hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
sequence | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
sequence | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
sequence | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
sequence | cell_types
sequence | cell_type_groups
sequence |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
d0e0c81388f09e85c66891e10c5dcb0db369d375 | 680,528 | ipynb | Jupyter Notebook | pymc3/bayesian_neural_network_wiecki.ipynb | jackbenn/probabilistic-programming-intro | b68f141481052dd53ebd5a2675b053740d745239 | [
"BSD-3-Clause"
] | 9 | 2017-02-08T04:18:46.000Z | 2018-09-10T14:33:13.000Z | pymc3/bayesian_neural_network_wiecki.ipynb | jackbenn/probabilistic-programming-intro | b68f141481052dd53ebd5a2675b053740d745239 | [
"BSD-3-Clause"
] | null | null | null | pymc3/bayesian_neural_network_wiecki.ipynb | jackbenn/probabilistic-programming-intro | b68f141481052dd53ebd5a2675b053740d745239 | [
"BSD-3-Clause"
] | 26 | 2017-02-09T01:06:04.000Z | 2018-02-09T06:09:29.000Z | 909.796791 | 298,672 | 0.939839 | [
[
[
"## Neural Networks in PyMC3 estimated with Variational Inference\n(c) 2016 by Thomas Wiecki",
"_____no_output_____"
],
[
"## Current trends in Machine Learning\n\nThere are currently three big trends in machine learning: **Probabilistic Programming**, **Deep Learning** and \"**Big Data**\". Inside of PP, a lot of innovation is in making things scale using **Variational Inference**. In this blog post, I will show how to use **Variational Inference** in [PyMC3](http://pymc-devs.github.io/pymc3/) to fit a simple Bayesian Neural Network. I will also discuss how bridging Probabilistic Programming and Deep Learning can open up very interesting avenues to explore in future research.\n\n### Probabilistic Programming at scale\n**Probabilistic Programming** allows very flexible creation of custom probabilistic models and is mainly concerned with **insight** and learning from your data. The approach is inherently **Bayesian** so we can specify **priors** to inform and constrain our models and get uncertainty estimation in form of a **posterior** distribution. Using [MCMC sampling algorithms](http://twiecki.github.io/blog/2015/11/10/mcmc-sampling/) we can draw samples from this posterior to very flexibly estimate these models. [PyMC3](http://pymc-devs.github.io/pymc3/) and [Stan](http://mc-stan.org/) are the current state-of-the-art tools to consruct and estimate these models. One major drawback of sampling, however, is that it's often very slow, especially for high-dimensional models. That's why more recently, **variational inference** algorithms have been developed that are almost as flexible as MCMC but much faster. Instead of drawing samples from the posterior, these algorithms instead fit a distribution (e.g. normal) to the posterior turning a sampling problem into and optimization problem. [ADVI](http://arxiv.org/abs/1506.03431) -- Automatic Differentation Variational Inference -- is implemented in [PyMC3](http://pymc-devs.github.io/pymc3/) and [Stan](http://mc-stan.org/), as well as a new package called [Edward](https://github.com/blei-lab/edward/) which is mainly concerned with Variational Inference. \n\nUnfortunately, when it comes traditional ML problems like classification or (non-linear) regression, Probabilistic Programming often plays second fiddle (in terms of accuracy and scalability) to more algorithmic approaches like [ensemble learning](https://en.wikipedia.org/wiki/Ensemble_learning) (e.g. [random forests](https://en.wikipedia.org/wiki/Random_forest) or [gradient boosted regression trees](https://en.wikipedia.org/wiki/Boosting_(machine_learning)).\n\n### Deep Learning\n\nNow in its third renaissance, deep learning has been making headlines repeatadly by dominating almost any object recognition benchmark, [kicking ass at Atari games](https://www.cs.toronto.edu/~vmnih/docs/dqn.pdf), and [beating the world-champion Lee Sedol at Go](http://www.nature.com/nature/journal/v529/n7587/full/nature16961.html). From a statistical point, Neural Networks are extremely good non-linear function approximators and representation learners. While mostly known for classification, they have been extended to unsupervised learning with [AutoEncoders](https://arxiv.org/abs/1312.6114) and in all sorts of other interesting ways (e.g. [Recurrent Networks](https://en.wikipedia.org/wiki/Recurrent_neural_network), or [MDNs](http://cbonnett.github.io/MDN_EDWARD_KERAS_TF.html) to estimate multimodal distributions). Why do they work so well? No one really knows as the statistical properties are still not fully understood.\n\nA large part of the innoviation in deep learning is the ability to train these extremely complex models. This rests on several pillars:\n* Speed: facilitating the GPU allowed for much faster processing.\n* Software: frameworks like [Theano](http://deeplearning.net/software/theano/) and [TensorFlow](https://www.tensorflow.org/) allow flexible creation of abstract models that can then be optimized and compiled to CPU or GPU.\n* Learning algorithms: training on sub-sets of the data -- stochastic gradient descent -- allows us to train these models on massive amounts of data. Techniques like drop-out avoid overfitting.\n* Architectural: A lot of innovation comes from changing the input layers, like for convolutional neural nets, or the output layers, like for [MDNs](http://cbonnett.github.io/MDN_EDWARD_KERAS_TF.html).\n\n### Bridging Deep Learning and Probabilistic Programming\nOn one hand we Probabilistic Programming which allows us to build rather small and focused models in a very principled and well-understood way to gain insight into our data; on the other hand we have deep learning which uses many heuristics to train huge and highly complex models that are amazing at prediction. Recent innovations in variational inference allow probabilistic programming to scale model complexity as well as data size. We are thus at the cusp of being able to combine these two approaches to hopefully unlock new innovations in Machine Learning. For more motivation, see also [Dustin Tran's](https://twitter.com/dustinvtran) recent [blog post](http://dustintran.com/blog/a-quick-update-edward-and-some-motivations/).\n\nWhile this would allow Probabilistic Programming to be applied to a much wider set of interesting problems, I believe this bridging also holds great promise for innovations in Deep Learning. Some ideas are:\n* **Uncertainty in predictions**: As we will see below, the Bayesian Neural Network informs us about the uncertainty in its predictions. I think uncertainty is an underappreciated concept in Machine Learning as it's clearly important for real-world applications. But it could also be useful in training. For example, we could train the model specifically on samples it is most uncertain about.\n* **Uncertainty in representations**: We also get uncertainty estimates of our weights which could inform us about the stability of the learned representations of the network.\n* **Regularization with priors**: Weights are often L2-regularized to avoid overfitting, this very naturally becomes a Gaussian prior for the weight coefficients. We could, however, imagine all kinds of other priors, like spike-and-slab to enforce sparsity (this would be more like using the L1-norm).\n* **Transfer learning with informed priors**: If we wanted to train a network on a new object recognition data set, we could bootstrap the learning by placing informed priors centered around weights retrieved from other pre-trained networks, like [GoogLeNet](https://arxiv.org/abs/1409.4842). \n* **Hierarchical Neural Networks**: A very powerful approach in Probabilistic Programming is hierarchical modeling that allows pooling of things that were learned on sub-groups to the overall population (see my tutorial on [Hierarchical Linear Regression in PyMC3](http://twiecki.github.io/blog/2014/03/17/bayesian-glms-3/)). Applied to Neural Networks, in hierarchical data sets, we could train individual neural nets to specialize on sub-groups while still being informed about representations of the overall population. For example, imagine a network trained to classify car models from pictures of cars. We could train a hierarchical neural network where a sub-neural network is trained to tell apart models from only a single manufacturer. The intuition being that all cars from a certain manufactures share certain similarities so it would make sense to train individual networks that specialize on brands. However, due to the individual networks being connected at a higher layer, they would still share information with the other specialized sub-networks about features that are useful to all brands. Interestingly, different layers of the network could be informed by various levels of the hierarchy -- e.g. early layers that extract visual lines could be identical in all sub-networks while the higher-order representations would be different. The hierarchical model would learn all that from the data.\n* **Other hybrid architectures**: We can more freely build all kinds of neural networks. For example, Bayesian non-parametrics could be used to flexibly adjust the size and shape of the hidden layers to optimally scale the network architecture to the problem at hand during training. Currently, this requires costly hyper-parameter optimization and a lot of tribal knowledge.",
"_____no_output_____"
],
[
"## Bayesian Neural Networks in PyMC3",
"_____no_output_____"
],
[
"### Generating data\n\nFirst, lets generate some toy data -- a simple binary classification problem that's not linearly separable.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport pymc3 as pm\nimport theano.tensor as T\nimport theano\nimport sklearn\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nsns.set_style('white')\nfrom sklearn import datasets\nfrom sklearn.preprocessing import scale\nfrom sklearn.cross_validation import train_test_split\nfrom sklearn.datasets import make_moons",
"/usr/lib/python3/dist-packages/matplotlib/__init__.py:874: UserWarning: axes.color_cycle is deprecated and replaced with axes.prop_cycle; please use the latter.\n warnings.warn(self.msg_depr % (key, alt_key))\n"
],
[
"X, Y = make_moons(noise=0.2, random_state=0, n_samples=1000)\nX = scale(X)\nX_train, X_test, Y_train, Y_test = train_test_split(X, Y, test_size=.5)",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\nax.scatter(X[Y==0, 0], X[Y==0, 1], label='Class 0')\nax.scatter(X[Y==1, 0], X[Y==1, 1], color='r', label='Class 1')\nsns.despine(); ax.legend()\nax.set(xlabel='X', ylabel='Y', title='Toy binary classification data set');",
"_____no_output_____"
]
],
[
[
"### Model specification\n\nA neural network is quite simple. The basic unit is a [perceptron](https://en.wikipedia.org/wiki/Perceptron) which is nothing more than [logistic regression](http://pymc-devs.github.io/pymc3/notebooks/posterior_predictive.html#Prediction). We use many of these in parallel and then stack them up to get hidden layers. Here we will use 2 hidden layers with 5 neurons each which is sufficient for such a simple problem.",
"_____no_output_____"
]
],
[
[
"# Trick: Turn inputs and outputs into shared variables. \n# It's still the same thing, but we can later change the values of the shared variable \n# (to switch in the test-data later) and pymc3 will just use the new data. \n# Kind-of like a pointer we can redirect.\n# For more info, see: http://deeplearning.net/software/theano/library/compile/shared.html\nann_input = theano.shared(X_train)\nann_output = theano.shared(Y_train)\n\nn_hidden = 5\n\n# Initialize random weights between each layer\ninit_1 = np.random.randn(X.shape[1], n_hidden)\ninit_2 = np.random.randn(n_hidden, n_hidden)\ninit_out = np.random.randn(n_hidden)\n \nwith pm.Model() as neural_network:\n # Weights from input to hidden layer\n weights_in_1 = pm.Normal('w_in_1', 0, sd=1, \n shape=(X.shape[1], n_hidden), \n testval=init_1)\n \n # Weights from 1st to 2nd layer\n weights_1_2 = pm.Normal('w_1_2', 0, sd=1, \n shape=(n_hidden, n_hidden), \n testval=init_2)\n \n # Weights from hidden layer to output\n weights_2_out = pm.Normal('w_2_out', 0, sd=1, \n shape=(n_hidden,), \n testval=init_out)\n \n # Build neural-network using tanh activation function\n act_1 = T.tanh(T.dot(ann_input, \n weights_in_1))\n act_2 = T.tanh(T.dot(act_1, \n weights_1_2))\n act_out = T.nnet.sigmoid(T.dot(act_2, \n weights_2_out))\n \n # Binary classification -> Bernoulli likelihood\n out = pm.Bernoulli('out', \n act_out,\n observed=ann_output)",
"_____no_output_____"
]
],
[
[
"That's not so bad. The `Normal` priors help regularize the weights. Usually we would add a constant `b` to the inputs but I omitted it here to keep the code cleaner.",
"_____no_output_____"
],
[
"### Variational Inference: Scaling model complexity\n\nWe could now just run a MCMC sampler like [`NUTS`](http://pymc-devs.github.io/pymc3/api.html#nuts) which works pretty well in this case but as I already mentioned, this will become very slow as we scale our model up to deeper architectures with more layers.\n\nInstead, we will use the brand-new [ADVI](http://pymc-devs.github.io/pymc3/api.html#advi) variational inference algorithm which was recently added to `PyMC3`. This is much faster and will scale better. Note, that this is a mean-field approximation so we ignore correlations in the posterior.",
"_____no_output_____"
]
],
[
[
"%%time\n\nwith neural_network:\n # Run ADVI which returns posterior means, standard deviations, and the evidence lower bound (ELBO)\n v_params = pm.variational.advi(n=50000)",
"Average ELBO = -167.9: 100%|██████████| 50000/50000 [00:17<00:00, 2860.61it/s]\nFinished [100%]: Average ELBO = -138.57\n"
]
],
[
[
"< 40 seconds on my older laptop. That's pretty good considering that NUTS is having a really hard time. Further below we make this even faster. To make it really fly, we probably want to run the Neural Network on the GPU.\n\nAs samples are more convenient to work with, we can very quickly draw samples from the variational posterior using `sample_vp()` (this is just sampling from Normal distributions, so not at all the same like MCMC):",
"_____no_output_____"
]
],
[
[
"with neural_network:\n trace = pm.variational.sample_vp(v_params, draws=5000)",
"100%|██████████| 5000/5000 [00:00<00:00, 12175.62it/s]\n"
]
],
[
[
"Plotting the objective function (ELBO) we can see that the optimization slowly improves the fit over time.",
"_____no_output_____"
]
],
[
[
"plt.plot(v_params.elbo_vals)\nplt.ylabel('ELBO')\nplt.xlabel('iteration')",
"_____no_output_____"
]
],
[
[
"Now that we trained our model, lets predict on the hold-out set using a posterior predictive check (PPC). We use [`sample_ppc()`](http://pymc-devs.github.io/pymc3/api.html#pymc3.sampling.sample_ppc) to generate new data (in this case class predictions) from the posterior (sampled from the variational estimation).",
"_____no_output_____"
]
],
[
[
"# Replace shared variables with testing set\nann_input.set_value(X_test)\nann_output.set_value(Y_test)\n\n# Creater posterior predictive samples\nppc = pm.sample_ppc(trace, model=neural_network, samples=500)\n\n# Use probability of > 0.5 to assume prediction of class 1\npred = ppc['out'].mean(axis=0) > 0.5",
"100%|██████████| 500/500 [00:04<00:00, 116.76it/s]\n"
],
[
"fig, ax = plt.subplots()\nax.scatter(X_test[pred==0, 0], X_test[pred==0, 1])\nax.scatter(X_test[pred==1, 0], X_test[pred==1, 1], color='r')\nsns.despine()\nax.set(title='Predicted labels in testing set', xlabel='X', ylabel='Y');\nplt.savefig(\"nn-0.png\",dpi=400)",
"_____no_output_____"
],
[
"print('Accuracy = {}%'.format((Y_test == pred).mean() * 100))",
"Accuracy = 94.6%\n"
]
],
[
[
"Hey, our neural network did all right!",
"_____no_output_____"
],
[
"## Lets look at what the classifier has learned\n\nFor this, we evaluate the class probability predictions on a grid over the whole input space.",
"_____no_output_____"
]
],
[
[
"grid = np.mgrid[-3:3:100j,-3:3:100j]\ngrid_2d = grid.reshape(2, -1).T\nX, Y = grid\ndummy_out = np.ones(grid.shape[1], dtype=np.int8)",
"_____no_output_____"
],
[
"ann_input.set_value(grid_2d)\nann_output.set_value(dummy_out)\n\n# Creater posterior predictive samples\nppc = pm.sample_ppc(trace, model=neural_network, samples=500)",
"100%|██████████| 500/500 [00:04<00:00, 113.79it/s]\n"
]
],
[
[
"### Probability surface",
"_____no_output_____"
]
],
[
[
"cmap = sns.diverging_palette(250, 12, s=85, l=25, as_cmap=True)\nfig, ax = plt.subplots(figsize=(10, 6))\ncontour = ax.contourf(X, Y, ppc['out'].mean(axis=0).reshape(100, 100), cmap=cmap)\nax.scatter(X_test[pred==0, 0], X_test[pred==0, 1])\nax.scatter(X_test[pred==1, 0], X_test[pred==1, 1], color='r')\ncbar = plt.colorbar(contour, ax=ax)\n_ = ax.set(xlim=(-3, 3), ylim=(-3, 3), xlabel='X', ylabel='Y');\ncbar.ax.set_ylabel('Posterior predictive mean probability of class label = 0');\nplt.savefig(\"nn-1.png\",dpi=400)",
"_____no_output_____"
]
],
[
[
"### Uncertainty in predicted value\n\nSo far, everything I showed we could have done with a non-Bayesian Neural Network. The mean of the posterior predictive for each class-label should be identical to maximum likelihood predicted values. However, we can also look at the standard deviation of the posterior predictive to get a sense for the uncertainty in our predictions. Here is what that looks like:",
"_____no_output_____"
]
],
[
[
"cmap = sns.cubehelix_palette(light=1, as_cmap=True)\nfig, ax = plt.subplots(figsize=(10, 6))\ncontour = ax.contourf(X, Y, ppc['out'].std(axis=0).reshape(100, 100), cmap=cmap)\nax.scatter(X_test[pred==0, 0], X_test[pred==0, 1])\nax.scatter(X_test[pred==1, 0], X_test[pred==1, 1], color='r')\ncbar = plt.colorbar(contour, ax=ax)\n_ = ax.set(xlim=(-3, 3), ylim=(-3, 3), xlabel='X', ylabel='Y');\ncbar.ax.set_ylabel('Uncertainty (posterior predictive standard deviation)');\nplt.savefig(\"nn-2.png\",dpi=400)",
"_____no_output_____"
]
],
[
[
"We can see that very close to the decision boundary, our uncertainty as to which label to predict is highest. You can imagine that associating predictions with uncertainty is a critical property for many applications like health care. To further maximize accuracy, we might want to train the model primarily on samples from that high-uncertainty region.",
"_____no_output_____"
],
[
"## Mini-batch ADVI: Scaling data size\n\nSo far, we have trained our model on all data at once. Obviously this won't scale to something like ImageNet. Moreover, training on mini-batches of data (stochastic gradient descent) avoids local minima and can lead to faster convergence.\n\nFortunately, ADVI can be run on mini-batches as well. It just requires some setting up:",
"_____no_output_____"
]
],
[
[
"from six.moves import zip\n\n# Set back to original data to retrain\nann_input.set_value(X_train)\nann_output.set_value(Y_train)\n\n# Tensors and RV that will be using mini-batches\nminibatch_tensors = [ann_input, ann_output]\nminibatch_RVs = [out]\n\n# Generator that returns mini-batches in each iteration\ndef create_minibatch(data):\n rng = np.random.RandomState(0)\n \n while True:\n # Return random data samples of set size 100 each iteration\n ixs = rng.randint(len(data), size=50)\n yield data[ixs]\n\nminibatches = zip(\n create_minibatch(X_train), \n create_minibatch(Y_train),\n)\n\ntotal_size = len(Y_train)",
"_____no_output_____"
]
],
[
[
"While the above might look a bit daunting, I really like the design. Especially the fact that you define a generator allows for great flexibility. In principle, we could just pool from a database there and not have to keep all the data in RAM. \n\nLets pass those to `advi_minibatch()`:",
"_____no_output_____"
]
],
[
[
"%%time\n\nwith neural_network:\n # Run advi_minibatch\n v_params = pm.variational.advi_minibatch(\n n=50000, minibatch_tensors=minibatch_tensors, \n minibatch_RVs=minibatch_RVs, minibatches=minibatches, \n total_size=total_size, learning_rate=1e-2, epsilon=1.0\n )",
"Average ELBO = -111.71: 100%|██████████| 50000/50000 [00:14<00:00, 3441.73it/s]\nFinished minibatch ADVI: ELBO = -137.60\n"
],
[
"with neural_network: \n trace = pm.variational.sample_vp(v_params, draws=5000)",
"100%|██████████| 5000/5000 [00:00<00:00, 15032.53it/s]\n"
],
[
"plt.plot(v_params.elbo_vals)\nplt.ylabel('ELBO')\nplt.xlabel('iteration')\nsns.despine()",
"_____no_output_____"
]
],
[
[
"As you can see, mini-batch ADVI's running time is much lower. It also seems to converge faster.\n\nFor fun, we can also look at the trace. The point is that we also get uncertainty of our Neural Network weights.",
"_____no_output_____"
]
],
[
[
"pm.traceplot(trace);",
"_____no_output_____"
]
],
[
[
"## Summary\n\nHopefully this blog post demonstrated a very powerful new inference algorithm available in [PyMC3](http://pymc-devs.github.io/pymc3/): [ADVI](http://pymc-devs.github.io/pymc3/api.html#advi). I also think bridging the gap between Probabilistic Programming and Deep Learning can open up many new avenues for innovation in this space, as discussed above. Specifically, a hierarchical neural network sounds pretty bad-ass. These are really exciting times.\n\n## Next steps\n\n[`Theano`](http://deeplearning.net/software/theano/), which is used by `PyMC3` as its computational backend, was mainly developed for estimating neural networks and there are great libraries like [`Lasagne`](https://github.com/Lasagne/Lasagne) that build on top of `Theano` to make construction of the most common neural network architectures easy. Ideally, we wouldn't have to build the models by hand as I did above, but use the convenient syntax of `Lasagne` to construct the architecture, define our priors, and run ADVI. \n\nWhile we haven't successfully run `PyMC3` on the GPU yet, it should be fairly straight forward (this is what `Theano` does after all) and further reduce the running time significantly. If you know some `Theano`, this would be a great area for contributions!\n\nYou might also argue that the above network isn't really deep, but note that we could easily extend it to have more layers, including convolutional ones to train on more challenging data sets.\n\nI also presented some of this work at PyData London, view the video below:\n<iframe width=\"560\" height=\"315\" src=\"https://www.youtube.com/embed/LlzVlqVzeD8\" frameborder=\"0\" allowfullscreen></iframe>\n\nFinally, you can download this NB [here](https://github.com/twiecki/WhileMyMCMCGentlySamples/blob/master/content/downloads/notebooks/bayesian_neural_network.ipynb). Leave a comment below, and [follow me on twitter](https://twitter.com/twiecki).\n\n## Acknowledgements\n\n[Taku Yoshioka](https://github.com/taku-y) did a lot of work on ADVI in PyMC3, including the mini-batch implementation as well as the sampling from the variational posterior. I'd also like to the thank the Stan guys (specifically Alp Kucukelbir and Daniel Lee) for deriving ADVI and teaching us about it. Thanks also to Chris Fonnesbeck, Andrew Campbell, Taku Yoshioka, and Peadar Coyle for useful comments on an earlier draft.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0e0ced362875d75ecb0897d2cae86fdb7090554 | 72,910 | ipynb | Jupyter Notebook | numpy-random.ipynb | mattshiel/emerging-technologies | 1c5fd119676ab9471deeb094904c3bac98cbec38 | [
"MIT"
] | null | null | null | numpy-random.ipynb | mattshiel/emerging-technologies | 1c5fd119676ab9471deeb094904c3bac98cbec38 | [
"MIT"
] | 5 | 2018-10-08T16:07:08.000Z | 2018-10-25T15:41:25.000Z | numpy-random.ipynb | mattshiel/emerging-technologies | 1c5fd119676ab9471deeb094904c3bac98cbec38 | [
"MIT"
] | null | null | null | 59.372964 | 13,996 | 0.771088 | [
[
[
"This notebook is broken into two sections, firstly an introduction to the NumPy package and secondly a breakdown of the NumPy random package.\n\n\n# The NumPy Package\n\n## What is it?\n\nNumPy is a Python package used for scientific computing. Often cited as a fundamental package in this area, this is for good reason. It provides a high performance multidimensional array object, and a variety of routines for operations on arrays. NumPy is not part of a basic Python installation. It has to be installed after the Python installation.\n\nThe implemented multi-dimensional arrays are very efficient as NumPy was mostly written in C; the precompiled mathematical and numerical functions and functionalities of NumPy guarantee excellent execution speed.\n\nAt its core NumPy provides the ndarray (multidimensional/n-dimensional array) object. This is a grid of values of the same type indexed by a tuple of non-negative integers. This is explained in further detail in the basics section below.\n\n## The Basics\n\nIn Numpy, axes are called dimensions, lets take a hypothetical set of coordinates and see what exactly this means.\n\nThe coordinates of a point in 3D space [1, 4, 5] has one axis that contains 3 elements ie. has a length of 3.\nLet's take another example to reinforce this idea.\n\nUsing the illustrated graph below we can see that the set of coordinates has 2 axes. Axis 1 has a length (shown in colour) of 2, and axis 2 has a length of 3.\n\n<mark>FIGURE 1</mark>\n\n\n\nOne of NumPy's most powerful features is it's array class: ```ndarray``` This is not to be confused with the Python's array class ```array.array``` which has less functionality and only handles one-dimensional arrays.\n\n##### Let's use some of ndarray's functions to prove and reinforce what we learnt above.\n\n#### A Simple Example\n\nBefore we use NumPy we have to import it.\nIt is very common to see NumPy renamed to \"np\".\n\n",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
]
],
[
[
"From here let's create an array of values that represent distances eg. in metres",
"_____no_output_____"
]
],
[
[
"mvalues = [45.26, 10.9, 26.3, 80.5, 24.1, 66.1, 19.8, 3.0, 8.8, 132.5]",
"_____no_output_____"
]
],
[
[
"Great, now let's do some NumPy stuff on that. We're going to turn our array of values into a one-dimensional NumPy array and print it to the screen.",
"_____no_output_____"
]
],
[
[
"M = np.array(mvalues)\nprint(M)",
"[ 45.26 10.9 26.3 80.5 24.1 66.1 19.8 3. 8.8 132.5 ]\n"
]
],
[
[
"Now let's say that we wanted to convert all of our values in metres to centimetres. This is easily achieved using a NumPy array and scalar multiplication.",
"_____no_output_____"
]
],
[
[
"print(M*100)",
"[ 4526. 1090. 2630. 8050. 2410. 6610. 1980. 300. 880. 13250.]\n"
]
],
[
[
"If we print out M again you can see that the values have not been changed.",
"_____no_output_____"
]
],
[
[
"print(M)",
"[ 45.26 10.9 26.3 80.5 24.1 66.1 19.8 3. 8.8 132.5 ]\n"
]
],
[
[
"Now if you wanted to do the same thing using standard Python, as shown below, then hopefully you can clearly see the advantage in using NumPy instead.",
"_____no_output_____"
]
],
[
[
"mvalues = [ i*100 for i in mvalues] \nprint(mvalues)",
"[4526.0, 1090.0, 2630.0, 8050.0, 2410.0, 6609.999999999999, 1980.0, 300.0, 880.0000000000001, 13250.0]\n"
]
],
[
[
"The values for mvalues have also all been permanently changed now, whereas we didn't need to alter them when using NumPy ",
"_____no_output_____"
],
[
"Earlier I explained that NumPy provides the ndarray object. \"M\" is an instance of the class ```numpy.ndarray```, proven below:",
"_____no_output_____"
]
],
[
[
"type(M)",
"_____no_output_____"
]
],
[
[
"# The NumPy Random Package",
"_____no_output_____"
],
[
"The NumPy random package is used for sampling and generating random data. Across this section I've split the descriptions and examples into 4 sections:\n\n Simple Random Data\n Permutations\n Distributions\n Random Generator\n\nIn order to use the random package the name of package must be specified, followed by the function. \neg. ```np.random.rand(3,3)``` (It's implied that NumPy has been imported as np)\n\nAs a precursor, you will notation that looks like this **[num, num]** across this notebook.\nThis is known as interval notation. This brief explanation from Wikipedia nicely describes the notations found in this notebook.\n\n \"In mathematics, a (real) interval is a set of real numbers with the property that any number that lies \n between two numbers in the set is also included in the set.\"\n\n \"For example, (0,1) means greater than 0 and less than 1. A closed interval is an interval which includes all \n its limit points, and is denoted with square brackets. For example, [0,1] means greater than or equal to 0 and \n less than or equal to 1. A half-open interval includes only one of its endpoints, and is denoted by mixing the \n notations for open and closed intervals. (0,1] means greater than 0 and less than or equal to 1, while [0,1) \n means greater than or equal to 0 and less than 1.\"\n",
"_____no_output_____"
],
[
"## Simple Random Data\nLet's go over some of the functions that NumPy provides to help us deal with simple random data. \n\n### rand\n`numpy.random.rand` \n\nUsed to generate random values in a given shape. The dimensions of the input should be positive and if they are not or they're empty a float will be returned. The example below creates an array of specified shape and populates it with random samples from a uniform distribution over [0, 1).\n\nLet's use that example and construct a 2D array of random values. If you wanted a 3D array or greater you would simply add an additional parameter to the function.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nnp.random.rand(3,3)",
"_____no_output_____"
]
],
[
[
"### randn",
"_____no_output_____"
],
[
"`numpy.random.randn` \n\nReturns a sample (or samples) from the “standard normal” distribution.\n\nA standard normal distribution is a normal distribution where the average value is 0, and the variance is 1. \n\nWhen the function is provided a positive \"int_like or int-convertible arguments\", randn generates an array of specified shape (d0, d1, ..., dn), filled with random floats sampled from this distribution. Like ```rand```, a single float is returned if no argument is provided.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nnp.random.randn(3,2)",
"_____no_output_____"
]
],
[
[
"Additional computations can be added for greater specificity",
"_____no_output_____"
]
],
[
[
"np.random.randn(3,2) + 8",
"_____no_output_____"
]
],
[
[
"### randint\n\n",
"_____no_output_____"
],
[
"`numpy.random.randint` \n\nReturns random integers from low (inclusive) to high (exclusive).\n\nthe parameters for ```randint``` are **(low, high=None, size=None, dtype='l')**\n\n* **low and high** simply refer to the lowest and highest numbers that can be drawn from the distribution,\n\n* **size** refers to the output shape,\n\n* **dtype** is optional but if you have a desired output type you may specifiy it here.\n\nThe example below generates a 4 x 4 array of ints between 0 and 4, inclusive:",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nnp.random.randint(5, size=(4, 4))",
"_____no_output_____"
]
],
[
[
"`randint` is often used for more simple operations eg. generate a random number between 1-10",
"_____no_output_____"
]
],
[
[
"np.random.randint(1,10)",
"_____no_output_____"
]
],
[
[
"### random_integers",
"_____no_output_____"
],
[
"`numpy.random.random_integers` \n\nReturns random integers of type np.int from the “discrete uniform” distribution in the closed interval [low, high].\n\n`random_integers` is similar to ```randint```, only for the closed interval [low, high]. \n\n0 is the lowest value if high is omitted when using `random_integers`, whereas it is 1 in the case that high is omitted when using `randint`. This function however has been deprecated so it is advised you use ```randint``` instead ie. `np.random.random_integers(10) --> randint(1, 10 + 1)`\n\n### random_sample",
"_____no_output_____"
],
[
"`numpy.random.random_sample` \n\nReturns random floats in the half-open interval [0.0, 1.0). \nThe inputs may be an int or a tuple of ints denoting the shape of the array.\n\nThis example shows how to draw a random sample from 0-5. ",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nx = 5 * np.random.random_sample((250, 250)) + 0\nx",
"_____no_output_____"
]
],
[
[
"Below is a plot of the interval above which shows the continuous uniform distribution produced by `random_sample`, in the form of a histogram",
"_____no_output_____"
]
],
[
[
"# Import matplot in order to generate a histogram\nimport matplotlib.pyplot as plt\n\n# Define the amount of bins (series of intervals) for the histogram\nnum_bins = [0,1,2,3,4,5]\n\n# Plot and display the histogram\nn, bins, patches = plt.hist(x, num_bins, facecolor = 'green', alpha = 0.8)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### random, ranf and sample",
"_____no_output_____"
],
[
"An interesting point to note is the homogeniety between the `random_sample`, `random`, `ranf` and `sample` functions. As noted in the official NumPy documentation, all of these functions,\n\n \"Return random floats in the half-open interval [0.0, 1.0).\"\n\nTo prove that the functions are the same and can be used interchangeably we can use the `is` keyword to compare object identity ie. \"Do I have two names for the same object?\"",
"_____no_output_____"
]
],
[
[
"np.random.random_sample is np.random.random",
"_____no_output_____"
],
[
"np.random.random_sample is np.random.ranf",
"_____no_output_____"
],
[
"np.random.random_sample is np.random.sample",
"_____no_output_____"
]
],
[
[
"### choice\n\n`numpy.random.choice` \n\nGenerates a random sample from a given 1-D array. If you use a ndarray as the input then a random number will be selected from the elements provided. This may be useful for situations where you have a large pool of numbers to choose from and need a random selection eg.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nj = [2,4,9,125,3,19,8,62,1004,203]\n\n# Convert array to a NumPy array\no = np.array(j)\n\n# Output a random choice\nnp.random.choice(o)",
"_____no_output_____"
]
],
[
[
"The sample assumes a uniform distribution over all entries, however, if we wanted to change this and account for different probabilities we can use a parameter to set the probability for each individual entry. Remember that probability must add up to 1.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\n# Apply probabilities for each individual entry \nnp.random.choice(o, p = [0.1, 0.025, 0.025, 0.025, 0.025, 0.1, 0.1, 0.3, 0.2, 0.1])",
"_____no_output_____"
]
],
[
[
"### bytes",
"_____no_output_____"
],
[
"`numpy.random.bytes` \n\nThis a useful function that simply return a string of bytes of given length eg.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\n# Output a string of 15 random bytes \nnp.random.bytes(15)",
"_____no_output_____"
]
],
[
[
"## Permutations",
"_____no_output_____"
],
[
"This definition from Wikipedia explains nicely what a permutation is:\n \n In mathematics, the notion of permutation relates to the act of arranging all the members of a set into some \n sequence or order, or if the set is already ordered, rearranging its elements, a process called permuting.\n \n The number of permutations on a set of n elements is given by n! (factorial)\n \n For example:\n \n 3! = 3x2x1 = 6\n the six possible rearrangements would be (1x2x3), (2x3x1), (3x1x2), (1x3x2), (2x1x3), (3x2x1)\n \n\n\n### shuffle \n`numpy.random.shuffle` \n\n`shuffle` as the name implies, shuffles an array or list. This function only shuffles the array along the first axis of a multi-dimensional array. The order of sub-arrays is changed but their contents remains the same.\n\nShown below is an example of creating a 1D array, reshaping it and shuffling the contents.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\n# Create an array of values between 0-15, reshape to 4x4 array\narr = np.arange(16).reshape((4,4))\n\n# Shuffle the array\nnp.random.shuffle(arr)\narr\n",
"_____no_output_____"
]
],
[
[
"But what if I want to shuffle a 2D array? If this is the case then you can use the `ravel` function to flatten a multi-dimensional array like so:",
"_____no_output_____"
]
],
[
[
"# Create the NumPy array\ne = np.array([[20,30,40],[50,60,70],[80,90,100]])\ne",
"_____no_output_____"
],
[
"# Flatten the array, shuffle and output result\nnp.random.shuffle(e.ravel())\ne",
"_____no_output_____"
]
],
[
[
"### permutation\n`numpy.random.permutation` \n\nRandomly permutes a sequence or returns a permuted range of values. In the case the the value is an array, `permutation` will copy and shuffle the elements randomly. If x is an integer, np.arange(x) is called and randomly permuted.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\n# Return a random permutation from an array of values in the range of 0-5 \n# (np.arange uses the half-open interval of [0,1))\nnp.random.permutation(6)",
"_____no_output_____"
],
[
"# Return a shuffled copy of the permutation of the array\nnp.random.permutation([1, 4, 9, 12, 15])",
"_____no_output_____"
],
[
"# Permute \narr = np.arange(4).reshape((2, 2))\nnp.random.permutation(arr)",
"_____no_output_____"
]
],
[
[
"## Distributions\n\nThe distribution of a statistical data set (or a population) is a listing or function showing all the possible values (or intervals) of the data and how often they occur. When a distribution of categorical data is organized, you see the number or percentage of individuals in each group. When a distribution of numerical data is organized, they’re often ordered from smallest to largest, broken into reasonably sized groups (if appropriate), and then put into graphs and charts to examine the shape, center, and amount of variability in the data.\n\nThe world of statistics includes dozens of different distributions for categorical and numerical data; the most common ones have their own names. One of the most well-known distributions is called the normal distribution, also known as the bell-shaped curve. The normal distribution is based on numerical data that is continuous; its possible values lie on the entire real number line. Its overall shape, when the data are organized in graph form, is a symmetric bell-shape. In other words, most (around 68%) of the data are centered around the mean (giving you the middle part of the bell), and as you move farther out on either side of the mean, you find fewer and fewer values (representing the downward sloping sides on either side of the bell).\n\nDue to symmetry, the mean and the median lie at the same point, directly in the center of the normal distribution. The standard deviation is measured by the distance from the mean to the inflection point (where the curvature of the bell changes from concave up to concave down). \n\n### Normal Distribution\n\n`np.random.normal`\n\nThe probability density function of the normal distribution, is often called the bell curve because of its characteristic shape. \n\nWhat this example does is it creates the data for a normal distribution plot centered around a mean of 100, showing 3 standard deviations of the data, and creates 30,000 random data points that are of normal distribution. Remember this is the normal() function, which means that the data will be normalized. 68% of the data values will generated will be (or very close to) ±1 standard deviations of the mean. 95% of the data values generated will be within ±2 standard deviations of the mean. 99.7% of the data values generated will be within ±3 standard deviations of the mean. ",
"_____no_output_____"
]
],
[
[
"# Import required packages\nimport matplotlib.pyplot as plt\nimport numpy as np\n\n# Set the mean and standard deviation\nmu, sigma = 100, 3\n\n# \ns = np.random.normal(mu, sigma, 30000)\n\n# \ncount, bins, ignored = plt.hist(s, 25, density=True, color='blue')\n\n# Plot a line that shows the probability density function for the normal distribution\nplt.plot(bins, 1/(sigma * np.sqrt(2 * np.pi)) *\n np.exp( - (bins - mu)**2 / (2 * sigma**2) ),\n linewidth=2, color='r')\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"## The Binomial Distribution\n\n`np.random.binomial`\n\nIt can be used to obtain the number of successes from N Bernoulli trials.\n\n Samples are drawn from a binomial distribution with specified parameters, n trials and p probability of \n success where n an integer >= 0 and p is in the interval [0,1]. (n may be input as a float, but it is \n truncated to an integer in use)\n \nHere we're going to use Seaborn, a Python data visualization library based on matplotlib, to graph the distribution.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n# Import the Seaborn package for graphing\nimport seaborn as sb\n\n# Number of trials, probability of each trial\nn, p = 100, .1\n\n# Generate binomial distribution\ns = np.random.binomial(n, p, 30000)\n\n# Plot the graph with Seaborn\nax = sb.distplot(s,\n kde=False,\n color='blue',\n hist_kws={\"linewidth\": 30,'alpha':0.8}) \nax.set(xlabel='Binomial', ylabel='Frequency')\n\n\n\np = 4/15 = 27%. 0.27*15 = 4,",
"/anaconda3/lib/python3.6/site-packages/matplotlib/axes/_axes.py:6462: UserWarning: The 'normed' kwarg is deprecated, and has been replaced by the 'density' kwarg.\n warnings.warn(\"The 'normed' kwarg is deprecated, and has been \"\n"
]
],
[
[
"What I've shown here is that a binomial distribution is very different from a normal distribution, and yet if the sample size is large enough, the shapes will be quite similar as can be seen here. I used 30,000 as the size of both distributions and both produced a bell shaped curve.\n\nThe main difference between the two is that the binomial distribution is discrete, whereas the normal distribution is continuous. Continuous simply means that (in theory) it's possible to find a data value between any two data values. Things like the heights of people vary continuously, there are lots of random influences, generally their overall distribution is close to normal.\n\nA discrete probability distribution on the other hand is one where the possible outcomes are distinct and non-overlapping. eg. rolling a dice, you can roll a 5 or a 6 but not 5.5. As a result the graphed distribution looks \"stepped\".\n",
"_____no_output_____"
],
[
"## Pareto Distribution\n\n`numpy.random.pareto`\n\nThis draw samples from a Pareto II or Lomax distribution with specified shape.\n\nThe original Pareto distribution, is a power-law probability distribution that was originally applied to describing the distribution of wealth in a society, showing the trend that a large portion of the wealth is held by a small percentage of the population and that this is observable across a number of natural phenomena ie. that a few items account for a lot of subject and a lot of items account for a little of it.\n\n\nThe Lomax or Pareto II distribution is a shifted Pareto distribution. We can obtain the original Pareto distribution from the Lomax distribution by adding 1 and multiplying by the scale parameter 2.0",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\n# shape and mode\na, m = 3., 2. \n\n# Calculate original pareto\ns = (np.random.pareto(a, 1000) + 1) * m\n\ncount, bins, _ = plt.hist(s, 100, density=True, color=\"blue\")\nfit = a*m**a / bins**(a+1)\nplt.plot(bins, max(count)*fit/max(fit), linewidth=2, color='red')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Random Generators\n\n### RandomState\n\n`numpy.random.RandomState` \n\nThe container for the Mersenne Twister pseudo-random number generator. \n\nThis class is what is primarily used in NumPy to generate random numbers. If no seed is provided, the generator will try to use the system clock as a seed.\n\n### seed\n`numpy.random.seed`\n\nWe can use `seed` to seed the RandomState generator. The only requirement is that it must be convertible to 32 bit unsigned integers.\n\nOne of the main uses a seed is when testing a program you've written and you want to make sure you generate the same pseudo-random numbers. This works as a seed is actually the starting point in the sequence. If we use same seed every time, it will yield same sequence of random numbers.\n\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\n# Set seed\nnp.random.seed(25)\n\n# Generate random numbers\na = np.random.rand(5)\n\n# Change seed\nnp.random.seed(55)\n\n# Generate random numbers again\nb = np.random.rand(5)\n\n# Output results\nprint(\"a: \", a)\nprint(\"b: \", b)",
"a: [0.87012414 0.58227693 0.27883894 0.18591123 0.41110013]\nb: [0.09310829 0.97165592 0.48385998 0.2425227 0.53112383]\n"
]
],
[
[
"## References:\n\n\nhttps://docs.scipy.org/doc/numpy-1.15.1/reference/routines.random.html\n\nhttps://www.quora.com/What-is-seed-in-random-number-generation\n\nhttps://www.dummies.com/education/math/statistics/what-the-distribution-tells-you-about-a-statistical-data-set/\n\nhttps://magoosh.com/statistics/understanding-binomial-distribution/\n\nhttp://wiki.stat.ucla.edu/socr/index.php/AP_Statistics_Curriculum_2007_Pareto\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0e0d9c95c47a7a4a7138a8332b4e2c245ac83a6 | 160,288 | ipynb | Jupyter Notebook | search/services/search-enrichment/jupyter-notebooks/ActivitiesUsersAnalysis.ipynb | bcc-code/bcc-search | b2e0bbb437f88bf2c704597f9157492d5f98446e | [
"MIT"
] | null | null | null | search/services/search-enrichment/jupyter-notebooks/ActivitiesUsersAnalysis.ipynb | bcc-code/bcc-search | b2e0bbb437f88bf2c704597f9157492d5f98446e | [
"MIT"
] | 15 | 2021-07-07T17:36:42.000Z | 2022-02-27T16:04:32.000Z | search/services/search-enrichment/jupyter-notebooks/ActivitiesUsersAnalysis.ipynb | bcc-code/bcc-search | b2e0bbb437f88bf2c704597f9157492d5f98446e | [
"MIT"
] | null | null | null | 46.419925 | 12,592 | 0.41164 | [
[
[
"import pandas as pd\n\nactivities = pd.read_csv('../data/activities.csv')\nregistrations = pd.read_csv('../data/registrations.csv')\nusers = pd.read_csv(\"../data/users.csv\")",
"/Users/admin/anaconda3/lib/python3.7/site-packages/IPython/core/interactiveshell.py:3020: DtypeWarning: Columns (62,71,76,78) have mixed types.Specify dtype option on import or set low_memory=False.\n interactivity=interactivity, compiler=compiler, result=result)\n"
],
[
"activities",
"_____no_output_____"
],
[
"activitiesIn2021 = activities[activities['Start'] > '2021-01-01']",
"_____no_output_____"
],
[
"activitiesIn2021",
"_____no_output_____"
],
[
"from collections import Counter\n\nCounter(\" \".join(map(str, activitiesIn2021[\"Description\"])).split()).most_common(100)",
"_____no_output_____"
],
[
"activitiesIn2021['Description'] = activitiesIn2021['Description'].astype(str)",
"/Users/admin/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:1: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n \"\"\"Entry point for launching an IPython kernel.\n"
],
[
"def parser(text):\n return re.sub('<[^<]+?>', '', str(text))\n\nresult=activitiesIn2021['Description'].apply(parser)\n\n\n",
"_____no_output_____"
],
[
"result",
"_____no_output_____"
],
[
"from collections import Counter\n\nCounter(\" \".join(map(str, result)).split()).most_common(100)",
"_____no_output_____"
],
[
"from collections import Counter\n\nCounter(\" \".join(map(str, activitiesIn2021[\"Name\"])).split()).most_common(100)",
"_____no_output_____"
],
[
"activitiesIn2021",
"_____no_output_____"
],
[
"#correlation between description and name\nactivitiesIn2021[\"Name\"]\nactivitiesIn2021[\"Description\"]",
"_____no_output_____"
],
[
"activitiesIn2021GroupedByNeededParticipant = activitiesIn2021.groupby(by=[\"NeededParticipants\"], dropna=False).count().reset_index();",
"_____no_output_____"
],
[
"activitiesIn2021",
"_____no_output_____"
],
[
"activities['NeededParticipants'].plot().area(figsize=(20, 12), subplots=True)",
"_____no_output_____"
],
[
"activitiesIn2021['NeededParticipants'].plot().area(figsize=(20, 12), subplots=True)",
"_____no_output_____"
],
[
"#adding random rating\nimport random\nactivitiesIn2021['Rating'] = random.randint(1,5)\n",
"/Users/admin/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:3: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n This is separate from the ipykernel package so we can avoid doing imports until\n"
],
[
"activitiesIn2021['Rating']",
"_____no_output_____"
],
[
"import numpy as np\nactivitiesIn2021['Rating'] = np.random.randint(1, 6, activitiesIn2021.shape[0])",
"/Users/admin/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:2: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n \n"
],
[
"activitiesIn2021",
"_____no_output_____"
],
[
"users",
"_____no_output_____"
],
[
"registrations",
"_____no_output_____"
],
[
"registrationsIn2021 = registrations[registrations['RegistrationDate'] > '2015-01-01']",
"_____no_output_____"
],
[
"registrationsIn2021[['RegistrationId','ActivityId', 'UserId']]\nusers[['UserId']]",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e0df5308021295f78b8fbf36d96d349a882e4e | 12,756 | ipynb | Jupyter Notebook | notebooks/signac_101_Getting_Started.ipynb | sbkashif/signac-examples | c2cd08b0c6059c6ff8c4fac7c6a8275bc8b6c695 | [
"BSD-3-Clause"
] | 10 | 2019-01-31T01:37:38.000Z | 2021-11-09T18:49:45.000Z | notebooks/signac_101_Getting_Started.ipynb | sbkashif/signac-examples | c2cd08b0c6059c6ff8c4fac7c6a8275bc8b6c695 | [
"BSD-3-Clause"
] | 32 | 2020-03-05T00:38:08.000Z | 2021-10-04T14:13:52.000Z | notebooks/signac_101_Getting_Started.ipynb | glotzerlab/signac-examples | 36295ce9da8024af747dd6702088c914d84a19d5 | [
"BSD-3-Clause"
] | 7 | 2020-01-10T08:46:12.000Z | 2022-03-09T14:57:52.000Z | 26.854737 | 272 | 0.561853 | [
[
[
"# 1.1 Getting started\n\n## Prerequisites\n\n### Installation\n\nThis tutorial requires **signac**, so make sure to install the package before starting.\nThe easiest way to do so is using conda:\n\n```$ conda config --add channels conda-forge```\n\n```$ conda install signac```\n\nor pip:\n\n```pip install signac --user```\n\n\nPlease refer to the [documentation](https://docs.signac.io/en/latest/installation.html#installation) for detailed instructions on how to install signac.\n\nAfter successful installation, the following cell should execute without error:",
"_____no_output_____"
]
],
[
[
"import signac",
"_____no_output_____"
]
],
[
[
"We start by removing all data which might be left-over from previous executions of this tutorial.",
"_____no_output_____"
]
],
[
[
"%rm -rf projects/tutorial/workspace",
"_____no_output_____"
]
],
[
[
"## A minimal example\n\nFor this tutorial we want to compute the volume of an ideal gas as a function of its pressure and thermal energy using the ideal gas equation\n\n$p V = N kT$, where\n\n$N$ refers to the system size, $p$ to the pressure, $kT$ to the thermal energy and $V$ is the volume of the system.",
"_____no_output_____"
]
],
[
[
"def V_idg(N, kT, p):\n return N * kT / p",
"_____no_output_____"
]
],
[
[
"We can execute the complete study in just a few lines of code.\nFirst, we initialize the project directory and get a project handle:",
"_____no_output_____"
]
],
[
[
"import signac\n\nproject = signac.init_project(name=\"TutorialProject\", root=\"projects/tutorial\")",
"_____no_output_____"
]
],
[
[
"We iterate over the variable of interest *p* and construct a complete state point *sp* which contains all the meta data associated with our data.\nIn this simple example the meta data is very compact, but in principle the state point may be highly complex.\n\nNext, we obtain a *job* handle and store the result of the calculation within the *job document*.\nThe *job document* is a persistent dictionary for storage of simple key-value pairs.\nHere, we exploit that the state point dictionary *sp* can easily be passed into the `V_idg()` function using the [keyword expansion syntax](https://docs.python.org/dev/tutorial/controlflow.html#keyword-arguments) (`**sp`).",
"_____no_output_____"
]
],
[
[
"for p in 0.1, 1.0, 10.0:\n sp = {\"p\": p, \"kT\": 1.0, \"N\": 1000}\n job = project.open_job(sp)\n job.document[\"V\"] = V_idg(**sp)",
"_____no_output_____"
]
],
[
[
"We can then examine our results by iterating over the data space:",
"_____no_output_____"
]
],
[
[
"for job in project:\n print(job.sp.p, job.document[\"V\"])",
"0.1 10000.0\n10.0 100.0\n1.0 1000.0\n"
]
],
[
[
"That's it.\n\n...\n\nOk, there's more...\nLet's have a closer look at the individual components.\n\n## The Basics\n\nThe **signac** data management framework assists the user in managing the data space of individual *projects*.\nAll data related to one or multiple projects is stored in a *workspace*, which by default is a directory called `workspace` within the project's root directory.",
"_____no_output_____"
]
],
[
[
"print(project.root_directory())\nprint(project.workspace())",
"notebooks/projects/tutorial\nnotebooks/projects/tutorial/workspace\n"
]
],
[
[
"The core idea is to tightly couple state points, unique sets of parameters, with their associated data.\nIn general, the parameter space needs to contain all parameters that will affect our data.\n\nFor the ideal gas that is a 3-dimensional space spanned by the thermal energy *kT*, the pressure *p* and the system size *N*.\nThese are the **input parameters** for our calculations, while the calculated volume *V* is the **output data**.\nIn terms of **signac** this relationship is represented by an instance of `Job`.\n\nWe use the `open_job()` method to get a *job handle* for a specific set of input parameters.",
"_____no_output_____"
]
],
[
[
"job = project.open_job({\"p\": 1.0, \"kT\": 1.0, \"N\": 1000})",
"_____no_output_____"
]
],
[
[
"The *job* handle tightly couples our input parameters (*p*, *kT*, *N*) with the storage location of the output data.\nYou can inspect both the input parameters and the storage location explicitly:",
"_____no_output_____"
]
],
[
[
"print(job.statepoint())\nprint(job.workspace())",
"{'p': 1.0, 'kT': 1.0, 'N': 1000}\nnotebooks/projects/tutorial/workspace/ee617ad585a90809947709a7a45dda9a\n"
]
],
[
[
"For convenience, a job's *state point* may also be accessed via the short-hand `sp` attribute.\nFor example, to access the pressure value `p` we can use either of the two following expressions:",
"_____no_output_____"
]
],
[
[
"print(job.statepoint()[\"p\"])\nprint(job.sp.p)",
"1.0\n1.0\n"
]
],
[
[
"Each *job* has a **unique id** representing the state point.\nThis means opening a job with the exact same input parameters is guaranteed to have the **exact same id**.",
"_____no_output_____"
]
],
[
[
"job2 = project.open_job({\"kT\": 1.0, \"N\": 1000, \"p\": 1.0})\nprint(job.id, job2.id)",
"ee617ad585a90809947709a7a45dda9a ee617ad585a90809947709a7a45dda9a\n"
]
],
[
[
"The *job id* is used to uniquely identify data associated with a specific state point.\nThink of the *job* as a container that is used to store all data associated with the state point.\nFor example, it should be safe to assume that all files that are stored within the job's workspace directory are tightly coupled to the job's statepoint.",
"_____no_output_____"
]
],
[
[
"print(job.workspace())",
"notebooks/projects/tutorial/workspace/ee617ad585a90809947709a7a45dda9a\n"
]
],
[
[
"Let's store the volume calculated for each state point in a file called `V.txt` within the job's workspace.",
"_____no_output_____"
]
],
[
[
"import os\n\nfn_out = os.path.join(job.workspace(), \"V.txt\")\nwith open(fn_out, \"w\") as file:\n V = V_idg(**job.statepoint())\n file.write(str(V) + \"\\n\")",
"_____no_output_____"
]
],
[
[
"Because this is such a common pattern, **signac** signac allows you to short-cut this with the `job.fn()` method.",
"_____no_output_____"
]
],
[
[
"with open(job.fn(\"V.txt\"), \"w\") as file:\n V = V_idg(**job.statepoint())\n file.write(str(V) + \"\\n\")",
"_____no_output_____"
]
],
[
[
"Sometimes it is easier to temporarily switch the *current working directory* while storing data for a specific job.\nFor this purpose, we can use the `Job` object as [context manager](https://docs.python.org/3/reference/compound_stmts.html#with).\nThis means that we switch into the workspace directory associated with the job after entering, and switch back into the original working directory after exiting.",
"_____no_output_____"
]
],
[
[
"with job:\n with open(\"V.txt\", \"w\") as file:\n file.write(str(V) + \"\\n\")",
"_____no_output_____"
]
],
[
[
"Another alternative to store light-weight data is the *job document* as shown in the minimal example.\nThe *job document* is a persistent JSON storage file for simple key-value pairs.",
"_____no_output_____"
]
],
[
[
"job.document[\"V\"] = V_idg(**job.statepoint())\nprint(job.statepoint(), job.document)",
"{'p': 1.0, 'kT': 1.0, 'N': 1000} {'V': 1000.0}\n"
]
],
[
[
"Since we are usually interested in more than one state point, the standard operation is to iterate over all variable(s) of interest, construct the full state point, get the associated job handle, and then either just initialize the job or perform the full operation.",
"_____no_output_____"
]
],
[
[
"for pressure in 0.1, 1.0, 10.0:\n statepoint = {\"p\": pressure, \"kT\": 1.0, \"N\": 1000}\n job = project.open_job(statepoint)\n job.document[\"V\"] = V_idg(**job.statepoint())",
"_____no_output_____"
]
],
[
[
"Let's verify our result by inspecting the data.",
"_____no_output_____"
]
],
[
[
"for job in project:\n print(job.statepoint(), job.document)",
"{'p': 0.1, 'kT': 1.0, 'N': 1000} {'V': 10000.0}\n{'p': 10.0, 'kT': 1.0, 'N': 1000} {'V': 100.0}\n{'p': 1.0, 'kT': 1.0, 'N': 1000} {'V': 1000.0}\n"
]
],
[
[
"Those are the basics for using **signac** for data management.\nThe [next section](signac_102_Exploring_Data.ipynb) demonstrates how to explore an existing data space.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0e0f83235fe36a2044e78b23827e3cba2ac11cb | 49,648 | ipynb | Jupyter Notebook | notebooks/tf-intro/2020-01-rnn-basics.ipynb | DJCordhose/ml-workshop | 6af8861becf6f33e7a5a10d4bf072312090f972d | [
"MIT"
] | 3 | 2020-02-17T13:35:56.000Z | 2020-10-22T13:15:28.000Z | notebooks/tf-intro/2020-01-rnn-basics.ipynb | DJCordhose/ml-workshop | 6af8861becf6f33e7a5a10d4bf072312090f972d | [
"MIT"
] | 7 | 2020-02-09T17:52:44.000Z | 2020-02-09T17:52:53.000Z | notebooks/tf-intro/2020-01-rnn-basics.ipynb | DJCordhose/ml-workshop | 6af8861becf6f33e7a5a10d4bf072312090f972d | [
"MIT"
] | 4 | 2019-07-22T17:05:52.000Z | 2020-01-23T12:17:59.000Z | 66.731183 | 28,362 | 0.734914 | [
[
[
"<a href=\"https://colab.research.google.com/github/DJCordhose/ml-workshop/blob/master/notebooks/tf-intro/2020-01-rnn-basics.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Sequences Basics\n\nExample, some code and a lot of inspiration taken from: https://machinelearningmastery.com/how-to-develop-lstm-models-for-time-series-forecasting/\n",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"import matplotlib as mpl\nmpl.rcParams['figure.figsize'] = (20, 8)",
"_____no_output_____"
],
[
"try:\n # %tensorflow_version only exists in Colab.\n %tensorflow_version 2.x\nexcept Exception:\n pass",
"_____no_output_____"
],
[
"import tensorflow as tf\nprint(tf.__version__)",
"2.2.0\n"
]
],
[
[
"## Univariate Sequences\n\njust one variable per time step\n\n### Challenge\n\nWe have a known series of events, possibly in time and you want to know what is the next event. Like this\n\n[10, 20, 30, 40, 50, 60, 70, 80, 90]",
"_____no_output_____"
]
],
[
[
"# univariate data preparation\nimport numpy as np\n\n# split a univariate sequence into samples\ndef split_sequence(sequence, n_steps):\n\tX, y = list(), list()\n\tfor i in range(len(sequence)):\n\t\t# find the end of this pattern\n\t\tend_ix = i + n_steps\n\t\t# check if we are beyond the sequence\n\t\tif end_ix > len(sequence)-1:\n\t\t\tbreak\n\t\t# gather input and output parts of the pattern\n\t\tseq_x, seq_y = sequence[i:end_ix], sequence[end_ix]\n\t\tX.append(seq_x)\n\t\ty.append(seq_y)\n\treturn np.array(X), np.array(y)",
"_____no_output_____"
],
[
"#@title Prediction from n past steps\n\n# https://colab.research.google.com/notebooks/forms.ipynb\n\nn_steps = 3 #@param {type:\"slider\", min:1, max:10, step:1}",
"_____no_output_____"
],
[
"# define input sequence\nraw_seq = [10, 20, 30, 40, 50, 60, 70, 80, 90]\n\n# split into samples\nX, y = split_sequence(raw_seq, n_steps)\n\n# summarize the data\nlist(zip(X, y))",
"_____no_output_____"
],
[
"X",
"_____no_output_____"
]
],
[
[
"### Converting shapes\n\n* one of the most frequent, yet most tedious steps\n* match between what you have and what an interface needs\n* expected input of RNN: 3D tensor with shape (samples, timesteps, input_dim)\n* we have: (samples, timesteps)\n* reshape on np arrays can do all that",
"_____no_output_____"
]
],
[
[
"# reshape from [samples, timesteps] into [samples, timesteps, features]\nn_features = 1\nX = X.reshape((X.shape[0], X.shape[1], n_features))\nX",
"_____no_output_____"
],
[
"import tensorflow as tf\nfrom tensorflow import keras\nfrom tensorflow.keras.layers import Dense, LSTM, GRU, SimpleRNN, Bidirectional\nfrom tensorflow.keras.models import Sequential, Model\n\nmodel = Sequential()\nmodel.add(SimpleRNN(units=50, activation='relu', input_shape=(n_steps, n_features), name=\"RNN_Input\"))\nmodel.add(Dense(units=1, name=\"Linear_Output\"))\nmodel.compile(optimizer='adam', loss='mse')\nmodel.summary()",
"Model: \"sequential\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nRNN_Input (SimpleRNN) (None, 50) 2600 \n_________________________________________________________________\nLinear_Output (Dense) (None, 1) 51 \n=================================================================\nTotal params: 2,651\nTrainable params: 2,651\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"EPOCHS = 1000\n\n%time history = model.fit(X, y, epochs=EPOCHS, verbose=0)",
"CPU times: user 3.27 s, sys: 170 ms, total: 3.44 s\nWall time: 2.65 s\n"
],
[
"loss = model.evaluate(X, y, verbose=0)\nloss",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n\nplt.yscale('log')\nplt.ylabel(\"loss\")\nplt.xlabel(\"epochs\")\n\nplt.plot(history.history['loss']);",
"_____no_output_____"
]
],
[
[
"### Let's try this on a few examples",
"_____no_output_____"
]
],
[
[
"# this does not look too bad\nX_sample = np.array([[10, 20, 30], [70, 80, 90]]).astype(np.float32)\nX_sample = X_sample.reshape((X_sample.shape[0], X_sample.shape[1], n_features))\nX_sample",
"_____no_output_____"
],
[
"y_pred = model.predict(X_sample)\ny_pred",
"_____no_output_____"
],
[
"def predict(model, samples, n_features=1):\n input = np.array(samples).astype(np.float32)\n input = input.reshape((input.shape[0], input.shape[1], n_features))\n y_pred = model.predict(input)\n return y_pred",
"_____no_output_____"
],
[
"# do not look too close, though\npredict(model, [[100, 110, 120], [200, 210, 220], [200, 300, 400]])",
"_____no_output_____"
]
],
[
[
"## Exercise\n\n* go through the notebook as it is\n* Try to improve the model\n * Change the number of values used as input\n * Change activation function\n * More nodes? less nodes?\n * What else might help improving the results?\n",
"_____no_output_____"
],
[
"# STOP HERE",
"_____no_output_____"
],
[
"### Input and output of an RNN layer",
"_____no_output_____"
]
],
[
[
"# https://keras.io/layers/recurrent/\n# input: (samples, timesteps, input_dim)\n# output: (samples, units)\n\n# let's have a look at the actual output for an example\nrnn_layer = model.get_layer(\"RNN_Input\")\nmodel_stub = Model(inputs = model.input, outputs = rnn_layer.output)\nhidden = predict(model_stub, [[10, 20, 30]])\nhidden.shape, hidden",
"_____no_output_____"
]
],
[
[
"#### What do we see?\n* each unit (50) has a single output\n* as a sidenote you nicely see the RELU nature of the output\n* so the timesteps of the input are lost\n* we are only looking at the final output\n* still with each timestep, the layer does produce a unique output we could potentially use",
"_____no_output_____"
],
[
"### We need to look into RNNs a bit more deeply now\n\n#### RNNs - Networks with Loops\n<img src='https://djcordhose.github.io/ai/img/nlp/colah/RNN-rolled.png' height=200>\n\nhttp://colah.github.io/posts/2015-08-Understanding-LSTMs/\n#### Unrolling the loop\n<img src='https://djcordhose.github.io/ai/img/nlp/colah/RNN-unrolled.png'>\n\nhttp://colah.github.io/posts/2015-08-Understanding-LSTMs/\n#### Simple RNN internals\n\n<img src='https://djcordhose.github.io/ai/img/nlp/fchollet_rnn.png'>\n\n## $output_t = \\tanh(W input_t + U output_{t-1} + b)$\n\nFrom Deep Learning with Python, Chapter 6, François Chollet, Manning: https://livebook.manning.com/#!/book/deep-learning-with-python/chapter-6/129\n\n#### Activation functions\n\n<img src='https://djcordhose.github.io/ai/img/sigmoid-activation.png' height=200>\n\nSigmoid compressing between 0 and 1\n\n<img src='https://djcordhose.github.io/ai/img/tanh-activation.png' height=200>\n\nHyperbolic tangent, like sigmoind, but compressing between -1 and 1, thus allowing for negative values as well",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
d0e0ff0439f4a5bf33d258cd7ef806ce12951897 | 90,401 | ipynb | Jupyter Notebook | nasws/cnn/search_space/nasbench101/test-nasbench-partial.ipynb | kcyu2014/nas-landmarkreg | a00c3619bf4042e446e1919087f0b09fe9fa3a65 | [
"MIT"
] | 8 | 2021-04-13T01:52:11.000Z | 2022-03-30T03:53:12.000Z | nasws/cnn/search_space/nasbench101/test-nasbench-partial.ipynb | kcyu2014/nas-landmarkreg | a00c3619bf4042e446e1919087f0b09fe9fa3a65 | [
"MIT"
] | 4 | 2021-05-29T01:41:00.000Z | 2021-08-24T09:40:43.000Z | nasws/cnn/search_space/nasbench101/test-nasbench-partial.ipynb | kcyu2014/nas-landmarkreg | a00c3619bf4042e446e1919087f0b09fe9fa3a65 | [
"MIT"
] | null | null | null | 132.74743 | 66,059 | 0.548799 | [
[
[
"%load_ext autoreload\n%autoreload 2\n!nvidia-smi\n\nfrom argparse import Namespace\nimport sys\nimport os\nhome = os.environ['HOME']\nos.environ['CUDA_VISIBLE_DEVICES'] = '0,1'\nprint(os.environ['CUDA_VISIBLE_DEVICES'])\nos.chdir(f'{home}/pycharm/automl')\n# os.chdir(f'{home}/pycharm/automl/search_policies/rnn')\nsys.path.append(f'{home}/pycharm/nasbench')\nsys.path.append(f'{home}/pycharm/automl')\nimport torch \nimport torch.nn as nn",
"0,1\n"
],
[
"import utils\nfrom image_dataloader import load_dataset\nfrom search_policies.cnn.search_space.nasbench101.nasbench_api_v2 import NASBench_v2\nfrom search_policies.cnn.search_space.nasbench101.util import load_nasbench_checkpoint, transfer_model_key_to_search_key, nasbench_zero_padding_load_state_dict_pre_hook \nfrom search_policies.cnn.procedures.train_search_procedure import darts_model_validation\nfrom search_policies.cnn.search_space.nasbench101.model_search import NasBenchNetSearch",
"Cannot import graphviz package\n"
],
[
"utils.torch_random_seed(10)\nprint('loading cifar ...')\ntrain_queue, valid_queue, test_queue = load_dataset()\nprint(\"loading Nasbench dataset...\")\nnasbench = NASBench_v2('data/nasbench/nasbench_only108.tfrecord', only_hash=True)",
"loading cifar ...\nFiles already downloaded and verified\nFiles already downloaded and verified\nloading Nasbench dataset...\n"
],
[
"sanity_check = True",
"_____no_output_____"
],
[
"# load one model and \nMODEL_FOLDER='/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_100002-arch_97a390a2cb02fdfbc505f6ac44a37228-eval-runid-0'\nCKPT_PATH=MODEL_FOLDER + '/checkpoint.pt'\nmodel_folder = MODEL_FOLDER\nckpt_path = CKPT_PATH\n\nprint(\"load model and test ...\")\nhash = model_folder.split('arch_')[1].split('-eval')[0]\nprint('model_hash ', hash)\nspec = nasbench.hash_to_model_spec(hash)\nprint('model spec', spec)\n# model, ckpt = load_nasbench_checkpoint(ckpt_path, spec, legacy=True)\n\nfrom search_policies.cnn.cnn_search_configs import build_default_args\n# def project the weights \ndefault_args = build_default_args()\ndefault_args.model_spec = spec\nspec = nasbench.hash_to_model_spec(hash)\nmodel, ckpt = load_nasbench_checkpoint(ckpt_path, spec, legacy=True)\nmodel = model.cuda()\nmodel.eval()\nmodel_search = NasBenchNetSearch(args=default_args)\nmodel_search.eval()\nmodel_search = model_search.cuda()\nsource_dict = model.state_dict()\ntarget_dict = model_search.state_dict()",
"load model and test ...\nmodel_hash 97a390a2cb02fdfbc505f6ac44a37228\nmodel spec Adjacent matrix: [[0, 1, 0, 1, 0, 0, 0], [0, 0, 1, 1, 0, 0, 1], [0, 0, 0, 0, 0, 0, 1], [0, 0, 0, 0, 1, 0, 0], [0, 0, 0, 0, 0, 1, 0], [0, 0, 0, 0, 0, 0, 1], [0, 0, 0, 0, 0, 0, 0]] \nOps: ['input', 'conv3x3-bn-relu', 'maxpool3x3', 'maxpool3x3', 'maxpool3x3', 'maxpool3x3', 'output']\n\n"
],
[
"# Checking the loaded model state dict.\nif sanity_check:\n for ind, k in enumerate(source_dict.keys()):\n if ind > 5:\n break\n print(k, ((source_dict[k] - target_dict[k]).sum()))\n\ntrans_dict = dict()\n\nfor k in model.state_dict().keys():\n kk = transfer_model_key_to_search_key(k, spec)\n if kk not in model_search.state_dict().keys():\n print('not found ', kk)\n continue\n # if sanity_check:\n # print(f'map {k}', source_dict[k].size()) \n # print(f'to {kk}' ,target_dict[kk].size())\n \n trans_dict[kk] = source_dict[k]\n\npadded_dict = nasbench_zero_padding_load_state_dict_pre_hook(trans_dict, model_search.state_dict())",
"stem.0.weight tensor(3.7924, device='cuda:0')\nstem.1.weight tensor(-42.6739, device='cuda:0')\nstem.1.bias tensor(5.2125, device='cuda:0')\nstem.1.running_mean tensor(0.2281, device='cuda:0')\nstem.1.running_var tensor(-110.5854, device='cuda:0')\nstem.1.num_batches_tracked tensor(313121, device='cuda:0')\n"
],
[
"if sanity_check:\n target_dict = model_search.state_dict()\n for ind, k in enumerate(padded_dict.keys()):\n if ind > 5:\n break\n print(k)\n print((padded_dict[k] - target_dict[k]).sum().item())\n print((ckpt['model_state'][k] - target_dict[k]).sum().item())\n # print((padded_dict[k][trans_dict[k].size()[0]] - trans_dict[k]).sum())",
"stem.0.weight\n0.0\n0.0\nstem.1.weight\n0.0\n0.0\nstem.1.bias\n0.0\n0.0\nstem.1.running_mean\n0.0\n0.0\nstem.1.running_var\n0.0\n0.0\nstem.1.num_batches_tracked\n0\n0\n"
],
[
"# loaded the padded dict and test the results.\nmodel_search.load_state_dict(padded_dict)\nmodel_search = model_search.change_model_spec(spec)\nmodel_search = model_search.eval()",
"_____no_output_____"
],
[
"res = darts_model_validation(test_queue, model, nn.CrossEntropyLoss(),Namespace(debug=False, report_freq=50))\nprint('original model evaluation on test split', res)\n\nsearch_res = darts_model_validation(test_queue, model_search, nn.CrossEntropyLoss(),Namespace(debug=False, report_freq=50))\nprint('Reload to NasBenchSearch, evaluation results ' ,search_res)\n# results get worse after padding to original node. Somethings goes wrong.",
"original model evaluation on test split (92.95, 0.40452058811187747)\nReload to NasBenchSearch, evaluation results (92.95, 0.40452058811187747)\n"
],
[
"# Further sanity checking, by loading part of the model and continue.",
"_____no_output_____"
],
[
"# print(model.stacks['stack0']['module0'].vertex_ops.keys())\n# print(spec.ops)\n# m0 = model.stacks['stack0']['module0']\n# print([k for k in m0.state_dict().keys() if 'vertex_1' not in k and 'proj' not in k ])\n# # print([k for k in m0.state_dict().keys() if 'vertex_1' not in k and 'proj' not in k ])\n# load_keys = sorted([transfer_model_key_to_search_key(k, spec) for k in k1])\n# train_param_keys = sorted(train_param_keys)\n# # for a1, a2 in zip(load_keys, train_param_keys):\n# # print(a1)\n# # print(a2)\n# for a1 in load_keys:\n# if a1 not in train_param_keys:\n# print(a1)\n# print(len(load_keys), len(train_param_keys))\n\n",
"_____no_output_____"
],
[
"model.eval()\nmodel_search.load_state_dict(padded_dict)\nmodel_search.eval()",
"_____no_output_____"
],
[
"# This debugs the entire network's output.\nimg, lab = iter(test_queue).__next__()\nimg = img.cuda()\nlab = lab.cuda()\nbz = 32\n\nwith torch.no_grad():\n m1 = model.forward_debug(img[:bz, :, : ,:])\n m2 = model_search.forward_debug(img[:bz, :, :, :])\n for ind,(a,b) in enumerate(zip(m1, m2)):\n print(ind, (a - b).sum().item())",
"0 0.0\n1 0.0\n2 0.0\n3 0.0\n4 0.0\n5 0.0\n6 0.0\n7 0.0\n8 0.0\n9 0.0\n10 0.0\n11 0.0\n12 0.0\n"
],
[
"# model_search.stem[1].eval()\n# model.stem[1].eval()\n# m1stem_out = model.stem(img)\n# model_search.stem.load_state_dict(model.stem.state_dict())\n# m2stem_out = model_search.stem(img)\n# for k, v in model.stem.state_dict().items():\n# print(k, (v - model_search.stem.state_dict()[k]).sum().item())\n# print((m1stem_out - m2stem_out).sum().item())\n\n",
"_____no_output_____"
],
[
"s1input = m1[0].cuda()\nm1stack = model.stacks['stack0']['module0']\nm2stack = model_search.stacks['stack0']['module0']\nprint(m1stack.__class__.__name__)\nprint(m2stack.__class__.__name__)\n\nprint((m1stack(s1input) - m2stack(s1input)).sum().item())",
"NasBenchCell\nNasBenchCellSearch\n-29079.44921875\n"
],
[
"m1out = m1stack.forward_debug(s1input)\nm2out = m2stack.forward_debug(s1input)",
"_____no_output_____"
],
[
"# Cell level, not true after 1 iters\nfor k, v in m1out.items():\n print(k, (v - m2out[k]).sum().item())\n \nprint(m1stack.execution_order.items())",
"input 0.0\noutput -29079.44921875\nvertex_1 4053.396728515625\nvertex_3 -13347.875\nvertex_4 -22021.6953125\nvertex_5 -28074.6796875\nvertex_2 -5058.1689453125\nodict_items([('vertex_1', ['input']), ('vertex_3', ['input', 'vertex_1']), ('vertex_4', ['vertex_3']), ('vertex_5', ['vertex_4']), ('vertex_2', ['vertex_1']), ('output', ['vertex_1', 'vertex_2', 'vertex_5'])])\n"
],
[
"# vertex level\nm1vertex = m1stack.vertex_ops['vertex_1']\nm2vertex = m2stack.vertex_ops['vertex_1']\n\nprint('vertex_1 difference', (m1vertex([s1input,]) - m2vertex([s1input,])).sum().item())\nprint(m1vertex)\nprint(m2vertex)",
"vertex_1 difference 4053.396728515625\nVertex(\n (proj_ops): ModuleList(\n (0): Sequential(\n (0): Conv2d(128, 86, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (1): BatchNorm2d(86, eps=1e-05, momentum=0.997, affine=True, track_running_stats=True)\n (2): ReLU()\n )\n )\n (op): Sequential(\n (0): Conv2d(86, 86, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (1): BatchNorm2d(86, eps=1e-05, momentum=0.997, affine=True, track_running_stats=True)\n (2): ReLU()\n )\n)\nMixedVertex(\n (proj_ops): ModuleList(\n (0): DynamicReLUConvBN(\n (relu): ReLU()\n (conv): DynamicConv2d(128, 256, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (bn): BatchNorm2d(256, eps=1e-05, momentum=0.997, affine=True, track_running_stats=True)\n (channel_drop): ChannelDropout()\n )\n )\n (ops): ModuleDict(\n (conv1x1-bn-relu): DynamicReLUConvBN(\n (relu): ReLU()\n (conv): DynamicConv2d(256, 256, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (bn): BatchNorm2d(256, eps=1e-05, momentum=0.997, affine=True, track_running_stats=True)\n (channel_drop): ChannelDropout()\n )\n (conv3x3-bn-relu): DynamicReLUConvBN(\n (relu): ReLU()\n (conv): DynamicConv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (bn): BatchNorm2d(256, eps=1e-05, momentum=0.997, affine=True, track_running_stats=True)\n (channel_drop): ChannelDropout()\n )\n (maxpool3x3): MaxPool2d(kernel_size=3, stride=1, padding=1, dilation=1, ceil_mode=False)\n )\n)\n"
],
[
"# inside vertex level, proj_ops and vertex_op\nm1_proj = m1vertex.proj_ops[0](s1input)\nm1_out = m1vertex.op(m1_proj)\n\nm2_proj = m2vertex.current_proj_ops[0](s1input)\nm2_out = m2vertex.current_op(m2_proj)\n\nprint('after v1 proj diff',(m1_proj - m2_proj).sum().item())\nprint('after v1 diff',(m1_out - m2_out).sum().item())",
"after v1 proj diff -12232.4296875\nafter v1 diff 4053.396728515625\n"
],
[
"# proj level\nm1projop = m1vertex.proj_ops[0]\nm2projop = m2vertex.current_proj_ops[0]\nprint(m1projop)\nprint(m2projop)",
"Sequential(\n (0): Conv2d(128, 86, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (1): BatchNorm2d(86, eps=1e-05, momentum=0.997, affine=True, track_running_stats=True)\n (2): ReLU()\n)\nDynamicReLUConvBN(\n (relu): ReLU()\n (conv): DynamicConv2d(128, 256, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (bn): BatchNorm2d(256, eps=1e-05, momentum=0.997, affine=True, track_running_stats=True)\n (channel_drop): ChannelDropout()\n)\n"
],
[
"print(m1projop[1].training)\nprint(m2projop.bn.training)",
"False\nFalse\n"
],
[
"# m2vertex.train()\nmodel_search.eval()\nprint(m1projop[1].training)\nprint(m2projop.bn.training)",
"False\nFalse\n"
],
[
"import glob\nmodel_lists = glob.glob('/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/*')\nprint(model_lists)\nfor model_folder in model_lists:\n if len(glob.glob(model_folder + '/checkpoint.pt')) > 0:\n hash = model_folder.split('arch_')[1].split('-eval')[0]\n rank = model_folder.split('rank_')[1].split('-arch')[0]\n print(rank, hash)\n \n\n",
"['/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_300004-arch_f2ff48f97a4f7b340b10f7600a062b5e-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_200001-arch_6c678b4d5c0bb786d69ba399c0ee6759-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_423000-arch_789eb4f52941c786ac64242606d379fb-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_200003-arch_bbbcaeddb4d0782013a4aa63864f46d9-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_400001-arch_1119da5413faccc8a6aec77d55d26fc7-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_4000-arch_523ac64c85d50533bbbb695d6f3c583f-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_5000-arch_81a7f132d0d93078bf6f58ae660985e4-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_10-arch_ef11a63e3dec4177d65648771c3689aa-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_400002-arch_f8167cf8d4c0926fc9436a7a058b2b54-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_100001-arch_018b46ecbd2d0cdc420cb4574383b694-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_100000-arch_538464bec1502be99fbfbd5200080caa-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_2000-arch_172b3b3d8163eb2f2ad63b5b6d7e214a-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_300001-arch_ffa05e224fdf80254991a0b91865f593-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_423004-arch_b7c8fc1e4501f140434e1854284b43f4-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_200000-arch_dbb82f92411e951be3c3249eb118328b-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_100004-arch_7b7924641b6ee467f82f37b3b7eb51dd-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_100002-arch_97a390a2cb02fdfbc505f6ac44a37228-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_100003-arch_d885d701640a7d5fb1904bb5212b3200-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_300000-arch_ebea8be67d25fc825ce6ffe4276479f2-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_1000-arch_a2d021f626023241ae7268eb8260240c-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_400000-arch_73a74dee1d13b18a4d8bc03eb1156e5c-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_423003-arch_208476d048a5551955622243d379048e-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_300002-arch_bd617b04c723cf1b7e63afae9b1963c2-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_423001-arch_0a597cbfd44a59343135083788bd3c94-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_200002-arch_42795b0b09cb57d2fab0f97d576d0ed3-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_423002-arch_b94e6efc2afbbcfb86607e534ffc1015-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_300003-arch_a84b1ca6851ea9ea913b7fbedcb47a94-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_400004-arch_c98011952d1679b2a7e3999338ff4a13-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_400003-arch_7411b9803ffe264b406bb658440c8f63-eval-runid-0', '/home/yukaiche/pycharm/automl/experiments/reproduce-nasbench/rank_200004-arch_253224d103e3c1099761405f2c965959-eval-runid-0']\n300004 f2ff48f97a4f7b340b10f7600a062b5e\n200003 bbbcaeddb4d0782013a4aa63864f46d9\n400001 1119da5413faccc8a6aec77d55d26fc7\n4000 523ac64c85d50533bbbb695d6f3c583f\n5000 81a7f132d0d93078bf6f58ae660985e4\n10 ef11a63e3dec4177d65648771c3689aa\n400002 f8167cf8d4c0926fc9436a7a058b2b54\n100001 018b46ecbd2d0cdc420cb4574383b694\n100000 538464bec1502be99fbfbd5200080caa\n2000 172b3b3d8163eb2f2ad63b5b6d7e214a\n300001 ffa05e224fdf80254991a0b91865f593\n200000 dbb82f92411e951be3c3249eb118328b\n100004 7b7924641b6ee467f82f37b3b7eb51dd\n100002 97a390a2cb02fdfbc505f6ac44a37228\n100003 d885d701640a7d5fb1904bb5212b3200\n300000 ebea8be67d25fc825ce6ffe4276479f2\n1000 a2d021f626023241ae7268eb8260240c\n300002 bd617b04c723cf1b7e63afae9b1963c2\n200002 42795b0b09cb57d2fab0f97d576d0ed3\n300003 a84b1ca6851ea9ea913b7fbedcb47a94\n400004 c98011952d1679b2a7e3999338ff4a13\n200004 253224d103e3c1099761405f2c965959\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e100a84e2de2643275e99e4b4a97c1f4eb15b7 | 8,204 | ipynb | Jupyter Notebook | docs/prototype/2020-05-18-recent-schema.ipynb | tonyfast/qhub | 3ada432fcfa85f65ba9385edcb650190dfa30eb8 | [
"BSD-3-Clause"
] | null | null | null | docs/prototype/2020-05-18-recent-schema.ipynb | tonyfast/qhub | 3ada432fcfa85f65ba9385edcb650190dfa30eb8 | [
"BSD-3-Clause"
] | null | null | null | docs/prototype/2020-05-18-recent-schema.ipynb | tonyfast/qhub | 3ada432fcfa85f65ba9385edcb650190dfa30eb8 | [
"BSD-3-Clause"
] | null | null | null | 42.729167 | 99 | 0.478303 | [
[
[
" __import__('qhapi').schema.DigitalOcean.schema()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code"
]
] |
d0e109836037a5ed76eb1990d5106f58cae38dee | 23,872 | ipynb | Jupyter Notebook | Big-Data-Clusters/CU6/Public/content/install/sop036-install-kubectl.ipynb | gantz-at-incomm/tigertoolbox | 9ea80d39a3c5e0c77553fc851c5ee787fbf9291d | [
"MIT"
] | 541 | 2019-05-07T11:41:25.000Z | 2022-03-29T17:33:19.000Z | Big-Data-Clusters/CU6/Public/content/install/sop036-install-kubectl.ipynb | gantz-at-incomm/tigertoolbox | 9ea80d39a3c5e0c77553fc851c5ee787fbf9291d | [
"MIT"
] | 89 | 2019-05-09T14:23:52.000Z | 2022-01-13T20:21:04.000Z | Big-Data-Clusters/CU6/Public/content/install/sop036-install-kubectl.ipynb | gantz-at-incomm/tigertoolbox | 9ea80d39a3c5e0c77553fc851c5ee787fbf9291d | [
"MIT"
] | 338 | 2019-05-08T05:45:16.000Z | 2022-03-28T15:35:03.000Z | 57.384615 | 203 | 0.402396 | [
[
[
"SOP036 - Install kubectl command line interface\n===============================================\n\nSteps\n-----\n\n### Common functions\n\nDefine helper functions used in this notebook.",
"_____no_output_____"
]
],
[
[
"# Define `run` function for transient fault handling, suggestions on error, and scrolling updates on Windows\n\nimport sys\nimport os\nimport re\nimport json\nimport platform\nimport shlex\nimport shutil\nimport datetime\n\nfrom subprocess import Popen, PIPE\nfrom IPython.display import Markdown\n\nretry_hints = {} # Output in stderr known to be transient, therefore automatically retry\nerror_hints = {} # Output in stderr where a known SOP/TSG exists which will be HINTed for further help\ninstall_hint = {} # The SOP to help install the executable if it cannot be found\n\nfirst_run = True\nrules = None\ndebug_logging = False\n\ndef run(cmd, return_output=False, no_output=False, retry_count=0, base64_decode=False, return_as_json=False):\n \"\"\"Run shell command, stream stdout, print stderr and optionally return output\n\n NOTES:\n\n 1. Commands that need this kind of ' quoting on Windows e.g.:\n\n kubectl get nodes -o jsonpath={.items[?(@.metadata.annotations.pv-candidate=='data-pool')].metadata.name}\n\n Need to actually pass in as '\"':\n\n kubectl get nodes -o jsonpath={.items[?(@.metadata.annotations.pv-candidate=='\"'data-pool'\"')].metadata.name}\n\n The ' quote approach, although correct when pasting into Windows cmd, will hang at the line:\n \n `iter(p.stdout.readline, b'')`\n\n The shlex.split call does the right thing for each platform, just use the '\"' pattern for a '\n \"\"\"\n MAX_RETRIES = 5\n output = \"\"\n retry = False\n\n global first_run\n global rules\n\n if first_run:\n first_run = False\n rules = load_rules()\n\n # When running `azdata sql query` on Windows, replace any \\n in \"\"\" strings, with \" \", otherwise we see:\n #\n # ('HY090', '[HY090] [Microsoft][ODBC Driver Manager] Invalid string or buffer length (0) (SQLExecDirectW)')\n #\n if platform.system() == \"Windows\" and cmd.startswith(\"azdata sql query\"):\n cmd = cmd.replace(\"\\n\", \" \")\n\n # shlex.split is required on bash and for Windows paths with spaces\n #\n cmd_actual = shlex.split(cmd)\n\n # Store this (i.e. kubectl, python etc.) to support binary context aware error_hints and retries\n #\n user_provided_exe_name = cmd_actual[0].lower()\n\n # When running python, use the python in the ADS sandbox ({sys.executable})\n #\n if cmd.startswith(\"python \"):\n cmd_actual[0] = cmd_actual[0].replace(\"python\", sys.executable)\n\n # On Mac, when ADS is not launched from terminal, LC_ALL may not be set, which causes pip installs to fail\n # with:\n #\n # UnicodeDecodeError: 'ascii' codec can't decode byte 0xc5 in position 4969: ordinal not in range(128)\n #\n # Setting it to a default value of \"en_US.UTF-8\" enables pip install to complete\n #\n if platform.system() == \"Darwin\" and \"LC_ALL\" not in os.environ:\n os.environ[\"LC_ALL\"] = \"en_US.UTF-8\"\n\n # When running `kubectl`, if AZDATA_OPENSHIFT is set, use `oc`\n #\n if cmd.startswith(\"kubectl \") and \"AZDATA_OPENSHIFT\" in os.environ:\n cmd_actual[0] = cmd_actual[0].replace(\"kubectl\", \"oc\")\n\n # To aid supportability, determine which binary file will actually be executed on the machine\n #\n which_binary = None\n\n # Special case for CURL on Windows. The version of CURL in Windows System32 does not work to\n # get JWT tokens, it returns \"(56) Failure when receiving data from the peer\". If another instance\n # of CURL exists on the machine use that one. (Unfortunately the curl.exe in System32 is almost\n # always the first curl.exe in the path, and it can't be uninstalled from System32, so here we\n # look for the 2nd installation of CURL in the path)\n if platform.system() == \"Windows\" and cmd.startswith(\"curl \"):\n path = os.getenv('PATH')\n for p in path.split(os.path.pathsep):\n p = os.path.join(p, \"curl.exe\")\n if os.path.exists(p) and os.access(p, os.X_OK):\n if p.lower().find(\"system32\") == -1:\n cmd_actual[0] = p\n which_binary = p\n break\n\n # Find the path based location (shutil.which) of the executable that will be run (and display it to aid supportability), this\n # seems to be required for .msi installs of azdata.cmd/az.cmd. (otherwise Popen returns FileNotFound) \n #\n # NOTE: Bash needs cmd to be the list of the space separated values hence shlex.split.\n #\n if which_binary == None:\n which_binary = shutil.which(cmd_actual[0])\n\n # Display an install HINT, so the user can click on a SOP to install the missing binary\n #\n if which_binary == None:\n if user_provided_exe_name in install_hint and install_hint[user_provided_exe_name] is not None:\n display(Markdown(f'HINT: Use [{install_hint[user_provided_exe_name][0]}]({install_hint[user_provided_exe_name][1]}) to resolve this issue.'))\n\n raise FileNotFoundError(f\"Executable '{cmd_actual[0]}' not found in path (where/which)\")\n else: \n cmd_actual[0] = which_binary\n\n start_time = datetime.datetime.now().replace(microsecond=0)\n\n print(f\"START: {cmd} @ {start_time} ({datetime.datetime.utcnow().replace(microsecond=0)} UTC)\")\n print(f\" using: {which_binary} ({platform.system()} {platform.release()} on {platform.machine()})\")\n print(f\" cwd: {os.getcwd()}\")\n\n # Command-line tools such as CURL and AZDATA HDFS commands output\n # scrolling progress bars, which causes Jupyter to hang forever, to\n # workaround this, use no_output=True\n #\n\n # Work around a infinite hang when a notebook generates a non-zero return code, break out, and do not wait\n #\n wait = True \n\n try:\n if no_output:\n p = Popen(cmd_actual)\n else:\n p = Popen(cmd_actual, stdout=PIPE, stderr=PIPE, bufsize=1)\n with p.stdout:\n for line in iter(p.stdout.readline, b''):\n line = line.decode()\n if return_output:\n output = output + line\n else:\n if cmd.startswith(\"azdata notebook run\"): # Hyperlink the .ipynb file\n regex = re.compile(' \"(.*)\"\\: \"(.*)\"') \n match = regex.match(line)\n if match:\n if match.group(1).find(\"HTML\") != -1:\n display(Markdown(f' - \"{match.group(1)}\": \"{match.group(2)}\"'))\n else:\n display(Markdown(f' - \"{match.group(1)}\": \"[{match.group(2)}]({match.group(2)})\"'))\n\n wait = False\n break # otherwise infinite hang, have not worked out why yet.\n else:\n print(line, end='')\n if rules is not None:\n apply_expert_rules(line)\n\n if wait:\n p.wait()\n except FileNotFoundError as e:\n if install_hint is not None:\n display(Markdown(f'HINT: Use {install_hint} to resolve this issue.'))\n\n raise FileNotFoundError(f\"Executable '{cmd_actual[0]}' not found in path (where/which)\") from e\n\n exit_code_workaround = 0 # WORKAROUND: azdata hangs on exception from notebook on p.wait()\n\n if not no_output:\n for line in iter(p.stderr.readline, b''):\n try:\n line_decoded = line.decode()\n except UnicodeDecodeError:\n # NOTE: Sometimes we get characters back that cannot be decoded(), e.g.\n #\n # \\xa0\n #\n # For example see this in the response from `az group create`:\n #\n # ERROR: Get Token request returned http error: 400 and server \n # response: {\"error\":\"invalid_grant\",# \"error_description\":\"AADSTS700082: \n # The refresh token has expired due to inactivity.\\xa0The token was \n # issued on 2018-10-25T23:35:11.9832872Z\n #\n # which generates the exception:\n #\n # UnicodeDecodeError: 'utf-8' codec can't decode byte 0xa0 in position 179: invalid start byte\n #\n print(\"WARNING: Unable to decode stderr line, printing raw bytes:\")\n print(line)\n line_decoded = \"\"\n pass\n else:\n\n # azdata emits a single empty line to stderr when doing an hdfs cp, don't\n # print this empty \"ERR:\" as it confuses.\n #\n if line_decoded == \"\":\n continue\n \n print(f\"STDERR: {line_decoded}\", end='')\n\n if line_decoded.startswith(\"An exception has occurred\") or line_decoded.startswith(\"ERROR: An error occurred while executing the following cell\"):\n exit_code_workaround = 1\n\n # inject HINTs to next TSG/SOP based on output in stderr\n #\n if user_provided_exe_name in error_hints:\n for error_hint in error_hints[user_provided_exe_name]:\n if line_decoded.find(error_hint[0]) != -1:\n display(Markdown(f'HINT: Use [{error_hint[1]}]({error_hint[2]}) to resolve this issue.'))\n\n # apply expert rules (to run follow-on notebooks), based on output\n #\n if rules is not None:\n apply_expert_rules(line_decoded)\n\n # Verify if a transient error, if so automatically retry (recursive)\n #\n if user_provided_exe_name in retry_hints:\n for retry_hint in retry_hints[user_provided_exe_name]:\n if line_decoded.find(retry_hint) != -1:\n if retry_count < MAX_RETRIES:\n print(f\"RETRY: {retry_count} (due to: {retry_hint})\")\n retry_count = retry_count + 1\n output = run(cmd, return_output=return_output, retry_count=retry_count)\n\n if return_output:\n if base64_decode:\n import base64\n\n return base64.b64decode(output).decode('utf-8')\n else:\n return output\n\n elapsed = datetime.datetime.now().replace(microsecond=0) - start_time\n\n # WORKAROUND: We avoid infinite hang above in the `azdata notebook run` failure case, by inferring success (from stdout output), so\n # don't wait here, if success known above\n #\n if wait: \n if p.returncode != 0:\n raise SystemExit(f'Shell command:\\n\\n\\t{cmd} ({elapsed}s elapsed)\\n\\nreturned non-zero exit code: {str(p.returncode)}.\\n')\n else:\n if exit_code_workaround !=0 :\n raise SystemExit(f'Shell command:\\n\\n\\t{cmd} ({elapsed}s elapsed)\\n\\nreturned non-zero exit code: {str(exit_code_workaround)}.\\n')\n\n print(f'\\nSUCCESS: {elapsed}s elapsed.\\n')\n\n if return_output:\n if base64_decode:\n import base64\n\n return base64.b64decode(output).decode('utf-8')\n else:\n return output\n\ndef load_json(filename):\n \"\"\"Load a json file from disk and return the contents\"\"\"\n\n with open(filename, encoding=\"utf8\") as json_file:\n return json.load(json_file)\n\ndef load_rules():\n \"\"\"Load any 'expert rules' from the metadata of this notebook (.ipynb) that should be applied to the stderr of the running executable\"\"\"\n\n # Load this notebook as json to get access to the expert rules in the notebook metadata.\n #\n try:\n j = load_json(\"sop036-install-kubectl.ipynb\")\n except:\n pass # If the user has renamed the book, we can't load ourself. NOTE: Is there a way in Jupyter, to know your own filename?\n else:\n if \"metadata\" in j and \\\n \"azdata\" in j[\"metadata\"] and \\\n \"expert\" in j[\"metadata\"][\"azdata\"] and \\\n \"expanded_rules\" in j[\"metadata\"][\"azdata\"][\"expert\"]:\n\n rules = j[\"metadata\"][\"azdata\"][\"expert\"][\"expanded_rules\"]\n\n rules.sort() # Sort rules, so they run in priority order (the [0] element). Lowest value first.\n\n # print (f\"EXPERT: There are {len(rules)} rules to evaluate.\")\n\n return rules\n\ndef apply_expert_rules(line):\n \"\"\"Determine if the stderr line passed in, matches the regular expressions for any of the 'expert rules', if so\n inject a 'HINT' to the follow-on SOP/TSG to run\"\"\"\n\n global rules\n\n for rule in rules:\n notebook = rule[1]\n cell_type = rule[2]\n output_type = rule[3] # i.e. stream or error\n output_type_name = rule[4] # i.e. ename or name \n output_type_value = rule[5] # i.e. SystemExit or stdout\n details_name = rule[6] # i.e. evalue or text \n expression = rule[7].replace(\"\\\\*\", \"*\") # Something escaped *, and put a \\ in front of it!\n\n if debug_logging:\n print(f\"EXPERT: If rule '{expression}' satisfied', run '{notebook}'.\")\n\n if re.match(expression, line, re.DOTALL):\n\n if debug_logging:\n print(\"EXPERT: MATCH: name = value: '{0}' = '{1}' matched expression '{2}', therefore HINT '{4}'\".format(output_type_name, output_type_value, expression, notebook))\n\n match_found = True\n\n display(Markdown(f'HINT: Use [{notebook}]({notebook}) to resolve this issue.'))\n",
"_____no_output_____"
]
],
[
[
"### Install Kubernetes CLI\n\nTo get the latest version number for `kubectl` for Windows, open this\nfile:\n\n- https://storage.googleapis.com/kubernetes-release/release/stable.txt\n\nNOTE: For Windows, `kubectl.exe` is installed in the folder containing\nthe `python.exe` (`sys.executable`), which will be in the path for\nnotebooks run in ADS.",
"_____no_output_____"
]
],
[
[
"import os\nimport sys\nimport platform\n\nfrom pathlib import Path\n\nif platform.system() == \"Darwin\":\n run('brew update')\n run('brew install kubernetes-cli')\nelif platform.system() == \"Windows\":\n path = Path(sys.executable)\n cwd = os.getcwd()\n os.chdir(path.parent)\n run('curl -L https://storage.googleapis.com/kubernetes-release/release/v1.17.0/bin/windows/amd64/kubectl.exe -o kubectl.exe')\n os.chdir(cwd)\nelif platform.system() == \"Linux\":\n run('sudo apt-get update')\n run('sudo apt-get install -y kubectl')\nelse:\n raise SystemExit(f\"Platform '{platform.system()}' is not recognized, must be 'Darwin', 'Windows' or 'Linux'\")",
"_____no_output_____"
],
[
"print('Notebook execution complete.')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d0e10b4983abf07ba36c26f8912911d70fd1488f | 69,670 | ipynb | Jupyter Notebook | notebooks/fastai/fastai-mnist-sample.ipynb | DevScope/ai-lab | 6d4838284a1c3341624fef1270f89d53fe65c3a1 | [
"MIT"
] | 20 | 2018-08-21T09:26:51.000Z | 2021-10-08T18:51:31.000Z | notebooks/fastai/fastai-mnist-sample.ipynb | DevScope/ai-lab | 6d4838284a1c3341624fef1270f89d53fe65c3a1 | [
"MIT"
] | 19 | 2020-03-24T18:05:28.000Z | 2022-02-01T10:24:30.000Z | notebooks/fastai/fastai-mnist-sample.ipynb | DevScope/ai-lab | 6d4838284a1c3341624fef1270f89d53fe65c3a1 | [
"MIT"
] | 4 | 2019-03-17T04:51:26.000Z | 2020-12-20T01:53:42.000Z | 99.956958 | 28,372 | 0.838395 | [
[
[
"# Fastai Mnist sample (for binder)\n- (adapted from fastai v3 https://github.com/fastai/course-v3/blob/master/nbs/dl1/lesson1-pets.ipynb)",
"_____no_output_____"
],
[
"In this lesson we will build our first image classifier from scratch. Let's dive in!\n\nEvery notebook starts with the following three lines; they ensure that any edits to libraries you make are reloaded here automatically, and also that any charts or images displayed are shown in this notebook.",
"_____no_output_____"
]
],
[
[
"%reload_ext autoreload\n%autoreload 2\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"We import all the necessary packages. We are going to work with the [fastai V1 library](http://www.fast.ai/2018/10/02/fastai-ai/) which sits on top of [Pytorch 1.0](https://hackernoon.com/pytorch-1-0-468332ba5163). The fastai library provides many useful functions that enable us to quickly and easily build neural networks and train our models.",
"_____no_output_____"
]
],
[
[
"from fastai.vision import *\nfrom fastai.metrics import error_rate",
"_____no_output_____"
],
[
"bs = 64\n# bs = 16 # uncomment this line if you run out of memory even after clicking Kernel->Restart",
"_____no_output_____"
]
],
[
[
"## Looking at the data",
"_____no_output_____"
],
[
"We are going to use the `untar_data` function to which we must pass a URL as an argument and which will download and extract the data.",
"_____no_output_____"
]
],
[
[
"help(untar_data)",
"Help on function untar_data in module fastai.datasets:\n\nuntar_data(url:str, fname:Union[pathlib.Path, str]=None, dest:Union[pathlib.Path, str]=None, data=True, force_download=False) -> pathlib.Path\n Download `url` to `fname` if it doesn't exist, and un-tgz to folder `dest`.\n\n"
]
],
[
[
"We'll use MNIST sample version, with two classes: 3 and 7",
"_____no_output_____"
]
],
[
[
"path = untar_data(URLs.MNIST_SAMPLE); path",
"_____no_output_____"
],
[
"path.ls()",
"_____no_output_____"
]
],
[
[
"## Binder/docker/fastai changes",
"_____no_output_____"
]
],
[
[
"#Binder doesn't allow for GPU, switch to CPU\ndefaults.device = 'cpu'",
"_____no_output_____"
],
[
"data = ImageDataBunch.from_folder(path)",
"_____no_output_____"
],
[
"data",
"_____no_output_____"
],
[
"data.show_batch(rows=3, figsize=(7,6))",
"_____no_output_____"
],
[
"print(data.classes)\nlen(data.classes),data.c",
"['3', '7']\n"
]
],
[
[
"## Training",
"_____no_output_____"
],
[
"Now we will start training our model. We will use a [convolutional neural network](http://cs231n.github.io/convolutional-networks/) backbone and a fully connected head with a single hidden layer as a classifier. \n\nWe will train for 2 epochs (2 cycles through all our data).",
"_____no_output_____"
]
],
[
[
"learn = create_cnn(data, models.resnet18, metrics=[accuracy])",
"_____no_output_____"
],
[
"learn.model",
"_____no_output_____"
],
[
"learn.fit_one_cycle(2)",
"_____no_output_____"
],
[
"learn.save('stage-1')",
"_____no_output_____"
]
],
[
[
"## Results",
"_____no_output_____"
]
],
[
[
"interp = ClassificationInterpretation.from_learner(learn)\n\nlosses,idxs = interp.top_losses()\n\nlen(data.valid_ds)==len(losses)==len(idxs)",
"_____no_output_____"
],
[
"interp.plot_top_losses(9, figsize=(15,11))",
"_____no_output_____"
],
[
"doc(interp.plot_top_losses)",
"_____no_output_____"
],
[
"interp.plot_confusion_matrix(figsize=(12,12), dpi=60)",
"_____no_output_____"
],
[
"interp.most_confused(min_val=2)",
"_____no_output_____"
]
],
[
[
"## Check fastai v3 course for more videos & tutorials!\n- https://course.fast.ai/",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
d0e1137af9e7bd873ae7e9fb8a8ae245aa425c55 | 14,595 | ipynb | Jupyter Notebook | module1-rnn-and-lstm/LS_DS_431_RNN_and_LSTM_Assignment.ipynb | nchibana/DS-Unit-4-Sprint-3-Deep-Learning | 33ec2a7c02b4a2c5d7bb3f028d8795309f9f43ad | [
"MIT"
] | null | null | null | module1-rnn-and-lstm/LS_DS_431_RNN_and_LSTM_Assignment.ipynb | nchibana/DS-Unit-4-Sprint-3-Deep-Learning | 33ec2a7c02b4a2c5d7bb3f028d8795309f9f43ad | [
"MIT"
] | null | null | null | module1-rnn-and-lstm/LS_DS_431_RNN_and_LSTM_Assignment.ipynb | nchibana/DS-Unit-4-Sprint-3-Deep-Learning | 33ec2a7c02b4a2c5d7bb3f028d8795309f9f43ad | [
"MIT"
] | null | null | null | 37.423077 | 318 | 0.587187 | [
[
[
"<img align=\"left\" src=\"https://lever-client-logos.s3.amazonaws.com/864372b1-534c-480e-acd5-9711f850815c-1524247202159.png\" width=200>\n<br></br>\n<br></br>\n\n## *Data Science Unit 4 Sprint 3 Assignment 1*\n\n# Recurrent Neural Networks and Long Short Term Memory (LSTM)\n\n\n\nIt is said that [infinite monkeys typing for an infinite amount of time](https://en.wikipedia.org/wiki/Infinite_monkey_theorem) will eventually type, among other things, the complete works of Wiliam Shakespeare. Let's see if we can get there a bit faster, with the power of Recurrent Neural Networks and LSTM.\n\nThis text file contains the complete works of Shakespeare: https://www.gutenberg.org/files/100/100-0.txt\n\nUse it as training data for an RNN - you can keep it simple and train character level, and that is suggested as an initial approach.\n\nThen, use that trained RNN to generate Shakespearean-ish text. Your goal - a function that can take, as an argument, the size of text (e.g. number of characters or lines) to generate, and returns generated text of that size.\n\nNote - Shakespeare wrote an awful lot. It's OK, especially initially, to sample/use smaller data and parameters, so you can have a tighter feedback loop when you're trying to get things running. Then, once you've got a proof of concept - start pushing it more!",
"_____no_output_____"
]
],
[
[
"import sys\nimport numpy\nfrom keras.models import Sequential\nfrom keras.layers import Dense\nfrom keras.layers import Dropout\nfrom keras.layers import LSTM\nfrom keras.callbacks import ModelCheckpoint\nfrom keras.utils import np_utils\n# load ascii text and covert to lowercase\nfilename = \"content/shakespeare.txt\"\nraw_text = open(filename, 'r', encoding='utf-8').read()\nraw_text = raw_text.lower()\nbad_chars = ['#', '*', '@', '_', '\\ufeff']\nfor i in range(len(bad_chars)):\n raw_text = raw_text.replace(bad_chars[i],\"\")",
"_____no_output_____"
],
[
"# create mapping of unique chars to integers, and a reverse mapping\nchars = sorted(list(set(raw_text)))\nchar_to_int = dict((c, i) for i, c in enumerate(chars))\nint_to_char = dict((i, c) for i, c in enumerate(chars))",
"_____no_output_____"
],
[
"# summarize the loaded data\nn_chars = len(raw_text)\nn_vocab = len(chars)\nprint(\"Total Characters: \", n_chars)\nprint(\"Total Vocab: \", n_vocab)",
"_____no_output_____"
],
[
"# prepare the dataset of input to output pairs encoded as integers\nseq_length = 100\ndataX = []\ndataY = []\nfor i in range(0, n_chars - seq_length, 1):\n seq_in = raw_text[i:i + seq_length]\n seq_out = raw_text[i + seq_length]\n dataX.append([char_to_int[char] for char in seq_in])\n dataY.append(char_to_int[seq_out])\nn_patterns = len(dataX)",
"_____no_output_____"
],
[
"# reshape X to be [samples, time steps, features]\nX = numpy.reshape(dataX, (n_patterns, seq_length, 1))\n# normalize\nX = X / float(n_vocab)\n# one hot encode the output variable\ny = np_utils.to_categorical(dataY)",
"_____no_output_____"
],
[
"# define the LSTM model\nmodel = Sequential()\nmodel.add(LSTM(256, input_shape=(X.shape[1], X.shape[2]), return_sequences=True))\nmodel.add(Dropout(0.2))\nmodel.add(LSTM(256))\nmodel.add(Dropout(0.2))\nmodel.add(Dense(y.shape[1], activation='softmax'))\nmodel.compile(loss='categorical_crossentropy', optimizer='adam')\n# define the checkpoint\nfilepath=\"weights-improvement-{epoch:02d}-{loss:.4f}-bigger.hdf5\"\ncheckpoint = ModelCheckpoint(filepath, monitor='loss', verbose=1, save_best_only=True, mode='min')\ncallbacks_list = [checkpoint]\n# fit the model\nmodel.fit(X, y, epochs=50, batch_size=64, callbacks=callbacks_list)",
"_____no_output_____"
],
[
"import numpy as np\nimport random\nimport sys",
"_____no_output_____"
],
[
"filename = \"content/shakespeare.txt\"\ntext = open(filename, 'r', encoding='utf-8').read()\ntext = text.lower()\nbad_chars = ['#', '*', '@', '_', '\\ufeff']\nfor i in range(len(bad_chars)):\n text = text.replace(bad_chars[i],\"\")",
"_____no_output_____"
],
[
"characters = sorted(list(set(text)))",
"_____no_output_____"
],
[
"char2indices = dict((c, i) for i, c in enumerate(characters))\nindices2char = dict((i, c) for i, c in enumerate(characters))",
"_____no_output_____"
],
[
"maxlen = 40\nstep = 3\nsentences = []\nnext_chars = []\nfor i in range(0, len(text) - maxlen, step):\n sentences.append(text[i: i + maxlen])\n next_chars.append(text[i + maxlen])",
"_____no_output_____"
],
[
"# Converting indices into vectorized format\nX = np.zeros((len(sentences), maxlen, len(characters)), dtype=np.bool)\ny = np.zeros((len(sentences), len(characters)), dtype=np.bool)\nfor i, sentence in enumerate(sentences):\n for t, char in enumerate(sentence):\n X[i, t, char2indices[char]] = 1\n y[i, char2indices[next_chars[i]]] = 1",
"_____no_output_____"
],
[
"from keras.models import Sequential\nfrom keras.layers import Dense, LSTM,Activation,Dropout\nfrom keras.optimizers import RMSprop",
"Using TensorFlow backend.\n"
],
[
"#Model Building\nmodel = Sequential()\nmodel.add(LSTM(128, input_shape=(maxlen, len(characters))))\nmodel.add(Dense(len(characters)))\nmodel.add(Activation('softmax'))\nmodel.compile(loss='categorical_crossentropy', optimizer=RMSprop(lr=0.01))\nprint (model.summary())",
"WARNING: Logging before flag parsing goes to stderr.\nW0917 02:24:34.408209 8040 deprecation_wrapper.py:119] From C:\\Users\\nchib\\Anaconda3\\envs\\U4-S2-NN\\lib\\site-packages\\keras\\backend\\tensorflow_backend.py:66: The name tf.get_default_graph is deprecated. Please use tf.compat.v1.get_default_graph instead.\n\nW0917 02:24:34.433205 8040 deprecation_wrapper.py:119] From C:\\Users\\nchib\\Anaconda3\\envs\\U4-S2-NN\\lib\\site-packages\\keras\\backend\\tensorflow_backend.py:541: The name tf.placeholder is deprecated. Please use tf.compat.v1.placeholder instead.\n\nW0917 02:24:34.435205 8040 deprecation_wrapper.py:119] From C:\\Users\\nchib\\Anaconda3\\envs\\U4-S2-NN\\lib\\site-packages\\keras\\backend\\tensorflow_backend.py:4432: The name tf.random_uniform is deprecated. Please use tf.random.uniform instead.\n\nW0917 02:24:34.657202 8040 deprecation_wrapper.py:119] From C:\\Users\\nchib\\Anaconda3\\envs\\U4-S2-NN\\lib\\site-packages\\keras\\optimizers.py:793: The name tf.train.Optimizer is deprecated. Please use tf.compat.v1.train.Optimizer instead.\n\nW0917 02:24:34.668214 8040 deprecation_wrapper.py:119] From C:\\Users\\nchib\\Anaconda3\\envs\\U4-S2-NN\\lib\\site-packages\\keras\\backend\\tensorflow_backend.py:3576: The name tf.log is deprecated. Please use tf.math.log instead.\n\n"
],
[
"def pred_indices(preds, metric=1.0):\n preds = np.asarray(preds).astype('float64')\n preds = np.log(preds) / metric\n exp_preds = np.exp(preds)\n preds = exp_preds/np.sum(exp_preds)\n probs = np.random.multinomial(1, preds, 1)\n return np.argmax(probs)",
"_____no_output_____"
],
[
"# Train and Evaluate the Model\n\nfor iteration in range(1, 10):\n print('-' * 40)\n print('Iteration', iteration)\n model.fit(X, y,batch_size=128,epochs=1)\n\n start_index = random.randint(0, len(text) - maxlen - 1)\n\n for diversity in [0.2, 0.7,1.2]:\n\n print('\\n----- diversity:', diversity)\n\n generated = ''\n sentence = text[start_index: start_index + maxlen]\n generated += sentence\n print('----- Generating with seed: \"' + sentence + '\"')\n sys.stdout.write(generated)\n\n for i in range(400):\n x = np.zeros((1, maxlen, len(characters)))\n for t, char in enumerate(sentence):\n x[0, t, char2indices[char]] = 1.\n\n preds = model.predict(x, verbose=0)[0]\n next_index = pred_indices(preds, diversity)\n pred_char = indices2char[next_index]\n\n generated += pred_char\n sentence = sentence[1:] + pred_char\n\n sys.stdout.write(pred_char)\n sys.stdout.flush()\n print(\"\\nOne combination completed \\n\")",
"----------------------------------------\nIteration 1\nEpoch 1/1\n 49280/1859890 [..............................] - ETA: 19:51 - loss: 1.5709"
]
],
[
[
"## Stretch goals:\n- Refine the training and generation of text to be able to ask for different genres/styles of Shakespearean text (e.g. plays versus sonnets)\n- Train a classification model that takes text and returns which work of Shakespeare it is most likely to be from\n- Make it more performant! Many possible routes here - lean on Keras, optimize the code, and/or use more resources (AWS, etc.)\n- Revisit the news example from class, and improve it - use categories or tags to refine the model/generation, or train a news classifier\n- Run on bigger, better data\n\n## Resources:\n- [The Unreasonable Effectiveness of Recurrent Neural Networks](https://karpathy.github.io/2015/05/21/rnn-effectiveness/) - a seminal writeup demonstrating a simple but effective character-level NLP RNN\n- [Simple NumPy implementation of RNN](https://github.com/JY-Yoon/RNN-Implementation-using-NumPy/blob/master/RNN%20Implementation%20using%20NumPy.ipynb) - Python 3 version of the code from \"Unreasonable Effectiveness\"\n- [TensorFlow RNN Tutorial](https://github.com/tensorflow/models/tree/master/tutorials/rnn) - code for training a RNN on the Penn Tree Bank language dataset\n- [4 part tutorial on RNN](http://www.wildml.com/2015/09/recurrent-neural-networks-tutorial-part-1-introduction-to-rnns/) - relates RNN to the vanishing gradient problem, and provides example implementation\n- [RNN training tips and tricks](https://github.com/karpathy/char-rnn#tips-and-tricks) - some rules of thumb for parameterizing and training your RNN",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
d0e1155c0bffbbc2fe15be83f37f08116e2a5375 | 4,811 | ipynb | Jupyter Notebook | 06Model_Save_Load/Model_Save_Load.ipynb | aonekoda/Sklearn | 4266799c22f41dfd302f164b07fc4b7b110a2929 | [
"MIT"
] | 2 | 2020-08-14T07:34:33.000Z | 2020-09-09T07:57:59.000Z | 06Model_Save_Load/Model_Save_Load.ipynb | aonekoda/Sklearn | 4266799c22f41dfd302f164b07fc4b7b110a2929 | [
"MIT"
] | null | null | null | 06Model_Save_Load/Model_Save_Load.ipynb | aonekoda/Sklearn | 4266799c22f41dfd302f164b07fc4b7b110a2929 | [
"MIT"
] | 2 | 2020-09-03T01:08:01.000Z | 2020-09-09T06:53:41.000Z | 25.321053 | 97 | 0.431719 | [
[
[
"# 훈련된 모형의 저장과 복원\n*모형을 훈련한 후 모형을 저장하고 애플리케이션에 필요할 때 이를 다시 복원 할 수 있어야 한다.\n* 모형을 피클(pickle)로 저장한다.\n* 피클이란? 파이썬 객체 자체를 binary 파일로 저장하는 것을 말한다.\n*사이킷런에는 피클 기능을 확장한 joblib 모듈을 제공한다.\n* 주의 할 점 – 저장된 모델이 사이킷런 버전 간에 호환되지 않는지 주의해야 한다. 피클을 사용하기 때문에 버전이 맞지 않으면 로드시 에러가 발생한다.\n",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import RandomForestClassifier\nfrom sklearn import datasets\nimport joblib",
"_____no_output_____"
],
[
"iris = datasets.load_iris()\nfeatures = iris.data\ntarget = iris.target",
"_____no_output_____"
],
[
"classifier = RandomForestClassifier()\nmodel = classifier.fit(features, target)",
"_____no_output_____"
]
],
[
[
"## Model save",
"_____no_output_____"
]
],
[
[
"joblib.dump(model, 'model.pkl')",
"_____no_output_____"
]
],
[
[
"## Model Load",
"_____no_output_____"
]
],
[
[
"loaded_model = joblib.load(\"model.pkl\")\nprint(loaded_model)",
"RandomForestClassifier(bootstrap=True, ccp_alpha=0.0, class_weight=None,\n criterion='gini', max_depth=None, max_features='auto',\n max_leaf_nodes=None, max_samples=None,\n min_impurity_decrease=0.0, min_impurity_split=None,\n min_samples_leaf=1, min_samples_split=2,\n min_weight_fraction_leaf=0.0, n_estimators=100,\n n_jobs=None, oob_score=False, random_state=None,\n verbose=0, warm_start=False)\n"
],
[
"new_obs = [[5.2, 3.2, 1.1, 0.1]]\nloaded_model.predict(new_obs)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d0e12262201f0909d770455f8e9b41fa4047d6c7 | 186,526 | ipynb | Jupyter Notebook | 01b_random-network-sequence-learning.ipynb | sagacitysite/pelene | e8d4112264acb44954c52053b4e3f9d63b46bdd6 | [
"MIT"
] | 10 | 2021-02-09T16:42:37.000Z | 2022-01-10T07:37:00.000Z | 01b_random-network-sequence-learning.ipynb | sagacitysite/pelene | e8d4112264acb44954c52053b4e3f9d63b46bdd6 | [
"MIT"
] | null | null | null | 01b_random-network-sequence-learning.ipynb | sagacitysite/pelene | e8d4112264acb44954c52053b4e3f9d63b46bdd6 | [
"MIT"
] | 3 | 2021-02-10T18:12:31.000Z | 2021-09-13T07:40:01.000Z | 683.245421 | 55,268 | 0.9473 | [
[
[
"\"\"\"\nA randomly connected network learning a sequence\n\nThis example contains a reservoir network of 500 neurons.\n400 neurons are excitatory and 100 neurons are inhibitory.\nThe weights are initialized randomly, based on a log-normal distribution.\n\nThe network activity is stimulated with three different inputs (A, B, C).\nThe inputs are given in i a row (A -> B -> C -> A -> ...)\n\nThe experiment is defined in 'pelenet/experiments/sequence.py' file.\nA log file, parameters, and plot figures are stored in the 'log' folder for every run of the simulation.\n\nNOTE: The main README file contains some more information about the structure of pelenet\n\"\"\"\n\n# Load pelenet modules\nfrom pelenet.utils import Utils\nfrom pelenet.experiments.sequence import SequenceExperiment\n\n# Official modules\nimport numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"# Overwrite default parameters (pelenet/parameters/ and pelenet/experiments/sequence.py)\nparameters = {\n # Experiment\n 'seed': 1, # Random seed\n 'trials': 10, # Number of trials\n 'stepsPerTrial': 60, # Number of simulation steps for every trial\n # Neurons\n 'refractoryDelay': 2, # Refactory period\n 'voltageTau': 100, # Voltage time constant\n 'currentTau': 5, # Current time constant\n 'thresholdMant': 1200, # Spiking threshold for membrane potential\n # Network\n 'reservoirExSize': 400, # Number of excitatory neurons\n 'reservoirConnPerNeuron': 35, # Number of connections per neuron\n 'isLearningRule': True, # Apply a learning rule\n 'learningRule': '2^-2*x1*y0 - 2^-2*y1*x0 + 2^-4*x1*y1*y0 - 2^-3*y0*w*w', # Defines the learning rule\n # Input\n 'inputIsSequence': True, # Activates sequence input\n 'inputSequenceSize': 3, # Number of input clusters in sequence\n 'inputSteps': 20, # Number of steps the trace input should drive the network\n 'inputGenSpikeProb': 0.8, # Probability of spike for the generator\n 'inputNumTargetNeurons': 40, # Number of neurons activated by the input\n # Probes\n 'isExSpikeProbe': True, # Probe excitatory spikes\n 'isInSpikeProbe': True, # Probe inhibitory spikes\n 'isWeightProbe': True # Probe weight matrix at the end of the simulation\n}",
"_____no_output_____"
],
[
"# Initilizes the experiment, also initializes the log\n# Creating a new object results in a new log entry in the 'log' folder\n# The name is optional, it is extended to the folder in the log directory\nexp = SequenceExperiment(name='random-network-sequence-learning', parameters=parameters)",
"_____no_output_____"
],
[
"# Instantiate the utils singleton\nutils = Utils.instance()",
"_____no_output_____"
],
[
"# Build the network, in this function the weight matrix, inputs, probes, etc. are defined and created\nexp.build()",
"_____no_output_____"
],
[
"# Run the network simulation, afterwards the probes are postprocessed to nice arrays\nexp.run()",
"\u001b[1;30m\u001b[1;30mINFO\u001b[0m\u001b[0m:\u001b[34m\u001b[34mDRV\u001b[0m\u001b[0m: Connecting to 127.0.0.1:37979\n\u001b[1;30m\u001b[1;30mINFO\u001b[0m\u001b[0m:\u001b[34m\u001b[34mDRV\u001b[0m\u001b[0m: Host server up..............Done 0.18s\n\u001b[1;30m\u001b[1;30mINFO\u001b[0m\u001b[0m:\u001b[34m\u001b[34mDRV\u001b[0m\u001b[0m: Compiling Embedded snips....Done 0.13s\n\u001b[1;30m\u001b[1;30mINFO\u001b[0m\u001b[0m:\u001b[34m\u001b[34mDRV\u001b[0m\u001b[0m: Encoding axons/synapses.....Done 0.38s\n\u001b[1;30m\u001b[1;30mINFO\u001b[0m\u001b[0m:\u001b[34m\u001b[34mDRV\u001b[0m\u001b[0m: Booting up..................Done 0.92s\n\u001b[1;30m\u001b[1;30mINFO\u001b[0m\u001b[0m:\u001b[34m\u001b[34mDRV\u001b[0m\u001b[0m: Encoding probes.............Done 0.03s\n\u001b[1;30m\u001b[1;30mINFO\u001b[0m\u001b[0m:\u001b[34m\u001b[34mDRV\u001b[0m\u001b[0m: Transferring probes.........Done 0.07s\n\u001b[1;30m\u001b[1;30mINFO\u001b[0m\u001b[0m:\u001b[34m\u001b[34mDRV\u001b[0m\u001b[0m: Configuring registers.......Done 0.53s\n\u001b[1;30m\u001b[1;30mINFO\u001b[0m\u001b[0m:\u001b[34m\u001b[34mDRV\u001b[0m\u001b[0m: Transferring spikes.........Done 0.28s\n\u001b[1;30m\u001b[1;30mINFO\u001b[0m\u001b[0m:\u001b[34m\u001b[34mDRV\u001b[0m\u001b[0m: Executing...................Done 14.36s\n\u001b[1;30m\u001b[1;30mINFO\u001b[0m\u001b[0m:\u001b[34m\u001b[34mDRV\u001b[0m\u001b[0m: Processing timeseries.......Done 0.88s\n\u001b[1;30m\u001b[1;30mINFO\u001b[0m\u001b[0m:\u001b[34m\u001b[34mDRV\u001b[0m\u001b[0m: Executor: 600 timesteps.........Done 17.91s\n\u001b[1;30m\u001b[1;30mINFO\u001b[0m\u001b[0m:\u001b[34m\u001b[34mHST\u001b[0m\u001b[0m: Using Kapoho Bay serial number 308\n\u001b[1;30m\u001b[1;30mINFO\u001b[0m\u001b[0m:\u001b[34m\u001b[34mHST\u001b[0m\u001b[0m: Args chip=0 cpu=0 /home/seq/.local/share/virtualenvs/seq-r0_WvjRo/lib/python3.5/site-packages/nxsdk/driver/compilers/../../../temp/1605893204.9922104/launcher_chip0_lmt0.bin --chips=1 --remote-relay=0 --epoch=4 \n\u001b[1;30m\u001b[1;30mINFO\u001b[0m\u001b[0m:\u001b[34m\u001b[34mHST\u001b[0m\u001b[0m: Lakemont_driver...\n\u001b[1;30m\u001b[1;30mINFO\u001b[0m\u001b[0m:\u001b[34m\u001b[34mHST\u001b[0m\u001b[0m: chip=0 cpu=0 halted, status=0x0\n"
],
[
"# Weight matrix before learning (randomly initialized)\nexp.net.plot.initialExWeightMatrix()",
"_____no_output_____"
],
[
"# Plot distribution of weights\nexp.net.plot.initialExWeightDistribution(figsize=(12,3))",
"_____no_output_____"
],
[
"# Plot spike trains of the excitatory (red) and inhibitory (blue) neurons\nexp.net.plot.reservoirSpikeTrain(figsize=(12,6), to=600)",
"_____no_output_____"
],
[
"# Weight matrix after learning\nexp.net.plot.trainedExWeightMatrix()",
"_____no_output_____"
],
[
"# Sorted weight matrix after learning\nsupportMask = utils.getSupportWeightsMask(exp.net.trainedWeightsExex)\nexp.net.plot.weightsSortedBySupport(supportMask)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e12dc23cd7c6b6ad6b63d2f42cc460f9aae8c1 | 4,259 | ipynb | Jupyter Notebook | cs229/Linear Regression (Julia).ipynb | vikasgorur/notebooks | f8b7cd6f9f8be840c6637af9e457be063ad5b50e | [
"MIT"
] | null | null | null | cs229/Linear Regression (Julia).ipynb | vikasgorur/notebooks | f8b7cd6f9f8be840c6637af9e457be063ad5b50e | [
"MIT"
] | null | null | null | cs229/Linear Regression (Julia).ipynb | vikasgorur/notebooks | f8b7cd6f9f8be840c6637af9e457be063ad5b50e | [
"MIT"
] | null | null | null | 23.530387 | 482 | 0.45128 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d0e130b2ffbd306fd002f06f9160d887881dd61b | 41,077 | ipynb | Jupyter Notebook | program_design/lesson1 .ipynb | NickChilling/notes | dca6756873303cae96833e6d1475fd38440bba8f | [
"Apache-2.0"
] | null | null | null | program_design/lesson1 .ipynb | NickChilling/notes | dca6756873303cae96833e6d1475fd38440bba8f | [
"Apache-2.0"
] | null | null | null | program_design/lesson1 .ipynb | NickChilling/notes | dca6756873303cae96833e6d1475fd38440bba8f | [
"Apache-2.0"
] | null | null | null | 35.350258 | 300 | 0.432627 | [
[
[
"## shuffle\n### 计算机如何生成素数\n\n只简单介绍一种方法,**线性同余法**\n\n$$N_{j+1} = (A*N_j + B) mod M $$ \n\n其中A,B,M是设定的常数。$N_0$被叫做随机种子,是计算机主板的计数器在内存中的记数值。\n\n从公式中可以看出$N_{j+1}$ 与 $N_j$ 存在线性。\n\n线性同余法生成的是均匀分布,如果需要生成其他分布则是据此进行采样。\n",
"_____no_output_____"
],
[
"### 那么如何随机打乱一个数组\n\n1. 考虑一种方法:对于一个长度为n的数组,每次随机选取两个index,交换这两个index对应的数字。并将他们标记。循环直到数组中所有元素被处理",
"_____no_output_____"
]
],
[
[
"import random\ndef shuffle_1(array):\n N = len(array)\n swaped = [False]*N\n while not all(swaped):\n a = random.randrange(N)\n b = random.randrange(N)\n array[a],array[b] = array[b],array[a]\n swaped[b] = True\n swaped[a] = True\ntest_case = [1,2,3]\nshuffle_1(test_case)\nprint(test_case)",
"[1, 3, 2]\n"
]
],
[
[
"这个打乱算法是**有问题**的。\n- 不一定能退出循环\n- 时间复杂度为$O(n)$ \n- 各种组合的概率不均匀",
"_____no_output_____"
],
[
"时间复杂度为什么为$O(n)$ \n这个问题实际上可以抽象为在袋子里有n个球的情况下,每次抽2个球,放回抽样,需要抽多少次才能全部抽过一遍。求这个次数的期望\n而这个随机变量的期望值与 $n^2$ 有关",
"_____no_output_____"
]
],
[
[
"def shuffle_2(array):\n \"Knuth's algorithm P\"\n N = len(array)\n for i in range(N-1): # 这里N 或者N-1都没有问题。N只是多做一次计算\n j = random.randrange(i,N)\n array[i],array[j] = array[j],array[i]\nshuffle_2(test_case)",
"_____no_output_____"
]
],
[
[
"显然,这个算法的时间复杂度为$O(n)$",
"_____no_output_____"
]
],
[
[
"from collections import defaultdict\n\ndef test_shuffler(shuffler,case=\"abc\",n=100000):\n counter = defaultdict(int)\n for i in range(n):\n case_array = list(case)\n shuffler(case_array)\n counter[\"\".join(case_array)] += 1\n e = n*1.0/factorial(len(case))\n test_ok = all(0.9 <= e/counter[j] <= 1.1 for j in counter)\n name = shuffler.__name__\n if test_ok:\n print(\"%s ok\" %name)\n else:\n print(\"%s failed\" %name)\n for k,v in sorted(counter.items()):\n print(k,v/n)\ndef factorial(n):\n if n<= 1: return 1\n else:\n return n*factorial(n-1)",
"_____no_output_____"
],
[
"test_shuffler(shuffle_1)",
"shuffle_1 failed\nabc 0.04993\nacb 0.14037\nbac 0.13798\nbca 0.26658\ncab 0.26639\ncba 0.13875\n"
],
[
"test_shuffler(shuffle_2)",
"shuffle_2 ok\nabc 0.16595\nacb 0.16526\nbac 0.16748\nbca 0.1649\ncab 0.1682\ncba 0.16821\n"
],
[
"def shuffle_3(array):\n N = len(array)\n swaped = [False]*N\n while not all(swaped):\n a = random.randrange(N)\n b = random.randrange(N)\n array[a],array[b] = array[b],array[a]\n swaped[a] = True\ndef shuffle_4(array):\n N = len(array)\n for i in range(N):\n j = random.randrange(N)\n array[i],array[j] = array[j],array[i]",
"_____no_output_____"
],
[
"test_shuffler(shuffle_3)\ntest_shuffler(shuffle_4)",
"shuffle_3 ok\nabc 0.16181\nacb 0.16803\nbac 0.16925\nbca 0.16721\ncab 0.16778\ncba 0.16592\nshuffle_4 failed\nabc 0.14805\nacb 0.18433\nbac 0.18513\nbca 0.18578\ncab 0.14766\ncba 0.14905\n"
]
],
[
[
"## 纸牌游戏\n",
"_____no_output_____"
]
],
[
[
"def hand_rank(hand):\n ranks = card_ranks(hand)\n if straight(ranks) and flush(hand):\n return (8,max(ranks))\n elif kind(4,ranks):\n return (7,kind(4,ranks),kind(1,ranks))\n elif kind(3,ranks) and kind(2,ranks):\n return (6,kind(3,ranks),kind(2,ranks))\n elif flush(hand):\n return (5,ranks)\n elif straight(ranks):\n return (4,max(ranks))\n elif kind(3,ranks):\n return(3,kind(3,ranks),ranks)\n elif two_pair(ranks):\n return(2,two_pair(ranks),ranks)\n elif kind(2,ranks):\n return(1,kind(2,ranks),ranks)\n else:\n return(0,ranks)\n\ndef card_ranks(hand):\n ranks = ['--23456789TJQKA'.index(r) for r, s in hand]\n ranks.sort(reverse=True)\n return [5,4,3,2,1] if (ranks==[14,5,4,3,2]) else ranks\n\ndef flush(hand):\n suits = [s for r,s in hand]\n return len(set(suits))==1\n\ndef straight(ranks):\n return (max(ranks)-min(ranks)==4) and len(set(ranks))==5\n\ndef kind(n,ranks):\n for r in ranks:\n if ranks.count(r) == n: return r\n\ndef two_pair(ranks):\n pair = kind(2,ranks)\n lowpair = kind(2,list(reversed(ranks)))\n if pair and lowpair!= pair:\n return (pair,lowpair)\n else:\n return None",
"_____no_output_____"
]
],
[
[
"### homework\n1. Seven Card Stud\n\nWrite a function best_hand(hand) that takes a seven\n\ncard hand as input and returns the best possible 5\n\ncard hand. The itertools library has some functions\n\nthat may help you solve this problem.",
"_____no_output_____"
]
],
[
[
"def test_best_hand():\n assert (sorted(best_hand(\"6C 7C 8C 9C TC 5C JS\".split()))==[\"6C\",\"7C\",\"8C\",\"9C\",\"TC\"])\n assert (sorted(best_hand(\"TD TC TH 7C 7D 8C 8S\".split()))\n == ['8C', '8S', 'TC', 'TD', 'TH'])\n assert (sorted(best_hand(\"JD TC TH 7C 7D 7S 7H\".split()))\n == ['7C', '7D', '7H', '7S', 'JD'])\n return 'test_best_hand passes'\n",
"_____no_output_____"
],
[
"import itertools\nhelp(itertools)",
"Help on built-in module itertools:\n\nNAME\n itertools - Functional tools for creating and using iterators.\n\nDESCRIPTION\n Infinite iterators:\n count(start=0, step=1) --> start, start+step, start+2*step, ...\n cycle(p) --> p0, p1, ... plast, p0, p1, ...\n repeat(elem [,n]) --> elem, elem, elem, ... endlessly or up to n times\n \n Iterators terminating on the shortest input sequence:\n accumulate(p[, func]) --> p0, p0+p1, p0+p1+p2\n chain(p, q, ...) --> p0, p1, ... plast, q0, q1, ... \n chain.from_iterable([p, q, ...]) --> p0, p1, ... plast, q0, q1, ... \n compress(data, selectors) --> (d[0] if s[0]), (d[1] if s[1]), ...\n dropwhile(pred, seq) --> seq[n], seq[n+1], starting when pred fails\n groupby(iterable[, keyfunc]) --> sub-iterators grouped by value of keyfunc(v)\n filterfalse(pred, seq) --> elements of seq where pred(elem) is False\n islice(seq, [start,] stop [, step]) --> elements from\n seq[start:stop:step]\n starmap(fun, seq) --> fun(*seq[0]), fun(*seq[1]), ...\n tee(it, n=2) --> (it1, it2 , ... itn) splits one iterator into n\n takewhile(pred, seq) --> seq[0], seq[1], until pred fails\n zip_longest(p, q, ...) --> (p[0], q[0]), (p[1], q[1]), ... \n \n Combinatoric generators:\n product(p, q, ... [repeat=1]) --> cartesian product\n permutations(p[, r])\n combinations(p, r)\n combinations_with_replacement(p, r)\n\nCLASSES\n builtins.object\n accumulate\n chain\n combinations\n combinations_with_replacement\n compress\n count\n cycle\n dropwhile\n filterfalse\n groupby\n islice\n permutations\n product\n repeat\n starmap\n takewhile\n zip_longest\n \n class accumulate(builtins.object)\n | accumulate(iterable[, func]) --> accumulate object\n | \n | Return series of accumulated sums (or other binary function results).\n | \n | Methods defined here:\n | \n | __getattribute__(self, name, /)\n | Return getattr(self, name).\n | \n | __iter__(self, /)\n | Implement iter(self).\n | \n | __next__(self, /)\n | Implement next(self).\n | \n | __reduce__(...)\n | Return state information for pickling.\n | \n | __setstate__(...)\n | Set state information for unpickling.\n | \n | ----------------------------------------------------------------------\n | Static methods defined here:\n | \n | __new__(*args, **kwargs) from builtins.type\n | Create and return a new object. See help(type) for accurate signature.\n \n class chain(builtins.object)\n | chain(*iterables) --> chain object\n | \n | Return a chain object whose .__next__() method returns elements from the\n | first iterable until it is exhausted, then elements from the next\n | iterable, until all of the iterables are exhausted.\n | \n | Methods defined here:\n | \n | __getattribute__(self, name, /)\n | Return getattr(self, name).\n | \n | __iter__(self, /)\n | Implement iter(self).\n | \n | __next__(self, /)\n | Implement next(self).\n | \n | __reduce__(...)\n | Return state information for pickling.\n | \n | __setstate__(...)\n | Set state information for unpickling.\n | \n | ----------------------------------------------------------------------\n | Class methods defined here:\n | \n | from_iterable(...) from builtins.type\n | chain.from_iterable(iterable) --> chain object\n | \n | Alternate chain() constructor taking a single iterable argument\n | that evaluates lazily.\n | \n | ----------------------------------------------------------------------\n | Static methods defined here:\n | \n | __new__(*args, **kwargs) from builtins.type\n | Create and return a new object. See help(type) for accurate signature.\n \n class combinations(builtins.object)\n | combinations(iterable, r) --> combinations object\n | \n | Return successive r-length combinations of elements in the iterable.\n | \n | combinations(range(4), 3) --> (0,1,2), (0,1,3), (0,2,3), (1,2,3)\n | \n | Methods defined here:\n | \n | __getattribute__(self, name, /)\n | Return getattr(self, name).\n | \n | __iter__(self, /)\n | Implement iter(self).\n | \n | __next__(self, /)\n | Implement next(self).\n | \n | __reduce__(...)\n | Return state information for pickling.\n | \n | __setstate__(...)\n | Set state information for unpickling.\n | \n | __sizeof__(...)\n | Returns size in memory, in bytes.\n | \n | ----------------------------------------------------------------------\n | Static methods defined here:\n | \n | __new__(*args, **kwargs) from builtins.type\n | Create and return a new object. See help(type) for accurate signature.\n \n class combinations_with_replacement(builtins.object)\n | combinations_with_replacement(iterable, r) --> combinations_with_replacement object\n | \n | Return successive r-length combinations of elements in the iterable\n | allowing individual elements to have successive repeats.\n | combinations_with_replacement('ABC', 2) --> AA AB AC BB BC CC\n | \n | Methods defined here:\n | \n | __getattribute__(self, name, /)\n | Return getattr(self, name).\n | \n | __iter__(self, /)\n | Implement iter(self).\n | \n | __next__(self, /)\n | Implement next(self).\n | \n | __reduce__(...)\n | Return state information for pickling.\n | \n | __setstate__(...)\n | Set state information for unpickling.\n | \n | __sizeof__(...)\n | Returns size in memory, in bytes.\n | \n | ----------------------------------------------------------------------\n | Static methods defined here:\n | \n | __new__(*args, **kwargs) from builtins.type\n | Create and return a new object. See help(type) for accurate signature.\n \n class compress(builtins.object)\n | compress(data, selectors) --> iterator over selected data\n | \n | Return data elements corresponding to true selector elements.\n | Forms a shorter iterator from selected data elements using the\n | selectors to choose the data elements.\n | \n | Methods defined here:\n | \n | __getattribute__(self, name, /)\n | Return getattr(self, name).\n | \n | __iter__(self, /)\n | Implement iter(self).\n | \n | __next__(self, /)\n | Implement next(self).\n | \n | __reduce__(...)\n | Return state information for pickling.\n | \n | ----------------------------------------------------------------------\n | Static methods defined here:\n | \n | __new__(*args, **kwargs) from builtins.type\n | Create and return a new object. See help(type) for accurate signature.\n \n class count(builtins.object)\n | count(start=0, step=1) --> count object\n | \n | Return a count object whose .__next__() method returns consecutive values.\n | Equivalent to:\n | \n | def count(firstval=0, step=1):\n | x = firstval\n | while 1:\n | yield x\n | x += step\n | \n | Methods defined here:\n | \n | __getattribute__(self, name, /)\n | Return getattr(self, name).\n | \n | __iter__(self, /)\n | Implement iter(self).\n | \n | __next__(self, /)\n | Implement next(self).\n | \n | __reduce__(...)\n | Return state information for pickling.\n | \n | __repr__(self, /)\n | Return repr(self).\n | \n | ----------------------------------------------------------------------\n | Static methods defined here:\n | \n | __new__(*args, **kwargs) from builtins.type\n | Create and return a new object. See help(type) for accurate signature.\n \n class cycle(builtins.object)\n | cycle(iterable) --> cycle object\n | \n | Return elements from the iterable until it is exhausted.\n | Then repeat the sequence indefinitely.\n | \n | Methods defined here:\n | \n | __getattribute__(self, name, /)\n | Return getattr(self, name).\n | \n | __iter__(self, /)\n | Implement iter(self).\n | \n | __next__(self, /)\n | Implement next(self).\n | \n | __reduce__(...)\n | Return state information for pickling.\n | \n | __setstate__(...)\n | Set state information for unpickling.\n | \n | ----------------------------------------------------------------------\n | Static methods defined here:\n | \n | __new__(*args, **kwargs) from builtins.type\n | Create and return a new object. See help(type) for accurate signature.\n \n class dropwhile(builtins.object)\n | dropwhile(predicate, iterable) --> dropwhile object\n | \n | Drop items from the iterable while predicate(item) is true.\n | Afterwards, return every element until the iterable is exhausted.\n | \n | Methods defined here:\n | \n | __getattribute__(self, name, /)\n | Return getattr(self, name).\n | \n | __iter__(self, /)\n | Implement iter(self).\n | \n | __next__(self, /)\n | Implement next(self).\n | \n | __reduce__(...)\n | Return state information for pickling.\n | \n | __setstate__(...)\n | Set state information for unpickling.\n | \n | ----------------------------------------------------------------------\n | Static methods defined here:\n | \n | __new__(*args, **kwargs) from builtins.type\n | Create and return a new object. See help(type) for accurate signature.\n \n class filterfalse(builtins.object)\n | filterfalse(function or None, sequence) --> filterfalse object\n | \n | Return those items of sequence for which function(item) is false.\n | If function is None, return the items that are false.\n | \n | Methods defined here:\n | \n | __getattribute__(self, name, /)\n | Return getattr(self, name).\n | \n | __iter__(self, /)\n | Implement iter(self).\n | \n | __next__(self, /)\n | Implement next(self).\n | \n | __reduce__(...)\n | Return state information for pickling.\n | \n | ----------------------------------------------------------------------\n | Static methods defined here:\n | \n | __new__(*args, **kwargs) from builtins.type\n | Create and return a new object. See help(type) for accurate signature.\n \n class groupby(builtins.object)\n | groupby(iterable, key=None) -> make an iterator that returns consecutive\n | keys and groups from the iterable. If the key function is not specified or\n | is None, the element itself is used for grouping.\n | \n | Methods defined here:\n | \n | __getattribute__(self, name, /)\n | Return getattr(self, name).\n | \n | __iter__(self, /)\n | Implement iter(self).\n | \n | __next__(self, /)\n | Implement next(self).\n | \n | __reduce__(...)\n | Return state information for pickling.\n | \n | __setstate__(...)\n | Set state information for unpickling.\n | \n | ----------------------------------------------------------------------\n | Static methods defined here:\n | \n | __new__(*args, **kwargs) from builtins.type\n | Create and return a new object. See help(type) for accurate signature.\n \n class islice(builtins.object)\n | islice(iterable, stop) --> islice object\n | islice(iterable, start, stop[, step]) --> islice object\n | \n | Return an iterator whose next() method returns selected values from an\n | iterable. If start is specified, will skip all preceding elements;\n | otherwise, start defaults to zero. Step defaults to one. If\n | specified as another value, step determines how many values are \n | skipped between successive calls. Works like a slice() on a list\n | but returns an iterator.\n | \n | Methods defined here:\n | \n | __getattribute__(self, name, /)\n | Return getattr(self, name).\n | \n | __iter__(self, /)\n | Implement iter(self).\n | \n | __next__(self, /)\n | Implement next(self).\n | \n | __reduce__(...)\n | Return state information for pickling.\n | \n | __setstate__(...)\n | Set state information for unpickling.\n | \n | ----------------------------------------------------------------------\n | Static methods defined here:\n | \n | __new__(*args, **kwargs) from builtins.type\n | Create and return a new object. See help(type) for accurate signature.\n \n class permutations(builtins.object)\n | permutations(iterable[, r]) --> permutations object\n | \n | Return successive r-length permutations of elements in the iterable.\n | \n | permutations(range(3), 2) --> (0,1), (0,2), (1,0), (1,2), (2,0), (2,1)\n | \n | Methods defined here:\n | \n | __getattribute__(self, name, /)\n | Return getattr(self, name).\n | \n | __iter__(self, /)\n | Implement iter(self).\n | \n | __next__(self, /)\n | Implement next(self).\n | \n | __reduce__(...)\n | Return state information for pickling.\n | \n | __setstate__(...)\n | Set state information for unpickling.\n | \n | __sizeof__(...)\n | Returns size in memory, in bytes.\n | \n | ----------------------------------------------------------------------\n | Static methods defined here:\n | \n | __new__(*args, **kwargs) from builtins.type\n | Create and return a new object. See help(type) for accurate signature.\n \n class product(builtins.object)\n | product(*iterables, repeat=1) --> product object\n | \n | Cartesian product of input iterables. Equivalent to nested for-loops.\n | \n | For example, product(A, B) returns the same as: ((x,y) for x in A for y in B).\n | The leftmost iterators are in the outermost for-loop, so the output tuples\n | cycle in a manner similar to an odometer (with the rightmost element changing\n | on every iteration).\n | \n | To compute the product of an iterable with itself, specify the number\n | of repetitions with the optional repeat keyword argument. For example,\n | product(A, repeat=4) means the same as product(A, A, A, A).\n | \n | product('ab', range(3)) --> ('a',0) ('a',1) ('a',2) ('b',0) ('b',1) ('b',2)\n | product((0,1), (0,1), (0,1)) --> (0,0,0) (0,0,1) (0,1,0) (0,1,1) (1,0,0) ...\n | \n | Methods defined here:\n | \n | __getattribute__(self, name, /)\n | Return getattr(self, name).\n | \n | __iter__(self, /)\n | Implement iter(self).\n | \n | __next__(self, /)\n | Implement next(self).\n | \n | __reduce__(...)\n | Return state information for pickling.\n | \n | __setstate__(...)\n | Set state information for unpickling.\n | \n | __sizeof__(...)\n | Returns size in memory, in bytes.\n | \n | ----------------------------------------------------------------------\n | Static methods defined here:\n | \n | __new__(*args, **kwargs) from builtins.type\n | Create and return a new object. See help(type) for accurate signature.\n \n class repeat(builtins.object)\n | repeat(object [,times]) -> create an iterator which returns the object\n | for the specified number of times. If not specified, returns the object\n | endlessly.\n | \n | Methods defined here:\n | \n | __getattribute__(self, name, /)\n | Return getattr(self, name).\n | \n | __iter__(self, /)\n | Implement iter(self).\n | \n | __length_hint__(...)\n | Private method returning an estimate of len(list(it)).\n | \n | __next__(self, /)\n | Implement next(self).\n | \n | __reduce__(...)\n | Return state information for pickling.\n | \n | __repr__(self, /)\n | Return repr(self).\n | \n | ----------------------------------------------------------------------\n | Static methods defined here:\n | \n | __new__(*args, **kwargs) from builtins.type\n | Create and return a new object. See help(type) for accurate signature.\n \n class starmap(builtins.object)\n | starmap(function, sequence) --> starmap object\n | \n | Return an iterator whose values are returned from the function evaluated\n | with an argument tuple taken from the given sequence.\n | \n | Methods defined here:\n | \n | __getattribute__(self, name, /)\n | Return getattr(self, name).\n | \n | __iter__(self, /)\n | Implement iter(self).\n | \n | __next__(self, /)\n | Implement next(self).\n | \n | __reduce__(...)\n | Return state information for pickling.\n | \n | ----------------------------------------------------------------------\n | Static methods defined here:\n | \n | __new__(*args, **kwargs) from builtins.type\n | Create and return a new object. See help(type) for accurate signature.\n \n class takewhile(builtins.object)\n | takewhile(predicate, iterable) --> takewhile object\n | \n | Return successive entries from an iterable as long as the \n | predicate evaluates to true for each entry.\n | \n | Methods defined here:\n | \n | __getattribute__(self, name, /)\n | Return getattr(self, name).\n | \n | __iter__(self, /)\n | Implement iter(self).\n | \n | __next__(self, /)\n | Implement next(self).\n | \n | __reduce__(...)\n | Return state information for pickling.\n | \n | __setstate__(...)\n | Set state information for unpickling.\n | \n | ----------------------------------------------------------------------\n | Static methods defined here:\n | \n | __new__(*args, **kwargs) from builtins.type\n | Create and return a new object. See help(type) for accurate signature.\n \n class zip_longest(builtins.object)\n | zip_longest(iter1 [,iter2 [...]], [fillvalue=None]) --> zip_longest object\n | \n | Return a zip_longest object whose .__next__() method returns a tuple where\n | the i-th element comes from the i-th iterable argument. The .__next__()\n | method continues until the longest iterable in the argument sequence\n | is exhausted and then it raises StopIteration. When the shorter iterables\n | are exhausted, the fillvalue is substituted in their place. The fillvalue\n | defaults to None or can be specified by a keyword argument.\n | \n | Methods defined here:\n | \n | __getattribute__(self, name, /)\n | Return getattr(self, name).\n | \n | __iter__(self, /)\n | Implement iter(self).\n | \n | __next__(self, /)\n | Implement next(self).\n | \n | __reduce__(...)\n | Return state information for pickling.\n | \n | __setstate__(...)\n | Set state information for unpickling.\n | \n | ----------------------------------------------------------------------\n | Static methods defined here:\n | \n | __new__(*args, **kwargs) from builtins.type\n | Create and return a new object. See help(type) for accurate signature.\n\nFUNCTIONS\n tee(...)\n tee(iterable, n=2) --> tuple of n independent iterators.\n\nFILE\n (built-in)\n\n\n"
],
[
"a = itertools.combinations(\"6C 7C 8C 9C TC 5C JS\".split(),5)\nlist(a)",
"_____no_output_____"
],
[
"def best_hand(hand):\n combs = itertools.combinations(hand,5)\n comb_rank = [(comb,hand_rank(comb)) for comb in combs]\n comb_rank.sort(reverse=True,key=lambda x : x[1])\n return comb_rank[0][0]\n\ntest_best_hand()",
"_____no_output_____"
],
[
"## answer \ndef best_hand(hand):\n return max(itertools.combinations(hand,5), key=hand_rank)",
"_____no_output_____"
]
],
[
[
"2. wild jokers\n\nWrite a function best_wild_hand(hand) that takes as\ninput a 7-card hand and returns the best 5 card hand.\n\nIn this problem, it is possible for a hand to include\njokers. Jokers will be treated as 'wild cards' which\ncan take any rank or suit of the same color. \n\nThe \nblack joker, '?B', can be used as any spade or club\nand the red joker, '?R', can be used as any heart \nor diamond.",
"_____no_output_____"
]
],
[
[
"def best_wild_hand(hand):\n car_res = [hand.copy()]\n if \"?R\" in hand:\n car_res[0].remove(\"?R\")\n variation = [i+j for i in \"23456789TJQKA\" for j in \"HD\"]\n car_res = [j+[i] for i in variation for j in car_res] \n if \"?B\" in hand:\n for i in car_res:\n if \"?B\" in i:\n i.remove(\"?B\")\n variation = [i+j for i in \"23456789TJQKA\" for j in \"SC\"]\n car_res = [j+[i] for i in variation for j in car_res]\n results = [max(itertools.combinations(x,5),key= hand_rank) for x in car_res]\n return max(results,key= hand_rank)",
"_____no_output_____"
],
[
"def test_best_wild_hand():\n assert (sorted(best_wild_hand(\"6C 7C 8C 9C TC 5C ?B\".split()))\n == ['7C', '8C', '9C', 'JC', 'TC'])\n assert (sorted(best_wild_hand(\"TD TC 5H 5C 7C ?R ?B\".split()))\n == ['7C', 'TC', 'TD', 'TH', 'TS'])\n assert (sorted(best_wild_hand(\"JD TC TH 7C 7D 7S 7H\".split()))\n == ['7C', '7D', '7H', '7S', 'JD'])\n return 'test_best_wild_hand passes'",
"_____no_output_____"
],
[
"test_best_wild_hand()",
"_____no_output_____"
]
],
[
[
"## answer \n",
"_____no_output_____"
]
],
[
[
"def best_wild_card(hand):\n hands = set(best_hand(h) for h in itertools.product(*map(replacements,hand)))\n \n return max(hands,key=hand_rank)\n\ndef replacements(card):\n if card == \"?B\": return blackcards\n elif card == \"?R\": return readcards\n else: return [card]\n ",
"_____no_output_____"
]
],
[
[
"## Office Hours\n\n### how to trade-off time spent on research and writing own code ?\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"raw"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"raw"
]
] |
d0e1322cbf66c6f7c82f50ef081afc58f32fe5c0 | 54,245 | ipynb | Jupyter Notebook | day-1/session-2.ipynb | vchrombie/da-python | 11c23847e3bb2cb30112d894d7bf9b58a2af795d | [
"MIT"
] | 1 | 2019-05-23T16:45:39.000Z | 2019-05-23T16:45:39.000Z | day-1/session-2.ipynb | vchrombie/da-python | 11c23847e3bb2cb30112d894d7bf9b58a2af795d | [
"MIT"
] | null | null | null | day-1/session-2.ipynb | vchrombie/da-python | 11c23847e3bb2cb30112d894d7bf9b58a2af795d | [
"MIT"
] | 2 | 2019-05-23T12:03:23.000Z | 2019-05-27T05:43:25.000Z | 20.248227 | 1,062 | 0.476892 | [
[
[
"# Basics",
"_____no_output_____"
]
],
[
[
"print(\"Hello World!\")",
"Hello World!\n"
],
[
"# This is comment!",
"_____no_output_____"
],
[
"bread=10",
"_____no_output_____"
],
[
"print(bread)",
"10\n"
],
[
"bread=input()",
"12\n"
],
[
"bread",
"_____no_output_____"
],
[
"43+5",
"_____no_output_____"
],
[
"'43'+5",
"_____no_output_____"
]
],
[
[
"**Find out the reason behind the above error.**\n\n**Tip**: Copy the error and search in [Google](https://www.google.com/).",
"_____no_output_____"
]
],
[
[
"8",
"_____no_output_____"
],
[
"8+3",
"_____no_output_____"
]
],
[
[
"### Data Types \n\nIntegers: -2, -1, 0, 1, 2, 3, 4, 5\n\nFloating-point numbers: -1.25, -1.0, --0.5, 0.0, 0.5, 1.0, 1.25\n\nStrings: 'a', 'aa', 'aaa', 'Hello!', '11 cats'",
"_____no_output_____"
],
[
"**Math Operations**",
"_____no_output_____"
]
],
[
[
"#Addition\n2+3",
"_____no_output_____"
],
[
"#Subtraction\n8-3",
"_____no_output_____"
],
[
"#Multiplication\n2*4",
"_____no_output_____"
],
[
"#Division\n42/10",
"_____no_output_____"
],
[
"# Exponent\n2**4",
"_____no_output_____"
],
[
"#Modulus/Remainder\n42%10",
"_____no_output_____"
],
[
"#Floored Quotient\n42//10",
"_____no_output_____"
]
],
[
[
"**Note**: The precedence rule is BODMAS.\n\nThe ** operator is evaluated first; the *, /, //, and % operators are evaluated next, from left to right; and the + and - operators are evaluated last (also from left to right). You can use parentheses to override the usual precedence if you need to. ",
"_____no_output_____"
]
],
[
[
"2 + 3 * 6",
"_____no_output_____"
],
[
"(2 + 3) * 6",
"_____no_output_____"
],
[
"48565878 * 578453",
"_____no_output_____"
],
[
"2 ** 8",
"_____no_output_____"
],
[
"23 / 7",
"_____no_output_____"
],
[
"23 // 7",
"_____no_output_____"
],
[
"23 % 7",
"_____no_output_____"
],
[
"#extra spaces? no problem \n#python is chill :P\n2 + 2",
"_____no_output_____"
],
[
"(5 - 1) * ((7 + 1) / (3 - 1))",
"_____no_output_____"
]
],
[
[
"#### Explanation\n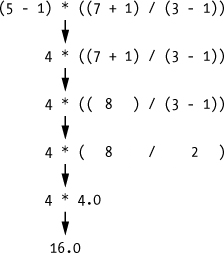",
"_____no_output_____"
]
],
[
[
"5 +",
"_____no_output_____"
],
[
"42 + 5 + * 2",
"_____no_output_____"
]
],
[
[
"The operations applicable for integers are also applicable for floating numbers too.",
"_____no_output_____"
]
],
[
[
"bread=10",
"_____no_output_____"
],
[
"bread",
"_____no_output_____"
],
[
"print(bread)",
"10\n"
],
[
"butter=3",
"_____no_output_____"
],
[
"bread+butter",
"_____no_output_____"
],
[
"print(bread+butter)",
"13\n"
],
[
"bread=bread+2\nbread",
"_____no_output_____"
],
[
"bread++",
"_____no_output_____"
]
],
[
[
"**Note**: Coding in python is entirely different from c/cpp.",
"_____no_output_____"
],
[
"### Strings",
"_____no_output_____"
]
],
[
[
"msg='Hello'",
"_____no_output_____"
],
[
"msg",
"_____no_output_____"
],
[
"print(msg)",
"Hello\n"
],
[
"msg='Goodbye'",
"_____no_output_____"
],
[
"print(msg)",
"Goodbye\n"
],
[
"len(msg)\nprint(len(msg))",
"7\n"
],
[
"msg+1",
"_____no_output_____"
],
[
"msg+'1'",
"_____no_output_____"
],
[
"msg+str(1)",
"_____no_output_____"
]
],
[
[
"### Variable Names\n\n**Valid** ✔️\n\nbalance\n\ncurrentBalance \n\ncurrent_balance \n\n_spam\n\nSPAM\n\naccount4\n\n----\n\n**Invalid** ❌\n\n'hello' (special characters like ' are not allowed)\n\ncurrent-balance (hyphens are not allowed)\n\n4account (can’t begin with a number)\n\n42 (can’t begin with a number)\n\ncurrent balance (spaces are not allowed)\n\ntotal_$um (special characters like $ are not allowed)",
"_____no_output_____"
]
],
[
[
"#INPUT: Hi! :)\nmsg=input()",
"Hi :)\n"
],
[
"print(msg)",
"Hi :)\n"
],
[
"print(type(msg))",
"<class 'str'>\n"
],
[
"#INPUT: 12\nmsg=input()",
"12\n"
],
[
"print(msg)",
"12\n"
],
[
"print(type(msg))",
"<class 'str'>\n"
],
[
"#integer input\n#INPUT : 12\nmsg=int(input())",
"12\n"
],
[
"print(msg)",
"12\n"
],
[
"print(type(msg))",
"<class 'int'>\n"
],
[
"#change the datatype\nmsg=str(msg)",
"_____no_output_____"
],
[
"print(msg)",
"12\n"
],
[
"print(type(msg))",
"<class 'str'>\n"
],
[
"print(int(msg)+1)",
"13\n"
],
[
"print(float(msg)+1)",
"13.0\n"
],
[
"print(int(7.7))",
"7\n"
],
[
"print(int(7.7) + 1)",
"8\n"
]
],
[
[
"Please understand this small program",
"_____no_output_____"
]
],
[
[
"print('Hello world!')\nprint('What is your name?') # ask for their name\nmyName = input()\nprint('It is good to meet you, ' + myName)\nprint('The length of your name is:')\nprint(len(myName))\nprint('What is your age?') # ask for their age\nmyAge = input()\nprint('You will be ' + str(int(myAge) + 1) + ' in a year.')",
"Hello world!\nWhat is your name?\nVenu\nIt is good to meet you, Venu\nThe length of your name is:\n4\nWhat is your age?\n21\nYou will be 22 in a year.\n"
]
],
[
[
"## Task 1\nWrite a program for polling booth. It should ask a name, his age and ask for vote (NDA/UPA).",
"_____no_output_____"
],
[
"# Flow Control",
"_____no_output_____"
]
],
[
[
"print(True, False)\nprint(1,0)",
"True False\n1 0\n"
]
],
[
[
"### Operators",
"_____no_output_____"
]
],
[
[
"#equal to\n2==3",
"_____no_output_____"
],
[
"#not equal to\n2!=3",
"_____no_output_____"
],
[
"#greater than\n2>3",
"_____no_output_____"
],
[
"#less than\n2<3",
"_____no_output_____"
],
[
"#less/greater than equal to\nprint(2>=3)\nprint(2<=3)",
"False\nTrue\n"
],
[
"1==True",
"_____no_output_____"
],
[
"print('hello' == 'hello')\nprint('hello' == 'Hello')",
"True\nFalse\n"
],
[
"True!=False",
"_____no_output_____"
],
[
"42==42.0",
"_____no_output_____"
],
[
"42=='42'",
"_____no_output_____"
],
[
"bread=21\nbread>10",
"_____no_output_____"
]
],
[
[
"### Binary and Boolean Operations",
"_____no_output_____"
]
],
[
[
"True and True",
"_____no_output_____"
],
[
"False or False",
"_____no_output_____"
],
[
"not False",
"_____no_output_____"
]
],
[
[
"### Conditional Statements",
"_____no_output_____"
]
],
[
[
"if(4<5):\n print(\"Yo!\")",
"Yo!\n"
],
[
"bread=int(input())\nif(bread>10):\n print(\"I have 10+ bread\")\nelif(bread==10):\n print(\"I have exactly 10 bread\")\nelse:\n print(\"I have less than 10 bread\")",
"12\nI have 10+ bread\n"
],
[
"if(True):\n print(\"This is how you overide a conditional statement.\")",
"This is how you overide a conditional statement.\n"
],
[
"if(False):\n print(\"This will not print anything.\")",
"_____no_output_____"
],
[
"if(input()):\n print(\"This will print anything.\")",
"False\nThis will print anything.\n"
]
],
[
[
"You may be wondering why the above code worked. It worked because `input()` always take the input as *string* format not *boolen* format.\n\nYou can convert it into *boolean* by modifying it as `bool(input())`",
"_____no_output_____"
]
],
[
[
"bread=12\nif(bread):\n print(\"Every value other than 0 is considered as True\")\n\nbread=0\nif(bread):\n print(\"I told you already. :P\")",
"Every value other than 0 is considered as True\n"
]
],
[
[
"## Taks 2\n\nImplement this following flowchart \nTake the initial values as\n```python\nname='Dracula'\nage=4000\n```\n",
"_____no_output_____"
],
[
"### Loops",
"_____no_output_____"
],
[
"### `while`",
"_____no_output_____"
]
],
[
[
"bread=0\nwhile(bread<5):\n print(\"need more bread\")\n bread=bread+1",
"need more bread\nneed more bread\nneed more bread\nneed more bread\nneed more bread\n"
],
[
"\"\"\"\nwhile True:\n print(\"This is an infinite loop!!\")\n\"\"\"",
"_____no_output_____"
]
],
[
[
"Please have a look at the below code and understand it. ",
"_____no_output_____"
]
],
[
[
"name = ''\nwhile name != 'your name':\n print('Please type your name.')\n name = input()\nprint('Thank you!')",
"Please type your name.\nVenu\nPlease type your name.\nVenu\nPlease type your name.\nugh, stop it\nPlease type your name.\nok, i got this\nPlease type your name.\nyour name\nThank you!\n"
]
],
[
[
"The above code will ask your input again and again until you give `your name` as input. You can build infinite loops accordingly.\n\nThe below code is a re-written version of the above. You can control infinite loops with valid `break` statements.",
"_____no_output_____"
]
],
[
[
"while True:\n print('Please type your name.')\n name = input()\n if name == 'your name':\n break\nprint('Thank you!')",
"Please type your name.\nVenu\nPlease type your name.\nyour name\nThank you!\n"
]
],
[
[
"The keyword `continue` skips all the statements which are below itself. Look at the following",
"_____no_output_____"
]
],
[
[
"bread=0\nwhile(bread<5):\n print(\"bread added\")\n bread=bread+1\n if(bread<3):\n print(\"I am hungry.\")\n continue\n print(\"I need more bread.\")",
"bread added\nI am hungry.\nbread added\nI am hungry.\nbread added\nbread added\nbread added\n"
]
],
[
[
"### `for`",
"_____no_output_____"
]
],
[
[
"for i in range(3):\n print(\"I need \"+str(i)+\" bread.\")",
"I need 0 bread.\nI need 1 bread.\nI need 2 bread.\n"
],
[
"#sum of n natural numbers program\ntotal=0\nn=int(input(\"Enter the n: \"))\nfor i in range(n+1):\n total=total+i\nprint(total)",
"Enter the n: 5\n15\n"
]
],
[
[
"**Note**: You can actually use a `while` loop to do the same thing as a `for` loop; `for` loops are just more concise.",
"_____no_output_____"
],
[
"`starting`, `stopping` and `stepping` arguments in `for` loop",
"_____no_output_____"
]
],
[
[
"for i in range(0,10):\n print(i)",
"0\n1\n2\n3\n4\n5\n6\n7\n8\n9\n"
],
[
"for i in range(0,10,3):\n print(i)",
"0\n3\n6\n9\n"
]
],
[
[
"## Task 3\nWrite a program for polling booth. It should ask a name, his age and ask for vote (NDA/UPA/Others). It should verify your age too. You need to ask for vote untill you see no voters left. Have seperate counters for the parties and increase the counter accordingly.",
"_____no_output_____"
],
[
"# Importing Modules",
"_____no_output_____"
],
[
"Python comes with many set of modules which can be used. Each module is a Python program that contains a related group of functions that can be embedded in your programs. For example, the math module has mathematics-related functions, the random module has random number–related functions, and so on.\n\nBefore you can use the functions in a module, you must import the module with an import statement.",
"_____no_output_____"
]
],
[
[
"#importing a module normally\nimport random\nfor i in range(5):\n print(random.randint(1, 10))",
"8\n4\n4\n9\n9\n"
],
[
"#importing a module using an object\nimport random as rd\nfor i in range(5):\n print(rd.randint(1, 10))",
"1\n4\n7\n4\n8\n"
],
[
"#importing one function from that module \n#here, other functions of that module doesn't work\nfrom random import randint\nfor i in range(5):\n print(randint(1, 10))",
"10\n4\n8\n1\n3\n"
],
[
"#importing one function using an object\nfrom random import randint as ri\nfor i in range(5):\n print(ri(1, 10))",
"10\n3\n8\n8\n3\n"
],
[
"#importing all functions from that module\nfrom random import *\nfor i in range(5):\n print(randint(1, 10))",
"9\n5\n9\n2\n9\n"
]
],
[
[
"# Functions",
"_____no_output_____"
]
],
[
[
"def getBread():\n print(\"bread!\")\n print(\"bread!!!\")\n\ngetBread()",
"bread!\nbread!!!\n"
],
[
"getBread()\ngetBread()",
"bread!\nbread!!!\nbread!\nbread!!!\n"
],
[
"def getBread(number):\n print(\"I want \"+str(number)+\" bread.\")\ngetBread(3)\ngetBread(5)",
"I want 3 bread.\nI want 5 bread.\n"
],
[
"getBread(8)\ngetBread()",
"I want 8 bread.\n"
]
],
[
[
"**Error**: python is not like C/C++. You can define only one funtion with one name. You cannot differentiate with arguments.",
"_____no_output_____"
]
],
[
[
"def makeItEven(n):\n return n*2\n\n\nresult=makeItEven(3)\nprint(result)\nresult=makeItEven(2)\nprint(result)",
"6\n4\n"
],
[
"def makeItEven(n):\n return n*2\n\n\nprint(makeItEven(3))\nprint(makeItEven(2))",
"6\n4\n"
]
],
[
[
"`None` represents absence of any value.",
"_____no_output_____"
]
],
[
[
"def getNothing():\n return \n\ngetNothing()\nprint(getNothing())",
"None\n"
],
[
"def getNone():\n return None\n\ngetNone()\nprint(getNone())",
"None\n"
]
],
[
[
"**Fact**: Behind the scenes, Python adds return `None` to the end of any function definition with no return statement. This is similar to how a `while` or `for` loop implicitly ends with a continue statement. ",
"_____no_output_____"
],
[
"### Scope (Global and Local)",
"_____no_output_____"
]
],
[
[
"def getDosa():\n dosa=8\n\ngetDosa()\nprint(dosa)",
"_____no_output_____"
]
],
[
[
"**Error**: The above error is because the variable `dosa` is defined only fro the `getDosa` function and out of the defined scope, a variable cannot be used.",
"_____no_output_____"
]
],
[
[
"dosa=3\n\ndef getDosa():\n print(\"dosa=\",dosa)\n\ngetDosa()\nprint(dosa)",
"dosa= 3\n3\n"
],
[
"dosa=3\n\ndef getDosa():\n dosa=8\n print(\"dosa=\",dosa)\n\ngetDosa()\nprint(\"dosa=\",dosa)",
"dosa= 8\ndosa= 3\n"
],
[
"def getDosa():\n global dosa\n dosa=8\n print(\"dosa=\",dosa)\n\ngetDosa()\nprint(\"dosa=\",dosa)",
"dosa= 8\ndosa= 8\n"
],
[
"def getDosa():\n print(\"dosa=\",dosa)\n dosa=8 #1\n\ndosa=3 #2\ngetDosa()\nprint(\"dosa=\",dosa)",
"_____no_output_____"
]
],
[
[
"**Error**: This error happens because Python sees that there is an assignment statement for `dosa` in the `getDosa` function #1 and therefore considers `dosa` to be local. But because `print(\"dosa=\",dosa)` is executed before `dosa` is assigned anything, the local variable `dosa` doesn’t exist. Python will not fall back to using the global `dosa` variable #2.",
"_____no_output_____"
],
[
"### Error Handling",
"_____no_output_____"
]
],
[
[
"def divideBy(n):\n return 4/n\n\nprint(divideBy(4))\nprint(divideBy(1))\nprint(divideBy(0))",
"1.0\n4.0\n"
]
],
[
[
"**Note**: The division by zero is not possible, so it raised error. Instead of stopping the program abruptly, we can handle the error (using `try-except`) such that an error message is displayed and the program finished the execution without any interruption.",
"_____no_output_____"
]
],
[
[
"def divideBy(n):\n try:\n return 4/n\n except ZeroDivisionError:\n print(\"Invalid argument.\")\n \nprint(divideBy(4))\nprint(divideBy(1))\nprint(divideBy(0))",
"1.0\n4.0\nInvalid argument.\nNone\n"
],
[
"def divideBy(n):\n return 4/n\n\ntry:\n print(divideBy(4))\n print(divideBy(1))\n print(divideBy(0))\nexcept ZeroDivisionError:\n print(\"Invalid argument.\")",
"1.0\n4.0\nInvalid argument.\n"
]
],
[
[
"## Task-4\n\nImplement the following scenario. It is *Guess the number* game. (need to be be the exact similar)\n```\nI am thinking of a number between 1 and 20.\nTake a guess.\n10\nYour guess is too low.\nTake a guess.\n15\nYour guess is too low.\nTake a guess.\n17\nYour guess is too high.\nTake a guess.\n16\nGood job! You guessed my number in 4 guesses!\n```",
"_____no_output_____"
],
[
"## Task-5\n\n### The Collatz Sequence\n\nWrite a function named `collatz()` that has one parameter named number. If number is even, then collatz() should print `number // 2` and return this value. If number is odd, then collatz() should print and `return 3 * number + 1`.\n\n*Hint*: An integer number is even if `number % 2 == 0`, and it’s odd if `number % 2 == 1`.\n\n",
"_____no_output_____"
],
[
"## Task-6\n\n### The Input Sequence\n\nAdd `try-except` statements to the previous project to detect whether the user types in a noninteger string. Normally, the `int()` function will raise a `ValueError` error if it is passed a noninteger string, as in `int('puppy')`. In the `except` clause, print a message to the user saying they must enter an integer.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
d0e13d0313d9343056053d9af83eea4f07b0e5e4 | 26,772 | ipynb | Jupyter Notebook | DR_colab.ipynb | tradman1/pytorch-pretrained-BERT | 836aa1791452224fac642cf29d3d748e82465415 | [
"Apache-2.0"
] | null | null | null | DR_colab.ipynb | tradman1/pytorch-pretrained-BERT | 836aa1791452224fac642cf29d3d748e82465415 | [
"Apache-2.0"
] | null | null | null | DR_colab.ipynb | tradman1/pytorch-pretrained-BERT | 836aa1791452224fac642cf29d3d748e82465415 | [
"Apache-2.0"
] | null | null | null | 26,772 | 26,772 | 0.755453 | [
[
[
"## Setup",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nDATA_PERCENT = 100",
"_____no_output_____"
]
],
[
[
"### Download data",
"_____no_output_____"
]
],
[
[
"!git clone https://gist.github.com/60c2bdb54d156a41194446737ce03e2e.git\n!mv 60c2bdb54d156a41194446737ce03e2e/download_glue_data.py .\n!rm -rf 60c2bdb54d156a41194446737ce03e2e",
"Cloning into '60c2bdb54d156a41194446737ce03e2e'...\nremote: Enumerating objects: 21, done.\u001b[K\nremote: Total 21 (delta 0), reused 0 (delta 0), pack-reused 21\u001b[K\nUnpacking objects: 100% (21/21), done.\n"
],
[
"%cd /content\n%run download_glue_data.py --data_dir glue_data --tasks SST",
"/content\nDownloading and extracting SST...\n\tCompleted!\n"
],
[
"task = \"SST-2\"\n\nwith open(f\"/content/glue_data/{task}/train.tsv\") as train_f:\n lines = train_f.read().splitlines()\n\nlines_np = np.array(lines)\nnp.random.shuffle(lines_np[1:])\n\n# take a percentage of shuffled dataset\nlines_np = lines_np[: int(lines_np.shape[0] * (DATA_PERCENT / 100.))]\n\n# rewrite train file with new reduced dataset\nwith open(f\"/content/glue_data/{task}/train.tsv\", \"w\") as train_f:\n train_f.write(\"\\n\".join(list(lines_np)))",
"_____no_output_____"
]
],
[
[
"### Install Pytorch BERT",
"_____no_output_____"
]
],
[
[
"#!git clone https://github.com/huggingface/pytorch-pretrained-BERT.git\n \nfrom google.colab import drive\ndrive.mount('/content/drive')\n# %cp -r /content/drive/My\\ Drive/dr/pytorch-pretrained-BERT /content\n\n!git clone https://github.com/tradman1/pytorch-pretrained-BERT.git",
"Go to this URL in a browser: https://accounts.google.com/o/oauth2/auth?client_id=947318989803-6bn6qk8qdgf4n4g3pfee6491hc0brc4i.apps.googleusercontent.com&redirect_uri=urn%3Aietf%3Awg%3Aoauth%3A2.0%3Aoob&scope=email%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fdocs.test%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fdrive%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fdrive.photos.readonly%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fpeopleapi.readonly&response_type=code\n\nEnter your authorization code:\n··········\nMounted at /content/drive\nCloning into 'pytorch-pretrained-BERT'...\nremote: Enumerating objects: 3358, done.\u001b[K\nremote: Total 3358 (delta 0), reused 0 (delta 0), pack-reused 3358\u001b[K\nReceiving objects: 100% (3358/3358), 1.56 MiB | 9.21 MiB/s, done.\nResolving deltas: 100% (2317/2317), done.\n"
],
[
"%cd /content/pytorch-pretrained-BERT",
"/content/pytorch-pretrained-BERT\n"
],
[
"!python setup.py install",
"running install\nrunning bdist_egg\nrunning egg_info\ncreating pytorch_pretrained_bert.egg-info\nwriting pytorch_pretrained_bert.egg-info/PKG-INFO\nwriting dependency_links to pytorch_pretrained_bert.egg-info/dependency_links.txt\nwriting entry points to pytorch_pretrained_bert.egg-info/entry_points.txt\nwriting requirements to pytorch_pretrained_bert.egg-info/requires.txt\nwriting top-level names to pytorch_pretrained_bert.egg-info/top_level.txt\nwriting manifest file 'pytorch_pretrained_bert.egg-info/SOURCES.txt'\nreading manifest template 'MANIFEST.in'\nwriting manifest file 'pytorch_pretrained_bert.egg-info/SOURCES.txt'\ninstalling library code to build/bdist.linux-x86_64/egg\nrunning install_lib\nrunning build_py\ncreating build\ncreating build/lib\ncreating build/lib/pytorch_pretrained_bert\ncopying pytorch_pretrained_bert/modeling.py -> build/lib/pytorch_pretrained_bert\ncopying pytorch_pretrained_bert/convert_transfo_xl_checkpoint_to_pytorch.py -> build/lib/pytorch_pretrained_bert\ncopying pytorch_pretrained_bert/convert_gpt2_checkpoint_to_pytorch.py -> build/lib/pytorch_pretrained_bert\ncopying pytorch_pretrained_bert/tokenization_transfo_xl.py -> build/lib/pytorch_pretrained_bert\ncopying pytorch_pretrained_bert/file_utils.py -> build/lib/pytorch_pretrained_bert\ncopying pytorch_pretrained_bert/convert_tf_checkpoint_to_pytorch.py -> build/lib/pytorch_pretrained_bert\ncopying pytorch_pretrained_bert/__main__.py -> build/lib/pytorch_pretrained_bert\ncopying pytorch_pretrained_bert/modeling_transfo_xl_utilities.py -> build/lib/pytorch_pretrained_bert\ncopying pytorch_pretrained_bert/optimization_openai.py -> build/lib/pytorch_pretrained_bert\ncopying pytorch_pretrained_bert/convert_openai_checkpoint_to_pytorch.py -> build/lib/pytorch_pretrained_bert\ncopying pytorch_pretrained_bert/tokenization.py -> build/lib/pytorch_pretrained_bert\ncopying pytorch_pretrained_bert/optimization.py -> build/lib/pytorch_pretrained_bert\ncopying pytorch_pretrained_bert/tokenization_openai.py -> build/lib/pytorch_pretrained_bert\ncopying pytorch_pretrained_bert/__init__.py -> build/lib/pytorch_pretrained_bert\ncopying pytorch_pretrained_bert/modeling_gpt2.py -> build/lib/pytorch_pretrained_bert\ncopying pytorch_pretrained_bert/modeling_transfo_xl.py -> build/lib/pytorch_pretrained_bert\ncopying pytorch_pretrained_bert/modeling_openai.py -> build/lib/pytorch_pretrained_bert\ncopying pytorch_pretrained_bert/tokenization_gpt2.py -> build/lib/pytorch_pretrained_bert\ncreating build/bdist.linux-x86_64\ncreating build/bdist.linux-x86_64/egg\ncreating build/bdist.linux-x86_64/egg/pytorch_pretrained_bert\ncopying build/lib/pytorch_pretrained_bert/modeling.py -> build/bdist.linux-x86_64/egg/pytorch_pretrained_bert\ncopying build/lib/pytorch_pretrained_bert/convert_transfo_xl_checkpoint_to_pytorch.py -> build/bdist.linux-x86_64/egg/pytorch_pretrained_bert\ncopying build/lib/pytorch_pretrained_bert/convert_gpt2_checkpoint_to_pytorch.py -> build/bdist.linux-x86_64/egg/pytorch_pretrained_bert\ncopying build/lib/pytorch_pretrained_bert/tokenization_transfo_xl.py -> build/bdist.linux-x86_64/egg/pytorch_pretrained_bert\ncopying build/lib/pytorch_pretrained_bert/file_utils.py -> build/bdist.linux-x86_64/egg/pytorch_pretrained_bert\ncopying build/lib/pytorch_pretrained_bert/convert_tf_checkpoint_to_pytorch.py -> build/bdist.linux-x86_64/egg/pytorch_pretrained_bert\ncopying build/lib/pytorch_pretrained_bert/__main__.py -> build/bdist.linux-x86_64/egg/pytorch_pretrained_bert\ncopying build/lib/pytorch_pretrained_bert/modeling_transfo_xl_utilities.py -> build/bdist.linux-x86_64/egg/pytorch_pretrained_bert\ncopying build/lib/pytorch_pretrained_bert/optimization_openai.py -> build/bdist.linux-x86_64/egg/pytorch_pretrained_bert\ncopying build/lib/pytorch_pretrained_bert/convert_openai_checkpoint_to_pytorch.py -> build/bdist.linux-x86_64/egg/pytorch_pretrained_bert\ncopying build/lib/pytorch_pretrained_bert/tokenization.py -> build/bdist.linux-x86_64/egg/pytorch_pretrained_bert\ncopying build/lib/pytorch_pretrained_bert/optimization.py -> build/bdist.linux-x86_64/egg/pytorch_pretrained_bert\ncopying build/lib/pytorch_pretrained_bert/tokenization_openai.py -> build/bdist.linux-x86_64/egg/pytorch_pretrained_bert\ncopying build/lib/pytorch_pretrained_bert/__init__.py -> build/bdist.linux-x86_64/egg/pytorch_pretrained_bert\ncopying build/lib/pytorch_pretrained_bert/modeling_gpt2.py -> build/bdist.linux-x86_64/egg/pytorch_pretrained_bert\ncopying build/lib/pytorch_pretrained_bert/modeling_transfo_xl.py -> build/bdist.linux-x86_64/egg/pytorch_pretrained_bert\ncopying build/lib/pytorch_pretrained_bert/modeling_openai.py -> build/bdist.linux-x86_64/egg/pytorch_pretrained_bert\ncopying build/lib/pytorch_pretrained_bert/tokenization_gpt2.py -> build/bdist.linux-x86_64/egg/pytorch_pretrained_bert\nbyte-compiling build/bdist.linux-x86_64/egg/pytorch_pretrained_bert/modeling.py to modeling.cpython-36.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/pytorch_pretrained_bert/convert_transfo_xl_checkpoint_to_pytorch.py to convert_transfo_xl_checkpoint_to_pytorch.cpython-36.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/pytorch_pretrained_bert/convert_gpt2_checkpoint_to_pytorch.py to convert_gpt2_checkpoint_to_pytorch.cpython-36.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/pytorch_pretrained_bert/tokenization_transfo_xl.py to tokenization_transfo_xl.cpython-36.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/pytorch_pretrained_bert/file_utils.py to file_utils.cpython-36.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/pytorch_pretrained_bert/convert_tf_checkpoint_to_pytorch.py to convert_tf_checkpoint_to_pytorch.cpython-36.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/pytorch_pretrained_bert/__main__.py to __main__.cpython-36.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/pytorch_pretrained_bert/modeling_transfo_xl_utilities.py to modeling_transfo_xl_utilities.cpython-36.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/pytorch_pretrained_bert/optimization_openai.py to optimization_openai.cpython-36.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/pytorch_pretrained_bert/convert_openai_checkpoint_to_pytorch.py to convert_openai_checkpoint_to_pytorch.cpython-36.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/pytorch_pretrained_bert/tokenization.py to tokenization.cpython-36.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/pytorch_pretrained_bert/optimization.py to optimization.cpython-36.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/pytorch_pretrained_bert/tokenization_openai.py to tokenization_openai.cpython-36.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/pytorch_pretrained_bert/__init__.py to __init__.cpython-36.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/pytorch_pretrained_bert/modeling_gpt2.py to modeling_gpt2.cpython-36.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/pytorch_pretrained_bert/modeling_transfo_xl.py to modeling_transfo_xl.cpython-36.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/pytorch_pretrained_bert/modeling_openai.py to modeling_openai.cpython-36.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/pytorch_pretrained_bert/tokenization_gpt2.py to tokenization_gpt2.cpython-36.pyc\ncreating build/bdist.linux-x86_64/egg/EGG-INFO\ncopying pytorch_pretrained_bert.egg-info/PKG-INFO -> build/bdist.linux-x86_64/egg/EGG-INFO\ncopying pytorch_pretrained_bert.egg-info/SOURCES.txt -> build/bdist.linux-x86_64/egg/EGG-INFO\ncopying pytorch_pretrained_bert.egg-info/dependency_links.txt -> build/bdist.linux-x86_64/egg/EGG-INFO\ncopying pytorch_pretrained_bert.egg-info/entry_points.txt -> build/bdist.linux-x86_64/egg/EGG-INFO\ncopying pytorch_pretrained_bert.egg-info/requires.txt -> build/bdist.linux-x86_64/egg/EGG-INFO\ncopying pytorch_pretrained_bert.egg-info/top_level.txt -> build/bdist.linux-x86_64/egg/EGG-INFO\nzip_safe flag not set; analyzing archive contents...\ncreating dist\ncreating 'dist/pytorch_pretrained_bert-0.6.2-py3.6.egg' and adding 'build/bdist.linux-x86_64/egg' to it\nremoving 'build/bdist.linux-x86_64/egg' (and everything under it)\nProcessing pytorch_pretrained_bert-0.6.2-py3.6.egg\nCopying pytorch_pretrained_bert-0.6.2-py3.6.egg to /usr/local/lib/python3.6/dist-packages\nAdding pytorch-pretrained-bert 0.6.2 to easy-install.pth file\nInstalling pytorch_pretrained_bert script to /usr/local/bin\n\nInstalled /usr/local/lib/python3.6/dist-packages/pytorch_pretrained_bert-0.6.2-py3.6.egg\nProcessing dependencies for pytorch-pretrained-bert==0.6.2\nSearching for regex\nReading https://pypi.org/simple/regex/\nDownloading https://files.pythonhosted.org/packages/6f/4e/1b178c38c9a1a184288f72065a65ca01f3154df43c6ad898624149b8b4e0/regex-2019.06.08.tar.gz#sha256=84daedefaa56320765e9c4d43912226d324ef3cc929f4d75fa95f8c579a08211\nBest match: regex 2019.6.8\nProcessing regex-2019.06.08.tar.gz\nWriting /tmp/easy_install-fexnv_or/regex-2019.06.08/setup.cfg\nRunning regex-2019.06.08/setup.py -q bdist_egg --dist-dir /tmp/easy_install-fexnv_or/regex-2019.06.08/egg-dist-tmp-z0lingzd\nBASE_DIR is /tmp/easy_install-fexnv_or/regex-2019.06.08\n/usr/local/lib/python3.6/dist-packages/setuptools/dist.py:472: UserWarning: Normalizing '2019.06.08' to '2019.6.8'\n normalized_version,\n\u001b[01m\u001b[Kregex_3/_regex.c:\u001b[m\u001b[K In function ‘\u001b[01m\u001b[Kfuzzy_match_group_fld\u001b[m\u001b[K’:\n\u001b[01m\u001b[Kregex_3/_regex.c:11341:10:\u001b[m\u001b[K \u001b[01;35m\u001b[Kwarning: \u001b[m\u001b[K‘\u001b[01m\u001b[Kdata.new_text_pos\u001b[m\u001b[K’ may be used uninitialized in this function [\u001b[01;35m\u001b[K-Wmaybe-uninitialized\u001b[m\u001b[K]\n if (!\u001b[01;35m\u001b[Krecord_fuzzy(state, data.fuzzy_type, data.new_text_pos - data.step)\u001b[m\u001b[K)\n \u001b[01;35m\u001b[K^~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\u001b[m\u001b[K\n\u001b[01m\u001b[Kregex_3/_regex.c:\u001b[m\u001b[K In function ‘\u001b[01m\u001b[Kfuzzy_match_string_fld\u001b[m\u001b[K’:\n\u001b[01m\u001b[Kregex_3/_regex.c:11116:10:\u001b[m\u001b[K \u001b[01;35m\u001b[Kwarning: \u001b[m\u001b[K‘\u001b[01m\u001b[Kdata.new_text_pos\u001b[m\u001b[K’ may be used uninitialized in this function [\u001b[01;35m\u001b[K-Wmaybe-uninitialized\u001b[m\u001b[K]\n if (!\u001b[01;35m\u001b[Krecord_fuzzy(state, data.fuzzy_type, data.new_text_pos - data.step)\u001b[m\u001b[K)\n \u001b[01;35m\u001b[K^~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\u001b[m\u001b[K\n\u001b[01m\u001b[Kregex_3/_regex.c:\u001b[m\u001b[K In function ‘\u001b[01m\u001b[Kbasic_match\u001b[m\u001b[K’:\n\u001b[01m\u001b[Kregex_3/_regex.c:11446:10:\u001b[m\u001b[K \u001b[01;35m\u001b[Kwarning: \u001b[m\u001b[K‘\u001b[01m\u001b[Kdata.new_text_pos\u001b[m\u001b[K’ may be used uninitialized in this function [\u001b[01;35m\u001b[K-Wmaybe-uninitialized\u001b[m\u001b[K]\n if (!\u001b[01;35m\u001b[Krecord_fuzzy(state, data.fuzzy_type, data.new_text_pos - data.step)\u001b[m\u001b[K)\n \u001b[01;35m\u001b[K^~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\u001b[m\u001b[K\n\u001b[01m\u001b[Kregex_3/_regex.c:11360:18:\u001b[m\u001b[K \u001b[01;36m\u001b[Knote: \u001b[m\u001b[K‘\u001b[01m\u001b[Kdata.new_text_pos\u001b[m\u001b[K’ was declared here\n RE_FuzzyData \u001b[01;36m\u001b[Kdata\u001b[m\u001b[K;\n \u001b[01;36m\u001b[K^~~~\u001b[m\u001b[K\n\u001b[01m\u001b[Kregex_3/_regex.c:11215:10:\u001b[m\u001b[K \u001b[01;35m\u001b[Kwarning: \u001b[m\u001b[K‘\u001b[01m\u001b[Kdata.new_text_pos\u001b[m\u001b[K’ may be used uninitialized in this function [\u001b[01;35m\u001b[K-Wmaybe-uninitialized\u001b[m\u001b[K]\n if (!\u001b[01;35m\u001b[Krecord_fuzzy(state, data.fuzzy_type, data.new_text_pos - data.step)\u001b[m\u001b[K)\n \u001b[01;35m\u001b[K^~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\u001b[m\u001b[K\n\u001b[01m\u001b[Kregex_3/_regex.c:11134:18:\u001b[m\u001b[K \u001b[01;36m\u001b[Knote: \u001b[m\u001b[K‘\u001b[01m\u001b[Kdata.new_text_pos\u001b[m\u001b[K’ was declared here\n RE_FuzzyData \u001b[01;36m\u001b[Kdata\u001b[m\u001b[K;\n \u001b[01;36m\u001b[K^~~~\u001b[m\u001b[K\nzip_safe flag not set; analyzing archive contents...\nregex.__pycache__._regex.cpython-36: module references __file__\ncreating /usr/local/lib/python3.6/dist-packages/regex-2019.6.8-py3.6-linux-x86_64.egg\nExtracting regex-2019.6.8-py3.6-linux-x86_64.egg to /usr/local/lib/python3.6/dist-packages\nAdding regex 2019.6.8 to easy-install.pth file\n\nInstalled /usr/local/lib/python3.6/dist-packages/regex-2019.6.8-py3.6-linux-x86_64.egg\nSearching for tqdm==4.28.1\nBest match: tqdm 4.28.1\nAdding tqdm 4.28.1 to easy-install.pth file\nInstalling tqdm script to /usr/local/bin\n\nUsing /usr/local/lib/python3.6/dist-packages\nSearching for requests==2.21.0\nBest match: requests 2.21.0\nAdding requests 2.21.0 to easy-install.pth file\n\nUsing /usr/local/lib/python3.6/dist-packages\nSearching for boto3==1.9.167\nBest match: boto3 1.9.167\nAdding boto3 1.9.167 to easy-install.pth file\n\nUsing /usr/local/lib/python3.6/dist-packages\nSearching for numpy==1.16.4\nBest match: numpy 1.16.4\nAdding numpy 1.16.4 to easy-install.pth file\nInstalling f2py script to /usr/local/bin\nInstalling f2py3 script to /usr/local/bin\nInstalling f2py3.6 script to /usr/local/bin\n\nUsing /usr/local/lib/python3.6/dist-packages\nSearching for torch==1.1.0\nBest match: torch 1.1.0\nAdding torch 1.1.0 to easy-install.pth file\nInstalling convert-caffe2-to-onnx script to /usr/local/bin\nInstalling convert-onnx-to-caffe2 script to /usr/local/bin\n\nUsing /usr/local/lib/python3.6/dist-packages\nSearching for idna==2.8\nBest match: idna 2.8\nAdding idna 2.8 to easy-install.pth file\n\nUsing /usr/local/lib/python3.6/dist-packages\nSearching for urllib3==1.24.3\nBest match: urllib3 1.24.3\nAdding urllib3 1.24.3 to easy-install.pth file\n\nUsing /usr/local/lib/python3.6/dist-packages\nSearching for certifi==2019.3.9\nBest match: certifi 2019.3.9\nAdding certifi 2019.3.9 to easy-install.pth file\n\nUsing /usr/local/lib/python3.6/dist-packages\nSearching for chardet==3.0.4\nBest match: chardet 3.0.4\nAdding chardet 3.0.4 to easy-install.pth file\nInstalling chardetect script to /usr/local/bin\n\nUsing /usr/local/lib/python3.6/dist-packages\nSearching for botocore==1.12.167\nBest match: botocore 1.12.167\nAdding botocore 1.12.167 to easy-install.pth file\n\nUsing /usr/local/lib/python3.6/dist-packages\nSearching for jmespath==0.9.4\nBest match: jmespath 0.9.4\nAdding jmespath 0.9.4 to easy-install.pth file\n\nUsing /usr/local/lib/python3.6/dist-packages\nSearching for s3transfer==0.2.1\nBest match: s3transfer 0.2.1\nAdding s3transfer 0.2.1 to easy-install.pth file\n\nUsing /usr/local/lib/python3.6/dist-packages\nSearching for python-dateutil==2.5.3\nBest match: python-dateutil 2.5.3\nAdding python-dateutil 2.5.3 to easy-install.pth file\n\nUsing /usr/local/lib/python3.6/dist-packages\nSearching for docutils==0.14\nBest match: docutils 0.14\nAdding docutils 0.14 to easy-install.pth file\n\nUsing /usr/local/lib/python3.6/dist-packages\nSearching for six==1.12.0\nBest match: six 1.12.0\nAdding six 1.12.0 to easy-install.pth file\n\nUsing /usr/local/lib/python3.6/dist-packages\nFinished processing dependencies for pytorch-pretrained-bert==0.6.2\n"
]
],
[
[
"## Run train",
"_____no_output_____"
]
],
[
[
"# without intermediate pretraining\n%run examples/run_classifier.py \\\n --task_name SST-2 \\\n --do_train \\\n --do_eval \\\n --do_lower_case \\\n --data_dir /content/glue_data/SST-2 \\\n --bert_model bert-base-uncased \\\n --max_seq_length 128 \\\n --train_batch_size 32 \\\n --learning_rate 2e-5 \\\n --num_train_epochs 3.0 \\\n --output_dir /content/drive/My\\ Drive/dr/models/SST_20",
"_____no_output_____"
],
[
"# with intermediate pretraining\n%run examples/run_classifier.py \\\n --task_name SST-2 \\\n --do_train \\\n --do_eval \\\n --do_lower_case \\\n --data_dir /content/glue_data/SST-2 \\\n --bert_model /content/drive/My\\ Drive/dr/models/MNLI_full \\\n --max_seq_length 128 \\\n --train_batch_size 32 \\\n --learning_rate 2e-5 \\\n --num_train_epochs 3.0 \\\n --output_dir /content/drive/My\\ Drive/dr/models/SST_20_MNLI \\\n --intermediate_task MNLI",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d0e150ac7c38a82487d22dd68aea05c6d195ce1d | 43,273 | ipynb | Jupyter Notebook | v1.52/Tests/1.3 Google form analysis visualizations.ipynb | CyberCRI/herocoli-metrics-redwire | abd2e73f3ec6a5088be3302074d5b4b2a1d65362 | [
"CC0-1.0"
] | 1 | 2017-01-18T13:46:31.000Z | 2017-01-18T13:46:31.000Z | v1.52/Tests/1.3 Google form analysis visualizations.ipynb | CyberCRI/herocoli-metrics-redwire | abd2e73f3ec6a5088be3302074d5b4b2a1d65362 | [
"CC0-1.0"
] | null | null | null | v1.52/Tests/1.3 Google form analysis visualizations.ipynb | CyberCRI/herocoli-metrics-redwire | abd2e73f3ec6a5088be3302074d5b4b2a1d65362 | [
"CC0-1.0"
] | null | null | null | 33.75429 | 149 | 0.586463 | [
[
[
"# Google form analysis visualizations",
"_____no_output_____"
],
[
"## Table of Contents\n\n\n['Google form analysis' functions checks](#funcchecks)\n\n['Google form analysis' functions tinkering](#functinkering)",
"_____no_output_____"
]
],
[
[
"%run \"../Functions/1. Google form analysis.ipynb\"",
"_____no_output_____"
]
],
[
[
"## 'Google form analysis' functions checks\n<a id=funcchecks />",
"_____no_output_____"
],
[
"## 'Google form analysis' functions tinkering\n<a id=functinkering />",
"_____no_output_____"
]
],
[
[
"binarizedAnswers = plotBasicStats(getSurveysOfBiologists(gform), 'non biologists', includeUndefined = True)",
"_____no_output_____"
],
[
"gform.loc[:, [localplayerguidkey, 'Temporality']].groupby('Temporality').count()",
"_____no_output_____"
],
[
"#sample = gform.copy()\nsamples = [\n [gform.copy(), 'complete set'],\n [gform[gform['Language'] == 'en'], 'English'],\n [gform[gform['Language'] == 'fr'], 'French'],\n [gform[gform['What is your gender?'] == 'Female'], 'female'],\n [gform[gform['What is your gender?'] == 'Male'], 'male'],\n [getSurveysOfUsersWhoAnsweredBoth(gform), 'answered both'],\n [getSurveysOfUsersWhoAnsweredBoth(gform[gform['Language'] == 'en']), 'answered both, en'],\n [getSurveysOfUsersWhoAnsweredBoth(gform[gform['Language'] == 'fr']), 'answered both, fr'],\n [getSurveysOfUsersWhoAnsweredBoth(gform[gform['What is your gender?'] == 'Female']), 'answered both, female'],\n [getSurveysOfUsersWhoAnsweredBoth(gform[gform['What is your gender?'] == 'Male']), 'answered both, male'],\n ]\n\n_progress = FloatProgress(min=0, max=len(samples))\ndisplay(_progress)\n\nincludeAll = False\nincludeBefore = True\nincludeAfter = True\nincludeUndefined = False\nincludeProgress = True\nincludeRelativeProgress = False\n\nfor sample, title in samples:\n\n ## basic stats:\n ### mean score\n ### median score\n ### std\n ## sample can be: all, those who answered both before and after,\n ## those who played between date1 and date2, ...\n #def plotBasicStats(sample, title, includeAll, includeBefore, includeAfter, includeUndefined, includeProgress, includeRelativeProgress):\n\n \n stepsPerInclude = 2\n includeCount = np.sum([includeAll, includeBefore, includeAfter, includeUndefined, includeProgress])\n stepsCount = stepsPerInclude*includeCount + 3\n \n #print(\"stepsPerInclude=\" + str(stepsPerInclude))\n #print(\"includeCount=\" + str(includeCount))\n #print(\"stepsCount=\" + str(stepsCount))\n \n __progress = FloatProgress(min=0, max=stepsCount)\n display(__progress)\n \n sampleBefore = sample[sample['Temporality'] == 'before']\n sampleAfter = sample[sample['Temporality'] == 'after']\n sampleUndefined = sample[sample['Temporality'] == 'undefined']\n\n #uniqueBefore = sampleBefore[localplayerguidkey]\n #uniqueAfter = \n #uniqueUndefined =\n\n scientificQuestions = correctAnswers.copy()\n allQuestions = correctAnswers + demographicAnswers\n \n categories = ['all', 'before', 'after', 'undefined', 'progress', 'rel. progress']\n data = {}\n \n sciBinarized = pd.DataFrame()\n allBinarized = pd.DataFrame()\n scoresAll = pd.DataFrame()\n \n sciBinarizedBefore = pd.DataFrame()\n allBinarizedBefore = pd.DataFrame()\n scoresBefore = pd.DataFrame()\n \n sciBinarizedAfter = pd.DataFrame()\n allBinarizedAfter = pd.DataFrame()\n scoresAfter = pd.DataFrame()\n \n sciBinarizedUndefined = pd.DataFrame()\n allBinarizedUndefined = pd.DataFrame()\n scoresUndefined = pd.DataFrame()\n\n scoresProgress = pd.DataFrame()\n\n ## basic stats:\n ### mean score\n ### median score\n ### std\n if includeAll:\n sciBinarized = getAllBinarized( _source = scientificQuestions, _form = sample)\n __progress.value += 1\n allBinarized = getAllBinarized( _source = allQuestions, _form = sample)\n __progress.value += 1\n scoresAll = pd.Series(np.dot(sciBinarized, np.ones(sciBinarized.shape[1])))\n \n data[categories[0]] = createStatSet(scoresAll, sample[localplayerguidkey])\n \n if includeBefore or includeProgress:\n sciBinarizedBefore = getAllBinarized( _source = scientificQuestions, _form = sampleBefore)\n __progress.value += 1\n allBinarizedBefore = getAllBinarized( _source = allQuestions, _form = sampleBefore)\n __progress.value += 1\n scoresBefore = pd.Series(np.dot(sciBinarizedBefore, np.ones(sciBinarizedBefore.shape[1])))\n temporaryStatSetBefore = createStatSet(scoresBefore, sampleBefore[localplayerguidkey])\n if includeBefore:\n data[categories[1]] = temporaryStatSetBefore\n \n if includeAfter or includeProgress:\n sciBinarizedAfter = getAllBinarized( _source = scientificQuestions, _form = sampleAfter)\n __progress.value += 1\n allBinarizedAfter = getAllBinarized( _source = allQuestions, _form = sampleAfter)\n __progress.value += 1\n scoresAfter = pd.Series(np.dot(sciBinarizedAfter, np.ones(sciBinarizedAfter.shape[1])))\n temporaryStatSetAfter = createStatSet(scoresAfter, sampleAfter[localplayerguidkey])\n if includeAfter:\n data[categories[2]] = temporaryStatSetAfter\n \n if includeUndefined:\n sciBinarizedUndefined = getAllBinarized( _source = scientificQuestions, _form = sampleUndefined)\n __progress.value += 1\n allBinarizedUndefined = getAllBinarized( _source = allQuestions, _form = sampleUndefined)\n __progress.value += 1\n scoresUndefined = pd.Series(np.dot(sciBinarizedUndefined, np.ones(sciBinarizedUndefined.shape[1])))\n \n data[categories[3]] = createStatSet(scoresUndefined, sampleUndefined[localplayerguidkey])\n\n if includeProgress:\n data[categories[4]] = {\n 'count' : min(temporaryStatSetAfter['count'], temporaryStatSetBefore['count']),\n 'unique' : min(temporaryStatSetAfter['unique'], temporaryStatSetBefore['unique']),\n 'median' : temporaryStatSetAfter['median']-temporaryStatSetBefore['median'],\n 'mean' : temporaryStatSetAfter['mean']-temporaryStatSetBefore['mean'],\n 'std' : temporaryStatSetAfter['std']-temporaryStatSetBefore['std'],\n }\n __progress.value += 2\n \n \n result = pd.DataFrame(data)\n __progress.value += 1\n\n print(title)\n print(result)\n if (includeBefore and includeAfter) or includeProgress:\n if (len(scoresBefore) > 2 and len(scoresAfter) > 2):\n ttest = ttest_ind(scoresBefore, scoresAfter)\n print(\"t test: statistic=\" + repr(ttest.statistic) + \" pvalue=\" + repr(ttest.pvalue))\n print()\n\n ## percentage correct\n ### percentage correct - max 5 columns\n percentagePerQuestionAll = pd.DataFrame()\n percentagePerQuestionBefore = pd.DataFrame()\n percentagePerQuestionAfter = pd.DataFrame()\n percentagePerQuestionUndefined = pd.DataFrame()\n percentagePerQuestionProgress = pd.DataFrame()\n \n tables = []\n\n if includeAll:\n percentagePerQuestionAll = getPercentagePerQuestion(allBinarized)\n tables.append([percentagePerQuestionAll, categories[0]])\n \n if includeBefore or includeProgress:\n percentagePerQuestionBefore = getPercentagePerQuestion(allBinarizedBefore)\n if includeBefore:\n tables.append([percentagePerQuestionBefore, categories[1]])\n \n if includeAfter or includeProgress:\n percentagePerQuestionAfter = getPercentagePerQuestion(allBinarizedAfter)\n if includeAfter:\n tables.append([percentagePerQuestionAfter, categories[2]])\n \n if includeUndefined:\n percentagePerQuestionUndefined = getPercentagePerQuestion(allBinarizedUndefined)\n tables.append([percentagePerQuestionUndefined, categories[3]])\n \n if includeProgress or includeRelativeProgress:\n percentagePerQuestionProgress = percentagePerQuestionAfter - percentagePerQuestionBefore\n \n if includeProgress:\n tables.append([percentagePerQuestionProgress, categories[4]])\n \n if includeRelativeProgress:\n # use temporaryStatSetAfter['count'], temporaryStatSetBefore['count']?\n percentagePerQuestionProgress2 = percentagePerQuestionProgress.copy()\n for index in range(0,len(percentagePerQuestionProgress.index)):\n if (0 == percentagePerQuestionBefore.iloc[index,0]):\n percentagePerQuestionProgress2.iloc[index,0] = 0\n else:\n percentagePerQuestionProgress2.iloc[index,0] = \\\n percentagePerQuestionProgress.iloc[index,0]/percentagePerQuestionBefore.iloc[index,0]\n tables.append([percentagePerQuestionProgress2, categories[5]])\n \n __progress.value += 1\n\n graphTitle = '% correct: '\n toConcat = []\n \n for table,category in tables:\n concat = (len(table.values) > 0)\n for elt in table.iloc[:,0].values:\n if np.isnan(elt):\n concat = False\n break\n if(concat):\n graphTitle = graphTitle + category + ' '\n toConcat.append(table)\n\n if (len(toConcat) > 0):\n percentagePerQuestionConcatenated = pd.concat(\n toConcat\n , axis=1)\n\n if(len(title) > 0):\n graphTitle = graphTitle + ' - ' + title\n\n _fig = plt.figure(figsize=(20,20))\n _ax1 = plt.subplot(111)\n _ax1.set_title(graphTitle)\n sns.heatmap(percentagePerQuestionConcatenated.round().astype(int),ax=_ax1,cmap=plt.cm.jet,square=True,annot=True,fmt='d')\n __progress.value += 1\n \n ### percentage cross correct\n ### percentage cross correct, conditionnally\n \n if(__progress.value != stepsCount):\n print(\"__progress.value=\" + str(__progress.value) + \" != stepsCount=\" + str(stepsCount))\n \n _progress.value += 1\n\nif(_progress.value != len(samples)):\n print(\"__progress.value=\" + str(__progress.value) + \" != len(samples)=\" + str(len(samples)))\n\n# sciBinarized, sciBinarizedBefore, sciBinarizedAfter, sciBinarizedUndefined, \\\n# allBinarized, allBinarizedBefore, allBinarizedAfter, allBinarizedUndefined",
"_____no_output_____"
],
[
"ttest = ttest_ind(scoresBefore, scoresAfter)\ntype(scoresBefore), len(scoresBefore),\\\ntype(scoresAfter), len(scoresAfter),\\\nttest",
"_____no_output_____"
],
[
"type(tables)",
"_____no_output_____"
],
[
"sciBinarized = getAllBinarized( _source = scientificQuestions, _form = sample)\nseries = pd.Series(np.dot(sciBinarized, np.ones(sciBinarized.shape[1])))\n#ids = pd.Series()\nids = sample[localplayerguidkey]\n\n#def createStatSet(series, ids):\nif(0 == len(ids)):\n ids = series.index\nresult = {\n 'count' : len(ids),\n 'unique' : len(ids.unique()),\n 'median' : series.median(),\n 'mean' : series.mean(),\n 'std' : series.std()}\nresult",
"_____no_output_____"
],
[
"## percentage correct\n### percentage correct - 3 columns\n### percentage cross correct\n### percentage cross correct, conditionnally",
"_____no_output_____"
],
[
"#_binarized = allBinarized\n#_binarized = allBinarizedUndefined\n_binarized = allBinarizedBefore\n#def getPercentagePerQuestion(_binarized):\ntotalPerQuestionDF = pd.DataFrame(data=np.dot(np.ones(_binarized.shape[0]), _binarized), index=_binarized.columns)\npercentagePerQuestion = totalPerQuestionDF*100 / _binarized.shape[0]\npercentagePerQuestion",
"_____no_output_____"
],
[
"#totalPerQuestion = np.dot(np.ones(allSciBinarized.shape[0]), allSciBinarized)\n#totalPerQuestion.shape\ntotalPerQuestionSci = np.dot(np.ones(sciBinarized.shape[0]), sciBinarized)\ntotalPerQuestionAll = np.dot(np.ones(allBinarized.shape[0]), allBinarized)\n\npercentagePerQuestionAll = getPercentagePerQuestion(allBinarized)\npercentagePerQuestionBefore = getPercentagePerQuestion(allBinarizedBefore)\npercentagePerQuestionAfter = getPercentagePerQuestion(allBinarizedAfter)\npercentagePerQuestionUndefined = getPercentagePerQuestion(allBinarizedUndefined)\n\npercentagePerQuestionConcatenated = pd.concat(\n [\n percentagePerQuestionAll,\n percentagePerQuestionBefore,\n percentagePerQuestionAfter,\n percentagePerQuestionUndefined,\n ]\n , axis=1)\n_fig = plt.figure(figsize=(20,20))\n_ax1 = plt.subplot(111)\n_ax1.set_title('percentage correct per question: all, before, after, undefined')\nsns.heatmap(percentagePerQuestionConcatenated.round().astype(int),ax=_ax1,cmap=plt.cm.jet,square=True,annot=True,fmt='d')",
"_____no_output_____"
],
[
"samples = [gform, gform[gform['Language'] == 'en'], gform[gform['Language'] == 'fr'],\n getSurveysOfUsersWhoAnsweredBoth(gform),\n getSurveysOfUsersWhoAnsweredBoth(gform[gform['Language'] == 'en']),\n getSurveysOfUsersWhoAnsweredBoth(gform[gform['Language'] == 'fr'])]\n\nfor sample in samples:\n sciBinarized, sciBinarizedBefore, sciBinarizedAfter, sciBinarizedUndefined, \\\n allBinarized, allBinarizedBefore, allBinarizedAfter, allBinarizedUndefined = plotBasicStats(sample)",
"_____no_output_____"
]
],
[
[
"### abandoned algorithms",
"_____no_output_____"
]
],
[
[
"#totalPerQuestion = np.dot(np.ones(sciBinarized.shape[0]), sciBinarized)\n#totalPerQuestion.shape\ntotalPerQuestionSci = np.dot(np.ones(sciBinarized.shape[0]), sciBinarized)\ntotalPerQuestionAll = np.dot(np.ones(allBinarized.shape[0]), allBinarized)\ntotalPerQuestionDFAll = pd.DataFrame(data=np.dot(np.ones(allBinarized.shape[0]), allBinarized), index=allBinarized.columns)\npercentagePerQuestionAll = totalPerQuestionDFAll*100 / allBinarized.shape[0]\n#totalPerQuestionDF\n#percentagePerQuestion\n\n#before\ntotalPerQuestionDFBefore = pd.DataFrame(\n data=np.dot(np.ones(allBinarizedBefore.shape[0]), allBinarizedBefore), index=allBinarizedBefore.columns\n)\npercentagePerQuestionBefore = totalPerQuestionDFBefore*100 / allBinarizedBefore.shape[0]\n\n#after\ntotalPerQuestionDFAfter = pd.DataFrame(\n data=np.dot(np.ones(allBinarizedAfter.shape[0]), allBinarizedAfter), index=allBinarizedAfter.columns\n)\npercentagePerQuestionAfter = totalPerQuestionDFAfter*100 / allBinarizedAfter.shape[0]\n\n_fig = plt.figure(figsize=(20,20))\nax1 = plt.subplot(131)\nax2 = plt.subplot(132)\nax3 = plt.subplot(133)\nax2.get_yaxis().set_visible(False)\nax3.get_yaxis().set_visible(False)\nsns.heatmap(percentagePerQuestionAll.round().astype(int),ax=ax1,cmap=plt.cm.jet,square=True,annot=True,fmt='d', cbar=False)\nsns.heatmap(percentagePerQuestionBefore.round().astype(int),ax=ax2,cmap=plt.cm.jet,square=True,annot=True,fmt='d', cbar=False)\nsns.heatmap(percentagePerQuestionAfter.round().astype(int),ax=ax3,cmap=plt.cm.jet,square=True,annot=True,fmt='d', cbar=True)\nax1.set_title('percentage correct per question - all')\nax2.set_title('percentage correct per question - before')\nax3.set_title('percentage correct per question - after')\n# Fine-tune figure; make subplots close to each other and hide x ticks for\n# all but bottom plot.\n_fig.tight_layout()\n\n_fig = plt.figure(figsize=(20,20))\nax1 = plt.subplot(131)\nax2 = plt.subplot(132)\nax3 = plt.subplot(133)\nax2.get_yaxis().set_visible(False)\nax3.get_yaxis().set_visible(False)\nsns.heatmap(percentagePerQuestionAll.round().astype(int),ax=ax1,cmap=plt.cm.jet,square=True,annot=True,fmt='d', cbar=False)\nsns.heatmap(percentagePerQuestionBefore.round().astype(int),ax=ax2,cmap=plt.cm.jet,square=True,annot=True,fmt='d', cbar=False)\nsns.heatmap(percentagePerQuestionAfter.round().astype(int),ax=ax3,cmap=plt.cm.jet,square=True,annot=True,fmt='d', cbar=True)\nax1.set_title('percentage correct per question - all')\nax2.set_title('percentage correct per question - before')\nax3.set_title('percentage correct per question - after')\n# Fine-tune figure; make subplots close to each other and hide x ticks for\n# all but bottom plot.\n_fig.tight_layout()",
"_____no_output_____"
],
[
"_fig = plt.figure(figsize=(20,20))\nax1 = plt.subplot(131)\nax2 = plt.subplot(132)\nax3 = plt.subplot(133)\nax2.get_yaxis().set_visible(False)\nax3.get_yaxis().set_visible(False)\nsns.heatmap(percentagePerQuestionAll.round().astype(int),ax=ax1,cmap=plt.cm.jet,square=True,annot=True,fmt='d', cbar=False)\nsns.heatmap(percentagePerQuestionBefore.round().astype(int),ax=ax2,cmap=plt.cm.jet,square=True,annot=True,fmt='d', cbar=False)\nsns.heatmap(percentagePerQuestionAfter.round().astype(int),ax=ax3,cmap=plt.cm.jet,square=True,annot=True,fmt='d', cbar=True)\nax1.set_title('percentage correct per question - all')\nax2.set_title('percentage correct per question - before')\nax3.set_title('percentage correct per question - after')\n# Fine-tune figure; make subplots close to each other and hide x ticks for\n# all but bottom plot.\n_fig.tight_layout()",
"_____no_output_____"
],
[
"percentagePerQuestionConcatenated = pd.concat([\n percentagePerQuestionAll,\n percentagePerQuestionBefore,\n percentagePerQuestionAfter]\n , axis=1)\n_fig = plt.figure(figsize=(20,20))\n_ax1 = plt.subplot(111)\n_ax1.set_title('percentage correct per question: all, before, after')\nsns.heatmap(percentagePerQuestionConcatenated.round().astype(int),ax=_ax1,cmap=plt.cm.jet,square=True,annot=True,fmt='d')",
"_____no_output_____"
]
],
[
[
"### sample getters tinkering",
"_____no_output_____"
]
],
[
[
"##### getRMAfter / Before tinkering\n#def getRMAfters(sample):\nafters = sample[sample['Temporality'] == 'after']\n#def getRMBefores(sample):\nbefores = sample[sample['Temporality'] == 'before']",
"_____no_output_____"
],
[
"QPlayed1 = 'Have you ever played an older version of Hero.Coli before?'\nQPlayed2 = 'Have you played the current version of Hero.Coli?'\nQPlayed3 = 'Have you played the arcade cabinet version of Hero.Coli?'\nQPlayed4 = 'Have you played the Android version of Hero.Coli?'",
"_____no_output_____"
]
],
[
[
"#### set operators",
"_____no_output_____"
]
],
[
[
"# equality tests\n#(sample1.columns == sample2.columns).all()\n#sample1.columns.duplicated().any() or sample2.columns.duplicated().any()\n#pd.concat([sample1, sample2], axis=1).columns.duplicated().any()",
"_____no_output_____"
]
],
[
[
"##### getUnionQuestionnaires tinkering",
"_____no_output_____"
]
],
[
[
"sample1 = befores\nsample2 = afters\n\n#def getUnionQuestionnaires(sample1, sample2):\nif (not (sample1.columns == sample2.columns).all()):\n print(\"warning: parameter columns are not the same\")\nresult = pd.concat([sample1, sample2]).drop_duplicates()",
"_____no_output_____"
]
],
[
[
"##### getIntersectionQuestionnaires tinkering",
"_____no_output_____"
]
],
[
[
"sample1 = befores[:15]\nsample2 = befores[10:]\n\n#def getIntersectionQuestionnaires(sample1, sample2):\nif (not (sample1.columns == sample2.columns).all()):\n print(\"warning: parameter columns are not the same\")\nresult = pd.merge(sample1, sample2, how = 'inner').drop_duplicates()",
"_____no_output_____"
]
],
[
[
"##### getIntersectionUsersSurveys tinkering",
"_____no_output_____"
]
],
[
[
"sample1 = befores\nsample2 = afters\n\n# get sample1 and sample2 rows where users are common to sample1 and sample2\n#def getIntersectionUsersSurveys(sample1, sample2):\nresult1 = sample1[sample1[localplayerguidkey].isin(sample2[localplayerguidkey])]\nresult2 = sample2[sample2[localplayerguidkey].isin(sample1[localplayerguidkey])]\nresult = getUnionQuestionnaires(result1,result2)",
"_____no_output_____"
],
[
"len(sample1), len(sample2), len(result)",
"_____no_output_____"
]
],
[
[
"##### getGFormBefores tinkering",
"_____no_output_____"
]
],
[
[
"sample = gform\n\n\n# returns users who declared that they have never played the game, whatever platform\n# previousPlayPositives is defined in '../Static data/English localization.ipynb'\n#def getGFormBefores(sample):\nbefores = sample[\n ~sample[QPlayed1].isin(previousPlayPositives)\n & ~sample[QPlayed2].isin(previousPlayPositives)\n & ~sample[QPlayed3].isin(previousPlayPositives)\n & ~sample[QPlayed4].isin(previousPlayPositives)\n ]\nlen(befores)",
"_____no_output_____"
]
],
[
[
"##### getGFormAfters tinkering",
"_____no_output_____"
]
],
[
[
"sample = gform\n\n# returns users who declared that they have already played the game, whatever platform\n# previousPlayPositives is defined in '../Static data/English localization.ipynb'\n#def getGFormAfters(sample):\nafters = sample[\n sample[QPlayed1].isin(previousPlayPositives)\n | sample[QPlayed2].isin(previousPlayPositives)\n | sample[QPlayed3].isin(previousPlayPositives)\n | sample[QPlayed4].isin(previousPlayPositives)\n ]\nlen(afters)",
"_____no_output_____"
]
],
[
[
"##### getGFormTemporality tinkering",
"_____no_output_____"
]
],
[
[
"_GFUserId = getSurveysOfBiologists(gform)[localplayerguidkey].iloc[3]\n_gformRow = gform[gform[localplayerguidkey] == _GFUserId].iloc[0]\nsample = gform",
"_____no_output_____"
],
[
"answerTemporalities[1]",
"_____no_output_____"
],
[
"#while result != 'after':\n_GFUserId = getRandomGFormGUID()\n_gformRow = gform[gform[localplayerguidkey] == _GFUserId].iloc[0]\n\n# returns an element of answerTemporalities\n# previousPlayPositives is defined in '../Static data/English localization.ipynb'\n#def getGFormRowGFormTemporality(_gformRow):\nresult = answerTemporalities[2]\nif (_gformRow[QPlayed1] in previousPlayPositives)\\\n or (_gformRow[QPlayed2] in previousPlayPositives)\\\n or (_gformRow[QPlayed3] in previousPlayPositives)\\\n or (_gformRow[QPlayed4] in previousPlayPositives):\n result = answerTemporalities[1]\nelse:\n result = answerTemporalities[0]\nresult",
"_____no_output_____"
]
],
[
[
"#### getSurveysOfUsersWhoAnsweredBoth tinkering",
"_____no_output_____"
]
],
[
[
"sample = gform\ngfMode = True\nrmMode = False\n\n#def getSurveysOfUsersWhoAnsweredBoth(sample, gfMode = True, rmMode = False):\n\nbefores = sample\nafters = sample\n\nif gfMode:\n befores = getGFormBefores(befores)\n afters = getGFormAfters(afters)\n\nif rmMode:\n befores = getRMBefores(befores)\n afters = getRMAfters(afters)\n\nresult = getIntersectionUsersSurveys(befores, afters)\n\n((len(getGFormBefores(sample)),\\\nlen(getRMBefores(sample)),\\\nlen(befores)),\\\n(len(getGFormAfters(sample)),\\\nlen(getRMAfters(sample)),\\\nlen(afters)),\\\nlen(result)),\\\n\\\n((getUniqueUserCount(getGFormBefores(sample)),\\\ngetUniqueUserCount(getRMBefores(sample)),\\\ngetUniqueUserCount(befores)),\\\n(getUniqueUserCount(getGFormAfters(sample)),\\\ngetUniqueUserCount(getRMAfters(sample)),\\\ngetUniqueUserCount(afters)),\\\ngetUniqueUserCount(result))\n\n",
"_____no_output_____"
],
[
"len(getSurveysOfUsersWhoAnsweredBoth(gform, gfMode = True, rmMode = True)[localplayerguidkey])",
"_____no_output_____"
]
],
[
[
"#### getSurveysThatAnswered tinkering",
"_____no_output_____"
]
],
[
[
"sample = gform\n\n#_GFUserId = getSurveysOfBiologists(gform)[localplayerguidkey].iloc[1]\n#sample = gform[gform[localplayerguidkey] == _GFUserId]\n\nhardPolicy = True\nquestionsAndPositiveAnswers = [[Q6BioEdu, biologyStudyPositives],\n [Q8SynBio, yesNoIdontknowPositives],\n [Q9BioBricks, yesNoIdontknowPositives]]\n\n#def getSurveysThatAnswered(sample, questionsAndPositiveAnswers, hardPolicy = True):\nfilterSeries = []\nif hardPolicy:\n filterSeries = pd.Series(True, sample.index)\n for question, positiveAnswers in questionsAndPositiveAnswers:\n filterSeries = filterSeries & (sample[question].isin(positiveAnswers))\nelse:\n filterSeries = pd.Series(False, sample.index)\n for question, positiveAnswers in questionsAndPositiveAnswers:\n filterSeries = filterSeries | (sample[question].isin(positiveAnswers))\nresult = sample[filterSeries]",
"_____no_output_____"
]
],
[
[
"#### getSurveysOfBiologists tinkering",
"_____no_output_____"
]
],
[
[
"sample = gform\nhardPolicy = True\n\n#def getSurveysOfBiologists(sample, hardPolicy = True):\nQ6BioEdu = 'How long have you studied biology?' #biologyStudyPositives\n#irrelevant QInterest 'Are you interested in biology?' #biologyInterestPositives\nQ8SynBio = 'Before playing Hero.Coli, had you ever heard about synthetic biology?' #yesNoIdontknowPositives\nQ9BioBricks = 'Before playing Hero.Coli, had you ever heard about BioBricks?' #yesNoIdontknowPositives\n\nquestionsAndPositiveAnswers = [[Q6BioEdu, biologyStudyPositives],\n [Q8SynBio, yesNoIdontknowPositives],\n [Q9BioBricks, yesNoIdontknowPositives]]\n\nresult = getSurveysThatAnswered(sample, questionsAndPositiveAnswers, hardPolicy)\nprint(len(result) > 0)",
"_____no_output_____"
],
[
"gform.index",
"_____no_output_____"
],
[
"len(result)",
"_____no_output_____"
],
[
"_GFUserId = getSurveysOfBiologists(gform)[localplayerguidkey].iloc[0]\nsample = gform[gform[localplayerguidkey] == _GFUserId]\nlen(getSurveysOfBiologists(sample)) > 0",
"_____no_output_____"
]
],
[
[
"#### getSurveysOfGamers tinkering",
"_____no_output_____"
]
],
[
[
"sample = gform\nhardPolicy = True\n\n#def getSurveysOfGamers(sample, hardPolicy = True):\nQ2Interest = 'Are you interested in video games?' #interestPositives\nQ3Play = 'Do you play video games?' #frequencyPositives\n\nquestionsAndPositiveAnswers = [[Q2Interest, interestPositives], [Q3Play, frequencyPositives]]\n\nresult = getSurveysThatAnswered(sample, questionsAndPositiveAnswers, hardPolicy)",
"_____no_output_____"
],
[
"len(result)",
"_____no_output_____"
],
[
"type(filterSeries)",
"_____no_output_____"
],
[
"len(afters[afters[QPlayed1].isin(previousPlayPositives)\n | afters[QPlayed2].isin(previousPlayPositives)\n | afters[QPlayed3].isin(previousPlayPositives)\n | afters[QPlayed4].isin(previousPlayPositives)\n ]),\\\nlen(afters[afters[QPlayed1].isin(previousPlayPositives)]),\\\nlen(afters[afters[QPlayed2].isin(previousPlayPositives)]),\\\nlen(afters[afters[QPlayed3].isin(previousPlayPositives)]),\\\nlen(afters[afters[QPlayed4].isin(previousPlayPositives)])",
"_____no_output_____"
]
],
[
[
"#### getSurveysWithMatchingAnswers tinkering",
"_____no_output_____"
]
],
[
[
"_GFUserId = getSurveysOfBiologists(gform)[localplayerguidkey].iloc[2]\n_gformRow = gform[gform[localplayerguidkey] == _GFUserId].iloc[0]\nsample = gform",
"_____no_output_____"
],
[
"sample = gform\n_gformRow = gform[gform[localplayerguidkey] == _GFUserId].iloc[0]\nhardPolicy = False\nQ4 = 'How old are you?'\nQ5 = 'What is your gender?'\nQ2Interest = 'Are you interested in video games?'\nQ3Play = 'Do you play video games?'\nQ6BioEdu = 'How long have you studied biology?'\nQ7BioInterest = 'Are you interested in biology?'\nQ8SynBio = 'Before playing Hero.Coli, had you ever heard about synthetic biology?'\nQ9BioBricks = 'Before playing Hero.Coli, had you ever heard about BioBricks?'\nQ42 = 'Language'\nstrictList = [Q4, Q5]\nextendedList = [Q2Interest, Q3Play, Q6BioEdu, Q8SynBio, Q9BioBricks, Q42]\n\n\n\n#def getSurveysWithMatchingAnswers(sample, _gformRow, strictList, extendedList = [], hardPolicy = False):\nquestions = strictList\n\nif (hardPolicy):\n questions += extendedList\n\nquestionsAndPositiveAnswers = []\nfor q in questions:\n questionsAndPositiveAnswers.append([q, [_gformRow[q]]])\n\ngetSurveysThatAnswered(sample, questionsAndPositiveAnswers, True)",
"_____no_output_____"
]
],
[
[
"#### getMatchingDemographics tinkering",
"_____no_output_____"
]
],
[
[
"sample = gform\n_gformRow = gform[gform[localplayerguidkey] == _GFUserId].iloc[0]\nhardPolicy = True\n\n#def getMatchingDemographics(sample, _gformRow, hardPolicy = False):\n# age and gender\nQ4 = 'How old are you?'\nQ5 = 'What is your gender?'\n\n# interests, hobbies, and knowledge - evaluation may vary after playing\nQ2Interest = 'Are you interested in video games?'\nQ3Play = 'Do you play video games?'\nQ6BioEdu = 'How long have you studied biology?'\nQ7BioInterest = 'Are you interested in biology?'\nQ8SynBio = 'Before playing Hero.Coli, had you ever heard about synthetic biology?'\nQ9BioBricks = 'Before playing Hero.Coli, had you ever heard about BioBricks?'\n\n# language may vary: players may have missed the opportunity to set it, or may want to try and change it\nQ42 = 'Language'\n\ngetSurveysWithMatchingAnswers(\n sample, \n _gformRow, [Q4, Q5], \n extendedList = [Q2Interest, Q3Play, Q6BioEdu, Q8SynBio, Q9BioBricks, Q42], \n hardPolicy = hardPolicy\n)",
"_____no_output_____"
],
[
"questionsAndPositiveAnswers",
"_____no_output_____"
]
],
[
[
"#### getGFormRowCorrection tinkering",
"_____no_output_____"
]
],
[
[
"_gformRow = gform[gform[localplayerguidkey] == _GFUserId].iloc[0]\n_source = correctAnswers\n\n#def getGFormRowCorrection( _gformRow, _source = correctAnswers):\nresult = _gformRow.copy()\n\nif(len(_gformRow) == 0):\n print(\"this gform row is empty\")\n\nelse:\n result = pd.Series(index = _gformRow.index, data = np.full(len(_gformRow), np.nan))\n\n for question in result.index:\n _correctAnswers = _source.loc[question]\n\n if(len(_correctAnswers) > 0):\n result.loc[question] = False\n for _correctAnswer in _correctAnswers:\n if str(_gformRow.loc[question]).startswith(str(_correctAnswer)):\n result.loc[question] = True\n break\nresult",
"_____no_output_____"
]
],
[
[
"#### getGFormRowScore tinkering",
"_____no_output_____"
]
],
[
[
"_gformRow = gform[gform[localplayerguidkey] == _GFUserId].iloc[0]\n_source = correctAnswers\n\n\n#def getGFormRowScore( _gformRow, _source = correctAnswers):\ncorrection = getGFormRowCorrection( _gformRow, _source = _source)\n_counts = correction.value_counts()\n_thisScore = 0\nif(True in _counts):\n _thisScore = _counts[True]\n_thisScore",
"_____no_output_____"
]
],
[
[
"#### getGFormDataPreview tinkering",
"_____no_output_____"
]
],
[
[
"_GFUserId = getSurveysOfBiologists(gform)[localplayerguidkey].iloc[2]\nsample = gform\n\n\n# for per-gform, manual analysis\n#def getGFormDataPreview(_GFUserId, sample):\ngforms = gform[gform[localplayerguidkey] == _GFUserId]\nresult = {}\nfor _ilocIndex in range(0, len(gforms)):\n gformsIndex = gforms.index[_ilocIndex]\n currentGForm = gforms.iloc[_ilocIndex]\n \n subresult = {}\n subresult['date'] = currentGForm['Timestamp']\n subresult['temporality RM'] = currentGForm['Temporality']\n subresult['temporality GF'] = getGFormRowGFormTemporality(currentGForm)\n subresult['score'] = getGFormRowScore(currentGForm)\n subresult['genderAge'] = [currentGForm['What is your gender?'], currentGForm['How old are you?']]\n \n # search for other users with similar demographics\n matchingDemographics = getMatchingDemographics(sample, currentGForm)\n matchingDemographicsIds = []\n #print(type(matchingDemographics))\n #print(matchingDemographics.index)\n for matchesIndex in matchingDemographics.index:\n matchingDemographicsIds.append([matchesIndex, matchingDemographics.loc[matchesIndex, localplayerguidkey]])\n \n subresult['demographic matches'] = matchingDemographicsIds\n \n result['survey' + str(_ilocIndex)] = subresult\n \nprint(result)",
"_____no_output_____"
],
[
"for match in result['survey0']['demographic matches']:\n print(match[0])",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d0e150e572b0082c0c8c769b2ec6ea60a7e1d999 | 31,236 | ipynb | Jupyter Notebook | jupyter/Generate_dave_xml_DNA_IDEX.ipynb | shiwei23/STORM6 | 669067503ebd164b575ce529fcc4a9a3f576b3d7 | [
"MIT"
] | 1 | 2022-03-29T22:40:38.000Z | 2022-03-29T22:40:38.000Z | jupyter/Generate_dave_xml_DNA_IDEX.ipynb | shiwei23/STORM6 | 669067503ebd164b575ce529fcc4a9a3f576b3d7 | [
"MIT"
] | null | null | null | jupyter/Generate_dave_xml_DNA_IDEX.ipynb | shiwei23/STORM6 | 669067503ebd164b575ce529fcc4a9a3f576b3d7 | [
"MIT"
] | 1 | 2021-08-01T20:20:09.000Z | 2021-08-01T20:20:09.000Z | 46.620896 | 125 | 0.611922 | [
[
[
"from xml.etree import ElementTree\nfrom xml.dom import minidom\nfrom xml.etree.ElementTree import Element, SubElement, Comment, indent\n\ndef prettify(elem):\n \"\"\"Return a pretty-printed XML string for the Element.\n \"\"\"\n rough_string = ElementTree.tostring(elem, encoding=\"ISO-8859-1\")\n reparsed = minidom.parseString(rough_string)\n return reparsed.toprettyxml(indent=\"\\t\")",
"_____no_output_____"
],
[
"import numpy as np\nimport os\n\nvalve_start = 1\n\nhyb_start = 51\n\nreg_start = 1\n\nnum_rounds = 14\n\n\ndata_type = 'U'\n\nvalve_ids = np.arange(valve_start, valve_start + num_rounds)\nhyb_ids = np.arange(hyb_start, hyb_start + num_rounds)\nreg_names = [f'{data_type}{_i}' for _i in np.arange(reg_start, reg_start + num_rounds)]",
"_____no_output_____"
],
[
"source_folder = r'D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229'\n#target_drive = r'\\\\KOLMOGOROV\\Chromatin_NAS_4'\ntarget_drive = r'\\\\10.245.74.158\\Chromatin_NAS_1'\n\n# imaging protocol\nimaging_protocol = r'Zscan_750_647_561_s50_n250_10Hz'\n# reference imaging protocol\nadd_ref_hyb = False\n\nref_imaging_protocol = r'Zscan_750_647_561_405_s50_n250_10Hz'\nref_hyb = 0\n# bleach protocol\nbleach = True\nbleach_protocol = r'Bleach_750_647_s5'",
"_____no_output_____"
],
[
"cmd_seq = Element('command_sequence')\n\nif add_ref_hyb:\n # add hyb 0\n # comments\n comment = Comment(f\"Hyb 0\")\n cmd_seq.append(comment)\n # flow imaging buffer\n imaging = SubElement(cmd_seq, 'valve_protocol')\n imaging.text = f\"Flow Imaging Buffer\"\n\n # change directory\n change_dir = SubElement(cmd_seq, 'change_directory')\n change_dir.text = os.path.join(source_folder, f\"H0C1\")\n # wakeup\n wakeup = SubElement(cmd_seq, 'wakeup')\n wakeup.text = \"5000\"\n\n # imaging loop\n _im_p = ref_imaging_protocol\n\n loop = SubElement(cmd_seq, 'loop', name='Position Loop Zscan', increment=\"name\")\n loop_item = SubElement(loop, 'item', name=_im_p)\n loop_item.text = \" \"\n # delay time\n delay = SubElement(cmd_seq, 'delay')\n delay.text = \"2000\"\n # copy folder\n copy_dir = SubElement(cmd_seq, 'copy_directory')\n source_dir = SubElement(copy_dir, 'source_path')\n source_dir.text = change_dir.text \n target_dir = SubElement(copy_dir, 'target_path')\n target_dir.text = os.path.join(target_drive, \n os.path.basename(os.path.dirname(source_dir.text)), \n os.path.basename(source_dir.text))\n del_source = SubElement(copy_dir, 'delete_source')\n del_source.text = \"True\"\n\n\nfor _i, (_vid, _hid, _rname) in enumerate(zip(valve_ids, hyb_ids, reg_names)):\n # select protocol\n _im_p = imaging_protocol\n # TCEP\n tcep = SubElement(cmd_seq, 'valve_protocol')\n tcep.text = \"Flow TCEP\"\n # wash tcep\n tcep_wash = SubElement(cmd_seq, 'valve_protocol')\n tcep_wash.text = \"Flow Wash Buffer\"\n \n # comments\n comment = Comment(f\"Hyb {_hid} with {_vid} for {_rname}\")\n cmd_seq.append(comment)\n # flow adaptor\n adt = SubElement(cmd_seq, 'valve_protocol')\n adt.text = f\"Hybridize {_vid}\"\n if bleach:\n # delay time\n delay = SubElement(cmd_seq, 'delay')\n delay.text = \"3000\"\n # change directory\n bleach_change_dir = SubElement(cmd_seq, 'change_directory')\n bleach_change_dir.text = os.path.join(source_folder, f\"Bleach\")\n # wakeup\n bleach_wakeup = SubElement(cmd_seq, 'wakeup')\n bleach_wakeup.text = \"5000\"\n # imaging loop\n bleach_loop = SubElement(cmd_seq, 'loop', name='Position Loop Zscan', increment=\"name\")\n bleach_loop_item = SubElement(bleach_loop, 'item', name=bleach_protocol)\n bleach_loop_item.text = \" \"\n # delay time\n delay = SubElement(cmd_seq, 'delay')\n delay.text = \"3000\"\n else:\n # delay time\n adt_incubation = SubElement(cmd_seq, 'valve_protocol')\n adt_incubation.text = f\"Incubate 10min\"\n # wash\n wash = SubElement(cmd_seq, 'valve_protocol')\n wash.text = \"Flow Wash Buffer\"\n \n # readouts\n readouts = SubElement(cmd_seq, 'valve_protocol')\n readouts.text = \"Flow RNA common readouts\"\n # incubate readouts\n readout_incubation = SubElement(cmd_seq, 'valve_protocol')\n readout_incubation.text = f\"Incubate 10min\"\n # wash readouts\n readout_wash = SubElement(cmd_seq, 'valve_protocol')\n readout_wash.text = f\"Flow Wash Buffer\"\n # flow imaging buffer\n imaging = SubElement(cmd_seq, 'valve_protocol')\n imaging.text = f\"Flow Imaging Buffer\"\n \n # change directory\n change_dir = SubElement(cmd_seq, 'change_directory')\n change_dir.text = os.path.join(source_folder, f\"H{_hid}{_rname.upper()}\")\n # wakeup\n wakeup = SubElement(cmd_seq, 'wakeup')\n wakeup.text = \"5000\"\n \n # imaging loop\n loop = SubElement(cmd_seq, 'loop', name='Position Loop Zscan', increment=\"name\")\n loop_item = SubElement(loop, 'item', name=_im_p)\n loop_item.text = \" \"\n # delay time\n delay = SubElement(cmd_seq, 'delay')\n delay.text = \"2000\"\n # copy folder\n copy_dir = SubElement(cmd_seq, 'copy_directory')\n source_dir = SubElement(copy_dir, 'source_path')\n source_dir.text = change_dir.text#cmd_seq.findall('change_directory')[-1].text\n target_dir = SubElement(copy_dir, 'target_path')\n target_dir.text = os.path.join(target_drive, \n os.path.basename(os.path.dirname(source_dir.text)), \n os.path.basename(source_dir.text))\n del_source = SubElement(copy_dir, 'delete_source')\n del_source.text = \"True\"\n # empty line\n indent(target_dir)\n\nfinal_str = prettify(cmd_seq)\n \nprint( final_str )",
"<?xml version=\"1.0\" ?>\n<command_sequence>\n\t<valve_protocol>Flow TCEP</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<!--Hyb 51 with 1 for U1-->\n\t<valve_protocol>Hybridize 1</valve_protocol>\n\t<delay>3000</delay>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\Bleach</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Bleach_750_647_s5\"> </item>\n\t</loop>\n\t<delay>3000</delay>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow RNA common readouts</valve_protocol>\n\t<valve_protocol>Incubate 10min</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow Imaging Buffer</valve_protocol>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H51U1</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Zscan_750_647_561_s50_n250_10Hz\"> </item>\n\t</loop>\n\t<delay>2000</delay>\n\t<copy_directory>\n\t\t<source_path>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H51U1</source_path>\n\t\t<target_path>\\\\10.245.74.158\\Chromatin_NAS_1\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H51U1</target_path>\n\t\t<delete_source>True</delete_source>\n\t</copy_directory>\n\t<valve_protocol>Flow TCEP</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<!--Hyb 52 with 2 for U2-->\n\t<valve_protocol>Hybridize 2</valve_protocol>\n\t<delay>3000</delay>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\Bleach</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Bleach_750_647_s5\"> </item>\n\t</loop>\n\t<delay>3000</delay>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow RNA common readouts</valve_protocol>\n\t<valve_protocol>Incubate 10min</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow Imaging Buffer</valve_protocol>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H52U2</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Zscan_750_647_561_s50_n250_10Hz\"> </item>\n\t</loop>\n\t<delay>2000</delay>\n\t<copy_directory>\n\t\t<source_path>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H52U2</source_path>\n\t\t<target_path>\\\\10.245.74.158\\Chromatin_NAS_1\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H52U2</target_path>\n\t\t<delete_source>True</delete_source>\n\t</copy_directory>\n\t<valve_protocol>Flow TCEP</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<!--Hyb 53 with 3 for U3-->\n\t<valve_protocol>Hybridize 3</valve_protocol>\n\t<delay>3000</delay>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\Bleach</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Bleach_750_647_s5\"> </item>\n\t</loop>\n\t<delay>3000</delay>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow RNA common readouts</valve_protocol>\n\t<valve_protocol>Incubate 10min</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow Imaging Buffer</valve_protocol>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H53U3</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Zscan_750_647_561_s50_n250_10Hz\"> </item>\n\t</loop>\n\t<delay>2000</delay>\n\t<copy_directory>\n\t\t<source_path>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H53U3</source_path>\n\t\t<target_path>\\\\10.245.74.158\\Chromatin_NAS_1\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H53U3</target_path>\n\t\t<delete_source>True</delete_source>\n\t</copy_directory>\n\t<valve_protocol>Flow TCEP</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<!--Hyb 54 with 4 for U4-->\n\t<valve_protocol>Hybridize 4</valve_protocol>\n\t<delay>3000</delay>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\Bleach</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Bleach_750_647_s5\"> </item>\n\t</loop>\n\t<delay>3000</delay>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow RNA common readouts</valve_protocol>\n\t<valve_protocol>Incubate 10min</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow Imaging Buffer</valve_protocol>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H54U4</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Zscan_750_647_561_s50_n250_10Hz\"> </item>\n\t</loop>\n\t<delay>2000</delay>\n\t<copy_directory>\n\t\t<source_path>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H54U4</source_path>\n\t\t<target_path>\\\\10.245.74.158\\Chromatin_NAS_1\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H54U4</target_path>\n\t\t<delete_source>True</delete_source>\n\t</copy_directory>\n\t<valve_protocol>Flow TCEP</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<!--Hyb 55 with 5 for U5-->\n\t<valve_protocol>Hybridize 5</valve_protocol>\n\t<delay>3000</delay>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\Bleach</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Bleach_750_647_s5\"> </item>\n\t</loop>\n\t<delay>3000</delay>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow RNA common readouts</valve_protocol>\n\t<valve_protocol>Incubate 10min</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow Imaging Buffer</valve_protocol>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H55U5</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Zscan_750_647_561_s50_n250_10Hz\"> </item>\n\t</loop>\n\t<delay>2000</delay>\n\t<copy_directory>\n\t\t<source_path>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H55U5</source_path>\n\t\t<target_path>\\\\10.245.74.158\\Chromatin_NAS_1\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H55U5</target_path>\n\t\t<delete_source>True</delete_source>\n\t</copy_directory>\n\t<valve_protocol>Flow TCEP</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<!--Hyb 56 with 6 for U6-->\n\t<valve_protocol>Hybridize 6</valve_protocol>\n\t<delay>3000</delay>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\Bleach</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Bleach_750_647_s5\"> </item>\n\t</loop>\n\t<delay>3000</delay>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow RNA common readouts</valve_protocol>\n\t<valve_protocol>Incubate 10min</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow Imaging Buffer</valve_protocol>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H56U6</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Zscan_750_647_561_s50_n250_10Hz\"> </item>\n\t</loop>\n\t<delay>2000</delay>\n\t<copy_directory>\n\t\t<source_path>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H56U6</source_path>\n\t\t<target_path>\\\\10.245.74.158\\Chromatin_NAS_1\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H56U6</target_path>\n\t\t<delete_source>True</delete_source>\n\t</copy_directory>\n\t<valve_protocol>Flow TCEP</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<!--Hyb 57 with 7 for U7-->\n\t<valve_protocol>Hybridize 7</valve_protocol>\n\t<delay>3000</delay>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\Bleach</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Bleach_750_647_s5\"> </item>\n\t</loop>\n\t<delay>3000</delay>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow RNA common readouts</valve_protocol>\n\t<valve_protocol>Incubate 10min</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow Imaging Buffer</valve_protocol>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H57U7</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Zscan_750_647_561_s50_n250_10Hz\"> </item>\n\t</loop>\n\t<delay>2000</delay>\n\t<copy_directory>\n\t\t<source_path>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H57U7</source_path>\n\t\t<target_path>\\\\10.245.74.158\\Chromatin_NAS_1\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H57U7</target_path>\n\t\t<delete_source>True</delete_source>\n\t</copy_directory>\n\t<valve_protocol>Flow TCEP</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<!--Hyb 58 with 8 for U8-->\n\t<valve_protocol>Hybridize 8</valve_protocol>\n\t<delay>3000</delay>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\Bleach</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Bleach_750_647_s5\"> </item>\n\t</loop>\n\t<delay>3000</delay>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow RNA common readouts</valve_protocol>\n\t<valve_protocol>Incubate 10min</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow Imaging Buffer</valve_protocol>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H58U8</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Zscan_750_647_561_s50_n250_10Hz\"> </item>\n\t</loop>\n\t<delay>2000</delay>\n\t<copy_directory>\n\t\t<source_path>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H58U8</source_path>\n\t\t<target_path>\\\\10.245.74.158\\Chromatin_NAS_1\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H58U8</target_path>\n\t\t<delete_source>True</delete_source>\n\t</copy_directory>\n\t<valve_protocol>Flow TCEP</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<!--Hyb 59 with 9 for U9-->\n\t<valve_protocol>Hybridize 9</valve_protocol>\n\t<delay>3000</delay>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\Bleach</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Bleach_750_647_s5\"> </item>\n\t</loop>\n\t<delay>3000</delay>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow RNA common readouts</valve_protocol>\n\t<valve_protocol>Incubate 10min</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow Imaging Buffer</valve_protocol>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H59U9</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Zscan_750_647_561_s50_n250_10Hz\"> </item>\n\t</loop>\n\t<delay>2000</delay>\n\t<copy_directory>\n\t\t<source_path>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H59U9</source_path>\n\t\t<target_path>\\\\10.245.74.158\\Chromatin_NAS_1\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H59U9</target_path>\n\t\t<delete_source>True</delete_source>\n\t</copy_directory>\n\t<valve_protocol>Flow TCEP</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<!--Hyb 60 with 10 for U10-->\n\t<valve_protocol>Hybridize 10</valve_protocol>\n\t<delay>3000</delay>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\Bleach</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Bleach_750_647_s5\"> </item>\n\t</loop>\n\t<delay>3000</delay>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow RNA common readouts</valve_protocol>\n\t<valve_protocol>Incubate 10min</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow Imaging Buffer</valve_protocol>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H60U10</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Zscan_750_647_561_s50_n250_10Hz\"> </item>\n\t</loop>\n\t<delay>2000</delay>\n\t<copy_directory>\n\t\t<source_path>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H60U10</source_path>\n\t\t<target_path>\\\\10.245.74.158\\Chromatin_NAS_1\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H60U10</target_path>\n\t\t<delete_source>True</delete_source>\n\t</copy_directory>\n\t<valve_protocol>Flow TCEP</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<!--Hyb 61 with 11 for U11-->\n\t<valve_protocol>Hybridize 11</valve_protocol>\n\t<delay>3000</delay>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\Bleach</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Bleach_750_647_s5\"> </item>\n\t</loop>\n\t<delay>3000</delay>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow RNA common readouts</valve_protocol>\n\t<valve_protocol>Incubate 10min</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow Imaging Buffer</valve_protocol>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H61U11</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Zscan_750_647_561_s50_n250_10Hz\"> </item>\n\t</loop>\n\t<delay>2000</delay>\n\t<copy_directory>\n\t\t<source_path>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H61U11</source_path>\n\t\t<target_path>\\\\10.245.74.158\\Chromatin_NAS_1\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H61U11</target_path>\n\t\t<delete_source>True</delete_source>\n\t</copy_directory>\n\t<valve_protocol>Flow TCEP</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<!--Hyb 62 with 12 for U12-->\n\t<valve_protocol>Hybridize 12</valve_protocol>\n\t<delay>3000</delay>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\Bleach</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Bleach_750_647_s5\"> </item>\n\t</loop>\n\t<delay>3000</delay>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow RNA common readouts</valve_protocol>\n\t<valve_protocol>Incubate 10min</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow Imaging Buffer</valve_protocol>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H62U12</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Zscan_750_647_561_s50_n250_10Hz\"> </item>\n\t</loop>\n\t<delay>2000</delay>\n\t<copy_directory>\n\t\t<source_path>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H62U12</source_path>\n\t\t<target_path>\\\\10.245.74.158\\Chromatin_NAS_1\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H62U12</target_path>\n\t\t<delete_source>True</delete_source>\n\t</copy_directory>\n\t<valve_protocol>Flow TCEP</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<!--Hyb 63 with 13 for U13-->\n\t<valve_protocol>Hybridize 13</valve_protocol>\n\t<delay>3000</delay>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\Bleach</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Bleach_750_647_s5\"> </item>\n\t</loop>\n\t<delay>3000</delay>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow RNA common readouts</valve_protocol>\n\t<valve_protocol>Incubate 10min</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow Imaging Buffer</valve_protocol>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H63U13</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Zscan_750_647_561_s50_n250_10Hz\"> </item>\n\t</loop>\n\t<delay>2000</delay>\n\t<copy_directory>\n\t\t<source_path>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H63U13</source_path>\n\t\t<target_path>\\\\10.245.74.158\\Chromatin_NAS_1\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H63U13</target_path>\n\t\t<delete_source>True</delete_source>\n\t</copy_directory>\n\t<valve_protocol>Flow TCEP</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<!--Hyb 64 with 14 for U14-->\n\t<valve_protocol>Hybridize 14</valve_protocol>\n\t<delay>3000</delay>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\Bleach</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Bleach_750_647_s5\"> </item>\n\t</loop>\n\t<delay>3000</delay>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow RNA common readouts</valve_protocol>\n\t<valve_protocol>Incubate 10min</valve_protocol>\n\t<valve_protocol>Flow Wash Buffer</valve_protocol>\n\t<valve_protocol>Flow Imaging Buffer</valve_protocol>\n\t<change_directory>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H64U14</change_directory>\n\t<wakeup>5000</wakeup>\n\t<loop name=\"Position Loop Zscan\" increment=\"name\">\n\t\t<item name=\"Zscan_750_647_561_s50_n250_10Hz\"> </item>\n\t</loop>\n\t<delay>2000</delay>\n\t<copy_directory>\n\t\t<source_path>D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H64U14</source_path>\n\t\t<target_path>\\\\10.245.74.158\\Chromatin_NAS_1\\20220102-CTP11-1000_CTP12-DNA_from_1229\\H64U14</target_path>\n\t\t<delete_source>True</delete_source>\n\t</copy_directory>\n</command_sequence>\n\n"
]
],
[
[
"# save this xml",
"_____no_output_____"
]
],
[
[
"save_filename = os.path.join(source_folder, f\"generated_dave_H{hyb_start}-{hyb_start+num_rounds-1}.txt\")\nwith open(save_filename, 'w') as _output_handle:\n print(save_filename)\n _output_handle.write(final_str)\n ",
"D:\\Pu\\20220102-CTP11-1000_CTP12-DNA_from_1229\\generated_dave_H51-64.txt\n"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0e159ab0214356424fe7b46700d79e5e90fda3b | 861,372 | ipynb | Jupyter Notebook | tutorials/W3D2_DynamicNetworks/W3D2_Tutorial1.ipynb | NinelK/course-content | 317b4cf153a89afbddbd194e7fbb15c91588b3c5 | [
"CC-BY-4.0"
] | null | null | null | tutorials/W3D2_DynamicNetworks/W3D2_Tutorial1.ipynb | NinelK/course-content | 317b4cf153a89afbddbd194e7fbb15c91588b3c5 | [
"CC-BY-4.0"
] | null | null | null | tutorials/W3D2_DynamicNetworks/W3D2_Tutorial1.ipynb | NinelK/course-content | 317b4cf153a89afbddbd194e7fbb15c91588b3c5 | [
"CC-BY-4.0"
] | null | null | null | 268.926631 | 111,608 | 0.915802 | [
[
[
"<a href=\"https://colab.research.google.com/github/NeuromatchAcademy/course-content/blob/master/tutorials/W3D2_DynamicNetworks/W3D2_Tutorial1.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Neuromatch Academy: Week 3, Day 2, Tutorial 1\n# Neuronal Network Dynamics: Neural Rate Models\n\n__Content creators:__ Qinglong Gu, Songtin Li, Arvind Kumar, John Murray, Julijana Gjorgjieva \n\n__Content reviewers:__ Spiros Chavlis, Lorenzo Fontolan, Richard Gao, Maryam Vaziri-Pashkam,Michael Waskom\n",
"_____no_output_____"
],
[
"---\n# Tutorial Objectives\n\nThe brain is a complex system, not because it is composed of a large number of diverse types of neurons, but mainly because of how neurons are connected to each other. The brain is indeed a network of highly specialized neuronal networks. \n\nThe activity of a neural network constantly evolves in time. For this reason, neurons can be modeled as dynamical systems. The dynamical system approach is only one of the many modeling approaches that computational neuroscientists have developed (other points of view include information processing, statistical models, etc.). \n\nHow the dynamics of neuronal networks affect the representation and processing of information in the brain is an open question. However, signatures of altered brain dynamics present in many brain diseases (e.g., in epilepsy or Parkinson's disease) tell us that it is crucial to study network activity dynamics if we want to understand the brain.\n\nIn this tutorial, we will simulate and study one of the simplest models of biological neuronal networks. Instead of modeling and simulating individual excitatory neurons (e.g., LIF models that you implemented yesterday), we will treat them as a single homogeneous population and approximate their dynamics using a single one-dimensional equation describing the evolution of their average spiking rate in time.\n\nIn this tutorial, we will learn how to build a firing rate model of a single population of excitatory neurons. \n\n**Steps:**\n- Write the equation for the firing rate dynamics of a 1D excitatory population.\n- Visualize the response of the population as a function of parameters such as threshold level and gain, using the frequency-current (F-I) curve.\n- Numerically simulate the dynamics of the excitatory population and find the fixed points of the system. \n- Investigate the stability of the fixed points by linearizing the dynamics around them.\n \n",
"_____no_output_____"
],
[
"---\n# Setup",
"_____no_output_____"
]
],
[
[
"# Imports\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport scipy.optimize as opt # root-finding algorithm",
"_____no_output_____"
],
[
"# @title Figure Settings\nimport ipywidgets as widgets # interactive display\n%config InlineBackend.figure_format = 'retina'\nplt.style.use(\"https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/nma.mplstyle\")",
"_____no_output_____"
],
[
"# @title Helper functions\n\n\ndef plot_fI(x, f):\n plt.figure(figsize=(6, 4)) # plot the figure\n plt.plot(x, f, 'k')\n plt.xlabel('x (a.u.)', fontsize=14)\n plt.ylabel('F(x)', fontsize=14)\n plt.show()\n\n\ndef plot_dr_r(r, drdt):\n plt.figure()\n plt.plot(r, drdt, 'k')\n plt.plot(r, 0. * r, 'k--')\n plt.xlabel(r'$r$')\n plt.ylabel(r'$\\frac{dr}{dt}$', fontsize=20)\n plt.ylim(-0.1, 0.1)\n\n\ndef plot_dFdt(x, dFdt):\n plt.figure()\n plt.plot(x, dFdt, 'r')\n plt.xlabel('x (a.u.)', fontsize=14)\n plt.ylabel('dF(x)', fontsize=14)\n plt.show()",
"_____no_output_____"
]
],
[
[
"# Section 1: Neuronal network dynamics",
"_____no_output_____"
]
],
[
[
"# @title Video 1: Dynamic networks\nfrom IPython.display import YouTubeVideo\nvideo = YouTubeVideo(id=\"p848349hPyw\", width=854, height=480, fs=1)\nprint(\"Video available at https://youtube.com/watch?v=\" + video.id)\nvideo",
"Video available at https://youtube.com/watch?v=p848349hPyw\n"
]
],
[
[
"## Section 1.1: Dynamics of a single excitatory population\n\nIndividual neurons respond by spiking. When we average the spikes of neurons in a population, we can define the average firing activity of the population. In this model, we are interested in how the population-averaged firing varies as a function of time and network parameters. Mathematically, we can describe the firing rate dynamic as:\n\n\\begin{align}\n\\tau \\frac{dr}{dt} &= -r + F(w\\cdot r + I_{\\text{ext}}) \\quad\\qquad (1)\n\\end{align}\n\n$r(t)$ represents the average firing rate of the excitatory population at time $t$, $\\tau$ controls the timescale of the evolution of the average firing rate, $w$ denotes the strength (synaptic weight) of the recurrent input to the population, $I_{\\text{ext}}$ represents the external input, and the transfer function $F(\\cdot)$ (which can be related to f-I curve of individual neurons described in the next sections) represents the population activation function in response to all received inputs.\n\nTo start building the model, please execute the cell below to initialize the simulation parameters.",
"_____no_output_____"
]
],
[
[
"# @title Default parameters for a single excitatory population model\n\n\ndef default_pars_single(**kwargs):\n pars = {}\n\n # Excitatory parameters\n pars['tau'] = 1. # Timescale of the E population [ms]\n pars['a'] = 1.2 # Gain of the E population\n pars['theta'] = 2.8 # Threshold of the E population\n\n # Connection strength\n pars['w'] = 0. # E to E, we first set it to 0\n\n # External input\n pars['I_ext'] = 0.\n\n # simulation parameters\n pars['T'] = 20. # Total duration of simulation [ms]\n pars['dt'] = .1 # Simulation time step [ms]\n pars['r_init'] = 0.2 # Initial value of E\n\n # External parameters if any\n for k in kwargs:\n pars[k] = kwargs[k]\n\n # Vector of discretized time points [ms]\n pars['range_t'] = np.arange(0, pars['T'], pars['dt'])\n\n return pars\n",
"_____no_output_____"
]
],
[
[
"You can use:\n- `pars = default_pars_single()` to get all the parameters, and then you can execute `print(pars)` to check these parameters. \n- `pars = default_pars_single(T=T_sim, dt=time_step)` to set new simulation time and time step\n- After `pars = default_pars_single()`, use `pars['New_para'] = value` to add an new parameter with its value",
"_____no_output_____"
],
[
"## Section 1.2: F-I curves\nIn electrophysiology, a neuron is often characterized by its spike rate output in response to input currents. This is often called the **F-I** curve, denoting the output spike frequency (**F**) in response to different injected currents (**I**). We estimated this for an LIF neuron in yesterday's tutorial.\n\nThe transfer function $F(\\cdot)$ in Equation $1$ represents the gain of the population as a function of the total input. The gain is often modeled as a sigmoidal function, i.e., more input drive leads to a nonlinear increase in the population firing rate. The output firing rate will eventually saturate for high input values. \n\nA sigmoidal $F(\\cdot)$ is parameterized by its gain $a$ and threshold $\\theta$.\n\n$$ F(x;a,\\theta) = \\frac{1}{1+\\text{e}^{-a(x-\\theta)}} - \\frac{1}{1+\\text{e}^{a\\theta}} \\quad(2)$$\n\nThe argument $x$ represents the input to the population. Note that the second term is chosen so that $F(0;a,\\theta)=0$.\n\nMany other transfer functions (generally monotonic) can be also used. Examples are the rectified linear function $ReLU(x)$ or the hyperbolic tangent $tanh(x)$.",
"_____no_output_____"
],
[
"### Exercise 1: Implement F-I curve \n\nLet's first investigate the activation functions before simulating the dynamics of the entire population. \n\nIn this exercise, you will implement a sigmoidal **F-I** curve or transfer function $F(x)$, with gain $a$ and threshold level $\\theta$ as parameters.",
"_____no_output_____"
]
],
[
[
"def F(x, a, theta):\n \"\"\"\n Population activation function.\n\n Args:\n x (float): the population input\n a (float): the gain of the function\n theta (float): the threshold of the function\n\n Returns:\n float: the population activation response F(x) for input x\n \"\"\"\n #################################################\n ## TODO for students: compute f = F(x) ##\n # Fill out function and remove\n raise NotImplementedError(\"Student excercise: implement the f-I function\")\n #################################################\n # add the expression of f = F(x)\n f = ...\n\n return f\n\n\npars = default_pars_single() # get default parameters\nx = np.arange(0, 10, .1) # set the range of input\n\n# Uncomment below to test your function\n# f = F(x, pars['a'], pars['theta'])\n\n# plot_fI(x, f)",
"_____no_output_____"
],
[
"# to_remove solution\n\n\ndef F(x, a, theta):\n \"\"\"\n Population activation function.\n\n Args:\n x (float): the population input\n a (float): the gain of the function\n theta (float): the threshold of the function\n\n Returns:\n float: the population activation response F(x) for input x\n \"\"\"\n\n # add the expression of f = F(x)\n f = (1 + np.exp(-a * (x - theta)))**-1 - (1 + np.exp(a * theta))**-1\n\n return f\n\n\npars = default_pars_single() # get default parameters\nx = np.arange(0, 10, .1) # set the range of input\n\n# Uncomment below to test your function\nf = F(x, pars['a'], pars['theta'])\n\nwith plt.xkcd():\n plot_fI(x, f)",
"_____no_output_____"
]
],
[
[
"### Interactive Demo: Parameter exploration of F-I curve\nHere's an interactive demo that shows how the F-I curve is changing for different values of the gain and threshold parameters.\n\n\n**Remember to enable the demo by running the cell.**",
"_____no_output_____"
]
],
[
[
"# @title\n\n# @markdown Make sure you execute this cell to enable the widget!\n\n\ndef interactive_plot_FI(a, theta):\n \"\"\"\n Population activation function.\n\n Expecxts:\n a : the gain of the function\n theta : the threshold of the function\n\n Returns:\n plot the F-I curve with give parameters\n \"\"\"\n\n # set the range of input\n x = np.arange(0, 10, .1)\n plt.figure()\n plt.plot(x, F(x, a, theta), 'k')\n plt.xlabel('x (a.u.)', fontsize=14)\n plt.ylabel('F(x)', fontsize=14)\n plt.show()\n\n\n_ = widgets.interact(interactive_plot_FI, a=(0.3, 3, 0.3), theta=(2, 4, 0.2))",
"_____no_output_____"
]
],
[
[
"## Section 1.3: Simulation scheme of E dynamics\n\nBecause $F(\\cdot)$ is a nonlinear function, the exact solution of Equation $1$ can not be determined via analytical methods. Therefore, numerical methods must be used to find the solution. In practice, the derivative on the left-hand side of Equation $1$ can be approximated using the Euler method on a time-grid of stepsize $\\Delta t$:\n\n\\begin{align}\n&\\frac{dr}{dt} \\approx \\frac{r[k+1]-r[k]}{\\Delta t} \n\\end{align}\nwhere $r[k] = r(k\\Delta t)$. \n\nThus,\n\n$$\\Delta r[k] = \\frac{\\Delta t}{\\tau}[-r[k] + F(w\\cdot r[k] + I_{\\text{ext}}(k;a,\\theta))]$$\n\n\nHence, Equation (1) is updated at each time step by:\n\n$$r[k+1] = r[k] + \\Delta r[k]$$\n\n**_Please execute the following cell to enable the WC simulator_**",
"_____no_output_____"
]
],
[
[
"# @title Single population rate model simulator: `simulate_single`\n\n\ndef simulate_single(pars):\n \"\"\"\n Simulate an excitatory population of neurons\n\n Args:\n pars : Parameter dictionary\n\n Returns:\n rE : Activity of excitatory population (array)\n\n Example:\n pars = default_pars_single()\n r = simulate_single(pars)\n \"\"\"\n\n # Set parameters\n tau, a, theta = pars['tau'], pars['a'], pars['theta']\n w = pars['w']\n I_ext = pars['I_ext']\n r_init = pars['r_init']\n dt, range_t = pars['dt'], pars['range_t']\n Lt = range_t.size\n\n # Initialize activity\n r = np.zeros(Lt)\n r[0] = r_init\n I_ext = I_ext * np.ones(Lt)\n\n # Update the E activity\n for k in range(Lt - 1):\n dr = dt / tau * (-r[k] + F(w * r[k] + I_ext[k], a, theta))\n r[k+1] = r[k] + dr\n\n return r\n\n\nprint(help(simulate_single))",
"Help on function simulate_single in module __main__:\n\nsimulate_single(pars)\n Simulate an excitatory population of neurons\n \n Args:\n pars : Parameter dictionary\n \n Returns:\n rE : Activity of excitatory population (array)\n \n Example:\n pars = default_pars_single()\n r = simulate_single(pars)\n\nNone\n"
]
],
[
[
"### Interactive Demo: Parameter Exploration of single population dynamics\n\nNote that $w=0$, as in the default setting, means no recurrent input to the neuron population in Equation (1). Hence, the dynamics is entirely determined by the external input $I_{\\text{ext}}$. Try to explore how $r_{\\text{sim}}(t)$ changes with different $I_{\\text{ext}}$ and $\\tau$ parameter values, and investigate the relationship between $F(I_{\\text{ext}}; a, \\theta)$ and the steady value of $r(t)$. Note that, $r_{\\rm ana}(t)$ denotes the analytical solution.",
"_____no_output_____"
]
],
[
[
"# @title\n\n# @markdown Make sure you execute this cell to enable the widget!\n\n# get default parameters\npars = default_pars_single(T=20.)\n\n\ndef Myplot_E_diffI_difftau(I_ext, tau):\n # set external input and time constant\n pars['I_ext'] = I_ext\n pars['tau'] = tau\n\n # simulation\n r = simulate_single(pars)\n\n # Analytical Solution\n r_ana = (pars['r_init']\n + (F(I_ext, pars['a'], pars['theta'])\n - pars['r_init']) * (1. - np.exp(-pars['range_t'] / pars['tau'])))\n\n # plot\n plt.figure()\n plt.plot(pars['range_t'], r, 'b', label=r'$r_{\\mathrm{sim}}$(t)', alpha=0.5,\n zorder=1)\n plt.plot(pars['range_t'], r_ana, 'b--', lw=5, dashes=(2, 2),\n label=r'$r_{\\mathrm{ana}}$(t)', zorder=2)\n plt.plot(pars['range_t'],\n F(I_ext, pars['a'], pars['theta']) * np.ones(pars['range_t'].size),\n 'k--', label=r'$F(I_{\\mathrm{ext}})$')\n plt.xlabel('t (ms)', fontsize=16.)\n plt.ylabel('Activity r(t)', fontsize=16.)\n plt.legend(loc='best', fontsize=14.)\n plt.show()\n\n\n_ = widgets.interact(Myplot_E_diffI_difftau, I_ext=(0.0, 10., 1.),\n tau=(1., 5., 0.2))",
"_____no_output_____"
]
],
[
[
"## Think!\nAbove, we have numerically solved a system driven by a positive input and that, if $w_{EE} \\neq 0$, receives an excitatory recurrent input (**try changing the value of $w_{EE}$ to a positive number**). Yet, $r_E(t)$ either decays to zero or reaches a fixed non-zero value.\n- Why doesn't the solution of the system \"explode\" in a finite time? In other words, what guarantees that $r_E$(t) stays finite? \n- Which parameter would you change in order to increase the maximum value of the response? ",
"_____no_output_____"
],
[
"# Section 2: Fixed points of the single population system\n",
"_____no_output_____"
]
],
[
[
"# @title Video 2: Fixed point\nfrom IPython.display import YouTubeVideo\nvideo = YouTubeVideo(id=\"Ox3ELd1UFyo\", width=854, height=480, fs=1)\nprint(\"Video available at https://youtube.com/watch?v=\" + video.id)\nvideo",
"Video available at https://youtube.com/watch?v=Ox3ELd1UFyo\n"
]
],
[
[
"As you varied the two parameters in the last Interactive Demo, you noticed that, while at first the system output quickly changes, with time, it reaches its maximum/minimum value and does not change anymore. The value eventually reached by the system is called the **steady state** of the system, or the **fixed point**. Essentially, in the steady states the derivative with respect to time of the activity ($r$) is zero, i.e. $\\displaystyle \\frac{dr}{dt}=0$. \n\nWe can find that the steady state of the Equation. (1) by setting $\\displaystyle{\\frac{dr}{dt}=0}$ and solve for $r$:\n\n$$-r_{\\text{steady}} + F(w\\cdot r_{\\text{steady}} + I_{\\text{ext}};a,\\theta) = 0, \\qquad (3)$$\n\nWhen it exists, the solution of Equation. (3) defines a **fixed point** of the dynamical system Equation. (1). Note that if $F(x)$ is nonlinear, it is not always possible to find an analytical solution, but the solution can be found via numerical simulations, as we will do later.\n\nFrom the Interactive Demo, one could also notice that the value of $\\tau$ influences how quickly the activity will converge to the steady state from its initial value. \n\nIn the specific case of $w=0$, we can also analytically compute the solution of Equation (1) (i.e., the thick blue dashed line) and deduce the role of $\\tau$ in determining the convergence to the fixed point: \n\n$$\\displaystyle{r(t) = \\big{[}F(I_{\\text{ext}};a,\\theta) -r(t=0)\\big{]} (1-\\text{e}^{-\\frac{t}{\\tau}})} + r(t=0)$$ \\\\\n\nWe can now numerically calculate the fixed point with the `scipy.optimize.root` function.\n\n<font size=3><font color='gray'>_(Recall that at the very beginning, we `import scipy.optimize as opt` )_</font></font>.\n\n\\\\\n\nPlease execute the cell below to define the functions `my_fp_single` and `check_fp_single`",
"_____no_output_____"
]
],
[
[
"# @title Function of calculating the fixed point\n\n# @markdown Make sure you execute this cell to enable the function!\n\n\ndef my_fp_single(pars, r_init):\n \"\"\"\n Calculate the fixed point through drE/dt=0\n\n Args:\n pars : Parameter dictionary\n r_init : Initial value used for scipy.optimize function\n\n Returns:\n x_fp : value of fixed point\n \"\"\"\n\n # get the parameters\n a, theta = pars['a'], pars['theta']\n w = pars['w']\n I_ext = pars['I_ext']\n\n # define the right hand of E dynamics\n def my_WCr(x):\n r = x\n drdt = (-r + F(w * r + I_ext, a, theta))\n y = np.array(drdt)\n\n return y\n\n x0 = np.array(r_init)\n x_fp = opt.root(my_WCr, x0).x\n\n return x_fp\n\n\nprint(help(my_fp_single))\n\n\ndef check_fp_single(pars, x_fp, mytol=1e-4):\n \"\"\"\n Verify |dr/dt| < mytol\n\n Args:\n pars : Parameter dictionary\n fp : value of fixed point\n mytol : tolerance, default as 10^{-4}\n\n Returns :\n Whether it is a correct fixed point: True/False\n \"\"\"\n\n a, theta = pars['a'], pars['theta']\n w = pars['w']\n I_ext = pars['I_ext']\n\n # calculate Equation(3)\n y = x_fp - F(w * x_fp + I_ext, a, theta)\n\n # Here we set tolerance as 10^{-4}\n return np.abs(y) < mytol\n\n\nprint(help(check_fp_single))",
"Help on function my_fp_single in module __main__:\n\nmy_fp_single(pars, r_init)\n Calculate the fixed point through drE/dt=0\n \n Args:\n pars : Parameter dictionary\n r_init : Initial value used for scipy.optimize function\n \n Returns:\n x_fp : value of fixed point\n\nNone\nHelp on function check_fp_single in module __main__:\n\ncheck_fp_single(pars, x_fp, mytol=0.0001)\n Verify |dr/dt| < mytol\n \n Args:\n pars : Parameter dictionary\n fp : value of fixed point\n mytol : tolerance, default as 10^{-4}\n \n Returns :\n Whether it is a correct fixed point: True/False\n\nNone\n"
]
],
[
[
"## Exercise 2: Visualization of the fixed point\n\nWhen it is not possible to find the solution for Equation (3) analytically, a graphical approach can be taken. To that end, it is useful to plot $\\displaystyle{\\frac{dr}{dt}=0}$ as a function of $r$. The values of $r$ for which the plotted function crosses zero on the y axis correspond to fixed points. \n\nHere, let us, for example, set $w=5.0$ and $I^{\\text{ext}}=0.5$. From Equation (1), you can obtain\n\n$$\\frac{dr}{dt} = [-r + F(w\\cdot r + I^{\\text{ext}})]/\\tau $$\n\nThen, plot the $dr/dt$ as a function of $r$, and check for the presence of fixed points. ",
"_____no_output_____"
],
[
"Finally, try to find the fixed points using the previously defined function `my_fp_single(pars, r_init)` with proper initial values ($r_{\\text{init}}$). You can use the previously defined function `check_fp_single(pars, x_fp)` to verify that the values of $r{\\rm fp}$ for which $\\displaystyle{\\frac{dr}{dt}} = 0$ are the true fixed points. From the line $\\displaystyle{\\frac{dr}{dt}}$ plotted above, the proper initial values can be chosen as the values close to where the line crosses zero on the y axis (real fixed point).",
"_____no_output_____"
]
],
[
[
"pars = default_pars_single() # get default parameters\n\n# set your external input and wEE\npars['I_ext'] = 0.5\npars['w'] = 5.0\n\nr = np.linspace(0, 1, 1000) # give the values of r\n\n# Calculate drEdt\n# drdt = ...\n\n# Uncomment this to plot the drdt across r\n# plot_dr_r(r, drdt)\n\n################################################################\n# TODO for students:\n# Find the values close to the intersections of drdt and y=0\n# as your initial values\n# Calculate the fixed point with your initial value, verify them,\n# and plot the corret ones\n# check if x_fp is the intersection of the lines with the given function\n# check_fpE(pars, x_fp)\n# vary different initial values to find the correct fixed point (Should be 3)\n# Use blue, red and yellow colors, respectively ('b', 'r', 'y' codenames)\n################################################################\n\n# Calculate the first fixed point with your initial value\n# x_fp_1 = my_fp_single(pars, ...)\n# if check_fp_single(pars, x_fp_1):\n# plt.plot(x_fp_1, 0, 'bo', ms=8)\n\n# Calculate the second fixed point with your initial value\n# x_fp_2 = my_fp_single(pars, ...)\n# if check_fp_single(pars, x_fp_2):\n# plt.plot(x_fp_2, 0, 'ro', ms=8)\n\n# Calculate the third fixed point with your initial value\n# x_fp_3 = my_fp_single(pars, ...)\n# if check_fp_single(pars, x_fp_3):\n# plt.plot(x_fp_3, 0, 'yo', ms=8)",
"_____no_output_____"
],
[
"# to_remove solution\npars = default_pars_single() # get default parameters\n\n# set your external input and wEE\npars['I_ext'] = 0.5\npars['w'] = 5.0\n\nr = np.linspace(0, 1, 1000) # give the values of r\n\n# Calculate drEdt\ndrdt = (-r + F(pars['w'] * r + pars['I_ext'],\n pars['a'], pars['theta'])) / pars['tau']\n\nwith plt.xkcd():\n plot_dr_r(r, drdt)\n\n # Calculate the first fixed point with your initial value\n x_fp_1 = my_fp_single(pars, 0.)\n if check_fp_single(pars, x_fp_1):\n plt.plot(x_fp_1, 0, 'bo', ms=8)\n\n # Calculate the second fixed point with your initial value\n x_fp_2 = my_fp_single(pars, 0.4)\n if check_fp_single(pars, x_fp_2):\n plt.plot(x_fp_2, 0, 'ro', ms=8)\n\n # Calculate the third fixed point with your initial value\n x_fp_3 = my_fp_single(pars, 0.9)\n if check_fp_single(pars, x_fp_3):\n plt.plot(x_fp_3, 0, 'yo', ms=8)\n\n plt.show()",
"_____no_output_____"
]
],
[
[
"## Interactive Demo: fixed points as a function of recurrent and external inputs.\n\nYou can now explore how the previous plot changes when the recurrent coupling $w$ and the external input $I_{\\text{ext}}$ take different values.",
"_____no_output_____"
]
],
[
[
"# @title\n\n# @markdown Make sure you execute this cell to enable the widget!\n\n\ndef plot_intersection_single(w, I_ext):\n # set your parameters\n pars['w'] = w\n pars['I_ext'] = I_ext\n\n # note that wEE!=0\n if w > 0:\n # find fixed point\n x_fp_1 = my_fp_single(pars, 0.)\n x_fp_2 = my_fp_single(pars, 0.4)\n x_fp_3 = my_fp_single(pars, 0.9)\n\n plt.figure()\n r = np.linspace(0, 1., 1000)\n drdt = (-r + F(w * r + I_ext, pars['a'], pars['theta'])) / pars['tau']\n\n plt.plot(r, drdt, 'k')\n plt.plot(r, 0. * r, 'k--')\n\n if check_fp_single(pars, x_fp_1):\n plt.plot(x_fp_1, 0, 'bo', ms=8)\n if check_fp_single(pars, x_fp_2):\n plt.plot(x_fp_2, 0, 'ro', ms=8)\n if check_fp_single(pars, x_fp_3):\n plt.plot(x_fp_3, 0, 'yo', ms=8)\n\n plt.xlabel(r'$r$', fontsize=14.)\n plt.ylabel(r'$\\frac{dr}{dt}$', fontsize=20.)\n\n plt.show()\n\n\n_ = widgets.interact(plot_intersection_single, w=(1, 7, 0.2),\n I_ext=(0, 3, 0.1))",
"_____no_output_____"
]
],
[
[
"---\n# Summary\n\nIn this tutorial, we have investigated the dynamics of a rate-based single population of neurons.\n\nWe learned about:\n- The effect of the input parameters and the time constant of the network on the dynamics of the population.\n- How to find the fixed point(s) of the system.\n\nNext, we have two Bonus, but important concepts in dynamical system analysis and simulation. If you have time left, watch the next video and proceed to solve the exercises. You will learn:\n\n- How to determine the stability of a fixed point by linearizing the system.\n- How to add realistic inputs to our model.",
"_____no_output_____"
],
[
"---\n# Bonus 1: Stability of a fixed point",
"_____no_output_____"
]
],
[
[
"# @title Video 3: Stability of fixed points\nfrom IPython.display import YouTubeVideo\nvideo = YouTubeVideo(id=\"KKMlWWU83Jg\", width=854, height=480, fs=1)\nprint(\"Video available at https://youtube.com/watch?v=\" + video.id)\nvideo",
"Video available at https://youtube.com/watch?v=KKMlWWU83Jg\n"
]
],
[
[
"#### Initial values and trajectories\n\nHere, let us first set $w=5.0$ and $I_{\\text{ext}}=0.5$, and investigate the dynamics of $r(t)$ starting with different initial values $r(0) \\equiv r_{\\text{init}}$. We will plot the trajectories of $r(t)$ with $r_{\\text{init}} = 0.0, 0.1, 0.2,..., 0.9$.",
"_____no_output_____"
]
],
[
[
"# @title Initial values\n\n# @markdown Make sure you execute this cell to see the trajectories!\n\npars = default_pars_single()\npars['w'] = 5.0\npars['I_ext'] = 0.5\n\nplt.figure(figsize=(8, 5))\nfor ie in range(10):\n pars['r_init'] = 0.1 * ie # set the initial value\n r = simulate_single(pars) # run the simulation\n\n # plot the activity with given initial\n plt.plot(pars['range_t'], r, 'b', alpha=0.1 + 0.1 * ie,\n label=r'r$_{\\mathrm{init}}$=%.1f' % (0.1 * ie))\n\nplt.xlabel('t (ms)')\nplt.title('Two steady states?')\nplt.ylabel(r'$r$(t)')\nplt.legend(loc=[1.01, -0.06], fontsize=14)\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Interactive Demo: dynamics as a function of the initial value\n\nLet's now set $r_{\\rm init}$ to a value of your choice in this demo. How does the solution change? What do you observe?",
"_____no_output_____"
]
],
[
[
"# @title\n\n# @markdown Make sure you execute this cell to enable the widget!\n\npars = default_pars_single()\npars['w'] = 5.0\npars['I_ext'] = 0.5\n\n\ndef plot_single_diffEinit(r_init):\n pars['r_init'] = r_init\n r = simulate_single(pars)\n\n plt.figure()\n plt.plot(pars['range_t'], r, 'b', zorder=1)\n plt.plot(0, r[0], 'bo', alpha=0.7, zorder=2)\n plt.xlabel('t (ms)', fontsize=16)\n plt.ylabel(r'$r(t)$', fontsize=16)\n plt.ylim(0, 1.0)\n plt.show()\n\n\n_ = widgets.interact(plot_single_diffEinit, r_init=(0, 1, 0.02))",
"_____no_output_____"
]
],
[
[
"### Stability analysis via linearization of the dynamics\n\nJust like Equation $1$ in the case ($w=0$) discussed above, a generic linear system \n$$\\frac{dx}{dt} = \\lambda (x - b),$$ \nhas a fixed point for $x=b$. The analytical solution of such a system can be found to be:\n$$x(t) = b + \\big{(} x(0) - b \\big{)} \\text{e}^{\\lambda t}.$$ \nNow consider a small perturbation of the activity around the fixed point: $x(0) = b+ \\epsilon$, where $|\\epsilon| \\ll 1$. Will the perturbation $\\epsilon(t)$ grow with time or will it decay to the fixed point? The evolution of the perturbation with time can be written, using the analytical solution for $x(t)$, as:\n $$\\epsilon (t) = x(t) - b = \\epsilon \\text{e}^{\\lambda t}$$\n\n- if $\\lambda < 0$, $\\epsilon(t)$ decays to zero, $x(t)$ will still converge to $b$ and the fixed point is \"**stable**\".\n\n- if $\\lambda > 0$, $\\epsilon(t)$ grows with time, $x(t)$ will leave the fixed point $b$ exponentially, and the fixed point is, therefore, \"**unstable**\" .",
"_____no_output_____"
],
[
"### Compute the stability of Equation $1$\n\nSimilar to what we did in the linear system above, in order to determine the stability of a fixed point $r^{*}$ of the excitatory population dynamics, we perturb Equation (1) around $r^{*}$ by $\\epsilon$, i.e. $r = r^{*} + \\epsilon$. We can plug in Equation (1) and obtain the equation determining the time evolution of the perturbation $\\epsilon(t)$:\n\n\\begin{align}\n\\tau \\frac{d\\epsilon}{dt} \\approx -\\epsilon + w F'(w\\cdot r^{*} + I_{\\text{ext}};a,\\theta) \\epsilon \n\\end{align}\n\nwhere $F'(\\cdot)$ is the derivative of the transfer function $F(\\cdot)$. We can rewrite the above equation as:\n\n\\begin{align}\n\\frac{d\\epsilon}{dt} \\approx \\frac{\\epsilon}{\\tau }[-1 + w F'(w\\cdot r^* + I_{\\text{ext}};a,\\theta)] \n\\end{align}\n\nThat is, as in the linear system above, the value of\n\n$$\\lambda = [-1+ wF'(w\\cdot r^* + I_{\\text{ext}};a,\\theta)]/\\tau \\qquad (4)$$\n\ndetermines whether the perturbation will grow or decay to zero, i.e., $\\lambda$ defines the stability of the fixed point. This value is called the **eigenvalue** of the dynamical system.",
"_____no_output_____"
],
[
"## Exercise 3: Compute $dF$ and Eigenvalue\n\nThe derivative of the sigmoid transfer function is:\n\\begin{align} \n\\frac{dF}{dx} & = \\frac{d}{dx} (1+\\exp\\{-a(x-\\theta)\\})^{-1} \\\\\n& = a\\exp\\{-a(x-\\theta)\\} (1+\\exp\\{-a(x-\\theta)\\})^{-2}. \\qquad (5)\n\\end{align}\n\nLet's now find the expression for the derivative $\\displaystyle{\\frac{dF}{dx}}$ in the following cell and plot it.",
"_____no_output_____"
]
],
[
[
"def dF(x, a, theta):\n \"\"\"\n Population activation function.\n\n Args:\n x : the population input\n a : the gain of the function\n theta : the threshold of the function\n\n Returns:\n dFdx : the population activation response F(x) for input x\n \"\"\"\n\n #################################################\n # TODO for students: compute dFdx ##\n raise NotImplementedError(\"Student excercise: compute the deravitive of F\")\n #################################################\n # Calculate the population activation\n dFdx = ...\n\n return dFdx\n\n\npars = default_pars_single() # get default parameters\nx = np.arange(0, 10, .1) # set the range of input\n\n# Uncomment below to test your function\n# df = dF(x, pars['a'], pars['theta'])\n\n# plot_dFdt(x, df)",
"_____no_output_____"
],
[
"# to_remove solution\ndef dF(x, a, theta):\n \"\"\"\n Population activation function.\n\n Args:\n x : the population input\n a : the gain of the function\n theta : the threshold of the function\n\n Returns:\n dFdx : the population activation response F(x) for input x\n \"\"\"\n\n # Calculate the population activation\n dFdx = a * np.exp(-a * (x - theta)) * (1 + np.exp(-a * (x - theta)))**-2\n\n return dFdx\n\n\npars = default_pars_single() # get default parameters\nx = np.arange(0, 10, .1) # set the range of input\n\ndf = dF(x, pars['a'], pars['theta'])\n\nwith plt.xkcd():\n plot_dFdt(x, df)",
"_____no_output_____"
]
],
[
[
"## Exercise 4: Compute eigenvalues\n\nAs discussed above, for the case with $w=5.0$ and $I_{\\text{ext}}=0.5$, the system displays **three** fixed points. However, when we simulated the dynamics and varied the initial conditions $r_{\\rm init}$, we could only obtain **two** steady states. In this exercise, we will now check the stability of each of the three fixed points by calculating the corresponding eigenvalues with the function `eig_single`. Check the sign of each eigenvalue (i.e., stability of each fixed point). How many of the fixed points are stable?\n\nNote that the expression of the eigenvalue at fixed point $r^*$\n$$\\lambda = [-1+ wF'(w\\cdot r^* + I_{\\text{ext}};a,\\theta)]/\\tau$$",
"_____no_output_____"
]
],
[
[
"def eig_single(pars, fp):\n \"\"\"\n Args:\n pars : Parameter dictionary\n fp : fixed point r_fp\n\n Returns:\n eig : eigevalue of the linearized system\n \"\"\"\n\n # get the parameters\n tau, a, theta = pars['tau'], pars['a'], pars['theta']\n w, I_ext = pars['w'], pars['I_ext']\n print(tau, a, theta, w, I_ext)\n\n #################################################\n ## TODO for students: compute eigenvalue ##\n raise NotImplementedError(\"Student excercise: compute the eigenvalue\")\n #################################################\n # Compute the eigenvalue\n eig = ...\n\n return eig\n\n\npars = default_pars_single()\npars['w'] = 5.0\npars['I_ext'] = 0.5\n\n# Uncomment below lines after completing the eig_single function.\n# Find the eigenvalues for all fixed points of Exercise 2\n\n# x_fp_1 = my_fp_single(pars, 0.).item()\n# eig_fp_1 = eig_single(pars, x_fp_1).item()\n# print(f'Fixed point1 at {x_fp_1:.3f} with Eigenvalue={eig_fp_1:.3f}')\n\n# x_fp_2 = my_fp_single(pars, 0.4).item()\n# eig_fp_2 = eig_single(pars, x_fp_2).item()\n# print(f'Fixed point2 at {x_fp_2:.3f} with Eigenvalue={eig_fp_2:.3f}')\n\n# x_fp_3 = my_fp_single(pars, 0.9).item()\n# eig_fp_3 = eig_single(pars, x_fp_3).item()\n# print(f'Fixed point3 at {x_fp_3:.3f} with Eigenvalue={eig_fp_3:.3f}')",
"_____no_output_____"
]
],
[
[
"**SAMPLE OUTPUT**\n\n```\nFixed point1 at 0.042 with Eigenvalue=-0.583\nFixed point2 at 0.447 with Eigenvalue=0.498\nFixed point3 at 0.900 with Eigenvalue=-0.626\n```",
"_____no_output_____"
]
],
[
[
"# to_remove solution\ndef eig_single(pars, fp):\n \"\"\"\n Args:\n pars : Parameter dictionary\n fp : fixed point r_fp\n\n Returns:\n eig : eigevalue of the linearized system\n \"\"\"\n\n # get the parameters\n tau, a, theta = pars['tau'], pars['a'], pars['theta']\n w, I_ext = pars['w'], pars['I_ext']\n print(tau, a, theta, w, I_ext)\n\n # Compute the eigenvalue\n eig = (-1. + w * dF(w * fp + I_ext, a, theta)) / tau\n\n return eig\n\n\npars = default_pars_single()\npars['w'] = 5.0\npars['I_ext'] = 0.5\n\n# Find the eigenvalues for all fixed points of Exercise 2\n\nx_fp_1 = my_fp_single(pars, 0.).item()\neig_fp_1 = eig_single(pars, x_fp_1).item()\nprint(f'Fixed point1 at {x_fp_1:.3f} with Eigenvalue={eig_fp_1:.3f}')\n\nx_fp_2 = my_fp_single(pars, 0.4).item()\neig_fp_2 = eig_single(pars, x_fp_2).item()\nprint(f'Fixed point2 at {x_fp_2:.3f} with Eigenvalue={eig_fp_2:.3f}')\n\nx_fp_3 = my_fp_single(pars, 0.9).item()\neig_fp_3 = eig_single(pars, x_fp_3).item()\nprint(f'Fixed point3 at {x_fp_3:.3f} with Eigenvalue={eig_fp_3:.3f}')",
"1.0 1.2 2.8 5.0 0.5\nFixed point1 at 0.042 with Eigenvalue=-0.583\n1.0 1.2 2.8 5.0 0.5\nFixed point2 at 0.447 with Eigenvalue=0.498\n1.0 1.2 2.8 5.0 0.5\nFixed point3 at 0.900 with Eigenvalue=-0.626\n"
]
],
[
[
"## Think! \nThroughout the tutorial, we have assumed $w> 0 $, i.e., we considered a single population of **excitatory** neurons. What do you think will be the behavior of a population of inhibitory neurons, i.e., where $w> 0$ is replaced by $w< 0$? ",
"_____no_output_____"
],
[
"---\n# Bonus 2: Noisy input drives the transition between two stable states\n\n",
"_____no_output_____"
],
[
"## Ornstein-Uhlenbeck (OU) process\n\nAs discussed in several previous tutorials, the OU process is usually used to generate a noisy input into the neuron. The OU input $\\eta(t)$ follows: \n\n$$\\tau_\\eta \\frac{d}{dt}\\eta(t) = -\\eta (t) + \\sigma_\\eta\\sqrt{2\\tau_\\eta}\\xi(t)$$\n\nExecute the following function `my_OU(pars, sig, myseed=False)` to generate an OU process.",
"_____no_output_____"
]
],
[
[
"# @title OU process `my_OU(pars, sig, myseed=False)`\n\n# @markdown Make sure you execute this cell to visualize the noise!\n\n\ndef my_OU(pars, sig, myseed=False):\n \"\"\"\n A functions that generates Ornstein-Uhlenback process\n\n Args:\n pars : parameter dictionary\n sig : noise amplitute\n myseed : random seed. int or boolean\n\n Returns:\n I : Ornstein-Uhlenbeck input current\n \"\"\"\n\n # Retrieve simulation parameters\n dt, range_t = pars['dt'], pars['range_t']\n Lt = range_t.size\n tau_ou = pars['tau_ou'] # [ms]\n\n # set random seed\n if myseed:\n np.random.seed(seed=myseed)\n else:\n np.random.seed()\n\n # Initialize\n noise = np.random.randn(Lt)\n I_ou = np.zeros(Lt)\n I_ou[0] = noise[0] * sig\n\n # generate OU\n for it in range(Lt - 1):\n I_ou[it + 1] = (I_ou[it]\n + dt / tau_ou * (0. - I_ou[it])\n + np.sqrt(2 * dt / tau_ou) * sig * noise[it + 1])\n\n return I_ou\n\n\npars = default_pars_single(T=100)\npars['tau_ou'] = 1. # [ms]\nsig_ou = 0.1\nI_ou = my_OU(pars, sig=sig_ou, myseed=2020)\nplt.figure(figsize=(10, 4))\nplt.plot(pars['range_t'], I_ou, 'r')\nplt.xlabel('t (ms)')\nplt.ylabel(r'$I_{\\mathrm{OU}}$')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Example: Up-Down transition\n\nIn the presence of two or more fixed points, noisy inputs can drive a transition between the fixed points! Here, we stimulate an E population for 1,000 ms applying OU inputs.",
"_____no_output_____"
]
],
[
[
"# @title Simulation of an E population with OU inputs\n\n# @markdown Make sure you execute this cell to spot the Up-Down states!\n\npars = default_pars_single(T=1000)\npars['w'] = 5.0\nsig_ou = 0.7\npars['tau_ou'] = 1. # [ms]\npars['I_ext'] = 0.56 + my_OU(pars, sig=sig_ou, myseed=2020)\n\nr = simulate_single(pars)\n\nplt.figure(figsize=(10, 4))\nplt.plot(pars['range_t'], r, 'b', alpha=0.8)\nplt.xlabel('t (ms)')\nplt.ylabel(r'$r(t)$')\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0e15bee3b601aa9327929451660a788487c9a85 | 2,036 | ipynb | Jupyter Notebook | networks/autoencoder/Untitled.ipynb | PiotrGrzybowski/NeuralNetworks | 4fe4295761bc1a1df5d52e69ce574256626833c6 | [
"Apache-2.0"
] | null | null | null | networks/autoencoder/Untitled.ipynb | PiotrGrzybowski/NeuralNetworks | 4fe4295761bc1a1df5d52e69ce574256626833c6 | [
"Apache-2.0"
] | null | null | null | networks/autoencoder/Untitled.ipynb | PiotrGrzybowski/NeuralNetworks | 4fe4295761bc1a1df5d52e69ce574256626833c6 | [
"Apache-2.0"
] | null | null | null | 22.622222 | 80 | 0.510806 | [
[
[
"import pickle\n\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport sys\nsys.path.append(\"../\") # go to parent dir\nfrom autoencoder.mlp import Network\n\ntest_data = np.load('test.npy')\ntrain_data = np.load('train.npy')",
"_____no_output_____"
],
[
"batch_size = 32\nlearning_rate = 0.2\nl2 = 0.0\nepochs = 5\nhidden=100\n\nresults = []\nfor size in [1,2,4,5,8,10,16,20,32,40,60]:\n print(\"Size = {}\".format(size * 1000))\n mlp = Network(100)\n mlp.drop = 0\n train = train_data[:, :size * 1000]\n mlp.train(train, learning_rate, epochs, batch_size, l2, test_data)\n results.append(mlp)\n \n mlp.hidden_weights = np.random.randn(hidden, 784) / np.sqrt(784)\n mlp.output_weights = np.random.randn(784, hidden) / np.sqrt(hidden)\n\n mlp.hidden_bias = np.ones(shape=(hidden, 1))\n mlp.output_bias = np.ones(shape=(784, 1))\n \n ",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
d0e16c36402efb412b866c7bdef608ad184cf352 | 15,552 | ipynb | Jupyter Notebook | documentos/.ipynb_checkpoints/estructuraweb2-checkpoint.ipynb | psnti/SubRepositorio | 3ddab49cc7d9d02667f83a7bb9e3485cf1984243 | [
"MIT"
] | null | null | null | documentos/.ipynb_checkpoints/estructuraweb2-checkpoint.ipynb | psnti/SubRepositorio | 3ddab49cc7d9d02667f83a7bb9e3485cf1984243 | [
"MIT"
] | 1 | 2020-04-28T08:43:20.000Z | 2020-04-28T08:45:04.000Z | documentos/.ipynb_checkpoints/estructuraweb2-checkpoint.ipynb | psnti/WebJellyfishForecast | 3ddab49cc7d9d02667f83a7bb9e3485cf1984243 | [
"MIT"
] | null | null | null | 33.019108 | 153 | 0.434799 | [
[
[
"import xarray as xr\nimport pandas as pd\nfrom joblib import load\nimport os\nimport math\nfrom datetime import datetime,timedelta\nfrom sklearn import preprocessing",
"_____no_output_____"
],
[
"def redondeo(coordenadas, base=1/12):\n \"\"\"\n Devuelve las coordenadas pasadas redondeadas\n \n Parametros:\n coordenadas -- lista de latitud y longitud\n base -- base del redondeo\n \"\"\"\n return base * round(coordenadas/base)\n",
"_____no_output_____"
],
[
"fecha = '2018-11-16'\ncoordenadas = '[-26.99053888888889, -70.78993333333334]'\nsalto = 1/12\nvar = ['mlotst','zos','bottomT','thetao','so','uo','vo']",
"_____no_output_____"
],
[
"separacion = coordenadas.index(', ')\nfinal = coordenadas.index(']')\ncoordenadas = [float(coordenadas[1:separacion]),float(coordenadas[separacion+2:final])]\ncoordenadas = [redondeo(coordenadas[0]),redondeo(coordenadas[1])]\ncoordenadas",
"_____no_output_____"
],
[
"def genera_resultados(fecha, coordenadas):\n # se cogerian los datos de copernicus\n # generar dataframe\n #normalizarlo\n # meterlo al modelo\n modelo = load('C:\\\\Users\\pablo\\Desktop\\medusas\\static\\modelo.joblib')\n df,fechas = genera_estructura(fecha,coordenadas)\n df = normaliza_min_max(df)\n salida = modelo.predict(df)\n print(salida,fechas)\n \n# lista = [{'y': '2020-1-1', 'v': 1},\n# {'y': '2020-2-1', 'v': 10},\n# {'y': '2020-1-5', 'v': 5},\n# {'y': '2020-3-5', 'v': 2}]\n# return lista\n return df",
"_____no_output_____"
],
[
"def genera_estructura(f,c):\n dataframe = pd.DataFrame(columns=list(range(231)))\n fechas = genera_fechas(f)\n for index,dia in enumerate(fechas):\n listado_variables = []\n # cargo el dataset\n ds =busca_archivo(dia) # cambiar para cada dia\n c = comprueba_datos(c[0],c[1],ds)\n coord = dame_coordenadas(c)\n\n for j in coord:\n variables1 = ds.sel({'latitude':coord[0][0],'longitude': coord[0][1], 'depth' : 0 },method='nearest').to_dataframe()\n l1 = dame_lista(variables1)[0]\n variables2 = ds.sel({'latitude':coord[1][0],'longitude': coord[1][1], 'depth' : 5 },method='nearest').to_dataframe()\n l2 = dame_lista(variables2)[0]\n variables3 = ds.sel({'latitude':coord[2][0],'longitude': coord[2][1], 'depth' : 10},method='nearest').to_dataframe()\n l3 = dame_lista(variables3)[0]\n l1+=l2\n l1+=l3\n listado_variables+=(l1)\n dataframe.loc[index] = listado_variables\n return dataframe,fechas",
"_____no_output_____"
],
[
"def normaliza_min_max(df_atributos):\n \"\"\"\n Normaliza los datos del dataframe pasado\n \"\"\"\n X = df_atributos.values.tolist()\n n = load('normalizador.pkl') \n x_normalizado_2 = n.transform(X)\n df_norm = pd.DataFrame(x_normalizado_2,columns=list(range(231)))\n return df_norm",
"_____no_output_____"
],
[
"def genera_fechas(f):\n lista_fechas = []\n for i in range(5):\n fecha= datetime.strptime(f, '%Y-%m-%d')\n fecha += timedelta(days=i)\n lista_fechas.append(str(fecha))\n return lista_fechas\n\ndef dame_lista(df):\n Row_list = []\n for index, rows in df.iterrows(): \n my_list =[rows.mlotst, rows.zos, rows.bottomT, rows.thetao, rows.so,\n rows.uo, rows.vo] \n Row_list.append(my_list) \n\n # Print the list \n return Row_list\n\ndef busca_archivo(fecha):\n \"\"\"\n Devuelve el archivo .nc de la fecha pasada por parametro\n \n Parametros:\n fecha -- fecha en formato AñoMesDia (20140105)\n \"\"\"\n listado_archivos = os.listdir('C:\\\\Users\\pablo\\Desktop\\medusas\\documentos\\copernicus') # Listo todos los archivos de Copernicus\n texto ='_{}_'.format(str(fecha).split()[0].replace('-',''))\n archivo = [x for x in listado_archivos if str(texto) in x]\n data = xr.open_dataset('C:\\\\Users\\pablo\\Desktop\\medusas\\documentos\\copernicus\\{}'.format(archivo[0])) # cargo el archivo\n return data # devuelvo dataset\n\ndef dame_coordenadas(c):\n paso = 1/12\n return [[c[0],c[1]],\n [c[0],c[1]-paso],[c[0]+paso,c[1]-paso],[c[0]+(paso*2),c[1]-paso],[c[0]-paso,c[1]-paso],[c[0]-(paso*2),c[1]-paso],\n [c[0],c[1]-(2*paso)],[c[0]+paso,c[1]-(2*paso)],[c[0]+(paso*2),c[1]-(2*paso)],[c[0]-paso,c[1]-(2*paso)],[c[0]-(paso*2),c[1]-(2*paso)]]\n\ndef comprueba_datos(latitud,longitud,ds):\n \"\"\"\n Comprueba si el dataset contiene valores en las coordenadas pasadas\n \n Devuelve las coordenadas mas cercanas con datos \n \n Parametros:\n latitud -- latitud\n longitud -- longitud\n ds -- dataset del que extraer los valores\n \"\"\"\n valor = dame_datos(latitud,longitud,ds)\n while math.isnan(valor.mlotst[0]):\n longitud = longitud - salto\n valor = dame_datos(latitud,longitud,ds)\n return latitud,longitud # devuelvo las coordenadas con datos\n\ndef dame_datos(latitud,longitud,ds):\n \"\"\"\n Devuelve los datos del dataset en las coordenadas pasadas\n \n Parametros:\n latitud -- latitud \n longitud -- longitud\n ds -- dataset del que extraer los valores\n \"\"\"\n return ds.sel({'latitude':latitud,'longitude': longitud})",
"_____no_output_____"
],
[
"d = genera_resultados(fecha,coordenadas)\nd",
"[100. 5. 23. 23. 17.]\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e1739e482dc5ef19c44ba9f7d89d24e541b4aa | 11,417 | ipynb | Jupyter Notebook | tpfinal/s5_model_explorer.ipynb | oussamagh97/tac | 7203d88fcbcb5560ebf13751f7c07bd81a4b10b0 | [
"MIT"
] | null | null | null | tpfinal/s5_model_explorer.ipynb | oussamagh97/tac | 7203d88fcbcb5560ebf13751f7c07bd81a4b10b0 | [
"MIT"
] | null | null | null | tpfinal/s5_model_explorer.ipynb | oussamagh97/tac | 7203d88fcbcb5560ebf13751f7c07bd81a4b10b0 | [
"MIT"
] | null | null | null | 25.037281 | 101 | 0.516861 | [
[
[
"from gensim.models import Word2Vec\nfrom pprint import pprint",
"_____no_output_____"
]
],
[
[
"# Charger le modèle que nous venons de créer",
"_____no_output_____"
]
],
[
[
"model = Word2Vec.load(\"../tpfinal/bulletins.model\")",
"_____no_output_____"
]
],
[
[
"# Comment est représenté le mot \"rue\" ?",
"_____no_output_____"
]
],
[
[
"model[\"hospices\"]",
"_____no_output_____"
]
],
[
[
"# Et fleuriste ?",
"_____no_output_____"
]
],
[
[
"model['medicaux']",
"_____no_output_____"
]
],
[
[
"# Quel est leur similarité ?",
"_____no_output_____"
]
],
[
[
"model.wv.similarity(\"budget\", \"lois\")",
"_____no_output_____"
],
[
"model.wv.similarity(\"services\", \"revenus\")",
"_____no_output_____"
],
[
"model.wv.similarity(\"ville\",\"politique\")",
"_____no_output_____"
],
[
"model.wv.similarity(\"hospices\",\"travaux\")",
"_____no_output_____"
]
],
[
[
"# Quel mot est le plus proche de rue ?",
"_____no_output_____"
]
],
[
[
"model.wv.most_similar(\"hospices\", topn=30)",
"_____no_output_____"
]
],
[
[
"# Et de Bruxelles ?",
"_____no_output_____"
]
],
[
[
"model.wv.most_similar(\"hopitaux\")",
"_____no_output_____"
],
[
"model.wv.most_similar(\"medicaux\")",
"_____no_output_____"
],
[
"model.wv.most_similar(\"medical\")",
"_____no_output_____"
],
[
"model.wv.most_similar(\"malade\")",
"_____no_output_____"
],
[
"model.wv.most_similar(\"politique\")",
"_____no_output_____"
]
],
[
[
"# Comment obtenir une ville de France grâce à notre modèle ?",
"_____no_output_____"
]
],
[
[
"model.wv.most_similar(positive=['bruxelles', 'france'], negative=['belgique'], topn=1)",
"_____no_output_____"
],
[
"model.wv.most_similar(positive=['bruxelles', 'liege', 'france'], negative=['belgique'], topn=1)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d0e17c0e121c668238115791b71c4f5f7aaf3bfa | 47,409 | ipynb | Jupyter Notebook | binder/sourmash-tax.ipynb | sourmash-bio/sourmash-recipes | 2ec4a111aa2c6dff0b7e946e4b5c43c06af99f4b | [
"BSD-3-Clause"
] | null | null | null | binder/sourmash-tax.ipynb | sourmash-bio/sourmash-recipes | 2ec4a111aa2c6dff0b7e946e4b5c43c06af99f4b | [
"BSD-3-Clause"
] | 1 | 2021-06-11T17:30:32.000Z | 2021-06-11T17:30:32.000Z | binder/sourmash-tax.ipynb | sourmash-bio/sourmash-recipes | 2ec4a111aa2c6dff0b7e946e4b5c43c06af99f4b | [
"BSD-3-Clause"
] | null | null | null | 42.141333 | 500 | 0.630408 | [
[
[
"# `sourmash tax` submodule\n### for integrating taxonomic information",
"_____no_output_____"
],
[
"The sourmash tax (alias `taxonomy`) commands integrate taxonomic information into the results of sourmash gather. tax commands require a properly formatted taxonomy csv file that corresponds to the database used for gather. For supported databases (e.g. GTDB), we provide these files, but they can also be generated for user-generated databases. For more information, see the [databases documentation](https://sourmash.readthedocs.io/en/latest/databases.html).\n\nThese commands rely upon the fact that gather results are non-overlapping: the fraction match for gather on each query will be between 0 (no database matches) and 1 (100% of query matched). We use this property to aggregate gather matches at the desired taxonomic rank. For example, if the gather results for a metagenome include results for 30 different strains of a given species, we can sum the fraction match to each strain to obtain the fraction match to this species.\n\nAs with all reference-based analysis, results can be affected by the completeness of the reference database. However, summarizing taxonomic results from gather minimizes the impact of reference database issues that can derail standard k-mer LCA approaches. See the [blog post]() for a full explanation, and the [`sourmash tax` documentation](https://sourmash.readthedocs.io/en/latest/command-line.html#sourmash-tax-prepare-prepare-and-or-combine-taxonomy-files) for additional usage details.",
"_____no_output_____"
],
[
"## Download example inputs for `sourmash tax`\n\nIn this example, we'll be using a small test dataset run against both the `GTDB-rs202` database\nand our legacy `Genbank` database. (New genbank databases coming soon, please bear with us :).\n",
"_____no_output_____"
],
[
"#### download and look at the gtdb-rs202 lineage file\n\nThis is the taxonomy file in `csv` format.\nThe column headers for `GTDB` are the accession (`ident`), and the taxonomic ranks `superkingdom` --> `species`.",
"_____no_output_____"
]
],
[
[
"%%bash\n\nmkdir -p lineages\ncurl -L https://osf.io/p6z3w/download -o lineages/gtdb-rs202.taxonomy.csv",
" % Total % Received % Xferd Average Speed Time Time Time Current\n Dload Upload Total Spent Left Speed\n100 483 100 483 0 0 273 0 0:00:01 0:00:01 --:--:-- 273\n100 35.2M 100 35.2M 0 0 5497k 0 0:00:06 0:00:06 --:--:-- 9.8M\n"
],
[
"%%bash\nhead lineages/gtdb-rs202.taxonomy.csv",
"ident,superkingdom,phylum,class,order,family,genus,species\nGCF_014075335.1,d__Bacteria,p__Proteobacteria,c__Gammaproteobacteria,o__Enterobacterales,f__Enterobacteriaceae,g__Escherichia,s__Escherichia flexneri\nGCF_002310555.1,d__Bacteria,p__Proteobacteria,c__Gammaproteobacteria,o__Enterobacterales,f__Enterobacteriaceae,g__Escherichia,s__Escherichia flexneri\nGCF_900013275.1,d__Bacteria,p__Proteobacteria,c__Gammaproteobacteria,o__Enterobacterales,f__Enterobacteriaceae,g__Escherichia,s__Escherichia flexneri\nGCF_000168095.1,d__Bacteria,p__Proteobacteria,c__Gammaproteobacteria,o__Enterobacterales,f__Enterobacteriaceae,g__Escherichia,s__Escherichia flexneri\nGCF_002459845.1,d__Bacteria,p__Proteobacteria,c__Gammaproteobacteria,o__Enterobacterales,f__Enterobacteriaceae,g__Escherichia,s__Escherichia flexneri\nGCF_001614695.1,d__Bacteria,p__Proteobacteria,c__Gammaproteobacteria,o__Enterobacterales,f__Enterobacteriaceae,g__Escherichia,s__Escherichia flexneri\nGCF_000356585.2,d__Bacteria,p__Proteobacteria,c__Gammaproteobacteria,o__Enterobacterales,f__Enterobacteriaceae,g__Escherichia,s__Escherichia flexneri\nGCF_014528595.1,d__Bacteria,p__Proteobacteria,c__Gammaproteobacteria,o__Enterobacterales,f__Enterobacteriaceae,g__Escherichia,s__Escherichia flexneri\nGCF_900196445.1,d__Bacteria,p__Proteobacteria,c__Gammaproteobacteria,o__Enterobacterales,f__Enterobacteriaceae,g__Escherichia,s__Escherichia flexneri\n"
]
],
[
[
"#### download NCBI lineage files\n\nNow let's go ahead and grab the Genbank taxonomy files as well. ",
"_____no_output_____"
]
],
[
[
"%%bash\ncurl -L https://osf.io/cbhgd/download -o lineages/bacteria_genbank_lineages.csv\ncurl -L https://osf.io/urtfx/download -o lineages/protozoa_genbank_lineages.csv",
" % Total % Received % Xferd Average Speed Time Time Time Current\n Dload Upload Total Spent Left Speed\n100 459 100 459 0 0 1267 0 --:--:-- --:--:-- --:--:-- 1267\n100 83.1M 100 83.1M 0 0 16.5M 0 0:00:05 0:00:05 --:--:-- 20.6M\n % Total % Received % Xferd Average Speed Time Time Time Current\n Dload Upload Total Spent Left Speed\n100 459 100 459 0 0 803 0 --:--:-- --:--:-- --:--:-- 803\n100 107k 100 107k 0 0 64622 0 0:00:01 0:00:01 --:--:-- 474k\n"
]
],
[
[
"If you do a `head` on these files, you'll notice they have an extra `taxid` column, but otherwise follow the same format.",
"_____no_output_____"
]
],
[
[
"%%bash\nhead lineages/protozoa_genbank_lineages.csv",
"accession,taxid,superkingdom,phylum,class,order,family,genus,species,strain\nGCA_004431415,2762,Eukaryota,,Glaucocystophyceae,,Cyanophoraceae,Cyanophora,Cyanophora paradoxa,\nGCA_000150955,556484,Eukaryota,Bacillariophyta,Bacillariophyceae,Naviculales,Phaeodactylaceae,Phaeodactylum,Phaeodactylum tricornutum,Phaeodactylum tricornutum CCAP 1055/1\nGCA_000310025,2880,Eukaryota,,Phaeophyceae,Ectocarpales,Ectocarpaceae,Ectocarpus,Ectocarpus siliculosus,\nGCA_000194455,2898,Eukaryota,,Cryptophyceae,Cryptomonadales,Cryptomonadaceae,Cryptomonas,Cryptomonas paramecium,\nGCA_000372725,280463,Eukaryota,Haptista,Haptophyta,Isochrysidales,Noelaerhabdaceae,Emiliania,Emiliania huxleyi,Emiliania huxleyi CCMP1516\nGCA_001939145,2951,Eukaryota,,Dinophyceae,Suessiales,Symbiodiniaceae,Symbiodinium,Symbiodinium microadriaticum,\nGCA_900617105,2996,Eukaryota,,Chrysophyceae,Hydrurales,Hydruraceae,Hydrurus,Hydrurus foetidus,\nGCA_001638955,158060,Eukaryota,Euglenozoa,Euglenida,Euglenales,Euglenaceae,Euglena,Euglena gracilis,\nGCA_900893395,3039,Eukaryota,Euglenozoa,Euglenida,Euglenales,Euglenaceae,Euglena,Euglena gracilis,\n"
]
],
[
[
"## Combining taxonomies with `sourmash tax prepare`",
"_____no_output_____"
],
[
"All sourmash tax commands must be given one or more taxonomy files as parameters to the `--taxonomy` argument.\n\n`sourmash tax prepare` is a utility function that can ingest and validate multiple CSV files or sqlite3\ndatabases, and output a CSV file or a sqlite3 database. It can be used to combine multiple taxonomies\ninto a single file, as well as change formats between CSV and sqlite3.\n\n> Note: `--taxonomy` files can be either CSV files or (as of sourmash 4.2.1) sqlite3 databases.\n> sqlite3 databases are much faster for large taxonomies, while CSV files are easier to view\n> and modify using spreadsheet software.\n\nLet's use `tax prepare` to combine the downloaded taxonomies and output into a sqlite3 database:",
"_____no_output_____"
]
],
[
[
"# to see the arguments, run the `--help` like so:\n! sourmash tax prepare --help",
"\u001b[K\n== This is sourmash version 4.2.1. ==\n\u001b[K== Please cite Brown and Irber (2016), doi:10.21105/joss.00027. ==\n\nusage: \n\n sourmash tax prepare --taxonomy-csv <taxonomy_file> [ ... ] -o <output>\n\nThe 'tax prepare' command reads in one or more taxonomy databases\nand saves them into a new database. It can be used to combine databases\nin the desired order, as well as output different database formats.\n\nPlease see the 'tax prepare' documentation for more details:\n https://sourmash.readthedocs.io/en/latest/command-line.html#sourmash-tax-prepare-prepare-and-or-combine-taxonomy-files\n\noptional arguments:\n -h, --help show this help message and exit\n -q, --quiet suppress non-error output\n -t FILE [FILE ...], --taxonomy-csv FILE [FILE ...], --taxonomy FILE [FILE ...]\n database lineages\n -o OUTPUT, --output OUTPUT\n output file\n -F {csv,sql}, --database-format {csv,sql}\n format of output file; default is 'sql')\n --keep-full-identifiers\n do not split identifiers on whitespace\n --keep-identifier-versions\n after splitting identifiers, do not remove accession\n versions\n --fail-on-missing-taxonomy\n fail quickly if taxonomy is not available for an\n identifier\n -f, --force continue past errors in file and taxonomy loading\n"
],
[
"%%bash\n\nsourmash tax prepare --taxonomy lineages/bacteria_genbank_lineages.csv \\\n lineages/protozoa_genbank_lineages.csv \\\n lineages/gtdb-rs202.taxonomy.csv \\\n -o lineages/gtdb-rs202_genbank.taxonomy.db",
"\u001b[K\n== This is sourmash version 4.2.1. ==\n\u001b[K== Please cite Brown and Irber (2016), doi:10.21105/joss.00027. ==\n\n\u001b[Kloading taxonomies...\n\u001b[K...loaded 882562 entries.\n\u001b[Ksaving to 'lineages/gtdb-rs202_genbank.taxonomy.db', format sql...\n\u001b[Kdone!\n"
]
],
[
[
"> Note that the **order is important if the databases contain overlapping\naccession identifiers**. In this case, GTDB contains only a subset of all identifiers\nin the NCBI taxonomy. Putting GTDB last here will allow the GTDB lineage information\nto override the lineage information provided in the NCBI file, thus utilizing GTDB\ntaxonomy when available, and NCBI lienages for all other accessions.",
"_____no_output_____"
]
],
[
[
"%%bash\nls -lsrht lineages/",
"total 566776\n 73856 -rw-r--r-- 1 tessa staff 35M Aug 2 15:16 gtdb-rs202.taxonomy.csv\n196736 -rw-r--r-- 1 tessa staff 83M Aug 2 15:16 bacteria_genbank_lineages.csv\n 256 -rw-r--r-- 1 tessa staff 107K Aug 2 15:16 protozoa_genbank_lineages.csv\n295928 -rw-r--r-- 1 tessa staff 139M Aug 2 15:17 gtdb-rs202_genbank.taxonomy.db\n"
]
],
[
[
"We'll use this prepared database in each of the commands below.",
"_____no_output_____"
],
[
"## `sourmash tax metagenome`",
"_____no_output_____"
]
],
[
[
"# to see the arguments, run the `--help` like so:\n! sourmash tax metagenome --help",
"\u001b[K\n== This is sourmash version 4.2.1. ==\n\u001b[K== Please cite Brown and Irber (2016), doi:10.21105/joss.00027. ==\n\nusage: \n\n sourmash tax metagenome --gather-csv <gather_csv> [ ... ] --taxonomy-csv <taxonomy-csv> [ ... ]\n\nThe 'tax metagenome' command reads in metagenome gather result CSVs and\nsummarizes by taxonomic lineage.\n\nThe default output format consists of four columns,\n `query_name,rank,fraction,lineage`, where `fraction` is the fraction\n of the query matched to that reported rank and lineage. The summarization\n is reported for each taxonomic rank.\n\nAlternatively, you can output results at a specific rank (e.g. species)\nin `krona` or `lineage_summary` formats.\n\nPlease see the 'tax metagenome' documentation for more details:\n https://sourmash.readthedocs.io/en/latest/command-line.html#sourmash-tax-metagenome-summarize-metagenome-content-from-gather-results\n\noptional arguments:\n -h, --help show this help message and exit\n -g [GATHER_CSV ...], --gather-csv [GATHER_CSV ...]\n CSVs from sourmash gather\n --from-file FILE input many gather results as a text file, with one\n gather CSV per line\n -q, --quiet suppress non-error output\n -o OUTPUT_BASE, --output-base OUTPUT_BASE\n base filepath for output file(s) (default stdout)\n --output-dir OUTPUT_DIR\n directory for output files\n -t FILE [FILE ...], --taxonomy-csv FILE [FILE ...], --taxonomy FILE [FILE ...]\n database lineages CSV\n --keep-full-identifiers\n do not split identifiers on whitespace\n --keep-identifier-versions\n after splitting identifiers, do not remove accession\n versions\n --fail-on-missing-taxonomy\n fail quickly if taxonomy is not available for an\n identifier\n --output-format {csv_summary,krona,lineage_summary} [{csv_summary,krona,lineage_summary} ...]\n choose output format(s)\n -r {strain,species,genus,family,order,class,phylum,superkingdom}, --rank {strain,species,genus,family,order,class,phylum,superkingdom}\n For non-default output formats: Summarize genome\n taxonomy at this rank and above. Note that the\n taxonomy CSV must contain lineage information at this\n rank.\n -f, --force continue past errors in taxonomy database loading\n"
]
],
[
[
"#### Download a small demo `sourmash gather` output file from metagenome `HSMA33MX`.\nThis `gather` was run at a DNA ksize of 31 against both GTDB and our legacy Genbank database.",
"_____no_output_____"
]
],
[
[
"%%bash\nmkdir -p gather\ncurl -L https://osf.io/xb8jg/download -o gather/HSMA33MX_gather_x_gtdbrs202_genbank_k31.csv",
" % Total % Received % Xferd Average Speed Time Time Time Current\n Dload Upload Total Spent Left Speed\n100 459 100 459 0 0 1201 0 --:--:-- --:--:-- --:--:-- 1201\n100 2662 100 2662 0 0 1928 0 0:00:01 0:00:01 --:--:-- 0\n"
]
],
[
[
"Take a look at this gather file:",
"_____no_output_____"
]
],
[
[
"%%bash\nhead gather/HSMA33MX_gather_x_gtdbrs202_genbank_k31.csv",
"intersect_bp,f_orig_query,f_match,f_unique_to_query,f_unique_weighted,average_abund,median_abund,std_abund,name,filename,md5,f_match_orig,unique_intersect_bp,gather_result_rank,remaining_bp,query_filename,query_name,query_md5,query_bp\n442000,0.08815317112086159,0.08438335242458954,0.08815317112086159,0.05815279361459521,1.6153846153846154,1.0,1.1059438185997785,\"GCF_001881345.1 Escherichia coli strain=SF-596, ASM188134v1\",/group/ctbrowngrp/gtdb/databases/ctb/gtdb-rs202.genomic.k31.sbt.zip,683df1ec13872b4b98d59e98b355b52c,0.042779713511420826,442000,0,4572000,outputs/abundtrim/HSMA33MX.abundtrim.fq.gz,HSMA33MX,9687eeed,5014000\n390000,0.07778220981252493,0.10416666666666667,0.07778220981252493,0.050496823586903404,1.5897435897435896,1.0,0.8804995294906566,\"GCF_009494285.1 Prevotella copri strain=iAK1218, ASM949428v1\",/group/ctbrowngrp/gtdb/databases/ctb/gtdb-rs202.genomic.k31.sbt.zip,1266c86141e3a5603da61f57dd863ed0,0.052236806857755155,390000,1,4182000,outputs/abundtrim/HSMA33MX.abundtrim.fq.gz,HSMA33MX,9687eeed,5014000\n206000,0.041084962106102914,0.007403148134837921,0.041084962106102914,0.2215344518651246,13.20388349514563,3.0,69.69466823965065,\"GCA_002754635.1 Plasmodium vivax strain=CMB-1, CMB-1_v2\",/home/irber/sourmash_databases/outputs/sbt/genbank-protozoa-x1e6-k31.sbt.zip,8125e7913e0d0b88deb63c9ad28f827c,0.0037419167332703625,206000,2,3976000,outputs/abundtrim/HSMA33MX.abundtrim.fq.gz,HSMA33MX,9687eeed,5014000\n138000,0.027522935779816515,0.024722321748477247,0.027522935779816515,0.015637726014008795,1.391304347826087,1.0,0.5702120455914782,\"GCF_013368705.1 Bacteroides vulgatus strain=B33, ASM1336870v1\",/group/ctbrowngrp/gtdb/databases/ctb/gtdb-rs202.genomic.k31.sbt.zip,7d5f4ba1d01c8c3f7a520d19faded7cb,0.012648945921173235,138000,3,3838000,outputs/abundtrim/HSMA33MX.abundtrim.fq.gz,HSMA33MX,9687eeed,5014000\n338000,0.06741124850418827,0.013789581205311542,0.010769844435580374,0.006515719172503665,1.4814814814814814,1.0,0.738886568268889,\"GCF_003471795.1 Prevotella copri strain=AM16-54, ASM347179v1\",/group/ctbrowngrp/gtdb/databases/ctb/gtdb-rs202.genomic.k31.sbt.zip,0ebd36ff45fc2810808789667f4aad84,0.04337782340862423,54000,4,3784000,outputs/abundtrim/HSMA33MX.abundtrim.fq.gz,HSMA33MX,9687eeed,5014000\n110000,0.021938571998404467,0.000842978957948319,0.010370961308336658,0.023293696041700604,5.5,2.5,7.417494911978758,\"GCA_000256725.2 Toxoplasma gondii TgCatPRC2 strain=TgCatPRC2, TGCATPRC2 v2\",/home/irber/sourmash_databases/outputs/sbt/genbank-protozoa-x1e6-k31.sbt.zip,2a3b1804cf5ea5fe75dde3e153294548,0.0008909768346023004,52000,5,3732000,outputs/abundtrim/HSMA33MX.abundtrim.fq.gz,HSMA33MX,9687eeed,5014000\n"
]
],
[
[
"### summarize this metagenome and produce `krona` output at the `species` level:",
"_____no_output_____"
]
],
[
[
"%%bash\nsourmash tax metagenome --gather-csv gather/HSMA33MX_gather_x_gtdbrs202_genbank_k31.csv \\\n --taxonomy lineages/gtdb-rs202_genbank.taxonomy.db \\\n --output-format csv_summary krona --rank species \\\n --output-base HSMA33MX.gather-tax",
"\u001b[K\n== This is sourmash version 4.2.1. ==\n\u001b[K== Please cite Brown and Irber (2016), doi:10.21105/joss.00027. ==\n\n\u001b[Kloaded 6 gather results.\n\u001b[Kof 6, missed 0 lineage assignments.\n\u001b[Kloaded results from 1 gather CSVs\n\u001b[Ksaving `csv_summary` output to HSMA33MX.gather-tax.summarized.csv.\n\u001b[Ksaving `krona` output to HSMA33MX.gather-tax.krona.tsv.\n"
],
[
"# build krona plot\n!ktImportText HSMA33MX.gather-tax.krona.tsv",
"Writing text.krona.html...\n"
]
],
[
[
"#### This will produce both `csv_summary` and `krona` output files, with the basename `HSMA33MX.gather-tax`:",
"_____no_output_____"
]
],
[
[
"%%bash\nls HSMA33MX.gather-tax*",
"HSMA33MX.gather-tax.krona.tsv\nHSMA33MX.gather-tax.summarized.csv\n"
],
[
"%%bash\nhead HSMA33MX.gather-tax.summarized.csv",
"query_name,rank,fraction,lineage,query_md5,query_filename,f_weighted_at_rank,bp_match_at_rank\nHSMA33MX,superkingdom,0.2042281611487834,d__Bacteria,9687eeed,outputs/abundtrim/HSMA33MX.abundtrim.fq.gz,0.13080306238801107,1024000\nHSMA33MX,superkingdom,0.051455923414439574,Eukaryota,9687eeed,outputs/abundtrim/HSMA33MX.abundtrim.fq.gz,0.24482814790682522,258000\nHSMA33MX,superkingdom,0.7443159154367771,unclassified,9687eeed,outputs/abundtrim/HSMA33MX.abundtrim.fq.gz,0.6243687897051637,3732000\nHSMA33MX,phylum,0.11607499002792182,d__Bacteria;p__Bacteroidota,9687eeed,outputs/abundtrim/HSMA33MX.abundtrim.fq.gz,0.07265026877341586,582000\nHSMA33MX,phylum,0.08815317112086159,d__Bacteria;p__Proteobacteria,9687eeed,outputs/abundtrim/HSMA33MX.abundtrim.fq.gz,0.05815279361459521,442000\nHSMA33MX,phylum,0.051455923414439574,Eukaryota;Apicomplexa,9687eeed,outputs/abundtrim/HSMA33MX.abundtrim.fq.gz,0.24482814790682522,258000\nHSMA33MX,phylum,0.7443159154367771,unclassified,9687eeed,outputs/abundtrim/HSMA33MX.abundtrim.fq.gz,0.6243687897051637,3732000\nHSMA33MX,class,0.11607499002792182,d__Bacteria;p__Bacteroidota;c__Bacteroidia,9687eeed,outputs/abundtrim/HSMA33MX.abundtrim.fq.gz,0.07265026877341586,582000\nHSMA33MX,class,0.08815317112086159,d__Bacteria;p__Proteobacteria;c__Gammaproteobacteria,9687eeed,outputs/abundtrim/HSMA33MX.abundtrim.fq.gz,0.05815279361459521,442000\n"
],
[
"%%bash\nhead HSMA33MX.gather-tax.krona.tsv",
"fraction\tsuperkingdom\tphylum\tclass\torder\tfamily\tgenus\tspecies\n0.0885520542481053\td__Bacteria\tp__Bacteroidota\tc__Bacteroidia\to__Bacteroidales\tf__Bacteroidaceae\tg__Prevotella\ts__Prevotella copri\n0.08815317112086159\td__Bacteria\tp__Proteobacteria\tc__Gammaproteobacteria\to__Enterobacterales\tf__Enterobacteriaceae\tg__Escherichia\ts__Escherichia coli\n0.041084962106102914\tEukaryota\tApicomplexa\tAconoidasida\tHaemosporida\tPlasmodiidae\tPlasmodium\tPlasmodium vivax\n0.027522935779816515\td__Bacteria\tp__Bacteroidota\tc__Bacteroidia\to__Bacteroidales\tf__Bacteroidaceae\tg__Phocaeicola\ts__Phocaeicola vulgatus\n0.010370961308336658\tEukaryota\tApicomplexa\tConoidasida\tEucoccidiorida\tSarcocystidae\tToxoplasma\tToxoplasma gondii\n0.7443159154367771\tunclassified\tunclassified\tunclassified\tunclassified\tunclassified\tunclassified\tunclassified\n"
],
[
"# generate krona html\n!ktImportText HSMA33MX.gather-tax.krona.tsv",
"Writing text.krona.html...\n"
]
],
[
[
"### comparing metagenomes with `sourmash tax metagenome`:\n\nWe can also download a second metagenome `gather` csv and use `metagenome` to generate a\n`lineage_summary` output to compare these samples.\n\n\n> The lineage summary format is most useful when comparing across metagenome queries.\n> Each row is a lineage at the desired reporting rank. The columns are each query used for\n> gather, with the fraction match reported for each lineage. This format is commonly used\n> as input for many external multi-sample visualization tools.",
"_____no_output_____"
]
],
[
[
"%%bash\n\ncurl -L https://osf.io/nqtgs/download -o gather/PSM7J4EF_gather_x_gtdbrs202_genbank_k31.csv",
" % Total % Received % Xferd Average Speed Time Time Time Current\n Dload Upload Total Spent Left Speed\n100 459 100 459 0 0 1180 0 --:--:-- --:--:-- --:--:-- 1182\n100 2312 100 2312 0 0 1202 0 0:00:01 0:00:01 --:--:-- 1742\n"
],
[
"%%bash\nsourmash tax metagenome --gather-csv gather/HSMA33MX_gather_x_gtdbrs202_genbank_k31.csv \\\n gather/PSM7J4EF_gather_x_gtdbrs202_genbank_k31.csv \\\n --taxonomy lineages/gtdb-rs202_genbank.taxonomy.db \\\n --output-format lineage_summary --rank species \\\n --output-base HSMA33MX-PSM7J4EF.gather-tax",
"\u001b[K\n== This is sourmash version 4.2.1. ==\n\u001b[K== Please cite Brown and Irber (2016), doi:10.21105/joss.00027. ==\n\n\u001b[Kloaded 6 gather results.\n\u001b[Kof 6, missed 0 lineage assignments.\n\u001b[Kloaded 5 gather results.\n\u001b[Kof 5, missed 0 lineage assignments.\n\u001b[Kloaded results from 2 gather CSVs\n\u001b[Ksaving `lineage_summary` output to HSMA33MX-PSM7J4EF.gather-tax.lineage_summary.tsv.\n"
],
[
"%%bash\nhead HSMA33MX-PSM7J4EF.gather-tax.lineage_summary.tsv",
"lineage\tHSMA33MX\tPSM7J4EF\nEukaryota;Apicomplexa;Aconoidasida;Haemosporida;Plasmodiidae;Plasmodium;Plasmodium vivax\t0.041084962106102914\t0.004553734061930784\nEukaryota;Apicomplexa;Conoidasida;Eucoccidiorida;Sarcocystidae;Toxoplasma;Toxoplasma gondii\t0.010370961308336658\t0.0011275912915257177\nd__Bacteria;p__Bacteroidota;c__Bacteroidia;o__Bacteroidales;f__Bacteroidaceae;g__Bacteroides;s__Bacteroides fragilis\t0\t0.05134877266024807\nd__Bacteria;p__Bacteroidota;c__Bacteroidia;o__Bacteroidales;f__Bacteroidaceae;g__Bacteroides;s__Bacteroides ovatus\t0\t0.056726515742909184\nd__Bacteria;p__Bacteroidota;c__Bacteroidia;o__Bacteroidales;f__Bacteroidaceae;g__Phocaeicola;s__Phocaeicola dorei\t0\t0.10625379477838494\nd__Bacteria;p__Bacteroidota;c__Bacteroidia;o__Bacteroidales;f__Bacteroidaceae;g__Phocaeicola;s__Phocaeicola vulgatus\t0.027522935779816515\t0\nd__Bacteria;p__Bacteroidota;c__Bacteroidia;o__Bacteroidales;f__Bacteroidaceae;g__Prevotella;s__Prevotella copri\t0.0885520542481053\t0\nd__Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Enterobacterales;f__Enterobacteriaceae;g__Escherichia;s__Escherichia coli\t0.08815317112086159\t0\nunclassified\t0.7443159154367771\t0.7799895914650012\n"
]
],
[
[
"Note, these are mini gather results, so your unclassified fraction will hopefully be much smaller!",
"_____no_output_____"
],
[
"## Classifying genomes with `sourmash tax genome`",
"_____no_output_____"
],
[
"To illustrate the utility of genome, let’s consider a signature consisting of two different\nShewanella strains, Shewanella baltica OS185 strain=OS185 and Shewanella baltica OS223 strain=OS223.\nFor simplicity, we gave this query the name “Sb47+63”.",
"_____no_output_____"
],
[
"When we gather this signature against the gtdb-rs202 representatives database, we see 66% matches to one strain, and 33% to the other:\n\nabbreviated `gather_csv`:\n\n```\nf_match,f_unique_to_query,name,query_name\n0.664,0.664,\"GCF_000021665.1 Shewanella baltica OS223 strain=OS223, ASM2166v1\",Sb47+63\n0.656,0.335,\"GCF_000017325.1 Shewanella baltica OS185 strain=OS185, ASM1732v1\",Sb47+63\n```\n> Here, f_match shows that independently, both strains match ~65% percent of this mixed query.\n> The f_unique_to_query column has the results of gather-style decomposition. As the OS223 strain\n> had a slightly higher f_match (66%), it was the first match. The remaining 33% of the query\n> matched to strain OS185.",
"_____no_output_____"
],
[
"#### download the gather results",
"_____no_output_____"
]
],
[
[
"%%bash\n\ncurl -L https://osf.io/pgsc2/download -o gather/Sb47+63_x_gtdb-rs202.gather.csv",
" % Total % Received % Xferd Average Speed Time Time Time Current\n Dload Upload Total Spent Left Speed\n100 459 100 459 0 0 1267 0 --:--:-- --:--:-- --:--:-- 1267\n100 820 100 820 0 0 738 0 0:00:01 0:00:01 --:--:-- 0\n"
],
[
"%%bash\n\nhead gather/Sb47+63_x_gtdb-rs202.gather.csv",
"intersect_bp,f_orig_query,f_match,f_unique_to_query,f_unique_weighted,average_abund,median_abund,std_abund,name,filename,md5,f_match_orig,unique_intersect_bp,gather_result_rank,remaining_bp,query_filename,query_name,query_md5,query_bp\n5238000,0.6642150646715699,1.0,0.6642150646715699,0.6642150646715699,,,,\"GCF_000021665.1 Shewanella baltica OS223 strain=OS223, ASM2166v1\",/group/ctbrowngrp/gtdb/databases/ctb/gtdb-rs202.genomic.k31.sbt.zip,38729c6374925585db28916b82a6f513,1.0,5238000,0,2648000,,Sb47+63,491c0a81,7886000\n5177000,0.6564798376870403,0.5114931427467645,0.3357849353284301,0.3357849353284301,,,,\"GCF_000017325.1 Shewanella baltica OS185 strain=OS185, ASM1732v1\",/group/ctbrowngrp/gtdb/databases/ctb/gtdb-rs202.genomic.k31.sbt.zip,09a08691ce52952152f0e866a59f6261,1.0,2648000,1,0,,Sb47+63,491c0a81,7886000\n"
]
],
[
[
"#### Now, let's run `tax genome` classification:",
"_____no_output_____"
]
],
[
[
"%%bash\n# to see the arguments, run the `--help` like so:\nsourmash tax genome --help",
"usage: \n\n sourmash tax genome --gather-csv <gather_csv> [ ... ] --taxonomy-csv <taxonomy-csv> [ ... ]\n\nThe 'tax genome' command reads in genome gather result CSVs and reports likely\nclassification for each query genome.\n\nBy default, classification uses a containment threshold of 0.1, meaning at least\n10 percent of the query was covered by matches with the reported taxonomic rank and lineage.\nYou can specify an alternate classification threshold or force classification by\ntaxonomic rank instead, e.g. at species or genus-level.\n\nThe default output format consists of five columns,\n `query_name,status,rank,fraction,lineage`, where `fraction` is the fraction\n of the query matched to the reported rank and lineage. The `status` column\n provides additional information on the classification, and can be:\n - `match` - this query was classified\n - `nomatch`- this query could not be classified\n - `below_threshold` - this query was classified at the specified rank,\n but the query fraction matched was below the containment threshold\n\nOptionally, you can report classifications in `krona` format, but note\nthat this forces classification by rank, rather than containment threshold.\n\nPlease see the 'tax genome' documentation for more details:\n https://sourmash.readthedocs.io/en/latest/command-line.html#sourmash-tax-genome-classify-a-genome-using-gather-results\n\noptional arguments:\n -h, --help show this help message and exit\n -g [GATHER_CSV ...], --gather-csv [GATHER_CSV ...]\n CSVs output by sourmash gather for this sample\n --from-file FILE input many gather results as a text file, with one\n gather CSV per line\n -q, --quiet suppress non-error output\n -t FILE [FILE ...], --taxonomy-csv FILE [FILE ...], --taxonomy FILE [FILE ...]\n database lineages CSV\n -o OUTPUT_BASE, --output-base OUTPUT_BASE\n base filepath for output file(s) (default stdout)\n --output-dir OUTPUT_DIR\n directory for output files\n -r {strain,species,genus,family,order,class,phylum,superkingdom}, --rank {strain,species,genus,family,order,class,phylum,superkingdom}\n Summarize genome taxonomy at this rank and above. Note\n that the taxonomy CSV must contain lineage information\n at this rank.\n --keep-full-identifiers\n do not split identifiers on whitespace\n --keep-identifier-versions\n after splitting identifiers, do not remove accession\n versions\n --fail-on-missing-taxonomy\n fail quickly if taxonomy is not available for an\n identifier\n --output-format {csv_summary,krona} [{csv_summary,krona} ...]\n choose output format(s)\n -f, --force continue past survivable errors in loading taxonomy\n database or gather results\n --containment-threshold CONTAINMENT_THRESHOLD\n minimum containment threshold for classification;\n default=0.1\n"
],
[
"%%bash\n\nsourmash tax genome --gather-csv gather/Sb47+63_x_gtdb-rs202.gather.csv \\\n --taxonomy lineages/gtdb-rs202_genbank.taxonomy.db \\\n --output-base Sb47+63.gather-tax",
"\u001b[K\n== This is sourmash version 4.2.1. ==\n\u001b[K== Please cite Brown and Irber (2016), doi:10.21105/joss.00027. ==\n\n\u001b[Kloaded 2 gather results.\n\u001b[Kof 2, missed 0 lineage assignments.\n\u001b[Kloaded results from 1 gather CSVs\n\u001b[Ksaving `classification` output to Sb47+63.gather-tax.classifications.csv.\n"
]
],
[
[
"The default output format is `csv_summary`.\n\nThis outputs a csv with taxonomic classification for each query genome. This output currently consists of six columns:\n`query_name`,`rank`,`fraction`,`lineage`,`query_md5`,`query_filename`, where `fraction` is the fraction of the query matched to\nthe reported rank and lineage. The `status` column provides additional information on the classification:\n\n- `match` - this query was classified\n- `nomatch` - this query could not be classified\n- `below_threshold` - this query was classified at the specified rank,\nbut the query fraction matched was below the containment threshold",
"_____no_output_____"
]
],
[
[
"!ls Sb47+63.gather-tax*",
"Sb47+63.gather-tax.classifications.csv\n"
],
[
"!head Sb47+63.gather-tax.classifications.csv",
"query_name,status,rank,fraction,lineage,query_md5,query_filename,f_weighted_at_rank,bp_match_at_rank\nSb47+63,match,species,1.0,d__Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Enterobacterales;f__Shewanellaceae;g__Shewanella;s__Shewanella baltica,491c0a81,,1.0,7886000.0\n"
]
],
[
[
"> Here, we see that the match percentages to both strains have been aggregated, and we have 100% species-level\n> Shewanella baltica annotation (fraction = 1.0).",
"_____no_output_____"
],
[
"## `sourmash tax annotate`",
"_____no_output_____"
],
[
"`sourmash tax annotate` adds a column with taxonomic lineage information for each database match to gather output.\nIt does not do any LCA summarization or classification. The results from `annotate` are not required for any other\n`tax` command, but may be useful if you're doing your own exploration of `gather` results.",
"_____no_output_____"
],
[
"Let's annotate a previously downloaded `gather` file",
"_____no_output_____"
]
],
[
[
"%%bash\nsourmash tax annotate --gather-csv gather/Sb47+63_x_gtdb-rs202.gather.csv \\\n --taxonomy lineages/gtdb-rs202_genbank.taxonomy.db",
"\u001b[K\n== This is sourmash version 4.2.1. ==\n\u001b[K== Please cite Brown and Irber (2016), doi:10.21105/joss.00027. ==\n\n\u001b[Kloaded 2 gather results.\n\u001b[Kof 2, missed 0 lineage assignments.\n\u001b[Kloaded results from 1 gather CSVs\n\u001b[Ksaving `annotate` output to Sb47+63_x_gtdb-rs202.gather.with-lineages.csv.\n"
],
[
"!head Sb47+63_x_gtdb-rs202.gather.with-lineages.csv",
"intersect_bp,f_orig_query,f_match,f_unique_to_query,f_unique_weighted,average_abund,median_abund,std_abund,name,filename,md5,f_match_orig,unique_intersect_bp,gather_result_rank,remaining_bp,query_filename,query_name,query_md5,query_bp,lineage\n5238000,0.6642150646715699,1.0,0.6642150646715699,0.6642150646715699,,,,\"GCF_000021665.1 Shewanella baltica OS223 strain=OS223, ASM2166v1\",/group/ctbrowngrp/gtdb/databases/ctb/gtdb-rs202.genomic.k31.sbt.zip,38729c6374925585db28916b82a6f513,1.0,5238000,0,2648000,,Sb47+63,491c0a81,7886000,d__Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Enterobacterales;f__Shewanellaceae;g__Shewanella;s__Shewanella baltica\n5177000,0.6564798376870403,0.5114931427467645,0.3357849353284301,0.3357849353284301,,,,\"GCF_000017325.1 Shewanella baltica OS185 strain=OS185, ASM1732v1\",/group/ctbrowngrp/gtdb/databases/ctb/gtdb-rs202.genomic.k31.sbt.zip,09a08691ce52952152f0e866a59f6261,1.0,2648000,1,0,,Sb47+63,491c0a81,7886000,d__Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Enterobacterales;f__Shewanellaceae;g__Shewanella;s__Shewanella baltica\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
d0e18c57b8bb336cf3b72e4ca627776167b7546c | 25,982 | ipynb | Jupyter Notebook | cryptography_and_network_security/lab_assignment_5/lab_assignment_5_q1_and_q2_complete.ipynb | piyushmoolchandani/Lab_codes | 5b478f31b41856b10c0df9f847b4a7794556fea5 | [
"MIT"
] | null | null | null | cryptography_and_network_security/lab_assignment_5/lab_assignment_5_q1_and_q2_complete.ipynb | piyushmoolchandani/Lab_codes | 5b478f31b41856b10c0df9f847b4a7794556fea5 | [
"MIT"
] | null | null | null | cryptography_and_network_security/lab_assignment_5/lab_assignment_5_q1_and_q2_complete.ipynb | piyushmoolchandani/Lab_codes | 5b478f31b41856b10c0df9f847b4a7794556fea5 | [
"MIT"
] | 2 | 2020-04-09T08:52:51.000Z | 2021-09-24T13:45:04.000Z | 41.771704 | 161 | 0.411708 | [
[
[
"def preprocess_string(s):\n plaintext = ''\n for i in s:\n temp = ord(i)\n if ((temp < 123 and temp > 96) or (temp < 91 and temp > 64)\n or (temp < 58 and temp > 47) or temp == 32):\n plaintext += i\n return plaintext",
"_____no_output_____"
],
[
"def char_to_bits(s, char_size = 4):\n if (char_size == 4):\n ans = bin(int(s, 2 ** char_size))[2:]\n ans = '0' * (char_size - len(ans)) + ans \n return ans\n elif (char_size == 8):\n ans = hex_to_bits(hex(ord(s)))\n ans = '0' * (char_size - len(ans)) + ans\n return ans",
"_____no_output_____"
],
[
"def char_chunk_to_bits(s, char_size = 4):\n ans = ''\n for i in s:\n ans += char_to_bits(i, char_size)\n return ans",
"_____no_output_____"
],
[
"def create_chunks(s, chunk_size, char_size = 4, bogus = 'F'):\n char_count = chunk_size // char_size\n increment = len(s) % char_count\n if increment:\n s += ((len(s) // char_count + 1) * char_count - len(s)) * bogus\n chunks = [s[i:i + char_count] for i in range(0, len(s), char_count)]\n return chunks",
"_____no_output_____"
],
[
"def get_bit_chunks(s, chunk_size, char_size = 4, bogus = 'F'):\n s = preprocess_string(s)\n chunks = create_chunks(s, chunk_size, char_size, bogus)\n bit_chunks = [char_chunk_to_bits(i, char_size) for i in chunks]\n return bit_chunks",
"_____no_output_____"
]
],
[
[
"## 1. DES",
"_____no_output_____"
]
],
[
[
"def shift_left(s, n):\n for i in range(n):\n s = s[1:] + s[0]\n return s\n\ndef permute(s, key):\n ans = []\n for i in key:\n ans.append(s[i - 1])\n return ''.join(ans)\n\ndef create_keys(key_with_parity_bits):\n parity_bit_drop_table = [57, 49, 41, 33, 25, 17, 9,\n 1, 58, 50, 42, 34, 26, 18,\n 10, 2, 59, 51, 43, 35, 27,\n 19, 11, 3, 60, 52, 44, 36,\n 63, 55, 47, 39, 31, 23, 15,\n 7, 62, 54, 46, 38, 30, 22,\n 14, 6, 61, 53, 45, 37, 29,\n 21, 13, 5, 28, 20, 12, 4]\n \n number_of_bits_shifts = [1, 1, 2, 2, 2, 2, 2, 2, 1, 2, 2, 2, 2, 2, 2, 1]\n \n key_compression_table = [14, 17, 11, 24, 1, 5, 3, 28,\n 15, 6, 21, 10, 23, 19, 12, 4,\n 26, 8, 16, 7, 27, 20, 13, 2,\n 41, 52, 31, 37, 47, 55, 30, 40,\n 51, 45, 33, 48, 44, 49, 39, 56,\n 34, 53, 46, 42, 50, 36, 29, 32]\n \n cipher_key = permute(key_with_parity_bits, parity_bit_drop_table)\n \n left_key = cipher_key[0:28]\n right_key = cipher_key[28:56]\n \n keys = []\n \n for i in range(16):\n left_key = shift_left(left_key, number_of_bits_shifts[i])\n right_key = shift_left(right_key, number_of_bits_shifts[i])\n keys.append(permute(left_key + right_key, key_compression_table))\n \n return keys",
"_____no_output_____"
],
[
"def permute(s, key):\n ans = []\n for i in key:\n ans.append(s[i - 1])\n return ''.join(ans)\n\ndef substitute(s, key):\n answer = ''\n for i in range(0, 48, 6):\n vertical_entry = s[i] + s[i + 5]\n horizontal_entry = s[i + 1:i + 5]\n temp = key[i // 6][int(vertical_entry, 2)][int(horizontal_entry, 2)]\n temp = bin(temp)[2:]\n temp = '0' * (4 - len(temp)) + temp\n answer += temp\n return answer\n \ndef round_normal_without_swapping(P, key):\n \n left_plaintext = P[0:32]\n right_plaintext = P[32:64]\n \n right_expansion_p_box_table = [32, 1, 2, 3, 4, 5,\n 4, 5, 6, 7, 8, 9,\n 8, 9, 10, 11, 12, 13,\n 12, 13, 14, 15, 16, 17,\n 16, 17, 18, 19, 20, 21,\n 20, 21, 22, 23, 24, 25,\n 24, 25, 26, 27, 28, 29,\n 28, 29, 30, 31, 32, 1]\n\n \n right_expanded_plaintext = permute(right_plaintext, right_expansion_p_box_table)\n \n xored_right_text = ''\n for i, j in zip(right_expanded_plaintext, key):\n xored_right_text += str(int(i) ^ int(j))\n \n substitution_table = [\n [[14, 4, 13, 1, 2, 15, 11, 8, 3, 10, 6, 12, 5, 9, 0, 7],\n [0, 15, 7, 4, 14, 2, 13, 1, 10, 6, 12, 11, 9, 5, 3, 8],\n [4, 1, 14, 8, 13, 6, 2, 11, 15, 12, 9, 7, 3, 10, 5, 0],\n [15, 12, 8, 2, 4, 9, 1, 7, 5, 11, 3, 14, 10, 0, 6, 13],\n ],\n [[15, 1, 8, 14, 6, 11, 3, 4, 9, 7, 2, 13, 12, 0, 5, 10],\n [3, 13, 4, 7, 15, 2, 8, 14, 12, 0, 1, 10, 6, 9, 11, 5],\n [0, 14, 7, 11, 10, 4, 13, 1, 5, 8, 12, 6, 9, 3, 2, 15],\n [13, 8, 10, 1, 3, 15, 4, 2, 11, 6, 7, 12, 0, 5, 14, 9],\n ],\n [[10, 0, 9, 14, 6, 3, 15, 5, 1, 13, 12, 7, 11, 4, 2, 8],\n [13, 7, 0, 9, 3, 4, 6, 10, 2, 8, 5, 14, 12, 11, 15, 1],\n [13, 6, 4, 9, 8, 15, 3, 0, 11, 1, 2, 12, 5, 10, 14, 7],\n [1, 10, 13, 0, 6, 9, 8, 7, 4, 15, 14, 3, 11, 5, 2, 12],\n ],\n [[7, 13, 14, 3, 0, 6, 9, 10, 1, 2, 8, 5, 11, 12, 4, 15],\n [13, 8, 11, 5, 6, 15, 0, 3, 4, 7, 2, 12, 1, 10, 14, 9],\n [10, 6, 9, 0, 12, 11, 7, 13, 15, 1, 3, 14, 5, 2, 8, 4],\n [3, 15, 0, 6, 10, 1, 13, 8, 9, 4, 5, 11, 12, 7, 2, 14],\n ], \n [[2, 12, 4, 1, 7, 10, 11, 6, 8, 5, 3, 15, 13, 0, 14, 9],\n [14, 11, 2, 12, 4, 7, 13, 1, 5, 0, 15, 10, 3, 9, 8, 6],\n [4, 2, 1, 11, 10, 13, 7, 8, 15, 9, 12, 5, 6, 3, 0, 14],\n [11, 8, 12, 7, 1, 14, 2, 13, 6, 15, 0, 9, 10, 4, 5, 3],\n ], \n [[12, 1, 10, 15, 9, 2, 6, 8, 0, 13, 3, 4, 14, 7, 5, 11],\n [10, 15, 4, 2, 7, 12, 9, 5, 6, 1, 13, 14, 0, 11, 3, 8],\n [9, 14, 15, 5, 2, 8, 12, 3, 7, 0, 4, 10, 1, 13, 11, 6],\n [4, 3, 2, 12, 9, 5, 15, 10, 11, 14, 1, 7, 6, 0, 8, 13],\n ], \n [[4, 11, 2, 14, 15, 0, 8, 13, 3, 12, 9, 7, 5, 10, 6, 1],\n [13, 0, 11, 7, 4, 9, 1, 10, 14, 3, 5, 12, 2, 15, 8, 6],\n [1, 4, 11, 13, 12, 3, 7, 14, 10, 15, 6, 8, 0, 5, 9, 2],\n [6, 11, 13, 8, 1, 4, 10, 7, 9, 5, 0, 15, 14, 2, 3, 12],\n ],\n [[13, 2, 8, 4, 6, 15, 11, 1, 10, 9, 3, 14, 5, 0, 12, 7],\n [1, 15, 13, 8, 10, 3, 7, 4, 12, 5, 6, 11, 0, 14, 9, 2],\n [7, 11, 4, 1, 9, 12, 14, 2, 0, 6, 10, 13, 15, 3, 5, 8],\n [2, 1, 14, 7, 4, 10, 8, 13, 15, 12, 9, 0, 3, 5, 6, 11],\n ]\n ]\n \n straight_permutation_table = [16, 7, 20, 21, 29, 12, 28, 17,\n 1, 15, 23, 26, 5, 18, 31, 10,\n 2, 8, 24, 14, 32, 27, 3, 9,\n 19, 13, 30, 6, 22, 11, 4, 25]\n \n substituted_right_text = substitute(xored_right_text, substitution_table)\n des_function_out = permute(substituted_right_text, straight_permutation_table)\n xored_left_text = ''\n \n for i, j in zip(left_plaintext, des_function_out):\n xored_left_text += str(int(i) ^ int(j))\n \n\n return xored_left_text + right_plaintext\n \ndef round_normal(P, key):\n ciphertext = round_normal_without_swapping(P, key)\n return ciphertext[32:] + ciphertext[0:32]\n\ndef round_last(P, key):\n return round_normal_without_swapping(P, key)",
"_____no_output_____"
],
[
"def DES(plaintext, K):\n \n keys = create_keys(K)\n \n initial_permutation_table = [58, 50, 42, 34, 26, 18, 10, 2,\n 60, 52, 44, 36, 28, 20, 12, 4,\n 62, 54, 46, 38, 30, 22, 14, 6,\n 64, 56, 48, 40, 32, 24, 16, 8,\n 57, 49, 41, 33, 25, 17, 9, 1,\n 59, 51, 43, 35, 27, 19, 11, 3,\n 61, 53, 45, 37, 29, 21, 13, 5,\n 63, 55, 47, 39, 31, 23, 15, 7]\n \n final_permutation_table = [40, 8, 48, 16, 56, 24, 64, 32,\n 39, 7, 47, 15, 55, 23, 63, 31,\n 38, 6, 46, 14, 54, 22, 62, 30,\n 37, 5, 45, 13, 53, 21, 61, 29,\n 36, 4, 44, 12, 52, 20, 60, 28,\n 35, 3, 43, 11, 51, 19, 59, 27,\n 34, 2, 42, 10, 50, 18, 58, 26,\n 33, 1, 41, 9, 49, 17, 57, 25]\n \n blocks = get_bit_chunks(P, 64)\n \n encrypted_blocks = []\n for i in blocks:\n permuted_text = permute(i, initial_permutation_table)\n cipher_text = permuted_text\n for i in range(15):\n cipher_text = round_normal(cipher_text, keys[i])\n cipher_text = round_last(cipher_text, keys[15])\n \n cipher_text_final = permute(cipher_text, final_permutation_table)\n encrypted_blocks.append(cipher_text_final)\n \n return ''.join(encrypted_blocks)",
"_____no_output_____"
],
[
"# P = input(\"Enter Plaintext: \")\n# K = input(\"Enter DES Key: \")\n\nP = \"0123456789ABCDEF\"\nK = \"0001001100110100010101110111100110011011101111001101111111110001\"\n\nciphertext = DES(P, K)\nprint(ciphertext)",
"1000010111101000000100110101010000001111000010101011010000000101\n"
],
[
"P = \"123456ABCD132536\"\nK = get_bit_chunks(\"AABB09182736CCDD\", 64)[0]\nciphertext = DES(P, K)\nprint(ciphertext)",
"1100000010110111101010001101000001011111001110101000001010011100\n"
]
],
[
[
"## 2. AES : 10 Round, 128 bit input, 128 bit key",
"_____no_output_____"
]
],
[
[
"def hex_to_bits(a):\n x = bin(int(a, 16))[2:]\n x = '0' * (8 - len(x)) + x\n return x\n \ndef left_shift(s):\n return s[1:] + s[0]\n\ndef sub_word(s):\n \n subbytes_table = [['0x63', '0x7c', '0x77', '0x7b', '0xf2', '0x6b', '0x6f', '0xc5', '0x30', '0x1', '0x67', '0x2b', '0xfe', '0xd7', '0xab', '0x76'], \n ['0xca', '0x82', '0xc9', '0x7d', '0xfa', '0x59', '0x47', '0xf0', '0xad', '0xd4', '0xa2', '0xaf', '0x9c', '0xa4', '0x72', '0xc0'], \n ['0xb7', '0xfd', '0x93', '0x26', '0x36', '0x3f', '0xf7', '0xcc', '0x34', '0xa5', '0xe5', '0xf1', '0x71', '0xd8', '0x31', '0x15'], \n ['0x4', '0xc7', '0x23', '0xc3', '0x18', '0x96', '0x5', '0x9a', '0x7', '0x12', '0x80', '0xe2', '0xeb', '0x27', '0xb2', '0x75'], \n ['0x9', '0x83', '0x2c', '0x1a', '0x1b', '0x6e', '0x5a', '0xa0', '0x52', '0x3b', '0xd6', '0xb3', '0x29', '0xe3', '0x2f', '0x84'], \n ['0x53', '0xd1', '0x0', '0xed', '0x20', '0xfc', '0xb1', '0x5b', '0x6a', '0xcb', '0xbe', '0x39', '0x4a', '0x4c', '0x58', '0xcf'], \n ['0xd0', '0xef', '0xaa', '0xfb', '0x43', '0x4d', '0x33', '0x85', '0x45', '0xf9', '0x2', '0x7f', '0x50', '0x3c', '0x9f', '0xa8'], \n ['0x51', '0xa3', '0x40', '0x8f', '0x92', '0x9d', '0x38', '0xf5', '0xbc', '0xb6', '0xda', '0x21', '0x10', '0xff', '0xf3', '0xd2'], \n ['0xcd', '0xc', '0x13', '0xec', '0x5f', '0x97', '0x44', '0x17', '0xc4', '0xa7', '0x7e', '0x3d', '0x64', '0x5d', '0x19', '0x73'], \n ['0x60', '0x81', '0x4f', '0xdc', '0x22', '0x2a', '0x90', '0x88', '0x46', '0xee', '0xb8', '0x14', '0xde', '0x5e', '0xb', '0xdb'], \n ['0xe0', '0x32', '0x3a', '0xa', '0x49', '0x6', '0x24', '0x5c', '0xc2', '0xd3', '0xac', '0x62', '0x91', '0x95', '0xe4', '0x79'], \n ['0xe7', '0xc8', '0x37', '0x6d', '0x8d', '0xd5', '0x4e', '0xa9', '0x6c', '0x56', '0xf4', '0xea', '0x65', '0x7a', '0xae', '0x8'], \n ['0xba', '0x78', '0x25', '0x2e', '0x1c', '0xa6', '0xb4', '0xc6', '0xe8', '0xdd', '0x74', '0x1f', '0x4b', '0xbd', '0x8b', '0x8a'], \n ['0x70', '0x3e', '0xb5', '0x66', '0x48', '0x3', '0xf6', '0xe', '0x61', '0x35', '0x57', '0xb9', '0x86', '0xc1', '0x1d', '0x9e'], \n ['0xe1', '0xf8', '0x98', '0x11', '0x69', '0xd9', '0x8e', '0x94', '0x9b', '0x1e', '0x87', '0xe9', '0xce', '0x55', '0x28', '0xdf'], \n ['0x8c', '0xa1', '0x89', '0xd', '0xbf', '0xe6', '0x42', '0x68', '0x41', '0x99', '0x2d', '0xf', '0xb0', '0x54', '0xbb', '0x16']]\n \n ans = ''\n for i in range(0, 32, 8):\n s_i = s[i:i+8]\n ans += hex_to_bits(subbytes_table[int(s_i[0:4], 2)][int(s_i[4:8], 2)])\n return ans\n\ndef rot_word(s):\n return s[8:] + s[:8]\n\ndef add_bit_array(a, b):\n ans = ''\n for i, j in zip(a, b):\n ans += str(int(i) ^ int(j))\n return ans\n\ndef create_keys(key):\n \n Rcons = ['01', '02', '04', '08', '10', '20', '40', '80', '1B', '36']\n words = [key[0:32], key[32:64], key[64:96], key[96:128]]\n for i in range(4, 44):\n if i % 4:\n words.append(add_bit_array(words[i - 1], words[i - 4]))\n else:\n Rcon = Rcons[(i - 4) // 4] + '000000'\n bit_rcon = ''\n for j in range(4):\n bit_rcon += hex_to_bits('0x' + Rcon[2*j : 2*(j + 1)])\n t = add_bit_array(sub_word(rot_word(words[i - 1])), bit_rcon)\n words.append(add_bit_array(t, words[i - 4]))\n return words\n",
"_____no_output_____"
],
[
"\ndef plaintext_to_matrix_state(plaintext):\n ans = [['', '', '', ''], ['', '', '', ''], ['', '', '', ''], ['', '', '', '']]\n for i in range(16):\n ans[i % 4][i // 4] = plaintext[8 * i : 8 * (i + 1)]\n return ans\n\ndef add_matrix(A, B, n = 4):\n ans = [['', '', '', ''], ['', '', '', ''], ['', '', '', ''], ['', '', '', '']]\n for i in range(n):\n for j in range(n):\n ans[i][j] = add_bit_array(A[i][j], B[i][j])\n return ans\n\ndef bit_matrix_to_hex(A):\n B = [['', '', '', ''], ['', '', '', ''], ['', '', '', ''], ['', '', '', '']]\n for i in range(len(A)):\n for j in range(len(A)):\n B[i][j] = hex(int(A[i][j], 2))\n return B",
"_____no_output_____"
],
[
"def sub_bytes(state):\n \n sub_bytes_table = [['0x63', '0x7c', '0x77', '0x7b', '0xf2', '0x6b', '0x6f', '0xc5', '0x30', '0x1', '0x67', '0x2b', '0xfe', '0xd7', '0xab', '0x76'], \n ['0xca', '0x82', '0xc9', '0x7d', '0xfa', '0x59', '0x47', '0xf0', '0xad', '0xd4', '0xa2', '0xaf', '0x9c', '0xa4', '0x72', '0xc0'], \n ['0xb7', '0xfd', '0x93', '0x26', '0x36', '0x3f', '0xf7', '0xcc', '0x34', '0xa5', '0xe5', '0xf1', '0x71', '0xd8', '0x31', '0x15'], \n ['0x4', '0xc7', '0x23', '0xc3', '0x18', '0x96', '0x5', '0x9a', '0x7', '0x12', '0x80', '0xe2', '0xeb', '0x27', '0xb2', '0x75'], \n ['0x9', '0x83', '0x2c', '0x1a', '0x1b', '0x6e', '0x5a', '0xa0', '0x52', '0x3b', '0xd6', '0xb3', '0x29', '0xe3', '0x2f', '0x84'], \n ['0x53', '0xd1', '0x0', '0xed', '0x20', '0xfc', '0xb1', '0x5b', '0x6a', '0xcb', '0xbe', '0x39', '0x4a', '0x4c', '0x58', '0xcf'], \n ['0xd0', '0xef', '0xaa', '0xfb', '0x43', '0x4d', '0x33', '0x85', '0x45', '0xf9', '0x2', '0x7f', '0x50', '0x3c', '0x9f', '0xa8'], \n ['0x51', '0xa3', '0x40', '0x8f', '0x92', '0x9d', '0x38', '0xf5', '0xbc', '0xb6', '0xda', '0x21', '0x10', '0xff', '0xf3', '0xd2'], \n ['0xcd', '0xc', '0x13', '0xec', '0x5f', '0x97', '0x44', '0x17', '0xc4', '0xa7', '0x7e', '0x3d', '0x64', '0x5d', '0x19', '0x73'], \n ['0x60', '0x81', '0x4f', '0xdc', '0x22', '0x2a', '0x90', '0x88', '0x46', '0xee', '0xb8', '0x14', '0xde', '0x5e', '0xb', '0xdb'], \n ['0xe0', '0x32', '0x3a', '0xa', '0x49', '0x6', '0x24', '0x5c', '0xc2', '0xd3', '0xac', '0x62', '0x91', '0x95', '0xe4', '0x79'], \n ['0xe7', '0xc8', '0x37', '0x6d', '0x8d', '0xd5', '0x4e', '0xa9', '0x6c', '0x56', '0xf4', '0xea', '0x65', '0x7a', '0xae', '0x8'], \n ['0xba', '0x78', '0x25', '0x2e', '0x1c', '0xa6', '0xb4', '0xc6', '0xe8', '0xdd', '0x74', '0x1f', '0x4b', '0xbd', '0x8b', '0x8a'], \n ['0x70', '0x3e', '0xb5', '0x66', '0x48', '0x3', '0xf6', '0xe', '0x61', '0x35', '0x57', '0xb9', '0x86', '0xc1', '0x1d', '0x9e'], \n ['0xe1', '0xf8', '0x98', '0x11', '0x69', '0xd9', '0x8e', '0x94', '0x9b', '0x1e', '0x87', '0xe9', '0xce', '0x55', '0x28', '0xdf'], \n ['0x8c', '0xa1', '0x89', '0xd', '0xbf', '0xe6', '0x42', '0x68', '0x41', '0x99', '0x2d', '0xf', '0xb0', '0x54', '0xbb', '0x16']]\n \n ans = [['', '', '', ''], ['', '', '', ''], ['', '', '', ''], ['', '', '', '']]\n n = len(state)\n for i in range(n):\n for j in range(n):\n ans[i][j] = hex_to_bits(sub_bytes_table[int(state[i][j][0:4], 2)][int(state[i][j][4:8], 2)])\n return ans\n\ndef rot_word_for_shift_rows(s):\n return s[1:] + s[:1]\n\ndef shift_rows(state):\n for i in range(4):\n for j in range(i):\n state[i] = rot_word_for_shift_rows(state[i])\n return state\n\ndef shift_and_reduce(s):\n irreducible = '100011011'\n if (s[0] == '1'):\n return add_bit_array(s + '0', irreducible)[1:]\n else:\n return s[1:] + '0'\n \ndef one_term_multiply(s, n):\n for i in range(n):\n s = shift_and_reduce(s)\n return s\n\ndef multiply(a, b):\n a = '0' * (8 - len(a)) + a\n temp = '0' * 8\n n = len(b)\n for i in range(n):\n if (b[-(i + 1)] == '1'):\n temp = add_bit_array(temp, one_term_multiply(a, i))\n return temp\n\ndef multiply_for_mix_columns(a, b):\n return multiply(a, bin(b)[2:])\n\ndef mix_columns(state):\n \n mix_columns_constant_matrix = [[2, 3, 1, 1], \n [1, 2, 3, 1], \n [1, 1, 2, 3], \n [3, 1, 1, 2]]\n \n \n ans = [['', '', '', ''], ['', '', '', ''], ['', '', '', ''], ['', '', '', '']]\n for i in range(4):\n temp = []\n for j in range(4):\n temp.append(state[j][i])\n \n for j in range(4):\n temp_2 = '00000000'\n for k in range(4):\n x = multiply_for_mix_columns(temp[k], mix_columns_constant_matrix[j][k])\n temp_2 = add_bit_array(temp_2, x)\n ans[j][i] = temp_2\n return ans\n\ndef add_round_key(A, key):\n return add_matrix(A, plaintext_to_matrix_state(key))\n\ndef round_normal(plaintext, key):\n x = add_round_key(mix_columns(shift_rows(sub_bytes(plaintext))), key)\n return x\n\ndef round_final(plaintext, key):\n x = add_round_key(shift_rows(sub_bytes(plaintext)), key)\n return x",
"_____no_output_____"
],
[
"def AES(plaintext, key):\n \n blocks = create_keys(key)\n keys = [blocks[4 * i] + blocks[4 * i + 1] + blocks[4 * i + 2] + blocks[4 * i + 3] for i in range(11)]\n\n plaintext_blocks = get_bit_chunks(plaintext, 128, 8)\n cipher_text_blocks = []\n for block in plaintext_blocks:\n initial_state = plaintext_to_matrix_state(block)\n pre_round_transformed_state = add_matrix(initial_state, plaintext_to_matrix_state(keys[0]))\n cipher_text_state = pre_round_transformed_state\n for i in range(1, 10):\n cipher_text_state = round_normal(cipher_text_state, keys[i])\n cipher_text_state = round_final(cipher_text_state, keys[10])\n cipher_text = ''\n for j in range(4):\n for i in range(4):\n cipher_text += cipher_text_state[i][j]\n cipher_text_blocks.append(cipher_text)\n \n hex_ciphertext = bit_matrix_to_hex(cipher_text_state)\n for i in hex_ciphertext:\n print(i)\n \n return ''.join(cipher_text_blocks)",
"_____no_output_____"
],
[
"plaintext = \"Two One Nine Two\"\nkey = get_bit_chunks(\"5468617473206D79204B756E67204675\", 128)[0] # please keep only necessary chars in key, no space\n\nciphertext = AES(plaintext, key)\nprint(ciphertext)",
"['0x29', '0x57', '0x40', '0x1a']\n['0xc3', '0x14', '0x22', '0x2']\n['0x50', '0x20', '0x99', '0xd7']\n['0x5f', '0xf6', '0xb3', '0x3a']\n00101001110000110101000001011111010101110001010000100000111101100100000000100010100110011011001100011010000000101101011100111010\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
d0e18ebe91b903b8f99828b65ba2030e8664de5f | 4,182 | ipynb | Jupyter Notebook | 00_deck.ipynb | fnauman/binary_classifier | 806fbd121119c77d7b666c4554377a811aca6778 | [
"Apache-2.0"
] | null | null | null | 00_deck.ipynb | fnauman/binary_classifier | 806fbd121119c77d7b666c4554377a811aca6778 | [
"Apache-2.0"
] | null | null | null | 00_deck.ipynb | fnauman/binary_classifier | 806fbd121119c77d7b666c4554377a811aca6778 | [
"Apache-2.0"
] | null | null | null | 25.656442 | 177 | 0.458154 | [
[
[
"#default_exp deck",
"_____no_output_____"
]
],
[
[
"# Deck\n\n> Deck of playing cards.",
"_____no_output_____"
]
],
[
[
"from nbdev import *\nfrom binary_classifier.card import Card",
"_____no_output_____"
],
[
"# export\nclass Deck:\n \"\"\"Represents a deck of cards.\n Attributes:\n cards: list of Card objects.\n \"\"\"\n \n def __init__(self):\n \"\"\"Initializes the Deck with 52 cards.\n \"\"\"\n self.cards = []\n for suit in range(4):\n for rank in range(1, 14):\n card = Card(suit, rank)\n self.cards.append(card)\n\n def __str__(self):\n \"\"\"Returns a string representation of the deck.\n \"\"\"\n res = []\n for card in self.cards:\n res.append(str(card))\n return '\\n'.join(res)\n \n def __repr__(self):\n return self.__str__()\n\n def add_card(self, card):\n \"\"\"Adds a card to the deck.\n card: Card\n \"\"\"\n self.cards.append(card)\n\n def remove_card(self, card):\n \"\"\"Removes a card from the deck or raises exception if it is not there.\n \n card: Card\n \"\"\"\n self.cards.remove(card)\n\n def pop_card(self, i=-1):\n \"\"\"Removes and returns a card from the deck.\n i: index of the card to pop; by default, pops the last card.\n \"\"\"\n return self.cards.pop(i)\n\n def shuffle(self):\n \"\"\"Shuffles the cards in this deck.\"\"\"\n random.shuffle(self.cards)\n\n def sort(self):\n \"\"\"Sorts the cards in ascending order.\"\"\"\n self.cards.sort()\n\n def move_cards(self, hand, num):\n \"\"\"Moves the given number of cards from the deck into the Hand.\n hand: destination Hand object\n num: integer number of cards to move\n \"\"\"\n for i in range(num):\n hand.add_card(self.pop_card())",
"_____no_output_____"
]
],
[
[
"A deck of cards",
"_____no_output_____"
]
],
[
[
"deck = Deck()\nassert isinstance(deck.pop_card(), Card)",
"_____no_output_____"
],
[
"show_doc(Deck.move_cards)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d0e1ae0786740f2cad95a4fee9d1e58bbbbc860a | 40,063 | ipynb | Jupyter Notebook | machine_learning/notebooks/accuracy_report.ipynb | cyberj0g/verification-classifier | efb19a3864e27a7f149a1c27ee8e13eaa19f96eb | [
"MIT"
] | 8 | 2019-06-06T08:16:45.000Z | 2021-06-26T11:53:48.000Z | machine_learning/notebooks/accuracy_report.ipynb | cyberj0g/verification-classifier | efb19a3864e27a7f149a1c27ee8e13eaa19f96eb | [
"MIT"
] | 95 | 2019-03-27T08:36:01.000Z | 2022-02-10T00:15:20.000Z | machine_learning/notebooks/accuracy_report.ipynb | cyberj0g/verification-classifier | efb19a3864e27a7f149a1c27ee8e13eaa19f96eb | [
"MIT"
] | 8 | 2019-02-28T11:21:46.000Z | 2022-03-21T07:34:20.000Z | 32.153291 | 121 | 0.410154 | [
[
[
"# Import Libraries",
"_____no_output_____"
]
],
[
[
"import sys\nimport pandas as pd\nimport numpy as np\n\nfrom sklearn import preprocessing\nfrom sklearn.decomposition import PCA\nfrom sklearn import random_projection\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.metrics import fbeta_score, roc_curve, auc\nfrom sklearn import svm\n\nimport pprint\n\nsys.path.insert(0, '../../scripts/modeling_toolbox/')\n# load the autoreload extension\n%load_ext autoreload\n# Set extension to reload modules every time before executing code\n%autoreload 2\n\nfrom metric_processor import MetricProcessor\nimport evaluation\n\n%matplotlib inline",
"Using TensorFlow backend.\n"
]
],
[
[
"# Data preparation",
"_____no_output_____"
]
],
[
[
"features = ['dimension',\n 'size',\n 'temporal_dct-mean', \n 'temporal_gaussian_mse-mean', \n 'temporal_gaussian_difference-mean',\n 'temporal_threshold_gaussian_difference-mean'\n ]\n\n\npath = '../../machine_learning/cloud_functions/data-large.csv'\n\nmetric_processor = MetricProcessor(features,'UL', path, reduced=False, bins=0)\ndf = metric_processor.read_and_process_data()\ndf.shape",
"dimension\nsize\ntemporal_dct-mean\ntemporal_gaussian_mse-mean\ntemporal_gaussian_difference-mean\ntemporal_threshold_gaussian_difference-mean\n"
],
[
"display(df.head())",
"_____no_output_____"
],
[
"# We remove the low bitrates since we are only focused on tampering attacks. The rotation attacks are also\n# removed since they will be detected by the pre-verifier just by checking output dimensions\ndf = df[~(df['attack'].str.contains('low_bitrate')) & ~(df['attack'].str.contains('rotate'))]",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"(X_train, X_test, X_attacks), (df_train, df_test, df_attacks) = metric_processor.split_test_and_train(df)\n\nprint('Shape of train: {}'.format(X_train.shape))\nprint('Shape of test: {}'.format(X_test.shape))\nprint('Shape of attacks: {}'.format(X_attacks.shape))",
"Shape of train: (18014, 6)\nShape of test: (4504, 6)\nShape of attacks: (111778, 6)\n"
]
],
[
[
"The train and test are **only** composed by legit assets",
"_____no_output_____"
]
],
[
[
"# Scaling the data\nss = StandardScaler()\nx_train = ss.fit_transform(X_train)\nx_test = ss.transform(X_test)\nx_attacks = ss.transform(X_attacks)",
"_____no_output_____"
]
],
[
[
"# One Class SVM",
"_____no_output_____"
]
],
[
[
"# Train the model\nOCSVM = svm.OneClassSVM(kernel='rbf',gamma='auto', nu=0.01, cache_size=5000)\nOCSVM.fit(x_train)",
"_____no_output_____"
],
[
"fb, area, tnr, tpr_train, tpr_test = evaluation.unsupervised_evaluation(OCSVM, x_train, x_test, x_attacks)",
"_____no_output_____"
],
[
"# Show global results of classification\nprint('TNR: {}\\nTPR_test: {}\\nTPR_train: {}\\n'.format(tnr, tpr_test, tpr_train))\nprint('F20: {}\\nAUC: {}'.format(fb, area))",
"TNR: 0.8769972624308898\nTPR_test: 0.9824600355239786\nTPR_train: 0.984845120461863\n\nF20: 0.9750798725556029\nAUC: 0.9297286489774341\n"
],
[
"# Show mean distances to the decision function. A negative distance means that the data is classified as\n# an attack\ntrain_scores = OCSVM.decision_function(x_train)\ntest_scores = OCSVM.decision_function(x_test)\nattack_scores = OCSVM.decision_function(x_attacks)\n\nprint('Mean score values:\\n-Train: {}\\n-Test: {}\\n-Attacks: {}'.format(np.mean(train_scores),\n np.mean(test_scores),\n np.mean(attack_scores)))",
"Mean score values:\n-Train: 0.697270521920494\n-Test: 0.6871460213667405\n-Attacks: -4.114003380259059\n"
],
[
"train_preds = OCSVM.predict(x_train)\ntest_preds = OCSVM.predict(x_test)\nattack_preds = OCSVM.predict(x_attacks)",
"_____no_output_____"
],
[
"df_train['dist_to_dec_funct'] = train_scores\ndf_test['dist_to_dec_funct'] = test_scores\ndf_attacks['dist_to_dec_funct'] = attack_scores",
"_____no_output_____"
],
[
"df_train['prediction'] = train_preds\ndf_test['prediction'] = test_preds\ndf_attacks['prediction'] = attack_preds",
"_____no_output_____"
]
],
[
[
"# Report",
"_____no_output_____"
]
],
[
[
"# Zoom in in the mean distances of the test set to the decision function by resolution. Percentiles, standard \n# deviation, min and max values are shown too.\ndisplay(df_test[['dist_to_dec_funct', 'dimension']].groupby('dimension').describe())",
"_____no_output_____"
],
[
"# Zoom in in the mean distances of the attack set to the decision function by resolution. Percentiles, standard \n# deviation, min and max values are shown too.\ndisplay(df_attacks[['dist_to_dec_funct', 'dimension']].groupby('dimension').describe())",
"_____no_output_____"
],
[
"# Zoom in in the mean distances of the test set to the decision function by attack type. Percentiles, standard \n# deviation, min and max values are shown too.\ndf_attacks['attack_'] = df_attacks['attack'].apply(lambda x: x[x.find('p') + 2:])\ndisplay(df_attacks[['dist_to_dec_funct', 'attack_']].groupby(['attack_']).describe())",
"_____no_output_____"
],
[
"resolutions = sorted(df_attacks['dimension'].unique())",
"_____no_output_____"
],
[
"pp = pprint.PrettyPrinter()",
"_____no_output_____"
],
[
"# Accuracy of the test set by resolution\nresults = {}\nfor res in resolutions:\n selection = df_test[df_test['dimension'] == res]\n count = sum(selection['prediction'] == 1)\n results[res] = count/len(selection)\npp.pprint(results)",
"{144: 0.968,\n 240: 0.9906790945406125,\n 360: 0.9893475366178429,\n 480: 0.9946737683089214,\n 720: 0.9946737683089214,\n 1080: 0.9573333333333334}\n"
],
[
"# Accuracy on the attack set by resolution\nresults = {}\nfor res in resolutions:\n selection = df_attacks[df_attacks['dimension'] == res]\n count = sum(selection['prediction'] == -1)\n results[res] = count/len(selection)\npp.pprint(results)",
"{144: 0.7895866881374127,\n 240: 0.8008588298443371,\n 360: 0.8459391271673198,\n 480: 0.8863062966342798,\n 720: 0.9447128287707998,\n 1080: 0.9945786366076221}\n"
],
[
"attacks = df_attacks['attack_'].unique()",
"_____no_output_____"
],
[
"# Accuracy on the attack set by attack type\nresults = {}\nfor attk in attacks:\n selection = df_attacks[df_attacks['attack_'] == attk]\n count = sum(selection['prediction'] == -1)\n results[attk] = count/len(selection)\npp.pprint(results)",
"{'black_and_white': 0.9810373923083755,\n 'flip_vertical': 1.0,\n 'vignette': 0.999333866240341,\n 'watermark': 0.8812896922325354,\n 'watermark-345x114': 0.5208218168417174,\n 'watermark-856x856': 0.9893122034366794}\n"
],
[
"\n# Accuracy on the attack set by attack type\nresults = {}\nfor res in resolutions:\n results[res] = {}\n for attk in attacks:\n selection = df_attacks[(df_attacks['attack_'] == attk) & (df_attacks['dimension'] == res)]\n count = sum(selection['prediction'] == -1)\n results[res][attk] = count/len(selection)\npp.pprint(results)",
"{144: {'black_and_white': 0.9661604050093259,\n 'flip_vertical': 1.0,\n 'vignette': 0.9992006394884093,\n 'watermark': 0.7503330668798295,\n 'watermark-345x114': 0.2269209508015478,\n 'watermark-856x856': 0.9845218352681039},\n 240: {'black_and_white': 0.9757527311484147,\n 'flip_vertical': 1.0,\n 'vignette': 0.9992006394884093,\n 'watermark': 0.7679189981348254,\n 'watermark-345x114': 0.2517965726920951,\n 'watermark-856x856': 0.9894969596462134},\n 360: {'black_and_white': 0.980548894217959,\n 'flip_vertical': 1.0,\n 'vignette': 0.9992006394884093,\n 'watermark': 0.8723687716493472,\n 'watermark-345x114': 0.37092316196793806,\n 'watermark-856x856': 0.989217583632845},\n 480: {'black_and_white': 0.9845456967759126,\n 'flip_vertical': 1.0,\n 'vignette': 0.9992006394884093,\n 'watermark': 0.9224413646055437,\n 'watermark-345x114': 0.5226644555002764,\n 'watermark-856x856': 0.9892205638474295},\n 720: {'black_and_white': 0.988009592326139,\n 'flip_vertical': 1.0,\n 'vignette': 0.9994670929922729,\n 'watermark': 0.9746869171329603,\n 'watermark-345x114': 0.7653399668325042,\n 'watermark-856x856': 0.9892205638474295},\n 1080: {'black_and_white': 0.991207034372502,\n 'flip_vertical': 1.0,\n 'vignette': 0.9997335464961364,\n 'watermark': 1.0,\n 'watermark-345x114': 0.9872857932559425,\n 'watermark-856x856': 0.9941956882255389}}\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e1c2f0fe9d3d0be253f42e2c0017efbee27224 | 5,378 | ipynb | Jupyter Notebook | template/colab.ipynb | ampl/amplcolab | 3bb80f4650adfe3047aeaa821fcd1bd3580349c2 | [
"MIT"
] | null | null | null | template/colab.ipynb | ampl/amplcolab | 3bb80f4650adfe3047aeaa821fcd1bd3580349c2 | [
"MIT"
] | null | null | null | template/colab.ipynb | ampl/amplcolab | 3bb80f4650adfe3047aeaa821fcd1bd3580349c2 | [
"MIT"
] | null | null | null | 2,689 | 5,377 | 0.629602 | [
[
[
"# AMPL Model Colaboratory Template\n[](https://github.com/ampl/amplcolab/blob/master/template/colab.ipynb) [](https://colab.research.google.com/github/ampl/amplcolab/blob/master/template/colab.ipynb) [](https://kaggle.com/kernels/welcome?src=https://github.com/ampl/amplcolab/blob/master/template/colab.ipynb) [](https://console.paperspace.com/github/ampl/amplcolab/blob/master/template/colab.ipynb) [](https://studiolab.sagemaker.aws/import/github/ampl/amplcolab/blob/master/template/colab.ipynb)\n\nDescription: Basic notebook templace for the AMPL Colab repository\n\nTags: ampl-only, template, industry\n\nNotebook author: Filipe Brandão\n\nModel author: Gilmore, P. and Gomory, R.\n\nReferences:\n1. Gilmore, P. and Gomory, R. (1963). A linear programming approach to the cutting stock\nproblem–part II.",
"_____no_output_____"
]
],
[
[
"# Install dependencies\n!pip install -q amplpy ampltools",
"_____no_output_____"
],
[
"# Google Colab & Kaggle interagration\nMODULES=['ampl', 'coin']\nfrom ampltools import cloud_platform_name, ampl_notebook\nfrom amplpy import AMPL, register_magics\nif cloud_platform_name() is None:\n ampl = AMPL() # Use local installation of AMPL\nelse:\n ampl = ampl_notebook(modules=MODULES) # Install AMPL and use it\nregister_magics(ampl_object=ampl) # Evaluate %%ampl_eval cells with ampl.eval()",
"_____no_output_____"
]
],
[
[
"### Use `%%ampl_eval` to evaluate AMPL commands",
"_____no_output_____"
]
],
[
[
"%%ampl_eval\noption version;",
"_____no_output_____"
]
],
[
[
"### Use %%writeifile to create files",
"_____no_output_____"
]
],
[
[
"%%writefile cut2.mod\n\n problem Cutting_Opt;\n# ----------------------------------------\n\nparam nPAT integer >= 0, default 0;\nparam roll_width;\n\nset PATTERNS = 1..nPAT;\nset WIDTHS;\n\nparam orders {WIDTHS} > 0;\nparam nbr {WIDTHS,PATTERNS} integer >= 0;\n\n check {j in PATTERNS}: sum {i in WIDTHS} i * nbr[i,j] <= roll_width;\n\nvar Cut {PATTERNS} integer >= 0;\n\nminimize Number: sum {j in PATTERNS} Cut[j];\n\nsubject to Fill {i in WIDTHS}:\n sum {j in PATTERNS} nbr[i,j] * Cut[j] >= orders[i];\n\n\n problem Pattern_Gen;\n# ----------------------------------------\n\nparam price {WIDTHS} default 0;\n\nvar Use {WIDTHS} integer >= 0;\n\nminimize Reduced_Cost:\n 1 - sum {i in WIDTHS} price[i] * Use[i];\n\nsubject to Width_Limit:\n sum {i in WIDTHS} i * Use[i] <= roll_width;",
"_____no_output_____"
],
[
"%%writefile cut.dat\ndata;\n\nparam roll_width := 110 ;\n\nparam: WIDTHS: orders :=\n 20 48\n 45 35\n 50 24\n 55 10\n 75 8 ;",
"_____no_output_____"
],
[
"%%writefile cut2.run\n# ----------------------------------------\n# GILMORE-GOMORY METHOD FOR\n# CUTTING STOCK PROBLEM\n# ----------------------------------------\n\noption solver cbc;\noption solution_round 6;\n\nmodel cut2.mod;\ndata cut.dat;\n\nproblem Cutting_Opt;\n option relax_integrality 1;\n option presolve 0;\n\nproblem Pattern_Gen;\n option relax_integrality 0;\n option presolve 1;\n\nlet nPAT := 0;\n\nfor {i in WIDTHS} {\n let nPAT := nPAT + 1;\n let nbr[i,nPAT] := floor (roll_width/i);\n let {i2 in WIDTHS: i2 <> i} nbr[i2,nPAT] := 0;\n };\n\nrepeat {\n solve Cutting_Opt;\n\n let {i in WIDTHS} price[i] := Fill[i].dual;\n\n solve Pattern_Gen;\n\n if Reduced_Cost < -0.00001 then {\n let nPAT := nPAT + 1;\n let {i in WIDTHS} nbr[i,nPAT] := Use[i];\n }\n else break;\n };\n\ndisplay nbr; \ndisplay Cut;\n\noption Cutting_Opt.relax_integrality 0;\noption Cutting_Opt.presolve 10;\nsolve Cutting_Opt;\n\ndisplay Cut;",
"_____no_output_____"
]
],
[
[
"### Use `%%ampl_eval` to run the script cut2.run",
"_____no_output_____"
]
],
[
[
"%%ampl_eval\ncommands cut2.run;",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0e1d8b688b6caf3b5ce43c52c2c8649e9c416b3 | 26,143 | ipynb | Jupyter Notebook | .ipynb_checkpoints/demo-checkpoint.ipynb | esandford/forecaster | 2f0ef37ce2990515549122ce9968fe27dad362b2 | [
"MIT"
] | 16 | 2016-03-30T13:22:21.000Z | 2021-09-27T21:49:46.000Z | .ipynb_checkpoints/demo-checkpoint.ipynb | esandford/forecaster | 2f0ef37ce2990515549122ce9968fe27dad362b2 | [
"MIT"
] | 4 | 2016-12-08T17:27:02.000Z | 2021-03-14T17:02:26.000Z | .ipynb_checkpoints/demo-checkpoint.ipynb | esandford/forecaster | 2f0ef37ce2990515549122ce9968fe27dad362b2 | [
"MIT"
] | 21 | 2016-03-28T19:24:10.000Z | 2022-02-17T03:14:59.000Z | 97.185874 | 14,334 | 0.856902 | [
[
[
"WARNING:\n\n\"fitting_parameters.h5\" need to be in the directory you are working on\nor there will be an error for importing mr_forecast in the next cell.\n\nIf you don't want the file in this directory,\nchange the mr_forecast.py line 16\n\nhyper_file = 'fitting_parameters.h5' \n\n->\n\nhyper_file = [directory of fitting parameter file]+'fitting_parameters.h5'",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport mr_forecast as mr\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"================================\n\npredict the mean and std of radius given those of the mass",
"_____no_output_____"
]
],
[
[
"Rmedian, Rplus, Rminus = mr.Mstat2R(mean=1.0, std=0.1, unit='Earth', sample_size=100, classify='Yes')",
"Terran 97.0 %, Neptunian 3.0 %, Jovian 0.0 %, Star 0.0 %\n"
],
[
"print 'R = %.2f (+ %.2f - %.2f) REarth' % (Rmedian, Rplus, Rminus)",
"R = 1.00 (+ 0.12 - 0.10) REarth\n"
]
],
[
[
"================================\n\npredict a vector of radius given a vector of mass",
"_____no_output_____"
]
],
[
[
"M1 = np.loadtxt('demo_mass.dat')\nR1 = mr.Mpost2R(M1, unit='Earth', classify='Yes')",
"Terran 100.0 %, Neptunian 0.0 %, Jovian 0.0 %, Star 0.0 %\n"
],
[
"plt.plot(np.log10(M1), np.log10(R1), 'bx')\nplt.xlabel(r'$log_{10}\\ M/M_{\\oplus}$')\nplt.ylabel(r'$log_{10}\\ R/R_{\\oplus}$')\nplt.show()",
"_____no_output_____"
]
],
[
[
"================================\n\npredict the mean and std of mass given those of the radius",
"_____no_output_____"
]
],
[
[
"Mmedian, Mplus, Mminus = mr.Rstat2M(mean=0.1, std=0.01, unit='Jupiter', sample_size=100, grid_size=1e3, classify='Yes')",
"Terran 69.0 %, Neptunian 31.0 %, Jovian 0.0 %, Star 0.0 %\n"
],
[
"print 'M = %.3f (+ %.3f - %.3f) MEarth' % (Mmedian, Mplus, Mminus)",
"M = 0.005 (+ 0.004 - 0.002) MEarth\n"
]
],
[
[
"================================\n\npredict a vector of mass given a vector of radius",
"_____no_output_____"
]
],
[
[
"R2 = np.loadtxt('demo_radius.dat')\nM2 = mr.Rpost2M(R2, unit='Earth', grid_size=1e3, classify='Yes')",
"Terran 72.0 %, Neptunian 28.0 %, Jovian 0.0 %, Star 0.0 %\n"
],
[
"plt.hist(np.log10(M2))\nplt.xlabel(r'$log_{10}\\ M/M_{\\odot}$')\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d0e1da9bfd54a9a961bcb97c221843774170432b | 41,455 | ipynb | Jupyter Notebook | project-3-tv-script-generation/dlnd_tv_script_generation.ipynb | vwiart/deeplearning-udacity | 669ed5402b2da5a589d4586f90a8a1ddc1e6d866 | [
"Apache-2.0"
] | null | null | null | project-3-tv-script-generation/dlnd_tv_script_generation.ipynb | vwiart/deeplearning-udacity | 669ed5402b2da5a589d4586f90a8a1ddc1e6d866 | [
"Apache-2.0"
] | null | null | null | project-3-tv-script-generation/dlnd_tv_script_generation.ipynb | vwiart/deeplearning-udacity | 669ed5402b2da5a589d4586f90a8a1ddc1e6d866 | [
"Apache-2.0"
] | null | null | null | 33.377617 | 556 | 0.558027 | [
[
[
"# TV Script Generation\nIn this project, you'll generate your own [Simpsons](https://en.wikipedia.org/wiki/The_Simpsons) TV scripts using RNNs. You'll be using part of the [Simpsons dataset](https://www.kaggle.com/wcukierski/the-simpsons-by-the-data) of scripts from 27 seasons. The Neural Network you'll build will generate a new TV script for a scene at [Moe's Tavern](https://simpsonswiki.com/wiki/Moe's_Tavern).\n## Get the Data\nThe data is already provided for you. You'll be using a subset of the original dataset. It consists of only the scenes in Moe's Tavern. This doesn't include other versions of the tavern, like \"Moe's Cavern\", \"Flaming Moe's\", \"Uncle Moe's Family Feed-Bag\", etc..",
"_____no_output_____"
]
],
[
[
"\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL\n\"\"\"\nimport helper\n\ndata_dir = './data/simpsons/moes_tavern_lines.txt'\ntext = helper.load_data(data_dir)\n# Ignore notice, since we don't use it for analysing the data\ntext = text[81:]",
"_____no_output_____"
]
],
[
[
"## Explore the Data\nPlay around with `view_sentence_range` to view different parts of the data.",
"_____no_output_____"
]
],
[
[
"view_sentence_range = (20, 30)\n\n\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL\n\"\"\"\nimport numpy as np\n\nprint('Dataset Stats')\nprint('Roughly the number of unique words: {}'.format(len({word: None for word in text.split()})))\nscenes = text.split('\\n\\n')\nprint('Number of scenes: {}'.format(len(scenes)))\nsentence_count_scene = [scene.count('\\n') for scene in scenes]\nprint('Average number of sentences in each scene: {}'.format(np.average(sentence_count_scene)))\n\nsentences = [sentence for scene in scenes for sentence in scene.split('\\n')]\nprint('Number of lines: {}'.format(len(sentences)))\nword_count_sentence = [len(sentence.split()) for sentence in sentences]\nprint('Average number of words in each line: {}'.format(np.average(word_count_sentence)))\n\nprint()\nprint('The sentences {} to {}:'.format(*view_sentence_range))\nprint('\\n'.join(text.split('\\n')[view_sentence_range[0]:view_sentence_range[1]]))",
"Dataset Stats\nRoughly the number of unique words: 11492\nNumber of scenes: 262\nAverage number of sentences in each scene: 15.251908396946565\nNumber of lines: 4258\nAverage number of words in each line: 11.50164396430249\n\nThe sentences 20 to 30:\n\n\nMoe_Szyslak: Looks like this is the end.\nBarney_Gumble: That's all right. I couldn't have led a richer life.\nBarney_Gumble: So the next time somebody tells you county folk are good, honest people, you can spit in their faces for me!\nLisa_Simpson: I will, Mr. Gumbel. But if you'll excuse me, I'm profiling my dad for the school paper. I thought it would be neat to follow him around for a day to see what makes him tick.\nBarney_Gumble: Oh, that's sweet. I used to follow my dad to a lot of bars too. (BELCH)\nMoe_Szyslak: Here you go. One beer, one chocolate milk.\nLisa_Simpson: Uh, excuse me, I have the chocolate milk.\nMoe_Szyslak: Oh.\n"
]
],
[
[
"## Implement Preprocessing Functions\nThe first thing to do to any dataset is preprocessing. Implement the following preprocessing functions below:\n- Lookup Table\n- Tokenize Punctuation\n\n### Lookup Table\nTo create a word embedding, you first need to transform the words to ids. In this function, create two dictionaries:\n- Dictionary to go from the words to an id, we'll call `vocab_to_int`\n- Dictionary to go from the id to word, we'll call `int_to_vocab`\n\nReturn these dictionaries in the following tuple `(vocab_to_int, int_to_vocab)`",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport problem_unittests as tests\n\ndef create_lookup_tables(text):\n \"\"\"\n Create lookup tables for vocabulary\n :param text: The text of tv scripts split into words\n :return: A tuple of dicts (vocab_to_int, int_to_vocab)\n \"\"\"\n filtered_out = ['']\n tokenized_text = set([t.lower() for t in text if t not in filtered_out])\n \n vocab_to_int = {t: i for i, t in enumerate(tokenized_text)}\n int_to_vocab = {i: t for i, t in enumerate(tokenized_text)}\n return vocab_to_int, int_to_vocab\n\n\n\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE\n\"\"\"\ntests.test_create_lookup_tables(create_lookup_tables)",
"Tests Passed\n"
]
],
[
[
"### Tokenize Punctuation\nWe'll be splitting the script into a word array using spaces as delimiters. However, punctuations like periods and exclamation marks make it hard for the neural network to distinguish between the word \"bye\" and \"bye!\".\n\nImplement the function `token_lookup` to return a dict that will be used to tokenize symbols like \"!\" into \"||Exclamation_Mark||\". Create a dictionary for the following symbols where the symbol is the key and value is the token:\n- Period ( . )\n- Comma ( , )\n- Quotation Mark ( \" )\n- Semicolon ( ; )\n- Exclamation mark ( ! )\n- Question mark ( ? )\n- Left Parentheses ( ( )\n- Right Parentheses ( ) )\n- Dash ( -- )\n- Return ( \\n )\n\nThis dictionary will be used to token the symbols and add the delimiter (space) around it. This separates the symbols as it's own word, making it easier for the neural network to predict on the next word. Make sure you don't use a token that could be confused as a word. Instead of using the token \"dash\", try using something like \"||dash||\".",
"_____no_output_____"
]
],
[
[
"def token_lookup():\n \"\"\"\n Generate a dict to turn punctuation into a token.\n :return: Tokenize dictionary where the key is the punctuation and the value is the token\n \"\"\"\n return {\n '.': '||period||',\n ',': '||comma||',\n '\"': '||quotation_mark||',\n ';': '||semicolon||',\n '!': '||exclamation_mark||',\n '?': '||question_mark||',\n '(': '||left_parentheses||',\n ')': '||right_parentheses||',\n '--': '||dash||',\n '\\n': '||line_break||',\n }\n\n\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE\n\"\"\"\ntests.test_tokenize(token_lookup)",
"Tests Passed\n"
]
],
[
[
"## Preprocess all the data and save it\nRunning the code cell below will preprocess all the data and save it to file.",
"_____no_output_____"
]
],
[
[
"\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL\n\"\"\"\n# Preprocess Training, Validation, and Testing Data\nhelper.preprocess_and_save_data(data_dir, token_lookup, create_lookup_tables)",
"_____no_output_____"
]
],
[
[
"# Check Point\nThis is your first checkpoint. If you ever decide to come back to this notebook or have to restart the notebook, you can start from here. The preprocessed data has been saved to disk.",
"_____no_output_____"
]
],
[
[
"\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL\n\"\"\"\nimport helper\nimport numpy as np\nimport problem_unittests as tests\n\nint_text, vocab_to_int, int_to_vocab, token_dict = helper.load_preprocess()",
"_____no_output_____"
]
],
[
[
"## Build the Neural Network\nYou'll build the components necessary to build a RNN by implementing the following functions below:\n- get_inputs\n- get_init_cell\n- get_embed\n- build_rnn\n- build_nn\n- get_batches\n\n### Check the Version of TensorFlow and Access to GPU",
"_____no_output_____"
]
],
[
[
"\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL\n\"\"\"\nfrom distutils.version import LooseVersion\nimport warnings\nimport tensorflow as tf\n\n# Check TensorFlow Version\nassert LooseVersion(tf.__version__) >= LooseVersion('1.0'), 'Please use TensorFlow version 1.0 or newer'\nprint('TensorFlow Version: {}'.format(tf.__version__))\n\n# Check for a GPU\nif not tf.test.gpu_device_name():\n warnings.warn('No GPU found. Please use a GPU to train your neural network.')\nelse:\n print('Default GPU Device: {}'.format(tf.test.gpu_device_name()))",
"TensorFlow Version: 1.0.0\nDefault GPU Device: /gpu:0\n"
]
],
[
[
"### Input\nImplement the `get_inputs()` function to create TF Placeholders for the Neural Network. It should create the following placeholders:\n- Input text placeholder named \"input\" using the [TF Placeholder](https://www.tensorflow.org/api_docs/python/tf/placeholder) `name` parameter.\n- Targets placeholder\n- Learning Rate placeholder\n\nReturn the placeholders in the following the tuple `(Input, Targets, LearingRate)`",
"_____no_output_____"
]
],
[
[
"def get_inputs():\n \"\"\"\n Create TF Placeholders for input, targets, and learning rate.\n :return: Tuple (input, targets, learning rate)\n \"\"\"\n inputs = tf.placeholder(tf.int32,\n shape=[None, None],\n name=\"input\")\n targets = tf.placeholder(tf.int32,\n shape=[None, None],\n name=\"target\")\n learning_rate = tf.placeholder(tf.float32,\n name=\"learning_rate\")\n return inputs, targets, learning_rate\n\n\n\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE\n\"\"\"\ntests.test_get_inputs(get_inputs)",
"Tests Passed\n"
]
],
[
[
"### Build RNN Cell and Initialize\nStack one or more [`BasicLSTMCells`](https://www.tensorflow.org/api_docs/python/tf/contrib/rnn/BasicLSTMCell) in a [`MultiRNNCell`](https://www.tensorflow.org/api_docs/python/tf/contrib/rnn/MultiRNNCell).\n- The Rnn size should be set using `rnn_size`\n- Initalize Cell State using the MultiRNNCell's [`zero_state()`](https://www.tensorflow.org/api_docs/python/tf/contrib/rnn/MultiRNNCell#zero_state) function\n - Apply the name \"initial_state\" to the initial state using [`tf.identity()`](https://www.tensorflow.org/api_docs/python/tf/identity)\n\nReturn the cell and initial state in the following tuple `(Cell, InitialState)`",
"_____no_output_____"
]
],
[
[
"keep_prob = 0.8\nlstm_layers = 16\n\ndef get_init_cell(batch_size, rnn_size):\n \"\"\"\n Create an RNN Cell and initialize it.\n :param batch_size: Size of batches\n :param rnn_size: Size of RNNs\n :return: Tuple (cell, initialize state)\n \"\"\"\n lstm = tf.contrib.rnn.BasicLSTMCell(rnn_size)\n drop = tf.contrib.rnn.DropoutWrapper(lstm, output_keep_prob=keep_prob)\n cell = tf.contrib.rnn.MultiRNNCell([drop], lstm_layers)\n initial_state = tf.identity(cell.zero_state(batch_size, tf.float32), \"initial_state\")\n return cell, initial_state\n\n\n\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE\n\"\"\"\ntests.test_get_init_cell(get_init_cell)",
"Tests Passed\n"
]
],
[
[
"### Word Embedding\nApply embedding to `input_data` using TensorFlow. Return the embedded sequence.",
"_____no_output_____"
]
],
[
[
"def get_embed(input_data, vocab_size, embed_dim):\n \"\"\"\n Create embedding for <input_data>.\n :param input_data: TF placeholder for text input.\n :param vocab_size: Number of words in vocabulary.\n :param embed_dim: Number of embedding dimensions\n :return: Embedded input.\n \"\"\"\n embedding = tf.Variable(tf.random_uniform((vocab_size, embed_dim), -1, 1))\n embedding_lookup = tf.nn.embedding_lookup(embedding, input_data)\n return embedding_lookup\n\n\n\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE\n\"\"\"\ntests.test_get_embed(get_embed)",
"Tests Passed\n"
]
],
[
[
"### Build RNN\nYou created a RNN Cell in the `get_init_cell()` function. Time to use the cell to create a RNN.\n- Build the RNN using the [`tf.nn.dynamic_rnn()`](https://www.tensorflow.org/api_docs/python/tf/nn/dynamic_rnn)\n - Apply the name \"final_state\" to the final state using [`tf.identity()`](https://www.tensorflow.org/api_docs/python/tf/identity)\n\nReturn the outputs and final_state state in the following tuple `(Outputs, FinalState)` ",
"_____no_output_____"
]
],
[
[
"def build_rnn(cell, inputs):\n \"\"\"\n Create a RNN using a RNN Cell\n :param cell: RNN Cell\n :param inputs: Input text data\n :return: Tuple (Outputs, Final State)\n \"\"\"\n outputs, final_state = tf.nn.dynamic_rnn(cell, inputs, dtype=tf.float32)\n final_state = tf.identity(final_state, \"final_state\")\n \n return outputs, final_state\n\n\n\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE\n\"\"\"\ntests.test_build_rnn(build_rnn)",
"Tests Passed\n"
]
],
[
[
"### Build the Neural Network\nApply the functions you implemented above to:\n- Apply embedding to `input_data` using your `get_embed(input_data, vocab_size, embed_dim)` function.\n- Build RNN using `cell` and your `build_rnn(cell, inputs)` function.\n- Apply a fully connected layer with a linear activation and `vocab_size` as the number of outputs.\n\nReturn the logits and final state in the following tuple (Logits, FinalState) ",
"_____no_output_____"
]
],
[
[
"def build_nn(cell, rnn_size, input_data, vocab_size):\n \"\"\"\n Build part of the neural network\n :param cell: RNN cell\n :param rnn_size: Size of rnns\n :param input_data: Input data\n :param vocab_size: Vocabulary size\n :return: Tuple (Logits, FinalState)\n \"\"\"\n embeds = get_embed(input_data, vocab_size, rnn_size)\n outputs, final_state = build_rnn(cell, embeds)\n \n w_initializer = tf.truncated_normal_initializer(stddev= 0.1)\n b_initializer = tf.zeros_initializer()\n logits = tf.contrib.layers.fully_connected(outputs,\n vocab_size,\n activation_fn=None,\n weights_initializer=w_initializer,\n biases_initializer=b_initializer)\n\n return logits, final_state\n\n\n\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE\n\"\"\"\ntests.test_build_nn(build_nn)",
"Tests Passed\n"
]
],
[
[
"### Batches\nImplement `get_batches` to create batches of input and targets using `int_text`. The batches should be a Numpy array with the shape `(number of batches, 2, batch size, sequence length)`. Each batch contains two elements:\n- The first element is a single batch of **input** with the shape `[batch size, sequence length]`\n- The second element is a single batch of **targets** with the shape `[batch size, sequence length]`\n\nIf you can't fill the last batch with enough data, drop the last batch.\n\nFor exmple, `get_batches([1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15], 2, 3)` would return a Numpy array of the following:\n```\n[\n # First Batch\n [\n # Batch of Input\n [[ 1 2 3], [ 7 8 9]],\n # Batch of targets\n [[ 2 3 4], [ 8 9 10]]\n ],\n \n # Second Batch\n [\n # Batch of Input\n [[ 4 5 6], [10 11 12]],\n # Batch of targets\n [[ 5 6 7], [11 12 13]]\n ]\n]\n```",
"_____no_output_____"
]
],
[
[
"def shuffle(arr, seq_length, n_batches):\n reshape = arr.reshape(-1, seq_length)\n splits = [[] for _ in range(n_batches)]\n for i, a in enumerate(reshape):\n splits[i % n_batches].append(a)\n return splits\n\ndef get_batches(int_text, batch_size, seq_length):\n \"\"\"\n Return batches of input and target\n :param int_text: Text with the words replaced by their ids\n :param batch_size: The size of batch\n :param seq_length: The length of sequence\n :return: Batches as a Numpy array\n \"\"\"\n \n n_batches = len(int_text) // (batch_size * seq_length)\n max_length = n_batches * batch_size * seq_length\n\n inputs = np.array(int_text[:max_length]) # drop last batch\n targets = np.array(int_text[1:max_length+1]) # shift\n\n s_inputs = shuffle(inputs, seq_length, n_batches)\n s_targets = shuffle(targets, seq_length, n_batches)\n\n return np.array(list(zip(s_inputs, s_targets)))\n\n\n\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE\n\"\"\"\ntests.test_get_batches(get_batches)",
"Tests Passed\n"
]
],
[
[
"## Neural Network Training\n### Hyperparameters\nTune the following parameters:\n\n- Set `num_epochs` to the number of epochs.\n- Set `batch_size` to the batch size.\n- Set `rnn_size` to the size of the RNNs.\n- Set `seq_length` to the length of sequence.\n- Set `learning_rate` to the learning rate.\n- Set `show_every_n_batches` to the number of batches the neural network should print progress.",
"_____no_output_____"
]
],
[
[
"# Number of Epochs\nnum_epochs = 200\n# Batch Size\nbatch_size = 64\n# RNN Size\nrnn_size = 256\n# Sequence Length\nseq_length = 256\n# Learning Rate\nlearning_rate = 0.01\n# Show stats for every n number of batches\nshow_every_n_batches = 10\n\n\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE\n\"\"\"\nsave_dir = './save'",
"_____no_output_____"
]
],
[
[
"### Build the Graph\nBuild the graph using the neural network you implemented.",
"_____no_output_____"
]
],
[
[
"\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL\n\"\"\"\nfrom tensorflow.contrib import seq2seq\n\ntrain_graph = tf.Graph()\nwith train_graph.as_default():\n vocab_size = len(int_to_vocab)\n input_text, targets, lr = get_inputs()\n input_data_shape = tf.shape(input_text)\n cell, initial_state = get_init_cell(input_data_shape[0], rnn_size)\n logits, final_state = build_nn(cell, rnn_size, input_text, vocab_size)\n\n # Probabilities for generating words\n probs = tf.nn.softmax(logits, name='probs')\n\n # Loss function\n cost = seq2seq.sequence_loss(\n logits,\n targets,\n tf.ones([input_data_shape[0], input_data_shape[1]]))\n\n # Optimizer\n optimizer = tf.train.AdamOptimizer(lr)\n\n # Gradient Clipping\n gradients = optimizer.compute_gradients(cost)\n capped_gradients = [(tf.clip_by_value(grad, -1., 1.), var) for grad, var in gradients]\n train_op = optimizer.apply_gradients(capped_gradients)",
"_____no_output_____"
]
],
[
[
"## Train\nTrain the neural network on the preprocessed data. If you have a hard time getting a good loss, check the [forms](https://discussions.udacity.com/) to see if anyone is having the same problem.",
"_____no_output_____"
]
],
[
[
"\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL\n\"\"\"\nbatches = get_batches(int_text, batch_size, seq_length)\n\nwith tf.Session(graph=train_graph) as sess:\n sess.run(tf.global_variables_initializer())\n\n for epoch_i in range(num_epochs):\n state = sess.run(initial_state, {input_text: batches[0][0]})\n\n for batch_i, (x, y) in enumerate(batches):\n feed = {\n input_text: x,\n targets: y,\n initial_state: state,\n lr: learning_rate}\n train_loss, state, _ = sess.run([cost, final_state, train_op], feed)\n\n # Show every <show_every_n_batches> batches\n if (epoch_i * len(batches) + batch_i) % show_every_n_batches == 0:\n print('Epoch {:>3} Batch {:>4}/{} train_loss = {:.3f}'.format(\n epoch_i,\n batch_i,\n len(batches),\n train_loss))\n\n # Save Model\n saver = tf.train.Saver()\n saver.save(sess, save_dir)\n print('Model Trained and Saved')",
"Epoch 0 Batch 0/4 train_loss = 8.849\nEpoch 2 Batch 2/4 train_loss = 5.364\nEpoch 5 Batch 0/4 train_loss = 4.697\nEpoch 7 Batch 2/4 train_loss = 4.241\nEpoch 10 Batch 0/4 train_loss = 3.752\nEpoch 12 Batch 2/4 train_loss = 3.367\nEpoch 15 Batch 0/4 train_loss = 2.984\nEpoch 17 Batch 2/4 train_loss = 2.704\nEpoch 20 Batch 0/4 train_loss = 2.461\nEpoch 22 Batch 2/4 train_loss = 2.216\nEpoch 25 Batch 0/4 train_loss = 1.982\nEpoch 27 Batch 2/4 train_loss = 1.811\nEpoch 30 Batch 0/4 train_loss = 1.678\nEpoch 32 Batch 2/4 train_loss = 1.532\nEpoch 35 Batch 0/4 train_loss = 1.413\nEpoch 37 Batch 2/4 train_loss = 1.317\nEpoch 40 Batch 0/4 train_loss = 1.179\nEpoch 42 Batch 2/4 train_loss = 1.082\nEpoch 45 Batch 0/4 train_loss = 1.006\nEpoch 47 Batch 2/4 train_loss = 0.948\nEpoch 50 Batch 0/4 train_loss = 0.881\nEpoch 52 Batch 2/4 train_loss = 0.809\nEpoch 55 Batch 0/4 train_loss = 0.755\nEpoch 57 Batch 2/4 train_loss = 0.698\nEpoch 60 Batch 0/4 train_loss = 0.651\nEpoch 62 Batch 2/4 train_loss = 0.605\nEpoch 65 Batch 0/4 train_loss = 0.568\nEpoch 67 Batch 2/4 train_loss = 0.539\nEpoch 70 Batch 0/4 train_loss = 0.501\nEpoch 72 Batch 2/4 train_loss = 0.489\nEpoch 75 Batch 0/4 train_loss = 0.462\nEpoch 77 Batch 2/4 train_loss = 0.410\nEpoch 80 Batch 0/4 train_loss = 0.390\nEpoch 82 Batch 2/4 train_loss = 0.361\nEpoch 85 Batch 0/4 train_loss = 0.339\nEpoch 87 Batch 2/4 train_loss = 0.327\nEpoch 90 Batch 0/4 train_loss = 0.302\nEpoch 92 Batch 2/4 train_loss = 0.294\nEpoch 95 Batch 0/4 train_loss = 0.281\nEpoch 97 Batch 2/4 train_loss = 0.279\nEpoch 100 Batch 0/4 train_loss = 0.257\nEpoch 102 Batch 2/4 train_loss = 0.246\nEpoch 105 Batch 0/4 train_loss = 0.229\nEpoch 107 Batch 2/4 train_loss = 0.215\nEpoch 110 Batch 0/4 train_loss = 0.208\nEpoch 112 Batch 2/4 train_loss = 0.196\nEpoch 115 Batch 0/4 train_loss = 0.188\nEpoch 117 Batch 2/4 train_loss = 0.191\nEpoch 120 Batch 0/4 train_loss = 0.184\nEpoch 122 Batch 2/4 train_loss = 0.179\nEpoch 125 Batch 0/4 train_loss = 0.177\nEpoch 127 Batch 2/4 train_loss = 0.167\nEpoch 130 Batch 0/4 train_loss = 0.157\nEpoch 132 Batch 2/4 train_loss = 0.159\nEpoch 135 Batch 0/4 train_loss = 0.150\nEpoch 137 Batch 2/4 train_loss = 0.142\nEpoch 140 Batch 0/4 train_loss = 0.141\nEpoch 142 Batch 2/4 train_loss = 0.134\nEpoch 145 Batch 0/4 train_loss = 0.128\nEpoch 147 Batch 2/4 train_loss = 0.129\nEpoch 150 Batch 0/4 train_loss = 0.130\nEpoch 152 Batch 2/4 train_loss = 0.127\nEpoch 155 Batch 0/4 train_loss = 0.129\nEpoch 157 Batch 2/4 train_loss = 0.121\nEpoch 160 Batch 0/4 train_loss = 0.121\nEpoch 162 Batch 2/4 train_loss = 0.118\nEpoch 165 Batch 0/4 train_loss = 0.117\nEpoch 167 Batch 2/4 train_loss = 0.111\nEpoch 170 Batch 0/4 train_loss = 0.115\nEpoch 172 Batch 2/4 train_loss = 0.114\nEpoch 175 Batch 0/4 train_loss = 0.107\nEpoch 177 Batch 2/4 train_loss = 0.111\nEpoch 180 Batch 0/4 train_loss = 0.105\nEpoch 182 Batch 2/4 train_loss = 0.107\nEpoch 185 Batch 0/4 train_loss = 0.102\nEpoch 187 Batch 2/4 train_loss = 0.099\nEpoch 190 Batch 0/4 train_loss = 0.099\nEpoch 192 Batch 2/4 train_loss = 0.100\nEpoch 195 Batch 0/4 train_loss = 0.101\nEpoch 197 Batch 2/4 train_loss = 0.101\nModel Trained and Saved\n"
]
],
[
[
"## Save Parameters\nSave `seq_length` and `save_dir` for generating a new TV script.",
"_____no_output_____"
]
],
[
[
"\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL\n\"\"\"\n# Save parameters for checkpoint\nhelper.save_params((seq_length, save_dir))",
"_____no_output_____"
]
],
[
[
"# Checkpoint",
"_____no_output_____"
]
],
[
[
"\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL\n\"\"\"\nimport tensorflow as tf\nimport numpy as np\nimport helper\nimport problem_unittests as tests\n\n_, vocab_to_int, int_to_vocab, token_dict = helper.load_preprocess()\nseq_length, load_dir = helper.load_params()",
"_____no_output_____"
]
],
[
[
"## Implement Generate Functions\n### Get Tensors\nGet tensors from `loaded_graph` using the function [`get_tensor_by_name()`](https://www.tensorflow.org/api_docs/python/tf/Graph#get_tensor_by_name). Get the tensors using the following names:\n- \"input:0\"\n- \"initial_state:0\"\n- \"final_state:0\"\n- \"probs:0\"\n\nReturn the tensors in the following tuple `(InputTensor, InitialStateTensor, FinalStateTensor, ProbsTensor)` ",
"_____no_output_____"
]
],
[
[
"def get_tensors(loaded_graph):\n \"\"\"\n Get input, initial state, final state, and probabilities tensor from <loaded_graph>\n :param loaded_graph: TensorFlow graph loaded from file\n :return: Tuple (InputTensor, InitialStateTensor, FinalStateTensor, ProbsTensor)\n \"\"\"\n # TODO: Implement Function\n input_tensor = loaded_graph.get_tensor_by_name(\"input:0\")\n initial_state_tensor = loaded_graph.get_tensor_by_name(\"initial_state:0\")\n final_state_tensor = loaded_graph.get_tensor_by_name(\"final_state:0\")\n probs_tensor = loaded_graph.get_tensor_by_name(\"probs:0\")\n \n return input_tensor, initial_state_tensor, final_state_tensor, probs_tensor\n\n\n\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE\n\"\"\"\ntests.test_get_tensors(get_tensors)",
"Tests Passed\n"
]
],
[
[
"### Choose Word\nImplement the `pick_word()` function to select the next word using `probabilities`.",
"_____no_output_____"
]
],
[
[
"def pick_word(probabilities, int_to_vocab):\n \"\"\"\n Pick the next word in the generated text\n :param probabilities: Probabilites of the next word\n :param int_to_vocab: Dictionary of word ids as the keys and words as the values\n :return: String of the predicted word\n \"\"\"\n word = np.random.choice(list(int_to_vocab.values()), p=probabilities)\n return word\n\n\n\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE\n\"\"\"\ntests.test_pick_word(pick_word)",
"Tests Passed\n"
]
],
[
[
"## Generate TV Script\nThis will generate the TV script for you. Set `gen_length` to the length of TV script you want to generate.",
"_____no_output_____"
]
],
[
[
"gen_length = 200\n# homer_simpson, moe_szyslak, or Barney_Gumble\nprime_word = 'moe_szyslak'\n\n\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE\n\"\"\"\nloaded_graph = tf.Graph()\nwith tf.Session(graph=loaded_graph) as sess:\n # Load saved model\n loader = tf.train.import_meta_graph(load_dir + '.meta')\n loader.restore(sess, load_dir)\n\n # Get Tensors from loaded model\n input_text, initial_state, final_state, probs = get_tensors(loaded_graph)\n\n # Sentences generation setup\n gen_sentences = [prime_word + ':']\n prev_state = sess.run(initial_state, {input_text: np.array([[1]])})\n\n # Generate sentences\n for n in range(gen_length):\n # Dynamic Input\n dyn_input = [[vocab_to_int[word] for word in gen_sentences[-seq_length:]]]\n dyn_seq_length = len(dyn_input[0])\n\n # Get Prediction\n probabilities, prev_state = sess.run(\n [probs, final_state],\n {input_text: dyn_input, initial_state: prev_state})\n \n pred_word = pick_word(probabilities[dyn_seq_length-1], int_to_vocab)\n\n gen_sentences.append(pred_word)\n \n # Remove tokens\n tv_script = ' '.join(gen_sentences)\n for key, token in token_dict.items():\n ending = ' ' if key in ['\\n', '(', '\"'] else ''\n tv_script = tv_script.replace(' ' + token.lower(), key)\n tv_script = tv_script.replace('\\n ', '\\n')\n tv_script = tv_script.replace('( ', '(')\n \n print(tv_script)",
"moe_szyslak:(lifts index finger) to the scooter store!\nmoe_szyslak: i did not think this through.\nkemi: no.\n\n\ncarl_carlson: oh, now we're talkin'. but why me? you still not?\nhomer_simpson: what? i'm not where anything for you, crazy.\nkent_brockman: this is kent brockman with a channel 6 exclusive: the evil ned.\nkent_brockman: seven forty-two evergreen terrace. a tiny slice of suburban heaven. but like dating an actress, what seemed like heaven soon turned to hell.\n\n\nhomer_simpson:(loud whisper) let's try what chapter beautiful mayor_joe_quimby:?\nhomer_simpson:(amazed) kids, eh, just gotta be dressed as a civil war nurse / and then when i'm finished, i'll dead...\nhomer_simpson: caveman brow...\nlenny_leonard: don't forget that fish snout.\nmoe_szyslak:(\" back off\") okay, i get it. i ain't pleasant to look at.\nlenny_leonard: or listen to.\ncarl_carlson: i find a new sports with a frenchman, and spoon an english\n"
]
],
[
[
"# The TV Script is Nonsensical\nIt's ok if the TV script doesn't make any sense. We trained on less than a megabyte of text. In order to get good results, you'll have to use a smaller vocabulary or get more data. Luckly there's more data! As we mentioned in the begging of this project, this is a subset of [another dataset](https://www.kaggle.com/wcukierski/the-simpsons-by-the-data). We didn't have you train on all the data, because that would take too long. However, you are free to train your neural network on all the data. After you complete the project, of course.\n# Submitting This Project\nWhen submitting this project, make sure to run all the cells before saving the notebook. Save the notebook file as \"dlnd_tv_script_generation.ipynb\" and save it as a HTML file under \"File\" -> \"Download as\". Include the \"helper.py\" and \"problem_unittests.py\" files in your submission.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0e1e1cc90f760c16b4579123f831f1e9c55c3bc | 21,802 | ipynb | Jupyter Notebook | random_signals_LTI_systems/acoustic_impulse_response_measurement.ipynb | ZeroCommits/digital-signal-processing-lecture | e1e65432a5617a309ec02327a14962e37a0f7ec5 | [
"MIT"
] | 630 | 2016-01-05T17:11:43.000Z | 2022-03-30T07:48:27.000Z | random_signals_LTI_systems/acoustic_impulse_response_measurement.ipynb | patel999jay/digital-signal-processing-lecture | eea6f46284a903297452d2c6fc489cb4d26a4a54 | [
"MIT"
] | 12 | 2016-11-07T15:49:55.000Z | 2022-03-10T13:05:50.000Z | random_signals_LTI_systems/acoustic_impulse_response_measurement.ipynb | patel999jay/digital-signal-processing-lecture | eea6f46284a903297452d2c6fc489cb4d26a4a54 | [
"MIT"
] | 172 | 2015-12-26T21:05:40.000Z | 2022-03-10T23:13:30.000Z | 123.875 | 15,856 | 0.856527 | [
[
[
"# Random Signals and LTI-Systems\n\n*This jupyter notebook is part of a [collection of notebooks](../index.ipynb) on various topics of Digital Signal Processing. Please direct questions and suggestions to [[email protected]](mailto:[email protected]).*",
"_____no_output_____"
],
[
"## Measurement of Acoustic Impulse Responses\n\nThe propagation of sound from one position (e.g. transmitter) to another (e.g. receiver) conforms reasonable well to the properties of a linear time-invariant (LTI) system. Consequently, the impulse response $h[k]$ characterizes the propagation of sound between theses two positions. Impulse responses have various applications in acoustics. For instance as [head-related impulse responses](https://en.wikipedia.org/wiki/Head-related_transfer_function) (HRIRs) or room impulse responses (RIRs) for the characterization of room acoustics. \n\nThe following example demonstrates how an acoustic impulse response can be estimated with [correlation-based system identification techniques](correlation_functions.ipynb#System-Identification) using the soundcard of a computer. The module [`sounddevice`](http://python-sounddevice.readthedocs.org/) provides access to the soundcard via [`portaudio`](http://www.portaudio.com/).",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport scipy.signal as sig\nimport sounddevice as sd",
"_____no_output_____"
]
],
[
[
"### Generation of the Measurement Signal\n\nWe generate white noise with a uniform distribution between $\\pm 0.5$ as the excitation signal $x[k]$",
"_____no_output_____"
]
],
[
[
"fs = 44100 # sampling rate\nT = 5 # length of the measurement signal in sec\nTr = 2 # length of the expected system response in sec\n\nx = np.random.uniform(-.5, .5, size=T*fs)",
"_____no_output_____"
]
],
[
[
"### Playback of Measurement Signal and Recording of Room Response\n\nThe measurement signal $x[k]$ is played through the output of the soundcard and the response $y[k]$ is captured synchronously by the input of the soundcard. The length of the played/captured signal has to be of equal length when using the soundcard. The measurement signal $x[k]$ is zero-padded so that the captured signal $y[k]$ includes the complete system response.\n\nBe sure not to overdrive the speaker and the microphone by keeping the input level well below 0 dB.",
"_____no_output_____"
]
],
[
[
"x = np.concatenate((x, np.zeros(Tr*fs)))\ny = sd.playrec(x, fs, channels=1)\nsd.wait()\ny = np.squeeze(y)\n\nprint('Playback level: ', 20*np.log10(max(x)), ' dB')\nprint('Input level: ', 20*np.log10(max(y)), ' dB')",
"Playback level: -6.02060087394 dB\nInput level: -2.23183822753 dB\n"
]
],
[
[
"### Estimation of the Acoustic Impulse Response\n\nThe acoustic impulse response is estimated by cross-correlation $\\varphi_{yx}[\\kappa]$ of the output with the input signal. Since the cross-correlation function (CCF) for finite-length signals is given as $\\varphi_{yx}[\\kappa] = \\frac{1}{K} \\cdot y[\\kappa] * x[-\\kappa]$, the computation of the CCF can be speeded up with the fast convolution method.",
"_____no_output_____"
]
],
[
[
"h = 1/len(y) * sig.fftconvolve(y, x[::-1], mode='full')\nh = h[fs*(T+Tr):fs*(T+2*Tr)]",
"_____no_output_____"
],
[
"plt.figure(figsize=(10, 5))\nt = 1/fs * np.arange(len(h))\nplt.plot(t, h)\nplt.axis([0.0, 1.0, -1.1*np.max(np.abs(h)), 1.1*np.max(np.abs(h))])\nplt.xlabel(r'$t$ in s')\nplt.ylabel(r'$\\hat{h}[k]$')",
"_____no_output_____"
]
],
[
[
"**Copyright**\n\nThis notebook is provided as [Open Educational Resource](https://en.wikipedia.org/wiki/Open_educational_resources). Feel free to use the notebook for your own purposes. The text is licensed under [Creative Commons Attribution 4.0](https://creativecommons.org/licenses/by/4.0/), the code of the IPython examples under the [MIT license](https://opensource.org/licenses/MIT). Please attribute the work as follows: *Sascha Spors, Digital Signal Processing - Lecture notes featuring computational examples*.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
d0e1e96500298cd5bd4c4ff478e148db9e682ea0 | 322,892 | ipynb | Jupyter Notebook | scripts/PrepareMyfoodRepo-Data.ipynb | salathegroup/myfoodrepo-mask-rcnn | 200ca9f24613c80463815efab270b2257ff3537f | [
"MIT"
] | null | null | null | scripts/PrepareMyfoodRepo-Data.ipynb | salathegroup/myfoodrepo-mask-rcnn | 200ca9f24613c80463815efab270b2257ff3537f | [
"MIT"
] | null | null | null | scripts/PrepareMyfoodRepo-Data.ipynb | salathegroup/myfoodrepo-mask-rcnn | 200ca9f24613c80463815efab270b2257ff3537f | [
"MIT"
] | null | null | null | 987.437309 | 313,120 | 0.942098 | [
[
[
"import json\nimport os\nimport re\nimport tqdm\nimport random\nfrom PIL import Image\n\n%matplotlib inline\nfrom pycocotools.coco import COCO\nfrom pycocotools import mask as cocomask\nimport numpy as np\nimport skimage.io as io\nimport matplotlib.pyplot as plt\nimport pylab\nimport random\nimport os\npylab.rcParams['figure.figsize'] = (8.0, 10.0)",
"_____no_output_____"
],
[
"IMAGES_FOLDER_PATH=\"/mount/SDB/myfoodrepo-seth/myfoodrepo-images/images\"\nANNOTATIONS_PATH=\"/mount/SDB/myfoodrepo-seth/prepared_annotations/coco_annotations_assignments_2018_08_02_14-29-36.json\"\n\nOUTPUT_DIRECCTORY = \"/mount/SDE/myfoodrepo/mohanty/myfoodrepo-mask-rcnn/data\"\n\nTRAIN_PERCENT = 0.8\nVAL_PERCENT = 0.1\nTEST_PERCENT = 1 - (TRAIN_PERCENT + VAL_PERCENT)",
"_____no_output_____"
],
[
"# Build index of image_id to filepath\nimage_path_map = {}\nrootDir = IMAGES_FOLDER_PATH\nfor dirName, subdirList, fileList in tqdm.tqdm(os.walk(rootDir, topdown=False)):\n for fname in fileList:\n if len(re.findall(\"^\\d+.jpg$\", fname))>0:\n #print('.', end='', flush=True)\n image_path_map[fname.replace(\".jpg\", \"\")] = os.path.join(dirName, fname)\n ",
"595it [00:00, 786.92it/s]\n"
],
[
"annotations = json.loads(open(ANNOTATIONS_PATH).read())",
"_____no_output_____"
],
[
"# Index annotations by image_id\nannotations_by_image_id = {}\nfor item in annotations:\n try:\n annotations_by_image_id[item[\"image_id\"]].append(item)\n except:\n annotations_by_image_id[item[\"image_id\"]] = [item]",
"_____no_output_____"
],
[
"images_in_annotations = list(annotations_by_image_id.keys())\nrandom.shuffle(images_in_annotations)\n\nTRAIN_IDX = int(len(images_in_annotations) * TRAIN_PERCENT)\nVAL_IDX = TRAIN_IDX + int(len(images_in_annotations) * VAL_PERCENT)\nTEST_IDX = len(images_in_annotations)\n\nTRAIN_IMAGES = images_in_annotations[0:TRAIN_IDX]\nVAL_IMAGES = images_in_annotations[TRAIN_IDX:VAL_IDX]\nTEST_IMAGES = images_in_annotations[VAL_IDX:TEST_IDX]",
"_____no_output_____"
],
[
"def generate_image_object(_image, IMAGES_DIR):\n image_path = image_path_map[_image]\n target_filename = \"{}.jpg\".format(_image.zfill(12))\n _image_object = {}\n _image_object[\"id\"] = int(_image)\n _image_object[\"file_name\"] = target_filename\n im = Image.open(image_path)\n width, height = im.size\n _image_object[\"width\"] = width\n _image_object[\"height\"] = height\n im.save(os.path.join(IMAGES_DIR, target_filename))\n im.close()\n return _image_object\n \ndef generate_annotations_file(IMAGES, OUTPUT_DIRECTORY):\n d = {}\n d[\"info\"] = {'contributor': 'spMohanty', 'about': 'My Food Repo dataset', 'date_created': '04/08/2018', 'description': 'Annotations on myfoodrepo dataset', 'version': '1.0', 'year': 2018}\n d[\"categories\"] = [{'id': 100, 'name': 'food', 'supercategory': 'food'}]\n d[\"images\"] = []\n d[\"annotations\"] = []\n \n IMAGES_DIR = os.path.join(OUTPUT_DIRECTORY, \"images\")\n if not os.path.exists(IMAGES_DIR):\n os.makedirs(IMAGES_DIR)\n \n annotation_count = 0\n for _image in tqdm.tqdm(IMAGES):\n try:\n image_path = image_path_map[_image]\n except:\n print(\"Errorr Processing : \", _image)\n continue\n \n _image_object = generate_image_object(_image, IMAGES_DIR)\n assert _image_object != None\n d[\"images\"].append(_image_object)\n for _annotation in annotations_by_image_id[_image]:\n _annotation[\"id\"] = annotation_count\n annotation_count += 1\n _annotation[\"image_id\"] = int(_image)\n d[\"annotations\"].append(_annotation)\n fp = open(os.path.join(OUTPUT_DIRECTORY, \"annotations.json\"), \"w\")\n fp.write(json.dumps(d))\n fp.close()\n print(len(d[\"images\"]))\n print(d[\"images\"][0])\n\n \ngenerate_annotations_file(TRAIN_IMAGES, os.path.join(OUTPUT_DIRECCTORY, \"myfoodrepo-train\"))\ngenerate_annotations_file(VAL_IMAGES, os.path.join(OUTPUT_DIRECCTORY, \"myfoodrepo-val\"))\ngenerate_annotations_file(TEST_IMAGES, os.path.join(OUTPUT_DIRECCTORY, \"myfoodrepo-test\"))",
" 86%|████████▋ | 7935/9176 [03:47<00:35, 34.94it/s]"
],
[
"\"\"\"\nTest if the annotations work properly\n!pip install git+https://github.com/crowdai/coco.git#subdirectory=PythonAPI\n\"\"\"\n\ndef test_annotations(subdir_name):\n IMAGES_DIRECTORY = \"../data/{subdir_name}/images\".format(subdir_name=subdir_name)\n ANNOTATIONS_PATH = \"../data/{subdir_name}/annotations.json\".format(subdir_name=subdir_name)\n \n coco = COCO(ANNOTATIONS_PATH)\n category_ids = coco.loadCats(coco.getCatIds())\n print(category_ids)\n image_ids = coco.getImgIds(catIds=coco.getCatIds())\n random_image_id = random.choice(image_ids)\n img = coco.loadImgs(random_image_id)[0]\n print(img)\n image_path = os.path.join(IMAGES_DIRECTORY, img[\"file_name\"])\n I = io.imread(image_path)\n \n annotation_ids = coco.getAnnIds(imgIds=img['id'])\n annotations = coco.loadAnns(annotation_ids)\n plt.imshow(I); plt.axis('off')\n coco.showAnns(annotations)\n\n#test_annotations(\"myfoodrepo-train\")\n#test_annotations(\"myfoodrepo-val\")\ntest_annotations(\"myfoodrepo-test\")",
"loading annotations into memory...\nDone (t=0.07s)\ncreating index...\nindex created!\n[{'id': 100, 'name': 'food', 'supercategory': 'food'}]\n{'id': 1252372, 'file_name': '000001252372.jpg', 'width': 512, 'height': 384}\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e1fbc5711c19618521eca11a405e65ede05dac | 183,420 | ipynb | Jupyter Notebook | Lab 01.ipynb | shipstone98/AdvCompLabs | 1bbe0997076b61618117ec430cf0bde8ab228d35 | [
"MIT"
] | null | null | null | Lab 01.ipynb | shipstone98/AdvCompLabs | 1bbe0997076b61618117ec430cf0bde8ab228d35 | [
"MIT"
] | null | null | null | Lab 01.ipynb | shipstone98/AdvCompLabs | 1bbe0997076b61618117ec430cf0bde8ab228d35 | [
"MIT"
] | null | null | null | 229.561952 | 39,976 | 0.714235 | [
[
[
"# Lab 01 - Modelling and Systems Dynamics",
"_____no_output_____"
]
],
[
[
"import math, pandas\nfrom matplotlib import pyplot",
"_____no_output_____"
]
],
[
[
"## Section 01 - Starting Jupyter Notebook",
"_____no_output_____"
]
],
[
[
"print(math.sin(4))",
"-0.7568024953079282\n"
]
],
[
[
"## Section 02 - Number and Arithmetic",
"_____no_output_____"
]
],
[
[
"print(5 / 8)\nprint(5 / 8.0) # or print(5 / float(8))\nprint(5 * 8)\nprint((1 / 2.0) ** 2)",
"0.625\n0.625\n40\n0.25\n"
]
],
[
[
"### Review Question",
"_____no_output_____"
]
],
[
[
"print(math.exp(10))",
"22026.465794806718\n"
]
],
[
[
"## Section 03 - Variables and Assignments",
"_____no_output_____"
]
],
[
[
"ans = 5 * math.pi\nans += 1\nprint(ans + 2)",
"18.707963267948966\n"
]
],
[
[
"### Review Question",
"_____no_output_____"
]
],
[
[
"vel = 5\nprint((7 + (vel / 3)) ** 2)",
"75.1111111111111\n"
]
],
[
[
"## Section 04 - User Defined Functions",
"_____no_output_____"
]
],
[
[
"def f(x):\n return x ** 2 + 1\n\nprint(f(4))\nprint(f(2.5))\n\ndef sum_squares(x, y):\n return x ** 2 + y ** 2\n\nprint(sum_squares(3, 4))",
"17\n7.25\n25\n"
]
],
[
[
"### Review Question\n#### Part A",
"_____no_output_____"
]
],
[
[
"def p(t):\n return 100 * math.exp(0.1 * t)\n\nprint(p(12))",
"332.01169227365483\n"
]
],
[
[
"#### Part B",
"_____no_output_____"
]
],
[
[
"def pop(init_pop, r, t):\n return init_pop * math.exp(r * t)\n\nprint(pop(1000, 0.15, 12))",
"6049.647464412945\n"
]
],
[
[
"## Section 05 - Printing",
"_____no_output_____"
]
],
[
[
"x = 3.0\nprint(\"x =\", x, \"and its square is\", x ** 2)",
"x = 3.0 and its square is 9.0\n"
]
],
[
[
"## Section 06 - Loops and Lists",
"_____no_output_____"
]
],
[
[
"list_1 = [1, 3, 5.0]\nlist_2 = [1, \"one\", 2, \"two\"]\nlist_3 = []\nprint(list_1, list_2, list_3)\n\nlist_4 = list(range(15))\nlist_5 = list(range(5, 15))\nlist_6 = list(range(4, 15, 3))\nprint(list_4, list_5, list_6)",
"[1, 3, 5.0] [1, 'one', 2, 'two'] []\n[0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14] [5, 6, 7, 8, 9, 10, 11, 12, 13, 14] [4, 7, 10, 13]\n"
]
],
[
[
"### Review Question 01",
"_____no_output_____"
]
],
[
[
"print(list(range(3, 16, 4)))\n\ndist = 0\n\nfor i in range(7):\n dist += 0.25\n print(dist)\n\nfor i in range(10):\n print(i, \" \", i ** 2)",
"[3, 7, 11, 15]\n0.25\n0.5\n0.75\n1.0\n1.25\n1.5\n1.75\n0 0\n1 1\n2 4\n3 9\n4 16\n5 25\n6 36\n7 49\n8 64\n9 81\n"
]
],
[
[
"### Review Question 02",
"_____no_output_____"
]
],
[
[
"dist = 1\n\nfor i in range(20):\n dist *= 2\n print(dist)",
"2\n4\n8\n16\n32\n64\n128\n256\n512\n1024\n2048\n4096\n8192\n16384\n32768\n65536\n131072\n262144\n524288\n1048576\n"
]
],
[
[
"## Section 07 - Supplementary Python Programming Exercises\n### Exercise 01",
"_____no_output_____"
]
],
[
[
"def sim(initial_pop, growth_rate_unit, sim_len, time_step):\n pops = []\n times = []\n pop = initial_pop\n time = 0\n pops.append(pop)\n times.append(time)\n GROWTH_RATE = growth_rate_unit * time_step\n\n while (time <= sim_len):\n time += time_step\n pop += pop * GROWTH_RATE\n pops.append(pop)\n times.append(time)\n \n ACTUAL_POP = initial_pop * ((1 + growth_rate_unit) ** sim_len)\n END_POP = pops[len(pops) - 1]\n RELATIVE_ERR = abs((ACTUAL_POP - END_POP) / END_POP)\n return pops, times, ACTUAL_POP, RELATIVE_ERR",
"_____no_output_____"
]
],
[
[
"#### Part A",
"_____no_output_____"
]
],
[
[
"INITAL_POP = 100\nGROWTH_RATE_PER_HOUR = 0.1\nSIM_LEN = 200\npops_0_0001, times_0_0001, actual_pop, relative_err = sim(INITAL_POP, GROWTH_RATE_PER_HOUR, SIM_LEN, 0.0001)\npops_0_025, times_0_025, actual_pop, relative_err = sim(INITAL_POP, GROWTH_RATE_PER_HOUR, SIM_LEN, 0.025)\npops_0_5, times_0_5, actual_pop, relative_err = sim(INITAL_POP, GROWTH_RATE_PER_HOUR, SIM_LEN, 0.5)\nprint(\"The number of bacteria at the end of one week =\", pops_0_5[168 * 2])\nINITAL_POP_DOUBLE = INITAL_POP * 2\nindex = -1",
"The number of bacteria at the end of one week = 1317056747.0776892\n"
]
],
[
[
"#### Part B",
"_____no_output_____"
]
],
[
[
"for i in range(len(pops_0_0001)):\n if pops_0_0001[i] < INITAL_POP_DOUBLE:\n continue\n\n index = i\n break\n\nif index != -1:\n print(\"The time taken for the population to double lies between\", round(times_0_0001[index - 1], 4), \"hrs and\", round(times_0_0001[index], 4), \"hrs\")",
"The time taken for the population to double lies between 6.9315 hrs and 6.9316 hrs\n"
]
],
[
[
"#### Part C",
"_____no_output_____"
]
],
[
[
"pyplot.title(\"Population growth simulation\")\npyplot.ylabel(\"Population\")\npyplot.xlabel(\"Time (hrs)\")\npyplot.plot(times_0_0001, pops_0_0001, label=\"0.0001hrs\")\npyplot.plot(times_0_025, pops_0_025, label=\"0.025hrs\")\npyplot.plot(times_0_5, pops_0_5, label=\"0.5hrs\")\npyplot.legend()\npyplot.show()",
"_____no_output_____"
]
],
[
[
"### Exercise 02\n#### Part A",
"_____no_output_____"
]
],
[
[
"INITAL_POP = 15000\nINITAL_POP_DOUBLE = INITAL_POP * 2\nGROWTH_RATE_PER_YEAR = 0.02\nTIME_STEP = 0.25\nSIM_LEN = 20\nACTUAL_POP_20_YRS = INITAL_POP * (GROWTH_RATE_PER_YEAR ** SIM_LEN)\n\npops, times, actual_pop, relative_err = sim(INITAL_POP, GROWTH_RATE_PER_YEAR, SIM_LEN, TIME_STEP)\nprint(\"Actual population after\", SIM_LEN, \"yrs is\", actual_pop)",
"Actual population after 20 yrs is 22289.210939675322\n"
]
],
[
[
"#### Part B",
"_____no_output_____"
]
],
[
[
"pyplot.title(\"Animal population growth simulation\")\npyplot.ylabel(\"Population\")\npyplot.xlabel(\"Time (yrs)\")\npyplot.plot(times, pops)\npyplot.show()",
"_____no_output_____"
]
],
[
[
"#### Part C",
"_____no_output_____"
]
],
[
[
"TIME_PAD = 10\nPOP_PAD = 20\nprint(\"Time (yrs)\".ljust(TIME_PAD), \"Population\".ljust(POP_PAD), \"Growth\")\nprint(\"0\".ljust(TIME_PAD), str(INITAL_POP).ljust(POP_PAD), \"INTERVAL\")\n\nfor i in range(1, len(pops)):\n GROWTH = str(100 * round((pops[i] - pops[i - 1]) / pops[i - 1], 8)) + \"%\"\n print(str(times[i]).ljust(TIME_PAD), str(pops[i]).ljust(POP_PAD), GROWTH)",
"Time (yrs) Population Growth\n0 15000 INTERVAL\n0.25 15075.0 0.5%\n0.5 15150.375 0.5%\n0.75 15226.126875 0.5%\n1.0 15302.257509375 0.5%\n1.25 15378.768796921875 0.5%\n1.5 15455.662640906485 0.5%\n1.75 15532.940954111018 0.5%\n2.0 15610.605658881574 0.5%\n2.25 15688.658687175981 0.5%\n2.5 15767.10198061186 0.5%\n2.75 15845.93749051492 0.5%\n3.0 15925.167177967494 0.5%\n3.25 16004.793013857332 0.5%\n3.5 16084.81697892662 0.5%\n3.75 16165.241063821253 0.5%\n4.0 16246.06726914036 0.5%\n4.25 16327.297605486061 0.5%\n4.5 16408.93409351349 0.5%\n4.75 16490.978763981057 0.5%\n5.0 16573.43365780096 0.5%\n5.25 16656.300826089966 0.5%\n5.5 16739.582330220415 0.5%\n5.75 16823.28024187152 0.5%\n6.0 16907.396643080876 0.5%\n6.25 16991.93362629628 0.5%\n6.5 17076.89329442776 0.5%\n6.75 17162.2777608999 0.5%\n7.0 17248.0891497044 0.5%\n7.25 17334.32959545292 0.5%\n7.5 17421.001243430186 0.5%\n7.75 17508.10624964734 0.5%\n8.0 17595.646780895575 0.5%\n8.25 17683.625014800054 0.5%\n8.5 17772.043139874055 0.5%\n8.75 17860.903355573424 0.5%\n9.0 17950.20787235129 0.5%\n9.25 18039.958911713045 0.5%\n9.5 18130.15870627161 0.5%\n9.75 18220.809499802966 0.5%\n10.0 18311.91354730198 0.5%\n10.25 18403.47311503849 0.5%\n10.5 18495.490480613684 0.5%\n10.75 18587.967933016753 0.5%\n11.0 18680.907772681836 0.5%\n11.25 18774.312311545244 0.5%\n11.5 18868.18387310297 0.5%\n11.75 18962.524792468485 0.5%\n12.0 19057.337416430826 0.5%\n12.25 19152.62410351298 0.5%\n12.5 19248.387224030543 0.5%\n12.75 19344.629160150696 0.5%\n13.0 19441.35230595145 0.5%\n13.25 19538.559067481205 0.5%\n13.5 19636.25186281861 0.5%\n13.75 19734.433122132705 0.5%\n14.0 19833.10528774337 0.5%\n14.25 19932.270814182084 0.5%\n14.5 20031.932168252995 0.5%\n14.75 20132.09182909426 0.5%\n15.0 20232.75228823973 0.5%\n15.25 20333.91604968093 0.5%\n15.5 20435.585629929337 0.5%\n15.75 20537.763558078983 0.5%\n16.0 20640.45237586938 0.5%\n16.25 20743.654637748725 0.5%\n16.5 20847.37291093747 0.5%\n16.75 20951.609775492157 0.5%\n17.0 21056.36782436962 0.5%\n17.25 21161.64966349147 0.5%\n17.5 21267.457911808928 0.5%\n17.75 21373.79520136797 0.5%\n18.0 21480.664177374812 0.5%\n18.25 21588.067498261687 0.5%\n18.5 21696.007835752996 0.5%\n18.75 21804.48787493176 0.5%\n19.0 21913.51031430642 0.5%\n19.25 22023.077865877953 0.5%\n19.5 22133.19325520734 0.5%\n19.75 22243.859221483377 0.5%\n20.0 22355.078517590795 0.5%\n20.25 22466.85391017875 0.5%\n"
]
],
[
[
"#### Part D",
"_____no_output_____"
]
],
[
[
"INDEX = int(1 / TIME_STEP)\npop = pops[INDEX]\nprint(\"Population at end of 1 year:\", pop, \"growth of\", pop - INITAL_POP, \"(\" + str(100 * round((pop - INITAL_POP) / INITAL_POP, 8)) + \"%)\")\npop = pops[2 * INDEX]\nprint(\"Population at end of 2 years:\", pop, \"growth of\", pop - INITAL_POP, \"(\" + str(100 * round((pop - INITAL_POP) / INITAL_POP, 8)) + \"%)\")\npop = pops[3 * INDEX]\nprint(\"Population at end of 3 years:\", pop, \"growth of\", pop - INITAL_POP, \"(\" + str(100 * round((pop - INITAL_POP) / INITAL_POP, 8)) + \"%)\")",
"Population at end of 1 year: 15302.257509375 growth of 302.25750937500015 (2.01505%)\nPopulation at end of 2 years: 15610.605658881574 growth of 610.6056588815736 (4.070704%)\nPopulation at end of 3 years: 15925.167177967494 growth of 925.1671779674944 (6.167781%)\n"
]
],
[
[
"### Exercise 03\n#### Part A",
"_____no_output_____"
]
],
[
[
"INITIAL_SAMPLE = 20\nGROWTH_RATE = -0.000120968\nSIM_LEN = 25000\n\ndef carbon_date(sample, proportion, sim_len, time_step):\n carbons = []\n times = []\n carbon = sample\n time = 0\n carbons.append(carbon)\n times.append(time)\n\n while (time <= sim_len):\n time += time_step\n carbon = 20 * math.exp(proportion * time)\n carbons.append(carbon)\n times.append(time)\n\n return carbons, times\n\ncarbons, carbon_times = carbon_date(INITIAL_SAMPLE, GROWTH_RATE, SIM_LEN, 0.25)\npyplot.title(\"Carbon dating simulation\")\npyplot.ylabel(\"Carbon\")\npyplot.xlabel(\"Time (yrs)\")\npyplot.plot(carbon_times, carbons)\npyplot.show()",
"_____no_output_____"
]
],
[
[
"#### Part B",
"_____no_output_____"
]
],
[
[
"print(\"Estimation of carbon-14 remaining after 25,000yrs =\", str(carbons[25000 * 4]) + \"g\")",
"Estimation of carbon-14 remaining after 25,000yrs = 0.9719336613934643g\n"
]
],
[
[
"### Exercise 04\n#### Part A",
"_____no_output_____"
],
[
"dT / dt is directly proportional to T - 25. Therefore, dT / dt = k(T - 25).",
"_____no_output_____"
],
[
"#### Part B",
"_____no_output_____"
],
[
" dT / dt = k(T - 25) \n \n du / dt = ku, where u = T - 25 \n\n (1 / u) du = k dt \n\n ln(u) = kt + c \n\n u = e ^ (kt + c) \n\n u = Ae ^ kt \n \n T - 25 = Ae ^ kt \n \n T = Ae ^ kt + 25",
"_____no_output_____"
],
[
"#### Part C",
"_____no_output_____"
],
[
" T(0) = A + 25 \n\n A = -19 \n\n\n \n T(1) = -19e ^ k + 25 \n\n 20 = -19e ^ k + 25 \n\n 5 = 19e ^ k \n \n k = ln(5 / 19) ",
"_____no_output_____"
],
[
"#### Part D",
"_____no_output_____"
],
[
" T(0.25) = -19e ^ (0.25 * ln(5 / 19)) + 25 \n\n T(0.25) = 25 - 19 * (5 / 19)^0.25 \n \n T(0.25) = 11.4 ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d0e21fd51c2df5422dab158470eab6ad0afea1f0 | 16,264 | ipynb | Jupyter Notebook | vizualize_real_data.ipynb | MichaelHopwood/MLMassSpectrom | 45add1688a96b3947174754deb2b359a72460c9e | [
"MIT"
] | null | null | null | vizualize_real_data.ipynb | MichaelHopwood/MLMassSpectrom | 45add1688a96b3947174754deb2b359a72460c9e | [
"MIT"
] | null | null | null | vizualize_real_data.ipynb | MichaelHopwood/MLMassSpectrom | 45add1688a96b3947174754deb2b359a72460c9e | [
"MIT"
] | null | null | null | 30.571429 | 153 | 0.426955 | [
[
[
"# Visualize real data\n\nGet to know the real data",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport os\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns",
"E:\\Users\\MichaelHopwood\\miniconda3\\lib\\site-packages\\numpy\\_distributor_init.py:30: UserWarning: loaded more than 1 DLL from .libs:\nE:\\Users\\MichaelHopwood\\miniconda3\\lib\\site-packages\\numpy\\.libs\\libopenblas.EL2C6PLE4ZYW3ECEVIV3OXXGRN2NRFM2.gfortran-win_amd64.dll\nE:\\Users\\MichaelHopwood\\miniconda3\\lib\\site-packages\\numpy\\.libs\\libopenblas.NOIJJG62EMASZI6NYURL6JBKM4EVBGM7.gfortran-win_amd64.dll\n warnings.warn(\"loaded more than 1 DLL from .libs:\\n%s\" %\n"
],
[
"savefolder = 'figures_real_data'\nfolder = 'data'\ndf = pd.read_excel(os.path.join(folder, '20211128 - Full DART Data (Model & Test).xlsx'), header=2)\ndf.head()",
"_____no_output_____"
],
[
"import pylab\n\nlabeled_df = df[df['Sample Types'] == 'Model']\nrecol = [(float('.'.join(col.split('.')[:2])) if isinstance(col, str) else col) for col in df.columns[4:]]\n\nunique_classes = labeled_df['Class'].unique()\n\nNUM_COLORS = len(unique_classes)\ncm = pylab.get_cmap('gist_rainbow')\ncgen = (cm(1.*i/NUM_COLORS) for i in range(NUM_COLORS))\n\nfor label in unique_classes:\n plt.figure(figsize=(12,8))\n mask = labeled_df['Class'] == label\n color = next(cgen)\n for ind, row in labeled_df.loc[mask, :].iterrows():\n data = row[df.columns[4:]].values\n data[data<25] = 0\n plt.plot(recol, data, color=color)\n plt.ylabel('%')\n plt.xlabel('Mass')\n plt.savefig(os.path.join(savefolder, f'sample_class_{label}_ind_{ind}.png'))\n #plt.show()\n plt.close()",
"_____no_output_____"
],
[
"# Rename numerical columns\n\nrecol = [(float('.'.join(col.split('.')[:2])) if isinstance(col, str) else col) for col in df.columns[4:]]\nplt.plot(recol)",
"_____no_output_____"
],
[
"recol",
"_____no_output_____"
]
],
[
[
"It appears there are more than 3 measurement iterations? Why?",
"_____no_output_____"
]
],
[
[
"Xcols = [f'Col{i}' for i in range(len(recol))]\ndf.columns = list(df.columns[:4]) + Xcols\ndf.head(n=10)",
"_____no_output_____"
],
[
"df[df.columns[4:]].iloc[0:3]",
"_____no_output_____"
]
],
[
[
"## Data formatting",
"_____no_output_____"
]
],
[
[
"def build_data_structure(df, labels, column_name_mass_locations):\n \"\"\"\n Convert dataframe which has columns of the mass locations and values of the intensities\n to array format where the sample is a list of peaks and the samples are grouped by the number of peaks.\n\n Input:\n df: dataframe with columns of the mass locations and values of the intensities\n \tCol0 Col1 Col2 ...\n 0\t0.0\t 0.0\t0.0\n\n column_name_mass_locations: column names of the mass locations\n [59.00498,\n 72.00792,\n 74.00967,\n ...\n ]\n\n We split it up just in case duplicate mass locations are present since multiple scans can be represented.\n\n Output:\n [ [samples with smallest number of peaks],\n [samples with 2nd smallest number peaks], \n ...,\n [samples with largest number of peaks]\n ]\n\n where each sample is an array of lists (mass, intensity):\n array([[peak_location1, peak_intensity1], [peak_location2, peak_intensity2], ...])\n \"\"\"\n df = df.copy()\n # Get nonzero values (aka \"peaks\")\n data = df.apply(lambda x: np.asarray([[column_name_mass_locations[i], val] for i, (val, b) in enumerate(zip(x, x.gt(0))) if b]), axis=1) \n\n X = []\n Y = []\n # Group so we have groups of the same number of peaks\n lengths = np.array([len(x) for x in data])\n unique_lengths = np.unique(lengths)\n for length in unique_lengths:\n mask_length = lengths == length\n mask_idx = np.where(mask_length)\n y = labels[mask_idx]\n x = np.stack(data.loc[mask_length].values.tolist())\n X.append(x)\n Y.append(y)\n return X, Y\n\nlabeled_df = df[df['Sample Types'] == 'Model']\nfrom sklearn import preprocessing\nlb = preprocessing.LabelBinarizer()\nlb.fit(labeled_df['Class'].unique())\nX, Y = build_data_structure(labeled_df[labeled_df.columns[4:]], labeled_df['Class'].values, recol)\ny = [lb.transform(y) for y in Y]\nlen(X), X[0].shape, len(y), y[0].shape",
"_____no_output_____"
]
],
[
[
"## Investigate test-train partition",
"_____no_output_____"
]
],
[
[
"df['Sample Types'].value_counts()",
"_____no_output_____"
],
[
"df['Class'].value_counts()",
"_____no_output_____"
],
[
"df[df['Sample Types'] == 'Model']['Class'].value_counts()",
"_____no_output_____"
],
[
"df[df['Sample Types'] == 'Test']['Class'].value_counts()",
"_____no_output_____"
]
],
[
[
"## Seeing no labels in the Test split, we must split the train for a supervised evaluation",
"_____no_output_____"
]
],
[
[
"labeled_df = df[df['Sample Types'] == 'Model']\n\nX = build_data_structure(df[df.columns[4:]])\n\nfrom sklearn import model_selection\nfrom sklearn import preprocessing\nlb = preprocessing.LabelBinarizer()\nlb.fit(labeled_df['Class'])\n\ntrain_labeled_df, test_labeled_df = model_selection.train_test_split(labeled_df, train_size=None, shuffle=True, stratify=labeled_df['Class'])\n\nX_train = build_data_structure(train_labeled_df[train_labeled_df.columns[4:]])\nX_test = build_data_structure(test_labeled_df[test_labeled_df.columns[4:]])\n\ny_train = lb.transform(train_labeled_df['Class'])\ny_test = lb.transform(test_labeled_df['Class'])",
"_____no_output_____"
],
[
"print(len(X_train), len(X_test))\n\nprint(X_train[12][0].shape, y_train[0].shape, X_test[12][0].shape, y_test[0].shape)",
"_____no_output_____"
],
[
"from core import RealDataGenerator\n\ngen = RealDataGenerator(\n X_train, y_train,\n X_test, y_test,\n X_test, y_test\n)\n\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d0e228b167969a76249d56356342425b43817de3 | 203,749 | ipynb | Jupyter Notebook | Machine Learning/Day 7/.ipynb_checkpoints/K-means example-checkpoint.ipynb | Mayankjh/Python_ML_Training | 87002cf98c7dd0833d81f96a6efae48a37e04dd2 | [
"MIT"
] | 2 | 2018-07-28T17:40:57.000Z | 2018-08-28T17:17:33.000Z | Machine Learning/Day 7/.ipynb_checkpoints/K-means example-checkpoint.ipynb | Mayankjh/Python_ML_Training | 87002cf98c7dd0833d81f96a6efae48a37e04dd2 | [
"MIT"
] | null | null | null | Machine Learning/Day 7/.ipynb_checkpoints/K-means example-checkpoint.ipynb | Mayankjh/Python_ML_Training | 87002cf98c7dd0833d81f96a6efae48a37e04dd2 | [
"MIT"
] | null | null | null | 749.077206 | 101,690 | 0.939774 | [
[
[
"%matplotlib inline\nfrom copy import deepcopy\nimport numpy as np\nimport pandas as pd\nfrom matplotlib import pyplot as plt\nplt.rcParams['figure.figsize'] = (16,9)\nplt.style.use('ggplot')",
"_____no_output_____"
],
[
"# importing dataset\ndata = pd.read_csv('xclara.csv')\nprint(data.shape)\ndata.head()",
"(3000, 2)\n"
],
[
"# getting the values nad plotiing it\nf1 = data['V1'].values\nf2 = data['V2'].values\n# print(zip(f1,f2))\nX = np.array(list(zip(f1,f2)))\nplt.scatter(f1,f2,c='purple',s=7)",
"_____no_output_____"
],
[
"# Ecuclidian Distance calculator\ndef dist(a,b,ax=1):\n return np.linalg.norm(a-b,axis=ax)",
"_____no_output_____"
],
[
"# number of clusters\nk=3\n# x cordinates of random centeroids\nC_x = np.random.randint(0,np.max(X)-20,size=k)\n# Y cordinates of random centeroids\nC_y = np.random.randint(0,np.max(X)-20,size=k)\nC = np.array(list(zip(C_x,C_y)),dtype=np.float32)\nprint(C)\n",
"[[ 54. 29.]\n [ 71. 58.]\n [ 81. 63.]]\n"
],
[
"plt.scatter(f1,f2,c='#595959',s=7)\nplt.scatter(C_x,C_y,marker='*',s=200,c='g')",
"_____no_output_____"
],
[
"# To store the values of centeriod when it updates\nC_old = np.zeros(C.shape)\n# cluster labels(0,1,2)\nclusters = np.zeros(len(X))\n# error func, - Distance between new centeroids and old centeroids\nerror = dist(C,C_old,None)\n# Loop will run till the error becomes zero\nfor i in range(len(X)):\n distances = dist(X[i],C)\n cluster = np.argmin(distances)\n cluster[i] = cluster\n# storing the old centeroid values\nC_old = deepcopy(C)\n# Finding the new centeroids by taking the average values\nfor i in range(1):\n points = [X[j] for j in range(len(X)) if clusters[j] ==i]\n C[i] = np.mean(points,axis=0)\nerror = dist(C,C_old,None)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e22bd8d31176ee34a58cace72dfa6d48cf8ee5 | 1,790 | ipynb | Jupyter Notebook | 2020-02-practioneer/scripts/03/.ipynb_checkpoints/scraper-checkpoint.ipynb | denistsoi/python-code-for-hk | ebc2d072e6f55f00bbe98f334a4eb97aa54334fa | [
"BSD-Source-Code"
] | null | null | null | 2020-02-practioneer/scripts/03/.ipynb_checkpoints/scraper-checkpoint.ipynb | denistsoi/python-code-for-hk | ebc2d072e6f55f00bbe98f334a4eb97aa54334fa | [
"BSD-Source-Code"
] | 2 | 2020-03-31T11:20:18.000Z | 2021-02-02T22:31:54.000Z | 2020-02-practioneer/scripts/03/.ipynb_checkpoints/scraper-checkpoint.ipynb | denistsoi/python-code-for-hk | ebc2d072e6f55f00bbe98f334a4eb97aa54334fa | [
"BSD-Source-Code"
] | null | null | null | 33.148148 | 556 | 0.59162 | [
[
[
"from selenium import webdriver\ndriver = webdriver.Chrome()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code"
]
] |
d0e23795e7ededa18c0c0470d44ac21135aedb69 | 167,995 | ipynb | Jupyter Notebook | Ahmed_Notebook.ipynb | ameliadogan/Philly-Police-Data | 24608c39651830928b1f0019c4afad82c2cc8884 | [
"MIT"
] | null | null | null | Ahmed_Notebook.ipynb | ameliadogan/Philly-Police-Data | 24608c39651830928b1f0019c4afad82c2cc8884 | [
"MIT"
] | null | null | null | Ahmed_Notebook.ipynb | ameliadogan/Philly-Police-Data | 24608c39651830928b1f0019c4afad82c2cc8884 | [
"MIT"
] | null | null | null | 71.124047 | 71,056 | 0.661288 | [
[
[
"import pandas as pd\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"ppd_df = pd.read_csv(\"policedata/ppd_complaints.csv\")",
"_____no_output_____"
],
[
"display(ppd_df.head())",
"_____no_output_____"
],
[
"# Add date of complaint\ndate_pattern = r\"(\\d{1,2}-\\d{1,2}-\\d{2,4})\" #parentheses is to treat all characters as a \"group\", making extract work\ncomplaint_str_series = ppd_df['summary'].str.extract(date_pattern, expand=False) #expand=False to get series\ncomplaint_str_series.replace(\"2-29-18\",\"03-01-18\",inplace=True) #replace faulty leap year day\nppd_df['date_of_complaint'] = pd.to_datetime(complaint_str_series)",
"_____no_output_____"
],
[
"# Basic information and visualizations\nprint(\"Number of complaints: {}\\n\".format(ppd_df.shape[0]))\n\nprint(\"Complaints over time\")\n# optionally, exclude the first few years\ncomplaints_over_time_series = ppd_df[ppd_df['date_of_complaint']>\"01-01-2014\"].groupby('date_of_complaint').size()\n#complaints_over_time_series = ppd_df.groupby('date_of_complaint').size().asfreq(freq=\"d\",fill_value=0)\ndisplay(complaints_over_time_series.groupby(pd.Grouper(freq=\"M\")).sum().plot(figsize=(20,10)))\nplt.show()\n\nprint(\"Complaints by district\")\ndisplay(ppd_df['district_occurrence'].value_counts())\n\nprint(\"Complaints by classification\")\nprint(ppd_df['general_cap_classification'].value_counts())",
"Number of complaints: 3317\n\nComplaints over time\n"
],
[
"disciplines_df = pd.read_csv(\"policedata/ppd_complaint_disciplines.csv\")",
"_____no_output_____"
],
[
"disciplines_df.head()",
"_____no_output_____"
],
[
"# Basic counts\nfor col in ['po_race','po_sex','allegations_investigated','investigative_findings','disciplinary_findings']:\n print(col)\n display(disciplines_df[col].value_counts())",
"po_race\n"
],
[
"# Merge the datasets, keeping rows with a complaint id a \ncomplaint_key = ['complaint_id','officer_id','allegations_investigated']\nmerged_df = ppd_df.merge(disciplines_df,on='complaint_id',how='inner').drop_duplicates()",
"_____no_output_____"
],
[
"# Add column for # of rws that appeared for each complaint key\nnum_complaints_df = disciplines_df.drop_duplicates()\\\n .groupby(complaint_key).size()\nprint(\"Number of unique allegations made (specific complaint, officer and allegation): \",str(num_complaints_df.shape[0]))\nmultiple_complaints_df = num_complaints_df.rename(\"num_complaint_rows\")\n\n# Make dataframe which will contain final investigative outcome for each complaint key\noutcome_df = merged_df.merge(multiple_complaints_df, \n how='inner',\n left_on=complaint_key,\n right_index=True)\nassert(all(~outcome_df.duplicated()))\n# display(outcome_df)\n\n# Find all complaints where at least one row had a Sustained Finding, and store those keys into a set\nsustained_findings_df = outcome_df[outcome_df['investigative_findings']=='Sustained Finding']\nprint('Number of unique allegations made with investigative outcome \"Sustained Findings\":',\n str(sustained_findings_df.shape[0]))\n#display(sustained_findings_df)\nassert(all(~sustained_findings_df.duplicated()))\nassert(all(~sustained_findings_df[complaint_key].duplicated()))\nsustained_findings_keys = {\",\".join(str(x) for x in arr) for arr in sustained_findings_df[complaint_key].values}\n\n# Find all complaints where at least one row had a Guilty Finding disciplinary finding \nguilty_df = outcome_df[outcome_df['disciplinary_findings'] == 'Guilty Finding']\nprint('Number of unique allegations made with disciplinary outcome \"Guilty Finding\":',\n str(guilty_df.shape[0]))\nguilty_finding_keys = {\",\".join(str(x) for x in arr) for arr in guilty_df[complaint_key].values}\n\n# Find all complaints where at least one row was Guilty Finding or Training/Counseling\nsome_punishment_df = outcome_df[(outcome_df['disciplinary_findings'] == 'Guilty Finding') | \n (outcome_df['disciplinary_findings'] == 'Training/Counseling')]\nprint('Number of unique allegations made with disciplinary outcome \"Guilty Finding\" or \"Training/Counseling\":',\n str(some_punishment_df.shape[0]))\nsome_punishment_keys = {\",\".join(str(x) for x in arr) for arr in some_punishment_df[complaint_key].values}\n\n# Remove duplicate rows for each complaint in the outcome dataframe, then add column with \n# True if the investigation had sustained findings\noutcome_df = outcome_df.drop_duplicates(subset=complaint_key)\\\n .drop(['investigative_findings','disciplinary_findings'],axis=1)\nkey_column = [\",\".join(str(x) for x in arr) for arr in outcome_df[complaint_key].values]\noutcome_df['complaint_key'] = key_column\noutcome_df['sustained_investigative_finding'] = outcome_df['complaint_key'].isin(sustained_findings_keys)\noutcome_df['guilty_disciplinary_finding'] = outcome_df['complaint_key'].isin(guilty_finding_keys)\noutcome_df['training_counseling_or_guilty_disciplinary_finding'] = outcome_df['complaint_key'].isin(some_punishment_keys)\ndisplay(outcome_df)",
"Number of unique allegations made (specific complaint, officer and allegation): 6701\nNumber of unique allegations made with investigative outcome \"Sustained Findings\": 1088\nNumber of unique allegations made with disciplinary outcome \"Guilty Finding\": 123\nNumber of unique allegations made with disciplinary outcome \"Guilty Finding\" or \"Training/Counseling\": 835\n"
],
[
"# Add complainant demographics columns\ncomplainant_demographics_df = pd.read_csv('policedata/ppd_complainant_demographics.csv').set_index('complaint_id')\ndisplay(complainant_demographics_df.head())\noutcome_df = outcome_df.merge(complainant_demographics_df, left_on='complaint_id',right_index=True)",
"_____no_output_____"
],
[
"outcome_df",
"_____no_output_____"
],
[
"outcome_df.to_csv(\"outputdata/outcome_df.csv\")",
"_____no_output_____"
],
[
"#outcome_to_test = \"guilty_disciplinary_finding\"\noutcome_to_test = 'training_counseling_or_guilty_disciplinary_finding'\nprint('Outcome to test: {}'.format(outcome_to_test))\nprint(\"Total number of complaints: {0}\\nTotal number of successful complaints: {1}\\nPercent of complaints which succeeded: {2:.2f}% \"\\\n .format(outcome_df.shape[0],outcome_df[outcome_to_test].sum(),\n 100*outcome_df[outcome_to_test].sum()/outcome_df.shape[0]))\nfor column in ['complainant_sex','complainant_race','po_race','po_sex','allegations_investigated']:\n print(\"Fraction of complaints with true outcome by {}\".format(column))\n print(outcome_df[outcome_df[outcome_to_test]==True][column].value_counts(normalize=True))\n print(\"Fraction of complaints with false outcome by {}\".format(column))\n print(outcome_df[outcome_df[outcome_to_test]==False][column].value_counts(normalize=True))\n ",
"Outcome to test: training_counseling_or_guilty_disciplinary_finding\nTotal number of complaints: 7052\nTotal number of successful complaints: 884\nPercent of complaints which succeeded: 12.54% \nFraction of complaints with true outcome by complainant_sex\nmale 0.570755\nfemale 0.429245\nName: complainant_sex, dtype: float64\nFraction of complaints with false outcome by complainant_sex\nmale 0.562449\nfemale 0.437551\nName: complainant_sex, dtype: float64\nFraction of complaints with true outcome by complainant_race\nblack 0.659060\nwhite 0.214765\nlatino 0.103356\nasian 0.009396\nother 0.006711\nmulti ethnic 0.006711\nName: complainant_race, dtype: float64\nFraction of complaints with false outcome by complainant_race\nblack 0.682210\nwhite 0.199927\nlatino 0.087571\nasian 0.018542\nother 0.006793\nindian 0.003121\nmulti ethnic 0.001285\nmiddle east 0.000551\nName: complainant_race, dtype: float64\nFraction of complaints with true outcome by po_race\nwhite 0.565611\nblack 0.315611\nlatino 0.098416\nasian 0.018100\nother 0.002262\nName: po_race, dtype: float64\nFraction of complaints with false outcome by po_race\nwhite 0.573606\nblack 0.300584\nlatino 0.091115\nasian 0.024805\nother 0.009079\nindian 0.000811\nName: po_race, dtype: float64\nFraction of complaints with true outcome by po_sex\nmale 0.839367\nfemale 0.160633\nName: po_sex, dtype: float64\nFraction of complaints with false outcome by po_sex\nmale 0.846628\nfemale 0.153372\nName: po_sex, dtype: float64\nFraction of complaints with true outcome by allegations_investigated\nDepartmental Violation 0.726244\nLack of Service 0.177602\nVerbal Abuse 0.037330\nUnprofessional Conduct 0.022624\nPhysical Abuse 0.009050\nDisciplinary Code Violation 0.007919\nFalsification 0.006787\nCriminal Allegation 0.004525\nHarassment 0.004525\nDomestic 0.003394\nName: allegations_investigated, dtype: float64\nFraction of complaints with false outcome by allegations_investigated\nDepartmental Violation 0.222276\nLack of Service 0.200713\nPhysical Abuse 0.134890\nVerbal Abuse 0.134728\nInvestigation OnGoing 0.081226\nUnprofessional Conduct 0.077821\nHarassment 0.035830\nNo C.A.P. Investigation 0.032912\nCriminal Allegation 0.029994\nCivil Rights Complaint 0.025454\nReferred to Other Agency/C.A.P. Investigation 0.009890\nDomestic 0.005026\nOther Misconduct 0.002918\nSexual Crime/Misconduct 0.002594\nFalsification 0.002108\nDisciplinary Code Violation 0.000973\nDrugs 0.000649\nName: allegations_investigated, dtype: float64\n"
],
[
"for column in ['complainant_sex','complainant_race','po_race','po_sex','allegations_investigated']:\n print(\"% of successful complaints by {}\".format(column))\n for column_possibility in outcome_df[column].unique():\n print(\"When {} = {}\".format(column,column_possibility))\n if pd.isna(column_possibility):\n print(outcome_df[outcome_df[column].isna()][outcome_to_test].value_counts())\n print(outcome_df[outcome_df[column].isna()][outcome_to_test].value_counts(normalize=True))\n else:\n print(outcome_df[outcome_df[column]==column_possibility][outcome_to_test].value_counts())\n print(outcome_df[outcome_df[column]==column_possibility][outcome_to_test].value_counts(normalize=True))",
"% of successful complaints by complainant_sex\nWhen complainant_sex = male\nFalse 3833\nTrue 69\nName: guilty_disciplinary_finding, dtype: int64\nFalse 0.982317\nTrue 0.017683\nName: guilty_disciplinary_finding, dtype: float64\nWhen complainant_sex = female\nFalse 2964\nTrue 59\nName: guilty_disciplinary_finding, dtype: int64\nFalse 0.980483\nTrue 0.019517\nName: guilty_disciplinary_finding, dtype: float64\nWhen complainant_sex = nan\nFalse 123\nTrue 4\nName: guilty_disciplinary_finding, dtype: int64\nFalse 0.968504\nTrue 0.031496\nName: guilty_disciplinary_finding, dtype: float64\n% of successful complaints by complainant_race\nWhen complainant_race = black\nFalse 4152\nTrue 55\nName: guilty_disciplinary_finding, dtype: int64\nFalse 0.986927\nTrue 0.013073\nName: guilty_disciplinary_finding, dtype: float64\nWhen complainant_race = white\nFalse 1215\nTrue 34\nName: guilty_disciplinary_finding, dtype: int64\nFalse 0.972778\nTrue 0.027222\nName: guilty_disciplinary_finding, dtype: float64\nWhen complainant_race = nan\nFalse 836\nTrue 24\nName: guilty_disciplinary_finding, dtype: int64\nFalse 0.972093\nTrue 0.027907\nName: guilty_disciplinary_finding, dtype: float64\nWhen complainant_race = latino\nFalse 540\nTrue 14\nName: guilty_disciplinary_finding, dtype: int64\nFalse 0.974729\nTrue 0.025271\nName: guilty_disciplinary_finding, dtype: float64\nWhen complainant_race = asian\nFalse 108\nName: guilty_disciplinary_finding, dtype: int64\nFalse 1.0\nName: guilty_disciplinary_finding, dtype: float64\nWhen complainant_race = other\nFalse 42\nName: guilty_disciplinary_finding, dtype: int64\nFalse 1.0\nName: guilty_disciplinary_finding, dtype: float64\nWhen complainant_race = indian\nFalse 17\nName: guilty_disciplinary_finding, dtype: int64\nFalse 1.0\nName: guilty_disciplinary_finding, dtype: float64\nWhen complainant_race = middle east\nFalse 3\nName: guilty_disciplinary_finding, dtype: int64\nFalse 1.0\nName: guilty_disciplinary_finding, dtype: float64\nWhen complainant_race = multi ethnic\nFalse 7\nTrue 5\nName: guilty_disciplinary_finding, dtype: int64\nFalse 0.583333\nTrue 0.416667\nName: guilty_disciplinary_finding, dtype: float64\n% of successful complaints by po_race\nWhen po_race = white\nFalse 3982\nTrue 56\nName: guilty_disciplinary_finding, dtype: int64\nFalse 0.986132\nTrue 0.013868\nName: guilty_disciplinary_finding, dtype: float64\nWhen po_race = black\nFalse 2087\nTrue 46\nName: guilty_disciplinary_finding, dtype: int64\nFalse 0.978434\nTrue 0.021566\nName: guilty_disciplinary_finding, dtype: float64\nWhen po_race = latino\nFalse 623\nTrue 26\nName: guilty_disciplinary_finding, dtype: int64\nFalse 0.959938\nTrue 0.040062\nName: guilty_disciplinary_finding, dtype: float64\nWhen po_race = asian\nFalse 165\nTrue 4\nName: guilty_disciplinary_finding, dtype: int64\nFalse 0.976331\nTrue 0.023669\nName: guilty_disciplinary_finding, dtype: float64\nWhen po_race = other\nFalse 58\nName: guilty_disciplinary_finding, dtype: int64\nFalse 1.0\nName: guilty_disciplinary_finding, dtype: float64\nWhen po_race = indian\nFalse 5\nName: guilty_disciplinary_finding, dtype: int64\nFalse 1.0\nName: guilty_disciplinary_finding, dtype: float64\n% of successful complaints by po_sex\nWhen po_sex = male\nFalse 5857\nTrue 107\nName: guilty_disciplinary_finding, dtype: int64\nFalse 0.982059\nTrue 0.017941\nName: guilty_disciplinary_finding, dtype: float64\nWhen po_sex = female\nFalse 1063\nTrue 25\nName: guilty_disciplinary_finding, dtype: int64\nFalse 0.977022\nTrue 0.022978\nName: guilty_disciplinary_finding, dtype: float64\n% of successful complaints by allegations_investigated\nWhen allegations_investigated = Departmental Violation\nFalse 1942\nTrue 71\nName: guilty_disciplinary_finding, dtype: int64\nFalse 0.964729\nTrue 0.035271\nName: guilty_disciplinary_finding, dtype: float64\nWhen allegations_investigated = Verbal Abuse\nFalse 851\nTrue 13\nName: guilty_disciplinary_finding, dtype: int64\nFalse 0.984954\nTrue 0.015046\nName: guilty_disciplinary_finding, dtype: float64\nWhen allegations_investigated = Lack of Service\nFalse 1379\nTrue 16\nName: guilty_disciplinary_finding, dtype: int64\nFalse 0.98853\nTrue 0.01147\nName: guilty_disciplinary_finding, dtype: float64\nWhen allegations_investigated = Criminal Allegation\nFalse 188\nTrue 1\nName: guilty_disciplinary_finding, dtype: int64\nFalse 0.994709\nTrue 0.005291\nName: guilty_disciplinary_finding, dtype: float64\nWhen allegations_investigated = Harassment\nFalse 224\nTrue 1\nName: guilty_disciplinary_finding, dtype: int64\nFalse 0.995556\nTrue 0.004444\nName: guilty_disciplinary_finding, dtype: float64\nWhen allegations_investigated = Physical Abuse\nFalse 835\nTrue 5\nName: guilty_disciplinary_finding, dtype: int64\nFalse 0.994048\nTrue 0.005952\nName: guilty_disciplinary_finding, dtype: float64\nWhen allegations_investigated = Unprofessional Conduct\nFalse 489\nTrue 11\nName: guilty_disciplinary_finding, dtype: int64\nFalse 0.978\nTrue 0.022\nName: guilty_disciplinary_finding, dtype: float64\nWhen allegations_investigated = Sexual Crime/Misconduct\nFalse 16\nName: guilty_disciplinary_finding, dtype: int64\nFalse 1.0\nName: guilty_disciplinary_finding, dtype: float64\nWhen allegations_investigated = No C.A.P. Investigation\nFalse 203\nName: guilty_disciplinary_finding, dtype: int64\nFalse 1.0\nName: guilty_disciplinary_finding, dtype: float64\nWhen allegations_investigated = Civil Rights Complaint\nFalse 157\nName: guilty_disciplinary_finding, dtype: int64\nFalse 1.0\nName: guilty_disciplinary_finding, dtype: float64\nWhen allegations_investigated = Drugs\nFalse 4\nName: guilty_disciplinary_finding, dtype: int64\nFalse 1.0\nName: guilty_disciplinary_finding, dtype: float64\nWhen allegations_investigated = Other Misconduct\nFalse 18\nName: guilty_disciplinary_finding, dtype: int64\nFalse 1.0\nName: guilty_disciplinary_finding, dtype: float64\nWhen allegations_investigated = Domestic\nFalse 33\nTrue 1\nName: guilty_disciplinary_finding, dtype: int64\nFalse 0.970588\nTrue 0.029412\nName: guilty_disciplinary_finding, dtype: float64\nWhen allegations_investigated = Falsification\nFalse 13\nTrue 6\nName: guilty_disciplinary_finding, dtype: int64\nFalse 0.684211\nTrue 0.315789\nName: guilty_disciplinary_finding, dtype: float64\nWhen allegations_investigated = Disciplinary Code Violation\nTrue 7\nFalse 6\nName: guilty_disciplinary_finding, dtype: int64\nTrue 0.538462\nFalse 0.461538\nName: guilty_disciplinary_finding, dtype: float64\nWhen allegations_investigated = Referred to Other Agency/C.A.P. Investigation\nFalse 61\nName: guilty_disciplinary_finding, dtype: int64\nFalse 1.0\nName: guilty_disciplinary_finding, dtype: float64\nWhen allegations_investigated = Investigation OnGoing\nFalse 501\nName: guilty_disciplinary_finding, dtype: int64\nFalse 1.0\nName: guilty_disciplinary_finding, dtype: float64\n"
],
[
"outcome_df['officer_id'].value_counts()",
"_____no_output_____"
],
[
"outcome_df",
"_____no_output_____"
],
[
"print(\"\\n\".join(outcome_df[outcome_df['officer_id']==29180642.0]['summary']))\nprint(outcome_df[outcome_df['officer_id']==29180642.0]['sustained_investigative_finding'].sum())\nprint(outcome_df[outcome_df['officer_id']==29180642.0]['guilty_disciplinary_finding'].sum())",
"According to the complainant, on 4-2-16 at 12:30am, they were harassed by officers assigned to the 35th District.\nAccording to the complainant, on 4-2-16 at 12:30am, they were harassed by officers assigned to the 35th District.\nAccording to the complainant, on 4-29-16 at 7:20am, they were harassed by officers assigned to the 35th District.\nAccording to the complainant, on 6-6-16 at 1:15am, they were verbally abused and threatened by an officer assigned to the 35th District.\nAccording to the complainant, on 9-7-16 at 12:45am, they were verbally abused by officers assigned to the 35th District.\nAccording to the complainant, on 9-7-16 at 12:45am, they were verbally abused by officers assigned to the 35th District.\nAccording to the complainant, on 10-21-17 at 10:00pm, they were improperly stopped by officers assigned to the 35th District.\nAccording to the complainant, on 1-30-18 at 7:00pm, they were verbally abused by officers assigned to the 35th District.\nAccording to the complainant, on 1-30-18 at 7:00pm, they were verbally abused by officers assigned to the 35th District.\nAccording to the complainant, on 3-11-18 at 8:45pm, they were verbally abused and falsely detained by officers assigned to the 35th District.\nAccording to the complainant, on 3-11-18 at 8:45pm, they were verbally abused and falsely detained by officers assigned to the 35th District.\nAccording to the complainant, on 3-11-18 at 8:45pm, they were verbally abused and falsely detained by officers assigned to the 35th District.\nAccording to the complainant, on 3-24-18 at 9:30pm, they were physically abused by officers assigned to the 35th District.\nAccording to the complainant, on 3-24-18 at 9:30pm, they were physically abused by officers assigned to the 35th District.\nAccording to the complainant, on 7-25-18 at 6:40pm, they were physically and verbally abused by an Officer assigned to the 35th District.\nAccording to the complainant, on 7-25-18 at 6:40pm, they were physically and verbally abused by an Officer assigned to the 35th District.\nAccording to the complainant, on 7-25-18 at 6:40pm, they were physically and verbally abused by an Officer assigned to the 35th District.\nAccording to the complainant, on 8-28-18 at 11:51pm, they were verbally abused by officers assigned to the 35th District.\nAccording to the complainant, on 8-28-18 at 11:51pm, they were verbally abused by officers assigned to the 35th District.\nAccording to the complainants, on 9-11-19 at 10:40 PM, they were physically and verbally abused by officers assigned to the 35th District.\nAccording to the complainant, on 10-13-19 at 9:00 PM, they were verbally abused, harassed and improperly ticketed by an officer assigned to the 35th District.\nAccording to the complainant, on 9-21-19 at 10:45 PM, they were treated unprofessionally and verbally abused by officers while in the confines of the 35th District.\n4\n1\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e243bf3e4ffef928249a1445d6aaa1b4e5488b | 9,713 | ipynb | Jupyter Notebook | python/Untitled.ipynb | valdirsjr/learning.data | a4b72dfd27f55f2f04120644b73232bf343f71e3 | [
"MIT"
] | null | null | null | python/Untitled.ipynb | valdirsjr/learning.data | a4b72dfd27f55f2f04120644b73232bf343f71e3 | [
"MIT"
] | null | null | null | python/Untitled.ipynb | valdirsjr/learning.data | a4b72dfd27f55f2f04120644b73232bf343f71e3 | [
"MIT"
] | null | null | null | 26.322493 | 292 | 0.541027 | [
[
[
"f = [0,1]\nf[1]\nf[-1]\nf[-2]",
"_____no_output_____"
],
[
"f.append(f[-1])",
"_____no_output_____"
],
[
"f",
"_____no_output_____"
],
[
"fibo = [0,1]\nfor i in range(100):\n fibo.append(fibo[-1]+fibo[-2])\n print(fibo)\n del fibo[0]",
"[0, 1, 1]\n[1, 1, 2]\n[1, 2, 3]\n[2, 3, 5]\n[3, 5, 8]\n[5, 8, 13]\n[8, 13, 21]\n[13, 21, 34]\n[21, 34, 55]\n[34, 55, 89]\n[55, 89, 144]\n[89, 144, 233]\n[144, 233, 377]\n[233, 377, 610]\n[377, 610, 987]\n[610, 987, 1597]\n[987, 1597, 2584]\n[1597, 2584, 4181]\n[2584, 4181, 6765]\n[4181, 6765, 10946]\n[6765, 10946, 17711]\n[10946, 17711, 28657]\n[17711, 28657, 46368]\n[28657, 46368, 75025]\n[46368, 75025, 121393]\n[75025, 121393, 196418]\n[121393, 196418, 317811]\n[196418, 317811, 514229]\n[317811, 514229, 832040]\n[514229, 832040, 1346269]\n[832040, 1346269, 2178309]\n[1346269, 2178309, 3524578]\n[2178309, 3524578, 5702887]\n[3524578, 5702887, 9227465]\n[5702887, 9227465, 14930352]\n[9227465, 14930352, 24157817]\n[14930352, 24157817, 39088169]\n[24157817, 39088169, 63245986]\n[39088169, 63245986, 102334155]\n[63245986, 102334155, 165580141]\n[102334155, 165580141, 267914296]\n[165580141, 267914296, 433494437]\n[267914296, 433494437, 701408733]\n[433494437, 701408733, 1134903170]\n[701408733, 1134903170, 1836311903]\n[1134903170, 1836311903, 2971215073]\n[1836311903, 2971215073, 4807526976]\n[2971215073, 4807526976, 7778742049]\n[4807526976, 7778742049, 12586269025]\n[7778742049, 12586269025, 20365011074]\n[12586269025, 20365011074, 32951280099]\n[20365011074, 32951280099, 53316291173]\n[32951280099, 53316291173, 86267571272]\n[53316291173, 86267571272, 139583862445]\n[86267571272, 139583862445, 225851433717]\n[139583862445, 225851433717, 365435296162]\n[225851433717, 365435296162, 591286729879]\n[365435296162, 591286729879, 956722026041]\n[591286729879, 956722026041, 1548008755920]\n[956722026041, 1548008755920, 2504730781961]\n[1548008755920, 2504730781961, 4052739537881]\n[2504730781961, 4052739537881, 6557470319842]\n[4052739537881, 6557470319842, 10610209857723]\n[6557470319842, 10610209857723, 17167680177565]\n[10610209857723, 17167680177565, 27777890035288]\n[17167680177565, 27777890035288, 44945570212853]\n[27777890035288, 44945570212853, 72723460248141]\n[44945570212853, 72723460248141, 117669030460994]\n[72723460248141, 117669030460994, 190392490709135]\n[117669030460994, 190392490709135, 308061521170129]\n[190392490709135, 308061521170129, 498454011879264]\n[308061521170129, 498454011879264, 806515533049393]\n[498454011879264, 806515533049393, 1304969544928657]\n[806515533049393, 1304969544928657, 2111485077978050]\n[1304969544928657, 2111485077978050, 3416454622906707]\n[2111485077978050, 3416454622906707, 5527939700884757]\n[3416454622906707, 5527939700884757, 8944394323791464]\n[5527939700884757, 8944394323791464, 14472334024676221]\n[8944394323791464, 14472334024676221, 23416728348467685]\n[14472334024676221, 23416728348467685, 37889062373143906]\n[23416728348467685, 37889062373143906, 61305790721611591]\n[37889062373143906, 61305790721611591, 99194853094755497]\n[61305790721611591, 99194853094755497, 160500643816367088]\n[99194853094755497, 160500643816367088, 259695496911122585]\n[160500643816367088, 259695496911122585, 420196140727489673]\n[259695496911122585, 420196140727489673, 679891637638612258]\n[420196140727489673, 679891637638612258, 1100087778366101931]\n[679891637638612258, 1100087778366101931, 1779979416004714189]\n[1100087778366101931, 1779979416004714189, 2880067194370816120]\n[1779979416004714189, 2880067194370816120, 4660046610375530309]\n[2880067194370816120, 4660046610375530309, 7540113804746346429]\n[4660046610375530309, 7540113804746346429, 12200160415121876738]\n[7540113804746346429, 12200160415121876738, 19740274219868223167]\n[12200160415121876738, 19740274219868223167, 31940434634990099905]\n[19740274219868223167, 31940434634990099905, 51680708854858323072]\n[31940434634990099905, 51680708854858323072, 83621143489848422977]\n[51680708854858323072, 83621143489848422977, 135301852344706746049]\n[83621143489848422977, 135301852344706746049, 218922995834555169026]\n[135301852344706746049, 218922995834555169026, 354224848179261915075]\n[218922995834555169026, 354224848179261915075, 573147844013817084101]\n"
],
[
"print(fibo)",
"[2, 3, 5]\n"
],
[
"for i in range(100):\n print(i,\": \",hex(i))",
"0x0\n0x1\n0x2\n0x3\n0x4\n0x5\n0x6\n0x7\n0x8\n0x9\n0xa\n0xb\n0xc\n0xd\n0xe\n0xf\n0x10\n0x11\n0x12\n0x13\n0x14\n0x15\n0x16\n0x17\n0x18\n0x19\n0x1a\n0x1b\n0x1c\n0x1d\n0x1e\n0x1f\n0x20\n0x21\n0x22\n0x23\n0x24\n0x25\n0x26\n0x27\n0x28\n0x29\n0x2a\n0x2b\n0x2c\n0x2d\n0x2e\n0x2f\n0x30\n0x31\n0x32\n0x33\n0x34\n0x35\n0x36\n0x37\n0x38\n0x39\n0x3a\n0x3b\n0x3c\n0x3d\n0x3e\n0x3f\n0x40\n0x41\n0x42\n0x43\n0x44\n0x45\n0x46\n0x47\n0x48\n0x49\n0x4a\n0x4b\n0x4c\n0x4d\n0x4e\n0x4f\n0x50\n0x51\n0x52\n0x53\n0x54\n0x55\n0x56\n0x57\n0x58\n0x59\n0x5a\n0x5b\n0x5c\n0x5d\n0x5e\n0x5f\n0x60\n0x61\n0x62\n0x63\n"
],
[
"import hashlib as hash",
"_____no_output_____"
],
[
"sha256(5)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e24d7ffd111dcdf462f9f2a021e85899eec3db | 123,007 | ipynb | Jupyter Notebook | HeroesOfPymoli/HeroesOfPymoli.ipynb | liliana-ilut/pandas-challenge | bda424f45dc09f09819d8e63ca88ee8058b77184 | [
"ADSL"
] | null | null | null | HeroesOfPymoli/HeroesOfPymoli.ipynb | liliana-ilut/pandas-challenge | bda424f45dc09f09819d8e63ca88ee8058b77184 | [
"ADSL"
] | null | null | null | HeroesOfPymoli/HeroesOfPymoli.ipynb | liliana-ilut/pandas-challenge | bda424f45dc09f09819d8e63ca88ee8058b77184 | [
"ADSL"
] | null | null | null | 32.653836 | 196 | 0.343094 | [
[
[
"# Dependencies and Setup\nimport pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"# Load the file and set up the path for it\npurchase_data = \"Resources/purchase_data.csv\"",
"_____no_output_____"
],
[
"# Read Purchasing File, store it in Panda frame, read the head of the file (first 5 items)\npurchase_data_df = pd.read_csv(purchase_data)\npurchase_data_df.head()",
"_____no_output_____"
],
[
"purchase_data_df.describe()",
"_____no_output_____"
],
[
"###PLAYER COUNT",
"_____no_output_____"
],
[
"TotalNrPlayer = len(purchase_data_df[\"SN\"].value_counts())\nTotalNrPlayer",
"_____no_output_____"
],
[
"#create a data frame to show the total players \nTotalNrPlayer = pd.DataFrame({\"Total Number of Players\" : [TotalNrPlayer]})\nTotalNrPlayer",
"_____no_output_____"
],
[
"###PURCHASING ANALYSIS (TOTAL)",
"_____no_output_____"
],
[
"#number of unique items\nNrUniqueItems = len((purchase_data_df[\"Item ID\"]).unique())\nNrUniqueItems",
"_____no_output_____"
],
[
"#calculate average price\nAveragePrice = float((purchase_data_df[\"Price\"]).mean())\nAveragePrice",
"_____no_output_____"
],
[
"#calculate total number of purchases\nTotalPurchases = len((purchase_data_df[\"Purchase ID\"]).value_counts())\nTotalPurchases",
"_____no_output_____"
],
[
"#calculate total revenue \nTotalRevenue = float((purchase_data_df[\"Price\"]).sum())\nTotalRevenue",
"_____no_output_____"
],
[
"#create data frame with the values we just calculated\ntotal_purchasing_analysis_df = pd.DataFrame ({\n \"Number of Unique Items\" : [NrUniqueItems],\n \"Average Purchase Price\" : [AveragePrice],\n \"Total Number of Purchases\" : [TotalPurchases],\n \"Total Revenue\" : [TotalRevenue]\n })\ntotal_purchasing_analysis_df",
"_____no_output_____"
],
[
"#format for $ sign and 2 decimals\ntotal_purchasing_analysis_df[\"Average Purchase Price\"] = total_purchasing_analysis_df[\"Average Purchase Price\"].map(\"${:.2f}\".format)\ntotal_purchasing_analysis_df[\"ATotal Revenue\"] = total_purchasing_analysis_df[\"Total Revenue\"].map(\"${:.2f}\".format)\ntotal_purchasing_analysis_df",
"_____no_output_____"
],
[
"### GENDER DEMOGRAPHICS ",
"_____no_output_____"
],
[
"#group by gender status, using groupby function\nGenderStatus_df = purchase_data_df.groupby(\"Gender\")\nGenderStatus_df.head()",
"_____no_output_____"
],
[
"#calculate the total number of gender type \nGenderCount_df = GenderStatus_df[\"SN\"].nunique()\nGenderCount_df",
"_____no_output_____"
],
[
"#calculate percentage of gender type\nGenderPercentage_df = purchase_data_df[\"Gender\"].value_counts(normalize=True) * 100\nGenderPercentage_df",
"_____no_output_____"
],
[
"#create data frame for the Gender Demographics table\nGender_Demographics_df = pd.DataFrame({\n \"percentage of players\" : GenderPercentage_df,\n \"count of players\" : GenderCount_df\n })\nGender_Demographics_df",
"_____no_output_____"
],
[
"#format table for % sign and 2 decimals\nGender_Demographics_df[\"percentage of players\"] = Gender_Demographics_df[\"percentage of players\"].map(\"{:.2f} %\".format)\nGender_Demographics_df",
"_____no_output_____"
],
[
"### PURCHASING ANALYSIS (GENDER) ",
"_____no_output_____"
],
[
"#Calculate Purchase Count by Gender\nPurchaseCount_df = GenderStatus_df[\"Purchase ID\"].count()\nPurchaseCount_df",
"_____no_output_____"
],
[
"#Calculate Average Purchase Price by Gender\nAveragePurchase_df = GenderStatus_df[\"Price\"].mean()\nAveragePurchase_df",
"_____no_output_____"
],
[
"#Calculate Total Purchase Value by Gender\nTotalValue_df = GenderStatus_df[\"Price\"].sum()\nTotalValue_df",
"_____no_output_____"
],
[
"#Calculate Average Purchase Total per Person by Gender\nAverage_Person_Gender_df = TotalValue_df/GenderCount_df\nAverage_Person_Gender_df",
"_____no_output_____"
],
[
"#Create Data Frame based on the calculations we just made \nGender_Demographics_df = pd.DataFrame({\n \"Purchase Count\": PurchaseCount_df, \n \"Average Purchase Price\": AveragePurchase_df,\n \"Average Purchase Value\":TotalValue_df,\n \"Avg Purchase Total per Person\": Average_Person_Gender_df\n })\n\nGender_Demographics_df",
"_____no_output_____"
],
[
"#Format the table to $ sign and 2 decimals\nGender_Demographics_df[\"Average Purchase Price\"] = Gender_Demographics_df[\"Average Purchase Price\"].map(\"${:.2f}\".format)\nGender_Demographics_df[\"Average Purchase Value\"] = Gender_Demographics_df[\"Average Purchase Value\"].map(\"${:.2f}\".format)\nGender_Demographics_df[\"Avg Purchase Total per Person\"] = Gender_Demographics_df[\"Avg Purchase Total per Person\"].map(\"${:.2f}\".format)\n\nGender_Demographics_df",
"_____no_output_____"
],
[
"### AGE DEMOGRAPHICS ",
"_____no_output_____"
],
[
"# Create bins for ages \nBinsAge = [0, 9, 14, 19, 24, 29, 34, 39, 99]\nBinsAge",
"_____no_output_____"
],
[
"# Create the names for the bins\nBinNames = [\"<10\", \"10-14\", \"15-19\", \"20-24\", \"25-29\", \"30-34\", \"35-39\", \"40+\"]\nBinNames",
"_____no_output_____"
],
[
"# Segment and sort age values into bins established above\npurchase_data_df[\"Age Group\"] = pd.cut(purchase_data_df[\"Age\"], BinsAge, labels = BinNames)\npurchase_data_df",
"_____no_output_____"
],
[
"# Group by \"Age Group\"\nGroupedAge = purchase_data_df.groupby(\"Age Group\")\nGroupedAge",
"_____no_output_____"
],
[
"# Calculate total players by age category\nTotalCount_age = GroupedAge [\"SN\"].nunique()\nTotalCount_age",
"_____no_output_____"
],
[
"# Calculate percentages by age category \nPercentage_age = (TotalCount_age/576) * 100\nPercentage_age",
"_____no_output_____"
],
[
"# Create data frame with the values we just calculated\nAge_Demographics_df = pd.DataFrame ({\n \"Total Count\" : TotalCount_age,\n \"Percentage of Players\" : Percentage_age,\n })\nAge_Demographics_df",
"_____no_output_____"
],
[
"#Format the table to % sign and 2 decimals\nAge_Demographics_df[\"Percentage of Players\"] = Age_Demographics_df[\"Percentage of Players\"].map(\"{:.2f} %\".format)\nAge_Demographics_df",
"_____no_output_____"
],
[
"#Calculate purchases count by age group\nPurchaseCountAge = GroupedAge[\"Purchase ID\"].count()\nPurchaseCountAge",
"_____no_output_____"
],
[
"# Calculate average purchase count by age\nAveragePriceAge = GroupedAge[\"Price\"].mean()\nAveragePriceAge",
"_____no_output_____"
],
[
"# Calculate total purchase value by age\nTotalPurchases_age = GroupedAge[\"Price\"].sum()\nTotalPurchases_age",
"_____no_output_____"
],
[
"# Calculate Average Purchase Total per Person by Age Group\nAverageTotal_person_age = TotalPurchases_age/TotalCount_age\nAverageTotal_person_age",
"_____no_output_____"
],
[
"#create data frame with the values we just calculated\nAge_Demographics_Analysis_df = pd.DataFrame ({\n \"Purchase Count\" : PurchaseCountAge,\n \"Average Purchase Price\" : AveragePriceAge,\n \"Total Purchase Value\" : TotalPurchases_age,\n \"Average Purchase Total per Person by Age Group\" : AverageTotal_person_age\n })\nAge_Demographics_Analysis_df",
"_____no_output_____"
],
[
"#Format the table for $ sign and 2 decimals\nAge_Demographics_Analysis_df[\"Average Purchase Price\"] = Age_Demographics_Analysis_df[\"Average Purchase Price\"].map(\"${:.2f}\".format)\nAge_Demographics_Analysis_df[\"Total Purchase Value\"] = Age_Demographics_Analysis_df[\"Total Purchase Value\"].map(\"${:.2f}\".format)\nAge_Demographics_Analysis_df[\"Average Purchase Total per Person by Age Group\"] = Age_Demographics_Analysis_df[\"Average Purchase Total per Person by Age Group\"].map(\"${:.2f}\".format)\nAge_Demographics_Analysis_df",
"_____no_output_____"
],
[
"###Top Spenders",
"_____no_output_____"
],
[
"#Create a new group by SN\nspender_group = purchase_data_df.groupby(\"SN\")\nspender_group",
"_____no_output_____"
],
[
"# Calculate purchases count by spender group\nPurchaseCountSpender = spender_group[\"Purchase ID\"].count()\nPurchaseCountSpender",
"_____no_output_____"
],
[
"# Calculate average purchase price by spender\nAveragePriceSpender = spender_group[\"Price\"].mean()\nAveragePriceSpender",
"_____no_output_____"
],
[
"# Calculate total purchase value by spender\nTotalPurchases_spender = spender_group[\"Price\"].sum()\nTotalPurchases_spender ",
"_____no_output_____"
],
[
"# Create data frame with obtained values\ntop_spender_group_df = pd.DataFrame({\n \"Purchase Count\": PurchaseCountSpender,\n \"Average Purchase Price\": AveragePriceSpender,\n \"Total Purchase Value\":TotalPurchases_spender \n })\ntop_spender_group_df",
"_____no_output_____"
],
[
"#Format table by \"Total Purchase Value\" Descending order \nspender_gropu_formatted_df = top_spender_group_df.sort_values([\"Total Purchase Value\"], ascending=False).head()\nspender_gropu_formatted_df ",
"_____no_output_____"
],
[
"#format the table for $ sign and 2 decimals\nspender_gropu_formatted_df[\"Average Purchase Price\"] = spender_gropu_formatted_df[\"Average Purchase Price\"].map(\"${:.2f}\".format)\nspender_gropu_formatted_df[\"Total Purchase Value\"] = spender_gropu_formatted_df[\"Total Purchase Value\"].map(\"${:.2f}\".format)\nspender_gropu_formatted_df",
"_____no_output_____"
],
[
"###MOST POPULAR ITEMS",
"_____no_output_____"
],
[
"# Create new data for most popular items \nPopularItems = purchase_data_df.set_index([\"Item ID\", \"Item Name\"])\nPopularItems",
"_____no_output_____"
],
[
"# Group by Item ID, Item Name\nGrouped_PopularItems = PopularItems.groupby([\"Item ID\",\"Item Name\"])\nGrouped_PopularItems",
"_____no_output_____"
],
[
"# Calculate the number of times an item has been purchased \nItem_purchase_count = Grouped_PopularItems[\"Price\"].count()\nItem_purchase_count",
"_____no_output_____"
],
[
"# Calcualte the average value per item \nItem_average_value = Grouped_PopularItems[\"Price\"].mean()\nItem_average_value",
"_____no_output_____"
],
[
"# Calcualte the total purchase value per item\nItem_purchase_value = Grouped_PopularItems[\"Price\"].sum()\nItem_purchase_value",
"_____no_output_____"
],
[
"# Create data frame with obtained values\nMostPopularItem_df = pd.DataFrame({\n \"Purchase Count\": Item_purchase_count,\n \"Item Price\": Item_average_value,\n \"Total Purchase Value\":Item_purchase_value \n })\nMostPopularItem_df",
"_____no_output_____"
],
[
"#Format table by \" Purchase Count\" Descending order \nMostPopularItem_formatted_df = MostPopularItem_df.sort_values([\"Purchase Count\"], ascending=False).head()\nMostPopularItem_formatted_df",
"_____no_output_____"
],
[
"# Format the table for $ sign and 2 decimals\nMostPopularItem_formatted_df[\"Item Price\"] = MostPopularItem_formatted_df[\"Item Price\"].map(\"${:.2f}\".format)\nMostPopularItem_formatted_df[\"Total Purchase Value\"] = MostPopularItem_formatted_df[\"Total Purchase Value\"].map(\"${:.2f}\".format)\nMostPopularItem_formatted_df",
"_____no_output_____"
],
[
"###MOST PROFITABLE ITEMS",
"_____no_output_____"
],
[
"#Format table by \" Total Purchase Value\" Descending order \nMostProfitableItem_formatted_df = MostPopularItem_df.sort_values([\"Total Purchase Value\"], ascending=False).head()\nMostProfitableItem_formatted_df",
"_____no_output_____"
],
[
"#Format the table for $ sign and 2 decimals\nMostProfitableItem_formatted_df[\"Item Price\"] = MostProfitableItem_formatted_df[\"Item Price\"].map(\"${:.2f}\".format)\nMostProfitableItem_formatted_df[\"Total Purchase Value\"] = MostProfitableItem_formatted_df[\"Total Purchase Value\"].map(\"${:.2f}\".format)\nMostProfitableItem_formatted_df",
"_____no_output_____"
],
[
"### WRITTEN DESCRIPTION OF 3 OBSERVABLE TRENDS BASED ON THE DATA \n#1. If we look at the Age Group we observe that 46% of the total values were purchased by people in between 20 and 24 years old. \n#2. If we look at the Gender Group, the Male population is taking the lead. They purchased 83% of the entire purcahses\n#3. Final Critic and Oathbreaker, Last Hope of the Breaking Storm were the Most Popular and the Most Profitable games",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e2697cee8110fb4b0a822c865817daf87e9f76 | 4,252 | ipynb | Jupyter Notebook | svm.ipynb | eaganjm1102/Machine-learning-challenge | 311dba4ecd95fe1fe8b91236e20b8f48b3b64d29 | [
"ADSL"
] | null | null | null | svm.ipynb | eaganjm1102/Machine-learning-challenge | 311dba4ecd95fe1fe8b91236e20b8f48b3b64d29 | [
"ADSL"
] | null | null | null | svm.ipynb | eaganjm1102/Machine-learning-challenge | 311dba4ecd95fe1fe8b91236e20b8f48b3b64d29 | [
"ADSL"
] | null | null | null | 21.154229 | 91 | 0.541157 | [
[
[
"# Update sklearn to prevent version mismatches\n!conda install scikit-learn\n!conda update scikit-learn\n!conda install joblib \n!conda update joblib ",
"_____no_output_____"
],
[
"import pandas as pd",
"_____no_output_____"
]
],
[
[
"# Read the CSV and Perform Basic Data Cleaning",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv(\"exoplanet_data.csv\")\n# Drop the null columns where all values are null\ndf = df.dropna(axis='columns', how='all')\n# Drop the null rows\ndf = df.dropna()\ndf.head()",
"_____no_output_____"
]
],
[
[
"# Create a Train Test Split\n\nUse `koi_disposition` for the y values",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\ny = df[\"koi_disposition\"]\nX = df.drop(columns=[\"koi_disposition\"])\n\nX_train, X_test, y_train, y_test = train_test_split(X, y, random_state=1, stratify=y)",
"_____no_output_____"
]
],
[
[
"# Pre-processing\n\nScale the data using the MinMaxScaler",
"_____no_output_____"
]
],
[
[
"from sklearn.preprocessing import MinMaxScaler\nX_scaler = MinMaxScaler().fit(X_train)\n\nX_train_scaled = X_scaler.transform(X_train)\nX_test_scaled = X_scaler.transform(X_test)",
"_____no_output_____"
]
],
[
[
"# Train the Model",
"_____no_output_____"
]
],
[
[
"# Support vector machine linear classifier\nfrom sklearn.svm import SVC \nmodel2 = SVC(kernel='linear')\nmodel2.fit(X_train_scaled, y_train)",
"_____no_output_____"
],
[
"print(f\"Training Data Score: {model2.score(X_train_scaled, y_train)}\")\nprint(f\"Testing Data Score: {model2.score(X_test_scaled, y_test)}\")",
"_____no_output_____"
]
],
[
[
"# Hyperparameter Tuning\n\nUse `GridSearchCV` to tune model's parameters",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import GridSearchCV\nparam_grid = {'C': [1, 5, 10],\n 'gamma': [0.0001, 0.001, 0.01]}\ngrid = GridSearchCV(model2, param_grid, verbose=3)",
"_____no_output_____"
],
[
"grid.fit(X_train_scaled, y_train)\n",
"_____no_output_____"
],
[
"print(grid.best_params_)\nprint(grid.best_score_)",
"_____no_output_____"
]
],
[
[
"# Save the Model",
"_____no_output_____"
]
],
[
[
"# save the fitted model to file\nimport joblib\nfilename = 'svm.sav'\njoblib.dump(grid, filename)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0e26dcc4548df5f3d052dcc75a5100df646bd20 | 637,768 | ipynb | Jupyter Notebook | coco2yolov5.ipynb | pylabelalpha/pylabel | 023723297aeffd7f5568f9b00fdf0afc1e7a9d72 | [
"MIT"
] | null | null | null | coco2yolov5.ipynb | pylabelalpha/pylabel | 023723297aeffd7f5568f9b00fdf0afc1e7a9d72 | [
"MIT"
] | 1 | 2021-09-20T02:57:18.000Z | 2021-11-16T15:26:00.000Z | coco2yolov5.ipynb | pylabelalpha/notebook | fc383e81e81cdf37f4e8ee45b873d4d6738161bb | [
"MIT"
] | null | null | null | 1,339.848739 | 319,676 | 0.950832 | [
[
[
"# Pylabel End to End Example \n## Coco JSON to VOC XML\nUse this notebook to try out importing, analyzing, and exporting datasets of image annotations. ",
"_____no_output_____"
]
],
[
[
"!pip install -i https://test.pypi.org/simple/ pylabel==0.1.1",
"Looking in indexes: https://test.pypi.org/simple/\nCollecting pylabelalphatest==0.1.5\n Downloading https://test-files.pythonhosted.org/packages/be/e4/4f66e7037d571e40f61ff8f26f29f361a9616bec88bcd32a52ec2398fbe2/pylabelalphatest-0.1.5-py3-none-any.whl (12 kB)\nCollecting bbox-visualizer\n Downloading https://test-files.pythonhosted.org/packages/a2/44/0d4339621b7251486b7ab3feaf6e60a63a0cb4e47d89764fd40b545797b4/bbox_visualizer-0.1.1-py2.py3-none-any.whl (6.2 kB)\nRequirement already satisfied: pandas in ./penv/lib/python3.8/site-packages (from pylabelalphatest==0.1.5) (1.3.4)\nRequirement already satisfied: numpy>=1.17.3 in ./penv/lib/python3.8/site-packages (from pandas->pylabelalphatest==0.1.5) (1.21.2)\nRequirement already satisfied: python-dateutil>=2.7.3 in ./penv/lib/python3.8/site-packages (from pandas->pylabelalphatest==0.1.5) (2.8.2)\nRequirement already satisfied: pytz>=2017.3 in ./penv/lib/python3.8/site-packages (from pandas->pylabelalphatest==0.1.5) (2021.3)\nRequirement already satisfied: six>=1.5 in ./penv/lib/python3.8/site-packages (from python-dateutil>=2.7.3->pandas->pylabelalphatest==0.1.5) (1.16.0)\nInstalling collected packages: bbox-visualizer, pylabelalphatest\nSuccessfully installed bbox-visualizer-0.1.1 pylabelalphatest-0.1.5\n"
],
[
"from pylabel import importer",
"_____no_output_____"
]
],
[
[
"## Import coco annotations \nFirst we will import annotations from the coco dataset, which are in coco json format. ",
"_____no_output_____"
]
],
[
[
"import os \nimport zipfile\n\n#Download sample coco dataset \nos.makedirs(\"data\", exist_ok=True)\n!wget \"https://github.com/pylabelalpha/notebook/blob/main/BCCD_coco.zip?raw=true\" -O data/BCCD_coco.zip\nwith zipfile.ZipFile(\"data/BCCD_coco.zip\", 'r') as zip_ref:\n zip_ref.extractall(\"data\")\n\n#Specify path to the coco.json file\npath_to_annotations = \"data/BCCD_Dataset.json\"\n#Specify the path to the images (if they are in a different folder than the annotations)\npath_to_images = \"\"\n\n#Import the dataset into the pylable schema \ndataset = importer.ImportCoco(path_to_annotations, path_to_images=path_to_images, name=\"BCCD_coco\")\ndataset.df.head(5)\n",
"--2021-10-18 20:42:09-- https://github.com/pylabelalpha/notebook/blob/main/BCCD_coco.zip?raw=true\nResolving github.com (github.com)... 192.30.255.112\nConnecting to github.com (github.com)|192.30.255.112|:443... connected.\nHTTP request sent, awaiting response... 302 Found\nLocation: https://github.com/pylabelalpha/notebook/raw/main/BCCD_coco.zip [following]\n--2021-10-18 20:42:09-- https://github.com/pylabelalpha/notebook/raw/main/BCCD_coco.zip\nReusing existing connection to github.com:443.\nHTTP request sent, awaiting response... 302 Found\nLocation: https://raw.githubusercontent.com/pylabelalpha/notebook/main/BCCD_coco.zip [following]\n--2021-10-18 20:42:09-- https://raw.githubusercontent.com/pylabelalpha/notebook/main/BCCD_coco.zip\nResolving raw.githubusercontent.com (raw.githubusercontent.com)... 185.199.108.133, 185.199.109.133, 185.199.110.133, ...\nConnecting to raw.githubusercontent.com (raw.githubusercontent.com)|185.199.108.133|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 7625693 (7.3M) [application/zip]\nSaving to: ‘data/BCCD_coco.zip’\n\ndata/BCCD_coco.zip 100%[===================>] 7.27M 25.2MB/s in 0.3s \n\n2021-10-18 20:42:10 (25.2 MB/s) - ‘data/BCCD_coco.zip’ saved [7625693/7625693]\n\n"
]
],
[
[
"## Analyze annotations\nPylabel can calculate basic summary statisticts about the dataset such as the number of files and the classes. \nThe dataset is stored as a pandas frame so the developer can do additional exploratory analysis on the dataset. ",
"_____no_output_____"
]
],
[
[
"print(f\"Number of images: {dataset.analyze.num_images}\")\nprint(f\"Number of classes: {dataset.analyze.num_classes}\")\nprint(f\"Classes:{dataset.analyze.classes}\")\nprint(f\"Class counts:\\n{dataset.analyze.class_counts}\")\nprint(f\"Path to annotations:\\n{dataset.path_to_annotations}\")\n",
"Number of images: 364\nNumber of classes: 3\nClasses:['RBC' 'WBC' 'Platelets']\nClass counts:\nRBC 4155\nWBC 372\nPlatelets 361\nName: cat_name, dtype: int64\nPath to annotations:\ndata\n"
]
],
[
[
"## Visualize Annotations \nYou can render the bounding boxes for your image to inspect them and confirm that they imported correctly. ",
"_____no_output_____"
]
],
[
[
"from IPython.display import Image, display\ndisplay(dataset.visualize.ShowBoundingBoxes(dataset, 100))\ndisplay(dataset.visualize.ShowBoundingBoxes(dataset, \"BloodImage_00315.jpg\"))",
"data/BloodImage_00407.jpg\n"
]
],
[
[
"# Export to Yolo v5\nThe PyLabel exporter will export all of the annotations in the dataframe to the desired target format.\nYolo creates one text file for each image in the dataset. ",
"_____no_output_____"
]
],
[
[
"# Inspect one of the files\ndataset.path_to_annotations = \"data/yolo\"\ndataset.export.ExportToYoloV5(dataset)",
"data/yolo\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0e2710f53b2c0f1244cc66dabd429b4b1ff3946 | 351,706 | ipynb | Jupyter Notebook | TCLab Help.ipynb | monocilindro/data_science | 36098bb6b3e731a11315e58e0b09628fae86069d | [
"MIT"
] | 41 | 2020-01-25T21:23:59.000Z | 2022-02-22T19:48:15.000Z | TCLab Help.ipynb | ichit/data_science | 36098bb6b3e731a11315e58e0b09628fae86069d | [
"MIT"
] | null | null | null | TCLab Help.ipynb | ichit/data_science | 36098bb6b3e731a11315e58e0b09628fae86069d | [
"MIT"
] | 38 | 2020-01-27T18:57:46.000Z | 2022-03-05T00:33:45.000Z | 1,803.620513 | 320,472 | 0.963032 | [
[
[
"## TCLab Function Help\n\n\n\n#### Connect/Disconnect\n```lab = tclab.TCLab()``` Connect and create new lab object, ```lab.close()``` disconnects lab.\n\n#### LED\n```lab.LED()``` Percentage of output light for __Hot__ Light.\n\n#### Heaters\n```lab.Q1()``` and ```lab.Q2()``` Percentage of power to heaters.\n\n#### Temperatures\n```lab.T1``` and ```lab.T2``` Value of current heater tempertures in Celsius.\n\n| **TCLab Function** | **Example** | **Description** |\n| ----------- | ----------- | ----------- |\n| `TCLab()` | `tclab.TCLab()` | Create new lab object and connect |\n| `LED` | `lab.LED(45)` | Turn on the LED to 45%. Valid range is 0-100% |\n| `Q1` | `lab.Q1(63)` | Turn on heater 1 (`Q1`) to 63%. Valid range is 0-100% |\n| `Q2` | `lab.Q2(28)` | Turn on heater 2 (`Q2`) to 28%. Valid range is 0-100% |\n| `T1` | `print(lab.T1)` | Read temperature 1 (`T1`) in °C. valid range is -40 to 150°C (TMP36 sensor) |\n| `T2` | `print(lab.T2)` | Read temperature 2 (`T2`) in °C. with +/- 1°C accuracy (TMP36 sensor) |\n| `close()` | `lab.close()` | Close serial USB connection to TCLab - not needed if using `with` to open |",
"_____no_output_____"
],
[
"### Errors\n\n\n\nSubmit an error to us at [email protected] so we can fix the problem and add it to this list. There is also a list of [troubleshooting items with frequently asked questions](https://apmonitor.com/pdc/index.php/Main/ArduinoSetup). If you get the error about already having an open connection to the TCLab, try restarting the **kernel**. Go to the top of the page to the **Kernel** tab, click it, then click restart.",
"_____no_output_____"
],
[
"### How to Run a Cell\nUse the symbol to run the needed cell, left of the program. If you don't see a symbol, try selecting the cell and holding `Ctrl`, then pressing `Enter`. Another option is clicking the \"Run\" button at the top of the page when the cell is selected.",
"_____no_output_____"
],
[
"### Install Temperature Control Lab\n\n\n\nThis code trys to import `tclab`. If it if it fails to find the package, it installs the program with `pip`. The `pip` installation is required once to let you use the Temperature Control Lab for all of the exercises. It does not need to be installed again, even if the IPython session is restarted.",
"_____no_output_____"
]
],
[
[
"# install tclab\ntry:\n import tclab\nexcept:\n # Needed to communicate through usb port\n !pip install --user pyserial\n # The --user is put in for accounts without admin privileges\n !pip install --user tclab \n # restart kernel if this doesn't import\n import tclab",
"_____no_output_____"
]
],
[
[
"### Update Temperature Control Lab\nUpdates package to latest version.",
"_____no_output_____"
]
],
[
[
"!pip install tclab --upgrade --user",
"_____no_output_____"
]
],
[
[
"### Connection Check\n\n\n\nThe following code connects, reads temperatures 1 and 2, sets heaters 1 and 2, turns on the LED to 45%, and then disconnects from the TCLab. If an error is shown, try unplugging your lab and/or restarting Jupyter's kernel from the Jupyter notebook menu.",
"_____no_output_____"
]
],
[
[
"import tclab\nlab = tclab.TCLab()\nprint(lab.T1,lab.T2)\nlab.Q1(50); lab.Q2(40)\nlab.LED(45)\nlab.close()",
"_____no_output_____"
]
],
[
[
"\n\nAnother way to connect to the TCLab is the `with` statement. The advantage of the `with` statement is that the connection automatically closes if there is a fatal error (bug) in the code and the program never reaches the `lab.close()` statement. This prevents a kernel restart to reset the USB connection to the TCLab Arduino.",
"_____no_output_____"
]
],
[
[
"import tclab\nimport time\nwith tclab.TCLab() as lab:\n print(lab.T1,lab.T2)\n lab.Q1(50); lab.Q2(40)\n lab.LED(45)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0e275d3c8ca8e4ceb9c52049b4a0de6d481fcae | 198,668 | ipynb | Jupyter Notebook | face_generation/.ipynb_checkpoints/dlnd_face_generation-floyd-checkpoint.ipynb | syednasar/deep-learning | 5d8ee53e6a3d2a1b4281c931ec93d698d8d45a72 | [
"MIT"
] | 2 | 2017-06-24T18:10:30.000Z | 2017-09-29T11:01:02.000Z | face_generation/.ipynb_checkpoints/dlnd_face_generation-floyd-checkpoint.ipynb | syednasar/deep-learning | 5d8ee53e6a3d2a1b4281c931ec93d698d8d45a72 | [
"MIT"
] | null | null | null | face_generation/.ipynb_checkpoints/dlnd_face_generation-floyd-checkpoint.ipynb | syednasar/deep-learning | 5d8ee53e6a3d2a1b4281c931ec93d698d8d45a72 | [
"MIT"
] | null | null | null | 225.502838 | 84,460 | 0.886142 | [
[
[
"# Face Generation\nIn this project, you'll use generative adversarial networks to generate new images of faces.\n### Get the Data\nYou'll be using two datasets in this project:\n- MNIST\n- CelebA\n\nSince the celebA dataset is complex and you're doing GANs in a project for the first time, we want you to test your neural network on MNIST before CelebA. Running the GANs on MNIST will allow you to see how well your model trains sooner.\n\nIf you're using [FloydHub](https://www.floydhub.com/), set `data_dir` to \"/input\" and use the [FloydHub data ID](http://docs.floydhub.com/home/using_datasets/) \"R5KrjnANiKVhLWAkpXhNBe\".",
"_____no_output_____"
]
],
[
[
"data_dir = './data'\n\n# FloydHub - Use with data ID \"R5KrjnANiKVhLWAkpXhNBe\"\ndata_dir = '/input'\n\n\n\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL\n\"\"\"\nimport helper\n\nhelper.download_extract('mnist', data_dir)\nhelper.download_extract('celeba', data_dir)",
"Downloading mnist: 9.92MB [00:02, 4.02MB/s] \nExtracting mnist: 100%|██████████| 60.0K/60.0K [00:24<00:00, 2.46KFile/s]\nDownloading celeba: 1.44GB [02:28, 9.73MB/s] \n"
]
],
[
[
"## Explore the Data\n### MNIST\nAs you're aware, the [MNIST](http://yann.lecun.com/exdb/mnist/) dataset contains images of handwritten digits. You can view the first number of examples by changing `show_n_images`. ",
"_____no_output_____"
]
],
[
[
"show_n_images = 25\n\n\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL\n\"\"\"\n%matplotlib inline\nimport os\nfrom glob import glob\nfrom matplotlib import pyplot\n\nmnist_images = helper.get_batch(glob(os.path.join(data_dir, 'mnist/*.jpg'))[:show_n_images], 28, 28, 'L')\npyplot.imshow(helper.images_square_grid(mnist_images, 'L'), cmap='gray')",
"_____no_output_____"
]
],
[
[
"### CelebA\nThe [CelebFaces Attributes Dataset (CelebA)](http://mmlab.ie.cuhk.edu.hk/projects/CelebA.html) dataset contains over 200,000 celebrity images with annotations. Since you're going to be generating faces, you won't need the annotations. You can view the first number of examples by changing `show_n_images`.",
"_____no_output_____"
]
],
[
[
"show_n_images = 25\n\n\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL\n\"\"\"\nmnist_images = helper.get_batch(glob(os.path.join(data_dir, 'img_align_celeba/*.jpg'))[:show_n_images], 28, 28, 'RGB')\npyplot.imshow(helper.images_square_grid(mnist_images, 'RGB'))",
"_____no_output_____"
]
],
[
[
"## Preprocess the Data\nSince the project's main focus is on building the GANs, we'll preprocess the data for you. The values of the MNIST and CelebA dataset will be in the range of -0.5 to 0.5 of 28x28 dimensional images. The CelebA images will be cropped to remove parts of the image that don't include a face, then resized down to 28x28.\n\nThe MNIST images are black and white images with a single [color channel](https://en.wikipedia.org/wiki/Channel_(digital_image%29) while the CelebA images have [3 color channels (RGB color channel)](https://en.wikipedia.org/wiki/Channel_(digital_image%29#RGB_Images).\n## Build the Neural Network\nYou'll build the components necessary to build a GANs by implementing the following functions below:\n- `model_inputs`\n- `discriminator`\n- `generator`\n- `model_loss`\n- `model_opt`\n- `train`\n\n### Check the Version of TensorFlow and Access to GPU\nThis will check to make sure you have the correct version of TensorFlow and access to a GPU",
"_____no_output_____"
]
],
[
[
"\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL\n\"\"\"\nfrom distutils.version import LooseVersion\nimport warnings\nimport tensorflow as tf\n\n# Check TensorFlow Version\nassert LooseVersion(tf.__version__) >= LooseVersion('1.0'), 'Please use TensorFlow version 1.0 or newer. You are using {}'.format(tf.__version__)\nprint('TensorFlow Version: {}'.format(tf.__version__))\n\n# Check for a GPU\nif not tf.test.gpu_device_name():\n warnings.warn('No GPU found. Please use a GPU to train your neural network.')\nelse:\n print('Default GPU Device: {}'.format(tf.test.gpu_device_name()))",
"TensorFlow Version: 1.0.0\n"
]
],
[
[
"### Input\nImplement the `model_inputs` function to create TF Placeholders for the Neural Network. It should create the following placeholders:\n- Real input images placeholder with rank 4 using `image_width`, `image_height`, and `image_channels`.\n- Z input placeholder with rank 2 using `z_dim`.\n- Learning rate placeholder with rank 0.\n\nReturn the placeholders in the following the tuple (tensor of real input images, tensor of z data)",
"_____no_output_____"
]
],
[
[
"import problem_unittests as tests\n\ndef model_inputs(image_width, image_height, image_channels, z_dim):\n \"\"\"\n Create the model inputs\n :param image_width: The input image width\n :param image_height: The input image height\n :param image_channels: The number of image channels\n :param z_dim: The dimension of Z\n :return: Tuple of (tensor of real input images, tensor of z data, learning rate)\n \"\"\"\n # TODO: Implement Function\n input_real = tf.placeholder(tf.float32, (None, image_width, image_height, image_channels), name=\"input_real\")\n input_z = tf.placeholder(tf.float32, (None, z_dim), name=\"input_z\")\n l_rate = tf.placeholder(tf.float32, name='l_rate')\n\n return input_real, input_z, l_rate\n\n\n\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE\n\"\"\"\ntests.test_model_inputs(model_inputs)",
"Tests Passed\n"
]
],
[
[
"### Discriminator\nImplement `discriminator` to create a discriminator neural network that discriminates on `images`. This function should be able to reuse the variabes in the neural network. Use [`tf.variable_scope`](https://www.tensorflow.org/api_docs/python/tf/variable_scope) with a scope name of \"discriminator\" to allow the variables to be reused. The function should return a tuple of (tensor output of the generator, tensor logits of the generator).",
"_____no_output_____"
]
],
[
[
"def discriminator(images, reuse=False):\n \"\"\"\n Create the discriminator network\n :param image: Tensor of input image(s)\n :param reuse: Boolean if the weights should be reused\n :return: Tuple of (tensor output of the discriminator, tensor logits of the discriminator)\n \"\"\"\n # TODO: Implement Function\n alpha = 0.2\n with tf.variable_scope('discriminator', reuse=reuse):\n # Input layer is 28x28x3\n x1 = tf.layers.conv2d(images, 64, 5, strides=2, padding='same')\n relu1 = tf.maximum(alpha * x1, x1)\n # 14x14x64\n \n x2 = tf.layers.conv2d(relu1, 128, 5, strides=1, padding='same')\n bn2= tf.layers.batch_normalization(x2, training=True)\n relu2 = tf.maximum(alpha * bn2, bn2)\n # 14x14x128\n \n x3 = tf.layers.conv2d(relu2, 128, 5, strides=2, padding='same')\n bn3= tf.layers.batch_normalization(x3, training=True)\n relu3 = tf.maximum(alpha * bn3, bn3)\n # 7x7x256\n\n # Flatten it\n flat = tf.reshape(relu2, (-1, 7*7*256))\n logits = tf.layers.dense(flat, 1)\n output = tf.sigmoid(logits)\n\n return output, logits\n\n\n\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE\n\"\"\"\ntests.test_discriminator(discriminator, tf)",
"Tests Passed\n"
]
],
[
[
"### Generator\nImplement `generator` to generate an image using `z`. This function should be able to reuse the variabes in the neural network. Use [`tf.variable_scope`](https://www.tensorflow.org/api_docs/python/tf/variable_scope) with a scope name of \"generator\" to allow the variables to be reused. The function should return the generated 28 x 28 x `out_channel_dim` images.",
"_____no_output_____"
]
],
[
[
"def generator(z, out_channel_dim, is_train=True):\n \"\"\"\n Create the generator network\n :param z: Input z\n :param out_channel_dim: The number of channels in the output image\n :param is_train: Boolean if generator is being used for training\n :return: The tensor output of the generator\n \"\"\"\n # TODO: Implement Function\n alpha = 0.2\n with tf.variable_scope('generator', reuse=not is_train):\n # First fully connected layer\n x1 = tf.layers.dense(z, 7*7*256)\n # Reshape it to start the convolutional stack\n x1 = tf.reshape(x1, (-1, 7, 7, 256))\n x1 = tf.layers.batch_normalization(x1, training=is_train)\n x1 = tf.maximum(alpha * x1, x1)\n # 7x7x256 now\n \n x2 = tf.layers.conv2d_transpose(x1, 128, 5, strides=2, padding='same')\n x2 = tf.layers.batch_normalization(x2, training=is_train)\n x2 = tf.maximum(alpha * x2, x2)\n # 14x14x128 now\n \n x3 = tf.layers.conv2d_transpose(x2, 64, 5, strides=2, padding='same')\n x3 = tf.layers.batch_normalization(x3, training=is_train)\n x3 = tf.maximum(alpha * x3, x3)\n # 28x28x64 now\n \n # Output layer\n logits = tf.layers.conv2d_transpose(x3, out_channel_dim, 5, strides=1, padding='same')\n # 28 x 28 x out_channel_dim\n \n output = tf.tanh(logits)\n return output\n\n\n\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE\n\"\"\"\ntests.test_generator(generator, tf)",
"Tests Passed\n"
]
],
[
[
"### Loss\nImplement `model_loss` to build the GANs for training and calculate the loss. The function should return a tuple of (discriminator loss, generator loss). Use the following functions you implemented:\n- `discriminator(images, reuse=False)`\n- `generator(z, out_channel_dim, is_train=True)`",
"_____no_output_____"
]
],
[
[
"def model_loss(input_real, input_z, out_channel_dim):\n \"\"\"\n Get the loss for the discriminator and generator\n :param input_real: Images from the real dataset\n :param input_z: Z input\n :param out_channel_dim: The number of channels in the output image\n :return: A tuple of (discriminator loss, generator loss)\n \"\"\"\n # TODO: Implement Function\n smooth = 0.1\n \n g_model = generator(input_z, out_channel_dim, is_train=True)\n d_model_real, d_logits_real = discriminator(input_real)\n d_model_fake, d_logits_fake = discriminator(g_model, reuse=True)\n\n d_loss_real = tf.reduce_mean(\n tf.nn.sigmoid_cross_entropy_with_logits(logits=d_logits_real, \n labels=tf.ones_like(d_model_real) * (1 - smooth)))\n d_loss_fake = tf.reduce_mean(\n tf.nn.sigmoid_cross_entropy_with_logits(logits=d_logits_fake, labels=tf.zeros_like(d_model_fake)))\n g_loss = tf.reduce_mean(\n tf.nn.sigmoid_cross_entropy_with_logits(logits=d_logits_fake, labels=tf.ones_like(d_model_fake)))\n\n d_loss = d_loss_real + d_loss_fake\n\n return d_loss, g_loss\n\n\n\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE\n\"\"\"\ntests.test_model_loss(model_loss)",
"Tests Passed\n"
]
],
[
[
"### Optimization\nImplement `model_opt` to create the optimization operations for the GANs. Use [`tf.trainable_variables`](https://www.tensorflow.org/api_docs/python/tf/trainable_variables) to get all the trainable variables. Filter the variables with names that are in the discriminator and generator scope names. The function should return a tuple of (discriminator training operation, generator training operation).",
"_____no_output_____"
]
],
[
[
"def model_opt(d_loss, g_loss, learning_rate, beta1):\n \"\"\"\n Get optimization operations\n :param d_loss: Discriminator loss Tensor\n :param g_loss: Generator loss Tensor\n :param learning_rate: Learning Rate Placeholder\n :param beta1: The exponential decay rate for the 1st moment in the optimizer\n :return: A tuple of (discriminator training operation, generator training operation)\n \"\"\"\n # TODO: Implement Function\n \n t_vars = tf.trainable_variables()\n d_vars = [var for var in t_vars if var.name.startswith('discriminator')]\n g_vars = [var for var in t_vars if var.name.startswith('generator')]\n\n # Optimize\n d_train_opt = tf.train.AdamOptimizer(learning_rate, beta1=beta1).minimize(d_loss, var_list=d_vars)\n g_train_opt = tf.train.AdamOptimizer(learning_rate, beta1=beta1).minimize(g_loss, var_list=g_vars)\n return d_train_opt, g_train_opt\n\n\n\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE\n\"\"\"\ntests.test_model_opt(model_opt, tf)",
"Tests Passed\n"
]
],
[
[
"## Neural Network Training\n### Show Output\nUse this function to show the current output of the generator during training. It will help you determine how well the GANs is training.",
"_____no_output_____"
]
],
[
[
"\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL\n\"\"\"\nimport numpy as np\n\ndef show_generator_output(sess, n_images, input_z, out_channel_dim, image_mode):\n \"\"\"\n Show example output for the generator\n :param sess: TensorFlow session\n :param n_images: Number of Images to display\n :param input_z: Input Z Tensor\n :param out_channel_dim: The number of channels in the output image\n :param image_mode: The mode to use for images (\"RGB\" or \"L\")\n \"\"\"\n cmap = None if image_mode == 'RGB' else 'gray'\n z_dim = input_z.get_shape().as_list()[-1]\n example_z = np.random.uniform(-1, 1, size=[n_images, z_dim])\n\n samples = sess.run(\n generator(input_z, out_channel_dim, False),\n feed_dict={input_z: example_z})\n\n images_grid = helper.images_square_grid(samples, image_mode)\n pyplot.imshow(images_grid, cmap=cmap)\n pyplot.show()",
"_____no_output_____"
]
],
[
[
"### Train\nImplement `train` to build and train the GANs. Use the following functions you implemented:\n- `model_inputs(image_width, image_height, image_channels, z_dim)`\n- `model_loss(input_real, input_z, out_channel_dim)`\n- `model_opt(d_loss, g_loss, learning_rate, beta1)`\n\nUse the `show_generator_output` to show `generator` output while you train. Running `show_generator_output` for every batch will drastically increase training time and increase the size of the notebook. It's recommended to print the `generator` output every 100 batches.",
"_____no_output_____"
]
],
[
[
"def train(epoch_count, batch_size, z_dim, learning_rate, beta1, get_batches, data_shape, data_image_mode):\n \"\"\"\n Train the GAN\n :param epoch_count: Number of epochs\n :param batch_size: Batch Size\n :param z_dim: Z dimension\n :param learning_rate: Learning Rate\n :param beta1: The exponential decay rate for the 1st moment in the optimizer\n :param get_batches: Function to get batches\n :param data_shape: Shape of the data\n :param data_image_mode: The image mode to use for images (\"RGB\" or \"L\")\n \"\"\"\n # TODO: Build Model\n class GAN:\n def __init__(self, data_shape, z_size, learning_rate, alpha=0.2, beta1=0.5):\n #tf.reset_default_graph()\n _, image_width, image_height, image_channels = data_shape\n self.input_real, self.input_z, self.learning_rate = model_inputs(image_width, image_height, \n image_channels, \n z_size)\n\n self.d_loss, self.g_loss = model_loss(self.input_real, self.input_z, image_channels)\n\n self.d_opt, self.g_opt = model_opt(self.d_loss, self.g_loss, learning_rate, 0.5)\n\n net = GAN(data_shape, z_dim, learning_rate, alpha=0.2, beta1=beta1)\n\n saver = tf.train.Saver()\n #sample_z = np.random.uniform(-1, 1, size=(50, z_dim))\n\n samples, losses = [], []\n batches = 0\n\n with tf.Session() as sess:\n sess.run(tf.global_variables_initializer())\n for e in range(epoch_count):\n for batch_images in get_batches(batch_size):\n batches += 1\n #dataset range from -0.5 to 0.5\n #batch_images = batch_images.reshape((batch_size, 784 * data_shape[3]))\n batch_images = batch_images * 2\n\n # Sample random noise for G\n batch_z = np.random.uniform(-1, 1, size=(batch_size, z_dim))\n\n # Run optimizers\n _ = sess.run(net.d_opt, feed_dict={net.input_real: batch_images, net.input_z: batch_z})\n _ = sess.run(net.g_opt, feed_dict={net.input_z: batch_z})\n\n if batches % 10 == 0:\n # At the end of each epoch, get the losses and print them out\n train_loss_d = net.d_loss.eval({net.input_z: batch_z, net.input_real: batch_images})\n train_loss_g = net.g_loss.eval({net.input_z: batch_z})\n\n print(\"Epoch {}/{}...\".format(e+1, epoch_count),\n \"Discriminator Loss: {:.4f}...\".format(train_loss_d),\n \"Generator Loss: {:.4f}\".format(train_loss_g))\n # Save losses to view after training\n losses.append((train_loss_d, train_loss_g))\n\n if batches % 100 == 0:\n _ = show_generator_output(sess, 16, net.input_z, data_shape[3], data_image_mode)\n\n saver.save(sess, './checkpoints/generator.ckpt')\n\n with open('samples.pkl', 'wb') as f:\n pkl.dump(samples, f)\n\n return losses, samples\n \n ",
"_____no_output_____"
]
],
[
[
"### MNIST\nTest your GANs architecture on MNIST. After 2 epochs, the GANs should be able to generate images that look like handwritten digits. Make sure the loss of the generator is lower than the loss of the discriminator or close to 0.",
"_____no_output_____"
]
],
[
[
"batch_size = 128\nz_dim = 100\nlearning_rate = 0.0001\nbeta1 = 0.5\n\n\n\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE\n\"\"\"\nepochs = 2\n\nmnist_dataset = helper.Dataset('mnist', glob(os.path.join(data_dir, 'mnist/*.jpg')))\nwith tf.Graph().as_default():\n train(epochs, batch_size, z_dim, learning_rate, beta1, mnist_dataset.get_batches,\n mnist_dataset.shape, mnist_dataset.image_mode)",
"Epoch 1/2... Discriminator Loss: 0.9857... Generator Loss: 0.9119\nEpoch 1/2... Discriminator Loss: 1.1420... Generator Loss: 0.9622\nEpoch 1/2... Discriminator Loss: 1.4581... Generator Loss: 0.7679\nEpoch 1/2... Discriminator Loss: 1.4356... Generator Loss: 0.7823\nEpoch 1/2... Discriminator Loss: 1.2925... Generator Loss: 0.9096\nEpoch 1/2... Discriminator Loss: 1.1594... Generator Loss: 1.0686\nEpoch 1/2... Discriminator Loss: 1.1845... Generator Loss: 1.0039\nEpoch 1/2... Discriminator Loss: 1.1674... Generator Loss: 0.9817\nEpoch 1/2... Discriminator Loss: 1.3653... Generator Loss: 0.8339\nEpoch 1/2... Discriminator Loss: 1.2765... Generator Loss: 0.8822\n"
]
],
[
[
"### CelebA\nRun your GANs on CelebA. It will take around 20 minutes on the average GPU to run one epoch. You can run the whole epoch or stop when it starts to generate realistic faces.",
"_____no_output_____"
]
],
[
[
"batch_size = 128\nz_dim = 100\nlearning_rate = 0.0001\nbeta1 = 0.5\n\n\n\"\"\"\nDON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE\n\"\"\"\nepochs = 1\n\nceleba_dataset = helper.Dataset('celeba', glob(os.path.join(data_dir, 'img_align_celeba/*.jpg')))\nwith tf.Graph().as_default():\n train(epochs, batch_size, z_dim, learning_rate, beta1, celeba_dataset.get_batches,\n celeba_dataset.shape, celeba_dataset.image_mode)",
"_____no_output_____"
]
],
[
[
"### Submitting This Project\nWhen submitting this project, make sure to run all the cells before saving the notebook. Save the notebook file as \"dlnd_face_generation.ipynb\" and save it as a HTML file under \"File\" -> \"Download as\". Include the \"helper.py\" and \"problem_unittests.py\" files in your submission.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0e29fd6dd5e9f59b1be0ccd71ebb9ee57ea18e1 | 241,843 | ipynb | Jupyter Notebook | code/soln/chap21soln.ipynb | rsmxingu/ModSimPy | 3eeb081d534a9e3943583c8d40944625ee562e43 | [
"MIT"
] | null | null | null | code/soln/chap21soln.ipynb | rsmxingu/ModSimPy | 3eeb081d534a9e3943583c8d40944625ee562e43 | [
"MIT"
] | null | null | null | code/soln/chap21soln.ipynb | rsmxingu/ModSimPy | 3eeb081d534a9e3943583c8d40944625ee562e43 | [
"MIT"
] | null | null | null | 104.19776 | 45,416 | 0.831792 | [
[
[
"# Modeling and Simulation in Python\n\nChapter 21\n\nCopyright 2017 Allen Downey\n\nLicense: [Creative Commons Attribution 4.0 International](https://creativecommons.org/licenses/by/4.0)\n",
"_____no_output_____"
]
],
[
[
"# Configure Jupyter so figures appear in the notebook\n%matplotlib inline\n\n# Configure Jupyter to display the assigned value after an assignment\n%config InteractiveShell.ast_node_interactivity='last_expr_or_assign'\n\n# import functions from the modsim.py module\nfrom modsim import *",
"_____no_output_____"
]
],
[
[
"### With air resistance",
"_____no_output_____"
],
[
"Next we'll add air resistance using the [drag equation](https://en.wikipedia.org/wiki/Drag_equation)",
"_____no_output_____"
],
[
"I'll start by getting the units we'll need from Pint.",
"_____no_output_____"
]
],
[
[
"m = UNITS.meter\ns = UNITS.second\nkg = UNITS.kilogram",
"_____no_output_____"
]
],
[
[
"Now I'll create a `Params` object to contain the quantities we need. Using a Params object is convenient for grouping the system parameters in a way that's easy to read (and double-check).",
"_____no_output_____"
]
],
[
[
"params = Params(height = 381 * m,\n v_init = 0 * m / s,\n g = 9.8 * m/s**2,\n mass = 2.5e-3 * kg,\n diameter = 19e-3 * m,\n rho = 1.2 * kg/m**3,\n v_term = 18 * m / s)",
"_____no_output_____"
]
],
[
[
"Now we can pass the `Params` object `make_system` which computes some additional parameters and defines `init`.\n\n`make_system` uses the given radius to compute `area` and the given `v_term` to compute the drag coefficient `C_d`.",
"_____no_output_____"
]
],
[
[
"def make_system(params):\n \"\"\"Makes a System object for the given conditions.\n \n params: Params object\n \n returns: System object\n \"\"\"\n unpack(params)\n \n area = np.pi * (diameter/2)**2\n C_d = 2 * mass * g / (rho * area * v_term**2)\n init = State(y=height, v=v_init)\n t_end = 30 * s\n \n return System(params, area=area, C_d=C_d, \n init=init, t_end=t_end)",
"_____no_output_____"
]
],
[
[
"Let's make a `System`",
"_____no_output_____"
]
],
[
[
"system = make_system(params)",
"_____no_output_____"
]
],
[
[
"Here's the slope function, including acceleration due to gravity and drag.",
"_____no_output_____"
]
],
[
[
"def slope_func(state, t, system):\n \"\"\"Compute derivatives of the state.\n \n state: position, velocity\n t: time\n system: System object\n \n returns: derivatives of y and v\n \"\"\"\n y, v = state\n unpack(system)\n \n f_drag = rho * v**2 * C_d * area / 2\n a_drag = f_drag / mass\n \n dydt = v\n dvdt = -g + a_drag\n \n return dydt, dvdt",
"_____no_output_____"
]
],
[
[
"As always, let's test the slope function with the initial conditions.",
"_____no_output_____"
]
],
[
[
"slope_func(system.init, 0, system)",
"_____no_output_____"
]
],
[
[
"We can use the same event function as last time.",
"_____no_output_____"
]
],
[
[
"def event_func(state, t, system):\n \"\"\"Return the height of the penny above the sidewalk.\n \"\"\"\n y, v = state\n return y",
"_____no_output_____"
]
],
[
[
"And then run the simulation.",
"_____no_output_____"
]
],
[
[
"results, details = run_ode_solver(system, slope_func, events=event_func)\ndetails.message",
"_____no_output_____"
]
],
[
[
"Here are the results.",
"_____no_output_____"
]
],
[
[
"results",
"_____no_output_____"
]
],
[
[
"The final height is close to 0, as expected.\n\nInterestingly, the final velocity is not exactly terminal velocity, which suggests that there are some numerical errors.\n\nWe can get the flight time from `results`.",
"_____no_output_____"
]
],
[
[
"t_sidewalk = get_last_label(results)",
"_____no_output_____"
]
],
[
[
"Here's the plot of position as a function of time.",
"_____no_output_____"
]
],
[
[
"def plot_position(results):\n plot(results.y)\n decorate(xlabel='Time (s)',\n ylabel='Position (m)')\n \nplot_position(results)\nsavefig('figs/chap09-fig02.pdf')",
"Saving figure to file figs/chap09-fig02.pdf\n"
]
],
[
[
"And velocity as a function of time:",
"_____no_output_____"
]
],
[
[
"def plot_velocity(results):\n plot(results.v, color='C1', label='v')\n \n decorate(xlabel='Time (s)',\n ylabel='Velocity (m/s)')\n \nplot_velocity(results)",
"_____no_output_____"
]
],
[
[
"From an initial velocity of 0, the penny accelerates downward until it reaches terminal velocity; after that, velocity is constant.",
"_____no_output_____"
],
[
"**Exercise:** Run the simulation with an initial velocity, downward, that exceeds the penny's terminal velocity. Hint: You can create a new `Params` object based on an existing one, like this:\n\n`params = Params(params, v_init = -30 * m / s)`\n\nWhat do you expect to happen? Plot velocity and position as a function of time, and see if they are consistent with your prediction.",
"_____no_output_____"
]
],
[
[
"# Solution\n\nparams = Params(params, v_init = -30 * m / s)\nsystem = make_system(params)\nresults, details = run_ode_solver(system, slope_func, events=event_func)\ndetails.message",
"_____no_output_____"
],
[
"plot_position(results)",
"_____no_output_____"
],
[
"# Solution\n\nplot_velocity(results)",
"_____no_output_____"
]
],
[
[
"**Exercise:** Suppose we drop a quarter from the Empire State Building and find that its flight time is 19.1 seconds. Use this measurement to estimate the terminal velocity.\n\n1. You can get the relevant dimensions of a quarter from https://en.wikipedia.org/wiki/Quarter_(United_States_coin).\n\n2. Create a `Params` object with the system parameters. We don't know `v_term`, so we'll start with the inital guess `v_term = 18 * m / s`.\n\n3. Use `make_system` to create a `System` object.\n\n4. Call `run_ode_solver` to simulate the system. How does the flight time of the simulation compare to the measurement?\n\n5. Try a few different values of `t_term` and see if you can get the simulated flight time close to 19.1 seconds.\n\n6. Optionally, write an error function and use `fsolve` to improve your estimate.\n\n7. Use your best estimate of `v_term` to compute `C_d`.\n\nNote: I fabricated the observed flight time, so don't take the results of this exercise too seriously.",
"_____no_output_____"
]
],
[
[
"# Solution\n\n# Here's a `Params` object with the dimensions of a quarter,\n# the observed flight time and our initial guess for `v_term`\n\nparams = Params(params,\n mass = 5.67e-3 * kg,\n diameter = 24.26e-3 * m,\n v_term = 18 * m / s,\n flight_time = 19.1 * s)",
"_____no_output_____"
],
[
"# Solution\n\n# Now we can make a `System` object\n\nsystem = make_system(params)",
"_____no_output_____"
],
[
"# Solution\n\n# And run the simulation\n\nresults, details = run_ode_solver(system, slope_func, events=event_func)\ndetails",
"_____no_output_____"
],
[
"# Solution\n\n# And get the flight time\n\nflight_time = get_last_label(results) * s",
"_____no_output_____"
],
[
"# Solution\n\n# The flight time is a little long, so we could increase `v_term` and try again.\n\n# Or we could write an error function\n\ndef error_func(v_term, params):\n \"\"\"Final height as a function of C_d.\n \n C_d: drag coefficient\n params: Params object\n \n returns: height in m\n \"\"\"\n params = Params(params, v_term=v_term)\n system = make_system(params)\n results, details = run_ode_solver(system, slope_func, events=event_func)\n flight_time = get_last_label(results) * s\n return flight_time - params.flight_time",
"_____no_output_____"
],
[
"# Solution\n\n# We can test the error function like this\n\nguess = 18 * m / s\nerror_func(guess, params)",
"_____no_output_____"
],
[
"# Solution\n\n# Now we can use `fsolve` to find the value of `v_term` that yields the measured flight time.\n\nsolution = fsolve(error_func, guess, params)\nv_term_solution = solution[0] * m/s",
"_____no_output_____"
],
[
"# Solution\n\n# Plugging in the estimated value, we can use `make_system` to compute `C_d`\n\nparams = Params(params, v_term=v_term_solution)\nsystem = make_system(params)\nsystem.C_d",
"_____no_output_____"
]
],
[
[
"### Bungee jumping",
"_____no_output_____"
],
[
"Suppose you want to set the world record for the highest \"bungee dunk\", [as shown in this video](https://www.youtube.com/watch?v=UBf7WC19lpw). Since the record is 70 m, let's design a jump for 80 m.\n\nWe'll make the following modeling assumptions:\n\n1. Initially the bungee cord hangs from a crane with the attachment point 80 m above a cup of tea.\n\n2. Until the cord is fully extended, it applies no force to the jumper. It turns out this might not be a good assumption; we will revisit it.\n\n3. After the cord is fully extended, it obeys [Hooke's Law](https://en.wikipedia.org/wiki/Hooke%27s_law); that is, it applies a force to the jumper proportional to the extension of the cord beyond its resting length.\n\n4. The jumper is subject to drag force proportional to the square of their velocity, in the opposite of their direction of motion.\n\nOur objective is to choose the length of the cord, `L`, and its spring constant, `k`, so that the jumper falls all the way to the tea cup, but no farther! \n\nFirst I'll create a `Param` object to contain the quantities we'll need:\n\n1. Let's assume that the jumper's mass is 75 kg.\n\n2. With a terminal velocity of 60 m/s.\n\n3. The length of the bungee cord is `L = 40 m`.\n\n4. The spring constant of the cord is `k = 20 N / m` when the cord is stretched, and 0 when it's compressed.\n",
"_____no_output_____"
]
],
[
[
"m = UNITS.meter\ns = UNITS.second\nkg = UNITS.kilogram\nN = UNITS.newton",
"_____no_output_____"
],
[
"params = Params(y_attach = 80 * m,\n v_init = 0 * m / s,\n g = 9.8 * m/s**2,\n mass = 75 * kg,\n area = 1 * m**2,\n rho = 1.2 * kg/m**3,\n v_term = 60 * m / s,\n L = 25 * m,\n k = 40 * N / m)",
"_____no_output_____"
]
],
[
[
"Now here's a version of `make_system` that takes a `Params` object as a parameter.\n\n`make_system` uses the given value of `v_term` to compute the drag coefficient `C_d`.",
"_____no_output_____"
]
],
[
[
"def make_system(params):\n \"\"\"Makes a System object for the given params.\n \n params: Params object\n \n returns: System object\n \"\"\"\n unpack(params)\n \n C_d = 2 * mass * g / (rho * area * v_term**2)\n init = State(y=y_attach, v=v_init)\n t_end = 30 * s\n\n return System(params, C_d=C_d, \n init=init, t_end=t_end)",
"_____no_output_____"
]
],
[
[
"Let's make a `System`",
"_____no_output_____"
]
],
[
[
"system = make_system(params)\nsystem",
"_____no_output_____"
]
],
[
[
"`spring_force` computes the force of the cord on the jumper:",
"_____no_output_____"
]
],
[
[
"def spring_force(y, system):\n \"\"\"Computes the force of the bungee cord on the jumper:\n \n y: height of the jumper\n \n Uses these variables from system|\n y_attach: height of the attachment point\n L: resting length of the cord\n k: spring constant of the cord\n \n returns: force in N\n \"\"\"\n unpack(system)\n distance_fallen = y_attach - y\n if distance_fallen <= L:\n return 0 * N\n \n extension = distance_fallen - L\n f_spring = k * extension\n return f_spring",
"_____no_output_____"
]
],
[
[
"The spring force is 0 until the cord is fully extended. When it is extended 1 m, the spring force is 40 N. ",
"_____no_output_____"
]
],
[
[
"spring_force(80*m, system)",
"_____no_output_____"
],
[
"spring_force(55*m, system)",
"_____no_output_____"
],
[
"spring_force(54*m, system)",
"_____no_output_____"
]
],
[
[
"`drag_force` computes drag as a function of velocity:",
"_____no_output_____"
]
],
[
[
"def drag_force(v, system):\n \"\"\"Computes drag force in the opposite direction of `v`.\n \n v: velocity\n \n returns: drag force\n \"\"\"\n unpack(system)\n f_drag = -np.sign(v) * rho * v**2 * C_d * area / 2\n return f_drag",
"_____no_output_____"
]
],
[
[
"Now here's the slope function:",
"_____no_output_____"
]
],
[
[
"def slope_func(state, t, system):\n \"\"\"Compute derivatives of the state.\n \n state: position, velocity\n t: time\n system: System object containing g, rho,\n C_d, area, and mass\n \n returns: derivatives of y and v\n \"\"\"\n y, v = state\n unpack(system)\n \n a_drag = drag_force(v, system) / mass\n a_spring = spring_force(y, system) / mass\n dvdt = -g + a_drag + a_spring\n \n return v, dvdt",
"_____no_output_____"
]
],
[
[
"As always, let's test the slope function with the initial params.",
"_____no_output_____"
]
],
[
[
"slope_func(system.init, 0, system)",
"_____no_output_____"
]
],
[
[
"And then run the simulation.",
"_____no_output_____"
]
],
[
[
"ts = linspace(0, system.t_end, 301)\nresults, details = run_ode_solver(system, slope_func, t_eval=ts)\ndetails",
"_____no_output_____"
]
],
[
[
"Here's the plot of position as a function of time.",
"_____no_output_____"
]
],
[
[
"plot_position(results)",
"_____no_output_____"
]
],
[
[
"After reaching the lowest point, the jumper springs back almost to almost 70 m, and oscillates several times. That looks like more osciallation that we expect from an actual jump, which suggests that there some dissipation of energy in the real world that is not captured in our model. To improve the model, that might be a good thing to investigate.\n\nBut since we are primarily interested in the initial descent, the model might be good enough for now.\n\nWe can use `min` to find the lowest point:",
"_____no_output_____"
]
],
[
[
"min(results.y)",
"_____no_output_____"
]
],
[
[
"At the lowest point, the jumper is still too high, so we'll need to increase `L` or decrease `k`.",
"_____no_output_____"
],
[
"Here's velocity as a function of time:",
"_____no_output_____"
]
],
[
[
"plot_velocity(results)",
"_____no_output_____"
],
[
"subplot(1, 2, 1)\nplot_position(results)\n\nsubplot(1, 2, 2)\nplot_velocity(results)\n\nsavefig('figs/chap09-fig03.pdf')",
"Saving figure to file figs/chap09-fig03.pdf\n"
]
],
[
[
"Although we compute acceleration inside the slope function, we don't get acceleration as a result from `run_ode_solver`.\n\nWe can approximate it by computing the numerical derivative of `ys`:",
"_____no_output_____"
]
],
[
[
"a = gradient(results.v)\nplot(a)\ndecorate(xlabel='Time (s)',\n ylabel='Acceleration (m/$s^2$)')",
"_____no_output_____"
]
],
[
[
"And we can compute the maximum acceleration the jumper experiences:",
"_____no_output_____"
]
],
[
[
"max_acceleration = max(a) * m/s**2",
"_____no_output_____"
]
],
[
[
"Relative to the acceleration of gravity, the jumper \"pulls\" about \"1.7 g's\".",
"_____no_output_____"
]
],
[
[
"max_acceleration / g",
"_____no_output_____"
]
],
[
[
"### Solving for length\n\nAssuming that `k` is fixed, let's find the length `L` that makes the minimum altitude of the jumper exactly 0.\n\nHere's the error function:",
"_____no_output_____"
]
],
[
[
"def error_func(L, params):\n \"\"\"Minimum height as a function of length.\n \n length: length in m\n params: Params object\n \n returns: height in m\n \"\"\"\n params = Params(params, L=L)\n system = make_system(params)\n\n ts = linspace(0, system.t_end, 201)\n results, details = run_ode_solver(system, slope_func, t_eval=ts)\n min_height = min(results.y)\n return min_height",
"_____no_output_____"
]
],
[
[
"Let's test it with the same initial guess, `L = 100 m`:",
"_____no_output_____"
]
],
[
[
"guess = 150 * m\nerror_func(guess, params)",
"_____no_output_____"
]
],
[
[
"And find the value of `L` we need for the world record jump:",
"_____no_output_____"
]
],
[
[
"solution = fsolve(error_func, guess, params)",
"_____no_output_____"
]
],
[
[
"**Optional:** Search for the combination of length and spring constant that yields minimum height 0 while minimizing peak acceleration.",
"_____no_output_____"
]
],
[
[
"# Solution\n\nks = np.linspace(1, 31, 11) * N / m\n\nfor k in ks:\n guess = 250 * m\n params = Params(params, k=k)\n solution = fsolve(error_func, guess, params)\n L = solution[0] * m\n params = Params(params, L=L)\n system = make_system(params)\n results, details = run_ode_solver(system, slope_func, t_eval=ts)\n a = gradient(results.v)\n g_max = max(a) * m/s**2 / g\n print(k, L, g_max)",
"1.0 newton / meter -483.1429396908503 meter 0.17584604083333497 dimensionless\n4.0 newton / meter 70.54780511680855 meter 0.6912121734141764 dimensionless\n7.0 newton / meter 161.1541432460714 meter 1.0914235802924046 dimensionless\n10.0 newton / meter 202.17405449658656 meter 1.435864054193015 dimensionless\n13.0 newton / meter 227.86005853679828 meter 1.7065736435639631 dimensionless\n16.0 newton / meter 243.97203228964753 meter 1.9887775175403402 dimensionless\n19.0 newton / meter 257.30496940959637 meter 2.204636494160503 dimensionless\n22.0 newton / meter 265.7902447183497 meter 2.4453581682255328 dimensionless\n25.0 newton / meter 274.91575508305107 meter 2.607142045194042 dimensionless\n28.0 newton / meter 281.1809127600765 meter 2.799398587830593 dimensionless\n31.0 newton / meter 286.360055638525 meter 2.994252693587456 dimensionless\n"
]
],
[
[
"**Optional exercise:** This model neglects the weight of the bungee cord, which might be non-negligible. Implement the [model described here](http://iopscience.iop.org/article/10.1088/0031-9120/45/1/007) and see how different it is from our simplified model. ",
"_____no_output_____"
],
[
"### Under the hood\n\nThe gradient function in `modsim.py` adapts the NumPy function of the same name so it works with `Series` objects.\n",
"_____no_output_____"
]
],
[
[
"%psource gradient",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
d0e2a0c6f4a3fa82489003c4d0edb2e0fe262af9 | 7,892 | ipynb | Jupyter Notebook | Analysis/Notebooks/Spiral Dataset/2_Measure_Curvature.ipynb | brouhardlab/kappa | 2e74530e54fc02e08e03e56a4a33ece6c7ce9941 | [
"MIT"
] | 7 | 2017-11-29T15:09:50.000Z | 2020-12-04T11:08:17.000Z | Analysis/Notebooks/Spiral Dataset/2_Measure_Curvature.ipynb | brouhardlab/kappa | 2e74530e54fc02e08e03e56a4a33ece6c7ce9941 | [
"MIT"
] | 18 | 2017-11-17T19:20:10.000Z | 2020-08-16T16:33:31.000Z | Analysis/Notebooks/Spiral Dataset/2_Measure_Curvature.ipynb | brouhardlab/kappa | 2e74530e54fc02e08e03e56a4a33ece6c7ce9941 | [
"MIT"
] | 5 | 2017-10-13T02:24:17.000Z | 2022-03-16T01:14:59.000Z | 34.313043 | 182 | 0.562215 | [
[
[
"**Important**: This notebook is different from the other as it directly calls **ImageJ Kappa plugin** using the [`scyjava` ImageJ brige](https://github.com/scijava/scyjava).\n\nSince Kappa uses ImageJ1 features, you might not be able to run this notebook on an headless machine (need to be tested).",
"_____no_output_____"
]
],
[
[
"from pathlib import Path\nimport pandas as pd\nimport numpy as np\nfrom tqdm.auto import tqdm\n\nimport sys; sys.path.append(\"../../\")\nimport pykappa\n\n# Init ImageJ with Fiji plugins\n# It can take a while if Java artifacts are not yet cached.\nimport imagej\njava_deps = []\njava_deps.append('org.scijava:Kappa:1.7.1')\nij = imagej.init(\"+\".join(java_deps), headless=False)\n\nimport jnius\n\n# Load Java classes\nKappaFrame = jnius.autoclass('sc.fiji.kappa.gui.KappaFrame')\nCurvesExporter = jnius.autoclass('sc.fiji.kappa.gui.CurvesExporter')\n\n# Load ImageJ services\ndsio = ij.context.getService(jnius.autoclass('io.scif.services.DatasetIOService'))\ndsio = jnius.cast('io.scif.services.DatasetIOService', dsio)\n\n# Set data path\ndata_dir = Path(\"/home/hadim/.data/Postdoc/Kappa/spiral_curve_SDM/\")\n\n# Pixel size used when fixed\nfixed_pixel_size = 0.16\n\n# Used to select pixels around the initialization curves\nbase_radius_um = 1.6\n\nenable_control_points_adjustment = True\n\n# \"Point Distance Minimization\" or \"Squared Distance Minimization\"\nif '_SDM' in data_dir.name:\n fitting_algorithm = \"Squared Distance Minimization\"\nelse:\n fitting_algorithm = \"Point Distance Minimization\"\nfitting_algorithm",
"_____no_output_____"
],
[
"experiment_names = ['variable_snr', 'variable_initial_position', 'variable_pixel_size', 'variable_psf_size']\nexperiment_names = ['variable_psf_size']\n\nfor experiment_name in tqdm(experiment_names, total=len(experiment_names)):\n \n experiment_path = data_dir / experiment_name\n fnames = sorted(list(experiment_path.glob(\"*.tif\")))\n n = len(fnames)\n\n for fname in tqdm(fnames, total=n, leave=False):\n \n tqdm.write(str(fname))\n \n kappa_path = fname.with_suffix(\".kapp\")\n assert kappa_path.exists(), f'{kappa_path} does not exist.'\n\n curvatures_path = fname.with_suffix(\".csv\")\n \n if not curvatures_path.is_file():\n\n frame = KappaFrame(ij.context)\n frame.getKappaMenubar().openImageFile(str(fname))\n frame.resetCurves()\n frame.getKappaMenubar().loadCurveFile(str(kappa_path))\n frame.getCurves().setAllSelected()\n\n # Compute threshold according to the image\n dataset = dsio.open(str(fname))\n mean = ij.op().stats().mean(dataset).getRealDouble() \n std = ij.op().stats().stdDev(dataset).getRealDouble()\n threshold = int(mean + std * 2)\n\n # Used fixed pixel size or the one in the filename\n if fname.stem.startswith('pixel_size'):\n pixel_size = float(fname.stem.split(\"_\")[-2])\n if experiment_name == 'variable_psf_size':\n pixel_size = 0.01\n else:\n pixel_size = fixed_pixel_size\n \n base_radius = int(np.round(base_radius_um / pixel_size))\n\n # Set curve fitting parameters\n frame.setEnableCtrlPtAdjustment(enable_control_points_adjustment)\n frame.setFittingAlgorithm(fitting_algorithm)\n frame.getInfoPanel().thresholdRadiusSpinner.setValue(ij.py.to_java(base_radius))\n frame.getInfoPanel().thresholdSlider.setValue(threshold)\n frame.getInfoPanel().updateConversionField(str(pixel_size))\n \n # Fit the curves\n frame.fitCurves()\n \n # Save fitted curves\n frame.getKappaMenubar().saveCurveFile(str(fname.with_suffix(\".FITTED.kapp\")))\n\n # Export results\n exporter = CurvesExporter(frame)\n exporter.exportToFile(str(curvatures_path), False)\n\n # Remove duplicate rows during CSV export.\n # See https://github.com/brouhardlab/Kappa/issues/12\n df = pd.read_csv(curvatures_path)\n df = df.drop_duplicates()\n df.to_csv(curvatures_path)",
"_____no_output_____"
],
[
"0.13**2",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d0e2a8cc3ddc3dded5f3bbf0e3feb62937c7bb6f | 1,373 | ipynb | Jupyter Notebook | Concepts/Fundamentals/turtleview.ipynb | RobInLabUJI/ROSIN-Tutorials | 8886865aab83c32cafa73b0ae0e54b27fa4dc210 | [
"Apache-2.0"
] | 11 | 2019-08-16T08:34:22.000Z | 2021-09-21T20:34:18.000Z | Concepts/Fundamentals/turtleview.ipynb | RobInLabUJI/ROSIN-Tutorials | 8886865aab83c32cafa73b0ae0e54b27fa4dc210 | [
"Apache-2.0"
] | null | null | null | Concepts/Fundamentals/turtleview.ipynb | RobInLabUJI/ROSIN-Tutorials | 8886865aab83c32cafa73b0ae0e54b27fa4dc210 | [
"Apache-2.0"
] | 2 | 2020-09-16T12:17:56.000Z | 2021-04-16T03:54:05.000Z | 21.123077 | 199 | 0.569556 | [
[
[
"### turtleview",
"_____no_output_____"
],
[
"You need to run the following code for viewing the simulator output in the notebook:",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport nbturtle\nnbturtle.plot()",
"_____no_output_____"
]
],
[
[
"NOTE: each time, the turtle may look different. Don't worry about it - there are [many types of turtle](http://wiki.ros.org/Distributions#Current_Distribution_Releases) and yours is a surprise!",
"_____no_output_____"
],
[
"No turtle shown? We are working on this bug, in the meantime you may try to [reset the simulation](reset.ipynb).",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d0e2a8d8752d11250e96a6de9aa50778f369eb6b | 125,248 | ipynb | Jupyter Notebook | tutorials/W1D1_BasicsAndPytorch/student/W1D1_Tutorial1.ipynb | tmachnitzki/course-content-dl | d84b06d42338f294c8a3d293c4f631ac33769518 | [
"CC-BY-4.0",
"BSD-3-Clause"
] | 1 | 2021-08-04T03:06:23.000Z | 2021-08-04T03:06:23.000Z | tutorials/W1D1_BasicsAndPytorch/student/W1D1_Tutorial1.ipynb | tmachnitzki/course-content-dl | d84b06d42338f294c8a3d293c4f631ac33769518 | [
"CC-BY-4.0",
"BSD-3-Clause"
] | null | null | null | tutorials/W1D1_BasicsAndPytorch/student/W1D1_Tutorial1.ipynb | tmachnitzki/course-content-dl | d84b06d42338f294c8a3d293c4f631ac33769518 | [
"CC-BY-4.0",
"BSD-3-Clause"
] | null | null | null | 32.405692 | 571 | 0.565518 | [
[
[
"<a href=\"https://colab.research.google.com/github/NeuromatchAcademy/course-content-dl/blob/main/tutorials/W1D1_BasicsAndPytorch/student/W1D1_Tutorial1.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Tutorial 1: PyTorch\n**Week 1, Day 1: Basics and PyTorch**\n\n**By Neuromatch Academy**\n\n\n__Content creators:__ Shubh Pachchigar, Vladimir Haltakov, Matthew Sargent, Konrad Kording\n\n__Content reviewers:__ Deepak Raya, Siwei Bai, Kelson Shilling-Scrivo\n\n__Content editors:__ Anoop Kulkarni, Spiros Chavlis\n\n__Production editors:__ Arush Tagade, Spiros Chavlis",
"_____no_output_____"
],
[
"**Our 2021 Sponsors, including Presenting Sponsor Facebook Reality Labs**\n\n<p align='center'><img src='https://github.com/NeuromatchAcademy/widgets/blob/master/sponsors.png?raw=True'/></p>",
"_____no_output_____"
],
[
"---\n# Tutorial Objectives\n\nThen have a few specific objectives for this tutorial:\n* Learn about PyTorch and tensors\n* Tensor Manipulations\n* Data Loading\n* GPUs and Cuda Tensors\n* Train NaiveNet\n* Get to know your pod\n* Start thinking about the course as a whole",
"_____no_output_____"
]
],
[
[
"# @title Tutorial slides\n\n# @markdown These are the slides for the videos in this tutorial today\nfrom IPython.display import IFrame\nIFrame(src=f\"https://mfr.ca-1.osf.io/render?url=https://osf.io/wcjrv/?direct%26mode=render%26action=download%26mode=render\", width=854, height=480)",
"_____no_output_____"
]
],
[
[
"---\n# Setup",
"_____no_output_____"
],
[
"Throughout your Neuromatch tutorials, most (probably all!) notebooks contain setup cells. These cells will import the required Python packages (e.g., PyTorch, NumPy); set global or environment variables, and load in helper functions for things like plotting. In some tutorials, you will notice that we install some dependencies even if they are preinstalled on google colab or kaggle. This happens because we have added automation to our repository through [GitHub Actions](https://docs.github.com/en/actions/learn-github-actions/introduction-to-github-actions).\n\nBe sure to run all of the cells in the setup section. Feel free to expand them and have a look at what you are loading in, but you should be able to fulfill the learning objectives of every tutorial without having to look at these cells.\n\nIf you start building your own projects built on this code base we highly recommend looking at them in more detail.",
"_____no_output_____"
]
],
[
[
"# @title Install dependencies\n!pip install pandas --quiet\n!pip install git+https://github.com/NeuromatchAcademy/evaltools --quiet\n\nfrom evaltools.airtable import AirtableForm",
"_____no_output_____"
],
[
"# Imports\nimport time\nimport torch\nimport random\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n\nfrom torch import nn\nfrom torchvision import datasets\nfrom torchvision.transforms import ToTensor\nfrom torch.utils.data import DataLoader",
"_____no_output_____"
],
[
"# @title Figure Settings\nimport ipywidgets as widgets\n%config InlineBackend.figure_format = 'retina'\nplt.style.use(\"https://raw.githubusercontent.com/NeuromatchAcademy/content-creation/main/nma.mplstyle\")",
"_____no_output_____"
],
[
"# @title Helper Functions\n\natform = AirtableForm('appn7VdPRseSoMXEG','W1D1_T1','https://portal.neuromatchacademy.org/api/redirect/to/97e94a29-0b3a-4e16-9a8d-f6838a5bd83d')\n\n\ndef checkExercise1(A, B, C, D):\n \"\"\"\n Helper function for checking exercise.\n\n Args:\n A: torch.Tensor\n B: torch.Tensor\n C: torch.Tensor\n D: torch.Tensor\n Returns:\n Nothing.\n \"\"\"\n errors = []\n # TODO better errors and error handling\n if not torch.equal(A.to(int),torch.ones(20, 21).to(int)):\n errors.append(f\"Got: {A} \\n Expected: {torch.ones(20, 21)} (shape: {torch.ones(20, 21).shape})\")\n if not np.array_equal( B.numpy(),np.vander([1, 2, 3], 4)):\n errors.append(\"B is not a tensor containing the elements of Z \")\n if C.shape != (20, 21):\n errors.append(\"C is not the correct shape \")\n if not torch.equal(D, torch.arange(4, 41, step=2)):\n errors.append(\"D does not contain the correct elements\")\n\n if errors == []:\n print(\"All correct!\")\n\n else:\n [print(e) for e in errors]\n\n\ndef timeFun(f, dim, iterations, device='cpu'):\n iterations = iterations\n t_total = 0\n for _ in range(iterations):\n start = time.time()\n f(dim, device)\n end = time.time()\n t_total += end - start\n\n if device == 'cpu':\n print(f\"time taken for {iterations} iterations of {f.__name__}({dim}, {device}): {t_total:.5f}\")\n else:\n print(f\"time taken for {iterations} iterations of {f.__name__}({dim}, {device}): {t_total:.5f}\")",
"_____no_output_____"
]
],
[
[
"**Important note: Google Colab users**\n\n*Scratch Code Cells*\n\nIf you want to quickly try out something or take a look at the data you can use scratch code cells. They allow you to run Python code, but will not mess up the structure of your notebook.\n\nTo open a new scratch cell go to *Insert* → *Scratch code cell*.",
"_____no_output_____"
],
[
"# Section 1: Welcome to Neuromatch Deep learning course\n\n*Time estimate: ~25mins*",
"_____no_output_____"
]
],
[
[
"# @title Video 1: Welcome and History\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1Av411n7oL\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"ca21SNqt78I\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\n# add timing\natform.add_event('Video 1: Welcome and History')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"*This will be an intensive 3 week adventure. We will all learn Deep Learning. In a group. Groups need standards. Read our \n[Code of Conduct](https://docs.google.com/document/d/1eHKIkaNbAlbx_92tLQelXnicKXEcvFzlyzzeWjEtifM/edit?usp=sharing).\n",
"_____no_output_____"
]
],
[
[
"# @title Video 2: Why DL is cool\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1gf4y1j7UZ\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"l-K6495BN-4\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\n# add timing to airtable\natform.add_event('Video 2: Why DL is cool')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"**Describe what you hope to get out of this course in about 100 words.**",
"_____no_output_____"
],
[
"---\n# Section 2: The Basics of PyTorch\n\n*Time estimate: ~2 hours 05 mins*",
"_____no_output_____"
],
[
"PyTorch is a Python-based scientific computing package targeted at two sets of\naudiences:\n\n- A replacement for NumPy to use the power of GPUs\n- A deep learning platform that provides significant flexibility\n and speed\n\nAt its core, PyTorch provides a few key features:\n\n- A multidimensional [Tensor](https://pytorch.org/docs/stable/tensors.html) object, similar to [NumPy Array](https://numpy.org/doc/stable/reference/generated/numpy.ndarray.html) but with GPU acceleration.\n- An optimized **autograd** engine for automatically computing derivatives.\n- A clean, modular API for building and deploying **deep learning models**.\n\nYou can find more information about PyTorch in the appendix.",
"_____no_output_____"
],
[
"## Section 2.1: Creating Tensors\n",
"_____no_output_____"
]
],
[
[
"# @title Video 3: Making Tensors\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1Rw411d7Uy\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"jGKd_4tPGrw\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\n# add timing to airtable\natform.add_event('Video 3: Making Tensors')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"There are various ways of creating tensors, and when doing any real deep learning project we will usually have to do so.",
"_____no_output_____"
],
[
"**Construct tensors directly:**\n\n---\n\n",
"_____no_output_____"
]
],
[
[
"# we can construct a tensor directly from some common python iterables,\n# such as list and tuple nested iterables can also be handled as long as the\n# dimensions make sense\n\n# tensor from a list\na = torch.tensor([0, 1, 2])\n\n#tensor from a tuple of tuples\nb = ((1.0, 1.1), (1.2, 1.3))\nb = torch.tensor(b)\n\n# tensor from a numpy array\nc = np.ones([2, 3])\nc = torch.tensor(c)\n\nprint(f\"Tensor a: {a}\")\nprint(f\"Tensor b: {b}\")\nprint(f\"Tensor c: {c}\")",
"_____no_output_____"
]
],
[
[
"**Some common tensor constructors:**\n\n---",
"_____no_output_____"
]
],
[
[
"# the numerical arguments we pass to these constructors\n# determine the shape of the output tensor\n\nx = torch.ones(5, 3)\ny = torch.zeros(2)\nz = torch.empty(1, 1, 5)\nprint(f\"Tensor x: {x}\")\nprint(f\"Tensor y: {y}\")\nprint(f\"Tensor z: {z}\")",
"_____no_output_____"
]
],
[
[
"Notice that ```.empty()``` does not return zeros, but seemingly random small numbers. Unlike ```.zeros()```, which initialises the elements of the tensor with zeros, ```.empty()``` just allocates the memory. It is hence a bit faster if you are looking to just create a tensor.",
"_____no_output_____"
],
[
"**Creating random tensors and tensors like other tensors:**\n\n---",
"_____no_output_____"
]
],
[
[
"# there are also constructors for random numbers\n\n# uniform distribution\na = torch.rand(1, 3)\n\n# normal distribution\nb = torch.randn(3, 4)\n\n# there are also constructors that allow us to construct\n# a tensor according to the above constructors, but with\n# dimensions equal to another tensor\n\nc = torch.zeros_like(a)\nd = torch.rand_like(c)\n\nprint(f\"Tensor a: {a}\")\nprint(f\"Tensor b: {b}\")\nprint(f\"Tensor c: {c}\")\nprint(f\"Tensor d: {d}\")",
"_____no_output_____"
]
],
[
[
"*Reproducibility*: \n\n- PyTorch random number generator: You can use `torch.manual_seed()` to seed the RNG for all devices (both CPU and CUDA)\n\n```python\nimport torch\ntorch.manual_seed(0)\n```\n- For custom operators, you might need to set python seed as well:\n\n```python\nimport random\nrandom.seed(0)\n```\n\n- Random number generators in other libraries\n\n```python\nimport numpy as np\nnp.random.seed(0)\n```\n",
"_____no_output_____"
],
[
"Here, we define for you a function called `set_seed` that does the job for you!",
"_____no_output_____"
]
],
[
[
"def set_seed(seed=None, seed_torch=True):\n \"\"\"\n Function that controls randomness. NumPy and random modules must be imported.\n\n Args:\n seed : Integer\n A non-negative integer that defines the random state. Default is `None`.\n seed_torch : Boolean\n If `True` sets the random seed for pytorch tensors, so pytorch module\n must be imported. Default is `True`.\n\n Returns:\n Nothing.\n \"\"\"\n if seed is None:\n seed = np.random.choice(2 ** 32)\n random.seed(seed)\n np.random.seed(seed)\n if seed_torch:\n torch.manual_seed(seed)\n torch.cuda.manual_seed_all(seed)\n torch.cuda.manual_seed(seed)\n torch.backends.cudnn.benchmark = False\n torch.backends.cudnn.deterministic = True\n\n print(f'Random seed {seed} has been set.')",
"_____no_output_____"
]
],
[
[
"Now, let's use the `set_seed` function in the previous example. Execute the cell multiple times to verify that the numbers printed are always the same.",
"_____no_output_____"
]
],
[
[
"def simplefun(seed=True, my_seed=None):\n if seed:\n set_seed(seed=my_seed)\n\n # uniform distribution\n a = torch.rand(1, 3)\n # normal distribution\n b = torch.randn(3, 4)\n\n print(\"Tensor a: \", a)\n print(\"Tensor b: \", b)",
"_____no_output_____"
],
[
"simplefun(seed=True, my_seed=0) # Turn `seed` to `False` or change `my_seed`",
"_____no_output_____"
]
],
[
[
"**Numpy-like number ranges:**\n---\nThe ```.arange()``` and ```.linspace()``` behave how you would expect them to if you are familar with numpy.",
"_____no_output_____"
]
],
[
[
"a = torch.arange(0, 10, step=1)\nb = np.arange(0, 10, step=1)\n\nc = torch.linspace(0, 5, steps=11)\nd = np.linspace(0, 5, num=11)\n\nprint(f\"Tensor a: {a}\\n\")\nprint(f\"Numpy array b: {b}\\n\")\nprint(f\"Tensor c: {c}\\n\")\nprint(f\"Numpy array d: {d}\\n\")",
"_____no_output_____"
]
],
[
[
"### Coding Exercise 2.1: Creating Tensors\n\nBelow you will find some incomplete code. Fill in the missing code to construct the specified tensors.\n\nWe want the tensors: \n\n$A:$ 20 by 21 tensor consisting of ones\n\n$B:$ a tensor with elements equal to the elements of numpy array $Z$\n\n$C:$ a tensor with the same number of elements as $A$ but with values $\n\\sim U(0,1)$\n\n$D:$ a 1D tensor containing the even numbers between 4 and 40 inclusive.\n",
"_____no_output_____"
]
],
[
[
"def tensor_creation(Z):\n \"\"\"A function that creates various tensors.\n\n Args:\n Z (numpy.ndarray): An array of shape\n\n Returns:\n A : 20 by 21 tensor consisting of ones\n B : a tensor with elements equal to the elements of numpy array Z\n C : a tensor with the same number of elements as A but with values ∼U(0,1)\n D : a 1D tensor containing the even numbers between 4 and 40 inclusive.\n \"\"\"\n #################################################\n ## TODO for students: fill in the missing code\n ## from the first expression\n raise NotImplementedError(\"Student exercise: say what they should have done\")\n #################################################\n A = ...\n B = ...\n C = ...\n D = ...\n\n return A, B, C, D\n\n\n# add timing to airtable\natform.add_event('Coding Exercise 2.1: Creating Tensors')\n\n\n\n# numpy array to copy later\nZ = np.vander([1, 2, 3], 4)\n\n# Uncomment below to check your function!\n# A, B, C, D = tensor_creation(Z)\n# checkExercise1(A, B, C, D)",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content-dl/tree/main//tutorials/W1D1_BasicsAndPytorch/solutions/W1D1_Tutorial1_Solution_ad4f6c0f.py)\n\n",
"_____no_output_____"
],
[
"```\nAll correct!\n```",
"_____no_output_____"
],
[
"## Section 2.2: Operations in PyTorch\n\n**Tensor-Tensor operations**\n\nWe can perform operations on tensors using methods under ```torch.``` \n\n",
"_____no_output_____"
]
],
[
[
"# @title Video 4: Tensor Operators\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1G44y127As\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"R1R8VoYXBVA\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\n# add timing to airtable\natform.add_event('Video 4: Tensor Operators')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"**Tensor-Tensor operations**\n\nWe can perform operations on tensors using methods under ```torch.``` ",
"_____no_output_____"
]
],
[
[
"a = torch.ones(5, 3)\nb = torch.rand(5, 3)\nc = torch.empty(5, 3)\nd = torch.empty(5, 3)\n\n# this only works if c and d already exist\ntorch.add(a, b, out=c)\n#Pointwise Multiplication of a and b\ntorch.multiply(a, b, out=d)\nprint(c)\nprint(d)",
"_____no_output_____"
]
],
[
[
"However, in PyTorch most common Python operators are overridden.\nThe common standard arithmetic operators (+, -, *, /, and **) have all been lifted to elementwise operations",
"_____no_output_____"
]
],
[
[
"x = torch.tensor([1, 2, 4, 8])\ny = torch.tensor([1, 2, 3, 4])\nx + y, x - y, x * y, x / y, x**y # The ** operator is exponentiation",
"_____no_output_____"
]
],
[
[
"**Tensor Methods**",
"_____no_output_____"
],
[
"Tensors also have a number of common arithmetic operations built in. A full list of **all** methods can be found in the appendix (there are a lot!) \n\nAll of these operations should have similar syntax to their numpy equivalents.(Feel free to skip if you already know this!)",
"_____no_output_____"
]
],
[
[
"x = torch.rand(3, 3)\nprint(x)\nprint(\"\\n\")\n# sum() - note the axis is the axis you move across when summing\nprint(f\"Sum of every element of x: {x.sum()}\")\nprint(f\"Sum of the columns of x: {x.sum(axis=0)}\")\nprint(f\"Sum of the rows of x: {x.sum(axis=1)}\")\nprint(\"\\n\")\n\nprint(f\"Mean value of all elements of x {x.mean()}\")\nprint(f\"Mean values of the columns of x {x.mean(axis=0)}\")\nprint(f\"Mean values of the rows of x {x.mean(axis=1)}\")",
"_____no_output_____"
]
],
[
[
"**Matrix Operations**\n\nThe ```@``` symbol is overridden to represent matrix multiplication. You can also use ```torch.matmul()``` to multiply tensors. For dot multiplication, you can use ```torch.dot()```, or manipulate the axes of your tensors and do matrix multiplication (we will cover that in the next section). \n\nTransposes of 2D tensors are obtained using ```torch.t()``` or ```Tensor.T```. Note the lack of brackets for ```Tensor.T``` - it is an attribute, not a method.",
"_____no_output_____"
],
[
"### Coding Exercise 2.2 : Simple tensor operations\n\nBelow are two expressions involving operations on matrices. \n\n$$ \\textbf{A} = \n\\begin{bmatrix}2 &4 \\\\5 & 7 \n\\end{bmatrix} \n\\begin{bmatrix} 1 &1 \\\\2 & 3\n\\end{bmatrix} \n + \n\\begin{bmatrix}10 & 10 \\\\ 12 & 1 \n\\end{bmatrix} \n$$\n\n\nand\n\n\n$$ b = \n\\begin{bmatrix} 3 \\\\ 5 \\\\ 7\n\\end{bmatrix} \\cdot \n\\begin{bmatrix} 2 \\\\ 4 \\\\ 8\n\\end{bmatrix}\n$$\n\nThe code block below that computes these expressions using PyTorch is incomplete - fill in the missing lines.\n\n",
"_____no_output_____"
]
],
[
[
"def simple_operations(a1: torch.Tensor, a2: torch.Tensor, a3: torch.Tensor):\n ################################################\n ## TODO for students: complete the first computation using the argument matricies\n raise NotImplementedError(\"Student exercise: fill in the missing code to complete the operation\")\n ################################################\n # multiplication of tensor a1 with tensor a2 and then add it with tensor a3\n answer = ...\n return answer\n\n# add timing to airtable\natform.add_event('Coding Exercise 2.2 : Simple tensor operations-simple_operations')\n\n# Computing expression 1:\n\n# init our tensors\na1 = torch.tensor([[2, 4], [5, 7]])\na2 = torch.tensor([[1, 1], [2, 3]])\na3 = torch.tensor([[10, 10], [12, 1]])\n## uncomment to test your function\n# A = simple_operations(a1, a2, a3)\n# print(A)",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content-dl/tree/main//tutorials/W1D1_BasicsAndPytorch/solutions/W1D1_Tutorial1_Solution_5562ea1d.py)\n\n",
"_____no_output_____"
],
[
"```\ntensor([[20, 24],\n [31, 27]])\n```",
"_____no_output_____"
]
],
[
[
"def dot_product(b1: torch.Tensor, b2: torch.Tensor):\n ###############################################\n ## TODO for students: complete the first computation using the argument matricies\n raise NotImplementedError(\"Student exercise: fill in the missing code to complete the operation\")\n ###############################################\n # Use torch.dot() to compute the dot product of two tensors\n product = ...\n return product\n\n# add timing to airtable\natform.add_event('Coding Exercise 2.2 : Simple tensor operations-dot_product')\n\n\n# Computing expression 2:\nb1 = torch.tensor([3, 5, 7])\nb2 = torch.tensor([2, 4, 8])\n## Uncomment to test your function\n# b = dot_product(b1, b2)\n# print(b)",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content-dl/tree/main//tutorials/W1D1_BasicsAndPytorch/solutions/W1D1_Tutorial1_Solution_00491ea4.py)\n\n",
"_____no_output_____"
],
[
"```\ntensor(82)\n```",
"_____no_output_____"
],
[
"## Section 2.3 Manipulating Tensors in Pytorch\n\n",
"_____no_output_____"
]
],
[
[
"# @title Video 5: Tensor Indexing\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1BM4y1K7pD\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"0d0KSJ3lJbg\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\n# add timing to airtable\natform.add_event('Video 5: Tensor Indexing')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"**Indexing**\n\nJust as in numpy, elements in a tensor can be accessed by index. As in any numpy array, the first element has index 0 and ranges are specified to include the first but before the last element. We can access elements according to their relative position to the end of the list by using negative indices. Indexing is also referred to as slicing.\n\nFor example, [-1] selects the last element; [1:3] selects the second and the third elements, and [:-2] will select all elements excluding the last and second-to-last elements.",
"_____no_output_____"
]
],
[
[
"x = torch.arange(0, 10)\nprint(x)\nprint(x[-1])\nprint(x[1:3])\nprint(x[:-2])",
"_____no_output_____"
]
],
[
[
"When we have multidimensional tensors, indexing rules work the same way as numpy.",
"_____no_output_____"
]
],
[
[
"# make a 5D tensor\nx = torch.rand(1, 2, 3, 4, 5)\n\nprint(f\" shape of x[0]:{x[0].shape}\")\nprint(f\" shape of x[0][0]:{x[0][0].shape}\")\nprint(f\" shape of x[0][0][0]:{x[0][0][0].shape}\")",
"_____no_output_____"
]
],
[
[
"**Flatten and reshape**\n\nThere are various methods for reshaping tensors. It is common to have to express 2D data in 1D format. Similarly, it is also common to have to reshape a 1D tensor into a 2D tensor. We can achieve this with the ```.flatten()``` and ```.reshape()``` methods.",
"_____no_output_____"
]
],
[
[
"z = torch.arange(12).reshape(6, 2)\nprint(f\"Original z: \\n {z}\")\n\n# 2D -> 1D\nz = z.flatten()\nprint(f\"Flattened z: \\n {z}\")\n\n# and back to 2D\nz = z.reshape(3, 4)\nprint(f\"Reshaped (3x4) z: \\n {z}\")",
"_____no_output_____"
]
],
[
[
"You will also see the ```.view()``` methods used a lot to reshape tensors. There is a subtle difference between ```.view()``` and ```.reshape()```, though for now we will just use ```.reshape()```. The documentation can be found in the appendix.",
"_____no_output_____"
],
[
"**Squeezing tensors**\n\nWhen processing batches of data, you will quite often be left with singleton dimensions. e.g. [1,10] or [256, 1, 3]. This dimension can quite easily mess up your matrix operations if you don't plan on it being there...\n\nIn order to compress tensors along their singleton dimensions we can use the ```.squeeze()``` method. We can use the ```.unsqueeze()``` method to do the opposite. \n",
"_____no_output_____"
]
],
[
[
"x = torch.randn(1, 10)\n# printing the zeroth element of the tensor will not give us the first number!\n\nprint(x.shape)\nprint(f\"x[0]: {x[0]}\")",
"_____no_output_____"
]
],
[
[
" Because of that pesky singleton dimension, x[0] gave us the first row instead!\n\n",
"_____no_output_____"
]
],
[
[
"# lets get rid of that singleton dimension and see what happens now\nx = x.squeeze(0)\nprint(x.shape)\nprint(f\"x[0]: {x[0]}\")",
"_____no_output_____"
],
[
"# adding singleton dimensions works a similar way, and is often used when tensors\n# being added need same number of dimensions\n\ny = torch.randn(5, 5)\nprint(f\"shape of y: {y.shape}\")\n\n# lets insert a singleton dimension\ny = y.unsqueeze(1)\nprint(f\"shape of y: {y.shape}\")",
"_____no_output_____"
]
],
[
[
"**Permutation**\nSometimes our dimensions will be in the wrong order! For example, we may be dealing with RGB images with dim [3x48x64], but our pipeline expects the colour dimension to be the last dimension i.e. [48x64x3]. To get around this we can use ```.permute()```\n",
"_____no_output_____"
]
],
[
[
"# `x` has dimensions [color,image_height,image_width]\nx = torch.rand(3, 48, 64)\n\n# we want to permute our tensor to be [ image_height , image_width , color ]\nx = x.permute(1, 2, 0)\n# permute(1,2,0) means:\n# the 0th dim of my new tensor = the 1st dim of my old tensor\n# the 1st dim of my new tensor = the 2nd\n# the 2nd dim of my new tensor = the 0th\nprint(x.shape)",
"_____no_output_____"
]
],
[
[
"You may also see ```.transpose()``` used. This works in a similar way as permute, but can only swap two dimensions at once.",
"_____no_output_____"
],
[
"**Concatenation**",
"_____no_output_____"
],
[
"In this example, we concatenate two matrices along rows (axis 0, the first element of the shape) vs. columns (axis 1, the second element of the shape). We can see that the first output tensor’s axis-0 length ( 6 ) is the sum of the two input tensors’ axis-0 lengths ( 3+3 ); while the second output tensor’s axis-1 length ( 8 ) is the sum of the two input tensors’ axis-1 lengths ( 4+4 ).",
"_____no_output_____"
]
],
[
[
"# Create two tensors of the same shape\nx = torch.arange(12, dtype=torch.float32).reshape((3, 4))\ny = torch.tensor([[2.0, 1, 4, 3], [1, 2, 3, 4], [4, 3, 2, 1]])\n\n\n#concatenate them along rows\ncat_rows = torch.cat((x, y), dim=0)\n\n# concatenate along columns\ncat_cols = torch.cat((x, y), dim=1)\n\n# printing outputs\nprint('Concatenated by rows: shape{} \\n {}'.format(list(cat_rows.shape), cat_rows))\nprint('\\n Concatenated by colums: shape{} \\n {}'.format(list(cat_cols.shape), cat_cols))",
"_____no_output_____"
]
],
[
[
"**Conversion to Other Python Objects**\n\nConverting to a NumPy tensor, or vice versa, is easy. The converted result does not share memory. This minor inconvenience is actually quite important: when you perform operations on the CPU or on GPUs, you do not want to halt computation, waiting to see whether the NumPy package of Python might want to be doing something else with the same chunk of memory.\n\nWhen converting to a numpy array, the information being tracked by the tensor will be lost i.e. the computational graph. This will be covered in detail when you are introduced to autograd tomorrow! ",
"_____no_output_____"
]
],
[
[
"x = torch.randn(5)\nprint(f\"x: {x} | x type: {x.type()}\")\n\ny = x.numpy()\nprint(f\"y: {y} | y type: {type(y)}\")\n\nz = torch.tensor(y)\nprint(f\"z: {z} | z type: {z.type()}\")",
"_____no_output_____"
]
],
[
[
"To convert a size-1 tensor to a Python scalar, we can invoke the item function or Python’s built-in functions.",
"_____no_output_____"
]
],
[
[
"a = torch.tensor([3.5])\na, a.item(), float(a), int(a)",
"_____no_output_____"
]
],
[
[
"### Coding Exercise 2.3: Manipulating Tensors\nUsing a combination of the methods discussed above, complete the functions below.",
"_____no_output_____"
],
[
"**Function A** \n\nThis function takes in two 2D tensors $A$ and $B$ and returns the column sum of A multiplied by the sum of all the elmements of $B$ i.e. a scalar, e.g.,:\n\n$ A = \\begin{bmatrix}\n1 & 1 \\\\\n1 & 1 \n\\end{bmatrix} \\,$\nand\n$ B = \\begin{bmatrix}\n1 & 2 & 3\\\\\n1 & 2 & 3 \n\\end{bmatrix} \\,$\nso\n$ \\, Out = \\begin{bmatrix} 2 & 2 \\\\\n\\end{bmatrix} \\cdot 12 = \\begin{bmatrix}\n24 & 24\\\\\n\\end{bmatrix}$\n\n**Function B** \n\nThis function takes in a square matrix $C$ and returns a 2D tensor consisting of a flattened $C$ with the index of each element appended to this tensor in the row dimension, e.g.,:\n\n$ C = \\begin{bmatrix}\n2 & 3 \\\\\n-1 & 10 \n\\end{bmatrix} \\,$\nso\n$ \\, Out = \\begin{bmatrix}\n0 & 2 \\\\\n1 & 3 \\\\\n2 & -1 \\\\\n3 & 10\n\\end{bmatrix}$\n\n**Hint:** pay close attention to singleton dimensions\n\n**Function C**\n\nThis function takes in two 2D tensors $D$ and $E$. If the dimensions allow it, this function returns the elementwise sum of $D$-shaped $E$, and $D$; else this function returns a 1D tensor that is the concatenation of the two tensors, e.g.,:\n\n$ D = \\begin{bmatrix}\n1 & -1 \\\\\n-1 & 3 \n\\end{bmatrix} \\,$\nand \n$ E = \\begin{bmatrix}\n2 & 3 & 0 & 2 \\\\\n\\end{bmatrix} \\, $\nso\n$ \\, Out = \\begin{bmatrix}\n3 & 2 \\\\\n-1 & 5 \n\\end{bmatrix}$\n\n$ D = \\begin{bmatrix}\n1 & -1 \\\\\n-1 & 3 \n\\end{bmatrix}$\nand\n$ \\, E = \\begin{bmatrix}\n2 & 3 & 0 \\\\\n\\end{bmatrix} \\,$\nso\n$ \\, Out = \\begin{bmatrix}\n1 & -1 & -1 & 3 & 2 & 3 & 0 \n\\end{bmatrix}$\n\n**Hint:** `torch.numel()` is an easy way of finding the number of elements in a tensor\n",
"_____no_output_____"
]
],
[
[
"def functionA(my_tensor1, my_tensor2):\n \"\"\"\n This function takes in two 2D tensors `my_tensor1` and `my_tensor2`\n and returns the column sum of\n `my_tensor1` multiplied by the sum of all the elmements of `my_tensor2`,\n i.e., a scalar.\n\n Args:\n my_tensor1: torch.Tensor\n my_tensor2: torch.Tensor\n Retuns:\n output: torch.Tensor\n The multiplication of the column sum of `my_tensor1` by the sum of\n `my_tensor2`.\n \"\"\"\n ################################################\n ## TODO for students: complete functionA\n raise NotImplementedError(\"Student exercise: complete function A\")\n ################################################\n # TODO multiplication the sum of the tensors\n output = ...\n\n return output\n\n\ndef functionB(my_tensor):\n \"\"\"\n This function takes in a square matrix `my_tensor` and returns a 2D tensor\n consisting of a flattened `my_tensor` with the index of each element\n appended to this tensor in the row dimension.\n\n Args:\n my_tensor: torch.Tensor\n Retuns:\n output: torch.Tensor\n Concatenated tensor.\n \"\"\"\n ################################################\n ## TODO for students: complete functionB\n raise NotImplementedError(\"Student exercise: complete function B\")\n ################################################\n # TODO flatten the tensor `my_tensor`\n my_tensor = ...\n # TODO create the idx tensor to be concatenated to `my_tensor`\n idx_tensor = ...\n # TODO concatenate the two tensors\n output = ...\n\n return output\n\n\ndef functionC(my_tensor1, my_tensor2):\n \"\"\"\n This function takes in two 2D tensors `my_tensor1` and `my_tensor2`.\n If the dimensions allow it, it returns the\n elementwise sum of `my_tensor1`-shaped `my_tensor2`, and `my_tensor2`;\n else this function returns a 1D tensor that is the concatenation of the\n two tensors.\n\n Args:\n my_tensor1: torch.Tensor\n my_tensor2: torch.Tensor\n Retuns:\n output: torch.Tensor\n Concatenated tensor.\n \"\"\"\n ################################################\n ## TODO for students: complete functionB\n raise NotImplementedError(\"Student exercise: complete function C\")\n ################################################\n # TODO check we can reshape `my_tensor2` into the shape of `my_tensor1`\n if ...:\n # TODO reshape `my_tensor2` into the shape of `my_tensor1`\n my_tensor2 = ...\n # TODO sum the two tensors\n output = ...\n else:\n # TODO flatten both tensors\n my_tensor1 = ...\n my_tensor2 = ...\n # TODO concatenate the two tensors in the correct dimension\n output = ...\n\n return output\n\n# add timing to airtable\natform.add_event('Coding Exercise 2.3: Manipulating Tensors')\n\n\n\n## Implement the functions above and then uncomment the following lines to test your code\n# print(functionA(torch.tensor([[1, 1], [1, 1]]), torch.tensor([[1, 2, 3], [1, 2, 3]])))\n# print(functionB(torch.tensor([[2, 3], [-1, 10]])))\n# print(functionC(torch.tensor([[1, -1], [-1, 3]]), torch.tensor([[2, 3, 0, 2]])))\n# print(functionC(torch.tensor([[1, -1], [-1, 3]]), torch.tensor([[2, 3, 0]])))",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content-dl/tree/main//tutorials/W1D1_BasicsAndPytorch/solutions/W1D1_Tutorial1_Solution_ea1718cb.py)\n\n",
"_____no_output_____"
],
[
"```\ntensor([24, 24])\ntensor([[ 0, 2],\n [ 1, 3],\n [ 2, -1],\n [ 3, 10]])\ntensor([[ 3, 2],\n [-1, 5]])\ntensor([ 1, -1, -1, 3, 2, 3, 0])\n```",
"_____no_output_____"
],
[
"## Section 2.4: GPUs \n",
"_____no_output_____"
]
],
[
[
"# @title Video 6: GPU vs CPU\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1nM4y1K7qx\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"9Mc9GFUtILY\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\n# add timing to airtable\natform.add_event('Video 6: GPU vs CPU')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"\nBy default, when we create a tensor it will *not* live on the GPU! ",
"_____no_output_____"
]
],
[
[
"x = torch.randn(10)\nprint(x.device)",
"_____no_output_____"
]
],
[
[
"When using Colab notebooks by default will not have access to a GPU. In order to start using GPUs we need to request one. We can do this by going to the runtime tab at the top of the page. \n\nBy following Runtime -> Change runtime type and selecting \"GPU\" from the Hardware Accelerator dropdown list, we can start playing with sending tensors to GPUs.\n\nOnce you have done this your runtime will restart and you will need to rerun the first setup cell to reimport PyTorch. Then proceed to the next cell.\n\n(For more information on the GPU usage policy you can view in the appendix)",
"_____no_output_____"
],
[
"**Now we have a GPU**\n",
"_____no_output_____"
],
[
"The cell below should return True.",
"_____no_output_____"
]
],
[
[
"print(torch.cuda.is_available())",
"_____no_output_____"
]
],
[
[
"CUDA is an API developed by Nvidia for interfacing with GPUs. PyTorch provides us with a layer of abstraction, and allows us to launch CUDA kernels using pure Python. *NOTE I am assuming that GPU stuff might be covered in more detail on another day but there could be a bit more detail here.*\n\nIn short, we get the power of parallising our tensor computations on GPUs, whilst only writing (relatively) simple Python!\n\nHere, we define the function `set_device`, which returns the device use in the notebook, i.e., `cpu` or `cuda`. Unless otherwise specified, we use this function on top of every tutorial, and we store the device variable such as\n\n```python\nDEVICE = set_device()\n```\n\nLet's define the function using the PyTorch package `torch.cuda`, which is lazily initialized, so we can always import it, and use `is_available()` to determine if our system supports CUDA.",
"_____no_output_____"
]
],
[
[
"def set_device():\n device = \"cuda\" if torch.cuda.is_available() else \"cpu\"\n if device != \"cuda\":\n print(\"GPU is not enabled in this notebook. \\n\"\n \"If you want to enable it, in the menu under `Runtime` -> \\n\"\n \"`Hardware accelerator.` and select `GPU` from the dropdown menu\")\n else:\n print(\"GPU is enabled in this notebook. \\n\"\n \"If you want to disable it, in the menu under `Runtime` -> \\n\"\n \"`Hardware accelerator.` and select `None` from the dropdown menu\")\n\n return device",
"_____no_output_____"
]
],
[
[
"Let's make some CUDA tensors!",
"_____no_output_____"
]
],
[
[
"# common device agnostic way of writing code that can run on cpu OR gpu\n# that we provide for you in each of the tutorials\nDEVICE = set_device()\n\n# we can specify a device when we first create our tensor\nx = torch.randn(2, 2, device=DEVICE)\nprint(x.dtype)\nprint(x.device)\n\n# we can also use the .to() method to change the device a tensor lives on\ny = torch.randn(2, 2)\nprint(f\"y before calling to() | device: {y.device} | dtype: {y.type()}\")\n\ny = y.to(DEVICE)\nprint(f\"y after calling to() | device: {y.device} | dtype: {y.type()}\")",
"_____no_output_____"
]
],
[
[
"**Operations between cpu tensors and cuda tensors**\n\nNote that the type of the tensor changed after calling ```.to()```. What happens if we try and perform operations on tensors on devices?\n\n\n",
"_____no_output_____"
]
],
[
[
"x = torch.tensor([0, 1, 2], device=DEVICE)\ny = torch.tensor([3, 4, 5], device=\"cpu\")\n\n# Uncomment the following line and run this cell\n# z = x + y",
"_____no_output_____"
]
],
[
[
"We cannot combine cuda tensors and cpu tensors in this fashion. If we want to compute an operation that combines tensors on different devices, we need to move them first! We can use the `.to()` method as before, or the `.cpu()` and `.cuda()` methods. Note that using the `.cuda()` will throw an error if CUDA is not enabled in your machine.\n\nGenrally in this course all Deep learning is done on the GPU and any computation is done on the CPU, so sometimes we have to pass things back and forth so you'll see us call.",
"_____no_output_____"
]
],
[
[
"x = torch.tensor([0, 1, 2], device=DEVICE)\ny = torch.tensor([3, 4, 5], device=\"cpu\")\nz = torch.tensor([6, 7, 8], device=DEVICE)\n\n# moving to cpu\nx = x.to(\"cpu\") # alternatively, you can use x = x.cpu()\nprint(x + y)\n\n# moving to gpu\ny = y.to(DEVICE) # alternatively, you can use y = y.cuda()\nprint(y + z)",
"_____no_output_____"
]
],
[
[
"### Coding Exercise 2.4: Just how much faster are GPUs?\n\nBelow is a simple function `simpleFun`. Complete this function, such that it performs the operations:\n\n- elementwise multiplication\n\n- matrix multiplication\n\nThe operations should be able to perfomed on either the CPU or GPU specified by the parameter `device`. We will use the helper function `timeFun(f, dim, iterations, device)`.",
"_____no_output_____"
]
],
[
[
"dim = 10000\niterations = 1",
"_____no_output_____"
],
[
"def simpleFun(dim, device):\n \"\"\"\n Args:\n dim: integer\n device: \"cpu\" or \"cuda\"\n Returns:\n Nothing.\n \"\"\"\n ###############################################\n ## TODO for students: recreate the function, but\n ## ensure all computations happens on the `device`\n raise NotImplementedError(\"Student exercise: fill in the missing code to create the tensors\")\n ###############################################\n # 2D tensor filled with uniform random numbers in [0,1), dim x dim\n x = ...\n # 2D tensor filled with uniform random numbers in [0,1), dim x dim\n y = ...\n # 2D tensor filled with the scalar value 2, dim x dim\n z = ...\n\n # elementwise multiplication of x and y\n a = ...\n # matrix multiplication of x and y\n b = ...\n\n del x\n del y\n del z\n del a\n del b\n\n\n## TODO: Implement the function above and uncomment the following lines to test your code\n# timeFun(f=simpleFun, dim=dim, iterations=iterations)\n# timeFun(f=simpleFun, dim=dim, iterations=iterations, device=DEVICE)",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content-dl/tree/main//tutorials/W1D1_BasicsAndPytorch/solutions/W1D1_Tutorial1_Solution_232a94a4.py)\n\n",
"_____no_output_____"
],
[
"Sample output (depends on your hardware)\n\n```\ntime taken for 1 iterations of simpleFun(10000, cpu): 23.74070\ntime taken for 1 iterations of simpleFun(10000, cuda): 0.87535\n```",
"_____no_output_____"
],
[
"**Discuss!**\n\nTry and reduce the dimensions of the tensors and increase the iterations. You can get to a point where the cpu only function is faster than the GPU function. Why might this be?\n",
"_____no_output_____"
],
[
"## Section 2.5: Datasets and Dataloaders\n\n",
"_____no_output_____"
]
],
[
[
"# @title Video 7: Getting Data\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1744y127SQ\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"LSkjPM1gFu0\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\n# add timing to airtable\natform.add_event('Video 7: Getting Data')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"When training neural network models you will be working with large amounts of data. Fortunately, PyTorch offers some great tools that help you organize and manipulate your data samples.\n",
"_____no_output_____"
]
],
[
[
"# Import dataset and dataloaders related packages\nfrom torchvision import datasets\nfrom torchvision.transforms import ToTensor\nfrom torch.utils.data import DataLoader\nfrom torchvision.transforms import Compose, Grayscale",
"_____no_output_____"
]
],
[
[
"**Datasets**\n\nThe `torchvision` package gives you easy access to many of the publicly available datasets. Let's load the [CIFAR10](https://www.cs.toronto.edu/~kriz/cifar.html) dataset, which contains color images of 10 different classes, like vehicles and animals.\n\nCreating an object of type `datasets.CIFAR10` will automatically download and load all images from the dataset. The resulting data structure can be treated as a list containing data samples and their corresponding labels.",
"_____no_output_____"
]
],
[
[
"# Download and load the images from the CIFAR10 dataset\ncifar10_data = datasets.CIFAR10(\n root=\"data\", # path where the images will be stored\n download=True, # all images should be downloaded\n transform=ToTensor() # transform the images to tensors\n )\n\n# Print the number of samples in the loaded dataset\nprint(f\"Number of samples: {len(cifar10_data)}\")\nprint(f\"Class names: {cifar10_data.classes}\")",
"_____no_output_____"
]
],
[
[
"We have 50000 samples loaded. Now let's take a look at one of them in detail. Each sample consists of an image and its corresponding label.",
"_____no_output_____"
]
],
[
[
"# Choose a random sample\nrandom.seed(2021)\nimage, label = cifar10_data[random.randint(0, len(cifar10_data))]\nprint(f\"Label: {cifar10_data.classes[label]}\")\nprint(f\"Image size: {image.shape}\")",
"_____no_output_____"
]
],
[
[
"Color images are modeled as 3 dimensional tensors. The first dimension corresponds to the channels (C) of the image (in this case we have RGB images). The second dimensions is the height (H) of the image and the third is the width (W). We can denote this image format as C × H × W.",
"_____no_output_____"
],
[
"### Coding Exercise 2.5: Display an image from the dataset\n\nLet's try to display the image using `matplotlib`. The code below will not work, because `imshow` expects to have the image in a different format - $H \\times W \\times C$.\n\nYou need to reorder the dimensions of the tensor using the `permute` method of the tensor. PyTorch `torch.permute(*dims)` rearranges the original tensor according to the desired ordering and returns a new multidimensional rotated tensor. The size of the returned tensor remains the same as that of the original.\n\n**Code hint:**\n\n```python\n# create a tensor of size 2 x 4\ninput_var = torch.randn(2, 4)\n# print its size and the tensor\nprint(input_var.size())\nprint(input_var)\n\n# dimensions permuted\ninput_var = input_var.permute(1, 0)\n# print its size and the permuted tensor\nprint(input_var.size())\nprint(input_var)\n```",
"_____no_output_____"
]
],
[
[
"# TODO: Uncomment the following line to see the error that arises from the current image format\n# plt.imshow(image)\n\n# TODO: Comment the above line and fix this code by reordering the tensor dimensions\n# plt.imshow(image.permute(...))\n# plt.show()",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content-dl/tree/main//tutorials/W1D1_BasicsAndPytorch/solutions/W1D1_Tutorial1_Solution_b04bd357.py)\n\n*Example output:*\n\n<img alt='Solution hint' align='left' width=835.0 height=827.0 src=https://raw.githubusercontent.com/NeuromatchAcademy/course-content-dl/main/tutorials/W1D1_BasicsAndPytorch/static/W1D1_Tutorial1_Solution_b04bd357_0.png>\n\n",
"_____no_output_____"
]
],
[
[
"#@title Video 8: Train and Test\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1rV411H7s5\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"JokSIuPs-ys\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\n# add timing to airtable\natform.add_event('Video 8: Train and Test')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"**Training and Test Datasets**\n\nWhen loading a dataset, you can specify if you want to load the training or the test samples using the `train` argument. We can load the training and test datasets separately. For simplicity, today we will not use both datasets separately, but this topic will be adressed in the next days.",
"_____no_output_____"
]
],
[
[
"# Load the training samples\ntraining_data = datasets.CIFAR10(\n root=\"data\",\n train=True,\n download=True,\n transform=ToTensor()\n )\n\n# Load the test samples\ntest_data = datasets.CIFAR10(\n root=\"data\",\n train=False,\n download=True,\n transform=ToTensor()\n )",
"_____no_output_____"
],
[
"# @title Video 9: Data Augmentation - Transformations\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV19B4y1N77t\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"sjegA9OBUPw\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\n# add timing to airtable\natform.add_event('Video 9: Data Augmentation - Transformations')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"**Dataloader**\n\nAnother important concept is the `Dataloader`. It is a wrapper around the `Dataset` that splits it into minibatches (important for training the neural network) and makes the data iterable. The `shuffle` argument is used to shuffle the order of the samples across the minibatches.\n",
"_____no_output_____"
]
],
[
[
"# Create dataloaders with\ntrain_dataloader = DataLoader(training_data, batch_size=64, shuffle=True)\ntest_dataloader = DataLoader(test_data, batch_size=64, shuffle=True)",
"_____no_output_____"
]
],
[
[
"*Reproducibility:* DataLoader will reseed workers following Randomness in multi-process data loading algorithm. Use `worker_init_fn()` and a `generator` to preserve reproducibility:\n\n\n```python\ndef seed_worker(worker_id):\n worker_seed = torch.initial_seed() % 2**32\n numpy.random.seed(worker_seed)\n random.seed(worker_seed)\n\n\ng_seed = torch.Generator()\ng_seed.manual_seed(my_seed)\n\nDataLoader(\n train_dataset,\n batch_size=batch_size,\n num_workers=num_workers,\n worker_init_fn=seed_worker,\n generator=g_seed\n )\n```",
"_____no_output_____"
],
[
"**Note:** For the `seed_worker` to have an effect, `num_workers` should be 2 or more.",
"_____no_output_____"
],
[
"We can now query the next batch from the data loader and inspect it. For this we need to convert the dataloader object to a Python iterator using the function `iter` and then we can query the next batch using the function `next`.\n\nWe can now see that we have a 4D tensor. This is because we have a 64 images in the batch ($B$) and each image has 3 dimensions: channels ($C$), height ($H$) and width ($W$). So, the size of the 4D tensor is $B \\times C \\times H \\times W$.",
"_____no_output_____"
]
],
[
[
"# Load the next batch\nbatch_images, batch_labels = next(iter(train_dataloader))\nprint('Batch size:', batch_images.shape)\n\n# Display the first image from the batch\nplt.imshow(batch_images[0].permute(1, 2, 0))\nplt.show()",
"_____no_output_____"
]
],
[
[
"**Transformations**\n\nAnother useful feature when loading a dataset is applying transformations on the data - color conversions, normalization, cropping, rotation etc. There are many predefined transformations in the `torchvision.transforms` package and you can also combine them using the `Compose` transform. Checkout the [pytorch documentation](https://pytorch.org/vision/stable/transforms.html) for details.",
"_____no_output_____"
],
[
"### Coding Exercise 2.6: Load the CIFAR10 dataset as grayscale images\n\nThe goal of this excercise is to load the images from the CIFAR10 dataset as grayscale images. Note that we rerun the `set_seed` function to ensure reproducibility.",
"_____no_output_____"
]
],
[
[
"def my_data_load():\n ###############################################\n ## TODO for students: load the CIFAR10 data,\n ## but as grayscale images and not as RGB colored.\n raise NotImplementedError(\"Student exercise: fill in the missing code to load the data\")\n ###############################################\n ## TODO Load the CIFAR10 data using a transform that converts the images to grayscale tensors\n data = datasets.CIFAR10(...,\n transform=...)\n # Display a random grayscale image\n image, label = data[random.randint(0, len(data))]\n plt.imshow(image.squeeze(), cmap=\"gray\")\n plt.show()\n\n return data\n\n\nset_seed(seed=2021)\n## After implementing the above code, uncomment the following lines to test your code\n# data = my_data_load()",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content-dl/tree/main//tutorials/W1D1_BasicsAndPytorch/solutions/W1D1_Tutorial1_Solution_6052d728.py)\n\n*Example output:*\n\n<img alt='Solution hint' align='left' width=835.0 height=827.0 src=https://raw.githubusercontent.com/NeuromatchAcademy/course-content-dl/main/tutorials/W1D1_BasicsAndPytorch/static/W1D1_Tutorial1_Solution_6052d728_1.png>\n\n",
"_____no_output_____"
],
[
"---\n# Section 3: Neural Networks\n\n*Time estimate: ~1 hour 30 mins (excluding video)*",
"_____no_output_____"
],
[
"Now it's time for you to create your first neural network using PyTorch. This section will walk you through the process of:\n- Creating a simple neural network model\n- Training the network\n- Visualizing the results of the network\n- Tweeking the network",
"_____no_output_____"
]
],
[
[
"# @title Video 10: CSV Files\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1xy4y1T7kv\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"JrC_UAJWYKU\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\n# add timing to airtable\natform.add_event('Video 10: CSV Files')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"## Section 3.1: Data Loading\n\nFirst we need some sample data to train our network on. You can use the function below to generate an example dataset consisting of 2D points along two interleaving half circles. The data will be stored in a file called `sample_data.csv`. You can inspect the file directly in Colab by going to Files on the left side and opening the CSV file.",
"_____no_output_____"
]
],
[
[
"# @title Generate sample data\n# @markdown we used `scikit-learn` module\nfrom sklearn.datasets import make_moons\n\n# Create a dataset of 256 points with a little noise\nX, y = make_moons(256, noise=0.1)\n\n# Store the data as a Pandas data frame and save it to a CSV file\ndf = pd.DataFrame(dict(x0=X[:,0], x1=X[:,1], y=y))\ndf.to_csv('sample_data.csv')",
"_____no_output_____"
]
],
[
[
"Now we can load the data from the CSV file using the Pandas library. Pandas provides many functions for reading files in various formats. When loading data from a CSV file, we can reference the columns directly by their names.",
"_____no_output_____"
]
],
[
[
"# Load the data from the CSV file in a Pandas DataFrame\ndata = pd.read_csv(\"sample_data.csv\")\n\n# Create a 2D numpy array from the x0 and x1 columns\nX_orig = data[[\"x0\", \"x1\"]].to_numpy()\n\n# Create a 1D numpy array from the y column\ny_orig = data[\"y\"].to_numpy()\n\n# Print the sizes of the generated 2D points X and the corresponding labels Y\nprint(f\"Size X:{X_orig.shape}\")\nprint(f\"Size y:{y_orig.shape}\")\n\n# Visualize the dataset. The color of the points is determined by the labels `y_orig`.\nplt.scatter(X_orig[:, 0], X_orig[:, 1], s=40, c=y_orig)\nplt.show()",
"_____no_output_____"
]
],
[
[
"**Prepare Data for PyTorch**\n\nNow let's prepare the data in a format suitable for PyTorch - convert everything into tensors.",
"_____no_output_____"
]
],
[
[
"# Initialize the device variable\nDEVICE = set_device()\n\n# Convert the 2D points to a float32 tensor\nX = torch.tensor(X_orig, dtype=torch.float32)\n\n# Upload the tensor to the device\nX = X.to(DEVICE)\n\nprint(f\"Size X:{X.shape}\")\n\n# Convert the labels to a long interger tensor\ny = torch.from_numpy(y_orig).type(torch.LongTensor)\n# Upload the tensor to the device\ny = y.to(DEVICE)\n\nprint(f\"Size y:{y.shape}\")",
"_____no_output_____"
]
],
[
[
"## Section 3.2: Create a Simple Neural Network",
"_____no_output_____"
]
],
[
[
"# @title Video 11: Generating the Neural Network\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1fK4y1M74a\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"PwSzRohUvck\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\n# add timing to airtable\natform.add_event('Video 11: Generating the Neural Network')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"For this example we want to have a simple neural network consisting of 3 layers:\n- 1 input layer of size 2 (our points have 2 coordinates)\n- 1 hidden layer of size 16 (you can play with different numbers here)\n- 1 output layer of size 2 (we want the have the scores for the two classes)\n\nDuring the course you will deal with differend kinds of neural networks. On Day 2 we will focus on linear networks, but you will work with some more complicated architectures in the next days. The example here is meant to demonstrate the process of creating and training a neural network end-to-end.\n\n**Programing the Network**\n\nPyTorch provides a base class for all neural network modules called [`nn.Module`](https://pytorch.org/docs/stable/generated/torch.nn.Module.html). You need to inherit from `nn.Module` and implement some important methods:\n\n`__init__`\n\nIn the `__init__` method you need to define the structure of your network. Here you will specify what layers will the network consist of, what activation functions will be used etc.\n\n`forward`\n\nAll neural network modules need to implement the `forward` method. It specifies the computations the network needs to do when data is passed through it.\n\n`predict`\n\nThis is not an obligatory method of a neural network module, but it is a good practice if you want to quickly get the most likely label from the network. It calls the `forward` method and chooses the label with the highest score.\n\n`train`\n\nThis is also not an obligatory method, but it is a good practice to have. The method will be used to train the network parameters and will be implemented later in the notebook.\n\n\n> Note that you can use the `__call__` method of a module directly and it will invoke the `forward` method: `net()` does the same as `net.forward()`.",
"_____no_output_____"
]
],
[
[
"# Inherit from nn.Module - the base class for neural network modules provided by Pytorch\nclass NaiveNet(nn.Module):\n\n # Define the structure of your network\n def __init__(self):\n super(NaiveNet, self).__init__()\n\n # The network is defined as a sequence of operations\n self.layers = nn.Sequential(\n nn.Linear(2, 16), # Transformation from the input to the hidden layer\n nn.ReLU(), # Activation function (ReLU) is a non-linearity which is widely used because it reduces computation. The function returns 0 if it receives any\n # negative input, but for any positive value x, it returns that value back.\n nn.Linear(16, 2), # Transformation from the hidden to the output layer\n )\n\n # Specify the computations performed on the data\n def forward(self, x):\n # Pass the data through the layers\n return self.layers(x)\n\n # Choose the most likely label predicted by the network\n def predict(self, x):\n # Pass the data through the networks\n output = self.forward(x)\n\n # Choose the label with the highest score\n return torch.argmax(output, 1)\n\n # Train the neural network (will be implemented later)\n def train(self, X, y):\n pass",
"_____no_output_____"
]
],
[
[
"**Check that your network works**\n\nCreate an instance of your model and visualize it",
"_____no_output_____"
]
],
[
[
"# Create new NaiveNet and transfer it to the device\nmodel = NaiveNet().to(DEVICE)\n\n# Print the structure of the network\nprint(model)",
"_____no_output_____"
]
],
[
[
"### Coding Exercise 3.2: Classify some samples\n\nNow let's pass some of the points of our dataset through the network and see if it works. You should not expect the network to actually classify the points correctly, because it has not been trained yet. \n\nThe goal here is just to get some experience with the data structures that are passed to the forward and predict methods and their results.",
"_____no_output_____"
]
],
[
[
"## Get the samples\n# X_samples = ...\n# print(\"Sample input:\\n\", X_samples)\n\n## Do a forward pass of the network\n# output = ...\n# print(\"\\nNetwork output:\\n\", output)\n\n## Predict the label of each point\n# y_predicted = ...\n# print(\"\\nPredicted labels:\\n\", y_predicted)",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content-dl/tree/main//tutorials/W1D1_BasicsAndPytorch/solutions/W1D1_Tutorial1_Solution_af8ae0ff.py)\n\n",
"_____no_output_____"
],
[
"```\nSample input:\n tensor([[ 0.9066, 0.5052],\n [-0.2024, 1.1226],\n [ 1.0685, 0.2809],\n [ 0.6720, 0.5097],\n [ 0.8548, 0.5122]], device='cuda:0')\n\nNetwork output:\n tensor([[ 0.1543, -0.8018],\n [ 2.2077, -2.9859],\n [-0.5745, -0.0195],\n [ 0.1924, -0.8367],\n [ 0.1818, -0.8301]], device='cuda:0', grad_fn=<AddmmBackward>)\n\nPredicted labels:\n tensor([0, 0, 1, 0, 0], device='cuda:0')\n```",
"_____no_output_____"
],
[
"## Section 3.3: Train Your Neural Network\n\n",
"_____no_output_____"
]
],
[
[
"# @title Video 12: Train the Network\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1v54y1n7CS\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"4MIqnE4XPaA\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\n# add timing to airtable\natform.add_event('Video 12: Train the Network')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"Now it is time to train your network on your dataset. Don't worry if you don't fully understand everything yet - we will cover training in much more details in the next days. For now, the goal is just to see your network in action!\n\nYou will usually implement the `train` method directly when implementing your class `NaiveNet`. Here, we will implement it as a function outside of the class in order to have it in a ceparate cell.",
"_____no_output_____"
]
],
[
[
"# @title Helper function to plot the decision boundary\n\n# Code adapted from this notebook: https://jonchar.net/notebooks/Artificial-Neural-Network-with-Keras/\n\nfrom pathlib import Path\n\ndef plot_decision_boundary(model, X, y, device):\n # Transfer the data to the CPU\n X = X.cpu().numpy()\n y = y.cpu().numpy()\n\n # Check if the frames folder exists and create it if needed\n frames_path = Path(\"frames\")\n if not frames_path.exists():\n frames_path.mkdir()\n\n # Set min and max values and give it some padding\n x_min, x_max = X[:, 0].min() - .5, X[:, 0].max() + .5\n y_min, y_max = X[:, 1].min() - .5, X[:, 1].max() + .5\n h = 0.01\n\n # Generate a grid of points with distance h between them\n xx, yy = np.meshgrid(np.arange(x_min, x_max, h), np.arange(y_min, y_max, h))\n\n # Predict the function value for the whole gid\n grid_points = np.c_[xx.ravel(), yy.ravel()]\n grid_points = torch.from_numpy(grid_points).type(torch.FloatTensor)\n Z = model.predict(grid_points.to(device)).cpu().numpy()\n Z = Z.reshape(xx.shape)\n\n # Plot the contour and training examples\n plt.contourf(xx, yy, Z, cmap=plt.cm.Spectral)\n plt.scatter(X[:, 0], X[:, 1], c=y, cmap=plt.cm.binary)",
"_____no_output_____"
],
[
"# Implement the train function given a training dataset X and correcsponding labels y\ndef train(model, X, y):\n # The Cross Entropy Loss is suitable for classification problems\n loss_function = nn.CrossEntropyLoss()\n\n # Create an optimizer (Stochastic Gradient Descent) that will be used to train the network\n learning_rate = 1e-2\n optimizer = torch.optim.SGD(model.parameters(), lr=learning_rate)\n\n # Number of epochs\n epochs = 15000\n\n # List of losses for visualization\n losses = []\n\n for i in range(epochs):\n # Pass the data through the network and compute the loss\n # We'll use the whole dataset during the training instead of using batches\n # in to order to keep the code simple for now.\n y_logits = model.forward(X)\n loss = loss_function(y_logits, y)\n\n # Clear the previous gradients and compute the new ones\n optimizer.zero_grad()\n loss.backward()\n\n # Adapt the weights of the network\n optimizer.step()\n\n # Store the loss\n losses.append(loss.item())\n\n # Print the results at every 1000th epoch\n if i % 1000 == 0:\n print(f\"Epoch {i} loss is {loss.item()}\")\n\n plot_decision_boundary(model, X, y, DEVICE)\n plt.savefig('frames/{:05d}.png'.format(i))\n\n return losses\n\n\n# Create a new network instance a train it\nmodel = NaiveNet().to(DEVICE)\nlosses = train(model, X, y)",
"_____no_output_____"
]
],
[
[
"**Plot the loss during training**\n\nPlot the loss during the training to see how it reduces and converges.",
"_____no_output_____"
]
],
[
[
"plt.plot(np.linspace(1, len(losses), len(losses)), losses)\nplt.xlabel(\"Epoch\")\nplt.ylabel(\"Loss\")",
"_____no_output_____"
],
[
"# @title Visualize the training process\n# @markdown ### Execute this cell!\n!pip install imageio --quiet\n!pip install pathlib --quiet\n\nimport imageio\nfrom IPython.core.interactiveshell import InteractiveShell\nfrom IPython.display import Image, display\nfrom pathlib import Path\n\nInteractiveShell.ast_node_interactivity = \"all\"\n\n# Make a list with all images\nimages = []\nfor i in range(10):\n filename = \"frames/0\"+str(i)+\"000.png\"\n images.append(imageio.imread(filename))\n# Save the gif\nimageio.mimsave('frames/movie.gif', images)\ngifPath = Path(\"frames/movie.gif\")\nwith open(gifPath,'rb') as f:\n display(Image(data=f.read(), format='png'))",
"_____no_output_____"
],
[
"# @title Video 13: Play with it\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1Cq4y1W7BH\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"_GGkapdOdSY\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\n# add timing to airtable\natform.add_event('Video 13: Play with it')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"### Exercise 3.3: Tweak your Network\n\nYou can now play around with the network a little bit to get a feeling of what different parameters are doing. Here are some ideas what you could try:\n- Increase or decrease the number of epochs for training\n- Increase or decrease the size of the hidden layer\n- Add one additional hidden layer\n\nCan you get the network to better fit the data?",
"_____no_output_____"
]
],
[
[
"# @title Video 14: XOR Widget\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1mB4y1N7QS\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"oTr1nE2rCWg\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\n# add timing to airtable\natform.add_event('Video 14: XOR Widget')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"Exclusive OR (XOR) logical operation gives a true (`1`) output when the number of true inputs is odd. That is, a true output result if one, and only one, of the inputs to the gate is true. If both inputs are false (`0`) or both are true or false output results. Mathematically speaking, XOR represents the inequality function, i.e., the output is true if the inputs are not alike; otherwise, the output is false.\n\nIn case of two inputs ($X$ and $Y$) the following truth table is applied:\n\n\\begin{array}{ccc}\nX & Y & \\text{XOR} \\\\\n\\hline\n0 & 0 & 0 \\\\\n0 & 1 & 1 \\\\\n1 & 0 & 1 \\\\\n1 & 1 & 0 \\\\\n\\end{array}\n\nHere, with `0`, we denote `False`, and with `1` we denote `True` in boolean terms.",
"_____no_output_____"
],
[
"### Interactive Demo 3.3: Solving XOR\n\nHere we use an open source and famous visualization widget developed by Tensorflow team available [here](https://github.com/tensorflow/playground).\n* Play with the widget and observe that you can not solve the continuous XOR dataset.\n* Now add one hidden layer with three units, play with the widget, and set weights by hand to solve this dataset perfectly.\n\nFor the second part, you should set the weights by clicking on the connections and either type the value or use the up and down keys to change it by one increment. You could also do the same for the biases by clicking on the tiny square to each neuron's bottom left.\nEven though there are infinitely many solutions, a neat solution when $f(x)$ is ReLU is: \n\n\\begin{equation}\n y = f(x_1)+f(x_2)-f(x_1+x_2)\n\\end{equation}\n\nTry to set the weights and biases to implement this function after you played enough :)",
"_____no_output_____"
]
],
[
[
"# @markdown ###Play with the parameters to solve XOR\nfrom IPython.display import HTML\nHTML('<iframe width=\"1020\" height=\"660\" src=\"https://playground.arashash.com/#activation=relu&batchSize=10&dataset=xor®Dataset=reg-plane&learningRate=0.03®ularizationRate=0&noise=0&networkShape=&seed=0.91390&showTestData=false&discretize=false&percTrainData=90&x=true&y=true&xTimesY=false&xSquared=false&ySquared=false&cosX=false&sinX=false&cosY=false&sinY=false&collectStats=false&problem=classification&initZero=false&hideText=false\" allowfullscreen></iframe>')",
"_____no_output_____"
],
[
"# @markdown Do you think we can solve the discrete XOR (only 4 possibilities) with only 2 hidden units?\nw1_min_xor = 'Select' #@param ['Select', 'Yes', 'No']\nif w1_min_xor == 'No':\n print(\"Correct!\")\nelse:\n print(\"How about giving it another try?\")",
"_____no_output_____"
]
],
[
[
"---\n# Section 4: Ethics And Course Info\n",
"_____no_output_____"
]
],
[
[
"# @title Video 15: Ethics\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1Hw41197oB\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"Kt6JLi3rUFU\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\ndisplay(out)",
"_____no_output_____"
],
[
"# @title Video 16: Be a group\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1j44y1272h\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"Sfp6--d_H1A\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\ndisplay(out)",
"_____no_output_____"
],
[
"# @title Video 17: Syllabus\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1iB4y1N7uQ\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"cDvAqG_hAvQ\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"Meet our lecturers:\n\nWeek 1: the building blocks\n* [Konrad Kording](https://kordinglab.com)\n* [Andrew Saxe](https://www.saxelab.org/)\n* [Surya Ganguli](https://ganguli-gang.stanford.edu/)\n* [Ioannis Mitliagkas](http://mitliagkas.github.io/)\n* [Lyle Ungar](https://www.cis.upenn.edu/~ungar/)\n\nWeek 2: making things work\n* [Alona Fyshe](https://webdocs.cs.ualberta.ca/~alona/)\n* [Alexander Ecker](https://eckerlab.org/)\n* [James Evans](https://sociology.uchicago.edu/directory/james-evans)\n* [He He](https://hhexiy.github.io/)\n* [Vikash Gilja](https://tnel.ucsd.edu/bio) and [Akash Srivastava](https://akashgit.github.io/)\n\nWeek 3: more magic\n* [Tim Lillicrap](https://contrastiveconvergence.net/~timothylillicrap/index.php) and [Blake Richards](https://www.mcgill.ca/neuro/blake-richards-phd)\n* [Jane Wang](http://www.janexwang.com/) and [Feryal Behbahani](https://feryal.github.io/)\n* [Tim Lillicrap](https://contrastiveconvergence.net/~timothylillicrap/index.php) and [Blake Richards](https://www.mcgill.ca/neuro/blake-richards-phd)\n* [Josh Vogelstein](https://jovo.me/) and [Vincenzo Lamonaco](https://www.vincenzolomonaco.com/)\n\nNow, go to the [visualization of ICLR papers](https://iclr.cc/virtual/2021/paper_vis.html). Read a few abstracts. Look at the various clusters. Where do you see yourself in this map?\n",
"_____no_output_____"
],
[
"---\n# Submit to Airtable",
"_____no_output_____"
]
],
[
[
"# @title Video 18: Submission info\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = \"https://player.bilibili.com/player.html?bvid={0}&page={1}\".format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=f\"BV1e44y127ti\", width=854, height=480, fs=1)\n print(\"Video available at https://www.bilibili.com/video/{0}\".format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=f\"JwTn7ej2dq8\", width=854, height=480, fs=1, rel=0)\n print(\"Video available at https://youtube.com/watch?v=\" + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"This is Darryl, the Deep Learning Dapper Lion, and he's here to teach you about content submission to airtable.\n<br>\n<img src=\"https://raw.githubusercontent.com/NeuromatchAcademy/course-content-dl/main/tutorials/static/DapperLion.png\" alt=\"Darryl\"> \n<br><br>\nAt the end of each tutorial there will be an <b>Airtable Submission Cell</b>. Run the cell to generate the airtable submission button and click on it to submit your information to airtable.\n<br><br>\nif it is the last tutorial of the day your button will look like this and take you to the end of day survey: \n<br>\n<img src=\"https://raw.githubusercontent.com/NeuromatchAcademy/course-content-dl/main/tutorials/static/SurveyButton.png?raw=1\" alt=\"Survey Button\">\n\notherwise it look like this: \n<br>\n\n<img src=\"https://raw.githubusercontent.com/NeuromatchAcademy/course-content-dl/main/tutorials/static/AirtableSubmissionButton.png?raw=1\" alt=\"Submission Button\"> \n<br><br>\n\nIt is critical that you push the submit button for every tutorial you run. <b> even if you don't finish the tutorial, still submit!</b>\nSubmitting is the only way we can verify that you attempted each tutorial, which is critical for us to be able to track your progress.",
"_____no_output_____"
],
[
"### TL;DR: Basic tutorial workflow\n1. work through the tutorial, answering Think! questions and code exercises\n\n2. at end each tutorial, (even if tutorial incomplete) run the airtable submission code cell\n\n3. Push the submission button\n\n4. if the last tutorial of the day, Submission button will also take you to the end of the day survey on a new page. complete that and submit it.",
"_____no_output_____"
],
[
"### Submission FAQs: \n\n1. What if I want to change my answers to previous discussion questions? \n> you are free to change and resubmit any of the answers and Think! questions as many times as you like. However, <b> please only run the airtable submission code and click on the link once you are ready to submit</b>.\n\n2. Okay, but what if I submitted my airtable anyway and reallly want to resubmit?\n> After making changes, you can re-run the airtable submission cell code cell. This will result in a second submission from you for the data. This will make Darryl sad as it will be more work for him to clean up the data later. \n\n3. HELP! I accidentally ran the code to generate the airtable submission button before I was ready to submit! what do I do?\n> If you run the code to generate the link, anything that happens afterwards will not be captured. Complete the tutorial and make sure to re-run the airtable submission again when you are finished before pressing the submission button. \n4. What if I want to work on this on my own later, should I wait to submit until I'm finished?\n> Please submit wherever you are at the end of the day. It's graet that you want to keep working on this, but it's important to see the places where we tried things that didn't quite work out, so we can fix them for next year. \n\nFinally, we try to keep the airtable code as hidden as possible, but if you ever see any calls to `atform` such as `atform.add_event()` in the coding exercises, just know that is for saving airtable information only.<b> It will not affect the code that is being run around it in any way</b> , so please do not modify, comment out, or worry about any of those lines of code.\n<br><br><br>\nNow, lets try submitting today's course to Airtable by running the next cell and clicking the button when it appears. ",
"_____no_output_____"
]
],
[
[
"# @title Airtable Submission Link\nfrom IPython import display as IPyDisplay\nIPyDisplay.HTML(\n f\"\"\"\n <div>\n <a href= \"{atform.url()}\" target=\"_blank\">\n <img src=\"https://github.com/NeuromatchAcademy/course-content-dl/blob/main/tutorials/static/SurveyButton.png?raw=1\"\n alt=\"button link to survey\" style=\"width:410px\"></a>\n </div>\"\"\" )",
"_____no_output_____"
]
],
[
[
"---\n# Bonus - 60 years of Machine Learning Research in one Plot\n\nby [Hendrik Strobelt](http://hendrik.strobelt.com) (MIT-IBM Watson AI Lab) with support from Benjamin Hoover.\n\nIn this notebook we visualize a subset* of 3,300 articles retreived from the AllenAI [S2ORC dataset](https://github.com/allenai/s2orc). We represent each paper by a position that is output of a dimensionality reduction method applied to a vector representation of each paper. The vector representation is output of a neural network.\n\n*The selection is very biased on the keywords and methodology we used to filter. Please see the details section to learn about what we did. ",
"_____no_output_____"
]
],
[
[
"# @title Import `altair` and load the data\n!pip install altair vega_datasets --quiet\nimport requests\nimport altair as alt # altair is defining data visualizations\n\n# Source data files\n# Position data file maps ID to x,y positions\n# original link: http://gltr.io/temp/ml_regexv1_cs_ma_citation+_99perc.pos_umap_cosine_100_d0.1.json\nPOS_FILE = 'https://osf.io/qyrfn/download'\n# original link: http://gltr.io/temp/ml_regexv1_cs_ma_citation+_99perc_clean.csv\n# Metadata file maps ID to title, abstract, author,....\nMETA_FILE = 'https://osf.io/vfdu6/download'\n\n# data loading and wrangling\ndef load_data():\n positions = pd.read_json(POS_FILE)\n positions[['x', 'y']] = positions['pos'].to_list()\n meta = pd.read_csv(META_FILE)\n return positions.merge(meta, left_on='id', right_on='paper_id')\n\n# load data\ndata = load_data()",
"_____no_output_____"
],
[
"# @title Define Visualization using ALtair\nYEAR_PERIOD = \"quinquennial\" # @param\nselection = alt.selection_multi(fields=[YEAR_PERIOD], bind='legend')\ndata[YEAR_PERIOD] = (data[\"year\"] / 5.0).apply(np.floor) * 5\nchart = alt.Chart(data[[\"x\", \"y\", \"authors\", \"title\", YEAR_PERIOD, \"citation_count\"]], width=800,\n height=800).mark_circle(radius=2, opacity=0.2).encode(\n alt.Color(YEAR_PERIOD+':O',\n scale=alt.Scale(scheme='viridis', reverse=False, clamp=True, domain=list(range(1955,2020,5))),\n # legend=alt.Legend(title='Total Records')\n ),\n alt.Size('citation_count',\n scale=alt.Scale(type=\"pow\", exponent=1, range=[15, 300])\n ),\n alt.X('x:Q',\n scale=alt.Scale(zero=False), axis=alt.Axis(labels=False)\n ),\n alt.Y('y:Q',\n scale=alt.Scale(zero=False), axis=alt.Axis(labels=False)\n ),\n tooltip=['title', 'authors'],\n # size='citation_count',\n # color=\"decade:O\",\n opacity=alt.condition(selection, alt.value(.8), alt.value(0.2)),\n\n).add_selection(\n selection\n).interactive()",
"_____no_output_____"
]
],
[
[
"Lets look at the Visualization. Each dot represents one paper. Close dots mean that the respective papers are closer related than distant ones. The color indicates the 5-year period of when the paper was published. The dot size indicates the citation count (within S2ORC corpus) as of July 2020. \n\nThe view is **interactive** and allows for three main interactions. Try them and play around.\n1. hover over a dot to see a tooltip (title, author)\n2. select a year in the legend (right) to filter dots\n2. zoom in/out with scroll -- double click resets view ",
"_____no_output_____"
]
],
[
[
"chart",
"_____no_output_____"
]
],
[
[
"## Questions\n\nBy playing around, can you find some answers to the follwing questions?\n\n1. Can you find topical clusters? What cluster might occur because of a filtering error?\n2. Can you see a temporal trend in the data and clusters?\n2. Can you determine when deep learning methods started booming ?\n3. Can you find the key papers that where written before the DL \"winter\" that define milestones for a cluster? (tipp: look for large dots of different color)",
"_____no_output_____"
],
[
"## Methods\n\nHere is what we did:\n1. Filtering of all papers who fullfilled the criterria:\n - are categorized as `Computer Science` or `Mathematics` \n - one of the following keywords appearing in title or abstract: `\"machine learning|artificial intelligence|neural network|(machine|computer) vision|perceptron|network architecture| RNN | CNN | LSTM | BLEU | MNIST | CIFAR |reinforcement learning|gradient descent| Imagenet \"`\n2. per year, remove all papers that are below the 99 percentile of citation count in that year\n3. embed each paper by using abstract+title in SPECTER model\n4. project based on embedding using UMAP\n5. visualize using Altair",
"_____no_output_____"
],
[
"### Find Authors",
"_____no_output_____"
]
],
[
[
"# @title Edit the `AUTHOR_FILTER` variable to full text search for authors.\n\nAUTHOR_FILTER = \"Rush \" # @param space at the end means \"word border\"\n\n### Don't ignore case when searching...\nFLAGS = 0\n### uncomment do ignore case\n# FLAGS = re.IGNORECASE\n\n## --- FILTER CODE.. make it your own ---\nimport re\ndata['issel'] = data['authors'].str.contains(AUTHOR_FILTER, na=False, flags=FLAGS, )\nif data['issel'].mean()<0.0000000001:\n print('No match found')\n\n## --- FROM HERE ON VIS CODE ---\nalt.Chart(data[[\"x\", \"y\", \"authors\", \"title\", YEAR_PERIOD, \"citation_count\", \"issel\"]], width=800,\n height=800) \\\n .mark_circle(stroke=\"black\", strokeOpacity=1).encode(\n alt.Color(YEAR_PERIOD+':O',\n scale=alt.Scale(scheme='viridis', reverse=False),\n # legend=alt.Legend(title='Total Records')\n ),\n alt.Size('citation_count',\n scale=alt.Scale(type=\"pow\", exponent=1, range=[15, 300])\n ),\n alt.StrokeWidth('issel:Q', scale=alt.Scale(type=\"linear\", domain=[0,1], range=[0, 2]), legend=None),\n\n alt.Opacity('issel:Q', scale=alt.Scale(type=\"linear\", domain=[0,1], range=[.2, 1]), legend=None),\n alt.X('x:Q',\n scale=alt.Scale(zero=False), axis=alt.Axis(labels=False)\n ),\n alt.Y('y:Q',\n scale=alt.Scale(zero=False), axis=alt.Axis(labels=False)\n ),\n tooltip=['title', 'authors'],\n).interactive()",
"_____no_output_____"
]
],
[
[
"---\n# Appendix\n\n## Official PyTorch resources:\n### Tutorials\nhttps://pytorch.org/tutorials/\n\n### Documentation\n\n https://pytorch.org/docs/stable/tensors.html (tensor methods)\n\n https://pytorch.org/docs/stable/tensors.html#torch.Tensor.view (The view method in particular)\n\n https://pytorch.org/vision/stable/datasets.html (pre-loaded image datasets)\n\n ## Google Colab Resources:\n https://research.google.com/colaboratory/faq.html (FAQ including guidance on GPU usage)\n\n ## Books for reference:\n\nhttps://www.deeplearningbook.org/ (Deep Learning by Ian Goodfellow, Yoshua Bengio and Aaron Courville)\n\n ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0e2b858140becbbb4c1c97112fa075a2be4fe20 | 11,288 | ipynb | Jupyter Notebook | Laborategia/.ipynb_checkpoints/Laborategia 1-checkpoint.ipynb | mpenagar/Programazioaren-Oinarriak | 831dd1c1ec6cbd290f958328acc0132185b89e96 | [
"MIT"
] | null | null | null | Laborategia/.ipynb_checkpoints/Laborategia 1-checkpoint.ipynb | mpenagar/Programazioaren-Oinarriak | 831dd1c1ec6cbd290f958328acc0132185b89e96 | [
"MIT"
] | null | null | null | Laborategia/.ipynb_checkpoints/Laborategia 1-checkpoint.ipynb | mpenagar/Programazioaren-Oinarriak | 831dd1c1ec6cbd290f958328acc0132185b89e96 | [
"MIT"
] | null | null | null | 24.275269 | 238 | 0.534904 | [
[
[
"### 1 - e-posta baten histograma\n\nAhalik eta funtzio gehien sortuko ditugu, azpiproblema bakoitza funtzio beten bidez adierazteko. Adibidez:\n\n* e-posta batetako testua, ilara zerrenda moduan (goiburua deskartatu)\n* hitz batetako ertzetatik puntuazio zeinuak kendu\n* hitz bat soilik karaktere alfabetikoez osotua ote dagoen",
"_____no_output_____"
]
],
[
[
"def eposta(bideizena,kodifikazioa=\"utf-8\"):\n with open(bideizena,encoding=kodifikazioa) as f:\n for ilara in f:\n if ilara == \"\\n\" :\n break\n return list(f)",
"_____no_output_____"
]
],
[
[
"`string` modulua erabil dezakegu puntuazio zeinu guztiak lortzeko (seguruenik batenbat ahaztuko genuke...):",
"_____no_output_____"
]
],
[
[
"import string\nstring.punctuation",
"_____no_output_____"
],
[
"def hitza_garbitu(w):\n \n return ...",
"_____no_output_____"
]
],
[
[
"Prozesatzera goazen testuak ingelesez daudenez, `string.ascii_letters` erabil dezakegu:",
"_____no_output_____"
]
],
[
[
"string.ascii_letters",
"_____no_output_____"
],
[
"def soilik_alfa(w):\n \n return True/False",
"_____no_output_____"
]
],
[
[
"Aurreko funtzioak erabiliz, `histograma` funtzioa askozaz errazagoa izango da:",
"_____no_output_____"
]
],
[
[
"def histograma(bideizena):\n h = {}\n for ilara in eposta(bideizena):\n for w in ilara.split():\n w = hitza_garbitu(w)\n if soilik_alfa(w) :\n w = w.lower()\n h[w] ...\n return h",
"_____no_output_____"
]
],
[
[
"### 2 - histogramen *batura*\n\nBi histograma ditugularik, beraien batura histograma bat izango da. Histograma horretan, batutako bi histogrametako hitz guztiak agertuko dira eta balioak hi histogrametako balioen batura izango da. \n",
"_____no_output_____"
]
],
[
[
"def hist_batura(h1,h2):\n # h1 = h1 + h2\n # return h1 + h2",
"_____no_output_____"
]
],
[
[
"### 3 - histogramen normalizazioa\n\nAgerpen kopuruetatik maiztasunetara",
"_____no_output_____"
]
],
[
[
"def hist_norm(h):\n ...\n return hnorm",
"_____no_output_____"
],
[
"h = ...\nh_norm = hist_norm(h)\nsum(h_norm.values()) == 1",
"_____no_output_____"
]
],
[
[
"### 4 - Gai baten eredua sortu\n\nGai batetako eredua (histograma normalizatua) lortzeko, gai horretako korreo guztien histogramen batura kalkulatuko dugu eta azkenik histograma hori normalizatu:\n\n$$h_{gaia} = norm \\left( \\sum_{posta \\in gaia}{h_{posta}} \\right)$$\n\nHau lortzeko ondoko pausuak emango ditugu:\n\n* Gai batetako entrenamenduko fitxategi bideizenen zerrenda lortu\n* Bideizen zerrenda batetako korrreoen histogramen batura lortu\n* Histogramen baturaren normalizazioa egin\n",
"_____no_output_____"
],
[
"#### Entrenamenduko fitxategiak\n\nGai bat dugularik, adibidez `alt.atheism`, bere entrenamenduko fitxategien bideizen zerrenda `20news-bydate/listas_train/alt.atheism.txt` fitxategian egongo da. Gure kasuan:",
"_____no_output_____"
]
],
[
[
"karpeta = '/home/data/'\ngaia = 'alt.atheism'\nbideizena = karpeta + '20news-bydate/listas_train/' + gaia + '.txt'",
"_____no_output_____"
]
],
[
[
"Fitxategi horretan, ilara bakoitzean entrenamenduko korreo baten bideizen **erlatiboa** agertzen da:\n\n```\n20news-bydate-train/alt.atheism/49960\n20news-bydate-train/alt.atheism/51060\n20news-bydate-train/alt.atheism/51119\n20news-bydate-train/alt.atheism/51120\n20news-bydate-train/alt.atheism/51121\n...\n```\nOndoko kodeak, esaterako, bideizen guzti horiek dituen zerrenda lortuko du:",
"_____no_output_____"
]
],
[
[
"karpeta = '/home/data/'\ngaia = 'alt.atheism'\nbideizena = karpeta + '20news-bydate/listas_train/' + gaia + '.txt'\nwith open(bideizena,encoding=\"utf8\") as f:\n izenak = f.readlines()",
"_____no_output_____"
]
],
[
[
"Baina bideizen horiek ez dira absolutuak (aurretik '/home/data/20news-bydate/') gehitu behar zaie eta amaieran duten `\"\\n\"`-a kendu behar zaie. Sortu gai baten entrenamenduko bideizen absolutuen zerrenda bueltatuko duen funtzioa:",
"_____no_output_____"
]
],
[
[
"def gaiko_entrenamendu_bideizenak(gaia):\n z = []\n ...\n return z",
"_____no_output_____"
]
],
[
[
"Sortu gai batetako eredua, hau da, histogramen batura normalizatua lortuko duen funtzioa:",
"_____no_output_____"
]
],
[
[
"def eredua(gaia):\n H = {}\n for bideizena in gaiko_entrenamendu_bideizenak(gaia):\n # h = histograma(...)\n # joan H-n h guztiak batzen\n # H normalizatu\n # emaitza buetatu",
"_____no_output_____"
]
],
[
[
"### 5 - Gai guztien ereduak sortu\n\n`20news-bydate/gaiak.txt` fitxategian gai guztiak ditugu. Sortu gai guztien izen zerrenda bueltatuko duen funtzioa:",
"_____no_output_____"
]
],
[
[
"def gai_zerrenda():\n ...",
"_____no_output_____"
]
],
[
[
"Azkenik sortu gai bakoitzaren eredua.",
"_____no_output_____"
]
],
[
[
"def Ereduak():\n # Eredu guztiak gordetzen dituen (hiztegien) hiztegia sortu\n # E[gaia] = eredua(gaia)\n return E",
"_____no_output_____"
]
],
[
[
"### 6 - Histograma normalizatuen arteko distantzia\n\n#### Gainjartze distantzia\n\n$$d(h_{a},h_{b}) = 1 - \\sum_{w \\in a}{\\min(h_{a}(w),h_{b}(w))}$$\n\n\n#### Euklidesen distantzia\n\n$$d(h_{a},h_{b}) = \\sqrt{ \\sum_{w \\in a,b}{(h_{a}(w)-h_{b}(w))^2} }$$\n\n#### korrelazio distantzia\n\n$$d(h_{a},h_{b}) = 1 - \\sum_{w \\in a}{h_{a}(w) \\cdot h_{b}(w)}$$\n",
"_____no_output_____"
]
],
[
[
"def dist_gainjartze(ha,hb):",
"_____no_output_____"
],
[
"def dist_euklides(ha,hb):",
"_____no_output_____"
],
[
"def dist_korrelazio(ha,hb):",
"_____no_output_____"
]
],
[
[
"### 7 - Histograma normalizatua sailkatu\n",
"_____no_output_____"
]
],
[
[
"def sailkatu(h,E,dist=dist_gainjartze):\n #dist()\n z = [ (d,gaia) , (d,gaia) , (d,gaia), ... ]\n z.sort()\n return z",
"_____no_output_____"
]
],
[
[
"### 8 - Errore tasa neurtu\n\nTest-eko posta elektroniko bat sailkatzen dugunean lehenengo posizioan postari dagokion gaia ez badago, sailkapen errore bat dugu. Errore tasa, $k$ errore kopurua eta $N$ posta kopuru arteko zatidura izango da:\n\n$$errore\\_tasa = \\frac{k}{N}$$",
"_____no_output_____"
],
[
"#### test-eko fitxategiak\n\nGai bat dugularik, adibidez `alt.atheism`, bere test-eko fitxategien bideizen zerrenda `20news-bydate/listas_test/alt.atheism.txt` fitxategian egongo da. Gure kasuan:",
"_____no_output_____"
]
],
[
[
"karpeta = '/home/data/'\ngaia = 'alt.atheism'\n#frogak egiteko\n#bideizena = karpeta + '20news-bydate/listas_minitest/' + gaia + '.txt'\nbideizena = karpeta + '20news-bydate/listas_test/' + gaia + '.txt'",
"_____no_output_____"
],
[
"def gaiko_test_bideizenak(gaia):\n z = []\n ...\n return z",
"_____no_output_____"
]
],
[
[
"#### Errore tasa lortu",
"_____no_output_____"
]
],
[
[
"def errore_tasa(E,dist=dist_gainjartze):\n #gai bakoitza zeharkatu\n for gaia in ... :\n #gaiko test-eko bideizenak zeharkatu\n for bideizena in ...\n #histograma normalizatua lortu\n #sailkatu\n #lehenengo gaia zuzena ez bada, errorea kontabilizatu\n return k/N",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0e2d1c8ee98ba16217d38c4fc309d3c8a7db2a4 | 91,761 | ipynb | Jupyter Notebook | assets/nb-lessons/11_classification_metrics.ipynb | gklarenberg/ml-training-site | 91de94d684f62f5c2f028981196620b7a28ff2ff | [
"MIT"
] | null | null | null | assets/nb-lessons/11_classification_metrics.ipynb | gklarenberg/ml-training-site | 91de94d684f62f5c2f028981196620b7a28ff2ff | [
"MIT"
] | null | null | null | assets/nb-lessons/11_classification_metrics.ipynb | gklarenberg/ml-training-site | 91de94d684f62f5c2f028981196620b7a28ff2ff | [
"MIT"
] | null | null | null | 89.522927 | 26,316 | 0.813777 | [
[
[
"# Classification metrics\nAuthor: Geraldine Klarenberg\n\nBased on the Google Machine Learning Crash Course",
"_____no_output_____"
],
[
"## Tresholds\nIn previous lessons, we have talked about using regression models to predict values. But sometimes we are interested in **classifying** things: \"spam\" vs \"not spam\", \"bark\" vs \"not barking\", etc. \n\nLogistic regression is a great tool to use in ML classification models. We can use the outputs from these models by defining **classification thresholds**. For instance, if our model tells us there's a probability of 0.8 that an email is spam (based on some characteristics), the model classifies it as such. If the probability estimate is less than 0.8, the model classifies it as \"not spam\". The threshold allows us to map a logistic regression value to a binary category (the prediction).\n\nTresholds are problem-dependent, so they will have to be tuned for the specific problem you are dealing with.\n\nIn this lesson we will look at metrics you can use to evaluate a classification model's predictions, and what changing the threshold does to your model and predictions.",
"_____no_output_____"
],
[
"## True, false, positive, negative...\n\nNow, we could simply look at \"accuracy\": the ratio of all correct predictions to all predictions. This is simple, intuitive and straightfoward. \n\nBut there are some problems with this approach:\n* This approach does not work well if there is (class) imbalance; situations where certain negative or positive values or outcomes are rare; \n* and, most importantly: different kind of mistakes can have different costs...",
"_____no_output_____"
],
[
"### The boy who cried wolf...\n\nWe all know the story!\n\n\n\nFor this example, we define \"there actually is a wolf\" as a positive class, and \"there is no wolf\" as a negative class. The predictions that a model makes can be true or false for both classes, generating 4 outcomes:\n\n",
"_____no_output_____"
],
[
"This table is also called a *confusion matrix*.\n\nThere are 2 metrics we can derive from these outcomes: precision and recall.\n\n## Precision\nPrecision asks the question what proportion of the positive predictions was actually correct?\n\nTo calculate the precision of your model, take all true positives divided by *all* positive predictions:\n$$\\text{Precision} = \\frac{TP}{TP+FP}$$\n\nBasically: **did the model cry 'wolf' too often or too little?**\n\n**NB** If your model produces no negative positives, the value of the precision is 1.0. Too many negative positives gives values greater than 1, too few gives values less than 1.",
"_____no_output_____"
],
[
"### Exercise\nCalculate the precision of a model with the following outcomes\n\ntrue positives (TP): 1 | false positives (FP): 1 \n-------|--------\n**false negatives (FN): 8** | **true negatives (TN): 90** ",
"_____no_output_____"
],
[
"## Recall\nRecall tries to answer the question what proportion of actual positives was answered correctly?\n\nTo calculate recall, divide all true positives by the true positives plus the false negatives:\n$$\\text{Recall} = \\frac{TP}{TP+FN}$$\n\nBasically: **how many wolves that tried to get into the village did the model actually get?**\n\n**NB** If the model produces no false negative, recall equals 1.0",
"_____no_output_____"
],
[
"### Exercise\nFor the same confusion matrix as above, calculate the recall.",
"_____no_output_____"
],
[
"## Balancing precision and recall\nTo evaluate your model, should look at **both** precision and recall. They are often in tension though: improving one reduces the other.\nLowering the classification treshold improves recall (your model will call wolf at every little sound it hears) but will negatively affect precision (it will call wolf too often).",
"_____no_output_____"
],
[
"### Exercise\n#### Part 1\nLook at the outputs of a model that classifies incoming emails as \"spam\" or \"not spam\".\n\n\n\nThe confusion matrix looks as follows\n\ntrue positives (TP): 8 | false positives (FP): 2 \n-------|--------\n**false negatives (FN): 3** | **true negatives (TN): 17** \n\nCalculate the precision and recall for this model.\n\n#### Part 2\nNow see what happens to the outcomes (below) if we increase the threshold\n\n\n\nThe confusion matrix looks as follows\n\ntrue positives (TP): 7 | false positives (FP): 4 \n-------|--------\n**false negatives (FN): 1** | **true negatives (TN): 18** \n\nCalculate the precision and recall again.\n\n**Compare the precision and recall from the first and second model. What do you notice?** ",
"_____no_output_____"
],
[
"## Evaluate model performance\nWe can evaluate the performance of a classification model at all classification thresholds. For all different thresholds, calculate the *true positive rate* and the *false positive rate*. The true positive rate is synonymous with recall (and sometimes called *sensitivity*) and is thus calculated as\n\n$ TPR = \\frac{TP} {TP + FN} $\n\nFalse positive rate (sometimes called *specificity*) is:\n\n$ FPR = \\frac{FP} {FP + TN} $\n\nWhen you plot the pairs of TPR and FPR for all the different thresholds, you get a Receiver Operating Characteristics (ROC) curve. Below is a typical ROC curve.\n\n\n\nTo evaluate the model, we look at the area under the curve (AUC). The AUC has a probabilistic interpretation: it represents the probability that a random positive (green) example is positioned to the right of a random negative (red) example.\n\n\n\nSo if that AUC is 0.9, that's the probability the pair-wise prediction is correct. Below are a few visualizations of AUC results. On top are the distributions of the outcomes of the negative and positive outcomes at various thresholds. Below is the corresponding ROC.\n\n \n\n**This AUC suggests a perfect model** (which is suspicious!)\n",
"_____no_output_____"
],
[
"\n\n**This is what most AUCs look like**. In this case, AUC = 0.7 means that there is 70% chance the model will be able to distinguish between positive and negative classes.",
"_____no_output_____"
],
[
"\n\n**This is actually the worst case scenario.** This model has no discrimination capacity at all... ",
"_____no_output_____"
],
[
"## Prediction bias\nLogistic regression should be unbiased, meaning that the average of the predictions should be more or less equal to the average of the observations. **Prediction bias** is the difference between the average of the predictions and the average of the labels in a data set.\n\nThis approach is not perfect, e.g. if your model almost always predicts the average there will not be much bias. However, if there **is** bias (\"significant nonzero bias\"), that means there is something something going on that needs to be checked, specifically that the model is wrong about the frequency of positive labels.\n\nPossible root causes of prediction bias are:\n* Incomplete feature set\n* Noisy data set\n* Buggy pipeline\n* Biased training sample\n* Overly strong regularization\n\n### Buckets and prediction bias\nFor logistic regression, this process is a bit more involved, as the labels assigned to an examples are either 0 or 1. So you cannot accurately predict the prediction bias based on one example. You need to group data in \"buckets\" and examine the prediction bias on that. Prediction bias for logistic regression only makes sense when grouping enough examples together to be able to compare a predicted value (for example, 0.392) to observed values (for example, 0.394). \n\nYou can create buckets by linearly breaking up the target predictions, or create quantiles. \n\nThe plot below is a calibration plot. Each dot represents a bucket with 1000 values. On the x-axis we have the average value of the predictions for that bucket and on the y-axis the average of the actual observations. Note that the axes are on logarithmic scales.\n\n",
"_____no_output_____"
],
[
"## Coding\nRecall the logistic regression model we made in the previous lesson. That was a perfect fit, so not that useful when we look at the metrics we just discussed.\n\nIn the cloud plot with the sepal length and petal width plotted against each other, it is clear that the other two iris species are less separated. Let's use one of these as an example. We'll rework the example so we're classifying irises for being \"virginica\" or \"not virginica\". ",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nfrom sklearn.datasets import load_iris\nimport pandas as pd\n\niris = load_iris()\n\nX = iris.data\ny = iris.target\n\ndf = pd.DataFrame(X, \n columns = ['sepal_length(cm)',\n 'sepal_width(cm)',\n 'petal_length(cm)',\n 'petal_width(cm)'])\n\ndf['species_id'] = y\n\nspecies_map = {0: 'setosa', 1: 'versicolor', 2: 'virginica'}\n\ndf['species_name'] = df['species_id'].map(species_map)\n\ndf.head()",
"_____no_output_____"
]
],
[
[
"Now extract the data we need and create the necessary dataframes again.",
"_____no_output_____"
]
],
[
[
"X = np.c_[X[:,0], X[:,3]]",
"_____no_output_____"
],
[
"y = []\nfor i in range(len(X)):\n if i > 99:\n y.append(1)\n else:\n y.append(0)\n\ny = np.array(y)\n\nplt.scatter(X[:,0], X[:,1], c = y)",
"_____no_output_____"
]
],
[
[
"Create our test and train data, and run a model. The default classification threshold is 0.5. If the predicted probability is > 0.5, the predicted result is 'virgnica'. If it is < 0.5, the predicted result is 'not virginica'.",
"_____no_output_____"
]
],
[
[
"random = np.random.permutation(len(X))\nx_train = X[random][30:]\nx_test = X[random][:30]\n\ny_train= y[random][30:]\ny_test = y[random][:30]",
"_____no_output_____"
],
[
"from sklearn.linear_model import LogisticRegression\n\nlog_reg = LogisticRegression()\nlog_reg.fit(x_train,y_train)",
"_____no_output_____"
]
],
[
[
"Instead of looking at the probabilities and the plot, like in the last lesson, let's run some classification metrics on the training dataset. \n\nIf you use \".score\", you get the mean accuracy.",
"_____no_output_____"
]
],
[
[
"log_reg.score(x_train, y_train)",
"_____no_output_____"
]
],
[
[
"Let's predict values and see what this ouput means and how we can look at other metrics.",
"_____no_output_____"
]
],
[
[
"predictions = log_reg.predict(x_train)\npredictions, y_train",
"_____no_output_____"
]
],
[
[
"There is a way to look at the confusion matrix. The output that is generated has the same structure as the confusion matrices we showed earlier:\n\ntrue positives (TP) | false positives (FP) \n-------|--------\n**false negatives (FN)** | **true negatives (TN)** ",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import confusion_matrix\nconfusion_matrix(y_train, predictions)",
"_____no_output_____"
]
],
[
[
"Indeed, for the accuracy calculation: we predicted 81 + 33 = 114 correct (true positives and true negatives), and 114/120 (remember, our training data had 120 points) = 0.95.\n\nThere is also a function to calculate recall and precision:\n\nSince we also have a testing data set, let's see what the metrics look like for that.",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import recall_score\nrecall_score(y_train, predictions)",
"_____no_output_____"
],
[
"from sklearn.metrics import precision_score\nprecision_score(y_train, predictions)",
"_____no_output_____"
]
],
[
[
"And, of course, there are also built-in functions to check the ROC curve and AUC! For these functions, the inputs are the labels of the original dataset and the predicted probabilities (- not the predicted labels -> **why?**). Remember what the two columns mean?",
"_____no_output_____"
]
],
[
[
"proba_virginica = log_reg.predict_proba(x_train)\nproba_virginica[0:10]",
"_____no_output_____"
],
[
"from sklearn.metrics import roc_curve\nfpr_model, tpr_model, thresholds_model = roc_curve(y_train, proba_virginica[:,1])",
"_____no_output_____"
],
[
"fpr_model",
"_____no_output_____"
],
[
"tpr_model",
"_____no_output_____"
],
[
"thresholds_model",
"_____no_output_____"
]
],
[
[
"Plot the ROC curve as follows",
"_____no_output_____"
]
],
[
[
"plt.plot(fpr_model, tpr_model,label='our model')\nplt.plot([0,1],[0,1],label='random')\nplt.plot([0,0,1,1],[0,1,1,1],label='perfect')\nplt.xlabel('False Positive Rate')\nplt.ylabel('True Positive Rate')\nplt.legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"The AUC:",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import roc_auc_score\nauc_model = roc_auc_score(y_train, proba_virginica[:,1])\nauc_model",
"_____no_output_____"
]
],
[
[
"You can use the ROC and AUC metric to evaluate competing models. Many people prefer to use these metrics to analyze each model’s performance because it does not require selecting a threshold and helps balance true positive rate and false positive rate.",
"_____no_output_____"
],
[
"Now let's do the same thing for our test data (but again, this dataset is fairly small, and K-fold cross-validation is recommended).",
"_____no_output_____"
]
],
[
[
"log_reg.score(x_test, y_test)",
"_____no_output_____"
],
[
"predictions = log_reg.predict(x_test)\npredictions, y_test",
"_____no_output_____"
],
[
"confusion_matrix(y_test, predictions)",
"_____no_output_____"
],
[
"recall_score(y_test, predictions)",
"_____no_output_____"
],
[
"precision_score(y_test, predictions)",
"_____no_output_____"
],
[
"proba_virginica = log_reg.predict_proba(x_test)\nfpr_model, tpr_model, thresholds_model = roc_curve(y_test, proba_virginica[:,1])",
"_____no_output_____"
],
[
"plt.plot(fpr_model, tpr_model,label='our model')\nplt.plot([0,1],[0,1],label='random')\nplt.plot([0,0,1,1],[0,1,1,1],label='perfect')\nplt.xlabel('False Positive Rate')\nplt.ylabel('True Positive Rate')\nplt.legend()\nplt.show()",
"_____no_output_____"
],
[
"auc_model = roc_auc_score(y_test, proba_virginica[:,1])\nauc_model",
"_____no_output_____"
]
],
[
[
"Learn more about the logistic regression function and options at https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LogisticRegression.html",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
d0e2d29d448028491fdf8c493f6afbc9d076fb9e | 5,168 | ipynb | Jupyter Notebook | lectures_2020/lecture005/kmeans_animation.ipynb | sroet/chemometrics | c797505d07e366319ba1544e8a602be94b88fbb6 | [
"CC-BY-4.0"
] | 5 | 2020-02-04T12:09:19.000Z | 2021-08-06T12:28:24.000Z | lectures_2020/lecture005/kmeans_animation.ipynb | sroet/chemometrics | c797505d07e366319ba1544e8a602be94b88fbb6 | [
"CC-BY-4.0"
] | 72 | 2020-01-06T10:24:33.000Z | 2022-01-21T10:37:46.000Z | lectures_2020/lecture005/kmeans_animation.ipynb | sroet/chemometrics | c797505d07e366319ba1544e8a602be94b88fbb6 | [
"CC-BY-4.0"
] | 4 | 2020-01-09T12:04:26.000Z | 2021-01-19T10:06:14.000Z | 26.639175 | 85 | 0.508707 | [
[
[
"from matplotlib import pyplot as plt\nfrom matplotlib.colors import ListedColormap\nfrom matplotlib import animation\nfrom IPython.display import HTML\nimport numpy as np\nfrom sklearn.datasets import make_blobs\nfrom sklearn.cluster import KMeans\n%matplotlib inline",
"_____no_output_____"
],
[
"# Set up colors:\ncolor_map = ListedColormap(['#1b9e77', '#d95f02', '#7570b3'])",
"_____no_output_____"
],
[
"# Make raw data using blobs\nX, y = make_blobs(\n n_samples=200,\n n_features=2,\n centers=3,\n cluster_std=0.5,\n shuffle=True,\n random_state=0\n)",
"_____no_output_____"
],
[
"# Plot the raw data:\nplt.scatter(X[:,0], X[:, 1], c=y, cmap=color_map)",
"_____no_output_____"
],
[
"# Define some initial centers, these are designed to be \"bad\":\ninitial = np.array(\n [\n [-3.0, 0.0],\n [-2.0, 0.0],\n [-1.0, 0.0],\n ]\n)\ny_c = [0, 1, 2]",
"_____no_output_____"
],
[
"# Plot the initial situation:\nfig0, ax0 = plt.subplots(constrained_layout=True)\nax0.scatter(X[:,0], X[:, 1], c=y, cmap=color_map)\nax0.scatter(initial[:,0], initial[:, 1],\n c=y_c, cmap=color_map, marker='X', edgecolors='black', s=150)\nax0.text(-3, 5, 'Iteration: 0', fontsize='xx-large')\nxlim = ax0.get_xlim()\nylim = ax0.get_ylim()\nresults = [(np.full_like(y, 0), initial)]",
"_____no_output_____"
],
[
"# Run clustering for different number of maximum iterations:\nmax_clusters = 15\nfor iterations in range(1, 15):\n km = KMeans(n_clusters=3, init=initial, n_init=1, max_iter=iterations,\n random_state=0)\n y_km = km.fit_predict(X)\n results.append((y_km, km.cluster_centers_))\n figi, axi = plt.subplots(constrained_layout=True)\n axi.scatter(X[:, 0], X[:, 1], c=y_km, cmap=color_map)\n axi.set_xlim(xlim)\n axi.set_ylim(ylim)\n axi.text(-3, 5, 'Iteration: {}'.format(iterations), fontsize='xx-large')\n axi.scatter(km.cluster_centers_[:, 0],\n km.cluster_centers_[:, 1],\n c=y_c, cmap=color_map,\n marker='X', edgecolors='black', s=150)",
"_____no_output_____"
],
[
"%%capture\n# Create an animation:\nfig2, ax2 = plt.subplots(constrained_layout=True)\nax2.set_xlim(xlim)\nax2.set_ylim(ylim)\nyi, centers = results[0]\nscat = ax2.scatter(X[:, 0], X[:, 1])\ncent = ax2.scatter(centers[:, 0], centers[:, 1], c=y_c, cmap=color_map,\n marker='X', edgecolors='black', s=250)\ntext = ax2.text(-3, 5, 'Iteration: 0', fontsize='xx-large')",
"_____no_output_____"
],
[
"def animate(i):\n \"\"\"Update the animation.\"\"\"\n yi, centers = results[i]\n cent.set_offsets(centers)\n text.set_text('Iteration: {}'.format(i))\n if i == 0:\n colors = ['#377eb8' for _ in yi]\n else:\n colors = [color_map.colors[j] for j in yi]\n scat.set_facecolors(colors)\n return (cent, scat, text)",
"_____no_output_____"
],
[
"anim = animation.FuncAnimation(fig2, animate,\n frames=max_clusters,\n interval=300, blit=True, repeat=True)",
"_____no_output_____"
],
[
"HTML(anim.to_jshtml())",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e2e59bbe409e5620ccf523d0ca62d07a2938bf | 7,616 | ipynb | Jupyter Notebook | examples/example_analysis_data_creation.ipynb | MeighenBergerS/fourth_day | 57e7abb64b5fd7efb06d8a6ad7e886f91e993edc | [
"MIT"
] | null | null | null | examples/example_analysis_data_creation.ipynb | MeighenBergerS/fourth_day | 57e7abb64b5fd7efb06d8a6ad7e886f91e993edc | [
"MIT"
] | null | null | null | examples/example_analysis_data_creation.ipynb | MeighenBergerS/fourth_day | 57e7abb64b5fd7efb06d8a6ad7e886f91e993edc | [
"MIT"
] | null | null | null | 34.152466 | 136 | 0.506434 | [
[
[
"# Name: example_analysis_data_creation.ipynb\n# uthors: Stephan Meighen-Berger\n# Example how to construct a data set to use for later analyses",
"_____no_output_____"
],
[
"# General imports\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport sys\nfrom tqdm import tqdm\nimport pickle",
"_____no_output_____"
],
[
"# Adding path to module\nsys.path.append(\"../\")",
"_____no_output_____"
],
[
"# picture path\nPICS = '../pics/'",
"_____no_output_____"
],
[
"# Module imports\nfrom fourth_day import Fourth_Day, config",
"_____no_output_____"
],
[
"# Scenario Settings\n# These are general settings pertaining to the simulation run\nconfig['scenario']['population size'] = 10 # The starting population size\nconfig['scenario']['duration'] = 600 * 5 # Total simulation time in seconds\nconfig['scenario']['exclusion'] = True # If an exclusion zone should be used (the detector)\nconfig['scenario']['injection']['rate'] = 1e-2 # Injection rate in per second, a distribution is constructed from this value\nconfig['scenario']['injection']['y range'] = [5., 10.] # The y-range of injection\nconfig['scenario']['light prop'] = { # Where the emitted light should be propagated to (typically the detector location)\n \"switch\": True, # If light should be propagated\n \"x_pos\": 3., # The x-coordinates\n \"y_pos\": 0.5 * 15. - 0.15, # The y-coordinates\n }\nconfig['scenario']['detector'] = { # detector specific properties, positions are defined as offsets from the light prop values\n \"switch\": True, # If the detector should be modelled\n \"type\": \"PMTSpec\", # Detector name, implemented types are given in the config\n \"response\": True, # If a detector response should be used\n \"acceptance\": \"Flat\", # Flat acceptance\n \"mean detection prob\": 0.5 # Used for the acceptance calculation\n}\n# ---------------------------------------------\n# Organisms\n# Organisms properties are defined here\nconfig['organisms']['emission fraction'] = 0.2 # Amount of energy an organism uses per pulse\nconfig['organisms']['alpha'] = 1e1 # Proportionality factor for the emission probability\nconfig['organisms'][\"minimal shear stress\"] = 0.05 # The minimal amount of shear stress needed to emit (generic units)\nconfig[\"organisms\"][\"filter\"] = 'depth' # Method of filtering organisms (here depth)\nconfig[\"organisms\"][\"depth filter\"] = 1000. # Organisms need to exist below this depth\n# ---------------------------------------------\n# Geometry\n# These settings define the geometry of the system\n# Typically a box (simulation volume) with a spherical exclusion zone (detector)\nconfig['geometry']['volume'] = {\n 'function': 'rectangle',\n 'x_length': 30.,\n 'y_length': 15.,\n 'offset': None,\n}\n# Reduce the observation size to reduce the computational load\nconfig['geometry']['observation'] = {\n 'function': 'rectangle',\n 'x_length': 30.,\n 'y_length': 15.,\n \"offset\": np.array([0., 0.]),\n}\n# The detector volume\nconfig['geometry'][\"exclusion\"] = {\n \"function\": \"sphere\",\n \"radius\": 0.15,\n \"x_pos\": 3.,\n \"y_pos\": 0.5 * 15. - 0.15,\n}\n# ---------------------------------------------\n# Water\n# Properties of the current model\nconfig['water']['model']['name'] = 'custom' # Use a custom (non analytic) model\nconfig['water']['model']['off set'] = np.array([0., 0.]) # Offset of the custom model\nconfig['water']['model']['directory'] = \"../data/current/Parabola_5mm/run_2cm_npy/\" # The files used by the custom model\nconfig['water']['model']['time step'] = 5. # in Seconds",
"_____no_output_____"
],
[
"# Which detectors we want to use\nwavelengths = {\n \"Detector 1\": [\"1\", \"#4575b4\"],\n \"Detector 5\": [\"2\", \"#91bfdb\"],\n \"Detector 8\": [\"3\", \"#e0f3f8\"],\n \"Detector 3\": [\"4\", \"#fee090\"],\n \"Detector 10\": [\"5\", \"#fc8d59\"],\n}",
"_____no_output_____"
],
[
"# Launching multiple simulations to use in the analysis\nseeds = np.arange(100)[1:]\nprint(seeds)\ncounter = 0\nfor seed in tqdm(seeds):\n # General\n config[\"general\"][\"random state seed\"] = seed\n # Creating a fourth_day object\n fd = Fourth_Day()\n # Launching solver\n fd.sim()\n # Fetching relevant data\n # Totals\n total = fd.measured[\"Detector 1\"].values\n for detector in wavelengths.keys():\n if detector == \"Detector 1\":\n continue\n fd.measured[detector].values\n total += fd.measured[detector].values\n storage_dic = {\n \"time\": fd.t,\n \"data\": total\n }\n pickle.dump(storage_dic, open(\"../data/storage/vort_run_%d.p\" % counter, \"wb\"))\n counter += 1",
"\r 0%| | 0/99 [00:00<?, ?it/s]"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e2fb15922fbbc484039bc065a886ef8ffd83df | 51,756 | ipynb | Jupyter Notebook | C_Impact of p_negative.ipynb | GLopezMUZH/ABM_NurseSchedullingModel | 40411a45025d50f26e47344d1f0114712eb2bf73 | [
"MIT"
] | null | null | null | C_Impact of p_negative.ipynb | GLopezMUZH/ABM_NurseSchedullingModel | 40411a45025d50f26e47344d1f0114712eb2bf73 | [
"MIT"
] | null | null | null | C_Impact of p_negative.ipynb | GLopezMUZH/ABM_NurseSchedullingModel | 40411a45025d50f26e47344d1f0114712eb2bf73 | [
"MIT"
] | null | null | null | 241.850467 | 26,292 | 0.908938 | [
[
[
"# Case: Impact of p_negative",
"_____no_output_____"
],
[
"Situation: \n- 70% agent availability on weekdays and weekends\n\nTask:\n- Evaluate Shift Coverage over p_negative: [.001, .0025, .005, .0075, .01, .02, .03, .05, .1, .2]\n- Evaluate Agent Satisfaction over p_negative: [.001, .0025, .005, .0075, .01, .02, .03, .05, .1, .2]",
"_____no_output_____"
]
],
[
[
"import abm_scheduling\nfrom abm_scheduling import Schedule as Schedule\nfrom abm_scheduling import Nurse as Nurse",
"_____no_output_____"
],
[
"import time\nfrom datetime import datetime\n\nimport abm_scheduling.Log\nfrom abm_scheduling.Log import Log as Log\n\nimport matplotlib.pylab as plt\n%matplotlib inline\n\nlog = Log()",
"_____no_output_____"
]
],
[
[
"## Define situation",
"_____no_output_____"
]
],
[
[
"num_nurses_per_shift = 5\nnum_nurses = 25\ndegree_of_agent_availability = 0.7\nworks_weekends = True\n\np_negatives = [.001, .0025, .005, .0075, .01, .02, .03, .05, .1, .2]",
"_____no_output_____"
],
[
"schedule = Schedule(num_nurses_needed=num_nurses_per_shift, is_random=True)\nmodel = abm_scheduling.NSP_AB_Model()\nnurses = model.generate_nurses(num_nurses=num_nurses,\n degree_of_agent_availability=degree_of_agent_availability,\n works_weekends=works_weekends)\n\nschedule.print_schedule(schedule_name=\"Intial Situation\")",
"Week's Schedule Intial Situation\n+---------+----------+----------+----------+----------+----------+----------+----------+\n| | Mo | Tu | We | Th | Fr | Sa | So |\n+---------+----------+----------+----------+----------+----------+----------+----------+\n| shift 1 | need: 7 | need: 6 | need: 5 | need: 8 | need: 5 | need: 6 | need: 5 |\n| | nurses: | nurses: | nurses: | nurses: | nurses: | nurses: | nurses: |\n| shift 2 | need: 5 | need: 5 | need: 6 | need: 7 | need: 7 | need: 4 | need: 6 |\n| | nurses: | nurses: | nurses: | nurses: | nurses: | nurses: | nurses: |\n| shift 3 | need: 7 | need: 7 | need: 5 | need: 6 | need: 4 | need: 6 | need: 5 |\n| | nurses: | nurses: | nurses: | nurses: | nurses: | nurses: | nurses: |\n+---------+----------+----------+----------+----------+----------+----------+----------+\n"
],
[
"p_neg_reg_util_results = []\nfor p_neg in p_negatives:\n p_neg_reg_util_result = model.run(schedule_org=schedule,\n nurses_org=nurses,\n p_to_accept_negative_change=p_neg,\n utility_function_parameters=None,\n print_stats=False)\n p_neg_reg_util_results.append(p_neg_reg_util_result)",
"_____no_output_____"
],
[
"plt.figure()\nplt.plot(p_negatives, [r.shift_coverage for r in p_neg_reg_util_results], label=\"Shift Coverage\")\nplt.title(f'Shift coverage as a function of p_negative')\nplt.xlabel(\"p_negative\")\nplt.ylabel(\"Shift Coverage\")\nplt.legend()\nplt.show()",
"_____no_output_____"
],
[
"p_neg_as_util_results = []\nfor p_neg in p_negatives:\n p_neg_as_util_result = model.run(schedule_org=schedule,\n nurses_org=nurses,\n p_to_accept_negative_change=p_neg,\n utility_function_parameters=None,\n print_stats=False)\n p_neg_as_util_results.append(p_neg_as_util_result)",
"_____no_output_____"
],
[
"plt.figure()\nplt.plot(p_negatives, [r.total_agent_satisfaction for r in p_neg_as_util_results], label=\"Agent Satisfaction\")\nplt.title(f'Agent satisfaction as a function of p_negative')\nplt.xlabel(\"p_negative\")\nplt.ylabel(\"Agent Satisfaction\")\nplt.legend()\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e2fdd8a9db9f41c036ac2ed9ab7faefedfbca5 | 191,603 | ipynb | Jupyter Notebook | Classifiers/Day1/Model_0_Basic_Features_256_1e-2_swish_mult2_200E.ipynb | GilesStrong/Kaggle_HiggsML | 5c066554f87defb29275f4e6f1930a1641d542bc | [
"MIT"
] | null | null | null | Classifiers/Day1/Model_0_Basic_Features_256_1e-2_swish_mult2_200E.ipynb | GilesStrong/Kaggle_HiggsML | 5c066554f87defb29275f4e6f1930a1641d542bc | [
"MIT"
] | null | null | null | Classifiers/Day1/Model_0_Basic_Features_256_1e-2_swish_mult2_200E.ipynb | GilesStrong/Kaggle_HiggsML | 5c066554f87defb29275f4e6f1930a1641d542bc | [
"MIT"
] | null | null | null | 219.728211 | 139,414 | 0.896839 | [
[
[
"# First Attempt\nbatch size 256 lr 1e-3",
"_____no_output_____"
],
[
"### Import modules",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nfrom __future__ import division\nimport sys\nimport os\nos.environ['MKL_THREADING_LAYER']='GNU'\nsys.path.append('../')\nfrom Modules.Basics import *\nfrom Modules.Class_Basics import *",
"/home/giles/anaconda2/lib/python2.7/site-packages/statsmodels/compat/pandas.py:56: FutureWarning: The pandas.core.datetools module is deprecated and will be removed in a future version. Please use the pandas.tseries module instead.\n from pandas.core import datetools\nUsing Theano backend.\n"
]
],
[
[
"## Options",
"_____no_output_____"
]
],
[
[
"classTrainFeatures = basic_features\nclassModel = 'modelSwish'\nvarSet = \"basic_features\"\n\nnSplits = 10\nensembleSize = 10\nensembleMode = 'loss'\n\nmaxEpochs = 200\ncompileArgs = {'loss':'binary_crossentropy', 'optimizer':'adam'}\ntrainParams = {'epochs' : 1, 'batch_size' : 256, 'verbose' : 0}\nmodelParams = {'version':classModel, 'nIn':len(classTrainFeatures), 'compileArgs':compileArgs}\n\nprint \"\\nTraining on\", len(classTrainFeatures), \"features:\", [var for var in classTrainFeatures]",
"\nTraining on 30 features: ['DER_mass_MMC', 'DER_mass_transverse_met_lep', 'DER_mass_vis', 'PRI_tau_pt', 'DER_deltar_tau_lep', 'PRI_jet_leading_eta', 'PRI_lep_eta', 'PRI_tau_eta', 'DER_lep_eta_centrality', 'DER_mass_jet_jet', 'DER_deltaeta_jet_jet', 'DER_pt_tot', 'DER_met_phi_centrality', 'PRI_jet_num', 'PRI_met', 'DER_pt_h', 'DER_pt_ratio_lep_tau', 'DER_sum_pt', 'PRI_jet_leading_pt', 'PRI_jet_subleading_eta', 'PRI_jet_all_pt', 'PRI_met_sumet', 'DER_prodeta_jet_jet', 'PRI_lep_pt', 'PRI_jet_subleading_pt', 'PRI_met_phi', 'PRI_jet_leading_phi', 'PRI_lep_phi', 'PRI_jet_subleading_phi', 'PRI_tau_phi']\n"
]
],
[
[
"## Import data",
"_____no_output_____"
]
],
[
[
"trainData = h5py.File(dirLoc + 'train.hdf5', \"r+\")\nvalData = h5py.File(dirLoc + 'val.hdf5', \"r+\")",
"_____no_output_____"
]
],
[
[
"## Determine LR",
"_____no_output_____"
]
],
[
[
"lrFinder = batchLRFindClassifier(trainData, nSplits, getClassifier, modelParams, trainParams, lrBounds=[1e-5,0.1], trainOnWeights=False, verbose=0)",
"_____no_output_____"
],
[
"compileArgs['lr'] = 1e-2",
"_____no_output_____"
]
],
[
[
"## Train classifier",
"_____no_output_____"
]
],
[
[
"results, histories = batchTrainClassifier(trainData, nSplits, getClassifier, modelParams, trainParams, cosAnnealMult=2, trainOnWeights=False, patience=200, maxEpochs=maxEpochs, verbose=1)",
"Using cosine annealing\nRunning fold 1 / 10\n2 classes found, running in binary mode\n\n1 New best found: 0.400561252252\n3 New best found: 0.378149787368\n6 New best found: 0.371132547385\n7 New best found: 0.367546338621\n13 New best found: 0.365329898119\n14 New best found: 0.363622459261\n15 New best found: 0.362778083891\n27 New best found: 0.362323531985\n28 New best found: 0.361314095019\n29 New best found: 0.360359164307\n30 New best found: 0.360207265465\n31 New best found: 0.360053416153\n58 New best found: 0.359536564909\n59 New best found: 0.359524779017\n60 New best found: 0.359370332884\n61 New best found: 0.359236776003\n62 New best found: 0.359138851781\nEarly stopping after 262 epochs\nScore is: {'loss': 0.35913885178074928, 'AUC': 0.091134905531261579, 'wAUC': 0.068973002518172732}\nFold took 221.853s\n\nRunning fold 2 / 10\n1 New best found: 0.401903512347\n2 New best found: 0.391692429919\n3 New best found: 0.379573900817\n6 New best found: 0.375494268449\n7 New best found: 0.370313402707\n14 New best found: 0.366249649046\n15 New best found: 0.365609946229\n25 New best found: 0.365609822355\n27 New best found: 0.364753267433\n29 New best found: 0.362467366022\n30 New best found: 0.361972920443\n55 New best found: 0.361758306303\n57 New best found: 0.360882739564\n59 New best found: 0.360404335175\n61 New best found: 0.360233538717\n62 New best found: 0.360221377798\nEarly stopping after 262 epochs\nScore is: {'loss': 0.36022137779786606, 'AUC': 0.092260790390672476, 'wAUC': 0.069028090143481435}\nFold took 218.044s\n\nRunning fold 3 / 10\n1 New best found: 0.402499031298\n2 New best found: 0.393286604285\n3 New best found: 0.37831263848\n6 New best found: 0.371401502888\n7 New best found: 0.370405659064\n13 New best found: 0.368147329216\n14 New best found: 0.367712372741\n15 New best found: 0.366819790413\n27 New best found: 0.366649085637\n28 New best found: 0.364753157958\n29 New best found: 0.364111734193\n30 New best found: 0.363359704223\n31 New best found: 0.363291525586\n57 New best found: 0.361748802082\n62 New best found: 0.361733175117\n63 New best found: 0.361716643863\nEarly stopping after 263 epochs\nScore is: {'loss': 0.36171664386295471, 'AUC': 0.09273474547175109, 'wAUC': 0.068348602646494006}\nFold took 216.031s\n\nRunning fold 4 / 10\n1 New best found: 0.398489952104\n2 New best found: 0.397302892314\n3 New best found: 0.375507921179\n6 New best found: 0.368396755834\n7 New best found: 0.366812843585\n13 New best found: 0.366642022393\n14 New best found: 0.364470832075\n15 New best found: 0.364113589039\n26 New best found: 0.362805404758\n27 New best found: 0.361670011848\n29 New best found: 0.361307285078\n30 New best found: 0.360717426293\n31 New best found: 0.360642859631\n57 New best found: 0.360494245985\n58 New best found: 0.359723593938\n60 New best found: 0.359694371535\n61 New best found: 0.359585634852\n62 New best found: 0.359474920075\n63 New best found: 0.359459306769\nEarly stopping after 263 epochs\nScore is: {'loss': 0.3594593067691354, 'AUC': 0.091928152908256067, 'wAUC': 0.07414236343664693}\nFold took 219.483s\n\nRunning fold 5 / 10\n1 New best found: 0.400227668724\n2 New best found: 0.389430728501\n3 New best found: 0.378947721036\n6 New best found: 0.373907483201\n7 New best found: 0.368085665044\n13 New best found: 0.366634270497\n14 New best found: 0.364187235957\n15 New best found: 0.362443283766\n28 New best found: 0.362227589029\n29 New best found: 0.362017792269\n30 New best found: 0.361387037048\n31 New best found: 0.361315149108\n55 New best found: 0.360782400927\n57 New best found: 0.360621922463\n58 New best found: 0.360281934191\n59 New best found: 0.359434385129\n60 New best found: 0.359411212781\n61 New best found: 0.359217418431\n62 New best found: 0.359215865111\n63 New best found: 0.359199308138\nEarly stopping after 263 epochs\nScore is: {'loss': 0.35919930813815532, 'AUC': 0.091406327327032244, 'wAUC': 0.068798651604689898}\nFold took 217.892s\n\nRunning fold 6 / 10\n1 New best found: 0.395253763781\n2 New best found: 0.385610660231\n3 New best found: 0.371539680148\n6 New best found: 0.368604496993\n7 New best found: 0.362322907656\n13 New best found: 0.361033487181\n14 New best found: 0.359503528573\n15 New best found: 0.358836954246\n26 New best found: 0.358073462105\n27 New best found: 0.355962787903\n28 New best found: 0.354837585622\n54 New best found: 0.354306453143\n58 New best found: 0.354021308051\n59 New best found: 0.353561773015\n60 New best found: 0.353241695464\n61 New best found: 0.353175475386\n62 New best found: 0.353154634453\n63 New best found: 0.353122269688\nEarly stopping after 263 epochs\nScore is: {'loss': 0.35312226968797134, 'AUC': 0.088355160898867968, 'wAUC': 0.065134879372251486}\nFold took 218.049s\n\nRunning fold 7 / 10\n1 New best found: 0.403426565813\n2 New best found: 0.39652233875\n3 New best found: 0.381249469247\n6 New best found: 0.374146268316\n7 New best found: 0.371582792588\n12 New best found: 0.371481210009\n13 New best found: 0.368887275094\n14 New best found: 0.366502197667\n15 New best found: 0.365790644186\n27 New best found: 0.364874081445\n29 New best found: 0.363248953627\n30 New best found: 0.362993282529\n31 New best found: 0.362833528747\n58 New best found: 0.362225916699\n59 New best found: 0.361424808801\n60 New best found: 0.361389370525\n63 New best found: 0.361383996214\nEarly stopping after 263 epochs\nScore is: {'loss': 0.36138399621391465, 'AUC': 0.092800479575312456, 'wAUC': 0.071808333174538053}\nFold took 220.947s\n\nRunning fold 8 / 10\n1 New best found: 0.406740896777\n2 New best found: 0.390777788073\n3 New best found: 0.380913545721\n6 New best found: 0.373353284001\n7 New best found: 0.371314225214\n13 New best found: 0.370230202063\n14 New best found: 0.367118355099\n15 New best found: 0.366223674172\n26 New best found: 0.365735905008\n27 New best found: 0.364510371496\n29 New best found: 0.364136617414\n30 New best found: 0.363498089709\n31 New best found: 0.363410157922\n54 New best found: 0.362878268972\n57 New best found: 0.362152574128\n59 New best found: 0.361270689989\n61 New best found: 0.361149474211\n62 New best found: 0.36107059767\n63 New best found: 0.361065514653\nEarly stopping after 263 epochs\nScore is: {'loss': 0.36106551465312653, 'AUC': 0.092703151387366955, 'wAUC': 0.068813843933388297}\nFold took 220.481s\n\nRunning fold 9 / 10\n1 New best found: 0.398462290645\n2 New best found: 0.3982423496\n3 New best found: 0.380724895715\n6 New best found: 0.376076513037\n7 New best found: 0.371555348524\n13 New best found: 0.369503058937\n14 New best found: 0.367363461168\n15 New best found: 0.366514191866\n26 New best found: 0.366005328579\n28 New best found: 0.365103807572\n29 New best found: 0.364885870243\n30 New best found: 0.364425115426\n31 New best found: 0.364239335693\n57 New best found: 0.363272260145\n59 New best found: 0.362432865489\n60 New best found: 0.362228694808\n61 New best found: 0.362018548702\n62 New best found: 0.362017504298\n63 New best found: 0.362010562017\nEarly stopping after 263 epochs\nScore is: {'loss': 0.36201056201729931, 'AUC': 0.093165815117476236, 'wAUC': 0.069295056715387937}\nFold took 222.766s\n\nRunning fold 10 / 10\n1 New best found: 0.409788827698\n2 New best found: 0.400589831695\n3 New best found: 0.384564526186\n6 New best found: 0.37910742848\n7 New best found: 0.375741563249\n14 New best found: 0.373050904657\n15 New best found: 0.371901812666\n27 New best found: 0.371900530735\n28 New best found: 0.370728478325\n29 New best found: 0.370192815734\n30 New best found: 0.369489900123\n31 New best found: 0.369378158389\n59 New best found: 0.369373137743\n60 New best found: 0.369208775984\n61 New best found: 0.368851420849\n62 New best found: 0.368611306237\nEarly stopping after 262 epochs\nScore is: {'loss': 0.36861130623721172, 'AUC': 0.097215422373152505, 'wAUC': 0.072149497991575773}\nFold took 223.560s\n\n\n______________________________________\nTraining finished\nCross-validation took 2199.396s \n"
]
],
[
[
"## Construct ensemble",
"_____no_output_____"
]
],
[
[
"with open('train_weights/resultsFile.pkl', 'r') as fin: \n results = pickle.load(fin)",
"_____no_output_____"
],
[
"ensemble, weights = assembleEnsemble(results, ensembleSize, ensembleMode, compileArgs)",
"Choosing ensemble by loss\nModel 0 is 5 with loss = 0.353122269688\nModel 1 is 0 with loss = 0.359138851781\nModel 2 is 4 with loss = 0.359199308138\nModel 3 is 3 with loss = 0.359459306769\nModel 4 is 1 with loss = 0.360221377798\nModel 5 is 7 with loss = 0.361065514653\nModel 6 is 6 with loss = 0.361383996214\nModel 7 is 2 with loss = 0.361716643863\nModel 8 is 8 with loss = 0.362010562017\nModel 9 is 9 with loss = 0.368611306237\n"
]
],
[
[
"## Response on development data",
"_____no_output_____"
]
],
[
[
"batchEnsemblePredict(ensemble, weights, trainData, ensembleSize=10, verbose=1)",
"Predicting batch 1 out of 10\nPrediction took 0.000193505530107s per sample\n\nPredicting batch 2 out of 10\nPrediction took 0.000175661971811s per sample\n\nPredicting batch 3 out of 10\nPrediction took 0.000175414982512s per sample\n\nPredicting batch 4 out of 10\nPrediction took 0.000165658700466s per sample\n\nPredicting batch 5 out of 10\nPrediction took 0.000176144301891s per sample\n\nPredicting batch 6 out of 10\nPrediction took 0.000171178507805s per sample\n\nPredicting batch 7 out of 10\nPrediction took 0.000160073745251s per sample\n\nPredicting batch 8 out of 10\nPrediction took 0.000167666175877s per sample\n\nPredicting batch 9 out of 10\nPrediction took 0.000181970909134s per sample\n\nPredicting batch 10 out of 10\nPrediction took 0.000160533587058s per sample\n\n"
],
[
"print 'Training ROC AUC: unweighted {}, weighted {}'.format(roc_auc_score(getFeature('targets', trainData), getFeature('pred', trainData)),\n roc_auc_score(getFeature('targets', trainData), getFeature('pred', trainData), sample_weight=getFeature('weights', trainData)))",
"Training ROC AUC: unweighted 0.913288160645, weighted 0.934247941282\n"
]
],
[
[
"## Response on val data",
"_____no_output_____"
]
],
[
[
"batchEnsemblePredict(ensemble, weights, valData, ensembleSize=10, verbose=1)",
"Predicting batch 1 out of 10\nPrediction took 0.000180698823929s per sample\n\nPredicting batch 2 out of 10\nPrediction took 0.000184695577621s per sample\n\nPredicting batch 3 out of 10\nPrediction took 0.000198355388641s per sample\n\nPredicting batch 4 out of 10\nPrediction took 0.00016278181076s per sample\n\nPredicting batch 5 out of 10\nPrediction took 0.000156305646896s per sample\n\nPredicting batch 6 out of 10\nPrediction took 0.000252939605713s per sample\n\nPredicting batch 7 out of 10\nPrediction took 0.000158601379395s per sample\n\nPredicting batch 8 out of 10\nPrediction took 0.000162871599197s per sample\n\nPredicting batch 9 out of 10\nPrediction took 0.00016643781662s per sample\n\nPredicting batch 10 out of 10\nPrediction took 0.000167182588577s per sample\n\n"
],
[
"print 'Testing ROC AUC: unweighted {}, weighted {}'.format(roc_auc_score(getFeature('targets', valData), getFeature('pred', valData)),\n roc_auc_score(getFeature('targets', valData), getFeature('pred', valData), sample_weight=getFeature('weights', valData)))",
"Testing ROC AUC: unweighted 0.908779216407, weighted 0.929871759305\n"
]
],
[
[
"## Evaluation",
"_____no_output_____"
],
[
"### Import in dataframe",
"_____no_output_____"
]
],
[
[
"def convertToDF(datafile, columns={'gen_target', 'gen_weight', 'pred_class'}, nLoad=-1):\n data = pandas.DataFrame()\n data['gen_target'] = getFeature('targets', datafile, nLoad)\n data['gen_weight'] = getFeature('weights', datafile, nLoad)\n data['pred_class'] = getFeature('pred', datafile, nLoad)\n print len(data), \"candidates loaded\"\n return data",
"_____no_output_____"
],
[
"devData = convertToDF(trainData)\nvalData = convertToDF(valData)",
"200000 candidates loaded\n50000 candidates loaded\n"
],
[
"sigVal = (valData.gen_target == 1)\nbkgVal = (valData.gen_target == 0)",
"_____no_output_____"
]
],
[
[
"### MVA distributions",
"_____no_output_____"
]
],
[
[
"getClassPredPlot([valData[bkgVal], valData[sigVal]], weightName='gen_weight')",
"_____no_output_____"
],
[
"def AMS(s, b):\n \"\"\" Approximate Median Significance defined as:\n AMS = sqrt(\n 2 { (s + b + b_r) log[1 + (s/(b+b_r))] - s}\n ) \n where b_r = 10, b = background, s = signal, log is natural logarithm \"\"\"\n \n br = 10.0\n radicand = 2 *( (s+b+br) * math.log (1.0 + s/(b+br)) -s)\n if radicand < 0:\n print 'radicand is negative. Exiting'\n exit()\n else:\n return math.sqrt(radicand)\n \ndef amsScan(inData, res=0.0001):\n best = [0,-1]\n for i in np.linspace(0.,1.,1./res):\n ams = AMS(np.sum(inData.loc[(inData['pred_class'] >= i) & sigVal, 'gen_weight']),\n np.sum(inData.loc[(inData['pred_class'] >= i) & bkgVal, 'gen_weight']))\n if ams > best[1]:\n best = [i, ams]\n print best",
"_____no_output_____"
],
[
"amsScan(valData)",
"/home/giles/anaconda2/lib/python2.7/site-packages/ipykernel_launcher.py:18: DeprecationWarning: object of type <type 'float'> cannot be safely interpreted as an integer.\n"
],
[
"def scoreTest(ensemble, weights, features, cut, name):\n testData = pandas.read_csv('../Data/test.csv')\n with open(dirLoc + 'inputPipe.pkl', 'r') as fin:\n inputPipe = pickle.load(fin)\n\n testData['pred_class'] = ensemblePredict(inputPipe.transform(testData[features].values.astype('float32')), ensemble, weights) \t\n\n testData['Class'] = 'b'\n testData.loc[testData.pred_class >= cut, 'Class'] = 's'\n\n testData.sort_values(by=['pred_class'], inplace=True)\n testData['RankOrder']=range(1, len(testData)+1)\n testData.sort_values(by=['EventId'], inplace=True)\n\n testData.to_csv(dirLoc + name + '_test.csv', columns=['EventId', 'RankOrder', 'Class'], index=False)",
"_____no_output_____"
],
[
"scoreTest(ensemble, weights, classTrainFeatures, 0.83498349834983498, 'Model_0_Basic_Features_256_1e-2_swish_mult2_200E')",
"_____no_output_____"
]
],
[
[
"## Save classified data",
"_____no_output_____"
]
],
[
[
"name = dirLoc + signal + \"_\" + channel + \"_\" + varSet + '_' + classModel + '_classifiedData.csv'\nprint \"Saving data to\", name\nvalData.to_csv(name, columns=['gen_target', 'gen_weight', 'gen_sample', 'pred_class'])",
"_____no_output_____"
]
],
[
[
"## Save/load",
"_____no_output_____"
]
],
[
[
"name = \"weights/DNN_\" + signal + \"_\" + channel + \"_\" + varSet + '_' + classModel\nprint name",
"_____no_output_____"
]
],
[
[
"### Save",
"_____no_output_____"
]
],
[
[
"saveEnsemble(name, ensemble, weights, compileArgs, overwrite=1)",
"_____no_output_____"
]
],
[
[
"### Load",
"_____no_output_____"
]
],
[
[
"ensemble, weights, compileArgs, inputPipe, outputPipe = loadEnsemble(name)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0e31983b9e01dd5cb8258e435e353e254a773c1 | 27,067 | ipynb | Jupyter Notebook | course3/week4/CookingLDA_PA.ipynb | astarostin/MachineLearningSpecializationCoursera | aebecae1effd5e4b148354d28cdad8bc6bd3a415 | [
"Apache-2.0"
] | 1 | 2019-06-23T19:32:59.000Z | 2019-06-23T19:32:59.000Z | course3/week4/CookingLDA_PA.ipynb | astarostin/MachineLearningSpecializationCoursera | aebecae1effd5e4b148354d28cdad8bc6bd3a415 | [
"Apache-2.0"
] | null | null | null | course3/week4/CookingLDA_PA.ipynb | astarostin/MachineLearningSpecializationCoursera | aebecae1effd5e4b148354d28cdad8bc6bd3a415 | [
"Apache-2.0"
] | 4 | 2016-08-21T21:20:28.000Z | 2021-05-10T15:24:16.000Z | 32.729141 | 834 | 0.62748 | [
[
[
"# Programming Assignment\n## Готовим LDA по рецептам",
"_____no_output_____"
],
[
"Как вы уже знаете, в тематическом моделировании делается предположение о том, что для определения тематики порядок слов в документе не важен; об этом гласит гипотеза <<мешка слов>>. Сегодня мы будем работать с несколько нестандартной для тематического моделирования коллекцией, которую можно назвать <<мешком ингредиентов>>, потому что на состоит из рецептов блюд разных кухонь. Тематические модели ищут слова, которые часто вместе встречаются в документах, и составляют из них темы. Мы попробуем применить эту идею к рецептам и найти кулинарные <<темы>>. Эта коллекция хороша тем, что не требует предобработки. Кроме того, эта задача достаточно наглядно иллюстрирует принцип работы тематических моделей.\n\nДля выполнения заданий, помимо часто используемых в курсе библиотек, потребуются модули json и gensim. Первый входит в дистрибутив Anaconda, второй можно поставить командой \n\npip install gensim\n\nили\n\nconda install gensim\n\nПостроение модели занимает некоторое время. На ноутбуке с процессором Intel Core i7 и тактовой частотой 2400 МГц на построение одной модели уходит менее 10 минут.",
"_____no_output_____"
],
[
"### Загрузка данных",
"_____no_output_____"
],
[
"Коллекция дана в json-формате: для каждого рецепта известны его id, кухня (\"cuisine\") и список ингредиентов, в него входящих. Загрузить данные можно с помощью модуля json (он входит в дистрибутив Anaconda):",
"_____no_output_____"
]
],
[
[
"import json",
"_____no_output_____"
],
[
"with open(\"recipes.json\") as f:\n recipes = json.load(f)",
"_____no_output_____"
],
[
"print recipes[1]",
"{u'cuisine': u'southern_us', u'id': 25693, u'ingredients': [u'plain flour', u'ground pepper', u'salt', u'tomatoes', u'ground black pepper', u'thyme', u'eggs', u'green tomatoes', u'yellow corn meal', u'milk', u'vegetable oil']}\n"
]
],
[
[
"### Составление корпуса",
"_____no_output_____"
]
],
[
[
"from gensim import corpora, models\nimport numpy as np",
"Couldn't import dot_parser, loading of dot files will not be possible.\n"
]
],
[
[
"Наша коллекция небольшая и влезает в оперативную память. Gensim может работать с такими данными и не требует их сохранения на диск в специальном формате. Для этого коллекция должна быть представлена в виде списка списков, каждый внутренний список соответствует отдельному документу и состоит из его слов. Пример коллекции из двух документов: \n\n[[\"hello\", \"world\"], [\"programming\", \"in\", \"python\"]]\n\nПреобразуем наши данные в такой формат, а затем создадим объекты corpus и dictionary, с которыми будет работать модель.",
"_____no_output_____"
]
],
[
[
"texts = [recipe[\"ingredients\"] for recipe in recipes]\ndictionary = corpora.Dictionary(texts) # составляем словарь\ncorpus = [dictionary.doc2bow(text) for text in texts] # составляем корпус документов",
"_____no_output_____"
],
[
"corpus[0]",
"_____no_output_____"
],
[
"print texts[0]\nprint corpus[0]",
"[u'romaine lettuce', u'black olives', u'grape tomatoes', u'garlic', u'pepper', u'purple onion', u'seasoning', u'garbanzo beans', u'feta cheese crumbles']\n[(0, 1), (1, 1), (2, 1), (3, 1), (4, 1), (5, 1), (6, 1), (7, 1), (8, 1)]\n"
]
],
[
[
"У объекта dictionary есть две полезных переменных: dictionary.id2token и dictionary.token2id; эти словари позволяют находить соответствие между ингредиентами и их индексами.",
"_____no_output_____"
],
[
"### Обучение модели\nВам может понадобиться [документация](https://radimrehurek.com/gensim/models/ldamodel.html) LDA в gensim.",
"_____no_output_____"
],
[
"__Задание 1.__ Обучите модель LDA с 40 темами, установив количество проходов по коллекции 5 и оставив остальные параметры по умолчанию. Затем вызовите метод модели show_topics, указав количество тем 40 и количество токенов 10, и сохраните результат (топы ингредиентов в темах) в отдельную переменную. Если при вызове метода show_topics указать параметр formatted=True, то топы ингредиентов будет удобно выводить на печать, если formatted=False, будет удобно работать со списком программно. Выведите топы на печать, рассмотрите темы, а затем ответьте на вопрос:\n\nСколько раз ингредиенты \"salt\", \"sugar\", \"water\", \"mushrooms\", \"chicken\", \"eggs\" встретились среди топов-10 всех 40 тем? При ответе __не нужно__ учитывать составные ингредиенты, например, \"hot water\".\n\nПередайте 6 чисел в функцию save_answers1 и загрузите сгенерированный файл в форму.\n\nУ gensim нет возможности фиксировать случайное приближение через параметры метода, но библиотека использует numpy для инициализации матриц. Поэтому, по утверждению автора библиотеки, фиксировать случайное приближение нужно командой, которая написана в следующей ячейке. __Перед строкой кода с построением модели обязательно вставляйте указанную строку фиксации random.seed.__",
"_____no_output_____"
]
],
[
[
"np.random.seed(76543)\n# здесь код для построения модели:\nldamodel = models.ldamodel.LdaModel(corpus, id2word=dictionary, num_topics=40, passes=5)",
"_____no_output_____"
],
[
"topics = ldamodel.show_topics(num_topics=40, num_words=10, formatted=False)",
"_____no_output_____"
],
[
"c_salt, c_sugar, c_water, c_mushrooms, c_chicken, c_eggs = 0, 0, 0, 0, 0, 0\nfor topic in topics:\n for word2prob in topic[1]:\n word = word2prob[0]\n if word == 'salt':\n c_salt += 1\n elif word == 'sugar':\n c_sugar += 1\n elif word == 'water':\n c_water += 1\n elif word == 'mushrooms':\n c_mushrooms += 1\n elif word == 'chicken':\n c_chicken += 1\n elif word == 'eggs':\n c_eggs += 1",
"_____no_output_____"
],
[
"def save_answers1(c_salt, c_sugar, c_water, c_mushrooms, c_chicken, c_eggs):\n with open(\"cooking_LDA_pa_task1.txt\", \"w\") as fout:\n fout.write(\" \".join([str(el) for el in [c_salt, c_sugar, c_water, c_mushrooms, c_chicken, c_eggs]]))",
"_____no_output_____"
],
[
"save_answers1(c_salt, c_sugar, c_water, c_mushrooms, c_chicken, c_eggs)",
"_____no_output_____"
],
[
"print c_salt, c_sugar, c_water, c_mushrooms, c_chicken, c_eggs",
"20 7 8 1 1 1\n"
]
],
[
[
"### Фильтрация словаря\nВ топах тем гораздо чаще встречаются первые три рассмотренных ингредиента, чем последние три. При этом наличие в рецепте курицы, яиц и грибов яснее дает понять, что мы будем готовить, чем наличие соли, сахара и воды. Таким образом, даже в рецептах есть слова, часто встречающиеся в текстах и не несущие смысловой нагрузки, и поэтому их не желательно видеть в темах. Наиболее простой прием борьбы с такими фоновыми элементами - фильтрация словаря по частоте. Обычно словарь фильтруют с двух сторон: убирают очень редкие слова (в целях экономии памяти) и очень частые слова (в целях повышения интерпретируемости тем). Мы уберем только частые слова.",
"_____no_output_____"
]
],
[
[
"import copy\ndictionary2 = copy.deepcopy(dictionary)",
"_____no_output_____"
]
],
[
[
"__Задание 2.__ У объекта dictionary2 есть переменная dfs - это словарь, ключами которого являются id токена, а элементами - число раз, сколько слово встретилось во всей коллекции. Сохраните в отдельный список ингредиенты, которые встретились в коллекции больше 4000 раз. Вызовите метод словаря filter_tokens, подав в качестве первого аргумента полученный список популярных ингредиентов. Вычислите две величины: dict_size_before и dict_size_after - размер словаря до и после фильтрации.\n\nЗатем, используя новый словарь, создайте новый корпус документов, corpus2, по аналогии с тем, как это сделано в начале ноутбука. Вычислите две величины: corpus_size_before и corpus_size_after - суммарное количество ингредиентов в корпусе (иными словами, сумма длин всех документов коллекции) до и после фильтрации.\n\nПередайте величины dict_size_before, dict_size_after, corpus_size_before, corpus_size_after в функцию save_answers2 и загрузите сгенерированный файл в форму.",
"_____no_output_____"
]
],
[
[
"more4000 = [w for w, count in dictionary2.dfs.iteritems() if count > 4000]",
"_____no_output_____"
],
[
"dict_size_before = len(dictionary2.items())\ndictionary2.filter_tokens(bad_ids=more4000)\ndict_size_after = len(dictionary2.items())",
"_____no_output_____"
],
[
"def get_corpus_size(corp):\n res = 0\n for doc in corp:\n res += len(doc)\n #for w in doc:\n # res += w[1]\n return res",
"_____no_output_____"
],
[
"corpus_size_before = get_corpus_size(corpus)\ncorpus2 = [dictionary2.doc2bow(text) for text in texts] # составляем корпус документов\ncorpus_size_after = get_corpus_size(corpus2)",
"_____no_output_____"
],
[
"def save_answers2(dict_size_before, dict_size_after, corpus_size_before, corpus_size_after):\n with open(\"cooking_LDA_pa_task2.txt\", \"w\") as fout:\n fout.write(\" \".join([str(el) for el in [dict_size_before, dict_size_after, corpus_size_before, corpus_size_after]]))",
"_____no_output_____"
],
[
"save_answers2(dict_size_before, dict_size_after, corpus_size_before, corpus_size_after)",
"_____no_output_____"
]
],
[
[
"### Сравнение когерентностей\n__Задание 3.__ Постройте еще одну модель по корпусу corpus2 и словарю dictioanary2, остальные параметры оставьте такими же, как при первом построении модели. Сохраните новую модель в другую переменную (не перезаписывайте предыдущую модель). Не забудьте про фиксирование seed!\n\nЗатем воспользуйтесь методом top_topics модели, чтобы вычислить ее когерентность. Передайте в качестве аргумента соответствующий модели корпус. Метод вернет список кортежей (топ токенов, когерентность), отсортированных по убыванию последней. Вычислите среднюю по всем темам когерентность для каждой из двух моделей и передайте в функцию save_answers3. ",
"_____no_output_____"
]
],
[
[
"np.random.seed(76543)\n# здесь код для построения модели:\nldamodel2 = models.ldamodel.LdaModel(corpus2, id2word=dictionary2, num_topics=40, passes=5)",
"_____no_output_____"
],
[
"coherences = ldamodel.top_topics(corpus)\ncoherences2 = ldamodel2.top_topics(corpus2)",
"_____no_output_____"
],
[
"import numpy as np\nlist1 = np.array([])\nfor coh in coherences:\n list1 = np.append(list1, coh[1])\nlist2 = np.array([])\nfor coh in coherences2:\n list2 = np.append(list2, coh[1])\ncoherence = list1.mean()\ncoherence2 = list2.mean()",
"_____no_output_____"
],
[
"def save_answers3(coherence, coherence2):\n with open(\"cooking_LDA_pa_task3.txt\", \"w\") as fout:\n fout.write(\" \".join([\"%3f\"%el for el in [coherence, coherence2]]))",
"_____no_output_____"
],
[
"save_answers3(coherence, coherence2)",
"_____no_output_____"
]
],
[
[
"Считается, что когерентность хорошо соотносится с человеческими оценками интерпретируемости тем. Поэтому на больших текстовых коллекциях когерентность обычно повышается, если убрать фоновую лексику. Однако в нашем случае этого не произошло. ",
"_____no_output_____"
],
[
"### Изучение влияния гиперпараметра alpha",
"_____no_output_____"
],
[
"В этом разделе мы будем работать со второй моделью, то есть той, которая построена по сокращенному корпусу. \n\nПока что мы посмотрели только на матрицу темы-слова, теперь давайте посмотрим на матрицу темы-документы. Выведите темы для нулевого (или любого другого) документа из корпуса, воспользовавшись методом get_document_topics второй модели:",
"_____no_output_____"
],
[
"Также выведите содержимое переменной .alpha второй модели:",
"_____no_output_____"
],
[
"У вас должно получиться, что документ характеризуется небольшим числом тем. Попробуем поменять гиперпараметр alpha, задающий априорное распределение Дирихле для распределений тем в документах.",
"_____no_output_____"
],
[
"__Задание 4.__ Обучите третью модель: используйте сокращенный корпус (corpus2 и dictionary2) и установите параметр __alpha=1__, passes=5. Не забудьте задать количество тем и зафиксировать seed! Выведите темы новой модели для нулевого документа; должно получиться, что распределение над множеством тем практически равномерное. Чтобы убедиться в том, что во второй модели документы описываются гораздо более разреженными распределениями, чем в третьей, посчитайте суммарное количество элементов, __превосходящих 0.01__, в матрицах темы-документы обеих моделей. Другими словами, запросите темы модели для каждого документа с параметром minimum_probability=0.01 и просуммируйте число элементов в получаемых массивах. Передайте две суммы (сначала для модели с alpha по умолчанию, затем для модели в alpha=1) в функцию save_answers4.",
"_____no_output_____"
]
],
[
[
"def save_answers4(count_model2, count_model3):\n with open(\"cooking_LDA_pa_task4.txt\", \"w\") as fout:\n fout.write(\" \".join([str(el) for el in [count_model2, count_model3]]))",
"_____no_output_____"
]
],
[
[
"Таким образом, гиперпараметр alpha влияет на разреженность распределений тем в документах. Аналогично гиперпараметр eta влияет на разреженность распределений слов в темах.",
"_____no_output_____"
],
[
"### LDA как способ понижения размерности\nИногда распределения над темами, найденные с помощью LDA, добавляют в матрицу объекты-признаки как дополнительные, семантические, признаки, и это может улучшить качество решения задачи. Для простоты давайте просто обучим классификатор рецептов на кухни на признаках, полученных из LDA, и измерим точность (accuracy).\n\n__Задание 5.__ Используйте модель, построенную по сокращенной выборке с alpha по умолчанию (вторую модель). Составьте матрицу $\\Theta = p(t|d)$ вероятностей тем в документах; вы можете использовать тот же метод get_document_topics, а также вектор правильных ответов y (в том же порядке, в котором рецепты идут в переменной recipes). Создайте объект RandomForestClassifier со 100 деревьями, с помощью функции cross_val_score вычислите среднюю accuracy по трем фолдам (перемешивать данные не нужно) и передайте в функцию save_answers5.",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import RandomForestClassifier\nfrom sklearn.cross_validation import cross_val_score",
"_____no_output_____"
],
[
"def save_answers5(accuracy):\n with open(\"cooking_LDA_pa_task5.txt\", \"w\") as fout:\n fout.write(str(accuracy))",
"_____no_output_____"
]
],
[
[
"Для такого большого количества классов это неплохая точность. Вы можете попроовать обучать RandomForest на исходной матрице частот слов, имеющей значительно большую размерность, и увидеть, что accuracy увеличивается на 10-15%. Таким образом, LDA собрал не всю, но достаточно большую часть информации из выборки, в матрице низкого ранга.",
"_____no_output_____"
],
[
"### LDA --- вероятностная модель\nМатричное разложение, использующееся в LDA, интерпретируется как следующий процесс генерации документов.\n\nДля документа $d$ длины $n_d$:\n1. Из априорного распределения Дирихле с параметром alpha сгенерировать распределение над множеством тем: $\\theta_d \\sim Dirichlet(\\alpha)$\n1. Для каждого слова $w = 1, \\dots, n_d$:\n 1. Сгенерировать тему из дискретного распределения $t \\sim \\theta_{d}$\n 1. Сгенерировать слово из дискретного распределения $w \\sim \\phi_{t}$.\n \nПодробнее об этом в [Википедии](https://en.wikipedia.org/wiki/Latent_Dirichlet_allocation).\n\nВ контексте нашей задачи получается, что, используя данный генеративный процесс, можно создавать новые рецепты. Вы можете передать в функцию модель и число ингредиентов и сгенерировать рецепт :)",
"_____no_output_____"
]
],
[
[
"def generate_recipe(model, num_ingredients):\n theta = np.random.dirichlet(model.alpha)\n for i in range(num_ingredients):\n t = np.random.choice(np.arange(model.num_topics), p=theta)\n topic = model.show_topic(0, topn=model.num_terms)\n topic_distr = [x[1] for x in topic]\n terms = [x[0] for x in topic]\n w = np.random.choice(terms, p=topic_distr)\n print w",
"_____no_output_____"
]
],
[
[
"### Интерпретация построенной модели\nВы можете рассмотреть топы ингредиентов каждой темы. Большиснтво тем сами по себе похожи на рецепты; в некоторых собираются продукты одного вида, например, свежие фрукты или разные виды сыра.\n\nПопробуем эмпирически соотнести наши темы с национальными кухнями (cuisine). Построим матрицу A размера темы x кухни, ее элементы $a_{tc}$ - суммы p(t|d) по всем документам d, которые отнесены к кухне c. Нормируем матрицу на частоты рецептов по разным кухням, чтобы избежать дисбаланса между кухнями. Следующая функция получает на вход объект модели, объект корпуса и исходные данные и возвращает нормированную матрицу A. Ее удобно визуализировать с помощью seaborn.",
"_____no_output_____"
]
],
[
[
"import pandas\nimport seaborn\nfrom matplotlib import pyplot as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"def compute_topic_cuisine_matrix(model, corpus, recipes):\n # составляем вектор целевых признаков\n targets = list(set([recipe[\"cuisine\"] for recipe in recipes]))\n # составляем матрицу\n tc_matrix = pandas.DataFrame(data=np.zeros((model.num_topics, len(targets))), columns=targets)\n for recipe, bow in zip(recipes, corpus):\n recipe_topic = model.get_document_topics(bow)\n for t, prob in recipe_topic:\n tc_matrix[recipe[\"cuisine\"]][t] += prob\n # нормируем матрицу\n target_sums = pandas.DataFrame(data=np.zeros((1, len(targets))), columns=targets)\n for recipe in recipes:\n target_sums[recipe[\"cuisine\"]] += 1\n return pandas.DataFrame(tc_matrix.values/target_sums.values, columns=tc_matrix.columns)",
"_____no_output_____"
],
[
"def plot_matrix(tc_matrix):\n plt.figure(figsize=(10, 10))\n seaborn.heatmap(tc_matrix, square=True)",
"_____no_output_____"
],
[
"# Визуализируйте матрицу\n",
"_____no_output_____"
]
],
[
[
"Чем темнее квадрат в матрице, тем больше связь этой темы с данной кухней. Мы видим, что у нас есть темы, которые связаны с несколькими кухнями. Такие темы показывают набор ингредиентов, которые популярны в кухнях нескольких народов, то есть указывают на схожесть кухонь этих народов. Некоторые темы распределены по всем кухням равномерно, они показывают наборы продуктов, которые часто используются в кулинарии всех стран. ",
"_____no_output_____"
],
[
"Жаль, что в датасете нет названий рецептов, иначе темы было бы проще интерпретировать...",
"_____no_output_____"
],
[
"### Заключение\nВ этом задании вы построили несколько моделей LDA, посмотрели, на что влияют гиперпараметры модели и как можно использовать построенную модель. ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
d0e32a36ffe21cdfdd4bb819a57ada442ef1f161 | 10,661 | ipynb | Jupyter Notebook | notebooks/nb_lite.ipynb | jckantor/nbcollection | 37d75ddfb16b8cb4958ae963a6973aa428f5feee | [
"MIT"
] | null | null | null | notebooks/nb_lite.ipynb | jckantor/nbcollection | 37d75ddfb16b8cb4958ae963a6973aa428f5feee | [
"MIT"
] | null | null | null | notebooks/nb_lite.ipynb | jckantor/nbcollection | 37d75ddfb16b8cb4958ae963a6973aa428f5feee | [
"MIT"
] | null | null | null | 28.891599 | 212 | 0.52209 | [
[
[
"# Reading Notebooks\n\n* https://nbformat.readthedocs.io/en/latest/api.html",
"_____no_output_____"
],
[
"## Read a .ipynb file\n\nA notebook consists of metadata, format info, and a list of cells. Very simple.",
"_____no_output_____"
]
],
[
[
"import nbformat\nfrom nbformat.v4.nbbase import new_code_cell, new_markdown_cell, new_notebook\n\n# read notebook file\nfilename = \"02.01-Tagging.ipynb\"\nwith open(filename, \"r\") as fp:\n nb = nbformat.read(fp, as_version=4)\n\n# display file metadata\nprint(f\"nbformat = {nb.nbformat}.{nb.nbformat_minor}\")\ndisplay(nb.metadata)",
"nbformat = 4.4\n"
]
],
[
[
"## Loop over cells\n\nThe heavy lifting is in the list of cells. \n\nHere's how to loop over cells. The cell metadata is editable in JupyterLab, and has a 'tags' key where you manage a list of your own tags. If you wanted to remove cells, this would be the place to tag them.",
"_____no_output_____"
]
],
[
[
"for n, cell in enumerate(nb.cells):\n print(f\"\\nCell {n}\")\n print(\" metadata:\", cell.metadata)\n print(\" cell_type:\", cell.cell_type)\n print(\" keys():\", cell.keys())",
"\nCell 0\n metadata: {}\n cell_type: markdown\n keys(): dict_keys(['cell_type', 'metadata', 'source'])\n\nCell 1\n metadata: {}\n cell_type: markdown\n keys(): dict_keys(['cell_type', 'metadata', 'source'])\n\nCell 2\n metadata: {'tags': ['differential-equations', 'SIR-model', 'compartmental-model']}\n cell_type: markdown\n keys(): dict_keys(['cell_type', 'metadata', 'source'])\n\nCell 3\n metadata: {'tags': ['scipy.integrate.solve_ivp', 'differential-equations']}\n cell_type: code\n keys(): dict_keys(['cell_type', 'execution_count', 'metadata', 'outputs', 'source'])\n\nCell 4\n metadata: {}\n cell_type: markdown\n keys(): dict_keys(['cell_type', 'metadata', 'source'])\n\nCell 5\n metadata: {'tags': ['home-activity', 'differential-equations']}\n cell_type: markdown\n keys(): dict_keys(['cell_type', 'metadata', 'source'])\n\nCell 6\n metadata: {'tags': ['class-activity']}\n cell_type: markdown\n keys(): dict_keys(['cell_type', 'metadata', 'source'])\n\nCell 7\n metadata: {'tags': ['important-note']}\n cell_type: markdown\n keys(): dict_keys(['cell_type', 'metadata', 'source'])\n\nCell 8\n metadata: {}\n cell_type: markdown\n keys(): dict_keys(['cell_type', 'metadata', 'source'])\n\nCell 9\n metadata: {'tags': ['exercise']}\n cell_type: markdown\n keys(): dict_keys(['cell_type', 'metadata', 'source'])\n\nCell 10\n metadata: {'tags': ['exercise']}\n cell_type: code\n keys(): dict_keys(['cell_type', 'execution_count', 'metadata', 'outputs', 'source'])\n\nCell 11\n metadata: {}\n cell_type: code\n keys(): dict_keys(['cell_type', 'execution_count', 'metadata', 'outputs', 'source'])\n"
]
],
[
[
"## Remove Code Elements\n\nRemove code segments with specific tags. This uses regular expressions to identify code segments in code cells. This is actually a bit more general and would allow substitution as well.",
"_____no_output_____"
]
],
[
[
"import re\n\nSOLUTION_CODE = \"### BEGIN SOLUTION(.*)### END SOLUTION\"\nHIDDEN_TESTS = \"### BEGIN HIDDEN TESTS(.*)### END HIDDEN TESTS\"\n\n\ndef replace_code(pattern, replacement):\n regex = re.compile(pattern, re.DOTALL)\n for cell in nb.cells:\n if cell.cell_type == \"code\" and regex.findall(cell.source):\n cell.source = regex.sub(replacement, cell.source)\n print(f\" - {pattern} removed\")\n\n\nreplace_code(SOLUTION_CODE, \"\")\nreplace_code(HIDDEN_TESTS, \"\")",
" - ### BEGIN SOLUTION(.*)### END SOLUTION removed\n - ### BEGIN HIDDEN TESTS(.*)### END HIDDEN TESTS removed\n"
]
],
[
[
"## Remove cells with a specified tag\n\nNote the use of a generator. This keeps things fast, but does need an explicit `list` if you need a list of tagged cells.",
"_____no_output_____"
]
],
[
[
"# a example of an iterator that returns all cells satisfying certain conditions.\ndef get_cells(nb, tag):\n for cell in nb.cells:\n if cell.cell_type == \"markdown\":\n if \"tags\" in cell.metadata.keys():\n if tag in cell.metadata[\"tags\"]:\n yield cell\n\n\ntagged_cells = list(get_cells(nb, \"exercise\"))\ntagged_cells",
"_____no_output_____"
],
[
"# remove all cells with a specified tag\ndef remove_cells(nb, tag):\n tagged_cells = list(get_cells(nb, tag))\n if tagged_cells:\n print(f\" - removing cells with tag {tag}\")\n nb.cells = list(filter(lambda cell: cell not in tagged_cells, nb.cells))\n\n\nremove_cells(nb, \"exercise\")",
" - removing cells with tag exercise\n"
]
],
[
[
"## Write file out",
"_____no_output_____"
]
],
[
[
"with open(\"out.ipynb\", \"w\") as fp:\n nbformat.write(nb, fp)",
"_____no_output_____"
]
],
[
[
"## Execute notebooks\n\nhttps://nbconvert.readthedocs.io/en/latest/execute_api.html",
"_____no_output_____"
]
],
[
[
"import nbformat\nfrom nbconvert.preprocessors import CellExecutionError, ExecutePreprocessor\n\nnb_filename = \"out.ipynb\"\nnb_filename_out = \"executed_\" + nb_filename\n\n# read file\nwith open(\"out.ipynb\", \"r\") as fp:\n nb = nbformat.read(fp, as_version=4)\n\n# instantiate an execution processor allowing errors\nep = ExecutePreprocessor(timeout=600, kernel_name=\"python3\", allow_errors=True)\nep.preprocess(nb, {\"metadata\": {\"path\": \"./\"}})\n\nwith open(nb_filename_out, mode=\"w\", encoding=\"utf-8\") as f:\n nbformat.write(nb, f)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0e3321e6698c241a47dd2950ac83203344fb1ad | 28,693 | ipynb | Jupyter Notebook | analysis/pagination.ipynb | osso73/contabilidad | babdedfdb47b2b4fd01a09e2db9db5d21bbc88f0 | [
"MIT"
] | null | null | null | analysis/pagination.ipynb | osso73/contabilidad | babdedfdb47b2b4fd01a09e2db9db5d21bbc88f0 | [
"MIT"
] | 23 | 2021-12-29T21:41:37.000Z | 2022-03-31T10:01:54.000Z | analysis/pagination.ipynb | osso73/contabilidad | babdedfdb47b2b4fd01a09e2db9db5d21bbc88f0 | [
"MIT"
] | 1 | 2022-02-18T19:58:52.000Z | 2022-02-18T19:58:52.000Z | 50.694346 | 327 | 0.399888 | [
[
[
"# Paginación\n\nEl objetivo es crear una lista de páginas a mostrar. Se mostrará la página actual y las 3 siguientes y las 3 anteriores, y además la primera y la última. El algoritmo debe calcular qué numeros se muestran, y crear una lista con ellos. Además se pondrán espacios en blanco en el lugar donde irán los puntos suspensivos.\n\nSi el número total de páginas es inferior a 10, se mostrarán todas las páginas.",
"_____no_output_____"
]
],
[
[
"def lista_paginas(paginas, actual):\n if paginas < 10:\n pagination = list(range(paginas))\n else:\n if actual <= 4:\n pagination = (actual + 6)*['']\n pagination[-1] = paginas - 1\n pages = range(0, actual+4)\n for i, p in enumerate(pages):\n pagination[i] = p\n elif actual >= paginas - 5:\n pagination = (paginas - actual + 5)*['']\n pagination[0] = 0\n pages = range(actual-3, paginas)\n for i, p in enumerate(pages):\n pagination[i+2] = p\n else:\n pagination = 11*['']\n pagination[0] = 0\n pagination[-1] = paginas -1\n pages = range(actual-3, actual+4)\n for i, p in enumerate(pages):\n pagination[i+2] = p\n return pagination",
"_____no_output_____"
],
[
"# comprobación de que funciona\nfor pags, act in [\n (8, 4), (9, 4), (10, 2), (10, 3), (10, 4), (10, 5), (10, 6), (10, 7), (10, 8), (10, 9), (10, 10),\n (20, 3), (20, 4), (20, 5), (20, 6), (20, 7), (20, 13), (20, 14), (20, 15), (20, 16), (20, 17), (20, 18),\n ]:\n print(pags, act, lista_paginas(pags, act))",
"8 4 [0, 1, 2, 3, 4, 5, 6, 7]\n9 4 [0, 1, 2, 3, 4, 5, 6, 7, 8]\n10 2 [0, 1, 2, 3, 4, 5, '', 9]\n10 3 [0, 1, 2, 3, 4, 5, 6, '', 9]\n10 4 [0, 1, 2, 3, 4, 5, 6, 7, '', 9]\n10 5 [0, '', 2, 3, 4, 5, 6, 7, 8, 9]\n10 6 [0, '', 3, 4, 5, 6, 7, 8, 9]\n10 7 [0, '', 4, 5, 6, 7, 8, 9]\n10 8 [0, '', 5, 6, 7, 8, 9]\n10 9 [0, '', 6, 7, 8, 9]\n10 10 [0, '', 7, 8, 9]\n20 3 [0, 1, 2, 3, 4, 5, 6, '', 19]\n20 4 [0, 1, 2, 3, 4, 5, 6, 7, '', 19]\n20 5 [0, '', 2, 3, 4, 5, 6, 7, 8, '', 19]\n20 6 [0, '', 3, 4, 5, 6, 7, 8, 9, '', 19]\n20 7 [0, '', 4, 5, 6, 7, 8, 9, 10, '', 19]\n20 13 [0, '', 10, 11, 12, 13, 14, 15, 16, '', 19]\n20 14 [0, '', 11, 12, 13, 14, 15, 16, 17, '', 19]\n20 15 [0, '', 12, 13, 14, 15, 16, 17, 18, 19]\n20 16 [0, '', 13, 14, 15, 16, 17, 18, 19]\n20 17 [0, '', 14, 15, 16, 17, 18, 19]\n20 18 [0, '', 15, 16, 17, 18, 19]\n"
]
],
[
[
"Ahora personalizo, puedo escoger el número de páginas anteriores y posteriores a mostrar.",
"_____no_output_____"
]
],
[
[
"def lista_paginas_custom(paginas, actual, num=3):\n \"\"\"Calcula la lista de páginas a mostrar. Las cadenas \n nulas corresponden a puntos suspensivos.\n \n Argumentos\n ----------\n paginas: int\n Total de páginas que hay.\n actual: int\n página actual.\n num: int\n número de paginas anteriores y posteriores que queremos \n mostrar.\n \n \n Devuelve\n --------\n paginacion: list\n lista de páginas calculada.\n \"\"\"\n if paginas < 10:\n pagination = list(range(paginas))\n else:\n if actual <= num + 1:\n pagination = (actual + num + 3)*['']\n pagination[-1] = paginas - 1\n pages = range(0, actual+num+1)\n for i, p in enumerate(pages):\n pagination[i] = p\n elif actual >= paginas - num - 2:\n pagination = (paginas - actual + num + 2)*['']\n pagination[0] = 0\n pages = range(actual-num, paginas)\n for i, p in enumerate(pages):\n pagination[i+2] = p\n else:\n pagination = (num*2 + 5)*['']\n pagination[0] = 0\n pagination[-1] = paginas -1\n pages = range(actual-num, actual+num+1)\n for i, p in enumerate(pages):\n pagination[i+2] = p\n return pagination",
"_____no_output_____"
],
[
"# comprobación de que funciona\nfor pags, act in [\n (8, 4), (9, 4), (10, 2), (10, 3), (10, 4), (10, 5), (10, 6), (10, 7), (10, 8), (10, 9), (10, 10),\n (20, 3), (20, 4), (20, 5), (20, 6), (20, 7), (20, 13), (20, 14), (20, 15), (20, 16), (20, 17), (20, 18),\n ]:\n print(lista_paginas_custom(pags, act) == lista_paginas(pags, act))",
"True\nTrue\nTrue\nTrue\nTrue\nTrue\nTrue\nTrue\nTrue\nTrue\nTrue\nTrue\nTrue\nTrue\nTrue\nTrue\nTrue\nTrue\nTrue\nTrue\nTrue\nTrue\n"
],
[
"# comprobación de que funciona\nfor pags, act in [\n (8, 4), (9, 4), (10, 2), (10, 3), (10, 4), (10, 5), (10, 6), (10, 7), (10, 8), (10, 9), (10, 10),\n (20, 3), (20, 4), (20, 5), (20, 6), (20, 7), (20, 13), (20, 14), (20, 15), (20, 16), (20, 17), (20, 18),\n ]:\n print(f'pags={pags} act={act} num=1: {lista_paginas_custom(pags, act, 1)}')\n print(f'pags={pags} act={act} num=2: {lista_paginas_custom(pags, act, 2)}')\n print(f'pags={pags} act={act} num=3: {lista_paginas_custom(pags, act, 3)}')\n print(f'pags={pags} act={act} num=4: {lista_paginas_custom(pags, act, 4)}')\n print(f'pags={pags} act={act} num=5: {lista_paginas_custom(pags, act, 5)}')\n ",
"pags=8 act=4 num=1: [0, 1, 2, 3, 4, 5, 6, 7]\npags=8 act=4 num=2: [0, 1, 2, 3, 4, 5, 6, 7]\npags=8 act=4 num=3: [0, 1, 2, 3, 4, 5, 6, 7]\npags=8 act=4 num=4: [0, 1, 2, 3, 4, 5, 6, 7]\npags=8 act=4 num=5: [0, 1, 2, 3, 4, 5, 6, 7]\npags=9 act=4 num=1: [0, 1, 2, 3, 4, 5, 6, 7, 8]\npags=9 act=4 num=2: [0, 1, 2, 3, 4, 5, 6, 7, 8]\npags=9 act=4 num=3: [0, 1, 2, 3, 4, 5, 6, 7, 8]\npags=9 act=4 num=4: [0, 1, 2, 3, 4, 5, 6, 7, 8]\npags=9 act=4 num=5: [0, 1, 2, 3, 4, 5, 6, 7, 8]\npags=10 act=2 num=1: [0, 1, 2, 3, '', 9]\npags=10 act=2 num=2: [0, 1, 2, 3, 4, '', 9]\npags=10 act=2 num=3: [0, 1, 2, 3, 4, 5, '', 9]\npags=10 act=2 num=4: [0, 1, 2, 3, 4, 5, 6, '', 9]\npags=10 act=2 num=5: [0, 1, 2, 3, 4, 5, 6, 7, '', 9]\npags=10 act=3 num=1: [0, '', 2, 3, 4, '', 9]\npags=10 act=3 num=2: [0, 1, 2, 3, 4, 5, '', 9]\npags=10 act=3 num=3: [0, 1, 2, 3, 4, 5, 6, '', 9]\npags=10 act=3 num=4: [0, 1, 2, 3, 4, 5, 6, 7, '', 9]\npags=10 act=3 num=5: [0, 1, 2, 3, 4, 5, 6, 7, 8, '', 9]\npags=10 act=4 num=1: [0, '', 3, 4, 5, '', 9]\npags=10 act=4 num=2: [0, '', 2, 3, 4, 5, 6, '', 9]\npags=10 act=4 num=3: [0, 1, 2, 3, 4, 5, 6, 7, '', 9]\npags=10 act=4 num=4: [0, 1, 2, 3, 4, 5, 6, 7, 8, '', 9]\npags=10 act=4 num=5: [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, '', 9]\npags=10 act=5 num=1: [0, '', 4, 5, 6, '', 9]\npags=10 act=5 num=2: [0, '', 3, 4, 5, 6, 7, '', 9]\npags=10 act=5 num=3: [0, '', 2, 3, 4, 5, 6, 7, 8, 9]\npags=10 act=5 num=4: [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, '', 9]\npags=10 act=5 num=5: [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, '', 9]\npags=10 act=6 num=1: [0, '', 5, 6, 7, '', 9]\npags=10 act=6 num=2: [0, '', 4, 5, 6, 7, 8, 9]\npags=10 act=6 num=3: [0, '', 3, 4, 5, 6, 7, 8, 9]\npags=10 act=6 num=4: [0, '', 2, 3, 4, 5, 6, 7, 8, 9]\npags=10 act=6 num=5: [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, '', 9]\npags=10 act=7 num=1: [0, '', 6, 7, 8, 9]\npags=10 act=7 num=2: [0, '', 5, 6, 7, 8, 9]\npags=10 act=7 num=3: [0, '', 4, 5, 6, 7, 8, 9]\npags=10 act=7 num=4: [0, '', 3, 4, 5, 6, 7, 8, 9]\npags=10 act=7 num=5: [0, '', 2, 3, 4, 5, 6, 7, 8, 9]\npags=10 act=8 num=1: [0, '', 7, 8, 9]\npags=10 act=8 num=2: [0, '', 6, 7, 8, 9]\npags=10 act=8 num=3: [0, '', 5, 6, 7, 8, 9]\npags=10 act=8 num=4: [0, '', 4, 5, 6, 7, 8, 9]\npags=10 act=8 num=5: [0, '', 3, 4, 5, 6, 7, 8, 9]\npags=10 act=9 num=1: [0, '', 8, 9]\npags=10 act=9 num=2: [0, '', 7, 8, 9]\npags=10 act=9 num=3: [0, '', 6, 7, 8, 9]\npags=10 act=9 num=4: [0, '', 5, 6, 7, 8, 9]\npags=10 act=9 num=5: [0, '', 4, 5, 6, 7, 8, 9]\npags=10 act=10 num=1: [0, '', 9]\npags=10 act=10 num=2: [0, '', 8, 9]\npags=10 act=10 num=3: [0, '', 7, 8, 9]\npags=10 act=10 num=4: [0, '', 6, 7, 8, 9]\npags=10 act=10 num=5: [0, '', 5, 6, 7, 8, 9]\npags=20 act=3 num=1: [0, '', 2, 3, 4, '', 19]\npags=20 act=3 num=2: [0, 1, 2, 3, 4, 5, '', 19]\npags=20 act=3 num=3: [0, 1, 2, 3, 4, 5, 6, '', 19]\npags=20 act=3 num=4: [0, 1, 2, 3, 4, 5, 6, 7, '', 19]\npags=20 act=3 num=5: [0, 1, 2, 3, 4, 5, 6, 7, 8, '', 19]\npags=20 act=4 num=1: [0, '', 3, 4, 5, '', 19]\npags=20 act=4 num=2: [0, '', 2, 3, 4, 5, 6, '', 19]\npags=20 act=4 num=3: [0, 1, 2, 3, 4, 5, 6, 7, '', 19]\npags=20 act=4 num=4: [0, 1, 2, 3, 4, 5, 6, 7, 8, '', 19]\npags=20 act=4 num=5: [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, '', 19]\npags=20 act=5 num=1: [0, '', 4, 5, 6, '', 19]\npags=20 act=5 num=2: [0, '', 3, 4, 5, 6, 7, '', 19]\npags=20 act=5 num=3: [0, '', 2, 3, 4, 5, 6, 7, 8, '', 19]\npags=20 act=5 num=4: [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, '', 19]\npags=20 act=5 num=5: [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, '', 19]\npags=20 act=6 num=1: [0, '', 5, 6, 7, '', 19]\npags=20 act=6 num=2: [0, '', 4, 5, 6, 7, 8, '', 19]\npags=20 act=6 num=3: [0, '', 3, 4, 5, 6, 7, 8, 9, '', 19]\npags=20 act=6 num=4: [0, '', 2, 3, 4, 5, 6, 7, 8, 9, 10, '', 19]\npags=20 act=6 num=5: [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, '', 19]\npags=20 act=7 num=1: [0, '', 6, 7, 8, '', 19]\npags=20 act=7 num=2: [0, '', 5, 6, 7, 8, 9, '', 19]\npags=20 act=7 num=3: [0, '', 4, 5, 6, 7, 8, 9, 10, '', 19]\npags=20 act=7 num=4: [0, '', 3, 4, 5, 6, 7, 8, 9, 10, 11, '', 19]\npags=20 act=7 num=5: [0, '', 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, '', 19]\npags=20 act=13 num=1: [0, '', 12, 13, 14, '', 19]\npags=20 act=13 num=2: [0, '', 11, 12, 13, 14, 15, '', 19]\npags=20 act=13 num=3: [0, '', 10, 11, 12, 13, 14, 15, 16, '', 19]\npags=20 act=13 num=4: [0, '', 9, 10, 11, 12, 13, 14, 15, 16, 17, '', 19]\npags=20 act=13 num=5: [0, '', 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19]\npags=20 act=14 num=1: [0, '', 13, 14, 15, '', 19]\npags=20 act=14 num=2: [0, '', 12, 13, 14, 15, 16, '', 19]\npags=20 act=14 num=3: [0, '', 11, 12, 13, 14, 15, 16, 17, '', 19]\npags=20 act=14 num=4: [0, '', 10, 11, 12, 13, 14, 15, 16, 17, 18, 19]\npags=20 act=14 num=5: [0, '', 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19]\npags=20 act=15 num=1: [0, '', 14, 15, 16, '', 19]\npags=20 act=15 num=2: [0, '', 13, 14, 15, 16, 17, '', 19]\npags=20 act=15 num=3: [0, '', 12, 13, 14, 15, 16, 17, 18, 19]\npags=20 act=15 num=4: [0, '', 11, 12, 13, 14, 15, 16, 17, 18, 19]\npags=20 act=15 num=5: [0, '', 10, 11, 12, 13, 14, 15, 16, 17, 18, 19]\npags=20 act=16 num=1: [0, '', 15, 16, 17, '', 19]\npags=20 act=16 num=2: [0, '', 14, 15, 16, 17, 18, 19]\npags=20 act=16 num=3: [0, '', 13, 14, 15, 16, 17, 18, 19]\npags=20 act=16 num=4: [0, '', 12, 13, 14, 15, 16, 17, 18, 19]\npags=20 act=16 num=5: [0, '', 11, 12, 13, 14, 15, 16, 17, 18, 19]\npags=20 act=17 num=1: [0, '', 16, 17, 18, 19]\npags=20 act=17 num=2: [0, '', 15, 16, 17, 18, 19]\npags=20 act=17 num=3: [0, '', 14, 15, 16, 17, 18, 19]\npags=20 act=17 num=4: [0, '', 13, 14, 15, 16, 17, 18, 19]\npags=20 act=17 num=5: [0, '', 12, 13, 14, 15, 16, 17, 18, 19]\npags=20 act=18 num=1: [0, '', 17, 18, 19]\npags=20 act=18 num=2: [0, '', 16, 17, 18, 19]\npags=20 act=18 num=3: [0, '', 15, 16, 17, 18, 19]\npags=20 act=18 num=4: [0, '', 14, 15, 16, 17, 18, 19]\npags=20 act=18 num=5: [0, '', 13, 14, 15, 16, 17, 18, 19]\n"
],
[
"a=3.8\nint(a)",
"_____no_output_____"
],
[
"# misma función, pero añadiendo 1 (e.g. las páginas van de 1 al número total de páginas)\ndef lista_paginas_más_uno(paginas, actual, num=3):\n \"\"\"Calcula la lista de páginas a mostrar. Las cadenas \n nulas corresponden a puntos suspensivos.\n \n Argumentos\n ----------\n paginas: int\n Total de páginas que hay.\n actual: int\n página actual.\n num: int\n número de paginas anteriores y posteriores que queremos \n mostrar.\n \n \n Devuelve\n --------\n paginacion: list\n lista de páginas calculada.\n \"\"\"\n if paginas < 10 or (actual <= num + 1 and actual >= paginas - num - 1):\n pagination = list(range(1, paginas+1))\n else:\n if actual <= num + 2:\n pagination = (actual + num + 2)*['']\n pagination[-1] = paginas\n pages = range(1, actual+num+1)\n for i, p in enumerate(pages):\n pagination[i] = p\n elif actual >= paginas - num - 1:\n pagination = (paginas - actual + num + 3)*['']\n pagination[0] = 1\n pages = range(actual-num, paginas+1)\n for i, p in enumerate(pages):\n pagination[i+2] = p\n else:\n pagination = (num*2 + 5)*['']\n pagination[0] = 1\n pagination[-1] = paginas\n pages = range(actual-num, actual+num+1)\n for i, p in enumerate(pages):\n pagination[i+2] = p\n return pagination",
"_____no_output_____"
],
[
"for pags, act in [\n (8, 4), (9, 4), (10, 2), (10, 3), (10, 4), (10, 5), (10, 6), (10, 7), (10, 8), (10, 9), (10, 10),\n (20, 3), (20, 4), (20, 5), (20, 6), (20, 7), (20, 13), (20, 14), (20, 15), (20, 16), (20, 17), (20, 18),\n (11, 6),\n ]:\n for n in range(1, 6):\n print(f'pags={pags} act={act} num={n}: {lista_paginas_custom(pags, act, n)} {lista_paginas_más_uno(pags, act, n)}')",
"pags=8 act=4 num=1: [0, 1, 2, 3, 4, 5, 6, 7] [1, 2, 3, 4, 5, 6, 7, 8]\npags=8 act=4 num=2: [0, 1, 2, 3, 4, 5, 6, 7] [1, 2, 3, 4, 5, 6, 7, 8]\npags=8 act=4 num=3: [0, 1, 2, 3, 4, 5, 6, 7] [1, 2, 3, 4, 5, 6, 7, 8]\npags=8 act=4 num=4: [0, 1, 2, 3, 4, 5, 6, 7] [1, 2, 3, 4, 5, 6, 7, 8]\npags=8 act=4 num=5: [0, 1, 2, 3, 4, 5, 6, 7] [1, 2, 3, 4, 5, 6, 7, 8]\npags=9 act=4 num=1: [0, 1, 2, 3, 4, 5, 6, 7, 8] [1, 2, 3, 4, 5, 6, 7, 8, 9]\npags=9 act=4 num=2: [0, 1, 2, 3, 4, 5, 6, 7, 8] [1, 2, 3, 4, 5, 6, 7, 8, 9]\npags=9 act=4 num=3: [0, 1, 2, 3, 4, 5, 6, 7, 8] [1, 2, 3, 4, 5, 6, 7, 8, 9]\npags=9 act=4 num=4: [0, 1, 2, 3, 4, 5, 6, 7, 8] [1, 2, 3, 4, 5, 6, 7, 8, 9]\npags=9 act=4 num=5: [0, 1, 2, 3, 4, 5, 6, 7, 8] [1, 2, 3, 4, 5, 6, 7, 8, 9]\npags=10 act=2 num=1: [0, 1, 2, 3, '', 9] [1, 2, 3, '', 10]\npags=10 act=2 num=2: [0, 1, 2, 3, 4, '', 9] [1, 2, 3, 4, '', 10]\npags=10 act=2 num=3: [0, 1, 2, 3, 4, 5, '', 9] [1, 2, 3, 4, 5, '', 10]\npags=10 act=2 num=4: [0, 1, 2, 3, 4, 5, 6, '', 9] [1, 2, 3, 4, 5, 6, '', 10]\npags=10 act=2 num=5: [0, 1, 2, 3, 4, 5, 6, 7, '', 9] [1, 2, 3, 4, 5, 6, 7, '', 10]\npags=10 act=3 num=1: [0, '', 2, 3, 4, '', 9] [1, 2, 3, 4, '', 10]\npags=10 act=3 num=2: [0, 1, 2, 3, 4, 5, '', 9] [1, 2, 3, 4, 5, '', 10]\npags=10 act=3 num=3: [0, 1, 2, 3, 4, 5, 6, '', 9] [1, 2, 3, 4, 5, 6, '', 10]\npags=10 act=3 num=4: [0, 1, 2, 3, 4, 5, 6, 7, '', 9] [1, 2, 3, 4, 5, 6, 7, '', 10]\npags=10 act=3 num=5: [0, 1, 2, 3, 4, 5, 6, 7, 8, '', 9] [1, 2, 3, 4, 5, 6, 7, 8, '', 10]\npags=10 act=4 num=1: [0, '', 3, 4, 5, '', 9] [1, '', 3, 4, 5, '', 10]\npags=10 act=4 num=2: [0, '', 2, 3, 4, 5, 6, '', 9] [1, 2, 3, 4, 5, 6, '', 10]\npags=10 act=4 num=3: [0, 1, 2, 3, 4, 5, 6, 7, '', 9] [1, 2, 3, 4, 5, 6, 7, '', 10]\npags=10 act=4 num=4: [0, 1, 2, 3, 4, 5, 6, 7, 8, '', 9] [1, 2, 3, 4, 5, 6, 7, 8, '', 10]\npags=10 act=4 num=5: [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, '', 9] [1, 2, 3, 4, 5, 6, 7, 8, 9, 10]\npags=10 act=5 num=1: [0, '', 4, 5, 6, '', 9] [1, '', 4, 5, 6, '', 10]\npags=10 act=5 num=2: [0, '', 3, 4, 5, 6, 7, '', 9] [1, '', 3, 4, 5, 6, 7, '', 10]\npags=10 act=5 num=3: [0, '', 2, 3, 4, 5, 6, 7, 8, 9] [1, 2, 3, 4, 5, 6, 7, 8, '', 10]\npags=10 act=5 num=4: [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, '', 9] [1, 2, 3, 4, 5, 6, 7, 8, 9, 10]\npags=10 act=5 num=5: [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, '', 9] [1, 2, 3, 4, 5, 6, 7, 8, 9, 10]\npags=10 act=6 num=1: [0, '', 5, 6, 7, '', 9] [1, '', 5, 6, 7, '', 10]\npags=10 act=6 num=2: [0, '', 4, 5, 6, 7, 8, 9] [1, '', 4, 5, 6, 7, 8, '', 10]\npags=10 act=6 num=3: [0, '', 3, 4, 5, 6, 7, 8, 9] [1, '', 3, 4, 5, 6, 7, 8, 9, 10]\npags=10 act=6 num=4: [0, '', 2, 3, 4, 5, 6, 7, 8, 9] [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, '', 10]\npags=10 act=6 num=5: [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, '', 9] [1, 2, 3, 4, 5, 6, 7, 8, 9, 10]\npags=10 act=7 num=1: [0, '', 6, 7, 8, 9] [1, '', 6, 7, 8, '', 10]\npags=10 act=7 num=2: [0, '', 5, 6, 7, 8, 9] [1, '', 5, 6, 7, 8, 9, 10]\npags=10 act=7 num=3: [0, '', 4, 5, 6, 7, 8, 9] [1, '', 4, 5, 6, 7, 8, 9, 10]\npags=10 act=7 num=4: [0, '', 3, 4, 5, 6, 7, 8, 9] [1, '', 3, 4, 5, 6, 7, 8, 9, 10]\npags=10 act=7 num=5: [0, '', 2, 3, 4, 5, 6, 7, 8, 9] [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, '', 10]\npags=10 act=8 num=1: [0, '', 7, 8, 9] [1, '', 7, 8, 9, 10]\npags=10 act=8 num=2: [0, '', 6, 7, 8, 9] [1, '', 6, 7, 8, 9, 10]\npags=10 act=8 num=3: [0, '', 5, 6, 7, 8, 9] [1, '', 5, 6, 7, 8, 9, 10]\npags=10 act=8 num=4: [0, '', 4, 5, 6, 7, 8, 9] [1, '', 4, 5, 6, 7, 8, 9, 10]\npags=10 act=8 num=5: [0, '', 3, 4, 5, 6, 7, 8, 9] [1, '', 3, 4, 5, 6, 7, 8, 9, 10]\npags=10 act=9 num=1: [0, '', 8, 9] [1, '', 8, 9, 10]\npags=10 act=9 num=2: [0, '', 7, 8, 9] [1, '', 7, 8, 9, 10]\npags=10 act=9 num=3: [0, '', 6, 7, 8, 9] [1, '', 6, 7, 8, 9, 10]\npags=10 act=9 num=4: [0, '', 5, 6, 7, 8, 9] [1, '', 5, 6, 7, 8, 9, 10]\npags=10 act=9 num=5: [0, '', 4, 5, 6, 7, 8, 9] [1, '', 4, 5, 6, 7, 8, 9, 10]\npags=10 act=10 num=1: [0, '', 9] [1, '', 9, 10]\npags=10 act=10 num=2: [0, '', 8, 9] [1, '', 8, 9, 10]\npags=10 act=10 num=3: [0, '', 7, 8, 9] [1, '', 7, 8, 9, 10]\npags=10 act=10 num=4: [0, '', 6, 7, 8, 9] [1, '', 6, 7, 8, 9, 10]\npags=10 act=10 num=5: [0, '', 5, 6, 7, 8, 9] [1, '', 5, 6, 7, 8, 9, 10]\npags=20 act=3 num=1: [0, '', 2, 3, 4, '', 19] [1, 2, 3, 4, '', 20]\npags=20 act=3 num=2: [0, 1, 2, 3, 4, 5, '', 19] [1, 2, 3, 4, 5, '', 20]\npags=20 act=3 num=3: [0, 1, 2, 3, 4, 5, 6, '', 19] [1, 2, 3, 4, 5, 6, '', 20]\npags=20 act=3 num=4: [0, 1, 2, 3, 4, 5, 6, 7, '', 19] [1, 2, 3, 4, 5, 6, 7, '', 20]\npags=20 act=3 num=5: [0, 1, 2, 3, 4, 5, 6, 7, 8, '', 19] [1, 2, 3, 4, 5, 6, 7, 8, '', 20]\npags=20 act=4 num=1: [0, '', 3, 4, 5, '', 19] [1, '', 3, 4, 5, '', 20]\npags=20 act=4 num=2: [0, '', 2, 3, 4, 5, 6, '', 19] [1, 2, 3, 4, 5, 6, '', 20]\npags=20 act=4 num=3: [0, 1, 2, 3, 4, 5, 6, 7, '', 19] [1, 2, 3, 4, 5, 6, 7, '', 20]\npags=20 act=4 num=4: [0, 1, 2, 3, 4, 5, 6, 7, 8, '', 19] [1, 2, 3, 4, 5, 6, 7, 8, '', 20]\npags=20 act=4 num=5: [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, '', 19] [1, 2, 3, 4, 5, 6, 7, 8, 9, '', 20]\npags=20 act=5 num=1: [0, '', 4, 5, 6, '', 19] [1, '', 4, 5, 6, '', 20]\npags=20 act=5 num=2: [0, '', 3, 4, 5, 6, 7, '', 19] [1, '', 3, 4, 5, 6, 7, '', 20]\npags=20 act=5 num=3: [0, '', 2, 3, 4, 5, 6, 7, 8, '', 19] [1, 2, 3, 4, 5, 6, 7, 8, '', 20]\npags=20 act=5 num=4: [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, '', 19] [1, 2, 3, 4, 5, 6, 7, 8, 9, '', 20]\npags=20 act=5 num=5: [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, '', 19] [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, '', 20]\npags=20 act=6 num=1: [0, '', 5, 6, 7, '', 19] [1, '', 5, 6, 7, '', 20]\npags=20 act=6 num=2: [0, '', 4, 5, 6, 7, 8, '', 19] [1, '', 4, 5, 6, 7, 8, '', 20]\npags=20 act=6 num=3: [0, '', 3, 4, 5, 6, 7, 8, 9, '', 19] [1, '', 3, 4, 5, 6, 7, 8, 9, '', 20]\npags=20 act=6 num=4: [0, '', 2, 3, 4, 5, 6, 7, 8, 9, 10, '', 19] [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, '', 20]\npags=20 act=6 num=5: [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, '', 19] [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, '', 20]\npags=20 act=7 num=1: [0, '', 6, 7, 8, '', 19] [1, '', 6, 7, 8, '', 20]\npags=20 act=7 num=2: [0, '', 5, 6, 7, 8, 9, '', 19] [1, '', 5, 6, 7, 8, 9, '', 20]\npags=20 act=7 num=3: [0, '', 4, 5, 6, 7, 8, 9, 10, '', 19] [1, '', 4, 5, 6, 7, 8, 9, 10, '', 20]\npags=20 act=7 num=4: [0, '', 3, 4, 5, 6, 7, 8, 9, 10, 11, '', 19] [1, '', 3, 4, 5, 6, 7, 8, 9, 10, 11, '', 20]\npags=20 act=7 num=5: [0, '', 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, '', 19] [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, '', 20]\npags=20 act=13 num=1: [0, '', 12, 13, 14, '', 19] [1, '', 12, 13, 14, '', 20]\npags=20 act=13 num=2: [0, '', 11, 12, 13, 14, 15, '', 19] [1, '', 11, 12, 13, 14, 15, '', 20]\npags=20 act=13 num=3: [0, '', 10, 11, 12, 13, 14, 15, 16, '', 19] [1, '', 10, 11, 12, 13, 14, 15, 16, '', 20]\npags=20 act=13 num=4: [0, '', 9, 10, 11, 12, 13, 14, 15, 16, 17, '', 19] [1, '', 9, 10, 11, 12, 13, 14, 15, 16, 17, '', 20]\npags=20 act=13 num=5: [0, '', 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19] [1, '', 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, '', 20]\npags=20 act=14 num=1: [0, '', 13, 14, 15, '', 19] [1, '', 13, 14, 15, '', 20]\npags=20 act=14 num=2: [0, '', 12, 13, 14, 15, 16, '', 19] [1, '', 12, 13, 14, 15, 16, '', 20]\npags=20 act=14 num=3: [0, '', 11, 12, 13, 14, 15, 16, 17, '', 19] [1, '', 11, 12, 13, 14, 15, 16, 17, '', 20]\npags=20 act=14 num=4: [0, '', 10, 11, 12, 13, 14, 15, 16, 17, 18, 19] [1, '', 10, 11, 12, 13, 14, 15, 16, 17, 18, '', 20]\npags=20 act=14 num=5: [0, '', 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19] [1, '', 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20]\npags=20 act=15 num=1: [0, '', 14, 15, 16, '', 19] [1, '', 14, 15, 16, '', 20]\npags=20 act=15 num=2: [0, '', 13, 14, 15, 16, 17, '', 19] [1, '', 13, 14, 15, 16, 17, '', 20]\npags=20 act=15 num=3: [0, '', 12, 13, 14, 15, 16, 17, 18, 19] [1, '', 12, 13, 14, 15, 16, 17, 18, '', 20]\npags=20 act=15 num=4: [0, '', 11, 12, 13, 14, 15, 16, 17, 18, 19] [1, '', 11, 12, 13, 14, 15, 16, 17, 18, 19, 20]\npags=20 act=15 num=5: [0, '', 10, 11, 12, 13, 14, 15, 16, 17, 18, 19] [1, '', 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20]\npags=20 act=16 num=1: [0, '', 15, 16, 17, '', 19] [1, '', 15, 16, 17, '', 20]\npags=20 act=16 num=2: [0, '', 14, 15, 16, 17, 18, 19] [1, '', 14, 15, 16, 17, 18, '', 20]\npags=20 act=16 num=3: [0, '', 13, 14, 15, 16, 17, 18, 19] [1, '', 13, 14, 15, 16, 17, 18, 19, 20]\npags=20 act=16 num=4: [0, '', 12, 13, 14, 15, 16, 17, 18, 19] [1, '', 12, 13, 14, 15, 16, 17, 18, 19, 20]\npags=20 act=16 num=5: [0, '', 11, 12, 13, 14, 15, 16, 17, 18, 19] [1, '', 11, 12, 13, 14, 15, 16, 17, 18, 19, 20]\npags=20 act=17 num=1: [0, '', 16, 17, 18, 19] [1, '', 16, 17, 18, '', 20]\npags=20 act=17 num=2: [0, '', 15, 16, 17, 18, 19] [1, '', 15, 16, 17, 18, 19, 20]\npags=20 act=17 num=3: [0, '', 14, 15, 16, 17, 18, 19] [1, '', 14, 15, 16, 17, 18, 19, 20]\npags=20 act=17 num=4: [0, '', 13, 14, 15, 16, 17, 18, 19] [1, '', 13, 14, 15, 16, 17, 18, 19, 20]\npags=20 act=17 num=5: [0, '', 12, 13, 14, 15, 16, 17, 18, 19] [1, '', 12, 13, 14, 15, 16, 17, 18, 19, 20]\npags=20 act=18 num=1: [0, '', 17, 18, 19] [1, '', 17, 18, 19, 20]\npags=20 act=18 num=2: [0, '', 16, 17, 18, 19] [1, '', 16, 17, 18, 19, 20]\npags=20 act=18 num=3: [0, '', 15, 16, 17, 18, 19] [1, '', 15, 16, 17, 18, 19, 20]\npags=20 act=18 num=4: [0, '', 14, 15, 16, 17, 18, 19] [1, '', 14, 15, 16, 17, 18, 19, 20]\npags=20 act=18 num=5: [0, '', 13, 14, 15, 16, 17, 18, 19] [1, '', 13, 14, 15, 16, 17, 18, 19, 20]\npags=11 act=6 num=1: [0, '', 5, 6, 7, '', 10] [1, '', 5, 6, 7, '', 11]\npags=11 act=6 num=2: [0, '', 4, 5, 6, 7, 8, '', 10] [1, '', 4, 5, 6, 7, 8, '', 11]\npags=11 act=6 num=3: [0, '', 3, 4, 5, 6, 7, 8, 9, 10] [1, '', 3, 4, 5, 6, 7, 8, 9, '', 11]\npags=11 act=6 num=4: [0, '', 2, 3, 4, 5, 6, 7, 8, 9, 10] [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, '', 11]\npags=11 act=6 num=5: [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, '', 10] [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11]\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e3438568d27c75a3cf385c07edd90cb946cc9c | 99,701 | ipynb | Jupyter Notebook | Text_Emotion_Data_Exp/text_data_exp.ipynb | Erenaliaslangiray/ENGR497_Emotion_Recog | cbd2ffd8999fa7b54d2b24217044209a23647923 | [
"MIT"
] | 4 | 2019-03-17T13:14:00.000Z | 2019-11-15T05:12:10.000Z | Text_Emotion_Data_Exp/text_data_exp.ipynb | Erenaliaslangiray/ENGR497_Emotion_Recog | cbd2ffd8999fa7b54d2b24217044209a23647923 | [
"MIT"
] | null | null | null | Text_Emotion_Data_Exp/text_data_exp.ipynb | Erenaliaslangiray/ENGR497_Emotion_Recog | cbd2ffd8999fa7b54d2b24217044209a23647923 | [
"MIT"
] | 1 | 2019-04-26T10:19:33.000Z | 2019-04-26T10:19:33.000Z | 34.64246 | 951 | 0.431189 | [
[
[
"#Author: Eren Ali Aslangiray, Meryem Şahin\n\nimport pandas as pd\nimport os\nimport time\nimport sys",
"_____no_output_____"
],
[
"path1 = \"/Users/erenmac/Desktop/NEW_DATA/Text/text_emotion.csv\"\npath2 = \"/Users/erenmac/Desktop/NEW_DATA/Text/primary-plutchik-wheel-DFE.csv\"\npath3 = \"/Users/erenmac/Desktop/NEW_DATA/Text/ssec-aggregated/train-combined-0.66.csv\"\npath4 = \"/Users/erenmac/Desktop/NEW_DATA/Text/twitter_emotion_SocialCom_Wang/dev.txt\"\npath5 = \"/Users/erenmac/Desktop/NEW_DATA/Text/twitter_emotion_SocialCom_Wang/train_2_10.txt\"\npath6 = \"/Users/erenmac/Desktop/NEW_DATA/Text/twitter_emotion_SocialCom_Wang/test.txt\"\npath7 = \"/Users/erenmac/Desktop/NEW_DATA/Text/GroundedEmotions/collected_data/collected_news_data.txt\"\npath8 = \"/Users/erenmac/Desktop/NEW_DATA/Text/GroundedEmotions/collected_data/collected_tweets.txt\"\npath9 = \"/Users/erenmac/Desktop/NEW_DATA/Text/GroundedEmotions/collected_data/collected_user_history_data.txt\"\npath10 = \"/Users/erenmac/Desktop/NEW_DATA/Text/GroundedEmotions/collected_data/collected_weather_dataset.txt\"\npath11 = \"/Users/erenmac/Desktop/NEW_DATA/Text/ssec-aggregated/test-combined-0.66.csv\"\npath12 = \"/Users/erenmac/Desktop/NEW_DATA/Text/twitter-emotion/train.csv\"\npath13 = \"/Users/erenmac/Desktop/NEW_DATA/Text/emotion.data\"\npath14 = \"/Users/erenmac/Desktop/NEW_DATA/Text/xmltext.txt\"\npath15 = \"/Users/erenmac/Desktop/NEW_DATA/Text/SentiWords_1.1.txt\"\npath16 = \"Users/erenmac/Desktop/NEW_DATA/Text/NRC-Emotion-Lexicon-Wordlevel-v0.92.txt\"\npathjson1 = \"/Users/erenmac/Desktop/NEW_DATA/Text/cybertroll.json\"",
"_____no_output_____"
],
[
"#mytask = [2,4,7,8,9,11,13,14,json]\n\n#df1 = pd.read_csv(path1)\n#df2 = pd.read_csv(path2)\n#df3 = pd.read_csv(path3, error_bad_lines=False, header = None )\n#df4 = pd.read_csv(path4, sep=\"\\t\", header = None) #twitter\ndf5 = pd.read_csv(path5, sep=\"\\t\", header = None)\ndf6 = pd.read_csv(path6, sep=\"\\t\", header = None)\n#df7 = pd.read_csv(path7, sep=\"|\", header = None)\n#df8 = pd.read_csv(path8, sep=\"|\", header = None) #twitter\n#df9 = pd.read_csv(path9, sep=\"|\", header = None) #twitter\n#df10 = pd.read_csv(path10, sep=\"|\", header = None)\n#df11 = pd.read_csv(path11, error_bad_lines=False, header = None )\n#df12 = pd.read_csv(path12,encoding = 'ISO-8859-1')\n#df13 = pd.read_csv (path13)\n#dfjson1 = pd.read_json(pathjson1, lines = True)",
"_____no_output_____"
],
[
"dfx = df5.append(df6, ignore_index=True)",
"_____no_output_____"
],
[
"dfx.to_json(\"/Users/erenmac/Desktop/NEW_DATA/tweets.json\")",
"_____no_output_____"
],
[
"template_dict = {\"anger\":[],\"disgust\":[],\"sad\":[],\"happy\":[],\"suprise\":[],\"fear\":[],\"neutral\":[]}",
"_____no_output_____"
]
],
[
[
"# DF2",
"_____no_output_____"
]
],
[
[
"df2 = df2.drop(columns = [\"emotion_gold\",\"emotion:confidence\",\"_unit_id\", \"_golden\",\"_unit_state\",\"_trusted_judgments\",\"_last_judgment_at\",\"id\",\"idiom_id\"])",
"_____no_output_____"
],
[
"unique_emoniotns = df2.emotion.unique()",
"_____no_output_____"
],
[
"unique_emoniotns",
"_____no_output_____"
],
[
"reverse_dict = {\"Anger\":\"anger\", \"Aggression\":\"anger\",\"Disgust\":\"disgust\",\"Contempt\":\"disgust\",\"Sadness\":\"sad\",\"Disapproval\":\"sad\",\"Remorse\":\"sad\",\"Optimism\":\"happy\",\"Love\":\"happy\",\"Joy\":\"happy\",\"Trust\":\"happy\",\"Surprise\":\"surprise\",\"Fear\":\"fear\",\"Awe\":\"fear\",\"Neutral\":\"neutral\",\"Ambiguous\":\"neutral\"}\n",
"_____no_output_____"
],
[
"dropemo = [\"Anticipation\",\"Submission\"]",
"_____no_output_____"
],
[
"df2 = df2[df2.emotion != \"Anticipation\"]",
"_____no_output_____"
],
[
"df2 = df2[df2.emotion != \"Submission\"]",
"_____no_output_____"
],
[
"df2[\"emotion\"] = df2[\"emotion\"].map(reverse_dict)",
"_____no_output_____"
],
[
"df2 = df2.reset_index(drop=True)",
"_____no_output_____"
],
[
"for i in range (2270):\n k = df2[\"sentence\"][i]\n if \".\" in k:\n k = k.replace(\".\",\"\")\n if \",\" in k:\n k = k.replace(\",\",\"\")\n if \"?\" in k:\n k = k.replace(\"?\",\"\")\n if \"!\" in k:\n k = k.replace(\"!\",\"\")\n df2[\"sentence\"][i] = k",
"_____no_output_____"
],
[
"df2final = df2",
"_____no_output_____"
],
[
"df2final.to_csv(\"/Users/erenmac/Desktop/NEW_DATA/Cleaned_Data/DF2.csv\")",
"_____no_output_____"
]
],
[
[
"# DF4",
"_____no_output_____"
]
],
[
[
"import tweepy\n\nauth = tweepy.OAuthHandler(\"wso6hl1mmqx5YlLuuiJ7apQci\", \"QQ1mGejEs70qBmpzODBGhMc2FLzVo30hctpaZjSVlBtlmsTiSZ\")\nauth.set_access_token(\"1055878812-fI40KKPy7zNOK8N18WROQ02AgnH820WFeOhTIDS\", \"q5WJXcqCWFRoU7KxR797fz4Ku3C6a876xdECJchupsnfY\")\n\ntry:\n redirect_url = auth.get_authorization_url()\nexcept tweepy.TweepError:\n print('Error! Failed to get request token.')\n \napi = tweepy.API(auth)",
"_____no_output_____"
],
[
"tweets = []\ncode8errorlist = []",
"_____no_output_____"
],
[
"def tweetminer(id):\n tweet = api.get_status(id)\n return tweet.text",
"_____no_output_____"
],
[
"#loaddf.to_json(\"/Users/erenmac/Desktop/NEW_DATA/semi_cleaned_data/loaddf.json\")",
"_____no_output_____"
],
[
"loaddf = pd.read_json(\"/Users/erenmac/Desktop/NEW_DATA/semi_cleaned_data/loaddf.json\")",
"_____no_output_____"
],
[
"for i in range (len(loaddf)):\n if i%100 == 0:\n print(i)\n if loaddf[2][i] == \"aslangiray\":\n try:\n x = (tweetminer(loaddf[0][i]))\n loaddf[2][i] = x\n except tweepy.TweepError as e:\n e1 = \"'code': 88\" in str(e)\n e2 = \"'code': 144\" in str(e)\n e3 = \"'code': 179\" in str(e)\n e4 = \"'code': 34\" in str(e)\n e5 = \"'code': 63\" in str(e)\n if e1 == True:\n print (\"Have code 88 error, waiting 5 min.\")\n time.sleep((60*5)+5)\n elif e2 == True:\n loaddf[2][i] = \"delet dis pls\"\n elif e3 == True:\n loaddf[2][i] = \"delet dis pls\"\n elif e4 == True:\n loaddf[2][i] = \"delet dis pls\"\n elif e5 == True:\n loaddf[2][i] = \"delet dis pls\"\n else:\n errorlist.append(i)\n errorlist.append(e)",
"_____no_output_____"
],
[
"len(errorlist)",
"_____no_output_____"
],
[
"loaddf = loaddf.drop(columns = [0])",
"_____no_output_____"
],
[
"loaddf = loaddf[loaddf[2] != \"delet dis pls\"]",
"_____no_output_____"
],
[
"loaddf = loaddf.reset_index(drop = True)",
"_____no_output_____"
],
[
"loaddf.to_json(\"/Users/erenmac/Desktop/NEW_DATA/Cleaned_Data/DF4.json\")",
"_____no_output_____"
]
],
[
[
"# DF7",
"_____no_output_____"
]
],
[
[
"None",
"_____no_output_____"
]
],
[
[
"# DF8",
"_____no_output_____"
]
],
[
[
"df8[3] = \"aslangiray\"",
"_____no_output_____"
],
[
"df8",
"_____no_output_____"
],
[
"tweets8 = []\nerrorlist8 = []\ncode88errorlist = []",
"_____no_output_____"
],
[
"for i in range (len(df8)):\n if i%100 == 0:\n print(i)\n if df8[3][i] == \"aslangiray\":\n try:\n x = (tweetminer(df8[0][i]))\n df8[3][i] = x\n except tweepy.TweepError as e:\n e1 = \"'code': 88\" in str(e)\n e2 = \"'code': 144\" in str(e)\n e3 = \"'code': 179\" in str(e)\n e4 = \"'code': 34\" in str(e)\n e5 = \"'code': 63\" in str(e)\n if e1 == True:\n print (\"Have code 88 error, waiting 5 min.\")\n time.sleep((60*5)+5)\n elif e2 == True:\n df8[3][i] = \"delet dis pls\"\n elif e3 == True:\n df8[3][i] = \"delet dis pls\"\n elif e4 == True:\n df8[3][i] = \"delet dis pls\"\n elif e5 == True:\n df8[3][i] = \"delet dis pls\"\n else:\n errorlist.append(i)\n errorlist.append(e)",
"0\n100\n200\n300\n400\n500\n600\n700\n800\n900\n"
],
[
"df8 = df8.drop(columns = [0,1])",
"_____no_output_____"
],
[
"df8 = df8[df8[3] != \"delet dis pls\"]",
"_____no_output_____"
],
[
"df8 = df8.reset_index(drop = True)",
"_____no_output_____"
],
[
"df8.to_csv(\"/Users/erenmac/Desktop/NEW_DATA/Cleaned_Data/DF8.csv\")",
"_____no_output_____"
]
],
[
[
"# DF9",
"_____no_output_____"
]
],
[
[
"None",
"_____no_output_____"
]
],
[
[
"# DF11",
"_____no_output_____"
]
],
[
[
"for i in range (1431):\n k = df11[0][i]\n if \"---\" in k:\n k = k.replace(\"---\",\"\")\n if \"\\t\" in k:\n k = k.replace(\"\\t\", \" \")\n if \".\" in k:\n k = k.replace(\".\",\"\")\n if \",\" in k:\n k = k.replace(\",\",\"\")\n if \"?\" in k:\n k = k.replace(\"?\",\"\")\n if \"!\" in k:\n k = k.replace(\"!\",\"\")\n if \"#\" in k:\n k = k.replace(\"#\",\" \")\n df11[0][i] = k",
"_____no_output_____"
],
[
"import re\ndef remove_mentions(input_text):\n return re.sub(r'@\\w+', '', input_text)",
"_____no_output_____"
],
[
"for i in range (1431):\n k = df11[0][i]\n k = remove_mentions(k)\n k = k.replace(\"SemST\",\"\")\n df11[0][i] = k",
"_____no_output_____"
],
[
"emotions_list = [\"Anger\",\"Anticipation\",\"Disgust\",\"Joy\",\"Fear\",\"Sadness\",\"Trust\",\"Surprise\"]",
"_____no_output_____"
],
[
"rowlist = []\nfor i in range (len(df11)):\n k = df11[0][i]\n k = k.split()\n rowlist.append(k)",
"_____no_output_____"
],
[
"dellist = []\nfor i in range (len(rowlist)):\n if len(rowlist[i]) < 5:\n dellist.append(i)\ndellist.remove(911)",
"_____no_output_____"
],
[
"i = 0\nfor item in dellist:\n rowlist.pop(item-i)\n i = i +1",
"_____no_output_____"
],
[
"dfdict = {}\nfor i in range (len(rowlist)):\n for k in range (3):\n if k == 0:\n if rowlist[i][k] in emotions_list:\n dfdict[i] = [rowlist[i][k]]\n else:\n dfdict[i] = []\n else:\n if rowlist[i][k] in emotions_list:\n dfdict[i] += [rowlist[i][k]]\n for q in range (len(dfdict[i])):\n rowlist[i].pop(0)",
"_____no_output_____"
],
[
"df11s = pd.DataFrame.from_dict(dfdict, orient='index')",
"_____no_output_____"
],
[
"df11s[3] = \"sad\"",
"_____no_output_____"
],
[
"for i in range (len(rowlist)):\n k = ' '.join(rowlist[i])\n df11s[3][i] = k",
"_____no_output_____"
],
[
"df11s = df11s.dropna(subset=[0])",
"_____no_output_____"
],
[
"df11final = df11s.reset_index(drop = True)",
"_____no_output_____"
],
[
"unique_emoniotns = df11final[1].unique()",
"_____no_output_____"
],
[
"unique_emoniotns",
"_____no_output_____"
],
[
"reverse_dict = {\"Trust\":\"happy\", \"Joy\":\"happy\",\"Anticipation\":\"neutral\",\"Anger\":\"anger\",\"fear\":\"Fear\",\"Sadness\":\"sad\",\"Disgust\":\"disgust\",\"Surprise\":\"surprise\"}",
"_____no_output_____"
],
[
"df11final[0] = df11final[0].map(reverse_dict)\ndf11final[1] = df11final[1].map(reverse_dict)\ndf11final[2] = df11final[2].map(reverse_dict)",
"_____no_output_____"
],
[
"df11final.to_csv(\"/Users/erenmac/Desktop/NEW_DATA/Cleaned_Data/DF11.csv\")",
"_____no_output_____"
]
],
[
[
"# DF13",
"_____no_output_____"
]
],
[
[
"df13 = df13.drop(columns = [\"Unnamed: 0\"])",
"_____no_output_____"
],
[
"unique_emoniotns = df13.emotions.unique()",
"_____no_output_____"
],
[
"unique_emoniotns",
"_____no_output_____"
],
[
"reverse_dict = {\"sadness\":\"sad\", \"joy\":\"happy\",\"love\":\"happy\",\"anger\":\"anger\",\"fear\":\"fear\",\"surprise\":\"surprise\"}",
"_____no_output_____"
],
[
"df13[\"emotions\"] = df13[\"emotions\"].map(reverse_dict)",
"_____no_output_____"
],
[
"df13final = df13",
"_____no_output_____"
],
[
"df13final.to_csv(\"/Users/erenmac/Desktop/NEW_DATA/Cleaned_Data/DF13.csv\")",
"_____no_output_____"
]
],
[
[
"# DFJSON1",
"_____no_output_____"
]
],
[
[
"dfjson1 = dfjson1.drop(columns = [\"extras\",\"metadata\"])",
"_____no_output_____"
],
[
"labellist = []\nfor i in range (len(dfjson1)):\n if dfjson1[\"annotation\"][i][\"label\"] == ['1']:\n labellist.append(\"negative\")\n else:\n labellist.append(\"del\")",
"_____no_output_____"
],
[
"for i in range (len(labellist)):\n k = labellist[i]\n dfjson1[\"annotation\"][i] = k",
"_____no_output_____"
],
[
"dfjson1 = dfjson1[dfjson1.annotation != \"del\"]",
"_____no_output_____"
],
[
"dfjson1final = dfjson1",
"_____no_output_____"
],
[
"dfjson1final.to_csv(\"/Users/erenmac/Desktop/NEW_DATA/Cleaned_Data/DF17.csv\")",
"_____no_output_____"
]
],
[
[
"# Finalize DB",
"_____no_output_____"
]
],
[
[
"meryem1 = pd.read_json(\"/Users/erenmac/Downloads/df12f.json\")\nmeryem2 = pd.read_json(\"/Users/erenmac/Downloads/result2.json\")",
"_____no_output_____"
],
[
"meryem1",
"_____no_output_____"
],
[
"erenfin = pd.read_json(\"/Users/erenmac/Desktop/NEW_DATA/Cleaned_Data/final_multilabel.json\")",
"_____no_output_____"
],
[
"erenfin2[0] = erenfin2.annotation\nerenfin2[1] = erenfin2.content\n\nerenfin2 = erenfin2.drop(columns= [\"annotation\",\"content\"])",
"_____no_output_____"
],
[
"erenfin2 = pd.read_csv(\"/Users/erenmac/Desktop/NEW_DATA/Cleaned_Data/df17.csv\")",
"_____no_output_____"
],
[
"erenfin2 = erenfin2.drop(columns = [\"Unnamed: 0\"])",
"_____no_output_____"
],
[
"frames = [erenfin2,meryem1]\nresult = pd.concat(frames)",
"_____no_output_____"
],
[
"result = result.reset_index(drop = True)",
"_____no_output_____"
],
[
"result.to_json(\"/Users/erenmac/Desktop/NEW_DATA/Finalized_Data/pos_neg_emotion_data.json\")",
"_____no_output_____"
],
[
"result",
"_____no_output_____"
],
[
"test1 = pd.read_json(\"/Users/erenmac/Desktop/NEW_DATA/Finalized_Data/pos_neg_emotion_data.json\")\ntest2 = pd.read_json(\"/Users/erenmac/Desktop/NEW_DATA/Finalized_Data/multilabel_emotion_data.json\")",
"_____no_output_____"
],
[
"test1",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e345e637a87b4648fadbbe458e5013843d5da9 | 4,165 | ipynb | Jupyter Notebook | .ipynb_checkpoints/t_mixture-checkpoint.ipynb | awjiang/mcmc-joint-tests | ce94b9070f8976ad2dc0731e46a5125db48c66b1 | [
"MIT"
] | null | null | null | .ipynb_checkpoints/t_mixture-checkpoint.ipynb | awjiang/mcmc-joint-tests | ce94b9070f8976ad2dc0731e46a5125db48c66b1 | [
"MIT"
] | null | null | null | .ipynb_checkpoints/t_mixture-checkpoint.ipynb | awjiang/mcmc-joint-tests | ce94b9070f8976ad2dc0731e46a5125db48c66b1 | [
"MIT"
] | null | null | null | 32.286822 | 144 | 0.518607 | [
[
[
"%matplotlib inline\nimport matplotlib\nfrom matplotlib import pyplot as plt\nimport seaborn as sns\nfrom IPython.display import set_matplotlib_formats\nset_matplotlib_formats('retina')\nsns.set(rc={'figure.figsize':(11.7,8.27)})\nsns.set_palette(sns.color_palette())\n\nimport pandas as pd\nimport pickle\n\nfrom mcmcjoint.samplers import *\nfrom mcmcjoint.tests import *",
"_____no_output_____"
],
[
"seed = 10000\nm_dim = 1\nnum_trials = 5\nnthreads = 5\nnum_samples = 5e5\nburn_in_samples = 0\ngeweke_thinning_samples = 50\nmmd_thinning_samples = 500\ntau = 1\nalpha = 0.05",
"_____no_output_____"
],
[
"for exper in range(1):\n if exper == 0:\n print(f'Experiment {exper}: no error')\n model = t_mixture_sampler\n else:\n print(f'Experiment {exper}: error')\n exec('model = t_mixture_sampler_error_'+str(exper))\n t_mix = model(D=m_dim, M=2, N=2, v=5,\n m_mu=[-1. * np.ones(m_dim), 1. * np.ones(m_dim)], \n S_mu=[10. * np.identity(m_dim), 10. * np.identity(m_dim)], \n v_Sigma=2.*np.array([3., 3.]), \n Psi_Sigma=2.*np.array([1. * np.identity(m_dim), 1. * np.identity(m_dim)]), \n alpha_p=np.array([2., 2.]))\n t_mix.set_nthreads(1)\n theta_indices = t_mix.theta_indices\n t_mix_exper = sample_experiment(num_trials=num_trials, nthreads=nthreads, seed=seed,\n sampler=t_mix, \n num_samples=num_samples, \n burn_in_samples=burn_in_samples,\n geweke_thinning_samples=geweke_thinning_samples,\n mmd_thinning_samples=mmd_thinning_samples, \n tau=tau, alpha=alpha, savedir = './results/t_mixture', experiment_name=exper_name)\n lst_saved = t_mix_exper.run()\n ",
"_____no_output_____"
],
[
"df_results = pd.DataFrame(index=np.arange(0, num_trials*3), columns=('experiment', 'test_type', 'result'))\ni=0\nfor exper in range(2):\n for thread in range(9):\n for trial in range(1):\n with open('./results/t_mixture/error_'+str(exper)+'_results_'+str(thread)+'_'+str(trial)+'.pkl', 'rb') as f:\n result = pickle.load(f)[0][0]\n df_results.loc[i] = [str(exper), 'geweke', onp.array(result['geweke'][0]).max()] # reject null if at least one test rejects\n df_results.loc[i+1] = [str(exper), 'backward', float(result['backward'][0])]\n df_results.loc[i+2] = [str(exper), 'wild', float(result['wild'][0])]\n i+=3\ndf_results['result'] = pd.to_numeric(df_results['result'])\ndf_results=df_results.groupby(['experiment', 'test_type']).mean()\ndf_results",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code"
]
] |
d0e35212ca3f7936d25e4b81661214b55d73c25b | 3,745 | ipynb | Jupyter Notebook | DS_Algo/Recursion/recursion_string.ipynb | ShivamBhosale/100DaysOfCode_V2 | 9a7a2bbc728c7014fea36bcb6ca0f6a02516e274 | [
"CC0-1.0"
] | null | null | null | DS_Algo/Recursion/recursion_string.ipynb | ShivamBhosale/100DaysOfCode_V2 | 9a7a2bbc728c7014fea36bcb6ca0f6a02516e274 | [
"CC0-1.0"
] | null | null | null | DS_Algo/Recursion/recursion_string.ipynb | ShivamBhosale/100DaysOfCode_V2 | 9a7a2bbc728c7014fea36bcb6ca0f6a02516e274 | [
"CC0-1.0"
] | null | null | null | 25.650685 | 287 | 0.526836 | [
[
[
"# Reverse a String\n<hr>\n\nThis interview question requires you to reverse a string using recursion. Make sure to think of the base case here.\n\nAgain, make sure you use recursion to accomplish this. Do not slice (e.g. string[::-1]) or use iteration, there muse be a recursive call for the function.\n\n## Solution\n\nIn order to reverse a string using recursion we need to consider what a base and recursive case would look like. Here we've set a base case to be when the length of the string we are passing through the function is length less than or equal to 1.\n\nDuring the recursive case we grab the first letter and add it on to the recursive call.",
"_____no_output_____"
]
],
[
[
"def rev(s):\n if len(s)==1:\n return s\n else:\n return rev(s[1:]) + rev(s[0])",
"_____no_output_____"
],
[
"rev(\"Hello my name is Shivam\")",
"_____no_output_____"
]
],
[
[
"## Extra Notes\n\nThe \"trick\" to this question was thinking about what a base case would look like when reversing a string recursively. It takes a lot of practice to be able to begin thinking like this, so don't worry if you're struggling! However it is important to fully understand the solution!",
"_____no_output_____"
],
[
"<hr>",
"_____no_output_____"
],
[
"# String Permutation\n\n## Problem Statement\n\nGiven a string, write a function that uses recursion to output a list of all the possible permutations of that string.\n\nFor example, given s='abc' the function should return [[ 'abc', 'acb', 'bac', 'bca', 'cab', 'cba' ]]\n\nNote: If a character is repeated, treat each occurence as distinct, for example an input of 'xxx' would return a list with 6 \"versions\" of 'xxx'",
"_____no_output_____"
]
],
[
[
"def permute(s):\n out = []\n\n if len(s) == 1:\n out = [s]\n else:\n # For every letter in string\n for i, let in enumerate(s):\n # For every permutation resulting from Step 2 and 3 described above\n for perm in permute(s[:i] + s[i+1:]):\n\n out += [let + perm]\n return out\n\n ",
"_____no_output_____"
],
[
"permute('hey')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
d0e377d9db22eeb51a62c1a17ed9ce4db32b8c74 | 6,016 | ipynb | Jupyter Notebook | misc_scripts/GetDogInfo.ipynb | Crazyglue/tinder-for-dogs | bed9b07d22d7e9d351d801c73e64273d58d02f60 | [
"MIT"
] | 1 | 2019-12-17T04:19:46.000Z | 2019-12-17T04:19:46.000Z | misc_scripts/GetDogInfo.ipynb | Crazyglue/tinder-for-dogs | bed9b07d22d7e9d351d801c73e64273d58d02f60 | [
"MIT"
] | 35 | 2019-12-17T04:15:14.000Z | 2020-01-07T06:29:50.000Z | misc_scripts/GetDogInfo.ipynb | Crazyglue/tinder-for-dogs | bed9b07d22d7e9d351d801c73e64273d58d02f60 | [
"MIT"
] | 1 | 2019-12-17T04:10:50.000Z | 2019-12-17T04:10:50.000Z | 29.635468 | 124 | 0.530419 | [
[
[
"import os, re\nimport pandas as pd\nimport numpy as np\nimport requests\nfrom bs4 import BeautifulSoup\nimport time\nimport json",
"_____no_output_____"
],
[
"URL_LIST_BASE = \"https://www.dogbreedslist.info/all-dog-breeds/list_1_{}.html\" # {} in [1, 19]",
"_____no_output_____"
],
[
"def get_dog_list_page(n):\n r = requests.get(URL_LIST_BASE.format(n))\n soup = BeautifulSoup(r.content, 'html.parser')\n divs = soup.find_all('div', {'class': 'left'})\n dog_urls = [div.find_all('a')[0][\"href\"] for div in divs]\n return [(re.findall(\"/all-dog-breeds/(.*)?\\.html\", url)[0], url) for url in dog_urls]\n\ndef load_dog_list_page():\n return json.load(open(\"./dog_url_list.json\", \"rb\"))",
"_____no_output_____"
],
[
"# URL_LIST_ALL_DOGS = []\n# for i in range(1, 20):\n# URL_LIST_ALL_DOGS.extend(get_dog_list_page(i))\n# time.sleep(2)\n\n# json.dump(URL_LIST_ALL_DOGS, open(\"./dog_url_list.json\", \"w+\"))",
"_____no_output_____"
],
[
"URL_LIST_ALL_DOGS_loaded = load_dog_list_page()",
"_____no_output_____"
],
[
"def get_dog_info(n):\n \"\"\"Gets breed-specific info. Unfortunately, numbered by index.\"\"\"\n dog = URL_LIST_ALL_DOGS_loaded[n]\n r = requests.get(dog[1])\n soup = BeautifulSoup(r.content, 'html.parser')\n\n info_table = soup.find_all('table', {'class': 'table-01'})\n characteristics_table = soup.find_all('table', {'class': 'table-02'})\n \n dog_classes = [[i for i in tr.contents if i != '\\n'] for tr in characteristics_table[0].find_all(\"tr\")][1:]\n characteristics = {dog_class[0].string: dog_class[1].p.contents[0][0] for dog_class in dog_classes}\n info = dict([[i.string for i in info_table[0].find_all(\"tr\")[2:][i].contents if i != '\\n'] for i in [0, 4]])\n dog_info = [info[\"Name\"], {**characteristics, **{'Size': info[\"Size\"]}}]\n\n return dog_info",
"_____no_output_____"
],
[
"# ALL_DOG_INFO = {}\n# for i in range(len(URL_LIST_ALL_DOGS_loaded)):\n# try:\n# time.sleep(1)\n# gdi = get_dog_info(i)\n# ALL_DOG_INFO[gdi[0]] = gdi[1]\n# except Exception as e:\n# print(i, e)",
"332 list index out of range\n369 list index out of range\n"
],
[
"## SPECIAL CASES DUE TO BAD HTML.\n\n# dog = URL_LIST_ALL_DOGS_loaded[332]\n# r = requests.get(dog[1])\n# soup = BeautifulSoup(r.content, 'html.parser')\n\n# info_table = soup.find_all('table', {'class': 'table-01'})\n# characteristics_table = soup.find_all('table', {'class': 'table-02'})\n\n# dog_classes = [[i for i in tr.contents if i != '\\n'] for tr in characteristics_table[0].find_all(\"tr\")][1:]\n# characteristics = {dog_class[0].string: dog_class[1].p.contents[0][0] for dog_class in dog_classes}\n# # info = dict([[i.string for i in info_table[0].find_all(\"tr\")[2:][i].contents if i != '\\n'] for i in [0, 4]])\n# dog_info = {**characteristics, **{\"Size\": \"Medium\"}}\n# ALL_DOG_INFO[\"American Husky\"] = dog_info\n\n# dog = URL_LIST_ALL_DOGS_loaded[369]\n# r = requests.get(dog[1])\n# soup = BeautifulSoup(r.content, 'html.parser')\n\n# info_table = soup.find_all('table', {'class': 'table-01'})\n# characteristics_table = soup.find_all('table', {'class': 'table-02'})\n\n# dog_classes = [[i for i in tr.contents if i != '\\n'] for tr in characteristics_table[0].find_all(\"tr\")][1:]\n# characteristics = {dog_class[0].string: dog_class[1].p.contents[0][0] for dog_class in dog_classes}\n# # info = dict([[i.string for i in info_table[0].find_all(\"tr\")[2:][i].contents if i != '\\n'] for i in [0, 4]])\n# dog_info = {**characteristics, **{\"Size\": \"Medium\"}}\n# ALL_DOG_INFO[\"Mountain Cur\"] = dog_info",
"_____no_output_____"
],
[
"json.dump(ALL_DOG_INFO, open(\"./dog_metadata.json\", \"w+\"))",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e383c262941cb3bae976cf83a378ee1a70408c | 694,451 | ipynb | Jupyter Notebook | 2021_JCAT_DeDonder_Solvents/Case Study 2.ipynb | jqbond/Research_Public | a6eb581e4e3e72f40fd6c7e900b6f4b30311076f | [
"MIT"
] | null | null | null | 2021_JCAT_DeDonder_Solvents/Case Study 2.ipynb | jqbond/Research_Public | a6eb581e4e3e72f40fd6c7e900b6f4b30311076f | [
"MIT"
] | null | null | null | 2021_JCAT_DeDonder_Solvents/Case Study 2.ipynb | jqbond/Research_Public | a6eb581e4e3e72f40fd6c7e900b6f4b30311076f | [
"MIT"
] | null | null | null | 177.472783 | 16,447 | 0.684044 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d0e3846911f71c8a4f7631a00887e3301476e5e7 | 31,754 | ipynb | Jupyter Notebook | Model backlog/Inference/239-tweet-inference-5fold-roberta-hf-cosine-wr.ipynb | dimitreOliveira/Tweet-Sentiment-Extraction | 0a775abe9a92c4bc2db957519c523be7655df8d8 | [
"MIT"
] | 11 | 2020-06-17T07:30:20.000Z | 2022-03-25T16:56:01.000Z | Model backlog/Inference/239-tweet-inference-5fold-roberta-hf-cosine-wr.ipynb | dimitreOliveira/Tweet-Sentiment-Extraction | 0a775abe9a92c4bc2db957519c523be7655df8d8 | [
"MIT"
] | null | null | null | Model backlog/Inference/239-tweet-inference-5fold-roberta-hf-cosine-wr.ipynb | dimitreOliveira/Tweet-Sentiment-Extraction | 0a775abe9a92c4bc2db957519c523be7655df8d8 | [
"MIT"
] | null | null | null | 33.744952 | 150 | 0.411917 | [
[
[
"## Dependencies",
"_____no_output_____"
]
],
[
[
"import json, glob\nfrom tweet_utility_scripts import *\nfrom tweet_utility_preprocess_roberta_scripts_aux import *\nfrom transformers import TFRobertaModel, RobertaConfig\nfrom tokenizers import ByteLevelBPETokenizer\nfrom tensorflow.keras import layers\nfrom tensorflow.keras.models import Model",
"_____no_output_____"
]
],
[
[
"# Load data",
"_____no_output_____"
]
],
[
[
"test = pd.read_csv('/kaggle/input/tweet-sentiment-extraction/test.csv')\n\nprint('Test samples: %s' % len(test))\ndisplay(test.head())",
"Test samples: 3534\n"
]
],
[
[
"# Model parameters",
"_____no_output_____"
]
],
[
[
"input_base_path = '/kaggle/input/239-robertabase/'\nwith open(input_base_path + 'config.json') as json_file:\n config = json.load(json_file)\n\nconfig",
"_____no_output_____"
],
[
"# vocab_path = input_base_path + 'vocab.json'\n# merges_path = input_base_path + 'merges.txt'\nbase_path = '/kaggle/input/qa-transformers/roberta/'\n\nvocab_path = base_path + 'roberta-base-vocab.json'\nmerges_path = base_path + 'roberta-base-merges.txt'\nconfig['base_model_path'] = base_path + 'roberta-base-tf_model.h5'\nconfig['config_path'] = base_path + 'roberta-base-config.json'\n\nmodel_path_list = glob.glob(input_base_path + '*.h5')\nmodel_path_list.sort()\n\nprint('Models to predict:')\nprint(*model_path_list, sep = '\\n')",
"Models to predict:\n/kaggle/input/239-robertabase/model_fold_1.h5\n/kaggle/input/239-robertabase/model_fold_2.h5\n/kaggle/input/239-robertabase/model_fold_3.h5\n/kaggle/input/239-robertabase/model_fold_4.h5\n/kaggle/input/239-robertabase/model_fold_5.h5\n"
]
],
[
[
"# Tokenizer",
"_____no_output_____"
]
],
[
[
"tokenizer = ByteLevelBPETokenizer(vocab_file=vocab_path, merges_file=merges_path, \n lowercase=True, add_prefix_space=True)",
"_____no_output_____"
]
],
[
[
"# Pre process",
"_____no_output_____"
]
],
[
[
"test['text'].fillna('', inplace=True)\ntest['text'] = test['text'].apply(lambda x: x.lower())\ntest['text'] = test['text'].apply(lambda x: x.strip())\n\nx_test, x_test_aux, x_test_aux_2 = get_data_test(test, tokenizer, config['MAX_LEN'], preprocess_fn=preprocess_roberta_test)",
"_____no_output_____"
]
],
[
[
"# Model",
"_____no_output_____"
]
],
[
[
"module_config = RobertaConfig.from_pretrained(config['config_path'], output_hidden_states=False)\n\ndef model_fn(MAX_LEN):\n input_ids = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='input_ids')\n attention_mask = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='attention_mask')\n \n base_model = TFRobertaModel.from_pretrained(config['base_model_path'], config=module_config, name=\"base_model\")\n last_hidden_state, _ = base_model({'input_ids': input_ids, 'attention_mask': attention_mask})\n \n logits = layers.Dense(2, name=\"qa_outputs\", use_bias=False)(last_hidden_state)\n \n start_logits, end_logits = tf.split(logits, 2, axis=-1)\n start_logits = tf.squeeze(start_logits, axis=-1)\n end_logits = tf.squeeze(end_logits, axis=-1)\n \n model = Model(inputs=[input_ids, attention_mask], outputs=[start_logits, end_logits])\n \n return model",
"_____no_output_____"
]
],
[
[
"# Make predictions",
"_____no_output_____"
]
],
[
[
"NUM_TEST_IMAGES = len(test)\ntest_start_preds = np.zeros((NUM_TEST_IMAGES, config['MAX_LEN']))\ntest_end_preds = np.zeros((NUM_TEST_IMAGES, config['MAX_LEN']))\n\nfor model_path in model_path_list:\n print(model_path)\n model = model_fn(config['MAX_LEN'])\n model.load_weights(model_path)\n \n test_preds = model.predict(get_test_dataset(x_test, config['BATCH_SIZE'])) \n test_start_preds += test_preds[0]\n test_end_preds += test_preds[1]",
"/kaggle/input/239-robertabase/model_fold_1.h5\n/kaggle/input/239-robertabase/model_fold_2.h5\n/kaggle/input/239-robertabase/model_fold_3.h5\n/kaggle/input/239-robertabase/model_fold_4.h5\n/kaggle/input/239-robertabase/model_fold_5.h5\n"
]
],
[
[
"# Post process",
"_____no_output_____"
]
],
[
[
"test['start'] = test_start_preds.argmax(axis=-1)\ntest['end'] = test_end_preds.argmax(axis=-1)\n\ntest['selected_text'] = test.apply(lambda x: decode(x['start'], x['end'], x['text'], config['question_size'], tokenizer), axis=1)\n\n# Post-process\ntest[\"selected_text\"] = test.apply(lambda x: ' '.join([word for word in x['selected_text'].split() if word in x['text'].split()]), axis=1)\ntest['selected_text'] = test.apply(lambda x: x['text'] if (x['selected_text'] == '') else x['selected_text'], axis=1)\ntest['selected_text'].fillna(test['text'], inplace=True)",
"_____no_output_____"
]
],
[
[
"# Visualize predictions",
"_____no_output_____"
]
],
[
[
"test['text_len'] = test['text'].apply(lambda x : len(x))\ntest['label_len'] = test['selected_text'].apply(lambda x : len(x))\ntest['text_wordCnt'] = test['text'].apply(lambda x : len(x.split(' ')))\ntest['label_wordCnt'] = test['selected_text'].apply(lambda x : len(x.split(' ')))\ntest['text_tokenCnt'] = test['text'].apply(lambda x : len(tokenizer.encode(x).ids))\ntest['label_tokenCnt'] = test['selected_text'].apply(lambda x : len(tokenizer.encode(x).ids))\ntest['jaccard'] = test.apply(lambda x: jaccard(x['text'], x['selected_text']), axis=1)\n\ndisplay(test.head(10))\ndisplay(test.describe())",
"_____no_output_____"
]
],
[
[
"# Test set predictions",
"_____no_output_____"
]
],
[
[
"submission = pd.read_csv('/kaggle/input/tweet-sentiment-extraction/sample_submission.csv')\nsubmission['selected_text'] = test['selected_text']\nsubmission.to_csv('submission.csv', index=False)\nsubmission.head(10)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0e385e71cb012010bce21e46e40e6216ad43797 | 1,845 | ipynb | Jupyter Notebook | MusicStreamingMachineLearning2.ipynb | ejrach/python-exercises | baaa02dff58652a9910d654be9bdd3e76dece9b7 | [
"MIT"
] | null | null | null | MusicStreamingMachineLearning2.ipynb | ejrach/python-exercises | baaa02dff58652a9910d654be9bdd3e76dece9b7 | [
"MIT"
] | null | null | null | MusicStreamingMachineLearning2.ipynb | ejrach/python-exercises | baaa02dff58652a9910d654be9bdd3e76dece9b7 | [
"MIT"
] | null | null | null | 24.932432 | 120 | 0.55122 | [
[
[
"import pandas as pd\nfrom sklearn.tree import DecisionTreeClassifier\nfrom sklearn.externals import joblib #persist models\n\n# music_data = pd.read_csv('music.csv')\n# X = music_data.drop(columns=['genre']) #this is the input data set, characterized by capital X\n# y = music_data['genre'] #this is the output data set, characterized by lowercase y\n\n# #train the model\n# model = DecisionTreeClassifier()\n# model.fit(X, y)\n\n#this stores the trained model in a file\n#joblib.dump(model, 'music-recommender.joblib')\n\n#this loads the trained model\nmodel = joblib.load('music-recommender.joblib')\npredictions = model.predict([[21,1]])\npredictions\n\n#predictions = model.predict([[21,1]]) #let's predict what a 21 y.o. male and 22 y.o. female would listen to\n#predictions",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code"
]
] |
d0e39615104fe64a0fad5f9822c031c967bc4ddf | 4,809 | ipynb | Jupyter Notebook | Src/Sandbox/SimpleCase.ipynb | denisuzhva/ML_task4 | 2eacf53ea53347a1ae0e91e3bbc98ba14386fb82 | [
"MIT"
] | null | null | null | Src/Sandbox/SimpleCase.ipynb | denisuzhva/ML_task4 | 2eacf53ea53347a1ae0e91e3bbc98ba14386fb82 | [
"MIT"
] | null | null | null | Src/Sandbox/SimpleCase.ipynb | denisuzhva/ML_task4 | 2eacf53ea53347a1ae0e91e3bbc98ba14386fb82 | [
"MIT"
] | null | null | null | 26.423077 | 108 | 0.422957 | [
[
[
"import numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"## Vars and params\n\nnodes = 5\nself_sim = 0.\nno_sim = -10.\n#s_mat = np.zeros((nodes, nodes))\ns_mat = np.full((nodes, nodes), no_sim)\n\n# fill S\nfor i in range(nodes):\n for j in range(nodes):\n if not i == j:\n p_put = np.random.rand()\n if p_put > 0.7:\n s_mat[i][j] = -1\n \nnp.fill_diagonal(s_mat, self_sim)\nprint(s_mat)",
"[[ 0. -10. -10. -1. -1.]\n [-10. 0. -10. -10. -10.]\n [-10. -1. 0. -10. -1.]\n [-10. -10. -10. 0. -10.]\n [ -1. -10. -10. -10. 0.]]\n"
],
[
"## Update R\n\ndef updateR(r_mat, damping=0):\n # For every column k, except for the column with the maximum value the max is the same.\n # So we can subtract the maximum for every row, \n # and only need to do something different for k == argmax\n\n v = S + A\n rows = np.arange(x.shape[0])\n # We only compare the current point to all other points, \n # so the diagonal can be filled with -infinity\n np.fill_diagonal(v, -np.inf)\n\n # max values\n idx_max = np.argmax(v, axis=1)\n first_max = v[rows, idx_max]\n\n # Second max values. For every column where k is the max value.\n v[rows, idx_max] = -np.inf\n second_max = v[rows, np.argmax(v, axis=1)]\n\n # Broadcast the maximum value per row over all the columns per row.\n max_matrix = np.zeros_like(R) + first_max[:, None]\n max_matrix[rows, idx_max] = second_max\n\n new_val = S - max_matrix\n\n R = R * damping + (1 - damping) * new_val ",
"_____no_output_____"
],
[
"## Update A \n\ndef updateA(a_mat, damping=0):\n for i in range(nodes):\n for k in range(nodes):\n a = np.array(r_mat[:, k]) # Select column k\n # a_matll indices but the diagonal\n if i != k:\n a[i] = -np.inf\n a[k] = - np.inf\n a[a < 0] = 0\n a_mat[i, k] = a_mat[i, k] * damping + (1 - damping) * min(0, r_mat[k, k] + a.sum())\n # The diagonal\n else:\n a[k] = -np.inf\n a[a < 0] = 0\n a_mat[k, k] = a_mat[k, k] * damping + (1 - damping) * a.sum()\n \n return a_mat",
"_____no_output_____"
],
[
"## Some simulations\nr_mat = np.zeros_like(s_mat)\na_mat = np.zeros_like(s_mat)\n\nprint(s_mat)\nprint(r_mat)\nr_mat = updateR(r_mat)\nprint(r_mat)",
"[[ 0. -10. -10. -1. -1.]\n [-10. 0. -10. -10. -10.]\n [-10. -1. 0. -10. -1.]\n [-10. -10. -10. 0. -10.]\n [ -1. -10. -10. -10. 0.]]\n[[0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0.]\n [0. 0. 0. 0. 0.]]\n[[ 1. -9. -9. 0. 0.]\n [ 0. 10. 0. 0. 0.]\n [-9. 0. 1. -9. 0.]\n [ 0. 0. 0. 10. 0.]\n [ 9. -9. -9. -9. 1.]]\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
d0e3a4f7b7e307f126348c6059d56e222a20247c | 403,252 | ipynb | Jupyter Notebook | tools/refer/pyReferDemo.ipynb | mikelmrtinez/ReferringExpression-COCO-VILBERT | 44e975f77ecca78486ae3793dfc9d7cdba158cb3 | [
"MIT"
] | null | null | null | tools/refer/pyReferDemo.ipynb | mikelmrtinez/ReferringExpression-COCO-VILBERT | 44e975f77ecca78486ae3793dfc9d7cdba158cb3 | [
"MIT"
] | null | null | null | tools/refer/pyReferDemo.ipynb | mikelmrtinez/ReferringExpression-COCO-VILBERT | 44e975f77ecca78486ae3793dfc9d7cdba158cb3 | [
"MIT"
] | null | null | null | 1,186.035294 | 194,392 | 0.949694 | [
[
[
"%matplotlib inline\nfrom refer import REFER\nimport numpy as np\nimport skimage.io as io\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"# Load Refer Dataset",
"_____no_output_____"
]
],
[
[
"data_root = '../../data' # contains refclef, refcoco, refcoco+, refcocog and images\ndataset = 'refcoco'\nsplitBy = 'unc'\nrefer = REFER(data_root, dataset, splitBy)",
"loading dataset refcoco into memory...\ncreating index...\nindex created.\nDONE (t=5.66s)\n"
]
],
[
[
"# Stats about the Dataset",
"_____no_output_____"
]
],
[
[
"# print stats about the given dataset\nprint ('dataset [%s_%s] contains: ' % (dataset, splitBy))\nref_ids = refer.getRefIds()\nimage_ids = refer.getImgIds()\nprint ('%s expressions for %s refs in %s images.' % (len(refer.Sents), len(ref_ids), len(image_ids)))\n\nprint ('\\nAmong them:')\nif dataset == 'refclef':\n if splitBy == 'unc':\n splits = ['train', 'val', 'testA', 'testB', 'testC']\n else:\n splits = ['train', 'val', 'test']\nelif dataset == 'refcoco':\n splits = ['train', 'val', 'test']\nelif dataset == 'refcoco+':\n splits = ['train', 'val', 'test']\nelif dataset == 'refcocog':\n splits = ['train', 'val'] # we don't have test split for refcocog right now.\n \nfor split in splits:\n ref_ids = refer.getRefIds(split=split)\n print ('%s refs are in split [%s].' % (len(ref_ids), split))",
"dataset [refcoco_unc] contains: \n142210 expressions for 50000 refs in 19994 images.\n\nAmong them:\n42404 refs are in split [train].\n3811 refs are in split [val].\n3785 refs are in split [test].\n"
],
[
"# randomly sample one ref\nref_ids = refer.getRefIds(split='test')\nref_id = ref_ids[np.random.randint(0, len(ref_ids))]\nprint(ref_id)\nprint(ref_ids[0])\nprint(refer.Refs[0])\nref = refer.Refs[ref_ids]",
"41479\n25\n{'sent_ids': [0, 1, 2], 'file_name': 'COCO_train2014_000000581857_16.jpg', 'ann_id': 1719310, 'ref_id': 0, 'image_id': 581857, 'split': 'train', 'sentences': [{'tokens': ['the', 'lady', 'with', 'the', 'blue', 'shirt'], 'raw': 'THE LADY WITH THE BLUE SHIRT', 'sent_id': 0, 'sent': 'the lady with the blue shirt'}, {'tokens': ['lady', 'with', 'back', 'to', 'us'], 'raw': 'lady w back to us', 'sent_id': 1, 'sent': 'lady with back to us'}, {'tokens': ['blue', 'shirt'], 'raw': 'blue shirt', 'sent_id': 2, 'sent': 'blue shirt'}], 'category_id': 1}\n"
],
[
"ref_ids = refer.getRefIds(split='test')\nref_id = ref_ids[0]\nprint('REF ID: ,' , ref_id)\nref = refer.Refs[ref_id]\nprint()\nprint(ref)\nprint()\n\n\nimg_id = refer.getImgIds(ref_id)\nimg = refer.loadImgs(img_id)[0]\nprint('IMG INFOS: ',img)\nprint('IMG ID: ',img_id)\n\nann_ids = refer.getAnnIds(img['id'])\nprint('ANN IDS: ',ann_ids)\n\nfor i in range(len(ann_ids)):\n ann = refer.loadAnns(ann_ids[i])[0]\n \n print('IMAGE ID ',ann['image_id'])\n print('ANN '+str(i+1)+': ', ann)\n\nprint('#################')\nprint(refer.loadAnns(ann_ids))",
"REF ID: , 25\n\n{'sent_ids': [71, 72, 73], 'file_name': 'COCO_train2014_000000581563_3.jpg', 'ann_id': 1345868, 'ref_id': 25, 'image_id': 581563, 'split': 'testB', 'sentences': [{'tokens': ['lower', 'left', 'corner', 'darkness'], 'raw': 'lower left corner darkness', 'sent_id': 71, 'sent': 'lower left corner darkness'}, {'tokens': ['bpttom', 'left', 'dark'], 'raw': 'bpttom left dark', 'sent_id': 72, 'sent': 'bpttom left dark'}, {'tokens': ['black', 'van', 'in', 'front', 'of', 'cab'], 'raw': 'black van in front of cab', 'sent_id': 73, 'sent': 'black van in front of cab'}], 'category_id': 3}\n\nIMG INFOS: {'license': 3, 'file_name': 'COCO_train2014_000000581563.jpg', 'coco_url': 'http://mscoco.org/images/581563', 'height': 500, 'width': 333, 'date_captured': '2013-11-16 17:51:55', 'flickr_url': 'http://farm1.staticflickr.com/125/337273203_7eb35b845b_z.jpg', 'id': 581563}\nIMG ID: [581563]\nANN IDS: [344837, 405760, 1213670, 1345868, 1735830, 2173204, 2232019, 2232030, 2232043]\nIMAGE ID 581563\nANN 1: {'segmentation': [[221.36, 381.55, 225.85, 383.8, 229.89, 390.53, 231.24, 396.37, 257.29, 398.17, 271.21, 398.17, 282.88, 398.62, 295.45, 398.62, 304.43, 399.51, 308.92, 399.96, 317.46, 399.96, 329.58, 408.05, 333.0, 411.64, 333.0, 426.9, 333.0, 441.72, 333.0, 460.58, 333.0, 469.56, 333.0, 479.44, 333.0, 486.18, 333.0, 500.0, 323.29, 500.0, 311.62, 500.0, 296.35, 500.0, 277.04, 500.0, 259.08, 500.0, 245.16, 500.0, 235.73, 500.0, 224.06, 500.0, 216.87, 500.0, 208.79, 500.0, 198.01, 500.0, 193.97, 500.0, 183.64, 500.0, 175.11, 500.0, 164.34, 500.0, 154.01, 500.0, 141.44, 500.0, 128.86, 500.0, 118.98, 500.0, 64.65, 500.0, 57.47, 500.0, 57.02, 494.71, 55.67, 485.73, 58.36, 471.81, 60.16, 464.62, 62.86, 458.34, 76.78, 452.95, 85.31, 449.8, 96.08, 447.11, 103.27, 447.11, 105.06, 441.72, 108.66, 434.99, 111.8, 434.54, 127.07, 419.27, 136.5, 409.84, 141.88, 404.9, 146.37, 401.31, 150.87, 400.86, 161.64, 396.82, 177.81, 395.47, 193.07, 395.47, 197.56, 385.59, 204.3, 382.45, 215.08, 381.55, 223.16, 383.35]], 'area': 25008.538849999994, 'iscrowd': 0, 'image_id': 581563, 'bbox': [55.67, 381.55, 277.33, 118.45], 'category_id': 3, 'id': 344837}\nIMAGE ID 581563\nANN 2: {'segmentation': [[234.85, 301.87, 256.9, 303.28, 252.21, 319.23, 249.86, 335.65, 238.13, 333.3, 233.91, 329.08, 233.91, 310.32, 232.04, 304.22]], 'area': 601.5432000000003, 'iscrowd': 0, 'image_id': 581563, 'bbox': [232.04, 301.87, 24.86, 33.78], 'category_id': 10, 'id': 405760}\nIMAGE ID 581563\nANN 3: {'segmentation': [[255.58, 424.43, 260.8, 412.71, 281.89, 410.37, 287.3, 423.53, 284.78, 423.89, 281.89, 419.02, 271.26, 417.94, 262.97, 422.09, 260.44, 424.25, 256.66, 424.43], [266.75, 391.81, 266.75, 397.03, 280.45, 397.75, 280.09, 394.87, 277.03, 390.72, 267.83, 391.08]], 'area': 309.6951999999996, 'iscrowd': 0, 'image_id': 581563, 'bbox': [255.58, 390.72, 31.72, 33.71], 'category_id': 1, 'id': 1213670}\nIMAGE ID 581563\nANN 4: {'segmentation': [[59.15, 500.0, 0.48, 500.0, 0.0, 375.26, 96.66, 373.89, 104.85, 380.71, 117.13, 384.81, 127.36, 395.04, 134.86, 403.91, 137.59, 410.04, 112.41, 433.82, 108.59, 435.48, 105.11, 441.62, 103.45, 447.26, 95.99, 447.09, 62.98, 457.54]], 'area': 12101.243650000002, 'iscrowd': 0, 'image_id': 581563, 'bbox': [0.0, 373.89, 137.59, 126.11], 'category_id': 3, 'id': 1345868}\nIMAGE ID 581563\nANN 5: {'segmentation': [[230.77, 389.54, 233.91, 388.06, 236.41, 387.97, 240.2, 389.08, 242.69, 393.7, 241.95, 395.27, 231.51, 395.09, 230.12, 390.93], [225.13, 410.06, 243.16, 409.14, 245.65, 409.51, 246.67, 412.65, 248.52, 422.08, 247.69, 424.12, 241.49, 421.16, 233.26, 416.26, 229.1, 415.06, 226.05, 413.58, 224.94, 412.75, 224.67, 412.1]], 'area': 267.77179999999987, 'iscrowd': 0, 'image_id': 581563, 'bbox': [224.67, 387.97, 23.85, 36.15], 'category_id': 1, 'id': 1735830}\nIMAGE ID 581563\nANN 6: {'segmentation': [[144.31, 193.05, 165.09, 192.68, 162.49, 254.27, 145.8, 255.01]], 'area': 1157.0466, 'iscrowd': 0, 'image_id': 581563, 'bbox': [144.31, 192.68, 20.78, 62.33], 'category_id': 10, 'id': 2173204}\nIMAGE ID 581563\nANN 7: {'segmentation': [[163.73, 230.33, 162.83, 254.27, 165.31, 254.04, 165.76, 241.85, 165.99, 233.27, 165.99, 232.36, 166.44, 229.88, 163.05, 230.1]], 'area': 59.33254999999996, 'iscrowd': 0, 'image_id': 581563, 'bbox': [162.83, 229.88, 3.61, 24.39], 'category_id': 10, 'id': 2232019}\nIMAGE ID 581563\nANN 8: {'segmentation': [[133.24, 261.14, 148.2, 260.79, 157.35, 262.2, 157.7, 277.15, 144.5, 279.09, 140.1, 277.15, 138.7, 276.8, 131.48, 277.15, 132.18, 262.02]], 'area': 425.35029999999927, 'iscrowd': 0, 'image_id': 581563, 'bbox': [131.48, 260.79, 26.22, 18.3], 'category_id': 10, 'id': 2232030}\nIMAGE ID 581563\nANN 9: {'segmentation': [[166.37, 258.23, 168.76, 195.52, 173.07, 195.52, 174.99, 202.22, 191.26, 202.22, 188.39, 254.88, 175.47, 255.35, 170.2, 260.14, 166.85, 260.14]], 'area': 1249.3459999999993, 'iscrowd': 0, 'image_id': 581563, 'bbox': [166.37, 195.52, 24.89, 64.62], 'category_id': 10, 'id': 2232043}\n#################\n[{'segmentation': [[221.36, 381.55, 225.85, 383.8, 229.89, 390.53, 231.24, 396.37, 257.29, 398.17, 271.21, 398.17, 282.88, 398.62, 295.45, 398.62, 304.43, 399.51, 308.92, 399.96, 317.46, 399.96, 329.58, 408.05, 333.0, 411.64, 333.0, 426.9, 333.0, 441.72, 333.0, 460.58, 333.0, 469.56, 333.0, 479.44, 333.0, 486.18, 333.0, 500.0, 323.29, 500.0, 311.62, 500.0, 296.35, 500.0, 277.04, 500.0, 259.08, 500.0, 245.16, 500.0, 235.73, 500.0, 224.06, 500.0, 216.87, 500.0, 208.79, 500.0, 198.01, 500.0, 193.97, 500.0, 183.64, 500.0, 175.11, 500.0, 164.34, 500.0, 154.01, 500.0, 141.44, 500.0, 128.86, 500.0, 118.98, 500.0, 64.65, 500.0, 57.47, 500.0, 57.02, 494.71, 55.67, 485.73, 58.36, 471.81, 60.16, 464.62, 62.86, 458.34, 76.78, 452.95, 85.31, 449.8, 96.08, 447.11, 103.27, 447.11, 105.06, 441.72, 108.66, 434.99, 111.8, 434.54, 127.07, 419.27, 136.5, 409.84, 141.88, 404.9, 146.37, 401.31, 150.87, 400.86, 161.64, 396.82, 177.81, 395.47, 193.07, 395.47, 197.56, 385.59, 204.3, 382.45, 215.08, 381.55, 223.16, 383.35]], 'area': 25008.538849999994, 'iscrowd': 0, 'image_id': 581563, 'bbox': [55.67, 381.55, 277.33, 118.45], 'category_id': 3, 'id': 344837}, {'segmentation': [[234.85, 301.87, 256.9, 303.28, 252.21, 319.23, 249.86, 335.65, 238.13, 333.3, 233.91, 329.08, 233.91, 310.32, 232.04, 304.22]], 'area': 601.5432000000003, 'iscrowd': 0, 'image_id': 581563, 'bbox': [232.04, 301.87, 24.86, 33.78], 'category_id': 10, 'id': 405760}, {'segmentation': [[255.58, 424.43, 260.8, 412.71, 281.89, 410.37, 287.3, 423.53, 284.78, 423.89, 281.89, 419.02, 271.26, 417.94, 262.97, 422.09, 260.44, 424.25, 256.66, 424.43], [266.75, 391.81, 266.75, 397.03, 280.45, 397.75, 280.09, 394.87, 277.03, 390.72, 267.83, 391.08]], 'area': 309.6951999999996, 'iscrowd': 0, 'image_id': 581563, 'bbox': [255.58, 390.72, 31.72, 33.71], 'category_id': 1, 'id': 1213670}, {'segmentation': [[59.15, 500.0, 0.48, 500.0, 0.0, 375.26, 96.66, 373.89, 104.85, 380.71, 117.13, 384.81, 127.36, 395.04, 134.86, 403.91, 137.59, 410.04, 112.41, 433.82, 108.59, 435.48, 105.11, 441.62, 103.45, 447.26, 95.99, 447.09, 62.98, 457.54]], 'area': 12101.243650000002, 'iscrowd': 0, 'image_id': 581563, 'bbox': [0.0, 373.89, 137.59, 126.11], 'category_id': 3, 'id': 1345868}, {'segmentation': [[230.77, 389.54, 233.91, 388.06, 236.41, 387.97, 240.2, 389.08, 242.69, 393.7, 241.95, 395.27, 231.51, 395.09, 230.12, 390.93], [225.13, 410.06, 243.16, 409.14, 245.65, 409.51, 246.67, 412.65, 248.52, 422.08, 247.69, 424.12, 241.49, 421.16, 233.26, 416.26, 229.1, 415.06, 226.05, 413.58, 224.94, 412.75, 224.67, 412.1]], 'area': 267.77179999999987, 'iscrowd': 0, 'image_id': 581563, 'bbox': [224.67, 387.97, 23.85, 36.15], 'category_id': 1, 'id': 1735830}, {'segmentation': [[144.31, 193.05, 165.09, 192.68, 162.49, 254.27, 145.8, 255.01]], 'area': 1157.0466, 'iscrowd': 0, 'image_id': 581563, 'bbox': [144.31, 192.68, 20.78, 62.33], 'category_id': 10, 'id': 2173204}, {'segmentation': [[163.73, 230.33, 162.83, 254.27, 165.31, 254.04, 165.76, 241.85, 165.99, 233.27, 165.99, 232.36, 166.44, 229.88, 163.05, 230.1]], 'area': 59.33254999999996, 'iscrowd': 0, 'image_id': 581563, 'bbox': [162.83, 229.88, 3.61, 24.39], 'category_id': 10, 'id': 2232019}, {'segmentation': [[133.24, 261.14, 148.2, 260.79, 157.35, 262.2, 157.7, 277.15, 144.5, 279.09, 140.1, 277.15, 138.7, 276.8, 131.48, 277.15, 132.18, 262.02]], 'area': 425.35029999999927, 'iscrowd': 0, 'image_id': 581563, 'bbox': [131.48, 260.79, 26.22, 18.3], 'category_id': 10, 'id': 2232030}, {'segmentation': [[166.37, 258.23, 168.76, 195.52, 173.07, 195.52, 174.99, 202.22, 191.26, 202.22, 188.39, 254.88, 175.47, 255.35, 170.2, 260.14, 166.85, 260.14]], 'area': 1249.3459999999993, 'iscrowd': 0, 'image_id': 581563, 'bbox': [166.37, 195.52, 24.89, 64.62], 'category_id': 10, 'id': 2232043}]\n"
]
],
[
[
"# Show Refered Object and its Expressions",
"_____no_output_____"
]
],
[
[
"# randomly sample one ref\nref_ids = refer.getRefIds()\nref_id = ref_ids[np.random.randint(0, len(ref_ids))]\nref = refer.Refs[ref_id]\nprint ('ref_id [%s] (ann_id [%s])' % (ref_id, refer.refToAnn[ref_id]['id']))\n# show the segmentation of the referred object\nplt.figure()\nrefer.showRef(ref, seg_box='seg')\nplt.show()",
"ref_id [26441] (ann_id [1717234])\n1. snowboarding buy in yellow\n2. man in yellow\n3. yellow guy\n"
],
[
"# or show the bounding box of the referred object\nrefer.showRef(ref, seg_box='box')\nplt.show()",
"1. snowboarding buy in yellow\n2. man in yellow\n3. yellow guy\n"
],
[
"# let's look at the details of each ref\nfor sent in ref['sentences']:\n print ('sent_id[%s]: %s' % (sent['sent_id'], sent['sent']))",
"sent_id[75185]: snowboarding buy in yellow\nsent_id[75186]: man in yellow\nsent_id[75187]: yellow guy\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d0e3b2c4685e2aebb87261e495b5c2f97141efc9 | 156,438 | ipynb | Jupyter Notebook | labs/trees/binary-trees.ipynb | testinggg-art/msds621 | 94335253ecf76cff7f36fc69880507fc99127c6d | [
"MIT"
] | 300 | 2019-07-23T17:30:45.000Z | 2022-03-28T18:45:16.000Z | labs/trees/binary-trees.ipynb | testinggg-art/msds621 | 94335253ecf76cff7f36fc69880507fc99127c6d | [
"MIT"
] | 1 | 2019-11-19T05:42:19.000Z | 2019-12-04T20:16:26.000Z | labs/trees/binary-trees.ipynb | testinggg-art/msds621 | 94335253ecf76cff7f36fc69880507fc99127c6d | [
"MIT"
] | 128 | 2019-08-02T20:11:35.000Z | 2022-03-27T19:12:24.000Z | 80.224615 | 21,468 | 0.605102 | [
[
[
"# Binary trees",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nfrom sklearn.tree import DecisionTreeClassifier, DecisionTreeRegressor\nfrom dtreeviz.trees import *\nfrom lolviz import *\n\nimport numpy as np\nimport pandas as pd\n\n%config InlineBackend.figure_format = 'retina'",
"_____no_output_____"
]
],
[
[
"## Setup",
"_____no_output_____"
],
[
"Make sure to install stuff:\n\n```\npip install -U dtreeviz\npip install -U lolviz\nbrew install graphviz\n```",
"_____no_output_____"
],
[
"## Binary tree class definition\n\nA binary tree has a payload (a value) and references to left and right children. One or both of the children references can be `None`. A reference to a node is the same thing as a reference to a tree as the tree is a self similar data structure. We don't distinguish between the two kinds of references. A reference to the root node is a reference to the entire tree.\n\nHere is a basic tree node class in Python. The constructor requires at least a value to store in the node.",
"_____no_output_____"
]
],
[
[
"class TreeNode:\n def __init__(self, value, left=None, right=None):\n self.value = value\n self.left = left\n self.right = right\n def __repr__(self):\n return str(self.value)\n def __str__(self):\n return str(self.value)",
"_____no_output_____"
]
],
[
[
"## Manual tree construction",
"_____no_output_____"
],
[
"Here's how to create and visualize a single node:",
"_____no_output_____"
]
],
[
[
"root = TreeNode(1)\ntreeviz(root)",
"_____no_output_____"
]
],
[
[
"**Given `left` and `right` nodes, create node `root` with those nodes as children.**",
"_____no_output_____"
]
],
[
[
"left = TreeNode(2)\nright = TreeNode(3)\nroot = ...\ntreeviz(root)",
"_____no_output_____"
]
],
[
[
"<details>\n<summary>Solution</summary>\n<pre>\nleft = TreeNode(2)\nright = TreeNode(3)\nroot = TreeNode(1,left,right)\ntreeviz(root)\n</pre>\n</details>",
"_____no_output_____"
],
[
"**Write code to create the following tree structure**\n\n<img src=\"3-level-tree.png\" width=\"30%\">",
"_____no_output_____"
]
],
[
[
"left = ...\nright = ...\nroot = ...\ntreeviz(root)",
"_____no_output_____"
]
],
[
[
"<details>\n<summary>Solution</summary>\n<pre>\nleft = TreeNode(2,TreeNode(4))\nright = TreeNode(3,TreeNode(5),TreeNode(6))\nroot = TreeNode(1,left,right)\ntreeviz(root)\n</pre>\n</details>",
"_____no_output_____"
],
[
"## Walking trees manually\n\nTo walk a tree, we simply follow the left and right children references, avoiding any `None` references.",
"_____no_output_____"
],
[
"**Q.** Given tree `r` shown here, what Python expressions refer to the nodes with 443 and 17 in them?",
"_____no_output_____"
]
],
[
[
"left = TreeNode(443,TreeNode(-34))\nright = TreeNode(17,TreeNode(99))\nr = TreeNode(10,left,right)\ntreeviz(r)",
"_____no_output_____"
]
],
[
[
"<details>\n<summary>Solution</summary>\n<pre>\nr.left, r.right\n</pre>\n</details>",
"_____no_output_____"
],
[
"**Q.** Given the same tree `r`, what Python expressions refer to the nodes with -34 and 99 in them?",
"_____no_output_____"
],
[
"<details>\n<summary>Solution</summary>\n<pre>\nr.left.left, r.right.left\n</pre>\n</details>",
"_____no_output_____"
],
[
"## Walking all nodes\n\nNow let's create a function to walk all nodes in a tree. Remember that our template for creating any recursive function looks like this:\n\n```\ndef f(input):\n 1. check termination condition\n 2. process the active input region / current node, etc…\n 3. invoke f on subregion(s)\n 4. combine and return results\n```",
"_____no_output_____"
]
],
[
[
"def walk(p:TreeNode):\n if p is None: return # step 1\n print(p.value) # step 2\n walk(p.left) # step 3\n walk(p.right) # step 3 (there is no step 4 for this problem)",
"_____no_output_____"
]
],
[
[
"Let's create the simple 3-level tree we had before:",
"_____no_output_____"
]
],
[
[
"left = TreeNode(2,TreeNode(4))\nright = TreeNode(3,TreeNode(5),TreeNode(6))\nroot = TreeNode(1,left,right)\ntreeviz(root)",
"_____no_output_____"
]
],
[
[
"**Q.** What is the output of running `walk(root)`?",
"_____no_output_____"
],
[
"<details>\n<summary>Solution</summary>\n We walk the tree depth first, from left to right<p>\n<pre>\n1\n2\n4\n3\n5\n6\n</pre>\n</details>",
"_____no_output_____"
],
[
"## Searching through nodes\n\nHere's how to search for an element as you walk, terminating as soon as the node with `x` is found:",
"_____no_output_____"
]
],
[
[
"def search(p:TreeNode, x:object):\n print(\"enter \",p)\n if p is None: return None\n print(p)\n if x==p.value:\n return p\n q = search(p.left, x)\n if q is not None:\n return q\n q = search(p.right, x)\n return q",
"_____no_output_____"
]
],
[
[
"**Q.** What is the output of running `search(root, 5)`?",
"_____no_output_____"
],
[
"<details>\n<summary>Solution</summary>\n We walk the tree depth first as before, but now we stop when we reach the node with 5:<p>\n<pre>\n1\n2\n4\n3\n5\n</pre>\n</details>",
"_____no_output_____"
],
[
"To see the recursion entering and exiting (or discovering and finishing) nodes, here is a variation that prints out its progress through the tree:",
"_____no_output_____"
]
],
[
[
"def search(p:TreeNode, x:object):\n if p is None: return None\n print(\"enter \",p)\n if x==p.value:\n print(\"exit \",p)\n return p\n q = search(p.left, x)\n if q is not None:\n print(\"exit \",p)\n return q\n q = search(p.right, x)\n print(\"exit \",p)\n return q",
"_____no_output_____"
],
[
"search(root, 5)",
"enter 1\nenter 2\nenter 4\nexit 4\nexit 2\nenter 3\nenter 5\nexit 5\nexit 3\nexit 1\n"
]
],
[
[
"## Creating (random) decision tree \"stumps\"\n\nA regression tree stump is a tree with a decision node at the root and two predictor leaves. These are used by gradient boosting machines as the \"weak learners.\"",
"_____no_output_____"
]
],
[
[
"class TreeNode: # acts as decision node and leaf. it's a leaf if split is None\n def __init__(self, split=None, prediction=None, left=None, right=None):\n self.split = split\n self.prediction = prediction\n self.left = left\n self.right = right\n def __repr__(self):\n return str(self.value)\n def __str__(self):\n return str(self.value)",
"_____no_output_____"
],
[
"df = pd.DataFrame()\ndf[\"sqfeet\"] = [750, 800, 850, 900,950]\ndf[\"rent\"] = [1160, 1200, 1280, 1450,1300]\ndf",
"_____no_output_____"
]
],
[
[
"The following code shows where sklearn would do a split with a normal decision tree.",
"_____no_output_____"
]
],
[
[
"X, y = df.sqfeet.values.reshape(-1,1), df.rent.values\nt = DecisionTreeRegressor(max_depth=1)\nt.fit(X,y)\n\nfig, ax = plt.subplots(1, 1, figsize=(3,1.5))\nt = rtreeviz_univar(t,\n X, y,\n feature_names='sqfeet',\n target_name='rent',\n fontsize=9,\n colors={'scatter_edge': 'black'},\n ax=ax)",
"_____no_output_____"
]
],
[
[
"Instead of picking the optimal split point, we can choose a random value in between the minimum and maximum x value, like extremely random forests do:",
"_____no_output_____"
]
],
[
[
"def stumpfit(x, y):\n if len(x)==1 or len(np.unique(x))==1: # if one x value, make leaf\n return TreeNode(prediction=y[0])\n split = np.round(np.random.uniform(min(x),max(x)))\n t = TreeNode(split)\n t.left = TreeNode(prediction=np.mean(y[x<split]))\n t.right = TreeNode(prediction=np.mean(y[x>=split]))\n return t",
"_____no_output_____"
]
],
[
[
"**Run the following code multiple times to see how it creates different y lists in the nodes, according to the split value.**",
"_____no_output_____"
]
],
[
[
"root = stumpfit(X.reshape(-1),y)\ntreeviz(root)",
"_____no_output_____"
]
],
[
[
"## Creating random decision trees (single variable)\n\nAnd now to demonstrate the magic of recursion. If we replace\n\n```\nt.left = TreeNode(prediction=np.mean(y[x<split]))\n```\n\nwith\n\n```\nt.left = treefit(x[x<split], y[x<split])\n```\n\n(and same for `t.right`) then all of the sudden we get a full decision tree, rather than just a stump!",
"_____no_output_____"
]
],
[
[
"def treefit(x, y):\n if len(x)==1 or len(np.unique(x))==1: # if one x value, make leaf\n return TreeNode(prediction=y[0])\n split = np.round(np.random.uniform(min(x),max(x)))\n t = TreeNode(split)\n t.left = treefit(x[x<split], y[x<split])\n t.right = treefit(x[x>=split], y[x>=split])\n return t",
"_____no_output_____"
],
[
"root = treefit(X.reshape(-1),y)\ntreeviz(root)",
"_____no_output_____"
]
],
[
[
"You can run that multiple times to see different tree layouts according to randomness.",
"_____no_output_____"
],
[
"**Q.** How would you modify `treefit()` so that it creates a typical decision tree rather than a randomized decision tree?",
"_____no_output_____"
],
[
"<details>\n<summary>Solution</summary>\nInstead of choosing a random split, we would pick the split value in $x$ that got the best average child $y$ purity/similarity. In other words, exhaustively test each $x$ value as candidate split point by computing the MSE for left and right $y$ subregions. The split point that gets the best weighted average for left and right MSE, is the optimal split point.\n</details>",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
d0e3b87ca9b49fd9203c0a934cdaebc6837e10e7 | 3,292 | ipynb | Jupyter Notebook | zh_CN_Jupyter_Learn/stdlib/SHA/docs/src/index.md.ipynb | aifact/JuliaZH.jl | 73a5eb3a7823bb95a37e8529be074a28d41cd78f | [
"MIT"
] | null | null | null | zh_CN_Jupyter_Learn/stdlib/SHA/docs/src/index.md.ipynb | aifact/JuliaZH.jl | 73a5eb3a7823bb95a37e8529be074a28d41cd78f | [
"MIT"
] | null | null | null | zh_CN_Jupyter_Learn/stdlib/SHA/docs/src/index.md.ipynb | aifact/JuliaZH.jl | 73a5eb3a7823bb95a37e8529be074a28d41cd78f | [
"MIT"
] | null | null | null | 22.243243 | 279 | 0.51367 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d0e3c66d08d4403d94f4acf1f12dd9d1f7f78989 | 2,697 | ipynb | Jupyter Notebook | fenics/test-dolfin-checkpoint.ipynb | fem-on-colab/fem-on-colab | d6d82e25425ee080a5562fd8825f4030cd8c4c35 | [
"MIT"
] | 18 | 2021-06-08T12:46:45.000Z | 2022-02-25T18:24:18.000Z | fenics/test-dolfin-checkpoint.ipynb | fem-on-colab/fem-on-colab | d6d82e25425ee080a5562fd8825f4030cd8c4c35 | [
"MIT"
] | 11 | 2021-05-31T14:32:44.000Z | 2022-02-16T13:25:08.000Z | fenics/test-dolfin-checkpoint.ipynb | fem-on-colab/fem-on-colab | 4a192c2482aee0e901b8c5f37b27d10e91eec584 | [
"MIT"
] | null | null | null | 22.475 | 149 | 0.539488 | [
[
[
"try:\n import google.colab # noqa: F401\nexcept ImportError:\n import dolfin\nelse:\n try:\n import dolfin\n except ImportError:\n !wget \"https://fem-on-colab.github.io/releases/fenics-install.sh\" -O \"/tmp/fenics-install.sh\" && bash \"/tmp/fenics-install.sh\"\n import dolfin",
"_____no_output_____"
],
[
"mesh = dolfin.UnitIntervalMesh(3)",
"_____no_output_____"
],
[
"V = dolfin.FunctionSpace(mesh, \"CG\", 1)\nassert V.dim() == 4",
"_____no_output_____"
],
[
"import numpy as np",
"_____no_output_____"
],
[
"solution = dolfin.Function(V)\nsolution.vector()[:] = np.arange(1, V.dim() + 1)",
"_____no_output_____"
],
[
"output_file = dolfin.XDMFFile(\"checkpoint.xdmf\")\noutput_file.parameters[\"flush_output\"] = True\noutput_file.write_checkpoint(solution, \"solution\", 0.0)\noutput_file.close()",
"_____no_output_____"
],
[
"solution_from_file = dolfin.Function(V)\ninput_file = dolfin.XDMFFile(\"checkpoint.xdmf\")\ninput_file.read_checkpoint(solution_from_file, \"solution\", 0)\ninput_file.close()",
"_____no_output_____"
],
[
"assert np.allclose(solution_from_file.vector().vec().getArray(), np.arange(1, V.dim() + 1))",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e3e44258ea8bc26c08dae06c106f5d3a559c17 | 5,442 | ipynb | Jupyter Notebook | how-to-guides/split-column-by-example.ipynb | Bhaskers-Blu-Org2/AMLDataPrepDocs | 61d1dc7e206d9032a6e5b5304598526c0516b5bb | [
"MIT"
] | null | null | null | how-to-guides/split-column-by-example.ipynb | Bhaskers-Blu-Org2/AMLDataPrepDocs | 61d1dc7e206d9032a6e5b5304598526c0516b5bb | [
"MIT"
] | null | null | null | how-to-guides/split-column-by-example.ipynb | Bhaskers-Blu-Org2/AMLDataPrepDocs | 61d1dc7e206d9032a6e5b5304598526c0516b5bb | [
"MIT"
] | 1 | 2020-07-30T12:35:49.000Z | 2020-07-30T12:35:49.000Z | 24.624434 | 220 | 0.587652 | [
[
[
"",
"_____no_output_____"
],
[
"# Split column by example\n",
"_____no_output_____"
],
[
"DataPrep also offers you a way to easily split a column into multiple columns.\nThe SplitColumnByExampleBuilder class lets you generate a proper split program that will work even when the cases are not trivial, like in example below.",
"_____no_output_____"
]
],
[
[
"import azureml.dataprep as dprep",
"_____no_output_____"
],
[
"dflow = dprep.read_lines(path='../data/crime.txt')\ndf = dflow.head(10)",
"_____no_output_____"
],
[
"df['Line'].iloc[0]",
"_____no_output_____"
]
],
[
[
"As you can see above, you can't split this particular file by space character as it will create too many columns.\nThat's where split_column_by_example could be quite useful.",
"_____no_output_____"
]
],
[
[
"builder = dflow.builders.split_column_by_example('Line', keep_delimiters=True)",
"_____no_output_____"
],
[
"builder.preview()",
"_____no_output_____"
]
],
[
[
"Couple things to take note of here. No examples were given, and yet DataPrep was able to generate quite reasonable split program. \nWe have passed keep_delimiters=True so we can see all the data split into columns. In practice, though, delimiters are rarely useful, so let's exclude them.",
"_____no_output_____"
]
],
[
[
"builder.keep_delimiters = False\nbuilder.preview()",
"_____no_output_____"
]
],
[
[
"This looks pretty good already, except that one case number is split into 2 columns. Taking the first row as an example, we want to keep case number as \"HY329907\" instead of \"HY\" and \"329907\" seperately. \nIf we request generation of suggested examples we will get a list of examples that require input.",
"_____no_output_____"
]
],
[
[
"suggestions = builder.generate_suggested_examples()\nsuggestions",
"_____no_output_____"
],
[
"suggestions.iloc[0]['Line']",
"_____no_output_____"
]
],
[
[
"Having retrieved source value we can now provide an example of desired split.\nNotice that we chose not to split date and time but rather keep them together in one column.",
"_____no_output_____"
]
],
[
[
"builder.add_example(example=(suggestions['Line'].iloc[0], ['10140490','HY329907','7/5/2015 23:50','050XX N NEWLAND AVE','820','THEFT']))",
"_____no_output_____"
],
[
"builder.preview()",
"_____no_output_____"
]
],
[
[
"As we can see from the preview, some of the crime types (`Line_6`) do not show up as expected. Let's try to add one more example. ",
"_____no_output_____"
]
],
[
[
"builder.add_example(example=(df['Line'].iloc[1],['10139776','HY329265','7/5/2015 23:30','011XX W MORSE AVE','460','BATTERY']))\nbuilder.preview()",
"_____no_output_____"
]
],
[
[
"This looks just like what we need. Let's get a dataflow with splited columns and drop original column.",
"_____no_output_____"
]
],
[
[
"dflow = builder.to_dataflow()\ndflow = dflow.drop_columns(['Line'])\ndflow.head(5)",
"_____no_output_____"
]
],
[
[
"Now we have successfully split the data into useful columns through examples.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0e3e464c39c9b12dc9c80bdc5d57f9567a630bf | 177,190 | ipynb | Jupyter Notebook | notebooks/dataset-projections/moons/moons-direct-embedding.ipynb | timsainb/ParametricUMAP_paper | 00b4d676647e45619552aec8f2663c0903a83e3f | [
"MIT"
] | 124 | 2020-09-27T23:59:01.000Z | 2022-03-22T06:27:35.000Z | notebooks/dataset-projections/moons/moons-direct-embedding.ipynb | kiminh/ParametricUMAP_paper | 00b4d676647e45619552aec8f2663c0903a83e3f | [
"MIT"
] | 2 | 2021-02-05T18:13:13.000Z | 2021-11-01T14:55:08.000Z | notebooks/dataset-projections/moons/moons-direct-embedding.ipynb | kiminh/ParametricUMAP_paper | 00b4d676647e45619552aec8f2663c0903a83e3f | [
"MIT"
] | 16 | 2020-09-28T07:43:21.000Z | 2022-03-21T00:31:34.000Z | 239.122807 | 62,328 | 0.922292 | [
[
[
"# reload packages\n%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
]
],
[
[
"### Choose GPU (this may not be needed on your computer)",
"_____no_output_____"
]
],
[
[
"%env CUDA_DEVICE_ORDER=PCI_BUS_ID\n%env CUDA_VISIBLE_DEVICES=''",
"env: CUDA_DEVICE_ORDER=PCI_BUS_ID\nenv: CUDA_VISIBLE_DEVICES=''\n"
]
],
[
[
"### load packages",
"_____no_output_____"
]
],
[
[
"from tfumap.umap import tfUMAP",
"/mnt/cube/tsainbur/conda_envs/tpy3/lib/python3.6/site-packages/tqdm/autonotebook/__init__.py:14: TqdmExperimentalWarning: Using `tqdm.autonotebook.tqdm` in notebook mode. Use `tqdm.tqdm` instead to force console mode (e.g. in jupyter console)\n \" (e.g. in jupyter console)\", TqdmExperimentalWarning)\n"
],
[
"import tensorflow as tf\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom tqdm.autonotebook import tqdm\nimport umap\nimport pandas as pd",
"_____no_output_____"
]
],
[
[
"### Load dataset",
"_____no_output_____"
]
],
[
[
"from sklearn.datasets import make_moons",
"_____no_output_____"
],
[
"X_train, Y_train = make_moons(1000, random_state=0, noise=0.1)\nX_train_flat = X_train",
"_____no_output_____"
],
[
"X_test, Y_test = make_moons(1000, random_state=1, noise=0.1)\nX_test_flat = X_test",
"_____no_output_____"
],
[
"X_valid, Y_valid = make_moons(1000, random_state=2, noise=0.1)",
"_____no_output_____"
],
[
"plt.scatter(X_test[:,0], X_test[:,1], c=Y_test)",
"_____no_output_____"
]
],
[
[
"### Create model and train",
"_____no_output_____"
]
],
[
[
"embedder = tfUMAP(direct_embedding=True, verbose=True, negative_sample_rate=5, training_epochs=100)",
"_____no_output_____"
],
[
"z = embedder.fit_transform(X_train_flat)",
"tfUMAP(direct_embedding=True, negative_sample_rate=5,\n optimizer=<tensorflow.python.keras.optimizer_v2.adadelta.Adadelta object at 0x7f56a22fbcc0>,\n tensorboard_logdir='/tmp/tensorboard/20200709-104443',\n training_epochs=100)\nConstruct fuzzy simplicial set\nThu Jul 9 10:44:47 2020 Finding Nearest Neighbors\nThu Jul 9 10:44:49 2020 Finished Nearest Neighbor Search\nThu Jul 9 10:44:51 2020 Embedding with TensorFlow\n"
]
],
[
[
"### Plot model output",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots( figsize=(8, 8))\nsc = ax.scatter(\n z[:, 0],\n z[:, 1],\n c=Y_train.astype(int)[:len(z)],\n cmap=\"tab10\",\n s=0.1,\n alpha=0.5,\n rasterized=True,\n)\nax.axis('equal')\nax.set_title(\"UMAP in Tensorflow embedding\", fontsize=20)\nplt.colorbar(sc, ax=ax);",
"_____no_output_____"
]
],
[
[
"### View loss",
"_____no_output_____"
]
],
[
[
"from tfumap.umap import retrieve_tensors\nimport seaborn as sns",
"_____no_output_____"
],
[
"loss_df = retrieve_tensors(embedder.tensorboard_logdir)\nloss_df[:3]",
"['umap_loss']\n[]\n"
],
[
"ax = sns.lineplot(x=\"step\", y=\"val\", hue=\"group\", data=loss_df[loss_df.variable=='umap_loss'])\nax.set_xscale('log')",
"_____no_output_____"
]
],
[
[
"### Save output",
"_____no_output_____"
]
],
[
[
"from tfumap.paths import ensure_dir, MODEL_DIR",
"_____no_output_____"
],
[
"output_dir = MODEL_DIR/'projections'/ 'moons' / 'direct'\nensure_dir(output_dir)",
"_____no_output_____"
],
[
"embedder.save(output_dir)",
"Pickle of model saved\n"
],
[
"loss_df.to_pickle(output_dir / 'loss_df.pickle')",
"_____no_output_____"
],
[
"np.save(output_dir / 'z.npy', z)",
"_____no_output_____"
]
],
[
[
"### Compare to direct embedding with base UMAP",
"_____no_output_____"
]
],
[
[
"from umap import UMAP",
"_____no_output_____"
],
[
"z_umap = UMAP(verbose=True).fit_transform(X_train_flat)",
"UMAP(dens_frac=0.0, dens_lambda=0.0, verbose=True)\nConstruct fuzzy simplicial set\nThu Jul 9 10:45:45 2020 Finding Nearest Neighbors\nThu Jul 9 10:45:45 2020 Finished Nearest Neighbor Search\nThu Jul 9 10:45:45 2020 Construct embedding\n\tcompleted 0 / 500 epochs\n\tcompleted 50 / 500 epochs\n\tcompleted 100 / 500 epochs\n\tcompleted 150 / 500 epochs\n\tcompleted 200 / 500 epochs\n\tcompleted 250 / 500 epochs\n\tcompleted 300 / 500 epochs\n\tcompleted 350 / 500 epochs\n\tcompleted 400 / 500 epochs\n\tcompleted 450 / 500 epochs\nThu Jul 9 10:45:49 2020 Finished embedding\n"
],
[
"### realign using procrustes \nfrom scipy.spatial import procrustes\nz_align, z_umap_align, disparity = procrustes(z, z_umap)\nprint(disparity)",
"0.03679125952869148\n"
],
[
"fig, axs = plt.subplots(ncols=2, figsize=(20, 8))\nax = axs[0]\nsc = ax.scatter(\n z_align[:, 0],\n z_align[:, 1],\n c=Y_train.astype(int)[:len(z)],\n cmap=\"tab10\",\n s=0.1,\n alpha=0.5,\n rasterized=True,\n)\nax.axis('equal')\nax.set_title(\"UMAP in Tensorflow\", fontsize=20)\n#plt.colorbar(sc, ax=ax);\n\nax = axs[1]\nsc = ax.scatter(\n z_umap_align[:, 0],\n z_umap_align[:, 1],\n c=Y_train.astype(int)[:len(z)],\n cmap=\"tab10\",\n s=0.1,\n alpha=0.5,\n rasterized=True,\n)\nax.axis('equal')\nax.set_title(\"UMAP with UMAP-learn\", fontsize=20)\n#plt.colorbar(sc, ax=ax);",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d0e3ef2252562b22bbccb1dc13efad84d4f92b75 | 18,673 | ipynb | Jupyter Notebook | azure/notebooks/.ipynb_checkpoints/HPO-RAPIDS-checkpoint.ipynb | miguelusque/cloud-ml-examples | daab428e12744267b97e05ed4c7b2f1115ac7a00 | [
"Apache-2.0"
] | null | null | null | azure/notebooks/.ipynb_checkpoints/HPO-RAPIDS-checkpoint.ipynb | miguelusque/cloud-ml-examples | daab428e12744267b97e05ed4c7b2f1115ac7a00 | [
"Apache-2.0"
] | null | null | null | azure/notebooks/.ipynb_checkpoints/HPO-RAPIDS-checkpoint.ipynb | miguelusque/cloud-ml-examples | daab428e12744267b97e05ed4c7b2f1115ac7a00 | [
"Apache-2.0"
] | null | null | null | 30.762768 | 581 | 0.611846 | [
[
[
"# Train and hyperparameter tune with RAPIDS",
"_____no_output_____"
],
[
"## Prerequisites",
"_____no_output_____"
],
[
"- Create an Azure ML Workspace and setup environmnet on local computer following the steps in [Azure README.md](https://gitlab-master.nvidia.com/drobison/aws-sagemaker-gtc-2020/tree/master/azure/README.md )",
"_____no_output_____"
]
],
[
[
"# verify installation and check Azure ML SDK version\nimport azureml.core\n\nprint('SDK version:', azureml.core.VERSION)",
"SDK version: 1.2.0\n"
]
],
[
[
"- Install [AzCopy](https://docs.microsoft.com/en-us/azure/storage/common/storage-use-azcopy-v10) to download dataset from [Azure Blob storage](https://docs.microsoft.com/en-us/azure/storage/blobs/storage-blobs-overview) to your local computer (or another storage account).",
"_____no_output_____"
],
[
"In this example, we will use 20 million rows (samples) of the [airline dataset](http://kt.ijs.si/elena_ikonomovska/data.html):",
"_____no_output_____"
]
],
[
[
"!./azcopy cp 'https://airlinedataset.blob.core.windows.net/airline-20m/airline_20m.parquet' '/home/jzedlewski/code/drobinson/aws-sagemaker-gtc-2020/azure/notebooks\n'",
"/bin/sh: 1: ./azcopy: not found\r\n"
]
],
[
[
"## Initialize workspace",
"_____no_output_____"
],
[
"Load and initialize a [Workspace](https://docs.microsoft.com/azure/machine-learning/service/concept-azure-machine-learning-architecture#workspace) object from the existing workspace you created in the prerequisites step. `Workspace.from_config()` creates a workspace object from the details stored in `config.json`",
"_____no_output_____"
]
],
[
[
"from azureml.core.workspace import Workspace\n\n# if a locally-saved configuration file for the workspace is not available, use the following to load workspace\n# ws = Workspace(subscription_id=subscription_id, resource_group=resource_group, workspace_name=workspace_name)\n\nws = Workspace.from_config()\nprint('Workspace name: ' + ws.name, \n 'Azure region: ' + ws.location, \n 'Subscription id: ' + ws.subscription_id, \n 'Resource group: ' + ws.resource_group, sep = '\\n')\n\ndatastore = ws.get_default_datastore()\nprint(\"Default datastore's name: {}\".format(datastore.name))",
"_____no_output_____"
]
],
[
[
"## Upload data",
"_____no_output_____"
],
[
"Upload the dataset to the workspace's default datastore:",
"_____no_output_____"
]
],
[
[
"path_on_datastore = 'data_airline'\ndatastore.upload(src_dir='/add/local/path', target_path=path_on_datastore, overwrite=False, show_progress=True)",
"_____no_output_____"
],
[
"ds_data = datastore.path(path_on_datastore)\nprint(ds_data)",
"_____no_output_____"
]
],
[
[
"## Create AML compute",
"_____no_output_____"
],
[
"You will need to create a [compute target](https://docs.microsoft.com/azure/machine-learning/service/concept-azure-machine-learning-architecture#compute-target) for training your model. In this notebook, we will use Azure ML managed compute ([AmlCompute](https://docs.microsoft.com/azure/machine-learning/service/how-to-set-up-training-targets#amlcompute)) for our remote training using a dynamically scalable pool of compute resources.\n\nThis notebook will use 10 nodes for hyperparameter optimization, you can modify `max_node` based on available quota in the desired region. Similar to other Azure services, there are limits on certain resources (e.g. AmlCompute) associated with the Azure Machine Learning service. [This article](https://docs.microsoft.com/en-us/azure/machine-learning/service/how-to-manage-quotas) includes details on the default limits and how to request more quota.",
"_____no_output_____"
],
[
"`vm_size` describes the virtual machine type and size that will be used in the cluster. RAPIDS requires NVIDIA Pascal or newer architecture, you will need to specify compute targets from one of `NC_v2`, `NC_v3`, `ND` or `ND_v2` [GPU virtual machines in Azure](https://docs.microsoft.com/en-us/azure/virtual-machines/sizes-gpu); these are VMs that are provisioned with P40 and V100 GPUs. Let's create an `AmlCompute` cluster of `Standard_NC6s_v3` GPU VMs:",
"_____no_output_____"
]
],
[
[
"from azureml.core.compute import ComputeTarget, AmlCompute\nfrom azureml.core.compute_target import ComputeTargetException\n\n# choose a name for your cluster\ngpu_cluster_name = 'gpu-cluster'\n\nif gpu_cluster_name in ws.compute_targets:\n gpu_cluster = ws.compute_targets[gpu_cluster_name]\n if gpu_cluster and type(gpu_cluster) is AmlCompute:\n print('Found compute target. Will use {0} '.format(gpu_cluster_name))\nelse:\n print('creating new cluster')\n # m_size parameter below could be modified to one of the RAPIDS-supported VM types\n provisioning_config = AmlCompute.provisioning_configuration(vm_size = 'Standard_NC6s_v3', max_nodes = 10, idle_seconds_before_scaledown = 300)\n\n # create the cluster\n gpu_cluster = ComputeTarget.create(ws, gpu_cluster_name, provisioning_config)\n \n # can poll for a minimum number of nodes and for a specific timeout \n # if no min node count is provided it uses the scale settings for the cluster\n gpu_cluster.wait_for_completion(show_output=True, min_node_count=None, timeout_in_minutes=20)\n \n# use get_status() to get a detailed status for the current cluster \nprint(gpu_cluster.get_status().serialize())",
"_____no_output_____"
]
],
[
[
"## Prepare training script",
"_____no_output_____"
],
[
"Create a project directory that will contain code from your local machine that you will need access to on the remote resource. This includes the training script and additional files your training script depends on. In this example, the training script is provided: \n<br>\n`train_rapids_RF.py` - entry script for RAPIDS Estimator that includes loading dataset into cuDF data frame, training with Random Forest and inference using cuML.",
"_____no_output_____"
]
],
[
[
"import os\n\nproject_folder = './train_rapids'\nos.makedirs(project_folder, exist_ok=True)",
"_____no_output_____"
]
],
[
[
"We will log some metrics by using the `Run` object within the training script:\n\n```python\nfrom azureml.core.run import Run\nrun = Run.get_context()\n```\n \nWe will also log the parameters and highest accuracy the model achieves:\n\n```python\nrun.log('Accuracy', np.float(accuracy))\n```\n\nThese run metrics will become particularly important when we begin hyperparameter tuning our model in the 'Tune model hyperparameters' section.",
"_____no_output_____"
],
[
"Copy the training script `train_rapids_RF.py` into your project directory:",
"_____no_output_____"
]
],
[
[
"import shutil\n\nshutil.copy('../code/train_rapids_RF.py', project_folder)",
"_____no_output_____"
]
],
[
[
"## Train model on the remote compute",
"_____no_output_____"
],
[
"Now that you have your data and training script prepared, you are ready to train on your remote compute.",
"_____no_output_____"
],
[
"### Create experiment",
"_____no_output_____"
],
[
"Create an [Experiment](https://docs.microsoft.com/azure/machine-learning/service/concept-azure-machine-learning-architecture#experiment) to track all the runs in your workspace.",
"_____no_output_____"
]
],
[
[
"from azureml.core import Experiment\n\nexperiment_name = 'train_rapids'\nexperiment = Experiment(ws, name=experiment_name)",
"_____no_output_____"
]
],
[
[
"### Create environment",
"_____no_output_____"
],
[
"The [Environment class](https://docs.microsoft.com/en-us/python/api/azureml-core/azureml.core.environment.environment?view=azure-ml-py) allows you to build a Docker image and customize the system that you will use for training. We will build a container image using a RAPIDS container as base image and install necessary packages. This build is necessary only the first time and will take about 15 minutes. The image will be added to your Azure Container Registry and the environment will be cached after the first run, as long as the environment definition remains the same.",
"_____no_output_____"
]
],
[
[
"from azureml.core import Environment\n\n# create the environment\nrapids_env = Environment('rapids_env')\n\n# create the environment inside a Docker container\nrapids_env.docker.enabled = True\n\n# specify docker steps as a string. Alternatively, load the string from a file\ndockerfile = \"\"\"\nFROM rapidsai/rapidsai-nightly:0.13-cuda10.0-runtime-ubuntu18.04-py3.7\nRUN source activate rapids && \\\npip install azureml-sdk && \\\npip install azureml-widgets\n\"\"\"\n#FROM nvcr.io/nvidia/rapidsai/rapidsai:0.12-cuda10.0-runtime-ubuntu18.04\n\n# set base image to None since the image is defined by dockerfile\nrapids_env.docker.base_image = None\nrapids_env.docker.base_dockerfile = dockerfile\n\n# use rapids environment in the container\nrapids_env.python.user_managed_dependencies = True",
"_____no_output_____"
],
[
"# from azureml.core.container_registry import ContainerRegistry\n\n# # this is an image available on Docker Hub\n# image_name = 'zronaghi/rapidsai-nightly:0.13-cuda10.0-runtime-ubuntu18.04-py3.7-azuresdk-030920'\n\n# # use rapids environment, don't build a new conda environment\n# user_managed_dependencies = True",
"_____no_output_____"
]
],
[
[
"### Create a RAPIDS Estimator",
"_____no_output_____"
],
[
"The [Estimator](https://docs.microsoft.com/en-us/python/api/azureml-train-core/azureml.train.estimator.estimator?view=azure-ml-py) class can be used with machine learning frameworks that do not have a pre-configure estimator. \n\n`script_params` is a dictionary of command-line arguments to pass to the training script.",
"_____no_output_____"
]
],
[
[
"from azureml.train.estimator import Estimator\n\nscript_params = {\n '--data_dir': ds_data.as_mount(),\n '--n_bins': 32,\n}\n\nestimator = Estimator(source_directory=project_folder,\n script_params=script_params,\n compute_target=gpu_cluster, \n entry_script='train_rapids_RF.py',\n environment_definition=rapids_env)\n# custom_docker_image=image_name,\n# user_managed=user_managed_dependencies",
"_____no_output_____"
]
],
[
[
"## Tune model hyperparameters",
"_____no_output_____"
],
[
"We can optimize our model's hyperparameters and improve the accuracy using Azure Machine Learning's hyperparameter tuning capabilities.",
"_____no_output_____"
],
[
"### Start a hyperparameter sweep",
"_____no_output_____"
],
[
"Let's define the hyperparameter space to sweep over. We will tune `n_estimators`, `max_depth` and `max_features` parameters. In this example we will use random sampling to try different configuration sets of hyperparameters and maximize `Accuracy`.",
"_____no_output_____"
]
],
[
[
"from azureml.train.hyperdrive.runconfig import HyperDriveConfig\nfrom azureml.train.hyperdrive.sampling import RandomParameterSampling\nfrom azureml.train.hyperdrive.run import PrimaryMetricGoal\nfrom azureml.train.hyperdrive.parameter_expressions import choice, loguniform, uniform\n\nparam_sampling = RandomParameterSampling( {\n '--n_estimators': choice(range(50, 500)),\n '--max_depth': choice(range(5, 19)),\n '--max_features': uniform(0.2, 1.0)\n }\n) \n\nhyperdrive_run_config = HyperDriveConfig(estimator=estimator,\n hyperparameter_sampling=param_sampling, \n primary_metric_name='Accuracy',\n primary_metric_goal=PrimaryMetricGoal.MAXIMIZE,\n max_total_runs=100,\n max_concurrent_runs=10)",
"_____no_output_____"
]
],
[
[
"This will launch the RAPIDS training script with parameters that were specified in the cell above.",
"_____no_output_____"
]
],
[
[
"# start the HyperDrive run\nhyperdrive_run = experiment.submit(hyperdrive_run_config)",
"_____no_output_____"
]
],
[
[
"## Monitor HyperDrive runs",
"_____no_output_____"
],
[
"Monitor and view the progress of the machine learning training run with a [Jupyter widget](https://docs.microsoft.com/en-us/python/api/azureml-widgets/azureml.widgets?view=azure-ml-py).The widget is asynchronous and provides live updates every 10-15 seconds until the job completes.",
"_____no_output_____"
]
],
[
[
"from azureml.widgets import RunDetails\n\nRunDetails(hyperdrive_run).show()",
"_____no_output_____"
],
[
"# hyperdrive_run.wait_for_completion(show_output=True)",
"_____no_output_____"
],
[
"# hyperdrive_run.cancel()",
"_____no_output_____"
]
],
[
[
"### Find and register best model",
"_____no_output_____"
]
],
[
[
"best_run = hyperdrive_run.get_best_run_by_primary_metric()\nprint(best_run.get_details()['runDefinition']['arguments'])",
"_____no_output_____"
]
],
[
[
"List the model files uploaded during the run:",
"_____no_output_____"
]
],
[
[
"print(best_run.get_file_names())",
"_____no_output_____"
]
],
[
[
"Register the folder (and all files in it) as a model named `train-rapids` under the workspace for deployment",
"_____no_output_____"
]
],
[
[
"# model = best_run.register_model(model_name='train-rapids', model_path='outputs/model-rapids.joblib')",
"_____no_output_____"
]
],
[
[
"## Delete cluster",
"_____no_output_____"
]
],
[
[
"# delete the cluster\n# gpu_cluster.delete()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0e3f32ce35d77596bbe08727472aba6102752d7 | 90,386 | ipynb | Jupyter Notebook | completed/Session 1 - Hello DL World.ipynb | tongni1975/tsundoku | 3f5d35bf28700f5b2fde671e4334a7ce49591902 | [
"MIT"
] | 18 | 2019-12-02T03:43:53.000Z | 2020-10-07T02:27:48.000Z | completed/Session 1 - Hello DL World.ipynb | tongni1975/tsundoku | 3f5d35bf28700f5b2fde671e4334a7ce49591902 | [
"MIT"
] | null | null | null | completed/Session 1 - Hello DL World.ipynb | tongni1975/tsundoku | 3f5d35bf28700f5b2fde671e4334a7ce49591902 | [
"MIT"
] | 18 | 2019-12-03T05:47:27.000Z | 2021-09-26T16:38:31.000Z | 42.414829 | 12,460 | 0.686334 | [
[
[
"import sys\nsys.executable",
"_____no_output_____"
]
],
[
[
"[Optional]: If you're using a Mac/Linux, you can check your environment with these commands:\n\n```\n!which pip3\n!which python3\n!ls -lah /usr/local/bin/python3\n```",
"_____no_output_____"
]
],
[
[
"!pip3 install -U pip\n!pip3 install torch==1.3.0\n!pip3 install seaborn",
"Requirement already up-to-date: pip in /Library/Frameworks/Python.framework/Versions/3.6/lib/python3.6/site-packages (19.3.1)\nCollecting torch==1.3.0\n Using cached https://files.pythonhosted.org/packages/8d/87/4e42d7ab7cb1e5ee9f2f81d9c5955a7c558894f11c90898fdf838ea40327/torch-1.3.0-cp36-none-macosx_10_7_x86_64.whl\nRequirement already satisfied: numpy in /Users/liling.tan/Library/Python/3.6/lib/python/site-packages (from torch==1.3.0) (1.18.1)\n\u001b[31mERROR: allennlp 0.9.0 requires pytorch-transformers==1.1.0, which is not installed.\u001b[0m\n\u001b[31mERROR: apes 1.0.0 requires transformers==2.3.0, which is not installed.\u001b[0m\n\u001b[31mERROR: opennmt-py 1.0.0 has requirement tqdm~=4.30.0, but you'll have tqdm 4.41.1 which is incompatible.\u001b[0m\n\u001b[31mERROR: onmt 1.0.0 has requirement numpy==1.17.4, but you'll have numpy 1.18.1 which is incompatible.\u001b[0m\n\u001b[31mERROR: onmt 1.0.0 has requirement torch==1.3.1, but you'll have torch 1.3.0 which is incompatible.\u001b[0m\n\u001b[31mERROR: onmt 1.0.0 has requirement tqdm==4.38.0, but you'll have tqdm 4.41.1 which is incompatible.\u001b[0m\n\u001b[31mERROR: apes 1.0.0 has requirement sacremoses==0.0.35, but you'll have sacremoses 0.0.38 which is incompatible.\u001b[0m\nInstalling collected packages: torch\n Found existing installation: torch 1.3.1\n Uninstalling torch-1.3.1:\n Successfully uninstalled torch-1.3.1\nSuccessfully installed torch-1.3.0\nRequirement already satisfied: seaborn in /Library/Frameworks/Python.framework/Versions/3.6/lib/python3.6/site-packages (0.9.0)\nRequirement already satisfied: matplotlib>=1.4.3 in /Library/Frameworks/Python.framework/Versions/3.6/lib/python3.6/site-packages (from seaborn) (3.1.1)\nRequirement already satisfied: scipy>=0.14.0 in /Users/liling.tan/Library/Python/3.6/lib/python/site-packages (from seaborn) (1.2.1)\nRequirement already satisfied: numpy>=1.9.3 in /Users/liling.tan/Library/Python/3.6/lib/python/site-packages (from seaborn) (1.18.1)\nRequirement already satisfied: pandas>=0.15.2 in /Users/liling.tan/Library/Python/3.6/lib/python/site-packages (from seaborn) (0.25.1)\nRequirement already satisfied: python-dateutil>=2.1 in /Users/liling.tan/Library/Python/3.6/lib/python/site-packages (from matplotlib>=1.4.3->seaborn) (2.8.0)\nRequirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in /Library/Frameworks/Python.framework/Versions/3.6/lib/python3.6/site-packages (from matplotlib>=1.4.3->seaborn) (2.2.0)\nRequirement already satisfied: cycler>=0.10 in /Library/Frameworks/Python.framework/Versions/3.6/lib/python3.6/site-packages (from matplotlib>=1.4.3->seaborn) (0.10.0)\nRequirement already satisfied: kiwisolver>=1.0.1 in /Library/Frameworks/Python.framework/Versions/3.6/lib/python3.6/site-packages (from matplotlib>=1.4.3->seaborn) (1.0.1)\nRequirement already satisfied: pytz>=2017.2 in /Users/liling.tan/Library/Python/3.6/lib/python/site-packages (from pandas>=0.15.2->seaborn) (2019.2)\nRequirement already satisfied: six>=1.5 in /Users/liling.tan/Library/Python/3.6/lib/python/site-packages (from python-dateutil>=2.1->matplotlib>=1.4.3->seaborn) (1.13.0)\nRequirement already satisfied: setuptools in /Users/liling.tan/Library/Python/3.6/lib/python/site-packages (from kiwisolver>=1.0.1->matplotlib>=1.4.3->seaborn) (41.4.0)\n"
],
[
"import torch\ntorch.cuda.is_available()",
"_____no_output_____"
],
[
"# IPython candies...\nfrom IPython.display import Image\nfrom IPython.core.display import HTML\n\nfrom IPython.display import clear_output",
"_____no_output_____"
],
[
"%%html\n<style> table {float:left} </style>",
"_____no_output_____"
]
],
[
[
"Perceptron\n=====\n\n**Perceptron** algorithm is a:\n\n> \"*system that depends on **probabilistic** rather than deterministic principles for its operation, gains its reliability from the **properties of statistical measurements obtain from a large population of elements***\"\n> \\- Frank Rosenblatt (1957)\n\nThen the news:\n\n> \"*[Perceptron is an] **embryo of an electronic computer** that [the Navy] expects will be **able to walk, talk, see, write, reproduce itself and be conscious of its existence.***\"\n> \\- The New York Times (1958)\n\nNews quote cite from Olazaran (1996) ",
"_____no_output_____"
],
[
"Perceptron in Bullets\n----\n\n - Perceptron learns to classify any linearly separable set of inputs. \n - Some nice graphics for perceptron with Go https://appliedgo.net/perceptron/ \n\nIf you've got some spare time: \n\n - There's a whole book just on perceptron: https://mitpress.mit.edu/books/perceptrons\n - For watercooler gossips on perceptron in the early days, read [Olazaran (1996)](https://pdfs.semanticscholar.org/f3b6/e5ef511b471ff508959f660c94036b434277.pdf?_ga=2.57343906.929185581.1517539221-1505787125.1517539221)\n \n \nPerceptron in Math\n----\n\nGiven a set of inputs $x$, the perceptron \n\n - learns $w$ vector to map the inputs to a real-value output between $[0,1]$\n - through the summation of the dot product of the $w·x$ with a transformation function\n \n \nPerceptron in Picture\n----\n",
"_____no_output_____"
]
],
[
[
"##Image(url=\"perceptron.png\", width=500)\nImage(url=\"https://ibin.co/4TyMU8AdpV4J.png\", width=500)",
"_____no_output_____"
]
],
[
[
"(**Note:** Usually, we use $x_1$ as the bias and fix the input to 1)",
"_____no_output_____"
],
[
"Perceptron as a Workflow Diagram\n----\n\nIf you're familiar with [mermaid flowchart](https://mermaidjs.github.io)\n\n```\n.. mermaid::\n\n graph LR\n subgraph Input\n x_1\n x_i \n x_n\n end\n subgraph Perceptron\n n1((s)) --> n2((\"f(s)\"))\n end\n x_1 --> |w_1| n1\n x_i --> |w_i| n1\n x_n --> |w_n| n1\n n2 --> y[\"[0,1]\"]\n\n```",
"_____no_output_____"
]
],
[
[
"##Image(url=\"perceptron-mermaid.svg\", width=500)\nImage(url=\"https://svgshare.com/i/AbJ.svg\", width=500)",
"_____no_output_____"
]
],
[
[
"Optimization Process\n====\n\nTo learn the weights, $w$, we use an **optimizer** to find the best-fit (optimal) values for $w$ such that the inputs correct maps to the outputs.\n\nTypically, process performs the following 4 steps iteratively.\n\n### **Initialization**\n\n - **Step 1**: Initialize weights vector\n \n### **Forward Propagation**\n\n \n - **Step 2a**: Multiply the weights vector with the inputs, sum the products, i.e. `s`\n - **Step 2b**: Put the sum through the sigmoid, i.e. `f()`\n \n### **Back Propagation**\n \n \n - **Step 3a**: Compute the errors, i.e. difference between expected output and predictions\n - **Step 3b**: Multiply the error with the **derivatives** to get the delta\n - **Step 3c**: Multiply the delta vector with the inputs, sum the product\n \n### **Optimizer takes a step**\n\n - **Step 4**: Multiply the learning rate with the output of Step 3c.\n \n\n\n",
"_____no_output_____"
]
],
[
[
"import math\nimport numpy as np\nnp.random.seed(0)",
"_____no_output_____"
],
[
"def sigmoid(x): # Returns values that sums to one.\n return 1 / (1 + np.exp(-x))\n\ndef sigmoid_derivative(sx): \n # See https://math.stackexchange.com/a/1225116\n # Hint: let sx = sigmoid(x)\n return sx * (1 - sx)",
"_____no_output_____"
],
[
"sigmoid(np.array([2.5, 0.32, -1.42])) # [out]: array([0.92414182, 0.57932425, 0.19466158])",
"_____no_output_____"
],
[
"sigmoid_derivative(np.array([2.5, 0.32, -1.42])) # [out]: array([0.07010372, 0.24370766, 0.15676845])",
"_____no_output_____"
],
[
"def cost(predicted, truth):\n return np.abs(truth - predicted)",
"_____no_output_____"
],
[
"gold = np.array([0.5, 1.2, 9.8])\npred = np.array([0.6, 1.0, 10.0])\ncost(pred, gold)",
"_____no_output_____"
],
[
"gold = np.array([0.5, 1.2, 9.8])\npred = np.array([9.3, 4.0, 99.0])\ncost(pred, gold)",
"_____no_output_____"
]
],
[
[
"Representing OR Boolean\n---\n\nLets consider the problem of the OR boolean and apply the perceptron with simple gradient descent. \n\n| x2 | x3 | y | \n|:--:|:--:|:--:|\n| 0 | 0 | 0 |\n| 0 | 1 | 1 | \n| 1 | 0 | 1 | \n| 1 | 1 | 1 | \n",
"_____no_output_____"
]
],
[
[
"X = or_input = np.array([[0,0], [0,1], [1,0], [1,1]])\nY = or_output = np.array([[0,1,1,1]]).T",
"_____no_output_____"
],
[
"or_input",
"_____no_output_____"
],
[
"or_output",
"_____no_output_____"
],
[
"# Define the shape of the weight vector.\nnum_data, input_dim = or_input.shape\n# Define the shape of the output vector. \noutput_dim = len(or_output.T)",
"_____no_output_____"
],
[
"print('Inputs\\n======')\nprint('no. of rows =', num_data) \nprint('no. of cols =', input_dim)\nprint('\\n')\nprint('Outputs\\n=======')\nprint('no. of cols =', output_dim)",
"Inputs\n======\nno. of rows = 4\nno. of cols = 2\n\n\nOutputs\n=======\nno. of cols = 1\n"
],
[
"# Initialize weights between the input layers and the perceptron\nW = np.random.random((input_dim, output_dim))\nW",
"_____no_output_____"
]
],
[
[
"Step 2a: Multiply the weights vector with the inputs, sum the products\n====\n\nTo get the output of step 2a, \n\n - Itrate through each row of the data, `X`\n - For each column in each row, find the product of the value and the respective weights\n - For each row, compute the sum of the products",
"_____no_output_____"
]
],
[
[
"# If we write it imperatively:\nsummation = []\nfor row in X:\n sum_wx = 0\n for feature, weight in zip(row, W):\n sum_wx += feature * weight\n summation.append(sum_wx)\nprint(np.array(summation))",
"[[0. ]\n [0.71518937]\n [0.5488135 ]\n [1.26400287]]\n"
],
[
"# If we vectorize the process and use numpy.\nnp.dot(X, W)",
"_____no_output_____"
]
],
[
[
"Train the Single-Layer Model\n====\n\n",
"_____no_output_____"
]
],
[
[
"num_epochs = 10000 # No. of times to iterate.\nlearning_rate = 0.03 # How large a step to take per iteration.\n\n# Lets standardize and call our inputs X and outputs Y\nX = or_input\nY = or_output\n\nfor _ in range(num_epochs):\n layer0 = X\n\n # Step 2a: Multiply the weights vector with the inputs, sum the products, i.e. s\n # Step 2b: Put the sum through the sigmoid, i.e. f()\n # Inside the perceptron, Step 2. \n layer1 = sigmoid(np.dot(X, W))\n\n # Back propagation.\n # Step 3a: Compute the errors, i.e. difference between expected output and predictions\n # How much did we miss?\n layer1_error = cost(layer1, Y)\n\n # Step 3b: Multiply the error with the derivatives to get the delta\n # multiply how much we missed by the slope of the sigmoid at the values in layer1\n layer1_delta = layer1_error * sigmoid_derivative(layer1)\n\n # Step 3c: Multiply the delta vector with the inputs, sum the product (use np.dot)\n # Step 4: Multiply the learning rate with the output of Step 3c.\n W += learning_rate * np.dot(layer0.T, layer1_delta)",
"_____no_output_____"
],
[
"layer1",
"_____no_output_____"
],
[
"# Expected output.\nY",
"_____no_output_____"
],
[
"# On the training data\n[[int(prediction > 0.5)] for prediction in layer1]",
"_____no_output_____"
]
],
[
[
"Lets try the XOR Boolean\n---\n\nLets consider the problem of the OR boolean and apply the perceptron with simple gradient descent. \n\n| x2 | x3 | y | \n|:--:|:--:|:--:|\n| 0 | 0 | 0 |\n| 0 | 1 | 1 | \n| 1 | 0 | 1 | \n| 1 | 1 | 0 | \n",
"_____no_output_____"
]
],
[
[
"X = xor_input = np.array([[0,0], [0,1], [1,0], [1,1]])\nY = xor_output = np.array([[0,1,1,0]]).T",
"_____no_output_____"
],
[
"xor_input",
"_____no_output_____"
],
[
"xor_output",
"_____no_output_____"
],
[
"num_epochs = 10000 # No. of times to iterate.\nlearning_rate = 0.003 # How large a step to take per iteration.\n\n# Lets drop the last row of data and use that as unseen test.\nX = xor_input\nY = xor_output\n\n# Define the shape of the weight vector.\nnum_data, input_dim = X.shape\n# Define the shape of the output vector. \noutput_dim = len(Y.T)\n# Initialize weights between the input layers and the perceptron\nW = np.random.random((input_dim, output_dim))\n\nfor _ in range(num_epochs):\n layer0 = X\n # Forward propagation.\n # Inside the perceptron, Step 2. \n layer1 = sigmoid(np.dot(X, W))\n\n # How much did we miss?\n layer1_error = cost(layer1, Y)\n\n # Back propagation.\n # multiply how much we missed by the slope of the sigmoid at the values in layer1\n layer1_delta = sigmoid_derivative(layer1) * layer1_error\n\n # update weights\n W += learning_rate * np.dot(layer0.T, layer1_delta)",
"_____no_output_____"
],
[
"# Expected output.\nY",
"_____no_output_____"
],
[
"# On the training data\n[int(prediction > 0.5) for prediction in layer1] # All correct.",
"_____no_output_____"
]
],
[
[
"You can't represent XOR with simple perceptron !!!\n====\n\nNo matter how you change the hyperparameters or data, the XOR function can't be represented by a single perceptron layer.\n \nThere's no way you can get all four data points to get the correct outputs for the XOR boolean operation.\n",
"_____no_output_____"
],
[
"Solving XOR (Add more layers)\n====",
"_____no_output_____"
]
],
[
[
"from itertools import chain\nimport numpy as np\nnp.random.seed(0)\n\ndef sigmoid(x): # Returns values that sums to one.\n return 1 / (1 + np.exp(-x))\n\ndef sigmoid_derivative(sx):\n # See https://math.stackexchange.com/a/1225116\n return sx * (1 - sx)\n\n# Cost functions.\ndef cost(predicted, truth):\n return truth - predicted\n\nxor_input = np.array([[0,0], [0,1], [1,0], [1,1]])\nxor_output = np.array([[0,1,1,0]]).T\n\n# Define the shape of the weight vector.\nnum_data, input_dim = X.shape\n# Lets set the dimensions for the intermediate layer.\nhidden_dim = 5\n# Initialize weights between the input layers and the hidden layer.\nW1 = np.random.random((input_dim, hidden_dim))\n\n# Define the shape of the output vector. \noutput_dim = len(Y.T)\n# Initialize weights between the hidden layers and the output layer.\nW2 = np.random.random((hidden_dim, output_dim))\n\n# Initialize weigh\nnum_epochs = 10000\nlearning_rate = 0.03\n\nfor epoch_n in range(num_epochs):\n layer0 = X\n # Forward propagation.\n \n # Inside the perceptron, Step 2. \n layer1 = sigmoid(np.dot(layer0, W1))\n layer2 = sigmoid(np.dot(layer1, W2))\n\n # Back propagation (Y -> layer2)\n \n # How much did we miss in the predictions?\n layer2_error = cost(layer2, Y)\n # In what direction is the target value?\n # Were we really close? If so, don't change too much.\n layer2_delta = layer2_error * sigmoid_derivative(layer2)\n\n \n # Back propagation (layer2 -> layer1)\n # How much did each layer1 value contribute to the layer2 error (according to the weights)?\n layer1_error = np.dot(layer2_delta, W2.T)\n layer1_delta = layer1_error * sigmoid_derivative(layer1)\n \n # update weights\n W2 += learning_rate * np.dot(layer1.T, layer2_delta)\n W1 += learning_rate * np.dot(layer0.T, layer1_delta)\n ##print(epoch_n, list((layer2)))",
"_____no_output_____"
],
[
"# Training input.\nX",
"_____no_output_____"
],
[
"# Expected output.\nY",
"_____no_output_____"
],
[
"layer2 # Our output layer",
"_____no_output_____"
],
[
"# On the training data\n[int(prediction > 0.5) for prediction in layer2] ",
"_____no_output_____"
]
],
[
[
"Now try adding another layer\n====\n\nUse the same process:\n \n 1. Initialize\n 2. Forward Propagate\n 3. Back Propagate \n 4. Update (aka step)\n \n",
"_____no_output_____"
]
],
[
[
"from itertools import chain\nimport numpy as np\nnp.random.seed(0)\n\ndef sigmoid(x): # Returns values that sums to one.\n return 1 / (1 + np.exp(-x))\n\ndef sigmoid_derivative(sx):\n # See https://math.stackexchange.com/a/1225116\n return sx * (1 - sx)\n\n# Cost functions.\ndef cost(predicted, truth):\n return truth - predicted\n\nxor_input = np.array([[0,0], [0,1], [1,0], [1,1]])\nxor_output = np.array([[0,1,1,0]]).T\n\n",
"_____no_output_____"
],
[
"X",
"_____no_output_____"
],
[
"Y",
"_____no_output_____"
],
[
"# Define the shape of the weight vector.\nnum_data, input_dim = X.shape\n# Lets set the dimensions for the intermediate layer.\nlayer0to1_hidden_dim = 5\nlayer1to2_hidden_dim = 5\n\n# Initialize weights between the input layers 0 -> layer 1\nW1 = np.random.random((input_dim, layer0to1_hidden_dim))\n\n# Initialize weights between the layer 1 -> layer 2\nW2 = np.random.random((layer0to1_hidden_dim, layer1to2_hidden_dim))\n\n# Define the shape of the output vector. \noutput_dim = len(Y.T)\n# Initialize weights between the layer 2 -> layer 3\nW3 = np.random.random((layer1to2_hidden_dim, output_dim))\n\n# Initialize weigh\nnum_epochs = 10000\nlearning_rate = 1.0\n\nfor epoch_n in range(num_epochs):\n layer0 = X\n # Forward propagation.\n \n # Inside the perceptron, Step 2. \n layer1 = sigmoid(np.dot(layer0, W1))\n layer2 = sigmoid(np.dot(layer1, W2))\n layer3 = sigmoid(np.dot(layer2, W3))\n\n # Back propagation (Y -> layer2)\n # How much did we miss in the predictions?\n layer3_error = cost(layer3, Y)\n # In what direction is the target value?\n # Were we really close? If so, don't change too much.\n layer3_delta = layer3_error * sigmoid_derivative(layer3)\n\n # Back propagation (layer2 -> layer1)\n # How much did each layer1 value contribute to the layer3 error (according to the weights)?\n layer2_error = np.dot(layer3_delta, W3.T)\n layer2_delta = layer3_error * sigmoid_derivative(layer2)\n \n # Back propagation (layer2 -> layer1)\n # How much did each layer1 value contribute to the layer2 error (according to the weights)?\n layer1_error = np.dot(layer2_delta, W2.T)\n layer1_delta = layer1_error * sigmoid_derivative(layer1)\n \n # update weights\n W3 += learning_rate * np.dot(layer2.T, layer3_delta)\n W2 += learning_rate * np.dot(layer1.T, layer2_delta)\n W1 += learning_rate * np.dot(layer0.T, layer1_delta)",
"_____no_output_____"
],
[
"Y",
"_____no_output_____"
],
[
"layer3",
"_____no_output_____"
],
[
"# On the training data\n[int(prediction > 0.5) for prediction in layer3] ",
"_____no_output_____"
]
],
[
[
"# Now, lets do it with PyTorch \n\nFirst lets try a single perceptron and see that we can't train a model that can represent XOR. \n\n\n",
"_____no_output_____"
]
],
[
[
"from tqdm import tqdm\n\nimport torch\nfrom torch import nn\nfrom torch.autograd import Variable\nfrom torch import FloatTensor\nfrom torch import optim\nuse_cuda = torch.cuda.is_available()",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nimport numpy as np\nimport seaborn as sns\n\nsns.set_style(\"darkgrid\")\nsns.set(rc={'figure.figsize':(15, 10)})",
"_____no_output_____"
],
[
"X # Original XOR X input in numpy array data structure.",
"_____no_output_____"
],
[
"Y # Original XOR Y output in numpy array data structure.",
"_____no_output_____"
],
[
"device = 'gpu' if torch.cuda.is_available() else 'cpu'\n# Converting the X to PyTorch-able data structure.\nX_pt = torch.tensor(X).float()\nX_pt = X_pt.to(device)\n# Converting the Y to PyTorch-able data structure.\nY_pt = torch.tensor(Y, requires_grad=False).float()\nY_pt = Y_pt.to(device)\nprint(X_pt)\nprint(Y_pt)",
"tensor([[0., 0.],\n [0., 1.],\n [1., 0.],\n [1., 1.]])\ntensor([[0.],\n [1.],\n [1.],\n [0.]])\n"
],
[
"# Use tensor.shape to get the shape of the matrix/tensor.\nnum_data, input_dim = X_pt.shape\nprint('Inputs Dim:', input_dim)\n\nnum_data, output_dim = Y_pt.shape\nprint('Output Dim:', output_dim)",
"Inputs Dim: 2\nOutput Dim: 1\n"
],
[
"# Use Sequential to define a simple feed-forward network.\nmodel = nn.Sequential(\n nn.Linear(input_dim, output_dim), # Use nn.Linear to get our simple perceptron\n nn.Sigmoid() # Use nn.Sigmoid to get our sigmoid non-linearity\n )\nmodel",
"_____no_output_____"
],
[
"# Remember we define as: cost = truth - predicted\n# If we take the absolute of cost, i.e.: cost = |truth - predicted|\n# we get the L1 loss function. \ncriterion = nn.L1Loss() \nlearning_rate = 0.03",
"_____no_output_____"
],
[
"# The simple weights/parameters update processes we did before\n# is call the gradient descent. SGD is the sochastic variant of\n# gradient descent. \noptimizer = optim.SGD(model.parameters(), lr=learning_rate)",
"_____no_output_____"
]
],
[
[
"(**Note**: Personally, I strongely encourage you to go through the [University of Washington course of machine learning regression](https://www.coursera.org/learn/ml-regression) to better understand the fundamentals of (i) ***gradient***, (ii) ***loss*** and (iii) ***optimizer***. But given that you know how to code it, the process of more complex variants of gradient/loss computation and optimizer's step is easy to grasp)",
"_____no_output_____"
],
[
"# Training a PyTorch model\n\nTo train a model using PyTorch, we simply iterate through the no. of epochs and imperatively state the computations we want to perform. \n\n## Remember the steps?\n\n 1. Initialize \n 2. Forward Propagation\n 3. Backward Propagation\n 4. Update Optimizer",
"_____no_output_____"
]
],
[
[
"num_epochs = 1000",
"_____no_output_____"
],
[
"# Step 1: Initialization. \n# Note: When using PyTorch a lot of the manual weights\n# initialization is done automatically when we define\n# the model (aka architecture)\nmodel = nn.Sequential(\n nn.Linear(input_dim, output_dim), \n nn.Sigmoid())\ncriterion = nn.MSELoss() \nlearning_rate = 1.0\noptimizer = optim.SGD(model.parameters(), lr=learning_rate)\nnum_epochs = 10000\n\nlosses = []\n\nfor i in tqdm(range(num_epochs)):\n # Reset the gradient after every epoch. \n optimizer.zero_grad() \n # Step 2: Foward Propagation\n predictions = model(X_pt)\n \n # Step 3: Back Propagation \n # Calculate the cost between the predictions and the truth.\n loss_this_epoch = criterion(predictions, Y_pt)\n # Note: The neat thing about PyTorch is it does the \n # auto-gradient computation, no more manually defining\n # derivative of functions and manually propagating\n # the errors layer by layer.\n loss_this_epoch.backward()\n \n # Step 4: Optimizer take a step. \n # Note: Previously, we have to manually update the \n # weights of each layer individually according to the\n # learning rate and the layer delta. \n # PyTorch does that automatically =)\n optimizer.step()\n \n # Log the loss value as we proceed through the epochs.\n losses.append(loss_this_epoch.data.item())\n \n# Visualize the losses\nplt.plot(losses)\nplt.show()\n",
"100%|██████████| 10000/10000 [00:02<00:00, 4373.51it/s]\n"
],
[
"for _x, _y in zip(X_pt, Y_pt):\n prediction = model(_x)\n print('Input:\\t', list(map(int, _x)))\n print('Pred:\\t', int(prediction))\n print('Ouput:\\t', int(_y))\n print('######')",
"Input:\t [0, 0]\nPred:\t 0\nOuput:\t 0\n######\nInput:\t [0, 1]\nPred:\t 0\nOuput:\t 1\n######\nInput:\t [1, 0]\nPred:\t 0\nOuput:\t 1\n######\nInput:\t [1, 1]\nPred:\t 0\nOuput:\t 0\n######\n"
]
],
[
[
"Now, try again with 2 layers using PyTorch\n====",
"_____no_output_____"
]
],
[
[
"%%time\n\nhidden_dim = 5\nnum_data, input_dim = X_pt.shape\nnum_data, output_dim = Y_pt.shape\n\nmodel = nn.Sequential(nn.Linear(input_dim, hidden_dim),\n nn.Sigmoid(), \n nn.Linear(hidden_dim, output_dim),\n nn.Sigmoid())\n\ncriterion = nn.MSELoss()\nlearning_rate = 0.3\noptimizer = optim.SGD(model.parameters(), lr=learning_rate)\nnum_epochs = 5000\n\nlosses = []\n\nfor _ in tqdm(range(num_epochs)):\n optimizer.zero_grad()\n predictions = model(X_pt)\n loss_this_epoch = criterion(predictions, Y_pt)\n loss_this_epoch.backward()\n optimizer.step()\n losses.append(loss_this_epoch.data.item())\n ##print([float(_pred) for _pred in predictions], list(map(int, Y_pt)), loss_this_epoch.data[0])\n \n# Visualize the losses\nplt.plot(losses)\nplt.show()",
"100%|██████████| 5000/5000 [00:02<00:00, 1698.25it/s]\n"
],
[
"for _x, _y in zip(X_pt, Y_pt):\n prediction = model(_x)\n print('Input:\\t', list(map(int, _x)))\n print('Pred:\\t', int(prediction > 0.5))\n print('Ouput:\\t', int(_y))\n print('######')",
"Input:\t [0, 0]\nPred:\t 0\nOuput:\t 0\n######\nInput:\t [0, 1]\nPred:\t 1\nOuput:\t 1\n######\nInput:\t [1, 0]\nPred:\t 1\nOuput:\t 1\n######\nInput:\t [1, 1]\nPred:\t 0\nOuput:\t 0\n######\n"
]
],
[
[
"MNIST: The \"Hello World\" of Neural Nets\n====\n\nLike any deep learning class, we ***must*** do the MNIST. \n\nThe MNIST dataset is \n\n - is made up of handwritten digits \n - 60,000 examples training set\n - 10,000 examples test set",
"_____no_output_____"
]
],
[
[
"# We're going to install tensorflow here because their dataset access is simpler =)\n!pip3 install torchvision",
"Requirement already satisfied: torchvision in /Library/Frameworks/Python.framework/Versions/3.6/lib/python3.6/site-packages (0.2.2.post3)\r\nRequirement already satisfied: six in /Users/liling.tan/Library/Python/3.6/lib/python/site-packages (from torchvision) (1.13.0)\r\nRequirement already satisfied: torch in /Library/Frameworks/Python.framework/Versions/3.6/lib/python3.6/site-packages (from torchvision) (1.3.0)\r\nRequirement already satisfied: pillow>=4.1.1 in /Library/Frameworks/Python.framework/Versions/3.6/lib/python3.6/site-packages (from torchvision) (5.2.0)\r\nRequirement already satisfied: numpy in /Users/liling.tan/Library/Python/3.6/lib/python/site-packages (from torchvision) (1.18.1)\r\n"
],
[
"from torchvision import datasets, transforms",
"_____no_output_____"
],
[
"mnist_train = datasets.MNIST('../data', train=True, download=True, \n transform=transforms.Compose([\n transforms.ToTensor(),\n transforms.Normalize((0.1307,), (0.3081,))\n ]))\n\nmnist_test = datasets.MNIST('../data', train=False, download=True, \n transform=transforms.Compose([\n transforms.ToTensor(),\n transforms.Normalize((0.1307,), (0.3081,))\n ]))",
"_____no_output_____"
],
[
"# Visualization Candies\nimport matplotlib.pyplot as plt\n\ndef show_image(mnist_x_vector, mnist_y_vector):\n pixels = mnist_x_vector.reshape((28, 28))\n label = np.where(mnist_y_vector == 1)[0]\n plt.title('Label is {}'.format(label))\n plt.imshow(pixels, cmap='gray')\n plt.show()",
"_____no_output_____"
],
[
"# Fifth image and label.\nshow_image(mnist_train.data[5], mnist_train.targets[5])",
"_____no_output_____"
]
],
[
[
"# Lets apply what we learn about multi-layered perceptron with PyTorch and apply it to the MNIST data. ",
"_____no_output_____"
]
],
[
[
"X_mnist = mnist_train.data.float()\nY_mnist = mnist_train.targets.float()\n\nX_mnist_test = mnist_test.data.float()\nY_mnist_test = mnist_test.targets.float()",
"_____no_output_____"
],
[
"Y_mnist.shape",
"_____no_output_____"
],
[
"# Use FloatTensor.shape to get the shape of the matrix/tensor.\nnum_data, *input_dim = X_mnist.shape\nprint('No. of images:', num_data)\nprint('Inputs Dim:', input_dim)\n\nnum_data, *output_dim = Y_mnist.shape\nnum_test_data, *output_dim = Y_mnist_test.shape\nprint('Output Dim:', output_dim)\n",
"No. of images: 60000\nInputs Dim: [28, 28]\nOutput Dim: []\n"
],
[
"# Flatten the dimensions of the images.\nX_mnist = mnist_train.data.float().view(num_data, -1)\nY_mnist = mnist_train.targets.float().unsqueeze(1)\n\nX_mnist_test = mnist_test.data.float().view(num_test_data, -1)\nY_mnist_test = mnist_test.targets.float().unsqueeze(1)",
"_____no_output_____"
],
[
"# Use FloatTensor.shape to get the shape of the matrix/tensor.\nnum_data, *input_dim = X_mnist.shape\nprint('No. of images:', num_data)\nprint('Inputs Dim:', input_dim)\n\nnum_data, *output_dim = Y_mnist.shape\nnum_test_data, *output_dim = Y_mnist_test.shape\nprint('Output Dim:', output_dim)",
"No. of images: 60000\nInputs Dim: [784]\nOutput Dim: [1]\n"
],
[
"hidden_dim = 500\n\nmodel = nn.Sequential(nn.Linear(784, 1),\n nn.Sigmoid())\n\ncriterion = nn.MSELoss()\nlearning_rate = 1.0\noptimizer = optim.SGD(model.parameters(), lr=learning_rate)\nnum_epochs = 10\n\n\nlosses = []\nplt.ion()\n\nfor _e in tqdm(range(num_epochs)):\n optimizer.zero_grad()\n predictions = model(X_mnist)\n loss_this_epoch = criterion(predictions, Y_mnist)\n loss_this_epoch.backward()\n optimizer.step()\n ##print([float(_pred) for _pred in predictions], list(map(int, Y_pt)), loss_this_epoch.data[0])\n losses.append(loss_this_epoch.data.item())\n\n clear_output(wait=True)\n plt.plot(losses)\n plt.pause(0.05)\n ",
"_____no_output_____"
],
[
"predictions = model(X_mnist_test)\npredictions",
"_____no_output_____"
],
[
"pred = np.array([np.argmax(_p) for _p in predictions.data.numpy()])\npred",
"_____no_output_____"
],
[
"truth = np.array([np.argmax(_p) for _p in Y_mnist_test.data.numpy()])\ntruth",
"_____no_output_____"
],
[
"(pred == truth).sum() / len(pred)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e419a5d9b329b194ef3930f10377fcc93db357 | 353,145 | ipynb | Jupyter Notebook | Time Series/Missing-values_Multistep_Multivariate.ipynb | AlexKH22/Finance | f15a65dadf80b55684a6d17b8fef1d57f2332f3f | [
"Apache-2.0"
] | null | null | null | Time Series/Missing-values_Multistep_Multivariate.ipynb | AlexKH22/Finance | f15a65dadf80b55684a6d17b8fef1d57f2332f3f | [
"Apache-2.0"
] | null | null | null | Time Series/Missing-values_Multistep_Multivariate.ipynb | AlexKH22/Finance | f15a65dadf80b55684a6d17b8fef1d57f2332f3f | [
"Apache-2.0"
] | null | null | null | 188.74666 | 231,144 | 0.863039 | [
[
[
"import pandas as pd\nimport numpy as np\nfrom math import sqrt\nfrom numpy import concatenate\nfrom matplotlib import pyplot\nfrom pandas import read_csv\nfrom pandas import DataFrame\nfrom pandas import concat\nfrom sklearn.preprocessing import MinMaxScaler\nfrom sklearn.preprocessing import LabelEncoder\nfrom sklearn.metrics import mean_squared_error\nfrom keras.models import Sequential\nfrom keras.layers import Dense\nfrom keras.layers import LSTM",
"/Library/Frameworks/Python.framework/Versions/3.6/lib/python3.6/site-packages/h5py/__init__.py:36: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n from ._conv import register_converters as _register_converters\nUsing TensorFlow backend.\n"
]
],
[
[
"### Interpolation",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame(\n {\n 'name': ['A','A', 'B','B','B','B', 'C','C','C'],\n 'value': [1, np.nan, np.nan, 2, 3, 1, 3, np.nan, 3],\n }\n)",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df.fillna(method=\"pad\")",
"_____no_output_____"
],
[
"df['value'].interpolate()",
"_____no_output_____"
],
[
"df['value'].interpolate(method=\"quadratic\")",
"_____no_output_____"
],
[
"# For cumsums\ndf['value'].interpolate(method=\"pchip\")",
"_____no_output_____"
]
],
[
[
"### LSTM",
"_____no_output_____"
]
],
[
[
"from pandas import read_csv\nfrom datetime import datetime\n# load data\ndef parse(x):\n return datetime.strptime(x, '%Y %m %d %H')\ndataset = read_csv('data/raw_pollution.csv', parse_dates = [['year', 'month', 'day', 'hour']], index_col=0, date_parser=parse)\ndataset.drop('No', axis=1, inplace=True)\n# manually specify column names\ndataset.columns = ['pollution', 'dew', 'temp', 'press', 'wnd_dir', 'wnd_spd', 'snow', 'rain']\ndataset.index.name = 'date'",
"_____no_output_____"
],
[
"# mark all NA values with 0\ndataset['pollution'] = dataset['pollution'].interpolate()\n# drop the first 24 hours\ndataset = dataset[24:]\n# summarize first 5 rows\nprint(dataset.head(5))\n# save to file\ndataset.to_csv('data/pollution.csv')",
" pollution dew temp press wnd_dir wnd_spd snow rain\ndate \n2010-01-02 00:00:00 129.0 -16 -4.0 1020.0 SE 1.79 0 0\n2010-01-02 01:00:00 148.0 -15 -4.0 1020.0 SE 2.68 0 0\n2010-01-02 02:00:00 159.0 -11 -5.0 1021.0 SE 3.57 0 0\n2010-01-02 03:00:00 181.0 -7 -5.0 1022.0 SE 5.36 1 0\n2010-01-02 04:00:00 138.0 -7 -5.0 1022.0 SE 6.25 2 0\n"
],
[
"from pandas import read_csv\nfrom matplotlib import pyplot\n# load dataset\ndataset = read_csv('pollution.csv', header=0, index_col=0)\nvalues = dataset.values\n# specify columns to plot\ngroups = [0, 1, 2, 3, 5, 6, 7]\ni = 1\n# plot each column\npyplot.figure(figsize=(20, 13))\nfor group in groups:\n pyplot.subplot(len(groups), 1, i)\n pyplot.plot(values[:, group])\n pyplot.title(dataset.columns[group], y=0.5, loc='right')\n pyplot.grid()\n i += 1\npyplot.show()",
"_____no_output_____"
],
[
"# convert series to supervised learning\ndef series_to_supervised(data, n_in=1, n_out=1, dropnan=True):\n n_vars = 1 if type(data) is list else data.shape[1]\n df = DataFrame(data)\n cols, names = list(), list()\n # input sequence (t-n, ... t-1)\n for i in range(n_in, 0, -1):\n cols.append(df.shift(i))\n names += [('var%d(t-%d)' % (j+1, i)) for j in range(n_vars)]\n # forecast sequence (t, t+1, ... t+n)\n for i in range(0, n_out):\n cols.append(df.shift(-i))\n if i == 0:\n names += [('var%d(t)' % (j+1)) for j in range(n_vars)]\n else:\n names += [('var%d(t+%d)' % (j+1, i)) for j in range(n_vars)]\n # put it all together\n agg = concat(cols, axis=1)\n agg.columns = names\n # drop rows with NaN values\n if dropnan:\n agg.dropna(inplace=True)\n return agg\n \n# load dataset\ndataset = read_csv('data/pollution.csv', header=0, index_col=0)\nvalues = dataset.values",
"_____no_output_____"
],
[
"dataset",
"_____no_output_____"
],
[
"values",
"_____no_output_____"
],
[
"# integer encode wind_direction\nencoder = LabelEncoder()\nvalues[:,4] = encoder.fit_transform(values[:,4])\n# ensure all data is float\nvalues = values.astype('float32')\n# normalize features\nscaler = MinMaxScaler(feature_range=(0, 1))\nscaled = scaler.fit_transform(values)\n# frame as supervised learning\nreframed = series_to_supervised(scaled, 1, 1)\n# drop columns we don't want to predict\nreframed.drop(reframed.columns[[9,10,11,12,13,14,15]], axis=1, inplace=True)\nprint(reframed.head())",
" var1(t-1) var2(t-1) var3(t-1) var4(t-1) var5(t-1) var6(t-1) \\\n1 0.129779 0.352941 0.245902 0.527273 0.666667 0.002290 \n2 0.148893 0.367647 0.245902 0.527273 0.666667 0.003811 \n3 0.159960 0.426471 0.229508 0.545454 0.666667 0.005332 \n4 0.182093 0.485294 0.229508 0.563637 0.666667 0.008391 \n5 0.138833 0.485294 0.229508 0.563637 0.666667 0.009912 \n\n var7(t-1) var8(t-1) var1(t) \n1 0.000000 0.0 0.148893 \n2 0.000000 0.0 0.159960 \n3 0.000000 0.0 0.182093 \n4 0.037037 0.0 0.138833 \n5 0.074074 0.0 0.109658 \n"
],
[
"# split into train and test sets\nvalues = reframed.values\nn_train_hours = 365 * 24\ntrain = values[:n_train_hours, :]\ntest = values[n_train_hours:, :]\n# split into input and outputs\ntrain_X, train_y = train[:, :-1], train[:, -1]\ntest_X, test_y = test[:, :-1], test[:, -1]\n# reshape input to be 3D [samples, timesteps, features]\ntrain_X = train_X.reshape((train_X.shape[0], 1, train_X.shape[1]))\ntest_X = test_X.reshape((test_X.shape[0], 1, test_X.shape[1]))\nprint(train_X.shape, train_y.shape, test_X.shape, test_y.shape)",
"(8760, 1, 8) (8760,) (35039, 1, 8) (35039,)\n"
]
],
[
[
"We will define the LSTM with 50 neurons in the first hidden layer and 1 neuron in the output layer for predicting pollution. The input shape will be 1 time step with 8 features.\n\nWe will use the Mean Absolute Error (MAE) loss function and the efficient Adam version of stochastic gradient descent.",
"_____no_output_____"
]
],
[
[
"# design network\nmodel = Sequential()\nmodel.add(LSTM(50, input_shape=(train_X.shape[1], train_X.shape[2])))\nmodel.add(Dense(1))\nmodel.compile(loss='mae', optimizer='adam')\n# fit network\nhistory = model.fit(train_X, train_y, epochs=50, batch_size=64, \n validation_data=(test_X, test_y), verbose=2, shuffle=False)\n# plot history\npyplot.plot(history.history['loss'], label='train')\npyplot.plot(history.history['val_loss'], label='test')\npyplot.legend()\npyplot.show()",
"Train on 8760 samples, validate on 35039 samples\nEpoch 1/50\n8760/8760 [==============================] - 3s 337us/step - loss: 0.0548 - val_loss: 0.0534\nEpoch 2/50\n8760/8760 [==============================] - 1s 170us/step - loss: 0.0332 - val_loss: 0.0570\nEpoch 3/50\n8760/8760 [==============================] - 2s 234us/step - loss: 0.0193 - val_loss: 0.0377\nEpoch 4/50\n8760/8760 [==============================] - 1s 161us/step - loss: 0.0151 - val_loss: 0.0237\nEpoch 5/50\n8760/8760 [==============================] - 1s 141us/step - loss: 0.0140 - val_loss: 0.0173\nEpoch 6/50\n8760/8760 [==============================] - 1s 121us/step - loss: 0.0139 - val_loss: 0.0157\nEpoch 7/50\n8760/8760 [==============================] - 1s 104us/step - loss: 0.0137 - val_loss: 0.0151\nEpoch 8/50\n8760/8760 [==============================] - 1s 110us/step - loss: 0.0138 - val_loss: 0.0150\nEpoch 9/50\n8760/8760 [==============================] - 1s 149us/step - loss: 0.0138 - val_loss: 0.0150\nEpoch 10/50\n8760/8760 [==============================] - 1s 148us/step - loss: 0.0137 - val_loss: 0.0147\nEpoch 11/50\n8760/8760 [==============================] - 1s 104us/step - loss: 0.0137 - val_loss: 0.0152\nEpoch 12/50\n8760/8760 [==============================] - 1s 141us/step - loss: 0.0138 - val_loss: 0.0152\nEpoch 13/50\n8760/8760 [==============================] - 1s 131us/step - loss: 0.0138 - val_loss: 0.0143\nEpoch 14/50\n8760/8760 [==============================] - 1s 120us/step - loss: 0.0137 - val_loss: 0.0144\nEpoch 15/50\n8760/8760 [==============================] - 1s 124us/step - loss: 0.0137 - val_loss: 0.0143\nEpoch 16/50\n8760/8760 [==============================] - 1s 108us/step - loss: 0.0138 - val_loss: 0.0145\nEpoch 17/50\n8760/8760 [==============================] - 1s 110us/step - loss: 0.0137 - val_loss: 0.0134\nEpoch 18/50\n8760/8760 [==============================] - 1s 137us/step - loss: 0.0138 - val_loss: 0.0133\nEpoch 19/50\n8760/8760 [==============================] - 1s 103us/step - loss: 0.0138 - val_loss: 0.0135\nEpoch 20/50\n8760/8760 [==============================] - 1s 104us/step - loss: 0.0138 - val_loss: 0.0135\nEpoch 21/50\n8760/8760 [==============================] - 1s 137us/step - loss: 0.0137 - val_loss: 0.0138\nEpoch 22/50\n8760/8760 [==============================] - 1s 104us/step - loss: 0.0138 - val_loss: 0.0134\nEpoch 23/50\n8760/8760 [==============================] - 1s 105us/step - loss: 0.0137 - val_loss: 0.0135\nEpoch 24/50\n8760/8760 [==============================] - 1s 105us/step - loss: 0.0136 - val_loss: 0.0137\nEpoch 25/50\n8760/8760 [==============================] - 1s 105us/step - loss: 0.0137 - val_loss: 0.0140\nEpoch 26/50\n8760/8760 [==============================] - 1s 105us/step - loss: 0.0137 - val_loss: 0.0141\nEpoch 27/50\n8760/8760 [==============================] - 1s 107us/step - loss: 0.0137 - val_loss: 0.0138\nEpoch 28/50\n8760/8760 [==============================] - 1s 106us/step - loss: 0.0136 - val_loss: 0.0137\nEpoch 29/50\n8760/8760 [==============================] - 1s 104us/step - loss: 0.0136 - val_loss: 0.0135\nEpoch 30/50\n8760/8760 [==============================] - 1s 114us/step - loss: 0.0135 - val_loss: 0.0131\nEpoch 31/50\n8760/8760 [==============================] - 1s 105us/step - loss: 0.0137 - val_loss: 0.0134\nEpoch 32/50\n8760/8760 [==============================] - 1s 105us/step - loss: 0.0135 - val_loss: 0.0132\nEpoch 33/50\n8760/8760 [==============================] - 1s 104us/step - loss: 0.0137 - val_loss: 0.0136\nEpoch 34/50\n8760/8760 [==============================] - 1s 113us/step - loss: 0.0136 - val_loss: 0.0133\nEpoch 35/50\n8760/8760 [==============================] - 1s 107us/step - loss: 0.0137 - val_loss: 0.0131\nEpoch 36/50\n8760/8760 [==============================] - 1s 111us/step - loss: 0.0136 - val_loss: 0.0137\nEpoch 37/50\n8760/8760 [==============================] - 1s 105us/step - loss: 0.0136 - val_loss: 0.0132\nEpoch 38/50\n8760/8760 [==============================] - 1s 106us/step - loss: 0.0136 - val_loss: 0.0132\nEpoch 39/50\n8760/8760 [==============================] - 1s 105us/step - loss: 0.0138 - val_loss: 0.0130\nEpoch 40/50\n8760/8760 [==============================] - 1s 107us/step - loss: 0.0137 - val_loss: 0.0132\nEpoch 41/50\n8760/8760 [==============================] - 1s 106us/step - loss: 0.0137 - val_loss: 0.0130\nEpoch 42/50\n8760/8760 [==============================] - 1s 103us/step - loss: 0.0136 - val_loss: 0.0130\nEpoch 43/50\n8760/8760 [==============================] - 1s 104us/step - loss: 0.0138 - val_loss: 0.0129\nEpoch 44/50\n8760/8760 [==============================] - 1s 107us/step - loss: 0.0137 - val_loss: 0.0129\nEpoch 45/50\n8760/8760 [==============================] - 1s 104us/step - loss: 0.0136 - val_loss: 0.0129\nEpoch 46/50\n8760/8760 [==============================] - 1s 124us/step - loss: 0.0136 - val_loss: 0.0130\nEpoch 47/50\n8760/8760 [==============================] - 1s 109us/step - loss: 0.0136 - val_loss: 0.0129\nEpoch 48/50\n8760/8760 [==============================] - 1s 123us/step - loss: 0.0138 - val_loss: 0.0130\nEpoch 49/50\n8760/8760 [==============================] - 1s 102us/step - loss: 0.0137 - val_loss: 0.0130\nEpoch 50/50\n8760/8760 [==============================] - 1s 103us/step - loss: 0.0136 - val_loss: 0.0129\n"
],
[
"# make a prediction\nyhat = model.predict(test_X)\ntest_X = test_X.reshape((test_X.shape[0], test_X.shape[2]))\n# invert scaling for forecast\ninv_yhat = concatenate((yhat, test_X[:, 1:]), axis=1)\ninv_yhat = scaler.inverse_transform(inv_yhat)\ninv_yhat = inv_yhat[:,0]\n# invert scaling for actual\ntest_y = test_y.reshape((len(test_y), 1))\ninv_y = concatenate((test_y, test_X[:, 1:]), axis=1)\ninv_y = scaler.inverse_transform(inv_y)\ninv_y = inv_y[:, 0]\n# calculate RMSE\nrmse = sqrt(mean_squared_error(inv_y, inv_yhat))\nprint('Test RMSE: %.3f' % rmse)",
"Test RMSE: 23.355\n"
],
[
"pyplot.plot(range(len(inv_yhat)), inv_yhat, alpha=0.5, label=\"Predicted\")\npyplot.show()\npyplot.plot(range(len(inv_y)), inv_y, alpha=0.5, label=\"Expected\")\npyplot.show()",
"_____no_output_____"
]
],
[
[
"### N previous lags",
"_____no_output_____"
]
],
[
[
"# load dataset\ndataset = read_csv('data/pollution.csv', header=0, index_col=0)\nvalues = dataset.values\n# integer encode direction\nencoder = LabelEncoder()\nvalues[:,4] = encoder.fit_transform(values[:,4])\n# ensure all data is float\nvalues = values.astype('float32')\n# normalize features\nscaler = MinMaxScaler(feature_range=(0, 1))\nscaled = scaler.fit_transform(values)\n# specify the number of lag hours\nn_hours = 3\nn_features = 8\n# frame as supervised learning\nreframed = series_to_supervised(scaled, n_hours, 1)\nprint(reframed.shape)",
"(43797, 32)\n"
]
],
[
[
"Next, we need to be more careful in specifying the column for input and output.\n\nWe have 3 * 8 + 8 columns in our framed dataset. We will take 3 * 8 or 24 columns as input for the obs of all features across the previous 3 hours. We will take just the pollution variable as output at the following hour, as follows:",
"_____no_output_____"
]
],
[
[
" # split into train and test sets\nvalues = reframed.values\nn_train_hours = 365 * 24\ntrain = values[:n_train_hours]\ntest = values[n_train_hours:]\n# split into input and outputs\nn_obs = n_hours * n_features\ntrain_X, train_y = train[:, :n_obs], train[:, -n_features]\ntest_X, test_y = test[:, :n_obs], test[:, -n_features]\nprint(train_X.shape, len(train_X), train_y.shape)",
"(8760, 24) 8760 (8760,)\n(8760, 3, 8) (8760,) (35037, 3, 8) (35037,)\n"
],
[
"# reshape input to be 3D [samples, timesteps, features]\ntrain_X = train_X.reshape((train_X.shape[0], n_hours, n_features))\ntest_X = test_X.reshape((test_X.shape[0], n_hours, n_features))\nprint(train_X.shape, train_y.shape, test_X.shape, test_y.shape)",
"_____no_output_____"
],
[
"# design network\nmodel = Sequential()\nmodel.add(LSTM(50, input_shape=(train_X.shape[1], train_X.shape[2])))\nmodel.add(Dense(1))\nmodel.compile(loss='mae', optimizer='adam')\n# fit network\nhistory = model.fit(train_X, train_y, epochs=50, \n batch_size=64, validation_data=(test_X, test_y), \n verbose=2, shuffle=False)\n# plot history\npyplot.plot(history.history['loss'], label='train')\npyplot.plot(history.history['val_loss'], label='test')\npyplot.legend()\npyplot.show()",
"Train on 8760 samples, validate on 35037 samples\nEpoch 1/50\n - 3s - loss: 0.0436 - val_loss: 0.0702\nEpoch 2/50\n - 2s - loss: 0.0261 - val_loss: 0.0502\nEpoch 3/50\n - 2s - loss: 0.0215 - val_loss: 0.0291\nEpoch 4/50\n - 2s - loss: 0.0201 - val_loss: 0.0205\nEpoch 5/50\n - 2s - loss: 0.0195 - val_loss: 0.0188\nEpoch 6/50\n - 2s - loss: 0.0185 - val_loss: 0.0175\nEpoch 7/50\n - 2s - loss: 0.0179 - val_loss: 0.0164\nEpoch 8/50\n - 2s - loss: 0.0176 - val_loss: 0.0161\nEpoch 9/50\n - 1s - loss: 0.0171 - val_loss: 0.0155\nEpoch 10/50\n - 1s - loss: 0.0167 - val_loss: 0.0152\nEpoch 11/50\n - 2s - loss: 0.0161 - val_loss: 0.0149\nEpoch 12/50\n - 1s - loss: 0.0159 - val_loss: 0.0141\nEpoch 13/50\n - 1s - loss: 0.0153 - val_loss: 0.0138\nEpoch 14/50\n - 1s - loss: 0.0150 - val_loss: 0.0133\nEpoch 15/50\n - 1s - loss: 0.0149 - val_loss: 0.0138\nEpoch 16/50\n - 1s - loss: 0.0147 - val_loss: 0.0137\nEpoch 17/50\n - 2s - loss: 0.0143 - val_loss: 0.0138\nEpoch 18/50\n - 2s - loss: 0.0140 - val_loss: 0.0129\nEpoch 19/50\n - 1s - loss: 0.0138 - val_loss: 0.0130\nEpoch 20/50\n - 2s - loss: 0.0139 - val_loss: 0.0127\nEpoch 21/50\n - 2s - loss: 0.0139 - val_loss: 0.0132\nEpoch 22/50\n - 1s - loss: 0.0137 - val_loss: 0.0127\nEpoch 23/50\n - 1s - loss: 0.0138 - val_loss: 0.0132\nEpoch 24/50\n - 1s - loss: 0.0137 - val_loss: 0.0127\nEpoch 25/50\n - 1s - loss: 0.0135 - val_loss: 0.0126\nEpoch 26/50\n - 1s - loss: 0.0136 - val_loss: 0.0130\nEpoch 27/50\n - 2s - loss: 0.0136 - val_loss: 0.0128\nEpoch 28/50\n - 2s - loss: 0.0134 - val_loss: 0.0126\nEpoch 29/50\n - 2s - loss: 0.0137 - val_loss: 0.0132\nEpoch 30/50\n - 2s - loss: 0.0134 - val_loss: 0.0126\nEpoch 31/50\n - 1s - loss: 0.0133 - val_loss: 0.0129\nEpoch 32/50\n - 1s - loss: 0.0133 - val_loss: 0.0129\nEpoch 33/50\n - 1s - loss: 0.0133 - val_loss: 0.0127\nEpoch 34/50\n - 1s - loss: 0.0133 - val_loss: 0.0128\nEpoch 35/50\n - 1s - loss: 0.0134 - val_loss: 0.0126\nEpoch 36/50\n - 1s - loss: 0.0134 - val_loss: 0.0127\nEpoch 37/50\n - 1s - loss: 0.0133 - val_loss: 0.0127\nEpoch 38/50\n - 2s - loss: 0.0134 - val_loss: 0.0127\nEpoch 39/50\n - 2s - loss: 0.0134 - val_loss: 0.0126\nEpoch 40/50\n - 2s - loss: 0.0133 - val_loss: 0.0126\nEpoch 41/50\n - 1s - loss: 0.0133 - val_loss: 0.0126\nEpoch 42/50\n - 1s - loss: 0.0134 - val_loss: 0.0125\nEpoch 43/50\n - 1s - loss: 0.0133 - val_loss: 0.0126\nEpoch 44/50\n - 1s - loss: 0.0133 - val_loss: 0.0125\nEpoch 45/50\n - 1s - loss: 0.0134 - val_loss: 0.0128\nEpoch 46/50\n - 1s - loss: 0.0133 - val_loss: 0.0127\nEpoch 47/50\n - 1s - loss: 0.0135 - val_loss: 0.0127\nEpoch 48/50\n - 1s - loss: 0.0134 - val_loss: 0.0126\nEpoch 49/50\n - 1s - loss: 0.0134 - val_loss: 0.0125\nEpoch 50/50\n - 1s - loss: 0.0134 - val_loss: 0.0127\n"
],
[
"# make a prediction\nyhat = model.predict(test_X)\ntest_X = test_X.reshape((test_X.shape[0], n_hours*n_features))\n# invert scaling for forecast\ninv_yhat = concatenate((yhat, test_X[:, -7:]), axis=1)\ninv_yhat = scaler.inverse_transform(inv_yhat)\ninv_yhat = inv_yhat[:,0]\n# invert scaling for actual\ntest_y = test_y.reshape((len(test_y), 1))\ninv_y = concatenate((test_y, test_X[:, -7:]), axis=1)\ninv_y = scaler.inverse_transform(inv_y)\ninv_y = inv_y[:,0]\n# calculate RMSE\nrmse = sqrt(mean_squared_error(inv_y, inv_yhat))\nprint('Test RMSE: %.3f' % rmse)",
"Test RMSE: 22.785\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d0e42e3f9d5949d1b7978b6748d6ccb80cd77c2a | 24,486 | ipynb | Jupyter Notebook | notebooks/generalPredictions4b.ipynb | UBC-MOAD/analysis-armaan | b270eaec72bd150db71e620c04dba4e58aa3f981 | [
"Apache-2.0"
] | null | null | null | notebooks/generalPredictions4b.ipynb | UBC-MOAD/analysis-armaan | b270eaec72bd150db71e620c04dba4e58aa3f981 | [
"Apache-2.0"
] | null | null | null | notebooks/generalPredictions4b.ipynb | UBC-MOAD/analysis-armaan | b270eaec72bd150db71e620c04dba4e58aa3f981 | [
"Apache-2.0"
] | null | null | null | 32.779116 | 213 | 0.529405 | [
[
[
"import matplotlib.pyplot as plt\nimport pandas as pd\nimport numpy as np\nimport xarray as xr\nimport scipy\nimport os\nfrom sklearn.decomposition import PCA\nfrom sklearn.linear_model import LinearRegression\nfrom sklearn.linear_model import QuantileRegressor\nfrom sklearn.metrics import mean_absolute_error\nfrom sklearn.metrics import mean_squared_error\nimport glob",
"_____no_output_____"
]
],
[
[
"Notebook for creating complete HRDPS file for 2007 from 2007 data, for every variable, all at once. For a more polished notebook, see generalPredictions4c. This notebook has been superceded by that notebook.",
"_____no_output_____"
],
[
"## Importing Training Data",
"_____no_output_____"
],
[
"| Description | HRDPS | CANRCM | \n| ----------- | ----------- | ----------- |\n| Near-Surface Air Temperature | tair | tas |\n| Precipitation | precip | pr |\n| Sea Level Pressure | atmpres | psl |\n| Near Surface Specific Humidity | qair | huss |\n| Shortwave radiation | solar | rsds |\n| Longwave radiation | therm_rad | rlds |",
"_____no_output_____"
]
],
[
[
"variables = [['tair', 'tas', 'Near-Surface Air Temperature'], \n ['precip', 'pr', 'Precipitation'], \n ['atmpres', 'psl', 'Sea Level Pressure'], \n ['qair', 'huss', 'Near Surface Specific Humidity'], \n ['solar', 'rsds', 'Shortwave radiation'], \n ['therm_rad', 'rlds', 'Longwave radiation']]",
"_____no_output_____"
],
[
"def import_data(variable):\n global name\n name = variable[2]\n global data_name_hr\n data_name_hr = variable[0]\n global data_name_can\n data_name_can = variable[1]\n \n ##2007 HRDPS import\n\n files = glob.glob('/results/forcing/atmospheric/GEM2.5/gemlam/gemlam_y2007m??d??.nc')\n files.sort()\n\n ## 3-hour averaged matrix\n global hr07\n hr07 = np.zeros( (8*len(files), 266, 256)) \n\n for i in range(len(files)):\n dayX = xr.open_dataset(files[i])\n ##adding 1 day of 3-hour averages to new data array\n hr07[8*i:8*i + 8,:,:] = np.array( dayX[ data_name_hr ] ).reshape(8, 3, 266, 256).mean(axis = 1) \n\n p_can07 = '/home/arandhawa/canrcm_' + data_name_can + '_2007.nc'\n ##CANRCM 2007 import\n global can07\n d1 = xr.open_dataset(p_can07)\n can07 = d1[data_name_can][16:,140:165,60:85] ##the first two days are removed to be consistent with 2007 HRDPS\n ",
"_____no_output_____"
],
[
"def import_wind_data():\n \n ##2007 HRDPS import\n\n files = glob.glob('/results/forcing/atmospheric/GEM2.5/gemlam/gemlam_y2007m??d??.nc')\n files.sort()\n\n ## 3-hour averaged matrix\n global hr07_u\n global hr07_v\n\n ## 3-hour averaged matrix\n\n hr07_u = np.zeros( (8*len(files), 266, 256)) \n hr07_v = np.zeros( (8*len(files), 266, 256)) \n\n for i in range(len(files)):\n dayX = xr.open_dataset(files[i])\n u = np.array( dayX['u_wind'] ).reshape(8, 3, 266, 256)\n v = np.array( dayX['v_wind'] ).reshape(8, 3, 266, 256)\n avg_spd = np.mean(np.sqrt(u**2 + v**2), axis = 1)\n avg_th = np.arctan2(v.mean(axis = 1), u.mean(axis = 1))\n avg_u = avg_spd*np.cos(avg_th)\n avg_v = avg_spd*np.sin(avg_th)\n hr07_u[8*i:8*i + 8, : , : ] = avg_u ##adding 3-hour average to new data array\n hr07_v[8*i:8*i + 8, : , : ] = avg_v\n \n del avg_u\n del avg_v\n del dayX\n del u\n del v\n del avg_spd\n del avg_th\n \n p_can07u = '/home/arandhawa/canrcm_uas_2007.nc'\n p_can07v = '/home/arandhawa/canrcm_vas_2007.nc'\n ##CANRCM 2007 import\n global can07_u\n global can07_v\n d1 = xr.open_dataset(p_can07u)\n can07_u = d1['uas'][16:,140:165,60:85] ##the first two days are removed to be consistent with 2007 HRDPS\n d2 = xr.open_dataset(p_can07v)\n can07_v = d2['vas'][16:,140:165,60:85] ##the first two days are removed to be consistent with 2007 HRDPS\n ",
"_____no_output_____"
]
],
[
[
"## PCA Functions",
"_____no_output_____"
]
],
[
[
"##transforms and concatenates two data sets\ndef transform2(data1, data2):\n A_mat = transform(data1)\n B_mat = transform(data2)\n return np.concatenate((A_mat, B_mat), axis=0) ",
"_____no_output_____"
],
[
"##inverse function of transform2 - splits data matrix and returns two data sets\ndef reverse2(matrix, orig_shape):\n split4 = int( matrix.shape[0]/2 )\n u_data = reverse(matrix[:split4,:], orig_shape) ##reconstructing u_winds from n PCs\n v_data = reverse(matrix[split4:,:], orig_shape) ##reconstructing v_winds from n PCs\n return (u_data, v_data)",
"_____no_output_____"
],
[
"##performs PCA analysis using sklearn.pca\ndef doPCA(comp, matrix):\n pca = PCA(n_components = comp) ##adjust the number of principle conponents to be calculated\n PCs = pca.fit_transform(matrix)\n eigvecs = pca.components_\n mean = pca.mean_\n return (PCs, eigvecs, mean)",
"_____no_output_____"
],
[
"##data must be converted into a 2D matrix for pca analysis\n##transform takes a 3D data array (time, a, b) -> (a*b, time)\n##(the data grid is flattened a column using numpy.flatten)\n\ndef transform(xarr):\n arr = np.array(xarr) ##converting to numpy array\n arr = arr.reshape(arr.shape[0], arr.shape[1]*arr.shape[2]) ##reshaping from size (a, b, c) to (a, b*c)\n arr = arr.transpose()\n return arr\n\ndef reverse(mat, orig_shape):\n arr = np.copy(mat)\n arr = arr.transpose()\n arr = arr.reshape(-1, orig_shape[1], orig_shape[2]) ##reshaping back to original array shape\n return arr",
"_____no_output_____"
],
[
"##graphing percentage of original data represented by the first n principle conponents\ndef graph_variance(matrix, n):\n pcaG = PCA(n_components = n) ##Number of principle conponents to show\n PCsG = pcaG.fit_transform(matrix)\n plt.plot(np.cumsum(pcaG.explained_variance_ratio_))\n plt.xlabel('number of components')\n plt.ylabel('cumulative explained variance');\n plt.show()\n del pcaG\n del PCsG",
"_____no_output_____"
],
[
"##can be used to visualize principle conponents for u/v winds\ndef graph_nPCs(PCs, eigvecs, n, orig_shape):\n fig, ax = plt.subplots(n, 3, figsize=(10, 3*n))\n \n ax[0, 0].set_title(\"u-conponent\")\n ax[0, 1].set_title(\"v-component\")\n ax[0, 2].set_title(\"time-loadings\")\n \n for i in range(n):\n mode_u, mode_v = get_mode(PCs, i, orig_shape)\n colors = ax[i, 0].pcolormesh(mode_u, cmap = 'bwr')\n fig.colorbar(colors, ax = ax[i,0])\n colors = ax[i, 1].pcolormesh(mode_v, cmap = 'bwr')\n fig.colorbar(colors, ax = ax[i,1])\n ax[i, 2].plot(eigvecs[i])\n\n plt.tight_layout()\n plt.show()\n\n##converts PCs (column vectors) to 2d conpoents for u and v wind\ndef get_mode(PCs, n, orig_shape): \n split = int(PCs.shape[0]/2)\n mode_u = PCs[:split, n].reshape(orig_shape[1], orig_shape[2])\n mode_v = PCs[split:, n].reshape(orig_shape[1], orig_shape[2])\n return (mode_u, mode_v)\n\n",
"_____no_output_____"
]
],
[
[
"## Multiple Linear Regression Functions",
"_____no_output_____"
]
],
[
[
"##functions that use multiple linear regression to fit eigenvectors\n##takes CANRCM eigenvectors (x1, x2, x3, x4...) and HRDPS eigenvectors (y1, y2, y3...)\n##For each y from 0:result_size, approximates yn = a0 + a1*x1 + a2*x2 + a3*x3 ... using num_vec x's\n##getCoefs returns (coeficients, intercept)\n##fit_modes returns each approximation and the R^2 value of each fit as (results, scores)\n\ndef getCoefs(vectors, num_vec, data, num_modes, type = 'LS'): \n \n X = vectors[0:num_vec,:].T\n coefs = np.zeros((num_modes, X.shape[1]))\n intercept = np.zeros(num_modes)\n \n if type == 'LS':\n for i in range(num_modes):\n y = data[i,:]\n reg = LinearRegression().fit(X, y)\n coefs[i] = reg.coef_[0:num_vec]\n intercept[i] = reg.intercept_\n elif type == 'MAE':\n for i in range(num_modes):\n y = data[i,:]\n reg = QuantileRegressor(quantile = 0.5, alpha = 0, solver = 'highs').fit(X, y)\n coefs[i] = reg.coef_[0:num_vec]\n intercept[i] = reg.intercept_\n \n return (coefs, intercept)\n\n\ndef fit_modes(vectors, num_vec, data, result_size, type = 'LS'): \n \n X = vectors[0:num_vec,:].T\n result = np.zeros((result_size, X.shape[0]))\n scores = np.zeros(result_size)\n \n if type == 'LS':\n for i in range(result_size):\n y = data[i,:]\n reg = LinearRegression().fit(X, y)\n result[i] = reg.predict(X)\n scores[i] = reg.score(X, y)\n \n elif type == 'MAE':\n for i in range(result_size):\n y = data[i,:]\n reg = QuantileRegressor(quantile = 0.5, alpha = 0, solver = 'highs').fit(X, y)\n result[i] = reg.predict(X)\n scores[i] = reg.score(X, y)\n \n return (result, scores)\n",
"_____no_output_____"
],
[
"##returns the ratio of the average energy between two sets of eigenvectors (element-wise)\n##\"energy\" is defined as value^2 - two sets of eigenvectors with the same \"energy\" would\n##recreate data with approximately the same kinetic energy (v^2)\n\ndef getEnergyCoefs(eigs, old_eigs):\n coefs = np.sqrt( (old_eigs[0:eigs.shape[0]]**2).mean(axis = 1)/(eigs**2).mean(axis = 1))\n return coefs",
"_____no_output_____"
]
],
[
[
"## Projection Function",
"_____no_output_____"
]
],
[
[
"##scalar projection of u onto v - with extra 1/norm factor (for math reasons)\n##projectData projects the data onto each principle conponent, at each time\n##output is a set of eigenvectors\n\ndef project(u, v): \n v_norm = np.sqrt(np.sum(v**2)) \n return np.dot(u, v)/v_norm**2\n\ndef projectData(data_mat, new_PCs, n):\n time = data_mat.shape[1]\n proj = np.empty((n, time))\n\n for j in range(n):\n for i in range(time):\n proj[j, i] = project(data_mat[:,i], new_PCs[:,j])\n \n return proj",
"_____no_output_____"
]
],
[
[
"## Overall Function",
"_____no_output_____"
]
],
[
[
"def reconstruct(downscale_mat, mean, can_PCs, can_me, hr_PCs, hr_me, n, r, method = 'LS', EB = 'False'):\n\n coefs = getCoefs(can_me, n + 1, hr_me, r + 1, type = method)\n proj = np.concatenate((mean.reshape(1, -1), projectData(downscale_mat - mean, can_PCs, n)), axis = 0)\n pred_eigs = np.matmul(coefs[0], proj) + coefs[1].reshape(-1, 1) ##multiple linear regression output\n if (EB == 'true'):\n energyCoefs = getEnergyCoefs( fit_modes(can_me, n + 1, hr_me, r + 1, type = method)[0], hr_me)\n energyCoefs = energyCoefs.reshape(-1, 1)\n pred_eigs = pred_eigs*energyCoefs ##energy balancing\n if (EB == 'function'):\n energyCoefs = getEnergyCoefs( fit_modes(can_me, n + 1, hr_me, r + 1, type = method)[0] , hr_me)\n def f(x):\n return np.exp(-x/50)\n for x in range(r + 1):\n energyCoefs = (energyCoefs - 1)*f(x) + 1\n energyCoefs = energyCoefs.reshape(-1, 1)\n pred_eigs = pred_eigs*energyCoefs ##energy balancing\n \n recon = np.matmul(hr_PCs[:,0:r], pred_eigs[1:r+1]) + pred_eigs[0]\n data_rec = reverse(recon, (-1, 266, 256))\n if (EB == 'constant'):\n data_rec *= 1.3\n \n return data_rec",
"_____no_output_____"
],
[
"def reconstruct2(downscale_mat, mean, can_PCs, can_me, hr_PCs, hr_me, n, r, method = 'LS', EB = 'f2alse'):\n\n coefs = getCoefs(can_me, n + 1, hr_me, r + 1, type = method)\n proj = np.concatenate((mean.reshape(1, -1), projectData(downscale_mat - mean, can_PCs, n)), axis = 0)\n pred_eigs = np.matmul(coefs[0], proj) + coefs[1].reshape(-1, 1) ##multiple linear regression output\n if (EB == 'true'):\n energyCoefs = getEnergyCoefs( fit_modes(can_me, n + 1, hr_me, r + 1, type = method)[0], hr_me)\n energyCoefs = energyCoefs.reshape(-1, 1)\n pred_eigs = pred_eigs*energyCoefs ##energy balancing\n if (EB == 'function'):\n energyCoefs = getEnergyCoefs( fit_modes(can_me, n + 1, hr_me, r + 1, type = method)[0] , hr_me)\n def f(x):\n return np.exp(-x/50)\n for x in range(r + 1):\n energyCoefs = (energyCoefs - 1)*f(x) + 1\n energyCoefs = energyCoefs.reshape(-1, 1)\n pred_eigs = pred_eigs*energyCoefs ##energy balancing\n \n recon = np.matmul(hr_PCs[:,0:r], pred_eigs[1:r+1]) + pred_eigs[0]\n u_data_rec, v_data_rec = reverse2(recon, (-1, 266, 256))\n if (EB == 'constant'):\n u_data_rec *= 1.3\n v_data_rec *= 1.3\n \n return (u_data_rec, v_data_rec)",
"_____no_output_____"
]
],
[
[
"## Reconstructing Data",
"_____no_output_____"
]
],
[
[
"data = ()\n\n##reconstructing u and v winds\nimport_wind_data()\n\n##PCA on CANRCM 2007\ncan07_mat = transform2(can07_u, can07_v)\ncan07_PCs, can07_eigs, can07_mean = doPCA(100, can07_mat)\n\n##PCA on HRDPS 2007\nhr07_mat = transform2(hr07_u, hr07_v)\nhr07_PCs, hr07_eigs, hr07_mean = doPCA(100, hr07_mat)\n\n## combining the eigenvectors and mean together in one array for analysis\ncan07_me = np.concatenate((can07_mean.reshape(1, -1), can07_eigs))\nhr07_me = np.concatenate((hr07_mean.reshape(1, -1), hr07_eigs))\n\n##calculating average of rows\nmean_2007 = can07_mat.mean(axis = 0)\n\nu_data_rec, v_data_rec = reconstruct2(can07_mat, mean_2007, can07_PCs, can07_me, hr07_PCs, hr07_me, 65, 65, method = 'LS')\nu_data_rec *= 1.25\nv_data_rec *= 1.25\ndata += (('u_wind', u_data_rec),)\ndata += (('v_wind', v_data_rec),)\nprint(\"u and v winds done\")\n\ndel can07_u\ndel can07_v\ndel hr07_u\ndel hr07_v\ndel can07_mat\ndel can07_PCs\ndel can07_eigs\ndel can07_me\ndel can07_mean\ndel u_data_rec\ndel v_data_rec\ndel hr07_mat\ndel hr07_PCs\ndel hr07_eigs\ndel hr07_me\ndel hr07_mean",
"u and v winds done\n"
],
[
"\n##reconstructing other variables\nfor i in variables:\n\n import_data(i)\n\n ##PCA on CANRCM 2007\n can07_mat = transform(can07)\n can07_PCs, can07_eigs, can07_mean = doPCA(100, can07_mat)\n\n ##PCA on HRDPS 2007\n hr07_mat = transform(hr07)\n hr07_PCs, hr07_eigs, hr07_mean = doPCA(100, hr07_mat)\n\n ## combining the eigenvectors and mean together in one array for analysis\n can07_me = np.concatenate((can07_mean.reshape(1, -1), can07_eigs))\n hr07_me = np.concatenate((hr07_mean.reshape(1, -1), hr07_eigs))\n\n ##calculating average of rows\n mean_2007 = can07_mat.mean(axis = 0)\n\n data_rec = reconstruct(can07_mat, mean_2007, can07_PCs, can07_me, hr07_PCs, hr07_me, 65, 65, method = 'LS')\n\n if data_name_hr == 'precip' or data_name_hr == 'qair' or data_name_hr == 'solar' or data_name_hr == 'therm_rad':\n avg = np.mean(data_rec, axis = 0)\n data_rec[data_rec < 0] = 0\n avg2 = np.mean(data_rec, axis = 0)\n data_rec *= avg/avg2\n \n data += ((data_name_hr, data_rec),)\n print(data_name_hr, \"done\")\ndel can07\ndel hr07\ndel can07_mat\ndel can07_PCs\ndel can07_eigs\ndel hr07_mat\ndel hr07_PCs\ndel hr07_eigs\ndel can07_me\ndel hr07_me\ndel data_rec",
"tair done\nprecip done\natmpres done\nqair done\nsolar done\ntherm_rad done\n"
],
[
"for j in data:\n print(j[0])",
"u_wind\nv_wind\ntair\ntair\nprecip\natmpres\nqair\nsolar\ntherm_rad\n"
],
[
"data_var = {}\ndims = ('time_counter', 'y', 'x')\ntimes = np.arange('2007-01-03T00:00', '2008-01-01T00:00', np.timedelta64(3, 'h'), dtype='datetime64[ns]')\n\nfor i in range(363):\n for j in data:\n data_var[ j[0] ] = (dims, j[1][8*i:8*i + 8], {})\n coords = {'time_counter': times[8*i:8*i + 8], 'y': range(266), 'x': range(256)}\n ds = xr.Dataset(data_var, coords)\n \n d = pd.to_datetime(times[8*i])\n if d.month < 10:\n month = '0' + str(d.month)\n else:\n month = str(d.month)\n if d.day < 10:\n day = '0' + str(d.day)\n else:\n day = str(d.day)\n path = '/ocean/arandhawa/reconstructed_data_2007_p2/recon_y2007m' + month + 'd' + day + '.nc'\n \n encoding = {var: {'zlib': True} for var in ds.data_vars}\n ds.to_netcdf(path, unlimited_dims=('time_counter'), encoding=encoding)",
"_____no_output_____"
],
[
"files = glob.glob('/ocean/arandhawa/reconstructed_data_2007/recon_*')\nlen(files)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
d0e434982b135d27626cad1a95bd74a6c5adbf45 | 2,644 | ipynb | Jupyter Notebook | notebooks/.ipynb_checkpoints/predict-checkpoint.ipynb | Lmath2001/Spam-Filtering | 3b1278b13981630929841911abc188b373631a4f | [
"MIT",
"Unlicense"
] | null | null | null | notebooks/.ipynb_checkpoints/predict-checkpoint.ipynb | Lmath2001/Spam-Filtering | 3b1278b13981630929841911abc188b373631a4f | [
"MIT",
"Unlicense"
] | null | null | null | notebooks/.ipynb_checkpoints/predict-checkpoint.ipynb | Lmath2001/Spam-Filtering | 3b1278b13981630929841911abc188b373631a4f | [
"MIT",
"Unlicense"
] | null | null | null | 25.180952 | 76 | 0.518533 | [
[
[
"import pickle\nimport nltk\nimport re\nnltk.download('wordnet')\nnltk.download('stopwords')\nfrom nltk.stem import WordNetLemmatizer\n\ndef predict_mail(mail):\n \n model = pickle.load(open(\"model.pckl\",mode=\"rb\"))\n vectorizer = pickle.load(open(\"vectorizer.pckl\",mode=\"rb\"))\n \n lemma = WordNetLemmatizer()\n \n stopwords = nltk.corpus.stopwords.words('english')\n \n mail = re.sub(r\"http\\S+\", \"\", mail)\n mail = re.sub(\"[^a-zA-Z0-9]\",\" \",mail)\n mail = mail.lower()\n mail = nltk.word_tokenize(mail)\n mail = [lemma.lemmatize(word) for word in mail]\n mail = [word for word in mail if word not in stopwords]\n mail = \" \".join(mail)\n \n vector = vectorizer.transform([mail])\n decision = model.predict(vector.toarray())\n \n return decision[0]",
"[nltk_data] Downloading package wordnet to\n[nltk_data] C:\\Users\\Warren\\AppData\\Roaming\\nltk_data...\n[nltk_data] Package wordnet is already up-to-date!\n[nltk_data] Downloading package stopwords to\n[nltk_data] C:\\Users\\Warren\\AppData\\Roaming\\nltk_data...\n[nltk_data] Package stopwords is already up-to-date!\n"
],
[
"predict_mail(\"It is out of deep concern and with great regret\")",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
d0e438ca6f45b47c776ab462398ab319e433c70e | 168,405 | ipynb | Jupyter Notebook | Code-Example-023-DBSCAN.ipynb | waltsco059/Elements-of-Data-Analytics | 13c27760474cf51a0aa5318d5b8518a03cddab99 | [
"MIT"
] | 8 | 2021-08-30T21:01:20.000Z | 2022-03-29T20:32:34.000Z | Code-Example-023-DBSCAN.ipynb | waltsco059/Elements-of-Data-Analytics | 13c27760474cf51a0aa5318d5b8518a03cddab99 | [
"MIT"
] | null | null | null | Code-Example-023-DBSCAN.ipynb | waltsco059/Elements-of-Data-Analytics | 13c27760474cf51a0aa5318d5b8518a03cddab99 | [
"MIT"
] | 9 | 2021-08-30T06:07:37.000Z | 2021-10-12T18:13:26.000Z | 437.415584 | 40,916 | 0.933808 | [
[
[
"# DBSCAN without Libraries ",
"_____no_output_____"
]
],
[
[
"import time\nimport warnings\nimport queue\n\nimport numpy as np\nimport pandas as pd\n\nfrom sklearn import cluster, datasets, mixture\nfrom sklearn.neighbors import kneighbors_graph\nfrom sklearn import datasets\n# from sklearn.datasets import make_blobs\nfrom sklearn.preprocessing import StandardScaler\n\nfrom scipy.stats import norm\n\nfrom matplotlib import pyplot\nimport matplotlib.cm as cm\nimport matplotlib.pyplot as plt\n\nfrom random import sample\n\nfrom itertools import cycle, islice",
"_____no_output_____"
],
[
"np.random.seed(0)\n\n# ============\n# Generate datasets. We choose the size big enough to see the scalability\n# of the algorithms, but not too big to avoid too long running times\n# ============\nn_samples = 1500\nn_components = 2 # the number of clusters\n\n\nX1, y1 = datasets.make_circles(n_samples=n_samples, factor=0.5, noise=0.05)\nX2, y2 = datasets.make_moons(n_samples=n_samples, noise=0.05)\nX3, y3 = datasets.make_blobs(n_samples=n_samples, random_state=8)\nX4, y4 = np.random.rand(n_samples, 2), None\n\n# Anisotropicly distributed data\nrandom_state = 170",
"_____no_output_____"
],
[
"# scatter plot, data points annotated by different colors\ndf = pd.DataFrame(dict(feature_1=X1[:,0], feature_2=X1[:,1], label=y1))\n\n\ncluster_name = set(y1)\ncolors = dict(zip(cluster_name, cm.rainbow(np.linspace(0, 1, len(cluster_name)))))\nfig, ax = pyplot.subplots()\n\ngrouped = df.groupby('label')\n\nfor key, group in grouped:\n# group.plot(ax=ax, kind='scatter', x='feature_1', y='feature_2', color=colors[key].reshape(1,-1))\n group.plot(ax=ax, kind='scatter', x='feature_1', y='feature_2')\n\n# pyplot.title('Original 2D Data from {} Clusters'.format(n_components))\npyplot.show()",
"_____no_output_____"
],
[
"# Based on Implementation of \n# https://github.com/kiat/Machine-Learning-Algorithms-from-Scratch/blob/master/DBSCAN.py\n\nclass CustomDBSCAN():\n def __init__(self):\n self.core = -1\n self.border = -2\n\n # Find all neighbour points at epsilon distance\n def neighbour_points(self, data, pointId, epsilon):\n points = []\n for i in range(len(data)):\n # Euclidian distance\n if np.linalg.norm([a_i - b_i for a_i, b_i in zip(data[i], data[pointId])]) <= epsilon:\n points.append(i)\n return points\n\n # Fit the data into the DBSCAN model\n def fit(self, data, Eps, MinPt):\n \n # initialize all points as outliers\n point_label = [0] * len(data)\n point_count = []\n\n # initilize list for core/border points\n core = []\n border = []\n\n # Find the neighbours of each individual point\n for i in range(len(data)):\n point_count.append(self.neighbour_points(data, i, Eps))\n\n # Find all the core points, border points and outliers\n for i in range(len(point_count)):\n if (len(point_count[i]) >= MinPt):\n point_label[i] = self.core\n core.append(i)\n else:\n border.append(i)\n\n for i in border:\n for j in point_count[i]:\n if j in core:\n point_label[i] = self.border\n break\n\n # Assign points to a cluster\n\n cluster = 1\n\n # Here we use a queue to find all the neighbourhood points of a core point and \n # find the indirectly reachable points\n # We are essentially performing Breadth First search of all points \n # which are within Epsilon distance for each other\n \n for i in range(len(point_label)):\n q = queue.Queue()\n if (point_label[i] == self.core):\n point_label[i] = cluster\n for x in point_count[i]:\n if(point_label[x] == self.core):\n q.put(x)\n point_label[x] = cluster\n elif(point_label[x] == self.border):\n point_label[x] = cluster\n while not q.empty():\n neighbors = point_count[q.get()]\n for y in neighbors:\n if (point_label[y] == self.core):\n point_label[y] = cluster\n q.put(y)\n if (point_label[y] == self.border):\n point_label[y] = cluster\n cluster += 1 # Move on to the next cluster\n\n return point_label, cluster\n\n # Visualize the clusters\n def visualize(self, data, cluster, numberOfClusters):\n N = len(data)\n\n colors = np.array(list(islice(cycle(['#FE4A49', '#2AB7CA']), 3)))\n\n for i in range(numberOfClusters):\n if (i == 0):\n # Plot all outliers point as black\n color = '#000000'\n else:\n color = colors[i % len(colors)]\n\n x, y = [], []\n for j in range(N):\n if cluster[j] == i:\n x.append(data[j, 0])\n y.append(data[j, 1])\n plt.scatter(x, y, c=color, alpha=1, marker='.')\n plt.show()",
"_____no_output_____"
],
[
"dataset = df.astype(float).values.tolist()\n\n# normalize dataset\nX = StandardScaler().fit_transform(dataset)\n\ncustom_DBSCAN = CustomDBSCAN()\npoint_labels, clusters = custom_DBSCAN.fit(X, 0.25, 4)\n\n# print(point_labels, clusters)\n\n\n\ncustom_DBSCAN.visualize(X, point_labels, clusters)",
"_____no_output_____"
],
[
"# Let us change the data and see the results \n\ndf = pd.DataFrame(dict(feature_1=X2[:,0], feature_2=X2[:,1], label=y2))\n\n\ndataset = df.astype(float).values.tolist()\n\n# normalize dataset\nX = StandardScaler().fit_transform(dataset)\n\ncustom_DBSCAN = CustomDBSCAN()\npoint_labels, clusters = custom_DBSCAN.fit(X, 0.25, 4)\n\n# print(point_labels, clusters)\n\ncustom_DBSCAN.visualize(X, point_labels, clusters)",
"_____no_output_____"
],
[
"# Let us change the data and see the results \n\ndf = pd.DataFrame(dict(feature_1=X3[:,0], feature_2=X3[:,1], label=y3))\n\n\ndataset = df.astype(float).values.tolist()\n\n# normalize dataset\nX = StandardScaler().fit_transform(dataset)\n\ncustom_DBSCAN = CustomDBSCAN()\npoint_labels, clusters = custom_DBSCAN.fit(X, 0.25, 4)\n\n# print(point_labels, clusters)\n\ncustom_DBSCAN.visualize(X, point_labels, clusters)",
"_____no_output_____"
],
[
"# Let us change the data and see the results \n\ndf = pd.DataFrame(dict(feature_1=X4[:,0], feature_2=X4[:,1], label=y4))\n\n\ndataset = df.astype(float).values.tolist()\n\n# normalize dataset\nX = StandardScaler().fit_transform(dataset)\n\ncustom_DBSCAN = CustomDBSCAN()\npoint_labels, clusters = custom_DBSCAN.fit(X, 0.25, 4)\n\n# print(point_labels, clusters)\n\ncustom_DBSCAN.visualize(X, point_labels, clusters)",
"/Users/kiat/opt/anaconda3/lib/python3.8/site-packages/sklearn/utils/extmath.py:770: RuntimeWarning: invalid value encountered in true_divide\n updated_mean = (last_sum + new_sum) / updated_sample_count\n/Users/kiat/opt/anaconda3/lib/python3.8/site-packages/sklearn/utils/extmath.py:711: RuntimeWarning: Degrees of freedom <= 0 for slice.\n result = op(x, *args, **kwargs)\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e439a8962a549f83feb43ac7117a69d7838b2a | 25,217 | ipynb | Jupyter Notebook | community/en/tensorflow_for_poets_reboot.ipynb | surabhithakare2128/examples | 365041d9ad0bb201629d4b9c0caae3b39d7c4476 | [
"Apache-2.0"
] | 1 | 2019-04-11T01:20:25.000Z | 2019-04-11T01:20:25.000Z | community/en/tensorflow_for_poets_reboot.ipynb | surabhithakare2128/examples | 365041d9ad0bb201629d4b9c0caae3b39d7c4476 | [
"Apache-2.0"
] | null | null | null | community/en/tensorflow_for_poets_reboot.ipynb | surabhithakare2128/examples | 365041d9ad0bb201629d4b9c0caae3b39d7c4476 | [
"Apache-2.0"
] | null | null | null | 31.879899 | 554 | 0.521077 | [
[
[
"##### Copyright 2019 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
],
[
"#@title MIT License\n#\n# Copyright (c) 2017 François Chollet # IGNORE_COPYRIGHT: cleared by OSS licensing\n#\n# Permission is hereby granted, free of charge, to any person obtaining a\n# copy of this software and associated documentation files (the \"Software\"),\n# to deal in the Software without restriction, including without limitation\n# the rights to use, copy, modify, merge, publish, distribute, sublicense,\n# and/or sell copies of the Software, and to permit persons to whom the\n# Software is furnished to do so, subject to the following conditions:\n#\n# The above copyright notice and this permission notice shall be included in\n# all copies or substantial portions of the Software.\n#\n# THE SOFTWARE IS PROVIDED \"AS IS\", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR\n# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,\n# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL\n# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER\n# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING\n# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER\n# DEALINGS IN THE SOFTWARE.",
"_____no_output_____"
]
],
[
[
"# Transfer Learning Using Pretrained ConvNets",
"_____no_output_____"
],
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/examples/community/en/tensorflow_for_poets_reboot.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/examples/community/en/tensorflow_for_poets_reboot.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View source on GitHub</a>\n </td>\n</table>",
"_____no_output_____"
]
],
[
[
"from __future__ import absolute_import, division, print_function\n\nimport tensorflow.compat.v2 as tf #nightly-gpu\ntf.enable_v2_behavior()\n\nimport os\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom functools import partial",
"_____no_output_____"
],
[
"tf.__version__",
"_____no_output_____"
]
],
[
[
"## Data preprocessing",
"_____no_output_____"
],
[
"### Data download",
"_____no_output_____"
]
],
[
[
"_URL = \"https://storage.googleapis.com/download.tensorflow.org/example_images/flower_photos.tgz\"\n\nzip_file = tf.keras.utils.get_file(origin=_URL, \n fname=\"flower_photos.tgz\", \n extract=True)\n\nbase_dir = os.path.join(os.path.dirname(zip_file), 'flower_photos')",
"_____no_output_____"
],
[
"IMAGE_SIZE = 224\nBATCH_SIZE = 64\n\ndatagen = tf.keras.preprocessing.image.ImageDataGenerator(\n rescale=1./255, \n validation_split=0.2)\n\ntrain_generator = datagen.flow_from_directory(\n base_dir,\n target_size=(IMAGE_SIZE, IMAGE_SIZE),\n batch_size=BATCH_SIZE, \n subset='training')\n\nval_generator = datagen.flow_from_directory(\n base_dir,\n target_size=(IMAGE_SIZE, IMAGE_SIZE),\n batch_size=BATCH_SIZE, \n subset='validation')",
"_____no_output_____"
],
[
"for image_batch, label_batch in train_generator:\n break\nimage_batch.shape, label_batch.shape",
"_____no_output_____"
],
[
"train_generator.class_indices",
"_____no_output_____"
]
],
[
[
"## Create the base model from the pre-trained convnets",
"_____no_output_____"
]
],
[
[
"IMG_SHAPE = (IMAGE_SIZE, IMAGE_SIZE, 3)\n\n# Create the base model from the pre-trained model MobileNet V2\nbase_model = tf.keras.applications.MobileNetV2(input_shape=IMG_SHAPE,\n include_top=False, \n weights='imagenet')",
"_____no_output_____"
]
],
[
[
"## Feature extraction\nYou will freeze the convolutional base created from the previous step and use that as a feature extractor, add a classifier on top of it and train the top-level classifier.",
"_____no_output_____"
]
],
[
[
"base_model.trainable = False",
"_____no_output_____"
]
],
[
[
"### Add a classification head",
"_____no_output_____"
]
],
[
[
"model = tf.keras.Sequential([\n base_model,\n tf.keras.layers.Conv2D(32, 3, activation='relu'),\n tf.keras.layers.Dropout(0.2),\n tf.keras.layers.GlobalAveragePooling2D(),\n tf.keras.layers.Dense(5, activation='softmax')\n])",
"_____no_output_____"
]
],
[
[
"### Compile the model\n\nYou must compile the model before training it. Since there are two classes, use a binary cross-entropy loss.",
"_____no_output_____"
]
],
[
[
"model.compile(optimizer=tf.keras.optimizers.Adam(), \n loss='categorical_crossentropy', \n metrics=['accuracy'])",
"_____no_output_____"
],
[
"model.summary()",
"_____no_output_____"
]
],
[
[
"The 2.5M parameters in MobileNet are frozen, but there are 1.2K _trainable_ parameters in the Dense layer. These are divided between two `tf.Variable` objects, the weights and biases.",
"_____no_output_____"
]
],
[
[
"len(model.trainable_variables)",
"_____no_output_____"
],
[
"train_generator.labels",
"_____no_output_____"
]
],
[
[
"### Train the model\n\n<!-- TODO(markdaoust): delete steps_per_epoch in TensorFlow r1.14/r2.0 -->",
"_____no_output_____"
]
],
[
[
"epochs = 10\n\nhistory = model.fit(train_generator, \n epochs=epochs, \n validation_data=val_generator)",
"_____no_output_____"
]
],
[
[
"### Learning curves\n\nLet's take a look at the learning curves of the training and validation accuracy/loss when using the MobileNet V2 base model as a fixed feature extractor. ",
"_____no_output_____"
]
],
[
[
"acc = history.history['accuracy']\nval_acc = history.history['val_accuracy']\n\nloss = history.history['loss']\nval_loss = history.history['val_loss']\n\nplt.figure(figsize=(8, 8))\nplt.subplot(2, 1, 1)\nplt.plot(acc, label='Training Accuracy')\nplt.plot(val_acc, label='Validation Accuracy')\nplt.legend(loc='lower right')\nplt.ylabel('Accuracy')\nplt.ylim([min(plt.ylim()),1])\nplt.title('Training and Validation Accuracy')\n\nplt.subplot(2, 1, 2)\nplt.plot(loss, label='Training Loss')\nplt.plot(val_loss, label='Validation Loss')\nplt.legend(loc='upper right')\nplt.ylabel('Cross Entropy')\nplt.ylim([0,1.0])\nplt.title('Training and Validation Loss')\nplt.xlabel('epoch')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Note: If you are wondering why the validation metrics are clearly better than the training metrics, the main factor is because layers like `tf.keras.layers.BatchNormalization` and `tf.keras.layers.Dropout` affect accuracy during training. They are turned off when calculating validation loss.\n\nTo a lesser extent, it is also because training metrics report the average for an epoch, while validation metrics are evaluated after the epoch, so validation metrics see a model that has trained slightly longer.",
"_____no_output_____"
],
[
"## Fine tuning\nIn our feature extraction experiment, you were only training a few layers on top of an MobileNet V2 base model. The weights of the pre-trained network were **not** updated during training.\n\nOne way to increase performance even further is to train (or \"fine-tune\") the weights of the top layers of the pre-trained model alongside the training of the classifier you added. The training process will force the weights to be tuned from generic features maps to features associated specifically to our dataset.\n\nNote: This should only be attempted after you have trained the top-level classifier with the pre-trained model set to non-trainable. If you add a randomly initialized classifier on top of a pre-trained model and attempt to train all layers jointly, the magnitude of the gradient updates will be too large (due to the random weights from the classifier) and your pre-trained model will forget what it has learned.\n\nAlso, you should try to fine-tune a small number of top layers rather than the whole MobileNet model. In most convolutional networks, the higher up a layer is, the more specialized it is. The first few layers learn very simple and generic features which generalize to almost all types of images. As you go higher up, the features are increasingly more specific to the dataset on which the model was trained. The goal of fine-tuning is to adapt these specialized features to work with the new dataset, rather than overwrite the generic learning.",
"_____no_output_____"
],
[
"### Un-freeze the top layers of the model\n",
"_____no_output_____"
],
[
"All you need to do is unfreeze the `base_model` and set the bottom layers be un-trainable. Then, you should recompile the model (necessary for these changes to take effect), and resume training.",
"_____no_output_____"
]
],
[
[
"base_model.trainable = True",
"_____no_output_____"
],
[
"# Let's take a look to see how many layers are in the base model\nprint(\"Number of layers in the base model: \", len(base_model.layers))\n\n# Fine tune from this layer onwards\nfine_tune_at = 100\n\n# Freeze all the layers before the `fine_tune_at` layer\nfor layer in base_model.layers[:fine_tune_at]:\n layer.trainable = False",
"_____no_output_____"
]
],
[
[
"### Compile the model\n\nCompile the model using a much lower training rate.",
"_____no_output_____"
]
],
[
[
"model.compile(loss='categorical_crossentropy',\n optimizer = tf.keras.optimizers.Adam(1e-5),\n metrics=['accuracy'])",
"_____no_output_____"
],
[
"model.summary()",
"_____no_output_____"
],
[
"len(model.trainable_variables)",
"_____no_output_____"
]
],
[
[
"### Continue Train the model",
"_____no_output_____"
],
[
"If you trained to convergence earlier, this will get you a few percent more accuracy.",
"_____no_output_____"
]
],
[
[
"history_fine = model.fit(train_generator, \n epochs=5,\n validation_data=val_generator)",
"_____no_output_____"
]
],
[
[
"Let's take a look at the learning curves of the training and validation accuracy/loss, when fine tuning the last few layers of the MobileNet V2 base model and training the classifier on top of it. The validation loss is much higher than the training loss, so you may get some overfitting.\n\nYou may also get some overfitting as the new training set is relatively small and similar to the original MobileNet V2 datasets.\n",
"_____no_output_____"
],
[
"## Convert to TFLite",
"_____no_output_____"
]
],
[
[
"saved_model_dir = 'save/fine_tuning'\ntf.saved_model.save(model, saved_model_dir)\nmodel = tf.saved_model.load(saved_model_dir)\n\nconcrete_func = model.signatures[\n tf.saved_model.DEFAULT_SERVING_SIGNATURE_DEF_KEY]\n\nconcrete_func.inputs[0].set_shape([1, 224, 224, 3])\n\nconverter = tf.lite.TFLiteConverter.from_concrete_function(concrete_func)\ntflite_model = converter.convert()\n\nwith open('model.tflite', 'wb') as f:\n f.write(tflite_model)",
"_____no_output_____"
]
],
[
[
"Download the converted model",
"_____no_output_____"
]
],
[
[
"from google.colab import files\n\nfiles.download('model.tflite')",
"_____no_output_____"
],
[
"acc += history_fine.history['accuracy']\nval_acc += history_fine.history['val_accuracy']\n\nloss += history_fine.history['loss']\nval_loss += history_fine.history['val_loss']",
"_____no_output_____"
],
[
"plt.figure(figsize=(8, 8))\nplt.subplot(2, 1, 1)\nplt.plot(acc, label='Training Accuracy')\nplt.plot(val_acc, label='Validation Accuracy')\nplt.ylim([0.8, 1])\nplt.plot([initial_epochs-1,initial_epochs-1], \n plt.ylim(), label='Start Fine Tuning')\nplt.legend(loc='lower right')\nplt.title('Training and Validation Accuracy')\n\nplt.subplot(2, 1, 2)\nplt.plot(loss, label='Training Loss')\nplt.plot(val_loss, label='Validation Loss')\nplt.ylim([0, 1.0])\nplt.plot([initial_epochs-1,initial_epochs-1], \n plt.ylim(), label='Start Fine Tuning')\nplt.legend(loc='upper right')\nplt.title('Training and Validation Loss')\nplt.xlabel('epoch')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Summary:\n\n* **Using a pre-trained model for feature extraction**: When working with a small dataset, it is common to take advantage of features learned by a model trained on a larger dataset in the same domain. This is done by instantiating the pre-trained model and adding a fully-connected classifier on top. The pre-trained model is \"frozen\" and only the weights of the classifier get updated during training.\nIn this case, the convolutional base extracted all the features associated with each image and you just trained a classifier that determines the image class given that set of extracted features.\n\n* **Fine-tuning a pre-trained model**: To further improve performance, one might want to repurpose the top-level layers of the pre-trained models to the new dataset via fine-tuning.\nIn this case, you tuned your weights such that your model learned high-level features specific to the dataset. This technique is usually recommended when the training dataset is large and very similar to the orginial dataset that the pre-trained model was trained on.\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
d0e43b64a275a033ca866722406761be3fa6bb11 | 928,990 | ipynb | Jupyter Notebook | tutorials/Certification_Trainings/Public/1.SparkNLP_Basics.ipynb | HashamUlHaq/spark-nlp-workshop | 52f372549941addc82c6b1311653cc6402ff4dda | [
"Apache-2.0"
] | null | null | null | tutorials/Certification_Trainings/Public/1.SparkNLP_Basics.ipynb | HashamUlHaq/spark-nlp-workshop | 52f372549941addc82c6b1311653cc6402ff4dda | [
"Apache-2.0"
] | null | null | null | tutorials/Certification_Trainings/Public/1.SparkNLP_Basics.ipynb | HashamUlHaq/spark-nlp-workshop | 52f372549941addc82c6b1311653cc6402ff4dda | [
"Apache-2.0"
] | null | null | null | 203.770564 | 373,639 | 0.859559 | [
[
[
"",
"_____no_output_____"
],
[
"[](https://colab.research.google.com/github/JohnSnowLabs/spark-nlp-workshop/blob/master/tutorials/Certification_Trainings/Public/1.SparkNLP_Basics.ipynb)",
"_____no_output_____"
],
[
"# Spark NLP Basics and Pretrained Pipelines",
"_____no_output_____"
],
[
"## 0. Colab Setup",
"_____no_output_____"
]
],
[
[
"import os\n\n# Install java\n! apt-get update -qq\n! apt-get install -y openjdk-8-jdk-headless -qq > /dev/null\n\nos.environ[\"JAVA_HOME\"] = \"/usr/lib/jvm/java-8-openjdk-amd64\"\nos.environ[\"PATH\"] = os.environ[\"JAVA_HOME\"] + \"/bin:\" + os.environ[\"PATH\"]\n! java -version\n\n# Install pyspark\n! pip install --ignore-installed -q pyspark==2.4.4\n! pip install --ignore-installed -q spark-nlp==2.7.1",
"openjdk version \"1.8.0_275\"\nOpenJDK Runtime Environment (build 1.8.0_275-8u275-b01-0ubuntu1~18.04-b01)\nOpenJDK 64-Bit Server VM (build 25.275-b01, mixed mode)\n\u001b[K |████████████████████████████████| 215.7MB 70kB/s \n\u001b[K |████████████████████████████████| 204kB 48.3MB/s \n\u001b[?25h Building wheel for pyspark (setup.py) ... \u001b[?25l\u001b[?25hdone\n\u001b[K |████████████████████████████████| 143kB 4.3MB/s \n\u001b[?25h"
]
],
[
[
"\n#### **How to prevent Google Colab from disconnecting?**\n\nGoogle Colab notebooks have an idle timeout of 90 minutes and absolute timeout of 12 hours. This means, if user does not interact with his Google Colab notebook for more than 90 minutes, its instance is automatically terminated. Also, maximum lifetime of a Colab instance is **12 hours.**\n\nSet a javascript interval to click on the connect button every 60 seconds. Open developer-settings (in your web-browser) with Ctrl+Shift+I then click on console tab and type this on the console prompt. (for mac press Option+Command+I)\n\n",
"_____no_output_____"
]
],
[
[
"''' \n\nfunction ConnectButton(){\n console.log(\"Connect pushed\"); \n document.querySelector(\"#top-toolbar > colab-connect-button\").shadowRoot.querySelector(\"#connect\").click() \n}\nsetInterval(ConnectButton,60000);\n\n'''",
"_____no_output_____"
]
],
[
[
"## 1. Start Spark Session",
"_____no_output_____"
]
],
[
[
"import sparknlp\n\nspark = sparknlp.start()\n\n# params =>> gpu=False, spark23=False (start with spark 2.3)\n\nprint(\"Spark NLP version\", sparknlp.version())\n\nprint(\"Apache Spark version:\", spark.version)\n",
"Spark NLP version 2.7.1\nApache Spark version: 2.4.4\n"
],
[
"! cd ~/.ivy2/cache/com.johnsnowlabs.nlp/spark-nlp_2.11/jars && ls -lt",
"total 39960\n-rw-r--r-- 1 root root 40918604 Jan 8 21:03 spark-nlp_2.11-2.7.1.jar\n"
]
],
[
[
"\n<b> if you want to work with Spark 2.3 </b>\n```\nimport os\n\n# Install java\n! apt-get update -qq\n! apt-get install -y openjdk-8-jdk-headless -qq > /dev/null\n\n!wget -q https://archive.apache.org/dist/spark/spark-2.3.0/spark-2.3.0-bin-hadoop2.7.tgz\n\n!tar xf spark-2.3.0-bin-hadoop2.7.tgz\n!pip install -q findspark\n\nos.environ[\"JAVA_HOME\"] = \"/usr/lib/jvm/java-8-openjdk-amd64\"\nos.environ[\"PATH\"] = os.environ[\"JAVA_HOME\"] + \"/bin:\" + os.environ[\"PATH\"]\nos.environ[\"SPARK_HOME\"] = \"/content/spark-2.3.0-bin-hadoop2.7\"\n! java -version\n\nimport findspark\nfindspark.init()\nfrom pyspark.sql import SparkSession\n\n! pip install --ignore-installed -q spark-nlp==2.5.5\nimport sparknlp\n\nspark = sparknlp.start(spark23=True)\n```",
"_____no_output_____"
],
[
"`sparknlp.start()` will start or get SparkSession with predefined parameters hardcoded in `spark-nlp/python/sparknlp/__init__.py`. here is what is going on under the hood when you run `sparknlp.start()` ",
"_____no_output_____"
]
],
[
[
"# https://github.com/JohnSnowLabs/spark-nlp/blob/master/python/sparknlp/__init__.py\n\nfrom pyspark.sql import SparkSession\n\ndef start(gpu=False, spark23=False):\n current_version=\"2.5.4\"\n maven_spark24 = \"com.johnsnowlabs.nlp:spark-nlp_2.11:{}\".format(current_version)\n maven_gpu_spark24 = \"com.johnsnowlabs.nlp:spark-nlp-gpu_2.11:{}\".format(current_version)\n maven_spark23 = \"com.johnsnowlabs.nlp:spark-nlp-spark23_2.11:{}\".format(current_version)\n maven_gpu_spark23 = \"com.johnsnowlabs.nlp:spark-nlp-gpu-spark23_2.11:{}\".format(current_version)\n\n builder = SparkSession.builder \\\n .appName(\"Spark NLP\") \\\n .master(\"local[*]\") \\\n .config(\"spark.driver.memory\", \"16G\") \\\n .config(\"spark.serializer\", \"org.apache.spark.serializer.KryoSerializer\") \\\n .config(\"spark.kryoserializer.buffer.max\", \"1000M\") \\\n .config(\"spark.driver.maxResultSize\", \"0\")\n if gpu and spark23:\n builder.config(\"spark.jars.packages\", maven_gpu_spark23)\n elif spark23:\n builder.config(\"spark.jars.packages\", maven_spark23)\n elif gpu:\n builder.config(\"spark.jars.packages\", maven_gpu_spark24)\n else:\n builder.config(\"spark.jars.packages\", maven_spark24)\n \n return builder.getOrCreate()\n",
"_____no_output_____"
]
],
[
[
"If you want to start `SparkSession` with your own parameters or you need to load the required jars/packages from your local disk, or you have no internet connection (that would be needed to pull the required packages from internet), you can skip `sparknlp.start()` and start your session manually as shown below.\n",
"_____no_output_____"
]
],
[
[
"\"\"\"\nfrom pyspark.sql import SparkSession\n\nspark = SparkSession.builder \\\n .appName(\"Spark NLP Enterprise 2.4.5\") \\\n .master(\"local[8]\") \\\n .config(\"spark.driver.memory\",\"12G\") \\\n .config(\"spark.driver.maxResultSize\", \"2G\") \\\n .config(\"spark.serializer\", \"org.apache.spark.serializer.KryoSerializer\") \\\n .config(\"spark.kryoserializer.buffer.max\", \"800M\")\\\n .config(\"spark.jars\", \"{}spark-nlp-2.4.5.jar,{}spark-nlp-jsl-2.4.5.jar\".format(jar_path,jar_path)) \\\n .getOrCreate()\n \n\"\"\"",
"_____no_output_____"
]
],
[
[
"CPU on Apache Spark 2.4.x: https://s3.amazonaws.com/auxdata.johnsnowlabs.com/public/spark-nlp-assembly-2.7.1.jar\n\nGPU on Apache Spark 2.4.x: https://s3.amazonaws.com/auxdata.johnsnowlabs.com/public/spark-nlp-gpu-assembly-2.7.1.jar\n\nCPU on Apache Spark 2.3.x: https://s3.amazonaws.com/auxdata.johnsnowlabs.com/public/spark-nlp-spark23-assembly-2.7.1.jar\n\nGPU on Apache Spark 2.3.x: https://s3.amazonaws.com/auxdata.johnsnowlabs.com/public/spark-nlp-spark23-gpu-assembly-2.7.1.jar",
"_____no_output_____"
],
[
"## 2. Using Pretrained Pipelines",
"_____no_output_____"
],
[
"https://github.com/JohnSnowLabs/spark-nlp-models (not maintained)\n\nhttps://nlp.johnsnowlabs.com/models\n",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"from sparknlp.pretrained import PretrainedPipeline",
"_____no_output_____"
],
[
"testDoc = '''\nPeter is a very good persn.\nMy life in Russia is very intersting.\nJohn and Peter are brothrs. However they don't support each other that much.\nLucas Nogal Dunbercker is no longer happy. He has a good car though.\nEurope is very culture rich. There are huge churches! and big houses!\n'''",
"_____no_output_____"
]
],
[
[
"### Explain Document ML",
"_____no_output_____"
],
[
"**Stages**\n- DocumentAssembler\n- SentenceDetector\n- Tokenizer\n- Lemmatizer\n- Stemmer\n- Part of Speech\n- SpellChecker (Norvig)\n\n\n",
"_____no_output_____"
]
],
[
[
"pipeline = PretrainedPipeline('explain_document_ml', lang='en')\n",
"explain_document_ml download started this may take some time.\nApprox size to download 9.4 MB\n[OK!]\n"
],
[
"pipeline.model.stages",
"_____no_output_____"
],
[
"# Load pretrained pipeline from local disk:\n\npipeline_local = PretrainedPipeline.from_disk('/root/cache_pretrained/explain_document_ml_en_2.4.0_2.4_1580252705962')",
"_____no_output_____"
],
[
"%%time\n\nresult = pipeline.annotate(testDoc)",
"CPU times: user 41 ms, sys: 7.51 ms, total: 48.5 ms\nWall time: 2.47 s\n"
],
[
"result.keys()",
"_____no_output_____"
],
[
"result['sentence']",
"_____no_output_____"
],
[
"result['token']",
"_____no_output_____"
],
[
"list(zip(result['token'], result['pos']))",
"_____no_output_____"
],
[
"list(zip(result['token'], result['lemmas'], result['stems'], result['spell']))",
"_____no_output_____"
],
[
"import pandas as pd\n\ndf = pd.DataFrame({'token':result['token'], \n 'corrected':result['spell'], 'POS':result['pos'],\n 'lemmas':result['lemmas'], 'stems':result['stems']})\ndf",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"### Explain Document DL",
"_____no_output_____"
],
[
"**Stages**\n- DocumentAssembler\n- SentenceDetector\n- Tokenizer\n- NER (NER with GloVe 100D embeddings, CoNLL2003 dataset)\n- Lemmatizer\n- Stemmer\n- Part of Speech\n- SpellChecker (Norvig)\n",
"_____no_output_____"
]
],
[
[
"pipeline_dl = PretrainedPipeline('explain_document_dl', lang='en')\n",
"explain_document_dl download started this may take some time.\nApprox size to download 168.4 MB\n[OK!]\n"
],
[
"pipeline_dl.model.stages",
"_____no_output_____"
],
[
"pipeline_dl.model.stages[-2].getStorageRef()",
"_____no_output_____"
],
[
"pipeline_dl.model.stages[-2].getClasses()",
"_____no_output_____"
],
[
"%%time\n\nresult = pipeline_dl.annotate(testDoc)\n\nresult.keys()",
"CPU times: user 61.3 ms, sys: 20.3 ms, total: 81.5 ms\nWall time: 1.62 s\n"
],
[
"result.keys()",
"_____no_output_____"
],
[
"result['entities']",
"_____no_output_____"
],
[
"df = pd.DataFrame({'token':result['token'], 'ner_label':result['ner'],\n 'spell_corrected':result['checked'], 'POS':result['pos'],\n 'lemmas':result['lemma'], 'stems':result['stem']})\n\ndf",
"_____no_output_____"
]
],
[
[
"### Recognize Entities DL",
"_____no_output_____"
]
],
[
[
"recognize_entities = PretrainedPipeline('recognize_entities_dl', lang='en')\n",
"recognize_entities_dl download started this may take some time.\nApprox size to download 159 MB\n[OK!]\n"
],
[
"recognize_entities.model.stages",
"_____no_output_____"
],
[
"recognize_entities.model.stages[3].getStorageRef()",
"_____no_output_____"
],
[
"recognize_entities.model.stages[4].getClasses()",
"_____no_output_____"
],
[
"testDoc = '''\nPeter is a very good persn.\nMy life in Russia is very intersting.\nJohn and Peter are brothrs. However they don't support each other that much.\nLucas Nogal Dunbercker is no longer happy. He has a good car though.\nEurope is very culture rich. There are huge churches! and big houses!\n'''\n\nresult = recognize_entities.annotate(testDoc)\n\nlist(zip(result['token'], result['ner']))",
"_____no_output_____"
]
],
[
[
"### Clean Stop Words",
"_____no_output_____"
]
],
[
[
"clean_stop = PretrainedPipeline('clean_stop', lang='en')\n",
"clean_stop download started this may take some time.\nApprox size to download 12.4 KB\n[OK!]\n"
],
[
"\nresult = clean_stop.annotate(testDoc)\nresult.keys()",
"_____no_output_____"
],
[
"\n' '.join(result['cleanTokens'])",
"_____no_output_____"
]
],
[
[
"### Clean Slang ",
"_____no_output_____"
]
],
[
[
"clean_slang = PretrainedPipeline('clean_slang', lang='en')\n\nresult = clean_slang.annotate(' Whatsup bro, call me ASAP')\n\n' '.join(result['normal'])",
"clean_slang download started this may take some time.\nApprox size to download 21.8 KB\n[OK!]\n"
],
[
"clean_slang.model.stages",
"_____no_output_____"
],
[
"clean_slang.model.stages[-1]",
"_____no_output_____"
]
],
[
[
"### Spell Checker \n\n(Norvig Algo)\n\nref: https://norvig.com/spell-correct.html",
"_____no_output_____"
]
],
[
[
"spell_checker = PretrainedPipeline('check_spelling', lang='en')\n",
"check_spelling download started this may take some time.\nApprox size to download 892.6 KB\n[OK!]\n"
],
[
"testDoc = '''\nPeter is a very good persn.\nMy life in Russia is very intersting.\nJohn and Peter are brothrs. However they don't support each other that much.\nLucas Nogal Dunbercker is no longer happy. He has a good car though.\nEurope is very culture rich. There are huge churches! and big houses!\n'''\n\nresult = spell_checker.annotate(testDoc)\n\nresult.keys()",
"_____no_output_____"
],
[
"list(zip(result['token'], result['checked']))",
"_____no_output_____"
]
],
[
[
"### Spell Checker DL\n\nhttps://medium.com/spark-nlp/applying-context-aware-spell-checking-in-spark-nlp-3c29c46963bc",
"_____no_output_____"
]
],
[
[
"spell_checker_dl = PretrainedPipeline('check_spelling_dl', lang='en')\n",
"check_spelling_dl download started this may take some time.\nApprox size to download 112.1 MB\n[OK!]\n"
],
[
"text = 'We will go to swimming if the ueather is nice.'\n\nresult = spell_checker_dl.annotate(text)\n\nlist(zip(result['token'], result['checked']))",
"_____no_output_____"
],
[
"result.keys()",
"_____no_output_____"
],
[
"# check for the different occurrences of the word \"ueather\"\nexamples = ['We will go to swimming if the ueather is nice.',\\\n \"I have a black ueather jacket, so nice.\",\\\n \"I introduce you to my sister, she is called ueather.\"]\n\nresults = spell_checker_dl.annotate(examples)\n\nfor result in results:\n print (list(zip(result['token'], result['checked'])))",
"[('We', 'We'), ('will', 'will'), ('go', 'go'), ('to', 'to'), ('swimming', 'swimming'), ('if', 'if'), ('the', 'the'), ('ueather', 'weather'), ('is', 'is'), ('nice', 'nice'), ('.', '.')]\n[('I', 'I'), ('have', 'have'), ('a', 'a'), ('black', 'black'), ('ueather', 'leather'), ('jacket', 'jacket'), (',', ','), ('so', 'so'), ('nice', 'nice'), ('.', '.')]\n[('I', 'I'), ('introduce', 'introduce'), ('you', 'you'), ('to', 'to'), ('my', 'my'), ('sister', 'sister'), (',', ','), ('she', 'she'), ('is', 'is'), ('called', 'called'), ('ueather', 'Heather'), ('.', '.')]\n"
],
[
"for result in results:\n print (result['document'],'>>',[pairs for pairs in list(zip(result['token'], result['checked'])) if pairs[0]!=pairs[1]])",
"['We will go to swimming if the ueather is nice.'] >> [('ueather', 'weather')]\n['I have a black ueather jacket, so nice.'] >> [('ueather', 'leather')]\n['I introduce you to my sister, she is called ueather.'] >> [('ueather', 'Heather')]\n"
],
[
"# if we had tried the same with spell_checker (previous version)\n\nresults = spell_checker.annotate(examples)\n\nfor result in results:\n print (list(zip(result['token'], result['checked'])))",
"[('We', 'We'), ('will', 'will'), ('go', 'go'), ('to', 'to'), ('swimming', 'swimming'), ('if', 'if'), ('the', 'the'), ('ueather', 'weather'), ('is', 'is'), ('nice', 'nice'), ('.', '.')]\n[('I', 'I'), ('have', 'have'), ('a', 'a'), ('black', 'black'), ('ueather', 'weather'), ('jacket', 'jacket'), (',', ','), ('so', 'so'), ('nice', 'nice'), ('.', '.')]\n[('I', 'I'), ('introduce', 'introduce'), ('you', 'you'), ('to', 'to'), ('my', 'my'), ('sister', 'sister'), (',', ','), ('she', 'she'), ('is', 'is'), ('called', 'called'), ('ueather', 'weather'), ('.', '.')]\n"
]
],
[
[
"### Parsing a list of texts",
"_____no_output_____"
]
],
[
[
"testDoc_list = ['French author who helped pioner the science-fiction genre.',\n'Verne wrate about space, air, and underwater travel before navigable aircrast',\n'Practical submarines were invented, and before any means of space travel had been devised.']\n\ntestDoc_list",
"_____no_output_____"
],
[
"pipeline = PretrainedPipeline('explain_document_ml', lang='en')\n",
"explain_document_ml download started this may take some time.\nApprox size to download 9.4 MB\n[OK!]\n"
],
[
"result_list = pipeline.annotate(testDoc_list)\n\nlen (result_list)",
"_____no_output_____"
],
[
"result_list[0]",
"_____no_output_____"
]
],
[
[
"### Using fullAnnotate to get more details\n\n",
"_____no_output_____"
],
[
"```\nannotatorType: String, \nbegin: Int, \nend: Int, \nresult: String, (this is what annotate returns)\nmetadata: Map[String, String], \nembeddings: Array[Float]\n```",
"_____no_output_____"
]
],
[
[
"text = 'Peter Parker is a nice guy and lives in New York'",
"_____no_output_____"
],
[
"# pipeline_dl >> explain_document_dl\n\ndetailed_result = pipeline_dl.fullAnnotate(text)",
"_____no_output_____"
],
[
"detailed_result",
"_____no_output_____"
],
[
"detailed_result[0]['entities']",
"_____no_output_____"
],
[
"detailed_result[0]['entities'][0].result",
"_____no_output_____"
],
[
"chunks=[]\nentities=[]\nfor n in detailed_result[0]['entities']:\n \n chunks.append(n.result)\n entities.append(n.metadata['entity']) \n \ndf = pd.DataFrame({'chunks':chunks, 'entities':entities})\ndf ",
"_____no_output_____"
],
[
"tuples = []\n\nfor x,y,z in zip(detailed_result[0][\"token\"], detailed_result[0][\"pos\"], detailed_result[0][\"ner\"]):\n\n tuples.append((int(x.metadata['sentence']), x.result, x.begin, x.end, y.result, z.result))\n\ndf = pd.DataFrame(tuples, columns=['sent_id','token','start','end','pos', 'ner'])\n\ndf\n",
"_____no_output_____"
]
],
[
[
"### Use pretrained match_chunk Pipeline for Individual Noun Phrase",
"_____no_output_____"
],
[
"**Stages**\n- DocumentAssembler\n- SentenceDetector\n- Tokenizer\n- Part of Speech\n- Chunker\n\nPipeline:\n\n- The pipeline uses regex `<DT>?<JJ>*<NN>+`\n- which states that whenever the chunk finds an optional determiner (DT) followed by any number of adjectives (JJ) and then a noun (NN) then the Noun Phrase(NP) chunk should be formed.",
"_____no_output_____"
]
],
[
[
"pipeline = PretrainedPipeline('match_chunks', lang='en')\n",
"match_chunks download started this may take some time.\nApprox size to download 4.3 MB\n[OK!]\n"
],
[
"pipeline.model.stages",
"_____no_output_____"
],
[
"result = pipeline.annotate(\"The book has many chapters\") # single noun phrase",
"_____no_output_____"
],
[
"result",
"_____no_output_____"
],
[
"result['chunk']",
"_____no_output_____"
],
[
"result = pipeline.annotate(\"the little yellow dog barked at the cat\") #multiple noune phrases",
"_____no_output_____"
],
[
"result",
"_____no_output_____"
],
[
"result['chunk']",
"_____no_output_____"
]
],
[
[
"### Extract exact dates from referential date phrases",
"_____no_output_____"
]
],
[
[
"pipeline = PretrainedPipeline('match_datetime', lang='en')\n",
"match_datetime download started this may take some time.\nApprox size to download 12.9 KB\n[OK!]\n"
],
[
"result = pipeline.annotate(\"I saw him yesterday and he told me that he will visit us next week\")\n\nresult",
"_____no_output_____"
],
[
"detailed_result = pipeline.fullAnnotate(\"I saw him yesterday and he told me that he will visit us next week\")\n\ndetailed_result",
"_____no_output_____"
],
[
"tuples = []\n\nfor x in detailed_result[0][\"token\"]:\n\n tuples.append((int(x.metadata['sentence']), x.result, x.begin, x.end))\n\ndf = pd.DataFrame(tuples, columns=['sent_id','token','start','end'])\n\ndf",
"_____no_output_____"
]
],
[
[
"### Sentiment Analysis\n",
"_____no_output_____"
],
[
"#### Vivek algo\n\npaper: `Fast and accurate sentiment classification using an enhanced Naive Bayes model`\n\nhttps://arxiv.org/abs/1305.6143\n\ncode `https://github.com/vivekn/sentiment`",
"_____no_output_____"
]
],
[
[
"sentiment = PretrainedPipeline('analyze_sentiment', lang='en')",
"analyze_sentiment download started this may take some time.\nApprox size to download 4.9 MB\n[OK!]\n"
],
[
"result = sentiment.annotate(\"The movie I watched today was not a good one\")\n\nresult['sentiment']",
"_____no_output_____"
]
],
[
[
"#### DL version (trained on imdb)",
"_____no_output_____"
]
],
[
[
"sentiment_imdb = PretrainedPipeline('analyze_sentimentdl_use_imdb', lang='en')",
"analyze_sentimentdl_use_imdb download started this may take some time.\nApprox size to download 935.8 MB\n[OK!]\n"
],
[
"sentiment_imdb_glove = PretrainedPipeline('analyze_sentimentdl_glove_imdb', lang='en')",
"analyze_sentimentdl_glove_imdb download started this may take some time.\nApprox size to download 154 MB\n[OK!]\n"
],
[
"comment = '''\nIt's a very scary film but what impressed me was how true the film sticks to the original's tricks; it isn't filled with loud in-your-face jump scares, in fact, a lot of what makes this film scary is the slick cinematography and intricate shadow play. The use of lighting and creation of atmosphere is what makes this film so tense, which is why it's perfectly suited for those who like Horror movies but without the obnoxious gore.\n'''\nresult = sentiment_imdb_glove.annotate(comment)\n\nresult['sentiment']",
"_____no_output_____"
],
[
"sentiment_imdb_glove.fullAnnotate(comment)[0]['sentiment']",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
d0e457a9e062d9f51ed4ffd139e4b85f388fbb2e | 280,461 | ipynb | Jupyter Notebook | jupyter/2018-09-20(Graph of non-orthogonal sequences example).ipynb | h-mayorquin/sequence_learning_paper | 0177ca52b144ea579e91e39f9ddd487a0c04726d | [
"MIT"
] | null | null | null | jupyter/2018-09-20(Graph of non-orthogonal sequences example).ipynb | h-mayorquin/sequence_learning_paper | 0177ca52b144ea579e91e39f9ddd487a0c04726d | [
"MIT"
] | null | null | null | jupyter/2018-09-20(Graph of non-orthogonal sequences example).ipynb | h-mayorquin/sequence_learning_paper | 0177ca52b144ea579e91e39f9ddd487a0c04726d | [
"MIT"
] | null | null | null | 509.00363 | 97,700 | 0.939828 | [
[
[
"<h1>Table of Contents<span class=\"tocSkip\"></span></h1>\n<div class=\"toc\"><ul class=\"toc-item\"><li><span><a href=\"#Preamble\" data-toc-modified-id=\"Preamble-1\"><span class=\"toc-item-num\">1 </span>Preamble</a></span><ul class=\"toc-item\"><li><span><a href=\"#Some-general-parameters\" data-toc-modified-id=\"Some-general-parameters-1.1\"><span class=\"toc-item-num\">1.1 </span>Some general parameters</a></span></li><li><span><a href=\"#Functions\" data-toc-modified-id=\"Functions-1.2\"><span class=\"toc-item-num\">1.2 </span>Functions</a></span><ul class=\"toc-item\"><li><ul class=\"toc-item\"><li><span><a href=\"#Create-overlap\" data-toc-modified-id=\"Create-overlap-1.2.0.1\"><span class=\"toc-item-num\">1.2.0.1 </span>Create overlap</a></span></li><li><span><a href=\"#Activity-representation\" data-toc-modified-id=\"Activity-representation-1.2.0.2\"><span class=\"toc-item-num\">1.2.0.2 </span>Activity representation</a></span></li></ul></li></ul></li></ul></li><li><span><a href=\"#The-example\" data-toc-modified-id=\"The-example-2\"><span class=\"toc-item-num\">2 </span>The example</a></span></li></ul></div>",
"_____no_output_____"
],
[
"# Preamble",
"_____no_output_____"
]
],
[
[
"import pprint\nimport subprocess \nimport sys \nsys.path.append('../')\n\nimport numpy as np\nimport scipy as sp\nimport statsmodels.api as sm\nimport matplotlib.pyplot as plt\nimport matplotlib\nimport matplotlib.gridspec as gridspec\nfrom mpl_toolkits.axes_grid1 import make_axes_locatable\nimport seaborn as sns\n\n%matplotlib inline\n\nnp.set_printoptions(suppress=True, precision=5)\n\nfrom network import Protocol, NetworkManager, Network\nfrom patterns_representation import PatternsRepresentation\nfrom analysis_functions import calculate_persistence_time, calculate_recall_quantities, calculate_triad_connectivity\nfrom plotting_functions import plot_weight_matrix, plot_network_activity_angle, plot_persistent_matrix",
"_____no_output_____"
]
],
[
[
"## Some general parameters",
"_____no_output_____"
]
],
[
[
"epsilon = 10e-80\nvmin = -3.0\nremove = 0.010\n\nstrict_maximum = True\n\ndt = 0.001\ntau_s = 0.010\ntau_a = 0.250\ng_I = 2.0\ng_a = 2.0\nG = 50.0\n\nsns.set(font_scale=3.5)\nsns.set_style(\"whitegrid\", {'axes.grid': False})\nplt.rcParams['figure.figsize'] = (12, 8)\nlw = 10\nms = 22\nalpha_graph = 0.3\ncolors = sns.color_palette()",
"_____no_output_____"
]
],
[
[
"## Functions",
"_____no_output_____"
],
[
"#### Create overlap",
"_____no_output_____"
]
],
[
[
"from copy import deepcopy\n\ndef create_overalaped_representation(manager, representation_overlap, sequence_overlap):\n x = deepcopy(manager.canonical_activity_representation)\n\n to_modify = int(representation_overlap * len(x[0]))\n sequence_size = int(0.5 * len(x))\n sequence_overlap_size = int(sequence_overlap * sequence_size)\n start_point = int(0.5 * sequence_size + sequence_size - np.floor(sequence_overlap_size/ 2.0))\n end_point = start_point + sequence_overlap_size\n\n for sequence_index in range(start_point, end_point):\n pattern = x[sequence_index]\n pattern[:to_modify] = manager.canonical_activity_representation[sequence_index - sequence_size][:to_modify]\n\n return x",
"_____no_output_____"
]
],
[
[
"#### Activity representation",
"_____no_output_____"
]
],
[
[
"activity_representation = np.array([[0, 0, 0], [1, 1, 1], [2, 2, 2], [3, 3, 3], [4, 4, 4], [5, 5, 5], \n [10, 10, 10], [11, 11, 11], [2, 2, 12], [3, 3, 13], [14, 14, 14], [15, 15, 15]])",
"_____no_output_____"
]
],
[
[
"# The example",
"_____no_output_____"
]
],
[
[
"sigma_out = 0.0\ntau_z_pre = 0.025\ntau_z_post = 0.020\n\nhypercolumns = 3\nminicolumns = 20\nn_patterns = 12\npatterns_per_sequence = 6\nrepresentation_overlap = 0.75\nsequence_overlap = 0.5\n\n# Training protocol\ntraining_times_base = 0.100\ntraining_times = [training_times_base for i in range(n_patterns)]\nipi_base = 0.0\ninter_pulse_intervals = [ipi_base for i in range(n_patterns)]\ninter_sequence_interval = 1.0\nresting_time = 1.0\nepochs = 1\nT_persistence = 1.0 / patterns_per_sequence\n\n# Manager properties\nvalues_to_save = ['o']\n\n\n# Neural Network\nnn = Network(hypercolumns, minicolumns, G=G, tau_s=tau_s, tau_z_pre=tau_z_pre, tau_z_post=tau_z_post,\n tau_a=tau_a, g_a=g_a, g_I=g_I, sigma_out=sigma_out, epsilon=epsilon, prng=np.random,\n strict_maximum=strict_maximum, perfect=False, normalized_currents=True)\n\n\n# Build the manager\nmanager = NetworkManager(nn=nn, dt=dt, values_to_save=values_to_save)\n\n# Build the representation\nrepresentation = PatternsRepresentation(activity_representation, minicolumns=minicolumns)\ninter_pulse_intervals[patterns_per_sequence - 1] = inter_sequence_interval\n\n\n# Build the protocol\nprotocol = Protocol()\nprotocol.simple_protocol(representation, training_times=training_times, inter_pulse_intervals=inter_pulse_intervals,\n inter_sequence_interval=inter_sequence_interval, epochs=epochs, resting_time=resting_time)\n\n# Run the protocol\ntimed_input = manager.run_network_protocol_offline(protocol=protocol)\n# Set the persistent time\nmanager.set_persistent_time_with_adaptation_gain(T_persistence=T_persistence, from_state=1, to_state=2)",
"../connectivity_functions.py:15: RuntimeWarning: invalid value encountered in true_divide\n x = P / outer\n"
],
[
"patterns_per_sequence = 6\nT_cue = 2.0 * manager.nn.tau_s\nT_recall = T_persistence * 6 + T_cue\n\nnr1 = representation.network_representation[:patterns_per_sequence]\nnr2 = representation.network_representation[patterns_per_sequence:]\n\n# Success 1\naux1 = calculate_recall_quantities(manager, nr1, T_recall, T_cue, remove=remove, reset=True, empty_history=True)\nsuccess1, pattern_sequence1, persistent_times1, timings1 = aux1\n\n# Success 2\naux2 = calculate_recall_quantities(manager, nr2, T_recall, T_cue, remove=remove, reset=False, empty_history=False)\nsuccess2, pattern_sequence2, persistent_times2, timings2 = aux2\n\ntotal_success = 0.5 * success1 + 0.5 * success2",
"_____no_output_____"
],
[
"cmap = matplotlib.cm.binary\nax1, ax2 = plot_network_activity_angle(manager, cmap=cmap, time_y=False)\nax1.axvline(T_recall - tau_s, ls='--', color='red')",
"_____no_output_____"
],
[
"nr1 = representation.network_representation[:patterns_per_sequence]\nnr2 = representation.network_representation[patterns_per_sequence:]\n\n# Success 1\naux1 = calculate_recall_quantities(manager, nr1, T_recall, T_cue, remove=remove, reset=True, empty_history=True)\nsuccess1, pattern_sequence1, persistent_times1, timings1 = aux1\no1 = manager.history['o']\n\n# Success 2\naux2 = calculate_recall_quantities(manager, nr2, T_recall, T_cue, remove=remove, reset=True, empty_history=True)\nsuccess2, pattern_sequence2, persistent_times2, timings2 = aux2\no2 = manager.history['o']",
"_____no_output_____"
],
[
"rect = plt.Rectangle((0.29, 0.2), 0.2, 0.72, linewidth=1, edgecolor='black',facecolor='gray', alpha=0.2)\nrect2 = plt.Rectangle((0.29, 0.2), 0.2, 0.72, linewidth=1, edgecolor='black',facecolor='gray', alpha=0.2)\nrect3 = plt.Rectangle((0.29, 0.2), 0.2, 0.72, linewidth=1, edgecolor='black',facecolor='gray', alpha=0.2)\n\n\nfactor_scale = 1.5\ns1 = int(16 * factor_scale)\ns2 = int(12 * factor_scale)\n\ngs = gridspec.GridSpec(2, 2)\nfig = plt.figure(figsize=(s1, s2))\nax = fig.add_subplot(gs[:, 0])\nax.set_ylim([0, minicolumns * hypercolumns])\n\nstart = T_persistence * 0.25\ndx = T_persistence\ndy = minicolumns * 0.4\ny1 = 28\ny2 = 2\n\nsequence1 = activity_representation[:patterns_per_sequence]\nsequence2 = activity_representation[patterns_per_sequence:]\n\nx = start\nfor pattern in sequence1:\n for index, unit in enumerate(pattern): \n ax.text(x, y1 + index*dy, str(pattern[index]))\n x += dx\n\nx = start \nfor pattern in sequence2:\n for index, unit in enumerate(pattern): \n ax.text(x, y2 + index*dy, str(pattern[hypercolumns - 1- index]))\n x += dx\n\nax.add_patch(rect)\n#ax.axis('off')\n\nax2 = fig.add_subplot(gs[0, 1])\n\n\ncmap = matplotlib.cm.binary\nextent = [0, manager.T_recall_total, 0, minicolumns * hypercolumns]\nax2.imshow(o1.T, origin='lower', cmap=cmap, aspect='auto', extent=extent)\n\nax2.add_patch(rect2)\n\n\nax3 = fig.add_subplot(gs[1, 1])\n\ncmap = matplotlib.cm.binary\nextent = [0, manager.T_recall_total, 0, minicolumns * hypercolumns]\nax3.imshow(o2.T, origin='lower', cmap=cmap, aspect='auto', extent=extent)\n\nax3.add_patch(rect3)\n",
"_____no_output_____"
],
[
"rect = plt.Rectangle((2*T_persistence, 0.2), 2*T_persistence, 55,\n linewidth=1, edgecolor='black',facecolor='gray', alpha=0.2)\nrect2 = plt.Rectangle((2*T_persistence, 0.2), 2*T_persistence, minicolumns * hypercolumns, \n linewidth=1, edgecolor='black',facecolor='gray', alpha=0.2)\n\nintersection = {2, 3}\n\nsave = True\nframe_recall = False\nsns.set(font_scale=3.5)\nsns.set_style(\"whitegrid\", {'axes.grid': False})\n\nfactor_scale = 1.6\ns1 = int(16 * factor_scale)\ns2 = int(12 * factor_scale)\n\ngs = gridspec.GridSpec(2, 2)\nfig = plt.figure(figsize=(s1, s2))\nax = fig.add_subplot(gs[:, 0])\nax.set_ylim([0, minicolumns * hypercolumns])\n\nstart = T_persistence * 0.25\ndx = T_persistence\ndy = minicolumns * 0.4\ny1 = 32\ny2 = 4\n\nsequence1 = activity_representation[:patterns_per_sequence]\nsequence2 = activity_representation[patterns_per_sequence:]\n\nx = start\nfor pattern in sequence2:\n for index, unit in enumerate(pattern): \n if pattern[index] in intersection:\n color = 'black'\n else:\n color = 'blue'\n ax.text(x, y1 + index*dy, str(pattern[index]), color=color)\n x += dx\n\nx = start \nfor pattern in sequence1:\n for index, unit in enumerate(pattern):\n if pattern[index] in intersection:\n color = 'black'\n else:\n color = 'red'\n ax.text(x, y2 + index*dy, str(pattern[index] + 1), color=color)\n x += dx\n\nax.add_patch(rect)\nax.axis('off')\n#ax.text(0.3, 25, r'{', rotation=270, fontsize=150)\n\n################\n# The sequence recall\n###############\nax2 = fig.add_subplot(gs[:, 1])\n\ncmap.set_under(color='red')\ncmap.set_over(color='blue')\n\ncmap = matplotlib.cm.binary\nextent = [0, manager.T_recall_total, 0, minicolumns * hypercolumns]\nax2.imshow((-1 * o1 + 2 * o2).T, origin='lower', cmap=cmap, aspect='auto', extent=extent, vmin=0.0, vmax=1.0)\nax2.axhline(minicolumns, ls='--', color='gray')\nax2.axhline(2 * minicolumns, ls='--', color='gray')\n\nax2.set_xlabel('Time (s)')\nax2.set_ylabel('Unit Id')\n\nax2.add_patch(rect2);\n\nif not frame_recall:\n ax2.spines['top'].set_visible(False)\n ax2.spines['right'].set_visible(False)\n ax2.spines['bottom'].set_visible(False)\n ax2.spines['left'].set_visible(False)\n \n \nfig.tight_layout()\nif save:\n directory = '../plot_producers/'\n file_name = 'rep_diagram' \n format_string = '.svg'\n string_to_save = directory + file_name + format_string\n fig.savefig(string_to_save, frameon=False, dpi=110, bbox_inches='tight', transparent=True)",
"_____no_output_____"
],
[
"2 in {2, 3}",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e45bd435a581495871625eb0d527bf9ef103b4 | 45,826 | ipynb | Jupyter Notebook | other/python_notebook.ipynb | nickwotton/MQP2019 | 8d7e7db5d47d605a605aeb394cf5b8af2b13e7fb | [
"MIT"
] | 1 | 2019-09-13T16:27:14.000Z | 2019-09-13T16:27:14.000Z | other/python_notebook.ipynb | nickwotton/MQP2019 | 8d7e7db5d47d605a605aeb394cf5b8af2b13e7fb | [
"MIT"
] | 40 | 2019-09-02T17:49:28.000Z | 2020-04-06T13:03:43.000Z | other/python_notebook.ipynb | nickwotton/MQP2019 | 8d7e7db5d47d605a605aeb394cf5b8af2b13e7fb | [
"MIT"
] | 3 | 2019-09-11T01:51:56.000Z | 2019-12-03T20:13:16.000Z | 177.620155 | 38,930 | 0.886396 | [
[
[
"## Abstract \n\n- Goal: Learn python basics on Jupyter notebook.\n- Ref: \n - Python crash course (Google for it)\n - the [naming section](https://github.com/google/styleguide/blob/gh-pages/pyguide.md#316-naming) of Google's python style guide",
"_____no_output_____"
],
[
"## Python on Jupyter notebook - Colab\n\nPython on Jupyter notebook allows one to perform a great deal of data analysis and statistical validation. cell by cell in a interactive way. A good online platform is colab, powered by google. Another good cloud platform is azure notebook, powered by microsoft. One can also install anaconda with python 3 on local computer to run Jupyter notebook.\n\n- open a new python3 notebook by visiting https://colab.research.google.com/\n- As you can see, each cell can be chosen either code or text from its top menu.\n- A code cell will be evaluated when you press play, or when you press the shortcut, shift-enter. ",
"_____no_output_____"
]
],
[
[
"2 + 2 #Executing a Command",
"_____no_output_____"
]
],
[
[
"## Importing Libraries\n\nThe vast majority of the time, you'll want to use functions from pre-built libraries. You can't import every library each time, but you can import most of the common scientific ones. Here I import numpy, pandas, and matplotlib, the most common and useful libraries. \n\nNotice that you can rename libraries to whatever you want after importing. The `as` statement allows this. Here we use `np` and `pd` as aliases for `numpy` and `pandas`. This is a very common aliasing and will be found in most code snippets around the web. The point behind this is to allow you to type fewer characters when you are frequently accessing these libraries.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"## Using numpy and matplotlib: A sampling example",
"_____no_output_____"
],
[
"Let's use numpy to select 100 standard normal radom variables, and make a plot.",
"_____no_output_____"
]
],
[
[
"X = np.random.normal(0, 1, 100)\n\nplt.plot(X);\nplt.xlabel('Time') \nplt.ylabel('Returns')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Let's use `numpy` to take some simple statistics.",
"_____no_output_____"
]
],
[
[
"np.mean(X)",
"_____no_output_____"
],
[
"np.std(X)",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d0e46390435ef3afbbb1d9a2af1a73fc02ca1cab | 2,241 | ipynb | Jupyter Notebook | keras/applications.ipynb | Rockung/sma | 4e2dad5bff698c78fbb4beea1cfe45534ed5ff1c | [
"Apache-2.0"
] | null | null | null | keras/applications.ipynb | Rockung/sma | 4e2dad5bff698c78fbb4beea1cfe45534ed5ff1c | [
"Apache-2.0"
] | null | null | null | keras/applications.ipynb | Rockung/sma | 4e2dad5bff698c78fbb4beea1cfe45534ed5ff1c | [
"Apache-2.0"
] | null | null | null | 27 | 155 | 0.589469 | [
[
[
"from keras.applications.resnet50 import ResNet50\nfrom keras.preprocessing import image\nfrom keras.applications.resnet50 import preprocess_input, decode_predictions\nimport numpy as np\n\nmodel = ResNet50(weights='imagenet')\n\nimg_path = 'elephant.jpg'\nimg = image.load_img(img_path, target_size=(224, 224))\nx = image.img_to_array(img)\nx = np.expand_dims(x, axis=0)\nx = preprocess_input(x)\n\npreds = model.predict(x)\n# decode the results into a list of tuples (class, description, probability)\n# (one such list for each sample in the batch)\nprint('Predicted:', decode_predictions(preds, top=3)[0])\n# Predicted: [(u'n02504013', u'Indian_elephant', 0.82658225), (u'n01871265', u'tusker', 0.1122357), (u'n02504458', u'African_elephant', 0.061040461)]",
"_____no_output_____"
],
[
"from keras.applications.vgg16 import VGG16\nfrom keras.preprocessing import image\nfrom keras.applications.vgg16 import preprocess_input\nimport numpy as np\n\nmodel = VGG16(weights='imagenet', include_top=False)\n\nimg_path = 'elephant.jpg'\nimg = image.load_img(img_path, target_size=(224, 224))\nx = image.img_to_array(img)\nx = np.expand_dims(x, axis=0)\nx = preprocess_input(x)\n\nfeatures = model.predict(x)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
d0e4671bd38da0e0b33964d00ffc68ec627d3b3a | 9,760 | ipynb | Jupyter Notebook | Deep Learning/NLP Tutorials/.ipynb_checkpoints/Conv_LSTM_Stack_in_keras-checkpoint.ipynb | surajch77/ML-Examples | 10f68c0a1b5367b63ec0505e74bfb40aff437125 | [
"Apache-2.0"
] | 2 | 2018-10-07T09:33:50.000Z | 2020-09-12T14:02:29.000Z | Deep Learning/NLP Tutorials/.ipynb_checkpoints/Conv_LSTM_Stack_in_keras-checkpoint.ipynb | surajch77/ML-Examples | 10f68c0a1b5367b63ec0505e74bfb40aff437125 | [
"Apache-2.0"
] | null | null | null | Deep Learning/NLP Tutorials/.ipynb_checkpoints/Conv_LSTM_Stack_in_keras-checkpoint.ipynb | surajch77/ML-Examples | 10f68c0a1b5367b63ec0505e74bfb40aff437125 | [
"Apache-2.0"
] | null | null | null | 23.746959 | 145 | 0.515061 | [
[
[
"# BiDirectional LSTM classifier in keras",
"_____no_output_____"
],
[
"#### Load dependencies",
"_____no_output_____"
]
],
[
[
"import keras\nfrom keras.datasets import imdb\nfrom keras.preprocessing.sequence import pad_sequences\nfrom keras.models import Sequential\nfrom keras.layers import Embedding, SpatialDropout1D, Dense, Flatten, Dropout, LSTM\nfrom keras.layers.wrappers import Bidirectional\nfrom keras.callbacks import ModelCheckpoint # new! \nimport os # new! \nfrom sklearn.metrics import roc_auc_score, roc_curve # new!\nimport matplotlib.pyplot as plt # new!\n%matplotlib inline",
"_____no_output_____"
],
[
"# output directory name:\noutput_dir = 'model_output/bilstm'\n\n# training:\nepochs = 6\nbatch_size = 128\n\n# vector-space embedding: \nn_dim = 64\nn_unique_words = 10000\nmax_review_length = 200\npad_type = trunc_type = 'pre'\ndrop_embed = 0.2\n\n# neural network architecture: \nn_lstm = 256\ndroput_lstm = 0.2",
"_____no_output_____"
]
],
[
[
"#### Load data",
"_____no_output_____"
],
[
"For a given data set: \n\n* the Keras text utilities [here](https://keras.io/preprocessing/text/) quickly preprocess natural language and convert it into an index\n* the `keras.preprocessing.text.Tokenizer` class may do everything you need in one line:\n * tokenize into words or characters\n * `num_words`: maximum unique tokens\n * filter out punctuation\n * lower case\n * convert words to an integer index",
"_____no_output_____"
]
],
[
[
"(x_train, y_train), (x_valid, y_valid) = imdb.load_data(num_words=n_unique_words)",
"_____no_output_____"
]
],
[
[
"#### Preprocess data",
"_____no_output_____"
]
],
[
[
"x_train = pad_sequences(x_train, maxlen=max_review_length, padding=pad_type, truncating=trunc_type, value=0)\nx_valid = pad_sequences(x_valid, maxlen=max_review_length, padding=pad_type, truncating=trunc_type, value=0)",
"_____no_output_____"
],
[
"x_train[:6]",
"_____no_output_____"
],
[
"for i in range(6):\n print len(x_train[i])",
"200\n200\n200\n200\n200\n200\n"
]
],
[
[
"#### Design neural network architecture",
"_____no_output_____"
]
],
[
[
"model = Sequential()\nmodel.add(Embedding(n_unique_words, n_dim, input_length=max_review_length)) \nmodel.add(SpatialDropout1D(drop_embed))\n# model.add(Conv1D(n_conv, k_conv, activation='relu'))\n# model.add(Conv1D(n_conv, k_conv, activation='relu'))\n# model.add(GlobalMaxPooling1D())\n# model.add(Dense(n_dense, activation='relu'))\n# model.add(Dropout(dropout))\nmodel.add(Bidirectional(LSTM(n_lstm, dropout=droput_lstm)))\nmodel.add(Dense(1, activation='sigmoid'))",
"_____no_output_____"
],
[
"model.summary()",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nembedding_1 (Embedding) (None, 200, 64) 640000 \n_________________________________________________________________\nspatial_dropout1d_1 (Spatial (None, 200, 64) 0 \n_________________________________________________________________\nbidirectional_1 (Bidirection (None, 512) 657408 \n_________________________________________________________________\ndense_1 (Dense) (None, 1) 513 \n=================================================================\nTotal params: 1,297,921\nTrainable params: 1,297,921\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"n_dim, n_unique_words, n_dim * n_unique_words",
"_____no_output_____"
],
[
"max_review_length, n_dim, n_dim * max_review_length",
"_____no_output_____"
]
],
[
[
"#### Configure Model",
"_____no_output_____"
]
],
[
[
"model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])",
"_____no_output_____"
],
[
"modelcheckpoint = ModelCheckpoint(filepath=output_dir+\"/weights.{epoch:02d}.hdf5\")",
"_____no_output_____"
],
[
"if not os.path.exists(output_dir):\n os.makedirs(output_dir)",
"_____no_output_____"
],
[
"model.fit(x_train, y_train, batch_size=batch_size, epochs=epochs, \n verbose=1, validation_data=(x_valid, y_valid), callbacks=[modelcheckpoint])",
"Train on 25000 samples, validate on 25000 samples\nEpoch 1/6\n25000/25000 [==============================] - 689s 28ms/step - loss: 0.5523 - acc: 0.7024 - val_loss: 0.3817 - val_acc: 0.8368\nEpoch 2/6\n 640/25000 [..............................] - ETA: 7:21 - loss: 0.2883 - acc: 0.8953"
]
],
[
[
"#### Evaluate",
"_____no_output_____"
]
],
[
[
"model.load_weights(output_dir+\"/weights.01.hdf5\") # zero-indexed",
"_____no_output_____"
],
[
"y_hat = model.predict_proba(x_valid)",
"_____no_output_____"
],
[
"len(y_hat)",
"_____no_output_____"
],
[
"y_hat[0]",
"_____no_output_____"
],
[
"plt.hist(y_hat)\n_ = plt.axvline(x=0.5, color='orange')",
"_____no_output_____"
],
[
"pct_auc = roc_auc_score(y_valid, y_hat)*100.0",
"_____no_output_____"
],
[
"\"{:0.2f}\".format(pct_auc)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e470a6f9ccd9942c4f8f8b7bb4188cd1b69218 | 40,421 | ipynb | Jupyter Notebook | notebooks/random_forest.ipynb | AshwathSalimath/amazon-lab | 27f9e179d85bf12552192fae5c495e1755f02536 | [
"Unlicense"
] | null | null | null | notebooks/random_forest.ipynb | AshwathSalimath/amazon-lab | 27f9e179d85bf12552192fae5c495e1755f02536 | [
"Unlicense"
] | null | null | null | notebooks/random_forest.ipynb | AshwathSalimath/amazon-lab | 27f9e179d85bf12552192fae5c495e1755f02536 | [
"Unlicense"
] | 1 | 2020-03-01T14:04:37.000Z | 2020-03-01T14:04:37.000Z | 51.755442 | 13,520 | 0.578734 | [
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport math\n\nfrom sklearn.model_selection import train_test_split",
"_____no_output_____"
]
],
[
[
"# Header1",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv('../data/raw/yellow_tripdata_2015-12-sample.csv')\ndf.head()",
"_____no_output_____"
],
[
"df['delta_lon'] = df.pickup_longitude - df.dropoff_longitude\ndf['delta_lat'] = df.pickup_latitude - df.dropoff_latitude\n\ndef calculate_direction(d_lon, d_lat):\n result = np.zeros(len(d_lon))\n l = np.sqrt(d_lon**2 + d_lat**2)\n result[d_lon>0] = (180/np.pi)*np.arcsin(d_lat[d_lon>0]/l[d_lon>0])\n idx = (d_lon<0) & (d_lat>0)\n result[idx] = 180 - (180/np.pi)*np.arcsin(d_lat[idx]/l[idx])\n idx = (d_lon<0) & (d_lat<0)\n result[idx] = -180 - (180/np.pi)*np.arcsin(d_lat[idx]/l[idx])\n return result\n\ndf['direction'] = calculate_direction(df.delta_lon, df.delta_lat)\n\ndf.head()",
"_____no_output_____"
],
[
"df['tpep_pickup_datetime'] = pd.to_datetime(df['tpep_pickup_datetime'])\ndf['day_of_week'] = df['tpep_pickup_datetime'].dt.weekday\n\ndf['hour_of_day'] = df['tpep_pickup_datetime'].dt.hour\n\ndf.head()",
"_____no_output_____"
],
[
"X = df[['passenger_count','trip_distance','trip_time','pickup_longitude','pickup_latitude','dropoff_longitude','dropoff_latitude','direction','day_of_week','hour_of_day']]\ny = df[['fare_amount']]\n\nX.head()",
"_____no_output_____"
],
[
"X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=42)\n",
"_____no_output_____"
],
[
"from sklearn.ensemble import RandomForestRegressor\nregr = RandomForestRegressor(max_depth=6, random_state=0,n_estimators=100)\n\nregr.fit(X_train,y_train)\nprint(regr.feature_importances_)\nplt.barh(range(len(X_train.columns)), regr.feature_importances_)\nplt.yticks(range(len(X_train.columns)), list(X_train));",
"C:\\Users\\Cathal\\Anaconda3\\lib\\site-packages\\ipykernel_launcher.py:4: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples,), for example using ravel().\n after removing the cwd from sys.path.\n"
],
[
"from sklearn.metrics import mean_squared_error\ny_pred = regr.predict(X_test)\n\n\nprint(np.sqrt(mean_squared_error(y_test, y_pred)))",
"4.7793882127762615\n"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e47b61efdc14d76a1171844f2a472633d5b41e | 8,923 | ipynb | Jupyter Notebook | ActiveFitContour.ipynb | YashGunjal/Active-shape-model-for-tooth-Segmentation | 8e22e1489fd84c221c8ed12c1ff1b6c78501c1c6 | [
"MIT"
] | 10 | 2020-07-08T08:57:17.000Z | 2021-11-25T14:18:42.000Z | ActiveFitContour.ipynb | YashGunjal/Active-shape-model-for-tooth-Segmentation | 8e22e1489fd84c221c8ed12c1ff1b6c78501c1c6 | [
"MIT"
] | 1 | 2018-12-28T13:55:53.000Z | 2018-12-28T13:55:53.000Z | ActiveFitContour.ipynb | YashGunjal/Active-shape-model-for-tooth-Segmentation | 8e22e1489fd84c221c8ed12c1ff1b6c78501c1c6 | [
"MIT"
] | 7 | 2018-07-09T03:13:27.000Z | 2019-12-20T04:17:39.000Z | 32.926199 | 92 | 0.501961 | [
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nfrom skimage.color import rgb2gray\nfrom skimage.filters import gaussian\nimport scipy\nimport cv2\nfrom scipy import ndimage\nimport Image_preperation as prep\nimport FitFunction as fit\nimport FileManager as fm\nimport Image_preperation as prep\n \ndef calc_mean(points):\n \n size = len(points)\n p1 = points[-1]\n p2 = points[0]\n mean_sum = scipy.spatial.distance.euclidean(p1,p2)\n \n for i in range(size-1):\n p1 = points[i]\n p2 = points[i+1]\n mean_sum += scipy.spatial.distance.euclidean(p1,p2)\n \n return mean_sum / size\n\n\ndef calc_internal2(p1,p2,mean_points):\n return np.sum( (p2 - p1)**2 ) / mean_points\n\ndef calc_internal(p1,p2,mean_points):\n return scipy.spatial.distance.euclidean(p1,p2) / mean_points\n\ndef calc_external_img2(img):\n median = prep.median_filter(img)\n edges = prep.edge_detection_low(median)\n return -edges\n\ndef calc_external_img(img): \n\n img = np.array(img, dtype=np.int16)\n kx = np.array([[-1,0,1],[-2,0,2],[-1,0,1]])\n Gx = cv2.filter2D(img,-1,kx)\n \n ky = np.array([[-1,-2,-1],[0,0,0],[1,2,1]])\n Gy = cv2.filter2D(img,-1,ky)\n \n G = np.sqrt(Gx**2 + Gy**2)\n \n return G\n \ndef calc_external(p, external_img):\n \n p = p.astype(int)\n max_value = np.abs(np.min(external_img))\n \n return external_img[p[1],p[0]] / max_value\n \ndef calc_energy(p1, p2, external_img, mean_points,alpha):\n \n internal = calc_internal(p1,p2, mean_points)\n external = calc_external(p1, external_img)\n \n return internal + alpha * external\n\n\ndef get_point_state(point, rad, number, pixel_width):\n \n positive = number // 2\n if(positive == 1):\n state = (number + 1) / 2\n else:\n state = -(number / 2)\n \n return fit.get_point_at_distance(point, state, rad)\n\ndef unpack(number, back_pointers, angles, points, pixel_width):\n \n size = len(points)\n new_points = np.empty((size,2))\n \n new_points[-1] = get_point_state(points[-1],angles[-1], number, pixel_width)\n pointer = back_pointers[-1,number]\n \n for i in range(size-2, -1, -1):\n \n new_points[i] = get_point_state(points[i],angles[i], pointer, pixel_width)\n pointer = back_pointers[i,pointer]\n \n return new_points\n\n#https://courses.engr.illinois.edu/cs447/fa2017/Slides/Lecture07.pdf\n#viterbi algo\ndef active_contour(points, edge_img, pixel_width, alpha):\n size = len(points)\n num_states = (2*pixel_width +1)\n \n trellis = np.zeros((size, num_states), dtype=np.float16)\n back_pointers = np.zeros((size, num_states), dtype=int)\n #external_img = calc_external_img(img)\n if(np.dtype('bool') == edge_img.dtype):\n external_img = -np.array(edge_img,dtype=np.int8)\n else:\n external_img = -edge_img \n mean_points = calc_mean(points)\n \n #init\n trellis[0,:] = np.zeros((num_states))\n back_pointers[0,:] = np.zeros((num_states))\n \n angles = get_angles_of(points)\n \n #recursion\n for i in range(1, size):\n for t in range(num_states):\n trellis[i,t] = np.inf\n for d in range(num_states):\n p1 = get_point_state(points[i-1], angles[i-1], d, pixel_width)\n p2 = get_point_state(points[i],angles[i], t, pixel_width)\n energy_trans = calc_energy(p1, p2, external_img,mean_points, alpha)\n\n tmp = trellis[i-1,d] + energy_trans\n\n if(tmp < trellis[i,t]):\n trellis[i,t] = tmp\n back_pointers[i,t] = d\n \n #find best\n t_best, vit_min = 0, np.inf\n for t in range(num_states):\n if(trellis[size-1, t] < vit_min):\n t_best = t\n vit_min = trellis[size-1, t]\n\n new_points = unpack(t_best, back_pointers,angles, points, pixel_width)\n return new_points\n\n\ndef active_contour_loop(points, img, max_loop, pixel_width, alpha):\n \n old_points = points\n for i in range(max_loop):\n new_points = active_contour(old_points, img, pixel_width, alpha)\n if np.array_equal(new_points, old_points):\n print(i)\n break\n \n #old_points = new_points\n head, tail = np.split(new_points, [6])\n old_points = np.append(tail, head).reshape(new_points.shape)\n \n return new_points\n\ndef resolution_scale(img, points, scale):\n new_points = resolution_scale_points(points, scale)\n new_img = resolution_downscale_img(img, scale)\n return new_img, new_points\n\ndef resolution_scale_points(points, scale): \n return np.around(points*scale)\n\ndef resolution_downscale_img(img, scale): \n x, y = img.shape\n xn = int(x*scale)\n yn = int(y*scale)\n\n return cv2.resize(img, (yn ,xn)) \n\ndef get_angles_of(points):\n \n size = len(points)\n angles = np.zeros(size)\n \n for i in range(size):\n \n if(i==size-1):\n p1, p2, p3 = points[i-1], points[i], points[0] \n else:\n p1, p2, p3 = points[i-1], points[i], points[i+1]\n\n angles[i] = fit.get_normal_angle(p1, p2, p3)\n \n return angles \n \n\ndef show_results():\n piece = fm.load_img_piece()\n edge_img = prep.canny(piece)\n tooth = fm.load_tooth_of_piece(2)\n fm.show_with_points(edge_img, tooth)\n\n new_tooth = active_contour(tooth, edge_img, 25, 1)\n fm.show_with_points(edge_img, new_tooth)\n \ndef show_influence_ext_int():\n new_piece, new_tooth = piece, tooth \n\n mean = calc_mean(new_tooth)\n ext = calc_external_img(new_piece)\n fm.show_with_points(ext, new_tooth[0:2])\n\n print(calc_external(new_tooth[0],ext))\n print(calc_internal(new_tooth[0], new_tooth[1], mean))\n print(calc_energy(new_tooth[0],new_tooth[1],ext,mean,10))\n",
"_____no_output_____"
],
[
"if __name__ == \"__main__\":\n\n piece = fm.load_img_piece()\n tooth = fm.load_tooth_of_piece()\n ext = prep.calc_external_img_active_contour(piece) \n fm.show_with_points(ext, tooth)\n ext2, stooth = fm.resolution_scale(ext, tooth, 1/6)\n fm.show_with_points(ext2, stooth)",
"\nKeyboardInterrupt\n\n"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
d0e4847635c81a1ec9aa0fe9f1cd2c170fe8c971 | 189,044 | ipynb | Jupyter Notebook | homework/homework_classification.ipynb | sathwikiyer/NEU-365P-385L-Spring-2021 | e0735657771caa12f052d82253d53b07d4ed128f | [
"Unlicense"
] | null | null | null | homework/homework_classification.ipynb | sathwikiyer/NEU-365P-385L-Spring-2021 | e0735657771caa12f052d82253d53b07d4ed128f | [
"Unlicense"
] | null | null | null | homework/homework_classification.ipynb | sathwikiyer/NEU-365P-385L-Spring-2021 | e0735657771caa12f052d82253d53b07d4ed128f | [
"Unlicense"
] | null | null | null | 266.634697 | 54,792 | 0.901494 | [
[
[
"# Homework (15 pts) - Classification",
"_____no_output_____"
]
],
[
[
"# Questions are based on the mouse cortex protein expression level dataset used in lecture.\n# The Data_Cortex_Nuclear.csv file is available in the same folder as this notebook\n# or at https://www.kaggle.com/ruslankl/mice-protein-expression\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.metrics import confusion_matrix, ConfusionMatrixDisplay\nfrom sklearn.metrics import classification_report\nfrom sklearn.metrics import roc_curve, auc\nfrom sklearn.preprocessing import label_binarize\nfrom sklearn.multiclass import OneVsRestClassifier\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.metrics import accuracy_score\n\nplt.rcParams['axes.labelsize'] = 14\nplt.rcParams['axes.titlesize'] = 14\nplt.rcParams['legend.fontsize'] = 12\nplt.rcParams['figure.figsize'] = (8, 5)\n%config InlineBackend.figure_format = 'retina'\n\nmice = pd.read_csv('Data_Cortex_Nuclear.csv')\nmice",
"_____no_output_____"
]
],
[
[
"---\n1. (3 pts) Remove mice/samples with missing measurements from the dataframe. Store the result in the mice variable for use with subsequent questions.",
"_____no_output_____"
]
],
[
[
"mice.isnull().sum()\nmice = mice.dropna()",
"_____no_output_____"
]
],
[
[
"---\n2. (3 pts) Use both logistic regression and a random forest to classify the mice as either memantine or saline treated based on their protein expression profiles. Contrast the overall accuracy of the two classifiers on a withheld test set. You can ignore any warnings similar to those seen in the lecture for the purposes of this assignment, not that you should ignore these for a real analysis.",
"_____no_output_____"
]
],
[
[
"X = mice.loc[:,'DYRK1A_N':'CaNA_N'] # just protein expression levels\ny = mice['Treatment']\n\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.4, random_state = 1)\n\nmodel = LogisticRegression()\nmodel.fit(X_train, y_train)\nLogisticRegression()\n\ny_test_pred = model.predict(X_test)\n\nlr_treatment_accuracy = model.score(X_test, y_test)\nlr_treatment_accuracy",
"/Users/sathwikiyer/opt/anaconda3/lib/python3.8/site-packages/sklearn/linear_model/_logistic.py:762: ConvergenceWarning: lbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\n n_iter_i = _check_optimize_result(\n"
],
[
"X = mice.loc[:,'DYRK1A_N':'CaNA_N'] # just protein expression levels\ny = mice['Treatment']\n\nX_train2, X_test2, y_train2, y_test2 = train_test_split(X, y, test_size = 0.4, random_state = 1)\n\nmodel2 = RandomForestClassifier(n_estimators = 100, random_state = 1)\nmodel2.fit(X_train2, y_train2)\n\ny_test_pred2 = model.predict(X_test2)\n\nrf_treatment_accuracy2 = accuracy_score(y_test2, y_test_pred2)\n\nrf_treatment_accuracy2\n\nprint(\"Random forests are a better predictor\")",
"Random forests are a better predictor\n"
]
],
[
[
"---\n3. (3 pts) For the two classifiers trained in #2 above, contrast their confusion matrices based on predictions for a withheld test set.",
"_____no_output_____"
]
],
[
[
"cm = confusion_matrix(y_test, y_test_pred)\n\nlr_treatment_cmd = ConfusionMatrixDisplay(confusion_matrix = cm, display_labels = model.classes_)\nlr_treatment_cmd.plot();\n\ncm = confusion_matrix(y_test, y_test_pred2)\n\nrf_treatment_cmd = ConfusionMatrixDisplay(confusion_matrix = cm, display_labels = model.classes_)\nrf_treatment_cmd.plot();",
"_____no_output_____"
]
],
[
[
"---\n4. (3 pts) For the two classifiers trained in #2 above, contrast their ROC curves based on predictions for Memantine treatment in withheld test set.",
"_____no_output_____"
]
],
[
[
"y_test_proba = model.predict_proba(X_test)\nfpr, tpr, thresholds = roc_curve(y_test, y_test_proba[:,1], pos_label = model.classes_[1])\n\nplt.plot(fpr, tpr, color = 'r', label = model.classes_[0])\nplt.plot([0, 1], [0, 1], color = 'k', linestyle = '--')\nplt.xlabel('False Positive Rate')\nplt.ylabel('True Positive Rate')\nplt.title('ROC Curve')\nplt.legend();\n\ny_test_proba2 = model2.predict_proba(X_test2)\nfpr2, tpr2, thresholds2 = roc_curve(y_test2, y_test_proba2[:,1], pos_label = model2.classes_[1])\n\nplt.plot(fpr2, tpr2, color = 'b', label = model2.classes_[0])\nplt.plot([0, 1], [0, 1], color = 'k', linestyle = '--')\nplt.xlabel('False Positive Rate')\nplt.ylabel('True Positive Rate')\nplt.title('ROC Curve')\nplt.legend();",
"_____no_output_____"
]
],
[
[
"---\n5. (3 pts) Use a random forest to classify the mice into each of the 8 classes defined by genotype, fear conditioning and treatment based on their protein expression profiles. Repeat this classification 77 separate times, where on the ith time you will use only the first i proteins. For example, the 1st time you will only use expression of DYRK1A_N, the 2nd time you will use both DYRK1A_N and ITSN1_N as predictive features, the 3rd time DYRK1A_N, ITSN1_N and BDNF_N, etc. until the 77th time you will use all 77 proteins from DYRK1A_N to CaNA_N. Plot the accuracy of the classifier on a withheld test set as a function of the number of protein features used as predictors.",
"_____no_output_____"
]
],
[
[
"proteins = X_train.shape[1]\naccuracies = np.zeros((proteins,))\n\nrFor = RandomForestClassifier(n_estimators = 100, random_state = 1)\n\nfor i in np.arange(proteins) + 1:\n pTrain = X_train.iloc[:, 0:i]\n pTest = X_test.iloc[:, 0:i]\n rFor.fit(pTrain, y_train)\n rForPred = rFor.predict(pTest)\n rForAcc = accuracy_score(y_test, rForPred)\n accuracies[i-1] = rForAcc\n\nplt.plot(np.arange(proteins) + 1, accuracies)\nplt.xlabel(\"Number of Features\")\nplt.ylabel(\"Accuracy\")\nplt.title(\"Accuracy vs Number of Features\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0e4858c706dc7a341639dd787b818a428e800cd | 76,978 | ipynb | Jupyter Notebook | Course 2 - Natural Language Processing with Probabilistic Models/Week 2/C2_W2_Assignment.ipynb | naveen-9697/Natural-Language-Specialisation---Coursera | 94ed5d8a357460bd8854b86be3d9667db5ddebad | [
"MIT"
] | null | null | null | Course 2 - Natural Language Processing with Probabilistic Models/Week 2/C2_W2_Assignment.ipynb | naveen-9697/Natural-Language-Specialisation---Coursera | 94ed5d8a357460bd8854b86be3d9667db5ddebad | [
"MIT"
] | null | null | null | Course 2 - Natural Language Processing with Probabilistic Models/Week 2/C2_W2_Assignment.ipynb | naveen-9697/Natural-Language-Specialisation---Coursera | 94ed5d8a357460bd8854b86be3d9667db5ddebad | [
"MIT"
] | null | null | null | 41.542364 | 528 | 0.551339 | [
[
[
"# Assignment 2: Parts-of-Speech Tagging (POS)\n\nWelcome to the second assignment of Course 2 in the Natural Language Processing specialization. This assignment will develop skills in part-of-speech (POS) tagging, the process of assigning a part-of-speech tag (Noun, Verb, Adjective...) to each word in an input text. Tagging is difficult because some words can represent more than one part of speech at different times. They are **Ambiguous**. Let's look at the following example: \n\n- The whole team played **well**. [adverb]\n- You are doing **well** for yourself. [adjective]\n- **Well**, this assignment took me forever to complete. [interjection]\n- The **well** is dry. [noun]\n- Tears were beginning to **well** in her eyes. [verb]\n\nDistinguishing the parts-of-speech of a word in a sentence will help you better understand the meaning of a sentence. This would be critically important in search queries. Identifying the proper noun, the organization, the stock symbol, or anything similar would greatly improve everything ranging from speech recognition to search. By completing this assignment, you will: \n\n- Learn how parts-of-speech tagging works\n- Compute the transition matrix A in a Hidden Markov Model\n- Compute the emission matrix B in a Hidden Markov Model\n- Compute the Viterbi algorithm \n- Compute the accuracy of your own model \n",
"_____no_output_____"
],
[
"## Outline\n\n- [0 Data Sources](#0)\n- [1 POS Tagging](#1)\n - [1.1 Training](#1.1)\n - [Exercise 01](#ex-01)\n - [1.2 Testing](#1.2)\n - [Exercise 02](#ex-02)\n- [2 Hidden Markov Models](#2)\n - [2.1 Generating Matrices](#2.1)\n - [Exercise 03](#ex-03)\n - [Exercise 04](#ex-04)\n- [3 Viterbi Algorithm](#3)\n - [3.1 Initialization](#3.1)\n - [Exercise 05](#ex-05)\n - [3.2 Viterbi Forward](#3.2)\n - [Exercise 06](#ex-06)\n - [3.3 Viterbi Backward](#3.3)\n - [Exercise 07](#ex-07)\n- [4 Predicting on a data set](#4)\n - [Exercise 08](#ex-08)",
"_____no_output_____"
]
],
[
[
"# Importing packages and loading in the data set \nfrom utils_pos import get_word_tag, preprocess \nimport pandas as pd\nfrom collections import defaultdict\nimport math\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"<a name='0'></a>\n## Part 0: Data Sources\nThis assignment will use two tagged data sets collected from the **Wall Street Journal (WSJ)**. \n\n[Here](http://relearn.be/2015/training-common-sense/sources/software/pattern-2.6-critical-fork/docs/html/mbsp-tags.html) is an example 'tag-set' or Part of Speech designation describing the two or three letter tag and their meaning. \n- One data set (**WSJ-2_21.pos**) will be used for **training**.\n- The other (**WSJ-24.pos**) for **testing**. \n- The tagged training data has been preprocessed to form a vocabulary (**hmm_vocab.txt**). \n- The words in the vocabulary are words from the training set that were used two or more times. \n- The vocabulary is augmented with a set of 'unknown word tokens', described below. \n\nThe training set will be used to create the emission, transmission and tag counts. \n\nThe test set (WSJ-24.pos) is read in to create `y`. \n- This contains both the test text and the true tag. \n- The test set has also been preprocessed to remove the tags to form **test_words.txt**. \n- This is read in and further processed to identify the end of sentences and handle words not in the vocabulary using functions provided in **utils_pos.py**. \n- This forms the list `prep`, the preprocessed text used to test our POS taggers.\n\nA POS tagger will necessarily encounter words that are not in its datasets. \n- To improve accuracy, these words are further analyzed during preprocessing to extract available hints as to their appropriate tag. \n- For example, the suffix 'ize' is a hint that the word is a verb, as in 'final-ize' or 'character-ize'. \n- A set of unknown-tokens, such as '--unk-verb--' or '--unk-noun--' will replace the unknown words in both the training and test corpus and will appear in the emission, transmission and tag data structures.\n\n\n<img src = \"DataSources1.PNG\" />",
"_____no_output_____"
],
[
"Implementation note: \n\n- For python 3.6 and beyond, dictionaries retain the insertion order. \n- Furthermore, their hash-based lookup makes them suitable for rapid membership tests. \n - If _di_ is a dictionary, `key in di` will return `True` if _di_ has a key _key_, else `False`. \n\nThe dictionary `vocab` will utilize these features.",
"_____no_output_____"
]
],
[
[
"# load in the training corpus\nwith open(\"WSJ_02-21.pos\", 'r') as f:\n training_corpus = f.readlines()\n\nprint(f\"A few items of the training corpus list\")\nprint(training_corpus[0:5])",
"A few items of the training corpus list\n['In\\tIN\\n', 'an\\tDT\\n', 'Oct.\\tNNP\\n', '19\\tCD\\n', 'review\\tNN\\n']\n"
],
[
"# read the vocabulary data, split by each line of text, and save the list\nwith open(\"hmm_vocab.txt\", 'r') as f:\n voc_l = f.read().split('\\n')\n\nprint(\"A few items of the vocabulary list\")\nprint(voc_l[0:50])\nprint()\nprint(\"A few items at the end of the vocabulary list\")\nprint(voc_l[-50:])",
"A few items of the vocabulary list\n['!', '#', '$', '%', '&', \"'\", \"''\", \"'40s\", \"'60s\", \"'70s\", \"'80s\", \"'86\", \"'90s\", \"'N\", \"'S\", \"'d\", \"'em\", \"'ll\", \"'m\", \"'n'\", \"'re\", \"'s\", \"'til\", \"'ve\", '(', ')', ',', '-', '--', '--n--', '--unk--', '--unk_adj--', '--unk_adv--', '--unk_digit--', '--unk_noun--', '--unk_punct--', '--unk_upper--', '--unk_verb--', '.', '...', '0.01', '0.0108', '0.02', '0.03', '0.05', '0.1', '0.10', '0.12', '0.13', '0.15']\n\nA few items at the end of the vocabulary list\n['yards', 'yardstick', 'year', 'year-ago', 'year-before', 'year-earlier', 'year-end', 'year-on-year', 'year-round', 'year-to-date', 'year-to-year', 'yearlong', 'yearly', 'years', 'yeast', 'yelled', 'yelling', 'yellow', 'yen', 'yes', 'yesterday', 'yet', 'yield', 'yielded', 'yielding', 'yields', 'you', 'young', 'younger', 'youngest', 'youngsters', 'your', 'yourself', 'youth', 'youthful', 'yuppie', 'yuppies', 'zero', 'zero-coupon', 'zeroing', 'zeros', 'zinc', 'zip', 'zombie', 'zone', 'zones', 'zoning', '{', '}', '']\n"
],
[
"# vocab: dictionary that has the index of the corresponding words\nvocab = {} \n\n# Get the index of the corresponding words. \nfor i, word in enumerate(sorted(voc_l)): \n vocab[word] = i \n \nprint(\"Vocabulary dictionary, key is the word, value is a unique integer\")\ncnt = 0\nfor k,v in vocab.items():\n print(f\"{k}:{v}\")\n cnt += 1\n if cnt > 20:\n break",
"Vocabulary dictionary, key is the word, value is a unique integer\n:0\n!:1\n#:2\n$:3\n%:4\n&:5\n':6\n'':7\n'40s:8\n'60s:9\n'70s:10\n'80s:11\n'86:12\n'90s:13\n'N:14\n'S:15\n'd:16\n'em:17\n'll:18\n'm:19\n'n':20\n"
],
[
"# load in the test corpus\nwith open(\"WSJ_24.pos\", 'r') as f:\n y = f.readlines()\n\nprint(\"A sample of the test corpus\")\nprint(y[0:10])",
"A sample of the test corpus\n['The\\tDT\\n', 'economy\\tNN\\n', \"'s\\tPOS\\n\", 'temperature\\tNN\\n', 'will\\tMD\\n', 'be\\tVB\\n', 'taken\\tVBN\\n', 'from\\tIN\\n', 'several\\tJJ\\n', 'vantage\\tNN\\n']\n"
],
[
"#corpus without tags, preprocessed\n_, prep = preprocess(vocab, \"test.words\") \n\nprint('The length of the preprocessed test corpus: ', len(prep))\nprint('This is a sample of the test_corpus: ')\nprint(prep[0:10])",
"The length of the preprocessed test corpus: 34199\nThis is a sample of the test_corpus: \n['The', 'economy', \"'s\", 'temperature', 'will', 'be', 'taken', 'from', 'several', '--unk--']\n"
]
],
[
[
"<a name='1'></a>\n# Part 1: Parts-of-speech tagging \n\n<a name='1.1'></a>\n## Part 1.1 - Training\nYou will start with the simplest possible parts-of-speech tagger and we will build up to the state of the art. \n\nIn this section, you will find the words that are not ambiguous. \n- For example, the word `is` is a verb and it is not ambiguous. \n- In the `WSJ` corpus, $86$% of the token are unambiguous (meaning they have only one tag) \n- About $14\\%$ are ambiguous (meaning that they have more than one tag)\n\n<img src = \"pos.png\" style=\"width:400px;height:250px;\"/>\n\nBefore you start predicting the tags of each word, you will need to compute a few dictionaries that will help you to generate the tables. ",
"_____no_output_____"
],
[
"#### Transition counts\n- The first dictionary is the `transition_counts` dictionary which computes the number of times each tag happened next to another tag. \n\nThis dictionary will be used to compute: \n$$P(t_i |t_{i-1}) \\tag{1}$$\n\nThis is the probability of a tag at position $i$ given the tag at position $i-1$.\n\nIn order for you to compute equation 1, you will create a `transition_counts` dictionary where \n- The keys are `(prev_tag, tag)`\n- The values are the number of times those two tags appeared in that order. ",
"_____no_output_____"
],
[
"#### Emission counts\n\nThe second dictionary you will compute is the `emission_counts` dictionary. This dictionary will be used to compute:\n\n$$P(w_i|t_i)\\tag{2}$$\n\nIn other words, you will use it to compute the probability of a word given its tag. \n\nIn order for you to compute equation 2, you will create an `emission_counts` dictionary where \n- The keys are `(tag, word)` \n- The values are the number of times that pair showed up in your training set. ",
"_____no_output_____"
],
[
"#### Tag counts\n\nThe last dictionary you will compute is the `tag_counts` dictionary. \n- The key is the tag \n- The value is the number of times each tag appeared.",
"_____no_output_____"
],
[
"<a name='ex-01'></a>\n### Exercise 01\n\n**Instructions:** Write a program that takes in the `training_corpus` and returns the three dictionaries mentioned above `transition_counts`, `emission_counts`, and `tag_counts`. \n- `emission_counts`: maps (tag, word) to the number of times it happened. \n- `transition_counts`: maps (prev_tag, tag) to the number of times it has appeared. \n- `tag_counts`: maps (tag) to the number of times it has occured. \n\nImplementation note: This routine utilises *defaultdict*, which is a subclass of *dict*. \n- A standard Python dictionary throws a *KeyError* if you try to access an item with a key that is not currently in the dictionary. \n- In contrast, the *defaultdict* will create an item of the type of the argument, in this case an integer with the default value of 0. \n- See [defaultdict](https://docs.python.org/3.3/library/collections.html#defaultdict-objects).",
"_____no_output_____"
]
],
[
[
"# UNQ_C1 (UNIQUE CELL IDENTIFIER, DO NOT EDIT)\n# GRADED FUNCTION: create_dictionaries\ndef create_dictionaries(training_corpus, vocab):\n \"\"\"\n Input: \n training_corpus: a corpus where each line has a word followed by its tag.\n vocab: a dictionary where keys are words in vocabulary and value is an index\n Output: \n emission_counts: a dictionary where the keys are (tag, word) and the values are the counts\n transition_counts: a dictionary where the keys are (prev_tag, tag) and the values are the counts\n tag_counts: a dictionary where the keys are the tags and the values are the counts\n \"\"\"\n \n # initialize the dictionaries using defaultdict\n emission_counts = defaultdict(int)\n transition_counts = defaultdict(int)\n tag_counts = defaultdict(int)\n \n # Initialize \"prev_tag\" (previous tag) with the start state, denoted by '--s--'\n prev_tag = '--s--' \n \n # use 'i' to track the line number in the corpus\n i = 0 \n \n # Each item in the training corpus contains a word and its POS tag\n # Go through each word and its tag in the training corpus\n for word_tag in training_corpus:\n \n # Increment the word_tag count\n i += 1\n \n # Every 50,000 words, print the word count\n if i % 50000 == 0:\n print(f\"word count = {i}\")\n \n ### START CODE HERE (Replace instances of 'None' with your code) ###\n # get the word and tag using the get_word_tag helper function (imported from utils_pos.py)\n word, tag = get_word_tag(line=word_tag, vocab=vocab)\n \n # Increment the transition count for the previous word and tag\n transition_counts[(prev_tag, tag)] += 1\n \n # Increment the emission count for the tag and word\n emission_counts[(tag, word)] += 1\n\n # Increment the tag count\n tag_counts[tag] += 1\n\n # Set the previous tag to this tag (for the next iteration of the loop)\n prev_tag = tag\n \n ### END CODE HERE ###\n \n return emission_counts, transition_counts, tag_counts",
"_____no_output_____"
],
[
"emission_counts, transition_counts, tag_counts = create_dictionaries(training_corpus, vocab)",
"word count = 50000\nword count = 100000\nword count = 150000\nword count = 200000\nword count = 250000\nword count = 300000\nword count = 350000\nword count = 400000\nword count = 450000\nword count = 500000\nword count = 550000\nword count = 600000\nword count = 650000\nword count = 700000\nword count = 750000\nword count = 800000\nword count = 850000\nword count = 900000\nword count = 950000\n"
],
[
"# get all the POS states\nstates = sorted(tag_counts.keys())\nprint(f\"Number of POS tags (number of 'states'): {len(states)}\")\nprint(\"View these POS tags (states)\")\nprint(states)",
"Number of POS tags (number of 'states'): 46\nView these POS tags (states)\n['#', '$', \"''\", '(', ')', ',', '--s--', '.', ':', 'CC', 'CD', 'DT', 'EX', 'FW', 'IN', 'JJ', 'JJR', 'JJS', 'LS', 'MD', 'NN', 'NNP', 'NNPS', 'NNS', 'PDT', 'POS', 'PRP', 'PRP$', 'RB', 'RBR', 'RBS', 'RP', 'SYM', 'TO', 'UH', 'VB', 'VBD', 'VBG', 'VBN', 'VBP', 'VBZ', 'WDT', 'WP', 'WP$', 'WRB', '``']\n"
]
],
[
[
"##### Expected Output\n\n```CPP\nNumber of POS tags (number of 'states'46\nView these states\n['#', '$', \"''\", '(', ')', ',', '--s--', '.', ':', 'CC', 'CD', 'DT', 'EX', 'FW', 'IN', 'JJ', 'JJR', 'JJS', 'LS', 'MD', 'NN', 'NNP', 'NNPS', 'NNS', 'PDT', 'POS', 'PRP', 'PRP$', 'RB', 'RBR', 'RBS', 'RP', 'SYM', 'TO', 'UH', 'VB', 'VBD', 'VBG', 'VBN', 'VBP', 'VBZ', 'WDT', 'WP', 'WP$', 'WRB', '``']\n```",
"_____no_output_____"
],
[
"The 'states' are the Parts-of-speech designations found in the training data. They will also be referred to as 'tags' or POS in this assignment. \n\n- \"NN\" is noun, singular, \n- 'NNS' is noun, plural. \n- In addition, there are helpful tags like '--s--' which indicate a start of a sentence.\n- You can get a more complete description at [Penn Treebank II tag set](https://www.clips.uantwerpen.be/pages/mbsp-tags). ",
"_____no_output_____"
]
],
[
[
"print(\"transition examples: \")\nfor ex in list(transition_counts.items())[:3]:\n print(ex)\nprint()\n\nprint(\"emission examples: \")\nfor ex in list(emission_counts.items())[200:203]:\n print (ex)\nprint()\n\nprint(\"ambiguous word example: \")\nfor tup,cnt in emission_counts.items():\n if tup[1] == 'back': print (tup, cnt) ",
"transition examples: \n(('--s--', 'IN'), 5050)\n(('IN', 'DT'), 32364)\n(('DT', 'NNP'), 9044)\n\nemission examples: \n(('DT', 'any'), 721)\n(('NN', 'decrease'), 7)\n(('NN', 'insider-trading'), 5)\n\nambiguous word example: \n('RB', 'back') 304\n('VB', 'back') 20\n('RP', 'back') 84\n('JJ', 'back') 25\n('NN', 'back') 29\n('VBP', 'back') 4\n"
]
],
[
[
"##### Expected Output\n\n```CPP\ntransition examples: \n(('--s--', 'IN'), 5050)\n(('IN', 'DT'), 32364)\n(('DT', 'NNP'), 9044)\n\nemission examples: \n(('DT', 'any'), 721)\n(('NN', 'decrease'), 7)\n(('NN', 'insider-trading'), 5)\n\nambiguous word example: \n('RB', 'back') 304\n('VB', 'back') 20\n('RP', 'back') 84\n('JJ', 'back') 25\n('NN', 'back') 29\n('VBP', 'back') 4\n```",
"_____no_output_____"
],
[
"<a name='1.2'></a>\n### Part 1.2 - Testing\n\nNow you will test the accuracy of your parts-of-speech tagger using your `emission_counts` dictionary. \n- Given your preprocessed test corpus `prep`, you will assign a parts-of-speech tag to every word in that corpus. \n- Using the original tagged test corpus `y`, you will then compute what percent of the tags you got correct. ",
"_____no_output_____"
],
[
"<a name='ex-02'></a>\n### Exercise 02\n\n**Instructions:** Implement `predict_pos` that computes the accuracy of your model. \n\n- This is a warm up exercise. \n- To assign a part of speech to a word, assign the most frequent POS for that word in the training set. \n- Then evaluate how well this approach works. Each time you predict based on the most frequent POS for the given word, check whether the actual POS of that word is the same. If so, the prediction was correct!\n- Calculate the accuracy as the number of correct predictions divided by the total number of words for which you predicted the POS tag.",
"_____no_output_____"
]
],
[
[
"# UNQ_C2 (UNIQUE CELL IDENTIFIER, DO NOT EDIT)\n# GRADED FUNCTION: predict_pos\n\ndef predict_pos(prep, y, emission_counts, vocab, states):\n '''\n Input: \n prep: a preprocessed version of 'y'. A list with the 'word' component of the tuples.\n y: a corpus composed of a list of tuples where each tuple consists of (word, POS)\n emission_counts: a dictionary where the keys are (tag,word) tuples and the value is the count\n vocab: a dictionary where keys are words in vocabulary and value is an index\n states: a sorted list of all possible tags for this assignment\n Output: \n accuracy: Number of times you classified a word correctly\n '''\n \n # Initialize the number of correct predictions to zero\n num_correct = 0\n \n # Get the (tag, word) tuples, stored as a set\n all_words = set(emission_counts.keys())\n \n # Get the number of (word, POS) tuples in the corpus 'y'\n total = len(y)\n for word, y_tup in zip(prep, y): \n\n # Split the (word, POS) string into a list of two items\n y_tup_l = y_tup.split()\n \n # Verify that y_tup contain both word and POS\n if len(y_tup_l) == 2:\n \n # Set the true POS label for this word\n true_label = y_tup_l[1]\n\n else:\n # If the y_tup didn't contain word and POS, go to next word\n continue\n \n count_final = 0\n pos_final = ''\n \n # If the word is in the vocabulary...\n if word in vocab:\n for pos in states:\n\n ### START CODE HERE (Replace instances of 'None' with your code) ###\n \n # define the key as the tuple containing the POS and word\n key = (pos, word)\n\n # check if the (pos, word) key exists in the emission_counts dictionary\n if key in emission_counts: # complete this line\n\n # get the emission count of the (pos,word) tuple \n count = emission_counts[key]\n\n # keep track of the POS with the largest count\n if count > count_final: # complete this line\n\n # update the final count (largest count)\n count_final = count\n\n # update the final POS\n pos_final = pos\n\n # If the final POS (with the largest count) matches the true POS:\n if pos_final == true_label: # complete this line\n \n # Update the number of correct predictions\n num_correct += 1\n \n ### END CODE HERE ###\n accuracy = num_correct / total\n \n return accuracy",
"_____no_output_____"
],
[
"accuracy_predict_pos = predict_pos(prep, y, emission_counts, vocab, states)\nprint(f\"Accuracy of prediction using predict_pos is {accuracy_predict_pos:.4f}\")",
"Accuracy of prediction using predict_pos is 0.8889\n"
]
],
[
[
"##### Expected Output\n```CPP\nAccuracy of prediction using predict_pos is 0.8889\n```\n\n88.9% is really good for this warm up exercise. With hidden markov models, you should be able to get **95% accuracy.**",
"_____no_output_____"
],
[
"<a name='2'></a>\n# Part 2: Hidden Markov Models for POS\n\nNow you will build something more context specific. Concretely, you will be implementing a Hidden Markov Model (HMM) with a Viterbi decoder\n- The HMM is one of the most commonly used algorithms in Natural Language Processing, and is a foundation to many deep learning techniques you will see in this specialization. \n- In addition to parts-of-speech tagging, HMM is used in speech recognition, speech synthesis, etc. \n- By completing this part of the assignment you will get a 95% accuracy on the same dataset you used in Part 1.\n\nThe Markov Model contains a number of states and the probability of transition between those states. \n- In this case, the states are the parts-of-speech. \n- A Markov Model utilizes a transition matrix, `A`. \n- A Hidden Markov Model adds an observation or emission matrix `B` which describes the probability of a visible observation when we are in a particular state. \n- In this case, the emissions are the words in the corpus\n- The state, which is hidden, is the POS tag of that word.",
"_____no_output_____"
],
[
"<a name='2.1'></a>\n## Part 2.1 Generating Matrices\n\n### Creating the 'A' transition probabilities matrix\nNow that you have your `emission_counts`, `transition_counts`, and `tag_counts`, you will start implementing the Hidden Markov Model. \n\nThis will allow you to quickly construct the \n- `A` transition probabilities matrix.\n- and the `B` emission probabilities matrix. \n\nYou will also use some smoothing when computing these matrices. \n\nHere is an example of what the `A` transition matrix would look like (it is simplified to 5 tags for viewing. It is 46x46 in this assignment.):\n\n\n|**A** |...| RBS | RP | SYM | TO | UH|...\n| --- ||---:-------------| ------------ | ------------ | -------- | ---------- |----\n|**RBS** |...|2.217069e-06 |2.217069e-06 |2.217069e-06 |0.008870 |2.217069e-06|...\n|**RP** |...|3.756509e-07 |7.516775e-04 |3.756509e-07 |0.051089 |3.756509e-07|...\n|**SYM** |...|1.722772e-05 |1.722772e-05 |1.722772e-05 |0.000017 |1.722772e-05|...\n|**TO** |...|4.477336e-05 |4.472863e-08 |4.472863e-08 |0.000090 |4.477336e-05|...\n|**UH** |...|1.030439e-05 |1.030439e-05 |1.030439e-05 |0.061837 |3.092348e-02|...\n| ... |...| ... | ... | ... | ... | ... | ...\n\nNote that the matrix above was computed with smoothing. \n\nEach cell gives you the probability to go from one part of speech to another. \n- In other words, there is a 4.47e-8 chance of going from parts-of-speech `TO` to `RP`. \n- The sum of each row has to equal 1, because we assume that the next POS tag must be one of the available columns in the table.\n\nThe smoothing was done as follows: \n\n$$ P(t_i | t_{i-1}) = \\frac{C(t_{i-1}, t_{i}) + \\alpha }{C(t_{i-1}) +\\alpha * N}\\tag{3}$$\n\n- $N$ is the total number of tags\n- $C(t_{i-1}, t_{i})$ is the count of the tuple (previous POS, current POS) in `transition_counts` dictionary.\n- $C(t_{i-1})$ is the count of the previous POS in the `tag_counts` dictionary.\n- $\\alpha$ is a smoothing parameter.",
"_____no_output_____"
],
[
"<a name='ex-03'></a>\n### Exercise 03\n\n**Instructions:** Implement the `create_transition_matrix` below for all tags. Your task is to output a matrix that computes equation 3 for each cell in matrix `A`. ",
"_____no_output_____"
]
],
[
[
"# UNQ_C3 (UNIQUE CELL IDENTIFIER, DO NOT EDIT)\n# GRADED FUNCTION: create_transition_matrix\ndef create_transition_matrix(alpha, tag_counts, transition_counts):\n ''' \n Input: \n alpha: number used for smoothing\n tag_counts: a dictionary mapping each tag to its respective count\n transition_counts: transition count for the previous word and tag\n Output:\n A: matrix of dimension (num_tags,num_tags)\n '''\n # Get a sorted list of unique POS tags\n all_tags = sorted(tag_counts.keys())\n \n # Count the number of unique POS tags\n num_tags = len(all_tags)\n \n # Initialize the transition matrix 'A'\n A = np.zeros((num_tags,num_tags))\n \n # Get the unique transition tuples (previous POS, current POS)\n trans_keys = set(transition_counts.keys())\n \n ### START CODE HERE (Replace instances of 'None' with your code) ### \n \n # Go through each row of the transition matrix A\n for i in range(num_tags):\n \n # Go through each column of the transition matrix A\n for j in range(num_tags):\n\n # Initialize the count of the (prev POS, current POS) to zero\n count = 0\n \n # Define the tuple (prev POS, current POS)\n # Get the tag at position i and tag at position j (from the all_tags list)\n key = (all_tags[i], all_tags[j])\n\n # Check if the (prev POS, current POS) tuple \n # exists in the transition counts dictionary\n if key in transition_counts: #complete this line\n \n # Get count from the transition_counts dictionary \n # for the (prev POS, current POS) tuple\n count = transition_counts[key]\n \n # Get the count of the previous tag (index position i) from tag_counts\n count_prev_tag = tag_counts[key[0]]\n \n # Apply smoothing using count of the tuple, alpha, \n # count of previous tag, alpha, and total number of tags\n A[i,j] = (count + alpha) / (count_prev_tag + (alpha * num_tags))\n\n ### END CODE HERE ###\n \n return A",
"_____no_output_____"
],
[
"alpha = 0.001\nA = create_transition_matrix(alpha, tag_counts, transition_counts)\n# Testing your function\nprint(f\"A at row 0, col 0: {A[0,0]:.9f}\")\nprint(f\"A at row 3, col 1: {A[3,1]:.4f}\")\n\nprint(\"View a subset of transition matrix A\")\nA_sub = pd.DataFrame(A[30:35,30:35], index=states[30:35], columns = states[30:35] )\nprint(A_sub)",
"A at row 0, col 0: 0.000007040\nA at row 3, col 1: 0.1691\nView a subset of transition matrix A\n RBS RP SYM TO UH\nRBS 2.217069e-06 2.217069e-06 2.217069e-06 0.008870 2.217069e-06\nRP 3.756509e-07 7.516775e-04 3.756509e-07 0.051089 3.756509e-07\nSYM 1.722772e-05 1.722772e-05 1.722772e-05 0.000017 1.722772e-05\nTO 4.477336e-05 4.472863e-08 4.472863e-08 0.000090 4.477336e-05\nUH 1.030439e-05 1.030439e-05 1.030439e-05 0.061837 3.092348e-02\n"
]
],
[
[
"##### Expected Output\n```CPP\nA at row 0, col 0: 0.000007040\nA at row 3, col 1: 0.1691\nView a subset of transition matrix A\n RBS RP SYM TO UH\nRBS 2.217069e-06 2.217069e-06 2.217069e-06 0.008870 2.217069e-06\nRP 3.756509e-07 7.516775e-04 3.756509e-07 0.051089 3.756509e-07\nSYM 1.722772e-05 1.722772e-05 1.722772e-05 0.000017 1.722772e-05\nTO 4.477336e-05 4.472863e-08 4.472863e-08 0.000090 4.477336e-05\nUH 1.030439e-05 1.030439e-05 1.030439e-05 0.061837 3.092348e-02\n```",
"_____no_output_____"
],
[
"### Create the 'B' emission probabilities matrix\n\nNow you will create the `B` transition matrix which computes the emission probability. \n\nYou will use smoothing as defined below: \n\n$$P(w_i | t_i) = \\frac{C(t_i, word_i)+ \\alpha}{C(t_{i}) +\\alpha * N}\\tag{4}$$\n\n- $C(t_i, word_i)$ is the number of times $word_i$ was associated with $tag_i$ in the training data (stored in `emission_counts` dictionary).\n- $C(t_i)$ is the number of times $tag_i$ was in the training data (stored in `tag_counts` dictionary).\n- $N$ is the number of words in the vocabulary\n- $\\alpha$ is a smoothing parameter. \n\nThe matrix `B` is of dimension (num_tags, N), where num_tags is the number of possible parts-of-speech tags. \n\nHere is an example of the matrix, only a subset of tags and words are shown: \n<p style='text-align: center;'> <b>B Emissions Probability Matrix (subset)</b> </p>\n\n|**B**| ...| 725 | adroitly | engineers | promoted | synergy| ...|\n|----|----|--------------|--------------|--------------|--------------|-------------|----|\n|**CD** | ...| **8.201296e-05** | 2.732854e-08 | 2.732854e-08 | 2.732854e-08 | 2.732854e-08| ...|\n|**NN** | ...| 7.521128e-09 | 7.521128e-09 | 7.521128e-09 | 7.521128e-09 | **2.257091e-05**| ...|\n|**NNS** | ...| 1.670013e-08 | 1.670013e-08 |**4.676203e-04** | 1.670013e-08 | 1.670013e-08| ...|\n|**VB** | ...| 3.779036e-08 | 3.779036e-08 | 3.779036e-08 | 3.779036e-08 | 3.779036e-08| ...|\n|**RB** | ...| 3.226454e-08 | **6.456135e-05** | 3.226454e-08 | 3.226454e-08 | 3.226454e-08| ...|\n|**RP** | ...| 3.723317e-07 | 3.723317e-07 | 3.723317e-07 | **3.723317e-07** | 3.723317e-07| ...|\n| ... | ...| ... | ... | ... | ... | ... | ...|\n\n",
"_____no_output_____"
],
[
"<a name='ex-04'></a>\n### Exercise 04\n**Instructions:** Implement the `create_emission_matrix` below that computes the `B` emission probabilities matrix. Your function takes in $\\alpha$, the smoothing parameter, `tag_counts`, which is a dictionary mapping each tag to its respective count, the `emission_counts` dictionary where the keys are (tag, word) and the values are the counts. Your task is to output a matrix that computes equation 4 for each cell in matrix `B`. ",
"_____no_output_____"
]
],
[
[
"# UNQ_C4 (UNIQUE CELL IDENTIFIER, DO NOT EDIT)\n# GRADED FUNCTION: create_emission_matrix\n\ndef create_emission_matrix(alpha, tag_counts, emission_counts, vocab):\n '''\n Input: \n alpha: tuning parameter used in smoothing \n tag_counts: a dictionary mapping each tag to its respective count\n emission_counts: a dictionary where the keys are (tag, word) and the values are the counts\n vocab: a dictionary where keys are words in vocabulary and value is an index.\n within the function it'll be treated as a list\n Output:\n B: a matrix of dimension (num_tags, len(vocab))\n '''\n \n # get the number of POS tag\n num_tags = len(tag_counts)\n \n # Get a list of all POS tags\n all_tags = sorted(tag_counts.keys())\n \n # Get the total number of unique words in the vocabulary\n num_words = len(vocab)\n \n # Initialize the emission matrix B with places for\n # tags in the rows and words in the columns\n B = np.zeros((num_tags, num_words))\n \n # Get a set of all (POS, word) tuples \n # from the keys of the emission_counts dictionary\n emis_keys = set(list(emission_counts.keys()))\n \n ### START CODE HERE (Replace instances of 'None' with your code) ###\n \n # Go through each row (POS tags)\n for i in range(num_tags): # complete this line\n \n # Go through each column (words)\n for j in range(num_words): # complete this line\n\n # Initialize the emission count for the (POS tag, word) to zero\n count = 0\n \n # Define the (POS tag, word) tuple for this row and column\n key = (all_tags[i], vocab[j])\n\n # check if the (POS tag, word) tuple exists as a key in emission counts\n if key in emission_counts: # complete this line\n \n # Get the count of (POS tag, word) from the emission_counts d\n count = emission_counts[key]\n \n # Get the count of the POS tag\n count_tag = tag_counts[key[0]]\n \n # Apply smoothing and store the smoothed value \n # into the emission matrix B for this row and column\n B[i,j] = (count + alpha) / (count_tag + (alpha * num_words))\n\n ### END CODE HERE ###\n return B",
"_____no_output_____"
],
[
"# creating your emission probability matrix. this takes a few minutes to run. \nB = create_emission_matrix(alpha, tag_counts, emission_counts, list(vocab))\n\nprint(f\"View Matrix position at row 0, column 0: {B[0,0]:.9f}\")\nprint(f\"View Matrix position at row 3, column 1: {B[3,1]:.9f}\")\n\n# Try viewing emissions for a few words in a sample dataframe\ncidx = ['725','adroitly','engineers', 'promoted', 'synergy']\n\n# Get the integer ID for each word\ncols = [vocab[a] for a in cidx]\n\n# Choose POS tags to show in a sample dataframe\nrvals =['CD','NN','NNS', 'VB','RB','RP']\n\n# For each POS tag, get the row number from the 'states' list\nrows = [states.index(a) for a in rvals]\n\n# Get the emissions for the sample of words, and the sample of POS tags\nB_sub = pd.DataFrame(B[np.ix_(rows,cols)], index=rvals, columns = cidx )\nprint(B_sub)",
"View Matrix position at row 0, column 0: 0.000006032\nView Matrix position at row 3, column 1: 0.000000720\n 725 adroitly engineers promoted synergy\nCD 8.201296e-05 2.732854e-08 2.732854e-08 2.732854e-08 2.732854e-08\nNN 7.521128e-09 7.521128e-09 7.521128e-09 7.521128e-09 2.257091e-05\nNNS 1.670013e-08 1.670013e-08 4.676203e-04 1.670013e-08 1.670013e-08\nVB 3.779036e-08 3.779036e-08 3.779036e-08 3.779036e-08 3.779036e-08\nRB 3.226454e-08 6.456135e-05 3.226454e-08 3.226454e-08 3.226454e-08\nRP 3.723317e-07 3.723317e-07 3.723317e-07 3.723317e-07 3.723317e-07\n"
]
],
[
[
"##### Expected Output\n\n```CPP\nView Matrix position at row 0, column 0: 0.000006032\nView Matrix position at row 3, column 1: 0.000000720\n 725 adroitly engineers promoted synergy\nCD 8.201296e-05 2.732854e-08 2.732854e-08 2.732854e-08 2.732854e-08\nNN 7.521128e-09 7.521128e-09 7.521128e-09 7.521128e-09 2.257091e-05\nNNS 1.670013e-08 1.670013e-08 4.676203e-04 1.670013e-08 1.670013e-08\nVB 3.779036e-08 3.779036e-08 3.779036e-08 3.779036e-08 3.779036e-08\nRB 3.226454e-08 6.456135e-05 3.226454e-08 3.226454e-08 3.226454e-08\nRP 3.723317e-07 3.723317e-07 3.723317e-07 3.723317e-07 3.723317e-07\n```",
"_____no_output_____"
],
[
"<a name='3'></a>\n# Part 3: Viterbi Algorithm and Dynamic Programming\n\nIn this part of the assignment you will implement the Viterbi algorithm which makes use of dynamic programming. Specifically, you will use your two matrices, `A` and `B` to compute the Viterbi algorithm. We have decomposed this process into three main steps for you. \n\n* **Initialization** - In this part you initialize the `best_paths` and `best_probabilities` matrices that you will be populating in `feed_forward`.\n* **Feed forward** - At each step, you calculate the probability of each path happening and the best paths up to that point. \n* **Feed backward**: This allows you to find the best path with the highest probabilities. \n\n<a name='3.1'></a>\n## Part 3.1: Initialization \n\nYou will start by initializing two matrices of the same dimension. \n\n- best_probs: Each cell contains the probability of going from one POS tag to a word in the corpus.\n\n- best_paths: A matrix that helps you trace through the best possible path in the corpus. ",
"_____no_output_____"
],
[
"<a name='ex-05'></a>\n### Exercise 05\n**Instructions**: \nWrite a program below that initializes the `best_probs` and the `best_paths` matrix. \n\nBoth matrices will be initialized to zero except for column zero of `best_probs`. \n- Column zero of `best_probs` is initialized with the assumption that the first word of the corpus was preceded by a start token (\"--s--\"). \n- This allows you to reference the **A** matrix for the transition probability\n\nHere is how to initialize column 0 of `best_probs`:\n- The probability of the best path going from the start index to a given POS tag indexed by integer $i$ is denoted by $\\textrm{best_probs}[s_{idx}, i]$.\n- This is estimated as the probability that the start tag transitions to the POS denoted by index $i$: $\\mathbf{A}[s_{idx}, i]$ AND that the POS tag denoted by $i$ emits the first word of the given corpus, which is $\\mathbf{B}[i, vocab[corpus[0]]]$.\n- Note that vocab[corpus[0]] refers to the first word of the corpus (the word at position 0 of the corpus). \n- **vocab** is a dictionary that returns the unique integer that refers to that particular word.\n\nConceptually, it looks like this:\n$\\textrm{best_probs}[s_{idx}, i] = \\mathbf{A}[s_{idx}, i] \\times \\mathbf{B}[i, corpus[0] ]$\n\n\nIn order to avoid multiplying and storing small values on the computer, we'll take the log of the product, which becomes the sum of two logs:\n\n$best\\_probs[i,0] = log(A[s_{idx}, i]) + log(B[i, vocab[corpus[0]]$\n\nAlso, to avoid taking the log of 0 (which is defined as negative infinity), the code itself will just set $best\\_probs[i,0] = float('-inf')$ when $A[s_{idx}, i] == 0$\n\n\nSo the implementation to initialize $best\\_probs$ looks like this:\n\n$ if A[s_{idx}, i] <> 0 : best\\_probs[i,0] = log(A[s_{idx}, i]) + log(B[i, vocab[corpus[0]]])$\n\n$ if A[s_{idx}, i] == 0 : best\\_probs[i,0] = float('-inf')$\n\nPlease use [math.log](https://docs.python.org/3/library/math.html) to compute the natural logarithm.",
"_____no_output_____"
],
[
"The example below shows the initialization assuming the corpus starts with the phrase \"Loss tracks upward\".\n\n<img src = \"Initialize4.PNG\"/>",
"_____no_output_____"
],
[
"Represent infinity and negative infinity like this:\n\n```CPP\nfloat('inf')\nfloat('-inf')\n```",
"_____no_output_____"
]
],
[
[
"# UNQ_C5 (UNIQUE CELL IDENTIFIER, DO NOT EDIT)\n# GRADED FUNCTION: initialize\ndef initialize(states, tag_counts, A, B, corpus, vocab):\n '''\n Input: \n states: a list of all possible parts-of-speech\n tag_counts: a dictionary mapping each tag to its respective count\n A: Transition Matrix of dimension (num_tags, num_tags)\n B: Emission Matrix of dimension (num_tags, len(vocab))\n corpus: a sequence of words whose POS is to be identified in a list \n vocab: a dictionary where keys are words in vocabulary and value is an index\n Output:\n best_probs: matrix of dimension (num_tags, len(corpus)) of floats\n best_paths: matrix of dimension (num_tags, len(corpus)) of integers\n '''\n # Get the total number of unique POS tags\n num_tags = len(tag_counts)\n \n # Initialize best_probs matrix \n # POS tags in the rows, number of words in the corpus as the columns\n best_probs = np.zeros((num_tags, len(corpus)))\n \n # Initialize best_paths matrix\n # POS tags in the rows, number of words in the corpus as columns\n best_paths = np.zeros((num_tags, len(corpus)), dtype=int)\n \n # Define the start token\n s_idx = states.index(\"--s--\")\n ### START CODE HERE (Replace instances of 'None' with your code) ###\n \n # Go through each of the POS tags\n for i in range(num_tags): # complete this line\n \n # Handle the special case when the transition from start token to POS tag i is zero\n if A[s_idx][i] == 0: # complete this line\n \n # Initialize best_probs at POS tag 'i', column 0, to negative infinity\n best_probs[i,0] = np.NINF\n \n # For all other cases when transition from start token to POS tag i is non-zero:\n else:\n \n # Initialize best_probs at POS tag 'i', column 0\n # Check the formula in the instructions above\n best_probs[i,0] = np.log(A[s_idx, i]) + np.log(B[i, vocab[corpus[0]]])\n \n ### END CODE HERE ### \n return best_probs, best_paths",
"_____no_output_____"
],
[
"best_probs, best_paths = initialize(states, tag_counts, A, B, prep, vocab)",
"_____no_output_____"
],
[
"# Test the function\nprint(f\"best_probs[0,0]: {best_probs[0,0]:.4f}\") \nprint(f\"best_paths[2,3]: {best_paths[2,3]:.4f}\")",
"best_probs[0,0]: -22.6098\nbest_paths[2,3]: 0.0000\n"
]
],
[
[
"##### Expected Output\n\n```CPP\nbest_probs[0,0]: -22.6098\nbest_paths[2,3]: 0.0000\n```\n",
"_____no_output_____"
],
[
"<a name='3.2'></a>\n## Part 3.2 Viterbi Forward\n\nIn this part of the assignment, you will implement the `viterbi_forward` segment. In other words, you will populate your `best_probs` and `best_paths` matrices.\n- Walk forward through the corpus.\n- For each word, compute a probability for each possible tag. \n- Unlike the previous algorithm `predict_pos` (the 'warm-up' exercise), this will include the path up to that (word,tag) combination. \n\nHere is an example with a three-word corpus \"Loss tracks upward\":\n- Note, in this example, only a subset of states (POS tags) are shown in the diagram below, for easier reading. \n- In the diagram below, the first word \"Loss\" is already initialized. \n- The algorithm will compute a probability for each of the potential tags in the second and future words. \n\nCompute the probability that the tag of the second work ('tracks') is a verb, 3rd person singular present (VBZ). \n- In the `best_probs` matrix, go to the column of the second word ('tracks'), and row 40 (VBZ), this cell is highlighted in light orange in the diagram below.\n- Examine each of the paths from the tags of the first word ('Loss') and choose the most likely path. \n- An example of the calculation for **one** of those paths is the path from ('Loss', NN) to ('tracks', VBZ).\n- The log of the probability of the path up to and including the first word 'Loss' having POS tag NN is $-14.32$. The `best_probs` matrix contains this value -14.32 in the column for 'Loss' and row for 'NN'.\n- Find the probability that NN transitions to VBZ. To find this probability, go to the `A` transition matrix, and go to the row for 'NN' and the column for 'VBZ'. The value is $4.37e-02$, which is circled in the diagram, so add $-14.32 + log(4.37e-02)$. \n- Find the log of the probability that the tag VBS would 'emit' the word 'tracks'. To find this, look at the 'B' emission matrix in row 'VBZ' and the column for the word 'tracks'. The value $4.61e-04$ is circled in the diagram below. So add $-14.32 + log(4.37e-02) + log(4.61e-04)$.\n- The sum of $-14.32 + log(4.37e-02) + log(4.61e-04)$ is $-25.13$. Store $-25.13$ in the `best_probs` matrix at row 'VBZ' and column 'tracks' (as seen in the cell that is highlighted in light orange in the diagram).\n- All other paths in best_probs are calculated. Notice that $-25.13$ is greater than all of the other values in column 'tracks' of matrix `best_probs`, and so the most likely path to 'VBZ' is from 'NN'. 'NN' is in row 20 of the `best_probs` matrix, so $20$ is the most likely path.\n- Store the most likely path $20$ in the `best_paths` table. This is highlighted in light orange in the diagram below.",
"_____no_output_____"
],
[
"The formula to compute the probability and path for the $i^{th}$ word in the $corpus$, the prior word $i-1$ in the corpus, current POS tag $j$, and previous POS tag $k$ is:\n\n$\\mathrm{prob} = \\mathbf{best\\_prob}_{k, i-1} + \\mathrm{log}(\\mathbf{A}_{k, j}) + \\mathrm{log}(\\mathbf{B}_{j, vocab(corpus_{i})})$\n\nwhere $corpus_{i}$ is the word in the corpus at index $i$, and $vocab$ is the dictionary that gets the unique integer that represents a given word.\n\n$\\mathrm{path} = k$\n\nwhere $k$ is the integer representing the previous POS tag.\n",
"_____no_output_____"
],
[
"<a name='ex-06'></a>\n\n### Exercise 06\n\nInstructions: Implement the `viterbi_forward` algorithm and store the best_path and best_prob for every possible tag for each word in the matrices `best_probs` and `best_tags` using the pseudo code below.\n\n`for each word in the corpus\n\n for each POS tag type that this word may be\n \n for POS tag type that the previous word could be\n \n compute the probability that the previous word had a given POS tag, that the current word has a given POS tag, and that the POS tag would emit this current word.\n \n retain the highest probability computed for the current word\n \n set best_probs to this highest probability\n \n set best_paths to the index 'k', representing the POS tag of the previous word which produced the highest probability `\n\nPlease use [math.log](https://docs.python.org/3/library/math.html) to compute the natural logarithm.",
"_____no_output_____"
],
[
"<img src = \"Forward4.PNG\"/>",
"_____no_output_____"
],
[
"<details> \n<summary>\n <font size=\"3\" color=\"darkgreen\"><b>Hints</b></font>\n</summary>\n<p>\n<ul>\n <li>Remember that when accessing emission matrix B, the column index is the unique integer ID associated with the word. It can be accessed by using the 'vocab' dictionary, where the key is the word, and the value is the unique integer ID for that word.</li>\n</ul>\n</p>\n",
"_____no_output_____"
]
],
[
[
"# UNQ_C6 (UNIQUE CELL IDENTIFIER, DO NOT EDIT)\n# GRADED FUNCTION: viterbi_forward\ndef viterbi_forward(A, B, test_corpus, best_probs, best_paths, vocab):\n '''\n Input: \n A, B: The transition and emission matrices respectively\n test_corpus: a list containing a preprocessed corpus\n best_probs: an initilized matrix of dimension (num_tags, len(corpus))\n best_paths: an initilized matrix of dimension (num_tags, len(corpus))\n vocab: a dictionary where keys are words in vocabulary and value is an index \n Output: \n best_probs: a completed matrix of dimension (num_tags, len(corpus))\n best_paths: a completed matrix of dimension (num_tags, len(corpus))\n '''\n # Get the number of unique POS tags (which is the num of rows in best_probs)\n num_tags = best_probs.shape[0]\n \n # Go through every word in the corpus starting from word 1\n # Recall that word 0 was initialized in `initialize()`\n for i in range(1, len(test_corpus)): \n \n # Print number of words processed, every 5000 words\n if i % 5000 == 0:\n print(\"Words processed: {:>8}\".format(i))\n \n ### START CODE HERE (Replace instances of 'None' with your code EXCEPT the first 'best_path_i = None') ###\n # For each unique POS tag that the current word can be\n for j in range(num_tags): # complete this line\n \n # Initialize best_prob for word i to negative infinity\n best_prob_i = np.NINF\n \n # Initialize best_path for current word i to None\n best_path_i = None\n\n # For each POS tag that the previous word can be:\n for k in range(num_tags): # complete this line\n \n # Calculate the probability = \n # best probs of POS tag k, previous word i-1 + \n # log(prob of transition from POS k to POS j) + \n # log(prob that emission of POS j is word i)\n prob = best_probs[k, i-1] + np.log(A[k, j]) + math.log(B[j, vocab[prep[i]]])\n\n # check if this path's probability is greater than\n # the best probability up to and before this point\n if prob > best_prob_i: # complete this line\n \n # Keep track of the best probability\n best_prob_i = prob\n \n # keep track of the POS tag of the previous word\n # that is part of the best path. \n # Save the index (integer) associated with \n # that previous word's POS tag\n best_path_i = k\n\n # Save the best probability for the \n # given current word's POS tag\n # and the position of the current word inside the corpus\n best_probs[j,i] = best_prob_i\n \n # Save the unique integer ID of the previous POS tag\n # into best_paths matrix, for the POS tag of the current word\n # and the position of the current word inside the corpus.\n best_paths[j,i] = best_path_i\n\n ### END CODE HERE ###\n return best_probs, best_paths",
"_____no_output_____"
]
],
[
[
"Run the `viterbi_forward` function to fill in the `best_probs` and `best_paths` matrices.\n\n**Note** that this will take a few minutes to run. There are about 30,000 words to process.",
"_____no_output_____"
]
],
[
[
"# this will take a few minutes to run => processes ~ 30,000 words\nbest_probs, best_paths = viterbi_forward(A, B, prep, best_probs, best_paths, vocab)",
"Words processed: 5000\nWords processed: 10000\nWords processed: 15000\nWords processed: 20000\nWords processed: 25000\nWords processed: 30000\n"
],
[
"# Test this function \nprint(f\"best_probs[0,1]: {best_probs[0,1]:.4f}\") \nprint(f\"best_probs[0,4]: {best_probs[0,4]:.4f}\") ",
"best_probs[0,1]: -24.7822\nbest_probs[0,4]: -49.5601\n"
]
],
[
[
"##### Expected Output\n\n```CPP\nbest_probs[0,1]: -24.7822\nbest_probs[0,4]: -49.5601\n```",
"_____no_output_____"
],
[
"<a name='3.3'></a>\n## Part 3.3 Viterbi backward\n\nNow you will implement the Viterbi backward algorithm.\n- The Viterbi backward algorithm gets the predictions of the POS tags for each word in the corpus using the `best_paths` and the `best_probs` matrices.\n\nThe example below shows how to walk backwards through the best_paths matrix to get the POS tags of each word in the corpus. Recall that this example corpus has three words: \"Loss tracks upward\".\n\nPOS tag for 'upward' is `RB`\n- Select the the most likely POS tag for the last word in the corpus, 'upward' in the `best_prob` table.\n- Look for the row in the column for 'upward' that has the largest probability.\n- Notice that in row 28 of `best_probs`, the estimated probability is -34.99, which is larger than the other values in the column. So the most likely POS tag for 'upward' is `RB` an adverb, at row 28 of `best_prob`. \n- The variable `z` is an array that stores the unique integer ID of the predicted POS tags for each word in the corpus. In array z, at position 2, store the value 28 to indicate that the word 'upward' (at index 2 in the corpus), most likely has the POS tag associated with unique ID 28 (which is `RB`).\n- The variable `pred` contains the POS tags in string form. So `pred` at index 2 stores the string `RB`.\n\n\nPOS tag for 'tracks' is `VBZ`\n- The next step is to go backward one word in the corpus ('tracks'). Since the most likely POS tag for 'upward' is `RB`, which is uniquely identified by integer ID 28, go to the `best_paths` matrix in column 2, row 28. The value stored in `best_paths`, column 2, row 28 indicates the unique ID of the POS tag of the previous word. In this case, the value stored here is 40, which is the unique ID for POS tag `VBZ` (verb, 3rd person singular present).\n- So the previous word at index 1 of the corpus ('tracks'), most likely has the POS tag with unique ID 40, which is `VBZ`.\n- In array `z`, store the value 40 at position 1, and for array `pred`, store the string `VBZ` to indicate that the word 'tracks' most likely has POS tag `VBZ`.\n\nPOS tag for 'Loss' is `NN`\n- In `best_paths` at column 1, the unique ID stored at row 40 is 20. 20 is the unique ID for POS tag `NN`.\n- In array `z` at position 0, store 20. In array `pred` at position 0, store `NN`.",
"_____no_output_____"
],
[
"<img src = \"Backwards5.PNG\"/>",
"_____no_output_____"
],
[
"<a name='ex-07'></a>\n### Exercise 07\nImplement the `viterbi_backward` algorithm, which returns a list of predicted POS tags for each word in the corpus.\n\n- Note that the numbering of the index positions starts at 0 and not 1. \n- `m` is the number of words in the corpus. \n - So the indexing into the corpus goes from `0` to `m - 1`.\n - Also, the columns in `best_probs` and `best_paths` are indexed from `0` to `m - 1`\n\n\n**In Step 1:** \nLoop through all the rows (POS tags) in the last entry of `best_probs` and find the row (POS tag) with the maximum value.\nConvert the unique integer ID to a tag (a string representation) using the list `states`. \n\nReferring to the three-word corpus described above:\n- `z[2] = 28`: For the word 'upward' at position 2 in the corpus, the POS tag ID is 28. Store 28 in `z` at position 2.\n- `states[28]` is 'RB': The POS tag ID 28 refers to the POS tag 'RB'.\n- `pred[2] = 'RB'`: In array `pred`, store the POS tag for the word 'upward'.\n\n**In Step 2:** \n- Starting at the last column of best_paths, use `best_probs` to find the most likely POS tag for the last word in the corpus.\n- Then use `best_paths` to find the most likely POS tag for the previous word. \n- Update the POS tag for each word in `z` and in `preds`.\n\nReferring to the three-word example from above, read best_paths at column 2 and fill in z at position 1. \n`z[1] = best_paths[z[2],2]` \n\nThe small test following the routine prints the last few words of the corpus and their states to aid in debug.",
"_____no_output_____"
]
],
[
[
"# UNQ_C7 (UNIQUE CELL IDENTIFIER, DO NOT EDIT)\n# GRADED FUNCTION: viterbi_backward\ndef viterbi_backward(best_probs, best_paths, corpus, states):\n '''\n This function returns the best path.\n \n '''\n # Get the number of words in the corpus\n # which is also the number of columns in best_probs, best_paths\n m = best_paths.shape[1] \n \n # Initialize array z, same length as the corpus\n z = [None] * m\n \n # Get the number of unique POS tags\n num_tags = best_probs.shape[0]\n \n # Initialize the best probability for the last word\n best_prob_for_last_word = float('-inf')\n \n # Initialize pred array, same length as corpus\n pred = [None] * m\n \n ### START CODE HERE (Replace instances of 'None' with your code) ###\n ## Step 1 ##\n \n # Go through each POS tag for the last word (last column of best_probs)\n # in order to find the row (POS tag integer ID) \n # with highest probability for the last word\n for k in range(num_tags): # complete this line\n\n # If the probability of POS tag at row k \n # is better than the previously best probability for the last word:\n if best_probs[k, m-1] > best_prob_for_last_word: # complete this line\n \n # Store the new best probability for the lsat word\n best_prob_for_last_word = best_probs[k, m-1]\n \n # Store the unique integer ID of the POS tag\n # which is also the row number in best_probs\n z[m - 1] = k\n \n # Convert the last word's predicted POS tag\n # from its unique integer ID into the string representation\n # using the 'states' dictionary\n # store this in the 'pred' array for the last word\n pred[m - 1] = states[z[m-1]]\n \n ## Step 2 ##\n # Find the best POS tags by walking backward through the best_paths\n # From the last word in the corpus to the 0th word in the corpus\n for i in range(len(corpus)-1, -1, -1): # complete this line\n \n # Retrieve the unique integer ID of\n # the POS tag for the word at position 'i' in the corpus\n pos_tag_for_word_i = z[i]\n \n # In best_paths, go to the row representing the POS tag of word i\n # and the column representing the word's position in the corpus\n # to retrieve the predicted POS for the word at position i-1 in the corpus\n z[i - 1] = best_paths[pos_tag_for_word_i, i]\n \n # Get the previous word's POS tag in string form\n # Use the 'states' dictionary, \n # where the key is the unique integer ID of the POS tag,\n # and the value is the string representation of that POS tag\n pred[i - 1] = states[z[i-1]]\n \n ### END CODE HERE ###\n return pred",
"_____no_output_____"
],
[
"# Run and test your function\npred = viterbi_backward(best_probs, best_paths, prep, states)\nm=len(pred)\nprint('The prediction for pred[-7:m-1] is: \\n', prep[-7:m-1], \"\\n\", pred[-7:m-1], \"\\n\")\nprint('The prediction for pred[0:8] is: \\n', pred[0:7], \"\\n\", prep[0:7])",
"The prediction for pred[-7:m-1] is: \n ['see', 'them', 'here', 'with', 'us', '.'] \n ['VB', 'PRP', 'RB', 'IN', 'PRP', '.'] \n\nThe prediction for pred[0:8] is: \n ['DT', 'NN', 'POS', 'NN', 'MD', 'VB', 'VBN'] \n ['The', 'economy', \"'s\", 'temperature', 'will', 'be', 'taken']\n"
]
],
[
[
"**Expected Output:** \n\n```CPP\nThe prediction for pred[-7:m-1] is: \n ['see', 'them', 'here', 'with', 'us', '.'] \n ['VB', 'PRP', 'RB', 'IN', 'PRP', '.'] \nThe prediction for pred[0:8] is: \n ['DT', 'NN', 'POS', 'NN', 'MD', 'VB', 'VBN'] \n ['The', 'economy', \"'s\", 'temperature', 'will', 'be', 'taken'] \n```\n\nNow you just have to compare the predicted labels to the true labels to evaluate your model on the accuracy metric!",
"_____no_output_____"
],
[
"<a name='4'></a>\n# Part 4: Predicting on a data set\n\nCompute the accuracy of your prediction by comparing it with the true `y` labels. \n- `pred` is a list of predicted POS tags corresponding to the words of the `test_corpus`. ",
"_____no_output_____"
]
],
[
[
"print('The third word is:', prep[3])\nprint('Your prediction is:', pred[3])\nprint('Your corresponding label y is: ', y[3])",
"The third word is: temperature\nYour prediction is: NN\nYour corresponding label y is: temperature\tNN\n\n"
]
],
[
[
"<a name='ex-08'></a>\n### Exercise 08\n\nImplement a function to compute the accuracy of the viterbi algorithm's POS tag predictions.\n- To split y into the word and its tag you can use `y.split()`. ",
"_____no_output_____"
]
],
[
[
"# UNQ_C8 (UNIQUE CELL IDENTIFIER, DO NOT EDIT)\n# GRADED FUNCTION: compute_accuracy\ndef compute_accuracy(pred, y):\n '''\n Input: \n pred: a list of the predicted parts-of-speech \n y: a list of lines where each word is separated by a '\\t' (i.e. word \\t tag)\n Output: \n \n '''\n num_correct = 0\n total = 0\n \n # Zip together the prediction and the labels\n for prediction, y in zip(pred, y):\n ### START CODE HERE (Replace instances of 'None' with your code) ###\n # Split the label into the word and the POS tag\n word_tag_tuple = tuple(y.rstrip().split('\\t'))\n \n # Check that there is actually a word and a tag\n # no more and no less than 2 items\n if len(word_tag_tuple) != 2: # complete this line\n continue \n\n # store the word and tag separately\n word, tag = word_tag_tuple\n \n # Check if the POS tag label matches the prediction\n if prediction == tag: # complete this line\n \n # count the number of times that the prediction\n # and label match\n num_correct += 1\n \n # keep track of the total number of examples (that have valid labels)\n total += 1\n \n ### END CODE HERE ###\n return num_correct/total",
"_____no_output_____"
],
[
"print(f\"Accuracy of the Viterbi algorithm is {compute_accuracy(pred, y):.4f}\")",
"Accuracy of the Viterbi algorithm is 0.9531\n"
]
],
[
[
"##### Expected Output\n\n```CPP\nAccuracy of the Viterbi algorithm is 0.9531\n```\n\nCongratulations you were able to classify the parts-of-speech with 95% accuracy. ",
"_____no_output_____"
],
[
"### Key Points and overview\n\nIn this assignment you learned about parts-of-speech tagging. \n- In this assignment, you predicted POS tags by walking forward through a corpus and knowing the previous word.\n- There are other implementations that use bidirectional POS tagging.\n- Bidirectional POS tagging requires knowing the previous word and the next word in the corpus when predicting the current word's POS tag.\n- Bidirectional POS tagging would tell you more about the POS instead of just knowing the previous word. \n- Since you have learned to implement the unidirectional approach, you have the foundation to implement other POS taggers used in industry.",
"_____no_output_____"
],
[
"### References\n\n- [\"Speech and Language Processing\", Dan Jurafsky and James H. Martin](https://web.stanford.edu/~jurafsky/slp3/)\n- We would like to thank Melanie Tosik for her help and inspiration",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
d0e487b3c0c670f0fd9f4571b39795a78ecdc098 | 38,946 | ipynb | Jupyter Notebook | spacy-examples/Introduction to Natural Language Processing with SpaCy - Working Files/Chapter 2/Working-with-SpaCy-for-the-first-time_Part_1.ipynb | peterwilliams97/spaCy_practice | 71fac240c045afffdbef75f80533fe57fb62583c | [
"MIT"
] | null | null | null | spacy-examples/Introduction to Natural Language Processing with SpaCy - Working Files/Chapter 2/Working-with-SpaCy-for-the-first-time_Part_1.ipynb | peterwilliams97/spaCy_practice | 71fac240c045afffdbef75f80533fe57fb62583c | [
"MIT"
] | null | null | null | spacy-examples/Introduction to Natural Language Processing with SpaCy - Working Files/Chapter 2/Working-with-SpaCy-for-the-first-time_Part_1.ipynb | peterwilliams97/spaCy_practice | 71fac240c045afffdbef75f80533fe57fb62583c | [
"MIT"
] | null | null | null | 50.843342 | 10,942 | 0.678683 | [
[
[
"### Dependencies for this notebook:\n* pip install spacy, pandas, matplotlib\n* python -m spacy.en.download ",
"_____no_output_____"
]
],
[
[
"from IPython.display import SVG, display\nimport spacy\nimport pandas as pd\nimport matplotlib.pyplot as plt\nfrom datetime import datetime\n%matplotlib inline",
"_____no_output_____"
],
[
"#encode some text as uncode\ntext = u\"I'm executing this code on an Apple Computer.\"\n\n#instantiate a language model\n#to download language model: python -m spacy.en.download \nnlp = spacy.load('en') # or spacy.en.English()\n\n#create a document\ndocument = nlp(text)",
"_____no_output_____"
],
[
"for function in nlp.pipeline:\n print(function)",
"<spacy.tagger.Tagger object at 0x116f18510>\n<spacy.pipeline.DependencyParser object at 0x110759f48>\n<spacy.matcher.Matcher object at 0x110743978>\n<spacy.pipeline.EntityRecognizer object at 0x116f9d188>\n"
],
[
"### Modifying the Language Model\ndef identify_starwars(doc):\n for token in doc:\n if token.text == u'starwars':\n token.tag_ = u'NNP'\n\ndef return_pipeline(nlp):\n return [nlp.tagger, nlp.parser, nlp.matcher, nlp.entity, identify_starwars]\n\ntext = u\"I loved all of the starwars movies\"\ncustom_nlp = spacy.load('en', create_pipeline=return_pipeline)\nnew_document = custom_nlp(text)\n\nfor function in custom_nlp.pipeline:\n print(function)",
"<spacy.tagger.Tagger object at 0x11968b5a0>\n<spacy.pipeline.DependencyParser object at 0x110759e08>\n<spacy.matcher.Matcher object at 0x1107437b8>\n<spacy.pipeline.EntityRecognizer object at 0x116f9d368>\n<function identify_starwars at 0x11075e488>\n"
],
[
"texts = [u'You have brains in your head.'] * 10000\n\nfor doc in nlp.pipe(texts,n_threads=4):\n doc.is_parsed",
"_____no_output_____"
],
[
"### Deploying Model on Many Texts with .pipe\nruntimes = {}\n\nfor thread_count in [1,2,3,4,8]:\n t0 = datetime.now() \n \n #Create generator of processed documents\n processed_documents = nlp.pipe(texts,n_threads=thread_count)\n \n #Iterate over generator\n for doc in processed_documents: \n \n #pipeline is only run once we access the generator\n doc.is_parsed \n \n t1 = datetime.now()\n runtimes[thread_count] = (t1 - t0).total_seconds()\n \nax = pd.Series(runtimes).plot(kind = 'bar')\nax.set_ylabel(\"Runtime (Seconds) with N Threads\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Accessing Tokens and Spans",
"_____no_output_____"
]
],
[
[
"import pandas as pd\ndef info(obj):\n return {'type':type(obj),'__str__': str(obj)}\n\ntext = u\"\"\"spaCy excels at large-scale information extraction tasks. \nIt's written from the ground up in carefully memory-managed Cython. \"\"\"\ndocument = nlp(text)\ntoken = document[0]\nspan = document[0:3]\n\npd.DataFrame(list(map(info, [token,span,document])))",
"_____no_output_____"
]
],
[
[
"### Sentence boundary detection",
"_____no_output_____"
]
],
[
[
"print(list(document.sents))\nprint()\nfor i, sent in enumerate(document.sents):\n print('%2d: \"%s\"' % (i, sent))",
"[spaCy excels at large-scale information extraction tasks. \n, It's written from the ground up in carefully memory-managed Cython.]\n\n 0: \"spaCy excels at large-scale information extraction tasks. \n\"\n 1: \"It's written from the ground up in carefully memory-managed Cython.\"\n"
]
],
[
[
"### Tokenization",
"_____no_output_____"
]
],
[
[
"for i, token in enumerate(document):\n print('%2d: \"%s\"' % (i, token))",
" 0: \"spaCy\"\n 1: \"excels\"\n 2: \"at\"\n 3: \"large\"\n 4: \"-\"\n 5: \"scale\"\n 6: \"information\"\n 7: \"extraction\"\n 8: \"tasks\"\n 9: \".\"\n10: \"\n\"\n11: \"It\"\n12: \"'s\"\n13: \"written\"\n14: \"from\"\n15: \"the\"\n16: \"ground\"\n17: \"up\"\n18: \"in\"\n19: \"carefully\"\n20: \"memory\"\n21: \"-\"\n22: \"managed\"\n23: \"Cython\"\n24: \".\"\n"
]
],
[
[
"### Morphological decomposition",
"_____no_output_____"
]
],
[
[
"token = document[13]\nprint(\"text: %s\" % token.text)\nprint(\"suffix: %s\" % token.suffix_)\nprint(\"lemma: %s\" % token.lemma_)",
"text: written\nsuffix: ten\nlemma: write\n"
]
],
[
[
"### Part of Speech Tagging",
"_____no_output_____"
]
],
[
[
"#Part of speech and Dependency tagging\nattrs = list(map(lambda token: {\n \"token\":token,\n \"part of speech\":token.pos_,\n \"Dependency\" : token.dep_},\n document))\npd.DataFrame(attrs)",
"_____no_output_____"
]
],
[
[
"### Noun Chunking",
"_____no_output_____"
]
],
[
[
"print(\"noun chunks: %s\" % list(document.noun_chunks))",
"noun chunks: [spaCy, large-scale information extraction tasks, It, the ground]\n"
]
],
[
[
"### Named Entity Recognition",
"_____no_output_____"
]
],
[
[
"ents = [(ent, ent.root.ent_type_) for ent in document.ents]\nprint(\"entities: %s\" % ents)",
"entities: [(Cython, 'ORG')]\n"
]
],
[
[
"### Text Similarity (Using Word Vectors)",
"_____no_output_____"
]
],
[
[
"#document, span, and token similarity\ndef plot_similarities(similarities, target):\n import matplotlib.pyplot as plt\n %matplotlib inline\n f, ax = plt.subplots(1)\n index = range(len(similarities))\n ax.barh(index, similarities)\n ax.set_yticks([i + 0. for i in index])\n ax.set_yticklabels(document2)\n ax.grid(axis='x')\n ax.set_title(\"Similarity to '{}'\".format(target))\n plt.show()\n return ax\n \ncomputer = nlp(u'computer')\ndocument2 = nlp(u'You might be using a machine running Windows')\nsimilarities = list(map(lambda token: token.similarity(computer), document2))\nax = plot_similarities(similarities, computer) ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0e49352bfed48e87a329ec9263879535b5cada0 | 5,074 | ipynb | Jupyter Notebook | Gumbel Softmax Exploration.ipynb | stefanthaler/gumbel-softmax-exploration | b307abfed3258a5fdd7d712d75bf2bc32b129ae1 | [
"MIT"
] | null | null | null | Gumbel Softmax Exploration.ipynb | stefanthaler/gumbel-softmax-exploration | b307abfed3258a5fdd7d712d75bf2bc32b129ae1 | [
"MIT"
] | null | null | null | Gumbel Softmax Exploration.ipynb | stefanthaler/gumbel-softmax-exploration | b307abfed3258a5fdd7d712d75bf2bc32b129ae1 | [
"MIT"
] | null | null | null | 28.994286 | 285 | 0.56957 | [
[
[
"from IPython.core.display import display, HTML\ndisplay(HTML(\"<style>.container { width:100% !important; }</style>\"))",
"_____no_output_____"
]
],
[
[
"# Unpacking the paper - CATEGORICAL REPARAMETERIZATION WITH GUMBEL-SOFTMAX\n\n* https://arxiv.org/pdf/1611.01144.pdf\n\n\n## Introduction\n* Not much to get here\n\n",
"_____no_output_____"
],
[
"## THE GUMBEL-SOFTMAX Distribution \n\n* Gumbel-Softmax distribution, a continuous distribution over the simplex that can approximate samples from a categorical distribution: Simplex means that it consist of variables that are all between 0 .. 1, and the sum of all these variables is 1. Example: [0.2,0.2,0.2,0.4] \n\n* z is a categorical variable with class probabilities π1, π2, ...πk\n* k is the number of classes\n* samples (e.g., z's) are encoded as k-dimensional 1-hot vector. So if you have five classes, an examples is: [0,0,0,1,0]\n\n\n### you can draw samples z efficiently by: \n\n* drawing k samples from a gumbel distribution $g_1...g_k$. The samples are independent and identically distributed drawn from a Gumbel Distribution $(\\mu=0,\\beta=1)^1$\n* calculating $argmax(g_i + log(\\pi_i))$ for all $k$ samples, with $\\pi_i$ being the class probability. \n* create a one hot encoded of that argmax. \n\n### if you use softmax as approximation of argmax, you'll get gumbel-softmax\n* additionally, they add $\\tau$ as a temperature parameter to their softmax\n* $y_i = exp(x_i) / sum of all exp(x_n) $ for $n = 1..k$ \n\n\n\n\n\n\n\n\n\nhttps://en.wikipedia.org/wiki/Gumbel_distribution",
"_____no_output_____"
],
[
"$$c = \\sqrt{a^2 + b^2}$$",
"_____no_output_____"
],
[
"## Sampling from a gumbel distribution\n* GIST: https://gist.github.com/ericjang/1001afd374c2c3b7752545ce6d9ed349\n\nFootnote 1 on page 2 ",
"_____no_output_____"
]
],
[
[
"def sample_gumbel(shape, eps=1e-20): \n U = tf.random_uniform(shape,minval=0,maxval=1)\n return -tf.log(-tf.log(U + eps) + eps)",
"_____no_output_____"
],
[
"def gumbel_softmax_sample(logits, temperature): \n y = logits + sample_gumbel(tf.shape(logits))\n return tf.nn.softmax( y / temperature)\n",
"_____no_output_____"
],
[
"def gumbel_softmax(logits, temperature, hard=False):\n \"\"\"Sample from the Gumbel-Softmax distribution and optionally discretize.\n Args:\n logits: [batch_size, n_class] unnormalized log-probs\n temperature: non-negative scalar\n hard: if True, take argmax, but differentiate w.r.t. soft sample y\n Returns:\n [batch_size, n_class] sample from the Gumbel-Softmax distribution.\n If hard=True, then the returned sample will be one-hot, otherwise it will\n be a probabilitiy distribution that sums to 1 across classes\n \"\"\"\n y = gumbel_softmax_sample(logits, temperature)\n if hard:\n k = tf.shape(logits)[-1]\n #y_hard = tf.cast(tf.one_hot(tf.argmax(y,1),k), y.dtype)\n y_hard = tf.cast(tf.equal(y,tf.reduce_max(y,1,keep_dims=True)),y.dtype)\n y = tf.stop_gradient(y_hard - y) + y\n return y",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
]
] |
d0e4a5268ac5a7d985ce9901edc66fa17559b21a | 76,614 | ipynb | Jupyter Notebook | Mini-Projects/IMDB Sentiment Analysis - XGBoost (Batch Transform).ipynb | Will282/mle-nanodegree | 336423b7fdef6a7bd0f3fbc94279de6297153f75 | [
"MIT"
] | null | null | null | Mini-Projects/IMDB Sentiment Analysis - XGBoost (Batch Transform).ipynb | Will282/mle-nanodegree | 336423b7fdef6a7bd0f3fbc94279de6297153f75 | [
"MIT"
] | null | null | null | Mini-Projects/IMDB Sentiment Analysis - XGBoost (Batch Transform).ipynb | Will282/mle-nanodegree | 336423b7fdef6a7bd0f3fbc94279de6297153f75 | [
"MIT"
] | null | null | null | 60.278521 | 631 | 0.650743 | [
[
[
"# Sentiment Analysis\n\n## Using XGBoost in SageMaker\n\n_Deep Learning Nanodegree Program | Deployment_\n\n---\n\nAs our first example of using Amazon's SageMaker service we will construct a random tree model to predict the sentiment of a movie review. You may have seen a version of this example in a pervious lesson although it would have been done using the sklearn package. Instead, we will be using the XGBoost package as it is provided to us by Amazon.\n\n## Instructions\n\nSome template code has already been provided for you, and you will need to implement additional functionality to successfully complete this notebook. You will not need to modify the included code beyond what is requested. Sections that begin with '**TODO**' in the header indicate that you need to complete or implement some portion within them. Instructions will be provided for each section and the specifics of the implementation are marked in the code block with a `# TODO: ...` comment. Please be sure to read the instructions carefully!\n\nIn addition to implementing code, there may be questions for you to answer which relate to the task and your implementation. Each section where you will answer a question is preceded by a '**Question:**' header. Carefully read each question and provide your answer below the '**Answer:**' header by editing the Markdown cell.\n\n> **Note**: Code and Markdown cells can be executed using the **Shift+Enter** keyboard shortcut. In addition, a cell can be edited by typically clicking it (double-click for Markdown cells) or by pressing **Enter** while it is highlighted.",
"_____no_output_____"
],
[
"## Step 1: Downloading the data\n\nThe dataset we are going to use is very popular among researchers in Natural Language Processing, usually referred to as the [IMDb dataset](http://ai.stanford.edu/~amaas/data/sentiment/). It consists of movie reviews from the website [imdb.com](http://www.imdb.com/), each labeled as either '**pos**itive', if the reviewer enjoyed the film, or '**neg**ative' otherwise.\n\n> Maas, Andrew L., et al. [Learning Word Vectors for Sentiment Analysis](http://ai.stanford.edu/~amaas/data/sentiment/). In _Proceedings of the 49th Annual Meeting of the Association for Computational Linguistics: Human Language Technologies_. Association for Computational Linguistics, 2011.\n\nWe begin by using some Jupyter Notebook magic to download and extract the dataset.",
"_____no_output_____"
]
],
[
[
"# %mkdir ../data\n# !wget -O ../data/aclImdb_v1.tar.gz http://ai.stanford.edu/~amaas/data/sentiment/aclImdb_v1.tar.gz\n# !tar -zxf ../data/aclImdb_v1.tar.gz -C ../data",
"_____no_output_____"
]
],
[
[
"## Step 2: Preparing the data\n\nThe data we have downloaded is split into various files, each of which contains a single review. It will be much easier going forward if we combine these individual files into two large files, one for training and one for testing.",
"_____no_output_____"
]
],
[
[
"import os\nimport glob\n\ndef read_imdb_data(data_dir='../data/aclImdb'):\n data = {}\n labels = {}\n \n for data_type in ['train', 'test']:\n data[data_type] = {}\n labels[data_type] = {}\n \n for sentiment in ['pos', 'neg']:\n data[data_type][sentiment] = []\n labels[data_type][sentiment] = []\n \n path = os.path.join(data_dir, data_type, sentiment, '*.txt')\n files = glob.glob(path)\n \n for f in files:\n with open(f) as review:\n data[data_type][sentiment].append(review.read())\n # Here we represent a positive review by '1' and a negative review by '0'\n labels[data_type][sentiment].append(1 if sentiment == 'pos' else 0)\n \n assert len(data[data_type][sentiment]) == len(labels[data_type][sentiment]), \\\n \"{}/{} data size does not match labels size\".format(data_type, sentiment)\n \n return data, labels",
"_____no_output_____"
],
[
"data, labels = read_imdb_data()\nprint(\"IMDB reviews: train = {} pos / {} neg, test = {} pos / {} neg\".format(\n len(data['train']['pos']), len(data['train']['neg']),\n len(data['test']['pos']), len(data['test']['neg'])))",
"IMDB reviews: train = 12500 pos / 12500 neg, test = 12500 pos / 12500 neg\n"
],
[
"from sklearn.utils import shuffle\n\ndef prepare_imdb_data(data, labels):\n \"\"\"Prepare training and test sets from IMDb movie reviews.\"\"\"\n \n #Combine positive and negative reviews and labels\n data_train = data['train']['pos'] + data['train']['neg']\n data_test = data['test']['pos'] + data['test']['neg']\n labels_train = labels['train']['pos'] + labels['train']['neg']\n labels_test = labels['test']['pos'] + labels['test']['neg']\n \n #Shuffle reviews and corresponding labels within training and test sets\n data_train, labels_train = shuffle(data_train, labels_train)\n data_test, labels_test = shuffle(data_test, labels_test)\n \n # Return a unified training data, test data, training labels, test labets\n return data_train, data_test, labels_train, labels_test",
"_____no_output_____"
],
[
"train_X, test_X, train_y, test_y = prepare_imdb_data(data, labels)\nprint(\"IMDb reviews (combined): train = {}, test = {}\".format(len(train_X), len(test_X)))",
"IMDb reviews (combined): train = 25000, test = 25000\n"
],
[
"train_X[100]",
"_____no_output_____"
]
],
[
[
"## Step 3: Processing the data\n\nNow that we have our training and testing datasets merged and ready to use, we need to start processing the raw data into something that will be useable by our machine learning algorithm. To begin with, we remove any html formatting that may appear in the reviews and perform some standard natural language processing in order to homogenize the data.",
"_____no_output_____"
]
],
[
[
"import nltk\nnltk.download(\"stopwords\")\nfrom nltk.corpus import stopwords\nfrom nltk.stem.porter import *\nstemmer = PorterStemmer()",
"[nltk_data] Downloading package stopwords to\n[nltk_data] /home/ec2-user/nltk_data...\n[nltk_data] Package stopwords is already up-to-date!\n"
],
[
"import re\nfrom bs4 import BeautifulSoup\n\ndef review_to_words(review):\n text = BeautifulSoup(review, \"html.parser\").get_text() # Remove HTML tags\n text = re.sub(r\"[^a-zA-Z0-9]\", \" \", text.lower()) # Convert to lower case\n words = text.split() # Split string into words\n words = [w for w in words if w not in stopwords.words(\"english\")] # Remove stopwords\n words = [PorterStemmer().stem(w) for w in words] # stem\n \n return words",
"_____no_output_____"
],
[
"import pickle\n\ncache_dir = os.path.join(\"../cache\", \"sentiment_analysis\") # where to store cache files\nos.makedirs(cache_dir, exist_ok=True) # ensure cache directory exists\n\ndef preprocess_data(data_train, data_test, labels_train, labels_test,\n cache_dir=cache_dir, cache_file=\"preprocessed_data.pkl\"):\n \"\"\"Convert each review to words; read from cache if available.\"\"\"\n\n # If cache_file is not None, try to read from it first\n cache_data = None\n if cache_file is not None:\n try:\n with open(os.path.join(cache_dir, cache_file), \"rb\") as f:\n cache_data = pickle.load(f)\n print(\"Read preprocessed data from cache file:\", cache_file)\n except:\n pass # unable to read from cache, but that's okay\n \n # If cache is missing, then do the heavy lifting\n if cache_data is None:\n # Preprocess training and test data to obtain words for each review\n #words_train = list(map(review_to_words, data_train))\n #words_test = list(map(review_to_words, data_test))\n words_train = [review_to_words(review) for review in data_train]\n words_test = [review_to_words(review) for review in data_test]\n \n # Write to cache file for future runs\n if cache_file is not None:\n cache_data = dict(words_train=words_train, words_test=words_test,\n labels_train=labels_train, labels_test=labels_test)\n with open(os.path.join(cache_dir, cache_file), \"wb\") as f:\n pickle.dump(cache_data, f)\n print(\"Wrote preprocessed data to cache file:\", cache_file)\n else:\n # Unpack data loaded from cache file\n words_train, words_test, labels_train, labels_test = (cache_data['words_train'],\n cache_data['words_test'], cache_data['labels_train'], cache_data['labels_test'])\n \n return words_train, words_test, labels_train, labels_test",
"_____no_output_____"
],
[
"# Preprocess data\ntrain_X, test_X, train_y, test_y = preprocess_data(train_X, test_X, train_y, test_y)",
"Read preprocessed data from cache file: preprocessed_data.pkl\n"
]
],
[
[
"### Extract Bag-of-Words features\n\nFor the model we will be implementing, rather than using the reviews directly, we are going to transform each review into a Bag-of-Words feature representation. Keep in mind that 'in the wild' we will only have access to the training set so our transformer can only use the training set to construct a representation.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom sklearn.feature_extraction.text import CountVectorizer\nfrom sklearn.externals import joblib\n# joblib is an enhanced version of pickle that is more efficient for storing NumPy arrays\n\ndef extract_BoW_features(words_train, words_test, vocabulary_size=5000,\n cache_dir=cache_dir, cache_file=\"bow_features.pkl\"):\n \"\"\"Extract Bag-of-Words for a given set of documents, already preprocessed into words.\"\"\"\n \n # If cache_file is not None, try to read from it first\n cache_data = None\n if cache_file is not None:\n try:\n with open(os.path.join(cache_dir, cache_file), \"rb\") as f:\n cache_data = joblib.load(f)\n print(\"Read features from cache file:\", cache_file)\n except:\n pass # unable to read from cache, but that's okay\n \n # If cache is missing, then do the heavy lifting\n if cache_data is None:\n # Fit a vectorizer to training documents and use it to transform them\n # NOTE: Training documents have already been preprocessed and tokenized into words;\n # pass in dummy functions to skip those steps, e.g. preprocessor=lambda x: x\n vectorizer = CountVectorizer(max_features=vocabulary_size,\n preprocessor=lambda x: x, tokenizer=lambda x: x) # already preprocessed\n features_train = vectorizer.fit_transform(words_train).toarray()\n\n # Apply the same vectorizer to transform the test documents (ignore unknown words)\n features_test = vectorizer.transform(words_test).toarray()\n \n # NOTE: Remember to convert the features using .toarray() for a compact representation\n \n # Write to cache file for future runs (store vocabulary as well)\n if cache_file is not None:\n vocabulary = vectorizer.vocabulary_\n cache_data = dict(features_train=features_train, features_test=features_test,\n vocabulary=vocabulary)\n with open(os.path.join(cache_dir, cache_file), \"wb\") as f:\n joblib.dump(cache_data, f)\n print(\"Wrote features to cache file:\", cache_file)\n else:\n # Unpack data loaded from cache file\n features_train, features_test, vocabulary = (cache_data['features_train'],\n cache_data['features_test'], cache_data['vocabulary'])\n \n # Return both the extracted features as well as the vocabulary\n return features_train, features_test, vocabulary",
"_____no_output_____"
],
[
"# Extract Bag of Words features for both training and test datasets\ntrain_X, test_X, vocabulary = extract_BoW_features(train_X, test_X)",
"Read features from cache file: bow_features.pkl\n"
]
],
[
[
"## Step 4: Classification using XGBoost\n\nNow that we have created the feature representation of our training (and testing) data, it is time to start setting up and using the XGBoost classifier provided by SageMaker.\n\n### (TODO) Writing the dataset\n\nThe XGBoost classifier that we will be using requires the dataset to be written to a file and stored using Amazon S3. To do this, we will start by splitting the training dataset into two parts, the data we will train the model with and a validation set. Then, we will write those datasets to a file and upload the files to S3. In addition, we will write the test set input to a file and upload the file to S3. This is so that we can use SageMakers Batch Transform functionality to test our model once we've fit it.",
"_____no_output_____"
]
],
[
[
"print(len(train_y))",
"25000\n"
],
[
"import pandas as pd\nfrom sklearn.model_selection import train_test_split\n\n# TODO: Split the train_X and train_y arrays into the DataFrames val_X, train_X and val_y, train_y. Make sure that\n# val_X and val_y contain 10 000 entires while train_X and train_y contain the remaining 15 000 entries.\ntrain_X, val_X, train_y, val_y = train_test_split(train_X, train_y, test_size=0.4)\n\ntrain_X = pd.DataFrame(train_X)\ntrain_y = pd.DataFrame(train_y)\nval_X = pd.DataFrame(val_X)\nval_y = pd.DataFrame(val_y)",
"_____no_output_____"
],
[
"print(train_X.shape)\nprint(val_X.shape)",
"(15000, 5000)\n(10000, 5000)\n"
]
],
[
[
"The documentation for the XGBoost algorithm in SageMaker requires that the saved datasets should contain no headers or index and that for the training and validation data, the label should occur first for each sample.\n\nFor more information about this and other algorithms, the SageMaker developer documentation can be found on __[Amazon's website.](https://docs.aws.amazon.com/sagemaker/latest/dg/)__",
"_____no_output_____"
]
],
[
[
"# First we make sure that the local directory in which we'd like to store the training and validation csv files exists.\ndata_dir = '../data/xgboost'\nif not os.path.exists(data_dir):\n os.makedirs(data_dir)",
"_____no_output_____"
],
[
"# First, save the test data to test.csv in the data_dir directory. Note that we do not save the associated ground truth\n# labels, instead we will use them later to compare with our model output.\n\npd.DataFrame(test_X).to_csv(os.path.join(data_dir, 'test.csv'), header=False, index=False)",
"_____no_output_____"
],
[
"# TODO: Save the training and validation data to train.csv and validation.csv in the data_dir directory.\n# Make sure that the files you create are in the correct format.\npd.concat([train_y, train_X], axis=1).to_csv(os.path.join(data_dir, 'train.csv'), header=False, index=False)\npd.concat([val_y, val_X], axis=1).to_csv(os.path.join(data_dir, 'validation.csv'), header=False, index=False)",
"_____no_output_____"
],
[
"# To save a bit of memory we can set text_X, train_X, val_X, train_y and val_y to None.\n\ntest_X = train_X = val_X = train_y = val_y = None",
"_____no_output_____"
]
],
[
[
"### (TODO) Uploading Training / Validation files to S3\n\nAmazon's S3 service allows us to store files that can be access by both the built-in training models such as the XGBoost model we will be using as well as custom models such as the one we will see a little later.\n\nFor this, and most other tasks we will be doing using SageMaker, there are two methods we could use. The first is to use the low level functionality of SageMaker which requires knowing each of the objects involved in the SageMaker environment. The second is to use the high level functionality in which certain choices have been made on the user's behalf. The low level approach benefits from allowing the user a great deal of flexibility while the high level approach makes development much quicker. For our purposes we will opt to use the high level approach although using the low-level approach is certainly an option.\n\nRecall the method `upload_data()` which is a member of object representing our current SageMaker session. What this method does is upload the data to the default bucket (which is created if it does not exist) into the path described by the key_prefix variable. To see this for yourself, once you have uploaded the data files, go to the S3 console and look to see where the files have been uploaded.\n\nFor additional resources, see the __[SageMaker API documentation](http://sagemaker.readthedocs.io/en/latest/)__ and in addition the __[SageMaker Developer Guide.](https://docs.aws.amazon.com/sagemaker/latest/dg/)__",
"_____no_output_____"
]
],
[
[
"import sagemaker\n\nsession = sagemaker.Session() # Store the current SageMaker session\n\n# S3 prefix (which folder will we use)\nprefix = 'sentiment-xgboost'\n\n# TODO: Upload the test.csv, train.csv and validation.csv files which are contained in data_dir to S3 using sess.upload_data().\ntest_location = session.upload_data(os.path.join(data_dir, 'test.csv'), key_prefix=prefix)\nval_location = session.upload_data(os.path.join(data_dir, 'train.csv'), key_prefix=prefix)\ntrain_location = session.upload_data(os.path.join(data_dir, 'validation.csv'), key_prefix=prefix)",
"_____no_output_____"
]
],
[
[
"### (TODO) Creating the XGBoost model\n\nNow that the data has been uploaded it is time to create the XGBoost model. To begin with, we need to do some setup. At this point it is worth discussing what a model is in SageMaker. It is easiest to think of a model of comprising three different objects in the SageMaker ecosystem, which interact with one another.\n\n- Model Artifacts\n- Training Code (Container)\n- Inference Code (Container)\n\nThe Model Artifacts are what you might think of as the actual model itself. For example, if you were building a neural network, the model artifacts would be the weights of the various layers. In our case, for an XGBoost model, the artifacts are the actual trees that are created during training.\n\nThe other two objects, the training code and the inference code are then used the manipulate the training artifacts. More precisely, the training code uses the training data that is provided and creates the model artifacts, while the inference code uses the model artifacts to make predictions on new data.\n\nThe way that SageMaker runs the training and inference code is by making use of Docker containers. For now, think of a container as being a way of packaging code up so that dependencies aren't an issue.",
"_____no_output_____"
]
],
[
[
"from sagemaker import get_execution_role\n\n# Our current execution role is require when creating the model as the training\n# and inference code will need to access the model artifacts.\nrole = get_execution_role()",
"_____no_output_____"
],
[
"# We need to retrieve the location of the container which is provided by Amazon for using XGBoost.\n# As a matter of convenience, the training and inference code both use the same container.\nfrom sagemaker.amazon.amazon_estimator import get_image_uri\n\ncontainer = get_image_uri(session.boto_region_name, 'xgboost')",
"WARNING:root:There is a more up to date SageMaker XGBoost image. To use the newer image, please set 'repo_version'='0.90-1'. For example:\n\tget_image_uri(region, 'xgboost', '0.90-1').\n"
],
[
"# TODO: Create a SageMaker estimator using the container location determined in the previous cell.\n# It is recommended that you use a single training instance of type ml.m4.xlarge. It is also\n# recommended that you use 's3://{}/{}/output'.format(session.default_bucket(), prefix) as the\n# output path.\n\nxgb = sagemaker.estimator.Estimator(container, # The image name of the training container\n role, # The IAM role to use (our current role in this case)\n train_instance_count=1, # The number of instances to use for training\n train_instance_type='ml.m4.xlarge', # The type of instance to use for training\n output_path='s3://{}/{}/output'.format(session.default_bucket(), prefix),\n # Where to save the output (the model artifacts)\n sagemaker_session=session) # The current SageMaker session\n\n\n",
"_____no_output_____"
],
[
"# TODO: Set the XGBoost hyperparameters in the xgb object. Don't forget that in this case we have a binary\n# label so we should be using the 'binary:logistic' objective.\nxgb.set_hyperparameters(max_depth=6,\n eta=0.2,\n gamma=0,\n min_child_weight=1,\n subsample=0.8,\n objective='binary:logistic',\n early_stopping_rounds=10,\n num_round=200)\n",
"_____no_output_____"
]
],
[
[
"### Fit the XGBoost model\n\nNow that our model has been set up we simply need to attach the training and validation datasets and then ask SageMaker to set up the computation.",
"_____no_output_____"
]
],
[
[
"s3_input_train = sagemaker.s3_input(s3_data=train_location, content_type='csv')\ns3_input_validation = sagemaker.s3_input(s3_data=val_location, content_type='csv')",
"_____no_output_____"
],
[
"xgb.fit({'train': s3_input_train, 'validation': s3_input_validation})",
"2020-05-02 12:55:23 Starting - Starting the training job...\n2020-05-02 12:55:25 Starting - Launching requested ML instances...\n2020-05-02 12:56:20 Starting - Preparing the instances for training......\n2020-05-02 12:57:09 Downloading - Downloading input data...\n2020-05-02 12:57:47 Training - Training image download completed. Training in progress..\u001b[34mArguments: train\u001b[0m\n\u001b[34m[2020-05-02:12:57:48:INFO] Running standalone xgboost training.\u001b[0m\n\u001b[34m[2020-05-02:12:57:48:INFO] File size need to be processed in the node: 238.47mb. Available memory size in the node: 8482.86mb\u001b[0m\n\u001b[34m[2020-05-02:12:57:48:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[12:57:48] S3DistributionType set as FullyReplicated\u001b[0m\n\u001b[34m[12:57:49] 10000x5000 matrix with 50000000 entries loaded from /opt/ml/input/data/train?format=csv&label_column=0&delimiter=,\u001b[0m\n\u001b[34m[2020-05-02:12:57:49:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[12:57:49] S3DistributionType set as FullyReplicated\u001b[0m\n\u001b[34m[12:57:51] 15000x5000 matrix with 75000000 entries loaded from /opt/ml/input/data/validation?format=csv&label_column=0&delimiter=,\u001b[0m\n\u001b[34m[12:57:53] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 76 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[0]#011train-error:0.2707#011validation-error:0.294867\u001b[0m\n\u001b[34mMultiple eval metrics have been passed: 'validation-error' will be used for early stopping.\n\u001b[0m\n\u001b[34mWill train until validation-error hasn't improved in 10 rounds.\u001b[0m\n\u001b[34m[12:57:54] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 80 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[1]#011train-error:0.2561#011validation-error:0.2804\u001b[0m\n\u001b[34m[12:57:55] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 78 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[2]#011train-error:0.2557#011validation-error:0.280267\u001b[0m\n\u001b[34m[12:57:57] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 84 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[3]#011train-error:0.2451#011validation-error:0.2744\u001b[0m\n\u001b[34m[12:57:58] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 58 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[4]#011train-error:0.2414#011validation-error:0.2712\u001b[0m\n\u001b[34m[12:57:59] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 76 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[5]#011train-error:0.2329#011validation-error:0.263133\u001b[0m\n\u001b[34m[12:58:00] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 82 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[6]#011train-error:0.2217#011validation-error:0.2488\u001b[0m\n\u001b[34m[12:58:01] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 76 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[7]#011train-error:0.2191#011validation-error:0.249067\u001b[0m\n\u001b[34m[12:58:02] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 80 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[8]#011train-error:0.207#011validation-error:0.242867\u001b[0m\n\u001b[34m[12:58:03] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 68 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[9]#011train-error:0.1984#011validation-error:0.2372\u001b[0m\n\u001b[34m[12:58:04] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 62 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[10]#011train-error:0.1911#011validation-error:0.230533\u001b[0m\n\u001b[34m[12:58:05] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 68 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[11]#011train-error:0.1843#011validation-error:0.227\u001b[0m\n\u001b[34m[12:58:06] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 70 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[12]#011train-error:0.1795#011validation-error:0.222867\u001b[0m\n\u001b[34m[12:58:07] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 62 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[13]#011train-error:0.1748#011validation-error:0.2212\u001b[0m\n\u001b[34m[12:58:08] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 78 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[14]#011train-error:0.1696#011validation-error:0.218733\u001b[0m\n\u001b[34m[12:58:09] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 64 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[15]#011train-error:0.166#011validation-error:0.216133\u001b[0m\n\u001b[34m[12:58:10] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 64 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[16]#011train-error:0.1623#011validation-error:0.214133\u001b[0m\n\u001b[34m[12:58:11] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 76 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[17]#011train-error:0.1605#011validation-error:0.2092\u001b[0m\n\u001b[34m[12:58:12] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 76 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[18]#011train-error:0.1543#011validation-error:0.2074\u001b[0m\n\u001b[34m[12:58:13] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 72 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[19]#011train-error:0.1498#011validation-error:0.204133\u001b[0m\n\u001b[34m[12:58:14] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 60 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[20]#011train-error:0.1471#011validation-error:0.201933\u001b[0m\n\u001b[34m[12:58:15] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 46 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[21]#011train-error:0.1446#011validation-error:0.200133\u001b[0m\n\u001b[34m[12:58:16] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 70 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[22]#011train-error:0.1402#011validation-error:0.1988\u001b[0m\n\u001b[34m[12:58:17] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 86 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[23]#011train-error:0.1382#011validation-error:0.1982\u001b[0m\n\u001b[34m[12:58:19] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 48 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[24]#011train-error:0.1343#011validation-error:0.196333\u001b[0m\n\u001b[34m[12:58:20] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 56 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[25]#011train-error:0.1312#011validation-error:0.192467\u001b[0m\n\u001b[34m[12:58:21] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 48 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[26]#011train-error:0.1294#011validation-error:0.190133\u001b[0m\n\u001b[34m[12:58:22] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 68 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[27]#011train-error:0.1286#011validation-error:0.189133\u001b[0m\n\u001b[34m[12:58:23] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 58 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[28]#011train-error:0.1263#011validation-error:0.187333\u001b[0m\n\u001b[34m[12:58:24] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 60 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[29]#011train-error:0.1244#011validation-error:0.186733\u001b[0m\n\u001b[34m[12:58:25] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 48 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[30]#011train-error:0.1231#011validation-error:0.186267\u001b[0m\n\u001b[34m[12:58:26] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 56 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[31]#011train-error:0.1218#011validation-error:0.186067\u001b[0m\n\u001b[34m[12:58:27] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 92 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[32]#011train-error:0.1176#011validation-error:0.185267\u001b[0m\n\u001b[34m[12:58:28] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 76 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[33]#011train-error:0.1167#011validation-error:0.1844\u001b[0m\n\u001b[34m[12:58:29] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 52 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[34]#011train-error:0.114#011validation-error:0.182667\u001b[0m\n\u001b[34m[12:58:30] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 64 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[35]#011train-error:0.1122#011validation-error:0.181467\u001b[0m\n\u001b[34m[12:58:31] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 60 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[36]#011train-error:0.1095#011validation-error:0.181067\u001b[0m\n\u001b[34m[12:58:32] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 44 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[37]#011train-error:0.1085#011validation-error:0.179933\u001b[0m\n\u001b[34m[12:58:33] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 66 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[38]#011train-error:0.1074#011validation-error:0.1798\u001b[0m\n\u001b[34m[12:58:34] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 46 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[39]#011train-error:0.107#011validation-error:0.178933\u001b[0m\n\u001b[34m[12:58:35] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 58 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[40]#011train-error:0.1059#011validation-error:0.178667\u001b[0m\n\u001b[34m[12:58:36] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 70 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[41]#011train-error:0.1037#011validation-error:0.177533\u001b[0m\n\u001b[34m[12:58:37] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 60 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[42]#011train-error:0.102#011validation-error:0.176133\u001b[0m\n\u001b[34m[12:58:38] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 46 extra nodes, 0 pruned nodes, max_depth=6\u001b[0m\n\u001b[34m[43]#011train-error:0.101#011validation-error:0.175867\u001b[0m\n"
]
],
[
[
"### (TODO) Testing the model\n\nNow that we've fit our XGBoost model, it's time to see how well it performs. To do this we will use SageMakers Batch Transform functionality. Batch Transform is a convenient way to perform inference on a large dataset in a way that is not realtime. That is, we don't necessarily need to use our model's results immediately and instead we can peform inference on a large number of samples. An example of this in industry might be peforming an end of month report. This method of inference can also be useful to us as it means to can perform inference on our entire test set. \n\nTo perform a Batch Transformation we need to first create a transformer objects from our trained estimator object.",
"_____no_output_____"
]
],
[
[
"# TODO: Create a transformer object from the trained model. Using an instance count of 1 and an instance type of ml.m4.xlarge\n# should be more than enough.\nxgb_transformer = xgb.transformer(instance_count = 1, instance_type = 'ml.m4.xlarge')",
"_____no_output_____"
]
],
[
[
"Next we actually perform the transform job. When doing so we need to make sure to specify the type of data we are sending so that it is serialized correctly in the background. In our case we are providing our model with csv data so we specify `text/csv`. Also, if the test data that we have provided is too large to process all at once then we need to specify how the data file should be split up. Since each line is a single entry in our data set we tell SageMaker that it can split the input on each line.",
"_____no_output_____"
]
],
[
[
"# TODO: Start the transform job. Make sure to specify the content type and the split type of the test data.\nxgb_transformer.transform(test_location, content_type='text/csv', split_type='Line')",
"_____no_output_____"
]
],
[
[
"Currently the transform job is running but it is doing so in the background. Since we wish to wait until the transform job is done and we would like a bit of feedback we can run the `wait()` method.",
"_____no_output_____"
]
],
[
[
"xgb_transformer.wait()",
".....................\u001b[34mArguments: serve\u001b[0m\n\u001b[34m[2020-05-02 13:07:37 +0000] [1] [INFO] Starting gunicorn 19.7.1\u001b[0m\n\u001b[34m[2020-05-02 13:07:37 +0000] [1] [INFO] Listening at: http://0.0.0.0:8080 (1)\u001b[0m\n\u001b[34m[2020-05-02 13:07:37 +0000] [1] [INFO] Using worker: gevent\u001b[0m\n\u001b[34m[2020-05-02 13:07:37 +0000] [38] [INFO] Booting worker with pid: 38\u001b[0m\n\u001b[34m[2020-05-02 13:07:37 +0000] [39] [INFO] Booting worker with pid: 39\u001b[0m\n\u001b[34m[2020-05-02 13:07:37 +0000] [40] [INFO] Booting worker with pid: 40\u001b[0m\n\u001b[34m[2020-05-02 13:07:37 +0000] [41] [INFO] Booting worker with pid: 41\u001b[0m\n\u001b[34m[2020-05-02:13:07:37:INFO] Model loaded successfully for worker : 38\u001b[0m\n\u001b[34m[2020-05-02:13:07:37:INFO] Model loaded successfully for worker : 40\u001b[0m\n\u001b[34m[2020-05-02:13:07:37:INFO] Model loaded successfully for worker : 39\u001b[0m\n\u001b[34m[2020-05-02:13:07:37:INFO] Model loaded successfully for worker : 41\u001b[0m\n\u001b[32m2020-05-02T13:08:05.699:[sagemaker logs]: MaxConcurrentTransforms=4, MaxPayloadInMB=6, BatchStrategy=MULTI_RECORD\u001b[0m\n\u001b[34m[2020-05-02:13:08:08:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:08:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:08:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:08:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:09:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:09:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:08:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:08:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:08:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:08:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:09:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:09:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:09:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:09:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:09:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:09:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:11:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:11:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:11:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:11:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:11:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:11:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:11:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:11:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:11:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:11:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:11:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:11:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:11:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:11:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:11:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:11:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:16:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:16:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:16:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:16:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:16:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:16:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:16:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:16:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:16:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:16:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:16:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:16:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:16:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:16:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:16:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:16:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:18:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:18:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:18:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:18:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:18:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:18:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:18:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:18:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:18:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:18:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:18:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:18:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:18:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:18:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:18:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:18:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:20:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:20:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:20:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:20:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:21:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:21:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:21:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:21:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:21:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:21:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:21:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:21:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:21:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:21:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:21:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:21:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:23:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:23:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:23:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:23:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:23:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:23:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:23:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:23:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:25:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:25:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:26:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:26:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:25:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:25:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:26:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:26:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:26:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:26:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:26:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:26:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:26:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:26:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:26:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:26:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:28:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:28:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:28:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:28:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:28:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:28:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:28:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:28:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:28:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:28:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:28:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:28:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:28:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:28:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:28:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:28:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:30:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:30:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:30:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:30:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:30:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:30:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:30:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:30:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:30:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:30:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:31:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-05-02:13:08:31:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:30:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:30:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:31:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-05-02:13:08:31:INFO] Determined delimiter of CSV input is ','\u001b[0m\n"
]
],
[
[
"Now the transform job has executed and the result, the estimated sentiment of each review, has been saved on S3. Since we would rather work on this file locally we can perform a bit of notebook magic to copy the file to the `data_dir`.",
"_____no_output_____"
]
],
[
[
"!aws s3 cp --recursive $xgb_transformer.output_path $data_dir",
"Completed 256.0 KiB/370.4 KiB (3.5 MiB/s) with 1 file(s) remaining\rCompleted 370.4 KiB/370.4 KiB (4.9 MiB/s) with 1 file(s) remaining\rdownload: s3://sagemaker-eu-west-2-463062221782/xgboost-2020-05-02-13-04-13-686/test.csv.out to ../data/xgboost/test.csv.out\r\n"
]
],
[
[
"The last step is now to read in the output from our model, convert the output to something a little more usable, in this case we want the sentiment to be either `1` (positive) or `0` (negative), and then compare to the ground truth labels.",
"_____no_output_____"
]
],
[
[
"predictions = pd.read_csv(os.path.join(data_dir, 'test.csv.out'), header=None)\npredictions = [round(num) for num in predictions.squeeze().values]",
"_____no_output_____"
],
[
"from sklearn.metrics import accuracy_score\naccuracy_score(test_y, predictions)",
"_____no_output_____"
]
],
[
[
"## Optional: Clean up\n\nThe default notebook instance on SageMaker doesn't have a lot of excess disk space available. As you continue to complete and execute notebooks you will eventually fill up this disk space, leading to errors which can be difficult to diagnose. Once you are completely finished using a notebook it is a good idea to remove the files that you created along the way. Of course, you can do this from the terminal or from the notebook hub if you would like. The cell below contains some commands to clean up the created files from within the notebook.",
"_____no_output_____"
]
],
[
[
"# First we will remove all of the files contained in the data_dir directory\n!rm $data_dir/*\n\n# And then we delete the directory itself\n!rmdir $data_dir\n\n# Similarly we will remove the files in the cache_dir directory and the directory itself\n!rm $cache_dir/*\n!rmdir $cache_dir",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0e4a606097b11940bba23b54b38a31e911e0a65 | 30,733 | ipynb | Jupyter Notebook | k1lib/callbacks/shorts.ipynb | 157239n/k1lib | 285520b8364ad5b21cb736b44471aa939e692e9b | [
"MIT"
] | 1 | 2021-08-11T19:10:08.000Z | 2021-08-11T19:10:08.000Z | k1lib/callbacks/shorts.ipynb | 157239n/k1lib | 285520b8364ad5b21cb736b44471aa939e692e9b | [
"MIT"
] | null | null | null | k1lib/callbacks/shorts.ipynb | 157239n/k1lib | 285520b8364ad5b21cb736b44471aa939e692e9b | [
"MIT"
] | null | null | null | 51.479062 | 132 | 0.65633 | [
[
[
"__name__ = \"k1lib.callbacks\"",
"_____no_output_____"
],
[
"#export\nfrom .callbacks import Callback, Callbacks, Cbs\nimport k1lib, os, torch\n__all__ = [\"Autosave\", \"DontTrainValid\", \"InspectLoss\", \"ModifyLoss\", \"Cpu\", \"Cuda\",\n \"DType\", \"InspectBatch\", \"ModifyBatch\", \"InspectOutput\", \"ModifyOutput\", \n \"Beep\"]",
"_____no_output_____"
],
[
"#export\[email protected](Cbs)\nclass Autosave(Callback):\n \"\"\"Autosaves 3 versions of the network to disk\"\"\"\n def __init__(self): super().__init__(); self.order = 23\n def endRun(self):\n os.system(\"mv autosave-1.pth autosave-0.pth\")\n os.system(\"mv autosave-2.pth autosave-1.pth\")\n self.l.save(\"autosave-2.pth\")",
"_____no_output_____"
],
[
"#export\[email protected](Cbs)\nclass DontTrainValid(Callback):\n \"\"\"If is not training, then don't run m.backward() and opt.step().\nThe core training loop in k1lib.Learner don't specifically do this,\ncause there may be some weird cases where you want to also train valid.\"\"\"\n def _common(self):\n if not self.l.model.training: return True\n def startBackward(self): return self._common()\n def startStep(self): return self._common()",
"_____no_output_____"
],
[
"#export\[email protected](Cbs)\nclass InspectLoss(Callback):\n \"\"\"Expected `f` to take in 1 float.\"\"\"\n def __init__(self, f): super().__init__(); self.f = f; self.order = 15\n def endLoss(self): self.f(self.loss.detach())",
"_____no_output_____"
],
[
"#export\[email protected](Cbs)\nclass ModifyLoss(Callback):\n \"\"\"Expected `f` to take in 1 float and return 1 float.\"\"\"\n def __init__(self, f): super().__init__(); self.f = f; self.order = 13\n def endLoss(self): self.l.loss = self.f(self.loss)",
"_____no_output_____"
],
[
"#export\[email protected](Cbs)\nclass Cuda(Callback):\n \"\"\"Moves batch and model to the default GPU\"\"\"\n def startRun(self): self.l.model.cuda()\n def startBatch(self):\n self.l.xb = self.l.xb.cuda()\n self.l.yb = self.l.yb.cuda()",
"_____no_output_____"
],
[
"#export\[email protected](Cbs)\nclass Cpu(Callback):\n \"\"\"Moves batch and model to CPU\"\"\"\n def startRun(self): self.l.model.cpu()\n def startBatch(self):\n self.l.xb = self.l.xb.cpu()\n self.l.yb = self.l.yb.cpu()",
"_____no_output_____"
],
[
"#export\[email protected](Cbs)\nclass DType(Callback):\n \"\"\"Moves batch and model to a specified data type\"\"\"\n def __init__(self, dtype): super().__init__(); self.dtype = dtype\n def startRun(self): self.l.model = self.l.model.to(self.dtype)\n def startBatch(self):\n self.l.xb = self.l.xb.to(self.dtype)\n self.l.yb = self.l.yb.to(self.dtype)",
"_____no_output_____"
],
[
"#export\[email protected](Cbs)\nclass InspectBatch(Callback):\n \"\"\"Expected `f` to take in 2 tensors.\"\"\"\n def __init__(self, f:callable): super().__init__(); self.f = f; self.order = 15\n def startBatch(self): self.f(self.l.xb, self.l.yb)",
"_____no_output_____"
],
[
"#export\[email protected](Cbs)\nclass ModifyBatch(Callback):\n \"\"\"Modifies xb and yb on the fly. Expected `f`\n to take in 2 tensors and return 2 tensors.\"\"\"\n def __init__(self, f): super().__init__(); self.f = f; self.order = 13\n def startBatch(self): self.l.xb, self.l.yb = self.f(self.l.xb, self.l.yb)",
"_____no_output_____"
],
[
"#export\[email protected](Cbs)\nclass InspectOutput(Callback):\n \"\"\"Expected `f` to take in 1 tensor.\"\"\"\n def __init__(self, f): super().__init__(); self.f = f; self.order = 15\n def endPass(self): self.f(self.y)",
"_____no_output_____"
],
[
"#export\[email protected](Cbs)\nclass ModifyOutput(Callback):\n \"\"\"Modifies output on the fly. Expected `f` to take\nin 1 tensor and return 1 tensor\"\"\"\n def __init__(self, f): super().__init__(); self.f = f; self.order = 13\n def endPass(self): self.l.y = self.f(self.y)",
"_____no_output_____"
],
[
"#export\[email protected](Cbs)\nclass Beep(Callback):\n \"\"\"Plays a beep sound when the run is over\"\"\"\n def endRun(self): k1lib.beep()",
"_____no_output_____"
],
[
"!../../export.py callbacks/shorts",
"Current dir: /home/kelvin/repos/labs/k1lib, ../../export.py\nrm: cannot remove '__pycache__': No such file or directory\nFound existing installation: k1lib 0.8\nUninstalling k1lib-0.8:\n Successfully uninstalled k1lib-0.8\nrunning install\nrunning bdist_egg\nrunning egg_info\ncreating k1lib.egg-info\nwriting k1lib.egg-info/PKG-INFO\nwriting dependency_links to k1lib.egg-info/dependency_links.txt\nwriting requirements to k1lib.egg-info/requires.txt\nwriting top-level names to k1lib.egg-info/top_level.txt\nwriting manifest file 'k1lib.egg-info/SOURCES.txt'\nreading manifest file 'k1lib.egg-info/SOURCES.txt'\nadding license file 'LICENSE'\nwriting manifest file 'k1lib.egg-info/SOURCES.txt'\ninstalling library code to build/bdist.linux-x86_64/egg\nrunning install_lib\nrunning build_py\ncreating build\ncreating build/lib\ncreating build/lib/k1lib\ncopying k1lib/_learner.py -> build/lib/k1lib\ncopying k1lib/fmt.py -> build/lib/k1lib\ncopying k1lib/selector.py -> build/lib/k1lib\ncopying k1lib/imports.py -> build/lib/k1lib\ncopying k1lib/_baseClasses.py -> build/lib/k1lib\ncopying k1lib/_basics.py -> build/lib/k1lib\ncopying k1lib/viz.py -> build/lib/k1lib\ncopying k1lib/_higher.py -> build/lib/k1lib\ncopying k1lib/__init__.py -> build/lib/k1lib\ncopying k1lib/_monkey.py -> build/lib/k1lib\ncopying k1lib/knn.py -> build/lib/k1lib\ncopying k1lib/graphEqn.py -> build/lib/k1lib\ncopying k1lib/schedule.py -> build/lib/k1lib\ncopying k1lib/_perlin.py -> build/lib/k1lib\ncopying k1lib/kdata.py -> build/lib/k1lib\ncopying k1lib/eqn.py -> build/lib/k1lib\ncreating build/lib/k1lib/_hidden\ncopying k1lib/_hidden/hiddenFile.py -> build/lib/k1lib/_hidden\ncopying k1lib/_hidden/__init__.py -> build/lib/k1lib/_hidden\ncreating build/lib/k1lib/cli\ncopying k1lib/cli/bio.py -> build/lib/k1lib/cli\ncopying k1lib/cli/structural.py -> build/lib/k1lib/cli\ncopying k1lib/cli/modifier.py -> build/lib/k1lib/cli\ncopying k1lib/cli/gb.py -> build/lib/k1lib/cli\ncopying k1lib/cli/output.py -> build/lib/k1lib/cli\ncopying k1lib/cli/kxml.py -> build/lib/k1lib/cli\ncopying k1lib/cli/inp.py -> build/lib/k1lib/cli\ncopying k1lib/cli/mgi.py -> build/lib/k1lib/cli\ncopying k1lib/cli/grep.py -> build/lib/k1lib/cli\ncopying k1lib/cli/sam.py -> build/lib/k1lib/cli\ncopying k1lib/cli/entrez.py -> build/lib/k1lib/cli\ncopying k1lib/cli/__init__.py -> build/lib/k1lib/cli\ncopying k1lib/cli/filt.py -> build/lib/k1lib/cli\ncopying k1lib/cli/utils.py -> build/lib/k1lib/cli\ncopying k1lib/cli/init.py -> build/lib/k1lib/cli\ncopying k1lib/cli/others.py -> build/lib/k1lib/cli\ncopying k1lib/cli/kcsv.py -> build/lib/k1lib/cli\ncreating build/lib/k1lib/callbacks\ncopying k1lib/callbacks/loss_accuracy.py -> build/lib/k1lib/callbacks\ncopying k1lib/callbacks/progress.py -> build/lib/k1lib/callbacks\ncopying k1lib/callbacks/limits.py -> build/lib/k1lib/callbacks\ncopying k1lib/callbacks/hookParam.py -> build/lib/k1lib/callbacks\ncopying k1lib/callbacks/profiler.py -> build/lib/k1lib/callbacks\ncopying k1lib/callbacks/callbacks.py -> build/lib/k1lib/callbacks\ncopying k1lib/callbacks/paramFinder.py -> build/lib/k1lib/callbacks\ncopying k1lib/callbacks/core.py -> build/lib/k1lib/callbacks\ncopying k1lib/callbacks/__init__.py -> build/lib/k1lib/callbacks\ncopying k1lib/callbacks/landscape.py -> build/lib/k1lib/callbacks\ncopying k1lib/callbacks/confusionMatrix.py -> build/lib/k1lib/callbacks\ncopying k1lib/callbacks/recorder.py -> build/lib/k1lib/callbacks\ncopying k1lib/callbacks/shorts.py -> build/lib/k1lib/callbacks\ncopying k1lib/callbacks/hookModule.py -> build/lib/k1lib/callbacks\ncreating build/lib/k1lib/callbacks/profilers\ncopying k1lib/callbacks/profilers/time.py -> build/lib/k1lib/callbacks/profilers\ncopying k1lib/callbacks/profilers/memory.py -> build/lib/k1lib/callbacks/profilers\ncopying k1lib/callbacks/profilers/__init__.py -> build/lib/k1lib/callbacks/profilers\ncopying k1lib/callbacks/profilers/io.py -> build/lib/k1lib/callbacks/profilers\ncopying k1lib/callbacks/profilers/computation.py -> build/lib/k1lib/callbacks/profilers\ncreating build/lib/k1lib/callbacks/lossFunctions\ncopying k1lib/callbacks/lossFunctions/accuracy.py -> build/lib/k1lib/callbacks/lossFunctions\ncopying k1lib/callbacks/lossFunctions/__init__.py -> build/lib/k1lib/callbacks/lossFunctions\ncopying k1lib/callbacks/lossFunctions/shorts.py -> build/lib/k1lib/callbacks/lossFunctions\ncreating build/lib/k1lib/_mo\ncopying k1lib/_mo/atom.py -> build/lib/k1lib/_mo\ncopying k1lib/_mo/parseM.py -> build/lib/k1lib/_mo\ncopying k1lib/_mo/substance.py -> build/lib/k1lib/_mo\ncopying k1lib/_mo/system.py -> build/lib/k1lib/_mo\ncopying k1lib/_mo/__init__.py -> build/lib/k1lib/_mo\ncreating build/bdist.linux-x86_64\ncreating build/bdist.linux-x86_64/egg\ncreating build/bdist.linux-x86_64/egg/k1lib\ncopying build/lib/k1lib/_learner.py -> build/bdist.linux-x86_64/egg/k1lib\ncopying build/lib/k1lib/fmt.py -> build/bdist.linux-x86_64/egg/k1lib\ncopying build/lib/k1lib/selector.py -> build/bdist.linux-x86_64/egg/k1lib\ncopying build/lib/k1lib/imports.py -> build/bdist.linux-x86_64/egg/k1lib\ncopying build/lib/k1lib/_baseClasses.py -> build/bdist.linux-x86_64/egg/k1lib\ncopying build/lib/k1lib/_basics.py -> build/bdist.linux-x86_64/egg/k1lib\ncreating build/bdist.linux-x86_64/egg/k1lib/cli\ncopying build/lib/k1lib/cli/bio.py -> build/bdist.linux-x86_64/egg/k1lib/cli\ncopying build/lib/k1lib/cli/structural.py -> build/bdist.linux-x86_64/egg/k1lib/cli\ncopying build/lib/k1lib/cli/modifier.py -> build/bdist.linux-x86_64/egg/k1lib/cli\ncopying build/lib/k1lib/cli/gb.py -> build/bdist.linux-x86_64/egg/k1lib/cli\ncopying build/lib/k1lib/cli/output.py -> build/bdist.linux-x86_64/egg/k1lib/cli\ncopying build/lib/k1lib/cli/kxml.py -> build/bdist.linux-x86_64/egg/k1lib/cli\ncopying build/lib/k1lib/cli/inp.py -> build/bdist.linux-x86_64/egg/k1lib/cli\ncopying build/lib/k1lib/cli/mgi.py -> build/bdist.linux-x86_64/egg/k1lib/cli\ncopying build/lib/k1lib/cli/grep.py -> build/bdist.linux-x86_64/egg/k1lib/cli\ncopying build/lib/k1lib/cli/sam.py -> build/bdist.linux-x86_64/egg/k1lib/cli\ncopying build/lib/k1lib/cli/entrez.py -> build/bdist.linux-x86_64/egg/k1lib/cli\ncopying build/lib/k1lib/cli/__init__.py -> build/bdist.linux-x86_64/egg/k1lib/cli\ncopying build/lib/k1lib/cli/filt.py -> build/bdist.linux-x86_64/egg/k1lib/cli\ncopying build/lib/k1lib/cli/utils.py -> build/bdist.linux-x86_64/egg/k1lib/cli\ncopying build/lib/k1lib/cli/init.py -> build/bdist.linux-x86_64/egg/k1lib/cli\ncopying build/lib/k1lib/cli/others.py -> build/bdist.linux-x86_64/egg/k1lib/cli\ncopying build/lib/k1lib/cli/kcsv.py -> build/bdist.linux-x86_64/egg/k1lib/cli\ncopying build/lib/k1lib/viz.py -> build/bdist.linux-x86_64/egg/k1lib\ncopying build/lib/k1lib/_higher.py -> build/bdist.linux-x86_64/egg/k1lib\ncopying build/lib/k1lib/__init__.py -> build/bdist.linux-x86_64/egg/k1lib\ncopying build/lib/k1lib/_monkey.py -> build/bdist.linux-x86_64/egg/k1lib\ncreating build/bdist.linux-x86_64/egg/k1lib/_mo\ncopying build/lib/k1lib/_mo/atom.py -> build/bdist.linux-x86_64/egg/k1lib/_mo\ncopying build/lib/k1lib/_mo/parseM.py -> build/bdist.linux-x86_64/egg/k1lib/_mo\ncopying build/lib/k1lib/_mo/substance.py -> build/bdist.linux-x86_64/egg/k1lib/_mo\ncopying build/lib/k1lib/_mo/system.py -> build/bdist.linux-x86_64/egg/k1lib/_mo\ncopying build/lib/k1lib/_mo/__init__.py -> build/bdist.linux-x86_64/egg/k1lib/_mo\ncopying build/lib/k1lib/knn.py -> build/bdist.linux-x86_64/egg/k1lib\ncopying build/lib/k1lib/graphEqn.py -> build/bdist.linux-x86_64/egg/k1lib\ncopying build/lib/k1lib/schedule.py -> build/bdist.linux-x86_64/egg/k1lib\ncreating build/bdist.linux-x86_64/egg/k1lib/callbacks\ncopying build/lib/k1lib/callbacks/loss_accuracy.py -> build/bdist.linux-x86_64/egg/k1lib/callbacks\ncopying build/lib/k1lib/callbacks/progress.py -> build/bdist.linux-x86_64/egg/k1lib/callbacks\ncopying build/lib/k1lib/callbacks/limits.py -> build/bdist.linux-x86_64/egg/k1lib/callbacks\ncopying build/lib/k1lib/callbacks/hookParam.py -> build/bdist.linux-x86_64/egg/k1lib/callbacks\ncopying build/lib/k1lib/callbacks/profiler.py -> build/bdist.linux-x86_64/egg/k1lib/callbacks\ncopying build/lib/k1lib/callbacks/callbacks.py -> build/bdist.linux-x86_64/egg/k1lib/callbacks\ncopying build/lib/k1lib/callbacks/paramFinder.py -> build/bdist.linux-x86_64/egg/k1lib/callbacks\ncopying build/lib/k1lib/callbacks/core.py -> build/bdist.linux-x86_64/egg/k1lib/callbacks\ncreating build/bdist.linux-x86_64/egg/k1lib/callbacks/profilers\ncopying build/lib/k1lib/callbacks/profilers/time.py -> build/bdist.linux-x86_64/egg/k1lib/callbacks/profilers\ncopying build/lib/k1lib/callbacks/profilers/memory.py -> build/bdist.linux-x86_64/egg/k1lib/callbacks/profilers\ncopying build/lib/k1lib/callbacks/profilers/__init__.py -> build/bdist.linux-x86_64/egg/k1lib/callbacks/profilers\ncopying build/lib/k1lib/callbacks/profilers/io.py -> build/bdist.linux-x86_64/egg/k1lib/callbacks/profilers\ncopying build/lib/k1lib/callbacks/profilers/computation.py -> build/bdist.linux-x86_64/egg/k1lib/callbacks/profilers\ncopying build/lib/k1lib/callbacks/__init__.py -> build/bdist.linux-x86_64/egg/k1lib/callbacks\ncopying build/lib/k1lib/callbacks/landscape.py -> build/bdist.linux-x86_64/egg/k1lib/callbacks\ncopying build/lib/k1lib/callbacks/confusionMatrix.py -> build/bdist.linux-x86_64/egg/k1lib/callbacks\ncopying build/lib/k1lib/callbacks/recorder.py -> build/bdist.linux-x86_64/egg/k1lib/callbacks\ncopying build/lib/k1lib/callbacks/shorts.py -> build/bdist.linux-x86_64/egg/k1lib/callbacks\ncopying build/lib/k1lib/callbacks/hookModule.py -> build/bdist.linux-x86_64/egg/k1lib/callbacks\ncreating build/bdist.linux-x86_64/egg/k1lib/callbacks/lossFunctions\ncopying build/lib/k1lib/callbacks/lossFunctions/accuracy.py -> build/bdist.linux-x86_64/egg/k1lib/callbacks/lossFunctions\ncopying build/lib/k1lib/callbacks/lossFunctions/__init__.py -> build/bdist.linux-x86_64/egg/k1lib/callbacks/lossFunctions\ncopying build/lib/k1lib/callbacks/lossFunctions/shorts.py -> build/bdist.linux-x86_64/egg/k1lib/callbacks/lossFunctions\ncopying build/lib/k1lib/_perlin.py -> build/bdist.linux-x86_64/egg/k1lib\ncopying build/lib/k1lib/kdata.py -> build/bdist.linux-x86_64/egg/k1lib\ncreating build/bdist.linux-x86_64/egg/k1lib/_hidden\ncopying build/lib/k1lib/_hidden/hiddenFile.py -> build/bdist.linux-x86_64/egg/k1lib/_hidden\ncopying build/lib/k1lib/_hidden/__init__.py -> build/bdist.linux-x86_64/egg/k1lib/_hidden\ncopying build/lib/k1lib/eqn.py -> build/bdist.linux-x86_64/egg/k1lib\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/_learner.py to _learner.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/fmt.py to fmt.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/selector.py to selector.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/imports.py to imports.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/_baseClasses.py to _baseClasses.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/_basics.py to _basics.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/cli/bio.py to bio.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/cli/structural.py to structural.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/cli/modifier.py to modifier.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/cli/gb.py to gb.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/cli/output.py to output.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/cli/kxml.py to kxml.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/cli/inp.py to inp.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/cli/mgi.py to mgi.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/cli/grep.py to grep.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/cli/sam.py to sam.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/cli/entrez.py to entrez.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/cli/__init__.py to __init__.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/cli/filt.py to filt.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/cli/utils.py to utils.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/cli/init.py to init.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/cli/others.py to others.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/cli/kcsv.py to kcsv.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/viz.py to viz.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/_higher.py to _higher.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/__init__.py to __init__.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/_monkey.py to _monkey.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/_mo/atom.py to atom.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/_mo/parseM.py to parseM.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/_mo/substance.py to substance.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/_mo/system.py to system.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/_mo/__init__.py to __init__.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/knn.py to knn.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/graphEqn.py to graphEqn.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/schedule.py to schedule.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/callbacks/loss_accuracy.py to loss_accuracy.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/callbacks/progress.py to progress.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/callbacks/limits.py to limits.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/callbacks/hookParam.py to hookParam.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/callbacks/profiler.py to profiler.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/callbacks/callbacks.py to callbacks.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/callbacks/paramFinder.py to paramFinder.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/callbacks/core.py to core.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/callbacks/profilers/time.py to time.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/callbacks/profilers/memory.py to memory.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/callbacks/profilers/__init__.py to __init__.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/callbacks/profilers/io.py to io.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/callbacks/profilers/computation.py to computation.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/callbacks/__init__.py to __init__.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/callbacks/landscape.py to landscape.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/callbacks/confusionMatrix.py to confusionMatrix.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/callbacks/recorder.py to recorder.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/callbacks/shorts.py to shorts.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/callbacks/hookModule.py to hookModule.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/callbacks/lossFunctions/accuracy.py to accuracy.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/callbacks/lossFunctions/__init__.py to __init__.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/callbacks/lossFunctions/shorts.py to shorts.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/_perlin.py to _perlin.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/kdata.py to kdata.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/_hidden/hiddenFile.py to hiddenFile.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/_hidden/__init__.py to __init__.cpython-38.pyc\nbyte-compiling build/bdist.linux-x86_64/egg/k1lib/eqn.py to eqn.cpython-38.pyc\ncreating build/bdist.linux-x86_64/egg/EGG-INFO\ncopying k1lib.egg-info/PKG-INFO -> build/bdist.linux-x86_64/egg/EGG-INFO\ncopying k1lib.egg-info/SOURCES.txt -> build/bdist.linux-x86_64/egg/EGG-INFO\ncopying k1lib.egg-info/dependency_links.txt -> build/bdist.linux-x86_64/egg/EGG-INFO\ncopying k1lib.egg-info/requires.txt -> build/bdist.linux-x86_64/egg/EGG-INFO\ncopying k1lib.egg-info/top_level.txt -> build/bdist.linux-x86_64/egg/EGG-INFO\nzip_safe flag not set; analyzing archive contents...\ncreating dist\ncreating 'dist/k1lib-0.8-py3.8.egg' and adding 'build/bdist.linux-x86_64/egg' to it\nremoving 'build/bdist.linux-x86_64/egg' (and everything under it)\nProcessing k1lib-0.8-py3.8.egg\nCopying k1lib-0.8-py3.8.egg to /home/kelvin/anaconda3/envs/torch/lib/python3.8/site-packages\nAdding k1lib 0.8 to easy-install.pth file\n\nInstalled /home/kelvin/anaconda3/envs/torch/lib/python3.8/site-packages/k1lib-0.8-py3.8.egg\nProcessing dependencies for k1lib==0.8\nSearching for dill==0.3.4\nBest match: dill 0.3.4\nAdding dill 0.3.4 to easy-install.pth file\n\nUsing /home/kelvin/anaconda3/envs/torch/lib/python3.8/site-packages\nSearching for matplotlib==3.3.2\nBest match: matplotlib 3.3.2\nAdding matplotlib 3.3.2 to easy-install.pth file\n\nUsing /home/kelvin/anaconda3/envs/torch/lib/python3.8/site-packages\nSearching for numpy==1.19.2\nBest match: numpy 1.19.2\nAdding numpy 1.19.2 to easy-install.pth file\nInstalling f2py script to /home/kelvin/anaconda3/envs/torch/bin\nInstalling f2py3 script to /home/kelvin/anaconda3/envs/torch/bin\nInstalling f2py3.8 script to /home/kelvin/anaconda3/envs/torch/bin\n\nUsing /home/kelvin/anaconda3/envs/torch/lib/python3.8/site-packages\nSearching for torch==1.10.0\nBest match: torch 1.10.0\nAdding torch 1.10.0 to easy-install.pth file\nInstalling convert-caffe2-to-onnx script to /home/kelvin/anaconda3/envs/torch/bin\nInstalling convert-onnx-to-caffe2 script to /home/kelvin/anaconda3/envs/torch/bin\nInstalling torchrun script to /home/kelvin/anaconda3/envs/torch/bin\n\nUsing /home/kelvin/anaconda3/envs/torch/lib/python3.8/site-packages\nSearching for python-dateutil==2.8.2\nBest match: python-dateutil 2.8.2\nAdding python-dateutil 2.8.2 to easy-install.pth file\n\nUsing /home/kelvin/anaconda3/envs/torch/lib/python3.8/site-packages\nSearching for kiwisolver==1.3.2\nBest match: kiwisolver 1.3.2\nAdding kiwisolver 1.3.2 to easy-install.pth file\n\nUsing /home/kelvin/anaconda3/envs/torch/lib/python3.8/site-packages\nSearching for Pillow==7.2.0\nBest match: Pillow 7.2.0\nAdding Pillow 7.2.0 to easy-install.pth file\n\nUsing /home/kelvin/anaconda3/envs/torch/lib/python3.8/site-packages\nSearching for pyparsing==2.4.7\nBest match: pyparsing 2.4.7\nAdding pyparsing 2.4.7 to easy-install.pth file\n\nUsing /home/kelvin/anaconda3/envs/torch/lib/python3.8/site-packages\nSearching for certifi==2021.10.8\nBest match: certifi 2021.10.8\nAdding certifi 2021.10.8 to easy-install.pth file\n\nUsing /home/kelvin/anaconda3/envs/torch/lib/python3.8/site-packages\nSearching for cycler==0.10.0\nBest match: cycler 0.10.0\nAdding cycler 0.10.0 to easy-install.pth file\n\nUsing /home/kelvin/anaconda3/envs/torch/lib/python3.8/site-packages\nSearching for typing-extensions==3.10.0.2\nBest match: typing-extensions 3.10.0.2\nAdding typing-extensions 3.10.0.2 to easy-install.pth file\n\nUsing /home/kelvin/anaconda3/envs/torch/lib/python3.8/site-packages\nSearching for six==1.16.0\nBest match: six 1.16.0\nAdding six 1.16.0 to easy-install.pth file\n\nUsing /home/kelvin/anaconda3/envs/torch/lib/python3.8/site-packages\nFinished processing dependencies for k1lib==0.8\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e4b0b1343853c919e368cf934d4300aa542155 | 83,567 | ipynb | Jupyter Notebook | Python/Learn/Kelas Persiapan/12. Data Manipulation with Pandas - Part 1.ipynb | Miadwicynthia/DQLab | 711e9650224683844a67963e76b10c1d4a38ebf4 | [
"MIT"
] | null | null | null | Python/Learn/Kelas Persiapan/12. Data Manipulation with Pandas - Part 1.ipynb | Miadwicynthia/DQLab | 711e9650224683844a67963e76b10c1d4a38ebf4 | [
"MIT"
] | null | null | null | Python/Learn/Kelas Persiapan/12. Data Manipulation with Pandas - Part 1.ipynb | Miadwicynthia/DQLab | 711e9650224683844a67963e76b10c1d4a38ebf4 | [
"MIT"
] | null | null | null | 41.888221 | 469 | 0.52699 | [
[
[
"# DataFrame & Series\n\nDi Pandas terdapat 2 kelas data baru yang digunakan sebagai struktur dari spreadsheet:\n\n1. Series: \nsatu kolom bagian dari tabel dataframe yang merupakan 1 dimensional numpy array sebagai basis datanya, terdiri dari 1 tipe data (integer, string, float, dll).\n2. DataFrame: \ngabungan dari Series, berbentuk rectangular data yang merupakan tabel spreadsheet itu sendiri (karena dibentuk dari banyak Series, tiap Series biasanya punya 1 tipe data, yang artinya 1 dataframe bisa memiliki banyak tipe data).",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n# Series\nnumber_list = pd.Series([1,2,3,4,5,6])\nprint(\"Series:\")\nprint(number_list)\n# DataFrame\nmatrix = [[1,2,3],\n ['a','b','c'],\n [3,4,5],\n ['d',4,6]]\nmatrix_list = pd.DataFrame(matrix)\nprint(\"DataFrame:\")\nprint(matrix_list)",
"Series:\n0 1\n1 2\n2 3\n3 4\n4 5\n5 6\ndtype: int64\nDataFrame:\n 0 1 2\n0 1 2 3\n1 a b c\n2 3 4 5\n3 d 4 6\n"
]
],
[
[
"# Atribut DataFrame & Series - Part 1\n\nDataframe dan Series memiliki sangat banyak atribut yang digunakan untuk transformasi data, tetapi ada beberapa attribute yang sering dipakai. Di sini series number_list dan dataframe matrix_list pada subbab sebelumnya digunakan kembali.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n# Series\nnumber_list = pd.Series([1,2,3,4,5,6])\n# DataFrame\nmatrix_list = pd.DataFrame([[1,2,3],\n\t\t\t\t ['a','b','c'],\n\t\t\t\t [3,4,5],\n\t\t\t\t ['d',4,6]])\n# [1] attribute .info()\nprint(\"[1] attribute .info()\")\nprint(matrix_list.info())\n# [2] attribute .shape\nprint(\"\\n[2] attribute .shape\")\nprint(\" Shape dari number_list:\", number_list.shape)\nprint(\" Shape dari matrix_list:\", matrix_list.shape)\n# [3] attribute .dtypes\nprint(\"\\n[3] attribute .dtypes\")\nprint(\" Tipe data number_list:\", number_list.dtypes)\nprint(\" Tipe data matrix_list:\", matrix_list.dtypes)\n# [4] attribute .astype()\nprint(\"\\n[4] attribute .astype()\")\nprint(\" Konversi number_list ke str:\", number_list.astype(\"str\"))\nprint(\" Konversi matrix_list ke str:\", matrix_list.astype(\"str\"))",
"[1] attribute .info()\n<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 4 entries, 0 to 3\nData columns (total 3 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 0 4 non-null object\n 1 1 4 non-null object\n 2 2 4 non-null object\ndtypes: object(3)\nmemory usage: 224.0+ bytes\nNone\n\n[2] attribute .shape\n Shape dari number_list: (6,)\n Shape dari matrix_list: (4, 3)\n\n[3] attribute .dtypes\n Tipe data number_list: int64\n Tipe data matrix_list: 0 object\n1 object\n2 object\ndtype: object\n\n[4] attribute .astype()\n Konversi number_list ke str: 0 1\n1 2\n2 3\n3 4\n4 5\n5 6\ndtype: object\n Konversi matrix_list ke str: 0 1 2\n0 1 2 3\n1 a b c\n2 3 4 5\n3 d 4 6\n"
]
],
[
[
"# Atribut DataFrame & Series - Part 2",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n# Series\nnumber_list = pd.Series([1,2,3,4,5,6])\n# DataFrame\nmatrix_list = pd.DataFrame([[1,2,3],\n\t\t\t\t ['a','b','c'],\n\t\t\t\t [3,4,5],\n\t\t\t\t ['d',4,6]])\n# [5] attribute .copy()\nprint(\"[5] attribute .copy()\")\nnum_list = number_list.copy()\nprint(\" Copy number_list ke num_list:\", num_list)\nmtr_list = matrix_list.copy()\nprint(\" Copy matrix_list ke mtr_list:\", mtr_list)\t\n# [6] attribute .to_list()\nprint(\"[6] attribute .to_list()\")\nprint(number_list.to_list())\n# [7] attribute .unique()\nprint(\"[7] attribute .unique()\")\nprint(number_list.unique())",
"[5] attribute .copy()\n Copy number_list ke num_list: 0 1\n1 2\n2 3\n3 4\n4 5\n5 6\ndtype: int64\n Copy matrix_list ke mtr_list: 0 1 2\n0 1 2 3\n1 a b c\n2 3 4 5\n3 d 4 6\n[6] attribute .to_list()\n[1, 2, 3, 4, 5, 6]\n[7] attribute .unique()\n[1 2 3 4 5 6]\n"
]
],
[
[
"# Atribut DataFrame & Series - Part 3",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n# Series\nnumber_list = pd.Series([1,2,3,4,5,6])\n# DataFrame\nmatrix_list = pd.DataFrame([[1,2,3],\n\t\t\t\t ['a','b','c'],\n\t\t\t\t [3,4,5],\n\t\t\t\t ['d',4,6]])\n# [8] attribute .index\nprint(\"[8] attribute .index\")\nprint(\" Index number_list:\", number_list.index)\nprint(\" Index matrix_list:\", matrix_list.index)\t\n# [9] attribute .columns\nprint(\"[9] attribute .columns\")\nprint(\" Column matrix_list:\", matrix_list.columns)\n# [10] attribute .loc\nprint(\"[10] attribute .loc\")\nprint(\" .loc[0:1] pada number_list:\", number_list.loc[0:1])\nprint(\" .loc[0:1] pada matrix_list:\", matrix_list.loc[0:1])\n# [11] attribute .iloc\nprint(\"[11] attribute .iloc\")\nprint(\" iloc[0:1] pada number_list:\", number_list.iloc[0:1])\nprint(\" iloc[0:1] pada matrix_list:\", matrix_list.iloc[0:1])\t",
"[8] attribute .index\n Index number_list: RangeIndex(start=0, stop=6, step=1)\n Index matrix_list: RangeIndex(start=0, stop=4, step=1)\n[9] attribute .columns\n Column matrix_list: RangeIndex(start=0, stop=3, step=1)\n[10] attribute .loc\n .loc[0:1] pada number_list: 0 1\n1 2\ndtype: int64\n .loc[0:1] pada matrix_list: 0 1 2\n0 1 2 3\n1 a b c\n[11] attribute .iloc\n iloc[0:1] pada number_list: 0 1\ndtype: int64\n iloc[0:1] pada matrix_list: 0 1 2\n0 1 2 3\n"
],
[
"matrix = [[1,2,3],\n ['a','b','c'],\n [3,4,5],\n ['d',4,6]]\nmatrix_list = pd.DataFrame(matrix)\nmatrix_list.iloc[0:2,2].to_list()",
"_____no_output_____"
]
],
[
[
"# Creating Series & Dataframe from List\nUntuk membuat Series atau Dataframe bisa dari berbagai macam tipe data container/mapping di python, seperti list dan dictionary, maupun dari numpy array.\n\nPada sub bagian ini, membuat Series dan Dataframe yang bersumber dari list. Sekadar meninjau bahwa list merupakan sebuah kumpulan data berbagai macam tipe data yang mutable (dapat diganti). ",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n# Creating series from list\nex_list = ['a',1,3,5,'c','d']\nex_series = pd.Series(ex_list)\nprint(ex_series)\n# Creating dataframe from list of list\nex_list_of_list = [[1 , 'a', 'b' , 'c'],\n [2.5, 'd', 'e' , 'f'],\n\t\t [5 , 'g', 'h' , 'i'],\n\t\t [7.5, 'j', 10.5, 'l']]\nindex = ['dq', 'lab', 'kar', 'lan']\ncols = ['float', 'char', 'obj', 'char']\nex_df = pd.DataFrame(ex_list_of_list, index=index, columns=cols)\nprint(ex_df)",
"0 a\n1 1\n2 3\n3 5\n4 c\n5 d\ndtype: object\n float char obj char\ndq 1.0 a b c\nlab 2.5 d e f\nkar 5.0 g h i\nlan 7.5 j 10.5 l\n"
]
],
[
[
"# Creating Series & Dataframe from Dictionary\nUntuk membuat Series atau Dataframe bisa dari berbagai macam tipe data container/mapping di python, seperti list dan dictionary, maupun dari numpy array.\n\nPada sub bagian ini, akan membuat Series dan Dataframe yang bersumber dari dictionary. Sekadar meninjau bahwa, dictionary merupakan kumpulan data yang strukturnya terdiri dari key dan value.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n# Creating series from dictionary\ndict_series = {'1':'a',\n\t\t\t '2':'b',\n\t\t\t '3':'c'}\nex_series = pd.Series(dict_series)\nprint(ex_series)\n# Creating dataframe from dictionary\ndf_series = {'1':['a','b','c'],\n '2':['b','c','d'],\n '4':[2,3,'z']}\nex_df = pd.DataFrame(df_series)\nprint(ex_df)",
"1 a\n2 b\n3 c\ndtype: object\n 1 2 4\n0 a b 2\n1 b c 3\n2 c d z\n"
]
],
[
[
"# reating Series & Dataframe from Numpy Array\nUntuk membuat Series atau Dataframe bisa dari berbagai macam tipe data container/mapping di python, seperti list dan dictionary, maupun dari numpy array.\n\nPada sub bagian ini, akan membuat Series dan Dataframe yang bersumber dari numpy array. Sekadar meninjau bahwa, numpy array kumpulan data yang terdiri atas berbagai macam tipe data, mutable, tapi dibungkus dalam array oleh library Numpy.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\n# Creating series from numpy array (1D)\narr_series = np.array([1,2,3,4,5,6,6,7])\nex_series = pd.Series(arr_series)\nprint(ex_series)\n# Creating dataframe from numpy array (2D)\narr_df = np.array([[1 ,2 ,3 ,5],\n [5 ,6 ,7 ,8],\n ['a','b','c',10]])\nex_df = pd.DataFrame(arr_df)\nprint(ex_df)",
"0 1\n1 2\n2 3\n3 4\n4 5\n5 6\n6 6\n7 7\ndtype: int32\n 0 1 2 3\n0 1 2 3 5\n1 5 6 7 8\n2 a b c 10\n"
]
],
[
[
"# Read Dataset - CSV dan TSV\nCSV dan TSV pada hakikatnya adalah tipe data text dengan perbedaan terletak pada pemisah antar data dalam satu baris. Pada file CSV, antar data dalam satu baris dipisahkan oleh comma, \",\". Namun, pada file TSV antar data dalam satu baris dipisahkan oleh \"Tab\".\n\nFungsi .read_csv() digunakan untuk membaca file yang value-nya dipisahkan oleh comma (default), terkadang pemisah value-nya bisa di set ‘\\t’ untuk file tsv (tab separated values).",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n# File CSV\ndf_csv = pd.read_csv(\"https://storage.googleapis.com/dqlab-dataset/sample_csv.csv\")\nprint(df_csv.head(3)) # Menampilkan 3 data teratas\n# File TSV\ndf_tsv = pd.read_csv(\"https://storage.googleapis.com/dqlab-dataset/sample_tsv.tsv\", sep='\\t')\nprint(df_tsv.head(3)) # Menampilkan 3 data teratas",
" order_id order_date customer_id city province product_id \\\n0 1612339 2019-01-01 18055 Jakarta Selatan DKI Jakarta P0648 \n1 1612339 2019-01-01 18055 Jakarta Selatan DKI Jakarta P3826 \n2 1612339 2019-01-01 18055 Jakarta Selatan DKI Jakarta P1508 \n\n brand quantity item_price \n0 BRAND_C 4 1934000 \n1 BRAND_V 8 604000 \n2 BRAND_G 12 747000 \n order_id order_date customer_id city province product_id \\\n0 1612339 2019-01-01 18055 Jakarta Selatan DKI Jakarta P0648 \n1 1612339 2019-01-01 18055 Jakarta Selatan DKI Jakarta P3826 \n2 1612339 2019-01-01 18055 Jakarta Selatan DKI Jakarta P1508 \n\n brand quantity item_price \n0 BRAND_C 4 1934000 \n1 BRAND_V 8 604000 \n2 BRAND_G 12 747000 \n"
]
],
[
[
"# Read Dataset - Excel\nFile Excel dengan ekstensi *.xls atau *.xlsx cukup banyak digunakan dalam menyimpan data. Pandas juga memiliki fitur untuk membaca file excel.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n# File xlsx dengan data di sheet \"test\"\ndf_excel = pd.read_excel(\"https://storage.googleapis.com/dqlab-dataset/sample_excel.xlsx\", sheet_name=\"test\")\nprint(df_excel.head(4)) # Menampilkan 4 data teratas",
" order_id order_date customer_id city province product_id \\\n0 1612339 2019-01-01 18055 Jakarta Selatan DKI Jakarta P0648 \n1 1612339 2019-01-01 18055 Jakarta Selatan DKI Jakarta P3826 \n2 1612339 2019-01-01 18055 Jakarta Selatan DKI Jakarta P1508 \n3 1612339 2019-01-01 18055 Jakarta Selatan DKI Jakarta P0520 \n\n brand quantity item_price \n0 BRAND_C 4 1934000 \n1 BRAND_V 8 604000 \n2 BRAND_G 12 747000 \n3 BRAND_B 12 450000 \n"
]
],
[
[
"# Read Dataset - JSON\nMethod .read_json() digunakan untuk membaca URL API yang formatnya JSON dan mengubahnya menjadi dataframe pandas.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n# File JSON\nurl = \"https://storage.googleapis.com/dqlab-dataset/covid2019-api-herokuapp-v2.json\"\ndf_json = pd.read_json(url)\nprint(df_json.head(10)) # Menampilkan 10 data teratas",
" data dt ts\n0 {'location': 'US', 'confirmed': 3363056, 'deat... 07-14-2020 1594684800\n1 {'location': 'Brazil', 'confirmed': 1884967, '... 07-14-2020 1594684800\n2 {'location': 'India', 'confirmed': 906752, 'de... 07-14-2020 1594684800\n3 {'location': 'Russia', 'confirmed': 732547, 'd... 07-14-2020 1594684800\n4 {'location': 'Peru', 'confirmed': 330123, 'dea... 07-14-2020 1594684800\n5 {'location': 'Chile', 'confirmed': 317657, 'de... 07-14-2020 1594684800\n6 {'location': 'Mexico', 'confirmed': 304435, 'd... 07-14-2020 1594684800\n7 {'location': 'United Kingdom', 'confirmed': 29... 07-14-2020 1594684800\n8 {'location': 'South Africa', 'confirmed': 2877... 07-14-2020 1594684800\n9 {'location': 'Iran', 'confirmed': 259652, 'dea... 07-14-2020 1594684800\n"
]
],
[
[
"# Read Dataset - SQL\nFungsi .read_sql() atau .read_sql_query() digunakan untuk membaca query dari database dan translate menjadi pandas dataframe, contoh case ini database sqlite.",
"_____no_output_____"
],
[
"# Read Dataset - Google BigQuery\nUntuk data yang besar (big data), umumnya digunakan Google BigQuery. Layanan ini dapat digunakan jika telah memiliki Google BigQuery account.\n\nFungsi .read_gbq() digunakan untuk membaca Google BigQuery table menjadi dataframe pandas.",
"_____no_output_____"
],
[
"# Write Dataset\nDalam bekerja sebagai data scientist/analis setelah dilakukan data cleaning dataset yang sudah rapi tentunya disimpan terlebih dahulu ke dalam media penyimpanan. \n\nPandas menyediakan fitur demikian secara ringkas melalui penerapan method pada dataframe/series yang ditabelkan berikut ini:\n- .to_csv()\n→ digunakan untuk export dataframe kembali ke csv atau tsv\nCSV\ndf.to_csv(\"csv1.csv\", index=False)\nTSV\ndf.to_csv(\"tsv1.tsv\", index=False, sep='\\t')\n\n- .to_clipboard()\n→ export dataframe menjadi bahan copy jadi nanti bisa tinggal klik paste di excel atau google sheets\ndf.to_clipboard()\n\n- .to_excel()\n→ export dataframe menjadi file excel\ndf_excel.to_excel(\"xlsx1.xlsx\", index=False)\n\n- .to_gbq()\n→ export dataframe menjadi table di Google BigQuery\ndf.to_gbq(\"temp.test\", project_id=\"XXXXXX\", if_exists=\"fail\")\n\ntemp: nama dataset,\ntest: nama table\nif_exists: ketika tabel dengan dataset.table_name yang sama sudah ada, apa action yang ingin dilakukan\n(\"fail\": tidak melakukan apa-apa,\n \"replace': membuang tabel yang sudah ada dan mengganti yang baru,\n \"append\": menambah baris di tabel tersebut dengan data yang baru\n)",
"_____no_output_____"
],
[
"# Head & Tail\nSeperti yang telah dipelajari sebelumnya bahwa ada method .head yang diterapkan pada suatu variabel bertipe pandas dataframe/series.\n\nMethod .head ditujukan untuk membatasi tampilan jumlah baris teratas dari dataset. Sementara itu, method .tail ditujukan untuk membatasi jumlah baris terbawah dari dataset. ",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n# Baca file sample_csv.csv\ndf = pd.read_csv(\"https://storage.googleapis.com/dqlab-dataset/sample_csv.csv\")\n# Tampilkan 3 data teratas\nprint(\"Tiga data teratas:\\n\", df.head(3))\n# Tampilkan 3 data terbawah\nprint(\"Tiga data terbawah:\\n\", df.tail(3))",
"Tiga data teratas:\n order_id order_date customer_id city province product_id \\\n0 1612339 2019-01-01 18055 Jakarta Selatan DKI Jakarta P0648 \n1 1612339 2019-01-01 18055 Jakarta Selatan DKI Jakarta P3826 \n2 1612339 2019-01-01 18055 Jakarta Selatan DKI Jakarta P1508 \n\n brand quantity item_price \n0 BRAND_C 4 1934000 \n1 BRAND_V 8 604000 \n2 BRAND_G 12 747000 \nTiga data terbawah:\n order_id order_date customer_id city province product_id \\\n98 1612390 2019-01-01 12681 Makassar Sulawesi Selatan P3354 \n99 1612390 2019-01-01 12681 Makassar Sulawesi Selatan P3357 \n100 1612390 2019-01-01 12681 Makassar Sulawesi Selatan P0422 \n\n brand quantity item_price \n98 BRAND_S 24 450000 \n99 BRAND_S 24 450000 \n100 BRAND_B 4 1325000 \n"
]
],
[
[
"# Indexing - Part 1\nIndex merupakan key identifier dari tiap row/column untuk Series atau Dataframe (sifatnya tidak mutable untuk masing-masing value tapi bisa diganti untuk semua value sekaligus).\n\nJika tidak disediakan, pandas akan membuat kolom index default secara otomatis sebagai bilangan bulat (integer) dari 0 sampai range jumlah baris data tersebut.\n\nKolom index dapat terdiri dari:\n1. satu kolom (single index), atau\n2. multiple kolom (disebut dengan hierarchical indexing).\n\nIndex dengan multiple kolom ini terjadi karena unique identifier tidak dapat dicapai hanya dengan set index di 1 kolom saja sehingga membutuhkan beberapa kolom yang menjadikan tiap row menjadi unique.",
"_____no_output_____"
],
[
"# Indexing - Part 2\nSecara default setelah suatu dataframe dibaca dari file dengan format tertentu, index-nya merupakan single index.\n\nPada sub bab ini akan mencetak index dan kolom yang dimiliki oleh file \"https://storage.googleapis.com/dqlab-dataset/sample_csv.csv\". Untuk menentukan index dan kolom yang dimiliki oleh dataset yang telah dinyatakan sebagai sebuah dataframe pandas dapat dilakukan dengan menggunakan atribut .index dan .columns.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n# Baca file TSV sample_tsv.tsv\ndf = pd.read_csv(\"https://storage.googleapis.com/dqlab-dataset/sample_tsv.tsv\", sep=\"\\t\")\n# Index dari df\nprint(\"Index:\", df.index)\n# Column dari df\nprint(\"Columns:\", df.columns)",
"Index: RangeIndex(start=0, stop=101, step=1)\nColumns: Index(['order_id', 'order_date', 'customer_id', 'city', 'province',\n 'product_id', 'brand', 'quantity', 'item_price'],\n dtype='object')\n"
]
],
[
[
"# Indexing - Part 3\nDi sub bab sebelumnya telah dibahas terkait single index, tentunya pada sub bab ini akan bahas multi index atau disebut juga dengan hierarchical indexing.\n\nUntuk membuat multi index (hierarchical indexing) dengan pandas diperlukan kolom-kolom mana saja yang perlu disusun agar index dari dataframe menjadi sebuah hirarki yang kemudian dapat dikenali. ",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n# Baca file TSV sample_tsv.tsv\ndf = pd.read_csv(\"https://storage.googleapis.com/dqlab-dataset/sample_tsv.tsv\", sep=\"\\t\")\n# Set multi index df\ndf_x = df.set_index(['order_date', 'city', 'customer_id'])\n# Print nama dan level dari multi index\nfor name, level in zip(df_x.index.names, df_x.index.levels):\n print(name,':',level)",
"order_date : Index(['2019-01-01'], dtype='object', name='order_date')\ncity : Index(['Bogor', 'Jakarta Pusat', 'Jakarta Selatan', 'Jakarta Utara',\n 'Makassar', 'Malang', 'Surabaya', 'Tangerang'],\n dtype='object', name='city')\ncustomer_id : Int64Index([12681, 13963, 15649, 17091, 17228, 17450, 17470, 17511, 17616,\n 18055],\n dtype='int64', name='customer_id')\n"
]
],
[
[
"# Indexing - Part 4\nTerdapat beberapa cara untuk membuat index, salah satunya adalah seperti yang telah dilakukan pada sub bab sebelumnya dengan menggunakan method .set_index().\n\nDi sub bab ini akan menggunakan assignment untuk menset index dari suatu dataframe. Untuk itu file \"sample_excel.xlsx\" yang digunakan ",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n# Baca file sample_tsv.tsv untuk 10 baris pertama saja\ndf = pd.read_csv(\"https://storage.googleapis.com/dqlab-dataset/sample_tsv.tsv\", sep=\"\\t\", nrows=10)\n# Cetak data frame awal\nprint(\"Dataframe awal:\\n\", df)\n# Set index baru\ndf.index = [\"Pesanan ke-\" + str(i) for i in range(1, 11)]\n# Cetak data frame dengan index baru\nprint(\"Dataframe dengan index baru:\\n\", df)",
"Dataframe awal:\n order_id order_date customer_id city province product_id \\\n0 1612339 2019-01-01 18055 Jakarta Selatan DKI Jakarta P0648 \n1 1612339 2019-01-01 18055 Jakarta Selatan DKI Jakarta P3826 \n2 1612339 2019-01-01 18055 Jakarta Selatan DKI Jakarta P1508 \n3 1612339 2019-01-01 18055 Jakarta Selatan DKI Jakarta P0520 \n4 1612339 2019-01-01 18055 Jakarta Selatan DKI Jakarta P1513 \n5 1612339 2019-01-01 18055 Jakarta Selatan DKI Jakarta P3911 \n6 1612339 2019-01-01 18055 Jakarta Selatan DKI Jakarta P1780 \n7 1612339 2019-01-01 18055 Jakarta Selatan DKI Jakarta P3132 \n8 1612339 2019-01-01 18055 Jakarta Selatan DKI Jakarta P1342 \n9 1612339 2019-01-01 18055 Jakarta Selatan DKI Jakarta P2556 \n\n brand quantity item_price \n0 BRAND_C 4 1934000 \n1 BRAND_V 8 604000 \n2 BRAND_G 12 747000 \n3 BRAND_B 12 450000 \n4 BRAND_G 3 1500000 \n5 BRAND_V 3 2095000 \n6 BRAND_H 3 2095000 \n7 BRAND_S 3 1745000 \n8 BRAND_F 6 1045000 \n9 BRAND_P 6 1045000 \nDataframe dengan index baru:\n order_id order_date customer_id city \\\nPesanan ke-1 1612339 2019-01-01 18055 Jakarta Selatan \nPesanan ke-2 1612339 2019-01-01 18055 Jakarta Selatan \nPesanan ke-3 1612339 2019-01-01 18055 Jakarta Selatan \nPesanan ke-4 1612339 2019-01-01 18055 Jakarta Selatan \nPesanan ke-5 1612339 2019-01-01 18055 Jakarta Selatan \nPesanan ke-6 1612339 2019-01-01 18055 Jakarta Selatan \nPesanan ke-7 1612339 2019-01-01 18055 Jakarta Selatan \nPesanan ke-8 1612339 2019-01-01 18055 Jakarta Selatan \nPesanan ke-9 1612339 2019-01-01 18055 Jakarta Selatan \nPesanan ke-10 1612339 2019-01-01 18055 Jakarta Selatan \n\n province product_id brand quantity item_price \nPesanan ke-1 DKI Jakarta P0648 BRAND_C 4 1934000 \nPesanan ke-2 DKI Jakarta P3826 BRAND_V 8 604000 \nPesanan ke-3 DKI Jakarta P1508 BRAND_G 12 747000 \nPesanan ke-4 DKI Jakarta P0520 BRAND_B 12 450000 \nPesanan ke-5 DKI Jakarta P1513 BRAND_G 3 1500000 \nPesanan ke-6 DKI Jakarta P3911 BRAND_V 3 2095000 \nPesanan ke-7 DKI Jakarta P1780 BRAND_H 3 2095000 \nPesanan ke-8 DKI Jakarta P3132 BRAND_S 3 1745000 \nPesanan ke-9 DKI Jakarta P1342 BRAND_F 6 1045000 \nPesanan ke-10 DKI Jakarta P2556 BRAND_P 6 1045000 \n"
]
],
[
[
"# Indexing - Part 5\nJika file yang akan dibaca melalui penggunaan library pandas dapat di-preview terlebih dahulu struktur datanya maka melalui fungsi yang ditujukan untuk membaca file dapat diset mana kolom yang akan dijadikan index.\n\nFitur ini telah dimiliki oleh setiap fungsi yang digunakan dalam membaca data dengan pandas, yaitu penggunaan argumen index_col pada fungsi yang dimaksud ",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n# Baca file sample_tsv.tsv dan set lah index_col sesuai instruksi\ndf = pd.read_csv(\"https://storage.googleapis.com/dqlab-dataset/sample_tsv.tsv\", sep=\"\\t\", index_col=[\"order_date\", \"order_id\"])\n# Cetak data frame untuk 8 data teratas\nprint(\"Dataframe:\\n\", df.head(8))",
"Dataframe:\n customer_id city province product_id \\\norder_date order_id \n2019-01-01 1612339 18055 Jakarta Selatan DKI Jakarta P0648 \n 1612339 18055 Jakarta Selatan DKI Jakarta P3826 \n 1612339 18055 Jakarta Selatan DKI Jakarta P1508 \n 1612339 18055 Jakarta Selatan DKI Jakarta P0520 \n 1612339 18055 Jakarta Selatan DKI Jakarta P1513 \n 1612339 18055 Jakarta Selatan DKI Jakarta P3911 \n 1612339 18055 Jakarta Selatan DKI Jakarta P1780 \n 1612339 18055 Jakarta Selatan DKI Jakarta P3132 \n\n brand quantity item_price \norder_date order_id \n2019-01-01 1612339 BRAND_C 4 1934000 \n 1612339 BRAND_V 8 604000 \n 1612339 BRAND_G 12 747000 \n 1612339 BRAND_B 12 450000 \n 1612339 BRAND_G 3 1500000 \n 1612339 BRAND_V 3 2095000 \n 1612339 BRAND_H 3 2095000 \n 1612339 BRAND_S 3 1745000 \n"
],
[
"df_week = pd.DataFrame({'day_number':[1,2,3,4,5,6,7],\n 'week_type':['weekday' for i in range(5)] + ['weekend' for i in range(2)]\n })\ndf_week_ix = ['Mon','Tue','Wed','Thu','Fri','Sat','Sun']\ndf_week.index = [df_week_ix, df_week['day_number'].to_list()]\ndf_week.index.names = ['name','num']",
"_____no_output_____"
]
],
[
[
"# Slicing - Part 1\nSeperti artinya slicing adalah cara untuk melakukan filter ke dataframe/series berdasarkan kriteria tertentu dari nilai kolomnya ataupun kriteria index-nya.\n\nTerdapat 2 cara paling terkenal untuk slicing dataframe, yaitu dengan menggunakan method .loc dan .iloc pada variabel bertipe pandas DataFrame/Series. Method .iloc ditujukan untuk proses slicing berdasarkan index berupa nilai integer tertentu. Akan tetapi akan lebih sering menggunakan dengan method .loc karena lebih fleksibel. ",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n# Baca file sample_csv.csv\ndf = pd.read_csv(\"https://storage.googleapis.com/dqlab-dataset/sample_csv.csv\")\n# Slice langsung berdasarkan kolom\ndf_slice = df.loc[(df[\"customer_id\"] == \"18055\") &\n\t\t (df[\"product_id\"].isin([\"P0029\", \"P0040\", \"P0041\", \"P0116\", \"P0117\"]))\n\t\t\t\t ]\nprint(\"Slice langsung berdasarkan kolom:\\n\", df_slice)",
"Slice langsung berdasarkan kolom:\n Empty DataFrame\nColumns: [order_id, order_date, customer_id, city, province, product_id, brand, quantity, item_price]\nIndex: []\n"
]
],
[
[
"# Slicing - Part 2\nDalam sub bab sebelumnya telah mempelajari bagaimana melakukan slicing/filtering dataset dengan menggunakan method .loc pada kolom dataset.\n\nSekarang, menerapkan berdasarkan index. Tentu syaratnya adalah dataset sudah dilakukan indexing terlebih dahulu melalui penerapan method .set_index ",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n# Baca file sample_csv.csv\ndf = pd.read_csv(\"https://storage.googleapis.com/dqlab-dataset/sample_csv.csv\")\n# Set index dari df sesuai instruksi\ndf = df.set_index([\"order_date\", \"order_id\", \"product_id\"])\n# Slice sesuai intruksi\ndf_slice = df.loc[(\"2019-01-01\", 1612339, [\"P2154\", \"P2159\"]),:]\nprint(\"Slice df:\\n\", df_slice)",
"Slice df:\n customer_id city province \\\norder_date order_id product_id \n2019-01-01 1612339 P2154 18055 Jakarta Selatan DKI Jakarta \n P2159 18055 Jakarta Selatan DKI Jakarta \n\n brand quantity item_price \norder_date order_id product_id \n2019-01-01 1612339 P2154 BRAND_M 4 1745000 \n P2159 BRAND_M 24 310000 \n"
]
],
[
[
"# Transforming - Part 1\nTransform adalah ketika mengubah dataset yang ada menjadi entitas baru, dapat dilakukan dengan:\n\nkonversi dari satu data type ke data type yang lain,\ntranspose dataframe,\natau yang lainnya.\nHal yang biasa dilakukan pertama kali setelah data dibaca adalah mengecek tipe data di setiap kolomnya apakah sesuai dengan representasinya. Untuk itu dapat menggunakan atribut .dtypes pada dataframe yang telah kita baca tadi,\n\n[nama_dataframe].dtypes \n \n\nUntuk konversi tipe data, secara default system akan mendeteksi data yang tidak bisa di render as date type or numeric type sebagai object yang basically string. Tidak bisa di render oleh system ini karena berbagai hal, mungkin karena formatnya asing dan tidak dikenali oleh python secara umum (misal: date type data → '2019Jan01').\n\nData contoh tersebut tidak bisa di render karena bulannya Jan tidak bisa di translate menjadi in form of number (00-12) dan tidak ada ‘-’ di antara tahun, bulan dan harinya. Jika seluruh data pada kolom di order_date sudah tertulis dalam bentuk 'YYYY-MM-DD' maka ketika dibaca, kolom order_date sudah langsung dinyatakan bertipe data datetime.\n\nUntuk merubah kolom date_order yang sebelumnya bertipe object menjadi kolom bertipe datetime, cara pertama yang dapat dilakukan adalah menggunakan:\n\npd.to_datetime(argumen) \ndengan argumen adalah isi kolom dari dataframe yang akan dirubah tipe datanya, misal dalam format umum.\n\nnama_dataframe[\"nama_kolom\"]\nSehingga lengkapnya dapat ditulis sebagai:\n\nnama_dataframe[\"nama_kolom\"] = ",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n# Baca file sample_csv.csv\ndf = pd.read_csv(\"https://storage.googleapis.com/dqlab-dataset/sample_csv.csv\")\n# Tampilkan tipe data\nprint(\"Tipe data df:\\n\", df.dtypes)\n# Ubah tipe data kolom order_date menjadi datetime\ndf[\"order_date\"] = pd.to_datetime(df[\"order_date\"])\n# Tampilkan tipe data df setelah transformasi\nprint(\"\\nTipe data df setelah transformasi:\\n\", df.dtypes)",
"Tipe data df:\n order_id int64\norder_date object\ncustomer_id int64\ncity object\nprovince object\nproduct_id object\nbrand object\nquantity int64\nitem_price int64\ndtype: object\n\nTipe data df setelah transformasi:\n order_id int64\norder_date datetime64[ns]\ncustomer_id int64\ncity object\nprovince object\nproduct_id object\nbrand object\nquantity int64\nitem_price int64\ndtype: object\n"
]
],
[
[
"# Transforming - Part 2\nPada sub bab ini akan mengubah tipe data pada kolom dataframe yang telah dibaca menjadi tipe data float (kolom quantity) dan tipe kategori (kolom city).\n\nSecara umum, untuk mengubah ke numerik dapat menggunakan pd.to_numeric(), yaitu:\n\nnama_dataframe[\"nama_kolom\"] = pd.to_numeric(nama_dataframe[\"nama_kolom\"], downcast=\"tipe_data_baru\")\nSedangkan untuk menjadi suatu kolom yang dapat dinyatakan sebagai kategori dapat menggunakan method .astype() pada dataframe, yaitu\n\nnama_dataframe[\"nama_kolom\"] = nama_dataframe[\"nama_kolom\"].astype(\"category\")",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n# Baca file sample_csv.csv\ndf = pd.read_csv(\"https://storage.googleapis.com/dqlab-dataset/sample_csv.csv\")\n# Tampilkan tipe data\nprint(\"Tipe data df:\\n\", df.dtypes)\n# Ubah tipe data kolom quantity menjadi tipe data numerik float\ndf[\"quantity\"] = pd.to_numeric(df[\"quantity\"], downcast=\"float\")\n# Ubah tipe data kolom city menjadi tipe data category\ndf[\"city\"] = df[\"city\"].astype(\"category\")\n# Tampilkan tipe data df setelah transformasi\nprint(\"\\nTipe data df setelah transformasi:\\n\", df.dtypes)",
"Tipe data df:\n order_id int64\norder_date object\ncustomer_id int64\ncity object\nprovince object\nproduct_id object\nbrand object\nquantity int64\nitem_price int64\ndtype: object\n\nTipe data df setelah transformasi:\n order_id int64\norder_date object\ncustomer_id int64\ncity category\nprovince object\nproduct_id object\nbrand object\nquantity float32\nitem_price int64\ndtype: object\n"
]
],
[
[
"# Transforming - Part 3\nSekarang akan mempelajari teknik/cara berikutnya dalam proses transformasi suatu dataframe. Di sub bab ini akan memakai method .apply() dan .map() pada suatu dataframe.\n\nMethod .apply() digunakan untuk menerapkan suatu fungsi python (yang dibuat dengan def atau anonymous dengan lambda) pada dataframe/series atau hanya kolom tertentu dari dataframe. \n\nBerikut ini adalah contohnya yaitu akan merubah setiap baris pada kolom brand menjadi lowercase. ",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n# Baca file sample_csv.csv\ndf = pd.read_csv(\"https://storage.googleapis.com/dqlab-dataset/sample_csv.csv\")\n# Cetak 5 baris teratas kolom brand\nprint(\"Kolom brand awal:\\n\", df[\"brand\"].head())\n# Gunakan method apply untuk merubah isi kolom menjadi lower case\ndf[\"brand\"] = df[\"brand\"].apply(lambda x: x.lower())\n# Cetak 5 baris teratas kolom brand\nprint(\"Kolom brand setelah apply:\\n\", df[\"brand\"].head())\n# Gunakan method map untuk mengambil kode brand yaitu karakter terakhirnya\ndf[\"brand\"] = df[\"brand\"].map(lambda x: x[-1])\n# Cetak 5 baris teratas kolom brand\nprint(\"Kolom brand setelah map:\\n\", df[\"brand\"].head())",
"Kolom brand awal:\n 0 BRAND_C\n1 BRAND_V\n2 BRAND_G\n3 BRAND_B\n4 BRAND_G\nName: brand, dtype: object\nKolom brand setelah apply:\n 0 brand_c\n1 brand_v\n2 brand_g\n3 brand_b\n4 brand_g\nName: brand, dtype: object\nKolom brand setelah map:\n 0 c\n1 v\n2 g\n3 b\n4 g\nName: brand, dtype: object\n"
]
],
[
[
"# Transforming - Part 4\nDi sub bab sebelumnya sudah mengetahui bahwa map hanya dapat digunakan untuk pandas series. Pada sub bab ini akan menggunakan method .applymap pada dataframe.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\n# number generator, set angka seed menjadi suatu angka, bisa semua angka, supaya hasil random nya selalu sama ketika kita run\nnp.random.seed(1234)\n# create dataframe 3 baris dan 4 kolom dengan angka random\ndf_tr = pd.DataFrame(np.random.rand(3,4))\n# Cetak dataframe\nprint(\"Dataframe:\\n\", df_tr)\n# Cara 1 dengan tanpa define function awalnya, langsung pake fungsi anonymous lambda x\ndf_tr1 = df_tr.applymap(lambda x: x**2 + 3*x + 2) \nprint(\"\\nDataframe - cara 1:\\n\", df_tr1)\n# Cara 2 dengan define function \ndef qudratic_fun(x):\n\treturn x**2 + 3*x + 2\ndf_tr2 = df_tr.applymap(qudratic_fun)\nprint(\"\\nDataframe - cara 2:\\n\", df_tr2)",
"Dataframe:\n 0 1 2 3\n0 0.191519 0.622109 0.437728 0.785359\n1 0.779976 0.272593 0.276464 0.801872\n2 0.958139 0.875933 0.357817 0.500995\n\nDataframe - cara 1:\n 0 1 2 3\n0 2.611238 4.253346 3.504789 4.972864\n1 4.948290 2.892085 2.905825 5.048616\n2 5.792449 5.395056 3.201485 3.753981\n\nDataframe - cara 2:\n 0 1 2 3\n0 2.611238 4.253346 3.504789 4.972864\n1 4.948290 2.892085 2.905825 5.048616\n2 5.792449 5.395056 3.201485 3.753981\n"
]
],
[
[
"# Inspeksi Missing Value\nValue yang hilang/tidak lengkap dari dataframe akan membuat analisis atau model prediksi yang dibuat menjadi tidak akurat dan mengakibatkan keputusan salah yang diambil. Terdapat beberapa cara untuk mengatasi data yang hilang/tidak lengkap tersebut.\n\nData COVID-19 yang akan digunakan ini diambil dari google big query, tetapi sudah disediakan datasetnya dalam format csv dengan nama \"public data covid19 jhu csse eu.csv\". Ini adalah studi kasus untuk meng-handle missing value. Bagaimanakah langkah-langkahnya?\n\nDi pandas data yang hilang umumnya direpresentasikan dengan NaN.\n\nLangkah pertama, harus tahu kolom mana yang terdapat data hilang dan berapa banyak dengan cara:\n\nCara 1: menerapkan method .info() pada dataframe",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n# Baca file \"public data covid19 jhu csse eu.csv\"\ndf = pd.read_csv(\"https://storage.googleapis.com/dqlab-dataset/CHAPTER%204%20-%20missing%20value%20-%20public%20data%20covid19%20.csv\")\n# Cetak info dari df\nprint(df.info())\n# Cetak jumlah missing value di setiap kolom\nmv = df.isna().sum()\nprint(\"\\nJumlah missing value per kolom:\\n\", mv)",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1000 entries, 0 to 999\nData columns (total 13 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 province_state 960 non-null object \n 1 country_region 1000 non-null object \n 2 date 1000 non-null object \n 3 latitude 874 non-null float64\n 4 longitude 874 non-null float64\n 5 location_geom 874 non-null object \n 6 confirmed 1000 non-null int64 \n 7 deaths 999 non-null float64\n 8 recovered 999 non-null float64\n 9 active 949 non-null float64\n 10 fips 949 non-null float64\n 11 admin2 842 non-null object \n 12 combined_key 0 non-null float64\ndtypes: float64(7), int64(1), object(5)\nmemory usage: 101.7+ KB\nNone\n\nJumlah missing value per kolom:\n province_state 40\ncountry_region 0\ndate 0\nlatitude 126\nlongitude 126\nlocation_geom 126\nconfirmed 0\ndeaths 1\nrecovered 1\nactive 51\nfips 51\nadmin2 158\ncombined_key 1000\ndtype: int64\n"
]
],
[
[
"# Treatment untuk Missing Value - Part 1\nTerdapat beberapa cara untuk mengatasi missing value, antara lain:\n\ndibiarkan saja,\nhapus value itu, atau\nisi value tersebut dengan value yang lain (biasanya interpolasi, mean, median, etc)",
"_____no_output_____"
],
[
"# Treatment untuk Missing Value - Part 2\nSekarang dapat menerapkan dua aksi yaitu\n\nMembiarkannya saja\nMenghapus kolom\n \n\nTreatment pertama (membiarkannya saja) seperti pada kolom confirmed, death, dan recovered. Akan tetapi jika tidak ada yang terkonfirmasi, meninggal dan sembuh sebenarnya dapat menukar value ini dengan angka nol. Meskipun ini lebih make sense dalam representasi datanya, tetapi untuk sub bab ini ketiga kolom tersebut diasumsikan dibiarkan memiliki nilai missing value.\n\n \n\nTreatment kedua yaitu dengan menghapus kolom, yang mana ini digunakan jika seluruh kolom dari dataset yang dipunyai semua barisnya adalah missing value. Untuk itu dapat menerapkan method .dropna() pada dataframe, bagaimana caranya?\n\nnama_dataframe.dropna(axis=1, how=\"all\")\nPada method .dropna() ada dua keyword argumen yang harus diisikan yaitu axis dan how. Keyword axis digunakan untuk menentukan arah dataframe yang akan dibuang angka 1 untuk menyatakan kolom (column-based) atau dapat ditulis dalam string \"column\". Jika digunakan angka 0 berarti itu dalam searah index (row-based) atau dapat ditulis dalam string \"index\".\n\nSementara, keyword how digunakan untuk bagaimana cara membuangnya. Opsi yang dapat diterimanya (dalam string) adalah\n\n\"all\" artinya jika seluruh data di satu/beberapa kolom atau di satu/beberapa baris adalah missing value.\n\"any\" artinya jika memiliki 1 saja data yang hilang maka buanglah baris/kolom tersebut.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n# Baca file \"public data covid19 jhu csse eu.csv\"\ndf = pd.read_csv(\"https://storage.googleapis.com/dqlab-dataset/CHAPTER%204%20-%20missing%20value%20-%20public%20data%20covid19%20.csv\")\n# Cetak ukuran awal dataframe\nprint(\"Ukuran awal df: %d baris, %d kolom.\" % df.shape)\n# Drop kolom yang seluruhnya missing value dan cetak ukurannya\ndf = df.dropna(axis=1, how=\"all\")\nprint(\"Ukuran df setelah buang kolom dengan seluruh data missing: %d baris, %d kolom.\" % df.shape)\n# Drop baris jika ada satu saja data yang missing dan cetak ukurannya\ndf = df.dropna(axis=0, how=\"any\")\nprint(\"Ukuran df setelah dibuang baris yang memiliki sekurangnya 1 missing value: %d baris, %d kolom.\" % df.shape)",
"Ukuran awal df: 1000 baris, 13 kolom.\nUkuran df setelah buang kolom dengan seluruh data missing: 1000 baris, 12 kolom.\nUkuran df setelah dibuang baris yang memiliki sekurangnya 1 missing value: 746 baris, 12 kolom.\n"
]
],
[
[
"# Treatment untuk Missing Value - Part 3\nSekarang, akan melakukan treatment ketiga untuk melakukan handle missing value pada dataframe. Treatment ini dilakukan dengan cara mengisi missing value dengan nilai lain, yang dapat berupa :\n\nnilai statistik seperti mean atau median\ninterpolasi data\ntext tertentu\n \n\nAkan mulai pada kolom yang missing yang tipe datanya adalah berupa object. Kolom tersebut adalah province_state, karena tidak tahu secara persis province_state mana yang dimaksud, bisa menempatkan string \"unknown\" sebagai substitusi missing value. Meskipun keduanya berarti sama-sama tidak tahu tetapi berbeda dalam representasi datanya.\n\nUntuk melakukan hal demikian dapat menggunakan method .fillna() pada kolom dataframe yang dimaksud.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n# Baca file \"public data covid19 jhu csse eu.csv\"\ndf = pd.read_csv(\"https://storage.googleapis.com/dqlab-dataset/CHAPTER%204%20-%20missing%20value%20-%20public%20data%20covid19%20.csv\")\n# Cetak unique value pada kolom province_state\nprint(\"Unique value awal:\\n\", df[\"province_state\"].unique())\n# Ganti missing value dengan string \"unknown_province_state\"\ndf[\"province_state\"] = df[\"province_state\"].fillna(\"unknown_province_state\")\n# Cetak kembali unique value pada kolom province_state\nprint(\"Unique value setelah fillna:\\n\", df[\"province_state\"].unique())",
"Unique value awal:\n [nan 'US' 'Guam' 'Iowa']\nUnique value setelah fillna:\n ['unknown_province_state' 'US' 'Guam' 'Iowa']\n"
]
],
[
[
"# Treatment untuk Missing Value - Part 4\nMasih melanjutkan bagaimana melakukan handle missing value tentunya dengan jalan mengganti missing value dengan nilai lainnya. Pada bab sebelumnya telah mengganti kolom bertipe objek dengan sesuatu string/teks.\n\nDalam sub bab ini akan mengganti missing value dengan nilai statistik kolom bersangkutan, baik median atau mean (nilai rata-rata). Misalnya akan menggunakan kolom active. Dengan mengabaikan terlebih dahulu sebaran berdasarkan negara (univariate), jika mengisi dengan nilai rata-rata maka harus melihat terlebih dahulu data apakah memiliki outliers atau tidak. Jika ada outliers dari data maka menggunakan nilai tengah (median) data adalah cara yang lebih safe.\n\nUntuk itu diputuskan dengan mengecek nilai median dan nilai mean kolom active juga nilai min dan max-nya. Jika data pada kolom active terdistribusi normal maka nilai mean dan median akan hampir sama.\n\nTerlihat data memiliki distribusi yang skewness, karena nilai mean dan median yang cukup jauh serta range data yang cukup lebar. Di sini pada kolom active data memiliki outliers. Jadi akan mengisi missing value dengan median.\n",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n# Baca file \"https://dqlab-dataset.s3-ap-southeast-1.amazonaws.com/CHAPTER+4+-+missing+value+-+public+data+covid19+.csv\"\ndf = pd.read_csv(\"https://storage.googleapis.com/dqlab-dataset/CHAPTER%204%20-%20missing%20value%20-%20public%20data%20covid19%20.csv\")\n# Cetak nilai mean dan median awal\nprint(\"Awal: mean = %f, median = %f.\" % (df[\"active\"].mean(), df[\"active\"].median()))\n# Isi missing value kolom active dengan median\ndf_median = df[\"active\"].fillna(df[\"active\"].median())\n# Cetak nilai mean dan median awal setelah diisi dengan median\nprint(\"Fillna median: mean = %f, median = %f.\" % (df_median.mean(), df_median.median()))\n# Isi missing value kolom active dengan mean\ndf_mean = df[\"active\"].fillna(df[\"active\"].mean())\n# Cetak nilai mean dan median awal setelah diisi dengan mean\nprint(\"Fillna mean: mean = %f, median = %f.\" % (df_mean.mean(), df_mean.median()))",
"Awal: mean = 192.571128, median = 41.000000.\nFillna median: mean = 184.841000, median = 41.000000.\nFillna mean: mean = 192.571128, median = 49.000000.\n"
]
],
[
[
"# Treatment untuk Missing Value - Part 5\nDi bagian ini akan menggunakan teknik interpolasi dalam mengisi nilai missing value pada suatu dataset.\n\nData yang menggunakan interpolasi untuk mengisi data yang hilang adalah time series data, yang secara default akan diisi dengan interpolasi linear.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\n# Data\nts = pd.Series({\n \"2020-01-01\":9,\n \"2020-01-02\":np.nan,\n \"2020-01-05\":np.nan,\n \"2020-01-07\":24,\n \"2020-01-10\":np.nan,\n \"2020-01-12\":np.nan,\n \"2020-01-15\":33,\n \"2020-01-17\":np.nan,\n \"2020-01-16\":40,\n \"2020-01-20\":45,\n \"2020-01-22\":52,\n \"2020-01-25\":75,\n \"2020-01-28\":np.nan,\n \"2020-01-30\":np.nan\n})\n# Isi missing value menggunakan interpolasi linier\nts = ts.interpolate()\n# Cetak time series setelah interpolasi linier\nprint(\"Setelah diisi missing valuenya:\\n\", ts)\n",
"Setelah diisi missing valuenya:\n 2020-01-01 9.0\n2020-01-02 14.0\n2020-01-05 19.0\n2020-01-07 24.0\n2020-01-10 27.0\n2020-01-12 30.0\n2020-01-15 33.0\n2020-01-17 36.5\n2020-01-16 40.0\n2020-01-20 45.0\n2020-01-22 52.0\n2020-01-25 75.0\n2020-01-28 75.0\n2020-01-30 75.0\ndtype: float64\n"
]
],
[
[
"# Project \nDiberikan dataset ‘retail_raw_test.csv’\n1. Baca dataset\n2. Tipe data diubah menjadi tipe yang seharusnya\n- customer_id dari string ke int64,\n- quantity dari string ke int64,\n- item_price dari string ke int64\n3. transform product_value supaya bentuknya seragam dengan format PXXXX, assign ke kolom baru \"product_id\", dan drop kolom \"product_value\", jika terdapat nan gantilah dengan \"unknown\".\n4. trasnform order_date menjadi value dengan format YYYY-mm-dd\n5. cek data hilang dari tiap kolom dan kemudian isi missing value\n- di brand dengan \"no_brand\", dan\n- cek dulu bagaimana missing value di city & province - isi missing value di city dan province dengan \"unknown\"\n6. create column city/province dari gabungan city & province\n7. membuat index berdasarkan city_provice, order_date, customer_id, order_id, product_id (cek index)\n8. membuat kolom \"total_price\" sebagai hasil perkalian quantity dengan item_price\n9. slice data hanya untuk Jan 2019",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\n# 1. Baca dataset\nprint(\"[1] BACA DATASET\")\ndf = pd.read_csv(\"https://storage.googleapis.com/dqlab-dataset/retail_raw_test.csv\", low_memory=False)\nprint(\" Dataset:\\n\", df.head())\nprint(\" Info:\\n\", df.info())\n\n# 2. Ubah tipe data\nprint(\"\\n[2] UBAH TIPE DATA\")\ndf[\"customer_id\"] = df[\"customer_id\"].apply(lambda x: x.split(\"'\")[1]).astype(\"int64\")\ndf[\"quantity\"] = df[\"quantity\"].apply(lambda x: x.split(\"'\")[1]).astype(\"int64\")\ndf[\"item_price\"] = df[\"item_price\"].apply(lambda x: x.split(\"'\")[1]).astype(\"int64\")\nprint(\" Tipe data:\\n\", df.dtypes)\n\n# 3. Transform \"product_value\" supaya bentuknya seragam dengan format \"PXXXX\", assign ke kolom baru \"product_id\", dan drop kolom \"product_value\", jika terdapat nan gantilah dengan \"unknown\"\nprint(\"\\n[3] TRANSFORM product_value MENJADI product_id\")\n# Buat fungsi\nimport math\ndef impute_product_value(val):\n if math.isnan(val):\n return \"unknown\"\n else:\n return 'P' + '{:0>4}'.format(str(val).split('.')[0])\n# Buat kolom \"product_id\"\ndf[\"product_id\"] = df[\"product_value\"].apply(lambda x: impute_product_value(x))\n# Hapus kolom \"product_value\"\ndf.drop([\"product_value\"], axis=1, inplace=True)\n# Cetak 5 data teratas\nprint(df.head())\n\n# 4. Tranform order_date menjadi value dengan format \"YYYY-mm-dd\"\nprint(\"\\n[4] TRANSFORM order_date MENJADI FORMAT YYYY-mm-dd\")\nmonths_dict = {\n \"Jan\":\"01\",\n \"Feb\":\"02\",\n \"Mar\":\"03\",\n \"Apr\":\"04\",\n \"May\":\"05\",\n \"Jun\":\"06\",\n \"Jul\":\"07\",\n \"Aug\":\"08\",\n \"Sep\":\"09\",\n \"Oct\":\"10\",\n \"Nov\":\"11\",\n \"Dec\":\"12\"\n}\ndf[\"order_date\"] = pd.to_datetime(df[\"order_date\"].apply(lambda x: str(x)[-4:] + \"-\" + months_dict[str(x)[:3]] + \"-\" + str(x)[4:7]))\nprint(\" Tipe data:\\n\", df.dtypes)\n\n# 5. Mengatasi data yang hilang di beberapa kolom\nprint(\"\\n[5] HANDLING MISSING VALUE\")\n# Kolom \"city\" dan \"province\" masih memiliki missing value, nilai yang hilang di kedua kolom ini diisi saja dengan \"unknown\"\ndf[[\"city\",\"province\"]] = df[[\"city\",\"province\"]].fillna(\"unknown\")\n# Kolom brand juga masih memiliki missing value, Ganti value NaN menjadi \"no_brand\"\ndf[\"brand\"] = df[\"brand\"].fillna(\"no_brand\")\n# Cek apakah masih terdapat missing value di seluruh kolom \nprint(\" Info:\\n\", df.info())\n\n# 6. Membuat kolom baru \"city/province\" dengan menggabungkan kolom \"city\" dan kolom \"province\" dan delete kolom asalnya\nprint(\"\\n[6] MEMBUAT KOLOM BARU city/province\")\ndf[\"city/province\"] = df[\"city\"] + \"/\" + df[\"province\"]\n# drop kolom \"city\" dan \"province\" karena telah digabungkan\ndf.drop([\"city\",\"province\"], axis=1, inplace=True)\n# Cetak 5 data teratas\nprint(df.head())\n\n# 7. Membuat hierarchical index yang terdiri dari kolom \"city/province\", \"order_date\", \"customer_id\", \"order_id\", \"product_id\"\nprint(\"\\n[7] MEMBUAT HIERACHICAL INDEX\")\ndf = df.set_index([\"city/province\",\"order_date\",\"customer_id\",\"order_id\",\"product_id\"])\n# urutkanlah berdasarkan index yang baru\ndf = df.sort_index()\n# Cetak 5 data teratas\nprint(df.head())\n\n# 8. Membuat kolom \"total_price\" yang formula nya perkalian antara kolom \"quantity\" dan kolom \"item_price\"\nprint(\"\\n[8] MEMBUAT KOLOM total_price\")\ndf[\"total_price\"] = df[\"quantity\"] * df[\"item_price\"]\n# Cetak 5 data teratas\nprint(df.head())\n\n# 9. Slice dataset agar hanya terdapat data bulan Januari 2019\nprint(\"\\n[9] SLICE DATASET UNTUK BULAN JANUARI 2019 SAJA\")\nidx = pd.IndexSlice\ndf_jan2019 = df.loc[idx[:, \"2019-01-01\":\"2019-01-31\"], :]\nprint(\"Dataset akhir:\\n\", df_jan2019)\n\n# END OF PROJECT",
"[1] BACA DATASET\n Dataset:\n order_id order_date customer_id city province brand \\\n0 1730350 Dec 11, 2019 '13447 Surakarta Jawa Tengah BRAND_F \n1 1677490 Jul 31, 2019 '0 NaN NaN BRAND_F \n2 1704211 Oct 18, 2019 '16128 Jakarta Pusat DKI Jakarta BRAND_H \n3 1679695 Aug 07, 2019 '16225 Yogyakarta Yogyakarta BRAND_H \n4 1679080 Aug 05, 2019 '0 NaN NaN BRAND_E \n\n quantity item_price product_value \n0 '24 '113000 1374.0 \n1 '1 '1164000 1370.0 \n2 '12 '747000 1679.0 \n3 '6 '590000 1708.0 \n4 '2 '740000 1201.0 \n<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 5000 entries, 0 to 4999\nData columns (total 9 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 order_id 5000 non-null int64 \n 1 order_date 5000 non-null object \n 2 customer_id 5000 non-null object \n 3 city 3802 non-null object \n 4 province 3802 non-null object \n 5 brand 4995 non-null object \n 6 quantity 5000 non-null object \n 7 item_price 5000 non-null object \n 8 product_value 4995 non-null float64\ndtypes: float64(1), int64(1), object(7)\nmemory usage: 351.7+ KB\n Info:\n None\n\n[2] UBAH TIPE DATA\n Tipe data:\n order_id int64\norder_date object\ncustomer_id int64\ncity object\nprovince object\nbrand object\nquantity int64\nitem_price int64\nproduct_value float64\ndtype: object\n\n[3] TRANSFORM product_value MENJADI product_id\n order_id order_date customer_id city province brand \\\n0 1730350 Dec 11, 2019 13447 Surakarta Jawa Tengah BRAND_F \n1 1677490 Jul 31, 2019 0 NaN NaN BRAND_F \n2 1704211 Oct 18, 2019 16128 Jakarta Pusat DKI Jakarta BRAND_H \n3 1679695 Aug 07, 2019 16225 Yogyakarta Yogyakarta BRAND_H \n4 1679080 Aug 05, 2019 0 NaN NaN BRAND_E \n\n quantity item_price product_id \n0 24 113000 P1374 \n1 1 1164000 P1370 \n2 12 747000 P1679 \n3 6 590000 P1708 \n4 2 740000 P1201 \n\n[4] TRANSFORM order_date MENJADI FORMAT YYYY-mm-dd\n Tipe data:\n order_id int64\norder_date datetime64[ns]\ncustomer_id int64\ncity object\nprovince object\nbrand object\nquantity int64\nitem_price int64\nproduct_id object\ndtype: object\n\n[5] HANDLING MISSING VALUE\n<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 5000 entries, 0 to 4999\nData columns (total 9 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 order_id 5000 non-null int64 \n 1 order_date 5000 non-null datetime64[ns]\n 2 customer_id 5000 non-null int64 \n 3 city 5000 non-null object \n 4 province 5000 non-null object \n 5 brand 5000 non-null object \n 6 quantity 5000 non-null int64 \n 7 item_price 5000 non-null int64 \n 8 product_id 5000 non-null object \ndtypes: datetime64[ns](1), int64(4), object(4)\nmemory usage: 351.7+ KB\n Info:\n None\n\n[6] MEMBUAT KOLOM BARU city/province\n order_id order_date customer_id brand quantity item_price product_id \\\n0 1730350 2019-12-11 13447 BRAND_F 24 113000 P1374 \n1 1677490 2019-07-31 0 BRAND_F 1 1164000 P1370 \n2 1704211 2019-10-18 16128 BRAND_H 12 747000 P1679 \n3 1679695 2019-08-07 16225 BRAND_H 6 590000 P1708 \n4 1679080 2019-08-05 0 BRAND_E 2 740000 P1201 \n\n city/province \n0 Surakarta/Jawa Tengah \n1 unknown/unknown \n2 Jakarta Pusat/DKI Jakarta \n3 Yogyakarta/Yogyakarta \n4 unknown/unknown \n\n[7] MEMBUAT HIERACHICAL INDEX\n brand \\\ncity/province order_date customer_id order_id product_id \nBanda Aceh/Aceh 2019-04-17 12818 1642480 P1936 BRAND_K \n 2019-11-12 12360 1715116 P0758 BRAND_C \n P3042 BRAND_R \n 2019-12-09 12374 1729036 P1660 BRAND_G \nBandar Lampung/Lampung 2019-01-15 12515 1619257 P0628 BRAND_C \n\n quantity \\\ncity/province order_date customer_id order_id product_id \nBanda Aceh/Aceh 2019-04-17 12818 1642480 P1936 24 \n 2019-11-12 12360 1715116 P0758 8 \n P3042 12 \n 2019-12-09 12374 1729036 P1660 4 \nBandar Lampung/Lampung 2019-01-15 12515 1619257 P0628 12 \n\n item_price \ncity/province order_date customer_id order_id product_id \nBanda Aceh/Aceh 2019-04-17 12818 1642480 P1936 450000 \n 2019-11-12 12360 1715116 P0758 695000 \n P3042 310000 \n 2019-12-09 12374 1729036 P1660 2795000 \nBandar Lampung/Lampung 2019-01-15 12515 1619257 P0628 695000 \n\n[8] MEMBUAT KOLOM total_price\n brand \\\ncity/province order_date customer_id order_id product_id \nBanda Aceh/Aceh 2019-04-17 12818 1642480 P1936 BRAND_K \n 2019-11-12 12360 1715116 P0758 BRAND_C \n P3042 BRAND_R \n 2019-12-09 12374 1729036 P1660 BRAND_G \nBandar Lampung/Lampung 2019-01-15 12515 1619257 P0628 BRAND_C \n\n quantity \\\ncity/province order_date customer_id order_id product_id \nBanda Aceh/Aceh 2019-04-17 12818 1642480 P1936 24 \n 2019-11-12 12360 1715116 P0758 8 \n P3042 12 \n 2019-12-09 12374 1729036 P1660 4 \nBandar Lampung/Lampung 2019-01-15 12515 1619257 P0628 12 \n\n item_price \\\ncity/province order_date customer_id order_id product_id \nBanda Aceh/Aceh 2019-04-17 12818 1642480 P1936 450000 \n 2019-11-12 12360 1715116 P0758 695000 \n P3042 310000 \n 2019-12-09 12374 1729036 P1660 2795000 \nBandar Lampung/Lampung 2019-01-15 12515 1619257 P0628 695000 \n\n total_price \ncity/province order_date customer_id order_id product_id \nBanda Aceh/Aceh 2019-04-17 12818 1642480 P1936 10800000 \n 2019-11-12 12360 1715116 P0758 5560000 \n P3042 3720000 \n 2019-12-09 12374 1729036 P1660 11180000 \nBandar Lampung/Lampung 2019-01-15 12515 1619257 P0628 8340000 \n\n[9] SLICE DATASET UNTUK BULAN JANUARI 2019 SAJA\nDataset akhir:\n brand \\\ncity/province order_date customer_id order_id product_id \nBandar Lampung/Lampung 2019-01-15 12515 1619257 P0628 BRAND_C \nBandung/Jawa Barat 2019-01-09 16134 1617055 P1597 BRAND_G \n 2019-01-10 17392 1617952 P2137 BRAND_M \n 2019-01-14 15527 1618828 P3115 BRAND_S \n 2019-01-29 13253 1620289 P0099 BRAND_A \n... ... \nunknown/unknown 2019-01-30 0 1620766 P3070 BRAND_R \n P3483 BRAND_S \n 2019-01-31 0 1621057 P1298 BRAND_F \n P1773 BRAND_H \n P2877 BRAND_R \n\n quantity \\\ncity/province order_date customer_id order_id product_id \nBandar Lampung/Lampung 2019-01-15 12515 1619257 P0628 12 \nBandung/Jawa Barat 2019-01-09 16134 1617055 P1597 9 \n 2019-01-10 17392 1617952 P2137 2 \n 2019-01-14 15527 1618828 P3115 1 \n 2019-01-29 13253 1620289 P0099 12 \n... ... \nunknown/unknown 2019-01-30 0 1620766 P3070 1 \n P3483 3 \n 2019-01-31 0 1621057 P1298 1 \n P1773 5 \n P2877 1 \n\n item_price \\\ncity/province order_date customer_id order_id product_id \nBandar Lampung/Lampung 2019-01-15 12515 1619257 P0628 695000 \nBandung/Jawa Barat 2019-01-09 16134 1617055 P1597 520000 \n 2019-01-10 17392 1617952 P2137 1062000 \n 2019-01-14 15527 1618828 P3115 1045000 \n 2019-01-29 13253 1620289 P0099 450000 \n... ... \nunknown/unknown 2019-01-30 0 1620766 P3070 593000 \n P3483 593000 \n 2019-01-31 0 1621057 P1298 296000 \n P1773 593000 \n P2877 1486000 \n\n total_price \ncity/province order_date customer_id order_id product_id \nBandar Lampung/Lampung 2019-01-15 12515 1619257 P0628 8340000 \nBandung/Jawa Barat 2019-01-09 16134 1617055 P1597 4680000 \n 2019-01-10 17392 1617952 P2137 2124000 \n 2019-01-14 15527 1618828 P3115 1045000 \n 2019-01-29 13253 1620289 P0099 5400000 \n... ... \nunknown/unknown 2019-01-30 0 1620766 P3070 593000 \n P3483 1779000 \n 2019-01-31 0 1621057 P1298 296000 \n P1773 2965000 \n P2877 1486000 \n\n[334 rows x 4 columns]\n"
]
],
[
[
"# Evaluasi Project \nMengapa di langkah ketiga, dicari tahu terlebih dulu nilai null. Mengapa tidak diconvert to string secara langsung?”\n\nkarena di df.info()nya masih ada yang kosong di kolom ‘product_value’. Kalau langsung convert to string, value NaN akan berubah menjadi string ‘nan’, kemudian ketika pad 0 di depan dan concat dengan char ‘P’, hasilnya akan menjadi ‘P0nan’ yang aneh sekali.\n\nKenapa kamu memakai langkah ke-4? Mengapa tidak langsung menggunakan kolom date yang sudah ada. Bukankah format waktunya sudah ideal?\n\nTidak semua format datetime yang ideal pada umumnya akan ideal di dalam pandas environment. Seenggaknya harus translate dulu menjadi format yang ideal di dalam pandas sehingga pandas bisa mengenali.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0e4cce80107c99ef1e465c2106f46e9fbf7dbe0 | 8,635 | ipynb | Jupyter Notebook | Data.ipynb | tracypoon/Web_Design_Challenge | 11008ebc56825a1a9cc61a28a03d74d4c91c804b | [
"ADSL"
] | null | null | null | Data.ipynb | tracypoon/Web_Design_Challenge | 11008ebc56825a1a9cc61a28a03d74d4c91c804b | [
"ADSL"
] | null | null | null | Data.ipynb | tracypoon/Web_Design_Challenge | 11008ebc56825a1a9cc61a28a03d74d4c91c804b | [
"ADSL"
] | null | null | null | 30.298246 | 91 | 0.316734 | [
[
[
"# Python program to convert\n# CSV to HTML Table\n \nimport pandas as pd\n \n# to read csv file named \"cities\"\ndata = pd.read_csv(\"Resources/cities.csv\")\ndata\n",
"_____no_output_____"
],
[
"# to save as html file\n# named as \"Table\"\ndata.to_html(\"Table.html\")\n \n",
"_____no_output_____"
],
[
"# assign it to data\n# variable (string)\nhtml_file = data.to_html()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
d0e4cffd77d585c7a7f6a448c23f65a85b58afce | 257,034 | ipynb | Jupyter Notebook | notebooks/D2_L5_Visualization/02-airports.ipynb | highrain2/myTestProjs | edc3c8bd041110ce79fe2a6dd134dc068205ca52 | [
"MIT"
] | null | null | null | notebooks/D2_L5_Visualization/02-airports.ipynb | highrain2/myTestProjs | edc3c8bd041110ce79fe2a6dd134dc068205ca52 | [
"MIT"
] | 5 | 2020-01-28T23:05:25.000Z | 2022-02-10T00:22:11.000Z | notebooks/D2_L5_Visualization/02-airports.ipynb | highrain2/myTestProjs | edc3c8bd041110ce79fe2a6dd134dc068205ca52 | [
"MIT"
] | null | null | null | 75.398651 | 58,096 | 0.665348 | [
[
[
"# Drawing flight routes with NetworkX",
"_____no_output_____"
]
],
[
[
"import math\nimport json\nimport numpy as np\nimport pandas as pd\nimport networkx as nx\nimport cartopy.crs as ccrs\nimport matplotlib.pyplot as plt\nfrom IPython.display import Image\n%matplotlib inline",
"_____no_output_____"
],
[
"names = ('airline,airline_id,source,source_id,dest,dest_id,codeshare,stops,equipment').split(',')",
"_____no_output_____"
],
[
"routes = pd.read_csv(\n 'data/routes.dat',\n names=names,\n header=None)\nroutes",
"_____no_output_____"
],
[
"names = ('id,name,city,country,iata,icao,lat,lon,alt,timezone,dst,tz,type,source').split(',')",
"_____no_output_____"
],
[
"airports = pd.read_csv(\n 'data/airports.dat',\n header=None,\n names=names,\n index_col=4,\n na_values='\\\\N')\nairports_us = airports[airports['country'] == 'United States']\nairports_us",
"_____no_output_____"
],
[
"routes_us = routes[\n routes['source'].isin(airports_us.index) &\n routes['dest'].isin(airports_us.index)]\nroutes_us",
"_____no_output_____"
],
[
"edges = routes_us[['source', 'dest']].values\nedges",
"_____no_output_____"
],
[
"g = nx.from_edgelist(edges)",
"_____no_output_____"
],
[
"len(g.nodes()), len(g.edges())",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(1, 1, figsize=(6, 6))\nnx.draw_networkx(g, ax=ax, node_size=5,\n font_size=6, alpha=.5,\n width=.5)\nax.set_axis_off()",
"C:\\ProgramData\\Anaconda3\\lib\\site-packages\\networkx\\drawing\\nx_pylab.py:579: MatplotlibDeprecationWarning: \nThe iterable function was deprecated in Matplotlib 3.1 and will be removed in 3.3. Use np.iterable instead.\n if not cb.iterable(width):\n"
],
[
"sg = next(nx.connected_component_subgraphs(g))",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(1, 1, figsize=(6, 6))\nnx.draw_networkx(sg, ax=ax, with_labels=False,\n node_size=5, width=.5)\nax.set_axis_off()",
"_____no_output_____"
],
[
"pos = {airport: (v['lon'], v['lat'])\n for airport, v in\n airports_us.to_dict('index').items()}",
"_____no_output_____"
],
[
"deg = nx.degree(sg)\nsizes = [5 * deg[iata] for iata in sg.nodes]",
"_____no_output_____"
],
[
"altitude = airports_us['alt']\naltitude = [altitude[iata] for iata in sg.nodes]",
"_____no_output_____"
],
[
"labels = {iata: iata if deg[iata] >= 20 else ''\n for iata in sg.nodes}",
"_____no_output_____"
],
[
"# Map projection\ncrs = ccrs.PlateCarree()\nfig, ax = plt.subplots(\n 1, 1, figsize=(12, 8),\n subplot_kw=dict(projection=crs))\nax.coastlines()\n# Extent of continental US.\nax.set_extent([-128, -62, 20, 50])\nnx.draw_networkx(sg, ax=ax,\n font_size=16,\n alpha=.5,\n width=.075,\n node_size=sizes,\n labels=labels,\n pos=pos,\n node_color=altitude,\n cmap=plt.cm.autumn)",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e4dd0c80bef60ff9a9a5ad4591a86694c203da | 33,512 | ipynb | Jupyter Notebook | Previous Final Projects/2017/1 Liter per Second Plants in Parallel/.ipynb_checkpoints/Team_CHANCEUX_1_LPS_Plants_in_Parallel_Report-checkpoint.ipynb | CEE-4540/python-tutorial-juandirection | e9a75b3ac3ab745244ff7798b5f86c6f9769bb6f | [
"MIT"
] | 10 | 2019-01-31T20:17:09.000Z | 2020-01-16T03:58:05.000Z | Previous Final Projects/2017/1 Liter per Second Plants in Parallel/.ipynb_checkpoints/Team_CHANCEUX_1_LPS_Plants_in_Parallel_Report-checkpoint.ipynb | CEE-4540/python-tutorial-juandirection | e9a75b3ac3ab745244ff7798b5f86c6f9769bb6f | [
"MIT"
] | 51 | 2019-01-24T17:32:24.000Z | 2019-12-05T17:29:22.000Z | Previous Final Projects/2017/1 Liter per Second Plants in Parallel/.ipynb_checkpoints/Team_CHANCEUX_1_LPS_Plants_in_Parallel_Report-checkpoint.ipynb | CEE-4540/python-tutorial-juandirection | e9a75b3ac3ab745244ff7798b5f86c6f9769bb6f | [
"MIT"
] | 5 | 2019-02-01T21:38:03.000Z | 2019-09-01T22:21:45.000Z | 62.405959 | 1,043 | 0.748568 | [
[
[
"from aide_design.play import*\nfrom aide_design import floc_model as floc\nfrom aide_design import cdc_functions as cdc\nfrom aide_design.unit_process_design.prefab import lfom_prefab_functional as lfom\nfrom pytexit import py2tex\nimport math",
"_____no_output_____"
]
],
[
[
"# 1 L/s Plants in Parallel ",
"_____no_output_____"
],
[
"# CHANCEUX\n## Priya Aggarwal, Sung Min Kim, Felix Yang\n\n",
"_____no_output_____"
],
[
"AguaClara has been designing and building water treatment plants since 2005 in Honduras, 2013 in India, and 2017 in Nicaragua. It has been providing gravity powered water treatment systems to thousands of people in rural communities. However, there were populations that could not be targeted due to the technology only being scaled up from 6 L/s. For towns and rural areas with populations with smaller needs, AguaClara technologies were out of their reach. \n\nRecently a one liter per second (1 LPS) plant was developed based on traditional AguaClara technology, to bring sustainable water treatment to towns with populations of 300 people per plant. \n\nThe first 1 LPS plant fabricated was sent to Cuatro Comunidades in Honduras, where a built in place plant already exists, and is currently operating without the filter attachment, also known as Enclosed Stacked Rapid Sand Filter (EStaRS). EStaRS is the last step in the 1 LPS plant processes before chlorination and completes the 4 step water treatment process: flocculation, sedimentation, filtration, and chlorination.\nHaving water treatment plants for smaller flow rates would increase AguaClara’s reach and allow it to help more people. Despite being in its initial stages, the demand for this technology is increasing. Three 1 LPS plants were recently ordered for a town that did not have water treatment. However, the implementation of 1 LPS plants is a problem that has not yet been solved. \n\nThis project has stemmed from the possibility of implementing AguaClara technologies to be helpful in Puerto Rico’s post hurricane rebuild effort. The goal of this project is to assess whether the portable 1 L/s plant could be a viable option to help rural communities have safe drinking water. The project models multiple 1 L/s plants working in parallel to provide for the community and plans for the future when communities would need to add capacity. For this project, the team has set 6 L/s as the design constraint. We need experience building and deploying 1 LPS plants to determine the economics and ease of operation to compare to those of built in place plants. For example, if we need 12 L/s, it could still be reasonable to use the 1 LPS plants in parallel or select a 16 L/s built in place plant if more than 12 L/s is needed. Because the dividing line between the modular prefabricated 1 LPS plants and the build in place plants is unknown, the team chose 6 L/s because it is the smallest built in place plant capacity. \n\nOur model is based on the following: \n* Standardization modular designs for each plant (1 plant has one EStaRs and Flocculator)\n* One entrance tank and chemical dosing controller\n* Entrance Tank accomodates to 6 L/s flow\n* Coagulant/ Chlorine dosing according to flow by operator\n* Parallel Layout for convenience \n* Extendable shelter walls to add capacity using chain-link fencing \n* Manifolds connecting up to 3 plants (accounting for 3 L/s) from the sedimentation tank to the ESTaRS and after filtration for chlorination (using Ts and fernco caps)\n* Manifolds to prevent flow to other filters being cut off if filters need to be backwashed and lacks enough flow \n* Equal flow to the filters and chlorination from the manifolds\n\n\nCalculations follow below. ",
"_____no_output_____"
],
[
"### Chemical Dosing Flow Rates",
"_____no_output_____"
],
[
"Below the functions for calculating the flow rate of the coagulant and chlorine based on the target plant flow rate are shown. The Q_Plant and concentrations of PACL and Cl can be set by the engineer and is set to 3 L/s in this sample calculation. \n\nChlorine would be ideally done at the end of the filtration where flow recombines so that the operator would only have to administer chlorine at one point. However our drafts did not account for that and instead lack piping that unites the top and bottom 1 L/s plants. Only the 6 L/s draft reflects this optimal design for chlorination.",
"_____no_output_____"
]
],
[
[
"#using Q_plant as the target variable, sample values of what a plant conditions might be are included below\nTemperature = u.Quantity(20,u.degC)\nDesired_PACl_Concentration=3*u.mg/u.L\nDesired_Cl_Concentration=3*u.mg/u.L\nC_stock_PACl=70*u.gram/u.L\nC_stock_Cl=51.4*u.gram/u.L \nNuPaCl = cdc.viscosity_kinematic_pacl(C_stock_PACl,Temperature)\nRatioError = 0.1\nKMinor = 2\n\nQ_Plant= 3*u.L/u.s\n\ndef CDC(Q_Plant, DesiredCl_Concentration,C_stock_PACl):\n Q_CDC=(Q_Plant*Desired_PACl_Concentration/C_stock_PACl).to(u.mL/u.s)\n return (Q_CDC)\ndef Chlorine_Dosing(Q_Plant,Desired_Cl_Concentration,C_stock_Cl):\n Q_Chlorine=(Q_Plant*Desired_Cl_Concentration/C_stock_Cl).to(u.mL/u.s)\n return (Q_Chlorine)\n\nprint('The flow rate of coagulant is ',CDC(Q_Plant, Desired_PACl_Concentration, C_stock_PACl).to(u.L/u.hour))\nprint('The flow rate of chlorine is ',Chlorine_Dosing(Q_Plant, Desired_Cl_Concentration, C_stock_Cl).to(u.L/u.hour))",
"The flow rate of coagulant is 0.4629 liter / hour\nThe flow rate of chlorine is 0.6304 liter / hour\n"
]
],
[
[
"### SPACE CONSTRAINTS \n\nIn the code below the team is calculating the floor plan area. The X distance and Y distance are the length and width of the floor plan respectively. The dimensions of the sedimentation tank, flocculator, and entrance tank are accounted for in this calculation. ",
"_____no_output_____"
]
],
[
[
"#Claculting the Y distance For the Sed Tank\n\n#properties of sedimentation tank \nSed_Tank_Diameter=0.965*u.m\nLength_Top_Half=1.546*u.m #See image for clearer understanding\n\nY_Sed_Top_Half=Length_Top_Half*math.cos(60*u.degrees)\nprint(Y_Sed_Top_Half)\nY_Sed_Total=Sed_Tank_Diameter+Y_Sed_Top_Half\nprint(Y_Sed_Total)",
"0.773 meter\n1.738 meter\n"
]
],
[
[
"SED TANK: Based on the calculation above, the space the Sedimentation tank would take on the floor plant is found to be 1.738 m. \n\n\n\nThis is a picture of the sedimentation tank with dimensions showing the distance jutting out from the sedimentation tank. This distance of 0.773 m is added to the sedimentation tank diameter totalling 1.738 meters.\n\nESTaRS: The dimensions of the ESTaRS are set and did not need to be calculated. A single manifold can collect water from the sed tanks and send it to the EStaRS. There will be valves on the manifold system installed before the entrance to the ESTaRS to allow for backwashing. These valves can be shut to allow for enough flow to provide backwashing capacity. There will be a manifold connecting flow after filtration to equate the flow for chlorination. \n\nFLOCCULATOR: We want symmetrical piping systems coming out of the flocculator. There is a flocculator for each plant so that available head going into the parallel sedimentation tanks will be the same. We will have an asymmetrical exit lauder system coming out of the sedimentation tanks going into the ESTaRS (diagram). \n\nENTRANCE TANK: The entrance tank is set to be at the front of the plant. The focus of this project is to calculate the dimensions and design the plant. The entrance tank dimensions should be left to be designed in the future. An estimated dimension was set in the drawing included in this project. There will be a grit chamber included after the entrance tank. The traditional design for rapid mix that is used in AguaClara plants will be included in the entrance tank. \n\nCONTACT CHAMBER: A contact chamber is included after the entrance tank to ensure that the coagulant is mixed with all of the water before it separates into the multiple treatment trains. Like the entrance tank, contact chamber dimensions should be left to be designed in the future. An estimated dimension was set in the drawing included in this project. \n\nWOODEN PLATFORM: The wooden platform is 4m long, 0.8m wide, and is 1.4 meters high allowing for the operator to be able to access the top of the sedimentation tank, flocculator, and ESTaRS. It would be placed in between every sedimentation tank. In the case of only a single sedimentation tank it would go on the right of the tank because the plant expands to the right. ",
"_____no_output_____"
]
],
[
[
"#Spacing due to Entrance Tank and contact chamber #estimated values\nSpace_ET=0.5*u.m\nCC_Diameter=0.4*u.m\nSpace_Between_ET_CC=0.1*u.m\nSpace_CC_ET=1*u.m\n\n#Spacing due to Manifold between contact chamber and flocculators\nSpace_CC_Floc=1.116*u.m\n\n#Spacing due to Flocculator \nSpace_Flocc_Sed=.1*u.m\nSpace_Flocculator=0.972*u.m\n\n#Spacing due to the Manifold\nSpace_Manifold=0.40*u.m\n\n#Spacing due to ESTaRS\nSpace_ESTaRS=0.607*u.m\n\n#Spacing for ESTaRS Manifold to the wall\nSpace_ESTaRS_Wall=0.962*u.m\n",
"_____no_output_____"
]
],
[
[
"The Y distance below 3 L/s is set to be as a sum of the total Y distance of the flocculator, sedimentation tank, and ESTaRS. An additional 2 meters of Y distance is added for operator access around the plant. The lengths between the sedimentation tank, flocculator and ESTarS were kept minimal but additional Y distance can be taken off between the sedimentation tank and ESTaRS. This is because the ESTaRS can hide under the sloping half of the sedimentation tank but this orientation would not account for the manifold drawn in the picture.",
"_____no_output_____"
],
[
"The total Y distance is calculated below.",
"_____no_output_____"
]
],
[
[
"Y_Length_Top=(Space_CC_ET+Space_CC_Floc+Space_Flocc_Sed\n +Space_Flocculator+Y_Sed_Total+Space_Manifold+Space_ESTaRS+\n Space_ESTaRS_Wall)\n\nY_Length_Bottom=Y_Length_Top-0.488*u.m",
"_____no_output_____"
]
],
[
[
"Below are functions that can be used to design a plant based on the desired flow rate",
"_____no_output_____"
]
],
[
[
"def X(Q):\n if Q>3*u.L/u.s:\n X_Distance_Bottom=X(Q-3*u.L/u.s)\n X_Distance_Top=6.9*u.m\n return(X_Distance_Top,X_Distance_Bottom)\n \n else:\n Q_Plant=Q.to(u.L/u.s)\n Extra_Space=2*u.m\n X_Distance=(Q_Plant.magnitude-1)*1*u.m+(Q_Plant.magnitude)*.965*u.m+Extra_Space \n return(X_Distance.to(u.m))\n\ndef Y(Q):\n if Q>3*u.L/u.s:\n return(Y_Length_Top+Y_Length_Bottom)\n else:\n return(Y_Length_Top)\n\nprint(X(Q_Plant_2).to(u.m))\ndef Area_Plant(Q):\n if Q>3*u.L/u.s:\n X_Distance_Bottom=X(Q-3*u.L/u.s)\n Area_Bottom=X_Distance_Bottom*Y_Length_Bottom*((Q/u.L*u.s-3))\n Area_Top=X(3*u.L/u.s)*Y_Length_Top\n Area_Total=Area_Top+Area_Bottom\n return(Area_Total)\n else:\n H_Distance=X(Q_Plant)\n Y_Distance=Y_Length_Top\n Area_Total=H_Distance*Y_Distance\n return(Area.to(u.m**2))\n",
"2.965 meter\n"
]
],
[
[
"\n\n\nThis is a layout of a sample plant for three 1 L/s tanks running in parallel. Check bottom of document for additional drafts. \n\nA platform will be in between sedimentation tanks as a way for the operators to have access to the sedimentation tanks. The platform height will be 1.4m to allow the plant operators 0.5m to view the sedimentation tanks just as in the built in place plants. The operator access requirements influence the optimal plant layout because the operators movements and safety have to be considered. The manifold will be built underneath the platform for space efficiency. The image below shows the wooden platform with 4m of length but could be truncated or extended to depending on the situation. The last image shows the platform in between the sedimentation tanks in the 3 L/s sample plant.\n\n\n\nWooden platforms would fill up the space in between sedimentation tanks to allow for operator access to the top of sedimentation tanks, ESTaRS and flocculators.\n\n\n",
"_____no_output_____"
],
[
"### Adding Capacity\nWhen adding capacity up to plant flow rates of 3 L/s, vertical distance will be constant so adding capacity will only change the horizontal distance of the plant. We define a set of flocculator, sedimentation tank, and EStARS as a 1 L/s plant. \n\nAfter the capacity of the plant reaches 3 L/s, additional 1 L/s plants will be added to the bottom half of the building, using a mirrored layout as the top half of the building. The only difference between the spacing is that the additions no longer need another entrance tank so the width of the bottom half of the plant is shorter than the width of the top half. This was done instead of simply increasing the length of the plant each time capacity was added because the length of the pipe between the contact chamber and the farthest flocculator would become increasingly large. This addition of major losses would cause different flow rates between the farther 1LPS plant and the closest one. \n\nThe following function will only account for adding 1 L/s plants one at at a time\n",
"_____no_output_____"
]
],
[
[
"def Additional_Area(Q_Plant,Q_Extra):\n if (Q_Plant+Q_Extra>3*u.L/u.s): \n X_Distance_Extra=X(Q_Extra)\n Y_Distance_Extra=Y_Length_Bottom\n Area=(X_Distance_Extra*Y_Distance_Extra).to(u.m**2)\n return(Area)\n else:\n Q=Q_Extra.to(u.L/u.s)\n Horizontal=(Q_Extra.magnitude)*.965*u.m+(Q_Extra.magnitude)*1*u.m\n Vertical=5.512*u.m\n Extra_Area=Vertical*Horizontal\n print('Extra length that has to be added is '+ut.sig(Horizontal,4))\n return(ut.sig(Extra_Area,2))",
"_____no_output_____"
]
],
[
[
"### ESTaRS\nThe total surface area required for the ESTaRS filter is simply a product of the area one ESTaRS requires and the flow rate coming into the plant. The surface area required for an ESTaRS was measured in the DeFrees lab. It is approximately a square meter.",
"_____no_output_____"
]
],
[
[
"ESTaRS_SA=1*u.m**2/u.L\ndef Area_ESTaRS(Q):\n Q_Plant=Q.to(u.L/u.s)\n Surface_Area_ESTaRS=(ESTaRS_SA.magnitude)*Q_Plant.magnitude\n return(Surface_Area_ESTaRS*u.m**2)\n\nprint('Surface area required for the ESTaR(s) is',Area_ESTaRS(Q_Plant))",
"_____no_output_____"
]
],
[
[
"### Flocculators\nThe total surface area required for the ESTaRS filter is simply a product of the area one ESTaRS requires and the flow rate coming into the plant. The surface area required for an ESTaRS was measured in the DeFrees lab. It is approximately a square meter.",
"_____no_output_____"
]
],
[
[
"Flocc_SA=0.972*u.m*0.536*u.m*u.L/u.s\ndef Area_Flocc(Q):\n Q_Plant=Q.to(u.L/u.s)\n Surface_Area_Flocc=(Flocc_SA.magnitude)*Q_Plant.magnitude\n return(Surface_Area_Flocc*u.m**2)\nprint('Surface area required for the flocculator(s) is',Area_Flocc(Q_Plant))\n ",
"_____no_output_____"
]
],
[
[
"### Manifold Headloss Calculations",
"_____no_output_____"
],
[
"The amount of headloss has to be minimal so that the amount of head availible coming out of the exit launder is enough to drive water fast enough to fluidize the sand bed in ESTaRS. This fluidization is required for backwashing the filter. Since the calculation for 4 inch pipe has a headloss for less than 1mm we conclude that the manifold is economically feasible. Any futher increase in the diameter of the manifold would become increasingly expensive. \n",
"_____no_output_____"
]
],
[
[
"SDR = 26\nSF=1.33\nQ_Manifold = 1 * u.L/u.s #max flowrate for a manifold \nQ_UpperBound=SF*Q_Manifold\nL_pipe = 5*u.m #Length sed tank to it’s ESTaRS\nK_minor_bend=0.3\nK_minor_branch=1\nK_minor_contractions=1\nK_minor_total= 2*(K_minor_bend+K_minor_branch+K_minor_contractions) # (two bends, two dividing branch, and two contractions)\n# The maximum viscosity will occur at the lowest temperature.\nT_crit = u.Quantity(10,u.degC)\nnu = pc.viscosity_kinematic(T_crit)\ne = 0.1 * u.mm\nManifold_1LPS_ID=4*u.inch\nHeadloss_Max=10*u.mm\n\nManifold_Diam=pc.diam_pipe(Q_Manifold,Headloss_Max,L_pipe,nu,e,K_minor_total).to(u.inch)\nprint(Manifold_Diam.to(u.inch))\nprint('The minimum pipe inner diameter is '+ ut.sig(Manifold_Diam,2)+'.')\nManifold_ND = pipe.ND_SDR_available(Manifold_Diam,SDR)\nprint('The nominal diameter of the manifold is '+ut.sig(Manifold_ND,2)+' ('+ut.sig(Manifold_ND.to(u.inch),2)+').')\n\nHLCheck = pc.headloss(Q_UpperBound,Manifold_1LPS_ID,L_pipe,nu,e,K_minor_total)\nprint('The head loss is',HLCheck.to(u.mm))",
"_____no_output_____"
]
],
[
[
"# Drafts of 1 L/s Plant to 6 L/s Plant\n\n\n\nA sample 1 L/s plant. The red t's indicate where the piping system can be expanded to include more 1 L/s systems. Additionally the t would be covered by a fernco cap so that the pipe can be removed when the plant is adding capacity. See cell below for pictures of manifolds with caps. \n\n\n\nThe sample 2 L/s plant. The red t's indicate where the piping system can be expanded to include more 1 L/s plants. Additionally the t exit that isn't connected to any piping would be covered by a fernco cap so that the pipe can be removed when the plant is adding capacity. \n\n\n\nThe sample 3 L/s plant. There are now no more t's that can be extended because the manifold was designed for up to 3 1 L/s plants. Further capacities are added on the bottom half of the plant like in the next 3 pictures.\n\n\n\nThe sample 4 L/s plant. Like in the 1 L/s plant the red t's indicate the start of the 2nd manifold system.\n\n\n\nThe sample 5 L/s plant. The length (x-direction) is extended by the diameter of a sedimentation tank and one meter like in the top half of the plant. However the width of the bottom half is slightly lower because it can use the already built entrance tank used for the first 3 L/s systems. \n\n\n\nThe sample 6 L/s plant. The building is now at full capacity with an area of 91.74 m^2. The flow reunites to allow for chlorination to be administered at one point.\n\n\n",
"_____no_output_____"
],
[
"# Manifold Drafts\n\n\n\nThe manifold with 1 L/s. The elbow at the top is connected to the exit launder of the sedimentation tank. The flow goes from the exit launder into the reducer which is not yet connected in the draft to the ESTaRS. The removeable fernco cap that allows for further expansions of the manifold pipe system can be seen in the picture.\n\n\n\nThe manifold with 2 L/s. Again a fernco cap is used to allow for future expansions up to 3 L/s.\n\n\n\n\nThe manifold with 3 L/s. Here there are no fernco caps because the manifold is designed for up to connections with three 1 L/s plants. \n",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d0e4e1035f7c792c43fea4da0ce3ca8a2fa6ae77 | 984,324 | ipynb | Jupyter Notebook | notebooks/.ipynb_checkpoints/Profiles and classification-checkpoint.ipynb | aasensio/SolarnetGranada | fe3e437198441fd61b2205ac5bbc4d37729d6bf0 | [
"MIT"
] | 1 | 2016-05-21T18:25:03.000Z | 2016-05-21T18:25:03.000Z | notebooks/Profiles and classification.ipynb | aasensio/SolarnetGranada | fe3e437198441fd61b2205ac5bbc4d37729d6bf0 | [
"MIT"
] | null | null | null | notebooks/Profiles and classification.ipynb | aasensio/SolarnetGranada | fe3e437198441fd61b2205ac5bbc4d37729d6bf0 | [
"MIT"
] | null | null | null | 2,349.221957 | 325,464 | 0.947123 | [
[
[
"import numpy as np\nimport matplotlib.pyplot as pl\nimport seaborn as sn\nfrom sklearn.cluster import MiniBatchKMeans\nfrom sklearn import metrics\n%matplotlib inline",
"_____no_output_____"
],
[
"stI=np.load('../data/stokesI_sunspot.npy')\nstV=np.load('../data/stokesV_sunspot.npy')\nwave = np.loadtxt('../data/wavelengthHinode.txt')\nstI.shape",
"_____no_output_____"
]
],
[
[
"### Index",
"_____no_output_____"
],
[
"[Contrast and velocity fields](#contrast)\n\n[Classification](#classification)",
"_____no_output_____"
],
[
"### Contrast and velocity fields",
"_____no_output_____"
],
[
"<a id='contrast'></a>",
"_____no_output_____"
]
],
[
[
"sn.set_style(\"dark\")\nf, ax = pl.subplots(figsize=(9,9))\nax.imshow(stI[:,:,0], aspect='auto', cmap=pl.cm.gray)",
"_____no_output_____"
]
],
[
[
"Let us compute simple things like the contrast and the Doppler velocity field",
"_____no_output_____"
]
],
[
[
"contrastFull = np.std(stI[:,:,0]) / np.mean(stI[:,:,0])\ncontrastQuiet = np.std(stI[400:,100:300,0]) / np.mean(stI[400:,100:300,0])\nprint(\"Contrast in the image : {0}%\".format(contrastFull * 100.0))\nprint(\"Contrast in the quiet Sun : {0}%\".format(contrastQuiet * 100.0))",
"Contrast in the image : 24.5026469231%\nContrast in the quiet Sun : 6.59198462963%\n"
]
],
[
[
"Now let us compute the velocity field. To this end, we compute the location of the core of the line in velocity units for each pixel.",
"_____no_output_____"
]
],
[
[
"v = np.zeros((512,512))\nfor i in range(512):\n for j in range(512):\n pos = np.argmin(stI[i,j,20:40]) + 20\n res = np.polyfit(wave[pos-2:pos+2], stI[i,j,pos-2:pos+2], 2) \n w = -res[1] / (2.0 * res[0])\n v[i,j] = (w-6301.5) / 6301.5 * 3e5",
"_____no_output_____"
],
[
"f, ax = pl.subplots(figsize=(9,9))\nax.imshow(np.clip(v,-5,5))\nf.savefig('velocities.png')",
"_____no_output_____"
],
[
"f, ax = pl.subplots(nrows=1, ncols=2, figsize=(15,9))\nax[0].imshow(stI[:,0,:], aspect='auto', cmap=pl.cm.gray)\nax[1].imshow(stV[:,0,:], aspect='auto', cmap=pl.cm.gray)",
"_____no_output_____"
],
[
"f.savefig('exampleStokes.png')",
"_____no_output_____"
]
],
[
[
"### Classification",
"_____no_output_____"
],
[
"<a id='classification'></a>",
"_____no_output_____"
]
],
[
[
"X = stV[50:300,200:450,:].reshape((250*250,112))",
"_____no_output_____"
],
[
"maxV = np.max(np.abs(X), axis=1)\nX = X / maxV[:,None]",
"_____no_output_____"
],
[
"nClusters = 9\nkm = MiniBatchKMeans(init='k-means++', n_clusters=nClusters, n_init=10, batch_size=500)",
"_____no_output_____"
],
[
"km.fit(X)",
"_____no_output_____"
],
[
"out = km.predict(X)",
"_____no_output_____"
],
[
"avg = np.zeros((nClusters,112))\nfor i in range(nClusters):\n avg[i,:] = np.mean(X[out==i,:], axis=0)",
"_____no_output_____"
],
[
"f, ax = pl.subplots(ncols=3, nrows=3, figsize=(12,9))\nloop = 0\nfor i in range(3):\n for j in range(3):\n percentage = X[out==i,:].shape[0] / (250*250.) * 100.0 \n ax[i,j].plot(km.cluster_centers_[loop,:])\n ax[i,j].set_title('Class {0} - {1}%'.format(loop, percentage))\n loop += 1\npl.tight_layout()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e4e47d24a3bb641e6f859c7e2aecde0360adbd | 7,957 | ipynb | Jupyter Notebook | src/old_files/Untitled2.ipynb | Data-is-life/lots-of-data | 932e605ca2b880c7aa9e23e3ea0861c802d00b80 | [
"Unlicense"
] | null | null | null | src/old_files/Untitled2.ipynb | Data-is-life/lots-of-data | 932e605ca2b880c7aa9e23e3ea0861c802d00b80 | [
"Unlicense"
] | null | null | null | src/old_files/Untitled2.ipynb | Data-is-life/lots-of-data | 932e605ca2b880c7aa9e23e3ea0861c802d00b80 | [
"Unlicense"
] | null | null | null | 27.25 | 113 | 0.47078 | [
[
[
"from pandas import read_csv\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import classification_report, confusion_matrix, roc_auc_score\nfrom sklearn.preprocessing import MaxAbsScaler\nfrom sklearn.feature_selection import SelectPercentile, f_classif\nimport statsmodels.api as sm\nfrom scipy import stats",
"_____no_output_____"
],
[
"df = read_csv('../data/use_for_predictions.csv')\ndf = df[df['result'] != 0.5]",
"_____no_output_____"
],
[
"y = df['result'].values\nX = df.drop(columns=['result', 'day', 'day_game_num', 'weekday', 'elo', 'start_time']).values",
"_____no_output_____"
],
[
"X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.05, random_state=5, shuffle=False)\nmmx = MaxAbsScaler().fit(X_train)\nX_train = mmx.transform(X_train)\nX_test = mmx.transform(X_test)",
"_____no_output_____"
],
[
"model = RandomForestClassifier(n_estimators=500, max_depth=10,\n n_jobs=-1)\nmodel.fit(X_train, y_train)\ny_pred = model.predict(X_test)",
"_____no_output_____"
],
[
"print(classification_report(y_test, y_pred))\nprint(confusion_matrix(y_test, y_pred))",
" precision recall f1-score support\n\n 0.0 0.64 0.89 0.74 63\n 1.0 0.82 0.49 0.61 63\n\n micro avg 0.69 0.69 0.69 126\n macro avg 0.73 0.69 0.68 126\nweighted avg 0.73 0.69 0.68 126\n\n[[56 7]\n [32 31]]\n"
],
[
"y_pred = model.predict_proba(X_test)\nprint(round(roc_auc_score(y_test, y_pred[:, 1])*100, 2))",
"73.38\n"
],
[
"model.feature_importances_",
"_____no_output_____"
],
[
"est = sm.Logit(y, X)\nest2 = est.fit(maxiter=3500)\nprint(est2.summary())",
"Optimization terminated successfully.\n Current function value: 0.582180\n Iterations 7\n Logit Regression Results \n==============================================================================\nDep. Variable: y No. Observations: 2514\nModel: Logit Df Residuals: 2510\nMethod: MLE Df Model: 3\nDate: Wed, 10 Apr 2019 Pseudo R-squ.: 0.1601\nTime: 17:57:18 Log-Likelihood: -1463.6\nconverged: True LL-Null: -1742.5\n LLR p-value: 1.389e-120\n==============================================================================\n coef std err z P>|z| [0.025 0.975]\n------------------------------------------------------------------------------\nx1 0.0187 0.001 17.041 0.000 0.017 0.021\nx2 -0.0006 0.000 -2.999 0.003 -0.001 -0.000\nx3 0.0008 0.000 2.377 0.017 0.000 0.001\nx4 0.3318 0.089 3.737 0.000 0.158 0.506\n==============================================================================\n"
],
[
"fs = SelectPercentile(f_classif, 10).fit(X, y)\nprint(fs.pvalues_)\nprint(fs.scores_)",
"[1.26919956e-68 2.43730194e-41 3.86856974e-06 6.62896057e-03]\n[326.04166139 188.11362772 21.42397691 7.38314543]\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e4e5a6376965f3b0d88a01fc7e11b816bf8218 | 3,106 | ipynb | Jupyter Notebook | notebook/1basic_inline_plots.ipynb | ma-laforge/CMDimData.jl | f543265841b81dc20f5dca18a37cd642371a0a34 | [
"MIT"
] | null | null | null | notebook/1basic_inline_plots.ipynb | ma-laforge/CMDimData.jl | f543265841b81dc20f5dca18a37cd642371a0a34 | [
"MIT"
] | null | null | null | notebook/1basic_inline_plots.ipynb | ma-laforge/CMDimData.jl | f543265841b81dc20f5dca18a37cd642371a0a34 | [
"MIT"
] | null | null | null | 23.179104 | 81 | 0.551513 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d0e4eae6663f8360f34b531373b17edd48150433 | 24,400 | ipynb | Jupyter Notebook | DatabaseNormalization.ipynb | ganprad/jupyter_notebooks | 61dbcf9169ac2ab99980d22bbe9fbb03579466e0 | [
"MIT"
] | null | null | null | DatabaseNormalization.ipynb | ganprad/jupyter_notebooks | 61dbcf9169ac2ab99980d22bbe9fbb03579466e0 | [
"MIT"
] | null | null | null | DatabaseNormalization.ipynb | ganprad/jupyter_notebooks | 61dbcf9169ac2ab99980d22bbe9fbb03579466e0 | [
"MIT"
] | null | null | null | 24.4 | 881 | 0.504385 | [
[
[
"import sqlite3",
"_____no_output_____"
],
[
"conn = sqlite3.connect('nominations.db')",
"_____no_output_____"
],
[
"#Checking Schema\nschema = conn.execute(\"pragma table_info(nominations)\")\nschema.fetchall()",
"_____no_output_____"
],
[
"first_ten = conn.execute(\"select * from nominations limit 10\").fetchall()\n\nfor i in first_ten:\n print(i)",
"(1, 'Actor -- Leading Role', 'Javier Bardem', 'Biutiful ', \"'Uxbal'\", 0, 1)\n(2, 'Actor -- Leading Role', 'Jeff Bridges', 'True Grit ', \"'Rooster Cogburn'\", 0, 1)\n(3, 'Actor -- Leading Role', 'Jesse Eisenberg', 'The Social Network ', \"'Mark Zuckerberg'\", 0, 1)\n(4, 'Actor -- Leading Role', 'Colin Firth', \"The King's Speech \", \"'King George VI'\", 1, 1)\n(5, 'Actor -- Leading Role', 'James Franco', '127 Hours ', \"'Aron Ralston'\", 0, 1)\n(6, 'Actor -- Supporting Role', 'Christian Bale', 'The Fighter ', \"'Dicky Eklund'\", 1, 1)\n(7, 'Actor -- Supporting Role', 'John Hawkes', \"Winter's Bone \", \"'Teardrop'\", 0, 1)\n(8, 'Actor -- Supporting Role', 'Jeremy Renner', 'The Town ', \"'James Coughlin'\", 0, 1)\n(9, 'Actor -- Supporting Role', 'Mark Ruffalo', 'The Kids Are All Right ', \"'Paul'\", 0, 1)\n(10, 'Actor -- Supporting Role', 'Geoffrey Rush', \"The King's Speech \", \"'Lionel Logue'\", 0, 1)\n"
],
[
"#Creating a ceremonies table that contains the list of tuples connecting ceremonial hosts corresponding to year \n#List of tuples\nyears_hosts = [(2010, \"Steve Martin\"),\n (2009, \"Hugh Jackman\"),\n (2008, \"Jon Stewart\"),\n (2007, \"Ellen DeGeneres\"),\n (2006, \"Jon Stewart\"),\n (2005, \"Chris Rock\"),\n (2004, \"Billy Crystal\"),\n (2003, \"Steve Martin\"),\n (2002, \"Whoopi Goldberg\"),\n (2001, \"Steve Martin\"),\n (2000, \"Billy Crystal\"),\n ]",
"_____no_output_____"
],
[
"#Creating table\n\nquery = '''create table ceremonies(\nid integer primary key,\nYear integer,\nHost text\n);'''\n\nf = conn.execute(query)",
"_____no_output_____"
],
[
"#Inserting years_host list into ceremonies table\n\ninsert_query = ''' insert into ceremonies (Year,Host) values (?,?)'''\nconn.executemany(insert_query,years_hosts)\n",
"_____no_output_____"
],
[
"f = conn.execute(\"pragma table_info(ceremonies)\").fetchall()\nf",
"_____no_output_____"
],
[
"f = conn.execute('select * from ceremonies limit 10').fetchall()\nf",
"_____no_output_____"
],
[
"f",
"_____no_output_____"
]
],
[
[
"# Fixes\n\nFixing the schema when the column names are incorrect.\nRename the table(with errors) to tempName and then recreate the original tableName and then insert the values into newly created table.\n\n\nhttps://stackoverflow.com/questions/805363/how-do-i-rename-a-column-in-a-sqlite-database-table\n\n fix1 = '''alter table ceremonies rename to tmp_table'''\n conn.execute(fix1)\n\nRemoving the old tmp table:\n\n fix2 = \"drop table tmp_table\"\n conn.execute(fix2)",
"_____no_output_____"
],
[
"# Foreign Keys",
"_____no_output_____"
],
[
" PRAGMA foreign_keys = ON;\n\nThe above query needs to be run every time you connect to a database where you'll be inserting foreign keys.",
"_____no_output_____"
]
],
[
[
"conn.execute('pragma foreign_keys = on')",
"_____no_output_____"
],
[
"#Creating an append listfrom old table to insert into new table.\n\nquery = '''SELECT ceremonies.id, nominations.category, nominations.nominee, nominations.movie, \nnominations.charachter, nominations.won\nFROM nominations\nINNER JOIN ceremonies ON\nnominations.year == ceremonies.year\n;'''\n",
"_____no_output_____"
],
[
"conn.execute(\"pragma table_info(nominations)\").fetchall()",
"_____no_output_____"
],
[
"join_list = conn.execute(query).fetchall()\nfor i in join_list[:10]: \n print(i)",
"_____no_output_____"
],
[
"fix1 = '''alter table nominations rename to tmp_table2'''\nconn.execute(fix1)",
"_____no_output_____"
],
[
"#Creating the new table with desired schema\n\ncreate_table = '''create table nominations(\nid integer primary key,\ncategory text,\nnominee text,\nmovie text,\ncharachter text,\nWon integer,\nceremony_id integer,\nforeign key(ceremony_id) references ceremonies(id)\n);\n'''",
"_____no_output_____"
],
[
"conn.execute(create_table)",
"_____no_output_____"
],
[
"insert_query = 'insert into nominations (ceremony_id,category,nominee,movie,charachter,Won) values (?,?,?,?,?,?)'",
"_____no_output_____"
],
[
"conn.executemany(insert_query,join_list)",
"_____no_output_____"
],
[
"f = conn.execute('pragma table_info(nominations)').fetchall()\nf",
"_____no_output_____"
],
[
"f = conn.execute('select * from nominations limit 10').fetchall()",
"_____no_output_____"
],
[
"#Drop tmp_table\n\nfix1 = '''alter table actors rename to tmp_table'''\nconn.execute(fix1)",
"_____no_output_____"
],
[
"fix2 = \"drop table movies_actors\"\nconn.execute(fix2)",
"_____no_output_____"
]
],
[
[
"# Many to Many relations through a join table",
"_____no_output_____"
]
],
[
[
"#Creating the tables\n\nactors = '''create table actors(\nid integer primary key,\nactor text\n)'''\n\nconn.execute(actors)",
"_____no_output_____"
],
[
"movies = '''create table movies(\nid integer primary key,\nmovie text\n)\n'''\nconn.execute(movies)",
"_____no_output_____"
],
[
"movies_actors = '''create table movies_actors(\nid integer primary key,\nmovie_id integer references movies(id),\nactor_id integer references actors(id)\n);'''\n\nconn.execute(movies_actors)",
"_____no_output_____"
],
[
"actor_sql= \"select distinct nominations.nominee from nominations;\"\nactor_list = conn.execute(actor_sql).fetchall()\nactor_list[:5]",
"_____no_output_____"
],
[
"conn.executemany(\"insert into actors (actor) values (?);\",actor_list)",
"_____no_output_____"
],
[
"movie_sql = \"select distinct nominations.movie from nominations;\"\nmovie_list = conn.execute(movie_sql).fetchall()\nmovie_list[:5]",
"_____no_output_____"
],
[
"conn.executemany(\"insert into movies (movie) values(?)\",movie_list)",
"_____no_output_____"
],
[
"query = \"select * from movies\"\n\nf = conn.execute(query).fetchall()\nf[:5]",
"_____no_output_____"
],
[
"query = \"select * from actors\"\n\nf = conn.execute(query).fetchall()\nf[:5]",
"_____no_output_____"
],
[
"joint_query = '''select * from actors\ninner join nominations on actors.actor == nominations.nominee'''",
"_____no_output_____"
],
[
"f = conn.execute(joint_query)\nfor i in f.fetchall()[:10]:\n print(i)\nf.fetchall()",
"_____no_output_____"
],
[
"f = conn.execute(\"pragma table_info(movies_actors)\").fetchall()\nf",
"_____no_output_____"
],
[
"query = \"select * from movies_actors\"\n\nf = conn.execute(query).fetchall()\nf[:5]",
"_____no_output_____"
],
[
"query = '''select ceremonies.year, nominations.movie from nominations \ninner join ceremonies on \nnominations.ceremony_id == ceremonies.id \nwhere nominations.nominee == 'Natalie Portman';\n'''\n\nf = conn.execute(query)\n\nportman_movies = f.fetchall()\nprint(portman_movies)",
"[]\n"
],
[
"query = '''SELECT * FROM movies\nINNER JOIN movies_actors ON movies.id == movies_actors.movie_id'''\n\nf = conn.execute(query)\nf.fetchall()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e50327cb8096401e74c221f94d68416369db24 | 6,232 | ipynb | Jupyter Notebook | jypter_legacy/cnn_wavelet.ipynb | passive-radio/find-peaks | b08eb3dea51217130bef86d179b5c717e831f576 | [
"MIT"
] | null | null | null | jypter_legacy/cnn_wavelet.ipynb | passive-radio/find-peaks | b08eb3dea51217130bef86d179b5c717e831f576 | [
"MIT"
] | 2 | 2021-12-10T12:16:09.000Z | 2022-01-17T01:40:58.000Z | jypter_legacy/cnn_wavelet.ipynb | passive-radio/find-peaks | b08eb3dea51217130bef86d179b5c717e831f576 | [
"MIT"
] | null | null | null | 38.708075 | 146 | 0.543485 | [
[
[
"'''\r\n# IoU_old\r\nusage of loss func\r\n\r\ncriterion = nn.NLLLoss()\r\n\r\ndef step(x, y, is_train=True):\r\n x = x.reshape(-1, 28 * 28)\r\n y_pred = model(x)\r\n loss = criterion(y_pred, y)\r\n if is_train:\r\n opt.zero_grad()\r\n loss.backward()\r\n opt.step()\r\n return loss, y_pred\r\n\r\n'''\r\ntorch.nn.Module\r\nclass IoU_old(nn.Module):\r\n def __init__(self, thresh: float = 0.5):\r\n super().__init__()\r\n self.thresh = thresh\r\n\r\n def forward(self, inputs: torch.Tensor, targets:torch.Tensor, weights: Optional[torch.Tensor] = None, smooth: float = 0.0) -> Tensor:\r\n inputs = torch.where(inputs < self.thresh, 0, 1)\r\n batch_size = targets.shape[0]\r\n\r\n intersect = torch.logical_and(inputs, targets)\r\n intersect = intersect.view(batch_size, -1).sum(-1)\r\n union = torch.logical_or(inputs, targets)\r\n union = union.view(batch_size, -1).sum(-1)\r\n\r\n print(intersect)\r\n print(union)\r\n print(union.shape)\r\n\r\n IoU = (intersect + smooth) / (union + smooth)\r\n IoU = torch.nan_to_num(IoU)\r\n return IoU",
"_____no_output_____"
],
[
"def iou2(outputs: torch.Tensor, labels: torch.Tensor, smooth: float=0, threshold: float = 0.5):\r\n\r\n # intersection = (outputs & labels).sum((1, 2)) # Will be zero if Truth=0 or Prediction=0\r\n # union = (outputs | labels).sum((1, 2)) # Will be zzero if both are 0\r\n # iou = (intersection + SMOOTH) / (union + SMOOTH) # We smooth our devision to avoid 0/0\r\n # # thresholded = torch.clamp(20 * (iou - 0.5), 0, 10).ceil() / 10 # This is equal to comparing with thresolds\r\n\r\n # return iou\r\n outputs = torch.where(outputs < threshold, 0, 1)\r\n\r\n print(f\"num of labels: {len(torch.nonzero(labels))}\")\r\n intersect = torch.logical_and(outputs, labels)\r\n intersect = intersect.long()\r\n print(intersect)\r\n # intersect = torch.where(outputs == labels, 1, 0)\r\n # intersect = torch.where(outputs == intersect, 1, 0)\r\n print(f\"num of interesect: {len(torch.nonzero(intersect))}\")\r\n union = torch.logical_or(outputs, labels)\r\n\r\n print(f\"num of union: {len(torch.nonzero(union))}\")\r\n\r\n # print(f\"intersect sum: {intersect.sum()}\")\r\n # print(f\"union sum: {union.sum()}\")\r\n iou_score = torch.sum(intersect)/torch.sum(union)\r\n print(iou_score)\r\n # # You can comment out this line if you are passing tensors of equal shape\r\n # # But if you are passing output from UNet or something it will most probably\r\n # # be with the BATCH x 1 x H x W shape\r\n # # outputs = outputs.squeeze(1) # BATCH x 1 x H x W => BATCH x H x W\r\n \r\n # # intersection = (outputs & labels).sum((1, 2)) # Will be zero if Truth=0 or Prediction=0\r\n # # union = (outputs | labels).float().sum((1, 2)) # Will be zzero if both are 0\r\n \r\n iou = (intersect + smooth) / (union + smooth) # We smooth our devision to avoid 0/0\r\n iou_score = torch.sum(iou)\r\n print(iou_score)\r\n \r\n return iou_score # Or thresholded.mean() if you are interested in average across the batch",
"_____no_output_____"
],
[
"'''\r\n# IoU soft\r\nnote: \"hard\" probabilistic IoU calculation is not differentiable, \r\nso be sure to use soft probabilistic loss func\r\n'''\r\ndef iou_soft(inputs: torch.Tensor, labels: torch.Tensor, \r\n smooth: float=0.1, threshold: float = 0.5, alpha: float = 1.0):\r\n \r\n '''\r\n - alpha: a parameter that sharpen the thresholding.\r\n if alpha = 1 -> thresholded input is the same as raw input.\r\n '''\r\n\r\n thresholded_inputs = inputs**alpha / (inputs**alpha + (1 - inputs)**alpha)\r\n # inputs = torch.where(thresholded_inputs < threshold, 0, 1)\r\n\r\n thresholded_inputs = torch.where(thresholded_inputs < threshold, 0, 1)\r\n inputs = (inputs + thresholded_inputs) - inputs.detach()\r\n\r\n batch_size = inputs.shape[0]\r\n\r\n # instead of hard prob calc: intersect = torch.logical_and(outputs, labels)\r\n intersect_tensor = (inputs * labels).view(batch_size, -1)\r\n intersect = intersect_tensor.sum(-1)\r\n\r\n # insetad of using the hard prob: union = torch.logical_or(outputs, labels) \r\n union_tensor = torch.max(inputs, labels).view(batch_size, -1)\r\n union = union_tensor.sum(-1)\r\n\r\n\r\n iou = (intersect + smooth) / (union + smooth) # We smooth our devision to avoid 0/0\r\n iou_score = iou.mean()\r\n\r\n # print(f\"num of labels: {len(torch.nonzero(labels))}\")\r\n # print(f\"intersection tensor: {intersect_tensor}\")\r\n # print(f\"union tensor: {union_tensor}\\n\")\r\n # print(f\"intersect sum: {intersect}\")\r\n # print(f\"union sum: {union}\\n\")\r\n # print(f\"iou error: {1- iou_score}\")\r\n \r\n return 1- iou_score\r\n",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
d0e503e89bba3e72ba59104f1b004b5773cb5f1c | 22,651 | ipynb | Jupyter Notebook | elsa/elsa_sentence_train_pt.ipynb | hiropppe/ELSA | 1ece189564e3827a4a7a44cd5f87e0ae4fca5c88 | [
"MIT"
] | null | null | null | elsa/elsa_sentence_train_pt.ipynb | hiropppe/ELSA | 1ece189564e3827a4a7a44cd5f87e0ae4fca5c88 | [
"MIT"
] | null | null | null | elsa/elsa_sentence_train_pt.ipynb | hiropppe/ELSA | 1ece189564e3827a4a7a44cd5f87e0ae4fca5c88 | [
"MIT"
] | null | null | null | 46.132383 | 237 | 0.500464 | [
[
[
"import h5py\nimport keras\nimport numpy as np\nimport json\nimport os\nimport uuid\nimport yaml\n\nfrom attlayer import AttentionWeightedAverage\n#from avglayer import MaskAverage\nfrom copy import deepcopy\n#from finetuning import (sampling_generator, finetuning_callbacks)\nfrom operator import itemgetter\n#from global_variables import NB_TOKENS, NB_EMOJI_CLASSES\nfrom keras.layers import *\nfrom keras.layers.merge import concatenate\nfrom keras.layers import Input, Bidirectional, Embedding, Dense, Dropout, SpatialDropout1D, LSTM, Activation\nfrom keras.models import Model, Sequential\nfrom keras.optimizers import Adam\nfrom keras.regularizers import L1L2 \nfrom pathlib import Path\nfrom sklearn.metrics import classification_report, recall_score, precision_score, f1_score\nfrom os.path import exists",
"Using TensorFlow backend.\n"
],
[
"def elsa_architecture(nb_classes, nb_tokens, maxlen, feature_output=False, embed_dropout_rate=0, final_dropout_rate=0, embed_dim=300,\n embed_l2=1E-6, return_attention=False, load_embedding=False, pre_embedding=None, high=False, LSTM_hidden=512, LSTM_drop=0.5):\n \"\"\"\n Returns the DeepMoji architecture uninitialized and\n without using the pretrained model weights.\n # Arguments:\n nb_classes: Number of classes in the dataset.\n nb_tokens: Number of tokens in the dataset (i.e. vocabulary size).\n maxlen: Maximum length of a token.\n feature_output: If True the model returns the penultimate\n feature vector rather than Softmax probabilities\n (defaults to False).\n embed_dropout_rate: Dropout rate for the embedding layer.\n final_dropout_rate: Dropout rate for the final Softmax layer.\n embed_l2: L2 regularization for the embedding layerl.\n high: use or not the highway network\n # Returns:\n Model with the given parameters.\n \"\"\"\n class NonMasking(Layer): \n def __init__(self, **kwargs): \n self.supports_masking = True \n super(NonMasking, self).__init__(**kwargs) \n\n def build(self, input_shape): \n input_shape = input_shape \n\n def compute_mask(self, input, input_mask=None): \n # do not pass the mask to the next layers \n return None \n\n def call(self, x, mask=None): \n return x \n\n def get_output_shape_for(self, input_shape): \n return input_shape \n # define embedding layer that turns word tokens into vectors\n # an activation function is used to bound the values of the embedding\n model_input = Input(shape=(maxlen,), dtype='int32')\n embed_reg = L1L2(l2=embed_l2) if embed_l2 != 0 else None\n\n if not load_embedding and pre_embedding is None:\n embed = Embedding(input_dim=nb_tokens, output_dim=embed_dim, mask_zero=True,input_length=maxlen,embeddings_regularizer=embed_reg,\n name='embedding')\n else:\n embed = Embedding(input_dim=nb_tokens, output_dim=embed_dim, mask_zero=True,input_length=maxlen, weights=[pre_embedding],\n embeddings_regularizer=embed_reg,trainable=True, name='embedding')\n if high:\n x = NonMasking()(embed(model_input))\n else:\n x = embed(model_input)\n x = Activation('tanh')(x)\n\n # entire embedding channels are dropped out instead of the\n # normal Keras embedding dropout, which drops all channels for entire words\n # many of the datasets contain so few words that losing one or more words can alter the emotions completely\n if embed_dropout_rate != 0:\n embed_drop = SpatialDropout1D(embed_dropout_rate, name='embed_drop')\n x = embed_drop(x)\n\n # skip-connection from embedding to output eases gradient-flow and allows access to lower-level features\n # ordering of the way the merge is done is important for consistency with the pretrained model\n lstm_0_output = Bidirectional(LSTM(LSTM_hidden, return_sequences=True, dropout=LSTM_drop), name=\"bi_lstm_0\" )(x)\n lstm_1_output = Bidirectional(LSTM(LSTM_hidden, return_sequences=True, dropout=LSTM_drop), name=\"bi_lstm_1\" )(lstm_0_output)\n x = concatenate([lstm_1_output, lstm_0_output, x])\n if high:\n x = TimeDistributed(Highway(activation='tanh', name=\"high\"))(x)\n # if return_attention is True in AttentionWeightedAverage, an additional tensor\n # representing the weight at each timestep is returned\n weights = None\n x = AttentionWeightedAverage(name='attlayer', return_attention=return_attention)(x)\n #x = MaskAverage(name='attlayer', return_attention=return_attention)(x)\n if return_attention:\n x, weights = x\n\n if not feature_output:\n # output class probabilities\n if final_dropout_rate != 0:\n x = Dropout(final_dropout_rate)(x)\n\n if nb_classes > 2:\n outputs = [Dense(nb_classes, activation='softmax', name='softmax')(x)]\n else:\n outputs = [Dense(1, activation='sigmoid', name='softmax')(x)]\n else:\n # output penultimate feature vector\n outputs = [x]\n\n if return_attention:\n # add the attention weights to the outputs if required\n outputs.append(weights)\n\n return Model(inputs=[model_input], outputs=outputs)",
"_____no_output_____"
],
[
"os.environ['CUDA_VISIBLE_DEVICES'] = \"2\"\ncur_lan = \"elsa_pt\"\nmaxlen = 20\nbatch_size = 250\nlr = 3e-4\nepoch_size = 25000\nnb_epochs = 1000\npatience = 5\ncheckpoint_weight_path = \"./ckpt\"\nloss = \"categorical_crossentropy\"\noptim = \"adam\"\nvocab_path = \"/data/elsa\"\nnb_classes=64",
"_____no_output_____"
],
[
"LSTM_hidden = 512\nLSTM_drop = 0.5\nfinal_dropout_rate = 0.5\nembed_dropout_rate = 0.0\nhigh = False\nload_embedding = True\nembed_dim = 200",
"_____no_output_____"
],
[
"steps = int(epoch_size/batch_size)\n\nwv_path = Path(vocab_path).joinpath(\"{:s}_wv.npy\".format(cur_lan)).as_posix()\nX_path = Path(vocab_path).joinpath(\"{:s}_X.npy\".format(cur_lan)).as_posix()\ny_path = Path(vocab_path).joinpath(\"{:s}_y.npy\".format(cur_lan)).as_posix()\n\nword_vec = np.load(wv_path, allow_pickle=True)\ninput_vec, input_label = np.load(X_path, allow_pickle=True), np.load(y_path, allow_pickle=True)\nnb_tokens, input_len = len(word_vec), len(input_label)\n\ntrain_end = int(input_len*0.7)\nval_end = int(input_len*0.9)\n\n(X_train, y_train) = (input_vec[:train_end], input_label[:train_end])\n(X_val, y_val) = (input_vec[train_end:val_end], input_label[train_end:val_end])\n(X_test, y_test) = (input_vec[val_end:], input_label[val_end:])",
"_____no_output_____"
],
[
"input_vec.shape",
"_____no_output_____"
],
[
"model = elsa_architecture(nb_classes=nb_classes, nb_tokens=nb_tokens, maxlen=maxlen, final_dropout_rate=final_dropout_rate, embed_dropout_rate=embed_dropout_rate, \n load_embedding=True, pre_embedding=word_vec, high=high, embed_dim=embed_dim)\nmodel.summary()",
"WARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow/python/framework/op_def_library.py:263: colocate_with (from tensorflow.python.framework.ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nColocations handled automatically by placer.\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:3445: calling dropout (from tensorflow.python.ops.nn_ops) with keep_prob is deprecated and will be removed in a future version.\nInstructions for updating:\nPlease use `rate` instead of `keep_prob`. Rate should be set to `rate = 1 - keep_prob`.\n__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_1 (InputLayer) (None, 20) 0 \n__________________________________________________________________________________________________\nembedding (Embedding) (None, 20, 200) 14168400 input_1[0][0] \n__________________________________________________________________________________________________\nactivation_1 (Activation) (None, 20, 200) 0 embedding[0][0] \n__________________________________________________________________________________________________\nbi_lstm_0 (Bidirectional) (None, 20, 1024) 2920448 activation_1[0][0] \n__________________________________________________________________________________________________\nbi_lstm_1 (Bidirectional) (None, 20, 1024) 6295552 bi_lstm_0[0][0] \n__________________________________________________________________________________________________\nconcatenate_1 (Concatenate) (None, 20, 2248) 0 bi_lstm_1[0][0] \n bi_lstm_0[0][0] \n activation_1[0][0] \n__________________________________________________________________________________________________\nattlayer (AttentionWeightedAver (None, 2248) 2248 concatenate_1[0][0] \n__________________________________________________________________________________________________\ndropout_1 (Dropout) (None, 2248) 0 attlayer[0][0] \n__________________________________________________________________________________________________\nsoftmax (Dense) (None, 64) 143936 dropout_1[0][0] \n==================================================================================================\nTotal params: 23,530,584\nTrainable params: 23,530,584\nNon-trainable params: 0\n__________________________________________________________________________________________________\n"
],
[
"if optim == 'adam':\n adam = Adam(clipnorm=1, lr=lr)\n model.compile(loss=loss, optimizer=adam, metrics=['accuracy'])\nelif optim == 'rmsprop':\n model.compile(loss=loss, optimizer='rmsprop', metrics=['accuracy'])\n\nmodel.fit(X_train,\n y_train,\n batch_size=batch_size,\n epochs=nb_epochs,\n validation_data=(X_val, y_val),\n callbacks=[keras.callbacks.EarlyStopping(monitor='val_loss', min_delta=0, patience=patience, verbose=0, mode='auto')],\n verbose=True)",
"WARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow/python/ops/math_ops.py:3066: to_int32 (from tensorflow.python.ops.math_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse tf.cast instead.\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow/python/ops/math_grad.py:102: div (from tensorflow.python.ops.math_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nDeprecated in favor of operator or tf.math.divide.\nTrain on 165309 samples, validate on 47232 samples\nEpoch 1/1000\n165309/165309 [==============================] - 154s 932us/step - loss: 3.0842 - acc: 0.2431 - val_loss: 2.6527 - val_acc: 0.3230\nEpoch 2/1000\n165309/165309 [==============================] - 136s 822us/step - loss: 2.5657 - acc: 0.3475 - val_loss: 2.4475 - val_acc: 0.3737\nEpoch 3/1000\n165309/165309 [==============================] - 135s 815us/step - loss: 2.3612 - acc: 0.3938 - val_loss: 2.3536 - val_acc: 0.4006\nEpoch 4/1000\n165309/165309 [==============================] - 136s 822us/step - loss: 2.2149 - acc: 0.4283 - val_loss: 2.2977 - val_acc: 0.4182\nEpoch 5/1000\n165309/165309 [==============================] - 132s 796us/step - loss: 2.0970 - acc: 0.4590 - val_loss: 2.2757 - val_acc: 0.4263\nEpoch 6/1000\n165309/165309 [==============================] - 134s 812us/step - loss: 1.9884 - acc: 0.4846 - val_loss: 2.2750 - val_acc: 0.4301\nEpoch 7/1000\n165309/165309 [==============================] - 135s 817us/step - loss: 1.8876 - acc: 0.5069 - val_loss: 2.2865 - val_acc: 0.4350\nEpoch 8/1000\n165309/165309 [==============================] - 134s 811us/step - loss: 1.7921 - acc: 0.5322 - val_loss: 2.3092 - val_acc: 0.4357\nEpoch 9/1000\n165309/165309 [==============================] - 140s 848us/step - loss: 1.7023 - acc: 0.5538 - val_loss: 2.3461 - val_acc: 0.4354\nEpoch 10/1000\n165309/165309 [==============================] - 142s 862us/step - loss: 1.6201 - acc: 0.5742 - val_loss: 2.3867 - val_acc: 0.4372\nEpoch 11/1000\n165309/165309 [==============================] - 135s 815us/step - loss: 1.5446 - acc: 0.5932 - val_loss: 2.4570 - val_acc: 0.4364\n"
],
[
"_, acc = model.evaluate(X_test, y_test, batch_size=batch_size, verbose=0)\n\nprint(acc)",
"0.43381605716043614\n"
],
[
"token2index = json.loads(open(\"/data/elsa/elsa_pt_vocab.txt\", \"r\").read())\n\nfreq = {line.split()[0]: int(line.split()[1]) for line in open(\"/data/elsa/elsa_pt_emoji.txt\").readlines()}\nfreq_topn = sorted(freq.items(), key=itemgetter(1), reverse=True)[:nb_classes]",
"_____no_output_____"
],
[
"y_pred = model.predict(X_test)\nprint(classification_report(y_test.argmax(axis=1), y_pred.argmax(axis=1), target_names=[e[0] for e in freq_topn]))",
" precision recall f1-score support\n\n ❤ 0.36 0.60 0.45 2777\n 😂 0.36 0.52 0.42 1967\n 😍 0.32 0.39 0.35 1791\n 💜 0.76 0.74 0.75 1023\n 😭 0.41 0.34 0.37 948\n 🤣 0.35 0.27 0.31 624\n 🙏 0.46 0.58 0.51 818\n 💕 0.38 0.14 0.21 312\n 👏 0.51 0.51 0.51 379\n 💙 0.65 0.49 0.56 347\n 🥰 0.38 0.18 0.25 580\n ♀ 0.73 0.73 0.73 127\n 🤦 0.66 0.69 0.67 698\n 💛 0.56 0.35 0.43 177\n 💚 0.60 0.50 0.55 211\n 🥺 0.78 0.70 0.74 746\n 😔 0.28 0.30 0.29 562\n 🔥 0.52 0.47 0.50 437\n 💖 0.46 0.13 0.20 279\n 🎶 0.40 0.54 0.46 532\n 💗 0.37 0.09 0.14 116\n ♂ 0.58 0.69 0.63 86\n 👍 0.44 0.43 0.44 421\n 🙌 0.42 0.23 0.30 361\n 🤔 0.38 0.48 0.43 435\n 🤪 0.38 0.28 0.32 378\n ♥ 0.47 0.13 0.20 326\n 🤤 0.38 0.35 0.36 333\n 🙄 0.20 0.16 0.18 358\n 😅 0.39 0.22 0.28 329\n 🤷 0.58 0.46 0.51 297\n 😋 0.34 0.23 0.27 269\n 😘 0.62 0.32 0.42 231\n 💔 0.35 0.23 0.28 279\n ✨ 0.61 0.50 0.55 234\n 😈 0.50 0.49 0.49 220\n 🤩 0.27 0.11 0.15 228\n 🌕 1.00 1.00 1.00 68\n 🌒 0.00 0.00 0.00 2\n 🌗 1.00 0.50 0.67 2\n 🌖 1.00 1.00 1.00 1\n 😎 0.42 0.28 0.33 247\n 😡 0.33 0.31 0.32 213\n 🤧 0.33 0.14 0.19 197\n 💓 1.00 0.04 0.07 56\n 🌘 0.00 0.00 0.00 1\n 🌑 1.00 0.60 0.75 5\n 😱 0.55 0.44 0.49 162\n 🌓 1.00 0.33 0.50 3\n 🌔 1.00 1.00 1.00 1\n 💦 0.72 0.70 0.71 73\n 😉 0.41 0.34 0.37 255\n ⚠ 0.80 0.77 0.79 159\n 😪 0.39 0.14 0.20 212\n 🔴 0.88 0.80 0.84 182\n 😏 0.41 0.14 0.21 207\n 😴 0.27 0.30 0.29 228\n 💪 0.44 0.34 0.38 169\n 😢 0.38 0.19 0.25 190\n 😩 0.25 0.13 0.17 178\n 💞 0.65 0.28 0.40 113\n ✌ 0.42 0.26 0.32 168\n 🤭 0.60 0.23 0.33 157\n ⚽ 0.60 0.47 0.52 131\n\n accuracy 0.43 23616\n macro avg 0.52 0.40 0.43 23616\nweighted avg 0.45 0.43 0.42 23616\n\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0e5055b5ccddc87751517936cbea9671fa23e08 | 116,330 | ipynb | Jupyter Notebook | code/main.ipynb | hadinej/DL-Domain-adaptation-on-PACS-dataset | 43866369e64ab62343b6f78c16a9b8a000b7cec0 | [
"MIT"
] | null | null | null | code/main.ipynb | hadinej/DL-Domain-adaptation-on-PACS-dataset | 43866369e64ab62343b6f78c16a9b8a000b7cec0 | [
"MIT"
] | null | null | null | code/main.ipynb | hadinej/DL-Domain-adaptation-on-PACS-dataset | 43866369e64ab62343b6f78c16a9b8a000b7cec0 | [
"MIT"
] | null | null | null | 113.492683 | 33,924 | 0.804204 | [
[
[
"**Import libraries**",
"_____no_output_____"
]
],
[
[
"import os\nimport logging\n\nimport torch\nimport torch.nn as nn\nimport torch.optim as optim\nfrom torch.utils.data import Subset, DataLoader\nfrom torch.backends import cudnn\n\nimport torch.utils.data\nimport torchvision\nfrom torchvision import transforms\nfrom torchvision.models import alexnet\nfrom torchvision.datasets import ImageFolder\nfrom torchvision import datasets\n\nfrom PIL import Image\nfrom tqdm import tqdm\n\nimport pandas as pd\nimport numpy as np\n\nfrom matplotlib import pyplot as plt\n%matplotlib inline\n\nimport pretrainedmodels\nimport pickle",
"_____no_output_____"
]
],
[
[
"**Definitions**",
"_____no_output_____"
]
],
[
[
"from torch.autograd import Function\n\n\nclass ReverseLayerF(Function):\n\n @staticmethod\n def forward(ctx, x, alpha):\n ctx.alpha = alpha\n\n return x.view_as(x)\n\n @staticmethod\n def backward(ctx, grad_output):\n output = grad_output.neg() * ctx.alpha\n\n return output, None",
"_____no_output_____"
]
],
[
[
"**Set Arguments**",
"_____no_output_____"
]
],
[
[
"device = torch.device('cuda' if torch.cuda.is_available() else 'cpu') # 'cuda' or 'cpu'\n \nDATA_DIR = 'PACS\\\\' # directory of dataset\n\nNUM_CLASSES = 7 \nNUM_DOMAINS = 2 \n\n\nimage_size = 224 # the size required by AlexNet \nBATCH_SIZE = 256 # Higher batch sizes allows for larger learning rates. An empirical heuristic suggests that, when changing\n # the batch size, learning rate should change by the same factor to have comparable results\n\nLR = 1e-3 # The initial Learning Rate\n\nNUM_EPOCHS = 35 # Total number of training epochs (iterations over dataset)",
"_____no_output_____"
]
],
[
[
"**Define Data Preprocessing**",
"_____no_output_____"
]
],
[
[
"# Define transforms for training phase \nsource_transform = transforms.Compose([\n transforms.ColorJitter(),\n transforms.RandomHorizontalFlip(),\n transforms.Resize((224, 224)), \n transforms.ToTensor(), \n transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225])]) # Normalizes tensor with mean and standard deviation of ImageNet\n# Define transforms for the evaluation phase\ntarget_transform = transforms.Compose([\n transforms.Resize((224, 224)),\n transforms.ToTensor(),\n transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225])]) ",
"_____no_output_____"
]
],
[
[
"**Prepare Dataset**",
"_____no_output_____"
]
],
[
[
"source_dataset = ImageFolder(DATA_DIR+'photo//', transform=source_transform)\ntarget_dataset = ImageFolder(DATA_DIR+'art_painting//', transform=target_transform)\nprint('source Dataset: {}'.format(len(source_dataset)))\nprint('target Dataset: {}'.format(len(target_dataset)))",
"source Dataset: 1670\ntarget Dataset: 2048\n"
]
],
[
[
"<!-- # xx=Caltech(DATA_DIR, split='train')\n# xx.__getitem__(36) -->",
"_____no_output_____"
],
[
"**Prepare Dataloaders**",
"_____no_output_____"
]
],
[
[
"# Dataloaders iterate over pytorch datasets and transparently provide useful functions (e.g. parallelization and shuffling)\nsource_dataloader = DataLoader(source_dataset, batch_size=BATCH_SIZE, shuffle=True)\ntarget_dataloader = DataLoader(target_dataset, batch_size=BATCH_SIZE, shuffle=True)",
"_____no_output_____"
]
],
[
[
"**Define Network**",
"_____no_output_____"
]
],
[
[
"import torch\nimport torch.nn as nn\nfrom torch.hub import load_state_dict_from_url\n\n__all__ = ['AlexNet', 'alexnet']\n\nmodel_urls = {\n 'alexnet': 'https://download.pytorch.org/models/alexnet-owt-4df8aa71.pth',\n}\n\nclass AlexNetDANN(nn.Module):\n\n def __init__(self, num_classes=1000, num_domains=2):\n super(AlexNetDANN, self).__init__()\n \n self.features = nn.Sequential(\n nn.Conv2d(3, 64, kernel_size=11, stride=4, padding=2),\n nn.ReLU(inplace=True),\n nn.MaxPool2d(kernel_size=3, stride=2),\n nn.Conv2d(64, 192, kernel_size=5, padding=2),\n nn.ReLU(inplace=True),\n nn.MaxPool2d(kernel_size=3, stride=2),\n nn.Conv2d(192, 384, kernel_size=3, padding=1),\n nn.ReLU(inplace=True),\n nn.Conv2d(384, 256, kernel_size=3, padding=1),\n nn.ReLU(inplace=True),\n nn.Conv2d(256, 256, kernel_size=3, padding=1),\n nn.ReLU(inplace=True),\n nn.MaxPool2d(kernel_size=3, stride=2),\n )\n \n self.class_classifier = nn.Sequential(\n nn.Dropout(),\n nn.Linear(256 * 6 * 6, 4096),\n nn.ReLU(inplace=True),\n nn.Dropout(),\n nn.Linear(4096, 4096),\n nn.ReLU(inplace=True),\n nn.Linear(4096, num_classes),\n )\n \n self.domain_classifier = nn.Sequential(\n nn.Dropout(),\n nn.Linear(256 * 6 * 6, 4096),\n nn.ReLU(inplace=True),\n nn.Dropout(),\n nn.Linear(4096, 4096),\n nn.ReLU(inplace=True),\n nn.Linear(4096, num_domains),\n )\n\n def forward(self, input_data, alpha):\n input_data = input_data.expand(input_data.data.shape[0], 3, 224, 224)\n feature = self.features(input_data)\n feature = feature.view(feature.size(0), -1)\n reverse_feature = ReverseLayerF.apply(feature, alpha)\n class_output = self.class_classifier(feature)\n domain_output = self.domain_classifier(reverse_feature)\n\n return class_output, domain_output\n\n\ndef alexnetdann(pretrained=False, progress=True, **kwargs):\n r\"\"\"AlexNet model architecture from the\n `\"One weird trick...\" <https://arxiv.org/abs/1404.5997>`_ paper.\n Args:\n pretrained (bool): If True, returns a model pre-trained on ImageNet\n progress (bool): If True, displays a progress bar of the download to stderr\n \"\"\"\n model = AlexNetDANN(**kwargs)\n if pretrained:\n state_dict = load_state_dict_from_url(model_urls['alexnet'],\n progress=progress)\n model.load_state_dict(state_dict, strict=False)\n r\"\"\"\n something of great importace:\n Whether you are loading from a partial state_dict, which is missing some keys, or loading a state_dict with more keys than the model that you are loading into, you can set the strict argument to False in the load_state_dict() function to ignore non-matching keys.\n \"\"\"\n return model",
"_____no_output_____"
]
],
[
[
"**Prepare Network Training Functions**",
"_____no_output_____"
]
],
[
[
"# Define model\nmy_net = alexnetdann(pretrained=True,progress=True, num_classes=NUM_CLASSES, num_domains=NUM_DOMAINS)\n\n# Define loss function\nloss_class = nn.CrossEntropyLoss()\nloss_domain = nn.CrossEntropyLoss()\n\n# Choose parameters to optimize\nparameters_to_optimize = my_net.parameters() # In this case we optimize over all the parameters of AlexNet\n\n# Define optimizer\n# An optimizer updates the weights based on loss\noptimizer = optim.SGD(parameters_to_optimize, lr=LR)\n\n\n#move network to GPU\nmy_net = my_net.to(device)\nloss_class = loss_class.to(device)\nloss_domain = loss_domain.to(device)\n\n#to train\nfor p in my_net.parameters():\n p.requires_grad = True",
"_____no_output_____"
]
],
[
[
"**Test Function**",
"_____no_output_____"
]
],
[
[
"def test(dataset_name,epoch):\n\n batch_size = BATCH_SIZE\n\n if dataset_name == source_dataloader:\n dataloader = source_dataloader\n else:\n dataloader = target_dataloader \n \n \n\n global my_net\n my_net = my_net.to(device)\n my_net = my_net.eval()\n \n len_dataloader = len(dataloader)\n data_target_iter = iter(dataloader)\n\n i = 0\n n_total = 0\n n_correct = 0\n\n while i < len_dataloader:\n \n data_target = data_target_iter.next()\n t_img, t_label = data_target\n\n batch_size = len(t_label)\n\n input_img = torch.FloatTensor(batch_size, 3, image_size, image_size)\n class_label = torch.LongTensor(batch_size)\n\n t_img = t_img.to(device)\n t_label = t_label.to(device)\n input_img = input_img.to(device)\n class_label = class_label.to(device)\n\n input_img.resize_as_(t_img).copy_(t_img)\n class_label.resize_as_(t_label).copy_(t_label)\n\n class_output, _ = my_net(input_data=input_img, alpha=alpha)\n pred = class_output.data.max(1, keepdim=True)[1]\n n_correct += pred.eq(class_label.data.view_as(pred)).cpu().sum()\n n_total += batch_size\n\n i += 1\n\n accu = n_correct.data.numpy() * 1.0 / n_total\n\n if dataset_name == source_dataloader: \n print ('epoch: %d, accuracy of the source_dataset: %f' % (epoch+1, accu))\n else:\n print ('epoch: %d, accuracy of the target_dataset: %f' % (epoch+1, accu))\n \n return accu\n",
"_____no_output_____"
]
],
[
[
"**Train & Test**",
"_____no_output_____"
]
],
[
[
"cudnn.benchmark # Calling this optimizes runtime\n\nall_err_s_label=[]\nall_err_s_domain=[]\nall_err_t_domain=[]\nall_acc_source_dataset=[]\nall_acc_target_dataset=[]\n# Start iterating over the epochs\nfor epoch in range(NUM_EPOCHS):\n \n it_err_s_label=[]\n it_err_s_domain=[]\n it_err_t_domain=[]\n\n len_dataloader = min(len(source_dataloader), len(target_dataloader))\n data_source_iter = iter(source_dataloader)\n data_target_iter = iter(target_dataloader)\n\n i = 0\n while i < len_dataloader:\n\n p = float(i + epoch * len_dataloader) / NUM_EPOCHS / len_dataloader\n alpha = 2. / (1. + np.exp(-10 * p)) - 1\n\n # training model using source data\n data_source = data_source_iter.next()\n s_img, s_label = data_source\n\n my_net.zero_grad()\n my_net.train()\n batch_size = len(s_label)\n\n input_img = torch.FloatTensor(batch_size, 3, image_size, image_size)\n class_label = torch.LongTensor(batch_size)\n domain_label = torch.zeros(batch_size)\n domain_label = domain_label.long()\n\n s_img = s_img.to(device)\n s_label = s_label.to(device)\n input_img = input_img.to(device)\n class_label = class_label.to(device)\n domain_label = domain_label.to(device)\n\n input_img.resize_as_(s_img).copy_(s_img)\n class_label.resize_as_(s_label).copy_(s_label)\n\n class_output, domain_output = my_net(input_data=input_img, alpha=alpha)\n err_s_label = loss_class(class_output, class_label)\n err_s_domain = loss_domain(domain_output, domain_label)\n\n # training model using target data\n data_target = data_target_iter.next()\n t_img, _ = data_target\n\n batch_size = len(t_img)\n\n input_img = torch.FloatTensor(batch_size, 3, image_size, image_size)\n domain_label = torch.ones(batch_size)\n domain_label = domain_label.long()\n\n t_img = t_img.to(device)\n input_img = input_img.to(device)\n domain_label = domain_label.to(device)\n\n input_img.resize_as_(t_img).copy_(t_img)\n\n _, domain_output = my_net(input_data=input_img, alpha=alpha)\n err_t_domain = loss_domain(domain_output, domain_label)\n err = err_t_domain + err_s_domain + err_s_label\n err.backward()\n optimizer.step()\n\n i += 1\n\n print ('epoch: %d, [iter: %d / %d], err_s_label: %f, err_s_domain: %f, err_t_domain: %f' \\\n % (epoch+1, i, len_dataloader, err_s_label.data.cpu().numpy(),\n err_s_domain.data.cpu().numpy(), err_t_domain.data.cpu().item()))\n \n it_err_s_label.append(err_s_label.data.cpu().numpy())\n it_err_s_domain.append(err_s_domain.data.cpu().numpy())\n it_err_t_domain.append(err_t_domain.data.cpu().item())\n \n all_err_s_label.append(min(it_err_s_label))\n all_err_s_domain.append(min(it_err_s_domain))\n all_err_t_domain.append(min(it_err_t_domain))\n\n all_acc_source_dataset.append(test(source_dataloader,epoch))\n all_acc_target_dataset.append(test(target_dataloader,epoch))",
"epoch: 1, [iter: 1 / 7], err_s_label: 2.016973, err_s_domain: 0.953112, err_t_domain: 0.519965\nepoch: 1, [iter: 2 / 7], err_s_label: 2.006701, err_s_domain: 0.749304, err_t_domain: 0.677271\nepoch: 1, [iter: 3 / 7], err_s_label: 1.958613, err_s_domain: 0.702738, err_t_domain: 0.698293\nepoch: 1, [iter: 4 / 7], err_s_label: 1.943361, err_s_domain: 0.702946, err_t_domain: 0.705443\nepoch: 1, [iter: 5 / 7], err_s_label: 1.888852, err_s_domain: 0.684455, err_t_domain: 0.670706\nepoch: 1, [iter: 6 / 7], err_s_label: 1.858400, err_s_domain: 0.695379, err_t_domain: 0.670126\nepoch: 1, [iter: 7 / 7], err_s_label: 1.874049, err_s_domain: 0.648646, err_t_domain: 0.661750\nepoch: 1, accuracy of the source_dataset: 0.416168\nepoch: 1, accuracy of the target_dataset: 0.246582\nepoch: 2, [iter: 1 / 7], err_s_label: 1.843632, err_s_domain: 0.671109, err_t_domain: 0.649514\nepoch: 2, [iter: 2 / 7], err_s_label: 1.839545, err_s_domain: 0.624539, err_t_domain: 0.647396\nepoch: 2, [iter: 3 / 7], err_s_label: 1.806576, err_s_domain: 0.616874, err_t_domain: 0.619375\nepoch: 2, [iter: 4 / 7], err_s_label: 1.739179, err_s_domain: 0.622366, err_t_domain: 0.634799\nepoch: 2, [iter: 5 / 7], err_s_label: 1.810307, err_s_domain: 0.638970, err_t_domain: 0.597006\nepoch: 2, [iter: 6 / 7], err_s_label: 1.728578, err_s_domain: 0.615580, err_t_domain: 0.616941\nepoch: 2, [iter: 7 / 7], err_s_label: 1.765443, err_s_domain: 0.611065, err_t_domain: 0.612035\nepoch: 2, accuracy of the source_dataset: 0.449102\nepoch: 2, accuracy of the target_dataset: 0.273438\nepoch: 3, [iter: 1 / 7], err_s_label: 1.748060, err_s_domain: 0.586070, err_t_domain: 0.593181\nepoch: 3, [iter: 2 / 7], err_s_label: 1.730242, err_s_domain: 0.590553, err_t_domain: 0.590888\nepoch: 3, [iter: 3 / 7], err_s_label: 1.745961, err_s_domain: 0.591154, err_t_domain: 0.562244\nepoch: 3, [iter: 4 / 7], err_s_label: 1.686607, err_s_domain: 0.559914, err_t_domain: 0.600836\nepoch: 3, [iter: 5 / 7], err_s_label: 1.676150, err_s_domain: 0.549846, err_t_domain: 0.583566\nepoch: 3, [iter: 6 / 7], err_s_label: 1.680283, err_s_domain: 0.569786, err_t_domain: 0.559359\nepoch: 3, [iter: 7 / 7], err_s_label: 1.625631, err_s_domain: 0.564431, err_t_domain: 0.547076\nepoch: 3, accuracy of the source_dataset: 0.508982\nepoch: 3, accuracy of the target_dataset: 0.305176\nepoch: 4, [iter: 1 / 7], err_s_label: 1.637074, err_s_domain: 0.516593, err_t_domain: 0.571097\nepoch: 4, [iter: 2 / 7], err_s_label: 1.582006, err_s_domain: 0.513584, err_t_domain: 0.555336\nepoch: 4, [iter: 3 / 7], err_s_label: 1.573785, err_s_domain: 0.543423, err_t_domain: 0.528820\nepoch: 4, [iter: 4 / 7], err_s_label: 1.582957, err_s_domain: 0.513608, err_t_domain: 0.547084\nepoch: 4, [iter: 5 / 7], err_s_label: 1.674368, err_s_domain: 0.546198, err_t_domain: 0.520523\nepoch: 4, [iter: 6 / 7], err_s_label: 1.631911, err_s_domain: 0.550603, err_t_domain: 0.544045\nepoch: 4, [iter: 7 / 7], err_s_label: 1.598608, err_s_domain: 0.485782, err_t_domain: 0.510271\nepoch: 4, accuracy of the source_dataset: 0.573653\nepoch: 4, accuracy of the target_dataset: 0.332520\nepoch: 5, [iter: 1 / 7], err_s_label: 1.601600, err_s_domain: 0.511025, err_t_domain: 0.515688\nepoch: 5, [iter: 2 / 7], err_s_label: 1.499426, err_s_domain: 0.465956, err_t_domain: 0.515227\nepoch: 5, [iter: 3 / 7], err_s_label: 1.545951, err_s_domain: 0.470147, err_t_domain: 0.500341\nepoch: 5, [iter: 4 / 7], err_s_label: 1.535792, err_s_domain: 0.498536, err_t_domain: 0.501819\nepoch: 5, [iter: 5 / 7], err_s_label: 1.480569, err_s_domain: 0.474470, err_t_domain: 0.496956\nepoch: 5, [iter: 6 / 7], err_s_label: 1.400137, err_s_domain: 0.444521, err_t_domain: 0.491441\nepoch: 5, [iter: 7 / 7], err_s_label: 1.442505, err_s_domain: 0.479597, err_t_domain: 0.509100\nepoch: 5, accuracy of the source_dataset: 0.628144\nepoch: 5, accuracy of the target_dataset: 0.347168\nepoch: 6, [iter: 1 / 7], err_s_label: 1.413393, err_s_domain: 0.468799, err_t_domain: 0.478612\nepoch: 6, [iter: 2 / 7], err_s_label: 1.472482, err_s_domain: 0.497971, err_t_domain: 0.452059\nepoch: 6, [iter: 3 / 7], err_s_label: 1.409759, err_s_domain: 0.435614, err_t_domain: 0.486160\nepoch: 6, [iter: 4 / 7], err_s_label: 1.415383, err_s_domain: 0.467968, err_t_domain: 0.447569\nepoch: 6, [iter: 5 / 7], err_s_label: 1.400301, err_s_domain: 0.430779, err_t_domain: 0.469757\nepoch: 6, [iter: 6 / 7], err_s_label: 1.419525, err_s_domain: 0.476563, err_t_domain: 0.493821\nepoch: 6, [iter: 7 / 7], err_s_label: 1.462294, err_s_domain: 0.415302, err_t_domain: 0.453413\nepoch: 6, accuracy of the source_dataset: 0.695808\nepoch: 6, accuracy of the target_dataset: 0.368652\nepoch: 7, [iter: 1 / 7], err_s_label: 1.421276, err_s_domain: 0.440430, err_t_domain: 0.444318\nepoch: 7, [iter: 2 / 7], err_s_label: 1.305704, err_s_domain: 0.408990, err_t_domain: 0.443797\nepoch: 7, [iter: 3 / 7], err_s_label: 1.391359, err_s_domain: 0.451011, err_t_domain: 0.412836\nepoch: 7, [iter: 4 / 7], err_s_label: 1.394546, err_s_domain: 0.429910, err_t_domain: 0.459945\nepoch: 7, [iter: 5 / 7], err_s_label: 1.365983, err_s_domain: 0.409501, err_t_domain: 0.480032\nepoch: 7, [iter: 6 / 7], err_s_label: 1.268306, err_s_domain: 0.404479, err_t_domain: 0.431149\nepoch: 7, [iter: 7 / 7], err_s_label: 1.388116, err_s_domain: 0.424651, err_t_domain: 0.394723\nepoch: 7, accuracy of the source_dataset: 0.744311\nepoch: 7, accuracy of the target_dataset: 0.384766\nepoch: 8, [iter: 1 / 7], err_s_label: 1.348285, err_s_domain: 0.405091, err_t_domain: 0.453557\nepoch: 8, [iter: 2 / 7], err_s_label: 1.307196, err_s_domain: 0.417189, err_t_domain: 0.435097\nepoch: 8, [iter: 3 / 7], err_s_label: 1.260960, err_s_domain: 0.381459, err_t_domain: 0.402690\nepoch: 8, [iter: 4 / 7], err_s_label: 1.319447, err_s_domain: 0.416914, err_t_domain: 0.422249\nepoch: 8, [iter: 5 / 7], err_s_label: 1.239590, err_s_domain: 0.382485, err_t_domain: 0.443774\nepoch: 8, [iter: 6 / 7], err_s_label: 1.227018, err_s_domain: 0.394228, err_t_domain: 0.397989\nepoch: 8, [iter: 7 / 7], err_s_label: 1.274644, err_s_domain: 0.363543, err_t_domain: 0.417326\nepoch: 8, accuracy of the source_dataset: 0.762275\nepoch: 8, accuracy of the target_dataset: 0.404297\nepoch: 9, [iter: 1 / 7], err_s_label: 1.263348, err_s_domain: 0.432271, err_t_domain: 0.413685\nepoch: 9, [iter: 2 / 7], err_s_label: 1.260342, err_s_domain: 0.389225, err_t_domain: 0.408134\nepoch: 9, [iter: 3 / 7], err_s_label: 1.197175, err_s_domain: 0.381348, err_t_domain: 0.375230\nepoch: 9, [iter: 4 / 7], err_s_label: 1.231898, err_s_domain: 0.387302, err_t_domain: 0.404238\nepoch: 9, [iter: 5 / 7], err_s_label: 1.174647, err_s_domain: 0.369771, err_t_domain: 0.377372\nepoch: 9, [iter: 6 / 7], err_s_label: 1.244384, err_s_domain: 0.386802, err_t_domain: 0.412414\nepoch: 9, [iter: 7 / 7], err_s_label: 1.274358, err_s_domain: 0.382575, err_t_domain: 0.393697\nepoch: 9, accuracy of the source_dataset: 0.790419\nepoch: 9, accuracy of the target_dataset: 0.412598\nepoch: 10, [iter: 1 / 7], err_s_label: 1.220906, err_s_domain: 0.359031, err_t_domain: 0.375389\nepoch: 10, [iter: 2 / 7], err_s_label: 1.168681, err_s_domain: 0.375113, err_t_domain: 0.396369\nepoch: 10, [iter: 3 / 7], err_s_label: 1.217747, err_s_domain: 0.361663, err_t_domain: 0.392944\nepoch: 10, [iter: 4 / 7], err_s_label: 1.215102, err_s_domain: 0.367760, err_t_domain: 0.373483\nepoch: 10, [iter: 5 / 7], err_s_label: 1.125806, err_s_domain: 0.344977, err_t_domain: 0.439451\nepoch: 10, [iter: 6 / 7], err_s_label: 1.052310, err_s_domain: 0.328862, err_t_domain: 0.385855\nepoch: 10, [iter: 7 / 7], err_s_label: 1.235539, err_s_domain: 0.438889, err_t_domain: 0.373131\nepoch: 10, accuracy of the source_dataset: 0.809581\nepoch: 10, accuracy of the target_dataset: 0.427734\nepoch: 11, [iter: 1 / 7], err_s_label: 1.101981, err_s_domain: 0.378486, err_t_domain: 0.379997\nepoch: 11, [iter: 2 / 7], err_s_label: 1.092054, err_s_domain: 0.339491, err_t_domain: 0.361604\nepoch: 11, [iter: 3 / 7], err_s_label: 1.188677, err_s_domain: 0.387163, err_t_domain: 0.367242\nepoch: 11, [iter: 4 / 7], err_s_label: 1.147236, err_s_domain: 0.372867, err_t_domain: 0.369229\nepoch: 11, [iter: 5 / 7], err_s_label: 1.105009, err_s_domain: 0.330834, err_t_domain: 0.401387\nepoch: 11, [iter: 6 / 7], err_s_label: 1.072930, err_s_domain: 0.329688, err_t_domain: 0.375511\nepoch: 11, [iter: 7 / 7], err_s_label: 1.072036, err_s_domain: 0.364242, err_t_domain: 0.371819\nepoch: 11, accuracy of the source_dataset: 0.826946\nepoch: 11, accuracy of the target_dataset: 0.439941\nepoch: 12, [iter: 1 / 7], err_s_label: 1.098573, err_s_domain: 0.386761, err_t_domain: 0.344731\nepoch: 12, [iter: 2 / 7], err_s_label: 1.049066, err_s_domain: 0.298639, err_t_domain: 0.375087\nepoch: 12, [iter: 3 / 7], err_s_label: 1.039566, err_s_domain: 0.324032, err_t_domain: 0.357408\nepoch: 12, [iter: 4 / 7], err_s_label: 1.145818, err_s_domain: 0.386581, err_t_domain: 0.327931\nepoch: 12, [iter: 5 / 7], err_s_label: 1.001768, err_s_domain: 0.340898, err_t_domain: 0.392067\nepoch: 12, [iter: 6 / 7], err_s_label: 1.047605, err_s_domain: 0.328640, err_t_domain: 0.370711\nepoch: 12, [iter: 7 / 7], err_s_label: 1.110372, err_s_domain: 0.362784, err_t_domain: 0.363499\nepoch: 12, accuracy of the source_dataset: 0.839521\nepoch: 12, accuracy of the target_dataset: 0.441895\nepoch: 13, [iter: 1 / 7], err_s_label: 1.045457, err_s_domain: 0.329734, err_t_domain: 0.384564\nepoch: 13, [iter: 2 / 7], err_s_label: 1.130854, err_s_domain: 0.356106, err_t_domain: 0.325828\nepoch: 13, [iter: 3 / 7], err_s_label: 0.974066, err_s_domain: 0.283998, err_t_domain: 0.375355\nepoch: 13, [iter: 4 / 7], err_s_label: 1.048728, err_s_domain: 0.379799, err_t_domain: 0.303322\nepoch: 13, [iter: 5 / 7], err_s_label: 0.968934, err_s_domain: 0.308299, err_t_domain: 0.380323\nepoch: 13, [iter: 6 / 7], err_s_label: 1.025390, err_s_domain: 0.309448, err_t_domain: 0.326013\nepoch: 13, [iter: 7 / 7], err_s_label: 0.967331, err_s_domain: 0.341489, err_t_domain: 0.334397\nepoch: 13, accuracy of the source_dataset: 0.840120\nepoch: 13, accuracy of the target_dataset: 0.453125\nepoch: 14, [iter: 1 / 7], err_s_label: 0.965886, err_s_domain: 0.303297, err_t_domain: 0.364607\nepoch: 14, [iter: 2 / 7], err_s_label: 1.030872, err_s_domain: 0.361949, err_t_domain: 0.355905\nepoch: 14, [iter: 3 / 7], err_s_label: 1.025075, err_s_domain: 0.319672, err_t_domain: 0.345718\nepoch: 14, [iter: 4 / 7], err_s_label: 0.986508, err_s_domain: 0.312751, err_t_domain: 0.369813\nepoch: 14, [iter: 5 / 7], err_s_label: 0.972301, err_s_domain: 0.362270, err_t_domain: 0.347547\nepoch: 14, [iter: 6 / 7], err_s_label: 0.998557, err_s_domain: 0.301598, err_t_domain: 0.326858\nepoch: 14, [iter: 7 / 7], err_s_label: 0.960262, err_s_domain: 0.326565, err_t_domain: 0.313907\nepoch: 14, accuracy of the source_dataset: 0.863473\nepoch: 14, accuracy of the target_dataset: 0.455566\nepoch: 15, [iter: 1 / 7], err_s_label: 0.966974, err_s_domain: 0.294919, err_t_domain: 0.335576\nepoch: 15, [iter: 2 / 7], err_s_label: 0.920078, err_s_domain: 0.302298, err_t_domain: 0.318146\nepoch: 15, [iter: 3 / 7], err_s_label: 1.029996, err_s_domain: 0.400218, err_t_domain: 0.322583\nepoch: 15, [iter: 4 / 7], err_s_label: 0.901816, err_s_domain: 0.226799, err_t_domain: 0.390164\nepoch: 15, [iter: 5 / 7], err_s_label: 0.961456, err_s_domain: 0.334749, err_t_domain: 0.312849\nepoch: 15, [iter: 6 / 7], err_s_label: 0.976521, err_s_domain: 0.341206, err_t_domain: 0.299012\nepoch: 15, [iter: 7 / 7], err_s_label: 0.901868, err_s_domain: 0.244202, err_t_domain: 0.346626\nepoch: 15, accuracy of the source_dataset: 0.866467\nepoch: 15, accuracy of the target_dataset: 0.468262\nepoch: 16, [iter: 1 / 7], err_s_label: 0.875097, err_s_domain: 0.327953, err_t_domain: 0.300015\nepoch: 16, [iter: 2 / 7], err_s_label: 0.930037, err_s_domain: 0.285870, err_t_domain: 0.341392\nepoch: 16, [iter: 3 / 7], err_s_label: 0.930809, err_s_domain: 0.353910, err_t_domain: 0.317914\nepoch: 16, [iter: 4 / 7], err_s_label: 0.928839, err_s_domain: 0.307928, err_t_domain: 0.341378\nepoch: 16, [iter: 5 / 7], err_s_label: 0.917337, err_s_domain: 0.301805, err_t_domain: 0.366616\nepoch: 16, [iter: 6 / 7], err_s_label: 0.851960, err_s_domain: 0.332049, err_t_domain: 0.314535\nepoch: 16, [iter: 7 / 7], err_s_label: 0.826500, err_s_domain: 0.239989, err_t_domain: 0.341516\nepoch: 16, accuracy of the source_dataset: 0.870659\nepoch: 16, accuracy of the target_dataset: 0.473145\nepoch: 17, [iter: 1 / 7], err_s_label: 0.792548, err_s_domain: 0.296159, err_t_domain: 0.306754\nepoch: 17, [iter: 2 / 7], err_s_label: 0.893546, err_s_domain: 0.328343, err_t_domain: 0.311362\nepoch: 17, [iter: 3 / 7], err_s_label: 0.905596, err_s_domain: 0.321932, err_t_domain: 0.328081\nepoch: 17, [iter: 4 / 7], err_s_label: 0.911837, err_s_domain: 0.300020, err_t_domain: 0.334114\nepoch: 17, [iter: 5 / 7], err_s_label: 0.888555, err_s_domain: 0.283410, err_t_domain: 0.324528\nepoch: 17, [iter: 6 / 7], err_s_label: 0.852029, err_s_domain: 0.300626, err_t_domain: 0.319645\nepoch: 17, [iter: 7 / 7], err_s_label: 0.782541, err_s_domain: 0.269756, err_t_domain: 0.299578\nepoch: 17, accuracy of the source_dataset: 0.873054\nepoch: 17, accuracy of the target_dataset: 0.482910\nepoch: 18, [iter: 1 / 7], err_s_label: 0.795490, err_s_domain: 0.296559, err_t_domain: 0.292586\nepoch: 18, [iter: 2 / 7], err_s_label: 0.871690, err_s_domain: 0.332933, err_t_domain: 0.317478\nepoch: 18, [iter: 3 / 7], err_s_label: 0.830925, err_s_domain: 0.306968, err_t_domain: 0.353154\nepoch: 18, [iter: 4 / 7], err_s_label: 0.874403, err_s_domain: 0.304290, err_t_domain: 0.301310\nepoch: 18, [iter: 5 / 7], err_s_label: 0.879852, err_s_domain: 0.276177, err_t_domain: 0.301399\nepoch: 18, [iter: 6 / 7], err_s_label: 0.814115, err_s_domain: 0.295968, err_t_domain: 0.306269\nepoch: 18, [iter: 7 / 7], err_s_label: 0.793346, err_s_domain: 0.241446, err_t_domain: 0.294381\nepoch: 18, accuracy of the source_dataset: 0.877844\nepoch: 18, accuracy of the target_dataset: 0.479004\nepoch: 19, [iter: 1 / 7], err_s_label: 0.822033, err_s_domain: 0.278200, err_t_domain: 0.339458\nepoch: 19, [iter: 2 / 7], err_s_label: 0.765337, err_s_domain: 0.319974, err_t_domain: 0.284192\nepoch: 19, [iter: 3 / 7], err_s_label: 0.782650, err_s_domain: 0.269660, err_t_domain: 0.310526\nepoch: 19, [iter: 4 / 7], err_s_label: 0.799926, err_s_domain: 0.299665, err_t_domain: 0.334611\nepoch: 19, [iter: 5 / 7], err_s_label: 0.817362, err_s_domain: 0.315373, err_t_domain: 0.273783\nepoch: 19, [iter: 6 / 7], err_s_label: 0.834446, err_s_domain: 0.259153, err_t_domain: 0.325824\nepoch: 19, [iter: 7 / 7], err_s_label: 0.802022, err_s_domain: 0.316240, err_t_domain: 0.304728\nepoch: 19, accuracy of the source_dataset: 0.882036\nepoch: 19, accuracy of the target_dataset: 0.483398\nepoch: 20, [iter: 1 / 7], err_s_label: 0.860349, err_s_domain: 0.339715, err_t_domain: 0.297814\nepoch: 20, [iter: 2 / 7], err_s_label: 0.831741, err_s_domain: 0.251926, err_t_domain: 0.376361\nepoch: 20, [iter: 3 / 7], err_s_label: 0.763444, err_s_domain: 0.305646, err_t_domain: 0.281218\nepoch: 20, [iter: 4 / 7], err_s_label: 0.717219, err_s_domain: 0.276297, err_t_domain: 0.323980\nepoch: 20, [iter: 5 / 7], err_s_label: 0.793929, err_s_domain: 0.300649, err_t_domain: 0.299523\nepoch: 20, [iter: 6 / 7], err_s_label: 0.741730, err_s_domain: 0.285963, err_t_domain: 0.304678\nepoch: 20, [iter: 7 / 7], err_s_label: 0.696370, err_s_domain: 0.310940, err_t_domain: 0.274453\nepoch: 20, accuracy of the source_dataset: 0.888623\nepoch: 20, accuracy of the target_dataset: 0.487793\nepoch: 21, [iter: 1 / 7], err_s_label: 0.820124, err_s_domain: 0.330818, err_t_domain: 0.315074\nepoch: 21, [iter: 2 / 7], err_s_label: 0.776975, err_s_domain: 0.274746, err_t_domain: 0.375955\nepoch: 21, [iter: 3 / 7], err_s_label: 0.754121, err_s_domain: 0.289164, err_t_domain: 0.321892\nepoch: 21, [iter: 4 / 7], err_s_label: 0.668952, err_s_domain: 0.274380, err_t_domain: 0.296007\nepoch: 21, [iter: 5 / 7], err_s_label: 0.688795, err_s_domain: 0.234383, err_t_domain: 0.328634\nepoch: 21, [iter: 6 / 7], err_s_label: 0.763450, err_s_domain: 0.299210, err_t_domain: 0.276722\nepoch: 21, [iter: 7 / 7], err_s_label: 0.759604, err_s_domain: 0.342027, err_t_domain: 0.278099\nepoch: 21, accuracy of the source_dataset: 0.886228\nepoch: 21, accuracy of the target_dataset: 0.487793\nepoch: 22, [iter: 1 / 7], err_s_label: 0.702863, err_s_domain: 0.280571, err_t_domain: 0.334275\nepoch: 22, [iter: 2 / 7], err_s_label: 0.768291, err_s_domain: 0.279846, err_t_domain: 0.312605\nepoch: 22, [iter: 3 / 7], err_s_label: 0.754109, err_s_domain: 0.273857, err_t_domain: 0.317683\nepoch: 22, [iter: 4 / 7], err_s_label: 0.754074, err_s_domain: 0.325817, err_t_domain: 0.344589\nepoch: 22, [iter: 5 / 7], err_s_label: 0.726676, err_s_domain: 0.275591, err_t_domain: 0.269645\nepoch: 22, [iter: 6 / 7], err_s_label: 0.650434, err_s_domain: 0.242501, err_t_domain: 0.304022\nepoch: 22, [iter: 7 / 7], err_s_label: 0.680657, err_s_domain: 0.312181, err_t_domain: 0.268257\nepoch: 22, accuracy of the source_dataset: 0.895808\nepoch: 22, accuracy of the target_dataset: 0.491699\nepoch: 23, [iter: 1 / 7], err_s_label: 0.719517, err_s_domain: 0.240564, err_t_domain: 0.299067\nepoch: 23, [iter: 2 / 7], err_s_label: 0.679984, err_s_domain: 0.300666, err_t_domain: 0.264786\nepoch: 23, [iter: 3 / 7], err_s_label: 0.630339, err_s_domain: 0.242883, err_t_domain: 0.337255\nepoch: 23, [iter: 4 / 7], err_s_label: 0.669175, err_s_domain: 0.336703, err_t_domain: 0.305431\nepoch: 23, [iter: 5 / 7], err_s_label: 0.662295, err_s_domain: 0.248101, err_t_domain: 0.280339\nepoch: 23, [iter: 6 / 7], err_s_label: 0.715453, err_s_domain: 0.308214, err_t_domain: 0.310301\nepoch: 23, [iter: 7 / 7], err_s_label: 0.664669, err_s_domain: 0.301210, err_t_domain: 0.343790\nepoch: 23, accuracy of the source_dataset: 0.885030\nepoch: 23, accuracy of the target_dataset: 0.495117\nepoch: 24, [iter: 1 / 7], err_s_label: 0.686867, err_s_domain: 0.294407, err_t_domain: 0.267800\nepoch: 24, [iter: 2 / 7], err_s_label: 0.633689, err_s_domain: 0.261191, err_t_domain: 0.320785\nepoch: 24, [iter: 3 / 7], err_s_label: 0.709318, err_s_domain: 0.290389, err_t_domain: 0.260650\nepoch: 24, [iter: 4 / 7], err_s_label: 0.680888, err_s_domain: 0.283037, err_t_domain: 0.309168\nepoch: 24, [iter: 5 / 7], err_s_label: 0.704849, err_s_domain: 0.286151, err_t_domain: 0.295278\nepoch: 24, [iter: 6 / 7], err_s_label: 0.578898, err_s_domain: 0.288721, err_t_domain: 0.264734\nepoch: 24, [iter: 7 / 7], err_s_label: 0.632983, err_s_domain: 0.245066, err_t_domain: 0.317848\nepoch: 24, accuracy of the source_dataset: 0.893413\nepoch: 24, accuracy of the target_dataset: 0.496582\nepoch: 25, [iter: 1 / 7], err_s_label: 0.592443, err_s_domain: 0.301478, err_t_domain: 0.242632\nepoch: 25, [iter: 2 / 7], err_s_label: 0.638555, err_s_domain: 0.288422, err_t_domain: 0.280256\nepoch: 25, [iter: 3 / 7], err_s_label: 0.737095, err_s_domain: 0.243268, err_t_domain: 0.314675\nepoch: 25, [iter: 4 / 7], err_s_label: 0.688460, err_s_domain: 0.309289, err_t_domain: 0.271316\nepoch: 25, [iter: 5 / 7], err_s_label: 0.585144, err_s_domain: 0.261817, err_t_domain: 0.354838\nepoch: 25, [iter: 6 / 7], err_s_label: 0.622304, err_s_domain: 0.265257, err_t_domain: 0.260274\nepoch: 25, [iter: 7 / 7], err_s_label: 0.646024, err_s_domain: 0.265205, err_t_domain: 0.289917\nepoch: 25, accuracy of the source_dataset: 0.887425\nepoch: 25, accuracy of the target_dataset: 0.491699\nepoch: 26, [iter: 1 / 7], err_s_label: 0.602159, err_s_domain: 0.267929, err_t_domain: 0.272255\nepoch: 26, [iter: 2 / 7], err_s_label: 0.620510, err_s_domain: 0.297581, err_t_domain: 0.288045\nepoch: 26, [iter: 3 / 7], err_s_label: 0.608833, err_s_domain: 0.310141, err_t_domain: 0.293771\nepoch: 26, [iter: 4 / 7], err_s_label: 0.651094, err_s_domain: 0.274941, err_t_domain: 0.282898\nepoch: 26, [iter: 5 / 7], err_s_label: 0.595068, err_s_domain: 0.266347, err_t_domain: 0.341013\nepoch: 26, [iter: 6 / 7], err_s_label: 0.609245, err_s_domain: 0.264535, err_t_domain: 0.326431\nepoch: 26, [iter: 7 / 7], err_s_label: 0.578238, err_s_domain: 0.280917, err_t_domain: 0.239750\nepoch: 26, accuracy of the source_dataset: 0.896407\nepoch: 26, accuracy of the target_dataset: 0.496582\nepoch: 27, [iter: 1 / 7], err_s_label: 0.571688, err_s_domain: 0.239098, err_t_domain: 0.286999\nepoch: 27, [iter: 2 / 7], err_s_label: 0.609199, err_s_domain: 0.270370, err_t_domain: 0.258303\nepoch: 27, [iter: 3 / 7], err_s_label: 0.595798, err_s_domain: 0.292287, err_t_domain: 0.333910\nepoch: 27, [iter: 4 / 7], err_s_label: 0.576865, err_s_domain: 0.239606, err_t_domain: 0.327447\nepoch: 27, [iter: 5 / 7], err_s_label: 0.542654, err_s_domain: 0.302625, err_t_domain: 0.269186\nepoch: 27, [iter: 6 / 7], err_s_label: 0.645830, err_s_domain: 0.356997, err_t_domain: 0.254960\nepoch: 27, [iter: 7 / 7], err_s_label: 0.592112, err_s_domain: 0.201448, err_t_domain: 0.309333\nepoch: 27, accuracy of the source_dataset: 0.894611\nepoch: 27, accuracy of the target_dataset: 0.497070\nepoch: 28, [iter: 1 / 7], err_s_label: 0.561699, err_s_domain: 0.256288, err_t_domain: 0.257007\nepoch: 28, [iter: 2 / 7], err_s_label: 0.585144, err_s_domain: 0.295792, err_t_domain: 0.263150\nepoch: 28, [iter: 3 / 7], err_s_label: 0.551236, err_s_domain: 0.313753, err_t_domain: 0.295037\nepoch: 28, [iter: 4 / 7], err_s_label: 0.589875, err_s_domain: 0.252434, err_t_domain: 0.297009\nepoch: 28, [iter: 5 / 7], err_s_label: 0.513780, err_s_domain: 0.299580, err_t_domain: 0.315401\nepoch: 28, [iter: 6 / 7], err_s_label: 0.611404, err_s_domain: 0.334664, err_t_domain: 0.295299\nepoch: 28, [iter: 7 / 7], err_s_label: 0.611564, err_s_domain: 0.257753, err_t_domain: 0.279676\nepoch: 28, accuracy of the source_dataset: 0.900000\nepoch: 28, accuracy of the target_dataset: 0.494141\nepoch: 29, [iter: 1 / 7], err_s_label: 0.569339, err_s_domain: 0.307565, err_t_domain: 0.240690\nepoch: 29, [iter: 2 / 7], err_s_label: 0.573364, err_s_domain: 0.257280, err_t_domain: 0.322713\nepoch: 29, [iter: 3 / 7], err_s_label: 0.496106, err_s_domain: 0.264480, err_t_domain: 0.338439\nepoch: 29, [iter: 4 / 7], err_s_label: 0.585562, err_s_domain: 0.324296, err_t_domain: 0.237467\nepoch: 29, [iter: 5 / 7], err_s_label: 0.497246, err_s_domain: 0.254488, err_t_domain: 0.293323\nepoch: 29, [iter: 6 / 7], err_s_label: 0.629726, err_s_domain: 0.284013, err_t_domain: 0.334936\nepoch: 29, [iter: 7 / 7], err_s_label: 0.595831, err_s_domain: 0.284793, err_t_domain: 0.287715\nepoch: 29, accuracy of the source_dataset: 0.892216\nepoch: 29, accuracy of the target_dataset: 0.492676\nepoch: 30, [iter: 1 / 7], err_s_label: 0.574720, err_s_domain: 0.316564, err_t_domain: 0.256874\nepoch: 30, [iter: 2 / 7], err_s_label: 0.535031, err_s_domain: 0.246776, err_t_domain: 0.367707\nepoch: 30, [iter: 3 / 7], err_s_label: 0.524217, err_s_domain: 0.326047, err_t_domain: 0.261240\nepoch: 30, [iter: 4 / 7], err_s_label: 0.559679, err_s_domain: 0.276420, err_t_domain: 0.322626\nepoch: 30, [iter: 5 / 7], err_s_label: 0.529012, err_s_domain: 0.274724, err_t_domain: 0.297331\nepoch: 30, [iter: 6 / 7], err_s_label: 0.520827, err_s_domain: 0.266807, err_t_domain: 0.257031\nepoch: 30, [iter: 7 / 7], err_s_label: 0.518005, err_s_domain: 0.287191, err_t_domain: 0.285307\nepoch: 30, accuracy of the source_dataset: 0.897006\nepoch: 30, accuracy of the target_dataset: 0.492676\nepoch: 31, [iter: 1 / 7], err_s_label: 0.519722, err_s_domain: 0.318713, err_t_domain: 0.253605\nepoch: 31, [iter: 2 / 7], err_s_label: 0.522369, err_s_domain: 0.225385, err_t_domain: 0.356496\nepoch: 31, [iter: 3 / 7], err_s_label: 0.517982, err_s_domain: 0.313076, err_t_domain: 0.276535\nepoch: 31, [iter: 4 / 7], err_s_label: 0.464997, err_s_domain: 0.268339, err_t_domain: 0.293741\nepoch: 31, [iter: 5 / 7], err_s_label: 0.481274, err_s_domain: 0.263774, err_t_domain: 0.301084\nepoch: 31, [iter: 6 / 7], err_s_label: 0.491202, err_s_domain: 0.293632, err_t_domain: 0.286175\nepoch: 31, [iter: 7 / 7], err_s_label: 0.563209, err_s_domain: 0.325694, err_t_domain: 0.281357\nepoch: 31, accuracy of the source_dataset: 0.895808\nepoch: 31, accuracy of the target_dataset: 0.494629\nepoch: 32, [iter: 1 / 7], err_s_label: 0.470510, err_s_domain: 0.245862, err_t_domain: 0.337900\nepoch: 32, [iter: 2 / 7], err_s_label: 0.553035, err_s_domain: 0.355526, err_t_domain: 0.301713\nepoch: 32, [iter: 3 / 7], err_s_label: 0.495609, err_s_domain: 0.288856, err_t_domain: 0.291788\nepoch: 32, [iter: 4 / 7], err_s_label: 0.515739, err_s_domain: 0.248252, err_t_domain: 0.342551\nepoch: 32, [iter: 5 / 7], err_s_label: 0.495953, err_s_domain: 0.332028, err_t_domain: 0.243237\nepoch: 32, [iter: 6 / 7], err_s_label: 0.501151, err_s_domain: 0.272157, err_t_domain: 0.303186\nepoch: 32, [iter: 7 / 7], err_s_label: 0.472207, err_s_domain: 0.296628, err_t_domain: 0.273031\nepoch: 32, accuracy of the source_dataset: 0.898802\nepoch: 32, accuracy of the target_dataset: 0.493652\nepoch: 33, [iter: 1 / 7], err_s_label: 0.530294, err_s_domain: 0.276754, err_t_domain: 0.294868\nepoch: 33, [iter: 2 / 7], err_s_label: 0.460581, err_s_domain: 0.281258, err_t_domain: 0.311186\nepoch: 33, [iter: 3 / 7], err_s_label: 0.454957, err_s_domain: 0.227576, err_t_domain: 0.311866\nepoch: 33, [iter: 4 / 7], err_s_label: 0.462616, err_s_domain: 0.295974, err_t_domain: 0.276058\nepoch: 33, [iter: 5 / 7], err_s_label: 0.508472, err_s_domain: 0.341496, err_t_domain: 0.264863\nepoch: 33, [iter: 6 / 7], err_s_label: 0.532044, err_s_domain: 0.283554, err_t_domain: 0.284515\nepoch: 33, [iter: 7 / 7], err_s_label: 0.473331, err_s_domain: 0.363960, err_t_domain: 0.326544\nepoch: 33, accuracy of the source_dataset: 0.901796\nepoch: 33, accuracy of the target_dataset: 0.493652\nepoch: 34, [iter: 1 / 7], err_s_label: 0.471347, err_s_domain: 0.269379, err_t_domain: 0.285377\nepoch: 34, [iter: 2 / 7], err_s_label: 0.521298, err_s_domain: 0.336305, err_t_domain: 0.293786\nepoch: 34, [iter: 3 / 7], err_s_label: 0.463993, err_s_domain: 0.224777, err_t_domain: 0.334311\nepoch: 34, [iter: 4 / 7], err_s_label: 0.450019, err_s_domain: 0.337906, err_t_domain: 0.266503\nepoch: 34, [iter: 5 / 7], err_s_label: 0.479113, err_s_domain: 0.291901, err_t_domain: 0.237073\nepoch: 34, [iter: 6 / 7], err_s_label: 0.457264, err_s_domain: 0.249432, err_t_domain: 0.358888\nepoch: 34, [iter: 7 / 7], err_s_label: 0.447618, err_s_domain: 0.401679, err_t_domain: 0.319231\nepoch: 34, accuracy of the source_dataset: 0.902395\nepoch: 34, accuracy of the target_dataset: 0.490234\nepoch: 35, [iter: 1 / 7], err_s_label: 0.435890, err_s_domain: 0.232333, err_t_domain: 0.346902\nepoch: 35, [iter: 2 / 7], err_s_label: 0.458509, err_s_domain: 0.291508, err_t_domain: 0.248252\nepoch: 35, [iter: 3 / 7], err_s_label: 0.493248, err_s_domain: 0.266078, err_t_domain: 0.308161\nepoch: 35, [iter: 4 / 7], err_s_label: 0.406230, err_s_domain: 0.299905, err_t_domain: 0.308751\nepoch: 35, [iter: 5 / 7], err_s_label: 0.464608, err_s_domain: 0.277129, err_t_domain: 0.253502\nepoch: 35, [iter: 6 / 7], err_s_label: 0.490028, err_s_domain: 0.328929, err_t_domain: 0.407178\nepoch: 35, [iter: 7 / 7], err_s_label: 0.466044, err_s_domain: 0.344588, err_t_domain: 0.274054\nepoch: 35, accuracy of the source_dataset: 0.905988\nepoch: 35, accuracy of the target_dataset: 0.487793\n"
]
],
[
[
"**Saving & Plotting Results**",
"_____no_output_____"
]
],
[
[
"# Error plots\nplt.figure(figsize=(10, 7))\nplt.plot(all_err_s_label, color='green', label='err_s_label')\nplt.plot(all_err_s_domain, color='blue', label='err_s_domain')\nplt.plot(all_err_t_domain, color='orange', label='err_t_domain')\n \nplt.xlabel('Epochs')\nplt.ylabel('Error')\nplt.legend()\nplt.savefig('Errors.png')\n\n# accuracy plots\nplt.figure(figsize=(10, 7))\nplt.plot(all_acc_source_dataset, color='green', label='accuracy of the source_dataset')\nplt.plot(all_acc_target_dataset, color='blue', label='accuracy of the target_dataset')\nplt.xlabel('Epochs')\nplt.ylabel('Accuracy')\nplt.legend()\nplt.savefig('accuracy.png')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.