hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
sequence | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
sequence | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
sequence | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
sequence | cell_types
sequence | cell_type_groups
sequence |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
d0cdf8b97a171f024fb8f2d26952ae3a3b8a9705 | 22,557 | ipynb | Jupyter Notebook | New York Taxi Trip Duration.ipynb | lithiumdenis/Kaggle | 49049e20b58302cfd434b064179aa575bb05720a | [
"MIT"
] | null | null | null | New York Taxi Trip Duration.ipynb | lithiumdenis/Kaggle | 49049e20b58302cfd434b064179aa575bb05720a | [
"MIT"
] | null | null | null | New York Taxi Trip Duration.ipynb | lithiumdenis/Kaggle | 49049e20b58302cfd434b064179aa575bb05720a | [
"MIT"
] | null | null | null | 35.635071 | 808 | 0.415259 | [
[
[
"import datetime\nimport lightgbm as lgb\nimport numpy as np\nimport os\nimport pandas as pd\nimport random\nfrom tqdm import tqdm\nfrom sklearn.model_selection import train_test_split\nimport haversine\n\nimport catboost as cb",
"_____no_output_____"
],
[
"random_seed = 174\nrandom.seed(random_seed)\nnp.random.seed(random_seed)",
"_____no_output_____"
],
[
"# Load data\ntrain = pd.read_csv('data/taxi/train.csv')\ntest = pd.read_csv('data/taxi/test.csv')\nss = pd.read_csv('data/taxi/sample_submission.csv')",
"_____no_output_____"
],
[
"train.head()",
"_____no_output_____"
],
[
"def rmsle(y_true, y_pred):\n assert len(y_true) == len(y_pred)\n return np.sqrt(np.mean(np.power(np.log1p(y_true + 1) - np.log1p(y_pred + 1), 2)))",
"_____no_output_____"
],
[
"def extract_features(df):\n df['hdistance'] = df.apply(lambda r: haversine.haversine((r['pickup_latitude'],r['pickup_longitude']),(r['dropoff_latitude'], r['dropoff_longitude'])), axis=1)\n df['distance'] = np.sqrt(np.power(df['dropoff_longitude'] - df['pickup_longitude'], 2) + np.power(df['dropoff_latitude'] - df['pickup_latitude'], 2))\n df['log_distance'] = np.log(df['distance'] + 1)\n df['month'] = df['pickup_datetime'].apply(lambda x: int(x.split(' ')[0].split('-')[1]))\n df['day'] = df['pickup_datetime'].apply(lambda x: int(x.split(' ')[0].split('-')[2]))\n df['hour'] = df['pickup_datetime'].apply(lambda x: int(x.split(' ')[1].split(':')[0]))\n df['minutes'] = df['pickup_datetime'].apply(lambda x: int(x.split(' ')[1].split(':')[1]))\n df['is_weekend'] = ((df.pickup_datetime.astype('datetime64[ns]').dt.dayofweek) // 4 == 1).astype(float)\n df['weekday'] = df.pickup_datetime.astype('datetime64[ns]').dt.dayofweek\n df['is_holyday'] = df.apply(lambda row: 1 if (row['month']==1 and row['day']==1) or (row['month']==7 and row['day']==4) or (row['month']==11 and row['day']==11) or (row['month']==12 and row['day']==25) or (row['month']==1 and row['day'] >= 15 and row['day'] <= 21 and row['weekday'] == 0) or (row['month']==2 and row['day'] >= 15 and row['day'] <= 21 and row['weekday'] == 0) or (row['month']==5 and row['day'] >= 25 and row['day'] <= 31 and row['weekday'] == 0) or (row['month']==9 and row['day'] >= 1 and row['day'] <= 7 and row['weekday'] == 0) or (row['month']==10 and row['day'] >= 8 and row['day'] <= 14 and row['weekday'] == 0) or (row['month']==11 and row['day'] >= 22 and row['day'] <= 28 and row['weekday'] == 3) else 0, axis=1)\n df['is_day_before_holyday'] = df.apply(lambda row: 1 if (row['month']==12 and row['day']==31) or (row['month']==7 and row['day']==3) or (row['month']==11 and row['day']==10) or (row['month']==12 and row['day']==24) or (row['month']==1 and row['day'] >= 14 and row['day'] <= 20 and row['weekday'] == 6) or (row['month']==2 and row['day'] >= 14 and row['day'] <= 20 and row['weekday'] == 6) or (row['month']==5 and row['day'] >= 24 and row['day'] <= 30 and row['weekday'] == 6) or ((row['month']==9 and row['day'] >= 1 and row['day'] <= 6) or (row['month']==8 and row['day'] == 31) and row['weekday'] == 6) or (row['month']==10 and row['day'] >= 7 and row['day'] <= 13 and row['weekday'] == 6) or (row['month']==11 and row['day'] >= 21 and row['day'] <= 27 and row['weekday'] == 2) else 0, axis=1)\n df['store_and_fwd_flag'] = df['store_and_fwd_flag'].map(lambda x: 0 if x =='N' else 1)\n df.drop('day', axis=1, inplace=True)",
"_____no_output_____"
],
[
"# Extract features\nprint('Extracting train features')\nextract_features(train)\nprint('Extracting test features')\nextract_features(test)\n\ntrain.head()",
"Extracting train features\nExtracting test features\n"
],
[
"# Prepare data\nX = np.array(train.drop(['id', 'pickup_datetime', 'dropoff_datetime', 'store_and_fwd_flag', 'trip_duration'], axis=1))\ny = np.log(train['trip_duration'].values)\nmedian_trip_duration = np.median(train['trip_duration'].values)\n\nprint('X.shape = ' + str(X.shape))\nprint('y.shape = ' + str(y.shape))\n\nX_test = np.array(test.drop(['id', 'pickup_datetime', 'store_and_fwd_flag'], axis=1))\n\nprint('X_test.shape = ' + str(X_test.shape))",
"X.shape = (1458644, 16)\ny.shape = (1458644,)\nX_test.shape = (625134, 16)\n"
],
[
"#Сделаем скейлинг данных\nfrom sklearn.preprocessing import StandardScaler\nscaler = StandardScaler()\n\nscaler.fit(X)\n#Преобразуем тренировочные данные\nX_scaled = scaler.transform(X)\n#Преобразуем тестовые данные\nX_te_scaled = scaler.transform(X_test)",
"_____no_output_____"
],
[
"\n\n\nprint('Training and making predictions')\n\nparams = {\n 'boosting_type': 'gbdt',\n 'objective': 'regression',\n 'metric': 'rmsle',\n 'max_depth': 6, \n 'learning_rate': 0.4,\n 'num_leaves': 45,\n 'max_bin': 250\n}\n\nn_estimators = 300\nn_iters = 5\npreds_buf = []\nerr_buf = []\nfor i in range(n_iters): #[9, 52, 100, 145, 174, 176, 184]: подборка с 0.38\n x_train, x_valid, y_train, y_valid = train_test_split(X_scaled, y, test_size=0.1, random_state=i) ############\n d_train = lgb.Dataset(x_train, label=y_train)\n d_valid = lgb.Dataset(x_valid, label=y_valid)\n watchlist = [d_valid]\n\n #model = cb.CatBoostRegressor()\n #model.fit(x_train, y_train)\n\n model = lgb.train(params, d_train, n_estimators, watchlist, verbose_eval=1)\n\n preds = model.predict(x_valid)\n preds = np.exp(preds)\n preds[preds < 0] = median_trip_duration\n err = rmsle(np.exp(y_valid), preds)\n err_buf.append(err)\n print(str(i) + ' random_state, ' + ' RMSLE = ' + str(err))\n \n preds = model.predict(X_te_scaled)#################################\n preds = np.exp(preds)\n preds[preds < 0] = median_trip_duration\n preds_buf.append(preds)\n\nprint('Mean RMSLE = ' + str(np.mean(err_buf)) + ' +/- ' + str(np.std(err_buf)))\n# Average predictions\npreds = np.mean(preds_buf, axis=0)",
"Training and making predictions\n0 random_state, RMSLE = 0.39609022303\n1 random_state, RMSLE = 0.392778933156\n2 random_state, RMSLE = 0.392264182691\n3 random_state, RMSLE = 0.397135250209\n4 random_state, RMSLE = 0.39514687773\nMean RMSLE = 0.394683093363 +/- 0.00188069869823\n"
],
[
"# Prepare submission\nsubm = pd.DataFrame()\nsubm['id'] = test.id.values\nsubm['trip_duration'] = preds\nsubm.to_csv('submission_taxi_lgbm.csv', index=False)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0cdfbb107de5fe466814408718a9f4701a3c09e | 1,480 | ipynb | Jupyter Notebook | nbs/_template.ipynb | arampacha/generative_models | 34f5a2fc760bbd7f9f9a956d8d8670c9746e5152 | [
"Apache-2.0"
] | null | null | null | nbs/_template.ipynb | arampacha/generative_models | 34f5a2fc760bbd7f9f9a956d8d8670c9746e5152 | [
"Apache-2.0"
] | null | null | null | nbs/_template.ipynb | arampacha/generative_models | 34f5a2fc760bbd7f9f9a956d8d8670c9746e5152 | [
"Apache-2.0"
] | null | null | null | 17.209302 | 77 | 0.510135 | [
[
[
"# default_exp layers",
"_____no_output_____"
],
[
"#hide\n%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
]
],
[
[
"# Layers\n\n> Common layers, blocks and utils.",
"_____no_output_____"
]
],
[
[
"#hide\nfrom nbdev.export import notebook2script; notebook2script()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0cdff6d06becbc1cd324599c52644776b0c2736 | 21,503 | ipynb | Jupyter Notebook | how-to-use-azureml/automated-machine-learning/remote-execution-with-datastore/auto-ml-remote-execution-with-datastore.ipynb | waith/MachineLearningNotebooks | 23bdf58e998e1a4880c22a87a011d44fac991ba4 | [
"MIT"
] | null | null | null | how-to-use-azureml/automated-machine-learning/remote-execution-with-datastore/auto-ml-remote-execution-with-datastore.ipynb | waith/MachineLearningNotebooks | 23bdf58e998e1a4880c22a87a011d44fac991ba4 | [
"MIT"
] | null | null | null | how-to-use-azureml/automated-machine-learning/remote-execution-with-datastore/auto-ml-remote-execution-with-datastore.ipynb | waith/MachineLearningNotebooks | 23bdf58e998e1a4880c22a87a011d44fac991ba4 | [
"MIT"
] | 1 | 2021-12-27T17:33:08.000Z | 2021-12-27T17:33:08.000Z | 36.261383 | 354 | 0.546807 | [
[
[
"Copyright (c) Microsoft Corporation. All rights reserved.\n\nLicensed under the MIT License.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"# Automated Machine Learning\n_**Remote Execution with DataStore**_\n\n## Contents\n1. [Introduction](#Introduction)\n1. [Setup](#Setup)\n1. [Data](#Data)\n1. [Train](#Train)\n1. [Results](#Results)\n1. [Test](#Test)",
"_____no_output_____"
],
[
"## Introduction\nThis sample accesses a data file on a remote DSVM through DataStore. Advantages of using data store are:\n1. DataStore secures the access details.\n2. DataStore supports read, write to blob and file store\n3. AutoML natively supports copying data from DataStore to DSVM\n\nMake sure you have executed the [configuration](../../../configuration.ipynb) before running this notebook.\n\nIn this notebook you would see\n1. Storing data in DataStore.\n2. get_data returning data from DataStore.",
"_____no_output_____"
],
[
"## Setup\n\nAs part of the setup you have already created a <b>Workspace</b>. For AutoML you would need to create an <b>Experiment</b>. An <b>Experiment</b> is a named object in a <b>Workspace</b>, which is used to run experiments.",
"_____no_output_____"
]
],
[
[
"import logging\nimport os\nimport time\n\nimport numpy as np\nimport pandas as pd\n\nimport azureml.core\nfrom azureml.core.compute import DsvmCompute\nfrom azureml.core.experiment import Experiment\nfrom azureml.core.workspace import Workspace\nfrom azureml.train.automl import AutoMLConfig",
"_____no_output_____"
],
[
"ws = Workspace.from_config()\n\n# choose a name for experiment\nexperiment_name = 'automl-remote-datastore-file'\n# project folder\nproject_folder = './sample_projects/automl-remote-datastore-file'\n\nexperiment=Experiment(ws, experiment_name)\n\noutput = {}\noutput['SDK version'] = azureml.core.VERSION\noutput['Subscription ID'] = ws.subscription_id\noutput['Workspace'] = ws.name\noutput['Resource Group'] = ws.resource_group\noutput['Location'] = ws.location\noutput['Project Directory'] = project_folder\noutput['Experiment Name'] = experiment.name\npd.set_option('display.max_colwidth', -1)\noutputDf = pd.DataFrame(data = output, index = [''])\noutputDf.T",
"_____no_output_____"
]
],
[
[
"### Create a Remote Linux DSVM\nNote: If creation fails with a message about Marketplace purchase eligibilty, go to portal.azure.com, start creating DSVM there, and select \"Want to create programmatically\" to enable programmatic creation. Once you've enabled it, you can exit without actually creating VM.\n\n**Note**: By default SSH runs on port 22 and you don't need to specify it. But if for security reasons you can switch to a different port (such as 5022), you can append the port number to the address. [Read more](https://docs.microsoft.com/en-us/azure/virtual-machines/troubleshooting/detailed-troubleshoot-ssh-connection) on this.",
"_____no_output_____"
]
],
[
[
"compute_target_name = 'mydsvmc'\n\ntry:\n while ws.compute_targets[compute_target_name].provisioning_state == 'Creating':\n time.sleep(1)\n \n dsvm_compute = DsvmCompute(workspace=ws, name=compute_target_name)\n print('found existing:', dsvm_compute.name)\nexcept:\n dsvm_config = DsvmCompute.provisioning_configuration(vm_size=\"Standard_D2_v2\")\n dsvm_compute = DsvmCompute.create(ws, name=compute_target_name, provisioning_configuration=dsvm_config)\n dsvm_compute.wait_for_completion(show_output=True)\n print(\"Waiting one minute for ssh to be accessible\")\n time.sleep(90) # Wait for ssh to be accessible",
"_____no_output_____"
]
],
[
[
"## Data\n\n### Copy data file to local\n\nDownload the data file.\n",
"_____no_output_____"
]
],
[
[
"if not os.path.isdir('data'):\n os.mkdir('data') ",
"_____no_output_____"
],
[
"from sklearn.datasets import fetch_20newsgroups\nimport csv\n\nremove = ('headers', 'footers', 'quotes')\ncategories = [\n 'alt.atheism',\n 'talk.religion.misc',\n 'comp.graphics',\n 'sci.space',\n ]\ndata_train = fetch_20newsgroups(subset = 'train', categories = categories,\n shuffle = True, random_state = 42,\n remove = remove)\n \npd.DataFrame(data_train.data).to_csv(\"data/X_train.tsv\", index=False, header=False, quoting=csv.QUOTE_ALL, sep=\"\\t\")\npd.DataFrame(data_train.target).to_csv(\"data/y_train.tsv\", index=False, header=False, sep=\"\\t\")",
"_____no_output_____"
]
],
[
[
"### Upload data to the cloud",
"_____no_output_____"
],
[
"Now make the data accessible remotely by uploading that data from your local machine into Azure so it can be accessed for remote training. The datastore is a convenient construct associated with your workspace for you to upload/download data, and interact with it from your remote compute targets. It is backed by Azure blob storage account.\n\nThe data.tsv files are uploaded into a directory named data at the root of the datastore.",
"_____no_output_____"
]
],
[
[
"#blob_datastore = Datastore(ws, blob_datastore_name)\nds = ws.get_default_datastore()\nprint(ds.datastore_type, ds.account_name, ds.container_name)",
"_____no_output_____"
],
[
"# ds.upload_files(\"data.tsv\")\nds.upload(src_dir='./data', target_path='data', overwrite=True, show_progress=True)",
"_____no_output_____"
]
],
[
[
"### Configure & Run\n\nFirst let's create a DataReferenceConfigruation object to inform the system what data folder to download to the compute target.\nThe path_on_compute should be an absolute path to ensure that the data files are downloaded only once. The get_data method should use this same path to access the data files.",
"_____no_output_____"
]
],
[
[
"from azureml.core.runconfig import DataReferenceConfiguration\ndr = DataReferenceConfiguration(datastore_name=ds.name, \n path_on_datastore='data', \n path_on_compute='/tmp/azureml_runs',\n mode='download', # download files from datastore to compute target\n overwrite=False)",
"_____no_output_____"
],
[
"from azureml.core.runconfig import RunConfiguration\nfrom azureml.core.conda_dependencies import CondaDependencies\nimport pkg_resources\n\n# create a new RunConfig object\nconda_run_config = RunConfiguration(framework=\"python\")\n\n# Set compute target to the Linux DSVM\nconda_run_config.target = dsvm_compute\n# set the data reference of the run coonfiguration\nconda_run_config.data_references = {ds.name: dr}\n\npandas_dependency = 'pandas==' + pkg_resources.get_distribution(\"pandas\").version\n\ncd = CondaDependencies.create(pip_packages=['azureml-sdk[automl]'], conda_packages=['numpy','py-xgboost<=0.80',pandas_dependency])\nconda_run_config.environment.python.conda_dependencies = cd",
"_____no_output_____"
]
],
[
[
"### Create Get Data File\nFor remote executions you should author a get_data.py file containing a get_data() function. This file should be in the root directory of the project. You can encapsulate code to read data either from a blob storage or local disk in this file.\n\nThe *get_data()* function returns a [dictionary](README.md#getdata).\n\nThe read_csv uses the path_on_compute value specified in the DataReferenceConfiguration call plus the path_on_datastore folder and then the actual file name.",
"_____no_output_____"
]
],
[
[
"if not os.path.exists(project_folder):\n os.makedirs(project_folder)",
"_____no_output_____"
],
[
"%%writefile $project_folder/get_data.py\n\nimport pandas as pd\n\ndef get_data():\n X_train = pd.read_csv(\"/tmp/azureml_runs/data/X_train.tsv\", delimiter=\"\\t\", header=None, quotechar='\"')\n y_train = pd.read_csv(\"/tmp/azureml_runs/data/y_train.tsv\", delimiter=\"\\t\", header=None, quotechar='\"')\n\n return { \"X\" : X_train.values, \"y\" : y_train[0].values }",
"_____no_output_____"
]
],
[
[
"## Train\n\nYou can specify automl_settings as **kwargs** as well. Also note that you can use the get_data() symantic for local excutions too. \n\n<i>Note: For Remote DSVM and Batch AI you cannot pass Numpy arrays directly to AutoMLConfig.</i>\n\n|Property|Description|\n|-|-|\n|**primary_metric**|This is the metric that you want to optimize. Classification supports the following primary metrics: <br><i>accuracy</i><br><i>AUC_weighted</i><br><i>average_precision_score_weighted</i><br><i>norm_macro_recall</i><br><i>precision_score_weighted</i>|\n|**iteration_timeout_minutes**|Time limit in minutes for each iteration|\n|**iterations**|Number of iterations. In each iteration Auto ML trains a specific pipeline with the data|\n|**n_cross_validations**|Number of cross validation splits|\n|**max_concurrent_iterations**|Max number of iterations that would be executed in parallel. This should be less than the number of cores on the DSVM\n|**preprocess**| *True/False* <br>Setting this to *True* enables Auto ML to perform preprocessing <br>on the input to handle *missing data*, and perform some common *feature extraction*|\n|**enable_cache**|Setting this to *True* enables preprocess done once and reuse the same preprocessed data for all the iterations. Default value is True.|\n|**max_cores_per_iteration**| Indicates how many cores on the compute target would be used to train a single pipeline.<br> Default is *1*, you can set it to *-1* to use all cores|",
"_____no_output_____"
]
],
[
[
"automl_settings = {\n \"iteration_timeout_minutes\": 60,\n \"iterations\": 4,\n \"n_cross_validations\": 5,\n \"primary_metric\": 'AUC_weighted',\n \"preprocess\": True,\n \"max_cores_per_iteration\": 1,\n \"verbosity\": logging.INFO\n}\nautoml_config = AutoMLConfig(task = 'classification',\n debug_log = 'automl_errors.log',\n path=project_folder,\n run_configuration=conda_run_config,\n #compute_target = dsvm_compute,\n data_script = project_folder + \"/get_data.py\",\n **automl_settings\n )",
"_____no_output_____"
]
],
[
[
"For remote runs the execution is asynchronous, so you will see the iterations get populated as they complete. You can interact with the widgets/models even when the experiment is running to retreive the best model up to that point. Once you are satisfied with the model you can cancel a particular iteration or the whole run.",
"_____no_output_____"
]
],
[
[
"remote_run = experiment.submit(automl_config, show_output=False)",
"_____no_output_____"
],
[
"remote_run",
"_____no_output_____"
]
],
[
[
"## Results\n#### Widget for monitoring runs\n\nThe widget will sit on \"loading\" until the first iteration completed, then you will see an auto-updating graph and table show up. It refreshed once per minute, so you should see the graph update as child runs complete.\n\nYou can click on a pipeline to see run properties and output logs. Logs are also available on the DSVM under /tmp/azureml_run/{iterationid}/azureml-logs\n\nNOTE: The widget displays a link at the bottom. This links to a web-ui to explore the individual run details.",
"_____no_output_____"
]
],
[
[
"from azureml.widgets import RunDetails\nRunDetails(remote_run).show() ",
"_____no_output_____"
],
[
"# Wait until the run finishes.\nremote_run.wait_for_completion(show_output = True)",
"_____no_output_____"
]
],
[
[
"\n#### Retrieve All Child Runs\nYou can also use sdk methods to fetch all the child runs and see individual metrics that we log. ",
"_____no_output_____"
]
],
[
[
"children = list(remote_run.get_children())\nmetricslist = {}\nfor run in children:\n properties = run.get_properties()\n metrics = {k: v for k, v in run.get_metrics().items() if isinstance(v, float)} \n metricslist[int(properties['iteration'])] = metrics\n\nrundata = pd.DataFrame(metricslist).sort_index(1)\nrundata",
"_____no_output_____"
]
],
[
[
"### Canceling Runs\nYou can cancel ongoing remote runs using the *cancel()* and *cancel_iteration()* functions",
"_____no_output_____"
]
],
[
[
"# Cancel the ongoing experiment and stop scheduling new iterations\n# remote_run.cancel()\n\n# Cancel iteration 1 and move onto iteration 2\n# remote_run.cancel_iteration(1)",
"_____no_output_____"
]
],
[
[
"### Pre-process cache cleanup\nThe preprocess data gets cache at user default file store. When the run is completed the cache can be cleaned by running below cell",
"_____no_output_____"
]
],
[
[
"remote_run.clean_preprocessor_cache()",
"_____no_output_____"
]
],
[
[
"### Retrieve the Best Model\n\nBelow we select the best pipeline from our iterations. The *get_output* method returns the best run and the fitted model. There are overloads on *get_output* that allow you to retrieve the best run and fitted model for *any* logged metric or a particular *iteration*.",
"_____no_output_____"
]
],
[
[
"best_run, fitted_model = remote_run.get_output()",
"_____no_output_____"
]
],
[
[
"#### Best Model based on any other metric",
"_____no_output_____"
]
],
[
[
"# lookup_metric = \"accuracy\"\n# best_run, fitted_model = remote_run.get_output(metric=lookup_metric)",
"_____no_output_____"
]
],
[
[
"#### Model from a specific iteration",
"_____no_output_____"
]
],
[
[
"# iteration = 1\n# best_run, fitted_model = remote_run.get_output(iteration=iteration)",
"_____no_output_____"
]
],
[
[
"## Test\n",
"_____no_output_____"
]
],
[
[
"# Load test data.\nfrom pandas_ml import ConfusionMatrix\n\ndata_test = fetch_20newsgroups(subset = 'test', categories = categories,\n shuffle = True, random_state = 42,\n remove = remove)\n\nX_test = np.array(data_test.data).reshape((len(data_test.data),1))\ny_test = data_test.target\n\n# Test our best pipeline.\n\ny_pred = fitted_model.predict(X_test)\ny_pred_strings = [data_test.target_names[i] for i in y_pred]\ny_test_strings = [data_test.target_names[i] for i in y_test]\n\ncm = ConfusionMatrix(y_test_strings, y_pred_strings)\nprint(cm)\ncm.plot()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0ce0903242e50ee4bdeaac7b56655014670e036 | 43,163 | ipynb | Jupyter Notebook | Course 4 - Natural Language Processing with Attention Models/Week 3/.ipynb_checkpoints/C4_W3_Assignment-checkpoint.ipynb | TRoboto/Natural-Language-Processing-Specialization | 9c22e94738fac49dcb02859a837e5b0dddd99d41 | [
"MIT"
] | 362 | 2020-10-08T07:34:25.000Z | 2022-03-30T05:11:30.000Z | 4. Natural Language Processing with Attention Models/Week 3 Question-Answering with Transformer(BERT) Models/Assignment Question Answering .ipynb | aqafridi/Natural-Language-Processing | b8a327b94034eadd7140df935fab33f815afe0f1 | [
"MIT"
] | 7 | 2020-07-07T16:10:23.000Z | 2021-06-04T08:17:55.000Z | Course 4 - Natural Language Processing with Attention Models/Week 3/.ipynb_checkpoints/C4_W3_Assignment-checkpoint.ipynb | TRoboto/Natural-Language-Processing-Specialization | 9c22e94738fac49dcb02859a837e5b0dddd99d41 | [
"MIT"
] | 238 | 2020-10-08T12:01:31.000Z | 2022-03-25T08:10:42.000Z | 35.177669 | 784 | 0.551097 | [
[
[
"# Assignment 3: Question Answering\n\nWelcome to this week's assignment of course 4. In this you will explore question answering. You will implement the \"Text to Text Transfer from Transformers\" (better known as T5). Since you implemented transformers from scratch last week you will now be able to use them. \n\n<img src = \"qa.png\"> ",
"_____no_output_____"
],
[
"## Outline\n\n- [Overview](#0)\n- [Part 0: Importing the Packages](#0)\n- [Part 1: C4 Dataset](#1)\n - [1.1 Pre-Training Objective](#1.1)\n - [1.2 Process C4](#1.2)\n - [1.2.1 Decode to natural language](#1.2.1)\n - [1.3 Tokenizing and Masking](#1.3)\n - [Exercise 01](#ex01)\n - [1.4 Creating the Pairs](#1.4)\n- [Part 2: Transfomer](#2)\n - [2.1 Transformer Encoder](#2.1)\n - [2.1.1 The Feedforward Block](#2.1.1)\n - [Exercise 02](#ex02)\n - [2.1.2 The Encoder Block](#2.1.2)\n - [Exercise 03](#ex03)\n - [2.1.3 The Transformer Encoder](#2.1.3) \n - [Exercise 04](#ex04)",
"_____no_output_____"
],
[
"<a name='0'></a>\n### Overview\n\nThis assignment will be different from the two previous ones. Due to memory and time constraints of this environment you will not be able to train a model and use it for inference. Instead you will create the necessary building blocks for the transformer encoder model and will use a pretrained version of the same model in two ungraded labs after this assignment.\n\nAfter completing these 3 (1 graded and 2 ungraded) labs you will:\n* Implement the code neccesary for Bidirectional Encoder Representation from Transformer (BERT).\n* Understand how the C4 dataset is structured.\n* Use a pretrained model for inference.\n* Understand how the \"Text to Text Transfer from Transformers\" or T5 model works. ",
"_____no_output_____"
],
[
"<a name='0'></a>\n# Part 0: Importing the Packages",
"_____no_output_____"
]
],
[
[
"import ast\nimport string\nimport textwrap\nimport itertools\nimport numpy as np\n\nimport trax \nfrom trax import layers as tl\nfrom trax.supervised import decoding\n\n# Will come handy later.\nwrapper = textwrap.TextWrapper(width=70)\n\n# Set random seed\nnp.random.seed(42)",
"_____no_output_____"
]
],
[
[
"<a name='1'></a>\n## Part 1: C4 Dataset\n\nThe [C4](https://www.tensorflow.org/datasets/catalog/c4) is a huge data set. For the purpose of this assignment you will use a few examples out of it which are present in `data.txt`. C4 is based on the [common crawl](https://commoncrawl.org/) project. Feel free to read more on their website. \n\nRun the cell below to see how the examples look like. ",
"_____no_output_____"
]
],
[
[
"# load example jsons\nexample_jsons = list(map(ast.literal_eval, open('data.txt')))",
"_____no_output_____"
],
[
"# Printing the examples to see how the data looks like\nfor i in range(5):\n print(f'example number {i+1}: \\n\\n{example_jsons[i]} \\n')",
"_____no_output_____"
]
],
[
[
"Notice the `b` before each string? This means that this data comes as bytes rather than strings. Strings are actually lists of bytes so for the rest of the assignments the name `strings` will be used to describe the data. \n\nTo check this run the following cell:",
"_____no_output_____"
]
],
[
[
"type(example_jsons[0].get('text'))",
"_____no_output_____"
]
],
[
[
"<a name='1.1'></a>\n### 1.1 Pre-Training Objective\n\n**Note:** The word \"mask\" will be used throughout this assignment in context of hiding/removing word(s)\n\nYou will be implementing the BERT loss as shown in the following image. \n\n<img src = \"loss.png\" width=\"600\" height = \"400\">\n\nAssume you have the following text: <span style = \"color:blue\"> **Thank you <span style = \"color:red\">for inviting </span> me to your party <span style = \"color:red\">last</span> week** </span> \n\n\nNow as input you will mask the words in red in the text: \n\n<span style = \"color:blue\"> **Input:**</span> Thank you **X** me to your party **Y** week.\n\n<span style = \"color:blue\">**Output:**</span> The model should predict the words(s) for **X** and **Y**. \n\n**Z** is used to represent the end.",
"_____no_output_____"
],
[
"<a name='1.2'></a>\n### 1.2 Process C4\n\nC4 only has the plain string `text` field, so you will tokenize and have `inputs` and `targets` out of it for supervised learning. Given your inputs, the goal is to predict the targets during training. \n\nYou will now take the `text` and convert it to `inputs` and `targets`.",
"_____no_output_____"
]
],
[
[
"# Grab text field from dictionary\nnatural_language_texts = [example_json['text'] for example_json in example_jsons]",
"_____no_output_____"
],
[
"# First text example\nnatural_language_texts[4]",
"_____no_output_____"
]
],
[
[
"<a name='1.2.1'></a>\n#### 1.2.1 Decode to natural language\n\nThe following functions will help you `detokenize` and`tokenize` the text data. \n\nThe `sentencepiece` vocabulary was used to convert from text to ids. This vocabulary file is loaded and used in this helper functions.\n\n`natural_language_texts` has the text from the examples we gave you. \n\nRun the cells below to see what is going on. ",
"_____no_output_____"
]
],
[
[
"# Special tokens\nPAD, EOS, UNK = 0, 1, 2\n\ndef detokenize(np_array):\n return trax.data.detokenize(\n np_array,\n vocab_type='sentencepiece',\n vocab_file='sentencepiece.model',\n vocab_dir='.')\n\ndef tokenize(s):\n # The trax.data.tokenize function operates on streams,\n # that's why we have to create 1-element stream with iter\n # and later retrieve the result with next.\n return next(trax.data.tokenize(\n iter([s]),\n vocab_type='sentencepiece',\n vocab_file='sentencepiece.model',\n vocab_dir='.'))",
"_____no_output_____"
],
[
"# printing the encoding of each word to see how subwords are tokenized\ntokenized_text = [(tokenize(word).tolist(), word) for word in natural_language_texts[0].split()]\nprint(tokenized_text, '\\n')",
"_____no_output_____"
],
[
"# We can see that detokenize successfully undoes the tokenization\nprint(f\"tokenized: {tokenize('Beginners')}\\ndetokenized: {detokenize(tokenize('Beginners'))}\")",
"_____no_output_____"
]
],
[
[
"As you can see above, you were able to take a piece of string and tokenize it. \n\nNow you will create `input` and `target` pairs that will allow you to train your model. T5 uses the ids at the end of the vocab file as sentinels. For example, it will replace: \n - `vocab_size - 1` by `<Z>`\n - `vocab_size - 2` by `<Y>`\n - and so forth. \n \nIt assigns every word a `chr`.\n\nThe `pretty_decode` function below, which you will use in a bit, helps in handling the type when decoding. Take a look and try to understand what the function is doing.\n\n\nNotice that:\n```python\nstring.ascii_letters = 'abcdefghijklmnopqrstuvwxyzABCDEFGHIJKLMNOPQRSTUVWXYZ'\n```\n\n**NOTE:** Targets may have more than the 52 sentinels we replace, but this is just to give you an idea of things.",
"_____no_output_____"
]
],
[
[
"vocab_size = trax.data.vocab_size(\n vocab_type='sentencepiece',\n vocab_file='sentencepiece.model',\n vocab_dir='.')\n\ndef get_sentinels(vocab_size=vocab_size, display=False):\n sentinels = {}\n for i, char in enumerate(reversed(string.ascii_letters), 1):\n decoded_text = detokenize([vocab_size - i]) \n \n # Sentinels, ex: <Z> - <a>\n sentinels[decoded_text] = f'<{char}>' \n \n if display:\n print(f'The sentinel is <{char}> and the decoded token is:', decoded_text)\n\n return sentinels",
"_____no_output_____"
],
[
"sentinels = get_sentinels(vocab_size, display=True)",
"_____no_output_____"
],
[
"def pretty_decode(encoded_str_list, sentinels=sentinels):\n # If already a string, just do the replacements.\n if isinstance(encoded_str_list, (str, bytes)):\n for token, char in sentinels.items():\n encoded_str_list = encoded_str_list.replace(token, char)\n return encoded_str_list\n \n # We need to decode and then prettyfy it.\n return pretty_decode(detokenize(encoded_str_list))",
"_____no_output_____"
],
[
"pretty_decode(\"I want to dress up as an Intellectual this halloween.\")",
"_____no_output_____"
]
],
[
[
"The functions above make your `inputs` and `targets` more readable. For example, you might see something like this once you implement the masking function below. \n\n- <span style=\"color:red\"> Input sentence: </span> Younes and Lukasz were working together in the lab yesterday after lunch. \n- <span style=\"color:red\">Input: </span> Younes and Lukasz **Z** together in the **Y** yesterday after lunch.\n- <span style=\"color:red\">Target: </span> **Z** were working **Y** lab.\n",
"_____no_output_____"
],
[
"<a name='1.3'></a>\n### 1.3 Tokenizing and Masking\n\nYou will now implement the `tokenize_and_mask` function. This function will allow you to tokenize and mask input words with a noise probability. We usually mask 15% of the words.",
"_____no_output_____"
],
[
"<a name='ex01'></a>\n### Exercise 01",
"_____no_output_____"
]
],
[
[
"# UNQ_C1\n# GRADED FUNCTION: tokenize_and_mask\ndef tokenize_and_mask(text, vocab_size=vocab_size, noise=0.15, \n randomizer=np.random.uniform, tokenize=tokenize):\n \"\"\"Tokenizes and masks a given input.\n\n Args:\n text (str or bytes): Text input.\n vocab_size (int, optional): Size of the vocabulary. Defaults to vocab_size.\n noise (float, optional): Probability of masking a token. Defaults to 0.15.\n randomizer (function, optional): Function that generates random values. Defaults to np.random.uniform.\n tokenize (function, optional): Tokenizer function. Defaults to tokenize.\n\n Returns:\n tuple: Tuple of lists of integers associated to inputs and targets.\n \"\"\"\n \n # current sentinel number (starts at 0)\n cur_sentinel_num = 0\n # inputs\n inps = []\n # targets\n targs = []\n \n ### START CODE HERE (REPLACE INSTANCES OF 'None' WITH YOUR CODE) ###\n \n # prev_no_mask is True if the previous token was NOT masked, False otherwise\n # set prev_no_mask to True\n prev_no_mask = None\n \n # loop through tokenized `text`\n for token in tokenize(text):\n # check if the `noise` is greater than a random value (weighted coin flip)\n if randomizer() < noise:\n # check to see if the previous token was not masked\n if prev_no_mask==True: # add new masked token at end_id\n # number of masked tokens increases by 1\n cur_sentinel_num += None\n # compute `end_id` by subtracting current sentinel value out of the total vocabulary size\n end_id = None - None\n # append `end_id` at the end of the targets\n targs.append(None)\n # append `end_id` at the end of the inputs\n inps.append(None)\n # append `token` at the end of the targets\n targs.append(None)\n # set prev_no_mask accordingly\n prev_no_mask = None\n \n else: # don't have two masked tokens in a row\n # append `token ` at the end of the inputs\n inps.append(None)\n # set prev_no_mask accordingly\n prev_no_mask = None\n \n ### END CODE HERE ###\n \n return inps, targs",
"_____no_output_____"
],
[
"# Some logic to mock a np.random value generator\n# Needs to be in the same cell for it to always generate same output\ndef testing_rnd():\n def dummy_generator():\n vals = np.linspace(0, 1, 10)\n cyclic_vals = itertools.cycle(vals)\n for _ in range(100):\n yield next(cyclic_vals)\n\n dumr = itertools.cycle(dummy_generator())\n\n def dummy_randomizer():\n return next(dumr)\n \n return dummy_randomizer\n\ninput_str = natural_language_texts[0]\nprint(f\"input string:\\n\\n{input_str}\\n\")\ninps, targs = tokenize_and_mask(input_str, randomizer=testing_rnd())\nprint(f\"tokenized inputs:\\n\\n{inps}\\n\")\nprint(f\"targets:\\n\\n{targs}\")",
"_____no_output_____"
]
],
[
[
"#### **Expected Output:**\n```CPP\nb'Beginners BBQ Class Taking Place in Missoula!\\nDo you want to get better at making delicious BBQ? You will have the opportunity, put this on your calendar now. Thursday, September 22nd join World Class BBQ Champion, Tony Balay from Lonestar Smoke Rangers. He will be teaching a beginner level class for everyone who wants to get better with their culinary skills.\\nHe will teach you everything you need to know to compete in a KCBS BBQ competition, including techniques, recipes, timelines, meat selection and trimming, plus smoker and fire information.\\nThe cost to be in the class is $35 per person, and for spectators it is free. Included in the cost will be either a t-shirt or apron and you will be tasting samples of each meat that is prepared.'\n\ntokenized inputs:\n\n[31999, 15068, 4501, 3, 12297, 3399, 16, 5964, 7115, 31998, 531, 25, 241, 12, 129, 394, 44, 492, 31997, 58, 148, 56, 43, 8, 1004, 6, 474, 31996, 39, 4793, 230, 5, 2721, 6, 1600, 1630, 31995, 1150, 4501, 15068, 16127, 6, 9137, 2659, 5595, 31994, 782, 3624, 14627, 15, 12612, 277, 5, 216, 31993, 2119, 3, 9, 19529, 593, 853, 21, 921, 31992, 12, 129, 394, 28, 70, 17712, 1098, 5, 31991, 3884, 25, 762, 25, 174, 12, 214, 12, 31990, 3, 9, 3, 23405, 4547, 15068, 2259, 6, 31989, 6, 5459, 6, 13618, 7, 6, 3604, 1801, 31988, 6, 303, 24190, 11, 1472, 251, 5, 37, 31987, 36, 16, 8, 853, 19, 25264, 399, 568, 31986, 21, 21380, 7, 34, 19, 339, 5, 15746, 31985, 8, 583, 56, 36, 893, 3, 9, 3, 31984, 9486, 42, 3, 9, 1409, 29, 11, 25, 31983, 12246, 5977, 13, 284, 3604, 24, 19, 2657, 31982]\n\ntargets:\n\n[31999, 12847, 277, 31998, 9, 55, 31997, 3326, 15068, 31996, 48, 30, 31995, 727, 1715, 31994, 45, 301, 31993, 56, 36, 31992, 113, 2746, 31991, 216, 56, 31990, 5978, 16, 31989, 379, 2097, 31988, 11, 27856, 31987, 583, 12, 31986, 6, 11, 31985, 26, 16, 31984, 17, 18, 31983, 56, 36, 31982, 5]\n```",
"_____no_output_____"
],
[
"You will now use the inputs and the targets from the `tokenize_and_mask` function you implemented above. Take a look at the masked sentence using your `inps` and `targs` from the sentence above. ",
"_____no_output_____"
]
],
[
[
"print('Inputs: \\n\\n', pretty_decode(inps))\nprint('\\nTargets: \\n\\n', pretty_decode(targs))",
"_____no_output_____"
]
],
[
[
"<a name='1.4'></a>\n### 1.4 Creating the Pairs\n\nYou will now create pairs using your dataset. You will iterate over your data and create (inp, targ) pairs using the functions that we have given you. ",
"_____no_output_____"
]
],
[
[
"# Apply tokenize_and_mask\ninputs_targets_pairs = [tokenize_and_mask(text) for text in natural_language_texts]",
"_____no_output_____"
],
[
"def display_input_target_pairs(inputs_targets_pairs):\n for i, inp_tgt_pair in enumerate(inputs_targets_pairs, 1):\n inps, tgts = inp_tgt_pair\n inps, tgts = pretty_decode(inps), pretty_decode(tgts)\n print(f'[{i}]\\n\\n'\n f'inputs:\\n{wrapper.fill(text=inps)}\\n\\n'\n f'targets:\\n{wrapper.fill(text=tgts)}\\n\\n\\n\\n')",
"_____no_output_____"
],
[
"display_input_target_pairs(inputs_targets_pairs)",
"_____no_output_____"
]
],
[
[
"<a name='2'></a>\n# Part 2: Transfomer\n\nWe now load a Transformer model checkpoint that has been pre-trained using the above C4 dataset and decode from it. This will save you a lot of time rather than have to train your model yourself. Later in this notebook, we will show you how to fine-tune your model.\n\n<img src = \"fulltransformer.png\" width=\"300\" height=\"600\">\n\nStart by loading in the model. We copy the checkpoint to local dir for speed, otherwise initialization takes a very long time. Last week you implemented the decoder part for the transformer. Now you will implement the encoder part. Concretely you will implement the following. \n\n\n<img src = \"encoder.png\" width=\"300\" height=\"600\">\n\n",
"_____no_output_____"
],
[
"<a name='2.1'></a>\n### 2.1 Transformer Encoder\n\nYou will now implement the transformer encoder. Concretely you will implement two functions. The first function is `FeedForwardBlock`.\n\n<a name='2.1.1'></a>\n#### 2.1.1 The Feedforward Block\n\nThe `FeedForwardBlock` function is an important one so you will start by implementing it. To do so, you need to return a list of the following: \n\n- [`tl.LayerNorm()`](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.normalization.LayerNorm) = layer normalization.\n- [`tl.Dense(d_ff)`](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.core.Dense) = fully connected layer.\n- [`activation`](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.activation_fns.Relu) = activation relu, tanh, sigmoid etc. \n- `dropout_middle` = we gave you this function (don't worry about its implementation).\n- [`tl.Dense(d_model)`](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.core.Dense) = fully connected layer with same dimension as the model.\n- `dropout_final` = we gave you this function (don't worry about its implementation).\n\nYou can always take a look at [trax documentation](https://trax-ml.readthedocs.io/en/latest/) if needed.\n\n**Instructions**: Implement the feedforward part of the transformer. You will be returning a list. ",
"_____no_output_____"
],
[
"<a name='ex02'></a>\n### Exercise 02",
"_____no_output_____"
]
],
[
[
"# UNQ_C2\n# GRADED FUNCTION: FeedForwardBlock\ndef FeedForwardBlock(d_model, d_ff, dropout, dropout_shared_axes, mode, activation):\n \"\"\"Returns a list of layers implementing a feed-forward block.\n Args:\n d_model: int: depth of embedding\n d_ff: int: depth of feed-forward layer\n dropout: float: dropout rate (how much to drop out)\n dropout_shared_axes: list of integers, axes to share dropout mask\n mode: str: 'train' or 'eval'\n activation: the non-linearity in feed-forward layer\n Returns:\n A list of layers which maps vectors to vectors.\n \"\"\"\n \n dropout_middle = tl.Dropout(rate=dropout,\n shared_axes=dropout_shared_axes, \n mode=mode)\n \n dropout_final = tl.Dropout(rate=dropout, \n shared_axes=dropout_shared_axes, \n mode=mode)\n\n ### START CODE HERE (REPLACE INSTANCES OF 'None' WITH YOUR CODE) ###\n \n ff_block = [ \n # trax Layer normalization \n None,\n # trax Dense layer using `d_ff`\n None,\n # activation() layer - you need to call (use parentheses) this func!\n None,\n # dropout middle layer\n None,\n # trax Dense layer using `d_model`\n None,\n # dropout final layer\n None,\n ]\n \n ### END CODE HERE ###\n \n return ff_block",
"_____no_output_____"
],
[
"# Print the block layout\nfeed_forward_example = FeedForwardBlock(d_model=512, d_ff=2048, dropout=0.8, dropout_shared_axes=0, mode = 'train', activation = tl.Relu)\nprint(feed_forward_example)",
"_____no_output_____"
]
],
[
[
"#### **Expected Output:**\n```CPP\n [LayerNorm, Dense_2048, Relu, Dropout, Dense_512, Dropout]\n```",
"_____no_output_____"
],
[
"<a name='2.1.2'></a>\n#### 2.1.2 The Encoder Block\n\nThe encoder block will use the `FeedForwardBlock`. \n\nYou will have to build two residual connections. Inside the first residual connection you will have the `tl.layerNorm()`, `attention`, and `dropout_` layers. The second residual connection will have the `feed_forward`. \n\nYou will also need to implement `feed_forward`, `attention` and `dropout_` blocks. \n\nSo far you haven't seen the [`tl.Attention()`](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.attention.Attention) and [`tl.Residual()`](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.combinators.Residual) layers so you can check the docs by clicking on them.",
"_____no_output_____"
],
[
"<a name='ex03'></a>\n### Exercise 03",
"_____no_output_____"
]
],
[
[
"# UNQ_C3\n# GRADED FUNCTION: EncoderBlock\ndef EncoderBlock(d_model, d_ff, n_heads, dropout, dropout_shared_axes,\n mode, ff_activation, FeedForwardBlock=FeedForwardBlock):\n \"\"\"\n Returns a list of layers that implements a Transformer encoder block.\n The input to the layer is a pair, (activations, mask), where the mask was\n created from the original source tokens to prevent attending to the padding\n part of the input.\n \n Args:\n d_model (int): depth of embedding.\n d_ff (int): depth of feed-forward layer.\n n_heads (int): number of attention heads.\n dropout (float): dropout rate (how much to drop out).\n dropout_shared_axes (int): axes on which to share dropout mask.\n mode (str): 'train' or 'eval'.\n ff_activation (function): the non-linearity in feed-forward layer.\n FeedForwardBlock (function): A function that returns the feed forward block.\n Returns:\n list: A list of layers that maps (activations, mask) to (activations, mask).\n \n \"\"\"\n \n ### START CODE HERE (REPLACE INSTANCES OF 'None' WITH YOUR CODE) ###\n \n # Attention block\n attention = tl.Attention( \n # Use dimension of the model\n d_feature=None,\n # Set it equal to number of attention heads\n n_heads=None,\n # Set it equal `dropout`\n dropout=None,\n # Set it equal `mode`\n mode=None\n )\n \n # Call the function `FeedForwardBlock` (implemented before) and pass in the parameters\n feed_forward = FeedForwardBlock( \n None,\n None,\n None,\n None,\n None,\n None\n )\n \n # Dropout block\n dropout_ = tl.Dropout( \n # set it equal to `dropout`\n rate=None,\n # set it equal to the axes on which to share dropout mask\n shared_axes=None,\n # set it equal to `mode`\n mode=None\n )\n \n encoder_block = [ \n # add `Residual` layer\n tl.Residual(\n # add norm layer\n None,\n # add attention\n None,\n # add dropout\n None,\n ),\n # add another `Residual` layer\n tl.Residual(\n # add feed forward\n None,\n ),\n ]\n \n ### END CODE HERE ###\n \n return encoder_block",
"_____no_output_____"
],
[
"# Print the block layout\nencoder_example = EncoderBlock(d_model=512, d_ff=2048, n_heads=6, dropout=0.8, dropout_shared_axes=0, mode = 'train', ff_activation=tl.Relu)\nprint(encoder_example)",
"_____no_output_____"
]
],
[
[
"#### **Expected Output:**\n```CPP\n[Serial_in2_out2[\n Branch_in2_out3[\n None\n Serial_in2_out2[\n LayerNorm\n Serial_in2_out2[\n Dup_out2\n Dup_out2\n Serial_in4_out2[\n Parallel_in3_out3[\n Dense_512\n Dense_512\n Dense_512\n ]\n PureAttention_in4_out2\n Dense_512\n ]\n ]\n Dropout\n ]\n ]\n Add_in2\n], Serial[\n Branch_out2[\n None\n Serial[\n LayerNorm\n Dense_2048\n Relu\n Dropout\n Dense_512\n Dropout\n ]\n ]\n Add_in2\n]]\n```",
"_____no_output_____"
],
[
"<a name='2.1.3'></a>\n### 2.1.3 The Transformer Encoder\n\nNow that you have implemented the `EncoderBlock`, it is time to build the full encoder. BERT, or Bidirectional Encoder Representations from Transformers is one such encoder. \n\nYou will implement its core code in the function below by using the functions you have coded so far. \n\nThe model takes in many hyperparameters, such as the `vocab_size`, the number of classes, the dimension of your model, etc. You want to build a generic function that will take in many parameters, so you can use it later. At the end of the day, anyone can just load in an API and call transformer, but we think it is important to make sure you understand how it is built. Let's get started. \n\n**Instructions:** For this encoder you will need a `positional_encoder` first (which is already provided) followed by `n_layers` encoder blocks, which are the same encoder blocks you previously built. Once you store the `n_layers` `EncoderBlock` in a list, you are going to encode a `Serial` layer with the following sublayers: \n\n- [`tl.Branch`](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.combinators.Branch): helps with the branching and has the following sublayers:\n - `positional_encoder`.\n - [`tl.PaddingMask()`](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.attention.PaddingMask): layer that maps integer sequences to padding masks.\n- Your list of `EncoderBlock`s\n- [`tl.Select([0], n_in=2)`](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.combinators.Select): Copies, reorders, or deletes stack elements according to indices.\n- [`tl.LayerNorm()`](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.normalization.LayerNorm).\n- [`tl.Mean()`](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.core.Mean): Mean along the first axis.\n- `tl.Dense()` with n_units set to n_classes. \n- `tl.LogSoftmax()` \n\nPlease refer to the [trax documentation](https://trax-ml.readthedocs.io/en/latest/) for further information. ",
"_____no_output_____"
],
[
"<a name='ex04'></a>\n### Exercise 04",
"_____no_output_____"
]
],
[
[
"# UNQ_C4\n# GRADED FUNCTION: TransformerEncoder\ndef TransformerEncoder(vocab_size=vocab_size,\n n_classes=10,\n d_model=512,\n d_ff=2048,\n n_layers=6,\n n_heads=8,\n dropout=0.1,\n dropout_shared_axes=None,\n max_len=2048,\n mode='train',\n ff_activation=tl.Relu,\n EncoderBlock=EncoderBlock):\n \n \"\"\"\n Returns a Transformer encoder model.\n The input to the model is a tensor of tokens.\n \n Args:\n vocab_size (int): vocab size. Defaults to vocab_size.\n n_classes (int): how many classes on output. Defaults to 10.\n d_model (int): depth of embedding. Defaults to 512.\n d_ff (int): depth of feed-forward layer. Defaults to 2048.\n n_layers (int): number of encoder/decoder layers. Defaults to 6.\n n_heads (int): number of attention heads. Defaults to 8.\n dropout (float): dropout rate (how much to drop out). Defaults to 0.1.\n dropout_shared_axes (int): axes on which to share dropout mask. Defaults to None.\n max_len (int): maximum symbol length for positional encoding. Defaults to 2048.\n mode (str): 'train' or 'eval'. Defaults to 'train'.\n ff_activation (function): the non-linearity in feed-forward layer. Defaults to tl.Relu.\n EncoderBlock (function): Returns the encoder block. Defaults to EncoderBlock.\n \n Returns:\n trax.layers.combinators.Serial: A Transformer model as a layer that maps\n from a tensor of tokens to activations over a set of output classes.\n \"\"\"\n \n positional_encoder = [\n tl.Embedding(vocab_size, d_model),\n tl.Dropout(rate=dropout, shared_axes=dropout_shared_axes, mode=mode),\n tl.PositionalEncoding(max_len=max_len)\n ]\n \n ### START CODE HERE (REPLACE INSTANCES OF 'None' WITH YOUR CODE) ###\n \n # Use the function `EncoderBlock` (implemented above) and pass in the parameters over `n_layers`\n encoder_blocks = [None for _ in range(None)]\n\n # Assemble and return the model.\n return tl.Serial(\n # Encode\n tl.Branch(\n # Use `positional_encoder`\n None,\n # Use trax padding mask\n None,\n ),\n # Use `encoder_blocks`\n None,\n # Use select layer\n None,\n # Use trax layer normalization\n None,\n # Map to output categories.\n # Use trax mean. set axis to 1\n None,\n # Use trax Dense using `n_classes`\n None,\n # Use trax log softmax\n None,\n )\n\n ### END CODE HERE ###",
"_____no_output_____"
],
[
"# Run this cell to see the structure of your model\n# Only 1 layer is used to keep the output readable\nTransformerEncoder(n_layers=1)",
"_____no_output_____"
]
],
[
[
"#### **Expected Output:**\n```CPP\nSerial[\n Branch_out2[\n [Embedding_32000_512, Dropout, PositionalEncoding]\n PaddingMask(0)\n ]\n Serial_in2_out2[\n Branch_in2_out3[\n None\n Serial_in2_out2[\n LayerNorm\n Serial_in2_out2[\n Dup_out2\n Dup_out2\n Serial_in4_out2[\n Parallel_in3_out3[\n Dense_512\n Dense_512\n Dense_512\n ]\n PureAttention_in4_out2\n Dense_512\n ]\n ]\n Dropout\n ]\n ]\n Add_in2\n ]\n Serial[\n Branch_out2[\n None\n Serial[\n LayerNorm\n Dense_2048\n Relu\n Dropout\n Dense_512\n Dropout\n ]\n ]\n Add_in2\n ]\n Select[0]_in2\n LayerNorm\n Mean\n Dense_10\n LogSoftmax\n]\n```",
"_____no_output_____"
],
[
"**NOTE Congratulations! You have completed all of the graded functions of this assignment.** Since the rest of the assignment takes a lot of time and memory to run we are providing some extra ungraded labs for you to see this model in action.\n\n**Keep it up!**\n\nTo see this model in action continue to the next 2 ungraded labs. **We strongly recommend you to try the colab versions of them as they will yield a much smoother experience.** The links to the colabs can be found within the ungraded labs or if you already know how to open files within colab here are some shortcuts (if not, head to the ungraded labs which contain some extra instructions):\n\n[BERT Loss Model Colab](https://drive.google.com/file/d/1EHAbMnW6u-GqYWh5r3Z8uLbz4KNpKOAv/view?usp=sharing)\n\n[T5 SQuAD Model Colab](https://drive.google.com/file/d/1c-8KJkTySRGqCx_JjwjvXuRBTNTqEE0N/view?usp=sharing)\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
d0ce237a259aa5c7b1adf29d4d40baaf2ac40ee3 | 10,312 | ipynb | Jupyter Notebook | dataset_torch.ipynb | Pernilleww/recsys_slates_dataset | f490972d9f9f5e210a545c2bae6c8162715632e8 | [
"Apache-2.0"
] | 32 | 2021-07-09T10:33:57.000Z | 2022-02-26T12:17:37.000Z | dataset_torch.ipynb | Pernilleww/recsys_slates_dataset | f490972d9f9f5e210a545c2bae6c8162715632e8 | [
"Apache-2.0"
] | 5 | 2021-07-01T12:59:47.000Z | 2022-02-26T12:10:45.000Z | dataset_torch.ipynb | Pernilleww/recsys_slates_dataset | f490972d9f9f5e210a545c2bae6c8162715632e8 | [
"Apache-2.0"
] | 3 | 2021-07-01T10:06:18.000Z | 2021-12-15T12:03:49.000Z | 45.427313 | 296 | 0.577095 | [
[
[
"#default_exp dataset_torch",
"_____no_output_____"
]
],
[
[
"# dataset_torch\n\n> Module to load the slates dataset into a Pytorch Dataset and Dataloaders with default train/valid test splits.",
"_____no_output_____"
]
],
[
[
"#export\nimport torch\nimport recsys_slates_dataset.data_helper as data_helper\nfrom torch.utils.data import Dataset, DataLoader\nimport torch\nimport json\nimport numpy as np\nimport logging\nlogging.basicConfig(format='%(asctime)s %(message)s', level='INFO')\n\nclass SequentialDataset(Dataset):\n ''' A Pytorch Dataset for the FINN Recsys Slates Dataset.\n Attributes:\n data: [Dict] A dictionary with tensors of the dataset. First dimension in each tensor must be the batch dimension. Requires the keys \"click\" and \"slate\". Additional elements can be added.\n sample_candidate_items: [int] Number of negative item examples sampled from the item universe for each interaction. If positive, the dataset provide an additional dictionary item \"allitem\". Often also called uniform candidate sampling. See Eide et. al. 2021 for more information.\n '''\n def __init__(self, data, sample_candidate_items=0):\n\n self.data = data\n self.num_items = self.data['slate'].max()+1\n self.sample_candidate_items = sample_candidate_items\n self.mask2ind = {'train' : 1, 'valid' : 2, 'test' : 3}\n\n logging.info(\n \"Loading dataset with slate size={} and number of negative samples={}\"\n .format(self.data['slate'].size(), self.sample_candidate_items))\n\n # Performs some checks on the dataset to make sure it is valid:\n assert \"slate\" in data.keys(), \"Slate tensor is not in dataset. This is required.\"\n assert \"click\" in data.keys(), \"Click tensor is not in dataset. This is required.\"\n assert all([val.size(0)==data['slate'].size(0) for key, val in data.items()]), \"Not all data tensors have the same batch dimension\"\n\n def __getitem__(self, idx):\n batch = {key: val[idx] for key, val in self.data.items()}\n\n if self.sample_candidate_items:\n # Sample actions uniformly (3 is the first non-special item)\n batch['allitem'] = torch.randint(\n size=(batch['click'].size(0), self.sample_candidate_items), \n low=3, high=self.num_items, device = batch['click'].device\n )\n \n return batch\n\n def __len__(self):\n return len(self.data['click'])",
"_____no_output_____"
],
[
"#export\ndef load_dataloaders(data_dir= \"dat\",\n batch_size=1024,\n num_workers= 0,\n sample_candidate_items=False,\n valid_pct= 0.05,\n test_pct= 0.05,\n t_testsplit= 5,\n limit_num_users=None,\n seed=0):\n \"\"\"\n Loads pytorch dataloaders to be used in training. If used with standard settings, the train/val/test split is equivalent to Eide et. al. 2021.\n\n Attributes:\n data_dir: [str] where download and store data if not already downloaded.\n batch_size: [int] Batch size given by dataloaders.\n num_workers: [int] How many threads should be used to prepare batches of data.\n sample_candidate_items: [int] Number of negative item examples sampled from the item universe for each interaction. If positive, the dataset provide an additional dictionary item \"allitem\". Often also called uniform candidate sampling. See Eide et. al. 2021 for more information.\n valid_pct: [float] Percentage of users allocated to validation dataset.\n test_pct: [float] Percentage of users allocated to test dataset.\n t_testsplit: [int] For users allocated to validation and test datasets, how many initial interactions should be part of the training dataset.\n limit_num_users: [int] For debugging purposes, only return some users.\n seed: [int] Seed used to sample users/items.\n\n \"\"\"\n \n logging.info(\"Download data if not in data folder..\")\n data_helper.download_data_files(data_dir=data_dir)\n\n logging.info('Load data..')\n with np.load(\"{}/data.npz\".format(data_dir)) as data_np:\n data = {key: torch.tensor(val) for key, val in data_np.items()}\n \n if limit_num_users is not None:\n logging.info(\"Limiting dataset to only return the first {} users.\".format(limit_num_users))\n data = {key : val[:limit_num_users] for key, val in data.items()}\n\n with open('{}/ind2val.json'.format(data_dir), 'rb') as handle:\n # Use string2int object_hook found here: https://stackoverflow.com/a/54112705\n ind2val = json.load(\n handle, \n object_hook=lambda d: {\n int(k) if k.lstrip('-').isdigit() else k: v \n for k, v in d.items()\n }\n )\n\n num_users = len(data['click'])\n num_validusers = int(num_users * valid_pct)\n num_testusers = int(num_users * test_pct)\n torch.manual_seed(seed)\n perm_user = torch.randperm(num_users)\n valid_user_idx = perm_user[:num_validusers]\n test_user_idx = perm_user[num_validusers:(num_validusers+num_testusers)]\n train_user_idx = perm_user[(num_validusers+num_testusers):]\n\n # Split dictionary into train/valid/test with a phase mask that shows which interactions are in different sets \n # (as some users have both train and valid data)\n data_train = data\n data_train['phase_mask'] = torch.ones_like(data['click']).bool()\n data_train['phase_mask'][test_user_idx,t_testsplit:]=False\n data_train['phase_mask'][valid_user_idx,t_testsplit:]=False\n\n data_valid = {key: val[valid_user_idx] for key, val in data.items()}\n data_valid['phase_mask'] = torch.zeros_like(data_valid['click']).bool()\n data_valid['phase_mask'][:,t_testsplit:] = True\n\n data_test = {key: val[test_user_idx] for key, val in data.items()}\n data_test['phase_mask'] = torch.zeros_like(data_test['click']).bool()\n data_test['phase_mask'][:,t_testsplit:] = True\n\n data_dicts = {\n \"train\" : data_train,\n \"valid\" : data_valid,\n \"test\" : data_test}\n\n datasets = {\n phase : SequentialDataset(data, sample_candidate_items) \n for phase, data in data_dicts.items()\n }\n \n\n # Build dataloaders for each data subset:\n dataloaders = {\n phase: DataLoader(ds, batch_size=batch_size, shuffle=(phase==\"train\"), num_workers=num_workers)\n for phase, ds in datasets.items()\n }\n for key, dl in dataloaders.items():\n logging.info(\n \"In {}: num_users: {}, num_batches: {}\".format(key, len(dl.dataset), len(dl))\n )\n \n # Load item attributes:\n with np.load('{}/itemattr.npz'.format(data_dir), mmap_mode=None) as itemattr_file:\n itemattr = {key : val for key, val in itemattr_file.items()}\n\n return ind2val, itemattr, dataloaders",
"_____no_output_____"
],
[
"#slow\nind2val, itemattr, dataloaders = load_dataloaders()",
"2021-08-13 10:15:07,665 Download data if not in data folder..\n2021-08-13 10:15:07,666 Downloading data.npz\n2021-08-13 10:15:07,667 Downloading ind2val.json\n2021-08-13 10:15:07,667 Downloading itemattr.npz\n2021-08-13 10:15:07,668 Done downloading all files.\n2021-08-13 10:15:07,668 Load data..\n2021-08-13 10:15:31,565 Loading dataset with slate size=torch.Size([2277645, 20, 25]) and uniform candidate sampling=False\n2021-08-13 10:15:31,834 Loading dataset with slate size=torch.Size([2277645, 20, 25]) and uniform candidate sampling=False\n2021-08-13 10:15:31,839 Loading dataset with slate size=torch.Size([113882, 20, 25]) and uniform candidate sampling=False\n2021-08-13 10:15:31,844 Loading dataset with slate size=torch.Size([113882, 20, 25]) and uniform candidate sampling=False\n2021-08-13 10:15:31,845 In train: num_users: 2277645, num_batches: 2225\n2021-08-13 10:15:31,846 In valid: num_users: 113882, num_batches: 112\n2021-08-13 10:15:31,846 In test: num_users: 113882, num_batches: 112\n"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d0ce2c57e5c5703b5fe5ccea3030cd2ccd1316e6 | 45,324 | ipynb | Jupyter Notebook | courses/machine_learning/deepdive2/recommendation_systems/solutions/als_bqml.ipynb | jonesevan007/training-data-analyst | 774446719316599cf221bdc5a67b00ec4c0b3ad0 | [
"Apache-2.0"
] | 2 | 2019-11-10T04:09:25.000Z | 2019-11-16T14:55:13.000Z | courses/machine_learning/deepdive2/recommendation_systems/solutions/als_bqml.ipynb | jonesevan007/training-data-analyst | 774446719316599cf221bdc5a67b00ec4c0b3ad0 | [
"Apache-2.0"
] | 10 | 2019-11-20T07:24:52.000Z | 2022-03-12T00:06:02.000Z | courses/machine_learning/deepdive2/recommendation_systems/solutions/als_bqml.ipynb | jonesevan007/training-data-analyst | 774446719316599cf221bdc5a67b00ec4c0b3ad0 | [
"Apache-2.0"
] | 4 | 2020-05-15T06:23:05.000Z | 2021-12-20T06:00:15.000Z | 31.628751 | 638 | 0.439921 | [
[
[
"# Collaborative filtering on the MovieLense Dataset",
"_____no_output_____"
],
[
"###### This notebook is based on part of Chapter 9 of [BigQuery: The Definitive Guide](https://www.oreilly.com/library/view/google-bigquery-the/9781492044451/ \"http://shop.oreilly.com/product/0636920207399.do\") by Lakshmanan and Tigani.\n### MovieLens dataset\nTo illustrate recommender systems in action, let’s use the MovieLens dataset. This is a dataset of movie reviews released by GroupLens, a research lab in the Department of Computer Science and Engineering at the University of Minnesota, through funding by the US National Science Foundation.\n\nDownload the data and load it as a BigQuery table using:",
"_____no_output_____"
]
],
[
[
"import os\nPROJECT = \"your-project-here\" # REPLACE WITH YOUR PROJECT ID\nBUCKET = \"your-bucket-here\" # REPLACE WITH YOUR BUCKET NAME\nREGION = \"us-central1\" # REPLACE WITH YOUR BUCKET REGION e.g. us-central1\n\n# Do not change these\nos.environ[\"PROJECT\"] = PROJECT\nos.environ[\"BUCKET\"] = BUCKET\nos.environ[\"REGION\"] = REGION\nos.environ[\"TFVERSION\"] = \"1.14\"",
"_____no_output_____"
],
[
"%%bash\nrm -r bqml_data\nmkdir bqml_data\ncd bqml_data\ncurl -O 'http://files.grouplens.org/datasets/movielens/ml-20m.zip'\nunzip ml-20m.zip\nbq --location=US mk --dataset \\\n --description 'Movie Recommendations' \\\n $PROJECT:movielens\nbq --location=US load --source_format=CSV \\\n --autodetect movielens.ratings ml-20m/ratings.csv\nbq --location=US load --source_format=CSV \\\n --autodetect movielens.movies_raw ml-20m/movies.csv",
"Archive: ml-20m.zip\n creating: ml-20m/\n inflating: ml-20m/genome-scores.csv \n inflating: ml-20m/genome-tags.csv \n inflating: ml-20m/links.csv \n inflating: ml-20m/movies.csv \n inflating: ml-20m/ratings.csv \n inflating: ml-20m/README.txt \n inflating: ml-20m/tags.csv \nDataset 'cloud-training-demos:movielens' successfully created.\n\n\n\n\n"
]
],
[
[
"## Exploring the data\nTwo tables should now be available in <a href=\"https://console.cloud.google.com/bigquery\">BigQuery</a>.\n\nCollaborative filtering provides a way to generate product recommendations for users, or user targeting for products. The starting point is a table, <b>movielens.ratings</b>, with three columns: a user id, an item id, and the rating that the user gave the product. This table can be sparse -- users don’t have to rate all products. Then, based on just the ratings, the technique finds similar users and similar products and determines the rating that a user would give an unseen product. Then, we can recommend the products with the highest predicted ratings to users, or target products at users with the highest predicted ratings.",
"_____no_output_____"
]
],
[
[
"%%bigquery --project $PROJECT\nSELECT *\nFROM movielens.ratings\nLIMIT 10",
"_____no_output_____"
]
],
[
[
"A quick exploratory query yields that the dataset consists of over 138 thousand users, nearly 27 thousand movies, and a little more than 20 million ratings, confirming that the data has been loaded successfully.",
"_____no_output_____"
]
],
[
[
"%%bigquery --project $PROJECT\nSELECT \n COUNT(DISTINCT userId) numUsers,\n COUNT(DISTINCT movieId) numMovies,\n COUNT(*) totalRatings\nFROM movielens.ratings",
"_____no_output_____"
]
],
[
[
"On examining the first few movies using the query following query, we can see that the genres column is a formatted string:",
"_____no_output_____"
]
],
[
[
"%%bigquery --project $PROJECT\nSELECT *\nFROM movielens.movies_raw\nWHERE movieId < 5",
"_____no_output_____"
]
],
[
[
"We can parse the genres into an array and rewrite the table as follows:",
"_____no_output_____"
]
],
[
[
"%%bigquery --project $PROJECT\nCREATE OR REPLACE TABLE movielens.movies AS\n SELECT * REPLACE(SPLIT(genres, \"|\") AS genres)\n FROM movielens.movies_raw",
"_____no_output_____"
],
[
"%%bigquery --project $PROJECT\nSELECT *\nFROM movielens.movies\nWHERE movieId < 5",
"_____no_output_____"
]
],
[
[
"## Matrix factorization\nMatrix factorization is a collaborative filtering technique that relies on factorizing the ratings matrix into two vectors called the user factors and the item factors. The user factors is a low-dimensional representation of a user_id and the item factors similarly represents an item_id.\n\nWe can create the recommender model using (<b>Optional</b>, takes 30 minutes. Note: we have a model we already trained if you want to skip this step):\n",
"_____no_output_____"
]
],
[
[
"%%bigquery --project $PROJECT\nCREATE OR REPLACE MODEL movielens.recommender\noptions(model_type='matrix_factorization',\n user_col='userId', item_col='movieId', rating_col='rating')\nAS\n\nSELECT \nuserId, movieId, rating\nFROM movielens.ratings",
"Executing query with job ID: 3b814912-7050-47db-8bf2-ada580e81a12\nQuery executing: 825.49s"
],
[
"%%bigquery --project $PROJECT\nSELECT *\n-- Note: remove cloud-training-demos if you are using your own model: \nFROM ML.TRAINING_INFO(MODEL `cloud-training-demos.movielens.recommender`)",
"_____no_output_____"
]
],
[
[
"Note that we create a model as usual, except that the model_type is matrix_factorization and that we have to identify which columns play what roles in the collaborative filtering setup.\n\nWhat did you get? Our model took an hour to train, and the training loss starts out extremely bad and gets driven down to near-zero over next the four iterations:\n\n<table>\n <tr>\n <th>Iteration</th>\n <th>Training Data Loss</th>\n <th>Evaluation Data Loss</th>\n <th>Duration (seconds)</th>\n </tr>\n <tr>\n <td>4</td>\n <td>0.5734</td>\n <td>172.4057</td>\n <td>180.99</td>\n </tr>\n <tr>\n <td>3</td>\n <td>0.5826</td>\n <td>187.2103</td>\n <td>1,040.06</td>\n </tr>\n <tr>\n <td>2</td>\n <td>0.6531</td>\n <td>4,758.2944</td>\n <td>219.46</td>\n </tr>\n <tr>\n <td>1</td>\n <td>1.9776</td>\n <td>6,297.2573</td>\n <td>1,093.76</td>\n </tr>\n <tr>\n <td>0</td>\n <td>63,287,833,220.5795</td>\n <td>168,995,333.0464</td>\n <td>1,091.21</td>\n </tr>\n</table>",
"_____no_output_____"
],
[
"However, the evaluation data loss is quite high, and much higher than the training data loss. This indicates that overfitting is happening, and so we need to add some regularization. Let’s do that next. Note the added l2_reg=0.2 (<b>Optional</b>, takes 30 minutes):",
"_____no_output_____"
]
],
[
[
"%%bigquery --project $PROJECT\nCREATE OR REPLACE MODEL movielens.recommender_l2\noptions(model_type='matrix_factorization',\n user_col='userId', item_col='movieId', \n rating_col='rating', l2_reg=0.2)\nAS\n\nSELECT \nuserId, movieId, rating\nFROM movielens.ratings",
"_____no_output_____"
],
[
"%%bigquery --project $PROJECT\nSELECT *\n-- Note: remove cloud-training-demos if you are using your own model: \nFROM ML.TRAINING_INFO(MODEL `cloud-training-demos.movielens.recommender_l2`)",
"_____no_output_____"
]
],
[
[
"Now, we get faster convergence (three iterations instead of five), and a lot less overfitting. Here are our results:\n\n<table>\n <tr>\n <th>Iteration</th>\n <th>Training Data Loss</th>\n <th>Evaluation Data Loss</th>\n <th>Duration (seconds)</th>\n </tr>\n <tr>\n <td>2</td>\n <td>0.6509</td>\n <td>1.4596</td>\n <td>198.17</td>\n </tr>\n <tr>\n <td>1</td>\n <td>1.9829</td>\n <td>33,814.3017</td>\n <td>1,066.06</td>\n </tr>\n <tr>\n <td>0</td>\n <td>481,434,346,060.7928</td>\n <td>2,156,993,687.7928</td>\n <td>1,024.59</td>\n </tr>\n</table>\n\nBy default, BigQuery sets the number of factors to be the log2 of the number of rows. In our case, since we have 20 million rows in the table, the number of factors would have been chosen to be 24. As with the number of clusters in K-Means clustering, this is a reasonable default but it is often worth experimenting with a number about 50% higher (36) and a number that is about a third lower (16):",
"_____no_output_____"
]
],
[
[
"%%bigquery --project $PROJECT\nCREATE OR REPLACE MODEL movielens.recommender_16\noptions(model_type='matrix_factorization',\n user_col='userId', item_col='movieId', \n rating_col='rating', l2_reg=0.2, num_factors=16)\nAS\n\nSELECT \nuserId, movieId, rating\nFROM movielens.ratings",
"_____no_output_____"
],
[
"%%bigquery --project $PROJECT\nSELECT *\n-- Note: remove cloud-training-demos if you are using your own model:\nFROM ML.TRAINING_INFO(MODEL `cloud-training-demos.movielens.recommender_16`)",
"_____no_output_____"
]
],
[
[
"When we did that, we discovered that the evaluation loss was lower (0.97) with num_factors=16 than with num_factors=36 (1.67) or num_factors=24 (1.45). We could continue experimenting, but we are likely to see diminishing returns with further experimentation. So, let’s pick this as the final matrix factorization model and move on.\n\n## Making recommendations\n\nWith the trained model, we can now provide recommendations. For example, let’s find the best comedy movies to recommend to the user whose userId is 903. In the query below, we are calling ML.PREDICT passing in the trained recommendation model and providing a set of movieId and userId to carry out the predictions on. In this case, it’s just one userId (903), but all movies whose genre includes Comedy.",
"_____no_output_____"
]
],
[
[
"%%bigquery --project $PROJECT\nSELECT * FROM\nML.PREDICT(MODEL `cloud-training-demos.movielens.recommender_16`, (\n SELECT \n movieId, title, 903 AS userId\n FROM movielens.movies, UNNEST(genres) g\n WHERE g = 'Comedy'\n))\nORDER BY predicted_rating DESC\nLIMIT 5",
"_____no_output_____"
]
],
[
[
"## Filtering out already rated movies\nOf course, this includes movies the user has already seen and rated in the past. Let’s remove them:\n",
"_____no_output_____"
]
],
[
[
"%%bigquery --project $PROJECT\nSELECT * FROM\nML.PREDICT(MODEL `cloud-training-demos.movielens.recommender_16`, (\n WITH seen AS (\n SELECT ARRAY_AGG(movieId) AS movies \n FROM movielens.ratings\n WHERE userId = 903\n )\n SELECT \n movieId, title, 903 AS userId\n FROM movielens.movies, UNNEST(genres) g, seen\n WHERE g = 'Comedy' AND movieId NOT IN UNNEST(seen.movies)\n))\nORDER BY predicted_rating DESC\nLIMIT 5",
"_____no_output_____"
]
],
[
[
"For this user, this happens to yield the same set of movies -- the top predicted ratings didn’t include any of the movies the user has already seen.\n\n## Customer targeting\n\nIn the previous section, we looked at how to identify the top-rated movies for a specific user. Sometimes, we have a product and have to find the customers who are likely to appreciate it. Suppose, for example, we wish to get more reviews for movieId=96481 which has only one rating and we wish to send coupons to the 5 users who are likely to rate it the highest. We can identify those users using:",
"_____no_output_____"
]
],
[
[
"%%bigquery --project $PROJECT\nSELECT * FROM\nML.PREDICT(MODEL `cloud-training-demos.movielens.recommender_16`, (\n WITH allUsers AS (\n SELECT DISTINCT userId\n FROM movielens.ratings\n )\n SELECT \n 96481 AS movieId, \n (SELECT title FROM movielens.movies WHERE movieId=96481) title,\n userId\n FROM\n allUsers\n))\nORDER BY predicted_rating DESC\nLIMIT 5",
"_____no_output_____"
]
],
[
[
"### Batch predictions for all users and movies\nWhat if we wish to carry out predictions for every user and movie combination? Instead of having to pull distinct users and movies as in the previous query, a convenience function is provided to carry out batch predictions for all movieId and userId encountered during training. A limit is applied here, otherwise, all user-movie predictions will be returned and will crash the notebook.",
"_____no_output_____"
]
],
[
[
"%%bigquery --project $PROJECT\nSELECT *\nFROM ML.RECOMMEND(MODEL `cloud-training-demos.movielens.recommender_16`)\nLIMIT 10",
"_____no_output_____"
]
],
[
[
"As seen in a section above, it is possible to filter out movies the user has already seen and rated in the past. The reason already seen movies aren’t filtered out by default is that there are situations (think of restaurant recommendations, for example) where it is perfectly expected that we would need to recommend restaurants the user has liked in the past.",
"_____no_output_____"
],
[
"Copyright 2019 Google Inc. Licensed under the Apache License, Version 2.0 (the \"License\"); you may not use this file except in compliance with the License. You may obtain a copy of the License at http://www.apache.org/licenses/LICENSE-2.0 Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an \"AS IS\" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d0ce2f94d54a28778b43071b2c55d1ea9adbda7b | 38,749 | ipynb | Jupyter Notebook | notebooks/coarse_registration.ipynb | chunglabmit/phathom | 304db7a95e898e9b03d6b2640172752d21a7e3ed | [
"MIT"
] | 1 | 2018-04-18T11:54:29.000Z | 2018-04-18T11:54:29.000Z | notebooks/coarse_registration.ipynb | chunglabmit/phathom | 304db7a95e898e9b03d6b2640172752d21a7e3ed | [
"MIT"
] | 2 | 2018-04-05T20:53:52.000Z | 2018-11-01T16:37:39.000Z | notebooks/coarse_registration.ipynb | chunglabmit/phathom | 304db7a95e898e9b03d6b2640172752d21a7e3ed | [
"MIT"
] | null | null | null | 45.214702 | 6,492 | 0.671553 | [
[
[
"import neuroglancer\n# Use this in IPython to allow external viewing\n# neuroglancer.set_server_bind_address(bind_address='192.168.158.128',\n# bind_port=80)\nfrom nuggt.utils import ngutils\n\nviewer = neuroglancer.Viewer()\nviewer",
"_____no_output_____"
],
[
"import numpy as np\nimport zarr\nimport os\n\n# working_dir = '/media/jswaney/Drive/Justin/coregistration/whole_brain_tde'\nworking_dir = '/home/jswaney/coregistration'\n\nfixed_path = 'fixed/zarr_stack/8_8_8'\nmoving_path = 'moving/zarr_stack/8_8_8'\n\nfixed_store = zarr.NestedDirectoryStore(os.path.join(working_dir, fixed_path))\nmoving_store = zarr.NestedDirectoryStore(os.path.join(working_dir, moving_path))\n\nfixed_img = zarr.open(fixed_store, mode='r')\nmoving_img = zarr.open(moving_store, mode='r')\n\nprint(fixed_img.shape)\nprint(moving_img.shape)",
"(564, 1547, 918)\n(508, 1549, 914)\n"
],
[
"normalization = 2000\n\ndef plot_image(img, viewer, layer, shader):\n with viewer.txn() as txn:\n source = neuroglancer.LocalVolume(img.astype(np.float32))\n txn.layers[layer] = neuroglancer.ImageLayer(source=source, shader=shader)\n\ndef plot_fixed(fixed_img, viewer):\n fixed_shader = ngutils.red_shader % (1 / normalization)\n plot_image(fixed_img, viewer, 'fixed', fixed_shader)\n \ndef plot_moving(moving_img, viewer):\n moving_shader = ngutils.green_shader % (1 / normalization)\n plot_image(moving_img, viewer, 'moving', moving_shader)\n \ndef plot_both(fixed_img, moving_img, viewer):\n plot_fixed(fixed_img, viewer)\n plot_moving(moving_img, viewer)\n \nplot_both(fixed_img, moving_img, viewer)",
"_____no_output_____"
]
],
[
[
"# Downsampling Zarr",
"_____no_output_____"
]
],
[
[
"from phathom.io.zarr import downsample_zarr",
"_____no_output_____"
],
[
"factors = (8, 8, 8)\noutput_path = os.path.join(working_dir, 'fixed/zarr_down8')\nnb_workers = 1\n\ndownsample_zarr(fixed_img, factors, output_path, nb_workers)",
" 11%|█ | 43623/399446 [04:51<39:34, 149.88it/s]"
],
[
"factors = (8, 8, 8)\noutput_path = os.path.join(working_dir, 'moving/zarr_down8')\nnb_workers = 1\n\ndownsample_zarr(moving_img, factors, output_path, nb_workers)",
"_____no_output_____"
]
],
[
[
"# Downsampling ndarray",
"_____no_output_____"
]
],
[
[
"from skimage.measure import block_reduce\n\nfactors = (16, 16, 16)\n\ndef downsample_mean(img, factors):\n return block_reduce(img, factors, np.mean, 0)\n\ndef downsample_max(img, factors):\n return block_reduce(img, factors, np.max, 0)\n\nfixed_downsample = downsample_mean(fixed_img, factors)\nmoving_downsample = downsample_mean(moving_img, factors)\n\nplot_both(fixed_downsample, moving_downsample, viewer)",
"_____no_output_____"
]
],
[
[
"# Gaussian smoothing",
"_____no_output_____"
]
],
[
[
"from skimage.filters import gaussian\n\nsigma = 1.0\n\nfixed_smooth = gaussian(fixed_downsample, sigma, preserve_range=True)\nmoving_smooth = gaussian(moving_downsample, sigma, preserve_range=True)\n\nplot_both(fixed_smooth, moving_smooth, viewer)",
"_____no_output_____"
]
],
[
[
"# Destriping",
"_____no_output_____"
]
],
[
[
"import pystripe\nimport multiprocessing\nimport tqdm\n\nbandwidth = [64, 64]\nwavelet = 'db4'\n\ndef _filter_streaks(img):\n return pystripe.filter_streaks(img, sigma=bandwidth, wavelet=wavelet)\n\nwith multiprocessing.Pool(12) as pool:\n rf = list(tqdm.tqdm(pool.imap(_filter_streaks, fixed_smooth), total=len(fixed_smooth)))\n rm = list(tqdm.tqdm(pool.imap(_filter_streaks, moving_smooth), total=len(moving_smooth)))\n\nfixed_destripe = np.array(rf).T\nmoving_destripe = np.array(rm).T\n\nwith multiprocessing.Pool(12) as pool:\n rf = list(tqdm.tqdm(pool.imap(_filter_streaks, fixed_destripe), total=len(fixed_smooth)))\n rm = list(tqdm.tqdm(pool.imap(_filter_streaks, moving_destripe), total=len(moving_smooth)))\n\nfixed_destripe = np.array(rf).T\nmoving_destripe = np.array(rm).T\n \nplot_both(fixed_destripe, moving_destripe, viewer)",
"100%|██████████| 6/6 [00:00<00:00, 870.73it/s]\n100%|██████████| 7/7 [00:00<00:00, 151.45it/s]\n9it [00:00, 1582.42it/s] \n9it [00:00, 14855.86it/s] \n"
]
],
[
[
"# Rigid transformation",
"_____no_output_____"
]
],
[
[
"from phathom.registration import coarse, pcloud\nfrom phathom import utils\nfrom scipy.ndimage import map_coordinates\n\nt = np.array([0, 0, 0])\nthetas = np.array([np.pi/4, 0, 0])\n\ndef rigid_warp(img, t, thetas, center, output_shape):\n r = pcloud.rotation_matrix(thetas)\n idx = np.indices(output_shape)\n pts = np.reshape(idx, (idx.shape[0], idx.size//idx.shape[0])).T\n warped_pts = coarse.rigid_transformation(t, r, pts, center)\n interp_values = map_coordinates(img, warped_pts.T)\n transformed = np.reshape(interp_values, output_shape)\n return transformed\n\ntransformed = rigid_warp(fixed_downsample, \n t, \n thetas, \n np.zeros(3), \n moving_downsample.shape)\n\nplot_fixed(transformed, viewer)",
"_____no_output_____"
],
[
"from scipy.ndimage.measurements import center_of_mass\n\ndef center_mass(img):\n return np.asarray(center_of_mass(img))\n\nfixed_com = center_mass(fixed_downsample)\nmoving_com = center_mass(moving_downsample)\n\nprint(fixed_com)\nprint(moving_com)",
"[13.95046336 40.30643648 26.72860826]\n[14.57307261 38.4847799 25.10686021]\n"
]
],
[
[
"# Optimization",
"_____no_output_____"
]
],
[
[
"def ncc(fixed, transformed, nonzero=False):\n if nonzero:\n idx = np.where(transformed)\n a = fixed[idx]\n b = transformed[idx]\n else:\n a = fixed\n b = transformed\n return np.sum((a-a.mean())*(b-b.mean())/((a.size-1)*a.std()*b.std()))\n\ndef ssd(fixed, transformed):\n return np.mean((fixed-transformed)**2)\n\ndef registration_objective(x, fixed_img, moving_img, t):\n transformed_img = rigid_warp(moving_img, \n t=t, \n thetas=x,\n center=fixed_com,\n output_shape=moving_img.shape)\n return ssd(moving_img, transformed_img)\n\ndef callback(x, f, accept):\n pass",
"_____no_output_____"
],
[
"from scipy.optimize import basinhopping\n\nniter = 4\n\nt_star = moving_com-fixed_com\nbounds = [(0, np.pi/2) for _ in range(3)]\n\nres = basinhopping(registration_objective,\n x0=np.zeros(3),\n niter=niter,\n T=1.0,\n stepsize=1.0,\n interval=5,\n minimizer_kwargs={\n 'method': 'L-BFGS-B',\n# 'method': 'Nelder-Mead',\n 'args': (fixed_smooth, \n moving_smooth, \n t_star),\n 'bounds': bounds,\n 'tol': 0.01,\n 'options': {'disp': False}\n },\n disp=True)\n\ntheta_star = res.x\nprint(res)",
"basinhopping step 0: f -0.580548\nbasinhopping step 1: f -0.584613 trial_f -0.584613 accepted 1 lowest_f -0.584613\nfound new global minimum on step 1 with function value -0.584613\nbasinhopping step 2: f -0.584613 trial_f -0.454478 accepted 0 lowest_f -0.584613\nbasinhopping step 3: f -0.580548 trial_f -0.580548 accepted 1 lowest_f -0.584613\nbasinhopping step 4: f -0.584938 trial_f -0.584938 accepted 1 lowest_f -0.584938\nfound new global minimum on step 4 with function value -0.584938\n fun: -0.5849382125048649\n lowest_optimization_result: fun: -0.5849382125048649\n hess_inv: <3x3 LbfgsInvHessProduct with dtype=float64>\n jac: array([0.00121516, 0.16166298, 0.02240939])\n message: b'CONVERGENCE: REL_REDUCTION_OF_F_<=_FACTR*EPSMCH'\n nfev: 20\n nit: 4\n status: 0\n success: True\n x: array([0.00256195, 0. , 0.24721074])\n message: ['requested number of basinhopping iterations completed successfully']\n minimization_failures: 0\n nfev: 76\n nit: 4\n x: array([0.00256195, 0. , 0.24721074])\n"
],
[
"registered = rigid_warp(fixed_smooth, t_star, theta_star, fixed_com, moving_destripe.shape)\n\nplot_fixed(registered, viewer)",
"_____no_output_____"
]
],
[
[
"# Contour",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n\nplt.hist(fixed_downsample.ravel(), bins=100)\nplt.xlim([0, 1000])\nplt.ylim([0, 100000])\nplt.show()\n\nplt.hist(moving_downsample.ravel(), bins=100)\nplt.xlim([0, 1000])\nplt.ylim([0, 100000])\nplt.show()",
"_____no_output_____"
],
[
"threshold = 150\n\nfixed_mask = fixed_downsample > threshold\nmoving_mask = moving_downsample > threshold\n\nplot_both(1000*fixed_mask, 1000*moving_mask, viewer)",
"_____no_output_____"
]
],
[
[
"# Convex hull",
"_____no_output_____"
]
],
[
[
"from skimage.morphology import convex_hull_image\nimport tqdm\n\nfixed_hull = np.zeros_like(fixed_mask)\n\nfor i, f in enumerate(tqdm.tqdm(fixed_mask)):\n if not np.all(f == 0):\n fixed_hull[i] = convex_hull_image(f)\n\nmoving_hull = np.zeros_like(moving_mask)\n \nfor i, m in enumerate(tqdm.tqdm(moving_mask)):\n if not np.all(m == 0):\n moving_hull[i] = convex_hull_image(m)\n\nplot_both(1000*fixed_hull, 1000*moving_hull, viewer)",
"100%|██████████| 71/71 [00:00<00:00, 571.26it/s]\n100%|██████████| 64/64 [00:00<00:00, 631.17it/s]\n"
],
[
"from scipy.ndimage.morphology import distance_transform_edt\n\nfixed_distance = distance_transform_edt(fixed_mask)\nmoving_distance = distance_transform_edt(moving_mask)\n\nplot_both(100*fixed_distance, 100*moving_distance, viewer)",
"_____no_output_____"
],
[
"niter = 3\n\nfrom scipy.optimize import basinhopping\n\nfixed_com = center_mass(fixed_mask)\nmoving_com = center_mass(moving_mask)\n\nt0 = moving_com-fixed_com\nbounds = [(-s, s) for s in moving_distance.shape]+[(-np.pi, np.pi) for _ in range(3)]\n# bounds = [(-np.pi, np.pi) for _ in range(3)]\n\ndef absolute_difference(img1, img2):\n return np.mean(np.abs(img1-img2))\n\ndef registration_objective(x, fixed_img, moving_img):\n transformed_img = rigid_warp(moving_img, \n t= x[:3], \n thetas= x[3:],\n center=fixed_com,\n output_shape=fixed_img.shape)\n return absolute_difference(fixed_img, transformed_img)\n# return ssd(fixed_img, transformed_img)\n\n\ndef callback(x, f, accept):\n print(x)\n\nres = basinhopping(registration_objective,\n x0=np.concatenate((t0, np.zeros(3))),\n niter=niter,\n T=0.5,\n stepsize=0.5,\n interval=5,\n minimizer_kwargs={\n 'method': 'L-BFGS-B',\n 'args': (fixed_distance, \n moving_distance),\n 'bounds': bounds,\n 'tol': 0.001,\n 'options': {'disp': False}\n },\n callback=callback,\n disp=True)\n\nt_star = res.x[:3]\ntheta_star = res.x[3:]\nprint(res)\n\nreg_distance = rigid_warp(moving_distance, \n t_star, \n theta_star, \n fixed_com, \n moving_distance.shape)\n\nplot_moving(100*reg_distance, viewer)",
"basinhopping step 0: f 0.126898\nbasinhopping step 1: f 0.130302 trial_f 0.130302 accepted 1 lowest_f 0.126898\n[ 0.02877251 -1.75668613 -1.94956175 0.32351345 0.0456788 0.01215395]\nbasinhopping step 2: f 0.131835 trial_f 0.131835 accepted 1 lowest_f 0.126898\n[-0.39748378 -1.37928118 -1.67215733 0.32702666 0.02950279 0.00710573]\nbasinhopping step 3: f 0.130997 trial_f 0.130997 accepted 1 lowest_f 0.126898\n[ 0.06094032 -1.64562144 -1.33690395 0.32847584 0.01498152 0.006094 ]\n fun: 0.12689757603303484\n lowest_optimization_result: fun: 0.12689757603303484\n hess_inv: <6x6 LbfgsInvHessProduct with dtype=float64>\n jac: array([ 0.02752217, 0.0092912 , -0.01756193, -0.0355355 , -0.02404413,\n 0.06360822])\n message: b'CONVERGENCE: REL_REDUCTION_OF_F_<=_FACTR*EPSMCH'\n nfev: 70\n nit: 7\n status: 0\n success: True\n x: array([-0.00914047, -1.67295537, -1.83032612, 0.32819386, 0.03238862,\n 0.00964872])\n message: ['requested number of basinhopping iterations completed successfully']\n minimization_failures: 0\n nfev: 357\n nit: 3\n x: array([-0.00914047, -1.67295537, -1.83032612, 0.32819386, 0.03238862,\n 0.00964872])\n"
]
],
[
[
"Sum of squared differences seems to provide slightly better registration than Normalized cross-correlation in the case of distance transformed convex hulls. This might be because NCC is indifferent to intensity difference and only considers correlations in the intensities, whereas SSD will penalize for any difference in intensity. In a multi-modal setting, this is usually not desired, but since we are dealing with the same brain in both images, the overall shape (and therefore distance transforms) should take similar values (not just correlated).\n\nAlso, it was necessary to include the translation component in the optimization procedure because our center of mass estimate for the center of rotation is not accurate. This causes the optimization for our rigid transformation to be partially constrained to inaccurate values, making it hard to converge to a rotation",
"_____no_output_____"
],
[
"# Coarse Registration",
"_____no_output_____"
]
],
[
[
"registered = rigid_warp(moving_downsample, \n t_star, \n theta_star, \n fixed_com, \n moving_downsample.shape)\n\nplot_both(fixed_downsample, registered, viewer)",
"_____no_output_____"
]
],
[
[
"We need to convert the downsampled transformation into an approprate transformation for the original resolution image. The rotation matrix is scale invariant, but we need to make sure the center of rotation and translation are upsampled by the same amount that we downsampled",
"_____no_output_____"
]
],
[
[
"print('Converged rigid transformation for downsampled image')\nprint('Rotation (deg):', theta_star*180/np.pi)\nprint('Center (px):', fixed_com)\nprint('Translation (px):', t_star)",
"Converged rigid transformation for downsampled image\nRotation (deg): [18.80412319 1.8557313 0.55283072]\nCenter (px): [14.20678094 41.99777965 27.26826236]\nTranslation (px): [-0.00914047 -1.67295537 -1.83032612]\n"
],
[
"fixed_fullres_path = os.path.join(working_dir, 'fixed/zarr_stack/8_8_8')\nfixed_fullres_store = zarr.NestedDirectoryStore(fixed_fullres_path)\nfixed_fullres = zarr.open(fixed_fullres_store, mode='r')\n\ntheta = theta_star\ntrue_factors = np.array(fixed_fullres.shape) / np.array(fixed_downsample.shape)\nt, center = coarse._scale_rigid_params(t_star, \n fixed_com, \n true_factors)\n\nprint('Converged rigid transformation for original image')\nprint('Rotation (deg):', theta*180/np.pi)\nprint('Center (px):', center)\nprint('Translation (px):', t)",
"Converged rigid transformation for original image\nRotation (deg): [18.80412319 1.8557313 0.55283072]\nCenter (px): [222.5729014 669.79964044 431.59077316]\nTranslation (px): [ -0.14320069 -26.68105113 -28.96964445]\n"
],
[
"plot_both(fixed_img, moving_img, viewer)",
"_____no_output_____"
],
[
"registered = rigid_warp(moving_img, \n t, \n theta, \n center, \n fixed_img.shape)\n\nplot_moving(registered, viewer)",
"_____no_output_____"
],
[
"np.save('rigid_8_8_8.npy', registered)",
"_____no_output_____"
]
],
[
[
"# Save the transformation",
"_____no_output_____"
]
],
[
[
"from phathom.utils import pickle_save\n\n\ntransformation_dict = {'t': t,\n 'center': center,\n 'theta': theta}\n\npickle_save(os.path.join(working_dir, 'rigid_transformation.pkl'), \n transformation_dict)",
"_____no_output_____"
],
[
"from phathom.utils import pickle_load\n\ntransformation_dict = pickle_load(os.path.join(working_dir, 'rigid_transformation.pkl'))",
"_____no_output_____"
],
[
"transformation_dict",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d0ce3ae20d76ff4dd5a4082a3ec7ab2f67147535 | 421,506 | ipynb | Jupyter Notebook | build/visualisation/Periodic Gaussian Plasma - large truncated lattice.ipynb | nimachm81/YEEFDTD | b6a757d574ad5475193c24c0bf83af445568a3fa | [
"MIT"
] | null | null | null | build/visualisation/Periodic Gaussian Plasma - large truncated lattice.ipynb | nimachm81/YEEFDTD | b6a757d574ad5475193c24c0bf83af445568a3fa | [
"MIT"
] | null | null | null | build/visualisation/Periodic Gaussian Plasma - large truncated lattice.ipynb | nimachm81/YEEFDTD | b6a757d574ad5475193c24c0bf83af445568a3fa | [
"MIT"
] | null | null | null | 780.566667 | 213,940 | 0.951436 | [
[
[
"## plot plasma density\n\n%pylab inline\nimport numpy as np\nfrom matplotlib import pyplot as plt\n\nfrom ReadBinary import *\n\nfileSuffix = \"-10\"\nfolder = \"../data/LargePeriodicLattice-GaussianPlasma/fp=1THz/\"\n#folder = \"../data/2D/\"\nfilename = folder+\"Wp2-x{}.data\".format(fileSuffix)\narrayInfo = GetArrayInfo(filename)\n\nprint(\"typeCode: \", arrayInfo[\"typeCode\"])\nprint(\"typeSize: \", arrayInfo[\"typeSize\"])\nprint(\"shape: \", arrayInfo[\"shape\"])\nprint(\"numOfArrays: \", arrayInfo[\"numOfArrays\"])\n\nWp2 = GetArrays(filename, 0, 1)[0,0,:,:]\nprint(\"shape: \", Wp2.shape)\n\nshape = Wp2.shape\n\nplt.figure(figsize=(6, 6*(shape[0]/shape[1])))\nplt.imshow(np.real(Wp2[:,:]), cmap=\"rainbow\", origin='lower', aspect='auto')\nplt.contour(np.real(Wp2[:,:]), cmap=\"Greys\", linewidths=0.5)\nplt.show()\n",
"Populating the interactive namespace from numpy and matplotlib\ntypeCode: 1\ntypeSize: 4\nshape: (1, 1001, 1001)\nnumOfArrays: 4\nshape: (1001, 1001)\n"
],
[
"## animate Electric field\n\n%pylab tk\nimport numpy as np\nfrom matplotlib import pyplot as plt\nimport matplotlib.animation as animation\nfrom ReadBinary import *\n\nfilename = folder+\"E-x{}.data\".format(fileSuffix)\narrayInfo = GetArrayInfo(filename)\n\nprint(\"typeCode: \", arrayInfo[\"typeCode\"])\nprint(\"typeSize: \", arrayInfo[\"typeSize\"])\nprint(\"shape: \", arrayInfo[\"shape\"])\nprint(\"numOfArrays: \", arrayInfo[\"numOfArrays\"])\n\nE = GetArrays(filename, indStart=0, indEnd=None)[:, 0, :, :]\nprint(\"shape: \", E.shape)\n\nshape = E.shape[1:]\nprint(\"Max E: \", np.max(np.abs(E)))\n\nanimate = True\nsave_animation = True\n\nif animate:\n def animate_E(n):\n plt.clf()\n fig = plt.imshow(np.real(E[n, :,:]), cmap=\"rainbow\", origin='lower', aspect='auto')\n plt.colorbar()\n plt.contour(np.real(Wp2[:,:]), cmap=\"Greys\", linewidths=0.5)\n plt.xticks([])\n plt.yticks([])\n plt.pause(0.05)\n \n return fig\n \n if not save_animation:\n plt.ion()\n plt.figure(figsize=(7,6*(shape[0]/shape[1])))\n\n for n in range(E.shape[0]):\n animate_E(n)\n else:\n fig = plt.figure(figsize=(7,6*(shape[0]/shape[1])))\n anim = animation.FuncAnimation(fig, animate_E, frames=E.shape[0], interval=1, repeat=False)\n anim.save(folder + 'Efield-anim.mp4', writer=\"ffmpeg\", fps=15, dpi=200)\n",
"Populating the interactive namespace from numpy and matplotlib\ntypeCode: 1\ntypeSize: 4\nshape: (1, 1001, 1001)\nnumOfArrays: 25\n"
],
[
"%pylab inline\n\nE = GetArrays(folder+\"E-x-slice{}.data\".format(fileSuffix), indStart=0, indEnd=None)[:, 0, :, :]\nprint(\"shape: \", E.shape)\n\nshape = E.shape[1:]\nE_ty = E[:, :, 0]\n\nplt.imshow(np.real(E_ty).T, cmap=\"rainbow\", origin='lower', aspect='auto')\nplt.colorbar()\nplt.show()\n\nNt, Ny = E_ty.shape\nN_pts = 50\nE_f_ty = np.fft.fftshift(np.fft.fft2(E_ty))[Nt//2-N_pts:Nt//2+N_pts, Ny//2-N_pts:Ny//2+N_pts]\n\nplt.imshow(np.abs(E_f_ty), cmap=\"rainbow\", origin='lower', aspect='auto')\nplt.show()\n",
"Populating the interactive namespace from numpy and matplotlib\n"
],
[
"%pylab inline\n\nplt.figure()\nE_t = np.sum(E_ty, axis=1)[700:]\nplt.plot(E_t)\nplt.show()\n\nNt = len(E_t)\nN_pts = 30\n\nplt.figure()\nE_f_t = np.fft.fftshift(np.fft.fft(E_t))[Nt//2-N_pts:Nt//2+N_pts]\nplt.plot(np.abs(E_f_t))\nplt.show()\n",
"Populating the interactive namespace from numpy and matplotlib\n"
],
[
"Nw = 200\nw_max = 5\n\nw = np.linspace(0, w_max, Nw)\ndt = 0.01*0.95/np.sqrt(2.0)\n\nNt = len(E_t)\nt = np.linspace(0, Nt*dt, Nt)\n\nE_f_t = np.zeros(Nw, dtype=complex)\n\nfor i in range(len(w)):\n w_i = w[i]\n E_f_t[i] = np.sum(E_t * np.exp(-1j*w_i*t))/Nt\n\n\nplot(w, np.abs(E_f_t))\nshow()\n\nfrom scipy import constants\npitch = 124\nplot(w*(constants.c/(pitch*constants.micro))/constants.tera/(2.0*np.pi), np.abs(E_f_t))\nshow()\n",
"/home/nima/.local/lib/python3.6/site-packages/ipykernel_launcher.py:14: ComplexWarning: Casting complex values to real discards the imaginary part\n \n"
],
[
"%pylab inline\n\nimport numpy as np\nfrom matplotlib import pyplot as plt\nimport os\nfrom scipy import constants\nfrom ReadBinary import *\n\nfolder = \"../data/LargePeriodicLattice-GaussianPlasma/fp=1THz/\"\nfile_list = os.listdir(folder)\nsuffixes = sorted([f[9:f.find(\".data\")] for f in file_list if \"slice\" in f])\nprint(suffixes)\n\nangles = [float(sfx[1:]) for sfx in suffixes]\nprint(angles)\n\nsorted_keys = sorted(range(len(angles)), key=lambda k: angles[k])\nsuffixes = [suffixes[i] for i in sorted_keys]\nprint(suffixes)\n\nfilename = folder+\"E-x-slice{}.data\".format(suffixes[0])\narrayInfo = GetArrayInfo(filename)\nNy = arrayInfo[\"shape\"][1]\nNt = arrayInfo[\"numOfArrays\"]\n\nprint(\"Nt: {}, Ny: {}\".format(Nt, Ny))\n\nNw = 200\ndef get_spectrum_si(E_t):\n w_max = 5\n\n w = np.linspace(0, w_max, Nw)\n dt = 0.01*0.95/np.sqrt(2.0)\n\n Nt = len(E_t)\n t = np.linspace(0, Nt*dt, Nt)\n\n E_f_t = np.zeros(Nw, dtype=complex)\n for i in range(len(w)):\n w_i = w[i]\n E_f_t[i] = np.sum(E_t * np.exp(-1j*w_i*t))/Nt\n\n pitch = 124\n f_si = w*(constants.c/(pitch*constants.micro))/constants.tera/(2.0*np.pi)\n return f_si, E_f_t\n\nE = GetArrays(folder+\"E-x-slice{}.data\".format(suffixes[0]), indStart=0, indEnd=None)[:, 0, :, :]\nE_ty = E[:, :, 0]\nE_t = np.sum(E_ty, axis=1)[700:]\n\nf_si, E_f_t = get_spectrum_si(E_t)\n\nplot(f_si, np.abs(E_f_t))\nshow()\n\nn_files = len(suffixes)\ngap_spec = np.zeros((Nw, n_files), dtype=complex)\n\nfor i in range(n_files):\n E = GetArrays(folder+\"E-x-slice{}.data\".format(suffixes[i]), indStart=0, indEnd=None)[:, 0, :, :]\n E_ty = E[:, :, 0]\n E_t = np.sum(E_ty, axis=1)[700:]\n\n f_si, E_f_t = get_spectrum_si(E_t)\n gap_spec[:, i] = E_f_t\n print(suffixes[i], end=\" \")\n\nimshow(np.abs(gap_spec), cmap=\"rainbow\", origin='lower', aspect='auto')\n",
"Populating the interactive namespace from numpy and matplotlib\n['-0', '-1', '-10', '-11', '-12', '-13', '-14', '-15', '-16', '-17', '-18', '-19', '-2', '-20', '-21', '-22', '-23', '-24', '-25', '-26', '-27', '-28', '-29', '-3', '-30', '-31', '-32', '-33', '-34', '-35', '-36', '-37', '-38', '-39', '-4', '-40', '-41', '-42', '-43', '-44', '-5', '-6', '-7', '-8', '-9']\n[0.0, 1.0, 10.0, 11.0, 12.0, 13.0, 14.0, 15.0, 16.0, 17.0, 18.0, 19.0, 2.0, 20.0, 21.0, 22.0, 23.0, 24.0, 25.0, 26.0, 27.0, 28.0, 29.0, 3.0, 30.0, 31.0, 32.0, 33.0, 34.0, 35.0, 36.0, 37.0, 38.0, 39.0, 4.0, 40.0, 41.0, 42.0, 43.0, 44.0, 5.0, 6.0, 7.0, 8.0, 9.0]\n['-0', '-1', '-2', '-3', '-4', '-5', '-6', '-7', '-8', '-9', '-10', '-11', '-12', '-13', '-14', '-15', '-16', '-17', '-18', '-19', '-20', '-21', '-22', '-23', '-24', '-25', '-26', '-27', '-28', '-29', '-30', '-31', '-32', '-33', '-34', '-35', '-36', '-37', '-38', '-39', '-40', '-41', '-42', '-43', '-44']\nNt: 2401, Ny: 1001\n"
],
[
"font = {'family' : 'serif', 'weight' : 'normal', 'size' : 14}\nmatplotlib.rc('font', **font)\n\nimshow(np.abs(gap_spec[0:100,:]), cmap=\"rainbow\", interpolation=None, origin='lower', aspect='auto', \\\n extent=[0, 45, 0, f_si[100]], vmax=None)\n#contour(np.abs(gap_spec[0:100,:]), extent=[0, 45, 0, f_si[100]])\n\nxlabel(r\"$\\theta$ (degrees)\")\nylabel(r\"f (THz)\")\n\nsavefig(folder+'E-r-spectrum.png', bbox_inches='tight', pad_inches=0.5)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0ce41bdd7a5dc8b0d48a4632db13531ccbe0548 | 31,254 | ipynb | Jupyter Notebook | DBSCAN - class.ipynb | WyTho/MachineLearning | 8f1e1efe9eb8f1e9e14c47665fc167876539313e | [
"MIT"
] | null | null | null | DBSCAN - class.ipynb | WyTho/MachineLearning | 8f1e1efe9eb8f1e9e14c47665fc167876539313e | [
"MIT"
] | null | null | null | DBSCAN - class.ipynb | WyTho/MachineLearning | 8f1e1efe9eb8f1e9e14c47665fc167876539313e | [
"MIT"
] | null | null | null | 40.908377 | 677 | 0.451174 | [
[
[
"import pandas as pd\nfrom pylab import rcParams\nimport seaborn as sb\nimport matplotlib.pyplot as plt\n\nimport sklearn\nfrom sklearn.cluster import DBSCAN\nfrom collections import Counter\nimport datetime\n\nfrom sklearn.preprocessing import LabelEncoder\nfrom collections import defaultdict\nfrom functools import reduce\n\nimport math",
"_____no_output_____"
],
[
"to_milliseconds = lambda seconds : seconds * 60 * 1000\n\nclass BinaryDataAnalysis:\n \"\"\"Convert non-nummeric values in the dataframe to numbers so that the dataframe can be used to fit a model\n\n Args (all optional):\n eps: The epsilon in minutes (starting minimum distance between datapoints to cluster them together)\n cluster_degregation: The next epsilon divider to use if clusters are too large \n (if eps=5 and cluster_degregation=2 then the next eps will be 2.5, and the next 1.25 etc.)\n max_cluster_distance: the maximum size of a cluster in minutes\n weeks: the amount of weeks to analyze, \n a minimum of 1 needed, \n a minimum of 2 is recommended\n decay_strength: how much the next week counts for predicting relevant groups\n e.g. with a decay_strength of 0.5, ech week before last week will \n count half as strong for predicting if the groups are still relevant\n cluster_threshold: from how many occourences (in one week) should it get 'self.threshold_percentage' \n as percentage that it is a group...\n More occourences will result in a higher persentage than 'self.threshold_percentage'\n Less occourences will result in a lower persentage\n threshold_percentage: the persentage to give a group if the amount of occourences is 'self.cluster_threshold'\n \"\"\"\n def __init__(self,\n eps=5,\n cluster_degregation=2,\n max_cluster_distance=7.5, #minutes\n weeks=5,\n decay_strength=0.5,\n cluster_threshold=25,\n threshold_percentage=90):\n self.eps = eps\n self.cluster_degregation = cluster_degregation\n self.max_cluster_distance = max_cluster_distance\n self.weeks = weeks\n self.decay_strength = decay_strength\n self.cluster_threshold = cluster_threshold\n self.threshold_percentage = threshold_percentage\n \n def analyze(self, df):\n \"\"\"Analyze a dataframe and return a list of predicted groups & relevant groups\n\n Args:\n df: the dataframe to analyze\n\n Returns:\n result: an array of predicted groups in the following format:\n [\n {\n item_ids: a list of item-id's that are predicted to be a group,\n is_predicted_group_percentage: the percentage chance that this is a group,\n is_relevant_group_percentage: the percentage chance that this group is still relevant \n (depending on how much it has been used lately)\n },\n {...},\n {...}\n ]\n \"\"\"\n # todo? cut off trailing days?\n \n self.lookup_table = self.create_lookup_table(\n df=df\n )\n df_fit = self.clean_dataframe(\n df=df\n )\n week_hashcodes = self.get_week_clusters_hash_codes(\n df=df_fit\n )\n hashcode_occurances = self.get_hashcode_occurances_per_week(\n week_hashcodes=week_hashcodes\n )\n predicted_groups = self.calculate_groups(\n hashcode_occurances_per_week=hashcode_occurances\n )\n \n result = []\n for key in predicted_groups:\n items = self.get_lookup_values(\n hashcode=key\n )\n result.append({\n 'item_ids': items,\n 'is_predicted_group_percentage': predicted_groups[key]['is_predicted_group_percentage'],\n 'is_relevant_group_percentage': predicted_groups[key]['is_relevant_group_percentage']\n })\n \n return result\n \n def create_lookup_table(self, df):\n \"\"\"Creates a lookup table for all unique row-id's\n\n Args:\n df: the dataframe containing an id column with several diffrent devices creating events\n\n Returns:\n lookup_dict: a dictionary where each id corresponds to an index e.g.\n { 0: 1743, 1: 1749, 2: 1803, 3: 1890, 4: 1911}\n \"\"\"\n df_lookup = pd.DataFrame(data={ 'id': pd.Series(df['id']).unique() })\n \n df_lookup['hashcode'] = self.clean_dataframe(\n df=df_lookup\n )['id']\n lookup_dict = dict()\n for index, row in df_lookup.iterrows():\n lookup_dict[row['hashcode']] = row['id']\n return lookup_dict\n \n def clean_dataframe(self, df):\n \"\"\"Convert non-nummeric values in the dataframe to numbers so that the dataframe can be used to fit a model\n\n Args:\n df: The dataframe to clean.\n\n Returns:\n df_fit: The dataframe with nummeric values\n \"\"\"\n d = defaultdict(LabelEncoder)\n df_fit = df.apply(lambda x: d[x.name].fit_transform(x))\n if 'state' in df.columns:\n df_fit['state'] = df['state']\n if 'time' in df.columns:\n df_fit['time'] = df['time']\n return df_fit\n \n def get_week_clusters_hash_codes(self, df):\n \"\"\"Get Cluster for a dataframe per week\n\n Args:\n df: The dataframe with more than one week of timestamps to cluster.\n\n Returns:\n week_hashcodes: A multidimentional array where each array is one week, and in one week array \n are a list of clusters represented by a hashcode.\n \n A hashcode is the reversed binary representation of a cluster, \n e.g. \n hashcode 3\n is binary 00000011\n is reversed 11000000\n means devices with index 0 and 1 (from the lookup table) are grouped\n \n Example output:\n [[3, 5, 20], [3, 3, 20]]\n means:\n amount of weeks: 2\n clusters in week 1:\n 3 (00000011) = a group with device 0 & 1\n 5 (00000101) = a group with device 0 & 2\n 20 (00010100) = a group with device 2 & 4\n clusters in week 2:\n 3 (00000011) = a group with device 0 & 1\n 3 (00000011) = another group with device 0 & 1\n 21 (00010101) = a group with device 0, 2 & 4\n \"\"\"\n one_week_in_milliseconds = (1000 * 60 * 60 * 24 * 7)\n last_timestamp = df['time'].max()\n week_hashcodes = []\n for week in range(self.weeks):\n week_hashcodes.append([])\n df_week = df[df['time'] >= last_timestamp - ((week + 1) * one_week_in_milliseconds)]\n df_week = df_week[df_week['time'] < last_timestamp - (week * one_week_in_milliseconds)]\n\n if not df_week.empty:\n cluster_arr = self.split_dataframe_on_state_and_get_cluster_arr(\n df=df_week, \n starting_eps=self.eps\n )\n for idx, df_week in enumerate(cluster_arr):\n cluster = []\n for row in df_week.iterrows():\n index, data = row\n cluster.append(data['id'].tolist())\n\n cluster = list(set(cluster))\n\n hashcode = 0\n for lamp in cluster:\n hashcode += pow(2, lamp)\n\n if(len(cluster) > 1):\n week_hashcodes[week].append(hashcode)\n else:\n print(\n 'WARNING!!! There are not', \n self.weeks, \n 'weeks in the dataset... amount_of_weeks HAS BEEN CHANGED TO', \n week\n )\n self.weeks = week\n break\n return week_hashcodes\n \n def split_dataframe_on_state_and_get_cluster_arr(self, df, starting_eps):\n \"\"\"Split a dataframe into 2 seperate dataframes (one with state=0, the other with state=1) \n and get the clusters for both of the dataframes\n\n Args:\n df: The dataframe to split & get clusters from.\n\n Returns:\n cluster_arr: an array that holds 0 or more dataframes (clusters)\n \"\"\"\n df_1 = df.loc[df['state'] == 1]\n df_0 = df.loc[df['state'] == 0]\n cluster_arr1 = self.get_clusters_recursive(df=df_1.copy(), eps=self.eps)\n cluster_arr2 = self.get_clusters_recursive(df=df_0.copy(), eps=self.eps)\n cluster_arr = cluster_arr1 + cluster_arr2\n return cluster_arr\n \n def get_clusters_recursive(self, df, eps, iteration=0, cluster_arr=None):\n \"\"\"Get clusters for a single dataframe\n \n Args:\n df: The dataframe\n eps: the epsilon to start with (maximum distance between two datapoints)\n\n Returns:\n cluster_arr: An array of dataframes (each one represents a cluster)e.g.\n [DataFrame, DataFrame, DataFrame, ...]\n \"\"\"\n if cluster_arr is None:\n cluster_arr = []\n \n model = self.fit_model(df, eps)\n cluster_dict = self.get_clusters(df=df, model=model)\n \n for idx, df in cluster_dict['too_large'].items():\n cluster_arr + self.get_clusters_recursive(\n df=cluster_dict['too_large'][idx], \n eps=eps / self.cluster_degregation, \n iteration=iteration + 1, \n cluster_arr=cluster_arr\n )\n \n for idx, df in cluster_dict['perfect_size'].items():\n cluster_arr.append(df)\n return cluster_arr\n \n \n def fit_model(self, df, eps):\n \"\"\"Fit the dataframe in the DBSCAN algoritm and return the model\n \n more information: https://scikit-learn.org/stable/modules/generated/sklearn.cluster.DBSCAN.html\n \n Args:\n df: The dataframe to run the algorithm on\n eps: the epsilon (maximum distance between two datapoints)\n\n Returns:\n model: The fitted DBSCAN model\n \"\"\"\n model = DBSCAN(\n eps=to_milliseconds(eps),\n min_samples=2\n ).fit(df)\n return model\n \n \n def get_clusters(self, df, model):\n \"\"\"Get clusters for a single dataframe\n \n Args:\n df: The dataframe\n model: the fitted model\n\n Returns:\n dict: A dictionary with 2 keys, each of wich is another dictionary which contains all dataframes (one per cluster)\n e.g.\n {\n 'too_large': {\n 0: DataFrame,\n 1: DataFrame,\n 2: DataFrame\n },\n 'perfect_size': {\n 0: DataFrame,\n 1: DataFrame\n }\n }\n \"\"\"\n df['cluster'] = model.labels_\n \n cluster_dict_too_large = {}\n cluster_dict_perfect_size = {}\n \n \n # Calculate amount of clusters\n cluster_data_count = Counter(model.labels_)\n if -1 in cluster_data_count:\n cluster_data_count.pop(-1) # don't count outliers as a cluster\n if (bool(cluster_data_count)):\n amount_of_clusters = max(cluster_data_count) + 1\n else:\n amount_of_clusters = 0;\n \n \n for idx in range(amount_of_clusters):\n cluster_df = df.loc[df['cluster'] == idx].drop(columns=['cluster'])\n \n first_time = cluster_df['time'].iloc[0]\n last_time = cluster_df['time'].iloc[cluster_df['time'].size - 1]\n diffrence_in_miliseconds = last_time - first_time\n if diffrence_in_miliseconds > to_milliseconds(self.max_cluster_distance):\n cluster_dict_too_large[idx] = cluster_df\n else:\n cluster_dict_perfect_size[idx] = cluster_df\n \n return {\n 'too_large': cluster_dict_too_large,\n 'perfect_size': cluster_dict_perfect_size\n }\n \n \n def get_hashcode_occurances_per_week(self, week_hashcodes):\n \"\"\"Count all occourences of hashcodes per week\n \n Args:\n week_hashcodes: The week_hashcodes (generated from self.get_week_clusters_hash_codes())\n\n Returns:\n count_dict: A dictionary with an index for each hashcode, with all\n occourences per week (last week = 0, the week before that = 1).\n e.g.\n {\n '3': {\n 'occurance_week': {\n '0': 24,\n '1': 56,\n '2': 32,\n '3': 12\n }\n },\n '5': { 'occurance_week': { ... } },\n '20': { 'occurance_week': { ... } },\n ...\n }\n \"\"\"\n count_dict = {}\n for week, hashcodes_arr in enumerate(week_hashcodes):\n for i in hashcodes_arr:\n if i in count_dict:\n count_dict[i]['occurance_week'][str(week)] += 1\n else:\n count_dict[i] = {}\n count_dict[i]['occurance_week'] = {}\n for w in range(self.weeks):\n count_dict[i]['occurance_week'][str(w)] = 0\n return count_dict\n \n def calculate_groups(self, hashcode_occurances_per_week):\n \"\"\"Calculate the predicted groups & relevant groups persentages from the amount of occourences.\n \n Args:\n hashcode_occurances_per_week: The hashcode occurances per week \n (generated from self.get_hashcode_occurances_per_week())\n\n Returns:\n count_dict: A dictionary with an index for each hashcode and the predicted groups & relevant groups persentages\n e.g.\n {\n '3': {\n 'is_predicted_group_percentage': 92.3,\n 'is_relevant_group_percentage': 72.1,\n },\n '5': { \n 'is_predicted_group_percentage': 42.9,\n 'is_relevant_group_percentage': 51.8,\n },\n '20': { ... },\n ...\n }\n \"\"\"\n count_dict = hashcode_occurances_per_week\n for key,val in count_dict.items():\n threshold = self.cluster_threshold * self.weeks\n\n total_occurances = 0\n for week in range(self.weeks):\n total_occurances += val['occurance_week'][str(week)]\n\n if total_occurances >= threshold:\n div = (total_occurances / threshold)\n count = 1\n perc = self.threshold_percentage\n\n while div > 1:\n div /= 2\n perc += ((100 - self.threshold_percentage) / 2) * (1 / count)\n count *= 2\n\n else:\n perc = (total_occurances / threshold) * self.threshold_percentage\n\n count_dict[key]['is_predicted_group_percentage'] = round(perc, 2)\n\n\n for key,val in count_dict.items():\n total = 0\n current = 0\n for week in range(self.weeks):\n\n perc = 0\n if val['occurance_week'][str(week)] >= self.cluster_threshold:\n div = (val['occurance_week'][str(week)] / self.cluster_threshold)\n count = 1\n perc = self.threshold_percentage\n while div > 1:\n div /= 2\n perc += ((100 - self.threshold_percentage) / 2) * (1 / count)\n count *= 2\n else:\n perc = (val['occurance_week'][str(week)] / self.cluster_threshold) * self.threshold_percentage\n\n total += 100 * (0.5) / pow(2, week * self.decay_strength)\n current += perc * (0.5) / pow(2, week * self.decay_strength)\n\n count_dict[key]['is_relevant_group_percentage'] = round((current / total) * 100, 2)\n count_dict[key].pop('occurance_week', None)\n return count_dict\n \n def get_lookup_values(self, hashcode):\n \"\"\"Get the individual item-indexes for a given hashcode\n\n Args:\n hashcode: The dataframe hashcode\n e.g. 21\n\n Returns:\n items: An array of items\n e.g. [0, 2, 4]\n \n e.g.\n hashcode 21 \n = 00010101 in binary \n = 10101000 reversed \n = item 0 = true,\n item 1 = false\n item 2 = true\n item 3 = false\n item 4 = true\n item 5 = false\n item 6 = false\n item 7 = false\n = a group with device 0, 2 & 4\n \"\"\"\n def bitfield(n):\n return [int(digit) for digit in bin(n)[2:]]\n \n bits = bitfield(hashcode)[::-1]\n \n items = []\n for idx, bit in enumerate(bits):\n if bit == 1:\n items.append(self.lookup_table[idx])\n return items",
"_____no_output_____"
],
[
"class DataFrameValidator:\n \"\"\"Validate a Dataframe for use in BinaryDataAnalisys\n Be aware: the values for the time column are expected to be timestamps in milliseconds\n \"\"\"\n time_column = 'time'\n expected_columns = ['id', 'state', time_column]\n minimum_days_of_data_needed = 7\n \n def validate(self, df):\n \"\"\"Validate a dataframe with the values specified above\n\n Args:\n df: the dataframe to validate\n\n Returns:\n boolean: if it's valid or not\n \"\"\"\n columns_valid = self.validate_columns(df)\n if not columns_valid:\n return False\n \n min_amount_of_data_valid = self.validate_minimum_days_of_data_needed(df)\n if not min_amount_of_data_valid:\n return False\n \n return True\n \n def validate_columns(self, df):\n \"\"\"Validate a dataframe's columns with the values specified above\n\n Args:\n df: the dataframe to validate\n\n Returns:\n boolean: if the columns are valid or not\n \"\"\"\n expected_df_columns = pd.DataFrame(columns=self.expected_columns)\n \n columns_too_many = df.columns.difference(expected_df_columns.columns)\n if not len(columns_too_many) == 0:\n print('The provided dataframe has too many columns:', *columns_too_many, sep='\\n')\n \n columns_too_few = expected_df_columns.columns.difference(df.columns)\n if not len(columns_too_few) == 0:\n print('The provided dataframe is missing the following columns:', *columns_too_few, sep='\\n')\n\n return len(columns_too_many) + len(columns_too_few) == 0\n \n def validate_minimum_days_of_data_needed(self, df):\n \"\"\"Validate a dataframe's amount of data with the values specified above\n\n Args:\n df: the dataframe to validate\n\n Returns:\n boolean: if the data is valid or not\n \"\"\"\n df_time = df.sort_values(by=[self.time_column])[self.time_column]\n first_timestamp = df_time.values[0]\n last_timestamp = df_time.values[-1]\n diff = last_timestamp - first_timestamp\n days = diff / 1000 / 60 / 60 / 24\n enough_data = days > self.minimum_days_of_data_needed\n \n if not enough_data:\n print(\n 'There is a minimum of ' + \n str(self.minimum_days_of_data_needed) + \n ' days of data needed, only ' + \n str(math.floor(days * 100) / 100) + \n ' days of data was given!'\n )\n \n return enough_data\n ",
"_____no_output_____"
]
],
[
[
"### get data & transform 'name' column into 'id' column",
"_____no_output_____"
]
],
[
[
"address = './datasets/staandelamp_realistic_huge.json'\ndf_data = pd.read_json(address)\ndf_data = df_data.sort_values(by=['time'])\ndf_data['id'] = df_data['name']\ndf_data = df_data.drop(columns=['name'])\nprint(df_data.shape)\ndf_data.head()",
"(175000, 3)\n"
]
],
[
[
"### Validate Dataframe",
"_____no_output_____"
]
],
[
[
"validator = DataFrameValidator()\ndataframe_is_valid = validator.validate(df_data)\n\nif dataframe_is_valid:\n print('Valid!')\nelse:\n print('WARNING! Dataframe validation failed!')",
"Valid!\n"
],
[
"if dataframe_is_valid:\n BDASCAN = BinaryDataAnalysis()\n result = BDASCAN.analyze(df_data)\n print(result[:5])",
"[{'item_ids': ['Staande_Lamp_1', 'Staande_Lamp_3'], 'is_predicted_group_percentage': 95.0, 'is_relevant_group_percentage': 93.27}, {'item_ids': ['Staande_Lamp_3', 'Staande_Lamp_4'], 'is_predicted_group_percentage': 95.0, 'is_relevant_group_percentage': 91.43}, {'item_ids': ['Staande_Lamp_2', 'Staande_Lamp_3'], 'is_predicted_group_percentage': 95.0, 'is_relevant_group_percentage': 85.54}, {'item_ids': ['Staande_Lamp_1', 'Staande_Lamp_3', 'Staande_Lamp_4'], 'is_predicted_group_percentage': 7.92, 'is_relevant_group_percentage': 7.76}, {'item_ids': ['Staande_Lamp_4', 'Staande_Lamp_5'], 'is_predicted_group_percentage': 28.08, 'is_relevant_group_percentage': 26.0}]\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d0ce4af3ab736f27caa51eea68698ced235f75a6 | 347,925 | ipynb | Jupyter Notebook | codes/create_circuit/GHZinitstate.ipynb | vutuanhai237/QuantumTomographyProject | 78058e3faece2209e46c9f9e16a1c38cdb33e7e2 | [
"MIT"
] | null | null | null | codes/create_circuit/GHZinitstate.ipynb | vutuanhai237/QuantumTomographyProject | 78058e3faece2209e46c9f9e16a1c38cdb33e7e2 | [
"MIT"
] | null | null | null | codes/create_circuit/GHZinitstate.ipynb | vutuanhai237/QuantumTomographyProject | 78058e3faece2209e46c9f9e16a1c38cdb33e7e2 | [
"MIT"
] | null | null | null | 1,353.793774 | 126,242 | 0.958494 | [
[
[
"# Quantum tomography for n-qubit\n\nInit state: general GHZ\nTarget state: 1 layer\n\nHere is the case for n-qubit with $n>1$.\n\nThe state that need to reconstruct is GHZ state:\n\n$\n|G H Z\\rangle=\\frac{1}{\\sqrt{2}}(|0 \\ldots 0\\rangle+|1 \\ldots 1\\rangle)=\\frac{1}{\\sqrt{2}}\\left[\\begin{array}{c}\n1 \\\\\n0 \\\\\n\\ldots \\\\\n1\n\\end{array}\\right]\n$\n\n$\n|G H Z\\rangle\\langle G H Z|=\\frac{1}{2}\\left[\\begin{array}{cccc}\n1 & 0 & \\ldots & 1 \\\\\n0 & \\ldots & \\ldots & 0 \\\\\n\\ldots & \\ldots & \\ldots & \\ldots \\\\\n1 & 0 & \\ldots & 1\n\\end{array}\\right]\n$\n\nIn general, the elements that have value 1 can be lower or greater base on $\\theta$. The below image is the construct GHZ state circuit in case of 4-qubits:\n\n<img src=\"../../images/general_ghz.png\" width=500px/>\n\nThe reconstructed circuit will be include $R_X, R_Z$ and $CNOT$ gates:\n\n<img src=\"../../images/1layer.png\"/>",
"_____no_output_____"
]
],
[
[
"import qiskit\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport sys\nsys.path.insert(1, '../')\nimport qtm.base, qtm.constant, qtm.ansatz",
"_____no_output_____"
],
[
"num_qubits = 3\nthetas = np.zeros((2*num_qubits*3))\ntheta = np.random.uniform(0, 2*np.pi)\n# Init quantum tomography n qubit\nqc = qiskit.QuantumCircuit(num_qubits, num_qubits)\n# qc = qtm.ansatz.create_ghz_state(qc, theta)\nqc = qtm.ansatz.u_cluster_nqubit(qc, thetas)\nqc.draw('mpl', scale = 6)",
"_____no_output_____"
],
[
"# Init parameters\nnum_qubits = 3\nthetas = np.zeros((2*num_qubits*3))\ntheta = np.random.uniform(0, 2*np.pi)\n# Init quantum tomography n qubit\nqc = qiskit.QuantumCircuit(num_qubits, num_qubits)\nqc = qtm.ansatz.create_ghz_state(qc)\n# Reduce loss value in 100 steps\nthetas, loss_values = qtm.base.fit(\n qc, num_steps = 200, thetas = thetas, \n create_circuit_func = qtm.ansatz.u_cluster_nqubit, \n grad_func = qtm.base.grad_loss,\n loss_func = qtm.loss.loss_basis,\n optimizer = qtm.optimizer.sgd,\n verbose = 1\n)\n# Plot loss value in 100 steps\nplt.plot(loss_values)\nplt.xlabel(\"Step\")\nplt.ylabel(\"Loss value\")\nplt.show()",
"Step: 100%|██████████| 200/200 [01:51<00:00, 1.79it/s]\n"
],
[
"plt.plot(loss_values)\nplt.xlabel(\"Step\")\nplt.ylabel(\"Loss value\")\nplt.savefig('ghz_init2', dpi = 600)",
"_____no_output_____"
],
[
"# Get statevector from circuit\npsi = qiskit.quantum_info.Statevector.from_instruction(qc)\nrho_psi = qiskit.quantum_info.DensityMatrix(psi)\npsi_hat = qiskit.quantum_info.Statevector(qtm.base.get_u_hat(\n thetas = thetas, \n create_circuit_func = qtm.ansatz.u_cluster_nqubit, \n num_qubits = qc.num_qubits\n))\nrho_psi_hat = qiskit.quantum_info.DensityMatrix(psi_hat)\n# Calculate the metrics\ntrace, fidelity = qtm.base.get_metrics(psi, psi_hat)\nprint(\"Trace: \", trace)\nprint(\"Fidelity: \", fidelity)",
"Trace: 0.008715837036442003\nFidelity: (0.9999240341854119-6.280715422572747e-16j)\n"
],
[
"qiskit.visualization.plot_state_city(rho_psi, title = 'rho_psi')",
"_____no_output_____"
],
[
"qiskit.visualization.plot_state_city(rho_psi_hat, title = 'rho_psi_hat')",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0ce60d9d0e9882419325dfe3827b615487324cd | 52,980 | ipynb | Jupyter Notebook | customer_journey_05.ipynb | quosi/doodl | 5965c9baa7ea929d2c88e690bf92c671865f31e8 | [
"MIT"
] | 1 | 2019-09-02T13:25:03.000Z | 2019-09-02T13:25:03.000Z | customer_journey_05.ipynb | quosi/doodl | 5965c9baa7ea929d2c88e690bf92c671865f31e8 | [
"MIT"
] | null | null | null | customer_journey_05.ipynb | quosi/doodl | 5965c9baa7ea929d2c88e690bf92c671865f31e8 | [
"MIT"
] | null | null | null | 28.438003 | 131 | 0.378728 | [
[
[
"import pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport numpy as np\nimport warnings\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
],
[
"df1 = pd.read_csv('monday.csv', sep = \";\")\ndf2 = pd.read_csv('tuesday.csv', sep = \";\")\ndf3 = pd.read_csv('wednesday.csv', sep = \";\")\ndf4 = pd.read_csv('thursday.csv', sep = \";\")\ndf5 = pd.read_csv('friday.csv', sep = \";\")",
"_____no_output_____"
],
[
"df = pd.concat([df1, df2, df3, df4, df5], ignore_index=True)",
"_____no_output_____"
],
[
"df_unique = pd.read_csv('df_unique.csv', index_col=None)\ndf_unique.drop('Unnamed: 0', axis=1, inplace=True)",
"_____no_output_____"
],
[
"# 5. Simulate a single customer\n# 5.1 create transition probability matrix",
"_____no_output_____"
],
[
"# create table with transition for each timestamp \n# two columns (from-->to)\n# concat/append this yourney for each customer\n# count individual transitions from each location \n# (from fruit -> drinks (87 people in total)\n# (from fruit -> spices (45 people in total) ...\n# those are the probabilities for each location)",
"_____no_output_____"
],
[
"df_journey = df_unique.groupby(['customer_unique', 'location', 'date']).count().sort_values(by = ['customer_unique', 'date'])",
"_____no_output_____"
],
[
"entry_point = df_journey.reset_index(1).index.get_level_values(level=0).unique()",
"_____no_output_____"
],
[
"entry_list= []\nfor i in entry_point:\n entry_list.append(df_journey.reset_index(1).xs(i)['location'][0])",
"_____no_output_____"
],
[
"df_journey.reset_index(inplace=True)",
"_____no_output_____"
],
[
"# count entry location\nimport collections\nentry_dict = collections.Counter(entry_list)",
"_____no_output_____"
],
[
"entry_p = []\nfor i in list(entry_dict.values()):\n entry_p.append(i/(sum(entry_dict.values())))",
"_____no_output_____"
],
[
"states_ = ['fruit', 'dairy', 'spices', 'drinks']",
"_____no_output_____"
],
[
"entry_dic = zip(states_,entry_p)\nlist(entry_dic)",
"_____no_output_____"
],
[
"sum(entry_p)",
"_____no_output_____"
],
[
"df_trans = df_journey[['location']]",
"_____no_output_____"
],
[
"df_trans['_to'] = df_journey[['location']].shift(periods=-1, fill_value='checkout')",
"_____no_output_____"
],
[
"indexlist = df_trans[df_trans['location'] == 'checkout'].index\ndf_trans.drop(indexlist, inplace=True)\ndf_trans['transition'] = df_trans['location'] + \"_\" + df_trans['_to']",
"_____no_output_____"
],
[
"df_trans",
"_____no_output_____"
],
[
"labels, trans = pd.factorize(df_trans['transition'])\ndf_trans['trans_fac'] = labels",
"_____no_output_____"
],
[
"len(labels)",
"_____no_output_____"
],
[
"trans_dict = df_trans['transition'].value_counts(ascending=True)",
"_____no_output_____"
],
[
"values = [value for (key, value) in sorted(trans_dict.items())]\nkeys = [key for (key, value) in sorted(trans_dict.items())]\nsorted_trans_dict = dict(zip(keys, values))\nsorted_trans_dict",
"_____no_output_____"
],
[
"df_unique.head()",
"_____no_output_____"
],
[
"trans_matrix = pd.DataFrame.from_dict({'entrance': [0,0,0,0,0,0],\n 'fruit': [0,0,0,0,0,0], \n 'spices': [0,0,0,0,0,0],\n 'dairy': [0,0,0,0,0,0],\n 'drinks': [0,0,0,0,0,0],\n 'checkout': [0,0,0,0,0,0]})\ntrans_matrix.set_index([pd.Index(['entrance', 'fruit', 'spices', 'dairy', 'drinks', 'checkout'])], inplace=True)",
"_____no_output_____"
],
[
"trans_matrix",
"_____no_output_____"
],
[
"for f in sorted_trans_dict.items():\n origin = str(f[0].split('_')[0])\n dest = str(f[0].split('_')[1])\n count_ = int(f[1])\n trans_matrix.loc[origin][dest] = count_",
"_____no_output_____"
],
[
"row_sum = trans_matrix.sum(axis=1)",
"_____no_output_____"
],
[
"prob_matrix = trans_matrix.T/row_sum\nprob_matrix.loc['checkout']['checkout']=1\n#prob_matrix.loc['entrance']['entrance']=1\nprob_matrix",
"_____no_output_____"
],
[
"prob_matrix.fillna(0, inplace=True)",
"_____no_output_____"
],
[
"prob_dict = dict(prob_matrix)\n# check:\nprob_matrix.T.loc['spices'].sum()",
"_____no_output_____"
],
[
"prob_matrix",
"_____no_output_____"
],
[
"# add 0 probability for going to checkout from entrance\nentry_p.append(0)\nentry_p.insert(0,0)",
"_____no_output_____"
],
[
"entry_p",
"_____no_output_____"
],
[
"# adding entrance probability\nprob_matrix['entrance'] = entry_p",
"_____no_output_____"
]
],
[
[
"# sort order of columns\ncols = list(prob_matrix.columns)\ncols = [cols[-1]] + cols[:-1]\nprob_matrix = prob_matrix[cols]",
"_____no_output_____"
]
],
[
[
"prob_matrix",
"_____no_output_____"
],
[
"# Average number of steps during journey",
"_____no_output_____"
],
[
"# counting every stage (including ideling customers)\n(df_journey.shape[0]-1)/df_journey['customer_unique'].nunique()",
"_____no_output_____"
],
[
"df_journey['customer_unique'].nunique()",
"_____no_output_____"
],
[
"len(set(df_journey['customer_unique'].values))",
"_____no_output_____"
],
[
"# counting every different stage",
"_____no_output_____"
],
[
"df.shape[0]/df_journey['customer_unique'].nunique()",
"_____no_output_____"
],
[
"# calculate probabilities at entrance\n# by counting first stage of all unique customers\n# cal. percentage value\n# use this number in markov.next_state('XXX') function ",
"_____no_output_____"
],
[
"x = prob_dict.keys()\nlist(x)[5]",
"_____no_output_____"
]
],
[
[
"_______________________________",
"_____no_output_____"
]
],
[
[
"class Customer(object):\n def __init__(self, transition_prob):\n \"\"\"\n Initialize the MarkovChain instance.\n Parameters\n ----------\n transition_prob: dict\n A dict object representing the transition\n probabilities in Markov Chain.\n Should be of the form:\n {'state1': {'state1': 0.1, 'state2': 0.4},\n 'state2': {...}}\n \"\"\"\n self.transition_prob = transition_prob\n self.states = list(transition_prob.keys())\n self.first_state = 'entrance'\n\n def next_state(self, current_state):\n return np.random.choice(\n self.states,\n p=[self.transition_prob[current_state][next_state]\n for next_state in self.states])\n\n def generate_states(self, current_state='entrance', no=50):\n future_states = []\n for i in range(no):\n next_state = self.next_state(current_state)\n future_states.append(next_state)\n current_state = next_state\n if future_states[-1] == self.states[-1]:\n break\n return future_states",
"_____no_output_____"
]
],
[
[
"# namespace\ndir(list)",
"_____no_output_____"
]
],
[
[
"markow = Customer(prob_dict)",
"_____no_output_____"
],
[
"markow.generate_states()",
"_____no_output_____"
],
[
"markov.transition_prob",
"_____no_output_____"
]
]
] | [
"code",
"raw",
"code",
"markdown",
"code",
"raw",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"raw"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"raw"
],
[
"code",
"code",
"code"
]
] |
d0ce63ff49ce00c4b72a7316d1c5d643594317fd | 3,367 | ipynb | Jupyter Notebook | notebooks/n02_problem_introduction.ipynb | pydy/pydy-tutorial-human-standing | 72b1d8513e339e9b10e501bd3490caa3fa997bc4 | [
"CC-BY-4.0"
] | 134 | 2015-05-19T15:24:18.000Z | 2022-03-12T09:39:03.000Z | notebooks/n02_problem_introduction.ipynb | pydy/pydy-tutorial-human-standing | 72b1d8513e339e9b10e501bd3490caa3fa997bc4 | [
"CC-BY-4.0"
] | 46 | 2015-05-05T18:08:20.000Z | 2022-01-28T11:12:42.000Z | notebooks/n02_problem_introduction.ipynb | pydy/pydy-tutorial-pycon-2014 | 72b1d8513e339e9b10e501bd3490caa3fa997bc4 | [
"CC-BY-4.0"
] | 62 | 2015-06-16T01:50:51.000Z | 2022-02-26T07:39:41.000Z | 36.204301 | 774 | 0.622513 | [
[
[
"In this tutorial we are going to make a simple model of a human (or humaniod robot) that is capable of balancing on its own. We will make several assumptions:\n\n- The human's motion is limited to a 2D plane (i.e. leaning backwards and forwards).\n- We only have three degrees of freedom: rotation at the ankle, knee, and hip.\n- The two legs do not move independently, but as one body (i.e. left and right thigh are one rigid body).\n- The forces generated by the muscles will be modeled as ideal torques between the adjacent body segments.\n\nThe following diagram shows the model and all of the parameters.",
"_____no_output_____"
]
],
[
[
"from IPython.display import Image",
"_____no_output_____"
],
[
"Image('figures/human_balance_diagram.png')",
"_____no_output_____"
]
],
[
[
"Reference Frames\n----------------\n\nThere are four reference frames and six important points. The orange inertial reference frame, $I$, is attached to the foot which is rigidly attached to the ground. The blue lower leg reference frame, $L$, is attached to the foot by a pin joint at the ankle point $A$ and rotates relative to the foot through $\\theta_1$. The green upper leg reference frame, $U$, is attached to the lower leg by a pin joint at the knee point $K$ and rotates relative to the lower leg through angle $\\theta_2$. The red torso reference frame, $T$, is pinned to the upper leg at the hip point, $H$, and rotates relative to the upper leg through the angle $\\theta_3$. Note that all rotations are about the $z$ axis and that they are relative to the orientation of the preceding body.\n\nGeometry\n--------\n\nThe lower and upper legs' lengths are defined as $l_L$ and $l_U$.\n\nMass Centers\n-------------\n\nThe three points $L_o$, $U_o$, $T_o$ are the mass centers of the body segments. These are each located on the line connecting the proximal and distal joints of each body segment and are located by the dimensions: $d_L$, $d_U$, and $d_T$.\n\nGravity\n-------\n\nGravity is directed downwards ($-y$) and applies a force with a magnitude of $m_Lg,m_Ug,m_Tg$ at each mass center, respectively.\n\nTorques\n-------\n\nThree torques represent the forces due to muscles contracting. The ankle $T_A$, knee $T_K$, and hip $T_H$ torques apply equal and opposite torques to the adjoining body segments.\n\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
d0ce78625dc45057aab17ea0f221a8ab12ee17ea | 43,378 | ipynb | Jupyter Notebook | ch04_classification/Concept03_logistic2d.ipynb | superizer/TensorFlow-Book | a751ada159c38f63d028943b6bb4e02f61c367d3 | [
"MIT"
] | null | null | null | ch04_classification/Concept03_logistic2d.ipynb | superizer/TensorFlow-Book | a751ada159c38f63d028943b6bb4e02f61c367d3 | [
"MIT"
] | null | null | null | ch04_classification/Concept03_logistic2d.ipynb | superizer/TensorFlow-Book | a751ada159c38f63d028943b6bb4e02f61c367d3 | [
"MIT"
] | null | null | null | 206.561905 | 38,376 | 0.912214 | [
[
[
"# Ch `04`: Concept `03`",
"_____no_output_____"
],
[
"## Logistic regression in higher dimensions",
"_____no_output_____"
],
[
"Set up the imports and hyper-parameters",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport numpy as np\nimport tensorflow as tf\nimport matplotlib.pyplot as plt\n\nlearning_rate = 0.1\ntraining_epochs = 2000",
"_____no_output_____"
]
],
[
[
"Define positive and negative to classify 2D data points:",
"_____no_output_____"
]
],
[
[
"x1_label1 = np.random.normal(3, 1, 1000)\nx2_label1 = np.random.normal(2, 1, 1000)\nx1_label2 = np.random.normal(7, 1, 1000)\nx2_label2 = np.random.normal(6, 1, 1000)\nx1s = np.append(x1_label1, x1_label2)\nx2s = np.append(x2_label1, x2_label2)\nys = np.asarray([0.] * len(x1_label1) + [1.] * len(x1_label2))",
"_____no_output_____"
]
],
[
[
"Define placeholders, variables, model, and the training op:",
"_____no_output_____"
]
],
[
[
"X1 = tf.placeholder(tf.float32, shape=(None,), name=\"x1\")\nX2 = tf.placeholder(tf.float32, shape=(None,), name=\"x2\")\nY = tf.placeholder(tf.float32, shape=(None,), name=\"y\")\nw = tf.Variable([0., 0., 0.], name=\"w\", trainable=True)\n\ny_model = tf.sigmoid(-(w[2] * X2 + w[1] * X1 + w[0]))\ncost = tf.reduce_mean(-tf.log(y_model * Y + (1 - y_model) * (1 - Y)))\ntrain_op = tf.train.GradientDescentOptimizer(learning_rate).minimize(cost)",
"_____no_output_____"
]
],
[
[
"Train the model on the data in a session:",
"_____no_output_____"
]
],
[
[
"with tf.Session() as sess:\n sess.run(tf.global_variables_initializer())\n prev_err = 0\n for epoch in range(training_epochs):\n err, _ = sess.run([cost, train_op], {X1: x1s, X2: x2s, Y: ys})\n if epoch % 100 == 0:\n print(epoch, err)\n if abs(prev_err - err) < 0.0001:\n break\n prev_err = err\n\n w_val = sess.run(w, {X1: x1s, X2: x2s, Y: ys})",
"0 0.6931461\n100 0.36477563\n200 0.26468396\n300 0.20803659\n400 0.17227423\n500 0.14782287\n600 0.13008825\n700 0.11664067\n"
]
],
[
[
"Here's one hacky, but simple, way to figure out the decision boundary of the classifier: ",
"_____no_output_____"
]
],
[
[
"x1_boundary, x2_boundary = [], []\nwith tf.Session() as sess:\n for x1_test in np.linspace(0, 10, 20):\n for x2_test in np.linspace(0, 10, 20):\n z = sess.run(tf.sigmoid(-x2_test*w_val[2] - x1_test*w_val[1] - w_val[0]))\n if abs(z - 0.5) < 0.05:\n x1_boundary.append(x1_test)\n x2_boundary.append(x2_test)",
"_____no_output_____"
]
],
[
[
"Ok, enough code. Let's see some a pretty plot:",
"_____no_output_____"
]
],
[
[
"plt.scatter(x1_boundary, x2_boundary, c='b', marker='o', s=20)\nplt.scatter(x1_label1, x2_label1, c='r', marker='x', s=20)\nplt.scatter(x1_label2, x2_label2, c='g', marker='1', s=20)\n\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0ce8bd6d5032ab139e49166f3aafcc4e9a8eedd | 397,220 | ipynb | Jupyter Notebook | Chapter2_Small_vs_Large_Worlds.ipynb | JoseJuan98/Statistical_Rethinking_Exercices | 03366df001193261fb429d422f477e156413ce88 | [
"Apache-2.0"
] | null | null | null | Chapter2_Small_vs_Large_Worlds.ipynb | JoseJuan98/Statistical_Rethinking_Exercices | 03366df001193261fb429d422f477e156413ce88 | [
"Apache-2.0"
] | null | null | null | Chapter2_Small_vs_Large_Worlds.ipynb | JoseJuan98/Statistical_Rethinking_Exercices | 03366df001193261fb429d422f477e156413ce88 | [
"Apache-2.0"
] | null | null | null | 447.31982 | 257,288 | 0.933926 | [
[
[
"# Chapter 2 - Small Worlds vs Large Wolrds\n\n[Recorded Classes 2019 Chap2 by Richard McElreath](https://www.youtube.com/watch?v=XoVtOAN0htU&list=PLDcUM9US4XdNM4Edgs7weiyIguLSToZRI&index=2)\n\nThe **Small World** represents the scientific model itself, and the **Large World**\nrepresents the broader context in which one deploys a model.\n\n**Bayesian inference** is just counting and comparing of possibilities. Consider\nby analogy Jorge Luis Borges’ short story “The Garden of Forking Paths.”\nIn order to make good inference about what actually happened, it helps to consider\n everything that could have happened. A Bayesian analysis is a garden of forking data,\n in which alternative sequences of events are cultivated.\n\n**The approach cannot guarantee a correct answer**, on large world terms. But it can\nguarantee the best possible answer, on small world terms, that could be derived\nfrom the information fed into it.\n\nThe goal of the Bayesian approach is to figure out which of the conjectures for a\ncertain context is **the most plausible**, given some evidence (data).",
"_____no_output_____"
],
[
"By comparing these counts, we have part of a solution\nfor a way to rate the relative plausibility of each conjecture.\nBut it’s only a part of a solution, because in order to compare these counts\nwe first have to decide how many ways each conjecture could itself be realized.\nWe might argue that when we have no reason to assume otherwise, we can just consider\n each conjecture equally plausible and compare the counts directly, **Principle of Indifference**.\n But often we do have reason to assume otherwise.\n\n> ***Principle of indifference***: When there is no reason to say that one conjecture is more plausible\n> than another, weigh all of the conjectures equally.\n\nTo grasp a solution, suppose we’re willing to say each conjecture is equally plausible\nat the start. Then, we just compare the counts of ways\nin which each conjecture is compatible with the observed data. So, comparing them can suggest\nthat ones are more plausible, than others. Since these are our initial counts, and\nprobably they are going to update later, they are labeled **prior**.\n\nThen when we get more evidence or observations, we can update the conjectures' plausibility.\nOnly if they new data is independent of the previous data,\n> To update the plausibility ***p*** of a conjecture ***C*** that is produced in ***W<sub>prior</sub>***\n> ways based on previous data ***D<sub>prior</sub>*** after providing more evidence ***D<sub>new</sub>***\n> is as follows:\n>\n> $\\Large P_c \\propto W_{prior} \\times W_{new} $\n\nWhy multiplication? Because it's a shortcut for counting all possible paths.\n\n",
"_____no_output_____"
],
[
"## From counting to probability\n\nIt’s hard to use these counts though, so almost always they are standardized in a way that\ntransforms them into probabilities.\n\nThe meaning would be the same, it’s just the relative values that matter. Second,\nas the amount of data grows, the counts will very quickly grow very large and become difficult\nto manipulate.\n\nThen, for any value p can take, we judge the plausibility of that value p\nas proportional to the number of ways it can get through the garden of forking data.\nFinally, we construct probabilities by standardizing the plausibility so that the sum of\nthe plausibilities for all possible conjectures will be one. All you need to do in order to\nstandardize is to add up all of the products, one for each value p can take, and then divide each\nproduct by the sum of products:",
"_____no_output_____"
],
[
"Being ***p*** the proportion of a feature,\n\n\\begin{align*}\n\\Large P_p={\\frac {W_{{p}_{new}} \\times P_{prior}}{\\sum \\small products}}\n\\end{align*}",
"_____no_output_____"
],
[
"## Example 2.1\nThere is a bag with four marbles, and we only know that they are <span style=\"color:blue\">blue [B]</span> and\n<span style=\"color:grey\">white [W]</span>. A marble is picked from the bag putting it back after finishing, after\ndoing this four times we got the sequence [<span style=\"color:blue\">B</span> <span style=\"color:grey\">W</span> <span style=\"color:blue\">B</span>] .\n\nSo if ***p*** is defined as the proportion of marbles that are blue, for [<span style=\"color:blue\">B </span><span style=\"color:grey\">W W W</span>]\nwith ***D<sub>new</sub>*** = [<span style=\"color:blue\">B</span> <span style=\"color:grey\">W</span> <span style=\"color:blue\">B</span>],\nwe can say that:\n\n> plausability of ***p*** after ***D<sub>new</sub>*** $\\propto$ was ***p*** can produce\n> ***D<sub>new</sub>*** $\\times$ prior plausability of ***p***\n\nThe above just means that for any value p can take, we judge the plausibility of that value p\nas proportional to the number of ways it can get through the garden of forking data.\n\n| Composition | p (prop.) | Ways (W) | Plausability (P) |\n| --- | --- | --- | --- |\n| [ <span style=\"color:grey\">W W W W</span> ] | 0 | 0 | 0 |\n| [ <span style=\"color:blue\">B </span><span style=\"color:grey\">W W W</span> ] | 0.25 | 3 | 0.15 |\n| [ <span style=\"color:blue\">B B</span><span style=\"color:grey\"> W W</span> ] | 0.5 | 8 | 0.4 |\n| [ <span style=\"color:blue\">B B B </span><span style=\"color:grey\">W</span> ] | 0.75 | 9 | 0.45 |\n| [ <span style=\"color:blue\">B B B B</span> ] | 1 | 0 | 0 |\n\n* A conjectured proportion of blue marbles, p, is usually called a ***parameter*** value.\nIt’s just a way of indexing possible explanations of the data.\n* The relative number of ways that a value p can produce the data is usually called\na ***likelihood***. It is derived by enumerating all the possible data sequences that\ncould have happened and then eliminating those sequences inconsistent with the\ndata.\n* The prior plausibility of any specific p is usually called the ***prior probability***.\n* The new, updated plausibility of any specific p is usually called the ***posterior\nprobability***.\n## Libraries import",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom scipy import stats\nimport random\nimport matplotlib.pyplot as plt\n%matplotlib inline\nnp.set_printoptions(suppress=True)",
"_____no_output_____"
]
],
[
[
"*2.1.1- How to calculate this plausibilities of the example 2.1 in Python?*",
"_____no_output_____"
]
],
[
[
"Ways=np.array([0,3,8,9,0])\n# Prior plausibility of p is 1 (it didn't change). So,\nWays/Ways.sum()",
"_____no_output_____"
]
],
[
[
"## 2.1 Building a model\nBy working with probabilities instead of raw counts, Bayesian inference is made much\neasier, but it looks much harder.\n\nTo get the logic moving, we need to make assumptions, and these assumptions constitute\nthe model. Designing a simple Bayesian model benefits from a design loop with three steps.\n1. Data story: Motivate the model by narrating how the data might arise.\n2. Update: Educate your model by feeding it the data.\n3. Evaluate: All statistical models require supervision, leading possibly to model revision.",
"_____no_output_____"
],
[
"### 2.1.1. A data story\n\nBayesian data analysis usually means producing a story for how the data came to be. This story may be descriptive, specifying associations that can be used to\npredict outcomes, given observations. Or it may be causal, a theory of how some events produce other events.\n\nTypically, any story you intend to be causal may also be descriptive. But many descriptive stories are hard to interpret causally. But all data stories are complete,\nin the sense that they are sufficient for specifying an algorithm for simulating new data.\n\n### 2.1.2 Bayesian updating\n\nUsing the evidence to decide among different possible conjectures, like the marbles on the bag previously. Each possible proportion may be more or less plausible, given the evidence.\nA Bayesian model begins with one set of plausibilities assigned to each of these possibilities.\nThese are the prior plausibilities. Then it updates them in light of the data, to produce the\nposterior plausibilities. This updating process is a kind of learning, called ***Bayesian Updating***.\n\nNotice that every updated set of plausibilities becomes the initial plausibilities for the\nnext observation. Every conclusion is the starting point for future inference. However, this\nupdating process works backwards, as well as forwards.\n\nGiven the final set of plausibilities, it is possible\nto mathematically divide out the observation, to infer the previous plausibility curve. So the\ndata could be presented to your model in any order, or all at once even. In most cases, you\nwill present the data all at once, for the sake of convenience. But it’s important to realize that\nthis merely represents abbreviation of an ***Iterated Learning Process***.",
"_____no_output_____"
],
[
"### 2.1.3 Evaluate\nThe Bayesian model learns in a way that is demonstrably optimal, provided\nthat the real, large world is accurately described by the model. This is to say that your\nBayesian machine guarantees perfect inference, within the small world. No other way of\nusing the available information, and beginning with the same state of information, could do\nbetter.\n\nHowever, the calculations may malfunction, so results always have to be checked. And if\nthere are important differences between the model and reality, then there is no logical\nguarantee of large world performance. And even if the two worlds did match, any particular\nsample of data could still be misleading. So it’s worth keeping in mind at least two cautious principles:\n\n1. *First, the model’s certainty is no guarantee that the model is a good one.*\n2. *Supervise and critique your model’s work.*\n\nMoreover, models do not need to be exactly true in order to produce highly precise and useful inferences.\nThis is because models are essentially information processing machines, and there are some surprising aspects of\ninformation that cannot be easily captured by framing the problem in terms of the truth of\nassumptions.\n\nInstead, the objective is to check the model’s adequacy for some purpose. This usually\nmeans asking and answering additional questions, beyond those that originally constructed\nthe model. Both the questions and answers will depend upon the scientific context.",
"_____no_output_____"
],
[
"## 2.2 Components of the model\nConsider three different kinds of things we counted in the previous sections.\n1. The number of ways each conjecture could produce an observation\n2. The accumulated number of ways each conjecture could produce the entire data\n3. The initial plausibility of each conjectured cause of the data\n\nEach of these things has a direct analog in conventional probability theory. And so the usual way we build a statistical model involves\nchoosing distributions and devices for each that represent the relative numbers of ways things can happen.\n\n\n1. Variables. Variables are just symbols that can take on different values. In a scientific context, \nvariables include things we wish to infer, such as proportions and rates, as well as things we might observe, the data. The first variable is our target of inference, *p*, *e.g. the proportion of marbles in the bag*. This variable cannot be observed. Unobserved variables are usually called ***PARAMETERS***. But while *p* itself is unobserved, we can infer it from the other variables.",
"_____no_output_____"
],
[
"> *When we observe a sample of variables, we need to say how likely that exact sample is, out of the universe of potential samples of the same length.*",
"_____no_output_____"
],
[
"2. Definitions. Once we have all the variables we need to define each, we build a model that relates the variables to one to another. The\ngoal is count all the ways the data could arise, given the assumptions. \n 1. ***Observed variables***. Define how plausible any combination of this variables is. Each specific value of ***p*** corresponds to aspecific plausibility of the data. In conventional statistics, a distribution function assigned to an observed variable is usually landcalled a likelihood. That term has special meaning in non-Bayesian statistics, however.\n 2. ***Unobserved variables***. The distributions we assign to the observed variables typically have their own variables. In the binomial below, there is $p$, the probability of sampling water. Since p is not observed, we usually call it a ***PARAMETER***. or every parameter you intend your Bayesian machine to consider, you must provide a\ndistribution of prior plausibility, its ***PRIOR***.\n\n\n",
"_____no_output_____"
],
[
"### 2.2.1 Prior\n \nWhen you have a previous estimate to provide, that can become the prior. As a result, each estimate becomes the prior for the next step.\nBut this doesn’t resolve the problem of providing a prior, because at the dawn of time, when $N = 0$, the machine still had an initial state of information for the parameter $p$: *a flat line specifying equal plausibility for every possible value*.\n\n*So where do priors come from?* They are both engineering assumptions, chosen to help the machine learn, and scientific assumptions, chosen to reflect what we know about a phenomenon. The flat prior is very common, but it is hardly ever the best prior.\n\nThere is a school of Bayesian inference that emphasizes choosing priors based upon the personal beliefs of the analyst. While this subjective Bayesian approach thrives in some statistics and philosophy and economics programs, it is rare in the sciences. Within Bayesian data analysis in the natural and social sciences, the prior is considered to be just part of the model. As such it should be chosen, evaluated, and revised just like all of the other components of the model. In practice, the subjectivist and the non-subjectivist will often analyze data in nearly the same way.\n\nBeyond all of the above, there’s no law mandating we use only one prior. If you don’t have a strong argument for any particular prior, then try different ones. Because the prior is an assumption, it should be interrogated like other assumptions: by altering it and checking how sensitive inference is to the assumption.",
"_____no_output_____"
],
[
"## Example 2.2\nSuppose you have a globe representing our planet, the Earth. This version of the world is small enough to hold in your hands. You are curious how much of the surface is covered in water. You adopt the following strategy: You will toss the globe up in the air. When you catch it, you will record whether or not the surface under your right index finger is water or land. Then you toss the globe up in the air again and repeat the procedure. This strategy generates a sequence of surface samples from the globe, where *W* indicates water and *L* indicates land.\n\nIn this case, once we add our assumptions that (1) every toss is independent of the other tosses and (2) the probability of W is the same on every toss, probability theory provides a unique answer, known as the binomial distribution. This is the common “coin tossing” distribution. And so the probability of observing W waters and L lands, with a probability p of water on each toss, is:",
"_____no_output_____"
],
[
"\\begin{align*}\n\\Large Pr(W, L|p)={\\frac {(W + L)!}{W!L!}}p^W(1-p)^L\n\\end{align*}\n\n> The counts of “water” W and “land’ L are distributed binomially, with prob-\nability p of “water” on each toss.",
"_____no_output_____"
],
[
"### Binom probability mass function \n\n\\begin{align*}\n\\Large f(k)={{n}\\choose{k}}p^k(1-p)^{n-k},\n\\end{align*}\n\n\\begin{align*} k \\in \\{0, 1,..., n\\} , 0 \\leq p \\leq 1 \n\\end{align*}",
"_____no_output_____"
],
[
"> Being $n$ the size of the sample or the *#samples*, $k$ the #times that a value has been selected in the sample, and $p$ the probability of that variable.\n\nBinom takes $n$ and $p$ as shape parameters, where $p$ is the probability of a single success $1 - p$ and is the probability of a single failure.\n\nThe probability mass function above is defined in the “standardized” form. To shift distribution use the loc parameter. Specifically, ```binom.pmf(k, n, p, loc)``` is identically equivalent to ```binom.pmf(k - loc, n, p)```",
"_____no_output_____"
],
[
"*How compute compute the likelihood of the data—six W’s in nine tosses—under any value of p with?*",
"_____no_output_____"
]
],
[
[
"stats.binom.pmf(k=6, n=9, p=0.5)",
"_____no_output_____"
]
],
[
[
"That number is the relative number of ways to get 6 water (in our globe-tossing model, holding $p$ at 0.5 and $N = W + L$ at nine.",
"_____no_output_____"
],
[
"With all the above work, we can now summarize out model. The observed variables $W$ and $L$ are given relative counts through the binomial distribution. So, we can write, as a shortcut:",
"_____no_output_____"
],
[
"\\begin{align*}\n\\Large W \\sim Binomial(N,p)\n\\end{align*}",
"_____no_output_____"
],
[
"where $N = W + L$. The above is just a convention for communicating the assumption that the relative counts of ways to realize W in N trials with probability p on each trial comes from the binomial distribution. And the unobserved parameter p similarly gets:\n\n\\begin{align*}\n\\Large p \\sim Uniform(0,1)\n\\end{align*}\n\nThis means that p has a uniform—flat—prior over its entire possible range, from zero to one.",
"_____no_output_____"
],
[
"## 2.4 Making the model\n\nOnce you have named all the variables and chosen definitions for each, a Bayesian model can update all of the prior distributions to their purely logical consequences: the ***POSTERIOR DISTRIBUTION***.\nFor every unique combination of data, likelihood, parameters, and prior, there is a unique posterior distribution.The posterior distribution takes the form of the probability of the parameters, conditional on the data. \n\nFor the proportion of water case case, it would be $Pr(p|W, L)$, the probability of each possible value of p, conditional on the specific $W$ and $L$ that we observed.",
"_____no_output_____"
],
[
"## 2.4.1 ***Baye's Theorem***\n\nThe mathematical definition of the posterior distribution arises from ***BAYES’ THEOREM***. \n\n\\begin{align*}\n\\Large Posterior={\\frac {Probability_{data} \\times Prior}{Average \\space probability_{data}}}\n\\end{align*}",
"_____no_output_____"
],
[
"Bayes’ theorem postulates that the probability of any particular value of p, considering the data, is equal to the product of the relative plausibility of the data, conditional on p, and the prior plausibility of p, divided by this thing $Pr(W, L)$, which I’ll call the ***average probability of the data***, sometimes called *“evidence”* or the *“average likelihood”*.",
"_____no_output_____"
],
[
"The posterior is proportional to the product of the prior and the probability of the data. Because for each specific value of $p$, the number of paths through the garden of forking data is the product of the prior number of paths and the new number of paths. Multiplication is just compressed counting.",
"_____no_output_____"
],
[
"### 2.4.2 Motors\n\nVarious numerical techniques are needed to approximate the mathematics that follows from the definition of Bayes’ theorem. One is the ***Grid Aprroximation***.\n\n## Grid Approximation\n\nWhile most parameters are continuous, capable of taking on an infinite number of values, it turns out that we can achieve an excellent approximation of the continuous posterior distribution by considering only a finite grid of parameter values.\n\nAt any particular value of a parameter, $p'$ , it’s a simple matter to compute the posterior probability: just multiply the prior probability of $p'$ by the likelihood at $p'$ . Repeating this procedure for each value in the grid generates an approximate picture of the exact posterior distribution.\n\n> *In most of real modeling, grid approximation isn’t practical. The reason is that it scales very poorly, as the number of parameters increases.*\n\nSummarizing:\n\n- The posterior probability is the *standarized product* of (1) probability of the data $\\times$ (2) prior probability\n- *Standarized* means: add up all the products and divide each by this sum: \n\n\\begin{align*}\n\\Large posterior\\{0...n\\} = \\sum_{i=1}^n {posterior}_i \\times {probability}_{data_i}\n\\end{align*}\n\n\\begin{align*}\n\\Large post\\_standarized_{i} = {\\frac {{posterior}_i} {\\sum_{j=1}^n posteriors_j}}\n\\end{align*}\n\n- Grid approximation uses *finite grid* of parameter values instead of continuous space\n- Too expensive with more than a few parameters\n\nIn the context of the globe tossing problem, grid approximation works extremely well. So let’s build a grid approximation for the model we’ve constructed so far. Here is the recipe:\n1. Define the grid. This means you decide how many points to use in estimating the\nposterior, and then you make a list of the parameter values on the grid.\n2. Compute the value of the prior at each parameter value on the grid.\n3. Compute the likelihood at each parameter value.\n4. Compute the unstandardized posterior at each parameter value, by multiplying the\nprior by the likelihood.\n5. Finally, standardize the posterior, by dividing each value by the sum of all values.\n\n",
"_____no_output_____"
],
[
"## Example 2.3",
"_____no_output_____"
],
[
"Following the example of globe tossing, we have as data:\n```python\n ['W','L','W','W','W','L','W','L','W']\n````",
"_____no_output_____"
],
[
"So that means $k=7$, $n=9$",
"_____no_output_____"
]
],
[
[
"def grid_approximation(k, n, points, prior, showDesc=False):\n #k=#success, n=#samples, points=#of point to aprroximate\n #Define grid\n grid = np.linspace(0, 1, points)\n #Define prior. For uniform=1, bc we assume all values are equally probable when N=0\n if type(prior) is int or type(prior) is float:\n prior = np.repeat(prior, points)\n #Compute likelihood at each value in the grid\n likelihood = stats.binom.pmf(k, n, grid)\n #Compute product of likelihood and prior\n posterior = likelihood * prior\n #Standarize the posterior, so it sums to 1\n posterior_std = posterior / np.sum(posterior)\n \n if(showDesc):\n print('Likelihood matrix {}'.format(likelihood))\n print('Posterior matrix {}'.format(posterior))\n print('Posterior standarize matrix {}'.format(posterior_std))\n \n return grid, posterior_std",
"_____no_output_____"
],
[
"k, n = 6, 9\ngrid, posterior = grid_approximation(k, n, 100, 1)",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(1, 1, figsize=(16,10))\nax.plot(grid, posterior,'-', label='binom pmf', color='grey')\nax.fill_between(grid, posterior, color='turquoise', alpha=.5)\nax.set_title('Grid Aprrox After 6/9 Successes with 100 points of estimation', fontsize=20)\nax.set_xlabel('Proportion of Earth is Water', fontsize=15)\nax.set_ylabel('Posterior Probability', fontsize=15);",
"_____no_output_____"
],
[
"def plot_likelihood(k, n, points, xlabel, ylabel, prior, showPrevious=False, showGridDesc=False):\n cols_per_row = 3\n from math import ceil\n if type(points) is list:\n m = len(points)\n if len(points) > cols_per_row:\n rows = ceil(m/cols_per_row)\n cols = cols_per_row\n else:\n cols = len(points)\n cols_per_row = len(points)\n rows = 1\n fig, axs = plt.subplots(nrows=rows, ncols=cols, sharey=False, sharex=False)\n if rows == 1:\n axs = np.array(axs).reshape(1, cols)\n fig.set_size_inches(12*cols,8*rows)\n else:\n m = 1\n fig, axs = plt.subplots(1, m, sharey=False, sharex=False)\n fig.set_size_inches(15,8)\n \n if type(points) is list:\n j = 0\n l = 0\n for i in points:\n grid, posterior = grid_approximation(k, n, i, prior, showDesc=showGridDesc)\n axs[l][j].plot(grid, posterior,'-', label='binom pmf', color='grey')\n axs[l][j].set_title(str(i)+' estimation points', fontsize=25)\n if j == 0 and l==0:\n axs[l][j].set_ylabel(ylabel+'\\n', fontsize=25)\n axs[l][j].legend(['Probability'])\n axs[l][j].fill_between(grid, posterior, color='turquoise')\n if showPrevious and (j,l) != (0,0):\n axs[l][j].legend(['Previous Probability','Current Probility'])\n j+=1\n if j == cols_per_row:\n j=0\n l+=1\n if showPrevious and (l*cols_per_row+j) < m:\n axs[l][j].plot(grid, posterior,':', label='binom pmf', color='red')\n if rows > 1:\n axs[l][j-1].set_xlabel(xlabel+'\\n\\n\\n ', fontsize=25)\n while j < cols_per_row:\n axs[l][j].set_visible(False)\n j+=1\n fig.suptitle('Grid Aprroximation After {}/{} Successes'.format(k,n), fontsize=35);\n else:\n if points < 3:\n points = 50\n print('Number of aprroximation points is to low. \\nSetting default #points of approximation to 50')\n grid, posterior = grid_approximation(k, n, points, prior, showDesc=showGridDesc)\n axs.plot(grid, posterior,'-', label='binom pmf', color='grey')\n axs.fill_between(grid, posterior, color='turquoise', alpha=.5)\n fig.suptitle('Grid Aprroximation After {}/{} Successes'.format(k,n), fontsize=35)\n axs.set_xlabel(xlabel+'\\n\\n\\n ', fontsize=25)\n axs.set_ylabel(ylabel+'\\n', fontsize=25);",
"_____no_output_____"
],
[
"point_lst = [4,10,50,75,100,200,1000]\nk, n = 6, 9\n\nplot_likelihood(k, n, point_lst,'Proportion of Earth is Water','Posterior Probability',1, showPrevious=True)",
"_____no_output_____"
]
],
[
[
"> After a certain amount of points the difference between approximation is minimal. \n\nMore points means more precision, but at certain amount of points there won’t be much change in inference, in this case after the first 100.",
"_____no_output_____"
],
[
"### Example 2.5\nLet's try the prior where we assume a 0% chance of the true proportion of water being less than 50%.",
"_____no_output_____"
]
],
[
[
"points = 100\nk, n = 6, 9\nprior = (np.linspace(0, 1, points) >= .5).astype(int)\n\nplot_likelihood(k, n, points, 'Proportion of Earth is Water','Posterior Probability', prior, showPrevious=True)",
"_____no_output_____"
]
],
[
[
"### 2.4.4 Quadratic approximation\n\nIt's a model that makes stronger assumptions than the *Grid Approximation*, because the main disadvatange of the Grid Approximation is that the number of unique values to consider in the grid grows rapidly as the number of parameters in your model increases. For two parameters approximated by 100 values each, that’s\nalready 100 2 = 10000 values to compute. For 10 parameters, the grid becomes many billions of values. These days, it’s routine to have models with hundreds or thousands of parameters. The grid approximation strategy scales very poorly with model complexity, so it won’t get very far.\n\nA useful approach is ***QUADRATIC APPROXIMATION***. Under quite general conditions, the region near the peak of the posterior distribution will be nearly Gaussian—or “normal”—in shape. This means the posterior distribution can be usefully approximated by a Gaussian distribution. A Gaussian distribution is convenient, because it can be completely described by only two numbers: the location of its center (mean) and its spread (variance). A Gaussian approximation is called “quadratic approximation” because the logarithm of a Gaussian distribution forms a parabola. And a parabola is a quadratic function. So this approximation essentially represents any log-posterior with a parabola.\n\nFor many of the most common procedures in applied statistics—linear regression, for example—the approximation works very well. Often, it is even exactly correct, not actually an approximation at all. Computationally, quadratic approximation is very inexpensive, at least compared to grid approximation and MCMC (discussed next).",
"_____no_output_____"
],
[
"# Problems\n## Easy\n\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d0ce95275ed00d80532259091078407c156739d5 | 300,783 | ipynb | Jupyter Notebook | Traffic_Sign_Classifier.ipynb | JohnGee96/CarND-Traffic-Sign-Classifier | 8bbb81bbfd57a6b4f4fe4937e4d9d92d2f923bc0 | [
"MIT"
] | null | null | null | Traffic_Sign_Classifier.ipynb | JohnGee96/CarND-Traffic-Sign-Classifier | 8bbb81bbfd57a6b4f4fe4937e4d9d92d2f923bc0 | [
"MIT"
] | null | null | null | Traffic_Sign_Classifier.ipynb | JohnGee96/CarND-Traffic-Sign-Classifier | 8bbb81bbfd57a6b4f4fe4937e4d9d92d2f923bc0 | [
"MIT"
] | null | null | null | 326.938043 | 119,038 | 0.9092 | [
[
[
"# Self-Driving Car Engineer Nanodegree\n\n## Deep Learning\n\n## Project: Build a Traffic Sign Recognition Classifier\n\nIn this notebook, a template is provided for you to implement your functionality in stages, which is required to successfully complete this project. If additional code is required that cannot be included in the notebook, be sure that the Python code is successfully imported and included in your submission if necessary. \n\n> **Note**: Once you have completed all of the code implementations, you need to finalize your work by exporting the iPython Notebook as an HTML document. Before exporting the notebook to html, all of the code cells need to have been run so that reviewers can see the final implementation and output. You can then export the notebook by using the menu above and navigating to \\n\",\n \"**File -> Download as -> HTML (.html)**. Include the finished document along with this notebook as your submission. \n\nIn addition to implementing code, there is a writeup to complete. The writeup should be completed in a separate file, which can be either a markdown file or a pdf document. There is a [write up template](https://github.com/udacity/CarND-Traffic-Sign-Classifier-Project/blob/master/writeup_template.md) that can be used to guide the writing process. Completing the code template and writeup template will cover all of the [rubric points](https://review.udacity.com/#!/rubrics/481/view) for this project.\n\nThe [rubric](https://review.udacity.com/#!/rubrics/481/view) contains \"Stand Out Suggestions\" for enhancing the project beyond the minimum requirements. The stand out suggestions are optional. If you decide to pursue the \"stand out suggestions\", you can include the code in this Ipython notebook and also discuss the results in the writeup file.\n\n\n>**Note:** Code and Markdown cells can be executed using the **Shift + Enter** keyboard shortcut. In addition, Markdown cells can be edited by typically double-clicking the cell to enter edit mode.",
"_____no_output_____"
],
[
"---\n## Step 0: Load The Data",
"_____no_output_____"
]
],
[
[
"# Load pickled data\nimport pickle\n\n# TODO: Fill this in based on where you saved the training and testing data\n\ntraining_file = './traffic-signs-data/train.p'\nvalidation_file = './traffic-signs-data/valid.p'\ntesting_file = './traffic-signs-data/test.p'\n\nwith open(training_file, mode='rb') as f:\n train = pickle.load(f)\nwith open(validation_file, mode='rb') as f:\n valid = pickle.load(f)\nwith open(testing_file, mode='rb') as f:\n test = pickle.load(f)\n \nX_train, y_train = train['features'], train['labels']\nX_valid, y_valid = valid['features'], valid['labels']\nX_test, y_test = test['features'], test['labels']",
"_____no_output_____"
]
],
[
[
"---\n\n## Step 1: Dataset Summary & Exploration\n\nThe pickled data is a dictionary with 4 key/value pairs:\n\n- `'features'` is a 4D array containing raw pixel data of the traffic sign images, (num examples, width, height, channels).\n- `'labels'` is a 1D array containing the label/class id of the traffic sign. The file `signnames.csv` contains id -> name mappings for each id.\n- `'sizes'` is a list containing tuples, (width, height) representing the original width and height the image.\n- `'coords'` is a list containing tuples, (x1, y1, x2, y2) representing coordinates of a bounding box around the sign in the image. **THESE COORDINATES ASSUME THE ORIGINAL IMAGE. THE PICKLED DATA CONTAINS RESIZED VERSIONS (32 by 32) OF THESE IMAGES**\n\nComplete the basic data summary below. Use python, numpy and/or pandas methods to calculate the data summary rather than hard coding the results. For example, the [pandas shape method](http://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.shape.html) might be useful for calculating some of the summary results. ",
"_____no_output_____"
],
[
"### Provide a Basic Summary of the Data Set Using Python, Numpy and/or Pandas",
"_____no_output_____"
]
],
[
[
"### Replace each question mark with the appropriate value. \n### Use python, pandas or numpy methods rather than hard coding the results\n\n# TODO: Number of training examples\nn_train = X_train.shape[0] # shape is a tuple: (num_samples, width, height, channel)\n\n# TODO: Number of validation examples\nn_validation = X_valid.shape[0]\n\n# TODO: Number of testing examples.\nn_test = X_test.shape[0]\n\n# TODO: What's the shape of an traffic sign image?\nimage_shape = X_train[0].shape\n\n# TODO: How many unique classes/labels there are in the dataset.\nn_classes = len(set(y_train))\n\nprint(\"Number of training examples =\", n_train)\nprint(\"Number of testing examples =\", n_test)\nprint(\"Image data shape =\", image_shape)\nprint(\"Number of classes =\", n_classes)",
"Number of training examples = 34799\nNumber of testing examples = 12630\nImage data shape = (32, 32, 3)\nNumber of classes = 43\n"
]
],
[
[
"### Include an exploratory visualization of the dataset",
"_____no_output_____"
],
[
"Visualize the German Traffic Signs Dataset using the pickled file(s). This is open ended, suggestions include: plotting traffic sign images, plotting the count of each sign, etc. \n\nThe [Matplotlib](http://matplotlib.org/) [examples](http://matplotlib.org/examples/index.html) and [gallery](http://matplotlib.org/gallery.html) pages are a great resource for doing visualizations in Python.\n\n**NOTE:** It's recommended you start with something simple first. If you wish to do more, come back to it after you've completed the rest of the sections. It can be interesting to look at the distribution of classes in the training, validation and test set. Is the distribution the same? Are there more examples of some classes than others?",
"_____no_output_____"
]
],
[
[
"### Data exploration visualization code goes here.\n### Feel free to use as many code cells as needed.\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport random\n\n# Visualizations will be shown in the notebook.\n%matplotlib inline\n\nROW = 4\nCOL = 6\n\ndef plot_data(data, label, channel=None, row=ROW, col=COL):\n fig, axes = plt.subplots(row, col, figsize=(12, 10))\n fig.subplots_adjust(hspace=0.01, wspace=0.1)\n axes = axes.flatten()\n for i in range(row * col):\n index = random.randint(0, len(data))\n if channel == 'gray':\n gray_image = data[index].squeeze()\n axes[i].imshow(gray_image, cmap='gray')\n else:\n axes[i].imshow(data[index])\n axes[i].set_title(label[index])\n axes[i].set_axis_off()\n \nplot_data(X_train, y_train)",
"_____no_output_____"
],
[
"# histogram of label frequency (once again, before data augmentation)\nhist, bins = np.histogram(y_train, bins=n_classes)\nwidth = 0.7 * (bins[1] - bins[0])\ncenter = (bins[:-1] + bins[1:]) / 2\nplt.bar(center, hist, align='center', width=width)\nplt.show()",
"_____no_output_____"
]
],
[
[
"----\n\n## Step 2: Design and Test a Model Architecture\n\nDesign and implement a deep learning model that learns to recognize traffic signs. Train and test your model on the [German Traffic Sign Dataset](http://benchmark.ini.rub.de/?section=gtsrb&subsection=dataset).\n\nThe LeNet-5 implementation shown in the [classroom](https://classroom.udacity.com/nanodegrees/nd013/parts/fbf77062-5703-404e-b60c-95b78b2f3f9e/modules/6df7ae49-c61c-4bb2-a23e-6527e69209ec/lessons/601ae704-1035-4287-8b11-e2c2716217ad/concepts/d4aca031-508f-4e0b-b493-e7b706120f81) at the end of the CNN lesson is a solid starting point. You'll have to change the number of classes and possibly the preprocessing, but aside from that it's plug and play! \n\nWith the LeNet-5 solution from the lecture, you should expect a validation set accuracy of about 0.89. To meet specifications, the validation set accuracy will need to be at least 0.93. It is possible to get an even higher accuracy, but 0.93 is the minimum for a successful project submission. \n\nThere are various aspects to consider when thinking about this problem:\n\n- Neural network architecture (is the network over or underfitting?)\n- Play around preprocessing techniques (normalization, rgb to grayscale, etc)\n- Number of examples per label (some have more than others).\n- Generate fake data.\n\nHere is an example of a [published baseline model on this problem](http://yann.lecun.com/exdb/publis/pdf/sermanet-ijcnn-11.pdf). It's not required to be familiar with the approach used in the paper but, it's good practice to try to read papers like these.",
"_____no_output_____"
],
[
"### Pre-process the Data Set (normalization, grayscale, etc.)",
"_____no_output_____"
],
[
"Minimally, the image data should be normalized so that the data has mean zero and equal variance. For image data, `(pixel - 128)/ 128` is a quick way to approximately normalize the data and can be used in this project. \n\nOther pre-processing steps are optional. You can try different techniques to see if it improves performance. \n\nUse the code cell (or multiple code cells, if necessary) to implement the first step of your project.",
"_____no_output_____"
]
],
[
[
"### Preprocess the data here. It is required to normalize the data. Other preprocessing steps could include \n### converting to grayscale, etc.\n### Feel free to use as many code cells as needed.\n\n# shuffle dataset\ndef unison_shuffle(x, y, seed=101):\n \"\"\"\n In-place unison shuffling\n \"\"\"\n rand_state = np.random.RandomState(seed)\n rand_state.shuffle(x)\n # re-seeding to get the same ordering\n rand_state.seed(seed)\n rand_state.shuffle(y)\n \ndef normalize(x):\n return (x - 128) / 128\n\ndef grayscale(x):\n return np.sum(x/3, axis=3, keepdims=True)\n \ndef preprocess(x):\n return normalize(grayscale(x))\n\n# Testing grayscaling\ngray = grayscale(X_valid)\nplot_data(gray, y_valid, channel='gray')",
"_____no_output_____"
],
[
"### Preprocessing Data \nprev_mean = np.mean(X_train)\nprint(\"Training data shape before preprocessing: \", X_train.shape)\nprint(\"Training data mean before preprocessing: \", prev_mean)\nX_train = preprocess(X_train)\nX_valid = preprocess(X_valid)\nX_test = preprocess(X_test)\nproc_mean = np.mean(X_train)\n\nprint(\"Training data shape after preprocessing: \", X_train.shape)\nprint(\"Training data mean after preprocessing: \", proc_mean)",
"Training data shape before preprocessing: (34799, 32, 32, 3)\nTraining data mean before preprocessing: 82.677589037\nTraining data shape after preprocessing: (34799, 32, 32, 1)\nTraining data mean after preprocessing: -0.354081335648\n"
]
],
[
[
"### Labeling",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nimport csv\nNUM_CHANNEL = 1\n\nwith open( './signnames.csv', 'rt') as f:\n reader = csv.reader(f)\n label_name = list(reader)\n\nx = tf.placeholder(tf.float32, (None, 32, 32, NUM_CHANNEL))\ny = tf.placeholder(tf.int32, (None))\nkeep_prob = tf.placeholder(tf.float32)\none_hot_y = tf.one_hot(y, n_classes)",
"_____no_output_____"
]
],
[
[
"### Model Architecture",
"_____no_output_____"
]
],
[
[
"from tensorflow.contrib.layers import flatten\n\ndef LeNet(x, mu=0, sigma= 0.1): \n # Layer 1: Convolutional. Input = 32x32x3. Output = 28x28x6.\n conv1_W = tf.Variable(tf.truncated_normal(shape=(5, 5, NUM_CHANNEL, 6), mean = mu, stddev = sigma))\n conv1_b = tf.Variable(tf.zeros(6))\n conv1 = tf.nn.conv2d(x, conv1_W, strides=[1, 1, 1, 1], padding='VALID') + conv1_b\n\n # Activation.\n conv1 = tf.nn.relu(conv1)\n\n # Pooling. Input = 28x28x6. Output = 14x14x6.\n conv1 = tf.nn.max_pool(conv1, ksize=[1, 2, 2, 1], strides=[1, 2, 2, 1], padding='VALID')\n\n # Layer 2: Convolutional. Output = 10x10x16.\n conv2_W = tf.Variable(tf.truncated_normal(shape=(5, 5, 6, 16), mean = mu, stddev = sigma))\n conv2_b = tf.Variable(tf.zeros(16))\n conv2 = tf.nn.conv2d(conv1, conv2_W, strides=[1, 1, 1, 1], padding='VALID') + conv2_b\n \n # Activation.\n conv2 = tf.nn.relu(conv2)\n\n # Pooling. Input = 10x10x16. Output = 5x5x16.\n conv2 = tf.nn.max_pool(conv2, ksize=[1, 2, 2, 1], strides=[1, 2, 2, 1], padding='VALID')\n\n # Flatten. Input = 5x5x16. Output = 400.\n fc0 = flatten(conv2)\n \n # Layer 3: Fully Connected. Input = 400. Output = 120.\n fc1_W = tf.Variable(tf.truncated_normal(shape=(400, 120), mean = mu, stddev = sigma))\n fc1_b = tf.Variable(tf.zeros(120))\n fc1 = tf.matmul(fc0, fc1_W) + fc1_b\n \n # Activation and drop out\n fc1 = tf.nn.relu(fc1)\n fc1 = tf.nn.dropout(fc1, keep_prob)\n\n # Layer 4: Fully Connected. Input = 120. Output = 86.\n fc2_W = tf.Variable(tf.truncated_normal(shape=(120, 86), mean = mu, stddev = sigma))\n fc2_b = tf.Variable(tf.zeros(86))\n fc2 = tf.matmul(fc1, fc2_W) + fc2_b\n \n # Activation and drop out\n fc2 = tf.nn.relu(fc2)\n fc2 = tf.nn.dropout(fc2, keep_prob)\n\n # Layer 5: Fully Connected. Input = 86. Output = 43.\n fc3_W = tf.Variable(tf.truncated_normal(shape=(86, n_classes), mean = mu, stddev = sigma))\n fc3_b = tf.Variable(tf.zeros(n_classes))\n logits = tf.matmul(fc2, fc3_W) + fc3_b\n \n return logits",
"_____no_output_____"
]
],
[
[
"### Train, Validate and Test the Model",
"_____no_output_____"
],
[
"A validation set can be used to assess how well the model is performing. A low accuracy on the training and validation\nsets imply underfitting. A high accuracy on the training set but low accuracy on the validation set implies overfitting.",
"_____no_output_____"
],
[
"### Setting up Pipeline",
"_____no_output_____"
]
],
[
[
"LR = 0.001\nEPOCHS = 50\nBATCH_SIZE = 128\nKEEP_PROB = 0.5\n# Arguments used for tf.truncated_normal, randomly defines variables for the weights and biases for each layer\nMU = 0\nSIGMA = 0.1\n# Saving output graphs in local directory\nGRAPH_FILENAME = './img/curr_training_curve.png'\n\nlogits = LeNet(x, MU, SIGMA)\ncross_entropy = tf.nn.softmax_cross_entropy_with_logits(labels=one_hot_y, logits=logits)\nloss_operation = tf.reduce_mean(cross_entropy)\noptimizer = tf.train.AdamOptimizer(learning_rate = LR)\ntraining_operation = optimizer.minimize(loss_operation)",
"_____no_output_____"
]
],
[
[
"### Validating",
"_____no_output_____"
]
],
[
[
"correct_prediction = tf.equal(tf.argmax(logits, 1), tf.argmax(one_hot_y, 1))\naccuracy_operation = tf.reduce_mean(tf.cast(correct_prediction, tf.float32))\nsaver = tf.train.Saver()\n \ndef evaluate(X_data, y_data):\n num_examples = len(X_data)\n total_accuracy = 0\n sess = tf.get_default_session()\n for offset in range(0, num_examples, BATCH_SIZE):\n end = offset + BATCH_SIZE\n batch_x, batch_y = X_data[offset:end], y_data[offset:end]\n accuracy = sess.run(accuracy_operation, feed_dict={x: batch_x, y: batch_y, keep_prob: 1.0})\n total_accuracy += (accuracy * len(batch_x))\n return total_accuracy / num_examples",
"_____no_output_____"
]
],
[
[
"### Training",
"_____no_output_____"
]
],
[
[
"class Plot(object):\n def __init__(self):\n self.train_accuracy_over_epoch = []\n self.valid_accuracy_over_epoch = [] \n \n def plot_graph(self):\n assert len(self.train_accuracy_over_epoch) == len(self.valid_accuracy_over_epoch)\n x = range(len(self.valid_accuracy_over_epoch))\n y1 = self.train_accuracy_over_epoch\n y2 = self.valid_accuracy_over_epoch\n \n plt.plot(x, y1,'b-', label='Training Accuray')\n plt.plot(x, y2,'r:', label='Validation Accuray')\n # Create empty plot with blank marker containing the extra label\n plt.plot([],[], ' ', label=\"LEARNING_RATE: {}\".format(LR))\n plt.plot([],[], ' ', label=\"BATCH_SIZE: {}\".format(BATCH_SIZE))\n plt.plot([],[], ' ', label=\"KEEP_PROB: {}\".format(KEEP_PROB))\n plt.plot([],[], ' ', label=\"Mu: {}\".format(MU))\n plt.plot([],[], ' ', label=\"Sigma: {}\".format(SIGMA))\n \n plt.title('Accuracy Over Time', fontsize=12)\n plt.xlabel('Epoch', fontsize=10)\n plt.ylabel('Accuracy', fontsize=10)\n plt.legend(loc='lower right', fontsize=6)\n plt.savefig(GRAPH_FILENAME)\n plt.show()\n \np = Plot()",
"_____no_output_____"
],
[
"with tf.Session() as sess:\n sess.run(tf.global_variables_initializer())\n num_examples = len(X_train)\n steps_per_epoch = int(num_examples / BATCH_SIZE)\n accum_entropy_loss = 0\n \n print(\"Training...\\n\")\n for i in range(EPOCHS):\n unison_shuffle(X_train, y_train)\n acc_training_accuracy = 0\n for offset in range(0, num_examples, BATCH_SIZE):\n end = offset + BATCH_SIZE\n batch_x, batch_y = X_train[offset:end], y_train[offset:end]\n _, batch_accuracy = sess.run([training_operation, accuracy_operation], \n feed_dict={x: batch_x, y: batch_y, keep_prob: KEEP_PROB})\n acc_training_accuracy += batch_accuracy\n \n avg_training_accuray = acc_training_accuracy / steps_per_epoch\n validation_accuracy = evaluate(X_valid, y_valid)\n p.train_accuracy_over_epoch.append(avg_training_accuray)\n p.valid_accuracy_over_epoch.append(validation_accuracy)\n print(\"EPOCH {} : Training Accuray {:.1f}%, Validation Accuracy {:.1f}%\".format(i, 100*avg_training_accuray, 100*validation_accuracy))\n p.plot_graph()\n \n saver.save(sess, './lenet')\n print(\"Model saved\")",
"Training...\n\nEPOCH 0 : Training Accuray 25.3%, Validation Accuracy 56.8%\nEPOCH 1 : Training Accuray 58.7%, Validation Accuracy 78.5%\nEPOCH 2 : Training Accuray 72.5%, Validation Accuracy 84.8%\nEPOCH 3 : Training Accuray 78.7%, Validation Accuracy 87.8%\nEPOCH 4 : Training Accuray 82.1%, Validation Accuracy 89.6%\nEPOCH 5 : Training Accuray 84.7%, Validation Accuracy 91.0%\nEPOCH 6 : Training Accuray 86.2%, Validation Accuracy 92.4%\nEPOCH 7 : Training Accuray 87.7%, Validation Accuracy 92.3%\nEPOCH 8 : Training Accuray 89.1%, Validation Accuracy 93.7%\nEPOCH 9 : Training Accuray 90.0%, Validation Accuracy 94.0%\nEPOCH 10 : Training Accuray 90.8%, Validation Accuracy 93.7%\nEPOCH 11 : Training Accuray 91.6%, Validation Accuracy 94.8%\nEPOCH 12 : Training Accuray 92.1%, Validation Accuracy 95.2%\nEPOCH 13 : Training Accuray 92.6%, Validation Accuracy 95.1%\nEPOCH 14 : Training Accuray 93.1%, Validation Accuracy 95.7%\nEPOCH 15 : Training Accuray 93.4%, Validation Accuracy 94.9%\nEPOCH 16 : Training Accuray 94.3%, Validation Accuracy 96.1%\nEPOCH 17 : Training Accuray 94.4%, Validation Accuracy 95.8%\nEPOCH 18 : Training Accuray 94.5%, Validation Accuracy 95.4%\nEPOCH 19 : Training Accuray 94.7%, Validation Accuracy 95.9%\nEPOCH 20 : Training Accuray 95.1%, Validation Accuracy 95.8%\nEPOCH 21 : Training Accuray 95.4%, Validation Accuracy 95.8%\nEPOCH 22 : Training Accuray 95.5%, Validation Accuracy 96.6%\nEPOCH 23 : Training Accuray 95.5%, Validation Accuracy 96.6%\nEPOCH 24 : Training Accuray 95.7%, Validation Accuracy 95.3%\nEPOCH 25 : Training Accuray 95.8%, Validation Accuracy 96.2%\nEPOCH 26 : Training Accuray 95.9%, Validation Accuracy 96.6%\nEPOCH 27 : Training Accuray 95.9%, Validation Accuracy 96.2%\nEPOCH 28 : Training Accuray 96.2%, Validation Accuracy 97.0%\nEPOCH 29 : Training Accuray 96.6%, Validation Accuracy 96.4%\nEPOCH 30 : Training Accuray 96.4%, Validation Accuracy 96.7%\nEPOCH 31 : Training Accuray 96.7%, Validation Accuracy 97.0%\nEPOCH 32 : Training Accuray 96.6%, Validation Accuracy 96.8%\nEPOCH 33 : Training Accuray 96.7%, Validation Accuracy 96.6%\nEPOCH 34 : Training Accuray 96.9%, Validation Accuracy 96.5%\nEPOCH 35 : Training Accuray 96.8%, Validation Accuracy 96.2%\nEPOCH 36 : Training Accuray 96.9%, Validation Accuracy 97.0%\nEPOCH 37 : Training Accuray 97.0%, Validation Accuracy 97.1%\nEPOCH 38 : Training Accuray 97.0%, Validation Accuracy 97.0%\nEPOCH 39 : Training Accuray 97.4%, Validation Accuracy 96.6%\nEPOCH 40 : Training Accuray 97.2%, Validation Accuracy 96.3%\nEPOCH 41 : Training Accuray 97.1%, Validation Accuracy 96.6%\nEPOCH 42 : Training Accuray 97.1%, Validation Accuracy 96.5%\nEPOCH 43 : Training Accuray 97.2%, Validation Accuracy 96.8%\nEPOCH 44 : Training Accuray 97.3%, Validation Accuracy 97.0%\nEPOCH 45 : Training Accuray 97.5%, Validation Accuracy 96.6%\nEPOCH 46 : Training Accuray 97.5%, Validation Accuracy 96.6%\nEPOCH 47 : Training Accuray 97.6%, Validation Accuracy 96.7%\nEPOCH 48 : Training Accuray 97.6%, Validation Accuracy 96.9%\nEPOCH 49 : Training Accuray 97.6%, Validation Accuracy 96.9%\n"
]
],
[
[
"### Log:\n1. 5/18/2018 - Training accuracy 99.4%, Validation Accuracy 92.3%\n - preprocessing: simple normalization: (pixel - 128)/128\n - lr: 0.001, batch: 128, epoch: 50, mu: 0, sigma: 0.1\n - Seems to be overfitting\n2. 5/18/2018 - Training accuracy 89.9%, Validation Accuracy 91.1%\n - added drop out at the fully connected layers\n - lr: 0.0015, batch: 128, epoch: 50, mu: 0, sigma: 0.1\n3. 5/18/2018 - Training Accuray 94.2%, Validation Accuracy 92.6%\n - preprocessing: normalization and grayscaling\n - everything else remains the same\n4. 5/18/2018 - Training Accuray 94.9% Validation Accuracy 94.1%\n - preprocessing: same\n - hyperparameter: everything stays the same except batch becomes 200\n - Try increasing learning rate \n5. 5/19/2018 - Training Accuray 85.0%, Validation Accuracy 86.7%\n - preprocessing: same\n - lr: 0.003 and everything else is the same\n6. 5/19/2018 - Training Accuray 97.0%, Validation Accuracy 96.4%\n - preprocessing: find bugs in preprocessing: didn't divide properly\n - pixel - 128/128 instead of (pixel - 128)/128\n - lr: 0.001 and everything else is the same\n - Much better convergence\n ",
"_____no_output_____"
],
[
"### Test Model",
"_____no_output_____"
]
],
[
[
"with tf.Session() as sess:\n saver = tf.train.import_meta_graph('./lenet.meta')\n saver.restore(sess, tf.train.latest_checkpoint('.'))\n test_accuracy = evaluate(X_test, y_test)\n print(\"Test Set Accuracy {:.1f}%\".format(100*test_accuracy))",
"Test Set Accuracy 94.2%\n"
]
],
[
[
"### Test Result\nThe accuracy of the model on the test set is **94.2%**. Comparing to the training accuracy of 97.6% and validation accuracy of 96.9%, the model seems to be overfitting. This can be due to the fact that the data set has unequal distribution across all 43 traffic signs. We can improve by generating more images for the scarce labels in the data set.",
"_____no_output_____"
],
[
"---\n\n## Step 3: Test a Model on New Images\n\nTo give yourself more insight into how your model is working, download at least five pictures of German traffic signs from the web and use your model to predict the traffic sign type.\n\nYou may find `signnames.csv` useful as it contains mappings from the class id (integer) to the actual sign name.",
"_____no_output_____"
],
[
"### Load and Output the Images",
"_____no_output_____"
]
],
[
[
"### Load the images and plot them here.\n### Feel free to use as many code cells as needed.\nimport matplotlib.image as mpimg\nimport cv2\n\ndownloaded_img_names = ['30kph','children-crossing','keep-right','road-work','stop']\nX_download = []\nfor name in downloaded_img_names:\n filepath = './downloaded_img/' + name + '.jpg'\n image = (mpimg.imread(filepath))\n X_download.append(cv2.resize(image, (32,32), interpolation=cv2.INTER_AREA))\n\n \nfig, axes = plt.subplots(1, 5, figsize=(20, 13), subplot_kw={'xticks': [], 'yticks': []})\nfig.subplots_adjust(hspace=0.3, wspace=0.05)\n\nfor ax, i in zip(axes.flat, range(5)):\n ax.imshow(X_download[i])\n ax.set_title(downloaded_img_names[i])\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Predict the Sign Type for Each Image",
"_____no_output_____"
]
],
[
[
"### Run the predictions here and use the model to output the prediction for each image.\n### Make sure to pre-process the images with the same pre-processing pipeline used earlier.\n### Feel free to use as many code cells as needed.\n \nX_download = preprocess(np.array(X_download))\ny_download = np.array([1, 28, 38, 25, 14])\n\nX_download.shape\n\npredict_operation = tf.nn.top_k(tf.nn.softmax(logits), k=5)\n\nwith tf.Session() as sess:\n saver.restore(sess, tf.train.latest_checkpoint('.'))\n prob, indices = sess.run(predict_operation, feed_dict={x: X_download, y: y_download, keep_prob: 1.0})\n# print(scores, indices)\n for i, image in enumerate(X_download):\n corr_label = label_name[y_download[i]+1]\n print(\"Image\", i + 1, \"- correct label:\", corr_label)\n for j in range(5):\n percent = prob[i][j] * 100\n label = label_name[indices[i][j] + 1]\n print('{:9.2f}% {}'.format(percent, label[1]))\n# print(\"Accuracy on Downloaded Images {:.1f}%\".format(100*test_accuracy))",
"Image 1 - correct label: ['1', 'Speed limit (30km/h)']\n 100.00% Speed limit (30km/h)\n 0.00% Speed limit (20km/h)\n 0.00% Speed limit (70km/h)\n 0.00% Stop\n 0.00% End of all speed and passing limits\nImage 2 - correct label: ['28', 'Children crossing']\n 83.90% Dangerous curve to the right\n 14.15% Road work\n 1.10% Keep right\n 0.34% General caution\n 0.29% Children crossing\nImage 3 - correct label: ['38', 'Keep right']\n 100.00% Keep right\n 0.00% No passing\n 0.00% Speed limit (20km/h)\n 0.00% Speed limit (30km/h)\n 0.00% Speed limit (50km/h)\nImage 4 - correct label: ['25', 'Road work']\n 100.00% Road work\n 0.00% Dangerous curve to the right\n 0.00% Bicycles crossing\n 0.00% Slippery road\n 0.00% Right-of-way at the next intersection\nImage 5 - correct label: ['14', 'Stop']\n 49.67% Stop\n 24.19% Road work\n 6.29% Turn right ahead\n 6.17% Turn left ahead\n 3.38% Priority road\n"
]
],
[
[
"### Analyze Performance\n\nThe accuracy on the downloaded image is **80%**.",
"_____no_output_____"
],
[
"### Project Writeup\n\nOnce you have completed the code implementation, document your results in a project writeup using this [template](https://github.com/udacity/CarND-Traffic-Sign-Classifier-Project/blob/master/writeup_template.md) as a guide. The writeup can be in a markdown or pdf file. ",
"_____no_output_____"
],
[
"> **Note**: Once you have completed all of the code implementations and successfully answered each question above, you may finalize your work by exporting the iPython Notebook as an HTML document. You can do this by using the menu above and navigating to \\n\",\n \"**File -> Download as -> HTML (.html)**. Include the finished document along with this notebook as your submission.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
d0cea51ddadf7d07d37261f18a7da001a4295d57 | 352,805 | ipynb | Jupyter Notebook | examples/tutorials/example 1 - how to generate synthetic data.ipynb | bcebere/synthetic_data_generation | 84d34d00a5859d3db5d160ef8798b865a0c59fe7 | [
"MIT"
] | 4 | 2021-02-16T00:38:27.000Z | 2022-01-21T21:59:28.000Z | examples/tutorials/example 1 - how to generate synthetic data.ipynb | Daan0/synthetic_data_generation | 5a0d1818cba2bc8b629869773a2f86a156d25fd9 | [
"MIT"
] | 2 | 2021-08-20T14:32:59.000Z | 2022-03-18T10:15:40.000Z | examples/tutorials/example 1 - how to generate synthetic data.ipynb | Daan0/synthetic_data_generation | 5a0d1818cba2bc8b629869773a2f86a156d25fd9 | [
"MIT"
] | 5 | 2020-12-08T05:01:39.000Z | 2022-03-24T18:12:45.000Z | 511.311594 | 295,820 | 0.937121 | [
[
[
"# Example 1: How to Generate Synthetic Data (MarginalSynthesizer)\nIn this notebook we show you how to create a simple synthetic dataset.",
"_____no_output_____"
],
[
"# Environment",
"_____no_output_____"
],
[
"## Library Imports",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nfrom pathlib import Path\nimport os\nimport sys",
"_____no_output_____"
]
],
[
[
"## Jupyter-specific Imports and Settings",
"_____no_output_____"
]
],
[
[
"# set printing options\nnp.set_printoptions(threshold=sys.maxsize)\npd.set_option('display.max_rows', 500)\npd.set_option('display.max_columns', 500)\npd.set_option('display.width', 1000)\npd.set_option('display.expand_frame_repr', False)\n\n# Display all cell outputs\nfrom IPython.core.interactiveshell import InteractiveShell\nInteractiveShell.ast_node_interactivity = 'all'\n\nfrom IPython import get_ipython\nipython = get_ipython()\n\n# autoreload extension\nif 'autoreload' not in ipython.extension_manager.loaded:\n get_ipython().run_line_magic('load_ext', 'autoreload')\n\nget_ipython().run_line_magic('autoreload', '2')\nfrom importlib import reload",
"_____no_output_____"
]
],
[
[
"## Import Synthesizer\nFor this example we use the MarginalSynthesizer algorithm. As the name suggests, this algorithm generates data via the marginal distributions of each column in the input dataset. In other words, the output synthetic data will have similar counts for each column but the statistical patterns between columns are likely not preserved. While this method is rather naive, it will work with data of any shape or size - and run relatively quickly as well. ",
"_____no_output_____"
]
],
[
[
"from synthesis.synthesizers import MarginalSynthesizer",
"_____no_output_____"
]
],
[
[
"# Synthetic Data Generation\nLet's load a dataset to see how the generation process works.\n\nIn this case, we will use the adult dataset - source: https://archive.ics.uci.edu/ml/datasets/adult",
"_____no_output_____"
]
],
[
[
"df_original = pd.read_csv('../data/original/adult.csv')\ndf_original.head()",
"_____no_output_____"
]
],
[
[
"We will now import our synthesizer and fit it on the input data. \n\nAdditionally we can specify the 'epsilon' value, which according to the definition of differential privacy is used to quantify the privacy risk posed by releasing statistics computed on sensitive data. More on that here: https://www.cis.upenn.edu/~aaroth/Papers/privacybook.pdf\n\nIn short, a lower value of epsilon will result in more randomness and v.v.",
"_____no_output_____"
]
],
[
[
"epsilon = 1 # set to float(np.inf) if you'd like to compute without differential privacy.\n\nsynthesizer = MarginalSynthesizer(epsilon=epsilon)\nsynthesizer.fit(df_original)",
"Marginal fitted: age\nMarginal fitted: workclass\nMarginal fitted: fnlwgt\nMarginal fitted: education\nMarginal fitted: education-num\nMarginal fitted: marital-status\nMarginal fitted: occupation\nMarginal fitted: relationship\nMarginal fitted: race\nMarginal fitted: sex\nMarginal fitted: capital-gain\nMarginal fitted: capital-loss\nMarginal fitted: hours-per-week\nMarginal fitted: native-country\nMarginal fitted: income\n"
]
],
[
[
"After our synthesizer has fitted the structure of the original data source, we can now use it to generate a new dataset.",
"_____no_output_____"
]
],
[
[
"# we can specify the number of records by sample(n_records=...), \n# default it generates the same number of records as the input data\ndf_synthetic = synthesizer.sample()",
"Column sampled: age\nColumn sampled: workclass\nColumn sampled: fnlwgt\nColumn sampled: education\nColumn sampled: education-num\nColumn sampled: marital-status\nColumn sampled: occupation\nColumn sampled: relationship\nColumn sampled: race\nColumn sampled: sex\nColumn sampled: capital-gain\nColumn sampled: capital-loss\nColumn sampled: hours-per-week\nColumn sampled: native-country\nColumn sampled: income\n"
]
],
[
[
"We now obtained a new dataset which looks very similar to the original one.",
"_____no_output_____"
]
],
[
[
"df_synthetic.head()",
"_____no_output_____"
]
],
[
[
"# Evaluation\nWe can see that the synthetic data has a similar structure the original. We can also evaluate whether it has retained the statistical distributions of the original data. We use the SyntheticDataEvaluator class to compare the synthetic data to the original by applying various metrics.\n\nNote: for more elaborate evaluation techniques we refer to the example notebook on 'evaluating synthetic data'.",
"_____no_output_____"
]
],
[
[
"from synthesis.evaluation import SyntheticDataEvaluator",
"_____no_output_____"
],
[
"evaluator = SyntheticDataEvaluator()\nevaluator.fit(df_original, df_synthetic)\nevaluator.score()",
"_____no_output_____"
],
[
"evaluator.plot()",
"_____no_output_____"
]
],
[
[
"Observe that the marginal distributions are preserved quite well - especially for columns with low dimensionality. When using differentialy private algorithms (like MarginalSynthesizer) it is advised to reduce the dimensionality of the original data by generalizing columns. \n\nAlso observe that the last plot shows the synthetic data did not capture any of the correlations in the original data. This is expected as MarginalSynthesizer synthesizes data by columns independently. ",
"_____no_output_____"
],
[
"# Conclusion\n\nWe hope that gave you a quick introduction on synthetic data generation. Now go try it on your own data!\n\nIn the next example notebook we show how a more sophisticated algorithm is able to preserve statistical patterns between columns in the original data.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
d0ceaa5acba4d5501fb8c1038d9dd26d02dd7b9d | 127,933 | ipynb | Jupyter Notebook | CTC2019_tutorial.ipynb | kbroman/Teaching_CTC2019tutorial | b04eba36570d740f40c7c23188509d8587930c3e | [
"CC0-1.0"
] | null | null | null | CTC2019_tutorial.ipynb | kbroman/Teaching_CTC2019tutorial | b04eba36570d740f40c7c23188509d8587930c3e | [
"CC0-1.0"
] | 1 | 2019-06-09T18:00:39.000Z | 2019-06-09T18:59:17.000Z | CTC2019_tutorial.ipynb | kbroman/Teaching_CTC2019tutorial | b04eba36570d740f40c7c23188509d8587930c3e | [
"CC0-1.0"
] | 1 | 2019-06-09T14:06:02.000Z | 2019-06-09T14:06:02.000Z | 247.932171 | 48,388 | 0.89568 | [
[
[
"# Mapping QTL in BXD mice using R/qtl2\n\n[Karl Broman](https://kbroman.org)\n[<img style=\"display:inline-block;\" src=\"https://orcid.org/sites/default/files/images/orcid_16x16(1).gif\">](https://orcid.org/0000-0002-4914-6671),\n[Department of Biostatistics & Medical Informatics](https://www.biostat.wisc.edu), \n[University of Wisconsin–Madison](https://www.wisc.edu)\n\nOur aim in this tutorial is to demonstrate how to map quantitative trait loci (QTL) in the BXD mouse recombinant inbred lines using the [R/qtl2](https://kbroman.org/qtl2) software. We will first show how to download BXD phenotypes from [GeneNetwork2](http://gn2.genenetwork.org) using its API, via the R package [R/GNapi](https://github.com/rqtl/GNapi). At the end, we will use the [R/qtl2browse](https://github.com/rqtl/qtl2browse) package to display genome scan results using the [Genetics Genome Browser](https://github.com/chfi/purescript-genome-browser).",
"_____no_output_____"
],
[
"## Acquiring phenotypes with the GeneNetwork API\n\nWe will first use the [GeneNetwork2](http://gn2.genenetwork.org) API to acquire BXD phenotypes to use for mapping. We will use the R package [R/GNapi](https://github.com/rqtl/GNapi). \n\nWe first need to install the package, which is not available on [CRAN](https://cran.r-project.org), but is available via a private repository.\n\n```r\ninstall.packages(\"GNapi\", repos=\"http://rqtl.org/qtl2cran\")\n```\n\nWe then load the package using `library()`.",
"_____no_output_____"
]
],
[
[
"library(GNapi)",
"_____no_output_____"
]
],
[
[
"The [R/GNapi](https://github.com/kbroman/GNapi) has a variety of functions. For an overview, see [its vignette](http://kbroman.org/GNapi/GNapi.html). Here we will just do one thing: use the function `get_pheno()` to grab BXD phenotype data. You provide a data set and a phenotype. Phenotype 10038 concerns \"habituation\", measured as a difference in locomotor activity between day 1 and day 3 in a 5 minute test trial. ",
"_____no_output_____"
]
],
[
[
"phe <- get_pheno(\"BXD\", \"10038\")\nhead(phe)",
"_____no_output_____"
]
],
[
[
"We will use just the column \"value\", but we need to include the strain names so that R/qtl2 can line up these phenotypes with the genotypes.",
"_____no_output_____"
]
],
[
[
"pheno <- setNames(phe$value, phe$sample_name)\nhead(pheno)",
"_____no_output_____"
]
],
[
[
"## Acquiring genotype data with R/qtl2\n\nWe now want to get genotype data for the BXD panel. We first need to install the [R/qtl2](https://kbroman.org/qtl2) package. As with R/GNapi, it is not available on CRAN, but rather is distributed via a private repository.\n\n```r\ninstall.packages(\"qtl2\", repos=\"http://rqtl.org/qtl2cran\")\n```\n\nWe then load the package with `library()`.",
"_____no_output_____"
]
],
[
[
"library(qtl2)",
"_____no_output_____"
]
],
[
[
"R/qtl2 uses a special file format for QTL data ([described here](https://kbroman.org/qtl2/assets/vignettes/input_files.html)). There are a variety of sample datasets [on Github](https://github.com/rqtl/qtl2data), including genotypes for the [mouse BXD lines](https://github.com/rqtl/qtl2data/tree/master/BXD), taken from [GeneNetwork2](http://gn2.genenetwork.org). We'll load those data directly into R using the function `read_cross2()`.",
"_____no_output_____"
]
],
[
[
"bxd_file <- \"https://raw.githubusercontent.com/rqtl/qtl2data/master/BXD/bxd.zip\"\nbxd <- read_cross2(bxd_file)",
"Warning message in recode_geno(sheet, genotypes):\n“117497 genotypes treated as missing: \"H\"”"
]
],
[
[
"We get a warning message about heterozygous genotypes being omitted. A number of the newer BXD lines have considerable heterozygosity. But these lines weren't among those phenotyped in the data we downloaded above, and so we don't need to worry about it here.\n\nThe data are read into the object `bxd`, which has class `\"cross2\"`. It contains the genotypes and well as genetic and physical marker maps. There are also phenotype data (which we will ignore).\n\nWe can get a quick summary of the dataset with `summary()`. For reasons that I don't understand, it gets printed as a big mess within this Jupyter notebook, and so here we need to surround it with `print()` to get the intended output.",
"_____no_output_____"
]
],
[
[
"print( summary(bxd) )",
"Object of class cross2 (crosstype \"risib\")\n\nTotal individuals 198\nNo. genotyped individuals 198\nNo. phenotyped individuals 198\nNo. with both geno & pheno 198\n\nNo. phenotypes 5806\nNo. covariates 0\nNo. phenotype covariates 1\n\nNo. chromosomes 20\nTotal markers 7320\n\nNo. markers by chr:\n 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 X \n636 583 431 460 470 449 437 319 447 317 375 308 244 281 247 272 291 250 310 193 \n"
]
],
[
[
"## QTL mapping in R/qtl2\n\nThe first step in QTL analysis is to calculate genotype probabilities at putative QTL positions across the genome, conditional on the observed marker data. This allows us that consider positions between the genotyped markers and to allow for the presence of genotyping errors.\n\nFirst, we need to define the positions that we will consider. We will take the observed marker positions and insert a set of \"pseudomarkers\" (marker-like positions that are not actually markers). We do this with the function `insert_pseudomarkers()`. We pull the genetic map (`gmap`) out of the `bxd` data as our basic map; `step=0.2` and `stepwidth=\"max\"` mean to insert pseudomarkers so that no two adjacent markers or pseudomarkers are more than 0.2 cM apart. That is, in any marker interval that is greater than 0.2 cM, we will insert one or more evenly spaced pseudomarkers, so that the intervals between markers and pseudomarkers are no more than 0.2 cM.",
"_____no_output_____"
]
],
[
[
"gmap <- insert_pseudomarkers(bxd$gmap, step=0.2, stepwidth=\"max\")",
"_____no_output_____"
]
],
[
[
"We will be interested in results with respect to the physical map (in Mbp), and so we need to create a corresponding map that includes the pseudomarker positions. We do this with the function `interp_map()`, which uses linear interpolation to get estimated positions for the inserted pseudomarkers.",
"_____no_output_____"
]
],
[
[
"pmap <- interp_map(gmap, bxd$gmap, bxd$pmap)",
"_____no_output_____"
]
],
[
[
"We can now proceed with calculating genotype probabilities for all BXD strains at all markers and pseudomarkers, conditional on the observed marker genotypes and assuming a 0.2% genotyping error rate. We use the [Carter-Falconer](https://doi.org/10.1007/BF02996226) map function to convert between cM and recombination fractions; it assumes a high degree of crossover interference, appropriate for the mouse.",
"_____no_output_____"
]
],
[
[
"pr <- calc_genoprob(bxd, gmap, error_prob=0.002, map_function=\"c-f\")",
"_____no_output_____"
]
],
[
[
"In the QTL analysis, we will fit a linear mixed model to account for polygenic background effects. We will use the \"leave one chromosome out\" (LOCO) method for this. When we scan a chromosome for a QTL, we include a polygenic term with a kinship matrix derived from all other chromosomes. \n\nWe first need to calculate this set of kinship matrices, which we do with the function `calc_kinship()`. The second argument, `\"loco\"`, indicates that we want to calculate a vector of kinship matrices, each derived from the genotype probabilities but leaving one chromosome out.",
"_____no_output_____"
]
],
[
[
"k <- calc_kinship(pr, \"loco\")",
"_____no_output_____"
]
],
[
[
"Now, finally, we're ready to perform the genome scan, which we do with the function `scan1()`. It takes the genotype probabilities and a set of phenotypes (here, just one phenotype). If kinship matrices are provided (here, as `k`), the scan is performed using a linear mixed model. To make the calculations faster, the residual polygenic variance is first estimated without including any QTL effect and is then taking to be fixed and known during the scan.",
"_____no_output_____"
]
],
[
[
"out <- scan1(pr, pheno, k)",
"_____no_output_____"
]
],
[
[
"The output of `scan1()` is a matrix of LOD scores; the rows are marker/pseudomarker positions and the columns are phenotypes. We can plot the results using `plot.scan1()`, and we can just use `plot()` because it uses the class of its input to determine what plot to make.\n\nHere I'm using the package [repr](https://cran.r-project.org/package=repr) to control the height and width of the plot that's created. I installed it with `install.packages(\"repr\")`. You can ignore that part, if you want.",
"_____no_output_____"
]
],
[
[
"library(repr)\noptions(repr.plot.height=4, repr.plot.width=8)\npar(mar=c(5.1, 4.1, 0.6, 0.6))\nplot(out, pmap)",
"_____no_output_____"
]
],
[
[
"There's a clear QTL on chromosome 8. We can make a plot of just that chromosome with the argument `chr=15`.",
"_____no_output_____"
]
],
[
[
"par(mar=c(5.1, 4.1, 0.6, 0.6))\nplot(out, pmap, chr=15)",
"_____no_output_____"
]
],
[
[
"Let's create a plot of the phenotype vs the genotype at the inferred QTL. We first need to identify the QTL location, which we can do using `max()`. We then use `maxmarg()` to get inferred genotypes at the inferred QTL.",
"_____no_output_____"
]
],
[
[
"mx <- max(out, pmap)\ng_imp <- maxmarg(pr, pmap, chr=mx$chr, pos=mx$pos, return_char=TRUE)",
"_____no_output_____"
]
],
[
[
"We can use `plot_pxg()` to plot the phenotype as a function of QTL genotype. We use `swap_axes=TRUE` to have the phenotype on the x-axis and the genotype on the y-axis, rather than the other way around. Here we see that the BB and DD genotypes are completely separated, phenotypically. ",
"_____no_output_____"
]
],
[
[
"par(mar=c(5.1, 4.1, 0.6, 0.6))\nplot_pxg(g_imp, pheno, swap_axes=TRUE, xlab=\"Habituation phenotype\")",
"_____no_output_____"
]
],
[
[
"## Browsing genome scan results with the Genetics Genome Browser\n\nThe [Genetics Genome Browser](https://github.com/chfi/purescript-genome-browser) is a fast, lightweight, [purescript]-based genome browser developed for browsing GWAS or QTL analysis results. We'll use the R package [R/qtl2browse](https://github.com/rqtl/qtl2browse) to view our QTL mapping results in the GGB.\n\nWe first need to install the R/qtl2browse package, again from a private [CRAN](https://cran.r-project.org)-like repository.\n\n```r\ninstall.packages(\"qtl2browse\", repos=\"http://rqtl.org/qtl2cran\")\n```\n\nWe then load the package and use its one function, `browse()`, which takes the `scan1()` output and corresponding physical map (in Mbp). This will open the Genetics Genome Browser in a separate tab in your web browser.",
"_____no_output_____"
]
],
[
[
"library(qtl2browse)\nbrowse(out, pmap)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0cead24b68061fa8c28ea22db987ab7a0816100 | 19,722 | ipynb | Jupyter Notebook | .ipynb_checkpoints/HIVTransTool-checkpoint.ipynb | JudoWill/ResearchNotebooks | 35796f7ef07361eb2926c8770e623f4e9d48ab96 | [
"MIT"
] | 1 | 2019-02-03T03:45:29.000Z | 2019-02-03T03:45:29.000Z | .ipynb_checkpoints/HIVTransTool-checkpoint.ipynb | JudoWill/ResearchNotebooks | 35796f7ef07361eb2926c8770e623f4e9d48ab96 | [
"MIT"
] | null | null | null | .ipynb_checkpoints/HIVTransTool-checkpoint.ipynb | JudoWill/ResearchNotebooks | 35796f7ef07361eb2926c8770e623f4e9d48ab96 | [
"MIT"
] | null | null | null | 49.428571 | 1,256 | 0.531285 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d0cec6e963ea1c99040e837116fbdeea0b30bf9f | 3,152 | ipynb | Jupyter Notebook | notebooks/Untitled.ipynb | Reusek/Fast-Correlation | fcea88dadf8d03db74ce375803565409a9c2dc10 | [
"MIT"
] | null | null | null | notebooks/Untitled.ipynb | Reusek/Fast-Correlation | fcea88dadf8d03db74ce375803565409a9c2dc10 | [
"MIT"
] | null | null | null | notebooks/Untitled.ipynb | Reusek/Fast-Correlation | fcea88dadf8d03db74ce375803565409a9c2dc10 | [
"MIT"
] | null | null | null | 21.013333 | 92 | 0.44797 | [
[
[
"# Kovariance",
"_____no_output_____"
],
[
"## Vzorec Pearsonova korelačního koeficientu\n\nPrůměrná hodnota vektoru:\n\n$$\nE(X) = \\frac{\\sum_{i=1}^{n} x_i}{n}\n$$\n\n$$\n\\rho_{X,Y}=\\frac{cov(X,Y)}{\\sigma_X\\sigma_Y}\n$$\n\nKovarianci převedeme na:\n\n$$\n\\rho_{X,Y} = \\frac{E((X-\\mu_X)(Y-\\mu_Y))}{\\sigma_X\\sigma_Y}\n$$\n\nProtože $\\mu_X=E(X)$ tak:\n\n$$\n\\sigma^2_X = E(X^2) - E^2(X)\n$$\n\nPo úpravě:\n\n$$\n\\rho_{X,Y} = \\frac{E(XY)-E(X)E(Y)}{\\sqrt{E(X^2) - E^2(X)} \\sqrt{E(Y^2)-E^2(Y)}}\n$$",
"_____no_output_____"
]
],
[
[
"def E(a):\n s = 0\n for i in a:\n s += i\n return s / len(a)\nassert E([1, 2]) == 1.5",
"_____no_output_____"
],
[
"def mul(a, b):\n assert len(a) == len(b)\n r = []\n for i in range(len(a)):\n r.append(a[i] * b[i])\n return r\n \nassert mul([2,2], [2,3]) == [4,6]",
"_____no_output_____"
],
[
"def reduce(a, b):\n assert len(a) == len(b)\n r = []\n for i in range(len(a)):\n r.append(a[i] - b[i])\n return r\nassert reduce([1,2], [1,1]) == [0,1]\n ",
"_____no_output_____"
],
[
"def cov(a, b):\n return E(mul(a,b)) - (E(a) * E(b))",
"_____no_output_____"
],
[
"cov([1, 2, 4], [1, 2, 4])",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
d0cedc8b5118b74537a3da398d32391478bf22a3 | 14,950 | ipynb | Jupyter Notebook | examples/user_guide/Simplifying_Codebases.ipynb | ektar/param | 7d702adfb4ace592a9550a25e8ca49522813fef4 | [
"BSD-3-Clause"
] | 123 | 2019-12-11T17:53:05.000Z | 2022-03-16T14:21:15.000Z | examples/user_guide/Simplifying_Codebases.ipynb | ektar/param | 7d702adfb4ace592a9550a25e8ca49522813fef4 | [
"BSD-3-Clause"
] | 282 | 2019-11-28T12:40:18.000Z | 2022-03-31T13:25:44.000Z | examples/user_guide/Simplifying_Codebases.ipynb | ektar/param | 7d702adfb4ace592a9550a25e8ca49522813fef4 | [
"BSD-3-Clause"
] | 37 | 2019-12-27T09:10:25.000Z | 2022-02-22T16:28:42.000Z | 41.876751 | 977 | 0.609565 | [
[
[
"# Simplifying Codebases\n\nParam's just a Python library, and so anything you can do with Param you can do \"manually\". So, why use Param?\n\nThe most immediate benefit to using Param is that it allows you to greatly simplify your codebases, making them much more clear, readable, and maintainable, while simultaneously providing robust handling against error conditions.\n\nParam does this by letting a programmer explicitly declare the types and values of parameters accepted by the code. Param then ensures that only suitable values of those parameters ever make it through to the underlying code, removing the need to handle any of those conditions explicitly.\n\nTo see how this works, let's create a Python class with some attributes without using Param:",
"_____no_output_____"
]
],
[
[
"class OrdinaryClass(object):\n def __init__(self, a=2, b=3, title=\"sum\"):\n self.a = a\n self.b = b\n self.title = title\n \n def __call__(self):\n return self.title + \": \" + str(self.a + self.b)",
"_____no_output_____"
]
],
[
[
"As this is just standard Python, we can of course instantiate this class, modify its variables, and call it:",
"_____no_output_____"
]
],
[
[
"o1 = OrdinaryClass(b=4, title=\"Sum\")\no1.a=4\no1()",
"_____no_output_____"
]
],
[
[
"The same code written using Param would look like:",
"_____no_output_____"
]
],
[
[
"import param\n \nclass ParamClass(param.Parameterized):\n a = param.Integer(2, bounds=(0,1000), doc=\"First addend\")\n b = param.Integer(3, bounds=(0,1000), doc=\"Second addend\")\n title = param.String(default=\"sum\", doc=\"Title for the result\")\n \n def __call__(self):\n return self.title + \": \" + str(self.a + self.b)",
"_____no_output_____"
],
[
"o2 = ParamClass(b=4, title=\"Sum\")\no2()",
"_____no_output_____"
]
],
[
[
"As you can see, the Parameters here are used precisely like normal attributes once they are defined, so the code for `__call__` and for invoking the constructor are the same in both cases. It's thus generally quite straightforward to migrate an existing class into Param. So, why do that?\n\nWell, with fewer lines of code than the ordinary class, you've now unlocked a whole wealth of features and better behavior! For instance, what happens if a user tries to supply some inappropriate data? With Param, such errors will be caught immediately:",
"_____no_output_____"
]
],
[
[
"with param.exceptions_summarized(): \n o3 = ParamClass()\n o3.b = -5",
"_____no_output_____"
]
],
[
[
"Of course, you could always add more code to an ordinary Python class to check for errors like that, but it quickly gets unwieldy:",
"_____no_output_____"
]
],
[
[
"class OrdinaryClass2(object):\n def __init__(self, a=2, b=3, title=\"sum\"):\n if type(a) is not int:\n raise ValueError(\"'a' must be an integer\")\n if type(b) is not int:\n raise ValueError(\"'b' must be an integer\")\n if a<0:\n raise ValueError(\"'a' must be at least `0`\")\n if b<0:\n raise ValueError(\"'b' must be at least `0`\")\n if type(title) is not str:\n raise ValueError(\"'title' must be a string\") \n \n self.a = a\n self.b = b\n self.title = title\n \n def __call__(self):\n return self.title + \": \" + str(self.a + self.b)",
"_____no_output_____"
],
[
"with param.exceptions_summarized(): \n OrdinaryClass2(a=\"f\")",
"_____no_output_____"
]
],
[
[
"Unfortunately, catching errors in the constructor like that won't help if someone modifies the attribute directly, which won't be detected as an error:",
"_____no_output_____"
]
],
[
[
"o4 = OrdinaryClass2()\no4.a = \"four\"",
"_____no_output_____"
]
],
[
[
"Python will happily accept this incorrect value and will continue processing. It may only be much later, in a very different part of your code, that you see a mysterious error message that's then very difficult to relate back to the actual problem you need to fix:",
"_____no_output_____"
]
],
[
[
"with param.exceptions_summarized(): \n o4()",
"_____no_output_____"
]
],
[
[
"Here there's no problem with the code in the cell above; `o4()` is fully valid Python; the real problem is in the preceding cell, which could have been in a completely different file or library. The error message is also obscure and confusing at this level, because the user of `o4` may have no idea why strings and integers are getting concatenated.\n\nTo get a better error message, you _could_ move those checks into the `__call__` method, which would make sure that errors are always eventually detected:",
"_____no_output_____"
]
],
[
[
"class OrdinaryClass3(object):\n def __init__(self, a=2, b=3, title=\"sum\"): \n self.a = a\n self.b = b\n self.title = title\n \n def __call__(self):\n if type(self.a) is not int:\n raise ValueError(\"'a' must be an integer\")\n if type(self.b) is not int:\n raise ValueError(\"'b' must be an integer\")\n if self.a<0:\n raise ValueError(\"'a' must be at least `0`\")\n if self.b<0:\n raise ValueError(\"'b' must be at least `0`\")\n if type(self.title) is not str:\n raise ValueError(\"'title' must be a string\") \n\n return self.title + \": \" + str(self.a + self.b)",
"_____no_output_____"
],
[
"o5 = OrdinaryClass3()\no5.a = \"four\"",
"_____no_output_____"
],
[
"with param.exceptions_summarized(): \n o5()",
"_____no_output_____"
]
],
[
[
"But you'd now have to check for errors in _every_ _single_ _method_ that might use those parameters. Worse, you still only detect the problem very late, far from where it was first introduced. Any distance between the error and the error report makes it much more difficult to address, as the user then has to track down where in the code `a` might have gotten set to a non-integer.\n\nWith Param you can catch such problems at their start, as soon as an incorrect value is provided, when it is still simple to detect and correct it. To get those same features in hand-written Python code, you would need to provide explicit getters and setters, which is made easier with Python properties and decorators, but is still quite unwieldy:",
"_____no_output_____"
]
],
[
[
"class OrdinaryClass4(object):\n def __init__(self, a=2, b=3, title=\"sum\"):\n self.a = a\n self.b = b\n self.title = title\n \n @property\n def a(self): return self.__a\n @a.setter\n def a(self, a):\n if type(a) is not int:\n raise ValueError(\"'a' must be an integer\")\n if a < 0:\n raise ValueError(\"'a' must be at least `0`\")\n self.__a = a\n \n @property\n def b(self): return self.__b\n @b.setter\n def b(self, b):\n if type(b) is not int:\n raise ValueError(\"'a' must be an integer\")\n if b < 0:\n raise ValueError(\"'a' must be at least `0`\")\n self.__b = b\n\n @property\n def title(self): return self.__title\n def title(self, b):\n if type(title) is not string:\n raise ValueError(\"'title' must be a string\")\n self.__title = title\n\n def __call__(self):\n return self.title + \": \" + str(self.a + self.b)",
"_____no_output_____"
],
[
"o5=OrdinaryClass4()\no5()",
"_____no_output_____"
],
[
"with param.exceptions_summarized(): \n o5=OrdinaryClass4()\n o5.b=-6",
"_____no_output_____"
]
],
[
[
"Note that this code has an easily overlooked mistake in it, reporting `a` rather than `b` as the problem. This sort of error is extremely common in copy-pasted validation code of this type, because tests rarely exercise all of the error conditions involved.\n\nAs you can see, even getting close to the automatic validation already provided by Param requires 8 methods and >30 highly repetitive lines of code, even when using relatively esoteric Python features like properties and decorators, and still doesn't yet implement other Param features like automatic documentation, attribute inheritance, or dynamic values. With Param, the corresponding `ParamClass` code only requires 6 lines and no fancy techniques beyond Python classes. Most importantly, the Param version lets readers and program authors focus directly on what this code actually does, which is to compute a function from three provided parameters:\n\n```\nclass ParamClass(param.Parameterized):\n a = param.Integer(2, bounds=(0,1000), doc=\"First addend\")\n b = param.Integer(3, bounds=(0,1000), doc=\"Second addend\")\n title = param.String(default=\"sum\", doc=\"Title for the result\")\n \n def __call__(self):\n return self.title + \": \" + str(self.a + self.b)\n```\n\nEven a quick skim of this code reveals what parameters are available, what values they will accept, what the default values are, and how those parameters will be used in the method. Plus the actual code of the method stands out immediately, as all the code is either parameters or actual functionality. In contrast, users of OrdinaryClass3 will have to read through dozens of lines of code to discern even basic information about usage, or else authors of the code will need to create and maintain docstrings that may or may not match the actual code over time and will further increase the amount of text to write and maintain.",
"_____no_output_____"
],
[
"## Programming contracts\n\nIf you think about the examples above, you can see how Param makes it simple for programmers to make a contract with their users, being explicit and clear what will be accepted and rejected, while also allowing programmers to make safe assumptions about what inputs the code may ever receive. There is no need for `__call__` _ever_ to check for the type of one of its parameters, whether it's in the range allowed, or any other property that can be enforced by Param. Your custom code can then be much more linear and straightforward, getting right to work with the actual task at hand, without having to have reams of `if` statements and `asserts()` that disrupt the flow of the source file and make the reader get sidetracked in error-handling code. Param lets you once and for all declare what this code accepts, which is both clear documentation to the user and a guarantee that the programmer can forget about any other possible value a user might someday supply.\n\nCrucially, these contracts apply not just between the user and a given piece of code, but also between components of the system itself. When validation code is expensive, as in ordinary Python, programmers will typically do it only at the edges of the system, where input from the user is accepted. But expressing types and ranges is so easy in Param, it can be done for any major component in the system. The Parameter list declares very clearly what that component accepts, which lets the code for that component ignore all potential inputs that are disallowed by the Parameter specifications, while correctly advertising to the rest of the codebase what inputs are allowed. Programmers can thus focus on their particular components of interest, knowing precisely what inputs will ever be let through, without having to reason about the flow of configuration and data throughout the whole system.\n\nWithout Param, you should expect Python code to be full of confusing error checking and handling of different input types, while still only catching a small fraction of the possible incorrect inputs that could be provided. But Param-based code should be dramatically easier to read, easier to maintain, easier to develop, and nearly bulletproof against mistaken or even malicious usage. ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
d0cee7d95f2c32da6732bd05fa32a2e5d6ef6366 | 7,986 | ipynb | Jupyter Notebook | examples/notebook/examples/zebra_sat.ipynb | personalcomputer/or-tools | 2cb85b4eead4c38e1c54b48044f92087cf165bce | [
"Apache-2.0"
] | 1 | 2022-03-08T22:28:12.000Z | 2022-03-08T22:28:12.000Z | examples/notebook/examples/zebra_sat.ipynb | personalcomputer/or-tools | 2cb85b4eead4c38e1c54b48044f92087cf165bce | [
"Apache-2.0"
] | null | null | null | examples/notebook/examples/zebra_sat.ipynb | personalcomputer/or-tools | 2cb85b4eead4c38e1c54b48044f92087cf165bce | [
"Apache-2.0"
] | null | null | null | 37.669811 | 248 | 0.57438 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d0ceef1ad39e0e5d48024a906646ed2cc0df58ce | 6,191 | ipynb | Jupyter Notebook | lessons/ML Pipelines/clean_tokenize.ipynb | ZacksAmber/Udacity-Data-Scientist | b21595413f21a1200fe0b46f47e747ca9bff8d1f | [
"MIT"
] | null | null | null | lessons/ML Pipelines/clean_tokenize.ipynb | ZacksAmber/Udacity-Data-Scientist | b21595413f21a1200fe0b46f47e747ca9bff8d1f | [
"MIT"
] | null | null | null | lessons/ML Pipelines/clean_tokenize.ipynb | ZacksAmber/Udacity-Data-Scientist | b21595413f21a1200fe0b46f47e747ca9bff8d1f | [
"MIT"
] | null | null | null | 34.20442 | 262 | 0.566952 | [
[
[
"## Tokenization\nBefore we can classify any posts, we'll need to clean and tokenize the text data. Use what you remember from the last lesson on NLP to implement the function `tokenize`. This function should perform the following steps on the string, `text`, using nltk:\n\n1. Identify any urls in `text`, and replace each one with the word, `\"urlplaceholder\"`.\n2. Split `text` into tokens.\n3. For each token: lemmatize, normalize case, and strip leading and trailing white space.\n4. Return the tokens in a list!\n\nFor example, this:\n```python\ntext = 'Barclays CEO stresses the importance of regulatory and cultural reform in financial services at Brussels conference http://t.co/Ge9Lp7hpyG'\n\ntokenize(text)\n```\nshould return this:\n```txt\n['barclays', 'ceo', 'stress', 'the', 'importance', 'of', 'regulatory', 'and', 'cultural', 'reform', 'in', 'financial', 'service', 'at', 'brussels', 'conference', 'urlplaceholder']\n```\n\nHint: You'll have to add an import statement to use the `re` package (which supports regular expressions) and two import statements to use the appropriate functions from `nltk`! Add them to this first code cell.",
"_____no_output_____"
]
],
[
[
"# download necessary NLTK data\nimport nltk\nnltk.download(['punkt', 'wordnet'])\n\n# import statements\nimport pandas as pd\nimport re\nfrom nltk.tokenize import word_tokenize\nfrom nltk.stem.wordnet import WordNetLemmatizer",
"[nltk_data] Downloading package punkt to /Users/zacks/nltk_data...\n[nltk_data] Package punkt is already up-to-date!\n[nltk_data] Downloading package wordnet to /Users/zacks/nltk_data...\n[nltk_data] Package wordnet is already up-to-date!\n"
],
[
"def load_data():\n df = pd.read_csv('corporate_messaging.csv', encoding='latin-1')\n df = df[(df[\"category:confidence\"] == 1) & (df['category'] != 'Exclude')]\n X = df.text.values\n y = df.category.values\n return X, y",
"_____no_output_____"
]
],
[
[
"#### For step 1, the regular expression to detect a url is given below",
"_____no_output_____"
]
],
[
[
"url_regex = 'http[s]?://(?:[a-zA-Z]|[0-9]|[$-_@.&+]|[!*\\(\\),]|(?:%[0-9a-fA-F][0-9a-fA-F]))+'",
"_____no_output_____"
],
[
"def tokenize(text):\n # replace each url in text string with placeholder\n url_pattern = re.compile(url_regex)\n text = url_pattern.sub('urlplaceholder', text)\n\n # tokenize text\n tokens = word_tokenize(text)\n \n # initiate lemmatizer\n lemmatizer = WordNetLemmatizer()\n\n # iterate through each token\n clean_tokens = []\n for tok in tokens:\n \n # lemmatize, normalize case, and remove leading/trailing white space\n clean_tok = lemmatizer.lemmatize(tok).lower().strip()\n clean_tokens.append(clean_tok)\n\n return clean_tokens",
"_____no_output_____"
],
[
"# test out function\nX, y = load_data()\nfor message in X[:5]:\n tokens = tokenize(message)\n print(message)\n print(tokens, '\\n')",
"Barclays CEO stresses the importance of regulatory and cultural reform in financial services at Brussels conference http://t.co/Ge9Lp7hpyG\n['barclays', 'ceo', 'stress', 'the', 'importance', 'of', 'regulatory', 'and', 'cultural', 'reform', 'in', 'financial', 'service', 'at', 'brussels', 'conference', 'urlplaceholder'] \n\nBarclays announces result of Rights Issue http://t.co/LbIqqh3wwG\n['barclays', 'announces', 'result', 'of', 'rights', 'issue', 'urlplaceholder'] \n\nBarclays publishes its prospectus for its å£5.8bn Rights Issue: http://t.co/YZk24iE8G6\n['barclays', 'publishes', 'it', 'prospectus', 'for', 'it', 'å£5.8bn', 'rights', 'issue', ':', 'urlplaceholder'] \n\nBarclays Group Finance Director Chris Lucas is to step down at the end of the week due to ill health http://t.co/nkuHoAfnSD\n['barclays', 'group', 'finance', 'director', 'chris', 'lucas', 'is', 'to', 'step', 'down', 'at', 'the', 'end', 'of', 'the', 'week', 'due', 'to', 'ill', 'health', 'urlplaceholder'] \n\nBarclays announces that Irene McDermott Brown has been appointed as Group Human Resources Director http://t.co/c3fNGY6NMT\n['barclays', 'announces', 'that', 'irene', 'mcdermott', 'brown', 'ha', 'been', 'appointed', 'a', 'group', 'human', 'resources', 'director', 'urlplaceholder'] \n\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d0cef7f1eb35a2a5688f5124960367f9223dbb57 | 58,268 | ipynb | Jupyter Notebook | pytorch_ipynb/cnn/cnn-vgg19.ipynb | DeepSE/deeplearning-models | 7120d9fc40ac471136aeb30766c5f44cffc36070 | [
"MIT"
] | 69 | 2020-06-27T11:33:02.000Z | 2021-11-20T10:17:02.000Z | pytorch_ipynb/cnn/cnn-vgg19.ipynb | DeepSE/deeplearning-models | 7120d9fc40ac471136aeb30766c5f44cffc36070 | [
"MIT"
] | null | null | null | pytorch_ipynb/cnn/cnn-vgg19.ipynb | DeepSE/deeplearning-models | 7120d9fc40ac471136aeb30766c5f44cffc36070 | [
"MIT"
] | 24 | 2020-06-27T11:56:58.000Z | 2021-11-20T10:17:04.000Z | 50.844677 | 13,191 | 0.483387 | [
[
[
"Deep Learning Models -- A collection of various deep learning architectures, models, and tips for TensorFlow and PyTorch in Jupyter Notebooks.\n- Author: Sebastian Raschka\n- GitHub Repository: https://github.com/rasbt/deeplearning-models",
"_____no_output_____"
]
],
[
[
"!pip install -q IPython\n!pip install -q ipykernel\n!pip install -q watermark\n!pip install -q matplotlib\n!pip install -q sklearn\n!pip install -q pandas\n!pip install -q pydot\n!pip install -q hiddenlayer\n!pip install -q graphviz",
"_____no_output_____"
],
[
"%load_ext watermark\n%watermark -a 'Sebastian Raschka' -v -p torch",
"Sebastian Raschka \n\nCPython 3.6.9\nIPython 5.5.0\n\ntorch 1.5.1+cu101\n"
]
],
[
[
"- Runs on CPU (not recommended here) or GPU (if available)",
"_____no_output_____"
],
[
"# Model Zoo -- Convolutional Neural Network (VGG19 Architecture)",
"_____no_output_____"
],
[
"Implementation of the VGG-19 architecture on Cifar10. \n\n\nReference for VGG-19:\n \n- Simonyan, K., & Zisserman, A. (2014). Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556.\n\n\nThe following table (taken from Simonyan & Zisserman referenced above) summarizes the VGG19 architecture:\n\n",
"_____no_output_____"
],
[
"## Imports",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport time\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nfrom torchvision import datasets\nfrom torchvision import transforms\nfrom torch.utils.data import DataLoader",
"_____no_output_____"
]
],
[
[
"## Settings and Dataset",
"_____no_output_____"
]
],
[
[
"##########################\n### SETTINGS\n##########################\n\n# Device\nDEVICE = torch.device(\"cuda\" if torch.cuda.is_available() else \"cpu\")\nprint('Device:', DEVICE)\n\n# Hyperparameters\nrandom_seed = 1\nlearning_rate = 0.001\nnum_epochs = 20\nbatch_size = 128\n\n# Architecture\nnum_features = 784\nnum_classes = 10\n\n\n##########################\n### MNIST DATASET\n##########################\n\n# Note transforms.ToTensor() scales input images\n# to 0-1 range\ntrain_dataset = datasets.CIFAR10(root='data', \n train=True, \n transform=transforms.ToTensor(),\n download=True)\n\ntest_dataset = datasets.CIFAR10(root='data', \n train=False, \n transform=transforms.ToTensor())\n\n\ntrain_loader = DataLoader(dataset=train_dataset, \n batch_size=batch_size, \n shuffle=True)\n\ntest_loader = DataLoader(dataset=test_dataset, \n batch_size=batch_size, \n shuffle=False)\n\n# Checking the dataset\nfor images, labels in train_loader: \n print('Image batch dimensions:', images.shape)\n print('Image label dimensions:', labels.shape)\n break",
"Device: cuda\nDownloading https://www.cs.toronto.edu/~kriz/cifar-10-python.tar.gz to data/cifar-10-python.tar.gz\n"
]
],
[
[
"## Model",
"_____no_output_____"
]
],
[
[
"##########################\n### MODEL\n##########################\n\n\nclass VGG16(torch.nn.Module):\n\n def __init__(self, num_features, num_classes):\n super(VGG16, self).__init__()\n \n # calculate same padding:\n # (w - k + 2*p)/s + 1 = o\n # => p = (s(o-1) - w + k)/2\n \n self.block_1 = nn.Sequential(\n nn.Conv2d(in_channels=3,\n out_channels=64,\n kernel_size=(3, 3),\n stride=(1, 1),\n # (1(32-1)- 32 + 3)/2 = 1\n padding=1), \n nn.ReLU(),\n nn.Conv2d(in_channels=64,\n out_channels=64,\n kernel_size=(3, 3),\n stride=(1, 1),\n padding=1),\n nn.ReLU(),\n nn.MaxPool2d(kernel_size=(2, 2),\n stride=(2, 2))\n )\n \n self.block_2 = nn.Sequential(\n nn.Conv2d(in_channels=64,\n out_channels=128,\n kernel_size=(3, 3),\n stride=(1, 1),\n padding=1),\n nn.ReLU(),\n nn.Conv2d(in_channels=128,\n out_channels=128,\n kernel_size=(3, 3),\n stride=(1, 1),\n padding=1),\n nn.ReLU(),\n nn.MaxPool2d(kernel_size=(2, 2),\n stride=(2, 2))\n )\n \n self.block_3 = nn.Sequential( \n nn.Conv2d(in_channels=128,\n out_channels=256,\n kernel_size=(3, 3),\n stride=(1, 1),\n padding=1),\n nn.ReLU(),\n nn.Conv2d(in_channels=256,\n out_channels=256,\n kernel_size=(3, 3),\n stride=(1, 1),\n padding=1),\n nn.ReLU(), \n nn.Conv2d(in_channels=256,\n out_channels=256,\n kernel_size=(3, 3),\n stride=(1, 1),\n padding=1),\n nn.ReLU(),\n nn.Conv2d(in_channels=256,\n out_channels=256,\n kernel_size=(3, 3),\n stride=(1, 1),\n padding=1),\n nn.ReLU(),\n nn.MaxPool2d(kernel_size=(2, 2),\n stride=(2, 2))\n )\n \n \n self.block_4 = nn.Sequential( \n nn.Conv2d(in_channels=256,\n out_channels=512,\n kernel_size=(3, 3),\n stride=(1, 1),\n padding=1),\n nn.ReLU(), \n nn.Conv2d(in_channels=512,\n out_channels=512,\n kernel_size=(3, 3),\n stride=(1, 1),\n padding=1),\n nn.ReLU(), \n nn.Conv2d(in_channels=512,\n out_channels=512,\n kernel_size=(3, 3),\n stride=(1, 1),\n padding=1),\n nn.ReLU(),\n nn.Conv2d(in_channels=512,\n out_channels=512,\n kernel_size=(3, 3),\n stride=(1, 1),\n padding=1),\n nn.ReLU(), \n nn.MaxPool2d(kernel_size=(2, 2),\n stride=(2, 2))\n )\n \n self.block_5 = nn.Sequential(\n nn.Conv2d(in_channels=512,\n out_channels=512,\n kernel_size=(3, 3),\n stride=(1, 1),\n padding=1),\n nn.ReLU(), \n nn.Conv2d(in_channels=512,\n out_channels=512,\n kernel_size=(3, 3),\n stride=(1, 1),\n padding=1),\n nn.ReLU(), \n nn.Conv2d(in_channels=512,\n out_channels=512,\n kernel_size=(3, 3),\n stride=(1, 1),\n padding=1),\n nn.ReLU(),\n nn.Conv2d(in_channels=512,\n out_channels=512,\n kernel_size=(3, 3),\n stride=(1, 1),\n padding=1),\n nn.ReLU(), \n nn.MaxPool2d(kernel_size=(2, 2),\n stride=(2, 2)) \n )\n \n self.classifier = nn.Sequential(\n nn.Linear(512, 4096),\n nn.ReLU(True),\n nn.Linear(4096, 4096),\n nn.ReLU(True),\n nn.Linear(4096, num_classes)\n )\n \n \n for m in self.modules():\n if isinstance(m, torch.nn.Conv2d):\n #n = m.kernel_size[0] * m.kernel_size[1] * m.out_channels\n #m.weight.data.normal_(0, np.sqrt(2. / n))\n m.weight.detach().normal_(0, 0.05)\n if m.bias is not None:\n m.bias.detach().zero_()\n elif isinstance(m, torch.nn.Linear):\n m.weight.detach().normal_(0, 0.05)\n m.bias.detach().detach().zero_()\n \n \n def forward(self, x):\n\n x = self.block_1(x)\n x = self.block_2(x)\n x = self.block_3(x)\n x = self.block_4(x)\n x = self.block_5(x)\n logits = self.classifier(x.view(-1, 512))\n probas = F.softmax(logits, dim=1)\n\n return logits, probas\n\n \ntorch.manual_seed(random_seed)\nmodel = VGG16(num_features=num_features,\n num_classes=num_classes)\n\nmodel = model.to(DEVICE)\n\noptimizer = torch.optim.Adam(model.parameters(), lr=learning_rate) ",
"_____no_output_____"
],
[
"import hiddenlayer as hl\nhl.build_graph(model, torch.zeros([128, 3, 32, 32]).to(DEVICE))",
"_____no_output_____"
]
],
[
[
"## Training",
"_____no_output_____"
]
],
[
[
"def compute_accuracy(model, data_loader):\n model.eval()\n correct_pred, num_examples = 0, 0\n for i, (features, targets) in enumerate(data_loader):\n \n features = features.to(DEVICE)\n targets = targets.to(DEVICE)\n\n logits, probas = model(features)\n _, predicted_labels = torch.max(probas, 1)\n num_examples += targets.size(0)\n correct_pred += (predicted_labels == targets).sum()\n return correct_pred.float()/num_examples * 100\n\n\ndef compute_epoch_loss(model, data_loader):\n model.eval()\n curr_loss, num_examples = 0., 0\n with torch.no_grad():\n for features, targets in data_loader:\n features = features.to(DEVICE)\n targets = targets.to(DEVICE)\n logits, probas = model(features)\n loss = F.cross_entropy(logits, targets, reduction='sum')\n num_examples += targets.size(0)\n curr_loss += loss\n\n curr_loss = curr_loss / num_examples\n return curr_loss\n \n \n\nstart_time = time.time()\nfor epoch in range(num_epochs):\n \n model.train()\n for batch_idx, (features, targets) in enumerate(train_loader):\n \n features = features.to(DEVICE)\n targets = targets.to(DEVICE)\n \n ### FORWARD AND BACK PROP\n logits, probas = model(features)\n cost = F.cross_entropy(logits, targets)\n optimizer.zero_grad()\n \n cost.backward()\n \n ### UPDATE MODEL PARAMETERS\n optimizer.step()\n \n ### LOGGING\n if not batch_idx % 50:\n print ('Epoch: %03d/%03d | Batch %04d/%04d | Cost: %.4f' \n %(epoch+1, num_epochs, batch_idx, \n len(train_loader), cost))\n\n model.eval()\n with torch.set_grad_enabled(False): # save memory during inference\n print('Epoch: %03d/%03d | Train: %.3f%% | Loss: %.3f' % (\n epoch+1, num_epochs, \n compute_accuracy(model, train_loader),\n compute_epoch_loss(model, train_loader)))\n\n\n print('Time elapsed: %.2f min' % ((time.time() - start_time)/60))\n \nprint('Total Training Time: %.2f min' % ((time.time() - start_time)/60))",
"Epoch: 001/020 | Batch 0000/0391 | Cost: 1061.4154\nEpoch: 001/020 | Batch 0050/0391 | Cost: 2.3029\nEpoch: 001/020 | Batch 0100/0391 | Cost: 1.9784\nEpoch: 001/020 | Batch 0150/0391 | Cost: 1.9533\nEpoch: 001/020 | Batch 0200/0391 | Cost: 1.8397\nEpoch: 001/020 | Batch 0250/0391 | Cost: 1.7760\nEpoch: 001/020 | Batch 0300/0391 | Cost: 1.8241\nEpoch: 001/020 | Batch 0350/0391 | Cost: 1.7632\nEpoch: 001/020 | Train: 32.588% | Loss: 1.733\nTime elapsed: 4.61 min\nEpoch: 002/020 | Batch 0000/0391 | Cost: 1.8287\nEpoch: 002/020 | Batch 0050/0391 | Cost: 1.8992\nEpoch: 002/020 | Batch 0100/0391 | Cost: 1.5520\nEpoch: 002/020 | Batch 0150/0391 | Cost: 1.5686\nEpoch: 002/020 | Batch 0200/0391 | Cost: 1.4790\nEpoch: 002/020 | Batch 0250/0391 | Cost: 1.4339\nEpoch: 002/020 | Batch 0300/0391 | Cost: 1.5641\nEpoch: 002/020 | Batch 0350/0391 | Cost: 1.5211\nEpoch: 002/020 | Train: 49.878% | Loss: 1.369\nTime elapsed: 9.21 min\nEpoch: 003/020 | Batch 0000/0391 | Cost: 1.3988\nEpoch: 003/020 | Batch 0050/0391 | Cost: 1.2016\nEpoch: 003/020 | Batch 0100/0391 | Cost: 1.3821\nEpoch: 003/020 | Batch 0150/0391 | Cost: 1.3700\nEpoch: 003/020 | Batch 0200/0391 | Cost: 1.4340\nEpoch: 003/020 | Batch 0250/0391 | Cost: 1.4271\nEpoch: 003/020 | Batch 0300/0391 | Cost: 1.3866\nEpoch: 003/020 | Batch 0350/0391 | Cost: 1.4448\nEpoch: 003/020 | Train: 53.490% | Loss: 1.274\nTime elapsed: 13.82 min\nEpoch: 004/020 | Batch 0000/0391 | Cost: 1.2869\nEpoch: 004/020 | Batch 0050/0391 | Cost: 1.2983\nEpoch: 004/020 | Batch 0100/0391 | Cost: 1.2728\nEpoch: 004/020 | Batch 0150/0391 | Cost: 1.1564\nEpoch: 004/020 | Batch 0200/0391 | Cost: 1.0972\nEpoch: 004/020 | Batch 0250/0391 | Cost: 1.1532\nEpoch: 004/020 | Batch 0300/0391 | Cost: 1.1784\nEpoch: 004/020 | Batch 0350/0391 | Cost: 1.1819\nEpoch: 004/020 | Train: 56.634% | Loss: 1.204\nTime elapsed: 18.43 min\nEpoch: 005/020 | Batch 0000/0391 | Cost: 1.1228\nEpoch: 005/020 | Batch 0050/0391 | Cost: 1.1564\nEpoch: 005/020 | Batch 0100/0391 | Cost: 1.0147\nEpoch: 005/020 | Batch 0150/0391 | Cost: 1.1273\nEpoch: 005/020 | Batch 0200/0391 | Cost: 0.9425\nEpoch: 005/020 | Batch 0250/0391 | Cost: 1.0747\nEpoch: 005/020 | Batch 0300/0391 | Cost: 1.1276\nEpoch: 005/020 | Batch 0350/0391 | Cost: 1.2110\nEpoch: 005/020 | Train: 62.844% | Loss: 1.038\nTime elapsed: 23.03 min\nEpoch: 006/020 | Batch 0000/0391 | Cost: 0.9599\nEpoch: 006/020 | Batch 0050/0391 | Cost: 0.9764\nEpoch: 006/020 | Batch 0100/0391 | Cost: 0.9482\nEpoch: 006/020 | Batch 0150/0391 | Cost: 1.1014\nEpoch: 006/020 | Batch 0200/0391 | Cost: 1.0980\nEpoch: 006/020 | Batch 0250/0391 | Cost: 0.7146\nEpoch: 006/020 | Batch 0300/0391 | Cost: 1.0462\nEpoch: 006/020 | Batch 0350/0391 | Cost: 1.0740\nEpoch: 006/020 | Train: 68.248% | Loss: 0.882\nTime elapsed: 27.64 min\nEpoch: 007/020 | Batch 0000/0391 | Cost: 1.0146\nEpoch: 007/020 | Batch 0050/0391 | Cost: 1.0388\nEpoch: 007/020 | Batch 0100/0391 | Cost: 1.1543\nEpoch: 007/020 | Batch 0150/0391 | Cost: 1.0155\nEpoch: 007/020 | Batch 0200/0391 | Cost: 0.8197\nEpoch: 007/020 | Batch 0250/0391 | Cost: 1.0015\nEpoch: 007/020 | Batch 0300/0391 | Cost: 1.0565\nEpoch: 007/020 | Batch 0350/0391 | Cost: 0.9709\nEpoch: 007/020 | Train: 64.582% | Loss: 0.963\nTime elapsed: 32.24 min\nEpoch: 008/020 | Batch 0000/0391 | Cost: 1.0335\nEpoch: 008/020 | Batch 0050/0391 | Cost: 1.0126\nEpoch: 008/020 | Batch 0100/0391 | Cost: 0.7439\nEpoch: 008/020 | Batch 0150/0391 | Cost: 1.0409\nEpoch: 008/020 | Batch 0200/0391 | Cost: 1.0831\nEpoch: 008/020 | Batch 0250/0391 | Cost: 1.0905\nEpoch: 008/020 | Batch 0300/0391 | Cost: 0.9062\nEpoch: 008/020 | Batch 0350/0391 | Cost: 0.9048\nEpoch: 008/020 | Train: 69.984% | Loss: 0.847\nTime elapsed: 36.85 min\nEpoch: 009/020 | Batch 0000/0391 | Cost: 0.7430\nEpoch: 009/020 | Batch 0050/0391 | Cost: 0.7811\nEpoch: 009/020 | Batch 0100/0391 | Cost: 0.8621\nEpoch: 009/020 | Batch 0150/0391 | Cost: 0.8378\nEpoch: 009/020 | Batch 0200/0391 | Cost: 0.7797\nEpoch: 009/020 | Batch 0250/0391 | Cost: 0.8398\nEpoch: 009/020 | Batch 0300/0391 | Cost: 0.7331\nEpoch: 009/020 | Batch 0350/0391 | Cost: 0.7951\nEpoch: 009/020 | Train: 73.738% | Loss: 0.751\nTime elapsed: 41.46 min\nEpoch: 010/020 | Batch 0000/0391 | Cost: 0.6359\nEpoch: 010/020 | Batch 0050/0391 | Cost: 0.7745\nEpoch: 010/020 | Batch 0100/0391 | Cost: 0.7155\nEpoch: 010/020 | Batch 0150/0391 | Cost: 0.5714\nEpoch: 010/020 | Batch 0200/0391 | Cost: 0.7268\nEpoch: 010/020 | Batch 0250/0391 | Cost: 0.5820\nEpoch: 010/020 | Batch 0300/0391 | Cost: 0.5438\nEpoch: 010/020 | Batch 0350/0391 | Cost: 0.7152\nEpoch: 010/020 | Train: 77.590% | Loss: 0.648\nTime elapsed: 46.06 min\nEpoch: 011/020 | Batch 0000/0391 | Cost: 0.6412\nEpoch: 011/020 | Batch 0050/0391 | Cost: 0.7381\nEpoch: 011/020 | Batch 0100/0391 | Cost: 0.6997\nEpoch: 011/020 | Batch 0150/0391 | Cost: 0.8753\nEpoch: 011/020 | Batch 0200/0391 | Cost: 0.7835\nEpoch: 011/020 | Batch 0250/0391 | Cost: 0.6201\nEpoch: 011/020 | Batch 0300/0391 | Cost: 0.5879\nEpoch: 011/020 | Batch 0350/0391 | Cost: 0.6144\nEpoch: 011/020 | Train: 76.562% | Loss: 0.688\nTime elapsed: 50.67 min\nEpoch: 012/020 | Batch 0000/0391 | Cost: 0.5913\nEpoch: 012/020 | Batch 0050/0391 | Cost: 0.7521\nEpoch: 012/020 | Batch 0100/0391 | Cost: 0.5401\nEpoch: 012/020 | Batch 0150/0391 | Cost: 0.5681\nEpoch: 012/020 | Batch 0200/0391 | Cost: 0.7862\nEpoch: 012/020 | Batch 0250/0391 | Cost: 0.8259\nEpoch: 012/020 | Batch 0300/0391 | Cost: 0.8741\nEpoch: 012/020 | Batch 0350/0391 | Cost: 0.7649\nEpoch: 012/020 | Train: 80.436% | Loss: 0.572\nTime elapsed: 55.28 min\nEpoch: 013/020 | Batch 0000/0391 | Cost: 0.5254\nEpoch: 013/020 | Batch 0050/0391 | Cost: 0.5755\nEpoch: 013/020 | Batch 0100/0391 | Cost: 0.5229\nEpoch: 013/020 | Batch 0150/0391 | Cost: 0.6322\nEpoch: 013/020 | Batch 0200/0391 | Cost: 0.5999\nEpoch: 013/020 | Batch 0250/0391 | Cost: 0.5342\nEpoch: 013/020 | Batch 0300/0391 | Cost: 0.4578\nEpoch: 013/020 | Batch 0350/0391 | Cost: 0.5497\nEpoch: 013/020 | Train: 78.690% | Loss: 0.611\nTime elapsed: 59.89 min\nEpoch: 014/020 | Batch 0000/0391 | Cost: 0.6422\nEpoch: 014/020 | Batch 0050/0391 | Cost: 0.4704\nEpoch: 014/020 | Batch 0100/0391 | Cost: 0.5061\nEpoch: 014/020 | Batch 0150/0391 | Cost: 0.5354\nEpoch: 014/020 | Batch 0200/0391 | Cost: 0.5494\nEpoch: 014/020 | Batch 0250/0391 | Cost: 0.6403\nEpoch: 014/020 | Batch 0300/0391 | Cost: 0.6411\nEpoch: 014/020 | Batch 0350/0391 | Cost: 0.6156\nEpoch: 014/020 | Train: 84.104% | Loss: 0.477\nTime elapsed: 64.50 min\nEpoch: 015/020 | Batch 0000/0391 | Cost: 0.5557\nEpoch: 015/020 | Batch 0050/0391 | Cost: 0.3461\nEpoch: 015/020 | Batch 0100/0391 | Cost: 0.4598\nEpoch: 015/020 | Batch 0150/0391 | Cost: 0.6011\nEpoch: 015/020 | Batch 0200/0391 | Cost: 0.6190\nEpoch: 015/020 | Batch 0250/0391 | Cost: 0.5742\nEpoch: 015/020 | Batch 0300/0391 | Cost: 0.5892\nEpoch: 015/020 | Batch 0350/0391 | Cost: 0.6255\nEpoch: 015/020 | Train: 85.278% | Loss: 0.433\nTime elapsed: 69.11 min\nEpoch: 016/020 | Batch 0000/0391 | Cost: 0.4109\nEpoch: 016/020 | Batch 0050/0391 | Cost: 0.5074\nEpoch: 016/020 | Batch 0100/0391 | Cost: 0.4900\nEpoch: 016/020 | Batch 0150/0391 | Cost: 0.5656\nEpoch: 016/020 | Batch 0200/0391 | Cost: 0.5251\nEpoch: 016/020 | Batch 0250/0391 | Cost: 0.5022\nEpoch: 016/020 | Batch 0300/0391 | Cost: 0.6132\nEpoch: 016/020 | Batch 0350/0391 | Cost: 0.4384\nEpoch: 016/020 | Train: 83.444% | Loss: 0.494\nTime elapsed: 73.71 min\nEpoch: 017/020 | Batch 0000/0391 | Cost: 0.6602\nEpoch: 017/020 | Batch 0050/0391 | Cost: 0.3956\nEpoch: 017/020 | Batch 0100/0391 | Cost: 0.4275\nEpoch: 017/020 | Batch 0150/0391 | Cost: 0.4977\nEpoch: 017/020 | Batch 0200/0391 | Cost: 0.4128\nEpoch: 017/020 | Batch 0250/0391 | Cost: 0.4469\nEpoch: 017/020 | Batch 0300/0391 | Cost: 0.3838\nEpoch: 017/020 | Batch 0350/0391 | Cost: 0.4705\nEpoch: 017/020 | Train: 88.050% | Loss: 0.347\nTime elapsed: 78.32 min\nEpoch: 018/020 | Batch 0000/0391 | Cost: 0.2744\nEpoch: 018/020 | Batch 0050/0391 | Cost: 0.4708\nEpoch: 018/020 | Batch 0100/0391 | Cost: 0.6125\nEpoch: 018/020 | Batch 0150/0391 | Cost: 0.3646\nEpoch: 018/020 | Batch 0200/0391 | Cost: 0.4439\nEpoch: 018/020 | Batch 0250/0391 | Cost: 0.4294\nEpoch: 018/020 | Batch 0300/0391 | Cost: 0.4744\nEpoch: 018/020 | Batch 0350/0391 | Cost: 0.4866\nEpoch: 018/020 | Train: 86.888% | Loss: 0.393\nTime elapsed: 82.93 min\nEpoch: 019/020 | Batch 0000/0391 | Cost: 0.3776\nEpoch: 019/020 | Batch 0050/0391 | Cost: 0.3887\nEpoch: 019/020 | Batch 0100/0391 | Cost: 0.4949\nEpoch: 019/020 | Batch 0150/0391 | Cost: 0.2877\nEpoch: 019/020 | Batch 0200/0391 | Cost: 0.3119\nEpoch: 019/020 | Batch 0250/0391 | Cost: 0.5047\nEpoch: 019/020 | Batch 0300/0391 | Cost: 0.6944\nEpoch: 019/020 | Batch 0350/0391 | Cost: 0.5497\nEpoch: 019/020 | Train: 88.150% | Loss: 0.357\nTime elapsed: 87.53 min\nEpoch: 020/020 | Batch 0000/0391 | Cost: 0.2804\nEpoch: 020/020 | Batch 0050/0391 | Cost: 0.3506\nEpoch: 020/020 | Batch 0100/0391 | Cost: 0.6187\nEpoch: 020/020 | Batch 0150/0391 | Cost: 0.3379\nEpoch: 020/020 | Batch 0200/0391 | Cost: 0.4413\nEpoch: 020/020 | Batch 0250/0391 | Cost: 0.4734\nEpoch: 020/020 | Batch 0300/0391 | Cost: 0.4052\nEpoch: 020/020 | Batch 0350/0391 | Cost: 0.3718\nEpoch: 020/020 | Train: 87.506% | Loss: 0.393\nTime elapsed: 92.14 min\nTotal Training Time: 92.14 min\n"
]
],
[
[
"## Evaluation",
"_____no_output_____"
]
],
[
[
"with torch.set_grad_enabled(False): # save memory during inference\n print('Test accuracy: %.2f%%' % (compute_accuracy(model, test_loader)))",
"Test accuracy: 74.72%\n"
],
[
"%watermark -iv",
"numpy 1.18.5\ntorch 1.5.1+cu101\n\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d0cf216944d227c4d63701cfc202bee7d9134543 | 10,555 | ipynb | Jupyter Notebook | examples/notebooks/change-input-current.ipynb | anandmy/PyBaMM | dd8e5ebf85dc4324e163adad274ccb56c88f3698 | [
"BSD-3-Clause"
] | 1 | 2021-03-06T15:10:34.000Z | 2021-03-06T15:10:34.000Z | examples/notebooks/change-input-current.ipynb | simontoyabe/PyBaMM | d431b5cfa155f53e527a2e2847a301a10c420b55 | [
"BSD-3-Clause"
] | null | null | null | examples/notebooks/change-input-current.ipynb | simontoyabe/PyBaMM | d431b5cfa155f53e527a2e2847a301a10c420b55 | [
"BSD-3-Clause"
] | null | null | null | 31.41369 | 362 | 0.598579 | [
[
[
"# Changing the input current when solving PyBaMM models\n\nThis notebook shows you how to change the input current when solving PyBaMM models. It also explains how to load in current data from a file, and how to add a user-defined current function. For more examples of different drive cycles see [here](https://github.com/pybamm-team/PyBaMM/tree/master/results/drive_cycles).\n\n### Table of Contents\n1. [Constant current](#constant)\n1. [Loading in current data](#data)\n1. [Adding your own current function](#function)",
"_____no_output_____"
],
[
"## Constant current <a name=\"constant\"></a>\n\nIn this notebook we will use the SPM as the example model, and change the input current from the default option. If you are not familiar with running a model in PyBaMM, please see [this](./models/SPM.ipynb) notebook for more details.\n\nIn PyBaMM, the current function is set using the parameter \"Current function [A]\". Below we load the SPM with the default parameters, and then change the the current function to be an input parameter, so that we can change it easily later.",
"_____no_output_____"
]
],
[
[
"%pip install pybamm -q # install PyBaMM if it is not installed\nimport pybamm\nimport numpy as np\nimport os\nos.chdir(pybamm.__path__[0]+'/..')\n\n# create the model\nmodel = pybamm.lithium_ion.DFN()\n\n# set the default model parameters\nparam = model.default_parameter_values\n\n# change the current function to be an input parameter\nparam[\"Current function [A]\"] = \"[input]\"",
"Note: you may need to restart the kernel to use updated packages.\n"
]
],
[
[
"We can set up a simulation in the usual way, making sure we pass in our updated parameters. We choose to solve with a 1.6A current. In order to do this we must pass a dictionary of inputs whose keys are the parameter names and values are the values we want to use for that call to solve",
"_____no_output_____"
]
],
[
[
"# set up simlation\nsimulation = pybamm.Simulation(model, parameter_values=param)\n\n# solve the model at the given time points, passing the current as an input\nt_eval = np.linspace(0, 600, 300)\nsimulation.solve(t_eval, inputs={\"Current function [A]\": 1.6})\n\n# plot\nsimulation.plot()",
"_____no_output_____"
]
],
[
[
"PyBaMM can also simulate rest behaviour by setting the current function to zero:",
"_____no_output_____"
]
],
[
[
"# solve the model at the given time points\nsimulation.solve(t_eval, inputs={\"Current function [A]\": 0})\n\n# plot\nsimulation.plot()",
"_____no_output_____"
]
],
[
[
"## Loading in current data <a name=\"data\"></a>\n\nTo run drive cycles from data we can create an interpolant and pass it as the current function. ",
"_____no_output_____"
]
],
[
[
"import pandas as pd # needed to read the csv data file\n\nmodel = pybamm.lithium_ion.DFN()\n\n# import drive cycle from file\ndrive_cycle = pd.read_csv(\"pybamm/input/drive_cycles/US06.csv\", comment=\"#\", header=None).to_numpy()\n\n# load parameter values\nparam = model.default_parameter_values\n\n# create interpolant - must be a function of *dimensional* time\ntimescale = param.evaluate(model.timescale)\ncurrent_interpolant = pybamm.Interpolant(drive_cycle, timescale * pybamm.t)\n\n# set drive cycle\nparam[\"Current function [A]\"] = current_interpolant\n\n# set up simulation - for drive cycles we recommend using the CasadiSolver in \"fast\" mode\nsolver = pybamm.CasadiSolver(mode=\"fast\")\nsimulation = pybamm.Simulation(model, parameter_values=param, solver=solver)",
"_____no_output_____"
]
],
[
[
"Note that when simulating drive cycles there is no need to pass a list of times at which to return the solution, the results are automatically returned at the time points in the data. If you would like the solution returned at times different to those in the data then you can pass an array of times `t_eval` to `solve` in the usual way.",
"_____no_output_____"
]
],
[
[
"# simulate US06 drive cycle (duration 600 seconds)\nsimulation.solve()\n\n# plot\nsimulation.plot()",
"_____no_output_____"
]
],
[
[
"Note that some solvers try to evaluate the model equations at a very large value of `t` during the first step. This may raise a warning if the time requested by the solver is outside of the range of the data provided. However, this does not affect the solve since this large timestep is rejected by the solver, and a suitable shorter initial step is taken.",
"_____no_output_____"
],
[
"## Adding your own current function <a name=\"function\"></a>\n\nA user defined current function can be passed to any model by specifying either a function or a set of data points for interpolation.\n\nFor example, you may want to simulate a sinusoidal current with amplitude A and frequency omega. In order to do so you must first define the method",
"_____no_output_____"
]
],
[
[
"# create user-defined function\ndef my_fun(A, omega):\n def current(t):\n return A * pybamm.sin(2 * np.pi * omega * t)\n \n return current",
"_____no_output_____"
]
],
[
[
"Note that the function returns a function which takes the input time.\nThen the model may be loaded and the \"Current function\" parameter updated to `my_fun` called with a specific value of `A` and `omega`",
"_____no_output_____"
]
],
[
[
"model = pybamm.lithium_ion.SPM()\n\n# load default parameter values\nparam = model.default_parameter_values\n\n# set user defined current function\nA = model.param.I_typ\nomega = 0.1\nparam[\"Current function [A]\"] = my_fun(A,omega)\n",
"_____no_output_____"
]
],
[
[
"Note that when `my_fun` is evaluated with `A` and `omega`, this creates a new function `current(t)` which can then be used in the expression tree. The model may then be solved in the usual way",
"_____no_output_____"
]
],
[
[
"# set up simulation\nsimulation = pybamm.Simulation(model, parameter_values=param)\n\n# Example: simulate for 30 seconds\nsimulation_time = 30 # end time in seconds\nnpts = int(50 * simulation_time * omega) # need enough timesteps to resolve output\nt_eval = np.linspace(0, simulation_time, npts)\nsolution = simulation.solve(t_eval)\nlabel = [\"Frequency: {} Hz\".format(omega)]\n\n# plot current and voltage\noutput_variables = [\"Current [A]\", \"Terminal voltage [V]\"]\nsimulation.plot(output_variables, labels=label)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0cf271e45b3006916fc1d936bcf814880c6c2d4 | 99,208 | ipynb | Jupyter Notebook | Section-08-Discretisation/08.02-Equal-frequency-discretisation.ipynb | cym3509/FeatureEngineering | 8237dbdec803f5bf91543466cff011108fb2c935 | [
"BSD-3-Clause"
] | 1 | 2020-07-27T14:00:50.000Z | 2020-07-27T14:00:50.000Z | Section-08-Discretisation/08.02-Equal-frequency-discretisation.ipynb | cym3509/FeatureEngineering | 8237dbdec803f5bf91543466cff011108fb2c935 | [
"BSD-3-Clause"
] | null | null | null | Section-08-Discretisation/08.02-Equal-frequency-discretisation.ipynb | cym3509/FeatureEngineering | 8237dbdec803f5bf91543466cff011108fb2c935 | [
"BSD-3-Clause"
] | null | null | null | 76.726991 | 11,556 | 0.78911 | [
[
[
"## Discretisation\n\nDiscretisation is the process of transforming continuous variables into discrete variables by creating a set of contiguous intervals that span the range of the variable's values. Discretisation is also called **binning**, where bin is an alternative name for interval.\n\n\n### Discretisation helps handle outliers and may improve value spread in skewed variables\n\nDiscretisation helps handle outliers by placing these values into the lower or higher intervals, together with the remaining inlier values of the distribution. Thus, these outlier observations no longer differ from the rest of the values at the tails of the distribution, as they are now all together in the same interval / bucket. In addition, by creating appropriate bins or intervals, discretisation can help spread the values of a skewed variable across a set of bins with equal number of observations.\n\n\n### Discretisation approaches\n\nThere are several approaches to transform continuous variables into discrete ones. Discretisation methods fall into 2 categories: **supervised and unsupervised**. Unsupervised methods do not use any information, other than the variable distribution, to create the contiguous bins in which the values will be placed. Supervised methods typically use target information in order to create the bins or intervals.\n\n\n#### Unsupervised discretisation methods\n\n- Equal width discretisation\n- Equal frequency discretisation\n- K-means discretisation\n\n#### Supervised discretisation methods\n\n- Discretisation using decision trees\n\n\nIn this lecture, I will describe **equal frequency discretisation**.\n\n\n## Equal frequency discretisation\n\nEqual frequency discretisation divides the scope of possible values of the variable into N bins, where each bin carries the same amount of observations. This is particularly useful for skewed variables as it spreads the observations over the different bins equally. We find the interval boundaries by determining the quantiles.\n\nEqual frequency discretisation using quantiles consists of dividing the continuous variable into N quantiles, N to be defined by the user.\n\nEqual frequency binning is straightforward to implement and by spreading the values of the observations more evenly it may help boost the algorithm's performance. This arbitrary binning may also disrupt the relationship with the target.\n\n## In this demo\n\nWe will learn how to perform equal frequency discretisation using the Titanic dataset with\n\n- pandas and NumPy\n- Feature-engine\n- Scikit-learn",
"_____no_output_____"
],
[
"## Titanic dataset",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\n\nimport matplotlib.pyplot as plt\n\nfrom sklearn.model_selection import train_test_split\n\nfrom sklearn.preprocessing import KBinsDiscretizer\n\nfrom feature_engine.discretisers import EqualFrequencyDiscretiser",
"_____no_output_____"
],
[
"# load the numerical variables of the Titanic Dataset\n\ndata = pd.read_csv('../titanic.csv',\n usecols=['age', 'fare', 'survived'])\n\ndata.head()",
"_____no_output_____"
],
[
"# Let's separate into train and test set\n\nX_train, X_test, y_train, y_test = train_test_split(\n data[['age', 'fare']],\n data['survived'],\n test_size=0.3,\n random_state=0)\n\nX_train.shape, X_test.shape",
"_____no_output_____"
]
],
[
[
"The variables Age and Fare contain missing data, that I will fill by extracting a random sample of the variable.",
"_____no_output_____"
]
],
[
[
"def impute_na(data, variable):\n # function to fill NA with a random sample\n\n df = data.copy()\n\n # random sampling\n df[variable+'_random'] = df[variable]\n\n # extract the random sample to fill the na\n random_sample = X_train[variable].dropna().sample(\n df[variable].isnull().sum(), random_state=0)\n\n # pandas needs to have the same index in order to merge datasets\n random_sample.index = df[df[variable].isnull()].index\n df.loc[df[variable].isnull(), variable+'_random'] = random_sample\n\n return df[variable+'_random']",
"_____no_output_____"
],
[
"# replace NA in both train and test sets\n\nX_train['age'] = impute_na(data, 'age')\nX_test['age'] = impute_na(data, 'age')\n\nX_train['fare'] = impute_na(data, 'fare')\nX_test['fare'] = impute_na(data, 'fare')",
"_____no_output_____"
],
[
"# let's explore the distribution of age\n\nX_train[['age', 'fare']].hist(bins=30, figsize=(8,4))\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Equal frequency discretisation with pandas and NumPy\n\nThe interval limits are the quantile limits. We can find those out with pandas qcut.",
"_____no_output_____"
]
],
[
[
"# let's use pandas qcut (quantile cut) and I indicate that\n# we want 10 bins.\n\n# retbins = True indicates that I want to capture the limits\n# of each interval (so I can then use them to cut the test set)\n\nAge_disccretised, intervals = pd.qcut(\n X_train['age'], 10, labels=None, retbins=True, precision=3, duplicates='raise')\n\npd.concat([Age_disccretised, X_train['age']], axis=1).head(10)",
"_____no_output_____"
]
],
[
[
"We can see in the above output how by discretising using quantiles, we placed each Age observation within one interval. For example, age 29 was placed in the 26-30 interval, whereas age 63 was placed into the 49-80 interval. \n\nNote how the interval widths are different.\n\nWe can visualise the interval cut points below:",
"_____no_output_____"
]
],
[
[
"intervals",
"_____no_output_____"
]
],
[
[
"And because we generated the bins using the quantile cut method, we should have roughly the same amount of observations per bin. See below.",
"_____no_output_____"
]
],
[
[
"# roughly the same number of passengers per interval\n\nAge_disccretised.value_counts()",
"_____no_output_____"
],
[
"# we can also add labels instead of having the interval boundaries, to the bins, as follows:\n\nlabels = ['Q'+str(i) for i in range(1,11)]\nlabels",
"_____no_output_____"
],
[
"Age_disccretised, intervals = pd.qcut(X_train['age'], 10, labels=labels,\n retbins=True,\n precision=3, duplicates='raise')\n\nAge_disccretised.head()",
"_____no_output_____"
],
[
"# to transform the test set:\n# we use pandas cut method (instead of qcut) and\n# pass the quantile edges calculated in the training set\n\nX_test['Age_disc_label'] = pd.cut(x = X_test['age'], bins=intervals, labels=labels)\nX_test['Age_disc'] = pd.cut(x = X_test['age'], bins=intervals)\n\nX_test.head(10)",
"_____no_output_____"
],
[
"# let's check that we have equal frequency (equal number of observations per bin)\nX_test.groupby('Age_disc')['age'].count().plot.bar()",
"_____no_output_____"
]
],
[
[
"We can see that the top intervals have less observations. This may happen with skewed distributions if we try to divide in a high number of intervals. To make the value spread more homogeneous, we should discretise in less intervals.\n\n## Equal frequency discretisation with Feature-Engine",
"_____no_output_____"
]
],
[
[
"# Let's separate into train and test set\n\nX_train, X_test, y_train, y_test = train_test_split(\n data[['age', 'fare']],\n data['survived'],\n test_size=0.3,\n random_state=0)\n\nX_train.shape, X_test.shape",
"_____no_output_____"
],
[
"# replace NA in both train and test sets\n\nX_train['age'] = impute_na(data, 'age')\nX_test['age'] = impute_na(data, 'age')\n\nX_train['fare'] = impute_na(data, 'fare')\nX_test['fare'] = impute_na(data, 'fare')",
"_____no_output_____"
],
[
"# with feature engine we can automate the process for many variables\n# in one line of code\n\ndisc = EqualFrequencyDiscretiser(q=10, variables = ['age', 'fare'])\n\ndisc.fit(X_train)",
"_____no_output_____"
],
[
"# in the binner dict, we can see the limits of the intervals. Note\n# that the intervals have different widths\n\ndisc.binner_dict_",
"_____no_output_____"
],
[
"# transform train and text\n\ntrain_t = disc.transform(X_train)\ntest_t = disc.transform(X_test)",
"_____no_output_____"
],
[
"train_t.head()",
"_____no_output_____"
],
[
"# and now let's explore the number of observations per bucket\n\nt1 = train_t.groupby(['age'])['age'].count() / len(train_t)\nt2 = test_t.groupby(['age'])['age'].count() / len(test_t)\n\ntmp = pd.concat([t1, t2], axis=1)\ntmp.columns = ['train', 'test']\ntmp.plot.bar()\nplt.xticks(rotation=0)\nplt.ylabel('Number of observations per bin')",
"_____no_output_____"
],
[
"t1 = train_t.groupby(['fare'])['fare'].count() / len(train_t)\nt2 = test_t.groupby(['fare'])['fare'].count() / len(test_t)\n\ntmp = pd.concat([t1, t2], axis=1)\ntmp.columns = ['train', 'test']\ntmp.plot.bar()\nplt.xticks(rotation=0)\nplt.ylabel('Number of observations per bin')",
"_____no_output_____"
]
],
[
[
"Note how equal frequency discretisation obtains a better value spread across the different intervals.\n\n## Equal frequency discretisation with Scikit-learn",
"_____no_output_____"
]
],
[
[
"# Let's separate into train and test set\n\nX_train, X_test, y_train, y_test = train_test_split(\n data[['age', 'fare']],\n data['survived'],\n test_size=0.3,\n random_state=0)\n\nX_train.shape, X_test.shape",
"_____no_output_____"
],
[
"# replace NA in both train and test sets\n\nX_train['age'] = impute_na(data, 'age')\nX_test['age'] = impute_na(data, 'age')\n\nX_train['fare'] = impute_na(data, 'fare')\nX_test['fare'] = impute_na(data, 'fare')",
"_____no_output_____"
],
[
"disc = KBinsDiscretizer(n_bins=10, encode='ordinal', strategy='quantile')\n\ndisc.fit(X_train[['age', 'fare']])",
"_____no_output_____"
],
[
"disc.bin_edges_",
"_____no_output_____"
],
[
"train_t = disc.transform(X_train[['age', 'fare']])\n\ntrain_t = pd.DataFrame(train_t, columns = ['age', 'fare'])\n\ntrain_t.head()",
"_____no_output_____"
],
[
"test_t = disc.transform(X_test[['age', 'fare']])\n\ntest_t = pd.DataFrame(test_t, columns = ['age', 'fare'])",
"_____no_output_____"
],
[
"t1 = train_t.groupby(['age'])['age'].count() / len(train_t)\nt2 = test_t.groupby(['age'])['age'].count() / len(test_t)\n\ntmp = pd.concat([t1, t2], axis=1)\ntmp.columns = ['train', 'test']\ntmp.plot.bar()\nplt.xticks(rotation=0)\nplt.ylabel('Number of observations per bin')",
"_____no_output_____"
],
[
"t1 = train_t.groupby(['fare'])['fare'].count() / len(train_t)\nt2 = test_t.groupby(['fare'])['fare'].count() / len(test_t)\n\ntmp = pd.concat([t1, t2], axis=1)\ntmp.columns = ['train', 'test']\ntmp.plot.bar()\nplt.xticks(rotation=0)\nplt.ylabel('Number of observations per bin')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0cf2d722bf765a0f87f4ddfaa7554ce78aeafdd | 7,745 | ipynb | Jupyter Notebook | Simple baseline with XGBoost.ipynb | inzva/earthquake-prediction-kaggle | 6e0d4bed9120f2997ca7d46174062d29289bd710 | [
"MIT"
] | 6 | 2019-03-19T12:56:56.000Z | 2020-09-22T10:11:13.000Z | Simple baseline with XGBoost.ipynb | inzva/earthquake-prediction-kaggle | 6e0d4bed9120f2997ca7d46174062d29289bd710 | [
"MIT"
] | null | null | null | Simple baseline with XGBoost.ipynb | inzva/earthquake-prediction-kaggle | 6e0d4bed9120f2997ca7d46174062d29289bd710 | [
"MIT"
] | 1 | 2019-06-05T11:46:03.000Z | 2019-06-05T11:46:03.000Z | 7,745 | 7,745 | 0.660297 | [
[
[
"This baseline has reached Top %11 with rank of #457/4540 Teams at Private Leader Board (missed Bronze with only 2 places)",
"_____no_output_____"
]
],
[
[
"import numpy as np \nimport pandas as pd \nimport sys\nimport gc\nfrom scipy.signal import hilbert\nfrom scipy.signal import hann\nfrom scipy.signal import convolve\n\npd.options.display.precision = 15",
"_____no_output_____"
],
[
"train_set = pd.read_csv('../input/train.csv', dtype={'acoustic_data': np.int16, 'time_to_failure': np.float32})",
"_____no_output_____"
],
[
"segments = int(np.floor(train_set.shape[0] / 150000))",
"_____no_output_____"
],
[
"X_train = pd.DataFrame(index=range(segments), dtype=np.float64)\ny_train = pd.DataFrame(index=range(segments), dtype=np.float64, columns=['time_to_failure'])",
"_____no_output_____"
],
[
"def feature_generate(df,x,seg):\n df.loc[seg, 'ave'] = x.mean()\n df.loc[seg, 'std'] = x.std()\n df.loc[seg, 'max'] = x.max()\n df.loc[seg, 'min'] = x.min()\n df.loc[seg, 'sum'] = x.sum()\n df.loc[seg, 'mad'] = x.mad()\n df.loc[seg, 'kurtosis'] = x.kurtosis()\n df.loc[seg, 'skew'] = x.skew()\n df.loc[seg, 'quant0_01'] = np.quantile(x,0.01)\n df.loc[seg, 'quant0_05'] = np.quantile(x,0.05)\n df.loc[seg, 'quant0_95'] = np.quantile(x,0.95)\n df.loc[seg, 'quant0_99'] = np.quantile(x,0.99)\n df.loc[seg, 'abs_min'] = np.abs(x).min()\n df.loc[seg, 'abs_max'] = np.abs(x).max()\n df.loc[seg, 'abs_mean'] = np.abs(x).mean()\n df.loc[seg, 'abs_std'] = np.abs(x).std()\n df.loc[seg, 'mean_change_abs'] = np.mean(np.diff(x))\n df.loc[seg, 'max_to_min'] = x.max() / np.abs(x.min())\n df.loc[seg, 'max_to_min_diff'] = x.max() - np.abs(x.min())\n df.loc[seg, 'count_big'] = len(x[np.abs(x) > 500])\n df.loc[seg, 'average_first_10000'] = x[:10000].mean()\n df.loc[seg, 'average_last_10000'] = x[-10000:].mean()\n df.loc[seg, 'average_first_50000'] = x[:50000].mean()\n df.loc[seg, 'average_last_50000'] = x[-50000:].mean()\n df.loc[seg, 'std_first_10000'] = x[:10000].std()\n df.loc[seg, 'std_last_10000'] = x[-10000:].std()\n df.loc[seg, 'std_first_50000'] = x[:50000].std()\n df.loc[seg, 'std_last_50000'] = x[-50000:].std()\n df.loc[seg, '10q'] = np.percentile(x, 0.10)\n df.loc[seg, '25q'] = np.percentile(x, 0.25)\n df.loc[seg, '50q'] = np.percentile(x, 0.50)\n df.loc[seg, '75q'] = np.percentile(x, 0.75)\n df.loc[seg, '90q'] = np.percentile(x, 0.90)\n df.loc[seg, 'abs_1q'] = np.percentile(x, np.abs(0.01))\n df.loc[seg, 'abs_5q'] = np.percentile(x, np.abs(0.05))\n df.loc[seg, 'abs_30q'] = np.percentile(x, np.abs(0.30))\n df.loc[seg, 'abs_60q'] = np.percentile(x, np.abs(0.60))\n df.loc[seg, 'abs_95q'] = np.percentile(x, np.abs(0.95))\n df.loc[seg, 'abs_99q'] = np.percentile(x, np.abs(0.99))\n df.loc[seg, 'hilbert_mean'] = np.abs(hilbert(x)).mean()\n df.loc[seg, 'hann_window_mean'] = (convolve(x, hann(150), mode = 'same') / sum(hann(150))).mean() \n\n for windows in [10, 100, 1000]:\n x_roll_std = x.rolling(windows).std().dropna().values\n x_roll_mean = x.rolling(windows).mean().dropna().values\n df.loc[seg, 'avg_roll_std' + str(windows)] = x_roll_std.mean()\n df.loc[seg, 'std_roll_std' + str(windows)] = x_roll_std.std()\n df.loc[seg, 'max_roll_std' + str(windows)] = x_roll_std.max()\n df.loc[seg, 'min_roll_std' + str(windows)] = x_roll_std.min()\n df.loc[seg, '1q_roll_std' + str(windows)] = np.quantile(x_roll_std, 0.01)\n df.loc[seg, '5q_roll_std' + str(windows)] = np.quantile(x_roll_std, 0.05)\n df.loc[seg, '95q_roll_std' + str(windows)] = np.quantile(x_roll_std, 0.95)\n df.loc[seg, '99q_roll_std' + str(windows)] = np.quantile(x_roll_std, 0.99)\n df.loc[seg, 'av_change_abs_roll_std' + str(windows)] = np.mean(np.diff(x_roll_std))\n df.loc[seg, 'abs_max_roll_std' + str(windows)] = np.abs(x_roll_std).max()\n df.loc[seg, 'avg_roll_mean' + str(windows)] = x_roll_mean.mean()\n df.loc[seg, 'std_roll_mean' + str(windows)] = x_roll_mean.std()\n df.loc[seg, 'max_roll_mean' + str(windows)] = x_roll_mean.max()\n df.loc[seg, 'min_roll_mean' + str(windows)] = x_roll_mean.min()\n df.loc[seg, '1q_roll_mean' + str(windows)] = np.quantile(x_roll_mean, 0.01)\n df.loc[seg, '5q_roll_mean' + str(windows)] = np.quantile(x_roll_mean, 0.05)\n df.loc[seg, '95q_roll_mean' + str(windows)] = np.quantile(x_roll_mean, 0.95)\n df.loc[seg, '99q_roll_mean' + str(windows)] = np.quantile(x_roll_mean, 0.99)\n df.loc[seg, 'av_change_abs_roll_mean' + str(windows)] = np.mean(np.diff(x_roll_mean))\n df.loc[seg, 'abs_max_roll_mean' + str(windows)] = np.abs(x_roll_mean).max() \n return df\n",
"_____no_output_____"
],
[
"for s in range(segments):\n seg = train_set.iloc[s*150000:s*150000+150000]\n x = pd.Series(seg['acoustic_data'].values)\n y = seg['time_to_failure'].values[-1]\n y_train.loc[s, 'time_to_failure'] = y\n X_train = feature_generate(X_train,x,s)\ncolumns=X_train.columns \ndel train_set\ngc.collect()",
"_____no_output_____"
],
[
"from sklearn.preprocessing import StandardScaler\nscaler = StandardScaler()\nscaler.fit(X_train)\nX_train = scaler.transform(X_train)\ny_train = y_train.values.flatten()\ngc.collect()",
"_____no_output_____"
],
[
"import xgboost as xgb\nmodel = xgb.XGBRegressor(objective = 'reg:linear',\n metric = 'mae',\n tree_method = 'gpu_hist',\n verbosity = 0)",
"_____no_output_____"
],
[
"%%time\nmodel.fit(X_train,y_train)",
"_____no_output_____"
],
[
"from matplotlib import pyplot\nprint(model.feature_importances_)\npyplot.bar(range(len(model.feature_importances_)), model.feature_importances_)\npyplot.show()",
"_____no_output_____"
],
[
"from xgboost import plot_importance\nplot_importance(model)\npyplot.show()",
"_____no_output_____"
],
[
"submission = pd.read_csv('../input/sample_submission.csv', index_col='seg_id')\nX_test = pd.DataFrame(columns=columns, dtype=np.float64, index=submission.index)\nfor s in X_test.index:\n seg = pd.read_csv('../input/test/' + s + '.csv')\n x = pd.Series(seg['acoustic_data'].values)\n X_test = feature_generate(X_test,x,s)\nX_test = scaler.transform(X_test)\nsubmission['time_to_failure'] = model.predict(X_test).clip(0, 16)\nsubmission.to_csv('submission.csv')",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0cf39980d0bcd7ead617e6e09e0f61277c1957b | 8,516 | ipynb | Jupyter Notebook | JAVA/spring-workspace/DynamicHana/py/odbc.ipynb | Bonseong/KOPO_TIL | 28da91fc052632f5a504e07f7cebfd1bfcc8cbc1 | [
"MIT"
] | null | null | null | JAVA/spring-workspace/DynamicHana/py/odbc.ipynb | Bonseong/KOPO_TIL | 28da91fc052632f5a504e07f7cebfd1bfcc8cbc1 | [
"MIT"
] | null | null | null | JAVA/spring-workspace/DynamicHana/py/odbc.ipynb | Bonseong/KOPO_TIL | 28da91fc052632f5a504e07f7cebfd1bfcc8cbc1 | [
"MIT"
] | null | null | null | 29.264605 | 895 | 0.468178 | [
[
[
"import os\nimport sys\nimport jpype\nimport jaydebeapi as jp\nimport pandas.io.sql as pd_sql\nimport pandas as pd",
"_____no_output_____"
],
[
"from matplotlib import pyplot as plt\nimport pyodbc\nimport pandas as pd\nimport numpy as np\nimport json\nimport folium\n\nimport warnings\nwarnings.filterwarnings(action='ignore')\n\nfrom branca.element import Template, MacroElement",
"_____no_output_____"
],
[
"cnxn = pyodbc.connect(\"DSN=DSN2; uid=DA2103; pwd=da03\")",
"_____no_output_____"
],
[
"query=\"SELECT ACCOUNT_MGR, AVG(CREDIT_LIMIT) AS CREDIT_LIMIT FROM CUSTOMER C, ORDERS O WHERE C.ID = O.CUSTOMER_ID GROUP BY ACCOUNT_MGR\"",
"_____no_output_____"
],
[
"import time\nstart = time.time()\n\ndf = pd.read_sql(query, cnxn)\n\nprint(\"time :\", time.time() - start)",
"time : 11.984530687332153\n"
],
[
"df",
"_____no_output_____"
],
[
"cursor.execute('''CREATE TABLE AAA AS (SELECT * FROM EMP)''')",
"_____no_output_____"
],
[
"cursor = conn.cursor()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0cf39d8ed3dbea1bfa1c8630e37ab1bdf682dac | 2,778 | ipynb | Jupyter Notebook | notebooks/summer-school/2021/lab4.ipynb | InfamousPlatypus/platypus | 54a8eae3577513e9168a5700f0e4431fa8b46f83 | [
"Apache-2.0"
] | 2 | 2022-03-09T13:39:05.000Z | 2022-03-24T16:35:55.000Z | notebooks/summer-school/2021/lab4.ipynb | InfamousPlatypus/platypus | 54a8eae3577513e9168a5700f0e4431fa8b46f83 | [
"Apache-2.0"
] | 2 | 2022-03-07T16:25:36.000Z | 2022-03-10T09:28:52.000Z | notebooks/summer-school/2021/lab4.ipynb | InfamousPlatypus/platypus | 54a8eae3577513e9168a5700f0e4431fa8b46f83 | [
"Apache-2.0"
] | 1 | 2021-11-22T14:15:42.000Z | 2021-11-22T14:15:42.000Z | 43.40625 | 244 | 0.661267 | [
[
[
"# Lab 4: Introduction to Training Quantum Circuits\n\n<div class=\"youtube-wrapper\">\n <iframe src=\"https://www.youtube.com/embed/mEfyCvwcSkQ\" title=\"YouTube video player\" frameborder=\"0\" allow=\"accelerometer; autoplay; clipboard-write; encrypted-media; gyroscope; picture-in-picture\" allowfullscreen></iframe>\n</div>\n\n- Download the notebook: <a href=\"/content/summer-school/2021/resources/lab-notebooks/lab-4.ipynb\">[en]</a> <a href=\"/content/summer-school/2021/resources/lab-notebooks/lab-4-ja.ipynb\">[ja]</a>\n\nIn this lab, you will learn how to train circuit-based variational models, using different training techniques and see restrictions the models have and how they might be overcome.\n\nComputing Expectation Values:\n* Graded Exercise 4-1: By matrix multiplication\n* Graded Exercise 4-2: By simulation\n\nTraining A New Loss Function:\n* Graded Exercise 4-3: Define the Hamiltonian\n* Graded Exercise 4-4: Use the SPSA optimizer to find the minimum\n\nNatural Gradients:\n* Exploratory Exercise: Natural Gradients and Barren Plateaus\n\n\n### Suggested resources\n* Read Maria Schuld, Ville Bergholm, Christian Gogolin, Josh Izaac, Nathan Killoran on [Evaluating analytic gradients on quantum hardware](https://arxiv.org/abs/1811.11184)\n\n* Read James Stokes, Josh Izaac, Nathan Killoran, Giuseppe Carleo on [Quantum Natural Gradient](https://arxiv.org/abs/1909.02108)\n\n* Read Julien Gacon, Christa Zoufal, Giuseppe Carleo, Stefan Woerner on [Simultaneous Perturbation Stochastic Approximation of the Quantum Fisher Information](https://arxiv.org/abs/2103.09232)\n\n* Read Jarrod R. McClean, Sergio Boixo, Vadim N. Smelyanskiy, Ryan Babbush, Hartmut Neven on [Barren plateaus in quantum neural network training landscapes](https://arxiv.org/abs/1803.11173)\n\n* Read M. Cerezo, Akira Sone, Tyler Volkoff, Lukasz Cincio, Patrick J. Coles on [Cost Function Dependent Barren Plateaus in Shallow Parametrized Quantum Circuits](https://arxiv.org/abs/2001.00550)\n",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown"
]
] |
d0cf62922413dc43b018c2be67a47840492767f9 | 3,919 | ipynb | Jupyter Notebook | labs/lab-5/notebooks/lab5_2.ipynb | tomdewildt/data-engineering | bdfb6ab0676846f1f027db5bd825b5b7d16ae2c7 | [
"MIT"
] | null | null | null | labs/lab-5/notebooks/lab5_2.ipynb | tomdewildt/data-engineering | bdfb6ab0676846f1f027db5bd825b5b7d16ae2c7 | [
"MIT"
] | 7 | 2021-11-05T12:39:48.000Z | 2021-12-24T11:48:13.000Z | labs/lab-5/notebooks/lab5_2.ipynb | tomdewildt/data-engineering | bdfb6ab0676846f1f027db5bd825b5b7d16ae2c7 | [
"MIT"
] | null | null | null | 22.653179 | 101 | 0.501403 | [
[
[
"from pyspark.sql import SparkSession\nfrom pyspark import SparkConf",
"_____no_output_____"
],
[
"sparkConf = SparkConf()\nsparkConf.setMaster(\"spark://master:7077\")\nsparkConf.setAppName(\"MyFirstSparkAppV2\")\nsparkConf.set(\"spark.driver.memory\", \"2g\")\nsparkConf.set(\"spark.executor.cores\", \"1\")\nsparkConf.set(\"spark.driver.cores\", \"1\")",
"_____no_output_____"
],
[
"# create the spark session, which is the entry point to Spark SQL engine.\nspark = SparkSession.builder.config(conf=sparkConf).getOrCreate()",
"_____no_output_____"
],
[
"# create a dataframe by providing raw values and columns \ndf = spark.createDataFrame(\n [\n (\"Tom\", 33, \"footballer\"), # create your data here, be consistent in the types.\n (\"Jim\", 45, \"teacher\"),\n (\"Alice\", 25, \"teacher\"),\n ],\n [\"name\", \"age\", \"occupation\"], # add your column names here\n)",
"_____no_output_____"
],
[
"# check the schema of the table\ndf.printSchema()",
"root\n |-- name: string (nullable = true)\n |-- age: long (nullable = true)\n |-- occupation: string (nullable = true)\n\n"
],
[
"# apply filtering transformation\ndf = df.where(df.occupation == \"teacher\")",
"_____no_output_____"
],
[
"# print the result\ndf.show()\ndf.count()",
"+-----+---+----------+\n| name|age|occupation|\n+-----+---+----------+\n| Jim| 45| teacher|\n|Alice| 25| teacher|\n+-----+---+----------+\n\n"
],
[
"# Stop the spark context\nspark.stop()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0cf6dec7cbf855bf673228a555134f586b9dff6 | 20,675 | ipynb | Jupyter Notebook | examples/AzureML-Primers/04 - Optimizing Model Training.ipynb | kunZooberg/MLOps | 4d971db43a9d74123ea92e7d481e93091697782f | [
"MIT"
] | 1,068 | 2019-05-07T06:41:51.000Z | 2022-03-31T12:14:22.000Z | examples/AzureML-Primers/04 - Optimizing Model Training.ipynb | pankajkumar002/MLOps | e5bdb50005509710146d10a22a96002c5f8aaae0 | [
"MIT"
] | 27 | 2019-06-25T13:31:52.000Z | 2022-03-17T16:37:09.000Z | examples/AzureML-Primers/04 - Optimizing Model Training.ipynb | pankajkumar002/MLOps | e5bdb50005509710146d10a22a96002c5f8aaae0 | [
"MIT"
] | 373 | 2019-05-07T17:51:30.000Z | 2022-03-30T09:54:19.000Z | 85.433884 | 2,469 | 0.675453 | [
[
[
"# Exercise 4 - Optimizing Model Training\n\nIn [the previous exercise](./03%20-%20Compute%20Contexts.ipynb), you created cloud-based compute and used it when running a model training experiment. The benefit of cloud compute is that it offers a cost-effective way to scale out your experiment workflow and try different algorithms and parameters in order to optimize your model's performance; and that's what we'll explore in this exercise.\n\n> **Important**: This exercise assumes you have completed the previous exercises in this series - specifically, you must have:\n>\n> - Created an Azure ML Workspace.\n> - Uploaded the diabetes.csv data file to the workspace's default datastore.\n> - Registered a **Diabetes Dataset** dataset in the workspace.\n> - Provisioned an Azure ML Compute resource named **cpu-cluster**.\n>\n> If you haven't done that, now would be a good time - nobody's going to do it for you!\n\n## Task 1: Connect to Your Workspace\n\nThe first thing you need to do is to connect to your workspace using the Azure ML SDK. Let's start by ensuring you still have the latest version installed (if you ended and restarted your Azure Notebooks session, the environment may have been reset)",
"_____no_output_____"
]
],
[
[
"!pip install --upgrade azureml-sdk[notebooks,automl,explain]\n\nimport azureml.core\nprint(\"Ready to use Azure ML\", azureml.core.VERSION)",
"_____no_output_____"
]
],
[
[
"Now you're ready to connect to your workspace. When you created it in the previous exercise, you saved its configuration; so now you can simply load the workspace from its configuration file.\n\n> **Note**: If the authenticated session with your Azure subscription has expired since you completed the previous exercise, you'll be prompted to reauthenticate.",
"_____no_output_____"
]
],
[
[
"from azureml.core import Workspace\n\n# Load the workspace from the saved config file\nws = Workspace.from_config()\nprint('Ready to work with', ws.name)",
"_____no_output_____"
]
],
[
[
"Now let's get the Azure ML compute resource you created previously (or recreate it if you deleted it!)",
"_____no_output_____"
]
],
[
[
"from azureml.core.compute import ComputeTarget, AmlCompute\nfrom azureml.core.compute_target import ComputeTargetException\n\n# Choose a name for your CPU cluster\ncpu_cluster_name = \"cpu-cluster\"\n\n# Verify that cluster does not exist already\ntry:\n cpu_cluster = ComputeTarget(workspace=ws, name=cpu_cluster_name)\n print('Found existing cluster, use it.')\nexcept ComputeTargetException:\n # Create an AzureMl Compute resource (a container cluster)\n compute_config = AmlCompute.provisioning_configuration(vm_size='STANDARD_D2_V2', \n vm_priority='lowpriority', \n max_nodes=4)\n cpu_cluster = ComputeTarget.create(ws, cpu_cluster_name, compute_config)\n\ncpu_cluster.wait_for_completion(show_output=True)",
"_____no_output_____"
]
],
[
[
"## Task 2: Use *Hyperdrive* to Determine Optimal Parameter Values\n\nThe remote compute you created is a four-node cluster, and you can take advantage of this to execute multiple experiment runs in parallel. One key reason to do this is to try training a model with a range of different hyperparameter values.\n\nAzure ML includes a feature called *hyperdrive* that enables you to randomly try different values for one or more hyperparameters, and find the best performing trained model based on a metric that you specify - such as *Accuracy* or *Area Under the Curve (AUC)*.\n\n> **More Information**: For more information about Hyperdrive, see the [Azure ML documentation](https://docs.microsoft.com/en-us/azure/machine-learning/service/how-to-tune-hyperparameters).\n\nLet's run a Hyperdrive experiment on the remote compute you have provisioned. First, we'll create the experiment and its associated folder.",
"_____no_output_____"
]
],
[
[
"import os\nfrom azureml.core import Experiment\n\n# Create an experiment\nexperiment_name = 'diabetes_training'\nexperiment = Experiment(workspace = ws, name = experiment_name)\n\n# Create a folder for the experiment files\nexperiment_folder = './' + experiment_name\nos.makedirs(experiment_folder, exist_ok=True)\n\nprint(\"Experiment:\", experiment.name)",
"_____no_output_____"
]
],
[
[
"Now we'll create the Python script our experiment will run in order to train a model.",
"_____no_output_____"
]
],
[
[
"%%writefile $experiment_folder/diabetes_training.py\n# Import libraries\nimport argparse\nimport joblib\nfrom azureml.core import Workspace, Dataset, Experiment, Run\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.metrics import roc_auc_score\nfrom sklearn.metrics import roc_curve\n\n# Set regularization parameter\nparser = argparse.ArgumentParser()\nparser.add_argument('--regularization', type=float, dest='reg_rate', default=0.01, help='regularization rate')\nargs = parser.parse_args()\nreg = args.reg_rate\n\n# Get the experiment run context\nrun = Run.get_context()\n\n# load the diabetes dataset\ndataset_name = 'Diabetes Dataset'\nprint(\"Loading data from \" + dataset_name)\ndiabetes = Dataset.get_by_name(workspace=run.experiment.workspace, name=dataset_name).to_pandas_dataframe()\n\n# Separate features and labels\nX, y = diabetes[['Pregnancies','PlasmaGlucose','DiastolicBloodPressure','TricepsThickness','SerumInsulin','BMI','DiabetesPedigree','Age']].values, diabetes['Diabetic'].values\n\n# Split data into training set and test set\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.30, random_state=0)\n\n# Train a logistic regression model\nprint('Training a logistic regression model with regularization rate of', reg)\nrun.log('Regularization Rate', np.float(reg))\nmodel = LogisticRegression(C=1/reg, solver=\"liblinear\").fit(X_train, y_train)\n\n# calculate accuracy\ny_hat = model.predict(X_test)\nacc = np.average(y_hat == y_test)\nprint('Accuracy:', acc)\nrun.log('Accuracy', np.float(acc))\n\n# calculate AUC\ny_scores = model.predict_proba(X_test)\nauc = roc_auc_score(y_test,y_scores[:,1])\nprint('AUC: ' + str(auc))\nrun.log('AUC', np.float(auc))\n\n# plot ROC curve\nfpr, tpr, thresholds = roc_curve(y_test, y_scores[:,1])\nfig = plt.figure(figsize=(6, 4))\n# Plot the diagonal 50% line\nplt.plot([0, 1], [0, 1], 'k--')\n# Plot the FPR and TPR achieved by our model\nplt.plot(fpr, tpr)\nplt.xlabel('False Positive Rate')\nplt.ylabel('True Positive Rate')\nplt.title('ROC Curve')\nrun.log_image(name = \"ROC\", plot = fig)\nplt.show()\n\nos.makedirs('outputs', exist_ok=True)\n# note file saved in the outputs folder is automatically uploaded into experiment record\njoblib.dump(value=model, filename='outputs/diabetes_model.pkl')\n\nrun.complete()",
"_____no_output_____"
]
],
[
[
"Now, we'll use the *Hyperdrive* feature of Azure ML to run multiple experiments in parallel, using different values for the **regularization** parameter to find the optimal value for our data.",
"_____no_output_____"
]
],
[
[
"from azureml.train.hyperdrive import GridParameterSampling, BanditPolicy, HyperDriveConfig, PrimaryMetricGoal\nfrom azureml.train.hyperdrive import choice\nfrom azureml.widgets import RunDetails\nfrom azureml.train.sklearn import SKLearn\n\n# Sample a range of parameter values\nparams = GridParameterSampling(\n {\n # There's only one parameter, so grid sampling will try each value - with multiple parameters it would try every combination\n '--regularization': choice(0.001, 0.005, 0.01, 0.05, 0.1, 1.0)\n }\n)\n\n# Set evaluation policy to stop poorly performing training runs early\npolicy = BanditPolicy(evaluation_interval=2, slack_factor=0.1)\n\n# Create an estimator that uses the remote compute\nhyper_estimator = SKLearn(source_directory=experiment_folder,\n compute_target = cpu_cluster,\n conda_packages=['pandas','ipykernel','matplotlib'],\n pip_packages=['azureml-sdk','argparse','pyarrow'],\n entry_script='diabetes_training.py')\n\n# Configure hyperdrive settings\nhyperdrive = HyperDriveConfig(estimator=hyper_estimator, \n hyperparameter_sampling=params, \n policy=policy, \n primary_metric_name='AUC', \n primary_metric_goal=PrimaryMetricGoal.MAXIMIZE, \n max_total_runs=6,\n max_concurrent_runs=4)\n\n\n# Run the experiment\nrun = experiment.submit(config=hyperdrive)\n\n# Show the status in the notebook as the experiment runs\nRunDetails(run).show()",
"_____no_output_____"
]
],
[
[
"When all of the runs have finished, you can find the best one based on the performance metric you specified (in this case, the one with the best AUC).",
"_____no_output_____"
]
],
[
[
"best_run = run.get_best_run_by_primary_metric()\nbest_run_metrics = best_run.get_metrics()\nparameter_values = best_run.get_details() ['runDefinition']['arguments']\n\nprint('Best Run Id: ', best_run.id)\nprint(' -AUC:', best_run_metrics['AUC'])\nprint(' -Accuracy:', best_run_metrics['Accuracy'])\nprint(' -Regularization Rate:',parameter_values)",
"_____no_output_____"
]
],
[
[
"Since we've found the best run, we can register the model it trained.",
"_____no_output_____"
]
],
[
[
"from azureml.core import Model\n\n# Register model\nbest_run.register_model(model_path='outputs/diabetes_model.pkl', model_name='diabetes_model', tags={'Training context':'Hyperdrive'}, properties={'AUC': best_run_metrics['AUC'], 'Accuracy': best_run_metrics['Accuracy']})\n\n# List registered models\nfor model in Model.list(ws):\n print(model.name, 'version:', model.version)\n for tag_name in model.tags:\n tag = model.tags[tag_name]\n print ('\\t',tag_name, ':', tag)\n for prop_name in model.properties:\n prop = model.properties[prop_name]\n print ('\\t',prop_name, ':', prop)\n print('\\n')",
"_____no_output_____"
]
],
[
[
"## Task 3: Use *Auto ML* to Find the Best Model\n\nHyperparameter tuning has helped us find the optimal regularization rate for our logistic regression model, but we might get better results by trying a different algorithm, and by performing some basic feature-engineering, such as scaling numeric feature values. You could just create lots of different training scripts that apply various scikit-learn algorithms, and try them all until you find the best result; but Azure ML provides a feature called *Automated Machine Learning* (or *Auto ML*) that can do this for you.\n\nFirst, let's create a folder for a new experiment.",
"_____no_output_____"
]
],
[
[
"# Create a project folder if it doesn't exist\nautoml_folder = \"automl_experiment\"\nif not os.path.exists(automl_folder):\n os.makedirs(automl_folder)\nprint(automl_folder, 'folder created')",
"_____no_output_____"
]
],
[
[
"You don't need to create a training script (Auto ML will do that for you), but you do need to load the training data; and when using remote compute, this is best achieved by creating a script containing a **get_data** function.",
"_____no_output_____"
]
],
[
[
"%%writefile $automl_folder/get_data.py\n#Write the get_data file.\nfrom azureml.core import Run, Workspace, Dataset\nfrom sklearn.model_selection import train_test_split\nimport pandas as pd\nimport numpy as np\n\ndef get_data():\n\n # load the diabetes dataset\n run = Run.get_context()\n dataset_name = 'Diabetes Dataset'\n diabetes = Dataset.get_by_name(workspace=run.experiment.workspace, name=dataset_name).to_pandas_dataframe()\n\n # Separate features and labels\n X, y = diabetes[['Pregnancies','PlasmaGlucose','DiastolicBloodPressure','TricepsThickness','SerumInsulin','BMI','DiabetesPedigree','Age']].values, diabetes['Diabetic'].values\n \n # Split data into training set and test set\n X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.30, random_state=0)\n\n return { \"X\" : X_train, \"y\" : y_train, \"X_valid\" : X_test, \"y_valid\" : y_test }",
"_____no_output_____"
]
],
[
[
"Now you're ready to confifure the Auto ML experiment. To do this, you'll need a run configuration that includes the required packages for the experiment environment, and a set of configuration settings that tells Auto ML how many options to try, which metric to use when evaluating models, and so on.\n\n> **More Information**: For more information about options when using Auto ML, see the [Azure ML documentation](https://docs.microsoft.com/en-us/azure/machine-learning/service/how-to-configure-auto-train).",
"_____no_output_____"
]
],
[
[
"from azureml.core.runconfig import RunConfiguration\nfrom azureml.core.conda_dependencies import CondaDependencies\nfrom azureml.train.automl import AutoMLConfig\nimport time\nimport logging\n\n\nautoml_run_config = RunConfiguration(framework=\"python\")\nautoml_run_config.environment.docker.enabled = True\n\nauto_ml_dependencies = CondaDependencies.create(\n pip_packages=[\"azureml-sdk\", \"pyarrow\", \"pandas\", \"scikit-learn\", \"numpy\"])\nautoml_run_config.environment.python.conda_dependencies = auto_ml_dependencies\n\n\nautoml_settings = {\n \"name\": \"Diabetes_AutoML_{0}\".format(time.time()),\n \"iteration_timeout_minutes\": 10,\n \"iterations\": 10,\n \"primary_metric\": 'AUC_weighted',\n \"preprocess\": False,\n \"max_concurrent_iterations\": 4,\n \"verbosity\": logging.INFO\n}\n\nautoml_config = AutoMLConfig(task='classification',\n debug_log='automl_errors.log',\n path=automl_folder,\n compute_target=cpu_cluster,\n run_configuration=automl_run_config,\n data_script=automl_folder + \"/get_data.py\",\n model_explainability=True,\n **automl_settings,\n )",
"_____no_output_____"
]
],
[
[
"OK, we're ready to go. Let's start the Auto ML run, which will generate child runs for different algorithms.\n\n> **Note**: This will take some time. Progress will be displayed as each child run completes, and then a widget showing the results will be displayed.",
"_____no_output_____"
]
],
[
[
"from azureml.core.experiment import Experiment\nfrom azureml.widgets import RunDetails\n\nautoml_experiment = Experiment(ws, 'diabetes_automl')\nautoml_run = automl_experiment.submit(automl_config, show_output=True)\nRunDetails(automl_run).show()",
"_____no_output_____"
]
],
[
[
"View the output of the experiment in the widget, and click the run that produced the best result to see its details.\nThen click the link to view the experiment details in the Azure portal and view the overall experiment details before viewing the details for the individual run that produced the best result. There's lots of information here about the performance of the model generated and how its features were used.\n\nLet's get the best run and the model that was generated (you can ignore any warnings about Azure ML package versions that might appear).",
"_____no_output_____"
]
],
[
[
"best_run, fitted_model = automl_run.get_output()\nprint(best_run)\nprint(fitted_model)\nbest_run_metrics = best_run.get_metrics()\nfor metric_name in best_run_metrics:\n metric = best_run_metrics[metric_name]\n print(metric_name, metric)",
"_____no_output_____"
]
],
[
[
"One of the options you used was to include model *explainability*. This uses a test dataset to evaluate the importance of each feature. You can view this data in the notebook widget or the portal, and you can also retrieve it from the run.",
"_____no_output_____"
]
],
[
[
"from azureml.train.automl.automlexplainer import retrieve_model_explanation\n\nshap_values, expected_values, overall_summary, overall_imp, per_class_summary, per_class_imp = retrieve_model_explanation(best_run)\n\n# Overall feature importance (the Feature value is the column index in the training data)\nprint(\"Feature\\tImportance\")\nfor i in range(len(overall_imp)):\n print(overall_imp[i], '\\t', overall_summary[i])\n",
"_____no_output_____"
]
],
[
[
"Finally, having found the best performing model, you can register it.",
"_____no_output_____"
]
],
[
[
"# Register model\nbest_run.register_model(model_path='outputs/model.pkl', model_name='diabetes_model', tags={'Training context':'Auto ML'}, properties={'AUC': best_run_metrics['AUC_weighted'], 'Accuracy': best_run_metrics['accuracy']})\n\n# List registered models\nfor model in Model.list(ws):\n print(model.name, 'version:', model.version)\n for tag_name in model.tags:\n tag = model.tags[tag_name]\n print ('\\t',tag_name, ':', tag)\n for prop_name in model.properties:\n prop = model.properties[prop_name]\n print ('\\t',prop_name, ':', prop)\n print('\\n')",
"_____no_output_____"
]
],
[
[
"Now you've seen several ways to leverage the high-scale compute capabilities of the cloud to experiment with model training and find the best performing model for your data. In the next exerise, you'll deploy a registered model into production.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0cf89db4b26029a608afa1fc695db955d631a63 | 4,227 | ipynb | Jupyter Notebook | Python/2. Python Basics (cont.)/1. Regular Expressions (REGEX)/0. Start Learning Here/Regex-Course-Modular/5.Regex_Character_Sets.ipynb | okara83/Becoming-a-Data-Scientist | f09a15f7f239b96b77a2f080c403b2f3e95c9650 | [
"MIT"
] | 8 | 2021-10-31T09:50:49.000Z | 2021-11-20T20:09:05.000Z | Python/2. Python Basics (cont.)/1. Regular Expressions (REGEX)/0. Start Learning Here/Regex-Course-Modular/5.Regex_Character_Sets.ipynb | okara83/Becoming-a-Data-Scientist | f09a15f7f239b96b77a2f080c403b2f3e95c9650 | [
"MIT"
] | null | null | null | Python/2. Python Basics (cont.)/1. Regular Expressions (REGEX)/0. Start Learning Here/Regex-Course-Modular/5.Regex_Character_Sets.ipynb | okara83/Becoming-a-Data-Scientist | f09a15f7f239b96b77a2f080c403b2f3e95c9650 | [
"MIT"
] | 2 | 2021-10-31T18:50:07.000Z | 2021-11-19T06:21:17.000Z | 23.225275 | 86 | 0.461793 | [
[
[
"# Hello,\n> ## Welcome to Module-5\n> ## In this module, we'll discuss about Character Sets.\n\n----",
"_____no_output_____"
],
[
"> ## 1. [a-z] or [A-Z]\n>\n> ### [a-z] represent a single character from the set of lowercase alphabets.\n>\n> ### [A-Z] represent a single character from the set of uppercase alphabets.\n>\n> ### They all react similarly to quantifiers as of a single character in regex.",
"_____no_output_____"
]
],
[
[
"import re\n\npattern1 = '[a-z]'\npattern2 = '[a-z]+'\npattern3 = '[A-Z]'\npattern4 = '[a-zA-Z]+'\n\nstring1 = 'India55'\n\n#--------------CODE---------------\ndef findpattern(pattern, text):\n if re.search(pattern, text):\n return re.search(pattern, text)\n else:\n return 'Not Found!'\n\nprint(findpattern(pattern1, string1))\nprint(findpattern(pattern2, string1))\nprint(findpattern(pattern3, string1))\nprint(findpattern(pattern4, string1))",
"<re.Match object; span=(1, 2), match='n'>\n<re.Match object; span=(1, 5), match='ndia'>\n<re.Match object; span=(0, 1), match='I'>\n<re.Match object; span=(0, 5), match='India'>\n"
]
],
[
[
"----",
"_____no_output_____"
],
[
"> ## 2. [0-9]\n>\n> ### [0-9] represent a single character from the set of digits.\n>\n> ### They all react similarly to quantifiers as of a single character in regex.",
"_____no_output_____"
]
],
[
[
"import re\n\npattern1 = '[0-9]'\npattern2 = '[0-9]+'\n\n\nstring1 = 'India08112021'\n\n#--------------CODE---------------\ndef findpattern(pattern, text):\n if re.search(pattern, text):\n return re.search(pattern, text)\n else:\n return 'Not Found!'\n\nprint(findpattern(pattern1, string1))\nprint(findpattern(pattern2, string1))",
"<re.Match object; span=(5, 6), match='0'>\n<re.Match object; span=(5, 13), match='08112021'>\n"
]
],
[
[
"----",
"_____no_output_____"
],
[
"# Character Sets\n\n### Some abbreviations of character sets.\n\n\n| Symbol | Purpose |\n| :----: | :---------------: |\n| \\s | White Space |\n| \\S | Not White Space |\n| \\d | Digit |\n| \\D | Not Digit |\n| \\w | Word |\n| \\W | Not Word |\n| \\x | Hexadecimal Digit |\n| \\O | Octal Digit |",
"_____no_output_____"
],
[
"----\n# This the end of this Module\n-----",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
d0cfa95f7bf1026a268dd1b7939a01df0de32d51 | 98,090 | ipynb | Jupyter Notebook | jupyter/4.shallow_neural_network/nn_numpy.ipynb | BaiduOSS/PaddleTutorial | d71136c105b78b57c5df1fe98e19e2fd8ee65045 | [
"Apache-2.0"
] | 33 | 2018-03-18T08:56:11.000Z | 2022-01-06T14:43:54.000Z | jupyter/4.shallow_neural_network/nn_numpy.ipynb | CaesarXI/DeepLearningAndPaddleTutorial | d71136c105b78b57c5df1fe98e19e2fd8ee65045 | [
"Apache-2.0"
] | 5 | 2017-12-06T12:40:59.000Z | 2019-07-05T08:47:48.000Z | jupyter/4.shallow_neural_network/nn_numpy.ipynb | CaesarXI/DeepLearningAndPaddleTutorial | d71136c105b78b57c5df1fe98e19e2fd8ee65045 | [
"Apache-2.0"
] | 43 | 2017-12-06T12:51:16.000Z | 2022-01-06T14:44:00.000Z | 145.10355 | 40,582 | 0.868886 | [
[
[
"# Numpy实现浅层神经网络\n\n\n实践部分将搭建神经网络,包含一个隐藏层,实验将会展现出与Logistic回归的不同之处。\n\n实验将使用两层神经网络实现对“花”型图案的分类,如图所示,图中的点包含红点(y=0)和蓝点(y=1)还有点的坐标信息,实验将通过以下步骤完成对两种点的分类,使用Numpy实现。\n\n- 输入样本;\n\n- 搭建神经网络;\n\n- 初始化参数;\n\n- 训练,包括前向传播与后向传播(即BP算法);\n\n- 得出训练后的参数;\n\n- 根据训练所得参数,绘制两类点边界曲线。\n\n<img src=\"image/data.png\" style=\"width:400px;height:300px;\">\n\n该实验将使用Python原生库实现两层神经网络的搭建,完成分类。\n",
"_____no_output_____"
],
[
"## 1 - 引用库\n\n首先,载入几个需要用到的库,它们分别是:\n- numpy:一个python的基本库,用于科学计算\n- planar_utils:定义了一些工具函数\n- matplotlib.pyplot:用于生成图,在验证模型准确率和展示成本变化趋势时会使用到\n- sklearn:用于数据挖掘和数据分析",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport sklearn\nfrom planar_utils import plot_decision_boundary, sigmoid, load_planar_dataset, load_extra_datasets\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\nnp.random.seed(1) ",
"_____no_output_____"
]
],
[
[
"## 2 - 载入数据并观察纬度\n\n载入数据后,输出维度",
"_____no_output_____"
]
],
[
[
"#载入数据\ntrain_x, train_y, test_x, test_y = load_planar_dataset()\n#输出维度\nshape_X = train_x.shape\nshape_Y = train_y.shape\nprint ('The shape of X is: ' + str(shape_X))\nprint ('The shape of Y is: ' + str(shape_Y))\n",
"The shape of X is: (2, 320)\nThe shape of Y is: (1, 320)\n"
]
],
[
[
"由输出可知每组输入坐标包含两个值,包含一个值,共320组数据(测试集在训练集基础上增加80组数据,共400组)。",
"_____no_output_____"
],
[
"## 3 - 简单逻辑回归实验\n\n使用逻辑回归处理该数据,观察分类结果",
"_____no_output_____"
]
],
[
[
"#训练逻辑回归分类器\nclf = sklearn.linear_model.LogisticRegressionCV();\nclf.fit(train_x.T, train_y.T);\n#绘制逻辑回归分类边界\nplot_decision_boundary(lambda x: clf.predict(x), train_x, train_y)\nplt.title(\"Logistic Regression\")\n\n#输出准确率\nLR_predictions = clf.predict(train_x.T)\nprint ('Accuracy of logistic regression:%d ' % float((np.dot(train_y,LR_predictions) + np.dot(1-train_y,1-LR_predictions))/float(train_y.size)*100) +\n '% ' + \"(percentage of correctly labelled datapoints)\")\n",
"/Users/starwe/Desktop/WorkDir/anaconda/lib/python3.6/site-packages/sklearn/utils/validation.py:526: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\n"
]
],
[
[
"可以看出逻辑回归效果并不好,这是因为逻辑回归网络结构只包含输入层和输出层,无法拟合更为复杂的模型,下面尝试神经网络模型。\n",
"_____no_output_____"
],
[
"## 4 - 神经网络模型\n\n下面开始搭建神经网络模型,我们采用两层神经网络实验,隐藏层包含4个节点,使用tanh激活函数;输出层包含一个节点,使用Sigmoid激活函数,结果小于0.5即认为是0,否则认为是1。\n\n** 神经网络结构 **\n\n下面用代码实现神经网络结构,首先确定神经网络的结构,即获取相关数据维度,并设置隐藏层节点个数(本实验设置4个隐藏层节点),用以初始化参数",
"_____no_output_____"
]
],
[
[
"#定义各层规模函数\ndef layer_sizes(X, Y):\n \"\"\"\n 参数含义:\n X -- 输入的数据\n Y -- 输出值\n \n 返回值:\n n_x -- 输入层节点数\n n_h -- 隐藏层节点数\n n_y -- 输出层节点数\n \"\"\"\n \n n_x = X.shape[0] #输入层大小(节点数)\n n_h = 4\n n_y = Y.shape[0] #输出层大小(节点数)\n return (n_x, n_h, n_y)",
"_____no_output_____"
]
],
[
[
"** 初始化模型参数 **\n\n获取相关维度信息后,开始初始化参数,定义相关函数",
"_____no_output_____"
]
],
[
[
"# 定义函数:初始化参数\n\ndef initialize_parameters(n_x, n_h, n_y):\n \"\"\"\n 参数:\n n_x -- 输入层大小\n n_h -- 隐藏层大小\n n_y -- 输出层大小\n \n 返回值:\n params -- 一个包含所有参数的python字典:\n W1 -- (隐藏层)权重,维度是 (n_h, n_x)\n b1 -- (隐藏层)偏移量,维度是 (n_h, 1)\n W2 -- (输出层)权重,维度是 (n_y, n_h)\n b2 -- (输出层)偏移量,维度是 (n_y, 1)\n \"\"\"\n \n np.random.seed(2) # 设置随机种子\n \n #随机初始化参数\n W1 = np.random.randn(n_h, n_x) * 0.01\n b1 = np.zeros((n_h, 1))\n W2 = np.random.randn(n_y, n_h) * 0.01\n b2 = np.zeros((n_y, 1))\n \n \n assert (W1.shape == (n_h, n_x))\n assert (b1.shape == (n_h, 1))\n assert (W2.shape == (n_y, n_h))\n assert (b2.shape == (n_y, 1))\n \n parameters = {\"W1\": W1,\n \"b1\": b1,\n \"W2\": W2,\n \"b2\": b2}\n \n return parameters\n",
"_____no_output_____"
]
],
[
[
"** 前向传播与后向传播 **\n\n获取输入数据,参数初始化完成后,可以开始前向传播的计算",
"_____no_output_____"
]
],
[
[
"# 定义函数:前向传播\n\ndef forward_propagation(X, parameters):\n \"\"\"\n 参数:\n X -- 输入值 \n parameters -- 一个python字典,包含计算所需全部参数(是initialize_parameters函数的输出) \n 返回值:\n A2 -- 模型输出值\n cache -- 一个字典,包含 \"Z1\", \"A1\", \"Z2\" and \"A2\"\n \"\"\"\n \n W1 = parameters[\"W1\"]\n b1 = parameters[\"b1\"]\n W2 = parameters[\"W2\"]\n b2 = parameters[\"b2\"]\n \n #计算中间量和节点值 \n Z1 = np.dot(W1, X) + b1\n A1 = np.tanh(Z1)\n Z2 = np.dot(W2, A1) + b2\n A2 = 1/(1+np.exp(-Z2))\n \n \n assert(A2.shape == (1, X.shape[1]))\n \n cache = {\"Z1\": Z1,\n \"A1\": A1,\n \"Z2\": Z2,\n \"A2\": A2}\n \n return A2, cache",
"_____no_output_____"
]
],
[
[
"前向传播最后可得出模型输出值(即代码中的A2),即可计算成本函数cost",
"_____no_output_____"
]
],
[
[
"# 定义函数:成本函数\n\ndef compute_cost(A2, Y, parameters):\n \"\"\"\n 根据第三章给出的公式计算成本\n \n 参数:\nA2 -- 模型输出值 \nY -- 真实值\n parameters -- 一个python字典包含参数 W1, b1, W2和b2\n \n 返回值:\n cost -- 成本函数\n \"\"\"\n \n m = Y.shape[1] #样本个数\n\n #计算成本\n logprobs = np.multiply(np.log(A2), Y) + np.multiply(np.log(1 - A2), 1 - Y)\n cost = -1. / m * np.sum(logprobs)\n \n cost = np.squeeze(cost) # 确保维度的正确性 \n assert(isinstance(cost, float))\n \n return cost",
"_____no_output_____"
]
],
[
[
"计算了成本函数,可以开始后向传播的计算",
"_____no_output_____"
]
],
[
[
"# 定义函数:后向传播\n\ndef backward_propagation(parameters, cache, X, Y):\n \"\"\" \n 参数:\n parameters -- 一个python字典,包含所有参数 \n cache -- 一个python字典包含\"Z1\", \"A1\", \"Z2\"和\"A2\".\n X -- 输入值\n Y -- 真实值\n \n 返回值:\n grads -- 一个python字典包含所有参数的梯度\n \"\"\"\n m = X.shape[1]\n \n #首先从\"parameters\"获取W1,W2\n W1 = parameters[\"W1\"]\n W2 = parameters[\"W2\"]\n \n # 从\"cache\"中获取A1,A2\n A1 = cache[\"A1\"]\n A2 = cache[\"A2\"]\n \n #后向传播: 计算dW1, db1, dW2, db2. \n dZ2 = A2 - Y\n dW2 = 1. / m * np.dot(dZ2, A1.T)\n db2 = 1. / m * np.sum(dZ2, axis = 1, keepdims = True)\n dZ1 = np.dot(W2.T, dZ2) * (1 - np.power(A1, 2))\n dW1 = 1. / m * np.dot(dZ1, X.T)\n db1 = 1. / m * np.sum(dZ1, axis = 1, keepdims = True)\n \n grads = {\"dW1\": dW1,\n \"db1\": db1,\n \"dW2\": dW2,\n \"db2\": db2}\n \n return grads",
"_____no_output_____"
]
],
[
[
"通过后向传播获取梯度后,可以根据梯度下降公式更新参数",
"_____no_output_____"
]
],
[
[
"def update_parameters(parameters, grads, learning_rate = 1.2):\n \"\"\"\n 使用梯度更新参数\n \n 参数:\n parameters -- 包含所有参数的python字典 \n grads -- 包含所有参数梯度的python字典 \n \n 返回值:\n parameters -- 包含更新后参数的python \n \"\"\"\n #从\"parameters\"中读取全部参数\n W1 = parameters[\"W1\"]\n b1 = parameters[\"b1\"]\n W2 = parameters[\"W2\"]\n b2 = parameters[\"b2\"]\n \n # 从\"grads\"中读取全部梯度\n dW1 = grads[\"dW1\"]\n db1 = grads[\"db1\"]\n dW2 = grads[\"dW2\"]\n db2 = grads[\"db2\"]\n \n #更新参数\n W1 = W1 - learning_rate * dW1\n b1 = b1 - learning_rate * db1\n W2 = W2 - learning_rate * dW2\n b2 = b2 - learning_rate * db2\n \n parameters = {\"W1\": W1,\n \"b1\": b1,\n \"W2\": W2,\n \"b2\": b2}\n \n return parameters",
"_____no_output_____"
]
],
[
[
"** 神经网络模型 **\n\n前向传播、成本函数计算和后向传播构成一个完整的神经网络,将上述函数组合,构建一个神经网络模型",
"_____no_output_____"
]
],
[
[
"#定义函数:神经网络模型\ndef nn_model(X, Y, n_h, num_iterations = 10000, print_cost=False):\n \"\"\"\n 参数:\n X -- 输入值\n Y -- 真实值\n n_h -- 隐藏层大小/节点数\n num_iterations -- 训练次数\n print_cost -- 设置为True,则每1000次训练打印一次成本函数值\n \n 返回值:\nparameters -- 训练结束,更新后的参数值 \n\"\"\"\n \n np.random.seed(3)\n n_x = layer_sizes(X, Y)[0]\n n_y = layer_sizes(X, Y)[2]\n \n #根据n_x, n_h, n_y初始化参数,并取出W1,b1,W2,b2 \n parameters = initialize_parameters(n_x, n_h, n_y)\n W1 = parameters[\"W1\"]\n b1 = parameters[\"b1\"]\n W2 = parameters[\"W2\"]\n b2 = parameters[\"b2\"]\n \n \n for i in range(0, num_iterations): \n \n #前向传播, 输入: \"X, parameters\". 输出: \"A2, cache\".\n A2, cache = forward_propagation(X, parameters)\n \n #成本计算. 输入: \"A2, Y, parameters\". 输出: \"cost\".\n cost = compute_cost(A2, Y, parameters)\n \n #后向传播, 输入: \"parameters, cache, X, Y\". 输出: \"grads\".\n grads = backward_propagation(parameters, cache, X, Y)\n \n #参数更新. 输入: \"parameters, grads\". 输出: \"parameters\".\n parameters = update_parameters(parameters, grads)\n \n #每1000次训练打印一次成本函数值\n if print_cost and i % 1000 == 0:\n print (\"Cost after iteration %i: %f\" %(i, cost))\n\n return parameters\n",
"_____no_output_____"
]
],
[
[
"** 预测 ** \n\n通过上述模型可以训练得出最后的参数,此时需检测其准确率,用训练后的参数预测训练的输出,大于0.5的值视作1,否则视作0",
"_____no_output_____"
]
],
[
[
"#定义函数:预测\ndef predict(parameters, X):\n \"\"\"\n 使用训练所得参数,对每个训练样本进行预测\n \n 参数:\n parameters -- 保安所有参数的python字典 \n X -- 输入值\n \n 返回值:\n predictions -- 模型预测值向量(红色: 0 / 蓝色: 1)\n \"\"\"\n \n #使用训练所得参数进行前向传播计算,并将模型输出值转化为预测值(大于0.5视作1,即True)\n A2, cache = forward_propagation(X, parameters)\n predictions = A2 > 0.5\n\n return predictions",
"_____no_output_____"
]
],
[
[
"下面对获取的数据进行训练,并输出准确率",
"_____no_output_____"
]
],
[
[
"#建立神经网络模型\nparameters = nn_model(train_x, train_y, n_h = 4, num_iterations = 10000, print_cost=True)\n\n#绘制分类边界\nplot_decision_boundary(lambda x: predict(parameters, x.T), train_x, train_y)\nplt.title(\"Decision Boundary for hidden layer size \" + str(4))\n \npredictions = predict(parameters, train_x)\n# 预测训练集\nprint('Train Accuracy: %d' % float((np.dot(train_y, predictions.T) +\n np.dot(1 - train_y, 1 - predictions.T)) /\n float(train_y.size) * 100) + '%')\n# 预测测试集\npredictions = predict(parameters, test_x)\nprint('Test Accuracy: %d' % float((np.dot(test_y, predictions.T) +\n np.dot(1 - test_y, 1 - predictions.T)) /\n float(test_y.size) * 100) + '%')",
"Cost after iteration 0: 0.693049\nCost after iteration 1000: 0.272304\nCost after iteration 2000: 0.261430\nCost after iteration 3000: 0.260627\nCost after iteration 4000: 0.257842\nCost after iteration 5000: 0.255809\nCost after iteration 6000: 0.254249\nCost after iteration 7000: 0.253019\nCost after iteration 8000: 0.252028\nCost after iteration 9000: 0.251208\nTrain Accuracy: 90%\nTest Accuracy: 89%\n"
]
],
[
[
"对比逻辑回归47%的准确率和分类结果图,神经网络分类的结果提高了不少,这是因为神经网络增加的隐藏层,为模型训练提供了更多选择,使得神经网络能拟合更加复杂的模型,对于更加复杂的图案分类更加准确。\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0cfb344fa8e5d5e45232d6a1b8633bc91f5d749 | 88,223 | ipynb | Jupyter Notebook | Assignment_02/TensorFlow.ipynb | andrewlstewart/CS231n-Convolutional-Neural-Networks- | 3a6066d790bd654d5fe3ad670c2308e8b2c05d93 | [
"Unlicense"
] | null | null | null | Assignment_02/TensorFlow.ipynb | andrewlstewart/CS231n-Convolutional-Neural-Networks- | 3a6066d790bd654d5fe3ad670c2308e8b2c05d93 | [
"Unlicense"
] | 15 | 2020-11-18T23:05:49.000Z | 2022-03-12T00:35:03.000Z | Assignment_02/TensorFlow.ipynb | andrewlstewart/CS231n-Convolutional-Neural-Networks- | 3a6066d790bd654d5fe3ad670c2308e8b2c05d93 | [
"Unlicense"
] | null | null | null | 48.051743 | 542 | 0.582705 | [
[
[
"# What's this TensorFlow business?\n\nYou've written a lot of code in this assignment to provide a whole host of neural network functionality. Dropout, Batch Norm, and 2D convolutions are some of the workhorses of deep learning in computer vision. You've also worked hard to make your code efficient and vectorized.\n\nFor the last part of this assignment, though, we're going to leave behind your beautiful codebase and instead migrate to one of two popular deep learning frameworks: in this instance, TensorFlow (or PyTorch, if you choose to work with that notebook).",
"_____no_output_____"
],
[
"#### What is it?\nTensorFlow is a system for executing computational graphs over Tensor objects, with native support for performing backpropogation for its Variables. In it, we work with Tensors which are n-dimensional arrays analogous to the numpy ndarray.\n\n#### Why?\n\n* Our code will now run on GPUs! Much faster training. Writing your own modules to run on GPUs is beyond the scope of this class, unfortunately.\n* We want you to be ready to use one of these frameworks for your project so you can experiment more efficiently than if you were writing every feature you want to use by hand. \n* We want you to stand on the shoulders of giants! TensorFlow and PyTorch are both excellent frameworks that will make your lives a lot easier, and now that you understand their guts, you are free to use them :) \n* We want you to be exposed to the sort of deep learning code you might run into in academia or industry. ",
"_____no_output_____"
],
[
"## How will I learn TensorFlow?\n\nTensorFlow has many excellent tutorials available, including those from [Google themselves](https://www.tensorflow.org/get_started/get_started).\n\nOtherwise, this notebook will walk you through much of what you need to do to train models in TensorFlow. See the end of the notebook for some links to helpful tutorials if you want to learn more or need further clarification on topics that aren't fully explained here.\n\n**NOTE: This notebook is meant to teach you the latest version of Tensorflow 2.0. Most examples on the web today are still in 1.x, so be careful not to confuse the two when looking up documentation**.\n\n## Install Tensorflow 2.0\nTensorflow 2.0 is still not in a fully 100% stable release, but it's still usable and more intuitive than TF 1.x. Please make sure you have it installed before moving on in this notebook! Here are some steps to get started:\n\n1. Have the latest version of Anaconda installed on your machine.\n2. Create a new conda environment starting from Python 3.7. In this setup example, we'll call it `tf_20_env`.\n3. Run the command: `source activate tf_20_env`\n4. Then pip install TF 2.0 as described here: https://www.tensorflow.org/install/pip \n\nA guide on creating Anaconda enviornments: https://uoa-eresearch.github.io/eresearch-cookbook/recipe/2014/11/20/conda/\n\nThis will give you an new enviornemnt to play in TF 2.0. Generally, if you plan to also use TensorFlow in your other projects, you might also want to keep a seperate Conda environment or virtualenv in Python 3.7 that has Tensorflow 1.9, so you can switch back and forth at will. ",
"_____no_output_____"
],
[
"# Table of Contents\n\nThis notebook has 5 parts. We will walk through TensorFlow at **three different levels of abstraction**, which should help you better understand it and prepare you for working on your project.\n\n1. Part I, Preparation: load the CIFAR-10 dataset.\n2. Part II, Barebone TensorFlow: **Abstraction Level 1**, we will work directly with low-level TensorFlow graphs. \n3. Part III, Keras Model API: **Abstraction Level 2**, we will use `tf.keras.Model` to define arbitrary neural network architecture. \n4. Part IV, Keras Sequential + Functional API: **Abstraction Level 3**, we will use `tf.keras.Sequential` to define a linear feed-forward network very conveniently, and then explore the functional libraries for building unique and uncommon models that require more flexibility.\n5. Part V, CIFAR-10 open-ended challenge: please implement your own network to get as high accuracy as possible on CIFAR-10. You can experiment with any layer, optimizer, hyperparameters or other advanced features. \n\nWe will discuss Keras in more detail later in the notebook.\n\nHere is a table of comparison:\n\n| API | Flexibility | Convenience |\n|---------------|-------------|-------------|\n| Barebone | High | Low |\n| `tf.keras.Model` | High | Medium |\n| `tf.keras.Sequential` | Low | High |",
"_____no_output_____"
],
[
"# Part I: Preparation\n\nFirst, we load the CIFAR-10 dataset. This might take a few minutes to download the first time you run it, but after that the files should be cached on disk and loading should be faster.\n\nIn previous parts of the assignment we used CS231N-specific code to download and read the CIFAR-10 dataset; however the `tf.keras.datasets` package in TensorFlow provides prebuilt utility functions for loading many common datasets.\n\nFor the purposes of this assignment we will still write our own code to preprocess the data and iterate through it in minibatches. The `tf.data` package in TensorFlow provides tools for automating this process, but working with this package adds extra complication and is beyond the scope of this notebook. However using `tf.data` can be much more efficient than the simple approach used in this notebook, so you should consider using it for your project.",
"_____no_output_____"
]
],
[
[
"import os\nimport tensorflow as tf\nimport numpy as np\nimport math\nimport timeit\nimport matplotlib.pyplot as plt\n\n%matplotlib inline",
"_____no_output_____"
],
[
"def load_cifar10(num_training=49000, num_validation=1000, num_test=10000):\n \"\"\"\n Fetch the CIFAR-10 dataset from the web and perform preprocessing to prepare\n it for the two-layer neural net classifier. These are the same steps as\n we used for the SVM, but condensed to a single function.\n \"\"\"\n # Load the raw CIFAR-10 dataset and use appropriate data types and shapes\n cifar10 = tf.keras.datasets.cifar10.load_data()\n (X_train, y_train), (X_test, y_test) = cifar10\n X_train = np.asarray(X_train, dtype=np.float32)\n y_train = np.asarray(y_train, dtype=np.int32).flatten()\n X_test = np.asarray(X_test, dtype=np.float32)\n y_test = np.asarray(y_test, dtype=np.int32).flatten()\n\n # Subsample the data\n mask = range(num_training, num_training + num_validation)\n X_val = X_train[mask]\n y_val = y_train[mask]\n mask = range(num_training)\n X_train = X_train[mask]\n y_train = y_train[mask]\n mask = range(num_test)\n X_test = X_test[mask]\n y_test = y_test[mask]\n\n # Normalize the data: subtract the mean pixel and divide by std\n mean_pixel = X_train.mean(axis=(0, 1, 2), keepdims=True)\n std_pixel = X_train.std(axis=(0, 1, 2), keepdims=True)\n X_train = (X_train - mean_pixel) / std_pixel\n X_val = (X_val - mean_pixel) / std_pixel\n X_test = (X_test - mean_pixel) / std_pixel\n\n return X_train, y_train, X_val, y_val, X_test, y_test\n\n# If there are errors with SSL downloading involving self-signed certificates,\n# it may be that your Python version was recently installed on the current machine.\n# See: https://github.com/tensorflow/tensorflow/issues/10779\n# To fix, run the command: /Applications/Python\\ 3.7/Install\\ Certificates.command\n# ...replacing paths as necessary.\n\n# Invoke the above function to get our data.\nNHW = (0, 1, 2)\nX_train, y_train, X_val, y_val, X_test, y_test = load_cifar10()\nprint('Train data shape: ', X_train.shape)\nprint('Train labels shape: ', y_train.shape, y_train.dtype)\nprint('Validation data shape: ', X_val.shape)\nprint('Validation labels shape: ', y_val.shape)\nprint('Test data shape: ', X_test.shape)\nprint('Test labels shape: ', y_test.shape)",
"Train data shape: (49000, 32, 32, 3)\nTrain labels shape: (49000,) int32\nValidation data shape: (1000, 32, 32, 3)\nValidation labels shape: (1000,)\nTest data shape: (10000, 32, 32, 3)\nTest labels shape: (10000,)\n"
],
[
"class Dataset(object):\n def __init__(self, X, y, batch_size, shuffle=False):\n \"\"\"\n Construct a Dataset object to iterate over data X and labels y\n \n Inputs:\n - X: Numpy array of data, of any shape\n - y: Numpy array of labels, of any shape but with y.shape[0] == X.shape[0]\n - batch_size: Integer giving number of elements per minibatch\n - shuffle: (optional) Boolean, whether to shuffle the data on each epoch\n \"\"\"\n assert X.shape[0] == y.shape[0], 'Got different numbers of data and labels'\n self.X, self.y = X, y\n self.batch_size, self.shuffle = batch_size, shuffle\n\n def __iter__(self):\n N, B = self.X.shape[0], self.batch_size\n idxs = np.arange(N)\n if self.shuffle:\n np.random.shuffle(idxs)\n return iter((self.X[i:i+B], self.y[i:i+B]) for i in range(0, N, B))\n\n\ntrain_dset = Dataset(X_train, y_train, batch_size=64, shuffle=True)\nval_dset = Dataset(X_val, y_val, batch_size=64, shuffle=False)\ntest_dset = Dataset(X_test, y_test, batch_size=64)",
"_____no_output_____"
],
[
"# We can iterate through a dataset like this:\nfor t, (x, y) in enumerate(train_dset):\n print(t, x.shape, y.shape)\n if t > 5: break",
"0 (64, 32, 32, 3) (64,)\n1 (64, 32, 32, 3) (64,)\n2 (64, 32, 32, 3) (64,)\n3 (64, 32, 32, 3) (64,)\n4 (64, 32, 32, 3) (64,)\n5 (64, 32, 32, 3) (64,)\n6 (64, 32, 32, 3) (64,)\n"
]
],
[
[
"You can optionally **use GPU by setting the flag to True below**. It's not neccessary to use a GPU for this assignment; if you are working on Google Cloud then we recommend that you do not use a GPU, as it will be significantly more expensive.",
"_____no_output_____"
]
],
[
[
"# Set up some global variables\nUSE_GPU = False\n\nif USE_GPU:\n device = '/device:GPU:0'\nelse:\n device = '/cpu:0'\n\n# Constant to control how often we print when training models\nprint_every = 100\n\nprint('Using device: ', device)",
"Using device: /cpu:0\n"
]
],
[
[
"# Part II: Barebones TensorFlow\nTensorFlow ships with various high-level APIs which make it very convenient to define and train neural networks; we will cover some of these constructs in Part III and Part IV of this notebook. In this section we will start by building a model with basic TensorFlow constructs to help you better understand what's going on under the hood of the higher-level APIs.\n\n**\"Barebones Tensorflow\" is important to understanding the building blocks of TensorFlow, but much of it involves concepts from TensorFlow 1.x.** We will be working with legacy modules such as `tf.Variable`.\n\nTherefore, please read and understand the differences between legacy (1.x) TF and the new (2.0) TF.\n\n### Historical background on TensorFlow 1.x\n\nTensorFlow 1.x is primarily a framework for working with **static computational graphs**. Nodes in the computational graph are Tensors which will hold n-dimensional arrays when the graph is run; edges in the graph represent functions that will operate on Tensors when the graph is run to actually perform useful computation.\n\nBefore Tensorflow 2.0, we had to configure the graph into two phases. There are plenty of tutorials online that explain this two-step process. The process generally looks like the following for TF 1.x:\n1. **Build a computational graph that describes the computation that you want to perform**. This stage doesn't actually perform any computation; it just builds up a symbolic representation of your computation. This stage will typically define one or more `placeholder` objects that represent inputs to the computational graph.\n2. **Run the computational graph many times.** Each time the graph is run (e.g. for one gradient descent step) you will specify which parts of the graph you want to compute, and pass a `feed_dict` dictionary that will give concrete values to any `placeholder`s in the graph.\n\n### The new paradigm in Tensorflow 2.0\nNow, with Tensorflow 2.0, we can simply adopt a functional form that is more Pythonic and similar in spirit to PyTorch and direct Numpy operation. Instead of the 2-step paradigm with computation graphs, making it (among other things) easier to debug TF code. You can read more details at https://www.tensorflow.org/guide/eager.\n\nThe main difference between the TF 1.x and 2.0 approach is that the 2.0 approach doesn't make use of `tf.Session`, `tf.run`, `placeholder`, `feed_dict`. To get more details of what's different between the two version and how to convert between the two, check out the official migration guide: https://www.tensorflow.org/alpha/guide/migration_guide\n\nLater, in the rest of this notebook we'll focus on this new, simpler approach.",
"_____no_output_____"
],
[
"### TensorFlow warmup: Flatten Function\n\nWe can see this in action by defining a simple `flatten` function that will reshape image data for use in a fully-connected network.\n\nIn TensorFlow, data for convolutional feature maps is typically stored in a Tensor of shape N x H x W x C where:\n\n- N is the number of datapoints (minibatch size)\n- H is the height of the feature map\n- W is the width of the feature map\n- C is the number of channels in the feature map\n\nThis is the right way to represent the data when we are doing something like a 2D convolution, that needs spatial understanding of where the intermediate features are relative to each other. When we use fully connected affine layers to process the image, however, we want each datapoint to be represented by a single vector -- it's no longer useful to segregate the different channels, rows, and columns of the data. So, we use a \"flatten\" operation to collapse the `H x W x C` values per representation into a single long vector. \n\nNotice the `tf.reshape` call has the target shape as `(N, -1)`, meaning it will reshape/keep the first dimension to be N, and then infer as necessary what the second dimension is in the output, so we can collapse the remaining dimensions from the input properly.\n\n**NOTE**: TensorFlow and PyTorch differ on the default Tensor layout; TensorFlow uses N x H x W x C but PyTorch uses N x C x H x W.",
"_____no_output_____"
]
],
[
[
"def flatten(x):\n \"\"\" \n Input:\n - TensorFlow Tensor of shape (N, D1, ..., DM)\n \n Output:\n - TensorFlow Tensor of shape (N, D1 * ... * DM)\n \"\"\"\n N = tf.shape(x)[0]\n return tf.reshape(x, (N, -1))",
"_____no_output_____"
],
[
"def test_flatten():\n # Construct concrete values of the input data x using numpy\n x_np = np.arange(24).reshape((2, 3, 4))\n print('x_np:\\n', x_np, '\\n')\n # Compute a concrete output value.\n x_flat_np = flatten(x_np)\n print('x_flat_np:\\n', x_flat_np, '\\n')\n\ntest_flatten()",
"x_np:\n [[[ 0 1 2 3]\n [ 4 5 6 7]\n [ 8 9 10 11]]\n\n [[12 13 14 15]\n [16 17 18 19]\n [20 21 22 23]]] \n\nx_flat_np:\n tf.Tensor(\n[[ 0 1 2 3 4 5 6 7 8 9 10 11]\n [12 13 14 15 16 17 18 19 20 21 22 23]], shape=(2, 12), dtype=int32) \n\n"
]
],
[
[
"### Barebones TensorFlow: Define a Two-Layer Network\nWe will now implement our first neural network with TensorFlow: a fully-connected ReLU network with two hidden layers and no biases on the CIFAR10 dataset. For now we will use only low-level TensorFlow operators to define the network; later we will see how to use the higher-level abstractions provided by `tf.keras` to simplify the process.\n\nWe will define the forward pass of the network in the function `two_layer_fc`; this will accept TensorFlow Tensors for the inputs and weights of the network, and return a TensorFlow Tensor for the scores. \n\nAfter defining the network architecture in the `two_layer_fc` function, we will test the implementation by checking the shape of the output.\n\n**It's important that you read and understand this implementation.**",
"_____no_output_____"
]
],
[
[
"def two_layer_fc(x, params):\n \"\"\"\n A fully-connected neural network; the architecture is:\n fully-connected layer -> ReLU -> fully connected layer.\n Note that we only need to define the forward pass here; TensorFlow will take\n care of computing the gradients for us.\n \n The input to the network will be a minibatch of data, of shape\n (N, d1, ..., dM) where d1 * ... * dM = D. The hidden layer will have H units,\n and the output layer will produce scores for C classes.\n\n Inputs:\n - x: A TensorFlow Tensor of shape (N, d1, ..., dM) giving a minibatch of\n input data.\n - params: A list [w1, w2] of TensorFlow Tensors giving weights for the\n network, where w1 has shape (D, H) and w2 has shape (H, C).\n \n Returns:\n - scores: A TensorFlow Tensor of shape (N, C) giving classification scores\n for the input data x.\n \"\"\"\n w1, w2 = params # Unpack the parameters\n x = flatten(x) # Flatten the input; now x has shape (N, D)\n h = tf.nn.relu(tf.matmul(x, w1)) # Hidden layer: h has shape (N, H)\n scores = tf.matmul(h, w2) # Compute scores of shape (N, C)\n return scores",
"_____no_output_____"
],
[
"def two_layer_fc_test():\n hidden_layer_size = 42\n\n # Scoping our TF operations under a tf.device context manager \n # lets us tell TensorFlow where we want these Tensors to be\n # multiplied and/or operated on, e.g. on a CPU or a GPU.\n with tf.device(device): \n x = tf.zeros((64, 32, 32, 3))\n w1 = tf.zeros((32 * 32 * 3, hidden_layer_size))\n w2 = tf.zeros((hidden_layer_size, 10))\n\n # Call our two_layer_fc function for the forward pass of the network.\n scores = two_layer_fc(x, [w1, w2])\n\n print(scores.shape)\n\ntwo_layer_fc_test()",
"(64, 10)\n"
]
],
[
[
"### Barebones TensorFlow: Three-Layer ConvNet\nHere you will complete the implementation of the function `three_layer_convnet` which will perform the forward pass of a three-layer convolutional network. The network should have the following architecture:\n\n1. A convolutional layer (with bias) with `channel_1` filters, each with shape `KW1 x KH1`, and zero-padding of two\n2. ReLU nonlinearity\n3. A convolutional layer (with bias) with `channel_2` filters, each with shape `KW2 x KH2`, and zero-padding of one\n4. ReLU nonlinearity\n5. Fully-connected layer with bias, producing scores for `C` classes.\n\n**HINT**: For convolutions: https://www.tensorflow.org/versions/r2.0/api_docs/python/tf/nn/conv2d; be careful with padding!\n\n**HINT**: For biases: https://www.tensorflow.org/performance/xla/broadcasting",
"_____no_output_____"
]
],
[
[
"def three_layer_convnet(x, params):\n \"\"\"\n A three-layer convolutional network with the architecture described above.\n \n Inputs:\n - x: A TensorFlow Tensor of shape (N, H, W, 3) giving a minibatch of images\n - params: A list of TensorFlow Tensors giving the weights and biases for the\n network; should contain the following:\n - conv_w1: TensorFlow Tensor of shape (KH1, KW1, 3, channel_1) giving\n weights for the first convolutional layer.\n - conv_b1: TensorFlow Tensor of shape (channel_1,) giving biases for the\n first convolutional layer.\n - conv_w2: TensorFlow Tensor of shape (KH2, KW2, channel_1, channel_2)\n giving weights for the second convolutional layer\n - conv_b2: TensorFlow Tensor of shape (channel_2,) giving biases for the\n second convolutional layer.\n - fc_w: TensorFlow Tensor giving weights for the fully-connected layer.\n Can you figure out what the shape should be?\n - fc_b: TensorFlow Tensor giving biases for the fully-connected layer.\n Can you figure out what the shape should be?\n \"\"\"\n conv_w1, conv_b1, conv_w2, conv_b2, fc_w, fc_b = params\n scores = None\n ############################################################################\n # TODO: Implement the forward pass for the three-layer ConvNet. #\n ############################################################################\n # *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n\n # I know that padding = \"SAME\" ends up the same <i> for this examples </i> but the problem explicitely states: \"and zero-padding of two\"\n padding = [[0,0], [2,2], [2,2], [0,0]]\n x = tf.pad(x, padding)\n x = tf.nn.conv2d(x, conv_w1, strides=(1,1), padding=\"VALID\")\n x = tf.nn.relu(x + conv_b1)\n padding = [[0,0], [1,1], [1,1], [0,0]]\n x = tf.pad(x, padding)\n x = tf.nn.conv2d(x, conv_w2, strides=(1,1), padding=\"VALID\")\n x = tf.nn.relu(x + conv_b2)\n x = flatten(x)\n scores = tf.matmul(x, fc_w) + fc_b # Compute scores of shape (N, C)\n\n # *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n ############################################################################\n # END OF YOUR CODE #\n ############################################################################\n return scores",
"_____no_output_____"
]
],
[
[
"After defing the forward pass of the three-layer ConvNet above, run the following cell to test your implementation. Like the two-layer network, we run the graph on a batch of zeros just to make sure the function doesn't crash, and produces outputs of the correct shape.\n\nWhen you run this function, `scores_np` should have shape `(64, 10)`.",
"_____no_output_____"
]
],
[
[
"def three_layer_convnet_test():\n \n with tf.device(device):\n x = tf.zeros((64, 32, 32, 3))\n conv_w1 = tf.zeros((5, 5, 3, 6))\n conv_b1 = tf.zeros((6,))\n conv_w2 = tf.zeros((3, 3, 6, 9))\n conv_b2 = tf.zeros((9,))\n fc_w = tf.zeros((32 * 32 * 9, 10))\n fc_b = tf.zeros((10,))\n params = [conv_w1, conv_b1, conv_w2, conv_b2, fc_w, fc_b]\n scores = three_layer_convnet(x, params)\n\n # Inputs to convolutional layers are 4-dimensional arrays with shape\n # [batch_size, height, width, channels]\n print('scores_np has shape: ', scores.shape)\n\nthree_layer_convnet_test()",
"scores_np has shape: (64, 10)\n"
]
],
[
[
"### Barebones TensorFlow: Training Step\n\nWe now define the `training_step` function performs a single training step. This will take three basic steps:\n\n1. Compute the loss\n2. Compute the gradient of the loss with respect to all network weights\n3. Make a weight update step using (stochastic) gradient descent.\n\n\nWe need to use a few new TensorFlow functions to do all of this:\n- For computing the cross-entropy loss we'll use `tf.nn.sparse_softmax_cross_entropy_with_logits`: https://www.tensorflow.org/versions/r2.0/api_docs/python/tf/nn/sparse_softmax_cross_entropy_with_logits\n\n- For averaging the loss across a minibatch of data we'll use `tf.reduce_mean`:\nhttps://www.tensorflow.org/versions/r2.0/api_docs/python/tf/reduce_mean\n\n- For computing gradients of the loss with respect to the weights we'll use `tf.GradientTape` (useful for Eager execution): https://www.tensorflow.org/versions/r2.0/api_docs/python/tf/GradientTape\n\n- We'll mutate the weight values stored in a TensorFlow Tensor using `tf.assign_sub` (\"sub\" is for subtraction): https://www.tensorflow.org/api_docs/python/tf/assign_sub \n",
"_____no_output_____"
]
],
[
[
"def training_step(model_fn, x, y, params, learning_rate):\n with tf.GradientTape() as tape:\n scores = model_fn(x, params) # Forward pass of the model\n loss = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y, logits=scores)\n total_loss = tf.reduce_mean(loss)\n grad_params = tape.gradient(total_loss, params)\n\n # Make a vanilla gradient descent step on all of the model parameters\n # Manually update the weights using assign_sub()\n for w, grad_w in zip(params, grad_params):\n w.assign_sub(learning_rate * grad_w)\n \n return total_loss",
"_____no_output_____"
],
[
"def train_part2(model_fn, init_fn, learning_rate):\n \"\"\"\n Train a model on CIFAR-10.\n \n Inputs:\n - model_fn: A Python function that performs the forward pass of the model\n using TensorFlow; it should have the following signature:\n scores = model_fn(x, params) where x is a TensorFlow Tensor giving a\n minibatch of image data, params is a list of TensorFlow Tensors holding\n the model weights, and scores is a TensorFlow Tensor of shape (N, C)\n giving scores for all elements of x.\n - init_fn: A Python function that initializes the parameters of the model.\n It should have the signature params = init_fn() where params is a list\n of TensorFlow Tensors holding the (randomly initialized) weights of the\n model.\n - learning_rate: Python float giving the learning rate to use for SGD.\n \"\"\"\n \n \n params = init_fn() # Initialize the model parameters \n \n for t, (x_np, y_np) in enumerate(train_dset):\n # Run the graph on a batch of training data.\n loss = training_step(model_fn, x_np, y_np, params, learning_rate)\n \n # Periodically print the loss and check accuracy on the val set.\n if t % print_every == 0:\n print('Iteration %d, loss = %.4f' % (t, loss))\n check_accuracy(val_dset, x_np, model_fn, params)",
"_____no_output_____"
],
[
"def check_accuracy(dset, x, model_fn, params):\n \"\"\"\n Check accuracy on a classification model, e.g. for validation.\n \n Inputs:\n - dset: A Dataset object against which to check accuracy\n - x: A TensorFlow placeholder Tensor where input images should be fed\n - model_fn: the Model we will be calling to make predictions on x\n - params: parameters for the model_fn to work with\n \n Returns: Nothing, but prints the accuracy of the model\n \"\"\"\n num_correct, num_samples = 0, 0\n for x_batch, y_batch in dset:\n scores_np = model_fn(x_batch, params).numpy()\n y_pred = scores_np.argmax(axis=1)\n num_samples += x_batch.shape[0]\n num_correct += (y_pred == y_batch).sum()\n acc = float(num_correct) / num_samples\n print('Got %d / %d correct (%.2f%%)' % (num_correct, num_samples, 100 * acc))",
"_____no_output_____"
]
],
[
[
"### Barebones TensorFlow: Initialization\nWe'll use the following utility method to initialize the weight matrices for our models using Kaiming's normalization method.\n\n[1] He et al, *Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification\n*, ICCV 2015, https://arxiv.org/abs/1502.01852",
"_____no_output_____"
]
],
[
[
"def create_matrix_with_kaiming_normal(shape):\n if len(shape) == 2:\n fan_in, fan_out = shape[0], shape[1]\n elif len(shape) == 4:\n fan_in, fan_out = np.prod(shape[:3]), shape[3]\n return tf.keras.backend.random_normal(shape) * np.sqrt(2.0 / fan_in)",
"_____no_output_____"
]
],
[
[
"### Barebones TensorFlow: Train a Two-Layer Network\nWe are finally ready to use all of the pieces defined above to train a two-layer fully-connected network on CIFAR-10.\n\nWe just need to define a function to initialize the weights of the model, and call `train_part2`.\n\nDefining the weights of the network introduces another important piece of TensorFlow API: `tf.Variable`. A TensorFlow Variable is a Tensor whose value is stored in the graph and persists across runs of the computational graph; however unlike constants defined with `tf.zeros` or `tf.random_normal`, the values of a Variable can be mutated as the graph runs; these mutations will persist across graph runs. Learnable parameters of the network are usually stored in Variables.\n\nYou don't need to tune any hyperparameters, but you should achieve validation accuracies above 40% after one epoch of training.",
"_____no_output_____"
]
],
[
[
"def two_layer_fc_init():\n \"\"\"\n Initialize the weights of a two-layer network, for use with the\n two_layer_network function defined above. \n You can use the `create_matrix_with_kaiming_normal` helper!\n \n Inputs: None\n \n Returns: A list of:\n - w1: TensorFlow tf.Variable giving the weights for the first layer\n - w2: TensorFlow tf.Variable giving the weights for the second layer\n \"\"\"\n hidden_layer_size = 4000\n w1 = tf.Variable(create_matrix_with_kaiming_normal((3 * 32 * 32, 4000)))\n w2 = tf.Variable(create_matrix_with_kaiming_normal((4000, 10)))\n return [w1, w2]\n\nlearning_rate = 1e-2\ntrain_part2(two_layer_fc, two_layer_fc_init, learning_rate)",
"Iteration 0, loss = 3.0008\nGot 130 / 1000 correct (13.00%)\nIteration 100, loss = 1.8573\nGot 373 / 1000 correct (37.30%)\nIteration 200, loss = 1.4276\nGot 407 / 1000 correct (40.70%)\nIteration 300, loss = 1.9213\nGot 370 / 1000 correct (37.00%)\nIteration 400, loss = 1.8742\nGot 440 / 1000 correct (44.00%)\nIteration 500, loss = 1.7802\nGot 430 / 1000 correct (43.00%)\nIteration 600, loss = 1.8748\nGot 432 / 1000 correct (43.20%)\nIteration 700, loss = 2.0062\nGot 432 / 1000 correct (43.20%)\n"
]
],
[
[
"### Barebones TensorFlow: Train a three-layer ConvNet\nWe will now use TensorFlow to train a three-layer ConvNet on CIFAR-10.\n\nYou need to implement the `three_layer_convnet_init` function. Recall that the architecture of the network is:\n\n1. Convolutional layer (with bias) with 32 5x5 filters, with zero-padding 2\n2. ReLU\n3. Convolutional layer (with bias) with 16 3x3 filters, with zero-padding 1\n4. ReLU\n5. Fully-connected layer (with bias) to compute scores for 10 classes\n\nYou don't need to do any hyperparameter tuning, but you should see validation accuracies above 43% after one epoch of training.",
"_____no_output_____"
]
],
[
[
"def three_layer_convnet_init():\n \"\"\"\n Initialize the weights of a Three-Layer ConvNet, for use with the\n three_layer_convnet function defined above.\n You can use the `create_matrix_with_kaiming_normal` helper!\n \n Inputs: None\n \n Returns a list containing:\n - conv_w1: TensorFlow tf.Variable giving weights for the first conv layer\n - conv_b1: TensorFlow tf.Variable giving biases for the first conv layer\n - conv_w2: TensorFlow tf.Variable giving weights for the second conv layer\n - conv_b2: TensorFlow tf.Variable giving biases for the second conv layer\n - fc_w: TensorFlow tf.Variable giving weights for the fully-connected layer\n - fc_b: TensorFlow tf.Variable giving biases for the fully-connected layer\n \"\"\"\n params = None\n ############################################################################\n # TODO: Initialize the parameters of the three-layer network. #\n ############################################################################\n # *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n\n conv_w1 = tf.Variable(create_matrix_with_kaiming_normal((5, 5, 3, 32)))\n conv_w2 = tf.Variable(create_matrix_with_kaiming_normal((3, 3, 32, 16)))\n conv_b1 = tf.Variable(tf.zeros(32))\n conv_b2 = tf.Variable(tf.zeros(16))\n fc_w = tf.Variable(create_matrix_with_kaiming_normal((16 * 32 * 32, 10)))\n fc_b = tf.Variable(tf.zeros(10))\n params = (conv_w1, conv_b1, conv_w2, conv_b2, fc_w, fc_b)\n\n # *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n ############################################################################\n # END OF YOUR CODE #\n ############################################################################\n return params\n\nlearning_rate = 3e-3\ntrain_part2(three_layer_convnet, three_layer_convnet_init, learning_rate)",
"Iteration 0, loss = 2.9100\nGot 99 / 1000 correct (9.90%)\nIteration 100, loss = 1.8889\nGot 383 / 1000 correct (38.30%)\nIteration 200, loss = 1.5494\nGot 416 / 1000 correct (41.60%)\nIteration 300, loss = 1.6844\nGot 405 / 1000 correct (40.50%)\nIteration 400, loss = 1.7709\nGot 441 / 1000 correct (44.10%)\nIteration 500, loss = 1.6460\nGot 460 / 1000 correct (46.00%)\nIteration 600, loss = 1.6623\nGot 475 / 1000 correct (47.50%)\nIteration 700, loss = 1.6201\nGot 482 / 1000 correct (48.20%)\n"
]
],
[
[
"# Part III: Keras Model Subclassing API\n\nImplementing a neural network using the low-level TensorFlow API is a good way to understand how TensorFlow works, but it's a little inconvenient - we had to manually keep track of all Tensors holding learnable parameters. This was fine for a small network, but could quickly become unweildy for a large complex model.\n\nFortunately TensorFlow 2.0 provides higher-level APIs such as `tf.keras` which make it easy to build models out of modular, object-oriented layers. Further, TensorFlow 2.0 uses eager execution that evaluates operations immediately, without explicitly constructing any computational graphs. This makes it easy to write and debug models, and reduces the boilerplate code.\n\nIn this part of the notebook we will define neural network models using the `tf.keras.Model` API. To implement your own model, you need to do the following:\n\n1. Define a new class which subclasses `tf.keras.Model`. Give your class an intuitive name that describes it, like `TwoLayerFC` or `ThreeLayerConvNet`.\n2. In the initializer `__init__()` for your new class, define all the layers you need as class attributes. The `tf.keras.layers` package provides many common neural-network layers, like `tf.keras.layers.Dense` for fully-connected layers and `tf.keras.layers.Conv2D` for convolutional layers. Under the hood, these layers will construct `Variable` Tensors for any learnable parameters. **Warning**: Don't forget to call `super(YourModelName, self).__init__()` as the first line in your initializer!\n3. Implement the `call()` method for your class; this implements the forward pass of your model, and defines the *connectivity* of your network. Layers defined in `__init__()` implement `__call__()` so they can be used as function objects that transform input Tensors into output Tensors. Don't define any new layers in `call()`; any layers you want to use in the forward pass should be defined in `__init__()`.\n\nAfter you define your `tf.keras.Model` subclass, you can instantiate it and use it like the model functions from Part II.\n\n### Keras Model Subclassing API: Two-Layer Network\n\nHere is a concrete example of using the `tf.keras.Model` API to define a two-layer network. There are a few new bits of API to be aware of here:\n\nWe use an `Initializer` object to set up the initial values of the learnable parameters of the layers; in particular `tf.initializers.VarianceScaling` gives behavior similar to the Kaiming initialization method we used in Part II. You can read more about it here: https://www.tensorflow.org/versions/r2.0/api_docs/python/tf/initializers/VarianceScaling\n\nWe construct `tf.keras.layers.Dense` objects to represent the two fully-connected layers of the model. In addition to multiplying their input by a weight matrix and adding a bias vector, these layer can also apply a nonlinearity for you. For the first layer we specify a ReLU activation function by passing `activation='relu'` to the constructor; the second layer uses softmax activation function. Finally, we use `tf.keras.layers.Flatten` to flatten the output from the previous fully-connected layer.",
"_____no_output_____"
]
],
[
[
"class TwoLayerFC(tf.keras.Model):\n def __init__(self, hidden_size, num_classes):\n super(TwoLayerFC, self).__init__() \n initializer = tf.initializers.VarianceScaling(scale=2.0)\n self.fc1 = tf.keras.layers.Dense(hidden_size, activation='relu',\n kernel_initializer=initializer)\n self.fc2 = tf.keras.layers.Dense(num_classes, activation='softmax',\n kernel_initializer=initializer)\n self.flatten = tf.keras.layers.Flatten()\n \n def call(self, x, training=False):\n x = self.flatten(x)\n x = self.fc1(x)\n x = self.fc2(x)\n return x\n\n\ndef test_TwoLayerFC():\n \"\"\" A small unit test to exercise the TwoLayerFC model above. \"\"\"\n input_size, hidden_size, num_classes = 50, 42, 10\n x = tf.zeros((64, input_size))\n model = TwoLayerFC(hidden_size, num_classes)\n with tf.device(device):\n scores = model(x)\n print(scores.shape)\n \ntest_TwoLayerFC()",
"(64, 10)\n"
]
],
[
[
"### Keras Model Subclassing API: Three-Layer ConvNet\nNow it's your turn to implement a three-layer ConvNet using the `tf.keras.Model` API. Your model should have the same architecture used in Part II:\n\n1. Convolutional layer with 5 x 5 kernels, with zero-padding of 2\n2. ReLU nonlinearity\n3. Convolutional layer with 3 x 3 kernels, with zero-padding of 1\n4. ReLU nonlinearity\n5. Fully-connected layer to give class scores\n6. Softmax nonlinearity\n\nYou should initialize the weights of your network using the same initialization method as was used in the two-layer network above.\n\n**Hint**: Refer to the documentation for `tf.keras.layers.Conv2D` and `tf.keras.layers.Dense`:\n\nhttps://www.tensorflow.org/versions/r2.0/api_docs/python/tf/keras/layers/Conv2D\n\nhttps://www.tensorflow.org/versions/r2.0/api_docs/python/tf/keras/layers/Dense",
"_____no_output_____"
]
],
[
[
"class ThreeLayerConvNet(tf.keras.Model):\n def __init__(self, channel_1, channel_2, num_classes):\n super(ThreeLayerConvNet, self).__init__()\n ########################################################################\n # TODO: Implement the __init__ method for a three-layer ConvNet. You #\n # should instantiate layer objects to be used in the forward pass. #\n ########################################################################\n # *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n\n self.conv1 = tf.keras.layers.Conv2D(filters=channel_1, kernel_size=5, strides=(1,1), padding='valid', activation='relu')\n self.conv2 = tf.keras.layers.Conv2D(filters=channel_2, kernel_size=3, strides=(1,1), padding='valid', activation='relu')\n self.flatten = tf.keras.layers.Flatten()\n self.fc = tf.keras.layers.Dense(num_classes, activation='softmax')\n# self.softmax = tf.keras.layers.Activation(tf.keras.activations.softmax)\n \n # *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n ########################################################################\n # END OF YOUR CODE #\n ########################################################################\n \n def call(self, x, training=False):\n scores = None\n ########################################################################\n # TODO: Implement the forward pass for a three-layer ConvNet. You #\n # should use the layer objects defined in the __init__ method. #\n ########################################################################\n # *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n \n padding = [[0,0], [2,2], [2,2], [0,0]]\n x = tf.pad(x, padding)\n x = self.conv1(x)\n padding = [[0,0], [1,1], [1,1], [0,0]]\n x = tf.pad(x, padding)\n x = self.conv2(x)\n \n x = self.flatten(x)\n x = self.fc(x)\n \n scores = x\n\n # *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n ########################################################################\n # END OF YOUR CODE #\n ######################################################################## \n return scores",
"_____no_output_____"
]
],
[
[
"Once you complete the implementation of the `ThreeLayerConvNet` above you can run the following to ensure that your implementation does not crash and produces outputs of the expected shape.",
"_____no_output_____"
]
],
[
[
"def test_ThreeLayerConvNet(): \n channel_1, channel_2, num_classes = 12, 8, 10\n model = ThreeLayerConvNet(channel_1, channel_2, num_classes)\n with tf.device(device):\n x = tf.zeros((64, 3, 32, 32))\n scores = model(x)\n print(scores.shape)\n\ntest_ThreeLayerConvNet()",
"(64, 10)\n"
]
],
[
[
"### Keras Model Subclassing API: Eager Training\n\nWhile keras models have a builtin training loop (using the `model.fit`), sometimes you need more customization. Here's an example, of a training loop implemented with eager execution.\n\nIn particular, notice `tf.GradientTape`. Automatic differentiation is used in the backend for implementing backpropagation in frameworks like TensorFlow. During eager execution, `tf.GradientTape` is used to trace operations for computing gradients later. A particular `tf.GradientTape` can only compute one gradient; subsequent calls to tape will throw a runtime error. \n\nTensorFlow 2.0 ships with easy-to-use built-in metrics under `tf.keras.metrics` module. Each metric is an object, and we can use `update_state()` to add observations and `reset_state()` to clear all observations. We can get the current result of a metric by calling `result()` on the metric object.",
"_____no_output_____"
]
],
[
[
"def train_part34(model_init_fn, optimizer_init_fn, num_epochs=1, is_training=False):\n \"\"\"\n Simple training loop for use with models defined using tf.keras. It trains\n a model for one epoch on the CIFAR-10 training set and periodically checks\n accuracy on the CIFAR-10 validation set.\n \n Inputs:\n - model_init_fn: A function that takes no parameters; when called it\n constructs the model we want to train: model = model_init_fn()\n - optimizer_init_fn: A function which takes no parameters; when called it\n constructs the Optimizer object we will use to optimize the model:\n optimizer = optimizer_init_fn()\n - num_epochs: The number of epochs to train for\n \n Returns: Nothing, but prints progress during trainingn\n \"\"\" \n with tf.device(device):\n\n # Compute the loss like we did in Part II\n loss_fn = tf.keras.losses.SparseCategoricalCrossentropy()\n \n model = model_init_fn()\n optimizer = optimizer_init_fn()\n \n train_loss = tf.keras.metrics.Mean(name='train_loss')\n train_accuracy = tf.keras.metrics.SparseCategoricalAccuracy(name='train_accuracy')\n \n val_loss = tf.keras.metrics.Mean(name='val_loss')\n val_accuracy = tf.keras.metrics.SparseCategoricalAccuracy(name='val_accuracy')\n \n t = 0\n for epoch in range(num_epochs):\n \n # Reset the metrics - https://www.tensorflow.org/alpha/guide/migration_guide#new-style_metrics\n train_loss.reset_states()\n train_accuracy.reset_states()\n \n for x_np, y_np in train_dset:\n with tf.GradientTape() as tape:\n \n # Use the model function to build the forward pass.\n scores = model(x_np, training=is_training)\n loss = loss_fn(y_np, scores)\n \n gradients = tape.gradient(loss, model.trainable_variables)\n optimizer.apply_gradients(zip(gradients, model.trainable_variables))\n \n # Update the metrics\n train_loss.update_state(loss)\n train_accuracy.update_state(y_np, scores)\n \n if t % print_every == 0:\n val_loss.reset_states()\n val_accuracy.reset_states()\n for test_x, test_y in val_dset:\n # During validation at end of epoch, training set to False\n prediction = model(test_x, training=False)\n t_loss = loss_fn(test_y, prediction)\n\n val_loss.update_state(t_loss)\n val_accuracy.update_state(test_y, prediction)\n \n template = 'Iteration {}, Epoch {}, Loss: {}, Accuracy: {}, Val Loss: {}, Val Accuracy: {}'\n print (template.format(t, epoch+1,\n train_loss.result(),\n train_accuracy.result()*100,\n val_loss.result(),\n val_accuracy.result()*100))\n t += 1",
"_____no_output_____"
]
],
[
[
"### Keras Model Subclassing API: Train a Two-Layer Network\nWe can now use the tools defined above to train a two-layer network on CIFAR-10. We define the `model_init_fn` and `optimizer_init_fn` that construct the model and optimizer respectively when called. Here we want to train the model using stochastic gradient descent with no momentum, so we construct a `tf.keras.optimizers.SGD` function; you can [read about it here](https://www.tensorflow.org/versions/r2.0/api_docs/python/tf/optimizers/SGD).\n\nYou don't need to tune any hyperparameters here, but you should achieve validation accuracies above 40% after one epoch of training.",
"_____no_output_____"
]
],
[
[
"hidden_size, num_classes = 4000, 10\nlearning_rate = 1e-2\n\ndef model_init_fn():\n return TwoLayerFC(hidden_size, num_classes)\n\ndef optimizer_init_fn():\n return tf.keras.optimizers.SGD(learning_rate=learning_rate)\n\ntrain_part34(model_init_fn, optimizer_init_fn)",
"Iteration 0, Epoch 1, Loss: 3.164823532104492, Accuracy: 7.8125, Val Loss: 2.628608465194702, Val Accuracy: 12.399999618530273\nIteration 100, Epoch 1, Loss: 2.236602544784546, Accuracy: 28.46534538269043, Val Loss: 1.8742749691009521, Val Accuracy: 40.0\nIteration 200, Epoch 1, Loss: 2.0714330673217773, Accuracy: 32.5948371887207, Val Loss: 1.860308051109314, Val Accuracy: 39.5\nIteration 300, Epoch 1, Loss: 1.9990333318710327, Accuracy: 34.44767379760742, Val Loss: 1.8852967023849487, Val Accuracy: 37.599998474121094\nIteration 400, Epoch 1, Loss: 1.9306987524032593, Accuracy: 35.97646713256836, Val Loss: 1.7222708463668823, Val Accuracy: 44.80000305175781\nIteration 500, Epoch 1, Loss: 1.8855254650115967, Accuracy: 37.12574768066406, Val Loss: 1.6626540422439575, Val Accuracy: 43.099998474121094\nIteration 600, Epoch 1, Loss: 1.856279969215393, Accuracy: 38.056365966796875, Val Loss: 1.6962710618972778, Val Accuracy: 41.5\nIteration 700, Epoch 1, Loss: 1.8313978910446167, Accuracy: 38.61225128173828, Val Loss: 1.6491316556930542, Val Accuracy: 42.29999923706055\n"
]
],
[
[
"### Keras Model Subclassing API: Train a Three-Layer ConvNet\nHere you should use the tools we've defined above to train a three-layer ConvNet on CIFAR-10. Your ConvNet should use 32 filters in the first convolutional layer and 16 filters in the second layer.\n\nTo train the model you should use gradient descent with Nesterov momentum 0.9. \n\n**HINT**: https://www.tensorflow.org/versions/r2.0/api_docs/python/tf/optimizers/SGD\n\nYou don't need to perform any hyperparameter tuning, but you should achieve validation accuracies above 50% after training for one epoch.",
"_____no_output_____"
]
],
[
[
"learning_rate = 3e-3\nchannel_1, channel_2, num_classes = 32, 16, 10\n\ndef model_init_fn():\n model = None\n ############################################################################\n # TODO: Complete the implementation of model_fn. #\n ############################################################################\n # *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n\n model = ThreeLayerConvNet(channel_1, channel_2, num_classes)\n\n # *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n ############################################################################\n # END OF YOUR CODE #\n ############################################################################\n return model\n\ndef optimizer_init_fn():\n optimizer = None\n ############################################################################\n # TODO: Complete the implementation of model_fn. #\n ############################################################################\n # *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n\n optimizer = tf.keras.optimizers.SGD(learning_rate=learning_rate, momentum=0.9, nesterov=True)\n\n # *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n ############################################################################\n # END OF YOUR CODE #\n ############################################################################\n return optimizer\n\ntrain_part34(model_init_fn, optimizer_init_fn)",
"Iteration 0, Epoch 1, Loss: 2.300231456756592, Accuracy: 4.6875, Val Loss: 2.2966694831848145, Val Accuracy: 13.600000381469727\nIteration 100, Epoch 1, Loss: 1.800517201423645, Accuracy: 35.45792007446289, Val Loss: 1.5509482622146606, Val Accuracy: 45.20000076293945\nIteration 200, Epoch 1, Loss: 1.661733865737915, Accuracy: 40.46952819824219, Val Loss: 1.4937281608581543, Val Accuracy: 47.60000228881836\nIteration 300, Epoch 1, Loss: 1.5924314260482788, Accuracy: 43.236087799072266, Val Loss: 1.4149495363235474, Val Accuracy: 50.400001525878906\nIteration 400, Epoch 1, Loss: 1.5270615816116333, Accuracy: 45.6592903137207, Val Loss: 1.3045035600662231, Val Accuracy: 52.79999923706055\nIteration 500, Epoch 1, Loss: 1.481753945350647, Accuracy: 47.31162643432617, Val Loss: 1.299678087234497, Val Accuracy: 52.89999771118164\nIteration 600, Epoch 1, Loss: 1.4521290063858032, Accuracy: 48.46869659423828, Val Loss: 1.2669525146484375, Val Accuracy: 55.599998474121094\nIteration 700, Epoch 1, Loss: 1.4251292943954468, Accuracy: 49.594329833984375, Val Loss: 1.2142601013183594, Val Accuracy: 58.0\n"
]
],
[
[
"# Part IV: Keras Sequential API\nIn Part III we introduced the `tf.keras.Model` API, which allows you to define models with any number of learnable layers and with arbitrary connectivity between layers.\n\nHowever for many models you don't need such flexibility - a lot of models can be expressed as a sequential stack of layers, with the output of each layer fed to the next layer as input. If your model fits this pattern, then there is an even easier way to define your model: using `tf.keras.Sequential`. You don't need to write any custom classes; you simply call the `tf.keras.Sequential` constructor with a list containing a sequence of layer objects.\n\nOne complication with `tf.keras.Sequential` is that you must define the shape of the input to the model by passing a value to the `input_shape` of the first layer in your model.\n\n### Keras Sequential API: Two-Layer Network\nIn this subsection, we will rewrite the two-layer fully-connected network using `tf.keras.Sequential`, and train it using the training loop defined above.\n\nYou don't need to perform any hyperparameter tuning here, but you should see validation accuracies above 40% after training for one epoch.",
"_____no_output_____"
]
],
[
[
"learning_rate = 1e-2\n\ndef model_init_fn():\n input_shape = (32, 32, 3)\n hidden_layer_size, num_classes = 4000, 10\n initializer = tf.initializers.VarianceScaling(scale=2.0)\n layers = [\n tf.keras.layers.Flatten(input_shape=input_shape),\n tf.keras.layers.Dense(hidden_layer_size, activation='relu',\n kernel_initializer=initializer),\n tf.keras.layers.Dense(num_classes, activation='softmax', \n kernel_initializer=initializer),\n ]\n model = tf.keras.Sequential(layers)\n return model\n\ndef optimizer_init_fn():\n return tf.keras.optimizers.SGD(learning_rate=learning_rate) \n\ntrain_part34(model_init_fn, optimizer_init_fn)",
"_____no_output_____"
]
],
[
[
"### Abstracting Away the Training Loop\nIn the previous examples, we used a customised training loop to train models (e.g. `train_part34`). Writing your own training loop is only required if you need more flexibility and control during training your model. Alternately, you can also use built-in APIs like `tf.keras.Model.fit()` and `tf.keras.Model.evaluate` to train and evaluate a model. Also remember to configure your model for training by calling `tf.keras.Model.compile.\n\nYou don't need to perform any hyperparameter tuning here, but you should see validation and test accuracies above 42% after training for one epoch.",
"_____no_output_____"
]
],
[
[
"model = model_init_fn()\nmodel.compile(optimizer=tf.keras.optimizers.SGD(learning_rate=learning_rate),\n loss='sparse_categorical_crossentropy',\n metrics=[tf.keras.metrics.sparse_categorical_accuracy])\nmodel.fit(X_train, y_train, batch_size=64, epochs=1, validation_data=(X_val, y_val))\nmodel.evaluate(X_test, y_test)",
"_____no_output_____"
]
],
[
[
"### Keras Sequential API: Three-Layer ConvNet\nHere you should use `tf.keras.Sequential` to reimplement the same three-layer ConvNet architecture used in Part II and Part III. As a reminder, your model should have the following architecture:\n\n1. Convolutional layer with 32 5x5 kernels, using zero padding of 2\n2. ReLU nonlinearity\n3. Convolutional layer with 16 3x3 kernels, using zero padding of 1\n4. ReLU nonlinearity\n5. Fully-connected layer giving class scores\n6. Softmax nonlinearity\n\nYou should initialize the weights of the model using a `tf.initializers.VarianceScaling` as above.\n\nYou should train the model using Nesterov momentum 0.9.\n\nYou don't need to perform any hyperparameter search, but you should achieve accuracy above 45% after training for one epoch.",
"_____no_output_____"
]
],
[
[
"def model_init_fn():\n model = None\n ############################################################################\n # TODO: Construct a three-layer ConvNet using tf.keras.Sequential. #\n ############################################################################\n # *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n\n model = tf.keras.Sequential()\n model.add(tf.keras.layers.Conv2D(filters=32, kernel_size=5, padding=\"same\", activation='relu'))\n model.add(tf.keras.layers.Conv2D(filters=16, kernel_size=3, padding=\"same\", activation='relu'))\n model.add(tf.keras.layers.Flatten())\n model.add(tf.keras.layers.Dense(units=10, activation='softmax'))\n\n # *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n ############################################################################\n # END OF YOUR CODE #\n ############################################################################\n return model\n\nlearning_rate = 5e-4\ndef optimizer_init_fn():\n optimizer = None\n ############################################################################\n # TODO: Complete the implementation of model_fn. #\n ############################################################################\n # *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n\n optimizer = tf.keras.optimizers.SGD(learning_rate=learning_rate, momentum=0.9, nesterov=True)\n\n # *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n ############################################################################\n # END OF YOUR CODE #\n ############################################################################\n return optimizer\n\ntrain_part34(model_init_fn, optimizer_init_fn)",
"Iteration 0, Epoch 1, Loss: 2.3726930618286133, Accuracy: 14.0625, Val Loss: 2.371110439300537, Val Accuracy: 10.59999942779541\nIteration 100, Epoch 1, Loss: 2.1053175926208496, Accuracy: 24.42759895324707, Val Loss: 1.9350684881210327, Val Accuracy: 33.599998474121094\nIteration 200, Epoch 1, Loss: 1.98719322681427, Accuracy: 29.951805114746094, Val Loss: 1.7907942533493042, Val Accuracy: 40.0\nIteration 300, Epoch 1, Loss: 1.913374900817871, Accuracy: 32.68791580200195, Val Loss: 1.7151117324829102, Val Accuracy: 42.89999771118164\nIteration 400, Epoch 1, Loss: 1.8458291292190552, Accuracy: 35.317955017089844, Val Loss: 1.6581902503967285, Val Accuracy: 43.5\nIteration 500, Epoch 1, Loss: 1.7973949909210205, Accuracy: 36.991641998291016, Val Loss: 1.5900821685791016, Val Accuracy: 44.10000228881836\nIteration 600, Epoch 1, Loss: 1.76349675655365, Accuracy: 38.209754943847656, Val Loss: 1.5667246580123901, Val Accuracy: 45.69999694824219\nIteration 700, Epoch 1, Loss: 1.7330719232559204, Accuracy: 39.35003662109375, Val Loss: 1.521426796913147, Val Accuracy: 47.60000228881836\n"
]
],
[
[
"We will also train this model with the built-in training loop APIs provided by TensorFlow.",
"_____no_output_____"
]
],
[
[
"model = model_init_fn()\nmodel.compile(optimizer='sgd',\n loss='sparse_categorical_crossentropy',\n metrics=[tf.keras.metrics.sparse_categorical_accuracy])\nmodel.fit(X_train, y_train, batch_size=64, epochs=1, validation_data=(X_val, y_val))\nmodel.evaluate(X_test, y_test)",
"Train on 49000 samples, validate on 1000 samples\n49000/49000 [==============================] - 5s 94us/sample - loss: 1.5918 - sparse_categorical_accuracy: 0.4327 - val_loss: 1.3710 - val_sparse_categorical_accuracy: 0.5150\n10000/10000 [==============================] - 1s 73us/sample - loss: 1.3944 - sparse_categorical_accuracy: 0.4968\n"
]
],
[
[
"## Part IV: Functional API\n### Demonstration with a Two-Layer Network \n\nIn the previous section, we saw how we can use `tf.keras.Sequential` to stack layers to quickly build simple models. But this comes at the cost of losing flexibility.\n\nOften we will have to write complex models that have non-sequential data flows: a layer can have **multiple inputs and/or outputs**, such as stacking the output of 2 previous layers together to feed as input to a third! (Some examples are residual connections and dense blocks.)\n\nIn such cases, we can use Keras functional API to write models with complex topologies such as:\n\n 1. Multi-input models\n 2. Multi-output models\n 3. Models with shared layers (the same layer called several times)\n 4. Models with non-sequential data flows (e.g. residual connections)\n\nWriting a model with Functional API requires us to create a `tf.keras.Model` instance and explicitly write input tensors and output tensors for this model. ",
"_____no_output_____"
]
],
[
[
"def two_layer_fc_functional(input_shape, hidden_size, num_classes): \n initializer = tf.initializers.VarianceScaling(scale=2.0)\n inputs = tf.keras.Input(shape=input_shape)\n flattened_inputs = tf.keras.layers.Flatten()(inputs)\n fc1_output = tf.keras.layers.Dense(hidden_size, activation='relu',\n kernel_initializer=initializer)(flattened_inputs)\n scores = tf.keras.layers.Dense(num_classes, activation='softmax',\n kernel_initializer=initializer)(fc1_output)\n\n # Instantiate the model given inputs and outputs.\n model = tf.keras.Model(inputs=inputs, outputs=scores)\n return model\n\ndef test_two_layer_fc_functional():\n \"\"\" A small unit test to exercise the TwoLayerFC model above. \"\"\"\n input_size, hidden_size, num_classes = 50, 42, 10\n input_shape = (50,)\n \n x = tf.zeros((64, input_size))\n model = two_layer_fc_functional(input_shape, hidden_size, num_classes)\n \n with tf.device(device):\n scores = model(x)\n print(scores.shape)\n \ntest_two_layer_fc_functional()",
"(64, 10)\n"
]
],
[
[
"### Keras Functional API: Train a Two-Layer Network\nYou can now train this two-layer network constructed using the functional API.\n\nYou don't need to perform any hyperparameter tuning here, but you should see validation accuracies above 40% after training for one epoch.",
"_____no_output_____"
]
],
[
[
"input_shape = (32, 32, 3)\nhidden_size, num_classes = 4000, 10\nlearning_rate = 1e-2\n\ndef model_init_fn():\n return two_layer_fc_functional(input_shape, hidden_size, num_classes)\n\ndef optimizer_init_fn():\n return tf.keras.optimizers.SGD(learning_rate=learning_rate)\n\ntrain_part34(model_init_fn, optimizer_init_fn)",
"Iteration 0, Epoch 1, Loss: 3.025775909423828, Accuracy: 3.125, Val Loss: 3.0881335735321045, Val Accuracy: 11.90000057220459\nIteration 100, Epoch 1, Loss: 2.242359161376953, Accuracy: 28.418933868408203, Val Loss: 1.8806251287460327, Val Accuracy: 37.400001525878906\nIteration 200, Epoch 1, Loss: 2.085632085800171, Accuracy: 32.136192321777344, Val Loss: 1.8255318403244019, Val Accuracy: 40.29999923706055\nIteration 300, Epoch 1, Loss: 2.0071797370910645, Accuracy: 33.964908599853516, Val Loss: 1.8452262878417969, Val Accuracy: 38.70000076293945\nIteration 400, Epoch 1, Loss: 1.937016248703003, Accuracy: 35.750465393066406, Val Loss: 1.719841480255127, Val Accuracy: 41.400001525878906\nIteration 500, Epoch 1, Loss: 1.8926761150360107, Accuracy: 36.73902130126953, Val Loss: 1.6710996627807617, Val Accuracy: 43.39999771118164\nIteration 600, Epoch 1, Loss: 1.8619439601898193, Accuracy: 37.67418670654297, Val Loss: 1.6859104633331299, Val Accuracy: 41.5\nIteration 700, Epoch 1, Loss: 1.8349393606185913, Accuracy: 38.41610336303711, Val Loss: 1.6123147010803223, Val Accuracy: 45.10000228881836\n"
]
],
[
[
"# Part V: CIFAR-10 open-ended challenge\n\nIn this section you can experiment with whatever ConvNet architecture you'd like on CIFAR-10.\n\nYou should experiment with architectures, hyperparameters, loss functions, regularization, or anything else you can think of to train a model that achieves **at least 70%** accuracy on the **validation** set within 10 epochs. You can use the built-in train function, the `train_part34` function from above, or implement your own training loop.\n\nDescribe what you did at the end of the notebook.\n\n### Some things you can try:\n- **Filter size**: Above we used 5x5 and 3x3; is this optimal?\n- **Number of filters**: Above we used 16 and 32 filters. Would more or fewer do better?\n- **Pooling**: We didn't use any pooling above. Would this improve the model?\n- **Normalization**: Would your model be improved with batch normalization, layer normalization, group normalization, or some other normalization strategy?\n- **Network architecture**: The ConvNet above has only three layers of trainable parameters. Would a deeper model do better?\n- **Global average pooling**: Instead of flattening after the final convolutional layer, would global average pooling do better? This strategy is used for example in Google's Inception network and in Residual Networks.\n- **Regularization**: Would some kind of regularization improve performance? Maybe weight decay or dropout?\n\n### NOTE: Batch Normalization / Dropout\nIf you are using Batch Normalization and Dropout, remember to pass `is_training=True` if you use the `train_part34()` function. BatchNorm and Dropout layers have different behaviors at training and inference time. `training` is a specific keyword argument reserved for this purpose in any `tf.keras.Model`'s `call()` function. Read more about this here : https://www.tensorflow.org/versions/r2.0/api_docs/python/tf/keras/layers/BatchNormalization#methods\nhttps://www.tensorflow.org/versions/r2.0/api_docs/python/tf/keras/layers/Dropout#methods\n\n### Tips for training\nFor each network architecture that you try, you should tune the learning rate and other hyperparameters. When doing this there are a couple important things to keep in mind: \n\n- If the parameters are working well, you should see improvement within a few hundred iterations\n- Remember the coarse-to-fine approach for hyperparameter tuning: start by testing a large range of hyperparameters for just a few training iterations to find the combinations of parameters that are working at all.\n- Once you have found some sets of parameters that seem to work, search more finely around these parameters. You may need to train for more epochs.\n- You should use the validation set for hyperparameter search, and save your test set for evaluating your architecture on the best parameters as selected by the validation set.\n\n### Going above and beyond\nIf you are feeling adventurous there are many other features you can implement to try and improve your performance. You are **not required** to implement any of these, but don't miss the fun if you have time!\n\n- Alternative optimizers: you can try Adam, Adagrad, RMSprop, etc.\n- Alternative activation functions such as leaky ReLU, parametric ReLU, ELU, or MaxOut.\n- Model ensembles\n- Data augmentation\n- New Architectures\n - [ResNets](https://arxiv.org/abs/1512.03385) where the input from the previous layer is added to the output.\n - [DenseNets](https://arxiv.org/abs/1608.06993) where inputs into previous layers are concatenated together.\n - [This blog has an in-depth overview](https://chatbotslife.com/resnets-highwaynets-and-densenets-oh-my-9bb15918ee32)\n \n### Have fun and happy training! ",
"_____no_output_____"
]
],
[
[
"class CustomConvNet(tf.keras.Model):\n def __init__(self):\n super(CustomConvNet, self).__init__()\n ############################################################################\n # TODO: Construct a model that performs well on CIFAR-10 #\n ############################################################################\n # *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n\n # https://www.tensorflow.org/tutorials/images/cnn\n \n self.conv1 = tf.keras.layers.Conv2D(filters=32, kernel_size=(3, 3), strides=(1,1), padding='valid', activation='relu')\n self.dropout1 = tf.keras.layers.Dropout(0.3)\n self.conv2 = tf.keras.layers.Conv2D(filters=64, kernel_size=(3, 3), strides=(1,1), padding='valid', activation='relu')\n self.dropout2 = tf.keras.layers.Dropout(0.3)\n self.conv3 = tf.keras.layers.Conv2D(filters=64, kernel_size=(3, 3), strides=(1,1), padding='valid', activation='relu')\n self.dropout3 = tf.keras.layers.Dropout(0.3)\n self.flatten = tf.keras.layers.Flatten()\n self.bn1 = tf.keras.layers.BatchNormalization()\n self.fc1 = tf.keras.layers.Dense(64, activation='relu')\n self.bn2 = tf.keras.layers.BatchNormalization()\n self.fc2 = tf.keras.layers.Dense(num_classes, activation='softmax')\n self.maxpool = tf.keras.layers.MaxPool2D()\n\n # *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n ############################################################################\n # END OF YOUR CODE #\n ############################################################################\n \n def call(self, input_tensor, training=False):\n ############################################################################\n # TODO: Construct a model that performs well on CIFAR-10 #\n ############################################################################\n # *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n\n x = self.conv1(input_tensor)\n# if training:\n# x = self.dropout1(x, training=training)\n x = self.maxpool(x)\n \n x = self.conv2(x)\n# if training:\n# x = self.dropout2(x, training=training)\n x = self.maxpool(x)\n \n x = self.conv3(x)\n# if training:\n# x = self.dropout3(x, training=training)\n# x = self.maxpool(x)\n \n x = self.flatten(x)\n# if training:\n# x = self.bn1(x, training=training)\n x = self.fc1(x)\n# if training:\n# x = self.bn2(x, training=training)\n x = self.fc2(x)\n \n # *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****\n ############################################################################\n # END OF YOUR CODE #\n ############################################################################\n \n return x\n\ndevice = '/device:GPU:0' # Change this to a CPU/GPU as you wish!\n# device = '/cpu:0' # Change this to a CPU/GPU as you wish!\nprint_every = 700\nnum_epochs = 10\n\nmodel = CustomConvNet()\n\ndef model_init_fn():\n return CustomConvNet()\n\ndef optimizer_init_fn():\n learning_rate = 1e-3\n return tf.keras.optimizers.Adam(learning_rate) \n\ntrain_part34(model_init_fn, optimizer_init_fn, num_epochs=num_epochs, is_training=True)",
"Iteration 0, Epoch 1, Loss: 2.3782572746276855, Accuracy: 10.9375, Val Loss: 2.315579414367676, Val Accuracy: 8.699999809265137\nIteration 700, Epoch 1, Loss: 1.4779174327850342, Accuracy: 46.58746337890625, Val Loss: 1.2174204587936401, Val Accuracy: 57.099998474121094\nIteration 1400, Epoch 2, Loss: 1.08061945438385, Accuracy: 62.19488525390625, Val Loss: 1.0568606853485107, Val Accuracy: 63.900001525878906\nIteration 2100, Epoch 3, Loss: 0.9213600158691406, Accuracy: 67.82732391357422, Val Loss: 0.9454669952392578, Val Accuracy: 67.9000015258789\nIteration 2800, Epoch 4, Loss: 0.8220710158348083, Accuracy: 71.3251724243164, Val Loss: 0.9219656586647034, Val Accuracy: 68.5999984741211\nIteration 3500, Epoch 5, Loss: 0.7531884908676147, Accuracy: 73.78789520263672, Val Loss: 0.8871719837188721, Val Accuracy: 70.0999984741211\nIteration 4200, Epoch 6, Loss: 0.6915034055709839, Accuracy: 75.87600708007812, Val Loss: 0.9000189304351807, Val Accuracy: 69.4000015258789\nIteration 4900, Epoch 7, Loss: 0.638451337814331, Accuracy: 77.80738067626953, Val Loss: 0.8617967963218689, Val Accuracy: 71.4000015258789\nIteration 5600, Epoch 8, Loss: 0.6056898236274719, Accuracy: 78.76569366455078, Val Loss: 0.9214778542518616, Val Accuracy: 70.4000015258789\nIteration 6300, Epoch 9, Loss: 0.5574114322662354, Accuracy: 80.78034210205078, Val Loss: 0.8956850171089172, Val Accuracy: 72.39999389648438\nIteration 7000, Epoch 10, Loss: 0.5189455151557922, Accuracy: 82.33061218261719, Val Loss: 0.9447248578071594, Val Accuracy: 70.5\n"
]
],
[
[
"## Describe what you did \n\nIn the cell below you should write an explanation of what you did, any additional features that you implemented, and/or any graphs that you made in the process of training and evaluating your network.",
"_____no_output_____"
],
[
"TODO: Tell us what you did\n\n\nTried Dropout, BatchNorm but just regular conv + pool into fully connected layers worked best and produced a model with > 70% val accuracy after 5 epochs.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d0cfb669bd14622c74e8be6d6e87e5feca7f5434 | 481,649 | ipynb | Jupyter Notebook | 09-statistics/modelling_number_of_days_at_each_weight.ipynb | aarora79/30DayChartChallenge | 322e85031f8984a7b7ed38e0202fc7604caac2fa | [
"MIT"
] | 4 | 2021-04-20T04:52:17.000Z | 2022-03-18T01:22:45.000Z | 09-statistics/modelling_number_of_days_at_each_weight.ipynb | aarora79/30DayChartChallenge | 322e85031f8984a7b7ed38e0202fc7604caac2fa | [
"MIT"
] | null | null | null | 09-statistics/modelling_number_of_days_at_each_weight.ipynb | aarora79/30DayChartChallenge | 322e85031f8984a7b7ed38e0202fc7604caac2fa | [
"MIT"
] | 3 | 2021-04-22T13:32:23.000Z | 2021-05-16T19:43:33.000Z | 1,067.957871 | 314,698 | 0.941866 | [
[
[
"library(glue)\nlibrary(dplyr)\nlibrary(tidyr)\nlibrary(ggplot2)\nlibrary(lubridate)\nlibrary(tidyverse)\nlibrary(fitdistrplus)\n\n# read data\nDATA_URL <- \"https://raw.githubusercontent.com/aarora79/biomettracker/master/data/Amit.csv\"\nbodyweight <- read_csv(DATA_URL) %>%\n janitor::clean_names()\nhead(bodyweight)",
"\nAttaching package: ‘dplyr’\n\n\nThe following object is masked from ‘package:glue’:\n\n collapse\n\n\nThe following objects are masked from ‘package:stats’:\n\n filter, lag\n\n\nThe following objects are masked from ‘package:base’:\n\n intersect, setdiff, setequal, union\n\n\n\nAttaching package: ‘lubridate’\n\n\nThe following objects are masked from ‘package:base’:\n\n date, intersect, setdiff, union\n\n\nRegistered S3 method overwritten by 'rvest':\n method from\n read_xml.response xml2\n\n── \u001b[1mAttaching packages\u001b[22m ─────────────────────────────────────── tidyverse 1.2.1 ──\n\n\u001b[32m✔\u001b[39m \u001b[34mtibble \u001b[39m 3.0.6 \u001b[32m✔\u001b[39m \u001b[34mpurrr \u001b[39m 0.3.4\n\u001b[32m✔\u001b[39m \u001b[34mreadr \u001b[39m 1.3.1 \u001b[32m✔\u001b[39m \u001b[34mstringr\u001b[39m 1.4.0\n\u001b[32m✔\u001b[39m \u001b[34mtibble \u001b[39m 3.0.6 \u001b[32m✔\u001b[39m \u001b[34mforcats\u001b[39m 0.4.0\n\n── \u001b[1mConflicts\u001b[22m ────────────────────────────────────────── tidyverse_conflicts() ──\n\u001b[31m✖\u001b[39m \u001b[34mlubridate\u001b[39m::\u001b[32mas.difftime()\u001b[39m masks \u001b[34mbase\u001b[39m::as.difftime()\n\u001b[31m✖\u001b[39m \u001b[34mdplyr\u001b[39m::\u001b[32mcollapse()\u001b[39m masks \u001b[34mglue\u001b[39m::collapse()\n\u001b[31m✖\u001b[39m \u001b[34mlubridate\u001b[39m::\u001b[32mdate()\u001b[39m masks \u001b[34mbase\u001b[39m::date()\n\u001b[31m✖\u001b[39m \u001b[34mdplyr\u001b[39m::\u001b[32mfilter()\u001b[39m masks \u001b[34mstats\u001b[39m::filter()\n\u001b[31m✖\u001b[39m \u001b[34mlubridate\u001b[39m::\u001b[32mintersect()\u001b[39m masks \u001b[34mbase\u001b[39m::intersect()\n\u001b[31m✖\u001b[39m \u001b[34mdplyr\u001b[39m::\u001b[32mlag()\u001b[39m masks \u001b[34mstats\u001b[39m::lag()\n\u001b[31m✖\u001b[39m \u001b[34mlubridate\u001b[39m::\u001b[32msetdiff()\u001b[39m masks \u001b[34mbase\u001b[39m::setdiff()\n\u001b[31m✖\u001b[39m \u001b[34mlubridate\u001b[39m::\u001b[32munion()\u001b[39m masks \u001b[34mbase\u001b[39m::union()\n\nLoading required package: MASS\n\n\nAttaching package: ‘MASS’\n\n\nThe following object is masked from ‘package:dplyr’:\n\n select\n\n\nLoading required package: survival\n\nParsed with column specification:\ncols(\n Date = \u001b[34mcol_date(format = \"\")\u001b[39m,\n Weight = \u001b[32mcol_double()\u001b[39m,\n BMI = \u001b[32mcol_double()\u001b[39m,\n `Body Fat` = \u001b[32mcol_double()\u001b[39m,\n `Lean Mass` = \u001b[32mcol_double()\u001b[39m,\n `Muscle Percentage` = \u001b[32mcol_double()\u001b[39m,\n `Water Percentage` = \u001b[32mcol_double()\u001b[39m\n)\n\n"
],
[
"# filter out weights between min weight and min weight + 5, basically we are trying\n# to only include weights which we know have been passed reliably i.e. no more fluctuations\ndays_at_weight <- bodyweight %>%\n mutate(weight = as.integer(weight)) %>%\n filter(weight >= min(bodyweight$weight)+5 & weight <= max(bodyweight$weight)-5) %>%\n count(weight) %>%\n arrange(weight)\n\nhead(days_at_weight, 10)\n\noptions(repr.plot.width=20, repr.plot.height=8)\ndays_at_weight %>%\n ggplot(aes(x=weight, y=n)) +\n geom_bar(stat='identity')",
"_____no_output_____"
],
[
"# plot the number of days as a distribution\ndays_at_weight %>%\n ggplot(aes(x=n)) +\n geom_density()",
"_____no_output_____"
],
[
"days_at_weight %>%\n ggplot(aes(x = n)) +\n stat_ecdf(geom = \"step\", size = 1.5) ",
"_____no_output_____"
],
[
"# plot the empiricial density and CDF using built-in plotdist function from fitdistrplus package\noptions(repr.plot.width=20, repr.plot.height=8)\n\npar(mfrow = c(1, 2), cex.lab=1.5, cex.axis=1.5, cex.main=1.5, cex.sub=1.5)\n\nplotdist(days_at_weight$n, histo = TRUE, demp = TRUE)",
"_____no_output_____"
],
[
"options(repr.plot.width=20, repr.plot.height=15)\n# the distribution is somewhat long tailed so lets try fitting weibull, gamma and log normal distrubtions\nfw <- fitdist(days_at_weight$n, \"weibull\")\nfg <- fitdist(days_at_weight$n, \"gamma\")\nfln <- fitdist(days_at_weight$n, \"lnorm\")\npar(mfrow = c(2, 2), cex.lab=1.5, cex.axis=1.5, cex.main=1.5, cex.sub=1.5)\nplot.legend <- c(\"Weibull\", \"lognormal\", \"gamma\")\n\ndenscomp(list(fw, fln, fg), legendtext = plot.legend)\nqqcomp(list(fw, fln, fg), legendtext = plot.legend)\ncdfcomp(list(fw, fln, fg), legendtext = plot.legend)\nppcomp(list(fw, fln, fg), legendtext = plot.legend)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0cfb8d0f1ef0d0fb9ee3102d9aaa9e04f464466 | 573,678 | ipynb | Jupyter Notebook | Trump Tweets at Internet Archive.ipynb | edsu/notebooks | fc69c12818ada0274750cf91edafdf6afb0aeb15 | [
"CC-BY-4.0"
] | 6 | 2019-01-03T02:32:36.000Z | 2021-12-10T01:53:29.000Z | Trump Tweets at Internet Archive.ipynb | edsu/notebooks | fc69c12818ada0274750cf91edafdf6afb0aeb15 | [
"CC-BY-4.0"
] | null | null | null | Trump Tweets at Internet Archive.ipynb | edsu/notebooks | fc69c12818ada0274750cf91edafdf6afb0aeb15 | [
"CC-BY-4.0"
] | 1 | 2022-02-24T08:04:57.000Z | 2022-02-24T08:04:57.000Z | 70.841936 | 39,480 | 0.742329 | [
[
[
"# Trump Tweets at the Internet Archive\n\nSo Trump's Twitter account is gone. At least at twitter.com. But (fortunately for history) there has probably never been a more heavily archived social media account at the Internet Archive and elsewhere on the web. There are also a plethora of online \"archives\" like [The Trump Archive](https://www.thetrumparchive.com/) which have collected these tweets as data. But seeing the tweets as they appeared in the browser is important. Of course you can go view the account in the Wayback Machine and [browse around](https://web.archive.org/web/20210107055108/https://twitter.com/realDonaldTrump) but what if we wanted a list of all the Trump tweets? How many times were these tweets actually archived?\n\n## CDX API\n\nThe Wayback Machine (and many other web archives) have a service called the [CDX API](https://github.com/internetarchive/wayback/tree/master/wayback-cdx-server). Think of it as the index to the archive. You can give it a URL and it'll tell you what snapshots it has for it. You can also ask the CDX API to search for a *url prefix* and it will tell you what snapshots it has that start with that string. Lets use the handy [wayback](https://wayback.readthedocs.io/en/stable/usage.html) Python module to search for tweet URLs in the Wayback machine. So URLs that look like:\n\n https://twitter.com/realDonaldTrump/status/{id}",
"_____no_output_____"
]
],
[
[
"! pip install wayback",
"Requirement already satisfied: wayback in /home/ed/.local/share/virtualenvs/notebooks-7Xlq9Gbm/lib/python3.8/site-packages (0.2.5)\r\nRequirement already satisfied: requests in /home/ed/.local/share/virtualenvs/notebooks-7Xlq9Gbm/lib/python3.8/site-packages (from wayback) (2.24.0)\r\nRequirement already satisfied: urllib3 in /home/ed/.local/share/virtualenvs/notebooks-7Xlq9Gbm/lib/python3.8/site-packages (from wayback) (1.25.11)\r\nRequirement already satisfied: idna<3,>=2.5 in /home/ed/.local/share/virtualenvs/notebooks-7Xlq9Gbm/lib/python3.8/site-packages (from requests->wayback) (2.10)\r\nRequirement already satisfied: chardet<4,>=3.0.2 in /home/ed/.local/share/virtualenvs/notebooks-7Xlq9Gbm/lib/python3.8/site-packages (from requests->wayback) (3.0.4)\r\nRequirement already satisfied: certifi>=2017.4.17 in /home/ed/.local/share/virtualenvs/notebooks-7Xlq9Gbm/lib/python3.8/site-packages (from requests->wayback) (2020.12.5)\r\n"
]
],
[
[
"The search() method handles paging through the API results using a resumption token behind the scenes. Lets look at the first 100 results just to see what they look like.",
"_____no_output_____"
]
],
[
[
"from wayback import WaybackClient\n\nwb = WaybackClient()\n\ncount = 0\nfor result in wb.search('twitter.com/realDonaldTrump/status/', matchType='prefix'):\n print(result.url)\n count += 1\n if count > 100:\n break",
"https://twitter.com/realDonaldTrump/status/%22/6ixpercent/status/642559156784775168%22\nhttps://twitter.com/realDonaldTrump/status/%22/6ixpercent/status/645771399726919680%22\nhttps://twitter.com/realdonaldtrump/status/%22/BeauJohnson4/status/481467498248671233%22\nhttps://twitter.com/realDonaldTrump/status/%22/bobby_stokes/status/497930146968395776%22\nhttps://twitter.com/realDonaldTrump/status/%22/CharlesWoodso10/status/652578215400046592%22\nhttps://twitter.com/realDonaldTrump/status/%22/El_Grinchh/status/637030969338294272%22\nhttps://twitter.com/realdonaldtrump/status/%22/jefftiedrich/status/481830345814188034%22\nhttps://twitter.com/realDonaldTrump/status/%22/JesusLezz/status/651908920714371072%22\nhttps://twitter.com/realDonaldTrump/status/%22/jimmygarner/status/652537219694108673%22\nhttps://twitter.com/realDonaldTrump/status/%22/LittelCaesar/status/497929766230831104%22\nhttps://twitter.com/realDonaldTrump/status/%22/PattyDs50/status/637067362353704960%22\nhttps://twitter.com/realDonaldTrump/status/%22/roy_wr/status/649422085610057728%22\nhttps://twitter.com/realDonaldTrump/status/%22/saltythetrain/status/641699255636201472%22\nhttps://twitter.com/realDonaldTrump/status/%22/settings/account%22\nhttps://twitter.com/realDonaldTrump/status/%22/settings/account%22\nhttps://twitter.com/realDonaldTrump/status/%22/signup%22\nhttps://twitter.com/realDonaldTrump/status/%22/signup%22\nhttps://twitter.com/realdonaldtrump/status/%22//support.twitter.com/articles/20069937%22%3E\nhttps://twitter.com/realDonaldTrump/status/%22//support.twitter.com/articles/20069937%22%3E\nhttps://twitter.com/realDonaldTrump/status/%22/SyakibPutera/status/636601131339087872%22\nhttps://twitter.com/realDonaldTrump/status/%22//twitter.com/6ixpercent/status/642559156784775168/photo/1%22%3E\nhttps://twitter.com/realDonaldTrump/status/%22//twitter.com/6ixpercent/status/645771399726919680/photo/1%22%3E\nhttps://twitter.com/realdonaldtrump/status/%22//twitter.com/BeauJohnson4/status/481467498248671233/photo/1%22%3E\nhttps://twitter.com/realDonaldTrump/status/%22//twitter.com/bobby_stokes/status/497930146968395776/photo/1%22%3E\nhttps://twitter.com/realDonaldTrump/status/%22//twitter.com/CharlesWoodso10/status/652578215400046592/photo/1%22%3E\nhttps://twitter.com/realDonaldTrump/status/%22//twitter.com/El_Grinchh/status/637030969338294272/photo/1%22%3E\nhttps://twitter.com/realDonaldTrump/status/%22//twitter.com/LittelCaesar/status/497929766230831104/photo/1%22%3E\nhttps://twitter.com/realDonaldTrump/status/%22//twitter.com/PattyDs50/status/637067362353704960/photo/1%22%3E\nhttps://twitter.com/realDonaldTrump/status/%22//twitter.com/roy_wr/status/649422085610057728/photo/1%22%3E\nhttps://twitter.com/realDonaldTrump/status/%22//twitter.com/saltythetrain/status/641699255636201472/photo/1%22%3E\nhttps://twitter.com/realDonaldTrump/status/%22//twitter.com/SyakibPutera/status/636601131339087872/photo/1%22%3E\nhttps://twitter.com/realDonaldTrump/status/%22//twitter.com/WarmNewt/status/643614304391049216/photo/1%22%3E\nhttps://twitter.com/realDonaldTrump/status/%22/WarmNewt/status/643614304391049216%22\nhttps://twitter.com/realDonaldTrump/status/%22https://o.twimg.com/2/proxy.jpg?t=HBhPaHR0cDovL2kubW9sLmltL2kvcGl4LzIwMTIvMTAvMjMvYXJ0aWNsZS0yMjIyMDcwLTE1QTIzNERDMDAwMDA1REMtODE2Xzg3eDg0LmpwZxR4FHgcFAIUAgAAFgASAA&s=bHeI9yRE6I0akZqlGUD15yjeZZ3LKsAsfQNXHeF7dyc%22\nhttps://twitter.com/realDonaldTrump/status/%22https://pbs.twimg.com/media/Baq6H8yCIAML7zI.jpg%22\nhttps://twitter.com/realDonaldTrump/status/%22https://pbs.twimg.com/media/BNtTr7uCcAAV7OR.png%22\nhttps://twitter.com/realDonaldTrump/status/%22https://pbs.twimg.com/media/BNtTr7uCcAAV7OR.png%22\nhttps://twitter.com/realDonaldTrump/status/%22https://pbs.twimg.com/media/BNtTr7uCcAAV7OR.png%22\nhttps://twitter.com/realDonaldTrump/status/%22https://pbs.twimg.com/media/BNtTr7uCcAAV7OR.png%22\nhttps://twitter.com/realDonaldTrump/status/%22https://pbs.twimg.com/media/BNtTr7uCcAAV7OR.png%22\nhttps://twitter.com/realdonaldtrump/status/%22https://pbs.twimg.com/media/Bq6EIyHCMAAsX7N.jpg%22\nhttps://twitter.com/realDonaldTrump/status/%22https://pbs.twimg.com/media/Bq6EIyHCMAAsX7N.jpg%22\nhttps://twitter.com/realDonaldTrump/status/%22https://pbs.twimg.com/media/Bq6JS8cIQAA_TTv.png%22\nhttps://twitter.com/realDonaldTrump/status/%22https://pbs.twimg.com/media/Bq6JS8cIQAA_TTv.png%22\nhttps://twitter.com/realDonaldTrump/status/%22https://pbs.twimg.com/media/Bq6JS8cIQAA_TTv.png%22\nhttps://twitter.com/realDonaldTrump/status/%22https://pbs.twimg.com/media/BU3csjaCEAALxBW.jpg%22\nhttps://twitter.com/realDonaldTrump/status/%22https://pbs.twimg.com/media/BukA02nCQAA1hwu.jpg%22\nhttps://twitter.com/realDonaldTrump/status/%22https://pbs.twimg.com/media/BukAe3bCYAAgrdR.jpg%22\nhttps://twitter.com/realDonaldTrump/status/%22https://pbs.twimg.com/media/BXIJ5yjIYAAa7rj.jpg%22\nhttps://twitter.com/realDonaldTrump/status/%22https://pbs.twimg.com/media/CNcwSdkUcAELPJc.jpg%22\nhttps://twitter.com/realDonaldTrump/status/%22https://pbs.twimg.com/media/CNdRY5cUEAA3fk3.jpg%22\nhttps://twitter.com/realDonaldTrump/status/%22https://pbs.twimg.com/media/CNWpWFcVAAEQLU6.jpg%22\nhttps://twitter.com/realDonaldTrump/status/%22https://pbs.twimg.com/media/CO6TtqnWIAAj8Xc.jpg%22\nhttps://twitter.com/realDonaldTrump/status/%22https://pbs.twimg.com/media/COfGEjqUYAAPzkg.jpg%22\nhttps://twitter.com/realDonaldTrump/status/%22https://pbs.twimg.com/media/COrUJIUUAAAii6P.jpg%22\nhttps://twitter.com/realDonaldTrump/status/%22https://pbs.twimg.com/media/CPY9qdtUsAAf-OQ.jpg%22\nhttps://twitter.com/realDonaldTrump/status/%22https://pbs.twimg.com/media/CQ5sbaSUsAEHRkD.jpg%22\nhttps://twitter.com/realDonaldTrump/status/%22https://pbs.twimg.com/media/CQM18eKUAAA5vns.jpg%22\nhttps://twitter.com/realDonaldTrump/status/%22https://pbs.twimg.com/media/CRPh7KqUAAAEMIb.jpg%22\nhttps://twitter.com/realDonaldTrump/status/%22https://player.stickam.com/stickamPlayerSecure/vo/183893934%22\nhttps://twitter.com/realDonaldTrump/status/%22https://si0.twimg.com/profile_images/2828388301/bfdc4547b7901454cdc6d51d91bf99d6_normal.png%22\nhttps://twitter.com/realDonaldTrump/status/%22https://si0.twimg.com/profile_images/758286584/mol_icon_bigger_normal.png%22\nhttps://twitter.com/realDonaldTrump/status/%22https://support.twitter.com/articles/20169200-media-settings-and-best-practices%22\nhttps://twitter.com/realDonaldTrump/status/%22https://support.twitter.com/articles/20169200-media-settings-and-best-practices%22\nhttps://twitter.com/realDonaldTrump/status/%22https://www.youtube.com/embed/V9WSzMu-5jw%22\nhttps://twitter.com/realdonaldtrump/status/%20890193981585444864\nhttps://www.twitter.com/realDonaldTrump/status/'+twitter_id+\nhttps://twitter.com/realDonaldTrump/status/'+twitter_id+\nhttps://www.twitter.com/realDonaldTrump/status/'+twitter_id+\nhttps://twitter.com/realDonaldTrump/status/'+twitter_id+\nhttps://www.twitter.com/realDonaldTrump/status/'+twitter_id+\nhttps://twitter.com/realDonaldTrump/status/'+twitter_id+\nhttps://www.twitter.com/realDonaldTrump/status/'+twitter_id+\nhttps://twitter.com/realDonaldTrump/status/'+twitter_id+\nhttps://twitter.com/realdonaldtrump/status/%2A\nhttps://twitter.com/realdonaldtrump/status/%2A\nhttps://twitter.com/realDonaldTrump/status/...658142592667648\nhttps://twitter.com/realdonaldtrump/status/.login-responsive%20.nav%20.dropdown-signin,.nav%20li%20.dropdown-signup%7Bdisplay:none%7D.login-responsive%20.nav%20.dropdown-signup%7Bdisplay:block%7D.login-responsive%20.nav%20.dropdown-signup%20.emphasize%7Bcolor:\nhttps://twitter.com/realDonaldTrump/status/0?s=19\nhttps://twitter.com/realDonaldTrump/status/0n/7\nhttps://twitter.com/realDonaldTrump/status/0n/7\nhttp://twitter.com/realDonaldTrump/status/1000061992042975232\nhttps://twitter.com/realDonaldTrump/status/1000061992042975232\nhttps://twitter.com/realDonaldTrump/status/1000061992042975232\nhttps://twitter.com/realDonaldTrump/status/1000061992042975232\nhttps://twitter.com/realDonaldTrump/status/1000061992042975232\nhttps://twitter.com/realDonaldTrump/status/1000061992042975232\nhttps://twitter.com/realDonaldTrump/status/1000061992042975232\nhttps://twitter.com/realDonaldTrump/status/1000061992042975232\nhttps://twitter.com/realDonaldTrump/status/1000061992042975232\nhttps://twitter.com/realDonaldTrump/status/1000061992042975232\nhttps://twitter.com/realDonaldTrump/status/1000061992042975232\nhttps://twitter.com/realDonaldTrump/status/1000061992042975232\nhttps://twitter.com/realDonaldTrump/status/1000061992042975232\nhttps://twitter.com/realDonaldTrump/status/1000061992042975232\nhttps://twitter.com/realDonaldTrump/status/1000061992042975232\nhttps://twitter.com/realDonaldTrump/status/1000061992042975232\nhttps://twitter.com/realDonaldTrump/status/1000061992042975232\nhttps://twitter.com/realDonaldTrump/status/1000061992042975232\nhttps://twitter.com/realDonaldTrump/status/1000061992042975232\nhttps://twitter.com/realDonaldTrump/status/1000061992042975232\n"
]
],
[
[
"So there are some weird URLs in there, that look like the result of buggy automated archive processes that aren't constructing URLs properly?\n\n* https://twitter.com/realDonaldTrump/status/%22/SyakibPutera/status/636601131339087872%22\n* https://twitter.com/realDonaldTrump/status/'+twitter_id+\n\nAnd then we can see lots of results for the same URL such as https://twitter.com/realDonaldTrump/status/1000061992042975232 repeated over and over. This is because that URL was archived at multiple points in time. So lets improve on this to filter out the URLs that don't look like tweet URLs, and to only emit the unique ones. But still we'll just look at the first 100 results to make sure things are working properly.",
"_____no_output_____"
]
],
[
[
"import re\n\nseen = set()\n\nfor result in wb.search('twitter.com/realDonaldTrump/status/', matchType='prefix'):\n if re.search(r'/realDonaldTrump/status/\\d+', result.url):\n if result.url not in seen:\n print(result.url)\n seen.add(result.url)\n if len(seen) > 100:\n break",
"https://twitter.com/realDonaldTrump/status/0?s=19\nhttps://twitter.com/realDonaldTrump/status/0n/7\nhttp://twitter.com/realDonaldTrump/status/1000061992042975232\nhttps://twitter.com/realDonaldTrump/status/1000061992042975232\nhttps://www.twitter.com/realDonaldTrump/status/1000061992042975232\nhttps://twitter.com/realDonaldTrump/status/1000061992042975232?lang=en\nhttps://twitter.com/realDonaldTrump/status/1000061992042975232?lang=en-gb\nhttps://twitter.com/realDonaldTrump/status/1000061992042975232?ref_src=twcamp%5Ecopy%7Ctwsrc%5Eandroid%7Ctwgr%5Ecopy%7Ctwcon%5E7090%7Ctwterm%5E3\nhttps://twitter.com/realDonaldTrump/status/1000061992042975232?ref_src=twsrc%5Etfw\nhttp://twitter.com/realDonaldTrump/status/1000064605903876096\nhttps://twitter.com/realDonaldTrump/status/1000064605903876096\nhttps://www.twitter.com/realDonaldTrump/status/1000064605903876096\nhttps://twitter.com/realDonaldTrump/status/1000064605903876096?ref_src=twsrc%5Etfw\nhttp://twitter.com/realDonaldTrump/status/1000114139136606209\nhttps://twitter.com/realDonaldTrump/status/1000114139136606209\nhttps://www.twitter.com/realDonaldTrump/status/1000114139136606209\nhttps://twitter.com/realDonaldTrump/status/1000114139136606209/video/1\nhttps://twitter.com/realDonaldTrump/status/1000114139136606209?conversation_id=1000114139136606209\nhttps://twitter.com/realDonaldTrump/status/1000114139136606209?lang=en\nhttps://twitter.com/realDonaldTrump/status/1000114139136606209?lang=en-gb\nhttps://twitter.com/realDonaldTrump/status/1000114139136606209?ref_src=twsrc%5Etfw\nhttps://twitter.com/realDonaldTrump/status/1000114139136606209?s=20\nhttp://twitter.com/realDonaldTrump/status/1000138164923781121\nhttps://twitter.com/realDonaldTrump/status/1000138164923781121\nhttps://www.twitter.com/realDonaldTrump/status/1000138164923781121\nhttps://twitter.com/realDonaldTrump/status/1000138164923781121?ref_src=twsrc%5Etfw\nhttps://twitter.com/realDonaldTrump/status/1000138164923781121?s=19\nhttp://twitter.com/realDonaldTrump/status/1000145873274359809\nhttps://twitter.com/realDonaldTrump/status/1000145873274359809\nhttps://www.twitter.com/realDonaldTrump/status/1000145873274359809\nhttps://twitter.com/realDonaldTrump/status/1000145873274359809?conversation_id=1000145873274359809\nhttps://twitter.com/realDonaldTrump/status/1000145873274359809?ref_src=twsrc%5Etfw\nhttps://twitter.com/realDonaldTrump/status/1000145873274359809?s=19\nhttp://twitter.com/realDonaldTrump/status/1000151354701213696\nhttps://twitter.com/realDonaldTrump/status/1000151354701213696\nhttps://www.twitter.com/realDonaldTrump/status/1000151354701213696\nhttps://twitter.com/realDonaldTrump/status/1000151354701213696?conversation_id=1000151354701213696\nhttps://twitter.com/realDonaldTrump/status/1000151354701213696?ref_src=twsrc%5Etfw\nhttp://twitter.com/realDonaldTrump/status/1000152863035805696\nhttps://twitter.com/realDonaldTrump/status/1000152863035805696\nhttps://www.twitter.com/realDonaldTrump/status/1000152863035805696\nhttps://twitter.com/realDonaldTrump/status/1000152863035805696?ref_src=twsrc%5Etfw\nhttp://twitter.com/realDonaldTrump/status/1000174070061813761\nhttps://twitter.com/realDonaldTrump/status/1000174070061813761\nhttps://www.twitter.com/realDonaldTrump/status/1000174070061813761\nhttps://twitter.com/realDonaldTrump/status/1000174070061813761?conversation_id=1000174070061813761\nhttps://twitter.com/realDonaldTrump/status/1000174070061813761?ref_src=twsrc%5Egoogle%7Ctwcamp%5Enews%7Ctwgr%5Etweet\nhttps://twitter.com/realDonaldTrump/status/1000174070061813761?ref_src=twsrc%5Egoogle%7Ctwcamp%5Eserp%7Ctwgr%5Etweet\nhttps://twitter.com/realDonaldTrump/status/1000174070061813761?ref_src=twsrc%5Etfw\nhttps://twitter.com/realDonaldTrump/status/1000174070061813761?tfw_site=RT_russian&ref_src=twsrc%5Etfw&ref_url=https%3A%2F%2Frussian.rt.com%2Fworld%2Fnews%2F516773-tramp-peregovory-kndr\nhttps://twitter.com/realDonaldTrump/status/1000174070061813761?ref_src=twsrc%5Etfw&ref_url=https%3A%2F%2Fsputniknews.com%2Fus%2F201805261064828745-trump-still-talking-north-korea%2F&tfw_site=SputnikInt\nhttp://twitter.com/realDonaldTrump/status/1000366478846300165\nhttps://twitter.com/realDonaldTrump/status/1000366478846300165\nhttps://www.twitter.com/realDonaldTrump/status/1000366478846300165\nhttps://twitter.com/realDonaldTrump/status/1000366478846300165?ref_src=twsrc%5Egoogle%7Ctwcamp%5Enews%7Ctwgr%5Etweet\nhttps://twitter.com/realDonaldTrump/status/1000366478846300165?ref_src=twsrc%5Etfw\nhttp://twitter.com/realDonaldTrump/status/1000375761604370434\nhttps://twitter.com/realDonaldTrump/status/1000375761604370434\nhttps://www.twitter.com/realDonaldTrump/status/1000375761604370434\nhttps://twitter.com/realDonaldTrump/status/1000375761604370434?ref_src=twsrc%5Etfw\nhttps://twitter.com/realDonaldTrump/status/1000375761604370434?ref_src=twsrc^tfw\nhttps://twitter.com/realDonaldTrump/status/1000375761604370434?ref_src=twsrc%5Etfw&ref_url=http%3A%2F%2Fnymag.com%2Fdaily%2Fintelligencer%2F2018%2F05%2Ftrump-blames-dems-for-his-most-horrifying-immigration-policy.html\nhttps://twitter.com/realDonaldTrump/status/1000375761604370434?ref_src=twsrc%5Etfw&ref_url=http%3A%2F%2Fthehill.com%2Fhomenews%2Fsunday-talk-shows%2F389559-rubio-open-to-changing-law-that-separates-immigrant-children-from\nhttps://twitter.com/realDonaldTrump/status/1000375761604370434?ref_src=twsrc%5Etfw&ref_url=https%3A%2F%2Fwww.cnn.com%2F2018%2F05%2F26%2Fpolitics%2Ftrump-separating-parents-children-border-crossing%2Findex.html\nhttps://twitter.com/realDonaldTrump/status/1000375761604370434?tfw_creator=NBCNews&tfw_site=NBCNews&ref_src=twsrc%5Etfw&ref_url=https%3A%2F%2Fwww.nbcnews.com%2Fpolitics%2Fwhite-house%2Ftrump-blames-democrats-his-policy-separating-migrant-families-border-n878001\nhttps://twitter.com/realDonaldTrump/status/1000375761604370434?ref_src=twsrc%5Etfw&ref_url=https%3A%2F%2Fwww.npr.org%2F2018%2F05%2F29%2F615211215%2Ffact-check-are-democrats-responsible-for-dhs-separating-children-from-their-pare&tfw_site=NPR\nhttps://twitter.com/realDonaldTrump/status/1000375761604370434?tfw_site=nytimes&ref_src=twsrc%5Etfw&ref_url=https%3A%2F%2Fwww.nytimes.com%2F2018%2F05%2F28%2Fus%2Ftrump-immigrant-children-lost.html\nhttps://twitter.com/realDonaldTrump/status/1000375761604370434?ref_src=twsrc%5Etfw&ref_url=https%3A%2F%2Fwww.washingtonpost.com%2Fnews%2Ffact-checker%2Fwp%2F2018%2F05%2F30%2Ffact-checking-immigration-spin-on-separating-families-and-1500-lost-children%2F&tfw_creator=rizzoTK&tfw_site=WashingtonPost\nhttps://twitter.com/realDonaldTrump/status/1000375761604370434?ref_src=twsrc%5Etfw%7Ctwcamp%5Etweetembed%7Ctwterm%5E1000375761604370434&ref_url=http%3A%2F%2Fwww.foxnews.com%2Fpolitics%2F2018%2F05%2F26%2Ftrump-calls-for-democrats-to-end-horrible-law-that-says-separates-children-from-parents-at-border.html\nhttps://twitter.com/realDonaldTrump/status/1000375761604370434?ref_src=twsrc%5Etfw%7Ctwcamp%5Etweetembed%7Ctwterm%5E1000375761604370434&ref_url=https%3A%2F%2Fthehill.com%2Fblogs%2Fblog-briefing-room%2F389503-trump-calls-for-end-to-immigrant-family-separation-blames-democrats\nhttps://twitter.com/realDonaldTrump/status/1000375761604370434?s=20\nhttp://twitter.com/realDonaldTrump/status/1000385794115325952\nhttps://twitter.com/realDonaldTrump/status/1000385794115325952\nhttps://www.twitter.com/realDonaldTrump/status/1000385794115325952\nhttps://twitter.com/realDonaldTrump/status/1000385794115325952?ref_src=twsrc%5Egoogle%7Ctwcamp%5Enews%7Ctwgr%5Etweet\nhttps://twitter.com/realDonaldTrump/status/1000385794115325952?ref_src=twsrc%5Etfw\nhttp://twitter.com/realDonaldTrump/status/1000387893427351553\nhttps://twitter.com/realDonaldTrump/status/1000387893427351553\nhttps://www.twitter.com/realDonaldTrump/status/1000387893427351553\nhttps://twitter.com/realDonaldTrump/status/1000387893427351553?ref_src=twsrc%5Egoogle%7Ctwcamp%5Enews%7Ctwgr%5Etweet\nhttps://twitter.com/realDonaldTrump/status/1000387893427351553?ref_src=twsrc%5Etfw\nhttp://twitter.com/realDonaldTrump/status/1000391997969092608\nhttps://twitter.com/realDonaldTrump/status/1000391997969092608\nhttps://www.twitter.com/realDonaldTrump/status/1000391997969092608\nhttps://twitter.com/realDonaldTrump/status/1000391997969092608?ref_src=twsrc%5Egoogle%7Ctwcamp%5Enews%7Ctwgr%5Etweet\nhttps://twitter.com/realDonaldTrump/status/1000391997969092608?ref_src=twsrc%5Etfw\nhttps://twitter.com/realDonaldTrump/status/1000391997969092608?s=20\nhttp://twitter.com/realDonaldTrump/status/1000396430371106817\nhttps://twitter.com/realDonaldTrump/status/1000396430371106817\nhttps://www.twitter.com/realDonaldTrump/status/1000396430371106817\nhttps://twitter.com/realDonaldTrump/status/1000396430371106817?ref_src=twsrc%5Etfw\nhttps://twitter.com/realDonaldTrump/status/1000396430371106817?ref_src=twsrc%5Etfw&ref_url=https%3A%2F%2Fwww.nytimes.com%2F2018%2F05%2F26%2Fworld%2Fasia%2Ftrump-korea-summit-twitter.html&tfw_site=nytimes\nhttp://twitter.com/realDonaldTrump/status/1000458567147839488\nhttps://twitter.com/realDonaldTrump/status/1000458567147839488\nhttps://www.twitter.com/realDonaldTrump/status/1000458567147839488\nhttps://twitter.com/realDonaldTrump/status/1000458567147839488?ref_src=twsrc%5Etfw\nhttp://twitter.com/realDonaldTrump/status/1000462031500099584\nhttps://twitter.com/realDonaldTrump/status/1000462031500099584\nhttps://www.twitter.com/realDonaldTrump/status/1000462031500099584\nhttps://twitter.com/realDonaldTrump/status/1000462031500099584?ref_src=twsrc%5Etfw\nhttps://twitter.com/realDonaldTrump/status/1000462031500099584?ref_src=twsrc%5Etfw%7Ctwcamp%5Etweetembed\n"
]
],
[
[
"This list shows that some tweet URLs can have query strings, which modify the presentation of the tweet in various ways. For example to change the language of the user interface:\n\n* https://twitter.com/realDonaldTrump/status/1000114139136606209?lang=en-gb\n\nOr to highlight certain information:\n\n* https://twitter.com/realDonaldTrump/status/1000114139136606209?conversation_id=1000114139136606209\n\nThe query parameters are essential for finding the right view in the Wayback Machine. But the different variants don't really matter if we are simply wanting to count the number of tweets that are archived. Also it looks like some URLs aren't for the tweets themselves, but for components of the tweet, like video:\n\n* https://twitter.com/realDonaldTrump/status/1000114139136606209/video/1\n\nThe process can be adjusted to parse the URL to ensure the path is for an actual tweet, not a tweet component. The tweet id can also be extracted from the path in order to track whether it has been seen before.",
"_____no_output_____"
]
],
[
[
"from urllib.parse import urlparse\n\nseen = set()\n\nfor result in wb.search('twitter.com/realDonaldTrump/status/', matchType='prefix'):\n uri = urlparse(result.url)\n \n m = re.match(r'^/realDonaldTrump/status/(\\d+)/?$', uri.path, re.IGNORECASE)\n if not m:\n continue\n \n tweet_id = m.group(1)\n if tweet_id not in seen:\n print(result.url)\n seen.add(tweet_id)\n \n if len(seen) > 100:\n break",
"https://twitter.com/realDonaldTrump/status/0?s=19\nhttp://twitter.com/realDonaldTrump/status/1000061992042975232\nhttp://twitter.com/realDonaldTrump/status/1000064605903876096\nhttp://twitter.com/realDonaldTrump/status/1000114139136606209\nhttp://twitter.com/realDonaldTrump/status/1000138164923781121\nhttp://twitter.com/realDonaldTrump/status/1000145873274359809\nhttp://twitter.com/realDonaldTrump/status/1000151354701213696\nhttp://twitter.com/realDonaldTrump/status/1000152863035805696\nhttp://twitter.com/realDonaldTrump/status/1000174070061813761\nhttp://twitter.com/realDonaldTrump/status/1000366478846300165\nhttp://twitter.com/realDonaldTrump/status/1000375761604370434\nhttp://twitter.com/realDonaldTrump/status/1000385794115325952\nhttp://twitter.com/realDonaldTrump/status/1000387893427351553\nhttp://twitter.com/realDonaldTrump/status/1000391997969092608\nhttp://twitter.com/realDonaldTrump/status/1000396430371106817\nhttp://twitter.com/realDonaldTrump/status/1000458567147839488\nhttp://twitter.com/realDonaldTrump/status/1000462031500099584\nhttp://twitter.com/realDonaldTrump/status/1000465814192099330\nhttp://twitter.com/realDonaldTrump/status/1000554657859670016\nhttp://twitter.com/realDonaldTrump/status/1000718611688943616\nhttp://twitter.com/realDonaldTrump/status/1000726832843501570\nhttp://twitter.com/realDonaldTrump/status/1000741764565753856\nhttp://twitter.com/realDonaldTrump/status/1000831304836018176\nhttp://twitter.com/realDonaldTrump/status/1000837182297464832\nhttps://twitter.com/realDonaldTrump/status/100089437434477630\nhttp://twitter.com/realDonaldTrump/status/1001055932376387584\nhttp://twitter.com/realDonaldTrump/status/1001085207825534976\nhttp://twitter.com/realDonaldTrump/status/1001099771602456576\nhttp://twitter.com/realDonaldTrump/status/1001103028626345984\nhttp://twitter.com/realDonaldTrump/status/1001108964216537089\nhttp://twitter.com/realDonaldTrump/status/1001149367271686145\nhttp://twitter.com/realDonaldTrump/status/1001149793496977408\nhttp://twitter.com/realDonaldTrump/status/1001151001699803138\nhttp://twitter.com/realDonaldTrump/status/1001212077699149825\nhttp://twitter.com/realDonaldTrump/status/1001220050995511298\nhttp://twitter.com/realDonaldTrump/status/1001404640796336128\nhttp://twitter.com/realDonaldTrump/status/1001410457092218880\nhttp://twitter.com/realDonaldTrump/status/1001415199516254208\nhttp://twitter.com/realDonaldTrump/status/1001417880116891650\nhttp://twitter.com/realDonaldTrump/status/1001420270094168064\nhttp://twitter.com/realDonaldTrump/status/1001424695126880258\nhttp://twitter.com/realDonaldTrump/status/1001455721588969472\nhttp://twitter.com/realDonaldTrump/status/1001807174249713664\nhttp://twitter.com/realDonaldTrump/status/1001807204519997442\nhttp://twitter.com/realDonaldTrump/status/1001807216297627648\nhttp://twitter.com/realDonaldTrump/status/1001834394359877633\nhttps://twitter.com/realDonaldTrump/status/1001848460881035265\nhttps://twitter.com/realDonaldTrump/status/1001873279597064194\nhttps://twitter.com/realDonaldTrump/status/1001873868498325504\nhttps://twitter.com/realDonaldTrump/status/1001875884142362625\nhttps://twitter.com/realdonaldtrump/status/1001961235838103552\nhttp://twitter.com/realDonaldTrump/status/1001973090107248641\nhttp://twitter.com/realDonaldTrump/status/1001978774219644929\nhttp://twitter.com/realDonaldTrump/status/1002023477531070464\nhttp://twitter.com/realDonaldTrump/status/1002023935360339969\nhttps://twitter.com/realDonaldTrump/status/1002027245131661312\nhttp://twitter.com/realDonaldTrump/status/1002141448232493056\nhttp://twitter.com/realDonaldTrump/status/1002141779406270469\nhttp://twitter.com/realDonaldTrump/status/1002141845655351296\nhttp://twitter.com/realDonaldTrump/status/1002142304726061059\nhttp://twitter.com/realDonaldTrump/status/1002142585673146368\nhttp://twitter.com/realDonaldTrump/status/1002156016060989445\nhttp://twitter.com/realDonaldTrump/status/1002159087830827008\nhttp://twitter.com/realDonaldTrump/status/1002160516733853696\nhttp://twitter.com/realDonaldTrump/status/1002176736761655298\nhttp://twitter.com/realDonaldTrump/status/1002177521599860736\nhttp://twitter.com/realDonaldTrump/status/1002297565277237248\nhttp://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/100241982119714070\nhttp://twitter.com/realDonaldTrump/status/1002506360351846400\nhttp://twitter.com/realDonaldTrump/status/1002508937445478402\nhttp://twitter.com/realDonaldTrump/status/1002510522032541701\nhttp://twitter.com/realDonaldTrump/status/1002539852171304960\nhttp://twitter.com/realDonaldTrump/status/1002612676634259456\nhttp://twitter.com/realDonaldTrump/status/1002666307043647489\nhttps://twitter.com/realDonaldTrump/status/10027087487\nhttp://twitter.com/realDonaldTrump/status/1002877499448152065\nhttp://twitter.com/realDonaldTrump/status/1002881893963051009\nhttp://twitter.com/realDonaldTrump/status/1002884227753480193\nhttp://twitter.com/realDonaldTrump/status/1002892787803676673\nhttp://twitter.com/realDonaldTrump/status/1002946237652504576\nhttp://twitter.com/realDonaldTrump/status/1002947636436066304\nhttp://twitter.com/realDonaldTrump/status/1002950189668360192\nhttp://twitter.com/realDonaldTrump/status/1002951786301415424\nhttp://twitter.com/realDonaldTrump/status/1002954515941941249\nhttp://twitter.com/realDonaldTrump/status/1002965829175169025\nhttp://twitter.com/realDonaldTrump/status/1002968869043097600\nhttp://twitter.com/realDonaldTrump/status/1002971013313908738\nhttps://twitter.com/realDonaldTrump/status/1003012639642017794\nhttp://twitter.com/realDonaldTrump/status/1003017786355015680\nhttp://twitter.com/realDonaldTrump/status/1003019018683539458\nhttp://twitter.com/realDonaldTrump/status/1003019564379197446\nhttp://twitter.com/realDonaldTrump/status/1003020764906512385\nhttp://twitter.com/realDonaldTrump/status/1003024268756733952\nhttp://twitter.com/realDonaldTrump/status/1003259934417580033\nhttp://twitter.com/realDonaldTrump/status/1003266374473519105\nhttp://twitter.com/realDonaldTrump/status/1003268646070874113\nhttp://twitter.com/realDonaldTrump/status/1003326698228764672\nhttp://twitter.com/realDonaldTrump/status/1003328970069331968\nhttp://twitter.com/realDonaldTrump/status/1003601217111707649\nhttps://twitter.com/realDonaldTrump/status/1003611857272360960\n"
]
],
[
[
"It looks like this is actually working pretty good. For completeness we can store a mapping of the tweet id to all the results for that tweet id. This will allow us to track how many tweet have been archiving, while letting us examing how many times that tweet was archived, and what their precise URLs are for playback.\n\nThis time we can let it keep running to get all the results.",
"_____no_output_____"
]
],
[
[
"\nfrom collections import defaultdict\n\ntweets = defaultdict(list)\n\nfor result in wb.search('twitter.com/realDonaldTrump/status/', matchType='prefix'):\n uri = urlparse(result.url)\n \n m = re.match(r'^/realDonaldTrump/status/(\\d{8,})/?$', uri.path, re.IGNORECASE)\n if not m:\n continue\n \n tweet_id = m.group(1)\n tweets[tweet_id].append(result)",
"_____no_output_____"
]
],
[
[
"Now we can see the tweet ids. Instead of printing them all out we can just look at the first 100:",
"_____no_output_____"
]
],
[
[
"list(tweets.keys())[0:100]",
"_____no_output_____"
]
],
[
[
"And we can look at when a given tweet was archived too, irrespective of the various query strings that can be part of it. Here we get all the snapshots for tweet id 1002298565299965953 and print out the times that it was archived, in descending order. ",
"_____no_output_____"
]
],
[
[
"for result in sorted(tweets['1002298565299965953'], key=lambda r: r.timestamp, reverse=True):\n print(result.timestamp)",
"2021-01-08 13:24:02+00:00\n2020-11-25 02:50:00+00:00\n2020-11-15 17:00:34+00:00\n2020-11-09 02:37:14+00:00\n2020-11-09 02:37:14+00:00\n2020-11-06 08:20:49+00:00\n2020-11-06 05:32:52+00:00\n2020-10-11 09:07:44+00:00\n2020-07-10 17:13:56+00:00\n2020-06-30 18:27:42+00:00\n2020-06-30 18:26:24+00:00\n2020-06-16 20:52:42+00:00\n2020-06-07 07:43:55+00:00\n2019-11-10 07:27:23+00:00\n2019-09-11 04:17:42+00:00\n2019-09-11 04:17:25+00:00\n2019-08-25 17:20:01+00:00\n2019-08-25 10:14:36+00:00\n2019-06-13 09:14:18+00:00\n2019-05-13 13:45:24+00:00\n2019-05-05 01:44:53+00:00\n2018-12-29 05:30:13+00:00\n2018-11-27 19:12:03+00:00\n2018-08-22 14:21:16+00:00\n2018-06-05 13:07:07+00:00\n2018-06-05 08:38:29+00:00\n2018-06-05 08:38:27+00:00\n2018-06-05 08:38:24+00:00\n2018-06-05 08:38:22+00:00\n2018-06-05 08:38:20+00:00\n2018-06-05 08:38:18+00:00\n2018-06-05 08:38:16+00:00\n2018-06-05 08:38:14+00:00\n2018-06-05 08:38:12+00:00\n2018-06-05 08:38:10+00:00\n2018-06-05 08:38:09+00:00\n2018-06-05 08:38:07+00:00\n2018-06-05 08:38:05+00:00\n2018-06-05 08:38:03+00:00\n2018-06-05 08:38:01+00:00\n2018-06-05 08:37:59+00:00\n2018-06-05 08:37:57+00:00\n2018-06-05 08:37:56+00:00\n2018-06-05 08:37:54+00:00\n2018-06-05 08:37:52+00:00\n2018-06-05 08:37:50+00:00\n2018-06-05 08:37:48+00:00\n2018-06-05 08:37:46+00:00\n2018-06-05 08:37:44+00:00\n2018-06-05 08:37:42+00:00\n2018-06-05 08:37:40+00:00\n2018-06-05 08:37:38+00:00\n2018-06-05 08:37:37+00:00\n2018-06-05 08:37:35+00:00\n2018-06-05 08:37:33+00:00\n2018-06-05 08:37:31+00:00\n2018-06-05 08:37:30+00:00\n2018-06-05 08:37:28+00:00\n2018-06-05 08:37:26+00:00\n2018-06-05 08:37:23+00:00\n2018-06-05 08:37:21+00:00\n2018-06-05 08:37:20+00:00\n2018-06-05 08:37:18+00:00\n2018-06-05 08:37:16+00:00\n2018-06-05 08:37:14+00:00\n2018-06-05 08:37:13+00:00\n2018-06-05 08:37:11+00:00\n2018-06-05 08:37:09+00:00\n2018-06-05 08:37:07+00:00\n2018-06-05 08:37:05+00:00\n2018-06-05 08:37:03+00:00\n2018-06-05 08:37:01+00:00\n2018-06-05 08:36:59+00:00\n2018-06-05 02:44:15+00:00\n2018-06-05 01:23:49+00:00\n2018-06-04 08:24:31+00:00\n2018-06-04 08:24:30+00:00\n2018-06-04 08:24:28+00:00\n2018-06-04 08:24:26+00:00\n2018-06-04 08:24:24+00:00\n2018-06-04 08:24:22+00:00\n2018-06-04 08:24:20+00:00\n2018-06-04 08:24:18+00:00\n2018-06-04 08:24:17+00:00\n2018-06-04 08:24:15+00:00\n2018-06-04 08:24:13+00:00\n2018-06-04 08:24:11+00:00\n2018-06-04 08:24:09+00:00\n2018-06-04 08:24:07+00:00\n2018-06-04 08:24:06+00:00\n2018-06-04 08:24:04+00:00\n2018-06-04 08:24:02+00:00\n2018-06-04 08:24:00+00:00\n2018-06-04 08:23:58+00:00\n2018-06-04 08:23:56+00:00\n2018-06-04 08:23:55+00:00\n2018-06-04 08:23:53+00:00\n2018-06-04 08:23:51+00:00\n2018-06-04 08:23:49+00:00\n2018-06-04 08:23:47+00:00\n2018-06-04 08:23:45+00:00\n2018-06-04 08:23:43+00:00\n2018-06-04 08:23:41+00:00\n2018-06-04 08:23:39+00:00\n2018-06-04 08:23:37+00:00\n2018-06-04 08:23:35+00:00\n2018-06-04 08:23:33+00:00\n2018-06-04 08:23:32+00:00\n2018-06-04 08:23:29+00:00\n2018-06-04 08:23:28+00:00\n2018-06-04 08:23:25+00:00\n2018-06-04 08:23:24+00:00\n2018-06-04 08:23:22+00:00\n2018-06-04 08:23:20+00:00\n2018-06-04 08:23:18+00:00\n2018-06-04 08:23:16+00:00\n2018-06-04 08:23:14+00:00\n2018-06-04 08:23:12+00:00\n2018-06-04 08:23:10+00:00\n2018-06-04 08:23:08+00:00\n2018-06-04 08:23:06+00:00\n2018-06-04 08:23:05+00:00\n2018-06-04 08:23:03+00:00\n2018-06-04 02:08:13+00:00\n2018-06-02 15:41:04+00:00\n2018-06-02 06:48:41+00:00\n2018-06-02 06:48:39+00:00\n2018-06-02 06:48:36+00:00\n2018-06-02 06:48:33+00:00\n2018-06-02 06:48:31+00:00\n2018-06-02 06:48:29+00:00\n2018-06-02 06:48:26+00:00\n2018-06-02 06:48:23+00:00\n2018-06-02 06:48:21+00:00\n2018-06-02 06:48:19+00:00\n2018-06-02 06:48:16+00:00\n2018-06-02 06:48:14+00:00\n2018-06-02 06:48:12+00:00\n2018-06-02 06:48:09+00:00\n2018-06-02 06:48:07+00:00\n2018-06-02 06:48:04+00:00\n2018-06-02 06:48:02+00:00\n2018-06-02 06:47:59+00:00\n2018-06-02 06:47:57+00:00\n2018-06-02 06:47:55+00:00\n2018-06-02 06:47:52+00:00\n2018-06-02 06:47:49+00:00\n2018-06-02 06:47:47+00:00\n2018-06-02 06:47:45+00:00\n2018-06-02 06:47:43+00:00\n2018-06-02 06:47:40+00:00\n2018-06-02 06:47:38+00:00\n2018-06-02 06:47:36+00:00\n2018-06-02 06:47:34+00:00\n2018-06-02 06:47:31+00:00\n2018-06-02 06:47:29+00:00\n2018-06-02 06:47:27+00:00\n2018-06-02 06:47:24+00:00\n2018-06-02 06:47:22+00:00\n2018-06-02 06:47:19+00:00\n2018-06-02 06:47:17+00:00\n2018-06-02 06:47:14+00:00\n2018-06-02 06:47:12+00:00\n2018-06-02 06:47:10+00:00\n2018-06-02 06:47:07+00:00\n2018-06-02 06:47:04+00:00\n2018-06-02 06:47:02+00:00\n2018-06-02 06:47:00+00:00\n2018-06-02 06:46:58+00:00\n2018-06-02 06:46:55+00:00\n2018-06-02 06:46:53+00:00\n2018-06-02 06:46:50+00:00\n2018-06-02 06:46:48+00:00\n2018-06-02 05:37:35+00:00\n2018-06-02 05:34:35+00:00\n2018-06-02 02:05:58+00:00\n2018-06-02 02:05:56+00:00\n2018-06-02 02:05:54+00:00\n2018-06-02 02:05:52+00:00\n2018-06-02 02:05:50+00:00\n2018-06-02 02:05:48+00:00\n2018-06-02 02:05:46+00:00\n2018-06-02 02:05:44+00:00\n2018-06-02 02:05:42+00:00\n2018-06-02 02:05:40+00:00\n2018-06-02 02:05:37+00:00\n2018-06-02 02:05:35+00:00\n2018-06-02 02:05:33+00:00\n2018-06-02 02:05:31+00:00\n2018-06-02 02:05:29+00:00\n2018-06-02 02:05:27+00:00\n2018-06-02 02:05:25+00:00\n2018-06-02 02:05:23+00:00\n2018-06-02 02:05:20+00:00\n2018-06-02 02:05:18+00:00\n2018-06-02 02:05:16+00:00\n2018-06-02 02:05:13+00:00\n2018-06-02 02:05:11+00:00\n2018-06-02 02:05:09+00:00\n2018-06-02 02:05:07+00:00\n2018-06-02 02:05:05+00:00\n2018-06-02 02:05:03+00:00\n2018-06-02 02:05:01+00:00\n2018-06-02 02:04:59+00:00\n2018-06-02 02:04:57+00:00\n2018-06-02 02:04:55+00:00\n2018-06-02 02:04:53+00:00\n2018-06-02 02:04:51+00:00\n2018-06-02 02:04:49+00:00\n2018-06-02 02:04:47+00:00\n2018-06-02 02:04:45+00:00\n2018-06-02 02:04:43+00:00\n2018-06-02 02:04:41+00:00\n2018-06-02 02:04:38+00:00\n2018-06-02 02:04:36+00:00\n2018-06-02 02:04:34+00:00\n2018-06-02 02:04:32+00:00\n2018-06-02 02:04:30+00:00\n2018-06-02 02:04:27+00:00\n2018-06-02 02:04:25+00:00\n2018-06-02 02:04:23+00:00\n2018-06-02 02:04:21+00:00\n2018-06-02 02:04:19+00:00\n2018-06-02 01:09:19+00:00\n2018-06-01 23:59:15+00:00\n2018-06-01 22:15:11+00:00\n2018-06-01 21:50:35+00:00\n2018-06-01 20:46:37+00:00\n2018-06-01 19:05:17+00:00\n2018-06-01 18:36:21+00:00\n2018-06-01 15:47:20+00:00\n2018-06-01 14:51:15+00:00\n2018-06-01 14:27:36+00:00\n2018-06-01 13:11:35+00:00\n2018-06-01 10:46:01+00:00\n2018-06-01 09:35:34+00:00\n2018-06-01 07:18:33+00:00\n2018-06-01 07:14:17+00:00\n2018-06-01 07:00:38+00:00\n2018-06-01 06:06:34+00:00\n2018-06-01 04:10:45+00:00\n2018-06-01 01:58:20+00:00\n2018-06-01 01:10:27+00:00\n2018-06-01 00:25:32+00:00\n2018-05-31 23:53:40+00:00\n2018-05-31 23:43:21+00:00\n2018-05-31 23:25:27+00:00\n2018-05-31 22:23:43+00:00\n2018-05-31 21:35:57+00:00\n2018-05-31 21:31:19+00:00\n2018-05-31 21:20:08+00:00\n2018-05-31 21:19:37+00:00\n2018-05-31 21:19:36+00:00\n"
],
[
"len(tweets['1002298565299965953'])",
"_____no_output_____"
]
],
[
[
"So this particular URL was archived 252 times! The snapshots start on May 31, 2018 and most of the snapshots are from a few days of that. But there are also a handful of snapshots in 2019 and 2020. Examining [one of the snapshots] shows that it was sent on May 31st at 2:19 PM. It's hard to tell what time zone the display was generated for. But since the first snapshot was at May 31, 2018 at 21:19:36 UTC it is safe to assume that the display is for -07:00 UTC, or (given the time of year) Pacific Daylight Time.\n\nThe [overview](https://web.archive.org/web/20180101000000*/twitter.com/realDonaldTrump/status/1002298565299965953) gives a picture of some of these snapshots. But the nice thing about our index is that it factors in the way that the tweet ID is expressed in the URL. So we know more than what the URL specific overview shows. For example here are all the various URLs that were collected.",
"_____no_output_____"
]
],
[
[
"for result in sorted(tweets['1002298565299965953'], key=lambda r: r.timestamp, reverse=True):\n print(result.url)",
"https://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://www.twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953?ref_src=twsrc%5Etfw\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953?s=20\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953?ref_src=twsrc%5Etfw\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953?ref_src=twsrc%5Etfw\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953?ref_src=twsrc%5Etfw\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953?ref_src=twsrc%5Etfw\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=kn\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ta\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=mr\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=gu\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=sk\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=sr\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=bg\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=bn\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=vi\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=en-gb\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=hr\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ro\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=gl\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=cs\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=eu\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=el\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ga\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ca\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=uk\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=th\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ur\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=he\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=fa\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=hu\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=pl\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=da\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=fi\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=sv\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=no\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=hi\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=zh-cn\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=zh-tw\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ms\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=fil\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=nl\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ru\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=tr\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ko\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=pt\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=id\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=it\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=de\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=es\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ja\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ar\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=en\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=fr\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953?ref_src=twsrc%5Etfw\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=kn\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ta\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=mr\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=gu\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=sk\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=sr\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=bg\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=bn\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=vi\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=en-gb\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=hr\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ro\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=gl\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=cs\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=eu\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=el\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ga\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ca\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=uk\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=th\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ur\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=he\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=fa\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=hu\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=pl\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=da\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=fi\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=sv\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=no\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=hi\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=zh-cn\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=zh-tw\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ms\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=fil\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=nl\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ru\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=tr\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ko\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=pt\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=id\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=it\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=de\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=es\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ja\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ar\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=en\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=fr\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953?ref_src=twsrc%5Etfw\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=kn\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ta\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=mr\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=gu\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=sk\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=sr\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=bg\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=bn\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=vi\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=en-gb\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=hr\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ro\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=gl\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=cs\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=eu\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=el\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ga\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ca\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=uk\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=th\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ur\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=he\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=fa\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=hu\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=pl\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=da\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=fi\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=sv\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=no\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=hi\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=zh-cn\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=zh-tw\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ms\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=fil\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=nl\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ru\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=tr\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ko\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=pt\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=id\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=it\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=de\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=es\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ja\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ar\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=en\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=fr\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=kn\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ta\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=mr\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=gu\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=sk\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=sr\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=bg\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=bn\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=vi\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=en-gb\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=hr\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ro\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=gl\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=cs\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=eu\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=el\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ga\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ca\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=uk\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=th\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ur\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=he\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=fa\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=hu\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=pl\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=da\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=fi\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=sv\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=no\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=hi\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=zh-cn\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=zh-tw\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ms\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=fil\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=nl\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ru\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=tr\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ko\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=pt\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=id\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=it\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=de\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=es\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ja\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=ar\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=en\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953?ref_src=twsrc%5Etfw&lang=fr\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953?ref_src=twsrc%5Etfw\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953?ref_src=twsrc%5Etfw\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realdonaldtrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttps://twitter.com/realDonaldTrump/status/1002298565299965953\nhttp://twitter.com/realDonaldTrump/status/1002298565299965953\n"
]
],
[
[
"What was the most archived tweet?",
"_____no_output_____"
]
],
[
[
"sorted(tweets, key=lambda r: len(tweets[r]), reverse=True)[0]",
"_____no_output_____"
],
[
"len(tweets['1006837823469735936'])",
"_____no_output_____"
]
],
[
[
"So https://twitter.com/realDonaldTrump/status/1006837823469735936 was archived 23,419 times?! It's interesting that the [overview page](https://web.archive.org/web/*/twitter.com/realDonaldTrump/status/1006837823469735936) only says 595 times, because it is looking at that exact URL. Looking at [the content](https://web.archive.org/web/20180613095659/twitter.com/realDonaldTrump/status/1006837823469735936) of the tweet it is understandable why this one was archived so much.",
"_____no_output_____"
],
[
"## Missing Data?\n\nSo what does the coverage look like? Before Trump's account was suspended [his profile](https://web.archive.org/web/20210107045727/https://twitter.com/realDonaldTrump/) indicated he has sent 59.6K tweets. The [TrumpTweetArchive](https://www.thetrumparchive.com/) also shows 56,571 tweets. How many tweet IDs did we find?",
"_____no_output_____"
]
],
[
[
"len(tweets)",
"_____no_output_____"
]
],
[
[
"That is *a lot* less than what we should have found. So either there is a problem with my code, or the wayback module isn't paging results properly, or the CDX API isn't functioning properly, or not all of Trumps tweets have been archived?\n\nIn conversation with [Rob Brackett](https://robbrackett.com/) who is the principal author of the Python [wayback](https://pypi.org/project/wayback) library it seems that using the `limit` parameter can help return more results. So instead of doing:\n\n wb.search('twitter.com/realDonaldTrump/status/', matchType='prefix')\n\nthe `limit` parameter should be used:\n\n wb.search('twitter.com/realDonaldTrump/status/', matchType='prefix', limit=500000)\n\nHere's Rob's explanation, which kind of begs more questions:\n\n> Basically what’s happening here is that, without the `limit` parameter, the first page of results hits the maximum size and then, in that situation, does not include a resume key for moving on to the next page. Including a low enough limit (I think anything less than 1.5 million, but not sure) prevents you from hitting that ceiling and lets you successfully page through everything. When I do that, I get 64,329 tweet IDs across 16,253,658 CDX records (but the default Colab instance doesn’t have enough memory to store every record like you’re doing, so I had to just store the first record for each ID).\n\nSo lets give this a try. Rob noted that we're likely to consume all working memory storing all these CDX records in RAM. So lets persist them to a sqlite database instead.",
"_____no_output_____"
]
],
[
[
"import pathlib\n\ndata = pathlib.Path(\"data\")\ndb_path = data / \"trump-tweets.sqlite3\"",
"_____no_output_____"
],
[
"import sqlite3\n\n# only generate the sqlite db if it's not already there\n\nif not db_path.is_file():\n \n db = sqlite3.connect(db_path)\n db.execute(\n '''\n CREATE TABLE tweets (\n tweet_id TEXT,\n url TEXT,\n timestamp DATETIME,\n mime_type TEXT,\n status_code INTEGER,\n digest TEXT,\n length INTEGER\n )\n '''\n )\n\n count = 0\n for result in wb.search('twitter.com/realDonaldTrump/status/', matchType='prefix', limit=500000):\n uri = urlparse(result.url)\n\n m = re.match(r'^/realDonaldTrump/status/(\\d{8,})/?$', uri.path, re.IGNORECASE)\n if not m:\n continue\n\n tweet_id = m.group(1) \n db.execute('INSERT INTO tweets VALUES (?, ?, ?, ?, ?, ?, ?)', [\n tweet_id,\n result.url,\n result.timestamp,\n result.mime_type,\n result.status_code,\n result.digest,\n result.length\n ])\n\n count += 1\n if count % 1000 == 0:\n db.commit()\n\n\n db.close()",
"_____no_output_____"
]
],
[
[
"Unfortunately GitHub won't let you upload the 3GB sqlite file--even with git-lfs enabled.",
"_____no_output_____"
]
],
[
[
"db = sqlite3.connect(\"data/trump-tweets.sqlite3\")\n\ndb.execute('SELECT COUNT(DISTINCT(tweet_id)) FROM tweets').fetchall()",
"_____no_output_____"
]
],
[
[
"So 65,314 tweets were found. That's quite a bit more than the 59k suggested by the Twitter display and the 56,571 by the Trump Archive. Let's limit to snapshots that had a 200 OK HTTP response. As we saw above it's possible people tried to archive bogus tweet URLs.",
"_____no_output_____"
]
],
[
[
"db.execute(\n '''\n SELECT COUNT(DISTINCT(tweet_id))\n FROM tweets\n WHERE status_code = 200\n ''').fetchall()",
"_____no_output_____"
]
],
[
[
"That seems a lot more like it. So what were the most archived tweet? Let's get the top 10.",
"_____no_output_____"
]
],
[
[
"cursor = db.execute(\n '''\n SELECT tweet_id,\n COUNT(*) AS total\n FROM tweets\n WHERE status_code = 200\n GROUP by tweet_id\n ORDER BY total DESC\n LIMIT 10\n '''\n)\n\nfor row in cursor.fetchall():\n print(row)",
"('704834185471598592', 65486)\n('704756216157839360', 65353)\n('704818842153971712', 65350)\n('796315640307060738', 55391)\n('796182637622816768', 54854)\n('796055597594578944', 54258)\n('795733366842806272', 54243)\n('796099494442057728', 54230)\n('796126077647196160', 54180)\n('794259252613414915', 54088)\n"
]
],
[
[
"So the most archived URL was archived 56,571 times:\n \n https://web.archive.org/web/20200521045242/https://twitter.com/realDonaldTrump/status/704834185471598592\n\nThe interface indicates it was archived 1,616 times, but remember we factored in alternate forms of the tweet URL. Lets see what those were.",
"_____no_output_____"
]
],
[
[
"cursor = db.execute(\n '''\n SELECT url,\n COUNT(*) as total\n FROM tweets\n WHERE tweet_id = \"704834185471598592\" \n AND status_code = 200\n GROUP BY url\n ORDER BY total DESC\n '''\n)\n\nfor row in cursor.fetchall():\n print(row)",
"('https://twitter.com/realDonaldTrump/status/704834185471598592', 1612)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=zh-tw', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=vi', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=ur', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=uk', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=tr', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=th', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=ta', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=sv', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=sr', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=sk', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=ru', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=ro', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=pt', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=pl', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=no', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=nl', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=ms', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=mr', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=ko', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=kn', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=ja', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=it', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=id', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=hu', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=hr', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=hi', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=he', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=gu', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=gl', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=ga', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=fr', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=fil', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=fi', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=fa', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=eu', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=es', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=en-gb', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=en', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=el', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=de', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=da', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=cs', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=ca', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=bn', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=bg', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=ar', 1359)\n('https://twitter.com/realdonaldtrump/status/704834185471598592?lang=zh-cn', 1358)\n('https://twitter.com/realdonaldtrump/status/704834185471598592', 1)\n('https://twitter.com/realDonaldTrump/status/704834185471598592?ref_src=twsrc%5Etfw', 1)\n"
]
],
[
[
"Now that is kind of fascinating. Why would there be 1,349 captures of each of those language specific URLs for this tweet? This seems like some kind of automation?",
"_____no_output_____"
],
[
"## Missing from Trump Archive?\n\nSo what tweets were found in the Internet Archive that are not in the Trump Archive. To figure this out we can first load in the Trump Archive tweets. This is relatively easy to do using the Google Drive download from their FAQ page.",
"_____no_output_____"
]
],
[
[
"import pandas\n\ndf = pandas.read_csv('https://drive.google.com/uc?export=download&id=1xRKHaP-QwACMydlDnyFPEaFdtskJuBa6'\n)\n\ntrump_archive = set([str(tweet_id) for tweet_id in df['id']])\nlen(trump_archive)",
"_____no_output_____"
]
],
[
[
"Now we need the tweet ids from our sqlite db. Tweets will return a 200 OK and retweets will 301 Moved Permanently to the original tweet so we will include both here.",
"_____no_output_____"
]
],
[
[
"cursor = db.execute(\n '''\n SELECT DISTINCT(tweet_id)\n FROM tweets\n WHERE status_code in (200, 301)\n '''\n)\n\nwayback = set([r[0] for r in cursor.fetchall()])\n\nlen(wayback)",
"_____no_output_____"
]
],
[
[
"Now we can see what tweet ids are in the Wayback Machine but not in the Trump Archive.",
"_____no_output_____"
]
],
[
[
"len(wayback - trump_archive)",
"_____no_output_____"
]
],
[
[
"Wow. so 3592 tweets are in the Wayback Machine but not in the Trump Archive?! Let's spot check one of them to see if this is the case. Lets generate the Wayback URLs for the first 25 of these ids.\n\n https://web.archive.org/web/{datetime}/{url}",
"_____no_output_____"
]
],
[
[
"ids = list(wayback - trump_archive)[0:25]\n\ncursor = db.execute(\n '''\n SELECT \n \"https://web.archive.org/web/\" \n || STRFTIME('%Y%m%d%H%M%S', timestamp) \n || \"/\" \n || url\n FROM tweets\n WHERE tweet_id IN ({})\n '''.format(\",\".join([\"?\"] * 25)),\n ids\n)\n\nfor row in cursor.fetchall():\n print(row[0])",
"https://web.archive.org/web/20201108045542/https://twitter.com/realDonaldTrump/status/1080839175392321541\nhttps://web.archive.org/web/20200706132436/http://twitter.com/realDonaldTrump/status/1280130505829224448\nhttps://web.archive.org/web/20200706132436/https://twitter.com/realDonaldTrump/status/1280130505829224448\nhttps://web.archive.org/web/20200706132436/https://twitter.com/realDonaldTrump/status/1280130505829224448\nhttps://web.archive.org/web/20200706132437/https://twitter.com/realDonaldTrump/status/1280130505829224448\nhttps://web.archive.org/web/20200706132437/https://twitter.com/realdonaldtrump/status/1280130505829224448\nhttps://web.archive.org/web/20201115180905/https://twitter.com/realDonaldTrump/status/1280130505829224448\nhttps://web.archive.org/web/20180411191618/https://twitter.com/realDonaldTrump/status/151737838684221441\nhttps://web.archive.org/web/20200710183716/https://twitter.com/realDonaldTrump/status/151737838684221441\nhttps://web.archive.org/web/20201106113805/https://twitter.com/realDonaldTrump/status/151737838684221441\nhttps://web.archive.org/web/20180411191134/https://twitter.com/realDonaldTrump/status/190429718808510465\nhttps://web.archive.org/web/20200710184009/https://twitter.com/realDonaldTrump/status/190429718808510465\nhttps://web.archive.org/web/20201106113239/https://twitter.com/realDonaldTrump/status/190429718808510465\nhttps://web.archive.org/web/20180411190710/https://twitter.com/realDonaldTrump/status/220190663826878465\nhttps://web.archive.org/web/20200710184232/https://twitter.com/realDonaldTrump/status/220190663826878465\nhttps://web.archive.org/web/20201106112743/https://twitter.com/realDonaldTrump/status/220190663826878465\nhttps://web.archive.org/web/20180411184944/https://twitter.com/realDonaldTrump/status/268016770491756544\nhttps://web.archive.org/web/20200710185127/https://twitter.com/realDonaldTrump/status/268016770491756544\nhttps://web.archive.org/web/20201106111418/https://twitter.com/realDonaldTrump/status/268016770491756544\nhttps://web.archive.org/web/20180411184252/https://twitter.com/realDonaldTrump/status/288320622130507777\nhttps://web.archive.org/web/20200710185449/https://twitter.com/realDonaldTrump/status/288320622130507777\nhttps://web.archive.org/web/20201106110951/https://twitter.com/realDonaldTrump/status/288320622130507777\nhttps://web.archive.org/web/20180411182619/https://twitter.com/realDonaldTrump/status/315107115054276608\nhttps://web.archive.org/web/20200710190234/https://twitter.com/realDonaldTrump/status/315107115054276608\nhttps://web.archive.org/web/20201106105930/https://twitter.com/realDonaldTrump/status/315107115054276608\nhttps://web.archive.org/web/20180411173140/https://twitter.com/realDonaldTrump/status/441924030745292800\nhttps://web.archive.org/web/20200710192749/https://twitter.com/realDonaldTrump/status/441924030745292800\nhttps://web.archive.org/web/20201106102615/https://twitter.com/realDonaldTrump/status/441924030745292800\nhttps://web.archive.org/web/20180411170535/https://twitter.com/realDonaldTrump/status/516549737668169728\nhttps://web.archive.org/web/20200710193959/https://twitter.com/realDonaldTrump/status/516549737668169728\nhttps://web.archive.org/web/20201106100815/https://twitter.com/realDonaldTrump/status/516549737668169728\nhttps://web.archive.org/web/20200710195750/https://twitter.com/realDonaldTrump/status/58919218913099776\nhttps://web.archive.org/web/20201106114237/https://twitter.com/realDonaldTrump/status/58919218913099776\nhttps://web.archive.org/web/20150922170628/https://twitter.com/realDonaldTrump/status/646009823356690432\nhttps://web.archive.org/web/20151013221318/https://twitter.com/realDonaldTrump/status/646009823356690432\nhttps://web.archive.org/web/20151025160235/https://twitter.com/realDonaldTrump/status/646009823356690432\nhttps://web.archive.org/web/20151113140248/https://twitter.com/realDonaldTrump/status/646009823356690432\nhttps://web.archive.org/web/20151118170051/https://twitter.com/realDonaldTrump/status/646009823356690432\nhttps://web.archive.org/web/20151228212242/https://twitter.com/realDonaldTrump/status/646009823356690432\nhttps://web.archive.org/web/20160213192003/https://twitter.com/realDonaldTrump/status/646009823356690432\nhttps://web.archive.org/web/20160218083548/https://twitter.com/realDonaldTrump/status/646009823356690432\nhttps://web.archive.org/web/20160419215920/https://twitter.com/realDonaldTrump/status/646009823356690432\nhttps://web.archive.org/web/20160426184152/https://twitter.com/realDonaldTrump/status/646009823356690432\nhttps://web.archive.org/web/20180411160400/https://twitter.com/realDonaldTrump/status/646009823356690432\nhttps://web.archive.org/web/20190726092418/https://twitter.com/realdonaldtrump/status/646009823356690432\nhttps://web.archive.org/web/20190726092420/https://twitter.com/realDonaldTrump/status/646009823356690432\nhttps://web.archive.org/web/20190826070031/https://twitter.com/realDonaldTrump/status/646009823356690432\nhttps://web.archive.org/web/20200307194518/https://twitter.com/realdonaldtrump/status/646009823356690432\nhttps://web.archive.org/web/20200710200953/https://twitter.com/realDonaldTrump/status/646009823356690432\nhttps://web.archive.org/web/20201106092344/https://twitter.com/realDonaldTrump/status/646009823356690432\nhttps://web.archive.org/web/20151014084434/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=ar\nhttps://web.archive.org/web/20151014084537/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=bn\nhttps://web.archive.org/web/20151014084523/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=ca\nhttps://web.archive.org/web/20151014084530/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=cs\nhttps://web.archive.org/web/20151014084510/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=da\nhttps://web.archive.org/web/20151014084442/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=de\nhttps://web.archive.org/web/20151014084526/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=el\nhttps://web.archive.org/web/20151014084432/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=en\nhttps://web.archive.org/web/20151014084534/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=en-gb\nhttps://web.archive.org/web/20151014084441/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=es\nhttps://web.archive.org/web/20151014084528/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=eu\nhttps://web.archive.org/web/20151014084515/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=fa\nhttps://web.archive.org/web/20151014084508/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=fi\nhttps://web.archive.org/web/20151014084456/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=fil\nhttps://web.archive.org/web/20151014084431/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=fr\nhttps://web.archive.org/web/20151014084525/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=ga\nhttps://web.archive.org/web/20151014084531/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=gl\nhttps://web.archive.org/web/20151014084539/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=gu\nhttps://web.archive.org/web/20151014084516/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=he\nhttps://web.archive.org/web/20151014084503/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=hi\nhttps://web.archive.org/web/20151014084513/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=hu\nhttps://web.archive.org/web/20151014084446/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=id\nhttps://web.archive.org/web/20151014084444/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=it\nhttps://web.archive.org/web/20151014084439/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=ja\nhttps://web.archive.org/web/20151014084544/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=kn\nhttps://web.archive.org/web/20151014084449/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=ko\nhttps://web.archive.org/web/20151014084540/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=mr\nhttps://web.archive.org/web/20151014084458/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=ms\nhttps://web.archive.org/web/20151014084454/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=nl\nhttps://web.archive.org/web/20151014084504/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=no\nhttps://web.archive.org/web/20151014084511/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=pl\nhttps://web.archive.org/web/20151014084448/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=pt\nhttps://web.archive.org/web/20151014084533/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=ro\nhttps://web.archive.org/web/20151014084453/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=ru\nhttps://web.archive.org/web/20151014084506/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=sv\nhttps://web.archive.org/web/20151014084542/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=ta\nhttps://web.archive.org/web/20151014084520/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=th\nhttps://web.archive.org/web/20151014084451/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=tr\nhttps://web.archive.org/web/20151014084521/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=uk\nhttps://web.archive.org/web/20151014084518/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=ur\nhttps://web.archive.org/web/20151014084536/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=vi\nhttps://web.archive.org/web/20151014084501/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=zh-cn\nhttps://web.archive.org/web/20151014084459/https://twitter.com/realdonaldtrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg&lang=zh-tw\nhttps://web.archive.org/web/20151014084429/https://twitter.com/realDonaldTrump/status/646009823356690432?replies_view=true&cursor=BICWH04t9wg\nhttps://web.archive.org/web/20151014083015/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=ar\nhttps://web.archive.org/web/20151014083120/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=bn\nhttps://web.archive.org/web/20151014083104/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=ca\nhttps://web.archive.org/web/20151014083111/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=cs\nhttps://web.archive.org/web/20151014083050/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=da\nhttps://web.archive.org/web/20151014083021/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=de\nhttps://web.archive.org/web/20151014083107/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=el\nhttps://web.archive.org/web/20150921171519/https://twitter.com/realDonaldTrump/status/646009823356690432?lang=en\nhttps://web.archive.org/web/20151014083014/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=en\nhttps://web.archive.org/web/20151014083117/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=en-gb\nhttps://web.archive.org/web/20151014083019/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=es\nhttps://web.archive.org/web/20151014083109/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=eu\nhttps://web.archive.org/web/20151014083056/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=fa\nhttps://web.archive.org/web/20151014083048/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=fi\nhttps://web.archive.org/web/20151014083036/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=fil\nhttps://web.archive.org/web/20151014083012/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=fr\nhttps://web.archive.org/web/20151014083106/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=ga\nhttps://web.archive.org/web/20151014083113/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=gl\nhttps://web.archive.org/web/20151014083122/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=gu\nhttps://web.archive.org/web/20151014083057/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=he\nhttps://web.archive.org/web/20151014083042/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=hi\nhttps://web.archive.org/web/20151014083054/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=hu\nhttps://web.archive.org/web/20151014083024/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=id\nhttps://web.archive.org/web/20151014083023/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=it\nhttps://web.archive.org/web/20151014083017/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=ja\nhttps://web.archive.org/web/20151014083127/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=kn\nhttps://web.archive.org/web/20151014083028/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=ko\nhttps://web.archive.org/web/20151014083123/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=mr\nhttps://web.archive.org/web/20151014083037/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=ms\nhttps://web.archive.org/web/20151014083129/https://twitter.com/realDonaldTrump/status/646009823356690432?lang=msa\nhttps://web.archive.org/web/20151014083034/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=nl\nhttps://web.archive.org/web/20151014083044/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=no\nhttps://web.archive.org/web/20151014083052/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=pl\nhttps://web.archive.org/web/20151014083026/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=pt\nhttps://web.archive.org/web/20151014083114/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=ro\nhttps://web.archive.org/web/20151014083031/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=ru\nhttps://web.archive.org/web/20151014083046/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=sv\nhttps://web.archive.org/web/20151014083125/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=ta\nhttps://web.archive.org/web/20151014083100/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=th\nhttps://web.archive.org/web/20151014083029/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=tr\nhttps://web.archive.org/web/20151014083102/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=uk\nhttps://web.archive.org/web/20151014083059/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=ur\nhttps://web.archive.org/web/20151014083118/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=vi\nhttps://web.archive.org/web/20151014083041/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=zh-cn\nhttps://web.archive.org/web/20151014083039/https://twitter.com/realdonaldtrump/status/646009823356690432?lang=zh-tw\nhttps://web.archive.org/web/20180411155103/https://twitter.com/realDonaldTrump/status/667434942826156032\nhttps://web.archive.org/web/20190828013604/https://twitter.com/realDonaldTrump/status/667434942826156032\nhttps://web.archive.org/web/20200710201530/https://twitter.com/realDonaldTrump/status/667434942826156032\nhttps://web.archive.org/web/20201106091445/https://twitter.com/realDonaldTrump/status/667434942826156032\nhttps://web.archive.org/web/20201111223531/https://twitter.com/realDonaldTrump/status/877251410466934784\nhttps://web.archive.org/web/20201111202739/https://twitter.com/realDonaldTrump/status/878632059065032704\nhttps://web.archive.org/web/20201112034009/https://twitter.com/realDonaldTrump/status/879447209871388672\nhttps://web.archive.org/web/20201109024810/https://twitter.com/realDonaldTrump/status/881376258432659456\nhttps://web.archive.org/web/20201109040343/https://twitter.com/realDonaldTrump/status/885912479376842752\nhttps://web.archive.org/web/20201112024608/https://twitter.com/realDonaldTrump/status/897562795386048514\nhttps://web.archive.org/web/20201111211910/https://twitter.com/realDonaldTrump/status/899648797374119936\nhttps://web.archive.org/web/20201111223122/https://twitter.com/realDonaldTrump/status/905114056327081988\nhttps://web.archive.org/web/20201112023125/https://twitter.com/realDonaldTrump/status/905198462525956097\nhttps://web.archive.org/web/20201112013623/https://twitter.com/realDonaldTrump/status/908433296513695744\nhttps://web.archive.org/web/20201112025110/https://twitter.com/realDonaldTrump/status/913909265118703616\nhttps://web.archive.org/web/20201111183124/https://twitter.com/realDonaldTrump/status/920341449220284418\n"
]
],
[
[
"The first 5 of these seem to generate a *Something Went Wrong Page*. Perhaps there were tweets there and the Wayback Machine failed fetch them properly? Or maybe the data is there but failing to play back? It's hard to say with confidence.\n\nhttps://web.archive.org/web/20201108045542/https://twitter.com/realDonaldTrump/status/1080839175392321541\n\n<img src=\"images/twitter-something-went-wrong.png\">",
"_____no_output_____"
],
[
"But then at least some of these appear to work such as:\n\n* https://web.archive.org/web/20201106091445/https://twitter.com/realDonaldTrump/status/667434942826156032\n* https://web.archive.org/web/20200307194518/https://twitter.com/realdonaldtrump/status/646009823356690432\n\nThe Trump Archive API can tell if they have these two:\n\n https://www.thetrumparchive.com/tweets/{tweet-id}\n\n* https://www.thetrumparchive.com/tweets/667434942826156032\n* https://www.thetrumparchive.com/tweets/646009823356690432\n\nSo it looks like there are definitely some realDonaldTrump tweets in the Internet Archive's Wayback Machine that are not in the Trump Archive. Some number less than 3,592. It would be necessary to somehow verify these to be sure. Here's a CSV of all the tweet IDs to see if they can be curated.",
"_____no_output_____"
]
],
[
[
"import csv",
"_____no_output_____"
],
[
"out = csv.writer(open('data/trump-tweets-missing-from-archive.csv', 'w'))\n\nout.writerow(['tweet_url', 'archive_url'])\n\nfor tweet_id in wayback - trump_archive:\n sql = \"\"\"\n SELECT\n url, \n STRFTIME('%Y%m%d%H%M%S', timestamp) AS timestamp\n FROM tweets\n WHERE tweet_id = ?\n ORDER BY timestamp DESC\n LIMIT 1\n \"\"\"\n [tweet_url, timestamp] = db.execute(sql, [tweet_id]).fetchone()\n out.writerow([\n tweet_url,\n \"https://web.archive.org/web/{}/{}\".format(timestamp, tweet_url)\n ])\n print(tweet_url)",
"https://twitter.com/realDonaldTrump/status/918240536984801280\nhttps://twitter.com/realDonaldTrump/status/315107115054276608\nhttps://twitter.com/realDonaldTrump/status/455802609589583872\nhttps://twitter.com/realDonaldTrump/status/303870510951981056\nhttps://twitter.com/realdonaldtrump/status/1198672518833430534\nhttps://twitter.com/realDonaldTrump/status/918305600810946561\nhttps://twitter.com/realDonaldTrump/status/143769703230357504\nhttps://twitter.com/realDonaldTrump/status/892775255835639808\nhttps://twitter.com/realDonaldTrump/status/1140846462236073984\nhttps://twitter.com/realDonaldTrump/status/179291529855246336\nhttps://twitter.com/realDonaldTrump/status/443402398049251329\nhttps://twitter.com/realDonaldTrump/status/883335550983450625\nhttps://twitter.com/realDonaldTrump/status/296278885346209793\nhttps://twitter.com/realDonaldTrump/status/329042069807054849\nhttps://twitter.com/realDonaldTrump/status/900010432563433478\nhttps://twitter.com/realDonaldTrump/status/313356899510804480\nhttps://twitter.com/realDonaldTrump/status/919336178347888640\nhttps://twitter.com/realDonaldTrump/status/908330100889931776\nhttps://twitter.com/realDonaldTrump/status/264417734320857088\nhttps://twitter.com/realDonaldTrump/status/280813217415041024\nhttps://twitter.com/realDonaldTrump/status/528539008914452481\nhttps://twitter.com/realDonaldTrump/status/929812182782558208\nhttps://twitter.com/realDonaldTrump/status/434214732774776832\nhttps://twitter.com/realDonaldTrump/status/913411224586637312\nhttps://twitter.com/realDonaldTrump/status/873227797120024578\nhttps://twitter.com/realDonaldTrump/status/907375029289144322\nhttps://twitter.com/realDonaldTrump/status/278156452684787712\nhttps://twitter.com/realDonaldTrump/status/788592944785817600\nhttps://twitter.com/realDonaldTrump/status/313356619809431552\nhttps://twitter.com/realDonaldTrump/status/1280863277543800832\nhttps://twitter.com/realDonaldTrump/status/317279532585455616\nhttps://twitter.com/realDonaldTrump/status/914864957346508801\nhttps://twitter.com/realDonaldTrump/status/273108190655283200\nhttps://twitter.com/realDonaldTrump/status/886623215270977537\nhttps://twitter.com/realDonaldTrump/status/883392939140960260\nhttps://twitter.com/realDonaldTrump/status/890950735587721217\nhttps://twitter.com/realDonaldTrump/status/882984839820881921\nhttps://twitter.com/realDonaldTrump/status/276380156736720896\nhttps://twitter.com/realDonaldTrump/status/877558750009331712\nhttps://twitter.com/realDonaldTrump/status/913960409803231235\nhttps://twitter.com/realDonaldTrump/status/333057798285963266\nhttps://twitter.com/realDonaldTrump/status/908458879989817350\nhttps://twitter.com/realDonaldTrump/status/214733405894606850\nhttps://twitter.com/realDonaldTrump/status/268088953440202753\nhttps://twitter.com/realDonaldTrump/status/339461349463699456\nhttps://twitter.com/realDonaldTrump/status/498905672810258433\nhttps://twitter.com/realDonaldTrump/status/179235478283882496\nhttps://twitter.com/realDonaldTrump/status/918869698602659840\nhttps://twitter.com/realDonaldTrump/status/146332661848358912\nhttps://twitter.com/realDonaldTrump/status/914694575876395009\nhttps://twitter.com/realDonaldTrump/status/783107076327284737\nhttps://twitter.com/realDonaldTrump/status/915216650894442502\nhttps://twitter.com/realDonaldTrump/status/498754376362885120\nhttps://twitter.com/realDonaldTrump/status/557202946354839552\nhttps://twitter.com/realDonaldTrump/status/908722590147063811\nhttps://twitter.com/realDonaldTrump/status/916694643395432448\nhttps://twitter.com/realDonaldTrump/status/917839153278193666\nhttps://twitter.com/realDonaldTrump/status/188017774742867969\nhttps://twitter.com/realDonaldTrump/status/128925217622994944\nhttps://twitter.com/realDonaldTrump/status/1052609500950261760\nhttps://twitter.com/realDonaldTrump/status/886977047662297090\nhttps://twitter.com/realDonaldTrump/status/878632059065032704\nhttps://twitter.com/realDonaldTrump/status/291232782858981376\nhttps://twitter.com/realDonaldTrump/status/1258113511730884611\nhttps://twitter.com/realDonaldTrump/status/441924030745292800\nhttps://twitter.com/realDonaldTrump/status/166628357193211905\nhttps://twitter.com/realDonaldTrump/status/906952828110983171\nhttps://twitter.com/realDonaldTrump/status/918305216826609665\nhttps://twitter.com/realDonaldTrump/status/220205157089148929\nhttps://twitter.com/realDonaldTrump/status/910994803311173634\nhttps://twitter.com/realDonaldTrump/status/93409642994077696\nhttp://twitter.com/realDonaldTrump/status/788479634694246400\nhttps://twitter.com/realDonaldTrump/status/871805576220749828\nhttps://twitter.com/realDonaldTrump/status/910582331232137216\nhttps://twitter.com/realDonaldTrump/status/899706812919951360\nhttps://twitter.com/realdonaldtrump/status/761631028058333184\nhttps://twitter.com/realDonaldTrump/status/148871667345666048\nhttps://twitter.com/realDonaldTrump/status/159297839292354560\nhttps://twitter.com/realDonaldTrump/status/918852077484367872\nhttps://twitter.com/realDonaldTrump/status/166581444485984256\nhttps://twitter.com/realDonaldTrump/status/885898280776945664\nhttps://twitter.com/realDonaldTrump/status/698520189684596737\nhttps://twitter.com/realDonaldTrump/status/783103327764553728\nhttps://twitter.com/realDonaldTrump/status/884483874276999173\nhttps://twitter.com/realDonaldTrump/status/884880230556585985\nhttps://twitter.com/realDonaldTrump/status/266221664537305089\nhttps://twitter.com/realDonaldTrump/status/636051227193700352\nhttps://twitter.com/realDonaldTrump/status/235106470012805122\nhttps://twitter.com/realDonaldTrump/status/908760866308415488\nhttps://twitter.com/realDonaldTrump/status/896185057118957568\nhttps://twitter.com/realDonaldTrump/status/663330357601804292\nhttps://twitter.com/realDonaldTrump/status/1235034700827283456\nhttps://twitter.com/realDonaldTrump/status/724916060424667136\nhttps://twitter.com/realDonaldTrump/status/180370089034526721\nhttps://twitter.com/realDonaldTrump/status/280812392491937792\nhttps://twitter.com/realDonaldTrump/status/890009535183806465\nhttps://twitter.com/realDonaldTrump/status/1220887923786178560\nhttps://twitter.com/realDonaldTrump/status/892913409792016387\nhttps://twitter.com/realDonaldTrump/status/222685835554848769\nhttps://twitter.com/realDonaldTrump/status/290931233205129217\nhttps://twitter.com/realDonaldTrump/status/887102408249163776\nhttps://twitter.com/realDonaldTrump/status/908687359557951488\nhttps://twitter.com/realdonaldtrump/status/180375447572463618\nhttps://twitter.com/realDonaldTrump/status/908107837250981888\nhttps://twitter.com/realDonaldTrump/status/712236978977296384\nhttps://twitter.com/realDonaldTrump/status/245225092618006528\nhttps://twitter.com/realDonaldTrump/status/440351535302991872\nhttps://twitter.com/realDonaldTrump/status/562454957618917377\nhttps://twitter.com/realDonaldTrump/status/564798780974039041\nhttps://twitter.com/realDonaldTrump/status/905155577533353984\nhttps://twitter.com/realDonaldTrump/status/232568020814401536\nhttps://twitter.com/realDonaldTrump/status/897900755453186048\nhttps://twitter.com/realDonaldTrump/status/743523444831182848\nhttps://twitter.com/realDonaldTrump/status/906217845117272066\nhttps://twitter.com/realDonaldTrump/status/899798707876634625\nhttps://twitter.com/realDonaldTrump/status/585224565959294978\nhttps://twitter.com/realDonaldTrump/status/738951969042227200\nhttps://twitter.com/realDonaldTrump/status/913410415895437313\nhttps://twitter.com/realDonaldTrump/status/919975234387636225\nhttps://twitter.com/realDonaldTrump/status/640220766966034433\nhttps://twitter.com/realdonaldtrump/status/1189548311809839105\nhttps://twitter.com/realDonaldTrump/status/907375052076789760\nhttps://twitter.com/realDonaldTrump/status/875508715042082818\nhttps://twitter.com/realDonaldTrump/status/885150230919905281\nhttps://twitter.com/realDonaldTrump/status/564796741019766787\nhttps://twitter.com/realDonaldTrump/status/575003905324621824\nhttps://twitter.com/realDonaldTrump/status/913892144271822849\nhttps://twitter.com/realDonaldTrump/status/1119718338555523074\nhttps://twitter.com/realDonaldTrump/status/252875278949441536\nhttps://twitter.com/realDonaldTrump/status/900509972131401729\nhttps://twitter.com/realDonaldTrump/status/879895332062154752\nhttps://twitter.com/realDonaldTrump/status/718954883618054144\nhttps://twitter.com/realDonaldTrump/status/278535467304120321\n"
]
],
[
[
"## Missing from Internet Archive?\n\nHow about the other angle: are there any tweet ids in the Trump Archive that didn't come back from the CDX API?",
"_____no_output_____"
]
],
[
[
"len(trump_archive - wayback)",
"_____no_output_____"
]
],
[
[
"It appears yes?",
"_____no_output_____"
]
],
[
[
"trump_archive - wayback",
"_____no_output_____"
]
],
[
[
"Lets examing the first one: 1175115230457802752. Is it in the Trump Archive?\n\nhttps://www.thetrumparchive.com/tweets/1175115230457802752\n\nYes. It looks like a retweet of @FLOTUS:\n\n RT @FLOTUS: Welcome to the @WhiteHouse PM Morrison and Mrs. Morrison! 🇺🇸🇦🇺 https://t.co/kYznIkJf9H\n\nBut the redirect of the retweet is not in the Internet Archive:\n\nhttps://web.archive.org/web/*/https://twitter.com/realDonaldTrump/1175115230457802752\n\nThis in itself isn't too surprising because people wouldn't typically archive the retweet redirect. Are there any non-retweets in the Trump Archive but not in the Wayback Machine? To test that we need to examine the text of these tweets. Luckily we can look those up pretty easily using The Trump Archive API.",
"_____no_output_____"
]
],
[
[
"import requests\n\nfor tweet_id in trump_archive - wayback:\n url = \"https://www.thetrumparchive.com/tweets/{}\".format(tweet_id)\n resp = requests.get(url)\n if resp.status_code == 200:\n tweet = resp.json()\n if tweet['isRetweet'] == False:\n print(\"id: {}\\ndate: {}\\n{}\\n\".format(\n tweet['id'], \n tweet['date'], \n tweet['text']\n ))",
"id: 1237490722317746178\ndate: 2020-03-10T21:29:14.000Z\nIf you like automobils, how can you vote for a Democrat who all want to get rid of cars, as quickly as possible, especially if they are powered by gasoline. Remember also, no more than one car per family. I, on the other hand, have new plants being built all over Michigan, Plus!\n\nid: 1281926278845812736\ndate: 2020-07-11T12:20:16.000Z\nRoger Stone was targeted by an illegal Witch Hunt tha never should have taken place. It is the other side that are criminals, including the fact that Biden and Obama illegally spied on my campaign - AND GOT CAUGHT!\n\nid: 1182847457396310017\ndate: 2019-10-12T02:36:24.000Z\nRT @realDonaldTrump: Just landed in Louisiana! Vote against John Bel Edwards, he has the worst jobs record in the United States. Louisiana...\n\nid: 1331749395214311427\ndate: 2020-11-25T23:59:33.000Z\nhttps://t.co/FJ6UcmXq5G?\n\nid: 1181420692572692480\ndate: 2019-10-08T04:06:57.000Z\nRT @seanmdav: Lawmakers in both chambers have demanded that IC IG Michael Atkinson explain why he backdated to August secret changes he mad...\n\nid: 1193376950481489921\ndate: 2019-11-10T03:56:51.000Z\nRT @realDonaldTrump: Just finished reading my son Donald’s just out new book, “Triggered.” It is really good! He, along with many of us, wa...\n\nid: 1187936132241219584\ndate: 2019-10-26T03:36:59.000Z\nRT @CNNPolitics: JUST IN: Trump administration suspends US commercial flights to 9 destinations in Cuba except for Havana https://t.co/KPPQ...\n\n"
],
[
"for tweet_id in trump_archive - wayback:\n url = \"https://www.thetrumparchive.com/tweets/{}\".format(tweet_id)\n resp = requests.get(url)\n if resp.status_code == 200:\n tweet = resp.json()\n if not re.match(r'^\"?RT', tweet['text']):\n print(\"id: {}\\ndate: {}\\ndeleted: {}\\n{}\\n\".format(\n tweet['id'], \n tweet['date'],\n tweet['isDeleted'],\n tweet['text']\n ))",
"id: 1237490722317746178\ndate: 2020-03-10T21:29:14.000Z\ndeleted: True\nIf you like automobils, how can you vote for a Democrat who all want to get rid of cars, as quickly as possible, especially if they are powered by gasoline. Remember also, no more than one car per family. I, on the other hand, have new plants being built all over Michigan, Plus!\n\nid: 1281926278845812736\ndate: 2020-07-11T12:20:16.000Z\ndeleted: True\nRoger Stone was targeted by an illegal Witch Hunt tha never should have taken place. It is the other side that are criminals, including the fact that Biden and Obama illegally spied on my campaign - AND GOT CAUGHT!\n\nid: 1331749395214311427\ndate: 2020-11-25T23:59:33.000Z\ndeleted: True\nhttps://t.co/FJ6UcmXq5G?\n\n"
]
],
[
[
"We can verify by looking in our database for a tweet id like 1281926278845812736:",
"_____no_output_____"
]
],
[
[
"db.execute('SELECT * FROM tweets WHERE tweet_id = ?', [\"1281926278845812736\"]).fetchall()",
"_____no_output_____"
]
],
[
[
"Sure enough, it looks like Internet Archive wasn't quite quick enough to pick this one up. It's hard to say when the tweet was deleted, but it was archived on 2020-11-15 which was well after when it was sent on 2020-07-11. \n\nBut this is truly remarkable that the Wayback Machine only seems to be missing three original tweets (non-retweets), at least with respect with the Trump Archive. But since the Trump Archive appears to be missing at least some content that is present in the Wayback Machine its not exactly clear how accurate this is. In the end this highlights why it is important for Twitter to make an archival snapshot available.",
"_____no_output_____"
],
[
"## Archiving Activity\n\nWe can use our little SQLite database to plot the archiving activity related to Trump's tweets over time.",
"_____no_output_____"
]
],
[
[
"sql = \\\n '''\n SELECT\n STRFTIME('%Y%m%d', timestamp) AS day,\n COUNT(*) AS \"snapshots\"\n FROM tweets\n GROUP BY day\n ORDER BY day ASC\n '''\n\ndf = pandas.read_sql_query(sql, db, parse_dates=['day'])\ndf.head()",
"_____no_output_____"
]
],
[
[
"Lets fill in the blanks for days where there was no archiving of Trump's tweets.",
"_____no_output_____"
]
],
[
[
"dates = pandas.date_range(min(df.day), max(df.day))\ndf = df.set_index('day').reindex(dates).fillna(0)\ndf.head()",
"_____no_output_____"
]
],
[
[
"Now we can try a plot!",
"_____no_output_____"
]
],
[
[
"df.plot(\n kind='line',\n title=\"Archiving Trump's Tweets at the Internet Archive\",\n figsize=(10, 4),\n legend=False,\n xlabel='Time',\n ylabel='Snapshots per Day'\n)",
"_____no_output_____"
]
],
[
[
"Kinda noisy. Maybe it will look better as tweets-per-week?",
"_____no_output_____"
]
],
[
[
"df = df.resample('W').sum().rename_axis('time')\ndf.plot(\n kind='line',\n title=\"Archiving Trump's Tweets at the Internet Archive\",\n figsize=(10, 4),\n legend=False,\n xlabel='Time',\n ylabel='Snapshots per Week'\n)",
"_____no_output_____"
]
],
[
[
"## Trump Archive URLs\n\nTo help media organizations update their links to point at snapshots at the Internet Archive I thought it could be useful to create a CSV dataset of the tweet ids and links. It's important to limit to known good tweets (within a particular range) and ones that returned a 200 OK. The latest snapshot will provide a picture of what interaction with that tweet looked like when the tweets were removed.\n\nThe situation is a bit tricky because just because there is a 200 OK response for a tweet URL in the Internet Archive doesn't mean its a good one to link to. For example this one seems to be OK but doesn't render: because of playback issues.\n\nhttps://web.archive.org/web/20201112035441/https://twitter.com/realdonaldtrump/status/1698308935?s=21\n\nWhat we can do is create a little function to make sure that it renders:",
"_____no_output_____"
]
],
[
[
"import requests_html\n\nhttp = requests_html.AsyncHTMLSession()\n\nasync def response_ok(url, tries=10):\n global http\n try:\n resp = await http.get(url)\n await resp.html.arender(timeout=60)\n match = resp.html.search(\"Something went wrong\")\n if match:\n return False\n return True\n except Exception as e:\n if tries == 0:\n raise e\n else:\n http = requests_html.AsyncHTMLSession()\n return await response_ok(url, tries - 1)",
"_____no_output_____"
],
[
"await response_ok('https://web.archive.org/web/20201112035441/https://twitter.com/realdonaldtrump/status/1698308935?s=21')",
"_____no_output_____"
],
[
"await response_ok('https://web.archive.org/web/20201106114341/https://twitter.com/realDonaldTrump/status/1776419923')",
"_____no_output_____"
],
[
"sql = \\\n \"\"\"\n SELECT DISTINCT(CAST(tweet_id AS NUMERIC)) AS tweet_num\n FROM tweets\n WHERE tweet_num > 1698308934\n AND tweet_num < 1351984482019115009\n ORDER BY tweet_num ASC\n \"\"\"\n\nout = csv.writer(open('data/trump-tweet-archive.csv', 'w'))\nout.writerow(['tweet_url', 'archive_url'])\n\ncount = 0\nfor row in db.execute(sql):\n tweet_id = row[0]\n sql = \\\n \"\"\"\n SELECT url, STRFTIME('%Y%m%d%H%M%S', timestamp)\n FROM tweets\n WHERE tweet_id = ?\n AND status_code = 200\n ORDER BY timestamp DESC\n \"\"\"\n for [url, timestamp] in db.execute(sql, [tweet_id]):\n archive_url = 'https://web.archive.org/web/{}/{}'.format(timestamp, url)\n print('checking {}'.format(archive_url))\n if await response_ok(archive_url):\n tweet_url = 'https://twitter.com/realDonaldTrump/status/{}'.format(tweet_id)\n print('ok {} {}'.format(tweet_url, archive_url))\n out.writerow([tweet_url, archive_url])\n break",
"checking https://web.archive.org/web/20201112035441/https://twitter.com/realdonaldtrump/status/1698308935?s=21\nchecking https://web.archive.org/web/20201112032934/https://twitter.com/realdonaldtrump/status/1698308935\nchecking https://web.archive.org/web/20201112032721/https://twitter.com/realdonaldtrump/status/1698308935?lang=en\nchecking https://web.archive.org/web/20201108093337/https://twitter.com/realDonaldTrump/status/1698308935?ref_src=twsrc%5Etfw\nchecking https://web.archive.org/web/20201106114341/https://twitter.com/realDonaldTrump/status/1698308935\nok https://twitter.com/realDonaldTrump/status/1698308935 https://web.archive.org/web/20201106114341/https://twitter.com/realDonaldTrump/status/1698308935\nchecking https://web.archive.org/web/20201106114342/https://twitter.com/realDonaldTrump/status/1701461182\nok https://twitter.com/realDonaldTrump/status/1701461182 https://web.archive.org/web/20201106114342/https://twitter.com/realDonaldTrump/status/1701461182\nchecking https://web.archive.org/web/20201106114341/https://twitter.com/realDonaldTrump/status/1737479987\nok https://twitter.com/realDonaldTrump/status/1737479987 https://web.archive.org/web/20201106114341/https://twitter.com/realDonaldTrump/status/1737479987\nchecking https://web.archive.org/web/20201106114341/https://twitter.com/realDonaldTrump/status/1741160716\nok https://twitter.com/realDonaldTrump/status/1741160716 https://web.archive.org/web/20201106114341/https://twitter.com/realDonaldTrump/status/1741160716\nchecking https://web.archive.org/web/20201112023743/https://twitter.com/realDonaldTrump/status/1773561338?ref\nchecking https://web.archive.org/web/20201112003751/https://twitter.com/realDonaldTrump/status/1773561338?ref_src=twsrc%5Etfw%7Ctwcamp%5Etweetembed%7Ctwterm%5E1773561338\nchecking https://web.archive.org/web/20201109035432/https://twitter.com/realDonaldTrump/status/1773561338?ref_src=twsrc%5Etfw%7Ctwcamp%5Etweetembed%7Ctwterm%5E1773561338&ref_url=http%3A%2F%2Fdidtrumptweetit.com%2F1773561338-2%2F\nchecking https://web.archive.org/web/20201109033729/https://twitter.com/realDonaldTrump/status/1773561338?ref_src=twsrc%5Etfw%7Ctwcamp%5Etweetembed%7Ctwterm%5E1773561338&ref_url=http%3A%2F%2Fdidtrumptweetit.com%2F1773561338-2%2F\nchecking https://web.archive.org/web/20201108125104/https://twitter.com/realDonaldTrump/status/1773561338?s=09\nchecking https://web.archive.org/web/20201108111054/https://twitter.com/realDonaldTrump/status/1773561338/\nchecking https://web.archive.org/web/20201106114341/https://twitter.com/realDonaldTrump/status/1773561338\nok https://twitter.com/realDonaldTrump/status/1773561338 https://web.archive.org/web/20201106114341/https://twitter.com/realDonaldTrump/status/1773561338\nchecking https://web.archive.org/web/20201106114341/https://twitter.com/realDonaldTrump/status/1776419923\nok https://twitter.com/realDonaldTrump/status/1776419923 https://web.archive.org/web/20201106114341/https://twitter.com/realDonaldTrump/status/1776419923\nchecking https://web.archive.org/web/20201106114340/https://twitter.com/realDonaldTrump/status/1786560616\nok https://twitter.com/realDonaldTrump/status/1786560616 https://web.archive.org/web/20201106114340/https://twitter.com/realDonaldTrump/status/1786560616\nchecking https://web.archive.org/web/20201106114340/https://twitter.com/realDonaldTrump/status/1796477499\nok https://twitter.com/realDonaldTrump/status/1796477499 https://web.archive.org/web/20201106114340/https://twitter.com/realDonaldTrump/status/1796477499\nchecking https://web.archive.org/web/20201106114340/https://twitter.com/realDonaldTrump/status/1806258917\nok https://twitter.com/realDonaldTrump/status/1806258917 https://web.archive.org/web/20201106114340/https://twitter.com/realDonaldTrump/status/1806258917\nchecking https://web.archive.org/web/20201106114340/https://twitter.com/realDonaldTrump/status/1820624395\nok https://twitter.com/realDonaldTrump/status/1820624395 https://web.archive.org/web/20201106114340/https://twitter.com/realDonaldTrump/status/1820624395\nchecking https://web.archive.org/web/20201106114340/https://twitter.com/realDonaldTrump/status/1826225450\nok https://twitter.com/realDonaldTrump/status/1826225450 https://web.archive.org/web/20201106114340/https://twitter.com/realDonaldTrump/status/1826225450\nchecking https://web.archive.org/web/20201108122517/https://twitter.com/realDonaldTrump/status/1836131903\nchecking https://web.archive.org/web/20201106114339/https://twitter.com/realDonaldTrump/status/1836131903\nok https://twitter.com/realDonaldTrump/status/1836131903 https://web.archive.org/web/20201106114339/https://twitter.com/realDonaldTrump/status/1836131903\nchecking https://web.archive.org/web/20201106114340/https://twitter.com/realDonaldTrump/status/1849558306\nok https://twitter.com/realDonaldTrump/status/1849558306 https://web.archive.org/web/20201106114340/https://twitter.com/realDonaldTrump/status/1849558306\nchecking https://web.archive.org/web/20201106114338/https://twitter.com/realDonaldTrump/status/1859044981\nok https://twitter.com/realDonaldTrump/status/1859044981 https://web.archive.org/web/20201106114338/https://twitter.com/realDonaldTrump/status/1859044981\nchecking https://web.archive.org/web/20201106114339/https://twitter.com/realDonaldTrump/status/1864367186\nok https://twitter.com/realDonaldTrump/status/1864367186 https://web.archive.org/web/20201106114339/https://twitter.com/realDonaldTrump/status/1864367186\nchecking https://web.archive.org/web/20201111190555/https://twitter.com/realDonaldTrump/status/1878373267\nchecking https://web.archive.org/web/20201109040540/https://twitter.com/realdonaldtrump/status/1878373267\nchecking https://web.archive.org/web/20201106114339/https://twitter.com/realDonaldTrump/status/1878373267\nok https://twitter.com/realDonaldTrump/status/1878373267 https://web.archive.org/web/20201106114339/https://twitter.com/realDonaldTrump/status/1878373267\nchecking https://web.archive.org/web/20201106114338/https://twitter.com/realDonaldTrump/status/1884022748\nok https://twitter.com/realDonaldTrump/status/1884022748 https://web.archive.org/web/20201106114338/https://twitter.com/realDonaldTrump/status/1884022748\nchecking https://web.archive.org/web/20201106114337/https://twitter.com/realDonaldTrump/status/1894284587\nok https://twitter.com/realDonaldTrump/status/1894284587 https://web.archive.org/web/20201106114337/https://twitter.com/realDonaldTrump/status/1894284587\nchecking https://web.archive.org/web/20201106114337/https://twitter.com/realDonaldTrump/status/1924074459\nok https://twitter.com/realDonaldTrump/status/1924074459 https://web.archive.org/web/20201106114337/https://twitter.com/realDonaldTrump/status/1924074459\nchecking https://web.archive.org/web/20201106114339/https://twitter.com/realDonaldTrump/status/1936022874\nok https://twitter.com/realDonaldTrump/status/1936022874 https://web.archive.org/web/20201106114339/https://twitter.com/realDonaldTrump/status/1936022874\nchecking https://web.archive.org/web/20201109023507/https://twitter.com/realdonaldtrump/status/1949899014\nchecking https://web.archive.org/web/20201106114337/https://twitter.com/realDonaldTrump/status/1949899014\nok https://twitter.com/realDonaldTrump/status/1949899014 https://web.archive.org/web/20201106114337/https://twitter.com/realDonaldTrump/status/1949899014\nchecking https://web.archive.org/web/20201106114337/https://twitter.com/realDonaldTrump/status/2019316195\nok https://twitter.com/realDonaldTrump/status/2019316195 https://web.archive.org/web/20201106114337/https://twitter.com/realDonaldTrump/status/2019316195\nchecking https://web.archive.org/web/20201106114336/https://twitter.com/realDonaldTrump/status/2045871770\nok https://twitter.com/realDonaldTrump/status/2045871770 https://web.archive.org/web/20201106114336/https://twitter.com/realDonaldTrump/status/2045871770\nchecking https://web.archive.org/web/20201106114336/https://twitter.com/realDonaldTrump/status/2080633709\nok https://twitter.com/realDonaldTrump/status/2080633709 https://web.archive.org/web/20201106114336/https://twitter.com/realDonaldTrump/status/2080633709\nchecking https://web.archive.org/web/20201106114336/https://twitter.com/realDonaldTrump/status/2165353946\nok https://twitter.com/realDonaldTrump/status/2165353946 https://web.archive.org/web/20201106114336/https://twitter.com/realDonaldTrump/status/2165353946\n"
]
],
[
[
"## Top 10\n\nWhat were the top 10 most archived tweets?",
"_____no_output_____"
]
],
[
[
"import sqlite3\nimport pandas\ndb = sqlite3.connect('data/trump-tweets.sqlite3')\ndf = pandas.read_csv('data/trump-archive.csv')",
"_____no_output_____"
],
[
"def get_text(tweet_id):\n v = df[df['id'] == tweet_id].text.values\n if len(v) != 0:\n return v[0]\n else:\n return \"???\"\n\nget_text(1698308935)",
"_____no_output_____"
],
[
"sql = '''\n SELECT tweet_id, \n COUNT(*) AS total\n FROM tweets\n GROUP BY tweet_id\n ORDER By total DESC\n LIMIT 10\n '''\n\nfor [tweet_id, total] in db.execute(sql):\n print('* [{}]({}) {}'.format(\n get_text(int(tweet_id)),\n 'https://web.archive.org/web/*/https://twitter.com/realDonaldTrump/status/{}'.format(tweet_id),\n total\n ))",
"* [Thank you Alabama! #Trump2016#SuperTuesday](https://web.archive.org/web/*/https://twitter.com/realDonaldTrump/status/704834185471598592) 65489\n* [MAKE AMERICA GREAT AGAIN!](https://web.archive.org/web/*/https://twitter.com/realDonaldTrump/status/704756216157839360) 65360\n* [Thank you Georgia!#SuperTuesday #Trump2016](https://web.archive.org/web/*/https://twitter.com/realDonaldTrump/status/704818842153971712) 65358\n* [Such a beautiful and important evening! The forgotten man and woman will never be forgotten again. We will all come together as never before](https://web.archive.org/web/*/https://twitter.com/realDonaldTrump/status/796315640307060738) 55426\n* [Watching the returns at 9:45pm. #ElectionNight #MAGA🇺🇸 https://t.co/HfuJeRZbod](https://web.archive.org/web/*/https://twitter.com/realDonaldTrump/status/796182637622816768) 54889\n* [#ElectionDay https://t.co/MXrAxYnTjY https://t.co/FZhOncih21](https://web.archive.org/web/*/https://twitter.com/realDonaldTrump/status/796055597594578944) 54291\n* [I will be watching the election results from Trump Tower in Manhattan with my family and friends. Very exciting!](https://web.archive.org/web/*/https://twitter.com/realDonaldTrump/status/796099494442057728) 54263\n* [I love you North Carolina- thank you for your amazing support! Get out and https://t.co/HfihPERFgZ tomorrow!Watch:... https://t.co/jZzfqUZNYh](https://web.archive.org/web/*/https://twitter.com/realDonaldTrump/status/795733366842806272) 54252\n* [Still time to #VoteTrump! #iVoted #ElectionNight https://t.co/UZtYAY1Ba6](https://web.archive.org/web/*/https://twitter.com/realDonaldTrump/status/796126077647196160) 54224\n* [Watching my beautiful wife, Melania, speak about our love of country and family. We will make you all very proud.... https://t.co/DiKmSnTlC2](https://web.archive.org/web/*/https://twitter.com/realDonaldTrump/status/794259252613414915) 54100\n"
]
],
[
[
"## Politwoops\n\nJust as a last exercise its interesting to see which tweets in Politwoops for Trump are in (or not in the Internet Archive). We saw one of them above when we were analyzing The Trump Twitter Archive.\n\nFirst we need all the politwoops ids. We can use Politwoops API:",
"_____no_output_____"
]
],
[
[
"import requests\n\npolitwoops = set()\n\npage = 1\nwhile True:\n url = \"https://projects.propublica.org/politwoops/user/realDonaldTrump\"\n data = requests.get(url, params={\"format\": \"json\", \"page\": page}).json()\n if not data or len(data[\"tweets\"]) == 0:\n break\n for tweet in data[\"tweets\"]:\n politwoops.add(tweet[\"id\"])\n page += 1",
"_____no_output_____"
],
[
"len(politwoops)",
"_____no_output_____"
],
[
"wayback_missing = politwoops - wayback\nlen(wayback_missing)",
"_____no_output_____"
],
[
"len(wayback_missing) / len(politwoops)",
"_____no_output_____"
]
],
[
[
"So it looks like there are 179 tweets in Politwoops that are missing from Wayback Machine? Lets take a look at the URLs to spot check a few.",
"_____no_output_____"
]
],
[
[
"for tweet_id in wayback_missing:\n politwoops_url = \"https://projects.propublica.org/politwoops/tweet/{}\".format(tweet_id)\n wayback_url = \"https://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/{}\".format(tweet_id)\n print(politwoops_url)\n print(wayback_url)\n print()",
"https://projects.propublica.org/politwoops/tweet/1228437662094196743\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1228437662094196743\n\nhttps://projects.propublica.org/politwoops/tweet/1193376950481489921\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1193376950481489921\n\nhttps://projects.propublica.org/politwoops/tweet/779769798217592832\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/779769798217592832\n\nhttps://projects.propublica.org/politwoops/tweet/1249505460761362432\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1249505460761362432\n\nhttps://projects.propublica.org/politwoops/tweet/767173043185737728\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/767173043185737728\n\nhttps://projects.propublica.org/politwoops/tweet/708182558840737792\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/708182558840737792\n\nhttps://projects.propublica.org/politwoops/tweet/1035905551039909890\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1035905551039909890\n\nhttps://projects.propublica.org/politwoops/tweet/979109801694244866\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/979109801694244866\n\nhttps://projects.propublica.org/politwoops/tweet/769250207036944384\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/769250207036944384\n\nhttps://projects.propublica.org/politwoops/tweet/1168652410128076801\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1168652410128076801\n\nhttps://projects.propublica.org/politwoops/tweet/1084761040291282944\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1084761040291282944\n\nhttps://projects.propublica.org/politwoops/tweet/729774883387912194\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/729774883387912194\n\nhttps://projects.propublica.org/politwoops/tweet/1338329628616830977\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1338329628616830977\n\nhttps://projects.propublica.org/politwoops/tweet/1328882445886902272\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1328882445886902272\n\nhttps://projects.propublica.org/politwoops/tweet/1288503931224633345\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1288503931224633345\n\nhttps://projects.propublica.org/politwoops/tweet/1331749395214311427\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1331749395214311427\n\nhttps://projects.propublica.org/politwoops/tweet/914261144415633409\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/914261144415633409\n\nhttps://projects.propublica.org/politwoops/tweet/1315287703391477760\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1315287703391477760\n\nhttps://projects.propublica.org/politwoops/tweet/1023904015887880192\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1023904015887880192\n\nhttps://projects.propublica.org/politwoops/tweet/716077569171390464\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/716077569171390464\n\nhttps://projects.propublica.org/politwoops/tweet/1304117284647186432\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1304117284647186432\n\nhttps://projects.propublica.org/politwoops/tweet/781247769499402240\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/781247769499402240\n\nhttps://projects.propublica.org/politwoops/tweet/1003697618642448384\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1003697618642448384\n\nhttps://projects.propublica.org/politwoops/tweet/1014191947265773570\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1014191947265773570\n\nhttps://projects.propublica.org/politwoops/tweet/1270924796931592192\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1270924796931592192\n\nhttps://projects.propublica.org/politwoops/tweet/1254476088526413825\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1254476088526413825\n\nhttps://projects.propublica.org/politwoops/tweet/716019359597137920\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/716019359597137920\n\nhttps://projects.propublica.org/politwoops/tweet/1003697643770601472\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1003697643770601472\n\nhttps://projects.propublica.org/politwoops/tweet/887795354573434880\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/887795354573434880\n\nhttps://projects.propublica.org/politwoops/tweet/1195442170255302657\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1195442170255302657\n\nhttps://projects.propublica.org/politwoops/tweet/705419616869339137\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/705419616869339137\n\nhttps://projects.propublica.org/politwoops/tweet/1203128389466497027\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1203128389466497027\n\nhttps://projects.propublica.org/politwoops/tweet/1293293043978776583\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1293293043978776583\n\nhttps://projects.propublica.org/politwoops/tweet/962517130943004672\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/962517130943004672\n\nhttps://projects.propublica.org/politwoops/tweet/722444029288427520\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/722444029288427520\n\nhttps://projects.propublica.org/politwoops/tweet/777615824848912384\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/777615824848912384\n\nhttps://projects.propublica.org/politwoops/tweet/760537036608577537\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/760537036608577537\n\nhttps://projects.propublica.org/politwoops/tweet/903363022663671808\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/903363022663671808\n\nhttps://projects.propublica.org/politwoops/tweet/1031620305196867585\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1031620305196867585\n\nhttps://projects.propublica.org/politwoops/tweet/1038534852293873666\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1038534852293873666\n\nhttps://projects.propublica.org/politwoops/tweet/1058355789264535552\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1058355789264535552\n\nhttps://projects.propublica.org/politwoops/tweet/1213148993745760256\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1213148993745760256\n\nhttps://projects.propublica.org/politwoops/tweet/1268259684978495490\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1268259684978495490\n\nhttps://projects.propublica.org/politwoops/tweet/1326910731237928962\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1326910731237928962\n\nhttps://projects.propublica.org/politwoops/tweet/722510216768712705\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/722510216768712705\n\nhttps://projects.propublica.org/politwoops/tweet/716727234179031040\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/716727234179031040\n\nhttps://projects.propublica.org/politwoops/tweet/1254449897031548928\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1254449897031548928\n\nhttps://projects.propublica.org/politwoops/tweet/948548809579261953\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/948548809579261953\n\nhttps://projects.propublica.org/politwoops/tweet/1243343887294488576\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1243343887294488576\n\nhttps://projects.propublica.org/politwoops/tweet/1326909956994568194\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1326909956994568194\n\nhttps://projects.propublica.org/politwoops/tweet/1166355047409754112\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1166355047409754112\n\nhttps://projects.propublica.org/politwoops/tweet/704747986446913541\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/704747986446913541\n\nhttps://projects.propublica.org/politwoops/tweet/700405212540854272\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/700405212540854272\n\nhttps://projects.propublica.org/politwoops/tweet/1336344843765866496\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1336344843765866496\n\nhttps://projects.propublica.org/politwoops/tweet/1191510920612241414\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1191510920612241414\n\nhttps://projects.propublica.org/politwoops/tweet/1299378312133914626\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1299378312133914626\n\nhttps://projects.propublica.org/politwoops/tweet/735656129053609984\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/735656129053609984\n\nhttps://projects.propublica.org/politwoops/tweet/1346929192420704258\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1346929192420704258\n\nhttps://projects.propublica.org/politwoops/tweet/1190375141433389064\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1190375141433389064\n\nhttps://projects.propublica.org/politwoops/tweet/1210204495206064135\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1210204495206064135\n\nhttps://projects.propublica.org/politwoops/tweet/1065952653522452482\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1065952653522452482\n\nhttps://projects.propublica.org/politwoops/tweet/930417340705067008\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/930417340705067008\n\nhttps://projects.propublica.org/politwoops/tweet/1190375037100023812\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1190375037100023812\n\nhttps://projects.propublica.org/politwoops/tweet/1014525769182253057\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1014525769182253057\n\nhttps://projects.propublica.org/politwoops/tweet/1268344749091684352\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1268344749091684352\n\nhttps://projects.propublica.org/politwoops/tweet/1268344826505953281\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1268344826505953281\n\nhttps://projects.propublica.org/politwoops/tweet/1062445863207886848\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1062445863207886848\n\nhttps://projects.propublica.org/politwoops/tweet/1337152292596965376\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1337152292596965376\n\nhttps://projects.propublica.org/politwoops/tweet/1228437662178140160\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1228437662178140160\n\nhttps://projects.propublica.org/politwoops/tweet/1004162564278964224\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1004162564278964224\n\nhttps://projects.propublica.org/politwoops/tweet/946151438765445121\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/946151438765445121\n\nhttps://projects.propublica.org/politwoops/tweet/1317058851003420672\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1317058851003420672\n\nhttps://projects.propublica.org/politwoops/tweet/1221089928035041283\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1221089928035041283\n\nhttps://projects.propublica.org/politwoops/tweet/698528140767834113\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/698528140767834113\n\nhttps://projects.propublica.org/politwoops/tweet/1341906561682309122\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1341906561682309122\n\nhttps://projects.propublica.org/politwoops/tweet/1187936132241219584\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1187936132241219584\n\nhttps://projects.propublica.org/politwoops/tweet/699732175298625538\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/699732175298625538\n\nhttps://projects.propublica.org/politwoops/tweet/1280693411666898945\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1280693411666898945\n\nhttps://projects.propublica.org/politwoops/tweet/1242580387165069315\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1242580387165069315\n\nhttps://projects.propublica.org/politwoops/tweet/926727074248777728\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/926727074248777728\n\nhttps://projects.propublica.org/politwoops/tweet/1020001936614010880\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1020001936614010880\n\nhttps://projects.propublica.org/politwoops/tweet/1185925254658646022\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1185925254658646022\n\nhttps://projects.propublica.org/politwoops/tweet/1289556873402695686\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1289556873402695686\n\nhttps://projects.propublica.org/politwoops/tweet/708410492583288833\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/708410492583288833\n\nhttps://projects.propublica.org/politwoops/tweet/719647781414170625\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/719647781414170625\n\nhttps://projects.propublica.org/politwoops/tweet/1010569678232489984\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1010569678232489984\n\nhttps://projects.propublica.org/politwoops/tweet/1339910340537139200\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1339910340537139200\n\nhttps://projects.propublica.org/politwoops/tweet/867049454414823425\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/867049454414823425\n\nhttps://projects.propublica.org/politwoops/tweet/1020826224401231876\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1020826224401231876\n\nhttps://projects.propublica.org/politwoops/tweet/1233223434940952577\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1233223434940952577\n\nhttps://projects.propublica.org/politwoops/tweet/1022588302241816576\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1022588302241816576\n\nhttps://projects.propublica.org/politwoops/tweet/1213148769547669504\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1213148769547669504\n\nhttps://projects.propublica.org/politwoops/tweet/774400741872050176\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/774400741872050176\n\nhttps://projects.propublica.org/politwoops/tweet/712856605243650051\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/712856605243650051\n\nhttps://projects.propublica.org/politwoops/tweet/1243623845220831233\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1243623845220831233\n\nhttps://projects.propublica.org/politwoops/tweet/972496257032839169\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/972496257032839169\n\nhttps://projects.propublica.org/politwoops/tweet/1077002502422360070\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1077002502422360070\n\nhttps://projects.propublica.org/politwoops/tweet/900850478908669952\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/900850478908669952\n\nhttps://projects.propublica.org/politwoops/tweet/1013883352380780544\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1013883352380780544\n\nhttps://projects.propublica.org/politwoops/tweet/1241815411991228421\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1241815411991228421\n\nhttps://projects.propublica.org/politwoops/tweet/710138349818912768\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/710138349818912768\n\nhttps://projects.propublica.org/politwoops/tweet/748653033039482880\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/748653033039482880\n\nhttps://projects.propublica.org/politwoops/tweet/698511394245840897\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/698511394245840897\n\nhttps://projects.propublica.org/politwoops/tweet/1122179275644440581\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1122179275644440581\n\nhttps://projects.propublica.org/politwoops/tweet/697918761207201793\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/697918761207201793\n\nhttps://projects.propublica.org/politwoops/tweet/695801249732694017\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/695801249732694017\n\nhttps://projects.propublica.org/politwoops/tweet/701480395909496834\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/701480395909496834\n\nhttps://projects.propublica.org/politwoops/tweet/697038423429767169\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/697038423429767169\n\nhttps://projects.propublica.org/politwoops/tweet/1331289633662099457\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1331289633662099457\n\nhttps://projects.propublica.org/politwoops/tweet/1249101518243942400\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1249101518243942400\n\nhttps://projects.propublica.org/politwoops/tweet/1135614206936604673\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1135614206936604673\n\nhttps://projects.propublica.org/politwoops/tweet/1248473431512768512\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1248473431512768512\n\nhttps://projects.propublica.org/politwoops/tweet/984482786424905734\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/984482786424905734\n\nhttps://projects.propublica.org/politwoops/tweet/1247636428521197568\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1247636428521197568\n\nhttps://projects.propublica.org/politwoops/tweet/710924145584771072\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/710924145584771072\n\nhttps://projects.propublica.org/politwoops/tweet/712343134664060929\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/712343134664060929\n\nhttps://projects.propublica.org/politwoops/tweet/1253685287227703301\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1253685287227703301\n\nhttps://projects.propublica.org/politwoops/tweet/730452843983998976\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/730452843983998976\n\nhttps://projects.propublica.org/politwoops/tweet/1127897078183731200\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1127897078183731200\n\nhttps://projects.propublica.org/politwoops/tweet/1243620215310757890\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1243620215310757890\n\nhttps://projects.propublica.org/politwoops/tweet/973709204891484160\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/973709204891484160\n\nhttps://projects.propublica.org/politwoops/tweet/1190375037112635395\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1190375037112635395\n\nhttps://projects.propublica.org/politwoops/tweet/1306338627258114049\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1306338627258114049\n\nhttps://projects.propublica.org/politwoops/tweet/1189193274445185025\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1189193274445185025\n\nhttps://projects.propublica.org/politwoops/tweet/1003697586098921472\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1003697586098921472\n\nhttps://projects.propublica.org/politwoops/tweet/1281926278845812736\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1281926278845812736\n\nhttps://projects.propublica.org/politwoops/tweet/1241393590242283522\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1241393590242283522\n\nhttps://projects.propublica.org/politwoops/tweet/700096279389605889\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/700096279389605889\n\nhttps://projects.propublica.org/politwoops/tweet/997256977192509440\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/997256977192509440\n\nhttps://projects.propublica.org/politwoops/tweet/777614991121846278\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/777614991121846278\n\nhttps://projects.propublica.org/politwoops/tweet/1246453101265063937\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1246453101265063937\n\nhttps://projects.propublica.org/politwoops/tweet/698262930584965120\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/698262930584965120\n\nhttps://projects.propublica.org/politwoops/tweet/1151866148616626180\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1151866148616626180\n\nhttps://projects.propublica.org/politwoops/tweet/1336401395554127877\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1336401395554127877\n\nhttps://projects.propublica.org/politwoops/tweet/771299405563588608\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/771299405563588608\n\nhttps://projects.propublica.org/politwoops/tweet/1242580319397531649\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1242580319397531649\n\nhttps://projects.propublica.org/politwoops/tweet/933039443476451329\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/933039443476451329\n\nhttps://projects.propublica.org/politwoops/tweet/1195442170255282177\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1195442170255282177\n\nhttps://projects.propublica.org/politwoops/tweet/1260035250740051975\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1260035250740051975\n\nhttps://projects.propublica.org/politwoops/tweet/927855529455648768\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/927855529455648768\n\nhttps://projects.propublica.org/politwoops/tweet/700122361065009158\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/700122361065009158\n\nhttps://projects.propublica.org/politwoops/tweet/1336344876082995200\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1336344876082995200\n\nhttps://projects.propublica.org/politwoops/tweet/755810123327561728\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/755810123327561728\n\nhttps://projects.propublica.org/politwoops/tweet/1213148881514639361\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1213148881514639361\n\nhttps://projects.propublica.org/politwoops/tweet/1254476023502159873\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1254476023502159873\n\nhttps://projects.propublica.org/politwoops/tweet/1243880517763809280\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1243880517763809280\n\nhttps://projects.propublica.org/politwoops/tweet/1224170336234758145\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1224170336234758145\n\nhttps://projects.propublica.org/politwoops/tweet/986396478078947328\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/986396478078947328\n\nhttps://projects.propublica.org/politwoops/tweet/1303834261087301632\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1303834261087301632\n\nhttps://projects.propublica.org/politwoops/tweet/1168652391111122947\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1168652391111122947\n\nhttps://projects.propublica.org/politwoops/tweet/969532384285687809\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/969532384285687809\n\nhttps://projects.propublica.org/politwoops/tweet/1151866161199554560\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1151866161199554560\n\nhttps://projects.propublica.org/politwoops/tweet/936399765742538752\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/936399765742538752\n\nhttps://projects.propublica.org/politwoops/tweet/1334001254012497923\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1334001254012497923\n\nhttps://projects.propublica.org/politwoops/tweet/999351128260259856\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/999351128260259856\n\nhttps://projects.propublica.org/politwoops/tweet/983764752538185728\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/983764752538185728\n\nhttps://projects.propublica.org/politwoops/tweet/1207659180380823552\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1207659180380823552\n\nhttps://projects.propublica.org/politwoops/tweet/1235364334198575104\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1235364334198575104\n\nhttps://projects.propublica.org/politwoops/tweet/997083751220940800\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/997083751220940800\n\nhttps://projects.propublica.org/politwoops/tweet/697099036013281280\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/697099036013281280\n\nhttps://projects.propublica.org/politwoops/tweet/1022873468197584901\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1022873468197584901\n\nhttps://projects.propublica.org/politwoops/tweet/756319896431128577\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/756319896431128577\n\nhttps://projects.propublica.org/politwoops/tweet/700404921913335808\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/700404921913335808\n\nhttps://projects.propublica.org/politwoops/tweet/1013868536375205891\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1013868536375205891\n\nhttps://projects.propublica.org/politwoops/tweet/713005071664091136\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/713005071664091136\n\nhttps://projects.propublica.org/politwoops/tweet/1190375037217447936\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1190375037217447936\n\nhttps://projects.propublica.org/politwoops/tweet/1330238111419428865\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1330238111419428865\n\nhttps://projects.propublica.org/politwoops/tweet/1211775130134831104\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1211775130134831104\n\nhttps://projects.propublica.org/politwoops/tweet/774384873570054144\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/774384873570054144\n\nhttps://projects.propublica.org/politwoops/tweet/1062455493216612353\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1062455493216612353\n\nhttps://projects.propublica.org/politwoops/tweet/697276230060666881\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/697276230060666881\n\nhttps://projects.propublica.org/politwoops/tweet/1127897052040630274\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1127897052040630274\n\nhttps://projects.propublica.org/politwoops/tweet/962470280554864641\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/962470280554864641\n\nhttps://projects.propublica.org/politwoops/tweet/1209466101018628097\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1209466101018628097\n\nhttps://projects.propublica.org/politwoops/tweet/1246883274389159937\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1246883274389159937\n\nhttps://projects.propublica.org/politwoops/tweet/1035700678969237506\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1035700678969237506\n\nhttps://projects.propublica.org/politwoops/tweet/1127897033828962304\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1127897033828962304\n\nhttps://projects.propublica.org/politwoops/tweet/1316957729571864578\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1316957729571864578\n\nhttps://projects.propublica.org/politwoops/tweet/1202274283382362112\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1202274283382362112\n\n"
]
],
[
[
"Looking at some of these it becomes clear that politwoops are lumping together realDonaldTrump and POTUS. But we didn't collect Wayback data for POTUS. We can collect the Politwoops data again but filter out the POTUS data.",
"_____no_output_____"
]
],
[
[
"politwoops = set()\n\npage = 1\nwhile True:\n url = \"https://projects.propublica.org/politwoops/user/realDonaldTrump\"\n data = requests.get(url, params={\"format\": \"json\", \"page\": page}).json()\n if not data or len(data[\"tweets\"]) == 0:\n break\n for tweet in data[\"tweets\"]:\n # make sure the user is realdonaldtrump and not potus\n if tweet[\"user_name\"].lower() == \"realdonaldtrump\":\n politwoops.add(tweet[\"id\"])\n page += 1",
"_____no_output_____"
],
[
"len(politwoops)",
"_____no_output_____"
],
[
"wayback_missing = politwoops - wayback\nlen(wayback_missing)",
"_____no_output_____"
],
[
"for tweet_id in wayback_missing:\n politwoops_url = \"https://projects.propublica.org/politwoops/tweet/{}\".format(tweet_id)\n wayback_url = \"https://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/{}\".format(tweet_id)\n print(politwoops_url)\n print(wayback_url)\n print()",
"https://projects.propublica.org/politwoops/tweet/1326909956994568194\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1326909956994568194\n\nhttps://projects.propublica.org/politwoops/tweet/1336401395554127877\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1336401395554127877\n\nhttps://projects.propublica.org/politwoops/tweet/1193376950481489921\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1193376950481489921\n\nhttps://projects.propublica.org/politwoops/tweet/704747986446913541\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/704747986446913541\n\nhttps://projects.propublica.org/politwoops/tweet/700405212540854272\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/700405212540854272\n\nhttps://projects.propublica.org/politwoops/tweet/779769798217592832\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/779769798217592832\n\nhttps://projects.propublica.org/politwoops/tweet/767173043185737728\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/767173043185737728\n\nhttps://projects.propublica.org/politwoops/tweet/771299405563588608\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/771299405563588608\n\nhttps://projects.propublica.org/politwoops/tweet/708182558840737792\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/708182558840737792\n\nhttps://projects.propublica.org/politwoops/tweet/1336344843765866496\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1336344843765866496\n\nhttps://projects.propublica.org/politwoops/tweet/774400741872050176\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/774400741872050176\n\nhttps://projects.propublica.org/politwoops/tweet/712856605243650051\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/712856605243650051\n\nhttps://projects.propublica.org/politwoops/tweet/769250207036944384\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/769250207036944384\n\nhttps://projects.propublica.org/politwoops/tweet/700122361065009158\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/700122361065009158\n\nhttps://projects.propublica.org/politwoops/tweet/1084761040291282944\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1084761040291282944\n\nhttps://projects.propublica.org/politwoops/tweet/729774883387912194\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/729774883387912194\n\nhttps://projects.propublica.org/politwoops/tweet/735656129053609984\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/735656129053609984\n\nhttps://projects.propublica.org/politwoops/tweet/1338329628616830977\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1338329628616830977\n\nhttps://projects.propublica.org/politwoops/tweet/1336344876082995200\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1336344876082995200\n\nhttps://projects.propublica.org/politwoops/tweet/755810123327561728\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/755810123327561728\n\nhttps://projects.propublica.org/politwoops/tweet/1328882445886902272\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1328882445886902272\n\nhttps://projects.propublica.org/politwoops/tweet/1331749395214311427\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1331749395214311427\n\nhttps://projects.propublica.org/politwoops/tweet/710138349818912768\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/710138349818912768\n\nhttps://projects.propublica.org/politwoops/tweet/698511394245840897\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/698511394245840897\n\nhttps://projects.propublica.org/politwoops/tweet/969532384285687809\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/969532384285687809\n\nhttps://projects.propublica.org/politwoops/tweet/697918761207201793\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/697918761207201793\n\nhttps://projects.propublica.org/politwoops/tweet/716077569171390464\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/716077569171390464\n\nhttps://projects.propublica.org/politwoops/tweet/701480395909496834\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/701480395909496834\n\nhttps://projects.propublica.org/politwoops/tweet/695801249732694017\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/695801249732694017\n\nhttps://projects.propublica.org/politwoops/tweet/781247769499402240\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/781247769499402240\n\nhttps://projects.propublica.org/politwoops/tweet/697038423429767169\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/697038423429767169\n\nhttps://projects.propublica.org/politwoops/tweet/1331289633662099457\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1331289633662099457\n\nhttps://projects.propublica.org/politwoops/tweet/1334001254012497923\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1334001254012497923\n\nhttps://projects.propublica.org/politwoops/tweet/1248473431512768512\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1248473431512768512\n\nhttps://projects.propublica.org/politwoops/tweet/698262930584965120\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/698262930584965120\n\nhttps://projects.propublica.org/politwoops/tweet/697099036013281280\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/697099036013281280\n\nhttps://projects.propublica.org/politwoops/tweet/698528140767834113\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/698528140767834113\n\nhttps://projects.propublica.org/politwoops/tweet/1317058851003420672\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1317058851003420672\n\nhttps://projects.propublica.org/politwoops/tweet/716019359597137920\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/716019359597137920\n\nhttps://projects.propublica.org/politwoops/tweet/1341906561682309122\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1341906561682309122\n\nhttps://projects.propublica.org/politwoops/tweet/705419616869339137\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/705419616869339137\n\nhttps://projects.propublica.org/politwoops/tweet/710924145584771072\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/710924145584771072\n\nhttps://projects.propublica.org/politwoops/tweet/756319896431128577\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/756319896431128577\n\nhttps://projects.propublica.org/politwoops/tweet/700404921913335808\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/700404921913335808\n\nhttps://projects.propublica.org/politwoops/tweet/712343134664060929\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/712343134664060929\n\nhttps://projects.propublica.org/politwoops/tweet/1187936132241219584\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1187936132241219584\n\nhttps://projects.propublica.org/politwoops/tweet/699732175298625538\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/699732175298625538\n\nhttps://projects.propublica.org/politwoops/tweet/713005071664091136\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/713005071664091136\n\nhttps://projects.propublica.org/politwoops/tweet/722444029288427520\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/722444029288427520\n\nhttps://projects.propublica.org/politwoops/tweet/730452843983998976\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/730452843983998976\n\nhttps://projects.propublica.org/politwoops/tweet/777615824848912384\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/777615824848912384\n\nhttps://projects.propublica.org/politwoops/tweet/1330238111419428865\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1330238111419428865\n\nhttps://projects.propublica.org/politwoops/tweet/760537036608577537\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/760537036608577537\n\nhttps://projects.propublica.org/politwoops/tweet/774384873570054144\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/774384873570054144\n\nhttps://projects.propublica.org/politwoops/tweet/697276230060666881\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/697276230060666881\n\nhttps://projects.propublica.org/politwoops/tweet/708410492583288833\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/708410492583288833\n\nhttps://projects.propublica.org/politwoops/tweet/719647781414170625\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/719647781414170625\n\nhttps://projects.propublica.org/politwoops/tweet/1035700678969237506\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1035700678969237506\n\nhttps://projects.propublica.org/politwoops/tweet/1339910340537139200\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1339910340537139200\n\nhttps://projects.propublica.org/politwoops/tweet/1316957729571864578\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1316957729571864578\n\nhttps://projects.propublica.org/politwoops/tweet/1281926278845812736\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1281926278845812736\n\nhttps://projects.propublica.org/politwoops/tweet/1326910731237928962\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/1326910731237928962\n\nhttps://projects.propublica.org/politwoops/tweet/700096279389605889\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/700096279389605889\n\nhttps://projects.propublica.org/politwoops/tweet/722510216768712705\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/722510216768712705\n\nhttps://projects.propublica.org/politwoops/tweet/777614991121846278\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/777614991121846278\n\nhttps://projects.propublica.org/politwoops/tweet/716727234179031040\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/716727234179031040\n\nhttps://projects.propublica.org/politwoops/tweet/948548809579261953\nhttps://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/948548809579261953\n\n"
],
[
"out = csv.writer(open('data/trump-politwoops-wayback.csv', 'w'))\nout.writerow(['tweet_id', 'politwoops_url', 'wayback_url'])\n\nfor tweet_id in wayback_missing:\n politwoops_url = \"https://projects.propublica.org/politwoops/tweet/{}\".format(tweet_id)\n wayback_url = \"https://web.archive.org/web/*/http://twitter.com/realDonaldTrump/status/{}\".format(tweet_id)\n out.writerow([tweet_id, politwoops_url, wayback_url])",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
d0cfba084c6ca9275d4ce19abd5db1873d20943a | 42,530 | ipynb | Jupyter Notebook | Stock Sentiment Analysis.ipynb | FGpramodgupta/Stock-News-Sentiment-Analysis | 11fdcc346550fddf51c343cd2499376904c2fd65 | [
"Apache-2.0"
] | null | null | null | Stock Sentiment Analysis.ipynb | FGpramodgupta/Stock-News-Sentiment-Analysis | 11fdcc346550fddf51c343cd2499376904c2fd65 | [
"Apache-2.0"
] | null | null | null | Stock Sentiment Analysis.ipynb | FGpramodgupta/Stock-News-Sentiment-Analysis | 11fdcc346550fddf51c343cd2499376904c2fd65 | [
"Apache-2.0"
] | null | null | null | 43.531218 | 922 | 0.431554 | [
[
[
"## Stock Sentiment Analysis using News Headlines",
"_____no_output_____"
]
],
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"df=pd.read_csv('Data.csv', encoding = \"ISO-8859-1\")",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"train = df[df['Date'] < '20150101']\ntest = df[df['Date'] > '20141231']",
"_____no_output_____"
],
[
"# Removing punctuations\ndata=train.iloc[:,2:27]\ndata.replace(\"[^a-zA-Z]\",\" \",regex=True, inplace=True)\n\n# Renaming column names for ease of access\nlist1= [i for i in range(25)]\nnew_Index=[str(i) for i in list1]\ndata.columns= new_Index\ndata.head(5)\n\n",
"_____no_output_____"
],
[
"# Convertng headlines to lower case\nfor index in new_Index:\n data[index]=data[index].str.lower()\ndata.head(1)",
"_____no_output_____"
],
[
"' '.join(str(x) for x in data.iloc[1,0:25])",
"_____no_output_____"
],
[
"headlines = []\nfor row in range(0,len(data.index)):\n headlines.append(' '.join(str(x) for x in data.iloc[row,0:25]))",
"_____no_output_____"
],
[
"headlines[0]",
"_____no_output_____"
],
[
"from sklearn.feature_extraction.text import CountVectorizer\nfrom sklearn.ensemble import RandomForestClassifier",
"_____no_output_____"
],
[
"## implement BAG OF WORDS\ncountvector=CountVectorizer(ngram_range=(3,3))\ntraindataset=countvector.fit_transform(headlines)",
"_____no_output_____"
],
[
"# implement RandomForest Classifier\nrandomclassifier=RandomForestClassifier(n_estimators=200,criterion='entropy')\nrandomclassifier.fit(traindataset,train['Label'])",
"_____no_output_____"
],
[
"## Predict for the Test Dataset\ntest_transform= []\nfor row in range(0,len(test.index)):\n test_transform.append(' '.join(str(x) for x in test.iloc[row,2:27]))\ntest_dataset = countvector.transform(test_transform)\npredictions = randomclassifier.predict(test_dataset)",
"_____no_output_____"
],
[
"## Import library to check accuracy\nfrom sklearn.metrics import classification_report,confusion_matrix,accuracy_score",
"_____no_output_____"
],
[
"matrix=confusion_matrix(test['Label'],predictions)\nprint(matrix)\nscore=accuracy_score(test['Label'],predictions)\nprint(score)\nreport=classification_report(test['Label'],predictions)\nprint(report)",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0cfd74a02bd6707353a367075fde3bbc13e3f0c | 10,969 | ipynb | Jupyter Notebook | sequential_pipeline/profiling_sequential_code.ipynb | rohuba/PACS | 5645d21b15c9dad876015beb738c4b6687b148da | [
"MIT"
] | null | null | null | sequential_pipeline/profiling_sequential_code.ipynb | rohuba/PACS | 5645d21b15c9dad876015beb738c4b6687b148da | [
"MIT"
] | null | null | null | sequential_pipeline/profiling_sequential_code.ipynb | rohuba/PACS | 5645d21b15c9dad876015beb738c4b6687b148da | [
"MIT"
] | null | null | null | 54.572139 | 790 | 0.652019 | [
[
[
"## Profiling sequential code",
"_____no_output_____"
],
[
"I profiled the sequential code `count_spacers_with_ED.py` using the `cProfile` Python package. I ran `count_spacers_with_ED.py` with a control file of 100 sequences (*Genome-Pos-3T3-Unsorted_100_seqs.txt*) and an experimental file of 100 sequences (*Genome-Pos-3T3-Bot10_100_seqs.txt*). Each of these input files contained 75 sequencing reads that could be perfectly matched to my database of 80,000 guide sequences, and 25 sequencing reads that needed an edit distance calculation. This breakdown was representative of the proportion of sequencing reads in the full input files that require an edit distance calculation because ~25% of sequencing reads cannot be perfectly matched to one of the 80,000 guide sequences.\n\nThe exact command I ran was <br>\n`python -m cProfile -o 100_seq_stats.profile count_spacers_with_ED.py -g ../data/Brie_CRISPR_library_with_controls_FOR_ANALYSIS.csv -u ../data/Genome-Pos-3T3-Unsorted_100_seqs.txt -s ../data/Genome-Pos-3T3-Bot10_100_seqs.txt -o cProfile_test_output`\n \nThis code was run on my Macbook Pro, which has a 2.2 GHz Intel Core i7 processor with 6 cores.\n\nThe profiling information was saved in a file called *100_seq_stats.profile*. I will now use the `pstats` package to see what parts of my code are taking the longest and if they can be parallelized.\n\n\n```python\nprint\n```",
"_____no_output_____"
]
],
[
[
"import pstats\n\np = pstats.Stats('100_seq_stats.profile'); #read in profiling stats\np.strip_dirs(); #remove the extraneous path from all the module names",
"_____no_output_____"
],
[
"#sort according to time spent within each function, and then print the statistics for the top 20 functions. \np.sort_stats('time').print_stats(20)",
"Mon Apr 29 16:08:45 2019 100_seq_stats.profile\n\n 350132307 function calls (350126604 primitive calls) in 537.388 seconds\n\n Ordered by: internal time\n List reduced from 1999 to 20 due to restriction <20>\n\n ncalls tottime percall cumtime percall filename:lineno(function)\n 3981650 462.315 0.000 534.561 0.000 count_spacers_with_ED.py:35(editDistDP)\n333437825 69.985 0.000 69.985 0.000 {built-in method builtins.min}\n 3982102 1.819 0.000 1.819 0.000 {built-in method numpy.zeros}\n 2 1.414 0.707 535.981 267.990 count_spacers_with_ED.py:69(count_spacers)\n7992171/7992125 0.447 0.000 0.447 0.000 {built-in method builtins.len}\n 1 0.215 0.215 0.232 0.232 count_spacers_with_ED.py:7(createDictionaries)\n 81/79 0.153 0.002 0.156 0.002 {built-in method _imp.create_dynamic}\n 20674 0.115 0.000 0.365 0.000 stats.py:3055(fisher_exact)\n 348 0.100 0.000 0.100 0.000 {method 'read' of '_io.FileIO' objects}\n 41950 0.092 0.000 0.092 0.000 {method 'reduce' of 'numpy.ufunc' objects}\n 1 0.075 0.075 0.075 0.075 {method 'dot' of 'numpy.ndarray' objects}\n 348 0.056 0.000 0.155 0.000 <frozen importlib._bootstrap_external>:830(get_data)\n 28062 0.052 0.000 0.052 0.000 {built-in method numpy.array}\n 1604 0.035 0.000 0.035 0.000 {built-in method posix.stat}\n 348 0.034 0.000 0.034 0.000 {built-in method marshal.loads}\n 593/1 0.033 0.000 537.389 537.389 {built-in method builtins.exec}\n 1 0.033 0.033 0.405 0.405 count_spacers_with_ED.py:135(calcGeneEnrich)\n 81/65 0.024 0.000 0.077 0.001 {built-in method _imp.exec_dynamic}\n 21078 0.021 0.000 0.071 0.000 fromnumeric.py:69(_wrapreduction)\n 20675 0.020 0.000 0.020 0.000 {method 'writerow' of '_csv.writer' objects}\n\n\n"
]
],
[
[
"As you can see from the table above, most of the runtime for our sequential code is spent within the `editDistDP` function. 534 of the 537 seconds, which accounts for 99.4% of our runtime, are spent calculating the edit distance between 50 sequencing reads and 80,000 guides. Generally, the input files contain ~10M sequencing reads, and about 25% of the sequences cannot be matched perfectly to one of the 80,000 guides. Thus for two input files of ~10M sequencing reads (~20M reads total), there are ~4-5M sequencing reads for which the edit distance calculations must be performed. If this code was run sequentially, this would require 10,000 hours of runtime. Therefore, we need to parallelize this portion of the code.\n\nThe edit distance calculation is currently nested within the function `count_spacers`, which matches each sequencing read from the input files to one of the 80,000 guides. For 200 sequencing reads provided as input, 1.4 seconds are spent performing the matching, which is only 0.007 seconds per sequencing read (I am using the 1.4 seconds from the *tottime* column because the *cumtime* takes into account the edit distance calculation). This number grows large if we have 20M sequencing reads we need to match because it would take $\\dfrac{0.007\\text{seconds/read} \\cdot 20\\text{M reads}}{3600\\text{seconds/hour}} = 39\\text{hours}$. Thus, the entire matching process of our workflow needs to be parallelized.\n\nWe want to parallelize this matching process by using a Spark cluster to have access to as many cores as possible to perform both the matching process and edit distance calculation (if needed). We will partition each input file into many tasks, and each task will run on a single core of the Spark cluster so a single core will perform both the matching process and edit distance for the sequencing reads in a partition. From what we have determined, there is not an easy way to parallelize the actual edit distance calculation between two strings. However, for a given sequencing read, we should be able to parallelize the 80,000 edit distance calculations that need to be performed between the sequencing read and the 80,000 guides by using Python multi-threading or possibly a GPU.",
"_____no_output_____"
],
[
"## Overheads",
"_____no_output_____"
],
[
"Since we do not know which sequences from our input files we will need to perform edit distance calculations for, load-balancing is the main overhead we anticipate dealing with because we do not want one or two cores slowed down with having to compute too many edit distance calculations. We would like to spread the number of edit distance calculations out evenly between the cores by tuning the number of Spark tasks. It may be good to shuffle the order of the sequencing reads because sometimes many sequences that require edit distance calculations are adjacent to each other in the input file.\n\nIf we try to use a GPU to perform the 80,000 edit distance calculations in parallel, memory-transfer (input/output) to the GPU would also be an overhead. For a single sequencing read, multiple transfers would need to be performed because we would not be able to perform the 80,000 calculations in parallel because we are limited by the number of cores on the GPU. Currently, we do not have a good way of mitigating this overhead if we were to use GPUs.",
"_____no_output_____"
],
[
"## Scaling",
"_____no_output_____"
],
[
"The sequence matching and edit distance portion of our code accounts for 99.4% of the runtime in our small example. With larger problem sizes, this percentage should only increase because the number of operations performed after the sequence matching and edit distance section is constant.\n\nAmdahl's Law states that potential program speedup $S_t$ is defined by the fraction of code $c$ that can be parallelized, according to the formula\n$$\nS_t = \\dfrac{1}{(1-c)+\\frac{c}{p}}\n$$\nwhere $p$ is the number of processors/cores. In our case, $c=0.994$ and the table below shows the speed-ups for $2$, $4$, $8$, $64$, and $128$ processors/cores:\n\n|processors|speed-up|\n|----------|--------|\n|2|1.98x|\n|4|3.93x|\n|8|7.68x|\n|64|46.44x|\n|128|72.64x|\n\nThus, our strong-scaling is almost linear when $p$ is small, but we observe that this begins to break down because we only get 73x speed-up if we were to use 128 processors/cores.",
"_____no_output_____"
],
[
"Gustafson's Law states larger systems should be used to solve larger problems because there should ideally be a fixed amount of parallel work per processor. The speed-up $S_t$ is calculated by\n$$\nS_t = 1 - c +c\\cdot p\n$$\nwhere $p$ is the number of processors/cores. In our case, $c=0.994$ and the table below shows the speed-ups for $2$, $4$, $8$, $64$, and $128$ processors/cores:\n\n|processors|speed-up|\n|----------|--------|\n|2|1.994x|\n|4|3.98x|\n|8|7.96x|\n|64|63.62x|\n|128|127.23x|\n\nThus, we almost achieve perfect weak-scaling because we can split up larger problem-sizes (which would be larger input files in our case) over more processors to achieve about the same runtime.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d0cfda79cd5f33b94fd40ef2ef463829c55e139f | 51,287 | ipynb | Jupyter Notebook | Section-10-Scikit-Optimize/06-Parallelization.ipynb | ankitario/hyperparameter-optimization | 0dfb1abbe883a64352eafd2ca433ec8770051b2a | [
"BSD-3-Clause"
] | 34 | 2021-05-05T09:29:23.000Z | 2022-03-05T03:16:09.000Z | Section-10-Scikit-Optimize/06-Parallelization.ipynb | GLASSY-GAIA/ML_Hyperparameter_Optimization_Python | 2e1b8478c9cd68404c3b415d45748eb5e09fe6c9 | [
"BSD-3-Clause"
] | null | null | null | Section-10-Scikit-Optimize/06-Parallelization.ipynb | GLASSY-GAIA/ML_Hyperparameter_Optimization_Python | 2e1b8478c9cd68404c3b415d45748eb5e09fe6c9 | [
"BSD-3-Clause"
] | 57 | 2021-05-07T10:54:43.000Z | 2022-03-31T13:05:37.000Z | 40.479084 | 10,500 | 0.506737 | [
[
[
"## Bayesian Optimization with Scikit-Optimize\n\nIn this notebook, we will perform **Bayesian Optimization** with Gaussian Processes in Parallel, utilizing various CPUs, to speed up the search.\n\nThis is useful to reduce search times. \n\nhttps://scikit-optimize.github.io/stable/auto_examples/parallel-optimization.html#example",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n\nfrom sklearn.datasets import load_breast_cancer\nfrom sklearn.ensemble import GradientBoostingClassifier\nfrom sklearn.model_selection import cross_val_score, train_test_split\n\nfrom skopt import Optimizer # for the optimization\nfrom joblib import Parallel, delayed # for the parallelization\n\nfrom skopt.space import Real, Integer, Categorical\nfrom skopt.utils import use_named_args",
"_____no_output_____"
],
[
"# load dataset\n\nbreast_cancer_X, breast_cancer_y = load_breast_cancer(return_X_y=True)\nX = pd.DataFrame(breast_cancer_X)\ny = pd.Series(breast_cancer_y).map({0:1, 1:0})\n\nX.head()",
"_____no_output_____"
],
[
"# the target:\n# percentage of benign (0) and malign tumors (1)\n\ny.value_counts() / len(y)",
"_____no_output_____"
],
[
"# split dataset into a train and test set\n\nX_train, X_test, y_train, y_test = train_test_split(\n X, y, test_size=0.3, random_state=0)\n\nX_train.shape, X_test.shape",
"_____no_output_____"
]
],
[
[
"## Define the Hyperparameter Space\n\nScikit-optimize provides an utility function to create the range of values to examine for each hyperparameters. More details in [skopt.Space](https://scikit-optimize.github.io/stable/modules/generated/skopt.Space.html)",
"_____no_output_____"
]
],
[
[
"# determine the hyperparameter space\n\nparam_grid = [\n Integer(10, 120, name=\"n_estimators\"),\n Integer(1, 5, name=\"max_depth\"),\n Real(0.0001, 0.1, prior='log-uniform', name='learning_rate'),\n Real(0.001, 0.999, prior='log-uniform', name=\"min_samples_split\"),\n Categorical(['deviance', 'exponential'], name=\"loss\"),\n]\n\n# Scikit-optimize parameter grid is a list\ntype(param_grid)",
"_____no_output_____"
]
],
[
[
"## Define the model",
"_____no_output_____"
]
],
[
[
"# set up the gradient boosting classifier\n\ngbm = GradientBoostingClassifier(random_state=0)",
"_____no_output_____"
]
],
[
[
"## Define the objective function\n\nThis is the hyperparameter response space, the function we want to minimize.",
"_____no_output_____"
]
],
[
[
"# We design a function to maximize the accuracy, of a GBM,\n# with cross-validation\n\n# the decorator allows our objective function to receive the parameters as\n# keyword arguments. This is a requirement for scikit-optimize.\n\n@use_named_args(param_grid)\ndef objective(**params):\n \n # model with new parameters\n gbm.set_params(**params)\n\n # optimization function (hyperparam response function)\n value = np.mean(\n cross_val_score(\n gbm, \n X_train,\n y_train,\n cv=3,\n n_jobs=-4,\n scoring='accuracy')\n )\n\n # negate because we need to minimize\n return -value",
"_____no_output_____"
]
],
[
[
"## Optimization with Gaussian Process",
"_____no_output_____"
]
],
[
[
"# We use the Optimizer\n\noptimizer = Optimizer(\n dimensions = param_grid, # the hyperparameter space\n base_estimator = \"GP\", # the surrogate\n n_initial_points=10, # the number of points to evaluate f(x) to start of\n acq_func='EI', # the acquisition function\n random_state=0, \n n_jobs=4,\n)",
"_____no_output_____"
],
[
"# we will use 4 CPUs (n_points)\n# if we loop 10 times using 4 end points, we perform 40 searches in total\n\nfor i in range(10):\n x = optimizer.ask(n_points=4) # x is a list of n_points points\n y = Parallel(n_jobs=4)(delayed(objective)(v) for v in x) # evaluate points in parallel\n optimizer.tell(x, y)",
"_____no_output_____"
],
[
"# the evaluated hyperparamters\n\noptimizer.Xi",
"_____no_output_____"
],
[
"# the accuracy\n\noptimizer.yi",
"_____no_output_____"
],
[
"# all together in one dataframe, so we can investigate further\ndim_names = ['n_estimators', 'max_depth', 'min_samples_split', 'learning_rate', 'loss']\n\ntmp = pd.concat([\n pd.DataFrame(optimizer.Xi),\n pd.Series(optimizer.yi),\n], axis=1)\n\ntmp.columns = dim_names + ['accuracy']\ntmp.head()",
"_____no_output_____"
]
],
[
[
"## Evaluate convergence of the search",
"_____no_output_____"
]
],
[
[
"tmp['accuracy'].sort_values(ascending=False).reset_index(drop=True).plot()",
"_____no_output_____"
]
],
[
[
"The trade-off with parallelization, is that we will not optimize the search after each evaluation of f(x), instead after, in this case 4, evaluations of f(x). Thus, we may need to perform more evaluations to find the optima. But, because we do it in parallel, overall, we reduce wall time.",
"_____no_output_____"
]
],
[
[
"tmp.sort_values(by='accuracy', ascending=True)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0cfdb2c7ec6c3a7f92a90e70933744cf3b1946e | 4,759 | ipynb | Jupyter Notebook | src/Problem_1.ipynb | JiaminJIAN/20MA573 | a9bc42a769b112c9beb9b1fcafb742a768a2a18e | [
"MIT"
] | 2 | 2020-02-07T23:11:44.000Z | 2020-02-08T14:39:45.000Z | src/Problem_1.ipynb | JiaminJIAN/20MA573 | a9bc42a769b112c9beb9b1fcafb742a768a2a18e | [
"MIT"
] | null | null | null | src/Problem_1.ipynb | JiaminJIAN/20MA573 | a9bc42a769b112c9beb9b1fcafb742a768a2a18e | [
"MIT"
] | null | null | null | 27.830409 | 241 | 0.455558 | [
[
[
"<a href=\"https://colab.research.google.com/github/JiaminJIAN/20MA573/blob/master/src/Problem_1.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"### **Problem 1**\n\nYiyang wants to understand the following question: If he randomly chooses three points on the perimeter of a circle, what is the probability that the triangle made of three points contains the center?\n\n(1) Write a pseudocode to estimate the probability.\n\n(2) Write a python code and implement it.\n\n(3) Give me your guess on this probability. Can you justify? ",
"_____no_output_____"
],
[
"Solution:\n\n(1) If the inner triangle of a circle is a obtuse triangle, the triangle will contain the center of the circle. And If the inner triangle of a circle is an acute triangle, the triangle does not contain the center of the circle.\n\nPseudocode:\n\n[pseudocode](https://github.com/JiaminJIAN/20MA573/blob/master/src/problem1.pdf)",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"(2) Write a python code and implement it.\n\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom scipy import stats\nfrom scipy.stats import bernoulli",
"_____no_output_____"
],
[
"def circle(N):\n p = 0\n for i in range(N):\n x = np.zeros(3)\n y = np.zeros(3)\n for j in range(3):\n x[j] = np.random.uniform(low=-1, high=1) \n y[j] = np.sqrt(1 - x[j]**2) * (2 * bernoulli.rvs(0.5) - 1)\n a = (x[0]-x[1])**2 + (y[0]-y[1])**2\n b = (x[1]-x[2])**2 + (y[1]-y[2])**2\n c = (x[0]-x[2])**2 + (y[0]-y[2])**2\n if a+b-c>0 and a+c-b>0 and b+c-a>0:\n p = p + 1\n return p/N",
"_____no_output_____"
],
[
"circle(10000)",
"_____no_output_____"
]
],
[
[
"(3) Firstly we choose a point on the perimeter of a circle and denote it as A. And suppose the angle between point A and point B is $\\theta$. Then the probobility of the triangle contains the center is:\n\n$$\\int_{0}^{\\pi} \\frac{\\theta}{2 \\pi} \\frac{1}{\\pi} \\, d \\theta = \\frac{\\pi^{2}}{4 \\pi^{2}} = \\frac{1}{4}.$$\n\n\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
d0cfdfc6870e6eefc3d3f9cd68adaa270c8e7809 | 10,785 | ipynb | Jupyter Notebook | notebooks/01-05 Control Flow.ipynb | Mohitsharma44/ucsl17 | 389206c8bbd5f4f0dd6d27d954aff0bdc3395c7f | [
"MIT"
] | 17 | 2017-06-12T04:08:07.000Z | 2020-12-08T06:34:21.000Z | notebooks/01-05 Control Flow.ipynb | Mohitsharma44/ucsl17 | 389206c8bbd5f4f0dd6d27d954aff0bdc3395c7f | [
"MIT"
] | null | null | null | notebooks/01-05 Control Flow.ipynb | Mohitsharma44/ucsl17 | 389206c8bbd5f4f0dd6d27d954aff0bdc3395c7f | [
"MIT"
] | 18 | 2017-06-12T14:19:28.000Z | 2020-10-11T06:52:23.000Z | 24.345372 | 390 | 0.525359 | [
[
[
"## Control Flow\nGenerally, a program is executed sequentially and once executed it is not repeated again. There may be a situation when you need to execute a piece of code n number of times, or maybe even execute certain piece of code based on a particular condition.. this is where the control flow statements come in.\n\nIn this section, we will be covering:\n\n- Conditional statements -- if, else, and elif\n- Loop statements -- for, while\n- Loop control statements -- break, continue, pass\n\n### Conditional Statements\nConditionals statements are used to change the flow of execution. You can use the relational operators, logical operators and membership operators for performing condition checks",
"_____no_output_____"
]
],
[
[
"result = 1\nif result == 1:\n print(\"Best Match\")\nelif result <= 3:\n print(\"Close Enough\")\nelse:\n print(\"This is Blasphemy!\")",
"Best Match\n"
]
],
[
[
"The logic is very simple.. *`if`* < `condition_is_met` >, *`then`* do something; *`else`* do something else. \n\nPython adopts the `if`-`else` clause as it is used in many languages.. However the `elif` part is unique to python. `elif` simply is a contraction for `else if`.",
"_____no_output_____"
],
[
"### Loop Statements\nThese statements are used when we want to execute a piece of code multiple times. Python has two types of loops -- `for` loop and `while` loop.",
"_____no_output_____"
]
],
[
[
"for i in [0,1,2]:\n print(\"{}\".format(i))",
"0\n1\n2\n"
]
],
[
[
"In `for` loop, we specify the variable we want to use, the `iterator` we want to loop over, and use the `in` (membership) operator to link them together.",
"_____no_output_____"
]
],
[
[
"i = 2\nwhile i >= 0:\n print(\"{}\".format(i))\n i -= 1",
"2\n1\n0\n"
]
],
[
[
"As you can see, they both serve different purposes. For loop is used when you want to run something for fixed amount of times, whereas while loop can theoretically run forever (if you use something like `while True:` .. *dont!* ). \n\nOne of the most commonly used `iterator` with for loop is the `range` object which is used to generate the sequence of numbers",
"_____no_output_____"
]
],
[
[
"list(range(10))",
"_____no_output_____"
]
],
[
[
"The `range` requires the *stop* argument. It can also accept *start* (at first position) and *step* (at third position) as arguments but if not passed, it creates a sequence of numbers from `0` till `stop - 1`. Remember, the *stop* is not included in the output",
"_____no_output_____"
]
],
[
[
"# With start and stop\nlist(range(2, 20))",
"_____no_output_____"
],
[
"# With start, stop and step\nlist(range(2, 20, 2))",
"_____no_output_____"
]
],
[
[
"When you have an iterator of iterators .. for example a list of lists .. then you can use what is known as nested loops to flatten the list.",
"_____no_output_____"
]
],
[
[
"# This is not the best way.. but for the sake of completion of\n# topic, this example is included.\narr = [range(3), range(3, 6)]\nfor lists in arr:\n for elem in lists:\n print(elem)",
"0\n1\n2\n3\n4\n5\n"
]
],
[
[
"### Loop Control Statements\nLoop control statements change the executing of loop from its normal sequence.\n\n#### Break",
"_____no_output_____"
],
[
"It terminates the current loop and resumes the execution at the next statement. The most common use for break is when some external condition is triggered requiring a hasty exit from a loop. The break statement can be used in both while and for loops.",
"_____no_output_____"
]
],
[
[
"for i in range(1, 10):\n if i == 5:\n print('Condition satisfied')\n break\n print(i) # What would happen if this is placed before if condition?",
"1\n2\n3\n4\nCondition satisfied\n"
]
],
[
[
"#### Continue\nContinue statement returns the control to the beginning of the loop. The continue statement rejects all the remaining statements in the current iteration of the loop and moves the control back to the top of the loop.",
"_____no_output_____"
]
],
[
[
"for i in range(1, 10):\n if i == 5:\n print('Condition satisfied')\n continue\n print(\"whatever.. I won't get printed anyways.\")\n print(i)",
"1\n2\n3\n4\nCondition satisfied\n6\n7\n8\n9\n"
]
],
[
[
"#### Pass\nPass is used when a statement is required syntactically but performs a null operation i.e. nothing happens when the statement is executed.",
"_____no_output_____"
]
],
[
[
"for i in range(1, 10):\n if i == 5:\n print('Condition satisfied')\n pass\n print(i)",
"1\n2\n3\n4\nCondition satisfied\n5\n6\n7\n8\n9\n"
]
],
[
[
"As you can see execution of pass statement had no effect on the flow of the code. It wouldn't have mattered if it was not there. \n\nIt is generally used as a temporary placeholder for an unimplemented logic. For example lets say you have written a function (we'll learn about functions a little later) and want to test the remaining part of code without actually running your function.. You can use pass statement in such cases. Python interpreter will read that and skip that part and get on with further execution.",
"_____no_output_____"
],
[
"### Loops with else\nPython's Loop statements can be accompanies with an else block in cases where a certain block of code needs to be executed after the loop has successfully completed its execution i.e. iff the loop didn't `break` out in the middle of execution\n",
"_____no_output_____"
]
],
[
[
"best = 11\nfor i in range(10):\n if i >= best:\n print(\"Excellent\")\n break\n else:\n continue\nelse:\n print(\"Couldn't find the best match\")",
"Couldn't find the best match\n"
]
],
[
[
"Now if we change the `best` to something less than `10`",
"_____no_output_____"
]
],
[
[
"best = 9\nfor i in range(10):\n if i >= best:\n print(\"Excellent\")\n break\n else:\n continue\nelse:\n print(\"Couldn't find the best match\")",
"Excellent\n"
]
],
[
[
"You can implement similar functionality using the `while` loop.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0d0048719304da96c6b168a0888ff923e430c76 | 114,460 | ipynb | Jupyter Notebook | smart-queueing-system/Retail_Scenario.ipynb | jlgarcia75/move-mouse-pointer | c0199c5b5313a1b58944adad467ba84c46091dad | [
"CC0-1.0"
] | null | null | null | smart-queueing-system/Retail_Scenario.ipynb | jlgarcia75/move-mouse-pointer | c0199c5b5313a1b58944adad467ba84c46091dad | [
"CC0-1.0"
] | null | null | null | smart-queueing-system/Retail_Scenario.ipynb | jlgarcia75/move-mouse-pointer | c0199c5b5313a1b58944adad467ba84c46091dad | [
"CC0-1.0"
] | null | null | null | 48.851899 | 8,540 | 0.625983 | [
[
[
"# Smart Queue Monitoring System - Retail Scenario\n\n## Overview\nNow that you have your Python script and job submission script, you're ready to request an **IEI Tank-870** edge node and run inference on the different hardware types (CPU, GPU, VPU, FPGA).\n\nAfter the inference is completed, the output video and stats files need to be retrieved and stored in the workspace, which can then be viewed within the Jupyter Notebook.\n\n## Objectives\n* Submit inference jobs to Intel's DevCloud using the `qsub` command.\n* Retrieve and review the results.\n* After testing, go back to the proposal doc and update your original proposed hardware device.",
"_____no_output_____"
],
[
"## Step 0: Set Up\n\n#### IMPORTANT: Set up paths so we can run Dev Cloud utilities\nYou *must* run this every time you enter a Workspace session.\n(Tip: select the cell and use **Shift+Enter** to run the cell.)",
"_____no_output_____"
]
],
[
[
"%env PATH=/opt/conda/bin:/opt/spark-2.4.3-bin-hadoop2.7/bin:/opt/conda/bin:/usr/local/sbin:/usr/local/bin:/usr/sbin:/usr/bin:/sbin:/bin:/opt/intel_devcloud_support\nimport os\nimport sys\nsys.path.insert(0, os.path.abspath('/opt/intel_devcloud_support'))\nsys.path.insert(0, os.path.abspath('/opt/intel'))",
"env: PATH=/opt/conda/bin:/opt/spark-2.4.3-bin-hadoop2.7/bin:/opt/conda/bin:/usr/local/sbin:/usr/local/bin:/usr/sbin:/usr/bin:/sbin:/bin:/opt/intel_devcloud_support\n"
]
],
[
[
"### Step 0.1: (Optional-step): Original Video\n\nIf you are curious to see the input video, run the following cell to view the original video stream we'll be using for inference.",
"_____no_output_____"
]
],
[
[
"import videoHtml\nvideoHtml.videoHTML('Retail', ['original_videos/Retail.mp4'])",
"_____no_output_____"
]
],
[
[
"## Step 1 : Inference on a Video\n\nIn the next few cells, You'll submit your job using the `qsub` command and retrieving the results for each job. Each of the cells below should submit a job to different edge compute nodes.\n\nThe output of the cell is the `JobID` of your job, which you can use to track progress of a job with `liveQStat`.\n\nYou will need to submit a job for each of the following hardware types:\n* **CPU**\n* **GPU**\n* **VPU**\n* **FPGA**\n\n**Note** You will have to submit each job one at a time and retrieve their results. \n\nAfter submission, they will go into a queue and run as soon as the requested compute resources become available. \n(Tip: **shift+enter** will run the cell and automatically move you to the next cell.)\n\nIf your job successfully runs and completes, once you retrieve your results, it should output a video and a stats text file in the `results/retail/<DEVICE>` directory.\n\nFor example, your **CPU** job should output its files in this directory:\n> **results/retail/cpu**\n\n**Note**: To get the queue labels for the different hardware devices, you can go to [this link](https://devcloud.intel.com/edge/get_started/devcloud/).\n\nThe following arguments should be passed to the job submission script after the `-F` flag:\n* Model path - `/data/models/intel/person-detection-retail-0013/<MODEL PRECISION>/`. You will need to adjust this path based on the model precision being using on the hardware.\n* Device - `CPU`, `GPU`, `MYRIAD`, `HETERO:FPGA,CPU`\n* Manufacturing video path - `/data/resources/retail.mp4`\n* Manufacturing queue_param file path - `/data/queue_param/retail.npy`\n* Output path - `/output/results/retail/<DEVICE>` This should be adjusted based on the device used in the job.\n* Max num of people - This is the max number of people in queue before the system would redirect them to another queue.",
"_____no_output_____"
],
[
"## Step 1.1: Submit to an Edge Compute Node with an Intel CPU\nIn the cell below, write a script to submit a job to an <a \n href=\"https://software.intel.com/en-us/iot/hardware/iei-tank-dev-kit-core\">IEI \n Tank* 870-Q170</a> edge node with an <a \n href=\"https://ark.intel.com/products/88186/Intel-Core-i5-6500TE-Processor-6M-Cache-up-to-3-30-GHz-\">Intel® Core™ i5-6500TE processor</a>. The inference workload should run on the CPU.",
"_____no_output_____"
]
],
[
[
"#Submit job to the queue\ncpu_job_id = !qsub queue_job.sh -d . -l nodes=1:tank-870:i5-6500te -F \"/data/models/intel/person-detection-retail-0013/FP32/person-detection-retail-0013 CPU /data/resources/retail.mp4 /data/queue_param/retail.npy /output/results/retail/cpu 2\"\n\nprint(cpu_job_id[0])",
"qRXPc4Bbf9OekZQx8RsXgtXu45iJSSlt\n"
]
],
[
[
"#### Check Job Status\n\nTo check on the job that was submitted, use `liveQStat` to check the status of the job.\n\nColumn `S` shows the state of your running jobs.\n\nFor example:\n- If `JOB ID`is in Q state, it is in the queue waiting for available resources.\n- If `JOB ID` is in R state, it is running.",
"_____no_output_____"
]
],
[
[
"import liveQStat\nliveQStat.liveQStat()",
"_____no_output_____"
]
],
[
[
"#### Get Results\n\nRun the next cell to retrieve your job's results.",
"_____no_output_____"
]
],
[
[
"import get_results\nget_results.getResults(cpu_job_id[0], filename='output.tgz', blocking=True)",
"getResults() is blocking until results of the job (id:qRXPc4Bbf9OekZQx8RsXgtXu45iJSSlt) are ready.\nPlease wait...Success!\noutput.tgz was downloaded in the same folder as this notebook.\n"
]
],
[
[
"#### Unpack your output files and view stdout.log",
"_____no_output_____"
]
],
[
[
"!tar zxf output.tgz",
"_____no_output_____"
],
[
"!cat stdout.log",
"Total People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 2}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 0\r\nNumber of people in queue = {1: 0, 2: 0}\r\nresults/\r\nresults/retail/\r\nresults/retail/cpu/\r\nresults/retail/cpu/stats.txt\r\nresults/retail/cpu/output_video.mp4\r\nstderr.log\r\n"
]
],
[
[
"#### View stderr.log\nThis can be used for debugging",
"_____no_output_____"
]
],
[
[
"!cat stderr.log",
"person_detect.py:48: DeprecationWarning: Reading network using constructor is deprecated. Please, use IECore.read_network() method instead\r\n self.model=IENetwork(self.model_structure, self.model_weights)\r\n"
]
],
[
[
"#### View Output Video\nRun the cell below to view the output video. If inference was successfully run, you should see a video with bounding boxes drawn around each person detected.",
"_____no_output_____"
]
],
[
[
"import videoHtml\n\nvideoHtml.videoHTML('Retail CPU', ['results/retail/cpu/output_video.mp4'])",
"_____no_output_____"
]
],
[
[
"## Step 1.2: Submit to an Edge Compute Node with a CPU and IGPU\nIn the cell below, write a script to submit a job to an <a \n href=\"https://software.intel.com/en-us/iot/hardware/iei-tank-dev-kit-core\">IEI \n Tank* 870-Q170</a> edge node with an <a href=\"https://ark.intel.com/products/88186/Intel-Core-i5-6500TE-Processor-6M-Cache-up-to-3-30-GHz-\">Intel® Core i5-6500TE</a>. The inference workload should run on the **Intel® HD Graphics 530** integrated GPU.",
"_____no_output_____"
]
],
[
[
"#Submit job to the queue\ngpu_job_id = !qsub queue_job.sh -d . -l nodes=1:tank-870:i5-6500te:intel-hd-530 -F \"/data/models/intel/person-detection-retail-0013/FP16/person-detection-retail-0013 GPU /data/resources/retail.mp4 /data/queue_param/retail.npy /output/results/retail/gpu 2\"\n\nprint(gpu_job_id[0])",
"7IR3HnoBY5c5zoHANzetnnOvTyazyk3O\n"
]
],
[
[
"### Check Job Status\n\nTo check on the job that was submitted, use `liveQStat` to check the status of the job.\n\nColumn `S` shows the state of your running jobs.\n\nFor example:\n- If `JOB ID`is in Q state, it is in the queue waiting for available resources.\n- If `JOB ID` is in R state, it is running.",
"_____no_output_____"
]
],
[
[
"import liveQStat\nliveQStat.liveQStat()",
"_____no_output_____"
]
],
[
[
"#### Get Results\n\nRun the next cell to retrieve your job's results.",
"_____no_output_____"
]
],
[
[
"import get_results\nget_results.getResults(gpu_job_id[0], filename='output.tgz', blocking=True)",
"getResults() is blocking until results of the job (id:7IR3HnoBY5c5zoHANzetnnOvTyazyk3O) are ready.\nPlease wait............................................Success!\noutput.tgz was downloaded in the same folder as this notebook.\n"
]
],
[
[
"#### Unpack your output files and view stdout.log",
"_____no_output_____"
]
],
[
[
"!tar zxf output.tgz",
"_____no_output_____"
],
[
"!cat stdout.log",
"Total People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 2}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 0\r\nNumber of people in queue = {1: 0, 2: 0}\r\nresults/\r\nresults/retail/\r\nresults/retail/gpu/\r\nresults/retail/gpu/stats.txt\r\nresults/retail/gpu/output_video.mp4\r\nstderr.log\r\n"
]
],
[
[
"#### View stderr.log\nThis can be used for debugging",
"_____no_output_____"
]
],
[
[
"!cat stderr.log",
"person_detect.py:48: DeprecationWarning: Reading network using constructor is deprecated. Please, use IECore.read_network() method instead\r\n self.model=IENetwork(self.model_structure, self.model_weights)\r\n"
]
],
[
[
"#### View Output Video\nRun the cell below to view the output video. If inference was successfully run, you should see a video with bounding boxes drawn around each person detected.",
"_____no_output_____"
]
],
[
[
"import videoHtml\n\nvideoHtml.videoHTML('Retail GPU', ['results/retail/gpu/output_video.mp4'])",
"_____no_output_____"
]
],
[
[
"## Step 1.3: Submit to an Edge Compute Node with an Intel® Neural Compute Stick 2\nIn the cell below, write a script to submit a job to an <a \n href=\"https://software.intel.com/en-us/iot/hardware/iei-tank-dev-kit-core\">IEI \n Tank 870-Q170</a> edge node with an <a href=\"https://ark.intel.com/products/88186/Intel-Core-i5-6500TE-Processor-6M-Cache-up-to-3-30-GHz-\">Intel Core i5-6500te CPU</a>. The inference workload should run on an <a \n href=\"https://software.intel.com/en-us/neural-compute-stick\">Intel Neural Compute Stick 2</a> installed in this node.",
"_____no_output_____"
]
],
[
[
"#Submit job to the queue\nvpu_job_id = !qsub queue_job.sh -d . -l nodes=1:tank-870:i5-6500te:intel-ncs2 -F \"/data/models/intel/person-detection-retail-0013/FP16/person-detection-retail-0013 MYRIAD /data/resources/retail.mp4 /data/queue_param/retail.npy /output/results/retail/vpu 2\"\n\nprint(vpu_job_id[0])",
"MuBdzZOs7DKMkjmgfIQWnGrnQ6Ilc2on\n"
]
],
[
[
"### Check Job Status\n\nTo check on the job that was submitted, use `liveQStat` to check the status of the job.\n\nColumn `S` shows the state of your running jobs.\n\nFor example:\n- If `JOB ID`is in Q state, it is in the queue waiting for available resources.\n- If `JOB ID` is in R state, it is running.",
"_____no_output_____"
]
],
[
[
"import liveQStat\nliveQStat.liveQStat()",
"_____no_output_____"
]
],
[
[
"#### Get Results\n\nRun the next cell to retrieve your job's results.",
"_____no_output_____"
]
],
[
[
"import get_results\nget_results.getResults(vpu_job_id[0], filename='output.tgz', blocking=True)",
"getResults() is blocking until results of the job (id:MuBdzZOs7DKMkjmgfIQWnGrnQ6Ilc2on) are ready.\nPlease wait..........................Success!\noutput.tgz was downloaded in the same folder as this notebook.\n"
]
],
[
[
"#### Unpack your output files and view stdout.log",
"_____no_output_____"
]
],
[
[
"!tar zxf output.tgz",
"_____no_output_____"
],
[
"!cat stdout.log",
"Total People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 2}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 0\r\nNumber of people in queue = {1: 0, 2: 0}\r\nresults/\r\nresults/retail/\r\nresults/retail/vpu/\r\nresults/retail/vpu/stats.txt\r\nresults/retail/vpu/output_video.mp4\r\nstderr.log\r\n"
]
],
[
[
"#### View stderr.log\nThis can be used for debugging",
"_____no_output_____"
]
],
[
[
"!cat stderr.log",
"person_detect.py:48: DeprecationWarning: Reading network using constructor is deprecated. Please, use IECore.read_network() method instead\r\n self.model=IENetwork(self.model_structure, self.model_weights)\r\n"
]
],
[
[
"#### View Output Video\nRun the cell below to view the output video. If inference was successfully run, you should see a video with bounding boxes drawn around each person detected.",
"_____no_output_____"
]
],
[
[
"import videoHtml\n\nvideoHtml.videoHTML('Retail VPU', ['results/retail/vpu/output_video.mp4'])",
"_____no_output_____"
]
],
[
[
"## Step 1.4: Submit to an Edge Compute Node with IEI Mustang-F100-A10\nIn the cell below, write a script to submit a job to an <a \n href=\"https://software.intel.com/en-us/iot/hardware/iei-tank-dev-kit-core\">IEI \n Tank 870-Q170</a> edge node with an <a href=\"https://ark.intel.com/products/88186/Intel-Core-i5-6500TE-Processor-6M-Cache-up-to-3-30-GHz-\">Intel Core™ i5-6500te CPU</a> . The inference workload will run on the <a href=\"https://www.ieiworld.com/mustang-f100/en/\"> IEI Mustang-F100-A10 </a> FPGA card installed in this node.",
"_____no_output_____"
]
],
[
[
"#Submit job to the queue\nfpga_job_id = !qsub queue_job.sh -d . -l nodes=1:tank-870:i5-6500te:iei-mustang-f100-a10 -F \"/data/models/intel/person-detection-retail-0013/FP16/person-detection-retail-0013 HETERO:FPGA,CPU /data/resources/retail.mp4 /data/queue_param/retail.npy /output/results/retail/fpga 2\"\n\nprint(fpga_job_id[0])",
"klPhfgBWRQNWcmxC7NNmKQN374e7Pi0t\n"
]
],
[
[
"### Check Job Status\n\nTo check on the job that was submitted, use `liveQStat` to check the status of the job.\n\nColumn `S` shows the state of your running jobs.\n\nFor example:\n- If `JOB ID`is in Q state, it is in the queue waiting for available resources.\n- If `JOB ID` is in R state, it is running.",
"_____no_output_____"
]
],
[
[
"import liveQStat\nliveQStat.liveQStat()",
"_____no_output_____"
]
],
[
[
"#### Get Results\n\nRun the next cell to retrieve your job's results.",
"_____no_output_____"
]
],
[
[
"import get_results\nget_results.getResults(fpga_job_id[0], filename='output.tgz', blocking=True)",
"getResults() is blocking until results of the job (id:klPhfgBWRQNWcmxC7NNmKQN374e7Pi0t) are ready.\nPlease wait..........................Success!\noutput.tgz was downloaded in the same folder as this notebook.\n"
]
],
[
[
"#### Unpack your output files and view stdout.log",
"_____no_output_____"
]
],
[
[
"!tar zxf output.tgz",
"_____no_output_____"
],
[
"!cat stdout.log",
"INTELFPGAOCLSDKROOT is not set\r\nUsing script's current directory (/opt/altera/aocl-pro-rte/aclrte-linux64)\r\n\r\naoc was not found, but aocl was found. Assuming only RTE is installed.\r\n\r\nAOCL_BOARD_PACKAGE_ROOT is set to /opt/intel/openvino/bitstreams/a10_vision_design_sg2_bitstreams/BSP/a10_1150_sg2. Using that.\r\nAdding /opt/altera/aocl-pro-rte/aclrte-linux64/bin to PATH\r\nAdding /opt/altera/aocl-pro-rte/aclrte-linux64/linux64/lib to LD_LIBRARY_PATH\r\nAdding /opt/altera/aocl-pro-rte/aclrte-linux64/host/linux64/lib to LD_LIBRARY_PATH\r\nAdding /opt/intel/openvino/bitstreams/a10_vision_design_sg2_bitstreams/BSP/a10_1150_sg2/linux64/lib to LD_LIBRARY_PATH\r\naocl program: Running program from /opt/intel/openvino/bitstreams/a10_vision_design_sg2_bitstreams/BSP/a10_1150_sg2/linux64/libexec\r\nProgramming device: a10gx_2ddr : Intel Vision Accelerator Design with Intel Arria 10 FPGA (acla10_1150_sg20)\r\nProgram succeed. \r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 6\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 2, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 5\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 2}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 4\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 3\r\nNumber of people in queue = {1: 1, 2: 1}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 2\r\nNumber of people in queue = {1: 1, 2: 0}\r\nTotal People in frame = 0\r\nNumber of people in queue = {1: 0, 2: 0}\r\nresults/\r\nresults/retail/\r\nresults/retail/fpga/\r\nresults/retail/fpga/stats.txt\r\nresults/retail/fpga/output_video.mp4\r\nstderr.log\r\n"
]
],
[
[
"#### View stderr.log\nThis can be used for debugging",
"_____no_output_____"
]
],
[
[
"!cat stderr.log",
"person_detect.py:48: DeprecationWarning: Reading network using constructor is deprecated. Please, use IECore.read_network() method instead\r\n self.model=IENetwork(self.model_structure, self.model_weights)\r\n"
]
],
[
[
"#### View Output Video\nRun the cell below to view the output video. If inference was successfully run, you should see a video with bounding boxes drawn around each person detected.",
"_____no_output_____"
]
],
[
[
"import videoHtml\n\nvideoHtml.videoHTML('Retail FPGA', ['results/retail/fpga/output_video.mp4'])",
"_____no_output_____"
]
],
[
[
"***Wait!***\n\nPlease wait for all the inference jobs and video rendering to complete before proceeding to the next step.\n\n## Step 2: Assess Performance\n\nRun the cells below to compare the performance across all 4 devices. The following timings for the model are being comapred across all 4 devices:\n\n- Model Loading Time\n- Average Inference Time\n- FPS",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n\ndevice_list=['cpu', 'gpu', 'fpga', 'vpu']\ninference_time=[]\nfps=[]\nmodel_load_time=[]\n\nfor device in device_list:\n with open('results/retail/'+device+'/stats.txt', 'r') as f:\n inference_time.append(float(f.readline().split(\"\\n\")[0]))\n fps.append(float(f.readline().split(\"\\n\")[0]))\n model_load_time.append(float(f.readline().split(\"\\n\")[0]))",
"_____no_output_____"
],
[
"plt.bar(device_list, inference_time)\nplt.xlabel(\"Device Used\")\nplt.ylabel(\"Total Inference Time in Seconds\")\nplt.show()",
"_____no_output_____"
],
[
"plt.bar(device_list, fps)\nplt.xlabel(\"Device Used\")\nplt.ylabel(\"Frames per Second\")\nplt.show()",
"_____no_output_____"
],
[
"plt.bar(device_list, model_load_time)\nplt.xlabel(\"Device Used\")\nplt.ylabel(\"Model Loading Time in Seconds\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Step 3: Update Proposal Document\n\nNow that you've completed your hardware testing, you should go back to the proposal document and validate or update your originally proposed hardware. Once you've updated your proposal, you can move onto the next scenario.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
d0d005b24b0d8e850c77c449c71a5ce157210425 | 12,091 | ipynb | Jupyter Notebook | hail/python/hail/docs/tutorials/07-matrixtable.ipynb | saponas/hail | bafea6b18247a4279d6ef11015e8e9f3c2b5ecea | [
"MIT"
] | 789 | 2016-09-05T04:14:25.000Z | 2022-03-30T09:51:54.000Z | hail/python/hail/docs/tutorials/07-matrixtable.ipynb | saponas/hail | bafea6b18247a4279d6ef11015e8e9f3c2b5ecea | [
"MIT"
] | 5,724 | 2016-08-29T18:58:40.000Z | 2022-03-31T23:49:42.000Z | hail/python/hail/docs/tutorials/07-matrixtable.ipynb | johnc1231/hail | 3235d42aea8c1a682258493ff77b320025903df3 | [
"MIT"
] | 233 | 2016-08-31T20:42:38.000Z | 2022-02-17T16:42:39.000Z | 25.946352 | 378 | 0.57059 | [
[
[
"## MatrixTable Tutorial\n\nIf you've gotten this far, you're probably thinking:\n\n- \"Can't I do all of this in `pandas` or `R`?\" \n- \"What does this have to do with biology?\"\n\nThe two crucial features that Hail adds are _scalability_ and the _domain-specific primitives_ needed to work easily with biological data. Fear not! You've learned most of the basic concepts of Hail and now are ready for the bit that makes it possible to represent and compute on genetic matrices: the [MatrixTable](https://hail.is/docs/0.2/hail.MatrixTable.html).",
"_____no_output_____"
],
[
"In the last example of the [Table Joins Tutorial](https://hail.is/docs/0.2/tutorials/08-joins.html), the ratings table had a compound key: `movie_id` and `user_id`. The ratings were secretly a movie-by-user matrix!\n\nHowever, since this matrix is very sparse, it is reasonably represented in a so-called \"coordinate form\" `Table`, where each row of the table is an entry of the sparse matrix. For large and dense matrices (like sequencing data), the per-row overhead of coordinate reresentations is untenable. That's why we built `MatrixTable`, a 2-dimensional generalization of `Table`.",
"_____no_output_____"
],
[
"### MatrixTable Anatomy\n\nRecall that `Table` has two kinds of fields:\n\n- global fields\n- row fields\n\n`MatrixTable` has four kinds of fields:\n\n- global fields\n- row fields\n- column fields\n- entry fields",
"_____no_output_____"
],
[
"Row fields are fields that are stored once per row. These can contain information about the rows, or summary data calculated per row.\n\nColumn fields are stored once per column. These can contain information about the columns, or summary data calculated per column.\n\nEntry fields are the piece that makes this structure a matrix -- there is an entry for each (row, column) pair.",
"_____no_output_____"
],
[
"### Importing and Reading\n\nLike tables, matrix tables can be [imported](https://hail.is/docs/0.2/methods/impex.html) from a variety of formats: VCF, (B)GEN, PLINK, TSV, etc. Matrix tables can also be *read* from a \"native\" matrix table format. Let's read a sample of prepared [1KG](https://en.wikipedia.org/wiki/1000_Genomes_Project) data.",
"_____no_output_____"
]
],
[
[
"import hail as hl\nfrom bokeh.io import output_notebook, show\noutput_notebook()\n\nhl.utils.get_1kg('data/')",
"_____no_output_____"
],
[
"mt = hl.read_matrix_table('data/1kg.mt')\nmt.describe()",
"_____no_output_____"
]
],
[
[
"There are a few things to note:\n\n- There is a single column field `s`. This is the sample ID from the VCF. It is also the column key.\n- There is a compound row key: `locus` and `alleles`. \n - `locus` has type `locus<GRCh37>`\n - `alleles` has type `array<str>`\n- GT has type `call`. That's a genotype call!",
"_____no_output_____"
],
[
"Whereas table expressions could be indexed by nothing or indexed by rows, matrix table expression have four options: nothing, indexed by row, indexed by column, or indexed by row and column (the entries). Let's see some examples.",
"_____no_output_____"
]
],
[
[
"mt.s.describe()",
"_____no_output_____"
],
[
"mt.GT.describe()",
"_____no_output_____"
]
],
[
[
"### MatrixTable operations\nWe belabored the operations on tables because they all have natural analogs (sometimes several) on matrix tables. For example:\n\n - `count` => `count_{rows, cols}` (and `count` which returns both)\n - `filter` => `filter_{rows, cols, entries}`\n - `annotate` => `annotate_{rows, cols, entries}` (and globals for both)\n - `select` => `select_{rows, cols, entries}` (and globals for both)\n - `transmute` => `transmute_{rows, cols, entries}` (and globals for both)\n - `group_by` => `group_{rows, cols}_by`\n - `explode` => `expode_{rows, cols}`\n - `aggregate` => `aggregate_{rows, cols, entries}`",
"_____no_output_____"
],
[
"Some operations are unique to `MatrixTable`:\n\n- The row fields can be accessed as a `Table` with [rows](https://hail.is/docs/0.2/hail.MatrixTable.html#hail.MatrixTable.rows)\n- The column fields can be accessed as a `Table` with [cols](https://hail.is/docs/0.2/hail.MatrixTable.html#hail.MatrixTable.cols).\n- The entire field space of a `MatrixTable` can be accessed as a coordinate-form `Table` with [entries](https://hail.is/docs/0.2/hail.MatrixTable.html#hail.MatrixTable.entries). Be careful with this! While it's fast to aggregate or query, trying to write this `Table` to disk could produce files _thousands of times larger_ than the corresponding `MatrixTable`.\n\nLet's explore `mt` using these tools. Let's get the size of the dataset.",
"_____no_output_____"
]
],
[
[
"mt.count() # (rows, cols)",
"_____no_output_____"
]
],
[
[
"Let's look at the first few row keys (variants) and column keys (sample IDs).",
"_____no_output_____"
]
],
[
[
"mt.rows().select().show()",
"_____no_output_____"
],
[
"mt.s.show()",
"_____no_output_____"
]
],
[
[
"Let's investigate the genotypes and the call rate. Let's look at the first few genotypes:",
"_____no_output_____"
]
],
[
[
"mt.GT.show()",
"_____no_output_____"
]
],
[
[
"All homozygous reference, which is not surprising. Let's look at the distribution of genotype calls:",
"_____no_output_____"
]
],
[
[
"mt.aggregate_entries(hl.agg.counter(mt.GT.n_alt_alleles()))",
"_____no_output_____"
]
],
[
[
"Let's compute the overall call rate directly, and then plot the distribution of call rate per variant.",
"_____no_output_____"
]
],
[
[
"mt.aggregate_entries(hl.agg.fraction(hl.is_defined(mt.GT)))",
"_____no_output_____"
]
],
[
[
"Here's a nice trick: you can use an aggregator inside `annotate_rows` and it will aggregate over columns, that is, summarize the values in the row using the aggregator. Let's compute and plot call rate per variant.",
"_____no_output_____"
]
],
[
[
"mt2 = mt.annotate_rows(call_rate = hl.agg.fraction(hl.is_defined(mt.GT)))\nmt2.describe()",
"_____no_output_____"
],
[
"p = hl.plot.histogram(mt2.call_rate, range=(0,1.0), bins=100, \n title='Variant Call Rate Histogram', legend='Call Rate')\nshow(p)",
"_____no_output_____"
]
],
[
[
"### Exercise: GQ vs DP\n\nIn this exercise, you'll use Hail to investigate a strange property of sequencing datasets.\n\nThe `DP` field is the sequencing depth (the number of reads).\n\nLet's first plot a histogram of `DP`:",
"_____no_output_____"
]
],
[
[
"p = hl.plot.histogram(mt.DP, range=(0,40), bins=40, title='DP Histogram', legend='DP')\nshow(p)",
"_____no_output_____"
]
],
[
[
"Now, let's do the same thing for GQ.\n\nThe `GQ` field is the phred-scaled \"genotype quality\". The formula to convert to a linear-scale confidence (0 to 1) is `10 ** -(mt.GQ / 10)`. GQ is truncated to lie between 0 and 99.\n",
"_____no_output_____"
]
],
[
[
"p = hl.plot.histogram(mt.GQ, range=(0,100), bins=100, title='GQ Histogram', legend='GQ')\nshow(p)",
"_____no_output_____"
]
],
[
[
"Whoa! That's a strange distribution! There's a big spike at 100. The rest of the values have roughly the same shape as the DP distribution, but form a [Dimetrodon](https://en.wikipedia.org/wiki/Dimetrodon). Use Hail to figure out what's going on!",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0d005f8a66220ca5ad697176c6b1324193c16ba | 308,806 | ipynb | Jupyter Notebook | James's Logistic Regression.ipynb | nilebooks/Jadeogun | 2a9dea78f6deede91555dbb99c6ecd70407990f8 | [
"Apache-2.0"
] | 1 | 2021-05-28T18:55:05.000Z | 2021-05-28T18:55:05.000Z | James's Logistic Regression.ipynb | nilebooks/Jadeogun | 2a9dea78f6deede91555dbb99c6ecd70407990f8 | [
"Apache-2.0"
] | null | null | null | James's Logistic Regression.ipynb | nilebooks/Jadeogun | 2a9dea78f6deede91555dbb99c6ecd70407990f8 | [
"Apache-2.0"
] | null | null | null | 678.694505 | 204,220 | 0.948913 | [
[
[
"# 1. Loading Libraries\n# Importing NumPy and Panda\nimport pandas as pd\nimport numpy as np\n\n# ---------Import libraries & modules for data visualizaiton\nfrom pandas.plotting import scatter_matrix\nfrom matplotlib import pyplot\n\n# Importing scit-learn module to split the dataset into train/test sub-datasets\nfrom sklearn.model_selection import train_test_split\n\n# Importing scit-learn module for the algorith/model: Linear Regression\nfrom sklearn.linear_model import LogisticRegression\n\n# Importing sci-Learn module for K-fole cross-validation - algorithm/modle evaluation & validation\nfrom sklearn.model_selection import KFold\nfrom sklearn.model_selection import cross_val_score\n\n# Importing scit-learn module fro classification report\nfrom sklearn.metrics import classification_report",
"_____no_output_____"
],
[
"# 2. Specifing data file location\nfilename = 'C:/Data Sets/Iris.csv'\n\n# Loading the data into a Pandas DataFrame\ndf = pd.read_csv(filename)",
"_____no_output_____"
],
[
"# 4 Preprocess Dataset\n\n# 4.1 Cleaning Data: Find & Mark Missing Values\n# Zero values cannot be use in these columns\n\n# Marking and updating zero values as missing or NaN\ndf[['SepalLengthCm', 'SepalWidthCm', 'SepalWidthCm', 'PetalWidthCm']] \\\n= df[['SepalLengthCm', 'SepalWidthCm', 'SepalWidthCm', 'PetalWidthCm']].replace(0, np.NaN)\n\n# count the number of NaN values in each column\nprint(df.isnull().sum())",
"Id 0\nSepalLengthCm 0\nSepalWidthCm 0\nPetalLengthCm 0\nPetalWidthCm 0\nSpecies 0\ndtype: int64\n"
],
[
"# 5. Performing Exploratory Data Analysis on Dataset\n\n# Get the dimensions or Shape of the dataset\n\n# i.e. number of records/rows x number of variables/columns\n\nprint(df.shape)",
"(150, 6)\n"
],
[
"# Getting the data types of all the variables/attributes of the data set\n# The resutls shows\n\nprint(df.dtypes)",
"Id int64\nSepalLengthCm float64\nSepalWidthCm float64\nPetalLengthCm float64\nPetalWidthCm float64\nSpecies object\ndtype: object\n"
],
[
"# Getting several records/rows at he top fo the dataset\n# Get the first five records\n\nprint(df.head(5))",
" Id SepalLengthCm SepalWidthCm PetalLengthCm PetalWidthCm Species\n0 1 5.1 3.5 1.4 0.2 Iris-setosa\n1 2 4.9 3.0 1.4 0.2 Iris-setosa\n2 3 4.7 3.2 1.3 0.2 Iris-setosa\n3 4 4.6 3.1 1.5 0.2 Iris-setosa\n4 5 5.0 3.6 1.4 0.2 Iris-setosa\n"
],
[
"# Get the summary statistics of the numerica variables/attributes fo the dataset\n\nprint(df.describe())",
" Id SepalLengthCm SepalWidthCm PetalLengthCm PetalWidthCm\ncount 150.000000 150.000000 150.000000 150.000000 150.000000\nmean 75.500000 5.843333 3.054000 3.758667 1.198667\nstd 43.445368 0.828066 0.433594 1.764420 0.763161\nmin 1.000000 4.300000 2.000000 1.000000 0.100000\n25% 38.250000 5.100000 2.800000 1.600000 0.300000\n50% 75.500000 5.800000 3.000000 4.350000 1.300000\n75% 112.750000 6.400000 3.300000 5.100000 1.800000\nmax 150.000000 7.900000 4.400000 6.900000 2.500000\n"
],
[
"# class distribution\n# i.e. how many records for each class\n# This dataset is a good candidate for the classification issues\n\nprint(df.groupby('Species').size())",
"Species\nIris-setosa 50\nIris-versicolor 50\nIris-virginica 50\ndtype: int64\n"
],
[
"# Plot historgram for each numerica variable/attribute of the dataset\n# VIP NOTES: The first variable ID is also plotted. However, the plot should be ignored\n\ndf.hist(figsize=(12, 8))\npyplot.show()",
"_____no_output_____"
],
[
"# Density plots\n# IMPORTANT NOTES: 5 numerica variables -->> at least 5 plots -->> layout (2, 3): 2 rows, each row with 3 plots\n\ndf.plot(kind='density', subplots=True, layout=(3,3), sharex=False, legend=True, fontsize=1, figsize=(12, 16))\npyplot.show()",
"_____no_output_____"
],
[
"df.plot(kind='box', subplots=True, layout=(3, 3), sharex=False, sharey=False, figsize=(12, 8))\npyplot.show()",
"_____no_output_____"
],
[
"# scatter plot matirx\n\nscatter_matrix(df, alpha=0.8, figsize=(15,15))\n\npyplot.show()",
"_____no_output_____"
],
[
"# Store datafram values into a numpy array\narray = df.values\n# separate array into input and output components by slicign\n# For X (input)[:, 1:5] --> all the rows, columns from 1 -4 (5 - 1)\nX = array[:,1:5]\n\n# For Y (input)[:, 5] --> all the rows, column 5\nY = array[:,5]",
"_____no_output_____"
],
[
"# Splittling the dataset --> training sub-dataset: 67%; test sub-dataset: 33%\n\ntest_size = 0.33\n\n# Selection of records to include in which sub-dataset mush be done randomely\n# Use this seed for randomizzation\n\nseed = 7\n\n# Split the dataset (both input & output) into training/testing datasets\nX_train, X_test, Y_train, Y_test = train_test_split(X, Y, test_size=test_size, random_state=seed)",
"_____no_output_____"
],
[
"# Building the model\nmodel = LogisticRegression()\n\n# Train the model using the training sub-dataset\n\nmodel.fit(X_train, Y_train)\n\n# Print the classification report\n# Ref: Section 10.2.5 Book: Machine Learning Mastery with Python\n\npredicted = model.predict(X_test)\n\nreport = classification_report(Y_test, predicted)\n\nprint(report)",
" precision recall f1-score support\n\n Iris-setosa 1.00 1.00 1.00 14\nIris-versicolor 0.89 0.89 0.89 18\n Iris-virginica 0.89 0.89 0.89 18\n\n accuracy 0.92 50\n macro avg 0.93 0.93 0.93 50\n weighted avg 0.92 0.92 0.92 50\n\n"
],
[
"# Finding the Accuracy Leve\n# score the accuracy level\n\nresult = model.score(X_test, Y_test)\n\n# Print out the results\nprint((\"Accuracy: %.3f%%\") % (result*100))\n",
"Accuracy: 92.000%\n"
],
[
"# 10. Classify/Predict\n\nmodel.predict([[5.3, 3.0, 4.5, 1.5]])",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0d02a7ce470ebbe4eb928cd68883eb6b321b989 | 3,427 | ipynb | Jupyter Notebook | 15 - Advanced Statistical Methods in Python/7_K-Means Clustering/4_Clustering Categorical Data (2:50)/Categorical data.ipynb | olayinka04/365-data-science-courses | 7d71215432f0ef07fd3def559d793a6f1938d108 | [
"Apache-2.0"
] | null | null | null | 15 - Advanced Statistical Methods in Python/7_K-Means Clustering/4_Clustering Categorical Data (2:50)/Categorical data.ipynb | olayinka04/365-data-science-courses | 7d71215432f0ef07fd3def559d793a6f1938d108 | [
"Apache-2.0"
] | null | null | null | 15 - Advanced Statistical Methods in Python/7_K-Means Clustering/4_Clustering Categorical Data (2:50)/Categorical data.ipynb | olayinka04/365-data-science-courses | 7d71215432f0ef07fd3def559d793a6f1938d108 | [
"Apache-2.0"
] | null | null | null | 17.942408 | 131 | 0.512985 | [
[
[
"# Basics of cluster analysis",
"_____no_output_____"
],
[
"## Import the relevant libraries",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nsns.set()\nfrom sklearn.cluster import KMeans",
"_____no_output_____"
]
],
[
[
"## Load the data",
"_____no_output_____"
]
],
[
[
"data = pd.read_csv('3.01. Country clusters.csv')",
"_____no_output_____"
],
[
"data",
"_____no_output_____"
]
],
[
[
"## Map the data",
"_____no_output_____"
]
],
[
[
"data_mapped = data.copy()\ndata_mapped['Language']=data_mapped['Language'].map({'English':0,'French':1,'German':2})\ndata_mapped",
"_____no_output_____"
]
],
[
[
"## Select the features",
"_____no_output_____"
]
],
[
[
"x = data_mapped.iloc[:,1:4]",
"_____no_output_____"
],
[
"x",
"_____no_output_____"
]
],
[
[
"## Clustering",
"_____no_output_____"
]
],
[
[
"kmeans = KMeans(2)",
"_____no_output_____"
],
[
"kmeans.fit(x)",
"_____no_output_____"
]
],
[
[
"## Clustering results",
"_____no_output_____"
]
],
[
[
"identified_clusters = kmeans.fit_predict(x)\nidentified_clusters",
"_____no_output_____"
],
[
"data_with_clusters = data_mapped.copy()\ndata_with_clusters['Cluster'] = identified_clusters\ndata_with_clusters",
"_____no_output_____"
],
[
"plt.scatter(data_with_clusters['Longitude'],data_with_clusters['Latitude'],c=data_with_clusters['Cluster'],cmap='rainbow')\nplt.xlim(-180,180)\nplt.ylim(-90,90)\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d0d030d83fae44226df6158987df20189d481031 | 3,373 | ipynb | Jupyter Notebook | ScrapingData-Master/Scraping Twitter Data.ipynb | ATTO-DT/DATA-SCIENCE-PROJECT | ae192c3e6c6aa1daf9f5610a732822bdc76f0691 | [
"Apache-2.0"
] | null | null | null | ScrapingData-Master/Scraping Twitter Data.ipynb | ATTO-DT/DATA-SCIENCE-PROJECT | ae192c3e6c6aa1daf9f5610a732822bdc76f0691 | [
"Apache-2.0"
] | null | null | null | ScrapingData-Master/Scraping Twitter Data.ipynb | ATTO-DT/DATA-SCIENCE-PROJECT | ae192c3e6c6aa1daf9f5610a732822bdc76f0691 | [
"Apache-2.0"
] | 1 | 2021-10-03T06:39:27.000Z | 2021-10-03T06:39:27.000Z | 30.116071 | 226 | 0.571005 | [
[
[
"import json\nimport csv\nimport tweepy\nimport re",
"_____no_output_____"
],
[
"\"\"\"\nINPUTS:\n consumer_key, consumer_secret, access_token, access_token_secret: codes \n telling twitter that we are authorized to access this data\n hashtag_phrase: the combination of hashtags to search for\nOUTPUTS:\n none, simply save the tweet info to a spreadsheet\n\"\"\"\ndef search_for_hashtags(consumer_key, consumer_secret, access_token, access_token_secret, hashtag_phrase):\n \n #create authentication for accessing Twitter\n auth = tweepy.OAuthHandler(consumer_key, consumer_secret)\n auth.set_access_token(access_token, access_token_secret)\n\n #initialize Tweepy API\n api = tweepy.API(auth)\n \n #get the name of the spreadsheet we will write to\n fname = '_'.join(re.findall(r\"#(\\w+)\", hashtag_phrase))\n\n #open the spreadsheet we will write to\n with open('%s.csv' % (fname), 'wb') as file:\n\n w = csv.writer(file)\n\n #write header row to spreadsheet\n w.writerow(['timestamp', 'tweet_text', 'username', 'all_hashtags', 'followers_count'])\n\n #for each tweet matching our hashtags, write relevant info to the spreadsheet\n for tweet in tweepy.Cursor(api.search, q=hashtag_phrase+' -filter:retweets', \\\n lang=\"en\", tweet_mode='extended').items(100):\n w.writerow([tweet.created_at, tweet.full_text.replace('\\n',' ').encode('utf-8'), tweet.user.screen_name.encode('utf-8'), [e['text'] for e in tweet._json['entities']['hashtags']], tweet.user.followers_count])",
"_____no_output_____"
],
[
"consumer_key = raw_input('Consumer Key ')\nconsumer_secret = raw_input('Consumer Secret ')\naccess_token = raw_input('Access Token ')\naccess_token_secret = raw_input('Access Token Secret ')\n \nhashtag_phrase = raw_input('Hashtag Phrase ')\n\nif __name__ == '__main__':\n search_for_hashtags(consumer_key, consumer_secret, access_token, access_token_secret, hashtag_phrase)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
d0d03bf34fc3a1ed981fc3d9dc690c02dffcc4d0 | 8,040 | ipynb | Jupyter Notebook | Jupyter_Notebooks/ibm_db-queryStream.ipynb | ibmdb/jupyter-node-ibm_db | 588bf32955237aa2829aa796a9ce10865a5dfa99 | [
"Apache-2.0"
] | null | null | null | Jupyter_Notebooks/ibm_db-queryStream.ipynb | ibmdb/jupyter-node-ibm_db | 588bf32955237aa2829aa796a9ce10865a5dfa99 | [
"Apache-2.0"
] | 1 | 2019-06-12T10:23:03.000Z | 2019-06-12T10:23:03.000Z | Jupyter_Notebooks/ibm_db-queryStream.ipynb | ibmdb/jupyter-node-ibm_db | 588bf32955237aa2829aa796a9ce10865a5dfa99 | [
"Apache-2.0"
] | 2 | 2019-11-03T17:23:38.000Z | 2021-12-28T11:00:46.000Z | 35.892857 | 333 | 0.443781 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d0d040b9efe05b02913ede5da45b135dee419fbc | 179,332 | ipynb | Jupyter Notebook | docs/examples/general/data_loading/coco_reader.ipynb | a-sansanwal/DALI | 83aeb96792d053f60dd4252b8efa0fc8fdd9012a | [
"ECL-2.0",
"Apache-2.0"
] | 1 | 2020-11-02T07:05:28.000Z | 2020-11-02T07:05:28.000Z | docs/examples/general/data_loading/coco_reader.ipynb | MAKali4737/DALI | 3b114c6ebee38ff3815a9b4a234402e4d1affaa0 | [
"ECL-2.0",
"Apache-2.0"
] | null | null | null | docs/examples/general/data_loading/coco_reader.ipynb | MAKali4737/DALI | 3b114c6ebee38ff3815a9b4a234402e4d1affaa0 | [
"ECL-2.0",
"Apache-2.0"
] | null | null | null | 919.651282 | 173,496 | 0.951581 | [
[
[
"# COCO Reader\n\nReader operator that reads a COCO dataset (or subset of COCO), which consists of an annotation file and the images directory.\n\n`DALI_EXTRA_PATH` environment variable should point to the place where data from [DALI extra repository](https://github.com/NVIDIA/DALI_extra) is downloaded. Please make sure that the proper release tag is checked out.",
"_____no_output_____"
]
],
[
[
"from nvidia.dali.pipeline import Pipeline\nimport nvidia.dali.ops as ops\nimport nvidia.dali.types as types\nimport numpy as np\nfrom time import time\nimport os.path\n\ntest_data_root = os.environ['DALI_EXTRA_PATH']\nfile_root = os.path.join(test_data_root, 'db', 'coco', 'images')\nannotations_file = os.path.join(test_data_root, 'db', 'coco', 'instances.json')\n\nnum_gpus = 1\nbatch_size = 16",
"_____no_output_____"
],
[
"class COCOPipeline(Pipeline): \n def __init__(self, batch_size, num_threads, device_id): \n super(COCOPipeline, self).__init__(batch_size, num_threads, device_id, seed = 15) \n self.input = ops.COCOReader(file_root = file_root, annotations_file = annotations_file,\n shard_id = device_id, num_shards = num_gpus, ratio=True)\n self.decode = ops.ImageDecoder(device = \"mixed\", output_type = types.RGB) \n \n def define_graph(self): \n inputs, bboxes, labels = self.input() \n images = self.decode(inputs) \n return (images, bboxes, labels) ",
"_____no_output_____"
],
[
"start = time()\npipes = [COCOPipeline(batch_size=batch_size, num_threads=2, device_id = device_id) for device_id in range(num_gpus)]\nfor pipe in pipes:\n pipe.build()\ntotal_time = time() - start\nprint(\"Computation graph built and dataset loaded in %f seconds.\" % total_time)",
"Computation graph built and dataset loaded in 0.307431 seconds.\n"
],
[
"pipe_out = [pipe.run() for pipe in pipes] \n\nimages_cpu = pipe_out[0][0].as_cpu()\nbboxes_cpu = pipe_out[0][1]\nlabels_cpu = pipe_out[0][2]",
"_____no_output_____"
]
],
[
[
"Bounding boxes returned by the operator are lists of floats containing composed of **\\[x, y, width, height]** (`ltrb` is set to `False` by default).",
"_____no_output_____"
]
],
[
[
"bboxes = bboxes_cpu.at(4)\nbboxes",
"_____no_output_____"
]
],
[
[
"Let's see the ground truth bounding boxes drawn on the image.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport matplotlib.patches as patches\nimport random\n\nimg_index = 4\n\nimg = images_cpu.at(img_index)\n\nH = img.shape[0]\nW = img.shape[1]\n\nfig,ax = plt.subplots(1)\n\nax.imshow(img)\nbboxes = bboxes_cpu.at(img_index)\nlabels = labels_cpu.at(img_index)\ncategories_set = set()\nfor label in labels:\n categories_set.add(label[0])\n\ncategory_id_to_color = dict([ (cat_id , [random.uniform(0, 1) ,random.uniform(0, 1), random.uniform(0, 1)]) for cat_id in categories_set])\n\nfor bbox, label in zip(bboxes, labels):\n rect = patches.Rectangle((bbox[0]*W,bbox[1]*H),bbox[2]*W,bbox[3]*H,linewidth=1,edgecolor=category_id_to_color[label[0]],facecolor='none')\n ax.add_patch(rect)\n\nplt.show()\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0d0438406fd2f0124f75238e4ecdfcdf0c0eea5 | 11,810 | ipynb | Jupyter Notebook | generative_net_with_trained_decoder.ipynb | jdespraz/deep_generative_networks | 5627c46ba40926113a2431e7217b1c5eee84ff50 | [
"MIT"
] | null | null | null | generative_net_with_trained_decoder.ipynb | jdespraz/deep_generative_networks | 5627c46ba40926113a2431e7217b1c5eee84ff50 | [
"MIT"
] | null | null | null | generative_net_with_trained_decoder.ipynb | jdespraz/deep_generative_networks | 5627c46ba40926113a2431e7217b1c5eee84ff50 | [
"MIT"
] | null | null | null | 36.791277 | 139 | 0.536918 | [
[
[
"import numpy as np\nnp.random.seed(123)\n\nimport os\nfrom keras.models import Model\nfrom keras.layers import Input, Convolution2D, MaxPooling2D, BatchNormalization\nfrom keras.layers import Flatten, Dense, Dropout, ZeroPadding2D, Reshape, UpSampling2D\nfrom keras.layers.local import LocallyConnected1D\nfrom keras.layers.noise import GaussianDropout\nfrom keras.optimizers import SGD\nfrom keras.regularizers import l2\nfrom keras import backend as K\nfrom keras.utils.layer_utils import print_summary\n\nimport tensorflow as tf\n\nimport cv2\nimport h5py\n\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"#os.environ[\"CUDA_VISIBLE_DEVICES\"] = \"\" # uncomment this line to run the code on the CPU",
"_____no_output_____"
],
[
"filter_id = 470 # candle class\nN = 3200 # feature vector size",
"_____no_output_____"
],
[
"def max_loss(y_true, y_pred):\n return (1.-K.sum(tf.mul(y_true,y_pred),axis=-1))\n\ndef max_metric(y_true, y_pred):\n return (1.-max_loss(y_true,y_pred))\n\ndef get_model(): \n # generator\n inputs = Input(shape=(N,), name='input')\n \n g0 = Reshape((N,1))(inputs)\n g0 = GaussianDropout(0.05)(g0)\n g1 = LocallyConnected1D(nb_filter=1, filter_length=1,\n init='one', activation='relu', bias=False,\n border_mode='valid',W_regularizer=l2(0.1))(g0)\n g2 = Reshape((128,5,5))(g1)\n \n g3 = UpSampling2D(size=(2, 2))(g2) # 10x10\n g3 = Convolution2D(512,2,2,activation='relu',border_mode='valid')(g3) # 9x9\n g3 = BatchNormalization(mode = 0 , axis = 1)(g3)\n g3 = Convolution2D(512,2,2,activation='relu',border_mode='same')(g3) # 9x9\n g3 = BatchNormalization(mode = 0 , axis = 1)(g3)\n \n g4 = UpSampling2D(size=(2, 2))(g3) # 18x18\n g4 = Convolution2D(256,3,3,activation='relu',border_mode='valid')(g4) # 16x16\n g4 = BatchNormalization(mode = 0 , axis = 1)(g4)\n g4 = Convolution2D(256,3,3,activation='relu',border_mode='same')(g4) # 16x16\n g4 = BatchNormalization(mode = 0 , axis = 1)(g4)\n \n g5 = UpSampling2D(size=(2, 2))(g4) # 32x32\n g5 = Convolution2D(256,3,3,activation='relu',border_mode='valid')(g5)# 30x30\n g5 = BatchNormalization(mode = 0 , axis = 1)(g5)\n g5 = Convolution2D(256,3,3,activation='relu',border_mode='same')(g5) # 30x30\n g5 = BatchNormalization(mode = 0 , axis = 1)(g5)\n \n g6 = UpSampling2D(size=(2, 2))(g5) # 60x60\n g6 = Convolution2D(128,3,3,activation='relu',border_mode='valid')(g6) # 58x58\n g6 = BatchNormalization(mode = 0 , axis = 1)(g6)\n g6 = Convolution2D(128,3,3,activation='relu',border_mode='same')(g6) # 58x58\n g6 = BatchNormalization(mode = 0 , axis = 1)(g6)\n \n g7 = UpSampling2D(size=(2, 2))(g6) # 116x116\n g7 = Convolution2D(128,4,4,activation='relu',border_mode='valid')(g7) # 113x113\n g7 = BatchNormalization(mode = 0 , axis = 1)(g7)\n g7 = Convolution2D(128,4,4,activation='relu',border_mode='same')(g7) # 113x113\n g7 = BatchNormalization(mode = 0 , axis = 1)(g7)\n \n g8 = UpSampling2D(size=(2, 2))(g7) # 226x226\n g8 = Convolution2D(64,3,3,activation='relu',border_mode='valid')(g8) # 224x224\n g8 = BatchNormalization(mode = 0 , axis = 1)(g8)\n g8 = Convolution2D(64,3,3,activation='relu',border_mode='same')(g8) # 224x224\n g8 = BatchNormalization(mode = 0 , axis = 1)(g8)\n g8 = Convolution2D(3,3,3,activation='linear',border_mode='same')(g8) # 224x224\n g8 = BatchNormalization(mode = 0, axis = 1, name='image')(g8)\n \n temp = Model(input=inputs, output=g8)\n offset = len(temp.layers)\n \n # discriminator \n vgg1 = ZeroPadding2D((1,1),input_shape=(3,224,224))(g8)\n vgg2 = Convolution2D(64, 3, 3, activation='relu')(vgg1)\n vgg3 = ZeroPadding2D((1,1))(vgg2)\n vgg4 = Convolution2D(64, 3, 3, activation='relu')(vgg3)\n vgg5 = MaxPooling2D((2,2), strides=(2,2))(vgg4)\n\n vgg6 = ZeroPadding2D((1,1))(vgg5)\n vgg7 = Convolution2D(128, 3, 3, activation='relu')(vgg6)\n vgg8 = ZeroPadding2D((1,1))(vgg7)\n vgg9 = Convolution2D(128, 3, 3, activation='relu')(vgg8)\n vgg10 = MaxPooling2D((2,2), strides=(2,2))(vgg9)\n\n vgg11 = ZeroPadding2D((1,1))(vgg10)\n vgg12 = Convolution2D(256, 3, 3, activation='relu')(vgg11)\n vgg13 = ZeroPadding2D((1,1))(vgg12)\n vgg14 = Convolution2D(256, 3, 3, activation='relu')(vgg13)\n vgg15 = ZeroPadding2D((1,1))(vgg14)\n vgg16 = Convolution2D(256, 3, 3, activation='relu')(vgg15)\n vgg17 = MaxPooling2D((2,2), strides=(2,2))(vgg16)\n\n vgg18 = ZeroPadding2D((1,1))(vgg17)\n vgg19 = Convolution2D(512, 3, 3, activation='relu')(vgg18)\n vgg20 = ZeroPadding2D((1,1))(vgg19)\n vgg21 = Convolution2D(512, 3, 3, activation='relu')(vgg20)\n vgg22 = ZeroPadding2D((1,1))(vgg21)\n vgg23 = Convolution2D(512, 3, 3, activation='relu')(vgg22)\n vgg24 = MaxPooling2D((2,2), strides=(2,2))(vgg23)\n\n vgg25 = ZeroPadding2D((1,1))(vgg24)\n vgg26 = Convolution2D(512, 3, 3, activation='relu')(vgg25)\n vgg27 = ZeroPadding2D((1,1))(vgg26)\n vgg28 = Convolution2D(512, 3, 3, activation='relu')(vgg27)\n vgg29 = ZeroPadding2D((1,1))(vgg28)\n vgg30 = Convolution2D(512, 3, 3, activation='relu')(vgg29)\n vgg31 = MaxPooling2D((2,2), strides=(2,2))(vgg30)\n\n vgg32 = Flatten()(vgg31)\n vgg33 = Dense(4096, activation='relu')(vgg32)\n vgg34 = Dropout(0.5)(vgg33)\n vgg35 = Dense(4096, activation='relu')(vgg34)\n vgg36 = Dropout(0.5)(vgg35)\n vgg37 = Dense(1000, activation='relu', name='vgg_class')(vgg36)\n \n # create model\n model = Model(input=inputs, output=[vgg37,g8])\n \n # set generator weights\n enc_size = 30\n f = h5py.File('decoder_weights.h5')\n for k, l in enumerate(f.attrs['layer_names']):\n if(k<enc_size):\n continue\n g = f[l]\n weight_names = [n.decode('utf8') for n in g.attrs['weight_names']]\n weights = [g[weight_name] for weight_name in weight_names]\n model.layers[k-enc_size+4].set_weights(weights)\n model.layers[k-enc_size+4].trainable = False\n f.close()\n \n # set discriminator weights (vgg)\n f = h5py.File('vgg16_weights.h5')\n for k in range(f.attrs['nb_layers']):\n g = f['layer_{}'.format(k)]\n weights = [g['param_{}'.format(p)] for p in range(g.attrs['nb_params'])]\n model.layers[k+offset].set_weights(weights)\n model.layers[k+offset].trainable = False\n f.close()\n \n # set the locally connected layer weights to trainable\n model.layers[3].trainable = True\n \n # compile model\n sgd = SGD(lr=0.01, decay=0.0, momentum=0.1, nesterov=True)\n model.compile(optimizer=sgd, loss=[max_loss, 'mse'], metrics=['mse'], loss_weights=[1.,0.])\n\n return model",
"_____no_output_____"
],
[
"# create neural network\nmodel = get_model()\nprint_summary(model.layers)",
"_____no_output_____"
],
[
"def reconstruct_image(im):\n im2 = np.squeeze(im)*1\n im2 = im2.transpose((1,2,0))\n im2[:,:,0] += 103.939\n im2[:,:,1] += 116.779\n im2[:,:,2] += 123.68\n im2 = im2.astype(np.uint8)\n return cv2.cvtColor(im2,cv2.COLOR_BGR2RGB)\n\ndef print_img(model,z=None):\n if(z is None):\n z = np.random.uniform(0,1,size=(1,N))\n out = model.predict(z, batch_size=z.shape[0])\n \n activ = out[0][0]\n img = out[1][0]\n\n # change to RGB colors and rescale image\n img -= np.min(img)\n img /= np.max(img)\n img *= 256.\n img = cv2.cvtColor(img.astype('uint8').transpose(1,2,0), cv2.COLOR_BGR2RGB)\n\n plt.figure(figsize=(6,6))\n plt.imshow(np.flipud(img))\n plt.title('filter activation: '+str(activ[filter_id]))\n plt.axis('off')\n plt.show()\n return img\n\n_ = print_img(model)",
"_____no_output_____"
],
[
"# training the model\nbatch_size = 1\nn_samples = 40\ndummy_labels2 = np.zeros(shape=(n_samples,3,224,224))\nvgg_nclasses = 1000\n\nz = np.ones(shape=(n_samples,N))\nIMG = np.zeros((30,224,224,3))\nfor k in np.arange(0,30):\n dummy_labels1 = np.ones(shape=(n_samples,vgg_nclasses))*(-10./vgg_nclasses) # put a penalty to the other classes\n dummy_labels1[:,filter_id] = 1. # give a positive unit weight for the target class\n out = model.fit(z, [dummy_labels1,dummy_labels2], batch_size=batch_size, nb_epoch=1, verbose=1)\n \n IMG[k,:,:,:] = print_img(model, z[0:1])\n\n# plotting the median of the last 10 iterations gives a smoother final image\nplt.figure()\nplt.imshow(np.flipud(np.median(IMG[20:,:,:,:],axis=0).astype('uint8')))\nplt.axis('off')\nplt.show()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0d04fa7f6e2eb0ba1f3424a7033a90226d99532 | 46,154 | ipynb | Jupyter Notebook | _sources/curriculum-notebooks/Mathematics/PatternsAndRelations/patterns-and-relations.ipynb | mlamoureux/CallystoJBook | 058080e1bb6370c0a072ed1c0ded96fe1e2947de | [
"CC0-1.0"
] | 21 | 2018-08-23T20:41:26.000Z | 2021-04-21T02:27:13.000Z | _sources/curriculum-notebooks/Mathematics/PatternsAndRelations/patterns-and-relations.ipynb | mlamoureux/CallystoJBook | 058080e1bb6370c0a072ed1c0ded96fe1e2947de | [
"CC0-1.0"
] | 113 | 2018-07-23T21:05:00.000Z | 2022-03-16T23:40:08.000Z | _sources/curriculum-notebooks/Mathematics/PatternsAndRelations/patterns-and-relations.ipynb | mlamoureux/CallystoJBook | 058080e1bb6370c0a072ed1c0ded96fe1e2947de | [
"CC0-1.0"
] | 11 | 2018-11-20T16:36:59.000Z | 2021-08-03T12:58:55.000Z | 46,154 | 46,154 | 0.578303 | [
[
[
"\n\n<a href=\"https://hub.callysto.ca/jupyter/hub/user-redirect/git-pull?repo=https%3A%2F%2Fgithub.com%2Fcallysto%2Fcurriculum-notebooks&branch=master&subPath=Mathematics/PatternsAndRelations/patterns-and-relations.ipynb&depth=1\" target=\"_parent\"><img src=\"https://raw.githubusercontent.com/callysto/curriculum-notebooks/master/open-in-callysto-button.svg?sanitize=true\" width=\"123\" height=\"24\" alt=\"Open in Callysto\"/></a>",
"_____no_output_____"
]
],
[
[
"from IPython.display import HTML\n\nHTML('''<script>\ncode_show=true; \nfunction code_toggle() {\n if (code_show){\n $('div.input').hide();\n } else {\n $('div.input').show();\n }\n code_show = !code_show\n} \n$( document ).ready(code_toggle);\n</script>\nThe raw code for this IPython notebook is by default hidden for easier reading.\nTo toggle on/off the raw code, click <a href=\"javascript:code_toggle()\">here</a>.''')\n",
"_____no_output_____"
],
[
"# Modules\n\nimport string\nimport numpy as np\nimport pandas as pd\nimport qgrid as q\nimport matplotlib.pyplot as plt\n\n# Widgets & Display modules, etc..\n\nfrom ipywidgets import widgets as w\nfrom ipywidgets import Button, Layout, widgets\nfrom IPython.display import display, Javascript, Markdown\n\n# grid features for interactive grids \n\ngrid_features = { 'fullWidthRows': True,\n 'syncColumnCellResize': True,\n 'forceFitColumns': True,\n 'rowHeight': 40,\n 'enableColumnReorder': True,\n 'enableTextSelectionOnCells': True,\n 'editable': True,\n 'filterable': False,\n 'sortable': False,\n 'highlightSelectedRow': True}",
"_____no_output_____"
],
[
"from ipywidgets import Button , Layout , interact,widgets\nfrom IPython.display import Javascript, display\n\n# Function: executes previous cell on button widget click event and hides achievement indicators message\n\ndef run_current(ev):\n \n display(Javascript('IPython.notebook.execute_cell_range(IPython.notebook.get_selected_index()+0,IPython.notebook.get_selected_index()+1)')) \n \n# Counter for toggling achievement indicator on/off\n\nbutton_ctr = 0\n\n# Achievement Indicators\n\nline_1 = \"#### Achievement Indicators\"\nline_2 = \"**General Outcome: **\"\nline_3 = \"* Create a table of values from a linear relation, graph the table of values, and analyze the graph to draw conclusions and solve problems\"\n\n# Use to print lines, then save in lines_list\n\ndef print_lines(n):\n \n lines_str = \"\"\n \n for i in range(1,n+1):\n lines_str = lines_str + \"line_\"+str(i)+\",\"\n \n lines_str = lines_str[:-1]\n\n print(lines_str)\n \nlines_list = [line_1,line_2,line_3]\n \n# Show/Hide buttons\n\nai_button_show = widgets.Button(button_style='info',description=\"Show Achievement Indicators\", layout=Layout(width='25%', height='30px') )\nai_button_hide = widgets.Button(button_style='info',description=\"Hide Achievement Indicators\", layout=Layout(width='25%', height='30px') )\n\ndisplay(Markdown(\"For instructors:\"))",
"_____no_output_____"
],
[
"button_ctr += 1\n\nif(button_ctr % 2 == 0):\n\n for line in lines_list:\n display(Markdown(line))\n \n display(ai_button_hide)\n ai_button_hide.on_click( run_current )\n \nelse:\n\n display(ai_button_show)\n ai_button_show.on_click( run_current )",
"_____no_output_____"
],
[
"# Import libraires\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport numpy as np\nimport operator\nimport qgrid as q\nfrom ipywidgets import widgets\nfrom ipywidgets import Button, Layout,interact_manual,interact\nfrom IPython.display import display, Javascript, Markdown\nimport numpy as np\nfrom mpl_toolkits.mplot3d import Axes3D\nfrom ipywidgets import widgets as w\nfrom ipywidgets import Button, Layout\nfrom IPython.display import display, Javascript, Markdown",
"_____no_output_____"
]
],
[
[
"<h1 align='center'>Patterns & Relations</h1>\n\n<h4 align = 'center'> $\\mid$ Grade 7 $\\mid$ Math $\\mid$</h4>\n\n<h2 align='center'>Introduction</h2>\n\nIn this notebook we will learn what an ordered pair is and how we can use a table of values to represent them. We will work with simple linear equations (relations) and tabulate values for them. \n\nWe will also learn what a plane and coordinate plane are and explore the relationship between an equation and a coordinate plane is. \n\nWe will then have an opportunity to practice the concepts we learned via a set of exercises that will help us build and plot a few points for a given linear relation. \n\nThis notebook is one in a series of notebooks that explore the use patterns to describe the world and to solve problems. Please refer to notebook CC-63 for specific outcome 1. ",
"_____no_output_____"
],
[
"We begin with a few definitions. \n\n<div class=\"alert alert-warning\">\n<font color=\"black\"><b>Definition.</b> An **ordered pair** $(n_1,n_2)$ is a pair of numbers where *order* matters. \n </font>\n</div>\n\nFor example, the pair $(1,2)$ is different from the pair $(2,1)$.\n\n<div class=\"alert alert-warning\">\n<font color=\"black\"><b>Definition.</b> An **equation** (also referred to as a **relation**) is an expression asserting that two quantities are equal.\n </font>\n</div>\n\nFor example, \n\n$y = x + 2$ \n\n$y = 3x$\n\n$y = 2$\n\nare all equations.\n\n<div class=\"alert alert-warning\">\n<font color=\"black\"><b>Definition.</b> An **linear equation** (or **linear relation**) is an equation of the form $$y = ax + b$$, where $a,b$ are fixed values. \n </font>\n</div>\n\nFor example,\n\n| a | b|Linear Relation |\n|---|--|-----------|\n|1|2|$$y = x + 2$$|\n| 3 |1|$$y = 3x + 1$$|\n|5|0|$$y = 5x$$ |\n|0|0|$$y = 0$$|\n\n<div class=\"alert alert-warning\">\n<font color=\"black\"><b>Definition.</b> A **table of values** is a set of ordered pairs usually resulting from substituting numbers into an equation. </font>\n</div>\n\nFor example, if we consider the equation \n\n$$y = x + 1$$\n\nand the values $x = 1,2,3$, the table of values corresponds to\n\n| Value for x | Value for y|Ordered Pair (x,y)|\n|---|--|-----|\n|1|2|(1,2)|\n|2|3|(2,3)|\n|3|4|(3,4)|\n\nLet us illustrate this with an example you can interact with.\n",
"_____no_output_____"
],
[
"<h2 align='center'>Interactive Example: Generating a table of values from a linear relation</h2>\n\nLet us take the relation\n\n$$y = x + 3$$\n\nand suppose that $x$ is an integer. We can then obtain different values for $y$, depending on the value of $x$. \n\nThen, if we consider the following values for x:\n\n| | | | |\n|---------|--|--|--|\n| x = ||0|1|2|3|4|5|\n\nWe can substitute each in the equation to obtain a new value of y. \n\n**Activity**\n\nLet us try all entries to illustrate. Using the widget below change the value of $x$. What is the value for $y$ as $x$ changes?\n",
"_____no_output_____"
]
],
[
[
"\n%matplotlib inline\nstyle = {'description_width': 'initial'}\n@interact(x_value=widgets.IntSlider(value=0,\n min=0,\n max=5,\n step=1,\n description='Value for x',\n disabled=False,\n continuous_update=False,\n orientation='horizontal',\n readout=True,\n readout_format='d',\n style =style\n))\n\ndef plug_and_play(x_value):\n fig = plt.figure(figsize=(16,5))\n\n ax1 = fig.add_subplot(1, 3, 1)\n ax2 = fig.add_subplot(1, 3, 2)\n ax3 = fig.add_subplot(1, 3, 3)\n\n ax1.text(0.4,0.5,\"x = \" + str(x_value),fontsize=30)\n ax2.text(0.34,0.7,\"y = x + 3\",fontsize=30)\n ax2.text(0.34,0.5,\"y =\" + str(x_value) + \" + 3\",fontsize=30)\n ax2.text(0.34,0.3,\"y =\" + str(x_value + 3),fontsize=30)\n ax3.text(0.4,0.5,\"(\" + str(x_value) + \",\" + str(x_value + 3) + \")\",fontsize=30)\n ax1.set_title(\"Value for x\",fontsize=30)\n ax2.set_title(\"Value for y\",fontsize=30)\n ax3.set_title(\"Ordered Pair\",fontsize=30)\n ax1.set_xticklabels([]),ax1.set_yticklabels([])\n ax2.set_xticklabels([]),ax2.set_yticklabels([])\n ax3.set_xticklabels([]),ax3.set_yticklabels([])\n ax1.axis(\"Off\"),ax2.axis(\"Off\"),ax3.axis(\"Off\")\n plt.show()\n",
"_____no_output_____"
]
],
[
[
"**Question**\n\nKnowing that the linear relation is $y = x + 3$, what is the value for y, when $x = 2$? Use the widget above to help you find the answer.",
"_____no_output_____"
]
],
[
[
"s = {'description_width': 'initial'} \n\nfrom ipywidgets import interact_manual\ndef question_q(answer):\n if answer==\"Select option\":\n print(\"Click on the correct value for y.\")\n \n elif answer==\"5\":\n ret=\"Correct!\"\n return ret\n elif answer != \"5\" or answer != \"Select Option\":\n ret = \"Not quite.Recall y = x + 3. We know x = 2. What does 2 + 3 equal to?\"\n return ret\nanswer_q = interact(question_q,answer=widgets.Select(\n options=[\"Select option\",\"2\",\\\n \"10\",\"3\",\\\n \"5\"],\n value='Select option',\n description=\"y value\",\n disabled=False,\n style=s\n))",
"_____no_output_____"
]
],
[
[
"**Question**\n\nUsing the correct answer above, what is the corresponding ordered pair? Recall that an ordered pair is of the form $(x,y)$.",
"_____no_output_____"
]
],
[
[
"s = {'description_width': 'initial'} \n\nfrom ipywidgets import interact_manual\ndef question_q(answer):\n if answer==\"Select option\":\n print(\"Click on the correct ordered pair (x,y).\")\n \n elif answer==\"(2,5)\":\n ret=\"Correct!\"\n return ret\n elif answer != \"(2,5)\" or answer != \"Select Option\":\n ret = \"Not quite.Recall x = 2, y = 5. The correct ordered pair is of the form (x,y).\"\n return ret\nanswer_q = interact(question_q,answer=widgets.Select(\n options=[\"Select option\",\"(2,5)\",\\\n \"(2,1)\",\"(5,2)\",\\\n \"(5,3)\"],\n value='Select option',\n description=\"Ordered pair (x,y)\",\n disabled=False,\n style=s\n))",
"_____no_output_____"
]
],
[
[
"Memorizing all different values for $x$ and $y$ is unnecessary. \n\nWe can organize the $x,y$ values along with the corresponding pairs $(x,y)$ in a table as follows.",
"_____no_output_____"
]
],
[
[
"### Create dataframe\n\n#df_num_rows = int(dropdown_widget.value)\n\ngrid_features = { 'fullWidthRows': False,\n 'syncColumnCellResize': True,\n 'forceFitColumns': True,\n 'rowHeight': 40,\n 'enableColumnReorder': True,\n 'enableTextSelectionOnCells': True,\n 'editable': False,\n 'filterable': False,\n 'sortable': False,\n 'highlightSelectedRow': True}\n\n\n# Set up data input for dataframe\n\nx_values = np.array([0,1,2,3,4])\ny_values = x_values + 3\nordered = [(x_values[i],y_values[i]) for i in range(len(x_values))]\ny_equals = [\"y = \" + str(x_values[i]) + \"+3\" for i in range(len(x_values))]\n\n\ndf_num_rows = len(x_values)\nempty_list = [ '' for i in range(df_num_rows) ] \ncategory_list = [ i+1 for i in range(df_num_rows) ] \n\n\ndf_dict = {'Entry Number':category_list,\\\n 'Values for x': empty_list, 'y = x + 3':empty_list,'Values for y': empty_list,\\\n 'Ordered pairs': empty_list}\nfeature_list = ['Entry Number','Values for x','y = x + 3','Values for y','Ordered pairs']\nstudent_df = pd.DataFrame(data = df_dict,columns=feature_list)\n\nstudent_df.set_index('Entry Number',inplace=True)\nstudent_df[\"Values for y\"] = y_values\nstudent_df[\"Values for x\"] = x_values\nstudent_df[\"y = x + 3\"] = y_equals\nstudent_df[\"Ordered pairs\"] = ordered\n\n# Set up & display as Qgrid\nq_student_df = q.show_grid( student_df , grid_options = grid_features )\ndisplay(q_student_df)\n",
"_____no_output_____"
]
],
[
[
"Once we compute a few ordered pairs, we can represent them visually. We define the following two concepts. \n\n\n<div class=\"alert alert-warning\">\n<font color=\"black\"><b>Definition.</b> A **plane** is a flat surface that extends infinitely in all directions.\n </font>\n</div>",
"_____no_output_____"
]
],
[
[
"\n\n\npoint = np.array([1, 1, 1])\nnormal = np.array([0, 0, 1])\n\n# a plane is a*x+b*y+c*z+d=0\n# [a,b,c] is the normal. Thus, we have to calculate\n# d and we're set\nd = -point.dot(normal)\n\n# create x,y\nxx, yy = np.meshgrid(range(10), range(10))\n\n# calculate corresponding z\nz = (-normal[0] * xx - normal[1] * yy - d) * 1. /normal[2]\n\n# plot the surface\nplt3d = plt.figure(figsize=(15,10)).gca(projection='3d')\n\n\nplt3d.plot_surface(xx, yy, z,color=\"#518900\",edgecolor=\"white\")\nplt3d.grid(False)\nplt3d.axis(\"Off\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-warning\">\n<font color=\"black\"><b>Definition.</b> A **coordinate plane** is a plane formed by a horizontal number line (the x-axis) and a vertical number line (the y-axis) that intersect at a point called the origin.\n </font>\n</div>\n\nWe can plot points on the coordinate plane. We use ordered pairs to encode information on where points are located. \n\nRecall that an ordered pair is of the form $(x,y)$. The first entry on the pair denotes how far from the origin along the x-axis the point is, the second entry denotes how far from the origin along the y-axis the point is. \n\nLet's see a simple example for the ordered pair $(1,4)$.\n",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(16,5))\nax1 = fig.add_subplot(1, 3, 1)\nax2 = fig.add_subplot(1, 3, 2)\nax3 = fig.add_subplot(1, 3, 3)\nax2.set_xticks(np.arange(-5,6)),ax2.set_yticks(np.arange(-5,6))\nax2.set_xlim(0,5)\nax2.set_ylim(0,5)\nax1.axis(\"Off\"),ax2.axis(\"On\"),ax3.axis(\"Off\")\nax2.axhline(y=0, color='blue')\nax2.axvline(x=0, color='blue')\nax2.text(5.1,0.1,\"x-axis\",fontsize=20)\nax2.text(0.1,5.1,\"y-axis\",fontsize=20)\nax2.grid(True)\nx_value,y_value = 1,4\nx_or,y_or = 0,0\nax2.scatter(x_value,y_value,color=\"black\",s=120)\nax2.scatter(x_or,y_or,color=\"black\",s=220)\nax2.text(x_value + 0.1,y_value + 0.5,\"(\" +str(x_value) + \",\" + str(y_value) + \")\")\nax2.text(x_or + 0.1,y_or + 0.3,\"origin\")\n \nax2.plot([-5,x_value], [y_value,y_value], color='green', marker='o', linestyle='dashed',\n linewidth=2, markersize=2)\nax2.plot([x_value,x_value], [-5,y_value], color='green', marker='o', linestyle='dashed',\n linewidth=2, markersize=2)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Notice why the order matters. Indeed, if we consider the pair $(4,1)$ we see that it is different. ",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(16,5))\nax1 = fig.add_subplot(1, 3, 1)\nax2 = fig.add_subplot(1, 3, 2)\nax3 = fig.add_subplot(1, 3, 3)\nax2.set_xticks(np.arange(-5,6)),ax2.set_yticks(np.arange(-5,6))\nax2.set_xlim(0,5)\nax2.set_ylim(0,5)\nax1.axis(\"Off\"),ax2.axis(\"On\"),ax3.axis(\"Off\")\nax2.axhline(y=0, color='blue')\nax2.axvline(x=0, color='blue')\nax2.text(5.1,0.1,\"x-axis\",fontsize=20)\nax2.text(0.1,5.1,\"y-axis\",fontsize=20)\nax2.grid(True)\nx_value,y_value = 4,1\nx_or,y_or = 0,0\nax2.scatter(x_value,y_value,color=\"black\",s=120)\nax2.scatter(x_or,y_or,color=\"black\",s=220)\nax2.text(x_value + 0.1,y_value + 0.5,\"(\" +str(x_value) + \",\" + str(y_value) + \")\")\nax2.text(x_or + 0.1,y_or + 0.3,\"origin\")\n \nax2.plot([-5,x_value], [y_value,y_value], color='green', marker='o', linestyle='dashed',\n linewidth=2, markersize=2)\nax2.plot([x_value,x_value], [-5,y_value], color='green', marker='o', linestyle='dashed',\n linewidth=2, markersize=2)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Let us take the table we computed previously for the relation\n\n$$y = x +3$$\n\nalong with the ordered pairs we computed. \n\nWe can then represent the ordered pairs in the coordinate plane. \n\n**Activity**\n\nUse the widget below to see the relationship between the different ordered pairs and the points on the coordinate plane. ",
"_____no_output_____"
]
],
[
[
"%matplotlib inline",
"_____no_output_____"
],
[
"@interact(x_value=widgets.IntSlider(value=0,\n min=0,\n max=5,\n step=1,\n description='Value for x',\n disabled=False,\n continuous_update=False,\n orientation='horizontal',\n readout=True,\n readout_format='d',\n style =style\n))\n\n\ndef show_points(x_value):\n x_values = np.array([0,1,2,3,4,5])\n y_values = x_values + 3\n\n fig = plt.figure()\n plt.subplots_adjust(left=14, bottom=0.2, right=16, top=1.5,\n wspace=0.1, hspace=0.2)\n ax1 = fig.add_subplot(1, 2, 1)\n\n\n ax1.text(0.1,0.8,\"x = \" + str(x_value),fontsize=20)\n ax1.text(0.1,0.6,\"y = \" + str(x_value) +\"+ 3 = \" + str(x_value + 3),fontsize=20)\n ax1.text(0.1,0.4,\"Ordered pair (\" + str(x_value) +\",\" + str(x_value + 3) + \")\",fontsize=20)\n ax1.set_title(\"Values for x and y\", fontsize=25)\n ax2 = fig.add_subplot(1, 2, 2)\n ax2.set_xticks(np.arange(-6,11)),ax2.set_yticks(np.arange(-6,11))\n ax2.set_xlim(0,6)\n ax2.set_ylim(0,9)\n ax1.axis(\"Off\"),ax2.axis(\"On\")\n ax2.axhline(y=0, color='blue')\n ax2.axvline(x=0, color='blue')\n ax2.text(6.5,0.2,\"x-axis\",fontsize=20)\n ax2.text(0.5,9.5,\"y-axis\",fontsize=20)\n ax2.grid(True)\n# for i in range(len(x_values)): \n# ax2.text(x_values[i] - 0.5,y_values[i]-0.7,\"(\" + str(x_values[i]) + \",\" + str(y_values[i]) + \")\") \n points = ax2.scatter(x_values,y_values,color=\"black\",s=60)\n ax2.scatter(x_value,x_value + 3,color=\"red\",s=120)\n #datacursor(points)\n\n plt.show()\n",
"_____no_output_____"
]
],
[
[
"### <h4>Conclusion</h4>\n\nFrom this graph we conclude that the relation between $x$ and $y$ is linear. This makes sense given the equation is of the form \n\n$$y = ax + b$$\n\nwhere $a,b$ are integers and in this particular case, $a = 1, b =3$. \n\nPoints which are of interest are the intersection between $y$ and the x-axis as well as $x$ and the $y$ axis. The former happens exactly when $y = 0$ while the latter occurs when $x=0$.\n\nWe observe that $y$ does not intersect the x axis for positive values of $x$. We also observe that $x$ intersects the y-axis when $x=0$. Such intersection can be observed in the ordered pair $(0,3)$.",
"_____no_output_____"
]
],
[
[
"# Create button and dropdown widget\ndef rerun_cell( b ):\n \n display(Javascript('IPython.notebook.execute_cell_range(IPython.notebook.get_selected_index()+1,IPython.notebook.get_selected_index()+2)')) \nstyle = {'description_width': 'initial'}\n\nnumber_of_cat = 13\ndropdown_options = [ str(i+1) for i in range(number_of_cat) ] \ndropdown_widget = widgets.Dropdown( options = dropdown_options , value = '3' , description = 'Number of entries' , disabled=False,style=style )\n\ncategories_button = widgets.Button(button_style='info',description=\"Enter\", layout=Layout(width='15%', height='30px'))\n\n# Display widgets\n\n#display(dropdown_widget)\n#display(categories_button)\n\n#categories_button.on_click( rerun_cell ) ",
"_____no_output_____"
]
],
[
[
"<h2 align='center'>Practice Area</h2>\n\n<h4>Exercise</h4>\n\nWe will repeat a similar exercise as above, only this time, we will use a different linear relation. \n\n$$y = 2x +4$$\n\nLet us begin by building a simple table. \n\nAnswer the questions below to complete a similar table. \n\n### Question 1\n\nKnowing that $y = 2x + 4$, what is the value of $y$ when $x = 3$? In other words, what does $2(3) + 4$ equal to?",
"_____no_output_____"
]
],
[
[
"s = {'description_width': 'initial'} \n\nfrom ipywidgets import interact_manual\ndef question_q(answer):\n if answer==\"Select option\":\n print(\"Click on the correct value of y.\")\n \n elif answer==\"10\":\n ret=\"Correct!\"\n return ret\n elif answer != \"10\" or answer != \"Select Option\":\n ret = \"You are close to the answer but need to improve your result.Recall 2(3) = 6. What does 6 + 4 equal to?\"\n return ret\nanswer_q = interact(question_q,answer=widgets.Select(\n options=[\"Select option\",\"1\",\\\n \"10\",\"3\",\\\n \"0\"],\n value='Select option',\n description=\"y value\",\n disabled=False,\n style=s\n))",
"_____no_output_____"
]
],
[
[
"### Question 2\n\nKnowing that $y = 2x + 4$, what is the value of $y$ when $x=0$?",
"_____no_output_____"
]
],
[
[
"s = {'description_width': 'initial'} \n\nfrom ipywidgets import interact_manual\ndef question_p(answer):\n if answer==\"Select option\":\n print(\"Click on the correct value of y.\")\n \n elif answer==\"4\":\n ret=\"Correct!\"\n return ret\n elif answer != \"4\" or answer != \"Select Option\":\n ret = \"You are close to the answer but need to improve your result.Recall y = x + 4. What does 0 + 4 equal to?\"\n return ret\nanswer_p = interact(question_p,answer=widgets.Select(\n options=[\"Select option\",\"-1\",\\\n \"10\",\"4\",\\\n \"0\"],\n value='Select option',\n description=\"y value\",\n disabled=False,\n style=s\n))",
"_____no_output_____"
]
],
[
[
"### Question 3\n\nWhat is the ordered pair obtained when $x = 2$? ",
"_____no_output_____"
]
],
[
[
"s = {'description_width': 'initial'} \n\nfrom ipywidgets import interact_manual\ndef question_s(answer):\n if answer==\"Select option\":\n print(\"Click on the correct ordered pair (x,y)\")\n \n elif answer==\"(2,8)\":\n ret=\"Correct!\"\n return ret\n elif answer != \"(2,8)\" or answer != \"Select Option\":\n ret = \"You are close to the answer but need to improve your result.We know y = 8 and x = 2. We also know an ordered pair is of the form (x,y).\"\n return ret\nanswer_s = interact(question_s,answer=widgets.Select(\n options=[\"Select option\",\"(2,6)\",\\\n \"(2,8)\",\"(8,2)\",\\\n \"(2,-2)\"],\n value='Select option',\n description=\"Ordered pair (x,y)\",\n disabled=False,\n style=s\n))",
"_____no_output_____"
],
[
"def math_function(relation,x_val):\n\n y_val = relation[\"+\"](relation[\"Coef1\"]*x_val,relation[\"Coef2\"])\n\n return y_val\n\n\ndef table_of_values_quad(range_val,relation):\n\n empty_list = [ '' for i in range(range_val + 1) ] \n category_list = [ i+1 for i in range(range_val + 1) ] \n\n# Set up data input for dataframe\n\n df_dict = {'Entry Number':category_list,\\\n 'Values for x': empty_list, \\\n 'y ='+ str(relation['Coef1']) + \"x + \" \\\n + str(relation['Coef2']):empty_list,\\\n 'Values for y': empty_list,\\\n 'Ordered pairs': empty_list}\n \n \n feature_list = ['Entry Number','Values for x',\\\n 'y ='+ str(relation['Coef1']) \\\n + \"x + \" + str(relation['Coef2']),\\\n 'Values for y','Ordered pairs']\n \n student_df = pd.DataFrame(data = df_dict,columns=feature_list)\n\n student_df.set_index('Entry Number',inplace=True)\n\n \n x_values = np.array(np.arange(range_val+1))\n y_values = math_function(relation,x_values)\n ordered = [(x_values[i],y_values[i]) for i in range(range_val+1)]\n \n \n y_equals = [\"y = \" + str(relation['Coef1']) +\"(\" + str(x_values[i]) + \")\" \\\n + \"+\" + str(relation['Coef2']) \n for i in range(len(x_values))]\n\n student_df[\"Values for y\"] = y_values\n student_df[\"Values for x\"] = x_values\n student_df['y ='+ str(relation['Coef1']) + \\\n \"x + \" + str(relation['Coef2'])] = y_equals\n student_df[\"Ordered pairs\"] = ordered\n\n q_student_df = q.show_grid( student_df , grid_options = grid_features )\n display(q_student_df)",
"_____no_output_____"
],
[
"\n\ndef generate_tab(value):\n if value==True:\n if \"Correct!\" in str(answer_p.widget.children)\\\n and \"Correct!\" in str(answer_q.widget.children)\\\n and \"Correct!\" in str(answer_s.widget.children):\n relation_ar = {\"Coef1\":2,\"Coef2\":4,\"+\": operator.add}\n table_of_values_quad(4,relation_ar)\n else:\n print(\"At least one of your answers is not correct. Compare your answers with the table.\")\n relation_ar = {\"Coef1\":2,\"Coef2\":4,\"+\": operator.add}\n table_of_values_quad(4,relation_ar)\n\n ",
"_____no_output_____"
],
[
"interact(generate_tab,value = widgets.ToggleButton(\n value=False,\n description='Generate Table',\n disabled=False,\n button_style='info', # 'success', 'info', 'warning', 'danger' or ''\n tooltip='Description',\n icon='check'\n ));",
"_____no_output_____"
]
],
[
[
"### Question 4\n\nUsing the information on the table and the widget below, identify and select what ordered pairs belong to the relation\n\n$$y = 2x + 4$$\n\nSelect one of the four following options. The correct answer will plot all points, the incorrect answer will print a message. ",
"_____no_output_____"
]
],
[
[
"def plot_answer(relation):\n x_values = np.array([0,1,2,3,4])\n y_values = relation[\"Coef1\"]*x_values + relation[\"Coef2\"]\n\n fig = plt.figure()\n plt.subplots_adjust(left=14, bottom=0.2, right=16, top=1.5,\n wspace=0.1, hspace=0.2)\n ax2 = fig.add_subplot(1, 1, 1)\n\n ax2.set_xticks(np.arange(-6,11))\n ax2.set_yticks(np.arange(-6,relation[\"Coef1\"]*x_values[-1] + relation[\"Coef2\"]+2))\n ax2.set_xlim(0,5)\n ax2.set_ylim(0,relation[\"Coef1\"]*x_values[-1] + relation[\"Coef2\"]+1)\n ax2.text(x_values[-1] + 1,0.001,\"x-axis\",fontsize=20)\n ax2.text(0.1,y_values[-1] + 1,\"y-axis\",fontsize=20)\n ax2.grid(True)\n# for i in range(len(x_values)): \n# ax2.text(x_values[i] - 0.5,y_values[i]-0.7,\"(\" + str(x_values[i]) + \",\" + str(y_values[i]) + \")\") \n points = ax2.scatter(x_values,y_values,color=\"black\",s=60)\n #ax2.scatter(x_value,x_value + 3,color=\"red\",s=120)\n #datacursor(points)\n\n plt.show()\ndef choose_points(value):\n if value==\"(3,10),(5,14),(0,4)\":\n print(\"Correct!\")\n rel = {\"Coef1\":2,\"Coef2\":4,\"+\": operator.add}\n plot_answer(rel)\n else:\n print(\"Those do not look like the ordered pairs in our table. Try again.\")\n\ninteract(choose_points,\n value = widgets.RadioButtons(\n options=[\n \"(3,11),(5,11),(2,8)\",\\\n \"(0,0),(1,2),(2,2)\",\\\n \"(3,10),(5,14),(0,4)\",\\\n \"(10,10),(10,8),(1,6)\"],\n# value='pineapple',\n description='Ordered Pairs:',\n disabled=False,\n style = style\n));",
"_____no_output_____"
]
],
[
[
"### Question 5: Conclusions\n\nWhat can you conclude from the table above? Use the following statements to guide your answer and add any other observations you make. \n\n| Statement |\n|-----------|\n|The relation between $x$ and $y$ is linear|\n|There is an intersection between the y-axis and $x$ at the ordered pair ... |\n|There is an intersection between the x-axis and $y$ at the ordered pair ... |\n",
"_____no_output_____"
]
],
[
[
"\nemma1_text = widgets.Textarea( value='', placeholder='Write your answer here. Press Record Answer when you finish.', description='', disabled=False , layout=Layout(width='100%', height='75px') )\nemma1_button = widgets.Button(button_style='info',description=\"Record Answer\", layout=Layout(width='15%', height='30px'))\n\ndisplay(emma1_text)\ndisplay(emma1_button)\n\nemma1_button.on_click( rerun_cell ) ",
"_____no_output_____"
],
[
"emma1_input = emma1_text.value\n\nif(emma1_input != ''):\n \n emma1_text.close()\n emma1_button.close()\n display(Markdown(\"### Your answer for Question 6: Conclusions\"))\n display(Markdown(emma1_input))",
"_____no_output_____"
]
],
[
[
"<h2 align='center'>Experiment</h2>\n\nIn this section you will have an opportunity to explore linear relations parameterized by you, to create their respective tables of values and to plot the ordered pairs. In the end, use what you learned in this notebook to make observations about your findings.\n\nRecall that a linear equation is of the form\n\n$$y = ax + b$$\n\nUse the widget below to choose new values for $a,b$. ",
"_____no_output_____"
]
],
[
[
"def choose(a,b):\n print(\"Equation: \" + str(a) + \"x + \" + str(b))\n return [a,b]\n\ncoeff = interact(choose,a=widgets.IntSlider(value=0,\n min=0,\n max=15,\n step=1,\n description='Value for a',\n disabled=False,\n continuous_update=False,\n orientation='horizontal',\n readout=True,\n readout_format='d',\n style =style)\n,b=widgets.IntSlider(value=0,\n min=0,\n max=15,\n step=1,\n description='Value for b',\n disabled=False,\n continuous_update=False,\n orientation='horizontal',\n readout=True,\n readout_format='d',\n style =style));",
"_____no_output_____"
],
[
"\n\ndef rerun_cell( b ):\n \n display(Javascript('IPython.notebook.execute_cell_range(IPython.notebook.get_selected_index()+1,IPython.notebook.get_selected_index()+3)')) \ntable_button = widgets.Button(button_style='info',description=\"Generate Table of Values and Plot\", layout=Layout(width='25%', height='30px'))\ndisplay(table_button)\n\ntable_button.on_click( rerun_cell ) ",
"_____no_output_____"
],
[
"relation_ar = {\"Coef1\":coeff.widget.kwargs['a'],\"Coef2\":coeff.widget.kwargs['b'],\"+\": operator.add}\ntable_of_values_quad(4,relation_ar)",
"_____no_output_____"
],
[
"plot_answer(relation_ar)",
"_____no_output_____"
]
],
[
[
"<h2 align='center'>Interactive Example: Find the relation from a table of values</h2>\n\nWhat if, instead of knowing what the relation is, we are only given a table of values or a plot? \n\nIf we know that the values belong to a linear relation, this along with the values is enough to determine what the relation is. \n\nConsider the table and the plotted ordered pairs below. ",
"_____no_output_____"
]
],
[
[
"def tabulate_to_eq(relation):\n x_values = np.array([0,1,2,3,4])\n y_values = relation[\"Coef1\"]*x_values + relation[\"Coef2\"]\n ordered = [(x_values[i],y_values[i]) for i in range(len(x_values))]\n\n\n df_num_rows = len(x_values)\n empty_list = [ '' for i in range(df_num_rows) ] \n category_list = [ i+1 for i in range(df_num_rows) ] \n\n\n df_dict_2 = {'Entry Number':category_list,\\\n 'Values for x': empty_list,'Values for y': empty_list,\\\n 'Ordered pairs': empty_list}\n feature_list = ['Entry Number','Values for x','Values for y','Ordered pairs']\n student_df_2 = pd.DataFrame(data = df_dict_2,columns=feature_list)\n\n student_df_2.set_index('Entry Number',inplace=True)\n student_df_2[\"Values for y\"] = y_values\n student_df_2[\"Values for x\"] = x_values\n student_df_2[\"Ordered pairs\"] = ordered\n\n# Set up & display as Qgrid\n q_student_df_2 = q.show_grid( student_df_2 , grid_options = grid_features )\n display(q_student_df_2)\nrels = {\"Coef1\":2,\"Coef2\":1,\"+\": operator.add}\ntabulate_to_eq(rels)",
"_____no_output_____"
],
[
"plot_answer(rels)",
"_____no_output_____"
]
],
[
[
"Can you determine what the equation is based on the ordered pairs? \n\nIn the questions below we will walk towards the solution. ",
"_____no_output_____"
],
[
"## Observation #1\n\nUsing the table or the plot, find what the value of $y$ is when $x = 0$. Enter your answer in the box. When you think you have the correct answer, press the Run Interact button.",
"_____no_output_____"
]
],
[
[
"\n\n\n \ns = {'description_width': 'initial'} \n@interact_manual(answer =widgets.Textarea(\n value=' ',\n placeholder='Type something',\n description='Your Answer:',\n disabled=False,\n style=s))\ndef get_answer_one(answer):\n if \"1\" in answer:\n print(\"Correct!\")\n else:\n print(\"HINT: Look at Entry Number 1 in the table. What is the value for y?\")\n",
"_____no_output_____"
]
],
[
[
"### Observation #2\n\nRecall that a linear relation is of the form $$y = ax + b$$\n\nUse this information along with the answer to Observation #1, to deduce the value of $b$.\n\nEnter your answer in the box below. When you think you have found an answer, press the Run Interact button.",
"_____no_output_____"
]
],
[
[
"\n\n\n \ns = {'description_width': 'initial'} \n@interact_manual(answer =widgets.Textarea(\n value=' ',\n placeholder='Type something',\n description='Your Answer:',\n disabled=False,\n style=s))\ndef get_answer_one(answer):\n if \"1\" in answer:\n print(\"Correct!\")\n else:\n print(\"HINT: y = ax + b. When x = 0, y = 0 + b = 1. This means that 0 + b = 1. What is the value of b?\")\n",
"_____no_output_____"
]
],
[
[
"From the observation above, we determined that the value of $b = 1$, as \n\n$$y = ax + b$$\n\nand when $x =0$, we observe $y = 1$. Via algebraic manipulation, this means that $ 0 +b = 1$ which means $b = 1$. \n\nWe now know our equation is of the form\n\n$$ y =ax + 1$$\n\nThere is only one loose end. We want to get the value of $a$. \n\n### Observation #3\n\nObserve Entry Number 2. In there we see that the ordered pair is $(1,3)$. This means that if $x = 1$, then $y = 3$. \n\nIn our equation, this looks as follows:\n\n$$y = a(1) + 1 = 3$$\n\nWhich is equivalent to \n\n$$y = a + 1 = 3$$\n\nWhat is the value of $a$?",
"_____no_output_____"
]
],
[
[
"\n\n \ns = {'description_width': 'initial'} \n@interact_manual(answer =widgets.Textarea(\n value=' ',\n placeholder='Type something',\n description='Your Answer:',\n disabled=False,\n style=s))\ndef get_answer_one(answer):\n if \"2\" in answer:\n print(\"Correct!\")\n else:\n print(\"HINT: a + 1 = 3. What value, when added to 1, results in 3? \")\n",
"_____no_output_____"
]
],
[
[
"### Recap\n\nObserve that all we needed to find the linear equation were the first two entries in the table. \n\nIndeed, we used Entry Number 1, x = 0, y = 1 to determine that b = 1. \n\nWe then used this, along with Entry Number 2 x = 1, y = 3, to determine that a = 2. \n\nThis yields to the linear equation \n\n$$y = 2x + 1$$\n\nUse the widget below to verify that this linear equation generates the adequate table of values.",
"_____no_output_____"
]
],
[
[
"@interact(x_value=widgets.IntSlider(value=0,\n min=0,\n max=4,\n step=1,\n description='Value for x',\n disabled=False,\n continuous_update=False,\n orientation='horizontal',\n readout=True,\n readout_format='d',\n style =style\n))\n\n\ndef verify_points(x_value):\n relation = {\"Coef1\":2,\"Coef2\":1,\"+\": operator.add}\n x_values = np.array([0,1,2,3,4])\n y_values = relation[\"Coef1\"]*x_values + relation[\"Coef2\"]\n\n fig = plt.figure()\n plt.subplots_adjust(left=14, bottom=0.2, right=16, top=1.5,\n wspace=0.1, hspace=0.2)\n ax1 = fig.add_subplot(1, 2, 1)\n\n\n ax1.text(0.1,0.8,\"x = \" + str(x_value),fontsize=20)\n ax1.text(0.1,0.6,\"y = \" + str(relation[\"Coef1\"]) + \"x + \"+ str(relation[\"Coef2\"]),fontsize=20)\n ax1.text(0.1,0.4,\"y = 2(\" + str(x_value) + \") + 1 = \" + str(2*x_value)+ \" + 1 = \" + str(relation[\"Coef1\"]*x_value+ relation[\"Coef2\"]),fontsize=20)\n ax1.text(0.1,0.2,\"Ordered pair (\" +str(x_value) + \",\" + str(relation[\"Coef1\"]*x_value+ relation[\"Coef2\"]) + \")\",fontsize=20)\n ax1.set_title(\"Values for x and y\", fontsize=25)\n ax2 = fig.add_subplot(1, 2, 2)\n ax2.set_xticks(np.arange(-6,x_values[-1]+2)),ax2.set_yticks(np.arange(-6,y_values[-1]+2))\n ax2.set_xlim(0,x_values[-1]+1)\n ax2.set_ylim(0,y_values[-1]+1)\n ax1.axis(\"Off\"),ax2.axis(\"On\")\n ax2.axhline(y=0, color='blue')\n ax2.axvline(x=0, color='blue')\n ax2.text(x_values[-1]+1,0.2,\"x-axis\",fontsize=20)\n ax2.text(0.1,y_values[-1]+1,\"y-axis\",fontsize=20)\n ax2.grid(True)\n# for i in range(len(x_values)): \n# ax2.text(x_values[i] - 0.5,y_values[i]-0.7,\"(\" + str(x_values[i]) + \",\" + str(y_values[i]) + \")\") \n points = ax2.scatter(x_values,y_values,color=\"black\",s=60)\n ax2.scatter(x_value,relation[\"Coef1\"]*x_value+ relation[\"Coef2\"] ,color=\"red\",s=120)\n #datacursor(points)\n\n plt.show()",
"_____no_output_____"
],
[
"tabulate_to_eq({\"Coef1\":2,\"Coef2\":1,\"+\": operator.add})",
"_____no_output_____"
]
],
[
[
"Why do you think equations of the form\n\n$$y = ax + b$$\n\nare called \"linear\"? \n\nUse the box below to enter your answer. ",
"_____no_output_____"
]
],
[
[
"\n\nemma1_text = w.Textarea( value='', placeholder='Write your answer here. Press Record Answer when you finish.', description='', disabled=False , layout=Layout(width='100%', height='75px') )\nemma1_button = w.Button(button_style='info',description=\"Record Answer\", layout=Layout(width='15%', height='30px'))\n\ndisplay(emma1_text)\ndisplay(emma1_button)\n\nemma1_button.on_click( rerun_cell ) ",
"_____no_output_____"
]
],
[
[
"<h2 align='center'>Conclusion</h2>\n\nIn this notebook we learned what an ordered pair is. We also learned what a table of values is as well as a plane and a coordinate plane. Furthermore, we learned that given a relation between x and y, we can track and represent the relation between x and y via a table of values or a coordinate plane.\n\nWe analyzed basic linear relations, tabulated their values and plotted on a coordinate plane. We explored the pairs that intersected the y and x axis and made remarks based on our observations. ",
"_____no_output_____"
],
[
"[](https://github.com/callysto/curriculum-notebooks/blob/master/LICENSE.md)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d0d051b764321f173c5ac26b76a5d85a080788a8 | 7,663 | ipynb | Jupyter Notebook | teaching_material/session_10/gruppe_17/Case 2 DOA.ipynb | tlh957/DO2021 | 20c615451240a80bc5a2100e15828dfc4163fd49 | [
"MIT"
] | 20 | 2021-09-08T12:14:32.000Z | 2021-11-19T11:57:39.000Z | teaching_material/session_10/gruppe_17/Case 2 DOA.ipynb | tlh957/DO2021 | 20c615451240a80bc5a2100e15828dfc4163fd49 | [
"MIT"
] | 10 | 2021-08-12T14:41:18.000Z | 2021-11-27T12:41:34.000Z | teaching_material/session_10/gruppe_17/Case 2 DOA.ipynb | tlh957/DO2021 | 20c615451240a80bc5a2100e15828dfc4163fd49 | [
"MIT"
] | 20 | 2021-09-12T22:13:22.000Z | 2021-12-07T19:27:05.000Z | 23.291793 | 117 | 0.552917 | [
[
[
"# Python code til udregning af data fra ATP\n",
"_____no_output_____"
]
],
[
[
"#Imports \n\n",
"_____no_output_____"
]
],
[
[
"# Udregninger",
"_____no_output_____"
],
[
"## Alder for at kunne blive tilbudt tidlig pension",
"_____no_output_____"
]
],
[
[
"#Årgange født i 1955-1960 har adgang til at søge i 2021. \n#Man skal være fyldt 61 for at søge. \nprint(2021-61, \"kan anmode om tidlig pension\")\n#Der indgår personer fra 6 1⁄2 årgange\nprint(2021-66, \"sidste år inden folkepension indtræder\")\n#da personer født 1.halvår 1955 har nået folkepensionsalderen inden 1. januar 2022.",
"1960 kan anmode om tidlig pension\n1955 sidste år inden folkepension indtræder\n"
]
],
[
[
"## Forventede ansøgning",
"_____no_output_____"
]
],
[
[
"#Omkring 38.000 helårspersoner forventes at opnå ret til at gå på tidligt pension i 2022. \n#Det dækker over muligheden for at 1, 2 eller 3 år. \n#22.000 vil benytte sig af retten i 2022, 6000 fra beskæftigelse. \nansøgning = 38100 #personer\nbenyttelse = 22000 #benytter tidlig pension\nprint(\"Det vil sige, at dem der afstår retten til tidlig pension er\", ansøgning-benyttelse, \"personer\")\n",
"Det vil sige, at dem der afstår retten til tidlig pension er 16100 personer\n"
]
],
[
[
"## Personer med 44+ på arbejdsmarkedet",
"_____no_output_____"
]
],
[
[
"#Personer med 44+ år på arbejdsmarkedet har automatisk fuld anciennitet.\nautomatisk_behandling = 1.7+7.1\nberettiget = 33.9\nprocent = round(automatisk_behandling/berettiget*100)\n\nprint(automatisk_behandling, \"af de\", round(automatisk_behandling/berettiget*100), \n \"% af ansøgerne har automatisk fuld anciennitet er\", round(ansøgning/100*procent), \"personer\")\n\n#8,8 ud af 33,9 = 26 % af ansøgerne har automatisk fuld anciennitet. 26% af 38.100 = 9906 personer.",
"8.799999999999999 af de 26 % af ansøgerne har automatisk fuld anciennitet er 9906 personer\n"
]
],
[
[
"## Manuel håndtering",
"_____no_output_____"
]
],
[
[
"#Personer der har 42 eller 43 års anciennitet på arbejdsmarkedet kræver manuel håndtering.\nresterende = 74\n#74% af 38.100 ansøgere = 28.194 ansøgere kræver manuel håndtering\nprint(round(ansøgning/100*resterende),\"ansøgere kræver manuel håndtering\")",
"28194 ansøgere kræver manuel håndtering\n"
]
],
[
[
"## Supplerende dokumentation",
"_____no_output_____"
]
],
[
[
"#Vi forventer, at 50% af ansøgerne der kræver manuel behandling indsender supplerende dokumentation. \nhalvdelen = 2\nmanuel_håndtering = 28194\n#50% af 28.194 = 14.097 personer.\nprint(round(manuel_håndtering/halvdelen))",
"14097\n"
]
],
[
[
"## Uger i ansøgningsperioden",
"_____no_output_____"
]
],
[
[
"#Hvor mange uger er der i perioden d. 1. august – 31. december?\nantal_uger = 52 #uger\nuger_i_perioden = 31 #uger\nprint(antal_uger-uger_i_perioden,\"uger ansøgningsperioden\")\n",
"21 uger ansøgningsperioden\n"
]
],
[
[
"## Arbejdstimer",
"_____no_output_____"
]
],
[
[
"#Hvor mange arbejdstimer er der behov for i perioden?\n#ATP har et erfaringsbaseret estimat for tidsforbruget ifm. manuel behandling af en typisk ansøgning, \n#svarende til 30 minutter.\n\nsupplerende = 14097 #antal ansøgning der kræver \nhalvtime = 2 #halv time\n\nprint(supplerende/halvtime,\" antal arbejdstimer\")\n\n",
"7048.5 antal arbejdstimer\n"
]
],
[
[
"## antal medarbejder",
"_____no_output_____"
]
],
[
[
"#Hvor mange timer om ugen er det rimeligt at sige at hver medarbejder bruger på sagsbehandling? \n#Der går også timer på administration, interne møder og andet.\nuger_ansøgning = 21 #uger\nårsværk_ATP = 1356 #timer\nprocent_år = 40.38 #procent\nprint(uger_ansøgning/antal_uger*100, \"% af et år, af de 1356 timer =\", round(årsværk_ATP*0.404),\"timer\")\nprint(round((supplerende/halvtime)/548),\"medarbejder\")",
"40.38461538461539 % af et år, af de 1356 timer = 548 timer\n13 medarbejder\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0d053a5f0aa9dd81b5c6f134abed9f386946b76 | 3,080 | ipynb | Jupyter Notebook | 04 Build and operate machine learning solutions with Azure Databricks/mslearn-dp100/02 - Get AutoML Prediction.ipynb | raj713335/DP-100 | 11c2550975f0e9ecec3fa6502fdb5d54ac54c714 | [
"Apache-2.0"
] | 1 | 2022-03-28T07:56:58.000Z | 2022-03-28T07:56:58.000Z | 04 Build and operate machine learning solutions with Azure Databricks/mslearn-dp100/02 - Get AutoML Prediction.ipynb | raj713335/DP-100 | 11c2550975f0e9ecec3fa6502fdb5d54ac54c714 | [
"Apache-2.0"
] | null | null | null | 04 Build and operate machine learning solutions with Azure Databricks/mslearn-dp100/02 - Get AutoML Prediction.ipynb | raj713335/DP-100 | 11c2550975f0e9ecec3fa6502fdb5d54ac54c714 | [
"Apache-2.0"
] | null | null | null | 29.902913 | 226 | 0.437338 | [
[
[
"# Get Diabetes Prediction from Automated ML Endpoint\n\nModify the code in the cell below to use the ***ENDPOINT*** and ***PRIMARY_KEY*** for your **auto-predict-diabetes** service. Then run the cell to submit new patient data and retrieve the predicted classifications. ",
"_____no_output_____"
]
],
[
[
"endpoint = 'ENDPOINT' #Replace with your endpoint\nkey = 'PRIMARY_KEY' #Replace with your key\n\nimport json\nimport requests\n\n#Features for a patient\ndata = {\n \"Inputs\": {\n \"data\": [\n {\"PatientID\": 1,\n \"Pregnancies\": 5,\n \"PlasmaGlucose\": 181.0,\n \"DiastolicBloodPressure\": 90.6,\n \"TricepsThickness\": 34.0,\n \"SerumInsulin\": 23.0,\n \"BMI\": 43.51,\n \"DiabetesPedigree\": 1.21,\n \"Age\": 21.0\n }\n ]\n }\n}\n\n#Create a \"data\" JSON object\nbody = str.encode(json.dumps(data))\n\n#Set the content type and authentication for the request\nheaders = {\"Content-Type\":\"application/json\",\n \"Authorization\":\"Bearer \" + key}\n\n#Send the request\nresponse = requests.post(endpoint, body, headers=headers)\n\n#If we got a valid response, display the predictions\nif response.status_code == 200:\n y = response.json()\n #Get the first prediction in the results\n if y[\"Results\"][0] == 1:\n print('Diabetic')\n else:\n print(\"Not Diabetic\")\nelse:\n print(response)",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
]
] |
d0d06a94361b547504200fd59004360c5f8b0ca3 | 16,209 | ipynb | Jupyter Notebook | supplementary/notebooks/2. Python reimplementation/2.3 Differences in code - Python.ipynb | bebatut/pypairs | cea1a1589dda178fcc0f33c38f1495b494460428 | [
"MIT"
] | null | null | null | supplementary/notebooks/2. Python reimplementation/2.3 Differences in code - Python.ipynb | bebatut/pypairs | cea1a1589dda178fcc0f33c38f1495b494460428 | [
"MIT"
] | null | null | null | supplementary/notebooks/2. Python reimplementation/2.3 Differences in code - Python.ipynb | bebatut/pypairs | cea1a1589dda178fcc0f33c38f1495b494460428 | [
"MIT"
] | null | null | null | 36.180804 | 1,168 | 0.559689 | [
[
[
"# Speed comparison between PyPairs and the R verison - PyPairs\n\nHere we ran the sandbag part of the original Pairs method on the oscope dataset for a growing subset of genes. Taking note of the required execution time. Single cored time is taken. For the result please see: [2.3 Differences in code - Python](./2.3%20Differences%20in%20code%20-%20R.ipynb)",
"_____no_output_____"
],
[
"<div id=\"toc\"></div>",
"_____no_output_____"
],
[
"## Neccessary Imports",
"_____no_output_____"
]
],
[
[
"%%javascript\n$.getScript('https://kmahelona.github.io/ipython_notebook_goodies/ipython_notebook_toc.js')",
"_____no_output_____"
],
[
"import sys\ncode = \"./../../code/\"\ndata = \"./../../data/\"\nsys.path.append(code)\nimport pandas\nimport pypairs as pairs\nfrom plotly.offline import download_plotlyjs, init_notebook_mode, plot, iplot\nimport plotly.graph_objs as go\nimport numpy as np\nfrom pathlib import Path\nfrom tqdm import tqdm_notebook as tqdm\nimport helper\nimport timeit\n\ninit_notebook_mode(connected=True)",
"_____no_output_____"
]
],
[
[
"## Loading Oscope Dataset",
"_____no_output_____"
]
],
[
[
"# Load matrix\noscope_gencounts = pandas.read_csv(Path(data + \"data/GSE64016_H1andFUCCI_normalized_EC_human.csv\"))\n\n# Set index right\noscope_gencounts.set_index(\"Unnamed: 0\", inplace=True)\n\n# Subset sorted\noscope_gencounts_sorted = oscope_gencounts.iloc[:, [oscope_gencounts.columns.get_loc(c) for c in oscope_gencounts.columns if \"G1_\" in c or \"G2_\" in c or \"S_\" in c]]\n\n# Define annotation\nis_G1 = [oscope_gencounts_sorted.columns.get_loc(c) for c in oscope_gencounts_sorted.columns if \"G1_\" in c]\nis_S = [oscope_gencounts_sorted.columns.get_loc(c) for c in oscope_gencounts_sorted.columns if \"S_\" in c]\nis_G2M = [oscope_gencounts_sorted.columns.get_loc(c) for c in oscope_gencounts_sorted.columns if \"G2_\" in c]\n\nannotation = {\n \"G1\": list(is_G1),\n \"S\": list(is_S),\n \"G2M\": list(is_G2M)\n}\n\nno_genes = len(oscope_gencounts_sorted.index) - 1\n\nprint(\"Total number of genes in oscope dataset {}\".format(no_genes))",
"Total number of genes in oscope dataset 19083\n"
]
],
[
[
"## Running sandbag with increasing number of genes",
"_____no_output_____"
],
[
"Notice: Long runtime, result stored in magic please see [Results](#Results)",
"_____no_output_____"
]
],
[
[
"t = []\ngenes = [10,100,500,1000,5000,10000,19000]\nfor g in tqdm(genes):\n \n sub = helper.random_subset(range(0, no_genes), g)\n subset = oscope_gencounts_sorted.iloc[sub, :]\n \n start = timeit.default_timer()\n oscope_marker_pairs = pairs.sandbag(x=subset, phases=annotation, fraction=0.65, processes=1, verbose=True)\n time_sandbag = timeit.default_timer() - start\n t.append(time_sandbag)\n",
"_____no_output_____"
],
[
"%store t",
"Stored 't' (list)\n"
]
],
[
[
"## Results\n\nPython times are feched from store magic, R times were copied manually ",
"_____no_output_____"
]
],
[
[
"%store -r",
"_____no_output_____"
],
[
"t_python = t\nt_r = [0.01, 0.08, 1.49, 6.37, 180.56, 803.64, 2761.00]",
"_____no_output_____"
],
[
"# Create traces\ntrace0 = go.Scatter(\n x= [10,100,500,1000,5000,10000,19000],\n y= t_python,\n mode='markers+lines',\n marker=dict(\n symbol='circle',\n size=10,\n color='green',\n ),\n name='PyPairs'\n)\n\ntrace1 = go.Scatter(\n x= [10,100,500,1000,5000,10000,19000],\n y= t_r,\n mode='markers+lines',\n marker=dict(\n symbol='square',\n size=10,\n color='blue',\n ),\n name='R Version'\n)\n\nlayout = go.Layout(\n title='Speed comparison: R implementation vs PyPairs',\n xaxis=dict(\n title='No. of genes',\n ),\n yaxis=dict(\n title='Time in ms',\n )\n)\n\ndata = go.Figure(data=[trace0, trace1], layout=layout)\n\niplot(data)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d0d06f8fc469bb0cb64844ed24d1598b15404fb8 | 372,796 | ipynb | Jupyter Notebook | notebooks/CORD-19-viewer.ipynb | dartar/habeas-corpus | 12b150691ea20b8f5e1aa1fa2d1df822e99fe64c | [
"MIT"
] | null | null | null | notebooks/CORD-19-viewer.ipynb | dartar/habeas-corpus | 12b150691ea20b8f5e1aa1fa2d1df822e99fe64c | [
"MIT"
] | null | null | null | notebooks/CORD-19-viewer.ipynb | dartar/habeas-corpus | 12b150691ea20b8f5e1aa1fa2d1df822e99fe64c | [
"MIT"
] | null | null | null | 739.674603 | 335,328 | 0.946384 | [
[
[
"# CORD-19 Software Mentions\n\nA notebook to explore the dataset and output some lightly processed data\n\nCollaborations Workshop 2021 HackDay Project \"Habeas Corpus\"\n\nhttps://github.com/softwaresaved/habeas-corpus\n",
"_____no_output_____"
],
[
"## Setup imports and files",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport csv\nimport ast\nimport collections\nimport matplotlib.pyplot as plt\nfrom wordcloud import WordCloud",
"_____no_output_____"
],
[
"CORD19_CSVFILE = '../data/cord-19/CORD19_software_mentions.csv'\nPOPULARITY_CSVFILE = '../data/output/CORD19_software_popularity.csv'",
"_____no_output_____"
]
],
[
[
"## Have a quick look at the data using pandas",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv(CORD19_CSVFILE)\ndf.head()",
"_____no_output_____"
]
],
[
[
"## Load the data and extract the software mentions",
"_____no_output_____"
]
],
[
[
"software = []\n\nwith open(CORD19_CSVFILE, newline='') as csvfile:\n reader = csv.DictReader(csvfile)\n for row in reader:\n mentions = set(ast.literal_eval(row['software']))\n for mention in mentions:\n software.append(mention)\n ",
"_____no_output_____"
],
[
"len(software)",
"_____no_output_____"
]
],
[
[
"## Take a quick look at the statistics",
"_____no_output_____"
]
],
[
[
"occurrences = collections.Counter(software)\n\nlen(occurrences)",
"_____no_output_____"
],
[
"common_software = occurrences.most_common(20)",
"_____no_output_____"
],
[
"common_software",
"_____no_output_____"
],
[
"labels, ys = zip(*common_software)\nxs = np.arange(len(labels)) \nwidth = 1\n\nplt.bar(xs, ys, width, align='center')\nplt.title('Frequency of software mentions in CORD-19 data')\nplt.xlabel('Software Title')\nplt.ylabel('Number of Mentions')\n\nplt.xticks(xs, labels, rotation='vertical')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Display the world cloud",
"_____no_output_____"
]
],
[
[
"wordcloud = WordCloud(max_font_size=50).generate_from_frequencies(occurrences)\nplt.figure(figsize=(15,8))\nplt.imshow(wordcloud)\nplt.axis(\"off\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Output the slightly processed data",
"_____no_output_____"
]
],
[
[
"with open(POPULARITY_CSVFILE, 'w', newline='') as csvfile:\n writer = csv.writer(csvfile)\n for row in occurrences.most_common(len(occurrences)):\n writer.writerow(row) ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0d0802665c24b3735e4aa194978d307380f7e4d | 121,924 | ipynb | Jupyter Notebook | Test_PolicyGuidedTreeSearch.ipynb | reymond-group/CASP-and-dataset-performance | ee335a955a42318da24f2a18d7a5709b5fa03c5c | [
"MIT"
] | 18 | 2019-11-13T10:05:30.000Z | 2022-03-22T09:58:43.000Z | Test_PolicyGuidedTreeSearch.ipynb | reymond-group/CASP-and-dataset-performance | ee335a955a42318da24f2a18d7a5709b5fa03c5c | [
"MIT"
] | 2 | 2020-02-24T10:54:45.000Z | 2021-03-02T09:59:28.000Z | Test_PolicyGuidedTreeSearch.ipynb | reymond-group/CASP-and-dataset-performance | ee335a955a42318da24f2a18d7a5709b5fa03c5c | [
"MIT"
] | 11 | 2020-02-25T09:37:23.000Z | 2021-05-24T08:27:14.000Z | 367.240964 | 19,868 | 0.93482 | [
[
[
"#Use CPU\n# %env CUDA_DEVICE_ORDER=PCI_BUS_ID #remove if TF-CPU\n# %env CUDA_VISIBLE_DEVICES=-1 #remove if TF-CPU",
"_____no_output_____"
],
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport pickle\nimport hashlib\nimport itertools\nfrom rdkit import Chem",
"_____no_output_____"
],
[
"from aizynthfinder import AiZynthFinder",
"Using TensorFlow backend.\n"
],
[
"finder = AiZynthFinder()",
"_____no_output_____"
],
[
"finder.load_stock(\"test_stock\")",
"Loading Stockfile: /projects/mai/synthesisplanning/data/stocks/Enamine_BBstock_all.h5\n"
],
[
"policy_files = ( #Path to policy model file as .hdf,\n #Path to template library file as .hdf)\nfinder.policy_files = policy_files\nfinder.load_policy()\nfinder.policy.use_prior = True\nfinder.policy.cutoff_cumulative = 0.995\nfinder.policy.cutoff_number = 50 #number of templates to examine \nfinder.time_limit = 120 #in seconds",
"Loading Policy: /projects/mai/knwb390_thakkar/synthesis_planning/fulldata_03_05_19_rdchiral/rehashed_models/full_uspto_03_05_19_fp2048m3s0/checkpoints/weights.hdf5\nLoading Templates: /projects/mai/knwb390_thakkar/synthesis_planning/fulldata_03_05_19_rdchiral/rehashed_data/usptofp2048m3s0/full_uspto_03_05_19_rdchiral_template_library.csv.hdf\n"
],
[
"n = 100 #max 100\n \nsmiles = 'CC1=C(C(=CC=C1)C)N(CC(=O)NC2=CC=C(C=C2)C3=NOC=N3)C(=O)C4CCS(=O)(=O)CC4' #Amenamevir\nfinder.target_smiles = smiles\nfinder.prepare_tree()\nresult = finder.tree_search(stop_when_solved=False)\nroute = finder.extract_route()\nsteps = len(route[0])\nresult.append(steps)\nprint('---SMILES---')\nprint(str(smiles + '\\n'))\nprint('---Compounds to Purchase---')\nroute[1][0].state.display()\nprint('---Route---')\nprint(finder.route_to_text(route) + '\\n')\nprint('---Time to Solved---')\nprint(str(result[0]) + '\\n')\nprint('---Solved---')\nif result[1] == 1:\n print('Solved' + '\\n')\nelse:\n print('Unsolved' + '\\n')\nprint('---Number of Steps---')\nprint(str(result[-1]) + '\\n')",
"Defining tree root: Cc1cccc(C)c1N(CC(=O)Nc1ccc(-c2ncon2)cc1)C(=O)C1CCS(=O)(=O)CC1\nStarting search\n................................................................................ \n....................Search completed\nAnalyzing_routes\nBest Score 0.97\n---SMILES---\nCC1=C(C(=CC=C1)C)N(CC(=O)NC2=CC=C(C=C2)C3=NOC=N3)C(=O)C4CCS(=O)(=O)CC4\n\n---Compounds to Purchase---\n"
],
[
"finder.display_route(route)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0d0812519b071c3fbe5d4730894a065e356f61b | 6,429 | ipynb | Jupyter Notebook | Crash Course on Python/WEEK 5/Methods and Classes/ utf-8''C1M5L2_Methods_and_Classes_V3.ipynb | atharvpuranik/Google-IT-Automation-with-Python-Professional-Certificate | 4d8fd587fa85ea4db62db6142fbb58cd9c29bb69 | [
"MIT"
] | 42 | 2020-04-28T09:06:21.000Z | 2022-01-09T01:01:55.000Z | Crash Course on Python/WEEK 5/Methods and Classes/ utf-8''C1M5L2_Methods_and_Classes_V3.ipynb | vaquarkhan/Google-IT-Automation-with-Python-Professional-Certificate | d87dffe924de218f73d61d27689798646824ed6c | [
"MIT"
] | null | null | null | Crash Course on Python/WEEK 5/Methods and Classes/ utf-8''C1M5L2_Methods_and_Classes_V3.ipynb | vaquarkhan/Google-IT-Automation-with-Python-Professional-Certificate | d87dffe924de218f73d61d27689798646824ed6c | [
"MIT"
] | 52 | 2020-05-12T05:29:46.000Z | 2022-01-26T21:24:08.000Z | 26.899582 | 342 | 0.54783 | [
[
[
"# Practice Notebook: Methods and Classes",
"_____no_output_____"
],
[
"The code below defines an *Elevator* class. The elevator has a current floor, it also has a top and a bottom floor that are the minimum and maximum floors it can go to. Fill in the blanks to make the elevator go through the floors requested.",
"_____no_output_____"
]
],
[
[
"class Elevator:\n def __init__(self, bottom, top, current):\n \"\"\"Initializes the Elevator instance.\"\"\"\n self.bottom=bottom\n self.top=top\n self.current=current\n def __str__(self):\n \"\"\"Information about Current floor\"\"\"\n return \"Current floor: {}\".format(self.current)\n def up(self):\n \"\"\"Makes the elevator go up one floor.\"\"\"\n if self.current<10:\n self.current+=1\n def down(self):\n \"\"\"Makes the elevator go down one floor.\"\"\"\n if self.current > 0:\n self.current -= 1\n def go_to(self, floor):\n \"\"\"Makes the elevator go to the specific floor.\"\"\"\n if floor >= self.bottom and floor <= self.top:\n self.current = floor\n elif floor < 0:\n self.current = 0\n else:\n self.current = 10\n\nelevator = Elevator(-1, 10, 0)",
"_____no_output_____"
]
],
[
[
"This class is pretty empty and doesn't do much. To test whether your *Elevator* class is working correctly, run the code blocks below.",
"_____no_output_____"
]
],
[
[
"elevator.up() \nelevator.current #should output 1",
"_____no_output_____"
],
[
"elevator.down() \nelevator.current #should output 0",
"_____no_output_____"
],
[
"elevator.go_to(10) \nelevator.current #should output 10",
"_____no_output_____"
]
],
[
[
"If you get a **<font color =red>NameError</font>** message, be sure to run the *Elevator* class definition code block first. If you get an **<font color =red>AttributeError</font>** message, be sure to initialize *self.current* in your *Elevator* class.",
"_____no_output_____"
],
[
"Once you've made the above methods output 1, 0 and 10, you've successfully coded the *Elevator* class and its methods. Great work!\n<br><br>\nFor the up and down methods, did you take into account the top and bottom floors? Keep in mind that the elevator shouldn't go above the top floor or below the bottom floor. To check that out, try the code below and verify if it's working as expected. If it's not, then go back and modify the methods so that this code behaves correctly.",
"_____no_output_____"
]
],
[
[
"# Go to the top floor. Try to go up, it should stay. Then go down.\nelevator.go_to(10)\nelevator.up()\nelevator.down()\nprint(elevator.current) # should be 9\n# Go to the bottom floor. Try to go down, it should stay. Then go up.\nelevator.go_to(-1)\nelevator.down()\nelevator.down()\nelevator.up()\nelevator.up()\nprint(elevator.current) # should be 1",
"9\n1\n"
]
],
[
[
"Now add the __str__ method to your *Elevator* class definition above so that when printing the elevator using the **print( )** method, we get the current floor together with a message. For example, in the 5th floor it should say \"Current floor: 5\"",
"_____no_output_____"
]
],
[
[
"elevator.go_to(5)\nprint(elevator)",
"Current floor: 5\n"
]
],
[
[
"Remember, Python uses the default method, that prints the position where the object is stored in the computer’s memory. If your output is something like: <br>\n> <__main__.Elevator object at 0x7ff6a9ff3fd0>\n\nThen you will need to add the special __str__ method, which returns the string that you want to print. Try again until you get the desired output, \"Current floor: 5\".",
"_____no_output_____"
],
[
"Once you have successfully produced the desired output, you are all done with this practice notebook. Awesome!",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d0d08d801e780368c3147257d7956fe43f3a3bfe | 246,700 | ipynb | Jupyter Notebook | experiments/tuned_1v2/oracle.run2_limited/trials/9/trial.ipynb | stevester94/csc500-notebooks | 4c1b04c537fe233a75bed82913d9d84985a89177 | [
"MIT"
] | null | null | null | experiments/tuned_1v2/oracle.run2_limited/trials/9/trial.ipynb | stevester94/csc500-notebooks | 4c1b04c537fe233a75bed82913d9d84985a89177 | [
"MIT"
] | null | null | null | experiments/tuned_1v2/oracle.run2_limited/trials/9/trial.ipynb | stevester94/csc500-notebooks | 4c1b04c537fe233a75bed82913d9d84985a89177 | [
"MIT"
] | null | null | null | 92.500937 | 73,640 | 0.783895 | [
[
[
"# PTN Template\nThis notebook serves as a template for single dataset PTN experiments \nIt can be run on its own by setting STANDALONE to True (do a find for \"STANDALONE\" to see where) \nBut it is intended to be executed as part of a *papermill.py script. See any of the \nexperimentes with a papermill script to get started with that workflow. ",
"_____no_output_____"
]
],
[
[
"%load_ext autoreload\n%autoreload 2\n%matplotlib inline\n\n \nimport os, json, sys, time, random\nimport numpy as np\nimport torch\nfrom torch.optim import Adam\nfrom easydict import EasyDict\nimport matplotlib.pyplot as plt\n\nfrom steves_models.steves_ptn import Steves_Prototypical_Network\n\nfrom steves_utils.lazy_iterable_wrapper import Lazy_Iterable_Wrapper\nfrom steves_utils.iterable_aggregator import Iterable_Aggregator\nfrom steves_utils.ptn_train_eval_test_jig import PTN_Train_Eval_Test_Jig\nfrom steves_utils.torch_sequential_builder import build_sequential\nfrom steves_utils.torch_utils import get_dataset_metrics, ptn_confusion_by_domain_over_dataloader\nfrom steves_utils.utils_v2 import (per_domain_accuracy_from_confusion, get_datasets_base_path)\nfrom steves_utils.PTN.utils import independent_accuracy_assesment\n\nfrom steves_utils.stratified_dataset.episodic_accessor import Episodic_Accessor_Factory\n\nfrom steves_utils.ptn_do_report import (\n get_loss_curve,\n get_results_table,\n get_parameters_table,\n get_domain_accuracies,\n)\n\nfrom steves_utils.transforms import get_chained_transform",
"_____no_output_____"
]
],
[
[
"# Required Parameters\nThese are allowed parameters, not defaults\nEach of these values need to be present in the injected parameters (the notebook will raise an exception if they are not present)\n\nPapermill uses the cell tag \"parameters\" to inject the real parameters below this cell.\nEnable tags to see what I mean",
"_____no_output_____"
]
],
[
[
"required_parameters = {\n \"experiment_name\",\n \"lr\",\n \"device\",\n \"seed\",\n \"dataset_seed\",\n \"labels_source\",\n \"labels_target\",\n \"domains_source\",\n \"domains_target\",\n \"num_examples_per_domain_per_label_source\",\n \"num_examples_per_domain_per_label_target\",\n \"n_shot\",\n \"n_way\",\n \"n_query\",\n \"train_k_factor\",\n \"val_k_factor\",\n \"test_k_factor\",\n \"n_epoch\",\n \"patience\",\n \"criteria_for_best\",\n \"x_transforms_source\",\n \"x_transforms_target\",\n \"episode_transforms_source\",\n \"episode_transforms_target\",\n \"pickle_name\",\n \"x_net\",\n \"NUM_LOGS_PER_EPOCH\",\n \"BEST_MODEL_PATH\",\n \"torch_default_dtype\"\n}",
"_____no_output_____"
],
[
"\n\nstandalone_parameters = {}\nstandalone_parameters[\"experiment_name\"] = \"STANDALONE PTN\"\nstandalone_parameters[\"lr\"] = 0.0001\nstandalone_parameters[\"device\"] = \"cuda\"\n\nstandalone_parameters[\"seed\"] = 1337\nstandalone_parameters[\"dataset_seed\"] = 1337\n\n\nstandalone_parameters[\"num_examples_per_domain_per_label_source\"]=100\nstandalone_parameters[\"num_examples_per_domain_per_label_target\"]=100\n\nstandalone_parameters[\"n_shot\"] = 3\nstandalone_parameters[\"n_query\"] = 2\nstandalone_parameters[\"train_k_factor\"] = 1\nstandalone_parameters[\"val_k_factor\"] = 2\nstandalone_parameters[\"test_k_factor\"] = 2\n\n\nstandalone_parameters[\"n_epoch\"] = 100\n\nstandalone_parameters[\"patience\"] = 10\nstandalone_parameters[\"criteria_for_best\"] = \"target_accuracy\"\n\nstandalone_parameters[\"x_transforms_source\"] = [\"unit_power\"]\nstandalone_parameters[\"x_transforms_target\"] = [\"unit_power\"]\nstandalone_parameters[\"episode_transforms_source\"] = []\nstandalone_parameters[\"episode_transforms_target\"] = []\n\nstandalone_parameters[\"torch_default_dtype\"] = \"torch.float32\" \n\n\n\nstandalone_parameters[\"x_net\"] = [\n {\"class\": \"nnReshape\", \"kargs\": {\"shape\":[-1, 1, 2, 256]}},\n {\"class\": \"Conv2d\", \"kargs\": { \"in_channels\":1, \"out_channels\":256, \"kernel_size\":(1,7), \"bias\":False, \"padding\":(0,3), },},\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\":256}},\n\n {\"class\": \"Conv2d\", \"kargs\": { \"in_channels\":256, \"out_channels\":80, \"kernel_size\":(2,7), \"bias\":True, \"padding\":(0,3), },},\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\":80}},\n {\"class\": \"Flatten\", \"kargs\": {}},\n\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 80*256, \"out_features\": 256}}, # 80 units per IQ pair\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm1d\", \"kargs\": {\"num_features\":256}},\n\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 256, \"out_features\": 256}},\n]\n\n# Parameters relevant to results\n# These parameters will basically never need to change\nstandalone_parameters[\"NUM_LOGS_PER_EPOCH\"] = 10\nstandalone_parameters[\"BEST_MODEL_PATH\"] = \"./best_model.pth\"\n\n# uncomment for CORES dataset\nfrom steves_utils.CORES.utils import (\n ALL_NODES,\n ALL_NODES_MINIMUM_1000_EXAMPLES,\n ALL_DAYS\n)\n\n\nstandalone_parameters[\"labels_source\"] = ALL_NODES\nstandalone_parameters[\"labels_target\"] = ALL_NODES\n\nstandalone_parameters[\"domains_source\"] = [1]\nstandalone_parameters[\"domains_target\"] = [2,3,4,5]\n\nstandalone_parameters[\"pickle_name\"] = \"cores.stratified_ds.2022A.pkl\"\n\n\n# Uncomment these for ORACLE dataset\n# from steves_utils.ORACLE.utils_v2 import (\n# ALL_DISTANCES_FEET,\n# ALL_RUNS,\n# ALL_SERIAL_NUMBERS,\n# )\n# standalone_parameters[\"labels_source\"] = ALL_SERIAL_NUMBERS\n# standalone_parameters[\"labels_target\"] = ALL_SERIAL_NUMBERS\n# standalone_parameters[\"domains_source\"] = [8,20, 38,50]\n# standalone_parameters[\"domains_target\"] = [14, 26, 32, 44, 56]\n# standalone_parameters[\"pickle_name\"] = \"oracle.frame_indexed.stratified_ds.2022A.pkl\"\n# standalone_parameters[\"num_examples_per_domain_per_label_source\"]=1000\n# standalone_parameters[\"num_examples_per_domain_per_label_target\"]=1000\n\n# Uncomment these for Metahan dataset\n# standalone_parameters[\"labels_source\"] = list(range(19))\n# standalone_parameters[\"labels_target\"] = list(range(19))\n# standalone_parameters[\"domains_source\"] = [0]\n# standalone_parameters[\"domains_target\"] = [1]\n# standalone_parameters[\"pickle_name\"] = \"metehan.stratified_ds.2022A.pkl\"\n# standalone_parameters[\"n_way\"] = len(standalone_parameters[\"labels_source\"])\n# standalone_parameters[\"num_examples_per_domain_per_label_source\"]=200\n# standalone_parameters[\"num_examples_per_domain_per_label_target\"]=100\n\n\nstandalone_parameters[\"n_way\"] = len(standalone_parameters[\"labels_source\"])",
"_____no_output_____"
],
[
"# Parameters\nparameters = {\n \"experiment_name\": \"tuned_1v2:oracle.run2_limited\",\n \"device\": \"cuda\",\n \"lr\": 0.0001,\n \"labels_source\": [\n \"3123D52\",\n \"3123D65\",\n \"3123D79\",\n \"3123D80\",\n \"3123D54\",\n \"3123D70\",\n \"3123D7B\",\n \"3123D89\",\n \"3123D58\",\n \"3123D76\",\n \"3123D7D\",\n \"3123EFE\",\n \"3123D64\",\n \"3123D78\",\n \"3123D7E\",\n \"3124E4A\",\n ],\n \"labels_target\": [\n \"3123D52\",\n \"3123D65\",\n \"3123D79\",\n \"3123D80\",\n \"3123D54\",\n \"3123D70\",\n \"3123D7B\",\n \"3123D89\",\n \"3123D58\",\n \"3123D76\",\n \"3123D7D\",\n \"3123EFE\",\n \"3123D64\",\n \"3123D78\",\n \"3123D7E\",\n \"3124E4A\",\n ],\n \"episode_transforms_source\": [],\n \"episode_transforms_target\": [],\n \"domains_source\": [8, 32, 50],\n \"domains_target\": [14, 20, 26, 38, 44],\n \"num_examples_per_domain_per_label_source\": 2000,\n \"num_examples_per_domain_per_label_target\": 2000,\n \"n_shot\": 3,\n \"n_way\": 16,\n \"n_query\": 2,\n \"train_k_factor\": 3,\n \"val_k_factor\": 2,\n \"test_k_factor\": 2,\n \"torch_default_dtype\": \"torch.float32\",\n \"n_epoch\": 50,\n \"patience\": 3,\n \"criteria_for_best\": \"target_accuracy\",\n \"x_net\": [\n {\"class\": \"nnReshape\", \"kargs\": {\"shape\": [-1, 1, 2, 256]}},\n {\n \"class\": \"Conv2d\",\n \"kargs\": {\n \"in_channels\": 1,\n \"out_channels\": 256,\n \"kernel_size\": [1, 7],\n \"bias\": False,\n \"padding\": [0, 3],\n },\n },\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\": 256}},\n {\n \"class\": \"Conv2d\",\n \"kargs\": {\n \"in_channels\": 256,\n \"out_channels\": 80,\n \"kernel_size\": [2, 7],\n \"bias\": True,\n \"padding\": [0, 3],\n },\n },\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\": 80}},\n {\"class\": \"Flatten\", \"kargs\": {}},\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 20480, \"out_features\": 256}},\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm1d\", \"kargs\": {\"num_features\": 256}},\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 256, \"out_features\": 256}},\n ],\n \"NUM_LOGS_PER_EPOCH\": 10,\n \"BEST_MODEL_PATH\": \"./best_model.pth\",\n \"pickle_name\": \"oracle.Run2_10kExamples_stratified_ds.2022A.pkl\",\n \"x_transforms_source\": [\"unit_mag\"],\n \"x_transforms_target\": [\"unit_mag\"],\n \"dataset_seed\": 7,\n \"seed\": 7,\n}\n",
"_____no_output_____"
],
[
"# Set this to True if you want to run this template directly\nSTANDALONE = False\nif STANDALONE:\n print(\"parameters not injected, running with standalone_parameters\")\n parameters = standalone_parameters\n\nif not 'parameters' in locals() and not 'parameters' in globals():\n raise Exception(\"Parameter injection failed\")\n\n#Use an easy dict for all the parameters\np = EasyDict(parameters)\n\nsupplied_keys = set(p.keys())\n\nif supplied_keys != required_parameters:\n print(\"Parameters are incorrect\")\n if len(supplied_keys - required_parameters)>0: print(\"Shouldn't have:\", str(supplied_keys - required_parameters))\n if len(required_parameters - supplied_keys)>0: print(\"Need to have:\", str(required_parameters - supplied_keys))\n raise RuntimeError(\"Parameters are incorrect\")\n\n",
"_____no_output_____"
],
[
"###################################\n# Set the RNGs and make it all deterministic\n###################################\nnp.random.seed(p.seed)\nrandom.seed(p.seed)\ntorch.manual_seed(p.seed)\n\ntorch.use_deterministic_algorithms(True) ",
"_____no_output_____"
],
[
"###########################################\n# The stratified datasets honor this\n###########################################\ntorch.set_default_dtype(eval(p.torch_default_dtype))",
"_____no_output_____"
],
[
"###################################\n# Build the network(s)\n# Note: It's critical to do this AFTER setting the RNG\n# (This is due to the randomized initial weights)\n###################################\nx_net = build_sequential(p.x_net)",
"_____no_output_____"
],
[
"start_time_secs = time.time()",
"_____no_output_____"
],
[
"###################################\n# Build the dataset\n###################################\n\nif p.x_transforms_source == []: x_transform_source = None\nelse: x_transform_source = get_chained_transform(p.x_transforms_source) \n\nif p.x_transforms_target == []: x_transform_target = None\nelse: x_transform_target = get_chained_transform(p.x_transforms_target)\n\nif p.episode_transforms_source == []: episode_transform_source = None\nelse: raise Exception(\"episode_transform_source not implemented\")\n\nif p.episode_transforms_target == []: episode_transform_target = None\nelse: raise Exception(\"episode_transform_target not implemented\")\n\n\neaf_source = Episodic_Accessor_Factory(\n labels=p.labels_source,\n domains=p.domains_source,\n num_examples_per_domain_per_label=p.num_examples_per_domain_per_label_source,\n iterator_seed=p.seed,\n dataset_seed=p.dataset_seed,\n n_shot=p.n_shot,\n n_way=p.n_way,\n n_query=p.n_query,\n train_val_test_k_factors=(p.train_k_factor,p.val_k_factor,p.test_k_factor),\n pickle_path=os.path.join(get_datasets_base_path(), p.pickle_name),\n x_transform_func=x_transform_source,\n example_transform_func=episode_transform_source,\n \n)\ntrain_original_source, val_original_source, test_original_source = eaf_source.get_train(), eaf_source.get_val(), eaf_source.get_test()\n\n\neaf_target = Episodic_Accessor_Factory(\n labels=p.labels_target,\n domains=p.domains_target,\n num_examples_per_domain_per_label=p.num_examples_per_domain_per_label_target,\n iterator_seed=p.seed,\n dataset_seed=p.dataset_seed,\n n_shot=p.n_shot,\n n_way=p.n_way,\n n_query=p.n_query,\n train_val_test_k_factors=(p.train_k_factor,p.val_k_factor,p.test_k_factor),\n pickle_path=os.path.join(get_datasets_base_path(), p.pickle_name),\n x_transform_func=x_transform_target,\n example_transform_func=episode_transform_target,\n)\ntrain_original_target, val_original_target, test_original_target = eaf_target.get_train(), eaf_target.get_val(), eaf_target.get_test()\n\n\ntransform_lambda = lambda ex: ex[1] # Original is (<domain>, <episode>) so we strip down to episode only\n\ntrain_processed_source = Lazy_Iterable_Wrapper(train_original_source, transform_lambda)\nval_processed_source = Lazy_Iterable_Wrapper(val_original_source, transform_lambda)\ntest_processed_source = Lazy_Iterable_Wrapper(test_original_source, transform_lambda)\n\ntrain_processed_target = Lazy_Iterable_Wrapper(train_original_target, transform_lambda)\nval_processed_target = Lazy_Iterable_Wrapper(val_original_target, transform_lambda)\ntest_processed_target = Lazy_Iterable_Wrapper(test_original_target, transform_lambda)\n\ndatasets = EasyDict({\n \"source\": {\n \"original\": {\"train\":train_original_source, \"val\":val_original_source, \"test\":test_original_source},\n \"processed\": {\"train\":train_processed_source, \"val\":val_processed_source, \"test\":test_processed_source}\n },\n \"target\": {\n \"original\": {\"train\":train_original_target, \"val\":val_original_target, \"test\":test_original_target},\n \"processed\": {\"train\":train_processed_target, \"val\":val_processed_target, \"test\":test_processed_target}\n },\n})",
"_____no_output_____"
],
[
"# Some quick unit tests on the data\nfrom steves_utils.transforms import get_average_power, get_average_magnitude\n\nq_x, q_y, s_x, s_y, truth = next(iter(train_processed_source))\n\nassert q_x.dtype == eval(p.torch_default_dtype)\nassert s_x.dtype == eval(p.torch_default_dtype)\n\nprint(\"Visually inspect these to see if they line up with expected values given the transforms\")\nprint('x_transforms_source', p.x_transforms_source)\nprint('x_transforms_target', p.x_transforms_target)\nprint(\"Average magnitude, source:\", get_average_magnitude(q_x[0].numpy()))\nprint(\"Average power, source:\", get_average_power(q_x[0].numpy()))\n\nq_x, q_y, s_x, s_y, truth = next(iter(train_processed_target))\nprint(\"Average magnitude, target:\", get_average_magnitude(q_x[0].numpy()))\nprint(\"Average power, target:\", get_average_power(q_x[0].numpy()))\n",
"Visually inspect these to see if they line up with expected values given the transforms\nx_transforms_source ['unit_mag']\nx_transforms_target ['unit_mag']\nAverage magnitude, source: 1.0\nAverage power, source: 1.0878031\n"
],
[
"###################################\n# Build the model\n###################################\nmodel = Steves_Prototypical_Network(x_net, device=p.device, x_shape=(2,256))\noptimizer = Adam(params=model.parameters(), lr=p.lr)",
"(2, 256)\n"
],
[
"###################################\n# train\n###################################\njig = PTN_Train_Eval_Test_Jig(model, p.BEST_MODEL_PATH, p.device)\n\njig.train(\n train_iterable=datasets.source.processed.train,\n source_val_iterable=datasets.source.processed.val,\n target_val_iterable=datasets.target.processed.val,\n num_epochs=p.n_epoch,\n num_logs_per_epoch=p.NUM_LOGS_PER_EPOCH,\n patience=p.patience,\n optimizer=optimizer,\n criteria_for_best=p.criteria_for_best,\n)",
"epoch: 1, [batch: 1 / 2520], examples_per_second: 127.6511, train_label_loss: 2.7871, \n"
],
[
"total_experiment_time_secs = time.time() - start_time_secs",
"_____no_output_____"
],
[
"###################################\n# Evaluate the model\n###################################\nsource_test_label_accuracy, source_test_label_loss = jig.test(datasets.source.processed.test)\ntarget_test_label_accuracy, target_test_label_loss = jig.test(datasets.target.processed.test)\n\nsource_val_label_accuracy, source_val_label_loss = jig.test(datasets.source.processed.val)\ntarget_val_label_accuracy, target_val_label_loss = jig.test(datasets.target.processed.val)\n\nhistory = jig.get_history()\n\ntotal_epochs_trained = len(history[\"epoch_indices\"])\n\nval_dl = Iterable_Aggregator((datasets.source.original.val,datasets.target.original.val))\n\nconfusion = ptn_confusion_by_domain_over_dataloader(model, p.device, val_dl)\nper_domain_accuracy = per_domain_accuracy_from_confusion(confusion)\n\n# Add a key to per_domain_accuracy for if it was a source domain\nfor domain, accuracy in per_domain_accuracy.items():\n per_domain_accuracy[domain] = {\n \"accuracy\": accuracy,\n \"source?\": domain in p.domains_source\n }\n\n# Do an independent accuracy assesment JUST TO BE SURE!\n# _source_test_label_accuracy = independent_accuracy_assesment(model, datasets.source.processed.test, p.device)\n# _target_test_label_accuracy = independent_accuracy_assesment(model, datasets.target.processed.test, p.device)\n# _source_val_label_accuracy = independent_accuracy_assesment(model, datasets.source.processed.val, p.device)\n# _target_val_label_accuracy = independent_accuracy_assesment(model, datasets.target.processed.val, p.device)\n\n# assert(_source_test_label_accuracy == source_test_label_accuracy)\n# assert(_target_test_label_accuracy == target_test_label_accuracy)\n# assert(_source_val_label_accuracy == source_val_label_accuracy)\n# assert(_target_val_label_accuracy == target_val_label_accuracy)\n\nexperiment = {\n \"experiment_name\": p.experiment_name,\n \"parameters\": dict(p),\n \"results\": {\n \"source_test_label_accuracy\": source_test_label_accuracy,\n \"source_test_label_loss\": source_test_label_loss,\n \"target_test_label_accuracy\": target_test_label_accuracy,\n \"target_test_label_loss\": target_test_label_loss,\n \"source_val_label_accuracy\": source_val_label_accuracy,\n \"source_val_label_loss\": source_val_label_loss,\n \"target_val_label_accuracy\": target_val_label_accuracy,\n \"target_val_label_loss\": target_val_label_loss,\n \"total_epochs_trained\": total_epochs_trained,\n \"total_experiment_time_secs\": total_experiment_time_secs,\n \"confusion\": confusion,\n \"per_domain_accuracy\": per_domain_accuracy,\n },\n \"history\": history,\n \"dataset_metrics\": get_dataset_metrics(datasets, \"ptn\"),\n}",
"_____no_output_____"
],
[
"ax = get_loss_curve(experiment)\nplt.show()",
"_____no_output_____"
],
[
"get_results_table(experiment)",
"_____no_output_____"
],
[
"get_domain_accuracies(experiment)",
"_____no_output_____"
],
[
"print(\"Source Test Label Accuracy:\", experiment[\"results\"][\"source_test_label_accuracy\"], \"Target Test Label Accuracy:\", experiment[\"results\"][\"target_test_label_accuracy\"])\nprint(\"Source Val Label Accuracy:\", experiment[\"results\"][\"source_val_label_accuracy\"], \"Target Val Label Accuracy:\", experiment[\"results\"][\"target_val_label_accuracy\"])",
"Source Test Label Accuracy: 0.6043402777777778 Target Test Label Accuracy: 0.5004166666666666\nSource Val Label Accuracy: 0.6008680555555556 Target Val Label Accuracy: 0.5006770833333334\n"
],
[
"json.dumps(experiment)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0d097ba512747fb9891b870346127b2cf0f0734 | 12,306 | ipynb | Jupyter Notebook | day-2/0_week-0-worksheet-while-loops.ipynb | allegheny-college-cmpsc-100-jan-2021/week-0 | af436f431a783e67e06383a642be93bbc2015369 | [
"CC-BY-4.0"
] | 1 | 2021-01-19T15:14:46.000Z | 2021-01-19T15:14:46.000Z | day-2/0_week-0-worksheet-while-loops.ipynb | allegheny-college-cmpsc-100-jan-2021/week-0 | af436f431a783e67e06383a642be93bbc2015369 | [
"CC-BY-4.0"
] | null | null | null | day-2/0_week-0-worksheet-while-loops.ipynb | allegheny-college-cmpsc-100-jan-2021/week-0 | af436f431a783e67e06383a642be93bbc2015369 | [
"CC-BY-4.0"
] | null | null | null | 39.191083 | 484 | 0.613197 | [
[
[
"# Worksheet 0.1.2: Python syntax (`while` loops)\n\n<div class=\"alert alert-block alert-info\">\nThis worksheet will invite you to tinker with the examples, as they are live code cells. Instead of the normal fill-in-the-blank style of notebook, feel free to mess with the code directly. Remember that -- to test things out -- the <a href = \"../sandbox/CMPSC%20100%20-%20Week%2000%20-%20Sandbox.ipynb\"><b>Sandbox</b></a> is available to you as well.\n</div>\n\n<div class=\"alert alert-block alert-warning\" id = \"warning\">\nThe work this week also offers the opportunity to tie up the server in loopish operations. Should you be unable to run cells, simply locate the <b>Kernel</b> menu at the top of the screen and click <b>Interrupt Kernel</b>. This should jumpstart the kernel again and clear out the infinite loop behavior.</div>\n\n## Feeling a bit loopy?\n\nIf you're not, you might start to during this worksheet.\n\nWe've mostly covered cases in which events or calculations need to happen one at a time or just one time in total. Occasionally -- much more than occasionally, really -- programs need to repeat instructions more than once until some given condition is met. \n\nIf you read the word \"condition\" above and have started thinking `booleans` are involved: you're right yet again.\n\nWhile casually referred to as a \"loop\" structure, the technical term for this repetition is _iteration_ -- the process of repeating a set of statements until a given condition is no longer `True`. In the case of `while` loops, we can rephrase the statement above to read \"while the condition is true, repeat some statements\"\n\n`while` loops recall syntax similar to `if` statements:\n\n```python\nwhile CONDITION:\n # Repeat\n # these\n # statements\n```\n\nAgain, notice that indentation plays a part here: everything indented underneat the `while` statement \"belongs to\" that `while` loop, and will be subject to repetition.\n\nFor example, a simple countdown:",
"_____no_output_____"
]
],
[
[
"# Initialize starting number\nseconds = 10\n\n# Start while loop\nwhile seconds > 0:\n print(seconds)\n seconds -= 1\nprint(\"Liftoff!\")",
"10\n9\n8\n7\n6\n5\n4\n3\n2\n1\nLiftoff!\n"
]
],
[
[
"In the above block of code, we start by telling the program where to, well, start. Then we print that number followed by an instruction `seconds -= 1` to _decrement_ (decrease) that number by one _each time the loop runs_ (on each _iteration_). By the time we reach the end, the last run notices that `seconds == 0`, therefore `seconds` _is not_ greater than `0` anymore and it breaks out of the loop, executing the next statement after the loop.\n\nLike Worksheet 0 this week, we can use any combination of expressions that `boolean` values can muster, be they _relational operators_ testing `integers`, `floating point numbers`, `strings`, or _logical operators_ looking for combinations of `boolean` expressions.\n\n### `while` loops and the flow of control\n\nHere's that control topic back to haunt us.\n\nAgain, the technical flow of the program's instructions (i.e. code) doesn't change. It's still technically top-down. However, something interesting happens when we hit a loop or _iteration_. Consider our countdown above:\n\n\n\nAs the diagram points out, the flow of control changes when we encounter a `while` loop. Statements _in the loop_ execute from `top -> bottom -> top` until the condition cited is no longer true (in this case, until the moment that `seconds` dips below `1`.\n\n## Detour into user input\n\nWhy user input and why now? As we'll see in future weeks, other kinds of loops can fulfill the same purpose as `while` loops. However, there's something unique that `while` loops can do that others aren't so well-suited to: handing user input. \n\nThis relies on our understanding of _functions_ as we're about to learn a new one: `input()` -- a function which allows us to prompt users to enter data so that our programs can do an operation called \"parsing\" (understanding/reading) it.\n\nIt's always helpful to put some `string` value in the parenthesis as the _argument_, to give a user some sense of what they're being requested to type. In fact, the general format of the function is, like others we've seen:\n\n```python\n# Where ARGUMENT will evaluate to a string\ninput(ARGUMENT)\n```\n\nIn order to effectively use it, we need to _assign_ the result of the function to a variable to store it in memory. Run the following cell for an example:",
"_____no_output_____"
]
],
[
[
"name = input(\"What is your name: \")\nprint(\"Hello, \" + name + \".\")",
"_____no_output_____"
]
],
[
[
"_Because we stored the result_ in `name`, we can use it -- whether that's to `print` it or test it:",
"_____no_output_____"
]
],
[
[
"if name == \"The Professor\":\n print(\"The Professor is in the house!\")\nelse:\n print(\"Oh, you're not the professor. Forget it.\")",
"_____no_output_____"
]
],
[
[
"One thing of particular note:",
"_____no_output_____"
]
],
[
[
"# The identifier \"str_value\" is arbitrary here, no real significance\nstr_value = input(\"Enter a numeric value of any kind: \")\nprint(type(str_value))",
"_____no_output_____"
]
],
[
[
"No matter what we enter here, the result will always be a `string` type. That's because the Python language is rigged to do its best with _whatever_ a user writes in the prompt. This means handling characters/symbols (`!@#^!%#@$` -- I promise I'm not swearing here), letters (`a`,`b`,`c`), or numbers (either `integer` or `float`).\n\nThis means that, to make it useful, we might have to _convert_ it if we want a numeric value from it:",
"_____no_output_____"
]
],
[
[
"float_value = float(str_value)\nprint(type(float_value))",
"_____no_output_____"
]
],
[
[
"However, insofar as user input is concerned, we can test it _or_ we can use it as something called a \"sentinel\" value. Using word \"sentinel\" here means exactly what it means in normal speech -- something to watch out for, like the following:",
"_____no_output_____"
]
],
[
[
"# Setup choice\nchoice = \"\"\n\n# Do the loop\nwhile choice != \"E\":\n # Print message to relay the user's choice\n print(\"The loop is running.\")\n choice = input(\"[C]ontinue or [E]xit? \")\nprint(\"You chose to exit!\")",
"_____no_output_____"
]
],
[
[
"## Mean`while`\n\nThe conclusion we should draw from our little detour is this: `while` statements are exceptionally good at handling all kinds of `boolean` conditions. When it's merely simple counting, like our countdown example above, it's OK, too -- but, as we'll see in the near future, counting and other more complex tests are better suited by other loop types.\n\n## Infinite loops\n\n<div class=\"alert alert-block alert-danger\">\nMost programmers fall prey to an infinite loop from time to time. The examples below are not code cells, because if you were to run them, they would -- well -- loop infinitely. If you think you're stuck in an infinite loop infinite loop infinite loop infinite loop infinite loop infinite loop infinite loop infinite loop infinite loop infinite loop infinite loop infinite loop infinite loop, don't hesistate to take the advice in the <a href = \"#warning\">warning</a> above.\n</div>\n\nThere are times when conditions _can't_ be met. For example:\n\n```python\ncount = 0\n\nwhile True:\n count += 1\n```\n\nNow, there _is_ an application for something like this. However, note that if we say `while True`, the condition is literally hard-coded to be `True` _all the time_ -- it can never change. Another example:\n\n```python\nsum = 0\nlimit = 5\n\nwhile sum < limit:\n sum -=1\n```\n\nHere, we're actually _counting backwards_, so `sum` will never be `+5`. It might be `-5` (at some point), but it will continue on to `-∞`, and never stop. In essence, `sum` will always be less than `limit`.",
"_____no_output_____"
],
[
"## The sum of its parts\n\nFirst things first: this program is a one-trick pony: it only adds numbers.\n\nI'm asking you to use what we have learned in this worksheet to write a program whose sole purpose is to present the sum of a set of user-entered numbers. The user should be able to enter as many numbers as they want, provided that:\n\n* all of the numbers are integers\n* uses `number` to store user input\n* users can choose to quit by entering an `0` at the prompt\n* `if number = 0`, don't add the number to `count`\n * non-hint hint: there are at least 3 ways to do this\n* the program output the sum in the following format:\n\n```\nThe sum of these # numbers -> ###\n```\n\n* The \"proof\" of this program is to add the following numbers when you grade the worksheet:\n * `4`,`8`,`15`,`16`,`23`,`42`\n\nI'll start you out.",
"_____no_output_____"
]
],
[
[
"# NOTE: YOU MUST RUN THIS CELL TO MAKE THESE VARIABLES AVAILABLE\n# NOTE 2: RUN THIS CELL TO RESET ALL NUMBERS AS WELL\n\n# Setup variable to handle input\nnumber = \"\"\n\n# Setup variable to keep running total\nsum = 0\n\n# Setup a count variable to track the count\ncount = 0\n\n# TODO: Write code to complete activity using knowledge you've gained about while loops",
"_____no_output_____"
]
],
[
[
"## Finishing this activity\n\nIf the program above runs and you've finished the worksheet, be sure to run `gradle grade` to do one final check!",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0d09fc3b1935fbacf2e62f41cd6de792623fbcc | 676,792 | ipynb | Jupyter Notebook | examples/2020-04-20-backtest_with_grid_search.ipynb | johnmarkacala/fastquant | cbf25ef28a7c886b1b229cb944ed0886518a1182 | [
"MIT"
] | 6 | 2020-01-12T01:59:46.000Z | 2020-01-20T05:01:42.000Z | examples/2020-04-20-backtest_with_grid_search.ipynb | johnmarkacala/fastquant | cbf25ef28a7c886b1b229cb944ed0886518a1182 | [
"MIT"
] | 3 | 2020-01-14T12:49:21.000Z | 2020-01-15T10:48:04.000Z | examples/2020-04-20-backtest_with_grid_search.ipynb | johnmarkacala/fastquant | cbf25ef28a7c886b1b229cb944ed0886518a1182 | [
"MIT"
] | null | null | null | 161.718519 | 171,205 | 0.842309 | [
[
[
"# \"# backtesting with grid search\"\n> \"Easily backtest a grid of parameters in a given trading strategy\"\n\n- toc: true\n- branch: master\n- badges: true\n- comments: true\n- author: Jerome de Leon\n- categories: [grid search, backtest]",
"_____no_output_____"
],
[
"<a href=\"https://colab.research.google.com/github/enzoampil/fastquant/blob/master/examples/2020-04-20-backtest_with_grid_search.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"# uncomment to install in colab\n# !pip3 install fastquant",
"_____no_output_____"
]
],
[
[
"## backtest SMAC",
"_____no_output_____"
],
[
"`fastquant` offers a convenient way to backtest several trading strategies. To backtest using Simple Moving Average Crossover (`SMAC`), we do the following.\n\n```python\nbacktest('smac', dcv_data, fast_period=15, slow_period=40)\n```\n\n`fast_period` and `slow_period` are two `SMAC` parameters that can be changed depending on the user's preferences. A simple way to fine tune these parameters is to run `backtest` on a grid of values and find which combination of `fast_period` and `slow_period` yields the highest net profit.\n\nFirst, we fetch `JFC`'s historical data comprised of date, close price, and volume.",
"_____no_output_____"
]
],
[
[
"from fastquant import get_stock_data, backtest\n\nsymbol='JFC'\ndcv_data = get_stock_data(symbol, \n start_date='2018-01-01', \n end_date='2020-04-28',\n format='cv',\n )\ndcv_data.head()",
"849it [04:08, 4.04it/s]\n"
],
[
"import matplotlib.pyplot as pl\npl.style.use(\"default\")",
"_____no_output_____"
],
[
"from fastquant import backtest\n\nresults = backtest(\"smac\", \n dcv_data, \n fast_period=15, \n slow_period=40, \n verbose=False, \n plot=True\n )",
"===Global level arguments===\ninit_cash : 100000\nbuy_prop : 1\nsell_prop : 1\n===Strategy level arguments===\nfast_period : 15\nslow_period : 40\nFinal PnL: -31257.65\nTime used (seconds): 0.10944700241088867\nOptimal parameters: {'init_cash': 100000, 'buy_prop': 1, 'sell_prop': 1, 'execution_type': 'close', 'fast_period': 15, 'slow_period': 40}\nOptimal metrics: {'rtot': -0.37480465562458976, 'ravg': -0.0006645472617457265, 'rnorm': -0.15419454966091925, 'rnorm100': -15.419454966091925, 'sharperatio': -0.9821454406209409, 'pnl': -31257.65, 'final_value': 68742.35499999995}\n"
]
],
[
[
"The plot above is optional. `backtest` returns a dataframe of parameters and corresponding metrics:",
"_____no_output_____"
]
],
[
[
"results.head()",
"_____no_output_____"
]
],
[
[
"## define the search space",
"_____no_output_____"
],
[
"Second, we specify the range of reasonable values to explore for `fast_period` and `slow_period`. Let's take between 1 and 20 trading days (roughly a month) in steps of 1 day for `fast_period`, and between 21 and 240 trading days (roughly a year) in steps of 5 days for `slow_period`.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nfast_periods = np.arange(1,20,1, dtype=int)\nslow_periods = np.arange(20,241,5, dtype=int)\n\n# make a grid of 0's (placeholder)\nperiod_grid = np.zeros(shape=(len(fast_periods),len(slow_periods)))\nperiod_grid.shape",
"_____no_output_____"
]
],
[
[
"## run grid search",
"_____no_output_____"
],
[
"Third, we run backtest for each iteration over each pair of `fast_period` and `slow_period`, saving each time the net profit to the `period_grid` variable.",
"_____no_output_____"
],
[
"Note: Before running backtest over a large grid, try measuring how long it takes your machine to run one backtest instance.\n```python\n%timeit\nbacktest(...)\n```",
"_____no_output_____"
],
[
"In my machine with 8 cores, `backtest` takes\n```\n101 ms ± 8.3 ms per loop (mean ± std. dev. of 7 runs, 10 loops each)\n```",
"_____no_output_____"
]
],
[
[
"from time import time\n\ninit_cash=100000\n\nstart_time = time()\nfor i,fast_period in enumerate(fast_periods): \n for j,slow_period in enumerate(slow_periods):\n results = backtest('smac', \n dcv_data, \n fast_period=fast_period,\n slow_period=slow_period,\n init_cash=100000,\n verbose=False, \n plot=False\n )\n net_profit = results.final_value.values[0]-init_cash\n period_grid[i,j] = net_profit\nend_time = time()",
"_____no_output_____"
],
[
"time_basic = end_time-start_time\nprint(\"Basic grid search took {:.1f} sec\".format(time_basic))",
"Basic grid search took 100.4 sec\n"
]
],
[
[
"## visualize the period grid",
"_____no_output_____"
],
[
"Next, we visualize `period_grid` as a 2D matrix.",
"_____no_output_____"
]
],
[
[
"import matplotlib.colors as mcolors\nimport matplotlib.pyplot as pl\npl.style.use(\"default\")\n\nfig, ax = pl.subplots(1,1, figsize=(8,4))\nxmin, xmax = slow_periods[0],slow_periods[-1]\nymin, ymax = fast_periods[0],fast_periods[-1]\n\n#make a diverging color map such that profit<0 is red and blue otherwise\ncmap = pl.get_cmap('RdBu')\nnorm = mcolors.TwoSlopeNorm(vmin=period_grid.min(), \n vmax = period_grid.max(), \n vcenter=0\n )\n#plot matrix\ncbar = ax.imshow(period_grid, \n origin='lower', \n interpolation='none', \n extent=[xmin, xmax, ymin, ymax], \n cmap=cmap,\n norm=norm\n )\npl.colorbar(cbar, ax=ax, shrink=0.9,\n label='net profit', orientation=\"horizontal\")\n\n# search position with highest net profit\ny, x = np.unravel_index(np.argmax(period_grid), period_grid.shape)\nbest_slow_period = slow_periods[x]\nbest_fast_period = fast_periods[y]\n# mark position\n# ax.annotate(f\"max profit={period_grid[y, x]:.0f}@({best_slow_period}, {best_fast_period}) days\", \n# (best_slow_period+5,best_fast_period+1)\n# )\nax.axvline(best_slow_period, 0, 1, c='k', ls='--')\nax.axhline(best_fast_period+0.5, 0, 1, c='k', ls='--')\n\n# add labels\nax.set_aspect(5)\npl.setp(ax,\n xlim=(xmin,xmax),\n ylim=(ymin,ymax),\n xlabel='slow period (days)',\n ylabel='fast period (days)',\n title='JFC w/ SMAC',\n );",
"_____no_output_____"
],
[
"print(f\"max profit={period_grid[y, x]:.0f} @ ({best_slow_period},{best_fast_period}) days\")",
"max profit=7042 @ (105,3) days\n"
]
],
[
[
"From the plot above, there are only a few period combinations which we can guarantee non-negative net profit using SMAC strategy. The best result is achieved with (105,30) for period_slow and period_fast, respectively.\n\nIn fact SMAC strategy is so bad such that there is only 9% chance it will yield profit when using any random period combinations in our grid, which is smaller than the 12% chance it will yield break even at least.",
"_____no_output_____"
]
],
[
[
"percent_positive_profit=(period_grid>0).sum()/np.product(period_grid.shape)*100\npercent_positive_profit",
"_____no_output_____"
],
[
"percent_breakeven=(period_grid==0).sum()/np.product(period_grid.shape)*100\npercent_breakeven",
"_____no_output_____"
]
],
[
[
"Anyway, let's check the results of backtest using the `best_fast_period` and `best_slow_period`.",
"_____no_output_____"
]
],
[
[
"results = backtest('smac', \n dcv_data, \n fast_period=best_fast_period, \n slow_period=best_slow_period, \n verbose=True, \n plot=True\n )\nnet_profit = results.final_value.values[0]-init_cash\nnet_profit",
"Starting Portfolio Value: 100000.00\n===Global level arguments===\ninit_cash : 100000\nbuy_prop : 1\nsell_prop : 1\n===Strategy level arguments===\nfast_period : 3\nslow_period : 105\n2018-08-22, BUY CREATE, 286.00\n2018-08-22, Cash: 100000.0\n2018-08-22, Price: 286.0\n2018-08-22, Buy prop size: 346\n2018-08-22, Afforded size: 346\n2018-08-22, Final size: 346\n2018-08-23, BUY EXECUTED, Price: 286.00, Cost: 98956.00, Comm 742.17\n2018-09-12, SELL CREATE, 277.00\n2018-09-13, SELL EXECUTED, Price: 277.00, Cost: 98956.00, Comm 718.81\n2018-09-13, OPERATION PROFIT, GROSS -3114.00, NET -4574.98\n2018-10-23, BUY CREATE, 268.00\n2018-10-23, Cash: 95425.015\n2018-10-23, Price: 268.0\n2018-10-23, Buy prop size: 353\n2018-10-23, Afforded size: 353\n2018-10-23, Final size: 353\n2018-10-24, BUY EXECUTED, Price: 268.00, Cost: 94604.00, Comm 709.53\n2018-10-25, SELL CREATE, 270.00\n2018-10-26, SELL EXECUTED, Price: 270.00, Cost: 94604.00, Comm 714.83\n2018-10-26, OPERATION PROFIT, GROSS 706.00, NET -718.36\n2018-10-30, BUY CREATE, 264.00\n2018-10-30, Cash: 94706.66\n2018-10-30, Price: 264.0\n2018-10-30, Buy prop size: 355\n2018-10-30, Afforded size: 355\n2018-10-30, Final size: 355\n2018-10-31, BUY EXECUTED, Price: 264.00, Cost: 93720.00, Comm 702.90\n2019-04-17, SELL CREATE, 303.00\n2019-04-22, SELL EXECUTED, Price: 303.00, Cost: 93720.00, Comm 806.74\n2019-04-22, OPERATION PROFIT, GROSS 13845.00, NET 12335.36\nFinal PnL: 7042.02\n==================================================\n**************************************************\n--------------------------------------------------\n{'init_cash': 100000, 'buy_prop': 1, 'sell_prop': 1, 'execution_type': 'close', 'fast_period': 3, 'slow_period': 105}\nOrderedDict([('rtot', 0.06805130501900258), ('ravg', 0.00012065834223227409), ('rnorm', 0.03087288265827186), ('rnorm100', 3.087288265827186)])\nOrderedDict([('sharperatio', 0.7850452330792583)])\nTime used (seconds): 0.11643362045288086\nOptimal parameters: {'init_cash': 100000, 'buy_prop': 1, 'sell_prop': 1, 'execution_type': 'close', 'fast_period': 3, 'slow_period': 105}\nOptimal metrics: {'rtot': 0.06805130501900258, 'ravg': 0.00012065834223227409, 'rnorm': 0.03087288265827186, 'rnorm100': 3.087288265827186, 'sharperatio': 0.7850452330792583, 'pnl': 7042.02, 'final_value': 107042.0225}\n"
]
],
[
[
"There are only 6 cross-over events of which only the latest transaction yielded positive gains resulting to a 7% net profit. Is 7% profit over a ~two-year baseline better than the market benchmark?",
"_____no_output_____"
],
[
"## built-in grid search in fastquant",
"_____no_output_____"
],
[
"The good news is `backtest` provides a built-in grid search if strategy parameters are lists. Let's re-run `backtest` with a grid we used above.",
"_____no_output_____"
]
],
[
[
"from fastquant import backtest\n\nstart_time = time()\nresults = backtest(\"smac\", \n dcv_data, \n fast_period=fast_periods, \n slow_period=slow_periods, \n verbose=False, \n plot=False\n )\nend_time = time()\ntime_optimized = end_time-start_time",
"_____no_output_____"
],
[
"print(\"Optimized grid search took {:.1f} sec\".format(time_optimized))",
"Optimized grid search took 95.6 sec\n"
]
],
[
[
"`results` is automatically ranked based on `rnorm` which is a proxy for performance. In this case, the best `fast_period`,`slow_period`=(8,200) d.\n\nThe returned parameters are should have `len(fast_periods)`x`len(slow_periods)` (19x45=855 in this case). ",
"_____no_output_____"
]
],
[
[
"results.shape",
"_____no_output_____"
],
[
"results.head()",
"_____no_output_____"
]
],
[
[
"Now, we recreate the 2D matrix before, but this time using scatter plot.",
"_____no_output_____"
]
],
[
[
"fig, ax = pl.subplots(1,1, figsize=(8,4))\n\n#make a diverging color map such that profit<0 is red and blue otherwise\ncmap = pl.get_cmap('RdBu')\nnorm = mcolors.TwoSlopeNorm(vmin=period_grid.min(), \n vmax = period_grid.max(), \n vcenter=0\n )\n#plot scatter\nresults['net_profit'] = results['final_value']-results['init_cash']\ndf = results[['slow_period','fast_period','net_profit']]\nax2 = df.plot.scatter(x='slow_period', y='fast_period', c='net_profit',\n norm=norm, cmap=cmap, ax=ax\n )\nymin,ymax = df.fast_period.min(), df.fast_period.max()\nxmin,xmax = df.slow_period.min(), df.slow_period.max()\n\n# best performance (instead of highest profit)\nbest_fast_period, best_slow_period, net_profit = df.loc[0,['fast_period','slow_period','net_profit']]\n# mark position\n# ax.annotate(f\"max profit={net_profit:.0f}@({best_slow_period}, {best_fast_period}) days\", \n# (best_slow_period-100,best_fast_period+1), color='r'\n# )\nax.axvline(best_slow_period, 0, 1, c='r', ls='--')\nax.axhline(best_fast_period+0.5, 0, 1, c='r', ls='--')\n\nax.set_aspect(5)\npl.setp(ax,\n xlim=(xmin,xmax),\n ylim=(ymin,ymax),\n xlabel='slow period (days)',\n ylabel='fast period (days)',\n title='JFC w/ SMAC',\n );\n\n# fig.colorbar(ax2, orientation=\"horizontal\", shrink=0.9, label='net profit')",
"_____no_output_____"
],
[
"print(f\"max profit={net_profit:.0f} @ ({best_slow_period},{best_fast_period}) days\")",
"max profit=7042 @ (105.0,3.0) days\n"
]
],
[
[
"Note also that built-in grid search in `backtest` is optimized and slightly faster than the basic loop-based grid search.",
"_____no_output_____"
]
],
[
[
"#time\ntime_basic/time_optimized",
"_____no_output_____"
]
],
[
[
"## Final notes\n\nWhile it is tempting to do a grid search over larger search space and finer resolutions, it is computationally expensive, inefficient, and prone to overfitting. There are better methods than brute force grid search which we will tackle in the next example.\n\nAs an exercise, it is good to try the following:\n* Use different trading strategies and compare their results\n* Use a longer data baseline",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0d0af9e6aeedef21e7870d163b37b3c9fb7d2d3 | 1,761 | ipynb | Jupyter Notebook | IBM_CRS9_W1_Capstone_Notebook.ipynb | mcmasty/Coursera_Capstone | f385a6e7b87ec0aef7b442fa0bb9fa5335626546 | [
"CC-BY-4.0"
] | null | null | null | IBM_CRS9_W1_Capstone_Notebook.ipynb | mcmasty/Coursera_Capstone | f385a6e7b87ec0aef7b442fa0bb9fa5335626546 | [
"CC-BY-4.0"
] | null | null | null | IBM_CRS9_W1_Capstone_Notebook.ipynb | mcmasty/Coursera_Capstone | f385a6e7b87ec0aef7b442fa0bb9fa5335626546 | [
"CC-BY-4.0"
] | 1 | 2019-10-06T05:41:13.000Z | 2019-10-06T05:41:13.000Z | 29.847458 | 157 | 0.399773 | [
[
[
"# IBM Data Science Specialization\n## Course 9: Applied Data Science\n\nThis notebook will be mainly used for the capstone project.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"print ('Hello Capstone Project Course!')",
"Hello Capstone Project Course!\n"
],
[
"# The code was removed by Watson Studio for sharing.",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d0d0b697fdc518cf3d5712643421092af48c239b | 571 | ipynb | Jupyter Notebook | notebook.ipynb | pacara1/ejercicio | 25eb0d1c000d4b4b36e2bb6f27ba2a84d6d8661d | [
"MIT"
] | null | null | null | notebook.ipynb | pacara1/ejercicio | 25eb0d1c000d4b4b36e2bb6f27ba2a84d6d8661d | [
"MIT"
] | null | null | null | notebook.ipynb | pacara1/ejercicio | 25eb0d1c000d4b4b36e2bb6f27ba2a84d6d8661d | [
"MIT"
] | null | null | null | 16.314286 | 34 | 0.516637 | [
[
[
"1+1=2",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code"
]
] |
d0d0b6d09fd2ebdfcdb8412e1f28da13e476a858 | 24,876 | ipynb | Jupyter Notebook | support_vector_mc.ipynb | ak-cell/support-vector-machine | f5972207225f8f6aa49ba949be81c63221cc6f16 | [
"MIT"
] | null | null | null | support_vector_mc.ipynb | ak-cell/support-vector-machine | f5972207225f8f6aa49ba949be81c63221cc6f16 | [
"MIT"
] | null | null | null | support_vector_mc.ipynb | ak-cell/support-vector-machine | f5972207225f8f6aa49ba949be81c63221cc6f16 | [
"MIT"
] | null | null | null | 32.056701 | 87 | 0.347845 | [
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"bank=pd.read_csv('C:/Users/aksha/Desktop/ai/datasets/bill_authentication.csv')\nbank.shape",
"_____no_output_____"
],
[
"bank",
"_____no_output_____"
],
[
"X=bank.drop('Class',axis=1)\ny=bank['Class']",
"_____no_output_____"
],
[
"#Test train split\nfrom sklearn.model_selection import train_test_split\nX_train,X_test,y_train,y_test=train_test_split(X,y,test_size=0.3)",
"_____no_output_____"
],
[
"#Support vector classifier\nfrom sklearn.svm import SVC\nsvclassifier=SVC(kernel='linear')\nsvclassifier.fit(X_train,y_train)",
"_____no_output_____"
],
[
"# Making prediction\ny_pred=svclassifier.predict(X_test)\nprint(y_pred)",
"[0 0 1 0 1 0 0 0 1 0 0 1 1 0 1 0 1 0 1 1 1 0 1 0 0 1 0 0 0 0 0 1 1 1 1 0 0\n 1 1 1 0 1 0 1 1 1 1 0 1 1 1 1 0 1 1 0 1 0 0 0 0 0 1 0 0 0 1 1 0 0 1 0 0 1\n 1 0 0 1 0 1 1 0 0 0 0 1 1 1 1 0 1 1 1 1 0 0 0 1 1 0 0 1 0 1 1 1 1 1 0 0 0\n 1 1 0 0 0 0 1 0 1 1 0 1 1 1 1 1 1 0 0 1 1 0 0 0 1 0 1 1 0 0 1 1 0 1 1 0 0\n 0 1 0 1 0 1 0 0 0 0 0 0 0 1 0 0 1 1 1 1 1 0 1 1 0 1 0 0 1 0 0 0 0 1 1 1 0\n 1 0 0 0 0 0 1 0 1 0 0 1 0 0 1 0 0 0 1 0 1 1 0 0 0 0 0 1 1 1 0 0 0 1 0 0 0\n 0 0 1 1 1 0 0 1 1 0 0 1 0 1 1 0 1 0 0 1 0 1 1 0 0 1 1 0 0 0 0 0 0 1 0 1 1\n 1 0 0 1 0 0 0 0 1 0 0 1 1 1 0 1 0 1 0 1 0 0 0 1 0 1 1 0 0 1 1 0 0 1 1 0 0\n 1 0 0 0 0 0 1 0 1 0 1 1 0 0 1 0 0 1 0 0 0 0 1 0 0 1 0 1 1 0 1 0 0 0 0 1 1\n 1 0 0 0 0 0 0 0 1 0 0 1 0 0 1 0 0 0 0 0 0 0 0 0 0 1 0 1 0 1 1 0 1 1 1 1 0\n 1 1 0 0 1 0 1 0 1 0 0 0 0 0 0 1 1 0 1 1 0 0 1 0 0 1 1 1 1 1 0 0 0 1 0 1 1\n 0 0 1 0 0]\n"
],
[
"#ACCuracy\nfrom sklearn.metrics import classification_report,confusion_matrix\nprint(confusion_matrix(y_test,y_pred))\nprint(classification_report(y_test,y_pred))",
"[[229 4]\n [ 1 178]]\n precision recall f1-score support\n\n 0 1.00 0.98 0.99 233\n 1 0.98 0.99 0.99 179\n\n micro avg 0.99 0.99 0.99 412\n macro avg 0.99 0.99 0.99 412\nweighted avg 0.99 0.99 0.99 412\n\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0d0d737b514f8d31373b5646bce18d9537779a7 | 3,634 | ipynb | Jupyter Notebook | BAMM.101x/sql_and_python.ipynb | KECB/learn | 5b52c5c3ac640dd2a9064c33baaa9bc1885cf15f | [
"MIT"
] | 2 | 2017-09-25T04:29:59.000Z | 2018-11-04T09:53:59.000Z | BAMM.101x/sql_and_python.ipynb | KECB/learn | 5b52c5c3ac640dd2a9064c33baaa9bc1885cf15f | [
"MIT"
] | null | null | null | BAMM.101x/sql_and_python.ipynb | KECB/learn | 5b52c5c3ac640dd2a9064c33baaa9bc1885cf15f | [
"MIT"
] | null | null | null | 21.502959 | 235 | 0.503302 | [
[
[
"<h1>Python and MySQL</h1>",
"_____no_output_____"
],
[
"<h2>First import the python module containing the API</h2>",
"_____no_output_____"
]
],
[
[
"import pymysql",
"_____no_output_____"
]
],
[
[
"<h2>Set up a connection and create a cursor object</h2>",
"_____no_output_____"
]
],
[
[
"db = pymysql.connect(\"localhost\",\"root\",\"None\" ,database=\"schooldb\")\ncursor = db.cursor()\n",
"_____no_output_____"
]
],
[
[
"<h2>Execute a query and get the results</h2>",
"_____no_output_____"
]
],
[
[
"cursor.execute('show tables;')\ncursor.fetchall()\n ",
"_____no_output_____"
],
[
"query = \"\"\"\nSELECT course.name FROM student \nINNER JOIN enrolls_in ON student.ssn = enrolls_in.ssn \nINNER JOIN course ON course.number = enrolls_in.class\nWHERE f_name = \"JOHN\";\n\"\"\"\ncursor.execute(query)\ncursor.fetchall()",
"_____no_output_____"
],
[
"# 向数据库插入数据\ninsert = 'INSERT INTO Student VALUES (\"\", \"\")' \ncursor.execute(insert)\ncursor.fetchall()",
"_____no_output_____"
],
[
"# 同步到数据库\ndb.commit()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d0d102159bd8d68f021329f7c9a0518267a8b10c | 87,140 | ipynb | Jupyter Notebook | Starter_Code.ipynb | ichakrabarty/Deep-Learning-Homework-Charity-Funding-Predictor | 755822c448df18f261e05a5b3428ade572ef4a2a | [
"ADSL"
] | null | null | null | Starter_Code.ipynb | ichakrabarty/Deep-Learning-Homework-Charity-Funding-Predictor | 755822c448df18f261e05a5b3428ade572ef4a2a | [
"ADSL"
] | null | null | null | Starter_Code.ipynb | ichakrabarty/Deep-Learning-Homework-Charity-Funding-Predictor | 755822c448df18f261e05a5b3428ade572ef4a2a | [
"ADSL"
] | null | null | null | 43.788945 | 143 | 0.404269 | [
[
[
"## Preprocessing",
"_____no_output_____"
]
],
[
[
"# Import our dependencies\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.preprocessing import StandardScaler\nimport pandas as pd\nimport tensorflow as tf\n\n# Import and read the charity_data.csv.\nimport pandas as pd \napplication_df = pd.read_csv(\"Resources/charity_data.csv\")\napplication_df.head()",
"_____no_output_____"
],
[
"# Drop the non-beneficial ID columns, 'EIN' and 'NAME'.\n\napplication_df = application_df.drop(columns = ['EIN','NAME'])\napplication_df.head()",
"_____no_output_____"
],
[
"# Determine the number of unique values in each column.\n\napplication_df.nunique()",
"_____no_output_____"
],
[
"# Look at APPLICATION_TYPE value counts for binning\n\napp_type = application_df['APPLICATION_TYPE'].value_counts()\napp_type",
"_____no_output_____"
],
[
"# Choose a cutoff value and create a list of application types to be replaced\n# use the variable name `application_types_to_replace`\n\napplication_types_to_replace = list(app_type[ app_type < 500].index)\n\n# Replace in dataframe\nfor app in application_types_to_replace:\n application_df['APPLICATION_TYPE'] = application_df['APPLICATION_TYPE'].replace(app,\"Other\")\n\n# Check to make sure binning was successful\napplication_df['APPLICATION_TYPE'].value_counts()",
"_____no_output_____"
],
[
"# Look at CLASSIFICATION value counts for binning\n\nclass_type = application_df['CLASSIFICATION'].value_counts()\nclass_type",
"_____no_output_____"
],
[
"# You may find it helpful to look at CLASSIFICATION value counts >1\nclass_type_greaterthan1 = class_type[class_type > 1]\nclass_type_greaterthan1.value_counts()",
"_____no_output_____"
],
[
"# Choose a cutoff value and create a list of classifications to be replaced\n# use the variable name `classifications_to_replace`\n\nclassifications_to_replace = list(class_type[class_type < 100].index)\n\n# Replace in dataframe\nfor cls in classifications_to_replace:\n application_df['CLASSIFICATION'] = application_df['CLASSIFICATION'].replace(cls,\"Other\")\n \n# Check to make sure binning was successful\napplication_df['CLASSIFICATION'].value_counts()",
"_____no_output_____"
],
[
"# Convert categorical data to numeric with `pd.get_dummies`\ndummy_app_df = pd.get_dummies(application_df)\ndummy_app_df.head()",
"_____no_output_____"
],
[
"# Split our preprocessed data into our features and target arrays\ny = dummy_app_df['IS_SUCCESSFUL']\nX = dummy_app_df.drop(columns=['IS_SUCCESSFUL'])\n\n# Split the preprocessed data into a training and testing dataset\nX_train,X_test,y_train,y_test = train_test_split(X,y,random_state = 28)",
"_____no_output_____"
],
[
"# Create a StandardScaler instances\nscaler = StandardScaler()\n\n# Fit the StandardScaler\nX_scaler = scaler.fit(X_train)\n\n# Scale the data\nX_train_scaled = X_scaler.transform(X_train)\nX_test_scaled = X_scaler.transform(X_test)\n\nprint(X_train_scaled.shape)",
"(25724, 45)\n"
]
],
[
[
"## Compile, Train and Evaluate the Model",
"_____no_output_____"
]
],
[
[
"# Define the model - deep neural net, i.e., the number of input features and hidden nodes for each layer.\n\nnn = tf.keras.models.Sequential()\n\n# First hidden layer\nnn.add(tf.keras.layers.Dense(units=80, activation ='relu', input_dim = 45))\n\n# Second hidden layer\nnn.add(tf.keras.layers.Dense(units=30, activation ='relu', input_dim = 45))\n\n# Output layer\nnn.add(tf.keras.layers.Dense(units=1, activation ='sigmoid'))\n\n# Check the structure of the model\nnn.summary()",
"Model: \"sequential_8\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense_29 (Dense) (None, 80) 3680 \n_________________________________________________________________\ndense_30 (Dense) (None, 30) 2430 \n_________________________________________________________________\ndense_31 (Dense) (None, 1) 31 \n=================================================================\nTotal params: 6,141\nTrainable params: 6,141\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"# Compile the model\nnn.compile(loss = 'binary_crossentropy', optimizer = 'adam', metrics = [\"accuracy\"])",
"_____no_output_____"
],
[
"# Train the model\nnn_fit = nn.fit(X_train_scaled,y_train,epochs= 100) ",
"Epoch 1/100\n804/804 [==============================] - 1s 455us/step - loss: 0.5701 - accuracy: 0.7202\nEpoch 2/100\n804/804 [==============================] - 0s 447us/step - loss: 0.5549 - accuracy: 0.7275\nEpoch 3/100\n804/804 [==============================] - 0s 454us/step - loss: 0.5513 - accuracy: 0.7298\nEpoch 4/100\n804/804 [==============================] - 0s 456us/step - loss: 0.5507 - accuracy: 0.7308\nEpoch 5/100\n804/804 [==============================] - 0s 449us/step - loss: 0.5492 - accuracy: 0.7312\nEpoch 6/100\n804/804 [==============================] - 0s 445us/step - loss: 0.5484 - accuracy: 0.7314\nEpoch 7/100\n804/804 [==============================] - 0s 455us/step - loss: 0.5475 - accuracy: 0.7322\nEpoch 8/100\n804/804 [==============================] - 0s 453us/step - loss: 0.5473 - accuracy: 0.7318\nEpoch 9/100\n804/804 [==============================] - 0s 446us/step - loss: 0.5460 - accuracy: 0.7325\nEpoch 10/100\n804/804 [==============================] - 0s 450us/step - loss: 0.5455 - accuracy: 0.7323\nEpoch 11/100\n804/804 [==============================] - 0s 448us/step - loss: 0.5450 - accuracy: 0.7336\nEpoch 12/100\n804/804 [==============================] - 0s 443us/step - loss: 0.5442 - accuracy: 0.7336\nEpoch 13/100\n804/804 [==============================] - 0s 446us/step - loss: 0.5443 - accuracy: 0.7341\nEpoch 14/100\n804/804 [==============================] - 0s 448us/step - loss: 0.5433 - accuracy: 0.7334\nEpoch 15/100\n804/804 [==============================] - 0s 450us/step - loss: 0.5430 - accuracy: 0.7346\nEpoch 16/100\n804/804 [==============================] - 0s 446us/step - loss: 0.5426 - accuracy: 0.7352\nEpoch 17/100\n804/804 [==============================] - 0s 446us/step - loss: 0.5426 - accuracy: 0.7343\nEpoch 18/100\n804/804 [==============================] - 0s 452us/step - loss: 0.5417 - accuracy: 0.7353\nEpoch 19/100\n804/804 [==============================] - 0s 452us/step - loss: 0.5418 - accuracy: 0.7352\nEpoch 20/100\n804/804 [==============================] - 0s 446us/step - loss: 0.5414 - accuracy: 0.7355\nEpoch 21/100\n804/804 [==============================] - 0s 448us/step - loss: 0.5416 - accuracy: 0.7350\nEpoch 22/100\n804/804 [==============================] - 0s 448us/step - loss: 0.5408 - accuracy: 0.7361\nEpoch 23/100\n804/804 [==============================] - 0s 447us/step - loss: 0.5413 - accuracy: 0.7358\nEpoch 24/100\n804/804 [==============================] - 0s 452us/step - loss: 0.5407 - accuracy: 0.7359\nEpoch 25/100\n804/804 [==============================] - 0s 451us/step - loss: 0.5405 - accuracy: 0.7372\nEpoch 26/100\n804/804 [==============================] - 0s 460us/step - loss: 0.5405 - accuracy: 0.7348\nEpoch 27/100\n804/804 [==============================] - 0s 467us/step - loss: 0.5399 - accuracy: 0.7360\nEpoch 28/100\n804/804 [==============================] - 0s 448us/step - loss: 0.5402 - accuracy: 0.7370\nEpoch 29/100\n804/804 [==============================] - 0s 460us/step - loss: 0.5401 - accuracy: 0.7360\nEpoch 30/100\n804/804 [==============================] - 0s 468us/step - loss: 0.5397 - accuracy: 0.7367\nEpoch 31/100\n804/804 [==============================] - 0s 451us/step - loss: 0.5399 - accuracy: 0.7370\nEpoch 32/100\n804/804 [==============================] - 0s 463us/step - loss: 0.5398 - accuracy: 0.7363\nEpoch 33/100\n804/804 [==============================] - 0s 469us/step - loss: 0.5394 - accuracy: 0.7362\nEpoch 34/100\n804/804 [==============================] - 0s 471us/step - loss: 0.5391 - accuracy: 0.7366\nEpoch 35/100\n804/804 [==============================] - 0s 470us/step - loss: 0.5393 - accuracy: 0.7368\nEpoch 36/100\n804/804 [==============================] - 0s 457us/step - loss: 0.5386 - accuracy: 0.7370\nEpoch 37/100\n804/804 [==============================] - 0s 465us/step - loss: 0.5391 - accuracy: 0.7365\nEpoch 38/100\n804/804 [==============================] - 0s 465us/step - loss: 0.5386 - accuracy: 0.7375\nEpoch 39/100\n804/804 [==============================] - 0s 452us/step - loss: 0.5387 - accuracy: 0.7377\nEpoch 40/100\n804/804 [==============================] - 0s 452us/step - loss: 0.5379 - accuracy: 0.7383\nEpoch 41/100\n804/804 [==============================] - 0s 460us/step - loss: 0.5387 - accuracy: 0.7373\nEpoch 42/100\n804/804 [==============================] - 0s 447us/step - loss: 0.5379 - accuracy: 0.7379\nEpoch 43/100\n804/804 [==============================] - 0s 449us/step - loss: 0.5377 - accuracy: 0.7372\nEpoch 44/100\n804/804 [==============================] - 0s 451us/step - loss: 0.5381 - accuracy: 0.7375\nEpoch 45/100\n804/804 [==============================] - 0s 456us/step - loss: 0.5381 - accuracy: 0.7384\nEpoch 46/100\n804/804 [==============================] - 0s 457us/step - loss: 0.5377 - accuracy: 0.7376\nEpoch 47/100\n804/804 [==============================] - 0s 452us/step - loss: 0.5377 - accuracy: 0.7381\nEpoch 48/100\n804/804 [==============================] - 0s 447us/step - loss: 0.5376 - accuracy: 0.7379\nEpoch 49/100\n804/804 [==============================] - 0s 451us/step - loss: 0.5377 - accuracy: 0.7372\nEpoch 50/100\n804/804 [==============================] - 0s 447us/step - loss: 0.5376 - accuracy: 0.7384\nEpoch 51/100\n804/804 [==============================] - 0s 447us/step - loss: 0.5373 - accuracy: 0.7381\nEpoch 52/100\n804/804 [==============================] - 0s 457us/step - loss: 0.5373 - accuracy: 0.7383\nEpoch 53/100\n804/804 [==============================] - 0s 445us/step - loss: 0.5374 - accuracy: 0.7373\nEpoch 54/100\n804/804 [==============================] - 0s 451us/step - loss: 0.5370 - accuracy: 0.7374\nEpoch 55/100\n804/804 [==============================] - 0s 452us/step - loss: 0.5369 - accuracy: 0.7382\nEpoch 56/100\n804/804 [==============================] - 0s 449us/step - loss: 0.5370 - accuracy: 0.7370\nEpoch 57/100\n804/804 [==============================] - 0s 442us/step - loss: 0.5368 - accuracy: 0.7376\nEpoch 58/100\n804/804 [==============================] - 0s 453us/step - loss: 0.5365 - accuracy: 0.7374\nEpoch 59/100\n804/804 [==============================] - 0s 448us/step - loss: 0.5364 - accuracy: 0.7371\nEpoch 60/100\n804/804 [==============================] - 0s 450us/step - loss: 0.5367 - accuracy: 0.7380\nEpoch 61/100\n804/804 [==============================] - 0s 458us/step - loss: 0.5362 - accuracy: 0.7387\nEpoch 62/100\n804/804 [==============================] - 0s 461us/step - loss: 0.5366 - accuracy: 0.7392\nEpoch 63/100\n804/804 [==============================] - 0s 458us/step - loss: 0.5360 - accuracy: 0.7388\nEpoch 64/100\n804/804 [==============================] - 0s 456us/step - loss: 0.5361 - accuracy: 0.7382\nEpoch 65/100\n804/804 [==============================] - 0s 469us/step - loss: 0.5359 - accuracy: 0.7392\nEpoch 66/100\n804/804 [==============================] - 0s 467us/step - loss: 0.5363 - accuracy: 0.7385\nEpoch 67/100\n804/804 [==============================] - 0s 458us/step - loss: 0.5355 - accuracy: 0.7393\nEpoch 68/100\n804/804 [==============================] - 0s 467us/step - loss: 0.5362 - accuracy: 0.7387\nEpoch 69/100\n804/804 [==============================] - 0s 463us/step - loss: 0.5359 - accuracy: 0.7385\nEpoch 70/100\n804/804 [==============================] - 0s 502us/step - loss: 0.5358 - accuracy: 0.7389\nEpoch 71/100\n804/804 [==============================] - 0s 460us/step - loss: 0.5356 - accuracy: 0.7393\nEpoch 72/100\n804/804 [==============================] - 0s 460us/step - loss: 0.5365 - accuracy: 0.7379\nEpoch 73/100\n804/804 [==============================] - 0s 453us/step - loss: 0.5351 - accuracy: 0.7398\nEpoch 74/100\n804/804 [==============================] - 0s 459us/step - loss: 0.5352 - accuracy: 0.7388\nEpoch 75/100\n804/804 [==============================] - 0s 446us/step - loss: 0.5354 - accuracy: 0.7385\nEpoch 76/100\n804/804 [==============================] - 0s 453us/step - loss: 0.5355 - accuracy: 0.7386\nEpoch 77/100\n804/804 [==============================] - 0s 473us/step - loss: 0.5349 - accuracy: 0.7383\nEpoch 78/100\n804/804 [==============================] - 0s 466us/step - loss: 0.5352 - accuracy: 0.7386\nEpoch 79/100\n"
],
[
"# Evaluate the model using the test data\nmodel_loss, model_accuracy = nn.evaluate(X_test_scaled,y_test,verbose=2)\nprint(f\"Loss: {model_loss}, Accuracy: {model_accuracy}\")",
"268/268 - 0s - loss: 0.5600 - accuracy: 0.7297\nLoss: 0.5600452423095703, Accuracy: 0.72967928647995\n"
],
[
"# Export our model to HDF5 file\nnn.save('AlphabetSoupCharity.h5')",
"_____no_output_____"
]
],
[
[
"## Attempt 2",
"_____no_output_____"
]
],
[
[
"# Define the model - deep neural net, i.e., the number of input features and hidden nodes for each layer.\n\nnn = tf.keras.models.Sequential()\n\n# First hidden layer\nnn.add(tf.keras.layers.Dense(units=80, activation ='relu', input_dim = 45))\n\n# Second hidden layer\nnn.add(tf.keras.layers.Dense(units=30, activation ='relu', input_dim = 45))\n\n# Third hidden layer\nnn.add(tf.keras.layers.Dense(units=5, activation ='relu', input_dim = 45))\n\n# Output layer\nnn.add(tf.keras.layers.Dense(units=1, activation ='sigmoid'))\n\n# Check the structure of the model\nnn.summary()",
"Model: \"sequential_9\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense_32 (Dense) (None, 80) 3680 \n_________________________________________________________________\ndense_33 (Dense) (None, 30) 2430 \n_________________________________________________________________\ndense_34 (Dense) (None, 5) 155 \n_________________________________________________________________\ndense_35 (Dense) (None, 1) 6 \n=================================================================\nTotal params: 6,271\nTrainable params: 6,271\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"# Compile the model\nnn.compile(loss = 'binary_crossentropy', optimizer = 'adam', metrics = [\"accuracy\"])",
"_____no_output_____"
],
[
"nn_fit = nn.fit(X_train_scaled,y_train,epochs= 100) ",
"Epoch 1/100\n804/804 [==============================] - 1s 475us/step - loss: 0.5712 - accuracy: 0.7187\nEpoch 2/100\n804/804 [==============================] - 0s 466us/step - loss: 0.5545 - accuracy: 0.7240\nEpoch 3/100\n804/804 [==============================] - 0s 467us/step - loss: 0.5520 - accuracy: 0.7257\nEpoch 4/100\n804/804 [==============================] - 0s 488us/step - loss: 0.5504 - accuracy: 0.7266\nEpoch 5/100\n804/804 [==============================] - 0s 473us/step - loss: 0.5490 - accuracy: 0.7288\nEpoch 6/100\n804/804 [==============================] - 0s 470us/step - loss: 0.5480 - accuracy: 0.7306\nEpoch 7/100\n804/804 [==============================] - 0s 471us/step - loss: 0.5471 - accuracy: 0.7315\nEpoch 8/100\n804/804 [==============================] - 0s 467us/step - loss: 0.5471 - accuracy: 0.7325\nEpoch 9/100\n804/804 [==============================] - 0s 473us/step - loss: 0.5461 - accuracy: 0.7320\nEpoch 10/100\n804/804 [==============================] - 0s 467us/step - loss: 0.5458 - accuracy: 0.7315\nEpoch 11/100\n804/804 [==============================] - 0s 464us/step - loss: 0.5455 - accuracy: 0.7329\nEpoch 12/100\n804/804 [==============================] - 0s 476us/step - loss: 0.5450 - accuracy: 0.7329\nEpoch 13/100\n804/804 [==============================] - 0s 468us/step - loss: 0.5447 - accuracy: 0.7336\nEpoch 14/100\n804/804 [==============================] - 0s 468us/step - loss: 0.5444 - accuracy: 0.7327\nEpoch 15/100\n804/804 [==============================] - 0s 476us/step - loss: 0.5436 - accuracy: 0.7341\nEpoch 16/100\n804/804 [==============================] - 0s 475us/step - loss: 0.5438 - accuracy: 0.7343\nEpoch 17/100\n804/804 [==============================] - 0s 467us/step - loss: 0.5440 - accuracy: 0.7336\nEpoch 18/100\n804/804 [==============================] - 0s 469us/step - loss: 0.5435 - accuracy: 0.7335\nEpoch 19/100\n804/804 [==============================] - 0s 466us/step - loss: 0.5430 - accuracy: 0.7340\nEpoch 20/100\n804/804 [==============================] - 0s 472us/step - loss: 0.5425 - accuracy: 0.7346\nEpoch 21/100\n804/804 [==============================] - 0s 471us/step - loss: 0.5425 - accuracy: 0.7348\nEpoch 22/100\n804/804 [==============================] - 0s 467us/step - loss: 0.5419 - accuracy: 0.7350\nEpoch 23/100\n804/804 [==============================] - 0s 465us/step - loss: 0.5421 - accuracy: 0.7346\nEpoch 24/100\n804/804 [==============================] - 0s 469us/step - loss: 0.5412 - accuracy: 0.7354\nEpoch 25/100\n804/804 [==============================] - 0s 472us/step - loss: 0.5416 - accuracy: 0.7355\nEpoch 26/100\n804/804 [==============================] - 0s 476us/step - loss: 0.5414 - accuracy: 0.7354\nEpoch 27/100\n804/804 [==============================] - 0s 472us/step - loss: 0.5414 - accuracy: 0.7356\nEpoch 28/100\n804/804 [==============================] - 0s 462us/step - loss: 0.5406 - accuracy: 0.7354\nEpoch 29/100\n804/804 [==============================] - 0s 476us/step - loss: 0.5406 - accuracy: 0.7357\nEpoch 30/100\n804/804 [==============================] - 0s 473us/step - loss: 0.5405 - accuracy: 0.7357\nEpoch 31/100\n804/804 [==============================] - 0s 476us/step - loss: 0.5406 - accuracy: 0.7358\nEpoch 32/100\n804/804 [==============================] - 0s 470us/step - loss: 0.5403 - accuracy: 0.7364\nEpoch 33/100\n804/804 [==============================] - 0s 472us/step - loss: 0.5404 - accuracy: 0.7357\nEpoch 34/100\n804/804 [==============================] - 0s 476us/step - loss: 0.5403 - accuracy: 0.7360\nEpoch 35/100\n804/804 [==============================] - 0s 473us/step - loss: 0.5400 - accuracy: 0.7355\nEpoch 36/100\n804/804 [==============================] - 0s 469us/step - loss: 0.5398 - accuracy: 0.7360\nEpoch 37/100\n804/804 [==============================] - 0s 472us/step - loss: 0.5397 - accuracy: 0.7371\nEpoch 38/100\n804/804 [==============================] - 0s 472us/step - loss: 0.5396 - accuracy: 0.7364\nEpoch 39/100\n804/804 [==============================] - 0s 473us/step - loss: 0.5392 - accuracy: 0.7370\nEpoch 40/100\n804/804 [==============================] - 0s 471us/step - loss: 0.5391 - accuracy: 0.7364\nEpoch 41/100\n804/804 [==============================] - 0s 466us/step - loss: 0.5392 - accuracy: 0.7367\nEpoch 42/100\n804/804 [==============================] - 0s 481us/step - loss: 0.5389 - accuracy: 0.7364\nEpoch 43/100\n804/804 [==============================] - 0s 472us/step - loss: 0.5389 - accuracy: 0.7373\nEpoch 44/100\n804/804 [==============================] - 0s 482us/step - loss: 0.5386 - accuracy: 0.7371\nEpoch 45/100\n804/804 [==============================] - 0s 474us/step - loss: 0.5387 - accuracy: 0.7378\nEpoch 46/100\n804/804 [==============================] - 0s 467us/step - loss: 0.5385 - accuracy: 0.7375\nEpoch 47/100\n804/804 [==============================] - 0s 473us/step - loss: 0.5390 - accuracy: 0.7372\nEpoch 48/100\n804/804 [==============================] - 0s 466us/step - loss: 0.5385 - accuracy: 0.7365\nEpoch 49/100\n804/804 [==============================] - 0s 465us/step - loss: 0.5383 - accuracy: 0.7371\nEpoch 50/100\n804/804 [==============================] - 0s 475us/step - loss: 0.5382 - accuracy: 0.7371\nEpoch 51/100\n804/804 [==============================] - 0s 465us/step - loss: 0.5379 - accuracy: 0.7376\nEpoch 52/100\n804/804 [==============================] - 0s 464us/step - loss: 0.5386 - accuracy: 0.7370\nEpoch 53/100\n804/804 [==============================] - 0s 472us/step - loss: 0.5373 - accuracy: 0.7373\nEpoch 54/100\n804/804 [==============================] - 0s 463us/step - loss: 0.5384 - accuracy: 0.7371\nEpoch 55/100\n804/804 [==============================] - 0s 472us/step - loss: 0.5380 - accuracy: 0.7379\nEpoch 56/100\n804/804 [==============================] - 0s 483us/step - loss: 0.5375 - accuracy: 0.7376\nEpoch 57/100\n804/804 [==============================] - 0s 473us/step - loss: 0.5380 - accuracy: 0.7372\nEpoch 58/100\n804/804 [==============================] - 0s 477us/step - loss: 0.5375 - accuracy: 0.7378\nEpoch 59/100\n804/804 [==============================] - 0s 472us/step - loss: 0.5374 - accuracy: 0.7381\nEpoch 60/100\n804/804 [==============================] - 0s 470us/step - loss: 0.5376 - accuracy: 0.7365\nEpoch 61/100\n804/804 [==============================] - 0s 471us/step - loss: 0.5373 - accuracy: 0.7373\nEpoch 62/100\n804/804 [==============================] - 0s 468us/step - loss: 0.5368 - accuracy: 0.7379\nEpoch 63/100\n804/804 [==============================] - 0s 468us/step - loss: 0.5369 - accuracy: 0.7379\nEpoch 64/100\n804/804 [==============================] - 0s 473us/step - loss: 0.5371 - accuracy: 0.7382\nEpoch 65/100\n804/804 [==============================] - 0s 465us/step - loss: 0.5369 - accuracy: 0.7375\nEpoch 66/100\n804/804 [==============================] - 0s 466us/step - loss: 0.5371 - accuracy: 0.7380\nEpoch 67/100\n804/804 [==============================] - 0s 468us/step - loss: 0.5363 - accuracy: 0.7382\nEpoch 68/100\n804/804 [==============================] - 0s 466us/step - loss: 0.5368 - accuracy: 0.7374\nEpoch 69/100\n804/804 [==============================] - 0s 474us/step - loss: 0.5368 - accuracy: 0.7376\nEpoch 70/100\n804/804 [==============================] - 0s 466us/step - loss: 0.5364 - accuracy: 0.7372\nEpoch 71/100\n804/804 [==============================] - 0s 468us/step - loss: 0.5365 - accuracy: 0.7378\nEpoch 72/100\n804/804 [==============================] - 0s 472us/step - loss: 0.5365 - accuracy: 0.7380\nEpoch 73/100\n804/804 [==============================] - 0s 464us/step - loss: 0.5364 - accuracy: 0.7383\nEpoch 74/100\n804/804 [==============================] - 0s 463us/step - loss: 0.5362 - accuracy: 0.7372\nEpoch 75/100\n804/804 [==============================] - 0s 472us/step - loss: 0.5362 - accuracy: 0.7383\nEpoch 76/100\n804/804 [==============================] - 0s 486us/step - loss: 0.5372 - accuracy: 0.7383\nEpoch 77/100\n804/804 [==============================] - 0s 482us/step - loss: 0.5358 - accuracy: 0.7383\nEpoch 78/100\n804/804 [==============================] - 0s 472us/step - loss: 0.5360 - accuracy: 0.7376\nEpoch 79/100\n"
],
[
"# Evaluate the model using the test data\nmodel_loss, model_accuracy = nn.evaluate(X_test_scaled,y_test,verbose=2)\nprint(f\"Loss: {model_loss}, Accuracy: {model_accuracy}\")",
"268/268 - 0s - loss: 0.5587 - accuracy: 0.7329\nLoss: 0.558691143989563, Accuracy: 0.7329446077346802\n"
],
[
"# Export our model to HDF5 file\nnn.save('AlphabetSoupCharity_Optimization.h5')",
"_____no_output_____"
]
],
[
[
"## Attempt 3",
"_____no_output_____"
]
],
[
[
"# Define the model - deep neural net, i.e., the number of input features and hidden nodes for each layer.\n\nnn = tf.keras.models.Sequential()\n\n# First hidden layer\nnn.add(tf.keras.layers.Dense(units=80, activation ='relu', input_dim = 45))\n\n# Second hidden layer\nnn.add(tf.keras.layers.Dense(units=30, activation ='sigmoid', input_dim = 45))\n\n# Third hidden layer\nnn.add(tf.keras.layers.Dense(units=5, activation ='sigmoid', input_dim = 45))\n\n# Output layer\nnn.add(tf.keras.layers.Dense(units=1, activation ='sigmoid'))\n\n# Check the structure of the model\nnn.summary()",
"Model: \"sequential_10\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense_36 (Dense) (None, 80) 3680 \n_________________________________________________________________\ndense_37 (Dense) (None, 30) 2430 \n_________________________________________________________________\ndense_38 (Dense) (None, 5) 155 \n_________________________________________________________________\ndense_39 (Dense) (None, 1) 6 \n=================================================================\nTotal params: 6,271\nTrainable params: 6,271\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"# Compile the model\nnn.compile(loss = 'binary_crossentropy', optimizer = 'adam', metrics = [\"accuracy\"])",
"_____no_output_____"
],
[
"nn_fit = nn.fit(X_train_scaled,y_train,epochs= 200) ",
"Epoch 1/200\n804/804 [==============================] - 1s 483us/step - loss: 0.5940 - accuracy: 0.7141\nEpoch 2/200\n804/804 [==============================] - 0s 472us/step - loss: 0.5709 - accuracy: 0.7276\nEpoch 3/200\n804/804 [==============================] - 0s 472us/step - loss: 0.5627 - accuracy: 0.7283\nEpoch 4/200\n804/804 [==============================] - 0s 476us/step - loss: 0.5586 - accuracy: 0.7303\nEpoch 5/200\n804/804 [==============================] - 0s 476us/step - loss: 0.5558 - accuracy: 0.7301\nEpoch 6/200\n804/804 [==============================] - 0s 472us/step - loss: 0.5538 - accuracy: 0.7318\nEpoch 7/200\n804/804 [==============================] - 0s 484us/step - loss: 0.5525 - accuracy: 0.7313\nEpoch 8/200\n804/804 [==============================] - 0s 476us/step - loss: 0.5504 - accuracy: 0.7311\nEpoch 9/200\n804/804 [==============================] - 0s 477us/step - loss: 0.5496 - accuracy: 0.7317\nEpoch 10/200\n804/804 [==============================] - 0s 496us/step - loss: 0.5489 - accuracy: 0.7317\nEpoch 11/200\n804/804 [==============================] - 0s 471us/step - loss: 0.5479 - accuracy: 0.7322\nEpoch 12/200\n804/804 [==============================] - 0s 478us/step - loss: 0.5473 - accuracy: 0.7324\nEpoch 13/200\n804/804 [==============================] - 0s 472us/step - loss: 0.5467 - accuracy: 0.7322\nEpoch 14/200\n804/804 [==============================] - 0s 476us/step - loss: 0.5462 - accuracy: 0.7336\nEpoch 15/200\n804/804 [==============================] - 0s 497us/step - loss: 0.5458 - accuracy: 0.7341\nEpoch 16/200\n804/804 [==============================] - 0s 488us/step - loss: 0.5453 - accuracy: 0.7332\nEpoch 17/200\n804/804 [==============================] - 0s 481us/step - loss: 0.5449 - accuracy: 0.7333\nEpoch 18/200\n804/804 [==============================] - 0s 475us/step - loss: 0.5446 - accuracy: 0.7339\nEpoch 19/200\n804/804 [==============================] - 0s 476us/step - loss: 0.5440 - accuracy: 0.7337\nEpoch 20/200\n804/804 [==============================] - 0s 482us/step - loss: 0.5438 - accuracy: 0.7338\nEpoch 21/200\n804/804 [==============================] - 0s 477us/step - loss: 0.5432 - accuracy: 0.7344\nEpoch 22/200\n804/804 [==============================] - 0s 475us/step - loss: 0.5432 - accuracy: 0.7345\nEpoch 23/200\n804/804 [==============================] - 0s 487us/step - loss: 0.5429 - accuracy: 0.7345\nEpoch 24/200\n804/804 [==============================] - 0s 486us/step - loss: 0.5425 - accuracy: 0.7341\nEpoch 25/200\n804/804 [==============================] - 0s 476us/step - loss: 0.5424 - accuracy: 0.7350\nEpoch 26/200\n804/804 [==============================] - 0s 476us/step - loss: 0.5423 - accuracy: 0.7347\nEpoch 27/200\n804/804 [==============================] - 0s 476us/step - loss: 0.5418 - accuracy: 0.7345\nEpoch 28/200\n804/804 [==============================] - 0s 478us/step - loss: 0.5416 - accuracy: 0.7347\nEpoch 29/200\n804/804 [==============================] - 0s 475us/step - loss: 0.5415 - accuracy: 0.7352\nEpoch 30/200\n804/804 [==============================] - 0s 486us/step - loss: 0.5412 - accuracy: 0.7346\nEpoch 31/200\n804/804 [==============================] - 0s 482us/step - loss: 0.5409 - accuracy: 0.7357\nEpoch 32/200\n804/804 [==============================] - 0s 482us/step - loss: 0.5407 - accuracy: 0.7359\nEpoch 33/200\n804/804 [==============================] - 0s 484us/step - loss: 0.5404 - accuracy: 0.7351\nEpoch 34/200\n804/804 [==============================] - 0s 475us/step - loss: 0.5403 - accuracy: 0.7359\nEpoch 35/200\n804/804 [==============================] - 0s 477us/step - loss: 0.5399 - accuracy: 0.7362\nEpoch 36/200\n804/804 [==============================] - 0s 480us/step - loss: 0.5397 - accuracy: 0.7364\nEpoch 37/200\n804/804 [==============================] - 0s 475us/step - loss: 0.5400 - accuracy: 0.7352\nEpoch 38/200\n804/804 [==============================] - 0s 480us/step - loss: 0.5396 - accuracy: 0.7360\nEpoch 39/200\n804/804 [==============================] - 0s 484us/step - loss: 0.5392 - accuracy: 0.7372\nEpoch 40/200\n804/804 [==============================] - 0s 484us/step - loss: 0.5392 - accuracy: 0.7353\nEpoch 41/200\n804/804 [==============================] - 0s 486us/step - loss: 0.5389 - accuracy: 0.7369\nEpoch 42/200\n804/804 [==============================] - 0s 487us/step - loss: 0.5388 - accuracy: 0.7372\nEpoch 43/200\n804/804 [==============================] - 0s 490us/step - loss: 0.5387 - accuracy: 0.7366\nEpoch 44/200\n804/804 [==============================] - 0s 477us/step - loss: 0.5385 - accuracy: 0.7368\nEpoch 45/200\n804/804 [==============================] - 0s 475us/step - loss: 0.5383 - accuracy: 0.7364\nEpoch 46/200\n804/804 [==============================] - 0s 476us/step - loss: 0.5382 - accuracy: 0.7373\nEpoch 47/200\n804/804 [==============================] - 0s 481us/step - loss: 0.5378 - accuracy: 0.7369\nEpoch 48/200\n804/804 [==============================] - 0s 479us/step - loss: 0.5378 - accuracy: 0.7373\nEpoch 49/200\n804/804 [==============================] - 0s 479us/step - loss: 0.5379 - accuracy: 0.7367\nEpoch 50/200\n804/804 [==============================] - 0s 476us/step - loss: 0.5379 - accuracy: 0.7364\nEpoch 51/200\n804/804 [==============================] - 0s 475us/step - loss: 0.5376 - accuracy: 0.7367\nEpoch 52/200\n804/804 [==============================] - 0s 478us/step - loss: 0.5373 - accuracy: 0.7369\nEpoch 53/200\n804/804 [==============================] - 0s 479us/step - loss: 0.5373 - accuracy: 0.7377\nEpoch 54/200\n804/804 [==============================] - 0s 481us/step - loss: 0.5371 - accuracy: 0.7376\nEpoch 55/200\n804/804 [==============================] - 0s 521us/step - loss: 0.5370 - accuracy: 0.7374\nEpoch 56/200\n804/804 [==============================] - 0s 485us/step - loss: 0.5368 - accuracy: 0.7377\nEpoch 57/200\n804/804 [==============================] - 0s 501us/step - loss: 0.5368 - accuracy: 0.7378\nEpoch 58/200\n804/804 [==============================] - 0s 472us/step - loss: 0.5367 - accuracy: 0.7373\nEpoch 59/200\n804/804 [==============================] - 0s 477us/step - loss: 0.5365 - accuracy: 0.7371\nEpoch 60/200\n804/804 [==============================] - 0s 486us/step - loss: 0.5366 - accuracy: 0.7381\nEpoch 61/200\n804/804 [==============================] - 0s 479us/step - loss: 0.5362 - accuracy: 0.7372\nEpoch 62/200\n804/804 [==============================] - 0s 480us/step - loss: 0.5363 - accuracy: 0.7378\nEpoch 63/200\n804/804 [==============================] - 0s 485us/step - loss: 0.5361 - accuracy: 0.7374\nEpoch 64/200\n804/804 [==============================] - 0s 478us/step - loss: 0.5360 - accuracy: 0.7373\nEpoch 65/200\n804/804 [==============================] - 0s 499us/step - loss: 0.5357 - accuracy: 0.7383\nEpoch 66/200\n804/804 [==============================] - 0s 481us/step - loss: 0.5358 - accuracy: 0.7379\nEpoch 67/200\n804/804 [==============================] - 0s 479us/step - loss: 0.5357 - accuracy: 0.7384\nEpoch 68/200\n804/804 [==============================] - 0s 493us/step - loss: 0.5355 - accuracy: 0.7379\nEpoch 69/200\n804/804 [==============================] - 0s 475us/step - loss: 0.5354 - accuracy: 0.7379\nEpoch 70/200\n804/804 [==============================] - 0s 488us/step - loss: 0.5354 - accuracy: 0.7390\nEpoch 71/200\n804/804 [==============================] - 0s 475us/step - loss: 0.5354 - accuracy: 0.7378\nEpoch 72/200\n804/804 [==============================] - 0s 473us/step - loss: 0.5352 - accuracy: 0.7388\nEpoch 73/200\n804/804 [==============================] - 0s 477us/step - loss: 0.5352 - accuracy: 0.7381\nEpoch 74/200\n804/804 [==============================] - 0s 472us/step - loss: 0.5351 - accuracy: 0.7384\nEpoch 75/200\n804/804 [==============================] - 0s 478us/step - loss: 0.5347 - accuracy: 0.7389\nEpoch 76/200\n804/804 [==============================] - 0s 491us/step - loss: 0.5351 - accuracy: 0.7389\nEpoch 77/200\n804/804 [==============================] - 0s 478us/step - loss: 0.5348 - accuracy: 0.7381\nEpoch 78/200\n804/804 [==============================] - 0s 489us/step - loss: 0.5346 - accuracy: 0.7385\nEpoch 79/200\n"
],
[
"# Evaluate the model using the test data\nmodel_loss, model_accuracy = nn.evaluate(X_test_scaled,y_test,verbose=2)\nprint(f\"Loss: {model_loss}, Accuracy: {model_accuracy}\")",
"268/268 - 0s - loss: 0.5587 - accuracy: 0.7329\nLoss: 0.558691143989563, Accuracy: 0.7329446077346802\n"
],
[
"# Export our model to HDF5 file\nnn.save('AlphabetSoupCharity_Optimization2.h5')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
d0d10ca5bddd597ce07f8264d96c70415307adfe | 30,481 | ipynb | Jupyter Notebook | Week1/prerun_code/3_Q_learning_scratch_prerun.ipynb | ashwin-M-D/Northeastern_RL_Discussions | 85e984c7a4783b77f4015117d49de6ae7d7d990a | [
"MIT"
] | null | null | null | Week1/prerun_code/3_Q_learning_scratch_prerun.ipynb | ashwin-M-D/Northeastern_RL_Discussions | 85e984c7a4783b77f4015117d49de6ae7d7d990a | [
"MIT"
] | null | null | null | Week1/prerun_code/3_Q_learning_scratch_prerun.ipynb | ashwin-M-D/Northeastern_RL_Discussions | 85e984c7a4783b77f4015117d49de6ae7d7d990a | [
"MIT"
] | null | null | null | 45.023634 | 11,484 | 0.586825 | [
[
[
"# Tabular Q-Learning From Scratch",
"_____no_output_____"
],
[
"### Custom Environment to train our model on",
"_____no_output_____"
]
],
[
[
"import gym\nfrom gym import spaces\n\nimport numpy as np\nimport random\nfrom copy import deepcopy\n\n\nclass gridworld_custom(gym.Env):\n\n \"\"\"Custom Environment that follows gym interface\"\"\"\n metadata = {'render.modes': ['human']}\n\n def __init__(self, *args, **kwargs):\n super(gridworld_custom, self).__init__()\n\n self.current_step = 0\n\n self.reward_range = (-10, 100)\n\n self.action_space = spaces.Discrete(2)\n\n self.observation_space = spaces.Box(low=np.array(\n [0, 0]), high=np.array([4, 4]), dtype=np.int64)\n\n self.target_coord = (4, 4)\n self.death_coord = [(3, 1), (4, 2)]\n\n def Reward_Function(self, obs):\n\n if (obs[0] == self.target_coord[0] and obs[1] == self.target_coord[1]):\n return 20\n elif (obs[0] == self.death_coord[0][0] and obs[1] == self.death_coord[0][1]) or \\\n (obs[0] == self.death_coord[1][0] and obs[1] == self.death_coord[1][1]):\n return -10\n else:\n return -1\n\n return 0\n\n def reset(self):\n self.current_step = 0\n\n self.prev_obs = [random.randint(0, 4), random.randint(0, 4)]\n\n if (self.prev_obs[0] == self.target_coord[0] and self.prev_obs[1] == self.target_coord[1]):\n\n return self.reset()\n\n return self.prev_obs\n\n def step(self, action):\n\n action = int(action)\n\n self.current_step += 1\n\n obs = deepcopy(self.prev_obs)\n\n if(action == 0):\n if(self.prev_obs[0] < 4):\n obs[0] = obs[0] + 1\n else:\n obs[0] = obs[0]\n\n if(action == 1):\n if(self.prev_obs[0] > 0):\n obs[0] = obs[0] - 1\n else:\n obs[0] = obs[0]\n\n if(action == 2):\n if(self.prev_obs[1] < 4):\n obs[1] = obs[1] + 1\n else:\n obs[1] = obs[1]\n\n if(action == 3):\n if(self.prev_obs[1] > 0):\n obs[1] = obs[1] - 1\n else:\n obs[1] = obs[1]\n\n reward = self.Reward_Function(obs)\n\n if (obs[0] == self.target_coord[0] and obs[1] == self.target_coord[1]) or (self.current_step >= 250):\n done = True\n else:\n done = False\n\n self.prev_obs = obs\n\n return obs, reward, done, {}\n\n def render(self, mode='human', close=False):\n\n for i in range(0, 5):\n for j in range(0, 5):\n if i == self.prev_obs[0] and j == self.prev_obs[1]:\n print(\"*\", end=\" \")\n elif i == self.target_coord[0] and j == self.target_coord[1]:\n print(\"w\", end=\" \")\n elif (i == self.death_coord[0][0] and j == self.death_coord[0][1]) or \\\n (i == self.death_coord[1][0] and j == self.death_coord[1][1]):\n print(\"D\", end=\" \")\n else:\n print(\"_\", end=\" \")\n print()\n\n print()\n print()\n",
"_____no_output_____"
]
],
[
[
"### Import Required Packages",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n\nfrom copy import deepcopy\n\nimport pandas as pd\n#from tqdm.auto import tqdm\nfrom tqdm import tqdm",
"_____no_output_____"
]
],
[
[
"### Build the q-learning class which contains the table storing all the q values for all states and actions",
"_____no_output_____"
]
],
[
[
"class q_learning():\n \n def __init__(self):\n self.q_table = pd.DataFrame(columns=['state', 'q_val_0', 'q_val_1', 'q_val_2', 'q_val_3'])\n\n for i in range(5):\n for j in range(5):\n state_str = \"(\"+str(i)+\",\"+str(j)+\")\"\n X = pd.DataFrame([[state_str, 0, 0, 0, 0]], columns=['state', 'q_val_0', 'q_val_1', 'q_val_2', 'q_val_3'])\n self.q_table = self.q_table.append(X, ignore_index=True)\n \n self.q_table = self.q_table.set_index('state')\n\n self.gamma = 1\n self.step_size = 0.8\n \n def update_q_value(self, curr_state, prev_state, action, reward):\n\n curr_state_str = \"(\"+str(curr_state[0])+\",\"+str(curr_state[1])+\")\"\n prev_state_str = \"(\"+str(prev_state[0])+\",\"+str(prev_state[1])+\")\"\n action_str = \"q_val_\"+str(action)\n\n q_pred = self.q_table.loc[prev_state_str][action_str]\n\n q_target = reward + self.gamma * np.max(self.q_table.loc[curr_state_str].to_numpy())\n\n self.q_table.loc[prev_state_str][action_str] = q_pred + self.step_size * (q_target - q_pred)\n\n def choose_action(self, curr_state):\n curr_state_str = \"(\"+str(curr_state[0])+\",\"+str(curr_state[1])+\")\"\n action = np.argmax(self.q_table.loc[curr_state_str].to_numpy())\n return action",
"_____no_output_____"
]
],
[
[
"#### Check up the functionality of epsilon greedy. Just for reference.",
"_____no_output_____"
]
],
[
[
"epsilon = 1\nepsilon_decay = 0.9997\n\nepisodes = 10000\nepsilon_copy = deepcopy(epsilon)\neps = []\n\nfor i in range(episodes):\n epsilon_copy = epsilon_copy * epsilon_decay\n eps.append(epsilon_copy)\n\nplt.plot(eps)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Run everything.",
"_____no_output_____"
]
],
[
[
"env = gridworld_custom()\nagent = q_learning()\n\n\npbar = tqdm(range(episodes))\n\nfor episode in pbar:\n\n prev_obs = env.reset()\n done = False\n\n epsilon = epsilon * epsilon_decay\n\n while not(done):\n if(random.uniform(0, 1) > epsilon):\n action = agent.choose_action(prev_obs)\n else:\n action = random.randint(0,3)\n obs, reward, done, _ = env.step(action)\n agent.update_q_value(obs, prev_obs, action, reward)\n prev_obs = deepcopy(obs)",
"100%|███████████████████████████████████████████████████████████████████████████| 10000/10000 [00:23<00:00, 434.64it/s]\n"
]
],
[
[
"### Take a look at the Q Table after training. Gives us an understanding as to how the model might function",
"_____no_output_____"
]
],
[
[
"agent.q_table",
"_____no_output_____"
]
],
[
[
"### Test the trained model",
"_____no_output_____"
]
],
[
[
"prev_obs = env.reset()\ndone = False\nenv.render()\nwhile not(done):\n action = agent.choose_action(prev_obs)\n obs, reward, done, _ = env.step(action)\n prev_obs = obs\n env.render()",
"_ _ _ _ _ \n_ _ _ _ _ \n_ _ _ _ _ \n_ D _ _ _ \n* _ D _ w \n\n\n_ _ _ _ _ \n_ _ _ _ _ \n_ _ _ _ _ \n* D _ _ _ \n_ _ D _ w \n\n\n_ _ _ _ _ \n_ _ _ _ _ \n* _ _ _ _ \n_ D _ _ _ \n_ _ D _ w \n\n\n_ _ _ _ _ \n_ _ _ _ _ \n_ * _ _ _ \n_ D _ _ _ \n_ _ D _ w \n\n\n_ _ _ _ _ \n_ _ _ _ _ \n_ _ * _ _ \n_ D _ _ _ \n_ _ D _ w \n\n\n_ _ _ _ _ \n_ _ _ _ _ \n_ _ _ _ _ \n_ D * _ _ \n_ _ D _ w \n\n\n_ _ _ _ _ \n_ _ _ _ _ \n_ _ _ _ _ \n_ D _ * _ \n_ _ D _ w \n\n\n_ _ _ _ _ \n_ _ _ _ _ \n_ _ _ _ _ \n_ D _ _ _ \n_ _ D * w \n\n\n_ _ _ _ _ \n_ _ _ _ _ \n_ _ _ _ _ \n_ D _ _ _ \n_ _ D _ * \n\n\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0d11c575e59a2ba7d73006dd878433fb2f5d963 | 52,738 | ipynb | Jupyter Notebook | .ipynb_checkpoints/Summarization-checkpoint.ipynb | ParasAvkirkar/AbstractiveTextSummarization | 9278b6e3ea55806c161a6623250d0629ddc51311 | [
"MIT"
] | null | null | null | .ipynb_checkpoints/Summarization-checkpoint.ipynb | ParasAvkirkar/AbstractiveTextSummarization | 9278b6e3ea55806c161a6623250d0629ddc51311 | [
"MIT"
] | null | null | null | .ipynb_checkpoints/Summarization-checkpoint.ipynb | ParasAvkirkar/AbstractiveTextSummarization | 9278b6e3ea55806c161a6623250d0629ddc51311 | [
"MIT"
] | 1 | 2019-10-22T08:58:42.000Z | 2019-10-22T08:58:42.000Z | 33.828095 | 293 | 0.504304 | [
[
[
"# Abstractive Summarization",
"_____no_output_____"
],
[
"### Loading Pre-processed Dataset\n\nThe Data is preprocessed in [Data_Pre-Processing.ipynb](https://github.com/JRC1995/Abstractive-Summarization/blob/master/Data_Pre-Processing.ipynb)\n\nDataset source: https://www.kaggle.com/snap/amazon-fine-food-reviews",
"_____no_output_____"
]
],
[
[
"import json\n\nwith open('Processed_Data/Amazon_Reviews_Processed.json') as file:\n\n for json_data in file:\n saved_data = json.loads(json_data)\n\n vocab2idx = saved_data[\"vocab\"]\n embd = saved_data[\"embd\"]\n train_batches_text = saved_data[\"train_batches_text\"]\n test_batches_text = saved_data[\"test_batches_text\"]\n val_batches_text = saved_data[\"val_batches_text\"]\n train_batches_summary = saved_data[\"train_batches_summary\"]\n test_batches_summary = saved_data[\"test_batches_summary\"]\n val_batches_summary = saved_data[\"val_batches_summary\"]\n train_batches_true_text_len = saved_data[\"train_batches_true_text_len\"]\n val_batches_true_text_len = saved_data[\"val_batches_true_text_len\"]\n test_batches_true_text_len = saved_data[\"test_batches_true_text_len\"]\n train_batches_true_summary_len = saved_data[\"train_batches_true_summary_len\"]\n val_batches_true_summary_len = saved_data[\"val_batches_true_summary_len\"]\n test_batches_true_summary_len = saved_data[\"test_batches_true_summary_len\"]\n\n break\n \nidx2vocab = {v:k for k,v in vocab2idx.items()}",
"_____no_output_____"
]
],
[
[
"## Hyperparameters",
"_____no_output_____"
]
],
[
[
"hidden_size = 300\nlearning_rate = 0.001\nepochs = 5\nmax_summary_len = 16 # should be summary_max_len as used in data_preprocessing with +1 (+1 for <EOS>) \nD = 5 # D determines local attention window size\nwindow_len = 2*D+1\nl2=1e-6",
"_____no_output_____"
]
],
[
[
"## Tensorflow Placeholders",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf \n\nembd_dim = len(embd[0])\n\ntf_text = tf.placeholder(tf.int32, [None, None])\ntf_embd = tf.placeholder(tf.float32, [len(vocab2idx),embd_dim])\ntf_true_summary_len = tf.placeholder(tf.int32, [None])\ntf_summary = tf.placeholder(tf.int32,[None, None])\ntf_train = tf.placeholder(tf.bool)",
"_____no_output_____"
]
],
[
[
"## Embed vectorized text\n\nDropout used for regularization \n(https://www.cs.toronto.edu/~hinton/absps/JMLRdropout.pdf)",
"_____no_output_____"
]
],
[
[
"embd_text = tf.nn.embedding_lookup(tf_embd, tf_text)\nembd_text = tf.layers.dropout(embd_text,rate=0.3,training=tf_train)",
"_____no_output_____"
]
],
[
[
"## LSTM function\n\nMore info: \n<br>\nhttps://dl.acm.org/citation.cfm?id=1246450, \n<br>\nhttps://www.bioinf.jku.at/publications/older/2604.pdf,\n<br>\nhttps://en.wikipedia.org/wiki/Long_short-term_memory",
"_____no_output_____"
]
],
[
[
"def LSTM(x,hidden_state,cell,input_dim,hidden_size,scope):\n \n with tf.variable_scope(scope,reuse=tf.AUTO_REUSE):\n \n w = tf.get_variable(\"w\", shape=[4,input_dim,hidden_size],\n dtype=tf.float32,\n trainable=True,\n initializer=tf.glorot_uniform_initializer())\n \n u = tf.get_variable(\"u\", shape=[4,hidden_size,hidden_size],\n dtype=tf.float32,\n trainable=True,\n initializer=tf.glorot_uniform_initializer())\n \n b = tf.get_variable(\"bias\", shape=[4,1,hidden_size],\n dtype=tf.float32,\n trainable=True,\n initializer=tf.zeros_initializer())\n \n input_gate = tf.nn.sigmoid( tf.matmul(x,w[0]) + tf.matmul(hidden_state,u[0]) + b[0])\n forget_gate = tf.nn.sigmoid( tf.matmul(x,w[1]) + tf.matmul(hidden_state,u[1]) + b[1])\n output_gate = tf.nn.sigmoid( tf.matmul(x,w[2]) + tf.matmul(hidden_state,u[2]) + b[2])\n cell_ = tf.nn.tanh( tf.matmul(x,w[3]) + tf.matmul(hidden_state,u[3]) + b[3])\n cell = forget_gate*cell + input_gate*cell_\n hidden_state = output_gate*tf.tanh(cell)\n \n return hidden_state, cell\n ",
"_____no_output_____"
]
],
[
[
"## Bi-Directional LSTM Encoder\n\n(https://maxwell.ict.griffith.edu.au/spl/publications/papers/ieeesp97_schuster.pdf)\n\nMore Info: https://machinelearningmastery.com/develop-bidirectional-lstm-sequence-classification-python-keras/\n\nBi-directional LSTM encoder has a forward encoder and a backward encoder. The forward encoder encodes a text sequence from start to end, and the backward encoder encodes the text sequence from end to start.\nThe final output is a combination (in this case, a concatenation) of the forward encoded text and the backward encoded text\n \n",
"_____no_output_____"
],
[
"## Forward Encoding",
"_____no_output_____"
]
],
[
[
"S = tf.shape(embd_text)[1] #text sequence length\nN = tf.shape(embd_text)[0] #batch_size\n\ni=0\nhidden=tf.zeros([N, hidden_size], dtype=tf.float32)\ncell=tf.zeros([N, hidden_size], dtype=tf.float32)\nhidden_forward=tf.TensorArray(size=S, dtype=tf.float32)\n\n#shape of embd_text: [N,S,embd_dim]\nembd_text_t = tf.transpose(embd_text,[1,0,2]) \n#current shape of embd_text: [S,N,embd_dim]\n\ndef cond(i, hidden, cell, hidden_forward):\n return i < S\n\ndef body(i, hidden, cell, hidden_forward):\n x = embd_text_t[i]\n \n hidden,cell = LSTM(x,hidden,cell,embd_dim,hidden_size,scope=\"forward_encoder\")\n hidden_forward = hidden_forward.write(i, hidden)\n\n return i+1, hidden, cell, hidden_forward\n\n_, _, _, hidden_forward = tf.while_loop(cond, body, [i, hidden, cell, hidden_forward])",
"_____no_output_____"
]
],
[
[
"## Backward Encoding",
"_____no_output_____"
]
],
[
[
"i=S-1\nhidden=tf.zeros([N, hidden_size], dtype=tf.float32)\ncell=tf.zeros([N, hidden_size], dtype=tf.float32)\nhidden_backward=tf.TensorArray(size=S, dtype=tf.float32)\n\ndef cond(i, hidden, cell, hidden_backward):\n return i >= 0\n\ndef body(i, hidden, cell, hidden_backward):\n x = embd_text_t[i]\n hidden,cell = LSTM(x,hidden,cell,embd_dim,hidden_size,scope=\"backward_encoder\")\n hidden_backward = hidden_backward.write(i, hidden)\n\n return i-1, hidden, cell, hidden_backward\n\n_, _, _, hidden_backward = tf.while_loop(cond, body, [i, hidden, cell, hidden_backward])",
"_____no_output_____"
]
],
[
[
"## Merge Forward and Backward Encoder Hidden States",
"_____no_output_____"
]
],
[
[
"hidden_forward = hidden_forward.stack()\nhidden_backward = hidden_backward.stack()\n\nencoder_states = tf.concat([hidden_forward,hidden_backward],axis=-1)\nencoder_states = tf.transpose(encoder_states,[1,0,2])\n\nencoder_states = tf.layers.dropout(encoder_states,rate=0.3,training=tf_train)\n\nfinal_encoded_state = tf.layers.dropout(tf.concat([hidden_forward[-1],hidden_backward[-1]],axis=-1),rate=0.3,training=tf_train)\n",
"_____no_output_____"
]
],
[
[
"## Implementation of attention scoring function\n\nGiven a sequence of encoder states ($H_s$) and the decoder hidden state ($H_t$) of current timestep $t$, the equation for computing attention score is:\n\n$$Score = (H_s.W_a).H_t^T $$\n\n($W_a$ = trainable parameters)\n\n(https://nlp.stanford.edu/pubs/emnlp15_attn.pdf)",
"_____no_output_____"
]
],
[
[
"def attention_score(encoder_states,decoder_hidden_state,scope=\"attention_score\"):\n \n with tf.variable_scope(scope,reuse=tf.AUTO_REUSE):\n Wa = tf.get_variable(\"Wa\", shape=[2*hidden_size,2*hidden_size],\n dtype=tf.float32,\n trainable=True,\n initializer=tf.glorot_uniform_initializer())\n \n encoder_states = tf.reshape(encoder_states,[N*S,2*hidden_size])\n \n encoder_states = tf.reshape(tf.matmul(encoder_states,Wa),[N,S,2*hidden_size])\n decoder_hidden_state = tf.reshape(decoder_hidden_state,[N,2*hidden_size,1])\n \n return tf.reshape(tf.matmul(encoder_states,decoder_hidden_state),[N,S])\n",
"_____no_output_____"
]
],
[
[
"## Local Attention Function\n\nBased on: https://nlp.stanford.edu/pubs/emnlp15_attn.pdf",
"_____no_output_____"
]
],
[
[
"\ndef align(encoder_states, decoder_hidden_state,scope=\"attention\"):\n \n with tf.variable_scope(scope,reuse=tf.AUTO_REUSE):\n Wp = tf.get_variable(\"Wp\", shape=[2*hidden_size,125],\n dtype=tf.float32,\n trainable=True,\n initializer=tf.glorot_uniform_initializer())\n \n Vp = tf.get_variable(\"Vp\", shape=[125,1],\n dtype=tf.float32,\n trainable=True,\n initializer=tf.glorot_uniform_initializer())\n \n positions = tf.cast(S-window_len,dtype=tf.float32) # Maximum valid attention window starting position\n \n # Predict attention window starting position \n ps = positions*tf.nn.sigmoid(tf.matmul(tf.tanh(tf.matmul(decoder_hidden_state,Wp)),Vp))\n # ps = (soft-)predicted starting position of attention window\n pt = ps+D # pt = center of attention window where the whole window length is 2*D+1\n pt = tf.reshape(pt,[N])\n \n i = 0\n gaussian_position_based_scores = tf.TensorArray(size=S,dtype=tf.float32)\n sigma = tf.constant(D/2,dtype=tf.float32)\n \n def cond(i,gaussian_position_based_scores):\n \n return i < S\n \n def body(i,gaussian_position_based_scores):\n \n score = tf.exp(-((tf.square(tf.cast(i,tf.float32)-pt))/(2*tf.square(sigma)))) \n # (equation (10) in https://nlp.stanford.edu/pubs/emnlp15_attn.pdf)\n gaussian_position_based_scores = gaussian_position_based_scores.write(i,score)\n \n return i+1,gaussian_position_based_scores\n \n i,gaussian_position_based_scores = tf.while_loop(cond,body,[i,gaussian_position_based_scores])\n \n gaussian_position_based_scores = gaussian_position_based_scores.stack()\n gaussian_position_based_scores = tf.transpose(gaussian_position_based_scores,[1,0])\n gaussian_position_based_scores = tf.reshape(gaussian_position_based_scores,[N,S])\n \n scores = attention_score(encoder_states,decoder_hidden_state)*gaussian_position_based_scores\n scores = tf.nn.softmax(scores,axis=-1)\n \n return tf.reshape(scores,[N,S,1])",
"_____no_output_____"
]
],
[
[
"## LSTM Decoder With Local Attention",
"_____no_output_____"
]
],
[
[
"with tf.variable_scope(\"decoder\",reuse=tf.AUTO_REUSE):\n SOS = tf.get_variable(\"sos\", shape=[1,embd_dim],\n dtype=tf.float32,\n trainable=True,\n initializer=tf.glorot_uniform_initializer())\n \n # SOS represents starting marker \n # It tells the decoder that it is about to decode the first word of the output\n # I have set SOS as a trainable parameter\n \n Wc = tf.get_variable(\"Wc\", shape=[4*hidden_size,embd_dim],\n dtype=tf.float32,\n trainable=True,\n initializer=tf.glorot_uniform_initializer())\n \n\n\nSOS = tf.tile(SOS,[N,1]) #now SOS shape: [N,embd_dim]\ninp = SOS\nhidden=final_encoded_state\ncell=tf.zeros([N, 2*hidden_size], dtype=tf.float32)\ndecoder_outputs=tf.TensorArray(size=max_summary_len, dtype=tf.float32)\noutputs=tf.TensorArray(size=max_summary_len, dtype=tf.int32)\n\nfor i in range(max_summary_len):\n \n inp = tf.layers.dropout(inp,rate=0.3,training=tf_train)\n \n attention_scores = align(encoder_states,hidden)\n encoder_context_vector = tf.reduce_sum(encoder_states*attention_scores,axis=1)\n \n hidden,cell = LSTM(inp,hidden,cell,embd_dim,2*hidden_size,scope=\"decoder\")\n \n hidden_ = tf.layers.dropout(hidden,rate=0.3,training=tf_train)\n \n concated = tf.concat([hidden_,encoder_context_vector],axis=-1)\n \n linear_out = tf.nn.tanh(tf.matmul(concated,Wc))\n decoder_output = tf.matmul(linear_out,tf.transpose(tf_embd,[1,0])) \n # produce unnormalized probability distribution over vocabulary\n \n \n decoder_outputs = decoder_outputs.write(i,decoder_output)\n \n # Pick out most probable vocab indices based on the unnormalized probability distribution\n \n next_word_vec = tf.cast(tf.argmax(decoder_output,1),tf.int32)\n\n next_word_vec = tf.reshape(next_word_vec, [N])\n\n outputs = outputs.write(i,next_word_vec)\n\n next_word = tf.nn.embedding_lookup(tf_embd, next_word_vec)\n inp = tf.reshape(next_word, [N, embd_dim])\n \n \ndecoder_outputs = decoder_outputs.stack()\noutputs = outputs.stack()\n\ndecoder_outputs = tf.transpose(decoder_outputs,[1,0,2])\noutputs = tf.transpose(outputs,[1,0])\n\n \n ",
"_____no_output_____"
]
],
[
[
"## Define Cross Entropy Cost Function and L2 Regularization",
"_____no_output_____"
]
],
[
[
"filtered_trainables = [var for var in tf.trainable_variables() if\n not(\"Bias\" in var.name or \"bias\" in var.name\n or \"noreg\" in var.name)]\n\nregularization = tf.reduce_sum([tf.nn.l2_loss(var) for var\n in filtered_trainables])\n\nwith tf.variable_scope(\"loss\"):\n\n epsilon = tf.constant(1e-9, tf.float32)\n\n cross_entropy = tf.nn.sparse_softmax_cross_entropy_with_logits(\n labels=tf_summary, logits=decoder_outputs)\n\n pad_mask = tf.sequence_mask(tf_true_summary_len,\n maxlen=max_summary_len,\n dtype=tf.float32)\n\n masked_cross_entropy = cross_entropy*pad_mask\n\n cost = tf.reduce_mean(masked_cross_entropy) + \\\n l2*regularization\n\n cross_entropy = tf.reduce_mean(masked_cross_entropy)",
"_____no_output_____"
]
],
[
[
"## Accuracy",
"_____no_output_____"
]
],
[
[
"# Comparing predicted sequence with labels\ncomparison = tf.cast(tf.equal(outputs, tf_summary),\n tf.float32)\n\n# Masking to ignore the effect of pads while calculating accuracy\npad_mask = tf.sequence_mask(tf_true_summary_len,\n maxlen=max_summary_len,\n dtype=tf.bool)\n\nmasked_comparison = tf.boolean_mask(comparison, pad_mask)\n\n# Accuracy\naccuracy = tf.reduce_mean(masked_comparison)",
"_____no_output_____"
]
],
[
[
"## Define Optimizer",
"_____no_output_____"
]
],
[
[
"all_vars = tf.get_collection(tf.GraphKeys.TRAINABLE_VARIABLES)\n\noptimizer = tf.contrib.opt.NadamOptimizer(\n learning_rate=learning_rate)\n\ngvs = optimizer.compute_gradients(cost, var_list=all_vars)\n\ncapped_gvs = [(tf.clip_by_norm(grad, 5), var) for grad, var in gvs] # Gradient Clipping\n\ntrain_op = optimizer.apply_gradients(capped_gvs)",
"_____no_output_____"
]
],
[
[
"## Training and Validation",
"_____no_output_____"
]
],
[
[
"import pickle\nimport random\n\nwith tf.Session() as sess: # Start Tensorflow Session\n display_step = 100\n patience = 5\n\n load = input(\"\\nLoad checkpoint? y/n: \")\n print(\"\")\n saver = tf.train.Saver()\n\n if load.lower() == 'y':\n\n print('Loading pre-trained weights for the model...')\n\n saver.restore(sess, 'Model_Backup/Seq2seq_summarization.ckpt')\n sess.run(tf.global_variables())\n sess.run(tf.tables_initializer())\n\n with open('Model_Backup/Seq2seq_summarization.pkl', 'rb') as fp:\n train_data = pickle.load(fp)\n\n covered_epochs = train_data['covered_epochs']\n best_loss = train_data['best_loss']\n impatience = 0\n \n print('\\nRESTORATION COMPLETE\\n')\n\n else:\n best_loss = 2**30\n impatience = 0\n covered_epochs = 0\n\n init = tf.global_variables_initializer()\n sess.run(init)\n sess.run(tf.tables_initializer())\n\n epoch=0\n while (epoch+covered_epochs)<epochs:\n \n print(\"\\n\\nSTARTING TRAINING\\n\\n\")\n \n batches_indices = [i for i in range(0, len(train_batches_text))]\n random.shuffle(batches_indices)\n\n total_train_acc = 0\n total_train_loss = 0\n\n for i in range(0, len(train_batches_text)):\n \n j = int(batches_indices[i])\n\n cost,prediction,\\\n acc, _ = sess.run([cross_entropy,\n outputs,\n accuracy,\n train_op],\n feed_dict={tf_text: train_batches_text[j],\n tf_embd: embd,\n tf_summary: train_batches_summary[j],\n tf_true_summary_len: train_batches_true_summary_len[j],\n tf_train: True})\n \n total_train_acc += acc\n total_train_loss += cost\n\n if i % display_step == 0:\n print(\"Iter \"+str(i)+\", Cost= \" +\n \"{:.3f}\".format(cost)+\", Acc = \" +\n \"{:.2f}%\".format(acc*100))\n \n if i % 500 == 0:\n \n idx = random.randint(0,len(train_batches_text[j])-1)\n \n \n \n text = \" \".join([idx2vocab.get(vec,\"<UNK>\") for vec in train_batches_text[j][idx]])\n predicted_summary = [idx2vocab.get(vec,\"<UNK>\") for vec in prediction[idx]]\n actual_summary = [idx2vocab.get(vec,\"<UNK>\") for vec in train_batches_summary[j][idx]]\n \n print(\"\\nSample Text\\n\")\n print(text)\n print(\"\\nSample Predicted Summary\\n\")\n for word in predicted_summary:\n if word == '<EOS>':\n break\n else:\n print(word,end=\" \")\n print(\"\\n\\nSample Actual Summary\\n\")\n for word in actual_summary:\n if word == '<EOS>':\n break\n else:\n print(word,end=\" \")\n print(\"\\n\\n\")\n \n print(\"\\n\\nSTARTING VALIDATION\\n\\n\")\n \n total_val_loss=0\n total_val_acc=0\n \n for i in range(0, len(val_batches_text)):\n \n if i%100==0:\n print(\"Validating data # {}\".format(i))\n\n cost, prediction,\\\n acc = sess.run([cross_entropy,\n outputs,\n accuracy],\n feed_dict={tf_text: val_batches_text[i],\n tf_embd: embd,\n tf_summary: val_batches_summary[i],\n tf_true_summary_len: val_batches_true_summary_len[i],\n tf_train: False})\n \n total_val_loss += cost\n total_val_acc += acc\n \n avg_val_loss = total_val_loss/len(val_batches_text)\n \n print(\"\\n\\nEpoch: {}\\n\\n\".format(epoch+covered_epochs))\n print(\"Average Training Loss: {:.3f}\".format(total_train_loss/len(train_batches_text)))\n print(\"Average Training Accuracy: {:.2f}\".format(100*total_train_acc/len(train_batches_text)))\n print(\"Average Validation Loss: {:.3f}\".format(avg_val_loss))\n print(\"Average Validation Accuracy: {:.2f}\".format(100*total_val_acc/len(val_batches_text)))\n \n if (avg_val_loss < best_loss):\n best_loss = avg_val_loss\n save_data={'best_loss':best_loss,'covered_epochs':covered_epochs+epoch+1}\n impatience=0\n with open('Model_Backup/Seq2seq_summarization.pkl', 'wb') as fp:\n pickle.dump(save_data, fp)\n saver.save(sess, 'Model_Backup/Seq2seq_summarization.ckpt')\n print(\"\\nModel saved\\n\")\n \n else:\n impatience+=1\n \n if impatience > patience:\n break\n \n \n epoch+=1\n ",
"\nLoad checkpoint? y/n: n\n\n\n\nSTARTING TRAINING\n\n\nIter 0, Cost= 2.086, Acc = 0.00%\n\nSample Text\n\ni 'm not a big pretzel eater , but i love this little <UNK> nibblers . i like the low fat snack and how it fills you up .\n\nSample Predicted Summary\n\nmunicipality jackass municipality mongolian seats han han mongolian hah sus sus wat hah casbah dynasty province \n\nSample Actual Summary\n\ngreat pretzels \n\n\nIter 100, Cost= 0.985, Acc = 35.58%\nIter 200, Cost= 0.914, Acc = 33.33%\nIter 300, Cost= 0.928, Acc = 36.11%\nIter 400, Cost= 0.943, Acc = 35.19%\nIter 500, Cost= 0.676, Acc = 42.71%\n\nSample Text\n\nwe <UNK> this one , but the flavor could have been a tad stronger . very yummy tho , we will totally purchase again !\n\nSample Predicted Summary\n\ndelicious ! \n\nSample Actual Summary\n\nvery good ! \n\n\nIter 600, Cost= 0.878, Acc = 35.24%\nIter 700, Cost= 0.949, Acc = 33.04%\nIter 800, Cost= 1.074, Acc = 34.65%\nIter 900, Cost= 0.831, Acc = 44.21%\nIter 1000, Cost= 0.911, Acc = 36.36%\n\nSample Text\n\ntried this hoping for something better than the thick salsa that everyone else makes and it was great ! after making our own it gets time consuming so this is a good alternative .\n\nSample Predicted Summary\n\ngreat \n\nSample Actual Summary\n\ngreat salsa \n\n\nIter 1100, Cost= 1.081, Acc = 23.33%\nIter 1200, Cost= 1.018, Acc = 32.73%\nIter 1300, Cost= 0.902, Acc = 35.87%\nIter 1400, Cost= 0.946, Acc = 31.07%\nIter 1500, Cost= 0.798, Acc = 42.31%\n\nSample Text\n\ni had a coupon for this so it was a good value . otherwise it is to expense for what you get . my box had a couple of opened cereals in it so i did n't get the full value of all ...\n\nSample Predicted Summary\n\ngood \n\nSample Actual Summary\n\ngood value \n\n\nIter 1600, Cost= 0.871, Acc = 33.33%\nIter 1700, Cost= 0.943, Acc = 40.00%\nIter 1800, Cost= 0.876, Acc = 40.20%\nIter 1900, Cost= 0.973, Acc = 37.25%\nIter 2000, Cost= 0.978, Acc = 29.73%\n\nSample Text\n\nmy 4 dogs all had allergies and are just fine now that i switched to <UNK> the <UNK> one smell abit but <UNK> they still love it <UNK> the dried <UNK> canned r terrific <UNK> nooo grani !\n\nSample Predicted Summary\n\n<UNK> ! \n\nSample Actual Summary\n\ngreat food \n\n\nIter 2100, Cost= 0.907, Acc = 37.04%\nIter 2200, Cost= 0.928, Acc = 34.31%\nIter 2300, Cost= 0.906, Acc = 31.25%\nIter 2400, Cost= 0.903, Acc = 37.00%\nIter 2500, Cost= 0.811, Acc = 33.01%\n\nSample Text\n\nthe chocolate was a little crumbly , but the taste is very good . my hubby has <UNK> , and it is gluten free , so it is an excellent bar to stock in the pantry for whenever he does n't have time for breakfast .\n\nSample Predicted Summary\n\ngreat \n\nSample Actual Summary\n\nyum \n\n\nIter 2600, Cost= 0.839, Acc = 34.62%\nIter 2700, Cost= 0.927, Acc = 37.07%\nIter 2800, Cost= 0.853, Acc = 36.73%\nIter 2900, Cost= 0.805, Acc = 40.00%\nIter 3000, Cost= 0.855, Acc = 35.51%\n\nSample Text\n\ntea came packaged as expected , delivered quickly and with stash you can not go wrong . individually wrapped and stays fresh and very flavorful . highly recommended for the earl gray tea lover .\n\nSample Predicted Summary\n\ndelicious tea \n\nSample Actual Summary\n\ngreat tea \n\n\nIter 3100, Cost= 0.854, Acc = 36.63%\n\n\nSTARTING VALIDATION\n\n\nValidating data # 0\nValidating data # 100\nValidating data # 200\nValidating data # 300\n\n\nEpoch: 0\n\n\nAverage Training Loss: 0.907\nAverage Training Accuracy: 35.42\nAverage Validation Loss: 0.865\nAverage Validation Accuracy: 36.65\n\nModel saved\n\n\n\nSTARTING TRAINING\n\n\nIter 0, Cost= 0.808, Acc = 34.34%\n\nSample Text\n\nquaker oatmeal squares has been our family favorite for a couple of years now . ca n't get enough of it . just the right sweetness and crunch .\n\nSample Predicted Summary\n\ngreat \n\nSample Actual Summary\n\nfavorite cereal \n\n\nIter 100, Cost= 1.036, Acc = 34.26%\nIter 200, Cost= 0.934, Acc = 33.03%\nIter 300, Cost= 0.972, Acc = 35.85%\nIter 400, Cost= 0.926, Acc = 32.35%\nIter 500, Cost= 0.738, Acc = 41.05%\n\nSample Text\n\ngreat taste , nice smell , great <UNK> < br / > if you mix it with fresh ment you will get fantastic <UNK> < br / > i will buy it again .\n\nSample Predicted Summary\n\ngreat \n\nSample Actual Summary\n\nthe best \n\n\nIter 600, Cost= 0.858, Acc = 41.24%\nIter 700, Cost= 0.905, Acc = 36.45%\nIter 800, Cost= 0.795, Acc = 35.05%\nIter 900, Cost= 0.806, Acc = 37.50%\nIter 1000, Cost= 0.795, Acc = 35.64%\n\nSample Text\n\ni bought about 5 different kinds of <UNK> when i first got my coffee maker , which i love by the way , and i 'd have to say that this was my favorite one out of them all . it has the perfect balance of everything , i was really surprised .\n\nSample Predicted Summary\n\ngreat \n\nSample Actual Summary\n\nexcellent stuff \n\n\nIter 1100, Cost= 0.825, Acc = 39.42%\nIter 1200, Cost= 0.743, Acc = 38.78%\nIter 1300, Cost= 0.813, Acc = 41.84%\nIter 1400, Cost= 0.933, Acc = 29.66%\nIter 1500, Cost= 0.978, Acc = 33.61%\n\nSample Text\n\ni really wanted to like this , as it was organic , and came in a glass bottle , but there was hardly any flavor at all . i could barely smell it , and even when i poured a generous amount on my dish , it imparts little to no truffle <UNK> . my truffle salt is much more potent .\n\nSample Predicted Summary\n\ngood \n\nSample Actual Summary\n\nweak \n\n\nIter 1600, Cost= 0.778, Acc = 45.10%\nIter 1700, Cost= 0.855, Acc = 38.83%\nIter 1800, Cost= 0.815, Acc = 41.58%\nIter 1900, Cost= 0.853, Acc = 37.62%\nIter 2000, Cost= 1.003, Acc = 32.74%\n\nSample Text\n\ni love milk chocolate and do n't like dark <UNK> . my husband is the opposite , so i always buy him the dark stuff and it 's safe for him , haha ! until i happened to try this one . it 's awesome !\n\nSample Predicted Summary\n\n<UNK> ! \n\nSample Actual Summary\n\nit 's good ! ! \n\n\nIter 2100, Cost= 0.817, Acc = 37.74%\nIter 2200, Cost= 0.977, Acc = 33.33%\nIter 2300, Cost= 0.840, Acc = 35.96%\nIter 2400, Cost= 0.749, Acc = 31.58%\nIter 2500, Cost= 0.885, Acc = 31.73%\n\nSample Text\n\nthe best thing about this coffee is the sweet smell , just like a blueberry muffin . the taste is good , not as sweet as i was expecting but it was good nonetheless . its a nice treat when you 're craving something sweet but it wo n't replace my morning donut shop coffee : )\n\nSample Predicted Summary\n\ndelicious \n\nSample Actual Summary\n\nsmells yummy : ) \n\n\nIter 2600, Cost= 0.887, Acc = 32.73%\nIter 2700, Cost= 0.780, Acc = 44.94%\nIter 2800, Cost= 0.899, Acc = 35.71%\nIter 2900, Cost= 0.797, Acc = 38.24%\nIter 3000, Cost= 1.061, Acc = 33.33%\n\nSample Text\n\nthis tea is wonderful , one bag will make three cups for most people . i like my tea very strong so these were perfect . i bet they will be good for making a good ice tea .\n\nSample Predicted Summary\n\ngreat tea \n\nSample Actual Summary\n\none bag 3 cups \n\n\nIter 3100, Cost= 0.769, Acc = 37.86%\n\n\nSTARTING VALIDATION\n\n\nValidating data # 0\nValidating data # 100\nValidating data # 200\nValidating data # 300\n\n\nEpoch: 1\n\n\nAverage Training Loss: 0.863\nAverage Training Accuracy: 36.40\nAverage Validation Loss: 0.837\nAverage Validation Accuracy: 37.30\n\nModel saved\n\n\n\nSTARTING TRAINING\n\n\nIter 0, Cost= 0.959, Acc = 35.85%\n\nSample Text\n\nreally good bars . you could cut this baby in 1/2 and have 2 snacks out of it ! i bought 1 at the store first to see if i liked them and paid lots more for it . i do n't eat alot of meat so this caught my eye . i now have them on auto delivery ! !\n\nSample Predicted Summary\n\ngreat ! \n\nSample Actual Summary\n\ngreat bars ! \n\n\nIter 100, Cost= 0.792, Acc = 33.33%\nIter 200, Cost= 0.781, Acc = 35.29%\nIter 300, Cost= 0.825, Acc = 40.74%\nIter 400, Cost= 0.793, Acc = 40.19%\nIter 500, Cost= 0.860, Acc = 31.07%\n\nSample Text\n\ni always buy my coffee from amazon as the prices are cheaper and i love all the coffee . best price on line .\n\nSample Predicted Summary\n\ngreat coffee \n\nSample Actual Summary\n\npeggy \n\n\nIter 600, Cost= 0.990, Acc = 28.57%\nIter 700, Cost= 0.736, Acc = 41.41%\nIter 800, Cost= 0.826, Acc = 33.68%\nIter 900, Cost= 0.904, Acc = 35.24%\nIter 1000, Cost= 0.858, Acc = 35.71%\n\nSample Text\n\ni am very pleased with this product and the company sent it on a timely basis , well packed to prevent breakage .\n\nSample Predicted Summary\n\ngreat \n\nSample Actual Summary\n\ngood stuff \n\n\nIter 1100, Cost= 0.999, Acc = 30.36%\nIter 1200, Cost= 0.726, Acc = 44.79%\nIter 1300, Cost= 0.798, Acc = 36.73%\n"
]
],
[
[
"### Future Works\n\n* Beam Search\n* Pointer Mechanisms\n* BLEU\\ROUGE evaluation\n* Implement Testing\n* Complete Training and Optimize Hyperparameters",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0d1232fe68f67953ee49290673a0bcf62cdad53 | 19,734 | ipynb | Jupyter Notebook | (Example_1) BasicUsageAndParams.ipynb | VitaliyPavlyukov/AutoMLWhitebox | 4acd55624490707a7fbf036631533e29123bb1bd | [
"Apache-2.0"
] | null | null | null | (Example_1) BasicUsageAndParams.ipynb | VitaliyPavlyukov/AutoMLWhitebox | 4acd55624490707a7fbf036631533e29123bb1bd | [
"Apache-2.0"
] | null | null | null | (Example_1) BasicUsageAndParams.ipynb | VitaliyPavlyukov/AutoMLWhitebox | 4acd55624490707a7fbf036631533e29123bb1bd | [
"Apache-2.0"
] | null | null | null | 40.273469 | 653 | 0.600537 | [
[
[
"import pandas as pd\nimport numpy as np\nimport lightgbm as lgb\n\nfrom collections import OrderedDict\nfrom sklearn.metrics import roc_auc_score\nfrom tqdm import tqdm\nfrom copy import deepcopy\n\nfrom autowoe import ReportDeco, AutoWoE",
"_____no_output_____"
]
],
[
[
"### Чтение и подготовка обучающей выборки",
"_____no_output_____"
]
],
[
[
"train = pd.read_csv(\"./example_data/train_demo.csv\",\n low_memory=False,\n index_col=\"line_id\",\n parse_dates = [\"datetime_\" + str(i) for i in range(2)],)\n\ntrain = train.iloc[:, 50:100]\n\nnum_col = list(filter(lambda x: \"numb\" in x, train.columns))\nnum_feature_type = {x: \"real\" for x in num_col}\n\ndate_col = filter(lambda x: \"datetime\" in x, train.columns)\nfor col in date_col:\n train[col + \"_year\"] = train[col].map(lambda x: x.year)\n train[col + \"_weekday\"] = train[col].map(lambda x: x.weekday())\n train[col + \"_month\"] = train[col].map(lambda x: x.month)",
"_____no_output_____"
]
],
[
[
"### Чтение и подготовка тестовой выборки",
"_____no_output_____"
]
],
[
[
"test = pd.read_csv(\"./example_data/test_demo.csv\",\n index_col=\"line_id\", \n parse_dates = [\"datetime_\" + str(i) for i in range(2)])\n\ndate_col = filter(lambda x: \"datetime\" in x, test.columns)\nfor col in date_col:\n test[col + \"_year\"] = test[col].map(lambda x: x.year)\n test[col + \"_weekday\"] = test[col].map(lambda x: x.weekday())\n test[col + \"_month\"] = test[col].map(lambda x: x.month)\n \ntest_target = pd.read_csv(\"./example_data/test-target_demo.csv\")[\"target\"]\ntest[\"target\"] = test_target.values",
"_____no_output_____"
]
],
[
[
"### Параметры модели",
"_____no_output_____"
],
[
"Для обучения модели рекомендуется указать тип признаков для обучения.\nПоэтому создается словарь features_type с ключами: \n\n\n\"real\" -- вещественный признак,\n\n\"cat\" -- категориальный.\n\nДля признаков, которые не размечены, типы будут определены автоматом. Такой вариант будет работать, но качество порядочно просядет",
"_____no_output_____"
],
[
"#### features_type",
"_____no_output_____"
]
],
[
[
"cat_col = list(filter(lambda x: \"str\" in x, train.columns))\ncat_feature_type = {x: \"cat\" for x in cat_col}\n\nyear_col = list(filter(lambda x: \"_year\" in x, train.columns))\nyear_feature_type = {x: \"cat\" for x in year_col}\n\nweekday_col = list(filter(lambda x: \"_weekday\" in x, train.columns))\nweekday_feature_type = {x: \"cat\" for x in weekday_col}\n\nmonth_col = list(filter(lambda x: \"_month\" in x, train.columns))\nmonth_feature_type = {x: \"cat\" for x in month_col}",
"_____no_output_____"
],
[
"features = cat_col + year_col + weekday_col + month_col + num_col",
"_____no_output_____"
]
],
[
[
"#### Feature level constrains",
"_____no_output_____"
]
],
[
[
"features_type = dict(**num_feature_type,\n **cat_feature_type,\n **year_feature_type,\n **weekday_feature_type,\n **month_feature_type)",
"_____no_output_____"
]
],
[
[
"- `features_monotone_constraints` - также можно указать зависимость целевой переменной от признака. Если заранее известно, что при возрастании признака feature_1, то эту информацию можно учесть в модели, добавив в словарь пару {feature_1: \"1\"}. Если же зависимость признака от целевой переменной обратная, то можно указать {feature_1: \"-1\"} Если про зависимость ничего неизвестно, но хочется, чтобы она была монотонная, можно указать 'auto'. Можно указать {feature_1: \"0\"}, в случае, если установлено общее ограничение на монотонность, чтобы не распространять его на эту фичу. Если специальных условий нет, то можно не собирать этот дикт\n\n\nРекомендуемое использование:\n\n1) В случае, если задано общее условие на монотонность, то можно собрать дикт {feature_1: \"0\", feature_2: \"0\"}, чтобы игнорировать это ограничение для признаков feature_1, feature_2\n\n2) В случае, если не задано общее условие на монотонность, то можно собрать дикт {feature_1: \"auto\", feature_2: \"auto\"}, чтобы установить это ограничение для признаков feature_1, feature_2",
"_____no_output_____"
]
],
[
[
"features_monotone_constraints = {'number_74': 'auto', 'number_83': 'auto'} ",
"_____no_output_____"
]
],
[
[
"- `max_bin_count` - через словарь max_bin_count можно задать число бинов для WoE кодирования, если для какого-то признака оно отлично от общего. ",
"_____no_output_____"
]
],
[
[
"max_bin_count = {'number_47': 3, 'number_51': 2}",
"_____no_output_____"
]
],
[
[
"#### Рекомендация\nВ общем случае, в первый момент построения модели лучше не указывать специальных ограничений в features_monotone_constraints и max_bin_count. Если в результате анализа полученной модели разбиение оказалось неинтерпретируемым или нестабильным по отдельным признакам, но в целом по модели ок, то ограничить сложность разбиения отдельных призаков имеет смысл. Если разбивка большинства признаков в модели оказалась неудовлетворительная, то рекомендуется в первую очередь настраивать глобальные ограничения (см параметры модели max_bin_count, monotonic, min_bin_size и др ниже)",
"_____no_output_____"
],
[
"#### Общие параметры модели\n\n- `interpreted_model` - требуется ли интерпретируемость модели (условие на знак в коэффициентах логистической регрессии)\n\n- `monotonic` - Глобальное условие на монотонность. Если указано True, то для всех признаков по умолчанию будут строится только монотонные разбиения. Указать специальные условия для отдельных признаков можно используя features_monotone_constraints аргумент метода .fit\n\n- `max_bin_count` - Глобальное ограничение на число бинов. Указать специальные условия для отдельных признаков можно используя max_bin_count аргумент метода .fit\n\n- `select_type` - способ ПРЕДВАРИТЕЛЬНОГО!!! (ЭТО ВАЖНО) отбора признаков. Если указать None, то будут отобраны признаки, у которых importance больше imp_th. Если указвать, например 50, то после предварительного отобра останется только 50 признаков самых важных признаков. Крайне не рекомендуется сильно ограничивать\n\n- `pearson_th` - пороговое значение для корреляции Пирсона. Используется на финальной стадии отбора признаков.\nЕсли корреляция вух признаков по модулю больше pearson_th, то будет выброшен тот, у которого \nинформативность меньше\n\n- `auc_th` - пороговое значение для одномерной оценки качества признака\n\n- `vif_th` - пороговое значение для VIF признака\n\n- `imp_th` - порог по которому будет произведен отбор признаков, если указать select_type=None (см. ниже).\n\n- `th_const` порог по которому признак будет считаться константным. Все константные признаки в модели не учитываются. Если число валидных значений больше трешхолда, то колонка не константная (int). В случае указания float, трешхолд будет определяться как размер_выборки * th_const\n\n- `force_single_split` - иногда в силу ограничений на min_bin_size невозможно построить ни одной группировки на переменную. force_single_split=True заставит в этом случае построить единственно возмоджный сплит, в случае если при этом выделяется группа размера более чем th_const. False будет выкидывать этот признак\n\n\n- `th_nan` - порог по которому будет выделена отдельная категория для пропусков в данных.\nЕсли число пропусков меньше чем th_nan, то WoE значения для пропусков берется равным нулю.\nВ противном случае пропущенные значения будут выделены в отдельную группу и для них отдельно\nбудет рассчитано WoE значение.\nТак же влияет на редкие категории (менее th_cat). Если суммарно таких категорий будет менее th_nan, то обработка будет производиться по принципу отпределенному в `cat_merge_to`, иначе оценено по группе\n\n- `th_cat` - порог, по которой немногочисленные категории в категориальных признаках будут объединятся в отдельную группу\n\n\n- `woe_diff_th` - Возмодность смеджить наны и редкие категории с каким-то бином, если разница в вое менее woe_diff_th\n\n\n- `min_bin_size` - минимальный размер бина при группировке. Возможно int как число наблюдений и float как доля от выбрки\n\n- `min_bin_mults` - в ходе построения бинов будут протестированы возможные значения min_bin_size, \nmin_bin_size * min_bin_mults[0], min_bin_size * min_bin_mults[1] ... . Ждем float > 1. Дефолт - (2, 4), в принципе можно не трогать\n\n- `min_gains_to_split` - возможные значения регуляризатора, которые будут протестированы в ходе построения биннинга\n\n\n- `auc_tol` - Чувствительность к AUC. Считаем, что можем пожертвовать auc_tol качества от максимального, чтобы сделать модель проще\n\n\n- `cat_alpha` - Регуляризатор для кодировщика категорий\n\n\n\n- `cat_merge_to` - группа для редких (менее th_cat) категорий либо новых на тесте\n \"to_nan\" -- в группу nan, \n \"to_woe_0\" -- отдельная группа с WoE = 0,\n \"to_maxfreq\" - в самую большую группу,\n \"to_maxp\" - в группу с наибольшей вероятностью события,\n \"to_minp\" - в группу с наименьшей вероятностью события\n \n- `nan_merge_to` - группа для НаНов\n \"to_woe_0\" -- отдельная группа с WoE = 0,\n \"to_maxfreq\" - в самую большую группу,\n \"to_maxp\" - в группу с наибольшей вероятностью события,\n \"to_minp\" - в группу с наименьшей вероятностью события \n \n \n- `oof_woe` - если указать oof_woe=True, то WoE кодирование будет происходить по кросс-валидации. Если же False, то сразу на всей обучающей выборке.\n\n- `n_folds` - количество фолдов для внутренней кроссвалидации\n\n\n- `n_jobs` - число процессов, которое будет использовать модель \n\n- `l1_grid_size` - в данной модели на одном из шагов используется отбор признаков LASSO. l1_base_step -- размер сетки для перебора C\n\n- `l1_exp_scale` - шкала сетки для L1 отбора. 4 соответствует макс значению C порядка 3-4. Увеличивать, если необходимо сделать менее регуляризованную модель\n\n- `imp_type` - способ определения значимости признаков -- features importance (\"feature_imp\" - в общем случае более сложная модель) или permutation importance (\"perm_imp\" - в общем случае более простая модель)\n\n- `regularized_refit` - после отбора признаков полученная модель пересчитывается на всех данных. Стоит ли включать L1 при этом. Если нет, то в интерпретируемом режиме модель будет итеративно переобучаться, пока все веса не станут отрицательны. Если да - то аналогичное будет получаться закручиванием L1. Может быть полезно ставить False если нужна стат модель, те p-value на оценки\n\n- `p_val` - допустимый уровень p_value на оценки модели при условии обучении стат модели (regularized_refit=False)",
"_____no_output_____"
]
],
[
[
"auto_woe = AutoWoE(interpreted_model=True,\n monotonic=False,\n max_bin_count=5,\n select_type=None,\n pearson_th=0.9,\n auc_th=.505,\n vif_th=10.,\n imp_th=0,\n th_const=32,\n force_single_split=True,\n th_nan=0.01,\n th_cat=0.005,\n woe_diff_th=0.01,\n min_bin_size=0.01,\n min_bin_mults=(2, 4),\n min_gains_to_split=(0.0, 0.5, 1.0),\n auc_tol=1e-4,\n cat_alpha=100,\n cat_merge_to=\"to_woe_0\",\n nan_merge_to=\"to_woe_0\",\n oof_woe=True,\n n_folds=6,\n n_jobs=4,\n l1_grid_size=20,\n l1_exp_scale=6,\n imp_type=\"feature_imp\",\n regularized_refit=False,\n p_val=0.05,\n debug=False,\n verbose=0\n )\n\nauto_woe = ReportDeco(auto_woe)",
"_____no_output_____"
]
],
[
[
"- `train` обучающая выборка\n\n- `target_name` - название целевой переменной\n\n- `features_type` - см выше описание дикта features_type. Возможно указание None для автозаполнения, но не рекомендуется\n\n- `group_kf` - название колонки-группы для GroupKFold https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.GroupKFold.html\n\n- `max_bin_count` - см выше описание дикта max_bin_count. Можно ничего не передавать, если специальных условий не предусмотрено. Общее для всех условние задано в __init__\n\n- `features_monotone_constraints` - см выше описание дикта features_monotone_constraints. Можно ничего не передавать, если специальных условий не предусмотрено. Общее для всех условние задано в __init__\n\n- `validation` - возможность использовать валидацию в построении/отборе признаков. Можно не передавать. На текущий момент используется для 1) отбора признаков по p-value при построении стат модели\n",
"_____no_output_____"
]
],
[
[
"auto_woe.fit(train[features + ['target']], \n target_name=\"target\",\n features_type=features_type,\n group_kf=None,\n max_bin_count=max_bin_count,\n features_monotone_constraints=features_monotone_constraints,\n validation=test\n )",
"_____no_output_____"
],
[
"pred = auto_woe.predict_proba(test)\nroc_auc_score(test['target'], pred)",
"_____no_output_____"
],
[
"pred = auto_woe.predict_proba(test[['number_72']], report=False)\nroc_auc_score(test['target'], pred)\n",
"_____no_output_____"
],
[
"print(auto_woe.get_sql_inference_query('table'))",
"_____no_output_____"
]
],
[
[
"### Полезные методы модели",
"_____no_output_____"
],
[
"- `private_features_type` - типизация признаков\n- `get_woe` - рабиение на бины и WoE значения в них\n- `get_split` - границы разбиения. Особо полезен для категориальных признаков\n\n\n##### Замечание: \nReportDeco - обертка для построения отчета. Она не обязательна для обучения и применения модели, но обязательна для построения отчета (см последнюю ячейку).\nДля доступа к атрибутам самой модели необходимо обратится к атрибуту auto_woe.model декоратора\nВсе атрибуты объекта-модели так же доступны через объект-отчета.\nОднако в пикл отчета будет весить существенно больше, так что для сохранения модели на инференс стоит сохранять только auto_woe.model\n",
"_____no_output_____"
],
[
"### Формирование отчета",
"_____no_output_____"
]
],
[
[
"report_params = {\"automl_date_column\": \"report_month\", # колонка с датой в формате params['datetimeFormat']\n \"output_path\": \"./AUTOWOE_REPORT_1\", # папка, куда сгенерится отчет и сложатся нужные файлы\n \"report_name\": \"___НАЗВАНИЕ ОТЧЕТА___\",\n \"report_version_id\": 1,\n \"city\": \"Воронеж\",\n \"model_aim\": \"___ЦЕЛЬ ПОСТРОЕНИЯ МОДЕЛИ___\",\n \"model_name\": \"___НАЗВАНИЕ МОДЕЛИ___\",\n \"zakazchik\": \"___ЗАКАЗЧИК___\",\n \"high_level_department\": \"___ПОДРАЗДЕЛЕНИЕ___\",\n \"ds_name\": \"___РАЗРАБОТЧИК МОДЕЛИ___\",\n \"target_descr\": \"___ОПИСАНИЕ ЦЕЛЕВОГО СОБЫТИЯ___\",\n \"non_target_descr\": \"___ОПИСАНИЕ НЕЦЕЛЕВОГО СОБЫТИЯ___\"}\n\nauto_woe.generate_report(report_params)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
]
] |
d0d131aa694fcd30c550df1127f805e38d4407ac | 4,971 | ipynb | Jupyter Notebook | ItemBased_example.ipynb | denpo1022/movie-lens-dataset-analysis | bdd2ef0782907eaba2e006f6f7718b36ca1337f4 | [
"Apache-2.0"
] | null | null | null | ItemBased_example.ipynb | denpo1022/movie-lens-dataset-analysis | bdd2ef0782907eaba2e006f6f7718b36ca1337f4 | [
"Apache-2.0"
] | null | null | null | ItemBased_example.ipynb | denpo1022/movie-lens-dataset-analysis | bdd2ef0782907eaba2e006f6f7718b36ca1337f4 | [
"Apache-2.0"
] | null | null | null | 34.047945 | 145 | 0.533695 | [
[
[
"import numpy as np\nimport pandas as pd\nfrom sklearn.metrics import pairwise_distances\nfrom scipy.spatial.distance import cosine, correlation",
"_____no_output_____"
],
[
"#Loading movielens data\n\n#User's data\nusers_cols = ['user_id', 'age', 'sex', 'occupation', 'zip_code']\nusers = pd.read_csv('ml-100k\\\\u.user', sep='|', names=users_cols, parse_dates=True) \n#Ratings\nrating_cols = ['user_id', 'movie_id', 'rating', 'unix_timestamp']\nratings = pd.read_csv('ml-100k\\\\u.data', sep='\\t', names=rating_cols)\n#Movies\nmovie_cols = ['movie_id', 'title', 'release_date', 'video_release_date', 'imdb_url']\nmovies = pd.read_csv('ml-100k\\\\u.item', sep='|', names=movie_cols, usecols=range(5),encoding='latin-1')\n\n# Merging movie data with their ratings\nmovie_ratings = pd.merge(movies, ratings)\n\n# merging movie_ratings data with the User's dataframe\ndf = pd.merge(movie_ratings, users)\n\n# pre-processing\n# dropping colums that aren't needed\ndf.drop(df.columns[[3,4,7]], axis=1, inplace=True)\nratings.drop( \"unix_timestamp\", inplace = True, axis = 1 ) \nmovies.drop(movies.columns[[3,4]], inplace = True, axis = 1 )\n\n#Pivot Table(This creates a matrix of users and movie_ratings)\nratings_matrix = ratings.pivot_table(index=['movie_id'],columns=['user_id'],values='rating').reset_index(drop=True)\nratings_matrix.fillna( 0, inplace = True )\n\n#Cosine Similarity(Creates a cosine matrix of similaraties ..... which is the pairwise distances\n# between two items )\n\nmovie_similarity = 1 - pairwise_distances(ratings_matrix.values, metric=\"cosine\")\nnp.fill_diagonal(movie_similarity, 0) \nratings_matrix = pd.DataFrame(movie_similarity)",
"_____no_output_____"
],
[
"#Recommender\ntry:\n# user_inp=input('Enter the reference movie title based on which recommendations are to be made: ')\n user_inp=\"Speed (1994)\"\n inp=movies[movies['title']==user_inp].index.tolist()\n inp=inp[0]\n \n movies['similarity'] = ratings_matrix.iloc[inp]\n movies.columns = ['movie_id', 'title', 'release_date','similarity']\n movies.head(5)\n \nexcept:\n print(\"Sorry, the movie is not in the database!\")\n \nprint(\"Recommended movies based on your choice of \",user_inp ,\": \\n\", movies.sort_values( [\"similarity\"], ascending = False )[1:10])",
"Recommended movies based on your choice of Speed (1994) : \n movie_id title release_date \\\n384 385 True Lies (1994) 01-Jan-1994 \n160 161 Top Gun (1986) 01-Jan-1986 \n78 79 Fugitive, The (1993) 01-Jan-1993 \n95 96 Terminator 2: Judgment Day (1991) 01-Jan-1991 \n194 195 Terminator, The (1984) 01-Jan-1984 \n173 174 Raiders of the Lost Ark (1981) 01-Jan-1981 \n209 210 Indiana Jones and the Last Crusade (1989) 01-Jan-1989 \n549 550 Die Hard: With a Vengeance (1995) 01-Jan-1995 \n203 204 Back to the Future (1985) 01-Jan-1985 \n\n similarity \n384 0.719504 \n160 0.707567 \n78 0.696583 \n95 0.695556 \n194 0.676778 \n173 0.676664 \n209 0.674811 \n549 0.674143 \n203 0.665776 \n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
d0d144603d93886af3fb98a0c41fc4553634192d | 63,582 | ipynb | Jupyter Notebook | patient_responsibilities/discussion-analysis.ipynb | levon003/icwsm-cancer-journeys | f0b39f80380ace20912e989964475056be27ebc5 | [
"MIT"
] | null | null | null | patient_responsibilities/discussion-analysis.ipynb | levon003/icwsm-cancer-journeys | f0b39f80380ace20912e989964475056be27ebc5 | [
"MIT"
] | null | null | null | patient_responsibilities/discussion-analysis.ipynb | levon003/icwsm-cancer-journeys | f0b39f80380ace20912e989964475056be27ebc5 | [
"MIT"
] | null | null | null | 46.648569 | 1,009 | 0.391384 | [
[
[
"Discussion Analysis\n===\n\nNotebook for analysis of discussion done in Evidence and Reconsider tasks via the annotation web client.\n",
"_____no_output_____"
]
],
[
[
"import os\nimport re\nimport pandas as pd\nimport numpy as np\nimport sklearn\nimport sklearn.metrics\nfrom collections import Counter\nimport itertools\nimport sqlite3",
"_____no_output_____"
],
[
"import sys\nsys.path.append(\"../annotation_data\")",
"_____no_output_____"
],
[
"import responsibility as responsibility_utils\nfrom utils import get_webclient_url",
"_____no_output_____"
],
[
"annotation_web_client_database = \"/home/srivbane/shared/caringbridge/data/projects/qual-health-journeys/instance/cbAnnotator.sqlite\"\n\n\ndef get_annotation_db():\n db = sqlite3.connect(\n annotation_web_client_database,\n detect_types=sqlite3.PARSE_DECLTYPES\n )\n db.row_factory = sqlite3.Row\n return db",
"_____no_output_____"
],
[
"def get_discussion_entries(responsibility, phase, evidence_user, reconsider_user):\n try:\n db = get_annotation_db()\n cursor = db.execute(\n \"\"\"SELECT * FROM discussionEntry\n WHERE responsibility = ? AND phase = ? AND evidence_username = ? AND reconsider_username = ? \n GROUP BY site_id, journal_oid\n ORDER BY id DESC\"\"\",\n (responsibility, phase, evidence_user, reconsider_user)\n )\n results = cursor.fetchall()\n if results is None or len(results) == 0:\n return None\n \n data = []\n for result in results:\n site_id, journal_oid = result['site_id'], result['journal_oid']\n highlighted_text, additional_discussion = result['highlighted_text'], result['additional_discussion']\n is_annotation_changed = result['is_annotation_changed'] == 1\n if additional_discussion.startswith(\"You indicated that this post does not contain the responsibility.\"):\n if not is_annotation_changed:\n print(\"WARNING: Forcibly changed is_annotation_changed based on assumption of looping.\")\n is_annotation_changed = True\n data.append({\n \"phase\": phase,\n \"responsibility\": responsibility,\n \"site_id\": site_id,\n \"journal_oid\": journal_oid,\n \"highlighted_text\": highlighted_text,\n \"additional_discussion\": additional_discussion,\n \"is_annotation_changed\": is_annotation_changed,\n \"evidence_username\": evidence_user,\n \"reconsider_username\": reconsider_user\n })\n return data\n finally:\n db.close()",
"_____no_output_____"
]
],
[
[
"### Experiment metadata",
"_____no_output_____"
]
],
[
[
"responsibility_list = [\"coordinating_support\", \n \"symptom_management\", \n \"preparation\", \n \"managing_transitions\", \n \"info_filtering\", \n \"continued_monitoring\", \n \"clinical_decisions\"]\nuser1 = \"luoxx498\"\nuser2 = \"eriks074\"\nevidence_phase = \"evidence\"\nreconsider_phase = \"reconsider\"",
"_____no_output_____"
]
],
[
[
"### Load data",
"_____no_output_____"
]
],
[
[
"all_rows = []\nfor phase in [evidence_phase, reconsider_phase]:\n for users in [(user1, user2), (user2, user1)]:\n evidence_username, reconsider_username = users\n for responsibility in responsibility_list:\n new_rows = get_discussion_entries(responsibility, phase, evidence_username, reconsider_username)\n if new_rows is None:\n print(responsibility, phase, evidence_username, reconsider_username)\n continue\n all_rows += new_rows\nlen(all_rows)",
"managing_transitions reconsider luoxx498 eriks074\n"
],
[
"df = pd.DataFrame(all_rows)\ndf.head(n=4)",
"_____no_output_____"
],
[
"indices_to_drop = []\nfor key, group in df.groupby(by=(\"site_id\", \"journal_oid\", \"responsibility\")):\n assert len(group) <= 2, len(group)\n if len(group) == 2:\n evidence = group[group.phase == evidence_phase]\n assert len(evidence) == 1\n if evidence.iloc[0].is_annotation_changed:\n indices_to_drop.append(evidence.index.values[0])\nlen(indices_to_drop)",
"/home/srivbane/shared/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:2: FutureWarning: Interpreting tuple 'by' as a list of keys, rather than a single key. Use 'by=[...]' instead of 'by=(...)'. In the future, a tuple will always mean a single key.\n \n"
],
[
"orig_size = len(df)\ndf = df.drop(indices_to_drop)\nnew_size = len(df)\norig_size, new_size",
"_____no_output_____"
]
],
[
[
"### Analysis",
"_____no_output_____"
]
],
[
[
"# first, how many fall into the three conditions?\nprint(\"Evidence phase annotation changes\")\nfor u1, u2 in [(user1, user2), (user2, user1)]:\n print(\"Evidence tasks:\", u1)\n print(\"Reconsider tasks:\", u2)\n print(\"=\"*40)\n for responsibility in responsibility_list:\n df_subset = df[(df.phase == evidence_phase)\n & (df.responsibility == responsibility)\n & (df.evidence_username == u1)\n & (df.reconsider_username == u2)]\n total_changed = np.sum(df_subset.is_annotation_changed)\n total = len(df_subset)\n pct_changed = total_changed / total\n print(f\"{responsibility:20}{' ':10}{total_changed:2}/{total:2}{' ':10}{pct_changed*100:.1f}%\")\n print()",
"Evidence phase annotation changes\nEvidence tasks: luoxx498\nReconsider tasks: eriks074\n========================================\ncoordinating_support 5/20 25.0%\nsymptom_management 4/20 20.0%\npreparation 3/19 15.8%\nmanaging_transitions 4/ 4 100.0%\ninfo_filtering 10/20 50.0%\ncontinued_monitoring 3/ 6 50.0%\nclinical_decisions 0/ 8 0.0%\n\nEvidence tasks: eriks074\nReconsider tasks: luoxx498\n========================================\ncoordinating_support 6/20 30.0%\nsymptom_management 7/20 35.0%\npreparation 4/20 20.0%\nmanaging_transitions 4/20 20.0%\ninfo_filtering 3/11 27.3%\ncontinued_monitoring 8/20 40.0%\nclinical_decisions 5/10 50.0%\n\n"
],
[
"print(\"Reconsider phase annotation changes\")\nfor u1, u2 in [(user1, user2), (user2, user1)]:\n print(\"Evidence tasks:\", u1)\n print(\"Reconsider tasks:\", u2)\n print(\"=\"*40)\n for responsibility in responsibility_list:\n df_subset = df[(df.phase == reconsider_phase)\n & (df.responsibility == responsibility)\n & (df.evidence_username == u1)\n & (df.reconsider_username == u2)]\n total_changed = np.sum(df_subset.is_annotation_changed)\n total = len(df_subset)\n pct_changed = total_changed / total if total > 0 else 0\n print(f\"{responsibility:20}{' ':10}{total_changed:2}/{total:2}{' ':10}{pct_changed*100:.1f}%\")\n print()",
"Reconsider phase annotation changes\nEvidence tasks: luoxx498\nReconsider tasks: eriks074\n========================================\ncoordinating_support 12/14 85.7%\nsymptom_management 15/16 93.8%\npreparation 15/17 88.2%\nmanaging_transitions 0/ 0 0.0%\ninfo_filtering 4/10 40.0%\ncontinued_monitoring 2/ 3 66.7%\nclinical_decisions 11/11 100.0%\n\nEvidence tasks: eriks074\nReconsider tasks: luoxx498\n========================================\ncoordinating_support 14/14 100.0%\nsymptom_management 9/13 69.2%\npreparation 10/16 62.5%\nmanaging_transitions 9/16 56.2%\ninfo_filtering 4/ 8 50.0%\ncontinued_monitoring 4/ 9 44.4%\nclinical_decisions 4/ 5 80.0%\n\n"
]
],
[
[
"### Irresolvable case analysis",
"_____no_output_____"
]
],
[
[
"print(\"Irresolvable disagreements from reconsider phase\")\nprint(\"=\"*50)\nu1_all_irresolvable = 0\nu2_all_irresolvable = 0\nu1_all_total = 0\nu2_all_total = 0\nfor responsibility in responsibility_list:\n df_subset = df[(df.phase == reconsider_phase)\n & (df.responsibility == responsibility)\n & (df.evidence_username == user1)\n & (df.reconsider_username == user2)]\n u2_irresolvable_count = np.sum(~df_subset.is_annotation_changed)\n u2_total = len(df_subset)\n df_subset = df[(df.phase == reconsider_phase)\n & (df.responsibility == responsibility)\n & (df.evidence_username == user2)\n & (df.reconsider_username == user1)]\n u1_irresolvable_count = np.sum(~df_subset.is_annotation_changed)\n u1_total = len(df_subset)\n \n u1_all_irresolvable += u1_irresolvable_count\n u2_all_irresolvable += u2_irresolvable_count\n u1_all_total += u1_total\n u2_all_total += u2_total\n print(f\"{responsibility:20}{' ':5}{u2_irresolvable_count:2}/{u2_total:2}{' ':5}{u1_irresolvable_count:2}/{u1_total:2}{' ':5}{u1_irresolvable_count+u2_irresolvable_count:2}/{u1_total + u2_total:2}\")\n print()\npct_irresolvable = (u1_all_irresolvable+u2_all_irresolvable)/(u1_all_total + u2_all_total) * 100\nprint(f\"{'Total':20}{' ':5}{u2_all_irresolvable:2}/{u2_all_total:2}{' ':5}{u1_all_irresolvable:2}/{u1_all_total:2}{' ':5}{u1_all_irresolvable+u2_all_irresolvable:2}/{u1_all_total + u2_all_total:2} ({pct_irresolvable:.2f}%)\")\nprint()",
"Irresolvable disagreements from reconsider phase\n==================================================\ncoordinating_support 2/14 0/14 2/28\n\nsymptom_management 1/16 4/13 5/29\n\npreparation 2/17 6/16 8/33\n\nmanaging_transitions 0/ 0 7/16 7/16\n\ninfo_filtering 6/10 4/ 8 10/18\n\ncontinued_monitoring 1/ 3 5/ 9 6/12\n\nclinical_decisions 0/11 1/ 5 1/16\n\nTotal 12/71 27/81 39/152 (25.66%)\n\n"
],
[
"# the original submission draft reports 26.3% of the updates as irresolvable, \n# but I'm not actually sure where that number's coming from. Should probably be 25.7, so updated accordingly",
"_____no_output_____"
],
[
"df[(df.additional_discussion != \"\") & (df.phase == reconsider_phase) & (~df.is_annotation_changed)][[\"responsibility\", \"additional_discussion\", \"evidence_username\", \"reconsider_username\", \"is_annotation_changed\"]]",
"_____no_output_____"
]
],
[
[
"Themes in comments:\n - Evidence to me of a different responsibility (and thus not this one)\n Takeaway: A problem with soft boundaries? Evidence that lies in margins especially hard to interpret\n - Not clear enough (in other words, ambiguous)\n - An edge case that falls just outside the boundary\n \n Qualitative analysis of annotator comments in irresolvable cases reveals two primary themes: (1) disagreement about the directness of supporting evidence needed to assign a responsibility and (2) disagreement about which responsibility a piece of evidence indicates. These themes align with two significant dimensions of ambiguity identified by Chen et al.: (a) data ambiguity, meaning multiple reasonable interpretations, often due to missing or unclear context, and (b) human subjectivity, meaning distinct interpretations resulting from ''different levels of understanding or sets of experiences'' among annotators \\cite{chen_using_2018}. Chen et al. further utilize disagreement between coders as a proxy for ambiguity, and the lower IRR scores relative to the phases indicates a higher degree of ambiguity. Could the primary dimension of ambiguity leading to low IRR scores be human subjectivity? Because the annotators are the same for both phases and responsibilities, it is unlikely.\nIs data ambiguity excacerbated by soft boundaries in the codebook? The irresolvable cases suggest that it could, and that further attempts to clarify the boundaries between responsibilities and the types of evidence that constitute a responsibility could decrease ambiguity and improve IRR. However, expert feedback indicates that our operationalization is reasonable. These points of evidence suggest an inherent ambiguity to the classification task. Only real option is to choose a different classification task! (I think that's what I believe, sadly...)\n\nThese qualitative observations align with conceptualizations of ambiguity by Chen et al... and the low IRR indicates this!\nFurther, since the set of annotators for the phases and responsibilities are the same, along with comments from the coders, indicates that it may be primarily data ambiguity at play.\n\nCoders are bad?\n\tSame coders for phases and responsibilities, so they can't be!\nOperationalization is wrong?\n\tBut experts think its fine!\nAnswer: there's inherently ambiguity! Chen et al indicates that low IRR indicates this!",
"_____no_output_____"
],
[
"### Discussion analysis",
"_____no_output_____"
]
],
[
[
"pd.set_option('display.max_colwidth', 255)",
"_____no_output_____"
],
[
"df[(df.additional_discussion != \"\") & (df.phase == evidence_phase)][[\"responsibility\", \"highlighted_text\", \"additional_discussion\", \"evidence_username\", \"reconsider_username\", \"is_annotation_changed\"]]",
"_____no_output_____"
],
[
"df[(df.additional_discussion != \"\") & (df.phase == reconsider_phase)][[\"responsibility\", \"additional_discussion\", \"evidence_username\", \"reconsider_username\", \"is_annotation_changed\"]]",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
]
] |
d0d145981cf8efa57e246578a53ccc3b63f9feed | 36,281 | ipynb | Jupyter Notebook | Arxiv_stats.ipynb | macbuse/Arxiv-Api | 8014997ac0d5616bac1085694fc754b51dbe695f | [
"MIT"
] | null | null | null | Arxiv_stats.ipynb | macbuse/Arxiv-Api | 8014997ac0d5616bac1085694fc754b51dbe695f | [
"MIT"
] | null | null | null | Arxiv_stats.ipynb | macbuse/Arxiv-Api | 8014997ac0d5616bac1085694fc754b51dbe695f | [
"MIT"
] | null | null | null | 98.056757 | 10,764 | 0.866018 | [
[
[
"import re\n",
"_____no_output_____"
],
[
"with open('listeJournals.txt', 'r') as fp:\n data = fp.read()",
"_____no_output_____"
],
[
"print(data[:100])",
"['ACM Trans. Math. Software', 'acmms', 1984]\n['ACM Trans. Math. Software', 'acmms', 1985]\n['ACM Tran\n"
],
[
"row_p = re.compile(\"\\[(.*?), (.*?), (\\d+)\")",
"_____no_output_____"
],
[
"long, short, dates = zip(* row_p.findall(data) )",
"_____no_output_____"
],
[
"stuff = {x : [] for x in set(short)}",
"_____no_output_____"
],
[
"for a, b in zip(short, dates):\n stuff[a].append(int(b))",
"_____no_output_____"
],
[
"for a in stuff.keys():\n stuff[a] = sorted(stuff[a])",
"_____no_output_____"
],
[
"for a in sorted(stuff.keys()):\n continue\n print( a, stuff[a][0], stuff[a][-1])\n ",
"_____no_output_____"
],
[
"import pandas as pd\nimport pickle\n\nfrom datetime import timedelta, date\nimport time\n\n\nstart_day= date(2010, 1, 11)",
"_____no_output_____"
],
[
"fn = 'arx_math_2010-06-18_2010-12-31.pkl'\nfn = 'arx_math_2011-01-01_2011-12-31.pkl'\nwith open( fn,'rb') as fp:\n output = pickle.load(fp)",
"_____no_output_____"
]
],
[
[
"Note how the not operator is ~ and non **not**",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame(output)\nyy = '2011'\ndfc = df[ df['created'].str.contains(yy) ]\ndfu = df[ ~df['created'].str.contains(yy) ]",
"_____no_output_____"
],
[
"math_cat = [ x for x in set( ' '.join(list(df['categories'])).split(' ') ) if 'math' in x]",
"_____no_output_____"
],
[
"len(dfc)",
"_____no_output_____"
],
[
"def date2doy(x, yy=2010):\n if x == '' : return None\n yy,mm,dd = [int(y) for y in x.split('-')]\n return date(yy,mm,dd) - date(yy, 1, 1)\n\nC = [date2doy(x, yy=int(yy)) for x in dfc['created'] ]\nU = [date2doy(x, yy=int(yy)) for x in dfu['updated'] ]\nU = [x for x in U if x]",
"_____no_output_____"
],
[
"doyc = [x.days for x in C ]\ndoyu = [x.days for x in U ]",
"_____no_output_____"
],
[
"from collections import Counter\nfreqs = Counter(doyc)\nfreqs.most_common(10)",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nimport numpy as np ",
"_____no_output_____"
],
[
"xs,ys = zip(*freqs.most_common(100))\nplt.plot(xs, ys,'r.')",
"_____no_output_____"
],
[
"plt.hist(doyu,bins=26, alpha=1., label='updated')\nplt.hist(doy,bins=26, alpha=0.5, label='created')\nplt.legend(loc='upper left')\nplt.title('Arxiv Submissions 2010')\n",
"_____no_output_____"
],
[
"plt.hist(doyu,bins=26, alpha=1., label='updated')\nplt.hist(doyc,bins=26, alpha=0.5, label='created')\nplt.legend(loc='upper left')\nplt.title('Arxiv Submissions ' + yy)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0d15505d050386589fb03f326c8d1880efae29d | 3,125 | ipynb | Jupyter Notebook | 2015-10_Lecture/Lecture2/code/4_Homework_Exercises.ipynb | hlin117/deeplearning4nlp-tutorial | 65ada138c07d657748ed37b5f6178a2be991d661 | [
"Apache-2.0"
] | 593 | 2016-01-20T14:45:46.000Z | 2022-03-24T04:58:11.000Z | 2015-10_Lecture/Lecture2/code/4_Homework_Exercises.ipynb | BhuvaneshwaranK/deeplearning4nlp-tutorial | 2a36ecc39f13b683752483546441468fd7d734b3 | [
"Apache-2.0"
] | 3 | 2016-02-23T21:44:51.000Z | 2020-02-06T17:05:38.000Z | 2015-10_Lecture/Lecture2/code/4_Homework_Exercises.ipynb | BhuvaneshwaranK/deeplearning4nlp-tutorial | 2a36ecc39f13b683752483546441468fd7d734b3 | [
"Apache-2.0"
] | 292 | 2016-01-18T13:39:22.000Z | 2022-02-04T13:10:46.000Z | 33.602151 | 285 | 0.6384 | [
[
[
"# Exercises\nTo get you started with Theano and Lasage, here are some task which you can try at home. Please also see Theano tutorial (http://deeplearning.net/software/theano/tutorial/) and the Lasagne Docs (http://lasagne.readthedocs.org/en/latest/index.html)",
"_____no_output_____"
],
[
"## Understand Theano\nThe above linked Theano tutorial is great to get to know Theano. I can highly recommend the following articles.\n\nLogistic Function: http://deeplearning.net/software/theano/tutorial/examples.html#logistic-function\n\nComputing More than one Thing at the Same Time: http://deeplearning.net/software/theano/tutorial/examples.html#computing-more-than-one-thing-at-the-same-time\n\nUsing Shared Variables: http://deeplearning.net/software/theano/tutorial/examples.html#using-shared-variables\n\nComputing Gradients: http://deeplearning.net/software/theano/tutorial/gradients.html#tutcomputinggrads\n\nClassifying MNIST digits using Logistic Regression: http://www.deeplearning.net/tutorial/logreg.html\n\nMulti Layer Peceptron: http://www.deeplearning.net/tutorial/mlp.html#mlp\n \n",
"_____no_output_____"
],
[
"## Understand the MNIST Example\nOpen the MNIST example and try to understand it and to modify it.\n\n**Task**: Change the hyperparameters for number of hidden units and learning rate. How does the accurarcy and training time change?\n\n**Task:** Change the mini-batch sizes. Which impact does it have?\n\n**Task:** Change the activation function from tanh to sigmoid.\n\n**Task:** In the current implementation the learning rate is fixed. Try a decreasing learning rate, start in the first epoch with e.g. 0.1, then decrease it stepwise to 0.01. Hint: Your Theano computation graph needs an additional variable which will capute the learning rate.\n\n**Task:** Try to add a second hidden layer. What's the impact?",
"_____no_output_____"
],
[
"## Understand the Lasagne Example\n\n**Task:** Same as above, try different parameters (hidden units, learning rate, activation function, mini batch size).\n\n**Task:** Add a second hidden layer",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d0d15911ae0f0353c284e6c1a6e61b1b4b7e553c | 1,177 | ipynb | Jupyter Notebook | _productivity/learnyoubash.ipynb | aixpact/data-science | f04a54595fbc2d797918d450b979fd4c2eabac15 | [
"MIT"
] | 2 | 2020-07-22T23:12:39.000Z | 2020-07-25T02:30:48.000Z | _productivity/learnyoubash.ipynb | aixpact/data-science | f04a54595fbc2d797918d450b979fd4c2eabac15 | [
"MIT"
] | null | null | null | _productivity/learnyoubash.ipynb | aixpact/data-science | f04a54595fbc2d797918d450b979fd4c2eabac15 | [
"MIT"
] | null | null | null | 16.347222 | 69 | 0.491079 | [
[
[
"# Learn you bash",
"_____no_output_____"
]
],
[
[
"bash_lessons = !find ~ | grep -1 LearnYou/learnyoubash",
"_____no_output_____"
],
[
"bash_lessons[2] # = bash_lessons[1]",
"_____no_output_____"
],
[
"!cat '/Users/frank/Documents/JS/LearnYou/learnyoubash/arr.bash'",
"_____no_output_____"
]
],
[
[
"# Arrays",
"_____no_output_____"
]
],
[
[
"%%!\nfruits=(Apple Pear Plum)\necho ${fruits[*]:0:2}",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0d15d345771496a4375c182fdd5258528f0202c | 5,636 | ipynb | Jupyter Notebook | tricks/valores_nulos.ipynb | everton3x/python-tricks | 3fdbe741155ee8d0225890393af191dcd7a782a0 | [
"MIT"
] | null | null | null | tricks/valores_nulos.ipynb | everton3x/python-tricks | 3fdbe741155ee8d0225890393af191dcd7a782a0 | [
"MIT"
] | null | null | null | tricks/valores_nulos.ipynb | everton3x/python-tricks | 3fdbe741155ee8d0225890393af191dcd7a782a0 | [
"MIT"
] | null | null | null | 26.584906 | 92 | 0.354862 | [
[
[
"# Valores nulos em data frame",
"_____no_output_____"
]
],
[
[
"import pandas as pd \nimport numpy as np ",
"_____no_output_____"
],
[
"# Importando o dataset\ndf = pd.read_csv(\"data_sets/pima-data.csv\")\ndf.shape\ndf.head()",
"_____no_output_____"
],
[
"# Verificando se existem valores nulos\ndf.isnull().values.any()",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d0d187a3bf0ec4fde2ecd5f83be10648ebd0aeaf | 200,290 | ipynb | Jupyter Notebook | DS_Sprint_Challenge_8_Regression_2.ipynb | AndrewMarksArt/DS-Unit-2-Applied-Modeling | 58275a934c7af652d8606e8022895f123229b974 | [
"MIT"
] | null | null | null | DS_Sprint_Challenge_8_Regression_2.ipynb | AndrewMarksArt/DS-Unit-2-Applied-Modeling | 58275a934c7af652d8606e8022895f123229b974 | [
"MIT"
] | null | null | null | DS_Sprint_Challenge_8_Regression_2.ipynb | AndrewMarksArt/DS-Unit-2-Applied-Modeling | 58275a934c7af652d8606e8022895f123229b974 | [
"MIT"
] | null | null | null | 211.053741 | 48,768 | 0.897484 | [
[
[
"_Lambda School Data Science, Unit 2_\n \n# Regression 2 Sprint Challenge: Predict drugstore sales 🏥\n\nFor your Sprint Challenge, you'll use real-world sales data from a German drugstore chain, from Jan 2, 2013 — July 31, 2015.\n\nYou are given three dataframes:\n\n- `train`: historical sales data for 100 stores\n- `test`: historical sales data for 100 different stores\n- `store`: supplemental information about the stores\n\n\nThe train and test set do _not_ have different date ranges. But they _do_ have different store ids. Your task is _not_ to forecast future sales from past sales. **Your task is to predict sales at unknown stores, from sales at known stores.**",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport category_encoders as ce\nimport eli5\n\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import mean_squared_error, mean_squared_log_error\nfrom sklearn.model_selection import cross_val_score\nfrom sklearn.pipeline import make_pipeline\n\nfrom xgboost import XGBRegressor\nfrom pdpbox.pdp import pdp_isolate, pdp_plot\nfrom pdpbox.pdp import pdp_interact, pdp_interact_plot\nfrom eli5.sklearn import PermutationImportance\n\nimport warnings\nwarnings.simplefilter(action='ignore', category=FutureWarning)",
"_____no_output_____"
],
[
"import pandas as pd\ntrain = pd.read_csv('https://drive.google.com/uc?export=download&id=1E9rgiGf1f_WL2S4-V6gD7ZhB8r8Yb_lE')\ntest = pd.read_csv('https://drive.google.com/uc?export=download&id=1vkaVptn4TTYC9-YPZvbvmfDNHVR8aUml')\nstore = pd.read_csv('https://drive.google.com/uc?export=download&id=1rZD-V1mWydeytptQfr-NL7dBqre6lZMo')\nassert train.shape == (78400, 7)\nassert test.shape == (78400, 7)\nassert store.shape == (200, 10)",
"_____no_output_____"
]
],
[
[
"The dataframes have a variety of columns:\n\n- **Store** - a unique Id for each store\n- **DayOfWeek** - integer, 1-6\n- **Date** - the date, from Jan 2, 2013 — July 31, 2015.\n- **Sales** - the units of inventory sold on a given date (this is the target you are predicting)\n- **Customers** - the number of customers on a given date\n- **Promo** - indicates whether a store is running a promo on that day\n- **SchoolHoliday** - indicates the closure of public schools\n- **StoreType** - differentiates between 4 different store models: a, b, c, d\n- **Assortment** - describes an assortment level: a = basic, b = extra, c = extended\n- **CompetitionDistance** - distance in meters to the nearest competitor store\n- **CompetitionOpenSince[Month/Year]** - gives the approximate year and month of the time the nearest competitor was opened\n- **Promo2** - Promo2 is a continuing and consecutive promotion for some stores: 0 = store is not participating, 1 = store is participating\n- **Promo2Since[Year/Week]** - describes the year and calendar week when the store started participating in Promo2\n- **PromoInterval** - describes the consecutive intervals Promo2 is started, naming the months the promotion is started anew. E.g. \"Feb,May,Aug,Nov\" means each round starts in February, May, August, November of any given year for that store",
"_____no_output_____"
],
[
"This Sprint Challenge has three parts. To demonstrate mastery on each part, do all the required instructions. To earn a score of \"3\" for the part, also do the stretch goals.",
"_____no_output_____"
],
[
"## 1. Wrangle relational data, Log-transform the target\n- Merge the `store` dataframe with the `train` and `test` dataframes. \n- Arrange the X matrix and y vector for the train and test sets.\n- Log-transform the target for the train and test set.\n- Plot the target's distribution for the train set, before and after the transformation.\n\n#### Stretch goals\n- Engineer 3+ more features.",
"_____no_output_____"
]
],
[
[
"store.head()",
"_____no_output_____"
],
[
"train.head()",
"_____no_output_____"
],
[
"test.head()",
"_____no_output_____"
]
],
[
[
"### Merge train, test and store",
"_____no_output_____"
]
],
[
[
"# Wrangle train, validation, and test sets\ndef wrangle(X):\n X = X.copy()\n \n # Engineer date features\n X['Date'] = pd.to_datetime(X['Date'], infer_datetime_format=True)\n X['date_year'] = X['Date'].dt.year\n X['date_month'] = X['Date'].dt.month\n X['date_day'] = X['Date'].dt.day\n X = X.drop(columns='Date')\n \n # Merge data\n X = (X.merge(store, how='left').fillna(0))\n \n return X\n\ntrain = wrangle(train)\ntest = wrangle(test)\n\ntrain.shape, test.shape",
"_____no_output_____"
]
],
[
[
"### Arrange X matrix and y target for train and test",
"_____no_output_____"
]
],
[
[
"target = 'Sales'\n\nX_train = train.drop(columns=target)\nX_test = test.drop(columns=target)\n\ny_train = train[target]\ny_test = test[target]\n\nX_train.shape, X_test.shape, y_train.shape, y_test.shape",
"_____no_output_____"
]
],
[
[
"### log transform train and test target",
"_____no_output_____"
]
],
[
[
"y_train_log = np.log1p(y_train)\ny_test_log = np.log1p(y_test)",
"_____no_output_____"
]
],
[
[
"### plot distribution of train target before and after transformation",
"_____no_output_____"
]
],
[
[
"sns.distplot(y_train);",
"_____no_output_____"
],
[
"sns.distplot(y_train_log);",
"_____no_output_____"
]
],
[
[
"## 2. Fit and validate your model\n- **Use Gradient Boosting** or any type of regression model.\n- **Beat the baseline:** The estimated baseline Root Mean Squared Logarithmic Error is 0.90, if we guessed the mean sales for every prediction. Remember that RMSE with the log-transformed target is equivalent to RMSLE with the original target. Try to get your error below 0.20.\n- **To validate your model, choose any one of these options:**\n - Split the train dataframe into train and validation sets. Put all dates for a given store into the same set. Use xgboost `early_stopping_rounds` with the validation set. \n - Or, use scikit-learn `cross_val_score`. Put all dates for a given store into the same fold.\n - Or, use scikit-learn `RandomizedSearchCV` for hyperparameter optimization. Put all dates for a given store into the same fold.\n- **Get the Validation Error** (multiple times if you try multiple iterations) **and Test Error** (one time, at the end).\n \n#### Stretch goal\n- Optimize 3+ hyperparameters by searching 10+ \"candidates\" (possible combinations of hyperparameters). ",
"_____no_output_____"
]
],
[
[
"def rmse(y_true, y_pred):\n return np.sqrt(mean_squared_error(y_true, y_pred))\n\ndef rmsle(y_true, y_pred):\n return np.sqrt(mean_squared_log_error(y_true, y_pred))",
"_____no_output_____"
]
],
[
[
"### Build baseline model ",
"_____no_output_____"
]
],
[
[
"y_base = np.full_like(y_test_log, fill_value=y_train_log.mean())\n\nprint('Validation RMSLE, Mean Baseline:', rmse(y_test_log, y_base))",
"Validation RMSLE, Mean Baseline: 0.3948331905413732\n"
]
],
[
[
"### Split train data into train and validate, put all dates for a given store in the same set",
"_____no_output_____"
]
],
[
[
"stores = train['Store'].unique()\n\ntrain_stores, val_stores = train_test_split(\n stores, random_state=42\n)\n\ntrain_stores = train[train.Store.isin(train_stores)]\nval_stores = train[train.Store.isin(val_stores)]\n\nassert train_stores.shape[0] + val_stores.shape[0] == train.shape[0]",
"_____no_output_____"
],
[
"target = 'Sales'\n\nX_train = train_stores.drop(columns=target)\nX_val = val_stores.drop(columns=target)\n\ny_train = train_stores[target]\ny_val = val_stores[target]\n\ny_train_log = np.log1p(y_train)\ny_val_log = np.log1p(y_val)\n\nX_train.shape, X_val.shape",
"_____no_output_____"
]
],
[
[
"### Use XGBoost to predict target",
"_____no_output_____"
]
],
[
[
"# Make pipeline\npipeline = make_pipeline(\n ce.OrdinalEncoder(), \n XGBRegressor(\n n_estimators=1000, \n n_jobs=-1,\n eval_metric='rmse',\n early_stopping_rounds=10\n )\n)\n\n# Fit\npipeline.fit(X_train, y_train_log)\n\n# Validate\ny_pred_log = pipeline.predict(X_val)\nprint('Validation Error', rmse(y_val_log, y_pred_log))",
"[20:08:19] WARNING: C:/Jenkins/workspace/xgboost-win64_release_0.90/src/objective/regression_obj.cu:152: reg:linear is now deprecated in favor of reg:squarederror.\nValidation Error 0.1641765041445393\n"
]
],
[
[
"### Get the test error",
"_____no_output_____"
]
],
[
[
"y_pred_test = pipeline.predict(X_test)\nprint('Test Error', rmse(y_test_log, y_pred_test))",
"Test Error 0.15633498742026974\n"
]
],
[
[
"## 3. Plot model interpretation visualizations\n- Choose any one of these options:\n - Permutation Importances plot\n - Partial Dependency Plot, 1 feature isolation\n - Partial Dependency Plot, 2 feature interaction\n \n#### Stretch goals\n- Plot 2+ visualizations.\n- Use permutation importances for feature selection. ",
"_____no_output_____"
]
],
[
[
"X_val.columns",
"_____no_output_____"
],
[
"features = [\n 'date_month',\n 'date_day',\n 'DayOfWeek'\n]\n\nfor feature in features:\n isolated = pdp_isolate(\n model=pipeline, \n dataset=X_val, \n model_features=X_val.columns, \n feature=feature\n )\n\n pdp_plot(isolated, feature_name=feature);",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d0d1919351a42c662b6167e4c3d130601676d650 | 12,559 | ipynb | Jupyter Notebook | tutorials/Deutsch_Algorithm.ipynb | MadhavJivrajani/QCircuit | 14e12d09585157927be15ba8f72263a78b1417cb | [
"Apache-2.0"
] | null | null | null | tutorials/Deutsch_Algorithm.ipynb | MadhavJivrajani/QCircuit | 14e12d09585157927be15ba8f72263a78b1417cb | [
"Apache-2.0"
] | null | null | null | tutorials/Deutsch_Algorithm.ipynb | MadhavJivrajani/QCircuit | 14e12d09585157927be15ba8f72263a78b1417cb | [
"Apache-2.0"
] | null | null | null | 49.25098 | 1,648 | 0.534278 | [
[
[
"<a href=\"https://colab.research.google.com/github/olgOk/QCircuit/blob/master/tutorials/Deutsch_Algorithm.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Deutsch Algorithm\n\nby Olga Okrut",
"_____no_output_____"
],
[
"Install frameworks, and import libraries",
"_____no_output_____"
]
],
[
[
"!pip install tensornetwork jax jaxlib colorama qcircuit",
"Collecting tensornetwork\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/37/37/f74c2fcdc56df69786b545bf58a7690832a63f643e0516ac6a92b2d5f5ca/tensornetwork-0.3.0-py3-none-any.whl (216kB)\n\r\u001b[K |█▌ | 10kB 17.6MB/s eta 0:00:01\r\u001b[K |███ | 20kB 3.2MB/s eta 0:00:01\r\u001b[K |████▌ | 30kB 4.6MB/s eta 0:00:01\r\u001b[K |██████ | 40kB 3.0MB/s eta 0:00:01\r\u001b[K |███████▋ | 51kB 3.7MB/s eta 0:00:01\r\u001b[K |█████████ | 61kB 4.4MB/s eta 0:00:01\r\u001b[K |██████████▋ | 71kB 5.0MB/s eta 0:00:01\r\u001b[K |████████████▏ | 81kB 3.9MB/s eta 0:00:01\r\u001b[K |█████████████▋ | 92kB 4.3MB/s eta 0:00:01\r\u001b[K |███████████████▏ | 102kB 4.7MB/s eta 0:00:01\r\u001b[K |████████████████▋ | 112kB 4.7MB/s eta 0:00:01\r\u001b[K |██████████████████▏ | 122kB 4.7MB/s eta 0:00:01\r\u001b[K |███████████████████▊ | 133kB 4.7MB/s eta 0:00:01\r\u001b[K |█████████████████████▏ | 143kB 4.7MB/s eta 0:00:01\r\u001b[K |██████████████████████▊ | 153kB 4.7MB/s eta 0:00:01\r\u001b[K |████████████████████████▎ | 163kB 4.7MB/s eta 0:00:01\r\u001b[K |█████████████████████████▊ | 174kB 4.7MB/s eta 0:00:01\r\u001b[K |███████████████████████████▎ | 184kB 4.7MB/s eta 0:00:01\r\u001b[K |████████████████████████████▊ | 194kB 4.7MB/s eta 0:00:01\r\u001b[K |██████████████████████████████▎ | 204kB 4.7MB/s eta 0:00:01\r\u001b[K |███████████████████████████████▉| 215kB 4.7MB/s eta 0:00:01\r\u001b[K |████████████████████████████████| 225kB 4.7MB/s \n\u001b[?25hRequirement already satisfied: jax in /usr/local/lib/python3.6/dist-packages (0.1.64)\nRequirement already satisfied: jaxlib in /usr/local/lib/python3.6/dist-packages (0.1.45)\nCollecting colorama\n Downloading https://files.pythonhosted.org/packages/c9/dc/45cdef1b4d119eb96316b3117e6d5708a08029992b2fee2c143c7a0a5cc5/colorama-0.4.3-py2.py3-none-any.whl\nCollecting qcircuit\n Downloading https://files.pythonhosted.org/packages/5f/e9/bd5cb2a97948e7cb00034582a2f108281c59b77e1a81405474115de38215/qcircuit-1.0.1.tar.gz\nRequirement already satisfied: scipy>=1.1 in /usr/local/lib/python3.6/dist-packages (from tensornetwork) (1.4.1)\nRequirement already satisfied: numpy>=1.17 in /usr/local/lib/python3.6/dist-packages (from tensornetwork) (1.18.3)\nCollecting graphviz>=0.11.1\n Downloading https://files.pythonhosted.org/packages/83/cc/c62100906d30f95d46451c15eb407da7db201e30f42008f3643945910373/graphviz-0.14-py2.py3-none-any.whl\nRequirement already satisfied: h5py>=2.9.0 in /usr/local/lib/python3.6/dist-packages (from tensornetwork) (2.10.0)\nRequirement already satisfied: opt-einsum>=2.3.0 in /usr/local/lib/python3.6/dist-packages (from tensornetwork) (3.2.1)\nRequirement already satisfied: absl-py in /usr/local/lib/python3.6/dist-packages (from jax) (0.9.0)\nRequirement already satisfied: six in /usr/local/lib/python3.6/dist-packages (from h5py>=2.9.0->tensornetwork) (1.12.0)\nBuilding wheels for collected packages: qcircuit\n Building wheel for qcircuit (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for qcircuit: filename=qcircuit-1.0.1-cp36-none-any.whl size=5988 sha256=506a90f6dbd06114774149fbfe798d9bb83491e2361e49d43b4cf708f70d415c\n Stored in directory: /root/.cache/pip/wheels/a3/dd/39/d3dece77c172e493c47e48a0b28382d116ff2daad674a7270b\nSuccessfully built qcircuit\nInstalling collected packages: graphviz, tensornetwork, colorama, qcircuit\n Found existing installation: graphviz 0.10.1\n Uninstalling graphviz-0.10.1:\n Successfully uninstalled graphviz-0.10.1\nSuccessfully installed colorama-0.4.3 graphviz-0.14 qcircuit-1.0.1 tensornetwork-0.3.0\n"
],
[
"from qcircuit import QCircuit as qc",
"_____no_output_____"
]
],
[
[
"Now, after we have learned how quantum gates work and how to build a quantum circuit, we will jump to the first quantum algorithm. We begin with a very simple quantum algorithm - Deutsch algorithm, named after its inventor David Deutsch, which serves as an excellent proof of the supremacy of quantum computers and algorithms over classical.\n\n\nThe problem Deutsch algorithm tackles can now be stated as follows. Given a black box *Uf* implementing\nsome unknown binary function *f* that maps {0, 1} into {0, 1}.\nWe have to clasify *f* as “constant” or “balanced” function. \n\n\n\nHere, constant means function always outputs the same bit, i.e. f(0) = f(1) = 1 or f(0) = f(1) = 0:\n\n\n\n\nBalanced means function outputs different bits on different inputs, i.e. f(0) != f(1):\n\n\nThe circuit for Deutsch’s algoritm is given below. The steps for the Deutsch algorithm:\n\n1. Prepare two qubits, one in state `1|0> + 0|1>` and the other in state `0|0> + 1|1>` (apply *X* gate on the second qubit).\n\n2. Apply the Hadamard gate (*H*) on both qubits to bring them to superposition.\n\n3. The output after the Hadamard transformation will be send through the gate *Uf*. The values of the *Uf* matrix depends on the *f(x)* function. That means that the state vector after the gate *Uf* depends on the function, e.g. constant or balanced function.\n\n4. The output from the *Uf* transormation is send to the gates Hadarard again. It will collapse the state vector from the superposition to one of the possible state depending on the function *f(x)*. \n\n5. The output from the Hadamard transformation will be a two qubit register. If all four possible function values are tested, it is revealed that the final output will be either `(0, 0), (0, 1), (1, 0), or (1, 1)` with probability of 1. The output value will depend on *f(x)*. The two qubits are entangled in the end, so only one of their values can be measured. This prevents us from known exactly which *f(x)* is being used. However, the first qubit in the pair will always be 1 if the function *f(x)* is **balanced**. If *f(x)* is **constant**, the algorithm outputs 0.\n\n\n\n",
"_____no_output_____"
],
[
"Now, let's create the quantum circuit above. We will use built-in method \n```\nUf(function)\n```\nwhich translates a classcal binary function *f(x)* into a unitary matrix *U*, and applies it to the circuit. As a parameter, it takes a function that needs to be tested for being balanced or constant. I will use a set of predefined functions to show the validity of the algorithm.",
"_____no_output_____"
]
],
[
[
"# define binary functions. Some of them are constant, other balanced\ndef f1(x):\n return x\n\ndef f2(x):\n return 1\n\ndef f3(x):\n return 0\n\ndef f4(x):\n return not x\n\ndef f5(x):\n return x ** 2\n\ndef f6(x):\n return not (x ** 3)\n\ndef f7(x):\n return (x % 3 == 2)\n\ndef f8(x):\n return not (x % 3 == 2)\n\n# check if the function constant\ndef is_const(func):\n deutsch = qc.QCircuit(2)\n deutsch.X(0)\n deutsch.H(1)\n deutsch.H(0)\n deutsch.Uf(func)\n deutsch.H(0)\n deutsch.H(1)\n\n # get output state vector\n # decide if a function constanta or balanced\n output_state = deutsch.get_state_vector()\n if abs(output_state[0]) == 1.+0.j or abs(output_state[1]) == 1.+0.j:\n return True\n else:\n return False\n\nfunctions = [f1, f2, f3, f4, f5, f6, f7, f8]\n\nfor func in range(len(functions)):\n print('function f{} is {}'.format(func+1, 'constant' if is_const(functions[func]) else 'balansed'))",
"/usr/local/lib/python3.6/dist-packages/jax/lib/xla_bridge.py:123: UserWarning: No GPU/TPU found, falling back to CPU.\n warnings.warn('No GPU/TPU found, falling back to CPU.')\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
d0d195c111686717b41f61c9dcb75a57ec02a2ec | 24,946 | ipynb | Jupyter Notebook | read-cgns.ipynb | mdpiper/notebooks | 5f88f0d6e37eab7125cfded784fe64a07a2cfe73 | [
"MIT"
] | null | null | null | read-cgns.ipynb | mdpiper/notebooks | 5f88f0d6e37eab7125cfded784fe64a07a2cfe73 | [
"MIT"
] | null | null | null | read-cgns.ipynb | mdpiper/notebooks | 5f88f0d6e37eab7125cfded784fe64a07a2cfe73 | [
"MIT"
] | null | null | null | 101.406504 | 11,992 | 0.870119 | [
[
[
"# Read and display data from a CGNS file",
"_____no_output_____"
]
],
[
[
"import h5py\nimport numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"Access a CGNS file.",
"_____no_output_____"
]
],
[
[
"f = h5py.File('data/Test1.cgn', 'r')",
"_____no_output_____"
],
[
"list(f.keys())",
"_____no_output_____"
]
],
[
[
"Move through the tree to find the x and y coordinate grids.",
"_____no_output_____"
]
],
[
[
"grid = f['iRIC']['iRICZone']['GridCoordinates']\nx = grid['CoordinateX'][' data'] # yes, there's a space\ny = grid['CoordinateY'][' data']",
"_____no_output_____"
],
[
"x.shape, y.shape",
"_____no_output_____"
],
[
"x.value",
"_____no_output_____"
]
],
[
[
"Display the coordinate grid.",
"_____no_output_____"
]
],
[
[
"plt.scatter(x, y)",
"_____no_output_____"
]
],
[
[
"Zoom in to see individula nodes.",
"_____no_output_____"
]
],
[
[
"plt.scatter(x, y)\nplt.ylim([5,15])\nplt.xlim([0,10])",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0d19ff5f210ce65b0da5e76e517efdcb166a235 | 854,367 | ipynb | Jupyter Notebook | _Nightly.ipynb | DeanLa/dont_reinvent_pandas | e2c6e78988d90e8eeceea5fa0055676486cc47b1 | [
"MIT"
] | 10 | 2018-11-07T16:23:51.000Z | 2021-08-07T12:50:10.000Z | _Nightly.ipynb | DeanLa/dont_reinvent_pandas | e2c6e78988d90e8eeceea5fa0055676486cc47b1 | [
"MIT"
] | null | null | null | _Nightly.ipynb | DeanLa/dont_reinvent_pandas | e2c6e78988d90e8eeceea5fa0055676486cc47b1 | [
"MIT"
] | 4 | 2018-11-23T14:50:36.000Z | 2019-11-01T00:16:37.000Z | 119.911158 | 153,532 | 0.780944 | [
[
[
"<h1>Table of Contents<span class=\"tocSkip\"></span></h1>\n<div class=\"toc\"><ul class=\"toc-item\"><li><span><a href=\"#Stop-Reinventing-Pandas\" data-toc-modified-id=\"Stop-Reinventing-Pandas-1\"><span class=\"toc-item-num\">1 </span>Stop Reinventing Pandas</a></span></li><li><span><a href=\"#First-Hacks!\" data-toc-modified-id=\"First-Hacks!-2\"><span class=\"toc-item-num\">2 </span>First Hacks!</a></span><ul class=\"toc-item\"><li><span><a href=\"#Beautiful-pipes!\" data-toc-modified-id=\"Beautiful-pipes!-2.1\"><span class=\"toc-item-num\">2.1 </span>Beautiful pipes!</a></span></li><li><span><a href=\"#The-Penny-Drops\" data-toc-modified-id=\"The-Penny-Drops-2.2\"><span class=\"toc-item-num\">2.2 </span>The Penny Drops</a></span></li><li><span><a href=\"#Map-with-dict\" data-toc-modified-id=\"Map-with-dict-2.3\"><span class=\"toc-item-num\">2.3 </span>Map with dict</a></span></li></ul></li><li><span><a href=\"#Time-Series\" data-toc-modified-id=\"Time-Series-3\"><span class=\"toc-item-num\">3 </span>Time Series</a></span><ul class=\"toc-item\"><li><span><a href=\"#Resample\" data-toc-modified-id=\"Resample-3.1\"><span class=\"toc-item-num\">3.1 </span>Resample</a></span><ul class=\"toc-item\"><li><span><a href=\"#The-Old-Way\" data-toc-modified-id=\"The-Old-Way-3.1.1\"><span class=\"toc-item-num\">3.1.1 </span>The Old Way</a></span></li><li><span><a href=\"#A-Better-Way\" data-toc-modified-id=\"A-Better-Way-3.1.2\"><span class=\"toc-item-num\">3.1.2 </span>A Better Way</a></span></li></ul></li><li><span><a href=\"#Slice-Easily\" data-toc-modified-id=\"Slice-Easily-3.2\"><span class=\"toc-item-num\">3.2 </span>Slice Easily</a></span></li><li><span><a href=\"#Time-Windows:-Rolling,-Expanding,-EWM\" data-toc-modified-id=\"Time-Windows:-Rolling,-Expanding,-EWM-3.3\"><span class=\"toc-item-num\">3.3 </span>Time Windows: Rolling, Expanding, EWM</a></span><ul class=\"toc-item\"><li><span><a href=\"#With-Apply\" data-toc-modified-id=\"With-Apply-3.3.1\"><span class=\"toc-item-num\">3.3.1 </span>With Apply</a></span></li></ul></li><li><span><a href=\"#Combine-with-GroupBy-🤯\" data-toc-modified-id=\"Combine-with-GroupBy-🤯-3.4\"><span class=\"toc-item-num\">3.4 </span>Combine with GroupBy 🤯</a></span></li></ul></li><li><span><a href=\"#Sorting\" data-toc-modified-id=\"Sorting-4\"><span class=\"toc-item-num\">4 </span>Sorting</a></span><ul class=\"toc-item\"><li><span><a href=\"#By-Values\" data-toc-modified-id=\"By-Values-4.1\"><span class=\"toc-item-num\">4.1 </span>By Values</a></span></li><li><span><a href=\"#By-Index\" data-toc-modified-id=\"By-Index-4.2\"><span class=\"toc-item-num\">4.2 </span>By Index</a></span></li><li><span><a href=\"#By-Both-(New-in-0.23)\" data-toc-modified-id=\"By-Both-(New-in-0.23)-4.3\"><span class=\"toc-item-num\">4.3 </span>By Both <span style=\"color: red\">(New in 0.23)</span></a></span></li></ul></li><li><span><a href=\"#Stack,-Unstack\" data-toc-modified-id=\"Stack,-Unstack-5\"><span class=\"toc-item-num\">5 </span>Stack, Unstack</a></span><ul class=\"toc-item\"><li><span><a href=\"#Unstack\" data-toc-modified-id=\"Unstack-5.1\"><span class=\"toc-item-num\">5.1 </span>Unstack</a></span><ul class=\"toc-item\"><li><span><a href=\"#The-Old-way\" data-toc-modified-id=\"The-Old-way-5.1.1\"><span class=\"toc-item-num\">5.1.1 </span>The Old way</a></span></li><li><span><a href=\"#A-better-way\" data-toc-modified-id=\"A-better-way-5.1.2\"><span class=\"toc-item-num\">5.1.2 </span>A better way</a></span></li></ul></li><li><span><a href=\"#Unstack\" data-toc-modified-id=\"Unstack-5.2\"><span class=\"toc-item-num\">5.2 </span>Unstack</a></span><ul class=\"toc-item\"><li><span><a href=\"#Some-More-Hacks\" data-toc-modified-id=\"Some-More-Hacks-5.2.1\"><span class=\"toc-item-num\">5.2.1 </span>Some More Hacks</a></span></li></ul></li></ul></li><li><span><a href=\"#GroupBy\" data-toc-modified-id=\"GroupBy-6\"><span class=\"toc-item-num\">6 </span>GroupBy</a></span><ul class=\"toc-item\"><li><span><a href=\"#Old-Ways\" data-toc-modified-id=\"Old-Ways-6.1\"><span class=\"toc-item-num\">6.1 </span>Old Ways</a></span><ul class=\"toc-item\"><li><span><a href=\"#List-Aggregates\" data-toc-modified-id=\"List-Aggregates-6.1.1\"><span class=\"toc-item-num\">6.1.1 </span>List Aggregates</a></span></li><li><span><a href=\"#Dict-aggregate\" data-toc-modified-id=\"Dict-aggregate-6.1.2\"><span class=\"toc-item-num\">6.1.2 </span>Dict aggregate</a></span></li><li><span><a href=\"#With-Rename\" data-toc-modified-id=\"With-Rename-6.1.3\"><span class=\"toc-item-num\">6.1.3 </span>With Rename</a></span></li></ul></li><li><span><a href=\"#Named-Aggregations-(New-in-0.25)\" data-toc-modified-id=\"Named-Aggregations-(New-in-0.25)-6.2\"><span class=\"toc-item-num\">6.2 </span>Named Aggregations <span style=\"color: red\">(New in 0.25)</span></a></span></li></ul></li><li><span><a href=\"#Clip\" data-toc-modified-id=\"Clip-7\"><span class=\"toc-item-num\">7 </span>Clip</a></span><ul class=\"toc-item\"><li><span><a href=\"#The-Old-Way\" data-toc-modified-id=\"The-Old-Way-7.1\"><span class=\"toc-item-num\">7.1 </span>The Old Way</a></span></li><li><span><a href=\"#A-better-way\" data-toc-modified-id=\"A-better-way-7.2\"><span class=\"toc-item-num\">7.2 </span>A better way</a></span></li></ul></li><li><span><a href=\"#Reindex\" data-toc-modified-id=\"Reindex-8\"><span class=\"toc-item-num\">8 </span>Reindex</a></span></li><li><span><a href=\"#Method-Chaining\" data-toc-modified-id=\"Method-Chaining-9\"><span class=\"toc-item-num\">9 </span>Method Chaining</a></span><ul class=\"toc-item\"><li><span><a href=\"#Assign\" data-toc-modified-id=\"Assign-9.1\"><span class=\"toc-item-num\">9.1 </span>Assign</a></span><ul class=\"toc-item\"><li><span><a href=\"#With-a-callable\" data-toc-modified-id=\"With-a-callable-9.1.1\"><span class=\"toc-item-num\">9.1.1 </span>With a callable</a></span></li></ul></li><li><span><a href=\"#Pipe\" data-toc-modified-id=\"Pipe-9.2\"><span class=\"toc-item-num\">9.2 </span>Pipe</a></span></li></ul></li><li><span><a href=\"#Beautiful-Code-Tells-a-Story\" data-toc-modified-id=\"Beautiful-Code-Tells-a-Story-10\"><span class=\"toc-item-num\">10 </span>Beautiful Code Tells a Story</a></span></li><li><span><a href=\"#Bonus!\" data-toc-modified-id=\"Bonus!-11\"><span class=\"toc-item-num\">11 </span>Bonus!</a></span><ul class=\"toc-item\"><li><span><a href=\"#Percent-Change\" data-toc-modified-id=\"Percent-Change-11.1\"><span class=\"toc-item-num\">11.1 </span>Percent Change</a></span></li><li><span><a href=\"#Interval-Index\" data-toc-modified-id=\"Interval-Index-11.2\"><span class=\"toc-item-num\">11.2 </span>Interval Index</a></span></li><li><span><a href=\"#Split-Strings\" data-toc-modified-id=\"Split-Strings-11.3\"><span class=\"toc-item-num\">11.3 </span>Split Strings</a></span></li><li><span><a href=\"#Toy-Examples-with-Pandas-Testing\" data-toc-modified-id=\"Toy-Examples-with-Pandas-Testing-11.4\"><span class=\"toc-item-num\">11.4 </span>Toy Examples with Pandas Testing</a></span></li></ul></li><li><span><a href=\"#Research-with-Style!\" data-toc-modified-id=\"Research-with-Style!-12\"><span class=\"toc-item-num\">12 </span>Research with Style!</a></span><ul class=\"toc-item\"><li><span><a href=\"#Basic\" data-toc-modified-id=\"Basic-12.1\"><span class=\"toc-item-num\">12.1 </span>Basic</a></span></li><li><span><a href=\"#Gradient\" data-toc-modified-id=\"Gradient-12.2\"><span class=\"toc-item-num\">12.2 </span>Gradient</a></span></li><li><span><a href=\"#Custom\" data-toc-modified-id=\"Custom-12.3\"><span class=\"toc-item-num\">12.3 </span>Custom</a></span></li><li><span><a href=\"#Bars\" data-toc-modified-id=\"Bars-12.4\"><span class=\"toc-item-num\">12.4 </span>Bars</a></span></li></ul></li><li><span><a href=\"#You-don't-have-to-memorize-this\" data-toc-modified-id=\"You-don't-have-to-memorize-this-13\"><span class=\"toc-item-num\">13 </span>You don't have to memorize this</a></span></li><li><span><a href=\"#Resources\" data-toc-modified-id=\"Resources-14\"><span class=\"toc-item-num\">14 </span>Resources</a></span></li></ul></div>",
"_____no_output_____"
],
[
"# Stop Reinventing Pandas",
"_____no_output_____"
],
[
"The following post was presented as a talk for the [IE@DS](https://www.facebook.com/groups/173376299978861/) community, and for the [PyData meetup](https://www.meetup.com/PyData-Tel-Aviv/events/256232456/). \nAll the resources for this post, including a runable notebook, can be found in the [github repo](https://github.com/DeanLa/dont_reinvent_pandas) \nblog post version here: \n\n\n<span style=\"font-size:2em\"> [DeanLa.com](http://deanla.com/)</span> ",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"This notebook aims to show some nice ways modern Pandas makes your life easier. It is not about efficiency. I'm Pandas' built-in methods will be more efficient than reinventing pandas, but the main goal is to make the code easier to read, and more imoprtant - easier to write.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline\nplt.style.use(['classic', 'ggplot', 'seaborn-poster', 'dean.style'])\n%load_ext autoreload\n%autoreload 2\nimport my_utils\nimport warnings\nwarnings.simplefilter(\"ignore\")",
"_____no_output_____"
]
],
[
[
"# First Hacks!\n",
"_____no_output_____"
],
[
"Reading the data and a few housekeeping tasks. is the first place we can make our code more readable.",
"_____no_output_____"
]
],
[
[
"df_io = pd.read_csv('./bear_data.csv', index_col=0, parse_dates=['date_'])\ndf_io.head()",
"_____no_output_____"
],
[
"df = df_io.copy().sort_values('date_').set_index('date_').drop(columns='val_updated')\ndf.head()",
"_____no_output_____"
]
],
[
[
"## Beautiful pipes!\nOne line method chaining is hard to read and prone to human error, chaining each method in its own line makes it a lot more readable.",
"_____no_output_____"
]
],
[
[
"df_io\\\n .copy()\\\n .sort_values('date_')\\\n .set_index('date_')\\\n .drop(columns='val_updated')\\\n .head()",
"_____no_output_____"
]
],
[
[
"But it has a problem. You can't comment out and even comment in between",
"_____no_output_____"
]
],
[
[
"# This block will result in an error\ndf_io\\\n.copy()\\ # This is an inline comment\n# This is a regular comment\n.sort_values('date_')\\\n# .set_index('date_')\\\n.drop(columns='val_updated')\\ \n.head()",
"_____no_output_____"
]
],
[
[
"Even an unnoticeable space character may break everything",
"_____no_output_____"
]
],
[
[
"# This block will result in an error\ndf_io\\\n.copy()\\\n.sort_values('date_')\\\n.set_index('date_')\\\n.drop(columns='val_updated')\\ \n.head()",
"_____no_output_____"
]
],
[
[
"## The Penny Drops\nI like those \"penny dropping\" moments, when you realize you knew everything that is presented, yet it is presented in a new way you never thought of.",
"_____no_output_____"
]
],
[
[
"# We can split these value inside ()\nusers = (134856, 195373, 295817, 294003, 262166, 121066, 129678, 307120, 258759, 277922, 220794, 192312,\n 318486, 314631, 306448, 297059,206892, \n 169046, 181703, 146200, 199876, 247904, 250884, 282989, 234280, 202520, \n 138064, 133577, 301053, 242157)",
"_____no_output_____"
],
[
"# Penny Drop: We can also Split here\ndf = (df_io\n .copy() # This is an inline comment\n # This is a regular comment\n .sort_values('date_')\n .set_index('date_')\n .drop(columns='val_updated') \n)\n\ndf.head()",
"_____no_output_____"
]
],
[
[
"## Map with dict\nA dict is a callable with $f(key) = value$, there for you can call `.map` with it. In this example I want to make int key codes into letter.",
"_____no_output_____"
]
],
[
[
"df.bear_type.map(lambda x: x+3).head()",
"_____no_output_____"
],
[
"# A dict is also a callable\nbears = {\n 1: 'Grizzly',\n 2: 'Sun',\n 3: 'Pizzly',\n 4: 'Sloth',\n 5: 'Polar',\n 6: 'Cave',\n 7: 'Black',\n 8: 'Panda'\n}\ndf['bear_type'] = df.bear_type.map(bears)\ndf.head()",
"_____no_output_____"
]
],
[
[
"# Time Series\n",
"_____no_output_____"
],
[
"## Resample\nTask: How many events happen each hour?",
"_____no_output_____"
],
[
"### The Old Way",
"_____no_output_____"
]
],
[
[
"bad = df.copy()\nbad['day'] = bad.index.date\nbad['hour'] = bad.index.hour\n(bad\n.groupby(['day','hour'])\n.count()\n)",
"_____no_output_____"
]
],
[
[
"* Many lines of code\n* unneeded columns\n* Index is not a time anymore\n* **missing rows** (Did you notice?)",
"_____no_output_____"
],
[
"### A Better Way",
"_____no_output_____"
]
],
[
[
"df.resample('H').count() # H is for Hour",
"_____no_output_____"
]
],
[
[
"But it's even better on non-round intervals",
"_____no_output_____"
]
],
[
[
"rs = df.resample('10T').count()\n# T is for Minute, and pandas understands 10 T, it will also under stand 11T if you wonder\nrs.head()",
"_____no_output_____"
]
],
[
[
"[Complete list of Pandas' time abbrevations](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.Period.strftime.html)",
"_____no_output_____"
],
[
"## Slice Easily\nPandas will automatically make string into timestamps, and it will understand what you want it to do.",
"_____no_output_____"
]
],
[
[
"# Take only timestamp in the hour of 21:00.\nrs.loc['2018-10-09 21',:]",
"_____no_output_____"
],
[
"# Take all time stamps before 18:31\nrs.loc[:'2018-10-09 18:31',:]",
"_____no_output_____"
]
],
[
[
"## Time Windows: Rolling, Expanding, EWM\nIf your Dataframe is indexed on a time index (Which we have)",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots()\nrs.rename(columns = {'bear_type':'bears'}).plot(ax=ax,linestyle='--')\n(rs\n .rolling('90T')\n .mean()\n .rename(columns = {'bear_type':'rolling mean'})\n .plot(ax=ax)\n)\n\nrs.expanding().mean().rename(columns = {'bear_type':'expanding mean'}).plot(ax=ax)\nrs.ewm(6).mean().rename(columns = {'bear_type':'ewm mean'}).plot(ax=ax)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### With Apply\nIntuitively, windows are like GroupBy, so you can apply anything you want after the grouping, e.g.: geometric mean.",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots()\nrs.plot(ax=ax,linestyle='--')\n(rs\n .rolling(6)\n .apply(lambda x: np.power(np.product(x),1/len(x)),raw=True)\n .rename(columns = {'bear_type':'Rolling Geometric Mean'})\n .plot(ax=ax)\n)\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Combine with GroupBy 🤯\nPandas has no problem with groupby and resample together. It's as simple as `groupby[col1,col2]`. In our specific case, we want to cound events in an interval per event type.",
"_____no_output_____"
]
],
[
[
"per_bear = (df\n .groupby('bear_type')\n .resample('15T')\n .apply('count')\n .rename(columns={'bear_type':'amount'})\n )\nper_bear.groupby('bear_type').head(2)",
"_____no_output_____"
]
],
[
[
"# Sorting",
"_____no_output_____"
],
[
"## By Values",
"_____no_output_____"
]
],
[
[
"per_bear.sort_values(by=['amount'], ascending=False).head(10)",
"_____no_output_____"
]
],
[
[
"## By Index ",
"_____no_output_____"
]
],
[
[
"per_bear.sort_index().head(7)",
"_____no_output_____"
],
[
"per_bear.sort_index(level=1).head(7)",
"_____no_output_____"
]
],
[
[
"## By Both <span style=\"color:red\">(New in 0.23)</span>\n`Index` has a name. Modern Pandas knows to address this index by name just like a regular column.",
"_____no_output_____"
]
],
[
[
"per_bear.sort_values(['amount','bear_type'], ascending=(False, True)).head(10)",
"_____no_output_____"
]
],
[
[
"# Stack, Unstack",
"_____no_output_____"
],
[
"## Unstack \nIn this case, working with a wide format indexed on intervals, with event types as columns, will make a lot more sense.",
"_____no_output_____"
],
[
"### The Old way\nPivot table in modern pandas is more robust than it used to be. Still, it requires you to specify everything.",
"_____no_output_____"
]
],
[
[
"pt = pd.pivot_table(per_bear,values = 'amount',columns='bear_type',index='date_')\npt.head()",
"_____no_output_____"
]
],
[
[
"### A better way\nWhen you have just one column of values, unstack does the same easily",
"_____no_output_____"
]
],
[
[
"pt = per_bear.unstack('bear_type')\npt.columns = pt.columns.droplevel() # Unstack creates a multiindex on columns\npt.head()",
"_____no_output_____"
]
],
[
[
"## Unstack\nAnd some extra tricks",
"_____no_output_____"
]
],
[
[
"pt.stack().head()",
"_____no_output_____"
]
],
[
[
"This looks kind of what we had expected but:\n* It's a series, not a DataFrame\n* The levels of the index are \"reversed\" to before\n* The main sort is on the date, yet it used to be on the event type\n\n\n### Some More Hacks\n\n",
"_____no_output_____"
]
],
[
[
"stack_back = (pt\n .stack()\n .to_frame('amount') # Turn Series to DF without calling the DF constructor\n .swaplevel() # Swaps the levels of the index\n .sort_index()\n )\nstack_back.head()",
"_____no_output_____"
],
[
"stack_back.equals(per_bear)",
"_____no_output_____"
]
],
[
[
"# GroupBy",
"_____no_output_____"
],
[
"```sql\nselect min(B), avg(B), geometric_mean(B), min(C), max(C)\nfrom pt\ngroup by A\n```",
"_____no_output_____"
]
],
[
[
"pt",
"_____no_output_____"
]
],
[
[
"## Old Ways",
"_____no_output_____"
]
],
[
[
"pt.groupby('Grizzly')['Polar'].agg(['min','mean']).head()",
"_____no_output_____"
]
],
[
[
"### List Aggregates",
"_____no_output_____"
]
],
[
[
"pt.groupby('Grizzly')[['Polar','Black']].agg(['min','mean',lambda x: x.prod()/len(x),'max']).head()",
"_____no_output_____"
]
],
[
[
"* Not what we wanted\n* MultiIndex\n* Names are not unique\n* How do you access `<lambda_0>`",
"_____no_output_____"
],
[
"### Dict aggregate",
"_____no_output_____"
]
],
[
[
"pt.groupby('Grizzly').agg({'Polar':['min','mean',lambda x: x.prod()/len(x)],'Black':['min','max']})",
"_____no_output_____"
]
],
[
[
"### With Rename",
"_____no_output_____"
]
],
[
[
"pt.groupby('Grizzly').Polar.agg({'min_Polar':'min'})",
"_____no_output_____"
],
[
"warnings.simplefilter(\"ignore\")\npt.groupby('Grizzly').agg({\n 'Polar':{'min_Polar':'min','avg_Polar':'mean','geo_Polar':lambda x: x.prod()/len(x)},\n 'Black':{'min_Black':'min','max_Black':'max'}\n \n})\nwarnings.simplefilter(\"default\")\n",
"_____no_output_____"
]
],
[
[
"Still a MultiIndex",
"_____no_output_____"
],
[
"## Named Aggregations <span style=\"color:red\">(New in 0.25)</span>\nThis is also the way to go from `1.0.0` as others will be depracated",
"_____no_output_____"
]
],
[
[
"def geo(x):\n return x.prod()/len(x) ",
"_____no_output_____"
],
[
"pt.groupby('Grizzly').agg(\n min_Polar = pd.NamedAgg(column='Polar', aggfunc='min'),\n avg_Polar = pd.NamedAgg(column='Polar', aggfunc='mean'),\n geo_Polar = pd.NamedAgg('Polar', geo),\n # But actually NamedAgg is optional\n min_Black = ('Black','min'),\n max_Black = ('Black','max')\n \n)\n",
"_____no_output_____"
]
],
[
[
"# Clip\nLet's say, we know from domain knowledge the that an bear walks around a minimum of 3 and maximum of 12 times at each timestamp. We would like to fix that. \nIn a real world example, we many time want to turn negative numbers to zeroes or some truly big numbers to sum known max.",
"_____no_output_____"
],
[
"## The Old Way\nIterate over columns and change values that meet condition.",
"_____no_output_____"
]
],
[
[
"cl = pt.copy()\nlb = 3\nub = 12\n# Needed A loop of 3 lines\nfor col in ['Grizzly','Polar','Black']:\n cl['clipped_{}'.format(col)] = cl[col]\n cl.loc[cl[col] < lb,'clipped_{}'.format(col)] = lb\n cl.loc[cl[col] > ub,'clipped_{}'.format(col)] = ub\nmy_utils.plot_clipped(cl) # my_utils can be found in the github repo",
"_____no_output_____"
]
],
[
[
"## A better way\n`.clip(lb,ub)`",
"_____no_output_____"
]
],
[
[
"cl = pt.copy()\ncl['Grizzly'] = cl.Grizzly.clip(3,12)\n",
"_____no_output_____"
],
[
"cl = pt.copy()\n# Beutiful One Liner\ncl[['clipped_Grizzly','clipped_Polar','clipped_Black']] = cl.clip(5,12)\nmy_utils.plot_clipped(cl) # my_utils can be found in the github repo",
"_____no_output_____"
]
],
[
[
"# Reindex\nNow we have 3 types of bears 17:00 to 23:00. But we were at the the park from 16:00 to 00:00. We've also been told that this park as Panda bears and Cave bears. \nIn the old way we would have this column assignment with a loop, and for the rows we would have maybe create a columns and do some join. A lot of work.",
"_____no_output_____"
]
],
[
[
"etypes = ['Grizzly','Polar','Black','Panda','Cave'] # New columns\n# Define a date range - Pandas will automatically make this into an index\nidx = pd.date_range(start='2018-10-09 16:00:00',end='2018-10-09 23:59:00',freq=pt.index.freq,tz='UTC')\ntype(idx)",
"_____no_output_____"
],
[
"pt.reindex(index=idx, columns=etypes, fill_value=0).head(8)",
"_____no_output_____"
],
[
"### Let's put this in a function - This will help us later.\ndef get_all_types_and_timestamps(df, min_date='2018-10-09 16:00:00',\n max_date='2018-10-09 23:59:00',\n etypes=['Grizzly','Polar','Black','Panda','Cave']):\n ret = df.copy()\n time_idx = pd.date_range(start=min_date,end=max_date,freq='15T',tz='UTC')\n # Indices work like set. This is a good practive so we don't override our intended index\n idx = ret.index.union(time_idx)\n etypes = df.columns.union(set(etypes))\n ret = ret.reindex(idx, columns=etypes, fill_value=0)\n return ret",
"_____no_output_____"
]
],
[
[
"# Method Chaining",
"_____no_output_____"
],
[
"## Assign\nAssign is for creating new columns on the dataframes. This is instead of\n`df[new_col] = function(df[old_col])`. They are both one lines, but `.assign` doesn't break the flow.",
"_____no_output_____"
]
],
[
[
"pt.assign(mean_all = pt.mean(axis=1)).head()",
"_____no_output_____"
]
],
[
[
"### With a callable\nThis is good when we have a filtering phase before.",
"_____no_output_____"
]
],
[
[
"pt.assign(mean_all = lambda x: x.mean(axis=1)).head()",
"_____no_output_____"
]
],
[
[
"## Pipe\nThink R's `%>%`, `.pipe` is a method that accepts a function. `pipe`, by default, assumes the first argument of this function is a dataframe and passes the current dataframe down the pipeline. The function should return a dataframe also, if you want to continue with the chaining. Yet, it can also return any other value if you put it in the last step. \nThis is incredibly valueable because it takes you one step further from \"sql\" where you do things \"in reverse\". \n$f(g(h($ `df` $)))$ = `df.pipe(h).pipe(g).pipe(f)`",
"_____no_output_____"
]
],
[
[
"def add_to_col(df, col='Grizzly', n = 200):\n ret = df.copy()\n # A dataframe is mutable, if you don't copy it first, this is prone to many errors.\n # I always copy when I enter a function, even if I'm sure it shouldn't change anything.\n ret[col] = ret[col] + n\n return ret",
"_____no_output_____"
],
[
"add_to_col(add_to_col(add_to_col(pt), 'Polar', 100), 'Black',500).head()",
"_____no_output_____"
],
[
"(pt\n .pipe(add_to_col)\n .pipe(add_to_col, col='Polar', n=100)\n .pipe(add_to_col, col='Black', n=500) \n .head(5))",
"_____no_output_____"
]
],
[
[
"You can always do this with multiple lines of `df = do_something(df)` but I think this method is more elegant.",
"_____no_output_____"
],
[
"# Beautiful Code Tells a Story\nYour code is not just about making the computer do things. It's about telling a story of what you wish to happen. Sometimes other people will want to read you code. Most time, it is you 3 monhts in the future who will want to read it. Some say good code documents itself. I'm not that extreme, yet storytelling with code may save you from many lines of unnecessary comments.\nThe next and final block tells the story in one block. It's elegant, it tells a story. If you build utility functions and `pipe` them while following meaningful naming, they help tell a story. if you `assign` columns with meaningful names, they tell a story. you `drop`, you `apply`, you `read`, you `groupby` and you `resample` - they all tell a story.\n\n(Well... Maybe they could have gone with better naming for `resample`)",
"_____no_output_____"
]
],
[
[
"df = (pd\n .read_csv ('./bear_data.csv', index_col=0, parse_dates=['date_'])\n .assign (bear_type=lambda df: df.bear_type.map(bears))\n .sort_values ('date_')\n .set_index ('date_')\n .drop (columns='val_updated')\n .groupby ('bear_type')\n .resample ('15T')\n .apply ('count')\n .rename (columns={'bear_type': 'amount'})\n .unstack ('bear_type')\n .pipe (my_utils.remove_multi_index)\n .pipe (get_all_types_and_timestamps) # Remember this from before?\n .assign (mean_bears=lambda x: x.mean(axis=1))\n .loc [:, ['mean_bears']]\n .pipe (my_utils.make_sliding_time_windows, steps_back=6)\n .dropna ()\n )",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
]
],
[
[
"# Bonus! \nCool methods I've found but did not fit in the talk's flow. \n\n\n<span style=\"font-size:2em\"> [No Time?](#You-don't-have-to-memorize-this)</span> \n",
"_____no_output_____"
]
],
[
[
"src = df.copy().loc[:,['mean_bears']]",
"_____no_output_____"
]
],
[
[
"## Percent Change ",
"_____no_output_____"
]
],
[
[
"src.assign(pct = src.pct_change()).head(11)",
"_____no_output_____"
]
],
[
[
"## Interval Index\nHelps creating a \"common language\" when talking about time series aggregations.",
"_____no_output_____"
]
],
[
[
"src = df.copy()\nir = pd.interval_range(start=df.index.min(),\n end=df.index.max() + df.index.freq,\n freq=df.index.freq)\ntype(ir)",
"_____no_output_____"
],
[
"ir",
"_____no_output_____"
],
[
"try:\n df.loc['2018-10-09 18:37',:] # Datetime Index\nexcept Exception as e:\n print (type(e), e)\n# Will result error",
"<class 'KeyError'> '2018-10-09 18:37'\n"
],
[
"src.index = ir # Interval Index\nsrc.loc['2018-10-09 18:37',:]\n",
"_____no_output_____"
],
[
"src.loc['2018-10-09 18:37':'2018-10-09 19:03',:]\n",
"_____no_output_____"
]
],
[
[
"## Split Strings\nThe entire concept of strings is different in `1.0.0`",
"_____no_output_____"
]
],
[
[
"txt = pd.DataFrame({'text':['hello','dean langsam','diving into pandas is better than reinventing it']})\n\ntxt",
"_____no_output_____"
],
[
"txt.text.str.split()",
"_____no_output_____"
],
[
"txt.text.str.split(expand = True) # Expand to make it a dataframe",
"_____no_output_____"
]
],
[
[
"## Toy Examples with Pandas Testing ",
"_____no_output_____"
]
],
[
[
"import pandas.util.testing as tm\ntm.N, tm.K = 15, 10\nst = pd.util.testing.makeTimeDataFrame() * 100\nst",
"_____no_output_____"
]
],
[
[
"# Research with Style!\n",
"_____no_output_____"
]
],
[
[
"stnan = st.copy()\nstnan[np.random.rand(*stnan.shape) < 0.05] = np.nan # Put some nans in it",
"_____no_output_____"
]
],
[
[
"## Basic",
"_____no_output_____"
]
],
[
[
"(stnan\n .style\n .highlight_null('red')\n .highlight_max(color='steelblue', axis = 0) # Max each row\n .highlight_min(color ='gold', axis = 1) # Min each columns\n)\n",
"/Users/deanl/anaconda3/envs/dont/lib/python3.7/site-packages/jinja2/utils.py:485: DeprecationWarning: Using or importing the ABCs from 'collections' instead of from 'collections.abc' is deprecated, and in 3.8 it will stop working\n from collections import MutableMapping\n/Users/deanl/anaconda3/envs/dont/lib/python3.7/site-packages/jinja2/runtime.py:318: DeprecationWarning: Using or importing the ABCs from 'collections' instead of from 'collections.abc' is deprecated, and in 3.8 it will stop working\n from collections import Mapping\n"
]
],
[
[
"## Gradient ",
"_____no_output_____"
]
],
[
[
"st.clip(0,100).style.background_gradient( cmap='Purples')",
"_____no_output_____"
]
],
[
[
"## Custom",
"_____no_output_____"
]
],
[
[
"def custom_style(val):\n if val < -100:\n return 'background-color:red'\n elif val > 100:\n return 'background-color:green'\n elif abs(val) <20:\n return 'background-color:yellow'\n else:\n return ''\nst.style.applymap(custom_style)",
"_____no_output_____"
]
],
[
[
"## Bars",
"_____no_output_____"
]
],
[
[
"(st.style\n .bar(subset=['A','D'],color='steelblue')\n .bar(subset=['J'],color=['indianred','limegreen'], align='mid')\n)",
"_____no_output_____"
]
],
[
[
"# You don't have to memorize this\nJust put this in the back of your mind and remember that modern Pandas has so many superpowers. Just remember they exist, and google them when you actually need them.\nAlways, when I feel I'm insecure about Pandas, I go back to [Greg Reda](https://twitter.com/gjreda)'s [tweet](https://twitter.com/gjreda/status/1049694953687924737):",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"# Resources \n* [Modern Pandas](https://tomaugspurger.github.io/modern-1-intro.html) by Tom Augspurger\n* [Basic Time Series Manipulation with Pandas](https://towardsdatascience.com/basic-time-series-manipulation-with-pandas-4432afee64ea) by Laura Fedoruk\n* [Pandas Docs](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.clip.html). You don't have to thoroughly go over everything, just randomly open a page in the docs and you're sure to learn a new thing. ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
d0d1a7f0c5d1f93e517e08933f5f1f7992f99030 | 580,590 | ipynb | Jupyter Notebook | examples/interacting_with_protodash.ipynb | gAldeia/itea-python | 689cd1eff61cd475277d63ca9387c6f9b6b7ee53 | [
"BSD-3-Clause"
] | null | null | null | examples/interacting_with_protodash.ipynb | gAldeia/itea-python | 689cd1eff61cd475277d63ca9387c6f9b6b7ee53 | [
"BSD-3-Clause"
] | null | null | null | examples/interacting_with_protodash.ipynb | gAldeia/itea-python | 689cd1eff61cd475277d63ca9387c6f9b6b7ee53 | [
"BSD-3-Clause"
] | null | null | null | 1,168.189135 | 128,232 | 0.952965 | [
[
[
"# Interacting with ProtoDash",
"_____no_output_____"
],
[
"In this notebook we'll combine the ProtoDash and the Partial Effects to obtain feature importances on the digits classifications task.\n\nProtoDash was proposed in _Gurumoorthy, Karthik & Dhurandhar, Amit & Cecchi, Guillermo & Aggarwal, Charu. (2019). Efficient Data Representation by Selecting Prototypes with Importance Weights. 260-269. 10.1109/ICDM.2019.00036_.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\n\n# automatically differentiable implementation of numpy\nimport jax.numpy as jnp # v0.2.13\n\nimport shap #0.34.0\n\nfrom sklearn.metrics import classification_report\nfrom sklearn import datasets\n\nfrom sklearn.model_selection import train_test_split\nfrom IPython.display import display\n\nimport matplotlib.pyplot as plt\n\nfrom itea.classification import ITEA_classifier\nfrom itea.inspection import *\n\nfrom sklearn.preprocessing import OneHotEncoder\n\nfrom aix360.algorithms.protodash import ProtodashExplainer #0.2.1\n\nimport warnings\nwarnings.filterwarnings(action='ignore', module=r'itea')",
"_____no_output_____"
],
[
"digits_data = datasets.load_digits(n_class=10)\n\nX, y = digits_data['data'], digits_data['target']\nlabels = digits_data['feature_names']\ntargets = digits_data['target_names']\n\nX /= X.max(axis=1).reshape(-1, 1)\n\nX_train, X_test, y_train, y_test = train_test_split(\n X, y, test_size=0.33, random_state=42)\n\ntfuncs = {\n 'id' : lambda x: x,\n 'sin': jnp.sin,\n 'cos': jnp.cos,\n 'tan': jnp.tan\n}\n\nclf = ITEA_classifier(\n gens = 100,\n popsize = 100,\n max_terms = 40,\n expolim = (0, 2),\n verbose = 10,\n tfuncs = tfuncs,\n labels = labels,\n simplify_method = None,\n random_state = 42,\n fit_kw = {'max_iter' : 5}\n).fit(X_train, y_train)\n\nfinal_itexpr = clf.bestsol_\nfinal_itexpr.selected_features_",
"gen | smallest fitness | mean fitness | highest fitness | remaining time\n----------------------------------------------------------------------------\n 0 | 0.105569 | 0.105569 | 0.105569 | 9min40sec \n 10 | 0.105569 | 0.105569 | 0.105569 | 9min8sec \n 20 | 0.105569 | 0.105669 | 0.107232 | 7min4sec \n 30 | 0.107232 | 0.111380 | 0.133001 | 6min41sec \n 40 | 0.133001 | 0.146708 | 0.152120 | 6min11sec \n 50 | 0.152120 | 0.154530 | 0.227764 | 6min19sec \n 60 | 0.227764 | 0.227839 | 0.230258 | 5min34sec \n 70 | 0.324190 | 0.335553 | 0.351621 | 4min23sec \n 80 | 0.351621 | 0.396259 | 0.428928 | 5min27sec \n 90 | 0.444722 | 0.467548 | 0.517872 | 2min53sec \n"
],
[
"print(classification_report(\n y_test,\n final_itexpr.predict(X_test),\n target_names=[str(t) for t in targets]\n))",
" precision recall f1-score support\n\n 0 0.54 0.64 0.58 55\n 1 0.70 0.80 0.75 55\n 2 0.53 0.44 0.48 52\n 3 0.29 0.71 0.41 56\n 4 0.69 0.69 0.69 64\n 5 0.61 0.52 0.56 73\n 6 0.58 0.49 0.53 57\n 7 0.43 0.53 0.48 62\n 8 0.40 0.19 0.26 52\n 9 0.27 0.04 0.08 68\n\n accuracy 0.50 594\n macro avg 0.51 0.51 0.48 594\nweighted avg 0.51 0.50 0.48 594\n\n"
]
],
[
[
"We can use the ``ITEA_summarizer`` to inspect the convergence during the evolution. In the cell below, we'll create 3 plots, one for the fitness (classification accuracy), one for the complexity (number of nodes if the IT expression was converted to a symbolic tree) and number of terms (number of IT terms of the solutions in the population for each generation).",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots(3, 1, figsize=(10, 8), sharex=True)\n\nsummarizer = ITEA_summarizer(itea=clf).fit(X_train, y_train).plot_convergence(\n data=['fitness', 'complexity', 'n_terms'],\n ax=ax,\n show=False\n)\n\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
],
[
"# features are named pixel_x_y. Lets extract those coordinates and\n# paint in a figure to show the selected features\n\nselected_features = np.zeros((8, 8))\nfor feature_name, feature_importance in zip(\n final_itexpr.selected_features_,\n np.sum(final_itexpr.feature_importances_, axis=0)\n):\n x, y = feature_name[-3], feature_name[-1]\n selected_features[int(x), int(y)] = feature_importance\n\nfig, axs = plt.subplots(1, 1, figsize=(3,3))\n\naxs.imshow(selected_features, cmap='gray_r')\naxs.set_title(f\"Selected features\")\n\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
],
[
"onehot_encoder = OneHotEncoder(sparse=False)\nonehot_encoded = onehot_encoder.fit_transform(\n np.hstack( (X_train, y_train.reshape(-1, 1)) ) )\n\nexplainer = ProtodashExplainer()\n\n# call protodash explainer. We'll select 10 prototypes\n# S contains indices of the selected prototypes\n# W contains importance weights associated with the selected prototypes \n(W, S, _) = explainer.explain(onehot_encoded, onehot_encoded, m=10)",
"elementwise comparison failed; returning scalar instead, but in the future will perform elementwise comparison\nelementwise comparison failed; returning scalar instead, but in the future will perform elementwise comparison\nelementwise comparison failed; returning scalar instead, but in the future will perform elementwise comparison\nelementwise comparison failed; returning scalar instead, but in the future will perform elementwise comparison\n"
],
[
"from matplotlib import cm\n\nfig, axs = plt.subplots(2, 5, figsize=(12,5))\n\n# Showing 10 prototypes\nfor s, ax in zip(S, fig.axes):\n ax.imshow(X_train[s].reshape(8, 8), cmap='gray_r')\n ax.set_title(f\"Prototype of class {y_train[s]}\")\n \n Z = X_train[s].reshape(8, 8)\n levels = [0.1, 0.2, 0.4]\n\n norm = cm.colors.Normalize(vmax=abs(Z).max(), vmin=-abs(Z).max())\n cmap = cm.PRGn\n\n cset2 = ax.contour(Z, levels, colors='y')\n\n for c in cset2.collections:\n c.set_linestyle('solid')\n\n\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
],
[
"it_explainer = ITExpr_explainer(\n itexpr=final_itexpr,\n tfuncs=tfuncs\n).fit(X_train, y_train)\n\nfig, axs = plt.subplots(2, 5, figsize=(12,5))\n\nfor s, ax in zip(S, fig.axes):\n \n importances = it_explainer.average_partial_effects(X_train[s, :].reshape(1, -1))[y_train[s]]\n \n ax.imshow(importances.reshape(8, 8), cmap='gray_r')\n ax.set_title(f\"Feature importance\\nprototype of class {y_train[s]}\")\n \n Z = X_train[s].reshape(8, 8)\n levels = [0.1, 0.2, 0.4]\n\n norm = cm.colors.Normalize(vmax=abs(Z).max(), vmin=-abs(Z).max())\n cmap = cm.PRGn\n\n cset2 = ax.contour(Z, levels, colors='y')\n\n for c in cset2.collections:\n c.set_linestyle('solid')\n \nplt.tight_layout()\nplt.show()",
"_____no_output_____"
],
[
"shap_explainer = shap.KernelExplainer(\n final_itexpr.predict,\n shap.sample(pd.DataFrame(X_train, columns=labels), 100)\n)\n\nfig, axs = plt.subplots(2, 5, figsize=(12,5))\n\nfor s, ax in zip(S, fig.axes):\n \n importances = np.abs(shap_explainer.shap_values(\n X_train[s, :].reshape(1, -1), silent=True, l1_reg='num_features(10)'))\n \n ax.imshow(importances.reshape(8, 8), cmap='gray_r')\n ax.set_title(f\"Feature importance\\nprototype of class {y_train[s]}\")\n \n Z = X_train[s].reshape(8, 8)\n levels = [0.1, 0.2, 0.4]\n\n norm = cm.colors.Normalize(vmax=abs(Z).max(), vmin=-abs(Z).max())\n cmap = cm.PRGn\n\n cset2 = ax.contour(Z, levels, colors='y')\n\n for c in cset2.collections:\n c.set_linestyle('solid')\n \nplt.tight_layout()\nplt.show()",
"_____no_output_____"
],
[
"it_explainer = ITExpr_explainer(\n itexpr=final_itexpr,\n tfuncs=tfuncs\n).fit(X_train, y_train)\n\nfig, axs = plt.subplots(2, 5, figsize=(12,5))\n\nfor c, ax in zip(final_itexpr.classes_, fig.axes):\n \n c_idx = np.array([i for i in range(len(y_train)) if y_train[i]==c])\n \n importances = it_explainer.average_partial_effects(X_train[c_idx, :])[c]\n \n ax.imshow(importances.reshape(8, 8), cmap='gray_r')\n ax.set_title(f\"Feature importance\\nprototype of class {c}\")\n \n Z = X_train[c_idx, :].mean(axis=0).reshape(8, 8)\n levels = [0.1, 0.2, 0.4]\n\n norm = cm.colors.Normalize(vmax=abs(Z).max(), vmin=-abs(Z).max())\n cmap = cm.PRGn\n\n cset2 = ax.contour(Z, levels, colors='y')\n\n for c in cset2.collections:\n c.set_linestyle('solid')\n \nplt.tight_layout()\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0d1cb5959dace70fa1e04afe8c3d8843fc55e4c | 164,285 | ipynb | Jupyter Notebook | quests/data-science-on-gcp-edition1_tf2/06_dataproc/quantization.ipynb | Glairly/introduction_to_tensorflow | aa0a44d9c428a6eb86d1f79d73f54c0861b6358d | [
"Apache-2.0"
] | 2 | 2022-01-06T11:52:57.000Z | 2022-01-09T01:53:56.000Z | quests/data-science-on-gcp-edition1_tf2/06_dataproc/quantization.ipynb | Glairly/introduction_to_tensorflow | aa0a44d9c428a6eb86d1f79d73f54c0861b6358d | [
"Apache-2.0"
] | null | null | null | quests/data-science-on-gcp-edition1_tf2/06_dataproc/quantization.ipynb | Glairly/introduction_to_tensorflow | aa0a44d9c428a6eb86d1f79d73f54c0861b6358d | [
"Apache-2.0"
] | null | null | null | 275.184255 | 90,619 | 0.911027 | [
[
[
"<h2> 6. Bayes Classification </h2>\n\nThis notebook has the code for the charts in Chapter 6\n",
"_____no_output_____"
],
[
"### Install BigQuery module\n\nYou don't need this on AI Platform, but you need this on plain-old JupyterLab",
"_____no_output_____"
]
],
[
[
"!pip install google-cloud-bigquery",
"_____no_output_____"
],
[
"%load_ext google.cloud.bigquery",
"_____no_output_____"
]
],
[
[
"### Setup",
"_____no_output_____"
]
],
[
[
"import os\nPROJECT = 'data-science-on-gcp-180606' # REPLACE WITH YOUR PROJECT ID\nBUCKET = 'data-science-on-gcp' # REPLACE WITH YOUR BUCKET NAME\nREGION = 'us-central1' # REPLACE WITH YOUR BUCKET REGION e.g. us-central1\n\nos.environ['BUCKET'] = BUCKET",
"_____no_output_____"
]
],
[
[
"<h3> Exploration using BigQuery </h3>",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport seaborn as sns\nimport pandas as pd\nimport numpy as np\nimport google.cloud.bigquery as bigquery\n\nbq = bigquery.Client()",
"_____no_output_____"
],
[
"sql = \"\"\"\nSELECT DISTANCE, DEP_DELAY\nFROM `flights.tzcorr`\nWHERE RAND() < 0.001 AND dep_delay > -20 AND dep_delay < 30 AND distance < 2000\n\"\"\"\ndf = bq.query(sql).to_dataframe()",
"_____no_output_____"
],
[
"sns.set_style(\"whitegrid\")\ng = sns.jointplot(df['DISTANCE'], df['DEP_DELAY'], kind=\"hex\", size=10, joint_kws={'gridsize':20})",
"_____no_output_____"
]
],
[
[
"<h3> Set up views in Spark SQL </h3>\n\nStart a Spark Session if necessary and get a handle to it.",
"_____no_output_____"
]
],
[
[
"from pyspark.sql import SparkSession\nspark = SparkSession \\\n .builder \\\n .appName(\"Bayes classification using Spark\") \\\n .getOrCreate()\nprint(spark)",
"<pyspark.sql.session.SparkSession object at 0x7f6e9c2088d0>\n"
]
],
[
[
"Set up the schema to read in the CSV files on GCS",
"_____no_output_____"
]
],
[
[
"from pyspark.sql.types import StringType, FloatType, StructType, StructField\n\nheader = 'FL_DATE,UNIQUE_CARRIER,AIRLINE_ID,CARRIER,FL_NUM,ORIGIN_AIRPORT_ID,ORIGIN_AIRPORT_SEQ_ID,ORIGIN_CITY_MARKET_ID,ORIGIN,DEST_AIRPORT_ID,DEST_AIRPORT_SEQ_ID,DEST_CITY_MARKET_ID,DEST,CRS_DEP_TIME,DEP_TIME,DEP_DELAY,TAXI_OUT,WHEELS_OFF,WHEELS_ON,TAXI_IN,CRS_ARR_TIME,ARR_TIME,ARR_DELAY,CANCELLED,CANCELLATION_CODE,DIVERTED,DISTANCE,DEP_AIRPORT_LAT,DEP_AIRPORT_LON,DEP_AIRPORT_TZOFFSET,ARR_AIRPORT_LAT,ARR_AIRPORT_LON,ARR_AIRPORT_TZOFFSET,EVENT,NOTIFY_TIME'\n\ndef get_structfield(colname):\n if colname in ['ARR_DELAY', 'DEP_DELAY', 'DISTANCE']:\n return StructField(colname, FloatType(), True)\n else:\n return StructField(colname, StringType(), True)\n\nschema = StructType([get_structfield(colname) for colname in header.split(',')])\nprint(schema)",
"StructType(List(StructField(FL_DATE,StringType,true),StructField(UNIQUE_CARRIER,StringType,true),StructField(AIRLINE_ID,StringType,true),StructField(CARRIER,StringType,true),StructField(FL_NUM,StringType,true),StructField(ORIGIN_AIRPORT_ID,StringType,true),StructField(ORIGIN_AIRPORT_SEQ_ID,StringType,true),StructField(ORIGIN_CITY_MARKET_ID,StringType,true),StructField(ORIGIN,StringType,true),StructField(DEST_AIRPORT_ID,StringType,true),StructField(DEST_AIRPORT_SEQ_ID,StringType,true),StructField(DEST_CITY_MARKET_ID,StringType,true),StructField(DEST,StringType,true),StructField(CRS_DEP_TIME,StringType,true),StructField(DEP_TIME,StringType,true),StructField(DEP_DELAY,FloatType,true),StructField(TAXI_OUT,StringType,true),StructField(WHEELS_OFF,StringType,true),StructField(WHEELS_ON,StringType,true),StructField(TAXI_IN,StringType,true),StructField(CRS_ARR_TIME,StringType,true),StructField(ARR_TIME,StringType,true),StructField(ARR_DELAY,FloatType,true),StructField(CANCELLED,StringType,true),StructField(CANCELLATION_CODE,StringType,true),StructField(DIVERTED,StringType,true),StructField(DISTANCE,FloatType,true),StructField(DEP_AIRPORT_LAT,StringType,true),StructField(DEP_AIRPORT_LON,StringType,true),StructField(DEP_AIRPORT_TZOFFSET,StringType,true),StructField(ARR_AIRPORT_LAT,StringType,true),StructField(ARR_AIRPORT_LON,StringType,true),StructField(ARR_AIRPORT_TZOFFSET,StringType,true),StructField(EVENT,StringType,true),StructField(NOTIFY_TIME,StringType,true)))\n"
]
],
[
[
"Create a table definition (this is done lazily; the files won't be read until we issue a query):",
"_____no_output_____"
]
],
[
[
"inputs = 'gs://{}/flights/tzcorr/all_flights-00000-*'.format(BUCKET) # 1/30th\n#inputs = 'gs://{}/flights/tzcorr/all_flights-*'.format(BUCKET) # FULL\nflights = spark.read\\\n .schema(schema)\\\n .csv(inputs)\n\n# this view can now be queried ...\nflights.createOrReplaceTempView('flights')",
"_____no_output_____"
]
],
[
[
"Example query over the view (this will take a while; it's Spark SQL, not BigQuery):",
"_____no_output_____"
]
],
[
[
"results = spark.sql('SELECT COUNT(*) FROM flights WHERE dep_delay > -20 AND distance < 2000')\nresults.show()",
"+--------+\n|count(1)|\n+--------+\n| 8813|\n+--------+\n\n"
]
],
[
[
"<h2> Restrict to train days </h2>",
"_____no_output_____"
],
[
"Let's create a CSV file of the training days",
"_____no_output_____"
]
],
[
[
"sql = \"\"\"\nSELECT *\nFROM `flights.trainday`\n\"\"\"\ndf = bq.query(sql).to_dataframe()\ndf.to_csv('trainday.csv', index=False)",
"_____no_output_____"
],
[
"!head -3 trainday.csv",
"FL_DATE,is_train_day\n2015-01-01,True\n2015-01-04,True\n"
],
[
"%%bash\ngsutil cp trainday.csv gs://${BUCKET}/flights/trainday.csv",
"Copying file://trainday.csv [Content-Type=text/csv]...\n/ [1 files][ 6.3 KiB/ 6.3 KiB] \nOperation completed over 1 objects/6.3 KiB. \n"
]
],
[
[
"Create dataframe of traindays, but this time because the file has a header, and is a small file, we can have Spark infer the schema",
"_____no_output_____"
]
],
[
[
"traindays = spark.read \\\n .option(\"header\", \"true\") \\\n .option(\"inferSchema\", \"true\") \\\n .csv('gs://{}/flights/trainday.csv'.format(BUCKET))\n\ntraindays.createOrReplaceTempView('traindays')",
"_____no_output_____"
],
[
"results = spark.sql('SELECT * FROM traindays')\nresults.head(5)",
"_____no_output_____"
],
[
"statement = \"\"\"\nSELECT\n f.FL_DATE AS date,\n distance,\n dep_delay\nFROM flights f\nJOIN traindays t\nON f.FL_DATE == t.FL_DATE\nWHERE\n t.is_train_day AND\n f.dep_delay IS NOT NULL\nORDER BY\n f.dep_delay DESC\n\"\"\"\nflights = spark.sql(statement)",
"_____no_output_____"
]
],
[
[
"<h3> Hexbin plot </h3>\n\nCreate a hexbin plot using Spark (repeat of what we did in BigQuery, except that we are now restricting to train days only).",
"_____no_output_____"
]
],
[
[
"df = flights[(flights['distance'] < 2000) & (flights['dep_delay'] > -20) & (flights['dep_delay'] < 30)]\ndf.describe().show()",
"+-------+----------+-----------------+-------------------+\n|summary| date| distance| dep_delay|\n+-------+----------+-----------------+-------------------+\n| count| 8311| 8311| 8311|\n| mean| null|748.7310792925039|-1.4972927445554085|\n| stddev| null|441.1737503226965| 7.327984777613346|\n| min|2015-11-26| 31.0| -19.0|\n| max|2015-11-27| 1999.0| 29.0|\n+-------+----------+-----------------+-------------------+\n\n"
]
],
[
[
"Sample the dataframe so that it fits into memory (not a problem in development, but will be on full dataset); then plot it.",
"_____no_output_____"
]
],
[
[
"pdf = df.sample(False, 0.02, 20).toPandas() # to 100,000 rows approx on complete dataset\ng = sns.jointplot(pdf['distance'], pdf['dep_delay'], kind=\"hex\", size=10, joint_kws={'gridsize':20})",
"_____no_output_____"
]
],
[
[
"<h3> Quantization </h3>\n\nNow find the quantiles",
"_____no_output_____"
]
],
[
[
"distthresh = flights.approxQuantile('distance', list(np.arange(0, 1.0, 0.1)), 0.02)\ndistthresh",
"_____no_output_____"
],
[
"delaythresh = flights.approxQuantile('dep_delay', list(np.arange(0, 1.0, 0.1)), 0.05)\ndelaythresh",
"_____no_output_____"
],
[
"results = spark.sql('SELECT COUNT(*) FROM flights WHERE dep_delay >= 3 AND dep_delay < 8 AND distance >= 447 AND distance < 557')\nresults.show()",
"+--------+\n|count(1)|\n+--------+\n| 62|\n+--------+\n\n"
]
],
[
[
"<h2> Repeat, but on full dataset </h2>\n\nYou can launch the above processing on the full dataset from within JupyterLab if you want the statistics and graphs updated. I didn't, though, because this is not what I would have really done. Instead, \nI would have created a standalone Python script and submitted it to the cluster -- there is no need to put JupyterLab in the middle of a production process. We'll submit a standalone Pig program to the cluster in the next section.\n\nSteps:\n<ol>\n<li> Change the input variable to process all-flights-* </li>\n<li> Increase cluster size (bash increase_cluster.sh from CloudShell) </li>\n<li> Clear all cells from this notebook </li>\n<li> Run all cells </li>\n<li> Decrease cluster size (bash decrease_cluster.sh from CloudShell) </li>\n</ol>\n",
"_____no_output_____"
],
[
"Copyright 2019 Google Inc. Licensed under the Apache License, Version 2.0 (the \"License\"); you may not use this file except in compliance with the License. You may obtain a copy of the License at http://www.apache.org/licenses/LICENSE-2.0 Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an \"AS IS\" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
d0d1d29240034d8d7437de7dea7ecc33599998bf | 41,230 | ipynb | Jupyter Notebook | Repo_Data_v2.1.ipynb | Watwell/Repo_Code | f40cfdd000718e5a98b1e30382bf337322b45571 | [
"BSD-2-Clause"
] | 1 | 2021-03-06T09:59:53.000Z | 2021-03-06T09:59:53.000Z | Repo_Data_v2.1.ipynb | Watwell/Repo_Code | f40cfdd000718e5a98b1e30382bf337322b45571 | [
"BSD-2-Clause"
] | null | null | null | Repo_Data_v2.1.ipynb | Watwell/Repo_Code | f40cfdd000718e5a98b1e30382bf337322b45571 | [
"BSD-2-Clause"
] | null | null | null | 84.661191 | 23,380 | 0.741159 | [
[
[
"import pandas as pd\nfrom datetime import timedelta, date\nimport matplotlib.pyplot as plt\n\ndef append_it(date, amount,treasury,Agency,MBS, duration):\n append_data = {'Date':[date], 'Amount':[amount], 'Duration':[duration],'Treasury':[treasury],'Agency':[Agency], 'MBS':[MBS]}\n append_df = pd.DataFrame(append_data)\n return append_df",
"_____no_output_____"
],
[
"print(repos)",
"_____no_output_____"
],
[
"data = {'Date':[date(2019, 9, 17)], 'Amount':[53.15], 'Treasury':[40.85], 'Agency':[0.6], 'MBS':[11.7], 'Duration':[1]} \nrepos = pd.DataFrame(data)\nrepos = repos.append(append_it(date(2019, 9, 18),75,51.55,0.7,22.75,1))\nrepos = repos.append(append_it(date(2019, 9, 19),75,55.843,0,19.157,1))\nrepos = repos.append(append_it(date(2019, 9, 20),75,59.6,0.5,15.350,3))\nrepos = repos.append(append_it(date(2019, 9, 23),67.75,49.7,0.6,15.45,1))\nrepos = repos.append(append_it(date(2019, 9, 24),30,22.732,0,7.268,14))\nrepos = repos.append(append_it(date(2019, 9, 24),75,58.75,.36,15.49,1))\nrepos = repos.append(append_it(date(2019, 9, 25),75,44.35,1,29.65,1))\nrepos = repos.append(append_it(date(2019, 9, 26),60,35.75,0,24.25,14))\nrepos = repos.append(append_it(date(2019, 9, 26),50.1,34.55,0,15.55,1))\nrepos = repos.append(append_it(date(2019, 9, 27),49,34.55,0,14.45,14))\nrepos = repos.append(append_it(date(2019, 9, 27),22.7,14.45,0,8.25,3))\nrepos = repos.append(append_it(date(2019, 9, 30),63.5,49.75,0,13.75,1))\nrepos = repos.append(append_it(date(2019, 10, 1),54.85,50.0,0.1,4.75,1))\nrepos = repos.append(append_it(date(2019, 10, 2),42.05,35.0,0,7.05,1))\nrepos = repos.append(append_it(date(2019, 10, 3),33.55,28.0,0,5.55,1))\nrepos = repos.append(append_it(date(2019, 10, 4),38.55,29.5,0,9.05,3))\nrepos = repos.append(append_it(date(2019, 10, 7),38.85,36.0,0,11.05,1))\nrepos = repos.append(append_it(date(2019, 10, 8),38.85,29.3,0,9.55,14))\nrepos = repos.append(append_it(date(2019, 10, 8),37.5,31.75,0,5.75,1))\nrepos = repos.append(append_it(date(2019, 10, 9),30.8,26.25,0,4.55,1))\nrepos = repos.append(append_it(date(2019, 10, 10),42.6,30.7,0,11.9,14))\nrepos = repos.append(append_it(date(2019, 10, 10),45.5,37.6,0.5,7.4,1))\nrepos = repos.append(append_it(date(2019, 10, 11),21.15,13.15,0,8.0,6))\nrepos = repos.append(append_it(date(2019, 10, 11),61.55,58.35,0,3.2,4))\nrepos = repos.append(append_it(date(2019, 10, 15),20.1,10.6,0,9.5,14))\nrepos = repos.append(append_it(date(2019, 10, 15),67.6,59.95,0,7.65,1))\nrepos = repos.append(append_it(date(2019, 10, 16),75,72.592,0,2.408,1))\nrepos = repos.append(append_it(date(2019, 10, 17),30.65,18.15,0,12.5,15))\nrepos = repos.append(append_it(date(2019, 10, 17),73.5,67.7,0.1,5.7,1))\nrepos = repos.append(append_it(date(2019, 10, 18),56.65,47.95,0.5,8.2,3))\nrepos = repos.append(append_it(date(2019, 10, 21),58.15,50.95,0.5,6.7,1))\nrepos = repos.append(append_it(date(2019, 10, 22),35,31.141,0,3.859,14))\nrepos = repos.append(append_it(date(2019, 10, 22),64.904,54.404,0,9.5,1))\nrepos = repos.reset_index(drop=True)\nrepos.tail(10)",
"_____no_output_____"
],
[
"\n\ndef daterange(start_date, end_date):\n for n in range(int ((end_date - start_date).days)):\n yield start_date + timedelta(n)\n#repos_amount = pd.DataFrame(columns=['foo', 'bar'])\nrepos_amount = pd.DataFrame(columns=['Date', 'Amount','Treasury','Agency', 'MBS'])\nstart_date = date(2019, 9, 17)\n\n# ***** Make it one higher than you need *****\n# ***** Make it one higher than you need *****\n# ***** Make it one higher than you need *****\nend_date = date(2019, 10, 22+1) \nfor single_date in daterange(start_date, end_date):\n append_data = {'Date':[single_date], 'Amount': 0,'Treasury':0,'Agency':0, 'MBS':0}\n append_df = pd.DataFrame(append_data)\n #print(\"Append:\")\n #print(append_df)\n #print(repos_amount)\n repos_amount = repos_amount.append(append_df)\n\nrepos_amount.set_index('Date', inplace=True)\nprint(repos_amount)\n\ndef update_row(row, df):\n the_date = row['Date']\n the_amount = row['Amount']\n the_duration = row['Duration']\n the_treasury = row['Treasury']\n the_agency = row['Agency']\n the_MBS = row['MBS']\n end_date = the_date + timedelta(the_duration)\n the_date = date(the_date.year, the_date.month, the_date.day)\n #the_date = \n end_date = date(end_date.year, end_date.month, end_date.day)\n for date_var in daterange(the_date, end_date):\n date_lookup = date(date_var.year, date_var.month, date_var.day)\n last_date = df.tail(1).index[0]\n if last_date >= date_lookup:\n #current_amount = df.loc[date_lookup]['Amount']\n #new_amount = df.loc[date_lookup]['Amount'] + the_amount\n df.loc[date_lookup]['Amount'] = df.loc[date_lookup]['Amount'] + the_amount\n df.loc[date_lookup]['Treasury'] = df.loc[date_lookup]['Treasury'] + the_treasury\n df.loc[date_lookup]['Agency'] = df.loc[date_lookup]['Agency'] + the_agency\n df.loc[date_lookup]['MBS'] = df.loc[date_lookup]['MBS'] + the_MBS\n #current_treasury = df.loc[date_lookup]['Treasury']\n #new_treasury = current_treasury + the_treasury\n #df.loc[date_lookup]['Treasury'] = new_treasury\n #current_agency = df.loc[date_lookup]['Agency']\n #new_agency = current_agency + the_amount\n #df.loc[date_lookup]['Agency'] = new_agency\n #current_amount = df.loc[date_lookup]['Amount']\n #new_amount = current_amount + the_amount\n #df.loc[date_lookup]['Amount'] = new_amount\n return df",
" Agency Amount MBS Treasury\nDate \n2019-09-17 0 0 0 0\n2019-09-18 0 0 0 0\n2019-09-19 0 0 0 0\n2019-09-20 0 0 0 0\n2019-09-21 0 0 0 0\n2019-09-22 0 0 0 0\n2019-09-23 0 0 0 0\n2019-09-24 0 0 0 0\n2019-09-25 0 0 0 0\n2019-09-26 0 0 0 0\n2019-09-27 0 0 0 0\n2019-09-28 0 0 0 0\n2019-09-29 0 0 0 0\n2019-09-30 0 0 0 0\n2019-10-01 0 0 0 0\n2019-10-02 0 0 0 0\n2019-10-03 0 0 0 0\n2019-10-04 0 0 0 0\n2019-10-05 0 0 0 0\n2019-10-06 0 0 0 0\n2019-10-07 0 0 0 0\n2019-10-08 0 0 0 0\n2019-10-09 0 0 0 0\n2019-10-10 0 0 0 0\n2019-10-11 0 0 0 0\n2019-10-12 0 0 0 0\n2019-10-13 0 0 0 0\n2019-10-14 0 0 0 0\n2019-10-15 0 0 0 0\n2019-10-16 0 0 0 0\n2019-10-17 0 0 0 0\n2019-10-18 0 0 0 0\n2019-10-19 0 0 0 0\n2019-10-20 0 0 0 0\n2019-10-21 0 0 0 0\n2019-10-22 0 0 0 0\n"
],
[
"for index, a_row in repos.iterrows():\n repos_amount = update_row(a_row, repos_amount)\n #print(repos_amount)",
"_____no_output_____"
],
[
"#repos_amount.plot(kind='bar',y='Amount',color='red')\ncolors = [\"Green\", \"Red\",\"Blue\"]\nrepos_amount[['Agency','MBS','Treasury']].plot.bar(stacked=True, color=colors, figsize=(12,7))\nplt.title('Total Outstanding Fed Repos', fontsize=16)\nplt.ylabel('$ Billions', fontsize=12)\nplt.show()",
"_____no_output_____"
],
[
"print(repos_amount)",
" Agency Amount MBS Treasury\nDate \n2019-09-17 0.6 53.15 11.7 40.85\n2019-09-18 0.7 75 22.75 51.55\n2019-09-19 0 75 19.157 55.843\n2019-09-20 0.5 75 15.35 59.6\n2019-09-21 0.5 75 15.35 59.6\n2019-09-22 0.5 75 15.35 59.6\n2019-09-23 0.6 67.75 15.45 49.7\n2019-09-24 0.36 105 22.758 81.482\n2019-09-25 1 105 36.918 67.082\n2019-09-26 0 140.1 47.068 93.032\n2019-09-27 0 161.7 54.218 107.482\n2019-09-28 0 161.7 54.218 107.482\n2019-09-29 0 161.7 54.218 107.482\n2019-09-30 0 202.5 59.718 142.782\n2019-10-01 0.1 193.85 50.718 143.032\n2019-10-02 0 181.05 53.018 128.032\n2019-10-03 0 172.55 51.518 121.032\n2019-10-04 0 177.55 55.018 122.532\n2019-10-05 0 177.55 55.018 122.532\n2019-10-06 0 177.55 55.018 122.532\n2019-10-07 0 177.85 57.018 129.032\n2019-10-08 0 185.35 54 131.35\n2019-10-09 0 178.65 52.8 125.85\n2019-10-10 0.5 175.95 43.3 132.15\n2019-10-11 0 164.15 32.65 131.5\n2019-10-12 0 164.15 32.65 131.5\n2019-10-13 0 164.15 32.65 131.5\n2019-10-14 0 164.15 32.65 131.5\n2019-10-15 0 190.3 46.6 143.7\n2019-10-16 0 197.7 41.358 156.342\n2019-10-17 0.1 205.7 49.15 156.45\n2019-10-18 0.5 188.85 51.65 136.7\n2019-10-19 0.5 188.85 51.65 136.7\n2019-10-20 0.5 188.85 51.65 136.7\n2019-10-21 0.5 190.35 50.15 139.7\n"
],
[
"print(repos_amount.loc[date(2019, 10, 11)])",
"_____no_output_____"
],
[
"repos_amount.set_index('Date', inplace=True)",
"_____no_output_____"
],
[
"repos_amount.info()",
"_____no_output_____"
],
[
"#print(repos_amount.iloc[1, :]['Amount'])\namount = repos_amount.loc[date(2019, 10, 11)]['Amount']\namount = amount + 10\nrepos_amount.set_value(date(2019, 10, 11), 'Amount', amount)\nprint(repos_amount)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0d1dd493654fcbeea306afb07194b6b6fde4b90 | 3,463 | ipynb | Jupyter Notebook | Q-Table.ipynb | 4SkyNet/DeepRL-Agents | 77060c3216e28f08dcf696d077bc7328700c0827 | [
"MIT"
] | 14 | 2017-02-06T09:16:20.000Z | 2021-06-11T08:34:39.000Z | Q-Table.ipynb | hunkim/DeepRL-Agents | 8c5be0b74b2d63c60f0175b9721d1594747f0c2d | [
"MIT"
] | null | null | null | Q-Table.ipynb | hunkim/DeepRL-Agents | 8c5be0b74b2d63c60f0175b9721d1594747f0c2d | [
"MIT"
] | 12 | 2017-05-26T14:18:42.000Z | 2021-06-11T08:34:43.000Z | 24.216783 | 267 | 0.531042 | [
[
[
"# Simple Reinforcement Learning with Tensorflow: Part 0 - Q-Tables\nIn this iPython notebook we implement a Q-Table algorithm that solves the FrozenLake problem. To learn more, read here: https://medium.com/@awjuliani/simple-reinforcement-learning-with-tensorflow-part-0-q-learning-with-tables-and-neural-networks-d195264329d0\n\nFor more reinforcment learning tutorials, see:\nhttps://github.com/awjuliani/DeepRL-Agents",
"_____no_output_____"
]
],
[
[
"import gym\nimport numpy as np\nimport random\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"### Load the environment",
"_____no_output_____"
]
],
[
[
"env = gym.make('FrozenLake-v0')",
"_____no_output_____"
]
],
[
[
"### Implement Q-Table learning algorithm",
"_____no_output_____"
]
],
[
[
"#Initialize table with all zeros\nQ = np.zeros([env.observation_space.n,env.action_space.n])\n# Set learning parameters\nlr = .85\ny = .99\nnum_episodes = 2000\n#create lists to contain total rewards and steps per episode\n#jList = []\nrList = []\nfor i in range(num_episodes):\n #Reset environment and get first new observation\n s = env.reset()\n rAll = 0\n d = False\n j = 0\n #The Q-Table learning algorithm\n while j < 99:\n j+=1\n #Choose an action by greedily (with noise) picking from Q table\n a = np.argmax(Q[s,:] + np.random.randn(1,env.action_space.n)*(1./(i+1)))\n #Get new state and reward from environment\n s1,r,d,_ = env.step(a)\n #Update Q-Table with new knowledge\n Q[s,a] = Q[s,a] + lr*(r + y*np.max(Q[s1,:]) - Q[s,a])\n rAll += r\n s = s1\n if d == True:\n break\n #jList.append(j)\n rList.append(rAll)",
"_____no_output_____"
],
[
"print \"Score over time: \" + str(sum(rList)/num_episodes)",
"_____no_output_____"
],
[
"print \"Final Q-Table Values\"\nprint Q",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d0d20196fb458841874495a8dbee15d66c98471a | 860,870 | ipynb | Jupyter Notebook | Week05/06 - Time series analysis (Pandas).ipynb | koldunovn/HCU | ae4bf54a8dfa59a89496840e293b7a1d14a4adc1 | [
"CC-BY-3.0"
] | 9 | 2017-12-02T05:39:11.000Z | 2021-08-25T13:33:05.000Z | Week05/06 - Time series analysis (Pandas).ipynb | koldunovn/HCU | ae4bf54a8dfa59a89496840e293b7a1d14a4adc1 | [
"CC-BY-3.0"
] | null | null | null | Week05/06 - Time series analysis (Pandas).ipynb | koldunovn/HCU | ae4bf54a8dfa59a89496840e293b7a1d14a4adc1 | [
"CC-BY-3.0"
] | 8 | 2016-09-21T01:49:27.000Z | 2021-08-25T13:33:05.000Z | 398.920297 | 231,962 | 0.923628 | [
[
[
"# Time series analysis (Pandas)",
"_____no_output_____"
],
[
"Nikolay Koldunov\n\[email protected]",
"_____no_output_____"
],
[
"================",
"_____no_output_____"
],
[
"Here I am going to show just some basic [pandas](http://pandas.pydata.org/) stuff for time series analysis, as I think for the Earth Scientists it's the most interesting topic. If you find this small tutorial useful, I encourage you to watch [this video](http://pyvideo.org/video/1198/time-series-data-analysis-with-pandas), where Wes McKinney give extensive introduction to the time series data analysis with pandas.\n\nOn the official website you can find explanation of what problems pandas solve in general, but I can tell you what problem pandas solve for me. It makes analysis and visualisation of 1D data, especially time series, MUCH faster. Before pandas working with time series in python was a pain for me, now it's fun. Ease of use stimulate in-depth exploration of the data: why wouldn't you make some additional analysis if it's just one line of code? Hope you will also find this great tool helpful and useful. So, let's begin.",
"_____no_output_____"
],
[
"As an example we are going to use time series of [Arctic Oscillation (AO)](http://en.wikipedia.org/wiki/Arctic_oscillation) and [North Atlantic Oscillation (NAO)](http://en.wikipedia.org/wiki/North_Atlantic_oscillation) data sets.",
"_____no_output_____"
],
[
"## Module import",
"_____no_output_____"
],
[
"First we have to import necessary modules:",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\n%matplotlib inline\npd.set_option('max_rows',15) # this limit maximum numbers of rows\nnp.set_printoptions(precision=3 , suppress= True) # this is just to make the output look better",
"_____no_output_____"
],
[
"pd.__version__",
"_____no_output_____"
]
],
[
[
"## Loading data",
"_____no_output_____"
],
[
"Now, when we are done with preparations, let's get some data.",
"_____no_output_____"
],
[
"Pandas has very good IO capabilities, but we not going to use them in this tutorial in order to keep things simple. For now we open the file simply with numpy loadtxt:",
"_____no_output_____"
]
],
[
[
"temp = np.loadtxt('../Week03/Ham_3column.txt')",
"_____no_output_____"
]
],
[
[
"Every line in the file consist of three elements: year, month, value:",
"_____no_output_____"
]
],
[
[
"temp[-1]",
"_____no_output_____"
]
],
[
[
"And here is the shape of our array (note that shape of the file might differ in your case, since data updated monthly):",
"_____no_output_____"
]
],
[
[
"temp.shape",
"_____no_output_____"
]
],
[
[
"## Time Series",
"_____no_output_____"
],
[
"We would like to convert this data in to time series, that can be manipulated naturally and easily. First step, that we have to do is to create the range of dates for our time series. From the file it is clear, that record starts at January 1891 and ends at August 2014 (at the time I am writing this, of course). Frequency of the data is one day (freq='D'). ",
"_____no_output_____"
]
],
[
[
"dates = pd.date_range('1891-01-01', '2014-08-31', freq='D')",
"_____no_output_____"
]
],
[
[
"As you see syntax is quite simple, and this is one of the reasons why I love Pandas so much :) You can check if the range of dates is properly generated:",
"_____no_output_____"
]
],
[
[
"dates",
"_____no_output_____"
]
],
[
[
"Now we are ready to create our first time series. Dates from the *dates* variable will be our index, and `temp` values will be our, hm... values:",
"_____no_output_____"
]
],
[
[
"ham = pd.Series(temp[:,3]/10., index=dates)",
"_____no_output_____"
],
[
"ham",
"_____no_output_____"
]
],
[
[
"Now we can plot complete time series:",
"_____no_output_____"
]
],
[
[
"ham.plot()",
"_____no_output_____"
]
],
[
[
"or its part:",
"_____no_output_____"
]
],
[
[
"ham['1980':'1990'].plot()",
"_____no_output_____"
]
],
[
[
"or even smaller part:",
"_____no_output_____"
]
],
[
[
"ham['1980-05-02':'1981-03-17'].plot()",
"_____no_output_____"
]
],
[
[
"Reference to the time periods is done in a very natural way. You, of course, can also get individual values. By number: ",
"_____no_output_____"
]
],
[
[
"ham[120]",
"_____no_output_____"
]
],
[
[
"or by index (date in our case):",
"_____no_output_____"
]
],
[
[
"ham['1960-01']",
"_____no_output_____"
]
],
[
[
"And what if we choose only one year?",
"_____no_output_____"
]
],
[
[
"ham['1960']",
"_____no_output_____"
]
],
[
[
"Isn't that great? :)",
"_____no_output_____"
],
[
"##Exercise\n\nWhat was temperature in Hampurg at your burthsday?",
"_____no_output_____"
],
[
"## One bonus example :)",
"_____no_output_____"
]
],
[
[
"ham[ham > 0]['1990':'2000'].plot(style='r*')\nham[ham < 0]['1990':'2000'].plot(style='b*')",
"_____no_output_____"
]
],
[
[
"##Exercise\n\n- plot all positive temperatures (red stars) and negative temperatires (blue stars)\n- limit this plot by 1990-2000 period",
"_____no_output_____"
],
[
"## Data Frame",
"_____no_output_____"
],
[
"Now let's make live a bit more interesting and get more data. This will be TMIN time series.",
"_____no_output_____"
],
[
"We use pandas function `read_csv` to parse dates and create Data Frame",
"_____no_output_____"
]
],
[
[
"hamm = pd.read_csv('Ham_tmin.txt', parse_dates=True, index_col=0, names=['Time','tmin'])",
"_____no_output_____"
],
[
"hamm",
"_____no_output_____"
],
[
"type(hamm)",
"_____no_output_____"
]
],
[
[
"Time period is the same:",
"_____no_output_____"
]
],
[
[
"hamm.index",
"_____no_output_____"
]
],
[
[
"Now we create Data Frame, that will contain both TMAX and TMIN data. It is sort of an Excel table where the first row contain headers for the columns and firs column is an index:",
"_____no_output_____"
]
],
[
[
"tmp = pd.DataFrame({'TMAX':ham, 'TMIN':hamm.tmin/10})",
"_____no_output_____"
],
[
"tmp",
"_____no_output_____"
]
],
[
[
"One can plot the data straight away:",
"_____no_output_____"
]
],
[
[
"tmp.plot()",
"_____no_output_____"
]
],
[
[
"Or have a look at the first several rows:",
"_____no_output_____"
]
],
[
[
"tmp.head()",
"_____no_output_____"
]
],
[
[
"We can reference each column by its name:",
"_____no_output_____"
]
],
[
[
"tmp['TMIN']",
"_____no_output_____"
]
],
[
[
"or as method of the Data Frame variable (if name of the variable is a valid python name):",
"_____no_output_____"
]
],
[
[
"tmp.TMIN",
"_____no_output_____"
]
],
[
[
"We can simply add column to the Data Frame:",
"_____no_output_____"
]
],
[
[
"tmp['Diff'] = tmp['TMAX'] - tmp['TMIN']\ntmp.head()",
"_____no_output_____"
]
],
[
[
"##Exercise\nFind and plot all differences that are larger than 20",
"_____no_output_____"
]
],
[
[
"tmp['Diff'][tmp['Diff']>20].plot(style='r*')",
"_____no_output_____"
]
],
[
[
"And delete it:",
"_____no_output_____"
]
],
[
[
"del tmp['Diff']\ntmp.tail()",
"_____no_output_____"
]
],
[
[
"Slicing will also work:",
"_____no_output_____"
]
],
[
[
"tmp['1981-03'].plot()",
"_____no_output_____"
]
],
[
[
"## Statistics",
"_____no_output_____"
],
[
"Back to simple stuff. We can obtain statistical information over elements of the Data Frame. Default is column wise:",
"_____no_output_____"
]
],
[
[
"tmp.mean()",
"_____no_output_____"
],
[
"tmp.max()",
"_____no_output_____"
],
[
"tmp.min()",
"_____no_output_____"
]
],
[
[
"You can also do it row-wise:",
"_____no_output_____"
]
],
[
[
"tmp.mean(1)",
"_____no_output_____"
]
],
[
[
"Or get everything at once:",
"_____no_output_____"
]
],
[
[
"tmp.describe()",
"_____no_output_____"
]
],
[
[
"By the way getting correlation coefficients for members of the Data Frame is as simple as:",
"_____no_output_____"
]
],
[
[
"tmp.corr()",
"_____no_output_____"
]
],
[
[
"##Exercise\nFind mean of all temperatures larger than 5",
"_____no_output_____"
]
],
[
[
"tmp[tmp>5].mean()",
"_____no_output_____"
]
],
[
[
"## Resampling",
"_____no_output_____"
],
[
"Pandas provide easy way to resample data to different time frequency. Two main parameters for resampling is time period you resemple to and the method that you use. By default the method is mean. Following example calculates monthly ('M'):",
"_____no_output_____"
]
],
[
[
"tmp_mm = tmp.resample(\"M\")\ntmp_mm['2000':].plot()",
"_____no_output_____"
]
],
[
[
"median:",
"_____no_output_____"
]
],
[
[
"tmp_mm = tmp.resample(\"M\", how='median')\ntmp_mm['2000':].plot()",
"_____no_output_____"
]
],
[
[
"You can use your methods for resampling, for example np.max (in this case we change resampling frequency to 3 years):",
"_____no_output_____"
]
],
[
[
"tmp_mm = tmp.resample(\"3M\", how=np.max)\ntmp_mm['2000':].plot()",
"_____no_output_____"
]
],
[
[
"You can specify several functions at once as a list:",
"_____no_output_____"
]
],
[
[
"tmp_mm = tmp.resample(\"M\", how=['mean', np.min, np.max])\ntmp_mm['1900':'2020'].plot(subplots=True, figsize=(10,10))\ntmp_mm['2000':].plot(figsize=(10,10))",
"_____no_output_____"
]
],
[
[
"##Exercise\nDefine function that will find difference between maximum and minimum values of the time series, and resample our `tmp` variable with this function.",
"_____no_output_____"
]
],
[
[
"def satardays(x):\n xmin = x.min()\n xmax = x.max()\n diff = xmin - xmax\n return diff",
"_____no_output_____"
],
[
"tmp_mm = tmp.resample(\"A\", how=satardays)\ntmp_mm['2000':].plot()",
"_____no_output_____"
],
[
"tmp_mm",
"_____no_output_____"
]
],
[
[
"That's it. I hope you at least get a rough impression of what pandas can do for you. Comments are very welcome (below). If you have intresting examples of pandas usage in Earth Science, we would be happy to put them on [EarthPy](http://earthpy.org).",
"_____no_output_____"
],
[
"## Links",
"_____no_output_____"
],
[
"[Time Series Data Analysis with pandas (Video)](http://www.youtube.com/watch?v=0unf-C-pBYE)",
"_____no_output_____"
],
[
"[Data analysis in Python with pandas (Video)](http://www.youtube.com/watch?v=w26x-z-BdWQ)",
"_____no_output_____"
],
[
"[Python for Data Analysis](http://shop.oreilly.com/product/0636920023784.do)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d0d20a3e2985ca70d93592cea501c7cd9c0291b7 | 242,496 | ipynb | Jupyter Notebook | forecasting/4_lightgbm.ipynb | kawo123/learning-ai | 35fa7e5b477c2d6edf4dcc692d612457167d490f | [
"MIT"
] | null | null | null | forecasting/4_lightgbm.ipynb | kawo123/learning-ai | 35fa7e5b477c2d6edf4dcc692d612457167d490f | [
"MIT"
] | 7 | 2020-11-13T18:50:04.000Z | 2022-02-10T01:46:20.000Z | forecasting/4_lightgbm.ipynb | kawo123/learning-ai | 35fa7e5b477c2d6edf4dcc692d612457167d490f | [
"MIT"
] | null | null | null | 429.19646 | 219,888 | 0.926254 | [
[
[
"<i>Copyright (c) Microsoft Corporation.</i>\n\n<i>Licensed under the MIT License.</i>",
"_____no_output_____"
],
[
"# LightGBM: A Highly Efficient Gradient Boosting Decision Tree\n\nThis notebook gives an example of how to perform multiple rounds of training and testing of LightGBM models to generate point forecasts of product sales in retail. We will train the LightGBM models based on the Orange Juice dataset.\n\n[LightGBM](https://github.com/Microsoft/LightGBM) is a gradient boosting framework that uses tree-based learning algorithms. [Gradient boosting](https://en.wikipedia.org/wiki/Gradient_boosting) is an ensemble technique in which models are added to the ensemble sequentially and at each iteration a new model is trained with respect to the error of the whole ensemble learned so far. More detailed information about gradient boosting can be found in this [tutorial paper](https://www.frontiersin.org/articles/10.3389/fnbot.2013.00021/full). Using this technique, LightGBM achieves great accuracy in many applications. Apart from this, it is designed to be distributed and efficient with the following advantages:\n* Fast training speed and high efficiency.\n* Low memory usage.\n* Support of parallel and GPU learning.\n* Capable of handling large-scale data.\n\nDue to these advantages, LightGBM has been widely-used in a lot of [winning solutions](https://github.com/microsoft/LightGBM/blob/master/examples/README.md#machine-learning-challenge-winning-solutions) of machine learning competitions.",
"_____no_output_____"
],
[
"## Global Settings and Imports",
"_____no_output_____"
]
],
[
[
"%load_ext autoreload\n%autoreload 2\n%matplotlib inline",
"_____no_output_____"
],
[
"import os\nimport sys\nimport math\nimport datetime\nimport warnings\nimport numpy as np\nimport pandas as pd\nimport lightgbm as lgb\n\nfrom fclib.models.lightgbm import predict\nfrom fclib.evaluation.evaluation_utils import MAPE\nfrom fclib.common.plot import plot_predictions_with_history\nfrom fclib.dataset.ojdata import split_train_test, FIRST_WEEK_START\nfrom fclib.feature_engineering.feature_utils import (\n week_of_month,\n df_from_cartesian_product,\n combine_features,\n)\n\nwarnings.filterwarnings(\"ignore\")\n\nprint(\"System version: {}\".format(sys.version))\nprint(\"LightGBM version: {}\".format(lgb.__version__))",
"System version: 3.6.9 (default, Nov 7 2019, 10:44:02) \n[GCC 8.3.0]\nLightGBM version: 2.3.0\n"
]
],
[
[
"## Parameter Settings\n\nIn what follows, we define global settings related to the model and feature engineering. LightGBM supports both classification models and regression models. In our case, we set the objective function to `mape` which stands for mean-absolute-percentage-error (MAPE) since we will build a regression model to predict product sales and evaluate the accuracy of the model using MAPE.\n\nGenerally, we can adjust the number of leaves (`num_leaves`), the minimum number of data in each leaf (`min_data_in_leaf`), maximum number of boosting rounds (`num_rounds`), the learning rate of trees (`learning_rate`) and `early_stopping_rounds` (to avoid overfitting) in the model to get better performance. Besides, we can also adjust other supported parameters to optimize the results. [In this link](https://github.com/Microsoft/LightGBM/blob/master/docs/Parameters.rst), a list of all the parameters is given. In addition, advice on how to tune these parameters can be found [in this url](https://github.com/Microsoft/LightGBM/blob/master/docs/Parameters-Tuning.rst).\n\nWe will use historical weekly sales, date time information, and product information as input features to train the model. Prior sales are used as lag features and `lags` contains the lags where each number indicates the number of time steps (i.e., weeks) that we shift the data backwards to get the historical sales. We also use the average sales within a certain time window in the past as a moving average feature. `window_size` controls the size of the moving window. Apart from these parameters, we use `use_columns` and `categ_fea` to denote all other features that we leverage in the model and the categorical features, respectively.\n",
"_____no_output_____"
]
],
[
[
"# Data directory\nDATA_DIR = os.path.join(\"ojdata\")\n\n# Parameters of LightGBM model\nparams = {\n \"objective\": \"mape\",\n \"num_leaves\": 124,\n \"min_data_in_leaf\": 340,\n \"learning_rate\": 0.1,\n \"feature_fraction\": 0.65,\n \"bagging_fraction\": 0.87,\n \"bagging_freq\": 19,\n \"num_rounds\": 940,\n \"early_stopping_rounds\": 125,\n \"num_threads\": 16,\n \"seed\": 1,\n}\n\n# Lags, window size, and feature column names\nlags = np.arange(2, 20)\nwindow_size = 40\nused_columns = [\n \"store\",\n \"brand\",\n \"week\",\n \"week_of_month\",\n \"month\",\n \"deal\",\n \"feat\",\n \"move\",\n \"price\",\n \"price_ratio\",\n]\ncateg_fea = [\"store\", \"brand\", \"deal\"]\n\n# Forecasting settings\nN_SPLITS = 10\nHORIZON = 2\nGAP = 2\nFIRST_WEEK = 40\nLAST_WEEK = 156",
"_____no_output_____"
]
],
[
[
"## Feature Engineering\n\nNext we create a function to extract a number of features from the data for training the forecasting model. These features include\n* datetime features including week, week of the month, and month\n* historical weekly sales of each orange juice in recent weeks\n* average sales of each orange juice during recent weeks\n* other features including `store`, `brand`, `deal`, `feat` columns and price features\n\nNote that the logarithm of the unit sales is stored in a column named `logmove` both for `train_df` and `test_df`. We compute the unit sales `move` based on this quantity.",
"_____no_output_____"
]
],
[
[
"def create_features(pred_round, train_dir, lags, window_size, used_columns):\n \"\"\"Create input features for model training and testing.\n\n \n Args: \n pred_round (int): Prediction round (1, 2, ...)\n train_dir (str): Path of the training data directory \n lags (np.array): Numpy array including all the lags\n window_size (int): Maximum step for computing the moving average\n used_columns (list[str]): A list of names of columns used in model training (including target variable)\n\n Returns:\n pd.Dataframe: Dataframe including all the input features and target variable\n int: Last week of the training data \n \"\"\"\n\n # Load training data\n train_df = pd.read_csv(os.path.join(train_dir, \"train_\" + str(pred_round) + \".csv\"))\n train_df[\"move\"] = train_df[\"logmove\"].apply(lambda x: round(math.exp(x)))\n train_df = train_df[[\"store\", \"brand\", \"week\", \"move\"]]\n\n # Create a dataframe to hold all necessary data\n store_list = train_df[\"store\"].unique()\n brand_list = train_df[\"brand\"].unique()\n train_end_week = train_df[\"week\"].max()\n week_list = range(FIRST_WEEK, train_end_week + GAP + HORIZON)\n d = {\"store\": store_list, \"brand\": brand_list, \"week\": week_list}\n data_grid = df_from_cartesian_product(d)\n data_filled = pd.merge(data_grid, train_df, how=\"left\", on=[\"store\", \"brand\", \"week\"])\n\n # Get future price, deal, and advertisement info\n aux_df = pd.read_csv(os.path.join(train_dir, \"auxi_\" + str(pred_round) + \".csv\"))\n data_filled = pd.merge(data_filled, aux_df, how=\"left\", on=[\"store\", \"brand\", \"week\"])\n\n # Create relative price feature\n price_cols = [\n \"price1\",\n \"price2\",\n \"price3\",\n \"price4\",\n \"price5\",\n \"price6\",\n \"price7\",\n \"price8\",\n \"price9\",\n \"price10\",\n \"price11\",\n ]\n data_filled[\"price\"] = data_filled.apply(lambda x: x.loc[\"price\" + str(int(x.loc[\"brand\"]))], axis=1)\n data_filled[\"avg_price\"] = data_filled[price_cols].sum(axis=1).apply(lambda x: x / len(price_cols))\n data_filled[\"price_ratio\"] = data_filled[\"price\"] / data_filled[\"avg_price\"]\n data_filled.drop(price_cols, axis=1, inplace=True)\n\n # Fill missing values\n data_filled = data_filled.groupby([\"store\", \"brand\"]).apply(\n lambda x: x.fillna(method=\"ffill\").fillna(method=\"bfill\")\n )\n\n # Create datetime features\n data_filled[\"week_start\"] = data_filled[\"week\"].apply(\n lambda x: FIRST_WEEK_START + datetime.timedelta(days=(x - 1) * 7)\n )\n data_filled[\"year\"] = data_filled[\"week_start\"].apply(lambda x: x.year)\n data_filled[\"month\"] = data_filled[\"week_start\"].apply(lambda x: x.month)\n data_filled[\"week_of_month\"] = data_filled[\"week_start\"].apply(lambda x: week_of_month(x))\n data_filled[\"day\"] = data_filled[\"week_start\"].apply(lambda x: x.day)\n data_filled.drop(\"week_start\", axis=1, inplace=True)\n\n # Create other features (lagged features, moving averages, etc.)\n features = data_filled.groupby([\"store\", \"brand\"]).apply(\n lambda x: combine_features(x, [\"move\"], lags, window_size, used_columns)\n )\n\n # Drop rows with NaN values\n features.dropna(inplace=True)\n\n return features, train_end_week",
"_____no_output_____"
]
],
[
[
"## Model Training\n\nWe then perform a multi-round training by fitting a LightGBM model using the training data in each forecast round. After the models are trained, we apply them to generate forecasts for the target weeks. The paradigm of model training and testing is illustrated in the following diagram\n\n<img src=\"https://user-images.githubusercontent.com/20047467/77784194-84faee00-7030-11ea-83ee-6e2c33eb2434.png\">",
"_____no_output_____"
]
],
[
[
"# Train and predict for all forecast rounds\npred_all = []\nmetric_all = []\ntrain_dir = os.path.join(DATA_DIR, \"train\")\nfor r in range(1, N_SPLITS + 1):\n print(\"---------- Round \" + str(r) + \" ----------\")\n features, train_end_week = create_features(r, train_dir, lags, window_size, used_columns)\n train_fea = features[features.week <= train_end_week].reset_index(drop=True)\n print(\"Maximum training week number is {}\".format(max(train_fea[\"week\"])))\n\n # Create training set\n dtrain = lgb.Dataset(train_fea.drop(\"move\", axis=1, inplace=False), label=train_fea[\"move\"])\n\n # Train GBM model\n print(\"Training LightGBM model started.\")\n bst = lgb.train(params, dtrain, valid_sets=[dtrain], categorical_feature=categ_fea, verbose_eval=False,)\n print(\"Training LightGBM model finished.\")\n\n # Generate forecasts\n test_fea = features[features.week >= train_end_week + GAP].reset_index(drop=True)\n idx_cols = [\"store\", \"brand\", \"week\"]\n pred = predict(test_fea, bst, target_col=\"move\", idx_cols=idx_cols).sort_values(by=idx_cols).reset_index(drop=True)\n print(\"Prediction results:\")\n print(pred.head())\n print(\"\")\n\n # Keep the predictions\n pred[\"round\"] = r\n pred_all.append(pred)\n\npred_all = pd.concat(pred_all, axis=0)\npred_all.rename(columns={\"move\": \"prediction\"}, inplace=True)\npred_all = pred_all[[\"round\", \"week\", \"store\", \"brand\", \"prediction\"]]",
"---------- Round 1 ----------\nMaximum training week number is 145\nTraining LightGBM model started.\nTraining LightGBM model finished.\nPrediction results:\n store brand week move\n0 2 1 147 9404\n1 2 1 148 8103\n2 2 2 147 6705\n3 2 2 148 6120\n4 2 3 147 2214\n\n---------- Round 2 ----------\nMaximum training week number is 147\nTraining LightGBM model started.\nTraining LightGBM model finished.\nPrediction results:\n store brand week move\n0 2 1 149 7641\n1 2 1 150 5629\n2 2 2 149 6967\n3 2 2 150 11881\n4 2 3 149 2874\n\n---------- Round 3 ----------\nMaximum training week number is 149\nTraining LightGBM model started.\nTraining LightGBM model finished.\nPrediction results:\n store brand week move\n0 2 1 151 7452\n1 2 1 152 7124\n2 2 2 151 7331\n3 2 2 152 8968\n4 2 3 151 3210\n\n---------- Round 4 ----------\nMaximum training week number is 151\nTraining LightGBM model started.\nTraining LightGBM model finished.\nPrediction results:\n store brand week move\n0 2 1 153 5451\n1 2 1 154 9084\n2 2 2 153 6404\n3 2 2 154 6989\n4 2 3 153 2876\n\n---------- Round 5 ----------\nMaximum training week number is 153\nTraining LightGBM model started.\nTraining LightGBM model finished.\nPrediction results:\n store brand week move\n0 2 1 155 6761\n1 2 1 156 12385\n2 2 2 155 6264\n3 2 2 156 5950\n4 2 3 155 3332\n\n---------- Round 6 ----------\nMaximum training week number is 145\nTraining LightGBM model started.\nTraining LightGBM model finished.\nPrediction results:\n store brand week move\n0 2 1 147 9404\n1 2 1 148 8103\n2 2 2 147 6705\n3 2 2 148 6120\n4 2 3 147 2214\n\n---------- Round 7 ----------\nMaximum training week number is 147\nTraining LightGBM model started.\nTraining LightGBM model finished.\nPrediction results:\n store brand week move\n0 2 1 149 7641\n1 2 1 150 5629\n2 2 2 149 6967\n3 2 2 150 11881\n4 2 3 149 2874\n\n---------- Round 8 ----------\nMaximum training week number is 149\nTraining LightGBM model started.\nTraining LightGBM model finished.\nPrediction results:\n store brand week move\n0 2 1 151 7452\n1 2 1 152 7124\n2 2 2 151 7331\n3 2 2 152 8968\n4 2 3 151 3210\n\n---------- Round 9 ----------\nMaximum training week number is 151\nTraining LightGBM model started.\nTraining LightGBM model finished.\nPrediction results:\n store brand week move\n0 2 1 153 5451\n1 2 1 154 9084\n2 2 2 153 6404\n3 2 2 154 6989\n4 2 3 153 2876\n\n---------- Round 10 ----------\nMaximum training week number is 153\nTraining LightGBM model started.\nTraining LightGBM model finished.\nPrediction results:\n store brand week move\n0 2 1 155 6761\n1 2 1 156 12385\n2 2 2 155 6264\n3 2 2 156 5950\n4 2 3 155 3332\n\n"
]
],
[
[
"## Model Evaluation\n\nTo evaluate the model performance, we compute MAPE of the forecasts from all the forecast rounds below.",
"_____no_output_____"
]
],
[
[
"# Evaluate prediction accuracy\ntest_all = []\ntest_dir = os.path.join(DATA_DIR, \"test\")\nfor r in range(1, N_SPLITS + 1):\n test_df = pd.read_csv(os.path.join(test_dir, \"test_\" + str(r) + \".csv\"))\n test_all.append(test_df)\ntest_all = pd.concat(test_all, axis=0).reset_index(drop=True)\ntest_all[\"actual\"] = test_all[\"logmove\"].apply(lambda x: round(math.exp(x)))\ntest_all.drop(\"logmove\", axis=1, inplace=True)\ncombined = pd.merge(pred_all, test_all, on=[\"store\", \"brand\", \"week\"], how=\"left\")\nmetric_value = MAPE(combined[\"prediction\"], combined[\"actual\"]) * 100\nprint(\"MAPE of the predictions is {}\".format(metric_value))",
"MAPE of the predictions is 36.102958386230824\n"
]
],
[
[
"## Result Visualization\n\nFinally, we plot out the forecast results of a few sample store-brand combinations to visually check the forecasts. Note that there could be gaps in the curve of actual sales due to missing sales data.",
"_____no_output_____"
]
],
[
[
"results = combined[[\"week\", \"store\", \"brand\", \"prediction\"]]\nresults.rename(columns={\"prediction\": \"move\"}, inplace=True)\nactual = combined[[\"week\", \"store\", \"brand\", \"actual\"]]\nactual.rename(columns={\"actual\": \"move\"}, inplace=True)\nstore_list = combined[\"store\"].unique()\nbrand_list = combined[\"brand\"].unique()\n\nplot_predictions_with_history(\n results,\n actual,\n store_list,\n brand_list,\n \"week\",\n \"move\",\n grain1_name=\"store\",\n grain2_name=\"brand\",\n min_timestep=137,\n num_samples=6,\n predict_at_timestep=135,\n line_at_predict_time=False,\n title=\"Prediction results for a few sample time series\",\n x_label=\"time step\",\n y_label=\"target value\",\n random_seed=6,\n)",
"_____no_output_____"
]
],
[
[
"## Additional Reading\n\n\\[1\\] Guolin Ke, Qi Meng, Thomas Finley, Taifeng Wang, Wei Chen, Weidong Ma, Qiwei Ye, and Tie-Yan Liu. 2017. LightGBM: A highly efficient gradient boosting decision tree. In Advances in Neural Information Processing Systems. 3146–3154.<br>\n\\[2\\] Alexey Natekin and Alois Knoll. 2013. Gradient boosting machines, a tutorial. Frontiers in neurorobotics, 7 (21). <br>\n\\[3\\] The parameters of LightGBM: https://github.com/Microsoft/LightGBM/blob/master/docs/Parameters.rst <br>\n\\[4\\] Anna Veronika Dorogush, Vasily Ershov, and Andrey Gulin. 2018. CatBoost: gradient boosting with categorical features support. arXiv preprint arXiv:1810.11363 (2018).<br>\n\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0d218c7c8c3f97ba69e37f277fd5466b70e0846 | 45,117 | ipynb | Jupyter Notebook | results/04_summary_external_validation.ipynb | TomMonks/swast-benchmarking | 96964fb705a8b3cebbce8adcf03e42d4fc3dd05a | [
"MIT"
] | null | null | null | results/04_summary_external_validation.ipynb | TomMonks/swast-benchmarking | 96964fb705a8b3cebbce8adcf03e42d4fc3dd05a | [
"MIT"
] | null | null | null | results/04_summary_external_validation.ipynb | TomMonks/swast-benchmarking | 96964fb705a8b3cebbce8adcf03e42d4fc3dd05a | [
"MIT"
] | 1 | 2021-11-16T14:38:22.000Z | 2021-11-16T14:38:22.000Z | 58.215484 | 7,544 | 0.704391 | [
[
[
"# External Validation of SWAST Forecasting Model\n## Overall results summary.\n\nThis notebook generates the overall results summary for the MASE, and prediction intervals for LAS, YAS and WAST.",
"_____no_output_____"
]
],
[
[
"print('******************Summary of External validation results*****************\\n\\n')",
"******************Summary of External validation results*****************\n\n\n"
],
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\nimport os\nfrom os import listdir\nfrom os.path import isfile, join\n\nfrom scipy.stats import norm, t\n\nsns.set(style=\"whitegrid\")",
"_____no_output_____"
],
[
"cwd = os.getcwd()\ncwd",
"_____no_output_____"
],
[
"if cwd[-7:] != \"results\":\n mypath = './results/external_validation/'\n TABLE_PATH = './paper/tables/'\n FIGURE_PATH = './paper/figures/'\n APPENDIX_PATH = './paper/appendix/'\nelse:\n mypath = './external_validation/'\n TABLE_PATH = '../paper/tables/'\n FIGURE_PATH = '../paper/figures/'\n APPENDIX_PATH = '../paper/appendix/'",
"_____no_output_____"
],
[
"result_files = [os.path.join(dp, f) for dp, dn, filenames in os.walk(mypath) \n for f in filenames if os.path.splitext(f)[1] == '.csv']",
"_____no_output_____"
],
[
"results_mean = pd.DataFrame()\nresults_med = pd.DataFrame()\nresults_mean_std = pd.DataFrame()\nresults_all = pd.DataFrame()",
"_____no_output_____"
]
],
[
[
"## Point Estimate Results",
"_____no_output_____"
]
],
[
[
"error_measures = ['smape', 'rmse', 'mase', 'coverage_80', 'coverage_95']",
"_____no_output_____"
],
[
"mypath",
"_____no_output_____"
],
[
"#start = len('/external_validation/')\nstart = len(mypath) - 1\nfor metric in error_measures:\n to_read = [filename for filename in result_files if metric in filename]\n model_names = ['.' + name[start:name.index('_', start)] for name in to_read]\n \n for filename, model_name in zip(to_read, model_names):\n df = pd.read_csv(filename, index_col=0)\n if 'snaive' not in model_name:\n prefix = model_name + '_' + metric\n results_mean[prefix + '_mean'] = df.mean()\n results_mean[prefix + '_std'] = df.std()\n results_med[prefix + '_med'] = df.median()\n results_med[prefix + '_iqr'] = df.quantile(0.75) - df.quantile(0.25)\n\n results_mean_std[prefix] = results_mean[prefix + '_mean'].map('{:,.2f}'.format) \\\n + ' (' + results_mean[prefix + '_std'].map('{:,.2f}'.format) + ')'\n \n if 'mase' in filename:\n results_all[prefix] = df.to_numpy().flatten()",
"_____no_output_____"
]
],
[
[
"## Seperate dataframes for prediction intervals",
"_____no_output_____"
]
],
[
[
"summary_fa = results_mean.filter(\n like=\"coverage_95\").filter(like=\"fbp-arima\").filter(like='mean')\nsummary_fa2 = results_mean.filter(\n like=\"coverage_80\").filter(like=\"fbp-arima\").filter(like='mean')",
"_____no_output_____"
]
],
[
[
"# Overall statistics\n\n## Mean absolute scaled error",
"_____no_output_____"
]
],
[
[
"alpha = 0.05\n\n#overall MASE\nsummary_fa = results_mean.filter(\n like=\"mase\").filter(like=\"fbp-arima\").filter(like='mean')\n\nmean = summary_fa.to_numpy().flatten().mean()\nprint(f'\\nOverall External Evaluation Statistics for MASE')\n\n#sample std\nstd = summary_fa.to_numpy().flatten().std(ddof=1)\n\nn = summary_fa.to_numpy().flatten().shape[0]\n\n#Confidence interval calculation\nse = std / np.sqrt(n)\nz = np.abs(t.ppf(alpha / 2, n - 1))\nhw = z * se\nlower = mean - hw\nupper = mean + hw\n\n#lower and upper 95% CI\nprint(f'mean: {mean:.4f} 95% CI ({lower:.4f}-{upper:.4f})')\n\n#median\nmed = np.percentile(summary_fa.to_numpy().flatten(), 50)\nlq = np.percentile(summary_fa.to_numpy().flatten(), 25)\nuq = np.percentile(summary_fa.to_numpy().flatten(), 75)\nprint(f'median: {med:.3f}, IQR {uq - lq:.3f}')\n\n\n#middle 90% of data lies between\nfifth = np.percentile(summary_fa.to_numpy().flatten(), 5)\nninetyfifth = np.percentile(summary_fa.to_numpy().flatten(), 95)\nprint(f'middle 90%: {fifth:.3f} - {ninetyfifth:.3f}')\n\nplt.hist(summary_fa.to_numpy().flatten());",
"\nOverall External Evaluation Statistics for MASE\nmean: 0.7275 95% CI (0.7193-0.7357)\nmedian: 0.734, IQR 0.084\nmiddle 90%: 0.641 - 0.834\n"
]
],
[
[
"## 80% Prediction interval coverage",
"_____no_output_____"
]
],
[
[
"alpha = 0.05\n\n#overall 80% PI coverage\nmean = summary_fa2.to_numpy().flatten().mean()\nprint(f'\\nOverall External Evaluation Statistics for 80% PI coverage')\n\n#sample std\nstd = summary_fa2.to_numpy().flatten().std(ddof=1)\n\nn = summary_fa2.to_numpy().flatten().shape[0]\n\n#Confidence interval calculation\nse = std / np.sqrt(n)\nz = np.abs(t.ppf(alpha / 2, n - 1))\nhw = z * se\nlower = mean - hw\nupper = mean + hw\n\n#lower and upper 95% CI\nprint(f'mean: {mean:.4f} 95% CI ({lower:.4f}-{upper:.4f})')\n\n#median\nmed = np.percentile(summary_fa2.to_numpy().flatten(), 50)\nlq = np.percentile(summary_fa2.to_numpy().flatten(), 25)\nuq = np.percentile(summary_fa2.to_numpy().flatten(), 75)\nprint(f'median: {med:.3f}, IQR {uq - lq:.3f}')\n\n\n#middle 90% of data lies between\nfifth = np.percentile(summary_fa2.to_numpy().flatten(), 5)\nninetyfifth = np.percentile(summary_fa2.to_numpy().flatten(), 95)\nprint(f'middle 90%: {fifth:.3f} - {ninetyfifth:.3f}')\n\nplt.hist(summary_fa.to_numpy().flatten());",
"\nOverall External Evaluation Statistics for 80% PI coverage\nmean: 0.8332 95% CI (0.8282-0.8381)\nmedian: 0.837, IQR 0.051\nmiddle 90%: 0.773 - 0.878\n"
]
],
[
[
"## 95% prediction interval coverage",
"_____no_output_____"
]
],
[
[
"summary_fa = results_mean.filter(\n like=\"coverage_95\").filter(like=\"fbp-arima\").filter(like='mean')\n\nmean = summary_fa.to_numpy().flatten().mean()\nprint(f'\\nOverall External Evaluation Statistics for 95% PI Coverage')\n\n#sample std\nstd = summary_fa.to_numpy().flatten().std(ddof=1)\n\nn = summary_fa.to_numpy().flatten().shape[0]\n\n#Confidence interval calculation\nse = std / np.sqrt(n)\nz = np.abs(t.ppf(alpha / 2, n - 1))\nhw = z * se\nlower = mean - hw\nupper = mean + hw\n\n#lower and upper 95% CI\nprint(f'mean: {mean:.4f} 95% CI ({lower:.4f}-{upper:.4f})')\n\n#median\nmed = np.percentile(summary_fa.to_numpy().flatten(), 50)\nlq = np.percentile(summary_fa.to_numpy().flatten(), 25)\nuq = np.percentile(summary_fa.to_numpy().flatten(), 75)\nprint(f'median: {med:.3f}, IQR {uq - lq:.3f}')\n\n\n#middle 90% of data lies between\nfifth = np.percentile(summary_fa.to_numpy().flatten(), 5)\nninetyfifth = np.percentile(summary_fa.to_numpy().flatten(), 95)\nprint(f'middle 90%: {fifth:.3f} - {ninetyfifth:.3f}')\n\nplt.hist(summary_fa.to_numpy().flatten());",
"\nOverall External Evaluation Statistics for 95% PI Coverage\nmean: 0.9649 95% CI (0.9626-0.9671)\nmedian: 0.970, IQR 0.019\nmiddle 90%: 0.936 - 0.983\n"
]
],
[
[
"## Overall Results Summary Table",
"_____no_output_____"
]
],
[
[
"#filter for mase - mean results\nregion_means = results_mean.filter(\n like=\"mase\").filter(like='mean').filter(like='fbp-arima').mean().sort_index()",
"_____no_output_____"
],
[
"#filter for mase stdev\nregion_std = results_mean.filter(\n like=\"mase\").filter(like='std').filter(like='fbp-arima').mean().sort_index()",
"_____no_output_____"
],
[
"#coverage mean\nregion_95_mean = results_mean.filter(like=\"coverage_95\").filter(like='mean').filter(like='fbp-arima').mean().sort_index()\nregion_80_mean = results_mean.filter(like=\"coverage_80\").filter(like='mean').filter(like='fbp-arima').mean().sort_index()",
"_____no_output_____"
],
[
"#coverage stdev\nregion_95_std = results_mean.filter(like=\"coverage_95\").filter(like='std').filter(like='fbp-arima').mean().sort_index()\nregion_80_std = results_mean.filter(like=\"coverage_80\").filter(like='std').filter(like='fbp-arima').mean().sort_index()",
"_____no_output_____"
],
[
"#create index of dataframe\ncomparisons = list(region_means.index)\nidx = [i.replace('_mase_mean', '') for i in comparisons]",
"_____no_output_____"
],
[
"#construct table\ndf_regions = pd.DataFrame(region_means.to_numpy(), columns=['mean'])\ndf_regions['std'] = region_std.to_numpy()\ndf_regions['mean_80'] = region_80_mean.to_numpy()\ndf_regions['std_80'] = region_80_std.to_numpy()\ndf_regions['mean_95'] = region_95_mean.to_numpy()\ndf_regions['std_95'] = region_95_std.to_numpy()\ndf_regions['MASE'] = df_regions['mean'].map('{:,.2f}'.format) \\\n + ' (' + df_regions['std'].map('{:,.2f}'.format) + ')'\n\ndf_regions['Coverage 80'] = df_regions['mean_80'].map('{:,.2f}'.format) \\\n + ' (' + df_regions['std_80'].map('{:,.2f}'.format) + ')'\n\ndf_regions['Coverage 95'] = df_regions['mean_95'].map('{:,.2f}'.format) \\\n + ' (' + df_regions['std_95'].map('{:,.2f}'.format) + ')'\n\ndf_regions.index = idx\ndf_regions = df_regions.drop(['mean', 'std', 'mean_80', 'std_80', \n 'mean_95', 'std_95'], axis=1)",
"_____no_output_____"
],
[
"df_regions.shape",
"_____no_output_____"
],
[
"#trim out trust and region intro seperate columns\ndf_regions['trust'] = [i[2:i.find('/', 2)] for i in list(df_regions.index)]\ndf_regions['region'] = [i[i.find('/', 2)+1:] for i in list(df_regions.index)]",
"_____no_output_____"
],
[
"#summary frame of results\nprint(\"**Table 5: External validation: Point forecast and coverage performance by region.\")\n\ndf_regions['region'] = [str(i).replace('-fbp-arima', '') \n for i in list(df_regions.region)]\n\ndf_regions.index = pd.MultiIndex.from_frame(df_regions[['trust', 'region']])\ndf_regions.drop(['trust'], axis=1, inplace=True)\ndf_regions.drop(['region'], axis=1, inplace=True)\n\ndf_regions.drop(('york','Trust'), axis=0, inplace=True)\ndf_regions.drop(('wales','Trust'), axis=0, inplace=True)\ndf_regions",
"**Table 5: External validation: Point forecast and coverage performance by region.\n"
],
[
"df_regions.sort_index().to_latex(f'{TABLE_PATH}Table5.tex')",
"_____no_output_____"
],
[
"print(df_regions)",
" MASE Coverage 80 Coverage 95\ntrust region \nlondon North Central 0.76 (0.11) 0.84 (0.06) 0.94 (0.04)\n North East 0.74 (0.08) 0.87 (0.06) 0.97 (0.03)\n North West 0.75 (0.11) 0.86 (0.07) 0.97 (0.03)\n South East 0.70 (0.10) 0.85 (0.06) 0.98 (0.03)\n South West 0.68 (0.10) 0.86 (0.06) 0.97 (0.04)\nwales Control Central and West 0.69 (0.12) 0.82 (0.09) 0.96 (0.04)\n Control North 0.73 (0.14) 0.79 (0.10) 0.97 (0.04)\n Control South East 0.64 (0.10) 0.87 (0.06) 0.98 (0.04)\nyork ABL 0.67 (0.11) 0.86 (0.05) 0.98 (0.02)\n CKW 0.74 (0.09) 0.82 (0.07) 0.96 (0.03)\n Humb and ER 0.77 (0.11) 0.79 (0.08) 0.94 (0.04)\n North Yorks 0.83 (0.15) 0.77 (0.09) 0.94 (0.06)\n South 0.76 (0.11) 0.81 (0.08) 0.96 (0.04)\n"
]
],
[
[
"# End",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
d0d2302873fbe1357fc164f892f30fd8beb43eca | 3,323 | ipynb | Jupyter Notebook | docs/api/facts.ipynb | ShreyaBM/draco2 | 1b937b53c59a0821a4ed32a7380709e5b1798fe4 | [
"MIT"
] | 22 | 2020-11-17T18:46:59.000Z | 2022-02-22T23:15:45.000Z | docs/api/facts.ipynb | ShreyaBM/draco2 | 1b937b53c59a0821a4ed32a7380709e5b1798fe4 | [
"MIT"
] | 65 | 2020-12-02T21:51:48.000Z | 2022-03-24T20:23:11.000Z | docs/api/facts.ipynb | ShreyaBM/draco2 | 1b937b53c59a0821a4ed32a7380709e5b1798fe4 | [
"MIT"
] | 5 | 2020-11-20T05:36:28.000Z | 2022-01-11T21:12:49.000Z | 26.165354 | 354 | 0.542281 | [
[
[
"# Fact Utils\n\nGenerating facts in the expected format can be tedious. To make it easier to put data into and get data our of Draco, we an API to convert nested data to facts and vice versa.",
"_____no_output_____"
],
[
"## Available functions\n\n```{eval-rst}\n.. automodule:: draco.fact_utils\n :members:\n```",
"_____no_output_____"
],
[
"## Usage Example",
"_____no_output_____"
]
],
[
[
"from draco import dict_to_facts, answer_set_to_dict, run_clingo\nfrom pprint import pprint",
"_____no_output_____"
],
[
"facts = dict_to_facts({\n \"mark\": \"bar\",\n \"encoding\": [{\n \"channel\": \"x\",\n \"field\": \"condition\"\n },{\n \"channel\": \"y\",\n \"aggregate\": \"count\"\n }]\n})\nfacts",
"_____no_output_____"
],
[
"# we can run Clingo and convert the model back into the nested representation\n\nfor model in run_clingo(facts):\n answer_set = model.answer_set\n print(answer_set)\n pprint(answer_set_to_dict(answer_set))",
"[<clingo.symbol.Symbol object at 0x7f9c68a9a250>, <clingo.symbol.Symbol object at 0x7f9c68a9aca0>, <clingo.symbol.Symbol object at 0x7f9c68a9a580>, <clingo.symbol.Symbol object at 0x7f9c68a9ad00>, <clingo.symbol.Symbol object at 0x7f9c68a9a790>, <clingo.symbol.Symbol object at 0x7f9c68a9abb0>, <clingo.symbol.Symbol object at 0x7f9c68a9ab20>]\n{'encoding': [{'channel': 'x', 'field': 'condition'},\n {'aggregate': 'count', 'channel': 'y'}],\n 'mark': 'bar'}\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
]
] |
d0d2518b98221e34ec2c2dcd56898ca1b02eb858 | 778,627 | ipynb | Jupyter Notebook | examples/bayesian_regression_ii.ipynb | fehiepsi/website | 7b0165deebc1379105d8b9d8051187f7d9914446 | [
"MIT"
] | null | null | null | examples/bayesian_regression_ii.ipynb | fehiepsi/website | 7b0165deebc1379105d8b9d8051187f7d9914446 | [
"MIT"
] | null | null | null | examples/bayesian_regression_ii.ipynb | fehiepsi/website | 7b0165deebc1379105d8b9d8051187f7d9914446 | [
"MIT"
] | null | null | null | 1,326.451448 | 209,764 | 0.955253 | [
[
[
"# Bayesian Regression - Inference Algorithms (Part 2)",
"_____no_output_____"
],
[
"In [Part I](bayesian_regression.ipynb), we looked at how to perform inference on a simple Bayesian linear regression model using SVI. In this tutorial, we'll explore more expressive guides as well as exact inference techniques. We'll use the same dataset as before.",
"_____no_output_____"
]
],
[
[
"%reset -sf",
"_____no_output_____"
],
[
"import logging\nimport os\n\nimport torch\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\nimport seaborn as sns\nfrom torch.distributions import constraints\n\nimport pyro\nimport pyro.distributions as dist\nimport pyro.optim as optim\n\npyro.set_rng_seed(1)\nassert pyro.__version__.startswith('1.5.0')",
"_____no_output_____"
],
[
"%matplotlib inline\nplt.style.use('default')\n\nlogging.basicConfig(format='%(message)s', level=logging.INFO)\n# Enable validation checks\npyro.enable_validation(True)\nsmoke_test = ('CI' in os.environ)\npyro.set_rng_seed(1)\nDATA_URL = \"https://d2hg8soec8ck9v.cloudfront.net/datasets/rugged_data.csv\"\nrugged_data = pd.read_csv(DATA_URL, encoding=\"ISO-8859-1\")",
"_____no_output_____"
]
],
[
[
"## Bayesian Linear Regression\n\nOur goal is once again to predict log GDP per capita of a nation as a function of two features from the dataset - whether the nation is in Africa, and its Terrain Ruggedness Index, but we will explore more expressive guides.",
"_____no_output_____"
],
[
"## Model + Guide\n\nWe will write out the model again, similar to that in [Part I](bayesian_regression.ipynb), but explicitly without the use of `PyroModule`. We will write out each term in the regression, using the same priors. `bA` and `bR` are regression coefficients corresponding to `is_cont_africa` and `ruggedness`, `a` is the intercept, and `bAR` is the correlating factor between the two features.\n\nWriting down a guide will proceed in close analogy to the construction of our model, with the key difference that the guide parameters need to be trainable. To do this we register the guide parameters in the ParamStore using `pyro.param()`. Note the positive constraints on scale parameters.",
"_____no_output_____"
]
],
[
[
"def model(is_cont_africa, ruggedness, log_gdp):\n a = pyro.sample(\"a\", dist.Normal(0., 10.))\n b_a = pyro.sample(\"bA\", dist.Normal(0., 1.))\n b_r = pyro.sample(\"bR\", dist.Normal(0., 1.))\n b_ar = pyro.sample(\"bAR\", dist.Normal(0., 1.))\n sigma = pyro.sample(\"sigma\", dist.Uniform(0., 10.))\n mean = a + b_a * is_cont_africa + b_r * ruggedness + b_ar * is_cont_africa * ruggedness\n with pyro.plate(\"data\", len(ruggedness)):\n pyro.sample(\"obs\", dist.Normal(mean, sigma), obs=log_gdp)\n \ndef guide(is_cont_africa, ruggedness, log_gdp):\n a_loc = pyro.param('a_loc', torch.tensor(0.))\n a_scale = pyro.param('a_scale', torch.tensor(1.),\n constraint=constraints.positive)\n sigma_loc = pyro.param('sigma_loc', torch.tensor(1.),\n constraint=constraints.positive)\n weights_loc = pyro.param('weights_loc', torch.randn(3))\n weights_scale = pyro.param('weights_scale', torch.ones(3),\n constraint=constraints.positive)\n a = pyro.sample(\"a\", dist.Normal(a_loc, a_scale))\n b_a = pyro.sample(\"bA\", dist.Normal(weights_loc[0], weights_scale[0]))\n b_r = pyro.sample(\"bR\", dist.Normal(weights_loc[1], weights_scale[1]))\n b_ar = pyro.sample(\"bAR\", dist.Normal(weights_loc[2], weights_scale[2]))\n sigma = pyro.sample(\"sigma\", dist.Normal(sigma_loc, torch.tensor(0.05)))\n mean = a + b_a * is_cont_africa + b_r * ruggedness + b_ar * is_cont_africa * ruggedness",
"_____no_output_____"
],
[
"# Utility function to print latent sites' quantile information.\ndef summary(samples):\n site_stats = {}\n for site_name, values in samples.items():\n marginal_site = pd.DataFrame(values)\n describe = marginal_site.describe(percentiles=[.05, 0.25, 0.5, 0.75, 0.95]).transpose()\n site_stats[site_name] = describe[[\"mean\", \"std\", \"5%\", \"25%\", \"50%\", \"75%\", \"95%\"]]\n return site_stats\n\n# Prepare training data\ndf = rugged_data[[\"cont_africa\", \"rugged\", \"rgdppc_2000\"]]\ndf = df[np.isfinite(df.rgdppc_2000)]\ndf[\"rgdppc_2000\"] = np.log(df[\"rgdppc_2000\"])\ntrain = torch.tensor(df.values, dtype=torch.float)",
"_____no_output_____"
]
],
[
[
"## SVI\n\nAs before, we will use SVI to perform inference.",
"_____no_output_____"
]
],
[
[
"from pyro.infer import SVI, Trace_ELBO\n\n\nsvi = SVI(model, \n guide, \n optim.Adam({\"lr\": .05}), \n loss=Trace_ELBO())\n\nis_cont_africa, ruggedness, log_gdp = train[:, 0], train[:, 1], train[:, 2]\npyro.clear_param_store()\nnum_iters = 5000 if not smoke_test else 2\nfor i in range(num_iters):\n elbo = svi.step(is_cont_africa, ruggedness, log_gdp)\n if i % 500 == 0:\n logging.info(\"Elbo loss: {}\".format(elbo))",
"Elbo loss: 5795.467590510845\nElbo loss: 415.8169444799423\nElbo loss: 250.71916329860687\nElbo loss: 247.19457268714905\nElbo loss: 249.2004036307335\nElbo loss: 250.96484470367432\nElbo loss: 249.35092514753342\nElbo loss: 248.7831552028656\nElbo loss: 248.62140649557114\nElbo loss: 250.4274433851242\n"
],
[
"from pyro.infer import Predictive\n\n\nnum_samples = 1000\npredictive = Predictive(model, guide=guide, num_samples=num_samples)\nsvi_samples = {k: v.reshape(num_samples).detach().cpu().numpy()\n for k, v in predictive(log_gdp, is_cont_africa, ruggedness).items()\n if k != \"obs\"}",
"_____no_output_____"
]
],
[
[
"Let us observe the posterior distribution over the different latent variables in the model.",
"_____no_output_____"
]
],
[
[
"for site, values in summary(svi_samples).items():\n print(\"Site: {}\".format(site))\n print(values, \"\\n\")",
"Site: a\n mean std 5% 25% 50% 75% 95%\n0 9.177024 0.059607 9.07811 9.140463 9.178211 9.217098 9.27152 \n\nSite: bA\n mean std 5% 25% 50% 75% 95%\n0 -1.890622 0.122805 -2.08849 -1.979107 -1.887476 -1.803683 -1.700853 \n\nSite: bR\n mean std 5% 25% 50% 75% 95%\n0 -0.157847 0.039538 -0.22324 -0.183673 -0.157873 -0.133102 -0.091713 \n\nSite: bAR\n mean std 5% 25% 50% 75% 95%\n0 0.304515 0.067683 0.194583 0.259464 0.304907 0.348932 0.415128 \n\nSite: sigma\n mean std 5% 25% 50% 75% 95%\n0 0.902898 0.047971 0.824166 0.870317 0.901981 0.935171 0.981577 \n\n"
]
],
[
[
"## HMC\n\nIn contrast to using variational inference which gives us an approximate posterior over our latent variables, we can also do exact inference using [Markov Chain Monte Carlo](http://docs.pyro.ai/en/dev/mcmc.html) (MCMC), a class of algorithms that in the limit, allow us to draw unbiased samples from the true posterior. The algorithm that we will be using is called the No-U Turn Sampler (NUTS) \\[1\\], which provides an efficient and automated way of running Hamiltonian Monte Carlo. It is slightly slower than variational inference, but provides an exact estimate.",
"_____no_output_____"
]
],
[
[
"from pyro.infer import MCMC, NUTS\n\n\nnuts_kernel = NUTS(model)\n\nmcmc = MCMC(nuts_kernel, num_samples=1000, warmup_steps=200)\nmcmc.run(is_cont_africa, ruggedness, log_gdp)\n\nhmc_samples = {k: v.detach().cpu().numpy() for k, v in mcmc.get_samples().items()}",
"Sample: 100%|██████████| 1200/1200 [00:30, 38.99it/s, step size=2.76e-01, acc. prob=0.934]\n"
],
[
"for site, values in summary(hmc_samples).items():\n print(\"Site: {}\".format(site))\n print(values, \"\\n\")",
"Site: a\n mean std 5% 25% 50% 75% 95%\n0 9.182098 0.13545 8.958712 9.095588 9.181347 9.277673 9.402615 \n\nSite: bA\n mean std 5% 25% 50% 75% 95%\n0 -1.847651 0.217768 -2.19934 -1.988024 -1.846978 -1.70495 -1.481822 \n\nSite: bR\n mean std 5% 25% 50% 75% 95%\n0 -0.183031 0.078067 -0.311403 -0.237077 -0.185945 -0.131043 -0.051233 \n\nSite: bAR\n mean std 5% 25% 50% 75% 95%\n0 0.348332 0.127478 0.131907 0.266548 0.34641 0.427984 0.560221 \n\nSite: sigma\n mean std 5% 25% 50% 75% 95%\n0 0.952041 0.052024 0.869388 0.914335 0.949961 0.986266 1.038723 \n\n"
]
],
[
[
"## Comparing Posterior Distributions\n\nLet us compare the posterior distribution of the latent variables that we obtained from variational inference with those from Hamiltonian Monte Carlo. As can be seen below, for Variational Inference, the marginal distribution of the different regression coefficients is under-dispersed w.r.t. the true posterior (from HMC). This is an artifact of the *KL(q||p)* loss (the KL divergence of the true posterior from the approximate posterior) that is minimized by Variational Inference.\n\nThis can be better seen when we plot different cross sections from the joint posterior distribution overlaid with the approximate posterior from variational inference. Note that since our variational family has diagonal covariance, we cannot model any correlation between the latents and the resulting approximation is overconfident (under-dispersed)",
"_____no_output_____"
]
],
[
[
"sites = [\"a\", \"bA\", \"bR\", \"bAR\", \"sigma\"]\n\nfig, axs = plt.subplots(nrows=2, ncols=2, figsize=(12, 10))\nfig.suptitle(\"Marginal Posterior density - Regression Coefficients\", fontsize=16)\nfor i, ax in enumerate(axs.reshape(-1)):\n site = sites[i]\n sns.distplot(svi_samples[site], ax=ax, label=\"SVI (DiagNormal)\")\n sns.distplot(hmc_samples[site], ax=ax, label=\"HMC\")\n ax.set_title(site)\nhandles, labels = ax.get_legend_handles_labels()\nfig.legend(handles, labels, loc='upper right');",
"_____no_output_____"
],
[
"fig, axs = plt.subplots(nrows=1, ncols=2, figsize=(12, 6))\nfig.suptitle(\"Cross-section of the Posterior Distribution\", fontsize=16)\nsns.kdeplot(hmc_samples[\"bA\"], hmc_samples[\"bR\"], ax=axs[0], shade=True, label=\"HMC\")\nsns.kdeplot(svi_samples[\"bA\"], svi_samples[\"bR\"], ax=axs[0], label=\"SVI (DiagNormal)\")\naxs[0].set(xlabel=\"bA\", ylabel=\"bR\", xlim=(-2.5, -1.2), ylim=(-0.5, 0.1))\nsns.kdeplot(hmc_samples[\"bR\"], hmc_samples[\"bAR\"], ax=axs[1], shade=True, label=\"HMC\")\nsns.kdeplot(svi_samples[\"bR\"], svi_samples[\"bAR\"], ax=axs[1], label=\"SVI (DiagNormal)\")\naxs[1].set(xlabel=\"bR\", ylabel=\"bAR\", xlim=(-0.45, 0.05), ylim=(-0.15, 0.8))\nhandles, labels = axs[1].get_legend_handles_labels()\nfig.legend(handles, labels, loc='upper right');",
"_____no_output_____"
]
],
[
[
"## MultivariateNormal Guide\n\nAs comparison to the previously obtained results from Diagonal Normal guide, we will now use a guide that generates samples from a Cholesky factorization of a multivariate normal distribution. This allows us to capture the correlations between the latent variables via a covariance matrix. If we wrote this manually, we would need to combine all the latent variables so we could sample a Multivarite Normal jointly.",
"_____no_output_____"
]
],
[
[
"from pyro.infer.autoguide import AutoMultivariateNormal, init_to_mean\n\n\nguide = AutoMultivariateNormal(model, init_loc_fn=init_to_mean)\n\nsvi = SVI(model, \n guide, \n optim.Adam({\"lr\": .01}), \n loss=Trace_ELBO())\n\nis_cont_africa, ruggedness, log_gdp = train[:, 0], train[:, 1], train[:, 2]\npyro.clear_param_store()\nfor i in range(num_iters):\n elbo = svi.step(is_cont_africa, ruggedness, log_gdp)\n if i % 500 == 0:\n logging.info(\"Elbo loss: {}\".format(elbo))",
"Elbo loss: 703.0100790262222\nElbo loss: 444.6930855512619\nElbo loss: 258.20718491077423\nElbo loss: 249.05364602804184\nElbo loss: 247.2170884013176\nElbo loss: 247.28261297941208\nElbo loss: 246.61236548423767\nElbo loss: 249.86004841327667\nElbo loss: 249.1157277226448\nElbo loss: 249.86634194850922\n"
]
],
[
[
"Let's look at the shape of the posteriors again. You can see the multivariate guide is able to capture more of the true posterior.",
"_____no_output_____"
]
],
[
[
"predictive = Predictive(model, guide=guide, num_samples=num_samples)\nsvi_mvn_samples = {k: v.reshape(num_samples).detach().cpu().numpy() \n for k, v in predictive(log_gdp, is_cont_africa, ruggedness).items()\n if k != \"obs\"}\nfig, axs = plt.subplots(nrows=2, ncols=2, figsize=(12, 10))\nfig.suptitle(\"Marginal Posterior density - Regression Coefficients\", fontsize=16)\nfor i, ax in enumerate(axs.reshape(-1)):\n site = sites[i]\n sns.distplot(svi_mvn_samples[site], ax=ax, label=\"SVI (Multivariate Normal)\")\n sns.distplot(hmc_samples[site], ax=ax, label=\"HMC\")\n ax.set_title(site)\nhandles, labels = ax.get_legend_handles_labels()\nfig.legend(handles, labels, loc='upper right');",
"_____no_output_____"
]
],
[
[
"Now let's compare the posterior computed by the Diagonal Normal guide vs the Multivariate Normal guide. Note that the multivariate distribution is more dispresed than the Diagonal Normal.",
"_____no_output_____"
]
],
[
[
"fig, axs = plt.subplots(nrows=1, ncols=2, figsize=(12, 6))\nfig.suptitle(\"Cross-sections of the Posterior Distribution\", fontsize=16)\nsns.kdeplot(svi_samples[\"bA\"], svi_samples[\"bR\"], ax=axs[0], label=\"SVI (Diagonal Normal)\")\nsns.kdeplot(svi_mvn_samples[\"bA\"], svi_mvn_samples[\"bR\"], ax=axs[0], shade=True, label=\"SVI (Multivariate Normal)\")\naxs[0].set(xlabel=\"bA\", ylabel=\"bR\", xlim=(-2.5, -1.2), ylim=(-0.5, 0.1))\nsns.kdeplot(svi_samples[\"bR\"], svi_samples[\"bAR\"], ax=axs[1], label=\"SVI (Diagonal Normal)\")\nsns.kdeplot(svi_mvn_samples[\"bR\"], svi_mvn_samples[\"bAR\"], ax=axs[1], shade=True, label=\"SVI (Multivariate Normal)\")\naxs[1].set(xlabel=\"bR\", ylabel=\"bAR\", xlim=(-0.45, 0.05), ylim=(-0.15, 0.8))\nhandles, labels = axs[1].get_legend_handles_labels()\nfig.legend(handles, labels, loc='upper right');",
"_____no_output_____"
]
],
[
[
"and the Multivariate guide with the posterior computed by HMC. Note that the Multivariate guide better captures the true posterior.",
"_____no_output_____"
]
],
[
[
"fig, axs = plt.subplots(nrows=1, ncols=2, figsize=(12, 6))\nfig.suptitle(\"Cross-sections of the Posterior Distribution\", fontsize=16)\nsns.kdeplot(hmc_samples[\"bA\"], hmc_samples[\"bR\"], ax=axs[0], shade=True, label=\"HMC\")\nsns.kdeplot(svi_mvn_samples[\"bA\"], svi_mvn_samples[\"bR\"], ax=axs[0], label=\"SVI (Multivariate Normal)\")\naxs[0].set(xlabel=\"bA\", ylabel=\"bR\", xlim=(-2.5, -1.2), ylim=(-0.5, 0.1))\nsns.kdeplot(hmc_samples[\"bR\"], hmc_samples[\"bAR\"], ax=axs[1], shade=True, label=\"HMC\")\nsns.kdeplot(svi_mvn_samples[\"bR\"], svi_mvn_samples[\"bAR\"], ax=axs[1], label=\"SVI (Multivariate Normal)\")\naxs[1].set(xlabel=\"bR\", ylabel=\"bAR\", xlim=(-0.45, 0.05), ylim=(-0.15, 0.8))\nhandles, labels = axs[1].get_legend_handles_labels()\nfig.legend(handles, labels, loc='upper right');",
"_____no_output_____"
]
],
[
[
"## References\n[1] Hoffman, Matthew D., and Andrew Gelman. \"The No-U-turn sampler: adaptively setting path lengths in Hamiltonian Monte Carlo.\" Journal of Machine Learning Research 15.1 (2014): 1593-1623. https://arxiv.org/abs/1111.4246.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0d25338139e52b8437b6884e240904a93a9bb45 | 591,921 | ipynb | Jupyter Notebook | notebook.ipynb | akh20/The-Android-App-Market-on-Google-Play | f269c37b2a1c0c11b6638f52cec8eec99b610b6a | [
"MIT"
] | 1 | 2020-06-12T18:49:44.000Z | 2020-06-12T18:49:44.000Z | notebook.ipynb | akh20/The-Android-App-Market-on-Google-Play | f269c37b2a1c0c11b6638f52cec8eec99b610b6a | [
"MIT"
] | null | null | null | notebook.ipynb | akh20/The-Android-App-Market-on-Google-Play | f269c37b2a1c0c11b6638f52cec8eec99b610b6a | [
"MIT"
] | null | null | null | 591,921 | 591,921 | 0.763833 | [
[
[
"## 1. Google Play Store apps and reviews\n<p>Mobile apps are everywhere. They are easy to create and can be lucrative. Because of these two factors, more and more apps are being developed. In this notebook, we will do a comprehensive analysis of the Android app market by comparing over ten thousand apps in Google Play across different categories. We'll look for insights in the data to devise strategies to drive growth and retention.</p>\n<p><img src=\"https://assets.datacamp.com/production/project_619/img/google_play_store.png\" alt=\"Google Play logo\"></p>\n<p>Let's take a look at the data, which consists of two files:</p>\n<ul>\n<li><code>apps.csv</code>: contains all the details of the applications on Google Play. There are 13 features that describe a given app.</li>\n<li><code>user_reviews.csv</code>: contains 100 reviews for each app, <a href=\"https://www.androidpolice.com/2019/01/21/google-play-stores-redesigned-ratings-and-reviews-section-lets-you-easily-filter-by-star-rating/\">most helpful first</a>. The text in each review has been pre-processed and attributed with three new features: Sentiment (Positive, Negative or Neutral), Sentiment Polarity and Sentiment Subjectivity.</li>\n</ul>",
"_____no_output_____"
]
],
[
[
"# Read in dataset\nimport pandas as pd\napps_with_duplicates = pd.read_csv('datasets/apps.csv')\n\n# Drop duplicates\napps = apps_with_duplicates.drop_duplicates()\n\n# Print the total number of apps\nprint('Total number of apps in the dataset = ', len(apps['App']))\n\n# Have a look at a random sample of 5 rows\nn = 5\napps.sample(n)",
"Total number of apps in the dataset = 9659\n"
]
],
[
[
"## 2. Data cleaning\n<p>The four features that we will be working with most frequently henceforth are <code>Installs</code>, <code>Size</code>, <code>Rating</code> and <code>Price</code>. The <code>info()</code> function (from the previous task) told us that <code>Installs</code> and <code>Price</code> columns are of type <code>object</code> and not <code>int64</code> or <code>float64</code> as we would expect. This is because the column contains some characters more than just [0,9] digits. Ideally, we would want these columns to be numeric as their name suggests. <br>\nHence, we now proceed to data cleaning and prepare our data to be consumed in our analyis later. Specifically, the presence of special characters (<code>, $ +</code>) in the <code>Installs</code> and <code>Price</code> columns make their conversion to a numerical data type difficult.</p>",
"_____no_output_____"
]
],
[
[
"# List of characters to remove\nchars_to_remove = ['+' , ',' , '$']\n# List of column names to clean\ncols_to_clean = ['Installs' , 'Price']\n\n# Loop for each column\nfor col in cols_to_clean:\n # Replace each character with an empty string\n for char in chars_to_remove:\n apps[col] = apps[col].str.replace(char, '')\n # Convert col to numeric\n apps[col] = pd.to_numeric(apps[col])",
"_____no_output_____"
]
],
[
[
"## 3. Exploring app categories\n<p>With more than 1 billion active users in 190 countries around the world, Google Play continues to be an important distribution platform to build a global audience. For businesses to get their apps in front of users, it's important to make them more quickly and easily discoverable on Google Play. To improve the overall search experience, Google has introduced the concept of grouping apps into categories.</p>\n<p>This brings us to the following questions:</p>\n<ul>\n<li>Which category has the highest share of (active) apps in the market? </li>\n<li>Is any specific category dominating the market?</li>\n<li>Which categories have the fewest number of apps?</li>\n</ul>\n<p>We will see that there are <code>33</code> unique app categories present in our dataset. <em>Family</em> and <em>Game</em> apps have the highest market prevalence. Interestingly, <em>Tools</em>, <em>Business</em> and <em>Medical</em> apps are also at the top.</p>",
"_____no_output_____"
]
],
[
[
"import plotly\nplotly.offline.init_notebook_mode(connected=True)\nimport plotly.graph_objs as go\n\n# Print the total number of unique categories\nnum_categories = len(apps[\"Category\"].unique())\nprint('Number of categories = ', num_categories)\n\n# Count the number of apps in each 'Category' and sort them in descending order\nnum_apps_in_category = apps[\"Category\"].value_counts().sort_values(ascending = False)\n\ndata = [go.Bar(\n x = num_apps_in_category.index, # index = category name\n y = num_apps_in_category.values, # value = count\n)]\n\nplotly.offline.iplot(data)",
"_____no_output_____"
]
],
[
[
"## 4. Distribution of app ratings\n<p>After having witnessed the market share for each category of apps, let's see how all these apps perform on an average. App ratings (on a scale of 1 to 5) impact the discoverability, conversion of apps as well as the company's overall brand image. Ratings are a key performance indicator of an app.</p>\n<p>From our research, we found that the average volume of ratings across all app categories is <code>4.17</code>. The histogram plot is skewed to the right indicating that the majority of the apps are highly rated with only a few exceptions in the low-rated apps.</p>",
"_____no_output_____"
]
],
[
[
"# Average rating of apps\navg_app_rating = apps['Rating'].mean()\nprint('Average app rating = ', avg_app_rating)\n\n# Distribution of apps according to their ratings\ndata = [go.Histogram(\n x = apps['Rating']\n)]\n\n# Vertical dashed line to indicate the average app rating\nlayout = {'shapes': [{\n 'type' :'line',\n 'x0': avg_app_rating,\n 'y0': 0,\n 'x1': avg_app_rating,\n 'y1': 1000,\n 'line': { 'dash': 'dashdot'}\n }]\n }\n\nplotly.offline.iplot({'data': data, 'layout': layout})",
"Average app rating = 4.173243045387994\n"
]
],
[
[
"## 5. Size and price of an app\n<p>Let's now examine app size and app price. For size, if the mobile app is too large, it may be difficult and/or expensive for users to download. Lengthy download times could turn users off before they even experience your mobile app. Plus, each user's device has a finite amount of disk space. For price, some users expect their apps to be free or inexpensive. These problems compound if the developing world is part of your target market; especially due to internet speeds, earning power and exchange rates.</p>\n<p>How can we effectively come up with strategies to size and price our app?</p>\n<ul>\n<li>Does the size of an app affect its rating? </li>\n<li>Do users really care about system-heavy apps or do they prefer light-weighted apps? </li>\n<li>Does the price of an app affect its rating? </li>\n<li>Do users always prefer free apps over paid apps?</li>\n</ul>\n<p>We find that the majority of top rated apps (rating over 4) range from 2 MB to 20 MB. We also find that the vast majority of apps price themselves under \\$10.</p>",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport seaborn as sns\nsns.set_style(\"darkgrid\")\n\napps_with_size_and_rating_present = apps[(apps['Rating'].notnull()) & (apps[\"Size\"].notnull())]\n# Subset for categories with at least 250 apps\nlarge_categories = apps_with_size_and_rating_present.groupby('Category').filter(lambda x: len(x) >= 250).reset_index()\n\n# Plot size vs. rating\nplt1 = sns.jointplot(x = large_categories['Size'] , y = large_categories['Rating'] , kind = 'hex')\n\n# Subset out apps whose type is 'Paid'\npaid_apps = apps_with_size_and_rating_present[apps_with_size_and_rating_present['Type'] == 'Paid']\n\n# Plot price vs. rating\nplt2 = sns.jointplot(x = paid_apps['Price'] , y = paid_apps['Rating'] )",
"_____no_output_____"
]
],
[
[
"## 6. Relation between app category and app price\n<p>So now comes the hard part. How are companies and developers supposed to make ends meet? What monetization strategies can companies use to maximize profit? The costs of apps are largely based on features, complexity, and platform.</p>\n<p>There are many factors to consider when selecting the right pricing strategy for your mobile app. It is important to consider the willingness of your customer to pay for your app. A wrong price could break the deal before the download even happens. Potential customers could be turned off by what they perceive to be a shocking cost, or they might delete an app they’ve downloaded after receiving too many ads or simply not getting their money's worth.</p>\n<p>Different categories demand different price ranges. Some apps that are simple and used daily, like the calculator app, should probably be kept free. However, it would make sense to charge for a highly-specialized medical app that diagnoses diabetic patients. Below, we see that <em>Medical and Family</em> apps are the most expensive. Some medical apps extend even up to \\$80! All game apps are reasonably priced below \\$20.</p>",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nfig, ax = plt.subplots()\nfig.set_size_inches(15, 8)\n\n# Select a few popular app categories\npopular_app_cats = apps[apps.Category.isin(['GAME', 'FAMILY', 'PHOTOGRAPHY',\n 'MEDICAL', 'TOOLS', 'FINANCE',\n 'LIFESTYLE','BUSINESS'])]\n\n# Examine the price trend by plotting Price vs Category\nax = sns.stripplot(x = popular_app_cats['Price'], y = popular_app_cats['Category'], jitter=True, linewidth=1)\nax.set_title('App pricing trend across categories')\n\n# Apps whose Price is greater than 200\napps_above_200 = popular_app_cats[['Category', 'App', 'Price']][popular_app_cats['Price'] > 200]\napps_above_200",
"_____no_output_____"
]
],
[
[
"## 7. Filter out \"junk\" apps\n<p>It looks like a bunch of the really expensive apps are \"junk\" apps. That is, apps that don't really have a purpose. Some app developer may create an app called <em>I Am Rich Premium</em> or <em>most expensive app (H)</em> just for a joke or to test their app development skills. Some developers even do this with malicious intent and try to make money by hoping people accidentally click purchase on their app in the store.</p>\n<p>Let's filter out these junk apps and re-do our visualization. The distribution of apps under \\$20 becomes clearer.</p>",
"_____no_output_____"
]
],
[
[
"# Select apps priced below $100\napps_under_100 = popular_app_cats[popular_app_cats['Price'] < 100]\n\nfig, ax = plt.subplots()\nfig.set_size_inches(15, 8)\n\n# Examine price vs category with the authentic apps\nax = sns.stripplot(x=apps_under_100['Price'], y=apps_under_100['Category'], data=apps_under_100,\n jitter=True, linewidth=1)\nax.set_title('App pricing trend across categories after filtering for junk apps')",
"_____no_output_____"
]
],
[
[
"## 8. Popularity of paid apps vs free apps\n<p>For apps in the Play Store today, there are five types of pricing strategies: free, freemium, paid, paymium, and subscription. Let's focus on free and paid apps only. Some characteristics of free apps are:</p>\n<ul>\n<li>Free to download.</li>\n<li>Main source of income often comes from advertisements.</li>\n<li>Often created by companies that have other products and the app serves as an extension of those products.</li>\n<li>Can serve as a tool for customer retention, communication, and customer service.</li>\n</ul>\n<p>Some characteristics of paid apps are:</p>\n<ul>\n<li>Users are asked to pay once for the app to download and use it.</li>\n<li>The user can't really get a feel for the app before buying it.</li>\n</ul>\n<p>Are paid apps installed as much as free apps? It turns out that paid apps have a relatively lower number of installs than free apps, though the difference is not as stark as I would have expected!</p>",
"_____no_output_____"
]
],
[
[
"trace0 = go.Box(\n # Data for paid apps\n y=apps[apps['Type'] == 'Paid']['Installs'],\n name = 'Paid'\n)\n\ntrace1 = go.Box(\n # Data for free apps\n y=apps[apps['Type'] == 'Free']['Installs'],\n name = 'Free'\n)\n\nlayout = go.Layout(\n title = \"Number of downloads of paid apps vs. free apps\",\n yaxis = dict(\n type = 'log',\n autorange = True\n )\n)\n\n# Add trace0 and trace1 to a list for plotting\ndata = [trace0 , trace1]\nplotly.offline.iplot({'data': data, 'layout': layout})",
"_____no_output_____"
]
],
[
[
"## 9. Sentiment analysis of user reviews\n<p>Mining user review data to determine how people feel about your product, brand, or service can be done using a technique called sentiment analysis. User reviews for apps can be analyzed to identify if the mood is positive, negative or neutral about that app. For example, positive words in an app review might include words such as 'amazing', 'friendly', 'good', 'great', and 'love'. Negative words might be words like 'malware', 'hate', 'problem', 'refund', and 'incompetent'.</p>\n<p>By plotting sentiment polarity scores of user reviews for paid and free apps, we observe that free apps receive a lot of harsh comments, as indicated by the outliers on the negative y-axis. Reviews for paid apps appear never to be extremely negative. This may indicate something about app quality, i.e., paid apps being of higher quality than free apps on average. The median polarity score for paid apps is a little higher than free apps, thereby syncing with our previous observation.</p>\n<p>In this notebook, we analyzed over ten thousand apps from the Google Play Store. We can use our findings to inform our decisions should we ever wish to create an app ourselves.</p>",
"_____no_output_____"
]
],
[
[
"# Load user_reviews.csv\nreviews_df = pd.read_csv('datasets/user_reviews.csv')\n\n# Join and merge the two dataframe\nmerged_df = pd.merge(apps, reviews_df, on = 'App', how = \"inner\")\n\n# Drop NA values from Sentiment and Translated_Review columns\nmerged_df = merged_df.dropna(subset=['Sentiment', 'Translated_Review'])\n\nsns.set_style('ticks')\nfig, ax = plt.subplots()\nfig.set_size_inches(11, 8)\n\n# User review sentiment polarity for paid vs. free apps\nax = sns.boxplot(x = merged_df['Type'], y = merged_df['Sentiment_Polarity'], data = merged_df)\nax.set_title('Sentiment Polarity Distribution')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0d27305f12fe300a71626e5e166b89d10e4f2de | 396,513 | ipynb | Jupyter Notebook | 07 NLP/kaggle hw/solution.ipynb | ksetdekov/HSE_DS | 619d5b84f9d9e97b58ca1f12c5914ec65456c2c8 | [
"MIT"
] | 1 | 2020-09-26T18:48:11.000Z | 2020-09-26T18:48:11.000Z | 07 NLP/kaggle hw/solution.ipynb | ksetdekov/HSE_DS | 619d5b84f9d9e97b58ca1f12c5914ec65456c2c8 | [
"MIT"
] | null | null | null | 07 NLP/kaggle hw/solution.ipynb | ksetdekov/HSE_DS | 619d5b84f9d9e97b58ca1f12c5914ec65456c2c8 | [
"MIT"
] | null | null | null | 48.711671 | 67,563 | 0.564874 | [
[
[
"<a href=\"https://colab.research.google.com/github/ksetdekov/HSE_DS/blob/master/07%20NLP/kaggle%20hw/solution.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"# !pip3 install kaggle",
"_____no_output_____"
],
[
"from google.colab import files\nfiles.upload()",
"_____no_output_____"
],
[
"!mkdir ~/.kaggle\n!cp kaggle.json ~/.kaggle/\n!chmod 600 ~/.kaggle/kaggle.json\n",
"mkdir: cannot create directory ‘/root/.kaggle’: File exists\n"
],
[
"!kaggle competitions download -c toxic-comments-classification-apdl-2021",
"Warning: Looks like you're using an outdated API Version, please consider updating (server 1.5.12 / client 1.5.4)\nsample_submission.csv: Skipping, found more recently modified local copy (use --force to force download)\ntest_data.csv.zip: Skipping, found more recently modified local copy (use --force to force download)\ntrain_data.csv.zip: Skipping, found more recently modified local copy (use --force to force download)\n"
],
[
"!ls",
" bow_v3.csv\t kaggle.json sample_submission.csv train_data.csv.zip\n'kaggle (1).json' sample_data test_data.csv.zip\n"
],
[
"import pandas as pd\nimport numpy as np\nfrom sklearn.metrics import *\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.pipeline import Pipeline",
"_____no_output_____"
],
[
"train = pd.read_csv('train_data.csv.zip', compression='zip')\ntest = pd.read_csv('test_data.csv.zip', compression='zip')",
"_____no_output_____"
],
[
"train.toxic.describe()",
"_____no_output_____"
],
[
"train.sample(5)",
"_____no_output_____"
],
[
"test.sample(5)",
"_____no_output_____"
],
[
"x_train, x_test, y_train, y_test = train_test_split(train.comment, train.toxic, random_state=0, stratify=train.toxic)",
"_____no_output_____"
],
[
"y_train.describe()",
"_____no_output_____"
],
[
"y_test.describe()",
"_____no_output_____"
]
],
[
[
"## Bag of words\n",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LogisticRegression \nfrom sklearn.feature_extraction.text import CountVectorizer\nfrom nltk import ngrams",
"_____no_output_____"
],
[
"vec = CountVectorizer(ngram_range=(1, 2)) # строим BoW для слов\nbow = vec.fit_transform(x_train) \nvec2 = CountVectorizer(ngram_range=(1, 2)) # строим BoW для слов\nbow2 = vec2.fit_transform(train.comment) ",
"_____no_output_____"
],
[
"list(vec2.vocabulary_.items())[:10]",
"_____no_output_____"
],
[
"bow.mean()",
"_____no_output_____"
],
[
"clf = LogisticRegression(random_state=0, max_iter=500, class_weight='balanced')\nclf.fit(bow, y_train)",
"_____no_output_____"
],
[
"clf2 = LogisticRegression(random_state=0, max_iter=500, class_weight='balanced')\nclf2.fit(bow2, train.toxic)",
"_____no_output_____"
],
[
"pred = clf.predict(vec.transform(x_test))\nprint(classification_report(pred, y_test))",
" precision recall f1-score support\n\n 0.0 0.90 0.87 0.89 1858\n 1.0 0.74 0.79 0.76 845\n\n accuracy 0.85 2703\n macro avg 0.82 0.83 0.82 2703\nweighted avg 0.85 0.85 0.85 2703\n\n"
],
[
"",
"_____no_output_____"
],
[
"test",
"_____no_output_____"
],
[
"bow_test_pred = test.copy()\nbow_test_pred['toxic'] = clf.predict(vec.transform(test.comment))\nbow_test_pred['toxic'] = bow_test_pred['toxic'].astype(int)\nbow_test_pred.drop('comment', axis=1, inplace=True)\nbow_test_pred",
"_____no_output_____"
],
[
"bow_test_pred2 = test.copy()\nbow_test_pred2['toxic'] = clf2.predict(vec2.transform(test.comment))\nbow_test_pred2['toxic'] = bow_test_pred2['toxic'].astype(int)\nbow_test_pred2.drop('comment', axis=1, inplace=True)\nbow_test_pred2",
"_____no_output_____"
],
[
"bow_test_pred.to_csv('bow_v1.csv', index=False)\nbow_test_pred2.to_csv('bow_v2.csv', index=False)",
"_____no_output_____"
],
[
"confusion_matrix(bow_test_pred.toxic, bow_test_pred2.toxic)",
"_____no_output_____"
],
[
"# !kaggle competitions submit -c toxic-comments-classification-apdl-2021 -f bow_v2.csv -m \"kirill_setdekov first bow v2 submission all data\"",
"Warning: Looks like you're using an outdated API Version, please consider updating (server 1.5.12 / client 1.5.4)\n100% 23.6k/23.6k [00:09<00:00, 2.45kB/s]\nSuccessfully submitted to Toxic comments classification"
]
],
[
[
"## TF-IDF\n",
"_____no_output_____"
]
],
[
[
"from sklearn.feature_extraction.text import TfidfVectorizer",
"_____no_output_____"
],
[
"vec = TfidfVectorizer(ngram_range=(1, 1))\nbow = vec.fit_transform(x_train)\nclf2 = LogisticRegression(random_state=1, max_iter = 500)\nclf2.fit(bow, y_train)\npred = clf2.predict(vec.transform(x_test))\nprint(classification_report(pred, y_test))",
" precision recall f1-score support\n\n 0.0 0.97 0.79 0.87 2214\n 1.0 0.48 0.89 0.63 489\n\n accuracy 0.81 2703\n macro avg 0.73 0.84 0.75 2703\nweighted avg 0.88 0.81 0.83 2703\n\n"
],
[
"tf_idf = test.copy()\ntf_idf['toxic'] = clf2.predict(vec.transform(test.comment))\ntf_idf['toxic'] = tf_idf['toxic'].astype(int)\ntf_idf.drop('comment', axis=1, inplace=True)\ntf_idf",
"_____no_output_____"
],
[
"tf_idf.to_csv('tf_idf_v1.csv', index=False)",
"_____no_output_____"
],
[
"# !kaggle competitions submit -c toxic-comments-classification-apdl-2021 -f tf_idf_v1.csv -m \"kirill_setdekov tfidf v1 submission\"",
"_____no_output_____"
]
],
[
[
"## Symbol n-Grams",
"_____no_output_____"
]
],
[
[
"vec = CountVectorizer(analyzer='char', ngram_range=(1, 5))\nbowsimb = vec.fit_transform(x_train)\n",
"_____no_output_____"
],
[
"from sklearn.preprocessing import MaxAbsScaler \nscaler = MaxAbsScaler()\nscaler.fit(bowsimb)",
"_____no_output_____"
],
[
"bowsimb = scaler.transform(bowsimb)",
"_____no_output_____"
],
[
"clf3 = LogisticRegression(random_state=0, max_iter=1000)\nclf3.fit(bowsimb, y_train)\npred = clf3.predict(scaler.transform(vec.transform(x_test)))\nprint(classification_report(pred, y_test))",
" precision recall f1-score support\n\n 0.0 0.97 0.87 0.91 2000\n 1.0 0.71 0.91 0.80 703\n\n accuracy 0.88 2703\n macro avg 0.84 0.89 0.86 2703\nweighted avg 0.90 0.88 0.88 2703\n\n"
],
[
"importances = list(zip(vec.vocabulary_, clf.coef_[0]))\nimportances[0]",
"_____no_output_____"
],
[
"sorted_importances = sorted(importances, key = lambda x: -abs(x[1]))\nsorted_importances[:20]",
"_____no_output_____"
],
[
"symbol_ngrams = test.copy()\nsymbol_ngrams['toxic'] = clf3.predict(scaler.\n transform(vec.transform(test.comment)))\nsymbol_ngrams['toxic'] = tf_idf['toxic'].astype(int)\nsymbol_ngrams.drop('comment', axis=1, inplace=True)\nsymbol_ngrams",
"_____no_output_____"
],
[
"symbol_ngrams.to_csv('symbol_ngrams_v1.csv', index=False)",
"_____no_output_____"
],
[
"from sklearn.metrics import confusion_matrix",
"_____no_output_____"
],
[
"confusion_matrix(symbol_ngrams.toxic, tf_idf.toxic)",
"_____no_output_____"
],
[
"# !kaggle competitions submit -c toxic-comments-classification-apdl-2021 -f symbol_ngrams_v1.csv -m \"kirill_setdekov symbol_ngrams_v1 v1 submission\"",
"_____no_output_____"
]
],
[
[
"#FastText",
"_____no_output_____"
]
],
[
[
"!pip3 install fasttext\nimport fasttext",
"Collecting fasttext\n Downloading fasttext-0.9.2.tar.gz (68 kB)\n\u001b[?25l\r\u001b[K |████▊ | 10 kB 23.7 MB/s eta 0:00:01\r\u001b[K |█████████▌ | 20 kB 9.4 MB/s eta 0:00:01\r\u001b[K |██████████████▎ | 30 kB 8.1 MB/s eta 0:00:01\r\u001b[K |███████████████████ | 40 kB 7.5 MB/s eta 0:00:01\r\u001b[K |███████████████████████▉ | 51 kB 4.1 MB/s eta 0:00:01\r\u001b[K |████████████████████████████▋ | 61 kB 4.4 MB/s eta 0:00:01\r\u001b[K |████████████████████████████████| 68 kB 2.7 MB/s \n\u001b[?25hCollecting pybind11>=2.2\n Using cached pybind11-2.8.0-py2.py3-none-any.whl (207 kB)\nRequirement already satisfied: setuptools>=0.7.0 in /usr/local/lib/python3.7/dist-packages (from fasttext) (57.4.0)\nRequirement already satisfied: numpy in /usr/local/lib/python3.7/dist-packages (from fasttext) (1.19.5)\nBuilding wheels for collected packages: fasttext\n Building wheel for fasttext (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for fasttext: filename=fasttext-0.9.2-cp37-cp37m-linux_x86_64.whl size=3119085 sha256=735c99fbd5315605cd10b9a522e6fdfb760f3aa8f0ae77105b4d572b31285bd5\n Stored in directory: /root/.cache/pip/wheels/4e/ca/bf/b020d2be95f7641801a6597a29c8f4f19e38f9c02a345bab9b\nSuccessfully built fasttext\nInstalling collected packages: pybind11, fasttext\nSuccessfully installed fasttext-0.9.2 pybind11-2.8.0\n"
],
[
"with open('ft_train_data.txt', 'w') as f:\n for pair in list(zip(x_train, y_train)):\n text, label = pair\n f.write(f'__label__{int(label)} {text.lower()}\\n')\n \nwith open('ft_test_data.txt', 'w') as f:\n for pair in list(zip(x_test, y_test)):\n text, label = pair\n f.write(f'__label__{int(label)} {text.lower()}\\n')",
"_____no_output_____"
],
[
"with open('ft_all.txt', 'w') as f:\n for pair in list(zip(train.comment, train.toxic)):\n text, label = pair\n f.write(f'__label__{int(label)} {text.lower()}\\n')\n ",
"_____no_output_____"
],
[
"classifier = fasttext.train_supervised('ft_train_data.txt')#, 'model')\nresult = classifier.test('ft_test_data.txt')\nprint('P@1:', result[1])#.precision)\nprint('R@1:', result[2])#.recall)\nprint('Number of examples:', result[0])#.nexamples)",
"P@1: 0.8146503884572697\nR@1: 0.8146503884572697\nNumber of examples: 2703\n"
],
[
"classifier2 = fasttext.train_supervised('ft_all.txt')#, 'model')\n",
"_____no_output_____"
],
[
"k = 0\nfor item in [i.lower() for i in test.comment]:\n item = item.replace(\"\\n\",\" \")\n k +=1\nk",
"_____no_output_____"
],
[
"prediction = []\nfor item in [i.lower() for i in test.comment]:\n item = item.replace(\"\\n\",\" \")\n prediction.append(classifier.predict(item))",
"_____no_output_____"
],
[
"prediction2 = []\nfor item in [i.lower() for i in test.comment]:\n item = item.replace(\"\\n\",\" \")\n prediction2.append(classifier2.predict(item))",
"_____no_output_____"
],
[
"pred = [int(label[0][0].split('__')[2][0]) for label in prediction]\npred2 = [int(label[0][0].split('__')[2][0]) for label in prediction2]\n",
"_____no_output_____"
],
[
"fasttext_pred = test.copy()\nfasttext_pred['toxic'] = pred\nfasttext_pred.drop('comment', axis=1, inplace=True)\nfasttext_pred",
"_____no_output_____"
],
[
"fasttext_pred2 = test.copy()\nfasttext_pred2['toxic'] = pred2\nfasttext_pred2.drop('comment', axis=1, inplace=True)\nfasttext_pred2",
"_____no_output_____"
],
[
"confusion_matrix(symbol_ngrams.toxic, fasttext_pred.toxic)",
"_____no_output_____"
],
[
"confusion_matrix(fasttext_pred2.toxic, fasttext_pred.toxic)",
"_____no_output_____"
],
[
"fasttext_pred.to_csv('fasttext_pred_v1.csv', index=False)",
"_____no_output_____"
],
[
"fasttext_pred2.to_csv('fasttext_pred_v2.csv', index=False)",
"_____no_output_____"
],
[
"!kaggle competitions submit -c toxic-comments-classification-apdl-2021 -f fasttext_pred_v2.csv -m \"kirill_setdekov fasttext_pred v2 submission\"",
"Warning: Looks like you're using an outdated API Version, please consider updating (server 1.5.12 / client 1.5.4)\n100% 23.6k/23.6k [00:07<00:00, 3.36kB/s]\nSuccessfully submitted to Toxic comments classification"
]
],
[
[
"## CNN",
"_____no_output_____"
]
],
[
[
"from torchtext.legacy import data",
"_____no_output_____"
],
[
"pd.read_csv('train_data.csv.zip', compression='zip')",
"_____no_output_____"
],
[
"!unzip train_data.csv.zip\n!unzip test_data.csv.zip\n",
"Archive: train_data.csv.zip\n inflating: train_data.csv \nArchive: test_data.csv.zip\n inflating: test_data.csv \n"
],
[
"# классы Field и LabelField отвечают за то, как данные будут храниться и обрабатываться при считывании\nTEXT = data.Field(tokenize='spacy') # spacy -- значит, токенизацию будет делать модуль \nLABEL = data.LabelField()\n\nds = data.TabularDataset(\n path='train_data.csv', format='csv', \n skip_header=True,\n fields=[('comment', TEXT),\n ('toxic', LABEL)]\n)",
"_____no_output_____"
],
[
"pd.read_csv('test_data.csv')",
"_____no_output_____"
],
[
"test = data.TabularDataset(\n path='test_data.csv', format='csv', \n skip_header=True,\n fields=[('id', TEXT), ('comment', TEXT)]\n)",
"_____no_output_____"
],
[
"next(ds.comment)",
"_____no_output_____"
],
[
"next(ds.toxic)",
"_____no_output_____"
],
[
"TEXT.build_vocab(ds, max_size=25000, vectors=\"glove.6B.100d\")\nLABEL.build_vocab(ds)",
".vector_cache/glove.6B.zip: 862MB [02:42, 5.30MB/s] \n100%|█████████▉| 399999/400000 [00:21<00:00, 18835.77it/s]\n"
],
[
"TEXT.vocab.itos[:20]",
"_____no_output_____"
],
[
"len(TEXT.vocab.itos)",
"_____no_output_____"
],
[
"train, val = ds.split(split_ratio=0.9, stratified=True, strata_field='toxic') # дефолтное соотношение 0.7\n",
"_____no_output_____"
],
[
"print(len(train))\nprint(len(val))\nprint(len(test))",
"9728\n1081\n3603\n"
],
[
"BATCH_SIZE = 64\n\ntrain_iterator, valid_iterator, test_iterator = data.BucketIterator.splits(\n (train, val, test), \n batch_size=BATCH_SIZE, \n sort=True,\n sort_key=lambda x: len(x.comment), # сорируем тексты по длине, чтобы рядом оказывались предложения с одинаковой длиной и добавлялось меньше паддинга\n repeat=False)",
"_____no_output_____"
],
[
"for i, batch in enumerate(valid_iterator):\n print(batch.batch_size)\n # pass",
"64\n64\n64\n64\n64\n64\n64\n64\n64\n64\n64\n64\n64\n64\n64\n64\n57\n"
],
[
"batch.fields",
"_____no_output_____"
],
[
"batch.batch_size",
"_____no_output_____"
],
[
"batch.comment",
"_____no_output_____"
],
[
"batch.toxic",
"_____no_output_____"
],
[
"len(batch.toxic)",
"_____no_output_____"
],
[
"import torch.nn as nn",
"_____no_output_____"
],
[
"class CNN(nn.Module):\n def __init__(self, vocab_size, embedding_dim, n_filters, filter_sizes, output_dim, dropout_proba):\n super().__init__()\n \n self.embedding = nn.Embedding(vocab_size, embedding_dim)\n self.conv_0 = nn.Conv2d(in_channels=1, out_channels=n_filters, kernel_size=(filter_sizes[0], embedding_dim))\n self.conv_1 = nn.Conv2d(in_channels=1, out_channels=n_filters, kernel_size=(filter_sizes[1], embedding_dim))\n self.conv_2 = nn.Conv2d(in_channels=1, out_channels=n_filters, kernel_size=(filter_sizes[2], embedding_dim))\n self.fc = nn.Linear(len(filter_sizes) * n_filters, output_dim)\n self.dropout = nn.Dropout(dropout_proba)\n \n def forward(self, x):\n #x = [sent len, batch size]\n # print(x.shape)\n x = x.permute(1, 0)\n \n #x = [batch size, sent len]\n embedded = self.embedding(x)\n #print(embedded.shape)\n \n #embedded = [batch size, sent len, emb dim]\n embedded = embedded.unsqueeze(1)\n \n \n #embedded = [batch size, 1, sent len, emb dim]\n conv_0 = self.conv_0(embedded)\n #print(conv_0.shape)\n conv_0 = conv_0.squeeze(3)\n #print(conv_0.shape)\n conved_0 = F.relu(conv_0)\n conved_1 = F.relu(self.conv_1(embedded).squeeze(3))\n conved_2 = F.relu(self.conv_2(embedded).squeeze(3))\n \n #conv_n = [batch size, n_filters, sent len - filter_sizes[n]]\n # print(conved_0.shape)\n pool_0 = F.max_pool1d(conved_0, conved_0.shape[2])\n # print(pool_0.shape)\n\n pooled_0 = pool_0.squeeze(2)\n # print(pooled_0.shape)\n pooled_1 = F.max_pool1d(conved_1, conved_1.shape[2]).squeeze(2)\n pooled_2 = F.max_pool1d(conved_2, conved_2.shape[2]).squeeze(2)\n \n #pooled_n = [batch size, n_filters]\n cat = self.dropout(torch.cat((pooled_0, pooled_1, pooled_2), dim=1))\n\n #cat = [batch size, n_filters * len(filter_sizes)]\n return self.fc(cat)",
"_____no_output_____"
],
[
"import torch.nn.functional as F\n\ndef binary_accuracy(preds, y):\n rounded_preds = torch.round(F.sigmoid(preds))\n correct = (rounded_preds == y).float()\n acc = correct.sum() / len(correct)\n return acc",
"_____no_output_____"
],
[
"def train_func(model, iterator, optimizer, criterion):\n epoch_loss = 0\n epoch_acc = 0\n \n model.train()\n \n for batch in iterator:\n optimizer.zero_grad()\n \n predictions = model(batch.comment.cuda()).squeeze(1)\n\n loss = criterion(predictions.float(), batch.toxic.float().cuda())\n acc = binary_accuracy(predictions.float(), batch.toxic.float().cuda())\n \n loss.backward()\n optimizer.step()\n \n epoch_loss += loss\n epoch_acc += acc\n \n return epoch_loss / len(iterator), epoch_acc / len(iterator)",
"_____no_output_____"
],
[
"def evaluate_func(model, iterator, criterion):\n epoch_loss = 0\n epoch_acc = 0\n \n model.eval()\n \n with torch.no_grad():\n for batch in iterator:\n predictions = model(batch.comment.cuda()).squeeze(1)\n\n loss = criterion(predictions.float(), batch.toxic.float().cuda())\n acc = binary_accuracy(predictions.float(), batch.toxic.float().cuda())\n\n epoch_loss += loss\n epoch_acc += acc\n \n return epoch_loss / len(iterator), epoch_acc / len(iterator)",
"_____no_output_____"
],
[
"INPUT_DIM = len(TEXT.vocab)\nEMBEDDING_DIM = 100\nN_FILTERS = 100\nFILTER_SIZES = [2,3,4]\nOUTPUT_DIM = 1\nDROPOUT_PROBA = 0.5\n\nmodel = CNN(INPUT_DIM, EMBEDDING_DIM, N_FILTERS, FILTER_SIZES, OUTPUT_DIM, DROPOUT_PROBA)",
"_____no_output_____"
],
[
"INPUT_DIM",
"_____no_output_____"
],
[
"model",
"_____no_output_____"
],
[
"pretrained_embeddings = TEXT.vocab.vectors\nmodel.embedding.weight.data.copy_(pretrained_embeddings)",
"_____no_output_____"
],
[
"import torch.optim as optim",
"_____no_output_____"
],
[
"optimizer = optim.Adam(model.parameters()) # мы подали оптимизатору все параметры -- значит, эмбеддиги тоже будут дообучаться\ncriterion = nn.BCEWithLogitsLoss() # бинарная кросс-энтропия с логитами\n\nmodel = model.cuda() # будем учить на gpu! =)",
"_____no_output_____"
],
[
"model.embedding",
"_____no_output_____"
],
[
"from torchsummary import summary\n\n# summary(model, (14))\n",
"_____no_output_____"
],
[
"import torch",
"_____no_output_____"
],
[
"N_EPOCHS = 8\n\nfor epoch in range(N_EPOCHS):\n train_loss, train_acc = train_func(model, train_iterator, optimizer, criterion)\n valid_loss, valid_acc = evaluate_func(model, valid_iterator, criterion)\n \n print(f'Epoch: {epoch+1:02}, Train Loss: {train_loss:.3f}, Train Acc: {train_acc*100:.2f}%, Val. Loss: {valid_loss:.3f}, Val. Acc: {valid_acc*100:.2f}%')",
"/usr/local/lib/python3.7/dist-packages/torch/nn/functional.py:652: UserWarning: Named tensors and all their associated APIs are an experimental feature and subject to change. Please do not use them for anything important until they are released as stable. (Triggered internally at /pytorch/c10/core/TensorImpl.h:1156.)\n return torch.max_pool1d(input, kernel_size, stride, padding, dilation, ceil_mode)\n/usr/local/lib/python3.7/dist-packages/torch/nn/functional.py:1805: UserWarning: nn.functional.sigmoid is deprecated. Use torch.sigmoid instead.\n warnings.warn(\"nn.functional.sigmoid is deprecated. Use torch.sigmoid instead.\")\n"
],
[
"test.examples",
"_____no_output_____"
],
[
"model.eval()\ncnn_res = []\nwith torch.no_grad():\n for batch in test_iterator:\n predictions = model(batch.comment.cuda())\n cnn_res.append(predictions)\n ",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"testout = pd.read_csv('test_data.csv.zip', compression='zip')",
"_____no_output_____"
],
[
"cnnpred = testout.copy()\ncnnpred['toxic'] = [float(item) for sublist in cnn_res for item in sublist]\ncnnpred.drop('comment', axis=1, inplace=True)\ncnnpred",
"_____no_output_____"
],
[
"cnnpred['toxic'] = (cnnpred['toxic'] > 0).astype(int)\ncnnpred",
"_____no_output_____"
],
[
"cnnpred.to_csv('cnnpred_v4.csv', index=False)",
"_____no_output_____"
],
[
"!kaggle competitions submit -c toxic-comments-classification-apdl-2021 -f cnnpred_v4.csv -m \"kirill_setdekov cnn v4 with threshold 0\"",
"Warning: Looks like you're using an outdated API Version, please consider updating (server 1.5.12 / client 1.5.4)\n100% 23.6k/23.6k [00:04<00:00, 4.90kB/s]\nSuccessfully submitted to Toxic comments classification"
],
[
"",
"_____no_output_____"
]
],
[
[
"# word2vec\n\n\n> not done, skip this model\n\n",
"_____no_output_____"
]
],
[
[
"! wget https://nlp.stanford.edu/data/glove.6B.zip",
"_____no_output_____"
],
[
"with open(\"alice.txt\", 'r', encoding='utf-8') as f:\n text = f.read()\n\ntext = re.sub('\\n', ' ', text)\nsents = sent_tokenize(text)\n\npunct = '!\"#$%&()*+,-./:;<=>?@[\\]^_`{|}~„“«»†*—/\\-‘’'\nclean_sents = []\n\nfor sent in sents:\n s = [w.lower().strip(punct) for w in sent.split()]\n clean_sents.append(s)\n \nprint(clean_sents[:2])",
"_____no_output_____"
],
[
"model_path = \"movie_reviews.model\"\n\nprint(\"Saving model...\")\nmodel_en.save(model_path)",
"_____no_output_____"
],
[
"model = word2vec.Word2Vec.load(model_path)\n\nmodel.build_vocab(clean_sents, update=True)\nmodel.train(clean_sents, total_examples=model.corpus_count, epochs=5)",
"_____no_output_____"
]
],
[
[
"# bow on random forest\n\n",
"_____no_output_____"
]
],
[
[
"! pip install pymystem3",
"Requirement already satisfied: pymystem3 in /usr/local/lib/python3.7/dist-packages (0.2.0)\nRequirement already satisfied: requests in /usr/local/lib/python3.7/dist-packages (from pymystem3) (2.23.0)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.7/dist-packages (from requests->pymystem3) (2.10)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.7/dist-packages (from requests->pymystem3) (3.0.4)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.7/dist-packages (from requests->pymystem3) (2021.5.30)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.7/dist-packages (from requests->pymystem3) (1.24.3)\n"
],
[
"! pip install --force-reinstall pymorphy2 \n",
"Collecting pymorphy2\n Using cached pymorphy2-0.9.1-py3-none-any.whl (55 kB)\nCollecting dawg-python>=0.7.1\n Using cached DAWG_Python-0.7.2-py2.py3-none-any.whl (11 kB)\nCollecting docopt>=0.6\n Using cached docopt-0.6.2-py2.py3-none-any.whl\nCollecting pymorphy2-dicts-ru<3.0,>=2.4\n Using cached pymorphy2_dicts_ru-2.4.417127.4579844-py2.py3-none-any.whl (8.2 MB)\nInstalling collected packages: pymorphy2-dicts-ru, docopt, dawg-python, pymorphy2\n Attempting uninstall: pymorphy2-dicts-ru\n Found existing installation: pymorphy2-dicts-ru 2.4.417127.4579844\n Uninstalling pymorphy2-dicts-ru-2.4.417127.4579844:\n Successfully uninstalled pymorphy2-dicts-ru-2.4.417127.4579844\n Attempting uninstall: docopt\n Found existing installation: docopt 0.6.2\n Uninstalling docopt-0.6.2:\n Successfully uninstalled docopt-0.6.2\n Attempting uninstall: dawg-python\n Found existing installation: DAWG-Python 0.7.2\n Uninstalling DAWG-Python-0.7.2:\n Successfully uninstalled DAWG-Python-0.7.2\n Attempting uninstall: pymorphy2\n Found existing installation: pymorphy2 0.9.1\n Uninstalling pymorphy2-0.9.1:\n Successfully uninstalled pymorphy2-0.9.1\nSuccessfully installed dawg-python-0.7.2 docopt-0.6.2 pymorphy2-0.9.1 pymorphy2-dicts-ru-2.4.417127.4579844\n"
],
[
"!pip install pymorphy2-dicts-ru",
"Requirement already satisfied: pymorphy2-dicts-ru in /usr/local/lib/python3.7/dist-packages (2.4.417127.4579844)\n"
],
[
"import pymorphy2\nimport re\n\nmorph = pymorphy2.MorphAnalyzer()",
"_____no_output_____"
],
[
"\n# убираем все небуквенные символы\nregex = re.compile(\"[А-Яа-яA-z]+\")\n\ndef words_only(text, regex=regex):\n try:\n return regex.findall(text.lower())\n except:\n return []",
"_____no_output_____"
],
[
"for i in train.comment[10].split():\n lemmas = morph.parse(i)\n print(lemmas[0])",
"Parse(word='куколдыш', tag=OpencorporaTag('NOUN,anim,masc,Sgtm,Surn sing,nomn'), normal_form='куколдыш', score=0.14285714285714285, methods_stack=((FakeDictionary(), 'куколдыш', 84, 0), (KnownSuffixAnalyzer(min_word_length=4, score_multiplier=0.5), 'лдыш')))\nParse(word='иди', tag=OpencorporaTag('VERB,impf,intr sing,impr,excl'), normal_form='идти', score=1.0, methods_stack=((DictionaryAnalyzer(), 'иди', 1696, 13),))\nParse(word='значение', tag=OpencorporaTag('NOUN,inan,neut sing,accs'), normal_form='значение', score=0.625, methods_stack=((DictionaryAnalyzer(), 'значение', 77, 6),))\nParse(word='выучи,', tag=OpencorporaTag('UNKN'), normal_form='выучи,', score=1.0, methods_stack=((UnknAnalyzer(), 'выучи,'),))\nParse(word='мурло', tag=OpencorporaTag('NOUN,inan,neut,Sgtm sing,nomn'), normal_form='мурло', score=0.5, methods_stack=((DictionaryAnalyzer(), 'мурло', 110, 0),))\nParse(word='скоро', tag=OpencorporaTag('ADVB'), normal_form='скоро', score=0.964285, methods_stack=((DictionaryAnalyzer(), 'скоро', 3, 0),))\nParse(word='и', tag=OpencorporaTag('CONJ'), normal_form='и', score=0.998263, methods_stack=((DictionaryAnalyzer(), 'и', 20, 0),))\nParse(word='тебе', tag=OpencorporaTag('NPRO,2per sing,datv'), normal_form='ты', score=0.970731, methods_stack=((DictionaryAnalyzer(), 'тебе', 3049, 2),))\nParse(word='пиздарики', tag=OpencorporaTag('NOUN,inan,masc plur,nomn'), normal_form='пиздарик', score=0.2, methods_stack=((DictionaryAnalyzer(), 'дарики', 19, 6), (UnknownPrefixAnalyzer(score_multiplier=0.5), 'пиз')))\nParse(word='наступят.', tag=OpencorporaTag('UNKN'), normal_form='наступят.', score=1.0, methods_stack=((UnknAnalyzer(), 'наступят.'),))\nParse(word='что', tag=OpencorporaTag('CONJ'), normal_form='что', score=0.922033, methods_stack=((DictionaryAnalyzer(), 'что', 3185, 0),))\nParse(word='сделаешь,', tag=OpencorporaTag('UNKN'), normal_form='сделаешь,', score=1.0, methods_stack=((UnknAnalyzer(), 'сделаешь,'),))\nParse(word='насрешь', tag=OpencorporaTag('VERB,impf,tran sing,2per,pres,indc'), normal_form='насресть', score=0.33653846153846156, methods_stack=((DictionaryAnalyzer(), 'ешь', 1447, 3), (UnknownPrefixAnalyzer(score_multiplier=0.5), 'наср')))\nParse(word='под', tag=OpencorporaTag('PREP'), normal_form='под', score=0.977989, methods_stack=((DictionaryAnalyzer(), 'под', 393, 0),))\nParse(word='дверью,', tag=OpencorporaTag('UNKN'), normal_form='дверью,', score=1.0, methods_stack=((UnknAnalyzer(), 'дверью,'),))\nParse(word='чмо?', tag=OpencorporaTag('UNKN'), normal_form='чмо?', score=1.0, methods_stack=((UnknAnalyzer(), 'ЧМО?'),))\n"
],
[
"from functools import lru_cache\n",
"_____no_output_____"
],
[
"@lru_cache(maxsize=128)\ndef lemmatize_word(token, pymorphy=morph):\n return pymorphy.parse(token)[0].normal_form\n\ndef lemmatize_text(text):\n return [lemmatize_word(w) for w in text]",
"_____no_output_____"
],
[
"tokens = words_only(train.comment[10])\n\nprint(lemmatize_text(tokens))",
"['куколдыш', 'идти', 'значение', 'выучить', 'мурло', 'скоро', 'и', 'ты', 'пиздарик', 'наступить', 'что', 'сделать', 'насресть', 'под', 'дверь', 'чмо']\n"
],
[
"from nltk.corpus import stopwords\n",
"_____no_output_____"
],
[
"import nltk",
"_____no_output_____"
],
[
"nltk.download('stopwords')",
"[nltk_data] Downloading package stopwords to /root/nltk_data...\n[nltk_data] Package stopwords is already up-to-date!\n"
],
[
"mystopwords = stopwords.words('russian') \n\ndef remove_stopwords(lemmas, stopwords = mystopwords):\n return [w for w in lemmas if not w in stopwords]",
"_____no_output_____"
],
[
"lemmas = lemmatize_text(tokens)\n\nprint(*remove_stopwords(lemmas))",
"куколдыш идти значение выучить мурло скоро пиздарик наступить сделать насресть дверь чмо\n"
],
[
"def remove_stopwords(lemmas, stopwords = mystopwords):\n return [w for w in lemmas if not w in stopwords and len(w) > 3]",
"_____no_output_____"
],
[
"print(*remove_stopwords(lemmas))",
"куколдыш идти значение выучить мурло скоро пиздарик наступить сделать насресть дверь\n"
],
[
"def clean_text(text):\n tokens = words_only(text)\n lemmas = lemmatize_text(tokens)\n \n return remove_stopwords(lemmas)",
"_____no_output_____"
],
[
"for i in range(20):\n print(* clean_text(train.comment[i]))",
"преступление наказание\nименно неработающий весы показывать работать\nяпония панелька ебанько\nвыявлять трещина помощь белый краска магнитная краска прислонять большой магнит трещина проявляться знать метод называться труба проверять\nдочитать поезд норильск далёкий стать\nвесь нужно перестать бухать вывести остаток токсин организм внезапно жизнь начинать играть новый краска чистый физиология психология хотя отказ транк доступный транк легализовать наркотик скорее весь прич депрессия таковой существовать вообще принцип просто состояние который индивид начинать вгонять оправдявый вступление тропа саморазрушение депрессия край полежать дать вино дать гкий наркотик тяжёлый депрессия просто деструктивизм сознательный аморализация надежда найда пойма нужно выплакаться жилетка состояние просто щелчок палец выйти затягивать многий затягивать самый вчерашний каприз становиться настоящий личностный проблема просто делать нужно\nнезависимый оператор хохол пытаться промутить\nответ батя говорить аккале\nсхуй эсэсэр коммунистический режим дебилушко юмор китай ебета хвост грива хлипкий социализм сэсэр простоять сотня\nполетать красавец попадаться женщина наоборот\nкуколдыш идти значение выучить мурло скоро пиздарик наступить сделать насресть дверь\nстрашно крутой тачка принести счастие честно знать мочь принести\nсвынок укатываться кропивач твой ранимый анус никто тронуть\nорать десять жопа ебать начать хуесос хотя наверняка понравиться\nхотя марцинкевич блядь строка перчеркнуть выше написать\nкрасиво подробный\nсторона модератор никто звать картинка удалить\nмочь бугор давно пофига частность большой половина killing floor христианский demon hunter христианский металл living sacrifice правда назвать скорее металл факт факт\nнищий нацист коммунист гитлер немец очередь стоять негр араб помогать помогать мера\nупоминание давыдов голополосов вызывать баттхерта пикабушник правильно\n"
],
[
"from tqdm.auto import trange",
"_____no_output_____"
],
[
"new_comments = []\nfor i in trange(len(train.comment), desc='loop'):\n new_comments.append(\" \".join(clean_text(train.comment[i])))\n",
"_____no_output_____"
],
[
"new_comments[:10]",
"_____no_output_____"
],
[
"vec3 = CountVectorizer(ngram_range=(1, 2)) # строим BoW для слов\nbow3 = vec3.fit_transform(new_comments) ",
"_____no_output_____"
],
[
"list(vec3.vocabulary_.items())[100:120]",
"_____no_output_____"
],
[
"bow3",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"clf3 = LogisticRegression(random_state=0, max_iter=500, class_weight='balanced')\nclf3.fit(bow3, train.toxic)",
"_____no_output_____"
],
[
"pred = clf3.predict(bow3)\nprint(classification_report(pred, train.toxic))",
" precision recall f1-score support\n\n 0.0 0.99 1.00 1.00 7148\n 1.0 1.00 0.98 0.99 3661\n\n accuracy 0.99 10809\n macro avg 1.00 0.99 0.99 10809\nweighted avg 0.99 0.99 0.99 10809\n\n"
],
[
"",
"_____no_output_____"
],
[
"test",
"_____no_output_____"
],
[
"new_commentstest = []\nfor i in trange(len(test.comment), desc='loop'):\n new_commentstest.append(\" \".join(clean_text(test.comment[i])))",
"_____no_output_____"
],
[
"bow_test_pred3 = test.copy()\nbow_test_pred3['newcomment'] = new_commentstest\nbow_test_pred3.tail()",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"bow_test_pred3['toxic'] = clf3.predict(vec3.transform(bow_test_pred3.newcomment))\nbow_test_pred3['toxic'] = bow_test_pred3['toxic'].astype(int)\nbow_test_pred3.drop('comment', axis=1, inplace=True)\nbow_test_pred3.drop('newcomment', axis=1, inplace=True)\n\nbow_test_pred3",
"_____no_output_____"
],
[
"confusion_matrix(bow_test_pred2.toxic, bow_test_pred3.toxic)",
"_____no_output_____"
],
[
"bow_test_pred3.to_csv('bow_v3.csv', index=False)",
"_____no_output_____"
],
[
"# !kaggle competitions submit -c toxic-comments-classification-apdl-2021 -f bow_v3.csv -m \"kirill_setdekov bow3 with preprocessing\"",
"_____no_output_____"
],
[
"!pip install scikit-learn==0.24",
"Requirement already satisfied: scikit-learn==0.24 in /usr/local/lib/python3.7/dist-packages (0.24.0)\nRequirement already satisfied: joblib>=0.11 in /usr/local/lib/python3.7/dist-packages (from scikit-learn==0.24) (1.0.1)\nRequirement already satisfied: threadpoolctl>=2.0.0 in /usr/local/lib/python3.7/dist-packages (from scikit-learn==0.24) (3.0.0)\nRequirement already satisfied: scipy>=0.19.1 in /usr/local/lib/python3.7/dist-packages (from scikit-learn==0.24) (1.4.1)\nRequirement already satisfied: numpy>=1.13.3 in /usr/local/lib/python3.7/dist-packages (from scikit-learn==0.24) (1.19.5)\n"
],
[
"from sklearn.ensemble import RandomForestClassifier\nfrom sklearn.experimental import enable_halving_search_cv # noqa\nfrom sklearn.model_selection import HalvingGridSearchCV",
"_____no_output_____"
]
],
[
[
"nor run -too slow",
"_____no_output_____"
]
],
[
[
"# rnd_reg = RandomForestClassifier( )\n\n# # hyper-parameter space\n# param_grid_RF = {\n# 'n_estimators' : [10,20,50,100,200,500,1000],\n# 'max_features' : [0.6,0.8,\"auto\",\"sqrt\"],\n# }\n\n# search_two = HalvingGridSearchCV(rnd_reg, param_grid_RF, factor=5, scoring='accuracy',\n# n_jobs=-1, random_state=0, verbose=2).fit(bow3, train.toxic)\n\n# search_two.best_params_",
"_____no_output_____"
],
[
"rnd_reg_2 = RandomForestClassifier(n_estimators=1000, verbose=5, n_jobs=-1)",
"_____no_output_____"
],
[
"search_no = rnd_reg_2.fit(bow3, train.toxic)",
"[Parallel(n_jobs=-1)]: Using backend ThreadingBackend with 2 concurrent workers.\n"
],
[
"bow_test_pred4 = test.copy()\nbow_test_pred4['newcomment'] = new_commentstest\nbow_test_pred4.tail()",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"bow_test_pred4['toxic'] = search_no.predict(vec3.transform(bow_test_pred4.newcomment))\nbow_test_pred4['toxic'] = bow_test_pred4['toxic'].astype(int)\nbow_test_pred4.drop('comment', axis=1, inplace=True)\nbow_test_pred4.drop('newcomment', axis=1, inplace=True)\n\nbow_test_pred4",
"[Parallel(n_jobs=2)]: Using backend ThreadingBackend with 2 concurrent workers.\n[Parallel(n_jobs=2)]: Done 14 tasks | elapsed: 0.1s\n[Parallel(n_jobs=2)]: Done 68 tasks | elapsed: 0.4s\n[Parallel(n_jobs=2)]: Done 158 tasks | elapsed: 1.0s\n[Parallel(n_jobs=2)]: Done 284 tasks | elapsed: 1.7s\n[Parallel(n_jobs=2)]: Done 446 tasks | elapsed: 2.7s\n[Parallel(n_jobs=2)]: Done 644 tasks | elapsed: 3.8s\n[Parallel(n_jobs=2)]: Done 878 tasks | elapsed: 5.1s\n[Parallel(n_jobs=2)]: Done 1000 out of 1000 | elapsed: 5.8s finished\n"
],
[
"confusion_matrix(bow_test_pred4.toxic, bow_test_pred3.toxic)",
"_____no_output_____"
],
[
"bow_test_pred4.to_csv('bow_v4.csv', index=False)",
"_____no_output_____"
],
[
"!kaggle competitions submit -c toxic-comments-classification-apdl-2021 -f bow_v4.csv -m \"kirill_setdekov bow4 with preprocessing and RF\"",
"Warning: Looks like you're using an outdated API Version, please consider updating (server 1.5.12 / client 1.5.4)\n100% 23.6k/23.6k [00:04<00:00, 5.15kB/s]\nSuccessfully submitted to Toxic comments classification"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0d2762f7b3216681080715b29be532c26257474 | 3,374 | ipynb | Jupyter Notebook | notebooks/WhileLoop.ipynb | gcallah/IntroPython | 30c4bd19236b8ab2f97293ce3a786a45828fc2ca | [
"Apache-2.0"
] | 1 | 2020-11-09T16:13:06.000Z | 2020-11-09T16:13:06.000Z | notebooks/WhileLoop.ipynb | gcallah/IntroPython | 30c4bd19236b8ab2f97293ce3a786a45828fc2ca | [
"Apache-2.0"
] | null | null | null | notebooks/WhileLoop.ipynb | gcallah/IntroPython | 30c4bd19236b8ab2f97293ce3a786a45828fc2ca | [
"Apache-2.0"
] | 6 | 2020-10-19T15:07:49.000Z | 2020-12-18T22:57:44.000Z | 23.268966 | 123 | 0.463545 | [
[
[
"## Looping with While\n\nNow let us look at looping with `while` in Python, one of the ways in which we can repeatedly perform some action(s).",
"_____no_output_____"
],
[
"### While loops",
"_____no_output_____"
],
[
"#### continue",
"_____no_output_____"
]
],
[
[
"n = 43\nwhile n > 2:\n n -= 1\n if n % 2 == 0:\n continue\n print(\"n = \", n, \"; Testing if number is prime.\")",
"n = 41 ; Testing if number is prime.\nn = 39 ; Testing if number is prime.\nn = 37 ; Testing if number is prime.\nn = 35 ; Testing if number is prime.\nn = 33 ; Testing if number is prime.\nn = 31 ; Testing if number is prime.\nn = 29 ; Testing if number is prime.\nn = 27 ; Testing if number is prime.\nn = 25 ; Testing if number is prime.\nn = 23 ; Testing if number is prime.\nn = 21 ; Testing if number is prime.\nn = 19 ; Testing if number is prime.\nn = 17 ; Testing if number is prime.\nn = 15 ; Testing if number is prime.\nn = 13 ; Testing if number is prime.\nn = 11 ; Testing if number is prime.\nn = 9 ; Testing if number is prime.\nn = 7 ; Testing if number is prime.\nn = 5 ; Testing if number is prime.\nn = 3 ; Testing if number is prime.\n"
]
],
[
[
"#### else",
"_____no_output_____"
]
],
[
[
"n = 1\nwhile n < 8:\n if n == 5:\n break\n print(\"loop1\", n ** 2)\n n += 1\nprint(\"Got to 8\")\n\nn = 1\nwhile n < 8:\n if n == 5:\n break\n print(\"loop2\", n ** 2)\n n += 1\nelse:\n print(\"Got to 8\")\n ",
"_____no_output_____"
],
[
" \ndef process_station_data(station_no):\n return station_no % 123 == 121\n\nerror = False\nnum_stations = 100\ni = 0\nwhile i < num_stations:\n i += 1\n error = process_station_data(i)\n if error:\n print(\"Error in station number\", i)\n break\nelse:\n print(\"Let's have a party!\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d0d27c77246f28950dbce7d3ab15587c85b0cbff | 61,800 | ipynb | Jupyter Notebook | Sheet02.ipynb | skleinbo/StatPhys2016 | 076507efe050f841968fda58e4096ded23a43044 | [
"MIT"
] | null | null | null | Sheet02.ipynb | skleinbo/StatPhys2016 | 076507efe050f841968fda58e4096ded23a43044 | [
"MIT"
] | null | null | null | Sheet02.ipynb | skleinbo/StatPhys2016 | 076507efe050f841968fda58e4096ded23a43044 | [
"MIT"
] | null | null | null | 494.4 | 59,179 | 0.946667 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d0d28082ca271d3e902397c35396148283b955b3 | 5,694 | ipynb | Jupyter Notebook | surprise/surprise.ipynb | neoville/Kaggle | 9090207becacaa42ef3f9de69479be85aad34755 | [
"MIT"
] | 284 | 2017-12-28T14:24:23.000Z | 2022-03-20T13:25:39.000Z | surprise/surprise.ipynb | neoville/Kaggle | 9090207becacaa42ef3f9de69479be85aad34755 | [
"MIT"
] | 1 | 2017-12-29T13:11:30.000Z | 2018-01-20T11:54:23.000Z | surprise/surprise.ipynb | corazzon/KaggleStruggle | 39e10b006354c4b3ebfc8400911ebd190ea8a2e8 | [
"MIT"
] | 340 | 2017-12-29T12:53:05.000Z | 2022-03-20T13:25:40.000Z | 26.732394 | 94 | 0.533895 | [
[
[
"[Surprise · A Python scikit for recommender systems.](http://surpriselib.com/)",
"_____no_output_____"
]
],
[
[
"# !pip install scikit-surprise\n# !conda install -c conda-forge scikit-surprise",
"_____no_output_____"
],
[
"from surprise import SVD\nfrom surprise import Dataset\nfrom surprise.model_selection import cross_validate\n\n# Load the movielens-100k dataset (download it if needed).\ndata = Dataset.load_builtin('ml-100k')\n\n# Use the famous SVD algorithm.\nalgo = SVD()\n\n# Run 5-fold cross-validation and print results.\ncross_validate(algo, data, measures=['RMSE', 'MAE'], cv=5, verbose=True)",
"Evaluating RMSE, MAE of algorithm SVD on 5 split(s).\n\n Fold 1 Fold 2 Fold 3 Fold 4 Fold 5 Mean Std \nRMSE (testset) 0.9373 0.9381 0.9372 0.9367 0.9317 0.9362 0.0023 \nMAE (testset) 0.7406 0.7401 0.7386 0.7381 0.7348 0.7384 0.0020 \nFit time 4.52 4.62 4.61 4.29 4.06 4.42 0.22 \nTest time 0.19 0.18 0.17 0.12 0.15 0.16 0.02 \n"
],
[
"from surprise import KNNBasic\n\n# Retrieve the trainset.\ntrainset = data.build_full_trainset()\n\n# Build an algorithm, and train it.\nalgo = KNNBasic()\nalgo.fit(trainset)\n",
"Computing the msd similarity matrix...\nDone computing similarity matrix.\n"
],
[
"import pandas as pd\n\nfrom surprise import NormalPredictor\nfrom surprise import Dataset\nfrom surprise import Reader\nfrom surprise.model_selection import cross_validate\n\n\n# Creation of the dataframe. Column names are irrelevant.\nratings_dict = {'itemID': [1, 1, 1, 2, 2],\n 'userID': [9, 32, 2, 45, 'user_foo'],\n 'rating': [3, 2, 4, 3, 1]}\ndf = pd.DataFrame(ratings_dict)\n\n# A reader is still needed but only the rating_scale param is requiered.\nreader = Reader(rating_scale=(1, 5))\n\n# The columns must correspond to user id, item id and ratings (in that order).\ndata = Dataset.load_from_df(df[['userID', 'itemID', 'rating']], reader)\n\n# We can now use this dataset as we please, e.g. calling cross_validate\ncross_validate(NormalPredictor(), data, cv=2)",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d0d288c5f064670531bf67d0507761a72ff0110a | 2,320 | ipynb | Jupyter Notebook | HomeWork/Day_041_HW.ipynb | rugl/3rd-ML100Days | 4c0c0427f623421574a0012b9069c43c27418691 | [
"Apache-2.0"
] | null | null | null | HomeWork/Day_041_HW.ipynb | rugl/3rd-ML100Days | 4c0c0427f623421574a0012b9069c43c27418691 | [
"Apache-2.0"
] | null | null | null | HomeWork/Day_041_HW.ipynb | rugl/3rd-ML100Days | 4c0c0427f623421574a0012b9069c43c27418691 | [
"Apache-2.0"
] | null | null | null | 20 | 306 | 0.546983 | [
[
[
"## 作業\n\n\n\n閱讀以下兩篇文獻,了解決策樹原理,並試著回答後續的問題\n- [決策樹 (Decision Tree) - 中文](https://medium.com/@yehjames/%E8%B3%87%E6%96%99%E5%88%86%E6%9E%90-%E6%A9%9F%E5%99%A8%E5%AD%B8%E7%BF%92-%E7%AC%AC3-5%E8%AC%9B-%E6%B1%BA%E7%AD%96%E6%A8%B9-decision-tree-%E4%BB%A5%E5%8F%8A%E9%9A%A8%E6%A9%9F%E6%A3%AE%E6%9E%97-random-forest-%E4%BB%8B%E7%B4%B9-7079b0ddfbda)\n- [how decision tree works - 英文](http://dataaspirant.com/2017/01/30/how-decision-tree-algorithm-works/)",
"_____no_output_____"
],
[
"# 1. 在分類問題中,若沒有任何限制,決策樹有辦法在訓練時將 training loss 完全降成 0 嗎?",
"_____no_output_____"
],
[
"可以,當樹長得足夠深,有機交替訓練損失降成0,但經常會因會樹長得太深而產生過度擬合的問題",
"_____no_output_____"
],
[
"過度擬合",
"_____no_output_____"
],
[
"建立決策樹模型時,過度擬合是一個實際問題。當算法繼續深入研究以減少訓練集誤差,但是結果卻增加了測試集誤差,即模型的預測準確性下降時,就會考慮模型存在過擬合的問題。通常由於數據異常和不規則而建立許多分支時會發生這種情況。",
"_____no_output_____"
],
[
"我們可以使用兩種方法來避免過度擬合:",
"_____no_output_____"
],
[
"1.預修剪",
"_____no_output_____"
],
[
"2.修剪後",
"_____no_output_____"
],
[
"# 2. 決策樹只能用在分類問題嗎?還是可以用來解決回歸問題?",
"_____no_output_____"
],
[
"也可以用在回歸問題:",
"_____no_output_____"
],
[
"CART -分類和回歸樹分類與回歸樹,是二叉樹,可以用於分類,也可以用於回歸問題,最先由Breiman等提出。分類樹的輸出是樣本的類別,回歸樹的輸出是一個實數。",
"_____no_output_____"
],
[
"GBDT / GBRT(梯度提升決策/回歸樹)梯度提升回歸樹",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d0d289038d0ba776539a1184e7e3bd6c47b422cf | 607,471 | ipynb | Jupyter Notebook | sst_science/West_Coast_HeatWave.ipynb | sevfour/cloud_science | 7965c09bfb3ffbb986af24070e745fee9e319542 | [
"Apache-2.0"
] | null | null | null | sst_science/West_Coast_HeatWave.ipynb | sevfour/cloud_science | 7965c09bfb3ffbb986af24070e745fee9e319542 | [
"Apache-2.0"
] | null | null | null | sst_science/West_Coast_HeatWave.ipynb | sevfour/cloud_science | 7965c09bfb3ffbb986af24070e745fee9e319542 | [
"Apache-2.0"
] | null | null | null | 119.369424 | 83,424 | 0.730195 | [
[
[
"# Satellite sea surface temperatures along the West Coast of the United States \n# during the 2014–2016 northeast Pacific marine heat wave\n\nIn 2016 we published a [paper](https://agupubs.onlinelibrary.wiley.com/doi/10.1002/2016GL071039) on the heat wave in the ocean off the California coast\n\nThis analysis was the last time I used Matlab to process scientific data. To make Figure 1, here are the following steps:\n- Download 4 TB of data from NASA PO.DAAC data archive via FTP\n- Go through each day of data and subset to the West Coast Region to reduce size and save each subsetted day\n- Go through 2002-2012 and create a daily climatology and save all 365 days of the climatology\n- Go through each day of data and calculate the anomaly and save each day's anomaly\n\nThis whole process took about 1 month. Once the anomalies were calculated, then I could start to do analyses and explore the data.\nBelow we will do this using MUR SST data on AWS Open Data Program in a few minutes using Python.",
"_____no_output_____"
]
],
[
[
"import warnings\nimport numpy as np\nimport pandas as pd\nimport xarray as xr\nimport fsspec\nimport matplotlib.pyplot as plt\n\nwarnings.simplefilter('ignore') # filter some warning messages\nxr.set_options(display_style=\"html\") #display dataset nicely",
"_____no_output_____"
],
[
"dir_out = './../../data/zarr_testing/'\nfile_aws = 'https://mur-sst.s3.us-west-2.amazonaws.com/zarr-v1'\nfile_aws_time = 'https://mur-sst.s3.us-west-2.amazonaws.com/zarr'",
"_____no_output_____"
],
[
"%%time\nds_sst = xr.open_zarr(file_aws,consolidated=True)\nds_sst",
"CPU times: user 2.11 s, sys: 221 ms, total: 2.33 s\nWall time: 3.67 s\n"
],
[
"#region for figure 1\nxlat1,xlat2 = 33,48\nxlon1,xlon2 = -132, -118, \ndate1,date2 = '2002-01-01','2013-01-01'\n\nsubset = ds_sst.sel(lat=slice(xlat1,xlat2),lon=slice(xlon1,xlon2))\nsubset",
"_____no_output_____"
]
],
[
[
"# Just plot a random day to make sure it looks correct",
"_____no_output_____"
]
],
[
[
"subset.analysed_sst[0,:,:].plot()",
"_____no_output_____"
]
],
[
[
"# How big is this dataset?\n- Because xarray uses lazy loading, we have access to this entire dataset but it only loads what it needs to for calculations",
"_____no_output_____"
]
],
[
[
"print('GB data = ',subset.nbytes/(1024 * 1024 * 1024))",
"GB data = 201.89575985074043\n"
]
],
[
[
"# Caluculate the Monthly Sea Surface Temperature Anomalies",
"_____no_output_____"
]
],
[
[
"sst_monthly = subset.resample(time='1MS').mean('time',keep_attrs=True,skipna=False)\n\nclimatology_mean_monthly = sst_monthly.sel(time=slice(date1,date2)).groupby('time.month').mean('time',keep_attrs=True,skipna=False)\n\nsst_anomaly_monthly = sst_monthly.groupby('time.month')-climatology_mean_monthly #take out annual mean to remove trends\n\nsst_anomaly_monthly",
"_____no_output_____"
],
[
"sst_anomaly_monthly.analysed_sst[0,:,:].plot(vmin=-3,vmax=3,cmap='RdYlBu_r')",
"_____no_output_____"
],
[
"sst_anomaly_monthly.analysed_sst.sel(time='2015-03').plot(vmin=-3,vmax=3,cmap='RdYlBu_r')",
"_____no_output_____"
],
[
"#plt.pcolormesh(tem.lon,tem.lat,tem.analysed_sst,transform=ccrs.PlateCarree(),cmap=vik_map,vmin=-2,vmax=2)\n#ax.coastlines(resolution='50m', color='black', linewidth=1)\n#ax.add_feature(cfeature.LAND)\n#ax.add_feature(cfeature.STATES.with_scale('10m'))\n#ax.set_extent([-132.27,-117,32,48])\n#plt.colorbar(ax=ax,label='SST Anomaly (K)')\n#tt=plt.text(-122,47,tstr,fontsize=16)\n",
"_____no_output_____"
]
],
[
[
"# Let's try and re-do figure 2 which uses 5-day average SST anomalies",
"_____no_output_____"
]
],
[
[
"sst_5day = subset.resample(time='5D').mean('time',keep_attrs=True,skipna=False)\n\nclimatology_mean_5day = sst_5day.sel(time=slice(date1,date2)).groupby('time.day').mean('time',keep_attrs=True,skipna=False)\n\nsst_anomaly_5day = sst_5day.groupby('time.day')-climatology_mean_5day #take out annual mean to remove trends\n\nsst_anomaly_5day",
"_____no_output_____"
],
[
"%%time\nmax_5day = sst_anomaly_5day.analysed_sst.sel(time=slice('2012','2016')).max(\"time\")\nmax_5day",
"CPU times: user 24.8 ms, sys: 0 ns, total: 24.8 ms\nWall time: 24.2 ms\n"
],
[
"#running out of memory right now. maybe need to breakdown into yearly bits or something. could try using time arranged zarr file store\n#max_5day.plot(vmin=0,vmax=5,cmap='jet')",
"_____no_output_____"
]
],
[
[
"# Switch to same data, but it is chunked differently\n- it is optimized for timeseries rather than spatial analysis",
"_____no_output_____"
]
],
[
[
"ds_sst = xr.open_zarr(file_aws_time,consolidated=True)\nds_sst",
"_____no_output_____"
],
[
"%%time\nsst_newport_nearshore = ds_sst.analysed_sst.sel(lat=44.6,lon=-124.11,method='nearest').rolling(time=30, center=True).mean().load()\nsst_newport_offshore = ds_sst.analysed_sst.sel(lat=44.6,lon=-134.11,method='nearest').rolling(time=30, center=True).mean().load()",
"CPU times: user 562 ms, sys: 256 ms, total: 818 ms\nWall time: 5.61 s\n"
],
[
"plt.plot(sst_newport_nearshore.time.dt.dayofyear,sst_newport_nearshore)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d0d28b1be724f9a1b61f821cc5aa87edb24b83a9 | 235,699 | ipynb | Jupyter Notebook | examples/minimal_end_to_end_causal_analysis.ipynb | MLResearchAtOSRAM/cause2e | 9420e88802172b893d4029b741dfd3e5e718880b | [
"MIT"
] | 33 | 2021-05-18T13:03:54.000Z | 2022-02-17T16:50:48.000Z | examples/minimal_end_to_end_causal_analysis.ipynb | MLResearchAtOSRAM/cause2e | 9420e88802172b893d4029b741dfd3e5e718880b | [
"MIT"
] | 11 | 2021-09-17T07:27:38.000Z | 2022-03-29T07:04:33.000Z | examples/minimal_end_to_end_causal_analysis.ipynb | MLResearchAtOSRAM/cause2e | 9420e88802172b893d4029b741dfd3e5e718880b | [
"MIT"
] | 1 | 2021-11-15T12:22:51.000Z | 2021-11-15T12:22:51.000Z | 272.799769 | 40,158 | 0.902316 | [
[
[
"# Minimal end-to-end causal analysis with ```cause2e```\nThis notebook shows a minimal example of how ```cause2e``` can be used as a standalone package for end-to-end causal analysis. It illustrates how we can proceed in stringing together many causal techniques that have previously required fitting together various algorithms from separate sources with unclear interfaces. Additionally, the numerous techniques have been packed into only two easy-to-use functions for causal discovery and causal estimation. Hopefully, you will find this notebook helpful in guiding you through the process of setting up your own causal analyses for custom problems. The overall structure should always be the same regardless of the application domain. For more advanced features, check out the other notebooks.",
"_____no_output_____"
],
[
"### Imports\nBy the end of this notebook, you will probably be pleasantly surprised by the fact that we did not have to import lots of different packages to perform a full causal analysis consisting of different subtasks.",
"_____no_output_____"
]
],
[
[
"import os\nfrom cause2e import path_mgr, knowledge, discovery",
"_____no_output_____"
]
],
[
[
"## Set up paths to data and output directories\nThis step is conveniently handled by the ```PathManager``` class, which avoids having to wrestle with paths throughout the multistep causal analysis. If we want to perform the analysis in a directory ```'dirname'``` that contains ```'dirname/data'``` and ```'dirname/output'``` as subdirectories, we can also use ```PathManagerQuick``` for an even easier setup. The ```experiment_name``` argument is used for generating output files with meaningful names, in case we want to study multiple scenarios (e.g. with varying model parameters).\n\nFor this analysis, we use the sprinkler dataset. Unfortunately, there are still some problems to be sorted out with categorical data in the estimation step, but continuous and discrete data work fine. Therefore, we use a version of the dataset where only the seasons ```'Spring'``` and ```'Summer'``` are present, such that we can replace these values by 0 and 1.",
"_____no_output_____"
]
],
[
[
"cwd = os.getcwd()\nwd = os.path.dirname(cwd)\npaths = path_mgr.PathManagerQuick(experiment_name='sprinkler',\n data_name='sprinkler.csv',\n directory=wd\n )",
"_____no_output_____"
]
],
[
[
"## Learn the causal graph from data and domain knowledge\nModel-based causal inference leverages qualitative knowledge about pairwise causal connections to obtain unbiased estimates of quantitative causal effects. The qualitative knowledge is encoded in the causal graph, so we must recover this graph before we can start actually estimating the desired effects. For learning the graph from data and domain knowledge, we use the ```StructureLearner``` class.",
"_____no_output_____"
]
],
[
[
"learner = discovery.StructureLearner(paths)",
"_____no_output_____"
]
],
[
[
"### Read the data\nThe ```StructureLearner``` has reading methods for csv and parquet files.",
"_____no_output_____"
]
],
[
[
"learner.read_csv(index_col=0)",
"_____no_output_____"
]
],
[
[
"The first step in the analysis should be an assessment of which variables we are dealing with. In the sprinkler dataset, each sample tells us \n- the current season\n- whether it is raining\n- whether our lawn sprinkler is activated\n- whether our lawn is slippery\n- whether our lawn is wet.",
"_____no_output_____"
]
],
[
[
"learner.variables",
"_____no_output_____"
]
],
[
[
"### Preprocess the data\nAs mentioned above, currently there are problems in the estimation step with categorical data, so we use this occasion to showcase ```cause2e```'s built-in preprocessing functionalities. We define a function that replaces instances of ```'Summer'``` by 1, and instances of ```'Spring'``` by 0. Afterwards we apply it to our data and throw out the categorical ```'Season'``` column. For more preprocessing options, check out the pertaining notebook.",
"_____no_output_____"
]
],
[
[
"def is_summer(data, col_name):\n return (data[col_name] == 'Summer').apply(int)\n\nlearner.combine_variables(name='Season_binary', func=is_summer, input_cols=['Season'], keep_old=False)",
"_____no_output_____"
]
],
[
[
"It necessary to communicate to the ```StructureLearner``` if the variables are discrete, continuous, or both. We check how many unique values each variable takes on in our sample and deduce that all variables are discrete.",
"_____no_output_____"
]
],
[
[
"learner.data.nunique()",
"_____no_output_____"
]
],
[
[
"This information is passed to the ```StructureLearner``` by indicating the exact sets of discrete and continuous variables.",
"_____no_output_____"
]
],
[
[
"learner.discrete = learner.variables\nlearner.continuous = set()",
"_____no_output_____"
]
],
[
[
"### Provide domain knowledge\nHumans can often infer parts of the causal graph from domain knowledge. The nodes are always just the variables in the data, so the problem of finding the right graph comes down to selecting the right edges between them. \n\nAs a reminder: The correct causal graph has an edge from variable A to variable B if and only if variable A directly influences variable B (changing the value of variable A changes the value of variable B if we keep all other variables fixed).\n\nThere are three ways of passing domain knowledge for the graph search:\n- Indicate which edges must be present in the causal graph.\n- Indicate which edges must not be present in the causal graph.\n- Indicate a temporal order in which the variables have been created. This is then used to generate forbidden edges, since the future can never influence the past.\n\nIn this example, we use the ```knowledge.EdgeCreator``` to prescribe that\n- no variables are direct causes of the season,\n- the lawn being slippery is not a direct cause of any other variable\n- turning the sprinkler on or off directly affects the wetness of the lawn,\n- turning the sprinkler on or off does not directly affect the weather.",
"_____no_output_____"
]
],
[
[
"edge_creator = knowledge.EdgeCreator()\nedge_creator.forbid_edges_from_groups({'Season_binary'}, incoming=learner.variables)\nedge_creator.forbid_edges_from_groups({'Slippery'}, outgoing=learner.variables)\nedge_creator.require_edge('Sprinkler', 'Wet')\nedge_creator.forbid_edge('Sprinkler', 'Rain')",
"_____no_output_____"
]
],
[
[
"There is a fourth way of passing knowledge which is not used in learning the graph, but in validating the quantitative estimates resulting from our end-to-end causal analysis. We often know beforehand what some of the quantitative effects should look like, e.g.\n- turning the sprinkler on should have a positive overall effect (-> average treatment effect; read below if you are not familiar with types of causal effects) on the lawn being wet and \n- making the lawn wet should have a positive overall effect on the lawn being slippery.\n\nInstead of checking manually at the end if our expectations have been met, we can automate this validation by using the ```knowledge.ValidationCreator```. For instructiveness, we also add two more validations that should fail: \n- the sprinkler has a negative natural direct effect on the weather and\n- the natural indirect effect of the lawn being slippery on the season is between 0.2 and 0.4 (remember to normalize your data before such a validation if they are not measured on the same scale).",
"_____no_output_____"
]
],
[
[
"validation_creator = knowledge.ValidationCreator()\nvalidation_creator.add_expected_effect(('Sprinkler', 'Wet', 'nonparametric-ate'), ('greater', 0))\nvalidation_creator.add_expected_effect(('Wet', 'Slippery', 'nonparametric-ate'), ('greater', 0))\nvalidation_creator.add_expected_effect(('Sprinkler', 'Rain', 'nonparametric-nde'), ('less', 0))\nvalidation_creator.add_expected_effect(('Slippery', 'Season_binary', 'nonparametric-nie'), ('between', 0.2, 0.4))",
"_____no_output_____"
]
],
[
[
"We pass the knowledge to the ```StructureLearner``` and check if it has been correctly received.",
"_____no_output_____"
]
],
[
[
"learner.set_knowledge(edge_creator=edge_creator, validation_creator=validation_creator)",
"====================\nShowing knowledge for graph search.\n\nCausal graph constructed from domain knowledge:\n(Edges required by domain knowledge in red, remaining allowed edges dotted)\n\n"
]
],
[
[
"### Apply a structure learning algorithm\nNow that the ```StructureLearner``` has received the data and the domain knowledge, we can try to recover the original graph using causal discovery methods provided by the internally called ```py-causal``` package. There are many parameters that can be tuned (choice of algorithm, search score, independence test, hyperparameters, ...) and we can get an overview by calling some informative methods of the learner. Reasonable default arguments are provided (FGES with CG-BIC score for possibly mixed datatypes and respecting domain knowledge), so we use these for our minimal example.",
"_____no_output_____"
]
],
[
[
"learner.run_quick_search()",
"Remember to stop the JVM after you are completely done.\nProposed causal graph (edges required by domain knowledge in red, undirected edges in blue):\n\n"
]
],
[
[
"The output of the search is a proposed causal graph. We can ignore the warning about stopping the Java Virtual Machine (needed by ```py-causal``` which is a wrapper around the ```TETRAD``` software that is written in Java) if we do not run into any problems. If the algorithm cannot orient all edges, we need to do this manually. Therefore, the output includes a list of all undirected edges, so we do not miss them in complicated graphs with many variables and edges. In our case, all the edges are already oriented.\n\nThe result seems reasonable:\n- The weather depends on the season.\n- The sprinkler use also depends on the season.\n- The lawn will be wet if it rains or if the sprinkler is activated.\n- The lawn will be slippery if it is wet.",
"_____no_output_____"
],
[
"### Saving the graph\n```Cause2e``` allows us to save the result of our search to different file formats with the ```StructureLearner.save_graphs``` method. The name of the file is determined by the ```experiment_name``` parameter from the ```PathManager```. If the result of the graph search is already a directed acyclic graph that respects our domain knowledge, the graph is automatically saved, as we can see from the above output. Check out the graph postprocessing notebook for information on how to proceed when the result of the search needs further adjustments.",
"_____no_output_____"
],
[
"## Estimate causal effects from the graph and the data\nAfter we have successfully recovered the causal graph from data and domain knowledge, we can use it to estimate quantitative causal effects between the variables in the graph. It is pleasant that we can use the same graph and data to estimate multiple causal effects, e.g. the one that the Sprinkler has on the lawn being slippery, as well as the one that the season has on the rain probability, without having to repeat the previous steps. Once we have managed to qualitatively model the data generating process, we are already in a very good position. The remaining challenges can be tackled with the core functionality from the ```DoWhy``` package, which we have wrapped into a single easy-to-use convenience method. Usually, all estimation topics are handled by the ```estimator.Estimator```, but the ```StructureLearner``` has the possibility to run a quick analysis of all causal effects with preset parameters. For more detailed analyses, check out the other notebooks that describe the causal identification and estimation process step by step.",
"_____no_output_____"
]
],
[
[
"learner.run_all_quick_analyses()",
"Showing and saving heatmaps of the causal estimates.\n\n"
]
],
[
[
"The output consists of a detailed analysis of the causal effects in our system.\n### Heatmaps\nThe first three images are heatmaps, where the (i, j)-entry shows the causal effect of variable i on variable j. The three heatmaps differ in the type of causal effect that they are describing:\n - **Average Treatment Effect (ATE)**: Shows how the outcome variable varies if we vary the treatment variable. This comprises direct and indirect effects. The sprinkler influences the lawn being slippery, even if this does not happen directly, but via its influence on the lawn being wet.\n - **Natural Direct Effect (NDE)**: Shows how the outcome variable varies if we vary the treatment variable and keep all other variables fixed. This comprises only direct effects. The sprinkler does not directly influence the lawn being slippery, as we can read off from the heatmap.\n - **Natural Indirect Effect (NIE)**: Shows the difference between ATE and NDE. By definition, this comprises only indirect effects. The sprinkler has a strong indirect influence on the lawn being slippery, as we can read off from the heatmap.\n\nIn our example, we can easily identify from the graph if an effect is direct or indirect, but in examples where a variable simultaneously has a direct and an indirect influence on another variable, it is very challenging to separate the effects without resorting to the algebraic methods that ```cause2e``` uses internally.\n\n### Validations\nThe next output shows if our model has passed each of the validations, based on the expected causal effects that we have communicated before running the causal end-to-end analysis. If we are interested in a specific effect, say, the effect of the sprinkler on the lawn being slippery, the estimation of this effect by our learnt causal model can be trusted more if the estimation for other effects match our expectations. We see that the results of the validations turned out exactly as described above (in practice we would not want validations to fail, this was only for demonstrative purposes).\n\n### Numeric tables\nThe three numeric tables show the same information as the three previous heatmaps, only in quantitative instead of visual form.\n\n### PDF report\n```Cause2e``` automatically generates a pdf report that contains\n- the causal graph indicating all qualitative relationships,\n- the three heatmaps visualizing all quantitative causal effects,\n- the results of the validations,\n- the three numeric tables reporting all quantitative causal effects.\n\nThis is helpful if we want to communicate our findings to other people, or if we want to modify the analysis at a later time and compare the outcome for both methods.",
"_____no_output_____"
],
[
"## Discussion of the results\nThe heatmaps show the effects that we would expect given our causal graph: \n- There is less rain in summer than in spring.\n- Sprinklers are more often turned on in summer than in spring.\n- Rain increases the wetness of the lawn.\n- Turning the sprinkler on also increases the wetness of the lawn.\n- Wetting the lawn causes it to be slippery.\n\nIt is interesting to see that the first two effects roughly cancel each other out, resulting in a small ATE of 0.1 that ```'Season_binary'``` has on ```'Slippery'``` and ```'Wet'```. In general, it is a good strategy to look at the heatmaps for discovering the qualitative nature of the different causal effects and then inspect the numeric tables for the exact numbers if needed.\n\nAnother noteworthy entry is the overall effect of ```'Sprinkler'``` on ```'Wet'```. The result is 0.638, so turning on the sprinkler makes it more likely for the lawn to be wet, as it should be. However, we might ask ourselves: \"Why is the effect not 1? Whenever we turn on the sprinkler, the lawn will be wet!\" This can be explained by looking at the definition of our chosen effect type, the nonparametric average treatment effect (ATE): The ATE tells us how much (on average) we change the outcome by changing the treatment. In our case, we can distinguish between two possible scenarios:\nIf it is raining, then the lawn is wet anyway, so turning the sprinkler on does not change the outcome at all. Only if it is not raining, the lawn state is changed to wet by turning on the sprinkler.\n\nWe can convince ourselves that this is the correct explanation by looking at the proportion of samples where it is not raining.",
"_____no_output_____"
]
],
[
[
"1 - sum(learner.data['Rain']) / len(learner.data)",
"_____no_output_____"
]
],
[
[
"We recover the same number of 0.638. Additionally, we can change our data to consist only of the instances where it is not raining. If we now repeat the causal analysis, the effect is indeed 1 (skip after the warnings that are caused by the now degenerate dataset). This procedure can be generalized to analyzing other conditional causal effects.",
"_____no_output_____"
]
],
[
[
"learner.data = learner.data[learner.data['Rain']==0]\nlearner.run_all_quick_analyses()",
"C:\\Users\\D.Gruenbaum\\Anaconda3\\envs\\test\\lib\\site-packages\\statsmodels\\regression\\linear_model.py:1830: RuntimeWarning: divide by zero encountered in double_scalars\n return np.sqrt(eigvals[0]/eigvals[-1])\nC:\\Users\\D.Gruenbaum\\Anaconda3\\envs\\test\\lib\\site-packages\\statsmodels\\base\\model.py:1362: RuntimeWarning: invalid value encountered in true_divide\n return self.params / self.bse\nC:\\Users\\D.Gruenbaum\\Anaconda3\\envs\\test\\lib\\site-packages\\statsmodels\\regression\\linear_model.py:1685: RuntimeWarning: invalid value encountered in double_scalars\n return 1 - self.ssr/self.centered_tss\nC:\\Users\\D.Gruenbaum\\Anaconda3\\envs\\test\\lib\\site-packages\\statsmodels\\regression\\linear_model.py:1774: RuntimeWarning: invalid value encountered in double_scalars\n return self.mse_model/self.mse_resid\nC:\\Users\\D.Gruenbaum\\Anaconda3\\envs\\test\\lib\\site-packages\\statsmodels\\regression\\linear_model.py:889: RuntimeWarning: divide by zero encountered in log\n llf = -nobs2*np.log(2*np.pi) - nobs2*np.log(ssr / nobs) - nobs2\nC:\\Users\\D.Gruenbaum\\Anaconda3\\envs\\test\\lib\\site-packages\\statsmodels\\stats\\stattools.py:46: RuntimeWarning: invalid value encountered in double_scalars\n dw = np.sum(diff_resids**2, axis=axis) / np.sum(resids**2, axis=axis)\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0d292b3f2e3c0cc64a7112e1369e825c8d2f57f | 105,377 | ipynb | Jupyter Notebook | Raw Codes/.ipynb_checkpoints/basic -2-checkpoint.ipynb | masudurHimel/ML---APS-Failure-at-Scania-Trucks-Data-Set | 28ce0f07da1b72616107316f7e77f2f6d7b64673 | [
"MIT"
] | null | null | null | Raw Codes/.ipynb_checkpoints/basic -2-checkpoint.ipynb | masudurHimel/ML---APS-Failure-at-Scania-Trucks-Data-Set | 28ce0f07da1b72616107316f7e77f2f6d7b64673 | [
"MIT"
] | null | null | null | Raw Codes/.ipynb_checkpoints/basic -2-checkpoint.ipynb | masudurHimel/ML---APS-Failure-at-Scania-Trucks-Data-Set | 28ce0f07da1b72616107316f7e77f2f6d7b64673 | [
"MIT"
] | 1 | 2019-07-19T15:35:22.000Z | 2019-07-19T15:35:22.000Z | 38.083484 | 1,690 | 0.369132 | [
[
[
"The data and the description:\nhttps://archive.ics.uci.edu/ml/datasets/APS+Failure+at+Scania+Trucks\n\nAbstract: The datasets' positive class consists of component failures for a specific component of the APS system. The negative class consists of trucks with failures for components not related to the APS.\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.metrics import confusion_matrix\nfrom sklearn.preprocessing import LabelEncoder\npd.options.display.max_columns = None\n\nimport warnings\nwarnings.filterwarnings('ignore')\n",
"_____no_output_____"
],
[
"df_original = pd.read_csv('../Data/aps_failure_training_set.csv', dtype = 'str')\ndf_original = df_original.replace(r'na', np.nan, regex=True)",
"_____no_output_____"
],
[
"df_original.head()",
"_____no_output_____"
],
[
"#encode labels to 0 and 1\nle = LabelEncoder()\ndf_original['class'] = le.fit_transform(df_original['class'])\ndf = df_original.copy()\ndf.head()",
"_____no_output_____"
]
],
[
[
"The set is very unbalanced with one label (0) being more frequent than the other (1). The algorithm needs to adjust for that. \nIt is done using 'class_weight' hyperparameter which is the ratio of number of 0s to 1s in the label.",
"_____no_output_____"
]
],
[
[
"df = df_original.copy()\nfrom sklearn.model_selection import train_test_split\nX, y = df.iloc[:,1:], df.iloc[:,0]\nX_tr, X_t, y_tr, y_t = train_test_split(X, y, test_size = 0.2, random_state = 0)\n\nweight = sum(y_tr == 0)/sum(y_tr == 1)\nlr_full = LogisticRegression(C = 1, class_weight={1:weight}, random_state = 0)\nlr_full.fit(X_tr, y_tr)\ny_pred = lr_full.predict(X_t)\n\n#calculate the score using confusion matrix values\ndef score(cm):\n cm_score = cm[0][1] * 10 + cm[1][0] * 500\n cm_score = int(cm_score * 1.33) #1.33 is because the actual test set is 33% larger than this test set\n return cm_score\n#calculate confusion matrix\ncm = confusion_matrix(y_t, y_pred)\nscore(cm)",
"_____no_output_____"
]
],
[
[
"13632 is our basic score. We'll use it as a reference for further optimizations.\nThe data seemed to be scaled but let's apply scaling to the data just in case. ",
"_____no_output_____"
]
],
[
[
"#testing scaling\ndf = df_original.copy()\n\nfrom sklearn.preprocessing import MinMaxScaler\nscaler_minmax = MinMaxScaler()\nX, y = df.iloc[:,1:], df.iloc[:,0]\nX_scaled = scaler_minmax.fit_transform(X.values)\nX_tr, X_t, y_tr, y_t = train_test_split(X_scaled, y, test_size = 0.2, random_state = 0)\n\nweight = sum(y_tr == 0)/sum(y_tr == 1)\nlr_full = LogisticRegression(C = 1, class_weight={1:weight}, random_state = 0)\nlr_full.fit(X_tr, y_tr)\ny_pred = lr_full.predict(X_t)\n\n#calculate confusion matrix\ncm = confusion_matrix(y_t, y_pred)\nscore(cm)",
"_____no_output_____"
]
],
[
[
"MinMaxScaler slightly improved the result. \n\nLet's tune the 'C', a hyperparameter (parameter) of the Logistic Regression algorithm.",
"_____no_output_____"
]
],
[
[
"#tuning hyperparameters for Logistic Regression\ndf = df_original.copy()\nX, y = df.iloc[:,1:], df.iloc[:,0]\nX_tr, X_t, y_tr, y_t = train_test_split(X, y, test_size = 0.2, random_state = 0)\nCs = list(np.arange(0.1, 0.5, 0.1))\nweight = sum(y_tr == 0)/sum(y_tr == 1)\nfor C_ in Cs: \n lr_full = LogisticRegression(C = C_, class_weight={1:weight}, random_state = 0)\n lr_full.fit(X_tr, y_tr)\n y_pred = lr_full.predict(X_t)\n\n #calculate confusion matrix\n cm = confusion_matrix(y_t, y_pred)\n score(cm)\n print(\"C is {0}. Score is: {1}\".format(C_, score(cm)))",
"_____no_output_____"
]
],
[
[
"C = 0.1 gives the best score.",
"_____no_output_____"
],
[
"Let's try another algorithm. Maybe Random Forest will perfom better.",
"_____no_output_____"
]
],
[
[
"#check algorithm with all NAs replaced with mean column values (none rows/columns dropped)\ndf = df_original.copy()\nX, y = df.iloc[:,1:], df.iloc[:,0]\n#split into train and test\nfrom sklearn.model_selection import train_test_split\nX_tr, X_t, y_tr, y_t = train_test_split(X, y, test_size = 0.2, random_state = 0)\n\nfrom sklearn.ensemble import RandomForestClassifier\nrf = RandomForestClassifier(n_estimators = 200, oob_score = True, class_weight={1:weight}, random_state = 0, bootstrap = True)\nrf.fit(X_tr, y_tr)\ny_pred = rf.predict(X_t)\ncm = confusion_matrix(y_t, y_pred)\nscore(cm)",
"_____no_output_____"
]
],
[
[
"61938 is significanlty worse.",
"_____no_output_____"
],
[
"It seems Logistic Regression gives us the best score.\nWe need to train it on the full training data set and fit the actual test set to get the final score.",
"_____no_output_____"
]
],
[
[
"df = df_original.copy()\n\nX_train, y_train = df.iloc[:,1:], df.iloc[:,0]\nX_train_scaled = scaler_minmax.fit_transform(X_train.values)\n#calculation of the score for the actual test set\nweight = sum(y_train == 0)/sum(y_train == 1)\nlog_reg = LogisticRegression(class_weight = {1:weight}, C = 0.2, random_state=1)\nlog_reg.fit(X_train_scaled, y_train)\n\n#process the test data set\ndf_test = pd.read_csv('../input/aps_failure_test_set_processed_8bit.csv', dtype = 'str')\ndf_test = df_test.replace(r'na', np.nan, regex=True)\n \nle = LabelEncoder()\ndf_test['class'] = le.fit_transform(df_test['class'])\nX_test, y_test = df_test.iloc[:,1:], df_test.iloc[:,0]\n\nX_test_scaled = scaler_minmax.transform(X_test.values)\n#predict the class for the test set\ny_test_pred = log_reg.predict(X_test_scaled)\n\ncm = confusion_matrix(y_test, y_test_pred)\ndef score(cm):\n cm_score = cm[0][1] * 10 + cm[1][0] * 500\n cm_score = int(cm_score)\n return cm_score\nscore(cm)",
"_____no_output_____"
]
],
[
[
"The final score is 14520. It is not the best score, but a good one. Scheduling repairs according to the algorithm's predictions will significantly reduce the cost of truck repairs.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0d29c89ccf21babb4d4a1d600401861cf29f95f | 515,684 | ipynb | Jupyter Notebook | intermediate_data_visualization_with_seaborn/3_additional_plot_types.ipynb | vhsenna/datacamp-courses | dad9982bf7e90061efcbecc3cce97b7a5d14dd80 | [
"MIT"
] | null | null | null | intermediate_data_visualization_with_seaborn/3_additional_plot_types.ipynb | vhsenna/datacamp-courses | dad9982bf7e90061efcbecc3cce97b7a5d14dd80 | [
"MIT"
] | 1 | 2022-02-19T17:18:22.000Z | 2022-02-19T21:51:45.000Z | intermediate_data_visualization_with_seaborn/3_additional_plot_types.ipynb | vhsenna/datacamp-courses | dad9982bf7e90061efcbecc3cce97b7a5d14dd80 | [
"MIT"
] | null | null | null | 374.770349 | 74,552 | 0.92637 | [
[
[
"## stripplot() and swarmplot()\n\nMany datasets have categorical data and Seaborn supports several useful plot types for this data. In this example, we will continue to look at the 2010 School Improvement data and segment the data by the types of school improvement models used.\n\nAs a refresher, here is the KDE distribution of the Award Amounts:\n\n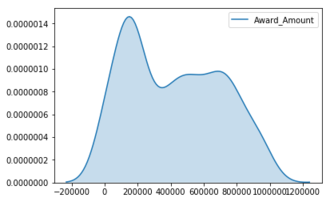\n\nWhile this plot is useful, there is a lot more we can learn by looking at the individual `Award_Amounts` and how they are distributed among the 4 categories.\n\nInstructions \n\n1. Create a `stripplot` of the `Award_Amount` with the `Model Selected` on the y axis with `jitter` enabled.\n2. Create a `swarmplot()` of the same data, but also include the `hue` by `Region`.",
"_____no_output_____"
]
],
[
[
"# Import packages\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\n# Import dataset\ndf = pd.read_csv('grant_file.csv')",
"_____no_output_____"
],
[
"# Create the stripplot\nsns.stripplot(data=df,\n x='Award_Amount',\n y='Model Selected',\n jitter=True)\n\nplt.show()",
"_____no_output_____"
],
[
"# Create and display a swarmplot with hue set to the Region\nsns.swarmplot(data=df,\n x='Award_Amount',\n y='Model Selected',\n hue='Region')\n\nplt.show()",
"/home/victor/.local/lib/python3.8/site-packages/seaborn/categorical.py:1296: UserWarning: 34.8% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.\n warnings.warn(msg, UserWarning)\n/home/victor/.local/lib/python3.8/site-packages/seaborn/categorical.py:1296: UserWarning: 9.3% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.\n warnings.warn(msg, UserWarning)\n"
]
],
[
[
"## boxplots, violinplots and lvplots\n\nSeaborn's categorical plots also support several abstract representations of data. The API for each of these is the same so it is very convenient to try each plot and see if the data lends itself to one over the other.\n\nIn this exercise, we will use the color palette options presented in Chapter 2 to show how colors can easily be included in the plots.\n\nInstructions\n\n1. Create and display a `boxplot` of the data with `Award_Amount` on the x axis and `Model Selected` on the y axis.\n2. Create and display a similar `violinplot` of the data, but use the `husl` palette for colors.\n3. Create and display an `lvplot` using the `Paired` palette and the `Region` column as the `hue`.\n\n_NOTE: lvplot function has been renamed to boxenplot._",
"_____no_output_____"
]
],
[
[
"# Create a boxplot\nsns.boxplot(data=df,\n x='Award_Amount',\n y='Model Selected')\n\nplt.show()\nplt.clf()",
"_____no_output_____"
],
[
"# Create a violinplot with the husl palette\nsns.violinplot(data=df,\n x='Award_Amount',\n y='Model Selected',\n palette='husl')\n\nplt.show()\nplt.clf()",
"_____no_output_____"
],
[
"# Create a lvplot with the Paired palette and the Region column as the hue\nsns.boxenplot(data=df,\n x='Award_Amount',\n y='Model Selected',\n palette='Paired',\n hue='Region')\n\nplt.show()\nplt.clf()",
"_____no_output_____"
]
],
[
[
"## barplots, pointplots and countplots\n\nThe final group of categorical plots are `barplots`, `pointplots` and `countplot` which create statistical summaries of the data. The plots follow a similar API as the other plots and allow further customization for the specific problem at hand.\n\nInstructions\n\n1. Create a `countplot` with the `df` dataframe and `Model Selected` on the y axis and the color varying by `Region`.\n2. Create a `pointplot` with the `df` dataframe and `Model Selected` on the x-axis and `Award_Amount` on the y-axis.\n3. Use a `capsize` in the `pointplot` in order to add caps to the error bars.\n4. Create a `barplot` with the same data on the x and y axis and change the color of each bar based on the `Region` column.",
"_____no_output_____"
]
],
[
[
"# Show a countplot with the number of models used with each region a different color\nsns.countplot(data=df,\n y='Model Selected',\n hue='Region')\n\nplt.show()\nplt.clf()",
"_____no_output_____"
],
[
"# Create a pointplot and include the capsize in order to show caps on the error bars\nsns.pointplot(data=df,\n x='Model Selected',\n y='Award_Amount',\n capsize=.1)\n\nplt.show()\nplt.clf()",
"_____no_output_____"
],
[
"# Create a barplot with each Region shown as a different color\nsns.barplot(data=df,\n x='Model Selected',\n y='Award_Amount',\n hue='Region')\n\nplt.show()\nplt.clf()",
"_____no_output_____"
]
],
[
[
"## Regression and residual plots\n\nLinear regression is a useful tool for understanding the relationship between numerical variables. Seaborn has simple but powerful tools for examining these relationships.\n\nFor these exercises, we will look at some details from the US Department of Education on 4 year college tuition information and see if there are any interesting insights into which variables might help predict tuition costs.\n\nFor these exercises, all data is loaded in the `college` variable.\n\nInstructions\n\n1. Plot a regression plot comparing `Tuition` and average SAT scores (`SAT_AVG_ALL`).\n2. Make sure the values are shown as green triangles.\n3. Use a residual plot to determine if the relationship looks linear.",
"_____no_output_____"
]
],
[
[
"# Import dataset\ncollege = pd.read_csv('college_datav3.csv')\n\n# Display a regression plot for Tuition\nsns.regplot(data=college,\n y='Tuition',\n x='SAT_AVG_ALL',\n marker='^',\n color='g')\n\nplt.show()\nplt.clf()",
"_____no_output_____"
],
[
"# Display the residual plot\nsns.residplot(data=college,\n y='Tuition',\n x=\"SAT_AVG_ALL\",\n color='g')\n\nplt.show()\nplt.clf()",
"_____no_output_____"
]
],
[
[
"## Regression plot parameters\n\nSeaborn's regression plot supports several parameters that can be used to configure the plots and drive more insight into the data.\n\nFor the next exercise, we can look at the relationship between tuition and the percent of students that receive Pell grants. A Pell grant is based on student financial need and subsidized by the US Government. In this data set, each University has some percentage of students that receive these grants. Since this data is continuous, using `x_bins` can be useful to break the percentages into categories in order to summarize and understand the data.\n\nInstructions\n\n1. Plot a regression plot of `Tuition` and `PCTPELL`.\n2. Create another plot that breaks the `PCTPELL` column into 5 different bins.\n3. Create a final regression plot that includes a 2nd `order` polynomial regression line.",
"_____no_output_____"
]
],
[
[
"# Plot a regression plot of Tuition and the Percentage of Pell Grants\nsns.regplot(data=college,\n y='Tuition',\n x='PCTPELL')\n\nplt.show()\nplt.clf()",
"_____no_output_____"
],
[
"# Create another plot that estimates the tuition by PCTPELL\nsns.regplot(data=college,\n y='Tuition',\n x='PCTPELL',\n x_bins=5)\n\nplt.show()\nplt.clf()",
"_____no_output_____"
],
[
"# The final plot should include a line using a 2nd order polynomial\nsns.regplot(data=college,\n y='Tuition',\n x='PCTPELL',\n x_bins=5,\n order=2)\n\nplt.show()\nplt.clf()",
"_____no_output_____"
]
],
[
[
"## Binning data\n\nWhen the data on the x axis is a continuous value, it can be useful to break it into different bins in order to get a better visualization of the changes in the data.\n\nFor this exercise, we will look at the relationship between tuition and the Undergraduate population abbreviated as `UG` in this data. We will start by looking at a scatter plot of the data and examining the impact of different bin sizes on the visualization.\n\nInstructions\n\n1. Create a `regplot` of `Tuition` and `UG` and set the `fit_reg` parameter to `False` to disable the regression line.\n2. Create another plot with the `UG` data divided into 5 bins.\n3. Create a `regplot()` with the data divided into 8 bins.",
"_____no_output_____"
]
],
[
[
"# Create a scatter plot by disabling the regression line\nsns.regplot(data=college,\n y='Tuition',\n x='UG',\n fit_reg=False)\n\nplt.show()\nplt.clf()",
"_____no_output_____"
],
[
"# Create a scatter plot and bin the data into 5 bins\nsns.regplot(data=college,\n y='Tuition',\n x='UG',\n x_bins=5)\n\nplt.show()\nplt.clf()",
"_____no_output_____"
],
[
"# Create a regplot and bin the data into 8 bins\nsns.regplot(data=college,\n y='Tuition',\n x='UG',\n x_bins=8)\n\nplt.show()\nplt.clf()",
"_____no_output_____"
]
],
[
[
"## Creating heatmaps\n\nA heatmap is a common matrix plot that can be used to graphically summarize the relationship between two variables. For this exercise, we will start by looking at guests of the Daily Show from 1999 - 2015 and see how the occupations of the guests have changed over time.\n\nThe data includes the date of each guest appearance as well as their occupation. For the first exercise, we need to get the data into the right format for Seaborn's `heatmap` function to correctly plot the data. All of the data has already been read into the `df` variable.\n\nInstructions\n\n1. Use pandas' `crosstab()` function to build a table of visits by `Group` and `Year`.\n2. Print the `pd_crosstab` DataFrame.\n3. Plot the data using Seaborn's `heatmap()`.",
"_____no_output_____"
]
],
[
[
"# Import dataset\ndaily_show = pd.read_csv('daily_show_guests_cleaned.csv')",
"_____no_output_____"
],
[
"# Create a crosstab table of the data\npd_crosstab = pd.crosstab(daily_show['Group'], daily_show['YEAR'])\npd_crosstab",
"_____no_output_____"
],
[
"# Plot a heatmap of the table\nsns.heatmap(pd_crosstab)\n\n# Rotate tick marks for visibility\nplt.yticks(rotation=0)\nplt.xticks(rotation=90)\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Customizing heatmaps\n\nSeaborn supports several types of additional customizations to improve the output of a heatmap. For this exercise, we will continue to use the Daily Show data that is stored in the `daily_show` variable but we will customize the output.\n\nInstructions\n\n1. Create a crosstab table of `Group` and `YEAR`.\n2. Create a heatmap of the data using the `BuGn` palette.\n3. Disable the `cbar` and increase the linewidth to `0.3`.",
"_____no_output_____"
]
],
[
[
"# Create the crosstab DataFrame\npd_crosstab = pd.crosstab(daily_show['Group'], daily_show['YEAR'])\n\n# Plot a heatmap of the table with no color bar and using the BuGn palette\nsns.heatmap(pd_crosstab, cbar=False, cmap='BuGn', linewidths=0.3)\n\n# Rotate tick marks for visibility\nplt.yticks(rotation=0)\nplt.xticks(rotation=90)\n\n#Show the plot\nplt.show()\nplt.clf()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0d2a3818b6ee652c093d9c69bc2c7a45459ec16 | 9,750 | ipynb | Jupyter Notebook | 7 Syllabification, prosody, phonetics.ipynb | cs145442/gsoc2018Practice | 434458aa7d7176387d2433d69bd94de4f8ddc410 | [
"MIT"
] | null | null | null | 7 Syllabification, prosody, phonetics.ipynb | cs145442/gsoc2018Practice | 434458aa7d7176387d2433d69bd94de4f8ddc410 | [
"MIT"
] | null | null | null | 7 Syllabification, prosody, phonetics.ipynb | cs145442/gsoc2018Practice | 434458aa7d7176387d2433d69bd94de4f8ddc410 | [
"MIT"
] | null | null | null | 34.946237 | 1,167 | 0.499795 | [
[
[
"# Syllables",
"_____no_output_____"
]
],
[
[
"# http://docs.cltk.org/en/latest/latin.html#syllabifier\nfrom cltk.stem.latin.syllabifier import Syllabifier\n\ncato_agri_praef = \"Est interdum praestare mercaturis rem quaerere, nisi tam periculosum sit, et item foenerari, si tam honestum. Maiores nostri sic habuerunt et ita in legibus posiverunt: furem dupli condemnari, foeneratorem quadrupli. Quanto peiorem civem existimarint foeneratorem quam furem, hinc licet existimare. Et virum bonum quom laudabant, ita laudabant: bonum agricolam bonumque colonum; amplissime laudari existimabatur qui ita laudabatur. Mercatorem autem strenuum studiosumque rei quaerendae existimo, verum, ut supra dixi, periculosum et calamitosum. At ex agricolis et viri fortissimi et milites strenuissimi gignuntur, maximeque pius quaestus stabilissimusque consequitur minimeque invidiosus, minimeque male cogitantes sunt qui in eo studio occupati sunt. Nunc, ut ad rem redeam, quod promisi institutum principium hoc erit.\"\n\nfrom cltk.tokenize.word import WordTokenizer\nword_tokenizer = WordTokenizer('latin')\ncato_cltk_word_tokens = word_tokenizer.tokenize(cato_agri_praef.lower())\ncato_cltk_word_tokens_no_punt = [token for token in cato_cltk_word_tokens if token not in ['.', ',', ':', ';']]\n\n# now you can see the word '-que'\nprint(cato_cltk_word_tokens_no_punt)",
"['est', 'interdum', 'praestare', 'mercaturis', 'rem', 'quaerere', 'nisi', 'tam', 'periculosum', 'sit', 'et', 'item', 'foenerari', 'si', 'tam', 'honestum', 'maiores', 'nostri', 'sic', 'habuerunt', 'et', 'ita', 'in', 'legibus', 'posiverunt', 'furem', 'dupli', 'condemnari', 'foeneratorem', 'quadrupli', 'quanto', 'peiorem', 'civem', 'existimarint', 'foeneratorem', 'quam', 'furem', 'hinc', 'licet', 'existimare', 'et', 'virum', 'bonum', 'quom', 'laudabant', 'ita', 'laudabant', 'bonum', 'agricolam', 'bonum', '-que', 'colonum', 'amplissime', 'laudari', 'existimabatur', 'qui', 'ita', 'laudabatur', 'mercatorem', 'autem', 'strenuum', 'studiosum', '-que', 'rei', 'quaerendae', 'existimo', 'verum', 'ut', 'supra', 'dixi', 'periculosum', 'et', 'calamitosum', 'at', 'ex', 'agricolis', 'et', 'viri', 'fortissimi', 'et', 'milites', 'strenuissimi', 'gignuntur', 'maxime', '-que', 'pius', 'quaestus', 'stabilissimus', '-que', 'consequitur', 'minime', '-que', 'invidiosus', 'minime', '-que', 'male', 'cogitantes', 'sunt', 'qui', 'in', 'eo', 'studio', 'occupati', 'sunt', 'nunc', 'ut', 'ad', 'rem', 'redeam', 'quod', 'promisi', 'institutum', 'principium', 'hoc', 'erit']\n"
],
[
"syllabifier = Syllabifier()\n\nfor word in cato_cltk_word_tokens_no_punt:\n syllables = syllabifier.syllabify(word)\n print(word, syllables)",
"est ['est']\ninterdum ['in', 'ter', 'dum']\npraestare ['praes', 'ta', 're']\nmercaturis ['mer', 'ca', 'tu', 'ris']\nrem ['rem']\nquaerere ['quae', 're', 're']\nnisi ['ni', 'si']\ntam ['tam']\npericulosum ['pe', 'ri', 'cu', 'lo', 'sum']\nsit ['sit']\net ['et']\nitem ['i', 'tem']\nfoenerari ['foe', 'ne', 'ra', 'ri']\nsi ['si']\ntam ['tam']\nhonestum ['ho', 'nes', 'tum']\nmaiores ['ma', 'io', 'res']\nnostri ['nos', 'tri']\nsic ['sic']\nhabuerunt ['ha', 'bu', 'e', 'runt']\net ['et']\nita ['i', 'ta']\nin ['in']\nlegibus ['le', 'gi', 'bus']\nposiverunt ['po', 'si', 've', 'runt']\nfurem ['fu', 'rem']\ndupli ['du', 'pli']\ncondemnari ['con', 'dem', 'na', 'ri']\nfoeneratorem ['foe', 'ne', 'ra', 'to', 'rem']\nquadrupli ['qua', 'dru', 'pli']\nquanto ['quan', 'to']\npeiorem ['peio', 'rem']\ncivem ['ci', 'vem']\nexistimarint ['ex', 'is', 'ti', 'ma', 'rint']\nfoeneratorem ['foe', 'ne', 'ra', 'to', 'rem']\nquam ['quam']\nfurem ['fu', 'rem']\nhinc ['hinc']\nlicet ['li', 'cet']\nexistimare ['ex', 'is', 'ti', 'ma', 're']\net ['et']\nvirum ['vi', 'rum']\nbonum ['bo', 'num']\nquom ['quom']\nlaudabant ['lau', 'da', 'bant']\nita ['i', 'ta']\nlaudabant ['lau', 'da', 'bant']\nbonum ['bo', 'num']\nagricolam ['a', 'gri', 'co', 'lam']\nbonum ['bo', 'num']\n-que ['-que']\ncolonum ['co', 'lo', 'num']\namplissime ['am', 'plis', 'si', 'me']\nlaudari ['lau', 'da', 'ri']\nexistimabatur ['ex', 'is', 'ti', 'ma', 'ba', 'tur']\nqui ['qui']\nita ['i', 'ta']\nlaudabatur ['lau', 'da', 'ba', 'tur']\nmercatorem ['mer', 'ca', 'to', 'rem']\nautem ['au', 'tem']\nstrenuum ['stre', 'nu', 'um']\nstudiosum ['stu', 'di', 'o', 'sum']\n-que ['-que']\nrei ['rei']\nquaerendae ['quae', 'ren', 'dae']\nexistimo ['ex', 'is', 'ti', 'mo']\nverum ['ve', 'rum']\nut ['ut']\nsupra ['su', 'pra']\ndixi ['di', 'xi']\npericulosum ['pe', 'ri', 'cu', 'lo', 'sum']\net ['et']\ncalamitosum ['ca', 'la', 'mi', 'to', 'sum']\nat ['at']\nex ['ex']\nagricolis ['a', 'gri', 'co', 'lis']\net ['et']\nviri ['vi', 'ri']\nfortissimi ['for', 'tis', 'si', 'mi']\net ['et']\nmilites ['mi', 'li', 'tes']\nstrenuissimi ['stre', 'nu', 'is', 'si', 'mi']\ngignuntur ['gig', 'nun', 'tur']\nmaxime ['ma', 'xi', 'me']\n-que ['-que']\npius ['pi', 'us']\nquaestus ['quaes', 'tus']\nstabilissimus ['sta', 'bi', 'lis', 'si', 'mus']\n-que ['-que']\nconsequitur ['con', 'se', 'qui', 'tur']\nminime ['mi', 'ni', 'me']\n-que ['-que']\ninvidiosus ['in', 'vi', 'di', 'o', 'sus']\nminime ['mi', 'ni', 'me']\n-que ['-que']\nmale ['ma', 'le']\ncogitantes ['co', 'gi', 'tan', 'tes']\nsunt ['sunt']\nqui ['qui']\nin ['in']\neo ['e', 'o']\nstudio ['stu', 'di', 'o']\noccupati ['oc', 'cu', 'pa', 'ti']\nsunt ['sunt']\nnunc ['nunc']\nut ['ut']\nad ['ad']\nrem ['rem']\nredeam ['re', 'de', 'am']\nquod ['quod']\npromisi ['pro', 'mi', 'si']\ninstitutum ['in', 'sti', 'tu', 'tum']\nprincipium ['prin', 'ci', 'pi', 'um']\nhoc ['hoc']\nerit ['e', 'rit']\n"
]
],
[
[
"# Prosody\n\nTakes two steps: first find long vowels, then scan actual meter",
"_____no_output_____"
]
],
[
[
"# macronizer\n# http://docs.cltk.org/en/latest/latin.html#macronizer\nfrom cltk.prosody.latin.macronizer import Macronizer\n\nmacronizer = Macronizer('tag_ngram_123_backoff')\n\ntext = 'Quo usque tandem, O Catilina, abutere nostra patientia?'\n\nscanned_text = macronizer.macronize_text(text)",
"_____no_output_____"
],
[
"# scanner\n# http://docs.cltk.org/en/latest/latin.html#prosody-scanning\n\nfrom cltk.prosody.latin.scanner import Scansion\n\nscanner = Scansion()\nprose_text = macronizer.macronize_tags(scanned_text)\nprint(prose_text)",
"[('quō', None, 'quō'), ('usque', 'd--------', 'usque'), ('tandem', 'd--------', 'tandem'), (',', 'u--------', ','), ('ō', None, 'ō'), ('catilīnā', None, 'catilīnā'), (',', 'u--------', ','), ('abūtēre', None, 'abūtēre'), ('nostrā', None, 'nostrā'), ('patientia', 'n-s---fn-', 'patientia'), ('?', None, '?')]\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d0d2ac2aeedb5441327e7d6675cf104da24a42ef | 16,840 | ipynb | Jupyter Notebook | module1-afirstlookatdata/LS_DSPT3_111_A_First_Look_at_Data.ipynb | elcoreano/DS-Unit-1-Sprint-1-Dealing-With-Data | 1734744535102348fc5dec3997365c196479fe44 | [
"MIT"
] | null | null | null | module1-afirstlookatdata/LS_DSPT3_111_A_First_Look_at_Data.ipynb | elcoreano/DS-Unit-1-Sprint-1-Dealing-With-Data | 1734744535102348fc5dec3997365c196479fe44 | [
"MIT"
] | null | null | null | module1-afirstlookatdata/LS_DSPT3_111_A_First_Look_at_Data.ipynb | elcoreano/DS-Unit-1-Sprint-1-Dealing-With-Data | 1734744535102348fc5dec3997365c196479fe44 | [
"MIT"
] | null | null | null | 30.233393 | 303 | 0.355463 | [
[
[
"<a href=\"https://colab.research.google.com/github/elcoreano/DS-Unit-1-Sprint-1-Dealing-With-Data/blob/master/module1-afirstlookatdata/LS_DSPT3_111_A_First_Look_at_Data.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Lambda School Data Science - A First Look at Data\n\n",
"_____no_output_____"
],
[
"## Lecture - let's explore Python DS libraries and examples!\n\nThe Python Data Science ecosystem is huge. You've seen some of the big pieces - pandas, scikit-learn, matplotlib. What parts do you want to see more of?",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport pandas as pd",
"_____no_output_____"
]
],
[
[
"## Assignment - now it's your turn\n\nPick at least one Python DS library, and using documentation/examples reproduce in this notebook something cool. It's OK if you don't fully understand it or get it 100% working, but do put in effort and look things up.",
"_____no_output_____"
]
],
[
[
"# TODO - your code here\n# Use what we did live in lecture as an example\n\nnp.random.randint(0, 10, size=10)",
"_____no_output_____"
],
[
"np.random.randint(0, 10, size=10)",
"_____no_output_____"
],
[
"x = [9, 4, 9, 9, 6, 2, 2, 5, 0, 3]\ny = [0, 5, 8, 8, 5, 2, 1, 0, 7, 9]",
"_____no_output_____"
],
[
"df = pd.DataFrame ({'set 1': x, \"set 2\": y})\ndf",
"_____no_output_____"
],
[
"df['set 1']",
"_____no_output_____"
],
[
"df.shape",
"_____no_output_____"
],
[
"df['set 3'] = df['set 1'] + 2*df['set 2']\ndf",
"_____no_output_____"
],
[
"df.shape",
"_____no_output_____"
]
],
[
[
"### Assignment questions\n\nAfter you've worked on some code, answer the following questions in this text block:\n\n1. Describe in a paragraph of text what you did and why, as if you were writing an email to somebody interested but nontechnical.\n\n*# I tinkered with three of the most used libraries for python. A library is a group of tools that makes life easier for a user of any coding program.*\n\n2. What was the most challenging part of what you did?\n\n*# The precourse helped in familiarizing me with the libraries and their hidden super powers, so, that wasn't too challenging. What was challenging was the lecture and getting used to Alex's teaching style, the Zoom/Slack environment, and using GitHub.*\n\n3. What was the most interesting thing you learned?\n\n*# GitHub is pretty damn powerful.*\n\n4. What area would you like to explore with more time?\n\n*# Everything covered in this lecture. However, with time, I'll become more fluent with all tools. Just a bit excited.*\n\n\n",
"_____no_output_____"
],
[
"## Stretch goals and resources\n\nFollowing are *optional* things for you to take a look at. Focus on the above assignment first, and make sure to commit and push your changes to GitHub (and since this is the first assignment of the sprint, open a PR as well).\n\n- [pandas documentation](https://pandas.pydata.org/pandas-docs/stable/)\n- [scikit-learn documentation](http://scikit-learn.org/stable/documentation.html)\n- [matplotlib documentation](https://matplotlib.org/contents.html)\n- [Awesome Data Science](https://github.com/bulutyazilim/awesome-datascience) - a list of many types of DS resources\n\nStretch goals:\n\n- Find and read blogs, walkthroughs, and other examples of people working through cool things with data science - and share with your classmates!\n- Write a blog post (Medium is a popular place to publish) introducing yourself as somebody learning data science, and talking about what you've learned already and what you're excited to learn more about.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
d0d2b0db91898329fe64e9a19aeb4cab560d050f | 35,138 | ipynb | Jupyter Notebook | 02_projects/keyword_spotting/2words/TrainKeywordSpottingModel2words.ipynb | KlausPuchner/tinyml | 6e0014dc70d2ef923f29be2a5dc0a48c0b21bc47 | [
"MIT"
] | 1 | 2021-04-05T20:11:36.000Z | 2021-04-05T20:11:36.000Z | 02_projects/keyword_spotting/2words/TrainKeywordSpottingModel2words.ipynb | KlausPuchner/tinyml | 6e0014dc70d2ef923f29be2a5dc0a48c0b21bc47 | [
"MIT"
] | null | null | null | 02_projects/keyword_spotting/2words/TrainKeywordSpottingModel2words.ipynb | KlausPuchner/tinyml | 6e0014dc70d2ef923f29be2a5dc0a48c0b21bc47 | [
"MIT"
] | null | null | null | 35.421371 | 498 | 0.601685 | [
[
[
"# Training Keyword Spotting\nThis notebook builds on the Colab in which we used the pre-trained [micro_speech](https://github.com/tensorflow/tensorflow/tree/master/tensorflow/lite/micro/examples/micro_speech) example as well as the HarvardX [3_5_18_TrainingKeywordSpotting.ipynb](https://github.com/tinyMLx/colabs) and [4_5_16_KWS_PretrainedModel](https://github.com/tinyMLx/colabs) from the [TinyML Specialization on edX](https://www.edx.org/professional-certificate/harvardx-tiny-machine-learning).\n\n\n",
"_____no_output_____"
],
[
"# Setup\n<font color='red'>**This Notebook only works on Tensorflow 1.15 and was tested with Tensorflow 1.15.5**</font>",
"_____no_output_____"
],
[
"### Prerequisites\nClone the TensorFlow Github Repository with the relevant base code.",
"_____no_output_____"
]
],
[
[
"%%bash\nrm -rf tensorflow log v2.4.1.zip logs models train dataset extract_loudest_section\napt-get update -qq && apt-get install -y wget unzip\nwget https://github.com/tensorflow/tensorflow/archive/v2.4.1.zip\nunzip v2.4.1.zip &> log\nmv tensorflow-2.4.1/ tensorflow/\nrm -rf v2.4.1.zip log",
"_____no_output_____"
]
],
[
[
"### Import Packages\nImport standard packages as well as the additional packages from the cloned Github Repo.",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nimport sys\n# We add this path so we can import the speech processing modules.\nsys.path.append(\"./tensorflow/tensorflow/examples/speech_commands/\")\nimport input_data\nimport models\nimport numpy as np\nimport pickle\nimport shutil\nimport os",
"_____no_output_____"
]
],
[
[
"### Check GPU availability\nThe code will also work without GPU acceleration, but it will be significantly slower.",
"_____no_output_____"
]
],
[
[
"tf.test.is_gpu_available(cuda_only=True, min_cuda_compute_capability=None)",
"_____no_output_____"
]
],
[
[
"### Configure Your Model!\nSelect your keywords and model settings with which to train!\n\n**This is where you need to make choices and input data!**\n\n```WANTED_WORDS``` = A comma-delimited string of the words you want to train for (e.g., \"yes,no\"). All the other words you do not select will be used to train an \"unknown\" label so that the model does not just recognize speech but your specific words. Audio data with no spoken words will be used to train a \"silence\" label. We suggest picking 2-4 words for best results.\n\nOptions for target words are (PICK FROM THIS LIST FOR BEST RESULTS): \"one\", \"two\", \"three\", \"four\", \"five\", \"six\", \"seven\", \"eight\", \"nine\", \"yes\", \"no\", \"up\", \"down\", \"left\", \"right\", \"on\", \"off\", \"stop\", \"go\", “backward”, “forward”, “follow”, “learn”,\n\nAdditional words that will be used to help train the \"unkown\" label are: \"bed\", \"bird\", \"cat\", \"dog\", \"happy\", \"house\", \"marvin\", \"sheila\", \"tree\", \"wow\"",
"_____no_output_____"
]
],
[
[
"WANTED_WORDS = \"stop,go\"",
"_____no_output_____"
]
],
[
[
"The number of training steps and learning rates can be specified as comma-separated strings to define the amount/rate at each stage. For example, ```TRAINING_STEPS=\"12000,3000\"``` and ```LEARNING_RATE=\"0.001,0.0001\"``` will run 12,000 training steps with a rate of 0.001 followed by 3,000 final steps with a learning rate of 0.0001. These are good default values to work off of when you choose your values as the course staff has gotten this to work well with those values in the past!",
"_____no_output_____"
]
],
[
[
"TRAINING_STEPS = \"12000,3000\"\nLEARNING_RATE = \"0.001,0.0001\"",
"_____no_output_____"
]
],
[
[
"We suggest you leave the ```MODEL_ARCHITECTURE``` as tiny_conv the first time but if you would like to do this again and explore additional models some options are: ```single_fc, conv, low_latency_conv, low_latency_svdf, tiny_embedding_conv```",
"_____no_output_____"
]
],
[
[
"MODEL_ARCHITECTURE = 'tiny_conv'",
"_____no_output_____"
],
[
"# Calculate the total number of steps, which is used to identify the checkpoint\n# file name.\nTOTAL_STEPS = str(sum(map(lambda string: int(string), TRAINING_STEPS.split(\",\"))))\n\n# Print the configuration to confirm it\nprint(\"Training these words: %s\" % WANTED_WORDS)\nprint(\"Training steps in each stage: %s\" % TRAINING_STEPS)\nprint(\"Learning rate in each stage: %s\" % LEARNING_RATE)\nprint(\"Total number of training steps: %s\" % TOTAL_STEPS)",
"_____no_output_____"
]
],
[
[
"**DO NOT MODIFY** the following constants as they include filepaths used in this notebook and data that is shared during training and inference.",
"_____no_output_____"
]
],
[
[
"# Calculate the percentage of 'silence' and 'unknown' training samples required\n# to ensure that we have equal number of samples for each label.\nnumber_of_labels = WANTED_WORDS.count(',') + 1\nnumber_of_total_labels = number_of_labels + 2 # for 'silence' and 'unknown' label\nequal_percentage_of_training_samples = int(100.0/(number_of_total_labels))\nSILENT_PERCENTAGE = equal_percentage_of_training_samples\nUNKNOWN_PERCENTAGE = equal_percentage_of_training_samples\n\n# Constants which are shared during training and inference\nPREPROCESS = 'micro'\nWINDOW_STRIDE = 20\n\n# Constants used during training only\nVERBOSITY = 'DEBUG'\nEVAL_STEP_INTERVAL = '1000'\nSAVE_STEP_INTERVAL = '1000'\n\n# Constants for training directories and filepaths\nDATASET_DIR = 'dataset/'\nLOGS_DIR = 'logs/'\nTRAIN_DIR = 'train/' # for training checkpoints and other files.\n\n# Constants for inference directories and filepaths\nimport os\nMODELS_DIR = 'models'\nif not os.path.exists(MODELS_DIR):\n os.mkdir(MODELS_DIR)\nMODEL_TF = os.path.join(MODELS_DIR, 'model.pb')\nMODEL_TFLITE = os.path.join(MODELS_DIR, 'model.tflite')\nFLOAT_MODEL_TFLITE = os.path.join(MODELS_DIR, 'float_model.tflite')\nMODEL_TFLITE_MICRO = os.path.join(MODELS_DIR, 'model.cc')\nSAVED_MODEL = os.path.join(MODELS_DIR, 'saved_model')\n\n# Constants for Quantization\nQUANT_INPUT_MIN = 0.0\nQUANT_INPUT_MAX = 26.0\nQUANT_INPUT_RANGE = QUANT_INPUT_MAX - QUANT_INPUT_MIN\n\n# Constants for audio process during Quantization and Evaluation\nSAMPLE_RATE = 16000\nCLIP_DURATION_MS = 1000\nWINDOW_SIZE_MS = 30.0\nFEATURE_BIN_COUNT = 40\nBACKGROUND_FREQUENCY = 0.8\nBACKGROUND_VOLUME_RANGE = 0.1\nTIME_SHIFT_MS = 100.0\n\n# URL for the dataset and train/val/test split\nDATA_URL = 'https://storage.googleapis.com/download.tensorflow.org/data/speech_commands_v0.02.tar.gz'\nVALIDATION_PERCENTAGE = 10\nTESTING_PERCENTAGE = 10",
"_____no_output_____"
],
[
"# Calculate the correct flattened input data shape for later use in model conversion\n# since the model takes a flattened version of the spectrogram. The shape is number of \n# overlapping windows times the number of frequency bins. For the default settings we have\n# 40 bins (as set above) times 49 windows (as calculated below) so the shape is (1,1960)\ndef window_counter(total_samples, window_size, stride):\n '''helper function to count the number of full-length overlapping windows'''\n window_count = 0\n sample_index = 0\n while True:\n window = range(sample_index,sample_index+stride)\n if window.stop < total_samples:\n window_count += 1\n else:\n break\n \n sample_index += stride\n return window_count\n\nOVERLAPPING_WINDOWS = window_counter(CLIP_DURATION_MS, int(WINDOW_SIZE_MS), WINDOW_STRIDE)\nFLATTENED_SPECTROGRAM_SHAPE = (1, OVERLAPPING_WINDOWS * FEATURE_BIN_COUNT)",
"_____no_output_____"
]
],
[
[
"# Train the model",
"_____no_output_____"
],
[
"### Load in TensorBoard to visulaize the training process.\n\nAs training progresses you should see the training status show up in the Tensorboard area. If this works it is very helpful for analyzing your training progress. Unfortunately, the staff has found that it sometimes doesn't start showing data for a while (~15 minutes) and sometimes doesn't show data until training completes (and instead shows ```No dashboards are active for the current data set```.). If it is working and then stops updating look to the top of the cell and click reconnect.",
"_____no_output_____"
]
],
[
[
"%load_ext tensorboard\nlogs_base_dir='./logs/'\nos.makedirs(logs_base_dir, exist_ok=True)\n%tensorboard --logdir {logs_base_dir} --host 0.0.0.0 --port 6006",
"_____no_output_____"
]
],
[
[
"### Launch Training\n\nIf you would like to get more information on the training script you can find the source code for the script [here](https://github.com/tensorflow/tensorflow/blob/master/tensorflow/examples/speech_commands/train.py). In short it sets up the optimizer and preprocessor based on all of the flags we pass in!\n\nFinally, by setting the ```VERBOSITY = 'DEBUG'``` above be aware that the training cell will print A LOT of information. Specifically you will get the accuracy and loss at each step as well as a confusion matrix every 1000 steps. We hope that is helpful in case TensorBoard fails to work. If you would like to run with less printouts you can change the setting to ```WARN``` or ```FATAL```. You will find this in the \"Configure Your Model!\" section.",
"_____no_output_____"
]
],
[
[
"!python tensorflow/tensorflow/examples/speech_commands/train.py \\\n--data_dir={DATASET_DIR} \\\n--wanted_words={WANTED_WORDS} \\\n--silence_percentage={SILENT_PERCENTAGE} \\\n--unknown_percentage={UNKNOWN_PERCENTAGE} \\\n--preprocess={PREPROCESS} \\\n--window_stride={WINDOW_STRIDE} \\\n--model_architecture={MODEL_ARCHITECTURE} \\\n--how_many_training_steps={TRAINING_STEPS} \\\n--learning_rate={LEARNING_RATE} \\\n--train_dir={TRAIN_DIR} \\\n--summaries_dir={LOGS_DIR} \\\n--verbosity={VERBOSITY} \\\n--eval_step_interval={EVAL_STEP_INTERVAL} \\\n--save_step_interval={SAVE_STEP_INTERVAL}",
"_____no_output_____"
]
],
[
[
"# Generating your Model\nJust like with the pre-trained model we will now take the final checkpoint and convert it into a quantized TensorFlow Lite model.",
"_____no_output_____"
],
[
"### Generate a TensorFlow Model for Inference\n\nCombine relevant training results (graph, weights, etc) into a single file for inference. This process is known as freezing a model and the resulting model is known as a frozen model/graph, as it cannot be further re-trained after this process.",
"_____no_output_____"
]
],
[
[
"!rm -rf {SAVED_MODEL}\n!python tensorflow/tensorflow/examples/speech_commands/freeze.py \\\n--wanted_words=$WANTED_WORDS \\\n--window_stride_ms=$WINDOW_STRIDE \\\n--preprocess=$PREPROCESS \\\n--model_architecture=$MODEL_ARCHITECTURE \\\n--start_checkpoint=$TRAIN_DIR$MODEL_ARCHITECTURE'.ckpt-'{TOTAL_STEPS} \\\n--save_format=saved_model \\\n--output_file={SAVED_MODEL}",
"_____no_output_____"
]
],
[
[
"### Generate a TensorFlow Lite Model\n\nConvert the frozen graph into a TensorFlow Lite model, which is fully quantized for use with embedded devices.\n\nThe following cell will also print the model size, which will be under 20 kilobytes.\n\nWe download the dataset to use as a representative dataset for more thoughtful post training quantization. ",
"_____no_output_____"
]
],
[
[
"model_settings = models.prepare_model_settings(\n len(input_data.prepare_words_list(WANTED_WORDS.split(','))),\n SAMPLE_RATE, CLIP_DURATION_MS, WINDOW_SIZE_MS,\n WINDOW_STRIDE, FEATURE_BIN_COUNT, PREPROCESS)\naudio_processor = input_data.AudioProcessor(\n DATA_URL, DATASET_DIR,\n SILENT_PERCENTAGE, UNKNOWN_PERCENTAGE,\n WANTED_WORDS.split(','), VALIDATION_PERCENTAGE,\n TESTING_PERCENTAGE, model_settings, LOGS_DIR)",
"_____no_output_____"
],
[
"with tf.Session() as sess:\n float_converter = tf.lite.TFLiteConverter.from_saved_model(SAVED_MODEL)\n float_tflite_model = float_converter.convert()\n float_tflite_model_size = open(FLOAT_MODEL_TFLITE, \"wb\").write(float_tflite_model)\n print(\"Float model is %d bytes\" % float_tflite_model_size)\n\n converter = tf.lite.TFLiteConverter.from_saved_model(SAVED_MODEL)\n converter.optimizations = [tf.lite.Optimize.DEFAULT]\n converter.inference_input_type = tf.lite.constants.INT8\n converter.inference_output_type = tf.lite.constants.INT8\n def representative_dataset_gen():\n for i in range(100):\n data, _ = audio_processor.get_data(1, i*1, model_settings,\n BACKGROUND_FREQUENCY, \n BACKGROUND_VOLUME_RANGE,\n TIME_SHIFT_MS,\n 'testing',\n sess)\n flattened_data = np.array(data.flatten(), dtype=np.float32).reshape(FLATTENED_SPECTROGRAM_SHAPE)\n yield [flattened_data]\n converter.representative_dataset = representative_dataset_gen\n tflite_model = converter.convert()\n tflite_model_size = open(MODEL_TFLITE, \"wb\").write(tflite_model)\n print(\"Quantized model is %d bytes\" % tflite_model_size)\n",
"_____no_output_____"
]
],
[
[
"### Testing the accuracy after Quantization\n\nVerify that the model we've exported is still accurate, using the TF Lite Python API and our test set.",
"_____no_output_____"
]
],
[
[
"# Helper function to run inference\ndef run_tflite_inference_testSet(tflite_model_path, model_type=\"Float\"):\n #\n # Load test data\n #\n np.random.seed(0) # set random seed for reproducible test results.\n with tf.Session() as sess:\n test_data, test_labels = audio_processor.get_data(\n -1, 0, model_settings, BACKGROUND_FREQUENCY, BACKGROUND_VOLUME_RANGE,\n TIME_SHIFT_MS, 'testing', sess)\n test_data = np.expand_dims(test_data, axis=1).astype(np.float32)\n\n #\n # Initialize the interpreter\n #\n interpreter = tf.lite.Interpreter(tflite_model_path)\n interpreter.allocate_tensors()\n input_details = interpreter.get_input_details()[0]\n output_details = interpreter.get_output_details()[0]\n \n #\n # For quantized models, manually quantize the input data from float to integer\n #\n if model_type == \"Quantized\":\n input_scale, input_zero_point = input_details[\"quantization\"]\n test_data = test_data / input_scale + input_zero_point\n test_data = test_data.astype(input_details[\"dtype\"])\n\n #\n # Evaluate the predictions\n #\n correct_predictions = 0\n for i in range(len(test_data)):\n interpreter.set_tensor(input_details[\"index\"], test_data[i])\n interpreter.invoke()\n output = interpreter.get_tensor(output_details[\"index\"])[0]\n top_prediction = output.argmax()\n correct_predictions += (top_prediction == test_labels[i])\n\n print('%s model accuracy is %f%% (Number of test samples=%d)' % (\n model_type, (correct_predictions * 100) / len(test_data), len(test_data)))",
"_____no_output_____"
],
[
"# Compute float model accuracy\nrun_tflite_inference_testSet(FLOAT_MODEL_TFLITE)\n\n# Compute quantized model accuracy\nrun_tflite_inference_testSet(MODEL_TFLITE, model_type='Quantized')",
"_____no_output_____"
]
],
[
[
"# Testing the model with your own data!\nNow comes the fun part. It's time to test your model with your own realworld data. We'll proceed in the same way we tested the pre-trained model. Have fun!",
"_____no_output_____"
],
[
"### Importing packages",
"_____no_output_____"
]
],
[
[
"!apt-get update -qqq && apt-get -y -qqq install apt-utils gcc libpq-dev libsndfile-dev git\n!python3 -m pip install --upgrade --no-cache-dir --quiet pip ffmpeg-python scipy librosa google-colab\nfrom IPython.display import HTML, Audio\nfrom google.colab.output import eval_js\nfrom base64 import b64decode\nimport numpy as np\nfrom scipy.io.wavfile import read as wav_read\nimport io\nimport ffmpeg\n#!pip install librosa\nimport librosa\nimport scipy.io.wavfile\n!git clone https://github.com/petewarden/extract_loudest_section.git\n!make -C extract_loudest_section/\nprint(\"Packages Imported, Extract_Loudest_Section Built\")",
"_____no_output_____"
]
],
[
[
"### Define the helper function to run inference",
"_____no_output_____"
]
],
[
[
"# Helper function to run inference (on a single input this time)\n# Note: this also includes additional manual pre-processing\nTF_SESS = tf.compat.v1.InteractiveSession()\ndef run_tflite_inference_singleFile(tflite_model_path, custom_audio, sr_custom_audio, model_type=\"Float\"):\n #\n # Preprocess the sample to get the features we pass to the model\n #\n # First re-sample to the needed rate (and convert to mono if needed)\n custom_audio_resampled = librosa.resample(librosa.to_mono(np.float64(custom_audio)), sr_custom_audio, SAMPLE_RATE)\n # Then extract the loudest one second\n scipy.io.wavfile.write('custom_audio.wav', SAMPLE_RATE, np.int16(custom_audio_resampled))\n !/tmp/extract_loudest_section/gen/bin/extract_loudest_section custom_audio.wav ./trimmed\n # Finally pass it through the TFLiteMicro preprocessor to produce the \n # spectrogram/MFCC input that the model expects\n custom_model_settings = models.prepare_model_settings(\n 0, SAMPLE_RATE, CLIP_DURATION_MS, WINDOW_SIZE_MS,\n WINDOW_STRIDE, FEATURE_BIN_COUNT, PREPROCESS)\n custom_audio_processor = input_data.AudioProcessor(None, None, 0, 0, '', 0, 0,\n model_settings, None)\n custom_audio_preprocessed = custom_audio_processor.get_features_for_wav(\n 'trimmed/custom_audio.wav', model_settings, TF_SESS)\n # Reshape the output into a 1,1960 matrix as that is what the model expects\n custom_audio_input = custom_audio_preprocessed[0].flatten()\n test_data = np.reshape(custom_audio_input,(1,len(custom_audio_input)))\n\n #\n # Initialize the interpreter\n #\n interpreter = tf.lite.Interpreter(tflite_model_path)\n interpreter.allocate_tensors()\n input_details = interpreter.get_input_details()[0]\n output_details = interpreter.get_output_details()[0]\n\n #\n # For quantized models, manually quantize the input data from float to integer\n #\n if model_type == \"Quantized\":\n input_scale, input_zero_point = input_details[\"quantization\"]\n test_data = test_data / input_scale + input_zero_point\n test_data = test_data.astype(input_details[\"dtype\"])\n\n #\n # Run the interpreter\n #\n interpreter.set_tensor(input_details[\"index\"], test_data)\n interpreter.invoke()\n output = interpreter.get_tensor(output_details[\"index\"])[0]\n top_prediction = output.argmax()\n\n #\n # Translate the output\n #\n top_prediction_str = ''\n if top_prediction >= 2:\n top_prediction_str = WANTED_WORDS.split(',')[top_prediction-2]\n elif top_prediction == 0:\n top_prediction_str = 'silence'\n else:\n top_prediction_str = 'unknown'\n\n print('%s model guessed the value to be %s' % (model_type, top_prediction_str))",
"_____no_output_____"
]
],
[
[
"### Define the audio importing function\nAdapted from: https://ricardodeazambuja.com/deep_learning/2019/03/09/audio_and_video_google_colab/ and https://colab.research.google.com/drive/1Z6VIRZ_sX314hyev3Gm5gBqvm1wQVo-a#scrollTo=RtMcXr3o6gxN",
"_____no_output_____"
]
],
[
[
"def get_audio():\n \"\"\"Records audio from your local microphone inside a colab notebook\n Returns\n -------\n tuple\n audio (numpy.ndarray), sample rate (int)\n Obs:\n To write this piece of code I took inspiration/code from a lot of places.\n It was late night, so I'm not sure how much I created or just copied o.O\n Here are some of the possible references:\n https://blog.addpipe.com/recording-audio-in-the-browser-using-pure-html5-and-minimal-javascript/\n https://stackoverflow.com/a/18650249\n https://hacks.mozilla.org/2014/06/easy-audio-capture-with-the-mediarecorder-api/\n https://air.ghost.io/recording-to-an-audio-file-using-html5-and-js/\n https://stackoverflow.com/a/49019356\n \"\"\"\n\n AUDIO_HTML = \"\"\"\n <script>\n var my_div = document.createElement(\"DIV\");\n var my_p = document.createElement(\"P\");\n var my_btn = document.createElement(\"BUTTON\");\n var t = document.createTextNode(\"Press to start recording\");\n my_btn.appendChild(t);\n //my_p.appendChild(my_btn);\n my_div.appendChild(my_btn);\n document.body.appendChild(my_div);\n var base64data = 0;\n var reader;\n var recorder, gumStream;\n var recordButton = my_btn;\n var handleSuccess = function(stream) {\n gumStream = stream;\n var options = {\n //bitsPerSecond: 8000, //chrome seems to ignore, always 48k\n mimeType : 'audio/webm;codecs=opus'\n //mimeType : 'audio/webm;codecs=pcm'\n }; \n //recorder = new MediaRecorder(stream, options);\n recorder = new MediaRecorder(stream);\n recorder.ondataavailable = function(e) { \n var url = URL.createObjectURL(e.data);\n var preview = document.createElement('audio');\n preview.controls = true;\n preview.src = url;\n document.body.appendChild(preview);\n reader = new FileReader();\n reader.readAsDataURL(e.data); \n reader.onloadend = function() {\n base64data = reader.result;\n //console.log(\"Inside FileReader:\" + base64data);\n }\n };\n recorder.start();\n };\n recordButton.innerText = \"Recording... press to stop\";\n navigator.mediaDevices.getUserMedia({audio: true}).then(handleSuccess);\n function toggleRecording() {\n if (recorder && recorder.state == \"recording\") {\n recorder.stop();\n gumStream.getAudioTracks()[0].stop();\n recordButton.innerText = \"Saving the recording... pls wait!\"\n }\n }\n // https://stackoverflow.com/a/951057\n function sleep(ms) {\n return new Promise(resolve => setTimeout(resolve, ms));\n }\n var data = new Promise(resolve=>{\n //recordButton.addEventListener(\"click\", toggleRecording);\n recordButton.onclick = ()=>{\n toggleRecording()\n sleep(2000).then(() => {\n // wait 2000ms for the data to be available...\n // ideally this should use something like await...\n //console.log(\"Inside data:\" + base64data)\n resolve(base64data.toString())\n });\n }\n });\n\n </script>\n \"\"\"\n\n display(HTML(AUDIO_HTML))\n data = eval_js(\"data\")\n binary = b64decode(data.split(',')[1])\n\n process = (ffmpeg\n .input('pipe:0')\n .output('pipe:1', format='wav')\n .run_async(pipe_stdin=True, pipe_stdout=True, pipe_stderr=True, quiet=True, overwrite_output=True)\n )\n output, err = process.communicate(input=binary)\n\n riff_chunk_size = len(output) - 8\n # Break up the chunk size into four bytes, held in b.\n q = riff_chunk_size\n b = []\n for i in range(4):\n q, r = divmod(q, 256)\n b.append(r)\n\n # Replace bytes 4:8 in proc.stdout with the actual size of the RIFF chunk.\n riff = output[:4] + bytes(b) + output[8:]\n\n sr, audio = wav_read(BytesIO(riff))\n\n return audio, sr",
"_____no_output_____"
]
],
[
[
"### Record your own audio and test the model!\nAfter you run the record cell wait for the stop button to appear then start recording and then press the button to stop the recording once you have said the word!",
"_____no_output_____"
]
],
[
[
"custom_audio, sr_custom_audio = get_audio()\nprint(\"DONE\")",
"_____no_output_____"
],
[
"# Then test the model\nrun_tflite_inference_singleFile(MODEL_TFLITE, custom_audio, sr_custom_audio, model_type=\"Quantized\")",
"_____no_output_____"
]
],
[
[
"### Generate a TensorFlow Lite for Microcontrollers Model\nTo convert the TensorFlow Lite quantized model into a C source file that can be loaded by TensorFlow Lite for Microcontrollers on Arduino we simply need to use the ```xxd``` tool to convert the ```.tflite``` file into a ```.cc``` file.",
"_____no_output_____"
]
],
[
[
"!apt-get update -qqq && apt-get -qqq install xxd",
"_____no_output_____"
],
[
"MODEL_TFLITE = './models/model.tflite'\nMODEL_TFLITE_MICRO = './models/model.cc'\n!xxd -i {MODEL_TFLITE} > {MODEL_TFLITE_MICRO}\nREPLACE_TEXT = MODEL_TFLITE.replace('/', '_').replace('.', '_')\n!sed -i 's/'{REPLACE_TEXT}'/g_model/g' {MODEL_TFLITE_MICRO}",
"_____no_output_____"
]
],
[
[
"The generated Tensorflow Lite for Microcontroller model can now be used in the Arduino IDE. There are **two options** to do this:\n\n1. Copy the screen output directly from the Jupyter Notebook into the **micro_features_model.cpp** file (in the Arduino IDE)\n2. Download the **model.cc** file for later use to copy its content into the **micro_features_model.cpp** file (in the Arduino IDE)\n\n### Option 1: Copy Output directly",
"_____no_output_____"
]
],
[
[
"!cat {MODEL_TFLITE_MICRO}",
"_____no_output_____"
]
],
[
[
"### Option 2: Download Model File",
"_____no_output_____"
]
],
[
[
"from IPython.display import FileLink\nlocal_file = FileLink('./models/model.cc', result_html_prefix=\"Click here to download: \")\ndisplay(local_file)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0d2b51086f288e3c1f1553103560ed4c696be25 | 580,602 | ipynb | Jupyter Notebook | bayes-opt.ipynb | chappers/chappers.github.io | 80f7fe93eaf97d4e77851a0bb5d1efbd1e9d5573 | [
"MIT"
] | null | null | null | bayes-opt.ipynb | chappers/chappers.github.io | 80f7fe93eaf97d4e77851a0bb5d1efbd1e9d5573 | [
"MIT"
] | null | null | null | bayes-opt.ipynb | chappers/chappers.github.io | 80f7fe93eaf97d4e77851a0bb5d1efbd1e9d5573 | [
"MIT"
] | null | null | null | 950.248773 | 253,640 | 0.953171 | [
[
[
"!pip install scikit-optimize",
"Requirement already satisfied: scikit-optimize in /home/chapman/anaconda3/envs/d4rl/lib/python3.8/site-packages (0.8.1)\nRequirement already satisfied: joblib>=0.11 in /home/chapman/anaconda3/envs/d4rl/lib/python3.8/site-packages (from scikit-optimize) (0.16.0)\nRequirement already satisfied: scikit-learn>=0.20.0 in /home/chapman/anaconda3/envs/d4rl/lib/python3.8/site-packages (from scikit-optimize) (0.23.2)\nRequirement already satisfied: numpy>=1.13.3 in /home/chapman/anaconda3/envs/d4rl/lib/python3.8/site-packages (from scikit-optimize) (1.19.0)\nRequirement already satisfied: scipy>=0.19.1 in /home/chapman/anaconda3/envs/d4rl/lib/python3.8/site-packages (from scikit-optimize) (1.5.1)\nRequirement already satisfied: pyaml>=16.9 in /home/chapman/anaconda3/envs/d4rl/lib/python3.8/site-packages (from scikit-optimize) (20.4.0)\nRequirement already satisfied: threadpoolctl>=2.0.0 in /home/chapman/anaconda3/envs/d4rl/lib/python3.8/site-packages (from scikit-learn>=0.20.0->scikit-optimize) (2.1.0)\nRequirement already satisfied: PyYAML in /home/chapman/anaconda3/envs/d4rl/lib/python3.8/site-packages (from pyaml>=16.9->scikit-optimize) (5.3.1)\n"
]
],
[
[
"Based on this: \n\n* https://scikit-optimize.github.io/stable/auto_examples/bayesian-optimization.html#sphx-glr-auto-examples-bayesian-optimization-py",
"_____no_output_____"
]
],
[
[
"import numpy as np\nnp.random.seed(1234)\nimport matplotlib.pyplot as plt\nfrom skopt.plots import plot_gaussian_process\nfrom skopt import Optimizer\n\nimport matplotlib.pyplot as plt\nimport matplotlib.image as mpimg\nfrom skopt import gp_minimize\nimport numpy as np\n%matplotlib inline\n\nfrom sklearn.gaussian_process import GaussianProcessRegressor\nfrom sklearn.gaussian_process.kernels import Matern",
"_____no_output_____"
],
[
"noise_level = 0.1\n\n# Our 1D toy problem, this is the function we are trying to\n# minimize\n\ndef objective(x, noise_level=noise_level):\n return np.sin(5 * x[0]) * (1 - np.tanh(x[0] ** 2))\\\n + np.random.randn() * noise_level\n\ndef objective_wo_noise(x):\n return objective(x, noise_level=0)",
"_____no_output_____"
],
[
"opt_gp = Optimizer([(-2.0, 2.0)], base_estimator=\"GP\", n_initial_points=5,\n acq_optimizer=\"sampling\", random_state=42)",
"_____no_output_____"
],
[
"# let's do this by hand first...\n\n\nX = np.linspace(-2, 2, 100)\ny = np.vectorize(lambda x: objective_wo_noise([x]))(X)\nplt.plot(X, y)",
"_____no_output_____"
],
[
"# Generate data and fit GP\nrng = np.random.RandomState(4)\nkernel = Matern(length_scale=1.0, nu=2.5)\ngp = GaussianProcessRegressor(kernel=kernel, alpha=0.0)\n# take 5 points...\nX = rng.uniform(-2, 2, 5)\nX = np.sort(X)\ny = np.vectorize(lambda x: objective_wo_noise([x]))(X)\ngp.fit(X.reshape(-1, 1), y)\n\n# how should we approach this? One curve?",
"_____no_output_____"
],
[
"X_ = np.linspace(-2, 2, 100)\ny_mean, y_std = gp.predict(X_.reshape(-1, 1), return_std=True)\n\ny_samples = gp.sample_y(X_.reshape(-1, 1), 1)\nplt.plot(X_, y_samples, 'r')\nplt.plot(X_, np.vectorize(lambda x: objective_wo_noise([x]))(X_))\nplt.plot(X, y, 'ro')",
"_____no_output_____"
],
[
"# if we add some noise...\nX_ = np.linspace(-2, 2, 100)\ny_mean, y_std = gp.predict(X_.reshape(-1, 1), return_std=True)\n\ny_samples = gp.sample_y(X_.reshape(-1, 1), 100)\nplt.plot(X_, y_samples)\n# plt.plot(X_, np.vectorize(lambda x: objective_wo_noise([x]))(X_))\nplt.plot(X, y, 'ro')",
"_____no_output_____"
]
],
[
[
"How do we pick the next point to evaluate?\n\n\nFrom here there are several way to pick the next point. Two common approaches are around:\n\n* Upper confidence bound (exploration vs exploitation)\n* Expected improvement\n\n",
"_____no_output_____"
]
],
[
[
"plt.plot(X_, y_mean, 'r', X, y, 'ro')\nplt.grid(True)\nplt.fill_between(X_, y_mean - y_std, y_mean + y_std,\n alpha=0.5, color='k')",
"_____no_output_____"
],
[
"# for example, let's just consider the lower bound\n# kappa controls the exploration/exploitation.\nkappa = 0.5\nplt.plot(X_, y_mean, 'r', X, y, 'ro', X_, y_mean - y_std, 'b', X_, y_mean - kappa*y_std, 'k')\nplt.grid(True)",
"_____no_output_____"
],
[
"# expected improvement\nfrom scipy.stats import norm\nbest_y = np.min(y)\nz = (y_mean - best_y + X_)/y_std\nei = (y_mean - best_y+X_)*norm.cdf(z) + y_std*norm.pdf(z)\nplt.plot(X_, y_mean, 'r', X, y, 'ro', X_, y_mean - y_std, 'b', X_, ei, 'k')\nplt.grid(True)",
"_____no_output_____"
]
],
[
[
"Let's use scikit optimise instead...",
"_____no_output_____"
]
],
[
[
"res = gp_minimize(objective_wo_noise, # the function to minimize\n [(-2.0, 2.0)], # the bounds on each dimension of x\n acq_func=\"EI\", # the acquisition function\n n_calls=10, # the number of evaluations of f\n n_random_starts=1, # the number of random initialization points\n x0 = [[x] for x in X],\n random_state=1234) # the random seed",
"_____no_output_____"
],
[
"from skopt.plots import plot_convergence\nplot_convergence(res);",
"_____no_output_____"
],
[
"plot_gaussian_process(res, n_calls=0,\n objective=objective_wo_noise,\n show_title=False)",
"_____no_output_____"
],
[
"plot_gaussian_process(res, n_calls=0,\n show_legend=True, show_title=False,\n show_mu=False, show_acq_func=True,\n show_observations=False,\n show_next_point=True)",
"_____no_output_____"
],
[
"plot_gaussian_process(res, n_calls=1,\n objective=objective_wo_noise,\n show_title=False)",
"_____no_output_____"
],
[
"plot_gaussian_process(res, n_calls=1,\n show_legend=True, show_title=False,\n show_mu=False, show_acq_func=True,\n show_observations=False,\n show_next_point=True)",
"_____no_output_____"
],
[
"plt.figure",
"_____no_output_____"
],
[
"plt.figure(figsize=(20,20))\nfor n_iter in range(5):\n # Plot true function.\n plt.subplot(5, 2, 2*n_iter+1)\n\n if n_iter == 0:\n show_legend = True\n else:\n show_legend = False\n\n ax = plot_gaussian_process(res, n_calls=n_iter,\n objective=objective_wo_noise,\n noise_level=noise_level,\n show_legend=show_legend, show_title=False,\n show_next_point=False, show_acq_func=False)\n ax.set_ylabel(\"\")\n ax.set_xlabel(\"\")\n # Plot EI(x)\n plt.subplot(5, 2, 2*n_iter+2)\n ax = plot_gaussian_process(res, n_calls=n_iter,\n show_legend=show_legend, show_title=False,\n show_mu=False, show_acq_func=True,\n show_observations=False,\n show_next_point=True)\n ax.set_ylabel(\"\")\n ax.set_xlabel(\"\")\n\nplt.show()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0d2bc0cbc76648791d871cc36c936eb9d502db5 | 350,556 | ipynb | Jupyter Notebook | notebooks/explore_models.ipynb | HofmannZ/global-ai-hackathon--truth-coin | 9f544cdb05de0811796d2465fba64875ee77cdab | [
"MIT"
] | 5 | 2017-06-24T22:54:13.000Z | 2020-02-13T17:23:12.000Z | notebooks/explore_models.ipynb | HofmannZ/global-ai-hackathon--truth-coin | 9f544cdb05de0811796d2465fba64875ee77cdab | [
"MIT"
] | 2 | 2017-06-24T12:07:22.000Z | 2017-06-25T18:12:24.000Z | notebooks/explore_models.ipynb | HofmannZ/global-ai-hackathon--truth-coin | 9f544cdb05de0811796d2465fba64875ee77cdab | [
"MIT"
] | 1 | 2017-08-02T12:37:52.000Z | 2017-08-02T12:37:52.000Z | 157.765977 | 160,386 | 0.848703 | [
[
[
"import pandas as pd\nimport numpy as np\nimport seaborn as sns\n%matplotlib inline",
"/Users/pokutnik/anaconda3/envs/ekans/lib/python2.7/site-packages/matplotlib/font_manager.py:273: UserWarning: Matplotlib is building the font cache using fc-list. This may take a moment.\n warnings.warn('Matplotlib is building the font cache using fc-list. This may take a moment.')\n"
],
[
"cc = pd.read_csv('./posts_ccompare_raw.csv', index_col=0, encoding='utf-8')\ncc['Timestamp'] = pd.to_datetime(cc['Timestamp'])",
"_____no_output_____"
]
],
[
[
"# Reaction features",
"_____no_output_____"
]
],
[
[
"features_reactions = pd.DataFrame(index=cc.index)\nfeatures_reactions['n_up'] = cc['Actions.Agree.Total']\nfeatures_reactions['n_down'] = cc['Actions.Disagree.Total']\nfeatures_reactions['n_reply'] = cc['Actions.Comment.Total']",
"_____no_output_____"
],
[
"sns.pairplot(features_reactions)",
"_____no_output_____"
]
],
[
[
"# Post date features",
"_____no_output_____"
]
],
[
[
"features_date = pd.DataFrame(index=cc.index)\nfeatures_date['t_week'] = cc.Timestamp.dt.week\nfeatures_date['t_dow'] = cc.Timestamp.dt.dayofweek\nfeatures_date['t_hour'] = cc.Timestamp.dt.hour\nfeatures_date['t_day'] = cc.Timestamp.dt.day",
"_____no_output_____"
],
[
"sns.pairplot(features_date)",
"_____no_output_____"
]
],
[
[
"# Spacy NLP ...",
"_____no_output_____"
]
],
[
[
"import spacy # See \"Installing spaCy\"\nnlp = spacy.load('en') # You are here.\n",
"_____no_output_____"
],
[
"spacy_docs = pd.DataFrame(index=cc.index)\ndocs = cc.Body.apply(nlp)\nvec = docs.apply(lambda x: x.vector)\nfeature_word_vec = pd.DataFrame(vec.tolist(), columns=['spacy_%s'%i for i in range(300)])",
"_____no_output_____"
],
[
"feature_word_vec['spacy_sent'] = docs.apply(lambda x: x.sentiment)",
"_____no_output_____"
],
[
"# tfidf ",
"_____no_output_____"
],
[
"'''\nAuthor: Giovanni Kastanja\nPython: 3.6.0\nDate: 24/6/2017\n'''\nimport pandas as pd\nimport numpy as np\nfrom nltk.corpus import stopwords\nfrom sklearn.feature_extraction.text import TfidfVectorizer\nfrom sklearn.cross_validation import train_test_split\nfrom sklearn.ensemble import RandomForestRegressor\nfrom sklearn.model_selection import cross_val_score\nfrom scipy.sparse import csr_matrix\ntext = cc['Body']\n# create a stopset (words that occur to many times)\nstopset = set(stopwords.words('english'))\nvectorizer = TfidfVectorizer(use_idf=True, lowercase=True, strip_accents='ascii', stop_words=stopset)\nfeatures_tfidf = pd.DataFrame(vectorizer.fit_transform(text).toarray())\n",
"_____no_output_____"
]
],
[
[
"# Target",
"_____no_output_____"
]
],
[
[
"targets = pd.read_csv('./btc-ind.csv')\ntargets['date'] = pd.to_datetime(targets['Date'])\ntargets = targets.set_index('date')\ndel targets['Date']\ntargets.tail()",
"_____no_output_____"
],
[
"join_by_date = pd.DataFrame(index=cc.index)\njoin_by_date['date'] = cc.Timestamp.dt.round(freq=\"d\")",
"_____no_output_____"
],
[
"Y_all = join_by_date.join(targets, on='date').dropna()\ngroups = Y_all['date']\ndel Y_all['date']\ncols = Y_all.columns\nindex = Y_all.index\n#Y_all = pd.DataFrame(normalize(Y_all, axis=1, norm='l2'), columns=cols, index=index)\nY_all = Y_all - Y_all.mean()\nY_all = Y_all/Y_all.std()\n#Y_all.plot()",
"_____no_output_____"
]
],
[
[
"# Combine features",
"_____no_output_____"
]
],
[
[
"#features = pd.concat([features_date, features_tfidf, features_reactions, feature_word_vec], axis=1)\nfeatures = pd.concat([features_date, features_reactions, feature_word_vec], axis=1)",
"_____no_output_____"
],
[
"X_all = features.ix[Y_all.index]\nX_all.shape",
"_____no_output_____"
],
[
"from sklearn.model_selection import cross_val_score\nfrom sklearn.ensemble import RandomForestRegressor\nfrom sklearn.preprocessing import normalize\nfrom xgboost.sklearn import XGBRegressor\nfrom sklearn.linear_model import LinearRegression, Lasso",
"_____no_output_____"
],
[
"rf = RandomForestRegressor(n_estimators=10, max_depth=3, criterion='mse')\nxgb = XGBRegressor(n_estimators=10)\nregressors = [rf, Lasso()]",
"_____no_output_____"
],
[
"target_scores = {}\nfor indicator in targets.columns:\n Y =Y_all[indicator] \n for reg in regressors:\n tag = indicator+':'+str(reg)[:15]\n scores = cross_val_score(reg, X_all, Y, cv=4, groups=groups, scoring='neg_mean_squared_error')\n print np.mean(scores), tag\n target_scores[tag] = scores\n\ncv_score = pd.DataFrame(target_scores)",
"-1.01510250221 BTC_pd_T0:RandomForestReg\n-1.00489834393 BTC_pd_T0:Lasso(alpha=1.0\n-1.01362175733 BTC_rpd_T0:RandomForestReg\n-1.00700271374 BTC_rpd_T0:Lasso(alpha=1.0\n-1.12117041142 BTC_v_T0:RandomForestReg\n-1.12333273979 BTC_v_T0:Lasso(alpha=1.0\n-1.0279897913 BTC_rpd_T1:RandomForestReg\n-1.02159575107 BTC_rpd_T1:Lasso(alpha=1.0\n-1.14329555947 BTC_v_T1:RandomForestReg\n-1.14257585964 BTC_v_T1:Lasso(alpha=1.0\n-1.03427313345 BTC_pd_T2:RandomForestReg\n-1.02477879379 BTC_pd_T2:Lasso(alpha=1.0\n-1.13260911269 BTC_v_T2:RandomForestReg\n-1.12544047748 BTC_v_T2:Lasso(alpha=1.0\n-1.51004848987 BTC_s_T2:RandomForestReg\n-1.50287169074 BTC_s_T2:Lasso(alpha=1.0\n-1.01129501027 BTC_dh_m3:RandomForestReg\n-1.00167162664 BTC_dh_m3:Lasso(alpha=1.0\n-1.00526912285 BTC_dl_m3:RandomForestReg\n-1.00073235397 BTC_dl_m3:Lasso(alpha=1.0\n-1.00164740181 BTC_do_m3:RandomForestReg\n-1.00238122689 BTC_do_m3:Lasso(alpha=1.0\n-1.00571974508 BTC_dp_m3:RandomForestReg\n-1.00041327712 BTC_dp_m3:Lasso(alpha=1.0\n-1.17915445144 BTC_log_v_T2:RandomForestReg\n-1.16959479508 BTC_log_v_T2:Lasso(alpha=1.0\n-1.5242905509 BTC_log_s_T2:RandomForestReg\n-1.5204661907 BTC_log_s_T2:Lasso(alpha=1.0\n-1.01681433113 BTC_cbrt_rpd_T0:RandomForestReg\n-1.00502746636 BTC_cbrt_rpd_T0:Lasso(alpha=1.0\n-1.01580076386 BTC_cbrt_pd_T1:RandomForestReg\n-1.00757264031 BTC_cbrt_pd_T1:Lasso(alpha=1.0\n-1.00875193424 BTC_cbrt_dv_T1:RandomForestReg\n-1.00127016481 BTC_cbrt_dv_T1:Lasso(alpha=1.0\n-1.51169196602 BTC_cbrt_hl_T1:RandomForestReg\n-1.51760727246 BTC_cbrt_hl_T1:Lasso(alpha=1.0\n-1.0315597114 BTC_cbrt_pd_T2:RandomForestReg\n-1.01853785493 BTC_cbrt_pd_T2:Lasso(alpha=1.0\n-0.99680604795 BTC_cbrt_dh_m3:RandomForestReg\n-1.00100385513 BTC_cbrt_dh_m3:Lasso(alpha=1.0\n-1.01059521017 BTC_cbrt_dl_m3:RandomForestReg\n-1.00360457359 BTC_cbrt_dl_m3:Lasso(alpha=1.0\n-1.00854660849 BTC_cbrt_do_m3:RandomForestReg\n-1.00367703909 BTC_cbrt_do_m3:Lasso(alpha=1.0\n-1.01929146151 BTC_cbrt_dp_m3:RandomForestReg\n-1.00361460398 BTC_cbrt_dp_m3:Lasso(alpha=1.0\n"
],
[
"ms = cv_score.mean(axis=0)\nms.sort_values(ascending=False)",
"_____no_output_____"
],
[
"indicator = 'BTC_cbrt_dv_T1:Lasso(alpha=1.0'\nindicator = indicator.split(\":\")[0]\nY = Y_all[indicator]\nreg = XGBRegressor(n_estimators=100)\nreg.fit(X_all, Y)\nY_t = reg.predict(X_all)\nerror = abs(Y - Y_t)",
"_____no_output_____"
],
[
"error.hist()",
"_____no_output_____"
],
[
"# DROP THE BULL$HIT\nitruth = error < error.quantile(0.3)\nX = X_all[itruth]\nY = Y_all[indicator][itruth]\nG = groups[itruth]",
"_____no_output_____"
],
[
"reg = XGBRegressor(n_estimators=100, max_depth=8)\nscores = cross_val_score(reg, X, Y, cv=4, groups=G, scoring='neg_mean_squared_error')\nprint sorted(scores)",
"[-0.23556982483024885, -0.21678759213304766, -0.18345071771185228, -0.16639456674108286]\n"
],
[
"ax = groups.hist(figsize=(12,5))\nG.hist(ax=ax)",
"_____no_output_____"
],
[
"reg = XGBRegressor(n_estimators=100, max_depth=8)\nreg.fit(X,Y)\nY_ = reg.predict(X)\ntruth_df = pd.DataFrame({'date': G, 'Y': Y_})",
"_____no_output_____"
],
[
"def get_stats(group):\n return {'min': group.min(), 'max': group.max(), 'count': group.count(), 'mean': group.mean()}\n\n",
"_____no_output_____"
],
[
"ax = targets.BTC_cbrt_dv_T1.plot()\ntruth.plot(ax=ax)",
"_____no_output_____"
],
[
"truth",
"_____no_output_____"
],
[
"def drop_bs(indicator, q=0.3):\n Y = Y_all[indicator]\n reg = XGBRegressor(n_estimators=100)\n reg.fit(X_all, Y)\n Y_t = reg.predict(X_all)\n error = abs(Y - Y_t)\n error.hist()\n itruth = error < error.quantile(q)\n X = X_all[itruth]\n Y = Y_all[indicator][itruth]\n G = groups[itruth]\n reg = XGBRegressor(n_estimators=30, max_depth=5)\n scores = cross_val_score(reg, X, Y, cv=4, groups=G, scoring='neg_mean_squared_error')\n print sorted(scores)\n print \"MEAN CV SCORE: \", np.mean(scores)\n \n reg = XGBRegressor(n_estimators=100, max_depth=8)\n reg.fit(X,Y)\n Y_ = reg.predict(X)\n agg = pd.Series(Y_).groupby(G)\n truthscore = agg.mean()\n impact_count = agg.count()\n truth_max = agg.max()\n return pd.DataFrame(dict(truthscore=truthscore, impact_count=impact_count, truth_max=truth_max, date=truthscore.index))",
"_____no_output_____"
],
[
"dv = drop_bs('BTC_cbrt_dv_T1', 0.4)",
"[-0.41367598429198782, -0.3770965620906167, -0.35208172797206533, -0.33427650602529679]\nMEAN CV SCORE: -0.369282695095\n"
],
[
"import json\ndef to_json(df, path):\n a = []\n for i,d in list(df.iterrows()):\n d = d.to_dict()\n d['date'] = str(d['date'])\n a.append(d) \n with open(path, 'w') as f:\n json.dump(a, f)\n \nto_json(dv, '../bitcoin-daily-bars/out-truth-volume.json')",
"_____no_output_____"
],
[
"impactfull = cc.ix[itruth.index][itruth]",
"_____no_output_____"
],
[
"impactfull.head()",
"_____no_output_____"
],
[
"f = 'Cryptopian.Name'\na = impactfull.groupby(f).size()\nb = cc.groupby(f).size()\nc = pd.DataFrame(dict(a=a,b=b))\nc = c[c.a>1]\nc['impact'] = c.a/c.b\nc.sort_values('impact', ascending=False)",
"_____no_output_____"
],
[
"dv.truthscore.plot()",
"_____no_output_____"
],
[
"target_sc",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.