prompt
stringlengths 19
1.03M
| completion
stringlengths 4
2.12k
| api
stringlengths 8
90
|
|---|---|---|
## 1. Introduction ##
import pandas as pd
happiness2015 = pd.read_csv("World_Happiness_2015.csv")
happiness2016 = pd.read_csv("World_Happiness_2016.csv")
happiness2017= pd.read_csv("World_Happiness_2017.csv")
happiness2015['Year'] = 2015
happiness2016['Year'] = 2016
happiness2017['Year'] = 2017
## 2. Combining Dataframes with the Concat Function ##
head_2015 = happiness2015[['Country','Happiness Score', 'Year']].head(3)
head_2016 = happiness2016[['Country','Happiness Score', 'Year']].head(3)
concat_axis0 = pd.concat([head_2015, head_2016], axis=0)
concat_axis1 = pd.concat([head_2015, head_2016], axis=1)
print(concat_axis0)
print(concat_axis1)
question1 = concat_axis0.shape[0]
print(question1)
question2 = concat_axis1.shape[0]
print(question2)
## 3. Combining Dataframes with the Concat Function Continued ##
head_2015 = happiness2015[['Year','Country','Happiness Score', 'Standard Error']].head(4)
head_2016 = happiness2016[['Country','Happiness Score', 'Year']].head(3)
concat_axis0 = pd.concat([head_2015, head_2016])
print(concat_axis0)
rows = concat_axis0.shape[0]
columns = concat_axis0.shape[1]
print(rows)
print(columns)
## 4. Combining Dataframes with Different Shapes Using the Concat Function ##
head_2015 = happiness2015[['Year','Country','Happiness Score', 'Standard Error']].head(4)
head_2016 = happiness2016[['Country','Happiness Score', 'Year']].head(3)
concat_update_index = pd.concat([head_2015, head_2016], ignore_index=True)
print(concat_update_index)
## 5. Joining Dataframes with the Merge Function ##
three_2015 = happiness2015[['Country','Happiness Rank','Year']].iloc[2:5]
three_2016 = happiness2016[['Country','Happiness Rank','Year']].iloc[2:5]
merged = pd.merge(left=three_2015, right= three_2016, on='Country')
print(merged)
## 6. Joining on Columns with the Merge Function ##
three_2015 = happiness2015[['Country','Happiness Rank','Year']].iloc[2:5]
three_2016 = happiness2016[['Country','Happiness Rank','Year']].iloc[2:5]
merged = pd.merge(left=three_2015, right=three_2016, on='Country')
merged_left = pd.merge(left=three_2015, right=three_2016, on='Country', how='left')
merged_left_updated = pd.merge(left=three_2016, right= three_2015, on='Country', how='left')
print(merged_left)
print(merged_left_updated)
## 7. Left Joins with the Merge Function ##
three_2015 = happiness2015[['Country','Happiness Rank','Year']].iloc[2:5]
three_2016 = happiness2016[['Country','Happiness Rank','Year']].iloc[2:5]
merged = pd.merge(left=three_2015, right=three_2016, how='left', on='Country')
merged_updated = pd.merge(left=three_2016, right=three_2015, how = 'left', on='Country')
merged_suffixes = pd.merge(left=three_2015, right=three_2016, how='left', on='Country', suffixes=('_2015','_2016'))
merged_updated_suffixes = pd.merge(left=three_2016, right=three_2015, on='Country', how='left', suffixes = ('_2016','_2015'))
print(merged_suffixes)
print(merged_updated_suffixes)
## 8. Join on Index with the Merge Function ##
import pandas as pd
four_2015 = happiness2015[['Country','Happiness Rank','Year']].iloc[2:6]
three_2016 = happiness2016[['Country','Happiness Rank','Year']].iloc[2:5]
merge_index = pd.merge(left = four_2015,right = three_2016, left_index = True, right_index = True, suffixes = ('_2015','_2016'))
rows = 4
columns = 6
merge_index_left = pd.merge(left=four_2015, right=three_2016, how='left', left_index=True, right_index=True, suffixes=('_2015','_2016'))
## 9. Challenge: Combine Data and Create a Visualization ##
happiness2017.rename(columns={'Happiness.Score': 'Happiness Score'}, inplace=True)
combined = | pd.concat([happiness2015, happiness2016, happiness2017]) | pandas.concat |
import wf_rdbms.utils
import pandas as pd
import logging
logger = logging.getLogger(__name__)
TYPES = {
'integer': {
'pandas_dtype': 'Int64',
'to_pandas_series': lambda x: pd.Series(x, dtype='Int64'),
'to_python_list': lambda x: wf_rdbms.utils.series_to_list(pd.Series(x, dtype='Int64'))
},
'float': {
'pandas_dtype': 'float',
'to_pandas_series': lambda x: pd.Series(x, dtype='float'),
'to_python_list': lambda x: wf_rdbms.utils.series_to_list(pd.Series(x, dtype='float'))
},
'string': {
'pandas_dtype': 'string',
'to_pandas_series': lambda x: | pd.Series(x, dtype='string') | pandas.Series |
import pandas as pd
def create_dataframe():
"""Create a sample Pandas dataframe used by the test functions.
Returns
-------
df : pandas.DataFrame
Pandas dataframe containing sample data.
"""
dti = pd.date_range("2018-01-01", periods=9, freq="H")
d = {'user': ['user_1','user_2','user_3','user_4','user_5','user_6','user_7','user_8','user_9'],
'group': ['group_1','group_1','group_1','group_2','group_2','group_2','group_3','group_3','group_3'],
'col_1': [1, 2, 3,4,5,6,7,8,9],
'col_2': [10, 11, 12, 13, 14, 15, 16, 17, 18]}
df = pd.DataFrame(data=d,index=dti)
return df
def create_categorical_dataframe():
"""Create a sample Pandas dataframe used by the test functions.
Returns
-------
df : pandas.DataFrame
Pandas dataframe containing sample data.
"""
dti = | pd.date_range("2018-01-01", periods=9, freq="H") | pandas.date_range |
# Modified from: https://www.kaggle.com/gauravs90/keras-bert-toxic-model-bert-fine-tuning-with-keras
from keras.callbacks import ModelCheckpoint
import keras as keras
from keras.layers import Input, concatenate
from keras_bert import load_trained_model_from_checkpoint, load_vocabulary
from keras_bert import Tokenizer
from keras_bert import AdamWarmup, calc_train_steps
from keras.callbacks import Callback, TensorBoard
from sklearn.metrics import confusion_matrix
from sklearn.preprocessing import OneHotEncoder
import pandas as pd
import os
import pickle
import numpy as np
import argparse
import time
TIME_START = time.time()
SEED = 2321598
RESULTS_FILE = 'results_fullcycle.csv'
for SEED in [SEED]:
ALL_SECTIONS = ['Kotimaa', 'Talous', 'Urheilu']
def get_arguments():
"""
Argument parser
"""
parser = argparse.ArgumentParser()
parser.add_argument("--neurons", action="store", dest="neurons", required=True, help="Number of neurons per layer." )
parser.add_argument("--bert_data_path", action="store", dest="bert_data_path", required=True, help="Path containing BERT data files.")
parser.add_argument("--features_file", action="store", dest="features_file", required=True, help="Path for the input data (pickle), similar as exampledata.pickle")
parser.add_argument("--use_sections", action='store', dest='use_sections', required=False, nargs='*', default=False, help="Use news article sections as additional input for the model.")
parser.add_argument("--epochs", action="store", dest='epochs', required=False, default=10, help="Number of training epochs")
parser.add_argument("--use_lemmatized_title", action="store_true", dest='use_lemmatized_title', required=False, help="Use titles in lemmatized format.")
parser.add_argument('--use_temporal_features', action='store_true', dest='use_temporal_features', required=False, help="Use publishing time as a feature")
parser.add_argument('--use_title_features', action='store', dest='use_title_features', required=False, choices=['binary', 'cont'], default=False)
parser.add_argument("--hidden_layers", action="store", dest='hidden_layers', required=False, default=1, help="Number of custom hidden layers.")
parser.add_argument("--calculate_response", action="store_true", required=False, default=False, help="Defines whether to calculate response variable or use the precalculated one on the input data.")
parser.add_argument("--no_premium", action="store_true", required=False, default=False, help="Ignore content behind a paywall.")
parser.add_argument("--prem_free_separate", action="store_true", required=False, default=False, help="Calculate response variable separately for free and premium content.")
parser.add_argument("--model_filename", action="store_true", required=False, default="model.h5", help="Filename where to save the trained model.")
return parser.parse_args()
def check_section_input(opts):
"""
To convert the section input.
If the flag is not used at all, boolean False is stored to it and nothing will be done
If the flag is used with no parameters, it will at first contain an empty list which is converted to True - all sections will be used
If the flag us used with an integer, we will use that many most usual sections
If the flag is used with string variables, they are assumed to be section names. We check whether the input is correct
"""
if isinstance(opts.use_sections, list):
if len(opts.use_sections) == 0:
opts.use_sections = True
return opts
elif len(opts.use_sections) == 1:
if opts.use_sections[0].isdigit():
opts.use_sections = int(opts.use_sections[0])
return opts
for section in opts.use_sections:
if section not in ALL_SECTIONS:
msg = "Input section '{}' does not exist. The sections available are: {}.".format(section, ALL_SECTIONS)
raise ValueError(msg)
return opts
def save_results(opts, res, results_file=None):
"""
To save results line by line to a file.
"""
if results_file is None:
results_file = 'neural_results.csv'
train_loss, train_acc, val_loss, val_acc, val_cmat, test_loss, test_acc, test_cmat = res
val_cmat = str([list(val_cmat[i]) for i in range(val_cmat.shape[1])])
test_cmat = str([list(test_cmat[i]) for i in range(test_cmat.shape[1])])
res_str = '{};{};{};{};{};{};{};{};{}\n'
res_str = res_str.format(opts, train_loss, train_acc, val_loss, val_acc, val_cmat, test_loss,
test_acc, test_cmat)
print("saving results {}".format(res_str))
with open(results_file, 'a') as fp:
fp.write(res_str)
def initialise_results_file(results_file=None):
"""Create results file and write headers to it if it does not already exist"""
if results_file is None:
results_file = 'neural_results.csv'
headers = 'opts;train_loss;train_acc;val_loss;val_acc;val_cmat;test_loss;test_acc;test_cmat\n'
if not os.path.isfile(results_file):
with open(results_file, 'w') as fp:
fp.write(headers)
def parse_title(opts):
if opts.use_lemmatized_title:
return 'orig_string_lemmatized'
else:
return 'orig_string'
def encode_time(arr, max_val=None):
if max_val is None:
max_val = arr.max()
max_val = len(np.arange(max_val))
ar1 = np.sin(2 * np.pi * arr / max_val)
ar2 = np.cos(2 * np.pi * arr / max_val)
return np.column_stack((ar1, ar2))
opts = get_arguments()
print('########################################')
print(opts)
print('########################################')
opts = check_section_input(opts)
# print("Section option is : {} and type {}".format(opts.use_sections, type(opts.use_sections)))
NUM_NEURONS = int(opts.neurons)
HIDDEN_LAYERS = int(opts.hidden_layers)
RESPONSE_VARIABLE='multiclass_response'
TITLE_VERSION = parse_title(opts)
TITLE_FEATURES_VARIABLE = 'title_features'
title_feature_names = ['n_words', 'n_chars_orig', 'n_chars_lemmas', 'mean_word_len', 'starts_with_identifier',
'n_sents_naive', 'n_sents_title', 'has_quotation', 'has_local_city_name', 'n_question', 'n_excl', 'n_dash',
'n_colon', 'n_comma', 'n_dot', 'n_semicolon', 'n_loc', 'n_ordinal', 'n_gpe', 'n_org', 'n_fac',
'n_norp', 'n_person', 'n_cardinal', 'n_date', 'n_work_of_art', 'n_quantity', 'n_product',
'n_money', 'n_time', 'n_event', 'n_percent', 'n_language', 'wordclass_N', 'wordclass_V',
'wordclass_C', 'wordclass_A', 'wordclass_Symb', 'wordclass_Punct', 'wordclass_Pron',
'wordclass_Num', 'wordclass_Interj', 'wordclass_Foreign', 'wordclass_Adv', 'wordclass_Adp']
if opts.use_title_features == 'binary':
TITLE_FEATURES_VARIABLE = 'title_features_binarised'
title_feature_names = ['n_words', 'n_dash', 'n_colon', 'n_comma', 'n_loc', 'n_ordinal', 'n_gpe', 'n_org',
'n_fac', 'n_norp', 'n_person', 'n_cardinal', 'n_date', 'n_work_of_art', 'n_quantity',
'n_product', 'n_money', 'n_time', 'n_event', 'n_percent', 'n_language', 'wordclass_N',
'wordclass_V', 'wordclass_C', 'wordclass_A', 'wordclass_Symb', 'wordclass_Punct',
'wordclass_Pron', 'wordclass_Num', 'wordclass_Interj', 'wordclass_Foreign',
'wordclass_Adv', 'wordclass_Adp', 'starts_with_identifier', 'has_quotation', 'has_local_city_name']
initialise_results_file(RESULTS_FILE)
SEQUENCE_LEN = 64
BATCH_SIZE = 128
EPOCHS = int(opts.epochs)
LR = 1e-4
# Language model related stuff
pretrained_path = opts.bert_data_path
ckpt_name = 'bert_model.ckpt'
config_path = os.path.join(pretrained_path, 'bert_config.json')
checkpoint_path = os.path.join(pretrained_path, ckpt_name)
vocab_path = os.path.join(pretrained_path, 'vocab.txt')
# Dataset location
FEATURES_FILE = opts.features_file
# Load dataset and combine news sections
with open(FEATURES_FILE, 'rb') as f:
features = pickle.load(f)
# Initialize tokenizer
token_dict = load_vocabulary(vocab_path)
tokenizer = Tokenizer(token_dict)
# collect blacklisted sections, method depends on which use_section option is used
BLACKLISTED_SECTIONS=['Urheilu']
if isinstance(opts.use_sections, list):
for section in ALL_SECTIONS:
if section not in opts.use_sections:
BLACKLISTED_SECTIONS.append(section)
elif isinstance(opts.use_sections, int) & (not isinstance(opts.use_sections, bool)):
sections = features.parsed_news_section
sections = list(sections.value_counts().head(opts.use_sections).index)
for section in ALL_SECTIONS:
if section not in sections:
BLACKLISTED_SECTIONS.append(section)
print("Blacklisted sections are {}".format(BLACKLISTED_SECTIONS))
# harvest the necessary data from the dataset
titles = []
title_targets = []
title_features = []
sections = []
days_of_week = []
hours_of_day = []
premiums = []
clicks = []
read_p = []
for key in features.index:
section = features.loc[key]['parsed_news_section']
if (section not in BLACKLISTED_SECTIONS) & (section in ALL_SECTIONS):
title = features[TITLE_FEATURES_VARIABLE][key][TITLE_VERSION]
premium = features.loc[key]['content_info']['access']
title_target = features[RESPONSE_VARIABLE][key]
if title is not None and title_target is not None:
titles.append(title)
title_targets.append(title_target)
tf = []
for feature_name in title_feature_names:
try:
val = float(features[TITLE_FEATURES_VARIABLE][key][feature_name])
tf.append(val)
except KeyError:
pass
title_features.append(tf)
sections.append(section)
premiums.append(premium)
days_of_week.append(features.loc[key]['temporal_features']['day_of_week'])
hours_of_day.append(features.loc[key]['temporal_features']['hour_of_day'])
clicks.append(features.loc[key]['clicks'])
read_p.append(features.loc[key]['read_percentage'])
# one hot encode sections
onehot_encoder = OneHotEncoder(sparse=False)
sections = np.asanyarray(sections).reshape(-1, 1)
sections = onehot_encoder.fit_transform(sections)
# fourier transform for temporal feats
days_of_week = encode_time(np.array(days_of_week), 7)
hours_of_day = encode_time(np.array(hours_of_day), 24)
def convert_data(titles, title_targets, title_features, sections, premiums, days_of_week, hours_of_day, clicks, read_p):
"""
Converts the data to the format expected by the neural network
"""
global tokenizer
indices, targets, orig_titles, title_features_filt, sections_filt, premiums_filt, \
days_of_week_filt, hours_of_day_filt, clicks_filt, read_p_filt = [], [], [], [], [], [], [], [], [], []
for i, (t, target, tf, sect, prem, day, hour, click, readp) in enumerate(zip(titles, title_targets, title_features, sections,
premiums, days_of_week, hours_of_day, clicks, read_p)):
if type(t)==str and target is not None:
if opts.no_premium:
if prem == 'free':
ids, segments = tokenizer.encode(t, max_len=SEQUENCE_LEN)
indices.append(ids)
targets.append(target)
orig_titles.append(t)
title_features_filt.append(tf)
sections_filt.append(sect)
premiums_filt.append(prem)
days_of_week_filt.append(day)
hours_of_day_filt.append(hour)
clicks_filt.append(click)
read_p_filt.append(readp)
else:
ids, segments = tokenizer.encode(t, max_len=SEQUENCE_LEN)
indices.append(ids)
targets.append(target)
orig_titles.append(t)
title_features_filt.append(tf)
sections_filt.append(sect)
premiums_filt.append(prem)
days_of_week_filt.append(day)
hours_of_day_filt.append(hour)
clicks_filt.append(click)
read_p_filt.append(readp)
indices = np.array(indices)
targets = np.array(targets)
orig_titles = np.array(orig_titles)
title_features_filt = np.array(title_features_filt)
sections_filt = np.array(sections_filt)
premiums_filt = np.array(premiums_filt)
days_of_week_filt = np.array(days_of_week_filt)
hours_of_day_filt = np.array(hours_of_day_filt)
clicks_filt = np.array(clicks_filt)
read_p_filt = np.array(read_p_filt)
return [indices, np.zeros_like(indices)], targets, orig_titles, title_features_filt, sections_filt, premiums_filt, days_of_week_filt, hours_of_day_filt, clicks_filt, read_p_filt
X, y, orig_titles, title_features, sections, premiums, days_of_week, hours_of_day, clicks, read_p = convert_data(titles, title_targets, title_features, sections, premiums, days_of_week, hours_of_day, clicks, read_p)
feats_to_include = list()
premiums = (premiums == 'premium').astype(int)
feats_to_include.append(premiums)
if opts.use_title_features:
if opts.use_title_features == 'cont':
feats_to_include.append(title_features)
else:
def onehot(a):
if len(np.unique(a)) > 2:
a = OneHotEncoder(sparse=False, categories='auto').fit_transform(a.reshape(-1, 1))
return a
tf = np.column_stack([onehot(title_features[:, i]) for i in range(title_features.shape[1])])
feats_to_include.append(tf)
if opts.use_sections:
dont = False
if isinstance(opts.use_sections, list):
if len(opts.use_sections) == 1:
dont = True
if not dont:
print("Adding sections to title_features")
section_names = [section for section in ALL_SECTIONS if section not in BLACKLISTED_SECTIONS]
print("There are {} sections, they are are {}".format(len(section_names), section_names))
feats_to_include.append(sections)
else:
print("Sections not used")
if opts.use_temporal_features:
print("Adding temporals to title_features")
feats_to_include.append(days_of_week)
feats_to_include.append(hours_of_day)
feats_to_include = np.column_stack(feats_to_include)
def generate_label(clicks, read_percentage, click_limits, read_limits):
click_mask = np.digitize(clicks, click_limits)
read_mask = np.digitize(read_percentage, read_limits)
y = np.zeros(len(clicks), dtype=int)
y[(click_mask == 1) | (read_mask == 1)] = -1
y[(click_mask == 2) & (read_mask == 0)] = 1
y[(click_mask == 0) & (read_mask == 2)] = 2
y[(click_mask == 2) & (read_mask == 2)] = 3
binary_y = np.zeros(len(clicks), dtype=int)
binary_y[y == -1] = -1
binary_y[y == 3] = 1
return y, binary_y
y_m = np.asarray(y)
# combine data
X.append(feats_to_include)
X.append(y_m)
X.append(orig_titles)
X.append(clicks)
X.append(read_p)
# shuffle
items = list(zip(X[0], X[1], X[2], X[3], X[4], X[5], X[6]))
np.random.seed(SEED)
np.random.shuffle(items)
X = list(zip(*items))
for i in range(len(X)):
X[i] = np.array(X[i])
# Split dataset into train and test
TEST_SPLIT_SIZE = 0.15
VAL_SPLIT_SIZE = 0.15
val_split_index = len(X[0]) - int(len(X[0]) * (VAL_SPLIT_SIZE + TEST_SPLIT_SIZE))
test_split_index = len(X[0]) - int(len(X[0]) * (TEST_SPLIT_SIZE))
X_train = [X[0][:val_split_index], X[1][:val_split_index], X[2][:val_split_index]]
y_train = X[3][:val_split_index]
X_val = [X[0][val_split_index:test_split_index], X[1][val_split_index:test_split_index], X[2][val_split_index:test_split_index]]
y_val = X[3][val_split_index:test_split_index]
X_test = [X[0][test_split_index:], X[1][test_split_index:], X[2][test_split_index:]]
y_test = X[3][test_split_index:]
orig_titles = X[4]
clicks = X[5]
read_p = X[6]
premiums = X[2][:, 0]
if opts.calculate_response:
if opts.prem_free_separate:
free = np.where(premiums == 0)[0]
prem = np.where(premiums == 1)[0]
else:
free = np.arange(len(premiums))
prem = []
y_m = np.repeat(np.nan, len(y))
if opts.response == 'clicks':
indsfree = [i for i in range(val_split_index) if i in free]
limsfree = list(np.quantile(clicks[indsfree], [0.45, 0.55]).round(0).astype(int))
y_m[free] = np.digitize(clicks[free], limsfree)
if len(prem) > 0:
indsprem = [i for i in range(val_split_index) if i in prem]
limsprem = list(np.quantile(clicks[indsprem], [0.45, 0.55]).round(0).astype(int))
y_m[prem] = np.digitize(clicks[prem], limsprem)
y_m[y_m == 1] = -1
y_m[y_m == 2] = 1
elif opts.response == 'read_percentage':
indsfree = [i for i in range(val_split_index) if i in free]
limsfree = list(np.quantile(read_p[indsfree], [0.45, 0.55]))
y_m[free] = np.digitize(read_p[free], limsfree)
if len(prem) > 0:
indsprem = [i for i in range(val_split_index) if i in prem]
limsprem = list(np.quantile(read_p[indsprem], [0.45, 0.55]))
y_m[prem] = np.digitize(read_p[prem], limsprem)
y_m[y_m == 1] = -1
y_m[y_m == 2] = 1
else:
indsfree = [i for i in range(val_split_index) if i in free]
click_limits = np.quantile(clicks[indsfree], [0.45, 0.55])
read_limits = np.quantile(read_p[indsfree], [0.45, 0.55])
y_m[free], _ = generate_label(clicks[free], read_p[free], click_limits, read_limits)
if len(prem) > 0:
indsprem = [i for i in range(val_split_index) if i in prem]
click_limits = np.quantile(clicks[indsprem], [0.45, 0.55])
read_limits = np.quantile(read_p[indsprem], [0.45, 0.55])
y_m[prem], _ = generate_label(clicks[prem], read_p[prem], click_limits, read_limits)
def remove_negs(xt, yt):
inds = np.where(yt > -1)[0]
for i in range(len(xt)):
xt[i] = xt[i][inds]
yt = yt[inds]
return xt, yt
y_train = y_m[:val_split_index]
X_train, y_train = remove_negs(X_train, y_train)
X, y = remove_negs(X, y_m)
test_split_index = len(y_train) + (len(y) - len(y_train)) // 2
val_split_index = len(y_train)
X_val = [X[0][val_split_index:test_split_index], X[1][val_split_index:test_split_index], X[2][val_split_index:test_split_index]]
y_val = y[val_split_index:test_split_index]
X_test = [X[0][test_split_index:], X[1][test_split_index:], X[2][test_split_index:]]
y_test = y[test_split_index:]
orig_titles = orig_titles[np.where(y_m > -1)[0]]
print("train: len {} \n {}".format(len(y_train), pd.Series(y_train).value_counts(normalize=True)))
print("val: len {} \n {}".format(len(y_val), pd.Series(y_val).value_counts(normalize=True)))
print("test: len {} \n {}".format(len(y_test), | pd.Series(y_test) | pandas.Series |
"""
Utilities for examining ABS NOM unit record
"""
import pickle
from pathlib import Path
import pandas as pd
import numpy as np
import matplotlib as mpl
from matplotlib import pyplot as plt
from IPython.display import display_html, display
from matplotlib.patches import Patch
from chris_utilities import adjust_chart
import file_paths
# the data storage
base_data_folder = file_paths.base_data_folder
abs_data_folder = file_paths.abs_data_folder
unit_record_folder = file_paths.unit_record_folder
individual_movements_folder = file_paths.individual_movements_folder
abs_nom_propensity = file_paths.abs_nom_propensity
abs_traveller_characteristics_folder = file_paths.abs_traveller_characteristics
grant_data_folder = file_paths.grant_data_folder
dict_data_folder = file_paths.dict_data_folder
program_data_folder = file_paths.program_data_folder
# local to current forecasting period folder
forecasting_data_folder = Path("data/forecasting")
forecasting_input_folder = forecasting_data_folder / "input"
### Utilities to read in raw ABS data:
def process_original_ABS_data(abs_original_data_folder, analysis_folder):
"""Process the SAS data, include removing previous preliminary parquet
and replace with final parquet, and add new preliminary parquet for latest quarter
Parameters
----------
abs_original_data_folder : Path ojbect
SAS data directory
analysis_folder : Path object
ABS Traveller characteristics folder pat
Returns
-------
None
Raises
------
ValueError
Check ABS NOM files must commence with p or f
This differentiates between preliminary and final NOM
Raise error to advice user that RTS file name convention not in place
"""
# TODO: read from the zip file rather than unzipped data
# variables to convert to ints or strings
ints_preliminary = [
"person_id",
"sex",
"country_of_birth",
"country_of_citizenship",
"country_of_stay",
"initial_erp_flag",
"final_erp_flag",
"duration_movement_sort_key",
"nom_direction",
"duration_in_australia_category",
"count_of_movements",
"initial_category_of_travel",
"age",
"status_flag",
"reason_for_journey",
"odb_time_code",
]
## For preliminary leave as floats: 'rky_val'
ints_final = [
"person_id",
"sex",
"country_of_birth",
"country_of_citizenship",
"country_of_stay",
"initial_erp_flag",
"final_erp_flag",
"duration_movement_sort_key",
"nom_direction",
"duration_in_australia_category",
"count_of_movements",
"initial_category_of_travel",
"age",
"status_flag",
"reason_for_journey",
"odb_time_code",
"net_erp_effect",
"nom_propensity",
]
# string vars are the same across preliminary and final
string_vars = [
"visa_group",
"visa_subclass",
"visa_applicant_type",
"visa_stream_code",
"stream_code_out",
"state",
"direction",
]
date_times = ["Duration_movement_date"]
### For unzipped sas data filess files
### Requires both options - older folders may not have the zipped version
# for abs_filepath in sorted(abs_original_data_folder.glob("*.sas7bdat")):
# print(abs_filepath.stem)
# df = pd.read_sas(abs_filepath, encoding="latin-1", format="sas7bdat").rename(
# columns=str.lower
# )
for abs_filepath in sorted(abs_original_data_folder.glob("*.sas7bdat")):
print(abs_filepath.stem)
df = pd.read_sas(abs_filepath, encoding="latin-1", format="sas7bdat").rename(
columns=str.lower
)
# for zip_filename in sorted(abs_original_data_folder.glob("*.zip")):
# zipped_file = zipfile.ZipFile(zip_filename, 'r')
# # There's only expected to be one file in each zip
# if len(zipped_file.namelist()) != 1:
# raise ValueError("Chris: zipped file has more than one file...recode!")
# sasfile = zipfile.open(zipped_file.namelist()[0])
# print(sasfile.stem)
# df = pd.read_sas(sasfile, encoding="latin-1", format="sas7bdat").rename(
# columns=str.lower
# )
### need to fix all abs_filepath below
# adjust datatypes and write out:
# string vars are the same across preliminary and final
for col in string_vars:
df[col] = df[col].astype("category")
# integer variables differ across final and preliminary data
if abs_filepath.stem[0] == "p": # preliminary NOM
for col in ints_preliminary:
df[col] = df[col].astype(int)
elif abs_filepath.stem[0] == "f": # final NOM
for col in ints_final:
df[col] = df[col].astype(int)
else:
raise ValueError(
"Chris - ABS NOM files must commence with p or f: {abs_filepath.stem} does not!"
)
write_outfile(df, abs_filepath, abs_original_data_folder, analysis_folder)
return None
def write_outfile(df, abs_filepath, abs_original_data_folder, analysis_folder):
"""
write out the processed ABS data to the ABS data folder and the analysis folder
Parameters
----------
df: pandas dataframe to write out
abs_filepath: Path object of original ABS file
abs_original_data_folder: Path object of path to ABS data folder
analysis_folder: Path to folder containing all NOM unit record parquet files
Returns
-------
None
"""
# ABS NOM filenames are of the type xxxx2018q1.sas...
# Want to extract the date compenent: 2018q1
date_start = abs_filepath.stem.find("2")
if date_start != -1: # if a '2' is found
filename_date = abs_filepath.stem[date_start:]
## append '_p' if it's a preliminary file
if abs_filepath.stem[0] == "p":
filename_date = filename_date + "_p"
else:
raise ValueError(
f"Chris - filename {abs_filepath.stem} does not appear to have a 20XXqY date in it"
)
filename = "traveller_characteristics" + filename_date + ".parquet"
# Write to original ABS folder:
# to keep as history for comparison with updated preliminary/final files
df.to_parquet(abs_original_data_folder / filename)
# Write to folder for analysis
df.to_parquet(analysis_folder / filename)
# if a final file replaces a preliminary file - delete it from the analysis file
if abs_filepath.stem[0] == "f":
preliminary_filename = (
"traveller_characteristics" + filename_date + "_p" + ".parquet"
)
preliminary_path = analysis_folder / preliminary_filename
if preliminary_path.exists():
preliminary_path.unlink()
return None
def get_visa_code_descriptions(vsc_list):
"""
get visa code descriptions
parameters
----------
vsc_list: list
visa suc codes as strings
returns
-------
a dictionary matching visa subcode to description
"""
with open(dict_data_folder / "dict_visa_code_descriptions.pickle", "rb") as pickle_file:
dict_visa_code_descriptions = pickle.load(pickle_file)
for vsc in vsc_list:
print(dict_visa_code_descriptions[vsc])
return dict_visa_code_descriptions
def get_monthly(
df, net_erp_effect, group_by=("Duration_movement_date", "Visa_subclass")
):
"""
Aggregate unit record NOM data to monthly by visa subclass
"""
summary = (
df[df.net_erp_effect == net_erp_effect]
.groupby(group_by)
.net_erp_effect.sum()
.unstack()
)
return summary.resample("M").sum()
def read_single_NOM_file(data_folder, file_name, field_list=None):
if field_list is None:
df = pd.read_parquet(data_folder / file_name)
else:
df = pd.read_parquet(data_folder / file_name, columns=field_list)
return df
def get_NOM_monthly_old(net_erp_effect, data_folder=Path("parquet")):
"""
A generator for returning NOM data selected for arrivals or departures
Parameters
----------
net_erp_effect: contribution to NOM: 1 = arrivals, -1 = departure
data_folder: a Path object to the folder containing ABS NOM unit record data
Yields:
-------
NOM_effect: a dataframe selected on net_erp_effect
"""
assert (net_erp_effect == 1) | (net_erp_effect == -1)
for p in sorted(data_folder.glob("*.parq")):
print(p.stem)
df = pd.read_parquet(p)
monthly_nom_outcomes = get_monthly(df, net_erp_effect)
yield monthly_nom_outcomes
def get_visa_groups_old(visa_groups, df_nom):
for group, idx in visa_groups.items():
df = df_nom[idx]
if group not in ["citizens", "student"]: # don't aggregate if in list:
if len(df.columns) > 1:
df = df.sum(axis=1)
df.name = group
if group == "student":
df.columns = [
s.lower().replace(" ", "_") for s in df.columns.droplevel(level=0)
]
# columns to breakout
idx_break_out = ["572", "573", "570"]
idx_break_outnames = ["higher_ed", "vet", "elicos", "student_other"]
df = pd.concat(
[df[idx_break_out], df.drop(columns=idx_break_out).sum(axis=1)], axis=1
)
df.columns = idx_break_outnames
if group == "citizens":
df.columns = [
s.lower().replace(" ", "_") for s in df.columns.droplevel(level=1)
]
yield df
def get_NOM(data_folder, abs_visa_group, nom_fields, abs_visagroup_exists=False):
"""
A generator to return unit records in an ABS visa group
Parameters:
-----------
data_folder: string, path object (pathlib.Path)
assumes contains parquet files
vsc: list
list of visa sub groups
nom_fields: list
list of nom fields to be extracts from ABS unit record file
"""
# abs_visa_group_current = ['AUST', 'NZLA', # Australian citizen, NZ citizen
# 'PSKL', 'PFAM', 'POTH', # skill, family, other
# 'TSKL', 'TSTD', 'TWRK', 'TOTH', 'TVIS' #still, student, WHM, other, visitor
# ]
# if not abs_visa_group in abs_visa_group_current:
# raise ValueError(f'Chris: {abs_visa_group} not legitimate ABS visa group.')
if not isinstance(nom_fields, (list, tuple)):
raise ValueError(
"Chris: get_NOM expects {nom_fields} to be a list of fields to extract."
)
for p in sorted(data_folder.glob("*.parquet")):
# Only loop over post 2011Q3 files
if abs_visagroup_exists:
if "ROADS" in p.stem:
continue
print(p.stem)
df = pd.read_parquet(p, columns=nom_fields)
yield df[(df.net_erp_effect != 0) & (df.visa_group == abs_visa_group)]
def append_nom_columns(df):
"""
Append each visa with a NOM column
Parameters
----------
df: data frame
the dataframe has hierarchical columns where:
level[0] has [arrival, departure]
level[1] has [visagroup, VSC, VSC etc]
"""
# set visa subclasses to level 0 & arrival, departure at levet 1)
df.columns = df.columns.swaplevel()
df = df.sort_index(axis="columns")
for col in df.columns.levels[0]:
df[(col, "nom")] = df[(col, "arrival")] - df[(col, "departure")]
df.columns = df.columns.swaplevel()
df = df.sort_index(axis="columns")
return df
def make_unique_movement_files(characteristcis_folder=abs_traveller_characteristics_folder, nom_final=True):
nom_fields = [
"person_id",
"duration_movement_date",
"visa_subclass",
"net_erp_effect",
]
# establish the generators
get_file_paths = gen_nom_files(
characteristcis_folder,
abs_visagroup_exists=False,
nom_final=nom_final)
df_get_fields = gen_nom_fields(get_file_paths, nom_fields)
df_visa_group = gen_get_visa_group(df_get_fields, vsc_list=None)
# build the NOM dataframe
df = (pd.concat(df_visa_group, axis="index", ignore_index=True, sort=False)
.rename({"duration_movement_date": "date"}, axis="columns")
.sort_values(["date", "person_id"])
)
if nom_final:
file_name = "NOM unique movement - final.parquet"
else:
file_name = "NOM unique movement - preliminary.parquet"
df.to_parquet(individual_movements_folder / file_name)
return df
# Dictionary utilities
def get_vsc_reference(file_path=None):
"""
Return a dataframe containing definitions and groupings for visa subclasses
The reference definitions and groupings is the sql table 'REF_VISA_SUBCLASS'
It is maintained by the visa stats team.
Parameters:
-----------
file_path: Path or str object
filepath to parquet file
Returns:
-------
dataframe
"""
if file_path == None:
file_path = dict_data_folder / "REF_VISA_SUBCLASS.parquet"
reference_visa_dict = (
pd.read_parquet(file_path)
.rename(columns=str.lower)
.rename(columns=lambda x: x.replace(" ", "_"))
)
return reference_visa_dict
def get_ABS_visa_grouping(file_path=None):
"""
Return a dataframe with ABS visa groupings (in cat no. 3412) by subclass
See ABS Migration unit for updated copies of excel file
Parameters:
-----------
file_path: None or Path object to 'ABS - Visacode3412mapping.xlsx'
Returns:
-------
dataframe
"""
if file_path is None:
file_path = dict_data_folder / "ABS - Visacode3412mapping.xlsx"
abs_3412 = (
| pd.read_excel(file_path) | pandas.read_excel |
import os
from mtsv.scripts.mtsv_analyze import *
import pytest
import datetime
import tables
import pandas as pd
@pytest.fixture(scope="function")
def existing_empty_datastore(tmpdir_factory):
fn = tmpdir_factory.mktemp("datastores").join("empty_datastore.h5")
store = pd.HDFStore(fn)
store.close()
return fn
@pytest.fixture(scope="function")
def existing_datastore(tmpdir_factory):
fn = tmpdir_factory.mktemp("datastores").join("datastore.h5")
ds = DataStore(fn)
ds.add_dataset(
"/datasets/t1", | pd.DataFrame([1,2,3,4]) | pandas.DataFrame |
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from tqdm import tqdm
from IPython.core.display import HTML
from fbprophet import Prophet
from fbprophet.plot import plot_plotly
import plotly.offline as py
import plotly.graph_objs as go
import plotly.express as px
class SalesForecaster:
"""This class creates 'easy to handle' forecaster objects
It will gather all the required variables to make the code more readable
- sales_clusters_df (pandas dataframe): The original sales dataframe
The columns are :
- product_code : string values such as CLA0 (CLA is the client and 0 is the product number)
- date : datetime64 (ns) the date of the sale such as pd.to_datetime("2018-01-02") : YYYY-MM-DD
- quantity : int64 an integer value: the number of products for this sale
- cluster : int64 an integer value The cluster the product is part of
- test_date (string : "2019-03-01" : YYYY-MM-DD): the training data is automatically all sales prior to this date
- max_waiting_time (string such as '7 days') : The maximum time a client is willing to wait :
required for grouping orders into batches)
- calendar_length (string such as '7 days'): The calendar length you want to zoom in
"""
def __init__(self,
sales_clusters_df,
test_date,
max_waiting_time,
detailed_view=False,
calendar_length='7 days'
):
self.sales_clusters_df = sales_clusters_df
self.test_date = test_date
self.max_waiting_time = max_waiting_time
self.detailed_view = detailed_view
self.calendar_length = calendar_length
self.optimal_batches = []
self.predicted_batches = []
self.predictions = []
def get_predicted_batches(self):
"""This function takes the original sales df,
computes the dates and quantities models at a product level using the test_date to split the dataset
into a training dataset and a testing dataset,
generates the predicted sales,
computes the associated "predicted" batches using the max waiting time value,
computes the optimal batches using the actual data using the max waiting time value,
outputs the optimal batches df and the predicted batches df,
and 2 graphs to visualize it:
- Input:
All the inputs are encapsulated in the SalesForecaster instance:
- sales_clusters_df
- test_date
- max_waiting_time
- calendar_length
- Output:
- Main graph with optimal batches vs predicted batches for the test data
- The same graph zoomed in the week following the test date
- 1 optimal batches df
- 1 predicted batches df
"""
clusters_list = self.sales_clusters_df['Cluster'].unique()
optimal_batches = []
predicted_batches = []
predictions = []
for cluster in clusters_list:
local_optimal_batches, local_predicted_batches, local_predictions = self.\
get_cluster_level_predicted_batches(cluster)
local_optimal_batches['Cluster'] = cluster
local_predicted_batches['Cluster'] = cluster
optimal_batches.append(local_optimal_batches)
predicted_batches.append(local_predicted_batches)
predictions.append(local_predictions)
optimal_batches = pd.concat(optimal_batches)
optimal_batches.reset_index(drop=True,
inplace=True)
optimal_batches['batch_date'] = optimal_batches.batch_date.str.split(' ').apply(lambda x: x[0])
predicted_batches = pd.concat(predicted_batches)
predicted_batches.reset_index(drop=True,
inplace=True)
predicted_batches['batch_date'] = predicted_batches.batch_date.str.split(' ').apply(lambda x: x[0])
predictions = pd.concat(predictions)
predictions.reset_index(drop=True,
inplace=True)
dark_map = px.colors.qualitative.Dark2
pastel_map = px.colors.qualitative.Pastel2
fig = go.Figure()
for (cluster, dark_color, pastel_color) in zip(clusters_list, dark_map, pastel_map):
local_optimal = optimal_batches[optimal_batches['Cluster'] == cluster]
local_predicted = predicted_batches[predicted_batches['Cluster'] == cluster]
fig.add_trace(go.Bar(x=pd.to_datetime(local_optimal[local_optimal['batch_date'] > self.test_date] \
['batch_date']) - pd.Timedelta('12 hours'),
y=local_optimal[local_optimal['batch_date'] > self.test_date] \
['quantities'],
name='Cluster #{}\nOptimized batches - actual values'.format(cluster),
width=1e3 * | pd.Timedelta('6 hours') | pandas.Timedelta |
import collections
import fnmatch
import os
from typing import Union
import tarfile
import pandas as pd
import numpy as np
from pandas.core.dtypes.common import is_string_dtype, is_numeric_dtype
from hydrodataset.data.data_base import DataSourceBase
from hydrodataset.data.stat import cal_fdc
from hydrodataset.utils import hydro_utils
from hydrodataset.utils.hydro_utils import download_one_zip, unzip_nested_zip
CAMELS_NO_DATASET_ERROR_LOG = (
"We cannot read this dataset now. Please check if you choose the correct dataset:\n"
' ["AUS", "BR", "CA", "CL", "GB", "US", "YR"]'
)
def time_intersect_dynamic_data(obs: np.array, date: np.array, t_range: list):
"""
chose data from obs in the t_range
Parameters
----------
obs
a np array
date
all periods for obs
t_range
the time range we need, such as ["1990-01-01","2000-01-01"]
Returns
-------
np.array
the chosen data
"""
t_lst = hydro_utils.t_range_days(t_range)
nt = t_lst.shape[0]
if len(obs) != nt:
out = np.full([nt], np.nan)
[c, ind1, ind2] = np.intersect1d(date, t_lst, return_indices=True)
out[ind2] = obs[ind1]
else:
out = obs
return out
class Camels(DataSourceBase):
def __init__(self, data_path, download=False, region: str = "US"):
"""
Initialization for CAMELS series dataset
Parameters
----------
data_path
where we put the dataset
download
if true, download
region
the default is CAMELS(-US), since it's the first CAMELS dataset.
Others now include: AUS, BR, CL, GB, YR
"""
super().__init__(data_path)
region_lst = ["AUS", "BR", "CA", "CE", "CL", "GB", "US", "YR"]
assert region in region_lst
self.region = region
self.data_source_description = self.set_data_source_describe()
if download:
self.download_data_source()
self.camels_sites = self.read_site_info()
def get_name(self):
return "CAMELS_" + self.region
def set_data_source_describe(self) -> collections.OrderedDict:
"""
Introduce the files in the dataset and list their location in the file system
Returns
-------
collections.OrderedDict
the description for a CAMELS dataset
"""
camels_db = self.data_source_dir
if self.region == "US":
# shp file of basins
camels_shp_file = os.path.join(
camels_db, "basin_set_full_res", "HCDN_nhru_final_671.shp"
)
# config of flow data
flow_dir = os.path.join(
camels_db,
"basin_timeseries_v1p2_metForcing_obsFlow",
"basin_dataset_public_v1p2",
"usgs_streamflow",
)
# forcing
forcing_dir = os.path.join(
camels_db,
"basin_timeseries_v1p2_metForcing_obsFlow",
"basin_dataset_public_v1p2",
"basin_mean_forcing",
)
forcing_types = ["daymet", "maurer", "nldas"]
# attr
attr_dir = os.path.join(
camels_db, "camels_attributes_v2.0", "camels_attributes_v2.0"
)
gauge_id_file = os.path.join(attr_dir, "camels_name.txt")
attr_key_lst = ["topo", "clim", "hydro", "vege", "soil", "geol"]
download_url_lst = [
"https://ral.ucar.edu/sites/default/files/public/product-tool/camels-catchment-attributes-and-meteorology-for-large-sample-studies-dataset-downloads/camels_attributes_v2.0.zip",
"https://ral.ucar.edu/sites/default/files/public/product-tool/camels-catchment-attributes-and-meteorology-for-large-sample-studies-dataset-downloads/basin_set_full_res.zip",
"https://ral.ucar.edu/sites/default/files/public/product-tool/camels-catchment-attributes-and-meteorology-for-large-sample-studies-dataset-downloads/basin_timeseries_v1p2_metForcing_obsFlow.zip",
]
return collections.OrderedDict(
CAMELS_DIR=camels_db,
CAMELS_FLOW_DIR=flow_dir,
CAMELS_FORCING_DIR=forcing_dir,
CAMELS_FORCING_TYPE=forcing_types,
CAMELS_ATTR_DIR=attr_dir,
CAMELS_ATTR_KEY_LST=attr_key_lst,
CAMELS_GAUGE_FILE=gauge_id_file,
CAMELS_BASINS_SHP_FILE=camels_shp_file,
CAMELS_DOWNLOAD_URL_LST=download_url_lst,
)
elif self.region == "AUS":
# id and name
gauge_id_file = os.path.join(
camels_db,
"01_id_name_metadata",
"01_id_name_metadata",
"id_name_metadata.csv",
)
# shp file of basins
camels_shp_file = os.path.join(
camels_db,
"02_location_boundary_area",
"02_location_boundary_area",
"shp",
"CAMELS_AUS_BasinOutlets_adopted.shp",
)
# config of flow data
flow_dir = os.path.join(camels_db, "03_streamflow", "03_streamflow")
# attr
attr_dir = os.path.join(camels_db, "04_attributes", "04_attributes")
# forcing
forcing_dir = os.path.join(
camels_db, "05_hydrometeorology", "05_hydrometeorology"
)
return collections.OrderedDict(
CAMELS_DIR=camels_db,
CAMELS_FLOW_DIR=flow_dir,
CAMELS_FORCING_DIR=forcing_dir,
CAMELS_ATTR_DIR=attr_dir,
CAMELS_GAUGE_FILE=gauge_id_file,
CAMELS_BASINS_SHP_FILE=camels_shp_file,
)
elif self.region == "BR":
# attr
attr_dir = os.path.join(
camels_db, "01_CAMELS_BR_attributes", "01_CAMELS_BR_attributes"
)
# we don't need the location attr file
attr_key_lst = [
"climate",
"geology",
"human_intervention",
"hydrology",
"land_cover",
"quality_check",
"soil",
"topography",
]
# id and name, there are two types stations in CAMELS_BR, and we only chose the 897-stations version
gauge_id_file = os.path.join(attr_dir, "camels_br_topography.txt")
# shp file of basins
camels_shp_file = os.path.join(
camels_db,
"14_CAMELS_BR_catchment_boundaries",
"14_CAMELS_BR_catchment_boundaries",
"camels_br_catchments.shp",
)
# config of flow data
flow_dir_m3s = os.path.join(
camels_db, "02_CAMELS_BR_streamflow_m3s", "02_CAMELS_BR_streamflow_m3s"
)
flow_dir_mm_selected_catchments = os.path.join(
camels_db,
"03_CAMELS_BR_streamflow_mm_selected_catchments",
"03_CAMELS_BR_streamflow_mm_selected_catchments",
)
flow_dir_simulated = os.path.join(
camels_db,
"04_CAMELS_BR_streamflow_simulated",
"04_CAMELS_BR_streamflow_simulated",
)
# forcing
forcing_dir_precipitation_chirps = os.path.join(
camels_db,
"05_CAMELS_BR_precipitation_chirps",
"05_CAMELS_BR_precipitation_chirps",
)
forcing_dir_precipitation_mswep = os.path.join(
camels_db,
"06_CAMELS_BR_precipitation_mswep",
"06_CAMELS_BR_precipitation_mswep",
)
forcing_dir_precipitation_cpc = os.path.join(
camels_db,
"07_CAMELS_BR_precipitation_cpc",
"07_CAMELS_BR_precipitation_cpc",
)
forcing_dir_evapotransp_gleam = os.path.join(
camels_db,
"08_CAMELS_BR_evapotransp_gleam",
"08_CAMELS_BR_evapotransp_gleam",
)
forcing_dir_evapotransp_mgb = os.path.join(
camels_db,
"09_CAMELS_BR_evapotransp_mgb",
"09_CAMELS_BR_evapotransp_mgb",
)
forcing_dir_potential_evapotransp_gleam = os.path.join(
camels_db,
"10_CAMELS_BR_potential_evapotransp_gleam",
"10_CAMELS_BR_potential_evapotransp_gleam",
)
forcing_dir_temperature_min_cpc = os.path.join(
camels_db,
"11_CAMELS_BR_temperature_min_cpc",
"11_CAMELS_BR_temperature_min_cpc",
)
forcing_dir_temperature_mean_cpc = os.path.join(
camels_db,
"12_CAMELS_BR_temperature_mean_cpc",
"12_CAMELS_BR_temperature_mean_cpc",
)
forcing_dir_temperature_max_cpc = os.path.join(
camels_db,
"13_CAMELS_BR_temperature_max_cpc",
"13_CAMELS_BR_temperature_max_cpc",
)
return collections.OrderedDict(
CAMELS_DIR=camels_db,
CAMELS_FLOW_DIR=[
flow_dir_m3s,
flow_dir_mm_selected_catchments,
flow_dir_simulated,
],
CAMELS_FORCING_DIR=[
forcing_dir_precipitation_chirps,
forcing_dir_precipitation_mswep,
forcing_dir_precipitation_cpc,
forcing_dir_evapotransp_gleam,
forcing_dir_evapotransp_mgb,
forcing_dir_potential_evapotransp_gleam,
forcing_dir_temperature_min_cpc,
forcing_dir_temperature_mean_cpc,
forcing_dir_temperature_max_cpc,
],
CAMELS_ATTR_DIR=attr_dir,
CAMELS_ATTR_KEY_LST=attr_key_lst,
CAMELS_GAUGE_FILE=gauge_id_file,
CAMELS_BASINS_SHP_FILE=camels_shp_file,
)
elif self.region == "CL":
# attr
attr_dir = os.path.join(camels_db, "1_CAMELScl_attributes")
attr_file = os.path.join(attr_dir, "1_CAMELScl_attributes.txt")
# shp file of basins
camels_shp_file = os.path.join(
camels_db,
"CAMELScl_catchment_boundaries",
"catchments_camels_cl_v1.3.shp",
)
# config of flow data
flow_dir_m3s = os.path.join(camels_db, "2_CAMELScl_streamflow_m3s")
flow_dir_mm = os.path.join(camels_db, "3_CAMELScl_streamflow_mm")
# forcing
forcing_dir_precip_cr2met = os.path.join(
camels_db, "4_CAMELScl_precip_cr2met"
)
forcing_dir_precip_chirps = os.path.join(
camels_db, "5_CAMELScl_precip_chirps"
)
forcing_dir_precip_mswep = os.path.join(
camels_db, "6_CAMELScl_precip_mswep"
)
forcing_dir_precip_tmpa = os.path.join(camels_db, "7_CAMELScl_precip_tmpa")
forcing_dir_tmin_cr2met = os.path.join(camels_db, "8_CAMELScl_tmin_cr2met")
forcing_dir_tmax_cr2met = os.path.join(camels_db, "9_CAMELScl_tmax_cr2met")
forcing_dir_tmean_cr2met = os.path.join(
camels_db, "10_CAMELScl_tmean_cr2met"
)
forcing_dir_pet_8d_modis = os.path.join(
camels_db, "11_CAMELScl_pet_8d_modis"
)
forcing_dir_pet_hargreaves = os.path.join(
camels_db,
"12_CAMELScl_pet_hargreaves",
)
forcing_dir_swe = os.path.join(camels_db, "13_CAMELScl_swe")
return collections.OrderedDict(
CAMELS_DIR=camels_db,
CAMELS_FLOW_DIR=[flow_dir_m3s, flow_dir_mm],
CAMELS_FORCING_DIR=[
forcing_dir_precip_cr2met,
forcing_dir_precip_chirps,
forcing_dir_precip_mswep,
forcing_dir_precip_tmpa,
forcing_dir_tmin_cr2met,
forcing_dir_tmax_cr2met,
forcing_dir_tmean_cr2met,
forcing_dir_pet_8d_modis,
forcing_dir_pet_hargreaves,
forcing_dir_swe,
],
CAMELS_ATTR_DIR=attr_dir,
CAMELS_GAUGE_FILE=attr_file,
CAMELS_BASINS_SHP_FILE=camels_shp_file,
)
elif self.region == "GB":
# shp file of basins
camels_shp_file = os.path.join(
camels_db,
"8344e4f3-d2ea-44f5-8afa-86d2987543a9",
"8344e4f3-d2ea-44f5-8afa-86d2987543a9",
"data",
"CAMELS_GB_catchment_boundaries",
"CAMELS_GB_catchment_boundaries.shp",
)
# flow and forcing data are in a same file
flow_dir = os.path.join(
camels_db,
"8344e4f3-d2ea-44f5-8afa-86d2987543a9",
"8344e4f3-d2ea-44f5-8afa-86d2987543a9",
"data",
"timeseries",
)
forcing_dir = flow_dir
# attr
attr_dir = os.path.join(
camels_db,
"8344e4f3-d2ea-44f5-8afa-86d2987543a9",
"8344e4f3-d2ea-44f5-8afa-86d2987543a9",
"data",
)
gauge_id_file = os.path.join(
attr_dir, "CAMELS_GB_hydrometry_attributes.csv"
)
attr_key_lst = [
"climatic",
"humaninfluence",
"hydrogeology",
"hydrologic",
"hydrometry",
"landcover",
"soil",
"topographic",
]
return collections.OrderedDict(
CAMELS_DIR=camels_db,
CAMELS_FLOW_DIR=flow_dir,
CAMELS_FORCING_DIR=forcing_dir,
CAMELS_ATTR_DIR=attr_dir,
CAMELS_ATTR_KEY_LST=attr_key_lst,
CAMELS_GAUGE_FILE=gauge_id_file,
CAMELS_BASINS_SHP_FILE=camels_shp_file,
)
elif self.region == "YR":
# shp files of basins
camels_shp_files_dir = os.path.join(
camels_db, "9_Normal_Camels_YR", "Normal_Camels_YR_basin_boundary"
)
# attr, flow and forcing data are all in the same dir. each basin has one dir.
flow_dir = os.path.join(
camels_db, "9_Normal_Camels_YR", "1_Normal_Camels_YR_basin_data"
)
forcing_dir = flow_dir
attr_dir = flow_dir
# no gauge id file for CAMELS_YR; natural_watersheds.txt showed unregulated basins in CAMELS_YR
gauge_id_file = os.path.join(
camels_db, "9_Normal_Camels_YR", "natural_watersheds.txt"
)
return collections.OrderedDict(
CAMELS_DIR=camels_db,
CAMELS_FLOW_DIR=flow_dir,
CAMELS_FORCING_DIR=forcing_dir,
CAMELS_ATTR_DIR=attr_dir,
CAMELS_GAUGE_FILE=gauge_id_file,
CAMELS_BASINS_SHP_DIR=camels_shp_files_dir,
)
elif self.region == "CA":
# shp file of basins
camels_shp_files_dir = os.path.join(camels_db, "CANOPEX_BOUNDARIES")
# config of flow data
flow_dir = os.path.join(
camels_db, "CANOPEX_NRCAN_ASCII", "CANOPEX_NRCAN_ASCII"
)
forcing_dir = flow_dir
# There is no attr data in CANOPEX, hence we use attr from HYSET -- https://osf.io/7fn4c/
attr_dir = camels_db
gauge_id_file = os.path.join(camels_db, "STATION_METADATA.xlsx")
return collections.OrderedDict(
CAMELS_DIR=camels_db,
CAMELS_FLOW_DIR=flow_dir,
CAMELS_FORCING_DIR=forcing_dir,
CAMELS_ATTR_DIR=attr_dir,
CAMELS_GAUGE_FILE=gauge_id_file,
CAMELS_BASINS_SHP_DIR=camels_shp_files_dir,
)
elif self.region == "CE":
# We use A_basins_total_upstrm
# shp file of basins
camels_shp_file = os.path.join(
camels_db,
"2_LamaH-CE_daily",
"A_basins_total_upstrm",
"3_shapefiles",
"Basins_A.shp",
)
# config of flow data
flow_dir = os.path.join(
camels_db, "2_LamaH-CE_daily", "D_gauges", "2_timeseries", "daily"
)
forcing_dir = os.path.join(
camels_db,
"2_LamaH-CE_daily",
"A_basins_total_upstrm",
"2_timeseries",
"daily",
)
attr_dir = os.path.join(
camels_db, "2_LamaH-CE_daily", "A_basins_total_upstrm", "1_attributes"
)
gauge_id_file = os.path.join(
camels_db,
"2_LamaH-CE_daily",
"D_gauges",
"1_attributes",
"Gauge_attributes.csv",
)
return collections.OrderedDict(
CAMELS_DIR=camels_db,
CAMELS_FLOW_DIR=flow_dir,
CAMELS_FORCING_DIR=forcing_dir,
CAMELS_ATTR_DIR=attr_dir,
CAMELS_GAUGE_FILE=gauge_id_file,
CAMELS_BASINS_SHP_FILE=camels_shp_file,
)
else:
raise NotImplementedError(CAMELS_NO_DATASET_ERROR_LOG)
def download_data_source(self) -> None:
"""
Download CAMELS dataset.
Now we only support CAMELS-US's downloading.
For others, please download it manually and put all files of a CAMELS dataset in one directory.
For example, all files of CAMELS_AUS should be put in "camels_aus" directory
Returns
-------
None
"""
camels_config = self.data_source_description
if self.region == "US":
if not os.path.isdir(camels_config["CAMELS_DIR"]):
os.makedirs(camels_config["CAMELS_DIR"])
[
download_one_zip(attr_url, camels_config["CAMELS_DIR"])
for attr_url in camels_config["CAMELS_DOWNLOAD_URL_LST"]
if not os.path.isfile(
os.path.join(camels_config["CAMELS_DIR"], attr_url.split("/")[-1])
)
]
print("The CAMELS_US data have been downloaded!")
print(
"Please download it manually and put all files of a CAMELS dataset in the CAMELS_DIR directory."
)
print("We unzip all files now.")
if self.region == "CE":
# We only use CE's dauly files now and it is tar.gz formatting
file = tarfile.open(
os.path.join(camels_config["CAMELS_DIR"], "2_LamaH-CE_daily.tar.gz")
)
# extracting file
file.extractall(
os.path.join(camels_config["CAMELS_DIR"], "2_LamaH-CE_daily")
)
file.close()
for f_name in os.listdir(camels_config["CAMELS_DIR"]):
if fnmatch.fnmatch(f_name, "*.zip"):
unzip_dir = os.path.join(camels_config["CAMELS_DIR"], f_name[0:-4])
file_name = os.path.join(camels_config["CAMELS_DIR"], f_name)
unzip_nested_zip(file_name, unzip_dir)
def read_site_info(self) -> pd.DataFrame:
"""
Read the basic information of gages in a CAMELS dataset
Returns
-------
pd.DataFrame
basic info of gages
"""
camels_file = self.data_source_description["CAMELS_GAUGE_FILE"]
if self.region == "US":
data = pd.read_csv(
camels_file, sep=";", dtype={"gauge_id": str, "huc_02": str}
)
elif self.region == "AUS":
data = pd.read_csv(camels_file, sep=",", dtype={"station_id": str})
elif self.region == "BR":
data = pd.read_csv(camels_file, sep="\s+", dtype={"gauge_id": str})
elif self.region == "CL":
data = pd.read_csv(camels_file, sep="\t", index_col=0)
elif self.region == "GB":
data = pd.read_csv(camels_file, sep=",", dtype={"gauge_id": str})
elif self.region == "YR":
dirs_ = os.listdir(self.data_source_description["CAMELS_ATTR_DIR"])
data = pd.DataFrame({"gauge_id": dirs_})
elif self.region == "CA":
data = pd.read_excel(camels_file)
elif self.region == "CE":
data = pd.read_csv(camels_file, sep=";")
else:
raise NotImplementedError(CAMELS_NO_DATASET_ERROR_LOG)
return data
def get_constant_cols(self) -> np.array:
"""
all readable attrs in CAMELS
Returns
-------
np.array
attribute types
"""
data_folder = self.data_source_description["CAMELS_ATTR_DIR"]
if self.region == "US":
var_dict = dict()
var_lst = list()
key_lst = self.data_source_description["CAMELS_ATTR_KEY_LST"]
for key in key_lst:
data_file = os.path.join(data_folder, "camels_" + key + ".txt")
data_temp = pd.read_csv(data_file, sep=";")
var_lst_temp = list(data_temp.columns[1:])
var_dict[key] = var_lst_temp
var_lst.extend(var_lst_temp)
return np.array(var_lst)
elif self.region == "AUS":
attr_all_file = os.path.join(
self.data_source_description["CAMELS_DIR"],
"CAMELS_AUS_Attributes-Indices_MasterTable.csv",
)
camels_aus_attr_indices_data = | pd.read_csv(attr_all_file, sep=",") | pandas.read_csv |
import hashlib
import json
import os
import random
import threading
import sys
import time
import uuid
import functools
import pandas as pd
import numpy as np
from fate_test._config import Config
from fate_test._io import echo, LOGGER
def import_fate():
from fate_arch import storage
from fate_flow.utils import data_utils
from fate_arch import session
from fate_arch.storage import StorageEngine
from fate_arch.common.conf_utils import get_base_config
from fate_arch.storage import EggRollStoreType
return storage, data_utils, session, StorageEngine, get_base_config, EggRollStoreType
storage, data_utils, session, StorageEngine, get_base_config, EggRollStoreType = import_fate()
sys.setrecursionlimit(1000000)
class data_progress:
def __init__(self, down_load, time_start):
self.time_start = time_start
self.down_load = down_load
self.time_percent = 0
self.switch = True
def set_switch(self, switch):
self.switch = switch
def get_switch(self):
return self.switch
def set_time_percent(self, time_percent):
self.time_percent = time_percent
def get_time_percent(self):
return self.time_percent
def progress(self, percent):
if percent > 100:
percent = 100
end = time.time()
if percent != 100:
print(f"\r{self.down_load} %.f%s [%s] running" % (percent, '%', self.timer(end - self.time_start)),
flush=True, end='')
else:
print(f"\r{self.down_load} %.f%s [%s] success" % (percent, '%', self.timer(end - self.time_start)),
flush=True, end='')
@staticmethod
def timer(times):
hours, rem = divmod(times, 3600)
minutes, seconds = divmod(rem, 60)
return "{:0>2}:{:0>2}:{:0>2}".format(int(hours), int(minutes), int(seconds))
def remove_file(path):
os.remove(path)
def id_encryption(encryption_type, start_num, end_num):
if encryption_type == 'md5':
return [hashlib.md5(bytes(str(value), encoding='utf-8')).hexdigest() for value in range(start_num, end_num)]
elif encryption_type == 'sha256':
return [hashlib.sha256(bytes(str(value), encoding='utf-8')).hexdigest() for value in range(start_num, end_num)]
else:
return [str(value) for value in range(start_num, end_num)]
def get_big_data(guest_data_size, host_data_size, guest_feature_num, host_feature_num, include_path, host_data_type,
conf: Config, encryption_type, match_rate, sparsity, force, split_host, output_path, parallelize):
global big_data_dir
def list_tag_value(feature_nums, head):
# data = ''
# for f in range(feature_nums):
# data += head[f] + ':' + str(round(np.random.randn(), 4)) + ";"
# return data[:-1]
return ";".join([head[k] + ':' + str(round(v, 4)) for k, v in enumerate(np.random.randn(feature_nums))])
def list_tag(feature_nums, data_list):
data = ''
for f in range(feature_nums):
data += random.choice(data_list) + ";"
return data[:-1]
def _generate_tag_value_data(data_path, start_num, end_num, feature_nums, progress):
data_num = end_num - start_num
section_data_size = round(data_num / 100)
iteration = round(data_num / section_data_size)
head = ['x' + str(i) for i in range(feature_nums)]
for batch in range(iteration + 1):
progress.set_time_percent(batch)
output_data = pd.DataFrame(columns=["id"])
if section_data_size * (batch + 1) <= data_num:
output_data["id"] = id_encryption(encryption_type, section_data_size * batch + start_num,
section_data_size * (batch + 1) + start_num)
slicing_data_size = section_data_size
elif section_data_size * batch < data_num:
output_data['id'] = id_encryption(encryption_type, section_data_size * batch + start_num, end_num)
slicing_data_size = data_num - section_data_size * batch
else:
break
feature = [list_tag_value(feature_nums, head) for i in range(slicing_data_size)]
output_data['feature'] = feature
output_data.to_csv(data_path, mode='a+', index=False, header=False)
def _generate_dens_data(data_path, start_num, end_num, feature_nums, label_flag, progress):
if label_flag:
head_1 = ['id', 'y']
else:
head_1 = ['id']
data_num = end_num - start_num
head_2 = ['x' + str(i) for i in range(feature_nums)]
df_data_1 = | pd.DataFrame(columns=head_1) | pandas.DataFrame |
import os
from datetime import date
from dask.dataframe import DataFrame as DaskDataFrame
from numpy import nan, ndarray
from numpy.testing import assert_allclose, assert_array_equal
from pandas import DataFrame, Series, Timedelta, Timestamp
from pandas.testing import assert_frame_equal, assert_series_equal
from pymove import (
DaskMoveDataFrame,
MoveDataFrame,
PandasDiscreteMoveDataFrame,
PandasMoveDataFrame,
read_csv,
)
from pymove.core.grid import Grid
from pymove.utils.constants import (
DATE,
DATETIME,
DAY,
DIST_PREV_TO_NEXT,
DIST_TO_PREV,
HOUR,
HOUR_SIN,
LATITUDE,
LOCAL_LABEL,
LONGITUDE,
PERIOD,
SITUATION,
SPEED_PREV_TO_NEXT,
TID,
TIME_PREV_TO_NEXT,
TRAJ_ID,
TYPE_DASK,
TYPE_PANDAS,
UID,
WEEK_END,
)
list_data = [
[39.984094, 116.319236, '2008-10-23 05:53:05', 1],
[39.984198, 116.319322, '2008-10-23 05:53:06', 1],
[39.984224, 116.319402, '2008-10-23 05:53:11', 2],
[39.984224, 116.319402, '2008-10-23 05:53:11', 2],
]
str_data_default = """
lat,lon,datetime,id
39.984093,116.319236,2008-10-23 05:53:05,4
39.9842,116.319322,2008-10-23 05:53:06,1
39.984222,116.319402,2008-10-23 05:53:11,2
39.984222,116.319402,2008-10-23 05:53:11,2
"""
str_data_different = """
latitude,longitude,time,traj_id
39.984093,116.319236,2008-10-23 05:53:05,4
39.9842,116.319322,2008-10-23 05:53:06,1
39.984222,116.319402,2008-10-23 05:53:11,2
39.984222,116.319402,2008-10-23 05:53:11,2
"""
str_data_missing = """
39.984093,116.319236,2008-10-23 05:53:05,4
39.9842,116.319322,2008-10-23 05:53:06,1
39.984222,116.319402,2008-10-23 05:53:11,2
39.984222,116.319402,2008-10-23 05:53:11,2
"""
def _default_move_df():
return MoveDataFrame(
data=list_data,
latitude=LATITUDE,
longitude=LONGITUDE,
datetime=DATETIME,
traj_id=TRAJ_ID,
)
def _default_pandas_df():
return DataFrame(
data=[
[39.984094, 116.319236, Timestamp('2008-10-23 05:53:05'), 1],
[39.984198, 116.319322, Timestamp('2008-10-23 05:53:06'), 1],
[39.984224, 116.319402, Timestamp('2008-10-23 05:53:11'), 2],
[39.984224, 116.319402, Timestamp('2008-10-23 05:53:11'), 2],
],
columns=['lat', 'lon', 'datetime', 'id'],
index=[0, 1, 2, 3],
)
def test_move_data_frame_from_list():
move_df = _default_move_df()
assert MoveDataFrame.has_columns(move_df)
try:
MoveDataFrame.validate_move_data_frame(move_df)
except Exception:
assert False
assert isinstance(move_df, PandasMoveDataFrame)
def test_move_data_frame_from_file(tmpdir):
d = tmpdir.mkdir('core')
file_default_columns = d.join('test_read_default.csv')
file_default_columns.write(str_data_default)
filename_default = os.path.join(
file_default_columns.dirname, file_default_columns.basename
)
move_df = read_csv(filename_default)
assert MoveDataFrame.has_columns(move_df)
try:
MoveDataFrame.validate_move_data_frame(move_df)
except Exception:
assert False
assert isinstance(move_df, PandasMoveDataFrame)
file_different_columns = d.join('test_read_different.csv')
file_different_columns.write(str_data_different)
filename_diferent = os.path.join(
file_different_columns.dirname, file_different_columns.basename
)
move_df = read_csv(
filename_diferent,
latitude='latitude',
longitude='longitude',
datetime='time',
traj_id='traj_id',
)
assert MoveDataFrame.has_columns(move_df)
try:
MoveDataFrame.validate_move_data_frame(move_df)
except Exception:
assert False
assert isinstance(move_df, PandasMoveDataFrame)
file_missing_columns = d.join('test_read_missing.csv')
file_missing_columns.write(str_data_missing)
filename_missing = os.path.join(
file_missing_columns.dirname, file_missing_columns.basename
)
move_df = read_csv(
filename_missing, names=[LATITUDE, LONGITUDE, DATETIME, TRAJ_ID]
)
assert MoveDataFrame.has_columns(move_df)
try:
MoveDataFrame.validate_move_data_frame(move_df)
except Exception:
assert False
assert isinstance(move_df, PandasMoveDataFrame)
def test_move_data_frame_from_dict():
dict_data = {
LATITUDE: [39.984198, 39.984224, 39.984094],
LONGITUDE: [116.319402, 116.319322, 116.319402],
DATETIME: [
'2008-10-23 05:53:11',
'2008-10-23 05:53:06',
'2008-10-23 05:53:06',
],
}
move_df = MoveDataFrame(
data=dict_data,
latitude=LATITUDE,
longitude=LONGITUDE,
datetime=DATETIME,
traj_id=TRAJ_ID,
)
assert MoveDataFrame.has_columns(move_df)
try:
MoveDataFrame.validate_move_data_frame(move_df)
except Exception:
assert False
assert isinstance(move_df, PandasMoveDataFrame)
def test_move_data_frame_from_data_frame():
df = _default_pandas_df()
move_df = MoveDataFrame(
data=df, latitude=LATITUDE, longitude=LONGITUDE, datetime=DATETIME
)
assert MoveDataFrame.has_columns(move_df)
try:
MoveDataFrame.validate_move_data_frame(move_df)
except Exception:
assert False
assert isinstance(move_df, PandasMoveDataFrame)
def test_attribute_error_from_data_frame():
df = DataFrame(
data=[
[39.984094, 116.319236, Timestamp('2008-10-23 05:53:05'), 1],
[39.984198, 116.319322, Timestamp('2008-10-23 05:53:06'), 1],
[39.984224, 116.319402, Timestamp('2008-10-23 05:53:11'), 2],
[39.984224, 116.319402, Timestamp('2008-10-23 05:53:11'), 2],
],
columns=['laterr', 'lon', 'datetime', 'id'],
index=[0, 1, 2, 3],
)
try:
MoveDataFrame(
data=df, latitude=LATITUDE, longitude=LONGITUDE, datetime=DATETIME
)
raise AssertionError(
'AttributeError error not raised by MoveDataFrame'
)
except KeyError:
pass
df = DataFrame(
data=[
[39.984094, 116.319236, Timestamp('2008-10-23 05:53:05'), 1],
[39.984198, 116.319322, Timestamp('2008-10-23 05:53:06'), 1],
[39.984224, 116.319402, Timestamp('2008-10-23 05:53:11'), 2],
[39.984224, 116.319402, Timestamp('2008-10-23 05:53:11'), 2],
],
columns=['lat', 'lonerr', 'datetime', 'id'],
index=[0, 1, 2, 3],
)
try:
MoveDataFrame(
data=df, latitude=LATITUDE, longitude=LONGITUDE, datetime=DATETIME
)
raise AssertionError(
'AttributeError error not raised by MoveDataFrame'
)
except KeyError:
pass
df = DataFrame(
data=[
[39.984094, 116.319236, Timestamp('2008-10-23 05:53:05'), 1],
[39.984198, 116.319322, Timestamp('2008-10-23 05:53:06'), 1],
[39.984224, 116.319402, Timestamp('2008-10-23 05:53:11'), 2],
[39.984224, 116.319402, Timestamp('2008-10-23 05:53:11'), 2],
],
columns=['lat', 'lon', 'datetimerr', 'id'],
index=[0, 1, 2, 3],
)
try:
MoveDataFrame(
data=df, latitude=LATITUDE, longitude=LONGITUDE, datetime=DATETIME
)
raise AssertionError(
'AttributeError error not raised by MoveDataFrame'
)
except KeyError:
pass
def test_lat():
move_df = _default_move_df()
lat = move_df.lat
srs = Series(
data=[39.984094, 39.984198, 39.984224, 39.984224],
index=[0, 1, 2, 3],
dtype='float64',
name='lat',
)
assert_series_equal(lat, srs)
def test_lon():
move_df = _default_move_df()
lon = move_df.lon
srs = Series(
data=[116.319236, 116.319322, 116.319402, 116.319402],
index=[0, 1, 2, 3],
dtype='float64',
name='lon',
)
assert_series_equal(lon, srs)
def test_datetime():
move_df = _default_move_df()
datetime = move_df.datetime
srs = Series(
data=[
'2008-10-23 05:53:05',
'2008-10-23 05:53:06',
'2008-10-23 05:53:11',
'2008-10-23 05:53:11',
],
index=[0, 1, 2, 3],
dtype='datetime64[ns]',
name='datetime',
)
assert_series_equal(datetime, srs)
def test_loc():
move_df = _default_move_df()
assert move_df.loc[0, TRAJ_ID] == 1
loc_ = move_df.loc[move_df[LONGITUDE] > 116.319321]
expected = DataFrame(
data=[
[39.984198, 116.319322, Timestamp('2008-10-23 05:53:06'), 1],
[39.984224, 116.319402, Timestamp('2008-10-23 05:53:11'), 2],
[39.984224, 116.319402, Timestamp('2008-10-23 05:53:11'), 2],
],
columns=['lat', 'lon', 'datetime', 'id'],
index=[1, 2, 3],
)
assert_frame_equal(loc_, expected)
def test_iloc():
move_df = _default_move_df()
expected = Series(
data=[39.984094, 116.319236, Timestamp('2008-10-23 05:53:05'), 1],
index=['lat', 'lon', 'datetime', 'id'],
dtype='object',
name=0,
)
assert_series_equal(move_df.iloc[0], expected)
def test_at():
move_df = _default_move_df()
assert move_df.at[0, TRAJ_ID] == 1
def test_values():
move_df = _default_move_df()
expected = [
[39.984094, 116.319236, | Timestamp('2008-10-23 05:53:05') | pandas.Timestamp |
"""Instantiate a Dash app."""
import datetime
import numpy as np
import pandas as pd
import dash
import dash_table
import dash_html_components as html
import dash_core_components as dcc
from dash.dependencies import Input, Output
from .layout import html_layout
import sqlite3
import plotly.express as px
app_colors = {
'background': '#0C0F0A',
'text': '#FFFFFF',
'sentiment-plot':'#41EAD4',
'volume-bar':'#FBFC74',
'someothercolor':'#FF206E',
}
def create_dashboard(server):
"""Create a Plotly Dash dashboard."""
dash_app = dash.Dash(server=server,
routes_pathname_prefix='/dashapp/',
external_stylesheets=['/static/dist/css/styles.css',
'https://fonts.googleapis.com/css?family=Lato']
)
#connect to the main database
conn = sqlite3.connect('data/alldata.db', isolation_level=None, check_same_thread=False)
c = conn.cursor()
# Prepare a youtube DataFrame------------------------------------------- ytdf
ytdf = pd.read_sql('select * from ytsentiment', conn)
ytdf['date'] = pd.to_datetime(ytdf['unix'])
num_entries = ytdf['id'].value_counts()
# Prepare a twitter DataFrame------------------------------------------- twdf
twdf = pd.read_sql('select * from twsentiment', conn)
twdf['date'] = | pd.to_datetime(twdf['timestamp']) | pandas.to_datetime |
"""
Module to work with result data
"""
import glob
import collections
import logging
import os
import shutil
from typing import Dict
import pandas as pd
import numpy as np
import tensorflow as tf
import settings
clinical = None
# Get the mixed results
def all_results(path, select_type, predictions=False, elem_folds=False):
"""
Get all the results when using multiple folds in training
:param path: Path where the results should be located. Folders starting with ``fold*`` will be searched
accordingly for results.
:param select_type: Type of results to be selected, it can be ``train``, ``test`` or ``mixed``.
:param predictions: Whether to load all the predictions or not to create a data frame with all the predictions.
Useful to create ROC curves.
:param elem_folds: Whether to load or not the predictions and store them in a dictionary where the key is the
most repeated key and the value is the pandas :class:`pandas.DataFrame` with the comparisons
:return:
"""
global clinical
if clinical is None:
clinical = pd.read_csv(settings.DATA_PATH_CLINICAL_PROCESSED)
logger = logging.getLogger(__name__)
logger.debug(f"Searching on {path} {select_type}")
files = glob.glob(path + f"/fold*/{select_type}*.csv")
logger.debug(f"Found {len(files)}")
df_list = []
elem_comparisons = {}
unique_ids = clinical['id'].unique()
for file in files:
df = pd.read_csv(file, index_col=0)
elem_right = len(df[df["labels"].astype(bool) == df["predictions"].astype(bool)])
elem_count = len(df)
ids = np.concatenate((df['pA'].values, df['pB'].values))
ids = collections.Counter(ids)
key, count = ids.most_common(1)[0]
# No LOOCV
if len(files) < len(clinical):
gather = np.array([0, 0])
is_censored = False
time = 0
else:
if key not in unique_ids:
continue
gather = df.loc[df['pA'] == key, 'gather_a'].values
gather = np.append(gather, df.loc[df['pB'] == key, 'gather_b'].values)
is_censored = not clinical.loc[clinical['id'] == key, 'event'].values[0]
time = clinical.loc[clinical['id'] == key, 'time'],
df_list.append(pd.DataFrame({
"id": [key],
"right": [elem_right],
"total": [elem_count],
"censored": [is_censored],
"time": [time],
"file": [file],
"gather": [gather.mean()]
}))
if elem_folds or predictions:
elem_comparisons[key] = df
if predictions:
predictions_df: pd.DataFrame = pd.concat(elem_comparisons.values(), ignore_index=True)
else:
predictions_df = None
results_df: pd.DataFrame = pd.concat(df_list, ignore_index=True)
if select_type == "mixed":
results_df['c-index'] = results_df['right']/results_df['total']
no_cens_results = results_df.loc[~results_df['censored']]
logger.info(f"Finished {path} {select_type}\n")
return (results_df, no_cens_results), predictions_df, elem_comparisons
def save_results(sess: tf.Session, results: Dict[str, pd.DataFrame], path: str, save_model: bool):
"""
Save the current results to disk. It creates a CSV file with the pairs and its values. Keeping in
mind that the results are pairs it uses the suffixes ``_a`` and ``_b`` to denote each member of the pair
- ``age_a``: Age of pair's member A
- ``age_b``: Age of pair's member B
- ``time_a``: Survival time of pair's member A
- ``time_b``: Survival time of pair's member B
- ``pairs_a``: Key of pair's member A
- ``pairs_b``: Key of pair's member B
- ``labels``: Labels that are true if :math:`T(p_a) < T(p_b)`
- ``predictions``: Predictions made by the current model
Moreover, the model is also saved into disk. It can be found in the ``path/weights/`` directory and can
loaded with Tensorflow using the following commands:
>>> import tensorflow as tf
>>> saver = tf.train.Saver()
>>> with tf.Session() as sess:
>>> saver.restore(sess, "<path>/weights/weights.ckpt")
:param sess: Current session that should be saved when saving the model
:param results: List with tuples with a name and a :class:`pandas.DataFrame` of results that should be saved.
the :class:`pandas.DataFrame` should contain at least the columns
``pairs_a``, ``pairs_b``, ``labels`` and ``predictions``.
:param path: Directory path where all the results should be saved
:param save_model: If :any:`True` save the model to disk
"""
weights_dir = os.path.join(path, 'weights')
# Always overwrite the previous weights
if os.path.exists(path):
shutil.rmtree(path)
os.makedirs(path)
os.makedirs(weights_dir)
if save_model:
saver = tf.train.Saver()
saver.save(sess, os.path.join(weights_dir, 'weights.ckpt'))
# Load clinical info
clinical_info = pd.read_csv(settings.DATA_PATH_CLINICAL_PROCESSED, index_col=0)
for name, result in results.items():
merged = _select_time_age(clinical_info, result)
merged.to_csv(os.path.join(path, f"{name}_results.csv"))
def _select_time_age(clinical_info: pd.DataFrame, results_df: pd.DataFrame) -> pd.DataFrame:
merge = | pd.merge(clinical_info, results_df, left_on='id', right_on='pA') | pandas.merge |
import os
import time
import csv
import torch
import torch.nn as nn
from mvcnn import Model
from args import get_parser
import torch.nn.functional as F
from dataset import MultiViewDataSet, preprocess
from torch.utils.data import DataLoader
# from helpers.logger import Logger
# import util
import numpy as np
from pathlib import Path
import pandas as pd
from tqdm import tqdm
from sklearn.metrics import classification_report, confusion_matrix, f1_score
# torch.use_deterministic_algorithms(True)
seed = 1
torch.manual_seed(seed)
import random
random.seed(seed)
np.random.seed(seed)
class Controller(object):
def __init__(self, args):
self.args = args
self.device = torch.device(args.device if torch.cuda.is_available() and args.device != 'cpu' else 'cpu')
self.model = nn.DataParallel(Model(args.model, args.pretrained, args.emb_dim, args.n_class))
self.model.to(self.device)
def train(self, train_loader, val_loader):
optimizer = torch.optim.Adam(self.model.parameters(), lr=self.args.lr)
scheduler = torch.optim.lr_scheduler.CosineAnnealingLR(optimizer, T_max=self.args.epoch)
best_acc, best_loss, patience_count, start_epoch = 0.0, 1e9, 0, 0
weights = train_loader.dataset.weights.to(self.device)
indices = torch.repeat_interleave(torch.arange(self.args.batch), self.args.views).to(self.device)
# logger = Logger(self.args.model_path)
if self.args.resume:
best_acc, start_epoch, optimizer = self.load()
for epoch in range(start_epoch, self.args.epoch):
epoch_loss = .0
total, correct = 0, 0
start = time.time()
self.model.train()
for x, yt in train_loader:
x, yt = x.to(self.device), yt.to(self.device)
xi, xm, yp = self.model(x)
if self.args.regime == 'supervised':
loss = Model.ce_loss(yp, yt, weights)
elif self.args.regime == 'contrastive':
loss = Model.jsd_loss(xi, xm, indices)
elif self.args.regime == 'hybrid':
loss = Model.ce_loss(yp, yt, weights) + Model.jsd_loss(xi, xm, indices)
epoch_loss += loss.item()
optimizer.zero_grad()
loss.backward()
optimizer.step()
if self.args.regime != 'contrastive':
_, yp = torch.max(yp.data, 1)
total += yt.size(0)
correct += (yp == yt).sum().item()
train_acc = 100 * correct / total if self.args.regime != 'contrastive' else .0
end = time.time()
self.model.eval()
val_acc, val_loss = self.eval(val_loader)
if self.args.regime != 'contrastive' and val_acc > best_acc:
best_acc = val_acc
if not os.path.exists(self.args.model_path):
os.mkdir(self.args.model_path)
torch.save(self.model.module.state_dict(), f'{self.args.model_path}/model-best.pth')
# torch.save(self.model.state_dict(), f'{self.args.model_path}/model-best.pth')
print(f'Epoch {epoch + 1}/{self.args.epoch} | Time: {end - start:.2f}s '
f'| Train Loss: {epoch_loss / len(train_loader): .4f} | Train Acc: {train_acc:.2f}% | '
f'Val Loss: {val_loss:.4f} | '
f'Val Acc: {val_acc:.2f}% | Best Acc: {best_acc:.2f}%')
# Log epoch to tensorboard
# See log using: tensorboard --logdir='args.model_path' --host localhost
# util.logEpoch(logger, self.model, epoch + 1, val_loss, val_acc)
if best_loss > val_loss:
best_loss = val_loss
patience_count = 0
if self.args.regime == 'contrastive':
if not os.path.exists(self.args.model_path):
os.mkdir(self.args.model_path)
# torch.save(self.model.state_dict(), f'{self.args.model_path}/model-best.pth')
torch.save(self.model.module.state_dict(), f'{self.args.model_path}/model-best.pth')
else:
patience_count += 1
if patience_count == self.args.patience:
print(f'Early stopping at epoch {epoch} ...')
break
scheduler.step()
# save model
if not os.path.exists(self.args.model_path):
os.mkdir(self.args.model_path)
# torch.save(self.model.state_dict(), f'{self.args.model_path}/model-last.pth')
torch.save(self.model.module.state_dict(), f'{self.args.model_path}/model-last.pth')
# save labels
labels = train_loader.dataset.classes
with open(f'{self.args.model_path}/labels.csv', 'w', newline='', encoding='utf-8') as csvfile:
writer = csv.writer(csvfile)
writer.writerow(labels)
# print out evaluation report
print("Validation report after training:")
try:
embeddings, predictions = self.encode(val_loader, self.args.model_path + "/model-last.pth")
gt_classes, pred_classes = self.print_classification_report(val_loader, predictions)
except Exception as e:
print(e)
@torch.no_grad()
def eval(self, data_loader, load_model=False):
weights = data_loader.dataset.weights.to(self.device)
total, correct = 0, 0
total_loss = 0.0
if load_model:
self.load()
# test
for x, yt in tqdm(data_loader, desc="Evaluating model"):
x, yt = x.to(self.device), yt.to(self.device)
xi, xm, yp = self.model(x)
if self.args.regime == 'supervised':
loss = Model.ce_loss(yp, yt, weights)
elif self.args.regime == 'contrastive':
indices = torch.repeat_interleave(torch.arange(x.size(0)), self.args.views).to(self.device)
loss = Model.jsd_loss(xi, xm, indices)
elif self.args.regime == 'hybrid':
indices = torch.repeat_interleave(torch.arange(x.size(0)), self.args.views).to(self.device)
loss = Model.ce_loss(yp, yt, weights) + Model.jsd_loss(xi, xm, indices)
total_loss += loss.item()
if self.args.regime != 'contrastive':
_, yp = torch.max(yp.data, 1)
total += yt.size(0)
correct += (yp == yt).sum().item()
val_acc = 100 * correct / total if self.args.regime != 'contrastive' else .0
val_loss = total_loss / len(data_loader)
return val_acc, val_loss
@torch.no_grad()
def encode(self, data_loader, model_path):
try:
self.model.load_state_dict(torch.load(model_path))
except:
state_dict = torch.load(model_path)
from collections import OrderedDict
new_state_dict = OrderedDict()
for k, v in state_dict.items():
if 'module' not in k:
k = 'module.' + k
else:
k = k.replace('features.module.', 'module.features.')
new_state_dict[k] = v
self.model.load_state_dict(new_state_dict)
self.model.eval()
emb, pred = [], []
for x, __ in tqdm(data_loader, desc='Embedding...'):
x = x.to(self.device)
__, x, y = self.model(x)
emb.append(x)
pred.append(y)
x = torch.cat(emb, 0).detach().cpu().numpy()
y = F.softmax(torch.cat(pred, 0), dim=-1)
return x, y
def save_embeddings(self, data_loader, embs, classes):
names = [Path(item).parts[-2] for item in data_loader.dataset.x]
embedding_df = pd.DataFrame(list(zip(classes, names, embs)), columns=["class_name", "part_name", "vector"])
dest = Path(self.args.model_path) / (Path(self.args.model_path).parts[-1] + '_embeddings')
os.makedirs(dest, exist_ok=True)
for class_name in tqdm(data_loader.dataset.classes, desc='Saving embeddings...'):
class_embedding = embedding_df[embedding_df['class_name'] == class_name].to_numpy()
np.save(dest / (class_name + "_embeddings"), class_embedding)
def load(self): # Does not work
print('\n==> Loading checkpoint..')
model_path = self.args.model_path + "/model-last.pth"
assert os.path.isfile(model_path), f'Error: no checkpoint file found in {model_path}!'
checkpoint = torch.load(model_path)
best_acc = checkpoint['best_acc']
start_epoch = checkpoint['epoch']
self.model.load_state_dict(checkpoint['state_dict'])
optimizer = torch.optim.Adam(self.model.parameters(), lr=self.args.lr)
optimizer.load_state_dict(checkpoint['optimizer'])
return best_acc, start_epoch, optimizer
def print_classification_report(self, encode_loader, predictions, top_k):
import matplotlib.pyplot as plt
import seaborn as sn
gt_classes = [encode_loader.dataset.classes[item] for item in encode_loader.dataset.y]
gt_classes_idx = [item for item in encode_loader.dataset.y]
if top_k == 1:
pred_classes_idx = np.argmax(predictions.detach().cpu().numpy(), axis=1)
pred_classes = [encode_loader.dataset.classes[item] for item in pred_classes_idx]
label = encode_loader.dataset.classes
print(f"f1 micro precision: {f1_score(gt_classes, pred_classes, average='micro')}")
print(classification_report(gt_classes, pred_classes, labels=label))
cf = confusion_matrix(gt_classes, pred_classes, normalize='true', labels=label)
if not os.path.exists('logs/'):
os.makedirs('logs/')
plt.figure(figsize=(24, 18))
sn.heatmap(cf, annot=False, fmt='.2f', cmap='Blues', xticklabels=label, yticklabels=label)
plt.xticks(size='xx-large', rotation=45)
plt.yticks(size='xx-large', rotation=45)
plt.tight_layout()
plt.savefig(fname=f'logs/{Path(self.args.model_path).parts[-1]}.pdf', format='pdf')
plt.show()
else:
log = | pd.DataFrame(columns=['f1 micro']) | pandas.DataFrame |
import numpy as np
import pandas as pd
import scipy as sp
import matplotlib.pyplot as plt
import seaborn as sns
from collections import OrderedDict
import pickle
from pystan import StanModel
import plot_coefficients as pc
import copy
import os
sns.set_context('notebook')
script_dir = os.path.dirname(os.path.abspath(__file__))
SAVE = True
save_model_1 = SAVE
save_fit_1 = SAVE
save_params_1 = SAVE
save_model_2 = SAVE
save_fit_2 = SAVE
save_params_2 = SAVE
"""Multilevel Modeling with Poststratification (MRP)"""
# Use multilevel regression to model individual survey responses as a function of demographic and geographic
# predictors, partially pooling respondents across regions to an extent determined by the data.
# The final step is poststratification.
"""Define dictionaries:"""
sex_categories = {'male': 1, 'female': 2}
ethnicity_categories = {'white': 1, 'mixed': 2, 'black': 3, 'asian': 4, 'other': 5}
age_categories = {'Age 18 to 19': 1, 'Age 20 to 24': 2, 'Age 25 to 29': 3, 'Age 30 to 34': 4, 'Age 35 to 39': 5,
'Age 40 to 44': 6, 'Age 45 to 49': 7, 'Age 50 to 54': 8, 'Age 55 to 59': 9, 'Age 60 to 64': 10,
'Age 65 to 69': 11, 'Age 70 to 74': 12, 'Age 75 to 79': 13, 'Age 80 to 84': 14, 'Age 85 and over': 15}
region_categories = {'East Midlands': 1, 'East of England': 2, 'London': 3, 'North East': 4, 'North West': 5,
'South East': 6, 'South West': 7, 'Wales': 8, 'West Midlands': 9, 'Yorkshire and the Humber': 10}
enddate_categories = {'2015-03-30': 1, '2015-03-31': 2, '2015-04-01': 3, '2015-04-02': 4, '2015-04-03': 5,
'2015-04-04': 6, '2015-04-05': 7, '2015-04-06': 8, '2015-04-07': 9, '2015-04-08': 10,
'2015-04-09': 11, '2015-04-10': 12, '2015-04-11': 13, '2015-04-12': 14, '2015-04-13': 15,
'2015-04-14': 16, '2015-04-15': 17, '2015-04-16': 18, '2015-04-17': 19, '2015-04-18': 20,
'2015-04-19': 21, '2015-04-20': 22, '2015-04-21': 23, '2015-04-22': 24, '2015-04-23': 25,
'2015-04-24': 26, '2015-04-25': 27, '2015-04-26': 28, '2015-04-27': 29, '2015-04-28': 30,
'2015-04-29': 31, '2015-04-30': 32, '2015-05-01': 33, '2015-05-02': 34, '2015-05-03': 35,
'2015-05-04': 36, '2015-05-05': 37, '2015-05-06': 38}
party_categories = {'Con': 1, 'Grn': 2, 'LD': 3, 'Lab': 4, 'Other': 5, 'PC': 6, 'UKIP': 7, "Don't vote": 8,
"Don't know": 9}
"""Step 1: gather national opinion polls (they need to include respondent information down to the level of
disaggregation the analysis is targetting) """
polls = pd.read_csv(script_dir + '/2015_ge_results_bes/bes_poll_data.csv')
# drop SNP voters in regions outside Scottland:
polls = polls[polls['vote'] != 'SNP']
polls['main'] = np.where(polls['vote'] == 'Con', 1, np.where(polls['vote'] == 'Lab', 1, 0))
# polls.shape
# (24579, 6)
n_age = len(age_categories)
n_sex = len(sex_categories)
n_ethnicity = len(ethnicity_categories)
n_region = len(region_categories)
n_party = len(party_categories)
n = polls.shape[0]
polls_numeric = copy.deepcopy(polls)
polls_numeric['sex'] = polls_numeric['sex'].apply(lambda x: sex_categories[x])
polls_numeric['age'] = polls_numeric['age'].apply(lambda x: age_categories[x])
polls_numeric['ethnicity'] = polls_numeric['ethnicity'].apply(lambda x: ethnicity_categories[x])
polls_numeric['region'] = polls_numeric['region'].apply(lambda x: region_categories[x])
polls_numeric['enddate'] = polls_numeric['enddate'].apply(lambda x: enddate_categories[x])
polls_numeric['vote'] = polls_numeric['vote'].apply(lambda x: party_categories[x])
"""Step 2: create a separate dataset of region-level predictors """
# load in 2010 election data as a region level predictor
# (http://www.electoralcommission.org.uk/our-work/our-research/electoral-data)
ge_10 = pd.read_csv(script_dir + '/2010_ge_results/GE2010.csv')
ge_10 = ge_10[['Press Association Reference', 'Constituency Name', 'Region', 'Electorate', 'Votes', 'Con', 'Lab', 'LD',
'Grn', 'UKIP', 'SNP', 'PC']]
ge_10['Other'] = ge_10['Votes']\
- ge_10.fillna(0)['Con']\
- ge_10.fillna(0)['Lab']\
- ge_10.fillna(0)['LD']\
- ge_10.fillna(0)['Grn']\
- ge_10.fillna(0)['UKIP']\
- ge_10.fillna(0)['SNP']\
- ge_10.fillna(0)['PC']
ge_10["Don't vote"] = ge_10['Electorate'] - ge_10['Votes']
ge_10_region = ge_10[['Region', 'Electorate', 'Votes', 'Con', 'Lab', 'LD', 'Grn', 'UKIP', 'SNP', 'PC', 'Other',
"Don't vote"]]
ge_10_region = ge_10_region.groupby('Region').sum()
ge_10_region = ge_10_region.drop(['SNP'], 1)
ge_10_region = ge_10_region.drop(['Northern Ireland', 'Scotland'], 0)
ge_10_region = ge_10_region.rename(index={'Eastern': 'East of England'})
ge_10_region_share = ge_10_region.div(ge_10_region['Votes'], axis=0)
ge_10_region_share['main'] = ge_10_region_share['Con'] + ge_10_region_share['Lab']
# UK parties:
# Conservatives (Con)
# Labour (Lab)
# Liberal Democrats (LD)
# Greens (Grn)
# UK Independence Party (UKIP)
# Scotish National Party (SNP) - Scottland only
# Democratic Unionists (DUP) - Northern Ireland only
# Sinn Fein (SF) - Nothern Ireland only
# Plaid Cymru (PC) - Wales only
# Social Democratic & Labour Party (SDLP) - Northern Ireland only
ge_10_region_share = ge_10_region_share.rename(index=region_categories)
ge_10_region_share = ge_10_region_share.rename(columns=party_categories)
""" Extra Step: Validation Data"""
# load in 2015 election data as a validation check
# (http://www.electoralcommission.org.uk/our-work/our-research/electoral-data)
ge_15 = pd.read_csv(script_dir + '/2015_ge_results/RESULTS_FOR_ANALYSIS.csv')
ge_15 = ge_15[['Press Association Reference', 'Constituency Name', 'Region', 'Electorate', 'Votes', 'Con', 'Lab', 'LD',
'Grn', 'UKIP', 'SNP', 'PC']]
ge_15['Votes'] = ge_15['Votes'].str.replace(',', '').astype(float)
ge_15['Electorate'] = ge_15['Electorate'].str.replace(',', '').astype(float)
ge_15['Other'] = ge_15['Votes']\
- ge_15.fillna(0)['Con']\
- ge_15.fillna(0)['Lab']\
- ge_15.fillna(0)['LD']\
- ge_15.fillna(0)['Grn']\
- ge_15.fillna(0)['UKIP']\
- ge_15.fillna(0)['SNP']\
- ge_15.fillna(0)['PC']
ge_15["Don't vote"] = ge_15['Electorate'] - ge_15['Votes']
ge_15_region = ge_15[['Region', 'Electorate', 'Votes', 'Con', 'Lab', 'LD', 'Grn', 'UKIP', 'SNP', 'PC', 'Other',
"Don't vote"]]
ge_15_region = ge_15_region.groupby('Region').sum()
ge_15_region = ge_15_region.drop(['SNP'], 1)
ge_15_region = ge_15_region.drop(['Northern Ireland', 'Scotland'], 0)
ge_15_region = ge_15_region.rename(index={'East': 'East of England',
'Yorkshire and The Humber': 'Yorkshire and the Humber'})
ge_15_region_share = ge_15_region.div(ge_15_region['Votes'], axis=0)
ge_15_region_share['main'] = ge_15_region_share['Con'] + ge_15_region_share['Lab']
ge_15_region_share = ge_15_region_share.rename(index=region_categories)
ge_15_region_share = ge_15_region_share.rename(columns=party_categories)
"""Step 3: Load 1988 census data to enable poststratification."""
# this is a full joint distribution; if only marginal distributions are available then use raking
census_11 = pd.read_csv(script_dir + '/2011_census/four_way_joint_distribution/four_way_joint_distribution_str.csv')
# age: categorical variable
# sex: indicator variable
# ethnicity: categorical variable
# region: categorical variable
# N: size of population in this cell
census_11_numeric = copy.deepcopy(census_11)
census_11_numeric['sex'] = census_11_numeric['sex'].apply(lambda x: sex_categories[x])
census_11_numeric['age'] = census_11_numeric['age'].apply(lambda x: age_categories[x])
census_11_numeric['ethnicity'] = census_11_numeric['ethnicity'].apply(lambda x: ethnicity_categories[x])
census_11_numeric['region'] = census_11_numeric['region'].apply(lambda x: region_categories[x])
# Test compatibility:
sorted(census_11.region.unique().tolist()) == sorted(polls.region.unique().tolist())
sorted(census_11.age.unique().tolist()) == sorted(polls.age.unique().tolist())
sorted(census_11.sex.unique().tolist()) == sorted(polls.sex.unique().tolist())
sorted(census_11.ethnicity.unique().tolist()) == sorted(polls.ethnicity.unique().tolist())
sorted(ge_10_region.index.tolist()) == sorted(ge_15_region.index.tolist())
sorted(ge_10_region.index.tolist()) == sorted(polls.region.unique().tolist())
"""Step 4: Fit a regression model for an individual survey response given demographics, geography etc."""
################################
# 1st model: Probability that a voter casts a vote on a main party candidate
################################
# Pr(Y_i \in {Option_1, Option_2}) = logit^{-1}(alpha[1] + alpha[2] * v_prev_j[i] + a^state_j[i] + a^edu_j[i]
# + a^sex_j[i] + a^age_j[i] + a^race_j[i] + a^partyID_j[i] + a^ideology_j[i] + a^lastvote_j[i])
# a^{}_j[i] are the varying coefficients associated with each categorical variable; with independent prior
# distributions:
# a^{}_j[i] ~ N(0,sigma^2_var)
# the variance parameters are assigned a hyper prior distribution:
# sigma^2_var ~ invX^2(v,sigma^2_0)
# with a weak prior specification for v and sigma^2_0
# Model description:
model_1 = """
data {
int<lower=0> N;
int<lower=0> n_region;
int<lower=0> n_sex;
int<lower=0> n_age;
int<lower=0> n_ethnicity;
real<lower=0,upper=1> region_v_prev[n_region];
int<lower=0,upper=n_region> region[N];
int<lower=0,upper=n_sex> sex[N];
int<lower=0,upper=n_age> age[N];
int<lower=0,upper=n_ethnicity> ethnicity[N];
int<lower=0,upper=1> y[N];
}
parameters {
vector[2] alpha;
vector[n_region] a;
vector[n_sex] b;
vector[n_age] c;
vector[n_ethnicity] d;
real<lower=0,upper=100> sigma_a;
real<lower=0,upper=100> sigma_b;
real<lower=0,upper=100> sigma_c;
real<lower=0,upper=100> sigma_d;
real<lower=0> mu;
real<lower=0,upper=100> sigma_0;
}
transformed parameters {
vector[N] y_hat;
for (i in 1:N)
y_hat[i] = alpha[1] + alpha[2] * region_v_prev[region[i]] + a[region[i]] + b[sex[i]] + c[age[i]]
+ d[ethnicity[i]];
}
model {
a ~ normal (0, sigma_a);
b ~ normal (0, sigma_b);
c ~ normal (0, sigma_c);
d ~ normal (0, sigma_d);
alpha ~ normal(0, 100);
sigma_a ~ scaled_inv_chi_square(mu,sigma_0);
sigma_b ~ scaled_inv_chi_square(mu,sigma_0);
sigma_c ~ scaled_inv_chi_square(mu,sigma_0);
sigma_d ~ scaled_inv_chi_square(mu,sigma_0);
mu ~ uniform(0, 100);
sigma_0 ~ uniform(0, 100);
y ~ bernoulli_logit(y_hat);
}
"""
# Model parameters and data:
model_1_data_dict = {'N': n, 'n_region': n_region, 'n_age': n_age, 'n_sex': n_sex, 'n_ethnicity': n_ethnicity,
'region': polls_numeric.region, 'age': polls_numeric.age, 'sex': polls_numeric.sex,
'ethnicity': polls_numeric.ethnicity, 'region_v_prev': ge_10_region_share['main'],
'y': polls_numeric.main}
# Fitting the model:
n_chains = 2
n_iter = 1000
# full_model_fit = pystan.stan(model_code=full_model, data=full_model_data_dict, iter=n_iter, chains=2)
if save_model_1:
sm = StanModel(model_code=model_1)
with open('pkl_objects/election15_model_1.pkl', 'wb') as f:
pickle.dump(sm, f)
else:
sm = pickle.load(open('pkl_objects/election15_model_1.pkl', 'rb'))
if save_fit_1:
model_1_fit = sm.sampling(data=model_1_data_dict, iter=n_iter, chains=n_chains)
with open('pkl_objects/election15_model_1_fitted.pkl', 'wb') as f:
pickle.dump(model_1_fit, f)
else:
model_1_fit = pickle.load(open('pkl_objects/election15_model_1_fitted.pkl', 'rb'))
if save_params_1:
params_m1 = model_1_fit.extract()
with open('pkl_objects/election15_model_1_params.pkl', 'wb') as f:
pickle.dump(params_m1, f)
else:
params_m1 = pickle.load(open('pkl_objects/election15_model_1_params.pkl', 'rb'))
# what to do about 'Don't know' voters?
# Extract and label parameters:
params_m1 = model_1_fit.extract()
params_m1_alpha_0 = pd.DataFrame({'Intercept': params_m1['alpha'][:, 0]})
params_m1_alpha_1 = pd.DataFrame({'Prev Vote': params_m1['alpha'][:, 1]})
o_dict = OrderedDict({'Region ' + str(i+1): params_m1['a'][:, i] for i in range(0, params_m1['a'].shape[1])})
params_m1_a = pd.DataFrame(o_dict)
o_dict = OrderedDict({'Sex ' + str(i+1): params_m1['b'][:, i] for i in range(0, params_m1['b'].shape[1])})
params_m1_b = pd.DataFrame(o_dict)
o_dict = OrderedDict({'Age ' + str(i+1): params_m1['c'][:, i] for i in range(0, params_m1['c'].shape[1])})
params_m1_c = pd.DataFrame(o_dict)
o_dict = OrderedDict({'Ethnicity ' + str(i+1): params_m1['d'][:, i] for i in range(0, params_m1['d'].shape[1])})
params_m1_d = pd.DataFrame(o_dict)
params_m1_demo = pd.concat([params_m1_alpha_0, params_m1_b, params_m1_c, params_m1_d], axis=1)
params_m1_region = pd.concat([params_m1_alpha_1, params_m1_a], axis=1)
# Plot demographic coefficients with confidence intervals:
pc.plot_coefficients(params=params_m1_demo,
ticks_list=list(params_m1_demo.columns.values),
title='Coefficients',
f_name='plots/DemoCoefficients_ConfidenceIntervals.png')
# Plot state coefficients with confidence intervals:
pc.plot_coefficients(params=params_m1_region,
ticks_list=list(params_m1_region.columns.values),
title='Region Intercepts',
f_name='plots/StateIntercepts_ConfidenceIntervals.png')
# Coefficient Distributions and Traceplots:
# model_1_fit.plot()
# plt.savefig('plots/ParameterDistributions_model_1.png')
################################
# 2nd model: Probability that a voter casts a vote for Option_1
################################
# 2nd model:
# Pr(Y_i = Option_1 | Y_i \in {Option_1, Option_2}) = logit^{-1}(beta_0 + beta_1 + b^state_j[i] + b^edu_j[i]
# + b^sex_j[i] + b^age_j[i] + b^race_j[i] + b^partyID_j[i] + b^ideology_j[i] + b^lastvote_j[i])
# b^{}_j[i] ~ N(0,eta^2_var)
# eta^2_var ~ invX^2(mu,eta^2_0)
# run daily with four-dat moving window(t, t-1, t-2, t-3)
polls_numeric_main = polls_numeric[polls_numeric['main'] == 1]
polls_numeric_main = polls_numeric_main.reset_index(drop=True)
polls_numeric_main['con'] = np.where(polls_numeric_main['vote'] == 1, 1, 0)
n = polls_numeric_main.shape[0]
# Model description:
model_2 = """
data {
int<lower=0> N;
int<lower=0> n_region;
int<lower=0> n_sex;
int<lower=0> n_age;
int<lower=0> n_ethnicity;
real<lower=0,upper=1> region_v_prev[n_region];
int<lower=0,upper=n_region> region[N];
int<lower=0,upper=n_sex> sex[N];
int<lower=0,upper=n_age> age[N];
int<lower=0,upper=n_ethnicity> ethnicity[N];
int<lower=0,upper=1> y[N];
}
parameters {
vector[2] alpha;
vector[n_region] a;
vector[n_sex] b;
vector[n_age] c;
vector[n_ethnicity] d;
real<lower=0,upper=100> sigma_a;
real<lower=0,upper=100> sigma_b;
real<lower=0,upper=100> sigma_c;
real<lower=0,upper=100> sigma_d;
real<lower=0> mu;
real<lower=0,upper=100> sigma_0;
}
transformed parameters {
vector[N] y_hat;
for (i in 1:N)
y_hat[i] = alpha[1] + alpha[2] * region_v_prev[region[i]] + a[region[i]] + b[sex[i]] + c[age[i]]
+ d[ethnicity[i]];
}
model {
a ~ normal (0, sigma_a);
b ~ normal (0, sigma_b);
c ~ normal (0, sigma_c);
d ~ normal (0, sigma_d);
alpha ~ normal(0, 100);
sigma_a ~ scaled_inv_chi_square(mu,sigma_0);
sigma_b ~ scaled_inv_chi_square(mu,sigma_0);
sigma_c ~ scaled_inv_chi_square(mu,sigma_0);
sigma_d ~ scaled_inv_chi_square(mu,sigma_0);
mu ~ uniform(0, 100);
sigma_0 ~ uniform(0, 100);
y ~ bernoulli_logit(y_hat);
}
"""
# Model parameters and data:
model_2_data_dict = {'N': n, 'n_region': n_region, 'n_sex': n_sex, 'n_age': n_age, 'n_ethnicity': n_ethnicity,
'region': polls_numeric_main.region, 'sex': polls_numeric_main.sex, 'age': polls_numeric_main.age,
'ethnicity': polls_numeric_main.ethnicity, 'region_v_prev': ge_10_region_share['main'],
'y': polls_numeric_main.con}
# Fitting the model:
n_chains = 2
n_iter = 1000
# full_model_fit = pystan.stan(model_code=full_model, data=full_model_data_dict, iter=n_iter, chains=2)
if save_model_2:
sm = StanModel(model_code=model_2)
with open('pkl_objects/election15_model_2.pkl', 'wb') as f:
pickle.dump(sm, f)
else:
sm = pickle.load(open('pkl_objects/election15_model_2.pkl', 'rb'))
if save_fit_2:
model_2_fit = sm.sampling(data=model_2_data_dict, iter=n_iter, chains=n_chains)
with open('pkl_objects/election15_model_2_fitted.pkl', 'wb') as f:
pickle.dump(model_2_fit, f)
else:
model_2_fit = pickle.load(open('pkl_objects/election15_model_2_fitted.pkl', 'rb'))
if save_params_2:
params_m2 = model_2_fit.extract()
with open('pkl_objects/election15_model_2_params.pkl', 'wb') as f:
pickle.dump(params_m2, f)
else:
params_m2 = pickle.load(open('pkl_objects/election15_model_2_params.pkl', 'rb'))
# Extract and label parameters:
params_m2 = model_2_fit.extract()
params_m2_alpha_0 = pd.DataFrame({'Intercept': params_m2['alpha'][:, 0]})
params_m2_alpha_1 = pd.DataFrame({'Prev Vote': params_m2['alpha'][:, 1]})
o_dict = OrderedDict({'Region ' + str(i+1): params_m2['a'][:, i] for i in range(0, params_m2['a'].shape[1])})
params_m2_a = | pd.DataFrame(o_dict) | pandas.DataFrame |
import pandas as pd
import geopandas
from geopandas import GeoDataFrame
from pandas import Series
from shapely.geometry import Point, MultiPoint, LineString, MultiLineString, Polygon
import itertools
import numpy as np
import math
import matplotlib.pyplot as plt
import time
import datetime
def ReadData(bestand):
kolommen = ['bird_name','date_time','longitude','latitude','direction','behaviour','calc_time_diff']
data = pd.read_csv(bestand,usecols=kolommen, sep=',' ,header=(0),low_memory=False, parse_dates=['date_time'], dayfirst=True)
return data
def MeeuwenDict(data):
dfdict = dict(tuple(data.groupby('bird_name')))
meeuwen = list(dfdict.keys())
return dfdict, meeuwen
def SyncMeeuwen(dictionary, syncfreq, limiet):
"""" Synchroniseer de meeuwen op een bepaalde frequentie"""
dfdict = dictionary
meeuwen = list(dfdict.keys())
for meeuw in meeuwen:
# voor iedere meeuw een eerste rij invoeren met afgeronde timestamp
# voorkomt problemen met synchronisatie, wanneer er seconden in de timestamp zitten
rond_tijdstip = dfdict[meeuw].iloc[0].date_time.floor(frequentie)
line = pd.DataFrame({'date_time':rond_tijdstip}, index=[-1])
dfdict[meeuw] = pd.concat([line, dfdict[meeuw].loc[:]], sort = True)
# timestamp als index zetten
dfdict[meeuw] = dfdict[meeuw].set_index('date_time')
#als frequentie instellen (gelijk aan .resamle('600S').asfreq() )
dfdict[meeuw] = dfdict[meeuw].asfreq(syncfreq)
#interpoleren van kolommen (series)
dfdict[meeuw][['longitude','latitude']] = dfdict[meeuw][['longitude','latitude']].interpolate(method='linear', limit = limiet, limit_area = 'inside')
# apart aangezien volgende lijn, error geeft: "Cannot interpolate with all NaNs."
# dfdict[meeuw][['bird_name','behaviour']] = dfdict[meeuw][['bird_name','behaviour']].interpolate(method='pad')
dfdict[meeuw]['bird_name'] = dfdict[meeuw]['bird_name'].interpolate(method='pad', limit = limiet, limit_area = 'inside')
dfdict[meeuw]['behaviour'] = dfdict[meeuw]['behaviour'].interpolate(method='pad', limit = limiet, limit_area = 'inside')
#geometrie toevoegen en onmiddelijk transformeren, kan ook nu pas omdat deze data niet geïnterpoleerd kan worden
punt= [Point(xy) for xy in zip(dfdict[meeuw].longitude, dfdict[meeuw].latitude)]
crs = {'init': 'epsg:4326'} #crs toekennen
dfdict[meeuw] = GeoDataFrame(dfdict[meeuw], crs=crs, geometry=punt)
dfdict[meeuw] = dfdict[meeuw].to_crs({'init': 'epsg:31370'})
dfdict[meeuw] = dfdict[meeuw].set_index(['bird_name'], append=True)
#eerste index naam van de meeuw
dfdict[meeuw] = dfdict[meeuw].swaplevel()
return dfdict
def Merge(dfdicts):
dfdict = dfdicts
meeuwen = list(dfdict.keys())
# alle data opnieuw samenbrengen
# eerste meeuw uit dictionary halen en aanvullen met de andere meeuwen
mergeddf = dfdict[meeuwen[0]]
for m in meeuwen[1:]:
mergeddf = mergeddf.append(dfdict[m])
#mergeddf = mergeddf.sort_index(sort=True)
return mergeddf
"""afstanden berekenen van alle meeuwen tot elkaar"""
def BerekenAfstanden(dataframe):
mergeddf = dataframe
mergeddf.index.get_level_values('bird_name').unique()
punten = mergeddf.loc[meeuwen[0]][['geometry']]
kolomnaam = 'geometry_'+meeuwen[0]
punten.columns = [kolomnaam]
for meeuw in meeuwen[1:]:
rsuf = '_'+meeuw
punten = punten.join(mergeddf.loc[meeuw][['geometry']], how='outer', rsuffix= rsuf)
punten.columns = meeuwen #naam veranderen van kolommen
koppels = list(itertools.combinations(meeuwen,2))
afstanden = pd.DataFrame(index=punten.index, columns= koppels)
for tijdstip, row in afstanden.iterrows():
for koppel in koppels:
if pd.notna(punten.loc[tijdstip, koppel[0]]) and pd.notna(punten.loc[tijdstip,koppel[1]]):
row[koppel] = punten.loc[tijdstip, koppel[0]].distance(punten.loc[tijdstip,koppel[1]])
afstanden.dropna(how='all', inplace=True)
return punten, afstanden
""" buren bepalen binnen afstand"""
def BepaalBuren(afstanden, meeuwen, buffer):
buren = pd.DataFrame(index=afstanden.index, columns= afstanden.columns)
koppels = list(itertools.combinations(meeuwen,2))
for koppel in koppels:
buren[koppel] = afstanden[koppel].apply(lambda x: True if x <= buffer else False)
return buren
""" tijdsintervallen bepalen wanneer meeuwen buren zijn"""
def GetBuurSequentiesDict(buren, meeuwen):
koppels = list(itertools.combinations(meeuwen,2))
sequenties = dict()
for koppel in koppels:
sequenties.update({koppel:[]})
start_sequentie = np.nan
einde_sequentie = np.nan
opzoek = True
for tijdstip, row in buren.iterrows():
if row[koppel] == True:
if opzoek == True:
start_sequentie = tijdstip
einde_sequentie = tijdstip
opzoek = False
else:
einde_sequentie = tijdstip
elif opzoek == False:
sequenties[koppel].append((start_sequentie, einde_sequentie))
start_sequentie = np.nan
einde_sequentie = np.nan
opzoek = True
return sequenties
""" sequenties naar dataframe zetten """
def SequentieDictToDF(sequenties_dict):
koppels = list(sequenties_dict.keys())
sequenties_df = pd.DataFrame()
for koppel in koppels:
length = len(sequenties_dict[koppel])
if sequenties_dict[koppel]:
koppel_seqdf = pd.DataFrame.from_dict(sequenties_dict[koppel])
koppel_seqdf.columns = ['begin','einde']
koppel_seqdf['koppel'] = Series([koppel]*length)
sequenties_df = sequenties_df.append(koppel_seqdf, ignore_index=True)
sequenties_df = sequenties_df[['koppel','begin','einde']]
sequenties_df["duur"] = sequenties_df["einde"]-sequenties_df["begin"]
return sequenties_df
"""momenteel enkel locatie eerste meeuw begintijdstip"""
class Locaties:
def BeginMeeuw1(row):
return mergeddf.loc[row.koppel[0], row.begin]['geometry']
def BeginMeeuw2(row):
return mergeddf.loc[row.koppel[1], row.begin]['geometry']
def EindeMeeuw1(row):
return mergeddf.loc[row.koppel[0], row.einde]['geometry']
def EindeMeeuw2(row):
return mergeddf.loc[row.koppel[1], row.einde]['geometry']
def BeginEind(dataframe):
dataframe['geometry'] = dataframe.apply(Locaties.BeginMeeuw1, axis=1)
# dataframe['beginlocatie2'] = dataframe.apply(Locaties.BeginMeeuw2, axis=1)
# dataframe['eindlocatie1'] = dataframe.apply(Locaties.EindeMeeuw1, axis=1)
# dataframe['eindlocatie2'] = dataframe.apply(Locaties.EindeMeeuw2, axis=1)
return dataframe
def FilterFrequentie(data, freq=5, kwartiel='25%'):
""" dictionary opstellen waarvoor de frequentie van de data op een bepaald kwartiel onder een maximale waarde valt.
Default moet 25% van de data een temporele frequentie hebben kleiner dan 5 seconden"""
hfreqdict = dict()
tempres_kwartiel = data.groupby('bird_name').calc_time_diff.describe()[kwartiel]
meeuwen_highfreq = list(tempres_kwartiel[tempres_kwartiel <= freq].index)
meeuwen_highfreq
for meeuw in meeuwen_highfreq:
hfreqdict[meeuw] = data.loc[(data['bird_name'] == meeuw) & (data['calc_time_diff'] <= freq)]
return hfreqdict
def FilterInterpolated(dfdictsynched):
dfdict = dfdictsynched
meeuwen = dfdict.keys()
for meeuw in meeuwen:
dfdict[meeuw].dropna(subset =['latitude'], inplace=True)
return dfdict
def Trajecten(dfdict):
lijnen_dfdict = dict()
meeuwen = list(dfdict.keys())
for meeuw in meeuwen:
#dfdict[meeuw] = dfdict[meeuw].set_index(pd.DatetimeIndex(dfdict[meeuw].date_time))
#geometry = [Point(xy) for xy in zip(dfdict[meeuw].longitude, dfdict[meeuw].latitude)]
punten_df = dfdict[meeuw] #GeoDataFrame(dfdict[meeuw], geometry=geometry)
crs = {'init': 'epsg:4326'}
grouped = punten_df.groupby([punten_df.index.year, punten_df.index.month, punten_df.index.day]).filter(lambda x: len(x) >1 )
grouped = grouped.groupby([grouped.index.year, grouped.index.month, grouped.index.day])['geometry'].apply(lambda x: LineString(x.tolist()))
grouped.index.rename(['jaar', 'maand', 'dag'], inplace = True)
lijnen_df = GeoDataFrame(grouped, crs = crs, geometry='geometry')
lijnen_df.reset_index(inplace=True)
lijnen_df = lijnen_df.to_crs({'init': 'epsg:31370'})
lijnen_df.to_file(r'C:\Users\maart\OneDrive\Master\Projectwerk Geo-ICT\TrajectenHighres\trajecten_{}'.format(meeuw), 'ESRI Shapefile')
lijnen_dfdict[meeuw] = lijnen_df
return lijnen_df
def PuntenShapefile(dfdict):
meeuwen = list(dfdict.keys())
df = dfdict
for meeuw in meeuwen:
df[meeuw].reset_index(inplace=True)
df[meeuw]['date']= str(df[meeuw])
df[meeuw] = df[meeuw].to_crs({'init': 'epsg:31370'})
print(df[meeuw].head())
df[meeuw].to_file(r'C:\Users\maart\OneDrive\Master\Projectwerk Geo-ICT\PuntenHighres\PuntenHighRes_{}'.format(meeuw), 'ESRI Shapefile')
return 'exported'
def ExportDataFrameCSV(dataframe):
dataframe.to_csv(r'C:\Users\maart\OneDrive\Master\Projectwerk Geo-ICT\PuntenHighres\puntenhighres.csv')
#ExportDataFrameCSV(mergeddf)
#data = ReadData(r'C:\Users\maart\OneDrive\Master\Projectwerk Geo-ICT\test\Hoogfrequentemeeuwen.csv')
#dfdict, meeuwen = MeeuwenDict(data)
#hfreqdict = FilterFrequentie(data, 5, '25%')
#dfdictsynched = SyncMeeuwen(hfreqdict, '1S', 5) #synchroniseren op 1 seconde, maximaal 5 waarden invullen
#dfdictsynchedfilterd = FilterInterpolated(dfdictsynched)
#mergeddf = Merge(dfdictsynchedfilterd)
#export shapefile van trajecten
#shplijn = Trajecten(dfdictsynchedfilterd)
#export shapefile van punten
#PuntenShapefile(dfdictsynchedfilterd)
#punten, afstandendf = BerekenAfstanden(mergeddf)
#buffer = 1000
#buren = BepaalBuren(afstandendf, meeuwen, buffer)
#sequenties = GetBuurSequentiesDict(buren, meeuwen)
#sequentiesdf = SequentieDictToDF(sequenties)
#sequentiesdfLoc = Locaties.BeginEind(sequentiesdf)
#sequentiesgdf = GeoDataFrame(sequentiesdfLoc, geometry=sequentiesdfLoc['geometry'], crs=31370)
def GetTrajectory(gull, t1, t2):
trackingpoints = mergeddf.loc[(gull, t1):(gull,t2)]
group = trackingpoints.groupby(level=0) #group by gull, even though there is only one gull, makes creating a line possible
lijn = group['geometry'].apply(lambda x: LineString(x.tolist()))
return lijn
def GetSequenceTrajectories():
trajectories = | pd.DataFrame(columns = ['gull pair', 'start', 'end','gull','geometry']) | pandas.DataFrame |
import MSfingerprinter.decoder as decoder
import MSfingerprinter.preprocessing as preprocessing
import MSfingerprinter.pysax as SAX
import MSfingerprinter.periodicityfinder as periodicityfinder
import MSfingerprinter.maxsubpatterntree as maxsubpatterntree
import MSfingerprinter.datacube as datacube
import MSfingerprinter.miningperiodicpatterns as miningperiodicpatterns
import MSfingerprinter.maxsubpatternhitset as maxsubpatternhitset
import MSfingerprinter.reactiontreecompletespace as reactiontreecompletespace
import MSfingerprinter.postprocessing as postprocessing
import MSfingerprinter.comparetrees as comparetrees
import matplotlib.pyplot as plt
import multiprocessing
import os
import math
import traceback
import sys
import ntpath
import csv
import itertools
import re
import sys
import pandas as pd
import treelib
import numpy as np
from collections import defaultdict
import gc
import timeit
# np.set_printoptions(threshold=np.inf)
class MSfingerprinter(object):
# MSfingerprinter object instance, constructor method
def __init__(self):
super(MSfingerprinter, self).__init__()
############################################################
#
# Postprocessing
#
###############################################################
def doPostProcessingSingleAlldiffs(self, resultfile1, repeatunitarray):
cwd = os.getcwd()
directory = cwd + '/MSfingerprinter/resultsPeriodicity/'
# outputfiles postprocessing
outfilemasspatterns = os.path.join(directory, 'postprocessedpatternsMASS' + resultfile1.rstrip('.txt') + 'MASSPATTERNS.csv')
# outfile massdifferences between and within patterns Mass and Freq
outfilepatternswithinmass = os.path.join('postprocessed' + resultfile1.rstrip('.txt') + 'DIFFWITHINPATTERNSMASS.txt')
# save similarity/dissimilarity of freq vs. mass space to file
periodicmasses, periodicfreqs = postprocessing.openresultssingle(resultfile1)
# gets from results periodicmasses and periodicfreqs
postperiodicmasses = postprocessing.retrieveperiodicmasses(periodicmasses)
return postperiodicmasses
def searchinTreesgroundtruthstart(self, meaningfuldiffpatterns, path, extensions):
# for later analysis saves patterns and rootnodes of trees found in initiatortrees
patternsplusrootnodes = []
nodesdetected = 0
treefilearray = list(comparetrees.find_files(path, extensions))
# for each tree
for tree in treefilearray:
nametree = ntpath.basename(tree).rstrip('.json')
data = comparetrees.getdata(tree)
nodesfound = []
counter = 0
parent = None
child = None
reactiontreeinstance = treelib.Tree()
reactiontreeinstance = comparetrees.retrievenodes(data, counter, parent, child, reactiontreeinstance)
for i in range(len(meaningfuldiffpatterns)):
# get for each meaningfulpattern the root and target
rootsubtree = round(meaningfuldiffpatterns[i][3], 6)
targetsubtree = round(meaningfuldiffpatterns[i][1],6)
pattern = meaningfuldiffpatterns[i][4]
try:
rootnode, stoichiometformula = comparetrees.searchMasspatterngroundtruth(reactiontreeinstance, rootsubtree, targetsubtree, pattern, nametree)
if rootnode != None and stoichiometformula != None:
print('original root value without rounding')
print(meaningfuldiffpatterns[i][3])
print('original target value without rounding')
print(meaningfuldiffpatterns[i][1])
patternsplusrootnodes.append([rootnode, rootsubtree, targetsubtree, stoichiometformula])
nodesfound.append(pattern)
except:
continue
print('all trees in directory : ' + path.rstrip('completeInitiatortrees/') + 'resultsubtrees/')
return patternsplusrootnodes
def searchinTreesgroundtruth(self, meaningfuldiffpatterns, path, extensions):
# for later analysis saves patterns and rootnodes of trees found in initiatortrees
patternsplusrootnodes = []
nodesdetected = 0
treefilearray = list(comparetrees.find_files(path, extensions))
# for each tree
for tree in treefilearray:
nametree = ntpath.basename(tree).rstrip('.json')
data = comparetrees.getdata(tree)
nodesfound = []
counter = 0
parent = None
child = None
reactiontreeinstance = treelib.Tree()
reactiontreeinstance = comparetrees.retrievenodes(data, counter, parent, child, reactiontreeinstance)
for i in range(len(meaningfuldiffpatterns)):
# get for each meaningfulpattern the root and target
rootsubtree = round(meaningfuldiffpatterns[i][1], 6)
targetsubtree = round(meaningfuldiffpatterns[i][3],6)
pattern = meaningfuldiffpatterns[i][4]
try:
rootnode, stoichiometformula = comparetrees.searchMasspatterngroundtruth(reactiontreeinstance, rootsubtree, targetsubtree, pattern, nametree)
if rootnode != None and stoichiometformula != None:
print('original root value without rounding')
print(meaningfuldiffpatterns[i][1])
print('original target value without rounding')
print(meaningfuldiffpatterns[i][3])
patternsplusrootnodes.append([rootnode, rootsubtree, targetsubtree, stoichiometformula])
nodesfound.append(pattern)
except:
continue
print('all trees in directory : ' + path.rstrip('completeInitiatortrees/') + 'resultsubtrees/')
return patternsplusrootnodes
def constructInitiatorTreescompletespace(self, Initiator, nodelist, massrange):
counter = 0
previousname = None
currenttrees = []
boolean = True
initiatorname = Initiator[0]
maxlevel, maxleveltwo = reactiontreecompletespace.getmaxtreelevel(nodelist, massrange)
treelen = len(Initiator)
for i in Initiator:
trees = reactiontreecompletespace.createRootsInitiators(Initiator)
counter += 1
while boolean == True:
for i in trees:
if len(currenttrees) == treelen:
trees = currenttrees
currenttrees = []
continue
else:
reactiontreeinstance, counter, previousname = reactiontreecompletespace.createnextlevelinitiator(nodelist, i, counter, maxlevel, maxleveltwo, previousname, initiatorname)
if reactiontreeinstance != None and treelen > 0:
currenttrees.append(reactiontreeinstance)
else:
treelen = treelen - 1
return
return
###############################################################################
#
# Preprocessing
#
#################################################################################3
# preprocessing data for Periodicityfindingalgorithms, returns function (i.e. intensity values)
# masspoints corresponding m/z values, Mass space
def preprocessMSSpectraMass(self, filename):
print('preprocessing raw data of ...'+ filename)
originaldata, sampleID = decoder.readdataframe(filename)
masspoints, function = preprocessing.FunctionMSMass(originaldata)
return function, masspoints
# preprocessing data for Periodicityfindingalgorithms, returns function (i.e. intensity values)
# Frequencyspace
def preprocessMSSpectraFreq(self, filename):
print('preprocessing raw data of ...'+ filename)
originaldata, sampleID = decoder.readdataframe(filename)
freqpoints, function = preprocessing.FunctionMSFreq(originaldata)
return function, freqpoints
def preprocess_directoryCSVfreqsampling(self, path, extensions, sampling, nprocesses=None):
filenames_to_preprocess = []
colnames = []
counter = 0
# Try to use the maximum amount of processes if not given.
try:
nprocesses = nprocesses or multiprocessing.cpu_count()
except NotImplementedError:
nprocesses = 1
else:
nprocesses = 1 if nprocesses <= 0 else nprocesses
for filename, _ in decoder.find_files(path, extensions):
filenames_to_preprocess.append(filename)
try:
filename = filename
except ValueError:
pass
# print('preprocessing raw data of ...'+ filename)
originaldata, sampleID = decoder.readdataframecsvfreqsampling(filename, sampling)
colnames.append(sampleID)
# three digits to make join possible
# originaldata['freq'] = originaldata['freq'].round(0)
if counter == 0:
originaldataframe = originaldata
counter+= 1
else:
originaldata = originaldata
merged = pd.concat([originaldataframe, originaldata], axis=1)
# merged = pd.merge(originaldataframe, originaldata, on='freq', how='outer')
originaldataframe = merged
originaldataframe.fillna(value=np.nan, inplace=True)
originaldataframe.columns = colnames
# originaldataframe.sort(columns='freq', inplace=True)
originaldataframe = originaldataframe.apply(np.log)
return originaldataframe
def preprocess_directoryCSVmasssampling(self, path, extensions, sampling, nprocesses=None):
filenames_to_preprocess = []
colnames = []
counter = 0
# Try to use the maximum amount of processes if not given.
try:
nprocesses = nprocesses or multiprocessing.cpu_count()
except NotImplementedError:
nprocesses = 1
else:
nprocesses = 1 if nprocesses <= 0 else nprocesses
for filename, _ in decoder.find_files(path, extensions):
filenames_to_preprocess.append(filename)
try:
filename = filename
except ValueError:
pass
# print('preprocessing raw data of ...'+ filename)
originaldata, sampleID = decoder.readdataframecsvmasssampling(filename, sampling)
colnames.append(sampleID)
# three digits to make join possible
# originaldata['mass'] = originaldata['mass'].round(3)
if counter == 0:
originaldataframe = originaldata
counter+= 1
else:
originaldata = originaldata
merged = pd.concat([originaldataframe, originaldata], axis=1)
# merged = pd.merge(originaldataframe, originaldata, on='freq', how='outer')
originaldataframe = merged
originaldataframe.fillna(value=999, inplace=True)
originaldataframe.columns = colnames
# originaldataframe.sort(columns='freq', inplace=True)
originaldataframe = originaldataframe.apply(np.log)
return originaldataframe
def getmaxminmass(self, path, extensions, nprocesses=None):
filenames_to_preprocess = []
minimum = 10000000000000000000000
maximum = 0
counter = 0
# Try to use the maximum amount of processes if not given.
try:
nprocesses = nprocesses or multiprocessing.cpu_count()
except NotImplementedError:
nprocesses = 1
else:
nprocesses = 1 if nprocesses <= 0 else nprocesses
for filename, _ in decoder.find_files(path, extensions):
filenames_to_preprocess.append(filename)
try:
filename = filename
except ValueError:
pass
print('preprocessing raw data of ...'+ filename)
originaldata, sampleID = decoder.readdataframe(filename)
# three digits to make join possible
originaldata['mass'] = originaldata['mass'].round(3)
maxmass = max(originaldata['mass'])
minmass = min(originaldata['mass'])
if maxmass > maximum:
maximum = maxmass
if minmass < minimum:
minimum = minmass
return maximum, minimum
#######################################################3
#
# Entropy based feature ranking
#
###############################################
# each feature is removed and total entropy is calculated
def RankFeatures(self, clusteringinputdata, rangenum):
featurenames = np.arange(len(clusteringinputdata))
bestfeatures = []
# produces a matrix with each column being one feature
X = np.array(clusteringinputdata)
totallengthfeatures = range(len(featurenames)/rangenum)
rangestart = 0
for i in totallengthfeatures:
rangeend = rangestart+rangenum
Xslice = X[rangestart:rangeend,0:10]
featurenamesslice = featurenames[rangestart:rangeend]
rankedfeatures = clustering.doRankfeatures(Xslice, featurenamesslice, rangestart, rangeend)
rangestart = rangeend
bestfeatureofsubset = rankedfeatures[-1]
bestfeatures.append(bestfeatureofsubset)
return bestfeatures
def doStats(self, entropyvectormass, entropyvectorfreq):
freqstats = clustering.statsentropy(entropyvectorfreq, 'freq')
massstats = clustering.statsentropy(entropyvectormass, 'mass')
correlatefreqmassentropy(rankedentropyvectormass, rankedentropyvectorfreq)
return freqstats, massstats
######################################################
#
# Time series standardization with SAX
#
########################################################
def standardizeTimeSeries(self, MSarray, timepoints):
# this uses the Symbolic aggregate approximation for time series discretization
# https://github.com/dolaameng/pysax/blob/master/Tutorial-SAX%20(Symbolic%20Aggregate%20Approximation).ipynb
# MS = decoder.readMStimedomaintransient(filename)
# MSarray = np.asarray(MS.columns.tolist(), dtype=float)
print('length original data')
print(len(MSarray))
# this does symbolization for each window, no overlap of windows
sax = SAX.SAXModel()
# standardizes the time series whiten accross windows
normalizedMSarray = sax.whiten(MSarray)
print('mean and standarddeviation of standardized MS: ')
print(normalizedMSarray.mean(), normalizedMSarray.std())
# saves MSarray (TS) to csv for later cube generation
dforiginalTS = | pd.DataFrame(normalizedMSarray) | pandas.DataFrame |
import copy
import re
from textwrap import dedent
import numpy as np
import pytest
import pandas as pd
from pandas import (
DataFrame,
MultiIndex,
)
import pandas._testing as tm
jinja2 = pytest.importorskip("jinja2")
from pandas.io.formats.style import ( # isort:skip
Styler,
)
from pandas.io.formats.style_render import (
_get_level_lengths,
_get_trimming_maximums,
maybe_convert_css_to_tuples,
non_reducing_slice,
)
@pytest.fixture
def mi_df():
return DataFrame(
[[1, 2], [3, 4]],
index=MultiIndex.from_product([["i0"], ["i1_a", "i1_b"]]),
columns=MultiIndex.from_product([["c0"], ["c1_a", "c1_b"]]),
dtype=int,
)
@pytest.fixture
def mi_styler(mi_df):
return Styler(mi_df, uuid_len=0)
@pytest.fixture
def mi_styler_comp(mi_styler):
# comprehensively add features to mi_styler
mi_styler = mi_styler._copy(deepcopy=True)
mi_styler.css = {**mi_styler.css, **{"row": "ROW", "col": "COL"}}
mi_styler.uuid_len = 5
mi_styler.uuid = "abcde"
mi_styler.set_caption("capt")
mi_styler.set_table_styles([{"selector": "a", "props": "a:v;"}])
mi_styler.hide(axis="columns")
mi_styler.hide([("c0", "c1_a")], axis="columns", names=True)
mi_styler.hide(axis="index")
mi_styler.hide([("i0", "i1_a")], axis="index", names=True)
mi_styler.set_table_attributes('class="box"')
mi_styler.format(na_rep="MISSING", precision=3)
mi_styler.format_index(precision=2, axis=0)
mi_styler.format_index(precision=4, axis=1)
mi_styler.highlight_max(axis=None)
mi_styler.applymap_index(lambda x: "color: white;", axis=0)
mi_styler.applymap_index(lambda x: "color: black;", axis=1)
mi_styler.set_td_classes(
DataFrame(
[["a", "b"], ["a", "c"]], index=mi_styler.index, columns=mi_styler.columns
)
)
mi_styler.set_tooltips(
DataFrame(
[["a2", "b2"], ["a2", "c2"]],
index=mi_styler.index,
columns=mi_styler.columns,
)
)
return mi_styler
@pytest.mark.parametrize(
"sparse_columns, exp_cols",
[
(
True,
[
{"is_visible": True, "attributes": 'colspan="2"', "value": "c0"},
{"is_visible": False, "attributes": "", "value": "c0"},
],
),
(
False,
[
{"is_visible": True, "attributes": "", "value": "c0"},
{"is_visible": True, "attributes": "", "value": "c0"},
],
),
],
)
def test_mi_styler_sparsify_columns(mi_styler, sparse_columns, exp_cols):
exp_l1_c0 = {"is_visible": True, "attributes": "", "display_value": "c1_a"}
exp_l1_c1 = {"is_visible": True, "attributes": "", "display_value": "c1_b"}
ctx = mi_styler._translate(True, sparse_columns)
assert exp_cols[0].items() <= ctx["head"][0][2].items()
assert exp_cols[1].items() <= ctx["head"][0][3].items()
assert exp_l1_c0.items() <= ctx["head"][1][2].items()
assert exp_l1_c1.items() <= ctx["head"][1][3].items()
@pytest.mark.parametrize(
"sparse_index, exp_rows",
[
(
True,
[
{"is_visible": True, "attributes": 'rowspan="2"', "value": "i0"},
{"is_visible": False, "attributes": "", "value": "i0"},
],
),
(
False,
[
{"is_visible": True, "attributes": "", "value": "i0"},
{"is_visible": True, "attributes": "", "value": "i0"},
],
),
],
)
def test_mi_styler_sparsify_index(mi_styler, sparse_index, exp_rows):
exp_l1_r0 = {"is_visible": True, "attributes": "", "display_value": "i1_a"}
exp_l1_r1 = {"is_visible": True, "attributes": "", "display_value": "i1_b"}
ctx = mi_styler._translate(sparse_index, True)
assert exp_rows[0].items() <= ctx["body"][0][0].items()
assert exp_rows[1].items() <= ctx["body"][1][0].items()
assert exp_l1_r0.items() <= ctx["body"][0][1].items()
assert exp_l1_r1.items() <= ctx["body"][1][1].items()
def test_mi_styler_sparsify_options(mi_styler):
with pd.option_context("styler.sparse.index", False):
html1 = mi_styler.to_html()
with pd.option_context("styler.sparse.index", True):
html2 = mi_styler.to_html()
assert html1 != html2
with pd.option_context("styler.sparse.columns", False):
html1 = mi_styler.to_html()
with pd.option_context("styler.sparse.columns", True):
html2 = mi_styler.to_html()
assert html1 != html2
@pytest.mark.parametrize(
"rn, cn, max_els, max_rows, max_cols, exp_rn, exp_cn",
[
(100, 100, 100, None, None, 12, 6), # reduce to (12, 6) < 100 elements
(1000, 3, 750, None, None, 250, 3), # dynamically reduce rows to 250, keep cols
(4, 1000, 500, None, None, 4, 125), # dynamically reduce cols to 125, keep rows
(1000, 3, 750, 10, None, 10, 3), # overwrite above dynamics with max_row
(4, 1000, 500, None, 5, 4, 5), # overwrite above dynamics with max_col
(100, 100, 700, 50, 50, 25, 25), # rows cols below given maxes so < 700 elmts
],
)
def test_trimming_maximum(rn, cn, max_els, max_rows, max_cols, exp_rn, exp_cn):
rn, cn = _get_trimming_maximums(
rn, cn, max_els, max_rows, max_cols, scaling_factor=0.5
)
assert (rn, cn) == (exp_rn, exp_cn)
@pytest.mark.parametrize(
"option, val",
[
("styler.render.max_elements", 6),
("styler.render.max_rows", 3),
],
)
def test_render_trimming_rows(option, val):
# test auto and specific trimming of rows
df = DataFrame(np.arange(120).reshape(60, 2))
with pd.option_context(option, val):
ctx = df.style._translate(True, True)
assert len(ctx["head"][0]) == 3 # index + 2 data cols
assert len(ctx["body"]) == 4 # 3 data rows + trimming row
assert len(ctx["body"][0]) == 3 # index + 2 data cols
@pytest.mark.parametrize(
"option, val",
[
("styler.render.max_elements", 6),
("styler.render.max_columns", 2),
],
)
def test_render_trimming_cols(option, val):
# test auto and specific trimming of cols
df = DataFrame(np.arange(30).reshape(3, 10))
with pd.option_context(option, val):
ctx = df.style._translate(True, True)
assert len(ctx["head"][0]) == 4 # index + 2 data cols + trimming col
assert len(ctx["body"]) == 3 # 3 data rows
assert len(ctx["body"][0]) == 4 # index + 2 data cols + trimming col
def test_render_trimming_mi():
midx = MultiIndex.from_product([[1, 2], [1, 2, 3]])
df = DataFrame(np.arange(36).reshape(6, 6), columns=midx, index=midx)
with pd.option_context("styler.render.max_elements", 4):
ctx = df.style._translate(True, True)
assert len(ctx["body"][0]) == 5 # 2 indexes + 2 data cols + trimming row
assert {"attributes": 'rowspan="2"'}.items() <= ctx["body"][0][0].items()
assert {"class": "data row0 col_trim"}.items() <= ctx["body"][0][4].items()
assert {"class": "data row_trim col_trim"}.items() <= ctx["body"][2][4].items()
assert len(ctx["body"]) == 3 # 2 data rows + trimming row
assert len(ctx["head"][0]) == 5 # 2 indexes + 2 column headers + trimming col
assert {"attributes": 'colspan="2"'}.items() <= ctx["head"][0][2].items()
def test_render_empty_mi():
# GH 43305
df = DataFrame(index=MultiIndex.from_product([["A"], [0, 1]], names=[None, "one"]))
expected = dedent(
"""\
>
<thead>
<tr>
<th class="index_name level0" > </th>
<th class="index_name level1" >one</th>
</tr>
</thead>
"""
)
assert expected in df.style.to_html()
@pytest.mark.parametrize("comprehensive", [True, False])
@pytest.mark.parametrize("render", [True, False])
@pytest.mark.parametrize("deepcopy", [True, False])
def test_copy(comprehensive, render, deepcopy, mi_styler, mi_styler_comp):
styler = mi_styler_comp if comprehensive else mi_styler
styler.uuid_len = 5
s2 = copy.deepcopy(styler) if deepcopy else copy.copy(styler) # make copy and check
assert s2 is not styler
if render:
styler.to_html()
excl = [
"na_rep", # deprecated
"precision", # deprecated
"cellstyle_map", # render time vars..
"cellstyle_map_columns",
"cellstyle_map_index",
"template_latex", # render templates are class level
"template_html",
"template_html_style",
"template_html_table",
]
if not deepcopy: # check memory locations are equal for all included attributes
for attr in [a for a in styler.__dict__ if (not callable(a) and a not in excl)]:
assert id(getattr(s2, attr)) == id(getattr(styler, attr))
else: # check memory locations are different for nested or mutable vars
shallow = [
"data",
"columns",
"index",
"uuid_len",
"uuid",
"caption",
"cell_ids",
"hide_index_",
"hide_columns_",
"hide_index_names",
"hide_column_names",
"table_attributes",
]
for attr in shallow:
assert id(getattr(s2, attr)) == id(getattr(styler, attr))
for attr in [
a
for a in styler.__dict__
if (not callable(a) and a not in excl and a not in shallow)
]:
if getattr(s2, attr) is None:
assert id(getattr(s2, attr)) == id(getattr(styler, attr))
else:
assert id(getattr(s2, attr)) != id(getattr(styler, attr))
def test_clear(mi_styler_comp):
# NOTE: if this test fails for new features then 'mi_styler_comp' should be updated
# to ensure proper testing of the 'copy', 'clear', 'export' methods with new feature
# GH 40675
styler = mi_styler_comp
styler._compute() # execute applied methods
clean_copy = Styler(styler.data, uuid=styler.uuid)
excl = [
"data",
"index",
"columns",
"uuid",
"uuid_len", # uuid is set to be the same on styler and clean_copy
"cell_ids",
"cellstyle_map", # execution time only
"cellstyle_map_columns", # execution time only
"cellstyle_map_index", # execution time only
"precision", # deprecated
"na_rep", # deprecated
"template_latex", # render templates are class level
"template_html",
"template_html_style",
"template_html_table",
]
# tests vars are not same vals on obj and clean copy before clear (except for excl)
for attr in [a for a in styler.__dict__ if not (callable(a) or a in excl)]:
res = getattr(styler, attr) == getattr(clean_copy, attr)
assert not (all(res) if (hasattr(res, "__iter__") and len(res) > 0) else res)
# test vars have same vales on obj and clean copy after clearing
styler.clear()
for attr in [a for a in styler.__dict__ if not (callable(a))]:
res = getattr(styler, attr) == getattr(clean_copy, attr)
assert all(res) if hasattr(res, "__iter__") else res
def test_export(mi_styler_comp, mi_styler):
exp_attrs = [
"_todo",
"hide_index_",
"hide_index_names",
"hide_columns_",
"hide_column_names",
"table_attributes",
"table_styles",
"css",
]
for attr in exp_attrs:
check = getattr(mi_styler, attr) == getattr(mi_styler_comp, attr)
assert not (
all(check) if (hasattr(check, "__iter__") and len(check) > 0) else check
)
export = mi_styler_comp.export()
used = mi_styler.use(export)
for attr in exp_attrs:
check = getattr(used, attr) == getattr(mi_styler_comp, attr)
assert all(check) if (hasattr(check, "__iter__") and len(check) > 0) else check
used.to_html()
def test_hide_raises(mi_styler):
msg = "`subset` and `level` cannot be passed simultaneously"
with pytest.raises(ValueError, match=msg):
mi_styler.hide(axis="index", subset="something", level="something else")
msg = "`level` must be of type `int`, `str` or list of such"
with pytest.raises(ValueError, match=msg):
mi_styler.hide(axis="index", level={"bad": 1, "type": 2})
@pytest.mark.parametrize("level", [1, "one", [1], ["one"]])
def test_hide_index_level(mi_styler, level):
mi_styler.index.names, mi_styler.columns.names = ["zero", "one"], ["zero", "one"]
ctx = mi_styler.hide(axis="index", level=level)._translate(False, True)
assert len(ctx["head"][0]) == 3
assert len(ctx["head"][1]) == 3
assert len(ctx["head"][2]) == 4
assert ctx["head"][2][0]["is_visible"]
assert not ctx["head"][2][1]["is_visible"]
assert ctx["body"][0][0]["is_visible"]
assert not ctx["body"][0][1]["is_visible"]
assert ctx["body"][1][0]["is_visible"]
assert not ctx["body"][1][1]["is_visible"]
@pytest.mark.parametrize("level", [1, "one", [1], ["one"]])
@pytest.mark.parametrize("names", [True, False])
def test_hide_columns_level(mi_styler, level, names):
mi_styler.columns.names = ["zero", "one"]
if names:
mi_styler.index.names = ["zero", "one"]
ctx = mi_styler.hide(axis="columns", level=level)._translate(True, False)
assert len(ctx["head"]) == (2 if names else 1)
@pytest.mark.parametrize("method", ["applymap", "apply"])
@pytest.mark.parametrize("axis", ["index", "columns"])
def test_apply_map_header(method, axis):
# GH 41893
df = DataFrame({"A": [0, 0], "B": [1, 1]}, index=["C", "D"])
func = {
"apply": lambda s: ["attr: val" if ("A" in v or "C" in v) else "" for v in s],
"applymap": lambda v: "attr: val" if ("A" in v or "C" in v) else "",
}
# test execution added to todo
result = getattr(df.style, f"{method}_index")(func[method], axis=axis)
assert len(result._todo) == 1
assert len(getattr(result, f"ctx_{axis}")) == 0
# test ctx object on compute
result._compute()
expected = {
(0, 0): [("attr", "val")],
}
assert getattr(result, f"ctx_{axis}") == expected
@pytest.mark.parametrize("method", ["apply", "applymap"])
@pytest.mark.parametrize("axis", ["index", "columns"])
def test_apply_map_header_mi(mi_styler, method, axis):
# GH 41893
func = {
"apply": lambda s: ["attr: val;" if "b" in v else "" for v in s],
"applymap": lambda v: "attr: val" if "b" in v else "",
}
result = getattr(mi_styler, f"{method}_index")(func[method], axis=axis)._compute()
expected = {(1, 1): [("attr", "val")]}
assert getattr(result, f"ctx_{axis}") == expected
def test_apply_map_header_raises(mi_styler):
# GH 41893
with pytest.raises(ValueError, match="No axis named bad for object type DataFrame"):
mi_styler.applymap_index(lambda v: "attr: val;", axis="bad")._compute()
class TestStyler:
def setup_method(self, method):
np.random.seed(24)
self.s = DataFrame({"A": np.random.permutation(range(6))})
self.df = DataFrame({"A": [0, 1], "B": np.random.randn(2)})
self.f = lambda x: x
self.g = lambda x: x
def h(x, foo="bar"):
return pd.Series(f"color: {foo}", index=x.index, name=x.name)
self.h = h
self.styler = Styler(self.df)
self.attrs = DataFrame({"A": ["color: red", "color: blue"]})
self.dataframes = [
self.df,
DataFrame(
{"f": [1.0, 2.0], "o": ["a", "b"], "c": pd.Categorical(["a", "b"])}
),
]
self.blank_value = " "
def test_init_non_pandas(self):
msg = "``data`` must be a Series or DataFrame"
with pytest.raises(TypeError, match=msg):
Styler([1, 2, 3])
def test_init_series(self):
result = Styler(pd.Series([1, 2]))
assert result.data.ndim == 2
def test_repr_html_ok(self):
self.styler._repr_html_()
def test_repr_html_mathjax(self):
# gh-19824 / 41395
assert "tex2jax_ignore" not in self.styler._repr_html_()
with pd.option_context("styler.html.mathjax", False):
assert "tex2jax_ignore" in self.styler._repr_html_()
def test_update_ctx(self):
self.styler._update_ctx(self.attrs)
expected = {(0, 0): [("color", "red")], (1, 0): [("color", "blue")]}
assert self.styler.ctx == expected
def test_update_ctx_flatten_multi_and_trailing_semi(self):
attrs = DataFrame({"A": ["color: red; foo: bar", "color:blue ; foo: baz;"]})
self.styler._update_ctx(attrs)
expected = {
(0, 0): [("color", "red"), ("foo", "bar")],
(1, 0): [("color", "blue"), ("foo", "baz")],
}
assert self.styler.ctx == expected
def test_render(self):
df = DataFrame({"A": [0, 1]})
style = lambda x: pd.Series(["color: red", "color: blue"], name=x.name)
s = Styler(df, uuid="AB").apply(style)
s.to_html()
# it worked?
def test_multiple_render(self):
# GH 39396
s = Styler(self.df, uuid_len=0).applymap(lambda x: "color: red;", subset=["A"])
s.to_html() # do 2 renders to ensure css styles not duplicated
assert (
'<style type="text/css">\n#T__row0_col0, #T__row1_col0 {\n'
" color: red;\n}\n</style>" in s.to_html()
)
def test_render_empty_dfs(self):
empty_df = DataFrame()
es = Styler(empty_df)
es.to_html()
# An index but no columns
DataFrame(columns=["a"]).style.to_html()
# A column but no index
DataFrame(index=["a"]).style.to_html()
# No IndexError raised?
def test_render_double(self):
df = DataFrame({"A": [0, 1]})
style = lambda x: pd.Series(
["color: red; border: 1px", "color: blue; border: 2px"], name=x.name
)
s = Styler(df, uuid="AB").apply(style)
s.to_html()
# it worked?
def test_set_properties(self):
df = DataFrame({"A": [0, 1]})
result = df.style.set_properties(color="white", size="10px")._compute().ctx
# order is deterministic
v = [("color", "white"), ("size", "10px")]
expected = {(0, 0): v, (1, 0): v}
assert result.keys() == expected.keys()
for v1, v2 in zip(result.values(), expected.values()):
assert sorted(v1) == sorted(v2)
def test_set_properties_subset(self):
df = DataFrame({"A": [0, 1]})
result = (
df.style.set_properties(subset=pd.IndexSlice[0, "A"], color="white")
._compute()
.ctx
)
expected = {(0, 0): [("color", "white")]}
assert result == expected
def test_empty_index_name_doesnt_display(self):
# https://github.com/pandas-dev/pandas/pull/12090#issuecomment-180695902
df = DataFrame({"A": [1, 2], "B": [3, 4], "C": [5, 6]})
result = df.style._translate(True, True)
assert len(result["head"]) == 1
expected = {
"class": "blank level0",
"type": "th",
"value": self.blank_value,
"is_visible": True,
"display_value": self.blank_value,
}
assert expected.items() <= result["head"][0][0].items()
def test_index_name(self):
# https://github.com/pandas-dev/pandas/issues/11655
df = DataFrame({"A": [1, 2], "B": [3, 4], "C": [5, 6]})
result = df.set_index("A").style._translate(True, True)
expected = {
"class": "index_name level0",
"type": "th",
"value": "A",
"is_visible": True,
"display_value": "A",
}
assert expected.items() <= result["head"][1][0].items()
def test_multiindex_name(self):
# https://github.com/pandas-dev/pandas/issues/11655
df = DataFrame({"A": [1, 2], "B": [3, 4], "C": [5, 6]})
result = df.set_index(["A", "B"]).style._translate(True, True)
expected = [
{
"class": "index_name level0",
"type": "th",
"value": "A",
"is_visible": True,
"display_value": "A",
},
{
"class": "index_name level1",
"type": "th",
"value": "B",
"is_visible": True,
"display_value": "B",
},
{
"class": "blank col0",
"type": "th",
"value": self.blank_value,
"is_visible": True,
"display_value": self.blank_value,
},
]
assert result["head"][1] == expected
def test_numeric_columns(self):
# https://github.com/pandas-dev/pandas/issues/12125
# smoke test for _translate
df = DataFrame({0: [1, 2, 3]})
df.style._translate(True, True)
def test_apply_axis(self):
df = DataFrame({"A": [0, 0], "B": [1, 1]})
f = lambda x: [f"val: {x.max()}" for v in x]
result = df.style.apply(f, axis=1)
assert len(result._todo) == 1
assert len(result.ctx) == 0
result._compute()
expected = {
(0, 0): [("val", "1")],
(0, 1): [("val", "1")],
(1, 0): [("val", "1")],
(1, 1): [("val", "1")],
}
assert result.ctx == expected
result = df.style.apply(f, axis=0)
expected = {
(0, 0): [("val", "0")],
(0, 1): [("val", "1")],
(1, 0): [("val", "0")],
(1, 1): [("val", "1")],
}
result._compute()
assert result.ctx == expected
result = df.style.apply(f) # default
result._compute()
assert result.ctx == expected
@pytest.mark.parametrize("axis", [0, 1])
def test_apply_series_return(self, axis):
# GH 42014
df = DataFrame([[1, 2], [3, 4]], index=["X", "Y"], columns=["X", "Y"])
# test Series return where len(Series) < df.index or df.columns but labels OK
func = lambda s: pd.Series(["color: red;"], index=["Y"])
result = df.style.apply(func, axis=axis)._compute().ctx
assert result[(1, 1)] == [("color", "red")]
assert result[(1 - axis, axis)] == [("color", "red")]
# test Series return where labels align but different order
func = lambda s: pd.Series(["color: red;", "color: blue;"], index=["Y", "X"])
result = df.style.apply(func, axis=axis)._compute().ctx
assert result[(0, 0)] == [("color", "blue")]
assert result[(1, 1)] == [("color", "red")]
assert result[(1 - axis, axis)] == [("color", "red")]
assert result[(axis, 1 - axis)] == [("color", "blue")]
@pytest.mark.parametrize("index", [False, True])
@pytest.mark.parametrize("columns", [False, True])
def test_apply_dataframe_return(self, index, columns):
# GH 42014
df = DataFrame([[1, 2], [3, 4]], index=["X", "Y"], columns=["X", "Y"])
idxs = ["X", "Y"] if index else ["Y"]
cols = ["X", "Y"] if columns else ["Y"]
df_styles = DataFrame("color: red;", index=idxs, columns=cols)
result = df.style.apply(lambda x: df_styles, axis=None)._compute().ctx
assert result[(1, 1)] == [("color", "red")] # (Y,Y) styles always present
assert (result[(0, 1)] == [("color", "red")]) is index # (X,Y) only if index
assert (result[(1, 0)] == [("color", "red")]) is columns # (Y,X) only if cols
assert (result[(0, 0)] == [("color", "red")]) is (index and columns) # (X,X)
@pytest.mark.parametrize(
"slice_",
[
pd.IndexSlice[:],
pd.IndexSlice[:, ["A"]],
pd.IndexSlice[[1], :],
pd.IndexSlice[[1], ["A"]],
pd.IndexSlice[:2, ["A", "B"]],
],
)
@pytest.mark.parametrize("axis", [0, 1])
def test_apply_subset(self, slice_, axis):
result = (
self.df.style.apply(self.h, axis=axis, subset=slice_, foo="baz")
._compute()
.ctx
)
expected = {
(r, c): [("color", "baz")]
for r, row in enumerate(self.df.index)
for c, col in enumerate(self.df.columns)
if row in self.df.loc[slice_].index and col in self.df.loc[slice_].columns
}
assert result == expected
@pytest.mark.parametrize(
"slice_",
[
pd.IndexSlice[:],
pd.IndexSlice[:, ["A"]],
pd.IndexSlice[[1], :],
pd.IndexSlice[[1], ["A"]],
pd.IndexSlice[:2, ["A", "B"]],
],
)
def test_applymap_subset(self, slice_):
result = (
self.df.style.applymap(lambda x: "color:baz;", subset=slice_)._compute().ctx
)
expected = {
(r, c): [("color", "baz")]
for r, row in enumerate(self.df.index)
for c, col in enumerate(self.df.columns)
if row in self.df.loc[slice_].index and col in self.df.loc[slice_].columns
}
assert result == expected
@pytest.mark.parametrize(
"slice_",
[
pd.IndexSlice[:, pd.IndexSlice["x", "A"]],
pd.IndexSlice[:, pd.IndexSlice[:, "A"]],
pd.IndexSlice[:, pd.IndexSlice[:, ["A", "C"]]], # missing col element
pd.IndexSlice[pd.IndexSlice["a", 1], :],
pd.IndexSlice[pd.IndexSlice[:, 1], :],
pd.IndexSlice[pd.IndexSlice[:, [1, 3]], :], # missing row element
pd.IndexSlice[:, ("x", "A")],
pd.IndexSlice[("a", 1), :],
],
)
def test_applymap_subset_multiindex(self, slice_):
# GH 19861
# edited for GH 33562
warn = None
msg = "indexing on a MultiIndex with a nested sequence of labels"
if (
isinstance(slice_[-1], tuple)
and isinstance(slice_[-1][-1], list)
and "C" in slice_[-1][-1]
):
warn = FutureWarning
elif (
isinstance(slice_[0], tuple)
and isinstance(slice_[0][1], list)
and 3 in slice_[0][1]
):
warn = FutureWarning
idx = MultiIndex.from_product([["a", "b"], [1, 2]])
col = MultiIndex.from_product([["x", "y"], ["A", "B"]])
df = DataFrame(np.random.rand(4, 4), columns=col, index=idx)
with tm.assert_produces_warning(warn, match=msg, check_stacklevel=False):
df.style.applymap(lambda x: "color: red;", subset=slice_).to_html()
def test_applymap_subset_multiindex_code(self):
# https://github.com/pandas-dev/pandas/issues/25858
# Checks styler.applymap works with multindex when codes are provided
codes = np.array([[0, 0, 1, 1], [0, 1, 0, 1]])
columns = MultiIndex(
levels=[["a", "b"], ["%", "#"]], codes=codes, names=["", ""]
)
df = DataFrame(
[[1, -1, 1, 1], [-1, 1, 1, 1]], index=["hello", "world"], columns=columns
)
pct_subset = pd.IndexSlice[:, pd.IndexSlice[:, "%":"%"]]
def color_negative_red(val):
color = "red" if val < 0 else "black"
return f"color: {color}"
df.loc[pct_subset]
df.style.applymap(color_negative_red, subset=pct_subset)
def test_empty(self):
df = DataFrame({"A": [1, 0]})
s = df.style
s.ctx = {(0, 0): [("color", "red")], (1, 0): [("", "")]}
result = s._translate(True, True)["cellstyle"]
expected = [
{"props": [("color", "red")], "selectors": ["row0_col0"]},
{"props": [("", "")], "selectors": ["row1_col0"]},
]
assert result == expected
def test_duplicate(self):
df = DataFrame({"A": [1, 0]})
s = df.style
s.ctx = {(0, 0): [("color", "red")], (1, 0): [("color", "red")]}
result = s._translate(True, True)["cellstyle"]
expected = [
{"props": [("color", "red")], "selectors": ["row0_col0", "row1_col0"]}
]
assert result == expected
def test_init_with_na_rep(self):
# GH 21527 28358
df = DataFrame([[None, None], [1.1, 1.2]], columns=["A", "B"])
ctx = Styler(df, na_rep="NA")._translate(True, True)
assert ctx["body"][0][1]["display_value"] == "NA"
assert ctx["body"][0][2]["display_value"] == "NA"
def test_caption(self):
styler = Styler(self.df, caption="foo")
result = styler.to_html()
assert all(["caption" in result, "foo" in result])
styler = self.df.style
result = styler.set_caption("baz")
assert styler is result
assert styler.caption == "baz"
def test_uuid(self):
styler = Styler(self.df, uuid="abc123")
result = styler.to_html()
assert "abc123" in result
styler = self.df.style
result = styler.set_uuid("aaa")
assert result is styler
assert result.uuid == "aaa"
def test_unique_id(self):
# See https://github.com/pandas-dev/pandas/issues/16780
df = DataFrame({"a": [1, 3, 5, 6], "b": [2, 4, 12, 21]})
result = df.style.to_html(uuid="test")
assert "test" in result
ids = re.findall('id="(.*?)"', result)
assert np.unique(ids).size == len(ids)
def test_table_styles(self):
style = [{"selector": "th", "props": [("foo", "bar")]}] # default format
styler = Styler(self.df, table_styles=style)
result = " ".join(styler.to_html().split())
assert "th { foo: bar; }" in result
styler = self.df.style
result = styler.set_table_styles(style)
assert styler is result
assert styler.table_styles == style
# GH 39563
style = [{"selector": "th", "props": "foo:bar;"}] # css string format
styler = self.df.style.set_table_styles(style)
result = " ".join(styler.to_html().split())
assert "th { foo: bar; }" in result
def test_table_styles_multiple(self):
ctx = self.df.style.set_table_styles(
[
{"selector": "th,td", "props": "color:red;"},
{"selector": "tr", "props": "color:green;"},
]
)._translate(True, True)["table_styles"]
assert ctx == [
{"selector": "th", "props": [("color", "red")]},
{"selector": "td", "props": [("color", "red")]},
{"selector": "tr", "props": [("color", "green")]},
]
def test_table_styles_dict_multiple_selectors(self):
# GH 44011
result = self.df.style.set_table_styles(
[{"selector": "th,td", "props": [("border-left", "2px solid black")]}]
)._translate(True, True)["table_styles"]
expected = [
{"selector": "th", "props": [("border-left", "2px solid black")]},
{"selector": "td", "props": [("border-left", "2px solid black")]},
]
assert result == expected
def test_maybe_convert_css_to_tuples(self):
expected = [("a", "b"), ("c", "d e")]
assert maybe_convert_css_to_tuples("a:b;c:d e;") == expected
assert maybe_convert_css_to_tuples("a: b ;c: d e ") == expected
expected = []
assert maybe_convert_css_to_tuples("") == expected
def test_maybe_convert_css_to_tuples_err(self):
msg = "Styles supplied as string must follow CSS rule formats"
with pytest.raises(ValueError, match=msg):
maybe_convert_css_to_tuples("err")
def test_table_attributes(self):
attributes = 'class="foo" data-bar'
styler = Styler(self.df, table_attributes=attributes)
result = styler.to_html()
assert 'class="foo" data-bar' in result
result = self.df.style.set_table_attributes(attributes).to_html()
assert 'class="foo" data-bar' in result
def test_apply_none(self):
def f(x):
return DataFrame(
np.where(x == x.max(), "color: red", ""),
index=x.index,
columns=x.columns,
)
result = DataFrame([[1, 2], [3, 4]]).style.apply(f, axis=None)._compute().ctx
assert result[(1, 1)] == [("color", "red")]
def test_trim(self):
result = self.df.style.to_html() # trim=True
assert result.count("#") == 0
result = self.df.style.highlight_max().to_html()
assert result.count("#") == len(self.df.columns)
def test_export(self):
f = lambda x: "color: red" if x > 0 else "color: blue"
g = lambda x, z: f"color: {z}" if x > 0 else f"color: {z}"
style1 = self.styler
style1.applymap(f).applymap(g, z="b").highlight_max()._compute() # = render
result = style1.export()
style2 = self.df.style
style2.use(result)
assert style1._todo == style2._todo
style2.to_html()
def test_bad_apply_shape(self):
df = DataFrame([[1, 2], [3, 4]], index=["A", "B"], columns=["X", "Y"])
msg = "resulted in the apply method collapsing to a Series."
with pytest.raises(ValueError, match=msg):
df.style._apply(lambda x: "x")
msg = "created invalid {} labels"
with pytest.raises(ValueError, match=msg.format("index")):
df.style._apply(lambda x: [""])
with pytest.raises(ValueError, match=msg.format("index")):
df.style._apply(lambda x: ["", "", "", ""])
with pytest.raises(ValueError, match=msg.format("index")):
df.style._apply(lambda x: pd.Series(["a:v;", ""], index=["A", "C"]), axis=0)
with pytest.raises(ValueError, match=msg.format("columns")):
df.style._apply(lambda x: ["", "", ""], axis=1)
with pytest.raises(ValueError, match=msg.format("columns")):
df.style._apply(lambda x: pd.Series(["a:v;", ""], index=["X", "Z"]), axis=1)
msg = "returned ndarray with wrong shape"
with pytest.raises(ValueError, match=msg):
df.style._apply(lambda x: np.array([[""], [""]]), axis=None)
def test_apply_bad_return(self):
def f(x):
return ""
df = DataFrame([[1, 2], [3, 4]])
msg = (
"must return a DataFrame or ndarray when passed to `Styler.apply` "
"with axis=None"
)
with pytest.raises(TypeError, match=msg):
df.style._apply(f, axis=None)
@pytest.mark.parametrize("axis", ["index", "columns"])
def test_apply_bad_labels(self, axis):
def f(x):
return DataFrame(**{axis: ["bad", "labels"]})
df = DataFrame([[1, 2], [3, 4]])
msg = f"created invalid {axis} labels."
with pytest.raises(ValueError, match=msg):
df.style._apply(f, axis=None)
def test_get_level_lengths(self):
index = MultiIndex.from_product([["a", "b"], [0, 1, 2]])
expected = {
(0, 0): 3,
(0, 3): 3,
(1, 0): 1,
(1, 1): 1,
(1, 2): 1,
(1, 3): 1,
(1, 4): 1,
(1, 5): 1,
}
result = _get_level_lengths(index, sparsify=True, max_index=100)
tm.assert_dict_equal(result, expected)
expected = {
(0, 0): 1,
(0, 1): 1,
(0, 2): 1,
(0, 3): 1,
(0, 4): 1,
(0, 5): 1,
(1, 0): 1,
(1, 1): 1,
(1, 2): 1,
(1, 3): 1,
(1, 4): 1,
(1, 5): 1,
}
result = _get_level_lengths(index, sparsify=False, max_index=100)
tm.assert_dict_equal(result, expected)
def test_get_level_lengths_un_sorted(self):
index = MultiIndex.from_arrays([[1, 1, 2, 1], ["a", "b", "b", "d"]])
expected = {
(0, 0): 2,
(0, 2): 1,
(0, 3): 1,
(1, 0): 1,
(1, 1): 1,
(1, 2): 1,
(1, 3): 1,
}
result = _get_level_lengths(index, sparsify=True, max_index=100)
tm.assert_dict_equal(result, expected)
expected = {
(0, 0): 1,
(0, 1): 1,
(0, 2): 1,
(0, 3): 1,
(1, 0): 1,
(1, 1): 1,
(1, 2): 1,
(1, 3): 1,
}
result = _get_level_lengths(index, sparsify=False, max_index=100)
tm.assert_dict_equal(result, expected)
def test_mi_sparse_index_names(self):
# TODO this test is verbose can be minimised to more directly target test
df = DataFrame(
{"A": [1, 2]},
index=MultiIndex.from_arrays(
[["a", "a"], [0, 1]], names=["idx_level_0", "idx_level_1"]
),
)
result = df.style._translate(True, True)
head = result["head"][1]
expected = [
{
"class": "index_name level0",
"value": "idx_level_0",
"type": "th",
"is_visible": True,
"display_value": "idx_level_0",
},
{
"class": "index_name level1",
"value": "idx_level_1",
"type": "th",
"is_visible": True,
"display_value": "idx_level_1",
},
{
"class": "blank col0",
"value": self.blank_value,
"type": "th",
"is_visible": True,
"display_value": self.blank_value,
},
]
assert head == expected
def test_mi_sparse_column_names(self):
df = DataFrame(
np.arange(16).reshape(4, 4),
index=MultiIndex.from_arrays(
[["a", "a", "b", "a"], [0, 1, 1, 2]],
names=["idx_level_0", "idx_level_1"],
),
columns=MultiIndex.from_arrays(
[["C1", "C1", "C2", "C2"], [1, 0, 1, 0]], names=["col_0", "col_1"]
),
)
result = Styler(df, cell_ids=False)._translate(True, True)
head = result["head"][1]
expected = [
{
"class": "blank",
"value": self.blank_value,
"display_value": self.blank_value,
"type": "th",
"is_visible": True,
},
{
"class": "index_name level1",
"value": "col_1",
"display_value": "col_1",
"is_visible": True,
"type": "th",
},
{
"class": "col_heading level1 col0",
"display_value": "1",
"is_visible": True,
"type": "th",
"value": 1,
"attributes": "",
},
{
"class": "col_heading level1 col1",
"display_value": "0",
"is_visible": True,
"type": "th",
"value": 0,
"attributes": "",
},
{
"class": "col_heading level1 col2",
"display_value": "1",
"is_visible": True,
"type": "th",
"value": 1,
"attributes": "",
},
{
"class": "col_heading level1 col3",
"display_value": "0",
"is_visible": True,
"type": "th",
"value": 0,
"attributes": "",
},
]
assert head == expected
def test_hide_column_headers(self):
ctx = self.styler.hide(axis="columns")._translate(True, True)
assert len(ctx["head"]) == 0 # no header entries with an unnamed index
self.df.index.name = "some_name"
ctx = self.df.style.hide(axis="columns")._translate(True, True)
assert len(ctx["head"]) == 1
# index names still visible, changed in #42101, reverted in 43404
def test_hide_single_index(self):
# GH 14194
# single unnamed index
ctx = self.df.style._translate(True, True)
assert ctx["body"][0][0]["is_visible"]
assert ctx["head"][0][0]["is_visible"]
ctx2 = self.df.style.hide(axis="index")._translate(True, True)
assert not ctx2["body"][0][0]["is_visible"]
assert not ctx2["head"][0][0]["is_visible"]
# single named index
ctx3 = self.df.set_index("A").style._translate(True, True)
assert ctx3["body"][0][0]["is_visible"]
assert len(ctx3["head"]) == 2 # 2 header levels
assert ctx3["head"][0][0]["is_visible"]
ctx4 = self.df.set_index("A").style.hide(axis="index")._translate(True, True)
assert not ctx4["body"][0][0]["is_visible"]
assert len(ctx4["head"]) == 1 # only 1 header levels
assert not ctx4["head"][0][0]["is_visible"]
def test_hide_multiindex(self):
# GH 14194
df = DataFrame(
{"A": [1, 2], "B": [1, 2]},
index=MultiIndex.from_arrays(
[["a", "a"], [0, 1]], names=["idx_level_0", "idx_level_1"]
),
)
ctx1 = df.style._translate(True, True)
# tests for 'a' and '0'
assert ctx1["body"][0][0]["is_visible"]
assert ctx1["body"][0][1]["is_visible"]
# check for blank header rows
assert len(ctx1["head"][0]) == 4 # two visible indexes and two data columns
ctx2 = df.style.hide(axis="index")._translate(True, True)
# tests for 'a' and '0'
assert not ctx2["body"][0][0]["is_visible"]
assert not ctx2["body"][0][1]["is_visible"]
# check for blank header rows
assert len(ctx2["head"][0]) == 3 # one hidden (col name) and two data columns
assert not ctx2["head"][0][0]["is_visible"]
def test_hide_columns_single_level(self):
# GH 14194
# test hiding single column
ctx = self.df.style._translate(True, True)
assert ctx["head"][0][1]["is_visible"]
assert ctx["head"][0][1]["display_value"] == "A"
assert ctx["head"][0][2]["is_visible"]
assert ctx["head"][0][2]["display_value"] == "B"
assert ctx["body"][0][1]["is_visible"] # col A, row 1
assert ctx["body"][1][2]["is_visible"] # col B, row 1
ctx = self.df.style.hide("A", axis="columns")._translate(True, True)
assert not ctx["head"][0][1]["is_visible"]
assert not ctx["body"][0][1]["is_visible"] # col A, row 1
assert ctx["body"][1][2]["is_visible"] # col B, row 1
# test hiding mulitiple columns
ctx = self.df.style.hide(["A", "B"], axis="columns")._translate(True, True)
assert not ctx["head"][0][1]["is_visible"]
assert not ctx["head"][0][2]["is_visible"]
assert not ctx["body"][0][1]["is_visible"] # col A, row 1
assert not ctx["body"][1][2]["is_visible"] # col B, row 1
def test_hide_columns_index_mult_levels(self):
# GH 14194
# setup dataframe with multiple column levels and indices
i1 = MultiIndex.from_arrays(
[["a", "a"], [0, 1]], names=["idx_level_0", "idx_level_1"]
)
i2 = MultiIndex.from_arrays(
[["b", "b"], [0, 1]], names=["col_level_0", "col_level_1"]
)
df = DataFrame([[1, 2], [3, 4]], index=i1, columns=i2)
ctx = df.style._translate(True, True)
# column headers
assert ctx["head"][0][2]["is_visible"]
assert ctx["head"][1][2]["is_visible"]
assert ctx["head"][1][3]["display_value"] == "1"
# indices
assert ctx["body"][0][0]["is_visible"]
# data
assert ctx["body"][1][2]["is_visible"]
assert ctx["body"][1][2]["display_value"] == "3"
assert ctx["body"][1][3]["is_visible"]
assert ctx["body"][1][3]["display_value"] == "4"
# hide top column level, which hides both columns
ctx = df.style.hide("b", axis="columns")._translate(True, True)
assert not ctx["head"][0][2]["is_visible"] # b
assert not ctx["head"][1][2]["is_visible"] # 0
assert not ctx["body"][1][2]["is_visible"] # 3
assert ctx["body"][0][0]["is_visible"] # index
# hide first column only
ctx = df.style.hide([("b", 0)], axis="columns")._translate(True, True)
assert not ctx["head"][0][2]["is_visible"] # b
assert ctx["head"][0][3]["is_visible"] # b
assert not ctx["head"][1][2]["is_visible"] # 0
assert not ctx["body"][1][2]["is_visible"] # 3
assert ctx["body"][1][3]["is_visible"]
assert ctx["body"][1][3]["display_value"] == "4"
# hide second column and index
ctx = df.style.hide([("b", 1)], axis=1).hide(axis=0)._translate(True, True)
assert not ctx["body"][0][0]["is_visible"] # index
assert len(ctx["head"][0]) == 3
assert ctx["head"][0][1]["is_visible"] # b
assert ctx["head"][1][1]["is_visible"] # 0
assert not ctx["head"][1][2]["is_visible"] # 1
assert not ctx["body"][1][3]["is_visible"] # 4
assert ctx["body"][1][2]["is_visible"]
assert ctx["body"][1][2]["display_value"] == "3"
# hide top row level, which hides both rows so body empty
ctx = df.style.hide("a", axis="index")._translate(True, True)
assert ctx["body"] == []
# hide first row only
ctx = df.style.hide(("a", 0), axis="index")._translate(True, True)
for i in [0, 1, 2, 3]:
assert "row1" in ctx["body"][0][i]["class"] # row0 not included in body
assert ctx["body"][0][i]["is_visible"]
def test_pipe(self):
def set_caption_from_template(styler, a, b):
return styler.set_caption(f"Dataframe with a = {a} and b = {b}")
styler = self.df.style.pipe(set_caption_from_template, "A", b="B")
assert "Dataframe with a = A and b = B" in styler.to_html()
# Test with an argument that is a (callable, keyword_name) pair.
def f(a, b, styler):
return (a, b, styler)
styler = self.df.style
result = styler.pipe((f, "styler"), a=1, b=2)
assert result == (1, 2, styler)
def test_no_cell_ids(self):
# GH 35588
# GH 35663
df = DataFrame(data=[[0]])
styler = Styler(df, uuid="_", cell_ids=False)
styler.to_html()
s = styler.to_html() # render twice to ensure ctx is not updated
assert s.find('<td class="data row0 col0" >') != -1
@pytest.mark.parametrize(
"classes",
[
DataFrame(
data=[["", "test-class"], [np.nan, None]],
columns=["A", "B"],
index=["a", "b"],
),
DataFrame(data=[["test-class"]], columns=["B"], index=["a"]),
DataFrame(data=[["test-class", "unused"]], columns=["B", "C"], index=["a"]),
],
)
def test_set_data_classes(self, classes):
# GH 36159
df = DataFrame(data=[[0, 1], [2, 3]], columns=["A", "B"], index=["a", "b"])
s = Styler(df, uuid_len=0, cell_ids=False).set_td_classes(classes).to_html()
assert '<td class="data row0 col0" >0</td>' in s
assert '<td class="data row0 col1 test-class" >1</td>' in s
assert '<td class="data row1 col0" >2</td>' in s
assert '<td class="data row1 col1" >3</td>' in s
# GH 39317
s = Styler(df, uuid_len=0, cell_ids=True).set_td_classes(classes).to_html()
assert '<td id="T__row0_col0" class="data row0 col0" >0</td>' in s
assert '<td id="T__row0_col1" class="data row0 col1 test-class" >1</td>' in s
assert '<td id="T__row1_col0" class="data row1 col0" >2</td>' in s
assert '<td id="T__row1_col1" class="data row1 col1" >3</td>' in s
def test_set_data_classes_reindex(self):
# GH 39317
df = DataFrame(
data=[[0, 1, 2], [3, 4, 5], [6, 7, 8]], columns=[0, 1, 2], index=[0, 1, 2]
)
classes = DataFrame(
data=[["mi", "ma"], ["mu", "mo"]],
columns=[0, 2],
index=[0, 2],
)
s = Styler(df, uuid_len=0).set_td_classes(classes).to_html()
assert '<td id="T__row0_col0" class="data row0 col0 mi" >0</td>' in s
assert '<td id="T__row0_col2" class="data row0 col2 ma" >2</td>' in s
assert '<td id="T__row1_col1" class="data row1 col1" >4</td>' in s
assert '<td id="T__row2_col0" class="data row2 col0 mu" >6</td>' in s
assert '<td id="T__row2_col2" class="data row2 col2 mo" >8</td>' in s
def test_chaining_table_styles(self):
# GH 35607
df = DataFrame(data=[[0, 1], [1, 2]], columns=["A", "B"])
styler = df.style.set_table_styles(
[{"selector": "", "props": [("background-color", "yellow")]}]
).set_table_styles(
[{"selector": ".col0", "props": [("background-color", "blue")]}],
overwrite=False,
)
assert len(styler.table_styles) == 2
def test_column_and_row_styling(self):
# GH 35607
df = DataFrame(data=[[0, 1], [1, 2]], columns=["A", "B"])
s = Styler(df, uuid_len=0)
s = s.set_table_styles({"A": [{"selector": "", "props": [("color", "blue")]}]})
assert "#T_ .col0 {\n color: blue;\n}" in s.to_html()
s = s.set_table_styles(
{0: [{"selector": "", "props": [("color", "blue")]}]}, axis=1
)
assert "#T_ .row0 {\n color: blue;\n}" in s.to_html()
@pytest.mark.parametrize("len_", [1, 5, 32, 33, 100])
def test_uuid_len(self, len_):
# GH 36345
df = DataFrame(data=[["A"]])
s = Styler(df, uuid_len=len_, cell_ids=False).to_html()
strt = s.find('id="T_')
end = s[strt + 6 :].find('"')
if len_ > 32:
assert end == 32
else:
assert end == len_
@pytest.mark.parametrize("len_", [-2, "bad", None])
def test_uuid_len_raises(self, len_):
# GH 36345
df = DataFrame(data=[["A"]])
msg = "``uuid_len`` must be an integer in range \\[0, 32\\]."
with pytest.raises(TypeError, match=msg):
| Styler(df, uuid_len=len_, cell_ids=False) | pandas.io.formats.style.Styler |
import os
import pandas as pd
from numpy import testing as npt
import pandas.util.testing as pdt
import ixmp
import pytest
from ixmp.default_path_constants import CONFIG_PATH
from testing_utils import (
test_mp,
test_mp_props,
test_mp_use_default_dbprops_file,
test_mp_use_db_config_path,
)
test_args = ('Douglas Adams', 'Hitchhiker')
can_args = ('canning problem', 'standard')
# string columns for timeseries checks
iamc_idx_cols = ['model', 'scenario', 'region', 'variable', 'unit']
cols_str = ['region', 'variable', 'unit', 'year']
def local_config_exists():
return os.path.exists(CONFIG_PATH)
@pytest.mark.skipif(local_config_exists(),
reason='will not overwrite local config files')
def test_default_dbprops_file(test_mp_use_default_dbprops_file):
test_mp = test_mp_use_default_dbprops_file
scenario = test_mp.scenario_list(model='Douglas Adams')['scenario']
assert scenario[0] == 'Hitchhiker'
@pytest.mark.skipif(local_config_exists(),
reason='will not overwrite local config files')
def test_db_config_path(test_mp_use_db_config_path):
test_mp = test_mp_use_db_config_path
scenario = test_mp.scenario_list(model='Douglas Adams')['scenario']
assert scenario[0] == 'Hitchhiker'
def test_platform_init_raises():
pytest.raises(ValueError, ixmp.Platform, dbtype='foo')
def test_scen_list(test_mp):
scenario = test_mp.scenario_list(model='Douglas Adams')['scenario']
assert scenario[0] == 'Hitchhiker'
def test_new_scen(test_mp):
scen = ixmp.Scenario(test_mp, *can_args, version='new')
assert scen.version == 0
def test_default_version(test_mp):
scen = ixmp.Scenario(test_mp, *can_args)
assert scen.version == 2
def test_init_par_35(test_mp):
scen = ixmp.Scenario(test_mp, *can_args, version='new')
scen.init_set('ii')
scen.init_par('new_par', idx_sets='ii')
def test_get_scalar(test_mp):
scen = ixmp.Scenario(test_mp, *can_args)
obs = scen.scalar('f')
exp = {'unit': 'USD/km', 'value': 90}
assert obs == exp
def test_init_scalar(test_mp):
scen = ixmp.Scenario(test_mp, *can_args)
scen2 = scen.clone(keep_solution=False)
scen2.check_out()
scen2.init_scalar('g', 90.0, 'USD/km')
scen2.commit("adding a scalar 'g'")
# make sure that changes to a scenario are copied over during clone
def test_add_clone(test_mp):
scen = ixmp.Scenario(test_mp, *can_args, version=1)
scen.check_out()
scen.init_set('h')
scen.add_set('h', 'test')
scen.commit("adding an index set 'h', wiht element 'test'")
scen2 = scen.clone(keep_solution=False)
obs = scen2.set('h')
npt.assert_array_equal(obs, ['test'])
# make sure that (only) the correct scenario is touched after cloning
def test_clone_edit(test_mp):
scen = ixmp.Scenario(test_mp, *can_args)
scen2 = scen.clone(keep_solution=False)
scen2.check_out()
scen2.change_scalar('f', 95.0, 'USD/km')
scen2.commit('change transport cost')
obs = scen.scalar('f')
exp = {'unit': 'USD/km', 'value': 90}
assert obs == exp
obs = scen2.scalar('f')
exp = {'unit': 'USD/km', 'value': 95}
assert obs == exp
def test_idx_name(test_mp):
scen = ixmp.Scenario(test_mp, *can_args)
df = scen.idx_names('d')
npt.assert_array_equal(df, ['i', 'j'])
def test_var_marginal(test_mp):
scen = ixmp.Scenario(test_mp, *can_args)
df = scen.var('x', filters={'i': ['seattle']})
npt.assert_array_almost_equal(df['mrg'], [0, 0, 0.036])
def test_var_level(test_mp):
scen = ixmp.Scenario(test_mp, *can_args)
df = scen.var('x', filters={'i': ['seattle']})
npt.assert_array_almost_equal(df['lvl'], [50, 300, 0])
def test_var_general_str(test_mp):
scen = ixmp.Scenario(test_mp, *can_args)
df = scen.var('x', filters={'i': ['seattle']})
npt.assert_array_equal(
df['j'], ['new-york', 'chicago', 'topeka'])
def test_unit_list(test_mp):
units = test_mp.units()
assert ('cases' in units) is True
def test_add_unit(test_mp):
test_mp.add_unit('test', 'just testing')
def test_par_filters_unit(test_mp):
scen = ixmp.Scenario(test_mp, *can_args)
df = scen.par('d', filters={'i': ['seattle']})
obs = df.loc[0, 'unit']
exp = 'km'
assert obs == exp
def test_new_timeseries(test_mp):
scen = ixmp.TimeSeries(test_mp, *test_args, version='new', annotation='fo')
df = {'year': [2010, 2020], 'value': [23.5, 23.6]}
df = pd.DataFrame.from_dict(df)
df['region'] = 'World'
df['variable'] = 'Testing'
df['unit'] = '???'
scen.add_timeseries(df)
scen.commit('importing a testing timeseries')
def test_new_timeseries_error(test_mp):
scen = ixmp.TimeSeries(test_mp, *test_args, version='new', annotation='fo')
df = {'year': [2010, 2020], 'value': [23.5, 23.6]}
df = pd.DataFrame.from_dict(df)
df['region'] = 'World'
df['variable'] = 'Testing'
pytest.raises(ValueError, scen.add_timeseries, df)
def test_get_timeseries(test_mp):
scen = ixmp.TimeSeries(test_mp, *test_args, version=2)
obs = scen.timeseries(regions='World', variables='Testing', units='???',
years=2020)
df = {'region': ['World'], 'variable': ['Testing'], 'unit': ['???'],
'year': [2020], 'value': [23.6]}
exp = pd.DataFrame.from_dict(df)
npt.assert_array_equal(exp[cols_str], obs[cols_str])
npt.assert_array_almost_equal(exp['value'], obs['value'])
def test_get_timeseries_iamc(test_mp):
scen = ixmp.TimeSeries(test_mp, *test_args, version=2)
obs = scen.timeseries(iamc=True, regions='World', variables='Testing')
df = {'year': [2010, 2020], 'value': [23.5, 23.6]}
df = pd.DataFrame.from_dict(df)
df['model'] = 'Douglas Adams'
df['scenario'] = 'Hitchhiker'
df['region'] = 'World'
df['variable'] = 'Testing'
df['unit'] = '???'
df = df.pivot_table(index=iamc_idx_cols, columns='year')['value']
df.reset_index(inplace=True)
exp = | pd.DataFrame.from_dict(df) | pandas.DataFrame.from_dict |
# --------------
# import the libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
import warnings
warnings.filterwarnings('ignore')
# Code starts here
df= | pd.read_json(path,lines=True) | pandas.read_json |
#!/usr/bin/env python
# coding: utf-8
# <div class="alert alert-block alert-info">
# <b><h1>ENGR 1330 Computational Thinking with Data Science </h1></b>
# </div>
#
# Copyright © 2021 <NAME> and <NAME>
#
# Last GitHub Commit Date: 4 Nov 2021
#
# # 26: Linear Regression
# - Purpose
# - Homebrew (using covariance formulas)
# - Homebrew using Matrix Algebra
# - Using packages: *numpy.linalg.lstsq* (future versions of this ebook)
# - Using packages: *statsmodel* package
# - Using packages: *sklearn* package
# - Why regression belongs to both statistics and machine learning
# - Learning algorithms used to create a linear regression model
# - Preparing data for linear regression
# In[ ]:
# ## What is Regression
#
# A **systematic** procedure to model the relationship between one dependent variable and one or more independent variables and quantify the uncertainty involved in *response predictions*.
#
# <!---->
#
# ### Objectives
#
# - Create linear regression models from data using primitive python
# - Create linear regression models from data using NumPy and Pandas tools
# - Create presentation-quality graphs and charts for reporting results
#
#
# ### Computational Thinking Concepts
#
# |Description|Computational Thinking Concept|
# |:---|:---|
# |Linear Model|Abstraction|
# |Response and Explanatory Variables|Decomposition|
# |Primitive arrays: vectors and matrices|Data Representation|
# |NumPy arrays: vectors and matrices|Data Representation|
#
# ### Textbook Resources
#
# [https://inferentialthinking.com/chapters/15/Prediction.html](https://inferentialthinking.com/chapters/15/Prediction.html)
#
# <hr>
#
# ## Data Modeling: Regression Approach
#
# Regression is a basic and commonly used type of predictive analysis.
#
# The overall idea of regression is to assess:
#
# - does a set of predictor/explainatory variables (features) do a good job in predicting an outcome (dependent/response) variable?
# - Which explainatory variables (features) in particular are significant predictors of the outcome variable, and in what way do they–indicated by the magnitude and sign of the beta estimates–impact the outcome variable?
# - What is the estimated(predicted) value of the response under various excitation (explainatory) variable values?
# - What is the uncertainty involved in the prediction?
#
# These regression estimates are used to explain the relationship between one dependent variable and one or more independent variables.
#
# The simplest form is a linear regression equation with one dependent(response) and one independent(explainatory) variable is defined by the formula
#
# $y_i = \beta_0 + \beta_1*x_i$, where $y_i$ = estimated dependent(response) variable value, $\beta_0$ = constant(intercept), $\beta_1$ = regression coefficient (slope), and $x_i$ = independent(predictor) variable value
#
# More complex forms involving non-linear (in the $\beta_{i}$) parameters and non-linear combinations of the independent(predictor) variables are also used - these are beyond the scope of this lesson.
#
# We have already explored the underlying computations involved (without explaination) by just solving a particular linear equation system; what follows is some background on the source of that equation system.
#
# <hr>
# ### Fundamental Questions
#
# - What is regression used for?
# - Why is it useful?
#
# Three major uses for regression analysis are (1) determining the strength of predictors, (2) forecasting an effect, and (3) trend forecasting.
#
# - First, the regression might be used to identify the strength of the effect that the independent variable(s) have on a dependent variable. Typical questions are what is the strength of relationship between dose and effect, sales and marketing spending, or age and income.
#
# - Second, it can be used to forecast effects or impact of changes. That is, the regression analysis helps us to understand how much the dependent variable changes with a change in one or more independent variables. A typical question is, “how much additional sales income do I get for each additional $1000 spent on marketing?”
#
# - Third, regression analysis predicts trends and future values. The regression analysis can be used to get point estimates. A typical question is, “what will the price of gold be in 6 months?”
# Consider the image below from a Texas Instruments Calculator user manual
#
# 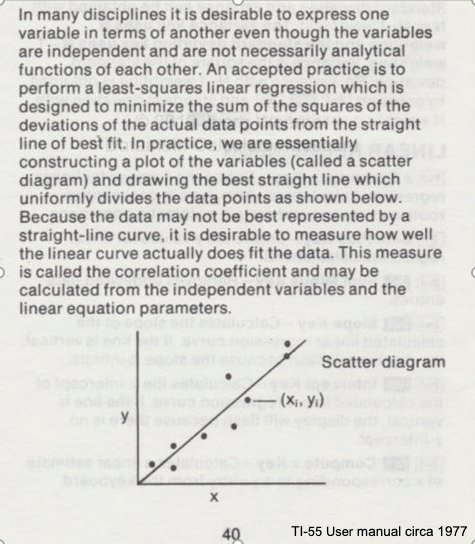
#
# In the context of our class, the straight solid line is the `Data Model` whose equation is
# $Y = \beta_0 + \beta_1*X$.
# The ordered pairs $(x_i,y_i)$ in the scatterplot are the observation (or training set).
# As depicted here $Y$ is the response to different values of the explainitory variable $X$. The typical convention is response on the up-down axis, but not always.
# The model parameters are $\beta_0$ and $\beta_1$ ; once known can estimate (predict) response to (as yet) unobserved values of $x$
#
# Classically, the normal equations are evaluated to find the model parameters:
#
# $\beta_1 = \frac{\sum x\sum y~-~N\sum xy}{(\sum x)^2~-~N\sum x^2}$ and $\beta_0 = \bar y - \beta_1 \bar x$
#
# These two equations are the solution to the "design matrix" linear system earlier, but presented as a set of discrete arithmetic operations.
# <hr><hr>
#
# ### Classical Regression by Normal Equations
#
# We will illustrate the classical approach to finding the slope and intercept using the normal equations first a plotting function, then we will use the values from the Texas Instruments TI-55 user manual.
#
# First a way to plot:
# In[1]:
### Lets Make a Plotting Function
def makeAbear(xvalues,yvalues,xleft,yleft,xright,yright,xlab,ylab,title):
# plotting function dependent on matplotlib installed above
# xvalues, yvalues == data pairs to scatterplot; FLOAT
# xleft,yleft == left endpoint line to draw; FLOAT
# xright,yright == right endpoint line to draw; FLOAT
# xlab,ylab == axis labels, STRINGS!!
# title == Plot title, STRING
import matplotlib.pyplot
matplotlib.pyplot.scatter(xvalues,yvalues)
matplotlib.pyplot.plot([xleft, xright], [yleft, yright], 'k--', lw=2, color="red")
matplotlib.pyplot.xlabel(xlab)
matplotlib.pyplot.ylabel(ylab)
matplotlib.pyplot.title(title)
matplotlib.pyplot.show()
return
# Now the two lists to process
# In[2]:
# Make two lists
sample_length = [101.3,103.7,98.6,99.9,97.2,100.1]
sample_weight = [609,626,586,594,579,605]
# We will assume weight is the explainatory variable, and it is to be used to predict length.
makeAbear(sample_weight, sample_length,580,96,630,106,'Weight (g)','Length (cm)','Length versus Weight for \n NASA CF0132 Fiber Reinforced Polymer')
# Notice the dashed line, we supplied only two (x,y) pairs to plot the line, so lets get a colonoscope and find where it came from.
# In[3]:
def myline(slope,intercept,value1,value2):
'''Returns a tuple ([x1,x2],[y1,y2]) from y=slope*value+intercept'''
listy = []
listx = []
listx.append(value1)
listx.append(value2)
listy.append(slope*listx[0]+intercept)
listy.append(slope*listx[1]+intercept)
return(listx,listy)
# The myline function returns a tuple, that we parse below to make the plot of the data model. This is useful if we wish to plot beyond the range of the observations data.
# In[4]:
slope = 0.13 #0.13
intercept = 23 # 23
xlow = 540 # here we define the lower bound of the model plot
xhigh = 640 # upper bound
object = myline(slope,intercept,xlow,xhigh)
xone = object[0][0]; xtwo = object[0][1]; yone = object[1][0]; ytwo = object[1][1]
makeAbear(sample_weight, sample_length,xone,yone,xtwo,ytwo,'Weight (g)','Length (cm)','Length versus Weight for \n NASA CF0132 Fiber Reinforced Polymer')
# In[5]:
print(xone,yone)
print(xtwo,ytwo)
# Now lets get "optimal" values of slope and intercept from the Normal equations
# In[6]:
# Evaluate the normal equations
sumx = 0.0
sumy = 0.0
sumxy = 0.0
sumx2 = 0.0
sumy2 = 0.0
for i in range(len(sample_weight)):
sumx = sumx + sample_weight[i]
sumx2 = sumx2 + sample_weight[i]**2
sumy = sumy + sample_length[i]
sumy2 = sumy2 + sample_length[i]**2
sumxy = sumxy + sample_weight[i]*sample_length[i]
b1 = (sumx*sumy - len(sample_weight)*sumxy)/(sumx**2-len(sample_weight)*sumx2)
b0 = sumy/len(sample_length) - b1* (sumx/len(sample_weight))
lineout = ("Linear Model is y=%.3f" % b1) + ("x + %.3f" % b0)
print("Linear Model is y=%.3f" % b1 ,"x + %.3f" % b0)
# In[7]:
slope = 0.129 #0.129
intercept = 22.813 # 22.813
xlow = 540
xhigh = 640
object = myline(slope,intercept,xlow,xhigh)
xone = object[0][0]; xtwo = object[0][1]; yone = object[1][0]; ytwo = object[1][1]
makeAbear(sample_weight, sample_length,xone,yone,xtwo,ytwo,'Weight (g)','Length (cm)','Length versus Weight for \n NASA CF0132 Fiber Reinforced Polymer')
# <hr>
#
# ### Where do these normal equations come from?
# Consider our linear model $y = \beta_0 + \beta_1 \cdot x + \epsilon$. Where $\epsilon$ is the error in the estimate. If we square each error and add them up (for our training set) we will have $\sum \epsilon^2 = \sum (y_i - \beta_0 - \beta_1 \cdot x_i)^2 $. Our goal is to minimize this error by our choice of $\beta_0 $ and $ \beta_1 $
#
# The necessary and sufficient conditions for a minimum is that the first partial derivatives of the `error` as a function must vanish (be equal to zero). We can leverage that requirement as
#
# $\frac{\partial(\sum \epsilon^2)}{\partial \beta_0} = \frac{\partial{\sum (y_i - \beta_0 - \beta_1 \cdot x_i)^2}}{\partial \beta_0} = - \sum 2[y_i - \beta_0 + \beta_1 \cdot x_i] = -2(\sum_{i=1}^n y_i - n \beta_0 - \beta_1 \sum_{i=1}^n x_i) = 0 $
#
# and
#
# $\frac{\partial(\sum \epsilon^2)}{\partial \beta_1} = \frac{\partial{\sum (y_i - \beta_0 - \beta_1 \cdot x_i)^2}}{\partial \beta_1} = - \sum 2[y_i - \beta_0 + \beta_1 \cdot x_i]x_i = -2(\sum_{i=1}^n x_i y_i - n \beta_0 \sum_{i=1}^n x_i - \beta_1 \sum_{i=1}^n x_i^2) = 0 $
#
# Solving the two equations for $\beta_0$ and $\beta_1$ produces the normal equations (for linear least squares), which leads to
#
# $\beta_1 = \frac{\sum x\sum y~-~n\sum xy}{(\sum x)^2~-~n\sum x^2}$
# $\beta_0 = \bar y - \beta_1 \bar x$
#
#
# Lets consider a more flexible way by fitting the data model using linear algebra instead of the summation notation.
#
# <hr><hr>
#
# ### Computational Linear Algebra
# We will start again with our linear data model'
#
# ```{note}
# The linear system below should be familiar, we used it in the Predictor-Response Data Model (without much background). Here we learn it is simply the matrix equivalent of minimizing the sumn of squares error for each equation
# ```
#
# $y_i = \beta_0 + \beta_1 \cdot x_i + \epsilon_i$ then replace with vectors as
#
# $$
# \begin{gather}
# \mathbf{Y}=
# \begin{pmatrix}
# y_1 \\
# y_2 \\
# \vdots \\
# y_n \\
# \end{pmatrix}
# \end{gather}
# $$
#
# $$\begin{gather}
# \mathbf{\beta}=
# \begin{pmatrix}
# \beta_0 \\
# \beta_1 \\
# \end{pmatrix}
# \end{gather}$$
#
# $$\begin{gather}
# \mathbf{X}=
# \begin{pmatrix}
# 1 & x_1 & x_1^2\\
# 1 & x_2 & x_2^2\\
# \vdots \\
# 1 & x_n & x_n^2\\
# \end{pmatrix}
# \end{gather}$$
#
# $$\begin{gather}
# \mathbf{\epsilon}=
# \begin{pmatrix}
# \epsilon_1 \\
# \epsilon_2 \\
# \vdots \\
# \epsilon_n \\
# \end{pmatrix}
# \end{gather}$$
#
# So our system can now be expressed in matrix-vector form as
#
# $\mathbf{Y}=\mathbf{X}\mathbf{\beta}+\mathbf{\epsilon}$ if we perfrom the same vector calculus as before we will end up with a result where pre-multiply by the transpose of $\mathbf{X}$ we will have a linear system in $\mathbf{\beta}$ which we can solve using Gaussian reduction, or LU decomposition or some other similar method.
#
# The resulting system (that minimizes $\mathbf{\epsilon^T}\mathbf{\epsilon}$) is
#
# $\mathbf{X^T}\mathbf{Y}=\mathbf{X^T}\mathbf{X}\mathbf{\beta}$ and solving for the parameters gives
# $\mathbf{\beta}=(\mathbf{X^T}\mathbf{X})^{-1}\mathbf{X^T}\mathbf{Y}$
#
# So lets apply it to our example - what follows is mostly in python primative
# In[8]:
# linearsolver with pivoting adapted from
# https://stackoverflow.com/questions/31957096/gaussian-elimination-with-pivoting-in-python/31959226
def linearsolver(A,b):
n = len(A)
M = A
i = 0
for x in M:
x.append(b[i])
i += 1
# row reduction with pivots
for k in range(n):
for i in range(k,n):
if abs(M[i][k]) > abs(M[k][k]):
M[k], M[i] = M[i],M[k]
else:
pass
for j in range(k+1,n):
q = float(M[j][k]) / M[k][k]
for m in range(k, n+1):
M[j][m] -= q * M[k][m]
# allocate space for result
x = [0 for i in range(n)]
# back-substitution
x[n-1] =float(M[n-1][n])/M[n-1][n-1]
for i in range (n-1,-1,-1):
z = 0
for j in range(i+1,n):
z = z + float(M[i][j])*x[j]
x[i] = float(M[i][n] - z)/M[i][i]
# return result
return(x)
#######
# In[9]:
# matrix multiply script
def mmult(amatrix,bmatrix,rowNumA,colNumA,rowNumB,colNumB):
result_matrix = [[0 for j in range(colNumB)] for i in range(rowNumA)]
for i in range(0,rowNumA):
for j in range(0,colNumB):
for k in range(0,colNumA):
result_matrix[i][j]=result_matrix[i][j]+amatrix[i][k]*bmatrix[k][j]
return(result_matrix)
# matrix vector multiply script
def mvmult(amatrix,bvector,rowNumA,colNumA):
result_v = [0 for i in range(rowNumA)]
for i in range(0,rowNumA):
for j in range(0,colNumA):
result_v[i]=result_v[i]+amatrix[i][j]*bvector[j]
return(result_v)
colNumX=2 #
rowNumX=len(sample_weight)
xmatrix = [[1 for j in range(colNumX)]for i in range(rowNumX)]
xtransp = [[1 for j in range(rowNumX)]for i in range(colNumX)]
yvector = [0 for i in range(rowNumX)]
for irow in range(rowNumX):
xmatrix[irow][1]=sample_weight[irow]
xtransp[1][irow]=sample_weight[irow]
yvector[irow] =sample_length[irow]
xtx = [[0 for j in range(colNumX)]for i in range(colNumX)]
xty = []
xtx = mmult(xtransp,xmatrix,colNumX,rowNumX,rowNumX,colNumX)
xty = mvmult(xtransp,yvector,colNumX,rowNumX)
beta = []
#solve XtXB = XtY for B
beta = linearsolver(xtx,xty) #Solve the linear system What would the numpy equivalent be?
slope = beta[1] #0.129
intercept = beta[0] # 22.813
xlow = 580
xhigh = 630
object = myline(slope,intercept,xlow,xhigh)
xone = object[0][0]; xtwo = object[0][1]; yone = object[1][0]; ytwo = object[1][1]
makeAbear(sample_weight, sample_length,xone,yone,xtwo,ytwo,'Weight (g)','Length (cm)','Length versus Weight for \n NASA CF0132 Fiber Reinforced Polymer')
# In[10]:
beta
# ---
#
# #### What's the Value of the Computational Linear Algebra ?
#
# The value comes when we have more explainatory variables, and we may want to deal with curvature.
#
# ```{note}
# The lists below are different that the example above!
# ```
# In[11]:
# Make two lists
yyy = [0,0,1,1,3]
xxx = [-2,-1,0,1,2]
slope = 0.5 #0.129
intercept = 1 # 22.813
xlow = -3
xhigh = 3
object = myline(slope,intercept,xlow,xhigh)
xone = object[0][0]; xtwo = object[0][1]; yone = object[1][0]; ytwo = object[1][1]
makeAbear(xxx, yyy,xone,yone,xtwo,ytwo,'xxx','yyy','yyy versus xxx')
# In[12]:
colNumX=2 #
rowNumX=len(xxx)
xmatrix = [[1 for j in range(colNumX)]for i in range(rowNumX)]
xtransp = [[1 for j in range(rowNumX)]for i in range(colNumX)]
yvector = [0 for i in range(rowNumX)]
for irow in range(rowNumX):
xmatrix[irow][1]=xxx[irow]
xtransp[1][irow]=xxx[irow]
yvector[irow] =yyy[irow]
xtx = [[0 for j in range(colNumX)]for i in range(colNumX)]
xty = []
xtx = mmult(xtransp,xmatrix,colNumX,rowNumX,rowNumX,colNumX)
xty = mvmult(xtransp,yvector,colNumX,rowNumX)
beta = []
#solve XtXB = XtY for B
beta = linearsolver(xtx,xty) #Solve the linear system
slope = beta[1] #0.129
intercept = beta[0] # 22.813
xlow = -3
xhigh = 3
object = myline(slope,intercept,xlow,xhigh)
xone = object[0][0]; xtwo = object[0][1]; yone = object[1][0]; ytwo = object[1][1]
makeAbear(xxx, yyy,xone,yone,xtwo,ytwo,'xxx','yyy','yyy versus xxx')
# In[13]:
colNumX=4 #
rowNumX=len(xxx)
xmatrix = [[1 for j in range(colNumX)]for i in range(rowNumX)]
xtransp = [[1 for j in range(rowNumX)]for i in range(colNumX)]
yvector = [0 for i in range(rowNumX)]
for irow in range(rowNumX):
xmatrix[irow][1]=xxx[irow]
xmatrix[irow][2]=xxx[irow]**2
xmatrix[irow][3]=xxx[irow]**3
xtransp[1][irow]=xxx[irow]
xtransp[2][irow]=xxx[irow]**2
xtransp[3][irow]=xxx[irow]**3
yvector[irow] =yyy[irow]
xtx = [[0 for j in range(colNumX)]for i in range(colNumX)]
xty = []
xtx = mmult(xtransp,xmatrix,colNumX,rowNumX,rowNumX,colNumX)
xty = mvmult(xtransp,yvector,colNumX,rowNumX)
beta = []
#solve XtXB = XtY for B
beta = linearsolver(xtx,xty) #Solve the linear system
# In[14]:
howMany = 20
xlow = -2
xhigh = 2
deltax = (xhigh - xlow)/howMany
xmodel = []
ymodel = []
for i in range(howMany+1):
xnow = xlow + deltax*float(i)
xmodel.append(xnow)
ymodel.append(beta[0]+beta[1]*xnow+beta[2]*xnow**2)
# Now plot the sample values and plotting position
import matplotlib.pyplot
myfigure = matplotlib.pyplot.figure(figsize = (10,5)) # generate a object from the figure class, set aspect ratio
# Built the plot
matplotlib.pyplot.scatter(xxx, yyy, color ='blue')
matplotlib.pyplot.plot(xmodel, ymodel, color ='red')
matplotlib.pyplot.ylabel("Y")
matplotlib.pyplot.xlabel("X")
mytitle = "YYY versus XXX"
matplotlib.pyplot.title(mytitle)
matplotlib.pyplot.show()
# ### Using packages
#
# So in core python, there is a fair amount of work involved to write script - how about an easier way? First lets get things into a dataframe. Using the lists from the example above we can build a dataframe using pandas.
# In[15]:
# Load the necessary packages
import numpy as np
import pandas as pd
import statistics
from matplotlib import pyplot as plt
# Create a dataframe:
data = pd.DataFrame({'X':xxx, 'Y':yyy})
data
# #### `statsmodel` package
#
# Now load in one of many modeling packages that have regression tools. Here we use **statsmodel** which is an API (applications programming interface) with a nice formula syntax. In the package call we use `Y~X` which is interpreted by the API as fit `Y` as a linear function of `X`, which interestingly is our design matrix from a few lessons ago.
# In[16]:
# repeat using statsmodel
import statsmodels.formula.api as smf
# Initialise and fit linear regression model using `statsmodels`
model = smf.ols('Y ~ X', data=data) # model object constructor syntax
model = model.fit()
# Now recover the parameters of the model
# In[17]:
model.params
# In[18]:
# Predict values
y_pred = model.predict()
# Plot regression against actual data
plt.figure(figsize=(12, 6))
plt.plot(data['X'], data['Y'], 'o') # scatter plot showing actual data
plt.plot(data['X'], y_pred, 'r', linewidth=2) # regression line
plt.xlabel('X')
plt.ylabel('Y')
plt.title('model vs observed')
plt.show();
# We could use our own plotting functions if we wished, and would obtain an identical plot
# In[19]:
slope = model.params[1] #0.7
intercept = model.params[0] # 1.0
xlow = -2
xhigh = 2
object = myline(slope,intercept,xlow,xhigh)
xone = object[0][0]; xtwo = object[0][1]; yone = object[1][0]; ytwo = object[1][1]
makeAbear(data['X'], data['Y'],xone,yone,xtwo,ytwo,'xxx','yyy','yyy versus xxx')
# Now lets add another column $x^2$ to introduce the ability to fit some curvature
# In[20]:
data['XX']=data['X']**2 # add a column of X^2
model = smf.ols('Y ~ X + XX', data=data) # model object constructor syntax
model = model.fit()
model.params
# In[21]:
# Predict values
y_pred = model.predict()
# Plot regression against actual data
plt.figure(figsize=(12, 6))
plt.plot(data['X'], data['Y'], 'o') # scatter plot showing actual data
plt.plot(data['X'], y_pred, 'r', linewidth=2) # regression line
plt.xlabel('X')
plt.ylabel('Y')
plt.title('model vs observed')
plt.show();
# Our homebrew plotting tool could be modified a bit (shown below just cause ...)
# In[22]:
myfigure = matplotlib.pyplot.figure(figsize = (10,5)) # generate a object from the figure class, set aspect ratio
# Built the plot
matplotlib.pyplot.scatter(data['X'], data['Y'], color ='blue')
matplotlib.pyplot.plot(data['X'], y_pred, color ='red')
matplotlib.pyplot.ylabel("Y")
matplotlib.pyplot.xlabel("X")
mytitle = "YYY versus XXX"
matplotlib.pyplot.title(mytitle)
matplotlib.pyplot.show()
# Another useful package is **sklearn** so repeat using that tool (same example)
#
# #### `sklearn` package
# In[23]:
# repeat using sklearn
# Multiple Linear Regression with scikit-learn:
from sklearn.linear_model import LinearRegression
# Build linear regression model using X,XX as predictors
# Split data into predictors X and output Y
predictors = ['X', 'XX']
X = data[predictors]
y = data['Y']
# Initialise and fit model
lm = LinearRegression() # This is the sklearn model tool here
model = lm.fit(X, y)
# In[24]:
print(f'alpha = {model.intercept_}')
print(f'betas = {model.coef_}')
# In[25]:
fitted = model.predict(X)
# Plot regression against actual data - What do we see?
plt.figure(figsize=(12, 6))
plt.plot(data['X'], data['Y'], 'o') # scatter plot showing actual data
plt.plot(data['X'], fitted,'r', linewidth=2) # regression line
plt.xlabel('x axis')
plt.ylabel('y axis')
plt.title('plot title')
plt.show();
# #### Now repeat using the original Texas Instruments example
#
# In[26]:
sample_length = [101.3,103.7,98.6,99.9,97.2,100.1]
sample_weight = [609,626,586,594,579,605]
data = | pd.DataFrame({'X':sample_weight, 'Y':sample_length}) | pandas.DataFrame |
import numpy as np
def rk4(x,ii,f,h):
for j in ii[1:]:
m1 = f(j,x[-1])
m2 = f(j+h/2,x[-1] + h/2*m1)
m3 =f(j+h/2,x[-1] + h/2*m2)
m4 = f(j+h,x[-1] + h*m3)
avg_slope = ( m1 +2*(m2 +m3) + m4 )/6
x.append(x[-1] + avg_slope*h)
def rk2(x,ii,f,h):
for j in ii[1:]:
x.append(x[-1] + (f(j,x[-1]) + f(j+h,x[-1] + h*f(j,x[-1])))/2*h)
def em(x,ii,f,h):
for j in ii[1:]:
x.append(x[-1] + f(j+h/2,x[-1] + (h/2)*(f(j,x[-1])))*h)
def ef(x,ii,f,h):
for j in ii[1:]:
x.append(x[-1] + f(j,x[-1])*h)
def ode_solver(f,d=(0,5),i=(0,0),N=7,method = "ef" ):
if method not in ["ef","em","rk2","rk4"]:
raise ValueError("Method not supported")
a,b = d
t0,x0 = i
ii = np.linspace(a,b,N+2)
h = np.diff(ii)[0]
x = [x0]
if method == "ef":
ef(x,ii,f,h)
elif method == "em":
em(x,ii,f,h)
elif method == "rk2":
rk2(x,ii,f,h)
elif method== "rk4" :
rk4(x,ii,f,h)
return(x[-1],np.array(x),ii)
def forall(f,g,dom,ini,d,N=100,yd=1):
nodes = np.linspace(dom[0],dom[1],N+2)
analytic = g(nodes)/yd
return(pd.DataFrame(np.array([nodes/d,ode_solver(f,dom,ini,N=N)[1]/yd,ode_solver(f,dom,ini,N=N,method="em")[1]/yd,ode_solver(f,dom,ini,N=N,method="rk2")[1]/yd,ode_solver(f,dom,ini,N=N,method="rk4")[1]/yd,analytic],dtype="double").T, columns=["nodes","euler_forward","euler_centered", "rk2","rk4","analytic"]))
def error_forall(y):
x = | pd.DataFrame(y) | pandas.DataFrame |
# coding=utf-8
# pylint: disable-msg=E1101,W0612
import numpy as np
import pandas as pd
from pandas import (Index, Series, DataFrame, isnull)
from pandas.compat import lrange
from pandas import compat
from pandas.util.testing import assert_series_equal
import pandas.util.testing as tm
from .common import TestData
class TestSeriesApply(TestData, tm.TestCase):
def test_apply(self):
with np.errstate(all='ignore'):
assert_series_equal(self.ts.apply(np.sqrt), np.sqrt(self.ts))
# elementwise-apply
import math
assert_series_equal(self.ts.apply(math.exp), np.exp(self.ts))
# how to handle Series result, #2316
result = self.ts.apply(lambda x: Series(
[x, x ** 2], index=['x', 'x^2']))
expected = DataFrame({'x': self.ts, 'x^2': self.ts ** 2})
tm.assert_frame_equal(result, expected)
# empty series
s = Series(dtype=object, name='foo', index=pd.Index([], name='bar'))
rs = s.apply(lambda x: x)
tm.assert_series_equal(s, rs)
# check all metadata (GH 9322)
self.assertIsNot(s, rs)
self.assertIs(s.index, rs.index)
self.assertEqual(s.dtype, rs.dtype)
self.assertEqual(s.name, rs.name)
# index but no data
s = Series(index=[1, 2, 3])
rs = s.apply(lambda x: x)
tm.assert_series_equal(s, rs)
def test_apply_same_length_inference_bug(self):
s = Series([1, 2])
f = lambda x: (x, x + 1)
result = s.apply(f)
expected = s.map(f)
assert_series_equal(result, expected)
s = Series([1, 2, 3])
result = s.apply(f)
expected = s.map(f)
assert_series_equal(result, expected)
def test_apply_dont_convert_dtype(self):
s = Series(np.random.randn(10))
f = lambda x: x if x > 0 else np.nan
result = s.apply(f, convert_dtype=False)
self.assertEqual(result.dtype, object)
def test_apply_args(self):
s = Series(['foo,bar'])
result = s.apply(str.split, args=(',', ))
self.assertEqual(result[0], ['foo', 'bar'])
tm.assertIsInstance(result[0], list)
def test_apply_box(self):
# ufunc will not be boxed. Same test cases as the test_map_box
vals = [pd.Timestamp('2011-01-01'), pd.Timestamp('2011-01-02')]
s = pd.Series(vals)
self.assertEqual(s.dtype, 'datetime64[ns]')
# boxed value must be Timestamp instance
res = s.apply(lambda x: '{0}_{1}_{2}'.format(x.__class__.__name__,
x.day, x.tz))
exp = pd.Series(['Timestamp_1_None', 'Timestamp_2_None'])
tm.assert_series_equal(res, exp)
vals = [pd.Timestamp('2011-01-01', tz='US/Eastern'),
pd.Timestamp('2011-01-02', tz='US/Eastern')]
s = pd.Series(vals)
self.assertEqual(s.dtype, 'datetime64[ns, US/Eastern]')
res = s.apply(lambda x: '{0}_{1}_{2}'.format(x.__class__.__name__,
x.day, x.tz))
exp = pd.Series(['Timestamp_1_US/Eastern', 'Timestamp_2_US/Eastern'])
tm.assert_series_equal(res, exp)
# timedelta
vals = [pd.Timedelta('1 days'), pd.Timedelta('2 days')]
s = | pd.Series(vals) | pandas.Series |
import os
import shutil
from attrdict import AttrDict
import numpy as np
import pandas as pd
from scipy.stats import gmean
from deepsense import neptune
from sklearn.metrics import roc_auc_score
from sklearn.model_selection import train_test_split, KFold, StratifiedKFold
from . import pipeline_config as cfg
from .pipelines import PIPELINES
from .utils import init_logger, read_params, set_seed, create_submission, verify_submission, calculate_rank
set_seed(cfg.RANDOM_SEED)
logger = init_logger()
ctx = neptune.Context()
params = read_params(ctx, fallback_file='neptune.yaml')
class PipelineManager():
def train(self, pipeline_name, dev_mode):
train(pipeline_name, dev_mode)
def evaluate(self, pipeline_name, dev_mode, ):
evaluate(pipeline_name, dev_mode)
def predict(self, pipeline_name, dev_mode, submit_predictions):
predict(pipeline_name, dev_mode, submit_predictions)
def train_evaluate_cv(self, pipeline_name, dev_mode):
train_evaluate_cv(pipeline_name, dev_mode)
def train_evaluate_predict_cv(self, pipeline_name, dev_mode, submit_predictions):
train_evaluate_predict_cv(pipeline_name, dev_mode, submit_predictions)
def train(pipeline_name, dev_mode):
logger.info('TRAINING')
if bool(params.clean_experiment_directory_before_training) and os.path.isdir(params.experiment_directory):
logger.info('Cleaning experiment_directory...')
shutil.rmtree(params.experiment_directory)
tables = _read_data(dev_mode, read_train=True, read_test=False)
logger.info('Shuffling and splitting into train and test...')
train_data_split, valid_data_split = train_test_split(tables.application_train,
test_size=params.validation_size,
random_state=cfg.RANDOM_SEED,
shuffle=params.shuffle)
logger.info('Target mean in train: {}'.format(train_data_split[cfg.TARGET_COLUMNS].mean()))
logger.info('Target mean in valid: {}'.format(valid_data_split[cfg.TARGET_COLUMNS].mean()))
logger.info('Train shape: {}'.format(train_data_split.shape))
logger.info('Valid shape: {}'.format(valid_data_split.shape))
train_data = {'application': {'X': train_data_split.drop(cfg.TARGET_COLUMNS, axis=1),
'y': train_data_split[cfg.TARGET_COLUMNS].values.reshape(-1),
'X_valid': valid_data_split.drop(cfg.TARGET_COLUMNS, axis=1),
'y_valid': valid_data_split[cfg.TARGET_COLUMNS].values.reshape(-1)
},
'bureau_balance': {'X': tables.bureau_balance},
'bureau': {'X': tables.bureau},
'credit_card_balance': {'X': tables.credit_card_balance},
'installments_payments': {'X': tables.installments_payments},
'pos_cash_balance': {'X': tables.pos_cash_balance},
'previous_application': {'X': tables.previous_application},
}
pipeline = PIPELINES[pipeline_name](config=cfg.SOLUTION_CONFIG, train_mode=True)
pipeline.clean_cache()
logger.info('Start pipeline fit and transform')
pipeline.fit_transform(train_data)
pipeline.clean_cache()
def evaluate(pipeline_name, dev_mode):
logger.info('EVALUATION')
logger.info('Reading data...')
tables = _read_data(dev_mode, read_train=True, read_test=False)
logger.info('Shuffling and splitting to get validation split...')
_, valid_data_split = train_test_split(tables.application_train,
test_size=params.validation_size,
random_state=cfg.RANDOM_SEED,
shuffle=params.shuffle)
logger.info('Target mean in valid: {}'.format(valid_data_split[cfg.TARGET_COLUMNS].mean()))
logger.info('Valid shape: {}'.format(valid_data_split.shape))
y_true = valid_data_split[cfg.TARGET_COLUMNS].values
eval_data = {'application': {'X': valid_data_split.drop(cfg.TARGET_COLUMNS, axis=1),
'y': None,
},
'bureau_balance': {'X': tables.bureau_balance},
'bureau': {'X': tables.bureau},
'credit_card_balance': {'X': tables.credit_card_balance},
'installments_payments': {'X': tables.installments_payments},
'pos_cash_balance': {'X': tables.pos_cash_balance},
'previous_application': {'X': tables.previous_application},
}
pipeline = PIPELINES[pipeline_name](config=cfg.SOLUTION_CONFIG, train_mode=False)
pipeline.clean_cache()
logger.info('Start pipeline transform')
output = pipeline.transform(eval_data)
pipeline.clean_cache()
y_pred = output['prediction']
logger.info('Calculating ROC_AUC on validation set')
score = roc_auc_score(y_true, y_pred)
logger.info('ROC_AUC score on validation is {}'.format(score))
ctx.channel_send('ROC_AUC', 0, score)
def predict(pipeline_name, dev_mode, submit_predictions):
logger.info('PREDICTION')
tables = _read_data(dev_mode, read_train=False, read_test=True)
test_data = {'application': {'X': tables.application_test,
'y': None,
},
'bureau_balance': {'X': tables.bureau_balance},
'bureau': {'X': tables.bureau},
'credit_card_balance': {'X': tables.credit_card_balance},
'installments_payments': {'X': tables.installments_payments},
'pos_cash_balance': {'X': tables.pos_cash_balance},
'previous_application': {'X': tables.previous_application},
}
pipeline = PIPELINES[pipeline_name](config=cfg.SOLUTION_CONFIG, train_mode=False)
pipeline.clean_cache()
logger.info('Start pipeline transform')
output = pipeline.transform(test_data)
pipeline.clean_cache()
y_pred = output['prediction']
if not dev_mode:
logger.info('creating submission file...')
submission = create_submission(tables.application_test, y_pred)
logger.info('verifying submission...')
sample_submission = pd.read_csv(params.sample_submission_filepath)
verify_submission(submission, sample_submission)
submission_filepath = os.path.join(params.experiment_directory, 'submission.csv')
submission.to_csv(submission_filepath, index=None, encoding='utf-8')
logger.info('submission persisted to {}'.format(submission_filepath))
logger.info('submission head \n\n{}'.format(submission.head()))
if submit_predictions and params.kaggle_api:
make_submission(submission_filepath)
def train_evaluate_cv(pipeline_name, dev_mode):
if bool(params.clean_experiment_directory_before_training) and os.path.isdir(params.experiment_directory):
logger.info('Cleaning experiment_directory...')
shutil.rmtree(params.experiment_directory)
tables = _read_data(dev_mode, read_train=True, read_test=False)
target_values = tables.application_train[cfg.TARGET_COLUMNS].values.reshape(-1)
fold_generator = _get_fold_generator(target_values)
fold_scores = []
for fold_id, (train_idx, valid_idx) in enumerate(fold_generator):
(train_data_split,
valid_data_split) = tables.application_train.iloc[train_idx], tables.application_train.iloc[valid_idx]
logger.info('Started fold {}'.format(fold_id))
logger.info('Target mean in train: {}'.format(train_data_split[cfg.TARGET_COLUMNS].mean()))
logger.info('Target mean in valid: {}'.format(valid_data_split[cfg.TARGET_COLUMNS].mean()))
logger.info('Train shape: {}'.format(train_data_split.shape))
logger.info('Valid shape: {}'.format(valid_data_split.shape))
score, _, _ = _fold_fit_evaluate_loop(train_data_split, valid_data_split, tables, fold_id, pipeline_name)
logger.info('Fold {} ROC_AUC {}'.format(fold_id, score))
ctx.channel_send('Fold {} ROC_AUC'.format(fold_id), 0, score)
fold_scores.append(score)
score_mean, score_std = np.mean(fold_scores), np.std(fold_scores)
logger.info('ROC_AUC mean {}, ROC_AUC std {}'.format(score_mean, score_std))
ctx.channel_send('ROC_AUC', 0, score_mean)
ctx.channel_send('ROC_AUC STD', 0, score_std)
def train_evaluate_predict_cv(pipeline_name, dev_mode, submit_predictions):
if bool(params.clean_experiment_directory_before_training) and os.path.isdir(params.experiment_directory):
logger.info('Cleaning experiment_directory...')
shutil.rmtree(params.experiment_directory)
tables = _read_data(dev_mode, read_train=True, read_test=True)
target_values = tables.application_train[cfg.TARGET_COLUMNS].values.reshape(-1)
fold_generator = _get_fold_generator(target_values)
fold_scores, out_of_fold_train_predictions, out_of_fold_test_predictions = [], [], []
for fold_id, (train_idx, valid_idx) in enumerate(fold_generator):
(train_data_split,
valid_data_split) = tables.application_train.iloc[train_idx], tables.application_train.iloc[valid_idx]
logger.info('Started fold {}'.format(fold_id))
logger.info('Target mean in train: {}'.format(train_data_split[cfg.TARGET_COLUMNS].mean()))
logger.info('Target mean in valid: {}'.format(valid_data_split[cfg.TARGET_COLUMNS].mean()))
logger.info('Train shape: {}'.format(train_data_split.shape))
logger.info('Valid shape: {}'.format(valid_data_split.shape))
score, out_of_fold_prediction, test_prediction = _fold_fit_evaluate_predict_loop(train_data_split,
valid_data_split,
tables,
fold_id, pipeline_name)
logger.info('Fold {} ROC_AUC {}'.format(fold_id, score))
ctx.channel_send('Fold {} ROC_AUC'.format(fold_id), 0, score)
out_of_fold_train_predictions.append(out_of_fold_prediction)
out_of_fold_test_predictions.append(test_prediction)
fold_scores.append(score)
out_of_fold_train_predictions = pd.concat(out_of_fold_train_predictions, axis=0)
out_of_fold_test_predictions = pd.concat(out_of_fold_test_predictions, axis=0)
test_prediction_aggregated = _aggregate_test_prediction(out_of_fold_test_predictions)
score_mean, score_std = np.mean(fold_scores), np.std(fold_scores)
logger.info('ROC_AUC mean {}, ROC_AUC std {}'.format(score_mean, score_std))
ctx.channel_send('ROC_AUC', 0, score_mean)
ctx.channel_send('ROC_AUC STD', 0, score_std)
logger.info('Saving predictions')
out_of_fold_train_predictions.to_csv(os.path.join(params.experiment_directory,
'{}_out_of_fold_train_predictions.csv'.format(pipeline_name)),
index=None)
out_of_fold_test_predictions.to_csv(os.path.join(params.experiment_directory,
'{}_out_of_fold_test_predictions.csv'.format(pipeline_name)),
index=None)
test_aggregated_file_path = os.path.join(params.experiment_directory,
'{}_test_predictions_{}.csv'.format(pipeline_name,
params.aggregation_method))
test_prediction_aggregated.to_csv(test_aggregated_file_path, index=None)
if not dev_mode:
logger.info('verifying submission...')
sample_submission = pd.read_csv(params.sample_submission_filepath)
verify_submission(test_prediction_aggregated, sample_submission)
if submit_predictions and params.kaggle_api:
make_submission(test_aggregated_file_path)
def make_submission(submission_filepath):
logger.info('making Kaggle submit...')
os.system('kaggle competitions submit -c home-credit-default-risk -f {} -m {}'
.format(submission_filepath, params.kaggle_message))
def _read_data(dev_mode, read_train=True, read_test=False):
logger.info('Reading data...')
if dev_mode:
nrows = cfg.DEV_SAMPLE_SIZE
logger.info('running in "dev-mode". Sample size is: {}'.format(cfg.DEV_SAMPLE_SIZE))
else:
nrows = None
raw_data = {}
if read_train:
raw_data['application_train'] = pd.read_csv(params.train_filepath, nrows=nrows)
if read_test:
raw_data['application_test'] = | pd.read_csv(params.test_filepath, nrows=nrows) | pandas.read_csv |
import pandas as pd
from tqdm import tqdm
import requests
import numpy as np
import sys
from typing import *
from time import sleep
class _UniProtClient:
def __init__(self, base_url):
self._base_url = base_url
@staticmethod
def _query(query_string) -> str:
for i in range(10):
try:
response = requests.get(query_string)
return response.text
except ConnectionResetError:
sleep(i*10)
@staticmethod
def _response2dictlist(response_string) -> List[dict]:
header_row = response_string.split("\n")[0]
header_items = header_row.split("\t")
r_dict_list = []
for line in response_string.split("\n")[1:]:
if not line:
continue
line_items = line.split("\t")
assert len(header_items) == len(line_items), (header_items, line_items)
r_dict_list.append(dict(zip(header_items, line_items)))
return r_dict_list
@staticmethod
def _chunkwise(iterables, chunk_size):
for i in range(0, len(iterables), chunk_size):
chunk = iterables[i:i + chunk_size]
yield chunk
class UniProtMapper(_UniProtClient):
def __init__(self, from_id: str, to_id: str):
"""For mapping of protein IDs to another ID. Uses UniProt API.
for valid parameters see: https://www.uniprot.org/help/api_idmapping
:param to_id:
Parameters
----------
from_id: origin ID string
to_id: target ID string
Examples
________
gi2uniprotmapping = UniProtMapper("P_GI", "ACC") # This class mapps form GI-number to Uniprot IDs
"""
super().__init__("https://www.uniprot.org/uploadlists/")
self._from_id = from_id
self._to_id = to_id
self._data_format = "tab"
def map_protein_ids(self, protein_list: List[str], chunk_size: int = 500) -> pd.DataFrame:
final_dict_list = []
pbar = tqdm(total=len(protein_list))
try:
for chunk in self._chunkwise(protein_list, chunk_size):
chunklist = "+".join(chunk)
server_query = f"?from={self._from_id}&to={self._to_id}&format={self._data_format}&query={chunklist}"
req = "".join([self._base_url, server_query])
server_response = self._query(req)
server_response_formatted = self._response2dictlist(server_response)
final_dict_list.extend(server_response_formatted)
pbar.update(len(chunk))
finally:
pbar.close()
valid_mappings = pd.DataFrame(final_dict_list)
invalid_ids = set(protein_list) - set(valid_mappings["From"].unique())
invalid_mapping = pd.DataFrame()
invalid_mapping["From"] = sorted(invalid_ids)
invalid_mapping["To"] = np.nan
return | pd.concat([valid_mappings, invalid_mapping]) | pandas.concat |
#!/usr/bin/env python
# -*- coding: utf-8 -*-
import os
import numpy as np
import pandas as pd
import datetime as dt
from scipy import stats
import pymannkendall as mk
from Modules import Read
from Modules.Utils import Listador, FindOutlier, FindOutlierMAD, Cycles
from Modules.Graphs import GraphSerieOutliers, GraphSerieOutliersMAD
from Modules.ENSO import ONIdata, OuliersENSOjust
ONI = ONIdata()
ONI = ONI['Anomalie'].astype(float)
ENSO = ONI[np.where((ONI.values<=-0.5)|(ONI.values>=0.5))[0]]
Path_out = os.path.abspath(os.path.join(os.path.dirname(__file__), 'Tests'))
def CompareNormStandar(Statistical, significance, tails=1):
"""
Compare an statistical of any test with the normal standar
INPUTS
Statistical : float of the value to compare in the normal standar
significance: level of confidence to acept o reject the test
tails : integer in [1,2] to use a test with one or two tails
OUTPUTS
test : boolean with the aceptance of rejection of the null hypothesis
"""
cuantil = 1-significance/tails
Z_norm = stats.norm.ppf(cuantil,loc=0,scale=1)
Pass = abs(Statistical)<Z_norm
return Pass
def CompareTdist(Statistical, DegreesFredom, significance, tails=1):
"""
Compare an statistical of any test with the t estudent distribution
INPUTS
Statistical : float of the value to compare in the normal standar
DegreesFredom : Degrees of fredom of the distirbution
significance : level of confidence to acept o reject the test
tails : integer in [1,2] to use a test with one or two tails
OUTPUTS
test : boolean with the aceptance of rejection of the null hypothesis
"""
cuantil = 1-significance/tails
t = stats.t.ppf(cuantil,df=DegreesFredom)
Pass = abs(Statistical)<t
return Pass
def SingChange(Serie):
"""
Count times where are sing change
INPUTS
Serie : list or array of with the data
"""
if isinstance(Serie, list) == True:
Serie = np.array(Serie)
sing = np.zeros(len(Serie),dtype=int) +1
sing[np.array(Serie)<0] = -1
# return sum((x ^ y)<0 for x, y in zip(Serie, Serie[1:])) # only works for integers
return sum((x ^ y)<0 for x, y in zip(sing, sing[1:]))
def PeaksValleys(Serie):
"""
Fin the peaks and valleys in a serie
INPUTS
Serie : list or array of with the data
"""
if isinstance(Serie, list) == True:
Serie = np.array(Serie)
diff = Serie[:-1]-Serie[1:]
sing = np.zeros(len(diff),dtype=int) +1
sing[np.array(diff)<0] = -1
return sum(((x ^ y)^(y ^ z))<0 for x, y, z in zip(sing, sing[1:], sing[2:]))
def RunsTest(Serie, significance=5E-2):
"""
Make run test (Rachas) for a series
INPUTS
Serie : list or array with the data
significance : level of significance to acept or reject the null hypothesis
OUTPUTS
test : boolean with the aceptance of rejection of the null hypothesis
"""
S_median = np.median(Serie)
runs = SingChange(Serie-S_median)
n1 = np.where(Serie>=S_median)[0].shape[0]
n2 = np.where(Serie< S_median)[0].shape[0]
runs_exp = ((2*n1*n2)/(n1+n2))+1
stan_dev = np.sqrt((2*n1*n2*(2*n1*n2-n1-n2))/ \
(((n1+n2)**2)*(n1+n2-1)))
z = (runs-runs_exp)/stan_dev
test = CompareNormStandar(z, significance,tails=2)
return test
def ChangePointTest(Serie, significance=5E-2):
"""
Make change point test for a serie
INPUTS
Serie : list or array with the data
significance : level of significance to acept or reject the null hypothesis
OUTPUTS
test : boolean with the aceptance of rejection of the null hypothesis
"""
N = len(Serie)
M = PeaksValleys(Serie)
U = abs((M-(2./3.)*(N-2))/np.sqrt((16*N-29)/90.))
test = CompareNormStandar(U, significance,tails=2)
return test
def SpearmanCoefTest(Serie, significance=5E-2):
"""
Make Spearman coeficient test
INPUTS
Serie : list or array with the data
significance : level of significance to acept or reject the null hypothesis
OUTPUTS
test : boolean with the aceptance of rejection of the null hypothesis
"""
if isinstance(Serie, list) == True:
Serie = np.array(Serie)
n = len(Serie)
S = Serie[Serie.argsort()]
R = 1-(6/(n*((n**2)-1)))* np.sum((Serie-S)**2 )
U = abs(R*np.sqrt(n-2)/np.sqrt(1-(R**2)))
test = CompareTdist(U,DegreesFredom=n-2,significance=significance,tails=2)
return test
def AndersonTest(Serie, rezagos=None, significance=5E-2, ):
"""
Make andreson independence test
INPUTS
"""
cuantil = 1-significance/2
Z_norm = stats.norm.ppf(cuantil,loc=0,scale=1)
N = len(Serie)
if rezagos is None:
rezagos = N -2
Mean = np.nanmean(Serie)
r = np.empty(len(Serie), dtype=float)
t = np.empty(len(Serie), dtype=bool)
for k in range(rezagos):
lim_up = (-1 + Z_norm*np.sqrt(N-k-1))/(N-k)
lim_dw = (-1 - Z_norm*np.sqrt(N-k-1))/(N-k)
r[k] = np.sum((Serie[:N-k]-Mean)*(Serie[k:]-Mean))/np.sum((Serie - Mean)**2)
if (r[k] > lim_dw)&(r[k]<lim_up):
t[k] = True
else:
t[k] = False
if t.sum() == N:
test = True
else:
test = False
return test
def MannKendall_modified(Serie, rezagos=None, significance=5E-2):
"""
This function checks the Modified Mann-Kendall (MK) test using Hamed and Rao (1998) method.
"""
MK = mk.hamed_rao_modification_test(Serie,alpha=significance,lag=rezagos)
test = CompareNormStandar(MK.z, significance,tails=2)
return test
# Est_path = os.path.abspath(os.path.join(os.path.dirname(__file__), 'CleanData/PPT'))
Est_path = os.path.abspath(os.path.join(os.path.dirname(__file__), 'CleanData/QDL'))
Estaciones = Listador(Est_path,final='.csv')
Pruebas = ['Rachas', 'PuntoCambio', 'Spearman', 'Anderson','MannKendall']
Test = pd.DataFrame([], columns=Pruebas)
Outl = | pd.DataFrame([], columns=['outlier_inf','outlier_sup']) | pandas.DataFrame |
# %% Imports
import pandas as pd
import numpy as np
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import LabelEncoder
import matplotlib.pyplot as plt
import tensorflow as tf
from tensorflow.keras import Sequential
from tensorflow.keras.layers import Dense
from geneticalgorithm import geneticalgorithm as ga
#%% General
datasetURL = './data/data.csv'
epochs = 8
#%% load the dataset
df = | pd.read_csv(datasetURL) | pandas.read_csv |
#!/usr/bin/env python
# coding: utf-8
# # Teaching your models to play fair
#
# In this notebook you will use `fairlearn` and the Fairness dashboard to generate predictors for the Census dataset. This dataset is a classification problem - given a range of data about 32,000 individuals, predict whether their annual income is above or below $50,000 \$ $ per year.
#
# For the purposes of this notebook, we will treat this as a loan decision problem. We will pretend that the label indicates whether or not each individual repaid a loan in the past. We will use the data to train a predictor to predict whether previously unseen individuals will repay a loan or not. The assumption is that the model predictions are used to decide whether an individual should be offered a loan.
#
# We will first train a fairness-unaware predictor and show that it leads to unfair decisions under a specific notion of fairness called *demographic parity*. We then mitigate unfairness by applying the `GridSearch` algorithm from `fairlearn` package.
# ## Setup
#
# We will first install `fairlearn`
# In[1]:
get_ipython().system('pip install fairlearn')
# We will import a few packages that would be required
# In[2]:
from fairlearn.reductions import GridSearch
from fairlearn.reductions import DemographicParity, ErrorRate
from sklearn import svm, neighbors, tree
from sklearn.preprocessing import LabelEncoder,StandardScaler
from sklearn.linear_model import LogisticRegression
import pandas as pd
import shap
import numpy as np
shap.initjs()
# ## Loading the data
#
# For simplicity, we import the data set from the `shap` package, which contains the data in a cleaned format.
# In[3]:
X_raw, Y = shap.datasets.adult()
X_raw.head()
# ## Performing data transformations
#
# We are going to treat the sex of each individual as a protected attribute (where 0 indicates female and 1 indicates male), and in this particular case we are going separate this attribute out and drop it from the main data. We then perform some standard data preprocessing steps to convert the data into a format suitable for the ML algorithms
# In[4]:
A = X_raw["Sex"]
X = X_raw.drop(labels=['Sex'],axis = 1)
X = pd.get_dummies(X)
sc = StandardScaler()
X_scaled = sc.fit_transform(X)
X_scaled = | pd.DataFrame(X_scaled, columns=X.columns) | pandas.DataFrame |
from hydroDL import kPath, utils
from hydroDL.app import waterQuality, wqRela
from hydroDL.data import gageII, usgs, gridMET
from hydroDL.master import basins
from hydroDL.post import axplot, figplot
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import os
import time
from astropy.timeseries import LombScargle
import scipy.signal as signal
import matplotlib.gridspec as gridspec
# pick out sites that are have relative large number of observations
dirInv = os.path.join(kPath.dirData, 'USGS', 'inventory')
fileSiteNo = os.path.join(dirInv, 'siteNoLst-1979')
siteNoLstAll = pd.read_csv(fileSiteNo, header=None, dtype=str)[0].tolist()
dfAll = pd.read_csv(os.path.join(dirInv, 'codeCount.csv'),
dtype={'siteNo': str}).set_index('siteNo')
# pick some sites
# codeLst = ['00915', '00940', '00955','00300']
codeLst = ['00915', '00945', '00955']
tempLst = list()
for code in codeLst:
temp = dfAll[dfAll[code] > 200].index.tolist()
tempLst.append(temp)
siteNoLst = tempLst[0]
for k in range(1, len(tempLst)):
siteNoLst = list(set(siteNoLst).intersection(tempLst[k]))
startDate = pd.datetime(1979, 1, 1)
endDate = pd.datetime(2019, 12, 31)
nc = len(codeLst)
ns = len(siteNoLst)
# cal dw
rMat = np.ndarray([ns, nc])
pdfArea = gageII.readData(varLst=['DRAIN_SQKM'], siteNoLst=siteNoLst)
unitConv = 0.3048**3*365*24*60*60/1000**2
for k, siteNo in enumerate(siteNoLst):
for i, code in enumerate(codeLst):
area = pdfArea.loc[siteNo]['DRAIN_SQKM']
dfC = usgs.readSample(siteNo, codeLst=codeLst, startDate=startDate)
dfQ = usgs.readStreamflow(siteNo, startDate=startDate)
df = dfC.join(dfQ)
t = df.index.values
q = df['00060_00003'].values/area*unitConv
c = df[code].values
(q, c), ind = utils.rmNan([q, c])
x = 10**np.linspace(np.log10(np.min(q[q > 0])),
np.log10(np.max(q[~np.isnan(q)])), 20)
ceq, dw, y = wqRela.kateModel(q, c, q)
corr = np.corrcoef(c, y)[0, 1]
rMat[k, i] = corr
dfCrd = gageII.readData(
varLst=['LAT_GAGE', 'LNG_GAGE'], siteNoLst=siteNoLst)
lat = dfCrd['LAT_GAGE'].values
lon = dfCrd['LNG_GAGE'].values
def funcMap():
figM, axM = plt.subplots(nc, 1, figsize=(8, 6))
for k in range(nc):
axplot.mapPoint(axM[k], lat, lon, rMat[:, k], s=12)
figP = plt.figure(constrained_layout=True)
spec = gridspec.GridSpec(ncols=3, nrows=2, figure=figP)
axLst = [figP.add_subplot(spec[0, :])] +\
[figP.add_subplot(spec[1, k]) for k in range(3)]
axP = np.array(axLst)
return figM, axM, figP, axP, lon, lat
def funcPoint(iP, axes):
kA = 0
siteNo = siteNoLst[iP]
startDate = pd.datetime(1979, 1, 1)
endDate = pd.datetime(2019, 12, 31)
ctR = pd.date_range(startDate, endDate)
dfData = | pd.DataFrame({'date': ctR}) | pandas.DataFrame |
import json
from datetime import datetime, timedelta
import requests
import pandas as pd
import numpy as np
from scipy.spatial.distance import euclidean, cityblock, cosine
import ssl
ssl._create_default_https_context = ssl._create_unverified_context
class Task(object):
def __init__(self, data):
self.df = pd.read_csv(data)
def t1(self, name):
df = self.df.copy()
sim_weights = {}
for user in df.columns[1:]:
df_subset = df[[str(name), user]][df[str(name)].notnull() & df[user].notnull()]
sim_weights[user] = 1 / (1 + cosine(df_subset[name].values.flatten(), df_subset[user].values.flatten()))
df = df[df[name].isna()]
del df[name]
result = []
for i in range(len(df)):
ratings = df.iloc[i][1:]
predicted_rating = 0.0
weights_sum = 0.0
for user in df.columns[1:]:
if not np.isnan(sim_weights[user]) and not np.isnan(ratings[user]):
predicted_rating += ratings[user] * sim_weights[user]
weights_sum += sim_weights[user]
predicted_rating /= weights_sum
result.append((df.iloc[i][0], predicted_rating))
return result
def t2(self, name):
df = self.df.copy().transpose()
weights = {}
for i in range(len(df.columns)):
j = 0
while j in range(len(df.columns)):
if i != j:
df_subset = df[[i, j]][df[i].notnull() & df[j].notnull()]
weights[i, j] = 1 / (1 + cosine(pd.to_numeric(df_subset[i].values.flatten()[1:]),
pd.to_numeric(df_subset[j].values.flatten()[1:])))
j += 1
i += 1
sim_weights = {}
key_list = []
for key, value in weights.items():
if (key[0], key[1]) not in key_list and (key[1], key[0]) not in key_list:
key_list.append(key)
sim_weights[key] = value
user_col = df.loc[name]
user_null_movies = []
user_valid_movies = []
for key, value in user_col.items():
if | pd.isna(value) | pandas.isna |
import os
import glob
import pandas as pd
game_files=glob.glob(os.path.join(os.getcwd(),'games','*.EVE'))
game_files.sort()
game_frames=[]
for game_file in game_files:
game_frame=pd.read_csv(game_file,names=['type','multi2','multi3','multi4','multi5','multi6','event'])
game_frames.append(game_frame)
games= pd.concat(game_frames)
games.loc[games['multi5']=='??','multi5']=''
identifiers=games['multi2'].str.extract(r'(.LS(\d{4})\d{5})')
identifiers=identifiers.fillna(method='ffill')
identifiers.columns=['game_id','year']
games = | pd.concat([games,identifiers], axis=1, sort=False) | pandas.concat |
# -*- coding: utf-8 -*-
"""
Created on Thu Sep 01 11:05:46 2019
@author: Neal
"""
import requests
from bs4 import BeautifulSoup
import pandas as pd
fake_header = { "user-agent": "Mozilla/5.0 (Windows NT 10.0; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/60.0.3112.113 Safari/537.36",
"Accept":"text/html,application/xhtml+xml,application/xml;q=0.9,image/webp,*/*;q=0.8",
"Accept-Encoding":"gzip, deflate, sdch",
"Accept-Language":"zh-TW,zh;q=0.8,en-US;q=0.6,en;q=0.4,zh-CN;q=0.2"
}
#send http request with fake http header
stocks = ['000001','000002','000004','000005']
base_url = 'http://vip.stock.finance.sina.com.cn/corp/view/vCB_AllMemordDetail.php?stockid='
#open file to with written permission
results = []
for stock in stocks:#process stock one by one
#prepare the request webpage with desired parameters
url = base_url+stock
print("Now we are crawling stock",url)
response = requests.get(url,headers = fake_header)
if response.status_code == 200:
response.encoding = 'gb2312'#because sina use
html = BeautifulSoup(response.text,"html.parser")
# search all the announcement tag by tag name and attribute/values
title_divs = html.find_all('div',attrs={'class':'title_cls'})
for div in title_divs:#get and store announcement title one by one
# writer.writerow([stock,div.get_text().strip()])
date_str="Missing"
for sibling in div.next_siblings:
if sibling.name=='center':
date_str= sibling.get_text().strip()
break
results.append([stock,date_str,div.get_text().strip()] )
print(results)
new_df = | pd.DataFrame(columns=['Stock', 'Date','Title'], data=results) | pandas.DataFrame |
import os
import pandas as pd
import csv
from sklearn.model_selection import train_test_split
import numpy as np
import random
import tensorflow as tf
import torch
#directory of tasks dataset
os.chdir("original_data")
#destination path to create tsv files, dipends on data cutting
path_0 = "mttransformer/data/0"
path_100_no_gan = "mttransformer/data/100/no_gan"
path_200_no_gan = "mttransformer/data/200/no_gan"
path_500_no_gan = "mttransformer/data/500/no_gan"
path_100_gan = "mttransformer/data/100/gan"
path_200_gan = "mttransformer/data/200/gan"
path_500_gan = "mttransformer/data/500/gan"
#if you use a model with gan the flag "apply_gan" is True, else False
apply_gan=False
#data cutting
number_labeled_examples=0 #0-100-200-500
#if you want activate balancing, that is used only in the model Multi-task, MT-DNN and MT-GANBERT
balancing=False
#path train and test dataset of the task
tsv_haspeede_train = 'haspeede_TW-train.tsv'
tsv_haspeede_test = 'haspeede_TW-reference.tsv'
tsv_AMI2018_train = 'AMI2018_it_training.tsv'
tsv_AMI2018_test = 'AMI2018_it_testing.tsv'
tsv_AMI2018_train = 'AMI2018_it_training.tsv'
tsv_AMI2018_test = 'AMI2018_it_testing.tsv'
tsv_DANKMEMES2020_train = 'dankmemes_task2_train.csv'
tsv_DANKMEMES2020_test = 'hate_test.csv'
tsv_SENTIPOLC2016_train = 'training_set_sentipolc16.csv'
tsv_SENTIPOLC2016_test = 'test_set_sentipolc16_gold2000.csv'
tsv_SENTIPOLC2016_train = 'training_set_sentipolc16.csv'
tsv_SENTIPOLC2016_test = 'test_set_sentipolc16_gold2000.csv'
#Upload the dataset of all task as dataframes
#haspeede_TW
df_train = pd.read_csv(tsv_haspeede_train, delimiter='\t', names=('id','sentence','label'))
df_train = df_train[['id']+['label']+['sentence']]
df_test = pd.read_csv(tsv_haspeede_test, delimiter='\t', names=('id','sentence','label'))
df_test = df_test[['id']+['label']+['sentence']]
#AMI2018A
df_train2 = pd.read_csv(tsv_AMI2018_train, delimiter='\t')
df_train2 = df_train2[['id']+['misogynous']+['text']]
df_test2 = pd.read_csv(tsv_AMI2018_test, delimiter='\t')
df_test2 = df_test2[['id']+['misogynous']+['text']]
#AMI2018B
df_train3 = pd.read_csv(tsv_AMI2018_train, delimiter='\t')
df = pd.DataFrame(columns=['id', 'misogyny_category', 'text'])
for ind in df_train3.index:
if df_train3.misogynous[ind]==1:
if df_train3.misogyny_category[ind] == 'stereotype':
df = df.append({'id' : df_train3['id'][ind], 'misogyny_category' : 0, 'text' : df_train3['text'][ind] }, ignore_index=True)
#elif df_train3.misogyny_category[ind] == 'dominance':
#df = df.append({'id' : df_train3['id'][ind], 'misogyny_category' : 1, 'text' : df_train3['text'][ind] }, ignore_index=True)
#elif df_train3.misogyny_category[ind] == 'derailing':
#df = df.append({'id' : df_train3['id'][ind], 'misogyny_category' : 2, 'text' : df_train3['text'][ind] }, ignore_index=True)
elif df_train3.misogyny_category[ind] == 'sexual_harassment':
df = df.append({'id' : df_train3['id'][ind], 'misogyny_category' : 1, 'text' : df_train3['text'][ind] }, ignore_index=True)
elif df_train3.misogyny_category[ind] == 'discredit':
df = df.append({'id' : df_train3['id'][ind], 'misogyny_category' : 2, 'text' : df_train3['text'][ind] }, ignore_index=True)
df_train3 = df
df_test3 = pd.read_csv(tsv_AMI2018_test, delimiter='\t')
df = pd.DataFrame(columns=['id', 'misogyny_category', 'text'])
for ind in df_test3.index:
if df_test3.misogynous[ind]==1:
if df_test3.misogyny_category[ind] == 'stereotype':
df = df.append({'id' : df_test3['id'][ind], 'misogyny_category' : 0, 'text' : df_test3['text'][ind] }, ignore_index=True)
#elif df_test3.misogyny_category[ind] == 'dominance':
#df = df.append({'id' : df_test3['id'][ind], 'misogyny_category' : 1, 'text' : df_test3['text'][ind] }, ignore_index=True)
#elif df_test3.misogyny_category[ind] == 'derailing':
#df = df.append({'id' : df_test3['id'][ind], 'misogyny_category' : 2, 'text' : df_test3['text'][ind] }, ignore_index=True)
elif df_test3.misogyny_category[ind] == 'sexual_harassment':
df = df.append({'id' : df_test3['id'][ind], 'misogyny_category' : 1, 'text' : df_test3['text'][ind] }, ignore_index=True)
elif df_test3.misogyny_category[ind] == 'discredit':
df = df.append({'id' : df_test3['id'][ind], 'misogyny_category' : 2, 'text' : df_test3['text'][ind] }, ignore_index=True)
df_test3 = df
#DANKMEMES2020
df_train4 = pd.read_csv(tsv_DANKMEMES2020_train, delimiter=',')
df_train4 = df_train4[['File']+['Hate Speech']+['Text']]
df_test4 = pd.read_csv(tsv_DANKMEMES2020_test, delimiter=',')
df_test4 = df_test4[['File']+['Hate Speech']+['Text']]
#SENTIPOLC20161
df_train5 = pd.read_csv(tsv_SENTIPOLC2016_train, delimiter=',')
df_train5 = df_train5[['idtwitter']+['subj']+['text']]
df_test5 = pd.read_csv(tsv_SENTIPOLC2016_test, delimiter=',')
df_test5 = df_test5[['idtwitter']+['subj']+['text']]
for ind in df_train5.index:
if "\t" in df_train5.text[ind]:
df_train5 = df_train5.replace(to_replace='\t', value='', regex=True)
#SENTIPOLC20162
df_train6 = pd.read_csv(tsv_SENTIPOLC2016_train, delimiter=',')
df = pd.DataFrame(columns=['idtwitter', 'polarity', 'text'])
for ind in df_train6.index:
if df_train6['subj'][ind] == 1:
if df_train6['opos'][ind] == 1 and df_train6['oneg'][ind] == 0:
df = df.append({'idtwitter' : df_train6['idtwitter'][ind], 'polarity' : 0, 'text' : df_train6['text'][ind] }, ignore_index=True)
elif df_train6['opos'][ind] == 0 and df_train6['oneg'][ind] == 1:
df = df.append({'idtwitter' : df_train6['idtwitter'][ind], 'polarity' : 1, 'text' : df_train6['text'][ind] }, ignore_index=True)
elif df_train6['opos'][ind] == 0 and df_train6['oneg'][ind] == 0:
df = df.append({'idtwitter' : df_train6['idtwitter'][ind], 'polarity' : 2, 'text' : df_train6['text'][ind] }, ignore_index=True)
else:
if df_train6['opos'][ind] == 0 and df_train6['oneg'][ind] == 0:
df = df.append({'idtwitter' : df_train6['idtwitter'][ind], 'polarity' : 2, 'text' : df_train6['text'][ind] }, ignore_index=True)
df_train6 = df
for ind in df_train6.index:
if "\t" in df_train6.text[ind]:
df_train6 = df_train6.replace(to_replace='\t', value='', regex=True)
df_test6 = | pd.read_csv(tsv_SENTIPOLC2016_test, delimiter=',') | pandas.read_csv |
# Licensed to Modin Development Team under one or more contributor license agreements.
# See the NOTICE file distributed with this work for additional information regarding
# copyright ownership. The Modin Development Team licenses this file to you under the
# Apache License, Version 2.0 (the "License"); you may not use this file except in
# compliance with the License. You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software distributed under
# the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF
# ANY KIND, either express or implied. See the License for the specific language
# governing permissions and limitations under the License.
import pytest
import numpy as np
import pandas
from pandas.testing import assert_index_equal
import matplotlib
import modin.pandas as pd
import sys
from modin.pandas.test.utils import (
NROWS,
RAND_LOW,
RAND_HIGH,
df_equals,
arg_keys,
name_contains,
test_data,
test_data_values,
test_data_keys,
axis_keys,
axis_values,
int_arg_keys,
int_arg_values,
create_test_dfs,
eval_general,
generate_multiindex,
extra_test_parameters,
)
from modin.config import NPartitions
NPartitions.put(4)
# Force matplotlib to not use any Xwindows backend.
matplotlib.use("Agg")
def eval_setitem(md_df, pd_df, value, col=None, loc=None):
if loc is not None:
col = pd_df.columns[loc]
value_getter = value if callable(value) else (lambda *args, **kwargs: value)
eval_general(
md_df, pd_df, lambda df: df.__setitem__(col, value_getter(df)), __inplace__=True
)
@pytest.mark.parametrize(
"dates",
[
["2018-02-27 09:03:30", "2018-02-27 09:04:30"],
["2018-02-27 09:03:00", "2018-02-27 09:05:00"],
],
)
@pytest.mark.parametrize("subset", ["a", "b", ["a", "b"], None])
def test_asof_with_nan(dates, subset):
data = {"a": [10, 20, 30, 40, 50], "b": [None, None, None, None, 500]}
index = pd.DatetimeIndex(
[
"2018-02-27 09:01:00",
"2018-02-27 09:02:00",
"2018-02-27 09:03:00",
"2018-02-27 09:04:00",
"2018-02-27 09:05:00",
]
)
modin_where = pd.DatetimeIndex(dates)
pandas_where = pandas.DatetimeIndex(dates)
compare_asof(data, index, modin_where, pandas_where, subset)
@pytest.mark.parametrize(
"dates",
[
["2018-02-27 09:03:30", "2018-02-27 09:04:30"],
["2018-02-27 09:03:00", "2018-02-27 09:05:00"],
],
)
@pytest.mark.parametrize("subset", ["a", "b", ["a", "b"], None])
def test_asof_without_nan(dates, subset):
data = {"a": [10, 20, 30, 40, 50], "b": [70, 600, 30, -200, 500]}
index = pd.DatetimeIndex(
[
"2018-02-27 09:01:00",
"2018-02-27 09:02:00",
"2018-02-27 09:03:00",
"2018-02-27 09:04:00",
"2018-02-27 09:05:00",
]
)
modin_where = pd.DatetimeIndex(dates)
pandas_where = pandas.DatetimeIndex(dates)
compare_asof(data, index, modin_where, pandas_where, subset)
@pytest.mark.parametrize(
"lookup",
[
[60, 70, 90],
[60.5, 70.5, 100],
],
)
@pytest.mark.parametrize("subset", ["col2", "col1", ["col1", "col2"], None])
def test_asof_large(lookup, subset):
data = test_data["float_nan_data"]
index = list(range(NROWS))
modin_where = pd.Index(lookup)
pandas_where = pandas.Index(lookup)
compare_asof(data, index, modin_where, pandas_where, subset)
def compare_asof(
data, index, modin_where: pd.Index, pandas_where: pandas.Index, subset
):
modin_df = pd.DataFrame(data, index=index)
pandas_df = pandas.DataFrame(data, index=index)
df_equals(
modin_df.asof(modin_where, subset=subset),
pandas_df.asof(pandas_where, subset=subset),
)
df_equals(
modin_df.asof(modin_where.values, subset=subset),
pandas_df.asof(pandas_where.values, subset=subset),
)
df_equals(
modin_df.asof(list(modin_where.values), subset=subset),
pandas_df.asof(list(pandas_where.values), subset=subset),
)
df_equals(
modin_df.asof(modin_where.values[0], subset=subset),
pandas_df.asof(pandas_where.values[0], subset=subset),
)
@pytest.mark.parametrize("data", test_data_values, ids=test_data_keys)
def test_first_valid_index(data):
modin_df = pd.DataFrame(data)
pandas_df = pandas.DataFrame(data)
assert modin_df.first_valid_index() == (pandas_df.first_valid_index())
@pytest.mark.parametrize("data", test_data_values, ids=test_data_keys)
@pytest.mark.parametrize("n", int_arg_values, ids=arg_keys("n", int_arg_keys))
def test_head(data, n):
# Test normal dataframe head
modin_df = pd.DataFrame(data)
pandas_df = pandas.DataFrame(data)
df_equals(modin_df.head(n), pandas_df.head(n))
df_equals(modin_df.head(len(modin_df) + 1), pandas_df.head(len(pandas_df) + 1))
# Test head when we call it from a QueryCompilerView
modin_result = modin_df.loc[:, ["col1", "col3", "col3"]].head(n)
pandas_result = pandas_df.loc[:, ["col1", "col3", "col3"]].head(n)
df_equals(modin_result, pandas_result)
@pytest.mark.skip(reason="Defaulting to Pandas")
@pytest.mark.parametrize("data", test_data_values, ids=test_data_keys)
def test_iat(data):
modin_df = pd.DataFrame(data)
pandas_df = pandas.DataFrame(data) # noqa F841
with pytest.raises(NotImplementedError):
modin_df.iat()
@pytest.mark.gpu
@pytest.mark.parametrize("data", test_data_values, ids=test_data_keys)
def test_iloc(request, data):
modin_df = pd.DataFrame(data)
pandas_df = pandas.DataFrame(data)
if not name_contains(request.node.name, ["empty_data"]):
# Scaler
np.testing.assert_equal(modin_df.iloc[0, 1], pandas_df.iloc[0, 1])
# Series
df_equals(modin_df.iloc[0], pandas_df.iloc[0])
df_equals(modin_df.iloc[1:, 0], pandas_df.iloc[1:, 0])
df_equals(modin_df.iloc[1:2, 0], pandas_df.iloc[1:2, 0])
# DataFrame
df_equals(modin_df.iloc[[1, 2]], pandas_df.iloc[[1, 2]])
# See issue #80
# df_equals(modin_df.iloc[[1, 2], [1, 0]], pandas_df.iloc[[1, 2], [1, 0]])
df_equals(modin_df.iloc[1:2, 0:2], pandas_df.iloc[1:2, 0:2])
# Issue #43
modin_df.iloc[0:3, :]
# Write Item
modin_df.iloc[[1, 2]] = 42
pandas_df.iloc[[1, 2]] = 42
df_equals(modin_df, pandas_df)
modin_df = pd.DataFrame(data)
pandas_df = pandas.DataFrame(data)
modin_df.iloc[0] = modin_df.iloc[1]
pandas_df.iloc[0] = pandas_df.iloc[1]
df_equals(modin_df, pandas_df)
modin_df = pd.DataFrame(data)
pandas_df = pandas.DataFrame(data)
modin_df.iloc[:, 0] = modin_df.iloc[:, 1]
pandas_df.iloc[:, 0] = pandas_df.iloc[:, 1]
df_equals(modin_df, pandas_df)
# From issue #1775
df_equals(
modin_df.iloc[lambda df: df.index.get_indexer_for(df.index[:5])],
pandas_df.iloc[lambda df: df.index.get_indexer_for(df.index[:5])],
)
else:
with pytest.raises(IndexError):
modin_df.iloc[0, 1]
@pytest.mark.gpu
@pytest.mark.parametrize("data", test_data_values, ids=test_data_keys)
def test_index(data):
modin_df = pd.DataFrame(data)
pandas_df = pandas.DataFrame(data)
df_equals(modin_df.index, pandas_df.index)
modin_df_cp = modin_df.copy()
pandas_df_cp = pandas_df.copy()
modin_df_cp.index = [str(i) for i in modin_df_cp.index]
pandas_df_cp.index = [str(i) for i in pandas_df_cp.index]
df_equals(modin_df_cp.index, pandas_df_cp.index)
@pytest.mark.gpu
@pytest.mark.parametrize("data", test_data_values, ids=test_data_keys)
def test_indexing_duplicate_axis(data):
modin_df = pd.DataFrame(data)
pandas_df = pandas.DataFrame(data)
modin_df.index = pandas_df.index = [i // 3 for i in range(len(modin_df))]
assert any(modin_df.index.duplicated())
assert any(pandas_df.index.duplicated())
df_equals(modin_df.iloc[0], pandas_df.iloc[0])
df_equals(modin_df.loc[0], pandas_df.loc[0])
df_equals(modin_df.iloc[0, 0:4], pandas_df.iloc[0, 0:4])
df_equals(
modin_df.loc[0, modin_df.columns[0:4]],
pandas_df.loc[0, pandas_df.columns[0:4]],
)
@pytest.mark.gpu
@pytest.mark.parametrize("data", test_data_values, ids=test_data_keys)
def test_keys(data):
modin_df = pd.DataFrame(data)
pandas_df = pandas.DataFrame(data)
df_equals(modin_df.keys(), pandas_df.keys())
@pytest.mark.parametrize("data", test_data_values, ids=test_data_keys)
def test_loc(data):
modin_df = pd.DataFrame(data)
pandas_df = pandas.DataFrame(data)
key1 = modin_df.columns[0]
key2 = modin_df.columns[1]
# Scaler
df_equals(modin_df.loc[0, key1], pandas_df.loc[0, key1])
# Series
df_equals(modin_df.loc[0], pandas_df.loc[0])
df_equals(modin_df.loc[1:, key1], pandas_df.loc[1:, key1])
df_equals(modin_df.loc[1:2, key1], pandas_df.loc[1:2, key1])
# DataFrame
df_equals(modin_df.loc[[1, 2]], pandas_df.loc[[1, 2]])
# List-like of booleans
indices = [i % 3 == 0 for i in range(len(modin_df.index))]
columns = [i % 5 == 0 for i in range(len(modin_df.columns))]
modin_result = modin_df.loc[indices, columns]
pandas_result = pandas_df.loc[indices, columns]
df_equals(modin_result, pandas_result)
modin_result = modin_df.loc[:, columns]
pandas_result = pandas_df.loc[:, columns]
df_equals(modin_result, pandas_result)
modin_result = modin_df.loc[indices]
pandas_result = pandas_df.loc[indices]
df_equals(modin_result, pandas_result)
# See issue #80
# df_equals(modin_df.loc[[1, 2], ['col1']], pandas_df.loc[[1, 2], ['col1']])
df_equals(modin_df.loc[1:2, key1:key2], pandas_df.loc[1:2, key1:key2])
# From issue #421
df_equals(modin_df.loc[:, [key2, key1]], pandas_df.loc[:, [key2, key1]])
df_equals(modin_df.loc[[2, 1], :], pandas_df.loc[[2, 1], :])
# From issue #1023
key1 = modin_df.columns[0]
key2 = modin_df.columns[-2]
df_equals(modin_df.loc[:, key1:key2], pandas_df.loc[:, key1:key2])
# Write Item
modin_df_copy = modin_df.copy()
pandas_df_copy = pandas_df.copy()
modin_df_copy.loc[[1, 2]] = 42
pandas_df_copy.loc[[1, 2]] = 42
df_equals(modin_df_copy, pandas_df_copy)
# From issue #1775
df_equals(
modin_df.loc[lambda df: df.iloc[:, 0].isin(list(range(1000)))],
pandas_df.loc[lambda df: df.iloc[:, 0].isin(list(range(1000)))],
)
# From issue #1374
with pytest.raises(KeyError):
modin_df.loc["NO_EXIST"]
def test_loc_multi_index():
modin_df = pd.read_csv(
"modin/pandas/test/data/blah.csv", header=[0, 1, 2, 3], index_col=0
)
pandas_df = pandas.read_csv(
"modin/pandas/test/data/blah.csv", header=[0, 1, 2, 3], index_col=0
)
df_equals(modin_df.loc[1], pandas_df.loc[1])
df_equals(modin_df.loc[1, "Presidents"], pandas_df.loc[1, "Presidents"])
df_equals(
modin_df.loc[1, ("Presidents", "Pure mentions")],
pandas_df.loc[1, ("Presidents", "Pure mentions")],
)
assert (
modin_df.loc[1, ("Presidents", "Pure mentions", "IND", "all")]
== pandas_df.loc[1, ("Presidents", "Pure mentions", "IND", "all")]
)
df_equals(modin_df.loc[(1, 2), "Presidents"], pandas_df.loc[(1, 2), "Presidents"])
tuples = [
("bar", "one"),
("bar", "two"),
("bar", "three"),
("bar", "four"),
("baz", "one"),
("baz", "two"),
("baz", "three"),
("baz", "four"),
("foo", "one"),
("foo", "two"),
("foo", "three"),
("foo", "four"),
("qux", "one"),
("qux", "two"),
("qux", "three"),
("qux", "four"),
]
modin_index = pd.MultiIndex.from_tuples(tuples, names=["first", "second"])
pandas_index = pandas.MultiIndex.from_tuples(tuples, names=["first", "second"])
frame_data = np.random.randint(0, 100, size=(16, 100))
modin_df = pd.DataFrame(
frame_data,
index=modin_index,
columns=["col{}".format(i) for i in range(100)],
)
pandas_df = pandas.DataFrame(
frame_data,
index=pandas_index,
columns=["col{}".format(i) for i in range(100)],
)
df_equals(modin_df.loc["bar", "col1"], pandas_df.loc["bar", "col1"])
assert modin_df.loc[("bar", "one"), "col1"] == pandas_df.loc[("bar", "one"), "col1"]
df_equals(
modin_df.loc["bar", ("col1", "col2")],
pandas_df.loc["bar", ("col1", "col2")],
)
# From issue #1456
transposed_modin = modin_df.T
transposed_pandas = pandas_df.T
df_equals(
transposed_modin.loc[transposed_modin.index[:-2], :],
transposed_pandas.loc[transposed_pandas.index[:-2], :],
)
# From issue #1610
df_equals(modin_df.loc[modin_df.index], pandas_df.loc[pandas_df.index])
df_equals(modin_df.loc[modin_df.index[:7]], pandas_df.loc[pandas_df.index[:7]])
@pytest.mark.parametrize("index", [["row1", "row2", "row3"]])
@pytest.mark.parametrize("columns", [["col1", "col2"]])
def test_loc_assignment(index, columns):
md_df, pd_df = create_test_dfs(index=index, columns=columns)
for i, ind in enumerate(index):
for j, col in enumerate(columns):
value_to_assign = int(str(i) + str(j))
md_df.loc[ind][col] = value_to_assign
pd_df.loc[ind][col] = value_to_assign
df_equals(md_df, pd_df)
@pytest.fixture
def loc_iter_dfs():
columns = ["col1", "col2", "col3"]
index = ["row1", "row2", "row3"]
return create_test_dfs(
{col: ([idx] * len(index)) for idx, col in enumerate(columns)},
columns=columns,
index=index,
)
@pytest.mark.parametrize("reverse_order", [False, True])
@pytest.mark.parametrize("axis", [0, 1])
def test_loc_iter_assignment(loc_iter_dfs, reverse_order, axis):
if reverse_order and axis:
pytest.xfail(
"Due to internal sorting of lookup values assignment order is lost, see GH-#2552"
)
md_df, pd_df = loc_iter_dfs
select = [slice(None), slice(None)]
select[axis] = sorted(pd_df.axes[axis][:-1], reverse=reverse_order)
select = tuple(select)
pd_df.loc[select] = pd_df.loc[select] + pd_df.loc[select]
md_df.loc[select] = md_df.loc[select] + md_df.loc[select]
df_equals(md_df, pd_df)
@pytest.mark.parametrize("reverse_order", [False, True])
@pytest.mark.parametrize("axis", [0, 1])
def test_loc_order(loc_iter_dfs, reverse_order, axis):
md_df, pd_df = loc_iter_dfs
select = [slice(None), slice(None)]
select[axis] = sorted(pd_df.axes[axis][:-1], reverse=reverse_order)
select = tuple(select)
df_equals(pd_df.loc[select], md_df.loc[select])
@pytest.mark.gpu
@pytest.mark.parametrize("data", test_data_values, ids=test_data_keys)
def test_loc_nested_assignment(data):
modin_df = pd.DataFrame(data)
pandas_df = pandas.DataFrame(data)
key1 = modin_df.columns[0]
key2 = modin_df.columns[1]
modin_df[key1].loc[0] = 500
pandas_df[key1].loc[0] = 500
df_equals(modin_df, pandas_df)
modin_df[key2].loc[0] = None
pandas_df[key2].loc[0] = None
df_equals(modin_df, pandas_df)
def test_iloc_assignment():
modin_df = pd.DataFrame(index=["row1", "row2", "row3"], columns=["col1", "col2"])
pandas_df = pandas.DataFrame(
index=["row1", "row2", "row3"], columns=["col1", "col2"]
)
modin_df.iloc[0]["col1"] = 11
modin_df.iloc[1]["col1"] = 21
modin_df.iloc[2]["col1"] = 31
modin_df.iloc[0]["col2"] = 12
modin_df.iloc[1]["col2"] = 22
modin_df.iloc[2]["col2"] = 32
pandas_df.iloc[0]["col1"] = 11
pandas_df.iloc[1]["col1"] = 21
pandas_df.iloc[2]["col1"] = 31
pandas_df.iloc[0]["col2"] = 12
pandas_df.iloc[1]["col2"] = 22
pandas_df.iloc[2]["col2"] = 32
df_equals(modin_df, pandas_df)
@pytest.mark.parametrize("data", test_data_values, ids=test_data_keys)
def test_iloc_nested_assignment(data):
modin_df = pd.DataFrame(data)
pandas_df = pandas.DataFrame(data)
key1 = modin_df.columns[0]
key2 = modin_df.columns[1]
modin_df[key1].iloc[0] = 500
pandas_df[key1].iloc[0] = 500
df_equals(modin_df, pandas_df)
modin_df[key2].iloc[0] = None
pandas_df[key2].iloc[0] = None
df_equals(modin_df, pandas_df)
def test_loc_series():
md_df, pd_df = create_test_dfs({"a": [1, 2], "b": [3, 4]})
pd_df.loc[pd_df["a"] > 1, "b"] = np.log(pd_df["b"])
md_df.loc[md_df["a"] > 1, "b"] = np.log(md_df["b"])
df_equals(pd_df, md_df)
@pytest.mark.parametrize("data", test_data_values, ids=test_data_keys)
def test_pop(request, data):
modin_df = pd.DataFrame(data)
pandas_df = | pandas.DataFrame(data) | pandas.DataFrame |
import logging
from datetime import datetime, timedelta
from logging import handlers
import numpy as np
from pandas import Series, to_datetime, Timedelta, Timestamp, date_range
from pandas.tseries.frequencies import to_offset
from scipy import interpolate
logger = logging.getLogger(__name__)
def frequency_is_supported(freq):
"""Method to determine if a frequency is supported for a pastas-model.
Parameters
----------
freq: str
Returns
-------
freq
String with the simulation frequency
Notes
-----
Possible frequency-offsets are listed in:
http://pandas.pydata.org/pandas-docs/stable/timeseries.html#offset-aliases
The frequency can be a multiple of these offsets, like '7D'. Because of the
use in convolution, only frequencies with an equidistant offset are
allowed. This means monthly ('M'), yearly ('Y') or even weekly ('W')
frequencies are not allowed. Use '7D' for a weekly simulation.
D calendar day frequency
H hourly frequency
T, min minutely frequency
S secondly frequency
L, ms milliseconds
U, us microseconds
N nanoseconds
TODO: Rename to get_frequency_string and change Returns-documentation
"""
offset = to_offset(freq)
if not hasattr(offset, 'delta'):
logger.error("Frequency %s not supported." % freq)
else:
if offset.n is 1:
freq = offset.name
else:
freq = str(offset.n) + offset.name
return freq
def get_stress_dt(freq):
"""Internal method to obtain a timestep in days from a frequency string.
Parameters
----------
freq: str
Returns
-------
dt: float
Approximate timestep in number of days.
Notes
-----
Used for comparison to determine if a time series needs to be up or
downsampled.
See http://pandas.pydata.org/pandas-docs/stable/timeseries.html#offset-aliases
for the offset_aliases supported by Pandas.
"""
# Get the frequency string and multiplier
offset = to_offset(freq)
if hasattr(offset, 'delta'):
dt = offset.delta / Timedelta(1, "D")
else:
num = offset.n
freq = offset.name
if freq in ['A', 'Y', 'AS', 'YS', 'BA', 'BY', 'BAS', 'BYS']:
# year
dt = num * 365
elif freq in ['BQ', 'BQS', 'Q', 'QS']:
# quarter
dt = num * 90
elif freq in ['BM', 'BMS', 'CBM', 'CBMS', 'M', 'MS']:
# month
dt = num * 30
elif freq in ['SM', 'SMS']:
# semi-month
dt = num * 15
elif freq in ['W']:
# week
dt = num * 7
elif freq in ['B', 'C']:
# day
dt = num
elif freq in ['BH', 'CBH']:
# hour
dt = num * 1 / 24
else:
raise (ValueError('freq of {} not supported'.format(freq)))
return dt
def get_dt(freq):
"""Method to obtain a timestep in DAYS from a frequency string.
Parameters
----------
freq: str
Returns
-------
dt: float
Number of days
"""
# Get the frequency string and multiplier
dt = to_offset(freq).delta / Timedelta(1, "D")
return dt
def get_time_offset(t, freq):
"""Internal method to calculate the time offset of a TimeStamp.
Parameters
----------
t: pandas.Timestamp
Timestamp to calculate the offset from the desired freq for.
freq: str
String with the desired frequency.
Returns
-------
offset: pandas.Timedelta
Timedelta with the offset for the timestamp t.
"""
return t - t.floor(freq)
def get_sample(tindex, ref_tindex):
"""Sample the index so that the frequency is not higher than the frequency
of ref_tindex.
Parameters
----------
tindex: pandas.index
Pandas index object
ref_tindex: pandas.index
Pandas index object
Returns
-------
series: pandas.index
Notes
-----
Find the index closest to the ref_tindex, and then return a selection
of the index.
"""
if len(tindex) == 1:
return tindex
else:
f = interpolate.interp1d(tindex.asi8, np.arange(0, tindex.size),
kind='nearest', bounds_error=False,
fill_value='extrapolate')
ind = np.unique(f(ref_tindex.asi8).astype(int))
return tindex[ind]
def timestep_weighted_resample(series0, tindex):
"""Resample a timeseries to a new tindex, using an overlapping period
weighted average.
The original series and the new tindex do not have to be equidistant. Also,
the timestep-edges of the new tindex do not have to overlap with the
original series.
It is assumed the series consists of measurements that describe an
intensity at the end of the period for which they apply. Therefore, when
upsampling, the values are uniformly spread over the new timestep (like
bfill).
Compared to the reample methods in Pandas, this method is more accurate for
non-equidistanct series. It is much slower however.
Parameters
----------
series0 : pandas.Series
The original series to be resampled
tindex : pandas.index
The index to which to resample the series
Returns
-------
series : pandas.Series
The resampled series
"""
# determine some arrays for the input-series
t0e = np.array(series0.index)
dt0 = np.diff(t0e)
dt0 = np.hstack((dt0[0], dt0))
t0s = t0e - dt0
v0 = series0.values
# determine some arrays for the output-series
t1e = np.array(tindex)
dt1 = np.diff(t1e)
dt1 = np.hstack((dt1[0], dt1))
t1s = t1e - dt1
v1 = []
for t1si, t1ei in zip(t1s, t1e):
# determine which periods within the series are within the new tindex
mask = (t0e > t1si) & (t0s < t1ei)
if np.any(mask):
# cut by the timestep-edges
ts = t0s[mask]
te = t0e[mask]
ts[ts < t1si] = t1si
te[te > t1ei] = t1ei
# determine timestep
dt = (te - ts).astype(float)
# determine timestep-weighted value
v1.append(np.sum(dt * v0[mask]) / np.sum(dt))
# replace all values in the series
series = Series(v1, index=tindex)
return series
def timestep_weighted_resample_fast(series0, freq):
"""Resample a time series to a new frequency, using an overlapping period
weighted average.
The original series does not have to be equidistant.
It is assumed the series consists of measurements that describe an
intensity at the end of the period for which they apply. Therefore, when
upsampling, the values are uniformly spread over the new timestep (like
bfill).
Compared to the resample methods in Pandas, this method is more accurate
for non-equidistant series. It is slower than Pandas (but faster then the
original timestep_weighted_resample).
Parameters
----------
series0 : pandas.Series
original series to be resampled
freq : str
a Pandas frequency string
Returns
-------
series : pandas.Series
resampled series
"""
series = series0.copy()
# first mutiply by the timestep in the unit of freq
dt = np.diff(series0.index) / Timedelta(1, freq)
series[1:] = series[1:] * dt
# get a new index
index = date_range(series.index[0].floor(freq), series.index[-1],
freq=freq)
# calculate the cumulative sum
series = series.cumsum()
# add NaNs at none-existing values in series at index
series = series.combine_first( | Series(np.NaN, index=index) | pandas.Series |
import os
import pandas as pd
def loadData(folder_path: str, date: str, start_time: str='9:30',
end_time: str='16:00') -> pd.DataFrame:
"""Function to load complete price data for a given asset, from a given
folder. This function loads all '*.csv' files from a given directory
corresponding to instruments on a specific asset. Given a date, this
returns a formatted DataFrame with 1 minute intervals from a start
time to end time of the day, with each of the aligned prices in columns
corresponding to the files they were sourced from. This function assumes
dates and times are in the first column of the CSV file (headers 'Dates'),
and that the prices are in the second column. The corresponding column in
the final DataFrame is the name of the file it was read from. This function
also forward and backward propagates prices from the last/first viable
value if one is not available for a given minute.
Arguments:
folder_path {str} -- Path from which CSV files are to be ingested.
date {str} -- Date the data was collected. This is encoded in the index
of the DataFrame (format: yyyy-mm-dd).
Keyword Arguments:
start_time {str} -- Start time (military time) (default: {'9:30'}).
end_time {str} -- End time (military time) (default: {'16:00'}).
Returns:
pd.DataFrame -- Formatted DataFrame with aligned prices.
"""
file_list = os.listdir(folder_path) # Getting files
# Removing non-CSV files from list (Assume one '.' in file name)
file_list = [x for x in file_list if x.split('.')[1] == 'csv']
# Defining full start and end time
start = date + ' ' + start_time
end = date + ' ' + end_time
# Building DataFrame with correct index
data_index = pd.DatetimeIndex(start=start, end=end, freq='1min')
# Creating empty DataFrame with index
data = pd.DataFrame(index=data_index)
for file_name in file_list:
# Isolating security name
asset_name = file_name.split('.')[0]
# Loading data to DataFrame, parsing dates
candidate_data = pd.read_csv(os.path.join(folder_path, file_name),
index_col='Dates', parse_dates=True)
# Renaming index
candidate_data.index.name = 'date'
# Renaming data column
candidate_data.columns = [asset_name]
# Re-indexing with correct index, propagate values forward
candidate_data.reindex(index=data_index)
# Add to main data
data = | pd.concat([data, candidate_data], axis=1) | pandas.concat |
import logging
import numpy as np
import pandas as pd
import pandas.testing as pdt
import pytest
import sentry_sdk
from solarforecastarbiter import utils
def _make_aggobs(obsid, ef=pd.Timestamp('20191001T1100Z'),
eu=None, oda=None):
return {
'observation_id': obsid,
'effective_from': ef,
'effective_until': eu,
'observation_deleted_at': oda
}
nindex = pd.date_range(start='20191004T0000Z',
freq='1h', periods=10)
@pytest.fixture()
def ids():
return ['f2844284-ea0a-11e9-a7da-f4939feddd82',
'f3e310ba-ea0a-11e9-a7da-f4939feddd82',
'09ed7cf6-ea0b-11e9-a7da-f4939feddd82',
'0fe9f2ba-ea0b-11e9-a7da-f4939feddd82',
'67ea9200-ea0e-11e9-832b-f4939feddd82']
@pytest.fixture()
def aggobs(ids):
return tuple([
_make_aggobs(ids[0]),
_make_aggobs(ids[1], pd.Timestamp('20191004T0501Z')),
_make_aggobs(ids[2], eu=pd.Timestamp('20191004T0400Z')),
_make_aggobs(ids[2], | pd.Timestamp('20191004T0700Z') | pandas.Timestamp |
# -*- coding: utf-8 -*-
from datetime import timedelta
from distutils.version import LooseVersion
import numpy as np
import pytest
import pandas as pd
import pandas.util.testing as tm
from pandas import (
DatetimeIndex, Int64Index, Series, Timedelta, TimedeltaIndex, Timestamp,
date_range, timedelta_range
)
from pandas.errors import NullFrequencyError
@pytest.fixture(params=[pd.offsets.Hour(2), timedelta(hours=2),
np.timedelta64(2, 'h'), Timedelta(hours=2)],
ids=str)
def delta(request):
# Several ways of representing two hours
return request.param
@pytest.fixture(params=['B', 'D'])
def freq(request):
return request.param
class TestTimedeltaIndexArithmetic(object):
# Addition and Subtraction Operations
# -------------------------------------------------------------
# TimedeltaIndex.shift is used by __add__/__sub__
def test_tdi_shift_empty(self):
# GH#9903
idx = pd.TimedeltaIndex([], name='xxx')
tm.assert_index_equal(idx.shift(0, freq='H'), idx)
tm.assert_index_equal(idx.shift(3, freq='H'), idx)
def test_tdi_shift_hours(self):
# GH#9903
idx = pd.TimedeltaIndex(['5 hours', '6 hours', '9 hours'], name='xxx')
tm.assert_index_equal(idx.shift(0, freq='H'), idx)
exp = pd.TimedeltaIndex(['8 hours', '9 hours', '12 hours'], name='xxx')
tm.assert_index_equal(idx.shift(3, freq='H'), exp)
exp = pd.TimedeltaIndex(['2 hours', '3 hours', '6 hours'], name='xxx')
tm.assert_index_equal(idx.shift(-3, freq='H'), exp)
def test_tdi_shift_minutes(self):
# GH#9903
idx = pd.TimedeltaIndex(['5 hours', '6 hours', '9 hours'], name='xxx')
tm.assert_index_equal(idx.shift(0, freq='T'), idx)
exp = pd.TimedeltaIndex(['05:03:00', '06:03:00', '9:03:00'],
name='xxx')
tm.assert_index_equal(idx.shift(3, freq='T'), exp)
exp = pd.TimedeltaIndex(['04:57:00', '05:57:00', '8:57:00'],
name='xxx')
tm.assert_index_equal(idx.shift(-3, freq='T'), exp)
def test_tdi_shift_int(self):
# GH#8083
trange = pd.to_timedelta(range(5), unit='d') + pd.offsets.Hour(1)
result = trange.shift(1)
expected = TimedeltaIndex(['1 days 01:00:00', '2 days 01:00:00',
'3 days 01:00:00',
'4 days 01:00:00', '5 days 01:00:00'],
freq='D')
tm.assert_index_equal(result, expected)
def test_tdi_shift_nonstandard_freq(self):
# GH#8083
trange = pd.to_timedelta(range(5), unit='d') + pd.offsets.Hour(1)
result = trange.shift(3, freq='2D 1s')
expected = TimedeltaIndex(['6 days 01:00:03', '7 days 01:00:03',
'8 days 01:00:03', '9 days 01:00:03',
'10 days 01:00:03'], freq='D')
tm.assert_index_equal(result, expected)
def test_shift_no_freq(self):
# GH#19147
tdi = TimedeltaIndex(['1 days 01:00:00', '2 days 01:00:00'], freq=None)
with pytest.raises(NullFrequencyError):
tdi.shift(2)
# -------------------------------------------------------------
def test_ufunc_coercions(self):
# normal ops are also tested in tseries/test_timedeltas.py
idx = TimedeltaIndex(['2H', '4H', '6H', '8H', '10H'],
freq='2H', name='x')
for result in [idx * 2, np.multiply(idx, 2)]:
assert isinstance(result, TimedeltaIndex)
exp = TimedeltaIndex(['4H', '8H', '12H', '16H', '20H'],
freq='4H', name='x')
tm.assert_index_equal(result, exp)
assert result.freq == '4H'
for result in [idx / 2, np.divide(idx, 2)]:
assert isinstance(result, TimedeltaIndex)
exp = TimedeltaIndex(['1H', '2H', '3H', '4H', '5H'],
freq='H', name='x')
tm.assert_index_equal(result, exp)
assert result.freq == 'H'
idx = TimedeltaIndex(['2H', '4H', '6H', '8H', '10H'],
freq='2H', name='x')
for result in [-idx, np.negative(idx)]:
assert isinstance(result, TimedeltaIndex)
exp = TimedeltaIndex(['-2H', '-4H', '-6H', '-8H', '-10H'],
freq='-2H', name='x')
tm.assert_index_equal(result, exp)
assert result.freq == '-2H'
idx = TimedeltaIndex(['-2H', '-1H', '0H', '1H', '2H'],
freq='H', name='x')
for result in [abs(idx), np.absolute(idx)]:
assert isinstance(result, TimedeltaIndex)
exp = TimedeltaIndex(['2H', '1H', '0H', '1H', '2H'],
freq=None, name='x')
tm.assert_index_equal(result, exp)
assert result.freq is None
# -------------------------------------------------------------
# Binary operations TimedeltaIndex and integer
def test_tdi_add_int(self, one):
# Variants of `one` for #19012
rng = timedelta_range('1 days 09:00:00', freq='H', periods=10)
result = rng + one
expected = timedelta_range('1 days 10:00:00', freq='H', periods=10)
tm.assert_index_equal(result, expected)
def test_tdi_iadd_int(self, one):
rng = timedelta_range('1 days 09:00:00', freq='H', periods=10)
expected = timedelta_range('1 days 10:00:00', freq='H', periods=10)
rng += one
tm.assert_index_equal(rng, expected)
def test_tdi_sub_int(self, one):
rng = timedelta_range('1 days 09:00:00', freq='H', periods=10)
result = rng - one
expected = timedelta_range('1 days 08:00:00', freq='H', periods=10)
tm.assert_index_equal(result, expected)
def test_tdi_isub_int(self, one):
rng = timedelta_range('1 days 09:00:00', freq='H', periods=10)
expected = timedelta_range('1 days 08:00:00', freq='H', periods=10)
rng -= one
tm.assert_index_equal(rng, expected)
# -------------------------------------------------------------
# __add__/__sub__ with integer arrays
@pytest.mark.parametrize('box', [np.array, pd.Index])
def test_tdi_add_integer_array(self, box):
# GH#19959
rng = timedelta_range('1 days 09:00:00', freq='H', periods=3)
other = box([4, 3, 2])
expected = TimedeltaIndex(['1 day 13:00:00'] * 3)
result = rng + other
tm.assert_index_equal(result, expected)
result = other + rng
tm.assert_index_equal(result, expected)
@pytest.mark.parametrize('box', [np.array, pd.Index])
def test_tdi_sub_integer_array(self, box):
# GH#19959
rng = timedelta_range('9H', freq='H', periods=3)
other = box([4, 3, 2])
expected = TimedeltaIndex(['5H', '7H', '9H'])
result = rng - other
tm.assert_index_equal(result, expected)
result = other - rng
tm.assert_index_equal(result, -expected)
@pytest.mark.parametrize('box', [np.array, pd.Index])
def test_tdi_addsub_integer_array_no_freq(self, box):
# GH#19959
tdi = TimedeltaIndex(['1 Day', 'NaT', '3 Hours'])
other = box([14, -1, 16])
with pytest.raises(NullFrequencyError):
tdi + other
with pytest.raises(NullFrequencyError):
other + tdi
with pytest.raises(NullFrequencyError):
tdi - other
with pytest.raises(NullFrequencyError):
other - tdi
# -------------------------------------------------------------
# Binary operations TimedeltaIndex and timedelta-like
# Note: add and sub are tested in tests.test_arithmetic
def test_tdi_iadd_timedeltalike(self, delta):
# only test adding/sub offsets as + is now numeric
rng = timedelta_range('1 days', '10 days')
expected = timedelta_range('1 days 02:00:00', '10 days 02:00:00',
freq='D')
rng += delta
tm.assert_index_equal(rng, expected)
def test_tdi_isub_timedeltalike(self, delta):
# only test adding/sub offsets as - is now numeric
rng = timedelta_range('1 days', '10 days')
expected = timedelta_range('0 days 22:00:00', '9 days 22:00:00')
rng -= delta
tm.assert_index_equal(rng, expected)
# -------------------------------------------------------------
def test_subtraction_ops(self):
# with datetimes/timedelta and tdi/dti
tdi = TimedeltaIndex(['1 days', pd.NaT, '2 days'], name='foo')
dti = date_range('20130101', periods=3, name='bar')
td = Timedelta('1 days')
dt = Timestamp('20130101')
pytest.raises(TypeError, lambda: tdi - dt)
pytest.raises(TypeError, lambda: tdi - dti)
pytest.raises(TypeError, lambda: td - dt)
pytest.raises(TypeError, lambda: td - dti)
result = dt - dti
expected = TimedeltaIndex(['0 days', '-1 days', '-2 days'], name='bar')
tm.assert_index_equal(result, expected)
result = dti - dt
expected = TimedeltaIndex(['0 days', '1 days', '2 days'], name='bar')
tm.assert_index_equal(result, expected)
result = tdi - td
expected = TimedeltaIndex(['0 days', pd.NaT, '1 days'], name='foo')
tm.assert_index_equal(result, expected, check_names=False)
result = td - tdi
expected = TimedeltaIndex(['0 days', pd.NaT, '-1 days'], name='foo')
tm.assert_index_equal(result, expected, check_names=False)
result = dti - td
expected = DatetimeIndex(
['20121231', '20130101', '20130102'], name='bar')
tm.assert_index_equal(result, expected, check_names=False)
result = dt - tdi
expected = DatetimeIndex(['20121231', pd.NaT, '20121230'], name='foo')
tm.assert_index_equal(result, expected)
def test_subtraction_ops_with_tz(self):
# check that dt/dti subtraction ops with tz are validated
dti = date_range('20130101', periods=3)
ts = Timestamp('20130101')
dt = ts.to_pydatetime()
dti_tz = date_range('20130101', periods=3).tz_localize('US/Eastern')
ts_tz = Timestamp('20130101').tz_localize('US/Eastern')
ts_tz2 = Timestamp('20130101').tz_localize('CET')
dt_tz = ts_tz.to_pydatetime()
td = Timedelta('1 days')
def _check(result, expected):
assert result == expected
assert isinstance(result, Timedelta)
# scalars
result = ts - ts
expected = Timedelta('0 days')
_check(result, expected)
result = dt_tz - ts_tz
expected = Timedelta('0 days')
_check(result, expected)
result = ts_tz - dt_tz
expected = Timedelta('0 days')
_check(result, expected)
# tz mismatches
pytest.raises(TypeError, lambda: dt_tz - ts)
pytest.raises(TypeError, lambda: dt_tz - dt)
pytest.raises(TypeError, lambda: dt_tz - ts_tz2)
pytest.raises(TypeError, lambda: dt - dt_tz)
pytest.raises(TypeError, lambda: ts - dt_tz)
pytest.raises(TypeError, lambda: ts_tz2 - ts)
pytest.raises(TypeError, lambda: ts_tz2 - dt)
pytest.raises(TypeError, lambda: ts_tz - ts_tz2)
# with dti
pytest.raises(TypeError, lambda: dti - ts_tz)
pytest.raises(TypeError, lambda: dti_tz - ts)
pytest.raises(TypeError, lambda: dti_tz - ts_tz2)
result = dti_tz - dt_tz
expected = TimedeltaIndex(['0 days', '1 days', '2 days'])
tm.assert_index_equal(result, expected)
result = dt_tz - dti_tz
expected = TimedeltaIndex(['0 days', '-1 days', '-2 days'])
tm.assert_index_equal(result, expected)
result = dti_tz - ts_tz
expected = TimedeltaIndex(['0 days', '1 days', '2 days'])
tm.assert_index_equal(result, expected)
result = ts_tz - dti_tz
expected = TimedeltaIndex(['0 days', '-1 days', '-2 days'])
tm.assert_index_equal(result, expected)
result = td - td
expected = Timedelta('0 days')
_check(result, expected)
result = dti_tz - td
expected = DatetimeIndex(
['20121231', '20130101', '20130102'], tz='US/Eastern')
tm.assert_index_equal(result, expected)
def test_dti_tdi_numeric_ops(self):
# These are normally union/diff set-like ops
tdi = TimedeltaIndex(['1 days', pd.NaT, '2 days'], name='foo')
dti = date_range('20130101', periods=3, name='bar')
# TODO(wesm): unused?
# td = Timedelta('1 days')
# dt = Timestamp('20130101')
result = tdi - tdi
expected = TimedeltaIndex(['0 days', pd.NaT, '0 days'], name='foo')
tm.assert_index_equal(result, expected)
result = tdi + tdi
expected = TimedeltaIndex(['2 days', pd.NaT, '4 days'], name='foo')
tm.assert_index_equal(result, expected)
result = dti - tdi # name will be reset
expected = DatetimeIndex(['20121231', pd.NaT, '20130101'])
tm.assert_index_equal(result, expected)
def test_addition_ops(self):
# with datetimes/timedelta and tdi/dti
tdi = TimedeltaIndex(['1 days', pd.NaT, '2 days'], name='foo')
dti = date_range('20130101', periods=3, name='bar')
td = Timedelta('1 days')
dt = Timestamp('20130101')
result = tdi + dt
expected = DatetimeIndex(['20130102', pd.NaT, '20130103'], name='foo')
tm.assert_index_equal(result, expected)
result = dt + tdi
expected = DatetimeIndex(['20130102', pd.NaT, '20130103'], name='foo')
tm.assert_index_equal(result, expected)
result = td + tdi
expected = TimedeltaIndex(['2 days', pd.NaT, '3 days'], name='foo')
tm.assert_index_equal(result, expected)
result = tdi + td
expected = TimedeltaIndex(['2 days', pd.NaT, '3 days'], name='foo')
tm.assert_index_equal(result, expected)
# unequal length
pytest.raises(ValueError, lambda: tdi + dti[0:1])
pytest.raises(ValueError, lambda: tdi[0:1] + dti)
# random indexes
pytest.raises(NullFrequencyError, lambda: tdi + Int64Index([1, 2, 3]))
# this is a union!
# pytest.raises(TypeError, lambda : Int64Index([1,2,3]) + tdi)
result = tdi + dti # name will be reset
expected = DatetimeIndex(['20130102', pd.NaT, '20130105'])
tm.assert_index_equal(result, expected)
result = dti + tdi # name will be reset
expected = DatetimeIndex(['20130102', pd.NaT, '20130105'])
tm.assert_index_equal(result, expected)
result = dt + td
expected = Timestamp('20130102')
assert result == expected
result = td + dt
expected = Timestamp('20130102')
assert result == expected
def test_ops_ndarray(self):
td = Timedelta('1 day')
# timedelta, timedelta
other = pd.to_timedelta(['1 day']).values
expected = pd.to_timedelta(['2 days']).values
tm.assert_numpy_array_equal(td + other, expected)
if LooseVersion(np.__version__) >= LooseVersion('1.8'):
tm.assert_numpy_array_equal(other + td, expected)
pytest.raises(TypeError, lambda: td + np.array([1]))
pytest.raises(TypeError, lambda: np.array([1]) + td)
expected = pd.to_timedelta(['0 days']).values
tm.assert_numpy_array_equal(td - other, expected)
if LooseVersion(np.__version__) >= LooseVersion('1.8'):
tm.assert_numpy_array_equal(-other + td, expected)
pytest.raises(TypeError, lambda: td - np.array([1]))
pytest.raises(TypeError, lambda: np.array([1]) - td)
expected = pd.to_timedelta(['2 days']).values
tm.assert_numpy_array_equal(td * np.array([2]), expected)
tm.assert_numpy_array_equal(np.array([2]) * td, expected)
pytest.raises(TypeError, lambda: td * other)
pytest.raises(TypeError, lambda: other * td)
tm.assert_numpy_array_equal(td / other,
np.array([1], dtype=np.float64))
if LooseVersion(np.__version__) >= LooseVersion('1.8'):
tm.assert_numpy_array_equal(other / td,
np.array([1], dtype=np.float64))
# timedelta, datetime
other = pd.to_datetime(['2000-01-01']).values
expected = pd.to_datetime(['2000-01-02']).values
tm.assert_numpy_array_equal(td + other, expected)
if LooseVersion(np.__version__) >= LooseVersion('1.8'):
tm.assert_numpy_array_equal(other + td, expected)
expected = pd.to_datetime(['1999-12-31']).values
tm.assert_numpy_array_equal(-td + other, expected)
if LooseVersion(np.__version__) >= LooseVersion('1.8'):
tm.assert_numpy_array_equal(other - td, expected)
def test_timedelta_ops_with_missing_values(self):
# setup
s1 = pd.to_timedelta(Series(['00:00:01']))
s2 = pd.to_timedelta(Series(['00:00:02']))
sn = pd.to_timedelta(Series([pd.NaT]))
df1 = pd.DataFrame(['00:00:01']).apply(pd.to_timedelta)
df2 = pd.DataFrame(['00:00:02']).apply(pd.to_timedelta)
dfn = pd.DataFrame([pd.NaT]).apply(pd.to_timedelta)
scalar1 = pd.to_timedelta('00:00:01')
scalar2 = pd.to_timedelta('00:00:02')
timedelta_NaT = pd.to_timedelta('NaT')
actual = scalar1 + scalar1
assert actual == scalar2
actual = scalar2 - scalar1
assert actual == scalar1
actual = s1 + s1
tm.assert_series_equal(actual, s2)
actual = s2 - s1
tm.assert_series_equal(actual, s1)
actual = s1 + scalar1
tm.assert_series_equal(actual, s2)
actual = scalar1 + s1
tm.assert_series_equal(actual, s2)
actual = s2 - scalar1
tm.assert_series_equal(actual, s1)
actual = -scalar1 + s2
tm.assert_series_equal(actual, s1)
actual = s1 + timedelta_NaT
tm.assert_series_equal(actual, sn)
actual = timedelta_NaT + s1
tm.assert_series_equal(actual, sn)
actual = s1 - timedelta_NaT
tm.assert_series_equal(actual, sn)
actual = -timedelta_NaT + s1
tm.assert_series_equal(actual, sn)
with pytest.raises(TypeError):
s1 + np.nan
with pytest.raises(TypeError):
np.nan + s1
with pytest.raises(TypeError):
s1 - np.nan
with pytest.raises(TypeError):
-np.nan + s1
actual = s1 + pd.NaT
| tm.assert_series_equal(actual, sn) | pandas.util.testing.assert_series_equal |
"""Tools to calculate mass balance and convert appropriately from volume."""
from __future__ import annotations
import json
import os
import pathlib
import warnings
from typing import Any, Callable
import geopandas as gpd
import numpy as np
import pandas as pd
import rasterio as rio
import shapely
from tqdm import tqdm
import terradem.dem_tools
import terradem.files
import terradem.metadata
ICE_DENSITY_CONVERSION = 0.85
ICE_DENSITY_ERROR = 0.06
STANDARD_START_YEAR = 1931
STANDARD_END_YEAR = 2016
def read_mb_index() -> pd.DataFrame:
data = pd.read_csv(
terradem.files.INPUT_FILE_PATHS["massbalance_index"],
delim_whitespace=True,
skiprows=2,
index_col=0,
)
data.index.name = "year"
return data
def match_zones() -> Callable[[float, float, float, float], tuple[float, str]]:
mb = read_mb_index().cumsum()
standard_mb = pd.Series(
index=mb.columns,
data=np.diff(mb.T[[STANDARD_START_YEAR, STANDARD_END_YEAR]], axis=1).ravel(),
)
zones = sorted(mb.columns, key=lambda x: len(x), reverse=True)
lk50_outlines = gpd.read_file(terradem.files.INPUT_FILE_PATHS["lk50_outlines"])
for zone in zones:
matches = []
for i, character in enumerate(zone):
matches.append(lk50_outlines[f"RivLevel{i}"] == str(character))
all_matches = np.all(matches, axis=0)
lk50_outlines.loc[all_matches, "zone"] = zone
# Zone A55 is not covered by the zones, so hardcode this to be A54 instead.
lk50_outlines.loc[
(lk50_outlines["RivLevel0"] == "A") & (lk50_outlines["RivLevel1"] == "5") & (lk50_outlines["RivLevel2"] == "5"),
"zone",
] = "A54"
lk50_outlines["easting"] = lk50_outlines.geometry.centroid.x
lk50_outlines["northing"] = lk50_outlines.geometry.centroid.y
def get_mb_factor(easting: float, northing: float, start_year: float, end_year: float) -> tuple[float, str]:
# Calculate the distance between the point and each lk50_outline centroid
distance = np.linalg.norm(
[lk50_outlines["easting"] - easting, lk50_outlines["northing"] - northing],
axis=0,
)
# Find the closest lk50 outline
min_distance_idx = np.argwhere(distance == distance.min()).ravel()[0]
# Extract the representative zone for the closest lk50 outline.
mb_zone = lk50_outlines.iloc[min_distance_idx]["zone"]
# Calculate the mass balance of that zone for the given start and end year
actual_mb = mb.loc[int(end_year), mb_zone] - mb.loc[int(start_year), mb_zone]
# Calculate the conversion factor to the STANDARD_START_YEAR--STANDARD_END_YEAR
factor = standard_mb[mb_zone] / actual_mb
return factor, zone
return get_mb_factor
def get_volume_change() -> None:
glacier_indices_ds = rio.open(terradem.files.TEMP_FILES["lk50_rasterized"])
ddem_versions = {
"non_interp": terradem.files.TEMP_FILES["ddem_coreg_tcorr"],
"norm-regional-national": terradem.files.TEMP_FILES["ddem_coreg_tcorr_national-interp-extrap"],
"norm-regional-sgi1-subregion": terradem.files.TEMP_FILES["ddem_coreg_tcorr_subregion1-interp-extrap"],
"norm-regional-sgi0-subregion": terradem.files.TEMP_FILES["ddem_coreg_tcorr_subregion0-interp-extrap"],
}
output = pd.DataFrame(
index=ddem_versions.keys(), columns=["mean", "median", "std", "area", "volume_change", "coverage"]
)
print("Reading glacier mask")
glacier_mask = glacier_indices_ds.read(1, masked=True).filled(0) > 0
total_area = np.count_nonzero(glacier_mask) * (glacier_indices_ds.res[0] * glacier_indices_ds.res[1])
for key in tqdm(ddem_versions):
ddem_ds = rio.open(ddem_versions[key])
ddem_values = ddem_ds.read(1, masked=True).filled(np.nan)[glacier_mask]
output.loc[key] = {
"mean": np.nanmean(ddem_values),
"median": np.nanmedian(ddem_values),
"std": np.nanstd(ddem_values),
"area": total_area,
"volume_change": np.nanmean(ddem_values) * total_area,
"coverage": np.count_nonzero(np.isfinite(ddem_values)) / np.count_nonzero(glacier_mask),
}
print(output)
output.to_csv("temp/volume_change.csv")
def get_corrections():
mb_index = read_mb_index().cumsum()
dirpath = pathlib.Path(terradem.files.TEMP_SUBDIRS["tcorr_meta_coreg"])
data_list: list[dict[str, Any]] = []
for filepath in dirpath.iterdir():
with open(filepath) as infile:
data = json.load(infile)
data["station"] = filepath.stem
data_list.append(data)
corrections = pd.DataFrame(data_list).set_index("station")
corrections["start_date"] = pd.to_datetime(corrections["start_date"])
for zone, data in corrections.groupby("sgi_zone", as_index=False):
corrections.loc[data.index, "masschange_standard"] = (
mb_index.loc[STANDARD_START_YEAR, zone] - mb_index.loc[STANDARD_END_YEAR, zone]
)
corrections.loc[data.index, "masschange_actual"] = (
mb_index.loc[data["start_date"].dt.year.values, zone].values
- mb_index.loc[data["end_year"].astype(int), zone].values
)
def get_masschanges(easting: float, northing: float) -> tuple[float, float]:
distances = np.argmin(
np.linalg.norm([corrections["easting"] - easting, corrections["northing"] - northing], axis=0)
)
return corrections.iloc[distances]["masschange_standard"], corrections.iloc[distances]["masschange_actual"]
return get_masschanges
def get_start_and_end_years():
mb_index = read_mb_index().cumsum()
dirpath = pathlib.Path(terradem.files.TEMP_SUBDIRS["tcorr_meta_coreg"])
data_list: list[dict[str, Any]] = []
for filepath in dirpath.iterdir():
with open(filepath) as infile:
data = json.load(infile)
data["station"] = filepath.stem
data_list.append(data)
corrections = pd.DataFrame(data_list).set_index("station")
corrections["start_date"] = pd.to_datetime(corrections["start_date"])
def get_start_and_end_year(easting: float, northing: float) -> tuple[float, float]:
distances = np.argmin(
np.linalg.norm([corrections["easting"] - easting, corrections["northing"] - northing], axis=0)
)
return (
corrections.iloc[distances]["start_date"].year
+ corrections.iloc[distances]["start_date"].month / 12
+ corrections.iloc[distances]["start_date"].day / 364.75,
corrections.iloc[distances]["end_year"],
)
return get_start_and_end_year
def temporal_corr_error_model():
stochastic_yearly_error = 0.2 # m/a w.e.
masschange_model = get_corrections()
def error_model(easting: float, northing: float):
standard, actual = masschange_model(easting, northing)
return np.sqrt(
(((2 * stochastic_yearly_error ** 2) / standard ** 2) + ((2 * stochastic_yearly_error ** 2) / actual ** 2))
* (standard / actual) ** 2
)
return error_model
def match_sgi_ids():
sgi_2016 = gpd.read_file(terradem.files.INPUT_FILE_PATHS["sgi_2016"])
sgi_2016["name_lower"] = sgi_2016["name"].str.lower().fillna("")
data_dir = pathlib.Path("data/external/mass_balance")
warnings.filterwarnings("ignore", category=shapely.errors.ShapelyDeprecationWarning)
result_data = []
ids = {
"seewijnen": "B52-22",
"corbassiere": "B83-03",
"murtel": "E23-16",
"gietro": "B82-14",
"findelen": "B56-03",
}
results = pd.DataFrame(columns=["sgi-id", "year", "dh", "dm"])
for filepath in filter(lambda s: "longterm" in str(s), data_dir.iterdir()):
name = filepath.stem.replace("_longterm", "")
if name in ids:
match = sgi_2016.loc[sgi_2016["sgi-id"] == ids[name]].iloc[0]
else:
name = {
"ugrindelwald": "<NAME>",
}.get(name, None) or name
try:
match = (
sgi_2016[sgi_2016["name_lower"].str.findall(f".*{name}.*").apply(len) > 0]
.sort_values("area_km2")
.iloc[-1]
)
except IndexError:
warnings.warn(f"Cannot find {name}")
continue
data = (
pd.read_csv(filepath, skiprows=1, delim_whitespace=True, na_values=[-99.0])
.rename(columns={"Year": "year", "B_a(mw.e.)": "dh"})
.ffill()
)
data["dm"] = (data["Area(km2)"] * 1e6) * data["dh"]
data["sgi-id"] = match["sgi-id"]
result_data.append(data[["sgi-id", "year", "dh", "dm"]])
continue
results = | pd.concat(result_data) | pandas.concat |
# Calculate cutting points for each case in the database, based on 2D scores.
# Generates computed/cuts.npy
import math
import json
import pandas as pd
import numpy as np
from sklearn.svm import SVC
import matplotlib.pyplot as plt
from tqdm import tqdm
def calculate(scores=None, votes=None, scdb=None, format="npy"):
global J, K, T
if not isinstance(scores, np.ndarray):
scores = np.load("computed/scores_2d.npy")
if not isinstance(votes, pd.DataFrame):
votes = | pd.read_csv("data/votes.csv") | pandas.read_csv |
"""
This script collects data from the Futurology and Science subreddits for NLP.
"""
############# DEPENDENCIES ##############
import requests
import pandas as pd
import time
import datetime
#########################################
# Save start time so time elapsed can be printed later.
t0 = time.time()
# General URL stub for Pushshift reddit API to retrieve comments.
url = 'https://api.pushshift.io/reddit/search/comment'
# Choose Futurology subreddit and maximum number of comments.
params = {
'subreddit' : 'Futurology',
'size' : 100
}
# Collect 100 comments and extract the 'data' portion.
fut_posts = requests.get(url, params).json()['data']
# Save the post time of the last comment collected so the next request can get the next 100 comments.
last_fut = fut_posts[-1]['created_utc']
# Repeat the above steps for the Science subreddit.
params = {
'subreddit' : 'science',
'size' : 100
}
sci_posts = requests.get(url, params).json()['data']
last_sci = sci_posts[-1]['created_utc']
"""
Below, j sets the number of files of comments to collect.
NOTES:
- I set j to 200, but this was much more than I needed.
- Collection was interrupted overnight after 17 files were collected.
- I adjusted j to range from 17 to 200 and manually set last_fut and last_sci
to the last posted comment to continue collection.
- This issue could have been avoided with try/except, but I already had plenty of data.
- I stopped collection after 52 files, which was already more than enough for this project.
- At one point, I accidentally over-wrote the first file of science comments.
- I ran a modified version of this script to reclaim the data from the same start time.
- Alignment with the original data wasn't perfect, but sufficient to have no likely impact on the analysis.
- The range of post times matched within minutes.
- This issue could be avoided in the future using Git.
"""
for j in range(200):
# Number of times to collect 100 comments. This setting gives me 10,000 comments per file.
if j == 0:
count = 99
else:
count = 100
for i in range(count):
# Slow request rate to avoid overburdening the server.
time.sleep(2)
# Collect comments as above, updating the start time so each 100 comments starts were the last left off.
params = {
'subreddit' : 'Futurology',
'size' : 100,
'before' : last_fut
}
fut_posts += requests.get(url, params).json()['data']
last_fut = fut_posts[-1]['created_utc']
time.sleep(2)
params = {
'subreddit' : 'science',
'size' : 100,
'before' : last_sci
}
sci_posts += requests.get(url, params).json()['data']
last_sci = sci_posts[-1]['created_utc']
# Print an update to the user after every 1000 comments are collected from each subreddit.
if (i+1) % 10 == 0:
# Referenced this resource to convert time from seconds: https://stackoverflow.com/questions/775049/how-do-i-convert-seconds-to-hours-minutes-and-seconds
print(f'Collected {(i+1)*100} comments for file {j+1} out of 200 files. {datetime.timedelta(seconds=time.time()-t0)} elapsed.')
# Save comments to a csv after each pass through the outer loop.
fut_df = pd.DataFrame(fut_posts)
sci_df = | pd.DataFrame(sci_posts) | pandas.DataFrame |
import pandas as pd
WINNING_PTS = 3
DRAWING_PTS = 1
LOOSING_PTS = 0
def single_match_points(match_row: pd.Series):
'''
Compute match points for a single match row.
:param match_row: Row representing a sigle match between two teams.
:return:
'''
if match_row["FTHG"] > match_row["FTAG"]:
return {"home_pts": WINNING_PTS, "away_pts": LOOSING_PTS}
elif match_row["FTHG"] == match_row["FTAG"]:
return {"home_pts": DRAWING_PTS, "away_pts": DRAWING_PTS}
else:
return {"home_pts": LOOSING_PTS, "away_pts": WINNING_PTS}
def matches_points(matches: pd.DataFrame):
'''
Compute all match points.
:param matches: Team(s) matches table.
:return:
'''
team_matches_points = matches.apply(single_match_points, axis=1).to_frame()
team_matches_points_rows = []
indexes = []
for i, team_match_points in team_matches_points.iterrows():
indexes.append(i)
team_matches_points_rows.append(team_match_points.item())
team_matches_points_df = | pd.DataFrame(team_matches_points_rows, index=indexes) | pandas.DataFrame |
import getpass, os, time, subprocess, math, pickle, queue, threading, argparse, time, backoff, shutil, gc
import numpy as np
import pandas as pd
import pyvo as vo
import traceback as tb
from timewise.general import main_logger, DATA_DIR_KEY, data_dir, bigdata_dir, backoff_hndlr
from timewise.wise_data_by_visit import WiseDataByVisit
logger = main_logger.getChild(__name__)
class WISEDataDESYCluster(WiseDataByVisit):
status_cmd = f'qstat -u {getpass.getuser()}'
# finding the file that contains the setup function tde_catalogue
BASHFILE = os.getenv('TIMEWISE_DESY_CLUSTER_BASHFILE', os.path.expanduser('~/.bashrc'))
def __init__(self, base_name, parent_sample_class, min_sep_arcsec, n_chunks):
super().__init__(base_name=base_name,
parent_sample_class=parent_sample_class,
min_sep_arcsec=min_sep_arcsec,
n_chunks=n_chunks)
# set up cluster stuff
self.job_id = None
self._n_cluster_jobs_per_chunk = None
self.cluster_jobID_map = None
self.clusterJob_chunk_map = None
self.cluster_info_file = os.path.join(self.cluster_dir, 'cluster_info.pkl')
self._overwrite = True
self._storage_dir = None
# status attributes
self.start_time = None
self._total_tasks = None
self._done_tasks = None
self._tap_queue = queue.Queue()
self._cluster_queue = queue.Queue()
self._io_queue = queue.PriorityQueue()
self._io_queue_done = queue.Queue()
def get_sample_photometric_data(self, max_nTAPjobs=8, perc=1, tables=None, chunks=None,
cluster_jobs_per_chunk=100, wait=5, remove_chunks=False,
query_type='positional', overwrite=True,
storage_directory=bigdata_dir):
"""
An alternative to `get_photometric_data()` that uses the DESY cluster and is optimised for large datasets.
:param max_nTAPjobs: The maximum number of TAP jobs active at the same time.
:type max_nTAPjobs: int
:param perc: The percentage of chunks to download
:type perc: float
:param tables: The tables to query
:type tables: str or list-like
:param chunks: chunks to download, default is all of the chunks
:type chunks: list-like
:param cluster_jobs_per_chunk: number of cluster jobs per chunk
:type cluster_jobs_per_chunk: int
:param wait: time in hours to wait after submitting TAP jobs
:type wait: float
:param remove_chunks: remove single chunk files after binning
:type remove_chunks: bool
:param query_type: 'positional': query photometry based on distance from object, 'by_allwise_id': select all photometry points within a radius of 50 arcsec with the corresponding AllWISE ID
:type query_type: str
:param overwrite: overwrite already existing lightcurves and metadata
:type overwrite: bool
:param storage_directory: move binned files and raw data here after work is done
:type storage_directory: str
"""
# --------------------- set defaults --------------------------- #
mag = True
flux = True
if tables is None:
tables = [
'AllWISE Multiepoch Photometry Table',
'NEOWISE-R Single Exposure (L1b) Source Table'
]
tables = np.atleast_1d(tables)
if chunks is None:
chunks = list(range(round(int(self.n_chunks * perc))))
else:
cm = [c not in self.chunk_map for c in chunks]
if np.any(cm):
raise ValueError(f"Chunks {np.array(chunks)[cm]} are not in chunk map. "
f"Probably they are larger than the set chunk number of {self._n_chunks}")
if remove_chunks:
raise NotImplementedError("Removing chunks is not implemented yet!")
if query_type not in self.query_types:
raise ValueError(f"Unknown query type {query_type}! Choose one of {self.query_types}")
service = 'tap'
# set up queue
self.queue = queue.Queue()
# set up dictionary to store jobs in
self.tap_jobs = {t: dict() for t in tables}
logger.debug(f"Getting {perc * 100:.2f}% of lightcurve chunks ({len(chunks)}) via {service} "
f"in {'magnitude' if mag else ''} {'flux' if flux else ''} "
f"from {tables}")
input('Correct? [hit enter] ')
# --------------------------- set up cluster info --------------------------- #
self.n_cluster_jobs_per_chunk = cluster_jobs_per_chunk
cluster_time_s = max(len(self.parent_sample.df) / self._n_chunks / self.n_cluster_jobs_per_chunk, 59 * 60)
if cluster_time_s > 24 * 3600:
raise ValueError(f"cluster time per job would be longer than 24h! "
f"Choose more than {self.n_cluster_jobs_per_chunk} jobs per chunk!")
cluster_time = time.strftime('%H:%M:%S', time.gmtime(cluster_time_s))
self.clear_cluster_log_dir()
self._save_cluster_info()
self._overwrite = overwrite
self._storage_dir = storage_directory
# --------------------------- starting threads --------------------------- #
tap_threads = [threading.Thread(target=self._tap_thread, daemon=True, name=f"TAPThread{_}")
for _ in range(max_nTAPjobs)]
cluster_threads = [threading.Thread(target=self._cluster_thread, daemon=True, name=f"ClusterThread{_}")
for _ in range(max_nTAPjobs)]
io_thread = threading.Thread(target=self._io_thread, daemon=True, name="IOThread")
status_thread = threading.Thread(target=self._status_thread, daemon=True, name='StatusThread')
for t in tap_threads + cluster_threads + [io_thread]:
logger.debug('starting thread')
t.start()
logger.debug(f'started {len(tap_threads)} TAP threads and {len(cluster_threads)} cluster threads.')
# --------------------------- filling queue with tasks --------------------------- #
self.start_time = time.time()
self._total_tasks = len(chunks)
self._done_tasks = 0
for c in chunks:
self._tap_queue.put((tables, c, wait, mag, flux, cluster_time, query_type))
status_thread.start()
# --------------------------- wait for completion --------------------------- #
logger.debug(f'added {self._tap_queue.qsize()} tasks to tap queue')
self._tap_queue.join()
logger.debug('TAP done')
self._cluster_queue.join()
logger.debug('cluster done')
@backoff.on_exception(
backoff.expo,
vo.dal.exceptions.DALServiceError,
giveup=WiseDataByVisit._give_up_tap,
max_tries=50,
on_backoff=backoff_hndlr
)
def _wait_for_job(self, t, i):
logger.info(f"Waiting on {i}th query of {t} ........")
_job = self.tap_jobs[t][i]
_job.wait()
logger.info(f'{i}th query of {t}: Done!')
def _get_results_from_job(self, t, i):
logger.debug(f"getting results for {i}th query of {t} .........")
_job = self.tap_jobs[t][i]
lightcurve = _job.fetch_result().to_table().to_pandas()
fn = self._chunk_photometry_cache_filename(t, i)
logger.debug(f"{i}th query of {t}: saving under {fn}")
cols = dict(self.photometry_table_keymap[t]['mag'])
cols.update(self.photometry_table_keymap[t]['flux'])
if 'allwise' in t:
cols['cntr_mf'] = 'allwise_cntr'
lightcurve.rename(columns=cols).to_csv(fn)
return
def _io_queue_hash(self, method_name, args):
return f"{method_name}_{args}"
def _wait_for_io_task(self, method_name, args):
h = self._io_queue_hash(method_name, args)
logger.debug(f"waiting on io-task {h}")
while True:
_io_queue_done = list(self._io_queue_done.queue)
if h in _io_queue_done:
break
time.sleep(30)
logger.debug(f"{h} done!")
def _io_thread(self):
logger.debug("started in-out thread")
while True:
priority, method_name, args = self._io_queue.get(block=True)
logger.debug(f"executing {method_name} with arguments {args} (priority {priority})")
try:
self.__getattribute__(method_name)(*args)
self._io_queue_done.put(self._io_queue_hash(method_name, args))
except Exception as e:
msg = (
f"#################################################################\n"
f" !!! ATTENTION !!! \n"
f" ----------------- {method_name}({args}) ---------------- \n"
f" AN ERROR OCCURED \n"
f"\n{''.join(tb.format_exception(None, e, e.__traceback__))}\n\n"
f"putting {method_name}({args}) back into IO-queue\n"
f"#################################################################\n"
)
logger.error(msg)
self._io_queue.put((priority, method_name, args))
finally:
self._io_queue.task_done()
gc.collect()
def _tap_thread(self):
logger.debug(f'started tap thread')
while True:
tables, chunk, wait, mag, flux, cluster_time, query_type = self._tap_queue.get(block=True)
logger.debug(f'querying IRSA for chunk {chunk}')
submit_to_cluster = True
for i in range(len(tables) + 1):
# ----------- submit jobs for chunk i via the IRSA TAP ---------- #
if i < len(tables):
t = tables[i]
submit_method = "_submit_job_to_TAP"
submit_args = [chunk, t, mag, flux, query_type]
self._io_queue.put((1, submit_method, submit_args))
self._wait_for_io_task(submit_method, submit_args)
# -------------- get results of TAP job for chunk i-1 ------------- #
if i > 0:
t_before = tables[i - 1]
if self.tap_jobs[t_before][chunk].phase == "COMPLETED":
result_method = "_get_results_from_job"
result_args = [t_before, chunk]
self._io_queue.put((2, result_method, result_args))
self._wait_for_io_task(result_method, result_args)
else:
logger.warning(
f"No completion for {chunk}th query of {t_before}! "
f"Phase is {self.tap_jobs[t_before][chunk].phase}!"
)
submit_to_cluster = False
# --------------- wait for the TAP job of chunk i -------------- #
if i < len(tables):
t = tables[i]
logger.info(f'waiting for {wait} hours')
time.sleep(wait * 3600)
try:
self._wait_for_job(t, chunk)
except vo.dal.exceptions.DALServiceError:
logger.warning(f"could not wait for {chunk}th query of {t}! Not submitting to cluster.")
# mark task as done and move on without submission to cluster
submit_to_cluster = False
continue
self._tap_queue.task_done()
if submit_to_cluster:
self._cluster_queue.put((cluster_time, chunk))
gc.collect()
def _move_file_to_storage(self, filename):
dst_fn = filename.replace(data_dir, self._storage_dir)
dst_dir = os.path.dirname(dst_fn)
if not os.path.isdir(dst_dir):
logger.debug(f"making directory {dst_dir}")
os.makedirs(dst_dir)
logger.debug(f"copy {filename} to {dst_fn}")
try:
shutil.copy2(filename, dst_fn)
if os.path.getsize(filename) == os.path.getsize(dst_fn):
logger.debug(f"copy successful, removing {filename}")
os.remove(filename)
else:
logger.warning(f"copy from {filename} to {dst_fn} gone wrong! Not removing source.")
except FileNotFoundError as e:
logger.warning(f"FileNotFoundError: {e}!")
def _cluster_thread(self):
logger.debug(f'started cluster thread')
while True:
cluster_time, chunk = self._cluster_queue.get(block=True)
logger.info(f'got all TAP results for chunk {chunk}. submitting to cluster')
job_id = self.submit_to_cluster(cluster_cpu=1,
cluster_h=cluster_time,
cluster_ram='40G',
tables=None,
service='tap',
single_chunk=chunk)
if not job_id:
logger.warning(f"could not submit {chunk} to cluster! Try later")
self._cluster_queue.put((cluster_time, chunk))
self._cluster_queue.task_done()
else:
logger.debug(f'waiting for chunk {chunk} (Cluster job {job_id})')
self.wait_for_job(job_id)
logger.debug(f'cluster done for chunk {chunk} (Cluster job {job_id}). Start combining')
try:
self._combine_lcs('tap', chunk_number=chunk, remove=True, overwrite=self._overwrite)
self._combine_metadata('tap', chunk_number=chunk, remove=True, overwrite=self._overwrite)
if self._storage_dir:
filenames_to_move = [
self._lightcurve_filename(service='tap', chunk_number=chunk),
self._metadata_filename(service='tap', chunk_number=chunk),
]
for t in self.photometry_table_keymap.keys():
filenames_to_move.append(self._chunk_photometry_cache_filename(t, chunk))
for fn in filenames_to_move:
self._move_file_to_storage(fn)
finally:
self._cluster_queue.task_done()
self._done_tasks += 1
gc.collect()
def _status_thread(self):
logger.debug('started status thread')
while True:
n_tap_tasks_queued = self._tap_queue.qsize()
n_cluster_tasks_queued = self._cluster_queue.qsize()
n_remaining = self._total_tasks - self._done_tasks
elapsed_time = time.time() - self.start_time
time_per_task = elapsed_time / self._done_tasks if self._done_tasks > 0 else np.nan
remaining_time = n_remaining * time_per_task
msg = f"\n----------------- STATUS -----------------\n" \
f"\ttasks in TAP queue:_______{n_tap_tasks_queued}\n" \
f"\ttasks in cluster queue:___{n_cluster_tasks_queued}\n" \
f"\tperformed io tasks:_______{len(list(self._io_queue_done.queue))}\n" \
f"\tdone total:_______________{self._done_tasks}/{self._total_tasks}\n" \
f"\truntime:__________________{elapsed_time/3600:.2f} hours\n" \
f"\tremaining:________________{remaining_time/3600:.2f} hours"
logger.info(msg)
time.sleep(5*3600)
# ----------------------------------------------------------------------------------- #
# START using cluster for downloading and binning #
# ---------------------------------------------------- #
@staticmethod
@backoff.on_exception(
backoff.expo,
OSError,
max_time=2*3600,
on_backoff=backoff_hndlr,
jitter=backoff.full_jitter,
)
def _execute_bash_command(cmd):
with subprocess.Popen(cmd, stdout=subprocess.PIPE, shell=True) as process:
msg = process.stdout.read().decode()
process.terminate()
return msg
@staticmethod
def _qstat_output(qstat_command):
"""return the output of the qstat_command"""
# start a subprocess to query the cluster
return str(WISEDataDESYCluster._execute_bash_command(qstat_command))
@staticmethod
def _get_ids(qstat_command):
"""Takes a command that queries the DESY cluster and returns a list of job IDs"""
st = WISEDataDESYCluster._qstat_output(qstat_command)
# If the output is an empty string there are no tasks left
if st == '':
ids = list()
else:
# Extract the list of job IDs
ids = np.array([int(s.split(' ')[2]) for s in st.split('\n')[2:-1]])
return ids
def _ntasks_from_qstat_command(self, qstat_command, job_id):
"""Returns the number of tasks from the output of qstat_command"""
# get the output of qstat_command
ids = self._get_ids(qstat_command)
ntasks = 0 if len(ids) == 0 else len(ids[ids == job_id])
return ntasks
def _ntasks_total(self, job_id):
"""Returns the total number of tasks"""
return self._ntasks_from_qstat_command(self.status_cmd, job_id)
def _ntasks_running(self, job_id):
"""Returns the number of running tasks"""
return self._ntasks_from_qstat_command(self.status_cmd + " -s r", job_id)
def wait_for_job(self, job_id=None):
"""
Wait until the cluster job is done
:param job_id: the ID of the cluster job, if `None` use `self.job_ID`
:type job_id: int
"""
_job_id = job_id if job_id else self.job_id
if _job_id:
logger.info(f'waiting on job {_job_id}')
time.sleep(10)
i = 31
j = 6
while self._ntasks_total(_job_id) != 0:
if i > 30:
logger.info(f'{time.asctime(time.localtime())} - Job{_job_id}:'
f' {self._ntasks_total(_job_id)} entries in queue. '
f'Of these, {self._ntasks_running(_job_id)} are running tasks, and '
f'{self._ntasks_total(_job_id) - self._ntasks_running(_job_id)} '
f'are tasks still waiting to be executed.')
i = 0
j += 1
if j > 7:
logger.info(self._qstat_output(self.status_cmd))
j = 0
time.sleep(30)
i += 1
logger.info('cluster is done')
else:
logger.info(f'No Job ID!')
@property
def n_cluster_jobs_per_chunk(self):
return self._n_cluster_jobs_per_chunk
@n_cluster_jobs_per_chunk.setter
def n_cluster_jobs_per_chunk(self, value):
self._n_cluster_jobs_per_chunk = value
if value:
n_jobs = self.n_chunks * int(value)
logger.debug(f'setting {n_jobs} jobs.')
self.cluster_jobID_map = np.zeros(len(self.parent_sample.df), dtype=int)
self.clusterJob_chunk_map = | pd.DataFrame(columns=['chunk_number']) | pandas.DataFrame |
from collections import Counter
from itertools import product
import os
import pickle
import numpy as np
import pandas as pd
from tqdm import tqdm
from sklearn.preprocessing import OneHotEncoder
import sys
sys.path.insert(0, '../myfunctions')
from prediction import fit_prediction_model
#from causal_inference import infer_mediation_3mediator
from causal_inference import infer_mediation, select_estimator
if __name__=='__main__':
## load data
res = pd.read_excel('../data/hiv-brain-age.xlsx')
A = res['HIV'].values.astype(float)
res.loc[res.Sex=='M', 'Sex'] = 1
res.loc[res.Sex=='F', 'Sex'] = 0
race = OneHotEncoder(sparse=False).fit_transform(res.Race.values.astype(str).reshape(-1,1))
L = np.c_[res[['Age', 'Sex', 'Tobacco use disorder', 'Alcoholism']].values.astype(float), race]
Y = res['BAI'].values.astype(float)
Mnames = ['obesity', 'heart disorder', 'sleep disorder']
M = res[Mnames].values.astype(float)
for mm in Mnames:
print(mm, Counter(res[mm]))
Mnames.append('avg')
n_mediator = len(Mnames)
sids = np.arange(len(A))
## set up numbers
random_state = 2020
prediction_methods = ['linear', 'rf']#, 'xgb']
ci_method = 'dr'
Nbt = 1000
np.random.seed(random_state)
## generate cv split
"""
cv_path = 'patients_cv_split_real_data2.pickle'
if os.path.exists(cv_path):
with open(cv_path, 'rb') as ff:
tr_sids, te_sids = pickle.load(ff)
else:
cvf = 5
sids2 = np.array(sids)
np.random.shuffle(sids2)
tr_sids = []
te_sids = []
cv_split = np.array_split(sids2, cvf)
for cvi in cv_split:
te_sids.append(np.sort(cvi))
tr_sids.append(np.sort(np.setdiff1d(sids2, cvi)))
with open(cv_path, 'wb') as ff:
pickle.dump([tr_sids, te_sids], ff)
"""
tr_sids = [sids]
te_sids = [sids]
## bootstrapping
#"""
res = []
for bti in tqdm(range(Nbt+1)):
np.random.seed(random_state+bti)
if bti==0:
# use the actual data
Abt = A
Ybt = Y
Lbt = L
Mbt = M
sidsbt = sids
prediction_methods_outcome = prediction_methods
prediction_methods_exposure = prediction_methods
else:
# use bootstrapped data
btids = np.random.choice(len(Y), len(Y), replace=True)
Abt = A[btids]
Ybt = Y[btids]
Lbt = L[btids]
Mbt = M[btids]
sidsbt = sids[btids]
prediction_methods_outcome = [best_pm_o]
prediction_methods_exposure = [best_pm_e]
# outer loop cross validation
for cvi in range(len(tr_sids)):
trid = np.in1d(sidsbt, tr_sids[cvi])
teid = np.in1d(sidsbt, te_sids[cvi])
Atr = Abt[trid]
Ytr = Ybt[trid]
Ltr = Lbt[trid]
Mtr = Mbt[trid]
Ate = Abt[teid]
Yte = Ybt[teid]
Lte = Lbt[teid]
Mte = Mbt[teid]
Lmean = Ltr.mean(axis=0)
Lstd = Ltr.std(axis=0)
Ltr = (Ltr-Lmean)/Lstd
Lte = (Lte-Lmean)/Lstd
#try:
for pi, pm in enumerate(product(prediction_methods_outcome, prediction_methods_exposure)):
if bti==0:
print(pm)
pm_outcome, pm_exposure = pm
# fit A|L
model_a_l, model_a_l_perf = fit_prediction_model(pm_exposure+':bclf', Ltr, Atr,
random_state=random_state+pi+1000)
# fit Y|A,L,M
model_y_alm, model_y_alm_perf = fit_prediction_model(pm_outcome+':reg', np.c_[Atr, Ltr, Mtr], Ytr,
random_state=random_state+pi*3000)
model_m_als = []
model_m_al_perfs = []
for mi, mediator_name in enumerate(Mnames[:-1]):
"""
if mi==0:
# fit M1|A,L
model_m_al, model_m_al_perf = fit_prediction_model(pm+':bclf', np.c_[Atr, Ltr], Mtr[:, mi],
save_path='models_real_data2/med_model_%s_cv%d_%s'%(mediator_name, cvi+1, pm) if bti==0 else None,
random_state=random_state+pi*2000+mi)
elif mi==1:
# fit M2|A,L,M1
model_m_al, model_m_al_perf = fit_prediction_model(pm+':bclf', np.c_[Atr, Ltr, Mtr[:,0]], Mtr[:, mi],
save_path='models_real_data2/med_model_%s_cv%d_%s'%(mediator_name, cvi+1, pm) if bti==0 else None,
random_state=random_state+pi*2000+mi)
elif mi==2:
# fit M3|A,L,M1,M2
model_m_al, model_m_al_perf = fit_prediction_model(pm+':bclf', np.c_[Atr, Ltr, Mtr[:,[0,1]]], Mtr[:, mi],
save_path='models_real_data2/med_model_%s_cv%d_%s'%(mediator_name, cvi+1, pm) if bti==0 else None,
random_state=random_state+pi*2000+mi)
"""
# fit Mi|A,L
model_m_al, model_m_al_perf = fit_prediction_model(pm_exposure+':bclf', np.c_[Atr, Ltr], Mtr[:, mi],
random_state=random_state+pi*2000+mi)
model_m_als.append(model_m_al)
model_m_al_perfs.append(model_m_al_perf)
# do causal inference
#cdes, scies, cies0, cies1 = infer_mediation_3mediator(cim, model_a_l, model_m_als, model_y_alms, Yte, Mte, Ate, Lte, random_state=random_state+pi*4000+ci)
cdes, scies, cies0, cies1 = infer_mediation(ci_method, model_a_l, model_m_als, model_y_alm,
Yte, Mte, Ate, Lte, random_state=random_state+pi*4000)
# add average performance
cdes.append(np.mean(cdes))
scies.append(np.mean(scies))
cies0.append(np.mean(cies0))
cies1.append(np.mean(cies1))
model_m_al_perfs.append(np.nan)
res.append([bti, cvi, pm_outcome, pm_exposure, ci_method, model_a_l_perf, model_y_alm_perf] + model_m_al_perfs + cdes + scies + cies0 + cies1)
#print(res[-1])
#except Exception as ee:
# print(str(ee))
if bti==0:
_,_,best_pm_o, best_pm_e = select_estimator(
np.array([x[1] for x in res if x[0]==bti]),
[(x[2],x[3]) for x in res if x[0]==bti],
np.array([x[-n_mediator*4+n_mediator-1]+x[-n_mediator*3+n_mediator-1] for x in res if x[0]==bti]))
print('best prediction model: outcome: %s; exposure: %s'%(best_pm_o, best_pm_e))
res = [x for x in res if x[2]==best_pm_o and x[3]==best_pm_e]
with open('results_real_data2.pickle', 'wb') as ff:
pickle.dump([res, best_pm_o, best_pm_e], ff, protocol=2)
#"""
#with open('results_real_data2.pickle', 'rb') as ff:
# res, best_pm_o, best_pm_e = pickle.load(ff)
res = np.array(res, dtype=object)
Nbt = res[:,0].max()
perf_A_L_cols = ['perf(A|L)']
perf_Y_ALM_cols = ['perf(Y|A,L,M)']
perf_M_AL_cols = ['perf(M|A,L) %s'%x for x in Mnames]
CDE_cols = ['CDE %s'%x for x in Mnames]
sCIE_cols = ['sCIE %s'%x for x in Mnames]
CIE0_cols = ['CIE0 %s'%x for x in Mnames]
CIE1_cols = ['CIE1 %s'%x for x in Mnames]
cols = perf_A_L_cols + perf_Y_ALM_cols + perf_M_AL_cols + CDE_cols + sCIE_cols + CIE0_cols + CIE1_cols
columns = ['bt', 'fold', 'outcome_prediction_model', 'exposure_prediction_model', 'causal_inference_model'] + cols
res = pd.DataFrame(data=res, columns=columns)
# take the average across folds
res2 = []
for bti in range(Nbt+1):
ids = res.bt==bti
if ids.sum()==0:
continue
res2.append([bti] + list(res[ids][cols].mean(axis=0)))
columns = ['bt'] + cols
res = pd.DataFrame(data=res2, columns=columns)
# add percentages
for m in Mnames:
total_effect = res['CDE %s'%m].values + res['sCIE %s'%m].values
res.insert(res.shape[1], 'TotalEffect %s'%m, total_effect)
cols.append('TotalEffect %s'%m)
for col in ['CDE', 'sCIE']:
for m in Mnames:
res_perc = res['%s %s'%(col,m)].values/res['TotalEffect %s'%m].values*100
res.insert(res.shape[1], '%%%s %s'%(col,m), res_perc)
cols.append('%%%s %s'%(col,m))
# add confidence interval
ids1 = np.where(res.bt==0)[0]
ids2 = np.where(res.bt>0)[0]
assert len(ids1)==1
vals = res.iloc[ids1[0]][cols].values
lb = np.percentile(res.iloc[ids2][cols].values, 2.5, axis=0)
ub = np.percentile(res.iloc[ids2][cols].values, 97.5, axis=0)
res2 = np.array([['%.3f [%.3f -- %.3f]'%(vals[ii], lb[ii], ub[ii]) for ii in range(len(vals))]])
columns = cols
res = pd.DataFrame(data=res2, columns=columns)
col_names2 = ['Mediator',
'%CDE', '%sCIE',
'CDE', 'sCIE', 'TotalEffect',
'CIE0', 'CIE1',
'perf(A|L)', 'perf(M|A,L)', 'perf(Y|A,L,M)',]
res2 = res[cols]
# get values that are the same for all mediators
a_l_perf = res2['perf(A|L)'].iloc[0]
y_alm_perf = res2['perf(Y|A,L,M)'].iloc[0]
# generate dataframe with each row being a mediator
res2 = res2.drop(columns=['perf(A|L)', 'perf(Y|A,L,M)'])
col_names = np.array(res2.columns).reshape(-1,n_mediator)[:,0]
col_names = ['Mediator', 'perf(A|L)', 'perf(Y|A,L,M)'] + [x.split(' ')[0] for x in col_names]
res2 = res2.values.reshape(-1,n_mediator).T
res2 = np.c_[Mnames, [a_l_perf]*n_mediator, [y_alm_perf]*n_mediator, res2]
df = | pd.DataFrame(data=res2, columns=col_names) | pandas.DataFrame |
import pandas as pd
import numpy as np
# 用于订单数据,用户数据,菜单数据的一系列操作
class Operate:
def __init__(self):
pass
#查询用户是否存在
def find_user(self,user):
data=pd.read_csv('用户.csv')
if user in np.array(data['username']).tolist():
return True
else:
return False
#查询管理员ID是否存在
def find_ID(self,id):
data=pd.read_csv('管理员.csv')
if id in np.array(data['管理员ID']).tolist():
return True
else:
return False
#验证用户名和密码
def verify_user(self,user,password):
data=pd.read_csv('用户.csv')
if [user,password] in np.array(data).tolist():
return True
else:
return False
#验证管理员的用户名和密码
def verify_Admin(self,user,password):
data=pd.read_csv('管理员.csv')
if [user,password] in np.array(data).tolist():
return True
else:
return False
#注册成功后,用户表插入新用户
def insert_user(self,user,password):
data=pd.read_csv('用户.csv')
data=data.append([{'username':user,'password':password}])
data.to_csv('用户.csv',index=0)
#管理员注册成功后,管理员表插入新管理员
def insert_admin(self,id,password):
data=pd.read_csv('管理员.csv')
data=data.append([{'管理员ID':id,'密码':password}])
data.to_csv('管理员.csv',index=0)
#获取菜单名称
def get_Menu_Name(self):
data=pd.read_csv('菜单.csv')
name=np.array(data['菜名']).tolist()
return name
#获取某个菜品的价格
def get_price(self,menuname):
data=pd.read_csv(r'菜单.csv')
price=np.array(data[data['菜名']==menuname]['价格'])[0]
return eval(price[0:-1])
#获取临时订单的编号
def get_temporary_No(self):
data=pd.read_csv('临时订单.csv')
result=data['订单编号'][0]
return result
#插入临时订单数据
def insert_temporary(self,data):
columns=['订单编号','订单人','时间','菜名','数量','价格','总计']
order=pd.DataFrame(data=data,columns=columns)
order.to_csv('临时订单.csv',index=0)
#更新临时订单
def update_temporary(self,data):
old=pd.read_csv(r'临时订单.csv')
columns=['订单编号','订单人','时间','菜名','数量','价格','总计']
new=pd.DataFrame(data=data,columns=columns)
old=old.append(new,ignore_index=True)
old.to_csv('临时订单.csv',index=0)
#清空临时订单
def clear_temporary(self):
columns=['订单编号','订单人','时间','菜名','数量','价格','总计']
order=pd.DataFrame(data=[],columns=columns)
order.to_csv('临时订单.csv',index=0)
#删除某个订单
def delete_temporary(self,menuname):
data=pd.read_csv('临时订单.csv')
data=data.drop(index=(data.loc[(data['菜名']==menuname)].index))
data.to_csv('临时订单.csv',index=0)
#判断临时订单是否为空
def judge_empty(self):
data= | pd.read_csv('临时订单.csv') | pandas.read_csv |
"""
STAT 656 HW-10
@author:<NAME>
@heavy_lifting_by: Dr. <NAME>
@date: 2020-07-29
"""
import pandas as pd
# Classes provided from AdvancedAnalytics ver 1.25
from AdvancedAnalytics.Text import text_analysis
from AdvancedAnalytics.Text import sentiment_analysis
from sklearn.feature_extraction.text import CountVectorizer
import numpy as np
from AdvancedAnalytics.Text import text_plot
def heading(headerstring):
"""
Centers headerstring on the page. For formatting to stdout
Parameters
----------
headerstring : string
String that you wish to center.
Returns
-------
Returns: None.
"""
tw = 70 # text width
lead = int(tw/2)-(int(len(headerstring)/2))-1
tail = tw-lead-len(headerstring)-2
print('\n' + ('*'*tw))
print(('*'*lead) + ' ' + headerstring + ' ' + ('*'*tail))
print(('*'*tw))
return
heading("READING DATA SOURCE...")
# Set Pandas Columns Width for Excel Columns
pd.set_option('max_colwidth', 32000)
df = pd.read_excel("hotels.xlsx")
text_col = 'Review' #Identify the Data Frame Text Target Column Name
# Check if any text was truncated
pd_width = | pd.get_option('max_colwidth') | pandas.get_option |
#!/usr/bin/env python
import os,sys
import pandas as pd
import argparse
daismdir = os.path.dirname(os.path.dirname(os.path.abspath(__file__)))
sys.path.insert(0,daismdir)
import daism.modules.simulation as simulation
import daism.modules.training as training
import daism.modules.prediction as prediction
#--------------------------------------
#--------------------------------------
# main()
parser = argparse.ArgumentParser(description='DAISM-XMBD deconvolution.')
subparsers = parser.add_subparsers(dest='subcommand', help='Select one of the following sub-commands')
# create the parser for the "one-stop DAISM-DNN" command
parser_a = subparsers.add_parser('DAISM', help='one-stop DAISM-XMBD',description="one-stop DAISM-XMBD")
parser_a.add_argument("-platform", type=str, help="Platform of calibration data, [R]: RNA-seq TPM, [S]: single cell RNA-seq", default="S")
parser_a.add_argument("-caliexp", type=str, help="Calibration samples expression file", default=None)
parser_a.add_argument("-califra", type=str, help="Calibration samples ground truth file", default=None)
parser_a.add_argument("-aug", type=str, help="Purified samples expression (h5ad)", default=None)
parser_a.add_argument("-N", type=int, help="Simulation samples number", default=16000)
parser_a.add_argument("-testexp", type=str, help="Test samples expression file", default=None)
parser_a.add_argument("-net", type=str, help="Network architecture used for training", default="coarse")
parser_a.add_argument("-outdir", type=str, help="Output result file directory", default="../output/")
# create the parser for the "DAISM simulation" command
parser_b = subparsers.add_parser('DAISM_simulation', help='training set simulation using DAISM strategy',description='training set simulation using DAISM strategy.')
parser_b.add_argument("-platform", type=str, help="Platform of calibration data, [R]: RNA-seq TPM, [S]: single cell RNA-seq", default="S")
parser_b.add_argument("-caliexp", type=str, help="Calibration samples expression file", default=None)
parser_b.add_argument("-califra", type=str, help="Calibration samples ground truth file", default=None)
parser_b.add_argument("-aug", type=str, help="Purified samples expression (h5ad)", default=None)
parser_b.add_argument("-testexp", type=str, help="Test samples expression file", default=None)
parser_b.add_argument("-N", type=int, help="Simulation samples number", default=16000)
parser_b.add_argument("-outdir", type=str, help="Output result file directory", default="../output/")
# create the parser for the "Generic simulation" command
parser_c = subparsers.add_parser('Generic_simulation', help='training set simulation using purified cells only',description='training set simulation using purified cells only.')
parser_c.add_argument("-platform", type=str, help="Platform of calibration data, [R]: RNA-seq TPM, [S]: single cell RNA-seq", default="S")
parser_c.add_argument("-aug", type=str, help="Purified samples expression (h5ad)", default=None)
parser_c.add_argument("-testexp", type=str, help="Test samples expression file", default=None)
parser_c.add_argument("-N", type=int, help="Simulation samples number", default=16000)
parser_c.add_argument("-outdir", type=str, help="Output result file directory", default="../output/")
# create the parser for the "training" command
parser_d = subparsers.add_parser('training', help='train DNN model',description='train DNN model.')
parser_d.add_argument("-trainexp", type=str, help="Simulated samples expression file", default=None)
parser_d.add_argument("-trainfra", type=str, help="Simulated samples ground truth file", default=None)
parser_d.add_argument("-net", type=str, help="Network architecture used for training", default="coarse")
parser_d.add_argument("-outdir", type=str, help="Output result file directory", default="../output/")
# create the parser for the "prediction" command
parser_e = subparsers.add_parser('prediction', help='predict using a trained model',description='predict using a trained model.')
parser_e.add_argument("-testexp", type=str, help="Test samples expression file", default=None)
parser_e.add_argument("-model", type=str, help="Deep-learing model file trained by DAISM", default="../output/DAISM_model.pkl")
parser_e.add_argument("-celltype", type=str, help="Model celltypes", default="../output/DAISM_model_celltypes.txt")
parser_e.add_argument("-feature", type=str, help="Model feature", default="../output/DAISM_model_feature.txt")
parser_e.add_argument("-net", type=str, help="Network architecture used for training", default="coarse")
parser_e.add_argument("-outdir", type=str, help="Output result file directory", default="../output/")
class Options:
random_seed = 777
min_f = 0.01
max_f = 0.99
lr = 1e-4
batchsize = 64
num_epoches = 500
ncuda = 0
def main():
# parse some argument lists
inputArgs = parser.parse_args()
if os.path.exists(inputArgs.outdir)==False:
os.mkdir(inputArgs.outdir)
#### DAISM modules ####
if (inputArgs.subcommand=='DAISM'):
# Load calibration data
caliexp = pd.read_csv(inputArgs.caliexp, sep="\t", index_col=0)
califra = pd.read_csv(inputArgs.califra, sep="\t", index_col=0)
# Load test data
test_sample = pd.read_csv(inputArgs.testexp, sep="\t", index_col=0)
# Preprocess purified data
mode = "daism"
commongenes,caliexp,C_all = simulation.preprocess_purified(inputArgs.aug,inputArgs.platform,mode,test_sample,caliexp,califra)
# Create training dataset
mixsam, mixfra, celltypes, feature = simulation.daism_simulation(caliexp,califra,C_all,Options.random_seed,inputArgs.N,inputArgs.platform,Options.min_f,Options.max_f)
# Save signature genes and celltype labels
if os.path.exists(inputArgs.outdir+"/output/")==False:
os.mkdir(inputArgs.outdir+"/output/")
pd.DataFrame(feature).to_csv(inputArgs.outdir+'/output/DAISM_feature.txt',sep='\t')
pd.DataFrame(celltypes).to_csv(inputArgs.outdir+'/output/DAISM_celltypes.txt',sep='\t')
print('Writing training data...')
# Save training data
mixsam.to_csv(inputArgs.outdir+'/output/DAISM_mixsam.txt',sep='\t')
mixfra.to_csv(inputArgs.outdir+'/output/DAISM_mixfra.txt',sep='\t')
# Training model
model = training.dnn_training(mixsam,mixfra,Options.random_seed,inputArgs.outdir+"/output/",Options.num_epoches,Options.lr,Options.batchsize,Options.ncuda,inputArgs.net)
# Save signature genes and celltype labels
pd.DataFrame(list(mixfra.index)).to_csv(inputArgs.outdir+'/output/DAISM_model_celltypes.txt',sep='\t')
pd.DataFrame(list(mixsam.index)).to_csv(inputArgs.outdir+'/output/DAISM_model_feature.txt',sep='\t')
# Prediction
result = prediction.dnn_prediction(model, test_sample, list(mixfra.index), list(mixsam.index),Options.ncuda)
# Save predicted result
result.to_csv(inputArgs.outdir+'/output/DAISM_result.txt',sep='\t')
############################
#### simulation modules ####
############################
#### DAISM simulation modules ####
if (inputArgs.subcommand=='DAISM_simulation'):
# Load calibration data
caliexp = pd.read_csv(inputArgs.caliexp, sep="\t", index_col=0)
califra = pd.read_csv(inputArgs.califra, sep="\t", index_col=0)
# Load test data
test_sample = pd.read_csv(inputArgs.testexp, sep="\t", index_col=0)
# Preprocess purified data
mode ="daism"
commongenes,caliexp,C_all = simulation.preprocess_purified(inputArgs.aug,inputArgs.platform,mode,test_sample,caliexp,califra)
# Create training dataset
mixsam, mixfra, celltypes, feature = simulation.daism_simulation(caliexp,califra,C_all,Options.random_seed,inputArgs.N,inputArgs.platform,Options.min_f,Options.max_f)
# Save signature genes and celltype labels
if os.path.exists(inputArgs.outdir+"/output/")==False:
os.mkdir(inputArgs.outdir+"/output/")
pd.DataFrame(feature).to_csv(inputArgs.outdir+'/output/DAISM_feature.txt',sep='\t')
pd.DataFrame(celltypes).to_csv(inputArgs.outdir+'/output/DAISM_celltypes.txt',sep='\t')
print('Writing training data...')
# Save training data
mixsam.to_csv(inputArgs.outdir+'/output/DAISM_mixsam.txt',sep='\t')
mixfra.to_csv(inputArgs.outdir+'/output/DAISM_mixfra.txt',sep='\t')
#### Generic simulation modules ####
if (inputArgs.subcommand=='Generic_simulation'):
# Load test data
test_sample = pd.read_csv(inputArgs.testexp, sep="\t", index_col=0)
# Preprocess purified data
mode = "generic"
commongenes,caliexp,C_all = simulation.preprocess_purified(inputArgs.aug,inputArgs.platform,mode,test_sample)
# Create training dataset
mixsam, mixfra, celltypes, feature = simulation.generic_simulation(C_all,Options.random_seed,inputArgs.N,inputArgs.platform,commongenes)
# Save signature genes and celltype labels
if os.path.exists(inputArgs.outdir+"/output/")==False:
os.mkdir(inputArgs.outdir+"/output/")
pd.DataFrame(feature).to_csv(inputArgs.outdir+'/output/Generic_feature.txt',sep='\t')
pd.DataFrame(celltypes).to_csv(inputArgs.outdir+'/output/Generic_celltypes.txt',sep='\t')
print('Writing training data...')
# Save training data
mixsam.to_csv(inputArgs.outdir+'/output/Generic_mixsam.txt',sep='\t')
mixfra.to_csv(inputArgs.outdir+'/output/Generic_mixfra.txt',sep='\t')
##########################
#### training modules ####
##########################
if (inputArgs.subcommand=='training'):
# Load training data
mixsam = pd.read_csv(inputArgs.trainexp, sep="\t", index_col=0)
mixfra = pd.read_csv(inputArgs.trainfra, sep="\t", index_col=0)
# Training model
model = training.dnn_training(mixsam,mixfra,Options.random_seed,inputArgs.outdir+"/output/",Options.num_epoches,Options.lr,Options.batchsize,Options.ncuda,inputArgs.net)
# Save signature genes and celltype labels
if os.path.exists(inputArgs.outdir+"/output/")==False:
os.mkdir(inputArgs.outdir+"/output/")
pd.DataFrame(list(mixfra.index)).to_csv(inputArgs.outdir+'/output/DAISM_model_celltypes.txt',sep='\t')
pd.DataFrame(list(mixsam.index)).to_csv(inputArgs.outdir+'/output/DAISM_model_feature.txt',sep='\t')
############################
#### prediction modules ####
############################
if (inputArgs.subcommand=='prediction'):
# Load test data
test_sample = pd.read_csv(inputArgs.testexp, sep="\t", index_col=0)
# Load signature genes and celltype labels
feature = | pd.read_csv(inputArgs.feature,sep='\t') | pandas.read_csv |
# pylint: disable=E1101,E1103,W0232
from datetime import datetime, timedelta
from pandas.compat import range, lrange, lzip, u, zip
import operator
import re
import nose
import warnings
import os
import numpy as np
from numpy.testing import assert_array_equal
from pandas import period_range, date_range
from pandas.core.index import (Index, Float64Index, Int64Index, MultiIndex,
InvalidIndexError, NumericIndex)
from pandas.tseries.index import DatetimeIndex
from pandas.tseries.tdi import TimedeltaIndex
from pandas.tseries.period import PeriodIndex
from pandas.core.series import Series
from pandas.util.testing import (assert_almost_equal, assertRaisesRegexp,
assert_copy)
from pandas import compat
from pandas.compat import long
import pandas.util.testing as tm
import pandas.core.config as cf
from pandas.tseries.index import _to_m8
import pandas.tseries.offsets as offsets
import pandas as pd
from pandas.lib import Timestamp
class Base(object):
""" base class for index sub-class tests """
_holder = None
_compat_props = ['shape', 'ndim', 'size', 'itemsize', 'nbytes']
def verify_pickle(self,index):
unpickled = self.round_trip_pickle(index)
self.assertTrue(index.equals(unpickled))
def test_pickle_compat_construction(self):
# this is testing for pickle compat
if self._holder is None:
return
# need an object to create with
self.assertRaises(TypeError, self._holder)
def test_numeric_compat(self):
idx = self.create_index()
tm.assertRaisesRegexp(TypeError,
"cannot perform __mul__",
lambda : idx * 1)
tm.assertRaisesRegexp(TypeError,
"cannot perform __mul__",
lambda : 1 * idx)
div_err = "cannot perform __truediv__" if compat.PY3 else "cannot perform __div__"
tm.assertRaisesRegexp(TypeError,
div_err,
lambda : idx / 1)
tm.assertRaisesRegexp(TypeError,
div_err,
lambda : 1 / idx)
tm.assertRaisesRegexp(TypeError,
"cannot perform __floordiv__",
lambda : idx // 1)
tm.assertRaisesRegexp(TypeError,
"cannot perform __floordiv__",
lambda : 1 // idx)
def test_boolean_context_compat(self):
# boolean context compat
idx = self.create_index()
def f():
if idx:
pass
tm.assertRaisesRegexp(ValueError,'The truth value of a',f)
def test_ndarray_compat_properties(self):
idx = self.create_index()
self.assertTrue(idx.T.equals(idx))
self.assertTrue(idx.transpose().equals(idx))
values = idx.values
for prop in self._compat_props:
self.assertEqual(getattr(idx, prop), getattr(values, prop))
# test for validity
idx.nbytes
idx.values.nbytes
class TestIndex(Base, tm.TestCase):
_holder = Index
_multiprocess_can_split_ = True
def setUp(self):
self.indices = dict(
unicodeIndex = tm.makeUnicodeIndex(100),
strIndex = | tm.makeStringIndex(100) | pandas.util.testing.makeStringIndex |
#
# Copyright (C) 2019 Databricks, Inc.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
#
import pandas as pd
import numpy as np
from databricks import koalas as ks
from databricks.koalas.config import set_option, reset_option, option_context
from databricks.koalas.plot import TopNPlotBase, SampledPlotBase, HistogramPlotBase
from databricks.koalas.exceptions import PandasNotImplementedError
from databricks.koalas.testing.utils import ReusedSQLTestCase
class DataFramePlotTest(ReusedSQLTestCase):
@classmethod
def setUpClass(cls):
super().setUpClass()
set_option("plotting.max_rows", 2000)
set_option("plotting.sample_ratio", None)
@classmethod
def tearDownClass(cls):
reset_option("plotting.max_rows")
reset_option("plotting.sample_ratio")
super().tearDownClass()
def test_missing(self):
kdf = ks.DataFrame(np.random.rand(2500, 4), columns=["a", "b", "c", "d"])
unsupported_functions = ["box", "hexbin"]
for name in unsupported_functions:
with self.assertRaisesRegex(
PandasNotImplementedError, "method.*DataFrame.*{}.*not implemented".format(name)
):
getattr(kdf.plot, name)()
def test_topn_max_rows(self):
pdf = pd.DataFrame(np.random.rand(2500, 4), columns=["a", "b", "c", "d"])
kdf = ks.from_pandas(pdf)
data = TopNPlotBase().get_top_n(kdf)
self.assertEqual(len(data), 2000)
def test_sampled_plot_with_ratio(self):
with option_context("plotting.sample_ratio", 0.5):
pdf = pd.DataFrame(np.random.rand(2500, 4), columns=["a", "b", "c", "d"])
kdf = ks.from_pandas(pdf)
data = SampledPlotBase().get_sampled(kdf)
self.assertEqual(round(len(data) / 2500, 1), 0.5)
def test_sampled_plot_with_max_rows(self):
# 'plotting.max_rows' is 2000
pdf = pd.DataFrame(np.random.rand(2000, 4), columns=["a", "b", "c", "d"])
kdf = ks.from_pandas(pdf)
data = SampledPlotBase().get_sampled(kdf)
self.assertEqual(round(len(data) / 2000, 1), 1)
def test_compute_hist_single_column(self):
kdf = ks.DataFrame(
{"a": [1, 2, 3, 4, 5, 6, 7, 8, 9, 15, 50],}, index=[0, 1, 3, 5, 6, 8, 9, 9, 9, 10, 10]
)
expected_bins = np.linspace(1, 50, 11)
bins = HistogramPlotBase.get_bins(kdf[["a"]].to_spark(), 10)
expected_histogram = np.array([5, 4, 1, 0, 0, 0, 0, 0, 0, 1])
histogram = HistogramPlotBase.compute_hist(kdf[["a"]], bins)[0]
self.assert_eq(pd.Series(expected_bins), | pd.Series(bins) | pandas.Series |
import pandas as pd
from sklearn.base import TransformerMixin
class Pruner(TransformerMixin):
"""Prune identifier columns, columns with numerous tokens (>100) and columns
with low information."""
def __init__(self):
self.pruned_columns = ['subjuct_id', 'row_id', 'hadm_id', 'cgid', 'itemid', 'icustay_id']
def fit(self, X):
X = pd.DataFrame(X)
for col in X.columns:
num = X[col].nunique()
# Remove columns with numerous tokens (>100)
if X[col].dtype == object and num > 100:
self.pruned_columns.append(col)
# Remove columns with low information (unique values <2)
elif num <= 1 or X[col].dropna().shape[0] == 0:
self.pruned_columns.append(col)
return self
def transform(self, X):
X = | pd.DataFrame(X) | pandas.DataFrame |
import numpy as np
import pandas as pd
import sidekick as sk
from scipy.integrate import cumtrapz
from sidekick import placeholder as _
from .hospitalization_with_delay import HospitalizationWithDelay
from ..utils import param_property, sliced
from ..mixins.info import Event
class HospitalizationWithOverflow(HospitalizationWithDelay):
"""
Hospitals have a maximum number of ICU and regular clinical beds.
Death rates increase when this number overflows.
"""
hospitalization_overflow_bias = param_property(default=0.0)
icu_capacity = param_property()
hospital_capacity = param_property()
icu_occupancy = param_property(default=0.75)
hospital_occupancy = param_property(default=0.75)
icu_surge_capacity = sk.lazy(_.icu_capacity * (1 - _.icu_occupancy))
hospital_surge_capacity = sk.lazy(_.hospital_capacity * (1 - _.hospital_occupancy))
def __init__(self, *args, occupancy=None, **kwargs):
if occupancy is not None:
kwargs.setdefault("icu_occupancy", occupancy)
kwargs.setdefault("hospital_occupancy", occupancy)
super().__init__(*args, **kwargs)
def _icu_capacity(self):
if self.region is not None:
capacity = self.region.icu_capacity
if np.isfinite(capacity):
return capacity
return self.population
def _hospital_capacity(self):
if self.region is not None:
capacity = self.region.hospital_capacity
if np.isfinite(capacity):
return capacity
return self.population
#
# Data methods
#
# Deaths
def get_data_deaths(self, idx):
return self["natural_deaths", idx] + self["overflow_deaths", idx]
def get_data_natural_deaths(self, idx):
"""
The number of deaths assuming healthcare system in full capacity.
"""
return super().get_data_deaths(idx)
def get_data_overflow_deaths(self, idx):
"""
The number of deaths caused by overflowing the healthcare system.
"""
return self["icu_overflow_deaths", idx] + self["hospital_overflow_deaths", idx]
def get_data_icu_overflow_deaths(self, idx):
"""
The number of deaths caused by overflowing ICUs.
"""
# We just want to comput the excess deaths, so we discount the
# contribution from natural ICUFR that is computed in natural deaths
scale = 1 - self.ICUFR
area = cumtrapz(self["critical_overflow"] * scale, self.times, initial=0)
data = pd.Series(area / self.critical_period, index=self.times)
return sliced(data, idx)
def get_data_hospital_overflow_deaths(self, idx):
"""
The number of deaths caused by overflowing regular hospital beds.
"""
area = cumtrapz(self["severe_overflow"], self.times, initial=0)
cases = area / self.severe_period
ratio = (self.Qcr / self.Qsv) * self.hospitalization_overflow_bias
deaths = cases * min(ratio, 1)
data = | pd.Series(deaths, index=self.times) | pandas.Series |
import os, re
import pandas as pd
_mag = None
def load(path='data/mag/', **kwargs):
global _mag
if 'target' in kwargs.keys() and kwargs['target']:
_mag = kwargs['target']
else:
_mag = {}
if 'file_regex' not in kwargs.keys():
kwargs['file_regex'] = re.compile(r"^(.*)\.csv$")
if 'nodes' not in _mag.keys():
frames = []
nodes_path = os.path.join(path, 'nodes')
for file in os.listdir(nodes_path):
if 'file_regex' not in kwargs.keys() or kwargs['file_regex'].match(file):
frames.append(pd.read_csv(os.path.join(nodes_path, file)))
_mag['nodes'] = pd.concat(frames, axis=0, ignore_index=True)
if 'edges' not in _mag.keys():
frames = []
edges_path = os.path.join(path, 'edges')
for file in os.listdir(edges_path):
if 'file_regex' not in kwargs.keys() or kwargs['file_regex'].match(file):
frames.append(pd.read_csv(os.path.join(edges_path, file)))
_mag['edges'] = pd.concat(frames, axis=0, ignore_index=True)
return _mag
def nodes_by(prop, value, **kwargs):
path = 'data/mag/' if 'mag_dir' not in kwargs.keys() else kwargs['mag_dir']
if not _mag:
load(path, **kwargs)
frame = _mag['nodes']
return frame[frame[prop] == value]
def get_simple_path(ids, start=None, end=None, **kwargs):
path = 'data/mag/' if 'mag_dir' not in kwargs.keys() else kwargs['mag_dir']
if not _mag:
load(path, **kwargs)
frame = _mag['edges']
edges = frame[frame['source'].isin(ids) | frame['target'].isin(ids)]
if not start or not end:
return edges
edges.to_csv('data/edges.csv')
target = edges[edges['source'] == start]
path = []
while (not target['source'].empty) and target['source'].values[0] != end:
source = target
path.append(source)
target = edges[edges['source'] == source['target'].values[0]]
if not target['source'].empty:
frame = pd.concat(path, axis=0, ignore_index=True)
frame = pd.concat([frame['source'], frame['target']], axis=1).stack().reset_index(drop=True)
frame.drop_duplicates(inplace=True)
return _mag['nodes'][_mag['nodes']['id'].isin(frame)]
target = edges[edges['target'] == start]
path = []
while (not target['target'].empty) and target['target'].values[0] != end:
source = target
path.append(source)
target = edges[edges['target'] == source['source'].values[0]]
if target['target'].empty:
return | pd.DataFrame() | pandas.DataFrame |
"""
Tests for the blaze interface to the pipeline api.
"""
from __future__ import division
from collections import OrderedDict
from datetime import timedelta
from unittest import TestCase
import warnings
import blaze as bz
from datashape import dshape, var, Record
from nose_parameterized import parameterized
import numpy as np
from numpy.testing.utils import assert_array_almost_equal
import pandas as pd
from pandas.util.testing import assert_frame_equal
from toolz import keymap, valmap, concatv
from toolz.curried import operator as op
from zipline.pipeline import Pipeline, CustomFactor
from zipline.pipeline.data import DataSet, BoundColumn
from zipline.pipeline.engine import SimplePipelineEngine
from zipline.pipeline.loaders.blaze import (
from_blaze,
BlazeLoader,
NoDeltasWarning,
NonNumpyField,
NonPipelineField,
)
from zipline.utils.numpy_utils import repeat_last_axis
from zipline.utils.test_utils import tmp_asset_finder, make_simple_asset_info
nameof = op.attrgetter('name')
dtypeof = op.attrgetter('dtype')
asset_infos = (
(make_simple_asset_info(
tuple(map(ord, 'ABC')),
pd.Timestamp(0),
pd.Timestamp('2015'),
),),
(make_simple_asset_info(
tuple(map(ord, 'ABCD')),
pd.Timestamp(0),
pd.Timestamp('2015'),
),),
)
with_extra_sid = parameterized.expand(asset_infos)
class BlazeToPipelineTestCase(TestCase):
@classmethod
def setUpClass(cls):
cls.dates = dates = pd.date_range('2014-01-01', '2014-01-03')
dates = cls.dates.repeat(3)
cls.sids = sids = ord('A'), ord('B'), ord('C')
cls.df = df = pd.DataFrame({
'sid': sids * 3,
'value': (0, 1, 2, 1, 2, 3, 2, 3, 4),
'asof_date': dates,
'timestamp': dates,
})
cls.dshape = dshape("""
var * {
sid: ?int64,
value: ?float64,
asof_date: datetime,
timestamp: datetime
}
""")
cls.macro_df = df[df.sid == 65].drop('sid', axis=1)
dshape_ = OrderedDict(cls.dshape.measure.fields)
del dshape_['sid']
cls.macro_dshape = var * Record(dshape_)
cls.garbage_loader = BlazeLoader()
def test_tabular(self):
name = 'expr'
expr = bz.Data(self.df, name=name, dshape=self.dshape)
ds = from_blaze(
expr,
loader=self.garbage_loader,
no_deltas_rule='ignore',
)
self.assertEqual(ds.__name__, name)
self.assertTrue(issubclass(ds, DataSet))
self.assertEqual(
{c.name: c.dtype for c in ds._columns},
{'sid': np.int64, 'value': np.float64},
)
for field in ('timestamp', 'asof_date'):
with self.assertRaises(AttributeError) as e:
getattr(ds, field)
self.assertIn("'%s'" % field, str(e.exception))
self.assertIn("'datetime'", str(e.exception))
# test memoization
self.assertIs(
from_blaze(
expr,
loader=self.garbage_loader,
no_deltas_rule='ignore',
),
ds,
)
def test_column(self):
exprname = 'expr'
expr = bz.Data(self.df, name=exprname, dshape=self.dshape)
value = from_blaze(
expr.value,
loader=self.garbage_loader,
no_deltas_rule='ignore',
)
self.assertEqual(value.name, 'value')
self.assertIsInstance(value, BoundColumn)
self.assertEqual(value.dtype, np.float64)
# test memoization
self.assertIs(
from_blaze(
expr.value,
loader=self.garbage_loader,
no_deltas_rule='ignore',
),
value,
)
self.assertIs(
from_blaze(
expr,
loader=self.garbage_loader,
no_deltas_rule='ignore',
).value,
value,
)
# test the walk back up the tree
self.assertIs(
from_blaze(
expr,
loader=self.garbage_loader,
no_deltas_rule='ignore',
),
value.dataset,
)
self.assertEqual(value.dataset.__name__, exprname)
def test_missing_asof(self):
expr = bz.Data(
self.df.loc[:, ['sid', 'value', 'timestamp']],
name='expr',
dshape="""
var * {
sid: ?int64,
value: float64,
timestamp: datetime,
}""",
)
with self.assertRaises(TypeError) as e:
from_blaze(
expr,
loader=self.garbage_loader,
no_deltas_rule='ignore',
)
self.assertIn("'asof_date'", str(e.exception))
self.assertIn(repr(str(expr.dshape.measure)), str(e.exception))
def test_auto_deltas(self):
expr = bz.Data(
{'ds': self.df,
'ds_deltas': | pd.DataFrame(columns=self.df.columns) | pandas.DataFrame |
from django.shortcuts import render
from plotly.offline import plot
import plotly.graph_objects as go
import plotly.express as px
import pandas as pd
import os
def home(chart):
return render(chart, "index.html")
def engage(chart):
directory = os.getcwd() + "/simulation/engage_sector.xlsx"
data = pd.read_excel(directory)
df = pd.DataFrame(data)
fig = px.sunburst(df, path=['Changes', 'Sector'],
values='Count', width=600, height=500)
eng_sector = plot(fig, output_type='div')
directory = os.getcwd() + "/simulation/engage_location.xlsx"
data = pd.read_excel(directory)
df = pd.DataFrame(data)
fig = px.sunburst(df, path=['Changes', 'Location'],
values='Count', width=600, height=500)
eng_loc = plot(fig, output_type='div')
directory = os.getcwd() + "/simulation/engage_owner.xlsx"
data = | pd.read_excel(directory) | pandas.read_excel |
# -*- coding: utf-8 -*-
# pylint: disable-msg=E1101,W0612
import nose
import numpy as np
from numpy import nan
import pandas as pd
from distutils.version import LooseVersion
from pandas import (Index, Series, DataFrame, Panel, isnull,
date_range, period_range)
from pandas.core.index import MultiIndex
import pandas.core.common as com
from pandas.compat import range, zip
from pandas import compat
from pandas.util.testing import (assert_series_equal,
assert_frame_equal,
assert_panel_equal,
assert_equal)
import pandas.util.testing as tm
def _skip_if_no_pchip():
try:
from scipy.interpolate import pchip_interpolate # noqa
except ImportError:
raise nose.SkipTest('scipy.interpolate.pchip missing')
# ----------------------------------------------------------------------
# Generic types test cases
class Generic(object):
_multiprocess_can_split_ = True
def setUp(self):
pass
@property
def _ndim(self):
return self._typ._AXIS_LEN
def _axes(self):
""" return the axes for my object typ """
return self._typ._AXIS_ORDERS
def _construct(self, shape, value=None, dtype=None, **kwargs):
""" construct an object for the given shape
if value is specified use that if its a scalar
if value is an array, repeat it as needed """
if isinstance(shape, int):
shape = tuple([shape] * self._ndim)
if value is not None:
if np.isscalar(value):
if value == 'empty':
arr = None
# remove the info axis
kwargs.pop(self._typ._info_axis_name, None)
else:
arr = np.empty(shape, dtype=dtype)
arr.fill(value)
else:
fshape = np.prod(shape)
arr = value.ravel()
new_shape = fshape / arr.shape[0]
if fshape % arr.shape[0] != 0:
raise Exception("invalid value passed in _construct")
arr = np.repeat(arr, new_shape).reshape(shape)
else:
arr = np.random.randn(*shape)
return self._typ(arr, dtype=dtype, **kwargs)
def _compare(self, result, expected):
self._comparator(result, expected)
def test_rename(self):
# single axis
for axis in self._axes():
kwargs = {axis: list('ABCD')}
obj = self._construct(4, **kwargs)
# no values passed
# self.assertRaises(Exception, o.rename(str.lower))
# rename a single axis
result = obj.rename(**{axis: str.lower})
expected = obj.copy()
setattr(expected, axis, list('abcd'))
self._compare(result, expected)
# multiple axes at once
def test_get_numeric_data(self):
n = 4
kwargs = {}
for i in range(self._ndim):
kwargs[self._typ._AXIS_NAMES[i]] = list(range(n))
# get the numeric data
o = self._construct(n, **kwargs)
result = o._get_numeric_data()
self._compare(result, o)
# non-inclusion
result = o._get_bool_data()
expected = self._construct(n, value='empty', **kwargs)
self._compare(result, expected)
# get the bool data
arr = np.array([True, True, False, True])
o = self._construct(n, value=arr, **kwargs)
result = o._get_numeric_data()
self._compare(result, o)
# _get_numeric_data is includes _get_bool_data, so can't test for
# non-inclusion
def test_get_default(self):
# GH 7725
d0 = "a", "b", "c", "d"
d1 = np.arange(4, dtype='int64')
others = "e", 10
for data, index in ((d0, d1), (d1, d0)):
s = Series(data, index=index)
for i, d in zip(index, data):
self.assertEqual(s.get(i), d)
self.assertEqual(s.get(i, d), d)
self.assertEqual(s.get(i, "z"), d)
for other in others:
self.assertEqual(s.get(other, "z"), "z")
self.assertEqual(s.get(other, other), other)
def test_nonzero(self):
# GH 4633
# look at the boolean/nonzero behavior for objects
obj = self._construct(shape=4)
self.assertRaises(ValueError, lambda: bool(obj == 0))
self.assertRaises(ValueError, lambda: bool(obj == 1))
self.assertRaises(ValueError, lambda: bool(obj))
obj = self._construct(shape=4, value=1)
self.assertRaises(ValueError, lambda: bool(obj == 0))
self.assertRaises(ValueError, lambda: bool(obj == 1))
self.assertRaises(ValueError, lambda: bool(obj))
obj = self._construct(shape=4, value=np.nan)
self.assertRaises(ValueError, lambda: bool(obj == 0))
self.assertRaises(ValueError, lambda: bool(obj == 1))
self.assertRaises(ValueError, lambda: bool(obj))
# empty
obj = self._construct(shape=0)
self.assertRaises(ValueError, lambda: bool(obj))
# invalid behaviors
obj1 = self._construct(shape=4, value=1)
obj2 = self._construct(shape=4, value=1)
def f():
if obj1:
com.pprint_thing("this works and shouldn't")
self.assertRaises(ValueError, f)
self.assertRaises(ValueError, lambda: obj1 and obj2)
self.assertRaises(ValueError, lambda: obj1 or obj2)
self.assertRaises(ValueError, lambda: not obj1)
def test_numpy_1_7_compat_numeric_methods(self):
# GH 4435
# numpy in 1.7 tries to pass addtional arguments to pandas functions
o = self._construct(shape=4)
for op in ['min', 'max', 'max', 'var', 'std', 'prod', 'sum', 'cumsum',
'cumprod', 'median', 'skew', 'kurt', 'compound', 'cummax',
'cummin', 'all', 'any']:
f = getattr(np, op, None)
if f is not None:
f(o)
def test_downcast(self):
# test close downcasting
o = self._construct(shape=4, value=9, dtype=np.int64)
result = o.copy()
result._data = o._data.downcast(dtypes='infer')
self._compare(result, o)
o = self._construct(shape=4, value=9.)
expected = o.astype(np.int64)
result = o.copy()
result._data = o._data.downcast(dtypes='infer')
self._compare(result, expected)
o = self._construct(shape=4, value=9.5)
result = o.copy()
result._data = o._data.downcast(dtypes='infer')
self._compare(result, o)
# are close
o = self._construct(shape=4, value=9.000000000005)
result = o.copy()
result._data = o._data.downcast(dtypes='infer')
expected = o.astype(np.int64)
self._compare(result, expected)
def test_constructor_compound_dtypes(self):
# GH 5191
# compound dtypes should raise not-implementederror
def f(dtype):
return self._construct(shape=3, dtype=dtype)
self.assertRaises(NotImplementedError, f, [("A", "datetime64[h]"),
("B", "str"),
("C", "int32")])
# these work (though results may be unexpected)
f('int64')
f('float64')
f('M8[ns]')
def check_metadata(self, x, y=None):
for m in x._metadata:
v = getattr(x, m, None)
if y is None:
self.assertIsNone(v)
else:
self.assertEqual(v, getattr(y, m, None))
def test_metadata_propagation(self):
# check that the metadata matches up on the resulting ops
o = self._construct(shape=3)
o.name = 'foo'
o2 = self._construct(shape=3)
o2.name = 'bar'
# TODO
# Once panel can do non-trivial combine operations
# (currently there is an a raise in the Panel arith_ops to prevent
# this, though it actually does work)
# can remove all of these try: except: blocks on the actual operations
# ----------
# preserving
# ----------
# simple ops with scalars
for op in ['__add__', '__sub__', '__truediv__', '__mul__']:
result = getattr(o, op)(1)
self.check_metadata(o, result)
# ops with like
for op in ['__add__', '__sub__', '__truediv__', '__mul__']:
try:
result = getattr(o, op)(o)
self.check_metadata(o, result)
except (ValueError, AttributeError):
pass
# simple boolean
for op in ['__eq__', '__le__', '__ge__']:
v1 = getattr(o, op)(o)
self.check_metadata(o, v1)
try:
self.check_metadata(o, v1 & v1)
except (ValueError):
pass
try:
self.check_metadata(o, v1 | v1)
except (ValueError):
pass
# combine_first
try:
result = o.combine_first(o2)
self.check_metadata(o, result)
except (AttributeError):
pass
# ---------------------------
# non-preserving (by default)
# ---------------------------
# add non-like
try:
result = o + o2
self.check_metadata(result)
except (ValueError, AttributeError):
pass
# simple boolean
for op in ['__eq__', '__le__', '__ge__']:
# this is a name matching op
v1 = getattr(o, op)(o)
v2 = getattr(o, op)(o2)
self.check_metadata(v2)
try:
self.check_metadata(v1 & v2)
except (ValueError):
pass
try:
self.check_metadata(v1 | v2)
except (ValueError):
pass
def test_head_tail(self):
# GH5370
o = self._construct(shape=10)
# check all index types
for index in [tm.makeFloatIndex, tm.makeIntIndex, tm.makeStringIndex,
tm.makeUnicodeIndex, tm.makeDateIndex,
tm.makePeriodIndex]:
axis = o._get_axis_name(0)
setattr(o, axis, index(len(getattr(o, axis))))
# Panel + dims
try:
o.head()
except (NotImplementedError):
raise nose.SkipTest('not implemented on {0}'.format(
o.__class__.__name__))
self._compare(o.head(), o.iloc[:5])
self._compare(o.tail(), o.iloc[-5:])
# 0-len
self._compare(o.head(0), o.iloc[0:0])
self._compare(o.tail(0), o.iloc[0:0])
# bounded
self._compare(o.head(len(o) + 1), o)
self._compare(o.tail(len(o) + 1), o)
# neg index
self._compare(o.head(-3), o.head(7))
self._compare(o.tail(-3), o.tail(7))
def test_sample(self):
# Fixes issue: 2419
o = self._construct(shape=10)
###
# Check behavior of random_state argument
###
# Check for stability when receives seed or random state -- run 10
# times.
for test in range(10):
seed = np.random.randint(0, 100)
self._compare(
o.sample(n=4, random_state=seed), o.sample(n=4,
random_state=seed))
self._compare(
o.sample(frac=0.7, random_state=seed), o.sample(
frac=0.7, random_state=seed))
self._compare(
o.sample(n=4, random_state=np.random.RandomState(test)),
o.sample(n=4, random_state=np.random.RandomState(test)))
self._compare(
o.sample(frac=0.7, random_state=np.random.RandomState(test)),
o.sample(frac=0.7, random_state=np.random.RandomState(test)))
# Check for error when random_state argument invalid.
with tm.assertRaises(ValueError):
o.sample(random_state='astring!')
###
# Check behavior of `frac` and `N`
###
# Giving both frac and N throws error
with tm.assertRaises(ValueError):
o.sample(n=3, frac=0.3)
# Check that raises right error for negative lengths
with tm.assertRaises(ValueError):
o.sample(n=-3)
with tm.assertRaises(ValueError):
o.sample(frac=-0.3)
# Make sure float values of `n` give error
with tm.assertRaises(ValueError):
o.sample(n=3.2)
# Check lengths are right
self.assertTrue(len(o.sample(n=4) == 4))
self.assertTrue(len(o.sample(frac=0.34) == 3))
self.assertTrue(len(o.sample(frac=0.36) == 4))
###
# Check weights
###
# Weight length must be right
with tm.assertRaises(ValueError):
o.sample(n=3, weights=[0, 1])
with tm.assertRaises(ValueError):
bad_weights = [0.5] * 11
o.sample(n=3, weights=bad_weights)
with tm.assertRaises(ValueError):
bad_weight_series = Series([0, 0, 0.2])
o.sample(n=4, weights=bad_weight_series)
# Check won't accept negative weights
with tm.assertRaises(ValueError):
bad_weights = [-0.1] * 10
o.sample(n=3, weights=bad_weights)
# Check inf and -inf throw errors:
with tm.assertRaises(ValueError):
weights_with_inf = [0.1] * 10
weights_with_inf[0] = np.inf
o.sample(n=3, weights=weights_with_inf)
with tm.assertRaises(ValueError):
weights_with_ninf = [0.1] * 10
weights_with_ninf[0] = -np.inf
o.sample(n=3, weights=weights_with_ninf)
# All zeros raises errors
zero_weights = [0] * 10
with tm.assertRaises(ValueError):
o.sample(n=3, weights=zero_weights)
# All missing weights
nan_weights = [np.nan] * 10
with tm.assertRaises(ValueError):
o.sample(n=3, weights=nan_weights)
# A few dataframe test with degenerate weights.
easy_weight_list = [0] * 10
easy_weight_list[5] = 1
df = pd.DataFrame({'col1': range(10, 20),
'col2': range(20, 30),
'colString': ['a'] * 10,
'easyweights': easy_weight_list})
sample1 = df.sample(n=1, weights='easyweights')
assert_frame_equal(sample1, df.iloc[5:6])
# Ensure proper error if string given as weight for Series, panel, or
# DataFrame with axis = 1.
s = Series(range(10))
with tm.assertRaises(ValueError):
s.sample(n=3, weights='weight_column')
panel = pd.Panel(items=[0, 1, 2], major_axis=[2, 3, 4],
minor_axis=[3, 4, 5])
with tm.assertRaises(ValueError):
panel.sample(n=1, weights='weight_column')
with tm.assertRaises(ValueError):
df.sample(n=1, weights='weight_column', axis=1)
# Check weighting key error
with tm.assertRaises(KeyError):
df.sample(n=3, weights='not_a_real_column_name')
# Check np.nan are replaced by zeros.
weights_with_nan = [np.nan] * 10
weights_with_nan[5] = 0.5
self._compare(
o.sample(n=1, axis=0, weights=weights_with_nan), o.iloc[5:6])
# Check None are also replaced by zeros.
weights_with_None = [None] * 10
weights_with_None[5] = 0.5
self._compare(
o.sample(n=1, axis=0, weights=weights_with_None), o.iloc[5:6])
# Check that re-normalizes weights that don't sum to one.
weights_less_than_1 = [0] * 10
weights_less_than_1[0] = 0.5
tm.assert_frame_equal(
df.sample(n=1, weights=weights_less_than_1), df.iloc[:1])
###
# Test axis argument
###
# Test axis argument
df = pd.DataFrame({'col1': range(10), 'col2': ['a'] * 10})
second_column_weight = [0, 1]
assert_frame_equal(
df.sample(n=1, axis=1, weights=second_column_weight), df[['col2']])
# Different axis arg types
assert_frame_equal(df.sample(n=1, axis='columns',
weights=second_column_weight),
df[['col2']])
weight = [0] * 10
weight[5] = 0.5
assert_frame_equal(df.sample(n=1, axis='rows', weights=weight),
df.iloc[5:6])
assert_frame_equal(df.sample(n=1, axis='index', weights=weight),
df.iloc[5:6])
# Check out of range axis values
with tm.assertRaises(ValueError):
df.sample(n=1, axis=2)
with tm.assertRaises(ValueError):
df.sample(n=1, axis='not_a_name')
with tm.assertRaises(ValueError):
s = pd.Series(range(10))
s.sample(n=1, axis=1)
# Test weight length compared to correct axis
with tm.assertRaises(ValueError):
df.sample(n=1, axis=1, weights=[0.5] * 10)
# Check weights with axis = 1
easy_weight_list = [0] * 3
easy_weight_list[2] = 1
df = pd.DataFrame({'col1': range(10, 20),
'col2': range(20, 30),
'colString': ['a'] * 10})
sample1 = df.sample(n=1, axis=1, weights=easy_weight_list)
assert_frame_equal(sample1, df[['colString']])
# Test default axes
p = pd.Panel(items=['a', 'b', 'c'], major_axis=[2, 4, 6],
minor_axis=[1, 3, 5])
assert_panel_equal(
p.sample(n=3, random_state=42), p.sample(n=3, axis=1,
random_state=42))
assert_frame_equal(
df.sample(n=3, random_state=42), df.sample(n=3, axis=0,
random_state=42))
# Test that function aligns weights with frame
df = DataFrame(
{'col1': [5, 6, 7],
'col2': ['a', 'b', 'c'], }, index=[9, 5, 3])
s = Series([1, 0, 0], index=[3, 5, 9])
assert_frame_equal(df.loc[[3]], df.sample(1, weights=s))
# Weights have index values to be dropped because not in
# sampled DataFrame
s2 = Series([0.001, 0, 10000], index=[3, 5, 10])
assert_frame_equal(df.loc[[3]], df.sample(1, weights=s2))
# Weights have empty values to be filed with zeros
s3 = Series([0.01, 0], index=[3, 5])
assert_frame_equal(df.loc[[3]], df.sample(1, weights=s3))
# No overlap in weight and sampled DataFrame indices
s4 = Series([1, 0], index=[1, 2])
with tm.assertRaises(ValueError):
df.sample(1, weights=s4)
def test_size_compat(self):
# GH8846
# size property should be defined
o = self._construct(shape=10)
self.assertTrue(o.size == np.prod(o.shape))
self.assertTrue(o.size == 10 ** len(o.axes))
def test_split_compat(self):
# xref GH8846
o = self._construct(shape=10)
self.assertTrue(len(np.array_split(o, 5)) == 5)
self.assertTrue(len(np.array_split(o, 2)) == 2)
def test_unexpected_keyword(self): # GH8597
from pandas.util.testing import assertRaisesRegexp
df = DataFrame(np.random.randn(5, 2), columns=['jim', 'joe'])
ca = pd.Categorical([0, 0, 2, 2, 3, np.nan])
ts = df['joe'].copy()
ts[2] = np.nan
with assertRaisesRegexp(TypeError, 'unexpected keyword'):
df.drop('joe', axis=1, in_place=True)
with assertRaisesRegexp(TypeError, 'unexpected keyword'):
df.reindex([1, 0], inplace=True)
with assertRaisesRegexp(TypeError, 'unexpected keyword'):
ca.fillna(0, inplace=True)
with assertRaisesRegexp(TypeError, 'unexpected keyword'):
ts.fillna(0, in_place=True)
class TestSeries(tm.TestCase, Generic):
_typ = Series
_comparator = lambda self, x, y: assert_series_equal(x, y)
def setUp(self):
self.ts = tm.makeTimeSeries() # Was at top level in test_series
self.ts.name = 'ts'
self.series = tm.makeStringSeries()
self.series.name = 'series'
def test_rename_mi(self):
s = Series([11, 21, 31],
index=MultiIndex.from_tuples(
[("A", x) for x in ["a", "B", "c"]]))
s.rename(str.lower)
def test_get_numeric_data_preserve_dtype(self):
# get the numeric data
o = Series([1, 2, 3])
result = o._get_numeric_data()
self._compare(result, o)
o = Series([1, '2', 3.])
result = o._get_numeric_data()
expected = Series([], dtype=object, index=pd.Index([], dtype=object))
self._compare(result, expected)
o = Series([True, False, True])
result = o._get_numeric_data()
self._compare(result, o)
o = Series([True, False, True])
result = o._get_bool_data()
self._compare(result, o)
o = Series(date_range('20130101', periods=3))
result = o._get_numeric_data()
expected = Series([], dtype='M8[ns]', index=pd.Index([], dtype=object))
self._compare(result, expected)
def test_nonzero_single_element(self):
# allow single item via bool method
s = Series([True])
self.assertTrue(s.bool())
s = Series([False])
self.assertFalse(s.bool())
# single item nan to raise
for s in [Series([np.nan]), Series([pd.NaT]), Series([True]),
Series([False])]:
self.assertRaises(ValueError, lambda: bool(s))
for s in [Series([np.nan]), Series([pd.NaT])]:
self.assertRaises(ValueError, lambda: s.bool())
# multiple bool are still an error
for s in [Series([True, True]), Series([False, False])]:
self.assertRaises(ValueError, lambda: bool(s))
self.assertRaises(ValueError, lambda: s.bool())
# single non-bool are an error
for s in [Series([1]), Series([0]), Series(['a']), Series([0.0])]:
self.assertRaises(ValueError, lambda: bool(s))
self.assertRaises(ValueError, lambda: s.bool())
def test_metadata_propagation_indiv(self):
# check that the metadata matches up on the resulting ops
o = Series(range(3), range(3))
o.name = 'foo'
o2 = Series(range(3), range(3))
o2.name = 'bar'
result = o.T
self.check_metadata(o, result)
# resample
ts = Series(np.random.rand(1000),
index=date_range('20130101', periods=1000, freq='s'),
name='foo')
result = ts.resample('1T').mean()
self.check_metadata(ts, result)
result = ts.resample('1T').min()
self.check_metadata(ts, result)
result = ts.resample('1T').apply(lambda x: x.sum())
self.check_metadata(ts, result)
_metadata = Series._metadata
_finalize = Series.__finalize__
Series._metadata = ['name', 'filename']
o.filename = 'foo'
o2.filename = 'bar'
def finalize(self, other, method=None, **kwargs):
for name in self._metadata:
if method == 'concat' and name == 'filename':
value = '+'.join([getattr(
o, name) for o in other.objs if getattr(o, name, None)
])
object.__setattr__(self, name, value)
else:
object.__setattr__(self, name, getattr(other, name, None))
return self
Series.__finalize__ = finalize
result = pd.concat([o, o2])
self.assertEqual(result.filename, 'foo+bar')
self.assertIsNone(result.name)
# reset
Series._metadata = _metadata
Series.__finalize__ = _finalize
def test_interpolate(self):
ts = Series(np.arange(len(self.ts), dtype=float), self.ts.index)
ts_copy = ts.copy()
ts_copy[5:10] = np.NaN
linear_interp = ts_copy.interpolate(method='linear')
self.assert_numpy_array_equal(linear_interp, ts)
ord_ts = Series([d.toordinal() for d in self.ts.index],
index=self.ts.index).astype(float)
ord_ts_copy = ord_ts.copy()
ord_ts_copy[5:10] = np.NaN
time_interp = ord_ts_copy.interpolate(method='time')
self.assert_numpy_array_equal(time_interp, ord_ts)
# try time interpolation on a non-TimeSeries
# Only raises ValueError if there are NaNs.
non_ts = self.series.copy()
non_ts[0] = np.NaN
self.assertRaises(ValueError, non_ts.interpolate, method='time')
def test_interp_regression(self):
tm._skip_if_no_scipy()
_skip_if_no_pchip()
ser = Series(np.sort(np.random.uniform(size=100)))
# interpolate at new_index
new_index = ser.index.union(Index([49.25, 49.5, 49.75, 50.25, 50.5,
50.75]))
interp_s = ser.reindex(new_index).interpolate(method='pchip')
# does not blow up, GH5977
interp_s[49:51]
def test_interpolate_corners(self):
s = Series([np.nan, np.nan])
assert_series_equal(s.interpolate(), s)
s = Series([]).interpolate()
assert_series_equal(s.interpolate(), s)
tm._skip_if_no_scipy()
s = Series([np.nan, np.nan])
assert_series_equal(s.interpolate(method='polynomial', order=1), s)
s = Series([]).interpolate()
assert_series_equal(s.interpolate(method='polynomial', order=1), s)
def test_interpolate_index_values(self):
s = Series(np.nan, index=np.sort(np.random.rand(30)))
s[::3] = np.random.randn(10)
vals = s.index.values.astype(float)
result = s.interpolate(method='index')
expected = s.copy()
bad = isnull(expected.values)
good = ~bad
expected = Series(np.interp(vals[bad], vals[good],
s.values[good]),
index=s.index[bad])
assert_series_equal(result[bad], expected)
# 'values' is synonymous with 'index' for the method kwarg
other_result = s.interpolate(method='values')
assert_series_equal(other_result, result)
assert_series_equal(other_result[bad], expected)
def test_interpolate_non_ts(self):
s = Series([1, 3, np.nan, np.nan, np.nan, 11])
with tm.assertRaises(ValueError):
s.interpolate(method='time')
# New interpolation tests
def test_nan_interpolate(self):
s = Series([0, 1, np.nan, 3])
result = s.interpolate()
expected = Series([0., 1., 2., 3.])
assert_series_equal(result, expected)
tm._skip_if_no_scipy()
result = s.interpolate(method='polynomial', order=1)
assert_series_equal(result, expected)
def test_nan_irregular_index(self):
s = Series([1, 2, np.nan, 4], index=[1, 3, 5, 9])
result = s.interpolate()
expected = Series([1., 2., 3., 4.], index=[1, 3, 5, 9])
assert_series_equal(result, expected)
def test_nan_str_index(self):
s = Series([0, 1, 2, np.nan], index=list('abcd'))
result = s.interpolate()
expected = Series([0., 1., 2., 2.], index=list('abcd'))
assert_series_equal(result, expected)
def test_interp_quad(self):
tm._skip_if_no_scipy()
sq = Series([1, 4, np.nan, 16], index=[1, 2, 3, 4])
result = sq.interpolate(method='quadratic')
expected = Series([1., 4., 9., 16.], index=[1, 2, 3, 4])
assert_series_equal(result, expected)
def test_interp_scipy_basic(self):
tm._skip_if_no_scipy()
s = Series([1, 3, np.nan, 12, np.nan, 25])
# slinear
expected = Series([1., 3., 7.5, 12., 18.5, 25.])
result = s.interpolate(method='slinear')
assert_series_equal(result, expected)
result = s.interpolate(method='slinear', downcast='infer')
assert_series_equal(result, expected)
# nearest
expected = Series([1, 3, 3, 12, 12, 25])
result = s.interpolate(method='nearest')
assert_series_equal(result, expected.astype('float'))
result = s.interpolate(method='nearest', downcast='infer')
assert_series_equal(result, expected)
# zero
expected = Series([1, 3, 3, 12, 12, 25])
result = s.interpolate(method='zero')
assert_series_equal(result, expected.astype('float'))
result = s.interpolate(method='zero', downcast='infer')
assert_series_equal(result, expected)
# quadratic
expected = Series([1, 3., 6.769231, 12., 18.230769, 25.])
result = s.interpolate(method='quadratic')
assert_series_equal(result, expected)
result = s.interpolate(method='quadratic', downcast='infer')
assert_series_equal(result, expected)
# cubic
expected = Series([1., 3., 6.8, 12., 18.2, 25.])
result = s.interpolate(method='cubic')
assert_series_equal(result, expected)
def test_interp_limit(self):
s = Series([1, 3, np.nan, np.nan, np.nan, 11])
expected = Series([1., 3., 5., 7., np.nan, 11.])
result = s.interpolate(method='linear', limit=2)
assert_series_equal(result, expected)
def test_interp_limit_forward(self):
s = Series([1, 3, np.nan, np.nan, np.nan, 11])
# Provide 'forward' (the default) explicitly here.
expected = Series([1., 3., 5., 7., np.nan, 11.])
result = s.interpolate(method='linear', limit=2,
limit_direction='forward')
assert_series_equal(result, expected)
result = s.interpolate(method='linear', limit=2,
limit_direction='FORWARD')
assert_series_equal(result, expected)
def test_interp_limit_bad_direction(self):
s = Series([1, 3, np.nan, np.nan, np.nan, 11])
self.assertRaises(ValueError, s.interpolate, method='linear', limit=2,
limit_direction='abc')
# raises an error even if no limit is specified.
self.assertRaises(ValueError, s.interpolate, method='linear',
limit_direction='abc')
def test_interp_limit_direction(self):
# These tests are for issue #9218 -- fill NaNs in both directions.
s = Series([1, 3, np.nan, np.nan, np.nan, 11])
expected = Series([1., 3., np.nan, 7., 9., 11.])
result = s.interpolate(method='linear', limit=2,
limit_direction='backward')
assert_series_equal(result, expected)
expected = Series([1., 3., 5., np.nan, 9., 11.])
result = s.interpolate(method='linear', limit=1,
limit_direction='both')
assert_series_equal(result, expected)
# Check that this works on a longer series of nans.
s = Series([1, 3, np.nan, np.nan, np.nan, 7, 9, np.nan, np.nan, 12,
np.nan])
expected = Series([1., 3., 4., 5., 6., 7., 9., 10., 11., 12., 12.])
result = s.interpolate(method='linear', limit=2,
limit_direction='both')
assert_series_equal(result, expected)
expected = Series([1., 3., 4., np.nan, 6., 7., 9., 10., 11., 12., 12.])
result = s.interpolate(method='linear', limit=1,
limit_direction='both')
assert_series_equal(result, expected)
def test_interp_limit_to_ends(self):
# These test are for issue #10420 -- flow back to beginning.
s = Series([np.nan, np.nan, 5, 7, 9, np.nan])
expected = Series([5., 5., 5., 7., 9., np.nan])
result = s.interpolate(method='linear', limit=2,
limit_direction='backward')
assert_series_equal(result, expected)
expected = Series([5., 5., 5., 7., 9., 9.])
result = s.interpolate(method='linear', limit=2,
limit_direction='both')
assert_series_equal(result, expected)
def test_interp_limit_before_ends(self):
# These test are for issue #11115 -- limit ends properly.
s = Series([np.nan, np.nan, 5, 7, np.nan, np.nan])
expected = Series([np.nan, np.nan, 5., 7., 7., np.nan])
result = s.interpolate(method='linear', limit=1,
limit_direction='forward')
assert_series_equal(result, expected)
expected = Series([np.nan, 5., 5., 7., np.nan, np.nan])
result = s.interpolate(method='linear', limit=1,
limit_direction='backward')
assert_series_equal(result, expected)
expected = Series([np.nan, 5., 5., 7., 7., np.nan])
result = s.interpolate(method='linear', limit=1,
limit_direction='both')
assert_series_equal(result, expected)
def test_interp_all_good(self):
# scipy
tm._skip_if_no_scipy()
s = Series([1, 2, 3])
result = s.interpolate(method='polynomial', order=1)
assert_series_equal(result, s)
# non-scipy
result = s.interpolate()
assert_series_equal(result, s)
def test_interp_multiIndex(self):
idx = MultiIndex.from_tuples([(0, 'a'), (1, 'b'), (2, 'c')])
s = Series([1, 2, np.nan], index=idx)
expected = s.copy()
expected.loc[2] = 2
result = s.interpolate()
assert_series_equal(result, expected)
tm._skip_if_no_scipy()
with tm.assertRaises(ValueError):
s.interpolate(method='polynomial', order=1)
def test_interp_nonmono_raise(self):
tm._skip_if_no_scipy()
s = Series([1, np.nan, 3], index=[0, 2, 1])
with tm.assertRaises(ValueError):
s.interpolate(method='krogh')
def test_interp_datetime64(self):
tm._skip_if_no_scipy()
df = Series([1, np.nan, 3], index=date_range('1/1/2000', periods=3))
result = df.interpolate(method='nearest')
expected = Series([1., 1., 3.],
index=date_range('1/1/2000', periods=3))
assert_series_equal(result, expected)
def test_interp_limit_no_nans(self):
# GH 7173
s = pd.Series([1., 2., 3.])
result = s.interpolate(limit=1)
expected = s
assert_series_equal(result, expected)
def test_describe(self):
self.series.describe()
self.ts.describe()
def test_describe_objects(self):
s = Series(['a', 'b', 'b', np.nan, np.nan, np.nan, 'c', 'd', 'a', 'a'])
result = s.describe()
expected = Series({'count': 7, 'unique': 4,
'top': 'a', 'freq': 3}, index=result.index)
assert_series_equal(result, expected)
dt = list(self.ts.index)
dt.append(dt[0])
ser = Series(dt)
rs = ser.describe()
min_date = min(dt)
max_date = max(dt)
xp = Series({'count': len(dt),
'unique': len(self.ts.index),
'first': min_date, 'last': max_date, 'freq': 2,
'top': min_date}, index=rs.index)
assert_series_equal(rs, xp)
def test_describe_empty(self):
result = pd.Series().describe()
self.assertEqual(result['count'], 0)
self.assertTrue(result.drop('count').isnull().all())
nanSeries = Series([np.nan])
nanSeries.name = 'NaN'
result = nanSeries.describe()
self.assertEqual(result['count'], 0)
self.assertTrue(result.drop('count').isnull().all())
def test_describe_none(self):
noneSeries = Series([None])
noneSeries.name = 'None'
expected = Series([0, 0], index=['count', 'unique'], name='None')
assert_series_equal(noneSeries.describe(), expected)
class TestDataFrame(tm.TestCase, Generic):
_typ = DataFrame
_comparator = lambda self, x, y: assert_frame_equal(x, y)
def test_rename_mi(self):
df = DataFrame([
11, 21, 31
], index=MultiIndex.from_tuples([("A", x) for x in ["a", "B", "c"]]))
df.rename(str.lower)
def test_nonzero_single_element(self):
# allow single item via bool method
df = DataFrame([[True]])
self.assertTrue(df.bool())
df = DataFrame([[False]])
self.assertFalse(df.bool())
df = DataFrame([[False, False]])
self.assertRaises(ValueError, lambda: df.bool())
self.assertRaises(ValueError, lambda: bool(df))
def test_get_numeric_data_preserve_dtype(self):
# get the numeric data
o = | DataFrame({'A': [1, '2', 3.]}) | pandas.DataFrame |
#!/usr/bin/env python3
import tempfile
import unittest
import warnings
import webbrowser
from copy import deepcopy
import matplotlib.pyplot as plt
import numpy as np
import numpy.testing as nptest
import pandas as pd
import pandas.testing as pdtest
from sklearn.model_selection import GridSearchCV
from sklearn.utils import estimator_html_repr
from datafold.appfold.edmd import EDMD, EDMDCV, EDMDWindowPrediction
from datafold.dynfold import DMDFull, gDMDFull
from datafold.dynfold.transform import (
TSCFeaturePreprocess,
TSCIdentity,
TSCPrincipalComponent,
TSCTakensEmbedding,
)
from datafold.pcfold import TSCDataFrame, TSCKfoldSeries, TSCKFoldTime
from datafold.pcfold.timeseries.collection import TSCException
from datafold.utils.general import is_df_same_index
from datafold.utils.plot import plot_eigenvalues
class EDMDTest(unittest.TestCase):
@staticmethod
def _setup_sine_wave_data() -> TSCDataFrame:
time = np.linspace(0, 2 * np.pi, 100)
df = pd.DataFrame(np.sin(time) + 10, index=time, columns=["sin"])
return TSCDataFrame.from_single_timeseries(df)
def _setup_multi_sine_wave_data(self) -> TSCDataFrame:
time = np.linspace(0, 4 * np.pi, 100)
omega = 1.5
for i in range(1, 11):
data = np.sin(i * omega * time)
df = pd.DataFrame(data=data, index=time, columns=["sin"])
if i == 1:
tsc = TSCDataFrame.from_single_timeseries(df)
else:
tsc = tsc.insert_ts(df)
self.assertTrue(tsc.is_same_time_values())
return tsc
def _setup_multi_sine_wave_data2(self) -> TSCDataFrame:
time = np.linspace(0, 2 * np.pi, 100)
omega = 1.5
for i in range(1, 11):
data = np.column_stack([np.sin(i * omega * time), np.cos(i * omega * time)])
df = pd.DataFrame(data=data, index=time, columns=["sin", "cos"])
if i == 1:
tsc = TSCDataFrame.from_single_timeseries(df)
else:
tsc = tsc.insert_ts(df)
self.assertTrue(tsc.is_same_time_values())
return tsc
def setUp(self) -> None:
self.sine_wave_tsc = self._setup_sine_wave_data()
self.multi_sine_wave_tsc = self._setup_multi_sine_wave_data()
self.multi_waves = self._setup_multi_sine_wave_data2()
def test_id_dict1(self):
_edmd = EDMD(
dict_steps=[("id", TSCIdentity())],
include_id_state=False,
use_transform_inverse=False,
verbose=2,
).fit(self.sine_wave_tsc)
pdtest.assert_frame_equal(
_edmd.transform(self.sine_wave_tsc), self.sine_wave_tsc
)
actual = _edmd.inverse_transform(_edmd.transform(self.sine_wave_tsc))
expected = self.sine_wave_tsc
pdtest.assert_frame_equal(actual, expected)
expected = _edmd.reconstruct(self.sine_wave_tsc)
is_df_same_index(expected, self.sine_wave_tsc)
self.assertEqual(_edmd.feature_names_in_, self.sine_wave_tsc.columns)
self.assertEqual(_edmd.feature_names_out_, self.sine_wave_tsc.columns)
self.assertEqual(_edmd.n_features_in_, self.sine_wave_tsc.shape[1])
self.assertEqual(_edmd.n_features_out_, self.sine_wave_tsc.shape[1])
def test_id_dict2(self):
_edmd = EDMD(
dict_steps=[("id", TSCIdentity())],
include_id_state=False,
use_transform_inverse=True, # different to test_id_dict1
verbose=2,
).fit(self.sine_wave_tsc)
pdtest.assert_frame_equal(
_edmd.transform(self.sine_wave_tsc), self.sine_wave_tsc
)
pdtest.assert_frame_equal(
_edmd.inverse_transform(self.sine_wave_tsc), self.sine_wave_tsc
)
expected = _edmd.reconstruct(self.sine_wave_tsc)
is_df_same_index(expected, self.sine_wave_tsc)
self.assertEqual(_edmd.feature_names_in_, self.sine_wave_tsc.columns)
self.assertEqual(_edmd.feature_names_out_, self.sine_wave_tsc.columns)
self.assertEqual(_edmd.n_features_in_, self.sine_wave_tsc.shape[1])
self.assertEqual(_edmd.n_features_out_, self.sine_wave_tsc.shape[1])
def test_id_dict3(self):
_edmd = EDMD(
dict_steps=[("id", TSCIdentity(include_const=True))],
include_id_state=False,
use_transform_inverse=False,
).fit(self.sine_wave_tsc)
actual = _edmd.inverse_transform(_edmd.transform(self.sine_wave_tsc))
expected = self.sine_wave_tsc
pdtest.assert_frame_equal(actual, expected)
expected = _edmd.reconstruct(self.sine_wave_tsc)
is_df_same_index(expected, self.sine_wave_tsc)
self.assertEqual(_edmd.feature_names_in_, self.sine_wave_tsc.columns)
self.assertEqual(_edmd.n_features_in_, self.sine_wave_tsc.shape[1])
self.assertEqual(_edmd.n_features_out_, self.sine_wave_tsc.shape[1] + 1)
def test_qoi_selection1(self):
X = self.multi_waves
# pre-selection
edmd = EDMD(dict_steps=[("id", TSCIdentity())], include_id_state=False).fit(X)
cos_values = edmd.predict(X.initial_states(), qois=["cos"])
sin_values = edmd.predict(X.initial_states(), qois=["sin"])
pdtest.assert_index_equal(X.loc[:, "cos"].columns, cos_values.columns)
pdtest.assert_index_equal(X.loc[:, "sin"].columns, sin_values.columns)
cos_values_reconstruct = edmd.reconstruct(X, qois=["cos"])
sin_values_reconstruct = edmd.reconstruct(X, qois=["sin"])
pdtest.assert_index_equal(
X.loc[:, "cos"].columns, cos_values_reconstruct.columns
)
pdtest.assert_index_equal(
X.loc[:, "sin"].columns, sin_values_reconstruct.columns
)
def test_qoi_selection2(self):
tsc = self.multi_waves
# pre-selection
edmd = EDMD(
dict_steps=[("id", TSCIdentity(include_const=False, rename_features=True))],
include_id_state=True,
).fit(tsc)
cos_values_predict = edmd.predict(tsc.initial_states(), qois=["cos"])
sin_values_predict = edmd.predict(tsc.initial_states(), qois=["sin"])
pdtest.assert_index_equal(tsc.loc[:, "cos"].columns, cos_values_predict.columns)
pdtest.assert_index_equal(tsc.loc[:, "sin"].columns, sin_values_predict.columns)
cos_values_reconstruct = edmd.reconstruct(tsc, qois=["cos"])
sin_values_reconstruct = edmd.reconstruct(tsc, qois=["sin"])
pdtest.assert_index_equal(
tsc.loc[:, "cos"].columns, cos_values_reconstruct.columns
)
pdtest.assert_index_equal(
tsc.loc[:, "sin"].columns, sin_values_reconstruct.columns
)
def test_qoi_selection3(self):
tsc = self.multi_waves
# pre-selection
edmd = EDMD(
dict_steps=[("id", TSCIdentity(include_const=False, rename_features=True))],
include_id_state=True,
).fit(tsc)
with self.assertRaises(ValueError):
edmd.predict(tsc.initial_states(), qois=["INVALID"])
def test_edmd_no_classifier(self):
# import from internal module -- subject to change without warning!
from sklearn.model_selection._validation import is_classifier
self.assertFalse(is_classifier(EDMD))
self.assertFalse(is_classifier(EDMDCV))
def test_n_samples_ic(self):
_edmd = EDMD(
dict_steps=[
("scale", TSCFeaturePreprocess.from_name(name="min-max")),
("delays", TSCTakensEmbedding(delays=10)),
("pca", TSCPrincipalComponent(n_components=2)),
],
include_id_state=True,
).fit(X=self.multi_waves)
actual = _edmd.transform(self.multi_waves.initial_states(_edmd.n_samples_ic_))
# each initial-condition time series must result into a single state in
# dictionary space
self.assertIsInstance(actual, pd.DataFrame)
# 2 ID states + 2 PCA components
self.assertEqual(actual.shape, (self.multi_waves.n_timeseries, 2 + 2))
# Take one sample more and transform the states
actual = _edmd.transform(
self.multi_waves.initial_states(_edmd.n_samples_ic_ + 1)
)
self.assertIsInstance(actual, TSCDataFrame)
# Having not enough samples must result into error
with self.assertRaises(TSCException):
_edmd.transform(self.multi_waves.initial_states(_edmd.n_samples_ic_ - 1))
def test_error_nonmatch_time_sample(self):
_edmd = EDMD(
dict_steps=[
("scale", TSCFeaturePreprocess.from_name(name="min-max")),
("delays", TSCTakensEmbedding(delays=10)),
("pca", TSCPrincipalComponent(n_components=2)),
],
include_id_state=True,
).fit(X=self.multi_waves)
initial_condition = self.multi_waves.initial_states(_edmd.n_samples_ic_)
# change time values to a different sampling interval
initial_condition.index = pd.MultiIndex.from_arrays(
[
initial_condition.index.get_level_values(TSCDataFrame.tsc_id_idx_name),
# change sample rate:
initial_condition.index.get_level_values(TSCDataFrame.tsc_time_idx_name)
* 2,
]
)
with self.assertRaises(TSCException):
_edmd.predict(initial_condition)
def test_access_koopman_triplet(self):
# triplet = eigenvalues, Koopman modes and eigenfunctions
_edmd = EDMD(
dict_steps=[
("scale", TSCFeaturePreprocess.from_name(name="min-max")),
("delays", TSCTakensEmbedding(delays=10)),
("pca", TSCPrincipalComponent(n_components=2)),
],
include_id_state=True,
).fit(X=self.multi_waves)
eval_waves = self.multi_waves.loc[pd.IndexSlice[0:1], :]
actual_modes = _edmd.koopman_modes
actual_eigvals = _edmd.koopman_eigenvalues
actual_eigfunc = _edmd.koopman_eigenfunction(X=eval_waves)
# 2 original states
# 4 eigenvectors in dictionary space (2 ID states + 2 PCA states)
expected = (2, 4)
self.assertEqual(actual_modes.shape, expected)
self.assertEqual(actual_eigvals.shape[0], expected[1])
self.assertEqual(
actual_eigfunc.shape,
(
eval_waves.shape[0]
# correct the output samples by number of samples required for
# initial condition
- eval_waves.n_timeseries * (_edmd.n_samples_ic_ - 1),
expected[1],
),
)
self.assertIsInstance(actual_modes, pd.DataFrame)
self.assertIsInstance(actual_eigvals, pd.Series)
self.assertIsInstance(actual_eigfunc, TSCDataFrame)
def test_sort_koopman_triplets(self):
_edmd_wo_sort = EDMD(
dict_steps=[
("scale", TSCFeaturePreprocess.from_name(name="min-max")),
("delays", TSCTakensEmbedding(delays=10)),
("pca", TSCPrincipalComponent(n_components=6)),
],
dmd_model=DMDFull(is_diagonalize=True),
include_id_state=True,
).fit(X=self.multi_waves)
_edmd_w_sort = EDMD(
dict_steps=[
("scale", TSCFeaturePreprocess.from_name(name="min-max")),
("delays", TSCTakensEmbedding(delays=10)),
("pca", TSCPrincipalComponent(n_components=6)),
],
dmd_model=DMDFull(is_diagonalize=True),
sort_koopman_triplets=True,
include_id_state=True,
).fit(X=self.multi_waves)
expected = _edmd_wo_sort.reconstruct(self.multi_waves)
actual = _edmd_w_sort.reconstruct(self.multi_waves)
pdtest.assert_frame_equal(expected, actual)
def test_koopman_eigenfunction_eval(self):
_edmd = EDMD(
dict_steps=[
("scale", TSCFeaturePreprocess.from_name(name="min-max")),
("delays", TSCTakensEmbedding(delays=10)),
("pca", TSCPrincipalComponent(n_components=2)),
],
include_id_state=True,
).fit(X=self.multi_waves)
actual = _edmd.koopman_eigenfunction(
self.multi_waves.initial_states(_edmd.n_samples_ic_ + 1)
)
self.assertIsInstance(actual, TSCDataFrame)
actual = _edmd.koopman_eigenfunction(
self.multi_waves.initial_states(_edmd.n_samples_ic_)
)
self.assertIsInstance(actual, pd.DataFrame)
with self.assertRaises(TSCException):
_edmd.koopman_eigenfunction(
self.multi_waves.initial_states(_edmd.n_samples_ic_ - 1)
)
def test_dmap_kernels(self, plot=False):
X = self._setup_multi_sine_wave_data2()
from datafold.dynfold import DiffusionMaps
from datafold.pcfold import (
ConeKernel,
ContinuousNNKernel,
GaussianKernel,
InverseMultiquadricKernel,
MultiquadricKernel,
)
kernels = [
ConeKernel(zeta=0.0, epsilon=0.5),
ConeKernel(zeta=0.9, epsilon=0.1),
GaussianKernel(epsilon=0.1),
ContinuousNNKernel(k_neighbor=20, delta=0.9),
# MultiquadricKernel(epsilon=1),
InverseMultiquadricKernel(epsilon=0.5),
]
f, ax = plt.subplots(nrows=len(kernels) + 1, ncols=1, sharex=True)
X.plot(ax=ax[0])
ax[0].set_title("original data")
for i, kernel in enumerate(kernels):
try:
X_predict = EDMD(
[
("takens", TSCTakensEmbedding(delays=20)),
("dmap", DiffusionMaps(kernel, n_eigenpairs=220)),
]
).fit_predict(X)
X_predict.plot(ax=ax[i + 1])
ax[i + 1].set_title(f"kernel={kernel}")
except Exception as e:
print(f"kernel={kernel} failed")
raise e
if plot:
plt.show()
def test_attach_illegal_id(self):
_edmd = EDMD(
dict_steps=[
("id", TSCIdentity()),
],
include_id_state=True, # cannot be attached to identity (same feature names)
)
with self.assertRaises(ValueError):
_edmd.fit(X=self.sine_wave_tsc)
def test_edmd_dict_sine_wave(self, plot=False):
_edmd = EDMD(
dict_steps=[
("scale", TSCFeaturePreprocess.from_name(name="min-max")),
("delays", TSCTakensEmbedding(delays=10)),
("pca", TSCPrincipalComponent(n_components=2)),
]
)
forward_dict = _edmd.fit_transform(X=self.sine_wave_tsc)
self.assertIsInstance(forward_dict, TSCDataFrame)
inverse_dict = _edmd.inverse_transform(X=forward_dict)
self.assertIsInstance(inverse_dict, TSCDataFrame)
# index not the same because of Takens, so only check column
pdtest.assert_index_equal(
self.sine_wave_tsc.columns,
inverse_dict.columns,
)
diff = inverse_dict - self.sine_wave_tsc
# sort out the removed rows from Takens (NaN values)
self.assertTrue((diff.dropna() < 1e-14).all().all())
if plot:
ax = self.sine_wave_tsc.plot()
inverse_dict.plot(ax=ax)
from datafold.utils.plot import plot_eigenvalues
f, ax = plt.subplots()
plot_eigenvalues(eigenvalues=_edmd.dmd_model.eigenvalues_, ax=ax)
plt.show()
def test_edmd_dict_sine_wave_generator(self, plot=False):
# Use a DMD model that approximates the generator matrix and not the Koopman
# operator
_edmd = EDMD(
dict_steps=[
("scale", TSCFeaturePreprocess.from_name(name="min-max")),
("delays", TSCTakensEmbedding(delays=10)),
("pca", TSCPrincipalComponent(n_components=2)),
],
dmd_model=gDMDFull(),
)
forward_dict = _edmd.fit_transform(
X=self.sine_wave_tsc, **dict(dmd__store_generator_matrix=True)
)
self.assertIsInstance(forward_dict, TSCDataFrame)
inverse_dict = _edmd.inverse_transform(X=forward_dict)
self.assertIsInstance(inverse_dict, TSCDataFrame)
# index not the same because of Takens, so only check column
pdtest.assert_index_equal(
self.sine_wave_tsc.columns,
inverse_dict.columns,
)
diff = inverse_dict - self.sine_wave_tsc
# sort out the removed rows from Takens (NaN values)
self.assertTrue((diff.dropna() < 1e-14).all().all())
# test that the fit_param dmd__store_generator_matrix was really passed to the
# DMD model.
self.assertTrue(hasattr(_edmd.dmd_model, "generator_matrix_"))
if plot:
ax = self.sine_wave_tsc.plot()
inverse_dict.plot(ax=ax)
f, ax = plt.subplots()
plot_eigenvalues(eigenvalues=_edmd.dmd_model.eigenvalues_, ax=ax)
plt.show()
def test_spectral_and_matrix_mode(self):
_edmd_spectral = EDMD(
dict_steps=[
("scale", TSCFeaturePreprocess.from_name(name="min-max")),
("delays", TSCTakensEmbedding(delays=10)),
("pca", TSCPrincipalComponent(n_components=2)),
],
dmd_model=DMDFull(sys_mode="spectral"),
)
_edmd_matrix = EDMD(
dict_steps=[
("scale", TSCFeaturePreprocess.from_name(name="min-max")),
("delays", TSCTakensEmbedding(delays=10)),
("pca", TSCPrincipalComponent(n_components=2)),
],
dmd_model=DMDFull(sys_mode="matrix"),
)
actual_spectral = _edmd_spectral.fit_predict(X=self.sine_wave_tsc)
actual_matrix = _edmd_matrix.fit_predict(X=self.sine_wave_tsc)
pdtest.assert_frame_equal(actual_spectral, actual_matrix)
self.assertTrue(_edmd_spectral.koopman_modes is not None)
self.assertTrue(_edmd_spectral.dmd_model.is_spectral_mode())
self.assertTrue(_edmd_matrix.koopman_modes is None)
self.assertTrue(_edmd_matrix.dmd_model.is_matrix_mode())
# use qois argument
actual_spectral = _edmd_spectral.reconstruct(X=self.sine_wave_tsc, qois=["sin"])
actual_matrix = _edmd_matrix.reconstruct(X=self.sine_wave_tsc, qois=["sin"])
pdtest.assert_frame_equal(actual_spectral, actual_matrix)
def test_edmd_with_composed_dict(self, display_html=False, plot=False):
from sklearn.compose import make_column_selector
from datafold.dynfold import TSCColumnTransformer
selector_sin = make_column_selector(pattern="sin")
selector_cos = make_column_selector(pattern="cos")
separate_transform = TSCColumnTransformer(
transformers=[
("sin_path", TSCPrincipalComponent(n_components=10), selector_sin),
("cos_path", TSCPrincipalComponent(n_components=10), selector_cos),
]
)
edmd = EDMD(
dict_steps=[
("delays", TSCTakensEmbedding(delays=15)),
("pca", separate_transform),
],
include_id_state=True,
).fit(self.multi_waves)
if display_html:
with tempfile.NamedTemporaryFile("w", suffix=".html") as fp:
fp.write(estimator_html_repr(edmd))
fp.flush()
webbrowser.open_new_tab(fp.name)
input("Press Enter to continue...")
f, ax = plt.subplots(nrows=2, sharex=True)
self.multi_waves.plot(ax=ax[0])
edmd.reconstruct(self.multi_waves).plot(ax=ax[1])
if plot:
plt.show()
def test_edmd_sine_wave(self):
edmd = EDMD(
dict_steps=[
("delays", TSCTakensEmbedding(delays=10)),
("pca", TSCPrincipalComponent(n_components=5)),
],
include_id_state=True,
)
case_one_edmd = deepcopy(edmd)
case_two_edmd = deepcopy(edmd)
case_one = case_one_edmd.fit(self.multi_sine_wave_tsc).reconstruct(
self.multi_sine_wave_tsc
)
case_two = case_two_edmd.fit_predict(self.multi_sine_wave_tsc)
pdtest.assert_frame_equal(case_one, case_two)
def test_edmd_cv_sine_wave(self):
# Tests a specific setting of EDMDCV compared to sklearn.GridSearchCV,
# where the results are expected to be the same. EDMDCV generalizes aspects
# that fail for GridSearchCV
edmd = EDMD(
dict_steps=[
("delays", TSCTakensEmbedding(delays=10)),
("pca", TSCPrincipalComponent(n_components=5)),
],
include_id_state=False,
)
# NOTE: cv only TSCKfoldSeries can be compared and is equal to sklearn. not
# E.g. TSCKfoldTime requires to adapt the internal data (setting the time
# series correctly for the DMD model
sklearn_cv = GridSearchCV(
estimator=edmd,
param_grid={"pca__n_components": [5, 7]},
cv=TSCKfoldSeries(2),
verbose=False,
return_train_score=True,
n_jobs=None,
)
edmdcv = EDMDCV(
estimator=edmd,
param_grid={"pca__n_components": [5, 7]},
cv=TSCKfoldSeries(2),
verbose=False,
return_train_score=True,
n_jobs=None,
)
sklearn_cv.fit(self.multi_sine_wave_tsc)
edmdcv.fit(self.multi_sine_wave_tsc)
# timings are very unlikely to be the same, so drop them for the comparison:
drop_rows = {
"mean_fit_time",
"std_fit_time",
"mean_score_time",
"std_score_time",
}
expected_results = pd.DataFrame(sklearn_cv.cv_results_).T.drop(
labels=drop_rows, axis=0
)
actual_results = pd.DataFrame(edmdcv.cv_results_).T.drop(
labels=drop_rows, axis=0
)
pdtest.assert_frame_equal(expected_results, actual_results)
def test_edmdcv_fail_all(self):
edmd = EDMD(
dict_steps=[
("timedelay", TSCTakensEmbedding(delays=1)),
]
)
with warnings.catch_warnings():
warnings.filterwarnings("ignore") # suppress error messages
edmdcv = EDMDCV(
estimator=edmd,
param_grid={"timedelay__delays": [9999999]}, # raises error
cv=TSCKfoldSeries(2),
verbose=2,
refit=False,
return_train_score=True,
error_score=np.nan,
n_jobs=1,
).fit(self.multi_sine_wave_tsc)
df = | pd.DataFrame(edmdcv.cv_results_) | pandas.DataFrame |
from copy import (
copy,
deepcopy,
)
import numpy as np
import pytest
from pandas.core.dtypes.common import is_scalar
from pandas import (
DataFrame,
Series,
)
import pandas._testing as tm
# ----------------------------------------------------------------------
# Generic types test cases
def construct(box, shape, value=None, dtype=None, **kwargs):
"""
construct an object for the given shape
if value is specified use that if its a scalar
if value is an array, repeat it as needed
"""
if isinstance(shape, int):
shape = tuple([shape] * box._AXIS_LEN)
if value is not None:
if is_scalar(value):
if value == "empty":
arr = None
dtype = np.float64
# remove the info axis
kwargs.pop(box._info_axis_name, None)
else:
arr = np.empty(shape, dtype=dtype)
arr.fill(value)
else:
fshape = np.prod(shape)
arr = value.ravel()
new_shape = fshape / arr.shape[0]
if fshape % arr.shape[0] != 0:
raise Exception("invalid value passed in construct")
arr = np.repeat(arr, new_shape).reshape(shape)
else:
arr = np.random.randn(*shape)
return box(arr, dtype=dtype, **kwargs)
class Generic:
@pytest.mark.parametrize(
"func",
[
str.lower,
{x: x.lower() for x in list("ABCD")},
Series({x: x.lower() for x in list("ABCD")}),
],
)
def test_rename(self, frame_or_series, func):
# single axis
idx = list("ABCD")
for axis in frame_or_series._AXIS_ORDERS:
kwargs = {axis: idx}
obj = construct(4, **kwargs)
# rename a single axis
result = obj.rename(**{axis: func})
expected = obj.copy()
setattr(expected, axis, list("abcd"))
tm.assert_equal(result, expected)
def test_get_numeric_data(self, frame_or_series):
n = 4
kwargs = {
frame_or_series._get_axis_name(i): list(range(n))
for i in range(frame_or_series._AXIS_LEN)
}
# get the numeric data
o = construct(n, **kwargs)
result = o._get_numeric_data()
| tm.assert_equal(result, o) | pandas._testing.assert_equal |
from unittest.mock import Mock, patch
import pandas as pd
import pytest
from faker import Faker
from faker.config import DEFAULT_LOCALE
from rdt.transformers.numerical import NumericalTransformer
from sdv.constraints.base import Constraint
from sdv.constraints.errors import MissingConstraintColumnError
from sdv.errors import ConstraintsNotMetError
from sdv.metadata import Table
class TestTable:
def test__get_faker_default_locale(self):
"""Test that ``_get_faker`` without locales parameter has default locale.
The ``_get_faker`` should return a Faker object localized to the default locale.
When no locales are specified explicitly.
Input:
- Field metadata from metadata dict.
Output:
- Faker object with default localization.
"""
# Setup
metadata_dict = {
'fields': {
'foo': {
'type': 'categorical',
'pii': True,
'pii_category': 'company'
}
}
}
# Run
faker = Table.from_dict(metadata_dict)._get_faker(metadata_dict['fields']['foo'])
# Assert
assert isinstance(faker, Faker)
assert faker.locales == [DEFAULT_LOCALE]
def test__get_faker_specified_locales_string(self):
"""Test that ``_get_faker`` with locales parameter sets localization correctly.
The ``_get_faker`` should return a Faker object localized to the specified locale.
Input:
- Field metadata from metadata dict.
Output:
- Faker object with specified localization string.
"""
# Setup
metadata_dict = {
'fields': {
'foo': {
'type': 'categorical',
'pii': True,
'pii_category': 'company',
'pii_locales': 'sv_SE'
}
}
}
# Run
faker = Table.from_dict(metadata_dict)._get_faker(metadata_dict['fields']['foo'])
# Assert
assert isinstance(faker, Faker)
assert faker.locales == ['sv_SE']
def test__get_faker_specified_locales_list(self):
"""Test that ``_get_faker`` with locales parameter sets localization correctly.
The ``_get_faker`` should return a Faker object localized to the specified locales.
Input:
- Field metadata from metadata dict.
Output:
- Faker object with specified list of localizations.
"""
# Setup
metadata_dict = {
'fields': {
'foo': {
'type': 'categorical',
'pii': True,
'pii_category': 'company',
'pii_locales': ['en_US', 'sv_SE']
}
}
}
# Run
faker = Table.from_dict(metadata_dict)._get_faker(metadata_dict['fields']['foo'])
# Assert
assert isinstance(faker, Faker)
assert faker.locales == ['en_US', 'sv_SE']
def test__get_faker_method_pass_args(self):
"""Test that ``_get_faker_method`` method utilizes parameters passed in category argument.
The ``_get_faker_method`` method uses the parameters passed to it in the category argument.
Input:
- Faker object to create faked values with.
- Category tuple of category name and parameters passed to the method creating fake values.
Output:
- Fake values created with the specified method from the Faker object.
Utilizing the arguments given to it.
"""
# Setup
metadata_dict = {
'fields': {
'foo': {
'type': 'categorical',
'pii': True,
'pii_category': 'ean'
}
}
}
metadata = Table.from_dict(metadata_dict)
# Run
fake_8_ean = metadata._get_faker_method(Faker(), ('ean', 8))
ean_8 = fake_8_ean()
fake_13_ean = metadata._get_faker_method(Faker(), ('ean', 13))
ean_13 = fake_13_ean()
# Assert
assert len(ean_8) == 8
assert len(ean_13) == 13
@patch('sdv.metadata.Table')
def test__make_anonymization_mappings(self, mock_table):
"""Test that ``_make_anonymization_mappings`` creates the expected mappings.
The ``_make_anonymization_mappings`` method should map values in the original
data to fake values for non-id fields that are labeled pii.
Setup:
- Create a Table that has metadata about three fields (one pii field, one id field,
and one non-pii field).
Input:
- Data that contains a pii field, an id field, and a non-pii field.
Side Effects:
- Expect ``_get_fake_values`` to be called with the number of unique values of the
pii field.
- Expect the resulting `_ANONYMIZATION_MAPPINGS` field to contain the pii field, with
the correct number of mappings and keys.
"""
# Setup
metadata = Mock()
metadata._ANONYMIZATION_MAPPINGS = {}
foo_metadata = {
'type': 'categorical',
'pii': True,
'pii_category': 'email',
}
metadata._fields_metadata = {
'foo': foo_metadata,
'bar': {
'type': 'categorical',
},
'baz': {
'type': 'id',
}
}
foo_values = ['<EMAIL>', '<EMAIL>', '<EMAIL>']
data = pd.DataFrame({
'foo': foo_values,
'bar': ['a', 'b', 'c'],
'baz': [1, 2, 3],
})
# Run
Table._make_anonymization_mappings(metadata, data)
# Assert
assert mock_table._get_fake_values.called_once_with(foo_metadata, 3)
mappings = metadata._ANONYMIZATION_MAPPINGS[id(metadata)]
assert len(mappings) == 1
foo_mappings = mappings['foo']
assert len(foo_mappings) == 3
assert list(foo_mappings.keys()) == foo_values
@patch('sdv.metadata.Table')
def test__make_anonymization_mappings_unique_faked_value_in_field(self, mock_table):
"""Test that ``_make_anonymization_mappings`` method creates mappings for anonymized values.
The ``_make_anonymization_mappings`` method should map equal values in the original data
to the same faked value.
Input:
- DataFrame with a field that should be anonymized based on the metadata description.
Side Effect:
- Mappings are created from the original values to faked values.
"""
# Setup
metadata = Mock()
metadata._ANONYMIZATION_MAPPINGS = {}
foo_metadata = {
'type': 'categorical',
'pii': True,
'pii_category': 'email'
}
metadata._fields_metadata = {
'foo': foo_metadata
}
data = pd.DataFrame({
'foo': ['<EMAIL>', '<EMAIL>', '<EMAIL>']
})
# Run
Table._make_anonymization_mappings(metadata, data)
# Assert
assert mock_table._get_fake_values.called_once_with(foo_metadata, 2)
mappings = metadata._ANONYMIZATION_MAPPINGS[id(metadata)]
assert len(mappings) == 1
foo_mappings = mappings['foo']
assert len(foo_mappings) == 2
assert list(foo_mappings.keys()) == ['<EMAIL>', '<EMAIL>']
@patch.object(Constraint, 'from_dict')
def test__prepare_constraints_sorts_constraints(self, from_dict_mock):
"""Test that ``_prepare_constraints`` method sorts constraints.
The ``_prepare_constraints`` method should sort constraints by putting
constraints with ``rebuild_columns`` before the ones without them.
Input:
- list of constraints with some having ``rebuild_columns``
before constraints without them.
Output:
- List of constraints sorted properly.
"""
# Setup
constraint1 = Constraint(handling_strategy='transform')
constraint2 = Constraint(handling_strategy='transform')
constraint3 = Constraint(handling_strategy='reject_sampling')
constraints = [constraint1, constraint2, constraint3]
constraint1.rebuild_columns = ['a']
constraint2.rebuild_columns = ['b']
constraint3.rebuild_columns = []
from_dict_mock.side_effect = [constraint1, constraint2, constraint3]
# Run
sorted_constraints = Table._prepare_constraints(constraints)
# Asserts
assert sorted_constraints == [constraint3, constraint1, constraint2]
@patch.object(Constraint, 'from_dict')
def test__prepare_constraints_sorts_constraints_none_rebuild_columns(self, from_dict_mock):
"""Test that ``_prepare_constraints`` method sorts constraints.
The ``_prepare_constraints`` method should sort constraints with None as
``rebuild_columns`` before those that have them.
Input:
- list of constraints with some having None as ``rebuild_columns``
listed after those with ``rebuild_columns``.
Output:
- List of constraints sorted properly.
"""
# Setup
constraint1 = Constraint(handling_strategy='transform')
constraint2 = Constraint(handling_strategy='transform')
constraint3 = Constraint(handling_strategy='reject_sampling')
constraints = [constraint1, constraint2, constraint3]
constraint1.rebuild_columns = ['a']
constraint2.rebuild_columns = ['b']
constraint3.rebuild_columns = None
from_dict_mock.side_effect = [constraint1, constraint2, constraint3]
# Run
sorted_constraints = Table._prepare_constraints(constraints)
# Asserts
assert sorted_constraints == [constraint3, constraint1, constraint2]
@patch.object(Constraint, 'from_dict')
def test__prepare_constraints_validates_constraint_order(self, from_dict_mock):
"""Test the ``_prepare_constraints`` method validates the constraint order.
If no constraint has ``rebuild_columns`` that are in a later
constraint's ``constraint_columns``, no exception should be raised.
Input:
- List of constraints with none having ``rebuild_columns``
that are in a later constraint's ``constraint_columns``.
Output:
- Sorted list of constraints.
"""
# Setup
constraint1 = Constraint(handling_strategy='reject_sampling')
constraint2 = Constraint(handling_strategy='reject_sampling')
constraint3 = Constraint(handling_strategy='transform')
constraint4 = Constraint(handling_strategy='transform')
constraints = [constraint1, constraint2, constraint3, constraint4]
constraint3.rebuild_columns = ['e', 'd']
constraint4.constraint_columns = ['a', 'b', 'c']
constraint4.rebuild_columns = ['a']
from_dict_mock.side_effect = [constraint1, constraint2, constraint3, constraint4]
# Run
sorted_constraints = Table._prepare_constraints(constraints)
# Assert
assert sorted_constraints == constraints
@patch.object(Constraint, 'from_dict')
def test__prepare_constraints_invalid_order_raises_exception(self, from_dict_mock):
"""Test the ``_prepare_constraints`` method validates the constraint order.
If one constraint has ``rebuild_columns`` that are in a later
constraint's ``constraint_columns``, an exception should be raised.
Input:
- List of constraints with some having ``rebuild_columns``
that are in a later constraint's ``constraint_columns``.
Side Effect:
- Exception should be raised.
"""
# Setup
constraint1 = Constraint(handling_strategy='reject_sampling')
constraint2 = Constraint(handling_strategy='reject_sampling')
constraint3 = Constraint(handling_strategy='transform')
constraint4 = Constraint(handling_strategy='transform')
constraints = [constraint1, constraint2, constraint3, constraint4]
constraint3.rebuild_columns = ['a', 'd']
constraint4.constraint_columns = ['a', 'b', 'c']
constraint4.rebuild_columns = ['a']
from_dict_mock.side_effect = [constraint1, constraint2, constraint3, constraint4]
# Run
with pytest.raises(Exception):
Table._prepare_constraints(constraints)
@patch('sdv.metadata.table.rdt.transformers.NumericalTransformer',
spec_set=NumericalTransformer)
def test___init__(self, transformer_mock):
"""Test that ``__init__`` method passes parameters.
The ``__init__`` method should pass the custom parameters
to the ``NumericalTransformer``.
Input:
- rounding set to an int
- max_value set to an int
- min_value set to an int
Side Effects:
- ``NumericalTransformer`` should receive the correct parameters
"""
# Run
Table(rounding=-1, max_value=100, min_value=-50)
# Asserts
assert len(transformer_mock.mock_calls) == 2
transformer_mock.assert_any_call(
dtype=int, rounding=-1, max_value=100, min_value=-50)
transformer_mock.assert_any_call(
dtype=float, rounding=-1, max_value=100, min_value=-50)
@patch.object(Table, '_prepare_constraints')
def test___init__calls_prepare_constraints(self, _prepare_constraints_mock):
"""Test that ``__init__`` method calls ``_prepare_constraints"""
# Run
Table(constraints=[])
# Assert
_prepare_constraints_mock.called_once_with([])
def test__make_ids(self):
"""Test whether regex is correctly generating expressions."""
metadata = {'subtype': 'string', 'regex': '[a-d]'}
keys = Table._make_ids(metadata, 3)
assert (keys == pd.Series(['a', 'b', 'c'])).all()
def test__make_ids_fail(self):
"""Test if regex fails with more requested ids than available unique values."""
metadata = {'subtype': 'string', 'regex': '[a-d]'}
with pytest.raises(ValueError):
Table._make_ids(metadata, 20)
def test__make_ids_unique_field_not_unique(self):
"""Test that id column is replaced with all unique values if not already unique."""
metadata_dict = {
'fields': {
'item 0': {'type': 'id', 'subtype': 'integer'},
'item 1': {'type': 'boolean'}
},
'primary_key': 'item 0'
}
metadata = Table.from_dict(metadata_dict)
data = pd.DataFrame({
'item 0': [0, 1, 1, 2, 3, 5, 5, 6],
'item 1': [True, True, False, False, True, False, False, True]
})
new_data = metadata.make_ids_unique(data)
assert new_data['item 1'].equals(data['item 1'])
assert new_data['item 0'].is_unique
def test__make_ids_unique_field_already_unique(self):
"""Test that id column is kept if already unique."""
metadata_dict = {
'fields': {
'item 0': {'type': 'id', 'subtype': 'integer'},
'item 1': {'type': 'boolean'}
},
'primary_key': 'item 0'
}
metadata = Table.from_dict(metadata_dict)
data = pd.DataFrame({
'item 0': [9, 1, 8, 2, 3, 7, 5, 6],
'item 1': [True, True, False, False, True, False, False, True]
})
new_data = metadata.make_ids_unique(data)
assert new_data['item 1'].equals(data['item 1'])
assert new_data['item 0'].equals(data['item 0'])
def test__make_ids_unique_field_index_out_of_order(self):
"""Test that updated id column is unique even if index is out of order."""
metadata_dict = {
'fields': {
'item 0': {'type': 'id', 'subtype': 'integer'},
'item 1': {'type': 'boolean'}
},
'primary_key': 'item 0'
}
metadata = Table.from_dict(metadata_dict)
data = pd.DataFrame({
'item 0': [0, 1, 1, 2, 3, 5, 5, 6],
'item 1': [True, True, False, False, True, False, False, True]
}, index=[0, 1, 1, 2, 3, 5, 5, 6])
new_data = metadata.make_ids_unique(data)
assert new_data['item 1'].equals(data['item 1'])
assert new_data['item 0'].is_unique
def test_transform_calls__transform_constraints(self):
"""Test that the `transform` method calls `_transform_constraints` with right parameters
The ``transform`` method is expected to call the ``_transform_constraints`` method
with the data and correct value for ``on_missing_column``.
Input:
- Table data
Side Effects:
- Calls _transform_constraints
"""
# Setup
data = pd.DataFrame({
'item 0': [0, 1, 2],
'item 1': [True, True, False]
}, index=[0, 1, 2])
dtypes = {'item 0': 'int', 'item 1': 'bool'}
table_mock = Mock()
table_mock.get_dtypes.return_value = dtypes
table_mock._transform_constraints.return_value = data
table_mock._anonymize.return_value = data
table_mock._hyper_transformer.transform.return_value = data
# Run
Table.transform(table_mock, data, 'error')
# Assert
expected_data = pd.DataFrame({
'item 0': [0, 1, 2],
'item 1': [True, True, False]
}, index=[0, 1, 2])
mock_calls = table_mock._transform_constraints.mock_calls
args = mock_calls[0][1]
assert len(mock_calls) == 1
assert args[0].equals(expected_data)
assert args[1] == 'error'
def test__transform_constraints(self):
"""Test that method correctly transforms data based on constraints
The ``_transform_constraints`` method is expected to loop through constraints
and call each constraint's ``transform`` method on the data.
Input:
- Table data
Output:
- Transformed data
"""
# Setup
data = pd.DataFrame({
'item 0': [0, 1, 2],
'item 1': [3, 4, 5]
}, index=[0, 1, 2])
transformed_data = pd.DataFrame({
'item 0': [0, 0.5, 1],
'item 1': [6, 8, 10]
}, index=[0, 1, 2])
first_constraint_mock = Mock()
second_constraint_mock = Mock()
first_constraint_mock.transform.return_value = transformed_data
second_constraint_mock.return_value = transformed_data
table_mock = Mock()
table_mock._constraints = [first_constraint_mock, second_constraint_mock]
# Run
result = Table._transform_constraints(table_mock, data)
# Assert
assert result.equals(transformed_data)
first_constraint_mock.transform.assert_called_once_with(data)
second_constraint_mock.transform.assert_called_once_with(transformed_data)
def test__transform_constraints_raises_error(self):
"""Test that method raises error when specified.
The ``_transform_constraints`` method is expected to raise ``MissingConstraintColumnError``
if the constraint transform raises one and ``on_missing_column`` is set to error.
Input:
- Table data
Side Effects:
- MissingConstraintColumnError
"""
# Setup
data = pd.DataFrame({
'item 0': [0, 1, 2],
'item 1': [3, 4, 5]
}, index=[0, 1, 2])
constraint_mock = Mock()
constraint_mock.transform.side_effect = MissingConstraintColumnError
table_mock = Mock()
table_mock._constraints = [constraint_mock]
# Run/Assert
with pytest.raises(MissingConstraintColumnError):
Table._transform_constraints(table_mock, data, 'error')
def test__transform_constraints_drops_columns(self):
"""Test that method drops columns when specified.
The ``_transform_constraints`` method is expected to drop columns associated with
a constraint its transform raises a MissingConstraintColumnError and ``on_missing_column``
is set to drop.
Input:
- Table data
Output:
- Table with dropped columns
"""
# Setup
data = pd.DataFrame({
'item 0': [0, 1, 2],
'item 1': [3, 4, 5]
}, index=[0, 1, 2])
constraint_mock = Mock()
constraint_mock.transform.side_effect = MissingConstraintColumnError
constraint_mock.constraint_columns = ['item 0']
table_mock = Mock()
table_mock._constraints = [constraint_mock]
# Run
result = Table._transform_constraints(table_mock, data, 'drop')
# Assert
expected_result = pd.DataFrame({
'item 1': [3, 4, 5]
}, index=[0, 1, 2])
assert result.equals(expected_result)
def test__validate_data_on_constraints(self):
"""Test the ``Table._validate_data_on_constraints`` method.
Expect that the method returns True when the constraint columns are in the given data,
and the constraint.is_valid method returns True.
Input:
- Table data
Output:
- None
Side Effects:
- No error
"""
# Setup
data = pd.DataFrame({
'a': [0, 1, 2],
'b': [3, 4, 5]
}, index=[0, 1, 2])
constraint_mock = Mock()
constraint_mock.is_valid.return_value = pd.Series([True, True, True])
constraint_mock.constraint_columns = ['a', 'b']
table_mock = Mock()
table_mock._constraints = [constraint_mock]
# Run
result = Table._validate_data_on_constraints(table_mock, data)
# Assert
assert result is None
def test__validate_data_on_constraints_invalid_input(self):
"""Test the ``Table._validate_data_on_constraints`` method.
Expect that the method returns False when the constraint columns are in the given data,
and the constraint.is_valid method returns False for any row.
Input:
- Table data contains an invalid row
Output:
- None
Side Effects:
- A ConstraintsNotMetError is thrown
"""
# Setup
data = pd.DataFrame({
'a': [0, 1, 2],
'b': [3, 4, 5]
}, index=[0, 1, 2])
constraint_mock = Mock()
constraint_mock.is_valid.return_value = | pd.Series([True, False, True]) | pandas.Series |
import pymongo
from finviz.screener import Screener
import logging
#client = pymongo.MongoClient("mongodb://xin:<EMAIL>/myFirstDatabase?ssl=true&authSource=admin")
#db = client.test
#https://github.com/peerchemist/finta
#https://medium.com/automation-generation/algorithmically-detecting-and-trading-technical-chart-patterns-with-python-c577b3a396ed
#https://github.com/twopirllc/pandas-ta#trend-15
#https://stackoverflow.com/questions/53965948/combine-aiohttp-with-multiprocessing
#fix price update making sure the price history match
#update a way to put more weight on a certain stock over another based on rsi more buying and selling
#add in timer for fail login in
from webull import paper_webull, endpoints # for real money trading, just import 'webull' instead
from webull.streamconn import StreamConn
import json
import trendln
import matplotlib
import matplotlib.pyplot as plt
import matplotlib.dates as mdates
import numpy as np
import pandas as pd
from datetime import datetime,timedelta
import sched
import time
import traceback
import yfinance as yf
from collections import defaultdict
from operator import attrgetter
from pandas_datareader import data as pdr
from moving import *
from finta import TA
import logging
from util import *
import traceback
#need day strat and daily strat
class Trades:
starting = 1000
instances = []
stock_count = 0
def __init__(self, stock_ticker, wb, id_value, cost, buy=0, sell=0, profitsl=None, realtime=False, islimit=False, finished=False):
self.wb = wb
self.stock_ticker=stock_ticker
self.profitsl = profitsl
self.id_value = id_value
self.cost = cost
self.buy = buy
self.sell = sell
self.realtime = realtime
self.islimit =islimit #checking if its limit sell
self.finished = finished #if positions has been sold
if realtime:
acc_data =self.wb.get_account()
Trades.starting = float(acc_data["accountMembers"][1]["value"])
Trades.stock_count = self.find_positions(acc_data["positions"])
self.sell_stock()
self.buy_stock()
else:
self.process_hist_buysell()
Trades.instances.append(self)
def process_hist_buysell(self):
if Trades.starting >= (self.cost*self.buy):#make sure enough money to buy
Trades.stock_count += self.buy #buy stock amount
Trades.starting -= (self.cost*self.buy)
else:
Trades.stock_count += (Trades.starting / self.cost)
Trades.starting = 0
if Trades.stock_count >= self.sell: #make sure enough stock to sell
Trades.stock_count -= self.sell
Trades.starting += (self.cost*self.sell)
else:
Trades.starting += (Trades.stock_count * self.cost)
Trades.stock_count = 0
def sell_stock(self): #given amount to buy and sell update the self variables
#self.wb.get_trade_token('19<PASSWORD>')
if (not Trades.stock_count) or (not self.sell):
print("No stocks to sell")
return
self.wb.get_trade_token('199694')
if Trades.stock_count >= self.sell: #make sure enough stock to sell
sucess_dict = self.wb.place_order(stock=self.stock_ticker, tId=None, action='SELL', orderType='MKT', enforce='DAY', quant=self.sell)
print(f"Im selling ${self.cost*self.sell} of {self.stock_ticker} at {self.cost} {self.sell} stocks Enough")
print(f"Finshed selling {self.stock_ticker}... {sucess_dict}")
Trades.stock_count -= self.sell
Trades.starting += (self.cost*self.sell)
else:
sucess_dict = self.wb.place_order(stock=self.stock_ticker, tId=None, action='SELL', orderType='MKT', enforce='DAY', quant=Trades.stock_count)
print(f"Im selling ${self.cost*Trades.stock_count} of {self.stock_ticker} at {self.cost} {Trades.stock_count} stocks not Enough")
print(f"Finshed selling {self.stock_ticker}... {sucess_dict}")
Trades.starting += (Trades.stock_count * self.cost)
Trades.stock_count = 0
def buy_stock(self):
if (not Trades.starting) or (not self.buy):
print("No money to buy")
return
self.wb.get_trade_token('<PASSWORD>')
if Trades.starting >= (self.cost*self.buy):#make sure enough money to buy
sucess_dict = self.wb.place_order(stock=self.stock_ticker, tId=None, action='BUY', orderType='MKT', enforce='DAY', quant=self.buy)
print(f"Im buying ${self.cost*self.buy} of {self.stock_ticker} at {self.cost} {self.buy} money Enough")
print(f"Finshed Buying {self.stock_ticker}...{sucess_dict}")
Trades.stock_count += self.buy #buy stock amount
Trades.starting -= (self.cost*self.buy)
else:
sucess_dict = self.wb.place_order(stock=self.stock_ticker, tId=None, action='BUY', orderType='MKT', enforce='DAY', quant=Trades.starting/self.cost)
print(f"Im buying ${Trades.starting} of {self.stock_ticker} at {self.cost} {Trades.starting/self.cost} money not Enough")
print(f"Finshed Buying {self.stock_ticker}... {sucess_dict}")
Trades.stock_count += (Trades.starting / self.cost)
Trades.starting = 0
def find_positions(self, position_data):
for position in position_data:
if position["ticker"]["symbol"] ==self.stock_ticker:
return float(position["position"])
return 0
class ProfitSL(object):
def __init__(self, profit=None, sl=None):
self.profit=profit
self.sl = sl
def __str__(self):
return f"profit: {self.profit} stopl: {self.sl}"
class PatternsandCandles(object): #this is use to determine enter and exit based on patterns and candles
def __init__(self):
self.buy = ["double bottom", "inverse head and shoulder"]
self.sell = ["double top", "head and shoulder"]
#it should have a probability for each of the me
#take in the current data,
#try multiple groups, increament the window size on one end
#1min -29
#30 - 59
#1h - 24
#1day-6day
#
self.smoothing = 5
self.window_range = 5
def double_bottom(self):
pass
#this needs to have acesss to ma object and df
def swing_low_price(self, row_value, number_up=2, period=60):
temp_focus = self.history_df.index.get_loc(row_value.name)
if temp_focus - period <0:
temp_focus=60
lookback = self.history_df.iloc[temp_focus-period:temp_focus+1, :]
max_min = self.get_max_min(lookback, target="min")
if max_min.empty:
return row_value.low, row_value.name
return max_min.iloc[-1], lookback.index[max_min.index[-1]]
def IHS(self):
pass
def get_max_min(self, hist, smoothing=None, window_range=None, target="both"):
if not smoothing:
smoothing =self.smoothing
if not window_range:
window_range = self.window_range
index_name = hist.index.name
smooth_prices = hist['close'].rolling(window=smoothing).mean().dropna()
local_max = argrelextrema(smooth_prices.values, np.greater)[0]
local_min = argrelextrema(smooth_prices.values, np.less)[0]
price_local_max_dt = []
for i in local_max:
if (i>window_range) and (i<len(hist)-window_range):
price_local_max_dt.append(hist.iloc[i-window_range:i+window_range]['close'].idxmax())
price_local_min_dt = []
for i in local_min:
if (i>window_range) and (i<len(hist)-window_range):
price_local_min_dt.append(hist.iloc[i-window_range:i+window_range]['close'].idxmin())
if target == "max":
max_min = pd.DataFrame(hist.loc[price_local_max_dt])
elif target == "min":
max_min = pd.DataFrame(hist.loc[price_local_min_dt])
else:
max_min = pd.concat([pd.DataFrame(hist.loc[price_local_max_dt]), pd.DataFrame(hist.loc[price_local_min_dt])]).sort_index()
max_min.index.name = 'date'
max_min = max_min.reset_index()
max_min = max_min[~max_min.date.duplicated()]
p = hist.reset_index()
max_min['day_num'] = p[p[index_name].isin(max_min.date)].index.values
max_min = max_min.set_index('day_num')['close']
#print(max_min)
#hist.reset_index()["close"].plot()
#plt.plot(max_min.index, max_min.values, "o")
#plt.show()
return max_min
def find_patterns(self, max_min):
patterns = defaultdict(list)
letter_locations = {}
# Window range is 5 units
for i in range(5, len(max_min)):
window = max_min.iloc[i-5:i]
# Pattern must play out in less than n units
if window.index[-1] - window.index[0] > 100:
continue
a, b, c, d, e = window.iloc[0:5]
# IHS
if a<b and c<a and c<e and c<d and e<d and abs(b-d)<=np.mean([b,d])*0.02 and abs(a-e)<=np.mean([a,e])*0.000001:
patterns['IHS'].append((window.index[0], window.index[-1]))
letter_locations[(window.index[0], window.index[-1])] = [a,b,c,d,e]
#print(letter_locations)
return patterns, letter_locations
def plot_minmax_patterns(self, max_min, patterns, stock, window, ema, letter_locations):
incr = str((self.hist.index[1] - self.hist.index[0]).seconds/60)
if len(patterns) == 0:
pass
else:
num_pat = len([x for x in patterns.items()][0][1])
f, axes = plt.subplots(1, 2, figsize=(16, 5))
axes = axes.flatten()
prices_ = self.hist.reset_index()['close']
axes[0].plot(prices_)
axes[0].scatter(max_min.index, max_min, s=100, alpha=.3, color='orange')
axes[1].plot(prices_)
for name, end_day_nums in patterns.items():
for i, tup in enumerate(end_day_nums):
sd = tup[0]
ed = tup[1]
axes[1].scatter(max_min.loc[sd:ed].index,
max_min.loc[sd:ed].values,
s=200, alpha=.3)
plt.yticks([])
plt.tight_layout()
plt.title('{}: {}: EMA {}, Window {} ({} patterns)'.format(stock, incr, ema, window, num_pat))
def get_results(self, max_min, pat, stock, ema_, window_):
incr = str((self.hist.index[1] - self.hist.index[0]).seconds/60)
#fw_list = [1, 12, 24, 36]
fw_list = [1, 2, 3]
results = []
if len(pat.items()) > 0:
end_dates = [v for k, v in pat.items()][0]
for date in end_dates:
param_res = {'stock': stock,
'increment': incr,
'ema': ema_,
'window': window_,
'date': date}
for x in fw_list:
returns = (self.hist['close'].pct_change(x).shift(-x).reset_index(drop=True).dropna())
try:
param_res['fw_ret_{}'.format(x)] = returns.loc[date[1]]
except Exception as e:
param_res['fw_ret_{}'.format(x)] = e
results.append(param_res)
else:
param_res = {'stock': stock,
'increment': incr,
'ema': ema_,
'window': window_,
'date': None}
for x in fw_list:
param_res['fw_ret_{}'.format(x)] = None
results.append(param_res)
return pd.DataFrame(results)
def screener(self, stock_data, ema_list, window_list, plot, results=False):
all_results = pd.DataFrame()
for stock in stock_data:
#prices = stock_data[stock]
yahoo_tick = yf.Ticker(stock)
self.hist = yahoo_tick.history(period=f"60d", interval="30m")
for ema_ in ema_list:
for window_ in window_list:
max_min = self.get_max_min(smoothing=ema_, window_range=window_)
pat,location = self.find_patterns(max_min)
if plot == True:
self.plot_minmax_patterns(max_min, pat, stock, window_, ema_, location)
if results == True:
all_results = pd.concat([all_results, self.get_results(max_min, pat, stock, ema_, window_)], axis=0)
if results == True:
return all_results.reset_index(drop=True)
class Strat(PatternsandCandles): #main object for main functions to apply sell and buy #should determine the peroid for computing sell and buy
def __init__(self, ticker, wb=None, extendTrading=0, backtest=False):
super(Strat, self).__init__()
self.ticker=ticker
self.starting_time = datetime.now().replace(hour=9, minute=30, second=0, microsecond=0)
self.ending_time = datetime.now().replace(hour=16, minute=0, second=0, microsecond=0)
#self.stock_count = 0 #when buy increase this
self.wb = wb
self.backtest = backtest
self.yahoo_tick = yf.Ticker(self.ticker)
self.ma = MA(self.yahoo_tick)
self.extendTrading=extendTrading
self.timeouttick = 0
self.timeoutmax = 200
self.required_period = None #this df is required for each strat as the dafult dict to perform analysis on. if its none then the stock can be used on real time data
self.required_timestamp = None
self.buy_count=0
self.sell_count=0
def process_df(self, hist, realtime=False):
#modi to hist
hist["buy"] = 0
hist["sell"] = 0
hist["Bottom"] = np.where(hist["close"] >= hist["open"], hist["open"], hist["close"])
hist["Top"] = np.where(hist["close"] >= hist["open"], hist["close"], hist["open"])
return hist
def preprocess(self, period, timestamp, count, local_csv=None):
#called from the plot main function to setup var before running
print(f"Running preprocess....")
if not self.required_period:
self.required_period = period
if not self.required_timestamp:
self.required_timestamp = timestamp
if local_csv:
#for when you have a static csv file set in place this will be set as the global df
self.history_df = | pd.read_csv(local_csv, index_col="timestamp") | pandas.read_csv |
from flask import g
from libs.extensions import Extension
import pandas as pd
from settings.gdxf.extensions.wxzb.base_sql_map import get_base_sql_map
from utils.qh_processor import get_qh_level
DEBUG = False
def get_sql_map(flag):
if flag:
base_sql_map = {
"1-1-全": "1_1",
}
else:
base_sql_map = get_base_sql_map()
sql_map = {
# 区划分布 --> 全部 --> 总体 【一个数】 省级 需求和
"wxzb_qh_all_xfbm_shej": (
f"select sum(a.wxsl)/sum(a.zs) as wxzb from {base_sql_map['1-1-全']} as a",
["wxzb"]),
# 区划分布 --> 全部 --> 总体 【一个数】 市级
"wxzb_qh_all_xfbm_shij": (
f"select a.wxsl/a.zs as wxzb from {base_sql_map['1-1-全']} as a where a.region_name='{{Cqh}}'",
["wxzb"]),
# 区划分布 --> 全部 --> 总体 【一个数】 县级
"wxzb_qh_all_xfbm_xj": (
f"select a.wxsl/a.zs as wxzb from {base_sql_map['1-2-全']} as a where a.region_name='{{Cqh}}'",
["wxzb"]),
# 区划分布 --> 全部 --> 地图 省级
"wxzb_qh_allshej": (
f"select a.region_name as qh,a.wxsl/a.zs as wxzb from {base_sql_map['1-1-全']} as a",
["qh", "wxzb"]),
# 区划分布 --> 全部 --> 地图 市级
"wxzb_qh_allshij": (
f"select a.region_name as qh,a.wxsl/a.zs as wxzb from {base_sql_map['1-2-全']} as a",
["qh", "wxzb"]),
# 信访部门 -->全部 --> 网信占比 【一个数】 用上面的
# "wxzb_xfbm_all": (
# f"select a.wxsl/a.zs as wxzb from {base_sql_map['1-1-全']} as a where a.region_name='{{Cqh}}'",
# ["wxzb"]),
# 信访部门 -->全部 --> 饼图 省级
"wxzb_xfbm_shejnums": (
f"SELECT case a.wxsl when a.wxsl then '网信' end as wxqk,sum(a.wxsl) as 'xfjc' FROM {base_sql_map['1-1-全']} as a UNION SELECT case a.wxsl when a.wxsl then '其他' end as wxqk ,(sum(a.zs) - sum(a.wxsl)) as 'xfjc' FROM {base_sql_map['1-1-全']} as a",
["wxqk", "xfjc"]),
# 信访部门 -->全部 --> 饼图 市级
"wxzb_xfbm_shijnums": (
f"SELECT case a.wxsl when a.wxsl then '网信' end as wxqk,a.wxsl as 'xfjc' FROM {base_sql_map['1-1-全']} as a WHERE a.region_name='{{Cqh}}' UNION SELECT case a.wxsl when a.wxsl then '其他' end as wxqk ,(a.zs - a.wxsl) as 'xfjc' FROM {base_sql_map['1-1-全']} as a WHERE a.region_name='{{Cqh}}'",
["wxqk", "xfjc"]),
# 信访部门 -->全部 --> 饼图 县级 待完成
"wxzb_xfbm_xjnums": (
f"SELECT case a.wxsl when a.wxsl then '网信' end as wxqk,a.wxsl as 'xfjc' FROM {base_sql_map['1-2-全']} as a WHERE a.region_name='{{Cqh}}' UNION SELECT case a.wxsl when a.wxsl then '其他' end as wxqk ,(a.zs - a.wxsl) as 'xfjc' FROM {base_sql_map['1-2-全']} as a WHERE a.region_name='{{Cqh}}'",
["wxqk", "xfjc"]),
}
return sql_map
class Wxzb(Extension):
"""网信占比"""
"""
计算公式:使用已提供的SQL语句进行计算
"""
def __init__(self, apis_copy, apis, *args, **kwargs):
self.sql, self.columns = None, None
super(Wxzb, self).__init__(apis_copy, apis) # 执行父类方法,获得self.apis/self.apis_copy
def before_search(self):
self.df = | pd.DataFrame([None], columns=["wxzb"]) | pandas.DataFrame |
# -*- coding: utf-8 -*-
try:
import json
except ImportError:
import simplejson as json
import math
import pytz
import locale
import pytest
import time
import datetime
import calendar
import re
import decimal
import dateutil
from functools import partial
from pandas.compat import range, StringIO, u
from pandas._libs.tslib import Timestamp
import pandas._libs.json as ujson
import pandas.compat as compat
import numpy as np
from pandas import DataFrame, Series, Index, NaT, DatetimeIndex, date_range
import pandas.util.testing as tm
json_unicode = (json.dumps if compat.PY3
else partial(json.dumps, encoding="utf-8"))
def _clean_dict(d):
"""
Sanitize dictionary for JSON by converting all keys to strings.
Parameters
----------
d : dict
The dictionary to convert.
Returns
-------
cleaned_dict : dict
"""
return {str(k): v for k, v in compat.iteritems(d)}
@pytest.fixture(params=[
None, # Column indexed by default.
"split",
"records",
"values",
"index"])
def orient(request):
return request.param
@pytest.fixture(params=[None, True])
def numpy(request):
return request.param
class TestUltraJSONTests(object):
@pytest.mark.skipif(compat.is_platform_32bit(),
reason="not compliant on 32-bit, xref #15865")
def test_encode_decimal(self):
sut = decimal.Decimal("1337.1337")
encoded = ujson.encode(sut, double_precision=15)
decoded = ujson.decode(encoded)
assert decoded == 1337.1337
sut = decimal.Decimal("0.95")
encoded = ujson.encode(sut, double_precision=1)
assert encoded == "1.0"
decoded = ujson.decode(encoded)
assert decoded == 1.0
sut = decimal.Decimal("0.94")
encoded = ujson.encode(sut, double_precision=1)
assert encoded == "0.9"
decoded = ujson.decode(encoded)
assert decoded == 0.9
sut = decimal.Decimal("1.95")
encoded = ujson.encode(sut, double_precision=1)
assert encoded == "2.0"
decoded = ujson.decode(encoded)
assert decoded == 2.0
sut = decimal.Decimal("-1.95")
encoded = ujson.encode(sut, double_precision=1)
assert encoded == "-2.0"
decoded = ujson.decode(encoded)
assert decoded == -2.0
sut = decimal.Decimal("0.995")
encoded = ujson.encode(sut, double_precision=2)
assert encoded == "1.0"
decoded = ujson.decode(encoded)
assert decoded == 1.0
sut = decimal.Decimal("0.9995")
encoded = ujson.encode(sut, double_precision=3)
assert encoded == "1.0"
decoded = ujson.decode(encoded)
assert decoded == 1.0
sut = decimal.Decimal("0.99999999999999944")
encoded = ujson.encode(sut, double_precision=15)
assert encoded == "1.0"
decoded = ujson.decode(encoded)
assert decoded == 1.0
@pytest.mark.parametrize("ensure_ascii", [True, False])
def test_encode_string_conversion(self, ensure_ascii):
string_input = "A string \\ / \b \f \n \r \t </script> &"
not_html_encoded = ('"A string \\\\ \\/ \\b \\f \\n '
'\\r \\t <\\/script> &"')
html_encoded = ('"A string \\\\ \\/ \\b \\f \\n \\r \\t '
'\\u003c\\/script\\u003e \\u0026"')
def helper(expected_output, **encode_kwargs):
output = ujson.encode(string_input,
ensure_ascii=ensure_ascii,
**encode_kwargs)
assert output == expected_output
assert string_input == json.loads(output)
assert string_input == ujson.decode(output)
# Default behavior assumes encode_html_chars=False.
helper(not_html_encoded)
# Make sure explicit encode_html_chars=False works.
helper(not_html_encoded, encode_html_chars=False)
# Make sure explicit encode_html_chars=True does the encoding.
helper(html_encoded, encode_html_chars=True)
@pytest.mark.parametrize("long_number", [
-4342969734183514, -12345678901234.56789012, -528656961.4399388
])
def test_double_long_numbers(self, long_number):
sut = {u("a"): long_number}
encoded = ujson.encode(sut, double_precision=15)
decoded = ujson.decode(encoded)
assert sut == decoded
def test_encode_non_c_locale(self):
lc_category = locale.LC_NUMERIC
# We just need one of these locales to work.
for new_locale in ("it_IT.UTF-8", "Italian_Italy"):
if tm.can_set_locale(new_locale, lc_category):
with tm.set_locale(new_locale, lc_category):
assert ujson.loads(ujson.dumps(4.78e60)) == 4.78e60
assert ujson.loads("4.78", precise_float=True) == 4.78
break
def test_decimal_decode_test_precise(self):
sut = {u("a"): 4.56}
encoded = ujson.encode(sut)
decoded = ujson.decode(encoded, precise_float=True)
assert sut == decoded
@pytest.mark.skipif(compat.is_platform_windows() and not compat.PY3,
reason="buggy on win-64 for py2")
def test_encode_double_tiny_exponential(self):
num = 1e-40
assert num == ujson.decode(ujson.encode(num))
num = 1e-100
assert num == ujson.decode(ujson.encode(num))
num = -1e-45
assert num == ujson.decode(ujson.encode(num))
num = -1e-145
assert np.allclose(num, ujson.decode(ujson.encode(num)))
@pytest.mark.parametrize("unicode_key", [
u("key1"), u("بن")
])
def test_encode_dict_with_unicode_keys(self, unicode_key):
unicode_dict = {unicode_key: u("value1")}
assert unicode_dict == ujson.decode(ujson.encode(unicode_dict))
@pytest.mark.parametrize("double_input", [
math.pi,
-math.pi # Should work with negatives too.
])
def test_encode_double_conversion(self, double_input):
output = ujson.encode(double_input)
assert round(double_input, 5) == round(json.loads(output), 5)
assert round(double_input, 5) == round(ujson.decode(output), 5)
def test_encode_with_decimal(self):
decimal_input = 1.0
output = ujson.encode(decimal_input)
assert output == "1.0"
def test_encode_array_of_nested_arrays(self):
nested_input = [[[[]]]] * 20
output = ujson.encode(nested_input)
assert nested_input == json.loads(output)
assert nested_input == ujson.decode(output)
nested_input = np.array(nested_input)
tm.assert_numpy_array_equal(nested_input, ujson.decode(
output, numpy=True, dtype=nested_input.dtype))
def test_encode_array_of_doubles(self):
doubles_input = [31337.31337, 31337.31337,
31337.31337, 31337.31337] * 10
output = ujson.encode(doubles_input)
assert doubles_input == json.loads(output)
assert doubles_input == ujson.decode(output)
tm.assert_numpy_array_equal(np.array(doubles_input),
ujson.decode(output, numpy=True))
def test_double_precision(self):
double_input = 30.012345678901234
output = ujson.encode(double_input, double_precision=15)
assert double_input == json.loads(output)
assert double_input == ujson.decode(output)
for double_precision in (3, 9):
output = ujson.encode(double_input,
double_precision=double_precision)
rounded_input = round(double_input, double_precision)
assert rounded_input == json.loads(output)
assert rounded_input == ujson.decode(output)
@pytest.mark.parametrize("invalid_val", [
20, -1, "9", None
])
def test_invalid_double_precision(self, invalid_val):
double_input = 30.12345678901234567890
expected_exception = (ValueError if isinstance(invalid_val, int)
else TypeError)
with pytest.raises(expected_exception):
ujson.encode(double_input, double_precision=invalid_val)
def test_encode_string_conversion2(self):
string_input = "A string \\ / \b \f \n \r \t"
output = ujson.encode(string_input)
assert string_input == json.loads(output)
assert string_input == ujson.decode(output)
assert output == '"A string \\\\ \\/ \\b \\f \\n \\r \\t"'
@pytest.mark.parametrize("unicode_input", [
"Räksmörgås اسامة بن محمد بن عوض بن لادن",
"\xe6\x97\xa5\xd1\x88"
])
def test_encode_unicode_conversion(self, unicode_input):
enc = ujson.encode(unicode_input)
dec = ujson.decode(enc)
assert enc == json_unicode(unicode_input)
assert dec == json.loads(enc)
def test_encode_control_escaping(self):
escaped_input = "\x19"
enc = | ujson.encode(escaped_input) | pandas._libs.json.encode |
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
# http://www.apache.org/licenses/LICENSE-2.0
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import pandas as pd
import numpy as np
import webbrowser
import warnings
from .file_download import update_index
from .file_tools import validate_version, get_version_files_paths
from .dataframe_tools import add_index_levels
from .exceptions import *
class DataSet:
"""
Note that all cancer datasets are class objects that inherit from cptac.dataset. Therefore
the same function calls exist for cptac.Endometrial, cptac.Colon, etc.
"""
def __init__(self, cancer_type, version, valid_versions, data_files):
"""Initialize variables for a DataSet object.
Parameters:
cancer_type (str): The cancer type requested for this dataset
version (str): The version number requested for this dataset
valid_versions (list of str): A list of all possible valid versions for this dataset
data_files (dict, keys of str, values of list of str): A dictionary where the keys are the existing version of the dataset, and the values are lists of the data file names for that version.
"""
# Initialize the _cancer_type instance variable
self._cancer_type = cancer_type.lower()
# Update the index, if possible. If there's no internet, that's fine.
try:
update_index(self._cancer_type)
except NoInternetError:
pass
# Validate the version
self._version = validate_version(version, self._cancer_type, use_context="init", valid_versions=valid_versions)
# Get the paths to the data files
version_data_files = data_files[self._version] # Get the data files for this version from the data files dictionary
self._data_files_paths = get_version_files_paths(self._cancer_type, self._version, version_data_files)
# Initialize dataframe and definitions dicts as empty for this parent class
self._data = {}
self._definitions = {}
# Assign the valid dfs lists, but make them instance variables so they're easy to override if needed
# These are the omics dataframes that are valid for use in the utilities functions
self._valid_omics_dfs = [
'acetylproteomics',
'circular_RNA',
'CNV',
'lincRNA',
'lipidomics',
'metabolomics',
'miRNA',
'phosphoproteomics',
'phosphoproteomics_gene',
'proteomics',
'somatic_mutation_binary',
'transcriptomics',
]
# These are the metadata dataframes that are valid for use in the utilities functions
self._valid_metadata_dfs = [
"clinical",
"derived_molecular",
"experimental_design",
#"followup", # Right now there are duplicate rows, so don't include follow up tables for joins.
] # We don't allow the treatment df, as in Ovarian, or medical_history df, as in Ccrcc, because they both have multiple rows for each sample.
# Methods to get metadata dataframes
def get_clinical(self):
"""Get the clinical dataframe."""
return self._get_dataframe("clinical")
def get_derived_molecular(self):
"""Get the derived_molecular dataframe."""
return self._get_dataframe("derived_molecular")
def get_experimental_design(self):
"""Get the experimental_design dataframe."""
return self._get_dataframe("experimental_design")
def get_medical_history(self):
"""Get the medical_history dataframe."""
return self._get_dataframe("medical_history")
def get_treatment(self):
"""Get the treatment dataframe."""
return self._get_dataframe("treatment")
def get_followup(self):
"""Get the followup dataframe."""
return self._get_dataframe("followup")
# Methods to get omics dataframes
def get_acetylproteomics(self):
"""Get the acetylproteomics dataframe."""
return self._get_dataframe("acetylproteomics")
def get_circular_RNA(self):
"""Get the circular_RNA dataframe."""
return self._get_dataframe("circular_RNA")
def get_CNV(self):
"""Get the CNV dataframe."""
return self._get_dataframe("CNV")
def get_lincRNA(self):
"""Get the lincRNA dataframe."""
return self._get_dataframe("lincRNA")
def get_lipidomics(self):
"""Get the lipidomics dataframe."""
return self._get_dataframe("lipidomics")
def get_metabolomics(self):
"""Get the metabolomics dataframe."""
return self._get_dataframe("metabolomics")
def get_methylation(self):
"""Get the methylation dataframe."""
return self._get_dataframe("methylation")
def get_miRNA(self):
"""Get the miRNA dataframe."""
return self._get_dataframe("miRNA")
def get_phosphoproteomics(self):
"""Get the phosphoproteomics dataframe."""
return self._get_dataframe("phosphoproteomics")
def get_phosphoproteomics_gene(self):
"""Get the phosphoproteomics_gene dataframe. The gene level phosphorylation measurement is an aggregate metric which potentially averages together individual measurements of different sites. Use get_phosphoproteomics() to view the data for individual sites."""
return self._get_dataframe("phosphoproteomics_gene")
def get_phosphosites(self, genes):
"""Returns dataframe with all phosphosites of specified gene or list of genes.
Parameters:
genes (str, or list or array-like of str): gene or list of genes to use to select phosphosites. str if single, list or array-like of str if multiple.
Returns:
pandas DataFrame: The phosphoproteomics for the specified gene(s).
"""
return self._get_omics_cols("phosphoproteomics", genes)
def get_proteomics(self):
"""Get the proteomics dataframe."""
return self._get_dataframe("proteomics")
def get_transcriptomics(self):
"""Get the transcriptomics dataframe."""
return self._get_dataframe("transcriptomics")
# Methods to get mutations dataframes
def get_gene_fusion(self):
"""Get the gene_fusion dataframe."""
return self._get_dataframe("gene_fusion")
def get_somatic_mutation(self):
"""Get the somatic_mutation dataframe."""
return self._get_dataframe("somatic_mutation")
def get_somatic_mutation_binary(self):
"""Get the somatic_mutation_binary dataframe, which has a binary value indicating, for each location on each gene, whether there was a mutation in that gene at that location, for each sample."""
return self._get_dataframe("somatic_mutation_binary")
# Help methods
def define(self, term):
"""Print the definition a term, if it is in the dataset's list of definitions.
Parameters:
term (str): term to be defined
Returns: None
"""
if len(self._definitions.keys()) == 0:
raise NoDefinitionsError("No definitions provided for this dataset.")
elif term in self._definitions.keys():
print(self._definitions[term])
else:
raise InvalidParameterError("{} not found in definitions. Check capitalization. Alternatively, the dataset's 'search(<your term>)' method can be used to perform a web search of the term provided.".format(term))
def get_cancer_type(self):
"""Return the cancer type for this dataset, as a string."""
return self._cancer_type
def version(self):
"""Return the dataset version of this instance, as a string."""
return self._version
def how_to_cite(self):
"""Print instructions for citing the data."""
print('Please include the following statement in publications using data accessed through this module:\n"Data used in this publication were generated by the Clinical Proteomic Tumor Analysis Consortium (NCI/NIH, <https://proteomics.cancer.gov/programs/cptac/>). Data were accessed through the Python module cptac, available at <https://pypi.org/project/cptac/>."')
def list_data(self):
"""Print list of loaded dataframes and dimensions."""
print("Below are the dataframes contained in this dataset:")
for name in sorted(self._data.keys(), key=str.lower):
df = self._data[name]
print("\t{}\n\t\tDimensions: {}".format(name, df.shape))
def list_definitions(self):
"""Print all terms defined in the dataset's list of definitions."""
if len(self._definitions.keys()) > 0:
for term in sorted(self._definitions.keys(), key=str.lower):
print(term)
else:
raise NoDefinitionsError("No definitions provided for this dataset.")
def search(self, term):
"""Search for a term in a web browser.
Parameters:
term (str): term to be searched
Returns: None
"""
url = "https://www.google.com/search?q=" + term
message = f"Searching for {term} in web browser..."
print(message, end='\r')
webbrowser.open(url)
print(" " * len(message), end='\r') # Erase the message
def reduce_multiindex(self, df, levels_to_drop=None, flatten=False, sep='_'):
"""Drop levels from and/or flatten the column axis of a dataframe with a column multiindex.
Parameters:
df (pandas DataFrame): The dataframe to make the changes to.
levels_to_drop (str, int, or list or array-like of str or int, optional): Levels, or indices of levels, to drop from the dataframe's column multiindex. These must match the names or indices of actual levels of the multiindex. Must be either all strings, or all ints. Default of None will drop no levels.
flatten (bool, optional): Whether or not to flatten the multiindex. Default of False will not flatten.
sep (str, optional): String to use to separate index levels when flattening. Default is underscore.
Returns:
pandas DataFrame: The dataframe, with the desired column index changes made.
"""
# Make a copy, so the original dataframe is preserved
df = df.copy(deep=True)
if levels_to_drop is not None:
if df.columns.nlevels < 2:
raise DropFromSingleIndexError("You attempted to drop level(s) from an index with only one level.")
if isinstance(levels_to_drop, (str, int)):
levels_to_drop = [levels_to_drop]
elif not isinstance(levels_to_drop, (list, pd.core.series.Series, pd.core.indexes.base.Index)):
raise InvalidParameterError(f"Parameter 'levels_to_drop' is of invalid type {type(levels_to_drop)}. Valid types: str, int, list or array-like of str or int, or NoneType.")
# Check that they're not trying to drop too many columns
existing_len = len(df.columns.names)
to_drop_len = len(levels_to_drop)
if to_drop_len >= existing_len:
raise InvalidParameterError(f"You tried to drop too many levels from the dataframe column index. The most levels you can drop is one less than however many exist. {existing_len} levels exist; you tried to drop {to_drop_len}.")
# Check that the levels they want to drop all exist
to_drop_set = set(levels_to_drop)
if all(isinstance(level, int) for level in to_drop_set):
existing_set_indices = set(range(len(df.columns.names)))
if not to_drop_set <= existing_set_indices:
raise InvalidParameterError(f"Some level indices in {levels_to_drop} do not exist in dataframe column index, so they cannot be dropped. Existing column level indices: {list(range(len(df.columns.names)))}")
else:
existing_set = set(df.columns.names)
if not to_drop_set <= existing_set:
raise InvalidParameterError(f"Some levels in {levels_to_drop} do not exist in dataframe column index, so they cannot be dropped. Existing column levels: {df.columns.names}")
df.columns = df.columns.droplevel(levels_to_drop)
num_dups = df.columns.duplicated(keep=False).sum()
if num_dups > 0:
warnings.warn(f"Due to dropping the specified levels, dataframe now has {num_dups} duplicated column headers.", DuplicateColumnHeaderWarning, stacklevel=2)
if flatten:
if df.columns.nlevels < 2:
warnings.warn("You tried to flatten an index that didn't have multiple levels, so we didn't actually change anything.", FlattenSingleIndexWarning, stacklevel=2)
return df
tuples = df.columns.to_flat_index() # Converts multiindex to an index of tuples
no_nan = tuples.map(lambda x: [item for item in x if pd.notnull(item)]) # Cut any NaNs out of tuples
joined = no_nan.map(lambda x: sep.join(x)) # Join each tuple
df.columns = joined
df.columns.name = "Name" # For consistency
return df
def get_genotype_all_vars(self, mutations_genes, mutations_filter=None, show_location=True, mutation_hotspot=None):
"""Return a dataframe that has the mutation type and wheather or not it is a multiple mutation
Parameters:
mutation_genes (str, or list or array-like of str): The gene(s) to get mutation data for.
mutations_filter (list, optional): List of mutations to prioritize when filtering out multiple mutations, in order of priority.
show_location (bool, optional): Whether to include the Location column from the mutation dataframe. Defaults to True.
mutation_hotspot (optional): a list of hotspots
"""
#If they don't give us a filter, this is the default.
if mutations_filter == None:
mutations_filter = ["Deletion", 'Frame_Shift_Del', 'Frame_Shift_Ins', 'Nonsense_Mutation', 'Missense_Mutation_hotspot',
'Missense_Mutation', 'Amplification', 'In_Frame_Del', 'In_Frame_Ins', 'Wildtype']
#combine the cnv and mutations dataframe
combined = self.join_omics_to_mutations(omics_df_name="CNV", mutations_genes=mutations_genes, omics_genes=mutations_genes)
#drop the database index from ccrcc
if self.get_cancer_type() == "ccrcc":
cc = self.get_CNV()
drop = ['Database_ID']
combined = self.reduce_multiindex(df=combined, levels_to_drop=drop)
#If there are hotspot mutations, append 'hotspot' to the mutation type so that it's prioritized correctly
def mark_hotspot_locations(row):
#iterate through each location in the current row
mutations = []
for location in row[mutations_genes+'_Location']:
if location in mutation_hotspot: #if it's a hotspot mutation
#get the position of the location
position = row[mutations_genes+'_Location'].index(location)
#use that to change the correct mutation
mutations.append(row[mutations_genes+"_Mutation"][position] + "_hotspot")
else:
# get the position of the location
position = row[mutations_genes+'_Location'].index(location)
mutations.append(row[mutations_genes+"_Mutation"][position])
return mutations
if mutation_hotspot is not None:
combined['hotspot'] = combined.apply(mark_hotspot_locations, axis=1)
combined[mutations_genes+"_Mutation"] = combined['hotspot']
combined = combined.drop(columns='hotspot')
# Based on cnv make a new column with mutation type that includes deletions and amplifications
def add_del_and_amp(row):
if row[mutations_genes+"_CNV"] <= -.2:
mutations = row[mutations_genes+"_Mutation"] + ['Deletion']
locations = row[mutations_genes+'_Location']+['Deletion']
elif row[mutations_genes+"_CNV"] >= .2:
mutations = row[mutations_genes+"_Mutation"] + ['Amplification']
locations = row[mutations_genes+'_Location']+['Amplification']
else:
mutations = row[mutations_genes+"_Mutation"]
locations = row[mutations_genes+"_Location"]
return mutations, locations
combined['mutations'], combined['locations'] = zip(*combined.apply(add_del_and_amp, axis=1))
#now that we have the deletion and amplifications, we need to prioritize the correct mutations.
def sort(row):
sortedcol = []
location = []
chosen_indices = []
sample_mutations_list = row['mutations']
sample_locations_list = row['locations']
if len(sample_mutations_list) == 1: #if there's only one mutation in the list
sortedcol.append(sample_mutations_list[0])
location.append(sample_locations_list[0])
else:
for filter_val in mutations_filter: # This will start at the beginning of the filter list, thus filters earlier in the list are prioritized, like we want
if filter_val in sample_mutations_list:
chosen_indices = [index for index, value in enumerate(sample_mutations_list) if value == filter_val]
if len(chosen_indices) > 0: # We found at least one mutation from the filter to prioritize, so we don't need to worry about later values in the filter priority list
break
if len(chosen_indices) == 0: # None of the mutations for the sample were in the filter
for mutation in sample_mutations_list:
if mutation in truncations:
chosen_indices += [index for index, value in enumerate(sample_mutations_list) if value == mutation]
soonest_mutation = sample_mutations_list[chosen_indices[0]]
soonest_location = sample_locations_list[chosen_indices[0]]
chosen_indices.clear()
sortedcol.append(soonest_mutation)
location.append(soonest_location)
return | pd.Series([sortedcol, location],index=['mutations', 'locations']) | pandas.Series |
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
from code import visualize
# Iteration 13
def split_3(df, test_size=0.1, oversampling_ratio=1):
print('\nSplit - Train&Dev Size = ', 1-test_size, ' , Test Size = ', test_size, '.', sep='')
vcs_one = df['city_id'].value_counts()
vcs_one = np.array(vcs_one[vcs_one==1].index)
df['city_id'][df['city_id'].isin(vcs_one)]=-1
df_train_dev, df_test = train_test_split(df, test_size=test_size, shuffle=True, stratify=df['city_id'], random_state=0)
n = round(oversampling_ratio*len(df_train_dev))
df_sampled = df_train_dev.sample(n=n, replace=True, weights=1+np.log1p(df_train_dev['n_clicks']), random_state=0)
df_train_dev = pd.concat([df_train_dev, df_sampled])
X_train_dev = np.array(df_train_dev.drop(columns=['n_clicks']), dtype=float)
y_train_dev = np.array(df_train_dev['n_clicks'], dtype=float)
X_test = np.array(df_test.drop(columns=['n_clicks']), dtype=float)
y_test = np.array(df_test['n_clicks'], dtype=float)
print('n_clicks summaries in train and test sets:', pd.DataFrame(y_train_dev).describe(), pd.DataFrame(y_test).describe(), sep='\n')
visualize.compare_last_quantiles(pd.Series(y_train_dev), pd.Series(y_test), 'n_clicks Balance Assessment', 'Training & Development Set', 'Hold-Out Test Set')
print('city_ids summaries in train and test sets:', pd.Series(X_train_dev[:, 0]).value_counts().describe(), pd.Series(X_test[:, 0]).value_counts().describe(), sep='\n')
visualize.compare_last_quantiles(pd.Series(X_train_dev[:, 0]).value_counts(), pd.Series(X_test[:, 0]).value_counts(), 'city_id Balance Assessment', 'Training & Development Set', 'Hold-Out Test Set')
return (X_train_dev, y_train_dev), (X_test, y_test)
# Iteration 10-12
def split_2(df, test_size=0.1, oversampling_ratio=1):
print('\nSplit - Train&Dev Size = ', 1-test_size, ' , Test Size = ', test_size, '.', sep='')
df_test = df.sample(frac=test_size, replace=False, random_state=0)
df_train_dev = df[~df.index.isin(df_test.index)]
n = round(oversampling_ratio*len(df_train_dev))
df_sampled = df_train_dev.sample(n=n, replace=True, weights=1+np.log1p(df_train_dev['n_clicks']), random_state=0)
df_train_dev = pd.concat([df_train_dev, df_sampled])
X_train_dev = np.array(df_train_dev.drop(columns=['n_clicks']), dtype=float)
y_train_dev = np.array(df_train_dev['n_clicks'], dtype=float)
X_test = np.array(df_test.drop(columns=['n_clicks']), dtype=float)
y_test = np.array(df_test['n_clicks'], dtype=float)
print('n_clicks summaries in train and test sets:', pd.DataFrame(y_train_dev).describe(), pd.DataFrame(y_test).describe(), sep='\n')
visualize.compare_last_quantiles(pd.Series(y_train_dev), pd.Series(y_test), 'n_clicks Balance Assessment', 'Training & Development Set', 'Hold-Out Test Set')
return (X_train_dev, y_train_dev), (X_test, y_test)
# Iteration 10
def split_1(df, test_size=0.1):
print('\nSplit - Train&Dev Size = ', 1-test_size, ' , Test Size = ', test_size, '.', sep='')
test_len = round(test_size*len(df))
bins = np.arange(6)
y_binned = np.digitize(np.log1p(df['n_clicks']), bins)
partitions = [df.loc[y_binned==b+1, :] for b in bins]
bin_sz = min([len(p) for p in partitions])
ss_df = pd.DataFrame(data=None, columns=df.columns)
for p in partitions[1:]:
ss_df = pd.concat([ss_df, p.sample(n=bin_sz, replace=False, random_state=0)])
X = np.array(ss_df.drop(columns=['n_clicks']), dtype=float)
y = np.array(ss_df['n_clicks'], dtype=float)
X_train_dev, X_test, y_train_dev, y_test = train_test_split(X, y, test_size=test_size, shuffle=True, random_state=0)
print('n_clicks summaries in train and test sets:', | pd.DataFrame(y_train_dev) | pandas.DataFrame |
import pytest
import warnings
import json
import numpy as np
import pandas as pd
from .. import infer_vegalite_type, sanitize_dataframe
def test_infer_vegalite_type():
def _check(arr, typ):
assert infer_vegalite_type(arr) == typ
_check(np.arange(5, dtype=float), "quantitative")
_check(np.arange(5, dtype=int), "quantitative")
_check(np.zeros(5, dtype=bool), "nominal")
_check(pd.date_range("2012", "2013"), "temporal")
_check(pd.timedelta_range(365, periods=12), "temporal")
nulled = pd.Series(np.random.randint(10, size=10))
nulled[0] = None
_check(nulled, "quantitative")
_check(["a", "b", "c"], "nominal")
if hasattr(pytest, "warns"): # added in pytest 2.8
with pytest.warns(UserWarning):
_check([], "nominal")
else:
with warnings.catch_warnings():
warnings.filterwarnings("ignore")
_check([], "nominal")
def test_sanitize_dataframe():
# create a dataframe with various types
df = pd.DataFrame(
{
"s": list("abcde"),
"f": np.arange(5, dtype=float),
"i": np.arange(5, dtype=int),
"b": np.array([True, False, True, True, False]),
"d": pd.date_range("2012-01-01", periods=5, freq="H"),
"c": pd.Series(list("ababc"), dtype="category"),
"c2": pd.Series([1, "A", 2.5, "B", None], dtype="category"),
"o": pd.Series([np.array(i) for i in range(5)]),
"p": pd.date_range("2012-01-01", periods=5, freq="H").tz_localize("UTC"),
}
)
# add some nulls
df.iloc[0, df.columns.get_loc("s")] = None
df.iloc[0, df.columns.get_loc("f")] = np.nan
df.iloc[0, df.columns.get_loc("d")] = pd.NaT
df.iloc[0, df.columns.get_loc("o")] = np.array(np.nan)
# JSON serialize. This will fail on non-sanitized dataframes
print(df[["s", "c2"]])
df_clean = sanitize_dataframe(df)
print(df_clean[["s", "c2"]])
print(df_clean[["s", "c2"]].to_dict())
s = json.dumps(df_clean.to_dict(orient="records"))
print(s)
# Re-construct pandas dataframe
df2 = pd.read_json(s)
# Re-order the columns to match df
df2 = df2[df.columns]
# Re-apply original types
for col in df:
if str(df[col].dtype).startswith("datetime"):
# astype(datetime) introduces time-zone issues:
# to_datetime() does not.
utc = isinstance(df[col].dtype, pd.core.dtypes.dtypes.DatetimeTZDtype)
df2[col] = pd.to_datetime(df2[col], utc=utc)
else:
df2[col] = df2[col].astype(df[col].dtype)
# pandas doesn't properly recognize np.array(np.nan), so change it here
df.iloc[0, df.columns.get_loc("o")] = np.nan
assert df.equals(df2)
def test_sanitize_dataframe_colnames():
df = pd.DataFrame(np.arange(12).reshape(4, 3))
# Test that RangeIndex is converted to strings
df = sanitize_dataframe(df)
assert [isinstance(col, str) for col in df.columns]
# Test that non-string columns result in an error
df.columns = [4, "foo", "bar"]
with pytest.raises(ValueError) as err:
sanitize_dataframe(df)
assert str(err.value).startswith("Dataframe contains invalid column name: 4.")
def test_sanitize_dataframe_timedelta():
df = pd.DataFrame({"r": pd.timedelta_range(start="1 day", periods=4)})
with pytest.raises(ValueError) as err:
sanitize_dataframe(df)
assert str(err.value).startswith('Field "r" has type "timedelta')
def test_sanitize_dataframe_infs():
df = pd.DataFrame({"x": [0, 1, 2, np.inf, -np.inf, np.nan]})
df_clean = sanitize_dataframe(df)
assert list(df_clean.dtypes) == [object]
assert list(df_clean["x"]) == [0, 1, 2, None, None, None]
@pytest.mark.skipif(
not hasattr(pd, "Int64Dtype"),
reason="Nullable integers not supported in pandas v{}".format(pd.__version__),
)
def test_sanitize_nullable_integers():
df = pd.DataFrame(
{
"int_np": [1, 2, 3, 4, 5],
"int64": pd.Series([1, 2, 3, None, 5], dtype="UInt8"),
"int64_nan": pd.Series([1, 2, 3, float("nan"), 5], dtype="Int64"),
"float": [1.0, 2.0, 3.0, 4, 5.0],
"float_null": [1, 2, None, 4, 5],
"float_inf": [1, 2, None, 4, (float("inf"))],
}
)
df_clean = sanitize_dataframe(df)
assert {col.dtype.name for _, col in df_clean.iteritems()} == {"object"}
result_python = {col_name: list(col) for col_name, col in df_clean.iteritems()}
assert result_python == {
"int_np": [1, 2, 3, 4, 5],
"int64": [1, 2, 3, None, 5],
"int64_nan": [1, 2, 3, None, 5],
"float": [1.0, 2.0, 3.0, 4.0, 5.0],
"float_null": [1.0, 2.0, None, 4.0, 5.0],
"float_inf": [1.0, 2.0, None, 4.0, None],
}
@pytest.mark.skipif(
not hasattr(pd, "StringDtype"),
reason="dedicated String dtype not supported in pandas v{}".format(pd.__version__),
)
def test_sanitize_string_dtype():
df = pd.DataFrame(
{
"string_object": ["a", "b", "c", "d"],
"string_string": | pd.array(["a", "b", "c", "d"], dtype="string") | pandas.array |
"""
The `frame` module allows working with StarTable tables as pandas dataframes.
This is implemented by providing both `Table` and `TableDataFrame` interfaces to the same object.
## Idea
The central idea is that as much as possible of the table information is stored as a pandas
dataframe, and that the remaining information is stored as a `ComplementaryTableInfo` object
attached to the dataframe as registered metadata. Further, access to the full table data
structure is provided through a facade object (of class `Table`). `Table` objects have no state
(except the underlying decorated dataframe) and are intended to be created when needed
and discarded afterwards:
```
dft = make_table_dataframe(...)
unit_height = Table(dft).height.unit
```
Advantages of this approach are that:
1. Code can be written for (and tested with) pandas dataframes and still operate on `TableDataFrame`
objects. This avoids unnecessary coupling to the StarTable project.
2. The table access methods of pandas are available for use by consumer code. This both saves
the work of making startable-specific access methods, and likely allows better performance
and documentation.
## Implementation details
The decorated dataframe objects are represented by the `TableDataFrame` class.
### Dataframe operations
Pandas allows us to hook into operations on dataframes via the `__finalize__` method.
This makes it possible to propagate table metadata over select dataframe operations.
See `TableDataFrame` documentation for details.
### Column metadata
Propagating metadata would be greatly simplified if column-specific metadata was stored with the
column. However, metadata attached to `Series` object is not retained within the dataframe, see
https://github.com/pandas-dev/pandas/issues/6923#issuecomment-41043225.
## Alternative approaches
It should be possible to maintain column metadata together with the column data through the use of
`ExtensionArray`. This option was discarded early due to performance concerns, but might be viable
and would the be preferable to the chosen approach.
"""
from warnings import warn
import pandas as pd
import warnings
from typing import Set, Dict, Optional, Iterable
from .table_metadata import TableMetadata, ColumnMetadata, ComplementaryTableInfo
from .table_origin import TableOrigin
_TABLE_INFO_FIELD_NAME = "_table_data"
class UnknownOperationError(Exception):
pass
class InvalidTableCombineError(Exception):
pass
def _combine_tables(
obj: "TableDataFrame", other, method, **kwargs
) -> Optional[ComplementaryTableInfo]:
"""
Called from dataframe.__finalize__ when dataframe operations have been performed
on the dataframe backing a table.
Implementation policy is that this will warn except for situations where
the metadata combination is safe.
For other cases, the operations should be implemented via the Table facade
if metadata is required, or by dropping to bare dataframes otherwise.
"""
if method is None or method in frozenset({"reindex", "take", "copy"}):
# method: None - copy, slicing (pandas <1.1)
src = [other]
elif method == "merge":
src = [other.left, other.right]
elif method == "concat":
src = other.objs
else:
# Unknown method - try to handle this as well as possible, but rather warn and drop units than break things.
src = [other]
warnings.warn(
f'While combining pdTable metadata an unknown __finalize__ method "{method}" was encountered. '
f"Will try to propagate metadata with generic methods, but please check outcome of this "
f"and notify pdTable maintainers."
)
data = [d for d in (getattr(s, _TABLE_INFO_FIELD_NAME, None) for s in src) if d is not None]
if not data:
return None
# 1: Create table metadata as combination of all
origin = TableOrigin(
operation=f"Pandas {method}",
parents=[d.metadata.origin for d in data]
)
meta = TableMetadata(
name=data[0].metadata.name,
destinations=data[0].metadata.destinations,
origin=origin,
)
# 2: Check that units match for columns that appear in more than one table
out_cols: Set[str] = set(obj.columns)
columns: Dict[str, ColumnMetadata] = dict()
for d in data:
for name, c in d.columns.items():
if name not in out_cols:
continue
col = columns.get(name, None)
if not col:
# not seen before in input
col = c.copy()
columns[name] = col
else:
if not col.unit == c.unit:
raise InvalidTableCombineError(
f'Column {name} appears with incompatible units "{col.unit}" and "{c.unit}".'
)
col.update_from(c)
return ComplementaryTableInfo(table_metadata=meta, columns=columns)
class TableDataFrame(pd.DataFrame):
"""
A pandas.DataFrame subclass with associated table metadata.
Behaves exactly as a pandas.DataFrame, and will try to retain metadata
through pandas operations. If this is not possible, the manipulations
will return a plain pandas.DataFrame.
No StarTable-specific methods are available directly on this class, and TableDataFrame
objects should not be created directly.
Instead, use either the methods in the this module, or the Table proxy
object, which can be constructed for a TableDataFrame object 'tdf' via Table(tdf).
"""
_metadata = [_TABLE_INFO_FIELD_NAME] # Register metadata fieldnames here
# If implemented, must handle metadata copying etc
# def __init__(self, *args, **kwargs):
# super().__init__(*args, **kwargs)
# These are implicit by inheritance from dataframe
# We could override _constructor_sliced to add metadata to exported columns
# but not clear if this would add value
# @property
# def _constructor_expanddim(self):
# return pd.DataFrame
# @property
# def _constructor_sliced(self):
# return pd.Series
@property
def _constructor(self):
return TableDataFrame
def __finalize__(self, other, method=None, **kwargs):
"""
Overrides pandas.core.generic.NDFrame.__finalize__()
This method is responsible for populating TableDataFrame metadata when creating a new Table object through
pandas operations. This may includes combining unit information and table origin for operations involving
more than one table.
"""
table_info = _combine_tables(self, other, method, **kwargs)
if table_info is None:
warn(
f"Unable to establish table metadata (units, origin, etc.). Will fall back to pd.DataFrame."
)
return | pd.DataFrame(self) | pandas.DataFrame |
import os
import json
import pandas as pd
import numpy as np
from segmenter.collectors.BaseCollector import BaseCollector
import glob
class MetricCollector(BaseCollector):
results = | pd.DataFrame() | pandas.DataFrame |
# import pandas, numpy, and matplotlib
import pandas as pd
from feature_engine.encoding import OneHotEncoder
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.ensemble import RandomForestClassifier
from sklearn.linear_model import LogisticRegression
from mlxtend.feature_selection import ExhaustiveFeatureSelector
from sklearn.metrics import accuracy_score
| pd.set_option('display.width', 75) | pandas.set_option |
#!/usr/bin/env python
from scipy import sparse
from sklearn.datasets import dump_svmlight_file
from sklearn.preprocessing import LabelEncoder
import argparse
import logging
import numpy as np
import os
import pandas as pd
logging.basicConfig(format='%(asctime)s %(levelname)s %(message)s',
level=logging.DEBUG)
def generate_feature(train_file, label_file, test_file, feature_dir,
feature_name):
# Load data files
logging.info('Loading training and test data')
trn = pd.read_csv(train_file, index_col=0)
tst = pd.read_csv(test_file, index_col=0)
label = pd.read_csv(label_file, index_col=0)
n_trn = trn.shape[0]
n_tst = tst.shape[0]
logging.info('Combining training and test data')
df = | pd.concat([trn, tst], ignore_index=True) | pandas.concat |
# -*- coding: utf-8 -*-
""" Overplot
Series of functions designed to help with charting in Plotly
"""
import pandas as pd
import plotly.express as px
import plotly.graph_objs as go
import plotly.io as pio
# %% PLOTLY EXPRESS STANLIB TEMPLATE
# Approximatation of STANLIB colour theme
COLOUR_MAP = {0:'purple',
1:'turquoise',
2:'grey',
3:'black',
4:'green',
5:'blue',
6:'crimson',
7:'orange',
8:'mediumvioletred'}
# Hack together basic template
fig = go.Figure(layout=dict(
font={'family':'Courier New', 'size':12},
plot_bgcolor= 'white',
colorway=['grey','turquoise', 'purple', 'lime', 'blue', 'black', 'brown', 'red', 'orange'],
showlegend=False,
legend={'orientation':'v'},
margin = {'l':75, 'r':50, 'b':50, 't':50},
xaxis= {'anchor': 'y1', 'title': '',
'showline':True, 'linecolor': 'gray',
'zeroline':True, 'zerolinewidth':1 , 'zerolinecolor':'whitesmoke',
'showgrid': True, 'gridcolor': 'whitesmoke',
},
yaxis= {'anchor': 'x1', 'title': '', 'hoverformat':'.1f', 'tickformat':'.1f',
'showline':True, 'linecolor':'gray',
'zeroline':True, 'zerolinewidth':1 , 'zerolinecolor':'whitesmoke',
'showgrid': True, 'gridcolor': 'whitesmoke'},
updatemenus= [dict(type='buttons',
active=-1, showactive = True,
direction='down',
y=0.5, x=1.1,
pad = {'l':0, 'r':0, 't':0, 'b':0},
buttons=[])],
annotations=[],))
# save it
templated_fig = pio.to_templated(fig)
pio.templates['multi_strat'] = templated_fig.layout.template
# %%
def line_stacker(df, template='multi_strat',
yaxis_title= '', source='', source_posn=[0.85, 0.08],
**kwargs):
""" Line plot with columns as lines
Plotly express does a cool thing called 'colours' where it loads multiple
traces which can be clicked on and off on the chart. It does however require
a f*cking stupid format where all data is in one long vector with repeated
dates AND a column of 'colours'... why they don't just let you use the
varname and have a logical df is beyond me'
INPUT:
df - dataframe with dates as index; column headers as legend titles
kwargs - ONLY use arguments you could normally pass to plotly express
"""
# set up the 3 columns we need
vn = ['date', 'value', 'index']
z = pd.DataFrame(columns = vn)
# itserate through df concatinating to silly long vector
for ticker in df:
i = df[ticker].reset_index()
i['value'] = ticker
i.columns = vn
z = | pd.concat([z, i]) | pandas.concat |
from netCDF4 import Dataset
import pandas as pd
import numpy as np
ncep_path = '/SubX/forecast/tas2m/daily/full/NCEP-CFSv2/' # the path where the raw data from NCEP-CFSv2 is saved
gmao_path = '/SubX/forecast/tas2m/daily/full/GMAO-GEOS_V2p1/'
for model in ['NCEP', 'GMAO']:
if model == 'NCEP':
path = ncep_path
NCEP_date = []
for date in pd.date_range('2017-07-01', '2019-12-31'):
data = Dataset(path + 'tas_2m_NCEP-CFSv2_{}.emean.daily.nc'.format(date.strftime('%Y%m%d')))
num = np.count_nonzero(np.isnan(data.variables['tas'][0]))
if num != 65160:
# print(date, num)
NCEP_date.append(date)
NCEP_date = pd.Series(NCEP_date)
NCEP_date.to_hdf('NCEP_date.h5', key='date')
elif model == 'GMAO':
path = gmao_path
GMAO_date = []
for date in | pd.date_range('2017-07-25', '2020-09-28') | pandas.date_range |
# -*- coding: utf-8 -*-
"""
Tools to collect Twitter data from specific accounts.
Part of the module is based on Twitter Scraper library:
https://github.com/bpb27/twitter_scraping
Author: <NAME> <<EMAIL>>
Part of https://github.com/crazyfrogspb/RedditScore project
Copyright (c) 2018 <NAME>. All rights reserved.
This work is licensed under the terms of the MIT license.
"""
import datetime
import math
import os.path as osp
import random
import warnings
from time import sleep
import pandas as pd
from dateutil import parser
import tweepy
from congress import Congress
try:
from selenium import webdriver
from selenium.common.exceptions import (NoSuchElementException,
TimeoutException,
StaleElementReferenceException,
WebDriverException)
except ImportError:
warnings.warn(('selenium library is not found, pulling tweets beyond'
' 3200 limit will be unavailable'))
def _format_day(date):
# convert date to required format
day = '0' + str(date.day) if len(str(date.day)) == 1 else str(date.day)
month = '0' + str(date.month) if len(str(date.month)
) == 1 else str(date.month)
year = str(date.year)
return '-'.join([year, month, day])
def _form_url(since, until, user):
# create url request
p1 = 'https://twitter.com/search?f=tweets&vertical=default&q=from%3A'
p2 = user + '%20since%3A' + since + '%20until%3A' + \
until + 'include%3Aretweets&src=typd'
return p1 + p2
def _increment_day(date, i):
# increment date by i days
return date + datetime.timedelta(days=i)
def _grab_tweet_by_ids(ids, api, delay=6.0):
# grab tweets by ids
full_tweets = []
start = 0
end = 100
limit = len(ids)
i = math.ceil(limit / 100)
for go in range(i):
sleep(delay)
id_batch = ids[start:end]
start += 100
end += 100
tweets = api.statuses_lookup(id_batch, tweet_mode='extended')
full_tweets.extend(tweets)
return full_tweets
def _grab_even_more_tweets(screen_name, dates, browser, delay=1.0):
# grab tweets beyond 3200 limit
startdate, enddate = dates
try:
if browser == 'Safari':
driver = webdriver.Safari()
elif browser == 'Firefox':
driver = webdriver.Firefox()
elif browser == 'Chrome':
driver = webdriver.Chrome()
else:
raise ValueError('{} browser is not supported')
except WebDriverException as e:
raise WebDriverException(('You need to download required driver'
' and add it to PATH')) from e
except AttributeError as e:
raise Exception('Check if the browser is installed') from e
except ValueError as e:
raise ValueError('{} browser is not supported') from e
days = (enddate - startdate).days + 1
id_selector = '.time a.tweet-timestamp'
tweet_selector = 'li.js-stream-item'
screen_name = screen_name.lower()
ids = []
for day in range(days):
d1 = _format_day(_increment_day(startdate, 0))
d2 = _format_day(_increment_day(startdate, 1))
url = _form_url(d1, d2, screen_name)
driver.get(url)
sleep(delay)
try:
found_tweets = driver.find_elements_by_css_selector(tweet_selector)
increment = 10
while len(found_tweets) >= increment:
driver.execute_script(
'window.scrollTo(0, document.body.scrollHeight);')
sleep(delay)
found_tweets = driver.find_elements_by_css_selector(
tweet_selector)
increment += 10
for tweet in found_tweets:
try:
id = tweet.find_element_by_css_selector(
id_selector).get_attribute('href').split('/')[-1]
ids.append(id)
except StaleElementReferenceException as e:
pass
except NoSuchElementException:
pass
except TimeoutException:
pass
startdate = _increment_day(startdate, 1)
return ids
def _handle_tweepy_error(e, user):
if e.api_code == 34:
warnings.warn("{} doesn't exist".format(user))
else:
warnings.warn('Error encountered for user {}: '.format(
user) + str(e))
return pd.DataFrame()
def generate_api(twitter_creds_list):
auths = []
for creds in twitter_creds_list:
try:
auth = tweepy.OAuthHandler(
creds['consumer_key'], creds['consumer_secret'])
auth.set_access_token(creds['access_key'], creds['access_secret'])
except KeyError as e:
raise Exception(("twitter_creds must contain cosnumer_key,"
" consumer_secret, access_key, and access_secret keys"))
auths.append(auth)
api = tweepy.API(
auths,
retry_count=3,
retry_delay=5,
retry_errors=set([401, 404, 500, 503]),
monitor_rate_limit=True,
wait_on_rate_limit=True,
wait_on_rate_limit_notify=True)
return api
def grab_tweets(twitter_creds=None, api=None, screen_name=None, user_id=None, timeout=0.1,
fields=None, get_more=False, browser='Firefox',
start_date=None):
"""
Get all tweets from the account
Parameters
----------
twitter_creds: dict
Dictionary or list with Twitter authentication credentials.
Has to contain consumer_key, consumer_secret, access_key, access_secret
screen_name : str, optional
Twitter handle to grab tweets for
user_id: int, optional
Twitter user_id to grab tweets for
timeout: float, optional
Sleeping time between requests
fields: iter, optional
Extra fields to pull from the tweets
get_more: bool, optional
If True, attempt to use Selenium to get more tweets after reaching
3200 tweets limit
browser: {'Firefox', 'Chrome', 'Safari'}, optional
Browser for Selenium to use. Corresponding browser and its webdriver
have to be installed
start_date: datetime.date, optional
The first date to start pulling extra tweets. If None, use 2016/01/01
Returns
----------
alltweets: pandas DataFrame
Pandas Dataframe with all collected tweets
"""
if not (bool(screen_name) != bool(user_id)):
raise ValueError('You have to provide either screen_name or user_id')
api = generate_api(list(twitter_creds))
if user_id:
try:
u = api.get_user(int(user_id))
screen_name = u.screen_name
reg_date = u.created_at.date()
sleep(timeout)
except tweepy.TweepError as e:
return _handle_tweepy_error(e, user_id)
except ValueError as e:
raise ValueError('{} is not a valid user_id'.format(user_id)) from e
else:
u = api.get_user(screen_name)
reg_date = u.created_at.date()
sleep(timeout)
if fields is None:
fields = []
if start_date is None or start_date < reg_date:
start_date = reg_date
alltweets = []
print("Now grabbing tweets for {}".format(screen_name))
try:
new_tweets = api.user_timeline(screen_name=screen_name,
user_id=user_id, count=200,
tweet_mode='extended')
except tweepy.TweepError as e:
return _handle_tweepy_error(e, screen_name)
alltweets.extend(new_tweets)
if not alltweets:
return pd.DataFrame()
oldest = alltweets[-1].id - 1
while len(new_tweets) > 0:
new_tweets = api.user_timeline(screen_name=screen_name, count=200,
max_id=oldest, tweet_mode='extended')
alltweets.extend(new_tweets)
oldest = alltweets[-1].id - 1
if new_tweets:
print('{} tweets downloaded'.format(len(alltweets)))
sleep(timeout)
if get_more and len(new_tweets) == 0 and len(alltweets) > 3200:
end_date = alltweets[-1].created_at.date()
print('Date of the last collected tweet: {}'.format(end_date))
if end_date > start_date:
print('Now grabbing tweets beyond 3200 limit')
dates = (start_date, end_date)
ids = _grab_even_more_tweets(screen_name, dates, browser)
tweets = _grab_tweet_by_ids(ids, api)
alltweets.extend(tweets)
full_tweets = []
for tweet in alltweets:
if hasattr(tweet, 'retweeted_status'):
text = tweet.retweeted_status.full_text
else:
text = tweet.full_text
retweet = False
if getattr(tweet, 'retweeted_status', None) is not None:
retweet = True
tweet_fields = [text, tweet.id, tweet.created_at, retweet]
for field in fields:
tweet_fields.append(getattr(tweet, field, None))
full_tweets.append(tweet_fields)
full_tweets = pd.DataFrame(
full_tweets, columns=(['text', 'id', 'created_at', 'retweet'] +
fields))
full_tweets['screen_name'] = screen_name
if user_id:
full_tweets['user_id'] = user_id
full_tweets.drop_duplicates('id', inplace=True)
return full_tweets
def collect_congress_tweets(congress_list, congress_tweets_file,
meta_info_file, start_date, twitter_creds,
chambers=None, propublica_api_key=None,
append_frequency=10, browser='Chrome',
fields=None, shuffle=False):
"""Collect tweets from American Congressmen.
Parameters
----------
congress_list : iterable
List with Congress numbers to collect data for.
congress_tweets_file : str
Path to the output file with tweets.
meta_info_file : str
Path to the output file with meta information about the Congress.
start_date : str
The first date to start pulling extra tweets.
twitter_creds : type
Dictionary or list with Twitter authentication credentials.
Has to contain consumer_key, consumer_secret, access_key, access_secret
chambers : iterable, optional
List of Chambers to collect tweets for (the default is Senate and House).
propublica_api_key : str, optional
API key for free Propublica Congress API (the default is None).
https://www.propublica.org/datastore/api/propublica-congress-api
append_frequency : int, optional
Frequency of dumping new tweets to CSV (the default is 10).
browser : str, optional
Browser for Selenium to use. Corresponding browser and its webdriver
have to be installed (the default is 'Chrome').
fields : iter, optional
Extra fields to pull from the tweets (the default is retweet_count and favorite_count).
shuffle: bool, optional
Whether to shuffle twitter handles before collecting.
"""
if chambers is None:
chambers = ['House', 'Senate']
if fields is None:
fields = ['retweet_count', 'favorite_count']
if osp.isfile(meta_info_file):
members = pd.read_csv(meta_info_file)
else:
congress = Congress(propublica_api_key)
all_members = []
for congress_num in congress_list:
for chamber in chambers:
members = pd.DataFrame(congress.members.filter(
chamber, congress=congress_num)[0]['members'])
members['chamber'] = chamber
members['congress_num'] = congress_num
all_members.append(members)
members = | pd.concat(all_members) | pandas.concat |
import itertools
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
datasets = [
'BPI11/f1/',
'BPI11/f2/',
'BPI11/f3/',
'BPI11/f4/',
'BPI15/f1/',
'BPI15/f2/',
'BPI15/f3/',
'Drift1/f1/',
'Drift2/f1/'
]
split_sizes = [
'0-40_80-100',
'0-60_80-100',
'0-80_80-100',
'40-60_80-100',
'60-80_80-100',
'40-55_80-100',
'0-55_80-100'
]
splits = {
'BPI11/f1/': {
'0-40_80-100': 138,
'0-80_80-100': 139,
'40-80_80-100': 140,
},
'BPI11/f2/': {
'0-40_80-100': 141,
'0-80_80-100': 142,
'40-80_80-100': 143,
},
'BPI11/f3/': {
'0-40_80-100': 144,
'0-80_80-100': 145,
'40-80_80-100': 146,
},
'BPI11/f4/': {
'0-40_80-100': 147,
'0-80_80-100': 148,
'40-80_80-100': 149,
},
'BPI15/f1/': {
'0-40_80-100': 150,
'0-80_80-100': 151,
'40-80_80-100': 152,
},
'BPI15/f2/': {
'0-40_80-100': 153,
'0-80_80-100': 154,
'40-80_80-100': 155,
},
'BPI15/f3/': {
'0-40_80-100': 156,
'0-80_80-100': 157,
'40-80_80-100': 158,
},
'Drift1/f1/': {
'0-40_80-100': 159,
'0-80_80-100': 160,
'40-80_80-100': 161,
'40-60_80-100': 1111,
'0-60_80-100': 1111,
'40-55_80-100': 1111,
'0-55_80-100': 1111
},
'Drift2/f1/': {
'0-40_80-100': 162,
'0-80_80-100': 163,
'40-80_80-100': 164,
'40-60_80-100': 1111,
'0-60_80-100': 1111,
'40-55_80-100': 1111,
'0-55_80-100': 1111
}
}
# }
# splits = {
# 'BPI11/f1/': {
# '0-40_80-100': 101,
# '0-80_80-100': 102,
# '40-80_80-100': 103,
# },
# 'BPI11/f2/': {
# '0-40_80-100': 104,
# '0-80_80-100': 105,
# '40-80_80-100': 106,
# },
# 'BPI11/f3/': {
# '0-40_80-100': 107,
# '0-80_80-100': 108,
# '40-80_80-100': 109,
# },
# 'BPI11/f4/': {
# '0-40_80-100': 110,
# '0-80_80-100': 111,
# '40-80_80-100': 112,
# },
# 'BPI15/f1/': {
# '0-40_80-100': 113,
# '0-80_80-100': 114,
# '40-80_80-100': 115,
# },
# 'BPI15/f2/': {
# '0-40_80-100': 116,
# '0-80_80-100': 117,
# '40-80_80-100': 118,
# },
# 'BPI15/f3/': {
# '0-40_80-100': 119,
# '0-80_80-100': 120,
# '40-80_80-100': 121,
# },
# 'Drift1/f1/': {
# '0-40_80-100': 122,
# '0-80_80-100': 123,
# '40-80_80-100': 124,
#
# '40-60_80-100': 1111,
# '0-60_80-100': 1111,
# '40-55_80-100': 1111,
# '0-55_80-100': 1111
# },
# 'Drift2/f1/': {
# '0-40_80-100': 125,
# '0-80_80-100': 126,
# '40-80_80-100': 127,
#
# '40-60_80-100': 1111,
# '0-60_80-100': 1111,
# '40-55_80-100': 1111,
# '0-55_80-100': 1111
# }
# }
# splits = {
# 'BPI11/f1/': {
# '0-40_80-100': 55,
# '0-80_80-100': 56,
# '40-80_80-100': 73,
# },
# 'BPI11/f2/': {
# '0-40_80-100': 57,
# '0-80_80-100': 58,
# '40-80_80-100': 74,
# },
# 'BPI11/f3/': {
# '0-40_80-100': 59,
# '0-80_80-100': 60,
# '40-80_80-100': 75,
# },
# 'BPI11/f4/': {
# '0-40_80-100': 61,
# '0-80_80-100': 62,
# '40-80_80-100': 76,
# },
# 'BPI15/f1/': {
# '0-40_80-100': 63,
# '0-80_80-100': 64,
# '40-80_80-100': 77,
# },
# 'BPI15/f2/': {
# '0-40_80-100': 65,
# '0-80_80-100': 66,
# '40-80_80-100': 78,
# },
# 'BPI15/f3/': {
# '0-40_80-100': 67,
# '0-80_80-100': 68,
# '40-80_80-100': 79,
# },
# 'Drift1/f1/': {
# '0-40_80-100': 69,
# '0-80_80-100': 70,
# '40-80_80-100': 80,
#
# '40-60_80-100': 1111,
# '0-60_80-100': 1111,
# '40-55_80-100': 1111,
# '0-55_80-100': 1111
# },
# 'Drift2/f1/': {
# '0-40_80-100': 90,#71,
# '0-80_80-100': 91,#72,
# '40-80_80-100': 92,#81,
#
# '40-60_80-100': 1111,
# '0-60_80-100': 1111,
# '40-55_80-100': 1111,
# '0-55_80-100': 1111
# }
# }
def get_row_metrics(table):
curr_row = table['evaluation_f1_score']
f1_score_mean, f1_score_std, f1_score_max = curr_row.mean(), curr_row.std(), curr_row.max()
curr_row = table['evaluation_accuracy']
accuracy_mean, accuracy_std, accuracy_max = curr_row.mean(), curr_row.std(), curr_row.max()
curr_row = table['evaluation_precision']
precision_mean, precision_std, precision_max = curr_row.mean(), curr_row.std(), curr_row.max()
curr_row = table['evaluation_recall']
recall_mean, recall_std, recall_max = curr_row.mean(), curr_row.std(), curr_row.max()
curr_row = table['evaluation_auc']
auc_mean, auc_std, auc_max = curr_row.mean(), curr_row.std(), curr_row.max()
curr_row = | pd.to_timedelta(table['evaluation_elapsed_time']) | pandas.to_timedelta |
import math
import re
import pandas as pd
from pandas.core.dtypes.inference import is_hashable
from .transformation import Transformation
from ..exceptions import IndexFilterException
class FilterMissing(Transformation):
filter = True
title = "Filter rows with missing values in {field}"
key = "Filter missing values"
fields = {
"field": {"name": "Input", "type": "string", "help": "The column to use as input",
"required": True, "input": "column", "multiple": False, "default": ""},
}
def __init__(self, arguments: dict, sample_size: int, example: dict = None):
super().__init__(arguments, sample_size, example)
self.field = arguments["field"]
def __call__(self, row, index: int):
if self.field not in row or | pd.isnull(row[self.field]) | pandas.isnull |
from datetime import timedelta
import numpy as np
import pytest
from pandas._libs import iNaT
import pandas as pd
from pandas import (
Categorical,
DataFrame,
Index,
IntervalIndex,
NaT,
Series,
Timestamp,
date_range,
isna,
)
import pandas._testing as tm
class TestSeriesMissingData:
def test_categorical_nan_equality(self):
cat = Series( | Categorical(["a", "b", "c", np.nan]) | pandas.Categorical |
from ieeg.auth import Session
from numpy.lib.function_base import select
import pandas as pd
import pickle
from pull_patient_localization import pull_patient_localization
from numbers import Number
import numpy as np
def get_iEEG_data(username, password, iEEG_filename, start_time_usec, stop_time_usec, select_electrodes=None, ignore_electrodes=None, outputfile=None):
""""
2020.04.06. Python 3.7
<NAME>, adapted by <NAME> (2021.06.23)
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
Purpose:
To get iEEG data from iEEG.org. Note, you must download iEEG python package from GitHub - instructions are below
1. Gets time series data and sampling frequency information. Specified electrodes are removed.
2. Saves as a pickle format
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
Input
username: your iEEG.org username
password: <PASSWORD>
iEEG_filename: The file name on iEEG.org you want to download from
start_time_usec: the start time in the iEEG_filename. In microseconds
stop_time_usec: the stop time in the iEEG_filename. In microseconds.
iEEG.org needs a duration input: this is calculated by stop_time_usec - start_time_usec
ignore_electrodes: the electrode/channel names you want to exclude. EXACT MATCH on iEEG.org. Caution: some may be LA08 or LA8
outputfile: the path and filename you want to save.
PLEASE INCLUDE EXTENSION .pickle.
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
Output:
Saves file outputfile as a pickle. For more info on pickling, see https://docs.python.org/3/library/pickle.html
Briefly: it is a way to save + compress data. it is useful for saving lists, as in a list of time series data and sampling frequency together along with channel names
List index 0: Pandas dataframe. T x C (rows x columns). T is time. C is channels.
List index 1: float. Sampling frequency. Single number
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
Example usage:
username = 'arevell'
password = 'password'
iEEG_filename='HUP138_phaseII'
start_time_usec = 248432340000
stop_time_usec = 248525740000
removed_channels = ['EKG1', 'EKG2', 'CZ', 'C3', 'C4', 'F3', 'F7', 'FZ', 'F4', 'F8', 'LF04', 'RC03', 'RE07', 'RC05', 'RF01', 'RF03', 'RB07', 'RG03', 'RF11', 'RF12']
outputfile = '/Users/andyrevell/mount/DATA/Human_Data/BIDS_processed/sub-RID0278/eeg/sub-RID0278_HUP138_phaseII_248432340000_248525740000_EEG.pickle'
get_iEEG_data(username, password, iEEG_filename, start_time_usec, stop_time_usec, removed_channels, outputfile)
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
To run from command line:
python3.6 -c 'import get_iEEG_data; get_iEEG_data.get_iEEG_data("arevell", "password", "<PASSWORD>", 248432340000, 248525740000, ["EKG1", "EKG2", "CZ", "C3", "C4", "F3", "F7", "FZ", "F4", "F8", "LF04", "RC03", "RE07", "RC05", "RF01", "RF03", "RB07", "RG03", "RF11", "RF12"], "/gdrive/public/DATA/Human_Data/BIDS_processed/sub-RID0278/eeg/sub-RID0278_HUP138_phaseII_D01_248432340000_248525740000_EEG.pickle")'
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
#How to get back pickled files
with open(outputfile, 'rb') as f: data, fs = pickle.load(f)
"""
# print("\n\nGetting data from iEEG.org:")
# print("iEEG_filename: {0}".format(iEEG_filename))
# print("start_time_usec: {0}".format(start_time_usec))
# print("stop_time_usec: {0}".format(stop_time_usec))
# print("ignore_electrodes: {0}".format(ignore_electrodes))
# if outputfile:
# print("Saving to: {0}".format(outputfile))
# else:
# print("Not saving, returning data and sampling frequency")
# Pull and format metadata from patient_localization_mat
start_time_usec = int(start_time_usec)
stop_time_usec = int(stop_time_usec)
duration = stop_time_usec - start_time_usec
s = Session(username, password)
ds = s.open_dataset(iEEG_filename)
all_channel_labels = ds.get_channel_labels()
assert((select_electrodes is not None) or (ignore_electrodes is not None))
if select_electrodes is not None:
if isinstance(select_electrodes[0], Number):
channel_ids = select_electrodes
channel_names = [all_channel_labels[e] for e in channel_ids]
elif isinstance(select_electrodes[0], str):
channel_ids = [i for i, e in enumerate(all_channel_labels) if e in select_electrodes]
channel_names = select_electrodes
else:
print("Electrodes not given as a list of ints or strings")
# if ignore_electrodes:
# if isinstance(ignore_electrodes[0], int):
# print('int')
# channel_ids = [i for i in np.arange(len(all_channel_labels)) if i not in ignore_electrodes]
# channel_names = [all_channel_labels[e] for e in channel_ids]
# elif isinstance(ignore_electrodes[0], str):
# print('str')
# channel_ids = [i for i, e in enumerate(all_channel_labels) if e not in ignore_electrodes]
# channel_names = select_electrodes
# else:
# print("Electrodes not given as a list of ints or strings")
# channel_ids = [i for i, e in enumerate(all_channel_labels) if e not in ignore_electrodes]
# channel_names = [e for e in all_channel_labels if e not in ignore_electrodes]
try:
data = ds.get_data(start_time_usec, duration, channel_ids)
except:
# clip is probably too big, pull chunks and concatenate
clip_size = 60 * 1e6
clip_start = start_time_usec
data = None
while clip_start + clip_size < stop_time_usec:
if data is None:
data = ds.get_data(clip_start, clip_size, channel_ids)
else:
data = np.concatenate(([data, ds.get_data(clip_start, clip_size, channel_ids)]), axis=0)
clip_start = clip_start + clip_size
data = np.concatenate(([data, ds.get_data(clip_start, stop_time_usec - clip_start, channel_ids)]), axis=0)
df = | pd.DataFrame(data, columns=channel_names) | pandas.DataFrame |
import config
import os
import pandas as pd
import numpy as np
from datetime import date, datetime, time, timedelta
import re
import warnings
warnings.filterwarnings('ignore')
CONF_CASES_THRESHOLD = 25000
# src_confirmed = config.confirmed_cases_global_online
# src_recovered = config.recovered_cases_global_online
# src_dead = config.deceased_cases_global_online
src_confirmed = os.path.join(config.base_data_dir, config.confirmed_cases_global_offline)
src_recovered = os.path.join(config.base_data_dir, config.recovered_cases_global_offline)
src_dead = os.path.join(config.base_data_dir, config.deceased_cases_global_offline)
def convert (df_src, col):
df = df_src.copy()
df = df.rename(columns={'Country/Region':'Country'})
df.drop(['Province/State', 'Lat', 'Long'], axis=1, inplace=True, errors='ignore')
df = df.melt(id_vars=["Country"],
var_name="Date",
value_name=col)
df['Date'] = pd.to_datetime(df['Date'], format='%m/%d/%y')
df_refined = pd.DataFrame(columns=['Country', col])
all_countries = df['Country'].unique()
for country in all_countries:
dfc = df.loc[df['Country']==country]
dfc_grouped = dfc.groupby('Date', as_index=False)[col].sum()
dfc_grouped['Country'] = country
dfc_grouped['Date_Copy'] = dfc_grouped['Date']
dfc_grouped['merge_col'] = dfc_grouped['Country'] + '-' + dfc_grouped['Date'].astype(str)
df_refined = pd.concat([df_refined, dfc_grouped], axis=0)
df_refined = df_refined.set_index('Date')
return df_refined
def load ():
try:
df_confirmed = | pd.read_csv(src_confirmed) | pandas.read_csv |
import pandas as pd
import numpy as np
"""
this file is crucial
data5 is the file that goes to R to create the forecast.
"""
data = pd.read_csv('Lokad_Orders.csv')
data['Quantity'] = data['Quantity'].astype('int')
data['Date'] =pd.to_datetime(data.Date) - pd.to_timedelta(7,unit = 'd')
data =data.groupby(['Id', | pd.Grouper(key='Date', freq='W-MON') | pandas.Grouper |
from nilearn import surface
import argparse
from braincoder.decoders import GaussianReceptiveFieldModel
from braincoder.utils import get_rsq
from bids import BIDSLayout
import pandas as pd
import os
import os.path as op
import numpy as np
import nibabel as nb
from nipype.interfaces.freesurfer.utils import SurfaceTransform
import subprocess
def main(subject,
sourcedata,
trialwise,
clip=(-100, 100)):
derivatives = op.join(sourcedata, 'derivatives')
if trialwise:
layout = BIDSLayout(op.join(derivatives, 'glm_stim1_trialwise_surf'), validate=False)
else:
layout = BIDSLayout(op.join(derivatives, 'glm_stim1_surf'), validate=False)
for hemi in ['L', 'R']:
pes = layout.get(subject=subject, suffix=hemi)
print(pes)
df = []
for pe in pes:
d = pd.DataFrame(np.clip(surface.load_surf_data(pe.path).T, clip[0], clip[1]))
df.append(d)
d['run'] = pe.run
d['number'] = np.log([5, 7, 10, 14, 20, 28])
df = pd.concat(df).set_index(['run', 'number'])
mask = ~df.isnull().any(0)
print('fitting {} time series'.format(mask.sum()))
for run in df.index.unique('run'):
train = df.drop(run)
test = df.loc[run]
model = GaussianReceptiveFieldModel()
costs, parameters, predictions = model.optimize(train.index.get_level_values('number').values,
train.loc[:, mask].values)
base_dir = op.join(derivatives, 'modelfit_surf_cv',
f'sub-{subject}', 'func')
if not op.exists(base_dir):
os.makedirs(base_dir)
parameters.columns = df.loc[:, mask].columns
pars_df = pd.DataFrame(columns=df.columns)
pars_df = pd.concat((pars_df, parameters))
par_fn = op.join(
base_dir, f'sub-{subject}_space-fsaverage6_desc-pars_hemi-{hemi}_cvrun-{run}.func.gii')
nb.gifti.GiftiImage(header=nb.load(pe.path).header, darrays=[nb.gifti.GiftiDataArray(data=p.astype(float)) for _,
p in pars_df.iterrows()]).to_filename(par_fn)
transformer = SurfaceTransform(source_subject='fsaverage6',
target_subject='fsaverage',
hemi={'L': 'lh', 'R': 'rh'}[hemi])
transformer.inputs.source_file = par_fn
transformer.inputs.out_file = par_fn.replace('fsaverage6', 'fsaverage')
# Disable on MAC OS X (SIP problem)
transformer.run()
r2 = get_rsq(test.loc[:, mask].values, predictions.values[:len(test), :]).to_frame('r2').T
r2.columns = test.loc[:, mask].columns
r2_df = pd.DataFrame(columns=test.columns)
r2_df = | pd.concat((r2_df, r2), axis=0) | pandas.concat |
#!/usr/bin/env python
# -*- coding: utf-8 -*-
import os
import pickle
import shutil
import sys
import tempfile
import numpy as np
from numpy import arange, nan
import pandas.testing as pdt
from pandas import DataFrame, MultiIndex, Series, to_datetime
# dependencies testing specific
import pytest
import recordlinkage
from recordlinkage.base import BaseCompareFeature
STRING_SIM_ALGORITHMS = [
'jaro', 'q_gram', 'cosine', 'jaro_winkler', 'dameraulevenshtein',
'levenshtein', 'lcs', 'smith_waterman'
]
NUMERIC_SIM_ALGORITHMS = ['step', 'linear', 'squared', 'exp', 'gauss']
FIRST_NAMES = [
u'Ronald', u'Amy', u'Andrew', u'William', u'Frank', u'Jessica', u'Kevin',
u'Tyler', u'Yvonne', nan
]
LAST_NAMES = [
u'Graham', u'Smith', u'Holt', u'Pope', u'Hernandez', u'Gutierrez',
u'Rivera', nan, u'Crane', u'Padilla'
]
STREET = [
u'<NAME>', nan, u'<NAME>', u'<NAME>', u'<NAME>',
u'<NAME>', u'Williams Trail', u'Durham Mountains', u'Anna Circle',
u'<NAME>'
]
JOB = [
u'Designer, multimedia', u'Designer, blown glass/stained glass',
u'Chiropractor', u'Engineer, mining', u'Quantity surveyor',
u'Phytotherapist', u'Teacher, English as a foreign language',
u'Electrical engineer', u'Research officer, government', u'Economist'
]
AGES = [23, 40, 70, 45, 23, 57, 38, nan, 45, 46]
# Run all tests in this file with:
# nosetests tests/test_compare.py
class TestData(object):
@classmethod
def setup_class(cls):
N_A = 100
N_B = 100
cls.A = DataFrame({
'age': np.random.choice(AGES, N_A),
'given_name': np.random.choice(FIRST_NAMES, N_A),
'lastname': np.random.choice(LAST_NAMES, N_A),
'street': np.random.choice(STREET, N_A)
})
cls.B = DataFrame({
'age': np.random.choice(AGES, N_B),
'given_name': np.random.choice(FIRST_NAMES, N_B),
'lastname': np.random.choice(LAST_NAMES, N_B),
'street': np.random.choice(STREET, N_B)
})
cls.A.index.name = 'index_df1'
cls.B.index.name = 'index_df2'
cls.index_AB = MultiIndex.from_arrays(
[arange(len(cls.A)), arange(len(cls.B))],
names=[cls.A.index.name, cls.B.index.name])
# Create a temporary directory
cls.test_dir = tempfile.mkdtemp()
@classmethod
def teardown_class(cls):
# Remove the test directory
shutil.rmtree(cls.test_dir)
class TestCompareApi(TestData):
"""General unittest for the compare API."""
def test_repr(self):
comp = recordlinkage.Compare()
comp.exact('given_name', 'given_name')
comp.string('given_name', 'given_name', method='jaro')
comp.numeric('age', 'age', method='step', offset=3, origin=2)
comp.numeric('age', 'age', method='step', offset=0, origin=2)
c_str = str(comp)
c_repr = repr(comp)
assert c_str == c_repr
start_str = '<{}'.format(comp.__class__.__name__)
assert c_str.startswith(start_str)
def test_instance_linking(self):
comp = recordlinkage.Compare()
comp.exact('given_name', 'given_name')
comp.string('given_name', 'given_name', method='jaro')
comp.numeric('age', 'age', method='step', offset=3, origin=2)
comp.numeric('age', 'age', method='step', offset=0, origin=2)
result = comp.compute(self.index_AB, self.A, self.B)
# returns a Series
assert isinstance(result, DataFrame)
# resulting series has a MultiIndex
assert isinstance(result.index, MultiIndex)
# indexnames are oke
assert result.index.names == [self.A.index.name, self.B.index.name]
assert len(result) == len(self.index_AB)
def test_instance_dedup(self):
comp = recordlinkage.Compare()
comp.string('given_name', 'given_name', method='jaro')
comp.numeric('age', 'age', method='step', offset=3, origin=2)
comp.numeric('age', 'age', method='step', offset=0, origin=2)
result = comp.compute(self.index_AB, self.A)
# returns a Series
assert isinstance(result, DataFrame)
# resulting series has a MultiIndex
assert isinstance(result.index, MultiIndex)
# indexnames are oke
assert result.index.names == [self.A.index.name, self.B.index.name]
assert len(result) == len(self.index_AB)
def test_label_linking(self):
comp = recordlinkage.Compare()
comp.compare_vectorized(
lambda s1, s2: np.ones(len(s1), dtype=np.int),
'given_name',
'given_name',
label='my_feature_label')
result = comp.compute(self.index_AB, self.A, self.B)
assert "my_feature_label" in result.columns.tolist()
def test_label_dedup(self):
comp = recordlinkage.Compare()
comp.compare_vectorized(
lambda s1, s2: np.ones(len(s1), dtype=np.int),
'given_name',
'given_name',
label='my_feature_label')
result = comp.compute(self.index_AB, self.A)
assert "my_feature_label" in result.columns.tolist()
def test_multilabel_none_linking(self):
def ones_np_multi(s1, s2):
return np.ones(len(s1)), np.ones((len(s1), 3))
def ones_pd_multi(s1, s2):
return (Series(np.ones(len(s1))), DataFrame(np.ones((len(s1), 3))))
comp = recordlinkage.Compare()
comp.string('given_name', 'given_name', method='jaro')
comp.compare_vectorized(
ones_np_multi,
'given_name',
'given_name')
comp.compare_vectorized(
ones_pd_multi,
'given_name',
'given_name')
result = comp.compute(self.index_AB, self.A, self.B)
assert [0, 1, 2, 3, 4, 5, 6, 7, 8] == \
result.columns.tolist()
def test_multilabel_linking(self):
def ones_np_multi(s1, s2):
return np.ones(len(s1)), np.ones((len(s1), 3))
def ones_pd_multi(s1, s2):
return (Series(np.ones(len(s1))), DataFrame(np.ones((len(s1), 3))))
comp = recordlinkage.Compare()
comp.string('given_name', 'given_name', method='jaro')
comp.compare_vectorized(
ones_np_multi,
'given_name',
'given_name',
label=['a', ['b', 'c', 'd']])
comp.compare_vectorized(
ones_pd_multi,
'given_name',
'given_name',
label=['e', ['f', 'g', 'h']])
result = comp.compute(self.index_AB, self.A, self.B)
assert [0, 'a', 'b', 'c', 'd', 'e', 'f', 'g', 'h'] == \
result.columns.tolist()
def test_multilabel_dedup(self):
def ones_np_multi(s1, s2):
return np.ones(len(s1)), np.ones((len(s1), 3))
def ones_pd_multi(s1, s2):
return (Series(np.ones(len(s1))), DataFrame(np.ones((len(s1), 3))))
comp = recordlinkage.Compare()
comp.string('given_name', 'given_name', method='jaro')
comp.compare_vectorized(
ones_np_multi,
'given_name',
'given_name',
label=['a', ['b', 'c', 'd']])
comp.compare_vectorized(
ones_pd_multi,
'given_name',
'given_name',
label=['e', ['f', 'g', 'h']])
result = comp.compute(self.index_AB, self.A)
assert [0, 'a', 'b', 'c', 'd', 'e', 'f', 'g', 'h'] == \
result.columns.tolist()
def test_multilabel_none_dedup(self):
def ones_np_multi(s1, s2):
return np.ones(len(s1)), np.ones((len(s1), 3))
def ones_pd_multi(s1, s2):
return (Series(np.ones(len(s1))), DataFrame(np.ones((len(s1), 3))))
comp = recordlinkage.Compare()
comp.string('given_name', 'given_name', method='jaro')
comp.compare_vectorized(
ones_np_multi,
'given_name',
'given_name')
comp.compare_vectorized(
ones_pd_multi,
'given_name',
'given_name')
result = comp.compute(self.index_AB, self.A)
assert [0, 1, 2, 3, 4, 5, 6, 7, 8] == \
result.columns.tolist()
def test_multilabel_error_dedup(self):
def ones(s1, s2):
return np.ones((len(s1), 2))
comp = recordlinkage.Compare()
comp.string('given_name', 'given_name', method='jaro')
comp.compare_vectorized(
ones, 'given_name', 'given_name', label=['a', 'b', 'c'])
with pytest.raises(ValueError):
comp.compute(self.index_AB, self.A)
def test_incorrect_collabels_linking(self):
comp = recordlinkage.Compare()
comp.string('given_name', 'given_name', method='jaro')
comp.compare_vectorized(lambda s1, s2: np.ones(len(s1), dtype=np.int),
"given_name", "not_existing_label")
with pytest.raises(KeyError):
comp.compute(self.index_AB, self.A, self.B)
def test_incorrect_collabels_dedup(self):
comp = recordlinkage.Compare()
comp.compare_vectorized(lambda s1, s2: np.ones(len(s1), dtype=np.int),
"given_name", "not_existing_label")
with pytest.raises(KeyError):
comp.compute(self.index_AB, self.A)
def test_compare_custom_vectorized_linking(self):
A = DataFrame({'col': ['abc', 'abc', 'abc', 'abc', 'abc']})
B = DataFrame({'col': ['abc', 'abd', 'abc', 'abc', '123']})
ix = MultiIndex.from_arrays([A.index.values, B.index.values])
# test without label
comp = recordlinkage.Compare()
comp.compare_vectorized(lambda s1, s2: np.ones(len(s1), dtype=np.int),
'col', 'col')
result = comp.compute(ix, A, B)
expected = DataFrame([1, 1, 1, 1, 1], index=ix)
pdt.assert_frame_equal(result, expected)
# test with label
comp = recordlinkage.Compare()
comp.compare_vectorized(
lambda s1, s2: np.ones(len(s1), dtype=np.int),
'col',
'col',
label='my_feature_label')
result = comp.compute(ix, A, B)
expected = DataFrame(
[1, 1, 1, 1, 1], index=ix, columns=['my_feature_label'])
pdt.assert_frame_equal(result, expected)
# def test_compare_custom_nonvectorized_linking(self):
# A = DataFrame({'col': [1, 2, 3, 4, 5]})
# B = DataFrame({'col': [1, 2, 3, 4, 5]})
# ix = MultiIndex.from_arrays([A.index.values, B.index.values])
# def custom_func(a, b):
# return np.int64(1)
# # test without label
# comp = recordlinkage.Compare()
# comp.compare_single(
# custom_func,
# 'col',
# 'col'
# )
# result = comp.compute(ix, A, B)
# expected = DataFrame([1, 1, 1, 1, 1], index=ix)
# pdt.assert_frame_equal(result, expected)
# # test with label
# comp = recordlinkage.Compare()
# comp.compare_single(
# custom_func,
# 'col',
# 'col',
# label='test'
# )
# result = comp.compute(ix, A, B)
# expected = DataFrame([1, 1, 1, 1, 1], index=ix, columns=['test'])
# pdt.assert_frame_equal(result, expected)
def test_compare_custom_instance_type(self):
A = DataFrame({'col': ['abc', 'abc', 'abc', 'abc', 'abc']})
B = DataFrame({'col': ['abc', 'abd', 'abc', 'abc', '123']})
ix = MultiIndex.from_arrays([A.index.values, B.index.values])
def call(s1, s2):
# this should raise on incorrect types
assert isinstance(s1, np.ndarray)
assert isinstance(s2, np.ndarray)
return np.ones(len(s1), dtype=np.int)
comp = recordlinkage.Compare()
comp.compare_vectorized(lambda s1, s2: np.ones(len(s1), dtype=np.int),
'col', 'col')
result = comp.compute(ix, A, B)
expected = DataFrame([1, 1, 1, 1, 1], index=ix)
pdt.assert_frame_equal(result, expected)
def test_compare_custom_vectorized_arguments_linking(self):
A = DataFrame({'col': ['abc', 'abc', 'abc', 'abc', 'abc']})
B = DataFrame({'col': ['abc', 'abd', 'abc', 'abc', '123']})
ix = MultiIndex.from_arrays([A.index.values, B.index.values])
# test without label
comp = recordlinkage.Compare()
comp.compare_vectorized(
lambda s1, s2, x: np.ones(len(s1), dtype=np.int) * x, 'col', 'col',
5)
result = comp.compute(ix, A, B)
expected = DataFrame([5, 5, 5, 5, 5], index=ix)
pdt.assert_frame_equal(result, expected)
# test with label
comp = recordlinkage.Compare()
comp.compare_vectorized(
lambda s1, s2, x: np.ones(len(s1), dtype=np.int) * x,
'col',
'col',
5,
label='test')
result = comp.compute(ix, A, B)
expected = DataFrame([5, 5, 5, 5, 5], index=ix, columns=['test'])
pdt.assert_frame_equal(result, expected)
# test with kwarg
comp = recordlinkage.Compare()
comp.compare_vectorized(
lambda s1, s2, x: np.ones(len(s1), dtype=np.int) * x,
'col',
'col',
x=5,
label='test')
result = comp.compute(ix, A, B)
expected = DataFrame([5, 5, 5, 5, 5], index=ix, columns=['test'])
pdt.assert_frame_equal(result, expected)
def test_compare_custom_vectorized_dedup(self):
A = DataFrame({'col': ['abc', 'abc', 'abc', 'abc', 'abc']})
ix = MultiIndex.from_arrays([[0, 1, 2, 3, 4], [1, 2, 3, 4, 0]])
# test without label
comp = recordlinkage.Compare()
comp.compare_vectorized(lambda s1, s2: np.ones(len(s1), dtype=np.int),
'col', 'col')
result = comp.compute(ix, A)
expected = DataFrame([1, 1, 1, 1, 1], index=ix)
pdt.assert_frame_equal(result, expected)
# test with label
comp = recordlinkage.Compare()
comp.compare_vectorized(
lambda s1, s2: np.ones(len(s1), dtype=np.int),
'col',
'col',
label='test')
result = comp.compute(ix, A)
expected = DataFrame([1, 1, 1, 1, 1], index=ix, columns=['test'])
pdt.assert_frame_equal(result, expected)
def test_compare_custom_vectorized_arguments_dedup(self):
A = DataFrame({'col': ['abc', 'abc', 'abc', 'abc', 'abc']})
ix = MultiIndex.from_arrays([[0, 1, 2, 3, 4], [1, 2, 3, 4, 0]])
# test without label
comp = recordlinkage.Compare()
comp.compare_vectorized(
lambda s1, s2, x: np.ones(len(s1), dtype=np.int) * x, 'col', 'col',
5)
result = comp.compute(ix, A)
expected = DataFrame([5, 5, 5, 5, 5], index=ix)
pdt.assert_frame_equal(result, expected)
# test with label
comp = recordlinkage.Compare()
comp.compare_vectorized(
lambda s1, s2, x: np.ones(len(s1), dtype=np.int) * x,
'col',
'col',
5,
label='test')
result = comp.compute(ix, A)
expected = DataFrame([5, 5, 5, 5, 5], index=ix, columns=['test'])
pdt.assert_frame_equal(result, expected)
def test_parallel_comparing_api(self):
# use single job
comp = recordlinkage.Compare(n_jobs=1)
comp.exact('given_name', 'given_name', label='my_feature_label')
result_single = comp.compute(self.index_AB, self.A, self.B)
result_single.sort_index(inplace=True)
# use two jobs
comp = recordlinkage.Compare(n_jobs=2)
comp.exact('given_name', 'given_name', label='my_feature_label')
result_2processes = comp.compute(self.index_AB, self.A, self.B)
result_2processes.sort_index(inplace=True)
# compare results
pdt.assert_frame_equal(result_single, result_2processes)
def test_parallel_comparing(self):
# use single job
comp = recordlinkage.Compare(n_jobs=1)
comp.exact('given_name', 'given_name', label='my_feature_label')
result_single = comp.compute(self.index_AB, self.A, self.B)
result_single.sort_index(inplace=True)
# use two jobs
comp = recordlinkage.Compare(n_jobs=2)
comp.exact('given_name', 'given_name', label='my_feature_label')
result_2processes = comp.compute(self.index_AB, self.A, self.B)
result_2processes.sort_index(inplace=True)
# use two jobs
comp = recordlinkage.Compare(n_jobs=4)
comp.exact('given_name', 'given_name', label='my_feature_label')
result_4processes = comp.compute(self.index_AB, self.A, self.B)
result_4processes.sort_index(inplace=True)
# compare results
pdt.assert_frame_equal(result_single, result_2processes)
pdt.assert_frame_equal(result_single, result_4processes)
def test_pickle(self):
# test if it is possible to pickle the Compare class
comp = recordlinkage.Compare()
comp.string('given_name', 'given_name')
comp.numeric('number', 'number')
comp.geo('lat', 'lng', 'lat', 'lng')
comp.date('before', 'after')
# do the test
pickle_path = os.path.join(self.test_dir, 'pickle_compare_obj.pickle')
pickle.dump(comp, open(pickle_path, 'wb'))
def test_manual_parallel_joblib(self):
# test if it is possible to pickle the Compare class
# This is only available for python 3. For python 2, it is not
# possible to pickle instancemethods. A workaround can be found at
# https://stackoverflow.com/a/29873604/8727928
if sys.version.startswith("3"):
# import joblib dependencies
from joblib import Parallel, delayed
# split the data into smaller parts
len_index = int(len(self.index_AB) / 2)
df_chunks = [self.index_AB[0:len_index], self.index_AB[len_index:]]
comp = recordlinkage.Compare()
comp.string('given_name', 'given_name')
comp.string('lastname', 'lastname')
comp.exact('street', 'street')
# do in parallel
Parallel(n_jobs=2)(
delayed(comp.compute)(df_chunks[i], self.A, self.B)
for i in [0, 1])
def test_indexing_types(self):
# test the two types of indexing
# this test needs improvement
A = DataFrame({'col': ['abc', 'abc', 'abc', 'abc', 'abc']})
B = DataFrame({'col': ['abc', 'abc', 'abc', 'abc', 'abc']})
B_reversed = B[::-1].copy()
ix = MultiIndex.from_arrays([np.arange(5), np.arange(5)])
# test with label indexing type
comp_label = recordlinkage.Compare(indexing_type='label')
comp_label.exact('col', 'col')
result_label = comp_label.compute(ix, A, B_reversed)
# test with position indexing type
comp_position = recordlinkage.Compare(indexing_type='position')
comp_position.exact('col', 'col')
result_position = comp_position.compute(ix, A, B_reversed)
assert (result_position.values == 1).all(axis=0)
pdt.assert_frame_equal(result_label, result_position)
def test_pass_list_of_features(self):
from recordlinkage.compare import FrequencyA, VariableA, VariableB
# setup datasets and record pairs
A = DataFrame({'col': ['abc', 'abc', 'abc', 'abc', 'abc']})
B = DataFrame({'col': ['abc', 'abc', 'abc', 'abc', 'abc']})
ix = MultiIndex.from_arrays([np.arange(5), np.arange(5)])
# test with label indexing type
features = [
VariableA('col', label='y1'),
VariableB('col', label='y2'),
FrequencyA('col', label='y3')
]
comp_label = recordlinkage.Compare(features=features)
result_label = comp_label.compute(ix, A, B)
assert list(result_label) == ["y1", "y2", "y3"]
class TestCompareFeatures(TestData):
def test_feature(self):
# test using classes and the base class
A = DataFrame({'col': ['abc', 'abc', 'abc', 'abc', 'abc']})
B = DataFrame({'col': ['abc', 'abd', 'abc', 'abc', '123']})
ix = MultiIndex.from_arrays([A.index.values, B.index.values])
feature = BaseCompareFeature('col', 'col')
feature._f_compare_vectorized = lambda s1, s2: np.ones(len(s1))
feature.compute(ix, A, B)
def test_feature_multicolumn_return(self):
# test using classes and the base class
A = DataFrame({'col': ['abc', 'abc', 'abc', 'abc', 'abc']})
B = DataFrame({'col': ['abc', 'abd', 'abc', 'abc', '123']})
ix = MultiIndex.from_arrays([A.index.values, B.index.values])
def ones(s1, s2):
return DataFrame(np.ones((len(s1), 3)))
feature = BaseCompareFeature('col', 'col')
feature._f_compare_vectorized = ones
result = feature.compute(ix, A, B)
assert result.shape == (5, 3)
def test_feature_multicolumn_input(self):
# test using classes and the base class
A = DataFrame({
'col1': ['abc', 'abc', 'abc', 'abc', 'abc'],
'col2': ['abc', 'abc', 'abc', 'abc', 'abc']
})
B = DataFrame({
'col1': ['abc', 'abd', 'abc', 'abc', '123'],
'col2': ['abc', 'abd', 'abc', 'abc', '123']
})
ix = MultiIndex.from_arrays([A.index.values, B.index.values])
feature = BaseCompareFeature(['col1', 'col2'], ['col1', 'col2'])
feature._f_compare_vectorized = \
lambda s1_1, s1_2, s2_1, s2_2: np.ones(len(s1_1))
feature.compute(ix, A, B)
class TestCompareExact(TestData):
"""Test the exact comparison method."""
def test_exact_str_type(self):
A = DataFrame({'col': ['abc', 'abc', 'abc', 'abc', 'abc']})
B = DataFrame({'col': ['abc', 'abd', 'abc', 'abc', '123']})
ix = MultiIndex.from_arrays([A.index.values, B.index.values])
expected = DataFrame([1, 0, 1, 1, 0], index=ix)
comp = recordlinkage.Compare()
comp.exact('col', 'col')
result = comp.compute(ix, A, B)
pdt.assert_frame_equal(result, expected)
def test_exact_num_type(self):
A = DataFrame({'col': [42, 42, 41, 43, nan]})
B = DataFrame({'col': [42, 42, 42, 42, 42]})
ix = MultiIndex.from_arrays([A.index.values, B.index.values])
expected = DataFrame([1, 1, 0, 0, 0], index=ix)
comp = recordlinkage.Compare()
comp.exact('col', 'col')
result = comp.compute(ix, A, B)
pdt.assert_frame_equal(result, expected)
def test_link_exact_missing(self):
A = DataFrame({'col': [u'a', u'b', u'c', u'd', nan]})
B = DataFrame({'col': [u'a', u'b', u'd', nan, nan]})
ix = MultiIndex.from_arrays([A.index.values, B.index.values])
comp = recordlinkage.Compare()
comp.exact('col', 'col', label='na_')
comp.exact('col', 'col', missing_value=0, label='na_0')
comp.exact('col', 'col', missing_value=9, label='na_9')
comp.exact('col', 'col', missing_value=nan, label='na_na')
comp.exact('col', 'col', missing_value='str', label='na_str')
result = comp.compute(ix, A, B)
# Missing values as default
expected = Series([1, 1, 0, 0, 0], index=ix, name='na_')
pdt.assert_series_equal(result['na_'], expected)
# Missing values as 0
expected = Series([1, 1, 0, 0, 0], index=ix, name='na_0')
pdt.assert_series_equal(result['na_0'], expected)
# Missing values as 9
expected = Series([1, 1, 0, 9, 9], index=ix, name='na_9')
pdt.assert_series_equal(result['na_9'], expected)
# Missing values as nan
expected = Series([1, 1, 0, nan, nan], index=ix, name='na_na')
pdt.assert_series_equal(result['na_na'], expected)
# Missing values as string
expected = Series([1, 1, 0, 'str', 'str'], index=ix, name='na_str')
pdt.assert_series_equal(result['na_str'], expected)
def test_link_exact_disagree(self):
A = DataFrame({'col': [u'a', u'b', u'c', u'd', nan]})
B = DataFrame({'col': [u'a', u'b', u'd', nan, nan]})
ix = MultiIndex.from_arrays([A.index.values, B.index.values])
comp = recordlinkage.Compare()
comp.exact('col', 'col', label='d_')
comp.exact('col', 'col', disagree_value=0, label='d_0')
comp.exact('col', 'col', disagree_value=9, label='d_9')
comp.exact('col', 'col', disagree_value=nan, label='d_na')
comp.exact('col', 'col', disagree_value='str', label='d_str')
result = comp.compute(ix, A, B)
# disagree values as default
expected = Series([1, 1, 0, 0, 0], index=ix, name='d_')
pdt.assert_series_equal(result['d_'], expected)
# disagree values as 0
expected = Series([1, 1, 0, 0, 0], index=ix, name='d_0')
pdt.assert_series_equal(result['d_0'], expected)
# disagree values as 9
expected = Series([1, 1, 9, 0, 0], index=ix, name='d_9')
pdt.assert_series_equal(result['d_9'], expected)
# disagree values as nan
expected = Series([1, 1, nan, 0, 0], index=ix, name='d_na')
pdt.assert_series_equal(result['d_na'], expected)
# disagree values as string
expected = Series([1, 1, 'str', 0, 0], index=ix, name='d_str')
pdt.assert_series_equal(result['d_str'], expected)
# tests/test_compare.py:TestCompareNumeric
class TestCompareNumeric(TestData):
"""Test the numeric comparison methods."""
def test_numeric(self):
A = DataFrame({'col': [1, 1, 1, nan, 0]})
B = DataFrame({'col': [1, 2, 3, nan, nan]})
ix = MultiIndex.from_arrays([A.index.values, B.index.values])
comp = recordlinkage.Compare()
comp.numeric('col', 'col', 'step', offset=2)
comp.numeric('col', 'col', method='step', offset=2)
comp.numeric('col', 'col', 'step', 2)
result = comp.compute(ix, A, B)
# Basics
expected = Series([1.0, 1.0, 1.0, 0.0, 0.0], index=ix, name=0)
pdt.assert_series_equal(result[0], expected)
# Basics
expected = Series([1.0, 1.0, 1.0, 0.0, 0.0], index=ix, name=1)
pdt.assert_series_equal(result[1], expected)
# Basics
expected = Series([1.0, 1.0, 1.0, 0.0, 0.0], index=ix, name=2)
pdt.assert_series_equal(result[2], expected)
def test_numeric_with_missings(self):
A = DataFrame({'col': [1, 1, 1, nan, 0]})
B = DataFrame({'col': [1, 1, 1, nan, nan]})
ix = MultiIndex.from_arrays([A.index.values, B.index.values])
comp = recordlinkage.Compare()
comp.numeric('col', 'col', scale=2)
comp.numeric('col', 'col', scale=2, missing_value=0)
comp.numeric('col', 'col', scale=2, missing_value=123.45)
comp.numeric('col', 'col', scale=2, missing_value=nan)
comp.numeric('col', 'col', scale=2, missing_value='str')
result = comp.compute(ix, A, B)
# Missing values as default
expected = Series([1.0, 1.0, 1.0, 0.0, 0.0], index=ix, name=0)
pdt.assert_series_equal(result[0], expected)
# Missing values as 0
expected = Series(
[1.0, 1.0, 1.0, 0.0, 0.0], index=ix, dtype=np.float64, name=1)
pdt.assert_series_equal(result[1], expected)
# Missing values as 123.45
expected = Series([1.0, 1.0, 1.0, 123.45, 123.45], index=ix, name=2)
pdt.assert_series_equal(result[2], expected)
# Missing values as nan
expected = Series([1.0, 1.0, 1.0, nan, nan], index=ix, name=3)
pdt.assert_series_equal(result[3], expected)
# Missing values as string
expected = Series(
[1, 1, 1, 'str', 'str'], index=ix, dtype=object, name=4)
pdt.assert_series_equal(result[4], expected)
@pytest.mark.parametrize("alg", NUMERIC_SIM_ALGORITHMS)
def test_numeric_algorithms(self, alg):
A = DataFrame({'col': [1, 1, 1, 1, 1]})
B = DataFrame({'col': [1, 2, 3, 4, 5]})
ix = MultiIndex.from_arrays([A.index.values, B.index.values])
comp = recordlinkage.Compare()
comp.numeric('col', 'col', method='step', offset=1, label='step')
comp.numeric(
'col', 'col', method='linear', offset=1, scale=2, label='linear')
comp.numeric(
'col', 'col', method='squared', offset=1, scale=2, label='squared')
comp.numeric(
'col', 'col', method='exp', offset=1, scale=2, label='exp')
comp.numeric(
'col', 'col', method='gauss', offset=1, scale=2, label='gauss')
result_df = comp.compute(ix, A, B)
result = result_df[alg]
# All values between 0 and 1.
assert (result >= 0.0).all()
assert (result <= 1.0).all()
if alg != 'step':
print(alg)
print(result)
# sim(scale) = 0.5
expected_bool = Series(
[False, False, False, True, False], index=ix, name=alg)
pdt.assert_series_equal(result == 0.5, expected_bool)
# sim(offset) = 1
expected_bool = Series(
[True, True, False, False, False], index=ix, name=alg)
pdt.assert_series_equal(result == 1.0, expected_bool)
# sim(scale) larger than 0.5
expected_bool = Series(
[False, False, True, False, False], index=ix, name=alg)
pdt.assert_series_equal((result > 0.5) & (result < 1.0),
expected_bool)
# sim(scale) smaller than 0.5
expected_bool = Series(
[False, False, False, False, True], index=ix, name=alg)
pdt.assert_series_equal((result < 0.5) & (result >= 0.0),
expected_bool)
@pytest.mark.parametrize("alg", NUMERIC_SIM_ALGORITHMS)
def test_numeric_algorithms_errors(self, alg):
# scale negative
if alg != "step":
with pytest.raises(ValueError):
comp = recordlinkage.Compare()
comp.numeric('age', 'age', method=alg, offset=2, scale=-2)
comp.compute(self.index_AB, self.A, self.B)
# offset negative
with pytest.raises(ValueError):
comp = recordlinkage.Compare()
comp.numeric('age', 'age', method=alg, offset=-2, scale=-2)
comp.compute(self.index_AB, self.A, self.B)
def test_numeric_does_not_exist(self):
# raise when algorithm doesn't exists
A = DataFrame({'col': [1, 1, 1, nan, 0]})
B = DataFrame({'col': [1, 1, 1, nan, nan]})
ix = MultiIndex.from_arrays([A.index.values, B.index.values])
comp = recordlinkage.Compare()
comp.numeric('col', 'col', method='unknown_algorithm')
pytest.raises(ValueError, comp.compute, ix, A, B)
# tests/test_compare.py:TestCompareDates
class TestCompareDates(TestData):
"""Test the exact comparison method."""
def test_dates(self):
A = DataFrame({
'col':
to_datetime(
['2005/11/23', nan, '2004/11/23', '2010/01/10', '2010/10/30'])
})
B = DataFrame({
'col':
to_datetime([
'2005/11/23', '2010/12/31', '2005/11/23', '2010/10/01',
'2010/9/30'
])
})
ix = MultiIndex.from_arrays([A.index.values, B.index.values])
comp = recordlinkage.Compare()
comp.date('col', 'col')
result = comp.compute(ix, A, B)[0]
expected = Series([1, 0, 0, 0.5, 0.5], index=ix, name=0)
pdt.assert_series_equal(result, expected)
def test_date_incorrect_dtype(self):
A = DataFrame({
'col':
['2005/11/23', nan, '2004/11/23', '2010/01/10', '2010/10/30']
})
B = DataFrame({
'col': [
'2005/11/23', '2010/12/31', '2005/11/23', '2010/10/01',
'2010/9/30'
]
})
ix = MultiIndex.from_arrays([A.index.values, B.index.values])
A['col1'] = to_datetime(A['col'])
B['col1'] = to_datetime(B['col'])
comp = recordlinkage.Compare()
comp.date('col', 'col1')
pytest.raises(ValueError, comp.compute, ix, A, B)
comp = recordlinkage.Compare()
comp.date('col1', 'col')
pytest.raises(ValueError, comp.compute, ix, A, B)
def test_dates_with_missings(self):
A = DataFrame({
'col':
to_datetime(
['2005/11/23', nan, '2004/11/23', '2010/01/10', '2010/10/30'])
})
B = DataFrame({
'col':
to_datetime([
'2005/11/23', '2010/12/31', '2005/11/23', '2010/10/01',
'2010/9/30'
])
})
ix = MultiIndex.from_arrays([A.index.values, B.index.values])
comp = recordlinkage.Compare()
comp.date('col', 'col', label='m_')
comp.date('col', 'col', missing_value=0, label='m_0')
comp.date('col', 'col', missing_value=123.45, label='m_float')
comp.date('col', 'col', missing_value=nan, label='m_na')
comp.date('col', 'col', missing_value='str', label='m_str')
result = comp.compute(ix, A, B)
# Missing values as default
expected = Series([1, 0, 0, 0.5, 0.5], index=ix, name='m_')
pdt.assert_series_equal(result['m_'], expected)
# Missing values as 0
expected = Series([1, 0, 0, 0.5, 0.5], index=ix, name='m_0')
pdt.assert_series_equal(result['m_0'], expected)
# Missing values as 123.45
expected = Series([1, 123.45, 0, 0.5, 0.5], index=ix, name='m_float')
pdt.assert_series_equal(result['m_float'], expected)
# Missing values as nan
expected = Series([1, nan, 0, 0.5, 0.5], index=ix, name='m_na')
pdt.assert_series_equal(result['m_na'], expected)
# Missing values as string
expected = Series(
[1, 'str', 0, 0.5, 0.5], index=ix, dtype=object, name='m_str')
pdt.assert_series_equal(result['m_str'], expected)
def test_dates_with_swap(self):
months_to_swap = [(9, 10, 123.45), (10, 9, 123.45), (1, 2, 123.45),
(2, 1, 123.45)]
A = DataFrame({
'col':
to_datetime(
['2005/11/23', nan, '2004/11/23', '2010/01/10', '2010/10/30'])
})
B = DataFrame({
'col':
to_datetime([
'2005/11/23', '2010/12/31', '2005/11/23', '2010/10/01',
'2010/9/30'
])
})
ix = MultiIndex.from_arrays([A.index.values, B.index.values])
comp = recordlinkage.Compare()
comp.date('col', 'col', label='s_')
comp.date(
'col', 'col', swap_month_day=0, swap_months='default', label='s_1')
comp.date(
'col',
'col',
swap_month_day=123.45,
swap_months='default',
label='s_2')
comp.date(
'col',
'col',
swap_month_day=123.45,
swap_months=months_to_swap,
label='s_3')
comp.date(
'col',
'col',
swap_month_day=nan,
swap_months='default',
missing_value=nan,
label='s_4')
comp.date('col', 'col', swap_month_day='str', label='s_5')
result = comp.compute(ix, A, B)
# swap_month_day as default
expected = Series([1, 0, 0, 0.5, 0.5], index=ix, name='s_')
pdt.assert_series_equal(result['s_'], expected)
# swap_month_day and swap_months as 0
expected = Series([1, 0, 0, 0, 0.5], index=ix, name='s_1')
pdt.assert_series_equal(result['s_1'], expected)
# swap_month_day 123.45 (float)
expected = Series([1, 0, 0, 123.45, 0.5], index=ix, name='s_2')
pdt.assert_series_equal(result['s_2'], expected)
# swap_month_day and swap_months 123.45 (float)
expected = Series([1, 0, 0, 123.45, 123.45], index=ix, name='s_3')
pdt.assert_series_equal(result['s_3'], expected)
# swap_month_day and swap_months as nan
expected = Series([1, nan, 0, nan, 0.5], index=ix, name='s_4')
pdt.assert_series_equal(result['s_4'], expected)
# swap_month_day as string
expected = Series(
[1, 0, 0, 'str', 0.5], index=ix, dtype=object, name='s_5')
pdt.assert_series_equal(result['s_5'], expected)
# tests/test_compare.py:TestCompareGeo
class TestCompareGeo(TestData):
"""Test the geo comparison method."""
def test_geo(self):
# Utrecht, Amsterdam, Rotterdam (Cities in The Netherlands)
A = DataFrame({
'lat': [52.0842455, 52.3747388, 51.9280573],
'lng': [5.0124516, 4.7585305, 4.4203581]
})
B = DataFrame({
'lat': [52.3747388, 51.9280573, 52.0842455],
'lng': [4.7585305, 4.4203581, 5.0124516]
})
ix = MultiIndex.from_arrays([A.index.values, B.index.values])
comp = recordlinkage.Compare()
comp.geo(
'lat', 'lng', 'lat', 'lng', method='step',
offset=50) # 50 km range
result = comp.compute(ix, A, B)
# Missing values as default [36.639460, 54.765854, 44.092472]
expected = Series([1.0, 0.0, 1.0], index=ix, name=0)
pdt.assert_series_equal(result[0], expected)
def test_geo_batch(self):
# Utrecht, Amsterdam, Rotterdam (Cities in The Netherlands)
A = DataFrame({
'lat': [52.0842455, 52.3747388, 51.9280573],
'lng': [5.0124516, 4.7585305, 4.4203581]
})
B = DataFrame({
'lat': [52.3747388, 51.9280573, 52.0842455],
'lng': [4.7585305, 4.4203581, 5.0124516]
})
ix = MultiIndex.from_arrays([A.index.values, B.index.values])
comp = recordlinkage.Compare()
comp.geo(
'lat', 'lng', 'lat', 'lng', method='step', offset=1, label='step')
comp.geo(
'lat',
'lng',
'lat',
'lng',
method='linear',
offset=1,
scale=2,
label='linear')
comp.geo(
'lat',
'lng',
'lat',
'lng',
method='squared',
offset=1,
scale=2,
label='squared')
comp.geo(
'lat',
'lng',
'lat',
'lng',
method='exp',
offset=1,
scale=2,
label='exp')
comp.geo(
'lat',
'lng',
'lat',
'lng',
method='gauss',
offset=1,
scale=2,
label='gauss')
result_df = comp.compute(ix, A, B)
print(result_df)
for alg in ['step', 'linear', 'squared', 'exp', 'gauss']:
result = result_df[alg]
# All values between 0 and 1.
assert (result >= 0.0).all()
assert (result <= 1.0).all()
def test_geo_does_not_exist(self):
# Utrecht, Amsterdam, Rotterdam (Cities in The Netherlands)
A = DataFrame({
'lat': [52.0842455, 52.3747388, 51.9280573],
'lng': [5.0124516, 4.7585305, 4.4203581]
})
B = DataFrame({
'lat': [52.3747388, 51.9280573, 52.0842455],
'lng': [4.7585305, 4.4203581, 5.0124516]
})
ix = MultiIndex.from_arrays([A.index.values, B.index.values])
comp = recordlinkage.Compare()
comp.geo('lat', 'lng', 'lat', 'lng', method='unknown')
pytest.raises(ValueError, comp.compute, ix, A, B)
class TestCompareStrings(TestData):
"""Test the exact comparison method."""
def test_defaults(self):
# default algorithm is levenshtein algorithm
# test default values are indentical to levenshtein
A = DataFrame({
'col': [u'str_abc', u'str_abc', u'str_abc', nan, u'hsdkf']
})
B = DataFrame({'col': [u'str_abc', u'str_abd', u'jaskdfsd', nan, nan]})
ix = MultiIndex.from_arrays([A.index.values, B.index.values])
comp = recordlinkage.Compare()
comp.string('col', 'col', label='default')
comp.string('col', 'col', method='levenshtein', label='with_args')
result = comp.compute(ix, A, B)
pdt.assert_series_equal(
result['default'].rename(None),
result['with_args'].rename(None)
)
def test_fuzzy(self):
A = DataFrame({
'col': [u'str_abc', u'str_abc', u'str_abc', nan, u'hsdkf']
})
B = DataFrame({'col': [u'str_abc', u'str_abd', u'jaskdfsd', nan, nan]})
ix = MultiIndex.from_arrays([A.index.values, B.index.values])
comp = recordlinkage.Compare()
comp.string('col', 'col', method='jaro', missing_value=0)
comp.string('col', 'col', method='q_gram', missing_value=0)
comp.string('col', 'col', method='cosine', missing_value=0)
comp.string('col', 'col', method='jaro_winkler', missing_value=0)
comp.string('col', 'col', method='dameraulevenshtein', missing_value=0)
comp.string('col', 'col', method='levenshtein', missing_value=0)
result = comp.compute(ix, A, B)
print(result)
assert result.notnull().all(1).all(0)
assert (result[result.notnull()] >= 0).all(1).all(0)
assert (result[result.notnull()] <= 1).all(1).all(0)
def test_threshold(self):
A = DataFrame({'col': [u"gretzky", u"gretzky99", u"gretzky", u"gretzky"]})
B = DataFrame({'col': [u"gretzky", u"gretzky", nan, u"wayne"]})
ix = MultiIndex.from_arrays([A.index.values, B.index.values])
comp = recordlinkage.Compare()
comp.string(
'col',
'col',
method="levenshtein",
threshold=0.5,
missing_value=2.0,
label="x_col1"
)
comp.string(
'col',
'col',
method="levenshtein",
threshold=1.0,
missing_value=0.5,
label="x_col2"
)
comp.string(
'col',
'col',
method="levenshtein",
threshold=0.0,
missing_value=nan,
label="x_col3"
)
result = comp.compute(ix, A, B)
expected = Series([1.0, 1.0, 2.0, 0.0], index=ix, name="x_col1")
pdt.assert_series_equal(result["x_col1"], expected)
expected = Series([1.0, 0.0, 0.5, 0.0], index=ix, name="x_col2")
pdt.assert_series_equal(result["x_col2"], expected)
expected = Series([1.0, 1.0, nan, 1.0], index=ix, name="x_col3")
pdt.assert_series_equal(result["x_col3"], expected)
@pytest.mark.parametrize("alg", STRING_SIM_ALGORITHMS)
def test_incorrect_input(self, alg):
A = DataFrame({'col': [1, 1, 1, nan, 0]})
B = DataFrame({'col': [1, 1, 1, nan, nan]})
ix = MultiIndex.from_arrays([A.index.values, B.index.values])
with pytest.raises(Exception):
comp = recordlinkage.Compare()
comp.string('col', 'col', method=alg)
comp.compute(ix, A, B)
@pytest.mark.parametrize("alg", STRING_SIM_ALGORITHMS)
def test_string_algorithms_nan(self, alg):
A = DataFrame({'col': [u"nan", nan, nan, nan, nan]})
B = DataFrame({'col': [u"nan", nan, nan, nan, nan]})
ix = MultiIndex.from_arrays([A.index.values, B.index.values])
comp = recordlinkage.Compare()
comp.string('col', 'col', method=alg)
result = comp.compute(ix, A, B)[0]
expected = Series([1.0, 0.0, 0.0, 0.0, 0.0], index=ix, name=0)
pdt.assert_series_equal(result, expected)
comp = recordlinkage.Compare()
comp.string('col', 'col', method=alg, missing_value=nan)
result = comp.compute(ix, A, B)[0]
expected = Series([1.0, nan, nan, nan, nan], index=ix, name=0)
pdt.assert_series_equal(result, expected)
comp = recordlinkage.Compare()
comp.string('col', 'col', method=alg, missing_value=9.0)
result = comp.compute(ix, A, B)[0]
expected = Series([1.0, 9.0, 9.0, 9.0, 9.0], index=ix, name=0)
| pdt.assert_series_equal(result, expected) | pandas.testing.assert_series_equal |
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
"""
Core classes and functions
============================
Entities & accounts
--------------------
The system is built upon the concept of *accounts* and
*transactions*.
An *account* is wehere the money goes to (or comes from) and
can be a person, company or a generic destination like for example 'expenses.'
Subaccounts are used for grouping
and better organisation.
The dot `.` sign is used to denote an entity and its (sub)accounts
Example of an entity with a subaccount:
`Equity.bank.savings`
Transactions
--------------
Transaction define money flow. In its basic form it is a transfer from A to B.
A tranaction may be taxed (with a VAT for example)
"""
from collections import UserList, UserDict
from typing import Tuple
import yaml
import wimm.utils as utils
import pandas as pd
import wimm
from dataclasses import dataclass, asdict
def parse_account(s):
""" parse entity and account string """
return s.strip().split('.')
def load_data(name, db_path):
""" load data from yaml """
import wimm.structure as structure
fcns = {'balance': load_start_balance,
'transactions': Transactions.from_yaml,
'invoices': Invoices.from_yaml}
p = db_path / structure.files[name]
assert p.exists(), f"File {p} not found"
return fcns[name](p)
def load_start_balance(yaml_file):
d = yaml.load(open(yaml_file), Loader=yaml.SafeLoader)
return pd.Series(d)
def get_account(item):
""" return account from an item. Can be a string or a dict """
try:
account = item['account'] # try value from dict
except TypeError:
account = item
return account
def balance(transactions, start_balance=None, invoices=None, depth=None):
""" calculate balance """
accounts = transactions.process()
if start_balance is not None:
accounts = accounts.add(start_balance, fill_value=0)
# if invoices is not None:
# inv_acc = invoices.to_accounts() # convert to series
# accounts = accounts.add(inv_acc, fill_value=0)
names = accounts.index.to_list()
if depth is None:
return accounts
else:
return accounts.groupby(utils.names_to_labels(names, depth)).sum()
class ListPlus(UserList):
""" base extensions for a list """
def __init__(self, *args, cls_factory=None, **kwargs):
super().__init__(*args, **kwargs)
self.cls_factory = cls_factory
if self.cls_factory is not None:
self.data = [self.cls_factory(**d) for d in self.data]
def to_yaml(self, yaml_file=None, confirm=False):
""" write to file or return string """
data = [utils.to_dict(obj) for obj in self.data]
if yaml_file:
utils.save_yaml(yaml_file, data, ask_confirmation=confirm)
return yaml.dump(data, sort_keys=False)
@classmethod
def from_yaml(cls, yaml_file):
""" create class from a yaml file """
data = yaml.load(open(yaml_file), Loader=yaml.SafeLoader)
# if cls.cls_factory is None:
return cls(data)
# else:
# return cls( [cls.cls_factory.from_dict(d) for d in data])
class Transaction(UserDict):
date: str
description: str
transfers: dict
def __init__(self, *args, **kwargs):
super().__init__(*args, **kwargs)
defaults = [('date', None),
('description', ''),
('transfers', {})]
for k, v in defaults:
self.setdefault(k, v)
self._check_totals()
def _check_totals(self):
total = 0
missing = None
for k, v in self.transfers.items():
if v is None:
if missing is None:
missing = k
else:
raise ValueError('More than one entry is missing')
else:
total += v
if missing:
self.transfers[missing] = -total
def __getattr__(self, name):
if name in self.data:
return self.data[name]
else:
raise AttributeError(name)
def to_records(self):
""" convert to simple account operations """
return [{'date': self.date, 'account': acct, 'amount': amount} for acct, amount in self.transfers.items()]
@classmethod
def from_dict(cls, d):
return cls(**d)
def to_dict(self, compact=False):
""" save to dict (dropping None values if `compact` is True) """
if compact:
return {k: v for k, v in self.items() if v is not None}
else:
return self
def to_yaml(self, compact=True):
return yaml.dump([self.to_dict(compact)], sort_keys=False)
@classmethod
def from_v1(cls, tr):
""" create from old (v1) dict type """
d = {'date': tr['date'],
'description': tr.get('description', None),
'transfers': {tr['from']: -tr['amount'],
tr['to']: tr['amount']}
}
return cls(**d)
class Transactions(ListPlus):
""" transactons class, extension of a list """
def __init__(self, *args, **kwargs):
super().__init__(*args, cls_factory=Transaction, **kwargs)
def to_records(self):
for tr in self.data:
for rec in tr.to_records():
yield rec
def process(self):
""" return accounts and their balances """
return self.to_df().groupby(['account']).sum()['amount']
def to_df(self, date_range: Tuple = None) -> pd.DataFrame:
""" transactions as DataFrame """
df = pd.DataFrame.from_records(self.to_records())
df['date'] = pd.to_datetime(df['date'])
if date_range is None:
return df
# select dates
mask = (df.date >= date_range[0]) & (df.date < date_range[1])
return df[mask]
class Invoice(UserDict):
def __init__(self, *args, **kwargs):
super().__init__(*args, **kwargs)
keys = ['id', 'amount', 'tax', 'date', 'from', 'to',
'description', 'attachment', 'due_date', 'ext_name']
vals = ['INV00_000', 0.0, None, None, 'Uncategorized',
'Uncategorized', None, None, None, 'ext_company_name']
for k, v in zip(keys, vals):
self.setdefault(k, v)
def __getattr__(self, name):
if name in self.data:
return self.data[name]
else:
raise AttributeError(name)
@property
def prefix(self):
return self['id'][:3]
def validate(self):
utils.validate(self.data['id'], "IN([A-Z]{1}[0-9]{2}_[0-9]{3})")
utils.validate(self.data['date'], "([0-9]{4}-[0-9]{2}-[0-9]{2})")
def set_accounts(self, accounts=None):
if accounts is None:
params = {'invoice_id': self.id, 'ext_name': self.ext_name}
accounts = utils.invoice_accounts(
self.prefix, params, 'invoice_accounts')
self.data['from'] = accounts['from']
self.data['to'] = accounts['to']
def transaction(self):
""" single transaction for an invoice """
# TODO: refactor. Ok, it's late and I don't have time to rewrite all the old code.
# so I'll just gently wrap it in a new function.
trs = [Transaction.from_v1(tr) for tr in self._transactions()]
tr = trs[0]
if len(trs) == 2:
tr.transfers = {**tr.transfers, **trs[1].transfers}
return tr
def _transactions(self):
""" return ivoice transactions as a list, `old style` transactions """
trs = Transactions()
# ----- invoice transaction
trs.append({'date': self.date,
'description': 'Invoice '+self.id,
'amount': self.amount,
'from': self['from'],
'to': self['to']})
# ----- tax transaction
if self.tax:
params = {'invoice_id': self.id, 'ext_name': self.ext_name}
accounts = utils.invoice_accounts(
self.prefix, params, 'tax_accounts')
trs.append({'date': self.date,
'description': 'Tax for invoice '+self.id,
'amount': self.tax,
**accounts})
return trs
def to_yaml(self):
return yaml.dump(self.data, sort_keys=False)
def rest_amount(self):
return self.amount - self.amount_payed
def __repr__(self):
return f"{self.id} amount:{self.amount:<10} {self['from']} --> {self.to}"
class Invoices(ListPlus):
def __init__(self, *args, **kwargs):
super().__init__(*args, cls_factory=Invoice, **kwargs)
def get_by_id(self, invoice_id):
""" get a invoice(s) by id
id may be a partial string, with a wildcard *. Example INS*
"""
if invoice_id[-1] == '*': # multiple matching
pat = invoice_id[:-1]
n = len(pat)
matches = []
for inv in self.data:
if inv['id'][:n] == pat:
matches.append(inv)
return Invoices(matches).get_sorted_by('id')
else:
for inv in self.data: # single matching
if inv['id'] == invoice_id:
return inv
raise KeyError('id not found')
def get_next_id(self, prefix):
""" get next available invoice number for a prefix """
try:
invoice_id = self.get_by_id(prefix+'*')[-1]['id']
except IndexError: # prefix not found, make new one
return f"{prefix}{utils.timestamp('%y')}_001"
nr = int(invoice_id[-3:]) + 1
return invoice_id[:-3] + '%03d' % nr
def get_sorted_by(self, key, reverse=False):
return sorted(self,
key=lambda x: x[key],
reverse=reverse)
def to_df(self):
""" convert to DataFrame """
return | pd.DataFrame.from_records(self.data) | pandas.DataFrame.from_records |
import numpy as np
import json
import os
import collections
import pandas as pd
import seaborn as sns; sns.set()
import matplotlib.pyplot as plt
import subprocess
import game_py
def count_last_positions(dir):
pos = set()
total_pos = []
for f in os.listdir(dir):
game = json.load(open(dir + f))
for p in game['history']:
pos.add(str(p))
total_pos.append(str(p))
print(len(total_pos))
print(len(pos))
def elo_plot(file, save_to):
elo = pd.read_csv(file, delimiter=r"\s+")
elo = elo.sort_values(by=['Name'], ascending=True)
ax = sns.catplot(x="Name", y="Elo", kind="point", data=elo)
ax.set(xlabel='Generation', ylabel='Elo', title= "Elo progression by generation with MCTS")
plt.gcf().set_size_inches(8, 5)
plt.savefig(save_to)
def agreement_plot(file, save_to):
agree = | pd.read_csv(file) | pandas.read_csv |
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
"""
Created on Mon Oct 15 16:41:37 2018
@author: krzysztof
This module contains utilities useful when performing data analysis and drug sensitivity prediction with
Genomics of Drug Sensitivity in Cancer (GDSC) database.
Main utilities are Drug classes and Experiment class. All classes beginning with a word "Drug" represent the compound
coming from GDSC. There is a separate class for every corresponding experiment setup and genomic feature space. All Drug
classes contain methods for extraction and storage of proper input data. Available data types include: gene expression, binary copy number and coding variants, and cell line tissue type. The set of considered genes is represented as "targets"
attribute of Drug classes.
The Experiment class is dedicated for storage and analysis of results coming from machine learning experiments. Actual
machine learning is done outside of a class. The Experiment class have methods for storage, analysis and visualisation
of results.
Classes:
Drug: Basic class representing a compound from GDSC.
DrugWithDrugBank: Inherits from Drug, accounts for target genes from DrugBank database.
DrugGenomeWide: Inherits from Drug, designed for using genome-wide gene exression as input data.
DrugDirectReactome: Inherits from DrugWithDrugBank, uses only input data related to target genes resulting
from direct compound-pathway matching from Reactome.
DrugWithGenesInSamePathways: Inherits from DrugWithDrugBank, uses only input data related to genes that belong in
the same pathways as target genes.
Experiment: Designed to store and analyze results coming from machine learning experiments.
"""
# Imports
import pandas as pd
import numpy as np
import time
import pickle
import matplotlib.pyplot as plt
import seaborn as sns
from scipy.stats import pearsonr
import collections
# Sklearn imports
from scipy.stats import pearsonr
from sklearn.linear_model import ElasticNet
from sklearn import model_selection
from sklearn import metrics
from sklearn import preprocessing
from sklearn.dummy import DummyRegressor
from sklearn.pipeline import Pipeline
from sklearn import feature_selection
from sklearn.svm import SVR
from sklearn.ensemble import RandomForestRegressor
from sklearn.base import clone
# General imports
import multiprocessing
import numpy as np
import pandas as pd
import time
import sys
import dill
import warnings
import matplotlib.pyplot as plt
import seaborn as sns
from scipy.stats import pearsonr
import collections
# Sklearn imports
from sklearn.preprocessing import StandardScaler
from sklearn.ensemble import RandomForestRegressor
from sklearn import model_selection
from sklearn.pipeline import Pipeline
from sklearn import metrics
from sklearn.dummy import DummyRegressor
from sklearn.linear_model import Lasso, ElasticNet
from stability_selection import StabilitySelection
#################################################################################################################
# Drug class
#################################################################################################################
class Drug(object):
"""Class representing compound from GDSC database.
This is the most basic, parent class. Different experimental settings will use more specific,
children classes. Main function of the class is to create and store input data corresponding to a given
drug. Five types of data are considered: gene expression, copy number variants, coding variants, gene expression
signatures, and tumor tissue type. Class instances are initialized with four basic drug properties: ID, name, gene
targets and target pathway. Data attributes are stored as pandas DataFrames and are filled using data files
from GDSC via corresponding methods.
Attributes:
gdsc_id (int): ID from GDSC website.
name (string): Drug name.
targets (list of strings): Drug's target gene names (HGNC).
target_pathway (string): Drug's target pathway as provided in GDSC annotations.
ensembl targets (list of strings): Drug's target genes ensembl IDs. Can have different length
than "targets" because some gene names may not be matched during mapping. Ensembl IDs are
needed for gene expression data.
map_from_hgnc_to_ensembl (dictionary): Dictionary mapping from gene names to ensembl IDs. Created
after calling the "load_mappings" method.
map_from_ensembl_to_hgnc (dictionary): Dictionary mapping from ensembl IDs to gene names. Created
after calling the "load_mappings" method.
total_no_samples_screened (int): Number of cell lines screened for that drug. Created after
calling the "extract_drug_response_data" method.
response_data (DataFrame): DataFrame with screened cell lines for that drug and corresponding AUC or
IC50 values. Created after calling the "extract_drug_response_data" method.
screened_cell_lines (list of ints): list containing COSMIC IDs representing cell lines screened for
that drug. Created after calling the "extract_screened_cell_lines" method.
gene_expression_data (DataFrame): DataFrame with gene expression data, considering only
target genes. Created after calling the "extract_gene_expression" method
mutation_data (DataFrame): DataFrame with binary calls for coding variants, considering only
target genes. Created after calling the "extract_mutation_data" method.
cnv_data (DataFrame): DataFrame with binary calls for copu number variants, considering only
target genes. Created after calling the "extract_cnv_data" method.
tissue_data (DataFrame): DataFrame with dummy encoded tumor tissue types in screened cell lines.
Dummy encoding results in 13 binary features. Created after calling the
"extract_tissue_data" method.
full_data (DataFrame): DataFrame with combined data coming from given set of genetic data
classes.
Methods:
Instance methods:
__init__: Initialize a Drug instance.
__repr__: Return string representation of an instance, as a command which can be used to create
this instance.
__str__: Return string representation of an instance.
extract_drug_response_data: Generate a DataFrame with drug-response data.
extract_screened_cell_lines: Generate a list of COSMIC IDs representing cell lines screened for that
drug.
extract_gene_expression: Generate a DataFrame with gene expression data for drug's screened cell lines
extract_mutation_data: Generate a DataFrame with binary calls for coding variants.
extract_cnv_data: Generate a DataFrame with binary calls for copy number variants.
extract_cnv_data_faster: Generate a DataFrame with binary calls for copy number variants.
extract_tissue_data: Generate a DataFrame with dummy encoded tissue types.
extract_merck_signatures_data: Generate a DataFrame with gene expression signatures provided by Merck.
concatenate_data: Generate a DataFrame containing all desired genetic data classes. Available data
classes are: gene expression, coding variants, cnv variants and tissue type.
create_full_data: Combines above data extraction methods in order to create desired input data
for the drug with one method call. Returns the full data and saves it in corresponding instance's
field.
return_full_data: Combines above data extraction methods in order to create desired input data
for the drug with one method call. Returns the full data but does not save it.
Class methods:
load_mappings: Load appropriate dictionaries mapping between ensembl and HGNC.
Static methods:
create_drugs: Create a dictionary of Drug class objects, each referenced by it's ID
(keys are drug GDSC ID's)
load_data: Load all needed data files as DataFrames with one function call.
"""
# Class variables
map_from_hgnc_to_ensembl = None
map_from_ensembl_to_hgnc = None
# Instance methods
def __init__(self, gdsc_id, name, targets, target_pathway):
"""Intiliaze the class instance with four basic attributes. "Targets" are gene names
and get mapped into Ensembl IDs using class mapping variable."""
self.gdsc_id = gdsc_id
self.name = name
self.targets = targets
self.target_pathway = target_pathway
self.ensembl_targets = []
for x in self.targets:
try:
self.ensembl_targets.append(self.map_from_hgnc_to_ensembl[x])
except KeyError:
pass
def extract_drug_response_data(self, sensitivity_profiles_df, metric="AUC"):
"""Generate a DataFrame containing reponses for every cell line screened for that drug.
Arguments:
sensitivity_profiles_df (DataFrame): DataFrame of drug response data from GDSC.
metric (string): Which statistic to use as a response metric (default "AUC").
Returns:
None
"""
df = sensitivity_profiles_df[sensitivity_profiles_df.DRUG_ID == self.gdsc_id][
["COSMIC_ID", metric]]
df.columns = ["cell_line_id", metric] # Insert column with samples ID
self.total_no_samples_screened = df.shape[0] # Record how many screened cell lines for drug
self.response_data = df # Put DataFrame into corresponding field
def extract_screened_cell_lines(self, sensitivity_profiles_df):
"""Generate set of cell lines screened for that drug.
Arguments:
sensitivity_profiles_df (DataFrame): DataFrame of drug response data from GDSC.
Returns:
None
"""
self.screened_cell_lines = list(
sensitivity_profiles_df[sensitivity_profiles_df.DRUG_ID == self.gdsc_id]["COSMIC_ID"])
def extract_gene_expression(self, gene_expression_df):
"""Generate DataFrame of gene expression data for cell lines screened for this drug, only
considering drug's target genes.
Arguments:
gene_expression_df (DataFrame): Original GDSC gene expression DataFrame.
sensitivity_profiles_df (DataFrame): DataFrame of drug response data from GDSC.
Returns:
None
"""
cell_lines_str = [] # Gene expressesion DF column names are strings
for x in self.screened_cell_lines:
cell_lines_str.append(str(x))
cl_to_extract = []
for x in cell_lines_str:
if x in list(gene_expression_df.columns):
cl_to_extract.append(x) # Extract only cell lines contained in gene expression data
gene_expr = gene_expression_df[
gene_expression_df.ensembl_gene.isin(self.ensembl_targets)][["ensembl_gene"] + cl_to_extract]
gene_expr_t = gene_expr.transpose()
columns = list(gene_expr_t.loc["ensembl_gene"])
gene_expr_t.columns = columns
gene_expr_t = gene_expr_t.drop(["ensembl_gene"])
rows = list(gene_expr_t.index)
gene_expr_t.insert(0, "cell_line_id", rows) # Insert columns with cell line IDs
gene_expr_t.reset_index(drop=True, inplace=True)
gene_expr_t["cell_line_id"] = pd.to_numeric(gene_expr_t["cell_line_id"])
self.gene_expression_data = gene_expr_t # Put DataFrame into corresponding field
def extract_mutation_data(self, mutation_df):
"""Generate a DataFrame with binary mutation calls for screened cell lines and target genes.
Arguments:
mutation_df: DataFrame with original mutation calls from GDSC.
Returns:
None
"""
targets = [x + "_mut" for x in self.targets]
df = mutation_df.copy()[
mutation_df.cosmic_sample_id.isin(self.screened_cell_lines)]
df = df[df.genetic_feature.isin(targets)][["cosmic_sample_id", "genetic_feature", "is_mutated"]]
cosmic_ids = []
genetic_features = {}
for feature in df.genetic_feature.unique():
genetic_features[feature] = []
for cl_id in df.cosmic_sample_id.unique():
cosmic_ids.append(cl_id)
df_cl = df[df.cosmic_sample_id == cl_id]
for feature in genetic_features:
mutation_status = df_cl[
df_cl.genetic_feature == feature]["is_mutated"].iloc[0]
genetic_features[feature].append(mutation_status)
df1 = pd.DataFrame()
df1.insert(0, "cell_line_id", cosmic_ids) # Insert column with samples IDs
for feature in genetic_features:
df1[feature] = genetic_features[feature]
self.mutation_data = df1 # Put DataFrame into corresponding field
def extract_cnv_data(self, cnv_binary_df):
"""Generate data containing binary CNV calls for cell lines screened for the drug.
Arguments:
cnv_binary_df: DataFrame from GDSC download tool with CNV data.
Returns:
None
"""
df = cnv_binary_df[cnv_binary_df.cosmic_sample_id.isin(self.screened_cell_lines)]
features_to_extract = [] # Map drug's targets to CNV features (segments)
for row in cnv_binary_df.drop_duplicates(subset="genetic_feature").itertuples():
feature_name = getattr(row, "genetic_feature")
genes_in_segment = getattr(row, "genes_in_segment").split(",")
for target in self.targets:
if target in genes_in_segment:
features_to_extract.append(feature_name) # If target is in any segment, add it to the list
features_to_extract = list(set(features_to_extract))
df = df[df.genetic_feature.isin(features_to_extract)]
cosmic_ids = []
feature_dict = {} # Separate lists for every column in final DataFrame
for feature in df.genetic_feature.unique():
feature_dict[feature] = []
for cl_id in df.cosmic_sample_id.unique():
cosmic_ids.append(cl_id)
for feature in feature_dict:
status = df[
(df.cosmic_sample_id == cl_id) & (df.genetic_feature == feature)]["is_mutated"].iloc[0]
feature_dict[feature].append(status)
new_df = pd.DataFrame()
for feature in feature_dict:
new_df[feature] = feature_dict[feature]
new_df.insert(0, "cell_line_id", cosmic_ids)
self.cnv_data = new_df
def extract_cnv_data_faster(self, cnv_binary_df, map_cl_id_and_feature_to_status):
"""Generate data containing binary CNV calls for cell lines screened for the drug.
Faster implementation than original "extract_cnv_data" by using mapping between genes and
genomic segments.
Arguments:
cnv_binary_df: DataFrame from GDSC download tool with CNV data.
Returns:
None
"""
df = cnv_binary_df[cnv_binary_df.cosmic_sample_id.isin(self.screened_cell_lines)]
features_to_extract = [] # Map drug's targets to CNV features (segments)
for row in cnv_binary_df.drop_duplicates(subset="genetic_feature").itertuples():
feature_name = getattr(row, "genetic_feature")
genes_in_segment = getattr(row, "genes_in_segment").split(",")
for target in self.targets:
if target in genes_in_segment:
features_to_extract.append(feature_name) # If target is in any segment, add it to the list
features_to_extract = list(set(features_to_extract))
df = df[df.genetic_feature.isin(features_to_extract)]
cosmic_ids = []
feature_dict = {} # Separate lists for every column in final DataFrame
for feature in features_to_extract:
feature_dict[feature] = []
for cl_id in df.cosmic_sample_id.unique():
cosmic_ids.append(cl_id)
for feature in feature_dict:
status = map_cl_id_and_feature_to_status[(cl_id, feature)]
feature_dict[feature].append(status)
new_df = pd.DataFrame()
for feature in feature_dict:
new_df[feature] = feature_dict[feature]
new_df.insert(0, "cell_line_id", cosmic_ids)
self.cnv_data = new_df
def extract_tissue_data(self, cell_line_list):
"""Generate (dummy encoded) data with cell line tissue type.
Arguments:
cell_line_list (DataFrame): Cell line list from GDSC.
Returns:
None
"""
df = cell_line_list[
cell_line_list["COSMIC_ID"].isin(self.screened_cell_lines)][["COSMIC_ID", "Tissue"]]
df.rename(columns={"COSMIC_ID": "cell_line_id"}, inplace=True)
self.tissue_data = pd.get_dummies(df, columns = ["Tissue"])
def extract_merck_signatures_data(self, signatures_df):
"""Generate data with gene expression signature scores for GDSC cell lines, provided by Merck.
Arguments:
signatures_df (DataFrame): DataFrame with gene signatures for cell lines.
Returns:
None
"""
# Compute list of screened cell lines as strings with prefix "X" in order to match
# signatures DataFrame columns
cell_lines_str = ["X" + str(cl) for cl in self.screened_cell_lines]
# Compute list of cell lines that are contained in signatures data
cls_to_extract = [cl for cl in cell_lines_str
if cl in list(signatures_df.columns)]
# Extract desired subset of signatures data
signatures_of_interest = signatures_df[cls_to_extract]
# Transpose the DataFrame
signatures_t = signatures_of_interest.transpose()
# Create a list of cell line IDs whose format matches rest of the data
cl_ids = pd.Series(signatures_t.index).apply(lambda x: int(x[1:]))
# Insert proper cell line IDs as a new column
signatures_t.insert(0, "cell_line_id", list(cl_ids))
# Drop the index and put computed DataFrame in an instance field
self.merck_signatures = signatures_t.reset_index(drop=True)
def concatenate_data(self, data_combination):
"""Generate data containing chosen combination of genetic data classes.
Arguments:
data_combination: List of strings containing data classes to be included. Available options are:
"mutation", "expression", "CNV", "tissue", "merck signatures".
Returns:
None
"""
# Create a list of DataFrames to include
objects = [self.response_data]
if "mutation" in data_combination and self.mutation_data.shape[0] > 0:
objects.append(self.mutation_data)
if "expression" in data_combination and self.gene_expression_data.shape[0] > 0:
objects.append(self.gene_expression_data)
if "CNV" in data_combination and self.cnv_data.shape[0] > 0:
objects.append(self.cnv_data)
if "tissue" in data_combination and self.tissue_data.shape[0] > 0:
objects.append(self.tissue_data)
if "merck signatures" in data_combination and self.merck_signatures.shape[0] > 0:
objects.append(self.merck_signatures)
# Find intersection in cell lines for all desirable DataFrames
cl_intersection = set(list(self.response_data["cell_line_id"]))
for obj in objects:
cl_intersection = cl_intersection.intersection(set(list(obj["cell_line_id"])))
objects_common = []
for obj in objects:
objects_common.append(obj[obj["cell_line_id"].isin(cl_intersection)])
# Check if all DataFrames have the same number of samples
no_samples = objects_common[0].shape[0]
for obj in objects_common:
assert obj.shape[0] == no_samples
obj.sort_values("cell_line_id", inplace=True)
obj.reset_index(drop=True, inplace=True)
cl_ids = objects_common[0]["cell_line_id"]
df_concatenated = pd.concat(objects_common, axis=1, ignore_index=False)
metric = self.response_data.columns[-1] # Extract the name of metric which was used for sensitivity
sensitivities = df_concatenated[metric]
df_concatenated = df_concatenated.drop(["cell_line_id", metric], axis=1)
df_concatenated.insert(0, "cell_line_id", cl_ids)
df_concatenated.insert(df_concatenated.shape[1], metric, sensitivities)
self.full_data = df_concatenated
def create_full_data(self, sensitivity_profiles_df, gene_expression_df=None, cnv_binary_df=None,
map_cl_id_and_feature_to_status=None,
cell_line_list=None, mutation_df=None, merck_signatures_df=None,
data_combination=None, metric="AUC"):
"""Combine extraction methods in one to generate a DataFrame with desired data.
When calling a function, original DataFrames parsed should match strings in
data_combination argument. If any of the "_df" arguments is None (default value),
the corresponding data is not included in the output DataFrame.
Arguments:
sensitivity_profiles_df (DataFrame): DataFrame of drug response data from GDSC.
gene_expression_df (DataFrame): Original GDSC gene expression DataFrame.
cnv_binary_df (DataFrame): DataFrame from GDSC download tool with CNV data.
cell_line_list (DataFrame): Cell line list from GDSC.
mutation_df (DataFrame): DataFrame with original mutation calls from GDSC.
data_combination (list): list of strings containing data classes to be included. Available
options are: "mutation", "expression", "CNV, "tissue", "merck signatures".
metric (string): Which statistic to use as a response metric (default "AUC").
Returns:
DataFrame containing desired data for the drug
"""
# Call separate methods for distinct data types
self.extract_screened_cell_lines(sensitivity_profiles_df)
self.extract_drug_response_data(sensitivity_profiles_df, metric)
if type(gene_expression_df) == type(pd.DataFrame()):
self.extract_gene_expression(gene_expression_df)
if type(cnv_binary_df) == type(pd.DataFrame()):
self.extract_cnv_data_faster(cnv_binary_df, map_cl_id_and_feature_to_status)
if type(cell_line_list) == type(pd.DataFrame()):
self.extract_tissue_data(cell_line_list)
if type(mutation_df) == type(pd.DataFrame()):
self.extract_mutation_data(mutation_df)
if type(merck_signatures_df) == type(pd.DataFrame()):
self.extract_merck_signatures_data(merck_signatures_df)
self.concatenate_data(data_combination)
return self.full_data
def return_full_data(self, sensitivity_profiles_df, gene_expression_df=None, cnv_binary_df=None,
map_cl_id_and_feature_to_status=None,
cell_line_list=None, mutation_df=None, merck_signatures_df=None,
data_combination=None, metric="AUC"):
"""Compute full data with desired data classes and return it, but after that delete data from
instance's data fields in order to save memory.
When calling a function, original DataFrames parsed should match strings in
data_combination argument. If any of the "_df" arguments is None (default value),
the corresponding data is not included in the output DataFrame.
Arguments:
sensitivity_profiles_df (DataFrame): DataFrame of drug response data from GDSC.
gene_expression_df (DataFrame): Original GDSC gene expression DataFrame.
cnv_binary_df (DataFrame): DataFrame from GDSC download tool with CNV data.
cell_line_list (DataFrame): Cell line list from GDSC.
mutation_df (DataFrame): DataFrame with original mutation calls from GDSC.
data_combination (list): list of strings containing data classes to be included. Available
options are: "mutation", "expression", "CNV, "tissue", "merck signatures".
metric (string): Which statistic to use as a response metric (default "AUC").
Returns:
DataFrame containing desired data for the drug
"""
full_df = self.create_full_data(sensitivity_profiles_df, gene_expression_df, cnv_binary_df,
map_cl_id_and_feature_to_status,
cell_line_list, mutation_df, merck_signatures_df,
data_combination, metric)
if type(gene_expression_df) == type(pd.DataFrame()):
self.gene_expression_data = None
if type(cnv_binary_df) == type(pd.DataFrame()):
self.cnv_data = None
if type(cell_line_list) == type(pd.DataFrame()):
self.tissue_data = None
if type(mutation_df) == type(pd.DataFrame()):
self.mutation_data = None
if type(merck_signatures_df) == type(pd.DataFrame()):
self.merck_signatures = None
self.full_data = None
return full_df
def __repr__(self):
"""Return string representation of an object, which can be used to create it."""
return 'Drug({}, "{}", {}, "{}")'.format(self.gdsc_id, self.name, self.targets, self.target_pathway)
def __str__(self):
"""Return string representation of an object"""
return "{} -- {}".format(self.name, self.gdsc_id)
# Class methods
@classmethod
def load_mappings(cls, filepath_hgnc_to_ensembl, filepath_ensembl_to_hgnc):
"""Load dictonaries with gene mappings between HGNC and Ensembl (from pickle files) and assign it
to corresponding class variables. Ensembl IDs are needed for gene expression data.
This method should be called on a Drug class before any other actions with the class.
Arguments:
filepath_hgnc_to_ensembl: file with accurate mapping
filepath_ensembl_to_hgnc: file with accurate mapping
Returns:
None
"""
cls.map_from_hgnc_to_ensembl = pickle.load(open(filepath_hgnc_to_ensembl, "rb"))
cls.map_from_ensembl_to_hgnc = pickle.load(open(filepath_ensembl_to_hgnc, "rb"))
# Static methods
@staticmethod
def create_drugs(drug_annotations_df):
"""Create a dictionary of Drug class objects, each referenced by it's ID (keys are drug GDSC ID's).
Arguments:
drug_annotations_df (DataFrame): DataFrame of drug annotations from GDSC website
Returns:
Dictionary of Drug objects as values and their ID's as keys
"""
drugs = {}
for row in drug_annotations_df.itertuples(index=True, name="Pandas"):
gdsc_id = getattr(row, "DRUG_ID")
name = getattr(row, "DRUG_NAME")
targets = getattr(row, "TARGET").split(", ")
target_pathway = getattr(row, "TARGET_PATHWAY")
drugs[gdsc_id] = Drug(gdsc_id, name, targets, target_pathway)
return drugs
@staticmethod
def load_data(drug_annotations, cell_line_list, gene_expr, cnv1, cnv2,
coding_variants, drug_response):
"""Load all needed files by calling one function and return data as tuple of DataFrames. All
argumenst are filepaths to corrresponding files."""
# Drug annotations
drug_annotations_df = pd.read_excel(drug_annotations)
# Cell line annotations
col_names = ["Name", "COSMIC_ID", "TCGA classification", "Tissue", "Tissue_subtype", "Count"]
cell_lines_list_df = pd.read_csv(cell_line_list, usecols=[1, 2, 3, 4, 5, 6], header=0, names=col_names)
# Gene expression
gene_expression_df = pd.read_table(gene_expr)
# CNV
d1 = pd.read_csv(cnv1)
d2 = pd.read_table(cnv2)
d2.columns = ["genes_in_segment"]
def f(s):
return s.strip(",")
cnv_binary_df = d1.copy()
cnv_binary_df["genes_in_segment"] = d2["genes_in_segment"].apply(f)
# Coding variants
coding_variants_df = pd.read_csv(coding_variants)
# Drug-response
drug_response_df = pd.read_excel(drug_response)
return (drug_annotations_df, cell_lines_list_df, gene_expression_df, cnv_binary_df, coding_variants_df,
drug_response_df)
#################################################################################################################
# DrugWithDrugBank class
##################################################################################################################
class DrugWithDrugBank(Drug):
"""Class representing drug from GDSC database.
Contrary to the parent class Drug, this class also incorporates data related to targets
derived from DrugBank, not only those from GDSC. Main function of the class is to create and store input data
corresponding to a given drug. Four types of data are considered: gene expression, copy number variants,
coding variants and tumor tissue type. Class instances are initialized with four basic drug properties:
ID, name, gene targets and target pathway. Data attributes are stored as pandas DataFrames and are filled
using data files from GDSC via corresponding methods.
In general, all utilities are the same as in parent Drug class, with an exception of "create_drugs"
method, which is overloaded in order to account for target genes data coming from DrugBank.
Attributes:
gdsc_id (int): ID from GDSC website.
name (string): Drug name.
targets (list of strings): Drug's target gene names (HGNC).
target_pathway (string): Drug's target pathway as provided in GDSC annotations.
ensembl targets (list of strings): Drug's target genes ensembl IDs. Can have different length
than "targets" because some gene names may not be matched during mapping. Ensembl IDs are
needed for gene expression data.
map_from_hgnc_to_ensembl (dictionary): Dictionary mapping from gene names to ensembl IDs. Created
after calling the "load_mappings" method.
map_from_ensembl_to_hgnc (dictionary): Dictionary mapping from ensembl IDs to gene names. Created
after calling the "load_mappings" method.
total_no_samples_screened (int): Number of cell lines screened for that drug. Created after
calling the "extract_drug_response_data" method.
response_data (DataFrame): DataFrame with screened cell lines for that drug and corresponding AUC or
IC50 values. Created after calling the "extract_drug_response_data" method.
screened_cell_lines (list of ints): list containing COSMIC IDs representing cell lines screened for
that drug. Created after calling the "extract_screened_cell_lines" method.
gene_expression_data (DataFrame): DataFrame with gene expression data, considering only
target genes. Created after calling the "extract_gene_expression" method
mutation_data (DataFrame): DataFrame with binary calls for coding variants, considering only
target genes. Created after calling the "extract_mutation_data" method.
cnv_data (DataFrame): DataFrame with binary calls for copu number variants, considering only
target genes. Created after calling the "extract_cnv_data" method.
tissue_data (DataFrame): DataFrame with dummy encoded tumor tissue types in screened cell lines.
Dummy encoding results in 13 binary features. Created after calling the
"extract_tissue_data" method.
full_data (DataFrame): DataFrame with combined data coming from given set of genetic data
classes.
Methods:
Instance methods:
__init__: Initialize a Drug instance.
__repr__: Return string representation of an instance, as a command which can be used to create
this instance.
__str__: Return string representation of an instance.
extract_drug_response_data: Generate a DataFrame with drug-response data.
extract_screened_cell_lines: Generate a list of COSMIC IDs representing cell lines screened for that
drug.
extract_gene_expression: Generate a DataFrame with gene expression data for drug's screened cell lines
extract_mutation_data: Generate a DataFrame with binary calls for coding variants.
extract_cnv_data: Generate a DataFrame with binary calls for copy number variants.
extract_tissue_data: Generate a DataFrame with dummy encoded tissue types.
concatenate_data: Generate a DataFrame containing all desired genetic data classes. Available data
classes are: gene expression, coding variants, cnv variants and tissue type.
create_full_data: Combines above data extraction methods in order to create desired input data
for the drug with one method call. Returns the full data.
Class methods:
load_mappings: Load appropriate dictionaries mapping between ensembl and HGNC.
Static methods:
create_drugs: Create a dictionary of DrugWithDrugBank class objects, each referenced by it's ID
(keys are drug GDSC ID's). Includes also target data coming from DrugBank.
load_data: Load all needed data files as DataFrames with one function call.
"""
def create_drugs(drug_annotations_df, drugbank_targets_mapping):
"""Create a dictionary of DrugWithDrugBank class objects, each referenced by it's ID. Add
also target data coming from DrugBank.
Arguments:
drug_annotations_df (DataFrame): DataFrame of drug annotations from GDSC website.
drugbank_targets_mapping (dictionary): Dictionary with mapping from drug ID to it's
targets from drugbank database.
Return:
Dictionary of DrugWithDrugBank objects as values and their ID's as keys.
"""
drugs = {}
for row in drug_annotations_df.itertuples(index=True, name="Pandas"):
name = getattr(row, "DRUG_NAME")
gdsc_id = getattr(row, "DRUG_ID")
targets = getattr(row, "TARGET").split(", ")
# Add targets from DrugBank (if drug is matched) and take a sum
if gdsc_id in drugbank_targets_mapping:
targets = list(set(targets + drugbank_targets_mapping[gdsc_id]))
target_pathway = getattr(row, "TARGET_PATHWAY")
# Create DrugWithDrugBank instance and put it into output dictionary
drugs[gdsc_id] = DrugWithDrugBank(gdsc_id, name, targets, target_pathway)
return drugs
def load_data(drug_annotations, cell_line_list, gene_expr, cnv1, cnv2,
coding_variants, drug_response, drugbank_targets):
"""Load all needed files by calling one function. All argumenst are filepaths to corrresponding files."""
# Drug annotations
drug_annotations_df = pd.read_excel(drug_annotations)
# Cell line annotations
col_names = ["Name", "COSMIC_ID", "TCGA classification", "Tissue", "Tissue_subtype", "Count"]
cell_lines_list_df = pd.read_csv(cell_line_list, usecols=[1, 2, 3, 4, 5, 6], header=0, names=col_names)
# Gene expression
gene_expression_df = pd.read_table(gene_expr)
# CNV
d1 = pd.read_csv(cnv1)
d2 = pd.read_table(cnv2)
d2.columns = ["genes_in_segment"]
def f(s):
return s.strip(",")
cnv_binary_df = d1.copy()
cnv_binary_df["genes_in_segment"] = d2["genes_in_segment"].apply(f)
# Coding variants
coding_variants_df = pd.read_csv(coding_variants)
# Drug-response
drug_response_df = pd.read_excel(drug_response)
# DrugBank targets
map_drugs_to_drugbank_targets = pickle.load(open(drugbank_targets, "rb"))
return (drug_annotations_df, cell_lines_list_df, gene_expression_df, cnv_binary_df, coding_variants_df,
drug_response_df, map_drugs_to_drugbank_targets)
#################################################################################################################
# DrugGenomeWide class
#################################################################################################################
class DrugGenomeWide(Drug):
"""Class designed to represent a drug with genome-wide input data.
Main function of the class is to create and store input data corresponding to a given
drug. Four types of data are considered: gene expression, copy number variants, coding variants and tumor
tissue type. Class instances are initialized with four basic drug properties: ID, name, gene targets and
target pathway. Data attributes are stored as pandas DataFrames and are filled using data files
from GDSC via corresponding methods.
In general, all the utilities are the same as in the parent Drug class, but with different input data.
When using this setting, we only use gene expression data as input, since it is recognized
as representative of the genome-wide cell line characterization. Therefore, other data extraction methods,
though available, should not be used when utilizing this class, for clarity. Two parent class
methods are overloaded: "extract_gene_expression" and "create_drugs".
Important note: in here, "create_full_data" method is not overloaded, but is supposed to be called
only parsing drug_response_df and gene_expression_df DataFrames and data_combination argument
set to "["expression"]".
--Example:
df = test_drug.create_full_data(drug_response_df, gene_expression_df, data_combination=["expression"])
Attributes:
gdsc_id (int): ID from GDSC website.
name (string): Drug name.
targets (list of strings): Drug's target gene names (HGNC).
target_pathway (string): Drug's target pathway as provided in GDSC annotations.
ensembl targets (list of strings): Drug's target genes ensembl IDs. Can have different length
than "targets" because some gene names may not be matched during mapping. Ensembl IDs are
needed for gene expression data.
map_from_hgnc_to_ensembl (dictionary): Dictionary mapping from gene names to ensembl IDs. Created
after calling the "load_mappings" method.
map_from_ensembl_to_hgnc (dictionary): Dictionary mapping from ensembl IDs to gene names. Created
after calling the "load_mappings" method.
total_no_samples_screened (int): Number of cell lines screened for that drug. Created after
calling the "extract_drug_response_data" method.
response_data (DataFrame): DataFrame with screened cell lines for that drug and corresponding AUC or
IC50 values. Created after calling the "extract_drug_response_data" method.
screened_cell_lines (list of ints): list containing COSMIC IDs representing cell lines screened for
that drug. Created after calling the "extract_screened_cell_lines" method.
gene_expression_data (DataFrame): DataFrame with gene expression data, considering all
available (genome-wide) genes. Created after calling the "extract_gene_expression"
method.
mutation_data (DataFrame): DataFrame with binary calls for coding variants, considering only
target genes. Created after calling the "extract_mutation_data" method.
cnv_data (DataFrame): DataFrame with binary calls for copu number variants, considering only
target genes. Created after calling the "extract_cnv_data" method.
tissue_data (DataFrame): DataFrame with dummy encoded tumor tissue types in screened cell lines.
Dummy encoding results in 13 binary features. Created after calling the
"extract_tissue_data" method.
full_data (DataFrame): DataFrame with combined data coming from given set of genetic data
classes.
Methods:
Instance methods:
__init__: Initialize a Drug instance.
__repr__: Return string representation of an instance, as a command which can be used to create
this instance.
__str__: Return string representation of an instance.
extract_drug_response_data: Generate a DataFrame with drug-response data.
extract_screened_cell_lines: Generate a list of COSMIC IDs representing cell lines screened for that
drug.
extract_gene_expression: Generate a DataFrame with gene expression data for drug's screened cell lines
extract_mutation_data: Generate a DataFrame with binary calls for coding variants.
extract_cnv_data: Generate a DataFrame with binary calls for copy number variants.
extract_tissue_data: Generate a DataFrame with dummy encoded tissue types.
concatenate_data: Generate a DataFrame containing all desired genetic data classes. Available data
classes are: gene expression, coding variants, cnv variants and tissue type.
create_full_data: Combines above data extraction methods in order to create desired input data
for the drug with one method call. Returns the full data. See the note above for correct
usage with DrugGenomeWide class.
Class methods:
load_mappings: Load appropriate dictionaries mapping between ensembl and HGNC.
Static methods:
create_drugs: Create a dictionary of Drug class objects, each referenced by it's ID
(keys are drug GDSC ID's)
load_data: Load all needed data files as DataFrames with one function call.
"""
def extract_gene_expression(self, gene_expression_df):
"""Generate DataFrame of gene expression data for cell lines screened for this drug,
genome-wide (all available genes).
Arguments:
gene_expression_df (DataFrame): original GDSC gene expression DataFrame
sensitivity_profiles_df (DataFrame): DataFrame of drug response data from GDSC
Return:
None
"""
cell_lines_str = [] # Gene expression DF column names are strings
for x in self.screened_cell_lines:
cell_lines_str.append(str(x))
cl_to_extract = []
for x in cell_lines_str:
if x in list(gene_expression_df.columns):
cl_to_extract.append(x) # Extract only cell lines contained in gene expression data
gene_expr = gene_expression_df[["ensembl_gene"] + cl_to_extract]
gene_expr_t = gene_expr.transpose()
columns = list(gene_expr_t.loc["ensembl_gene"])
gene_expr_t.columns = columns
gene_expr_t = gene_expr_t.drop(["ensembl_gene"])
rows = list(gene_expr_t.index)
gene_expr_t.insert(0, "cell_line_id", rows) # Insert columns with cell line IDs
gene_expr_t.reset_index(drop=True, inplace=True)
gene_expr_t["cell_line_id"] = pd.to_numeric(gene_expr_t["cell_line_id"])
# DataFrame should have same number of columns for each drug
assert gene_expr_t.shape[1] == 17738
self.gene_expression_data = gene_expr_t
def extract_mutation_data(self, mutation_df):
"""Generate a DataFrame with binary mutation calls for screened cell lines and target genes.
Arguments:
mutation_df: DataFrame with original mutation calls from GDSC.
Returns:
None
"""
targets = [x + "_mut" for x in self.targets]
df = mutation_df.copy()[
mutation_df.cosmic_sample_id.isin(self.screened_cell_lines)]
df = df[["cosmic_sample_id", "genetic_feature", "is_mutated"]]
cosmic_ids = []
genetic_features = {}
for feature in df.genetic_feature.unique():
genetic_features[feature] = []
for cl_id in df.cosmic_sample_id.unique():
cosmic_ids.append(cl_id)
df_cl = df[df.cosmic_sample_id == cl_id]
for feature in genetic_features:
mutation_status = df_cl[
df_cl.genetic_feature == feature]["is_mutated"].iloc[0]
genetic_features[feature].append(mutation_status)
df1 = pd.DataFrame()
df1.insert(0, "cell_line_id", cosmic_ids) # Insert column with samples IDs
for feature in genetic_features:
df1[feature] = genetic_features[feature]
self.mutation_data = df1 # Put DataFrame into corresponding field
def extract_cnv_data_faster(self, cnv_binary_df, map_cl_id_and_feature_to_status):
"""Generate data containing binary CNV calls for cell lines screened for the drug.
Faster implementation than original "extract_cnv_data" by using mapping between genes and
genomic segments.
Arguments:
cnv_binary_df: DataFrame from GDSC download tool with CNV data.
Returns:
None
"""
df = cnv_binary_df[cnv_binary_df.cosmic_sample_id.isin(self.screened_cell_lines)]
features_to_extract = [] # Map drug's targets to CNV features (segments)
cosmic_ids = []
feature_dict = {} # Separate lists for every column in final DataFrame
for feature in df.genetic_feature.unique():
feature_dict[feature] = []
for cl_id in df.cosmic_sample_id.unique():
cosmic_ids.append(cl_id)
for feature in feature_dict:
status = map_cl_id_and_feature_to_status[(cl_id, feature)]
feature_dict[feature].append(status)
new_df = pd.DataFrame()
for feature in feature_dict:
new_df[feature] = feature_dict[feature]
new_df.insert(0, "cell_line_id", cosmic_ids)
self.cnv_data = new_df
def create_drugs(drug_annotations_df):
"""Create a dictionary of DrugGenomeWide class objects, each referenced by it's ID.
Arguments:
drug_annotations_df (DataFrame): DataFrame of drug annotations from GDSC website
Returns:
Dictionary of DrugGenomeWide objects as values and their ID's as keys
"""
drugs = {}
for row in drug_annotations_df.itertuples(index=True, name="Pandas"):
gdsc_id = getattr(row, "DRUG_ID")
name = getattr(row, "DRUG_NAME")
targets = getattr(row, "TARGET").split(", ")
target_pathway = getattr(row, "TARGET_PATHWAY")
drugs[gdsc_id] = DrugGenomeWide(gdsc_id, name, targets, target_pathway)
return drugs
def load_data(drug_annotations, cell_line_list, gene_expr, cnv1, cnv2,
coding_variants, drug_response):
"""Load all needed files by calling one function. All argumenst are filepaths to corrresponding files."""
# Drug annotations
drug_annotations_df = pd.read_excel(drug_annotations)
# Cell line annotations
col_names = ["Name", "COSMIC_ID", "TCGA classification", "Tissue", "Tissue_subtype", "Count"]
cell_lines_list_df = pd.read_csv(cell_line_list, usecols=[1, 2, 3, 4, 5, 6], header=0, names=col_names)
# Gene expression
gene_expression_df = pd.read_table(gene_expr)
# CNV
d1 = pd.read_csv(cnv1)
d2 = pd.read_table(cnv2)
d2.columns = ["genes_in_segment"]
def f(s):
return s.strip(",")
cnv_binary_df = d1.copy()
cnv_binary_df["genes_in_segment"] = d2["genes_in_segment"].apply(f)
# Coding variants
coding_variants_df = pd.read_csv(coding_variants)
# Drug-response
drug_response_df = pd.read_excel(drug_response)
return (drug_annotations_df, cell_lines_list_df, gene_expression_df, cnv_binary_df, coding_variants_df,
drug_response_df)
#################################################################################################################
# DrugDirectReactome class
#################################################################################################################
class DrugDirectReactome(DrugWithDrugBank):
"""Class representing compound from GDSC database.
Main function of the class is to create and store input data corresponding to a given
drug. Four types of data are considered: gene expression, copy number variants, coding variants and tumor
tissue type. Class instances are initialized with four basic drug properties: ID, name, gene targets and
target pathway. Data attributes are stored as pandas DataFrames and are filled using data files
from GDSC via corresponding methods.
In this setting, drugs gene targets are derived not only from GDSC and DrugBank, but also using the direct
compound-pathway mapping from Reactome database. All genes belonging to corresponding Reactome target pathway
are considered when computing input data. The utilities are the same as in parent DrugWithDrugBank class with
an exception of "create_drugs" method which accounts for mappings coming from Reactome, and "load_data"
method.
Attributes:
gdsc_id (int): ID from GDSC website.
name (string): Drug name.
targets (list of strings): Drug's target gene names (HGNC).
target_pathway (string): Drug's target pathway as provided in GDSC annotations.
ensembl targets (list of strings): Drug's target genes ensembl IDs. Can have different length
than "targets" because some gene names may not be matched during mapping. Ensembl IDs are
needed for gene expression data.
map_from_hgnc_to_ensembl (dictionary): Dictionary mapping from gene names to ensembl IDs. Created
after calling the "load_mappings" method.
map_from_ensembl_to_hgnc (dictionary): Dictionary mapping from ensembl IDs to gene names. Created
after calling the "load_mappings" method.
total_no_samples_screened (int): Number of cell lines screened for that drug. Created after
calling the "extract_drug_response_data" method.
response_data (DataFrame): DataFrame with screened cell lines for that drug and corresponding AUC or
IC50 values. Created after calling the "extract_drug_response_data" method.
screened_cell_lines (list of ints): list containing COSMIC IDs representing cell lines screened for
that drug. Created after calling the "extract_screened_cell_lines" method.
gene_expression_data (DataFrame): DataFrame with gene expression data, considering only
target genes. Created after calling the "extract_gene_expression" method
mutation_data (DataFrame): DataFrame with binary calls for coding variants, considering only
target genes. Created after calling the "extract_mutation_data" method.
cnv_data (DataFrame): DataFrame with binary calls for copu number variants, considering only
target genes. Created after calling the "extract_cnv_data" method.
tissue_data (DataFrame): DataFrame with dummy encoded tumor tissue types in screened cell lines.
Dummy encoding results in 13 binary features. Created after calling the
"extract_tissue_data" method.
full_data (DataFrame): DataFrame with combined data coming from given set of genetic data
classes.
Methods:
Instance methods:
__init__: Initialize a Drug instance.
__repr__: Return string representation of an instance, as a command which can be used to create
this instance.
__str__: Return string representation of an instance.
extract_drug_response_data: Generate a DataFrame with drug-response data.
extract_screened_cell_lines: Generate a list of COSMIC IDs representing cell lines screened for that
drug.
extract_gene_expression: Generate a DataFrame with gene expression data for drug's screened cell lines
extract_mutation_data: Generate a DataFrame with binary calls for coding variants.
extract_cnv_data: Generate a DataFrame with binary calls for copy number variants.
extract_tissue_data: Generate a DataFrame with dummy encoded tissue types.
concatenate_data: Generate a DataFrame containing all desired genetic data classes. Available data
classes are: gene expression, coding variants, cnv variants and tissue type.
create_full_data: Combines above data extraction methods in order to create desired input data
for the drug with one method call. Returns the full data.
Class methods:
load_mappings: Load appropriate dictionaries mapping between ensembl and HGNC.
Static methods:
create_drugs: Create a dictionary of DrugWithDrugBank class objects, each referenced by it's ID
(keys are drug GDSC ID's). Includes also target data coming from DrugBank.
load_data: Load all needed data files as DataFrames with one function call.
"""
def create_drugs(drug_annotations_df, drugbank_targets_mapping, reactome_direct_mapping):
"""Create a dictionary of DrugWithDrugBank class objects, each referenced by it's ID.
Arguments:
drug_annotations_df (DataFrame): DataFrame of drug annotations from GDSC website
drugbank_targets_mapping (dictionary): dictionary with mapping from drug ID to it's
targets from drugbank database
reactome_direct_mapping:
Returns:
Dictionary of Drug objects as values and their ID's as keys
"""
drugs = {}
for row in drug_annotations_df.itertuples(index=True, name="Pandas"):
name = getattr(row, "DRUG_NAME")
gdsc_id = getattr(row, "DRUG_ID")
# Create an object only if it exists in Reactome mapping dictionary
if gdsc_id in reactome_direct_mapping:
targets = getattr(row, "TARGET").split(", ")
# If this ID exists in DrugBank mapping, take the sum of all three sets
if gdsc_id in drugbank_targets_mapping:
targets = list(set(targets + drugbank_targets_mapping[gdsc_id] + reactome_direct_mapping[gdsc_id]))
# Otherwise add just the Reactome targets
else:
targets = list(set(targets + reactome_direct_mapping[gdsc_id]))
target_pathway = getattr(row, "TARGET_PATHWAY")
drugs[gdsc_id] = DrugDirectReactome(gdsc_id, name, targets, target_pathway)
else:
continue
return drugs
def load_data(drug_annotations, cell_line_list, gene_expr, cnv1, cnv2,
coding_variants, drug_response, drugbank_targets, reactome_targets):
"""Load all needed files by calling one function. All argumenst are filepaths to
corrresponding files."""
# Drug annotations
drug_annotations_df = pd.read_excel(drug_annotations)
# Cell line annotations
col_names = ["Name", "COSMIC_ID", "TCGA classification", "Tissue", "Tissue_subtype", "Count"]
cell_lines_list_df = pd.read_csv(cell_line_list, usecols=[1, 2, 3, 4, 5, 6], header=0, names=col_names)
# Gene expression
gene_expression_df = pd.read_table(gene_expr)
# CNV
d1 = pd.read_csv(cnv1)
d2 = pd.read_table(cnv2)
d2.columns = ["genes_in_segment"]
def f(s):
return s.strip(",")
cnv_binary_df = d1.copy()
cnv_binary_df["genes_in_segment"] = d2["genes_in_segment"].apply(f)
# Coding variants
coding_variants_df = | pd.read_csv(coding_variants) | pandas.read_csv |
# -*- coding: utf-8 -*-
"""
Created on Fri Apr 2 09:20:37 2021
Compiles SNODAS data into SQLite DB
@author: buriona,tclarkin
"""
import sys
from pathlib import Path
import pandas as pd
import sqlalchemy as sql
import sqlite3
import zipfile
from zipfile import ZipFile
# Load directories and defaults
this_dir = Path(__file__).absolute().resolve().parent
#this_dir = Path('C:/Programs/shread_dash/database/SHREAD')
ZIP_IT = False
ZIP_FRMT = zipfile.ZIP_LZMA
DEFAULT_DATE_FIELD = 'Date'
DEFAULT_CSV_DIR = Path(this_dir, 'data')
DEFAULT_DB_DIR = this_dir
COL_TYPES = {
'Date': str, 'Type': str, 'OBJECTID': int, 'elev_ft': int, 'slope_d': int,
'aspct': int, 'nlcd': int, 'LOCAL_ID': str, 'LOCAL_NAME': str, 'mean': float
}
# Define functions
def get_dfs(data_dir=DEFAULT_CSV_DIR, verbose=False):
"""
Get and merge dataframes imported using shread.py
"""
swe_df_list = []
sd_df_list = []
print('Preparing .csv files for database creation...')
for data_file in data_dir.glob('snodas*.csv'):
if verbose:
print(f'Adding {data_file.name} to dataframe...')
df = pd.read_csv(
data_file,
usecols=COL_TYPES.keys(),
parse_dates=['Date'],
dtype=COL_TYPES
)
if not df.empty:
swe_df_list.append(
df[df['Type'] == 'swe'].drop(columns='Type').copy()
)
sd_df_list.append(
df[df['Type'] == 'snowdepth'].drop(columns='Type').copy()
)
df_swe = pd.concat(swe_df_list)
df_swe.name = 'swe'
df_sd = pd.concat(sd_df_list)
df_sd.name = 'sd'
print(' Success!!!\n')
return {'swe': df_swe, 'sd': df_sd}
def get_unique_dates(tbl_name, db_path, date_field=DEFAULT_DATE_FIELD):
"""
Get unique dates from shread data, to ensure no duplicates
"""
if not db_path.is_file():
return pd.DataFrame(columns=[DEFAULT_DATE_FIELD])
db_con_str = f'sqlite:///{db_path.as_posix()}'
eng = sql.create_engine(db_con_str)
with eng.connect() as con:
try:
unique_dates = pd.read_sql(
f'select distinct {date_field} from {tbl_name}',
con
).dropna()
except Exception:
return pd.DataFrame(columns=[DEFAULT_DATE_FIELD])
return pd.to_datetime(unique_dates[date_field])
def write_db(df, db_path=DEFAULT_DB_DIR, if_exists='replace', check_dups=False,
zip_db=ZIP_IT, zip_frmt=ZIP_FRMT, verbose=False):
"""
Write dataframe to database
"""
sensor = df.name
print(f'Creating sqlite db for {df.name}...\n')
print(' Getting unique basin names...')
basin_list = | pd.unique(df['LOCAL_NAME']) | pandas.unique |
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
"""
The fluorescence_extract() function performs the following:
1) Combines MEAN_INTENSITY values from the Spots Statistics TrackMate output files for a single neuron.
2) Subtracts the background, finds the maximal value for each timepoint
3) Calculates the change in fluorescence (∆F/F0) over time and plots the ∆F/F0.
4) Outputs a .csv with the ∆F/F0 for each time point and the plot of the
∆F/F0 over time as a .png in a separate “Neuron Plots” folder within the results folder.
The loop_fluorescence_extract() function performs the following:
1) Runs fluorescence_extract() on all the neurons in a folder. This requires specific file structure as an input. Refer to the README file
2) Outputs 2 files for each neuron into the set results folder: the .csv with the ∆F/F0 for each time point
and the plot of the ∆F/F0 over time as a .png in a separate “Neuron Plots” folder within the results folder
@author: alisasmacbook
"""
import os
import shutil
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
from matplotlib.ticker import MaxNLocator
def fluorescence_extract(working_dir,
results_folder = "results",
trial_name = "Neuron",
position_t=100,
background_averages=[1]):
os.chdir(working_dir)
output_path = os.path.join(working_dir, 'python_files')
if os.path.exists(output_path):
shutil.rmtree(output_path)
os.mkdir('python_files')
position_t_column = np.arange(0,position_t)
position_t_array = pd.DataFrame(position_t_column, columns = ['POSITION_T'])
POSITION_T_path = os.path.join(output_path, "POSITION_T.csv")
position_t_array.to_csv(POSITION_T_path,index=False)
for filename in os.listdir(working_dir):
if filename.endswith(".csv"):
df = pd.read_csv(filename, usecols=(["POSITION_T","MEAN_INTENSITY"]))
file_name = os.path.splitext(filename)[0]
df.columns = ['POSITION_T', file_name]
df.to_csv(f"python_files/{file_name}py.csv",index=False, na_rep = '')
for filename in os.listdir(output_path):
if filename.endswith("py.csv"):
POSITION_T = pd.read_csv(POSITION_T_path, na_values = '')
data = os.path.join(output_path, filename)
df = | pd.read_csv(data) | pandas.read_csv |
#!/usr/bin/env python3
"""
Trenco Modules for running scripts
"""
import sys
import re
import os
import glob
import subprocess
import json
import logging
import time
import shlex
import pandas as pd
import numpy as np
import seaborn as sb
import multiprocessing as mp
import matplotlib.pyplot as plt
plt.switch_backend('agg')
# Switch
debug = True
tf_enh_mx = None
# --------------------------------------------------------------------------- #
# General scripts that are used in trenco wrapper and in multiple places #
# throughout the core module #
# --------------------------------------------------------------------------- #
def build_dir():
dirs = ['log', 'process', 'results']
for d in dirs:
if not os.path.exists(d):
print("Building {}".format(d))
os.mkdir(d)
def plot_mtx(fnmtx):
mtx = pd.read_csv(fnmtx, sep='\t')
try:
g = sb.pairplot(mtx,diag_kind='kde')
except:
mtx.columns = mtx.columns.str.split("_").str[-2]
g = sb.pairplot(mtx, diag_kind='kde')
pass
for ax in g.axes.flat:
ax.set_xlabel(xlabel = ax.xaxis.get_label_text(), rotation=90)
ax.set_ylabel(ylabel = ax.yaxis.get_label_text(), rotation=80)
g.savefig("results/{}.png".format(fnmtx.split('/')[-1]))
plt.clf()
def plot_corr_heatmap(df, df_name, size):
""" Plot heatmap of numeric pandas DataFrame """
a4_dims = (size, size)
_, ax = plt.subplots(figsize=a4_dims)
sb.set_context('paper', font_scale=1.8)
g = sb.heatmap(df, yticklabels=False, cmap='Blues', annot=True, ax=ax)
fig = g.get_figure()
fig.savefig('{}-heatmap.png'.format(df_name))
plt.clf()
def files_exist(fnames = []):
for name in fnames:
if not os.path.exists(name):
return False
return True
def check_if_commons(commonfile, exact = True):
filepath = "/".join(os.path.realpath(__file__).split("/")[:-2]) \
+ "/common_files/*"
commons = glob.glob(filepath)
for file_full in commons:
file_name = file_full.split("/")[-1]
if exact:
if commonfile == file_name:
return file_full
else:
if commonfile in file_name:
return file_full
return None
def load_convert_name(fn):
converter = {}
with open(fn, "r") as ifi:
for line in ifi:
line = line.strip()
if line.startswith("#"):
continue
key, val = line.split()
converter[key] = val
return converter
def load_gtf_conversion(gtf, conv_from, conv_to):
if not os.path.exists(gtf):
print("File %s cannot be found" % gtf)
exit(1)
converter = {}
with open(gtf, "r") as ifi:
for line in ifi:
line = line.strip()
if line.startswith("#"):
continue
keys = line.split("\t")[-1]
keys = [k.strip().replace("\"", "") for k in keys.split(";")]
keyset = {}
for k in keys:
if not k:
continue
flag, entry = k.split()
keyset[flag] = entry
if keyset[conv_from] not in converter:
converter[keyset[conv_from]] = keyset[conv_to]
else:
if converter[keyset[conv_from]] == keyset[conv_to]:
continue
elif isinstance(converter[keyset[conv_from]], list):
converter[keyset[conv_from]].append(keyset[conv_to])
else:
converter[keyset[conv_from]] = [converter[keyset[conv_from]],
keyset[conv_to]]
return converter
# --------------------------------------------------------------------------- #
# Methods for Getting Transcripts from ANNOTATIONS gtf and requires #
# AWK script #
# --------------------------------------------------------------------------- #
def get_tss(annotation, biotypes, logger):
'''Get the TSS.'''
# Make biotypes file
biotypes_file = open('process/biotypes.txt', 'w')
biotypes_file.write("\n".join(biotypes))
biotypes_file.close()
# Get file basename
file_name = os.path.basename(annotation)
basename = file_name.rpartition('.gtf')[0]
# Get the gencode
rel_path = os.path.dirname(os.path.realpath(__file__))
make_gencode_tss = '{}/../trenco_tools'.format(rel_path)
tss_program = 'make_TSS_file_from_annotation_with_confidence_better.sh'
tss_command = '{}/{} {} process/biotypes.txt'.format(make_gencode_tss,
tss_program,
annotation)
logger.info("Running TSS caluclation %s", tss_command)
tss = subprocess.Popen(tss_command, shell=True)
out, err = tss.communicate()
# Remove extra files
os.remove('{}_exons_most5p.gff'.format(basename))
os.remove('{}_capped_sites_nr_with_confidence.gff'.format(basename))
def process_tss(version, organism, biotypes, fname, logger):
''' Process the TSS into a bed file that can be used '''
# If files don't exists
if fname:
annotations = fname
if not os.path.exists(annotations):
print("ERROR: %s not found" % annotations,
file=sys.stderr)
exit(1)
else:
repo_fn = "gencode.%s.annotation.gtf" % (version)
annotations = "process/" + repo_fn
if not os.path.exists(annotations):
cmds = ["wget ftp://ftp.ebi.ac.uk/pub/databases/"\
"gencode/Gencode_%s/release_%s/%s.gz -P process"
% (organism, version.replace('v',''), repo_fn),
"gunzip %s.gz" % (annotations)]
for cmd in cmds:
#os.system(cmd) # the code needs to wait at this point
subprocess.check_call(cmd, shell=True)
# this is a temporary fix to make sure the gunzip works and has
# time to finish
time.sleep(30)
if annotations.endswith('.gtf'):
tail = '.gtf'
else:
tail = '.gff'
cs_annot = annotations.replace(tail, '_capped_sites.gff').split('/')[-1]
ofn = "process/gene.bed"
if files_exist(['process/'+cs_annot, ofn]):
return 'process/'+cs_annot, ofn
# Generate Converstion table for gene names
convertion = load_gtf_conversion(annotations, "gene_id", "gene_name")
with open("process/converter.json", "w") as ofo:
json.dump(convertion, ofo, indent = 2)
# Create a file handler
handler = logging.FileHandler('log/get_transcript.log')
logger.addHandler(handler)
# Extract biotype genes
if os.path.exists("process/gene_txn.gtf"):
os.remove("process/gene_txn.gtf")
for biotype in biotypes:
grep_cmd = "grep -w gene {} | grep {} "\
">> process/gene_txn.gtf".format(annotations, biotype)
print(grep_cmd)
subprocess.check_call(grep_cmd, shell=True)
subprocess.check_call("gtf2bed < process/gene_txn.gtf "\
"> \process/gene_txn.bed", shell=True)
ofo = open(ofn, "w")
with open("process/gene_txn.bed", "r") as ifn:
for line in ifn:
line = line.strip().split('\t')
idx = line[-1]
idx = idx.split(';')[1].split("\"")[1]
nline = line[0:6]
nline[3] = idx
ofo.write("\t".join(nline) + "\n")
ofo.close()
os.system("rm process/gene_txn.gtf")
os.system("rm process/gene_txn.bed")
# Run get_tss
get_tss(annotations, biotypes, logger)
assert os.path.exists(cs_annot), 'cannot find %s' % cs_annot
os.system('mv %s process/' % cs_annot)
return 'process/'+cs_annot, ofn
# --------------------------------------------------------------------------- #
# Method for making Enhancer Boundries from PEAKS files and ensuring no #
# overlaps with TSS #
# --------------------------------------------------------------------------- #
def extend_tss(annotation,
sizes,
region,
prange,
logger):
'''Extend TSS for masking.'''
file_name = os.path.basename(annotation)
if 'gtf' in annotation:
basename = file_name.rpartition('.gtf')[0]
else:
if 'gff' not in annotation:
print("Please provide a GTF or GFF file for -t option",
file=sys.stderr)
exit(1)
basename = file_name.rpartition('.gff')[0]
# outputs
up, down = prange
promoter_file = 'process/{}_{}u{}d_promoter.bed'.format(basename, up, down)
flanking_file = 'process/{}_{}.bed'.format(basename, region)
if files_exist([promoter_file, flanking_file]):
return flanking_file, promoter_file
# Convert gff to bed file
bed_file = 'process/{}.bed'.format(basename)
extract = False
if not os.path.exists(bed_file):
extract = True
if 'gtf' in annotation:
convert_command = 'gtf2bed < %s > %s' % (annotation, bed_file)
else:
convert_command = 'gff2bed < %s | awk \'BEGIN{FS=OFS="\t"} '\
'{print $1,$2,$3,$5,$4,$6}\' > %s' \
% (annotation, bed_file)
else:
convert_command = "File Exists"
logger.info("Running convert with %s", convert_command)
if extract:
#subprocess.check_call(shlex.split(convert_command))
os.system(convert_command)
# Extend bed file
flanking_command = 'bedtools slop -i {} -g {} -b {}'.format(bed_file,
sizes,
region)
logger.info("Running flanking around TSS with %s", flanking_command)
subprocess.check_call(shlex.split(flanking_command),
stdout=open(flanking_file, 'w'))
#Generate promoter files
promoter_command = 'bedtools slop -i {} -g {} -l {} -r {} -s'.format(bed_file,
sizes,
up,
down)
logger.info("Running promoter around TSS with %s", promoter_command)
subprocess.check_call(shlex.split(promoter_command),
stdout=open(promoter_file, 'w'))
os.remove(bed_file)
return flanking_file, promoter_file
def merge_peaks(peaks, logger, universe=None):
'''Merge peak files.'''
tmp_file = 'process/tmp.bed'
new_universe = 'process/universe_peaks.bed'
# Merge peaks files and master file if present
if 'gz' not in peaks:
if universe is not None:
merge_command = 'bedops --everything {} {} '\
'> {}'.format(peaks,
universe,
tmp_file)
else:
merge_command = 'bedops --everything {} '\
'> {}'.format(peaks,
tmp_file)
else:
if universe is not None:
merge_command = 'gunzip -c {} '\
'| bedops --everything - {} '\
'> {}'.format(peaks,universe,
tmp_file)
else:
merge_command = 'gunzip -c {} '\
'| bedops --everything - '\
'> {}'.format(peaks,
tmp_file)
logger.info("Running merge with %s", merge_command)
subprocess.check_call(merge_command, shell=True)
os.rename(tmp_file, new_universe)
return new_universe
def exclude_tss(peaks,
tss,
distance,
logger):
'''Exclude peaks that overlap tss.'''
tmp_file = 'process/tmp.bed'
sort_file = 'process/sort.bed'
merge_file = 'process/merge.bed'
merge_peaks = 'process/merged_enhancer_peaks_{}.srt.bed'.format(distance)
if files_exist([merge_peaks]):
return merge_peaks
# Exclude peaks that overlap tss
exclude_command = "bedtools intersect -a {} -b {} -v".format(peaks, tss)
logger.info("Running exclude with %s", exclude_command)
subprocess.check_call(exclude_command, stdout=open(tmp_file, 'w'), shell=True)
# Sort and get unique rows
sort_command = "bedtools sort -i {} | uniq > {}".format(tmp_file, sort_file)
logger.info("Running exclude with %s", sort_command)
subprocess.check_call(sort_command, shell=True)
# Merge peaks
merge_command = "bedtools merge -i {} -d {}".format(sort_file, distance)
logger.info("Running merge with %s", merge_command)
subprocess.check_call(merge_command, stdout=open(merge_file, 'w'), shell=True)
# Sort merged file
sort_command = "bedtools sort -i {} > {}".format(merge_file, merge_peaks)
logger.info("Running sort with %s", sort_command)
subprocess.check_call(sort_command, shell=True)
os.remove(sort_file)
os.remove(tmp_file)
os.remove(merge_file)
return merge_peaks
def bed_distribution(bed, fname, flabel):
'''Explore the enhancer distribution on the merged peaks.'''
enh = pd.read_csv(bed, sep='\t', header=None)
distribution = 'results/{}_distribution_{}.png'.format(fname, flabel)
# Plot enhancer disctribtion
plt.hist(enh[2]-enh[1], bins=100)
plt.xlabel('size (bp)')
plt.ylabel('frequency')
plt.savefig(distribution)
plt.clf()
def enhancer_bounds(tss,
region,
sizes,
distance,
peaks,
prange,
logger):
if len(prange) > 2:
print("Please only use two numbers for Promoter range separated by a -",
file = sys.stderr)
exit(1)
# download chromosome sizes
if not os.path.exists(sizes):
if not os.path.exists('process/%s.chrom.sizes' % sizes):
os.system("wget hgdownload.cse.ucsc.edu/goldenPath/%s/"\
"bigZips/%s.chrom.sizes -P process/" % (sizes, sizes))
sizes = 'process/%s.chrom.sizes' % sizes
# Create a file handler
handler = logging.FileHandler('log/enhancers_boundries.log')
logger.addHandler(handler)
# Run extend_tss
extended_tss, promoter_file = extend_tss(tss,
sizes,
region,
prange,
logger)
# Merge all peak files
universe_peaks = merge_peaks(peaks[0], logger, None)
peaks = peaks[1:]
if len(peaks) > 1:
for p in peaks:
universe_peaks = merge_peaks(p, logger, universe_peaks)
# Exlcude out tss and merge merge peaks
merged = exclude_tss(universe_peaks,
extended_tss,
distance,
logger)
os.remove(universe_peaks)
# Plot distribution
bed_distribution(merged,
'enhancer',
'_exclude_{}_within_{}'.format(region, distance))
bed_distribution(promoter_file,
'promoter',
'_{}up_{}down'.format(prange[0], prange[1]))
return merged, promoter_file
# --------------------------------------------------------------------------- #
# Methods for Merging Transcript Expression and getting a normalized log2 #
# based TPM matrix of expression #
# --------------------------------------------------------------------------- #
def filter_txn_mx(tfmtx):
corrmtx = np.square(tfmtx.corr())
plot_corr_heatmap(corrmtx, "results/prefilter_tx_expression_log2_corr", 24)
mask = corrmtx.columns[corrmtx.mean() > 0.5].to_list()
tfmtx = tfmtx[mask]
corrmtx = np.square(tfmtx.corr())
plot_corr_heatmap(corrmtx, "results/postfilter_tx_expression_log2_corr", 24)
return tfmtx
def get_filtered_transcripts(df, name):
'''Filter for only ENS transcript.'''
transcript_list = list(df['gene_id'])
selected_genes = ['ENS' in c for c in transcript_list]
selected_df = pd.DataFrame(df[selected_genes][['gene_id', 'TPM']])
selected_df.columns = ['gene_id', name]
return selected_df
def merge_transcript(expression_files, logger):
for fname in expression_files:
if not os.path.exists(fname):
print("Path %s does not exist!" % (fname),
file=sys.stderr)
exit(1)
# Create a file handler
handler = logging.FileHandler('log/merge_transcript_expression.log')
logger.addHandler(handler)
# Get list of all expression matrixes
list_files = []
for files in expression_files:
list_files.extend(glob.glob(files))
# Loop through all files to add to dataframe
expression_df = pd.DataFrame()
for f in list_files:
# Read in file name
current_df = pd.read_csv(f, sep='\t')
file_name = os.path.basename(f)
basename = file_name.rpartition('.tsv')[0]
logger.info('Processing: %s', f)
# Filter genes
selected_current_df = get_filtered_transcripts(current_df, basename)
selected_current_df.set_index('gene_id', inplace=True)
del(current_df)
# Append to dataframe
if expression_df.shape == (0, 0):
expression_df = selected_current_df
else:
expression_df = expression_df.join(selected_current_df)
# Convert TPM to log2(TPM)
expression_nfile = 'process/transcript_expression_log2TPM_matrix.txt'
expression_df_log2TPM = np.log2(expression_df+1)
expression_df_log2TPM = filter_txn_mx(expression_df_log2TPM)
expression_df_log2TPM.to_csv(expression_nfile, sep='\t')
plot_mtx(expression_nfile)
return expression_nfile
# --------------------------------------------------------------------------- #
# Methods for Merging Enhancer Expression to get log2 TPM matrix #
# --------------------------------------------------------------------------- #
def enhancer_coverage(alignment_tup):
'''Get coverage for enhancers in sample.'''
enhancer, alignment, logger = alignment_tup
# Calculate coverage
file_name = os.path.basename(alignment)
basename = file_name.rpartition('.bam')[0]
coverage_bed = "process/{}_coverage.bed".format(basename)
coverage_command = "bedtools coverage -sorted "\
"-a {} -b {} > {}".format(enhancer,
alignment,
coverage_bed)
logger.info('Processing coverage using: %s', coverage_command)
subprocess.check_call(coverage_command, shell=True)
def merge_enhancer(enhancers, target, alignment_files, logger):
# Create a file handler
handler = logging.FileHandler('log/merge_enhancer_expression.log')
logger.addHandler(handler)
# Get list of all alignment files
list_files = []
for files in alignment_files:
list_files.extend(glob.glob(files))
# Calculate coverage on all files
all_files_tup = [(enhancers, x, logger) for x in list_files]
cpus = mp.cpu_count()
process = mp.Pool(cpus)
process.map(enhancer_coverage, all_files_tup)
process.close()
process.join()
# Loop through all files to add to dataframe
enhancers_df = pd.read_csv(enhancers, sep='\t', header=None)
expression_df = enhancers_df.iloc[:, 0:3]
expression_df.columns = ['chr', 'start', 'end']
del(enhancers_df)
# List of coverage files
list_of_coverage = glob.glob('{}/process/*_coverage.bed'.format(os.getcwd()))
for f in list_of_coverage:
# Read in file name
file_name = os.path.basename(f)
basename = file_name.rpartition('_coverage.bed')[0]
logger.info('Processing: %s', basename)
# Calculate TPM
counts_df = pd.read_csv(f, sep='\t', header=None)
enhancer_tpm = (counts_df[3]/counts_df[5])\
*(1/sum(counts_df[3]/counts_df[5]))\
*1e6
enhancer_signal = np.log2(enhancer_tpm+1)
expression_df[basename] = enhancer_signal
os.remove(f)
# Convert TPM to log2(TPM)
expression_df['name'] = expression_df['chr'] \
+ ':' \
+ expression_df['start'].map(str) \
+ '-' \
+ expression_df['end'].map(str)
columns = ['chr', 'start', 'end']
expression_df.drop(columns, inplace=True, axis=1)
expression_df.set_index('name', inplace=True)
expression_file="process/enhancer_{}_log2TPM_signal_matrix.txt".format(target)
expression_df.to_csv(expression_file, sep='\t')
bedenh = "process/enh.bed"
os.system("awk '{print $1}' %s "\
"| sed '1d' "\
"| tr ':' '\\t' "\
"| tr '-' '\\t' "\
"> %s" % (expression_file, bedenh))
plot_mtx(expression_file)
return expression_file, bedenh
# --------------------------------------------------------------------------- #
# Methods for Building Transcription Factor logodds matricies using FIMO #
# from meme suir of software #
# --------------------------------------------------------------------------- #
def download_meme(database, species):
if not os.path.exists("motif_databases"):
cmdpipe = ["wget http://meme-suite.org/meme-software/"\
"Databases/motifs/motif_databases.12.18.tgz -P process/",
"tar xzf process/motif_databases.12.18.tgz"]
for cmd in cmdpipe:
try:
# subprocess.check_call(shlex.split(cmd), shell=True)
os.system(cmd)
except:
pass
memels = glob.glob("motif_databases/{}/*".format(database.upper()))
species = '_'.join(species)
memedir = "motif_databases/{}/{}.meme".format(database.upper(), species)
if memedir not in memels:
print("MEME database {} not found in {}".format(species, database))
exit(1)
return memedir
def download_genome(orgname, gvers, logger):
path = '/'.join(os.path.realpath(__file__).split('/')[:-2])
cmd = [path + '/trenco_tools/get_genome.py', '--refseq', '-s', orgname]
genomes = subprocess.check_output(cmd + ['--get-versions'])
genomes = genomes.decode("utf-8").replace('\n','\t').split('\t')
genome = ''
print(genomes)
for i in range(4,len(genomes),4):
if gvers:
if gvers in genomes[i]:
genome = genomes[i]
else:
if genomes[i-1] == "latest":
genome = genomes[i]
assert genome
if os.path.exists(genome + '_genomic.fna.gz'):
subprocess.call(['gunzip', genome + '_genomic.fna.gz'])
elif os.path.exists(genome + '_genomic.fna'):
genome = genome + '_genomic.fna'
else:
if gvers:
cmd += ['-v', gvers]
gout = subprocess.check_output(cmd + ['--use-ucsc'])
if "Completed" not in str(gout):
logger.warning('Error: Download output - {}'.format(gout))
exit(1)
genome = gout.decode("utf-8")\
.replace('\n','')\
.split(':')[-1]\
.replace('.gz','')
return genome
def fimo_2_mtx(nfile,
name,
gff = False,
correction = None):
assert os.path.exists(nfile)
if '/' in name:
name = name.split('/')[-1]
tfmx = 'process/TFx{}_score_matrix.txt'.format(name)
if os.path.exists(tfmx):
return tfmx
if gff: # NOT RECOMMENDED: the score is different
melted_df = pd.read_csv(nfile,
sep="\t",
skiprows=1,
header=None,
engine='python')
melted_df.columns = ['seqname',
'source',
'feature',
'start',
'end',
'score',
'strand',
'frame',
'attribute']
# Function to parse the attribute column into multiple named
# columns of attributes VERY memory intensive
attributes = pd.DataFrame.from_dict(
[dict(
a.split('=') for a in b if a
) for b in melted_df['attribute'].str.split(";")
])
try:
melted_df['MotifID'] = attributes['Alias']
except:
melted_df['MotifID'] = attributes['Name']
melted_df = melted_df[['MotifID', 'seqname', 'score']]
else:
melted_df = pd.read_csv(nfile, sep="\t", skipfooter=4, engine='python')
melted_df = melted_df[['motif_alt_id', 'sequence_name', 'score']]
melted_df.columns = ['MotifID', 'seqname', 'score']
#plot the histogram of the scores
s = np.float64(melted_df[['score']])
plt.hist(s, bins=100)
plt.xlabel('FIMO score')
plt.ylabel('frequancy')
plt.savefig('results/fimo_score_freq_histi{}.png'.format(name))
plt.clf()
df_mtx = pd.pivot_table(melted_df,
index='MotifID',
columns='seqname',
values='score')
df_mtx.fillna(0, inplace=True)
if correction:
converter = load_convert_name(correction)
df_mtx = df_mtx.rename(converter, axis=0)
df_mtx.to_csv(tfmx, sep='\t')
return tfmx
def run_fimo(odir,
genome,
motif_file,
bed,
logger,
correction = None,
wiggle = None,
use_priors = False):
verscmd = ['bedtools',
'--version']
bedvers = subprocess.check_output(verscmd, shell=True).decode('utf-8')
bedvers = re.search(r'v([\d.]+)', bedvers).group(1).replace(".", "")
cmdpipe = [['bedtools',
'getfasta',
'-fi', genome,
'-bed', bed,
'-fo',
'{}.fasta'.format(bed)]]
if "promoter" in bed:
if int(bedvers) < 2290:
cmdpipe[-1].append("-name")
else:
cmdpipe[-1].append("-nameOnly")
fullout = '{}/{}.motif'.format(odir, bed)
if not os.path.exists(fullout):
if use_priors:
cmdpipe += [] # TODO finish priors script
else:
cmdpipe += [['fimo',
'-o',
fullout,
motif_file,
'{}.fasta'.format(bed)]]
files_needed = [genome, motif_file, bed]
for f in files_needed:
if not os.path.exists(f):
logger.warning('Cannot find file: {}'.format(f))
exit(1)
for cmd in cmdpipe:
print(" ".join(cmd))
subprocess.check_call(cmd)
fimo_fn = '{}/fimo.tsv'.format(fullout)
tfmx = fimo_2_mtx(fimo_fn, bed.split('/')[-1], correction = correction)
return tfmx
def build_tf_matrix(bed,
meme_db,
memedb,
orgname,
gvers,
logger,
vector = None):
correction = check_if_commons(meme_db, False)
meme_file = download_meme(meme_db, memedb)
genome = download_genome(orgname, gvers, logger)
fn_mx = run_fimo('.', genome, meme_file, bed, logger, correction)
logger.info("Using MEME: {}".format(meme_file))
logger.info("Using Genome: {}".format(genome))
logger.info("Built Motif file: {}".format(genome))
return fn_mx
# --------------------------------------------------------------------------- #
# Main methods for getting Enhancer Gene Networks and building small TAD #
# matricies of enhancers x genes #
# --------------------------------------------------------------------------- #
def init_e(_exp, _enh, _k):
"""To provide global variables shared across multiple processes."""
global s_gene_tpm
global s_enh_tpm
global sample
s_gene_tpm = _exp
s_enh_tpm = _enh
sample = _k
def get_distance_df(d):
"""To get distance between enh and gene for each TAD."""
enh_path = '{}/enh.bed'.format(d)
gene_path = '{}/gene.bed'.format(d)
if os.path.isfile(enh_path) and os.path.isfile(gene_path):
enh_df = pd.read_csv(enh_path, sep='\t', header=None)
gene_df = pd.read_csv(gene_path, sep='\t', header=None)
# generate enh identifiers
enh_pos = []
for r in enh_df.iterrows():
pos = "{}:{}-{}".format(r[1][0], r[1][1], r[1][2])
enh_pos.append(pos)
# get gene names
gene_names = list(gene_df[3])
out_d = pd.DataFrame(0, index=enh_pos, columns=gene_names)
# calculate enh gene distance
for g in gene_df.iterrows():
name = g[1][3]
if g[1][5] == '-':
g_start = g[1][2]
else:
g_start = g[1][1]
for e in enh_df.iterrows():
e_pos = "{}:{}-{}".format(e[1][0], e[1][1], e[1][2])
e_mid = e[1][1]+((e[1][2]-e[1][1])/2)
distance = np.abs(g_start - e_mid)
out_d.loc[e_pos, name] = int(distance)
out_d.to_csv('{}/enh_gene_distance.txt'.format(d), sep='\t')
scaled_out_d = get_distance_based_weight(out_d)
scaled_out_d.to_csv('{}/enh_gene_distance_scaled.txt'.format(d),
sep='\t')
def get_distance_based_weight(distance):
"""To scale the distance between enh and gene for each TAD."""
# set weight 0 for distance greater than 500 kb
# TODO clean this out of the code if we determine it to be unnecessary
# distance[distance > 500000] = 1e10000
dist_weight = 1/np.log2(distance)
dist_list = np.sort(list(set(dist_weight.values.flatten())))
if set(dist_weight.values.flatten()) == {0} or len(dist_list) == 1:
scaled_dist_weight = dist_weight
else:
wmin = np.sort(list(set(dist_weight.values.flatten())))[1]
wmax = np.sort(list(set(dist_weight.values.flatten())))[-1]
scaled_dist_weight = (dist_weight-wmin)/(wmax-wmin)
scaled_dist_weight[scaled_dist_weight < 0] = 0
return(scaled_dist_weight)
def get_enh_gene_weights(d):
"""To get the enh gene weight based on enh activity and distance."""
dist_path = '{}/enh_gene_distance_scaled.txt'.format(d)
weight_path = '{}/{}_enh_gene_weight.txt'.format(d, sample)
if os.path.exists(dist_path) and not os.path.exists(weight_path):
dist_df = pd.read_csv(dist_path, sep='\t', index_col=0)
tad_gene_tpm = s_gene_tpm.loc[
s_gene_tpm.index.intersection(dist_df.columns)
].reindex(dist_df.columns)
tad_enh_tpm = s_enh_tpm.loc[
s_enh_tpm.index.intersection(dist_df.index)
].reindex(dist_df.index)
try:
enh_gene_weight = np.multiply(
dist_df, np.sqrt(np.matmul(tad_enh_tpm, tad_gene_tpm.T)))
except:
enh_gene_weight = np.multiply(
dist_df, np.sqrt(tad_enh_tpm.dot(tad_gene_tpm.T)))
enh_gene_weight.to_csv(weight_path, sep='\t')
def plot_scatter(x_vals, y_vals, outdir, tad):
#To scatter plot the scaled distance as function of enh gene distance.
plt.scatter(x_vals,
y_vals,
edgecolors='#2374AB',
facecolors='none',
s=30,
linewidths=1,
alpha=0.4)
plt.xscale('log')
plt.vlines(500000, 0, 0.9, label='500kb')
plt.xlabel('enh-gene distance (log scale)')
plt.ylabel('weight')
plt.legend()
plt.savefig('{}/{}-scatter_enh_gene_scaled_distance.png'.format(outdir,tad))
plt.clf()
def plot_heatmap(df, outdir, tad):
"""To plot heatmap of the scaled distance between enh and gene."""
sb.set_context('paper', font_scale=1.8)
g = sb.heatmap(df, yticklabels=False, cmap='Blues')
fig = g.get_figure()
fig.savefig('{}/{}-heatmap_enh_gene_scaled_distance.png'.format(outdir,tad))
plt.clf()
def get_enh_networks(sample,
gene,
enh,
sample_out,
threads,
enhBED,
geneBED,
tadBED,
mask = []):
os.makedirs(os.path.dirname('log/{}_log_enh_network.txt'.format(sample)),
exist_ok=True)
# get the enhancer and gene quantifications
gene_tpm = | pd.read_csv(gene, sep='\t', index_col=0) | pandas.read_csv |
# -*- coding: utf-8 -*-
"""
@author: hkaneko
"""
import math
import sys
import numpy as np
import pandas as pd
import sample_functions
from sklearn import metrics, svm
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import cross_val_predict, train_test_split, GridSearchCV
from sklearn.neighbors import KNeighborsClassifier
method_name = 'rf' # 'knn' or 'svm' or 'rf'
add_nonlinear_terms_flag = False # True (二乗項・交差項を追加) or False (追加しない)
number_of_test_samples = 800
fold_number = 2 # N-fold CV の N
max_number_of_k = 20 # 使用する k の最大値
svm_cs = 2 ** np.arange(-5, 11, dtype=float)
svm_gammas = 2 ** np.arange(-20, 11, dtype=float)
rf_number_of_trees = 300 # RF における決定木の数
rf_x_variables_rates = np.arange(1, 11, dtype=float) / 10 # 1 つの決定木における説明変数の数の割合の候補
if method_name != 'knn' and method_name != 'svm' and method_name != 'rf':
sys.exit('\'{0}\' というクラス分類手法はありません。method_name を見直してください。'.format(method_name))
dataset = | pd.read_csv('unique_m.csv', index_col=-1) | pandas.read_csv |
from datetime import datetime, timedelta
import pandas as pd
class TuDataModel:
"""
Class Terminal Unit, never modify any variable direct. The idea is that all gets managed via functions
"""
def __init__(self, name):
"""
Constructor of the Tu as a holder for all the TG Tu Data. we will refer in the function descriptions as Tu
PLease never change parameters directly and use the functions and setters and getters
:param name: str, name of the instanced unit
"""
# name of the widget
self.name = name
# beginning of time from 1st connection
self.first_connection = -1
# counter for disconnections
self.disc_counter = 0
# management of disconnections event
self.disconnection_start = None # start of event
self.disconnection_end = None # end of event
self.connection_state = False # state of the connection
self.disconnection_last_event_time = timedelta(seconds=0) # the total time of last disconnection
# total disconnection time
self.disconnection_total_time = timedelta(seconds=0)
# total actual availability
self.availability = 100.0
# Variables to capture and show
self.local_sector = None
self.rssi = None
self.snr = None
self.rx_mcs = None
self.tx_mcs = None
self.rx_speed_num = None
self.tx_speed_num = None
self.rx_mcs_dr = None
self.tx_mcs_dr = None
self.tx_power_index = None
# disconnections dataframe to have the list
# we need to append with a pd.Series(data_in_a_dict, name=datetime.now()/or time)
self.disconnections = pd.DataFrame(columns=['Time End', 'Disconnection #', 'Downtime', 'Availability'])
self.parameters_df = pd.DataFrame(columns=['Power Index', 'RSSI', 'SNR', 'MCS-RX', 'MCS-TX', 'MCS-DR-RX',
'MCS-DR-TX', 'Local Sector'])
# setters and getters for the internal variables
def get_local_sector(self):
"""
Returns the local sector antenna index of the connected unit
:return: int, index of connected unit
"""
return self.local_sector
def set_local_sector(self, ls):
"""
Sets the local sector index
:param ls: int, local sector index of connected unit
:return: None
"""
self.local_sector = ls
def get_rssi(self):
"""
Gets the rssi value of the connected unit
:return: int, rssi value of the connected unit
"""
return self.rssi
def set_rssi(self, rssi):
"""
Sets the rssi
:param rssi: int, rssi to set to the object
:return: None
"""
self.rssi = rssi
def get_snr(self):
"""
Gets the CINR of the connected unit
:return: int, CINR of the connected unit
"""
return self.snr
def set_snr(self, snr):
"""
Sets the CINR value of the connected unit
:param snr: int, CINR value
:return: None
"""
self.snr = snr
def get_rxmcs(self):
"""
Gets the Rx Modulation Coding Scheme of the connected Unit
:return: int, Rx MCS value
"""
return self.rx_mcs
def set_rxmcs(self, rxmcs):
"""
Sets the Rx Modulation Coding Scheme of the connected Unit
:param rxmcs: int, Rx MCS value
:return: None
"""
self.rx_mcs = rxmcs
def get_txmcs(self):
"""
Gets the Tx Modulation Coding Scheme of the connected Unit
:return: int, Tx MCS value
"""
return self.tx_mcs
def set_txmcs(self, txmcs):
"""
Sets the Tx Modulation Coding Scheme of the connected Unit
:param txmcs: int, Tx MCS value
:return: None
"""
self.tx_mcs = txmcs
def get_rxspeednum(self):
"""
Gets the Rx capacity currently going in the Tu in Mbps
:return: float, Rx In capacity in Mbps
"""
return self.rx_speed_num
def set_rxspeednum(self, rxspeednum):
"""
Sets the Rx capacity currently going in the Tu in Mbps
:param rxspeednum: float, Rx In capacity in Mbps
:return: None
"""
self.rx_speed_num = rxspeednum
def get_txspeednum(self):
"""
Gets the Tx capacity currently going in the Tu in Mbps
:return: float, Tx In capacity in Mbps
"""
return self.tx_speed_num
def set_txspeednum(self, txspeednum):
"""
Sets the Tx capacity currently going in the Tu in Mbps
:param txspeednum: float, Rx In capacity in Mbps
:return: None
"""
self.tx_speed_num = txspeednum
def get_rxmcsdr(self):
"""
Gets the Rx Over the Air Data Rate
:return: int, Rx OTA DR
"""
return self.rx_mcs_dr
def set_rxmcsdr(self):
"""
Sets the Rx Over the Air Dara Rate. based on the RX-MCS
:param rxmcsdr: int, OTA DR value
:return: None
"""
value_rx = self.get_rxmcs()
if value_rx == '0':
self.rx_mcs_dr = '0'
elif value_rx == '2':
self.rx_mcs_dr = '620'
elif value_rx == '3':
self.rx_mcs_dr = '780'
elif value_rx == '4':
self.rx_mcs_dr = '950'
elif value_rx == '7':
self.rx_mcs_dr = '1580'
elif value_rx == '8':
self.rx_mcs_dr = '1900'
elif value_rx == '9':
self.rx_mcs_dr = '2050'
elif value_rx == '10':
self.rx_mcs_dr = '2500'
elif value_rx == '11':
self.rx_mcs_dr = '3150'
elif value_rx == '12':
self.rx_mcs_dr = '3800'
else:
self.rx_mcs_dr = '0'
def get_txmcsdr(self):
"""
Gets the Tx Over the Air Data Rate
:return: int, Tx OTA DR
"""
return self.tx_mcs_dr
def set_txmcsdr(self):
"""
Sets the Tx Over the Air Dara Rate. Based on TX-MCS
:param txmcsdr: int, OTA DR value
:return: None
"""
value_tx = self.get_txmcs()
if value_tx == '0':
self.tx_mcs_dr = '0'
elif value_tx == '2':
self.tx_mcs_dr = '620'
elif value_tx == '3':
self.tx_mcs_dr = '780'
elif value_tx == '4':
self.tx_mcs_dr = '950'
elif value_tx == '7':
self.tx_mcs_dr = '1580'
elif value_tx == '8':
self.tx_mcs_dr = '1900'
elif value_tx == '9':
self.tx_mcs_dr = '2050'
elif value_tx == '10':
self.tx_mcs_dr = '2500'
elif value_tx == '11':
self.tx_mcs_dr = '3150'
elif value_tx == '12':
self.tx_mcs_dr = '3800'
else:
self.tx_mcs_dr = '0'
def get_power_index(self):
"""
Gets the Power Index
:return: int, Power Index
"""
return self.tx_power_index
def set_power_index(self, power_index_):
"""
Sets the Power Index
:return: int, Power Index
"""
self.tx_power_index = power_index_
def get_availability(self):
"""
Gets the Availability
:return: float, calculated availability value
"""
return self.availability
def get_disconnection_counter(self):
return self.disc_counter
def get_disconnection_ldt(self):
return self.disconnection_last_event_time
def get_disconnection_lds(self):
return self.disconnection_start
def get_disconnection_tdt(self):
return self.disconnection_total_time
def get_connection_status(self):
return self.connection_state
# Automated behaviour of the object for connections and disconnections
def disconnected(self, time_disc):
"""
Function that sets the start of a disconnection. It will get a datetime time
:param time_disc: datetime, will set the time
:return: None
"""
if self.connection_state: # the Tu was connected and we will disconnect it
self.connection_state = False # Set the connection flag down
self.disconnection_start = time_disc # record the time of the disconnection time
self.disc_counter = self.increment_disconnections(self.disc_counter) # increment the counter of disconn.
# We update parameters to reflect the disconnection:
self.set_rssi(-100)
self.set_snr(0)
self.set_rxmcs(0)
self.set_txmcs(0)
else: # we enter the disconnected state but the unit was already disconnected
pass
def connected(self, time_con):
if not self.connection_state and self.first_connection != -1: # the Tu was disconnected and it got connected
self.disconnection_end = time_con # record the time the disconnection time ended
# calculate the total time of the disconnection
self.disconnection_last_event_time = self.calculate_disconnection_time(self.disconnection_start, time_con)
# calculate the total time of disconnection
self.disconnection_total_time = self.update_total_time(self.disconnection_total_time,
self.disconnection_last_event_time)
# calculate availability
availability = self.calculate_availability(self.disconnection_total_time, self.first_connection, time_con)
self.availability = availability
# update the disconnections dataframe
# update 1 : update time end
self.disconnections = self.update_record(self.disconnections, self.disconnection_start, 'Time End', time_con)
# update 2: update duration of the desconnection
self.disconnections = self.update_record(self.disconnections, self.disconnection_start, 'Downtime',
f'{self.disconnection_last_event_time}')
# update 3: update of the availability
self.disconnections = self.update_record(self.disconnections, self.disconnection_start, 'Availability',
availability)
# update 4: update of disconnection#
self.disconnections = self.update_record(self.disconnections, self.disconnection_start, 'Disconnection #',
self.disc_counter)
self.connection_state = True # change flag to connected
elif self.first_connection == -1: # the Tu was first connected
self.first_connection = time_con
self.connection_state = True
else:
# calculate availability
availability = self.calculate_availability(self.disconnection_total_time, self.first_connection, time_con)
self.availability = availability
@staticmethod
def calculate_availability(time_span, start_t, time_t):
"""
Calculate availability of time_span from start to time
:param time_span: datetime, time where we can to calculate availability
:param start_t: datetime, start time to calculate availability
:param time_t: datetime, time to calculate availability
:return: float, availability
"""
if start_t == -1: # the unit was never connected
return 0
return (1 - (time_span / (time_t - start_t))) * 100
@staticmethod
def update_total_time(total_time_counter, update):
"""
Updates the total_time_counter by update
:param total_time_counter: datetime, has the current total time
:param update: datetime, the value to update the total time
:return: total_time_counter + update
"""
return total_time_counter + update
@staticmethod
def calculate_disconnection_time(start, end):
"""
Calculates the total time of disconnection end - start
:param start: datetime, start time of the event
:param end: datetime, end time of the event
:return: end - start
"""
return end - start
@staticmethod
def update_record(df, find_variable, field, update_data):
df.loc[find_variable, field] = update_data
return df
def create_end(self, end_time):
end_ = pd.Series(
{'Time End': datetime.now(), 'Disconnection #': self.disc_counter,
'Downtime': f'{self.disconnection_total_time}',
'Availability': self.availability}, name='Total')
self.disconnections = self.disconnections.append(end_)
# Change type of columns to print in excel to proper value
self.disconnections['Disconnection #'] = self.disconnections['Disconnection #'].astype(int)
self.disconnections['Availability'] = self.disconnections['Availability'].astype(float)
@staticmethod
def increment_disconnections(counter):
"""
Function that will add counter + 1 and return it
:param counter: int, disconnections counter
:return: int, counter + 1
"""
return counter + 1
@staticmethod
def seconds_to_split(time_split):
"""
Function that will get a time (timedelta) range and will convert it to days minutes seconds. It will trunkate
the value to only days, hours minutes and seconds. if the time is not timedelta it will raise an exception
:return: days (int), hours (int), minutes (int), seconds (int)
"""
# validation that the time is timedelta
if isinstance(timedelta, time_split):
total_seconds = time_split.seconds
days = time_split.days
hours = total_seconds // 3600
total_seconds_wo_hours = total_seconds - (hours * 3600)
minutes = total_seconds_wo_hours // 60
seconds = total_seconds_wo_hours - (minutes * 60)
return set(days, hours, minutes, seconds)
else:
raise ValueError(f'The input to the function is not timedelta, it is {type(time_split)}')
def print(self):
print('*****Tu instance*****')
print(f'- name: {self.name}')
print(f'- first connected: {self.first_connection}')
print(f'-------conection status------------')
print(f'connection: {self.connection_state}')
print(f'-------disconnection info----------')
print(f'- diconnections: {self.disc_counter}')
print(f'- disconnection event-start: {self.disconnection_start}')
print(f'- disconnection event-end: {self.disconnection_end}')
print(f'- disconnection event time: {self.disconnection_last_event_time}')
print(f'----disconnection total time-------')
print(f'- total time disconnected: {self.disconnection_total_time}')
print(f'-----total availability at the time of print----')
print(f'- availability: {self.calculate_availability(self.disconnection_total_time, self.first_connection, datetime.now())}')
print(f'--------operation parameters-------')
print(f'- local sector: {self.local_sector}')
print(f'- rssi: {self.rssi}')
print(f'- srn: {self.rssi}')
print(f'- rx_mcs: {self.rx_mcs}')
print(f'- tx_mcs: {self.tx_mcs}')
print(f'- rx_speed_num: {self.rx_speed_num}')
print(f'- tx_speed_num: {self.tx_speed_num}')
print(f'- rx_mcs_dr: {self.rx_mcs_dr}')
print(f'- tx_mcs_dr: {self.tx_mcs_dr}')
print(f'- power_index: {self.tx_power_index}')
print(f'------------events dataframe-------------')
print(f'{self.disconnections}')
if __name__ == '__main__':
import time
# options to display the whole dataframe for checks
pd.set_option('display.max_rows', None)
pd.set_option('display.max_columns', None)
pd.set_option('display.width', None)
| pd.set_option('display.max_colwidth', -1) | pandas.set_option |
# Copyright 2022 <NAME>
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""Kraken public API."""
from datetime import datetime
from typing import Any
from typing import Iterable
import httpx
import numpy as np
import pandas as pd
from typeguard import typechecked
from ..client import Client
from .utils import content
from .utils import public_url
Pair = list[tuple[str, str]] | tuple[str, str] | list[str] | str
@typechecked
def _make_array(junk: Any) -> np.ndarray:
return np.asarray(junk).astype(np.float)
@typechecked
def _make_pair(pair: Pair) -> str:
if isinstance(pair, list):
pair = ",".join(pair)
if isinstance(pair, tuple):
pair = "".join(pair)
return pair
@typechecked
def _format_ohlc(
ohlc: pd.DataFrame, *, interval: int, ascending: bool = True
) -> pd.DataFrame:
if ohlc.empty:
return ohlc
ohlc["dtime"] = | pd.to_datetime(ohlc.time, unit="s") | pandas.to_datetime |
'''
Preprocessing Tranformers Based on sci-kit's API
By <NAME>
Created on June 12, 2017
'''
import copy
import pandas as pd
import numpy as np
import transforms3d as t3d
import scipy.ndimage.filters as filters
from sklearn.base import BaseEstimator, TransformerMixin
from analysis.pymo.rotation_tools import Rotation, euler2expmap, euler2expmap2, expmap2euler, euler_reorder, unroll
from analysis.pymo.Quaternions import Quaternions
from analysis.pymo.Pivots import Pivots
class MocapParameterizer(BaseEstimator, TransformerMixin):
def __init__(self, param_type = 'euler'):
'''
param_type = {'euler', 'quat', 'expmap', 'position', 'expmap2pos'}
'''
self.param_type = param_type
def fit(self, X, y=None):
return self
def transform(self, X, y=None):
print("MocapParameterizer: " + self.param_type)
if self.param_type == 'euler':
return X
elif self.param_type == 'expmap':
return self._to_expmap(X)
elif self.param_type == 'quat':
return X
elif self.param_type == 'position':
return self._to_pos(X)
elif self.param_type == 'expmap2pos':
return self._expmap_to_pos(X)
else:
raise 'param types: euler, quat, expmap, position, expmap2pos'
# return X
def inverse_transform(self, X, copy=None):
if self.param_type == 'euler':
return X
elif self.param_type == 'expmap':
return self._expmap_to_euler(X)
elif self.param_type == 'quat':
raise 'quat2euler is not supported'
elif self.param_type == 'position':
# raise 'positions 2 eulers is not supported'
print('positions 2 eulers is not supported')
return X
else:
raise 'param types: euler, quat, expmap, position'
def _to_pos(self, X):
'''Converts joints rotations in Euler angles to joint positions'''
Q = []
for track in X:
channels = []
titles = []
euler_df = track.values
# Create a new DataFrame to store the exponential map rep
pos_df = pd.DataFrame(index=euler_df.index)
# Copy the root rotations into the new DataFrame
# rxp = '%s_Xrotation'%track.root_name
# ryp = '%s_Yrotation'%track.root_name
# rzp = '%s_Zrotation'%track.root_name
# pos_df[rxp] = pd.Series(data=euler_df[rxp], index=pos_df.index)
# pos_df[ryp] = pd.Series(data=euler_df[ryp], index=pos_df.index)
# pos_df[rzp] = pd.Series(data=euler_df[rzp], index=pos_df.index)
# List the columns that contain rotation channels
rot_cols = [c for c in euler_df.columns if ('rotation' in c)]
# List the columns that contain position channels
pos_cols = [c for c in euler_df.columns if ('position' in c)]
# List the joints that are not end sites, i.e., have channels
joints = (joint for joint in track.skeleton)
tree_data = {}
for joint in track.traverse():
parent = track.skeleton[joint]['parent']
rot_order = track.skeleton[joint]['order']
#print("rot_order:" + joint + " :" + rot_order)
# Get the rotation columns that belong to this joint
rc = euler_df[[c for c in rot_cols if joint in c]]
# Get the position columns that belong to this joint
pc = euler_df[[c for c in pos_cols if joint in c]]
# Make sure the columns are organized in xyz order
if rc.shape[1] < 3:
euler_values = np.zeros((euler_df.shape[0], 3))
rot_order = "XYZ"
else:
euler_values = np.pi/180.0*np.transpose(np.array([track.values['%s_%srotation'%(joint, rot_order[0])], track.values['%s_%srotation'%(joint, rot_order[1])], track.values['%s_%srotation'%(joint, rot_order[2])]]))
if pc.shape[1] < 3:
pos_values = np.asarray([[0,0,0] for f in pc.iterrows()])
else:
pos_values =np.asarray([[f[1]['%s_Xposition'%joint],
f[1]['%s_Yposition'%joint],
f[1]['%s_Zposition'%joint]] for f in pc.iterrows()])
quats = Quaternions.from_euler(np.asarray(euler_values), order=rot_order.lower(), world=False)
tree_data[joint]=[
[], # to store the rotation matrix
[] # to store the calculated position
]
if track.root_name == joint:
tree_data[joint][0] = quats#rotmats
# tree_data[joint][1] = np.add(pos_values, track.skeleton[joint]['offsets'])
tree_data[joint][1] = pos_values
else:
# for every frame i, multiply this joint's rotmat to the rotmat of its parent
tree_data[joint][0] = tree_data[parent][0]*quats# np.matmul(rotmats, tree_data[parent][0])
# add the position channel to the offset and store it in k, for every frame i
k = pos_values + np.asarray(track.skeleton[joint]['offsets'])
# multiply k to the rotmat of the parent for every frame i
q = tree_data[parent][0]*k #np.matmul(k.reshape(k.shape[0],1,3), tree_data[parent][0])
# add q to the position of the parent, for every frame i
tree_data[joint][1] = tree_data[parent][1] + q #q.reshape(k.shape[0],3) + tree_data[parent][1]
# Create the corresponding columns in the new DataFrame
pos_df['%s_Xposition'%joint] = pd.Series(data=[e[0] for e in tree_data[joint][1]], index=pos_df.index)
pos_df['%s_Yposition'%joint] = pd.Series(data=[e[1] for e in tree_data[joint][1]], index=pos_df.index)
pos_df['%s_Zposition'%joint] = pd.Series(data=[e[2] for e in tree_data[joint][1]], index=pos_df.index)
new_track = track.clone()
new_track.values = pos_df
Q.append(new_track)
return Q
def _expmap2rot(self, expmap):
theta = np.linalg.norm(expmap, axis=1, keepdims=True)
nz = np.nonzero(theta)[0]
expmap[nz,:] = expmap[nz,:]/theta[nz]
nrows=expmap.shape[0]
x = expmap[:,0]
y = expmap[:,1]
z = expmap[:,2]
s = np.sin(theta*0.5).reshape(nrows)
c = np.cos(theta*0.5).reshape(nrows)
rotmats = np.zeros((nrows, 3, 3))
rotmats[:,0,0] = 2*(x*x-1)*s*s+1
rotmats[:,0,1] = 2*x*y*s*s-2*z*c*s
rotmats[:,0,2] = 2*x*z*s*s+2*y*c*s
rotmats[:,1,0] = 2*x*y*s*s+2*z*c*s
rotmats[:,1,1] = 2*(y*y-1)*s*s+1
rotmats[:,1,2] = 2*y*z*s*s-2*x*c*s
rotmats[:,2,0] = 2*x*z*s*s-2*y*c*s
rotmats[:,2,1] = 2*y*z*s*s+2*x*c*s
rotmats[:,2,2] = 2*(z*z-1)*s*s+1
return rotmats
def _expmap_to_pos(self, X):
'''Converts joints rotations in expmap notation to joint positions'''
Q = []
for track in X:
channels = []
titles = []
exp_df = track.values
# Create a new DataFrame to store the exponential map rep
pos_df = pd.DataFrame(index=exp_df.index)
# Copy the root rotations into the new DataFrame
# rxp = '%s_Xrotation'%track.root_name
# ryp = '%s_Yrotation'%track.root_name
# rzp = '%s_Zrotation'%track.root_name
# pos_df[rxp] = pd.Series(data=euler_df[rxp], index=pos_df.index)
# pos_df[ryp] = pd.Series(data=euler_df[ryp], index=pos_df.index)
# pos_df[rzp] = pd.Series(data=euler_df[rzp], index=pos_df.index)
# List the columns that contain rotation channels
exp_params = [c for c in exp_df.columns if ( any(p in c for p in ['alpha', 'beta','gamma']) and 'Nub' not in c)]
# List the joints that are not end sites, i.e., have channels
joints = (joint for joint in track.skeleton)
tree_data = {}
for joint in track.traverse():
parent = track.skeleton[joint]['parent']
if 'Nub' not in joint:
r = exp_df[[c for c in exp_params if joint in c]] # Get the columns that belong to this joint
expmap = r.values
#expmap = [[f[1]['%s_alpha'%joint], f[1]['%s_beta'%joint], f[1]['%s_gamma'%joint]] for f in r.iterrows()]
else:
expmap = np.zeros((exp_df.shape[0], 3))
# Convert the eulers to rotation matrices
#rotmats = np.asarray([Rotation(f, 'expmap').rotmat for f in expmap])
#angs = np.linalg.norm(expmap,axis=1, keepdims=True)
rotmats = self._expmap2rot(expmap)
tree_data[joint]=[
[], # to store the rotation matrix
[] # to store the calculated position
]
pos_values = np.zeros((exp_df.shape[0], 3))
if track.root_name == joint:
tree_data[joint][0] = rotmats
# tree_data[joint][1] = np.add(pos_values, track.skeleton[joint]['offsets'])
tree_data[joint][1] = pos_values
else:
# for every frame i, multiply this joint's rotmat to the rotmat of its parent
tree_data[joint][0] = np.matmul(rotmats, tree_data[parent][0])
# add the position channel to the offset and store it in k, for every frame i
k = pos_values + track.skeleton[joint]['offsets']
# multiply k to the rotmat of the parent for every frame i
q = np.matmul(k.reshape(k.shape[0],1,3), tree_data[parent][0])
# add q to the position of the parent, for every frame i
tree_data[joint][1] = q.reshape(k.shape[0],3) + tree_data[parent][1]
# Create the corresponding columns in the new DataFrame
pos_df['%s_Xposition'%joint] = pd.Series(data=tree_data[joint][1][:,0], index=pos_df.index)
pos_df['%s_Yposition'%joint] = pd.Series(data=tree_data[joint][1][:,1], index=pos_df.index)
pos_df['%s_Zposition'%joint] = pd.Series(data=tree_data[joint][1][:,2], index=pos_df.index)
new_track = track.clone()
new_track.values = pos_df
Q.append(new_track)
return Q
def _to_expmap(self, X):
'''Converts Euler angles to Exponential Maps'''
Q = []
for track in X:
channels = []
titles = []
euler_df = track.values
# Create a new DataFrame to store the exponential map rep
exp_df = euler_df.copy()# pd.DataFrame(index=euler_df.index)
# Copy the root positions into the new DataFrame
#rxp = '%s_Xposition'%track.root_name
#ryp = '%s_Yposition'%track.root_name
#rzp = '%s_Zposition'%track.root_name
#exp_df[rxp] = pd.Series(data=euler_df[rxp], index=exp_df.index)
#exp_df[ryp] = pd.Series(data=euler_df[ryp], index=exp_df.index)
#exp_df[rzp] = pd.Series(data=euler_df[rzp], index=exp_df.index)
# List the columns that contain rotation channels
rots = [c for c in euler_df.columns if ('rotation' in c and 'Nub' not in c)]
# List the joints that are not end sites, i.e., have channels
joints = (joint for joint in track.skeleton if 'Nub' not in joint)
for joint in joints:
#print(joint)
r = euler_df[[c for c in rots if joint in c]] # Get the columns that belong to this joint
rot_order = track.skeleton[joint]['order']
r1_col = '%s_%srotation'%(joint, rot_order[0])
r2_col = '%s_%srotation'%(joint, rot_order[1])
r3_col = '%s_%srotation'%(joint, rot_order[2])
exp_df.drop([r1_col, r2_col, r3_col], axis=1, inplace=True)
euler = [[f[1][r1_col], f[1][r2_col], f[1][r3_col]] for f in r.iterrows()]
#exps = [Rotation(f, 'euler', from_deg=True, order=rot_order).to_expmap() for f in euler] # Convert the eulers to exp maps
exps = unroll(np.array([euler2expmap(f, rot_order, True) for f in euler])) # Convert the exp maps to eulers
# exps = np.array([euler2expmap(f, rot_order, True) for f in euler]) # Convert the exp maps to eulers
#exps = euler2expmap2(euler, rot_order, True) # Convert the eulers to exp maps
# Create the corresponding columns in the new DataFrame
exp_df.insert(loc=0, column='%s_gamma'%joint, value=pd.Series(data=[e[2] for e in exps], index=exp_df.index))
exp_df.insert(loc=0, column='%s_beta'%joint, value=pd.Series(data=[e[1] for e in exps], index=exp_df.index))
exp_df.insert(loc=0, column='%s_alpha'%joint, value=pd.Series(data=[e[0] for e in exps], index=exp_df.index))
#print(exp_df.columns)
new_track = track.clone()
new_track.values = exp_df
Q.append(new_track)
return Q
def _expmap_to_euler(self, X):
Q = []
for track in X:
channels = []
titles = []
exp_df = track.values
# Create a new DataFrame to store the exponential map rep
#euler_df = pd.DataFrame(index=exp_df.index)
euler_df = exp_df.copy()
# Copy the root positions into the new DataFrame
#rxp = '%s_Xposition'%track.root_name
#ryp = '%s_Yposition'%track.root_name
#rzp = '%s_Zposition'%track.root_name
#euler_df[rxp] = pd.Series(data=exp_df[rxp], index=euler_df.index)
#euler_df[ryp] = pd.Series(data=exp_df[ryp], index=euler_df.index)
#euler_df[rzp] = pd.Series(data=exp_df[rzp], index=euler_df.index)
# List the columns that contain rotation channels
exp_params = [c for c in exp_df.columns if ( any(p in c for p in ['alpha', 'beta','gamma']) and 'Nub' not in c)]
# List the joints that are not end sites, i.e., have channels
joints = (joint for joint in track.skeleton if 'Nub' not in joint)
for joint in joints:
r = exp_df[[c for c in exp_params if joint in c]] # Get the columns that belong to this joint
euler_df.drop(['%s_alpha'%joint, '%s_beta'%joint, '%s_gamma'%joint], axis=1, inplace=True)
expmap = [[f[1]['%s_alpha'%joint], f[1]['%s_beta'%joint], f[1]['%s_gamma'%joint]] for f in r.iterrows()] # Make sure the columsn are organized in xyz order
rot_order = track.skeleton[joint]['order']
#euler_rots = [Rotation(f, 'expmap').to_euler(True, rot_order) for f in expmap] # Convert the exp maps to eulers
euler_rots = [expmap2euler(f, rot_order, True) for f in expmap] # Convert the exp maps to eulers
# Create the corresponding columns in the new DataFrame
euler_df['%s_%srotation'%(joint, rot_order[0])] = pd.Series(data=[e[0] for e in euler_rots], index=euler_df.index)
euler_df['%s_%srotation'%(joint, rot_order[1])] = pd.Series(data=[e[1] for e in euler_rots], index=euler_df.index)
euler_df['%s_%srotation'%(joint, rot_order[2])] = pd.Series(data=[e[2] for e in euler_rots], index=euler_df.index)
new_track = track.clone()
new_track.values = euler_df
Q.append(new_track)
return Q
class Mirror(BaseEstimator, TransformerMixin):
def __init__(self, axis="X", append=True):
"""
Mirrors the data
"""
self.axis = axis
self.append = append
def fit(self, X, y=None):
return self
def transform(self, X, y=None):
print("Mirror: " + self.axis)
Q = []
if self.append:
for track in X:
Q.append(track)
for track in X:
channels = []
titles = []
if self.axis == "X":
signs = np.array([1,-1,-1])
if self.axis == "Y":
signs = np.array([-1,1,-1])
if self.axis == "Z":
signs = np.array([-1,-1,1])
euler_df = track.values
# Create a new DataFrame to store the exponential map rep
new_df = pd.DataFrame(index=euler_df.index)
# Copy the root positions into the new DataFrame
rxp = '%s_Xposition'%track.root_name
ryp = '%s_Yposition'%track.root_name
rzp = '%s_Zposition'%track.root_name
new_df[rxp] = pd.Series(data=-signs[0]*euler_df[rxp], index=new_df.index)
new_df[ryp] = pd.Series(data=-signs[1]*euler_df[ryp], index=new_df.index)
new_df[rzp] = pd.Series(data=-signs[2]*euler_df[rzp], index=new_df.index)
# List the columns that contain rotation channels
rots = [c for c in euler_df.columns if ('rotation' in c and 'Nub' not in c)]
#lft_rots = [c for c in euler_df.columns if ('Left' in c and 'rotation' in c and 'Nub' not in c)]
#rgt_rots = [c for c in euler_df.columns if ('Right' in c and 'rotation' in c and 'Nub' not in c)]
lft_joints = (joint for joint in track.skeleton if 'Left' in joint and 'Nub' not in joint)
rgt_joints = (joint for joint in track.skeleton if 'Right' in joint and 'Nub' not in joint)
new_track = track.clone()
for lft_joint in lft_joints:
#lr = euler_df[[c for c in rots if lft_joint + "_" in c]]
#rot_order = track.skeleton[lft_joint]['order']
#lft_eulers = [[f[1]['%s_Xrotation'%lft_joint], f[1]['%s_Yrotation'%lft_joint], f[1]['%s_Zrotation'%lft_joint]] for f in lr.iterrows()]
rgt_joint = lft_joint.replace('Left', 'Right')
#rr = euler_df[[c for c in rots if rgt_joint + "_" in c]]
#rot_order = track.skeleton[rgt_joint]['order']
# rgt_eulers = [[f[1]['%s_Xrotation'%rgt_joint], f[1]['%s_Yrotation'%rgt_joint], f[1]['%s_Zrotation'%rgt_joint]] for f in rr.iterrows()]
# Create the corresponding columns in the new DataFrame
new_df['%s_Xrotation'%lft_joint] = pd.Series(data=signs[0]*track.values['%s_Xrotation'%rgt_joint], index=new_df.index)
new_df['%s_Yrotation'%lft_joint] = pd.Series(data=signs[1]*track.values['%s_Yrotation'%rgt_joint], index=new_df.index)
new_df['%s_Zrotation'%lft_joint] = pd.Series(data=signs[2]*track.values['%s_Zrotation'%rgt_joint], index=new_df.index)
new_df['%s_Xrotation'%rgt_joint] = pd.Series(data=signs[0]*track.values['%s_Xrotation'%lft_joint], index=new_df.index)
new_df['%s_Yrotation'%rgt_joint] = pd.Series(data=signs[1]*track.values['%s_Yrotation'%lft_joint], index=new_df.index)
new_df['%s_Zrotation'%rgt_joint] = pd.Series(data=signs[2]*track.values['%s_Zrotation'%lft_joint], index=new_df.index)
# List the joints that are not left or right, i.e. are on the trunk
joints = (joint for joint in track.skeleton if 'Nub' not in joint and 'Left' not in joint and 'Right' not in joint)
for joint in joints:
#r = euler_df[[c for c in rots if joint in c]] # Get the columns that belong to this joint
#rot_order = track.skeleton[joint]['order']
#eulers = [[f[1]['%s_Xrotation'%joint], f[1]['%s_Yrotation'%joint], f[1]['%s_Zrotation'%joint]] for f in r.iterrows()]
# Create the corresponding columns in the new DataFrame
new_df['%s_Xrotation'%joint] = pd.Series(data=signs[0]*track.values['%s_Xrotation'%joint], index=new_df.index)
new_df['%s_Yrotation'%joint] = pd.Series(data=signs[1]*track.values['%s_Yrotation'%joint], index=new_df.index)
new_df['%s_Zrotation'%joint] = pd.Series(data=signs[2]*track.values['%s_Zrotation'%joint], index=new_df.index)
new_track.values = new_df
Q.append(new_track)
return Q
def inverse_transform(self, X, copy=None, start_pos=None):
return X
class EulerReorder(BaseEstimator, TransformerMixin):
def __init__(self, new_order):
"""
Add a
"""
self.new_order = new_order
def fit(self, X, y=None):
self.orig_skeleton = copy.deepcopy(X[0].skeleton)
print(self.orig_skeleton)
return self
def transform(self, X, y=None):
Q = []
for track in X:
channels = []
titles = []
euler_df = track.values
# Create a new DataFrame to store the exponential map rep
new_df = pd.DataFrame(index=euler_df.index)
# Copy the root positions into the new DataFrame
rxp = '%s_Xposition'%track.root_name
ryp = '%s_Yposition'%track.root_name
rzp = '%s_Zposition'%track.root_name
new_df[rxp] = pd.Series(data=euler_df[rxp], index=new_df.index)
new_df[ryp] = pd.Series(data=euler_df[ryp], index=new_df.index)
new_df[rzp] = pd.Series(data=euler_df[rzp], index=new_df.index)
# List the columns that contain rotation channels
rots = [c for c in euler_df.columns if ('rotation' in c and 'Nub' not in c)]
# List the joints that are not end sites, i.e., have channels
joints = (joint for joint in track.skeleton if 'Nub' not in joint)
new_track = track.clone()
for joint in joints:
r = euler_df[[c for c in rots if joint in c]] # Get the columns that belong to this joint
rot_order = track.skeleton[joint]['order']
euler = [[f[1]['%s_Xrotation'%(joint)], f[1]['%s_Yrotation'%(joint)], f[1]['%s_Zrotation'%(joint)]] for f in r.iterrows()]
new_euler = [euler_reorder(f, rot_order, self.new_order, True) for f in euler]
#new_euler = euler_reorder2(np.array(euler), rot_order, self.new_order, True)
# Create the corresponding columns in the new DataFrame
new_df['%s_%srotation'%(joint, self.new_order[0])] = pd.Series(data=[e[0] for e in new_euler], index=new_df.index)
new_df['%s_%srotation'%(joint, self.new_order[1])] = pd.Series(data=[e[1] for e in new_euler], index=new_df.index)
new_df['%s_%srotation'%(joint, self.new_order[2])] = pd.Series(data=[e[2] for e in new_euler], index=new_df.index)
new_track.skeleton[joint]['order'] = self.new_order
new_track.values = new_df
Q.append(new_track)
return Q
def inverse_transform(self, X, copy=None, start_pos=None):
return X
# Q = []
#
# for track in X:
# channels = []
# titles = []
# euler_df = track.values
#
# # Create a new DataFrame to store the exponential map rep
# new_df = pd.DataFrame(index=euler_df.index)
#
# # Copy the root positions into the new DataFrame
# rxp = '%s_Xposition'%track.root_name
# ryp = '%s_Yposition'%track.root_name
# rzp = '%s_Zposition'%track.root_name
# new_df[rxp] = pd.Series(data=euler_df[rxp], index=new_df.index)
# new_df[ryp] = pd.Series(data=euler_df[ryp], index=new_df.index)
# new_df[rzp] = pd.Series(data=euler_df[rzp], index=new_df.index)
#
# # List the columns that contain rotation channels
# rots = [c for c in euler_df.columns if ('rotation' in c and 'Nub' not in c)]
#
# # List the joints that are not end sites, i.e., have channels
# joints = (joint for joint in track.skeleton if 'Nub' not in joint)
#
# new_track = track.clone()
# for joint in joints:
# r = euler_df[[c for c in rots if joint in c]] # Get the columns that belong to this joint
# rot_order = track.skeleton[joint]['order']
# new_order = self.orig_skeleton[joint]['order']
# print("rot_order:" + str(rot_order))
# print("new_order:" + str(new_order))
#
# euler = [[f[1]['%s_%srotation'%(joint, rot_order[0])], f[1]['%s_%srotation'%(joint, rot_order[1])], f[1]['%s_%srotation'%(joint, rot_order[2])]] for f in r.iterrows()]
# #new_euler = [euler_reorder(f, rot_order, new_order, True) for f in euler]
# new_euler = euler_reorder2(np.array(euler), rot_order, self.new_order, True)
#
# # Create the corresponding columns in the new DataFrame
# new_df['%s_%srotation'%(joint, new_order[0])] = pd.Series(data=[e[0] for e in new_euler], index=new_df.index)
# new_df['%s_%srotation'%(joint, new_order[1])] = pd.Series(data=[e[1] for e in new_euler], index=new_df.index)
# new_df['%s_%srotation'%(joint, new_order[2])] = pd.Series(data=[e[2] for e in new_euler], index=new_df.index)
#
# new_track.skeleton[joint]['order'] = new_order
#
# new_track.values = new_df
# Q.append(new_track)
# return Q
class JointSelector(BaseEstimator, TransformerMixin):
'''
Allows for filtering the mocap data to include only the selected joints
'''
def __init__(self, joints, include_root=False):
self.joints = joints
self.include_root = include_root
def fit(self, X, y=None):
selected_joints = []
selected_channels = []
if self.include_root:
selected_joints.append(X[0].root_name)
selected_joints.extend(self.joints)
for joint_name in selected_joints:
selected_channels.extend([o for o in X[0].values.columns if (joint_name + "_") in o and 'Nub' not in o])
self.selected_joints = selected_joints
self.selected_channels = selected_channels
self.not_selected = X[0].values.columns.difference(selected_channels)
self.not_selected_values = {c:X[0].values[c].values[0] for c in self.not_selected}
self.orig_skeleton = X[0].skeleton
return self
def transform(self, X, y=None):
print("JointSelector")
Q = []
for track in X:
t2 = track.clone()
for key in track.skeleton.keys():
if key not in self.selected_joints:
parent = t2.skeleton[key]['parent']
if parent in t2.skeleton:
t2.skeleton[parent]['children'].remove(key)
t2.skeleton.pop(key)
t2.values = track.values[self.selected_channels]
Q.append(t2)
return Q
def inverse_transform(self, X, copy=None):
Q = []
for track in X:
t2 = track.clone()
t2.skeleton = self.orig_skeleton
for d in self.not_selected:
t2.values[d] = self.not_selected_values[d]
Q.append(t2)
return Q
class Numpyfier(BaseEstimator, TransformerMixin):
'''
Just converts the values in a MocapData object into a numpy array
Useful for the final stage of a pipeline before training
'''
def __init__(self):
pass
def fit(self, X, y=None):
self.org_mocap_ = X[0].clone()
self.org_mocap_.values.drop(self.org_mocap_.values.index, inplace=True)
return self
def transform(self, X, y=None):
print("Numpyfier")
Q = []
for track in X:
Q.append(track.values.values)
#print("Numpyfier:" + str(track.values.columns))
return np.array(Q)
def inverse_transform(self, X, copy=None):
Q = []
for track in X:
new_mocap = self.org_mocap_.clone()
time_index = pd.to_timedelta([f for f in range(track.shape[0])], unit='s')
# print(self.org_mocap_.values.columns)
# import pdb;pdb.set_trace()
new_df = pd.DataFrame(data=track, index=time_index, columns=self.org_mocap_.values.columns)
new_mocap.values = new_df
Q.append(new_mocap)
return Q
class Slicer(BaseEstimator, TransformerMixin):
'''
Slice the data into intervals of equal size
'''
def __init__(self, window_size, overlap=0.5):
self.window_size = window_size
self.overlap = overlap
pass
def fit(self, X, y=None):
self.org_mocap_ = X[0].clone()
self.org_mocap_.values.drop(self.org_mocap_.values.index, inplace=True)
return self
def transform(self, X, y=None):
print("Slicer")
Q = []
for track in X:
vals = track.values.values
nframes = vals.shape[0]
overlap_frames = (int)(self.overlap*self.window_size)
n_sequences = (nframes-overlap_frames)//(self.window_size-overlap_frames)
if n_sequences>0:
y = np.zeros((n_sequences, self.window_size, vals.shape[1]))
# extract sequences from the input data
for i in range(0,n_sequences):
frameIdx = (self.window_size-overlap_frames) * i
Q.append(vals[frameIdx:frameIdx+self.window_size,:])
return np.array(Q)
def inverse_transform(self, X, copy=None):
Q = []
for track in X:
new_mocap = self.org_mocap_.clone()
time_index = pd.to_timedelta([f for f in range(track.shape[0])], unit='s')
new_df = pd.DataFrame(data=track, index=time_index, columns=self.org_mocap_.values.columns)
new_mocap.values = new_df
Q.append(new_mocap)
return Q
class RootTransformer(BaseEstimator, TransformerMixin):
def __init__(self, method, position_smoothing=0, rotation_smoothing=0):
"""
Accepted methods:
abdolute_translation_deltas
pos_rot_deltas
"""
self.method = method
self.position_smoothing=position_smoothing
self.rotation_smoothing=rotation_smoothing
def fit(self, X, y=None):
return self
def transform(self, X, y=None):
print("RootTransformer")
Q = []
for track in X:
if self.method == 'abdolute_translation_deltas':
new_df = track.values.copy()
xpcol = '%s_Xposition'%track.root_name
ypcol = '%s_Yposition'%track.root_name
zpcol = '%s_Zposition'%track.root_name
dxpcol = '%s_dXposition'%track.root_name
dzpcol = '%s_dZposition'%track.root_name
x=track.values[xpcol].copy()
z=track.values[zpcol].copy()
if self.position_smoothing>0:
x_sm = filters.gaussian_filter1d(x, self.position_smoothing, axis=0, mode='nearest')
z_sm = filters.gaussian_filter1d(z, self.position_smoothing, axis=0, mode='nearest')
dx = pd.Series(data=x_sm, index=new_df.index).diff()
dz = pd.Series(data=z_sm, index=new_df.index).diff()
new_df[xpcol] = x-x_sm
new_df[zpcol] = z-z_sm
else:
dx = x.diff()
dz = z.diff()
new_df.drop([xpcol, zpcol], axis=1, inplace=True)
dx[0] = dx[1]
dz[0] = dz[1]
new_df[dxpcol] = dx
new_df[dzpcol] = dz
new_track = track.clone()
new_track.values = new_df
# end of abdolute_translation_deltas
elif self.method == 'pos_rot_deltas':
new_track = track.clone()
# Absolute columns
xp_col = '%s_Xposition'%track.root_name
yp_col = '%s_Yposition'%track.root_name
zp_col = '%s_Zposition'%track.root_name
#rot_order = track.skeleton[track.root_name]['order']
#%(joint, rot_order[0])
rot_order = track.skeleton[track.root_name]['order']
r1_col = '%s_%srotation'%(track.root_name, rot_order[0])
r2_col = '%s_%srotation'%(track.root_name, rot_order[1])
r3_col = '%s_%srotation'%(track.root_name, rot_order[2])
# Delta columns
dxp_col = '%s_dXposition'%track.root_name
dzp_col = '%s_dZposition'%track.root_name
dxr_col = '%s_dXrotation'%track.root_name
dyr_col = '%s_dYrotation'%track.root_name
dzr_col = '%s_dZrotation'%track.root_name
positions = np.transpose(np.array([track.values[xp_col], track.values[yp_col], track.values[zp_col]]))
rotations = np.pi/180.0*np.transpose(np.array([track.values[r1_col], track.values[r2_col], track.values[r3_col]]))
""" Get Trajectory and smooth it"""
trajectory_filterwidth = self.position_smoothing
reference = positions.copy()*np.array([1,0,1])
if trajectory_filterwidth>0:
reference = filters.gaussian_filter1d(reference, trajectory_filterwidth, axis=0, mode='nearest')
""" Get Root Velocity """
velocity = np.diff(reference, axis=0)
velocity = np.vstack((velocity[0,:], velocity))
""" Remove Root Translation """
positions = positions-reference
""" Get Forward Direction along the x-z plane, assuming character is facig z-forward """
#forward = [Rotation(f, 'euler', from_deg=True, order=rot_order).rotmat[:,2] for f in rotations] # get the z-axis of the rotation matrix, assuming character is facig z-forward
#print("order:" + rot_order.lower())
quats = Quaternions.from_euler(rotations, order=rot_order.lower(), world=False)
forward = quats*np.array([[0,0,1]])
forward[:,1] = 0
""" Smooth Forward Direction """
direction_filterwidth = self.rotation_smoothing
if direction_filterwidth>0:
forward = filters.gaussian_filter1d(forward, direction_filterwidth, axis=0, mode='nearest')
forward = forward / np.sqrt((forward**2).sum(axis=-1))[...,np.newaxis]
""" Remove Y Rotation """
target = np.array([[0,0,1]]).repeat(len(forward), axis=0)
rotation = Quaternions.between(target, forward)[:,np.newaxis]
positions = (-rotation[:,0]) * positions
new_rotations = (-rotation[:,0]) * quats
velocity = (-rotation[:,0]) * velocity
""" Get Root Rotation """
#print(rotation[:,0])
rvelocity = Pivots.from_quaternions(rotation[1:] * -rotation[:-1]).ps
rvelocity = np.vstack((rvelocity[0], rvelocity))
eulers = np.array([t3d.euler.quat2euler(q, axes=('s'+rot_order.lower()[::-1]))[::-1] for q in new_rotations])*180.0/np.pi
new_df = track.values.copy()
root_pos_x = pd.Series(data=positions[:,0], index=new_df.index)
root_pos_y = pd.Series(data=positions[:,1], index=new_df.index)
root_pos_z = pd.Series(data=positions[:,2], index=new_df.index)
root_pos_x_diff = pd.Series(data=velocity[:,0], index=new_df.index)
root_pos_z_diff = pd.Series(data=velocity[:,2], index=new_df.index)
root_rot_1 = pd.Series(data=eulers[:,0], index=new_df.index)
root_rot_2 = pd.Series(data=eulers[:,1], index=new_df.index)
root_rot_3 = pd.Series(data=eulers[:,2], index=new_df.index)
root_rot_y_diff = pd.Series(data=rvelocity[:,0], index=new_df.index)
#new_df.drop([xr_col, yr_col, zr_col, xp_col, zp_col], axis=1, inplace=True)
new_df[xp_col] = root_pos_x
new_df[yp_col] = root_pos_y
new_df[zp_col] = root_pos_z
new_df[dxp_col] = root_pos_x_diff
new_df[dzp_col] = root_pos_z_diff
new_df[r1_col] = root_rot_1
new_df[r2_col] = root_rot_2
new_df[r3_col] = root_rot_3
#new_df[dxr_col] = root_rot_x_diff
new_df[dyr_col] = root_rot_y_diff
#new_df[dzr_col] = root_rot_z_diff
new_track.values = new_df
elif self.method == 'hip_centric':
new_track = track.clone()
# Absolute columns
xp_col = '%s_Xposition'%track.root_name
yp_col = '%s_Yposition'%track.root_name
zp_col = '%s_Zposition'%track.root_name
xr_col = '%s_Xrotation'%track.root_name
yr_col = '%s_Yrotation'%track.root_name
zr_col = '%s_Zrotation'%track.root_name
new_df = track.values.copy()
all_zeros = np.zeros(track.values[xp_col].values.shape)
new_df[xp_col] = pd.Series(data=all_zeros, index=new_df.index)
new_df[yp_col] = pd.Series(data=all_zeros, index=new_df.index)
new_df[zp_col] = pd.Series(data=all_zeros, index=new_df.index)
new_df[xr_col] = pd.Series(data=all_zeros, index=new_df.index)
new_df[yr_col] = pd.Series(data=all_zeros, index=new_df.index)
new_df[zr_col] = pd.Series(data=all_zeros, index=new_df.index)
new_track.values = new_df
#print(new_track.values.columns)
Q.append(new_track)
return Q
def inverse_transform(self, X, copy=None, start_pos=None):
Q = []
#TODO: simplify this implementation
startx = 0
startz = 0
if start_pos is not None:
startx, startz = start_pos
for track in X:
new_track = track.clone()
if self.method == 'abdolute_translation_deltas':
new_df = new_track.values
xpcol = '%s_Xposition'%track.root_name
ypcol = '%s_Yposition'%track.root_name
zpcol = '%s_Zposition'%track.root_name
dxpcol = '%s_dXposition'%track.root_name
dzpcol = '%s_dZposition'%track.root_name
dx = track.values[dxpcol].values
dz = track.values[dzpcol].values
recx = [startx]
recz = [startz]
for i in range(dx.shape[0]-1):
recx.append(recx[i]+dx[i+1])
recz.append(recz[i]+dz[i+1])
# recx = [recx[i]+dx[i+1] for i in range(dx.shape[0]-1)]
# recz = [recz[i]+dz[i+1] for i in range(dz.shape[0]-1)]
# recx = dx[:-1] + dx[1:]
# recz = dz[:-1] + dz[1:]
if self.position_smoothing > 0:
new_df[xpcol] = pd.Series(data=new_df[xpcol]+recx, index=new_df.index)
new_df[zpcol] = pd.Series(data=new_df[zpcol]+recz, index=new_df.index)
else:
new_df[xpcol] = | pd.Series(data=recx, index=new_df.index) | pandas.Series |
import pandas as pd
import helpers
import re
from datetime import datetime
from Bio import SeqIO
from .generate_viral_seq_dataset import get_viralseq_dataset
from .generate_viral_seq_datadownload import get_viralseq_downloads
DATADIR = "/Users/laurahughes/GitHub/cvisb_data/sample-viewer-api/src/static/data/"
alignment_file = f"{DATADIR}/input_data/expt_summary_data/viral_seq/clean_ebola_orfs_aln_2019.11.12.fasta"
metadata_file = f"{DATADIR}/input_data/expt_summary_data/viral_seq/survival_dataset_ebov_public_2019.11.12.csv"
def clean_ebola_viral_seq(export_dir, alignment_file, uncurated_file, metadata_file, alignments, expt_cols, patient_cols, sample_cols, download_cols, dateModified, version, updatedBy, saveFiles, verbose, virus="Ebola"):
# --- constants ---
today = datetime.today().strftime('%Y-%m-%d')
# Custom, extra properties specific to viral sequencing
exptCols = expt_cols.copy()
exptCols.extend(['genbankID', 'inAlignment', 'cvisb_data'])
# --- read in metadata ---
md = pd.read_csv(metadata_file)
# --- Initial checks ---
dupe_patientID = md[md.duplicated(
subset=["patientID", "patient_timepoint"], keep=False)]
if(len(dupe_patientID) > 0):
helpers.log_msg(
f"DATA ERROR: {len(dupe_patientID)} duplicate patient ids found in virus sequences:", verbose)
helpers.log_msg(dupe_patientID[['patientID']].sort_values(
"patientID"), verbose)
helpers.log_msg("-" * 50, verbose)
dupe_accession = md[md.duplicated(subset=["accession"], keep=False)]
if(len(dupe_accession) > 0):
helpers.log_msg(
f"DATA ERROR: {len(dupe_accession)} duplicate accession IDs found in virus sequences:", verbose)
helpers.log_msg(dupe_patientID[[
'patientID', 'patient_timepoint', 'accession']].sort_values("accession"), verbose)
helpers.log_msg("-" * 50, verbose)
helpers.log_msg("finished initial checks", verbose)
# --- clean up common properties, across patient/expt/sample/downloads/dataset ---
md['variableMeasured'] = f"{virus} virus sequence"
md['measurementTechnique'] = "Nucleic Acid Sequencing"
md['measurementCategory'] = "virus sequencing"
md['includedInDataset'] = f"{virus.lower()}-virus-seq"
md['creator'] = None
md['correction'] = None
source = [alignment_file.split("/")[-1], metadata_file.split("/")[-1]]
md['sourceFiles'] = md.apply(lambda x: source, axis=1)
md['version'] = version
md['dateModified'] = dateModified
md['updatedBy'] = updatedBy
md['releaseDate'] = today
md['dataStatus'] = "final"
md['publisher'] = md.apply(getPublisher, axis=1)
md['batchID'] = None
md['experimentDate'] = md.collection_date.apply(checkDate)
md['isControl'] = False
helpers.log_msg("finished chunk 1", verbose)
# --- clean up patient metadata ---
md['privatePatientID'] = md.patientID
md['KGH_id'] = md.patientID.apply(
helpers.checkIDstructure).apply(lambda x: not x)
md['outcome_copy'] = md.outcome
md['outcome'] = md.outcome.apply(helpers.cleanOutcome)
md['cohort'] = virus
md['alternateIdentifier'] = md.patientID.apply(helpers.listify)
md['country'] = md.country_iso3.apply(helpers.getCountry)
md['location'] = md.apply(getLocation, axis = 1)
md['locationPrivate'] = md.apply(getLocationPrivate, axis = 1)
md['countryName'] = md.country.apply(helpers.pullCountryName)
md['infectionYear'] = md.year
md['samplingDate'] = md.collection_date.apply(checkDate)
md['species'] = md.host.apply(helpers.convertSpecies)
# Patient timepoints
md['visitCode'] = md.patient_timepoint.apply(lambda x: str(x))
# Note: not technically true; if a KGH patient, could have patient / survivor data.
# But-- since only uploading the non-KGH patient data, should be fine.
md['hasPatientData'] = False
md['hasSurvivorData'] = False
helpers.log_msg("finished chunk 2", verbose)
# --- clean up experiment properties ---
md['inAlignment'] = md.curated.apply(bool)
md['cvisb_data'] = md.CViSB_data.apply(bool)
helpers.log_msg("finished chunk 3", verbose)
print("Is this going really slowly? Make sure your VPN is turned off when you're getting citations from NCBI.")
citation_dict = helpers.createCitationDict(md, "source_pmid")
md['citation'] = md["source_pmid"].apply(
lambda x: helpers.lookupCitation(x, citation_dict))
# Make sure arrays are arrays
md['citation'] = md.citation.apply(helpers.listify)
md['experimentID'] = md.apply(lambda x: getExptID(x, virus), axis=1)
md['genbankID'] = md.accession
helpers.log_msg("finished chunk 4 (citations done!)", verbose)
# --- clean up sample properties ---
md['sampleLabel'] = md.label
md['sampleType'] = "viralRNA"
md['creatorInitials'] = f"{(updatedBy.split(' ')[0][0] + updatedBy.split(' ')[1][0]).lower()}"
md['sampleID'] = md.apply(
lambda x: f"{x.creatorInitials}-{x.sampleLabel}_{x.sampleType}", axis=1)
helpers.log_msg("finished chunk 4", verbose)
# --- clean up download properties ---
md['name'] = md.apply(lambda x: x.patientID + "_" + x.accession, axis=1)
md['identifier'] = md.apply(lambda x: x.patientID + "_" + x.accession, axis=1)
md['contentUrl'] = md.apply(lambda x: "https://www.ncbi.nlm.nih.gov/nuccore/" + x.accession, axis=1)
md['additionalType'] = 'raw data'
md['experimentIDs'] = md.experimentID.apply(lambda x: [x])
md['contentUrlRepository'] = "GenBank"
md['contentUrlIdentifier'] = md.accession
helpers.log_msg("finished chunk 5", verbose)
# --- Merge together data and metadata ---
seqs = getDNAseq(alignment_file, uncurated_file, virus)
helpers.log_msg("finished chunk 6", verbose)
merged = pd.merge(md, seqs, on="sequenceID", how="outer", indicator=True)
no_seq = merged[merged._merge == "left_only"]
seq_only = merged[merged._merge == "right_only"]
if(len(no_seq) > 0):
helpers.log_msg(f"\tDATA ERROR: no sequence found in sequence alignment file for {len(no_seq)} patients:", verbose)
helpers.log_msg(no_seq ['patientID'], verbose)
helpers.log_msg("-" * 50, verbose)
if(len(seq_only) > 0):
helpers.log_msg(f"\tDATA ERROR: no patient found in sequence metadata file for {len(seq_only)} sequences:", verbose)
helpers.log_msg(seq_only['sequenceID'], verbose)
helpers.log_msg("-" * 50, verbose)
helpers.log_msg("finished chunk 7", verbose)
# Make sure arrays are arrays
# merged['data'] = merged.data.apply(helpers.listify)
# --- partition data to different endpoints ---
patients = md.loc[~ md.KGH_id, patient_cols]
# de-duplicate patients; some patients are timepoints of the same person
patients.drop_duplicates(subset=["patientID", "cohort", "outcome", "countryName", "infectionYear", "species"], inplace = True)
samples = md.loc[~ md.KGH_id, sample_cols]
experiments = merged[exptCols]
helpers.log_msg("finished chunk 8", verbose)
# --- Call to get data downloads, dataset ---
dwnlds = md[download_cols]
all_dwnlds = get_viralseq_downloads(alignments, dateModified, dwnlds, experiments, version, virus)
ds = get_viralseq_dataset(dateModified, dwnlds, md, version, virus)
helpers.log_msg("finished chunk 9", verbose)
# [Export] ----------------------------------------------------------------------------------------------------
if(saveFiles):
# --- experiments ---
experiments.to_json(
f"{output_dir}/experiments/virus_seq_experiments_{today}.json", orient="records")
# --- patients ---
patients.to_json(
f"{output_dir}/patients/virus_seq_patients_{today}.json", orient="records")
# --- samples ---
samples.to_json(
f"{output_dir}/samples/virus_seq_samples_{today}.json", orient="records")
helpers.log_msg("finished chunk 10", verbose)
return({"patient": patients, "sample": samples, "dataset": ds, "datadownload": all_dwnlds, "experiment": experiments})
# sum(md.duplicated(subset = "accession"))
def getPublisher(row, varName="CViSB_data"):
# Check binary if CVISB_data
if(row[varName]):
return([helpers.cvisb])
def getExptID(row, virus):
if(virus == "Ebola"):
expt_stub = "EBOV_seq_"
if(virus == "Lassa"):
expt_stub = "LASV_seq_"
return(expt_stub + row.patientID + row.visitCode)
def getDNAseq(alignment_file, uncurated_alignment, virus, seq_type="DNAsequence", segment=None, data_type="VirusSeqData"):
all_seq = list(SeqIO.parse(alignment_file, "fasta"))
if(uncurated_alignment is not None):
uncurated_seq = list(SeqIO.parse(uncurated_alignment, "fasta"))
else:
uncurated_seq = list()
# curated sequences
curated = pd.DataFrame(columns=['sequenceID', 'curated'])
for seq in all_seq:
seq_obj = [{
seq_type: str(seq.seq).upper(),
"@type": data_type,
"virus": virus,
"curated": True,
"virusSegment": segment}]
curated = curated.append(pd.DataFrame({'sequenceID': seq.id, 'curated': seq_obj}))
uncurated = | pd.DataFrame(columns=['sequenceID', 'uncurated']) | pandas.DataFrame |
"""
Script for plotting Figures 3, 5, 6
"""
import numpy as np
import pandas as pd
from datetime import datetime
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
from matplotlib.lines import Line2D
import matplotlib.ticker as mtick
import seaborn as sns
from datetime import timedelta
import plotly.graph_objects as go
countries = ['Brazil', 'Canada', 'England', 'France', 'Germany', 'India', 'Japan', 'Scotland', 'USA', 'Wales']
samp_entropy_df = {}
for country in countries:
df = pd.read_csv(f'data/entropy/monthly/fast_samp_entropy_monthly_{country}.csv')
df['Date'] = pd.to_datetime(df['Date'])
df = df[(df['Date'] >= '03-01-2020') & (df['Date'] <= '06-30-2021')]
samp_entropy_df[country] = df
colors = {'France':'darkblue', 'Germany':'dodgerblue', 'Northern-Ireland':'yellowgreen',
'USA':'orange', 'Canada':'red'}
########## Fig 3
fig, ax = plt.subplots(1,2, figsize=(16,4), sharey=True)
fig.subplots_adjust(hspace=0.2, wspace=0.1)
for country in ['France', 'Germany']:
v = samp_entropy_df[country].iloc[:,1:37].sum(axis=0)
if country == 'Germany':
ax[0].plot(v, color = colors[country], label=country, alpha=0.5)
else:
ax[0].plot(v, color = colors[country], label=country)
ax[0].legend(bbox_to_anchor=(1, 1), loc=1, borderaxespad=0, fontsize=12)
ax[0].set_xticks(list(range(0,36,2)))
ax[0].set_xticklabels(list(range(1,37,2)), fontsize=12)
ax[0].set_xlabel('Dimension', fontsize=13, labelpad=10)
for country in ['USA', 'Canada']:
v = samp_entropy_df[country].iloc[:,1:37].sum(axis=0)
ax[1].plot(v, color = colors[country], label=country)
ax[1].legend(bbox_to_anchor=(1, 1), loc=1, borderaxespad=0, fontsize=12)
ax[1].set_xticks(list(range(0,36,2)))
ax[1].set_xticklabels(list(range(1,37,2)), fontsize=12)
ax[1].set_xlabel('Dimension', fontsize=13, labelpad=10)
fig.text(0.09, 0.5, 'Sum of Entropies', va='center', rotation='vertical', fontsize=13)
plt.suptitle("Sum of Sample Entropies for each dimension", fontsize=16)
fig.savefig("figures/Fig2.png", dpi=500, bbox_inches = 'tight')
plt.show()
########## Fig 5
def get_melted_df(df, dim_nums):
melted_df = pd.DataFrame()
for dim in dim_nums:
entropy_dim = f'Entropy_{dim}'
subdf = df[['Date', entropy_dim]]
subdf = subdf.rename(columns = {entropy_dim: 'Entropy'})
melted_df = pd.concat([melted_df, subdf], axis=0, ignore_index=True)
return melted_df
cases_df = pd.read_csv('new_cases.csv')
cases_df = cases_df.rename(columns = {'Unnamed: 0': 'month'})
cases_df['month'] = pd.to_datetime(cases_df['month'])
cases_df = cases_df[(cases_df['month'] >= '03-01-2020') & (cases_df['month'] <= '06-30-2021')]
rf1 = | pd.read_csv('predictions/RF_preds.csv') | pandas.read_csv |
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
"""
Created on Thu Apr 4 06:03:28 2019
@author: tanujsinghal
"""
import numpy as np
import pandas as pd
import matplotlib
matplotlib.use('agg')
import matplotlib.pyplot as plt
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
import re
import copy
from sklearn.metrics import jaccard_similarity_score,confusion_matrix
from sklearn.externals import joblib
import os
import pathlib
import pickle
if __name__=='__main__':
hin=pd.read_csv("Hinglish_Profanity_List.csv")
train = pd.read_csv("train.csv")
test = pd.read_csv("test.csv")
columns = ['obscene','insult','toxic','severe_toxic','identity_hate','threat']
hin_bad_words = hin.iloc[:,0].values.tolist()
bad_words_to_english = hin.iloc[:,1].values.tolist()
hin = hin.iloc[:,:-1].values.tolist()
train, test = train_test_split(train, test_size=0.2)
labels = train.iloc[:,2:]
train_data = train.iloc[:,1]
test_data = test.iloc[:,1]
features = 5000
ngram = (1,2)
vectorizer = TfidfVectorizer(stop_words='english',\
token_pattern = "\w*[a-z]\w*",\
ngram_range=ngram,\
max_features=features)
train_features = vectorizer.fit_transform(train_data)
filename='vect'
pickle.dump(vectorizer, open(filename, 'wb'))
test_features = vectorizer.transform(test_data)
logreg = LogisticRegression(C=10,solver="liblinear")
models={}
logistic_results = pd.DataFrame(columns=columns)
cnt=0
for i in columns:
y = train[i]
models[i]=copy.copy(logreg.fit(train_features, y))
filename = "model_"+ str(cnt)
pickle.dump(models[i], open(filename, 'wb'))
ypred_X = logreg.predict(train_features)
testy_prob = logreg.predict_proba(test_features)[:,1]
logistic_results[i] = testy_prob
cnt+=1
def abusive_hinglish_to_english(data):
hin=pd.read_csv("Hinglish_Profanity_List.csv")
hin_bad_words = hin.iloc[:,0].values.tolist()
bad_words_to_english = hin.iloc[:,1].values.tolist()
hin = hin.iloc[:,:-1].values.tolist()
cnt=0
for sentence in data:
wordList = sentence.split()
for word in hin_bad_words:
if word in wordList:
x=wordList.index(word)
wordList[x]=bad_words_to_english[hin_bad_words.index(word)]
sentence = ' '.join(wordList)
data[cnt]=sentence
cnt+=1
return data
def check_dir(directory):
if not os.path.exists(directory):
os.makedirs(directory)
def myinput(vectorizer,model,val):
sent="Thank you for understanding. I think very highly of you and would not revert without discussion."
sent2="Yo bitch Ja Rule is more succesful then you'll ever be whats up with you and hating you sad mofuckas...i should bitch slap ur pethedic white faces and get you to kiss my ass you guys sicken me. Ja rule is about pride in da music man. dont diss that shit on him. and nothin is wrong bein like tupac he was a brother too...fuckin white boys get things right next time.,"
sen3="Explanation Why the edits made under my username Hardcore Metallica Fan were reverted? They weren't vandalisms, just closure on some GAs after I voted at New York Dolls FAC. And please don't remove the template from the talk page since I'm retired now.172.16.17.32"
sen4="COCKSUCKER BEFORE YOU PISS AROUND ON MY WORK"
sen5="While booking during rush hour, it is always advisable to check this box ON. It will deduct your FULL ticket amount first. After that it check whether confirmed ticket available or not. If confirmed ticket not available it will show ticket not available. Within 2 days will get your full amount Refund."
sen6="<NAME>"
sent7='F**K YOU!good day'
l=[sent,sent2,sen3,sen4,sen5,sen6,sent7]
l=abusive_hinglish_to_english(l)
df = pd.DataFrame(l)
user_data = vectorizer.transform(l)
results2 = pd.DataFrame(columns=columns)
for i in columns:
user_results = models[i].predict_proba(user_data)[:,1]
results2[i] = user_results
y=results2.iloc[val].values
x = columns
plt.ylim(0, 100)
plt.tight_layout()
plt.bar(x, height= y)
plt.show()
plt.savefig('foo.png')
return df,results2
def myinput_network(text):
columns = ['obscene','insult','toxic','severe_toxic','identity_hate','threat']
sent="Thank you for understanding. I think very highly of you and would not revert without discussion."
sent2="Yo bitch Ja Rule is more succesful then you'll ever be whats up with you and hating you sad mofuckas...i should bitch slap ur pethedic white faces and get you to kiss my ass you guys sicken me. Ja rule is about pride in da music man. dont diss that shit on him. and nothin is wrong bein like tupac he was a brother too...fuckin white boys get things right next time.,"
sen3="Explanation Why the edits made under my username Hardcore Metallica Fan were reverted? They weren't vandalisms, just closure on some GAs after I voted at New York Dolls FAC. And please don't remove the template from the talk page since I'm retired now.172.16.17.32"
sen4="COCKSUCKER BEFORE YOU PISS AROUND ON MY WORK"
sen5="While booking during rush hour, it is always advisable to check this box ON. It will deduct your FULL ticket amount first. After that it check whether confirmed ticket available or not. If confirmed ticket not available it will show ticket not available. Within 2 days will get your full amount Refund."
sen6="<NAME>"
sent7='F**K YOU!good day'
l=[sent,sent2,sen3,sen4,sen5,sen6,sent7,text]
l=[text,sent2]
if len(text)>1 and type(text) is list:
l=text
res,x=myinput_network2(l)
return res,x
l=abusive_hinglish_to_english(l)
df = | pd.DataFrame(l) | pandas.DataFrame |
#!/usr/bin/env python
# coding: utf-8
# In[74]:
import pandas as pd
import numpy as np
from pathlib import Path
import os
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.model_selection import RandomizedSearchCV
from sklearn.decomposition import PCA
from sklearn.metrics import roc_auc_score
from sklearn.metrics import roc_curve
from sklearn.metrics import precision_recall_curve
from sklearn.metrics import confusion_matrix
from sklearn.pipeline import Pipeline
from scipy import stats
from joblib import dump
from joblib import load
import xgboost as xgb
import matplotlib.pyplot as plt
from typing import Dict
from src.data import make_dataset
from kaggle.api.kaggle_api_extended import KaggleApi
from dotenv import find_dotenv, load_dotenv
# In[78]:
load_dotenv(find_dotenv())
api = KaggleApi()
api.authenticate()
# In[80]:
competition = os.environ['COMPETITION']
# # Set up directories
# In[65]:
project_dir = Path.cwd().parent
data_dir = project_dir / 'data'
raw_data_dir = data_dir / 'raw'
interim_data_dir = data_dir / 'interim'
processed_data_dir = data_dir / 'processed'
models_dir = project_dir / 'models'
# # Load data
# In[57]:
df_train = pd.read_csv(raw_data_dir / 'train.csv')
df_test = pd.read_csv(raw_data_dir / 'test.csv')
X_train = np.load(interim_data_dir / 'X_train.npy')
X_val = np.load(interim_data_dir / 'X_val.npy')
y_train = np.load(interim_data_dir / 'y_train.npy')
y_val = np.load(interim_data_dir / 'y_val.npy')
X_test = np.load(interim_data_dir / 'X_test.npy')
test_id = pd.read_csv(interim_data_dir / 'test_id.csv')
# # Baseline
#
# The base line prediction is simply to make them all negative.
# In[36]:
labels = 'Positive', 'Negative'
pos_count = (y_train == 1).sum()
neg_count = (y_train == 0).sum()
sizes = [pos_count, neg_count]
explode = (0, 0.1) # only "explode" the 2nd slice (i.e. 'Hogs')
fig1, ax1 = plt.subplots()
ax1.pie(sizes, explode=explode, labels=labels, autopct='%1.1f%%',
shadow=True, startangle=90)
ax1.axis('equal') # Equal aspect ratio ensures that pie is drawn as a circle.
plt.show()
# In[37]:
labels = 'Positive', 'Negative'
pos_count = (y_val == 1).sum()
neg_count = (y_val == 0).sum()
sizes = [pos_count, neg_count]
explode = (0, 0.1) # only "explode" the 2nd slice (i.e. 'Hogs')
fig1, ax1 = plt.subplots()
ax1.pie(sizes, explode=explode, labels=labels, autopct='%1.1f%%',
shadow=True, startangle=90)
ax1.axis('equal') # Equal aspect ratio ensures that pie is drawn as a circle.
plt.show()
# In[38]:
preds = [1] * len(y_val)
roc_auc_score(y_val, preds)
# # XGB
# In[39]:
clf_xgb = xgb.XGBClassifier()
# In[40]:
clf_xgb.fit(X_train, y_train)
# In[41]:
preds = clf_xgb.predict(X_val)
probs = clf_xgb.predict_proba(X_val)
# In[42]:
X_val.shape
# In[43]:
len(y_val)
# In[44]:
auc = roc_auc_score(y_val, probs[:, 1])
tpr, fpr, threshold = roc_curve(y_val, probs[:, 1])
auc
# In[ ]:
# # RandomizedSearchCV
# In[45]:
# test
df_train.info()
# In[46]:
pipe = Pipeline([
('scaler', StandardScaler()),
('pca', PCA()),
('classifier', xgb.XGBClassifier(
objective='binary:logistic',
use_label_encoder=False,
eval_metric='logloss'
))
])
param_dist = {
'pca__n_components': stats.randint(1, X_train.shape[1]),
'classifier__n_estimators': stats.randint(150, 1000),
'classifier__learning_rate': stats.uniform(0.01, 0.6),
'classifier__subsample': stats.uniform(0.3, 0.9),
'classifier__max_depth': [3, 4, 5, 6, 7, 8, 9],
'classifier__colsample_bytree': stats.uniform(0.5, 0.9),
'classifier__min_child_weight': [1, 2, 3, 4]
}
cv = RandomizedSearchCV(
estimator=pipe,
param_distributions=param_dist,
random_state=42,
n_iter=100,
cv=5,
n_jobs=7,
verbose=10
)
cv.fit(X_train, y_train)
# In[60]:
dump(cv, models_dir / 'randomised_xgb')
# In[47]:
| pd.DataFrame(cv.cv_results_) | pandas.DataFrame |
import csv
import json
import pysam
import numpy as np
import pandas as pd
import random
from .bamutils import fetch_upto_next_ID
from . import gtfutils
DEFAULT_ANNOT=['?','?','?','?',int(0)]
def model(file_ambiv_gps, file_expression_model):
#load ambivalence gps
with open(file_ambiv_gps) as f:
json_data=f.read()
ambiv_gps_tx = json.loads(json_data)[0]
ambiv_gps_tx_LUT = {v:k for k, v in ambiv_gps_tx.items()}
#load expression model (hash table)
if not file_expression_model is None:
expression_model=_load_expression_model_sf(file_expression_model)
#compute the probabilities
ambivalence_proba, tx_LUT, tx_ix_dict = _compute_proba(ambiv_gps_tx_LUT, expression_model)
else:
ambivalence_proba, tx_LUT, tx_ix_dict = _compute_proba(ambiv_gps_tx_LUT, {})
return ambivalence_proba, tx_LUT, tx_ix_dict
# self.ambivalence_proba=ambivalence_proba
# self.tx_LUT=tx_LUT
# self.tx_ix_dict=tx_ix_dict
def _load_expression_model_sf(file, ix=3): #load a salmon file model
with open(file,'r') as f:
myreader=csv.reader(f, delimiter='\t')
myreader.__next__() #get rid of header
expression_model={row[0]:float(row[ix]) for row in myreader if float(row[ix])>0}
return expression_model
def load_sf(file,ix): #load a salmon file model
with open(file,'r') as f:
myreader=csv.reader(f, delimiter='\t')
myreader.__next__() #get rid of header
expression_model={row[0]:float(row[ix]) for row in myreader if float(row[ix])>0}
return expression_model
def _compute_proba(ambiv_gps_LUT, expression_model, tx_ix_dict=None):
# this assign probabibilities to all tx within ambivalence groups
if tx_ix_dict is None:
tx_ix_dict={}
if len(tx_ix_dict)==0:
i=-1
else:
i=max(v for _, v in tx_ix_dict.items())
ambiv_proba=[[[],[]]]*(len(ambiv_gps_LUT)+1)
for k, v in ambiv_gps_LUT.items():
tx_all=v.split("|")
tx_all_proba=[expression_model.get(tx,0) for tx in tx_all]
tx_all_ix=[0]*len(tx_all_proba)
for ix, tx in enumerate(tx_all):
tx_ix=tx_ix_dict.get(tx,-1)
if tx_ix<0:
i+=1
tx_ix_dict[tx]=i
tx_all_ix[ix]=i
else:
tx_all_ix[ix]=tx_ix
ambiv_proba[k]=[tx_all_proba,tx_all_ix]
tx_LUT_dict={v:k for k,v in tx_ix_dict.items()} #{1:ENST*, 2:ENSTXXX,...}
tx_LUT=[""]*(i+1)
for k in range(i+1):
tx_LUT[k]=tx_LUT_dict.get(k)
return ambiv_proba, tx_LUT, tx_ix_dict
class Quantifier: #quantify .tag.bam file
def __init__(self, file_ambiv_gps, file_expression_model=None): #agi: ambivalence group index
ambivalence_proba, tx_LUT, tx_ix_dict=model(file_ambiv_gps, file_expression_model)
self.ambivalence_proba=ambivalence_proba
self.tx_LUT=tx_LUT
self.tx_ix_dict=tx_ix_dict
self.annot_dict=None
def set_annotations_dictionary(self, annot_file):
# Load annotation dictionnary
if isinstance(annot_file,str):
print("Loading annotations dictionary from %s"%annot_file)
self.annot_dict=gtfutils.make_tx_dict_fromCSV(annot_file)
else:
print('Annotation dictionnary passed directly as argument')
self.annot_dict=annot_file
def quant(self, qbam_in, todf=True, simplify=True, drop_zeros=True, allow_no_prior=True):
# dumps a quantification table
#prepares the output table for know tx, and an empty dict for unknown stuff
noprior=1.0 if allow_no_prior else 0
q=np.zeros((len(self.tx_ix_dict),10))
q_unknown={}
q_unknown2={}
n_ambiv_with_prior=0
n_ambiv_without_prior=0
n_unique_with_prior=0
n_unique_new=0
reader = pysam.AlignmentFile(qbam_in)
bamiter = reader.fetch(until_eof=True)
try:
r = bamiter.__next__()
except StopIteration:
r = None
while not(r==None):
ambiv_ix = r.get_tag("ai") if r.has_tag("ai") else 0
ambiv_ix_abs=abs(ambiv_ix)
ambiv_ix_gene = r.get_tag("aI") if r.has_tag("aI") else 0
ambiv_gene_offset = 0 if abs(ambiv_ix_gene)>1 else 3 #3 if NOT ambiv at gene level
offset=0 if ambiv_ix>0 else 5 #if negative ix, means there is other alignment
# q cols: 0 is ambiv_tx with prior, 1 is ambiv_tx no prior, 2 is no ambiv,
# 3 is ambiv at tx but not at gene, 4 same no prior
if ambiv_ix_abs>1: #this is an ambivalence group
Es, tx_ixs =self.ambivalence_proba[abs(ambiv_ix)] #expression values
Etotal=sum(Es)
if Etotal>0:
n_ambiv_with_prior+=1
for i, tx_ix in enumerate(tx_ixs):
q[tx_ix,0+ambiv_gene_offset+offset]+=Es[i]/Etotal
else:
n_ambiv_without_prior+=1
for i, tx_ix in enumerate(tx_ixs):
q[tx_ix,1+ambiv_gene_offset+offset]+=1.0/len(tx_ixs)
elif r.has_tag("ar"):
tx_name=r.get_tag("ar")
tx_ix=self.tx_ix_dict.get(tx_name,-1)
if tx_ix>-1: # this is a known one
n_unique_with_prior+=1
q[tx_ix,2+offset]+=1
else:
n_unique_new+=1
if offset==0:
if tx_name in q_unknown:
q_unknown[tx_name]+=1
else:
q_unknown[tx_name]=1
else:
if tx_name in q_unknown2:
q_unknown2[tx_name]+=1
else:
q_unknown2[tx_name]=1
try:
r = bamiter.__next__()
except StopIteration:
r = None
reader.close()
counts=(n_ambiv_with_prior,n_ambiv_without_prior,n_unique_with_prior,n_unique_new)
print("Found: %g ambiv_with_prior, %g ambiv_without_prior, %g unique known, %g unknown"%counts)
#Now consolidate
#create LUT dict for the new tx
tx_new_LUT_dict={i:k for i,(k,_) in enumerate(q_unknown.items())} #{1:ENST*, 2:ENSTXXX,...}
imax=len(tx_new_LUT_dict)
#add in the new tx that belong to the ai=-1 gp
for k, _ in q_unknown2.items():
if not k in q_unknown:
tx_new_LUT_dict[imax]=k
imax+=1
#transform the LUT dict into a list
tx_new_LUT=[""]*(len(tx_new_LUT_dict))
for k in range(len(tx_new_LUT_dict)):
tx_new_LUT[k]=tx_new_LUT_dict.get(k)
#transform the q_unknown dictionnary into a list
q_new=np.zeros((len(tx_new_LUT_dict),2))
for k, tx in enumerate(tx_new_LUT):
counts=q_unknown.get(tx,0)
counts2=q_unknown2.get(tx,0)
q_new[k,0]=counts
q_new[k,1]=counts2
if not todf:
return q, q_new, tx_new_LUT, counts
else:
#ambiv_tx is ambiv at tx level (not at gene)
#ambiv_gene is ambiv also at gene level
#no prior means no_prior proba for this ambivalence group
#2 means there is alignment to other areas
# df_known=pd.DataFrame(q,columns=['ambiv','ambiv_noprior','unique','ambiv2','ambiv_noprior2','unique2'], index=self.tx_LUT)
# df_unknown=pd.DataFrame(q_new,columns=['unique','unique2'], index=tx_new_LUT)
# df_all=pd.concat([df_known,df_unknown], axis=0)
# df_all.index.names=['Name']
# if simplify:
# df=pd.concat(
# [df_all.sum(axis=1),df_all["ambiv"]+df_all["ambiv2"],df_all["ambiv2"]+df_all["unique2"]],axis=1).drop(["ENST*"])
# df.columns=["N","N_ambiv","N_hasunannot"]
# if drop_zeros:
# df.fillna(0, inplace=True)
# df.drop(df[(df.N ==0)].index, inplace=True)
# return df
# else:
# return df_all
df_known= | pd.DataFrame(q,columns=['ambiv_gene','ambiv_gene_noprior','unique','ambiv_tx','ambiv_tx_noprior','ambiv_gene2','ambiv_gene_noprior2','unique2','ambiv_tx2','ambiv_tx_noprior2'], index=self.tx_LUT) | pandas.DataFrame |
import pandas as pd
import numpy as np
import requests
import time
import sqlite3
import matplotlib.pyplot as plt
from matplotlib.pyplot import figure
from bs4 import BeautifulSoup
column_names = ["Team", "ABV", "Schedule"]
abbreviations = pd.read_csv("schedule_urls.csv", names=column_names)
team = abbreviations.ABV.to_list()
teamb = sorted(team, reverse=True)
teamp = sorted(team, reverse=True)
team2=sorted(team)
graph_List = sorted(team, reverse=True)
team_df = pd.DataFrame(team)
# Goal is to scrape all teams in the MLB for pitching and batting statitics and find the leader in the categories of average and strikeouts(K's)
def Batting(): # Creates database table for every hitter in the major league with up to date statistics for 2021 from espn.com
start_time = time.time()
aggrigate_teams = pd.DataFrame()
while len(teamb) > 0:
#scrape data and create readable data
url = f'https://www.espn.com/mlb/team/stats/_/name/{teamb[-1]}/season/2021/seasontype/2'
page = requests.get(url)
soup = BeautifulSoup(page.text, 'html.parser')
# Creates elements from html that will be the base for the Data Frame
team_name_object = soup.find('span', attrs= {'class': 'flex flex-wrap'})
table = soup.find('div', attrs = {'class':'ResponsiveTable ResponsiveTable--fixed-left mt5 remove_capitalize'})
table2 = soup.find('div', attrs = {'class':'Table__Scroller'})
#Creates Pandas dataframes
df= pd.read_html(str(table))[0]
df2= pd.read_html(str(table2))[0]
# Combines tables to create readable data
resultB = df.join(df2)
index = resultB.index
resultB = resultB.drop(resultB.index[len(index) - 1])
resultB = resultB.assign(Team = teamb[-1])
aggrigate_teams = aggrigate_teams.append(resultB)
del teamb[-1]
if len(teamb) < 1:
break
#Sends data to database
con = sqlite3.connect("MLB_Stats.sqlite") # change to 'sqlite:///your_filename.db'
cur = con.cursor()
aggrigate_teams.to_sql("Batting_Stats", con, if_exists='replace', index=False)
con.commit()
con.close()
#ends timer for inquary
clock = "{:.3f}".format(time.time() - start_time)
print("Inquiry took:", clock ,"seconds.\n")
return aggrigate_teams
def Pitching():
start_time = time.time()
aggrigate_teams = pd.DataFrame()
while len(teamp) > 0:
#scrape data and create readable data
url = f'https://www.espn.com/mlb/team/stats/_/type/pitching/name/{teamp[-1]}/season/2021/seasontype/2'
page = requests.get(url)
soup = BeautifulSoup(page.text, 'html.parser')
# Creates elements from html that will be the base for the Data Frame
table = soup.find('div', attrs = {'class':'ResponsiveTable ResponsiveTable--fixed-left mt5 remove_capitalize'})
table2 = soup.find('div', attrs = {'class':'Table__Scroller'})
#Creates Pandas dataframes
df= pd.read_html(str(table))[0]
df2= pd.read_html(str(table2))[0]
# Combines tables to create readable data
resultP = df.join(df2)
index = resultP.index
resultP = resultP.drop(resultP.index[len(index) - 1])
resultP = resultP.assign(Team = teamp[-1])
aggrigate_teams = aggrigate_teams.append(resultP)
del teamp[-1]
if len(team) < 1:
break
#Sends data to database
con = sqlite3.connect("MLB_Stats.sqlite") # change to 'sqlite:///your_filename.db'
cur = con.cursor()
aggrigate_teams.to_sql("Pitching_Stats", con, if_exists='replace', index=False)
con.commit()
con.close()
# stops timer of the inquary and ouputs total time taken for pitching scrape
clock = "{:.3f}".format(time.time() - start_time)
#set this up because there has to be a way to make this more efficient, not sure how.
print("Inquiry took:", clock ,"seconds.\n")
return aggrigate_teams
def team_hitting(team):
con = sqlite3.connect("MLB_Stats.sqlite") # connecting to database
print("Hitting Statistics for you chosen team: \n")
pd.set_option('display.max_columns', None)
print(pd.read_sql_query("Select Name, AB, R, H , HR, RBI, BB, SO, AVG, OBP, OPS FROM Batting_Stats WHERE Team == ('%s')" % (team), con))
def team_pitching(team):
con = sqlite3.connect("MLB_Stats.sqlite") # connecting to database
print("Pitching Statistics for you chosen team: \n")
pd.set_option('display.max_columns', None)
print(pd.read_sql_query("Select Name, GP, W, L, SV, IP, H, ER, HR, BB, K, WHIP, ERA FROM Pitching_Stats WHERE Team == ('%s')" % (team), con))
def Average_query():
con = sqlite3.connect("MLB_Stats.sqlite") # connecting to database
print("Top 5 for Batting Average with at least 45 at bats:\n")
pd.set_option('display.max_columns', None)
print(pd.read_sql_query("SELECT Name, AB, AVG, Team FROM Batting_Stats WHERE AB >=45 ORDER BY AVG DESC LIMIT 5", con))
def Strikeout_query():
con = sqlite3.connect("MLB_Stats.sqlite") # connecting to database
print("Top 5 for Strikeouts: \n")
pd.set_option('display.max_columns', None)
print(pd.read_sql_query("SELECT Name, GP, GS, W, L, SV, IP, K, ERA, Team FROM Pitching_Stats ORDER BY K DESC LIMIT 5", con),"\n")
def ERA_query():
con = sqlite3.connect("MLB_Stats.sqlite") # connecting to database
print("ERA Leaders with at least 40 innings pitched: \n")
print(pd.read_sql_query("SELECT Name, GP, W, L, SV, IP, K, ERA, Team FROM Pitching_Stats WHERE IP >=40 ORDER BY ERA ASC LIMIT 5", con),"\n")
def Homerun_query():
con = sqlite3.connect("MLB_Stats.sqlite") # connecting to database
print("Top 5 for Home Runs: \n")
print(pd.read_sql_query("SELECT Name, AB, HR, Team FROM Batting_Stats ORDER BY HR DESC LIMIT 5",con),"\n")
def Hitting_league_leaders():
con = sqlite3.connect("MLB_Stats.sqlite") #connect to database
print("Hitting leaders from around the league:\n")
AVG = pd.read_sql_query("SELECT Name, AVG, Team FROM Batting_Stats WHERE AB >45 ORDER BY AVG DESC LIMIT 1",con)
H = pd.read_sql_query("SELECT Name, H, Team FROM Batting_Stats ORDER BY H DESC LIMIT 1",con)
Dbl = pd.read_sql_query('SELECT Name, "2B", Team FROM Batting_Stats ORDER BY "2B" DESC LIMIT 1',con)
Trip = pd.read_sql_query('SELECT Name, "3B", Team FROM Batting_Stats ORDER BY "3B" DESC LIMIT 1',con)
HR = pd.read_sql_query("SELECT Name, HR, Team FROM Batting_Stats ORDER BY HR DESC LIMIT 1",con)
RBI = pd.read_sql_query("SELECT Name, RBI, Team FROM Batting_Stats ORDER BY RBI DESC LIMIT 1",con)
| pd.set_option('display.max_colwidth', 40) | pandas.set_option |
# coding: utf-8
# # ------------- Logistics -------------
# In[1]:
from __future__ import division
import numpy
import os
import pandas
import sklearn
import sys
import sqlite3
import pickle
from operator import itemgetter
from collections import Counter
import itertools
import matplotlib
import matplotlib.pyplot as plt
from sklearn.svm import SVC, LinearSVC
from sklearn.naive_bayes import GaussianNB
from sklearn.ensemble import RandomForestClassifier, RandomForestRegressor, ExtraTreesClassifier, AdaBoostClassifier
from sklearn.metrics import confusion_matrix
from sklearn.preprocessing import normalize, scale, LabelEncoder
from sklearn import model_selection
from sklearn.feature_selection import VarianceThreshold, SelectFromModel, RFECV
#------------- Custom functions -------------#
def plot_recall(classifier_name, cm, output_directory):
#---Plot settings ---#
fig, ax = plt.subplots()
# the size of A4 paper
fig.set_size_inches(11.7, 8.27)
# Number of ticks in axes
plt.yticks(numpy.arange(0.0, 1.05, 0.05))
# Axes limit
axes = ax.axes
axes.set_ylim(0.0,1.05)
# Pad margins so that markers don't get clipped by the axes
plt.margins(0.2)
# Tweak spacing to prevent clipping of tick-labels
plt.subplots_adjust(bottom=0.15)
numpy.set_printoptions(precision=3, suppress=True)
#---Plot data ---#
row_sums = cm.sum(axis=1)
normalized_cm = cm / row_sums[:, numpy.newaxis]
cm_diag = normalized_cm.diagonal()
bar_labels = sorted(list(set(ground_truth)))
y_pos = numpy.arange(len(bar_labels))
plt.bar(y_pos,
cm_diag,
align='center',
color='blue')
plt.ylabel('Percent of cells correctly classifed (recall)')
plt.xticks(y_pos, bar_labels, rotation='vertical')
plt.title('Cell Classes, ' + classifier_name)
plt_name = classifier_name + '_plot.png'
plt.savefig(os.path.join(output_directory, plt_name))
plt.clf()
def plot_confusion_matrix(cm, output_directory, classifier_name, classes,
normalize=False,
title='Confusion matrix',
cmap=plt.cm.Blues
):
"""
This function prints and plots the confusion matrix.
Normalization can be applied by setting `normalize=True`.
"""
if normalize:
cm = cm.astype('float') / cm.sum(axis=1)[:, numpy.newaxis]
print("Normalized confusion matrix")
else:
print('Confusion matrix, without normalization')
print(cm)
cm_file_name = name + '_cm.txt'
cm_file = open(os.path.join(output_directory, cm_file_name), 'w+')
cm_file.write(str(cm))
cm_file.close()
plt.imshow(cm, interpolation='nearest', cmap=cmap)
plt.title(title)
plt.colorbar()
tick_marks = numpy.arange(len(classes))
plt.xticks(tick_marks, classes, rotation=90)
plt.yticks(tick_marks, classes)
fmt = '.2f' if normalize else 'd'
thresh = cm.max() / 2.
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(j, i, format(cm[i, j], fmt),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black")
plt.ylabel('True label')
plt.xlabel('Predicted label')
plt_name = classifier_name + '_confusion_matrix.png'
plt.savefig(os.path.join(output_directory, plt_name))
plt.clf()
def variance_threshold_select(df, thresh=0.0, na_replacement=-999):
df1 = df.copy(deep=True) # Make a deep copy of the dataframe
selector = VarianceThreshold(thresh)
selector.fit(df1.fillna(na_replacement)) # Fill NA values as VarianceThreshold cannot deal with those
df2 = df.loc[:,selector.get_support(indices=False)] # Get new dataframe with columns deleted that have NA values
return df2
def save_metadata(file, label_list):
with open(file, 'w') as f:
for i in label_list:
f.write('{}\n'.format( i ))
def plot_rank_importance(data, labels, output_directory):
model = RandomForestRegressor(n_estimators=20, max_features=2)
model = model.fit(data, labels)
model.feature_importances_
important_features = pandas.Series(data=model.feature_importances_, index=data.columns)
important_features.sort_values(ascending=False,inplace=True)
std = numpy.std([tree.feature_importances_ for tree in model.estimators_],axis=0)
# Plot the feature importances of the forest
plt.figure(figsize=(35, 40))
plt.title("Feature importances")
plt.bar(range(important_features.shape[0]), important_features, yerr = std, color="r", align="center")
feature_names = list(important_features.index)
plt.xticks(range(important_features.shape[0]), feature_names)
plt.xticks(rotation=90)
plt.xlim([-1, important_features.shape[0]])
matplotlib.rcParams.update({'font.size': 10})
plt_name = 'feature_ranking.png'
plt.savefig(os.path.join(output_directory, plt_name))
plt.clf()
# # ------------- User's settings -------------
# In[2]:
#----- The dot "." means "current directory" -----#
train_directory = "./train_results"
test_directory = "./test_results"
#----- Load CellProfiler output database -----#
database_train = sqlite3.connect('../CPOut/DefaultDB_train.db')
database_test = sqlite3.connect('../CPOut/DefaultDB_test.db')
#----- Define unwanted parameters -----#
not_wanted = [ "Number", "Location", "Center" ]
#----- Define the models to test. Multiple choices possible -----#
names_classifiers = []
names_classifiers.append(( 'NaiveBayes', GaussianNB() ) )
names_classifiers.append(( 'RandomForest', RandomForestClassifier() ) )
# names_classifiers.append(( 'KernelSVM', SVC() ) )
# names_classifiers.append(( 'LinearSVM', LinearSVC() ) )
#----- Logistic -----#
for directory in [train_directory, test_directory]:
if not os.path.exists(directory):
os.makedirs(directory)
# # ------------- Data loading and preprocessing -------------
# In[3]:
#----- Load training data -----#
query_objects = database_train.execute("SELECT * From Per_Object")
cols_objects = [column[0] for column in query_objects.description]
objects = pandas.DataFrame.from_records(data = query_objects.fetchall(), columns = cols_objects)
variables_object = [x for x in objects.columns.values if numpy.all([not z in x for z in not_wanted])]
objects_train = objects.loc[: , variables_object + ["ImageNumber"]]
print('Original data has shape (rows, columns) : ', objects_train.shape)
query_image = database_train.execute("SELECT * From Per_Image")
cols_image = [column[0] for column in query_image.description]
images = pandas.DataFrame.from_records(data = query_image.fetchall(), columns = cols_image)
variables_image = [col for col in images.columns if col.startswith(('ImageNumber','Image_Metadata_folder'))]
images_train = pandas.DataFrame(images, columns=variables_image)
#----- Remove any row that has NA -----#
objects_train = objects_train.dropna()
print('After removing NA rows, data has shape : ', objects_train.shape)
#----- Create ground-truth label for each cells -----#
merged_table = pandas.merge(images_train, objects_train, on=['ImageNumber'], how='inner')
ground_truth = list(merged_table.Image_Metadata_folder)
# Save labels, to be used as "metadata" on http://projector.tensorflow.org
save_metadata( os.path.join(train_directory, 'ground_truth_labels.tsv'), ground_truth )
#----- Remove all zero-variance features -----#
# i.e. features that have the same value in all samples.
objects_train = variance_threshold_select(objects_train.drop([objects_train.columns[-1]], axis=1))
print('After removing zero-variance features, data has shape: ', objects_train.shape)
#----- Logistic for training data -----#
le = LabelEncoder()
le.fit(ground_truth)
numeric_labels = le.fit_transform(ground_truth)
#----- Tree-based feature selection -----#
'''
When the dataset has two (or more) correlated features,
then any of these correlated features can be used as the predictor.
For the computer, there's no concrete preference of one over the others.
It makes sense to remove features that are mostly duplicated by other features (redundancy)
Tree-based feature selection will help us to (randomly)
keep only one of them, and remove others.
This is not an issue when we want to use feature selection to reduce overfitting.
But when interpreting the data, it can lead to the incorrect conclusion that
one of the variables is a strong predictor while the others in the same group are unimportant.
'''
clf = RandomForestRegressor(n_estimators=20, max_features=2)
clf = clf.fit(objects_train, numeric_labels)
clf.feature_importances_
df1 = objects_train.copy(deep=True) # Make a deep copy of the dataframe
selector = SelectFromModel(clf, prefit=True)
selector.transform(df1)
data_train = objects_train.loc[:,selector.get_support(indices=False)]
print('After feature selection, data has shape : ', data_train.shape)
# List of all the feature names
selected_features_names =list(data_train.columns.values)
plot_rank_importance(data_train, numeric_labels, train_directory)
data_train.to_csv( os.path.join(train_directory, 'after_feature_selection_data.txt' ))
#----- To be used as main data on http://projector.tensorflow.org -----#
numpy.savetxt( os.path.join(train_directory, 'after_feature_selection_scaled_data.txt' ), scale(data_train), delimiter='\t')
# ------------- Preprocessing testing data accordingly -------------
# In[4]:
#----- Load testing data -----#
query_objects = database_test.execute("SELECT * From Per_Object")
cols_objects = [column[0] for column in query_objects.description]
objects_test = pandas.DataFrame.from_records(data = query_objects.fetchall(), columns = cols_objects)
print('Original test data has shape (rows, columns) : ', objects_test.shape)
query_image = database_test.execute("SELECT * From Per_Image")
cols_image = [column[0] for column in query_image.description]
images_test = pandas.DataFrame.from_records(data = query_image.fetchall(), columns = cols_image)
variables_image = [col for col in images_test.columns if col.startswith(('ImageNumber','Image_Metadata_folder'))]
selected_var_images_test = | pandas.DataFrame(images_test, columns=variables_image) | pandas.DataFrame |
# Collection of preprocessing functions
from nltk.tokenize import word_tokenize
from transformers import CamembertTokenizer
from transformers import BertTokenizer
from tqdm import tqdm
import numpy as np
import pandas as pd
import re
import string
import unicodedata
import tensorflow as tf
import glob
import os
MAX_LEN = 500
IMG_SHAPE = 224
AUTO = tf.data.experimental.AUTOTUNE
model_bert = 'bert-base-multilingual-cased'
model_camembert = 'camembert-base'
tokenizer_bert = BertTokenizer.from_pretrained(model_bert, do_lowercase=False)
tokenizer_cam = CamembertTokenizer.from_pretrained(model_camembert, do_lowercase=False)
def preprocessing_csv(file):
file = pd.read_csv(file, index_col=0)
file = file.reset_index()
file['filename'] = file.apply(
lambda x: "datas/images/image_test/image_" + str(x['imageid']) + "_product_" + str(x['productid']) + ".jpg",
axis=1)
file['text'] = file.apply(lambda x: str(x['designation']) + ' ' + str(x['description']), axis=1)
file['text'] = file['text'].str.replace('nan', '')
file = file.drop(['designation', 'description', 'imageid', 'productid'], axis=1)
return file
# Converts the unicode file to ascii
def unicode_to_ascii(s):
return ''.join(c for c in unicodedata.normalize('NFD', s)
if unicodedata.category(c) != 'Mn')
# preprocess sentences
def preprocess_sentence(w):
w = unicode_to_ascii(w.lower().strip())
w = re.sub('https?://\S+|www\.\S+', '', w)
w = re.sub('[%s]' % re.escape(string.punctuation), '', w)
w = re.sub('\n', '', w)
w = re.sub(r"([?.!,¿])", r" \1 ", w)
w = re.sub(r"[^a-zA-Z?.!]+", " ", w)
mots = word_tokenize(w.strip())
return ' '.join(mots).strip()
# encode transfomers to tokenizer
def encode(sentences, tokenizer, maxlen=500):
input_ids = []
attention_masks = []
# Pour chaque sentences...
for sent in tqdm(sentences):
encoded_dict = tokenizer.encode_plus(
sent, # Sentence to encode. / des fois j'écris en Anglais
add_special_tokens=True, # Add '[CLS]' and '[SEP]'
max_length=maxlen, # Pad & truncate all sentences.
pad_to_max_length=True,
return_attention_mask=True, # Construct attn. masks.
return_tensors='np', # Return Numpy.
)
# Add the encoded sentence to the list.
input_ids.append(encoded_dict['input_ids'])
# And its attention mask (simply differentiates padding from non-padding).
attention_masks.append(encoded_dict['attention_mask'])
# Convert the lists into arrays.
input_ids = np.asarray(input_ids, dtype='int32')
attention_masks = np.asarray(attention_masks, dtype='int32')
input_ids = np.squeeze(input_ids)
attention_masks = np.squeeze(attention_masks)
return input_ids, attention_masks
@tf.function
# Fonction pour preprocessing des images
def preprocessing_test(img):
# Lecture et décodage des images:
img = tf.io.read_file(img)
img = tf.io.decode_jpeg(img, channels=3)
# Resize
img = tf.cast(img, dtype=tf.float32)
img = tf.image.resize(img, [IMG_SHAPE, IMG_SHAPE])
img = (img / 255)
return img
def make_test(x1, x2, x3, x4, x5):
dataset = tf.data.Dataset.from_tensor_slices((x1, x2, x3, x4, x5)) \
.map(lambda r, s, t, u, w: [(preprocessing_test(r), s, t, u, w)], num_parallel_calls=AUTO) \
.batch(1) \
.prefetch(AUTO)
return dataset
def fusion_features():
file_names = glob.glob('datas/images/upload_images/*')
df_image = | pd.DataFrame(file_names, columns=['filename']) | pandas.DataFrame |
# Copyright 2017-2021 QuantRocket LLC - All Rights Reserved
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# To run: python3 -m unittest discover -s tests/ -p test_*.py -t . -v
import os
import unittest
from unittest.mock import patch
import glob
import pickle
import joblib
import inspect
import pandas as pd
import numpy as np
from moonshot import MoonshotML
from moonshot.cache import TMP_DIR
from moonshot.exceptions import MoonshotError
from sklearn.tree import DecisionTreeClassifier
class SKLearnMachineLearningTestCase(unittest.TestCase):
def setUp(self):
"""
Trains a scikit-learn model.
"""
self.model = DecisionTreeClassifier()
# Predict Y will be same as X
X = np.array([[1,1],[0,0]])
Y = np.array([1,0])
self.model.fit(X, Y)
self.pickle_path = "{0}/decision_tree_model.pkl".format(TMP_DIR)
self.joblib_path = "{0}/decision_tree_model.joblib".format(TMP_DIR)
def tearDown(self):
"""
Remove cached files.
"""
for file in glob.glob("{0}/moonshot*.pkl".format(TMP_DIR)):
os.remove(file)
for file in (self.pickle_path, self.joblib_path):
if os.path.exists(file):
os.remove(file)
def test_complain_if_mix_dataframe_and_series(self):
"""
Tests error handling when the features list contains a mix of
DataFrames and Series.
"""
# pickle model
with open(self.pickle_path, "wb") as f:
pickle.dump(self.model, f)
class DecisionTreeML1(MoonshotML):
MODEL = self.pickle_path
def prices_to_features(self, prices):
features = []
# DataFrame then Series
features.append(prices.loc["Close"] > 10)
features.append(prices.loc["Close"]["FI12345"] > 10)
return features, None
class DecisionTreeML2(MoonshotML):
MODEL = self.pickle_path
def prices_to_features(self, prices):
features = []
# Series then DataFrame
features.append(prices.loc["Close"]["FI12345"] > 10)
features.append(prices.loc["Close"] > 10)
return features, None
def mock_get_prices(*args, **kwargs):
dt_idx = pd.DatetimeIndex(["2018-05-01","2018-05-02","2018-05-03", "2018-05-04"])
fields = ["Close"]
idx = pd.MultiIndex.from_product([fields, dt_idx], names=["Field", "Date"])
prices = pd.DataFrame(
{
"FI12345": [
# Close
9,
11,
10.50,
9.99,
],
"FI23456": [
# Close
9.89,
11,
8.50,
10.50,
],
},
index=idx
)
prices.columns.name = "Sid"
return prices
def mock_download_master_file(f, *args, **kwargs):
master_fields = ["Timezone", "Symbol", "SecType", "Currency", "PriceMagnifier", "Multiplier"]
securities = pd.DataFrame(
{
"FI12345": [
"America/New_York",
"ABC",
"STK",
"USD",
None,
None
],
"FI23456": [
"America/New_York",
"DEF",
"STK",
"USD",
None,
None,
]
},
index=master_fields
)
securities.columns.name = "Sid"
securities.T.to_csv(f, index=True, header=True)
f.seek(0)
with patch("moonshot.strategies.base.get_prices", new=mock_get_prices):
with patch("moonshot.strategies.base.download_master_file", new=mock_download_master_file):
with self.assertRaises(MoonshotError) as cm:
results = DecisionTreeML1().backtest()
self.assertIn(
"features should be either all DataFrames or all Series, not a mix of both",
repr(cm.exception))
# clear cache
for file in glob.glob("{0}/moonshot*.pkl".format(TMP_DIR)):
os.remove(file)
with self.assertRaises(MoonshotError) as cm:
results = DecisionTreeML2().backtest()
self.assertIn(
"features should be either all DataFrames or all Series, not a mix of both",
repr(cm.exception))
def test_complain_if_no_targets(self):
"""
Tests error handling when prices_to_features doesn't return a two-tuple.
"""
# pickle model
with open(self.pickle_path, "wb") as f:
pickle.dump(self.model, f)
class DecisionTreeML(MoonshotML):
MODEL = self.pickle_path
def prices_to_features(self, prices):
features = []
features.append(prices.loc["Close"] > 10)
features.append(prices.loc["Close"] > 100)
return features
def mock_get_prices(*args, **kwargs):
dt_idx = pd.DatetimeIndex(["2018-05-01","2018-05-02","2018-05-03", "2018-05-04"])
fields = ["Close"]
idx = pd.MultiIndex.from_product([fields, dt_idx], names=["Field", "Date"])
prices = pd.DataFrame(
{
"FI12345": [
# Close
9,
11,
10.50,
9.99,
],
"FI23456": [
# Close
9.89,
11,
8.50,
10.50,
],
},
index=idx
)
prices.columns.name = "Sid"
return prices
def mock_download_master_file(f, *args, **kwargs):
master_fields = ["Timezone", "Symbol", "SecType", "Currency", "PriceMagnifier", "Multiplier"]
securities = pd.DataFrame(
{
"FI12345": [
"America/New_York",
"ABC",
"STK",
"USD",
None,
None
],
"FI23456": [
"America/New_York",
"DEF",
"STK",
"USD",
None,
None,
]
},
index=master_fields
)
securities.columns.name = "Sid"
securities.T.to_csv(f, index=True, header=True)
f.seek(0)
with patch("moonshot.strategies.base.get_prices", new=mock_get_prices):
with patch("moonshot.strategies.base.download_master_file", new=mock_download_master_file):
with self.assertRaises(MoonshotError) as cm:
results = DecisionTreeML().backtest()
self.assertIn(
"prices_to_features should return a tuple of (features, targets)", repr(cm.exception))
def test_backtest_from_pickle(self):
"""
Tests that the resulting DataFrames are correct after running a basic
machine learning strategy and loading the model from a pickle.
"""
# pickle model
with open(self.pickle_path, "wb") as f:
pickle.dump(self.model, f)
class DecisionTreeML(MoonshotML):
MODEL = self.pickle_path
def prices_to_features(self, prices):
features = {}
features["feature1"] = prices.loc["Close"] > 10
features["feature2"] = prices.loc["Close"] > 10 # silly, duplicate feature
return features, None
def predictions_to_signals(self, predictions, prices):
# Go long when price is predicted to be below 10
signals = predictions == 0
return signals.astype(int)
def mock_get_prices(*args, **kwargs):
dt_idx = pd.DatetimeIndex(["2018-05-01","2018-05-02","2018-05-03", "2018-05-04"])
fields = ["Close"]
idx = pd.MultiIndex.from_product([fields, dt_idx], names=["Field", "Date"])
prices = pd.DataFrame(
{
"FI12345": [
# Close
9,
11,
10.50,
9.99,
],
"FI23456": [
# Close
9.89,
11,
8.50,
10.50,
],
},
index=idx
)
prices.columns.name = "Sid"
return prices
def mock_download_master_file(f, *args, **kwargs):
master_fields = ["Timezone", "Symbol", "SecType", "Currency", "PriceMagnifier", "Multiplier"]
securities = pd.DataFrame(
{
"FI12345": [
"America/New_York",
"ABC",
"STK",
"USD",
None,
None
],
"FI23456": [
"America/New_York",
"DEF",
"STK",
"USD",
None,
None,
]
},
index=master_fields
)
securities.columns.name = "Sid"
securities.T.to_csv(f, index=True, header=True)
f.seek(0)
with patch("moonshot.strategies.base.get_prices", new=mock_get_prices):
with patch("moonshot.strategies.base.download_master_file", new=mock_download_master_file):
results = DecisionTreeML().backtest()
self.assertSetEqual(
set(results.index.get_level_values("Field")),
{'Commission',
'AbsExposure',
'Signal',
'Return',
'Slippage',
'NetExposure',
'TotalHoldings',
'Turnover',
'AbsWeight',
'Weight'}
)
# replace nan with "nan" to allow equality comparisons
results = results.round(7)
results = results.where(results.notnull(), "nan")
signals = results.loc["Signal"].reset_index()
signals.loc[:, "Date"] = signals.Date.dt.strftime("%Y-%m-%dT%H:%M:%S%z")
self.assertDictEqual(
signals.to_dict(orient="list"),
{'Date': [
'2018-05-01T00:00:00',
'2018-05-02T00:00:00',
'2018-05-03T00:00:00',
'2018-05-04T00:00:00'],
"FI12345": [1.0,
0.0,
0.0,
1.0],
"FI23456": [1.0,
0.0,
1.0,
0.0]}
)
weights = results.loc["Weight"].reset_index()
weights.loc[:, "Date"] = weights.Date.dt.strftime("%Y-%m-%dT%H:%M:%S%z")
self.assertDictEqual(
weights.to_dict(orient="list"),
{'Date': [
'2018-05-01T00:00:00',
'2018-05-02T00:00:00',
'2018-05-03T00:00:00',
'2018-05-04T00:00:00'],
"FI12345": [0.5,
0.0,
0.0,
1.0],
"FI23456": [0.5,
0.0,
1.0,
0.0]}
)
abs_weights = results.loc["AbsWeight"].reset_index()
abs_weights.loc[:, "Date"] = abs_weights.Date.dt.strftime("%Y-%m-%dT%H:%M:%S%z")
self.assertDictEqual(
abs_weights.to_dict(orient="list"),
{'Date': [
'2018-05-01T00:00:00',
'2018-05-02T00:00:00',
'2018-05-03T00:00:00',
'2018-05-04T00:00:00'],
"FI12345": [0.5,
0.0,
0.0,
1.0],
"FI23456": [0.5,
0.0,
1.0,
0.0]}
)
net_positions = results.loc["NetExposure"].reset_index()
net_positions.loc[:, "Date"] = net_positions.Date.dt.strftime("%Y-%m-%dT%H:%M:%S%z")
self.assertDictEqual(
net_positions.to_dict(orient="list"),
{'Date': [
'2018-05-01T00:00:00',
'2018-05-02T00:00:00',
'2018-05-03T00:00:00',
'2018-05-04T00:00:00'],
"FI12345": ["nan",
0.5,
0.0,
0.0],
"FI23456": ["nan",
0.5,
0.0,
1.0]}
)
abs_positions = results.loc["AbsExposure"].reset_index()
abs_positions.loc[:, "Date"] = abs_positions.Date.dt.strftime("%Y-%m-%dT%H:%M:%S%z")
self.assertDictEqual(
abs_positions.to_dict(orient="list"),
{'Date': [
'2018-05-01T00:00:00',
'2018-05-02T00:00:00',
'2018-05-03T00:00:00',
'2018-05-04T00:00:00'],
"FI12345": ["nan",
0.5,
0.0,
0.0],
"FI23456": ["nan",
0.5,
0.0,
1.0]}
)
total_holdings = results.loc["TotalHoldings"].reset_index()
total_holdings.loc[:, "Date"] = total_holdings.Date.dt.strftime("%Y-%m-%dT%H:%M:%S%z")
self.assertDictEqual(
total_holdings.to_dict(orient="list"),
{'Date': [
'2018-05-01T00:00:00',
'2018-05-02T00:00:00',
'2018-05-03T00:00:00',
'2018-05-04T00:00:00'],
"FI12345": [0,
1.0,
0,
0],
"FI23456": [0,
1.0,
0,
1.0]}
)
turnover = results.loc["Turnover"].reset_index()
turnover.loc[:, "Date"] = turnover.Date.dt.strftime("%Y-%m-%dT%H:%M:%S%z")
self.assertDictEqual(
turnover.to_dict(orient="list"),
{'Date': [
'2018-05-01T00:00:00',
'2018-05-02T00:00:00',
'2018-05-03T00:00:00',
'2018-05-04T00:00:00'],
"FI12345": ["nan",
0.5,
0.5,
0.0],
"FI23456": ["nan",
0.5,
0.5,
1.0]}
)
commissions = results.loc["Commission"].reset_index()
commissions.loc[:, "Date"] = commissions.Date.dt.strftime("%Y-%m-%dT%H:%M:%S%z")
self.assertDictEqual(
commissions.to_dict(orient="list"),
{'Date': [
'2018-05-01T00:00:00',
'2018-05-02T00:00:00',
'2018-05-03T00:00:00',
'2018-05-04T00:00:00'],
"FI12345": [0.0,
0.0,
0.0,
0.0],
"FI23456": [0.0,
0.0,
0.0,
0.0]}
)
slippage = results.loc["Slippage"].reset_index()
slippage.loc[:, "Date"] = slippage.Date.dt.strftime("%Y-%m-%dT%H:%M:%S%z")
self.assertDictEqual(
slippage.to_dict(orient="list"),
{'Date': [
'2018-05-01T00:00:00',
'2018-05-02T00:00:00',
'2018-05-03T00:00:00',
'2018-05-04T00:00:00'],
"FI12345": [0.0,
0.0,
0.0,
0.0],
"FI23456": [0.0,
0.0,
0.0,
0.0]}
)
returns = results.loc["Return"]
returns = returns.reset_index()
returns.loc[:, "Date"] = returns.Date.dt.strftime("%Y-%m-%dT%H:%M:%S%z")
self.assertDictEqual(
returns.to_dict(orient="list"),
{'Date': [
'2018-05-01T00:00:00',
'2018-05-02T00:00:00',
'2018-05-03T00:00:00',
'2018-05-04T00:00:00'],
"FI12345": [0.0,
0.0,
-0.0227273, # (10.50 - 11)/11 * 0.5
-0.0],
"FI23456": [0.0,
0.0,
-0.1136364, # (8.50 - 11)/11 * 0.5
0.0]})
def test_backtest_from_joblib(self):
"""
Tests that the resulting DataFrames are correct after running a basic
machine learning strategy and loading the model from joblib.
"""
# save model
joblib.dump(self.model, self.joblib_path)
class DecisionTreeML(MoonshotML):
MODEL = self.joblib_path
def prices_to_features(self, prices):
features = {}
features["feature1"] = prices.loc["Close"] > 10
features["feature2"] = prices.loc["Close"] > 10 # silly, duplicate feature
return features, None
def predictions_to_signals(self, predictions, prices):
# Go long when price is predicted to be below 10
signals = predictions == 0
return signals.astype(int)
def mock_get_prices(*args, **kwargs):
dt_idx = | pd.DatetimeIndex(["2018-05-01","2018-05-02","2018-05-03", "2018-05-04"]) | pandas.DatetimeIndex |
# Licensed to Elasticsearch B.V. under one or more contributor
# license agreements. See the NOTICE file distributed with
# this work for additional information regarding copyright
# ownership. Elasticsearch B.V. licenses this file to you under
# the Apache License, Version 2.0 (the "License"); you may
# not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing,
# software distributed under the License is distributed on an
# "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY
# KIND, either express or implied. See the License for the
# specific language governing permissions and limitations
# under the License.
import re
import warnings
from enum import Enum
from typing import Any, Callable, Dict, List, Optional, Tuple, Union, cast
import numpy as np # type: ignore
import pandas as pd # type: ignore
from elasticsearch import Elasticsearch
# Default number of rows displayed (different to pandas where ALL could be displayed)
DEFAULT_NUM_ROWS_DISPLAYED = 60
DEFAULT_CHUNK_SIZE = 10000
DEFAULT_CSV_BATCH_OUTPUT_SIZE = 10000
DEFAULT_PROGRESS_REPORTING_NUM_ROWS = 10000
DEFAULT_ES_MAX_RESULT_WINDOW = 10000 # index.max_result_window
DEFAULT_PAGINATION_SIZE = 5000 # for composite aggregations
PANDAS_VERSION: Tuple[int, ...] = tuple(
int(part) for part in pd.__version__.split(".") if part.isdigit()
)[:2]
with warnings.catch_warnings():
warnings.simplefilter("ignore")
EMPTY_SERIES_DTYPE = pd.Series().dtype
def build_pd_series(
data: Dict[str, Any], dtype: Optional[np.dtype] = None, **kwargs: Any
) -> pd.Series:
"""Builds a pd.Series while squelching the warning
for unspecified dtype on empty series
"""
dtype = dtype or (EMPTY_SERIES_DTYPE if not data else dtype)
if dtype is not None:
kwargs["dtype"] = dtype
return pd.Series(data, **kwargs)
def docstring_parameter(*sub: Any) -> Callable[[Any], Any]:
def dec(obj: Any) -> Any:
obj.__doc__ = obj.__doc__.format(*sub)
return obj
return dec
class SortOrder(Enum):
ASC = 0
DESC = 1
@staticmethod
def reverse(order: "SortOrder") -> "SortOrder":
if order == SortOrder.ASC:
return SortOrder.DESC
return SortOrder.ASC
@staticmethod
def to_string(order: "SortOrder") -> str:
if order == SortOrder.ASC:
return "asc"
return "desc"
@staticmethod
def from_string(order: str) -> "SortOrder":
if order == "asc":
return SortOrder.ASC
return SortOrder.DESC
def elasticsearch_date_to_pandas_date(
value: Union[int, str], date_format: Optional[str]
) -> pd.Timestamp:
"""
Given a specific Elasticsearch format for a date datatype, returns the
'partial' `to_datetime` function to parse a given value in that format
**Date Formats: https://www.elastic.co/guide/en/elasticsearch/reference/current/mapping-date-format.html#built-in-date-formats
Parameters
----------
value: Union[int, str]
The date value.
date_format: str
The Elasticsearch date format (ex. 'epoch_millis', 'epoch_second', etc.)
Returns
-------
datetime: pd.Timestamp
From https://www.elastic.co/guide/en/elasticsearch/reference/current/date.html
Date formats can be customised, but if no format is specified then it uses the default:
"strict_date_optional_time||epoch_millis"
Therefore if no format is specified we assume either strict_date_optional_time
or epoch_millis.
"""
if date_format is None or isinstance(value, (int, float)):
try:
return pd.to_datetime(
value, unit="s" if date_format == "epoch_second" else "ms"
)
except ValueError:
return pd.to_datetime(value)
elif date_format == "epoch_millis":
return pd.to_datetime(value, unit="ms")
elif date_format == "epoch_second":
return pd.to_datetime(value, unit="s")
elif date_format == "strict_date_optional_time":
return pd.to_datetime(value, format="%Y-%m-%dT%H:%M:%S.%f%z", exact=False)
elif date_format == "basic_date":
return pd.to_datetime(value, format="%Y%m%d")
elif date_format == "basic_date_time":
return pd.to_datetime(value, format="%Y%m%dT%H%M%S.%f", exact=False)
elif date_format == "basic_date_time_no_millis":
return pd.to_datetime(value, format="%Y%m%dT%H%M%S%z")
elif date_format == "basic_ordinal_date":
return pd.to_datetime(value, format="%Y%j")
elif date_format == "basic_ordinal_date_time":
return pd.to_datetime(value, format="%Y%jT%H%M%S.%f%z", exact=False)
elif date_format == "basic_ordinal_date_time_no_millis":
return pd.to_datetime(value, format="%Y%jT%H%M%S%z")
elif date_format == "basic_time":
return pd.to_datetime(value, format="%H%M%S.%f%z", exact=False)
elif date_format == "basic_time_no_millis":
return pd.to_datetime(value, format="%H%M%S%z")
elif date_format == "basic_t_time":
return pd.to_datetime(value, format="T%H%M%S.%f%z", exact=False)
elif date_format == "basic_t_time_no_millis":
return pd.to_datetime(value, format="T%H%M%S%z")
elif date_format == "basic_week_date":
return pd.to_datetime(value, format="%GW%V%u")
elif date_format == "basic_week_date_time":
return pd.to_datetime(value, format="%GW%V%uT%H%M%S.%f%z", exact=False)
elif date_format == "basic_week_date_time_no_millis":
return pd.to_datetime(value, format="%GW%V%uT%H%M%S%z")
elif date_format == "strict_date":
return pd.to_datetime(value, format="%Y-%m-%d")
elif date_format == "date":
return pd.to_datetime(value, format="%Y-%m-%d")
elif date_format == "strict_date_hour":
return pd.to_datetime(value, format="%Y-%m-%dT%H")
elif date_format == "date_hour":
return pd.to_datetime(value, format="%Y-%m-%dT%H")
elif date_format == "strict_date_hour_minute":
return pd.to_datetime(value, format="%Y-%m-%dT%H:%M")
elif date_format == "date_hour_minute":
return pd.to_datetime(value, format="%Y-%m-%dT%H:%M")
elif date_format == "strict_date_hour_minute_second":
return pd.to_datetime(value, format="%Y-%m-%dT%H:%M:%S")
elif date_format == "date_hour_minute_second":
return pd.to_datetime(value, format="%Y-%m-%dT%H:%M:%S")
elif date_format == "strict_date_hour_minute_second_fraction":
return pd.to_datetime(value, format="%Y-%m-%dT%H:%M:%S.%f", exact=False)
elif date_format == "date_hour_minute_second_fraction":
return pd.to_datetime(value, format="%Y-%m-%dT%H:%M:%S.%f", exact=False)
elif date_format == "strict_date_hour_minute_second_millis":
return pd.to_datetime(value, format="%Y-%m-%dT%H:%M:%S.%f", exact=False)
elif date_format == "date_hour_minute_second_millis":
return pd.to_datetime(value, format="%Y-%m-%dT%H:%M:%S.%f", exact=False)
elif date_format == "strict_date_time":
return pd.to_datetime(value, format="%Y-%m-%dT%H:%M:%S.%f%z", exact=False)
elif date_format == "date_time":
return pd.to_datetime(value, format="%Y-%m-%dT%H:%M:%S.%f%z", exact=False)
elif date_format == "strict_date_time_no_millis":
return pd.to_datetime(value, format="%Y-%m-%dT%H:%M:%S%z")
elif date_format == "date_time_no_millis":
return pd.to_datetime(value, format="%Y-%m-%dT%H:%M:%S%z")
elif date_format == "strict_hour":
return pd.to_datetime(value, format="%H")
elif date_format == "hour":
return pd.to_datetime(value, format="%H")
elif date_format == "strict_hour_minute":
return pd.to_datetime(value, format="%H:%M")
elif date_format == "hour_minute":
return pd.to_datetime(value, format="%H:%M")
elif date_format == "strict_hour_minute_second":
return pd.to_datetime(value, format="%H:%M:%S")
elif date_format == "hour_minute_second":
return pd.to_datetime(value, format="%H:%M:%S")
elif date_format == "strict_hour_minute_second_fraction":
return pd.to_datetime(value, format="%H:%M:%S.%f", exact=False)
elif date_format == "hour_minute_second_fraction":
return | pd.to_datetime(value, format="%H:%M:%S.%f", exact=False) | pandas.to_datetime |
"""Testing calculate axis range"""
import pandas as pd
import pytest
from gov_uk_dashboards.axes import calc_axis_range
def test_given_positive_data_returns_min_range_zero():
"""Testing the axis range given values greater than zero returns zero for min range"""
df = | pd.DataFrame(data={"col1": [6, 8]}) | pandas.DataFrame |
# -*- coding: utf-8 -*-
from datetime import timedelta
from distutils.version import LooseVersion
import numpy as np
import pytest
import pandas as pd
import pandas.util.testing as tm
from pandas import (
DatetimeIndex, Int64Index, Series, Timedelta, TimedeltaIndex, Timestamp,
date_range, timedelta_range
)
from pandas.errors import NullFrequencyError
@pytest.fixture(params=[pd.offsets.Hour(2), timedelta(hours=2),
np.timedelta64(2, 'h'), Timedelta(hours=2)],
ids=str)
def delta(request):
# Several ways of representing two hours
return request.param
@pytest.fixture(params=['B', 'D'])
def freq(request):
return request.param
class TestTimedeltaIndexArithmetic(object):
# Addition and Subtraction Operations
# -------------------------------------------------------------
# TimedeltaIndex.shift is used by __add__/__sub__
def test_tdi_shift_empty(self):
# GH#9903
idx = pd.TimedeltaIndex([], name='xxx')
tm.assert_index_equal(idx.shift(0, freq='H'), idx)
tm.assert_index_equal(idx.shift(3, freq='H'), idx)
def test_tdi_shift_hours(self):
# GH#9903
idx = pd.TimedeltaIndex(['5 hours', '6 hours', '9 hours'], name='xxx')
tm.assert_index_equal(idx.shift(0, freq='H'), idx)
exp = pd.TimedeltaIndex(['8 hours', '9 hours', '12 hours'], name='xxx')
tm.assert_index_equal(idx.shift(3, freq='H'), exp)
exp = pd.TimedeltaIndex(['2 hours', '3 hours', '6 hours'], name='xxx')
tm.assert_index_equal(idx.shift(-3, freq='H'), exp)
def test_tdi_shift_minutes(self):
# GH#9903
idx = pd.TimedeltaIndex(['5 hours', '6 hours', '9 hours'], name='xxx')
tm.assert_index_equal(idx.shift(0, freq='T'), idx)
exp = pd.TimedeltaIndex(['05:03:00', '06:03:00', '9:03:00'],
name='xxx')
tm.assert_index_equal(idx.shift(3, freq='T'), exp)
exp = pd.TimedeltaIndex(['04:57:00', '05:57:00', '8:57:00'],
name='xxx')
tm.assert_index_equal(idx.shift(-3, freq='T'), exp)
def test_tdi_shift_int(self):
# GH#8083
trange = pd.to_timedelta(range(5), unit='d') + pd.offsets.Hour(1)
result = trange.shift(1)
expected = TimedeltaIndex(['1 days 01:00:00', '2 days 01:00:00',
'3 days 01:00:00',
'4 days 01:00:00', '5 days 01:00:00'],
freq='D')
tm.assert_index_equal(result, expected)
def test_tdi_shift_nonstandard_freq(self):
# GH#8083
trange = pd.to_timedelta(range(5), unit='d') + pd.offsets.Hour(1)
result = trange.shift(3, freq='2D 1s')
expected = TimedeltaIndex(['6 days 01:00:03', '7 days 01:00:03',
'8 days 01:00:03', '9 days 01:00:03',
'10 days 01:00:03'], freq='D')
tm.assert_index_equal(result, expected)
def test_shift_no_freq(self):
# GH#19147
tdi = TimedeltaIndex(['1 days 01:00:00', '2 days 01:00:00'], freq=None)
with pytest.raises(NullFrequencyError):
tdi.shift(2)
# -------------------------------------------------------------
def test_ufunc_coercions(self):
# normal ops are also tested in tseries/test_timedeltas.py
idx = TimedeltaIndex(['2H', '4H', '6H', '8H', '10H'],
freq='2H', name='x')
for result in [idx * 2, np.multiply(idx, 2)]:
assert isinstance(result, TimedeltaIndex)
exp = TimedeltaIndex(['4H', '8H', '12H', '16H', '20H'],
freq='4H', name='x')
tm.assert_index_equal(result, exp)
assert result.freq == '4H'
for result in [idx / 2, np.divide(idx, 2)]:
assert isinstance(result, TimedeltaIndex)
exp = TimedeltaIndex(['1H', '2H', '3H', '4H', '5H'],
freq='H', name='x')
tm.assert_index_equal(result, exp)
assert result.freq == 'H'
idx = TimedeltaIndex(['2H', '4H', '6H', '8H', '10H'],
freq='2H', name='x')
for result in [-idx, np.negative(idx)]:
assert isinstance(result, TimedeltaIndex)
exp = TimedeltaIndex(['-2H', '-4H', '-6H', '-8H', '-10H'],
freq='-2H', name='x')
tm.assert_index_equal(result, exp)
assert result.freq == '-2H'
idx = TimedeltaIndex(['-2H', '-1H', '0H', '1H', '2H'],
freq='H', name='x')
for result in [abs(idx), np.absolute(idx)]:
assert isinstance(result, TimedeltaIndex)
exp = TimedeltaIndex(['2H', '1H', '0H', '1H', '2H'],
freq=None, name='x')
tm.assert_index_equal(result, exp)
assert result.freq is None
# -------------------------------------------------------------
# Binary operations TimedeltaIndex and integer
def test_tdi_add_int(self, one):
# Variants of `one` for #19012
rng = timedelta_range('1 days 09:00:00', freq='H', periods=10)
result = rng + one
expected = timedelta_range('1 days 10:00:00', freq='H', periods=10)
tm.assert_index_equal(result, expected)
def test_tdi_iadd_int(self, one):
rng = timedelta_range('1 days 09:00:00', freq='H', periods=10)
expected = timedelta_range('1 days 10:00:00', freq='H', periods=10)
rng += one
tm.assert_index_equal(rng, expected)
def test_tdi_sub_int(self, one):
rng = timedelta_range('1 days 09:00:00', freq='H', periods=10)
result = rng - one
expected = timedelta_range('1 days 08:00:00', freq='H', periods=10)
tm.assert_index_equal(result, expected)
def test_tdi_isub_int(self, one):
rng = timedelta_range('1 days 09:00:00', freq='H', periods=10)
expected = timedelta_range('1 days 08:00:00', freq='H', periods=10)
rng -= one
tm.assert_index_equal(rng, expected)
# -------------------------------------------------------------
# __add__/__sub__ with integer arrays
@pytest.mark.parametrize('box', [np.array, pd.Index])
def test_tdi_add_integer_array(self, box):
# GH#19959
rng = timedelta_range('1 days 09:00:00', freq='H', periods=3)
other = box([4, 3, 2])
expected = TimedeltaIndex(['1 day 13:00:00'] * 3)
result = rng + other
tm.assert_index_equal(result, expected)
result = other + rng
tm.assert_index_equal(result, expected)
@pytest.mark.parametrize('box', [np.array, pd.Index])
def test_tdi_sub_integer_array(self, box):
# GH#19959
rng = timedelta_range('9H', freq='H', periods=3)
other = box([4, 3, 2])
expected = TimedeltaIndex(['5H', '7H', '9H'])
result = rng - other
tm.assert_index_equal(result, expected)
result = other - rng
tm.assert_index_equal(result, -expected)
@pytest.mark.parametrize('box', [np.array, pd.Index])
def test_tdi_addsub_integer_array_no_freq(self, box):
# GH#19959
tdi = TimedeltaIndex(['1 Day', 'NaT', '3 Hours'])
other = box([14, -1, 16])
with pytest.raises(NullFrequencyError):
tdi + other
with pytest.raises(NullFrequencyError):
other + tdi
with pytest.raises(NullFrequencyError):
tdi - other
with pytest.raises(NullFrequencyError):
other - tdi
# -------------------------------------------------------------
# Binary operations TimedeltaIndex and timedelta-like
# Note: add and sub are tested in tests.test_arithmetic
def test_tdi_iadd_timedeltalike(self, delta):
# only test adding/sub offsets as + is now numeric
rng = timedelta_range('1 days', '10 days')
expected = timedelta_range('1 days 02:00:00', '10 days 02:00:00',
freq='D')
rng += delta
tm.assert_index_equal(rng, expected)
def test_tdi_isub_timedeltalike(self, delta):
# only test adding/sub offsets as - is now numeric
rng = timedelta_range('1 days', '10 days')
expected = timedelta_range('0 days 22:00:00', '9 days 22:00:00')
rng -= delta
tm.assert_index_equal(rng, expected)
# -------------------------------------------------------------
def test_subtraction_ops(self):
# with datetimes/timedelta and tdi/dti
tdi = TimedeltaIndex(['1 days', pd.NaT, '2 days'], name='foo')
dti = date_range('20130101', periods=3, name='bar')
td = Timedelta('1 days')
dt = Timestamp('20130101')
pytest.raises(TypeError, lambda: tdi - dt)
pytest.raises(TypeError, lambda: tdi - dti)
pytest.raises(TypeError, lambda: td - dt)
pytest.raises(TypeError, lambda: td - dti)
result = dt - dti
expected = TimedeltaIndex(['0 days', '-1 days', '-2 days'], name='bar')
tm.assert_index_equal(result, expected)
result = dti - dt
expected = TimedeltaIndex(['0 days', '1 days', '2 days'], name='bar')
tm.assert_index_equal(result, expected)
result = tdi - td
expected = TimedeltaIndex(['0 days', pd.NaT, '1 days'], name='foo')
tm.assert_index_equal(result, expected, check_names=False)
result = td - tdi
expected = TimedeltaIndex(['0 days', pd.NaT, '-1 days'], name='foo')
tm.assert_index_equal(result, expected, check_names=False)
result = dti - td
expected = DatetimeIndex(
['20121231', '20130101', '20130102'], name='bar')
tm.assert_index_equal(result, expected, check_names=False)
result = dt - tdi
expected = DatetimeIndex(['20121231', pd.NaT, '20121230'], name='foo')
tm.assert_index_equal(result, expected)
def test_subtraction_ops_with_tz(self):
# check that dt/dti subtraction ops with tz are validated
dti = date_range('20130101', periods=3)
ts = Timestamp('20130101')
dt = ts.to_pydatetime()
dti_tz = date_range('20130101', periods=3).tz_localize('US/Eastern')
ts_tz = Timestamp('20130101').tz_localize('US/Eastern')
ts_tz2 = Timestamp('20130101').tz_localize('CET')
dt_tz = ts_tz.to_pydatetime()
td = Timedelta('1 days')
def _check(result, expected):
assert result == expected
assert isinstance(result, Timedelta)
# scalars
result = ts - ts
expected = Timedelta('0 days')
_check(result, expected)
result = dt_tz - ts_tz
expected = Timedelta('0 days')
_check(result, expected)
result = ts_tz - dt_tz
expected = Timedelta('0 days')
_check(result, expected)
# tz mismatches
pytest.raises(TypeError, lambda: dt_tz - ts)
pytest.raises(TypeError, lambda: dt_tz - dt)
pytest.raises(TypeError, lambda: dt_tz - ts_tz2)
pytest.raises(TypeError, lambda: dt - dt_tz)
pytest.raises(TypeError, lambda: ts - dt_tz)
pytest.raises(TypeError, lambda: ts_tz2 - ts)
pytest.raises(TypeError, lambda: ts_tz2 - dt)
pytest.raises(TypeError, lambda: ts_tz - ts_tz2)
# with dti
pytest.raises(TypeError, lambda: dti - ts_tz)
pytest.raises(TypeError, lambda: dti_tz - ts)
pytest.raises(TypeError, lambda: dti_tz - ts_tz2)
result = dti_tz - dt_tz
expected = TimedeltaIndex(['0 days', '1 days', '2 days'])
tm.assert_index_equal(result, expected)
result = dt_tz - dti_tz
expected = TimedeltaIndex(['0 days', '-1 days', '-2 days'])
tm.assert_index_equal(result, expected)
result = dti_tz - ts_tz
expected = TimedeltaIndex(['0 days', '1 days', '2 days'])
tm.assert_index_equal(result, expected)
result = ts_tz - dti_tz
expected = TimedeltaIndex(['0 days', '-1 days', '-2 days'])
tm.assert_index_equal(result, expected)
result = td - td
expected = Timedelta('0 days')
_check(result, expected)
result = dti_tz - td
expected = DatetimeIndex(
['20121231', '20130101', '20130102'], tz='US/Eastern')
tm.assert_index_equal(result, expected)
def test_dti_tdi_numeric_ops(self):
# These are normally union/diff set-like ops
tdi = TimedeltaIndex(['1 days', pd.NaT, '2 days'], name='foo')
dti = date_range('20130101', periods=3, name='bar')
# TODO(wesm): unused?
# td = Timedelta('1 days')
# dt = Timestamp('20130101')
result = tdi - tdi
expected = TimedeltaIndex(['0 days', pd.NaT, '0 days'], name='foo')
tm.assert_index_equal(result, expected)
result = tdi + tdi
expected = TimedeltaIndex(['2 days', pd.NaT, '4 days'], name='foo')
tm.assert_index_equal(result, expected)
result = dti - tdi # name will be reset
expected = DatetimeIndex(['20121231', pd.NaT, '20130101'])
tm.assert_index_equal(result, expected)
def test_addition_ops(self):
# with datetimes/timedelta and tdi/dti
tdi = TimedeltaIndex(['1 days', pd.NaT, '2 days'], name='foo')
dti = date_range('20130101', periods=3, name='bar')
td = Timedelta('1 days')
dt = Timestamp('20130101')
result = tdi + dt
expected = DatetimeIndex(['20130102', pd.NaT, '20130103'], name='foo')
tm.assert_index_equal(result, expected)
result = dt + tdi
expected = DatetimeIndex(['20130102', pd.NaT, '20130103'], name='foo')
tm.assert_index_equal(result, expected)
result = td + tdi
expected = TimedeltaIndex(['2 days', pd.NaT, '3 days'], name='foo')
tm.assert_index_equal(result, expected)
result = tdi + td
expected = TimedeltaIndex(['2 days', pd.NaT, '3 days'], name='foo')
tm.assert_index_equal(result, expected)
# unequal length
pytest.raises(ValueError, lambda: tdi + dti[0:1])
pytest.raises(ValueError, lambda: tdi[0:1] + dti)
# random indexes
pytest.raises(NullFrequencyError, lambda: tdi + Int64Index([1, 2, 3]))
# this is a union!
# pytest.raises(TypeError, lambda : Int64Index([1,2,3]) + tdi)
result = tdi + dti # name will be reset
expected = DatetimeIndex(['20130102', pd.NaT, '20130105'])
tm.assert_index_equal(result, expected)
result = dti + tdi # name will be reset
expected = DatetimeIndex(['20130102', pd.NaT, '20130105'])
tm.assert_index_equal(result, expected)
result = dt + td
expected = Timestamp('20130102')
assert result == expected
result = td + dt
expected = Timestamp('20130102')
assert result == expected
def test_ops_ndarray(self):
td = Timedelta('1 day')
# timedelta, timedelta
other = pd.to_timedelta(['1 day']).values
expected = pd.to_timedelta(['2 days']).values
tm.assert_numpy_array_equal(td + other, expected)
if LooseVersion(np.__version__) >= LooseVersion('1.8'):
tm.assert_numpy_array_equal(other + td, expected)
pytest.raises(TypeError, lambda: td + np.array([1]))
pytest.raises(TypeError, lambda: np.array([1]) + td)
expected = pd.to_timedelta(['0 days']).values
tm.assert_numpy_array_equal(td - other, expected)
if LooseVersion(np.__version__) >= LooseVersion('1.8'):
tm.assert_numpy_array_equal(-other + td, expected)
pytest.raises(TypeError, lambda: td - np.array([1]))
pytest.raises(TypeError, lambda: np.array([1]) - td)
expected = pd.to_timedelta(['2 days']).values
tm.assert_numpy_array_equal(td * np.array([2]), expected)
tm.assert_numpy_array_equal(np.array([2]) * td, expected)
pytest.raises(TypeError, lambda: td * other)
pytest.raises(TypeError, lambda: other * td)
tm.assert_numpy_array_equal(td / other,
np.array([1], dtype=np.float64))
if LooseVersion(np.__version__) >= LooseVersion('1.8'):
tm.assert_numpy_array_equal(other / td,
np.array([1], dtype=np.float64))
# timedelta, datetime
other = pd.to_datetime(['2000-01-01']).values
expected = pd.to_datetime(['2000-01-02']).values
tm.assert_numpy_array_equal(td + other, expected)
if LooseVersion(np.__version__) >= LooseVersion('1.8'):
tm.assert_numpy_array_equal(other + td, expected)
expected = pd.to_datetime(['1999-12-31']).values
tm.assert_numpy_array_equal(-td + other, expected)
if LooseVersion(np.__version__) >= LooseVersion('1.8'):
tm.assert_numpy_array_equal(other - td, expected)
def test_timedelta_ops_with_missing_values(self):
# setup
s1 = pd.to_timedelta(Series(['00:00:01']))
s2 = pd.to_timedelta(Series(['00:00:02']))
sn = pd.to_timedelta(Series([pd.NaT]))
df1 = pd.DataFrame(['00:00:01']).apply(pd.to_timedelta)
df2 = pd.DataFrame(['00:00:02']).apply(pd.to_timedelta)
dfn = pd.DataFrame([pd.NaT]).apply(pd.to_timedelta)
scalar1 = pd.to_timedelta('00:00:01')
scalar2 = pd.to_timedelta('00:00:02')
timedelta_NaT = pd.to_timedelta('NaT')
actual = scalar1 + scalar1
assert actual == scalar2
actual = scalar2 - scalar1
assert actual == scalar1
actual = s1 + s1
tm.assert_series_equal(actual, s2)
actual = s2 - s1
tm.assert_series_equal(actual, s1)
actual = s1 + scalar1
tm.assert_series_equal(actual, s2)
actual = scalar1 + s1
tm.assert_series_equal(actual, s2)
actual = s2 - scalar1
tm.assert_series_equal(actual, s1)
actual = -scalar1 + s2
tm.assert_series_equal(actual, s1)
actual = s1 + timedelta_NaT
tm.assert_series_equal(actual, sn)
actual = timedelta_NaT + s1
tm.assert_series_equal(actual, sn)
actual = s1 - timedelta_NaT
tm.assert_series_equal(actual, sn)
actual = -timedelta_NaT + s1
tm.assert_series_equal(actual, sn)
with pytest.raises(TypeError):
s1 + np.nan
with pytest.raises(TypeError):
np.nan + s1
with pytest.raises(TypeError):
s1 - np.nan
with pytest.raises(TypeError):
-np.nan + s1
actual = s1 + pd.NaT
tm.assert_series_equal(actual, sn)
actual = s2 - pd.NaT
tm.assert_series_equal(actual, sn)
actual = s1 + df1
tm.assert_frame_equal(actual, df2)
actual = s2 - df1
tm.assert_frame_equal(actual, df1)
actual = df1 + s1
tm.assert_frame_equal(actual, df2)
actual = df2 - s1
tm.assert_frame_equal(actual, df1)
actual = df1 + df1
tm.assert_frame_equal(actual, df2)
actual = df2 - df1
tm.assert_frame_equal(actual, df1)
actual = df1 + scalar1
tm.assert_frame_equal(actual, df2)
actual = df2 - scalar1
tm.assert_frame_equal(actual, df1)
actual = df1 + timedelta_NaT
tm.assert_frame_equal(actual, dfn)
actual = df1 - timedelta_NaT
tm.assert_frame_equal(actual, dfn)
with pytest.raises(TypeError):
df1 + np.nan
with pytest.raises(TypeError):
df1 - np.nan
actual = df1 + pd.NaT # NaT is datetime, not timedelta
tm.assert_frame_equal(actual, dfn)
actual = df1 - pd.NaT
tm.assert_frame_equal(actual, dfn)
def test_tdi_ops_attributes(self):
rng = timedelta_range('2 days', periods=5, freq='2D', name='x')
result = rng + 1
exp = timedelta_range('4 days', periods=5, freq='2D', name='x')
tm.assert_index_equal(result, exp)
assert result.freq == '2D'
result = rng - 2
exp = timedelta_range('-2 days', periods=5, freq='2D', name='x')
tm.assert_index_equal(result, exp)
assert result.freq == '2D'
result = rng * 2
exp = timedelta_range('4 days', periods=5, freq='4D', name='x')
tm.assert_index_equal(result, exp)
assert result.freq == '4D'
result = rng / 2
exp = timedelta_range('1 days', periods=5, freq='D', name='x')
tm.assert_index_equal(result, exp)
assert result.freq == 'D'
result = -rng
exp = timedelta_range('-2 days', periods=5, freq='-2D', name='x')
tm.assert_index_equal(result, exp)
assert result.freq == '-2D'
rng = pd.timedelta_range('-2 days', periods=5, freq='D', name='x')
result = abs(rng)
exp = TimedeltaIndex(['2 days', '1 days', '0 days', '1 days',
'2 days'], name='x')
tm.assert_index_equal(result, exp)
assert result.freq is None
# TODO: Needs more informative name, probably split up into
# more targeted tests
def test_timedelta(self, freq):
index = date_range('1/1/2000', periods=50, freq=freq)
shifted = index + timedelta(1)
back = shifted + timedelta(-1)
| tm.assert_index_equal(index, back) | pandas.util.testing.assert_index_equal |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.