index'
if self.is_classifier:
if pos_label is None: pos_label = self.pos_label
if not np.isnan(self.y[idx]):
y = 100*self.y_binary(pos_label)[idx]
else:
y = None
return plotly_rf_trees(self.model, self.X.iloc[[idx]], y,
highlight_tree=highlight_tree, round=round,
pos_label=pos_label, target=self.target)
else:
y = self.y[idx]
return plotly_rf_trees(self.model, self.X.iloc[[idx]], y,
highlight_tree=highlight_tree, round=round,
target=self.target, units=self.units)
def calculate_properties(self, include_interactions=True):
"""
Args:
include_interactions: If False do not calculate shap interaction value
(Default value = True)
Returns:
"""
_ = self.decision_trees
super().calculate_properties(include_interactions=include_interactions)
class XGBExplainer(BaseExplainer):
"""XGBExplainer allows for the analysis of individual DecisionTrees that
make up the xgboost model.
"""
@property
def is_tree_explainer(self):
"""this is either a RandomForestExplainer or XGBExplainer"""
return True
@property
def model_dump_list(self):
if not hasattr(self, "_model_dump_list"):
print("Generating model dump...", flush=True)
self._model_dump_list = self.model.get_booster().get_dump()
return self._model_dump_list
@property
def no_of_trees(self):
"""The number of trees in the RandomForest model"""
if self.is_classifier and len(self.labels) > 2:
return int(len(self.model_dump_list) / len(self.labels))
return len(self.model_dump_list)
@property
def graphviz_available(self):
""" """
if not hasattr(self, '_graphviz_available'):
try:
import graphviz.backend as be
cmd = ["dot", "-V"]
stdout, stderr = be.run(cmd, capture_output=True, check=True, quiet=True)
except:
print("""
WARNING: you don't seem to have graphviz in your path (cannot run 'dot -V'),
so no dtreeviz visualisation of decision trees will be shown on the shadow trees tab.
See https://github.com/parrt/dtreeviz for info on how to properly install graphviz
for dtreeviz.
""")
self._graphviz_available = False
else:
self._graphviz_available = True
return self._graphviz_available
@property
def decision_trees(self):
"""a list of ShadowDecTree objects"""
if not hasattr(self, '_decision_trees'):
#print("Calculating ShadowDecTree for each individual decision tree...", flush=True)
self._decision_trees = [
ShadowDecTree.get_shadow_tree(self.model.get_booster(),
self.X,
self.y,
feature_names=self.X.columns.tolist(),
target_name='target',
class_names = self.labels if self.is_classifier else None,
tree_index=i)
for i in range(len(self.model_dump_list))]
return self._decision_trees
def decisiontree_df(self, tree_idx, index, pos_label=None):
"""dataframe with all decision nodes of a particular decision tree
Args:
tree_idx: the n'th tree in the random forest
index: row index
round: (Default value = 2)
pos_label: positive class (Default value = None)
Returns:
dataframe with summary of the decision tree path
"""
assert tree_idx >= 0 and tree_idx < self.no_of_trees, \
f"tree index {tree_idx} outside 0 and number of trees ({len(self.decision_trees)}) range"
idx = self.get_int_idx(index)
assert idx >= 0 and idx < len(self.X), \
f"=index {idx} outside 0 and size of X ({len(self.X)}) range"
if self.is_classifier:
if pos_label is None:
pos_label = self.pos_label
if len(self.labels) > 2:
tree_idx = tree_idx * len(self.labels) + pos_label
return get_xgboost_path_df(self.model_dump_list[tree_idx], self.X.iloc[idx])
def decisiontree_summary_df(self, tree_idx, index, round=2, pos_label=None):
"""formats decisiontree_df in a slightly more human readable format.
Args:
tree_idx: the n'th tree in the random forest
index: row index
round: (Default value = 2)
pos_label: positive class (Default value = None)
Returns:
dataframe with summary of the decision tree path
"""
idx = self.get_int_idx(index)
return get_xgboost_path_summary_df(self.decisiontree_df(tree_idx, idx, pos_label=pos_label))
def decision_path_file(self, tree_idx, index, show_just_path=False, pos_label=None):
"""get a dtreeviz visualization of a particular tree in the random forest.
Args:
tree_idx: the n'th tree in the random forest
index: row index
show_just_path (bool, optional): show only the path not rest of the
tree. Defaults to False.
pos_label: for classifiers, positive label class
Returns:
the path where the .svg file is stored.
"""
if not self.graphviz_available:
print("No graphviz 'dot' executable available!")
return None
idx = self.get_int_idx(index)
if self.is_classifier:
if pos_label is None:
pos_label = self.pos_label
if len(self.labels) > 2:
tree_idx = tree_idx * len(self.labels) + pos_label
viz = dtreeviz(self.decision_trees[tree_idx],
X=self.X.iloc[idx],
fancy=False,
show_node_labels = False,
show_just_path=show_just_path)
return viz.save_svg()
def decision_path(self, tree_idx, index, show_just_path=False, pos_label=None):
"""get a dtreeviz visualization of a particular tree in the random forest.
Args:
tree_idx: the n'th tree in the random forest
index: row index
show_just_path (bool, optional): show only the path not rest of the
tree. Defaults to False.
Returns:
a IPython display SVG object for e.g. jupyter notebook.
"""
if not self.graphviz_available:
print("No graphviz 'dot' executable available!")
return None
from IPython.display import SVG
svg_file = self.decision_path_file(tree_idx, index, show_just_path, pos_label)
return SVG(open(svg_file,'rb').read())
def decision_path_encoded(self, tree_idx, index, show_just_path=False, pos_label=None):
"""get a dtreeviz visualization of a particular tree in the random forest.
Args:
tree_idx: the n'th tree in the random forest
index: row index
show_just_path (bool, optional): show only the path not rest of the
tree. Defaults to False.
Returns:
a base64 encoded image, for inclusion in websites (e.g. dashboard)
"""
if not self.graphviz_available:
print("No graphviz 'dot' executable available!")
return None
svg_file = self.decision_path_file(tree_idx, index, show_just_path, pos_label)
encoded = base64.b64encode(open(svg_file,'rb').read())
svg_encoded = 'data:image/svg+xml;base64,{}'.format(encoded.decode())
return svg_encoded
def plot_trees(self, index, highlight_tree=None, round=2,
higher_is_better=True, pos_label=None):
"""plot barchart predictions of each individual prediction tree
Args:
index: index to display predictions for
highlight_tree: tree to highlight in plot (Default value = None)
round: rounding of numbers in plot (Default value = 2)
higher_is_better (bool, optional): up is green, down is red. If False
flip the colors.
pos_label: positive class (Default value = None)
Returns:
"""
idx=self.get_int_idx(index)
assert idx is not None, 'invalid index'
if self.is_classifier:
if pos_label is None:
pos_label = self.pos_label
y = self.y_binary(pos_label)[idx]
xgboost_preds_df = get_xgboost_preds_df(
self.model, self.X.iloc[[idx]], pos_label=pos_label)
return plotly_xgboost_trees(xgboost_preds_df,
y=y,
highlight_tree=highlight_tree,
target=self.target,
higher_is_better=higher_is_better)
else:
y = self.y[idx]
xgboost_preds_df = get_xgboost_preds_df(
self.model, self.X.iloc[[idx]])
return plotly_xgboost_trees(xgboost_preds_df,
y=y, highlight_tree=highlight_tree,
target=self.target, units=self.units,
higher_is_better=higher_is_better)
def calculate_properties(self, include_interactions=True):
"""
Args:
include_interactions: If False do not calculate shap interaction value
(Default value = True)
Returns:
"""
_ = self.decision_trees, self.model_dump_list
super().calculate_properties(include_interactions=include_interactions)
class RandomForestClassifierExplainer(RandomForestExplainer, ClassifierExplainer):
"""RandomForestClassifierExplainer inherits from both RandomForestExplainer and
ClassifierExplainer.
"""
pass
class RandomForestRegressionExplainer(RandomForestExplainer, RegressionExplainer):
"""RandomForestRegressionExplainer inherits from both RandomForestExplainer and
RegressionExplainer.
"""
pass
class XGBClassifierExplainer(XGBExplainer, ClassifierExplainer):
"""RandomForestClassifierBunch inherits from both RandomForestExplainer and
ClassifierExplainer.
"""
pass
class XGBRegressionExplainer(XGBExplainer, RegressionExplainer):
"""XGBRegressionExplainer inherits from both XGBExplainer and
RegressionExplainer.
"""
pass
class ClassifierBunch:
""" """
def __init__(self, *args, **kwargs):
raise ValueError("ClassifierBunch has been deprecated, use ClassifierExplainer instead...")
class RegressionBunch:
""" """
def __init__(self, *args, **kwargs):
raise ValueError("RegressionBunch has been deprecated, use RegressionrExplainer instead...")
class RandomForestExplainerBunch:
""" """
def __init__(self, *args, **kwargs):
raise ValueError("RandomForestExplainerBunch has been deprecated, use RandomForestExplainer instead...")
class RandomForestClassifierBunch:
""" """
def __init__(self, *args, **kwargs):
raise ValueError("RandomForestClassifierBunch has been deprecated, use RandomForestClassifierExplainer instead...")
class RandomForestRegressionBunch:
""" """
def __init__(self, *args, **kwargs):
raise ValueError("RandomForestRegressionBunch has been deprecated, use RandomForestRegressionExplainer instead...")
from .AMLBID_Dashboard import *
def explain(model,conf, X_test, Y_test):
explainer = ClassifierExplainer(model,conf, X_test, Y_test)
db = ExplainerDashboard(explainer, mode='external',dev_tools_props_check=False)
return db | AMLBID | /Explainer/AMLBID_Explainer.py | AMLBID_Explainer.py |
__all__= [
'plotly_prediction_piechart',
'plotly_contribution_plot',
'plotly_precision_plot',
'plotly_classification_plot',
'plotly_lift_curve',
'plotly_cumulative_precision_plot',
'plotly_dependence_plot',
'plotly_shap_violin_plot',
'plotly_pdp',
'plotly_importances_plot',
'plotly_confusion_matrix',
'plotly_roc_auc_curve',
'plotly_pr_auc_curve',
'plotly_shap_scatter_plot',
'plotly_predicted_vs_actual',
'plotly_plot_residuals',
'plotly_residuals_vs_col',
'plotly_actual_vs_col',
'plotly_preds_vs_col',
'plotly_rf_trees',
'plotly_xgboost_trees'
]
import numpy as np
import pandas as pd
from pandas.api.types import is_numeric_dtype, is_string_dtype
import plotly.graph_objs as go
from plotly.subplots import make_subplots
from sklearn.metrics import (classification_report, confusion_matrix,
precision_recall_curve, roc_curve,
roc_auc_score, average_precision_score)
def plotly_prediction_piechart(predictions_df, showlegend=True, size=250):
"""Return piechart with predict_proba distributions for ClassifierExplainer
Args:
predictions_df (pd.DataFrame): generated with
ClassifierExplainer.prediction_summary_df(index)
showlegend (bool, optional): Show the legend. Defaults to True.
size (int): width and height of the plot
Returns:
plotly.Fig
"""
data = [
go.Pie(labels=predictions_df.label.values,
values=predictions_df.probability.values,
hole=0.3,
sort=False)
]
layout = dict(autosize=False, width=size, height=size,
margin=dict(l=20, r=20, b=20, t=30, pad=4),
showlegend=showlegend)
fig = go.Figure(data, layout)
return fig
def plotly_contribution_plot(contrib_df, target="",
model_output="raw", higher_is_better=True,
include_base_value=True, include_prediction=True,
orientation='vertical', round=2, units=""):
"""Generate a shap contributions waterfall plot from a contrib_df dataframe
Args:
contrib_df (pd.DataFrame): contrib_df generated with get_contrib_df(...)
target (str, optional): Target variable to be displayed. Defaults to "".
model_output ({"raw", "logodds", "probability"}, optional): Kind of
output of the model. Defaults to "raw".
higher_is_better (bool, optional): Display increases in shap as green,
decreases as red. Defaults to False.
include_base_value (bool, optional): Include shap base value in the plot.
Defaults to True.
include_prediction (bool, optional): Include the final prediction in
the plot. Defaults to True.
orientation ({'vertical', 'horizontal'}, optional): Display the plot
vertically or horizontally. Defaults to 'vertical'.
round (int, optional): Round of floats. Defaults to 2.
units (str, optional): Units of outcome variable. Defaults to "".
Returns:
plotly fig:
"""
if orientation not in ['vertical', 'horizontal']:
raise ValueError(f"orientation should be in ['vertical', 'horizontal'], but you passed orientation={orientation}")
if model_output not in ['raw', 'probability', 'logodds']:
raise ValueError(f"model_output should be in ['raw', 'probability', 'logodds'], but you passed orientation={model_output}")
contrib_df = contrib_df.copy()
try:
base_value = contrib_df.query("col=='_BASE'")['contribution'].item()
except:
base_value = None
if not include_base_value:
contrib_df = contrib_df[contrib_df.col != '_BASE']
if not include_prediction:
contrib_df = contrib_df[contrib_df.col != '_PREDICTION']
contrib_df = contrib_df.replace({'_BASE': 'Population<br>average',
'_REST': 'Other features combined',
'_PREDICTION': 'Final Prediction'})
multiplier = 100 if model_output=='probability' else 1
contrib_df['base'] = np.round(multiplier * contrib_df['base'].astype(float), round)
contrib_df['cumulative'] = np.round(multiplier * contrib_df['cumulative'].astype(float), round)
contrib_df['contribution'] = np.round(multiplier * contrib_df['contribution'].astype(float), round)
if not include_base_value:
contrib_df = contrib_df[contrib_df.col != '_BASE']
longest_feature_name = contrib_df['col'].str.len().max()
# prediction is the sum of all contributions:
prediction = contrib_df['cumulative'].values[-1]
cols = contrib_df['col'].values.tolist()
values = contrib_df.value.tolist()
bases = contrib_df.base.tolist()
contribs = contrib_df.contribution.tolist()
if 'value' in contrib_df.columns:
hover_text=[f"{col}={value}<BR>{'+' if contrib>0 else ''}{contrib} {units}"
for col, value, contrib in zip(
cols, values, contribs)]
else:
hover_text=[f"{col}=?<BR>{'+' if contrib>0 else ''}{contrib} {units}"
for col, contrib in zip(cols, contribs)]
green_fill, green_line = 'rgba(50, 200, 50, 1.0)', 'rgba(40, 160, 50, 1.0)'
yellow_fill, yellow_line = 'rgba(230, 230, 30, 1.0)', 'rgba(190, 190, 30, 1.0)'
blue_fill, blue_line = 'rgba(55, 128, 191, 0.7)', 'rgba(55, 128, 191, 1.0)'
red_fill, red_line = 'rgba(219, 64, 82, 0.7)', 'rgba(219, 64, 82, 1.0)'
fill_color_up = green_fill if higher_is_better else red_fill
fill_color_down = red_fill if higher_is_better else green_fill
line_color_up = green_line if higher_is_better else red_line
line_color_down = red_line if higher_is_better else green_line
fill_colors = [fill_color_up if y > 0 else fill_color_down for y in contribs]
line_colors = [line_color_up if y > 0 else line_color_down for y in contribs]
if include_base_value:
fill_colors[0] = yellow_fill
line_colors[0] = yellow_line
if include_prediction:
fill_colors[-1] = blue_fill
line_colors[-1] = blue_line
if orientation == 'horizontal':
cols = cols[::-1]
values = values[::-1]
contribs = contribs[::-1]
bases = bases[::-1]
fill_colors = fill_colors[::-1]
line_colors = line_colors[::-1]
# Base of each bar
trace0 = go.Bar(
x=bases if orientation=='horizontal' else cols,
y=cols if orientation=='horizontal' else bases,
hoverinfo='skip',
name="",
marker=dict(
color='rgba(1,1,1, 0.0)',
),
orientation='h' if orientation=='horizontal' else None
)
# top of each bar (base + contribution)
trace1 = go.Bar(
x=contribs if orientation=='horizontal' else cols,
y=cols if orientation=='horizontal' else contribs,
text=hover_text,
name="contribution",
hoverinfo="text",
marker=dict(
# blue if positive contribution, red if negative
color=fill_colors,
line=dict(
color=line_colors,
width=2,
)
),
orientation='h' if orientation=='horizontal' else None
)
if model_output == "probability":
title = f'Contribution to prediction probability = {prediction}%'
elif model_output == "logodds":
title = f'Contribution to prediction logodds = {prediction}'
else:
title = f'Contribution to prediction {target} = {prediction} {units}'
data = [trace0, trace1]
layout = go.Layout(
height=600 if orientation=='vertical' else 100+35*len(cols),
title=title,
barmode='stack',
plot_bgcolor = '#fff',
showlegend=False
)
fig = go.Figure(data=data, layout=layout)
if model_output=='probability' and base_value is not None and base_value > 0.3 and base_value < 0.7:
# stretch out probability axis to entire probability range (0-100)
if orientation=='vertical':
fig.update_yaxes(range=[0, 100])
elif orientation=='horizontal':
fig.update_xaxes(range=[0, 100])
fig.update_layout(margin=go.layout.Margin(
l=longest_feature_name*7 if orientation=='horizontal' else 50,
r=100,
b=50 if orientation=='horizontal' else longest_feature_name*6 ,
t=50,
pad=4
),
title_x=0.5)
if orientation == 'vertical':
fig.update_yaxes(title_text='Predicted ' + ('%' if model_output=="probability" else units))
else:
fig.update_xaxes(title_text='Predicted ' + ('%' if model_output=="probability" else units))
return fig
def plotly_precision_plot(precision_df, cutoff=None, labels=None, pos_label=None):
"""Returns a plotly figure with average predicted probability and
percentage positive per probability bin.
Args:
precision_df (pd.DataFrame): generated with get_precision_df(..)
cutoff (float, optional): Model cutoff to display in graph. Defaults to None.
labels (List[str], optional): Labels for prediction classes. Defaults to None.
pos_label (int, optional): For multiclass classifiers: which class to treat
as positive class. Defaults to None.
Returns:
Plotly fig
"""
label = labels[pos_label] if labels is not None and pos_label is not None else 'positive'
precision_df = precision_df.copy()
spacing = 0.1 / len(precision_df)
bin_widths = precision_df['bin_width'] - spacing
bin_widths[bin_widths<0.005] = 0.005
trace1 = go.Bar(
x=(0.5*(precision_df['p_min']+precision_df['p_max'])).values,
y=precision_df['count'].values,
width=bin_widths,
name='counts'
)
data = [trace1]
if 'precision_0' in precision_df.columns.tolist():
# if a pred_proba with probability for every class gets passed
# to get_precision_df, it generates a precision for every class
# in every bin as well.
precision_cols = [col for col in precision_df.columns.tolist()
if col.startswith('precision_')]
if labels is None: labels = ['class ' + str(i) for i in range(len(precision_cols))]
if pos_label is not None:
# add the positive class first with thick line
trace = go.Scatter(
x=precision_df['p_avg'].values.tolist(),
y=precision_df['precision_'+str(pos_label)].values.tolist(),
name=labels[pos_label] + '(positive class)',
line = dict(width=4),
yaxis='y2')
data.append(trace)
for i, precision_col in enumerate(precision_cols):
# add the rest of the classes with thin lines
if pos_label is None or i != pos_label:
trace = go.Scatter(
x=precision_df['p_avg'].values.tolist(),
y=precision_df[precision_col].values.tolist(),
name=labels[i],
line = dict(width=2),
yaxis='y2')
data.append(trace)
else:
trace2 = go.Scatter(
x=precision_df['p_avg'].values.tolist(),
y=precision_df['precision'].values.tolist(),
name='percentage ' + label,
yaxis='y2'
)
data = [trace1, trace2]
layout = go.Layout(
title=f'percentage {label} vs predicted probability',
yaxis=dict(
title='counts'
),
yaxis2=dict(
title='percentage',
titlefont=dict(
color='rgb(148, 103, 189)'
),
tickfont=dict(
color='rgb(148, 103, 189)'
),
overlaying='y',
side='right',
rangemode='tozero'
),
xaxis=dict(
title='predicted probability'
),
plot_bgcolor = '#fff',
)
if cutoff is not None:
layout['shapes'] = [dict(
type='line',
xref='x',
yref='y2',
x0=cutoff,
x1=cutoff,
y0=0,
y1=1.0,
)]
fig = go.Figure(data=data, layout=layout)
fig.update_layout(legend=dict(orientation="h",
xanchor="center",
y=-0.2,
x=0.5))
if cutoff is not None:
fig.update_layout(annotations=[
go.layout.Annotation(x=cutoff, y=0.1, yref='y2',
text=f"cutoff={cutoff}")])
return fig
def plotly_classification_plot(pred_probas, targets, labels=None, cutoff=0.5,
pos_label=1, percentage=False):
"""Displays bar plots showing label distributions above and below cutoff
value.
Args:
pred_probas (np.ndarray): array of predicted probabilities
targets (np.ndarray): array of actual target labels
(e.g. [0, 1, 1, 0,...,1])
labels (List[str], optional): List of labels for classes. Defaults to None.
cutoff (float, optional): Cutoff pred_proba. Defaults to 0.5.
pos_label (int, optional): Positive label class. Defaults to 1.
percentage (bool, optional): Display percentage instead of absolute
numbers. Defaults to False.
Returns:
Plotly fig
"""
if len(pred_probas.shape) == 2:
below = (pred_probas[:, pos_label] < cutoff)
else:
below = pred_probas < cutoff
below_threshold = (pred_probas[below], targets[below])
above_threshold = (pred_probas[~below], targets[~below])
x = ['below cutoff', 'above cutoff', 'all']
fig = go.Figure()
for i, label in enumerate(labels):
text = [f"<b>{sum(below_threshold[1]==i)}</b><br>({np.round(100*np.mean(below_threshold[1]==i), 1)}%)",
f"<b>{sum(above_threshold[1]==i)}</b><br>({np.round(100*np.mean(above_threshold[1]==i), 1)}%)",
f"<b>{sum(targets==i)}</b><br>({np.round(100*np.mean(targets==i), 1)}%)"]
if percentage:
fig.add_trace(go.Bar(
x=x,
y=[100*np.mean(below_threshold[1]==i),
100*np.mean(above_threshold[1]==i),
100*np.mean(targets==i)],
# text=[str(np.round(100*np.mean(below_threshold[1]==i), 2)) + '%',
# str(np.round(100*np.mean(above_threshold[1]==i), 2)) + '%',
# str(np.round(100*np.mean(targets==i), 2)) + '%'],
text=text,
textposition='auto',
hoverinfo="text",
name=label))
fig.update_layout(title='Percentage above and below cutoff')
else:
fig.add_trace(go.Bar(
x=x,
y=[sum(below_threshold[1]==i),
sum(above_threshold[1]==i),
sum(targets==i)],
# text = [sum(below_threshold[1]==i),
# sum(above_threshold[1]==i),
# sum(targets==i)],
text=text,
textposition='auto',
hoverinfo="text",
name=label))
fig.update_layout(title='Total above and below cutoff')
fig.update_layout(barmode='stack')
fig.update_layout(legend=dict(orientation="h",
xanchor="center",
y=-0.2,
x=0.5))
return fig
def plotly_lift_curve(lift_curve_df, cutoff=None, percentage=False, round=2):
"""returns a lift plot for values
Args:
lift_curve_df (pd.DataFrame): generated with get_lift_curve_df(pred_proba, y)
cutoff (float, optional): cutoff above which samples get classified as
positive. Defaults to None.
percentage (bool, optional): Display percentages instead of absolute
numbers along axis. Defaults to False.
round (int, optional): Rounding to apply to floats. Defaults to 2.
Returns:
Plotly fig
"""
if percentage:
model_text=[f"model selected {np.round(pos, round)}% of all positives in first {np.round(i, round)}% sampled<br>" \
+ f"precision={np.round(precision, 2)}% positives in sample<br>" \
+ f"lift={np.round(pos/exp, 2)}"
for (i, pos, exp, precision) in zip(lift_curve_df.index_percentage,
lift_curve_df.cumulative_percentage_pos,
lift_curve_df.random_cumulative_percentage_pos,
lift_curve_df.precision)]
random_text=[f"random selected {np.round(exp, round)}% of all positives in first {np.round(i, round)}% sampled<br>" \
+ f"precision={np.round(precision, 2)}% positives in sample"
for (i, pos, exp, precision) in zip(lift_curve_df.index_percentage,
lift_curve_df.cumulative_percentage_pos,
lift_curve_df.random_cumulative_percentage_pos,
lift_curve_df.random_precision)]
else:
model_text=[f"model selected {pos} positives out of {i}<br>" \
+ f"precision={np.round(precision, 2)}<br>" \
+ f"lift={np.round(pos/exp, 2)}"
for (i, pos, exp, precision) in zip(lift_curve_df['index'],
lift_curve_df.positives,
lift_curve_df.random_pos,
lift_curve_df.precision)]
random_text=[f"random selected {np.round(exp).astype(int)} positives out of {i}<br>" \
+ f"precision={np.round(precision, 2)}"
for (i, pos, exp, precision) in zip(lift_curve_df['index'],
lift_curve_df.positives,
lift_curve_df.random_pos,
lift_curve_df.random_precision)]
trace0 = go.Scatter(
x=lift_curve_df['index_percentage'].values if percentage else lift_curve_df['index'],
y=np.round(lift_curve_df.cumulative_percentage_pos.values, round) if percentage \
else np.round(lift_curve_df.positives.values, round),
name='model',
text=model_text,
hoverinfo="text",
)
trace1 = go.Scatter(
x=lift_curve_df['index_percentage'].values if percentage else lift_curve_df['index'],
y=np.round(lift_curve_df.random_cumulative_percentage_pos.values, round) if percentage \
else np.round(lift_curve_df.random_pos.values, round),
name='random',
text=random_text,
hoverinfo="text",
)
data = [trace0, trace1]
fig = go.Figure(data)
fig.update_layout(title=dict(text='Lift curve',
x=0.5,
font=dict(size=18)),
xaxis_title= 'Percentage sampled' if percentage else 'Number sampled',
yaxis_title='Percentage of positive' if percentage else 'Number of positives',
xaxis=dict(spikemode="across"),
hovermode="x",
plot_bgcolor = '#fff')
fig.update_layout(legend=dict(xanchor="center", y=0.9, x=0.1))
if cutoff is not None:
#cutoff_idx = max(0, (np.abs(lift_curve_df.pred_proba - cutoff)).argmin() - 1)
cutoff_idx = max(0, len(lift_curve_df[lift_curve_df.pred_proba >= cutoff])-1)
if percentage:
cutoff_x = lift_curve_df['index_percentage'].iloc[cutoff_idx]
else:
cutoff_x = lift_curve_df['index'].iloc[cutoff_idx]
cutoff_n = lift_curve_df['index'].iloc[cutoff_idx]
cutoff_pos = lift_curve_df['positives'].iloc[cutoff_idx]
cutoff_random_pos = int(lift_curve_df['random_pos'].iloc[cutoff_idx])
cutoff_lift = np.round(lift_curve_df['positives'].iloc[cutoff_idx] / lift_curve_df.random_pos.iloc[cutoff_idx], 1)
cutoff_precision = np.round(lift_curve_df['precision'].iloc[cutoff_idx], 2)
cutoff_random_precision = np.round(lift_curve_df['random_precision'].iloc[cutoff_idx], 2)
fig.update_layout(shapes = [dict(
type='line',
xref='x',
yref='y',
x0=cutoff_x,
x1=cutoff_x,
y0=0,
y1=100.0 if percentage else lift_curve_df.positives.max(),
)]
)
fig.update_layout(annotations=[
go.layout.Annotation(
x=cutoff_x,
y=5,
yref='y',
text=f"cutoff={np.round(cutoff,3)}"),
go.layout.Annotation(x=0.5, y=0.4,
text=f"Model: {cutoff_pos} out {cutoff_n} ({cutoff_precision}%)",
showarrow=False, align="right",
xref='paper', yref='paper',
xanchor='left', yanchor='top'
),
go.layout.Annotation(x=0.5, y=0.33,
text=f"Random: {cutoff_random_pos} out {cutoff_n} ({cutoff_random_precision}%)",
showarrow=False, align="right",
xref='paper', yref='paper',
xanchor='left', yanchor='top'
),
go.layout.Annotation(x=0.5, y=0.26,
text=f"Lift: {cutoff_lift}",
showarrow=False, align="right",
xref='paper', yref='paper',
xanchor='left', yanchor='top'
)
])
return fig
def plotly_cumulative_precision_plot(lift_curve_df, labels=None, percentile=None, pos_label=1):
"""Return cumulative precision plot showing the expected label distribution
if you cumulatively sample a more and more of the highest predicted samples.
Args:
lift_curve_df (pd.DataFrame): generated with get_liftcurve_df(...)
labels (List[str], optional): list of labels for classes. Defaults to None.
pos_label (int, optional): Positive class label. Defaults to 1.
Returns:
Plotly fig
"""
if labels is None:
labels = ['category ' + str(i) for i in range(lift_curve_df.y.max()+1)]
fig = go.Figure()
text = [f"percentage sampled = top {round(idx_perc,2)}%"
for idx_perc in lift_curve_df['index_percentage'].values]
fig = fig.add_trace(go.Scatter(x=lift_curve_df.index_percentage,
y=np.zeros(len(lift_curve_df)),
showlegend=False,
text=text,
hoverinfo="text"))
text = [f"percentage {labels[pos_label]}={round(perc, 2)}%"
for perc in lift_curve_df['precision_' +str(pos_label)].values]
fig = fig.add_trace(go.Scatter(x=lift_curve_df.index_percentage,
y=lift_curve_df['precision_' +str(pos_label)].values,
fill='tozeroy',
name=labels[pos_label],
text=text,
hoverinfo="text"))
cumulative_y = lift_curve_df['precision_' +str(pos_label)].values
for y_label in range(pos_label, lift_curve_df.y.max()+1):
if y_label != pos_label:
cumulative_y = cumulative_y + lift_curve_df['precision_' +str(y_label)].values
text = [f"percentage {labels[y_label]}={round(perc, 2)}%"
for perc in lift_curve_df['precision_' +str(y_label)].values]
fig=fig.add_trace(go.Scatter(x=lift_curve_df.index_percentage,
y=cumulative_y,
fill='tonexty',
name=labels[y_label],
text=text,
hoverinfo="text"))
for y_label in range(0, pos_label):
if y_label != pos_label:
cumulative_y = cumulative_y + lift_curve_df['precision_' +str(y_label)].values
text = [f"percentage {labels[y_label]}={round(perc, 2)}%"
for perc in lift_curve_df['precision_' +str(y_label)].values]
fig=fig.add_trace(go.Scatter(x=lift_curve_df.index_percentage,
y=cumulative_y,
fill='tonexty',
name=labels[y_label],
text=text,
hoverinfo="text"))
fig.update_layout(title=dict(text='Cumulative percentage per category when sampling top X%',
x=0.5,
font=dict(size=18)),
yaxis=dict(title='Cumulative precision per category'),
xaxis=dict(title='Top X% model scores', spikemode="across", range=[0, 100]),
hovermode="x",
plot_bgcolor = '#fff')
if percentile is not None:
fig.update_layout(shapes=[dict(
type='line',
xref='x',
yref='y',
x0=100*percentile,
x1=100*percentile,
y0=0,
y1=100.0,
)])
fig.update_layout(annotations=[
go.layout.Annotation(x=100*percentile, y=20,
yref='y', ax=60,
text=f"percentile={np.round(100*percentile, 2)}")])
fig.update_xaxes(nticks=10)
return fig
def plotly_dependence_plot(X, shap_values, col_name, interact_col_name=None,
interaction=False, na_fill=-999, round=2, units="",
highlight_index=None, idxs=None, index_name="index"):
"""Returns a dependence plot showing the relationship between feature col_name
and shap values for col_name. Do higher values of col_name increase prediction
or decrease them? Or some kind of U-shape or other?
Args:
X (pd.DataFrame): dataframe with rows of input data
shap_values (np.ndarray): shap values generated for X
col_name (str): column name for which to generate plot
interact_col_name (str, optional): Column name by which to color the
markers. Defaults to None.
interaction (bool, optional): Is this a plot of shap interaction values?
Defaults to False.
na_fill (int, optional): value used for filling missing values.
Defaults to -999.
round (int, optional): Rounding to apply to floats. Defaults to 2.
units (str, optional): Units of the target variable. Defaults to "".
highlight_index (str, int, optional): index row of X to highlight in t
he plot. Defaults to None.
idxs (list, optional): list of descriptors of the index, e.g.
names or other identifiers. Defaults to None.
index_name (str): identifier for idxs. Defaults to "index".
Returns:
Plotly fig
"""
assert col_name in X.columns.tolist(), f'{col_name} not in X.columns'
assert (interact_col_name is None and not interaction) or interact_col_name in X.columns.tolist(),\
f'{interact_col_name} not in X.columns'
if idxs is not None:
assert len(idxs)==X.shape[0]
idxs = [str(idx) for idx in idxs]
else:
idxs = [str(i) for i in range(X.shape[0])]
if highlight_index is not None:
if isinstance(highlight_index, int):
highlight_idx = highlight_index
highlight_name = idxs[highlight_idx]
elif isinstance(highlight_index, str):
assert highlight_index in idxs, f'highlight_index should be int or in idxs, {highlight_index} is neither!'
highlight_idx = np.where(idxs==highlight_index)[0].item()
highlight_name = highlight_index
x = X[col_name].replace({-999:np.nan})
if len(shap_values.shape)==2:
y = shap_values[:, X.columns.get_loc(col_name)]
elif len(shap_values.shape)==3 and interact_col_name is not None:
y = shap_values[:, X.columns.get_loc(col_name), X.columns.get_loc(interact_col_name)]
else:
raise Exception('Either provide shap_values or shap_interaction_values with an interact_col_name')
if interact_col_name is not None:
text = np.array([f'{index_name}={index}<br>{col_name}={col_val}<br>{interact_col_name}={col_col_val}<br>SHAP={shap_val}'
for index, col_val, col_col_val, shap_val in zip(idxs, x, X[interact_col_name], np.round(y, round))])
else:
text = np.array([f'{index_name}={index}<br>{col_name}={col_val}<br>SHAP={shap_val}'
for index, col_val, shap_val in zip(idxs, x, np.round(y, round))])
data = []
if interact_col_name is not None and is_string_dtype(X[interact_col_name]):
for onehot_col in X[interact_col_name].unique().tolist():
data.append(
go.Scattergl(
x=X[X[interact_col_name]==onehot_col][col_name].replace({-999:np.nan}),
y=shap_values[X[interact_col_name]==onehot_col, X.columns.get_loc(col_name)],
mode='markers',
marker=dict(
size=7,
showscale=False,
opacity=0.6,
),
showlegend=True,
opacity=0.8,
hoverinfo="text",
name=onehot_col,
text=[f'{index_name}={index}<br>{col_name}={col_val}<br>{interact_col_name}={col_col_val}<br>SHAP={shap_val}'
for index, col_val, col_col_val, shap_val in zip(idxs,
X[X[interact_col_name]==onehot_col][col_name],
X[X[interact_col_name]==onehot_col][interact_col_name],
np.round(shap_values[X[interact_col_name]==onehot_col, X.columns.get_loc(col_name)], round))],
)
)
elif interact_col_name is not None and is_numeric_dtype(X[interact_col_name]):
data.append(go.Scattergl(
x=x[X[interact_col_name]!=na_fill],
y=y[X[interact_col_name]!=na_fill],
mode='markers',
text=text[X[interact_col_name]!=na_fill],
hoverinfo="text",
marker=dict(size=7,
opacity=0.6,
color=X[interact_col_name][X[interact_col_name]!=na_fill],
colorscale='Bluered',
colorbar=dict(
title=interact_col_name
),
showscale=True),
))
data.append(go.Scattergl(
x=x[X[interact_col_name]==na_fill],
y=y[X[interact_col_name]==na_fill],
mode='markers',
text=text[X[interact_col_name]==na_fill],
hoverinfo="text",
marker=dict(size=7,
opacity=0.35,
color='grey'),
))
else:
data.append(go.Scattergl(
x=x,
y=y,
mode='markers',
text=text,
hoverinfo="text",
marker=dict(size=7,
opacity=0.6) ,
))
if interaction:
title = f'Interaction plot for {col_name} and {interact_col_name}'
else:
title = f'Dependence plot for {col_name}'
layout = go.Layout(
title=title,
paper_bgcolor='#fff',
plot_bgcolor = '#fff',
showlegend=False,
hovermode='closest',
xaxis=dict(title=col_name),
yaxis=dict(title=f"SHAP value ({units})" if units !="" else "SHAP value")
)
fig = go.Figure(data, layout)
if interact_col_name is not None and is_string_dtype(X[interact_col_name]):
fig.update_layout(showlegend=True)
if highlight_index is not None:
fig.add_trace(
go.Scattergl(
x=[x[highlight_idx]],
y=[y[highlight_idx]],
mode='markers',
marker=dict(
color='LightSkyBlue',
size=25,
opacity=0.5,
line=dict(
color='MediumPurple',
width=4
)
),
name=f"{index_name} {highlight_name}",
text=f"{index_name} {highlight_name}",
hoverinfo="text",
showlegend=False,
),
)
fig.update_traces(selector = dict(mode='markers'))
return fig
def plotly_shap_violin_plot(X, shap_values, col_name, color_col=None, points=False,
interaction=False, units="", highlight_index=None, idxs=None, index_name="index"):
"""Generates a violin plot for displaying shap value distributions for
categorical features.
Args:
X (pd.DataFrame): dataframe of input rows
shap_values (np.ndarray): shap values generated for X
col_name (str): Column of X to display violin plot for
color_col (str, optional): Column of X to color plot markers by.
Defaults to None.
points (bool, optional): display point cloud next to violin plot.
Defaults to False.
interaction (bool, optional): Is this a plot for shap_interaction_values?
Defaults to False.
units (str, optional): Units of target variable. Defaults to "".
highlight_index (int, str, optional): Row index to highligh. Defaults to None.
idxs (List[str], optional): List of identifiers for each row in X, e.g.
names or id's. Defaults to None.
index_name (str): identifier for idxs. Defaults to "index".
Returns:
Plotly fig
"""
assert is_string_dtype(X[col_name]), \
f'{col_name} is not categorical! Can only plot violin plots for categorical features!'
x = X[col_name].copy()
shaps = shap_values[:, X.columns.get_loc(col_name)]
n_cats = X[col_name].nunique()
if idxs is not None:
assert len(idxs)==X.shape[0]
idxs = np.array([str(idx) for idx in idxs])
else:
idxs = np.array([str(i) for i in range(X.shape[0])])
if highlight_index is not None:
if isinstance(highlight_index, int):
highlight_idx = highlight_index
highlight_name = idxs[highlight_idx]
elif isinstance(highlight_index, str):
assert highlight_index in idxs, f'highlight_index should be int or in idxs, {highlight_index} is neither!'
highlight_idx = np.where(idxs==highlight_index)[0].item()
highlight_name = highlight_index
if points or color_col is not None:
fig = make_subplots(rows=1, cols=2*n_cats, column_widths=[3, 1]*n_cats, shared_yaxes=True)
showscale = True
else:
fig = make_subplots(rows=1, cols=n_cats, shared_yaxes=True)
fig.update_yaxes(range=[shaps.min()*1.3 if shaps.min() < 0 else shaps.min()*0.76, shaps.max()*1.3])
for i, cat in enumerate(X[col_name].unique()):
col = 1+i*2 if points or color_col is not None else 1+i
fig.add_trace(go.Violin(
x=x[x == cat],
y=shaps[x == cat],
name=cat,
box_visible=True,
meanline_visible=True,
showlegend=False,
),
row=1, col=col)
if color_col is not None:
if is_numeric_dtype(X[color_col]):
fig.add_trace(go.Scattergl(
x=np.random.randn(len(x[x == cat])),
y=shaps[x == cat],
name=color_col,
mode='markers',
showlegend=False,
hoverinfo="text",
# hovertemplate =
# "<i>shap</i>: %{y:.2f}<BR>" +
# f"<i>{color_col}" + ": %{marker.color}",
text = [f"{index_name}: {index}<br>shap: {shap}<br>{color_col}: {col}"
for index, shap, col in zip(idxs[x==cat], shaps[x == cat], X[color_col][x==cat])],
marker=dict(size=7,
opacity=0.6,
cmin=X[color_col].min(),
cmax=X[color_col].max(),
color=X[color_col][x==cat],
colorscale='Bluered',
showscale=showscale,
colorbar=dict(title=color_col)),
),
row=1, col=col+1)
else:
n_color_cats = X[color_col].nunique()
colors = ['#636EFA', '#EF553B', '#00CC96', '#AB63FA', '#FFA15A', '#19D3F3', '#FF6692', '#B6E880', '#FF97FF', '#FECB52']
colors = colors * (1+int(n_color_cats / len(colors)))
colors = colors[:n_color_cats]
for color_cat, color in zip(X[color_col].unique(), colors):
fig.add_trace(go.Scattergl(
x=np.random.randn(len(x[(x == cat) & (X[color_col] == color_cat)])),
y=shaps[(x == cat) & (X[color_col] == color_cat)],
name=color_cat,
mode='markers',
showlegend=showscale,
hoverinfo="text",
text = [f"{index_name}: {index}<br>shap: {shap}<br>{color_col}: {col}"
for index, shap, col in zip(
idxs[(x == cat) & (X[color_col] == color_cat)],
shaps[(x == cat) & (X[color_col] == color_cat)],
X[color_col][(x == cat) & (X[color_col] == color_cat)])],
# hovertemplate =
# "<i>shap</i>: %{y:.2f}<BR>" +
# f"<i>{color_col}: {color_cat}",
marker=dict(size=7,
opacity=0.8,
color=color)
),
row=1, col=col+1)
showscale = False
elif points:
fig.add_trace(go.Scattergl(
x=np.random.randn(len(x[x == cat])),
y=shaps[x == cat],
mode='markers',
showlegend=False,
# hovertemplate =
# "<i>shap</i>: %{y:.2f}",
hoverinfo="text",
text = [f"{index_name}: {index}<br>shap: {shap}"
for index, shap in zip(idxs[(x == cat)], shaps[x == cat])],
marker=dict(size=7,
opacity=0.6,
color='blue'),
), row=1, col=col+1)
if highlight_index is not None and X[col_name][highlight_idx]==cat:
fig.add_trace(
go.Scattergl(
x=[0],
y=[shaps[highlight_idx]],
mode='markers',
marker=dict(
color='LightSkyBlue',
size=25,
opacity=0.5,
line=dict(
color='MediumPurple',
width=4
)
),
name = f"{index_name} {highlight_name}",
text=f"{index_name} {highlight_name}",
hoverinfo="text",
showlegend=False,
), row=1, col=col+1)
if points or color_col is not None:
for i in range(n_cats):
fig.update_xaxes(showgrid=False, zeroline=False, visible=False, row=1, col=2+i*2)
fig.update_yaxes(showgrid=False, zeroline=False, row=1, col=2+i*2)
fig.update_layout(
yaxis=dict(title=f"SHAP value ({units})" if units !="" else "SHAP value"),
hovermode='closest')
if color_col is not None and interaction:
fig.update_layout(title=f'Interaction plot for {col_name} and {color_col}')
elif color_col is not None:
fig.update_layout(title=f'Shap values for {col_name}<br>(colored by {color_col})')
else:
fig.update_layout(title=f'Shap values for {col_name}')
return fig
def plotly_pdp(pdp_result,
display_index=None, index_feature_value=None, index_prediction=None,
absolute=True, plot_lines=True, num_grid_lines=100, feature_name=None,
round=2, target="", units="", index_name="index"):
"""Display partial-dependence plot (pdp)
Args:
pdp_result (pdp_result): Generated from pdp.pdp_result()
display_index (int, str, optional): Index to highligh in plot.
Defaults to None.
index_feature_value (str, float, optional): value of feature for index.
Defaults to None.
index_prediction (float, optional): Final prediction for index.
Defaults to None.
absolute (bool, optional): Display absolute pdp lines. If false then
display relative to base. Defaults to True.
plot_lines (bool, optional): Display selection of individual pdp lines.
Defaults to True.
num_grid_lines (int, optional): Number of sample gridlines to display.
Defaults to 100.
feature_name (str, optional): Name of the feature that the pdp_result
was generated for. Defaults to None.
round (int, optional): Rounding to apply to floats. Defaults to 2.
target (str, optional): Name of target variables. Defaults to "".
units (str, optional): Units of target variable. Defaults to "".
index_name (str): identifier for idxs. Defaults to "index".
Returns:
Plotly fig
"""
if feature_name is None: feature_name = pdp_result.feature
trace0 = go.Scatter(
x = pdp_result.feature_grids,
y = pdp_result.pdp.round(round) if absolute else (
pdp_result.pdp - pdp_result.pdp[0]).round(round),
mode = 'lines+markers',
line = dict(color='grey', width = 4),
name = f'average prediction <br>for different values of <br>{pdp_result.feature}'
)
data = [trace0]
if display_index is not None:
trace1 = go.Scatter(
x = pdp_result.feature_grids,
y = pdp_result.ice_lines.iloc[display_index].round(round).values if absolute else \
pdp_result.ice_lines.iloc[display_index].round(round).values - pdp_result.ice_lines.iloc[display_index].round(round).values[0],
mode = 'lines+markers',
line = dict(color='blue', width = 4),
name = f'prediction for {index_name} {display_index} <br>for different values of <br>{pdp_result.feature}'
)
data.append(trace1)
if plot_lines:
x = pdp_result.feature_grids
ice_lines = pdp_result.ice_lines.sample(num_grid_lines)
ice_lines = ice_lines.values if absolute else\
ice_lines.values - np.expand_dims(ice_lines.iloc[:, 0].transpose().values, axis=1)
for y in ice_lines:
data.append(
go.Scatter(
x = x,
y = y,
mode='lines',
hoverinfo='skip',
line=dict(color='grey'),
opacity=0.1,
showlegend=False
)
)
layout = go.Layout(title = f'pdp plot for {feature_name}',
plot_bgcolor = '#fff',
yaxis=dict(title=f"Predicted {target}{f' ({units})' if units else ''}"),
xaxis=dict(title=feature_name))
fig = go.Figure(data=data, layout=layout)
shapes = []
annotations = []
if index_feature_value is not None:
if not isinstance(index_feature_value, str):
index_feature_value = np.round(index_feature_value, 2)
shapes.append(
dict(
type='line',
xref='x',
yref='y',
x0=index_feature_value,
x1=index_feature_value,
y0=np.min(ice_lines) if plot_lines else \
np.min(pdp_result.pdp),
y1=np.max(ice_lines) if plot_lines \
else np.max(pdp_result.pdp),
line=dict(
color="MediumPurple",
width=4,
dash="dot",
),
))
annotations.append(
go.layout.Annotation(x=index_feature_value,
y=np.min(ice_lines) if plot_lines else \
np.min(pdp_result.pdp),
text=f"baseline value = {index_feature_value}"))
if index_prediction is not None:
shapes.append(
dict(
type='line',
xref='x',
yref='y',
x0=pdp_result.feature_grids[0],
x1=pdp_result.feature_grids[-1],
y0=index_prediction,
y1=index_prediction,
line=dict(
color="MediumPurple",
width=4,
dash="dot",
),
))
annotations.append(
go.layout.Annotation(
x=pdp_result.feature_grids[
int(0.5*len(pdp_result.feature_grids))],
y=index_prediction,
text=f"baseline pred = {np.round(index_prediction,2)}"))
fig.update_layout(annotations=annotations)
fig.update_layout(shapes=shapes)
fig.update_layout(showlegend=False)
return fig
def plotly_importances_plot(importance_df, descriptions=None, round=3,
target="target" , units="", title=None, xaxis_title=None):
"""Return feature importance plot
Args:
importance_df (pd.DataFrame): generate with get_importance_df(...)
descriptions (dict, optional): dict of descriptions of each feature.
round (int, optional): Rounding to apply to floats. Defaults to 3.
target (str, optional): Name of target variable. Defaults to "target".
units (str, optional): Units of target variable. Defaults to "".
title (str, optional): Title for graph. Defaults to None.
xaxis_title (str, optional): Title for x-axis Defaults to None.
Returns:
Plotly fig
"""
importance_name = importance_df.columns[1] # can be "MEAN_ABS_SHAP", "Permutation Importance", etc
if title is None:
title = importance_name
longest_feature_name = importance_df['Feature'].str.len().max()
imp = importance_df.sort_values(importance_name)
feature_names = [str(len(imp)-i)+". "+col
for i, col in enumerate(imp.iloc[:, 0].astype(str).values.tolist())]
importance_values = imp.iloc[:,1]
data = [go.Bar(
y=feature_names,
x=importance_values,
#text=importance_values.round(round),
text=descriptions[::-1] if descriptions is not None else None, #don't know why, but order needs to be reversed
#textposition='inside',
#insidetextanchor='end',
hoverinfo="text",
orientation='h')]
layout = go.Layout(
title=title,
plot_bgcolor = '#fff',
showlegend=False
)
fig = go.Figure(data=data, layout=layout)
fig.update_yaxes(automargin=True)
if xaxis_title is None:
xaxis_title = units
fig.update_xaxes(automargin=True, title=xaxis_title)
left_margin = longest_feature_name*7
if np.isnan(left_margin):
left_margin = 100
fig.update_layout(height=200+len(importance_df)*20,
margin=go.layout.Margin(
l=left_margin,
r=50,
b=50,
t=50,
pad=4
))
return fig
def plotly_confusion_matrix(y_true, y_preds, labels = None, percentage=True):
"""Generates Plotly fig confusion matrix
Args:
y_true (np.ndarray): array of actual values
y_preds (np.ndarray): array of predicted labels
labels (List[str], optional): List of labels for classes. Defaults to None.
percentage (bool, optional): Display percentages instead of absolute number.
Defaults to True.
Returns:
Plotly fig
"""
cm = confusion_matrix(y_true, y_preds)
cm_normalized = np.round(100*cm / cm.sum(), 1)
if labels is None:
labels = [str(i) for i in range(cm.shape[0])]
zmax = len(y_true)
data=[go.Heatmap(
z=cm,
x=[f'predicted {lab}' if len(lab) < 5 else f'predicted<br>{lab}' for lab in labels],
y=[f'actual {lab}' if len(lab) < 5 else f'actual<br>{lab}' for lab in labels],
hoverinfo="skip",
zmin=0, zmax=zmax, colorscale='Blues',
showscale=False,
)]
layout = go.Layout(
title="Confusion Matrix",
xaxis=dict(side='top', constrain="domain"),
yaxis=dict(autorange="reversed", side='left',
scaleanchor='x', scaleratio=1),
plot_bgcolor = '#fff',
)
fig = go.Figure(data, layout)
annotations = []
for x in range(cm.shape[0]):
for y in range(cm.shape[1]):
top_text = f"{cm_normalized[x, y]}%" if percentage else f"{cm[x, y]}"
bottom_text = f"{cm_normalized[x, y]}%" if not percentage else f"{cm[x, y]}"
annotations.extend([
go.layout.Annotation(
x=fig.data[0].x[y],
y=fig.data[0].y[x],
text=top_text,
showarrow=False,
font=dict(size=20)
),
go.layout.Annotation(
x=fig.data[0].x[y],
y=fig.data[0].y[x],
text=f" <br> <br> <br>({bottom_text})",
showarrow=False,
font=dict(size=12)
)]
)
fig.update_layout(annotations=annotations)
return fig
def plotly_roc_auc_curve(true_y, pred_probas, cutoff=None):
"""Plot ROC AUC curve
Args:
true_y (np.ndarray): array of true labels
pred_probas (np.ndarray): array of predicted probabilities
cutoff (float, optional): Cutoff proba to display. Defaults to None.
Returns:
Plotly Fig:
"""
fpr, tpr, thresholds = roc_curve(true_y, pred_probas)
roc_auc = roc_auc_score(true_y, pred_probas)
trace0 = go.Scatter(x=fpr, y=tpr,
mode='lines',
name='ROC AUC CURVE',
text=[f"threshold: {np.round(th,2)} <br> FP: {np.round(fp,2)} <br> TP: {np.round(tp,2)}"
for fp, tp, th in zip(fpr, tpr, thresholds)],
hoverinfo="text"
)
data = [trace0]
layout = go.Layout(title='ROC AUC CURVE',
# width=450,
# height=450,
xaxis= dict(title='False Positive Rate', range=[0,1], constrain="domain"),
yaxis = dict(title='True Positive Rate', range=[0,1], constrain="domain",
scaleanchor='x', scaleratio=1),
hovermode='closest',
plot_bgcolor = '#fff',)
fig = go.Figure(data, layout)
shapes = [dict(
type='line',
xref='x',
yref='y',
x0=0,
x1=1,
y0=0,
y1=1,
line=dict(
color="darkslategray",
width=4,
dash="dot"),
)]
if cutoff is not None:
threshold_idx = np.argmin(np.abs(thresholds-cutoff))
shapes.append(
dict(type='line', xref='x', yref='y',
x0=0, x1=1, y0=tpr[threshold_idx], y1=tpr[threshold_idx],
line=dict(color="lightslategray",width=1)))
shapes.append(
dict(type='line', xref='x', yref='y',
x0=fpr[threshold_idx], x1=fpr[threshold_idx], y0=0, y1=1,
line=dict(color="lightslategray", width=1)))
rep = classification_report(true_y, np.where(pred_probas >= cutoff, 1,0),
output_dict=True)
annotations = [go.layout.Annotation(x=0.6, y=0.45,
text=f"Cutoff: {np.round(cutoff,3)}",
showarrow=False, align="right",
xanchor='left', yanchor='top'),
go.layout.Annotation(x=0.6, y=0.4,
text=f"Accuracy: {np.round(rep['accuracy'],3)}",
showarrow=False, align="right",
xanchor='left', yanchor='top'),
go.layout.Annotation(x=0.6, y=0.35,
text=f"Precision: {np.round(rep['1']['precision'], 3)}",
showarrow=False, align="right",
xanchor='left', yanchor='top'),
go.layout.Annotation(x=0.6, y=0.30,
text=f"Recall: {np.round(rep['1']['recall'], 3)}",
showarrow=False, align="right",
xanchor='left', yanchor='top'),
go.layout.Annotation(x=0.6, y=0.25,
text=f"F1-score: {np.round(rep['1']['f1-score'], 3)}",
showarrow=False, align="right",
xanchor='left', yanchor='top'),
go.layout.Annotation(x=0.6, y=0.20,
text=f"roc-auc-score: {np.round(roc_auc, 3)}",
showarrow=False, align="right",
xanchor='left', yanchor='top'),]
fig.update_layout(annotations=annotations)
fig.update_layout(shapes=shapes)
return fig
def plotly_pr_auc_curve(true_y, pred_probas, cutoff=None):
"""Generate Precision-Recall Area Under Curve plot
Args:
true_y (np.ndarray): array of tru labels
pred_probas (np.ndarray): array of predicted probabilities
cutoff (float, optional): model cutoff to display in graph. Defaults to None.
Returns:
Plotly fig:
"""
precision, recall, thresholds = precision_recall_curve(true_y, pred_probas)
pr_auc_score = average_precision_score(true_y, pred_probas)
trace0 = go.Scatter(x=precision, y=recall,
mode='lines',
name='PR AUC CURVE',
text=[f"threshold: {np.round(th,2)} <br>" +\
f"precision: {np.round(p,2)} <br>" +\
f"recall: {np.round(r,2)}"
for p, r, th in zip(precision, recall, thresholds)],
hoverinfo="text"
)
data = [trace0]
layout = go.Layout(title='PR AUC CURVE',
# width=450,
# height=450,
xaxis= dict(title='Precision', range=[0,1], constrain="domain"),
yaxis = dict(title='Recall', range=[0,1], constrain="domain",
scaleanchor='x', scaleratio=1),
hovermode='closest',
plot_bgcolor = '#fff',)
fig = go.Figure(data, layout)
shapes = []
if cutoff is not None:
threshold_idx = np.argmin(np.abs(thresholds-cutoff))
shapes.append(
dict(type='line', xref='x', yref='y',
x0=0, x1=1,
y0=recall[threshold_idx], y1=recall[threshold_idx],
line=dict(color="lightslategray",width=1)))
shapes.append(
dict(type='line', xref='x', yref='y',
x0=precision[threshold_idx], x1=precision[threshold_idx],
y0=0, y1=1,
line=dict(color="lightslategray", width=1)))
report = classification_report(
true_y, np.where(pred_probas > cutoff, 1,0),
output_dict=True)
annotations = [go.layout.Annotation(x=0.15, y=0.45,
text=f"Cutoff: {np.round(cutoff,3)}",
showarrow=False, align="right",
xanchor='left', yanchor='top'),
go.layout.Annotation(x=0.15, y=0.4,
text=f"Accuracy: {np.round(report['accuracy'],3)}",
showarrow=False, align="right",
xanchor='left', yanchor='top'),
go.layout.Annotation(x=0.15, y=0.35,
text=f"Precision: {np.round(report['1']['precision'], 3)}",
showarrow=False, align="right",
xanchor='left', yanchor='top'),
go.layout.Annotation(x=0.15, y=0.30,
text=f"Recall: {np.round(report['1']['recall'], 3)}",
showarrow=False, align="right",
xanchor='left', yanchor='top'),
go.layout.Annotation(x=0.15, y=0.25,
text=f"F1-score: {np.round(report['1']['f1-score'], 3)}",
showarrow=False, align="right",
xanchor='left', yanchor='top'),
go.layout.Annotation(x=0.15, y=0.20,
text=f"pr-auc-score: {np.round(pr_auc_score, 3)}",
showarrow=False, align="right",
xanchor='left', yanchor='top'),]
fig.update_layout(annotations=annotations)
fig.update_layout(shapes=shapes)
return fig
def plotly_shap_scatter_plot(shap_values, X, display_columns=None, title="Shap values",
idxs=None, highlight_index=None, na_fill=-999, index_name="index"):
"""Generate a shap values summary plot where features are ranked from
highest mean absolute shap value to lowest, with point clouds shown
for each feature.
Args:
shap_values (np.ndarray): shap_values
X (pd.DataFrame): dataframe of input features
display_columns (List[str]): list of feature to be displayed. If None
default to all columns in X.
title (str, optional): Title to display above graph.
Defaults to "Shap values".
idxs (List[str], optional): List of identifiers for each row in X.
Defaults to None.
highlight_index ({str, int}, optional): Index to highlight in graph.
Defaults to None.
na_fill (int, optional): Fill value used to fill missing values,
will be colored grey in the graph.. Defaults to -999.
index_name (str): identifier for idxs. Defaults to "index".
Returns:
Plotly fig
"""
if display_columns is None:
display_columns = X.columns.tolist()
if idxs is not None:
assert len(idxs)==X.shape[0]
idxs = np.array([str(idx) for idx in idxs])
else:
idxs = np.array([str(i) for i in range(X.shape[0])])
if highlight_index is not None:
if isinstance(highlight_index, int):
assert highlight_index >=0 and highlight_index < len(X), \
"if highlight_index is int, then should be between 0 and {len(X)}!"
highlight_idx = highlight_index
highlight_index = idxs[highlight_idx]
elif isinstance(highlight_index, str):
assert str(highlight_index) in idxs, f"{highlight_index} not found in idxs!"
highlight_idx = np.where(idxs == str(highlight_index))[0].item()
else:
raise ValueError("Please pass either int or str highlight_index!")
# make sure that columns are actually in X:
display_columns = [col for col in display_columns if col in X.columns.tolist()]
shap_df = pd.DataFrame(shap_values, columns=X.columns, index=X.index)
min_shap = np.round(shap_values.min()-0.01, 2)
max_shap = np.round(shap_values.max()+0.01, 2)
fig = make_subplots(rows=len(display_columns), cols=1,
subplot_titles=display_columns, shared_xaxes=True)
for i, col in enumerate(display_columns):
if is_string_dtype(X[col]):
# if str type then categorical variable,
# so plot each category in a different color:
for onehot_col in X[col].unique().tolist():
fig.add_trace(go.Scattergl(
x=shap_df[X[col]==onehot_col][col],
y=np.random.rand(len(shap_df[X[col]==onehot_col])),
mode='markers',
marker=dict(
size=5,
showscale=False,
opacity=0.3,
),
name=onehot_col,
showlegend=False,
opacity=0.8,
hoverinfo="text",
text=[f"{index_name}={i}<br>{col}={onehot_col}<br>shap={np.round(shap,3)}"
for i, shap in zip(idxs[X[col]==onehot_col], shap_df[X[col]==onehot_col][col])],
),
row=i+1, col=1);
else:
# numerical feature get a single bluered plot
fig.add_trace(go.Scattergl(x=shap_df[col],
y=np.random.rand(len(shap_df)),
mode='markers',
marker=dict(
size=5,
color=X[col].replace({na_fill:np.nan}),
colorscale='Bluered',
showscale=True,
opacity=0.3,
colorbar=dict(
title="feature value <br> (red is high)",
showticklabels=False),
),
name=col,
showlegend=False,
opacity=0.8,
hoverinfo="text",
text=[f"{index_name}={i}<br>{col}={value}<br>shap={np.round(shap,3)}"
for i, shap, value in zip(idxs, shap_df[col], X[col].replace({-999:np.nan}))],
),
row=i+1, col=1);
if highlight_index is not None:
fig.add_trace(
go.Scattergl(
x=[shap_df[col].iloc[highlight_idx]],
y=[0],
mode='markers',
marker=dict(
color='LightSkyBlue',
size=20,
opacity=0.5,
line=dict(
color='MediumPurple',
width=4
)
),
name = f"{index_name} {highlight_index}",
text=f"index={highlight_index}<br>{col}={X[col].iloc[highlight_idx]}<br>shap={shap_df[col].iloc[highlight_idx]}",
hoverinfo="text",
showlegend=False,
), row=i+1, col=1)
fig.update_xaxes(showgrid=False, zeroline=False,
range=[min_shap, max_shap], row=i+1, col=1)
fig.update_yaxes(showgrid=False, zeroline=False,
showticklabels=False, row=i+1, col=1)
fig.update_layout(title=title + "<br>",
height=100+len(display_columns)*50,
margin=go.layout.Margin(
l=50,
r=50,
b=50,
t=100,
pad=4
),
hovermode='closest',
plot_bgcolor = '#fff',)
return fig
def plotly_predicted_vs_actual(y, preds, target="" , units="", round=2,
logs=False, log_x=False, log_y=False, idxs=None, index_name="index"):
"""Generate graph showing predicted values from a regressor model vs actual
values.
Args:
y (np.ndarray): Actual values
preds (np.ndarray): Predicted values
target (str, optional): Label for target. Defaults to "".
units (str, optional): Units of target. Defaults to "".
round (int, optional): Rounding to apply to floats. Defaults to 2.
logs (bool, optional): Log both axis. Defaults to False.
log_x (bool, optional): Log x axis. Defaults to False.
log_y (bool, optional): Log y axis. Defaults to False.
idxs (List[str], optional): list of identifiers for each observation. Defaults to None.
index_name (str): identifier for idxs. Defaults to "index".
Returns:
Plotly fig
"""
if idxs is not None:
assert len(idxs)==len(preds)
idxs = [str(idx) for idx in idxs]
else:
idxs = [str(i) for i in range(len(preds))]
marker_text=[f"{index_name}: {idx}<br>Observed: {actual}<br>Prediction: {pred}"
for idx, actual, pred in zip(idxs,
np.round(y, round),
np.round(preds, round))]
trace0 = go.Scattergl(
x = y,
y = preds,
mode='markers',
name=f'predicted {target}' + f" ({units})" if units else "",
text=marker_text,
hoverinfo="text",
)
sorted_y = np.sort(y)
trace1 = go.Scattergl(
x = sorted_y,
y = sorted_y,
mode='lines',
name=f"observed {target}" + f" ({units})" if units else "",
hoverinfo="none",
)
data = [trace0, trace1]
layout = go.Layout(
title=f"Predicted {target} vs Observed {target}",
yaxis=dict(
title=f"Predicted {target}" + f" ({units})" if units else "",
),
xaxis=dict(
title=f"Observed {target}" + f" ({units})" if units else "",
),
plot_bgcolor = '#fff',
hovermode = 'closest',
)
fig = go.Figure(data, layout)
if logs:
fig.update_layout(xaxis_type='log', yaxis_type='log')
if log_x:
fig.update_layout(xaxis_type='log')
if log_y:
fig.update_layout(yaxis_type='log')
return fig
def plotly_plot_residuals(y, preds, vs_actual=False, target="", units="",
residuals='difference', round=2, idxs=None, index_name="index"):
"""generates a residual plot
Args:
y (np.array, pd.Series): Actual values
preds (np.array, pd.Series): Predictions
vs_actual (bool, optional): Put actual values (y) on the x-axis.
Defaults to False (i.e. preds on the x-axis)
target (str, optional): name of the target variable. Defaults to ""
units (str, optional): units of the axis. Defaults to "".
residuals (str, {'difference', 'ratio', 'log-ratio'} optional):
How to calcualte residuals. Defaults to 'difference'.
round (int, optional): [description]. Defaults to 2.
idxs ([type], optional): [description]. Defaults to None.
index_name (str): identifier for idxs. Defaults to "index".
Returns:
[type]: [description]
"""
if idxs is not None:
assert len(idxs)==len(preds)
idxs = [str(idx) for idx in idxs]
else:
idxs = [str(i) for i in range(len(preds))]
res= y - preds
res_ratio = y / preds
if residuals == 'log-ratio':
residuals_display = np.log(res_ratio)
residuals_name = 'residuals log ratio<br>(log(y/preds))'
elif residuals == 'ratio':
residuals_display = res_ratio
residuals_name = 'residuals ratio<br>(y/preds)'
elif residuals == 'difference':
residuals_display = res
residuals_name = 'residuals (y-preds)'
else:
raise ValueError(f"parameter residuals should be in ['difference', "
f"'ratio', 'log-ratio'] but is equal to {residuals}!")
residuals_text=[f"{index_name}: {idx}<br>Observed: {actual}<br>Prediction: {pred}<br>Residual: {residual}"
for idx, actual, pred, residual in zip(idxs,
np.round(y, round),
np.round(preds, round),
np.round(res, round))]
trace0 = go.Scattergl(
x=y if vs_actual else preds,
y=residuals_display,
mode='markers',
name=residuals_name,
text=residuals_text,
hoverinfo="text",
)
trace1 = go.Scattergl(
x=y if vs_actual else preds,
y=np.ones(len(preds)) if residuals=='ratio' else np.zeros(len(preds)),
mode='lines',
name=(f"Observed {target}" + f" ({units})" if units else "") if vs_actual \
else (f"Predicted {target}" + f" ({units})" if units else ""),
hoverinfo="none",
)
data = [trace0, trace1]
layout = go.Layout(
title=f"Residuals vs {'observed' if vs_actual else 'predicted'} {target}",
yaxis=dict(
title=residuals_name
),
xaxis=dict(
title=(f"Observed {target}" + f" ({units})" if units else "") if vs_actual \
else (f"Predicted {target}" + f" ({units})" if units else "")
),
plot_bgcolor = '#fff',
hovermode = 'closest',
)
fig = go.Figure(data, layout)
return fig
def plotly_residuals_vs_col(y, preds, col, col_name=None, residuals='difference',
idxs=None, round=2, points=True, winsor=0,
na_fill=-999, index_name="index"):
"""Generates a residuals plot vs a particular feature column.
Args:
y (np.ndarray): array of actual target values
preds (np.ndarray): array of predicted values
col (pd.Series): series of values to be used as x-axis
col_name (str, optional): feature name to display.
Defaults to None, in which case col.name gets used.
residuals ({'log-ratio', 'ratio', 'difference'}, optional):
type of residuals to display. Defaults to 'difference'.
idxs (List[str], optional): str identifiers for each sample.
Defaults to None.
round (int, optional): Rounding to apply to floats. Defaults to 2.
points (bool, optional): For categorical features display point cloud
next to violin plots. Defaults to True.
winsor (int, optional): Winsorize the outliers. Remove the top `winsor`
percent highest and lowest values. Defaults to 0.
na_fill (int, optional): Value used to fill missing values. Defaults to -999.
index_name (str): identifier for idxs. Defaults to "index".
Returns:
Plotly fig
"""
if col_name is None:
try:
col_name = col.name
except:
col_name = 'Feature'
if idxs is not None:
assert len(idxs)==len(preds)
idxs = [str(idx) for idx in idxs]
else:
idxs = [str(i) for i in range(len(preds))]
res = y - preds
res_ratio = y / preds
if residuals == 'log-ratio':
residuals_display = np.log(res_ratio)
residuals_name = 'residuals log ratio<br>(log(y/preds))'
elif residuals == 'ratio':
residuals_display = res_ratio
residuals_name = 'residuals ratio<br>(y/preds)'
elif residuals == 'difference':
residuals_display = res
residuals_name = 'residuals (y-preds)'
else:
raise ValueError(f"parameter residuals should be in ['difference', "
f"'ratio', 'log-ratio'] but is equal to {residuals}!")
residuals_text=[f"{index_name}: {idx}<br>Actual: {actual}<br>Prediction: {pred}<br>Residual: {residual}"
for idx, actual, pred, residual in zip(idxs,
np.round(y, round),
np.round(preds, round),
np.round(res, round))]
if is_string_dtype(col):
n_cats = col.nunique()
if points:
fig = make_subplots(rows=1, cols=2*n_cats, column_widths=[3, 1]*n_cats, shared_yaxes=True)
showscale = True
else:
fig = make_subplots(rows=1, cols=n_cats, shared_yaxes=True)
fig.update_yaxes(range=[np.percentile(residuals_display, winsor),
np.percentile(residuals_display, 100-winsor)])
for i, cat in enumerate(col.unique()):
column = 1+i*2 if points else 1+i
fig.add_trace(go.Violin(
x=col[col == cat],
y=residuals_display[col == cat],
name=cat,
box_visible=True,
meanline_visible=True,
showlegend=False),
row=1, col=column)
if points:
fig.add_trace(go.Scattergl(
x=np.random.randn(len(col[col == cat])),
y=residuals_display[col == cat],
mode='markers',
showlegend=False,
text=[t for t, b in zip(residuals_text, col == cat) if b],
hoverinfo="text",
marker=dict(size=7,
opacity=0.6,
color='blue'),
), row=1, col=column+1)
if points:
for i in range(n_cats):
fig.update_xaxes(showgrid=False, zeroline=False, visible=False, row=1, col=2+i*2)
fig.update_yaxes(showgrid=False, zeroline=False, row=1, col=2+i*2)
fig.update_layout(title=f'Residuals vs {col_name}',
hovermode = 'closest')
return fig
else:
col[col==na_fill] = np.nan
trace0 = go.Scattergl(
x=col,
y=residuals_display,
mode='markers',
name=residuals_name,
text=residuals_text,
hoverinfo="text",
)
trace1 = go.Scattergl(
x=col,
y=np.ones(len(preds)) if residuals=='ratio' else np.zeros(len(preds)),
mode='lines',
name=col_name,
hoverinfo="none",
)
data = [trace0, trace1]
layout = go.Layout(
title=f'Residuals vs {col_name}',
yaxis=dict(
title=residuals_name
),
xaxis=dict(
title=f'{col_name} value'
),
plot_bgcolor = '#fff',
hovermode = 'closest'
)
fig = go.Figure(data, layout)
fig.update_yaxes(range=[np.percentile(residuals_display, winsor),
np.percentile(residuals_display, 100-winsor)])
return fig
def plotly_actual_vs_col(y, preds, col, col_name=None,
idxs=None, round=2, points=True, winsor=0, na_fill=-999,
units="", target="", index_name="index"):
"""Generates a residuals plot vs a particular feature column.
Args:
y (np.ndarray): array of actual target values
preds (np.ndarray): array of predicted values
col (pd.Series): series of values to be used as x-axis
col_name (str, optional): feature name to display.
Defaults to None, in which case col.name gets used.
idxs (List[str], optional): str identifiers for each sample.
Defaults to None.
round (int, optional): Rounding to apply to floats. Defaults to 2.
points (bool, optional): For categorical features display point cloud
next to violin plots. Defaults to True.
winsor (int, optional): Winsorize the outliers. Remove the top `winsor`
percent highest and lowest values. Defaults to 0.
na_fill (int, optional): Value used to fill missing values. Defaults to -999.
index_name (str): identifier for idxs. Defaults to "index".
Returns:
Plotly fig
"""
if col_name is None:
try:
col_name = col.name
except:
col_name = 'Feature'
if idxs is not None:
assert len(idxs)==len(preds)
idxs = [str(idx) for idx in idxs]
else:
idxs = [str(i) for i in range(len(preds))]
y_text=[f"{index_name}: {idx}<br>Observed {target}: {actual}<br>Prediction: {pred}"
for idx, actual, pred in zip(idxs,
np.round(y, round),
np.round(preds, round))]
if is_string_dtype(col):
n_cats = col.nunique()
if points:
fig = make_subplots(rows=1, cols=2*n_cats, column_widths=[3, 1]*n_cats, shared_yaxes=True)
showscale = True
else:
fig = make_subplots(rows=1, cols=n_cats, shared_yaxes=True)
fig.update_yaxes(range=[np.percentile(y, winsor),
np.percentile(y, 100-winsor)])
for i, cat in enumerate(col.unique()):
column = 1+i*2 if points else 1+i
fig.add_trace(go.Violin(
x=col[col == cat],
y=y[col == cat],
name=cat,
box_visible=True,
meanline_visible=True,
showlegend=False),
row=1, col=column)
if points:
fig.add_trace(go.Scattergl(
x=np.random.randn(len(col[col == cat])),
y=y[col == cat],
mode='markers',
showlegend=False,
text=[t for t, b in zip(y_text, col == cat) if b],
hoverinfo="text",
marker=dict(size=7,
opacity=0.6,
color='blue'),
), row=1, col=column+1)
if points:
for i in range(n_cats):
fig.update_xaxes(showgrid=False, zeroline=False, visible=False, row=1, col=2+i*2)
fig.update_yaxes(showgrid=False, zeroline=False, row=1, col=2+i*2)
fig.update_layout(title=f'Observed {target} vs {col_name}',
yaxis=dict(
title=f"Observed {target} ({units})" if units else f"Observed {target}"),
hovermode = 'closest')
return fig
else:
col[col==na_fill] = np.nan
trace0 = go.Scattergl(
x=col,
y=y,
mode='markers',
name='Observed',
text=y_text,
hoverinfo="text",
)
data = [trace0]
layout = go.Layout(
title=f'Observed {target} vs {col_name}',
yaxis=dict(
title=f"Observed {target} ({units})" if units else f"Observed {target}"
),
xaxis=dict(
title=f'{col_name} value'
),
plot_bgcolor = '#fff',
hovermode = 'closest'
)
fig = go.Figure(data, layout)
fig.update_yaxes(range=[np.percentile(y, winsor),
np.percentile(y, 100-winsor)])
return fig
def plotly_preds_vs_col(y, preds, col, col_name=None,
idxs=None, round=2, points=True, winsor=0, na_fill=-999,
units="", target="", index_name="index"):
"""Generates plot of predictions vs a particular feature column.
Args:
y (np.ndarray): array of actual target values
preds (np.ndarray): array of predicted values
col (pd.Series): series of values to be used as x-axis
col_name (str, optional): feature name to display.
Defaults to None, in which case col.name gets used.
idxs (List[str], optional): str identifiers for each sample.
Defaults to None.
round (int, optional): Rounding to apply to floats. Defaults to 2.
points (bool, optional): For categorical features display point cloud
next to violin plots. Defaults to True.
winsor (int, optional): Winsorize the outliers. Remove the top `winsor`
percent highest and lowest values. Defaults to 0.
na_fill (int, optional): Value used to fill missing values. Defaults to -999.
index_name (str): identifier for idxs. Defaults to "index".
Returns:
Plotly fig
"""
if col_name is None:
try:
col_name = col.name
except:
col_name = 'Feature'
if idxs is not None:
assert len(idxs)==len(preds)
idxs = [str(idx) for idx in idxs]
else:
idxs = [str(i) for i in range(len(preds))]
preds_text=[f"{index_name}: {idx}<br>Predicted {target}: {pred}{units}<br>Observed {target}: {actual}{units}"
for idx, actual, pred in zip(idxs,np.round(y, round), np.round(preds, round))]
if is_string_dtype(col):
n_cats = col.nunique()
if points:
fig = make_subplots(rows=1, cols=2*n_cats, column_widths=[3, 1]*n_cats, shared_yaxes=True)
showscale = True
else:
fig = make_subplots(rows=1, cols=n_cats, shared_yaxes=True)
fig.update_yaxes(range=[np.percentile(preds, winsor),
np.percentile(preds, 100-winsor)])
for i, cat in enumerate(col.unique()):
column = 1+i*2 if points else 1+i
fig.add_trace(go.Violin(
x=col[col == cat],
y=preds[col == cat],
name=cat,
box_visible=True,
meanline_visible=True,
showlegend=False),
row=1, col=column)
if points:
fig.add_trace(go.Scattergl(
x=np.random.randn(len(col[col == cat])),
y=preds[col == cat],
mode='markers',
showlegend=False,
text=[t for t, b in zip(preds_text, col == cat) if b],
hoverinfo="text",
marker=dict(size=7,
opacity=0.6,
color='blue'),
), row=1, col=column+1)
if points:
for i in range(n_cats):
fig.update_xaxes(showgrid=False, zeroline=False, visible=False, row=1, col=2+i*2)
fig.update_yaxes(showgrid=False, zeroline=False, row=1, col=2+i*2)
fig.update_layout(title=f'Predicted {target} vs {col_name}',
yaxis=dict(
title=f"Predicted {target} ({units})" if units else f"Predicted {target}"),
hovermode = 'closest')
return fig
else:
col[col==na_fill] = np.nan
trace0 = go.Scattergl(
x=col,
y=preds,
mode='markers',
name='Predicted',
text=preds_text,
hoverinfo="text",
)
data = [trace0]
layout = go.Layout(
title=f'Predicted {target} vs {col_name}',
yaxis=dict(
title=f"Predicted {target} ({units})" if units else f"Predicted {target}"
),
xaxis=dict(
title=f'{col_name} value'
),
plot_bgcolor = '#fff',
hovermode = 'closest'
)
fig = go.Figure(data, layout)
fig.update_yaxes(range=[np.percentile(preds, winsor),
np.percentile(preds, 100-winsor)])
return fig
def plotly_rf_trees(model, observation, y=None, highlight_tree=None,
round=2, pos_label=1, target="", units=""):
"""Generate a plot showing the prediction of every single tree inside a RandomForest model
Args:
model ({RandomForestClassifier, RandomForestRegressor}): model to display trees for
observation (pd.DataFrame): row of input data, e.g. X.iloc[[0]]
y (np.ndarray, optional): Target values. Defaults to None.
highlight_tree (int, optional): DecisionTree to highlight in graph. Defaults to None.
round (int, optional): Apply rounding to floats. Defaults to 2.
pos_label (int, optional): For RandomForestClassifier: Class label
to generate graph for. Defaults to 1.
target (str, optional): Description of target variable. Defaults to "".
units (str, optional): Units of target variable. Defaults to "".
Returns:
Plotly fig
"""
assert (str(type(model)).endswith("RandomForestClassifier'>")
or str(type(model)).endswith("RandomForestRegressor'>")), \
f"model is of type {type(model)}, but should be either RandomForestClassifier or RandomForestRegressor"
colors = ['blue'] * len(model.estimators_)
if highlight_tree is not None:
assert highlight_tree >= 0 and highlight_tree <= len(model.estimators_), \
f"{highlight_tree} is out of range (0, {len(model.estimators_)})"
colors[highlight_tree] = 'red'
if (hasattr(model.estimators_[0], "classes_")
and model.estimators_[0].classes_[0] is not None): #if classifier
preds_df = (
pd.DataFrame({
'model' : range(len(model.estimators_)),
'prediction' : [
np.round(100*m.predict_proba(observation)[0, pos_label], round)
for m in model.estimators_],
'color' : colors
})
.sort_values('prediction')\
.reset_index(drop=True))
else:
preds_df = (
pd.DataFrame({
'model' : range(len(model.estimators_)),
'prediction' : [np.round(m.predict(observation)[0] , round)
for m in model.estimators_],
'color' : colors
})
.sort_values('prediction')\
.reset_index(drop=True))
trace0 = go.Bar(x=preds_df.index,
y=preds_df.prediction,
marker_color=preds_df.color,
text=[f"tree no {t}:<br> prediction={p}<br> click for detailed info"
for (t, p) in zip(preds_df.model.values, preds_df.prediction.values)],
hoverinfo="text")
if target:
title = f"Individual RandomForest decision trees predicting {target}"
yaxis_title = f"Predicted {target} {f'({units})' if units else ''}"
else:
title = f"Individual RandomForest decision trees"
yaxis_title = f"Predicted outcome ({units})" if units else "Predicted outcome"
layout = go.Layout(
title=title,
plot_bgcolor = '#fff',
yaxis=dict(title=yaxis_title),
xaxis=dict(title="decision trees (sorted by prediction")
)
fig = go.Figure(data = [trace0], layout=layout)
shapes = [dict(
type='line',
xref='x', yref='y',
x0=0, x1=preds_df.model.max(),
y0=preds_df.prediction.mean(), y1=preds_df.prediction.mean(),
line=dict(
color="lightgray",
width=4,
dash="dot"),
)]
annotations = [go.layout.Annotation(
x=1.2*preds_df.model.mean(),
y=preds_df.prediction.mean(),
text=f"Average prediction = {np.round(preds_df.prediction.mean(),2)}",
bgcolor="lightgrey",
arrowcolor="lightgrey",
startstandoff=0)]
if y is not None:
shapes.append(dict(
type='line',
xref='x', yref='y',
x0=0, x1=preds_df.model.max(),
y0=y, y1=y,
line=dict(
color="red",
width=4,
dash="dashdot"),
))
annotations.append(go.layout.Annotation(
x=0.8*preds_df.model.mean(),
y=y,
text=f"observed={y}",
bgcolor="red",
arrowcolor="red"))
fig.update_layout(shapes=shapes)
fig.update_layout(annotations=annotations)
return fig
def plotly_xgboost_trees(xgboost_preds_df, highlight_tree=None, y=None, round=2,
pos_label=1, target="", units="", higher_is_better=True):
"""Generate a plot showing the prediction of every single tree inside an XGBoost model
Args:
xgboost_preds_df (pd.DataFrame): generated with get_xgboost_preds_df(...)
highlight_tree (int, optional): DecisionTree to highlight in graph. Defaults to None.
y (np.ndarray, optional): Target values. Defaults to None.
round (int, optional): Apply rounding to floats. Defaults to 2.
pos_label (int, optional): For RandomForestClassifier: Class label
to generate graph for. Defaults to 1.
target (str, optional): Description of target variable. Defaults to "".
units (str, optional): Units of target variable. Defaults to "".
higher_is_better (bool, optional): up is green, down is red. If False then
flip the colors.
Returns:
Plotly fig
"""
xgboost_preds_df['color'] = 'blue'
xgboost_preds_df.loc[0, 'color'] = 'yellow'
if highlight_tree is not None:
xgboost_preds_df.loc[highlight_tree+1, 'color'] = 'red'
trees = xgboost_preds_df.tree.values[1:]
colors = xgboost_preds_df.color.values[1:]
is_classifier = True if 'pred_proba' in xgboost_preds_df.columns else False
colors = xgboost_preds_df.color.values
if is_classifier:
final_prediction = xgboost_preds_df.pred_proba.values[-1]
base_prediction = xgboost_preds_df.pred_proba.values[0]
preds = xgboost_preds_df.pred_proba.values[1:]
bases = xgboost_preds_df.pred_proba.values[:-1]
diffs = xgboost_preds_df.pred_proba_diff.values[1:]
texts=[f"tree no {t}:<br>change = {np.round(100*d, round)}%<br> click for detailed info"
for (t, d) in zip(trees, diffs)]
texts.insert(0, f"Base prediction: <br>proba = {np.round(100*base_prediction, round)}%")
texts.append(f"Final Prediction: <br>proba = {np.round(100*final_prediction, round)}%")
else:
final_prediction = xgboost_preds_df.pred.values[-1]
base_prediction = xgboost_preds_df.pred.values[0]
preds = xgboost_preds_df.pred.values[1:]
bases = xgboost_preds_df.pred.values[:-1]
diffs = xgboost_preds_df.pred_diff.values[1:]
texts=[f"tree no {t}:<br>change = {np.round(d, round)}<br> click for detailed info"
for (t, d) in zip(trees, diffs)]
texts.insert(0, f"Base prediction: <br>pred = {np.round(base_prediction, round)}")
texts.append(f"Final Prediction: <br>pred = {np.round(final_prediction, round)}")
green_fill, green_line = 'rgba(50, 200, 50, 1.0)', 'rgba(40, 160, 50, 1.0)'
yellow_fill, yellow_line = 'rgba(230, 230, 30, 1.0)', 'rgba(190, 190, 30, 1.0)'
blue_fill, blue_line = 'rgba(55, 128, 191, 0.7)', 'rgba(55, 128, 191, 1.0)'
red_fill, red_line = 'rgba(219, 64, 82, 0.7)', 'rgba(219, 64, 82, 1.0)'
if higher_is_better:
fill_color_up, line_color_up = green_fill, green_line
fill_color_down, line_color_down =red_fill, red_line
else:
fill_color_up, line_color_up = red_fill, red_line
fill_color_down, line_color_down = green_fill, green_line
fill_colors = [fill_color_up if diff > 0 else fill_color_down for diff in diffs]
line_colors = [line_color_up if diff > 0 else line_color_down for diff in diffs]
fill_colors.insert(0, yellow_fill)
line_colors.insert(0, yellow_line)
fill_colors.append(blue_fill)
line_colors.append(blue_line)
trees = np.append(trees, len(trees))
trees = np.insert(trees, 0, -1)
bases = np.insert(bases, 0, 0)
bases = np.append(bases, 0)
diffs = np.insert(diffs, 0, base_prediction)
diffs = np.append(diffs, final_prediction)
trace0 = go.Bar(x=trees,
y=bases,
hoverinfo='skip',
name="",
showlegend=False,
marker=dict(color='rgba(1,1,1, 0.0)'))
trace1 = go.Bar(x=trees,
y=diffs,
text=texts,
name="",
hoverinfo="text",
showlegend=False,
marker=dict(
color=fill_colors,
line=dict(
color=line_colors,
width=2,
)
),
)
if target:
title = f"Individual xgboost decision trees predicting {target}"
yaxis_title = f"Predicted {target} {f'({units})' if units else ''}"
else:
title = f"Individual xgboost decision trees"
yaxis_title = f"Predicted outcome ({units})" if units else "Predicted outcome"
layout = go.Layout(
title=title,
barmode='stack',
plot_bgcolor = '#fff',
yaxis=dict(title=yaxis_title),
xaxis=dict(title="decision trees")
)
fig = go.Figure(data = [trace0, trace1], layout=layout)
shapes = []
annotations = []
if y is not None:
shapes.append(dict(
type='line',
xref='x', yref='y',
x0=trees.min(), x1=trees.max(),
y0=y, y1=y,
line=dict(
color="black",
width=4,
dash="dashdot"),
))
annotations.append(go.layout.Annotation(
x=0.75*trees.max(),
y=y,
text=f"Observed={y}",
bgcolor="white"))
fig.update_layout(shapes=shapes)
fig.update_layout(annotations=annotations)
return fig | AMLBID | /Explainer/explainer_plots.py | explainer_plots.py |
__all__ = ['ExplainerTabsLayout',
'ExplainerPageLayout',
'ExplainerDashboard',
'ExplainerHub',
'JupyterExplainerDashboard',
'ExplainerTab',
'JupyterExplainerTab',
'InlineExplainer']
import sys
import re
import json
import inspect
import requests
from typing import List, Union
from pathlib import Path
from copy import copy, deepcopy
import oyaml as yaml
import shortuuid
import dash
import dash_auth
import dash_core_components as dcc
import dash_html_components as html
import dash_bootstrap_components as dbc
from flask import Flask
from flask_simplelogin import SimpleLogin, login_required
from werkzeug.security import check_password_hash, generate_password_hash
from jupyter_dash import JupyterDash
import plotly.io as pio
from .dashboard_components import *
from .dashboard_tabs import *
from .AMLBID_Explainer import BaseExplainer
def instantiate_component(component, explainer, name=None, **kwargs):
"""Returns an instantiated ExplainerComponent.
If the component input is just a class definition, instantiate it with
explainer and k**wargs.
If it is already an ExplainerComponent instance then return it.
If it is any other instance with layout and register_components methods,
then add a name property and return it.
Args:
component ([type]): Either a class definition or instance
explainer ([type]): An Explainer object that will be used to instantiate class definitions
kwargs: kwargs will be passed on to the instance
Raises:
ValueError: if component is not a subclass or instance of ExplainerComponent,
or is an instance without layout and register_callbacks methods
Returns:
[type]: instantiated component
"""
if inspect.isclass(component) and issubclass(component, ExplainerComponent):
component = component(explainer, name=name, **kwargs)
return component
elif isinstance(component, ExplainerComponent):
return component
elif (not inspect.isclass(component)
and hasattr(component, "layout")):
if not (hasattr(component, "name") and isinstance(component.name, str)):
if name is None:
name = shortuuid.ShortUUID().random(length=5)
print(f"Warning: setting {component}.name to {name}")
component.name = name
if not hasattr(component, "title"):
print(f"Warning: setting {component}.title to 'Custom'")
component.title = "Custom"
return component
else:
raise ValueError(f"{component} is not a valid component...")
class ExplainerTabsLayout:
def __init__(self, explainer, tabs,
title='Model Explainer',
description=None,
header_hide_title=False,
header_hide_selector=False,
block_selector_callbacks=False,
pos_label=None,
fluid=True,
**kwargs):
"""Generates a multi tab layout from a a list of ExplainerComponents.
If the component is a class definition, it gets instantiated first. If
the component is not derived from an ExplainerComponent, then attempt
with duck typing to nevertheless instantiate a layout.
Args:
explainer ([type]): explainer
tabs (list[ExplainerComponent class or instance]): list of
ExplainerComponent class definitions or instances.
title (str, optional): [description]. Defaults to 'Model Explainer'.
description (str, optional): description tooltip to add to the title.
header_hide_title (bool, optional): Hide the title. Defaults to False.
header_hide_selector (bool, optional): Hide the positive label selector.
Defaults to False.
block_selector_callbacks (bool, optional): block the callback of the
pos label selector. Useful to avoid clashes when you
have your own PosLabelSelector in your layout.
Defaults to False.
pos_label ({int, str}, optional): initial pos label.
Defaults to explainer.pos_label
fluid (bool, optional): Stretch layout to fill space. Defaults to False.
"""
self.title = title
self.description = description
self.header_hide_title = header_hide_title
self.header_hide_selector = header_hide_selector
self.block_selector_callbacks = block_selector_callbacks
if self.block_selector_callbacks:
self.header_hide_selector = True
self.fluid = fluid
self.selector = PosLabelSelector(explainer, name="0", pos_label=pos_label)
self.tabs = [instantiate_component(tab, explainer, name=str(i+1), **kwargs) for i, tab in enumerate(tabs)]
assert len(self.tabs) > 0, 'When passing a list to tabs, need to pass at least one valid tab!'
self.connector = PosLabelConnector(self.selector, self.tabs)
def layout(self):
"""returns a multitab layout plus ExplainerHeader"""
return dbc.Container([
dbc.Row([
make_hideable(
dbc.Col([
make_hideable(html.H1(self.title, id='dashboard-titlee'), hide=True),
html.Div(html.Img(src="./assets/title.png",style={"max-width":"150%", "height:":"auto"} ,id='dashboard-title'),
style={ "margin-left": "10px"}),
dbc.Tooltip(self.description, target='dashboard-title')
], md=6), hide=True),
make_hideable(
dbc.Col([
html.Br(),
self.selector.layout()
],md=6), hide=True),
], justify="start", style=dict(marginBottom=10)),
dcc.Tabs(id="tabs", value=self.tabs[0].name,
children=[dcc.Tab(label=tab.title, id=tab.name, value=tab.name,
children=tab.layout()) for tab in self.tabs]),
], fluid=self.fluid)
def register_callbacks(self, app):
"""Registers callbacks for all tabs"""
for tab in self.tabs:
try:
tab.register_callbacks(app)
except AttributeError:
print(f"Warning: {tab} does not have a register_callbacks method!")
if not self.block_selector_callbacks:
if any([tab.has_pos_label_connector() for tab in self.tabs]):
print("Warning: detected PosLabelConnectors already in the layout. "
"This may clash with the global pos label selector and generate duplicate callback errors. "
"If so set block_selector_callbacks=True.")
self.connector.register_callbacks(app)
def calculate_dependencies(self):
"""Calculates dependencies for all tabs"""
for tab in self.tabs:
try:
tab.calculate_dependencies()
except AttributeError:
print(f"Warning: {tab} does not have a calculate_dependencies method!")
class ExplainerPageLayout(ExplainerComponent):
def __init__(self, explainer, component,
title='Model Explainer',
description=None,
header_hide_title=False,
header_hide_selector=False,
block_selector_callbacks=False,
pos_label=None,
fluid=False,
**kwargs):
"""Generates a single page layout from a single ExplainerComponent.
If the component is a class definition, it gets instantiated.
If the component is not derived from an ExplainerComponent, then tries
with duck typing to nevertheless instantiate a layout.
Args:
explainer ([type]): explainer
component (ExplainerComponent class or instance): ExplainerComponent
class definition or instance.
title (str, optional): Defaults to 'Model Explainer'.
description (str, optional): Will be displayed as title tooltip.
header_hide_title (bool, optional): Hide the title. Defaults to False.
header_hide_selector (bool, optional): Hide the positive label selector.
Defaults to False.
block_selector_callbacks (bool, optional): block the callback of the
pos label selector. Useful to avoid clashes when you
have your own PosLabelSelector in your layout.
Defaults to False.
pos_label ({int, str}, optional): initial pos label.
Defaults to explainer.pos_label
fluid (bool, optional): Stretch layout to fill space. Defaults to False.
"""
self.title = title
self.description = description
self.header_hide_title = header_hide_title
self.header_hide_selector = header_hide_selector
self.block_selector_callbacks = block_selector_callbacks
if self.block_selector_callbacks:
self.header_hide_selector = True
self.fluid = fluid
self.selector = PosLabelSelector(explainer, name="0", pos_label=pos_label)
self.page = instantiate_component(component, explainer, name="1", **kwargs)
print(self.page.name, flush=True)
self.connector = PosLabelConnector(self.selector, self.page)
self.fluid = fluid
def layout(self):
"""returns single page layout with an ExplainerHeader"""
return dbc.Container([
dbc.Row([
make_hideable(
dbc.Col([
html.H1(self.title, id='dashboard-title'),
dbc.Tooltip(self.description, target='dashboard-title')
], width="auto"), hide=self.header_hide_title),
make_hideable(
dbc.Col([
self.selector.layout()
], md=3), hide=self.header_hide_selector),
], justify="start"),
self.page.layout()
], fluid=self.fluid)
def register_callbacks(self, app):
"""Register callbacks of page"""
try:
self.page.register_callbacks(app)
except AttributeError:
print(f"Warning: {self.page} does not have a register_callbacks method!")
if not self.block_selector_callbacks:
if hasattr(self.page, "has_pos_label_connector") and self.page.has_pos_label_connector():
print("Warning: detected PosLabelConnectors already in the layout. "
"This may clash with the global pos label selector and generate duplicate callback errors. "
"If so set block_selector_callbacks=True.")
self.connector.register_callbacks(app)
def calculate_dependencies(self):
"""Calculate dependencies of page"""
try:
self.page.calculate_dependencies()
except AttributeError:
print(f"Warning: {self.page} does not have a calculate_dependencies method!", flush=True)
class ExplainerDashboard:
def __init__(self, explainer=None, tabs=None,
title='AMLBID Explainer',
name=None,
description=None,
hide_header=False,
header_hide_title=False,
header_hide_selector=False,
block_selector_callbacks=False,
pos_label=None,
fluid=True,
mode="dash",
width=1000,
height=800,
bootstrap=None,
external_stylesheets=None,
server=True,
url_base_pathname=None,
responsive=True,
logins=None,
port=8889, #8050
importances=True,
model_summary=True,
contributions=True,
whatif=True,
shap_dependence=False,
shap_interaction=False,
decision_trees=True,
debug=False,
**kwargs):
"""Creates an explainerdashboard out of an Explainer object.
single page dashboard:
If tabs is a single ExplainerComponent class or instance, display it
as a standalone page without tabs.
Multi tab dashboard:
If tabs is a list of ExplainerComponent classes or instances, then construct
a layout with a tab per component. Instead of components you can also pass
the following strings: "importances", "model_summary", "contributions",
"shap_dependence", "shap_interaction" or "decision_trees". You can mix and
combine these different modularities, e.g.:
tabs=[ImportancesTab, "contributions", custom_tab]
If tabs is None, then construct tabs based on the boolean parameters:
importances, model_summary, contributions, shap_dependence,
shap_interaction and decision_trees, which all default to True.
You can select four different modes:
- 'dash': standard dash.Dash() app
- 'inline': JupyterDash app inline in a notebook cell output
- 'jupyterlab': JupyterDash app in jupyterlab pane
- 'external': JupyterDash app in external tab
You can switch off the title and positive label selector
with header_hide_title=True and header_hide_selector=True.
You run the dashboard
with e.g. ExplainerDashboard(explainer).run(port=8050)
Args:
explainer(): explainer object
tabs(): single component or list of components
title(str, optional): title of dashboard, defaults to 'Model Explainer'
name (str): name of the dashboard. Used for assigning url in ExplainerHub.
description (str): summary for dashboard. Gets used for title tooltip and
in description for ExplainerHub.
hide_header (bool, optional) hide the header (title+selector), defaults to False.
header_hide_title(bool, optional): hide the title, defaults to False
header_hide_selector(bool, optional): hide the positive class selector for classifier models, defaults, to False
block_selector_callbacks (bool, optional): block the callback of the
pos label selector. Useful to avoid clashes when you
have your own PosLabelSelector in your layout.
Defaults to False.
pos_label ({int, str}, optional): initial pos label.
Defaults to explainer.pos_label
mode(str, {'dash', 'inline' , 'jupyterlab', 'external'}, optional):
type of dash server to start. 'inline' runs in a jupyter notebook output cell.
'jupyterlab' runs in a jupyterlab pane. 'external' runs in an external tab
while keeping the notebook interactive.
fluid(bool, optional): whether to stretch the layout to available space.
Defaults to True.
width(int, optional): width of notebook output cell in pixels, defaults to 1000.
height(int, optional): height of notebookn output cell in pixels, defaults to 800.
bootstrap (str, optional): link to bootstrap url. Can use dbc.themese
to generate the url, e.g. bootstrap=dbc.themes.FLATLY. Defaults
to default bootstrap theme that is stored in the /assets folder
so that it works even behind a firewall.
external_stylesheets(list, optional): additional external stylesheets
to add. (for themes use the bootstrap parameter)
server (Flask instance or bool): either an instance of an existing Flask
server to tie the dashboard to, or True in which case a new Flask
server is created.
url_base_pathname (str): url_base_pathname for dashboard,
e.g. "/dashboard". Defaults to None.
responsive (bool): make layout responsive to viewport size
(i.e. reorganize bootstrap columns on small devices). Set to False
when e.g. testing with a headless browser. Defaults to True.
logins (list of lists): list of (hardcoded) logins, e.g.
[['login1', 'password1'], ['login2', 'password2']].
Defaults to None (no login required)
importances(bool, optional): include ImportancesTab, defaults to True.
model_summary(bool, optional): include ModelSummaryTab, defaults to True.
contributions(bool, optional): include ContributionsTab, defaults to True.
whatif (bool, optional): include WhatIfTab, defaults to True.
shap_dependence(bool, optional): include ShapDependenceTab, defaults to True.
shap_interaction(bool, optional): include InteractionsTab if model allows it, defaults to True.
decision_trees(bool, optional): include DecisionTreesTab if model allows it, defaults to True.
"""
print("Building ExplainerDashboard...", flush=True)
self._store_params(no_param=['explainer', 'tabs', 'server'])
self._stored_params['tabs'] = self._tabs_to_yaml(tabs)
if self.description is None:
self.description = """This dashboard shows the workings of the recommended, fitted
machine learning model, and explains its predictions"""
if self.hide_header:
self.header_hide_title = True
self.header_hide_selector = True
try:
ipython_kernel = str(get_ipython())
self.is_notebook = True
self.is_colab = True if 'google.colab' in ipython_kernel else False
except:
self.is_notebook, self.is_colab = False, False
if self.mode == 'dash' and self.is_colab:
print("Detected google colab environment, setting mode='external'", flush=True)
self.mode = 'external'
elif self.mode == 'dash' and self.is_notebook:
print("Detected notebook environment, consider setting "
"mode='external', mode='inline' or mode='jupyterlab' "
"to keep the notebook interactive while the dashboard "
"is running...", flush=True)
if self.bootstrap is not None:
bootstrap_theme = self.bootstrap if isinstance(self.bootstrap, str) else dbc.themes.BOOTSTRAP
if self.external_stylesheets is None:
self.external_stylesheets = [bootstrap_theme]
else:
self.external_stylesheets.append(bootstrap_theme)
self.app = self._get_dash_app()
if logins is not None:
if len(logins)==2 and isinstance(logins[0], str) and isinstance(logins[1], str):
logins = [logins]
assert isinstance(logins, list), \
("Parameter logins should be a list of lists of str pairs, e.g."
" logins=[['user1', 'password1'], ['user2', 'password2']]!")
for login in logins:
assert isinstance(login, list), \
("Parameter logins should be a list of lists of str pairs, "
"e.g. logins=[['user1', 'password1'], ['user2', 'password2']]!")
assert isinstance(login[0], str) and isinstance(login[1], str), \
("For logins such as [['user1', 'password1']] user1 and "
"password1 should be type(str)!")
self.auth = dash_auth.BasicAuth(self.app, logins)
self.app.title = title
assert 'BaseExplainer' in str(explainer.__class__.mro()), \
("explainer should be an instance of BaseExplainer, such as "
"ClassifierExplainer or RegressionExplainer!")
if kwargs:
pass
if tabs is None:
tabs = []
if model_summary and explainer.y_missing:
#print("No y labels were passed to the Explainer, so setting" " model_summary=False...", flush=True)
model_summary = False
if shap_interaction and not explainer.interactions_should_work:
#print("For this type of model and model_output interactions don't ""work, so setting shap_interaction=False...", flush=True)
shap_interaction = False
if decision_trees and not hasattr(explainer, 'is_tree_explainer'):
#print("The explainer object has no decision_trees property. so setting decision_trees=False...", flush=True)
decision_trees = False
tabs.append(Testcomposite)
#tabs.append(SuggestedModelComposite)
#tabs.append(Testcomposite)
if model_summary:
tabs.append(ClassifierModelStatsComposite if explainer.is_classifier else RegressionModelStatsComposite)
if importances:
tabs.append(ImportancesComposite)
if contributions:
pass
#tabs.append(IndividualPredictionsComposite)
if whatif:
tabs.append(WhatIfComposite)
if shap_dependence:
tabs.append(ShapDependenceComposite)
if shap_interaction:
print("Warning: calculating shap interaction values can be slow! "
"Pass shap_interaction=False to remove interactions tab.",
flush=True)
tabs.append(ShapInteractionsComposite)
if decision_trees:
tabs.append(DecisionTreesComposite)
tabs.append(RefinementComposite)
if isinstance(tabs, list) and len(tabs)==1:
tabs = tabs[0]
#print("Generating layout...")
if isinstance(tabs, list):
tabs = [self._convert_str_tabs(tab) for tab in tabs]
self.explainer_layout = ExplainerTabsLayout(explainer, tabs, title,
description=self.description,
**update_kwargs(kwargs,
header_hide_title=self.header_hide_title,
header_hide_selector=self.header_hide_selector,
block_selector_callbacks=self.block_selector_callbacks,
pos_label=self.pos_label,
fluid=fluid))
else:
tabs = self._convert_str_tabs(tabs)
self.explainer_layout = ExplainerPageLayout(explainer, tabs, title,
description=self.description,
**update_kwargs(kwargs,
header_hide_title=self.header_hide_title,
header_hide_selector=self.header_hide_selector,
block_selector_callbacks=self.block_selector_callbacks,
pos_label=self.pos_label,
fluid=self.fluid))
self.app.layout = self.explainer_layout.layout()
#print("Calculating dependencies...", flush=True)
self.explainer_layout.calculate_dependencies()
self.explainer_layout.register_callbacks(self.app)
@classmethod
def from_config(cls, arg1, arg2=None, **update_params):
"""Loading a dashboard from a configuration .yaml file. You can either
pass both an explainer and a yaml file generated with
ExplainerDashboard.to_yaml("dashboard.yaml"):
db = ExplainerDashboard.from_config(explainer, "dashboard.yaml")
When you specify an explainerfile in to_yaml with
ExplainerDashboard.to_yaml("dashboard.yaml", explainerfile="explainer.joblib"),
you can also pass just the .yaml:
db = ExplainerDashboard.from_config("dashboard.yaml")
You can also load the explainerfile seperately:
db = ExplainerDashboard.from_config("explainer.joblib", "dashboard.yaml")
Args:
arg1 (explainer or config): arg1 should either be a config (yaml or dict),
or an explainer (instance or str/Path).
arg2 ([type], optional): If arg1 is an explainer, arg2 should be config.
update_params (dict): You can override parameters in the the yaml
config by passing additional kwargs to .from_config()
Returns:
ExplainerDashboard
"""
if arg2 is None:
if isinstance(arg1, (Path, str)) and str(arg1).endswith(".yaml"):
config = yaml.safe_load(open(str(arg1), "r"))
elif isinstance(arg1, dict):
config = arg1
assert 'dashboard' in config, \
".yaml file does not have `dashboard` param."
assert 'explainerfile' in config['dashboard'], \
".yaml file does not have explainerfile param"
explainer = BaseExplainer.from_file(config['dashboard']['explainerfile'])
else:
if isinstance(arg1, BaseExplainer):
explainer = arg1
elif isinstance(arg1, (Path, str)) and (
str(arg1).endswith(".joblib") or
str(arg1).endswith(".pkl") or str(arg1).endswith(".dill")):
explainer = BaseExplainer.from_file(arg1)
else:
raise ValueError(
"When passing two arguments to ExplainerDashboard.from_config(arg1, arg2), "
"arg1 should either be an explainer or an explainer filename (e.g. 'explainer.joblib')!")
if isinstance(arg2, (Path, str)) and str(arg2).endswith(".yaml"):
config = yaml.safe_load(open(str(arg2), "r"))
elif isinstance(arg2, dict):
config = arg2
else:
raise ValueError(
"When passing two arguments to ExplainerDashboard.from_config(arg1, arg2), "
"arg2 should be a .yaml file or a dict!")
dashboard_params = config['dashboard']['params']
for k, v in update_params.items():
if k in dashboard_params:
dashboard_params[k] = v
elif 'kwargs' in dashboard_params:
dashboard_params['kwargs'][k] = v
else:
dashboard_params['kwargs'] = dict(k=v)
if 'kwargs' in dashboard_params:
kwargs = dashboard_params.pop('kwargs')
else:
kwargs = {}
if 'tabs' in dashboard_params:
tabs = cls._yamltabs_to_tabs(dashboard_params['tabs'], explainer)
del dashboard_params['tabs']
return cls(explainer, tabs, **dashboard_params, **kwargs)
else:
return cls(explainer, **dashboard_params, **kwargs)
def to_yaml(self, filepath=None, return_dict=False,
explainerfile=None, dump_explainer=False):
"""Returns a yaml configuration of the current ExplainerDashboard
that can be used by the explainerdashboard CLI. Recommended filename
is `dashboard.yaml`.
Args:
filepath ({str, Path}, optional): Filepath to dump yaml. If None
returns the yaml as a string. Defaults to None.
return_dict (bool, optional): instead of yaml return dict with
config.
explainerfile (str, optional): filename of explainer dump. Defaults
to `explainer.joblib`.
dump_explainer (bool, optional): dump the explainer along with the yaml.
You must pass explainerfile parameter for the filename. Defaults to False.
"""
import oyaml as yaml
dashboard_config = dict(
dashboard=dict(
explainerfile=str(explainerfile),
params=self._stored_params))
if dump_explainer:
if explainerfile is None:
raise ValueError("When you pass dump_explainer=True, then you "
"must pass an explainerfile filename parameter!")
print(f"Dumping explainer to {explainerfile}...", flush=True)
self.explainer.dump(explainerfile)
if return_dict:
return dashboard_config
if filepath is not None:
yaml.dump(dashboard_config, open(filepath, "w"))
return
return yaml.dump(dashboard_config)
def _store_params(self, no_store=None, no_attr=None, no_param=None):
"""Stores the parameter of the class to instance attributes and
to a ._stored_params dict. You can optionall exclude all or some
parameters from being stored.
Args:
no_store ({bool, List[str]}, optional): If True do not store any
parameters to either attribute or _stored_params dict. If
a list of str, then do not store parameters with those names.
Defaults to None.
no_attr ({bool, List[str]},, optional): . If True do not store any
parameters to class attribute. If
a list of str, then do not store parameters with those names.
Defaults to None.
no_param ({bool, List[str]},, optional): If True do not store any
parameters to _stored_params dict. If
a list of str, then do not store parameters with those names.
Defaults to None.
"""
if not hasattr(self, '_stored_params'):
self._stored_params = {}
frame = sys._getframe(1)
args = frame.f_code.co_varnames[1:frame.f_code.co_argcount]
args_dict = {arg: frame.f_locals[arg] for arg in args}
if 'kwargs' in frame.f_locals:
args_dict['kwargs'] = frame.f_locals['kwargs']
if isinstance(no_store, bool) and no_store:
return
else:
if no_store is None: no_store = tuple()
if isinstance(no_attr, bool) and no_attr: dont_attr = True
else:
if no_attr is None: no_attr = tuple()
dont_attr = False
if isinstance(no_param, bool) and no_param: dont_param = True
else:
if no_param is None: no_param = tuple()
dont_param = False
for name, value in args_dict.items():
if not dont_attr and name not in no_store and name not in no_attr:
setattr(self, name, value)
if not dont_param and name not in no_store and name not in no_param:
self._stored_params[name] = value
def _convert_str_tabs(self, component):
if isinstance(component, str):
if component == 'importances':
return ImportancesTab
elif component == 'model_summary':
return ModelSummaryTab
elif component == 'contributions':
return ContributionsTab
elif component == 'whatif':
return WhatIfTab
elif component == 'shap_dependence':
return ShapDependenceTab
elif component == 'shap_interaction':
return ShapInteractionsTab
elif component == 'decision_trees':
return DecisionTreesTab
return component
@staticmethod
def _tabs_to_yaml(tabs):
"""converts tabs to a yaml friendly format"""
if tabs is None:
return None
def get_name_and_module(component):
if inspect.isclass(component) and issubclass(component, ExplainerComponent):
return dict(
name=component.__name__,
module=component.__module__,
params=None
)
elif isinstance(component, ExplainerComponent):
component_imports = dict(component.component_imports)
del component_imports[component.__class__.__name__]
return dict(
name=component.__class__.__name__,
module=component.__class__.__module__,
params=component._stored_params,
component_imports = component_imports
)
else:
raise ValueError(f"Please only pass strings or ExplainerComponents to parameter `tabs`!"
"You passed {component.__class__}")
if not hasattr(tabs, "__iter__"):
return tabs if isinstance(tabs, str) else get_name_and_module(tabs)
return [tab if isinstance(tab, str) else get_name_and_module(tab) for tab in tabs]
@staticmethod
def _yamltabs_to_tabs(yamltabs, explainer):
"""converts a yaml tabs list back to ExplainerDashboard compatible original"""
from importlib import import_module
if yamltabs is None:
return None
def instantiate_tab(tab, explainer, name=None):
if isinstance(tab, str):
return tab
elif isinstance(tab, dict):
print(tab)
if 'component_imports' in tab and tab['component_imports'] is not None:
for class_name, module_name in tab['component_imports'].items():
if class_name not in globals():
import_module(class_module, class_name)
tab_class = getattr(import_module(tab['module']), tab['name'])
if tab['params'] is None:
return tab_class
else:
if not 'name' in tab['params'] or tab['params']['name'] is None:
tab['params']['name'] = name
tab_instance = tab_class(explainer, **tab['params'])
return tab_instance
else:
raise ValueError("yaml tab should be either string, e.g. 'importances', "
"or a dict(name=..,module=..,params=...)")
if not hasattr(yamltabs, "__iter__"):
return instantiate_tab(yamltabs, explainer, name="1")
tabs = [instantiate_tab(tab, explainer, name=str(i+1)) for i, tab in enumerate(yamltabs)]
print(tabs)
return tabs
def _get_dash_app(self):
if self.responsive:
meta_tags = [
{'name': 'viewport',
'content': 'width=device-width, initial-scale=1.0, maximum-scale=1.2, minimum-scale=0.5,'}
]
else:
meta_tags = None
if self.bootstrap is not None:
assets_ignore = '^bootstrap.min.css$'
else:
assets_ignore = ""
if self.mode=="dash":
app = dash.Dash(__name__,
server=self.server,
external_stylesheets=self.external_stylesheets,
assets_ignore=assets_ignore,
url_base_pathname=self.url_base_pathname,
meta_tags=meta_tags)
elif self.mode in ['inline', 'jupyterlab', 'external']:
app = JupyterDash(__name__,
external_stylesheets=self.external_stylesheets,
assets_ignore=assets_ignore,
meta_tags=meta_tags)
else:
raise ValueError(f"mode=={self.mode} but should be in "
"{'dash', 'inline', 'juypyterlab', 'external'}")
app.config['suppress_callback_exceptions'] = True
app.scripts.config.serve_locally = True
app.css.config.serve_locally = True
return app
def flask_server(self):
"""returns self.app.server so that it can be exposed to e.g. gunicorn"""
if self.mode != 'dash':
print("Warning: in production you should probably use mode='dash'...")
return self.app.server
def run(self, port=None, use_waitress=False, **kwargs):
"""Start ExplainerDashboard on port
Args:
port (int, optional): port to run on. If None, then use self.port.
use_waitress (bool, optional): use the waitress python web server
instead of the flask development server. Only works with mode='dash'.
Defaults to False.
Defaults to None.self.port defaults to 8050.
Raises:
ValueError: if mode is unknown
"""
pio.templates.default = "none"
if port is None:
port = self.port
if use_waitress and self.mode != 'dash':
print(f"Warning: waitress does not work with mode={self.mode}, "
"using JupyterDash server instead!", flush=True)
if self.mode == 'dash':
print(f"Starting ExplainerDashboard on http://localhost:{port}", flush=True)
if use_waitress:
from waitress import serve
serve(self.app.server, host='0.0.0.0', port=port)
else:
self.app.run_server(port=port, **kwargs)
elif self.mode == 'external':
if not self.is_colab:
#print(f"Starting ExplainerDashboard on http://localhost:{port}\n", flush=True)
pass
self.app.run_server(port=port,debug=False, mode=self.mode, **kwargs)
elif self.mode in ['inline', 'jupyterlab']:
print(f"Starting ExplainerDashboard inline (terminate it with "
f"ExplainerDashboard.terminate({port}))", flush=True)
self.app.run_server(port=port, mode=self.mode,debug=False,
width=self.width, height=self.height, **kwargs)
else:
raise ValueError(f"Unknown mode: {mode}...")
@classmethod
def terminate(cls, port, token=None):
"""
Classmethodd to terminate any JupyterDash dashboard (so started with
mode='inline', mode='external' or mode='jupyterlab') from any
ExplainerDashboard by specifying the right port.
Example:
ExplainerDashboard(explainer, mode='external').run(port=8050)
ExplainerDashboard.terminate(8050)
Args:
port (int): port on which the dashboard is running.
token (str, optional): JupyterDash._token class property.
Defaults to the _token of the JupyterDash in the current namespace.
Raises:
ValueError: if can't find the port to terminate.
"""
if token is None:
token = JupyterDash._token
shutdown_url = f"http://localhost:{port}/_shutdown_{token}"
print(f"Trying to shut down dashboard on port {port}...")
try:
response = requests.get(shutdown_url)
except Exception as e:
print(f"Something seems to have failed: {e}")
class ExplainerHub:
"""ExplainerHub is a way to host multiple dashboards in a single point,
and manage access through adding user accounts.
Example:
``hub = ExplainerHub([db1, db2], logins=[['user', 'password']], secret_key="SECRET")``
``hub.run()``
A frontend is hosted at e.g. ``localhost:8050``, with summaries and links to
each individual dashboard. Each ExplainerDashboard is hosted on its own url path,
so that you can also find it directly, e.g.:
``localhost:8050/dashboard1`` and ``localhost:8050/dashboard2``.
You can store the hub configuration, dashboard configurations, explainers
and user database with a single command: ``hub.to_yaml('hub.yaml')``.
You can restore the hub with ``hub2 = ExplainerHub.from_config('hub.yaml')``
You can start the hub from the command line using the ``explainerhub`` CLI
command: ``$ explainerhub run hub.yaml``. You can also use the CLI to
add and delete users.
"""
def __init__(self, dashboards:List[ExplainerDashboard], title:str="ExplainerHub",
description:str=None, masonry:bool=False, n_dashboard_cols:int=3,
user_json:str="users.json", logins:List[List]=None, db_users:dict=None,
port:int=8050, secret_key:str=None, **kwargs):
"""
Note:
Logins can be defined in multiple places: users.json, ExplainerHub.logins
and ExplainerDashboard.logins for each dashboard in dashboards.
When users with the same username are defined in multiple
locations then passwords are looked up in the following order:
hub.logins > dashboard.logins > user.json
Note:
**kwargs will be forwarded to each dashboard in dashboards.
Args:
dashboards (List[ExplainerDashboard]): list of ExplainerDashboard to
include in ExplainerHub.
title (str, optional): title to display. Defaults to "ExplainerHub".
description (str, optional): Short description of ExplainerHub.
Defaults to default text.
masonry (bool, optional): Lay out dashboard cards in fluid bootstrap
masonry responsive style. Defaults to False.
n_dashboard_cols (int, optional): If masonry is False, organize cards
in rows and columns. Defaults to 3 columns.
user_json (Path, optional): a .json file used to store user and (hashed)
password data. Defaults to 'users.json'.
logins (List[List[str, str]], optional): List of ['login', 'password'] pairs,
e.g. logins = [['user1', 'password1'], ['user2', 'password2']]
db_users (dict, optional): dictionary limiting access to certain
dashboards to a subset of users, e.g
dict(dashboard1=['user1', 'user2'], dashboard2=['user3']).
port (int, optional): Port to run hub on. Defaults to 8050.
secret_key (str): Flask secret key to pass to dashboard in order to persist
logins. Defaults to a new random uuid string every time you start
the dashboard. (i.e. no persistence) You should store the secret
key somewhere save, e.g. in a environmental variable.
**kwargs: all kwargs will be forwarded to the constructors of
each dashboard in dashboards dashboards.
"""
self._store_params(no_store=['dashboards', 'logins', 'secret_key'])
if self.description is None:
self.description = ("This ExplainerHub shows an overview of different "
"ExplainerDashboards generated for a number of different machine learning models."
"\n"
"These dashboards make the inner workings and predictions of the trained models "
"transparent and explainable.")
self._stored_params['description'] = self.description
if len(logins)==2 and isinstance(logins[0], str) and isinstance(logins[1], str):
logins = [logins]
self.logins = self._hash_logins(logins)
self.db_users = db_users if db_users is not None else {}
self._validate_user_json(self.user_json)
self.app = Flask(__name__)
if secret_key is not None:
self.app.config['SECRET_KEY'] = secret_key
SimpleLogin(self.app, login_checker=self._validate_user)
self.dashboards = []
for i, dashboard in enumerate(dashboards):
if dashboard.name is None:
print("Reminder, you can set ExplainerDashboard .name and .description "
"in order to control the url path of the dashboard. Now "
f"defaulting to name=dashboard{i+1} and default description...", flush=True)
dashboard_name = f"dashboard{i+1}"
else:
dashboard_name = dashboard.name
update_params = dict(
server=self.app,
name=dashboard_name,
url_base_pathname = f"/{dashboard_name}/"
)
if dashboard.logins is not None:
for user, password in dashboard.logins:
if user not in self.logins:
self.add_user(user, password)
else:
print(f"Warning: {user} in {dashboard.name} already in "
"ExplainerHub logins! So not adding to logins...")
self.add_user_to_dashboard(dashboard_name, user)
config = deepcopy(dashboard.to_yaml(return_dict=True))
config['dashboard']['params']['logins'] = None
self.dashboards.append(
ExplainerDashboard.from_config(
dashboard.explainer, config, **update_kwargs(kwargs, **update_params)))
dashboard_names = [db.name for db in self.dashboards]
assert len(set(dashboard_names)) == len(dashboard_names), \
f"All dashboard .name properties should be unique, but received the folowing: {dashboard_names}"
illegal_names = list(set(dashboard_names) & {'login', 'logout', 'admin'})
assert not illegal_names, \
f"The following .name properties for dashboards are not allowed: {illegal_names}!"
self.index_page = self._get_index_page()
if self.users:
self._protect_dashviews(self.index_page)
for dashboard in self.dashboards:
self._protect_dashviews(dashboard.app, username=self.get_dashboard_users(dashboard.name))
@classmethod
def from_config(cls, config:Union[dict, str, Path], **update_params):
"""Instantiate an ExplainerHub based on a config file.
Args:
config (Union[dict, str, Path]): either a dict or a .yaml config
file to load
update_params: additional kwargs to override stored settings.
Returns:
ExplainerHub: new instance of ExplainerHub according to the config.
"""
if isinstance(config, (Path, str)) and str(config).endswith(".yaml"):
config = yaml.safe_load(open(str(config), "r"))
elif isinstance(config, dict):
config = deepcopy(config)
assert 'explainerhub' in config, \
"Misformed yaml: explainerhub yaml file should start with 'explainerhub:'!"
config = config['explainerhub']
dashboards = [ExplainerDashboard.from_config(dashboard)
for dashboard in config['dashboards']]
del config['dashboards']
config.update(config.pop('kwargs'))
return cls(dashboards, **update_kwargs(config, **update_params))
def to_yaml(self, filepath:Path=None, dump_explainers=True,
return_dict=False, integrate_dashboard_yamls=False):
"""Store ExplainerHub to configuration .yaml, store the users to users.json
and dump the underlying dashboard .yamls and explainers.
If filepath is None, does not store yaml config to file, but simply
return config yaml string.
If filepath provided and dump_explainers=True, then store all underlying
explainers to disk.
Args:
filepath (Path, optional): .yaml file filepath. Defaults to None.
dump_explainers (bool, optional): Store the explainers to disk
along with the .yaml file. Defaults to True.
return_dict (bool, optional): Instead of returning or storing yaml
return a configuration dictionary. Returns a single dict as if
separate_dashboard_yamls=True. Defaults to False.
integrate_dashboard_yamls(bool, optional): Do not generate an individual
.yaml file for each dashboard, but integrate them in hub.yaml.
Returns:
{dict, yaml, None}
"""
for login in self.logins.values():
self._add_user_to_json(self.user_json,
login['username'], login['password'],
already_hashed=True)
for dashboard, users in self.db_users.items():
for user in users:
self._add_user_to_dashboard_json(self.user_json, dashboard, user)
if filepath is None or return_dict or integrate_dashboard_yamls:
hub_config = dict(
explainerhub=dict(
**self._stored_params,
dashboards=[dashboard.to_yaml(
return_dict=True,
explainerfile=dashboard.name+"_explainer.joblib",
dump_explainer=dump_explainers)
for dashboard in self.dashboards]))
else:
for dashboard in self.dashboards:
print(f"Storing {dashboard.name}_dashboard.yaml...")
dashboard.to_yaml(dashboard.name+"_dashboard.yaml",
explainerfile=dashboard.name+"_explainer.joblib",
dump_explainer=dump_explainers)
hub_config = dict(
explainerhub=dict(
**self._stored_params,
dashboards=[dashboard.name+"_dashboard.yaml"
for dashboard in self.dashboards]))
if return_dict:
return hub_config
if filepath is None:
return yaml.dump(hub_config)
filepath = Path(filepath)
print(f"Storing {filepath}...")
yaml.dump(hub_config, open(filepath, "w"))
return
def _store_params(self, no_store=None, no_attr=None, no_param=None):
"""Stores the parameter of the class to instance attributes and
to a ._stored_params dict. You can optionall exclude all or some
parameters from being stored.
Args:
no_store ({bool, List[str]}, optional): If True do not store any
parameters to either attribute or _stored_params dict. If
a list of str, then do not store parameters with those names.
Defaults to None.
no_attr ({bool, List[str]},, optional): . If True do not store any
parameters to class attribute. If
a list of str, then do not store parameters with those names.
Defaults to None.
no_param ({bool, List[str]},, optional): If True do not store any
parameters to _stored_params dict. If
a list of str, then do not store parameters with those names.
Defaults to None.
"""
if not hasattr(self, '_stored_params'):
self._stored_params = {}
frame = sys._getframe(1)
args = frame.f_code.co_varnames[1:frame.f_code.co_argcount]
args_dict = {arg: frame.f_locals[arg] for arg in args}
if 'kwargs' in frame.f_locals:
args_dict['kwargs'] = frame.f_locals['kwargs']
if isinstance(no_store, bool) and no_store:
return
else:
if no_store is None: no_store = tuple()
if isinstance(no_attr, bool) and no_attr: dont_attr = True
else:
if no_attr is None: no_attr = tuple()
dont_attr = False
if isinstance(no_param, bool) and no_param: dont_param = True
else:
if no_param is None: no_param = tuple()
dont_param = False
for name, value in args_dict.items():
if not dont_attr and name not in no_store and name not in no_attr:
setattr(self, name, value)
if not dont_param and name not in no_store and name not in no_param:
self._stored_params[name] = value
@staticmethod
def _validate_user_json(user_json:Path):
"""validat that user_json is a well formed .json file.
If it does not exist, then create an empty .json file.
"""
if user_json is not None:
if not Path(user_json).exists():
with open(user_json, 'w') as json_file:
json.dump(dict(users={}, dashboard_users={}), json_file)
users_db = json.load(open(user_json))
assert 'users' in users_db, \
f"{self.user_json} should contain a 'users' dict!"
assert 'dashboard_users' in users_db, \
f"{self.user_json} should contain a 'dashboard_users' dict!"
def _hash_logins(self, logins:List[List], add_to_user_json:bool=False):
"""Turn a list of [user, password] pairs into a Flask-Login style user
dictionary with hashed passwords. If passwords already in hash-form
then simply copy them.
Args:
logins (List[List]): List of logins e.g.
logins = [['user1', 'password1], ['user2', 'password2]]
add_to_user_json (bool, optional): Add the users to
users.json. Defaults to False.
Returns:
dict
"""
logins_dict = {}
if logins is None:
return logins_dict
regex=re.compile(r'^pbkdf2:sha256:[0-9]+\$[a-zA-Z0-9]+\$[a-z0-9]{64}$', re.IGNORECASE)
for username, password in logins:
if re.search(regex, password) is not None:
logins_dict[username] = dict(
username=username,
password=password
)
if add_to_user_json and self.user_json is not None:
self._add_user_to_json(self.user_json, user, password, already_hashed=True)
else:
logins_dict[username] = dict(
username=username,
password=generate_password_hash(password, method='pbkdf2:sha256')
)
if add_to_user_json and self.user_json is not None:
self._add_user_to_json(self.user_json, user, password)
return logins_dict
@staticmethod
def _add_user_to_json(user_json:Path, username:str, password:str, already_hashed=False):
"""Add a user to a user_json .json file.
Args:
user_json (Path): json file, e.g 'users.json'
username (str): username to add
password (str): password to add
already_hashed (bool, optional): If already hashed then do not hash
the password again. Defaults to False.
"""
users_db = json.load(open(Path(user_json)))
users_db['users'][username] = dict(
username=username,
password=password if already_hashed else generate_password_hash(password, method='pbkdf2:sha256')
)
json.dump(users_db, open(Path(user_json), 'w'))
@staticmethod
def _add_user_to_dashboard_json(user_json:Path, dashboard:str, user:str):
"""Add a user to dashboard_users inside a json file
Args:
user_json (Path): json file e.g. 'users.json'
dashboard (str): name of dashboard
user (str): username
"""
users_db = json.load(open(Path(user_json)))
dashboard_users = users_db['dashboard_users'].get(dashboard)
if dashboard_users is None:
dashboard_users = [user]
else:
dashboard_users = sorted(list(set(dashboard_users + [user])))
users_db['dashboard_users'][dashboard] = dashboard_users
json.dump(users_db, open(Path(user_json), 'w'))
@staticmethod
def _delete_user_from_json(user_json:Path, username:str):
"""delete user from user_json .json file.
Also removes user from all dashboard_user lists.
Args:
user_json (Path): json file e.g. 'users.json'
username (str): username to delete
"""
# TODO: also remove username from dashboard_users
users_db = json.load(open(Path(user_json)))
try:
del users_db['users'][username]
except:
pass
for dashboard in users_db['dashboard_users'].keys():
dashboard_users = users_db['dashboard_users'].get(dashboard)
if dashboard_users is not None:
dashboard_users = sorted(list(set(dashboard_users) ^{user}))
users_db['dashboard_users'][dashboard] = dashboard_users
json.dump(users_db, open(Path(user_json), 'w'))
@staticmethod
def _delete_user_from_dashboard_json(user_json:Path, dashboard:str, username:str):
"""remove a user from a specific dashboard_users list inside a users.json file
Args:
user_json (Path): json file, e.g. 'users.json'
dashboard (str): name of the dashboard
username (str): name of the user to remove
"""
users_db = json.load(open(Path(user_json)))
dashboard_users = users_db['dashboard_users'].get(dashboard)
if dashboard_users is not None:
dashboard_users = sorted(list(set(dashboard_users) ^{user}))
users_db['dashboard_users'][dashboard] = dashboard_users
json.dump(users_db, open(Path(user_json), 'w'))
def add_user(self, username:str, password:str, add_to_json:bool=False):
"""add a user with username and password.
Args:
username (str): username
password (str): password
add_to_json (bool, optional): Add the user to the .json file defined
in self.user_json instead of to self.logins. Defaults to False.
"""
if add_to_json and self.user_json is not None:
self._add_user_to_json(self.user_json, username, password)
else:
self.logins[username] = dict(
username=username,
password=generate_password_hash(password, method='pbkdf2:sha256')
)
def add_user_to_dashboard(self, dashboard:str, username:str, add_to_json:bool=False):
"""add a user to a specific dashboard. If
Args:
dashboard (str): name of dashboard
username (str): user to add to dashboard
add_to_json (bool, optional): add the user to the .json file defined
in self.user_json instead of to self.db_users. Defaults to False.
"""
if add_to_json and self.user_json is not None:
self._add_user_to_dashboard_json(self.user_json, dashboard, user)
else:
dashboard_users = self.db_users.get(dashboard)
dashboard_users = dashboard_users if dashboard_users is not None else []
dashboard_users = sorted(list(set(dashboard_users + [username])))
self.db_users[dashboard] = dashboard_users
@property
def users(self):
"""returns a list of all users, both in users.json and in self.logins"""
users = []
if self.user_json is not None:
users = list(json.load(open(self.user_json))['users'].keys())
if self.logins is not None:
users.extend(list(self.logins.keys()))
return users
@property
def dashboards_with_users(self):
"""returns a list of all dashboards that have a restricted list of users
that can access it"""
dashboards = []
if self.user_json is not None:
dashboards = list(json.load(open(self.user_json))['dashboard_users'].keys())
if self.logins is not None:
dashboards.extend(list(self.db_users.keys()))
return dashboards
@property
def dashboard_users(self):
"""return a dict with the list of users per dashboard"""
dashboard_users = {}
if self.user_json is not None:
dashboard_users.update(json.load(open(self.user_json))['dashboard_users'])
if self.db_users is not None:
for dashboard, users in self.db_users.items():
if not dashboard in dashboard_users:
dashboard_users[dashboard] = users
else:
dashboard_users[dashboard] = sorted(list(set(dashboard_users[dashboard] + users)))
return dashboard_users
def get_dashboard_users(self, dashboard:str):
"""return all users that have been approved to use a specific dashboard
Args:
dashboard (str): dashboard
Returns:
List
"""
dashboard_users = []
if self.user_json is not None:
json_users = json.load(open(self.user_json))['dashboard_users'].get(dashboard)
if json_users is not None:
dashboard_users = json_users
if self.db_users is not None:
param_users = self.db_users.get(dashboard)
if param_users is not None:
dashboard_users.extend(param_users)
dashboard_users = list(set(dashboard_users))
return dashboard_users
def _validate_user(self, user):
"""validation function for SimpleLogin. Returns True when user should
be given access (i.e. no users defined or password correct) and False
when user should be rejected.
Args:
user (dict(username, password)): dictionary with a username and
password key.
Returns:
bool
"""
if not self.users:
return True
users_db = json.load(open(self.user_json))['users'] if self.user_json is not None else {}
if not self.logins.get(user['username']) and not users_db.get(user['username']):
return False
if user['username'] in users_db:
stored_password = users_db[user['username']]['password']
else:
stored_password = self.logins[user['username']]['password']
if check_password_hash(stored_password, user['password']):
return True
return False
@staticmethod
def _protect_dashviews(dashapp:dash.Dash, username:List[str]=None):
"""Wraps a dash dashboard inside a login_required decorator to make sure
unauthorized viewers cannot access it.
Args:
dashapp (dash.Dash): [
username (List[str], optional): list of usernames that can access
this specific dashboard. Defaults to None (all registered users
can access)
"""
for view_func in dashapp.server.view_functions:
if view_func.startswith(dashapp.config.url_base_pathname):
dashapp.server.view_functions[view_func] = login_required(username=username)(
dashapp.server.view_functions[view_func])
def _get_index_page(self):
"""Returns the front end of ExplainerHub:
- title
- description
- links and description for each dashboard
Returns:
dbc.Container
"""
def dashboard_decks(dashboards, n_cols):
full_rows = int(len(dashboards)/ n_cols)
n_last_row = len(dashboards) % n_cols
card_decks = []
for i in range(0, full_rows*n_cols, n_cols):
card_decks.append(
[
dbc.Card([
dbc.CardHeader([
html.H3(dashboard.title, className='card-title'),
]),
dbc.CardBody([
html.H6(dashboard.description),
]),
dbc.CardFooter([
dbc.CardLink("Go to dashboard",
href=f"/{dashboard.name}",
external_link=True),
])
]) for dashboard in dashboards[i:i+n_cols]
]
)
if n_last_row > 0:
last_row = [
dbc.Card([
dbc.CardHeader([
html.H3(dashboard.title, className='card-title'),
]),
dbc.CardBody([
html.H6(dashboard.description),
]),
dbc.CardFooter([
dbc.CardLink("Go to dashboard",
href=f"/{dashboard.name}",
external_link=True),
])
]) for dashboard in dashboards[full_rows*n_cols:full_rows*n_cols+n_last_row]]
for i in range(len(last_row), n_cols):
last_row.append(dbc.Card([], style=dict(border="none")))
card_decks.append(last_row)
return card_decks
header = dbc.Jumbotron([
html.H1(self.title, className="display-3"),
html.Hr(className="my-2"),
html.P(self.description, className="lead"),
])
if self.masonry:
dashboard_rows = [
dbc.Row([
dbc.Col([
dbc.CardColumns(dashboard_cards(self.dashboards))
])
])
]
else:
dashboard_rows = [
dbc.Row([dbc.CardDeck(deck)], style=dict(marginBottom=30))
for deck in dashboard_decks(self.dashboards, self.n_dashboard_cols)]
index_page = dash.Dash(__name__, server=self.app, url_base_pathname="/")
index_page.title = self.title
index_page.layout = dbc.Container([
dbc.Row([dbc.Col([html.A("logout", href="/logout")], md=1)], justify="end"),
dbc.Row([dbc.Col([header])]),
dbc.Row([dbc.Col([html.H2("Dashboards:")])]),
*dashboard_rows
])
return index_page
def flask_server(self):
"""return the Flask server inside the class instance"""
return self.app
def run(self, port=None, host='0.0.0.0', use_waitress=False, **kwargs):
"""start the ExplainerHub.
Args:
port (int, optional): Override default port. Defaults to None.
host (str, optional): host name to run dashboard. Defaults to '0.0.0.0'.
use_waitress (bool, optional): Use the waitress python web server
instead of the Flask development server. Defaults to False.
**kwargs: will be passed forward to either waitress.serve() or app.run()
"""
if port is None:
port = self.port
if use_waitress:
import waitress
waitress.serve(self.app, host=host, port=port, **kwargs)
else:
self.app.run(host=host, port=port, **kwargs)
class InlineExplainer:
"""
Run a single tab inline in a Jupyter notebook using specific method calls.
"""
def __init__(self, explainer, mode='inline', width=1000, height=800,
port=8050, **kwargs):
"""
:param explainer: an Explainer object
:param mode: either 'inline', 'jupyterlab' or 'external'
:type mode: str, optional
:param width: width in pixels of inline iframe
:param height: height in pixels of inline iframe
:param port: port to run if mode='external'
"""
assert mode in ['inline', 'external', 'jupyterlab'], \
"mode should either be 'inline', 'external' or 'jupyterlab'!"
self._explainer = explainer
self._mode = mode
self._width = width
self._height = height
self._port = port
self._kwargs = kwargs
self.tab = InlineExplainerTabs(self, "tabs")
"""subclass with InlineExplainerTabs layouts, e.g. InlineExplainer(explainer).tab.modelsummary()"""
self.shap = InlineShapExplainer(self, "shap")
"""subclass with InlineShapExplainer layouts, e.g. InlineExplainer(explainer).shap.dependence()"""
self.classifier = InlineClassifierExplainer(self, "classifier")
"""subclass with InlineClassifierExplainer plots, e.g. InlineExplainer(explainer).classifier.confusion_matrix()"""
self.regression = InlineRegressionExplainer(self, "regression")
"""subclass with InlineRegressionExplainer plots, e.g. InlineExplainer(explainer).regression.residuals()"""
self.decisiontrees =InlineDecisionTreesExplainer(self, "decisiontrees")
"""subclass with InlineDecisionTreesExplainer plots, e.g. InlineExplainer(explainer).decisiontrees.decisiontrees()"""
def terminate(self, port=None, token=None):
"""terminate an InlineExplainer on particular port.
You can kill any JupyterDash dashboard from any ExplainerDashboard
by specifying the right port.
Args:
port (int, optional): port on which the InlineExplainer is running.
Defaults to the last port the instance had started on.
token (str, optional): JupyterDash._token class property.
Defaults to the _token of the JupyterDash in the current namespace.
Raises:
ValueError: if can't find the port to terminate.
"""
if port is None:
port = self._port
if token is None:
token = JupyterDash._token
shutdown_url = f"http://localhost:{port}/_shutdown_{token}"
print(f"Trying to shut down dashboard on port {port}...")
try:
response = requests.get(shutdown_url)
except Exception as e:
print(f"Something seems to have failed: {e}")
def _run_app(self, app, **kwargs):
"""Starts the dashboard either inline or in a seperate tab
:param app: the JupyterDash app to be run
:type mode: JupyterDash app instance
"""
pio.templates.default = "none"
if self._mode in ['inline', 'jupyterlab']:
app.run_server(mode=self._mode, width=self._width, height=self._height, port=self._port)
elif self._mode == 'external':
app.run_server(mode=self._mode, port=self._port, **self._kwargs)
else:
raise ValueError("mode should either be 'inline', 'jupyterlab' or 'external'!")
def _run_component(self, component, title):
app = JupyterDash(__name__)
app.title = title
app.layout = component.layout()
component.register_callbacks(app)
self._run_app(app)
@delegates_kwargs(ImportancesComponent)
@delegates_doc(ImportancesComponent)
def importances(self, title='Importances', **kwargs):
"""Runs model_summary tab inline in notebook"""
comp = ImportancesComponent(self._explainer, **kwargs)
self._run_component(comp, title)
def model_stats(self, title='Models Stats', **kwargs):
"""Runs model_stats inline in notebook"""
if self._explainer.is_classifier:
comp = ClassifierModelStatsComposite(self._explainer, **kwargs)
elif self._explainer.is_regression:
comp = RegressionModelStatsComposite(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(PredictionSummaryComponent)
@delegates_doc(PredictionSummaryComponent)
def prediction(self, title='Prediction', **kwargs):
"""Show contributions (permutation or shap) inline in notebook"""
comp = PredictionSummaryComponent(self._explainer, **kwargs)
self._run_component(comp, title)
def random_index(self, title='Random Index', **kwargs):
"""show random index selector inline in notebook"""
if self._explainer.is_classifier:
comp = ClassifierRandomIndexComponent(self._explainer, **kwargs)
elif self._explainer.is_regression:
comp = RegressionRandomIndexComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(PdpComponent)
@delegates_doc(PdpComponent)
def pdp(self, title="Partial Dependence Plots", **kwargs):
"""Show contributions (permutation or shap) inline in notebook"""
comp = PdpComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(WhatIfComponent)
@delegates_doc(WhatIfComponent)
def whatif(self, title="What if...", **kwargs):
"""Show What if... component inline in notebook"""
comp = WhatIfComponent(self._explainer, **kwargs)
self._run_component(comp, title)
class InlineExplainerComponent:
def __init__(self, inline_explainer, name):
self._inline_explainer = inline_explainer
self._explainer = inline_explainer._explainer
self._name = name
def _run_component(self, component, title):
self._inline_explainer._run_component(component, title)
def __repr__(self):
component_methods = [method_name for method_name in dir(self)
if callable(getattr(self, method_name)) and not method_name.startswith("_")]
return f"InlineExplainer.{self._name} has the following components: {', '.join(component_methods)}"
class InlineExplainerTabs(InlineExplainerComponent):
@delegates_kwargs(ImportancesTab)
@delegates_doc(ImportancesTab)
def importances(self, title='Importances', **kwargs):
"""Show contributions (permutation or shap) inline in notebook"""
tab = ImportancesTab(self._explainer, **kwargs)
self._run_component(tab, title)
@delegates_kwargs(ModelSummaryTab)
@delegates_doc(ModelSummaryTab)
def modelsummary(self, title='Model Summary', **kwargs):
"""Runs model_summary tab inline in notebook"""
tab = ModelSummaryTab(self._explainer, **kwargs)
self._run_component(tab, title)
@delegates_kwargs(ContributionsTab)
@delegates_doc(ContributionsTab)
def contributions(self, title='Contributions', **kwargs):
"""Show contributions (permutation or shap) inline in notebook"""
tab = ContributionsTab(self._explainer, **kwargs)
self._run_component(tab, title)
@delegates_kwargs(WhatIfTab)
@delegates_doc(WhatIfTab)
def whatif(self, title='What if...', **kwargs):
"""Show What if... tab inline in notebook"""
tab = WhatIfTab(self._explainer, **kwargs)
self._run_component(tab, title)
@delegates_kwargs(ShapDependenceTab)
@delegates_doc(ShapDependenceTab)
def dependence(self, title='Shap Dependence', **kwargs):
"""Runs shap_dependence tab inline in notebook"""
tab = ShapDependenceTab(self._explainer, **kwargs)
self._run_component(tab, title)
@delegates_kwargs(ShapInteractionsTab)
@delegates_doc(ShapInteractionsTab)
def interactions(self, title='Shap Interactions', **kwargs):
"""Runs shap_interactions tab inline in notebook"""
tab = ShapInteractionsTab(self._explainer, **kwargs)
self._run_component(tab, title)
@delegates_kwargs(DecisionTreesTab)
@delegates_doc(DecisionTreesTab)
def decisiontrees(self, title='Decision Trees', **kwargs):
"""Runs shap_interactions tab inline in notebook"""
tab = DecisionTreesTab(self._explainer, **kwargs)
self._run_component(tab, title)
class InlineShapExplainer(InlineExplainerComponent):
@delegates_kwargs(ShapDependenceComposite)
@delegates_doc(ShapDependenceComposite)
def overview(self, title='Shap Overview', **kwargs):
"""Runs shap_dependence tab inline in notebook"""
comp = ShapDependenceComposite(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(ShapSummaryComponent)
@delegates_doc(ShapSummaryComponent)
def summary(self, title='Shap Summary', **kwargs):
"""Show shap summary inline in notebook"""
comp = ShapSummaryComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(ShapDependenceComponent)
@delegates_doc(ShapDependenceComponent)
def dependence(self, title='Shap Dependence', **kwargs):
"""Show shap summary inline in notebook"""
comp = ShapDependenceComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(ShapInteractionsComposite)
@delegates_doc(ShapInteractionsComposite)
def interaction_overview(self, title='Interactions Overview', **kwargs):
"""Runs shap_dependence tab inline in notebook"""
comp = ShapInteractionsComposite(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(InteractionSummaryComponent)
@delegates_doc(InteractionSummaryComponent)
def interaction_summary(self, title='Shap Interaction Summary', **kwargs):
"""show shap interaction summary inline in notebook"""
comp =InteractionSummaryComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(InteractionDependenceComponent)
@delegates_doc(InteractionDependenceComponent)
def interaction_dependence(self, title='Shap Interaction Dependence', **kwargs):
"""show shap interaction dependence inline in notebook"""
comp =InteractionDependenceComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(ShapContributionsGraphComponent)
@delegates_doc(ShapContributionsGraphComponent)
def contributions_graph(self, title='Contributions', **kwargs):
"""Show contributions (permutation or shap) inline in notebook"""
comp = ShapContributionsGraphComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(ShapContributionsTableComponent)
@delegates_doc(ShapContributionsTableComponent)
def contributions_table(self, title='Contributions', **kwargs):
"""Show contributions (permutation or shap) inline in notebook"""
comp = ShapContributionsTableComponent(self._explainer, **kwargs)
self._run_component(comp, title)
class InlineClassifierExplainer(InlineExplainerComponent):
@delegates_kwargs(ClassifierModelStatsComposite)
@delegates_doc(ClassifierModelStatsComposite)
def model_stats(self, title='Models Stats', **kwargs):
"""Runs model_stats inline in notebook"""
comp = ClassifierModelStatsComposite(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(PrecisionComponent)
@delegates_doc(PrecisionComponent)
def precision(self, title="Precision Plot", **kwargs):
"""shows precision plot"""
assert self._explainer.is_classifier
comp = PrecisionComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(CumulativePrecisionComponent)
@delegates_doc(CumulativePrecisionComponent)
def cumulative_precision(self, title="Cumulative Precision Plot", **kwargs):
"""shows cumulative precision plot"""
assert self._explainer.is_classifier
comp = CumulativePrecisionComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(ConfusionMatrixComponent)
@delegates_doc(ConfusionMatrixComponent)
def confusion_matrix(self, title="Confusion Matrix", **kwargs):
"""shows precision plot"""
comp= ConfusionMatrixComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(LiftCurveComponent)
@delegates_doc(LiftCurveComponent)
def lift_curve(self, title="Lift Curve", **kwargs):
"""shows precision plot"""
assert self._explainer.is_classifier
comp = LiftCurveComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(ClassificationComponent)
@delegates_doc(ClassificationComponent)
def classification(self, title="Classification", **kwargs):
"""shows precision plot"""
assert self._explainer.is_classifier
comp = ClassificationComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(RocAucComponent)
@delegates_doc(RocAucComponent)
def roc_auc(self, title="ROC AUC Curve", **kwargs):
"""shows precision plot"""
assert self._explainer.is_classifier
comp = RocAucComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(PrAucComponent)
@delegates_doc(PrAucComponent)
def pr_auc(self, title="PR AUC Curve", **kwargs):
"""shows precision plot"""
assert self._explainer.is_classifier
comp = PrAucComponent(self._explainer, **kwargs)
self._run_component(comp, title)
class InlineRegressionExplainer(InlineExplainerComponent):
@delegates_kwargs(RegressionModelStatsComposite)
@delegates_doc(RegressionModelStatsComposite)
def model_stats(self, title='Models Stats', **kwargs):
"""Runs model_stats inline in notebook"""
comp = RegressionModelStatsComposite(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(PredictedVsActualComponent)
@delegates_doc(PredictedVsActualComponent)
def pred_vs_actual(self, title="Predicted vs Actual", **kwargs):
"shows predicted vs actual for regression"
assert self._explainer.is_regression
comp = PredictedVsActualComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(ResidualsComponent)
@delegates_doc(ResidualsComponent)
def residuals(self, title="Residuals", **kwargs):
"shows residuals for regression"
assert self._explainer.is_regression
comp = ResidualsComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(RegressionVsColComponent)
@delegates_doc(RegressionVsColComponent)
def plots_vs_col(self, title="Plots vs col", **kwargs):
"shows plots vs col for regression"
assert self._explainer.is_regression
comp = RegressionVsColComponent(self._explainer, **kwargs)
self._run_component(comp, title)
class InlineDecisionTreesExplainer(InlineExplainerComponent):
@delegates_kwargs(DecisionTreesComposite)
@delegates_doc(DecisionTreesComposite)
def overview(self, title="Decision Trees", **kwargs):
"""shap decision tree composite inline in notebook"""
comp = DecisionTreesComposite(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(DecisionTreesComponent)
@delegates_doc(DecisionTreesComponent)
def decisiontrees(self, title='Decision Trees', **kwargs):
"""Runs decision_trees tab inline in notebook"""
comp = DecisionTreesComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(DecisionPathTableComponent)
@delegates_doc(DecisionPathTableComponent)
def decisionpath_table(self, title='Decision path', **kwargs):
"""Runs decision_trees tab inline in notebook"""
comp = DecisionPathTableComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(DecisionPathTableComponent)
@delegates_doc(DecisionPathTableComponent)
def decisionpath_graph(self, title='Decision path', **kwargs):
"""Runs decision_trees tab inline in notebook"""
comp = DecisionPathTableComponent(self._explainer, **kwargs)
self._run_component(comp, title)
class JupyterExplainerDashboard(ExplainerDashboard):
def __init__(self, *args, **kwargs):
raise ValueError("JupyterExplainerDashboard has been deprecated. "
"Use e.g. ExplainerDashboard(mode='inline') instead.")
class ExplainerTab:
def __init__(self, *args, **kwargs):
raise ValueError("ExplainerTab has been deprecated. "
"Use e.g. ExplainerDashboard(explainer, ImportancesTab) instead.")
class JupyterExplainerTab(ExplainerTab):
def __init__(self, *args, **kwargs):
raise ValueError("ExplainerTab has been deprecated. "
"Use e.g. ExplainerDashboard(explainer, ImportancesTab, mode='inline') instead.") | AMLBID | /Explainer/AMLBID_Dashboard.py | AMLBID_Dashboard.py |
from functools import partial
import re
from collections import Counter
import numpy as np
import pandas as pd
from dtreeviz.trees import ShadowDecTree
from sklearn.metrics import make_scorer
from sklearn.base import clone
from sklearn.pipeline import Pipeline
from sklearn.model_selection import StratifiedKFold
from joblib import Parallel, delayed
def guess_shap(model):
"""guesses which SHAP explainer to use for a particular model, based
on str(type(model)). Returns 'tree' for tree based models such as
RandomForest and XGBoost that need shap.TreeExplainer, and 'linear'
for linear models such as LinearRegression or Elasticnet that can use
shap.LinearExplainer.
Args:
model: a fitted (sklearn-compatible) model
Returns:
str: {'tree', 'linear', None}
"""
tree_models = ['RandomForestClassifier', 'RandomForestRegressor',
'DecisionTreeClassifier', 'DecisionTreeRegressor',
'ExtraTreesClassifier', 'ExtraTreesRegressor',
'GradientBoostingClassifier', 'GradientBoostingRegressor',
'HistGradientBoostingClassifier', 'HistGradientBoostingRegressor',
'XGBClassifier', 'XGBRegressor',
'LGBMClassifier', 'LGBMRegressor',
'CatBoostClassifier', 'CatBoostRegressor',
'NGClassifier', 'NGBRegressor',
'GBTClassifier', ' GBTRegressor',
'IsolationForest'
]
linear_models = ['LinearRegression', 'LogisticRegression',
'Ridge', 'Lasso', 'ElasticNet']
for tree_model in tree_models:
if str(type(model)).endswith(tree_model + "'>"):
return 'tree'
for lin_model in linear_models:
if str(type(model)).endswith(lin_model + "'>"):
return 'linear'
return None
def parse_cats(X, cats, sep:str="_"):
"""parse onehot encoded columns to a cats_dict.
- cats can be a dict where you enumerate each individual onehot encoded column belonging to
each categorical feature, e.g. cats={
'Sex':['Sex_female', 'Sex_male'],
'Deck':['Deck_A', 'Deck_B', 'Deck_C', 'Deck_nan']
}
- if you encode your categorical features as Cat_Label, you can pass a list of the
original feature names: cats=["Sex", "Deck"]
- or a combination of the two: cats = ["Sex", {'Deck':['Deck_A', 'Deck_B', 'Deck_C', 'Deck_nan']}]
Asserts that all columns can be found in X.columns.
Asserts that all columns are only passed once.
"""
cols = X.columns
col_counter = Counter()
cats_dict = {}
if isinstance(cats, dict):
for k, v in cats.items():
assert set(v).issubset(set(cols)), \
f"These cats columns for {k} could not be found in X.columns: {set(v)-set(cols)}!"
col_counter.update(v)
cats_dict = cats
elif isinstance(cats, list):
for cat in cats:
if isinstance(cat, str):
cats_dict[cat] = [c for c in cols if c.startswith(cat + sep)]
col_counter.update(cats_dict[cat])
if isinstance(cat, dict):
for k, v in cat.items():
assert set(v).issubset(set(cols)), \
f"These cats columns for {k} could not be found in X.columns: {set(v)-set(cols)}!"
col_counter.update(v)
cats_dict[k] = v
multi_cols = [v for v, c in col_counter.most_common() if c > 1]
assert not multi_cols, \
(f"The following columns seem to have been passed to cats multiple times: {multi_cols}. "
"Please make sure that each onehot encoded column is only assigned to one cat column!")
assert not set(cats_dict.keys()) & set(cols), \
(f"These new cats columns are already in X.columns: {list(set(cats_dict.keys()) & set(cols))}! "
"Please select a different name for your new cats columns!")
for col, count in col_counter.most_common():
assert set(X[col].astype(int).unique()).issubset({0,1}), \
f"{col} is not a onehot encoded column (i.e. has values other than 0, 1)!"
cats_list = list(cats_dict.keys())
for col in [col for col in cols if col not in col_counter.keys()]:
cats_dict[col] = [col]
return cats_list, cats_dict
# Could be removed:
# def get_feature_dict(cols, cats=None, sep="_"):
# """helper function to get a dictionary with onehot-encoded columns
# grouped per category.
# Example:
# get_features_dict(["Age", "Sex_Male", "Sex_Female"], cats=["Sex"])
# will return {"Age": ["Age"], "Sex": ["Sex_Male", "Sex_Female"]}
# Args:
# cols (list of str): list of column names
# cats (list of str, optional): list of categorically encoded columns.
# All columns names starting with such a column name will be grouped together.
# Defaults to None.
# sep (str), seperator used between the category and encoding. Defaults to '_'
# Returns:
# dict
# """
# feature_dict = {}
# if cats is None:
# return {col: [col] for col in cols}
# for col in cats:
# cat_cols = [c for c in cols if c.startswith(col + sep)]
# if len(cat_cols) > 1:
# feature_dict[col] = cat_cols
# # add all the individual features
# other_cols = list(set(cols) - set([item for sublist in list(feature_dict.values())
# for item in sublist]))
# for col in other_cols:
# feature_dict[col] = [col]
# return feature_dict
def split_pipeline(pipeline, X, verbose=1):
"""Returns an X_transformed dataframe and model from a fitted
sklearn.pipelines.Pipeline and input dataframe X. Currently only supports
Pipelines that do not change or reorder the columns in the input dataframe.
Args:
pipeline (sklearn.Pipeline): a fitted pipeline with an estimator
with .predict method as the last step.
X (pd.DataFrame): input dataframe
Returns:
X_transformed, model
"""
if verbose:
print("Warning: there is currently limited support for sklearn.Pipelines in explainerdashboard. "
"Only pipelines that return the same number of columns in the same order are supported, "
"until sklearn properly implements a pipeline.get_feature_names() method.", flush=True)
assert hasattr(pipeline.steps[-1][1], 'predict'), \
("When passing an sklearn.Pipeline, the last step of the pipeline should be a model, "
f"but {pipeline.steps[-1][1]} does not have a .predict() function!")
model = pipeline.steps[-1][1]
if X is None:
return X, model
X_transformed, columns = Pipeline(pipeline.steps[:-1]).transform(X), None
if hasattr(pipeline, "get_feature_names"):
try:
columns = pipeline.get_feature_names()
except:
pass
else:
if len(columns) != X_transformed.shape[0]:
print(f"len(pipeline.get_feature_names())={len(columns)} does"
f" not equal X_transformed.shape[0]={X_transformed.shape[0]}!", flush=True)
columns = None
if columns is None and X_transformed.shape == X.values.shape:
for i, pipe in enumerate(pipeline):
if hasattr(pipe, "n_features_in_"):
assert pipe.n_features_in_ == len(X.columns), \
(f".n_features_in_ did not match len(X.columns)={len(X.columns)} for pipeline step {i}: {pipe}!"
"For now explainerdashboard only supports sklearn Pipelines that have a "
".get_feature_names() method or do not add/drop any columns...")
print("Note: sklearn.Pipeline output shape is equal to X input shape, "
f"so assigning column names from X.columns: {X.columns.tolist()}, so"
" make sure that your pipeline does not add, remove or reorders columns!", flush=True)
columns = X.columns
else:
raise ValueError("Pipeline does not return same number of columns as input, "
"nor does it have a proper .get_feature_names() method! "
"Try passing the final estimator in the pipeline seperately "
"together with an already transformed dataframe.")
X_transformed = pd.DataFrame(X_transformed, columns=columns)
return X_transformed, model
def retrieve_onehot_value(X, encoded_col, onehot_cols, sep="_"):
"""
Reverses a onehot encoding.
i.e. Finds the column name starting with encoded_col_ that has a value of 1.
if no such column exists (e.g. they are all 0), then return 'NOT_ENCODED'
Args:
X (pd.DataFrame): dataframe from which to retrieve onehot column
encoded_col (str): Name of the encoded col (e.g. 'Sex')
onehot_cols (list): list of onehot cols, e.g. ['Sex_female', 'Sex_male']
sep (str): seperator between category and value, e.g. '_' for Sex_Male.
Returns:
pd.Series with categories. If no category is found, coded as "NOT_ENCODED".
"""
feature_value = np.argmax(X[onehot_cols].values, axis=1)
# if not a single 1 then encoded feature must have been dropped
feature_value[np.max(X[onehot_cols].values, axis=1) == 0] = -1
mapping = {-1: "NOT_ENCODED"}
col_values = [col[len(encoded_col)+1:] if col.startswith(encoded_col+sep)
else col for col in onehot_cols]
mapping.update({i: col for i, col in enumerate(col_values)})
return pd.Series(feature_value).map(mapping).values
def merge_categorical_columns(X, cats_dict=None, sep="_"):
"""
Returns a new feature Dataframe X_cats where the onehotencoded
categorical features have been merged back with the old value retrieved
from the encodings.
Args:
X (pd.DataFrame): original dataframe with onehotencoded columns, e.g.
columns=['Age', 'Sex_Male', 'Sex_Female"].
cats_dict (dict): dict of features with lists for onehot-encoded variables,
e.g. {'Fare': ['Fare'], 'Sex' : ['Sex_male', 'Sex_Female']}
sep (str): separator used in the encoding, e.g. "_" for Sex_Male.
Defaults to "_".
Returns:
pd.DataFrame, with onehot encodings merged back into categorical columns.
"""
X_cats = X.copy()
for col_name, col_list in cats_dict.items():
if len(col_list) > 1:
X_cats[col_name] = retrieve_onehot_value(X, col_name, col_list, sep)
X_cats.drop(col_list, axis=1, inplace=True)
return X_cats
def X_cats_to_X(X_cats, cats_dict, X_columns, sep="_"):
"""
re-onehotencodes a dataframe where onehotencoded columns had previously
been merged with merge_categorical_columns(...)
Args:
X_cats (pd.DataFrame): dataframe with merged categorical columns cats
cats_dict (dict): dict of features with lists for onehot-encoded variables,
e.g. {'Fare': ['Fare'], 'Sex' : ['Sex_male', 'Sex_Female']}
X_columns: list of columns of original dataframe
Returns:
pd.DataFrame: dataframe X with same encoding as original
"""
non_cat_cols = [col for col in X_cats.columns if col in X_columns]
X_new = X_cats[non_cat_cols].copy()
for cat, labels in cats_dict.items():
if len(labels) > 1:
for label in labels:
if label.startswith(cat+sep):
label_val = label[len(cat)+len(sep):]
else:
label_val = label
X_new[label] = (X_cats[cat]==label_val).astype(int)
return X_new[X_columns]
def merge_categorical_shap_values(X, shap_values, cats_dict=None, sep="_"):
"""
Returns a new feature new shap values np.array
where the shap values of onehotencoded categorical features have been
added up.
Args:
X (pd.DataFrame): dataframe whose columns correspond to the columns
in the shap_values np.ndarray.
shap_values (np.ndarray): numpy array of shap values, output of
e.g. shap.TreeExplainer(X).shap_values()
cats_dict (dict): dict of features with lists for onehot-encoded variables,
e.g. {'Fare': ['Fare'], 'Sex' : ['Sex_male', 'Sex_Female']}
sep (str): seperator used between variable and category.
Defaults to "_".
"""
shap_df = pd.DataFrame(shap_values, columns=X.columns)
for col_name, col_list in cats_dict.items():
if len(col_list) > 1:
shap_df[col_name] = shap_df[col_list].sum(axis=1)
shap_df.drop(col_list, axis=1, inplace=True)
return shap_df.values
def merge_categorical_shap_interaction_values(shap_interaction_values,
old_columns, new_columns, cats_dict):
"""
Returns a 3d numpy array shap_interaction_values where the onehot-encoded
categorical columns have been added up together.
Warning:
Column names in new_columns that are not found in old_columns are
assumed to be categorical feature names.
Args:
shap_interaction_values (np.ndarray): shap_interaction output from
e.g. shap.TreeExplainer(X).shap_interaction_values().
old_columns (list of str): list of column names with onehotencodings,
e.g. ["Age", "Sex_Male", "Sex_Female"]
new_columns (list of str): list of column names without onehotencodings,
e.g. ["Age", "Sex"]
cats_dict (dict): dict of features with lists for onehot-encoded variables,
e.g. {'Fare': ['Fare'], 'Sex' : ['Sex_male', 'Sex_Female']}
Returns:
np.ndarray: shap_interaction values with all the onehot-encoded features
summed together.
"""
if isinstance(old_columns, pd.DataFrame):
old_columns = old_columns.columns.tolist()
if isinstance(new_columns, pd.DataFrame):
new_columns = new_columns.columns.tolist()
siv = np.zeros((shap_interaction_values.shape[0],
len(new_columns), len(new_columns)))
# note: given the for loops here, this code could probably be optimized.
# but only run once anyway
for new_col1 in new_columns:
for new_col2 in new_columns:
newcol_idx1 = new_columns.index(new_col1)
newcol_idx2 = new_columns.index(new_col2)
oldcol_idxs1 = [old_columns.index(col)
for col in cats_dict[new_col1]]
oldcol_idxs2 = [old_columns.index(col)
for col in cats_dict[new_col2]]
siv[:, newcol_idx1, newcol_idx2] = \
shap_interaction_values[:, oldcol_idxs1, :][:, :, oldcol_idxs2]\
.sum(axis=(1, 2))
return siv
def make_one_vs_all_scorer(metric, pos_label=1, greater_is_better=True):
"""
Returns a binary one vs all scorer for a single class('pos_label') of a
multiclass classifier metric.
Args:
metric (function): classification metric of the form metric(y_true, y_pred)
pos_label (int): index of the positive label. Defaults to 1.
greater_is_better (bool): does a higher metric correspond to a better model.
Defaults to True.
Returns:
a binary sklearn-compatible scorer function.
"""
def one_vs_all_metric(metric, pos_label, y_true, y_pred):
return metric((y_true == pos_label).astype(int), y_pred[:, pos_label])
partial_metric = partial(one_vs_all_metric, metric, pos_label)
sign = 1 if greater_is_better else -1
def _scorer(clf, X, y):
y_pred = clf.predict_proba(X)
score = sign * partial_metric(y, y_pred)
return score
return _scorer
def permutation_importances(model, X, y, metric, cats_dict=None,
greater_is_better=True, needs_proba=False,
pos_label=1, n_repeats=1, n_jobs=None, sort=True, verbose=0):
"""
adapted from rfpimp package, returns permutation importances, optionally grouping
onehot-encoded features together.
Args:
model: fitted model for which you'd like to calculate importances.
X (pd.DataFrame): dataframe of features
y (pd.Series): series of targets
metric: metric to be evaluated (usually R2 for regression, roc_auc for
classification)
cats_dict (dict): dict of features with lists for onehot-encoded variables,
e.g. {'Fare': ['Fare'], 'Sex' : ['Sex_male', 'Sex_Female']}
greater_is_better (bool): indicates whether the higher score on the metric
indicates a better model.
needs_proba (bool): does the metric need a classification probability
or direct prediction?
pos_label (int): for classification, the label to use a positive label.
Defaults to 1.
n_repeats (int): number of time to permute each column to take the average score.
Defaults to 1.
n_jobs (int): number of jobs for joblib parallel. Defaults to None.
sort (bool): sort the output from highest importances to lowest.
verbose (int): set to 1 to print output for debugging. Defaults to 0.
"""
X = X.copy()
if cats_dict is None:
cats_dict = {col:[col] for col in X.columns}
if isinstance(metric, str):
scorer = make_scorer(metric, greater_is_better=greater_is_better, needs_proba=needs_proba)
elif not needs_proba or pos_label is None:
scorer = make_scorer(metric, greater_is_better=greater_is_better, needs_proba=needs_proba)
else:
scorer = make_one_vs_all_scorer(metric, pos_label, greater_is_better)
baseline = scorer(model, X, y)
def _permutation_importance(model, X, y, scorer, col_name, col_list, baseline, n_repeats=1):
X = X.copy()
scores = []
for i in range(n_repeats):
old_cols = X[col_list].copy()
X[col_list] = np.random.permutation(X[col_list])
scores.append(scorer(model, X, y))
X[col_list] = old_cols
return col_name, np.mean(scores)
scores = Parallel(n_jobs=n_jobs)(delayed(_permutation_importance)(
model, X, y, scorer, col_name, col_list, baseline, n_repeats
) for col_name, col_list in cats_dict.items())
importances_df = pd.DataFrame(scores, columns=['Feature', 'Score'])
importances_df['Importance'] = baseline - importances_df['Score']
importances_df = importances_df[['Feature', 'Importance', 'Score']]
if sort:
return importances_df.sort_values('Importance', ascending=False)
else:
return importances_df
def cv_permutation_importances(model, X, y, metric, cats_dict=None, greater_is_better=True,
needs_proba=False, pos_label=None, cv=None,
n_repeats=1, n_jobs=None, verbose=0):
"""
Returns the permutation importances averages over `cv` cross-validated folds.
Args:
model: fitted model for which you'd like to calculate importances.
X (pd.DataFrame): dataframe of features
y (pd.Series): series of targets
metric: metric to be evaluated (usually R2 for regression, roc_auc for
classification)
cats_dict (dict): dict of features with lists for onehot-encoded variables,
e.g. {'Fare': ['Fare'], 'Sex' : ['Sex_male', 'Sex_Female']}
greater_is_better (bool): indicates whether the higher score on the metric
indicates a better model.
needs_proba (bool): does the metric need a classification probability
or direct prediction?
pos_label (int): for classification, the label to use a positive label.
Defaults to 1.
cv (int): number of cross-validation folds to apply.
sort (bool): sort the output from highest importances to lowest.
verbose (int): set to 1 to print output for debugging. Defaults to 0.
"""
if cv is None:
return permutation_importances(model, X, y, metric, cats_dict,
greater_is_better=greater_is_better,
needs_proba=needs_proba,
pos_label=pos_label,
n_repeats=n_repeats,
n_jobs=n_jobs,
sort=False,
verbose=verbose)
skf = StratifiedKFold(n_splits=cv, random_state=None, shuffle=False)
model = clone(model)
for i, (train_index, test_index) in enumerate(skf.split(X, y)):
X_train, X_test = X.iloc[train_index], X.iloc[test_index]
y_train, y_test = y.iloc[train_index], y.iloc[test_index]
model.fit(X_train, y_train)
imp = permutation_importances(model, X_test, y_test, metric, cats_dict,
greater_is_better=greater_is_better,
needs_proba=needs_proba,
pos_label=pos_label,
n_repeats=n_repeats,
n_jobs=n_jobs,
sort=False,
verbose=verbose)
if i == 0:
imps = imp
else:
imps = imps.merge(imp, on='Feature', suffixes=("", "_" + str(i)))
return pd.DataFrame(imps.mean(axis=1), columns=['Importance'])\
.sort_values('Importance', ascending=False)
def mean_absolute_shap_values(columns, shap_values, cats_dict=None):
"""
Returns a dataframe with the mean absolute shap values for each feature.
Args:
columns (list of str): list of column names
shap_values (np.ndarray): 2d array of SHAP values
cats_dict (dict): dict of features with lists for onehot-encoded variables,
e.g. {'Fare': ['Fare'], 'Sex' : ['Sex_male', 'Sex_Female']}
Returns:
pd.DataFrame with columns 'Feature' and 'MEAN_ABS_SHAP'.
"""
if cats_dict is None:
cats_dict = {col:[col] for col in columns}
shap_abs_mean_dict = {}
for col_name, col_list in cats_dict.items():
shap_abs_mean_dict[col_name] = np.absolute(
shap_values[:, [columns.index(col) for col in col_list]].sum(axis=1)
).mean()
shap_df = pd.DataFrame(
{
'Feature': list(shap_abs_mean_dict.keys()),
'MEAN_ABS_SHAP': list(shap_abs_mean_dict.values())
}).sort_values('MEAN_ABS_SHAP', ascending=False).reset_index(drop=True)
return shap_df
def get_precision_df(pred_probas, y_true, bin_size=None, quantiles=None,
round=3, pos_label=1):
"""
returns a pd.DataFrame with the predicted probabilities and
the observed frequency per bin_size or quantile.
If pred_probas has one dimension (i.e. only probabilities of positive class)
only returns a single precision. If pred_probas containts probabilities for
every class (typically a multiclass classifier), also returns precision
for every class in every bin.
Args:
pred_probas (np.ndarray): result of model.predict_proba(X). Can either
be probabilities of a single class or multiple classes.
y_true (np.ndarray): array of true class labels.
bin_size (float): bin sizes to bin by. E.g. 0.1 to bin all prediction
between 0 and 0.1, between 0.1 and 0.2, etc. If setting bin_size
you cannot set quantiles.
quantiles (int): number of quantiles to divide pred_probas in.
e.g. if quantiles=4, set bins such that the lowest 25% of pred_probas
go into first bin, next 25% go in second bin, etc. Each bin will
have (approximatly the same amount of observations). If setting
quantiles you cannot set bin_size.
round (int): the number of figures to round the output by. Defaults to 3.
pos_label (int): the label of the positive class. Defaults to 1.
Returns:
pd.DataFrame with columns ['p_min', 'p_max', 'p_avg', 'bin_width',
'precision', 'count']
"""
if bin_size is None and quantiles is None:
bin_size = 0.1
assert ((bin_size is not None and quantiles is None)
or (bin_size is None and quantiles is not None)), \
"either only pass bin_size or only pass quantiles!"
if len(pred_probas.shape) == 2:
# in case the full binary classifier pred_proba is passed,
# we only select the probability of the positive class
predictions_df = pd.DataFrame(
{'pred_proba': pred_probas[:, pos_label], 'target': y_true})
n_classes = pred_probas.shape[1]
else:
predictions_df = pd.DataFrame(
{'pred_proba': pred_probas, 'target': y_true})
n_classes = 1
predictions_df = predictions_df.sort_values('pred_proba')
# define a placeholder df:
columns = ['p_min', 'p_max', 'p_avg', 'bin_width', 'precision', 'count']
if n_classes > 1:
for i in range(n_classes):
columns.append('precision_' + str(i))
precision_df = pd.DataFrame(columns=columns)
if bin_size:
thresholds = np.arange(0.0, 1.0, bin_size).tolist()
# loop through prediction intervals, and compute
for bin_min, bin_max in zip(thresholds, thresholds[1:] + [1.0]):
if bin_min != bin_max:
new_row_dict = {
'p_min': [bin_min],
'p_max': [bin_max],
'p_avg': [bin_min + (bin_max - bin_min) / 2.0],
'bin_width': [bin_max - bin_min]
}
if bin_min == 0.0:
new_row_dict['p_avg'] = predictions_df[
(predictions_df.pred_proba >= bin_min)
& (predictions_df.pred_proba <= bin_max)
]['pred_proba'].mean()
new_row_dict['precision'] = (
predictions_df[
(predictions_df.pred_proba >= bin_min)
& (predictions_df.pred_proba <= bin_max)
].target == pos_label
).mean()
new_row_dict['count'] = predictions_df[
(predictions_df.pred_proba >= bin_min)
& (predictions_df.pred_proba <= bin_max)
].target.count()
if n_classes > 1:
for i in range(n_classes):
new_row_dict['precision_' + str(i)] = (
predictions_df[
(predictions_df.pred_proba >= bin_min)
& (predictions_df.pred_proba <= bin_max)
].target == i
).mean()
else:
new_row_dict['p_avg'] = predictions_df[
(predictions_df.pred_proba > bin_min)
& (predictions_df.pred_proba <= bin_max)
]['pred_proba'].mean()
new_row_dict['precision'] = (
predictions_df[
(predictions_df.pred_proba > bin_min)
& (predictions_df.pred_proba <= bin_max)
].target == pos_label
).mean()
new_row_dict['count'] = (
predictions_df[
(predictions_df.pred_proba > bin_min)
& (predictions_df.pred_proba <= bin_max)
].target == pos_label
).count()
if n_classes > 1:
for i in range(n_classes):
new_row_dict['precision_' + str(i)] = (
predictions_df[
(predictions_df.pred_proba > bin_min)
& (predictions_df.pred_proba <= bin_max)
].target == i
).mean()
new_row_df = pd.DataFrame(new_row_dict, columns=precision_df.columns)
precision_df = pd.concat([precision_df, new_row_df])
elif quantiles:
preds_quantiles = np.array_split(predictions_df.pred_proba.values, quantiles)
target_quantiles = np.array_split(predictions_df.target.values, quantiles)
last_p_max = 0.0
for preds, targets in zip(preds_quantiles, target_quantiles):
new_row_dict = {
'p_min': [last_p_max],
'p_max': [preds.max()],
'p_avg': [preds.mean()],
'bin_width': [preds.max() - last_p_max],
'precision': [np.mean(targets==pos_label)],
'count' : [len(preds)],
}
if n_classes > 1:
for i in range(n_classes):
new_row_dict['precision_' + str(i)] = np.mean(targets==i)
new_row_df = pd.DataFrame(new_row_dict, columns=precision_df.columns)
precision_df = pd.concat([precision_df, new_row_df])
last_p_max = preds.max()
precision_df[['p_avg', 'precision']] = precision_df[['p_avg', 'precision']]\
.astype(float).apply(partial(np.round, decimals=round))
if n_classes > 1:
precision_cols = ['precision_' + str(i) for i in range(n_classes)]
precision_df[precision_cols] = precision_df[precision_cols]\
.astype(float).apply(partial(np.round, decimals=round))
return precision_df
def get_lift_curve_df(pred_probas, y, pos_label=1):
"""returns a pd.DataFrame that can be used to generate a lift curve plot.
Args:
pred_probas (np.ndarray): predicted probabilities of the positive class
y (np.ndarray): the actual labels (y_true), encoded 0, 1 [, 2, 3, etc]
pos_label (int): label of the positive class. Defaults to 1.
Returns:
pd.DataFrame with columns=['pred_proba', 'y', 'index', 'index_percentage',
'positives', 'precision', 'cumulative_percentage_pos',
'random_pos', 'random_precision', 'random_cumulative_percentage_pos']
"""
lift_df = pd.DataFrame(
{
'pred_proba' : pred_probas,
'y' : y
}).sort_values('pred_proba', ascending=False).reset_index(drop=True)
lift_df['index'] = lift_df.index + 1
lift_df['index_percentage'] = 100*lift_df['index'] / len(lift_df)
lift_df['positives'] = (lift_df.y==pos_label).astype(int).cumsum()
lift_df['precision'] = 100 * (lift_df['positives'] / lift_df['index'])
lift_df['cumulative_percentage_pos'] = 100 * (lift_df['positives'] / (lift_df.y==pos_label).astype(int).sum())
lift_df['random_pos'] = (lift_df.y==pos_label).astype(int).mean() * lift_df['index']
lift_df['random_precision'] = 100 * (lift_df['random_pos'] / lift_df['index'])
lift_df['random_cumulative_percentage_pos'] = 100 * (lift_df['random_pos'] / (lift_df.y==pos_label).astype(int).sum())
for y_label in range(y.nunique()):
lift_df['precision_' + str(y_label)] = 100*(lift_df.y==y_label).astype(int).cumsum() / lift_df['index']
return lift_df
def get_contrib_df(shap_base_value, shap_values, X_row, topx=None, cutoff=None, sort='abs', cols=None):
"""
Return a contrib_df DataFrame that lists the SHAP contribution of each input
variable for a single prediction, formatted in a way that makes it easy to
plot a waterfall plot.
Args:
shap_base_value (float): the value of shap.Explainer.expected_value
shap_values (np.ndarray): single array of shap values for a specific
prediction, corresponding to X_row
X_row (pd.DataFrame): a single row of data, generated with e.g. X.iloc[[index]]
topx (int): only display the topx highest impact features.
cutoff (float): only display features with a SHAP value of at least
cutoff.
sort ({'abs', 'high-to-low', 'low-to-high'}), sort the shap value
contributions either from highest absolute shap to lowest absolute
shap ('abs'), or from most positive to most negative ('high-to-low')
or from most negative to most positive ('low-to-high'). Defaults
to 'abs'.
cols (list of str): particular list of columns to display, in that order. Will
override topx, cutoff, sort, etc.
Features below topx or cutoff are summed together under _REST. Final
prediction is added as _PREDICTION.
Returns:
pd.DataFrame with columns=['col', 'contribution', 'value', 'cumulative', 'base']
"""
assert isinstance(X_row, pd.DataFrame),\
'X_row should be a pd.DataFrame! Use X.iloc[[index]]'
assert len(X_row.iloc[[0]].values[0].shape) == 1,\
"""X is not the right shape: len(X.values[0]) should be 1.
Try passing X.iloc[[index]]"""
assert sort in {'abs', 'high-to-low', 'low-to-high', 'importance', None}
# start with the shap_base_value
base_df = pd.DataFrame(
{
'col': ['_BASE'],
'contribution': [shap_base_value],
'value': ['']
})
contrib_df = pd.DataFrame(
{
'col': X_row.columns,
'contribution': shap_values,
'value': X_row.values[0]
})
if cols is None:
if cutoff is None and topx is not None:
cutoff = contrib_df.contribution.abs().nlargest(topx).min()
elif cutoff is None and topx is None:
cutoff = 0
display_df = contrib_df[contrib_df.contribution.abs() >= cutoff]
if topx is not None and len(display_df) > topx:
# in case of ties around cutoff
display_df = display_df.reindex(
display_df.contribution.abs().sort_values(ascending=False).index).head(topx)
display_df_neg = display_df[display_df.contribution < 0]
display_df_pos = display_df[display_df.contribution >= 0]
rest_df = (contrib_df[~contrib_df.col.isin(display_df.col.tolist())]
.sum().to_frame().T
.assign(col="_REST", value=""))
# sort the df by absolute value from highest to lowest:
if sort=='abs':
display_df = display_df.reindex(
display_df.contribution.abs().sort_values(ascending=False).index)
contrib_df = pd.concat([base_df, display_df, rest_df], ignore_index=True)
if sort=='high-to-low':
display_df_pos = display_df_pos.reindex(
display_df_pos.contribution.abs().sort_values(ascending=False).index)
display_df_neg = display_df_neg.reindex(
display_df_neg.contribution.abs().sort_values().index)
contrib_df = pd.concat([base_df, display_df_pos, rest_df, display_df_neg], ignore_index=True)
if sort=='low-to-high':
display_df_pos = display_df_pos.reindex(
display_df_pos.contribution.abs().sort_values().index)
display_df_neg = display_df_neg.reindex(
display_df_neg.contribution.abs().sort_values(ascending=False).index)
contrib_df = pd.concat([base_df, display_df_neg, rest_df, display_df_pos], ignore_index=True)
else:
display_df = contrib_df[contrib_df.col.isin(cols)].set_index('col').reindex(cols).reset_index()
rest_df = (contrib_df[~contrib_df.col.isin(cols)]
.sum().to_frame().T
.assign(col="_REST", value=""))
contrib_df = pd.concat([base_df, display_df, rest_df], ignore_index=True)
# add cumulative contribution from top to bottom (for making bar chart):
contrib_df['cumulative'] = contrib_df.contribution.cumsum()
contrib_df['base']= contrib_df['cumulative'] - contrib_df['contribution']
pred_df = contrib_df[['contribution']].sum().to_frame().T.assign(
col='_PREDICTION',
value="",
cumulative=lambda df:df.contribution,
base=0)
return pd.concat([contrib_df, pred_df], ignore_index=True)
def get_contrib_summary_df(contrib_df, model_output="raw", round=2, units="", na_fill=None):
"""
returns a DataFrame that summarizes a contrib_df as a pair of
Reasons+Effect.
Args:
contrib_df (pd.DataFrame): output from get_contrib_df(...)
model_output (str, {'raw', 'probability', 'logodds'}): the type of
predictions that the model produces. 'probability' multiplies by 100
and adds '%'.
round (int): number of decimals to round the output to. Defaults to 1.
units (str): units to add to output. Defaults to "".
na_fill (int, str): if value equals na_fill replace with "MISSING".
"""
assert model_output in {'raw', 'probability', 'logodds'}
contrib_summary_df = pd.DataFrame(columns=['Reason', 'Effect'])
for _, row in contrib_df.iterrows():
if row['col'] == '_BASE':
reason = 'Average of population'
effect = ""
elif row['col'] == '_REST':
reason = 'Other features combined'
effect = f"{'+' if row['contribution'] >= 0 else ''}"
elif row['col'] == '_PREDICTION':
reason = 'Final prediction'
effect = ""
else:
if na_fill is not None and row['value']==na_fill:
reason = f"{row['col']} = MISSING"
else:
reason = f"{row['col']} = {row['value']}"
effect = f"{'+' if row['contribution'] >= 0 else ''}"
if model_output == "probability":
effect += str(np.round(100*row['contribution'], round))+'%'
elif model_output == 'logodds':
effect += str(np.round(row['contribution'], round))
else:
effect += str(np.round(row['contribution'], round)) + f" {units}"
contrib_summary_df = contrib_summary_df.append(
dict(Reason=reason, Effect=effect), ignore_index=True)
return contrib_summary_df.reset_index(drop=True)
def normalize_shap_interaction_values(shap_interaction_values, shap_values=None):
"""
Normalizes shap_interaction_values to make sure that the rows add up to
the shap_values.
This is a workaround for an apparant bug where the diagonals of
shap_interaction_values of a RandomForestClassifier are set equal to the
shap_values instead of the main effect.
I Opened an issue here: https://github.com/slundberg/shap/issues/723
(so far doesn't seem to be fixed)
Args:
shap_interaction_values (np.ndarray): output of shap.Explainer.shap_interaction_values()
shap_values (np.ndarray): output of shap.Explainer.shap_values()
"""
siv = shap_interaction_values.copy()
orig_diags = np.einsum('ijj->ij', siv)
row_sums = np.einsum('ijk->ij', siv)
row_diffs = row_sums - orig_diags # sum of rows excluding diagonal elements
if shap_values is not None:
diags = shap_values - row_diffs
else:
# if no shap_values provided assume that the original diagonal values
# were indeed equal to the shap values, and so simply
diags = orig_diags - row_diffs
s0, s1, s2 = siv.shape
# should have commented this bit of code earlier:
# (can't really explain it anymore, but it works! :)
# In any case, it assigns our new diagonal values to siv:
siv.reshape(s0,-1)[:,::s2+1] = diags
return siv
def get_decisiontree_df(decision_tree, observation, pos_label=1):
"""summarize the path through a DecisionTree for a specific observation.
Args:
decision_tree (DecisionTreeClassifier or DecisionTreeRegressor):
a fitted DecisionTree model.
observation ([type]): single row of data to display tree path for.
pos_label (int, optional): label of positive class. Defaults to 1.
Returns:
pd.DataFrame: columns=['node_id', 'average', 'feature',
'value', 'split', 'direction', 'left', 'right', 'diff']
"""
_, nodes = decision_tree.predict(observation)
decisiontree_df = pd.DataFrame(columns=['node_id', 'average', 'feature',
'value', 'split', 'direction',
'left', 'right', 'diff'])
if decision_tree.is_classifier():
def node_pred_proba(node):
return node.class_counts()[pos_label]/ sum(node.class_counts())
for node in nodes:
if not node.isleaf():
decisiontree_df = decisiontree_df.append({
'node_id' : node.id,
'average' : node_pred_proba(node),
'feature' : node.feature_name(),
'value' : observation[node.feature_name()],
'split' : node.split(),
'direction' : 'left' if observation[node.feature_name()] < node.split() else 'right',
'left' : node_pred_proba(node.left),
'right' : node_pred_proba(node.right),
'diff' : node_pred_proba(node.left) - node_pred_proba(node) \
if observation[node.feature_name()] < node.split() \
else node_pred_proba(node.right) - node_pred_proba(node)
}, ignore_index=True)
else:
def node_mean(node):
return decision_tree.tree_model.tree_.value[node.id].item()
for node in nodes:
if not node.isleaf():
decisiontree_df = decisiontree_df.append({
'node_id' : node.id,
'average' : node_mean(node),
'feature' : node.feature_name(),
'value' : observation[node.feature_name()],
'split' : node.split(),
'direction' : 'left' if observation[node.feature_name()] < node.split() else 'right',
'left' : node_mean(node.left),
'right' : node_mean(node.right),
'diff' : node_mean(node.left) - node_mean(node) \
if observation[node.feature_name()] < node.split() \
else node_mean(node.right) - node_mean(node)
}, ignore_index=True)
return decisiontree_df
def get_decisiontree_summary_df(decisiontree_df, classifier=False, round=2, units=""):
"""generate a pd.DataFrame with a more readable summary of a dataframe
generated with get_decisiontree_df(...)
Args:
decisiontree_df (pd.DataFrame): dataframe generated with get_decisiontree_df(...)
classifier (bool, optional): model is a classifier. Defaults to False.
round (int, optional): Rounding to apply to floats. Defaults to 2.
units (str, optional): units of target to display. Defaults to "".
Returns:
pd.DataFrame: columns=['Feature', 'Condition', 'Adjustment', 'New Prediction']
"""
if classifier:
base_value = np.round(100*decisiontree_df.iloc[[0]]['average'].item(), round)
prediction = np.round(100*(decisiontree_df.iloc[[-1]]['average'].item() \
+ decisiontree_df.iloc[[-1]]['diff'].item()), round)
else:
base_value = np.round(decisiontree_df.iloc[[0]]['average'].item(), round)
prediction = np.round(decisiontree_df.iloc[[-1]]['average'].item() \
+ decisiontree_df.iloc[[-1]]['diff'].item(), round)
decisiontree_summary_df = pd.DataFrame(columns=['Feature', 'Condition', 'Adjustment', 'New Prediction'])
decisiontree_summary_df = decisiontree_summary_df.append({
'Feature' : "",
'Condition' : "",
'Adjustment' : "Starting average",
'New Prediction' : str(np.round(base_value, round)) + ('%' if classifier else f' {units}')
}, ignore_index=True)
for _, row in decisiontree_df.iterrows():
if classifier:
decisiontree_summary_df = decisiontree_summary_df.append({
'Feature' : row['feature'],
'Condition' : str(row['value']) + str(' >= ' if row['direction'] == 'right' else ' < ') + str(row['split']).ljust(10),
'Adjustment' : str('+' if row['diff'] >= 0 else '') + str(np.round(100*row['diff'], round)) +'%',
'New Prediction' : str(np.round(100*(row['average']+row['diff']), round)) + '%'
}, ignore_index=True)
else:
decisiontree_summary_df = decisiontree_summary_df.append({
'Feature' : row['feature'],
'Condition' : str(row['value']) + str(' >= ' if row['direction'] == 'right' else ' < ') + str(row['split']).ljust(10),
'Adjustment' : str('+' if row['diff'] >= 0 else '') + str(np.round(row['diff'], round)),
'New Prediction' : str(np.round((row['average']+row['diff']), round)) + f" {units}"
}, ignore_index=True)
decisiontree_summary_df = decisiontree_summary_df.append({
'Feature' : "",
'Condition' : "",
'Adjustment' : "Final Prediction",
'New Prediction' : str(np.round(prediction, round)) + ('%' if classifier else '') + f" {units}"
}, ignore_index=True)
return decisiontree_summary_df
def get_xgboost_node_dict(xgboost_treedump):
"""Turns the output of a xgboostmodel.get_dump() into a dictionary
of nodes for easy parsing a prediction path through individual trees
in the model.
Args:
xgboost_treedump (str): a single element of the list output from
xgboost model.get_dump() that represents a single tree in the
ensemble.
Returns:
dict
"""
node_dict = {}
for row in xgboost_treedump.splitlines():
s = row.strip()
node = int(re.search("^(.*)\:", s).group(1))
is_leaf = re.search(":(.*)\=", s).group(1) == "leaf"
leaf_value = re.search("leaf=(.*)$", s).group(1) if is_leaf else None
feature = re.search('\[(.*)\<', s).group(1) if not is_leaf else None
cutoff = float(re.search('\<(.*)\]', s).group(1)) if not is_leaf else None
left_node = int(re.search('yes=(.*)\,no', s).group(1)) if not is_leaf else None
right_node = int(re.search('no=(.*)\,', s).group(1)) if not is_leaf else None
node_dict[node] = dict(
node=node,
is_leaf=is_leaf,
leaf_value=leaf_value,
feature=feature,
cutoff=cutoff,
left_node=left_node,
right_node=right_node
)
return node_dict
def get_xgboost_path_df(xgbmodel, X_row, n_tree=None):
"""returns a pd.DataFrame of the prediction path through
an individual tree in a xgboost ensemble.
Args:
xgbmodel: either a fitted xgboost model, or the output of a get_dump()
X_row: single row from a dataframe (e.g. X_test.iloc[0])
n_tree: the tree number to display:
Returns:
pd.DataFrame
"""
if isinstance(xgbmodel, str) and xgbmodel.startswith("0:"):
xgbmodel_treedump = xgbmodel
elif str(type(xgbmodel)).endswith("xgboost.core.Booster'>"):
xgbmodel_treedump = xgbmodel.get_dump()[n_tree]
elif str(type(xgbmodel)).endswith("XGBClassifier'>") or str(type(xgbmodel)).endswith("XGBRegressor'>"):
xgbmodel_treedump = xgbmodel.get_booster().get_dump()[n_tree]
else:
raise ValueError("Couldn't extract a treedump. Please pass a fitted xgboost model.")
node_dict = get_xgboost_node_dict(xgbmodel_treedump)
prediction_path_df = pd.DataFrame(columns = ['node', 'feature', 'cutoff', 'value'])
node = node_dict[0]
while not node['is_leaf']:
prediction_path_df = prediction_path_df.append(
dict(
node=node['node'],
feature=node['feature'],
cutoff=node['cutoff'],
value=float(X_row[node['feature']])
), ignore_index=True)
if np.isnan(X_row[node['feature']]) or X_row[node['feature']] < node['cutoff']:
node = node_dict[node['left_node']]
else:
node = node_dict[node['right_node']]
if node['is_leaf']:
prediction_path_df = prediction_path_df.append(dict(node=node['node'], feature="_PREDICTION", value=node['leaf_value']), ignore_index=True)
return prediction_path_df
def get_xgboost_path_summary_df(xgboost_path_df, output="margin"):
"""turn output of get_xgboost_path_df output into a formatted dataframe
Args:
xgboost_path_df (pd.DataFrame): output of get_xgboost_path_df
prediction (str, {'logodds', 'margin'}): Type of output prediction.
Defaults to "margin".
Returns:
pd.DataFrame: dataframe with nodes and split conditions
"""
xgboost_path_summary_df = pd.DataFrame(columns=['node', 'split_condition'])
for row in xgboost_path_df.itertuples():
if row.feature == "_PREDICTION":
xgboost_path_summary_df = xgboost_path_summary_df.append(
dict(
node=row.node,
split_condition=f"prediction ({output}) = {row.value}"
), ignore_index=True
)
elif row.value < row.cutoff:
xgboost_path_summary_df = xgboost_path_summary_df.append(
dict(
node=row.node,
split_condition=f"{row.feature} = {row.value} < {row.cutoff}"
), ignore_index=True
)
else:
xgboost_path_summary_df = xgboost_path_summary_df.append(
dict(
node=row.node,
split_condition=f"{row.feature} = {row.value} >= {row.cutoff}"
), ignore_index=True
)
return xgboost_path_summary_df
def get_xgboost_preds_df(xgbmodel, X_row, pos_label=1):
""" returns the marginal contributions of each tree in
an xgboost ensemble
Args:
xgbmodel: a fitted sklearn-comptaible xgboost model
(i.e. XGBClassifier or XGBRegressor)
X_row: a single row of data, e.g X_train.iloc[0]
pos_label: for classifier the label to be used as positive label
Defaults to 1.
Returns:
pd.DataFrame
"""
if str(type(xgbmodel)).endswith("XGBClassifier'>"):
is_classifier=True
n_classes = len(xgbmodel.classes_)
if n_classes == 2:
if pos_label==1:
base_proba = xgbmodel.get_params()['base_score']
elif pos_label==0:
base_proba = 1 - xgbmodel.get_params()['base_score']
else:
raise ValueError("pos_label should be either 0 or 1!")
n_trees = len(xgbmodel.get_booster().get_dump())
base_score = np.log(base_proba/(1-base_proba))
else:
base_proba = 1.0 / n_classes
base_score = xgbmodel.get_params()['base_score']
n_trees = int(len(xgbmodel.get_booster().get_dump()) / n_classes)
elif str(type(xgbmodel)).endswith("XGBRegressor'>"):
is_classifier=False
base_score = xgbmodel.get_params()['base_score']
n_trees = len(xgbmodel.get_booster().get_dump())
else:
raise ValueError("Pass either an XGBClassifier or XGBRegressor!")
if is_classifier:
if n_classes == 2:
if pos_label==1:
preds = [xgbmodel.predict(X_row, ntree_limit=i+1, output_margin=True)[0] for i in range(n_trees)]
elif pos_label==0:
preds = [-xgbmodel.predict(X_row, ntree_limit=i+1, output_margin=True)[0] for i in range(n_trees)]
pred_probas = (np.exp(preds)/(1+np.exp(preds))).tolist()
else:
margins = [xgbmodel.predict(X_row, ntree_limit=i+1, output_margin=True)[0] for i in range(n_trees)]
preds = [margin[pos_label] for margin in margins]
pred_probas = [(np.exp(margin)/ np.exp(margin).sum())[pos_label] for margin in margins]
else:
preds = [xgbmodel.predict(X_row, ntree_limit=i+1, output_margin=True)[0] for i in range(n_trees)]
xgboost_preds_df = pd.DataFrame(
dict(
tree=range(-1, n_trees),
pred=[base_score] + preds
)
)
xgboost_preds_df['pred_diff'] = xgboost_preds_df.pred.diff()
xgboost_preds_df.loc[0, "pred_diff"] = xgboost_preds_df.loc[0, "pred"]
if is_classifier:
xgboost_preds_df['pred_proba'] = [base_proba] + pred_probas
xgboost_preds_df['pred_proba_diff'] = xgboost_preds_df.pred_proba.diff()
xgboost_preds_df.loc[0, "pred_proba_diff"] = xgboost_preds_df.loc[0, "pred_proba"]
return xgboost_preds_df | AMLBID | /Explainer/explainer_methods.py | explainer_methods.py |
__all__ = [
'ImportancesTab',
'ModelSummaryTab',
'ContributionsTab',
'WhatIfTab',
'ShapDependenceTab',
'ShapInteractionsTab',
'DecisionTreesTab',
]
import dash_html_components as html
from .dashboard_components import *
class ImportancesTab(ExplainerComponent):
def __init__(self, explainer, title="Feature Importances", name=None,
hide_type=False, hide_depth=False, hide_cats=False,
hide_title=False, hide_selector=False,
pos_label=None, importance_type="shap", depth=None,
cats=True, disable_permutations=False, **kwargs):
"""Overview tab of feature importances
Can show both permutation importances and mean absolute shap values.
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"Feature Importances".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
hide_selector(bool, optional) Hide pos label selector. Defaults to False.
importance_type (str, {'permutation', 'shap'} optional):
Type of importance to describe. Defaults to "shap".
depth (int, optional): Number of features to display by default. Defaults to None.
cats (bool, optional): Group categoricals together. Defaults to True.
"""
super().__init__(explainer, title, name)
self.importances = ImportancesComponent(
explainer,
hide_selector=hide_selector,
importance_type=importance_type,
depth=depth,
cats=cats, hide_cats=hide_cats)
self.register_components(self.importances)
def layout(self):
return html.Div([
self.importances.layout(),
])
class ModelSummaryTab(ExplainerComponent):
def __init__(self, explainer, title="Model Performance", name=None,
bin_size=0.1, quantiles=10, cutoff=0.5,
logs=False, pred_or_actual="vs_pred", residuals='difference',
col=None, **kwargs):
"""Tab shows a summary of model performance.
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"Model Performance".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
bin_size (float, optional): precision plot bin_size. Defaults to 0.1.
quantiles (int, optional): precision plot number of quantiles. Defaults to 10.
cutoff (float, optional): cutoff for classifier plots. Defaults to 0.5.
logs (bool, optional): use logs for regression plots. Defaults to False.
pred_or_actual (str, optional): show residuals vs prediction or vs actual. Defaults to "vs_pred".
residuals (str, {'difference', 'ratio', 'log-ratio'} optional):
How to calcualte residuals. Defaults to 'difference'.
col ([type], optional): Feature to show residuals against. Defaults to None.
"""
super().__init__(explainer, title, name)
if self.explainer.is_classifier:
self.model_stats = ClassifierModelStatsComposite(explainer,
bin_size=bin_size, quantiles=quantiles, cutoff=cutoff, **kwargs)
elif explainer.is_regression:
self.model_stats = RegressionModelStatsComposite(explainer,
logs=logs, pred_or_actual=pred_or_actual, residuals=residuals, **kwargs)
self.register_components(self.model_stats)
def layout(self):
return html.Div([
self.model_stats.layout()
])
class ContributionsTab(ExplainerComponent):
def __init__(self, explainer, title="Individual Predictions", name=None,
**kwargs):
"""Tab showing individual predictions, the SHAP contributions that
add up to this predictions, in both graph and table form, and a pdp plot.
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"Individual Predictions".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure it's unique.
Defaults to None.
higher_is_better (bool, optional): in contributions plot, up is green
and down is red. (set to False to flip)
"""
super().__init__(explainer, title, name)
self.tab_id = "contributions"
self.contribs = IndividualPredictionsComposite(explainer,
#higher_is_better=higher_is_better,
**kwargs)
self.register_components(self.contribs)
def layout(self):
return html.Div([
self.contribs.layout()
])
class WhatIfTab(ExplainerComponent):
def __init__(self, explainer, title="What if...", name=None,
**kwargs):
"""Tab showing individual predictions and allowing edits
to the features...
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"Individual Predictions".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
"""
super().__init__(explainer, title, name)
self.tab_id = "whatif"
self.whatif = WhatIfComposite(explainer, **kwargs)
self.register_components(self.whatif)
def layout(self):
return html.Div([
self.whatif.layout()
])
class ShapDependenceTab(ExplainerComponent):
def __init__(self, explainer, title='Feature Dependence', name=None,
tab_id="shap_dependence",
depth=None, cats=True, **kwargs):
"""Tab showing both a summary of feature importance (aggregate or detailed).
for each feature, and a shap dependence graph.
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
'Feature Dependence'.
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
"""
super().__init__(explainer, title, name)
self.shap_overview = ShapDependenceComposite(
explainer, depth=depth, cats=cats, **kwargs)
self.register_components(self.shap_overview)
def layout(self):
return html.Div([
self.shap_overview.layout()
])
class ShapInteractionsTab(ExplainerComponent):
def __init__(self, explainer, title='Feature Interactions', name=None,
depth=None, cats=True, **kwargs):
"""[summary]
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
'Feature Interactions'.
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
depth (int, optional): default number of feature to display. Defaults to None.
cats (bool, optional): default grouping of cats. Defaults to True.
"""
super().__init__(explainer, title, name)
self.interaction_overview = ShapInteractionsComposite(
explainer, depth=depth, cats=cats, **kwargs)
self.register_components(self.interaction_overview)
def layout(self):
return html.Div([
self.interaction_overview.layout()
])
class DecisionTreesTab(ExplainerComponent):
def __init__(self, explainer, title="Decision Trees", name=None,
**kwargs):
"""Tab showing individual decision trees
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
'Decision Trees'.
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
"""
super().__init__(explainer, title, name)
self.trees = DecisionTreesComposite(explainer, **kwargs)
self.register_components(self.trees)
def layout(self):
return html.Div([
self.trees.layout()
]) | AMLBID | /Explainer/dashboard_tabs.py | dashboard_tabs.py |
__all__ = ['BaseExplainer',
'ClassifierExplainer',
'RegressionExplainer',
'RandomForestClassifierExplainer',
'RandomForestRegressionExplainer',
'XGBClassifierExplainer',
'XGBRegressionExplainer',
'ClassifierBunch', # deprecated
'RegressionBunch', # deprecated
'RandomForestClassifierBunch', # deprecated
'RandomForestRegressionBunch', # deprecated
]
from abc import ABC
import base64
from pathlib import Path
from typing import List, Union
import numpy as np
import pandas as pd
from pandas.api.types import is_numeric_dtype, is_string_dtype
import shap
from dtreeviz.trees import ShadowDecTree, dtreeviz
from sklearn.pipeline import Pipeline
from sklearn.metrics import roc_auc_score, accuracy_score, f1_score
from sklearn.metrics import precision_score, recall_score, log_loss
from sklearn.metrics import mean_squared_error, mean_absolute_error, r2_score
from sklearn.metrics import average_precision_score
from .explainer_methods import *
from .explainer_plots import *
from .make_callables import make_callable, default_list, default_2darray
import plotly.io as pio
pio.templates.default = "none"
class BaseExplainer(ABC):
""" """
def __init__(self, model, X, y=None, permutation_metric=r2_score,
shap="guess", X_background=None, model_output="raw",
cats=None, idxs=None, index_name=None, target=None,
descriptions=None,
n_jobs=None, permutation_cv=None, na_fill=-999):
"""Defines the basic functionality that is shared by both
ClassifierExplainer and RegressionExplainer.
Args:
model: a model with a scikit-learn compatible .fit and .predict methods
X (pd.DataFrame): a pd.DataFrame with your model features
y (pd.Series): Dependent variable of your model, defaults to None
permutation_metric (function or str): is a scikit-learn compatible
metric function (or string). Defaults to r2_score
shap (str): type of shap_explainer to fit: 'tree', 'linear', 'kernel'.
Defaults to 'guess'.
X_background (pd.DataFrame): background X to be used by shap
explainers that need a background dataset (e.g. shap.KernelExplainer
or shap.TreeExplainer with boosting models and
model_output='probability').
model_output (str): model_output of shap values, either 'raw',
'logodds' or 'probability'. Defaults to 'raw' for regression and
'probability' for classification.
cats ({dict, list}): dict of features that have been
onehotencoded. e.g. cats={'Sex':['Sex_male', 'Sex_female']}.
If all encoded columns are underscore-seperated (as above), can simply
pass a list of prefixes: cats=['Sex']. Allows to
group onehot encoded categorical variables together in
various plots. Defaults to None.
idxs (pd.Series): list of row identifiers. Can be names, id's, etc.
Defaults to X.index.
index_name (str): identifier for row indexes. e.g. index_name='Passenger'.
Defaults to X.index.name or idxs.name.
target: name of the predicted target, e.g. "Survival",
"Ticket price", etc. Defaults to y.name.
n_jobs (int): for jobs that can be parallelized using joblib,
how many processes to split the job in. For now only used
for calculating permutation importances. Defaults to None.
permutation_cv (int): If not None then permutation importances
will get calculated using cross validation across X.
This is for calculating permutation importances against
X_train. Defaults to None
na_fill (int): The filler used for missing values, defaults to -999.
"""
self._params_dict = dict(
shap=shap, model_output=model_output, cats=cats,
descriptions=descriptions, target=target, n_jobs=n_jobs,
permutation_cv=n_jobs, na_fill=na_fill)
if isinstance(model, Pipeline):
self.X, self.model = split_pipeline(model, X)
self.X_background, _ = split_pipeline(model, X_background, verbose=0)
else:
self.X, self.X_background = X, X_background
self.model = model
if not all([is_numeric_dtype(X[col]) for col in X.columns]):
self.cats_only = True
self.cats = [col for col in X.columns if not is_numeric_dtype(X[col])]
self.cats_dict = {col:self.X[col].unique().tolist() for col in self.cats}
print("Warning: detected non-numeric columns in X! "
f"Autodetecting the following categorical columns: {self.cats}. \n"
"Setting self.cats_only=True, which means that passing cats=False "
"to explainer methods will not work, and shap interaction values "
"will not work... ExplainerDashboard will disable these features "
" by default.", flush=True)
else:
self.cats_only = False
self.cats, self.cats_dict = parse_cats(self.X, cats)
if y is not None:
self.y = pd.Series(y)
self.y_missing = False
else:
self.y = pd.Series(np.full(len(X), np.nan))
self.y_missing = True
if self.y.name is None: self.y.name = 'Target'
self.metric = permutation_metric
if shap == "guess":
shap_guess = guess_shap(self.model)
if shap_guess is not None:
model_str = str(type(self.model))\
.replace("'", "").replace("<", "").replace(">", "")\
.split(".")[-1]
print(f"Note: shap=='guess' so guessing for {model_str}"
f" shap='{shap_guess}'...")
self.shap = shap_guess
else:
raise ValueError(
"Parameter shap='gues'', but failed to to guess the type of "
"shap explainer to use. "
"Please explicitly pass a `shap` parameter to the explainer, "
"e.g. shap='tree', shap='linear', etc.")
else:
assert shap in ['tree', 'linear', 'deep', 'kernel'], \
"Only shap='guess', 'tree', 'linear', 'deep', or ' kernel' allowed."
self.shap = shap
self.model_output = model_output
if idxs is not None:
assert len(idxs) == len(self.X) == len(self.y), \
("idxs should be same length as X but is not: "
f"len(idxs)={len(idxs)} but len(X)={len(self.X)}!")
self.idxs = pd.Index(idxs, dtype=str)
else:
self.idxs = X.index.astype(str)
self.X.index = self.idxs
self.y.index = self.idxs
if index_name is None:
if self.idxs.name is not None:
self.index_name = self.idxs.name.capitalize()
else:
self.index_name = "Index"
else:
self.index_name = index_name.capitalize()
self.descriptions = {} if descriptions is None else descriptions
self.target = target if target is not None else self.y.name
self.n_jobs = n_jobs
self.permutation_cv = permutation_cv
self.na_fill = na_fill
self.columns = self.X.columns.tolist()
self.pos_label = None
self.units = ""
self.is_classifier = False
self.is_regression = False
self.interactions_should_work = True
@classmethod
def from_file(cls, filepath):
"""Load an Explainer from file. Depending on the suffix of the filepath
will either load with pickle ('.pkl'), dill ('.dill') or joblib ('joblib').
If no suffix given, will try with joblib.
Args:
filepath {str, Path} the location of the stored Explainer
returns:
Explainer object
"""
filepath = Path(filepath)
if str(filepath).endswith(".pkl") or str(filepath).endswith(".pickle"):
import pickle
return pickle.load(open(filepath, "rb"))
elif str(filepath).endswith(".dill"):
import dill
return dill.load(open(filepath, "rb"))
else:
if not filepath.exists():
if (filepath.parent / (filepath.name + ".joblib")).exists():
filepath = filepath.parent / (filepath.name + ".joblib")
else:
raise ValueError(f"Cannot find file: {str(filepath)}")
import joblib
return joblib.load(filepath)
def dump(self, filepath):
"""
Dump the current Explainer to file. Depending on the suffix of the filepath
will either dump with pickle ('.pkl'), dill ('.dill') or joblib ('joblib').
If no suffix given, will dump with joblib and add '.joblib'
Args:
filepath (str, Path): filepath where to save the Explainer.
"""
filepath = Path(filepath)
if str(filepath).endswith(".pkl") or str(filepath).endswith(".pickle"):
import pickle
pickle.dump(self, open(str(filepath), "wb"))
elif str(filepath).endswith(".dill"):
import dill
dill.dump(self, open(str(filepath), "wb"))
elif str(filepath).endswith(".joblib"):
import joblib
joblib.dump(self, filepath)
else:
filepath = Path(filepath)
filepath = filepath.parent / (filepath.name + ".joblib")
import joblib
joblib.dump(self, filepath)
def to_yaml(self, filepath=None, return_dict=False,
modelfile="model.pkl",
datafile="data.csv",
index_col=None,
target_col=None,
explainerfile="explainer.joblib",
dashboard_yaml="dashboard.yaml"):
"""Returns a yaml configuration for the current Explainer
that can be used by the explainerdashboard CLI. Recommended filename
is `explainer.yaml`.
Args:
filepath ({str, Path}, optional): Filepath to dump yaml. If None
returns the yaml as a string. Defaults to None.
return_dict (bool, optional): instead of yaml return dict with config.
modelfile (str, optional): filename of model dump. Defaults to
`model.pkl`
datafile (str, optional): filename of datafile. Defaults to
`data.csv`.
index_col (str, optional): column to be used for idxs. Defaults to
self.idxs.name.
target_col (str, optional): column to be used for to split X and y
from datafile. Defaults to self.target.
explainerfile (str, optional): filename of explainer dump. Defaults
to `explainer.joblib`.
dashboard_yaml (str, optional): filename of the dashboard.yaml
configuration file. This will be used to determine which
properties to calculate before storing to disk.
Defaults to `dashboard.yaml`.
"""
import oyaml as yaml
yaml_config = dict(
explainer=dict(
modelfile=modelfile,
datafile=datafile,
explainerfile=explainerfile,
data_target=self.target,
data_index=self.idxs.name,
explainer_type="classifier" if self.is_classifier else "regression",
dashboard_yaml=dashboard_yaml,
params=self._params_dict))
if return_dict:
return yaml_config
if filepath is not None:
yaml.dump(yaml_config, open(filepath, "w"))
return
return yaml.dump(yaml_config)
def __len__(self):
return len(self.X)
def __contains__(self, index):
if self.get_int_idx(index) is not None:
return True
return False
def check_cats(self, col1, col2=None):
"""check whether should use cats=True based on col1 (and col2)
Args:
col1: First column
col2: Second column (Default value = None)
Returns:
Boolean whether cats should be True
"""
if self.cats_only:
return True
if col2 is None:
if col1 in self.columns:
return False
elif col1 in self.columns_cats:
return True
raise ValueError(f"Can't find {col1}.")
if col1 not in self.columns and col1 not in self.columns_cats:
raise ValueError(f"Can't find {col1}.")
if col2 not in self.columns and col2 not in self.columns_cats:
raise ValueError(f"Can't find {col2}.")
if col1 in self.columns and col2 in self.columns:
return False
if col1 in self.columns_cats and col2 in self.columns_cats:
return True
if col1 in self.columns_cats and not col2 in self.columns_cats:
raise ValueError(
f"{col1} is categorical but {col2} is not in columns_cats")
if col2 in self.columns_cats and not col1 in self.columns_cats:
raise ValueError(
f"{col2} is categorical but {col1} is not in columns_cats")
@property
def shap_explainer(self):
""" """
if not hasattr(self, '_shap_explainer'):
X_str = ", X_background" if self.X_background is not None else 'X'
NoX_str = ", X_background" if self.X_background is not None else ''
if self.shap == 'tree':
print("Generating self.shap_explainer = "
f"shap.TreeExplainer(model{NoX_str})")
self._shap_explainer = shap.TreeExplainer(self.model)
elif self.shap=='linear':
if self.X_background is None:
print(
"Warning: shap values for shap.LinearExplainer get "
"calculated against X_background, but paramater "
"X_background=None, so using X instead")
print(f"Generating self.shap_explainer = shap.LinearExplainer(model{X_str})...")
self._shap_explainer = shap.LinearExplainer(self.model,
self.X_background if self.X_background is not None else self.X)
elif self.shap=='deep':
print(f"Generating self.shap_explainer = "
f"shap.DeepExplainer(model{NoX_str})")
self._shap_explainer = shap.DeepExplainer(self.model)
elif self.shap=='kernel':
if self.X_background is None:
print(
"Warning: shap values for shap.LinearExplainer get "
"calculated against X_background, but paramater "
"X_background=None, so using X instead")
print("Generating self.shap_explainer = "
f"shap.KernelExplainer(model, {X_str})...")
self._shap_explainer = shap.KernelExplainer(self.model,
self.X_background if self.X_background is not None else self.X)
return self._shap_explainer
def get_int_idx(self, index):
"""Turn str index into an int index
Args:
index(str or int):
Returns:
int index
"""
if isinstance(index, int):
if index >= 0 and index < len(self):
return index
elif isinstance(index, str):
if self.idxs is not None and index in self.idxs:
return self.idxs.get_loc(index)
return None
def get_index(self, index):
"""Turn int index into a str index
Args:
index(str or int):
Returns:
str index
"""
if isinstance(index, int) and index >= 0 and index < len(self):
return self.idxs[index]
elif isinstance(index, str) and index in self.idxs:
return index
return None
def random_index(self, y_min=None, y_max=None, pred_min=None, pred_max=None,
return_str=False, **kwargs):
"""random index following constraints
Args:
y_min: (Default value = None)
y_max: (Default value = None)
pred_min: (Default value = None)
pred_max: (Default value = None)
return_str: (Default value = False)
**kwargs:
Returns:
if y_values is given select an index for which y in y_values
if return_str return str index from self.idxs
"""
if pred_min is None:
pred_min = self.preds.min()
if pred_max is None:
pred_max = self.preds.max()
if not self.y_missing:
if y_min is None: y_min = self.y.min()
if y_max is None: y_max = self.y.max()
potential_idxs = self.y[(self.y>=y_min) &
(self.y <= y_max) &
(self.preds>=pred_min) &
(self.preds <= pred_max)].index
else:
potential_idxs = self.y[(self.preds>=pred_min) &
(self.preds <= pred_max)].index
if len(potential_idxs) > 0:
idx = np.random.choice(potential_idxs)
else:
return None
if return_str:
return idx
return idxs.get_loc(idx)
@property
def preds(self):
"""returns model model predictions"""
if not hasattr(self, '_preds'):
print("Calculating predictions...", flush=True)
self._preds = self.model.predict(self.X).astype(np.float64)
return self._preds
@property
def pred_percentiles(self):
"""returns percentile rank of model predictions"""
if not hasattr(self, '_pred_percentiles'):
print("Calculating prediction percentiles...", flush=True)
self._pred_percentiles = (pd.Series(self.preds)
.rank(method='min')
.divide(len(self.preds))
.values)
return make_callable(self._pred_percentiles)
def columns_ranked_by_shap(self, cats=False, pos_label=None):
"""returns the columns of X, ranked by mean abs shap value
Args:
cats: Group categorical together (Default value = False)
pos_label: (Default value = None)
Returns:
list of columns
"""
if cats or self.cats_only:
return self.mean_abs_shap_cats(pos_label).Feature.tolist()
else:
return self.mean_abs_shap(pos_label).Feature.tolist()
def n_features(self, cats=False):
"""number of features with cats=True or cats=False
Args:
cats: (Default value = False)
Returns:
int, number of features
"""
if cats or self.cats_only:
return len(self.columns_cats)
else:
return len(self.columns)
def equivalent_col(self, col):
"""Find equivalent col in columns_cats or columns
if col in self.columns, return equivalent col in self.columns_cats,
e.g. equivalent_col('Gender_Male') -> 'Gender'
if col in self.columns_cats, return first one hot encoded col,
e.g. equivalent_col('Gender') -> 'Gender_Male'
(useful for switching between cats=True and cats=False, while
maintaining column selection)
Args:
col: col to get equivalent col for
Returns:
col
"""
if self.cats_only:
return col
if col in self.cats:
# first onehot-encoded columns
return self.cats_dict[col][0]
elif col in self.columns:
# the cat that the col belongs to
return [k for k, v in self.cats_dict.items() if col in v][0]
return None
def get_row_from_input(self, inputs:List, ranked_by_shap=False):
"""returns a single row pd.DataFrame from a given list of *inputs"""
if len(inputs)==1 and isinstance(inputs[0], list):
inputs = inputs[0]
elif len(inputs)==1 and isinstance(inputs[0], tuple):
inputs = list(inputs[0])
else:
inputs = list(inputs)
if len(inputs) == len(self.columns_cats):
cols = self.columns_ranked_by_shap(cats=True) if ranked_by_shap else self.columns_cats
df = pd.DataFrame(dict(zip(cols, inputs)), index=[0]).fillna(self.na_fill)
return df[self.columns_cats]
elif len(inputs) == len(self.columns):
cols = self.columns_ranked_by_shap() if ranked_by_shap else self.columns
df = pd.DataFrame(dict(zip(cols, inputs)), index=[0]).fillna(self.na_fill)
return df[self.columns]
else:
raise ValueError(f"len inputs {len(inputs)} should be the same length as either "
f"explainer.columns_cats ({len(self.columns_cats)}) or "
f"explainer.columns ({len(self.columns)})!")
def description(self, col):
"""returns the written out description of what feature col means
Args:
col(str): col to get description for
Returns:
str, description
"""
if col in self.descriptions.keys():
return self.descriptions[col]
elif self.equivalent_col(col) in self.descriptions.keys():
return self.descriptions[self.equivalent_col(col)]
return ""
def description_list(self, cols):
"""returns a list of descriptions of a list of cols
Args:
cols(list): cols to be converted to descriptions
Returns:
list of descriptions
"""
return [self.description(col) for col in cols]
def get_col(self, col):
"""return pd.Series with values of col
For categorical feature reverse engineers the onehotencoding.
Args:
col: column tof values to be returned
Returns:
pd.Series with values of col
"""
assert col in self.columns or col in self.cats, \
f"{col} not in columns!"
if col in self.X.columns:
return self.X[col]
elif col in self.cats:
return pd.Series(retrieve_onehot_value(
self.X, col, self.cats_dict[col]), name=col)
def get_col_value_plus_prediction(self, col, index=None, X_row=None, pos_label=None):
"""return value of col and prediction for either index or X_row
Args:
col: feature col
index (str or int, optional): index row
X_row (single row pd.DataFrame, optional): single row of features
Returns:
tupe(value of col, prediction for index)
"""
assert (col in self.X.columns) or (col in self.cats),\
f"{col} not in columns of dataset"
if index is not None:
assert index in self, f"index {index} not found"
idx = self.get_int_idx(index)
if col in self.X.columns:
col_value = self.X[col].iloc[idx]
elif col in self.cats:
col_value = retrieve_onehot_value(self.X, col, self.cats_dict[col])[idx]
if self.is_classifier:
if pos_label is None:
pos_label = self.pos_label
prediction = self.pred_probas(pos_label)[idx]
if self.model_output == 'probability':
prediction = 100*prediction
elif self.is_regression:
prediction = self.preds[idx]
return col_value, prediction
elif X_row is not None:
assert X_row.shape[0] == 1, "X_Row should be single row dataframe!"
if ((len(X_row.columns) == len(self.X_cats.columns)) and
(X_row.columns == self.X_cats.columns).all()):
X_row = X_cats_to_X(X_row, self.cats_dict, self.X.columns)
else:
assert (X_row.columns == self.X.columns).all(), \
"X_row should have the same columns as self.X or self.X_cats!"
if col in X_row.columns:
col_value = X_row[col].item()
elif col in self.cats:
col_value = retrieve_onehot_value(X_row, col, self.cats_dict[col]).item()
if self.is_classifier:
if pos_label is None:
pos_label = self.pos_label
prediction = self.model.predict_proba(X_row)[0][pos_label]
if self.model_output == 'probability':
prediction = 100*prediction
elif self.is_regression:
prediction = self.model.predict(X_row)[0]
return col_value, prediction
else:
raise ValueError("You need to pass either index or X_row!")
@property
def permutation_importances(self):
"""Permutation importances """
if not hasattr(self, '_perm_imps'):
print("Calculating importances...", flush=True)
self._perm_imps = cv_permutation_importances(
self.model, self.X, self.y, self.metric,
cv=self.permutation_cv,
n_jobs=self.n_jobs,
needs_proba=self.is_classifier)
return make_callable(self._perm_imps)
@property
def permutation_importances_cats(self):
"""permutation importances with categoricals grouped"""
if not hasattr(self, '_perm_imps_cats'):
if self.cats_only:
self._perm_imps_cats = cv_permutation_importances(
self.model, self.X, self.y, self.metric,
cv=self.permutation_cv,
n_jobs=self.n_jobs,
needs_proba=self.is_classifier)
else:
self._perm_imps_cats = cv_permutation_importances(
self.model, self.X, self.y, self.metric,
cats_dict=self.cats_dict,
cv=self.permutation_cv,
n_jobs=self.n_jobs,
needs_proba=self.is_classifier)
return make_callable(self._perm_imps_cats)
@property
def X_cats(self):
"""X with categorical variables grouped together"""
if not hasattr(self, '_X_cats'):
if self.cats_only:
self._X_cats = self.X
else:
self._X_cats = merge_categorical_columns(self.X, self.cats_dict)
return self._X_cats
@property
def columns_cats(self):
"""columns of X with categorical features grouped"""
if not hasattr(self, '_columns_cats'):
self._columns_cats = self.X_cats.columns.tolist()
return self._columns_cats
@property
def shap_base_value(self):
"""the intercept for the shap values.
(i.e. 'what would the prediction be if we knew none of the features?')
"""
if not hasattr(self, '_shap_base_value'):
# CatBoost needs shap values calculated before expected value
if not hasattr(self, "_shap_values"):
_ = self.shap_values
self._shap_base_value = self.shap_explainer.expected_value
if isinstance(self._shap_base_value, np.ndarray):
# shap library now returns an array instead of float
self._shap_base_value = self._shap_base_value.item()
return make_callable(self._shap_base_value)
@property
def shap_values(self):
"""SHAP values calculated using the shap library"""
if not hasattr(self, '_shap_values'):
print("Calculating shap values...", flush=True)
self._shap_values = self.shap_explainer.shap_values(self.X)
return make_callable(self._shap_values)
@property
def shap_values_cats(self):
"""SHAP values when categorical features have been grouped"""
if not hasattr(self, '_shap_values_cats'):
if self.cats_only:
self._shap_values_cats = self.shap_explainer.shap_values(self.X)
else:
self._shap_values_cats = merge_categorical_shap_values(
self.X, self.shap_values, self.cats_dict)
return make_callable(self._shap_values_cats)
@property
def shap_interaction_values(self):
"""SHAP interaction values calculated using shap library"""
assert self.shap != 'linear', \
"Unfortunately shap.LinearExplainer does not provide " \
"shap interaction values! So no interactions tab!"
if not hasattr(self, '_shap_interaction_values'):
print("Calculating shap interaction values...", flush=True)
if self.shap == 'tree':
print("Reminder: TreeShap computational complexity is O(TLD^2), "
"where T is the number of trees, L is the maximum number of"
" leaves in any tree and D the maximal depth of any tree. So "
"reducing these will speed up the calculation.",
flush=True)
self._shap_interaction_values = \
self.shap_explainer.shap_interaction_values(self.X)
return make_callable(self._shap_interaction_values)
@property
def shap_interaction_values_cats(self):
"""SHAP interaction values with categorical features grouped"""
if not hasattr(self, '_shap_interaction_values_cats'):
self._shap_interaction_values_cats = \
merge_categorical_shap_interaction_values(
self.shap_interaction_values, self.X, self.X_cats, self.cats_dict)
return make_callable(self._shap_interaction_values_cats)
@property
def mean_abs_shap(self):
"""Mean absolute SHAP values per feature."""
if not hasattr(self, '_mean_abs_shap'):
self._mean_abs_shap = mean_absolute_shap_values(
self.columns, self.shap_values)
return make_callable(self._mean_abs_shap)
@property
def mean_abs_shap_cats(self):
"""Mean absolute SHAP values with categoricals grouped."""
if not hasattr(self, '_mean_abs_shap_cats'):
self._mean_abs_shap_cats = mean_absolute_shap_values(
self.columns_cats, self.shap_values_cats)
return make_callable(self._mean_abs_shap_cats)
def calculate_properties(self, include_interactions=True):
"""Explicitely calculates all lazily calculated properties.
Useful so that properties are not calculate multiple times in
parallel when starting a dashboard.
Args:
include_interactions(bool, optional, optional): shap interaction values can take a long
time to compute for larger datasets with more features. Therefore you
can choose not to calculate these, defaults to True
Returns:
"""
_ = (self.preds, self.pred_percentiles,
self.shap_base_value, self.shap_values,
self.mean_abs_shap)
if not self.y_missing:
_ = self.permutation_importances
if self.cats is not None:
_ = (self.mean_abs_shap_cats, self.X_cats,
self.shap_values_cats)
if self.interactions_should_work and include_interactions:
_ = self.shap_interaction_values
if self.cats is not None:
_ = self.shap_interaction_values_cats
def metrics(self, *args, **kwargs):
"""returns a dict of metrics.
Implemented by either ClassifierExplainer or RegressionExplainer
"""
return {}
def mean_abs_shap_df(self, topx=None, cutoff=None, cats=False, pos_label=None):
"""sorted dataframe with mean_abs_shap
returns a pd.DataFrame with the mean absolute shap values per features,
sorted rom highest to lowest.
Args:
topx(int, optional, optional): Only return topx most importance features, defaults to None
cutoff(float, optional, optional): Only return features with mean abs shap of at least cutoff, defaults to None
cats(bool, optional, optional): group categorical variables, defaults to False
pos_label: (Default value = None)
Returns:
pd.DataFrame: shap_df
"""
if cats or self.cats_only:
shap_df = self.mean_abs_shap_cats(pos_label)
else:
shap_df = self.mean_abs_shap(pos_label)
if topx is None: topx = len(shap_df)
if cutoff is None: cutoff = shap_df['MEAN_ABS_SHAP'].min()
return (shap_df[shap_df['MEAN_ABS_SHAP'] >= cutoff]
.sort_values('MEAN_ABS_SHAP', ascending=False).head(topx))
def shap_top_interactions(self, col, topx=None, cats=False, pos_label=None):
"""returns the features that interact with feature col in descending order.
if shap interaction values have already been calculated, use those.
Otherwise use shap approximate_interactions or simply mean abs shap.
Args:
col(str): feature for which you want to get the interactions
topx(int, optional, optional): Only return topx features, defaults to None
cats(bool, optional, optional): Group categorical features, defaults to False
pos_label: (Default value = None)
Returns:
list: top_interactions
"""
if cats or self.cats_only:
if hasattr(self, '_shap_interaction_values'):
col_idx = self.X_cats.columns.get_loc(col)
top_interactions = self.X_cats.columns[
np.argsort(
-np.abs(self.shap_interaction_values_cats(
pos_label)[:, col_idx, :]).mean(0))].tolist()
else:
top_interactions = self.mean_abs_shap_cats(pos_label)\
.Feature.values.tolist()
top_interactions.insert(0, top_interactions.pop(
top_interactions.index(col))) #put col first
if topx is None: topx = len(top_interactions)
return top_interactions[:topx]
else:
if hasattr(self, '_shap_interaction_values'):
col_idx = self.X.columns.get_loc(col)
top_interactions = self.X.columns[np.argsort(-np.abs(
self.shap_interaction_values(
pos_label)[:, col_idx, :]).mean(0))].tolist()
else:
if hasattr(shap, "utils"):
interaction_idxs = shap.utils.approximate_interactions(
col, self.shap_values(pos_label), self.X)
elif hasattr(shap, "common"):
# shap < 0.35 has approximate interactions in common
interaction_idxs = shap.common.approximate_interactions(
col, self.shap_values(pos_label), self.X)
top_interactions = self.X.columns[interaction_idxs].tolist()
#put col first
top_interactions.insert(0, top_interactions.pop(-1))
if topx is None: topx = len(top_interactions)
return top_interactions[:topx]
def shap_interaction_values_by_col(self, col, cats=False, pos_label=None):
"""returns the shap interaction values[np.array(N,N)] for feature col
Args:
col(str): features for which you'd like to get the interaction value
cats(bool, optional, optional): group categorical, defaults to False
pos_label: (Default value = None)
Returns:
np.array(N,N): shap_interaction_values
"""
if cats or self.cats_only:
return self.shap_interaction_values_cats(pos_label)[:,
self.X_cats.columns.get_loc(col), :]
else:
return self.shap_interaction_values(pos_label)[:,
self.X.columns.get_loc(col), :]
def permutation_importances_df(self, topx=None, cutoff=None, cats=False,
pos_label=None):
"""dataframe with features ordered by permutation importance.
For more about permutation importances.
see https://explained.ai/rf-importance/index.html
Args:
topx(int, optional, optional): only return topx most important
features, defaults to None
cutoff(float, optional, optional): only return features with importance
of at least cutoff, defaults to None
cats(bool, optional, optional): Group categoricals, defaults to False
pos_label: (Default value = None)
Returns:
pd.DataFrame: importance_df
"""
if cats or self.cats_only:
importance_df = self.permutation_importances_cats(pos_label)
else:
importance_df = self.permutation_importances(pos_label)
if topx is None: topx = len(importance_df)
if cutoff is None: cutoff = importance_df.Importance.min()
return importance_df[importance_df.Importance >= cutoff].head(topx)
def importances_df(self, kind="shap", topx=None, cutoff=None, cats=False,
pos_label=None):
"""wrapper function for mean_abs_shap_df() and permutation_importance_df()
Args:
kind(str): 'shap' or 'permutations' (Default value = "shap")
topx: only display topx highest features (Default value = None)
cutoff: only display features above cutoff (Default value = None)
cats: Group categoricals (Default value = False)
pos_label: Positive class (Default value = None)
Returns:
pd.DataFrame
"""
assert kind=='shap' or kind=='permutation', \
"kind should either be 'shap' or 'permutation'!"
if kind=='permutation':
return self.permutation_importances_df(topx, cutoff, cats, pos_label)
elif kind=='shap':
return self.mean_abs_shap_df(topx, cutoff, cats, pos_label)
def contrib_df(self, index=None, X_row=None, cats=True, topx=None, cutoff=None, sort='abs',
pos_label=None):
"""shap value contributions to the prediction for index.
Used as input for the plot_contributions() method.
Args:
index(int or str): index for which to calculate contributions
X_row (pd.DataFrame, single row): single row of feature for which
to calculate contrib_df. Can us this instead of index
cats(bool, optional, optional): Group categoricals, defaults to True
topx(int, optional, optional): Only return topx features, remainder
called REST, defaults to None
cutoff(float, optional, optional): only return features with at least
cutoff contributions, defaults to None
sort({'abs', 'high-to-low', 'low-to-high', 'importance'}, optional): sort by
absolute shap value, or from high to low, low to high, or
ordered by the global shap importances.
Defaults to 'abs'.
pos_label: (Default value = None)
Returns:
pd.DataFrame: contrib_df
"""
if pos_label is None:
pos_label = self.pos_label
if sort =='importance':
if cutoff is None:
cols = self.columns_ranked_by_shap(cats)
else:
cols = self.mean_abs_shap_df(cats=cats).query(f"MEAN_ABS_SHAP > {cutoff}").Feature.tolist()
if topx is not None:
cols = cols[:topx]
else:
cols = None
if X_row is not None:
if ((len(X_row.columns) == len(self.X_cats.columns)) and
(X_row.columns == self.X_cats.columns).all()):
if cats or self.cats_only:
X_row_cats = X_row
if not self.cats_only:
X_row = X_cats_to_X(X_row, self.cats_dict, self.X.columns)
else:
assert (X_row.columns == self.X.columns).all(), \
"X_row should have the same columns as self.X or self.X_cats!"
X_row_cats = merge_categorical_columns(X_row, self.cats_dict)
shap_values = self.shap_explainer.shap_values(X_row)
if self.is_classifier:
if not isinstance(shap_values, list) and len(self.labels)==2:
shap_values = [-shap_values, shap_values]
shap_values = shap_values[self.get_pos_label_index(pos_label)]
if cats:
if not self.cats_only:
shap_values = merge_categorical_shap_values(X_row, shap_values, self.cats_dict)
return get_contrib_df(self.shap_base_value(pos_label), shap_values[0],
X_row_cats, topx, cutoff, sort, cols)
else:
return get_contrib_df(self.shap_base_value(pos_label), shap_values[0],
X_row, topx, cutoff, sort, cols)
elif index is not None:
idx = self.get_int_idx(index)
if cats:
return get_contrib_df(self.shap_base_value(pos_label),
self.shap_values_cats(pos_label)[idx],
self.X_cats.iloc[[idx]], topx, cutoff, sort, cols)
else:
return get_contrib_df(self.shap_base_value(pos_label),
self.shap_values(pos_label)[idx],
self.X.iloc[[idx]], topx, cutoff, sort, cols)
else:
raise ValueError("Either index or X_row should be passed!")
def contrib_summary_df(self, index=None, X_row=None, cats=True, topx=None, cutoff=None,
round=2, sort='abs', pos_label=None):
"""Takes a contrib_df, and formats it to a more human readable format
Args:
index: index to show contrib_summary_df for
X_row (pd.DataFrame, single row): single row of feature for which
to calculate contrib_df. Can us this instead of index
cats: Group categoricals (Default value = True)
topx: Only show topx highest features(Default value = None)
cutoff: Only show features above cutoff (Default value = None)
round: round figures (Default value = 2)
sort({'abs', 'high-to-low', 'low-to-high', 'importance'}, optional): sort by
absolute shap value, or from high to low, or low to high, or
ordered by the global shap importances.
Defaults to 'abs'.
pos_label: Positive class (Default value = None)
Returns:
pd.DataFrame
"""
idx = self.get_int_idx(index) # if passed str convert to int index
return get_contrib_summary_df(
self.contrib_df(idx, X_row, cats, topx, cutoff, sort, pos_label),
model_output=self.model_output, round=round, units=self.units, na_fill=self.na_fill)
def interactions_df(self, col, cats=False, topx=None, cutoff=None,
pos_label=None):
"""dataframe of mean absolute shap interaction values for col
Args:
col: Feature to get interactions_df for
cats: Group categoricals (Default value = False)
topx: Only display topx most important features (Default value = None)
cutoff: Only display features with mean abs shap of at least cutoff (Default value = None)
pos_label: Positive class (Default value = None)
Returns:
pd.DataFrame
"""
importance_df = mean_absolute_shap_values(
self.columns_cats if (cats or self.cats_only) else self.columns,
self.shap_interaction_values_by_col(col, cats, pos_label))
if topx is None: topx = len(importance_df)
if cutoff is None: cutoff = importance_df.MEAN_ABS_SHAP.min()
return importance_df[importance_df.MEAN_ABS_SHAP >= cutoff].head(topx)
def formatted_contrib_df(self, index, round=None, lang='en', pos_label=None):
"""contrib_df formatted in a particular idiosyncratic way.
Additional language option for output in Dutch (lang='nl')
Args:
index(str or int): index to return contrib_df for
round(int, optional, optional): rounding of continuous features, defaults to 2
lang(str, optional, optional): language to name the columns, defaults to 'en'
pos_label: (Default value = None)
Returns:
pd.DataFrame: formatted_contrib_df
"""
cdf = self.contrib_df(index, cats=True, pos_label=pos_label).copy()
cdf.reset_index(inplace=True)
cdf.loc[cdf.col=='base_value', 'value'] = np.nan
cdf['row_id'] = self.get_int_idx(index)
cdf['name_id'] = index
cdf['cat_value'] = np.where(cdf.col.isin(self.cats), cdf.value, np.nan)
cdf['cont_value'] = np.where(cdf.col.isin(self.cats), np.nan, cdf.value)
if round is not None:
rounded_cont = np.round(cdf['cont_value'].values.astype(float), round)
cdf['value'] = np.where(cdf.col.isin(self.cats), cdf.cat_value, rounded_cont)
cdf['type'] = np.where(cdf.col.isin(self.cats), 'cat', 'cont')
cdf['abs_contribution'] = np.abs(cdf.contribution)
cdf = cdf[['row_id', 'name_id', 'contribution', 'abs_contribution',
'col', 'value', 'cat_value', 'cont_value', 'type', 'index']]
if lang == 'nl':
cdf.columns = ['row_id', 'name_id', 'SHAP', 'ABS_SHAP', 'Variabele', 'Waarde',
'Cat_Waarde', 'Cont_Waarde', 'Waarde_Type', 'Variabele_Volgorde']
return cdf
cdf.columns = ['row_id', 'name_id', 'SHAP', 'ABS_SHAP', 'Feature', 'Value',
'Cat_Value', 'Cont_Value', 'Value_Type', 'Feature_Order']
return cdf
def pdp_df(self, col, index=None, X_row=None, drop_na=True,
sample=500, num_grid_points=20, pos_label=None):
assert col in self.X.columns or col in self.cats, \
f"{col} not in columns of dataset"
if col in self.cats and not self.cats_only:
features = self.cats_dict[col]
else:
features = col
if pos_label is None:
pos_label = self.pos_label
if index is not None:
index = self.get_index(index)
if isinstance(features, str) and drop_na: # regular col, not onehotencoded
sample_size=min(sample, len(self.X[(self.X[features] != self.na_fill)])-1)
sampleX = pd.concat([
self.X[self.X.index==index],
self.X[(self.X.index != index) & (self.X[features] != self.na_fill)]\
.sample(sample_size)],
ignore_index=True, axis=0)
else:
sample_size = min(sample, len(self.X)-1)
sampleX = pd.concat([
self.X[self.X.index==index],
self.X[(self.X.index!=index)].sample(sample_size)],
ignore_index=True, axis=0)
elif X_row is not None:
if ((len(X_row.columns) == len(self.X_cats.columns)) and
(X_row.columns == self.X_cats.columns).all()):
X_row = X_cats_to_X(X_row, self.cats_dict, self.X.columns)
else:
assert (X_row.columns == self.X.columns).all(), \
"X_row should have the same columns as self.X or self.X_cats!"
if isinstance(features, str) and drop_na: # regular col, not onehotencoded
sample_size=min(sample, len(self.X[(self.X[features] != self.na_fill)])-1)
sampleX = pd.concat([
X_row,
self.X[(self.X[features] != self.na_fill)]\
.sample(sample_size)],
ignore_index=True, axis=0)
else:
sample_size = min(sample, len(self.X)-1)
sampleX = pd.concat([
X_row,
self.X.sample(sample_size)],
ignore_index=True, axis=0)
else:
if isinstance(features, str) and drop_na: # regular col, not onehotencoded
sample_size=min(sample, len(self.X[(self.X[features] != self.na_fill)])-1)
sampleX = self.X[(self.X[features] != self.na_fill)]\
.sample(sample_size)
else:
sampleX = self.X.sample(min(sample, len(self.X)))
# if only a single value (i.e. not onehot encoded, take that value
# instead of list):
pdp_df = get_pdp_df(
model=self.model, X_sample=sampleX,
feature=features,
n_grid_points=num_grid_points, pos_label=pos_label)
if all([str(c).startswith(col+"_") for c in pdp_df.columns]):
pdp_df.columns = [str(c)[len(col)+1:] for c in pdp_df.columns]
if self.is_classifier:
pdp_df = pdp_df.multiply(100)
return pdp_df
def get_dfs(self, cats=True, round=None, lang='en', pos_label=None):
"""return three summary dataframes for storing main results
Returns three pd.DataFrames. The first with id, prediction, actual and
feature values, the second with only id and shap values. The third
is similar to contrib_df for every id.
These can then be used to build your own custom dashboard on these data,
for example using PowerBI.
Args:
cats(bool, optional, optional): group categorical variables, defaults to True
round(int, optional, optional): how to round shap values (Default value = None)
lang(str, optional, optional): language to format dfs in. Defaults to 'en', 'nl' also available
pos_label: (Default value = None)
Returns:
pd.DataFrame, pd.DataFrame, pd.DataFrame: cols_df, shap_df, contribs_df
"""
if cats or self.cats_only:
cols_df = self.X_cats.copy()
shap_df = pd.DataFrame(self.shap_values_cats(pos_label), columns = self.X_cats.columns)
else:
cols_df = self.X.copy()
shap_df = pd.DataFrame(self.shap_values(pos_label), columns = self.X.columns)
actual_str = 'Uitkomst' if lang == 'nl' else 'Actual'
prediction_str = 'Voorspelling' if lang == 'nl' else 'Prediction'
cols_df.insert(0, actual_str, self.y )
if self.is_classifier:
cols_df.insert(0, prediction_str, self.pred_probas)
else:
cols_df.insert(0, prediction_str, self.preds)
cols_df.insert(0, 'name_id', self.idxs)
cols_df.insert(0, 'row_id', range(len(self)))
shap_df.insert(0, 'SHAP_base', np.repeat(self.shap_base_value, len(self)))
shap_df.insert(0, 'name_id', self.idxs)
shap_df.insert(0, 'row_id', range(len(self)))
contribs_df = None
for idx in range(len(self)):
fcdf = self.formatted_contrib_df(idx, round=round, lang=lang)
if contribs_df is None: contribs_df = fcdf
else: contribs_df = pd.concat([contribs_df, fcdf])
return cols_df, shap_df, contribs_df
def to_sql(self, conn, schema, name, if_exists='replace',
cats=True, round=None, lang='en', pos_label=None):
"""Writes three dataframes generated by .get_dfs() to a sql server.
Tables will be called name_COLS and name_SHAP and name_CONTRBIB
Args:
conn(sqlalchemy.engine.Engine or sqlite3.Connection):
database connecter acceptable for pd.to_sql
schema(str): schema to write to
name(str): name prefix of tables
cats(bool, optional, optional): group categorical variables, defaults to True
if_exists({'fail’, ‘replace’, ‘append’}, default ‘replace’, optional):
How to behave if the table already exists. (Default value = 'replace')
round(int, optional, optional): how to round shap values (Default value = None)
lang(str, optional, optional): language to format dfs in. Defaults to 'en', 'nl' also available
pos_label: (Default value = None)
Returns:
"""
cols_df, shap_df, contribs_df = self.get_dfs(cats, round, lang, pos_label)
cols_df.to_sql(con=conn, schema=schema, name=name+"_COLS",
if_exists=if_exists, index=False)
shap_df.to_sql(con=conn, schema=schema, name=name+"_SHAP",
if_exists=if_exists, index=False)
contribs_df.to_sql(con=conn, schema=schema, name=name+"_CONTRIB",
if_exists=if_exists, index=False)
def plot_importances(self, kind='shap', topx=None, cats=False, round=3, pos_label=None):
"""plot barchart of importances in descending order.
Args:
type(str, optional): shap' for mean absolute shap values, 'permutation' for
permutation importances, defaults to 'shap'
topx(int, optional, optional): Only return topx features, defaults to None
cats(bool, optional, optional): Group categoricals defaults to False
kind: (Default value = 'shap')
round: (Default value = 3)
pos_label: (Default value = None)
Returns:
plotly.fig: fig
"""
importances_df = self.importances_df(kind=kind, topx=topx, cats=cats, pos_label=pos_label)
if kind=='shap':
if self.target:
title = f"Average impact on predicted {self.target}<br>(mean absolute SHAP value)"
else:
title = 'Average impact on prediction<br>(mean absolute SHAP value)'
units = self.units
else:
title = f"Permutation Importances <br>(decrease in metric '{self.metric.__name__}'' with randomized feature)"
units = ""
if self.descriptions:
descriptions = self.description_list(importances_df.Feature)
return plotly_importances_plot(importances_df, descriptions, round=round, units=units, title=title)
else:
return plotly_importances_plot(importances_df, round=round, units=units, title=title)
def plot_interactions(self, col, cats=False, topx=None, pos_label=None):
"""plot mean absolute shap interaction value for col.
Args:
col: column for which to generate shap interaction value
cats(bool, optional, optional): Group categoricals defaults to False
topx(int, optional, optional): Only return topx features, defaults to None
pos_label: (Default value = None)
Returns:
plotly.fig: fig
"""
if col in self.cats or self.cats_only:
cats = True
interactions_df = self.interactions_df(col, cats=cats, topx=topx, pos_label=pos_label)
title = f"Average interaction shap values for {col}"
return plotly_importances_plot(interactions_df, units=self.units, title=title)
def plot_shap_contributions(self, index=None, X_row=None, cats=True, topx=None, cutoff=None,
sort='abs', orientation='vertical', higher_is_better=True,
round=2, pos_label=None):
"""plot waterfall plot of shap value contributions to the model prediction for index.
Args:
index(int or str): index for which to display prediction
X_row (pd.DataFrame single row): a single row of a features to plot
shap contributions for. Can use this instead of index for
what-if scenarios.
cats(bool, optional, optional): Group categoricals, defaults to True
topx(int, optional, optional): Only display topx features,
defaults to None
cutoff(float, optional, optional): Only display features with at least
cutoff contribution, defaults to None
sort({'abs', 'high-to-low', 'low-to-high', 'importance'}, optional):
sort by absolute shap value, or from high to low,
or low to high, or by order of shap feature importance.
Defaults to 'abs'.
orientation({'vertical', 'horizontal'}): Horizontal or vertical bar chart.
Horizontal may be better if you have lots of features.
Defaults to 'vertical'.
higher_is_better (bool): if True, up=green, down=red. If false reversed.
Defaults to True.
round(int, optional, optional): round contributions to round precision,
defaults to 2
pos_label: (Default value = None)
Returns:
plotly.Fig: fig
"""
assert orientation in ['vertical', 'horizontal']
contrib_df = self.contrib_df(self.get_int_idx(index), X_row, cats, topx, cutoff, sort, pos_label)
return plotly_contribution_plot(contrib_df, model_output=self.model_output,
orientation=orientation, round=round, higher_is_better=higher_is_better,
target=self.target, units=self.units)
def plot_shap_summary(self, index=None, topx=None, cats=False, pos_label=None):
"""Plot barchart of mean absolute shap value.
Displays all individual shap value for each feature in a horizontal
scatter chart in descending order by mean absolute shap value.
Args:
index (str or int): index to highlight
topx(int, optional): Only display topx most important features, defaults to None
cats(bool, optional): Group categoricals , defaults to False
pos_label: positive class (Default value = None)
Returns:
plotly.Fig
"""
if self.is_classifier:
if pos_label is None:
pos_label = self.pos_label
pos_label_str = self.labels[self.get_pos_label_index(pos_label)]
if self.model_output == 'probability':
if self.target:
title = f"Impact of feature on predicted probability {self.target}={pos_label_str} <br> (SHAP values)"
else:
title = f"Impact of Feature on Prediction probability <br> (SHAP values)"
elif self.model_output == 'logodds':
title = f"Impact of Feature on predicted logodds <br> (SHAP values)"
elif self.is_regression:
if self.target:
title = f"Impact of Feature on Predicted {self.target} <br> (SHAP values)"
else:
title = f"Impact of Feature on Prediction<br> (SHAP values)"
if cats or self.cats_only:
return plotly_shap_scatter_plot(
self.shap_values_cats(pos_label),
self.X_cats,
self.importances_df(kind='shap', topx=topx, cats=True, pos_label=pos_label)\
['Feature'].values.tolist(),
idxs=self.idxs.values,
highlight_index=index,
title=title,
na_fill=self.na_fill,
index_name=self.index_name)
else:
return plotly_shap_scatter_plot(
self.shap_values(pos_label),
self.X,
self.importances_df(kind='shap', topx=topx, cats=False, pos_label=pos_label)\
['Feature'].values.tolist(),
idxs=self.idxs.values,
highlight_index=index,
title=title,
na_fill=self.na_fill,
index_name=self.index_name)
def plot_shap_interaction_summary(self, col, index=None, topx=None, cats=False, pos_label=None):
"""Plot barchart of mean absolute shap interaction values
Displays all individual shap interaction values for each feature in a
horizontal scatter chart in descending order by mean absolute shap value.
Args:
col(type]): feature for which to show interactions summary
index (str or int): index to highlight
topx(int, optional): only show topx most important features, defaults to None
cats: group categorical features (Default value = False)
pos_label: positive class (Default value = None)
Returns:
fig
"""
if col in self.cats or self.cats_only:
cats = True
interact_cols = self.shap_top_interactions(col, cats=cats, pos_label=pos_label)
if topx is None: topx = len(interact_cols)
title = f"Shap interaction values for {col}"
return plotly_shap_scatter_plot(
self.shap_interaction_values_by_col(col, cats=cats, pos_label=pos_label),
self.X_cats if cats else self.X, interact_cols[:topx], title=title,
idxs=self.idxs.values, highlight_index=index, na_fill=self.na_fill,
index_name=self.index_name)
def plot_shap_dependence(self, col, color_col=None, highlight_index=None, pos_label=None):
"""plot shap dependence
Plots a shap dependence plot:
- on the x axis the possible values of the feature `col`
- on the y axis the associated individual shap values
Args:
col(str): feature to be displayed
color_col(str): if color_col provided then shap values colored (blue-red)
according to feature color_col (Default value = None)
highlight_idx: individual observation to be highlighed in the plot.
(Default value = None)
pos_label: positive class (Default value = None)
Returns:
"""
cats = self.check_cats(col, color_col)
highlight_idx = self.get_int_idx(highlight_index)
if cats:
if col in self.cats:
return plotly_shap_violin_plot(
self.X_cats,
self.shap_values_cats(pos_label),
col,
color_col,
highlight_index=highlight_idx,
idxs=self.idxs.values,
index_name=self.index_name)
else:
return plotly_dependence_plot(
self.X_cats,
self.shap_values_cats(pos_label),
col,
color_col,
na_fill=self.na_fill,
units=self.units,
highlight_index=highlight_idx,
idxs=self.idxs.values,
index_name=self.index_name)
else:
return plotly_dependence_plot(
self.X,
self.shap_values(pos_label),
col,
color_col,
na_fill=self.na_fill,
units=self.units,
highlight_index=highlight_idx,
idxs=self.idxs.values,
index_name=self.index_name)
def plot_shap_interaction(self, col, interact_col, highlight_index=None,
pos_label=None):
"""plots a dependence plot for shap interaction effects
Args:
col(str): feature for which to find interaction values
interact_col(str): feature for which interaction value are displayed
highlight_idx(int, optional, optional): idx that will be highlighted, defaults to None
pos_label: (Default value = None)
Returns:
plotly.Fig: Plotly Fig
"""
cats = self.check_cats(col, interact_col)
highlight_idx = self.get_int_idx(highlight_index)
if cats and interact_col in self.cats:
return plotly_shap_violin_plot(
self.X_cats,
self.shap_interaction_values_by_col(col, cats, pos_label=pos_label),
interact_col, col, interaction=True, units=self.units,
highlight_index=highlight_idx, idxs=self.idxs.values,
index_name=self.index_name)
else:
return plotly_dependence_plot(self.X_cats if cats else self.X,
self.shap_interaction_values_by_col(col, cats, pos_label=pos_label),
interact_col, col, interaction=True, units=self.units,
highlight_index=highlight_idx, idxs=self.idxs.values,
index_name=self.index_name)
def plot_pdp(self, col, index=None, X_row=None, drop_na=True, sample=100,
gridlines=100, gridpoints=10, pos_label=None):
"""plot partial dependence plot (pdp)
returns plotly fig for a partial dependence plot showing ice lines
for num_grid_lines rows, average pdp based on sample of sample.
If index is given, display pdp for this specific index.
Args:
col(str): feature to display pdp graph for
index(int or str, optional, optional): index to highlight in pdp graph,
defaults to None
X_row (pd.Dataframe, single row, optional): a row of features to highlight
predictions for. Alternative to passing index.
drop_na(bool, optional, optional): if true drop samples with value
equal to na_fill, defaults to True
sample(int, optional, optional): sample size on which the average
pdp will be calculated, defaults to 100
gridlines(int, optional): number of ice lines to display,
defaults to 100
gridpoints(ints: int, optional): number of points on the x axis
to calculate the pdp for, defaults to 10
pos_label: (Default value = None)
Returns:
plotly.Fig: fig
"""
pdp_df = self.pdp_df(col, index, X_row,
drop_na=drop_na, sample=sample, num_grid_points=gridpoints, pos_label=pos_label)
units = "Predicted %" if self.model_output=='probability' else self.units
if index is not None:
col_value, pred = self.get_col_value_plus_prediction(col, index=index, pos_label=pos_label)
return plotly_pdp(pdp_df,
display_index=0, # the idx to be displayed is always set to the first row by self.pdp_df()
index_feature_value=col_value, index_prediction=pred,
feature_name=col,
num_grid_lines=min(gridlines, sample, len(self.X)),
target=self.target, units=units)
elif X_row is not None:
col_value, pred = self.get_col_value_plus_prediction(col, X_row=X_row, pos_label=pos_label)
return plotly_pdp(pdp_df,
display_index=0, # the idx to be displayed is always set to the first row by self.pdp_df()
index_feature_value=col_value, index_prediction=pred,
feature_name=col,
num_grid_lines=min(gridlines, sample, len(self.X)),
target=self.target, units=units)
else:
return plotly_pdp(pdp_df, feature_name=col,
num_grid_lines=min(gridlines, sample, len(self.X)),
target=self.target, units=units)
class ClassifierExplainer(BaseExplainer):
""" """
def __init__(self, model, X, y=None, permutation_metric=roc_auc_score,
shap='guess', X_background=None, model_output="probability",
cats=None, idxs=None, index_name=None, target=None,
descriptions=None, n_jobs=None, permutation_cv=None, na_fill=-999,
labels=None, pos_label=1):
"""
Explainer for classification models. Defines the shap values for
each possible class in the classification.
You assign the positive label class afterwards with e.g. explainer.pos_label=0
In addition defines a number of plots specific to classification problems
such as a precision plot, confusion matrix, roc auc curve and pr auc curve.
Compared to BaseExplainer defines two additional parameters
Args:
labels(list): list of str labels for the different classes,
defaults to e.g. ['0', '1'] for a binary classification
pos_label: class that should be used as the positive class,
defaults to 1
"""
super().__init__(model, X, y, permutation_metric,
shap, X_background, model_output,
cats, idxs, index_name, target, descriptions,
n_jobs, permutation_cv, na_fill)
assert hasattr(model, "predict_proba"), \
("for ClassifierExplainer, model should be a scikit-learn "
"compatible *classifier* model that has a predict_proba(...) "
f"method, so not a {type(model)}!")
self._params_dict = {**self._params_dict, **dict(
labels=labels, pos_label=pos_label)}
if self.cats_only and model_output == 'probability':
print("Warning: Models that deal with categorical features directly "
f"such as {self.model.__class__.__name__} are incompatible with model_output='probability'"
" for now. So setting model_output='logodds'...", flush=True)
self.model_output = 'logodds'
if labels is not None:
self.labels = labels
elif hasattr(self.model, 'classes_'):
self.labels = [str(cls) for cls in self.model.classes_]
else:
self.labels = [str(i) for i in range(self.y.nunique())]
self.pos_label = pos_label
self.is_classifier = True
if str(type(self.model)).endswith("RandomForestClassifier'>"):
print(f"Detected RandomForestClassifier model: "
"Changing class type to RandomForestClassifierExplainer...",
flush=True)
self.__class__ = RandomForestClassifierExplainer
if str(type(self.model)).endswith("XGBClassifier'>"):
print(f"Detected XGBClassifier model: "
"Changing class type to XGBClassifierExplainer...",
flush=True)
self.__class__ = XGBClassifierExplainer
_ = self.shap_explainer
@property
def shap_explainer(self):
"""Initialize SHAP explainer.
Taking into account model type and model_output
"""
if not hasattr(self, '_shap_explainer'):
model_str = str(type(self.model)).replace("'", "").replace("<", "").replace(">", "").split(".")[-1]
if self.shap == 'tree':
if safe_is_instance(self.model,
"XGBClassifier", "LGBMClassifier", "CatBoostClassifier",
"GradientBoostingClassifier", "HistGradientBoostingClassifier"):
if self.model_output == "probability":
if self.X_background is None:
print(
f"Note: model_output=='probability'. For {model_str} shap values normally get "
"calculated against X_background, but paramater X_background=None, "
"so using X instead")
print("Generating self.shap_explainer = shap.TreeExplainer(model, "
f"{'X_background' if self.X_background is not None else 'X'}"
", model_output='probability', feature_perturbation='interventional')...")
print("Note: Shap interaction values will not be available. "
"If shap values in probability space are not necessary you can "
"pass model_output='logodds' to get shap values in logodds without the need for "
"a background dataset and also working shap interaction values...")
self._shap_explainer = shap.TreeExplainer(
self.model,
self.X_background if self.X_background is not None else self.X,
model_output="probability",
feature_perturbation="interventional")
self.interactions_should_work = False
else:
self.model_output = "logodds"
print(f"Generating self.shap_explainer = shap.TreeExplainer(model{', X_background' if self.X_background is not None else ''})")
self._shap_explainer = shap.TreeExplainer(self.model, self.X_background)
else:
if self.model_output == "probability":
print(f"Note: model_output=='probability', so assuming that raw shap output of {model_str} is in probability space...")
print(f"Generating self.shap_explainer = shap.TreeExplainer(model{', X_background' if self.X_background is not None else ''})")
self._shap_explainer = shap.TreeExplainer(self.model, self.X_background)
elif self.shap=='linear':
if self.model_output == "probability":
print(
"Note: model_output='probability' is currently not supported for linear classifiers "
"models with shap. So defaulting to model_output='logodds' "
"If you really need probability outputs use shap='kernel' instead."
)
self.model_output = "logodds"
if self.X_background is None:
print(
"Note: shap values for shap='linear' get calculated against "
"X_background, but paramater X_background=None, so using X instead...")
print("Generating self.shap_explainer = shap.LinearExplainer(model, "
f"{'X_background' if self.X_background is not None else 'X'})...")
self._shap_explainer = shap.LinearExplainer(self.model,
self.X_background if self.X_background is not None else self.X)
elif self.shap=='deep':
print("Generating self.shap_explainer = shap.DeepExplainer(model{', X_background' if self.X_background is not None else ''})")
self._shap_explainer = shap.DeepExplainer(self.model, self.X_background)
elif self.shap=='kernel':
if self.X_background is None:
print(
"Note: shap values for shap='kernel' normally get calculated against "
"X_background, but paramater X_background=None, so using X instead...")
if self.model_output != "probability":
print(
"Note: for ClassifierExplainer shap='kernel' defaults to model_output='probability"
)
self.model_output = 'probability'
print("Generating self.shap_explainer = shap.KernelExplainer(model, "
f"{'X_background' if self.X_background is not None else 'X'}"
", link='identity')")
self._shap_explainer = shap.KernelExplainer(self.model.predict_proba,
self.X_background if self.X_background is not None else self.X,
link="identity")
return self._shap_explainer
@property
def pos_label(self):
return self._pos_label
@pos_label.setter
def pos_label(self, label):
if label is None or isinstance(label, int) and label >=0 and label <len(self.labels):
self._pos_label = label
elif isinstance(label, str) and label in self.labels:
self._pos_label = self.labels.index(label)
else:
raise ValueError(f"'{label}' not in labels")
@property
def pos_label_str(self):
"""return str label of self.pos_label"""
return self.labels[self.pos_label]
def get_pos_label_index(self, pos_label):
"""return int index of pos_label_str"""
if isinstance(pos_label, int):
assert pos_label <= len(self.labels), \
f"pos_label {pos_label} is larger than number of labels!"
return pos_label
elif isinstance(pos_label, str):
assert pos_label in self.labels, \
f"Unknown pos_label. {pos_label} not in self.labels!"
return self.labels.index(pos_label)
raise ValueError("pos_label should either be int or str in self.labels!")
def get_prop_for_label(self, prop:str, label):
"""return property for a specific pos_label
Args:
prop: property to get for a certain pos_label
label: pos_label
Returns:
property
"""
tmp = self.pos_label
self.pos_label = label
ret = getattr(self, prop)
self.pos_label = tmp
return ret
@property
def y_binary(self):
"""for multiclass problems returns one-vs-rest array of [1,0] pos_label"""
if not hasattr(self, '_y_binaries'):
if not self.y_missing:
self._y_binaries = [np.where(self.y.values==i, 1, 0)
for i in range(self.y.nunique())]
else:
self._y_binaries = [self.y.values for i in range(len(self.labels))]
return default_list(self._y_binaries, self.pos_label)
@property
def pred_probas_raw(self):
"""returns pred_probas with probability for each class"""
if not hasattr(self, '_pred_probas'):
print("Calculating prediction probabilities...", flush=True)
assert hasattr(self.model, 'predict_proba'), \
"model does not have a predict_proba method!"
self._pred_probas = self.model.predict_proba(self.X)
return self._pred_probas
@property
def pred_percentiles_raw(self):
""" """
if not hasattr(self, '_pred_percentiles_raw'):
print("Calculating pred_percentiles...", flush=True)
self._pred_percentiles_raw = (pd.DataFrame(self.pred_probas_raw)
.rank(method='min')
.divide(len(self.pred_probas_raw))
.values)
return self._pred_percentiles_raw
@property
def pred_probas(self):
"""returns pred_proba for pos_label class"""
return default_2darray(self.pred_probas_raw, self.pos_label)
@property
def pred_percentiles(self):
"""returns ranks for pos_label class"""
return default_2darray(self.pred_percentiles_raw, self.pos_label)
@property
def permutation_importances(self):
"""Permutation importances"""
if not hasattr(self, '_perm_imps'):
print("Calculating permutation importances (if slow, try setting n_jobs parameter)...", flush=True)
self._perm_imps = [cv_permutation_importances(
self.model, self.X, self.y, self.metric,
cv=self.permutation_cv,
needs_proba=self.is_classifier,
pos_label=label) for label in range(len(self.labels))]
return default_list(self._perm_imps, self.pos_label)
@property
def permutation_importances_cats(self):
"""permutation importances with categoricals grouped"""
if not hasattr(self, '_perm_imps_cats'):
print("Calculating categorical permutation importances (if slow, try setting n_jobs parameter)...", flush=True)
if self.cats_only:
_ = self.permutation_importances
self._perm_imps_cats = self._perm_imps
else:
self._perm_imps_cats = [cv_permutation_importances(
self.model, self.X, self.y, self.metric,
cats_dict=self.cats_dict,
cv=self.permutation_cv,
needs_proba=self.is_classifier,
pos_label=label) for label in range(len(self.labels))]
return default_list(self._perm_imps_cats, self.pos_label)
@property
def shap_base_value(self):
"""SHAP base value: average outcome of population"""
if not hasattr(self, '_shap_base_value'):
_ = self.shap_values() # CatBoost needs to have shap values calculated before expected value for some reason
self._shap_base_value = self.shap_explainer.expected_value
if isinstance(self._shap_base_value, np.ndarray) and len(self._shap_base_value) == 1:
self._shap_base_value = self._shap_base_value[0]
if isinstance(self._shap_base_value, np.ndarray):
self._shap_base_value = list(self._shap_base_value)
if len(self.labels)==2 and isinstance(self._shap_base_value, (np.floating, float)):
if self.model_output == 'probability':
self._shap_base_value = [1-self._shap_base_value, self._shap_base_value]
else: # assume logodds
self._shap_base_value = [-self._shap_base_value, self._shap_base_value]
assert len(self._shap_base_value)==len(self.labels),\
f"len(shap_explainer.expected_value)={len(self._shap_base_value)}"\
+ f"and len(labels)={len(self.labels)} do not match!"
if self.model_output == 'probability':
for shap_base_value in self._shap_base_value:
assert shap_base_value >= 0.0 and shap_base_value <= 1.0, \
(f"Shap base value does not look like a probability: {self._shap_base_value}. "
"Try setting model_output='logodds'.")
return default_list(self._shap_base_value, self.pos_label)
@property
def shap_values(self):
"""SHAP Values"""
if not hasattr(self, '_shap_values'):
print("Calculating shap values...", flush=True)
self._shap_values = self.shap_explainer.shap_values(self.X)
if not isinstance(self._shap_values, list) and len(self.labels)==2:
self._shap_values = [-self._shap_values, self._shap_values]
assert len(self._shap_values)==len(self.labels),\
f"len(shap_values)={len(self._shap_values)}"\
+ f"and len(labels)={len(self.labels)} do not match!"
if self.model_output == 'probability':
for shap_values in self._shap_values:
assert np.all(shap_values >= -1.0) , \
(f"model_output=='probability but some shap values are < 1.0!"
"Try setting model_output='logodds'.")
for shap_values in self._shap_values:
assert np.all(shap_values <= 1.0) , \
(f"model_output=='probability but some shap values are > 1.0!"
"Try setting model_output='logodds'.")
return default_list(self._shap_values, self.pos_label)
@property
def shap_values_cats(self):
"""SHAP values with categoricals grouped together"""
if not hasattr(self, '_shap_values_cats'):
if self.cats_only:
_ = self.shap_values
self._shap_values_cats = self._shap_values
else:
_ = self.shap_values
self._shap_values_cats = [
merge_categorical_shap_values(
self.X, sv, self.cats_dict) for sv in self._shap_values]
return default_list(self._shap_values_cats, self.pos_label)
@property
def shap_interaction_values(self):
"""SHAP interaction values"""
if not hasattr(self, '_shap_interaction_values'):
_ = self.shap_values #make sure shap values have been calculated
print("Calculating shap interaction values...", flush=True)
if self.shap == 'tree':
print("Reminder: TreeShap computational complexity is O(TLD^2), "
"where T is the number of trees, L is the maximum number of"
" leaves in any tree and D the maximal depth of any tree. So "
"reducing these will speed up the calculation.",
flush=True)
self._shap_interaction_values = self.shap_explainer.shap_interaction_values(self.X)
if not isinstance(self._shap_interaction_values, list) and len(self.labels)==2:
if self.model_output == "probability":
self._shap_interaction_values = [1-self._shap_interaction_values,
self._shap_interaction_values]
else: # assume logodds so logodds of negative class is -logodds of positive class
self._shap_interaction_values = [-self._shap_interaction_values,
self._shap_interaction_values]
self._shap_interaction_values = [
normalize_shap_interaction_values(siv, self.shap_values)
for siv, sv in zip(self._shap_interaction_values, self._shap_values)]
return default_list(self._shap_interaction_values, self.pos_label)
@property
def shap_interaction_values_cats(self):
"""SHAP interaction values with categoricals grouped together"""
if not hasattr(self, '_shap_interaction_values_cats'):
_ = self.shap_interaction_values
self._shap_interaction_values_cats = [
merge_categorical_shap_interaction_values(
siv, self.X, self.X_cats, self.cats_dict)
for siv in self._shap_interaction_values]
return default_list(self._shap_interaction_values_cats, self.pos_label)
@property
def mean_abs_shap(self):
"""mean absolute SHAP values"""
if not hasattr(self, '_mean_abs_shap'):
_ = self.shap_values
self._mean_abs_shap = [mean_absolute_shap_values(
self.columns, sv) for sv in self._shap_values]
return default_list(self._mean_abs_shap, self.pos_label)
@property
def mean_abs_shap_cats(self):
"""mean absolute SHAP values with categoricals grouped together"""
if not hasattr(self, '_mean_abs_shap_cats'):
_ = self.shap_values_cats
self._mean_abs_shap_cats = [
mean_absolute_shap_values(self.columns_cats, sv)
for sv in self._shap_values_cats]
return default_list(self._mean_abs_shap_cats, self.pos_label)
def cutoff_from_percentile(self, percentile, pos_label=None):
"""The cutoff equivalent to the percentile given
For example if you want the cutoff that splits the highest 20%
pred_proba from the lowest 80%, you would set percentile=0.8
and get the correct cutoff.
Args:
percentile(float): percentile to convert to cutoff
pos_label: positive class (Default value = None)
Returns:
cutoff
"""
if pos_label is None:
return pd.Series(self.pred_probas).nlargest(int((1-percentile)*len(self))).min()
else:
return pd.Series(self.pred_probas_raw[:, pos_label]).nlargest(int((1-percentile)*len(self))).min()
def percentile_from_cutoff(self, cutoff, pos_label=None):
"""The percentile equivalent to the cutoff given
For example if set the cutoff at 0.8, then what percentage
of pred_proba is above this cutoff?
Args:
cutoff (float): cutoff to convert to percentile
pos_label: positive class (Default value = None)
Returns:
percentile
"""
if cutoff is None:
return None
if pos_label is None:
return 1-(self.pred_probas < cutoff).mean()
else:
pos_label = self.get_pos_label_index(pos_label)
return 1-np.mean(self.pred_probas_raw[:, pos_label] < cutoff)
def metrics(self, cutoff=0.5, pos_label=None, **kwargs):
"""returns a dict with useful metrics for your classifier:
accuracy, precision, recall, f1, roc auc, pr auc, log loss
Args:
cutoff(float): cutoff used to calculate metrics (Default value = 0.5)
pos_label: positive class (Default value = None)
Returns:
dict
"""
if self.y_missing:
raise ValueError("No y was passed to explainer, so cannot calculate metrics!")
if pos_label is None: pos_label = self.pos_label
metrics_dict = {
'accuracy' : accuracy_score(self.y_binary(pos_label), np.where(self.pred_probas(pos_label) > cutoff, 1, 0)),
'precision' : precision_score(self.y_binary(pos_label), np.where(self.pred_probas(pos_label) > cutoff, 1, 0)),
'recall' : recall_score(self.y_binary(pos_label), np.where(self.pred_probas(pos_label) > cutoff, 1, 0)),
'f1' : f1_score(self.y_binary(pos_label), np.where(self.pred_probas(pos_label) > cutoff, 1, 0)),
'roc_auc_score' : roc_auc_score(self.y_binary(pos_label), self.pred_probas(pos_label)),
'pr_auc_score' : average_precision_score(self.y_binary(pos_label), self.pred_probas(pos_label)),
'log_loss' : log_loss(self.y_binary(pos_label), self.pred_probas(pos_label))
}
return metrics_dict
def metrics_descriptions(self, cutoff=0.5, pos_label=None):
metrics_dict = self.metrics(cutoff, pos_label)
metrics_descriptions_dict = {}
for k, v in metrics_dict.items():
if k == 'accuracy':
metrics_descriptions_dict[k] = f"{round(100*v, 2)}% of predicted labels was predicted correctly."
if k == 'precision':
metrics_descriptions_dict[k] = f"{round(100*v, 2)}% of predicted positive labels was predicted correctly."
if k == 'recall':
metrics_descriptions_dict[k] = f"{round(100*v, 2)}% of positive labels was predicted correctly."
if k == 'f1':
metrics_descriptions_dict[k] = f"The weighted average of precision and recall is {round(v, 2)}"
if k == 'roc_auc_score':
metrics_descriptions_dict[k] = f"The probability that a random positive label has a higher score than a random negative label is {round(100*v, 2)}%"
if k == 'pr_auc_score':
metrics_descriptions_dict[k] = f"The average precision score calculated for each recall threshold is {round(v, 2)}. This ignores true negatives."
if k == 'log_loss':
metrics_descriptions_dict[k] = f"A measure of how far the predicted label is from the true label on average in log space {round(v, 2)}"
return metrics_descriptions_dict
def random_index(self, y_values=None, return_str=False,
pred_proba_min=None, pred_proba_max=None,
pred_percentile_min=None, pred_percentile_max=None, pos_label=None):
"""random index satisfying various constraint
Args:
y_values: list of labels to include (Default value = None)
return_str: return str from self.idxs (Default value = False)
pred_proba_min: minimum pred_proba (Default value = None)
pred_proba_max: maximum pred_proba (Default value = None)
pred_percentile_min: minimum pred_proba percentile (Default value = None)
pred_percentile_max: maximum pred_proba percentile (Default value = None)
pos_label: positive class (Default value = None)
Returns:
index
"""
# if pos_label is None: pos_label = self.pos_label
if (y_values is None
and pred_proba_min is None and pred_proba_max is None
and pred_percentile_min is None and pred_percentile_max is None):
potential_idxs = self.idxs.values
else:
if pred_proba_min is None: pred_proba_min = self.pred_probas(pos_label).min()
if pred_proba_max is None: pred_proba_max = self.pred_probas(pos_label).max()
if pred_percentile_min is None: pred_percentile_min = 0.0
if pred_percentile_max is None: pred_percentile_max = 1.0
if not self.y_missing:
if y_values is None: y_values = self.y.unique().tolist()
if not isinstance(y_values, list): y_values = [y_values]
y_values = [y if isinstance(y, int) else self.labels.index(y) for y in y_values]
potential_idxs = self.idxs[(self.y.isin(y_values)) &
(self.pred_probas(pos_label) >= pred_proba_min) &
(self.pred_probas(pos_label) <= pred_proba_max) &
(self.pred_percentiles(pos_label) > pred_percentile_min) &
(self.pred_percentiles(pos_label) <= pred_percentile_max)].values
else:
potential_idxs = self.idxs[
(self.pred_probas(pos_label) >= pred_proba_min) &
(self.pred_probas(pos_label) <= pred_proba_max) &
(self.pred_percentiles(pos_label) > pred_percentile_min) &
(self.pred_percentiles(pos_label) <= pred_percentile_max)].values
if len(potential_idxs) > 0:
idx = np.random.choice(potential_idxs)
else:
return None
if return_str:
return idx
return self.idxs.get_loc(idx)
def prediction_result_df(self, index=None, X_row=None, add_star=True, logodds=False, round=3):
"""returns a table with the predicted probability for each label for index
Args:
index ({int, str}): index
add_star(bool): add a star to the observed label
round (int): rounding to apply to pred_proba float
Returns:
pd.DataFrame
"""
if index is None and X_row is None:
raise ValueError("You need to either pass an index or X_row!")
if index is not None:
int_idx = self.get_int_idx(index)
pred_probas = self.pred_probas_raw[int_idx, :]
elif X_row is not None:
if X_row.columns.tolist()==self.X_cats.columns.tolist():
X_row = X_cats_to_X(X_row, self.cats_dict, self.X.columns)
pred_probas = self.model.predict_proba(X_row)[0, :]
preds_df = pd.DataFrame(dict(
label=self.labels,
probability=pred_probas))
if logodds:
preds_df.loc[:, "logodds"] = \
preds_df.probability.apply(lambda p: np.log(p / (1-p)))
if index is not None and not self.y_missing and not np.isnan(self.y[int_idx]):
preds_df.iloc[self.y[int_idx], 0] = f"{preds_df.iloc[self.y[int_idx], 0]}*"
return preds_df.round(round)
def precision_df(self, bin_size=None, quantiles=None, multiclass=False,
round=3, pos_label=None):
"""dataframe with predicted probabilities and precision
Args:
bin_size(float, optional, optional): group predictions in bins of size bin_size, defaults to 0.1
quantiles(int, optional, optional): group predictions in evenly sized quantiles of size quantiles, defaults to None
multiclass(bool, optional, optional): whether to calculate precision for every class (Default value = False)
round: (Default value = 3)
pos_label: (Default value = None)
Returns:
pd.DataFrame: precision_df
"""
if self.y_missing:
raise ValueError("No y was passed to explainer, so cannot calculate precision_df!")
assert self.pred_probas is not None
if pos_label is None: pos_label = self.pos_label
if bin_size is None and quantiles is None:
bin_size=0.1 # defaults to bin_size=0.1
if multiclass:
return get_precision_df(self.pred_probas_raw, self.y,
bin_size, quantiles,
round=round, pos_label=pos_label)
else:
return get_precision_df(self.pred_probas(pos_label), self.y_binary(pos_label),
bin_size, quantiles, round=round)
def lift_curve_df(self, pos_label=None):
"""returns a pd.DataFrame with data needed to build a lift curve
Args:
pos_label: (Default value = None)
Returns:
"""
if pos_label is None: pos_label = self.pos_label
return get_lift_curve_df(self.pred_probas(pos_label), self.y, pos_label)
def prediction_result_markdown(self, index, include_percentile=True, round=2, pos_label=None):
"""markdown of result of prediction for index
Args:
index(int or str): the index of the row for which to generate the prediction
include_percentile(bool, optional, optional): include the rank
percentile of the prediction, defaults to True
round(int, optional, optional): rounding to apply to results, defaults to 2
pos_label: (Default value = None)
**kwargs:
Returns:
str: markdown string
"""
int_idx = self.get_int_idx(index)
if pos_label is None: pos_label = self.pos_label
def display_probas(pred_probas_raw, labels, model_output='probability', round=2):
assert (len(pred_probas_raw.shape)==1 and len(pred_probas_raw) ==len(labels))
def log_odds(p, round=2):
return np.round(np.log(p / (1-p)), round)
for i in range(len(labels)):
proba_str = f"{np.round(100*pred_probas_raw[i], round)}%"
logodds_str = f"(logodds={log_odds(pred_probas_raw[i], round)})"
yield f"* {labels[i]}: {proba_str} {logodds_str if model_output=='logodds' else ''}\n"
model_prediction = "###### Prediction:\n\n"
if (isinstance(self.y[0], int) or
isinstance(self.y[0], np.int64)):
model_prediction += f"Observed {self.target}: {self.labels[self.y[int_idx]]}\n\n"
model_prediction += "Prediction probabilities per label:\n\n"
for pred in display_probas(
self.pred_probas_raw[int_idx],
self.labels, self.model_output, round):
model_prediction += pred
if include_percentile:
percentile = np.round(100*(1-self.pred_percentiles(pos_label)[int_idx]))
model_prediction += f'\nIn top {percentile}% percentile probability {self.labels[pos_label]}'
return model_prediction
def plot_precision(self, bin_size=None, quantiles=None, cutoff=None, multiclass=False, pos_label=None):
"""plot precision vs predicted probability
plots predicted probability on the x-axis and observed precision (fraction of actual positive
cases) on the y-axis.
Should pass either bin_size fraction of number of quantiles, but not both.
Args:
bin_size(float, optional): size of the bins on x-axis (e.g. 0.05 for 20 bins)
quantiles(int, optional): number of equal sized quantiles to split
the predictions by e.g. 20, optional)
cutoff: cutoff of model to include in the plot (Default value = None)
multiclass: whether to display all classes or only positive class,
defaults to False
pos_label: positive label to display, defaults to self.pos_label
Returns:
Plotly fig
"""
if pos_label is None: pos_label = self.pos_label
if bin_size is None and quantiles is None:
bin_size=0.1 # defaults to bin_size=0.1
precision_df = self.precision_df(
bin_size=bin_size, quantiles=quantiles, multiclass=multiclass, pos_label=pos_label)
return plotly_precision_plot(precision_df,
cutoff=cutoff, labels=self.labels, pos_label=pos_label)
def plot_cumulative_precision(self, percentile=None, pos_label=None):
"""plot cumulative precision
returns a cumulative precision plot, which is a slightly different
representation of a lift curve.
Args:
pos_label: positive label to display, defaults to self.pos_label
Returns:
plotly fig
"""
if pos_label is None: pos_label = self.pos_label
return plotly_cumulative_precision_plot(
self.lift_curve_df(pos_label=pos_label), labels=self.labels,
percentile=percentile, pos_label=pos_label)
def plot_confusion_matrix(self, cutoff=0.5, normalized=False, binary=False, pos_label=None):
"""plot of a confusion matrix.
Args:
cutoff(float, optional, optional): cutoff of positive class to
calculate confusion matrix for, defaults to 0.5
normalized(bool, optional, optional): display percentages instead
of counts , defaults to False
binary(bool, optional, optional): if multiclass display one-vs-rest
instead, defaults to False
pos_label: positive label to display, defaults to self.pos_label
Returns:
plotly fig
"""
if self.y_missing:
raise ValueError("No y was passed to explainer, so cannot plot confusion matrix!")
if pos_label is None: pos_label = self.pos_label
pos_label_str = self.labels[pos_label]
if binary:
if len(self.labels)==2:
def order_binary_labels(labels, pos_label):
pos_index = labels.index(pos_label)
return [labels[1-pos_index], labels[pos_index]]
labels = order_binary_labels(self.labels, pos_label_str)
else:
labels = ['Not ' + pos_label_str, pos_label_str]
return plotly_confusion_matrix(
self.y_binary(pos_label), np.where(self.pred_probas(pos_label) > cutoff, 1, 0),
percentage=normalized, labels=labels)
else:
return plotly_confusion_matrix(
self.y, self.pred_probas_raw.argmax(axis=1),
percentage=normalized, labels=self.labels)
def plot_lift_curve(self, cutoff=None, percentage=False, add_wizard=True,
round=2, pos_label=None):
"""plot of a lift curve.
Args:
cutoff(float, optional): cutoff of positive class to calculate lift
(Default value = None)
percentage(bool, optional): display percentages instead of counts,
defaults to False
add_wizard (bool, optional): Add a line indicating how a perfect model
would perform ("the wizard"). Defaults to True.
round: number of digits to round to (Default value = 2)
pos_label: positive label to display, defaults to self.pos_label
Returns:
plotly fig
"""
return plotly_lift_curve(self.lift_curve_df(pos_label), cutoff=cutoff,
percentage=percentage, add_wizard=add_wizard, round=round)
def plot_classification(self, cutoff=0.5, percentage=True, pos_label=None):
"""plot showing a barchart of the classification result for cutoff
Args:
cutoff(float, optional): cutoff of positive class to calculate lift
(Default value = 0.5)
percentage(bool, optional): display percentages instead of counts,
defaults to True
pos_label: positive label to display, defaults to self.pos_label
Returns:
plotly fig
"""
return plotly_classification_plot(self.pred_probas(pos_label), self.y, self.labels, cutoff, percentage=percentage)
def plot_roc_auc(self, cutoff=0.5, pos_label=None):
"""plots ROC_AUC curve.
The TPR and FPR of a particular cutoff is displayed in crosshairs.
Args:
cutoff: cutoff value to be included in plot (Default value = 0.5)
pos_label: (Default value = None)
Returns:
"""
if self.y_missing:
raise ValueError("No y was passed to explainer, so cannot plot roc auc!")
return plotly_roc_auc_curve(self.y_binary(pos_label), self.pred_probas(pos_label), cutoff=cutoff)
def plot_pr_auc(self, cutoff=0.5, pos_label=None):
"""plots PR_AUC curve.
the precision and recall of particular cutoff is displayed in crosshairs.
Args:
cutoff: cutoff value to be included in plot (Default value = 0.5)
pos_label: (Default value = None)
Returns:
"""
if self.y_missing:
raise ValueError("No y was passed to explainer, so cannot plot PR AUC!")
return plotly_pr_auc_curve(self.y_binary(pos_label), self.pred_probas(pos_label), cutoff=cutoff)
def plot_prediction_result(self, index=None, X_row=None, showlegend=True):
"""Returns a piechart with the predicted probabilities distribution
Args:
index ({int, str}): Index for which to display prediction
X_row (pd.DataFrame): single row of an input dataframe, e.g.
explainer.X.iloc[[0]]
showlegend (bool, optional): Display legend. Defaults to False.
Returns:
plotly.fig
"""
preds_df = self.prediction_result_df(index, X_row)
return plotly_prediction_piechart(preds_df, showlegend=showlegend)
def calculate_properties(self, include_interactions=True):
"""calculate all lazily calculated properties of explainer
Args:
include_interactions: (Default value = True)
Returns:
None
"""
_ = self.pred_probas
super().calculate_properties(include_interactions=include_interactions)
class RegressionExplainer(BaseExplainer):
""" """
def __init__(self, model, X, y=None, permutation_metric=r2_score,
shap="guess", X_background=None, model_output="raw",
cats=None, idxs=None, index_name=None, target=None,
descriptions=None, n_jobs=None, permutation_cv=None,
na_fill=-999, units=""):
"""Explainer for regression models.
In addition to BaseExplainer defines a number of plots specific to
regression problems such as a predicted vs actual and residual plots.
Combared to BaseExplainerBunch defines two additional parameters.
Args:
units(str): units to display for regression quantity
"""
super().__init__(model, X, y, permutation_metric,
shap, X_background, model_output,
cats, idxs, index_name, target, descriptions,
n_jobs, permutation_cv, na_fill)
self._params_dict = {**self._params_dict, **dict(units=units)}
self.units = units
self.is_regression = True
if str(type(self.model)).endswith("RandomForestRegressor'>"):
print(f"Changing class type to RandomForestRegressionExplainer...", flush=True)
self.__class__ = RandomForestRegressionExplainer
if str(type(self.model)).endswith("XGBRegressor'>"):
print(f"Changing class type to XGBRegressionExplainer...", flush=True)
self.__class__ = XGBRegressionExplainer
_ = self.shap_explainer
@property
def residuals(self):
"""residuals: y-preds"""
if not hasattr(self, '_residuals'):
print("Calculating residuals...")
self._residuals = self.y-self.preds
return self._residuals
@property
def abs_residuals(self):
"""absolute residuals"""
if not hasattr(self, '_abs_residuals'):
print("Calculating absolute residuals...")
self._abs_residuals = np.abs(self.residuals)
return self._abs_residuals
def random_index(self, y_min=None, y_max=None, pred_min=None, pred_max=None,
residuals_min=None, residuals_max=None,
abs_residuals_min=None, abs_residuals_max=None,
return_str=False, **kwargs):
"""random index following to various exclusion criteria
Args:
y_min: (Default value = None)
y_max: (Default value = None)
pred_min: (Default value = None)
pred_max: (Default value = None)
residuals_min: (Default value = None)
residuals_max: (Default value = None)
abs_residuals_min: (Default value = None)
abs_residuals_max: (Default value = None)
return_str: return the str index from self.idxs (Default value = False)
**kwargs:
Returns:
a random index that fits the exclusion criteria
"""
if self.y_missing:
if pred_min is None:
pred_min = self.preds.min()
if pred_max is None:
pred_max = self.preds.max()
potential_idxs = self.idxs[
(self.preds >= pred_min) &
(self.preds <= pred_max)].values
else:
if y_min is None:
y_min = self.y.min()
if y_max is None:
y_max = self.y.max()
if pred_min is None:
pred_min = self.preds.min()
if pred_max is None:
pred_max = self.preds.max()
if residuals_min is None:
residuals_min = self.residuals.min()
if residuals_max is None:
residuals_max = self.residuals.max()
if abs_residuals_min is None:
abs_residuals_min = self.abs_residuals.min()
if abs_residuals_max is None:
abs_residuals_max = self.abs_residuals.max()
potential_idxs = self.idxs[(self.y >= y_min) &
(self.y <= y_max) &
(self.preds >= pred_min) &
(self.preds <= pred_max) &
(self.residuals >= residuals_min) &
(self.residuals <= residuals_max) &
(self.abs_residuals >= abs_residuals_min) &
(self.abs_residuals <= abs_residuals_max)].values
if len(potential_idxs) > 0:
idx = np.random.choice(potential_idxs)
else:
return None
if return_str:
return idx
return self.idxs.get_loc(idx)
def prediction_result_markdown(self, index, include_percentile=True, round=2):
"""markdown of prediction result
Args:
index: row index to be predicted
include_percentile (bool): include line about prediciton percentile
round: (Default value = 2)
Returns:
str: markdown summary of prediction for index
"""
int_idx = self.get_int_idx(index)
model_prediction = "###### Prediction:\n"
model_prediction += f"Predicted {self.target}: {np.round(self.preds[int_idx], round)} {self.units}\n\n"
if not self.y_missing:
model_prediction += f"Observed {self.target}: {np.round(self.y[int_idx], round)} {self.units}\n\n"
model_prediction += f"Residual: {np.round(self.residuals[int_idx], round)} {self.units}\n\n"
if include_percentile:
percentile = np.round(100*(1-self.pred_percentiles[int_idx]))
model_prediction += f"\nIn top {percentile}% percentile predicted {self.target}"
return model_prediction
def prediction_result_df(self, index=None, X_row=None, round=3):
"""prediction result in dataframe format
Args:
index: row index to be predicted
round (int): rounding applied to floats (defaults to 3)
Returns:
pd.DataFrame
"""
if index is None and X_row is None:
raise ValueError("You need to either pass an index or X_row!")
if index is not None:
int_idx = self.get_int_idx(index)
preds_df = pd.DataFrame(columns = ["", self.target])
preds_df = preds_df.append(
pd.Series(("Predicted", str(np.round(self.preds[int_idx], round)) + f" {self.units}"),
index=preds_df.columns), ignore_index=True)
if not self.y_missing:
preds_df = preds_df.append(
pd.Series(("Observed", str(np.round(self.y[int_idx], round)) + f" {self.units}"),
index=preds_df.columns), ignore_index=True)
preds_df = preds_df.append(
pd.Series(("Residual", str(np.round(self.residuals[int_idx], round)) + f" {self.units}"),
index=preds_df.columns), ignore_index=True)
elif X_row is not None:
if X_row.columns.tolist()==self.X_cats.columns.tolist():
X_row = X_cats_to_X(X_row, self.cats_dict, self.X.columns)
assert np.all(X_row.columns==self.X.columns), \
("The column names of X_row should match X! Instead X_row.columns"
f"={X_row.columns.tolist()}...")
prediction = self.model.predict(X_row)[0]
preds_df = pd.DataFrame(columns = ["", self.target])
preds_df = preds_df.append(
pd.Series(("Predicted", str(np.round(prediction, round)) + f" {self.units}"),
index=preds_df.columns), ignore_index=True)
return preds_df
def metrics(self):
"""dict of performance metrics: rmse, mae and R^2"""
if self.y_missing:
raise ValueError("No y was passed to explainer, so cannot calculate metrics!")
metrics_dict = {
'root_mean_squared_error' : np.sqrt(mean_squared_error(self.y, self.preds)),
'mean_absolute_error' : mean_absolute_error(self.y, self.preds),
'R-squared' : r2_score(self.y, self.preds),
}
return metrics_dict
def metrics_descriptions(self):
metrics_dict = self.metrics()
metrics_descriptions_dict = {}
for k, v in metrics_dict.items():
if k == 'root_mean_squared_error':
metrics_descriptions_dict[k] = f"A measure of how close predicted value fits true values, where large deviations are punished more heavily. So the lower this number the better the model."
if k == 'mean_absolute_error':
metrics_descriptions_dict[k] = f"On average predictions deviate {round(v, 2)} {self.units} off the observed value of {self.target} (can be both above or below)"
if k == 'R-squared':
metrics_descriptions_dict[k] = f"{round(100*v, 2)}% of all variation in {self.target} was explained by the model."
return metrics_descriptions_dict
def plot_predicted_vs_actual(self, round=2, logs=False, log_x=False, log_y=False, **kwargs):
"""plot with predicted value on x-axis and actual value on y axis.
Args:
round(int, optional): rounding to apply to outcome, defaults to 2
logs (bool, optional): log both x and y axis, defaults to False
log_y (bool, optional): only log x axis. Defaults to False.
log_x (bool, optional): only log y axis. Defaults to False.
**kwargs:
Returns:
Plotly fig
"""
if self.y_missing:
raise ValueError("No y was passed to explainer, so cannot plot predicted vs actual!")
return plotly_predicted_vs_actual(self.y, self.preds,
target=self.target, units=self.units, idxs=self.idxs.values,
logs=logs, log_x=log_x, log_y=log_y, round=round,
index_name=self.index_name)
def plot_residuals(self, vs_actual=False, round=2, residuals='difference'):
"""plot of residuals. x-axis is the predicted outcome by default
Args:
vs_actual(bool, optional): use actual value for x-axis,
defaults to False
round(int, optional): rounding to perform on values, defaults to 2
residuals (str, {'difference', 'ratio', 'log-ratio'} optional):
How to calcualte residuals. Defaults to 'difference'.
Returns:
Plotly fig
"""
if self.y_missing:
raise ValueError("No y was passed to explainer, so cannot plot residuals!")
return plotly_plot_residuals(self.y, self.preds, idxs=self.idxs.values,
vs_actual=vs_actual, target=self.target,
units=self.units, residuals=residuals,
round=round, index_name=self.index_name)
def plot_residuals_vs_feature(self, col, residuals='difference', round=2,
dropna=True, points=True, winsor=0):
"""Plot residuals vs individual features
Args:
col(str): Plot against feature col
residuals (str, {'difference', 'ratio', 'log-ratio'} optional):
How to calcualte residuals. Defaults to 'difference'.
round(int, optional): rounding to perform on residuals, defaults to 2
dropna(bool, optional): drop missing values from plot, defaults to True.
points (bool, optional): display point cloud next to violin plot.
Defaults to True.
winsor (int, 0-50, optional): percentage of outliers to winsor out of
the y-axis. Defaults to 0.
Returns:
plotly fig
"""
if self.y_missing:
raise ValueError("No y was passed to explainer, so cannot plot residuals!")
assert col in self.columns or col in self.columns_cats, \
f'{col} not in columns or columns_cats!'
col_vals = self.X_cats[col] if self.check_cats(col) else self.X[col]
na_mask = col_vals != self.na_fill if dropna else np.array([True]*len(col_vals))
return plotly_residuals_vs_col(
self.y[na_mask], self.preds[na_mask], col_vals[na_mask],
residuals=residuals, idxs=self.idxs.values[na_mask], points=points,
round=round, winsor=winsor, index_name=self.index_name)
def plot_y_vs_feature(self, col, residuals='difference', round=2,
dropna=True, points=True, winsor=0):
"""Plot y vs individual features
Args:
col(str): Plot against feature col
round(int, optional): rounding to perform on residuals, defaults to 2
dropna(bool, optional): drop missing values from plot, defaults to True.
points (bool, optional): display point cloud next to violin plot.
Defaults to True.
winsor (int, 0-50, optional): percentage of outliers to winsor out of
the y-axis. Defaults to 0.
Returns:
plotly fig
"""
if self.y_missing:
raise ValueError("No y was passed to explainer, so cannot plot y vs feature!")
assert col in self.columns or col in self.columns_cats, \
f'{col} not in columns or columns_cats!'
col_vals = self.X_cats[col] if self.check_cats(col) else self.X[col]
na_mask = col_vals != self.na_fill if dropna else np.array([True]*len(col_vals))
return plotly_actual_vs_col(self.y[na_mask], self.preds[na_mask], col_vals[na_mask],
idxs=self.idxs.values[na_mask], points=points, round=round, winsor=winsor,
units=self.units, target=self.target, index_name=self.index_name)
def plot_preds_vs_feature(self, col, residuals='difference', round=2,
dropna=True, points=True, winsor=0):
"""Plot y vs individual features
Args:
col(str): Plot against feature col
round(int, optional): rounding to perform on residuals, defaults to 2
dropna(bool, optional): drop missing values from plot, defaults to True.
points (bool, optional): display point cloud next to violin plot.
Defaults to True.
winsor (int, 0-50, optional): percentage of outliers to winsor out of
the y-axis. Defaults to 0.
Returns:
plotly fig
"""
assert col in self.columns or col in self.columns_cats, \
f'{col} not in columns or columns_cats!'
col_vals = self.X_cats[col] if self.check_cats(col) else self.X[col]
na_mask = col_vals != self.na_fill if dropna else np.array([True]*len(col_vals))
return plotly_preds_vs_col(self.y[na_mask], self.preds[na_mask], col_vals[na_mask],
idxs=self.idxs.values[na_mask], points=points, round=round, winsor=winsor,
units=self.units, target=self.target, index_name=self.index_name)
class RandomForestExplainer(BaseExplainer):
"""RandomForestBunch allows for the analysis of individual DecisionTrees that
make up the RandomForest.
"""
@property
def is_tree_explainer(self):
"""this is either a RandomForestExplainer or XGBExplainer"""
return True
@property
def no_of_trees(self):
"""The number of trees in the RandomForest model"""
return len(self.model.estimators_)
@property
def graphviz_available(self):
""" """
if not hasattr(self, '_graphviz_available'):
try:
import graphviz.backend as be
cmd = ["dot", "-V"]
stdout, stderr = be.run(cmd, capture_output=True, check=True, quiet=True)
except:
print("""
WARNING: you don't seem to have graphviz in your path (cannot run 'dot -V'),
so no dtreeviz visualisation of decision trees will be shown on the shadow trees tab.
See https://github.com/parrt/dtreeviz for info on how to properly install graphviz
for dtreeviz.
""")
self._graphviz_available = False
else:
self._graphviz_available = True
return self._graphviz_available
@property
def decision_trees(self):
"""a list of ShadowDecTree objects"""
if not hasattr(self, '_decision_trees'):
print("Calculating ShadowDecTree for each individual decision tree...", flush=True)
assert hasattr(self.model, 'estimators_'), \
"""self.model does not have an estimators_ attribute, so probably not
actually a sklearn RandomForest?"""
self._decision_trees = [
ShadowDecTree.get_shadow_tree(decision_tree,
self.X,
self.y,
feature_names=self.X.columns.tolist(),
target_name='target',
class_names = self.labels if self.is_classifier else None)
for decision_tree in self.model.estimators_]
return self._decision_trees
def decisiontree_df(self, tree_idx, index, pos_label=None):
"""dataframe with all decision nodes of a particular decision tree
Args:
tree_idx: the n'th tree in the random forest
index: row index
round: (Default value = 2)
pos_label: positive class (Default value = None)
Returns:
dataframe with summary of the decision tree path
"""
assert tree_idx >= 0 and tree_idx < len(self.decision_trees), \
f"tree index {tree_idx} outside 0 and number of trees ({len(self.decision_trees)}) range"
idx = self.get_int_idx(index)
assert idx >= 0 and idx < len(self.X), \
f"=index {idx} outside 0 and size of X ({len(self.X)}) range"
if self.is_classifier:
if pos_label is None: pos_label = self.pos_label
return get_decisiontree_df(self.decision_trees[tree_idx], self.X.iloc[idx],
pos_label=pos_label)
else:
return get_decisiontree_df(self.decision_trees[tree_idx], self.X.iloc[idx])
def decisiontree_summary_df(self, tree_idx, index, round=2, pos_label=None):
"""formats decisiontree_df in a slightly more human readable format.
Args:
tree_idx: the n'th tree in the random forest
index: row index
round: (Default value = 2)
pos_label: positive class (Default value = None)
Returns:
dataframe with summary of the decision tree path
"""
idx=self.get_int_idx(index)
return get_decisiontree_summary_df(self.decisiontree_df(tree_idx, idx, pos_label=pos_label),
classifier=self.is_classifier, round=round, units=self.units)
def decision_path_file(self, tree_idx, index, show_just_path=False):
"""get a dtreeviz visualization of a particular tree in the random forest.
Args:
tree_idx: the n'th tree in the random forest
index: row index
show_just_path (bool, optional): show only the path not rest of the
tree. Defaults to False.
Returns:
the path where the .svg file is stored.
"""
if not self.graphviz_available:
print("No graphviz 'dot' executable available!")
return None
idx = self.get_int_idx(index)
viz = dtreeviz(self.decision_trees[tree_idx],
X=self.X.iloc[idx, :],
fancy=False,
show_node_labels = False,
show_just_path=show_just_path)
return viz.save_svg()
def decision_path(self, tree_idx, index, show_just_path=False):
"""get a dtreeviz visualization of a particular tree in the random forest.
Args:
tree_idx: the n'th tree in the random forest
index: row index
show_just_path (bool, optional): show only the path not rest of the
tree. Defaults to False.
Returns:
a IPython display SVG object for e.g. jupyter notebook.
"""
if not self.graphviz_available:
print("No graphviz 'dot' executable available!")
return None
from IPython.display import SVG
svg_file = self.decision_path_file(tree_idx, index, show_just_path)
return SVG(open(svg_file,'rb').read())
def decision_path_encoded(self, tree_idx, index, show_just_path=False):
"""get a dtreeviz visualization of a particular tree in the random forest.
Args:
tree_idx: the n'th tree in the random forest
index: row index
show_just_path (bool, optional): show only the path not rest of the
tree. Defaults to False.
Returns:
a base64 encoded image, for inclusion in websites (e.g. dashboard)
"""
if not self.graphviz_available:
print("No graphviz 'dot' executable available!")
return None
svg_file = self.decision_path_file(tree_idx, index, show_just_path)
encoded = base64.b64encode(open(svg_file,'rb').read())
svg_encoded = 'data:image/svg+xml;base64,{}'.format(encoded.decode())
return svg_encoded
def plot_trees(self, index, highlight_tree=None, round=2,
higher_is_better=True, pos_label=None):
"""plot barchart predictions of each individual prediction tree
Args:
index: index to display predictions for
highlight_tree: tree to highlight in plot (Default value = None)
round: rounding of numbers in plot (Default value = 2)
higher_is_better (bool): flip red and green. Dummy bool for compatibility
with gbm plot_trees().
pos_label: positive class (Default value = None)
Returns:
"""
idx=self.get_int_idx(index)
assert idx is not None, 'invalid index'
if self.is_classifier:
if pos_label is None: pos_label = self.pos_label
if not np.isnan(self.y[idx]):
y = 100*self.y_binary(pos_label)[idx]
else:
y = None
return plotly_rf_trees(self.model, self.X.iloc[[idx]], y,
highlight_tree=highlight_tree, round=round,
pos_label=pos_label, target=self.target)
else:
y = self.y[idx]
return plotly_rf_trees(self.model, self.X.iloc[[idx]], y,
highlight_tree=highlight_tree, round=round,
target=self.target, units=self.units)
def calculate_properties(self, include_interactions=True):
"""
Args:
include_interactions: If False do not calculate shap interaction value
(Default value = True)
Returns:
"""
_ = self.decision_trees
super().calculate_properties(include_interactions=include_interactions)
class XGBExplainer(BaseExplainer):
"""XGBExplainer allows for the analysis of individual DecisionTrees that
make up the xgboost model.
"""
@property
def is_tree_explainer(self):
"""this is either a RandomForestExplainer or XGBExplainer"""
return True
@property
def model_dump_list(self):
if not hasattr(self, "_model_dump_list"):
print("Generating model dump...", flush=True)
self._model_dump_list = self.model.get_booster().get_dump()
return self._model_dump_list
@property
def no_of_trees(self):
"""The number of trees in the RandomForest model"""
if self.is_classifier and len(self.labels) > 2:
return int(len(self.model_dump_list) / len(self.labels))
return len(self.model_dump_list)
@property
def graphviz_available(self):
""" """
if not hasattr(self, '_graphviz_available'):
try:
import graphviz.backend as be
cmd = ["dot", "-V"]
stdout, stderr = be.run(cmd, capture_output=True, check=True, quiet=True)
except:
print("""
WARNING: you don't seem to have graphviz in your path (cannot run 'dot -V'),
so no dtreeviz visualisation of decision trees will be shown on the shadow trees tab.
See https://github.com/parrt/dtreeviz for info on how to properly install graphviz
for dtreeviz.
""")
self._graphviz_available = False
else:
self._graphviz_available = True
return self._graphviz_available
@property
def decision_trees(self):
"""a list of ShadowDecTree objects"""
if not hasattr(self, '_decision_trees'):
print("Calculating ShadowDecTree for each individual decision tree...", flush=True)
self._decision_trees = [
ShadowDecTree.get_shadow_tree(self.model.get_booster(),
self.X,
self.y,
feature_names=self.X.columns.tolist(),
target_name='target',
class_names = self.labels if self.is_classifier else None,
tree_index=i)
for i in range(len(self.model_dump_list))]
return self._decision_trees
def decisiontree_df(self, tree_idx, index, pos_label=None):
"""dataframe with all decision nodes of a particular decision tree
Args:
tree_idx: the n'th tree in the random forest
index: row index
round: (Default value = 2)
pos_label: positive class (Default value = None)
Returns:
dataframe with summary of the decision tree path
"""
assert tree_idx >= 0 and tree_idx < self.no_of_trees, \
f"tree index {tree_idx} outside 0 and number of trees ({len(self.decision_trees)}) range"
idx = self.get_int_idx(index)
assert idx >= 0 and idx < len(self.X), \
f"=index {idx} outside 0 and size of X ({len(self.X)}) range"
if self.is_classifier:
if pos_label is None:
pos_label = self.pos_label
if len(self.labels) > 2:
tree_idx = tree_idx * len(self.labels) + pos_label
return get_xgboost_path_df(self.model_dump_list[tree_idx], self.X.iloc[idx])
def decisiontree_summary_df(self, tree_idx, index, round=2, pos_label=None):
"""formats decisiontree_df in a slightly more human readable format.
Args:
tree_idx: the n'th tree in the random forest
index: row index
round: (Default value = 2)
pos_label: positive class (Default value = None)
Returns:
dataframe with summary of the decision tree path
"""
idx = self.get_int_idx(index)
return get_xgboost_path_summary_df(self.decisiontree_df(tree_idx, idx, pos_label=pos_label))
def decision_path_file(self, tree_idx, index, show_just_path=False, pos_label=None):
"""get a dtreeviz visualization of a particular tree in the random forest.
Args:
tree_idx: the n'th tree in the random forest
index: row index
show_just_path (bool, optional): show only the path not rest of the
tree. Defaults to False.
pos_label: for classifiers, positive label class
Returns:
the path where the .svg file is stored.
"""
if not self.graphviz_available:
print("No graphviz 'dot' executable available!")
return None
idx = self.get_int_idx(index)
if self.is_classifier:
if pos_label is None:
pos_label = self.pos_label
if len(self.labels) > 2:
tree_idx = tree_idx * len(self.labels) + pos_label
viz = dtreeviz(self.decision_trees[tree_idx],
X=self.X.iloc[idx],
fancy=False,
show_node_labels = False,
show_just_path=show_just_path)
return viz.save_svg()
def decision_path(self, tree_idx, index, show_just_path=False, pos_label=None):
"""get a dtreeviz visualization of a particular tree in the random forest.
Args:
tree_idx: the n'th tree in the random forest
index: row index
show_just_path (bool, optional): show only the path not rest of the
tree. Defaults to False.
Returns:
a IPython display SVG object for e.g. jupyter notebook.
"""
if not self.graphviz_available:
print("No graphviz 'dot' executable available!")
return None
from IPython.display import SVG
svg_file = self.decision_path_file(tree_idx, index, show_just_path, pos_label)
return SVG(open(svg_file,'rb').read())
def decision_path_encoded(self, tree_idx, index, show_just_path=False, pos_label=None):
"""get a dtreeviz visualization of a particular tree in the random forest.
Args:
tree_idx: the n'th tree in the random forest
index: row index
show_just_path (bool, optional): show only the path not rest of the
tree. Defaults to False.
Returns:
a base64 encoded image, for inclusion in websites (e.g. dashboard)
"""
if not self.graphviz_available:
print("No graphviz 'dot' executable available!")
return None
svg_file = self.decision_path_file(tree_idx, index, show_just_path, pos_label)
encoded = base64.b64encode(open(svg_file,'rb').read())
svg_encoded = 'data:image/svg+xml;base64,{}'.format(encoded.decode())
return svg_encoded
def plot_trees(self, index, highlight_tree=None, round=2,
higher_is_better=True, pos_label=None):
"""plot barchart predictions of each individual prediction tree
Args:
index: index to display predictions for
highlight_tree: tree to highlight in plot (Default value = None)
round: rounding of numbers in plot (Default value = 2)
higher_is_better (bool, optional): up is green, down is red. If False
flip the colors.
pos_label: positive class (Default value = None)
Returns:
"""
idx=self.get_int_idx(index)
assert idx is not None, 'invalid index'
if self.is_classifier:
if pos_label is None:
pos_label = self.pos_label
y = self.y_binary(pos_label)[idx]
xgboost_preds_df = get_xgboost_preds_df(
self.model, self.X.iloc[[idx]], pos_label=pos_label)
return plotly_xgboost_trees(xgboost_preds_df,
y=y,
highlight_tree=highlight_tree,
target=self.target,
higher_is_better=higher_is_better)
else:
y = self.y[idx]
xgboost_preds_df = get_xgboost_preds_df(
self.model, self.X.iloc[[idx]])
return plotly_xgboost_trees(xgboost_preds_df,
y=y, highlight_tree=highlight_tree,
target=self.target, units=self.units,
higher_is_better=higher_is_better)
def calculate_properties(self, include_interactions=True):
"""
Args:
include_interactions: If False do not calculate shap interaction value
(Default value = True)
Returns:
"""
_ = self.decision_trees, self.model_dump_list
super().calculate_properties(include_interactions=include_interactions)
class RandomForestClassifierExplainer(RandomForestExplainer, ClassifierExplainer):
"""RandomForestClassifierExplainer inherits from both RandomForestExplainer and
ClassifierExplainer.
"""
pass
class RandomForestRegressionExplainer(RandomForestExplainer, RegressionExplainer):
"""RandomForestRegressionExplainer inherits from both RandomForestExplainer and
RegressionExplainer.
"""
pass
class XGBClassifierExplainer(XGBExplainer, ClassifierExplainer):
"""RandomForestClassifierBunch inherits from both RandomForestExplainer and
ClassifierExplainer.
"""
pass
class XGBRegressionExplainer(XGBExplainer, RegressionExplainer):
"""XGBRegressionExplainer inherits from both XGBExplainer and
RegressionExplainer.
"""
pass
class ClassifierBunch:
""" """
def __init__(self, *args, **kwargs):
raise ValueError("ClassifierBunch has been deprecated, use ClassifierExplainer instead...")
class RegressionBunch:
""" """
def __init__(self, *args, **kwargs):
raise ValueError("RegressionBunch has been deprecated, use RegressionrExplainer instead...")
class RandomForestExplainerBunch:
""" """
def __init__(self, *args, **kwargs):
raise ValueError("RandomForestExplainerBunch has been deprecated, use RandomForestExplainer instead...")
class RandomForestClassifierBunch:
""" """
def __init__(self, *args, **kwargs):
raise ValueError("RandomForestClassifierBunch has been deprecated, use RandomForestClassifierExplainer instead...")
class RandomForestRegressionBunch:
""" """
def __init__(self, *args, **kwargs):
raise ValueError("RandomForestRegressionBunch has been deprecated, use RandomForestRegressionExplainer instead...") | AMLBID | /Explainer/explainerss.py | explainerss.py |
__all__ = ['ExplainerTabsLayout',
'ExplainerPageLayout',
'ExplainerDashboard',
'ExplainerHub',
'JupyterExplainerDashboard',
'ExplainerTab',
'JupyterExplainerTab',
'InlineExplainer']
import sys
import re
import json
import inspect
import requests
from typing import List, Union
from pathlib import Path
from copy import copy, deepcopy
import oyaml as yaml
import shortuuid
import dash
import dash_auth
import dash_core_components as dcc
import dash_html_components as html
import dash_bootstrap_components as dbc
from flask import Flask
from flask_simplelogin import SimpleLogin, login_required
from werkzeug.security import check_password_hash, generate_password_hash
from jupyter_dash import JupyterDash
import plotly.io as pio
from .dashboard_methods import instantiate_component
from .dashboard_components import *
from .dashboard_tabs import *
from .explainers import BaseExplainer
class ExplainerTabsLayout:
def __init__(self, explainer, tabs,
title='Model Explainer',
description=None,
header_hide_title=False,
header_hide_selector=False,
hide_poweredby=False,
block_selector_callbacks=False,
pos_label=None,
fluid=True,
**kwargs):
"""Generates a multi tab layout from a a list of ExplainerComponents.
If the component is a class definition, it gets instantiated first. If
the component is not derived from an ExplainerComponent, then attempt
with duck typing to nevertheless instantiate a layout.
Args:
explainer ([type]): explainer
tabs (list[ExplainerComponent class or instance]): list of
ExplainerComponent class definitions or instances.
title (str, optional): [description]. Defaults to 'Model Explainer'.
description (str, optional): description tooltip to add to the title.
header_hide_title (bool, optional): Hide the title. Defaults to False.
header_hide_selector (bool, optional): Hide the positive label selector.
Defaults to False.
hide_poweredby (bool, optional): hide the powered by footer
block_selector_callbacks (bool, optional): block the callback of the
pos label selector. Useful to avoid clashes when you
have your own PosLabelSelector in your layout.
Defaults to False.
pos_label ({int, str}, optional): initial pos label.
Defaults to explainer.pos_label
fluid (bool, optional): Stretch layout to fill space. Defaults to False.
"""
self.title = title
self.description = description
self.header_hide_title = header_hide_title
self.header_hide_selector = header_hide_selector
self.block_selector_callbacks = block_selector_callbacks
self.hide_poweredby = hide_poweredby
if self.block_selector_callbacks:
self.header_hide_selector = True
self.fluid = fluid
self.selector = PosLabelSelector(explainer, name="0", pos_label=pos_label)
self.tabs = [instantiate_component(tab, explainer, name=str(i+1), **kwargs) for i, tab in enumerate(tabs)]
assert len(self.tabs) > 0, 'When passing a list to tabs, need to pass at least one valid tab!'
self.connector = PosLabelConnector(self.selector, self.tabs)
def layout(self):
"""returns a multitab layout plus ExplainerHeader"""
return dbc.Container([
dbc.Row([
make_hideable(
dbc.Col([
html.H1(self.title, id='dashboard-title'),
dbc.Tooltip(self.description, target='dashboard-title')
], width="auto"), hide=self.header_hide_title),
make_hideable(
dbc.Col([
self.selector.layout()
], md=3), hide=self.header_hide_selector),
], justify="start", style=dict(marginBottom=10)),
dcc.Tabs(id="tabs", value=self.tabs[0].name,
children=[dcc.Tab(label=tab.title, id=tab.name, value=tab.name,
children=tab.layout()) for tab in self.tabs]),
dbc.Row([
make_hideable(
dbc.Col([
html.Div([
html.Small("powered by: "),
html.Small(html.A("explainerdashboard",
className="text-muted", target='_blank',
href="https://github.com/oegedijk/explainerdashboard"))
], className="text-right"),
]), hide=self.hide_poweredby),
], justify="end"),
], fluid=self.fluid)
def register_callbacks(self, app):
"""Registers callbacks for all tabs"""
for tab in self.tabs:
try:
tab.register_callbacks(app)
except AttributeError:
print(f"Warning: {tab} does not have a register_callbacks method!")
if not self.block_selector_callbacks:
if any([tab.has_pos_label_connector() for tab in self.tabs]):
print("Warning: detected PosLabelConnectors already in the layout. "
"This may clash with the global pos label selector and generate duplicate callback errors. "
"If so set block_selector_callbacks=True.")
self.connector.register_callbacks(app)
def calculate_dependencies(self):
"""Calculates dependencies for all tabs"""
for tab in self.tabs:
try:
tab.calculate_dependencies()
except AttributeError:
print(f"Warning: {tab} does not have a calculate_dependencies method!")
class ExplainerPageLayout(ExplainerComponent):
def __init__(self, explainer, component,
title='Model Explainer',
description=None,
header_hide_title=False,
header_hide_selector=False,
hide_poweredby=False,
block_selector_callbacks=False,
pos_label=None,
fluid=False,
**kwargs):
"""Generates a single page layout from a single ExplainerComponent.
If the component is a class definition, it gets instantiated.
If the component is not derived from an ExplainerComponent, then tries
with duck typing to nevertheless instantiate a layout.
Args:
explainer ([type]): explainer
component (ExplainerComponent class or instance): ExplainerComponent
class definition or instance.
title (str, optional): Defaults to 'Model Explainer'.
description (str, optional): Will be displayed as title tooltip.
header_hide_title (bool, optional): Hide the title. Defaults to False.
header_hide_selector (bool, optional): Hide the positive label selector.
Defaults to False.
hide_poweredby (bool, optional): hide the powered by footer
block_selector_callbacks (bool, optional): block the callback of the
pos label selector. Useful to avoid clashes when you
have your own PosLabelSelector in your layout.
Defaults to False.
pos_label ({int, str}, optional): initial pos label.
Defaults to explainer.pos_label
fluid (bool, optional): Stretch layout to fill space. Defaults to False.
"""
self.title = title
self.description = description
self.header_hide_title = header_hide_title
self.header_hide_selector = header_hide_selector
self.hide_poweredby = hide_poweredby
self.block_selector_callbacks = block_selector_callbacks
if self.block_selector_callbacks:
self.header_hide_selector = True
self.fluid = fluid
self.selector = PosLabelSelector(explainer, name="0", pos_label=pos_label)
self.page = instantiate_component(component, explainer, name="1", **kwargs)
self.connector = PosLabelConnector(self.selector, self.page)
self.fluid = fluid
def layout(self):
"""returns single page layout with an ExplainerHeader"""
return dbc.Container([
dbc.Row([
make_hideable(
dbc.Col([
html.H1(self.title, id='dashboard-title'),
dbc.Tooltip(self.description, target='dashboard-title')
], width="auto"), hide=self.header_hide_title),
make_hideable(
dbc.Col([
self.selector.layout()
], md=3), hide=self.header_hide_selector),
], justify="start"),
self.page.layout()
], fluid=self.fluid)
def register_callbacks(self, app):
"""Register callbacks of page"""
try:
self.page.register_callbacks(app)
except AttributeError:
print(f"Warning: {self.page} does not have a register_callbacks method!")
if not self.block_selector_callbacks:
if hasattr(self.page, "has_pos_label_connector") and self.page.has_pos_label_connector():
print("Warning: detected PosLabelConnectors already in the layout. "
"This may clash with the global pos label selector and generate duplicate callback errors. "
"If so set block_selector_callbacks=True.")
self.connector.register_callbacks(app)
def calculate_dependencies(self):
"""Calculate dependencies of page"""
try:
self.page.calculate_dependencies()
except AttributeError:
print(f"Warning: {self.page} does not have a calculate_dependencies method!", flush=True)
class ExplainerDashboard:
def __init__(self, explainer=None, tabs=None,
title='Model Explainer',
name=None,
description=None,
hide_header=False,
header_hide_title=False,
header_hide_selector=False,
hide_poweredby=False,
block_selector_callbacks=False,
pos_label=None,
fluid=True,
mode="dash",
width=1000,
height=800,
bootstrap=None,
external_stylesheets=None,
server=True,
url_base_pathname=None,
responsive=True,
logins=None,
port=8050,
importances=True,
model_summary=True,
contributions=True,
whatif=True,
shap_dependence=True,
shap_interaction=True,
decision_trees=True,
**kwargs):
"""Creates an explainerdashboard out of an Explainer object.
single page dashboard:
If tabs is a single ExplainerComponent class or instance, display it
as a standalone page without tabs.
Multi tab dashboard:
If tabs is a list of ExplainerComponent classes or instances, then construct
a layout with a tab per component. Instead of components you can also pass
the following strings: "importances", "model_summary", "contributions",
"shap_dependence", "shap_interaction" or "decision_trees". You can mix and
combine these different modularities, e.g.:
tabs=[ImportancesTab, "contributions", custom_tab]
If tabs is None, then construct tabs based on the boolean parameters:
importances, model_summary, contributions, shap_dependence,
shap_interaction and decision_trees, which all default to True.
You can select four different modes:
- 'dash': standard dash.Dash() app
- 'inline': JupyterDash app inline in a notebook cell output
- 'jupyterlab': JupyterDash app in jupyterlab pane
- 'external': JupyterDash app in external tab
You can switch off the title and positive label selector
with header_hide_title=True and header_hide_selector=True.
You run the dashboard
with e.g. ExplainerDashboard(explainer).run(port=8050)
Args:
explainer(): explainer object
tabs(): single component or list of components
title(str, optional): title of dashboard, defaults to 'Model Explainer'
name (str): name of the dashboard. Used for assigning url in ExplainerHub.
description (str): summary for dashboard. Gets used for title tooltip and
in description for ExplainerHub.
hide_header (bool, optional) hide the header (title+selector), defaults to False.
header_hide_title(bool, optional): hide the title, defaults to False
header_hide_selector(bool, optional): hide the positive class selector for classifier models, defaults, to False
hide_poweredby (bool, optional): hide the powered by footer
block_selector_callbacks (bool, optional): block the callback of the
pos label selector. Useful to avoid clashes when you
have your own PosLabelSelector in your layout.
Defaults to False.
pos_label ({int, str}, optional): initial pos label.
Defaults to explainer.pos_label
mode(str, {'dash', 'inline' , 'jupyterlab', 'external'}, optional):
type of dash server to start. 'inline' runs in a jupyter notebook output cell.
'jupyterlab' runs in a jupyterlab pane. 'external' runs in an external tab
while keeping the notebook interactive.
fluid(bool, optional): whether to stretch the layout to available space.
Defaults to True.
width(int, optional): width of notebook output cell in pixels, defaults to 1000.
height(int, optional): height of notebookn output cell in pixels, defaults to 800.
bootstrap (str, optional): link to bootstrap url. Can use dbc.themese
to generate the url, e.g. bootstrap=dbc.themes.FLATLY. Defaults
to default bootstrap theme that is stored in the /assets folder
so that it works even behind a firewall.
external_stylesheets(list, optional): additional external stylesheets
to add. (for themes use the bootstrap parameter)
server (Flask instance or bool): either an instance of an existing Flask
server to tie the dashboard to, or True in which case a new Flask
server is created.
url_base_pathname (str): url_base_pathname for dashboard,
e.g. "/dashboard". Defaults to None.
responsive (bool): make layout responsive to viewport size
(i.e. reorganize bootstrap columns on small devices). Set to False
when e.g. testing with a headless browser. Defaults to True.
logins (list of lists): list of (hardcoded) logins, e.g.
[['login1', 'password1'], ['login2', 'password2']].
Defaults to None (no login required)
importances(bool, optional): include ImportancesTab, defaults to True.
model_summary(bool, optional): include ModelSummaryTab, defaults to True.
contributions(bool, optional): include ContributionsTab, defaults to True.
whatif (bool, optional): include WhatIfTab, defaults to True.
shap_dependence(bool, optional): include ShapDependenceTab, defaults to True.
shap_interaction(bool, optional): include InteractionsTab if model allows it, defaults to True.
decision_trees(bool, optional): include DecisionTreesTab if model allows it, defaults to True.
"""
print("Building ExplainerDashboard..", flush=True)
self._store_params(no_param=['explainer', 'tabs', 'server'])
self._stored_params['tabs'] = self._tabs_to_yaml(tabs)
if self.description is None:
self.description = """This dashboard shows the workings of a fitted
machine learning model, and explains its predictions."""
self._stored_params['description'] = self.description
if self.hide_header:
self.header_hide_title = True
self.header_hide_selector = True
try:
ipython_kernel = str(get_ipython())
self.is_notebook = True
self.is_colab = True if 'google.colab' in ipython_kernel else False
except:
self.is_notebook, self.is_colab = False, False
if self.mode == 'dash' and self.is_colab:
print("Detected google colab environment, setting mode='external'", flush=True)
self.mode = 'external'
elif self.mode == 'dash' and self.is_notebook:
print("Detected notebook environment, consider setting "
"mode='external', mode='inline' or mode='jupyterlab' "
"to keep the notebook interactive while the dashboard "
"is running...", flush=True)
if self.bootstrap is not None:
bootstrap_theme = self.bootstrap if isinstance(self.bootstrap, str) else dbc.themes.BOOTSTRAP
if self.external_stylesheets is None:
self.external_stylesheets = [bootstrap_theme]
else:
self.external_stylesheets.append(bootstrap_theme)
self.app = self._get_dash_app()
if logins is not None:
if len(logins)==2 and isinstance(logins[0], str) and isinstance(logins[1], str):
logins = [logins]
assert isinstance(logins, list), \
("Parameter logins should be a list of lists of str pairs, e.g."
" logins=[['user1', 'password1'], ['user2', 'password2']]!")
for login in logins:
assert isinstance(login, list), \
("Parameter logins should be a list of lists of str pairs, "
"e.g. logins=[['user1', 'password1'], ['user2', 'password2']]!")
assert isinstance(login[0], str) and isinstance(login[1], str), \
("For logins such as [['user1', 'password1']] user1 and "
"password1 should be type(str)!")
self.auth = dash_auth.BasicAuth(self.app, logins)
self.app.title = title
assert 'BaseExplainer' in str(explainer.__class__.mro()), \
("explainer should be an instance of BaseExplainer, such as "
"ClassifierExplainer or RegressionExplainer!")
if self.explainer.cats_only:
print("Note: explainer contains a model and data that deal with "
"categorical features directly. Not all elements of the "
"ExplainerDashboard are compatible with such models, and "
"so setting the following **kwargs: "
"cats=True, hide_cats=True, shap_interaction=False", flush=True)
kwargs.update(dict(
cats=True, hide_cats=True, shap_interaction=False))
if kwargs:
print("**kwargs: Passing the following keyword arguments to all the dashboard"
f" ExplainerComponents: {', '.join([f'{k}={v}' for k,v in kwargs.items()])}...")
if tabs is None:
tabs = []
if model_summary and explainer.y_missing:
print("No y labels were passed to the Explainer, so setting"
" model_summary=False...", flush=True)
model_summary = False
if shap_interaction and (not explainer.interactions_should_work or self.explainer.cats_only):
print("For this type of model and model_output interactions don't "
"work, so setting shap_interaction=False...", flush=True)
shap_interaction = False
if decision_trees and not hasattr(explainer, 'is_tree_explainer'):
print("The explainer object has no decision_trees property. so "
"setting decision_trees=False...", flush=True)
decision_trees = False
if importances:
tabs.append(ImportancesComposite)
if model_summary:
tabs.append(ClassifierModelStatsComposite if explainer.is_classifier else RegressionModelStatsComposite)
if contributions:
tabs.append(IndividualPredictionsComposite)
if whatif:
tabs.append(WhatIfComposite)
if shap_dependence:
tabs.append(ShapDependenceComposite)
if shap_interaction:
print("Warning: calculating shap interaction values can be slow! "
"Pass shap_interaction=False to remove interactions tab.",
flush=True)
tabs.append(ShapInteractionsComposite)
if decision_trees:
tabs.append(DecisionTreesComposite)
if isinstance(tabs, list) and len(tabs)==1:
tabs = tabs[0]
print("Generating layout...")
if isinstance(tabs, list):
tabs = [self._convert_str_tabs(tab) for tab in tabs]
self.explainer_layout = ExplainerTabsLayout(explainer, tabs, title,
description=self.description,
**update_kwargs(kwargs,
header_hide_title=self.header_hide_title,
header_hide_selector=self.header_hide_selector,
hide_poweredby=self.hide_poweredby,
block_selector_callbacks=self.block_selector_callbacks,
pos_label=self.pos_label,
fluid=fluid))
else:
tabs = self._convert_str_tabs(tabs)
self.explainer_layout = ExplainerPageLayout(explainer, tabs, title,
description=self.description,
**update_kwargs(kwargs,
header_hide_title=self.header_hide_title,
header_hide_selector=self.header_hide_selector,
hide_poweredby=self.hide_poweredby,
block_selector_callbacks=self.block_selector_callbacks,
pos_label=self.pos_label,
fluid=self.fluid))
self.app.layout = self.explainer_layout.layout()
print("Calculating dependencies...", flush=True)
self.explainer_layout.calculate_dependencies()
print("Reminder: you can store the explainer (including calculated "
"dependencies) with explainer.dump('explainer.joblib') and "
"reload with e.g. ClassifierExplainer.from_file('explainer.joblib')",
flush=True)
print("Registering callbacks...", flush=True)
self.explainer_layout.register_callbacks(self.app)
@classmethod
def from_config(cls, arg1, arg2=None, **update_params):
"""Loading a dashboard from a configuration .yaml file. You can either
pass both an explainer and a yaml file generated with
ExplainerDashboard.to_yaml("dashboard.yaml"):
db = ExplainerDashboard.from_config(explainer, "dashboard.yaml")
When you specify an explainerfile in to_yaml with
ExplainerDashboard.to_yaml("dashboard.yaml", explainerfile="explainer.joblib"),
you can also pass just the .yaml:
db = ExplainerDashboard.from_config("dashboard.yaml")
You can also load the explainerfile seperately:
db = ExplainerDashboard.from_config("explainer.joblib", "dashboard.yaml")
Args:
arg1 (explainer or config): arg1 should either be a config (yaml or dict),
or an explainer (instance or str/Path).
arg2 ([type], optional): If arg1 is an explainer, arg2 should be config.
update_params (dict): You can override parameters in the the yaml
config by passing additional kwargs to .from_config()
Returns:
ExplainerDashboard
"""
if arg2 is None:
if isinstance(arg1, (Path, str)) and str(arg1).endswith(".yaml"):
config = yaml.safe_load(open(str(arg1), "r"))
elif isinstance(arg1, dict):
config = arg1
assert 'dashboard' in config, \
".yaml file does not have `dashboard` param."
assert 'explainerfile' in config['dashboard'], \
".yaml file does not have explainerfile param"
explainer = BaseExplainer.from_file(config['dashboard']['explainerfile'])
else:
if isinstance(arg1, BaseExplainer):
explainer = arg1
elif isinstance(arg1, (Path, str)) and (
str(arg1).endswith(".joblib") or
str(arg1).endswith(".pkl") or str(arg1).endswith(".dill")):
explainer = BaseExplainer.from_file(arg1)
else:
raise ValueError(
"When passing two arguments to ExplainerDashboard.from_config(arg1, arg2), "
"arg1 should either be an explainer or an explainer filename (e.g. 'explainer.joblib')!")
if isinstance(arg2, (Path, str)) and str(arg2).endswith(".yaml"):
config = yaml.safe_load(open(str(arg2), "r"))
elif isinstance(arg2, dict):
config = arg2
else:
raise ValueError(
"When passing two arguments to ExplainerDashboard.from_config(arg1, arg2), "
"arg2 should be a .yaml file or a dict!")
dashboard_params = config['dashboard']['params']
for k, v in update_params.items():
if k in dashboard_params:
dashboard_params[k] = v
elif 'kwargs' in dashboard_params:
dashboard_params['kwargs'][k] = v
else:
dashboard_params['kwargs'] = dict(k=v)
if 'kwargs' in dashboard_params:
kwargs = dashboard_params.pop('kwargs')
else:
kwargs = {}
if 'tabs' in dashboard_params:
tabs = cls._yamltabs_to_tabs(dashboard_params['tabs'], explainer)
del dashboard_params['tabs']
return cls(explainer, tabs, **dashboard_params, **kwargs)
else:
return cls(explainer, **dashboard_params, **kwargs)
def to_yaml(self, filepath=None, return_dict=False,
explainerfile=None, dump_explainer=False):
"""Returns a yaml configuration of the current ExplainerDashboard
that can be used by the explainerdashboard CLI. Recommended filename
is `dashboard.yaml`.
Args:
filepath ({str, Path}, optional): Filepath to dump yaml. If None
returns the yaml as a string. Defaults to None.
return_dict (bool, optional): instead of yaml return dict with
config.
explainerfile (str, optional): filename of explainer dump. Defaults
to `explainer.joblib`.
dump_explainer (bool, optional): dump the explainer along with the yaml.
You must pass explainerfile parameter for the filename. Defaults to False.
"""
import oyaml as yaml
dashboard_config = dict(
dashboard=dict(
explainerfile=str(explainerfile),
params=self._stored_params))
if dump_explainer:
if explainerfile is None:
raise ValueError("When you pass dump_explainer=True, then you "
"must pass an explainerfile filename parameter!")
print(f"Dumping explainer to {explainerfile}...", flush=True)
self.explainer.dump(explainerfile)
if return_dict:
return dashboard_config
if filepath is not None:
yaml.dump(dashboard_config, open(filepath, "w"))
return
return yaml.dump(dashboard_config)
def _store_params(self, no_store=None, no_attr=None, no_param=None):
"""Stores the parameter of the class to instance attributes and
to a ._stored_params dict. You can optionall exclude all or some
parameters from being stored.
Args:
no_store ({bool, List[str]}, optional): If True do not store any
parameters to either attribute or _stored_params dict. If
a list of str, then do not store parameters with those names.
Defaults to None.
no_attr ({bool, List[str]},, optional): . If True do not store any
parameters to class attribute. If
a list of str, then do not store parameters with those names.
Defaults to None.
no_param ({bool, List[str]},, optional): If True do not store any
parameters to _stored_params dict. If
a list of str, then do not store parameters with those names.
Defaults to None.
"""
if not hasattr(self, '_stored_params'):
self._stored_params = {}
frame = sys._getframe(1)
args = frame.f_code.co_varnames[1:frame.f_code.co_argcount]
args_dict = {arg: frame.f_locals[arg] for arg in args}
if 'kwargs' in frame.f_locals:
args_dict['kwargs'] = frame.f_locals['kwargs']
if isinstance(no_store, bool) and no_store:
return
else:
if no_store is None: no_store = tuple()
if isinstance(no_attr, bool) and no_attr: dont_attr = True
else:
if no_attr is None: no_attr = tuple()
dont_attr = False
if isinstance(no_param, bool) and no_param: dont_param = True
else:
if no_param is None: no_param = tuple()
dont_param = False
for name, value in args_dict.items():
if not dont_attr and name not in no_store and name not in no_attr:
setattr(self, name, value)
if not dont_param and name not in no_store and name not in no_param:
self._stored_params[name] = value
def _convert_str_tabs(self, component):
if isinstance(component, str):
if component == 'importances':
return ImportancesTab
elif component == 'model_summary':
return ModelSummaryTab
elif component == 'contributions':
return ContributionsTab
elif component == 'whatif':
return WhatIfTab
elif component == 'shap_dependence':
return ShapDependenceTab
elif component == 'shap_interaction':
return ShapInteractionsTab
elif component == 'decision_trees':
return DecisionTreesTab
return component
@staticmethod
def _tabs_to_yaml(tabs):
"""converts tabs to a yaml friendly format"""
if tabs is None:
return None
def get_name_and_module(component):
if inspect.isclass(component) and issubclass(component, ExplainerComponent):
return dict(
name=component.__name__,
module=component.__module__,
params=None
)
elif isinstance(component, ExplainerComponent):
component_imports = dict(component.component_imports)
del component_imports[component.__class__.__name__]
return dict(
name=component.__class__.__name__,
module=component.__class__.__module__,
params=component._stored_params,
component_imports = component_imports
)
else:
raise ValueError(f"Please only pass strings or ExplainerComponents to parameter `tabs`!"
"You passed {component.__class__}")
if not hasattr(tabs, "__iter__"):
return tabs if isinstance(tabs, str) else get_name_and_module(tabs)
return [tab if isinstance(tab, str) else get_name_and_module(tab) for tab in tabs]
@staticmethod
def _yamltabs_to_tabs(yamltabs, explainer):
"""converts a yaml tabs list back to ExplainerDashboard compatible original"""
from importlib import import_module
if yamltabs is None:
return None
def instantiate_tab(tab, explainer, name=None):
if isinstance(tab, str):
return tab
elif isinstance(tab, dict):
print(tab)
if 'component_imports' in tab and tab['component_imports'] is not None:
for class_name, module_name in tab['component_imports'].items():
if class_name not in globals():
import_module(class_module, class_name)
tab_class = getattr(import_module(tab['module']), tab['name'])
if tab['params'] is None:
return tab_class
else:
if not 'name' in tab['params'] or tab['params']['name'] is None:
tab['params']['name'] = name
tab_instance = tab_class(explainer, **tab['params'])
return tab_instance
else:
raise ValueError("yaml tab should be either string, e.g. 'importances', "
"or a dict(name=..,module=..,params=...)")
if not hasattr(yamltabs, "__iter__"):
return instantiate_tab(yamltabs, explainer, name="1")
tabs = [instantiate_tab(tab, explainer, name=str(i+1)) for i, tab in enumerate(yamltabs)]
print(tabs)
return tabs
def _get_dash_app(self):
if self.responsive:
meta_tags = [
{'name': 'viewport',
'content': 'width=device-width, initial-scale=1.0, maximum-scale=1.2, minimum-scale=0.5,'}
]
else:
meta_tags = None
if self.bootstrap is not None:
assets_ignore = '^bootstrap.min.css$'
else:
assets_ignore = ""
if self.mode=="dash":
app = dash.Dash(__name__,
server=self.server,
external_stylesheets=self.external_stylesheets,
assets_ignore=assets_ignore,
url_base_pathname=self.url_base_pathname,
meta_tags=meta_tags)
elif self.mode in ['inline', 'jupyterlab', 'external']:
app = JupyterDash(__name__,
external_stylesheets=self.external_stylesheets,
assets_ignore=assets_ignore,
meta_tags=meta_tags)
else:
raise ValueError(f"mode=={self.mode} but should be in "
"{'dash', 'inline', 'juypyterlab', 'external'}")
app.config['suppress_callback_exceptions'] = True
app.scripts.config.serve_locally = True
app.css.config.serve_locally = True
return app
def flask_server(self):
"""returns self.app.server so that it can be exposed to e.g. gunicorn"""
if self.mode != 'dash':
print("Warning: in production you should probably use mode='dash'...")
return self.app.server
def run(self, port=None, host='0.0.0.0', use_waitress=False, mode=None, **kwargs):
"""Start ExplainerDashboard on port
Args:
port (int, optional): port to run on. If None, then use self.port.
host (str, optional): host to run on. Defaults to '0.0.0.0'.
use_waitress (bool, optional): use the waitress python web server
instead of the flask development server. Only works with mode='dash'.
Defaults to False.
mode(str, {'dash', 'inline' , 'jupyterlab', 'external'}, optional):
Type of dash server to start. 'inline' runs in a jupyter notebook output cell.
'jupyterlab' runs in a jupyterlab pane. 'external' runs in an external tab
while keeping the notebook interactive. 'dash' is the default server.
Overrides self.mode, in which case the dashboard will get
rebuilt before running it with the right type of dash server.
(dash.Dash or JupyterDash). Defaults to None (i.e. self.mode)
Defaults to None.self.port defaults to 8050.
Raises:
ValueError: if mode is unknown
"""
pio.templates.default = "none"
if port is None:
port = self.port
if mode is None:
mode = self.mode
if use_waitress and mode != 'dash':
print(f"Warning: waitress does not work with mode={self.mode}, "
"using JupyterDash server instead!", flush=True)
if mode == 'dash':
if self.mode != 'dash':
print("Warning: Original ExplainerDashboard was not initialized "
"with mode='dash'. Rebuilding dashboard before launch:", flush=True)
app = ExplainerDashboard.from_config(
self.explainer, self.to_yaml(return_dict=True), mode='dash').app
else:
app = self.app
print(f"Starting ExplainerDashboard on http://localhost:{port}", flush=True)
if use_waitress:
from waitress import serve
serve(app.server, host=host, port=port)
else:
app.run_server(port=port, host=host, **kwargs)
else:
if self.mode == 'dash':
print("Warning: Original ExplainerDashboard was initialized "
"with mode='dash'. Rebuilding dashboard before launch:", flush=True)
app = ExplainerDashboard.from_config(
self.explainer, self.to_yaml(return_dict=True), mode=mode).app
else:
app = self.app
if mode == 'external':
if not self.is_colab:
print(f"Starting ExplainerDashboard on http://localhost:{port}\n"
"You can terminate the dashboard with "
f"ExplainerDashboard.terminate({port})", flush=True)
app.run_server(port=port, mode=mode, **kwargs)
elif mode in ['inline', 'jupyterlab']:
print(f"Starting ExplainerDashboard inline (terminate it with "
f"ExplainerDashboard.terminate({port}))", flush=True)
app.run_server(port=port, mode=mode,
width=self.width, height=self.height, **kwargs)
else:
raise ValueError(f"Unknown mode: mode='{mode}'!")
@classmethod
def terminate(cls, port, token=None):
"""
Classmethodd to terminate any JupyterDash dashboard (so started with
mode='inline', mode='external' or mode='jupyterlab') from any
ExplainerDashboard by specifying the right port.
Example:
ExplainerDashboard(explainer, mode='external').run(port=8050)
ExplainerDashboard.terminate(8050)
Args:
port (int): port on which the dashboard is running.
token (str, optional): JupyterDash._token class property.
Defaults to the _token of the JupyterDash in the current namespace.
Raises:
ValueError: if can't find the port to terminate.
"""
if token is None:
token = JupyterDash._token
shutdown_url = f"http://localhost:{port}/_shutdown_{token}"
print(f"Trying to shut down dashboard on port {port}...")
try:
response = requests.get(shutdown_url)
except Exception as e:
print(f"Something seems to have failed: {e}")
class ExplainerHub:
"""ExplainerHub is a way to host multiple dashboards in a single point,
and manage access through adding user accounts.
Example:
``hub = ExplainerHub([db1, db2], logins=[['user', 'password']], secret_key="SECRET")``
``hub.run()``
A frontend is hosted at e.g. ``localhost:8050``, with summaries and links to
each individual dashboard. Each ExplainerDashboard is hosted on its own url path,
so that you can also find it directly, e.g.:
``localhost:8050/dashboard1`` and ``localhost:8050/dashboard2``.
You can store the hub configuration, dashboard configurations, explainers
and user database with a single command: ``hub.to_yaml('hub.yaml')``.
You can restore the hub with ``hub2 = ExplainerHub.from_config('hub.yaml')``
You can start the hub from the command line using the ``explainerhub`` CLI
command: ``$ explainerhub run hub.yaml``. You can also use the CLI to
add and delete users.
"""
def __init__(self, dashboards:List[ExplainerDashboard], title:str="ExplainerHub",
description:str=None, masonry:bool=False, n_dashboard_cols:int=3,
users_file:str="users.yaml", user_json=None,
logins:List[List]=None, db_users:dict=None,
dbs_open_by_default:bool=False, port:int=8050,
min_height:int=3000, secret_key:str=None, no_index:bool=False,
bootstrap:str=None, fluid:bool=True, **kwargs):
"""
Note:
Logins can be defined in multiple places: users.json, ExplainerHub.logins
and ExplainerDashboard.logins for each dashboard in dashboards.
When users with the same username are defined in multiple
locations then passwords are looked up in the following order:
hub.logins > dashboard.logins > user.json
Note:
**kwargs will be forwarded to each dashboard in dashboards.
Args:
dashboards (List[ExplainerDashboard]): list of ExplainerDashboard to
include in ExplainerHub.
title (str, optional): title to display. Defaults to "ExplainerHub".
description (str, optional): Short description of ExplainerHub.
Defaults to default text.
masonry (bool, optional): Lay out dashboard cards in fluid bootstrap
masonry responsive style. Defaults to False.
n_dashboard_cols (int, optional): If masonry is False, organize cards
in rows and columns. Defaults to 3 columns.
users_file (Path, optional): a .yaml or .json file used to store user and (hashed)
password data. Defaults to 'users.yaml'.
user_json (Path, optional): Deprecated! A .json file used to store user and (hashed)
password data. Defaults to 'users.json'. Was replaced by users_file which
can also be a more readable .yaml.
logins (List[List[str, str]], optional): List of ['login', 'password'] pairs,
e.g. logins = [['user1', 'password1'], ['user2', 'password2']]
db_users (dict, optional): dictionary limiting access to certain
dashboards to a subset of users, e.g
dict(dashboard1=['user1', 'user2'], dashboard2=['user3']).
dbs_open_by_default (bool, optional): Only force logins for dashboard
with defined db_users. All other dashboards and index no login
required. Default to False,
port (int, optional): Port to run hub on. Defaults to 8050.
min_height (int, optional) size of the iframe the holds the dashboard.
Defaults to 3000 pixels.
secret_key (str): Flask secret key to pass to dashboard in order to persist
logins. Defaults to a new random uuid string every time you start
the dashboard. (i.e. no persistence) You should store the secret
key somewhere save, e.g. in a environmental variable.
no_index (bool, optional): do not add the "/" route and "/_dashboard1"
etc routes, but only mount the dashboards on e.g. /dashboard1. This
allows you to add your own custom front_end.
bootstrap (str, optional): url with custom bootstrap css, e.g.
bootstrap=dbc.themes.FLATLY. Defaults to static bootstrap css.
fluid (bool, optional): Let the bootstrap container ill the entire width
of the browser. Defaults to True.
**kwargs: all kwargs will be forwarded to the constructors of
each dashboard in dashboards dashboards.
"""
self._store_params(no_store=['dashboards', 'logins', 'secret_key'])
if user_json is not None:
print("Warning: user_json has been deprecated, use users_file parameter instead!")
self.users_file = user_json
if self.description is None:
self.description = ("This ExplainerHub shows a number of ExplainerDashboards.\n"
"Each dashboard makes the inner workings and predictions of a trained machine "
"learning model transparent and explainable.")
self._stored_params['description'] = self.description
if (logins is not None and len(logins)==2
and isinstance(logins[0], str) and isinstance(logins[1], str)):
# if logins=['user', 'password'] then add the outer list
logins = [logins]
self.logins = self._hash_logins(logins)
self.db_users = db_users if db_users is not None else {}
self._validate_users_file(self.users_file)
self.app = Flask(__name__)
if secret_key is not None:
self.app.config['SECRET_KEY'] = secret_key
SimpleLogin(self.app, login_checker=self._validate_user)
self.dashboards = self._instantiate_dashboards(dashboards, **kwargs)
dashboard_names = [db.name for db in self.dashboards]
assert len(set(dashboard_names)) == len(dashboard_names), \
f"All dashboard .name properties should be unique, but received the folowing: {dashboard_names}"
illegal_names = list(set(dashboard_names) & {'login', 'logout', 'admin'})
assert not illegal_names, \
f"The following .name properties for dashboards are not allowed: {illegal_names}!"
if self.users:
for dashboard in self.dashboards:
if not self.dbs_open_by_default or dashboard.name in self.dashboards_with_users:
self._protect_dashviews(dashboard.app, username=self.get_dashboard_users(dashboard.name))
if not self.no_index:
self.index_page = self._get_index_page()
if self.users and not self.dbs_open_by_default:
self._protect_dashviews(self.index_page)
self._add_flask_routes(self.app)
@classmethod
def from_config(cls, config:Union[dict, str, Path], **update_params):
"""Instantiate an ExplainerHub based on a config file.
Args:
config (Union[dict, str, Path]): either a dict or a .yaml config
file to load
update_params: additional kwargs to override stored settings.
Returns:
ExplainerHub: new instance of ExplainerHub according to the config.
"""
if isinstance(config, (Path, str)) and str(config).endswith(".yaml"):
filepath = Path(config).parent
config = yaml.safe_load(open(str(Path(config)), "r"))
elif isinstance(config, dict):
config = deepcopy(config)
assert 'explainerhub' in config, \
"Misformed yaml: explainerhub yaml file should start with 'explainerhub:'!"
config = config['explainerhub']
def convert_db(db, filepath=None):
if isinstance(db, dict): return db
elif Path(db).is_absolute():
return Path(db)
else:
filepath = Path(filepath or Path.cwd())
return filepath / db
dashboards = [ExplainerDashboard.from_config(convert_db(db, filepath))
for db in config['dashboards']]
del config['dashboards']
config.update(config.pop('kwargs'))
return cls(dashboards, **update_kwargs(config, **update_params))
def to_yaml(self, filepath:Path=None, dump_explainers=True,
return_dict=False, integrate_dashboard_yamls=False):
"""Store ExplainerHub to configuration .yaml, store the users to users.json
and dump the underlying dashboard .yamls and explainers.
If filepath is None, does not store yaml config to file, but simply
return config yaml string.
If filepath provided and dump_explainers=True, then store all underlying
explainers to disk.
Args:
filepath (Path, optional): .yaml file filepath. Defaults to None.
dump_explainers (bool, optional): Store the explainers to disk
along with the .yaml file. Defaults to True.
return_dict (bool, optional): Instead of returning or storing yaml
return a configuration dictionary. Returns a single dict as if
separate_dashboard_yamls=True. Defaults to False.
integrate_dashboard_yamls(bool, optional): Do not generate an individual
.yaml file for each dashboard, but integrate them in hub.yaml.
Returns:
{dict, yaml, None}
"""
filepath = Path(filepath)
self._dump_all_users_to_file(filepath.parent / str(self.users_file))
# for login in self.logins.values():
# self._add_user_to_file(self.users_file,
# login['username'], login['password'],
# already_hashed=True)
# for dashboard, users in self.db_users.items():
# for user in users:
# self._add_user_to_dashboard_file(self.users_file, dashboard, user)
if filepath is None or return_dict or integrate_dashboard_yamls:
hub_config = dict(
explainerhub=dict(
**self._stored_params,
dashboards=[dashboard.to_yaml(
return_dict=True,
explainerfile=dashboard.name+"_explainer.joblib",
dump_explainer=dump_explainers)
for dashboard in self.dashboards]))
else:
for dashboard in self.dashboards:
print(f"Storing {dashboard.name}_dashboard.yaml...")
dashboard.to_yaml(filepath.parent / (dashboard.name+"_dashboard.yaml"),
explainerfile=filepath.parent / (dashboard.name+"_explainer.joblib"),
dump_explainer=dump_explainers)
hub_config = dict(
explainerhub=dict(
**self._stored_params,
dashboards=[dashboard.name+"_dashboard.yaml"
for dashboard in self.dashboards]))
if return_dict:
return hub_config
if filepath is None:
return yaml.dump(hub_config)
filepath = Path(filepath)
print(f"Storing {filepath}...")
yaml.dump(hub_config, open(filepath, "w"))
return
def _store_params(self, no_store=None, no_attr=None, no_param=None):
"""Stores the parameter of the class to instance attributes and
to a ._stored_params dict. You can optionall exclude all or some
parameters from being stored.
Args:
no_store ({bool, List[str]}, optional): If True do not store any
parameters to either attribute or _stored_params dict. If
a list of str, then do not store parameters with those names.
Defaults to None.
no_attr ({bool, List[str]},, optional): . If True do not store any
parameters to class attribute. If
a list of str, then do not store parameters with those names.
Defaults to None.
no_param ({bool, List[str]},, optional): If True do not store any
parameters to _stored_params dict. If
a list of str, then do not store parameters with those names.
Defaults to None.
"""
if not hasattr(self, '_stored_params'):
self._stored_params = {}
frame = sys._getframe(1)
args = frame.f_code.co_varnames[1:frame.f_code.co_argcount]
args_dict = {arg: frame.f_locals[arg] for arg in args}
if 'kwargs' in frame.f_locals:
args_dict['kwargs'] = frame.f_locals['kwargs']
if isinstance(no_store, bool) and no_store:
return
else:
if no_store is None: no_store = tuple()
if isinstance(no_attr, bool) and no_attr: dont_attr = True
else:
if no_attr is None: no_attr = tuple()
dont_attr = False
if isinstance(no_param, bool) and no_param: dont_param = True
else:
if no_param is None: no_param = tuple()
dont_param = False
for name, value in args_dict.items():
if not dont_attr and name not in no_store and name not in no_attr:
setattr(self, name, value)
if not dont_param and name not in no_store and name not in no_param:
self._stored_params[name] = value
def _instantiate_dashboards(self, dashboards, **kwargs):
"""Instantiate a list of dashboards and copy to logins to the ExplainerHub self.logins.
"""
dashboard_list = []
for i, dashboard in enumerate(dashboards):
if dashboard.name is None:
print("Reminder, you can set ExplainerDashboard .name and .description "
"in order to control the url path of the dashboard. Now "
f"defaulting to name=dashboard{i+1} and default description...", flush=True)
dashboard_name = f"dashboard{i+1}"
else:
dashboard_name = dashboard.name
update_params = dict(
server=self.app,
name=dashboard_name,
url_base_pathname = f"/{dashboard_name}/",
mode='dash'
)
if dashboard.logins is not None:
for user, password in dashboard.logins:
if user not in self.logins:
self.add_user(user, password)
else:
print(f"Warning: {user} in {dashboard.name} already in "
"ExplainerHub logins! So not adding to logins...")
self.add_user_to_dashboard(dashboard_name, user)
config = deepcopy(dashboard.to_yaml(return_dict=True))
config['dashboard']['params']['logins'] = None
dashboard_list.append(
ExplainerDashboard.from_config(
dashboard.explainer, config, **update_kwargs(kwargs, **update_params)))
return dashboard_list
@staticmethod
def _validate_users_file(users_file:Path):
"""validat that user_json is a well formed .json file.
If it does not exist, then create an empty .json file.
"""
if users_file is not None:
if not Path(users_file).exists():
users_db = dict(users={}, dashboard_users={})
if str(users_file).endswith(".json"):
json.dump(users_db, open(Path(users_file), 'w'))
elif str(users_file).endswith(".yaml"):
yaml.dump(users_db, open(Path(users_file), "w"))
if str(users_file).endswith(".json"):
users_db = json.load(open(Path(users_file)))
elif str(users_file).endswith(".yaml"):
users_db = yaml.safe_load(open(str(users_file), "r"))
else:
raise ValueError("users_file should end with either .json or .yaml!")
assert 'users' in users_db, \
f"{self.users_file} should contain a 'users' dict!"
assert 'dashboard_users' in users_db, \
f"{self.users_file} should contain a 'dashboard_users' dict!"
def _hash_logins(self, logins:List[List], add_to_users_file:bool=False):
"""Turn a list of [user, password] pairs into a Flask-Login style user
dictionary with hashed passwords. If passwords already in hash-form
then simply copy them.
Args:
logins (List[List]): List of logins e.g.
logins = [['user1', 'password1], ['user2', 'password2]]
add_to_users_file (bool, optional): Add the users to
users.yaml. Defaults to False.
Returns:
dict
"""
logins_dict = {}
if logins is None:
return logins_dict
regex=re.compile(r'^pbkdf2:sha256:[0-9]+\$[a-zA-Z0-9]+\$[a-z0-9]{64}$', re.IGNORECASE)
for username, password in logins:
if re.search(regex, password) is not None:
logins_dict[username] = dict(
username=username,
password=password
)
if add_to_user_file and self.users_file is not None:
self._add_user_to_file(self.users_file, user, password, already_hashed=True)
else:
logins_dict[username] = dict(
username=username,
password=generate_password_hash(password, method='pbkdf2:sha256')
)
if add_to_users_file and self.users_jfile is not None:
self._add_user_to_file(self.users_file, user, password)
return logins_dict
@staticmethod
def _load_users_db(users_file:Path):
if str(users_file).endswith(".json"):
users_db = json.load(open(Path(users_file)))
elif str(users_file).endswith(".yaml"):
users_db = yaml.safe_load(open(str(users_file), "r"))
else:
raise ValueError("users_file should end with either .json or .yaml!")
return users_db
@staticmethod
def _dump_users_db(users_db, users_file:Path):
if str(users_file).endswith(".json"):
json.dump(users_db, open(Path(users_file), 'w'))
elif str(users_file).endswith(".yaml"):
yaml.dump(users_db, open(Path(users_file), "w"))
else:
raise ValueError("users_file should end with either .json or .yaml!")
def _dump_all_users_to_file(self, output_users_file:Path=None):
"""Stores all users (both on file and in the instance) to single users_file.
Users in the instance overwrite users in users_file.
Args:
output_users_file (Path, optional): users_file to store users in.
By default equal to self.users_file.
"""
users_db = ExplainerHub._load_users_db(self.users_file)
users_db['users'].update(self.logins)
for db, instance_users in self.db_users.items():
file_users = users_db['dashboard_users'].get(db) or []
dashboard_users = sorted(list(set(file_users + instance_users)))
users_db['dashboard_users'][db] = dashboard_users
if output_users_file is None:
output_users_file = self.users_file
ExplainerHub._dump_users_db(users_db, output_users_file)
@staticmethod
def _add_user_to_file(users_file:Path, username:str, password:str, already_hashed=False):
"""Add a user to a user_json .json file.
Args:
user_json (Path): json file, e.g 'users.json'
username (str): username to add
password (str): password to add
already_hashed (bool, optional): If already hashed then do not hash
the password again. Defaults to False.
"""
users_db = ExplainerHub._load_users_db(users_file)
users_db['users'][username] = dict(
username=username,
password=password if already_hashed else generate_password_hash(password, method='pbkdf2:sha256')
)
ExplainerHub._dump_users_db(users_db, users_file)
@staticmethod
def _add_user_to_dashboard_file(users_file:Path, dashboard:str, username:str):
"""Add a user to dashboard_users inside a json file
Args:
user_json (Path): json file e.g. 'users.json'
dashboard (str): name of dashboard
username (str): username
"""
users_db = ExplainerHub._load_users_db(users_file)
dashboard_users = users_db['dashboard_users'].get(dashboard)
if dashboard_users is None:
dashboard_users = [username]
else:
dashboard_users = sorted(list(set(dashboard_users + [username])))
users_db['dashboard_users'][dashboard] = dashboard_users
ExplainerHub._dump_users_db(users_db, users_file)
@staticmethod
def _delete_user_from_file(users_file:Path, username:str):
"""delete user from user_json .json file.
Also removes user from all dashboard_user lists.
Args:
user_json (Path): json file e.g. 'users.json'
username (str): username to delete
"""
users_db = ExplainerHub._load_users_db(users_file)
try:
del users_db['users'][username]
except:
pass
for dashboard in users_db['dashboard_users'].keys():
dashboard_users = users_db['dashboard_users'].get(dashboard)
if dashboard_users is not None:
dashboard_users = sorted(list(set(dashboard_users) - {username}))
users_db['dashboard_users'][dashboard] = dashboard_users
ExplainerHub._dump_users_db(users_db, users_file)
@staticmethod
def _delete_user_from_dashboard_file(users_file:Path, dashboard:str, username:str):
"""remove a user from a specific dashboard_users list inside a users.json file
Args:
user_json (Path): json file, e.g. 'users.json'
dashboard (str): name of the dashboard
username (str): name of the user to remove
"""
users_db = ExplainerHub._load_users_db(users_file)
dashboard_users = users_db['dashboard_users'].get(dashboard)
if dashboard_users is not None:
dashboard_users = sorted(list(set(dashboard_users) - {username}))
if not dashboard_users:
del users_db['dashboard_users'][dashboard]
else:
users_db['dashboard_users'][dashboard] = dashboard_users
ExplainerHub._dump_users_db(users_db, users_file)
def add_user(self, username:str, password:str, add_to_users_file:bool=False):
"""add a user with username and password.
Args:
username (str): username
password (str): password
add_to_users_file(bool, optional): Add the user to the .yaml file defined
in self.users_file instead of to self.logins. Defaults to False.
"""
if add_to_users_file and self.users_file is not None:
self._add_user_to_file(self.users_file, username, password)
else:
self.logins[username] = dict(
username=username,
password=generate_password_hash(password, method='pbkdf2:sha256')
)
def add_user_to_dashboard(self, dashboard:str, username:str, add_to_users_file:bool=False):
"""add a user to a specific dashboard. If
Args:
dashboard (str): name of dashboard
username (str): user to add to dashboard
add_to_users_file (bool, optional): add the user to the .yaml or .json file defined
in self.users_file instead of to self.db_users. Defaults to False.
"""
if add_to_users_file and self.users_file is not None:
self._add_user_to_dashboard_file(self.users_file, dashboard, username)
else:
dashboard_users = self.db_users.get(dashboard)
dashboard_users = dashboard_users if dashboard_users is not None else []
dashboard_users = sorted(list(set(dashboard_users + [username])))
self.db_users[dashboard] = dashboard_users
@property
def users(self):
"""returns a list of all users, both in users.json and in self.logins"""
users = []
if self.users_file is not None and Path(self.users_file).exists():
users = list(self._load_users_db(self.users_file)['users'].keys())
if self.logins is not None:
users.extend(list(self.logins.keys()))
return users
@property
def dashboards_with_users(self):
"""returns a list of all dashboards that have a restricted list of users
that can access it"""
dashboards = []
if self.users_file is not None and Path(self.users_file).exists():
dashboards = list(self._load_users_db(self.users_file)['dashboard_users'].keys())
if self.logins is not None:
dashboards.extend(list(self.db_users.keys()))
return dashboards
@property
def dashboard_users(self):
"""return a dict with the list of users per dashboard"""
dashboard_users = {}
if self.users_file is not None and Path(self.users_file).exists():
dashboard_users.update(self._load_users_db(self.users_file)['dashboard_users'])
if self.db_users is not None:
for dashboard, users in self.db_users.items():
if not dashboard in dashboard_users:
dashboard_users[dashboard] = users
else:
dashboard_users[dashboard] = sorted(list(set(dashboard_users[dashboard] + users)))
return dashboard_users
def get_dashboard_users(self, dashboard:str):
"""return all users that have been approved to use a specific dashboard
Args:
dashboard (str): dashboard
Returns:
List
"""
dashboard_users = []
if self.users_file is not None:
file_users = self._load_users_db(self.users_file)['dashboard_users'].get(dashboard)
if file_users is not None:
dashboard_users = file_users
if self.db_users is not None:
param_users = self.db_users.get(dashboard)
if param_users is not None:
dashboard_users.extend(param_users)
dashboard_users = list(set(dashboard_users))
return dashboard_users
def _validate_user(self, user):
"""validation function for SimpleLogin. Returns True when user should
be given access (i.e. no users defined or password correct) and False
when user should be rejected.
Args:
user (dict(username, password)): dictionary with a username and
password key.
Returns:
bool
"""
if not self.users:
return True
users_db = self._load_users_db(self.users_file)['users'] if self.users_file is not None else {}
if not self.logins.get(user['username']) and not users_db.get(user['username']):
return False
if user['username'] in users_db:
stored_password = users_db[user['username']]['password']
else:
stored_password = self.logins[user['username']]['password']
if check_password_hash(stored_password, user['password']):
return True
return False
@staticmethod
def _protect_dashviews(dashapp:dash.Dash, username:List[str]=None):
"""Wraps a dash dashboard inside a login_required decorator to make sure
unauthorized viewers cannot access it.
Args:
dashapp (dash.Dash): [
username (List[str], optional): list of usernames that can access
this specific dashboard. Defaults to None (all registered users
can access)
"""
for view_func in dashapp.server.view_functions:
if view_func.startswith(dashapp.config.url_base_pathname):
dashapp.server.view_functions[view_func] = login_required(username=username)(
dashapp.server.view_functions[view_func])
def _get_index_page(self):
"""Returns the front end of ExplainerHub:
- title
- description
- links and description for each dashboard
Returns:
dbc.Container
"""
def dashboard_decks(dashboards, n_cols):
full_rows = int(len(dashboards)/ n_cols)
n_last_row = len(dashboards) % n_cols
card_decks = []
for i in range(0, full_rows*n_cols, n_cols):
card_decks.append(
[dbc.Card([
dbc.CardHeader([
html.H3(dashboard.title, className='card-title'),
]),
dbc.CardBody([
html.H6(dashboard.description),
]),
dbc.CardFooter([
dbc.CardLink("Go to dashboard",
href=f"/{dashboard.name}",
external_link=True),
])]) for dashboard in dashboards[i:i+n_cols]
]
)
if n_last_row > 0:
last_row = [
dbc.Card([
dbc.CardHeader([
html.H3(dashboard.title, className='card-title'),
]),
dbc.CardBody([
html.H6(dashboard.description),
]),
dbc.CardFooter([
dbc.CardLink("Go to dashboard",
href=f"/{dashboard.name}",
external_link=True),
])
]) for dashboard in dashboards[full_rows*n_cols:full_rows*n_cols+n_last_row]]
for i in range(len(last_row), n_cols):
last_row.append(dbc.Card([], style=dict(border="none")))
card_decks.append(last_row)
return card_decks
header = dbc.Jumbotron([
html.H1(self.title, className="display-3"),
html.Hr(className="my-2"),
html.P(self.description, className="lead"),
], style=dict(marginTop=40))
if self.masonry:
dashboard_rows = [
dbc.Row([
dbc.Col([
dbc.CardColumns(dashboard_cards(self.dashboards))
])
])
]
else:
dashboard_rows = [
dbc.Row([dbc.CardDeck(deck)], style=dict(marginBottom=30))
for deck in dashboard_decks(self.dashboards, self.n_dashboard_cols)]
index_page = dash.Dash(__name__,
server=self.app,
url_base_pathname="/index/",
external_stylesheets=[self.bootstrap] if self.bootstrap is not None else None)
index_page.title = self.title
index_page.layout = dbc.Container([
dbc.Row([dbc.Col([header])]),
dbc.Row([dbc.Col([html.H2("Dashboards:")])]),
*dashboard_rows
], fluid=self.fluid)
return index_page
def _hub_page(self, route):
"""Returns a html bootstrap wrapper around a particular flask route (hosting an ExplainerDashbaord)
It contains:
- a NavBar with links to all dashboards
- an iframe containing the flask route
- a footer with a link to github.com/oegedijk/explainerdashboard
"""
return f"""
<script type="text/javascript" src="/static/jquery-3.5.1.slim.min.js"></script>
<script type="text/javascript" src="/static/bootstrap.min.js"></script>
<link type="text/css" rel="stylesheet" href="{'/static/bootstrap.min.css' if self.bootstrap is None else self.bootstrap}"/>
<link rel="shortcut icon" href="/static/favicon.ico">
<title>{self.title}</title>
<body class="d-flex flex-column min-vh-100">
<div class="container{'-fluid' if self.fluid else ''}">
<nav class="navbar navbar-expand-lg navbar-light bg-light">
<a class="navbar-brand" href="/">{self.title}</a>
<button class="navbar-toggler" type="button" data-toggle="collapse" data-target="#navbarSupportedContent" aria-controls="navbarSupportedContent" aria-expanded="false" aria-label="Toggle navigation">
<span class="navbar-toggler-icon"></span>
</button>
<div class="collapse navbar-collapse justify-content-end" id="navbarSupportedContent">
<ul class="navbar-nav ml-auto">
<li class="nav-item dropdown">
<a class="nav-link dropdown-toggle" href="#" id="navbarDropdownMenuLink2" data-toggle="dropdown" aria-haspopup="true" aria-expanded="false">
Dashboards
</a>
<div class="dropdown-menu" aria-labelledby="navbarDropdownMenuLink2">
{"".join([f'<a class="dropdown-item" href="/_{db.name}">{db.title}</a>'
for db in self.dashboards])}
</div>
</li>
<li class="nav-item">
<a class="nav-link" href="/logout">Logout</a>
</li>
</ul>
</div>
</nav>
<div class="embed-responsive" style="min-height: {self.min_height}px">
<iframe
src="{route}"
style="overflow-x: hidden; overflow-y: visible; position: absolute; width: 95%; height: 100%; background: transparent"
></iframe>
</div>
</div>
</body>
"""
def _add_flask_routes(self, app):
""" adds the index route "/" with the index_page
and routes for each dashboard with a navbar and an iframe, e.g. "/_dashboard1"
If you pass no_index to the contructor, this method does not get called.
Args:
app (flask.Flask): flask app to add routes to.
"""
if self.users and not self.dbs_open_by_default:
@app.route("/")
@login_required
def index_route():
return self._hub_page("/index")
def dashboard_route(dashboard):
def inner():
return self._hub_page(f"/{dashboard.name}/")
inner.__name__ = "return_dashboard_"+dashboard.name
return inner
for dashboard in self.dashboards:
app.route(f"/_{dashboard.name}")(login_required(dashboard_route(dashboard)))
else:
@app.route("/")
def index_route():
return self._hub_page("/index")
def dashboard_route(dashboard):
def inner():
return self._hub_page(f"/{dashboard.name}/")
inner.__name__ = "return_dashboard_"+dashboard.name
return inner
for dashboard in self.dashboards:
app.route(f"/_{dashboard.name}")(dashboard_route(dashboard))
def flask_server(self):
"""return the Flask server inside the class instance"""
return self.app
def run(self, port=None, host='0.0.0.0', use_waitress=False, **kwargs):
"""start the ExplainerHub.
Args:
port (int, optional): Override default port. Defaults to None.
host (str, optional): host name to run dashboard. Defaults to '0.0.0.0'.
use_waitress (bool, optional): Use the waitress python web server
instead of the Flask development server. Defaults to False.
**kwargs: will be passed forward to either waitress.serve() or app.run()
"""
if port is None:
port = self.port
print(f"Starting ExplainerHub on http://{host}:{port}", flush=True)
if use_waitress:
import waitress
waitress.serve(self.app, host=host, port=port, **kwargs)
else:
self.app.run(host=host, port=port, **kwargs)
class InlineExplainer:
"""
Run a single tab inline in a Jupyter notebook using specific method calls.
"""
def __init__(self, explainer, mode='inline', width=1000, height=800,
port=8050, **kwargs):
"""
:param explainer: an Explainer object
:param mode: either 'inline', 'jupyterlab' or 'external'
:type mode: str, optional
:param width: width in pixels of inline iframe
:param height: height in pixels of inline iframe
:param port: port to run if mode='external'
"""
assert mode in ['inline', 'external', 'jupyterlab'], \
"mode should either be 'inline', 'external' or 'jupyterlab'!"
self._explainer = explainer
self._mode = mode
self._width = width
self._height = height
self._port = port
self._kwargs = kwargs
self.tab = InlineExplainerTabs(self, "tabs")
"""subclass with InlineExplainerTabs layouts, e.g. InlineExplainer(explainer).tab.modelsummary()"""
self.shap = InlineShapExplainer(self, "shap")
"""subclass with InlineShapExplainer layouts, e.g. InlineExplainer(explainer).shap.dependence()"""
self.classifier = InlineClassifierExplainer(self, "classifier")
"""subclass with InlineClassifierExplainer plots, e.g. InlineExplainer(explainer).classifier.confusion_matrix()"""
self.regression = InlineRegressionExplainer(self, "regression")
"""subclass with InlineRegressionExplainer plots, e.g. InlineExplainer(explainer).regression.residuals()"""
self.decisiontrees =InlineDecisionTreesExplainer(self, "decisiontrees")
"""subclass with InlineDecisionTreesExplainer plots, e.g. InlineExplainer(explainer).decisiontrees.decisiontrees()"""
def terminate(self, port=None, token=None):
"""terminate an InlineExplainer on particular port.
You can kill any JupyterDash dashboard from any ExplainerDashboard
by specifying the right port.
Args:
port (int, optional): port on which the InlineExplainer is running.
Defaults to the last port the instance had started on.
token (str, optional): JupyterDash._token class property.
Defaults to the _token of the JupyterDash in the current namespace.
Raises:
ValueError: if can't find the port to terminate.
"""
if port is None:
port = self._port
if token is None:
token = JupyterDash._token
shutdown_url = f"http://localhost:{port}/_shutdown_{token}"
print(f"Trying to shut down dashboard on port {port}...")
try:
response = requests.get(shutdown_url)
except Exception as e:
print(f"Something seems to have failed: {e}")
def _run_app(self, app, **kwargs):
"""Starts the dashboard either inline or in a seperate tab
:param app: the JupyterDash app to be run
:type mode: JupyterDash app instance
"""
pio.templates.default = "none"
if self._mode in ['inline', 'jupyterlab']:
app.run_server(mode=self._mode, width=self._width, height=self._height, port=self._port)
elif self._mode == 'external':
app.run_server(mode=self._mode, port=self._port, **self._kwargs)
else:
raise ValueError("mode should either be 'inline', 'jupyterlab' or 'external'!")
def _run_component(self, component, title):
app = JupyterDash(__name__)
app.title = title
app.layout = component.layout()
component.register_callbacks(app)
self._run_app(app)
@delegates_kwargs(ImportancesComponent)
@delegates_doc(ImportancesComponent)
def importances(self, title='Importances', **kwargs):
"""Runs model_summary tab inline in notebook"""
comp = ImportancesComponent(self._explainer, **kwargs)
self._run_component(comp, title)
def model_stats(self, title='Models Stats', **kwargs):
"""Runs model_stats inline in notebook"""
if self._explainer.is_classifier:
comp = ClassifierModelStatsComposite(self._explainer, **kwargs)
elif self._explainer.is_regression:
comp = RegressionModelStatsComposite(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(PredictionSummaryComponent)
@delegates_doc(PredictionSummaryComponent)
def prediction(self, title='Prediction', **kwargs):
"""Show contributions (permutation or shap) inline in notebook"""
comp = PredictionSummaryComponent(self._explainer, **kwargs)
self._run_component(comp, title)
def random_index(self, title='Random Index', **kwargs):
"""show random index selector inline in notebook"""
if self._explainer.is_classifier:
comp = ClassifierRandomIndexComponent(self._explainer, **kwargs)
elif self._explainer.is_regression:
comp = RegressionRandomIndexComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(PdpComponent)
@delegates_doc(PdpComponent)
def pdp(self, title="Partial Dependence Plots", **kwargs):
"""Show contributions (permutation or shap) inline in notebook"""
comp = PdpComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(WhatIfComponent)
@delegates_doc(WhatIfComponent)
def whatif(self, title="What if...", **kwargs):
"""Show What if... component inline in notebook"""
comp = WhatIfComponent(self._explainer, **kwargs)
self._run_component(comp, title)
class InlineExplainerComponent:
def __init__(self, inline_explainer, name):
self._inline_explainer = inline_explainer
self._explainer = inline_explainer._explainer
self._name = name
def _run_component(self, component, title):
self._inline_explainer._run_component(component, title)
def __repr__(self):
component_methods = [method_name for method_name in dir(self)
if callable(getattr(self, method_name)) and not method_name.startswith("_")]
return f"InlineExplainer.{self._name} has the following components: {', '.join(component_methods)}"
class InlineExplainerTabs(InlineExplainerComponent):
@delegates_kwargs(ImportancesTab)
@delegates_doc(ImportancesTab)
def importances(self, title='Importances', **kwargs):
"""Show contributions (permutation or shap) inline in notebook"""
tab = ImportancesTab(self._explainer, **kwargs)
self._run_component(tab, title)
@delegates_kwargs(ModelSummaryTab)
@delegates_doc(ModelSummaryTab)
def modelsummary(self, title='Model Summary', **kwargs):
"""Runs model_summary tab inline in notebook"""
tab = ModelSummaryTab(self._explainer, **kwargs)
self._run_component(tab, title)
@delegates_kwargs(ContributionsTab)
@delegates_doc(ContributionsTab)
def contributions(self, title='Contributions', **kwargs):
"""Show contributions (permutation or shap) inline in notebook"""
tab = ContributionsTab(self._explainer, **kwargs)
self._run_component(tab, title)
@delegates_kwargs(WhatIfTab)
@delegates_doc(WhatIfTab)
def whatif(self, title='What if...', **kwargs):
"""Show What if... tab inline in notebook"""
tab = WhatIfTab(self._explainer, **kwargs)
self._run_component(tab, title)
@delegates_kwargs(ShapDependenceTab)
@delegates_doc(ShapDependenceTab)
def dependence(self, title='Shap Dependence', **kwargs):
"""Runs shap_dependence tab inline in notebook"""
tab = ShapDependenceTab(self._explainer, **kwargs)
self._run_component(tab, title)
@delegates_kwargs(ShapInteractionsTab)
@delegates_doc(ShapInteractionsTab)
def interactions(self, title='Shap Interactions', **kwargs):
"""Runs shap_interactions tab inline in notebook"""
tab = ShapInteractionsTab(self._explainer, **kwargs)
self._run_component(tab, title)
@delegates_kwargs(DecisionTreesTab)
@delegates_doc(DecisionTreesTab)
def decisiontrees(self, title='Decision Trees', **kwargs):
"""Runs shap_interactions tab inline in notebook"""
tab = DecisionTreesTab(self._explainer, **kwargs)
self._run_component(tab, title)
class InlineShapExplainer(InlineExplainerComponent):
@delegates_kwargs(ShapDependenceComposite)
@delegates_doc(ShapDependenceComposite)
def overview(self, title='Shap Overview', **kwargs):
"""Runs shap_dependence tab inline in notebook"""
comp = ShapDependenceComposite(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(ShapSummaryComponent)
@delegates_doc(ShapSummaryComponent)
def summary(self, title='Shap Summary', **kwargs):
"""Show shap summary inline in notebook"""
comp = ShapSummaryComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(ShapDependenceComponent)
@delegates_doc(ShapDependenceComponent)
def dependence(self, title='Shap Dependence', **kwargs):
"""Show shap summary inline in notebook"""
comp = ShapDependenceComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(ShapInteractionsComposite)
@delegates_doc(ShapInteractionsComposite)
def interaction_overview(self, title='Interactions Overview', **kwargs):
"""Runs shap_dependence tab inline in notebook"""
comp = ShapInteractionsComposite(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(InteractionSummaryComponent)
@delegates_doc(InteractionSummaryComponent)
def interaction_summary(self, title='Shap Interaction Summary', **kwargs):
"""show shap interaction summary inline in notebook"""
comp =InteractionSummaryComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(InteractionDependenceComponent)
@delegates_doc(InteractionDependenceComponent)
def interaction_dependence(self, title='Shap Interaction Dependence', **kwargs):
"""show shap interaction dependence inline in notebook"""
comp =InteractionDependenceComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(ShapContributionsGraphComponent)
@delegates_doc(ShapContributionsGraphComponent)
def contributions_graph(self, title='Contributions', **kwargs):
"""Show contributions (permutation or shap) inline in notebook"""
comp = ShapContributionsGraphComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(ShapContributionsTableComponent)
@delegates_doc(ShapContributionsTableComponent)
def contributions_table(self, title='Contributions', **kwargs):
"""Show contributions (permutation or shap) inline in notebook"""
comp = ShapContributionsTableComponent(self._explainer, **kwargs)
self._run_component(comp, title)
class InlineClassifierExplainer(InlineExplainerComponent):
@delegates_kwargs(ClassifierModelStatsComposite)
@delegates_doc(ClassifierModelStatsComposite)
def model_stats(self, title='Models Stats', **kwargs):
"""Runs model_stats inline in notebook"""
comp = ClassifierModelStatsComposite(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(PrecisionComponent)
@delegates_doc(PrecisionComponent)
def precision(self, title="Precision Plot", **kwargs):
"""shows precision plot"""
assert self._explainer.is_classifier
comp = PrecisionComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(CumulativePrecisionComponent)
@delegates_doc(CumulativePrecisionComponent)
def cumulative_precision(self, title="Cumulative Precision Plot", **kwargs):
"""shows cumulative precision plot"""
assert self._explainer.is_classifier
comp = CumulativePrecisionComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(ConfusionMatrixComponent)
@delegates_doc(ConfusionMatrixComponent)
def confusion_matrix(self, title="Confusion Matrix", **kwargs):
"""shows precision plot"""
comp= ConfusionMatrixComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(LiftCurveComponent)
@delegates_doc(LiftCurveComponent)
def lift_curve(self, title="Lift Curve", **kwargs):
"""shows precision plot"""
assert self._explainer.is_classifier
comp = LiftCurveComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(ClassificationComponent)
@delegates_doc(ClassificationComponent)
def classification(self, title="Classification", **kwargs):
"""shows precision plot"""
assert self._explainer.is_classifier
comp = ClassificationComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(RocAucComponent)
@delegates_doc(RocAucComponent)
def roc_auc(self, title="ROC AUC Curve", **kwargs):
"""shows precision plot"""
assert self._explainer.is_classifier
comp = RocAucComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(PrAucComponent)
@delegates_doc(PrAucComponent)
def pr_auc(self, title="PR AUC Curve", **kwargs):
"""shows precision plot"""
assert self._explainer.is_classifier
comp = PrAucComponent(self._explainer, **kwargs)
self._run_component(comp, title)
class InlineRegressionExplainer(InlineExplainerComponent):
@delegates_kwargs(RegressionModelStatsComposite)
@delegates_doc(RegressionModelStatsComposite)
def model_stats(self, title='Models Stats', **kwargs):
"""Runs model_stats inline in notebook"""
comp = RegressionModelStatsComposite(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(PredictedVsActualComponent)
@delegates_doc(PredictedVsActualComponent)
def pred_vs_actual(self, title="Predicted vs Actual", **kwargs):
"shows predicted vs actual for regression"
assert self._explainer.is_regression
comp = PredictedVsActualComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(ResidualsComponent)
@delegates_doc(ResidualsComponent)
def residuals(self, title="Residuals", **kwargs):
"shows residuals for regression"
assert self._explainer.is_regression
comp = ResidualsComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(RegressionVsColComponent)
@delegates_doc(RegressionVsColComponent)
def plots_vs_col(self, title="Plots vs col", **kwargs):
"shows plots vs col for regression"
assert self._explainer.is_regression
comp = RegressionVsColComponent(self._explainer, **kwargs)
self._run_component(comp, title)
class InlineDecisionTreesExplainer(InlineExplainerComponent):
@delegates_kwargs(DecisionTreesComposite)
@delegates_doc(DecisionTreesComposite)
def overview(self, title="Decision Trees", **kwargs):
"""shap decision tree composite inline in notebook"""
comp = DecisionTreesComposite(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(DecisionTreesComponent)
@delegates_doc(DecisionTreesComponent)
def decisiontrees(self, title='Decision Trees', **kwargs):
"""Runs decision_trees tab inline in notebook"""
comp = DecisionTreesComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(DecisionPathTableComponent)
@delegates_doc(DecisionPathTableComponent)
def decisionpath_table(self, title='Decision path', **kwargs):
"""Runs decision_trees tab inline in notebook"""
comp = DecisionPathTableComponent(self._explainer, **kwargs)
self._run_component(comp, title)
@delegates_kwargs(DecisionPathTableComponent)
@delegates_doc(DecisionPathTableComponent)
def decisionpath_graph(self, title='Decision path', **kwargs):
"""Runs decision_trees tab inline in notebook"""
comp = DecisionPathTableComponent(self._explainer, **kwargs)
self._run_component(comp, title)
class JupyterExplainerDashboard(ExplainerDashboard):
def __init__(self, *args, **kwargs):
raise ValueError("JupyterExplainerDashboard has been deprecated. "
"Use e.g. ExplainerDashboard(mode='inline') instead.")
class ExplainerTab:
def __init__(self, *args, **kwargs):
raise ValueError("ExplainerTab has been deprecated. "
"Use e.g. ExplainerDashboard(explainer, ImportancesTab) instead.")
class JupyterExplainerTab(ExplainerTab):
def __init__(self, *args, **kwargs):
raise ValueError("ExplainerTab has been deprecated. "
"Use e.g. ExplainerDashboard(explainer, ImportancesTab, mode='inline') instead.") | AMLBID | /Explainer/dashboardss.py | dashboardss.py |
__all__ = [
'delegates_kwargs',
'delegates_doc',
'DummyComponent',
'ExplainerComponent',
'PosLabelSelector',
'make_hideable',
'get_dbc_tooltips',
'update_params',
'update_kwargs',
'instantiate_component'
]
import sys
from abc import ABC
import inspect
import types
import dash
import dash_core_components as dcc
import dash_bootstrap_components as dbc
import dash_html_components as html
import shortuuid
# Stolen from https://www.fast.ai/2019/08/06/delegation/
# then extended to deal with multiple inheritance
def delegates_kwargs(to=None, keep=False):
"Decorator: replace `**kwargs` in signature with params from `to`"
def _f(f):
from_f = f.__init__ if to is None else f
sig = inspect.signature(from_f)
sigd = dict(sig.parameters)
k = sigd.pop('kwargs')
if to is None:
for base_cls in f.__bases__:
to_f = base_cls.__init__
s2 = {k: v for k, v in inspect.signature(to_f).parameters.items()
if v.default != inspect.Parameter.empty and k not in sigd}
sigd.update(s2)
else:
to_f = to
s2 = {k: v for k, v in inspect.signature(to_f).parameters.items()
if v.default != inspect.Parameter.empty and k not in sigd}
sigd.update(s2)
if keep:
sigd['kwargs'] = k
from_f.__signature__ = sig.replace(parameters=sigd.values())
return f
return _f
def delegates_doc(to=None, keep=False):
"Decorator: replace `__doc__` with `__doc__` from `to`"
def _f(f):
from_f = f.__init__ if to is None else f
if to is None:
for base_cls in f.__bases__:
to_f = base_cls.__init__
else:
if isinstance(to, types.FunctionType):
to_f = to
else:
to_f = to.__init__
from_f.__doc__ = to_f.__doc__
return f
return _f
def update_params(kwargs, **params):
"""kwargs override params"""
return dict(params, **kwargs)
def update_kwargs(kwargs, **params):
"""params override kwargs"""
return dict(kwargs, **params)
def get_dbc_tooltips(dbc_table, desc_dict, hover_id, name):
"""Return a dbc.Table and a list of dbc.Tooltips.
Args:
dbc_table (dbc.Table): Table with first column consisting of label
desc_dict (dict): dict that map labels to a description (str)
hover_id (str): dash component_id base: tooltips will have
component_id=f"{hover_id}-{label}-{name}"
name (str): name to be used in hover_id
Returns:
dbc.Table, List[dbc.Tooltip]
"""
tooltips_dict = {}
for tr in dbc_table.children[1].children:
tds = tr.children
label = tds[0].children
if label in desc_dict:
tr.id = f'{hover_id}-{label}-'+name
tooltips_dict[label] = desc_dict[label]
tooltips = [dbc.Tooltip(desc,
target=f'{hover_id}-{label}-'+name,
placement="top") for label, desc in tooltips_dict.items()]
return dbc_table, tooltips
def make_hideable(element, hide=False):
"""helper function to optionally not display an element in a layout.
This is used for all the hide_ flags in ExplainerComponent constructors.
e.g. hide_cutoff=True to hide a cutoff slider from a layout:
Example:
make_hideable(dbc.Col([cutoff.layout()]), hide=hide_cutoff)
Args:
hide(bool): wrap the element inside a hidden html.div. If the element
is a dbc.Col or a dbc.FormGroup, wrap element.children in
a hidden html.Div instead. Defaults to False.
"""
if hide:
if isinstance(element, dbc.Col) or isinstance(element, dbc.FormGroup):
return html.Div(element.children, style=dict(display="none"))
else:
return html.Div(element, style=dict(display="none"))
else:
return element
class DummyComponent:
def __init__(self):
pass
def layout(self):
return None
def register_callbacks(self, app):
pass
class ExplainerComponent(ABC):
"""ExplainerComponent is a bundle of a dash layout and callbacks that
make use of an Explainer object.
An ExplainerComponent can have ExplainerComponent subcomponents, that
you register with register_components(). If the component depends on
certain lazily calculated Explainer properties, you can register these
with register_dependencies().
ExplainerComponent makes sure that:
1. Callbacks of subcomponents are registered.
2. Lazily calculated dependencies (even of subcomponents) can be calculated.
3. Pos labels selector id's of all subcomponents can be calculated.
Each ExplainerComponent adds a unique uuid name string to all elements, so
that there is never a name clash even with multiple ExplanerComponents of
the same type in a layout.
Important:
define your callbacks in component_callbacks() and
ExplainerComponent will register callbacks of subcomponents in addition
to component_callbacks() when calling register_callbacks()
"""
def __init__(self, explainer, title=None, name=None):
"""initialize the ExplainerComponent
Args:
explainer (Explainer): explainer object constructed with e.g.
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to None.
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
"""
self._store_child_params(no_param=['explainer'])
if not hasattr(self, "name") or self.name is None:
self.name = "uuid"+shortuuid.ShortUUID().random(length=5)
if title is not None:
self.title = title
if not hasattr(self, "title"):
self.title = "Custom"
self._components = []
self._dependencies = []
def _store_child_params(self, no_store=None, no_attr=None, no_param=None):
if not hasattr(self, '_stored_params'):
self._stored_params = {}
child_frame = sys._getframe(2)
child_args = child_frame.f_code.co_varnames[1:child_frame.f_code.co_argcount]
child_dict = {arg: child_frame.f_locals[arg] for arg in child_args}
if isinstance(no_store, bool) and no_store:
return
else:
if no_store is None: no_store = tuple()
if isinstance(no_attr, bool) and no_attr: dont_attr = True
else:
if no_attr is None: no_attr = tuple()
dont_attr = False
if isinstance(no_param, bool) and no_param: dont_param = True
else:
if no_param is None: no_param = tuple()
dont_param = False
for name, value in child_dict.items():
if not dont_attr and name not in no_store and name not in no_attr:
setattr(self, name, value)
if not dont_param and name not in no_store and name not in no_param:
self._stored_params[name] = value
def exclude_callbacks(self, *components):
"""exclude certain subcomponents from the register_components scan
"""
if not hasattr(self, '_exclude_components'):
self._exclude_components = []
for comp in components:
if isinstance(comp, ExplainerComponent) and comp not in self._exclude_components:
self._exclude_components.append(comp)
def register_components(self, *components):
"""register subcomponents so that their callbacks will be registered
and dependencies can be tracked
Args:
scan_self (bool, optional): scan self.__dict__ and add all
ExplainerComponent attributes to _components. Defaults to True
"""
if not hasattr(self, '_components'):
self._components = []
if not hasattr(self, '_exclude_components'):
self._exclude_components = []
for comp in components:
if (isinstance(comp, ExplainerComponent)
and comp not in self._components
and comp not in self._exclude_components):
self._components.append(comp)
elif hasattr(comp, '__iter__'):
for subcomp in comp:
if (isinstance(subcomp, ExplainerComponent)
and subcomp not in self._components
and subcomp not in self._exclude_components):
self._components.append(subcomp)
else:
print(f"{subcomp.__name__} is not an ExplainerComponent so not adding to self.components")
else:
print(f"{comp.__name__} is not an ExplainerComponent so not adding to self.components")
for k, v in self.__dict__.items():
if (k != '_components'
and isinstance(v, ExplainerComponent)
and v not in self._components
and v not in self._exclude_components):
self._components.append(v)
def has_pos_label_connector(self):
if not hasattr(self, '_components'):
self._components = []
for comp in self._components:
if str(type(comp)).endswith("PosLabelConnector'>"):
return True
elif comp.has_pos_label_connector():
return True
return False
def register_dependencies(self, *dependencies):
"""register dependencies: lazily calculated explainer properties that
you want to calculate *before* starting the dashboard"""
for dep in dependencies:
if isinstance(dep, str):
self._dependencies.append(dep)
elif hasattr(dep, '__iter__'):
for subdep in dep:
if isinstance(subdep, str):
self._dependencies.append(subdep)
else:
print(f"{subdep.__name__} is not a str so not adding to self.dependencies")
else:
print(f"{dep.__name__} is not a str or list of str so not adding to self.dependencies")
@property
def dependencies(self):
"""returns a list of unique dependencies of the component
and all subcomponents"""
if not hasattr(self, '_dependencies'):
self._dependencies = []
self.register_components()
deps = self._dependencies
for comp in self._components:
deps.extend(comp.dependencies)
deps = list(set(deps))
return deps
@property
def component_imports(self):
"""returns a list of ComponentImport namedtuples("component", "module")
all components and and subcomponents"""
self.register_components()
_component_imports = [(self.__class__.__name__, self.__class__.__module__)]
for comp in self._components:
_component_imports.extend(comp.component_imports)
return list(set(_component_imports))
@property
def pos_labels(self):
"""returns a list of unique pos label selector elements
of the component and all subcomponents"""
self.register_components()
pos_labels = []
for k, v in self.__dict__.items():
if isinstance(v, PosLabelSelector) and v.name not in pos_labels:
pos_labels.append('pos-label-'+v.name)
# if hasattr(self, 'selector') and isinstance(self.selector, PosLabelSelector):
# pos_labels.append('pos-label-'+self.selector.name)
for comp in self._components:
pos_labels.extend(comp.pos_labels)
pos_labels = list(set(pos_labels))
return pos_labels
def calculate_dependencies(self):
"""calls all properties in self.dependencies so that they get calculated
up front. This is useful to do before starting a dashboard, so you don't
compute properties multiple times in parallel."""
for dep in self.dependencies:
try:
_ = getattr(self.explainer, dep)
except:
ValueError(f"Failed to generate dependency '{dep}': "
"Failed to calculate or retrieve explainer property explainer.{dep}...")
def layout(self):
"""layout to be defined by the particular ExplainerComponent instance.
All element id's should append +self.name to make sure they are unique."""
return None
def component_callbacks(self, app):
"""register callbacks specific to this ExplainerComponent."""
if hasattr(self, "_register_callbacks"):
print("Warning: the use of _register_callbacks() will be deprecated!"
" Use component_callbacks() from now on...")
self._register_callbacks(app)
def register_callbacks(self, app):
"""First register callbacks of all subcomponents, then call
component_callbacks(app)
"""
self.register_components()
for comp in self._components:
comp.register_callbacks(app)
self.component_callbacks(app)
class PosLabelSelector(ExplainerComponent):
"""For classifier models displays a drop down menu with labels to be selected
as the positive class.
"""
def __init__(self, explainer, title='Pos Label Selector', name=None,
pos_label=None):
"""Generates a positive label selector with element id 'pos_label-'+self.name
Args:
explainer (Explainer): explainer object constructed with e.g.
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to None.
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to 'Pos Label Selector'.
pos_label (int, optional): Initial pos label. Defaults to
explainer.pos_label.
"""
super().__init__(explainer, title, name)
if pos_label is not None:
self.pos_label = explainer.get_pos_label_index(pos_label)
else:
self.pos_label = explainer.pos_label
def layout(self):
if self.explainer.is_classifier:
return html.Div([dbc.Form([ dbc.FormGroup([
dbc.Label("Positive class",
html_for="pos-label-"+self.name,
id="pos-label-label-"+self.name),
dbc.Tooltip("Select the label to be set as the positive class",
target="pos-label-label-"+self.name),
dcc.Dropdown(
id='pos-label-'+self.name,
options = [{'label': label, 'value': i}
for i, label in enumerate(self.explainer.labels)],
value = self.pos_label,
clearable=False,style={"width":"140px"}
)
])],inline=True) ],style={"float":"right"})
else:
return html.Div([dcc.Input(id="pos-label-"+self.name)], style=dict(display="none"))
def instantiate_component(component, explainer, name=None, **kwargs):
"""Returns an instantiated ExplainerComponent.
If the component input is just a class definition, instantiate it with
explainer and k**wargs.
If it is already an ExplainerComponent instance then return it.
If it is any other instance with layout and register_components methods,
then add a name property and return it.
Args:
component ([type]): Either a class definition or instance
explainer ([type]): An Explainer object that will be used to instantiate class definitions
name (str): name to assign to ExplainerComponent
kwargs: kwargs will be passed on to the instance
Raises:
ValueError: if component is not a subclass or instance of ExplainerComponent,
or is an instance without layout and register_callbacks methods
Returns:
ExplainerComponent: instantiated component
"""
if inspect.isclass(component) and issubclass(component, ExplainerComponent):
init_argspec = inspect.getargspec(component.__init__)
if not init_argspec.keywords:
kwargs = {k:v for k,v in kwargs.items() if k in init_argspec.args}
if "name" in init_argspec.args:
component = component(explainer, name=name, **kwargs)
else:
print(f"ExplainerComponent {component} does not accept a name parameter, "
f"so cannot assign name={name}!"
"Make sure to set name explicitly yourself if you want to "
"deploy across multiple workers or a cluster, as otherwise "
"each instance in the cluster will generate its own random "
"uuid name!")
component = component(explainer, **kwargs)
return component
elif isinstance(component, ExplainerComponent):
return component
else:
raise ValueError(f"{component} is not a valid ExplainerComponent...") | AMLBID | /Explainer/dashboard_methods.py | dashboard_methods.py |
__all__ = ['titanic_survive',
'titanic_fare',
'titanic_embarked',
'titanic_names',
'feature_descriptions',
'train_csv',
'test_csv']
import numpy as np
import pandas as pd
from pathlib import Path
train_csv = Path(__file__).resolve().parent / 'datasets'/ 'titanic_train.csv'
test_csv = Path(__file__).resolve().parent / 'datasets'/'titanic_test.csv'
d_train = pd.read_csv(train_csv)
d_test = pd.read_csv(test_csv)
feature_descriptions = {
"Sex": "Gender of passenger",
"Gender": "Gender of passenger",
"Deck": "The deck the passenger had their cabin on",
"PassengerClass": "The class of the ticket: 1st, 2nd or 3rd class",
"Fare": "The amount of money people paid for their ticket",
"Embarked": "the port where the passenger boarded the Titanic. Either Southampton, Cherbourg or Queenstown",
"Age": "Age of the passenger",
"No_of_siblings_plus_spouses_on_board": "The sum of the number of siblings plus the number of spouses on board",
"No_of_parents_plus_children_on_board" : "The sum of the number of parents plus the number of children on board",
}
def titanic_survive():
X_train = d_train.drop(['Survival', 'Name'], axis=1)
X_train.index = d_train.Name
X_train.index.name = "Passenger"
y_train = d_train['Survival']
X_test = d_test.drop(['Survival', 'Name'], axis=1)
X_test.index = d_test.Name
X_test.index.name = "Passenger"
y_test = d_test['Survival']
return X_train, y_train, X_test, y_test
def titanic_fare():
X_train = d_train.drop(['Fare', 'Name'], axis=1)
X_train.index = d_train.Name
X_train.index.name = "Passenger"
y_train = d_train['Fare']
X_test = d_test.drop(['Fare', 'Name'], axis=1)
X_test.index = d_test.Name
X_test.index.name = "Passenger"
y_test = d_test['Fare']
return X_train, y_train, X_test, y_test
def titanic_embarked():
d_train2 = d_train.copy()
d_train2 = d_train2[d_train2.Embarked_Unknown==0]
X_train = d_train2.drop(['Embarked_Cherbourg', 'Embarked_Queenstown',
'Embarked_Southampton', 'Embarked_Unknown', 'Name'], axis=1)
X_train.index = d_train2.Name
X_train.index.name = "Passenger"
y_train = pd.Series(np.where(d_train2.Embarked_Queenstown==1, 0,
np.where(d_train2.Embarked_Southampton==1, 1,
np.where(d_train2.Embarked_Cherbourg==1, 2, 3))),
name="Embarked")
X_test = d_test.drop(['Embarked_Cherbourg', 'Embarked_Queenstown',
'Embarked_Southampton', 'Embarked_Unknown', 'Name'], axis=1)
X_test.index = d_test.Name
X_test.index.name = "Passenger"
y_test = pd.Series(np.where(d_test.Embarked_Queenstown==1, 0,
np.where(d_test.Embarked_Southampton==1, 1,
np.where(d_test.Embarked_Cherbourg==1, 2, 3))),
name="Embarked")
return X_train, y_train, X_test, y_test
def titanic_names(train_only=False, test_only=False):
if train_only:
return d_train['Name'].values.tolist()
if test_only:
return d_test['Name'].values.tolist()
return (d_train['Name'].values.tolist(), d_test['Name'].values.tolist()) | AMLBID | /Explainer/datasets.py | datasets.py |
import os
import webbrowser
from pathlib import Path
from importlib import import_module
from copy import deepcopy
import pickle
import oyaml as yaml
import pandas as pd
import joblib
import click
import waitress
from explainerdashboard import *
from explainerdashboard.explainers import BaseExplainer
from explainerdashboard.dashboards import ExplainerDashboard
explainer_ascii = """
_____ ___ __| |__ _(_)_ _ ___ _ _ __| |__ _ __| |_ | |__ ___ __ _ _ _ __| |
/ -_) \ / '_ \ / _` | | ' \/ -_) '_/ _` / _` (_-< ' \| '_ \/ _ \/ _` | '_/ _` |
\___/_\_\ .__/_\__,_|_|_||_\___|_| \__,_\__,_/__/_||_|_.__/\___/\__,_|_| \__,_|
|_|
"""
hub_ascii = """
_ _ _ _
_____ ___ __| |__ _(_)_ _ ___ _ _| |_ _ _| |__
/ -_) \ / '_ \ / _` | | ' \/ -_) '_| ' \ || | '_ \
\___/_\_\ .__/_\__,_|_|_||_\___|_| |_||_\_,_|_.__/
|_|
"""
def build_explainer(explainer_config):
if isinstance(explainer_config, (Path, str)) and str(explainer_config).endswith(".yaml"):
config = yaml.safe_load(open(str(explainer_config), "r"))
elif isinstance(explainer_config, dict):
config = explainer_config
assert 'explainer' in config, \
"Please pass a proper explainer.yaml config file that starts with `explainer:`!"
config = explainer_config['explainer']
print(f"explainerdashboard ===> Loading model from {config['modelfile']}")
model = pickle.load(open(config['modelfile'], "rb"))
print(f"explainerdashboard ===> Loading data from {config['datafile']}")
if str(config['datafile']).endswith('.csv'):
df = pd.read_csv(config['datafile'])
elif str(config['datafile']).endswith('.parquet'):
df = pd.read_parquet(config['datafile'])
else:
raise ValueError("datafile should either be a .csv or .parquet!")
print(f"explainerdashboard ===> Using column {config['data_target']} to generate X, y ")
target_col = config['data_target']
X = df.drop(target_col, axis=1)
y = df[target_col]
if config['data_index'] is not None:
print(f"explainerdashboard ===> Generating index from column {config['data_index']}")
assert config['data_index'] in X.columns, \
(f"Cannot find data_index column ({config['data_index']})"
f" in datafile ({config['datafile']})!"
"Please set it to the proper index column name, or set it to null")
X = X.set_index(config['data_index'])
params = config['params']
if config['explainer_type'] == "classifier":
print(f"explainerdashboard ===> Generating ClassifierExplainer...")
explainer = ClassifierExplainer(model, X, y, **params)
elif config['explainer_type'] == "regression":
print(f"explainerdashboard ===> Generating RegressionExplainer...")
explainer = ClassifierExplainer(model, X, y, **params)
return explainer
def build_and_dump_explainer(explainer_config, dashboard_config=None):
explainer = build_explainer(explainer_config)
click.echo(f"explainerdashboard ===> Calculating properties by building Dashboard...")
if dashboard_config is not None:
ExplainerDashboard.from_config(explainer, dashboard_config)
elif Path(explainer_config['explainer']['dashboard_yaml']).exists():
click.echo(f"explainerdashboard ===> Calculating properties by building Dashboard from {explainer_config['explainer']['dashboard_yaml']}...")
dashboard_config = yaml.safe_load(open(str(explainer_config['explainer']['dashboard_yaml']), "r"))
ExplainerDashboard.from_config(explainer, dashboard_config)
else:
click.echo(f"explainerdashboard ===> Calculating all properties")
explainer.calculate_properties()
click.echo(f"explainerdashboard ===> Saving explainer to {explainer_config['explainer']['explainerfile']}...")
if (dashboard_config is not None and
explainer_config['explainer']['explainerfile'] != dashboard_config['dashboard']['explainerfile']):
click.echo(f"explainerdashboard ===> Warning explainerfile in explainer config and dashboard config do not match!")
explainer.dump(explainer_config['explainer']['explainerfile'])
return
def launch_dashboard_from_pkl(explainer_filepath, no_browser, port, no_dashboard=False):
explainer = BaseExplainer.from_file(explainer_filepath)
if port is None:
click.echo(f"explainerdashboard ===> Setting port to 8050, override with e.g. --port 8051")
port = 8050
db = ExplainerDashboard(explainer, port=port)
if not no_browser and not os.environ.get("WERKZEUG_RUN_MAIN"):
webbrowser.open_new(f"http://127.0.0.1:{port}/")
if not no_dashboard:
waitress.serve(db.flask_server(), host='0.0.0.0', port=port)
return
def launch_dashboard_from_yaml(dashboard_config, no_browser, port, no_dashboard=False):
if isinstance(dashboard_config, (Path, str)) and str(dashboard_config).endswith(".yaml"):
config = yaml.safe_load(open(str(dashboard_config), "r"))
elif isinstance(dashboard_config, dict):
config = dashboard_config
else:
raise ValueError(f"dashboard_config should either be a .yaml filepath or a dict!")
if not Path(config['dashboard']['explainerfile']).exists():
click.echo(f"explainerdashboard ===> {config['dashboard']['explainerfile']} does not exist!")
click.echo(f"explainerdashboard ===> first generate {config['dashboard']['explainerfile']} with explainerdashboard build")
return
click.echo(f"explainerdashboard ===> Building dashboard from {config['dashboard']['explainerfile']}")
db = ExplainerDashboard.from_config(config)
if port is None:
port = config['dashboard']['params']['port']
if port is None:
port = 8050
click.echo(f"explainerdashboard ===> Setting port to {port}, override with e.g. --port 8051")
if not no_browser and not os.environ.get("WERKZEUG_RUN_MAIN"):
click.echo(f"explainerdashboard ===> launching browser at {f'http://localhost:{port}/'}")
webbrowser.open_new(f"http://localhost:{port}/")
click.echo(f"explainerdashboard ===> Starting dashboard:")
if not no_dashboard:
waitress.serve(db.flask_server(), host='0.0.0.0', port=port)
return
def launch_hub_from_yaml(hub_config, no_browser, port, no_dashboard=False):
hub = ExplainerHub.from_config(hub_config)
if port is None:
port = hub.port
if port is None:
port = 8050
click.echo(f"explainerhub ===> Setting port to {port}, override with e.g. --port 8051")
if not no_browser and not os.environ.get("WERKZEUG_RUN_MAIN"):
click.echo(f"explainerhub ===> launching browser at {f'http://localhost:{port}/'}")
webbrowser.open_new(f"http://localhost:{port}/")
click.echo(f"explainerhub ===> Starting dashboard:")
if not no_dashboard:
waitress.serve(hub.flask_server(), host='0.0.0.0', port=port)
return
@click.group()
@click.pass_context
def explainerdashboard_cli(ctx):
"""
explainerdashboard CLI tool. Used to launch an explainerdashboard from
the commandline.
\b
explainerdashboard run
----------------------
Run explainerdashboard and start browser directly from command line.
\b
Example use:
explainerdashboard run explainer.joblib
explainerdashboard run dashboard.yaml
explainerdashboard run dashboard.yaml --no-browser --port 8051
explainerdashboard run --help
If you pass an explainer.joblib file, will launch the full default dashboard.
Generate this file with explainer.dump("explainer.joblib")
If you pass a dashboard.yaml file, will launch a fully configured
explainerdashboard. Generate dashboard.yaml with
ExplainerDashboard.to_yaml('dashboard.yaml')
If no argument given, searches for either dashboard.yaml or
explainer.joblib, in that order, so if you keep that naming convention
you can simply start with:
\b
explainerdashboard run
\b
explainerdashboard build
------------------------
Build and store an explainer object, based on explainer.yaml file, that indicates
where to find stored model (e.g. model.pkl), stored datafile (e.g. data.csv),
and other explainer parameters.
\b
Example use:
explainerdashboard build explainer.yaml
explainerdashboard build explainer.yaml dashboard.yaml
explainerdashboard build --help
If given a second dashboard.yaml argument, will use that dashboard
configuration to calculate necessary properties for that specific dashboard
configuration before storing to disk. Otherwise will use dashboard_yaml
parameter in explainer.yaml to find configuration, or alternatively
simply calculate all properties.
explainer.yaml file can be generated with explainer.to_yaml("explainer.yaml")
If no argument given, searches for explainer.yaml, so if you keep that
naming convention you can simply start the build with:
\b
explainerdashboard build
"""
@explainerdashboard_cli.command(help="run dashboard and open browser")
@click.pass_context
@click.argument("explainer_filepath", nargs=1, required=False)
@click.option("--no-browser", "-nb", "no_browser", is_flag=True,
help="Launch a dashboard, but do not launch a browser.")
@click.option("--port", "-p", "port", default=None,
help="port to run dashboard on defaults.")
def run(ctx, explainer_filepath, no_browser, port):
click.echo(explainer_ascii)
if explainer_filepath is None:
if (Path().cwd() / "dashboard.yaml").exists():
explainer_filepath = Path().cwd() / "dashboard.yaml"
elif (Path().cwd() / "explainer.joblib").exists():
explainer_filepath = Path().cwd() / "explainer.joblib"
else:
click.echo("No argument given and could find neither a "
"dashboard.yaml nor a explainer.joblib. Aborting.")
return
if (str(explainer_filepath).endswith(".joblib") or
str(explainer_filepath).endswith(".pkl") or
str(explainer_filepath).endswith(".pickle") or
str(explainer_filepath).endswith(".dill")):
launch_dashboard_from_pkl(explainer_filepath, no_browser, port)
return
elif str(explainer_filepath).endswith(".yaml"):
launch_dashboard_from_yaml(explainer_filepath, no_browser, port)
else:
click.echo("Please pass a proper argument to explainerdashboard run"
"(i.e. either an explainer.joblib or a dashboard.yaml)")
return
@explainerdashboard_cli.command(help="build and save explainer object")
@click.pass_context
@click.argument("explainer_filepath", nargs=1, required=False)
@click.argument("dashboard_filepath", nargs=1, required=False)
def build(ctx, explainer_filepath, dashboard_filepath):
click.echo(explainer_ascii)
if explainer_filepath is None:
if (Path().cwd() / "explainer.yaml").exists():
explainer_filepath = Path().cwd() / "explainer.yaml"
else:
click.echo("No argument given to explainerdashboard build and "
"could not find an explainer.yaml. Aborting.")
return
if str(explainer_filepath).endswith(".yaml"):
explainer_config = yaml.safe_load(open(str(explainer_filepath), "r"))
click.echo(f"explainerdashboard ===> Building {explainer_config['explainer']['explainerfile']}")
if (dashboard_filepath is not None and
str(dashboard_filepath).endswith(".yaml")
and Path(dashboard_filepath).exists()):
click.echo(f"explainerdashboard ===> Using {dashboard_filepath} to calculate explainer properties")
dashboard_config = yaml.safe_load(open(str(dashboard_filepath), "r"))
else:
dashboard_config = None
print(f"explainerdashboard ===> Building {explainer_config['explainer']['explainerfile']}")
build_and_dump_explainer(explainer_config, dashboard_config)
print(f"explainerdashboard ===> Build finished!")
return
@explainerdashboard_cli.command(help="run without launching dashboard")
@click.pass_context
@click.argument("explainer_filepath", nargs=1, required=True)
@click.option("--port", "-p", "port", default=None,
help="port to run dashboard on defaults.")
def test(ctx, explainer_filepath, port):
if (str(explainer_filepath).endswith(".joblib") or
str(explainer_filepath).endswith(".pkl") or
str(explainer_filepath).endswith(".pickle") or
str(explainer_filepath).endswith(".dill")):
launch_dashboard_from_pkl(explainer_filepath,
no_browser=True, port=port, no_dashboard=True)
return
elif str(explainer_filepath).endswith(".yaml"):
launch_dashboard_from_yaml(explainer_filepath,
no_browser=True, port=port, no_dashboard=True)
return
else:
raise ValueError("Please pass a proper argument "
"(i.e. .joblib, .pkl, .dill or .yaml)!")
@click.group()
@click.pass_context
def explainerhub_cli(ctx):
"""
explainerhub CLI tool. Used to launch and manage explainerhub from
the commandline.
\b
explainerhub run
----------------------
Run explainerdashboard and start browser directly from command line.
\b
Example use:
explainerhub run hub.yaml
\b
If no argument given assumed argument is hub.yaml
\b
explainerhub user management
----------------------------
You can use the CLI to add and remove users from the users.json file that
stores the usernames and (hashed) passwords for the explainerhub. If no
filename is given, will look for either a hub.yaml or users.json file.
\b
If you don't provide the username or password on the commandline, you will get prompted.
\b
Examples use:
explainerhub add_user
explainerhub add_user users.json
explainerhub add_user users2.json
explainerhub add_user hub.yaml
explainerhub delete_user
explainerhub add_dashboard_user
explainerhub delete_dashboard_user
"""
@explainerhub_cli.command(help="run explainerhub and open browser")
@click.pass_context
@click.argument("hub_filepath", nargs=1, required=False)
@click.option("--no-browser", "-nb", "no_browser", is_flag=True,
help="Launch hub, but do not launch a browser.")
@click.option("--port", "-p", "port", default=None,
help="port to run hub on.")
def run(ctx, hub_filepath, no_browser, port):
if hub_filepath is None:
if (Path().cwd() / "hub.yaml").exists():
hub_filepath = Path().cwd() / "hub.yaml"
else:
click.echo("No argument given and could find neither a "
"hub.yaml. Aborting.")
return
click.echo(hub_ascii)
launch_hub_from_yaml(hub_filepath, no_browser, port)
@explainerhub_cli.command(help="add a user to users.json")
@click.argument("filepath", nargs=1, required=False)
@click.option('--username', "-u", required=True, prompt=True)
@click.option('--password', "-p", required=True, prompt=True, hide_input=True,
confirmation_prompt=True)
def add_user(filepath, username, password):
if filepath is None:
if (Path().cwd() / "hub.yaml").exists():
click.echo("explainerhub ===> Detected hub.yaml...")
filepath = Path().cwd() / "hub.yaml"
elif (Path().cwd() / "users.json").exists():
click.echo("explainerhub ===> Detected users.json...")
filepath = Path().cwd() / "users.json"
else:
click.echo("No argument given and could find neither a "
"hub.yaml nor users.json. Aborting.")
return
if str(filepath).endswith(".yaml"):
config = yaml.safe_load(open(str(filepath), "r"))
filepath = config['explainerhub']['user_json']
click.echo(f"explainerhub ===> Using {filepath} to add user...")
ExplainerHub._validate_user_json(filepath)
ExplainerHub._add_user_to_json(filepath, username=username, password=password)
click.echo(f'user added to {filepath}!')
@explainerhub_cli.command(help="remove a user from users.json")
@click.argument("filepath", nargs=1, required=False)
@click.option('--username', "-u", required=True, prompt=True)
def delete_user(filepath, username):
if filepath is None:
if (Path().cwd() / "hub.yaml").exists():
click.echo("explainerhub ===> Detected hub.yaml...")
filepath = Path().cwd() / "hub.yaml"
elif (Path().cwd() / "users.json").exists():
click.echo("explainerhub ===> Detected users.json...")
filepath = Path().cwd() / "users.json"
else:
click.echo("No argument given and could find neither a "
"hub.yaml nor users.json. Aborting.")
return
if str(filepath).endswith(".yaml"):
config = yaml.safe_load(open(str(filepath), "r"))
filepath = config['explainerhub']['user_json']
click.echo(f"explainerhub ===> Using {filepath} to add user...")
ExplainerHub._validate_user_json(filepath)
ExplainerHub._delete_user_from_json(filepath, username=username, password=password)
click.echo(f'user removed from {filepath}!')
@explainerhub_cli.command(help="add a username to a dashboard users.json")
@click.argument("filepath", nargs=1, required=False)
@click.option('--dashboard', "-d", required=True, prompt=True)
@click.option('--username', "-u", required=True, prompt=True)
def add_dashboard_user(filepath, dashboard, username):
if filepath is None:
if (Path().cwd() / "hub.yaml").exists():
click.echo("explainerhub ===> Detected hub.yaml...")
filepath = Path().cwd() / "hub.yaml"
elif (Path().cwd() / "users.json").exists():
click.echo("explainerhub ===> Detected users.json...")
filepath = Path().cwd() / "users.json"
else:
click.echo("No argument given and could find neither a "
"hub.yaml nor users.json. Aborting.")
return
if str(filepath).endswith(".yaml"):
config = yaml.safe_load(open(str(filepath), "r"))
filepath = config['explainerhub']['user_json']
click.echo(f"explainerhub ===> Using {filepath} to add user...")
ExplainerHub._validate_user_json(filepath)
ExplainerHub._add_user_to_dashboard_json(
filepath, dashboard=dashboard, username=username)
click.echo(f'user added to {dashboard} in {filepath}!')
@explainerhub_cli.command(help="remove a username from a dashboard in users.json")
@click.argument("filepath", nargs=1, required=False)
@click.option('--dashboard', "-d", required=True, prompt=True)
@click.option('--username', "-u", required=True, prompt=True)
def delete_dashboard_user(filepath, dashboard, username):
if filepath is None:
if (Path().cwd() / "hub.yaml").exists():
click.echo("explainerhub ===> Detected hub.yaml...")
filepath = Path().cwd() / "hub.yaml"
elif (Path().cwd() / "users.json").exists():
click.echo("explainerhub ===> Detected users.json...")
filepath = Path().cwd() / "users.json"
else:
click.echo("No argument given and could find neither a "
"hub.yaml nor users.json. Aborting.")
return
if str(filepath).endswith(".yaml"):
config = yaml.safe_load(open(str(filepath), "r"))
filepath = config['explainerhub']['user_json']
click.echo(f"explainerhub ===> Using {filepath} to remove user...")
ExplainerHub._validate_user_json(filepath)
ExplainerHub._delete_user_from_dashboard_json(
filepath, dashboard=dashboard, username=username)
click.echo(f'user removed from {dashboard} in {filepath}!')
if __name__ =="__main__":
explainerdashboard_cli() | AMLBID | /Explainer/cli.py | cli.py |
__all__ = [
'PredictionSummaryComponent',
'ImportancesComponent',
'FeatureInputComponent',
'PdpComponent',
'WhatIfComponent',
]
from math import ceil
import numpy as np
import pandas as pd
import dash
import dash_core_components as dcc
import dash_bootstrap_components as dbc
import dash_html_components as html
import dash_table
from dash.dependencies import Input, Output, State
from dash.exceptions import PreventUpdate
from ..dashboard_methods import *
class PredictionSummaryComponent(ExplainerComponent):
def __init__(self, explainer, title="Prediction Summary", name=None,
hide_index=False, hide_percentile=False,
hide_title=False, hide_subtitle=False, hide_selector=False,
pos_label=None, index=None, percentile=True,
description=None, **kwargs):
"""Shows a summary for a particular prediction
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"Prediction Summary".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
hide_index (bool, optional): hide index selector. Defaults to False.
hide_percentile (bool, optional): hide percentile toggle. Defaults to False.
hide_title (bool, optional): hide title. Defaults to False.
hide_subtitle (bool, optional): Hide subtitle. Defaults to False.
hide_selector (bool, optional): hide pos label selectors. Defaults to False.
pos_label ({int, str}, optional): initial pos label.
Defaults to explainer.pos_label
index ({int, str}, optional): Index to display prediction summary for. Defaults to None.
percentile (bool, optional): Whether to add the prediction percentile. Defaults to True.
"""
super().__init__(explainer, title, name)
self.index_name = 'modelprediction-index-'+self.name
self.selector = PosLabelSelector(explainer, name=self.name, pos_label=pos_label)
def layout(self):
return dbc.Card([
make_hideable(
dbc.CardHeader([
html.H3(self.title),
]), hide=self.hide_title),
dbc.CardBody([
dbc.Row([
make_hideable(
dbc.Col([
dbc.Label(f"{self.explainer.index_name}:"),
dcc.Dropdown(id='modelprediction-index-'+self.name,
options = [{'label': str(idx), 'value':idx}
for idx in self.explainer.idxs],
value=self.index)
], md=6), hide=self.hide_index),
make_hideable(
dbc.Col([self.selector.layout()
], width=3), hide=self.hide_selector),
make_hideable(
dbc.Col([
dbc.Label("Show Percentile:"),
dbc.FormGroup(
[
dbc.RadioButton(
id='modelprediction-percentile-'+self.name,
className="form-check-input",
checked=self.percentile),
dbc.Label("Show percentile",
html_for='modelprediction-percentile'+self.name,
className="form-check-label"),
], check=True)
], md=3), hide=self.hide_percentile),
]),
dbc.Row([
dbc.Col([
dcc.Markdown(id='modelprediction-'+self.name),
], md=12)
])
])
])
def component_callbacks(self, app):
@app.callback(
Output('modelprediction-'+self.name, 'children'),
[Input('modelprediction-index-'+self.name, 'value'),
Input('modelprediction-percentile-'+self.name, 'checked'),
Input('pos-label-'+self.name, 'value')])
def update_output_div(index, include_percentile, pos_label):
if index is not None:
return self.explainer.prediction_result_markdown(index, include_percentile=include_percentile, pos_label=pos_label)
raise PreventUpdate
class ImportancesComponent(ExplainerComponent):
def __init__(self, explainer, title="Feature Importances", name=None,
subtitle="Which features had the biggest impact?",
hide_type=False, hide_depth=False, hide_cats=False,
hide_title=False, hide_subtitle=False, hide_selector=False,
pos_label=None, importance_type="shap", depth=None,
cats=True, no_permutations=False,
description=None, **kwargs):
"""Display features importances component
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"Feature Importances".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
subtitle(str, optional): Subtitle.
hide_type (bool, optional): Hide permutation/shap selector toggle.
Defaults to False.
hide_depth (bool, optional): Hide number of features toggle.
Defaults to False.
hide_cats (bool, optional): Hide group cats toggle.
Defaults to False.
hide_title (bool, optional): hide title. Defaults to False.
hide_subtitle (bool, optional): Hide subtitle. Defaults to False.
hide_selector (bool, optional): hide pos label selectors.
Defaults to False.
pos_label ({int, str}, optional): initial pos label.
Defaults to explainer.pos_label
importance_type (str, {'permutation', 'shap'} optional):
initial importance type to display. Defaults to "shap".
depth (int, optional): Initial number of top features to display.
Defaults to None (=show all).
cats (bool, optional): Group categoricals. Defaults to True.
no_permutations (bool, optional): Do not use the permutation
importances for this component. Defaults to False.
description (str, optional): Tooltip to display when hover over
component title. When None default text is shown.
"""
super().__init__(explainer, title, name)
if not self.explainer.cats:
self.hide_cats = True
assert importance_type in ['shap', 'permutation'], \
"importance type must be either 'shap' or 'permutation'!"
if depth is not None:
self.depth = min(depth, len(explainer.columns_ranked_by_shap(cats)))
self.selector = PosLabelSelector(explainer, name=self.name, pos_label=pos_label)
if self.explainer.y_missing or self.no_permutations:
self.hide_type = True
self.importance_type = 'shap'
if self.description is None: self.description = f"""
Shows the features sorted from most important to least important. Can
be either sorted by absolute SHAP value (average absolute impact of
the feature on final prediction) or by permutation importance (how much
does the model get worse when you shuffle this feature, rendering it
useless?).
"""
self.register_dependencies('shap_values', 'shap_values_cats')
if not (self.hide_type and self.importance_type == 'shap'):
self.register_dependencies('permutation_importances', 'permutation_importances_cats')
def layout(self):
return dbc.Card([
make_hideable(
dbc.CardHeader([
html.Div([
html.H3(self.title, className="card-title", id='importances-title-'+self.name),
make_hideable(html.H6(self.subtitle, className='card-subtitle'), hide=self.hide_subtitle),
dbc.Tooltip(self.description, target='importances-title-'+self.name),
]),
]), hide=self.hide_title),
dbc.CardBody([
dbc.Row([
make_hideable(
dbc.Col([
dbc.FormGroup([
dbc.Label("Importances type:"),
dbc.Select(
options=[
{'label': 'Permutation Importances',
'value': 'permutation'},
{'label': 'SHAP values',
'value': 'shap'}
],
value=self.importance_type,
id='importances-permutation-or-shap-'+self.name,
#inline=True,
),
], id='importances-permutation-or-shap-form-'+self.name),
dbc.Tooltip("Select Feature importance type: \n"
"Permutation Importance: How much does performance metric decrease when shuffling this feature?\n"
"SHAP values: What is the average SHAP contribution (positive or negative) of this feature?",
target='importances-permutation-or-shap-form-'+self.name),
], md=3), self.hide_type),
make_hideable(
dbc.Col([
html.Label('Depth:', id='importances-depth-label-'+self.name),
dbc.Select(id='importances-depth-'+self.name,
options = [{'label': str(i+1), 'value':i+1}
for i in range(self.explainer.n_features(self.cats))],
value=self.depth),
dbc.Tooltip("Select how many features to display", target='importances-depth-label-'+self.name)
], md=2), self.hide_depth),
make_hideable(
dbc.Col([
dbc.FormGroup([
dbc.Label("Grouping:", id='importances-group-cats-label-'+self.name),
dbc.Tooltip("Group onehot encoded categorical variables together",
target='importances-group-cats-label-'+self.name),
dbc.Checklist(
options=[
{"label": "Group cats", "value": True},
],
value=[True] if self.cats else [],
id='importances-group-cats-'+self.name,
inline=True,
switch=True,
),
]),
]), self.hide_cats),
make_hideable(
dbc.Col([self.selector.layout()
], width=2), hide=self.hide_selector)
], form=True),
dbc.Row([
dbc.Col([
dcc.Loading(id='importances-graph-loading-'+self.name,
children=dcc.Graph(id='importances-graph-'+self.name,
config=dict(modeBarButtons=[['toImage']], displaylogo=False))),
]),
]),
])
])
def component_callbacks(self, app, **kwargs):
@app.callback(
[Output('importances-graph-'+self.name, 'figure'),
Output('importances-depth-'+self.name, 'options')],
[Input('importances-depth-'+self.name, 'value'),
Input('importances-group-cats-'+self.name, 'value'),
Input('importances-permutation-or-shap-'+self.name, 'value'),
Input('pos-label-'+self.name, 'value')],
)
def update_importances(depth, cats, permutation_shap, pos_label):
depth = None if depth is None else int(depth)
plot = self.explainer.plot_importances(
kind=permutation_shap, topx=depth,
cats=bool(cats), pos_label=pos_label)
trigger = dash.callback_context.triggered[0]['prop_id'].split('.')[0]
if trigger == 'importances-group-cats-'+self.name:
depth_options = [{'label': str(i+1), 'value': i+1}
for i in range(self.explainer.n_features(bool(cats)))]
return (plot, depth_options)
else:
return (plot, dash.no_update)
class PdpComponent(ExplainerComponent):
def __init__(self, explainer, title="Partial Dependence Plot", name=None,
subtitle="How does the prediction change if you change one feature?",
hide_col=False, hide_index=False, hide_cats=False,
hide_title=False, hide_subtitle=False,
hide_footer=False, hide_selector=False,
hide_dropna=False, hide_sample=False,
hide_gridlines=False, hide_gridpoints=False,
feature_input_component=None,
pos_label=None, col=None, index=None, cats=True,
dropna=True, sample=100, gridlines=50, gridpoints=10,
description=None, **kwargs):
"""Show Partial Dependence Plot component
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"Partial Dependence Plot".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
subtitle (str): subtitle
hide_col (bool, optional): Hide feature selector. Defaults to False.
hide_index (bool, optional): Hide index selector. Defaults to False.
hide_cats (bool, optional): Hide group cats toggle. Defaults to False.
hide_title (bool, optional): Hide title, Defaults to False.
hide_subtitle (bool, optional): Hide subtitle. Defaults to False.
hide_footer (bool, optional): hide the footer at the bottom of the component
hide_selector (bool, optional): hide pos label selectors. Defaults to False.
hide_dropna (bool, optional): Hide drop na's toggle Defaults to False.
hide_sample (bool, optional): Hide sample size input. Defaults to False.
hide_gridlines (bool, optional): Hide gridlines input. Defaults to False.
hide_gridpoints (bool, optional): Hide gridpounts input. Defaults to False.
feature_input_component (FeatureInputComponent): A FeatureInputComponent
that will give the input to the graph instead of the index selector.
If not None, hide_index=True. Defaults to None.
pos_label ({int, str}, optional): initial pos label.
Defaults to explainer.pos_label
col (str, optional): Feature to display PDP for. Defaults to None.
index ({int, str}, optional): Index to add ice line to plot. Defaults to None.
cats (bool, optional): Group categoricals for feature selector. Defaults to True.
dropna (bool, optional): Drop rows where values equal explainer.na_fill (usually -999). Defaults to True.
sample (int, optional): Sample size to calculate average partial dependence. Defaults to 100.
gridlines (int, optional): Number of ice lines to display in plot. Defaults to 50.
gridpoints (int, optional): Number of breakpoints on horizontal axis Defaults to 10.
description (str, optional): Tooltip to display when hover over
component title. When None default text is shown.
"""
super().__init__(explainer, title, name)
self.index_name = 'pdp-index-'+self.name
if self.col is None:
self.col = self.explainer.columns_ranked_by_shap(self.cats)[0]
if not self.explainer.cats:
self.hide_cats = True
if self.feature_input_component is not None:
self.exclude_callbacks(self.feature_input_component)
self.hide_index = True
if self.description is None: self.description = f"""
The partial dependence plot (pdp) show how the model prediction would
change if you change one particular feature. The plot shows you a sample
of observations and how these observations would change with this
feature (gridlines). The average effect is shown in grey. The effect
of changing the feature for a single {self.explainer.index_name} is
shown in blue. You can adjust how many observations to sample for the
average, how many gridlines to show, and how many points along the
x-axis to calculate model predictions for (gridpoints).
"""
self.selector = PosLabelSelector(explainer, name=self.name, pos_label=pos_label)
def layout(self):
return dbc.Card([
make_hideable(
dbc.CardHeader([
html.Div([
html.H3(self.title, id='pdp-title-'+self.name),
make_hideable(html.H6(self.subtitle, className='card-subtitle'), hide=self.hide_subtitle),
dbc.Tooltip(self.description, target='pdp-title-'+self.name),
]),
]), hide=self.hide_title),
dbc.CardBody([
dbc.Row([
make_hideable(
dbc.Col([
dbc.Label("Feature:",
html_for='pdp-col'+self.name, id='pdp-col-label-'+self.name),
dbc.Tooltip("Select the feature for which you want to see the partial dependence plot",
target='pdp-col-label-'+self.name),
dbc.Select(id='pdp-col-'+self.name,
options=[{'label': col, 'value':col}
for col in self.explainer.columns_ranked_by_shap(self.cats)],
value=self.col),
], md=4), hide=self.hide_col),
make_hideable(
dbc.Col([
dbc.Label(f"{self.explainer.index_name}:", id='pdp-index-label-'+self.name),
dbc.Tooltip(f"Select the {self.explainer.index_name} to display the partial dependence plot for",
target='pdp-index-label-'+self.name),
dcc.Dropdown(id='pdp-index-'+self.name,
options = [{'label': str(idx), 'value':idx}
for idx in self.explainer.idxs],
value=self.index)
], md=4), hide=self.hide_index),
make_hideable(
dbc.Col([self.selector.layout()
], width=2), hide=self.hide_selector),
make_hideable(
dbc.Col([
dbc.FormGroup([
dbc.Label("Grouping:", id='pdp-group-cats-label-'+self.name),
dbc.Tooltip("Group onehot encoded categorical variables together",
target='pdp-group-cats-label-'+self.name),
dbc.Checklist(
options=[
{"label": "Group cats", "value": True},
],
value=[True] if self.cats else [],
id='pdp-group-cats-'+self.name,
inline=True,
switch=True,
),
]),
], md=2), hide=self.hide_cats),
], form=True),
dbc.Row([
dbc.Col([
dcc.Loading(id='loading-pdp-graph-'+self.name,
children=[dcc.Graph(id='pdp-graph-'+self.name,
config=dict(modeBarButtons=[['toImage']], displaylogo=False))]),
])
]),
]),
make_hideable(
dbc.CardFooter([
dbc.Row([
make_hideable(
dbc.Col([
dbc.FormGroup([
dbc.Label("Drop fill:"),
dbc.Tooltip("Drop all observations with feature values "
f"equal to {self.explainer.na_fill} from the plot. "
"This prevents the filler values from ruining the x-axis.",
target='pdp-dropna-'+self.name),
dbc.Checklist(
options=[{"label": "Drop na_fill", "value": True}],
value=[True] if self.dropna else [],
id='pdp-dropna-'+self.name,
inline=True,
switch=True,
),
]),
]), hide=self.hide_dropna),
make_hideable(
dbc.Col([
dbc.Label("Pdp sample size:", id='pdp-sample-label-'+self.name ),
dbc.Tooltip("Number of observations to use to calculate average partial dependence",
target='pdp-sample-label-'+self.name ),
dbc.Input(id='pdp-sample-'+self.name, value=self.sample,
type="number", min=0, max=len(self.explainer), step=1),
]), hide=self.hide_sample),
make_hideable(
dbc.Col([ #gridlines
dbc.Label("Gridlines:", id='pdp-gridlines-label-'+self.name ),
dbc.Tooltip("Number of individual observations' partial dependences to show in plot",
target='pdp-gridlines-label-'+self.name),
dbc.Input(id='pdp-gridlines-'+self.name, value=self.gridlines,
type="number", min=0, max=len(self.explainer), step=1),
]), hide=self.hide_gridlines),
make_hideable(
dbc.Col([ #gridpoints
dbc.Label("Gridpoints:", id='pdp-gridpoints-label-'+self.name ),
dbc.Tooltip("Number of points to sample the feature axis for predictions."
" The higher, the smoother the curve, but takes longer to calculate",
target='pdp-gridpoints-label-'+self.name ),
dbc.Input(id='pdp-gridpoints-'+self.name, value=self.gridpoints,
type="number", min=0, max=100, step=1),
]), hide=self.hide_gridpoints),
], form=True),
]), hide=self.hide_footer)
])
def component_callbacks(self, app):
@app.callback(
Output('pdp-col-'+self.name, 'options'),
[Input('pdp-group-cats-'+self.name, 'value')],
[State('pos-label-'+self.name, 'value')]
)
def update_pdp_graph(cats, pos_label):
col_options = [{'label': col, 'value':col}
for col in self.explainer.columns_ranked_by_shap(bool(cats), pos_label=pos_label)]
return col_options
if self.feature_input_component is None:
@app.callback(
Output('pdp-graph-'+self.name, 'figure'),
[Input('pdp-index-'+self.name, 'value'),
Input('pdp-col-'+self.name, 'value'),
Input('pdp-dropna-'+self.name, 'value'),
Input('pdp-sample-'+self.name, 'value'),
Input('pdp-gridlines-'+self.name, 'value'),
Input('pdp-gridpoints-'+self.name, 'value'),
Input('pos-label-'+self.name, 'value')]
)
def update_pdp_graph(index, col, drop_na, sample, gridlines, gridpoints, pos_label):
return self.explainer.plot_pdp(col, index,
drop_na=bool(drop_na), sample=sample, gridlines=gridlines, gridpoints=gridpoints,
pos_label=pos_label)
else:
@app.callback(
Output('pdp-graph-'+self.name, 'figure'),
[Input('pdp-col-'+self.name, 'value'),
Input('pdp-dropna-'+self.name, 'value'),
Input('pdp-sample-'+self.name, 'value'),
Input('pdp-gridlines-'+self.name, 'value'),
Input('pdp-gridpoints-'+self.name, 'value'),
Input('pos-label-'+self.name, 'value'),
*self.feature_input_component._feature_callback_inputs]
)
def update_pdp_graph(col, drop_na, sample, gridlines, gridpoints, pos_label, *inputs):
X_row = self.explainer.get_row_from_input(inputs, ranked_by_shap=True)
return self.explainer.plot_pdp(col, X_row=X_row,
drop_na=bool(drop_na), sample=sample, gridlines=gridlines, gridpoints=gridpoints,
pos_label=pos_label)
class FeatureInputComponent(ExplainerComponent):
def __init__(self, explainer, title="Feature Input", name=None,
subtitle="Adjust the feature values to change the prediction",
hide_title=False, hide_subtitle=False, hide_index=False,
hide_range=False,
index=None, n_input_cols=2, description=None, **kwargs):
"""Interaction Dependence Component.
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"What if...".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
subtitle (str): subtitle
hide_title (bool, optional): hide the title
hide_subtitle (bool, optional): Hide subtitle. Defaults to False.
hide_index (bool, optional): hide the index selector
hide_range (bool, optional): hide the range label under the inputs
index (str, int, optional): default index
n_input_cols (int): number of columns to split features inputs in.
Defaults to 2.
description (str, optional): Tooltip to display when hover over
component title. When None default text is shown.
"""
super().__init__(explainer, title, name)
assert len(explainer.columns) == len(set(explainer.columns)), \
"Not all X column names are unique, so cannot launch FeatureInputComponent component/tab!"
self.index_name = 'feature-input-index-'+self.name
self._input_features = self.explainer.columns_ranked_by_shap(cats=True)
self._feature_inputs = [
self._generate_dash_input(
feature, self.explainer.cats, self.explainer.cats_dict)
for feature in self._input_features]
self._feature_callback_inputs = [Input('feature-input-'+feature+'-input-'+self.name, 'value') for feature in self._input_features]
self._feature_callback_outputs = [Output('feature-input-'+feature+'-input-'+self.name, 'value') for feature in self._input_features]
if self.description is None: self.description = """
Adjust the input values to see predictions for what if scenarios."""
def _generate_dash_input(self, col, cats, cats_dict):
if col in cats:
col_values = [
col_val[len(col)+1:] if col_val.startswith(col+"_") else col_val
for col_val in cats_dict[col]]
return dbc.FormGroup([
dbc.Label(col),
dcc.Dropdown(id='feature-input-'+col+'-input-'+self.name,
options=[dict(label=col_val, value=col_val) for col_val in col_values],
clearable=False),
#dbc.FormText(f"Select any {col}") if not self.hide_range else None,
])
else:
min_range = np.round(self.explainer.X[col][lambda x: x != self.explainer.na_fill].min(), 2)
max_range = np.round(self.explainer.X[col][lambda x: x != self.explainer.na_fill].max(), 2)
return dbc.FormGroup([
dbc.Label(col),
dbc.Input(id='feature-input-'+col+'-input-'+self.name, type="number"),
#dbc.FormText(f"Range: {min_range}-{max_range}") if not self.hide_range else None
])
def get_slices(self, n_inputs, n_cols=2):
"""returns a list of slices to divide n inputs into n_cols columns"""
if n_inputs < n_cols:
n_cols = n_inputs
rows_per_col = ceil(n_inputs / n_cols)
slices = []
for col in range(n_cols):
if col == n_cols-1 and n_inputs % rows_per_col > 0:
slices.append(slice(col*rows_per_col, col*rows_per_col+(n_inputs % rows_per_col)))
else:
slices.append(slice(col*rows_per_col, col*rows_per_col+rows_per_col))
return slices
def layout(self):
return html.Div([
make_hideable(
dbc.CardHeader([
html.Div([
html.H3(self.title, id='feature-input-title-'+self.name),
make_hideable(html.H6(self.subtitle, className='card-subtitle'), hide=self.hide_subtitle),
dbc.Tooltip(self.description, target='feature-input-title-'+self.name),
]),
]), hide=self.hide_title),
dbc.CardBody([
dbc.Row([
make_hideable(
dbc.Col([
dbc.Label(f"{self.explainer.index_name}:"),
dcc.Dropdown(id='feature-input-index-'+self.name,
options = [{'label': str(idx), 'value':idx}
for idx in self.explainer.idxs],
value=self.index)
], md=4), hide=self.hide_index),
], form=True),
dbc.Row([dbc.Col(self._feature_inputs[slicer])
for slicer in self.get_slices(len(self._feature_inputs), self.n_input_cols)]),
])
], style={"height":"190px","overflow":"auto","margin-bottom":"0px"})
def component_callbacks(self, app):
@app.callback(
[*self._feature_callback_outputs],
[Input('feature-input-index-'+self.name, 'value')]
)
def update_whatif_inputs(index):
idx = self.explainer.get_int_idx(index)
if idx is None:
raise PreventUpdate
feature_values = (self.explainer.X_cats
[self.explainer.columns_ranked_by_shap(cats=True)]
.iloc[[idx]].values[0].tolist())
return feature_values
class WhatIfComponent(ExplainerComponent):
def __init__(self, explainer, title="What if...", name=None,
hide_title=False, hide_subtitle=False, hide_index=False,
hide_selector=False, hide_contributions=False, hide_pdp=False,
index=None, pdp_col=None, pos_label=None, description=None,
**kwargs):
"""Interaction Dependence Component.
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"What if...".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
hide_title (bool, optional): hide the title
hide_subtitle (bool, optional): Hide subtitle. Defaults to False.
hide_index (bool, optional): hide the index selector
hide_selector (bool, optional): hide the pos_label selector
hide_contributions (bool, optional): hide the contributions graph
hide_pdp (bool, optional): hide the pdp graph
index (str, int, optional): default index
pdp_col (str, optional): default pdp feature col
pos_label ({int, str}, optional): initial pos label.
Defaults to explainer.pos_label
description (str, optional): Tooltip to display when hover over
component title. When None default text is shown.
"""
super().__init__(explainer, title, name)
assert len(explainer.columns) == len(set(explainer.columns)), \
"Not all column names are unique, so cannot launch whatif component/tab!"
if self.pdp_col is None:
self.pdp_col = self.explainer.columns_ranked_by_shap(cats=True)[0]
self.index_name = 'whatif-index-'+self.name
self._input_features = self.explainer.columns_cats
self._feature_inputs = [
self._generate_dash_input(
feature, self.explainer.cats, self.explainer.cats_dict)
for feature in self._input_features]
self._feature_callback_inputs = [Input('whatif-'+feature+'-input-'+self.name, 'value') for feature in self._input_features]
self._feature_callback_outputs = [Output('whatif-'+feature+'-input-'+self.name, 'value') for feature in self._input_features]
self.selector = PosLabelSelector(explainer, name=self.name, pos_label=pos_label)
self.register_dependencies('preds', 'shap_values')
def _generate_dash_input(self, col, cats, cats_dict):
if col in cats:
col_values = [
col_val[len(col)+1:] if col_val.startswith(col+"_") else col_val
for col_val in cats_dict[col]]
return html.Div([
html.P(col),
dcc.Dropdown(id='whatif-'+col+'-input-'+self.name,
options=[dict(label=col_val, value=col_val) for col_val in col_values],
clearable=False)
])
else:
return html.Div([
html.P(col),
dbc.Input(id='whatif-'+col+'-input-'+self.name, type="number"),
])
def layout(self):
return dbc.Card([
make_hideable(
dbc.CardHeader([
dbc.Row([
dbc.Col([
html.H1(self.title)
]),
]),
]), hide=self.hide_title),
dbc.CardBody([
dbc.Row([
make_hideable(
dbc.Col([
dbc.Label(f"{self.explainer.index_name}:"),
dcc.Dropdown(id='whatif-index-'+self.name,
options = [{'label': str(idx), 'value':idx}
for idx in self.explainer.idxs],
value=self.index)
], md=4), hide=self.hide_index),
make_hideable(
dbc.Col([self.selector.layout()
], md=2), hide=self.hide_selector),
], form=True),
dbc.Row([
dbc.Col([
html.H3("Edit Feature input:")
])
]),
dbc.Row([
dbc.Col([
*self._feature_inputs[:int((len(self._feature_inputs) + 1)/2)]
]),
dbc.Col([
*self._feature_inputs[int((len(self._feature_inputs) + 1)/2):]
]),
]),
dbc.Row([
make_hideable(
dbc.Col([
html.H3("Prediction and contributions:"),
dcc.Graph(id='whatif-contrib-graph-'+self.name,
config=dict(modeBarButtons=[['toImage']], displaylogo=False)),
]), hide=self.hide_contributions),
make_hideable(
dbc.Col([
html.H3("Partial dependence:"),
dcc.Dropdown(id='whatif-pdp-col-'+self.name,
options=[dict(label=col, value=col) for col in self._input_features],
value=self.pdp_col),
dcc.Graph(id='whatif-pdp-graph-'+self.name,
config=dict(modeBarButtons=[['toImage']], displaylogo=False)),
]), hide=self.hide_pdp),
])
])
])
def component_callbacks(self, app):
@app.callback(
[Output('whatif-contrib-graph-'+self.name, 'figure'),
Output('whatif-pdp-graph-'+self.name, 'figure')],
[Input('whatif-pdp-col-'+self.name, 'value'),
Input('pos-label-'+self.name, 'value'),
*self._feature_callback_inputs,
],
)
def update_whatif_plots(pdp_col, pos_label, *input_args):
X_row = pd.DataFrame(dict(zip(self._input_features, input_args)), index=[0]).fillna(0)
contrib_plot = self.explainer.plot_shap_contributions(X_row=X_row, pos_label=pos_label)
pdp_plot = self.explainer.plot_pdp(pdp_col, X_row=X_row, pos_label=pos_label)
return contrib_plot, pdp_plot
@app.callback(
[*self._feature_callback_outputs],
[Input('whatif-index-'+self.name, 'value')]
)
def update_whatif_inputs(index):
idx = self.explainer.get_int_idx(index)
if idx is None:
raise PreventUpdate
feature_values = self.explainer.X_cats.iloc[[idx]].values[0].tolist()
return feature_values | AMLBID | /Explainer/dashboard_components/overview_components.py | overview_components.py |
__all__ = [
'ClassifierRandomIndexComponent',
'ClassifierRandomIndexComponentPerso',
'ClassifierPredictionSummaryComponent',
'ClassifierPredictionSummaryComponentPerso',
'PrecisionComponent',
'ConfusionMatrixComponent',
'LiftCurveComponent',
'ClassificationComponent',
'RocAucComponent',
'PrAucComponent',
'CumulativePrecisionComponent',
'ClassifierModelSummaryComponent'
]
import numpy as np
import pandas as pd
import dash
import dash_core_components as dcc
import dash_bootstrap_components as dbc
import dash_html_components as html
from dash.dependencies import Input, Output, State
from dash.exceptions import PreventUpdate
import plotly.graph_objs as go
from ..dashboard_methods import *
class ClassifierRandomIndexComponentPerso(ExplainerComponent):
def __init__(self, explainer, title="Select Random Index", name=None,
subtitle="Select from list or pick at random",
hide_title=False, hide_subtitle=False,
hide_index=False, hide_slider=False,
hide_labels=False, hide_pred_or_perc=False,
hide_selector=False, hide_button=False,
pos_label=None, index=None, slider= None, labels=None,
pred_or_perc='predictions', description=None,
**kwargs):
"""Select a random index subject to constraints component
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"Select Random Index".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
subtitle (str): subtitle
hide_title (bool, optional): Hide title. Defaults to False.
hide_subtitle (bool, optional): Hide subtitle. Defaults to False.
hide_index (bool, optional): Hide index selector. Defaults to False.
hide_slider (bool, optional): Hide prediction/percentile slider.
Defaults to False.
hide_labels (bool, optional): Hide label selector Defaults to False.
hide_pred_or_perc (bool, optional): Hide prediction/percentiles
toggle. Defaults to False.
hide_selector (bool, optional): hide pos label selectors. Defaults to False.
hide_button (bool, optional): Hide button. Defaults to False.
pos_label ({int, str}, optional): initial pos label. Defaults to explainer.pos_label
index ({str, int}, optional): Initial index to display.
Defaults to None.
slider ([float,float], optional): initial slider position
[lower bound, upper bound]. Defaults to None.
labels ([str], optional): list of initial labels(str) to include.
Defaults to None.
pred_or_perc (str, optional): Whether to use prediction or
percentiles slider. Defaults to 'predictions'.
description (str, optional): Tooltip to display when hover over
component title. When None default text is shown.
"""
super().__init__(explainer, title, name)
assert self.explainer.is_classifier, \
("explainer is not a ClassifierExplainer ""so the ClassifierRandomIndexComponent "
" will not work. Try using the RegressionRandomIndexComponent instead.")
self.index_name = 'random-index-clas-index-'+self.name
if self.slider is None:
self.slider = [0.0, 1.0]
if self.labels is None:
self.labels = self.explainer.labels
if self.explainer.y_missing:
self.hide_labels = True
self.selector = PosLabelSelector(explainer, name=self.name, pos_label=pos_label)
assert (len(self.slider) == 2 and
self.slider[0] >= 0 and self.slider[0] <=1 and
self.slider[1] >= 0.0 and self.slider[1] <= 1.0 and
self.slider[0] <= self.slider[1]), \
"slider should be e.g. [0.5, 1.0]"
assert all([lab in self.explainer.labels for lab in self.labels]), \
f"These labels are not in explainer.labels: {[lab for lab in self.labels if lab not in explainer.labels]}!"
assert self.pred_or_perc in ['predictions', 'percentiles'], \
"pred_or_perc should either be `predictions` or `percentiles`!"
if self.description is None: self.description = f"""
You can select a {self.explainer.index_name} directly by choosing it
from the dropdown (if you start typing you can search inside the list),
or hit the Random {self.explainer.index_name} button to randomly select
a {self.explainer.index_name} that fits the constraints. For example
you can select a {self.explainer.index_name} where the observed
{self.explainer.target} is {self.explainer.labels[0]} but the
predicted probability of {self.explainer.labels[1]} is very high. This
allows you to for example sample only false positives or only false negatives.
"""
def layout(self):
return html.Div([
make_hideable(
dbc.CardHeader([
html.Div([
html.H3(f"Select {self.explainer.index_name}", id='random-index-clas-title-'+self.name),
make_hideable(make_hideable(html.H6(self.subtitle, className='card-subtitle'), hide=self.hide_subtitle), hide=self.hide_subtitle),
dbc.Tooltip(self.description, target='random-index-clas-title-'+self.name),
]),
]), hide=self.hide_title),
dbc.CardBody([
dbc.Row([
dbc.Col([
make_hideable(
dbc.Col([
self.selector.layout()
], md=2), hide=self.hide_selector),
])
]),
dbc.Row([
make_hideable(
dbc.Col([
dcc.Dropdown(id='random-index-clas-index-'+self.name,
options = [{'label': str(idx), 'value':idx}
for idx in self.explainer.idxs],
value=self.index),
], md=8), hide=self.hide_index),
make_hideable(
dbc.Col([
dbc.Button(f"Rand. {self.explainer.index_name}", color="primary", id='random-index-clas-button-'+self.name, block=True),
dbc.Tooltip(f"Select a random {self.explainer.index_name} according to the constraints",
target='random-index-clas-button-'+self.name),
], md=4), hide=self.hide_button),
], form=True, style=dict(marginBottom=0)),
dbc.Row([
make_hideable(
dbc.Col([
dbc.Label(f"Observed {self.explainer.target}:", id='random-index-clas-labels-label-'+self.name),
dbc.Tooltip(f"Only select a random {self.explainer.index_name} where the observed "
f"{self.explainer.target} is one of the selected labels:",
target='random-index-clas-labels-label-'+self.name),
dcc.Dropdown(
id='random-index-clas-labels-'+self.name,
options=[{'label': lab, 'value': lab} for lab in self.explainer.labels],
multi=True,
value=self.labels),
], md=8), hide=self.hide_labels),
make_hideable(
dbc.Col([
html.Div([
dbc.Label("Range:", html_for='random-index-clas-pred-or-perc-'+self.name),
dbc.Select(
id='random-index-clas-pred-or-perc-'+self.name,
options=[
{'label': 'probability', 'value': 'predictions'},
{'label': 'percentile', 'value': 'percentiles'},
],
value=self.pred_or_perc)
], id='random-index-clas-pred-or-perc-div-'+self.name),
dbc.Tooltip("Instead of selecting from a range of predicted probabilities "
"you can also select from a range of predicted percentiles. "
"For example if you set the slider to percentile (0.9-1.0) you would"
f" only sample random {self.explainer.index_name} from the top "
"10% highest predicted probabilities.",
target='random-index-clas-pred-or-perc-div-'+self.name),
], md=4), hide=self.hide_pred_or_perc),
], style=dict(marginBottom=0)),
dbc.Row([
make_hideable(
dbc.Col([
html.Div([
dbc.Label(id='random-index-clas-slider-label-'+self.name,
children="Predicted probability range:",
html_for='prediction-range-slider-'+self.name),
dbc.Tooltip(f"Only select a random {self.explainer.index_name} where the "
"predicted probability of positive label is in the following range:",
id='random-index-clas-slider-label-tooltip-'+self.name,
target='random-index-clas-slider-label-'+self.name),
dcc.RangeSlider(
id='random-index-clas-slider-'+self.name,
min=0.0, max=1.0, step=0.01,
value=self.slider, allowCross=False,
marks={0.0:'0.0', 0.2:'0.2', 0.4:'0.4', 0.6:'0.6',
0.8:'0.8', 1.0:'1.0'},
tooltip = {'always_visible' : False})
])
]), hide=self.hide_slider),
], justify="start"),
]),
])
def component_callbacks(self, app):
@app.callback(
Output('random-index-clas-index-'+self.name, 'value'),
[Input('random-index-clas-button-'+self.name, 'n_clicks')],
[State('random-index-clas-slider-'+self.name, 'value'),
State('random-index-clas-labels-'+self.name, 'value'),
State('random-index-clas-pred-or-perc-'+self.name, 'value'),
State('pos-label-'+self.name, 'value')])
def update_index(n_clicks, slider_range, labels, pred_or_perc, pos_label):
if n_clicks is None and self.index is not None:
raise PreventUpdate
if pred_or_perc == 'predictions':
return self.explainer.random_index(y_values=labels,
pred_proba_min=slider_range[0], pred_proba_max=slider_range[1],
return_str=True, pos_label=pos_label)
elif pred_or_perc == 'percentiles':
return self.explainer.random_index(y_values=labels,
pred_percentile_min=slider_range[0], pred_percentile_max=slider_range[1],
return_str=True, pos_label=pos_label)
@app.callback(
[Output('random-index-clas-slider-label-'+self.name, 'children'),
Output('random-index-clas-slider-label-tooltip-'+self.name, 'children')],
[Input('random-index-clas-pred-or-perc-'+self.name, 'value'),
Input('pos-label-'+self.name, 'value')]
)
def update_slider_label(pred_or_perc, pos_label):
if pred_or_perc == 'predictions':
return (
"Predicted probability range:",
f"Only select a random {self.explainer.index_name} where the "
f"predicted probability of {self.explainer.labels[pos_label]}"
" is in the following range:"
)
elif pred_or_perc == 'percentiles':
return (
"Predicted percentile range:",
f"Only select a random {self.explainer.index_name} where the "
f"predicted probability of {self.explainer.labels[pos_label]}"
" is in the following percentile range. For example you can "
"only sample from the top 10% highest predicted probabilities."
)
raise PreventUpdate
class ClassifierPredictionSummaryComponentPerso(ExplainerComponent):
def __init__(self, explainer, title="Prediction", name=None,
hide_index=True, hide_title=False, hide_subtitle=False,
hide_table=False, hide_piechart=False,
hide_star_explanation=False, hide_selector=False,
feature_input_component=None,
pos_label=None, index=None, round=3, description=None,
**kwargs):
"""Shows a summary for a particular prediction
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"Prediction Summary".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
hide_index (bool, optional): hide index selector. Defaults to False.
hide_title (bool, optional): hide title. Defaults to False.
hide_subtitle (bool, optional): Hide subtitle. Defaults to False.
hide_table (bool, optional): hide the results table
hide_piechart (bool, optional): hide the results piechart
hide_star_explanation (bool, optional): hide the `* indicates..`
Defaults to False.
hide_selector (bool, optional): hide pos label selectors.
Defaults to False.
feature_input_component (FeatureInputComponent): A FeatureInputComponent
that will give the input to the graph instead of the index selector.
If not None, hide_index=True. Defaults to None.
pos_label ({int, str}, optional): initial pos label.
Defaults to explainer.pos_label
index ({int, str}, optional): Index to display prediction summary
for. Defaults to None.
round (int, optional): rounding to apply to pred_proba float.
Defaults to 3.
description (str, optional): Tooltip to display when hover over
component title. When None default text is shown.
"""
super().__init__(explainer, title, name)
self.index_name = 'clas-prediction-index-'+self.name
self.selector = PosLabelSelector(explainer, name=self.name, pos_label=pos_label)
if self.feature_input_component is not None:
self.exclude_callbacks(self.feature_input_component)
self.hide_index = True
if self.description is None: self.description = f"""
Shows the predicted probability for each {self.explainer.target} label.
"""
def layout(self):
return html.Div([
make_hideable(
dbc.CardHeader([
html.Div([
html.H3(self.title, id='clas-prediction-index-title-'+self.name, className='card-title'),
dbc.Tooltip(self.description, target='clas-prediction-index-title-'+self.name),
]),
]), hide=self.hide_title),
dbc.CardBody([
dbc.Row([
make_hideable(
dbc.Col([
dbc.Label(f"{self.explainer.index_name}:"),
dcc.Dropdown(id='clas-prediction-index-'+self.name,
options = [{'label': str(idx), 'value':idx}
for idx in self.explainer.idxs],
value=self.index)
], md=6), hide=self.hide_index),
make_hideable(
dbc.Col([self.selector.layout()
], width=3), hide=self.hide_selector),
]),
dbc.Row([
make_hideable(
dbc.Col([
html.Div(id='clas-prediction-div-'+self.name),
make_hideable(
html.Div("* indicates observed label") if not self.explainer.y_missing else None,
hide=self.hide_star_explanation),
]), hide=self.hide_table),
make_hideable(
dbc.Col([
dcc.Graph(id='clas-prediction-graph-'+self.name,
config=dict(modeBarButtons=[['toImage']], displaylogo=False))
] ), hide=self.hide_piechart),
]),
])
])
def component_callbacks(self, app):
if self.feature_input_component is None:
@app.callback(
[Output('clas-prediction-div-'+self.name, 'children'),
Output('clas-prediction-graph-'+self.name, 'figure')],
[Input('clas-prediction-index-'+self.name, 'value'),
Input('pos-label-'+self.name, 'value')])
def update_output_div(index, pos_label):
if index is not None:
fig = self.explainer.plot_prediction_result(index, showlegend=False)
preds_df = self.explainer.prediction_result_df(index, round=self.round, logodds=True)
preds_df.probability = np.round(100*preds_df.probability.values, self.round).astype(str)
preds_df.probability = preds_df.probability + ' %'
preds_df.logodds = np.round(preds_df.logodds.values, self.round).astype(str)
if self.explainer.model_output!='logodds':
preds_df = preds_df[['label', 'probability']]
preds_table = dbc.Table.from_dataframe(preds_df,
striped=False, bordered=False, hover=False)
return preds_table, fig
raise PreventUpdate
else:
@app.callback(
[Output('clas-prediction-div-'+self.name, 'children'),
Output('clas-prediction-graph-'+self.name, 'figure')],
[Input('pos-label-'+self.name, 'value'),
*self.feature_input_component._feature_callback_inputs])
def update_output_div(pos_label, *inputs):
X_row = self.explainer.get_row_from_input(inputs, ranked_by_shap=True)
fig = self.explainer.plot_prediction_result(X_row=X_row, showlegend=False)
preds_df = self.explainer.prediction_result_df(X_row=X_row, round=self.round, logodds=True)
preds_df.probability = np.round(100*preds_df.probability.values, self.round).astype(str)
preds_df.probability = preds_df.probability + ' %'
preds_df.logodds = np.round(preds_df.logodds.values, self.round).astype(str)
if self.explainer.model_output!='logodds':
preds_df = preds_df[['label', 'probability']]
preds_table = dbc.Table.from_dataframe(preds_df,
striped=False, bordered=False, hover=False)
return preds_table, fig
class ClassifierRandomIndexComponent(ExplainerComponent):
def __init__(self, explainer, title="Select Random Index", name=None,
subtitle="Select from list or pick at random",
hide_title=False, hide_subtitle=False,
hide_index=False, hide_slider=False,
hide_labels=False, hide_pred_or_perc=False,
hide_selector=False, hide_button=False,
pos_label=None, index=None, slider= None, labels=None,
pred_or_perc='predictions', description=None,
**kwargs):
"""Select a random index subject to constraints component
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"Select Random Index".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
subtitle (str): subtitle
hide_title (bool, optional): Hide title. Defaults to False.
hide_subtitle (bool, optional): Hide subtitle. Defaults to False.
hide_index (bool, optional): Hide index selector. Defaults to False.
hide_slider (bool, optional): Hide prediction/percentile slider.
Defaults to False.
hide_labels (bool, optional): Hide label selector Defaults to False.
hide_pred_or_perc (bool, optional): Hide prediction/percentiles
toggle. Defaults to False.
hide_selector (bool, optional): hide pos label selectors. Defaults to False.
hide_button (bool, optional): Hide button. Defaults to False.
pos_label ({int, str}, optional): initial pos label. Defaults to explainer.pos_label
index ({str, int}, optional): Initial index to display.
Defaults to None.
slider ([float,float], optional): initial slider position
[lower bound, upper bound]. Defaults to None.
labels ([str], optional): list of initial labels(str) to include.
Defaults to None.
pred_or_perc (str, optional): Whether to use prediction or
percentiles slider. Defaults to 'predictions'.
description (str, optional): Tooltip to display when hover over
component title. When None default text is shown.
"""
super().__init__(explainer, title, name)
assert self.explainer.is_classifier, \
("explainer is not a ClassifierExplainer ""so the ClassifierRandomIndexComponent "
" will not work. Try using the RegressionRandomIndexComponent instead.")
self.index_name = 'random-index-clas-index-'+self.name
if self.slider is None:
self.slider = [0.0, 1.0]
if self.labels is None:
self.labels = self.explainer.labels
if self.explainer.y_missing:
self.hide_labels = True
self.selector = PosLabelSelector(explainer, name=self.name, pos_label=pos_label)
assert (len(self.slider) == 2 and
self.slider[0] >= 0 and self.slider[0] <=1 and
self.slider[1] >= 0.0 and self.slider[1] <= 1.0 and
self.slider[0] <= self.slider[1]), \
"slider should be e.g. [0.5, 1.0]"
assert all([lab in self.explainer.labels for lab in self.labels]), \
f"These labels are not in explainer.labels: {[lab for lab in self.labels if lab not in explainer.labels]}!"
assert self.pred_or_perc in ['predictions', 'percentiles'], \
"pred_or_perc should either be `predictions` or `percentiles`!"
if self.description is None: self.description = f"""
You can select a {self.explainer.index_name} directly by choosing it
from the dropdown (if you start typing you can search inside the list),
or hit the Random {self.explainer.index_name} button to randomly select
a {self.explainer.index_name} that fits the constraints. For example
you can select a {self.explainer.index_name} where the observed
{self.explainer.target} is {self.explainer.labels[0]} but the
predicted probability of {self.explainer.labels[1]} is very high. This
allows you to for example sample only false positives or only false negatives.
"""
def layout(self):
return dbc.Card([
make_hideable(
dbc.CardHeader([
html.Div([
html.H3(f"Select {self.explainer.index_name}", id='random-index-clas-title-'+self.name),
make_hideable(make_hideable(html.H6(self.subtitle, className='card-subtitle'), hide=self.hide_subtitle), hide=self.hide_subtitle),
dbc.Tooltip(self.description, target='random-index-clas-title-'+self.name),
]),
]), hide=self.hide_title),
dbc.CardBody([
dbc.Row([
dbc.Col([
make_hideable(
dbc.Col([
self.selector.layout()
], md=2), hide=self.hide_selector),
])
]),
dbc.Row([
make_hideable(
dbc.Col([
dcc.Dropdown(id='random-index-clas-index-'+self.name,
options = [{'label': str(idx), 'value':idx}
for idx in self.explainer.idxs],
value=self.index),
], md=8), hide=self.hide_index),
make_hideable(
dbc.Col([
dbc.Button(f"Random {self.explainer.index_name}", color="primary", id='random-index-clas-button-'+self.name, block=True),
dbc.Tooltip(f"Select a random {self.explainer.index_name} according to the constraints",
target='random-index-clas-button-'+self.name),
], md=4), hide=self.hide_button),
], form=True, style=dict(marginBottom=10)),
dbc.Row([
make_hideable(
dbc.Col([
dbc.Label(f"Observed {self.explainer.target}:", id='random-index-clas-labels-label-'+self.name),
dbc.Tooltip(f"Only select a random {self.explainer.index_name} where the observed "
f"{self.explainer.target} is one of the selected labels:",
target='random-index-clas-labels-label-'+self.name),
dcc.Dropdown(
id='random-index-clas-labels-'+self.name,
options=[{'label': lab, 'value': lab} for lab in self.explainer.labels],
multi=True,
value=self.labels),
], md=8), hide=self.hide_labels),
make_hideable(
dbc.Col([
html.Div([
dbc.Label("Range:", html_for='random-index-clas-pred-or-perc-'+self.name),
dbc.Select(
id='random-index-clas-pred-or-perc-'+self.name,
options=[
{'label': 'probability', 'value': 'predictions'},
{'label': 'percentile', 'value': 'percentiles'},
],
value=self.pred_or_perc)
], id='random-index-clas-pred-or-perc-div-'+self.name),
dbc.Tooltip("Instead of selecting from a range of predicted probabilities "
"you can also select from a range of predicted percentiles. "
"For example if you set the slider to percentile (0.9-1.0) you would"
f" only sample random {self.explainer.index_name} from the top "
"10% highest predicted probabilities.",
target='random-index-clas-pred-or-perc-div-'+self.name),
], md=4), hide=self.hide_pred_or_perc),
], style=dict(marginBottom=10)),
dbc.Row([
make_hideable(
dbc.Col([
html.Div([
dbc.Label(id='random-index-clas-slider-label-'+self.name,
children="Predicted probability range:",
html_for='prediction-range-slider-'+self.name),
dbc.Tooltip(f"Only select a random {self.explainer.index_name} where the "
"predicted probability of positive label is in the following range:",
id='random-index-clas-slider-label-tooltip-'+self.name,
target='random-index-clas-slider-label-'+self.name),
dcc.RangeSlider(
id='random-index-clas-slider-'+self.name,
min=0.0, max=1.0, step=0.01,
value=self.slider, allowCross=False,
marks={0.0:'0.0', 0.2:'0.2', 0.4:'0.4', 0.6:'0.6',
0.8:'0.8', 1.0:'1.0'},
tooltip = {'always_visible' : False})
])
]), hide=self.hide_slider),
], justify="start"),
]),
])
def component_callbacks(self, app):
@app.callback(
Output('random-index-clas-index-'+self.name, 'value'),
[Input('random-index-clas-button-'+self.name, 'n_clicks')],
[State('random-index-clas-slider-'+self.name, 'value'),
State('random-index-clas-labels-'+self.name, 'value'),
State('random-index-clas-pred-or-perc-'+self.name, 'value'),
State('pos-label-'+self.name, 'value')])
def update_index(n_clicks, slider_range, labels, pred_or_perc, pos_label):
if n_clicks is None and self.index is not None:
raise PreventUpdate
if pred_or_perc == 'predictions':
return self.explainer.random_index(y_values=labels,
pred_proba_min=slider_range[0], pred_proba_max=slider_range[1],
return_str=True, pos_label=pos_label)
elif pred_or_perc == 'percentiles':
return self.explainer.random_index(y_values=labels,
pred_percentile_min=slider_range[0], pred_percentile_max=slider_range[1],
return_str=True, pos_label=pos_label)
@app.callback(
[Output('random-index-clas-slider-label-'+self.name, 'children'),
Output('random-index-clas-slider-label-tooltip-'+self.name, 'children')],
[Input('random-index-clas-pred-or-perc-'+self.name, 'value'),
Input('pos-label-'+self.name, 'value')]
)
def update_slider_label(pred_or_perc, pos_label):
if pred_or_perc == 'predictions':
return (
"Predicted probability range:",
f"Only select a random {self.explainer.index_name} where the "
f"predicted probability of {self.explainer.labels[pos_label]}"
" is in the following range:"
)
elif pred_or_perc == 'percentiles':
return (
"Predicted percentile range:",
f"Only select a random {self.explainer.index_name} where the "
f"predicted probability of {self.explainer.labels[pos_label]}"
" is in the following percentile range. For example you can "
"only sample from the top 10% highest predicted probabilities."
)
raise PreventUpdate
class ClassifierPredictionSummaryComponent(ExplainerComponent):
def __init__(self, explainer, title="Prediction", name=None,
hide_index=False, hide_title=False, hide_subtitle=False,
hide_table=False, hide_piechart=False,
hide_star_explanation=False, hide_selector=False,
feature_input_component=None,
pos_label=None, index=None, round=3, description=None,
**kwargs):
"""Shows a summary for a particular prediction
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"Prediction Summary".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
hide_index (bool, optional): hide index selector. Defaults to False.
hide_title (bool, optional): hide title. Defaults to False.
hide_subtitle (bool, optional): Hide subtitle. Defaults to False.
hide_table (bool, optional): hide the results table
hide_piechart (bool, optional): hide the results piechart
hide_star_explanation (bool, optional): hide the `* indicates..`
Defaults to False.
hide_selector (bool, optional): hide pos label selectors.
Defaults to False.
feature_input_component (FeatureInputComponent): A FeatureInputComponent
that will give the input to the graph instead of the index selector.
If not None, hide_index=True. Defaults to None.
pos_label ({int, str}, optional): initial pos label.
Defaults to explainer.pos_label
index ({int, str}, optional): Index to display prediction summary
for. Defaults to None.
round (int, optional): rounding to apply to pred_proba float.
Defaults to 3.
description (str, optional): Tooltip to display when hover over
component title. When None default text is shown.
"""
super().__init__(explainer, title, name)
self.index_name = 'clas-prediction-index-'+self.name
self.selector = PosLabelSelector(explainer, name=self.name, pos_label=pos_label)
if self.feature_input_component is not None:
self.exclude_callbacks(self.feature_input_component)
self.hide_index = True
if self.description is None: self.description = f"""
Shows the predicted probability for each {self.explainer.target} label.
"""
def layout(self):
return dbc.Card([
make_hideable(
dbc.CardHeader([
html.Div([
html.H3(self.title, id='clas-prediction-index-title-'+self.name, className='card-title'),
dbc.Tooltip(self.description, target='clas-prediction-index-title-'+self.name),
]),
]), hide=self.hide_title),
dbc.CardBody([
dbc.Row([
make_hideable(
dbc.Col([
dbc.Label(f"{self.explainer.index_name}:"),
dcc.Dropdown(id='clas-prediction-index-'+self.name,
options = [{'label': str(idx), 'value':idx}
for idx in self.explainer.idxs],
value=self.index)
], md=6), hide=self.hide_index),
make_hideable(
dbc.Col([self.selector.layout()
], width=3), hide=self.hide_selector),
]),
dbc.Row([
make_hideable(
dbc.Col([
html.Div(id='clas-prediction-div-'+self.name),
make_hideable(
html.Div("* indicates observed label") if not self.explainer.y_missing else None,
hide=self.hide_star_explanation),
]), hide=self.hide_table),
make_hideable(
dbc.Col([
dcc.Graph(id='clas-prediction-graph-'+self.name,
config=dict(modeBarButtons=[['toImage']], displaylogo=False))
] ), hide=self.hide_piechart),
]),
])
])
def component_callbacks(self, app):
if self.feature_input_component is None:
@app.callback(
[Output('clas-prediction-div-'+self.name, 'children'),
Output('clas-prediction-graph-'+self.name, 'figure')],
[Input('clas-prediction-index-'+self.name, 'value'),
Input('pos-label-'+self.name, 'value')])
def update_output_div(index, pos_label):
if index is not None:
fig = self.explainer.plot_prediction_result(index, showlegend=False)
preds_df = self.explainer.prediction_result_df(index, round=self.round, logodds=True)
preds_df.probability = np.round(100*preds_df.probability.values, self.round).astype(str)
preds_df.probability = preds_df.probability + ' %'
preds_df.logodds = np.round(preds_df.logodds.values, self.round).astype(str)
if self.explainer.model_output!='logodds':
preds_df = preds_df[['label', 'probability']]
preds_table = dbc.Table.from_dataframe(preds_df,
striped=False, bordered=False, hover=False)
return preds_table, fig
raise PreventUpdate
else:
@app.callback(
[Output('clas-prediction-div-'+self.name, 'children'),
Output('clas-prediction-graph-'+self.name, 'figure')],
[Input('pos-label-'+self.name, 'value'),
*self.feature_input_component._feature_callback_inputs])
def update_output_div(pos_label, *inputs):
X_row = self.explainer.get_row_from_input(inputs, ranked_by_shap=True)
fig = self.explainer.plot_prediction_result(X_row=X_row, showlegend=False)
preds_df = self.explainer.prediction_result_df(X_row=X_row, round=self.round, logodds=True)
preds_df.probability = np.round(100*preds_df.probability.values, self.round).astype(str)
preds_df.probability = preds_df.probability + ' %'
preds_df.logodds = np.round(preds_df.logodds.values, self.round).astype(str)
if self.explainer.model_output!='logodds':
preds_df = preds_df[['label', 'probability']]
preds_table = dbc.Table.from_dataframe(preds_df,
striped=False, bordered=False, hover=False)
return preds_table, fig
class PrecisionComponent(ExplainerComponent):
def __init__(self, explainer, title="Precision Plot", name=None,
subtitle="Does fraction positive increase with predicted probability?",
hide_title=False, hide_subtitle=False, hide_footer=False,
hide_cutoff=False, hide_binsize=False, hide_binmethod=False,
hide_multiclass=False, hide_selector=False, pos_label=None,
bin_size=0.1, quantiles=10, cutoff=0.5,
quantiles_or_binsize='bin_size', multiclass=False, description=None,
**kwargs):
"""Shows a precision graph with toggles.
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"Precision Plot".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
subtitle (str): subtitle
hide_title (bool, optional): hide title
hide_subtitle (bool, optional): Hide subtitle. Defaults to False.
hide_footer (bool, optional): hide the footer at the bottom of the component
hide_cutoff (bool, optional): Hide cutoff slider. Defaults to False.
hide_binsize (bool, optional): hide binsize/quantiles slider. Defaults to False.
hide_selector(bool, optional): hide pos label selector. Defaults to False.
hide_binmethod (bool, optional): Hide binsize/quantiles toggle. Defaults to False.
hide_multiclass (bool, optional): Hide multiclass toggle. Defaults to False.
hide_selector (bool, optional): Hide pos label selector. Default to True.
pos_label ({int, str}, optional): initial pos label. Defaults to explainer.pos_label
bin_size (float, optional): Size of bins in probability space. Defaults to 0.1.
quantiles (int, optional): Number of quantiles to divide plot. Defaults to 10.
cutoff (float, optional): Cutoff to display in graph. Defaults to 0.5.
quantiles_or_binsize (str, {'quantiles', 'bin_size'}, optional): Default bin method. Defaults to 'bin_size'.
multiclass (bool, optional): Display all classes. Defaults to False.
description (str, optional): Tooltip to display when hover over
component title. When None default text is shown.
"""
super().__init__(explainer, title, name)
self.cutoff_name = 'precision-cutoff-' + self.name
self.selector = PosLabelSelector(explainer, name=self.name, pos_label=pos_label)
if self.description is None: self.description = f"""
On this plot you can see the relation between the predicted probability
that a {self.explainer.index_name} belongs to the positive class, and
the percentage of observed {self.explainer.index_name} in the positive class.
The observations get binned together in groups of roughly
equal predicted probabilities, and the percentage of positives is calculated
for each bin. A perfectly calibrated model would show a straight line
from the bottom left corner to the top right corner. A strong model would
classify most observations correctly and close to 0% or 100% probability.
"""
self.register_dependencies("preds", "pred_probas", "pred_percentiles")
def layout(self):
return dbc.Card([
make_hideable(
dbc.CardHeader([
html.Div([
html.H3(self.title, id='precision-title-'+self.name, className="card-title"),
make_hideable(html.H6(self.subtitle, className='card-subtitle'), hide=self.hide_subtitle),
dbc.Tooltip(self.description, target='precision-title-'+self.name),
]),
]), hide=self.hide_title),
dbc.CardBody([
dbc.Row([
make_hideable(
dbc.Col([self.selector.layout()], width=3), hide=self.hide_selector)
], justify="end"),
dbc.Row([
dbc.Col([
html.Div([
dcc.Graph(id='precision-graph-'+self.name,
config=dict(modeBarButtons=[['toImage']], displaylogo=False)),
], style={'margin': 0}),
])
]),
]),
make_hideable(
dbc.CardFooter([
dbc.Row([
dbc.Col([
make_hideable(
html.Div([
html.Div([
dbc.Label('Bin size:', html_for='precision-binsize-'+self.name),
html.Div([
dcc.Slider(id='precision-binsize-'+self.name,
min = 0.01, max = 0.5, step=0.01, value=self.bin_size,
marks={0.01: '0.01', 0.05: '0.05', 0.10: '0.10',
0.20: '0.20', 0.25: '0.25' , 0.33: '0.33',
0.5: '0.5'},
included=False,
tooltip = {'always_visible' : False})
], style={'margin-bottom': 5}),
], id='precision-bin-size-div-'+self.name, style=dict(margin=5)),
dbc.Tooltip("Size of the bins to divide prediction score by",
target='precision-bin-size-div-'+self.name,
placement='bottom'),
]), hide=self.hide_binsize),
make_hideable(
html.Div([
html.Div([
dbc.Label('Quantiles:', html_for='precision-quantiles-'+self.name),
html.Div([
dcc.Slider(id='precision-quantiles-'+self.name,
min = 1, max = 20, step=1, value=self.quantiles,
marks={1: '1', 5: '5', 10: '10', 15: '15', 20:'20'},
included=False,
tooltip = {'always_visible' : False}),
], style={'margin-bottom':5}),
], id='precision-quantiles-div-'+self.name),
dbc.Tooltip("Number of equally populated bins to divide prediction score by",
target='precision-quantiles-div-'+self.name,
placement='bottom'),
]), hide=self.hide_binsize),
make_hideable(
html.Div([
html.Div([
html.Label('Cutoff prediction probability:'),
dcc.Slider(id='precision-cutoff-'+self.name,
min = 0.01, max = 0.99, step=0.01, value=self.cutoff,
marks={0.01: '0.01', 0.25: '0.25', 0.50: '0.50',
0.75: '0.75', 0.99: '0.99'},
included=False,
tooltip = {'always_visible' : False}),
], id='precision-cutoff-div-'+self.name),
dbc.Tooltip(f"Scores above this cutoff will be labeled positive",
target='precision-cutoff-div-'+self.name,
placement='bottom'),
], style={'margin-bottom': 5}), hide=self.hide_cutoff),
]),
]),
dbc.Row([
make_hideable(
dbc.Col([
dbc.Label('Binning Method:', html_for='precision-binsize-or-quantiles-'+self.name),
dbc.Select(
id='precision-binsize-or-quantiles-'+self.name,
options=[
{'label': 'Bins',
'value': 'bin_size'},
{'label': 'Quantiles',
'value': 'quantiles'}
],
value=self.quantiles_or_binsize,
),
dbc.Tooltip("Divide the x-axis by equally sized ranges of prediction scores (bins),"
" or bins with the same number of observations (counts) in each bin: quantiles",
target='precision-binsize-or-quantiles-'+self.name),
], width=4), hide=self.hide_binmethod),
make_hideable(
dbc.Col([
dbc.FormGroup([
dbc.Label("Multi class:", id="precision-multiclass-label-"+self.name),
dbc.Tooltip("Display the observed proportion for all class"
" labels, not just positive label.",
target="precision-multiclass-"+self.name),
dbc.Checklist(
options=[{"label": "Display all classes", "value": True}],
value=[True] if self.multiclass else [],
id='precision-multiclass-'+self.name,
inline=True,
switch=True,
),
]),
], width=4), hide=self.hide_multiclass),
]),
]), hide=self.hide_footer)
])
def component_callbacks(self, app):
@app.callback(
[Output('precision-bin-size-div-'+self.name, 'style'),
Output('precision-quantiles-div-'+self.name, 'style')],
[Input('precision-binsize-or-quantiles-'+self.name, 'value')],
)
def update_div_visibility(bins_or_quantiles):
if self.hide_binsize:
return dict(display='none'), dict(display='none')
if bins_or_quantiles=='bin_size':
return {}, dict(display='none')
elif bins_or_quantiles=='quantiles':
return dict(display='none'), {}
raise PreventUpdate
@app.callback(
Output('precision-graph-'+self.name, 'figure'),
[Input('precision-binsize-'+self.name, 'value'),
Input('precision-quantiles-'+self.name, 'value'),
Input('precision-binsize-or-quantiles-'+self.name, 'value'),
Input('precision-cutoff-'+self.name, 'value'),
Input('precision-multiclass-'+self.name, 'value'),
Input('pos-label-'+self.name, 'value')],
#[State('tabs', 'value')],
)
def update_precision_graph(bin_size, quantiles, bins, cutoff, multiclass, pos_label):
if bins == 'bin_size':
return self.explainer.plot_precision(
bin_size=bin_size, cutoff=cutoff,
multiclass=bool(multiclass), pos_label=pos_label)
elif bins == 'quantiles':
return self.explainer.plot_precision(
quantiles=quantiles, cutoff=cutoff,
multiclass=bool(multiclass), pos_label=pos_label)
raise PreventUpdate
class ConfusionMatrixComponent(ExplainerComponent):
def __init__(self, explainer, title="Confusion Matrix", name=None,
subtitle="How many false positives and false negatives?",
hide_title=False, hide_subtitle=False, hide_footer=False,
hide_cutoff=False, hide_percentage=False, hide_binary=False,
hide_selector=False, pos_label=None,
cutoff=0.5, percentage=True, binary=True, description=None,
**kwargs):
"""Display confusion matrix component
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"Confusion Matrix".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
subtitle (str): subtitle
hide_title (bool, optional): hide title.
hide_subtitle (bool, optional): Hide subtitle. Defaults to False.
hide_footer (bool, optional): hide the footer at the bottom of the component
hide_cutoff (bool, optional): Hide cutoff slider. Defaults to False.
hide_percentage (bool, optional): Hide percentage toggle. Defaults to False.
hide_binary (bool, optional): Hide binary toggle. Defaults to False.
hide_selector(bool, optional): hide pos label selector. Defaults to False.
pos_label ({int, str}, optional): initial pos label. Defaults to explainer.pos_label
cutoff (float, optional): Default cutoff. Defaults to 0.5.
percentage (bool, optional): Display percentages instead of counts. Defaults to True.
binary (bool, optional): Show binary instead of multiclass confusion matrix. Defaults to True.
description (str, optional): Tooltip to display when hover over
component title. When None default text is shown.
"""
super().__init__(explainer, title, name)
self.cutoff_name = 'confusionmatrix-cutoff-' + self.name
if len(self.explainer.labels) <= 2:
self.hide_binary = True
if self.description is None: self.description = """
The confusion matrix shows the number of true negatives (predicted negative, observed negative),
true positives (predicted positive, observed positive),
false negatives (predicted negative, but observed positive) and
false positives (predicted positive, but observed negative). The amount
of false negatives and false positives determine the costs of deploying
and imperfect model. For different cutoffs you will get a different number
of false positives and false negatives. This plot can help you select
the optimal cutoff.
"""
self.selector = PosLabelSelector(explainer, name=self.name, pos_label=pos_label)
self.register_dependencies("preds", "pred_probas", "pred_percentiles")
def layout(self):
return dbc.Card([
make_hideable(
dbc.CardHeader([
html.Div([
html.H3(self.title, id='confusionmatrix-title-'+self.name),
make_hideable(html.H6(self.subtitle, className='card-subtitle'), hide=self.hide_subtitle),
dbc.Tooltip(self.description, target='confusionmatrix-title-'+self.name),
]),
]), hide=self.hide_title),
dbc.CardBody([
dbc.Row([
make_hideable(
dbc.Col([self.selector.layout()], width=3), hide=self.hide_selector)
], justify="end"),
dcc.Graph(id='confusionmatrix-graph-'+self.name,
config=dict(modeBarButtons=[['toImage']], displaylogo=False)),
]),
make_hideable(
dbc.CardFooter([
make_hideable(
html.Div([
html.Div([
html.Label('Cutoff prediction probability:'),
dcc.Slider(id='confusionmatrix-cutoff-'+self.name,
min = 0.01, max = 0.99, step=0.01, value=self.cutoff,
marks={0.01: '0.01', 0.25: '0.25', 0.50: '0.50',
0.75: '0.75', 0.99: '0.99'},
included=False,
tooltip = {'always_visible' : False},
updatemode='drag'),
], id='confusionmatrix-cutoff-div-'+self.name),
dbc.Tooltip(f"Scores above this cutoff will be labeled positive",
target='confusionmatrix-cutoff-div-'+self.name,
placement='bottom'),
], style={'margin-bottom': 25}), hide=True),
make_hideable(
html.Div([
dbc.FormGroup([
#dbc.Label("Percentage:", id='confusionmatrix-percentage-label-'+self.name),
dbc.Tooltip("Highlight the percentage in each cell instead of the absolute numbers",
target='confusionmatrix-percentage-'+self.name),
dbc.Checklist(
options=[{"label": "Highlight percentage", "value": True}],
value=[True] if self.percentage else [],
id='confusionmatrix-percentage-'+self.name,
inline=True,
switch=True,
),
]),
]), hide=self.hide_percentage),
make_hideable(
html.Div([
dbc.FormGroup([
dbc.Label("Binary:", id='confusionmatrix-binary-label-'+self.name),
dbc.Tooltip("display a binary confusion matrix of positive "
"class vs all other classes instead of a multi"
" class confusion matrix.",
target="confusionmatrix-binary-label-"+self.name),
dbc.Checklist(
options=[{"label": "Display one-vs-rest matrix", "value": True}],
value=[True] if self.binary else [],
id='confusionmatrix-binary-'+self.name,
inline=True,
switch=True,
),
]),
]), hide=self.hide_binary),
]), hide=self.hide_footer)
])
def component_callbacks(self, app):
@app.callback(
Output('confusionmatrix-graph-'+self.name, 'figure'),
[Input('confusionmatrix-cutoff-'+self.name, 'value'),
Input('confusionmatrix-percentage-'+self.name, 'value'),
Input('confusionmatrix-binary-'+self.name, 'value'),
Input('pos-label-'+self.name, 'value')],
)
def update_confusionmatrix_graph(cutoff, normalized, binary, pos_label):
return self.explainer.plot_confusion_matrix(
cutoff=cutoff, normalized=bool(normalized),
binary=bool(binary), pos_label=pos_label)
class LiftCurveComponent(ExplainerComponent):
def __init__(self, explainer, title="Lift Curve", name=None,
subtitle="Performance how much better than random?",
hide_title=False, hide_subtitle=False, hide_footer=False,
hide_cutoff=False, hide_percentage=False, hide_selector=False,
pos_label=None, cutoff=0.5, percentage=True, description=None,
**kwargs):
"""Show liftcurve component
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"Lift Curve".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
subtitle (str): subtitle
hide_title (bool, optional): hide title.
hide_subtitle (bool, optional): Hide subtitle. Defaults to False.
hide_footer (bool, optional): hide the footer at the bottom of the component
hide_cutoff (bool, optional): Hide cutoff slider. Defaults to False.
hide_percentage (bool, optional): Hide percentage toggle. Defaults to False.
hide_selector(bool, optional): hide pos label selector. Defaults to False.
pos_label ({int, str}, optional): initial pos label. Defaults to explainer.pos_label
cutoff (float, optional): Cutoff for lift curve. Defaults to 0.5.
percentage (bool, optional): Display percentages instead of counts. Defaults to True.
description (str, optional): Tooltip to display when hover over
component title. When None default text is shown.
"""
super().__init__(explainer, title, name)
self.cutoff_name = 'liftcurve-cutoff-' + self.name
self.selector = PosLabelSelector(explainer, name=self.name, pos_label=pos_label)
if self.description is None: self.description = """
The lift curve shows you the percentage of positive classes when you only
select observations with a score above cutoff vs selecting observations
randomly. This shows you how much better the model is than random (the lift).
"""
self.register_dependencies("preds", "pred_probas", "pred_percentiles")
def layout(self):
return dbc.Card([
make_hideable(
dbc.CardHeader([
html.Div([
html.H3(self.title, id='liftcurve-title-'+self.name),
make_hideable(html.H6(self.subtitle, className='card-subtitle'), hide=self.hide_subtitle),
dbc.Tooltip(self.description, target='liftcurve-title-'+self.name),
]),
]), hide=self.hide_title),
dbc.CardBody([
dbc.Row([
make_hideable(
dbc.Col([self.selector.layout()], width=3), hide=self.hide_selector)
], justify="end"),
html.Div([
dcc.Graph(id='liftcurve-graph-'+self.name,
config=dict(modeBarButtons=[['toImage']], displaylogo=False)),
], style={'margin': 0}),
]),
make_hideable(
dbc.CardFooter([
make_hideable(
html.Div([
html.Div([
html.Label('Cutoff prediction probability:'),
dcc.Slider(id='liftcurve-cutoff-'+self.name,
min = 0.01, max = 0.99, step=0.01, value=self.cutoff,
marks={0.01: '0.01', 0.25: '0.25', 0.50: '0.50',
0.75: '0.75', 0.99: '0.99'},
included=False,
tooltip = {'always_visible' : False},
updatemode='drag'),
], id='liftcurve-cutoff-div-'+self.name),
dbc.Tooltip(f"Scores above this cutoff will be labeled positive",
target='liftcurve-cutoff-div-'+self.name,
placement='bottom'),
], style={'margin-bottom': 25}), hide=self.hide_cutoff),
make_hideable(
html.Div([
dbc.FormGroup([
dbc.Tooltip("Display percentages positive and sampled"
" instead of absolute numbers",
target='liftcurve-percentage-'+self.name),
dbc.Checklist(
options=[{"label": "Display percentage", "value": True}],
value=[True] if self.percentage else [],
id='liftcurve-percentage-'+self.name,
inline=True,
switch=True,
),
]),
]), hide=self.hide_percentage),
]), hide=self.hide_footer)
])
def component_callbacks(self, app):
@app.callback(
Output('liftcurve-graph-'+self.name, 'figure'),
[Input('liftcurve-cutoff-'+self.name, 'value'),
Input('liftcurve-percentage-'+self.name, 'value'),
Input('pos-label-'+self.name, 'value')],
)
def update_precision_graph(cutoff, percentage, pos_label):
return self.explainer.plot_lift_curve(cutoff=cutoff,
percentage=bool(percentage), pos_label=pos_label)
class CumulativePrecisionComponent(ExplainerComponent):
def __init__(self, explainer, title="Cumulative Precision", name=None,
subtitle="Expected distribution for highest scores",
hide_title=False, hide_subtitle=False, hide_footer=False,
hide_selector=False, pos_label=None,
hide_cutoff=False, cutoff=None,
hide_percentile=False, percentile=None, description=None,
**kwargs):
"""Show cumulative precision component
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"Cumulative Precision".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
subtitle (str): subtitle
hide_title (bool, optional): hide the title.
hide_subtitle (bool, optional): Hide subtitle. Defaults to False.
hide_footer (bool, optional): hide the footer at the bottom of the component
hide_selector(bool, optional): hide pos label selector. Defaults to False.
pos_label ({int, str}, optional): initial pos label. Defaults to explainer.pos_label
"""
super().__init__(explainer, title, name)
self.selector = PosLabelSelector(explainer, name=self.name, pos_label=pos_label)
self.cutoff_name = 'cumulative-precision-cutoff-'+self.name
if self.description is None: self.description = """
This plot shows the percentage of each label that you can expect when you
only sample the top x% highest scores.
"""
self.register_dependencies("preds", "pred_probas", "pred_percentiles")
def layout(self):
return dbc.Card([
make_hideable(
dbc.CardHeader([
html.Div([
html.H3(self.title, id='cumulative-precision-title-'+self.name),
make_hideable(html.H6(self.subtitle, className='card-subtitle'), hide=self.hide_subtitle),
dbc.Tooltip(self.description, target='cumulative-precision-title-'+self.name),
]),
]), hide=self.hide_title),
dbc.CardBody([
html.Div([
dcc.Graph(id='cumulative-precision-graph-'+self.name,
config=dict(modeBarButtons=[['toImage']], displaylogo=False)),
], style={'margin': 0}),
]),
make_hideable(
dbc.CardFooter([
dbc.Row([
dbc.Col([
dbc.Row([
make_hideable(
dbc.Col([
html.Div([
html.Label('Sample top fraction:'),
dcc.Slider(id='cumulative-precision-percentile-'+self.name,
min = 0.01, max = 0.99, step=0.01, value=self.percentile,
marks={0.01: '0.01', 0.25: '0.25', 0.50: '0.50',
0.75: '0.75', 0.99: '0.99'},
included=False,
tooltip = {'always_visible' : False},
updatemode='drag')
], style={'margin-bottom': 15}, id='cumulative-precision-percentile-div-'+self.name),
dbc.Tooltip("Draw the line where you only sample the top x% fraction of all samples",
target='cumulative-precision-percentile-div-'+self.name)
]), hide=self.hide_percentile),
]),
dbc.Row([
make_hideable(
dbc.Col([
html.Div([
html.Label('Cutoff prediction probability:'),
dcc.Slider(id='cumulative-precision-cutoff-'+self.name,
min = 0.01, max = 0.99, step=0.01, value=self.cutoff,
marks={0.01: '0.01', 0.25: '0.25', 0.50: '0.50',
0.75: '0.75', 0.99: '0.99'},
included=False,
tooltip = {'always_visible' : False}),
], style={'margin-bottom': 15}, id='cumulative-precision-cutoff-div-'+self.name),
dbc.Tooltip(f"Scores above this cutoff will be labeled positive",
target='cumulative-precision-cutoff-div-'+self.name,
placement='bottom'),
]), hide=self.hide_cutoff),
]),
]),
make_hideable(
dbc.Col([
self.selector.layout()
], width=2), hide=self.hide_selector),
])
]), hide=self.hide_footer)
])
def component_callbacks(self, app):
@app.callback(
Output('cumulative-precision-graph-'+self.name, 'figure'),
[Input('cumulative-precision-percentile-'+self.name, 'value'),
Input('pos-label-'+self.name, 'value')],
)
def update_cumulative_precision_graph(percentile, pos_label):
return self.explainer.plot_cumulative_precision(percentile=percentile, pos_label=pos_label)
@app.callback(
Output('cumulative-precision-percentile-'+self.name, 'value'),
[Input('cumulative-precision-cutoff-'+self.name, 'value'),
Input('pos-label-'+self.name, 'value')],
)
def update_cumulative_precision_percentile(cutoff, pos_label):
return self.explainer.percentile_from_cutoff(cutoff, pos_label)
class ClassificationComponent(ExplainerComponent):
def __init__(self, explainer, title="Classification Plot", name=None,
subtitle="Distribution of labels above and below cutoff",
hide_title=False, hide_subtitle=False, hide_footer=False,
hide_cutoff=False, hide_percentage=False, hide_selector=False,
pos_label=None, cutoff=0.5, percentage=True, description=None,
**kwargs):
"""Shows a barchart of the number of classes above the cutoff and below
the cutoff.
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"Classification Plot".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
subtitle (str): subtitle
hide_title (bool, optional): hide the title.
hide_subtitle (bool, optional): Hide subtitle. Defaults to False.
hide_footer (bool, optional): hide the footer at the bottom of the component
hide_cutoff (bool, optional): Hide cutoff slider. Defaults to False.
hide_percentage (bool, optional): Hide percentage toggle. Defaults to False.
hide_selector(bool, optional): hide pos label selector. Defaults to False.
pos_label ({int, str}, optional): initial pos label. Defaults to explainer.pos_label
cutoff (float, optional): Cutoff for prediction. Defaults to 0.5.
percentage (bool, optional): Show percentage instead of counts. Defaults to True.
description (str, optional): Tooltip to display when hover over
component title. When None default text is shown.
"""
super().__init__(explainer, title, name)
self.cutoff_name = 'classification-cutoff-' + self.name
self.selector = PosLabelSelector(explainer, name=self.name, pos_label=pos_label)
if self.description is None: self.description = """
Plot showing the fraction of each class above and below the cutoff.
"""
self.register_dependencies("preds", "pred_probas", "pred_percentiles")
def layout(self):
return dbc.Card([
make_hideable(
dbc.CardHeader([
html.Div([
html.H3(self.title, id='classification-title-'+self.name),
make_hideable(html.H6(self.subtitle, className='card-subtitle'), hide=self.hide_subtitle),
dbc.Tooltip(self.description, target='classification-title-'+self.name),
]),
]), hide=self.hide_title),
dbc.CardBody([
dbc.Row([
make_hideable(
dbc.Col([self.selector.layout()], width=3), hide=self.hide_selector)
], justify="end"),
html.Div([
dcc.Graph(id='classification-graph-'+self.name,
config=dict(modeBarButtons=[['toImage']], displaylogo=False)),
], style={'margin': 0}),
]),
make_hideable(
dbc.CardFooter([
make_hideable(
html.Div([
html.Div([
html.Label('Cutoff prediction probability:'),
dcc.Slider(id='classification-cutoff-'+self.name,
min = 0.01, max = 0.99, step=0.01, value=self.cutoff,
marks={0.01: '0.01', 0.25: '0.25', 0.50: '0.50',
0.75: '0.75', 0.99: '0.99'},
included=False,
tooltip = {'always_visible' : False},
updatemode='drag'),
], id='classification-cutoff-div-'+self.name),
dbc.Tooltip(f"Scores above this cutoff will be labeled positive",
target='classification-cutoff-div-'+self.name,
placement='bottom'),
], style={'margin-bottom': 25}), hide=self.hide_cutoff),
make_hideable(
html.Div([
dbc.FormGroup([
dbc.Tooltip("Do not resize the bar chart by absolute number of observations",
target='classification-percentage-'+self.name),
dbc.Checklist(
options=[{"label": "Display percentage", "value": True}],
value=[True] if self.percentage else [],
id='classification-percentage-'+self.name,
inline=True,
switch=True,
),
]),
]), hide=self.hide_percentage),
]), hide=self.hide_footer)
])
def component_callbacks(self, app):
@app.callback(
Output('classification-graph-'+self.name, 'figure'),
[Input('classification-cutoff-'+self.name, 'value'),
Input('classification-percentage-'+self.name, 'value'),
Input('pos-label-'+self.name, 'value')],
)
def update_precision_graph(cutoff, percentage, pos_label):
return self.explainer.plot_classification(
cutoff=cutoff, percentage=bool(percentage), pos_label=pos_label)
class RocAucComponent(ExplainerComponent):
def __init__(self, explainer, title="ROC AUC Plot", name=None,
subtitle="Trade-off between False positives and false negatives",
hide_title=False, hide_subtitle=False, hide_footer=False,
hide_cutoff=False, hide_selector=False,
pos_label=None, cutoff=0.5, description=None,
**kwargs):
"""Show ROC AUC curve component
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"ROC AUC Plot".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
subtitle (str): subtitle
hide_title (bool, optional): hide title.
hide_subtitle (bool, optional): Hide subtitle. Defaults to False.
hide_footer (bool, optional): hide the footer at the bottom of the component
hide_cutoff (bool, optional): Hide cutoff slider. Defaults to False.
hide_selector(bool, optional): hide pos label selector. Defaults to False.
pos_label ({int, str}, optional): initial pos label. Defaults to explainer.pos_label
cutoff (float, optional): default cutoff. Defaults to 0.5.
"""
super().__init__(explainer, title, name)
self.cutoff_name = 'rocauc-cutoff-' + self.name
self.selector = PosLabelSelector(explainer, name=self.name, pos_label=pos_label)
if self.description is None: self.description = """
"""
self.register_dependencies("preds", "pred_probas", "pred_percentiles")
def layout(self):
return dbc.Card([
make_hideable(
dbc.CardHeader([
html.Div([
html.H3(self.title, id='rocauc-title-'+self.name),
make_hideable(html.H6(self.subtitle, className='card-subtitle'), hide=self.hide_subtitle),
dbc.Tooltip(self.description, target='rocauc-title-'+self.name),
]),
]), hide=self.hide_title),
dbc.CardBody([
dbc.Row([
make_hideable(
dbc.Col([self.selector.layout()], width=3), hide=self.hide_selector)
], justify="end"),
dcc.Graph(id='rocauc-graph-'+self.name,
config=dict(modeBarButtons=[['toImage']], displaylogo=False)),
]),
make_hideable(
dbc.CardFooter([
make_hideable(
html.Div([
html.Div([
html.Label('Cutoff prediction probability:'),
dcc.Slider(id='rocauc-cutoff-'+self.name,
min = 0.01, max = 0.99, step=0.01, value=self.cutoff,
marks={0.01: '0.01', 0.25: '0.25', 0.50: '0.50',
0.75: '0.75', 0.99: '0.99'},
included=False,
tooltip = {'always_visible' : False},
updatemode='drag' ),
] ,id='rocauc-cutoff-div-'+self.name),
dbc.Tooltip(f"Scores above this cutoff will be labeled positive",
target='rocauc-cutoff-div-'+self.name,
placement='bottom'),
], style={'margin-bottom': 25}), hide=self.hide_cutoff),
]), hide=self.hide_footer)
])
def component_callbacks(self, app):
@app.callback(
Output('rocauc-graph-'+self.name, 'figure'),
[Input('rocauc-cutoff-'+self.name, 'value'),
Input('pos-label-'+self.name, 'value')],
)
def update_precision_graph(cutoff, pos_label):
return self.explainer.plot_roc_auc(cutoff=cutoff, pos_label=pos_label)
class PrAucComponent(ExplainerComponent):
def __init__(self, explainer, title="PR AUC Plot", name=None,
subtitle="Trade-off between Precision and Recall",
hide_title=False, hide_subtitle=False, hide_footer=False,
hide_cutoff=False, hide_selector=False,
pos_label=None, cutoff=0.5, description=None,
**kwargs):
"""Display PR AUC plot component
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"PR AUC Plot".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
subtitle (str): subtitle
hide_title (bool, optional): hide title.
hide_subtitle (bool, optional): Hide subtitle. Defaults to False.
hide_footer (bool, optional): hide the footer at the bottom of the component
hide_cutoff (bool, optional): hide cutoff slider. Defaults to False.
hide_selector(bool, optional): hide pos label selector. Defaults to False.
pos_label ({int, str}, optional): initial pos label. Defaults to explainer.pos_label
cutoff (float, optional): default cutoff. Defaults to 0.5.
description (str, optional): Tooltip to display when hover over
component title. When None default text is shown.
"""
super().__init__(explainer, title, name)
self.cutoff_name = 'prauc-cutoff-' + self.name
self.selector = PosLabelSelector(explainer, name=self.name, pos_label=pos_label)
if self.description is None: self.description = """
Show the trade-off between Precision and Recall in one plot.
"""
self.register_dependencies("preds", "pred_probas", "pred_percentiles")
def layout(self):
return dbc.Card([
make_hideable(
dbc.CardHeader([
html.Div([
html.H3(self.title, id='prauc-title-'+self.name),
make_hideable(html.H6(self.subtitle, className='card-subtitle'), hide=self.hide_subtitle),
dbc.Tooltip(self.description, target='prauc-title-'+self.name),
]),
]), hide=self.hide_title),
dbc.CardBody([
dbc.Row([
make_hideable(
dbc.Col([self.selector.layout()], width=3), hide=self.hide_selector)
], justify="end"),
dcc.Graph(id='prauc-graph-'+self.name,
config=dict(modeBarButtons=[['toImage']], displaylogo=False)),
]),
make_hideable(
dbc.CardFooter([
make_hideable(
html.Div([
html.Div([
html.Label('Cutoff prediction probability:'),
dcc.Slider(id='prauc-cutoff-'+self.name,
min = 0.01, max = 0.99, step=0.01, value=self.cutoff,
marks={0.01: '0.01', 0.25: '0.25', 0.50: '0.50',
0.75: '0.75', 0.99: '0.99'},
included=False,
tooltip = {'always_visible' : False},
updatemode='drag'),
], id='prauc-cutoff-div-'+self.name),
dbc.Tooltip(f"Scores above this cutoff will be labeled positive",
target='prauc-cutoff-div-'+self.name,
placement='bottom'),
], style={'margin-bottom': 25}), hide=self.hide_cutoff),
]), hide=self.hide_footer)
])
def component_callbacks(self, app):
@app.callback(
Output('prauc-graph-'+self.name, 'figure'),
[Input('prauc-cutoff-'+self.name, 'value'),
Input('pos-label-'+self.name, 'value')],
)
def update_precision_graph(cutoff, pos_label):
return self.explainer.plot_pr_auc(cutoff=cutoff, pos_label=pos_label)
class ClassifierModelSummaryComponent(ExplainerComponent):
def __init__(self, explainer, title="Model performance metrics", name=None,
hide_title=False, hide_subtitle=False, hide_footer=True,
hide_cutoff=False, hide_selector=False,
pos_label=None, cutoff=0.5, round=3, description=None,
**kwargs):
"""Show model summary statistics (accuracy, precision, recall,
f1, roc_auc, pr_auc, log_loss) component.
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer()
title (str, optional): Title of tab or page. Defaults to
"Model performance metrics".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
hide_title (bool, optional): hide title.
hide_subtitle (bool, optional): Hide subtitle. Defaults to False.
hide_footer (bool, optional): hide the footer at the bottom of the component
hide_cutoff (bool, optional): hide cutoff slider. Defaults to False.
hide_selector(bool, optional): hide pos label selector. Defaults to False.
pos_label ({int, str}, optional): initial pos label. Defaults to explainer.pos_label
cutoff (float, optional): default cutoff. Defaults to 0.5.
round (int): round floats. Defaults to 3.
description (str, optional): Tooltip to display when hover over
component title. When None default text is shown.
"""
super().__init__(explainer, title, name)
self.cutoff_name = 'clas-model-summary-cutoff-' + self.name
self.selector = PosLabelSelector(explainer, name=self.name, pos_label=pos_label)
if self.description is None: self.description = """
Shows a list of various performance metrics.
"""
self.register_dependencies(['preds', 'pred_probas'])
def layout(self):
return dbc.Card([
make_hideable(
dbc.CardHeader([
html.Div([
html.H3(self.title, id='clas-model-summary-title-'+self.name),
dbc.Tooltip(self.description, target='clas-model-summary-title-'+self.name),
]),
]), hide=self.hide_title),
dbc.CardBody([
dbc.Row([
make_hideable(
dbc.Col([self.selector.layout()], width=3), hide=self.hide_selector)
], justify="end"),
html.Div(id='clas-model-summary-div-'+self.name),
]),
make_hideable(
dbc.CardFooter([
make_hideable(
html.Div([
html.Div([
html.Label('Cutoff prediction probability:'),
dcc.Slider(id='clas-model-summary-cutoff-'+self.name,
min = 0.01, max = 0.99, step=0.01, value=self.cutoff,
marks={0.01: '0.01', 0.25: '0.25', 0.50: '0.50',
0.75: '0.75', 0.99: '0.99'},
included=False,
tooltip = {'always_visible' : False},
updatemode='drag'),
], id='clas-model-summary-cutoff-div-'+self.name),
dbc.Tooltip(f"Scores above this cutoff will be labeled positive",
target='clas-model-summary-cutoff-div-'+self.name,
placement='bottom'),
], style={'margin-bottom': 25}), hide=self.hide_cutoff),
]), hide=self.hide_footer)
])
def component_callbacks(self, app):
@app.callback(
Output('clas-model-summary-div-'+self.name, 'children'),
[Input('clas-model-summary-cutoff-'+self.name, 'value'),
Input('pos-label-'+self.name, 'value')],
)
def update_classifier_summary(cutoff, pos_label):
metrics_dict = self.explainer.metrics_descriptions(cutoff, pos_label)
metrics_df = (pd.DataFrame(
self.explainer.metrics(cutoff=cutoff, pos_label=pos_label),
index=["Score"])
.T.rename_axis(index="metric").reset_index()
.round(self.round))
metrics_table = dbc.Table.from_dataframe(metrics_df, striped=False, bordered=False, hover=False)
metrics_table, tooltips = get_dbc_tooltips(metrics_table,
metrics_dict,
"clas-model-summary-div-hover",
self.name)
return html.Div([
metrics_table,
*tooltips
]) | AMLBID | /Explainer/dashboard_components/classifier_components.py | classifier_components.py |
__all__ = [
'CutoffPercentileComponent',
'PosLabelConnector',
'CutoffConnector',
'IndexConnector',
'HighlightConnector'
]
import numpy as np
import dash_core_components as dcc
import dash_bootstrap_components as dbc
import dash_html_components as html
from dash.dependencies import Input, Output, State
from dash.exceptions import PreventUpdate
from ..dashboard_methods import *
class CutoffPercentileComponent(ExplainerComponent):
def __init__(self, explainer, title="Global cutoff", name=None,
hide_title=False, hide_cutoff=False, hide_percentile=False,
hide_selector=False,
pos_label=None, cutoff=0.5, percentile=None,
description=None, **kwargs):
"""
Slider to set a cutoff for Classifier components, based on setting the
cutoff at a certain percentile of predictions, e.g.:
percentile=0.8 means "mark the 20% highest scores as positive".
This cutoff can then be conencted with other components like e.g.
RocAucComponent with a CutoffConnector.
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"Global Cutoff".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
hide_title (bool, optional): Hide title.
hide_cutoff (bool, optional): Hide the cutoff slider. Defaults to False.
hide_percentile (bool, optional): Hide percentile slider. Defaults to False.
hide_selector (bool, optional): hide pos label selectors. Defaults to False.
pos_label ({int, str}, optional): initial pos label.
Defaults to explainer.pos_label
cutoff (float, optional): Initial cutoff. Defaults to 0.5.
percentile ([type], optional): Initial percentile. Defaults to None.
description (str, optional): Tooltip to display when hover over
component title. When None default text is shown.
"""
super().__init__(explainer, title, name)
self.cutoff_name = 'cutoffconnector-cutoff-'+self.name
self.selector = PosLabelSelector(explainer, name=self.name, pos_label=pos_label)
if self.description is None: self.description = """
Select a model cutoff such that all predicted probabilities higher than
the cutoff will be labeled positive, and all predicted probabilities
lower than the cutoff will be labeled negative. You can also set
the cutoff as a percenntile of all observations. Setting the cutoff
here will automatically set the cutoff in multiple other connected
component.
"""
self.register_dependencies(['preds', 'pred_percentiles'])
def layout(self):
return dbc.Card([
make_hideable(
dbc.CardHeader([
html.H3(self.title, className="card-title", id='cutoffconnector-title-'+self.name),
dbc.Tooltip(self.description, target='cutoffconnector-title-'+self.name),
]), hide=self.hide_title),
dbc.CardBody([
dbc.Row([
dbc.Col([
dbc.Row([
make_hideable(
dbc.Col([
html.Div([
html.Label('Cutoff prediction probability:'),
dcc.Slider(id='cutoffconnector-cutoff-'+self.name,
min = 0.01, max = 0.99, step=0.01, value=self.cutoff,
marks={0.01: '0.01', 0.25: '0.25', 0.50: '0.50',
0.75: '0.75', 0.99: '0.99'},
included=False,
tooltip = {'always_visible' : False}),
], style={'margin-bottom': 15}, id='cutoffconnector-cutoff-div-'+self.name),
dbc.Tooltip(f"Scores above this cutoff will be labeled positive",
target='cutoffconnector-cutoff-div-'+self.name,
placement='bottom'),
]), hide=self.hide_cutoff),
]),
dbc.Row([
make_hideable(
dbc.Col([
html.Div([
html.Label('Cutoff percentile of samples:'),
dcc.Slider(id='cutoffconnector-percentile-'+self.name,
min = 0.01, max = 0.99, step=0.01, value=self.percentile,
marks={0.01: '0.01', 0.25: '0.25', 0.50: '0.50',
0.75: '0.75', 0.99: '0.99'},
included=False,
tooltip = {'always_visible' : False}),
], style={'margin-bottom': 15}, id='cutoffconnector-percentile-div-'+self.name),
dbc.Tooltip(f"example: if set to percentile=0.9: label the top 10% highest scores as positive, the rest negative.",
target='cutoffconnector-percentile-div-'+self.name,
placement='bottom'),
]), hide=self.hide_percentile),
])
]),
make_hideable(
dbc.Col([
self.selector.layout()
], width=2), hide=self.hide_selector),
])
])
])
def component_callbacks(self, app):
@app.callback(
Output('cutoffconnector-cutoff-'+self.name, 'value'),
[Input('cutoffconnector-percentile-'+self.name, 'value'),
Input('pos-label-'+self.name, 'value')]
)
def update_cutoff(percentile, pos_label):
if percentile is not None:
return np.round(self.explainer.cutoff_from_percentile(percentile, pos_label=pos_label), 2)
raise PreventUpdate
class PosLabelConnector(ExplainerComponent):
def __init__(self, input_pos_label, output_pos_labels):
self.input_pos_label_name = self._get_pos_label(input_pos_label)
self.output_pos_label_names = self._get_pos_labels(output_pos_labels)
# if self.input_pos_label_name in self.output_pos_label_names:
# # avoid circulat callbacks
# self.output_pos_label_names.remove(self.input_pos_label_name)
def _get_pos_label(self, input_pos_label):
if isinstance(input_pos_label, PosLabelSelector):
return 'pos-label-' + input_pos_label.name
elif hasattr(input_pos_label, 'selector') and isinstance(input_pos_label.selector, PosLabelSelector):
return 'pos-label-' + input_pos_label.selector.name
elif isinstance(input_pos_label, str):
return input_pos_label
else:
raise ValueError("input_pos_label should either be a str, "
"PosLabelSelector or an instance with a .selector property"
" that is a PosLabelSelector!")
def _get_pos_labels(self, output_pos_labels):
def get_pos_labels(o):
if isinstance(o, PosLabelSelector):
return ['pos-label-'+o.name]
elif isinstance(o, str):
return [str]
elif hasattr(o, 'pos_labels'):
return o.pos_labels
return []
if hasattr(output_pos_labels, '__iter__'):
pos_labels = []
for comp in output_pos_labels:
pos_labels.extend(get_pos_labels(comp))
return list(set(pos_labels))
else:
return get_pos_labels(output_pos_labels)
def component_callbacks(self, app):
if self.output_pos_label_names:
@app.callback(
[Output(pos_label_name, 'value') for pos_label_name in self.output_pos_label_names],
[Input(self.input_pos_label_name, 'value')]
)
def update_pos_labels(pos_label):
return tuple(pos_label for i in range(len(self.output_pos_label_names)))
class CutoffConnector(ExplainerComponent):
def __init__(self, input_cutoff, output_cutoffs):
"""Connect the cutoff selector of input_cutoff with those of output_cutoffs.
You can use this to connect a CutoffPercentileComponent with a
RocAucComponent for example,
When you change the cutoff in input_cutoff, all the cutoffs in output_cutoffs
will automatically be updated.
Args:
input_cutoff ([{str, ExplainerComponent}]): Either a str or an
ExplainerComponent. If str should be equal to the
name of the cutoff property. If ExplainerComponent then
should have a .cutoff_name property.
output_cutoffs (list(str, ExplainerComponent)): list of str of
ExplainerComponents.
"""
self.input_cutoff_name = self.cutoff_name(input_cutoff)
self.output_cutoff_names = self.cutoff_name(output_cutoffs)
if not isinstance(self.output_cutoff_names, list):
self.output_cutoff_names = [self.output_cutoff_names]
@staticmethod
def cutoff_name(cutoffs):
def get_cutoff_name(o):
if isinstance(o, str): return o
elif isinstance(o, ExplainerComponent):
if not hasattr(o, "cutoff_name"):
raise ValueError(f"{o} does not have an .cutoff_name property!")
return o.cutoff_name
raise ValueError(f"{o} is neither str nor an ExplainerComponent with an .cutoff_name property")
if hasattr(cutoffs, '__iter__'):
cutoff_name_list = []
for cutoff in cutoffs:
cutoff_name_list.append(get_cutoff_name(cutoff))
return cutoff_name_list
else:
return get_cutoff_name(cutoffs)
def component_callbacks(self, app):
@app.callback(
[Output(cutoff_name, 'value') for cutoff_name in self.output_cutoff_names],
[Input(self.input_cutoff_name, 'value')]
)
def update_cutoffs(cutoff):
return tuple(cutoff for i in range(len(self.output_cutoff_names)))
class IndexConnector(ExplainerComponent):
def __init__(self, input_index, output_indexes):
"""Connect the index selector of input_index with those of output_indexes.
You can use this to connect a RandomIndexComponent with a
PredictionSummaryComponent for example.
When you change the index in input_index, all the indexes in output_indexes
will automatically be updated.
Args:
input_index ([{str, ExplainerComponent}]): Either a str or an
ExplainerComponent. If str should be equal to the
name of the index property. If ExplainerComponent then
should have a .index_name property.
output_indexes (list(str, ExplainerComponent)): list of str of
ExplainerComponents.
"""
self.input_index_name = self.index_name(input_index)
self.output_index_names = self.index_name(output_indexes)
if not isinstance(self.output_index_names, list):
self.output_index_names = [self.output_index_names]
@staticmethod
def index_name(indexes):#, multi=False):
def get_index_name(o):
if isinstance(o, str): return o
elif isinstance(o, ExplainerComponent):
if not hasattr(o, "index_name"):
raise ValueError(f"{o} does not have an .index_name property!")
return o.index_name
raise ValueError(f"{o} is neither str nor an ExplainerComponent with an .index_name property")
if hasattr(indexes, '__iter__'):
index_name_list = []
for index in indexes:
index_name_list.append(get_index_name(index))
return index_name_list
else:
return get_index_name(indexes)
def component_callbacks(self, app):
@app.callback(
[Output(index_name, 'value') for index_name in self.output_index_names],
[Input(self.input_index_name, 'value')]
)
def update_indexes(index):
return tuple(index for i in range(len(self.output_index_names)))
class HighlightConnector(ExplainerComponent):
def __init__(self, input_highlight, output_highlights):
"""Connect the highlight selector of input_highlight with those of output_highlights.
You can use this to connect a DecisionTreesComponent component to a
DecisionPathGraphComponent for example.
When you change the highlight in input_highlight, all the highlights in output_highlights
will automatically be updated.
Args:
input_highlight ([{str, ExplainerComponent}]): Either a str or an
ExplainerComponent. If str should be equal to the
name of the highlight property. If ExplainerComponent then
should have a .highlight_name property.
output_highlights (list(str, ExplainerComponent)): list of str of
ExplainerComponents.
"""
self.input_highlight_name = self.highlight_name(input_highlight)
self.output_highlight_names = self.highlight_name(output_highlights)
if not isinstance(self.output_highlight_names, list):
self.output_highlight_names = [self.output_highlight_names]
@staticmethod
def highlight_name(highlights):
def get_highlight_name(o):
if isinstance(o, str): return o
elif isinstance(o, ExplainerComponent):
if not hasattr(o, "highlight_name"):
raise ValueError(f"{o} does not have an .highlight_name property!")
return o.highlight_name
raise ValueError(f"{o} is neither str nor an ExplainerComponent with an .highlight_name property")
if hasattr(highlights, '__iter__'):
highlight_name_list = []
for highlight in highlights:
highlight_name_list.append(get_highlight_name(highlight))
return highlight_name_list
else:
return get_highlight_name(highlights)
def component_callbacks(self, app):
@app.callback(
[Output(highlight_name, 'value') for highlight_name in self.output_highlight_names],
[Input(self.input_highlight_name, 'value')])
def update_highlights(highlight):
return tuple(highlight for i in range(len(self.output_highlight_names))) | AMLBID | /Explainer/dashboard_components/connectors.py | connectors.py |
__all__ = [
'RegressionRandomIndexComponent',
'RegressionPredictionSummaryComponent',
'PredictedVsActualComponent',
'ResidualsComponent',
'RegressionVsColComponent',
'RegressionModelSummaryComponent',
]
import numpy as np
import pandas as pd
import dash
import dash_core_components as dcc
import dash_bootstrap_components as dbc
import dash_html_components as html
from dash.dependencies import Input, Output, State
from dash.exceptions import PreventUpdate
from ..dashboard_methods import *
class RegressionRandomIndexComponent(ExplainerComponent):
def __init__(self, explainer, title="Select Random Index", name=None,
subtitle="Select from list or pick at random",
hide_title=False, hide_subtitle=False,
hide_index=False, hide_pred_slider=False,
hide_residual_slider=False, hide_pred_or_y=False,
hide_abs_residuals=False, hide_button=False,
index=None, pred_slider=None, y_slider=None,
residual_slider=None, abs_residual_slider=None,
pred_or_y="preds", abs_residuals=True, round=2,
description=None, **kwargs):
"""Select a random index subject to constraints component
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"Select Random Index".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
subtitle (str): subtitle
hide_title (bool, optional): hide title
hide_subtitle (bool, optional): Hide subtitle. Defaults to False.
hide_index (bool, optional): Hide index selector.
Defaults to False.
hide_pred_slider (bool, optional): Hide prediction slider.
Defaults to False.
hide_residual_slider (bool, optional): hide residuals slider.
Defaults to False.
hide_pred_or_y (bool, optional): hide prediction or actual toggle.
Defaults to False.
hide_abs_residuals (bool, optional): hide absolute residuals toggle.
Defaults to False.
hide_button (bool, optional): hide button. Defaults to False.
index ({str, int}, optional): Initial index to display.
Defaults to None.
pred_slider ([lb, ub], optional): Initial values for prediction
values slider [lowerbound, upperbound]. Defaults to None.
y_slider ([lb, ub], optional): Initial values for y slider
[lower bound, upper bound]. Defaults to None.
residual_slider ([lb, ub], optional): Initial values for residual slider
[lower bound, upper bound]. Defaults to None.
abs_residual_slider ([lb, ub], optional): Initial values for absolute
residuals slider [lower bound, upper bound]
Defaults to None.
pred_or_y (str, {'preds', 'y'}, optional): Initial use predictions
or y slider. Defaults to "preds".
abs_residuals (bool, optional): Initial use residuals or absolute
residuals. Defaults to True.
round (int, optional): rounding used for slider spacing. Defaults to 2.
description (str, optional): Tooltip to display when hover over
component title. When None default text is shown.
"""
super().__init__(explainer, title, name)
assert self.explainer.is_regression, \
("explainer is not a RegressionExplainer so the RegressionRandomIndexComponent "
"will not work. Try using the ClassifierRandomIndexComponent instead.")
self.index_name = 'random-index-reg-index-'+self.name
if self.explainer.y_missing:
self.hide_residual_slider = True
self.hide_pred_or_y = True
self.hide_abs_residuals = True
self.pred_or_y = "preds"
self.y_slider = [0.0, 1.0]
self.residual_slider = [0.0, 1.0]
self.abs_residual_slider = [0.0, 1.0]
if self.pred_slider is None:
self.pred_slider = [self.explainer.preds.min(), self.explainer.preds.max()]
if not self.explainer.y_missing:
if self.y_slider is None:
self.y_slider = [self.explainer.y.min(), self.explainer.y.max()]
if self.residual_slider is None:
self.residual_slider = [self.explainer.residuals.min(), self.explainer.residuals.max()]
if self.abs_residual_slider is None:
self.abs_residual_slider = [self.explainer.abs_residuals.min(), self.explainer.abs_residuals.max()]
assert (len(self.pred_slider)==2 and self.pred_slider[0]<=self.pred_slider[1]), \
"pred_slider should be a list of a [lower_bound, upper_bound]!"
assert (len(self.y_slider)==2 and self.y_slider[0]<=self.y_slider[1]), \
"y_slider should be a list of a [lower_bound, upper_bound]!"
assert (len(self.residual_slider)==2 and self.residual_slider[0]<=self.residual_slider[1]), \
"residual_slider should be a list of a [lower_bound, upper_bound]!"
assert (len(self.abs_residual_slider)==2 and self.abs_residual_slider[0]<=self.abs_residual_slider[1]), \
"abs_residual_slider should be a list of a [lower_bound, upper_bound]!"
self.y_slider = [float(y) for y in self.y_slider]
self.pred_slider = [float(p) for p in self.pred_slider]
self.residual_slider = [float(r) for r in self.residual_slider]
self.abs_residual_slider = [float(a) for a in self.abs_residual_slider]
assert self.pred_or_y in ['preds', 'y'], "pred_or_y should be in ['preds', 'y']!"
if self.description is None: self.description = f"""
You can select a {self.explainer.index_name} directly by choosing it
from the dropdown (if you start typing you can search inside the list),
or hit the Random {self.explainer.index_name} button to randomly select
a {self.explainer.index_name} that fits the constraints. For example
you can select a {self.explainer.index_name} with a very high predicted
{self.explainer.target}, or a very low observed {self.explainer.target},
or a {self.explainer.index_name} whose predicted {self.explainer.target}
was very far off from the observed {self.explainer.target} and so had a
high (absolute) residual.
"""
def layout(self):
return dbc.Card([
make_hideable(
dbc.CardHeader([
html.Div([
html.H3(f"Select {self.explainer.index_name}", id='random-index-reg-title-'+self.name),
make_hideable(html.H6(self.subtitle, className='card-subtitle'), hide=self.hide_subtitle),
dbc.Tooltip(self.description, target='random-index-reg-title-'+self.name),
]),
]), hide=self.hide_title),
dbc.CardBody([
dbc.Row([
make_hideable(
dbc.Col([
dcc.Dropdown(id='random-index-reg-index-'+self.name,
options = [{'label': str(idx), 'value':idx}
for idx in self.explainer.idxs],
value=self.index)
], md=8), hide=self.hide_index),
make_hideable(
dbc.Col([
dbc.Button(f"Random {self.explainer.index_name}", color="primary", id='random-index-reg-button-'+self.name, block=True),
dbc.Tooltip(f"Select a random {self.explainer.index_name} according to the constraints",
target='random-index-reg-button-'+self.name),
], md=4), hide=self.hide_button),
], form=True),
dbc.Row([
make_hideable(
dbc.Col([
html.Div([
dbc.Label("Predicted range:", id='random-index-reg-pred-slider-label-'+self.name,
html_for='random-index-reg-pred-slider-'+self.name),
dbc.Tooltip(f"Only select {self.explainer.index_name} where the "
f"predicted {self.explainer.target} was within the following range:",
target='random-index-reg-pred-slider-label-'+self.name),
dcc.RangeSlider(
id='random-index-reg-pred-slider-'+self.name,
min=float(self.explainer.preds.min()),
max=float(self.explainer.preds.max()),
step=np.float_power(10, -self.round),
value=[self.pred_slider[0], self.pred_slider[1]],
marks={float(self.explainer.preds.min()): str(np.round(self.explainer.preds.min(), self.round)),
float(self.explainer.preds.max()): str(np.round(self.explainer.preds.max(), self.round))},
allowCross=False,
tooltip = {'always_visible' : False}
)
], id='random-index-reg-pred-slider-div-'+self.name),
html.Div([
dbc.Label("Observed range:", id='random-index-reg-y-slider-label-'+self.name,
html_for='random-index-reg-y-slider-'+self.name),
dbc.Tooltip(f"Only select {self.explainer.index_name} where the "
f"observed {self.explainer.target} was within the following range:",
target='random-index-reg-y-slider-label-'+self.name),
dcc.RangeSlider(
id='random-index-reg-y-slider-'+self.name,
min=float(self.explainer.y.min()),
max=float(self.explainer.y.max()),
step=np.float_power(10, -self.round),
value=[self.y_slider[0], self.y_slider[1]],
marks={float(self.explainer.y.min()): str(np.round(self.explainer.y.min(), self.round)),
float(self.explainer.y.max()): str(np.round(self.explainer.y.max(), self.round))},
allowCross=False,
tooltip = {'always_visible' : False}
)
], id='random-index-reg-y-slider-div-'+self.name),
], md=8), hide=self.hide_pred_slider),
make_hideable(
dbc.Col([
dbc.Label("Range:", id='random-index-reg-preds-or-y-label-'+self.name, html_for='random-index-reg-preds-or-y-'+self.name),
dbc.Select(
id='random-index-reg-preds-or-y-'+self.name,
options=[
{'label': 'Predicted', 'value': 'preds'},
{'label': 'Observed', 'value': 'y'},
],
value=self.pred_or_y),
dbc.Tooltip(f"You can either only select a random {self.explainer.index_name}"
f"from within a certain range of observed {self.explainer.target} or"
f"from within a certain range of predicted {self.explainer.target}.",
target='random-index-reg-preds-or-y-label-'+self.name)
], md=4), hide=self.hide_pred_or_y),
]),
dbc.Row([
make_hideable(
dbc.Col([
html.Div([
dbc.Label("Residuals range:", id='random-index-reg-residual-slider-label-'+self.name,
html_for='random-index-reg-residual-slider-'+self.name),
dbc.Tooltip(f"Only select {self.explainer.index_name} where the "
f"residual (difference between observed {self.explainer.target} and predicted {self.explainer.target})"
" was within the following range:",
target='random-index-reg-residual-slider-label-'+self.name),
dcc.RangeSlider(
id='random-index-reg-residual-slider-'+self.name,
min=float(self.explainer.residuals.min()),
max=float(self.explainer.residuals.max()),
step=np.float_power(10, -self.round),
value=[self.residual_slider[0], self.residual_slider[1]],
marks={float(self.explainer.residuals.min()): str(np.round(self.explainer.residuals.min(), self.round)),
float(self.explainer.residuals.max()): str(np.round(self.explainer.residuals.max(), self.round))},
allowCross=False,
tooltip={'always_visible' : False}
)
], id='random-index-reg-residual-slider-div-'+self.name),
html.Div([
dbc.Label("Absolute residuals", id='random-index-reg-abs-residual-slider-label'+self.name,
html_for='random-index-reg-abs-residual-slider-'+self.name),
dbc.Tooltip(f"Only select {self.explainer.index_name} where the absolute "
f"residual (difference between observed {self.explainer.target} and predicted {self.explainer.target})"
" was within the following range:",
target='random-index-reg-abs-residual-slider-label'+self.name),
dcc.RangeSlider(
id='random-index-reg-abs-residual-slider-'+self.name,
min=float(self.explainer.abs_residuals.min()),
max=float(self.explainer.abs_residuals.max()),
step=np.float_power(10, -self.round),
value=[self.abs_residual_slider[0], self.abs_residual_slider[1]],
marks={float(self.explainer.abs_residuals.min()): str(np.round(self.explainer.abs_residuals.min(), self.round)),
float(self.explainer.abs_residuals.max()): str(np.round(self.explainer.abs_residuals.max(), self.round))},
allowCross=False,
tooltip={'always_visible' : False}
)
], id='random-index-reg-abs-residual-slider-div-'+self.name),
], md=8), hide=self.hide_residual_slider),
make_hideable(
dbc.Col([
dbc.Label("Residuals:", id='random-index-reg-abs-residual-label-'+self.name,
html_for='random-index-reg-abs-residual-'+self.name),
dbc.Select(
id='random-index-reg-abs-residual-'+self.name,
options=[
{'label': 'Residuals', 'value': 'relative'},
{'label': 'Absolute Residuals', 'value': 'absolute'},
],
value='absolute' if self.abs_residuals else 'relative'),
dbc.Tooltip(f"You can either only select random a {self.explainer.index_name} "
f"from within a certain range of residuals "
f"(difference between observed and predicted {self.explainer.target}), "
f"so for example only {self.explainer.index_name} for whom the prediction "
f"was too high or too low."
f"Or you can select only from a certain absolute residual range. So for "
f"example only select {self.explainer.index_name} for which the prediction was at "
f"least a certain amount of {self.explainer.units} off.",
target='random-index-reg-abs-residual-label-'+self.name),
], md=4), hide=self.hide_abs_residuals),
]),
# make_hideable(
# html.Div([
# html.Div([
# dbc.Row([
# dbc.Col([
# html.Div([
# dbc.Label("Residuals range:", id='random-index-reg-residual-slider-label-'+self.name,
# html_for='random-index-reg-residual-slider-'+self.name),
# dbc.Tooltip(f"Only select {self.explainer.index_name} where the "
# f"residual (difference between observed {self.explainer.target} and predicted {self.explainer.target})"
# " was within the following range:",
# target='random-index-reg-residual-slider-label-'+self.name),
# dcc.RangeSlider(
# id='random-index-reg-residual-slider-'+self.name,
# min=float(self.explainer.residuals.min()),
# max=float(self.explainer.residuals.max()),
# step=np.float_power(10, -self.round),
# value=[self.residual_slider[0], self.residual_slider[1]],
# marks={float(self.explainer.residuals.min()): str(np.round(self.explainer.residuals.min(), self.round)),
# float(self.explainer.residuals.max()): str(np.round(self.explainer.residuals.max(), self.round))},
# allowCross=False,
# tooltip={'always_visible' : False}
# )
# ], style={'margin-bottom':0})
# ], md=8)
# ]),
# ], id='random-index-reg-residual-slider-div-'+self.name),
# html.Div([
# dbc.Row([
# dbc.Col([
# html.Div([
# dbc.Label("Absolute residuals", id='random-index-reg-abs-residual-slider-label'+self.name,
# html_for='random-index-reg-abs-residual-slider-'+self.name),
# dbc.Tooltip(f"Only select {self.explainer.index_name} where the absolute "
# f"residual (difference between observed {self.explainer.target} and predicted {self.explainer.target})"
# " was within the following range:",
# target='random-index-reg-abs-residual-slider-label'+self.name),
# dcc.RangeSlider(
# id='random-index-reg-abs-residual-slider-'+self.name,
# min=float(self.explainer.abs_residuals.min()),
# max=float(self.explainer.abs_residuals.max()),
# step=np.float_power(10, -self.round),
# value=[self.abs_residual_slider[0], self.abs_residual_slider[1]],
# marks={float(self.explainer.abs_residuals.min()): str(np.round(self.explainer.abs_residuals.min(), self.round)),
# float(self.explainer.abs_residuals.max()): str(np.round(self.explainer.abs_residuals.max(), self.round))},
# allowCross=False,
# tooltip={'always_visible' : False}
# )
# ], style={'margin-bottom':0})
# ], md=8)
# ])
# ], id='random-index-reg-abs-residual-slider-div-'+self.name),
# ]), hide=self.hide_residual_slider),
# dbc.Row([
# make_hideable(
# dbc.Col([
# dbc.Label("Residuals:", id='random-index-reg-abs-residual-label-'+self.name,
# html_for='random-index-reg-abs-residual-'+self.name),
# dbc.Select(
# id='random-index-reg-abs-residual-'+self.name,
# options=[
# {'label': 'Residuals', 'value': 'relative'},
# {'label': 'Absolute Residuals', 'value': 'absolute'},
# ],
# value='absolute' if self.abs_residuals else 'relative'),
# dbc.Tooltip(f"You can either only select random a {self.explainer.index_name} "
# f"from within a certain range of residuals "
# f"(difference between observed and predicted {self.explainer.target}), "
# f"so for example only {self.explainer.index_name} for whom the prediction "
# f"was too high or too low."
# f"Or you can select only from a certain absolute residual range. So for "
# f"example only select {self.explainer.index_name} for which the prediction was at "
# f"least a certain amount of {self.explainer.units} off.",
# target='random-index-reg-abs-residual-label-'+self.name),
# ], md=4), hide=self.hide_pred_or_y),
# make_hideable(
# dbc.Col([
# html.Div([
# dbc.Select(
# id='random-index-reg-abs-residual-'+self.name,
# options=[
# {'label': 'Use Residuals', 'value': 'relative'},
# {'label': 'Use Absolute Residuals', 'value': 'absolute'},
# ],
# value='absolute' if self.abs_residuals else 'relative'),
# ], id='random-index-reg-abs-residual-div-'+self.name),
# dbc.Tooltip(f"You can either only select random a {self.explainer.index_name} "
# f"from within a certain range of residuals "
# f"(difference between observed and predicted {self.explainer.target}), "
# f"so for example only {self.explainer.index_name} for whom the prediction "
# f"was too high or too low."
# f"Or you can select only from a certain absolute residual range. So for "
# f"example only select {self.explainer.index_name} for which the prediction was at "
# f"least a certain amount of {self.explainer.units} off.",
# target='random-index-reg-abs-residual-div-'+self.name),
# ], md=4), hide=self.hide_abs_residuals),
]),
])
def component_callbacks(self, app):
@app.callback(
[Output('random-index-reg-pred-slider-div-'+self.name, 'style'),
Output('random-index-reg-y-slider-div-'+self.name, 'style')],
[Input('random-index-reg-preds-or-y-'+self.name, 'value')])
def update_reg_hidden_div_pred_sliders(preds_or_y):
if preds_or_y == 'preds':
return (None, dict(display="none"))
elif preds_or_y == 'y':
return (dict(display="none"), None)
raise PreventUpdate
@app.callback(
[Output('random-index-reg-residual-slider-div-'+self.name, 'style'),
Output('random-index-reg-abs-residual-slider-div-'+self.name, 'style')],
[Input('random-index-reg-abs-residual-'+self.name, 'value')])
def update_reg_hidden_div_pred_sliders(abs_residuals):
if abs_residuals == 'absolute':
return (dict(display="none"), None)
else:
return (None, dict(display="none"))
raise PreventUpdate
@app.callback(
[Output('random-index-reg-residual-slider-'+self.name, 'min'),
Output('random-index-reg-residual-slider-'+self.name, 'max'),
Output('random-index-reg-residual-slider-'+self.name, 'value'),
Output('random-index-reg-residual-slider-'+self.name, 'marks'),
Output('random-index-reg-abs-residual-slider-'+self.name, 'min'),
Output('random-index-reg-abs-residual-slider-'+self.name, 'max'),
Output('random-index-reg-abs-residual-slider-'+self.name, 'value'),
Output('random-index-reg-abs-residual-slider-'+self.name, 'marks'),],
[Input('random-index-reg-pred-slider-'+self.name, 'value'),
Input('random-index-reg-y-slider-'+self.name, 'value')],
[State('random-index-reg-preds-or-y-'+self.name, 'value'),
State('random-index-reg-residual-slider-'+self.name, 'value'),
State('random-index-reg-abs-residual-slider-'+self.name, 'value')])
def update_residual_slider_limits(pred_range, y_range, preds_or_y, residuals_range, abs_residuals_range):
if preds_or_y=='preds':
min_residuals = self.explainer.residuals[(self.explainer.preds >= pred_range[0]) & (self.explainer.preds <= pred_range[1])].min()
max_residuals = self.explainer.residuals[(self.explainer.preds >= pred_range[0]) & (self.explainer.preds <= pred_range[1])].max()
min_abs_residuals = self.explainer.abs_residuals[(self.explainer.preds >= pred_range[0]) & (self.explainer.preds <= pred_range[1])].min()
max_abs_residuals = self.explainer.abs_residuals[(self.explainer.preds >= pred_range[0]) & (self.explainer.preds <= pred_range[1])].max()
elif preds_or_y=='y':
min_residuals = self.explainer.residuals[(self.explainer.y >= y_range[0]) & (self.explainer.y <= y_range[1])].min()
max_residuals = self.explainer.residuals[(self.explainer.y >= y_range[0]) & (self.explainer.y <= y_range[1])].max()
min_abs_residuals = self.explainer.abs_residuals[(self.explainer.y >= y_range[0]) & (self.explainer.y <= y_range[1])].min()
max_abs_residuals = self.explainer.abs_residuals[(self.explainer.y >= y_range[0]) & (self.explainer.y <= y_range[1])].max()
new_residuals_range = [max(min_residuals, residuals_range[0]), min(max_residuals, residuals_range[1])]
new_abs_residuals_range = [max(min_abs_residuals, abs_residuals_range[0]), min(max_abs_residuals, abs_residuals_range[1])]
residuals_marks = {min_residuals: str(np.round(min_residuals, self.round)),
max_residuals: str(np.round(max_residuals, self.round))}
abs_residuals_marks = {min_abs_residuals: str(np.round(min_abs_residuals, self.round)),
max_abs_residuals: str(np.round(max_abs_residuals, self.round))}
return (min_residuals, max_residuals, new_residuals_range, residuals_marks,
min_abs_residuals, max_abs_residuals, new_abs_residuals_range, abs_residuals_marks)
@app.callback(
Output('random-index-reg-index-'+self.name, 'value'),
[Input('random-index-reg-button-'+self.name, 'n_clicks')],
[State('random-index-reg-pred-slider-'+self.name, 'value'),
State('random-index-reg-y-slider-'+self.name, 'value'),
State('random-index-reg-residual-slider-'+self.name, 'value'),
State('random-index-reg-abs-residual-slider-'+self.name, 'value'),
State('random-index-reg-preds-or-y-'+self.name, 'value'),
State('random-index-reg-abs-residual-'+self.name, 'value')])
def update_index(n_clicks, pred_range, y_range, residual_range, abs_residuals_range, preds_or_y, abs_residuals):
if n_clicks is None and self.index is not None:
raise PreventUpdate
if preds_or_y == 'preds':
if abs_residuals=='absolute':
return self.explainer.random_index(
pred_min=pred_range[0], pred_max=pred_range[1],
abs_residuals_min=abs_residuals_range[0],
abs_residuals_max=abs_residuals_range[1],
return_str=True)
else:
return self.explainer.random_index(
pred_min=pred_range[0], pred_max=pred_range[1],
residuals_min=residual_range[0],
residuals_max=residual_range[1],
return_str=True)
elif preds_or_y == 'y':
if abs_residuals=='absolute':
return self.explainer.random_index(
y_min=y_range[0], y_max=y_range[1],
abs_residuals_min=abs_residuals_range[0],
abs_residuals_max=abs_residuals_range[1],
return_str=True)
else:
return self.explainer.random_index(
y_min=pred_range[0], y_max=pred_range[1],
residuals_min=residual_range[0],
residuals_max=residual_range[1],
return_str=True)
class RegressionPredictionSummaryComponent(ExplainerComponent):
def __init__(self, explainer, title="Prediction", name=None,
hide_index=False, hide_title=False,
hide_subtitle=False, hide_table=False,
feature_input_component=None,
index=None, round=3, description=None,
**kwargs):
"""Shows a summary for a particular prediction
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"Prediction".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
hide_index (bool, optional): hide index selector. Defaults to False.
hide_title (bool, optional): hide title. Defaults to False.
hide_subtitle (bool, optional): Hide subtitle. Defaults to False.
hide_table (bool, optional): hide the results table
feature_input_component (FeatureInputComponent): A FeatureInputComponent
that will give the input to the graph instead of the index selector.
If not None, hide_index=True. Defaults to None.
index ({int, str}, optional): Index to display prediction summary for. Defaults to None.
description (str, optional): Tooltip to display when hover over
component title. When None default text is shown.
"""
super().__init__(explainer, title, name)
self.index_name = 'reg-prediction-index-'+self.name
if self.feature_input_component is not None:
self.exclude_callbacks(self.feature_input_component)
self.hide_index = True
if self.description is None: self.description = f"""
Shows the predicted {self.explainer.target} and the observed {self.explainer.target},
as well as the difference between the two (residual)
"""
def layout(self):
return dbc.Card([
make_hideable(
dbc.CardHeader([
html.H3(self.title, id='reg-prediction-title-'+self.name, className='card-title'),
dbc.Tooltip(self.description, target='reg-prediction-title-'+self.name),
]), hide=self.hide_title),
dbc.CardBody([
dbc.Row([
make_hideable(
dbc.Col([
dbc.Label(f"{self.explainer.index_name}:"),
dcc.Dropdown(id='reg-prediction-index-'+self.name,
options = [{'label': str(idx), 'value':idx}
for idx in self.explainer.idxs],
value=self.index)
], md=6), hide=self.hide_index),
]),
dbc.Row([
dbc.Col([
html.Div(id='reg-prediction-div-'+self.name)
])
])
])
])
def component_callbacks(self, app):
if self.feature_input_component is None:
@app.callback(
Output('reg-prediction-div-'+self.name, 'children'),
[Input('reg-prediction-index-'+self.name, 'value')])
def update_output_div(index):
if index is not None:
preds_df = self.explainer.prediction_result_df(index, round=self.round)
return make_hideable(
dbc.Table.from_dataframe(preds_df, striped=False, bordered=False, hover=False),
hide=self.hide_table)
raise PreventUpdate
else:
@app.callback(
Output('reg-prediction-div-'+self.name, 'children'),
[*self.feature_input_component._feature_callback_inputs])
def update_output_div(*inputs):
X_row = self.explainer.get_row_from_input(inputs, ranked_by_shap=True)
preds_df = self.explainer.prediction_result_df(X_row=X_row, round=self.round)
return make_hideable(
dbc.Table.from_dataframe(preds_df, striped=False, bordered=False, hover=False),
hide=self.hide_table)
class PredictedVsActualComponent(ExplainerComponent):
def __init__(self, explainer, title="Predicted vs Actual", name=None,
subtitle="How close is the predicted value to the observed?",
hide_title=False, hide_subtitle=False,
hide_log_x=False, hide_log_y=False,
logs=False, log_x=False, log_y=False, description=None,
**kwargs):
"""Shows a plot of predictions vs y.
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"Predicted vs Actual".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
subtitle (str): subtitle
hide_title (bool, optional) Hide the title. Defaults to False.
hide_subtitle (bool, optional): Hide subtitle. Defaults to False.
hide_log_x (bool, optional): Hide the log_x toggle. Defaults to False.
hide_log_y (bool, optional): Hide the log_y toggle. Defaults to False.
logs (bool, optional): Whether to use log axis. Defaults to False.
log_x (bool, optional): log only x axis. Defaults to False.
log_y (bool, optional): log only y axis. Defaults to False.
description (str, optional): Tooltip to display when hover over
component title. When None default text is shown.
"""
super().__init__(explainer, title, name)
self.logs, self.log_x, self.log_y = logs, log_x, log_y
if self.description is None: self.description = f"""
Plot shows the observed {self.explainer.target} and the predicted
{self.explainer.target} in the same plot. A perfect model would have
all the points on the diagonal (predicted matches observed). The further
away point are from the diagonal the worse the model is in predicting
{self.explainer.target}.
"""
self.register_dependencies(['preds'])
def layout(self):
return dbc.Card([
make_hideable(
dbc.CardHeader([
html.Div([
html.H3(self.title, id='pred-vs-actual-title-'+self.name),
make_hideable(html.H6(self.subtitle, className='card-subtitle'), hide=self.hide_subtitle),
dbc.Tooltip(self.description, target='pred-vs-actual-title-'+self.name),
]),
]), hide=self.hide_title),
dbc.CardBody([
dbc.Row([
make_hideable(
dbc.Col([
dbc.FormGroup(
[
# html.Label("Log y"),
dbc.RadioButton(
id='pred-vs-actual-logy-'+self.name,
className="form-check-input",
checked=self.log_y),
dbc.Tooltip("By using a log axis, it is easier to see relative "
"errors instead of absolute errors.",
target='pred-vs-actual-logy-'+self.name),
dbc.Label("Log y",
html_for='pred-vs-actual-logy-'+self.name,
className="form-check-label"),
], check=True),
], md=1, align="center"), hide=self.hide_log_y),
dbc.Col([
dcc.Graph(id='pred-vs-actual-graph-'+self.name,
config=dict(modeBarButtons=[['toImage']], displaylogo=False)),
], md=11)
]),
dbc.Row([
make_hideable(
dbc.Col([
dbc.FormGroup(
[
dbc.RadioButton(
id='pred-vs-actual-logx-'+self.name,
className="form-check-input",
checked=self.log_x),
dbc.Tooltip("By using a log axis, it is easier to see relative "
"errors instead of absolute errors.",
target='pred-vs-actual-logx-'+self.name),
dbc.Label("Log x",
html_for='pred-vs-actual-logx-'+self.name,
className="form-check-label"),
], check=True),
], md=2), hide=self.hide_log_x),
], justify="center"),
]),
])
def component_callbacks(self, app):
@app.callback(
Output('pred-vs-actual-graph-'+self.name, 'figure'),
[Input('pred-vs-actual-logx-'+self.name, 'checked'),
Input('pred-vs-actual-logy-'+self.name, 'checked')],
)
def update_predicted_vs_actual_graph(log_x, log_y):
return self.explainer.plot_predicted_vs_actual(log_x=log_x, log_y=log_y)
class ResidualsComponent(ExplainerComponent):
def __init__(self, explainer, title="Residuals", name=None,
subtitle="How much is the model off?",
hide_title=False, hide_subtitle=False, hide_footer=False,
hide_pred_or_actual=False, hide_ratio=False,
pred_or_actual="vs_pred", residuals="difference",
description=None, **kwargs):
"""Residuals plot component
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"Residuals".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
subtitle (str): subtitle
hide_title (bool, optional) Hide the title. Defaults to False.
hide_subtitle (bool, optional): Hide subtitle. Defaults to False.
hide_footer (bool, optional): hide the footer at the bottom of the component
hide_pred_or_actual (bool, optional): hide vs predictions or vs
actual for x-axis toggle. Defaults to False.
hide_ratio (bool, optional): hide residual type dropdown. Defaults to False.
pred_or_actual (str, {'vs_actual', 'vs_pred'}, optional): Whether
to plot actual or predictions on the x-axis.
Defaults to "vs_pred".
residuals (str, {'difference', 'ratio', 'log-ratio'} optional):
How to calcualte residuals. Defaults to 'difference'.
description (str, optional): Tooltip to display when hover over
component title. When None default text is shown.
"""
super().__init__(explainer, title, name)
assert residuals in ['difference', 'ratio', 'log-ratio'], \
("parameter residuals should in ['difference', 'ratio', 'log-ratio']"
f" but you passed residuals={residuals}")
if self.description is None: self.description = f"""
The residuals are the difference between the observed {self.explainer.target}
and predicted {self.explainer.target}. In this plot you can check if
the residuals are higher or lower for higher/lower actual/predicted outcomes.
So you can check if the model works better or worse for different {self.explainer.target}
levels.
"""
self.register_dependencies(['preds', 'residuals'])
def layout(self):
return dbc.Card([
make_hideable(
dbc.CardHeader([
html.Div([
html.H3(self.title, id='residuals-title-'+self.name),
make_hideable(html.H6(self.subtitle, className='card-subtitle'), hide=self.hide_subtitle),
dbc.Tooltip(self.description, target='residuals-title-'+self.name),
]),
]), hide=self.hide_title),
dbc.CardBody([
dbc.Row([
dbc.Col([
dcc.Graph(id='residuals-graph-'+self.name,
config=dict(modeBarButtons=[['toImage']], displaylogo=False)),
])
])
]),
make_hideable(
dbc.CardFooter([
dbc.Row([
make_hideable(
dbc.Col([
dbc.FormGroup(
[
dbc.Label("Horizontal axis:", html_for='residuals-pred-or-actual-'+self.name),
dbc.Select(
options=[
{"label": "Predicted", "value": "vs_pred"},
{"label": "Observed", "value": "vs_actual"},
],
value=self.pred_or_actual,
id='residuals-pred-or-actual-'+self.name,
),
], id='residuals-pred-or-actual-form-'+self.name),
dbc.Tooltip("Select what you would like to put on the x-axis:"
f" observed {self.explainer.target} or predicted {self.explainer.target}.",
target='residuals-pred-or-actual-form-'+self.name),
], md=3), hide=self.hide_pred_or_actual),
make_hideable(
dbc.Col([
html.Label('Residual type:', id='residuals-type-label-'+self.name),
dbc.Tooltip("Type of residuals to display: y-preds (difference), "
"y/preds (ratio) or log(y/preds) (logratio).",
target='residuals-type-label-'+self.name),
dbc.Select(id='residuals-type-'+self.name,
options = [{'label': 'Difference', 'value': 'difference'},
{'label': 'Ratio', 'value': 'ratio'},
{'label': 'Log ratio', 'value': 'log-ratio'}],
value=self.residuals),
], md=3), hide=self.hide_ratio),
]),
]), hide=self.hide_footer)
])
def register_callbacks(self, app):
@app.callback(
Output('residuals-graph-'+self.name, 'figure'),
[Input('residuals-pred-or-actual-'+self.name, 'value'),
Input('residuals-type-'+self.name, 'value')],
)
def update_residuals_graph(pred_or_actual, residuals):
vs_actual = pred_or_actual=='vs_actual'
return self.explainer.plot_residuals(vs_actual=vs_actual, residuals=residuals)
class RegressionVsColComponent(ExplainerComponent):
def __init__(self, explainer, title="Plot vs feature", name=None,
subtitle="Are predictions and residuals correlated with features?",
hide_title=False, hide_subtitle=False, hide_footer=False,
hide_col=False, hide_ratio=False, hide_cats=False,
hide_points=False, hide_winsor=False,
col=None, display='difference', cats=True,
points=True, winsor=0, description=None, **kwargs):
"""Show residuals, observed or preds vs a particular Feature component
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"Plot vs feature".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
subtitle (str): subtitle
hide_title (bool, optional) Hide the title. Defaults to False.
hide_subtitle (bool, optional): Hide subtitle. Defaults to False.
hide_footer (bool, optional): hide the footer at the bottom of the component
hide_col (bool, optional): Hide de column selector. Defaults to False.
hide_ratio (bool, optional): Hide the toggle. Defaults to False.
hide_cats (bool, optional): Hide group cats toggle. Defaults to False.
hide_points (bool, optional): Hide group points toggle. Defaults to False.
hide_winsor (bool, optional): Hide winsor input. Defaults to False.
col ([type], optional): Initial feature to display. Defaults to None.
display (str, {'observed', 'predicted', difference', 'ratio', 'log-ratio'} optional):
What to display on y axis. Defaults to 'difference'.
cats (bool, optional): group categorical columns. Defaults to True.
points (bool, optional): display point cloud next to violin plot
for categorical cols. Defaults to True
winsor (int, 0-50, optional): percentage of outliers to winsor out of
the y-axis. Defaults to 0.
description (str, optional): Tooltip to display when hover over
component title. When None default text is shown.
"""
super().__init__(explainer, title, name)
if self.col is None:
self.col = self.explainer.columns_ranked_by_shap(self.cats)[0]
assert self.display in {'observed', 'predicted', 'difference', 'ratio', 'log-ratio'}, \
("parameter display should in {'observed', 'predicted', 'difference', 'ratio', 'log-ratio'}"
f" but you passed display={self.display}!")
if self.description is None: self.description = f"""
This plot shows either residuals (difference between observed {self.explainer.target}
and predicted {self.explainer.target}) plotted against the values of different features,
or the observed or predicted {self.explainer.target}.
This allows you to inspect whether the model is more wrong for particular
range of feature values than others.
"""
self.register_dependencies(['preds', 'residuals'])
def layout(self):
return dbc.Card([
make_hideable(
dbc.CardHeader([
html.Div([
html.H3(self.title, id='reg-vs-col-title-'+self.name),
make_hideable(html.H6(self.subtitle, className='card-subtitle'), hide=self.hide_subtitle),
dbc.Tooltip(self.description, target='reg-vs-col-title-'+self.name),
]),
]), hide=self.hide_title),
dbc.CardBody([
dbc.Row([
make_hideable(
dbc.Col([
dbc.Label("Feature:", id='reg-vs-col-col-label-'+self.name),
dbc.Tooltip("Select the feature to display on the x-axis.",
target='reg-vs-col-col-label-'+self.name),
dbc.Select(id='reg-vs-col-col-'+self.name,
options=[{'label': col, 'value':col}
for col in self.explainer.columns_ranked_by_shap(self.cats)],
value=self.col),
], md=4), hide=self.hide_col),
make_hideable(
dbc.Col([
html.Label('Display:', id='reg-vs-col-display-type-label-'+self.name),
dbc.Tooltip(f"Select what to display on the y axis: observed {self.explainer.target}, "
f"predicted {self.explainer.target} or residuals. Residuals can either "
"be calculated by takind the difference (y-preds), "
"ratio (y/preds) or log ratio log(y/preds). The latter makes it easier to "
"see relative differences.",
target='reg-vs-col-display-type-label-'+self.name),
dbc.Select(id='reg-vs-col-display-type-'+self.name,
options = [{'label': 'Observed', 'value': 'observed'},
{'label': 'Predicted', 'value': 'predicted'},
{'label': 'Residuals: Difference', 'value': 'difference'},
{'label': 'Residuals: Ratio', 'value': 'ratio'},
{'label': 'Residuals: Log ratio', 'value': 'log-ratio'}],
value=self.display),
], md=4), hide=self.hide_ratio),
make_hideable(
dbc.Col([
dbc.FormGroup([
dbc.Label("Grouping:", id='reg-vs-col-group-cats-label-'+self.name),
dbc.Tooltip("Group onehot encoded categorical variables together",
target='reg-vs-col-group-cats-label-'+self.name),
dbc.Checklist(
options=[{"label": "Group cats", "value": True}],
value=[True] if self.cats else [],
id='reg-vs-col-group-cats-'+self.name,
inline=True,
switch=True,
),
]),
], md=2), self.hide_cats),
]),
dbc.Row([
dbc.Col([
dcc.Graph(id='reg-vs-col-graph-'+self.name,
config=dict(modeBarButtons=[['toImage']], displaylogo=False)),
])
]),
]),
make_hideable(
dbc.CardFooter([
dbc.Row([
make_hideable(
dbc.Col([
dbc.Label("Winsor:", id='reg-vs-col-winsor-label-'+self.name),
dbc.Tooltip("Excluded the highest and lowest y values from the plot. "
"When you have some real outliers it can help to remove them"
" from the plot so it is easier to see the overall pattern.",
target='reg-vs-col-winsor-label-'+self.name),
dbc.Input(id='reg-vs-col-winsor-'+self.name,
value=self.winsor,
type="number", min=0, max=49, step=1),
], md=4), hide=self.hide_winsor),
make_hideable(
dbc.Col([
html.Div([
dbc.FormGroup([
dbc.Label("Scatter:"),
dbc.Tooltip("For categorical features, display "
"a point cloud next to the violin plots.",
target='reg-vs-col-show-points-'+self.name),
dbc.Checklist(
options=[{"label": "Show point cloud", "value": True}],
value=[True] if self.points else [],
id='reg-vs-col-show-points-'+self.name,
inline=True,
switch=True,
),
]),
], id='reg-vs-col-show-points-div-'+self.name)
], md=4), self.hide_points),
])
]), hide=self.hide_footer)
])
def register_callbacks(self, app):
@app.callback(
[Output('reg-vs-col-graph-'+self.name, 'figure'),
Output('reg-vs-col-show-points-div-'+self.name, 'style')],
[Input('reg-vs-col-col-'+self.name, 'value'),
Input('reg-vs-col-display-type-'+self.name, 'value'),
Input('reg-vs-col-show-points-'+self.name, 'value'),
Input('reg-vs-col-winsor-'+self.name, 'value')],
)
def update_residuals_graph(col, display, points, winsor):
style = {} if col in self.explainer.cats else dict(display="none")
if display == 'observed':
return self.explainer.plot_y_vs_feature(
col, points=bool(points), winsor=winsor, dropna=True), style
elif display == 'predicted':
return self.explainer.plot_preds_vs_feature(
col, points=bool(points), winsor=winsor, dropna=True), style
else:
return self.explainer.plot_residuals_vs_feature(
col, residuals=display, points=bool(points),
winsor=winsor, dropna=True), style
@app.callback(
Output('reg-vs-col-col-'+self.name, 'options'),
[Input('reg-vs-col-group-cats-'+self.name, 'value')])
def update_dependence_shap_scatter_graph(cats):
return [{'label': col, 'value': col}
for col in self.explainer.columns_ranked_by_shap(bool(cats))]
class RegressionModelSummaryComponent(ExplainerComponent):
def __init__(self, explainer, title="Model Summary", name=None,
subtitle="Quantitative metrics for model performance",
hide_title=False, hide_subtitle=False,
round=3, description=None, **kwargs):
"""Show model summary statistics (RMSE, MAE, R2) component
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"Model Summary".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
subtitle (str): subtitle
hide_title (bool, optional): hide title
hide_subtitle (bool, optional): Hide subtitle. Defaults to False.
round (int): rounding to perform to metric floats.
description (str, optional): Tooltip to display when hover over
component title. When None default text is shown.
"""
super().__init__(explainer, title, name)
if self.description is None: self.description = f"""
In the table below you can find a number of regression performance
metrics that describe how well the model is able to predict
{self.explainer.target}.
"""
self.register_dependencies(['preds', 'residuals'])
def layout(self):
metrics_dict = self.explainer.metrics_descriptions()
metrics_df = (pd.DataFrame(self.explainer.metrics(), index=["Score"]).T
.rename_axis(index="metric").reset_index().round(self.round))
metrics_table = dbc.Table.from_dataframe(metrics_df, striped=False, bordered=False, hover=False)
metrics_table, tooltips = get_dbc_tooltips(metrics_table,
metrics_dict,
"reg-model-summary-div-hover",
self.name)
return dbc.Card([
make_hideable(
dbc.CardHeader([
html.Div([
html.H3(self.title, id='reg-model-summary-title-'+self.name),
make_hideable(html.H6(self.subtitle, className='card-subtitle'), hide=self.hide_subtitle),
dbc.Tooltip(self.description, target='reg-model-summary-title-'+self.name),
]),
]), hide=self.hide_title),
dbc.CardBody([
metrics_table,
*tooltips
]),
]) | AMLBID | /Explainer/dashboard_components/regression_components.py | regression_components.py |
__all__ = [
'DecisionTreesComponent',
'DecisionPathTableComponent',
'DecisionPathGraphComponent',
]
import dash
import dash_core_components as dcc
import dash_bootstrap_components as dbc
import dash_html_components as html
import dash_table
from dash.dependencies import Input, Output, State
from dash.exceptions import PreventUpdate
from ..AMLBID_Explainer import RandomForestExplainer, XGBExplainer
from ..dashboard_methods import *
from .classifier_components import ClassifierRandomIndexComponent
from .connectors import IndexConnector, HighlightConnector
class DecisionTreesComponent(ExplainerComponent):
def __init__(self, explainer, title="Decision Trees", name=None,
subtitle="Displaying individual decision trees",
hide_title=False, hide_subtitle=False,
hide_index=False, hide_highlight=False,
hide_selector=False,
pos_label=None, index=None, highlight=None,
higher_is_better=True, description=None, **kwargs):
"""Show prediction from individual decision trees inside RandomForest component
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"Decision Trees".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
subtitle (str): subtitle
hide_title (bool, optional): hide title, Defaults to False.
hide_subtitle (bool, optional): Hide subtitle. Defaults to False.
hide_index (bool, optional): Hide index selector. Defaults to False.
hide_highlight (bool, optional): Hide tree highlight selector. Defaults to False.
hide_selector (bool, optional): hide pos label selectors. Defaults to False.
pos_label ({int, str}, optional): initial pos label.
Defaults to explainer.pos_label
index ({str, int}, optional): Initial index to display. Defaults to None.
highlight (int, optional): Initial tree to highlight. Defaults to None.
higher_is_better (bool, optional): up is green, down is red. If False
flip the colors. (for gbm models only)
description (str, optional): Tooltip to display when hover over
component title. When None default text is shown.
"""
super().__init__(explainer, title, name)
self.index_name = 'decisiontrees-index-'+self.name
self.highlight_name = 'decisiontrees-highlight-'+self.name
self.selector = PosLabelSelector(explainer, name=self.name, pos_label=pos_label)
if isinstance(self.explainer, RandomForestExplainer):
if self.description is None: self.description = """
Show the prediction of every individul tree in a random forest.
This demonstrates how a random forest is simply an average of an
ensemble of decision trees.
"""
if self.subtitle == "Displaying individual decision trees":
self.subtitle += " inside Random Forest"
elif isinstance(self.explainer, XGBExplainer):
if self.description is None: self.description = """
Shows the marginal contributions of each decision tree in an
xgboost ensemble to the final prediction. This demonstrates that
an xgboost model is simply a sum of individual decision trees.
"""
if self.subtitle == "Displaying individual decision trees":
self.subtitle += " inside xgboost model"
else:
if self.description is None: self.description = ""
self.register_dependencies("preds", "pred_probas")
def layout(self):
return dbc.Card([
make_hideable(
dbc.CardHeader([
html.Div([
html.H3(self.title, id='decisiontrees-title-'+self.name),
make_hideable(html.H6(self.subtitle, className='card-subtitle'), hide=self.hide_subtitle),
dbc.Tooltip(self.description, target='decisiontrees-title-'+self.name),
]),
]), hide=self.hide_title),
dbc.CardBody([
dbc.Row([
make_hideable(
dbc.Col([
dbc.Label(f"{self.explainer.index_name}:", id='decisiontrees-index-label-'+self.name),
dbc.Tooltip(f"Select {self.explainer.index_name} to display decision trees for",
target='decisiontrees-index-label-'+self.name),
dcc.Dropdown(id='decisiontrees-index-'+self.name,
options = [{'label': str(idx), 'value':idx}
for idx in self.explainer.idxs],
value=self.index)
], md=4), hide=self.hide_index),
make_hideable(
dbc.Col([
dbc.Label("Highlight tree:", id='decisiontrees-tree-label-'+self.name),
dbc.Tooltip(f"Select a specific tree to highlight. You can also "
"highlight by clicking on a specifc bar in the bar chart.",
target='decisiontrees-tree-label-'+self.name),
dbc.Select(id='decisiontrees-highlight-'+self.name,
options = [{'label': str(tree), 'value': tree}
for tree in range(self.explainer.no_of_trees)],
value=self.highlight)
], md=2), hide=self.hide_highlight),
make_hideable(
dbc.Col([self.selector.layout()
], width=2), hide=self.hide_selector)
]),
dbc.Row([
dbc.Col([
dcc.Graph(id="decisiontrees-graph-"+self.name,
config=dict(modeBarButtons=[['toImage']], displaylogo=False)),
])
]),
])
])
def component_callbacks(self, app):
@app.callback(
Output("decisiontrees-graph-"+self.name, 'figure'),
[Input('decisiontrees-index-'+self.name, 'value'),
Input('decisiontrees-highlight-'+self.name, 'value'),
Input('pos-label-'+self.name, 'value')],
)
def update_tree_graph(index, highlight, pos_label):
if index is not None:
highlight = None if highlight is None else int(highlight)
return self.explainer.plot_trees(index,
highlight_tree=highlight, pos_label=pos_label,
higher_is_better=self.higher_is_better)
return {}
@app.callback(
Output('decisiontrees-highlight-'+self.name, 'value'),
[Input("decisiontrees-graph-"+self.name, 'clickData')])
def update_highlight(clickdata):
highlight_tree = int(clickdata['points'][0]['text'].split('tree no ')[1].split(':')[0]) if clickdata is not None else None
if highlight_tree is not None:
return highlight_tree
raise PreventUpdate
class DecisionPathTableComponent(ExplainerComponent):
def __init__(self, explainer, title="Decision path table", name=None,
subtitle="Decision path through decision tree",
hide_title=False, hide_subtitle=False,
hide_index=False, hide_highlight=False,
hide_selector=False,
pos_label=None, index=None, highlight=None, description=None,
**kwargs):
"""Display a table of the decision path through a particular decision tree
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"Decision path table".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
subtitle (str): subtitle
hide_title (bool, optional): hide title, Defaults to False.
hide_subtitle (bool, optional): Hide subtitle. Defaults to False.
hide_index (bool, optional): Hide index selector.
Defaults to False.
hide_highlight (bool, optional): Hide tree index selector.
Defaults to False.
hide_selector (bool, optional): hide pos label selectors.
Defaults to False.
pos_label ({int, str}, optional): initial pos label.
Defaults to explainer.pos_label
index ({str, int}, optional): Initial index to display decision
path for. Defaults to None.
highlight (int, optional): Initial tree idx to display decision
path for. Defaults to None.
description (str, optional): Tooltip to display when hover over
component title. When None default text is shown.
"""
super().__init__(explainer, title, name)
self.index_name = 'decisionpath-table-index-'+self.name
self.highlight_name = 'decisionpath-table-highlight-'+self.name
self.selector = PosLabelSelector(explainer, name=self.name, pos_label=pos_label)
if self.description is None: self.description = """
Shows the path that an observation took down a specific decision tree.
"""
self.register_dependencies("decision_trees")
def layout(self):
return dbc.Card([
make_hideable(
dbc.CardHeader([
html.Div([
html.H3(self.title, id='decisionpath-table-title-'+self.name),
make_hideable(html.H6(self.subtitle, className='card-subtitle'), hide=self.hide_subtitle),
dbc.Tooltip(self.description, target='decisionpath-table-title-'+self.name),
]),
]), hide=self.hide_title),
dbc.CardBody([
dbc.Row([
make_hideable(
dbc.Col([
dbc.Label(f"{self.explainer.index_name}:", id='decisionpath-table-index-label-'+self.name),
dbc.Tooltip(f"Select {self.explainer.index_name} to display decision tree for",
target='decisionpath-table-index-label-'+self.name),
dcc.Dropdown(id='decisionpath-table-index-'+self.name,
options = [{'label': str(idx), 'value':idx}
for idx in self.explainer.idxs],
value=self.index)
], md=4), hide=self.hide_index),
make_hideable(
dbc.Col([
dbc.Label("Show tree:", id='decisionpath-table-tree-label-'+self.name),
dbc.Tooltip(f"Select decision tree to display decision tree path for",
target='decisionpath-table-tree-label-'+self.name),
dbc.Select(id='decisionpath-table-highlight-'+self.name,
options = [{'label': str(tree), 'value': tree}
for tree in range(self.explainer.no_of_trees)],
value=self.highlight)
], md=2), hide=self.hide_highlight),
make_hideable(
dbc.Col([self.selector.layout()
], md=2), hide=self.hide_selector)
]),
dbc.Row([
dbc.Col([
html.Div(id="decisionpath-table-"+self.name),
]),
]),
]),
])
def component_callbacks(self, app):
@app.callback(
Output("decisionpath-table-"+self.name, 'children'),
[Input('decisionpath-table-index-'+self.name, 'value'),
Input('decisionpath-table-highlight-'+self.name, 'value'),
Input('pos-label-'+self.name, 'value')],
)
def update_decisiontree_table(index, highlight, pos_label):
if index is not None and highlight is not None:
decisionpath_df = self.explainer.decisiontree_summary_df(
int(highlight), index, pos_label=pos_label)
return dbc.Table.from_dataframe(decisionpath_df)
raise PreventUpdate
class DecisionPathGraphComponent(ExplainerComponent):
def __init__(self, explainer, title="Decision path", name=None,
subtitle="Visualizing individual predictions process",
hide_title=False, hide_subtitle=False, hide_index=False,
hide_highlight=False, hide_button=False,
hide_selector=False,
pos_label=None, index=None, highlight=None, description=None,
**kwargs):
"""Display dtreeviz decision path
Args:
explainer (Explainer): explainer object constructed with either
ClassifierExplainer() or RegressionExplainer()
title (str, optional): Title of tab or page. Defaults to
"Decision path graph".
name (str, optional): unique name to add to Component elements.
If None then random uuid is generated to make sure
it's unique. Defaults to None.
subtitle (str): subtitle
hide_title (bool, optional): hide title
hide_subtitle (bool, optional): Hide subtitle. Defaults to False.
hide_index (bool, optional): hide index selector. Defaults to False.
hide_highlight (bool, optional): hide tree idx selector. Defaults to False.
hide_button (bool, optional): hide the button, Defaults to False.
hide_selector (bool, optional): hide pos label selectors. Defaults to False.
pos_label ({int, str}, optional): initial pos label.
Defaults to explainer.pos_label
index ({str, int}, optional): Initial index to display. Defaults to None.
highlight ([type], optional): Initial tree idx to display. Defaults to None.
description (str, optional): Tooltip to display when hover over
component title. When None default text is shown.
"""
super().__init__(explainer, title, name)
# if explainer.is_regression:
# raise ValueError("DecisionPathGraphComponent only available for classifiers for now!")
self.index_name = 'decisionpath-index-'+self.name
self.highlight_name = 'decisionpath-highlight-'+self.name
if self.description is None: self.description = """
Visualizes the path that an observation took down a specific decision tree,
by showing the entire decision tree and the path that a specific observation
took down this tree.
"""
self.selector = PosLabelSelector(explainer, name=self.name, pos_label=pos_label)
def layout(self):
return dbc.Card([
make_hideable(
dbc.CardHeader([
html.Div([
html.H3(self.title, id='decisionpath-title-'+self.name),
make_hideable(html.H6(self.subtitle, className='card-subtitle'), hide=self.hide_subtitle),
dbc.Tooltip(self.description, target='decisionpath-title-'+self.name),
]),
]), hide=self.hide_title),
dbc.CardBody([
dbc.Row([
make_hideable(
dbc.Col([
dbc.Label(f"{self.explainer.index_name}:", id='decisionpath-index-label-'+self.name),
dbc.Tooltip(f"Select {self.explainer.index_name} to display decision tree for",
target='decisionpath-index-label-'+self.name),
dcc.Dropdown(id='decisionpath-index-'+self.name,
options = [{'label': str(idx), 'value':idx}
for idx in self.explainer.idxs],
value=self.index)
], md=4), hide=self.hide_index),
make_hideable(
dbc.Col([
dbc.Label("Show tree:", id='decisionpath-tree-label-'+self.name),
dbc.Tooltip(f"Select decision tree to display decision tree for",
target='decisionpath-tree-label-'+self.name),
dbc.Select(id='decisionpath-highlight-'+self.name,
options = [{'label': str(tree), 'value': tree}
for tree in range(self.explainer.no_of_trees)],
value=self.highlight)
], md=2), hide=self.hide_highlight),
make_hideable(
dbc.Col([self.selector.layout()
], width=2), hide=self.hide_selector),
make_hideable(
dbc.Col([
dbc.Button("Generate Tree Graph", color="primary",
id='decisionpath-button-'+self.name),
dbc.Tooltip("Generate visualisation of decision tree. "
"Only works if graphviz is properly installed,"
" and may take a while for large trees.",
target='decisionpath-button-'+self.name)
], md=2, align="end"), hide=self.hide_button),
]),
dbc.Row([
dbc.Col([
dcc.Loading(id="loading-decisionpath-"+self.name,
children=html.Img(id="decisionpath-svg-"+self.name)),
]),
]),
]),
])
def component_callbacks(self, app):
@app.callback(
Output("decisionpath-svg-"+self.name, 'src'),
[Input('decisionpath-button-'+self.name, 'n_clicks')],
[State('decisionpath-index-'+self.name, 'value'),
State('decisionpath-highlight-'+self.name, 'value'),
State('pos-label-'+self.name, 'value')]
)
def update_tree_graph(n_clicks, index, highlight, pos_label):
if n_clicks is not None and index is not None and highlight is not None:
return self.explainer.decision_path_encoded(int(highlight), index)
raise PreventUpdate | AMLBID | /Explainer/dashboard_components/decisiontree_components.py | decisiontree_components.py |
from sklearn.svm import SVC
from xgboost import XGBClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.linear_model import LogisticRegression, SGDClassifier
from sklearn.metrics import accuracy_score, f1_score, recall_score, precision_score, roc_auc_score
from sklearn.ensemble import RandomForestClassifier, AdaBoostClassifier, GradientBoostingClassifier, ExtraTreesClassifier
def generate_imports_code(algorithm):
"""Generate all library import calls.
Parameters
----------
algorithm: string
name of the recommended learner
Returns
-------
imports: String
The Python code that imports all required library used in the current
optimized pipeline
"""
clf_import= {'AdaBoostClassifier': "from sklearn.ensemble import AdaBoostClassifier\nfrom sklearn.tree import DecisionTreeClassifier\n",
'RandomForestClassifier': "from sklearn.ensemble import RandomForestClassifier\n",
'SVC': "from sklearn.svm import SVC\n",
'ExtraTreesClassifier': "from sklearn.ensemble import ExtraTreesClassifier\n",
'GradientBoostingClassifier': "from sklearn.ensemble import GradientBoostingClassifier\n",
'DecisionTreeClassifier': "from sklearn.tree import DecisionTreeClassifier\n",
'LogisticRegression': "from sklearn.linear_model import LogisticRegression\n",
'SGDClassifier': "from sklearn.linear_model import SGDClassifier\n"
}
imports_basic="""import numpy as np\nimport pandas as pd\nfrom sklearn.metrics import classification_report\nfrom sklearn.model_selection import train_test_split\n"""
imports=imports_basic+clf_import[algorithm]
return imports
def generate_pipeline_code(pipeline,DS_path):
"""Generate recommended pipeline code.
Parameters
----------
pipeline: tuple
name and recommended pipeline configuration
Returns
-------
code: String
The Python code recommended pipeline
"""
code= """
# NOTE: Make sure that the target column is labeled 'class' in the data file
data = pd.read_csv('{}')
X = data.drop('class', axis=1)
y = data['class']
X_train, X_test, Y_train, Y_test = train_test_split(X, y, test_size=0.3, random_state=42)
model= {}
model.fit(X_train, Y_train)
Y_pred = model.predict(X_test)
score = model.score(X_test, Y_test)
print(classification_report(Y_test, Y_pred))
print(' Pipeline test accuracy: %.3f' % score)
""".format(DS_path, pipeline)
return code
def get_pipeline(algorithm,config):
algorithms = {'AdaBoostClassifier': AdaBoostClassifier(base_estimator=DecisionTreeClassifier()),
'RandomForestClassifier': RandomForestClassifier(),
'SVC': SVC(),
'ExtraTreesClassifier': ExtraTreesClassifier(),
'GradientBoostingClassifier': GradientBoostingClassifier(),
'DecisionTreeClassifier': DecisionTreeClassifier(),
'LogisticRegression': LogisticRegression(),
'SGDClassifier': SGDClassifier()}
model=algorithms[algorithm]
for k, v in config.items():
model.set_params(**{k: v})
#print(model)
return model
def generate_pipeline_file(algorithm,config,DS_path):
model_conf=get_pipeline(algorithm,config)
imports=generate_imports_code(algorithm)
code=generate_pipeline_code(model_conf,DS_path)
All=imports+code
filename = 'Recommended_config_implementation' +'.py'
# open the file to be written
fo = open(filename, 'w')
fo.write('%s' % All)
fo.close()
return All | AMLBID | /Explainer/dashboard_components/ConfGenerator.py | ConfGenerator.py |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.